code
stringlengths 2.5k
6.36M
| kind
stringclasses 2
values | parsed_code
stringlengths 0
404k
| quality_prob
float64 0
0.98
| learning_prob
float64 0.03
1
|
|---|---|---|---|---|
# Chapter 5 - Core concepts of containers
In the next chapters, we will introduce the most important containers in the Python language: **lists**, **sets**, **tuples**, and **dictionaries**. However, before we introduce them, it's important that we present some things that they all share, which is the goal of this chapter.
**At the end of this chapter, you will be able to understand the following concepts:**
* positional parameters
* keyword parameters
* [positional-only arguments](https://deepsource.io/blog/python-positional-only-arguments/)
* mutability
**If you want to learn more about these topics, you might find the following links useful:**
* [the Python glossary](https://docs.python.org/3/glossary.html): please look for the terms *immutable*, *parameter*, and *argument*
* [What is the difference between arguments and parameters?](https://docs.python.org/3/faq/programming.html#faq-argument-vs-parameter)
If you have **questions** about this chapter, please contact us **(cltl.python.course@gmail.com)**.
## 1. Containers
When working with data, we use different Python objects (which we summarize **containers**) to order data in a way that is convenient for the task we are trying to solve. Each of the following container types has different advantages for storing and accessing data (which you will learn about in the following chapters):
* lists
* tuples
* sets
* dictionaries
Each container type can be manipulated using different methods and functions, for instance, allowing us to add, access, or remove data. It is important that you understand those.
```
# Some examples (you do not have to remember this now):
a_list = [1,2,3, "let's", "use", "containers"]
a_tuple = (1, 2, 3, "let's", "use", "containers")
a_set = {1, 2, 3, "let's", "use", "containers"}
a_dict = {1:"let's", 2:"use", 3: "containers"}
#print(a_list)
#print(a_tuple)
#print(a_set)
#print(a_dict)
```
## 2. Understanding class methods
Let's look at some string method examples from the last chapters:
```
a_string = 'hello world'
print('example 1. upper method:', a_string.upper())
print('example 2. count method:', a_string.count('l'))
print('example 3. replace method:', a_string.replace('l', 'b'))
print('example 4. split method:', a_string.split())
print('example 5. split method:', a_string.split(sep='o'))
```
In all of the examples above, a string method is called, e.g., *upper* or *count*.
However, they differ regarding their arguments:
* There are no arguments in the case of upper, i.e., no arguments between the round brackets.
* for count, we specify a string 'l' as an argument
* for replace, we specify two strings as arguments
* for split, we can specify an argument, but we do not have to
This might look a bit confusing. Luckily Python has a built-in function **help**, which provides us insight into how to use each method. We will guide you through understanding the information provided for the string method **replace**.
```
help(str.replace)
```
The method documentation contains three parts:
1. **data structure**: sentence starting with *Help on*. This simply indicates the data structure for which information is shown, which is a method in this case.
2. **parameters**: information about the parameters of the method, i.e., **replace(self, old, new, count=-1, /)**. This is the most important part of the documentation.
3. **docstring**: explanation about the method in free text
Let's go through the parameters of the string method **replace**:
* *self*: for now, the only thing to remember about *self* is that it tells you that replace is a method and that you should ignore it when calling the method!
* *old*: this is a positional parameter
* *new*: this is a positional parameter
* *count=-1*: this is a keyword parameter, meaning that it has a default value, i.e., -1
* */* (forward slash): for now, please ignore, we will come back to this.
In the enumeration above, we've used the terms **positional parameter** and **keyword parameter**. What are they, and in what do they differ?
* Positional parameters are **compulsory** to call a method. Without them, you will not successfully call the method.
* Keyword parameters are **optional**. They have a default value, e.g., -1 in the case of *count*, and are optional.
Let's put this to the test! Since **positional parameters** are needed to call our method, we should be able to call the method by specifying a value for *old* and *new*, but not for *count*. The value for *old* is 'r', and the value for *new* is 'c'.
```
a_string = 'rats are the best.'
result = a_string.replace('r', 'c')
print(result)
```
It worked! We've called the string method by only providing a value for the positional parameters. However, what if we are not happy with the provided default value, can we override it?
Let's try this. The keyword parameter *count* allows us to indicate how many times to replace a substring. Let's try to only replace 'r' to 'c' one time.
```
a_string = 'rats are the best.'
result = a_string.replace('r', 'c', 1)
print(result)
```
Yes! We've provided a value for *count*, e.g., 1, and now 'r' is only replaced once with 'c'. Luckily, the 'r' in 'are' has not been replaced.
We will now move on to the string method **split**.
```
help(str.split)
```
Let's go through the parameters of the string method **split**:
* *self*: for now, the only thing to remember about *self* is that it tells you that replace is a method and that you should ignore it in calling the method!
* */* (forward slash): for now, please ignore, we will come back to this.
* *sep=None*: this is a keyword parameter, meaning that it has a default value, i.e., None.
* *maxsplit=-1*: this is a keyword parameter, by which you can indicate how many times to split.
Since **split** has no positional parameters, we should be able to call the method without providing arguments.
```
a_string = 'USA Today has come out with a new survey: Apparently three out of four people make up 75 percent of the population.'
words = a_string.split()
print(words)
```
And that is correct! Of course, we can specify a value for the keyword parameters. We provide the a space ' ' for *sep* and 2 for *maxsplit*.
```
a_string = 'USA Today has come out with a new survey: Apparently three out of four people make up 75 percent of the population.'
words = a_string.split(' ', 2)
print(words)
```
Please note that we have splitted the string on a space ' ' two times.
Try and play with with the split function: (e.g. how does split(' ') differ from split()?)
## 2.1 The forward slash
So far, we have not explained the forward slash in the parameters. Here, we highlight its importance to calling a method. We show two examples. The main question is the following: why is the first call successful, and why does the second call result in error?
```
a_string = 'USA Today has come out with a new survey: Apparently three out of four people make up 75 percent of the population.'
words = a_string.split(sep=' ', maxsplit=2)
print(words)
a_string = 'rats are the best.'
result = a_string.replace('r', 'c', count=1)
```
For the answer, we need to go back to the function parameters:
* **replace**: replace(self, old, new, count=-1, /)
* **split**: split(self, /, sep=None, maxsplit=-1)
Please note that the difference is that *count* is to the **left** of the forward slash, and *sep* and *maxsplit* are to the **right** of the forward slash! We can call any parameter to the right of the forward slash using the name of the parameter. For any parameter to the left of the forward slash, we can only provide the value.
This does work:
```
a_string = 'rats are the best.'
result = a_string.replace('r', 'c', 1)
print(result)
```
This does not.
```
a_string = 'rats are the best.'
result = a_string.replace('r', 'c', count=1)
```
**Summary**:
* ignore **self**
* **positional parameters** are mandatory to call a method
* **keyword parameters** are optional since they have a default value
* any parameter to the right of the forward slash, we can call using the name of the parameter.
* any parameter to the left of the forward slash, we can only provide the value.
For those interested in understanding it in more detail, please check the link about **positional-only arguments** at the top of this notebook.
## 3. Mutability
Hopefully, it will become clear in the following chapters what we mean by **mutability**. For now, you can think of it in terms of 'can I change the data?'. Please remember the following categories for the subsequent chapters:
| **immutable** | **mutable** |
|-----------------|-------------|
| integer | list |
| string | set |
| - | dictionary |
You have already seen a little bit about strings and immutability in Chapter 3. To change a string, we have to create a new one. In contrast, you will learn that many containers can be modified.
# Exercises
Please find some exercises about core concepts of python containers below.
### Exercise 1:
Use the help function to figure out what the string methods below are doing. Then analyze how many positional and keyword parameters are used in the following examples:
```
print(a_string.lower())
print(a_string.strip())
print(a_string.strip('an'))
print(a_string.partition('and'))
```
### Exercise 2:
Please illustrate the difference between positional and keyword parameters using the example of string methods. Feel free to use dir(str) and the help function for inspiration.
```
# your examples here
```
|
github_jupyter
|
# Some examples (you do not have to remember this now):
a_list = [1,2,3, "let's", "use", "containers"]
a_tuple = (1, 2, 3, "let's", "use", "containers")
a_set = {1, 2, 3, "let's", "use", "containers"}
a_dict = {1:"let's", 2:"use", 3: "containers"}
#print(a_list)
#print(a_tuple)
#print(a_set)
#print(a_dict)
a_string = 'hello world'
print('example 1. upper method:', a_string.upper())
print('example 2. count method:', a_string.count('l'))
print('example 3. replace method:', a_string.replace('l', 'b'))
print('example 4. split method:', a_string.split())
print('example 5. split method:', a_string.split(sep='o'))
help(str.replace)
a_string = 'rats are the best.'
result = a_string.replace('r', 'c')
print(result)
a_string = 'rats are the best.'
result = a_string.replace('r', 'c', 1)
print(result)
help(str.split)
a_string = 'USA Today has come out with a new survey: Apparently three out of four people make up 75 percent of the population.'
words = a_string.split()
print(words)
a_string = 'USA Today has come out with a new survey: Apparently three out of four people make up 75 percent of the population.'
words = a_string.split(' ', 2)
print(words)
a_string = 'USA Today has come out with a new survey: Apparently three out of four people make up 75 percent of the population.'
words = a_string.split(sep=' ', maxsplit=2)
print(words)
a_string = 'rats are the best.'
result = a_string.replace('r', 'c', count=1)
a_string = 'rats are the best.'
result = a_string.replace('r', 'c', 1)
print(result)
a_string = 'rats are the best.'
result = a_string.replace('r', 'c', count=1)
print(a_string.lower())
print(a_string.strip())
print(a_string.strip('an'))
print(a_string.partition('and'))
# your examples here
| 0.158174 | 0.982591 |
## CS536: Computing Solutions
#### Done by: Vedant Choudhary, vc389
### Linear Regression
Consider data generated in the following way:
- $X_1$ through $X_{10}$ and $X_{16}$ through $X_{20}$ are i.i.d. standard normals
- $X_{11} = X_1 + X_2 + N (\mu=0, \sigma^2=0.1)$
- $X_{12} = X_3 + X_4 + N (\mu=0, \sigma^2=0.1)$
- $X_{13} = X_4 + X_5 + N (\mu=0, \sigma^2=0.1)$
- $X_{14} = 0.1X_7 + N (\mu=0, \sigma^2=0.1)$
- $X_{15} = 2X_2 - 10 +N (\mu=0, \sigma^2=0.1)$
The values $Y$ are generated according to the following linear model:
$$ Y = 10 + \sum_{i=1}^10{(0.6)^iX_i} $$
Note, the variables $X_{11}$ through $X_{20}$ are technically irrelevant
```
# Importing required libraries
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import pprint
from tqdm import tqdm
import seaborn as sns
from copy import copy
%matplotlib inline
```
#### 1. Generate a data set of size m = 1000. Solve the naive least squares regression model for the weights and bias that minimize the training error - how do they compare to the true weights and biases? What did your model conclude as the most significant and least significant features - was it able to prune anything? Simulate a large test set of data and estimate the ‘true’ error of your solved model.
```
# Creating X (feature) vectors for the data
def create_data(m):
X_1_10 = np.random.normal(0, 1, (m,11))
X_11 = np.asarray([X_1_10[i][1] + X_1_10[i][2] + np.random.normal(loc=0, scale=(0.1)**(0.5)) for i in range(m)]).reshape((-1,1))
X_12 = np.asarray([X_1_10[i][3] + X_1_10[i][4] + np.random.normal(loc=0, scale=(0.1)**(0.5)) for i in range(m)]).reshape((-1,1))
X_13 = np.asarray([X_1_10[i][4] + X_1_10[i][5] + np.random.normal(loc=0, scale=(0.1)**(0.5)) for i in range(m)]).reshape((-1,1))
X_14 = np.asarray([0.1*X_1_10[i][7] + np.random.normal(loc=0, scale=(0.1)**(0.5)) for i in range(m)]).reshape((-1,1))
X_15 = np.asarray([2*X_1_10[i][2] - 10 + np.random.normal(loc=0, scale=(0.1)**(0.5)) for i in range(m)]).reshape((-1,1))
X_16_20 = np.random.normal(0, 1, (m, 5))
return np.concatenate((X_1_10, X_11, X_12, X_13, X_14, X_15, X_16_20), axis=1)
# Creating target column for the data
def create_y(X, m):
y = []
for i in range(m):
temp = 10
for j in range(1, 11):
temp += ((0.6)**j)*X[i][j]
temp += np.random.normal(loc=0, scale=(0.1)**(0.5))
y.append(temp)
return np.asarray(y)
# Combining all the sub data points into a dataframe
def create_dataset(m):
X = create_data(m)
y = create_y(X, m).reshape((m,1))
# print(X)
# Training data is an appended version of X and y arrays
data = pd.DataFrame(np.append(X, y, axis=1), columns=["X" + str(i) for i in range(21)]+['Y'])
data['X0'] = 1
return data
m = 1000
train_data = create_dataset(m)
train_data.head()
class LinearRegression():
def __init__(self):
pass
def thresh(self, support, lmbda):
if support > 0.0 and lmbda < abs(support):
return (support - lmbda)
elif support < 0.0 and lmbda < abs(support):
return (support + lmbda)
else:
return 0.0
def naive_regression(self, X, y):
n_samples, n_features = X.shape
self.w = np.zeros(shape=(n_features,1))
self.w = np.dot(np.dot(np.linalg.inv(np.dot(X.T, X)), X.T), y)
return self.w
def ridge_regression(self, X, y, lmbda):
n_samples, n_features = X.shape
self.w = np.zeros(shape=(n_features,1))
self.w = np.dot(np.dot(np.linalg.inv(np.dot(X.T, X) + lmbda*np.identity(n_features)), X.T), y)
return self.w
def lasso_regression(self, X, y, lmbda, iterations):
n_samples, n_features = X.shape
self.w = np.zeros(shape=(n_features,1))
# Since bias is basically mean of original - predictions
self.w[0] = np.sum(y - np.dot(X[:, 1:], self.w[1:]))/n_samples
for i in range(iterations):
for j in range(1, n_features):
copy_w = self.w.copy()
copy_w[j] = 0.0
residue = y - np.dot(X, copy_w)
a1 = np.dot(X[:, j], residue)
a2 = lmbda*n_samples
self.w[j] = self.thresh(a1, a2)/(X[:, j]**2).sum()
return self.w
def predict(self, X, w):
return np.dot(X, w)
def error_calculation(self, X, y, w):
h_x = self.predict(X, w)
error = 0
for i in range(len(y)):
error += (y[i] - h_x[i])**2
error /= len(y)
return error
def plot_comparison(actual, predicted, features):
plt.figure(figsize=(8,6))
data_actual = pd.DataFrame(pd.Series(actual), columns=['weight'])
data_actual['type'] = pd.Series(['actual']*len(actual))
data_actual['feature'] = pd.Series(list(range(features)))
data_actual = data_actual.iloc[1:]
data_pred = pd.DataFrame(pd.Series(predicted), columns=['weight'])
data_pred['type'] = pd.Series(['pred']*len(predicted))
data_pred['feature'] = pd.Series(list(range(features)))
data_pred = data_pred.iloc[1:]
data = pd.concat([data_actual, data_pred])
sns.barplot(x="feature", y="weight", hue="type", data=data)
lin_reg = LinearRegression()
X = np.asarray(train_data.iloc[:,:-1])
y = np.asarray(train_data.iloc[:,-1:])
w_actual = [10]
for i in range(1, 21):
if i <= 10:
w_actual.append((0.6)**i)
else:
w_actual.append(0)
w_trained = lin_reg.naive_regression(X, y)
features = X.shape[1]
# print(w_trained)
plot_comparison(w_actual, w_trained.flatten(), features)
print("Error: ", float(lin_reg.error_calculation(X, y, w_trained)))
print("Trained Bias: ", w_trained[0])
```
- After training the model on naive least squares regression and comparing the predicted weights and bias with true ones, we see that the predicted weights are in line with the true weights but there is some weight associated to irrelevant variables too.
- The bias value has been printed instead of charting, to maintain the y-axis range. Trained bias is also similar to actual bias
- This model regards X1 as the most significant and X16 as least significant feature
- This naive model has no mechanism to perform pruning of features.
```
def estimated_error(w):
estimated_error = 0
for i in (range(10)):
data = create_dataset(10000)
X = np.asarray(data.iloc[:,:-1])
y = np.asarray(data.iloc[:,-1:])
estimated_error += lin_reg.error_calculation(X, y, w)
return float(estimated_error/10)
estimated_err = estimated_error(w_trained)
print("True error: ", estimated_err)
```
#### 2. Write a program to take a data set of size m and a parameter λ, and solve for the ridge regression model for that data. Write another program to take the solved model and estimate the true error by evaluating that model on a large test data set. For data sets of size m = 1000, plot estimated true error of the ridge regression model as a function of λ. What is the optimal λ to minimize testing error? What are the weights and biases ridge regression gives at this λ, and how do they compare to the true weights? What did your model conclude as the most significant and least significant features - was it able to prune anything? How does the optimal ridge regression model compare to the naive least squares model?
```
lmbda = 0.01
w_lambda = lin_reg.ridge_regression(X, y, lmbda)
estimated_err = estimated_error(w_lambda)
estimated_err
def varying_lambda(m):
lmbda = np.arange(0, 1, 0.03)
estimated_error_list = []
for i in lmbda:
print(round(i, 2), end='\t')
error = 0
data = create_dataset(m)
X = np.asarray(data.iloc[:,:-1])
y = np.asarray(data.iloc[:,-1:])
lin_reg = LinearRegression()
w = lin_reg.ridge_regression(X, y, i)
estimated_error_list.append(estimated_error(w))
plt.figure(figsize=(10,8))
plt.plot(lmbda, estimated_error_list, marker='x')
plt.title("True Error w.r.t. lambda")
plt.xlabel("Lambda")
plt.ylabel("True Error")
plt.show()
varying_lambda(1000)
```
As can be seen from the plot, as the value of lambda is increased, the true error of the ridge regression model also increases. For this example, we can see that an optimal lambda value is in the range [0.08, 0.1)
```
w_lambda = lin_reg.ridge_regression(X, y, 0.08)
estimated_err = estimated_error(w_lambda)
plot_comparison(w_actual, w_lambda.flatten(), features)
print("Error: ", float(lin_reg.error_calculation(X, y, w_lambda)))
print("Weights (Bias at zero index): \n", w_lambda)
```
- The above plots convey that on using the optimal value of lambda from ridge regression (0.08 in this case), we get weights and bias very similar to the actual weights and bias
- The ridge regression model considers X1 to be most significant, while X15 to be the least
- I do not believe that ridge was able to prune any variables since they have some weights associated to the features
#### 3. Write a program to take a data set of size m and a parameter λ, and solve for the Lasso regression model for that data. For a data set of size m = 1000, show that as λ increases, features are effectively eliminated from the model until all weights are set to zero.
```
lmbda = np.arange(0, 1.0, 0.01)
weights = []
lin_reg = LinearRegression()
for l in lmbda:
print(round(l,2), end='\t')
weight = lin_reg.lasso_regression(X, y, l, 100)
weights.append(weight.flatten())
weights_lasso = np.stack(weights).T
weights_lasso[1:].shape
n,_ = weights_lasso[1:].shape
plt.figure(figsize = (20,10))
for i in range(n):
plt.plot(lmbda, weights_lasso[1:][i], label = train_data.columns[1:][i])
# plt.xscale('log')
plt.xlabel('$\\lambda$')
plt.ylabel('Coefficients')
plt.title('Lasso Paths')
plt.legend()
plt.axis('tight')
plt.show()
```
- This plot shows that as the value of lambda is increased, it starts pruning more and more variables/features of the dataset.
- So, all feature weights tend towards 0 at some value of lambda.
#### 4. For data sets of size m = 1000, plot estimated true error of the lasso regression model as a function of λ. What is the optimal λ to minimize testing error? What are the weights and biases lasso regression gives at this λ, and how do they compare to the true weights? What did your model conclude as the most significant and least signficiant features - was it able to prune anything? How does the optimal regression model compare tot he naive least squares model?
```
def varying_lambda_lasso(m):
lmbda = np.arange(0, 1, 0.01)
estimated_error_list = []
for i in lmbda:
print(round(i,2), end="\t")
error = 0
data = create_dataset(m)
X = np.asarray(data.iloc[:,:-1])
y = np.asarray(data.iloc[:,-1:])
lin_reg = LinearRegression()
w = lin_reg.lasso_regression(X, y, i, 100)
estimated_error_list.append(estimated_error(w))
plt.figure(figsize=(10,8))
plt.plot(lmbda, estimated_error_list, marker='x')
plt.title("True Error w.r.t. lambda")
plt.xlabel("Lambda")
plt.ylabel("True Error")
plt.show()
varying_lambda_lasso(1000)
```
The minimum is occuring in the range [0, 0.01], so let us calculate the optimal lasso regression bias and weights by having a value of lambda between the range.
```
optimal_weight = lin_reg.lasso_regression(X, y, 0.001, 1000)
plot_comparison(w_actual, optimal_weight.flatten(), features)
print("Error: ", float(lin_reg.error_calculation(X, y, optimal_weight)))
```
- Optimal Lasso weights are almost equal to actual weights. However, the model makes feature X9 insignificant.
- The model is mostly recognizing the variables from X11 to X20 to be insignificant, with having X12, X13, X14 as 0
- The model prunes some of the insignificant weights
- The model is working almost like the naive regression model
#### 5. Consider using lasso as a means for feature selection: on a data set of size m = 1000, run lasso regression with the optimal regularization constant from the previous problems, and identify the set of relevant features; then run ridge regression to fit a model to only those features. How can you determine a good ridge regression regularization constant to use here? How does the resulting lasso-ridge combination model compare to the naive least squares model? What features does it conclude are significant or relatively insignificant? How do the testing errors of these two models compare?
```
new_data = create_dataset(1000)
linreg = LinearRegression()
X = np.asarray(new_data.iloc[:,:-1])
y = np.asarray(new_data.iloc[:,-1:])
new_weight = linreg.lasso_regression(X, y, 0.001, 100)
plot_comparison(w_actual, new_weight.flatten(), features)
print("Error: ", float(lin_reg.error_calculation(X, y, new_weight)))
feature_weights = dict(zip(*(new_data.drop(['Y'], 1).columns, new_weight)))
feature_weights
```
The relevant features are features with positive weights. So, according to the above output, the relevant features are:
[X1, ...X8, X10,..., X13, X15, X16]
Even though the weights for insignificant values is low, we'll be considering them since they are positive
```
relevant_features = ["X1", "X2", "X3", "X4", "X5", "X6", "X7", "X8", "X10", "X11", "X12", "X13", "X15", "X16", "Y"]
rel = [a[1:] for a in relevant_features[:-1]]
trim_data = new_data[relevant_features]
trim_data.head()
X_trim = np.asarray(trim_data.iloc[:,:-1])
y_trim = np.asarray(trim_data.iloc[:,-1:])
trim_actual_weights = []
for i in range(1, 21):
if str(i) in rel:
trim_actual_weights.append(w_actual[i])
else:
trim_actual_weights.append(0)
new_ridge = LinearRegression()
w_lambda_2 = new_ridge.ridge_regression(X_trim, y_trim, 0.001)
plot_comparison(trim_actual_weights, w_lambda_2.flatten(), features)
w_lambda_2 = new_ridge.ridge_regression(X_trim, y_trim, 0.01)
plot_comparison(trim_actual_weights, w_lambda_2.flatten(), features)
w_lambda_2 = new_ridge.ridge_regression(X_trim, y_trim, 0.1)
plot_comparison(trim_actual_weights, w_lambda_2.flatten(), features)
w_lambda_2 = new_ridge.ridge_regression(X_trim, y_trim, 0.3)
plot_comparison(trim_actual_weights, w_lambda_2.flatten(), features)
w_lambda_2 = new_ridge.ridge_regression(X_trim, y_trim, 0.7)
plot_comparison(trim_actual_weights, w_lambda_2.flatten(), features)
```
- We see that as the value of $\lambda$ increases, the insignifcant features are eliminated. Let us try to calculate true error
- Features after X12 have been considered as insignificant by this model. It has somehow also made X7 as insignificant
- For lambda close to 0.01, least true error is there in the model.
```
w_lambda_2 = new_ridge.ridge_regression(X_trim, y_trim, 0.01)
plot_comparison(trim_actual_weights, w_lambda_2.flatten(), features)
w_lambda_2 = new_ridge.naive_regression(X_trim, y_trim)
plot_comparison(trim_actual_weights, w_lambda_2.flatten(), features)
```
- We observe that the weights given by the models are almost the same.
### SVM
#### 1. Implement a barrier-method dual SVM solver. How can you (easily!) generate an initial feasible α solution away from the boundaries of the constraint region? How can you ensure that you do not step outside the constraint region in any update step? How do you choose your $\epsilon_t$? Be sure to return all $\alpha_i$ including $\alpha_i$ in the final answer.
Using Damped Newton's method for recentering in the Barrier method to solve dual SVM:
$$minimize \phi_t(x) = t(0.5x^TQx + p^Tx) + B(b-Ax)$$
where B is the logarithmic barrier defined as:
$$B(x) = - \sum_i{log(x_i)}$$
The steps followed below have been referred from this article https://github.com/lenassero/linear-svm
```
class SVM():
def __init__(self, tau=1, t_0=1, tol=0.0001, mu=15, sol="Dual", kernel=None, poly_d=2):
self.tau = tau
self.t_0 = t_0
self.tol = tol
self.mu = mu
self.kernel = kernel
self.poly_d = poly_d
self.sol = sol
def damp_newt_step(self, x, objective_func, grad, hess):
g = grad(x)
h = hess(x)
h_inverse = np.linalg.inv(h)
lmbda = np.sqrt((g.T.dot(h_inverse.dot(g))))
x_new = x - (1/(1+lmbda))*h_inverse.dot(g)
gap = 0.5*lmbda**2
return x_new, gap
def damp_newt_method(self, x, objective_func, grad, hess):
x, gap = self.damp_newt_step(x, objective_func, grad, hess)
x_histogram = [x]
if self.tol < (3-(5**0.5))/2:
while gap > self.tol:
x, gap = self.damp_newt_step(x, objective_func, grad, hess)
x_histogram.append(x)
x_star = x
else:
raise ValueError("tol should be less than the condition value")
return x_star, x_histogram
def transform_dual_svm(self, tau, X, y):
n_obs = X.shape[1]
if self.kernel == "Polynomial":
K = (np.identity(n=n_obs) + X.T.dot(X)) ** self.poly_d
Q = (y*y)*K
else:
Q = (X*y).T.dot(X*y)
p = -np.ones(n_obs)
A = np.zeros((2 * n_obs, n_obs))
A[:n_obs, :] = np.identity(n_obs)
A[n_obs:, :] = -np.identity(n_obs)
b = np.zeros(2 * n_obs)
b[:n_obs] = 1 / (tau * n_obs)
return Q, p, A, b
def transform_primal_svm(self, tau, X, y):
d = X.shape[0]
d_ = d - 1
n = X.shape[1]
Q = np.zeros((d+n, d+n))
Q[:d_, :d_] = np.identity(d_)
p = np.zeros(d + n)
p[d:] = 1/(tau*n)
A = np.zeros((2*n, d + n))
A[:n, :d] = -(X*y).T
A[:n, d:] = np.diag([-1]*n)
A[n:, d:] = np.diag([-1]*n)
b = np.zeros(2 * n)
b[:n] = -1
return Q, p, A, b
def barrier_method(self, Q, p, A, b, x_0, t_0, mu, tol):
o_iters = []
m = b.shape[0]
if np.sum(A.dot(x_0) < b) == m:
t = t_0
x = x_0
x_histogram = [x_0]
while m / t >= tol:
f = lambda x: t*(0.5*np.dot(x, Q.dot(x)) + p.dot(x)) - np.sum(np.log(b - A.dot(x)))
g = lambda x: t*(Q.dot(x) + p) + np.sum(np.divide(A.T, b - A.dot(x)), axis=1)
h = lambda x: t*Q + (np.divide(A.T, b - A.dot(x))).dot((np.divide(A.T, b - A.dot(x))).T)
x, x_hist_newton = self.damp_newt_method(x, f, g, h)
x_histogram += x_hist_newton
o_iters += [len(x_hist_newton)]
t *= mu
x_sol = x
else:
raise ValueError("x_0 is not feasible, cannot proceed")
return x_sol, x_histogram, o_iters
def train(self, X, y):
self.n = X.shape[0]
self.d = X.shape[1]
X = np.vstack((X.T, np.ones(self.n)))
if self.sol == 'Dual':
self.x_0 = (1/(100*self.tau*self.n))*np.ones(self.n)
self.Q, self.p, self.A, self.b = self.transform_dual_svm(self.tau, X, y)
self.x_sol, self.x_histogram, self.o_iters = self.barrier_method(self.Q,self.p, self.A, self.b, self.x_0, self.t_0,self.mu, self.tol)
self.w = self.x_sol.dot((X*y).T)
elif self.sol == 'Primal':
self.x_0 = np.zeros(self.d + 1 + self.n)
self.x_0[self.d + 1:] = 1.1
self.Q, self.p, self.A, self.b = self.transform_primal_svm(self.tau, X, y)
self.x_sol, self.x_histogram, self.o_iters = self.barrier_method(self.Q,self.p, self.A, self.b, self.x_0, self.t_0,self.mu, self.tol)
self.w = self.x_sol[:self.d + 1]
def predict(self, X_test, y_test):
n_test = X_test.shape[0]
X_test = np.vstack((X_test.T, np.ones(n_test)))
y_pred = np.sign(self.w.T.dot(X_test))
accuracy = self.compute_mean_accuracy(y_pred, y_test)
return y_pred, accuracy
def compute_mean_accuracy(self, y_pred, y_test):
accuracy = np.sum(y_pred == y_test)
accuracy /= np.shape(y_test)[0]
return accuracy
```
- The initial feasible points comes directly from the constraint on the dual SVM problem
- As t increases, $\epsilon$ decreases till we reach an optimal solution.
```
from sklearn import datasets
iris_data = datasets.load_iris()
X, y = iris_data.data, iris_data.target
X.shape, y.shape
svm = SVM(kernel = False, sol = "Dual")
svm.train(X, y)
print("The training Error is computed as : ", svm.predict(X, y)[1])
print(svm.w)
xor_dict = {'A': [0,0,1,1], 'B': [0,1,0,1], 'Y': [0,1,1,0]}
xor_data = pd.DataFrame(xor_dict)
xor_data.head()
svm = SVM(kernel=True, tol=0.0001)
svm.train(xor_data.drop('Y', 1).values, xor_data['Y'].values)
print("The training Error is computed as : ", svm.predict(xor_data.drop('Y', 1).values, xor_data['Y'].values)[1])
print(svm.w)
svm = SVM(kernel=False, sol="Primal")
svm.train(xor_data.drop('Y', 1).values, xor_data['Y'].values)
print("The training Error is computed as : ", svm.predict(xor_data.drop('Y', 1).values, xor_data['Y'].values)[1])
print(svm.w)
```
|
github_jupyter
|
# Importing required libraries
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import pprint
from tqdm import tqdm
import seaborn as sns
from copy import copy
%matplotlib inline
# Creating X (feature) vectors for the data
def create_data(m):
X_1_10 = np.random.normal(0, 1, (m,11))
X_11 = np.asarray([X_1_10[i][1] + X_1_10[i][2] + np.random.normal(loc=0, scale=(0.1)**(0.5)) for i in range(m)]).reshape((-1,1))
X_12 = np.asarray([X_1_10[i][3] + X_1_10[i][4] + np.random.normal(loc=0, scale=(0.1)**(0.5)) for i in range(m)]).reshape((-1,1))
X_13 = np.asarray([X_1_10[i][4] + X_1_10[i][5] + np.random.normal(loc=0, scale=(0.1)**(0.5)) for i in range(m)]).reshape((-1,1))
X_14 = np.asarray([0.1*X_1_10[i][7] + np.random.normal(loc=0, scale=(0.1)**(0.5)) for i in range(m)]).reshape((-1,1))
X_15 = np.asarray([2*X_1_10[i][2] - 10 + np.random.normal(loc=0, scale=(0.1)**(0.5)) for i in range(m)]).reshape((-1,1))
X_16_20 = np.random.normal(0, 1, (m, 5))
return np.concatenate((X_1_10, X_11, X_12, X_13, X_14, X_15, X_16_20), axis=1)
# Creating target column for the data
def create_y(X, m):
y = []
for i in range(m):
temp = 10
for j in range(1, 11):
temp += ((0.6)**j)*X[i][j]
temp += np.random.normal(loc=0, scale=(0.1)**(0.5))
y.append(temp)
return np.asarray(y)
# Combining all the sub data points into a dataframe
def create_dataset(m):
X = create_data(m)
y = create_y(X, m).reshape((m,1))
# print(X)
# Training data is an appended version of X and y arrays
data = pd.DataFrame(np.append(X, y, axis=1), columns=["X" + str(i) for i in range(21)]+['Y'])
data['X0'] = 1
return data
m = 1000
train_data = create_dataset(m)
train_data.head()
class LinearRegression():
def __init__(self):
pass
def thresh(self, support, lmbda):
if support > 0.0 and lmbda < abs(support):
return (support - lmbda)
elif support < 0.0 and lmbda < abs(support):
return (support + lmbda)
else:
return 0.0
def naive_regression(self, X, y):
n_samples, n_features = X.shape
self.w = np.zeros(shape=(n_features,1))
self.w = np.dot(np.dot(np.linalg.inv(np.dot(X.T, X)), X.T), y)
return self.w
def ridge_regression(self, X, y, lmbda):
n_samples, n_features = X.shape
self.w = np.zeros(shape=(n_features,1))
self.w = np.dot(np.dot(np.linalg.inv(np.dot(X.T, X) + lmbda*np.identity(n_features)), X.T), y)
return self.w
def lasso_regression(self, X, y, lmbda, iterations):
n_samples, n_features = X.shape
self.w = np.zeros(shape=(n_features,1))
# Since bias is basically mean of original - predictions
self.w[0] = np.sum(y - np.dot(X[:, 1:], self.w[1:]))/n_samples
for i in range(iterations):
for j in range(1, n_features):
copy_w = self.w.copy()
copy_w[j] = 0.0
residue = y - np.dot(X, copy_w)
a1 = np.dot(X[:, j], residue)
a2 = lmbda*n_samples
self.w[j] = self.thresh(a1, a2)/(X[:, j]**2).sum()
return self.w
def predict(self, X, w):
return np.dot(X, w)
def error_calculation(self, X, y, w):
h_x = self.predict(X, w)
error = 0
for i in range(len(y)):
error += (y[i] - h_x[i])**2
error /= len(y)
return error
def plot_comparison(actual, predicted, features):
plt.figure(figsize=(8,6))
data_actual = pd.DataFrame(pd.Series(actual), columns=['weight'])
data_actual['type'] = pd.Series(['actual']*len(actual))
data_actual['feature'] = pd.Series(list(range(features)))
data_actual = data_actual.iloc[1:]
data_pred = pd.DataFrame(pd.Series(predicted), columns=['weight'])
data_pred['type'] = pd.Series(['pred']*len(predicted))
data_pred['feature'] = pd.Series(list(range(features)))
data_pred = data_pred.iloc[1:]
data = pd.concat([data_actual, data_pred])
sns.barplot(x="feature", y="weight", hue="type", data=data)
lin_reg = LinearRegression()
X = np.asarray(train_data.iloc[:,:-1])
y = np.asarray(train_data.iloc[:,-1:])
w_actual = [10]
for i in range(1, 21):
if i <= 10:
w_actual.append((0.6)**i)
else:
w_actual.append(0)
w_trained = lin_reg.naive_regression(X, y)
features = X.shape[1]
# print(w_trained)
plot_comparison(w_actual, w_trained.flatten(), features)
print("Error: ", float(lin_reg.error_calculation(X, y, w_trained)))
print("Trained Bias: ", w_trained[0])
def estimated_error(w):
estimated_error = 0
for i in (range(10)):
data = create_dataset(10000)
X = np.asarray(data.iloc[:,:-1])
y = np.asarray(data.iloc[:,-1:])
estimated_error += lin_reg.error_calculation(X, y, w)
return float(estimated_error/10)
estimated_err = estimated_error(w_trained)
print("True error: ", estimated_err)
lmbda = 0.01
w_lambda = lin_reg.ridge_regression(X, y, lmbda)
estimated_err = estimated_error(w_lambda)
estimated_err
def varying_lambda(m):
lmbda = np.arange(0, 1, 0.03)
estimated_error_list = []
for i in lmbda:
print(round(i, 2), end='\t')
error = 0
data = create_dataset(m)
X = np.asarray(data.iloc[:,:-1])
y = np.asarray(data.iloc[:,-1:])
lin_reg = LinearRegression()
w = lin_reg.ridge_regression(X, y, i)
estimated_error_list.append(estimated_error(w))
plt.figure(figsize=(10,8))
plt.plot(lmbda, estimated_error_list, marker='x')
plt.title("True Error w.r.t. lambda")
plt.xlabel("Lambda")
plt.ylabel("True Error")
plt.show()
varying_lambda(1000)
w_lambda = lin_reg.ridge_regression(X, y, 0.08)
estimated_err = estimated_error(w_lambda)
plot_comparison(w_actual, w_lambda.flatten(), features)
print("Error: ", float(lin_reg.error_calculation(X, y, w_lambda)))
print("Weights (Bias at zero index): \n", w_lambda)
lmbda = np.arange(0, 1.0, 0.01)
weights = []
lin_reg = LinearRegression()
for l in lmbda:
print(round(l,2), end='\t')
weight = lin_reg.lasso_regression(X, y, l, 100)
weights.append(weight.flatten())
weights_lasso = np.stack(weights).T
weights_lasso[1:].shape
n,_ = weights_lasso[1:].shape
plt.figure(figsize = (20,10))
for i in range(n):
plt.plot(lmbda, weights_lasso[1:][i], label = train_data.columns[1:][i])
# plt.xscale('log')
plt.xlabel('$\\lambda$')
plt.ylabel('Coefficients')
plt.title('Lasso Paths')
plt.legend()
plt.axis('tight')
plt.show()
def varying_lambda_lasso(m):
lmbda = np.arange(0, 1, 0.01)
estimated_error_list = []
for i in lmbda:
print(round(i,2), end="\t")
error = 0
data = create_dataset(m)
X = np.asarray(data.iloc[:,:-1])
y = np.asarray(data.iloc[:,-1:])
lin_reg = LinearRegression()
w = lin_reg.lasso_regression(X, y, i, 100)
estimated_error_list.append(estimated_error(w))
plt.figure(figsize=(10,8))
plt.plot(lmbda, estimated_error_list, marker='x')
plt.title("True Error w.r.t. lambda")
plt.xlabel("Lambda")
plt.ylabel("True Error")
plt.show()
varying_lambda_lasso(1000)
optimal_weight = lin_reg.lasso_regression(X, y, 0.001, 1000)
plot_comparison(w_actual, optimal_weight.flatten(), features)
print("Error: ", float(lin_reg.error_calculation(X, y, optimal_weight)))
new_data = create_dataset(1000)
linreg = LinearRegression()
X = np.asarray(new_data.iloc[:,:-1])
y = np.asarray(new_data.iloc[:,-1:])
new_weight = linreg.lasso_regression(X, y, 0.001, 100)
plot_comparison(w_actual, new_weight.flatten(), features)
print("Error: ", float(lin_reg.error_calculation(X, y, new_weight)))
feature_weights = dict(zip(*(new_data.drop(['Y'], 1).columns, new_weight)))
feature_weights
relevant_features = ["X1", "X2", "X3", "X4", "X5", "X6", "X7", "X8", "X10", "X11", "X12", "X13", "X15", "X16", "Y"]
rel = [a[1:] for a in relevant_features[:-1]]
trim_data = new_data[relevant_features]
trim_data.head()
X_trim = np.asarray(trim_data.iloc[:,:-1])
y_trim = np.asarray(trim_data.iloc[:,-1:])
trim_actual_weights = []
for i in range(1, 21):
if str(i) in rel:
trim_actual_weights.append(w_actual[i])
else:
trim_actual_weights.append(0)
new_ridge = LinearRegression()
w_lambda_2 = new_ridge.ridge_regression(X_trim, y_trim, 0.001)
plot_comparison(trim_actual_weights, w_lambda_2.flatten(), features)
w_lambda_2 = new_ridge.ridge_regression(X_trim, y_trim, 0.01)
plot_comparison(trim_actual_weights, w_lambda_2.flatten(), features)
w_lambda_2 = new_ridge.ridge_regression(X_trim, y_trim, 0.1)
plot_comparison(trim_actual_weights, w_lambda_2.flatten(), features)
w_lambda_2 = new_ridge.ridge_regression(X_trim, y_trim, 0.3)
plot_comparison(trim_actual_weights, w_lambda_2.flatten(), features)
w_lambda_2 = new_ridge.ridge_regression(X_trim, y_trim, 0.7)
plot_comparison(trim_actual_weights, w_lambda_2.flatten(), features)
w_lambda_2 = new_ridge.ridge_regression(X_trim, y_trim, 0.01)
plot_comparison(trim_actual_weights, w_lambda_2.flatten(), features)
w_lambda_2 = new_ridge.naive_regression(X_trim, y_trim)
plot_comparison(trim_actual_weights, w_lambda_2.flatten(), features)
class SVM():
def __init__(self, tau=1, t_0=1, tol=0.0001, mu=15, sol="Dual", kernel=None, poly_d=2):
self.tau = tau
self.t_0 = t_0
self.tol = tol
self.mu = mu
self.kernel = kernel
self.poly_d = poly_d
self.sol = sol
def damp_newt_step(self, x, objective_func, grad, hess):
g = grad(x)
h = hess(x)
h_inverse = np.linalg.inv(h)
lmbda = np.sqrt((g.T.dot(h_inverse.dot(g))))
x_new = x - (1/(1+lmbda))*h_inverse.dot(g)
gap = 0.5*lmbda**2
return x_new, gap
def damp_newt_method(self, x, objective_func, grad, hess):
x, gap = self.damp_newt_step(x, objective_func, grad, hess)
x_histogram = [x]
if self.tol < (3-(5**0.5))/2:
while gap > self.tol:
x, gap = self.damp_newt_step(x, objective_func, grad, hess)
x_histogram.append(x)
x_star = x
else:
raise ValueError("tol should be less than the condition value")
return x_star, x_histogram
def transform_dual_svm(self, tau, X, y):
n_obs = X.shape[1]
if self.kernel == "Polynomial":
K = (np.identity(n=n_obs) + X.T.dot(X)) ** self.poly_d
Q = (y*y)*K
else:
Q = (X*y).T.dot(X*y)
p = -np.ones(n_obs)
A = np.zeros((2 * n_obs, n_obs))
A[:n_obs, :] = np.identity(n_obs)
A[n_obs:, :] = -np.identity(n_obs)
b = np.zeros(2 * n_obs)
b[:n_obs] = 1 / (tau * n_obs)
return Q, p, A, b
def transform_primal_svm(self, tau, X, y):
d = X.shape[0]
d_ = d - 1
n = X.shape[1]
Q = np.zeros((d+n, d+n))
Q[:d_, :d_] = np.identity(d_)
p = np.zeros(d + n)
p[d:] = 1/(tau*n)
A = np.zeros((2*n, d + n))
A[:n, :d] = -(X*y).T
A[:n, d:] = np.diag([-1]*n)
A[n:, d:] = np.diag([-1]*n)
b = np.zeros(2 * n)
b[:n] = -1
return Q, p, A, b
def barrier_method(self, Q, p, A, b, x_0, t_0, mu, tol):
o_iters = []
m = b.shape[0]
if np.sum(A.dot(x_0) < b) == m:
t = t_0
x = x_0
x_histogram = [x_0]
while m / t >= tol:
f = lambda x: t*(0.5*np.dot(x, Q.dot(x)) + p.dot(x)) - np.sum(np.log(b - A.dot(x)))
g = lambda x: t*(Q.dot(x) + p) + np.sum(np.divide(A.T, b - A.dot(x)), axis=1)
h = lambda x: t*Q + (np.divide(A.T, b - A.dot(x))).dot((np.divide(A.T, b - A.dot(x))).T)
x, x_hist_newton = self.damp_newt_method(x, f, g, h)
x_histogram += x_hist_newton
o_iters += [len(x_hist_newton)]
t *= mu
x_sol = x
else:
raise ValueError("x_0 is not feasible, cannot proceed")
return x_sol, x_histogram, o_iters
def train(self, X, y):
self.n = X.shape[0]
self.d = X.shape[1]
X = np.vstack((X.T, np.ones(self.n)))
if self.sol == 'Dual':
self.x_0 = (1/(100*self.tau*self.n))*np.ones(self.n)
self.Q, self.p, self.A, self.b = self.transform_dual_svm(self.tau, X, y)
self.x_sol, self.x_histogram, self.o_iters = self.barrier_method(self.Q,self.p, self.A, self.b, self.x_0, self.t_0,self.mu, self.tol)
self.w = self.x_sol.dot((X*y).T)
elif self.sol == 'Primal':
self.x_0 = np.zeros(self.d + 1 + self.n)
self.x_0[self.d + 1:] = 1.1
self.Q, self.p, self.A, self.b = self.transform_primal_svm(self.tau, X, y)
self.x_sol, self.x_histogram, self.o_iters = self.barrier_method(self.Q,self.p, self.A, self.b, self.x_0, self.t_0,self.mu, self.tol)
self.w = self.x_sol[:self.d + 1]
def predict(self, X_test, y_test):
n_test = X_test.shape[0]
X_test = np.vstack((X_test.T, np.ones(n_test)))
y_pred = np.sign(self.w.T.dot(X_test))
accuracy = self.compute_mean_accuracy(y_pred, y_test)
return y_pred, accuracy
def compute_mean_accuracy(self, y_pred, y_test):
accuracy = np.sum(y_pred == y_test)
accuracy /= np.shape(y_test)[0]
return accuracy
from sklearn import datasets
iris_data = datasets.load_iris()
X, y = iris_data.data, iris_data.target
X.shape, y.shape
svm = SVM(kernel = False, sol = "Dual")
svm.train(X, y)
print("The training Error is computed as : ", svm.predict(X, y)[1])
print(svm.w)
xor_dict = {'A': [0,0,1,1], 'B': [0,1,0,1], 'Y': [0,1,1,0]}
xor_data = pd.DataFrame(xor_dict)
xor_data.head()
svm = SVM(kernel=True, tol=0.0001)
svm.train(xor_data.drop('Y', 1).values, xor_data['Y'].values)
print("The training Error is computed as : ", svm.predict(xor_data.drop('Y', 1).values, xor_data['Y'].values)[1])
print(svm.w)
svm = SVM(kernel=False, sol="Primal")
svm.train(xor_data.drop('Y', 1).values, xor_data['Y'].values)
print("The training Error is computed as : ", svm.predict(xor_data.drop('Y', 1).values, xor_data['Y'].values)[1])
print(svm.w)
| 0.233444 | 0.917006 |
# Phase 4. Modeling
In this phase, various modeling techniques are selected and applied, and their parameters are
calibrated to optimal values. Typically, there are several techniques for the same data mining
problem type. Some techniques have specific requirements on the form of data. Therefore,
going back to the data preparation phase is often necessary.
## 4.1 Select modeling techniques
### 4.1.1 Task
As the first step in modeling, select the actual modeling technique that is to be used. Although
you may have already selected a tool during the Business Understanding phase, this task refers to
the specific modeling technique.
### 4.1.2 Output
#### 4.1.2.1 Modeling technique
Document the actual modeling technique that is to be used
#### 4.1.2.2 Modeling assumptions
Many models make assumptions about the data, recording any of such assumptions made.
## 4.2 Generate test design
### 4.2.1 Task
Before we actually build a model, we need to generate a procedure or mechanism to test the model’s
quality and validity.
### 4.2.2 Output
#### 4.2.2.1 Test design
Describe the intended plan for training, testing, and evaluating the models.
## 4.3 Build model
### 4.3.1 Task
Run the modeling tool on the prepared dataset to create one or more models.
### 4.3.2 Output
#### 4.3.2.1 Parameter settings
With any modeling tool, there are often a large number of parameters that can be adjusted. List the
parameters and their chosen values, along with the rationale for the choice of parameter settings.
#### 4.3.2.2 Models
These are the actual models produced by the modeling tool, not a report.
#### 4.3.2.3 Model descriptions
Describe the resulting models. Report on the interpretation of the models and document any
difficulties encountered with their meanings.
## 4.4 Assess model
### 4.4.1 Task
Interpret the models according to available domain knowledge, sucess criteria and the desired
test design. The success of the modeling and discovered techniques need to be judged technically.
The models need to be evaluated according to the evaluation critera, and ranked in relation to each
other.
### 4.4.2 Output
#### 4.4.2.1 Model assessment
Summarize results of this task, list qualities of generated models (e.g., in terms of accuracy),
and rank their quality in relation to each other.
#### 4.4.2.2 Revised parameter settings
Summarize results of this task, list qualities of generated models (e.g., in terms of accuracy),
and rank their quality in relation to each other.
|
github_jupyter
|
# Phase 4. Modeling
In this phase, various modeling techniques are selected and applied, and their parameters are
calibrated to optimal values. Typically, there are several techniques for the same data mining
problem type. Some techniques have specific requirements on the form of data. Therefore,
going back to the data preparation phase is often necessary.
## 4.1 Select modeling techniques
### 4.1.1 Task
As the first step in modeling, select the actual modeling technique that is to be used. Although
you may have already selected a tool during the Business Understanding phase, this task refers to
the specific modeling technique.
### 4.1.2 Output
#### 4.1.2.1 Modeling technique
Document the actual modeling technique that is to be used
#### 4.1.2.2 Modeling assumptions
Many models make assumptions about the data, recording any of such assumptions made.
## 4.2 Generate test design
### 4.2.1 Task
Before we actually build a model, we need to generate a procedure or mechanism to test the model’s
quality and validity.
### 4.2.2 Output
#### 4.2.2.1 Test design
Describe the intended plan for training, testing, and evaluating the models.
## 4.3 Build model
### 4.3.1 Task
Run the modeling tool on the prepared dataset to create one or more models.
### 4.3.2 Output
#### 4.3.2.1 Parameter settings
With any modeling tool, there are often a large number of parameters that can be adjusted. List the
parameters and their chosen values, along with the rationale for the choice of parameter settings.
#### 4.3.2.2 Models
These are the actual models produced by the modeling tool, not a report.
#### 4.3.2.3 Model descriptions
Describe the resulting models. Report on the interpretation of the models and document any
difficulties encountered with their meanings.
## 4.4 Assess model
### 4.4.1 Task
Interpret the models according to available domain knowledge, sucess criteria and the desired
test design. The success of the modeling and discovered techniques need to be judged technically.
The models need to be evaluated according to the evaluation critera, and ranked in relation to each
other.
### 4.4.2 Output
#### 4.4.2.1 Model assessment
Summarize results of this task, list qualities of generated models (e.g., in terms of accuracy),
and rank their quality in relation to each other.
#### 4.4.2.2 Revised parameter settings
Summarize results of this task, list qualities of generated models (e.g., in terms of accuracy),
and rank their quality in relation to each other.
| 0.865494 | 0.987092 |
## Motif analyses on CLIP/RIP binding sites and DRNs in diffBUM-HMM and deltaSHAPE data.
For this to work you need to have the pyCRAC package and MEME tool suite installed.
```
import os
import math
import sys
import glob
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from scipy.special import comb
from more_itertools import pairwise
from scipy.stats import hypergeom,chisquare,fisher_exact
from matplotlib import rcParams
from collections import defaultdict
from pyCRAC.Methods import sortbyvalue,contigousarray2Intervals
rcParams['font.family'] = 'sans-serif'
rcParams['font.sans-serif'] = ['Arial']
rcParams['pdf.fonttype'] = 42
def normalizeIntervalLength(start,end,chromosomelength,length=20):
""" Extends or trims the interval coordinates to a set length. Default = 20 bp. """
newstart = int()
newend = int()
if start - length < 0: start = 1 # to make sure that the start is always a positive number
if end + length > chromosomelength: end = chromosomelength # to make sure that interval doesn't go beyond the chromosome boundaries.
actlength = end - start
difference = length - actlength
if difference == 0:
return start,end
else:
newstart = round(float(start) - float(difference)/2.0)
if newstart < 0: newstart = 1
newend = round(float(end) + float(difference)/2.0)
if newend > chromosomelength: newend = chromosomelength
return int(newstart),int(newend) # convert back to integers
def intervalsFromClosePositions(positions,mindistance=5):
""" Merges positions that are within a specific distance
to their neighbours:
[1,3,5,15,20,30,35,36,37,69,70,80,90,91]
should become:
[(1, 5), (15, 20), (30, 37), (69, 70), (80, 80), (90, 91)]
"""
start = None
end = None
intervallist = list()
for nr,i in enumerate(sorted(list(positions))):
if not start:
start = i
try:
if positions[nr+1] - positions[nr] <= mindistance:
continue
elif positions[nr+1] - positions[nr] > mindistance:
end = i
intervallist.append((start,end))
start = positions[nr+1]
except IndexError:
if start:
end = i
intervallist.append((start,end))
break
return intervallist
```
### Loading the big dataframe:
```
data = pd.read_csv('../../New_data_table.txt',\
sep="\t",\
header=0,\
index_col=0)
```
### Masking positions not considered by deltaSHAPE:
Any positions that have -999 values should not be further considered as these were positions where there was insufficient coverage.
```
positionstomask = data[(data["SHAPE_reactivity_ex_vivo_1"] < -900) |
(data["SHAPE_reactivity_ex_vivo_2"] < -900) |
(data["SHAPE_reactivity_in_cell_1"] < -900) |
(data["SHAPE_reactivity_in_cell_2"] < -900)].index
print(len(positionstomask))
data.loc[positionstomask,data.columns[11:]] = np.nan
data.columns
data.head()
```
### Now doing Motif analyses:
### First I need to make fasta files for all the protein binding sites in Xist and find their motifs:
I will normalize the interval size to a fixed lenght so that MEME can deal better with it. I will group together those DRNs that are within 2nt distance from each other.
```
data.columns
data.head()
chromosomelength = len(data.index)
```
### Setting the length of each interval for motif analyses:
```
fixedlength = 30
```
### Setting the threshold for selecting DRNs in the diffBUM-HMM data:
```
threshold = 0.95
proteins = ["CELF1","FUS","HuR","PTBP1","RBFOX2","TARDBP"]
proteinfastas = list()
for protein in proteins:
outfilename = "%s_Xist_RNA_binding_sites.fa" % protein
#print(outfilename)
outfile = open(outfilename,"w")
proteinfastas.append(outfilename)
indices = data[data[protein] > 0].index
intervals = intervalsFromClosePositions(indices)
print("%s:\t%s" % (protein,len(intervals)))
count = 1
for (i,j) in intervals:
start,end = normalizeIntervalLength(i,j,chromosomelength,fixedlength)
sequence = data.loc[start:end+1,"nucleotide"].values
sequence = "".join(sequence)
outfile.write(">%s_%s\n%s\n" % (protein,count,sequence))
count += 1
outfile.close()
proteinfastas = " ".join(proteinfastas)
print(proteinfastas)
```
### Running meme on these sequences:
```
%%bash -s "$proteinfastas"
DIR=$HOME/meme/bin
for i in $1
do
echo $i
FILE="$(basename -- $i)"
FILE=${FILE%.fa}
PREFIX=MEME_
OUTFILE=$PREFIX$FILE
$DIR/meme-chip \
-meme-minw 4 \
-meme-maxw 10 \
-meme-nmotifs 3 \
-meme-p 8 \
-meme-mod anr \
-norc \
-rna \
-noecho \
-oc $OUTFILE $i &
done
```
### Now doing it for the diffBUM_HMM and deltaSHAPE sites. Fragment sizes = fixedlength:
All the DRNs that were located within a 5 nucleotide window from each other were grouped into a single interval and length of this interval was normalized (so either extended or trimmed) to the fixed length (30 bp for these analyses). The reason for doing this is because otherwise the analyes would generate for each DRN a sequence containing the DRN in the middle and many of these sequences could overlap if the DRNs are in quite close proximity. This would result in a much higher enrichment of motifs from regions that have a high concentration of DRNs. So this is why they were first grouped together.
```
intervallengthcounter = defaultdict(lambda: defaultdict(int))
```
### diffBUM_HMM:
```
sequence = "".join(data.nucleotide.values)
outfile = open("diffBUM_HMM_ex_vivo_%s_mers.fa" % fixedlength,"w")
### How many DRNs are there in the data?:
ex_vivo_pos = data[data.ex_vivo >= threshold].index
### intervalsFromClosePositions groups the DRNs together in intervals.
intervals = intervalsFromClosePositions(ex_vivo_pos,)
### This prints the number of intervals that were detected and how many DRNs there were in the data.
print(len(intervals))
print(len(ex_vivo_pos))
count = 0
for (i,j) in intervals:
length = (j-i)+1
intervallengthcounter["diffBUM_HMM_ex_vivo"][length] += 1
### These intervals are then set to a fixed length here:
start,end = normalizeIntervalLength(i,j,chromosomelength,fixedlength)
sequence = data.loc[start:end+1,"nucleotide"].values
sequence = "".join(sequence)
outfile.write(">diffBUM_HMM_ex_vivo_%s\n%s\n" % (count,sequence))
count += 1
outfile.close()
in_vivo_pos = data[data.in_vivo >= threshold].index
outfile = open("diffBUM_HMM_in_vivo_%s_mers.fa" % fixedlength,"w")
intervals = intervalsFromClosePositions(in_vivo_pos)
print(len(intervals))
print(len(in_vivo_pos))
count = 0
for (i,j) in intervals:
length = (j-i)+1
intervallengthcounter["diffBUM_HMM_in_vivo"][length] += 1
start,end = normalizeIntervalLength(i,j,chromosomelength,fixedlength)
sequence = data.loc[start:end+1,"nucleotide"].values
sequence = "".join(sequence)
outfile.write(">diffBUM_HMM_in_vivo_%s\n%s\n" % (count,sequence))
count += 1
outfile.close()
```
### deltaSHAPE rep 1:
```
sequence = "".join(data.nucleotide.values)
ex_vivo_pos = data[data.deltaSHAPE_rep1 > 0].index
outfile = open("deltaSHAPE_rep_1_ex_vivo_%s_mers.fa" % fixedlength,"w")
intervals = intervalsFromClosePositions(ex_vivo_pos)
print(len(intervals))
print(len(ex_vivo_pos))
count = 0
for (i,j) in intervals:
length = (j-i)+1
intervallengthcounter["deltaSHAPE_rep1_ex_vivo"][length] += 1
start,end = normalizeIntervalLength(i,j,chromosomelength,fixedlength)
sequence = data.loc[start:end+1,"nucleotide"].values
sequence = "".join(sequence)
outfile.write(">deltaSHAPE_rep1_ex_vivo_%s\n%s\n" % (count,sequence))
count += 1
outfile.close()
in_vivo_pos = data[data.deltaSHAPE_rep1 < 0].index
outfile = open("deltaSHAPE_rep_1_in_vivo_%s_mers.fa" % fixedlength,"w")
intervals = intervalsFromClosePositions(in_vivo_pos)
print(len(intervals))
print(len(in_vivo_pos))
count = 0
for (i,j) in intervals:
length = (j-i)+1
intervallengthcounter["deltaSHAPE_rep1_in_vivo"][length] += 1
start,end = normalizeIntervalLength(i,j,chromosomelength,fixedlength)
sequence = data.loc[start:end+1,"nucleotide"].values
sequence = "".join(sequence)
outfile.write(">deltaSHAPE_rep1_in_vivo_%s\n%s\n" % (count,sequence))
count += 1
outfile.close()
```
### deltaSHAPE rep 2:
```
sequence = "".join(data.nucleotide.values)
ex_vivo_pos = data[data.deltaSHAPE_rep2 > 0].index
outfile = open("deltaSHAPE_rep_2_ex_vivo_%s_mers.fa" % fixedlength,"w")
intervals = intervalsFromClosePositions(ex_vivo_pos)
print(len(intervals))
print(len(ex_vivo_pos))
count = 0
for (i,j) in intervals:
length = (j-i)+1
intervallengthcounter["deltaSHAPE_rep2_ex_vivo"][length] += 1
start,end = normalizeIntervalLength(i,j,chromosomelength,fixedlength)
sequence = data.loc[start:end+1,"nucleotide"].values
sequence = "".join(sequence)
outfile.write(">deltaSHAPE_rep2_ex_vivo_%s\n%s\n" % (count,sequence))
count += 1
outfile.close()
in_vivo_pos = data[data.deltaSHAPE_rep2 < 0].index
outfile = open("deltaSHAPE_rep_2_in_vivo_%s_mers.fa" % fixedlength,"w")
intervals = intervalsFromClosePositions(in_vivo_pos)
print(len(intervals))
print(len(in_vivo_pos))
count = 0
for (i,j) in intervals:
length = (j-i)+1
intervallengthcounter["deltaSHAPE_rep2_in_vivo"][length] += 1
start,end = normalizeIntervalLength(i,j,chromosomelength,fixedlength)
sequence = data.loc[start:end+1,"nucleotide"].values
sequence = "".join(sequence)
outfile.write(">deltaSHAPE_rep2_in_vivo_%s\n%s\n" % (count,sequence))
count += 1
outfile.close()
kmerfasta = " ".join(glob.glob("*%s_mers.fa" % fixedlength))
print(kmerfasta.split())
%%bash -s "$kmerfasta"
echo $1
DIR=$HOME/meme/bin
for i in $1
do
FILE="$(basename -- $i)"
FILE=${FILE%.fa}
PREFIX=MEME_V2
OUTFILE=$PREFIX$FILE
$DIR/meme-chip \
-meme-minw 4 \
-meme-maxw 10 \
-meme-nmotifs 20 \
-meme-p 8 \
-meme-mod anr \
-norc \
-rna \
-noecho \
-o $OUTFILE $i &
done
```
### Look at the lenghts of the intervals:
```
list(intervallengthcounter.keys())
intervallengthcounter
```
### This plot was generated to check on average how many DRNs were found in the 30bp sequences used for MEME analyses.
As you can see, the majority of the DRN intervals still consist of single-nucleotide DRNs, which means that most DRNs are actually quite far apart in the data. This is not the case for deltaSHAPE that consitenly picks up stretches of 3 DRNs.
```
numberofplots = len(intervallengthcounter.keys())
samples = list(intervallengthcounter.keys())
fig,ax = plt.subplots(numberofplots,figsize=[3,10],sharex=True)
for i in range(numberofplots):
sample = samples[i]
data = intervallengthcounter[sample]
x = list(data.keys())
y = list(data.values())
sns.barplot(x,y,ax=ax[i],color="blue")
ax[i].set_title(sample)
ax[i].set_ylabel('Counts')
plt.tight_layout()
fig.savefig("Distribution_of_stretches_of_diff_mod_nucleotides_v2.pdf",dpi=400)
```
### Now running everything through fimo to get the coordinates for the motifs identified by deltaSHAPE and diffBUM-HMM.
This would be a useful resource for people studying Xist and Xist RBPs.
```
directories = glob.glob("MEME_V2*")
directories
directories = " ".join(directories)
%%bash -s "$directories"
DIR=$HOME/meme/bin
for i in $1
do
$DIR/fimo \
--oc $i \
--verbosity 1 \
$i/meme_out/meme.txt \
../../Reference_sequences/Xist.fa &
done
```
|
github_jupyter
|
import os
import math
import sys
import glob
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from scipy.special import comb
from more_itertools import pairwise
from scipy.stats import hypergeom,chisquare,fisher_exact
from matplotlib import rcParams
from collections import defaultdict
from pyCRAC.Methods import sortbyvalue,contigousarray2Intervals
rcParams['font.family'] = 'sans-serif'
rcParams['font.sans-serif'] = ['Arial']
rcParams['pdf.fonttype'] = 42
def normalizeIntervalLength(start,end,chromosomelength,length=20):
""" Extends or trims the interval coordinates to a set length. Default = 20 bp. """
newstart = int()
newend = int()
if start - length < 0: start = 1 # to make sure that the start is always a positive number
if end + length > chromosomelength: end = chromosomelength # to make sure that interval doesn't go beyond the chromosome boundaries.
actlength = end - start
difference = length - actlength
if difference == 0:
return start,end
else:
newstart = round(float(start) - float(difference)/2.0)
if newstart < 0: newstart = 1
newend = round(float(end) + float(difference)/2.0)
if newend > chromosomelength: newend = chromosomelength
return int(newstart),int(newend) # convert back to integers
def intervalsFromClosePositions(positions,mindistance=5):
""" Merges positions that are within a specific distance
to their neighbours:
[1,3,5,15,20,30,35,36,37,69,70,80,90,91]
should become:
[(1, 5), (15, 20), (30, 37), (69, 70), (80, 80), (90, 91)]
"""
start = None
end = None
intervallist = list()
for nr,i in enumerate(sorted(list(positions))):
if not start:
start = i
try:
if positions[nr+1] - positions[nr] <= mindistance:
continue
elif positions[nr+1] - positions[nr] > mindistance:
end = i
intervallist.append((start,end))
start = positions[nr+1]
except IndexError:
if start:
end = i
intervallist.append((start,end))
break
return intervallist
data = pd.read_csv('../../New_data_table.txt',\
sep="\t",\
header=0,\
index_col=0)
positionstomask = data[(data["SHAPE_reactivity_ex_vivo_1"] < -900) |
(data["SHAPE_reactivity_ex_vivo_2"] < -900) |
(data["SHAPE_reactivity_in_cell_1"] < -900) |
(data["SHAPE_reactivity_in_cell_2"] < -900)].index
print(len(positionstomask))
data.loc[positionstomask,data.columns[11:]] = np.nan
data.columns
data.head()
data.columns
data.head()
chromosomelength = len(data.index)
fixedlength = 30
threshold = 0.95
proteins = ["CELF1","FUS","HuR","PTBP1","RBFOX2","TARDBP"]
proteinfastas = list()
for protein in proteins:
outfilename = "%s_Xist_RNA_binding_sites.fa" % protein
#print(outfilename)
outfile = open(outfilename,"w")
proteinfastas.append(outfilename)
indices = data[data[protein] > 0].index
intervals = intervalsFromClosePositions(indices)
print("%s:\t%s" % (protein,len(intervals)))
count = 1
for (i,j) in intervals:
start,end = normalizeIntervalLength(i,j,chromosomelength,fixedlength)
sequence = data.loc[start:end+1,"nucleotide"].values
sequence = "".join(sequence)
outfile.write(">%s_%s\n%s\n" % (protein,count,sequence))
count += 1
outfile.close()
proteinfastas = " ".join(proteinfastas)
print(proteinfastas)
%%bash -s "$proteinfastas"
DIR=$HOME/meme/bin
for i in $1
do
echo $i
FILE="$(basename -- $i)"
FILE=${FILE%.fa}
PREFIX=MEME_
OUTFILE=$PREFIX$FILE
$DIR/meme-chip \
-meme-minw 4 \
-meme-maxw 10 \
-meme-nmotifs 3 \
-meme-p 8 \
-meme-mod anr \
-norc \
-rna \
-noecho \
-oc $OUTFILE $i &
done
intervallengthcounter = defaultdict(lambda: defaultdict(int))
sequence = "".join(data.nucleotide.values)
outfile = open("diffBUM_HMM_ex_vivo_%s_mers.fa" % fixedlength,"w")
### How many DRNs are there in the data?:
ex_vivo_pos = data[data.ex_vivo >= threshold].index
### intervalsFromClosePositions groups the DRNs together in intervals.
intervals = intervalsFromClosePositions(ex_vivo_pos,)
### This prints the number of intervals that were detected and how many DRNs there were in the data.
print(len(intervals))
print(len(ex_vivo_pos))
count = 0
for (i,j) in intervals:
length = (j-i)+1
intervallengthcounter["diffBUM_HMM_ex_vivo"][length] += 1
### These intervals are then set to a fixed length here:
start,end = normalizeIntervalLength(i,j,chromosomelength,fixedlength)
sequence = data.loc[start:end+1,"nucleotide"].values
sequence = "".join(sequence)
outfile.write(">diffBUM_HMM_ex_vivo_%s\n%s\n" % (count,sequence))
count += 1
outfile.close()
in_vivo_pos = data[data.in_vivo >= threshold].index
outfile = open("diffBUM_HMM_in_vivo_%s_mers.fa" % fixedlength,"w")
intervals = intervalsFromClosePositions(in_vivo_pos)
print(len(intervals))
print(len(in_vivo_pos))
count = 0
for (i,j) in intervals:
length = (j-i)+1
intervallengthcounter["diffBUM_HMM_in_vivo"][length] += 1
start,end = normalizeIntervalLength(i,j,chromosomelength,fixedlength)
sequence = data.loc[start:end+1,"nucleotide"].values
sequence = "".join(sequence)
outfile.write(">diffBUM_HMM_in_vivo_%s\n%s\n" % (count,sequence))
count += 1
outfile.close()
sequence = "".join(data.nucleotide.values)
ex_vivo_pos = data[data.deltaSHAPE_rep1 > 0].index
outfile = open("deltaSHAPE_rep_1_ex_vivo_%s_mers.fa" % fixedlength,"w")
intervals = intervalsFromClosePositions(ex_vivo_pos)
print(len(intervals))
print(len(ex_vivo_pos))
count = 0
for (i,j) in intervals:
length = (j-i)+1
intervallengthcounter["deltaSHAPE_rep1_ex_vivo"][length] += 1
start,end = normalizeIntervalLength(i,j,chromosomelength,fixedlength)
sequence = data.loc[start:end+1,"nucleotide"].values
sequence = "".join(sequence)
outfile.write(">deltaSHAPE_rep1_ex_vivo_%s\n%s\n" % (count,sequence))
count += 1
outfile.close()
in_vivo_pos = data[data.deltaSHAPE_rep1 < 0].index
outfile = open("deltaSHAPE_rep_1_in_vivo_%s_mers.fa" % fixedlength,"w")
intervals = intervalsFromClosePositions(in_vivo_pos)
print(len(intervals))
print(len(in_vivo_pos))
count = 0
for (i,j) in intervals:
length = (j-i)+1
intervallengthcounter["deltaSHAPE_rep1_in_vivo"][length] += 1
start,end = normalizeIntervalLength(i,j,chromosomelength,fixedlength)
sequence = data.loc[start:end+1,"nucleotide"].values
sequence = "".join(sequence)
outfile.write(">deltaSHAPE_rep1_in_vivo_%s\n%s\n" % (count,sequence))
count += 1
outfile.close()
sequence = "".join(data.nucleotide.values)
ex_vivo_pos = data[data.deltaSHAPE_rep2 > 0].index
outfile = open("deltaSHAPE_rep_2_ex_vivo_%s_mers.fa" % fixedlength,"w")
intervals = intervalsFromClosePositions(ex_vivo_pos)
print(len(intervals))
print(len(ex_vivo_pos))
count = 0
for (i,j) in intervals:
length = (j-i)+1
intervallengthcounter["deltaSHAPE_rep2_ex_vivo"][length] += 1
start,end = normalizeIntervalLength(i,j,chromosomelength,fixedlength)
sequence = data.loc[start:end+1,"nucleotide"].values
sequence = "".join(sequence)
outfile.write(">deltaSHAPE_rep2_ex_vivo_%s\n%s\n" % (count,sequence))
count += 1
outfile.close()
in_vivo_pos = data[data.deltaSHAPE_rep2 < 0].index
outfile = open("deltaSHAPE_rep_2_in_vivo_%s_mers.fa" % fixedlength,"w")
intervals = intervalsFromClosePositions(in_vivo_pos)
print(len(intervals))
print(len(in_vivo_pos))
count = 0
for (i,j) in intervals:
length = (j-i)+1
intervallengthcounter["deltaSHAPE_rep2_in_vivo"][length] += 1
start,end = normalizeIntervalLength(i,j,chromosomelength,fixedlength)
sequence = data.loc[start:end+1,"nucleotide"].values
sequence = "".join(sequence)
outfile.write(">deltaSHAPE_rep2_in_vivo_%s\n%s\n" % (count,sequence))
count += 1
outfile.close()
kmerfasta = " ".join(glob.glob("*%s_mers.fa" % fixedlength))
print(kmerfasta.split())
%%bash -s "$kmerfasta"
echo $1
DIR=$HOME/meme/bin
for i in $1
do
FILE="$(basename -- $i)"
FILE=${FILE%.fa}
PREFIX=MEME_V2
OUTFILE=$PREFIX$FILE
$DIR/meme-chip \
-meme-minw 4 \
-meme-maxw 10 \
-meme-nmotifs 20 \
-meme-p 8 \
-meme-mod anr \
-norc \
-rna \
-noecho \
-o $OUTFILE $i &
done
list(intervallengthcounter.keys())
intervallengthcounter
numberofplots = len(intervallengthcounter.keys())
samples = list(intervallengthcounter.keys())
fig,ax = plt.subplots(numberofplots,figsize=[3,10],sharex=True)
for i in range(numberofplots):
sample = samples[i]
data = intervallengthcounter[sample]
x = list(data.keys())
y = list(data.values())
sns.barplot(x,y,ax=ax[i],color="blue")
ax[i].set_title(sample)
ax[i].set_ylabel('Counts')
plt.tight_layout()
fig.savefig("Distribution_of_stretches_of_diff_mod_nucleotides_v2.pdf",dpi=400)
directories = glob.glob("MEME_V2*")
directories
directories = " ".join(directories)
%%bash -s "$directories"
DIR=$HOME/meme/bin
for i in $1
do
$DIR/fimo \
--oc $i \
--verbosity 1 \
$i/meme_out/meme.txt \
../../Reference_sequences/Xist.fa &
done
| 0.290176 | 0.808408 |
**Brief Honor Code**. Do the homework on your own. You may discuss ideas with your classmates, but DO NOT copy the solutions from someone else or the Internet. If stuck, discuss with TA.
**1**. (10 points)
Rewrite the following code into functional form using lambdas, map, filter and reduce.
```
n = 10
s = 10
for i in range(n):
if i % 2:
s = s| i**2
s
from functools import reduce
reduce(lambda x, y: x | y, list(map(lambda x: x**2,list(filter(lambda x: x % 2, range(10))))),10)
```
**2**. (10 points)
Rewrite the code above as a `toolz` pipeline, using lambdas and curried or partially applied functions as necessary.
#### Method 1
```
import toolz as tz
import toolz.curried as c
res=tz.pipe(
range(10),
c.map(lambda x: x**2),
c.filter(lambda x: x % 2),
c.reduce(lambda x, y: x | y ),
lambda x: x|10
)
res
```
#### Method 2
```
res2=tz.pipe(
range(10),
c.map(lambda x: x**2),
c.filter(lambda x: x % 2),
list,
lambda x: x+[10],
c.reduce(lambda x, y: x | y ),
)
res2
```
**3**. (10 points)
Repeat the Buffon's needle simulation from Lab01 as a function that takes the number of needels `n` as input and returns the estimate of $\pi$. The function should use `numpy` and vectorization. What is $\pi$ for 1 million needles?
```
import random
from numpy import pi, sin
def buffon(L, D, N):
""" function to simulate the buffon's needle experiment
L is the length of the needle,
D is the difference between two lines
N is the number of times to drop the needle
"""
count= 0;
for loop in range(N) :
theta = pi*random.uniform(0,180)/180
if L * sin(theta) > random.uniform(0,D):
count += 1
est=count/N
Pi_est = (2*L) / (est*D)
return Pi_est
L = 1
D = 4
N = int(1e6)
buffon(L, D, N)
```
**4**. (20 points)
Simpsons rule is given by the follwoing approximation

- Write Simpsons rule as a function `simpsons(f, a, b, n=100)` where n is the number of equally spaced intervals from `a` to `b`. (10 points)
- Use this function to estimate the probability mass of the standard normal distribution between -1 and 1. Implement the PDF of the standard normal distribution $\psi(x)$ as a function. (10 points)
$$
\psi(x) = \frac{1}{\sqrt{2\pi}}e^{-\frac{1}{2}x^2}
$$
```
import numpy as np
def normal(x):
""" function to get the pdf of a normal distribution
"""
res=(1/np.sqrt(2*np.pi))*np.exp(-0.5*(x**2))
return(res)
def simpsons(f,a,b,n=100):
""" function for the using of simpsons methods to estimate the intergation values
"""
h=(b-a)
x=np.arange(a, b, h/n)
temp=f(x[0])+f(x[n-1])
even=0
odd=0
N=len(x)
for i in range(1,N):
if i % 2==0:
even=even+2*f(x[i])
else:
odd=odd+4*f(x[i])
res=(temp+even+odd)*h/(3*N)
return(res)
simpsons(normal,-1,1,n=100)
```
**5**. (50 points)
Write code to generate a plot similar to the following

using the explanation for generation of 1D Cellular Automata found [here](http://mathworld.wolfram.com/ElementaryCellularAutomaton.html). You should only need to use standard Python, `numpy` and `matplotllib`.
The input to the function making the plots should be a simple list of rules
```python
rules = [30, 54, 60, 62, 90, 94, 102, 110, 122, 126,
150, 158, 182, 188, 190, 220, 222, 250]
make_plots(rules, niter, ncols)
```
You may, of course, write other helper functions to keep your code modular.
```
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
def make_plots(rules, niter,ncols):
"""Function to generate of 1D Cellular Automata;
type in the rules, a list of number from 0 to 255;
niter is the number of rows in the plot
ncols is the number of columns in the plot
"""
fig, axes = plt.subplots(niter, ncols, figsize=(12, 18))
rule=np.array([1,1,1,1,1,0,1,0,1,1,0,0,0,1,1,0,1,0,0,0,1,0,0,0]).reshape((8,3))
rules_new=np.array(rules).reshape((niter,ncols))
for rows in range(niter):
for cols in range(ncols):
r=rules_new[rows][cols]
binary=list(np.binary_repr(r,width=8))
binary = list(map(int, binary))
def next_line(start):
"""Function to generate of 1D Cellular Automata;
Can get the numbers of each cell;
start determines the number of iterations.
"""
res=[]
res.append(np.array([start[30],start[0],start[1]]))
for i in range(29):
res.append(start[(i):(i+3)])
res.append(np.array([start[29],start[30],start[0]]))
point=[]
for i in range(31):
temp=res[i]
temp2= temp==rule
num=np.where(np.sum(temp2,axis=1)==3)[0][0]
point.append(binary[num])
return(point)
xs=np.zeros((16,31))
xs[0,15]=1
start=xs[0,]
res=list(start)
temp=start
for i in range(15):
temp=next_line(temp)
res=res+temp
xs=np.array(res).reshape((16,31))
name='Rule ' + str(r)
axes[rows][cols].imshow(xs, cmap='Greys', interpolation='nearest')
axes[rows][cols].set_title(name,fontsize=12)
axes[rows][cols].set_xticks(np.arange(0.5, 30.5, 1));
axes[rows][cols].set_yticks(np.arange(0.5, 15.5, 1));
axes[rows][cols].set_xticks(np.arange(-.5, 30.5, 1), minor=True);
axes[rows][cols].set_yticks(np.arange(-.5, 15.5, 1), minor=True);
axes[rows][cols].grid(which='minor', color='black', linestyle='-', linewidth=0.5)
axes[rows][cols].set_xticks([])
axes[rows][cols].set_yticks([])
pass
rules = [30, 54, 60, 62, 90, 94, 102, 110, 122, 126,
150, 158, 182, 188, 190, 220, 222, 250]
make_plots(rules, 6,3)
```
|
github_jupyter
|
n = 10
s = 10
for i in range(n):
if i % 2:
s = s| i**2
s
from functools import reduce
reduce(lambda x, y: x | y, list(map(lambda x: x**2,list(filter(lambda x: x % 2, range(10))))),10)
import toolz as tz
import toolz.curried as c
res=tz.pipe(
range(10),
c.map(lambda x: x**2),
c.filter(lambda x: x % 2),
c.reduce(lambda x, y: x | y ),
lambda x: x|10
)
res
res2=tz.pipe(
range(10),
c.map(lambda x: x**2),
c.filter(lambda x: x % 2),
list,
lambda x: x+[10],
c.reduce(lambda x, y: x | y ),
)
res2
import random
from numpy import pi, sin
def buffon(L, D, N):
""" function to simulate the buffon's needle experiment
L is the length of the needle,
D is the difference between two lines
N is the number of times to drop the needle
"""
count= 0;
for loop in range(N) :
theta = pi*random.uniform(0,180)/180
if L * sin(theta) > random.uniform(0,D):
count += 1
est=count/N
Pi_est = (2*L) / (est*D)
return Pi_est
L = 1
D = 4
N = int(1e6)
buffon(L, D, N)
import numpy as np
def normal(x):
""" function to get the pdf of a normal distribution
"""
res=(1/np.sqrt(2*np.pi))*np.exp(-0.5*(x**2))
return(res)
def simpsons(f,a,b,n=100):
""" function for the using of simpsons methods to estimate the intergation values
"""
h=(b-a)
x=np.arange(a, b, h/n)
temp=f(x[0])+f(x[n-1])
even=0
odd=0
N=len(x)
for i in range(1,N):
if i % 2==0:
even=even+2*f(x[i])
else:
odd=odd+4*f(x[i])
res=(temp+even+odd)*h/(3*N)
return(res)
simpsons(normal,-1,1,n=100)
rules = [30, 54, 60, 62, 90, 94, 102, 110, 122, 126,
150, 158, 182, 188, 190, 220, 222, 250]
make_plots(rules, niter, ncols)
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
def make_plots(rules, niter,ncols):
"""Function to generate of 1D Cellular Automata;
type in the rules, a list of number from 0 to 255;
niter is the number of rows in the plot
ncols is the number of columns in the plot
"""
fig, axes = plt.subplots(niter, ncols, figsize=(12, 18))
rule=np.array([1,1,1,1,1,0,1,0,1,1,0,0,0,1,1,0,1,0,0,0,1,0,0,0]).reshape((8,3))
rules_new=np.array(rules).reshape((niter,ncols))
for rows in range(niter):
for cols in range(ncols):
r=rules_new[rows][cols]
binary=list(np.binary_repr(r,width=8))
binary = list(map(int, binary))
def next_line(start):
"""Function to generate of 1D Cellular Automata;
Can get the numbers of each cell;
start determines the number of iterations.
"""
res=[]
res.append(np.array([start[30],start[0],start[1]]))
for i in range(29):
res.append(start[(i):(i+3)])
res.append(np.array([start[29],start[30],start[0]]))
point=[]
for i in range(31):
temp=res[i]
temp2= temp==rule
num=np.where(np.sum(temp2,axis=1)==3)[0][0]
point.append(binary[num])
return(point)
xs=np.zeros((16,31))
xs[0,15]=1
start=xs[0,]
res=list(start)
temp=start
for i in range(15):
temp=next_line(temp)
res=res+temp
xs=np.array(res).reshape((16,31))
name='Rule ' + str(r)
axes[rows][cols].imshow(xs, cmap='Greys', interpolation='nearest')
axes[rows][cols].set_title(name,fontsize=12)
axes[rows][cols].set_xticks(np.arange(0.5, 30.5, 1));
axes[rows][cols].set_yticks(np.arange(0.5, 15.5, 1));
axes[rows][cols].set_xticks(np.arange(-.5, 30.5, 1), minor=True);
axes[rows][cols].set_yticks(np.arange(-.5, 15.5, 1), minor=True);
axes[rows][cols].grid(which='minor', color='black', linestyle='-', linewidth=0.5)
axes[rows][cols].set_xticks([])
axes[rows][cols].set_yticks([])
pass
rules = [30, 54, 60, 62, 90, 94, 102, 110, 122, 126,
150, 158, 182, 188, 190, 220, 222, 250]
make_plots(rules, 6,3)
| 0.324235 | 0.929248 |
# Units and Quantities
## Objectives
- Use units
- Create functions that accept quantities as arguments
- Create new units
## Basics
How do we define a Quantity and which parts does it have?
```
from astropy import units as u
# Define a quantity length
# print it
# Type of quantity
# Type of unit
# Quantity
# value
# unit
# information
```
Quantities can be converted to other units systems or factors by using `to()`
```
# Convert it to: km, lyr
```
We can do arithmetic operations when the quantities have the compatible units:
```
# arithmetic with distances
```
Quantities can also be combined, for example to measure speed
```
# calculate a speed
# decompose it
```
<div style='background:#B1E0A8; padding:10px 10px 10px 10px;'>
<H2> Challenges </H2>
<ol>
<li> Convert the speed in imperial units (miles/hour) using: <br>
```from astropy.units import imperial```
</li>
<li> Calculate whether a pint is more than half litre<br>
<emph>You can compare quantities as comparing variables.</emph> <br>
Something strange? Check what deffinition of <a href='https://en.wikipedia.org/wiki/Pint'>pint</a> astropy is using.
</li>
<li> Does units work with areas? calculate the area of a rectangle of 3 km of side and 5 meter of width. Show them in m^2 and convert them to yards^2</li>
</div>
```
#1
#2
#3
```
## Composed units
Many units are compositions of others, for example, one could create new combinationes for ease of use:
```
# create a composite unit
# and in the imperial system
```
and others are already a composition:
```
# what can be converted from s-1?
# or Jules?
# Unity of R
```
Sometime we get *no units* quantitites
```
# no units
```
What happen if we add a number to this?
```
# arithmetic with no units
# final value of a no unit quantity
```
## Equivalencies
Some conversions are not done by a conversion factor as between miles and kilometers, for example converting between wavelength and frequency.
```
# converting spectral quantities
# but doing it right
```
Other built-in equivalencies are:
- `parallax()`
- Doppler (`dopplr_radio`, `doppler_optical`, `doppler_relativistic`)
- spectral flux density
- brigthness temperature
- temperature energy
- and you can [build your own](http://astropy.readthedocs.org/en/stable/units/equivalencies.html#writing-new-equivalencies)
```
# finding the equivalencies
# but also using other systems
```
## Printing the quantities
```
# Printing values with different formats
```
## Arrays
Quantities can also be applied to arrays
```
# different ways of defining a quantity for a single value
# now with lists
# and arrays
# and its arithmetics
# angles are smart!
```
## Plotting quantities
To work nicely with matplotlib we need to do as follows:
```
# allowing for plotting
from astropy.visualization import quantity_support
quantity_support()
# loading matplotlib
%matplotlib inline
from matplotlib import pyplot as plt
# Ploting the previous array
```
## Creating functions with quantities as units
We want to have functions that contain the information of the untis, and with them we can be sure that we will be always have the *right* result.
```
# Create a function for the Kinetic energy
# run with and without units
```
<div style='background:#B1E0A8; padding:10px 10px 10px 10px;'>
<H2> Challenges </H2>
<ol start=4>
<li> Create a function that calculates potential energy where *g* defaults to Earth value,
but could be used for different planets.
Test it for any of the *g* values for any other
<a href="http://www.physicsclassroom.com/class/circles/Lesson-3/The-Value-of-g">planet</a>.
</li>
</ol>
</div>
```
#4
# run it for some values
# on Mars:
```
## Create your own units
Some times we want to create our own units:
```
# Create units for a laugh scale
```
<div style='background:#B1E0A8; padding:10px 10px 10px 10px;'>
<H2> Challenges </H2>
<ol start=5>
<li> Convert the area calculated before `rectangle_area` in <a href="https://en.wikipedia.org/wiki/Hectare">Hectare</a>
(1 hectare = 100 ares; 1 are = 100 m2).
</li>
</ol>
</div>
```
#5
```
|
github_jupyter
|
from astropy import units as u
# Define a quantity length
# print it
# Type of quantity
# Type of unit
# Quantity
# value
# unit
# information
# Convert it to: km, lyr
# arithmetic with distances
# calculate a speed
# decompose it
</li>
<li> Calculate whether a pint is more than half litre<br>
<emph>You can compare quantities as comparing variables.</emph> <br>
Something strange? Check what deffinition of <a href='https://en.wikipedia.org/wiki/Pint'>pint</a> astropy is using.
</li>
<li> Does units work with areas? calculate the area of a rectangle of 3 km of side and 5 meter of width. Show them in m^2 and convert them to yards^2</li>
</div>
## Composed units
Many units are compositions of others, for example, one could create new combinationes for ease of use:
and others are already a composition:
Sometime we get *no units* quantitites
What happen if we add a number to this?
## Equivalencies
Some conversions are not done by a conversion factor as between miles and kilometers, for example converting between wavelength and frequency.
Other built-in equivalencies are:
- `parallax()`
- Doppler (`dopplr_radio`, `doppler_optical`, `doppler_relativistic`)
- spectral flux density
- brigthness temperature
- temperature energy
- and you can [build your own](http://astropy.readthedocs.org/en/stable/units/equivalencies.html#writing-new-equivalencies)
## Printing the quantities
## Arrays
Quantities can also be applied to arrays
## Plotting quantities
To work nicely with matplotlib we need to do as follows:
## Creating functions with quantities as units
We want to have functions that contain the information of the untis, and with them we can be sure that we will be always have the *right* result.
<div style='background:#B1E0A8; padding:10px 10px 10px 10px;'>
<H2> Challenges </H2>
<ol start=4>
<li> Create a function that calculates potential energy where *g* defaults to Earth value,
but could be used for different planets.
Test it for any of the *g* values for any other
<a href="http://www.physicsclassroom.com/class/circles/Lesson-3/The-Value-of-g">planet</a>.
</li>
</ol>
</div>
## Create your own units
Some times we want to create our own units:
<div style='background:#B1E0A8; padding:10px 10px 10px 10px;'>
<H2> Challenges </H2>
<ol start=5>
<li> Convert the area calculated before `rectangle_area` in <a href="https://en.wikipedia.org/wiki/Hectare">Hectare</a>
(1 hectare = 100 ares; 1 are = 100 m2).
</li>
</ol>
</div>
| 0.776199 | 0.987017 |
# Text classification
**Supervised learning: classification**
**Abstract:** Classifying text, e.g. assigning a topic to a text or document, is one of the canonical examples of problems we try to solve with machine learning. This notebook demonstrates a simple example where, after training a classification algorithm with a bunch of text examples and topic labels, we use it to tell us the topic of any previously unseen text.
**Topics covevered:** multinomial naive Bayes, Bayes Theorem, feature engineering, term frequency–inverse document frequency (TF–IDF), stopwords, sparse matrices, confusion matrix.
**Motivation.** Solving such a text classification problem can be extremely useful. Say, right now your digital customer care team is swamped with comments being left on your website. You want to be able to tell the relevant comments from the irrelevant ones, and you want to know what they are about. Are they complaints about servicing times? Is a critical point in the purchase flow of the site suddenly broken? Are they about a particular aspect of a rather complex product? You want to do this classification fast and at scale. Enter machine learning.
**A word on naive Bayes.** As given away in the name, naive Bayes (NB) relies on Bayes' Theorem. Given some examples the goal is to predict the probability of a label. Say we want to decide between two labels, let's call them $L_0$ and $L_1$. For that we observe some features $(x_0, x_1, \dots, x_p) \equiv X$. In order to decide what example has what label, we can compute the ratio of the posterior probabilities
$$
\frac{P\left(L_0 | X\right)}{P\left(L_1 | X\right)}
=
\frac{P\left(X | L_0\right)P\left(L_0\right)}{P\left(X | L_1\right)P\left(L_1\right)}
.
$$
This provides us with a measure of the uncertainty attached to every label-assignment decision we make.
The group of NB classification models are _generative_, meaning they try to specify the (hypothetical) distribution of each underlying class. The 'naive' part comes from the fact that we make very loose assumptions about the process that generated the data. Multinomial NB for one, assumes the data we observe was generated from a simple, run-of-the-mill multinomial distribution. NB algorithms are simple, fast and very effective when working with high-dimensional data.
**Setup & data.**
```
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns; sns.set()
from sklearn.feature_extraction.text import CountVectorizer, TfidfVectorizer
from sklearn.naive_bayes import MultinomialNB
from sklearn.pipeline import make_pipeline
from sklearn.metrics import confusion_matrix
from sklearn.metrics import classification_report
from sklearn.metrics import accuracy_score
import nltk
nltk.download('stopwords')
from nltk.corpus import stopwords
%matplotlib inline
# Fetch data
from sklearn.datasets import fetch_20newsgroups
data = fetch_20newsgroups()
# Load training and test sets
train = fetch_20newsgroups(subset='train', categories=data.target_names)
test = fetch_20newsgroups(subset='test', categories=data.target_names)
```
In this notebook we use the '20 newsgroups' dataset from `sklearn`. The data comprises around 18,000 newsgroups posts on 20 topics ranging from computer graphics, motorcycles to gun politics and religion.
```
print('Number of topics:', len(data.target_names))
# Display labels
data.target_names
```
Let's look at some excerpts:
```
print('Topic:', train.target_names[train.target[-1]],
'\n', ' ', '-'*len(train.target_names[train.target[-1]]), '\n')
print(train.data[-1])
print('='*40, '\n')
print('Topic:', train.target_names[train.target[-5]],
'\n', ' ', '-'*len(train.target_names[train.target[-5]]), '\n')
print(train.data[-5])
```
**Feature engineering.** Before we can have the computer work with the data we need to convert the text in the posts into a numerical representation. This process is known as feature engineering. We take whatever information we have and turn it into numbers; often a real-valued vector or matrix. This way we extract the relevant features so we can feed them to our algorithm.
A good starting point for converting text into a set of representative numerical values would be, for instance, to simply count the frequency of each word in the text. This would leave us with a matrix where each row represents a post and each column represents a word. The matrix entries represent the word counts. So each time a word appears in a post we add one to the respective row and column. As you can imagine, most of the entries in this matrix would be 0 or 'empty', for that reason it is not a dense matrix but a sparse one.
A simple example should develop our intuition. Let's assume we have the following three posts. We can quickly visualize the resulting sparse matrix of the word counts with off-the-shelf tools.
```
samples = ['you have to peel the onion',
'one day I am going to make an onion cry',
'an onion a day keeps everyone away']
# Compute sparse matrix with raw counts
v = CountVectorizer()
S = v.fit_transform(samples)
# Viz
plt.spy(S)
# Or as a table
tab = pd.DataFrame(S.toarray(), columns=v.get_feature_names())
pd.options.display.float_format = "{:,.1f}".format
display(tab)
```
Already we can see that there are some issues with this approach. For one, we count all words irrespective of how meaningful they are. For another, we assign weight to words only by their raw count. Take 'am', 'an', 'one' or 'to' for instance. These appear equally frequently or even more frequently than actual keywords. So by keeping these 'useless' words on and giving them equal or more weight than to actual useful words we end up with a lot of meaningless features in our data. And this only makes finding interesting patterns harder.
**A quick fix.** We can easily tackle both of these issues, though. First, we can remove those words that occur frequently but do not carry substantive meaning. These are known as _stopwords_. Removing stopwords is a common text-preprocessing step. We can get a comprehensive list of stopwords from the `nltk` module.
```
# Load English stopwords
stpw = stopwords.words('english')
# Display a few
print(stpw[-10:])
```
Second, we can calibrate the weights by how often words appear in all posts. This approach is known as term frequency–inverse document frequency (TF–IDF). The function `TfidfVectorizer` from `sklearn` has a built-in parameter that let's us parse stopwords as a list of strings.
```
# Compute sparse matrix with raw counts
v = TfidfVectorizer(stop_words=stpw)
S = v.fit_transform(samples)
# Viz
plt.spy(S)
tab = pd.DataFrame(S.toarray(), columns=v.get_feature_names())
keywords = list(({w for w in tab.columns if w not in stpw}))
pd.options.display.float_format = "{:,.1f}".format
display(tab[keywords])
```
This sparse matrix is now a good numerical representation of the important words in our posts. We will use it to train our multinomial NB algorithm.
**Train model & make prediction on test set.**
```
# Build model
mnb = make_pipeline(TfidfVectorizer(stop_words=stpw),
MultinomialNB())
# Train with training set
mnb.fit(train.data, train.target)
# Predict on test set
labels = mnb.predict(test.data)
```
As we can see at this point, a huge advantage of using a 'naive' model is that we don't need to bother with choosing and fine-tuning hyperparameters. So no need to do any cross-validation, grid searches or the likes. Let's see how well the multinomial NB model does in correctly telling the topic of a previously unseen text. For that we'll construct a confusion matrix.
**Confusion matrix.**
```
# Compute CM
cm = confusion_matrix(test.target, labels)
# Plot
plt.subplots(figsize=(12, 12))
sns.heatmap(cm.T, square=True, annot=True, cmap='YlGnBu', fmt='d', cbar=False,
xticklabels=train.target_names, yticklabels=train.target_names)
plt.xlabel('True Label')
plt.ylabel('Predicted Label')
plt.title('Confusion Matrix')
# Compute accuracy score
print('Overall accuracy of multinomial NB model: {:2.2%}'.format(accuracy_score(test.target, labels)))
```
We see that a naive algorithm such as this does very well in telling the topic of new posts. 81% of the time it will be correct! It does seem, however, to have a hard time separating atheist posts from christian ones, or general politics from gun chatter. It also confuses religious text with Christian and atheist texts. These confusions stem from the fact that many words appear with similar frequency in these topics so that the computer cannot really tell them apart.
**What now?** The cool thing now that we have a pretty accurate predictive model to classify text, is that we get to play around with it! :)
In an image classification setting it would be more cumbersome to provide the model with new images that are outside the test set. But with text classification, well, we can always write something semi-coherent with a topic in mind and see if the trained algorithm guesses it. And if you can't come up with anything yourself, well, there's always Google.
Let's first create a helper function to ease things along. Then some obvious examples below. If you are running this notebook live, you can type in whatever comes to your mind.
```
def guess_topic(s):
probs = mnb.predict_proba([s])[0]
top3 = np.argsort(probs)[:-4:-1]
r_0 = '1st guess is {} with {:2.2%} certainty.'.format(train.target_names[top3[0]],
probs[top3[0]])
r_1 = '2nd guess is {} with {:2.2%} certainty.'.format(train.target_names[top3[1]],
probs[top3[1]])
r_2 = '3rd guess is {} with {:2.2%} certainty.'.format(train.target_names[top3[2]],
probs[top3[2]])
print(r_0 + '\n' + r_1 + '\n' + r_2)
guess_topic('I think my screen is broken. The resolution is all off.')
guess_topic('Is you Harley just outside? Can we go for a ride?')
guess_topic('American Epidemic: One Nation Under Fire')
guess_topic('The Perseid meteor shower is often considered one of the best meteor showers of the year.')
```
The first three guesses were spot on! The last one not so much. But alas, the algorithm is correct 81% of the time. It's bound not to get all guesses right.
|
github_jupyter
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns; sns.set()
from sklearn.feature_extraction.text import CountVectorizer, TfidfVectorizer
from sklearn.naive_bayes import MultinomialNB
from sklearn.pipeline import make_pipeline
from sklearn.metrics import confusion_matrix
from sklearn.metrics import classification_report
from sklearn.metrics import accuracy_score
import nltk
nltk.download('stopwords')
from nltk.corpus import stopwords
%matplotlib inline
# Fetch data
from sklearn.datasets import fetch_20newsgroups
data = fetch_20newsgroups()
# Load training and test sets
train = fetch_20newsgroups(subset='train', categories=data.target_names)
test = fetch_20newsgroups(subset='test', categories=data.target_names)
print('Number of topics:', len(data.target_names))
# Display labels
data.target_names
print('Topic:', train.target_names[train.target[-1]],
'\n', ' ', '-'*len(train.target_names[train.target[-1]]), '\n')
print(train.data[-1])
print('='*40, '\n')
print('Topic:', train.target_names[train.target[-5]],
'\n', ' ', '-'*len(train.target_names[train.target[-5]]), '\n')
print(train.data[-5])
samples = ['you have to peel the onion',
'one day I am going to make an onion cry',
'an onion a day keeps everyone away']
# Compute sparse matrix with raw counts
v = CountVectorizer()
S = v.fit_transform(samples)
# Viz
plt.spy(S)
# Or as a table
tab = pd.DataFrame(S.toarray(), columns=v.get_feature_names())
pd.options.display.float_format = "{:,.1f}".format
display(tab)
# Load English stopwords
stpw = stopwords.words('english')
# Display a few
print(stpw[-10:])
# Compute sparse matrix with raw counts
v = TfidfVectorizer(stop_words=stpw)
S = v.fit_transform(samples)
# Viz
plt.spy(S)
tab = pd.DataFrame(S.toarray(), columns=v.get_feature_names())
keywords = list(({w for w in tab.columns if w not in stpw}))
pd.options.display.float_format = "{:,.1f}".format
display(tab[keywords])
# Build model
mnb = make_pipeline(TfidfVectorizer(stop_words=stpw),
MultinomialNB())
# Train with training set
mnb.fit(train.data, train.target)
# Predict on test set
labels = mnb.predict(test.data)
# Compute CM
cm = confusion_matrix(test.target, labels)
# Plot
plt.subplots(figsize=(12, 12))
sns.heatmap(cm.T, square=True, annot=True, cmap='YlGnBu', fmt='d', cbar=False,
xticklabels=train.target_names, yticklabels=train.target_names)
plt.xlabel('True Label')
plt.ylabel('Predicted Label')
plt.title('Confusion Matrix')
# Compute accuracy score
print('Overall accuracy of multinomial NB model: {:2.2%}'.format(accuracy_score(test.target, labels)))
def guess_topic(s):
probs = mnb.predict_proba([s])[0]
top3 = np.argsort(probs)[:-4:-1]
r_0 = '1st guess is {} with {:2.2%} certainty.'.format(train.target_names[top3[0]],
probs[top3[0]])
r_1 = '2nd guess is {} with {:2.2%} certainty.'.format(train.target_names[top3[1]],
probs[top3[1]])
r_2 = '3rd guess is {} with {:2.2%} certainty.'.format(train.target_names[top3[2]],
probs[top3[2]])
print(r_0 + '\n' + r_1 + '\n' + r_2)
guess_topic('I think my screen is broken. The resolution is all off.')
guess_topic('Is you Harley just outside? Can we go for a ride?')
guess_topic('American Epidemic: One Nation Under Fire')
guess_topic('The Perseid meteor shower is often considered one of the best meteor showers of the year.')
| 0.827445 | 0.990812 |
<a href="https://colab.research.google.com/github/joanby/python-ml-course/blob/master/T1%20-%201%20-%20Data%20Cleaning%20-%20Carga%20de%20datos-Colab.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
```
from google.colab import drive
drive.mount('/content/drive')
```
# Carga de datos a través de la función read_csv
```
import pandas as pd
import os
mainpath = "/content/drive/My Drive/Curso Machine Learning con Python/datasets/"
filename = "titanic/titanic3.csv"
fullpath = os.path.join(mainpath, filename)
data = pd.read_csv(fullpath)
data.head()
```
### Ejemplos de los parámetros de la función read_csv
```
read.csv(filepath="/Users/JuanGabriel/Developer/AnacondaProjects/python-ml-course/datasets/titanic/titanic3.csv",
sep = ",",
dtype={"ingresos":np.float64, "edad":np.int32},
header=0,names={"ingresos", "edad"},
skiprows=12, index_col=None,
skip_blank_lines=False, na_filter=False
)
```
```
data2 = pd.read_csv(mainpath + "/" + "customer-churn-model/Customer Churn Model.txt", sep=",") #CUIDADO: ES EL TXT; NO EL CSV
data2.head()
data2.columns.values
data_cols = pd.read_csv(mainpath + "/" + "customer-churn-model/Customer Churn Columns.csv")
data_col_list = data_cols["Column_Names"].tolist()
data2 = pd.read_csv(mainpath + "/" + "customer-churn-model/Customer Churn Model.txt",
header = None, names = data_col_list)
data2.columns.values
```
# Carga de datos a través de la función open
```
data3 = open(mainpath + "/" + "customer-churn-model/Customer Churn Model.txt",'r')
cols = data3.readline().strip().split(",")
n_cols = len(cols)
counter = 0
main_dict = {}
for col in cols:
main_dict[col] = []
for line in data3:
values = line.strip().split(",")
for i in range(len(cols)):
main_dict[cols[i]].append(values[i])
counter += 1
print("El data set tiene %d filas y %d columnas"%(counter, n_cols))
df3 = pd.DataFrame(main_dict)
df3.head()
```
## Lectura y escritura de ficheros
```
infile = mainpath + "/" + "customer-churn-model/Customer Churn Model.txt"
outfile = mainpath + "/" + "customer-churn-model/Table Customer Churn Model.txt"
with open(infile, "r") as infile1:
with open(outfile, "w") as outfile1:
for line in infile1:
fields = line.strip().split(",")
outfile1.write("\t".join(fields))
outfile1.write("\n")
df4 = pd.read_csv(outfile, sep = "\t")
df4.head()
```
# Leer datos desde una URL
```
medals_url = "http://winterolympicsmedals.com/medals.csv"
medals_data = pd.read_csv(medals_url)
medals_data.head()
```
#### Ejercicio de descarga de datos con urllib3
Vamos a hacer un ejemplo usando la librería urllib3 para leer los datos desde una URL externa, procesarlos y convertirlos a un data frame de *python* antes de guardarlos en un CSV local.
```
def downloadFromURL(url, filename, sep = ",", delim = "\n", encoding="utf-8",
mainpath = "/content/drive/My Drive/Curso Machine Learning con Python/datasets"):
#primero importamos la librería y hacemos la conexión con la web de los datos
import urllib3
http = urllib3.PoolManager()
r = http.request('GET', url)
print("El estado de la respuesta es %d" %(r.status))
response = r.data ## CORREGIDO: eliminado un doble decode que daba error
#El objeto reponse contiene un string binario, así que lo convertimos a un string descodificándolo en UTF-8
str_data = response.decode(encoding)
#Dividimos el string en un array de filas, separándolo por intros
lines = str_data.split(delim)
#La primera línea contiene la cabecera, así que la extraemos
col_names = lines[0].split(sep)
n_cols = len(col_names)
#Generamos un diccionario vacío donde irá la información procesada desde la URL externa
counter = 0
main_dict = {}
for col in col_names:
main_dict[col] = []
#Procesamos fila a fila la información para ir rellenando el diccionario con los datos como hicimos antes
for line in lines:
#Nos saltamos la primera línea que es la que contiene la cabecera y ya tenemos procesada
if(counter > 0):
#Dividimos cada string por las comas como elemento separador
values = line.strip().split(sep)
#Añadimos cada valor a su respectiva columna del diccionario
for i in range(len(col_names)):
main_dict[col_names[i]].append(values[i])
counter += 1
print("El data set tiene %d filas y %d columnas"%(counter, n_cols))
#Convertimos el diccionario procesado a Data Frame y comprobamos que los datos son correctos
df = pd.DataFrame(main_dict)
print(df.head())
#Elegimos donde guardarlo (en la carpeta athletes es donde tiene más sentido por el contexto del análisis)
fullpath = os.path.join(mainpath, filename)
#Lo guardamos en CSV, en JSON o en Excel según queramos
df.to_csv(fullpath+".csv")
df.to_json(fullpath+".json")
df.to_excel(fullpath+".xls")
print("Los ficheros se han guardado correctamente en: "+fullpath)
return df
medals_df = downloadFromURL(medals_url, "athletes/downloaded_medals")
medals_df.head()
```
## Ficheros XLS y XLSX
```
mainpath = "/content/drive/My Drive/Curso Machine Learning con Python/datasets"
filename = "titanic/titanic3.xls"
titanic2 = pd.read_excel(mainpath + "/" + filename, "titanic3")
titanic3 = pd.read_excel(mainpath + "/" + filename, "titanic3")
titanic3.to_csv(mainpath + "/titanic/titanic_custom.csv")
titanic3.to_excel(mainpath + "/titanic/titanic_custom.xls")
titanic3.to_json(mainpath + "/titanic/titanic_custom.json")
```
|
github_jupyter
|
from google.colab import drive
drive.mount('/content/drive')
import pandas as pd
import os
mainpath = "/content/drive/My Drive/Curso Machine Learning con Python/datasets/"
filename = "titanic/titanic3.csv"
fullpath = os.path.join(mainpath, filename)
data = pd.read_csv(fullpath)
data.head()
read.csv(filepath="/Users/JuanGabriel/Developer/AnacondaProjects/python-ml-course/datasets/titanic/titanic3.csv",
sep = ",",
dtype={"ingresos":np.float64, "edad":np.int32},
header=0,names={"ingresos", "edad"},
skiprows=12, index_col=None,
skip_blank_lines=False, na_filter=False
)
data2 = pd.read_csv(mainpath + "/" + "customer-churn-model/Customer Churn Model.txt", sep=",") #CUIDADO: ES EL TXT; NO EL CSV
data2.head()
data2.columns.values
data_cols = pd.read_csv(mainpath + "/" + "customer-churn-model/Customer Churn Columns.csv")
data_col_list = data_cols["Column_Names"].tolist()
data2 = pd.read_csv(mainpath + "/" + "customer-churn-model/Customer Churn Model.txt",
header = None, names = data_col_list)
data2.columns.values
data3 = open(mainpath + "/" + "customer-churn-model/Customer Churn Model.txt",'r')
cols = data3.readline().strip().split(",")
n_cols = len(cols)
counter = 0
main_dict = {}
for col in cols:
main_dict[col] = []
for line in data3:
values = line.strip().split(",")
for i in range(len(cols)):
main_dict[cols[i]].append(values[i])
counter += 1
print("El data set tiene %d filas y %d columnas"%(counter, n_cols))
df3 = pd.DataFrame(main_dict)
df3.head()
infile = mainpath + "/" + "customer-churn-model/Customer Churn Model.txt"
outfile = mainpath + "/" + "customer-churn-model/Table Customer Churn Model.txt"
with open(infile, "r") as infile1:
with open(outfile, "w") as outfile1:
for line in infile1:
fields = line.strip().split(",")
outfile1.write("\t".join(fields))
outfile1.write("\n")
df4 = pd.read_csv(outfile, sep = "\t")
df4.head()
medals_url = "http://winterolympicsmedals.com/medals.csv"
medals_data = pd.read_csv(medals_url)
medals_data.head()
def downloadFromURL(url, filename, sep = ",", delim = "\n", encoding="utf-8",
mainpath = "/content/drive/My Drive/Curso Machine Learning con Python/datasets"):
#primero importamos la librería y hacemos la conexión con la web de los datos
import urllib3
http = urllib3.PoolManager()
r = http.request('GET', url)
print("El estado de la respuesta es %d" %(r.status))
response = r.data ## CORREGIDO: eliminado un doble decode que daba error
#El objeto reponse contiene un string binario, así que lo convertimos a un string descodificándolo en UTF-8
str_data = response.decode(encoding)
#Dividimos el string en un array de filas, separándolo por intros
lines = str_data.split(delim)
#La primera línea contiene la cabecera, así que la extraemos
col_names = lines[0].split(sep)
n_cols = len(col_names)
#Generamos un diccionario vacío donde irá la información procesada desde la URL externa
counter = 0
main_dict = {}
for col in col_names:
main_dict[col] = []
#Procesamos fila a fila la información para ir rellenando el diccionario con los datos como hicimos antes
for line in lines:
#Nos saltamos la primera línea que es la que contiene la cabecera y ya tenemos procesada
if(counter > 0):
#Dividimos cada string por las comas como elemento separador
values = line.strip().split(sep)
#Añadimos cada valor a su respectiva columna del diccionario
for i in range(len(col_names)):
main_dict[col_names[i]].append(values[i])
counter += 1
print("El data set tiene %d filas y %d columnas"%(counter, n_cols))
#Convertimos el diccionario procesado a Data Frame y comprobamos que los datos son correctos
df = pd.DataFrame(main_dict)
print(df.head())
#Elegimos donde guardarlo (en la carpeta athletes es donde tiene más sentido por el contexto del análisis)
fullpath = os.path.join(mainpath, filename)
#Lo guardamos en CSV, en JSON o en Excel según queramos
df.to_csv(fullpath+".csv")
df.to_json(fullpath+".json")
df.to_excel(fullpath+".xls")
print("Los ficheros se han guardado correctamente en: "+fullpath)
return df
medals_df = downloadFromURL(medals_url, "athletes/downloaded_medals")
medals_df.head()
mainpath = "/content/drive/My Drive/Curso Machine Learning con Python/datasets"
filename = "titanic/titanic3.xls"
titanic2 = pd.read_excel(mainpath + "/" + filename, "titanic3")
titanic3 = pd.read_excel(mainpath + "/" + filename, "titanic3")
titanic3.to_csv(mainpath + "/titanic/titanic_custom.csv")
titanic3.to_excel(mainpath + "/titanic/titanic_custom.xls")
titanic3.to_json(mainpath + "/titanic/titanic_custom.json")
| 0.177811 | 0.808597 |
# Import dependencies
First, you may need to install
* NumPy - `pip install numpy`,
* Matplotlib - `pip install matplotlib`,
* PyTorch - `pip install torch torchvision`.
Then you should be able to import all required dependencies.
```
import os, sys
import numpy as np
import matplotlib.pyplot as plt
plt.set_cmap("Greys")
import torch
import torch.nn as nn
import torch.nn.functional as F
from torchvision import datasets, transforms
from torch.utils.data import ConcatDataset
from torch.optim import Adam
from torch.optim.lr_scheduler import MultiStepLR, ExponentialLR
from torch.distributions import MultivariateNormal
```
Add parent directory to path to access sibling modules
```
if '..' not in sys.path:
sys.path.append('..')
%load_ext autoreload
%autoreload 1
%aimport neural_ot.data_loading, neural_ot.model, neural_ot.train
from neural_ot.data_loading import ZipLoader, get_mean_covariance, CustomGaussian, DistributionDataset
from neural_ot.model import NeuralOT, Unflatten, Vector
from neural_ot.train import train
_ = torch.manual_seed(42)
```
# Global variables
First, we set `DEVICE` and `IS_CUDA` global variables.
```
if torch.cuda.is_available():
DEVICE = torch.device('cuda')
IS_CUDA = True
else:
DEVICE = torch.device('cpu')
IS_CUDA = False
```
Second, we download MNIST dataset using `torchvision.datasets`. We normalize intensities from the default interval $[0, 1]$ to the interval $[-1, 1]$ via linear transformation $I' = (I - 0.5) / 0.5$ (as it was done in the original paper). Also for the generative modeling task we concatenate train and test sets to the single dataset, held in global variable `MNIST`.
```
transform = transforms.Compose([
transforms.ToTensor(),
transforms.Normalize([.5], [.5]),
])
mnist_train = datasets.MNIST('data/mnist', download=True, transform=transform, train=True)
mnist_test = datasets.MNIST('data/mnist', download=True, transform=transform, train=False)
MNIST = ConcatDataset([mnist_train, mnist_test])
BATCH_SIZE = 300
N_BATCHES_PER_EPOCH = 10
N_WORKERS = 4 if os.name != 'nt' else 0 # no threads for windows :c
MNIST_MEAN, MNIST_COV = get_mean_covariance(MNIST)
if __name__ == "__main__":
gauss = CustomGaussian(MNIST_MEAN, MNIST_COV)
gauss_dset = DistributionDataset(gauss, transform=lambda x: x.reshape(1, 28, 28))
batch_generator = ZipLoader(gauss_dset, MNIST, batch_size=BATCH_SIZE, n_batches=N_BATCHES_PER_EPOCH,
pin_memory=IS_CUDA, return_idx=True, num_workers=N_WORKERS)
for (x_idx, x), (y_idx, y) in batch_generator:
print(x_idx.shape, x.shape, y_idx.shape, y.shape)
break
plt.imshow(y[0, 0].numpy())
source_dual_net = nn.Sequential(
nn.Flatten(),
nn.Linear(28 * 28, 1024),
nn.ReLU(),
nn.BatchNorm1d(1024),
nn.Linear(1024, 1024),
nn.ReLU(),
nn.BatchNorm1d(1024),
nn.Linear(1024, 1),
nn.Flatten(start_dim=0)
)
target_dual_net = nn.Sequential(
nn.Flatten(),
nn.Linear(28 * 28, 1024),
nn.ReLU(),
nn.BatchNorm1d(1024),
nn.Linear(1024, 1024),
nn.ReLU(),
nn.BatchNorm1d(1024),
nn.Linear(1024, 1),
nn.Flatten(start_dim=0)
)
# target_dual_net = Vector(initial=1e-2 * torch.randn(len(mnist)))
source_to_target_net = nn.Sequential(
nn.Flatten(),
nn.Linear(28 * 28, 1024),
nn.ReLU(),
nn.BatchNorm1d(1024),
nn.Linear(1024, 1024),
nn.ReLU(),
nn.BatchNorm1d(1024),
nn.Linear(1024, 28 * 28),
Unflatten(28, 28),
nn.Tanh()
)
# ot = torch.load('generative_model.pth')
ot = NeuralOT(source_dual_net, target_dual_net, source_to_target_net,
regularization_mode='l2', regularization_parameter=0.05,
from_discrete=False, to_discrete=False).to(DEVICE)
plan_optimizer = Adam(ot.parameters(), lr=1e-3)
plan_scheduler = MultiStepLR(plan_optimizer, [20, 75])
losses = train(ot.plan_criterion, plan_optimizer, batch_generator, n_epochs=300, device=DEVICE,
scheduler=plan_scheduler)
plt.plot(losses)
mapping_optimizer = Adam(ot.parameters(), lr=1e-4)
mapping_scheduler = None # MultiStepLR(plan_optimizer, [10])
mapping_losses = train(ot.mapping_criterion, mapping_optimizer, batch_generator, n_epochs=300, device=DEVICE,
scheduler=mapping_scheduler)
plt.plot(mapping_losses)
tmp_loader = ZipLoader(gauss_dset, batch_size=100, n_batches=1,
pin_memory=IS_CUDA, return_idx=False, num_workers=N_WORKERS)
for x in tmp_loader:
x = x[0]
x = x.to(DEVICE)
mapped = ot.map(x)
imgs = mapped[:, 0].detach().cpu().numpy()
fig, axes = plt.subplots(nrows=10, ncols=10, figsize=(15, 15))
for i, img in enumerate(imgs):
ax = axes[i // 10, i % 10]
ax.imshow(img)
ax.axis('off')
fig.savefig("generated.png")
torch.save(ot, 'generative_model.pth')
```
|
github_jupyter
|
import os, sys
import numpy as np
import matplotlib.pyplot as plt
plt.set_cmap("Greys")
import torch
import torch.nn as nn
import torch.nn.functional as F
from torchvision import datasets, transforms
from torch.utils.data import ConcatDataset
from torch.optim import Adam
from torch.optim.lr_scheduler import MultiStepLR, ExponentialLR
from torch.distributions import MultivariateNormal
if '..' not in sys.path:
sys.path.append('..')
%load_ext autoreload
%autoreload 1
%aimport neural_ot.data_loading, neural_ot.model, neural_ot.train
from neural_ot.data_loading import ZipLoader, get_mean_covariance, CustomGaussian, DistributionDataset
from neural_ot.model import NeuralOT, Unflatten, Vector
from neural_ot.train import train
_ = torch.manual_seed(42)
if torch.cuda.is_available():
DEVICE = torch.device('cuda')
IS_CUDA = True
else:
DEVICE = torch.device('cpu')
IS_CUDA = False
transform = transforms.Compose([
transforms.ToTensor(),
transforms.Normalize([.5], [.5]),
])
mnist_train = datasets.MNIST('data/mnist', download=True, transform=transform, train=True)
mnist_test = datasets.MNIST('data/mnist', download=True, transform=transform, train=False)
MNIST = ConcatDataset([mnist_train, mnist_test])
BATCH_SIZE = 300
N_BATCHES_PER_EPOCH = 10
N_WORKERS = 4 if os.name != 'nt' else 0 # no threads for windows :c
MNIST_MEAN, MNIST_COV = get_mean_covariance(MNIST)
if __name__ == "__main__":
gauss = CustomGaussian(MNIST_MEAN, MNIST_COV)
gauss_dset = DistributionDataset(gauss, transform=lambda x: x.reshape(1, 28, 28))
batch_generator = ZipLoader(gauss_dset, MNIST, batch_size=BATCH_SIZE, n_batches=N_BATCHES_PER_EPOCH,
pin_memory=IS_CUDA, return_idx=True, num_workers=N_WORKERS)
for (x_idx, x), (y_idx, y) in batch_generator:
print(x_idx.shape, x.shape, y_idx.shape, y.shape)
break
plt.imshow(y[0, 0].numpy())
source_dual_net = nn.Sequential(
nn.Flatten(),
nn.Linear(28 * 28, 1024),
nn.ReLU(),
nn.BatchNorm1d(1024),
nn.Linear(1024, 1024),
nn.ReLU(),
nn.BatchNorm1d(1024),
nn.Linear(1024, 1),
nn.Flatten(start_dim=0)
)
target_dual_net = nn.Sequential(
nn.Flatten(),
nn.Linear(28 * 28, 1024),
nn.ReLU(),
nn.BatchNorm1d(1024),
nn.Linear(1024, 1024),
nn.ReLU(),
nn.BatchNorm1d(1024),
nn.Linear(1024, 1),
nn.Flatten(start_dim=0)
)
# target_dual_net = Vector(initial=1e-2 * torch.randn(len(mnist)))
source_to_target_net = nn.Sequential(
nn.Flatten(),
nn.Linear(28 * 28, 1024),
nn.ReLU(),
nn.BatchNorm1d(1024),
nn.Linear(1024, 1024),
nn.ReLU(),
nn.BatchNorm1d(1024),
nn.Linear(1024, 28 * 28),
Unflatten(28, 28),
nn.Tanh()
)
# ot = torch.load('generative_model.pth')
ot = NeuralOT(source_dual_net, target_dual_net, source_to_target_net,
regularization_mode='l2', regularization_parameter=0.05,
from_discrete=False, to_discrete=False).to(DEVICE)
plan_optimizer = Adam(ot.parameters(), lr=1e-3)
plan_scheduler = MultiStepLR(plan_optimizer, [20, 75])
losses = train(ot.plan_criterion, plan_optimizer, batch_generator, n_epochs=300, device=DEVICE,
scheduler=plan_scheduler)
plt.plot(losses)
mapping_optimizer = Adam(ot.parameters(), lr=1e-4)
mapping_scheduler = None # MultiStepLR(plan_optimizer, [10])
mapping_losses = train(ot.mapping_criterion, mapping_optimizer, batch_generator, n_epochs=300, device=DEVICE,
scheduler=mapping_scheduler)
plt.plot(mapping_losses)
tmp_loader = ZipLoader(gauss_dset, batch_size=100, n_batches=1,
pin_memory=IS_CUDA, return_idx=False, num_workers=N_WORKERS)
for x in tmp_loader:
x = x[0]
x = x.to(DEVICE)
mapped = ot.map(x)
imgs = mapped[:, 0].detach().cpu().numpy()
fig, axes = plt.subplots(nrows=10, ncols=10, figsize=(15, 15))
for i, img in enumerate(imgs):
ax = axes[i // 10, i % 10]
ax.imshow(img)
ax.axis('off')
fig.savefig("generated.png")
torch.save(ot, 'generative_model.pth')
| 0.727589 | 0.921675 |
# NumPy Arrays
**python objects:**
1. high-level number objects: integers, floating point
2. containers: lists (costless insertion and append), dictionaries (fast lookup)
**Numpy provides:**
1. extension package to Python for multi-dimensional arrays
2. closer to hardware (efficiency)
3. designed for scientific computation (convenience)
4. Also known as array oriented computing
# 1. Creating arrays
```
#1-D
import numpy as np
a = np.array([0, 1, 2, 3])
a
a.ndim
a.shape
# 2-D, 3-D....
b = np.array([[0, 1, 2], [3, 4, 5]])
b
#print dimenstions
print(b.ndim)
#print shape of the array
print(b.shape)
```
** 1.2 Functions for creating arrays**
```
#using arrange function
# arange is an array-valued version of the built-in Python range function
a = np.arange(10) # 0.... n-1
a
#using linspace
a = np.linspace(0, 1, 6) #start, end, number of points
a
#common arrays
a = np.ones((3, 3))
a
b = np.zeros((3, 3))
b
c = np.eye(3) #Return a 2-D array with ones on the diagonal and zeros elsewhere.
c
d = np.eye(3, 2) #3 is number of rows, 2 is number of columns, index of diagonal start with 0
d
#create array using diag function
a = np.diag([1, 2, 3, 4]) #construct a diagonal array.
a
#create array using random
#Create an array of the given shape and populate it with random samples from a uniform distribution over [0, 1).
a = np.random.rand(4)
a
```
# 2. Basic DataTypes
You may have noticed that, in some instances, array elements are displayed with a **trailing dot (e.g. 2. vs 2)**. This is due to a difference in the **data-type** used:
```
a = np.arange(10)
a.dtype
#You can explicitly specify which data-type you want:
a = np.arange(10, dtype='float64')
a
#The default data type is float for zeros and ones function
a = np.zeros((3, 3))
print(a)
d = np.array([1+2j, 2+4j]) #Complex datatype
print(d.dtype)
b = np.array([True, False, True, False]) #Boolean datatype
print(b.dtype)
```
# Elementwise Operations
**1. Basic Operations**
**with scalars**
```
a = np.array([1, 2, 3, 4]) #create an array
a + 1
a ** 2
#multiplication
c = np.diag([1, 2, 3, 4])
c*c
```
**Logical Operations**
```
a = np.array([1, 1, 0, 0], dtype=bool)
b = np.array([1, 0, 1, 0], dtype=bool)
np.logical_or(a, b)
a = np.array([1, 1, 0, 0], dtype=bool)
b = np.array([1, 0, 1, 0], dtype=bool)
np.logical_and(a, b)
```
**Transcendental functions:**
```
a = np.arange(5)
np.sin(a)
np.log(1)
np.exp(a) #evaluates e^x for each element in a given input
```
# Basic Reductions
**computing sums**
```
x = np.array([1, 2, 3, 4])
np.sum(x)
#sum by rows and by columns
x = np.array([[1, 1], [2, 2]])
x
x.sum(axis=0)
```
**Other reductions**
```
x = np.array([[1, 3, 2], [0, 2, 4]])
x.min()
x.max()
x.argmin()# index of minimum element
x.argmax()
```
# Array Shape Manipulation
**Flattening**
```
a = np.array([[1, 2, 3], [4, 5, 6]])
a.ravel() #Return a contiguous flattened array. A 1-D array, containing the elements of the input, is returned. A copy is made only if needed.
a.T #Transpose
```
**Reshaping**
The inverse operation to flattening:
```
a.shape
b = a.ravel()
b = b.reshape((2, 3))
b
```
**Sorting Data**
```
#Sorting along an axis:
a = np.array([[5, 4, 6], [2, 3, 2]])
b = np.sort(a, axis=1)
b
#in-place sort
a.sort(axis=1)
a
#Finding indexes of minima and maxima:
a = np.array([4, 3, 1, 2])
j_max = np.argmax(a)
j_min = np.argmin(a)
j_max, j_min
```
|
github_jupyter
|
#1-D
import numpy as np
a = np.array([0, 1, 2, 3])
a
a.ndim
a.shape
# 2-D, 3-D....
b = np.array([[0, 1, 2], [3, 4, 5]])
b
#print dimenstions
print(b.ndim)
#print shape of the array
print(b.shape)
#using arrange function
# arange is an array-valued version of the built-in Python range function
a = np.arange(10) # 0.... n-1
a
#using linspace
a = np.linspace(0, 1, 6) #start, end, number of points
a
#common arrays
a = np.ones((3, 3))
a
b = np.zeros((3, 3))
b
c = np.eye(3) #Return a 2-D array with ones on the diagonal and zeros elsewhere.
c
d = np.eye(3, 2) #3 is number of rows, 2 is number of columns, index of diagonal start with 0
d
#create array using diag function
a = np.diag([1, 2, 3, 4]) #construct a diagonal array.
a
#create array using random
#Create an array of the given shape and populate it with random samples from a uniform distribution over [0, 1).
a = np.random.rand(4)
a
a = np.arange(10)
a.dtype
#You can explicitly specify which data-type you want:
a = np.arange(10, dtype='float64')
a
#The default data type is float for zeros and ones function
a = np.zeros((3, 3))
print(a)
d = np.array([1+2j, 2+4j]) #Complex datatype
print(d.dtype)
b = np.array([True, False, True, False]) #Boolean datatype
print(b.dtype)
a = np.array([1, 2, 3, 4]) #create an array
a + 1
a ** 2
#multiplication
c = np.diag([1, 2, 3, 4])
c*c
a = np.array([1, 1, 0, 0], dtype=bool)
b = np.array([1, 0, 1, 0], dtype=bool)
np.logical_or(a, b)
a = np.array([1, 1, 0, 0], dtype=bool)
b = np.array([1, 0, 1, 0], dtype=bool)
np.logical_and(a, b)
a = np.arange(5)
np.sin(a)
np.log(1)
np.exp(a) #evaluates e^x for each element in a given input
x = np.array([1, 2, 3, 4])
np.sum(x)
#sum by rows and by columns
x = np.array([[1, 1], [2, 2]])
x
x.sum(axis=0)
x = np.array([[1, 3, 2], [0, 2, 4]])
x.min()
x.max()
x.argmin()# index of minimum element
x.argmax()
a = np.array([[1, 2, 3], [4, 5, 6]])
a.ravel() #Return a contiguous flattened array. A 1-D array, containing the elements of the input, is returned. A copy is made only if needed.
a.T #Transpose
a.shape
b = a.ravel()
b = b.reshape((2, 3))
b
#Sorting along an axis:
a = np.array([[5, 4, 6], [2, 3, 2]])
b = np.sort(a, axis=1)
b
#in-place sort
a.sort(axis=1)
a
#Finding indexes of minima and maxima:
a = np.array([4, 3, 1, 2])
j_max = np.argmax(a)
j_min = np.argmin(a)
j_max, j_min
| 0.433742 | 0.987005 |
# Linear regression using scikit-learn
In the previous notebook, we presented the parametrization of a linear model.
During the exercise, you saw that varying parameters will give different models
that will fit better or worse the data. To evaluate quantitatively this
goodness of fit, you implemented a so-called metric.
When doing machine learning, you are interested in selecting the model which
will minimize the error on the data available the most.
From the previous exercise, we could implement a brute-force approach,
varying the weights and intercept and select the model with the lowest error.
Hopefully, this problem of finding the best parameters values (i.e. that
result in the lowest error) can be solved without the need to check every
potential parameter combination. Indeed, this problem has a closed-form
solution: the best parameter values can be found by solving an equation. This
avoids the need for brute-force search. This strategy is implemented in
scikit-learn.
```
import pandas as pd
penguins = pd.read_csv("../datasets/penguins_regression.csv")
feature_names = "Flipper Length (mm)"
target_name = "Body Mass (g)"
data, target = penguins[[feature_names]], penguins[target_name]
```
<div class="admonition note alert alert-info">
<p class="first admonition-title" style="font-weight: bold;">Note</p>
<p class="last">If you want a deeper overview regarding this dataset, you can refer to the
Appendix - Datasets description section at the end of this MOOC.</p>
</div>
```
from sklearn.linear_model import LinearRegression
linear_regression = LinearRegression()
linear_regression.fit(data, target)
```
The instance `linear_regression` will store the parameter values in the
attributes `coef_` and `intercept_`. We can check what the optimal model
found is:
```
weight_flipper_length = linear_regression.coef_[0]
weight_flipper_length
intercept_body_mass = linear_regression.intercept_
intercept_body_mass
```
We will use the weight and intercept to plot the model found using the
scikit-learn.
```
import numpy as np
flipper_length_range = np.linspace(data.min(), data.max(), num=300)
predicted_body_mass = (
weight_flipper_length * flipper_length_range + intercept_body_mass)
import matplotlib.pyplot as plt
import seaborn as sns
sns.scatterplot(x=data[feature_names], y=target, color="black", alpha=0.5)
plt.plot(flipper_length_range, predicted_body_mass)
_ = plt.title("Model using LinearRegression from scikit-learn")
```
In the solution of the previous exercise, we implemented a function to
compute the goodness of fit of a model. Indeed, we mentioned two metrics: (i)
the mean squared error and (ii) the mean absolute error. These metrics are
implemented in scikit-learn and we do not need to use our own implementation.
We can first compute the mean squared error.
```
from sklearn.metrics import mean_squared_error
inferred_body_mass = linear_regression.predict(data)
model_error = mean_squared_error(target, inferred_body_mass)
print(f"The mean squared error of the optimal model is {model_error:.2f}")
```
A linear regression model minimizes the mean squared error on the training
set. This means that the parameters obtained after the fit (i.e. `coef_` and
`intercept_`) are the optimal parameters that minimizes the mean squared
error. In other words, any other choice of parameters will yield a model with
a higher mean squared error on the training set.
However, the mean squared error is difficult to interpret. The mean absolute
error is more intuitive since it provides an error in the same unit as the
one of the target.
```
from sklearn.metrics import mean_absolute_error
model_error = mean_absolute_error(target, inferred_body_mass)
print(f"The mean absolute error of the optimal model is {model_error:.2f} g")
```
A mean absolute error of 313 means that in average, our model make an error
of +/- 313 grams when predicting the body mass of a penguin given its flipper
length.
In this notebook, you saw how to train a linear regression model using
scikit-learn.
|
github_jupyter
|
import pandas as pd
penguins = pd.read_csv("../datasets/penguins_regression.csv")
feature_names = "Flipper Length (mm)"
target_name = "Body Mass (g)"
data, target = penguins[[feature_names]], penguins[target_name]
from sklearn.linear_model import LinearRegression
linear_regression = LinearRegression()
linear_regression.fit(data, target)
weight_flipper_length = linear_regression.coef_[0]
weight_flipper_length
intercept_body_mass = linear_regression.intercept_
intercept_body_mass
import numpy as np
flipper_length_range = np.linspace(data.min(), data.max(), num=300)
predicted_body_mass = (
weight_flipper_length * flipper_length_range + intercept_body_mass)
import matplotlib.pyplot as plt
import seaborn as sns
sns.scatterplot(x=data[feature_names], y=target, color="black", alpha=0.5)
plt.plot(flipper_length_range, predicted_body_mass)
_ = plt.title("Model using LinearRegression from scikit-learn")
from sklearn.metrics import mean_squared_error
inferred_body_mass = linear_regression.predict(data)
model_error = mean_squared_error(target, inferred_body_mass)
print(f"The mean squared error of the optimal model is {model_error:.2f}")
from sklearn.metrics import mean_absolute_error
model_error = mean_absolute_error(target, inferred_body_mass)
print(f"The mean absolute error of the optimal model is {model_error:.2f} g")
| 0.759493 | 0.991187 |
# Modes of a Vibrating Building Under Sinusoidal Forcing
The equations of motion for the four story building take this form:
$$
\mathbf{M} \dot{\bar{s}} + \mathbf{K} \bar{c} = \bar{F}
$$
where
$$
\bar{c} = \begin{bmatrix} x_1 \\ x_2 \\ x_3 \\ x_4 \end{bmatrix},
\bar{s} = \begin{bmatrix} v_1 \\ v_2 \\ v_3 \\ v_4 \end{bmatrix}
$$
$$
\mathbf{M} =
\begin{bmatrix}
m_1 & 0 & 0 & 0 \\
0 & m_2 & 0 & 0 \\
0 & 0 & m_3 & 0 \\
0 & 0 & 0 & m_4
\end{bmatrix}
$$
$$
\mathbf{K} =
\begin{bmatrix}
k_1+k_2 & -k_2 & 0 & 0 \\
-k_2 & k_2 + k_3 & -k_3 \\
0 & -k_3 & k_3+k_4 & -k_4 \\
0 & 0 & -k_4 & k_4
\end{bmatrix}
$$
$$
\bar{F} = \begin{bmatrix} F_1(t) \\ F_2(t) \\ F_3(t) \\ F_4(t) \end{bmatrix}
$$
The forces $F_1, F_2, F_3, F_4$ are the lateral, arbitrary, forces applied to each floor.
```
import numpy as np
import matplotlib.pyplot as plt
from resonance.linear_systems import FourStoryBuildingSystem
```
This gives a bit nicer printing of large NumPy arrays.
```
np.set_printoptions(precision=5, linewidth=100, suppress=True)
%matplotlib widget
```
# Determine the model frequiences by calculating the eigenvalues
```
sys = FourStoryBuildingSystem()
M, C, K = sys.canonical_coefficients()
L = np.linalg.cholesky(M)
K_tilde = np.linalg.inv(L) @ K @ np.linalg.inv(L.T)
evals, evecs = np.linalg.eig(K_tilde)
```
These are the modal frequencies in radians per second:
```
ws = np.sqrt(evals)
ws
```
# Forcing the fourth floor at the largest natural frequency
Note that the initial state values are not all zero. Set them all to zero so we see only the effects of forcing.
```
sys.states
sys.coordinates['x1'] = 0.0
sys.coordinates['x2'] = 0.0
sys.coordinates['x3'] = 0.0
sys.coordinates['x4'] = 0.0
sys.states
```
Create two new constants that describe an amplitude for a sinusoidal forcing.
$$F(t) = A\sin(\omega T)$$
```
sys.constants['amplitude'] = 100 # N
sys.constants['frequency'] = np.deg2rad(10.0) # rad/s
```
Now define a function that takes constants and/or time as inputs and outputs the entries of $\bar{F}$ in the same order as the coordinates and speeds. The following definition applies a sinusoidal force only to the 4th floor.
```
def push_floor(amplitude, frequency, time):
F1 = 0.0 if np.isscalar(time) else np.zeros_like(time)
F2 = 0.0 if np.isscalar(time) else np.zeros_like(time)
F3 = 0.0 if np.isscalar(time) else np.zeros_like(time)
F4 = amplitude * np.sin(frequency * time)
return F1, F2, F3, F4
```
This function should work with scalar values of time and 1d arrays of time:
```
push_floor(1.0, 2.0, 3.0)
push_floor(1.0, 2.0, np.ones(5))
```
Now add the forcing function to the system:
```
sys.forcing_func = push_floor
```
The `forced_response()` function works like the `free_response()` function but it will apply the forcing in the simulation.
```
traj = sys.forced_response(100)
traj[sys.coordinates.keys()].plot(subplots=True);
sys.animate_configuration(fps=10, repeat=False)
```
# Exercise: Forcing at the modal frequencies
Update the `frequency` value to simulate the fourth floor being forced at each of the four natural frequencies and note your observations. Compare the obeserved motion to the mode shapes associated with that modal frequency. Use the animation to help visualize what is happening.
```
sys.constants['frequency'] = ws[0] # rad/s
traj = sys.forced_response(100)
traj[sys.coordinates.keys()].plot(subplots=True)
sys.animate_configuration(fps=10, repeat=False)
sys.constants['frequency'] = ws[1] # rad/s
traj = sys.forced_response(100)
traj[sys.coordinates.keys()].plot(subplots=True)
sys.animate_configuration(fps=10, repeat=False)
sys.constants['frequency'] = ws[2] # rad/s
traj = sys.forced_response(100)
traj[sys.coordinates.keys()].plot(subplots=True)
sys.animate_configuration(fps=10, repeat=False)
sys.constants['frequency'] = ws[3] # rad/s
traj = sys.forced_response(100)
traj[sys.coordinates.keys()].plot(subplots=True)
sys.animate_configuration(fps=10, repeat=False)
```
# Exercise: Forcing at a node
Recall that the 3rd mode shape has an eigenvector component that is zero:
```
evecs
```
This is called a "node". This node is associated with $x_3$, the third floor and it tells us that there is no motion at floor 3 if this mode is excited.
Adjust the forcing function to apply sinusoidal forcing at the third floor. Use the third modal frequency to apply forcing to the third floor, then use one of the other modal frequencies to force the third floor. Compare the results and discuss your observations.
```
sys.constants['frequency'] = ws[2] # rad/s
def push_floor(amplitude, frequency, time):
F1 = 0.0 if np.isscalar(time) else np.zeros_like(time)
F2 = 0.0 if np.isscalar(time) else np.zeros_like(time)
F3 = amplitude * np.sin(frequency * time)
F4 = 0.0 if np.isscalar(time) else np.zeros_like(time)
return F1, F2, F3, F4
sys.forcing_func = push_floor
traj = sys.forced_response(100)
traj[sys.coordinates.keys()].plot(subplots=True)
sys.animate_configuration(fps=10, repeat=False)
sys.constants['frequency'] = ws[0] # rad/s
def push_floor(amplitude, frequency, time):
F1 = 0.0 if np.isscalar(time) else np.zeros_like(time)
F2 = 0.0 if np.isscalar(time) else np.zeros_like(time)
F3 = amplitude * np.sin(frequency * time)
F4 = 0.0 if np.isscalar(time) else np.zeros_like(time)
return F1, F2, F3, F4
sys.forcing_func = push_floor
traj = sys.forced_response(100)
traj[sys.coordinates.keys()].plot(subplots=True)
sys.animate_configuration(fps=10, repeat=False)
```
|
github_jupyter
|
import numpy as np
import matplotlib.pyplot as plt
from resonance.linear_systems import FourStoryBuildingSystem
np.set_printoptions(precision=5, linewidth=100, suppress=True)
%matplotlib widget
sys = FourStoryBuildingSystem()
M, C, K = sys.canonical_coefficients()
L = np.linalg.cholesky(M)
K_tilde = np.linalg.inv(L) @ K @ np.linalg.inv(L.T)
evals, evecs = np.linalg.eig(K_tilde)
ws = np.sqrt(evals)
ws
sys.states
sys.coordinates['x1'] = 0.0
sys.coordinates['x2'] = 0.0
sys.coordinates['x3'] = 0.0
sys.coordinates['x4'] = 0.0
sys.states
sys.constants['amplitude'] = 100 # N
sys.constants['frequency'] = np.deg2rad(10.0) # rad/s
def push_floor(amplitude, frequency, time):
F1 = 0.0 if np.isscalar(time) else np.zeros_like(time)
F2 = 0.0 if np.isscalar(time) else np.zeros_like(time)
F3 = 0.0 if np.isscalar(time) else np.zeros_like(time)
F4 = amplitude * np.sin(frequency * time)
return F1, F2, F3, F4
push_floor(1.0, 2.0, 3.0)
push_floor(1.0, 2.0, np.ones(5))
sys.forcing_func = push_floor
traj = sys.forced_response(100)
traj[sys.coordinates.keys()].plot(subplots=True);
sys.animate_configuration(fps=10, repeat=False)
sys.constants['frequency'] = ws[0] # rad/s
traj = sys.forced_response(100)
traj[sys.coordinates.keys()].plot(subplots=True)
sys.animate_configuration(fps=10, repeat=False)
sys.constants['frequency'] = ws[1] # rad/s
traj = sys.forced_response(100)
traj[sys.coordinates.keys()].plot(subplots=True)
sys.animate_configuration(fps=10, repeat=False)
sys.constants['frequency'] = ws[2] # rad/s
traj = sys.forced_response(100)
traj[sys.coordinates.keys()].plot(subplots=True)
sys.animate_configuration(fps=10, repeat=False)
sys.constants['frequency'] = ws[3] # rad/s
traj = sys.forced_response(100)
traj[sys.coordinates.keys()].plot(subplots=True)
sys.animate_configuration(fps=10, repeat=False)
evecs
sys.constants['frequency'] = ws[2] # rad/s
def push_floor(amplitude, frequency, time):
F1 = 0.0 if np.isscalar(time) else np.zeros_like(time)
F2 = 0.0 if np.isscalar(time) else np.zeros_like(time)
F3 = amplitude * np.sin(frequency * time)
F4 = 0.0 if np.isscalar(time) else np.zeros_like(time)
return F1, F2, F3, F4
sys.forcing_func = push_floor
traj = sys.forced_response(100)
traj[sys.coordinates.keys()].plot(subplots=True)
sys.animate_configuration(fps=10, repeat=False)
sys.constants['frequency'] = ws[0] # rad/s
def push_floor(amplitude, frequency, time):
F1 = 0.0 if np.isscalar(time) else np.zeros_like(time)
F2 = 0.0 if np.isscalar(time) else np.zeros_like(time)
F3 = amplitude * np.sin(frequency * time)
F4 = 0.0 if np.isscalar(time) else np.zeros_like(time)
return F1, F2, F3, F4
sys.forcing_func = push_floor
traj = sys.forced_response(100)
traj[sys.coordinates.keys()].plot(subplots=True)
sys.animate_configuration(fps=10, repeat=False)
| 0.367497 | 0.98244 |
## Import modules
```
from __future__ import division
import os; os.chdir(os.path.join('..', '..', '..'))
print os.getcwd()
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.gridspec as gs
import seaborn as sns
import pandas as pd
from grr.cell_class import Cell
from grr.Trace import detectSpikes
from grr.Tools import stripNan
from grr.FrequencyInputCurve import FrequencyInputCurve
from ezephys.pltools import hide_border
plt.style.use(os.path.join('figs', 'scripts', 'publication_figure_style.dms'))
IMG_PATH = os.path.join('figs', 'ims', 'writeup', 'fI')
```
## Load data
First we'll just load the current step data.
```
DATA_PATH = os.path.join('data', 'raw', '5HT', 'current_steps')
fnames = pd.read_csv(os.path.join(DATA_PATH, 'index.csv'))
fnames
curr_steps = Cell().read_ABF([os.path.join(DATA_PATH, fn) for fn in fnames['Steps']])
curr_steps[0].plot()
for expt in curr_steps:
plt.plot(expt[1, :, 0])
plt.show()
```
Current step recordings have a similar structure, but differ in number of sweeps. Also possibly in spacing of current steps.
Automatically detect the start/end of the test pulse and current steps based on the first recording and then show whether this works for all cells.
```
change_threshold = 6. # pA threshold at which to detect a step.
tstpts = {'start': [], 'stop': []}
mainpts = {'start': [], 'stop': []}
for expt in curr_steps:
try:
falling = np.where(np.diff(expt[1, :, 0]) < -change_threshold)[0]
tstpts['start'].append(falling[0])
mainpts['start'].append(falling[1])
rising = np.where(np.diff(expt[1, :, 0]) > change_threshold)[0]
tstpts['stop'].append(rising[0])
mainpts['stop'].append(rising[1])
except ValueError:
print 'Too many or too few steps detected. Might need to adjust `change_threshold`.'
raise
del change_threshold
dt = 0.1 # ms. Assumed.
buffer_timesteps = 500
plt.figure()
tst_ax = plt.subplot(121)
tst_ax.set_title('Test pulse')
step_ax = plt.subplot(122)
step_ax.set_title('Current step')
for i, expt in enumerate(curr_steps):
tst_ax.plot(
expt[0, (tstpts['start'][i] - buffer_timesteps):(tstpts['stop'][i] + buffer_timesteps), :].mean(axis = 1),
'k-', lw = 0.5, alpha = 0.5
)
step_ax.plot(
expt[0, (mainpts['start'][i] - buffer_timesteps):(mainpts['stop'][i] + buffer_timesteps), 8],
'k-', lw = 0.5, alpha = 0.5
)
tst_ax.set_xlabel('Time (timesteps)')
tst_ax.set_ylabel('V (mV)')
step_ax.set_xlabel('Time (timesteps)')
step_ax.set_ylabel('')
plt.tight_layout()
plt.show()
qc_mask = []
for i, rec in enumerate(curr_steps):
if (np.abs(rec[1, :, :] - np.mean(rec[1, :, :])) < 1.).all() :
qc_mask.append(False)
rec.plot()
else:
qc_mask.append(True)
curr_steps = [curr_steps[i] for i in range(len(curr_steps)) if qc_mask[i]]
print '{} of {} cells passed quality control.'.format(len(curr_steps), len(qc_mask))
del qc_mask
```
## Generate f/I curves
f/I curves are usually rectified linear. However, in some cases non-monotonic f/I curves are observed, usually due to depolarization block.
```
# Detect spikes in all recordings.
spktimes = [detectSpikes(rec[0, :, :], 0., 3., 0, 0.1) for rec in curr_steps]
fi_summarizer = FrequencyInputCurve(lambda x_: np.all(np.nan_to_num(np.diff(x_) / x_[:-1]) > -0.25))
fi_data = {metric: [] for metric in fi_summarizer.summary_metrics}
fi_summaries = []
for i, (rec, times) in enumerate(zip(curr_steps, spktimes)):
fi_summarizer.fit(times, rec[1, ...], (mainpts['start'][i], mainpts['stop'][i]), dt)
fi_summaries.append(fi_summarizer.copy())
for metric in fi_summarizer.summary_metrics:
fi_data[metric].append(fi_summarizer.get_metric(metric))
fi_df = pd.DataFrame(fi_data)
fi_df.drop(columns=['CV', 'I', 'f'], inplace=True)
fi_df
fi_df.to_csv(os.path.join('data', 'processed', '5HT', 'current_steps_gain.csv'), index=False)
plt.figure(figsize=(1.5, 1))
first_fit = True
curves = plt.subplot(111)
for i, curve in enumerate(fi_summaries):
if curve.is_monotonic:
if first_fit:
curve.plot(fitted_pltargs={'color': 'r', 'ls': '--', 'label': 'Linear fit'}, ax=curves, color='k')
first_fit = False
else:
curve.plot(fitted_pltargs={'color': 'r', 'ls': '--'}, ax=curves, color='k')
else:
curve.plot(fitted=False, ax=curves, color='grey', alpha=0.8)
curves.set_xlim(-50, 170)
curves.set_ylim(-2, 40)
curves.legend(loc='upper left')
curves.set_xlabel('$I$ (pA)')
curves.set_ylabel('$f$ (Hz)')
hide_border('tr', ax=curves, trim=True)
plt.subplots_adjust(top=0.97, right=0.97, bottom=0.3, left=0.25)
if IMG_PATH is not None:
plt.savefig(os.path.join(IMG_PATH, 'ser_fi_curve_only.png'))
plt.savefig(os.path.join(IMG_PATH, 'ser_fi_curve_only.svg'))
plt.figure(figsize=(2, 1.5))
first_fit = True
curves = plt.subplot(111)
for i, curve in enumerate(fi_summaries):
if curve.is_monotonic:
if first_fit:
curve.plot(fitted_pltargs={'color': 'r', 'ls': '--', 'label': 'Linear fit'}, ax=curves, color='k')
first_fit = False
else:
curve.plot(fitted_pltargs={'color': 'r', 'ls': '--'}, ax=curves, color='k')
else:
curve.plot(fitted=False, ax=curves, color='grey', alpha=0.8d)
curves.set_xlim(-50, curves.get_xlim()[1])
curves.legend(loc='upper left')
curves.set_xlabel('$I$ (pA)')
curves.set_ylabel('$f$ (Hz)')
hide_border('tr', ax=curves, trim=True)
plt.subplots_adjust(top=0.97, right=0.97, bottom=0.3, left=0.25)
if IMG_PATH is not None:
plt.savefig(os.path.join(IMG_PATH, 'ser_fi_curve_only_unscaled.png'))
plt.savefig(os.path.join(IMG_PATH, 'ser_fi_curve_only_unscaled.svg'))
```
|
github_jupyter
|
from __future__ import division
import os; os.chdir(os.path.join('..', '..', '..'))
print os.getcwd()
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.gridspec as gs
import seaborn as sns
import pandas as pd
from grr.cell_class import Cell
from grr.Trace import detectSpikes
from grr.Tools import stripNan
from grr.FrequencyInputCurve import FrequencyInputCurve
from ezephys.pltools import hide_border
plt.style.use(os.path.join('figs', 'scripts', 'publication_figure_style.dms'))
IMG_PATH = os.path.join('figs', 'ims', 'writeup', 'fI')
DATA_PATH = os.path.join('data', 'raw', '5HT', 'current_steps')
fnames = pd.read_csv(os.path.join(DATA_PATH, 'index.csv'))
fnames
curr_steps = Cell().read_ABF([os.path.join(DATA_PATH, fn) for fn in fnames['Steps']])
curr_steps[0].plot()
for expt in curr_steps:
plt.plot(expt[1, :, 0])
plt.show()
change_threshold = 6. # pA threshold at which to detect a step.
tstpts = {'start': [], 'stop': []}
mainpts = {'start': [], 'stop': []}
for expt in curr_steps:
try:
falling = np.where(np.diff(expt[1, :, 0]) < -change_threshold)[0]
tstpts['start'].append(falling[0])
mainpts['start'].append(falling[1])
rising = np.where(np.diff(expt[1, :, 0]) > change_threshold)[0]
tstpts['stop'].append(rising[0])
mainpts['stop'].append(rising[1])
except ValueError:
print 'Too many or too few steps detected. Might need to adjust `change_threshold`.'
raise
del change_threshold
dt = 0.1 # ms. Assumed.
buffer_timesteps = 500
plt.figure()
tst_ax = plt.subplot(121)
tst_ax.set_title('Test pulse')
step_ax = plt.subplot(122)
step_ax.set_title('Current step')
for i, expt in enumerate(curr_steps):
tst_ax.plot(
expt[0, (tstpts['start'][i] - buffer_timesteps):(tstpts['stop'][i] + buffer_timesteps), :].mean(axis = 1),
'k-', lw = 0.5, alpha = 0.5
)
step_ax.plot(
expt[0, (mainpts['start'][i] - buffer_timesteps):(mainpts['stop'][i] + buffer_timesteps), 8],
'k-', lw = 0.5, alpha = 0.5
)
tst_ax.set_xlabel('Time (timesteps)')
tst_ax.set_ylabel('V (mV)')
step_ax.set_xlabel('Time (timesteps)')
step_ax.set_ylabel('')
plt.tight_layout()
plt.show()
qc_mask = []
for i, rec in enumerate(curr_steps):
if (np.abs(rec[1, :, :] - np.mean(rec[1, :, :])) < 1.).all() :
qc_mask.append(False)
rec.plot()
else:
qc_mask.append(True)
curr_steps = [curr_steps[i] for i in range(len(curr_steps)) if qc_mask[i]]
print '{} of {} cells passed quality control.'.format(len(curr_steps), len(qc_mask))
del qc_mask
# Detect spikes in all recordings.
spktimes = [detectSpikes(rec[0, :, :], 0., 3., 0, 0.1) for rec in curr_steps]
fi_summarizer = FrequencyInputCurve(lambda x_: np.all(np.nan_to_num(np.diff(x_) / x_[:-1]) > -0.25))
fi_data = {metric: [] for metric in fi_summarizer.summary_metrics}
fi_summaries = []
for i, (rec, times) in enumerate(zip(curr_steps, spktimes)):
fi_summarizer.fit(times, rec[1, ...], (mainpts['start'][i], mainpts['stop'][i]), dt)
fi_summaries.append(fi_summarizer.copy())
for metric in fi_summarizer.summary_metrics:
fi_data[metric].append(fi_summarizer.get_metric(metric))
fi_df = pd.DataFrame(fi_data)
fi_df.drop(columns=['CV', 'I', 'f'], inplace=True)
fi_df
fi_df.to_csv(os.path.join('data', 'processed', '5HT', 'current_steps_gain.csv'), index=False)
plt.figure(figsize=(1.5, 1))
first_fit = True
curves = plt.subplot(111)
for i, curve in enumerate(fi_summaries):
if curve.is_monotonic:
if first_fit:
curve.plot(fitted_pltargs={'color': 'r', 'ls': '--', 'label': 'Linear fit'}, ax=curves, color='k')
first_fit = False
else:
curve.plot(fitted_pltargs={'color': 'r', 'ls': '--'}, ax=curves, color='k')
else:
curve.plot(fitted=False, ax=curves, color='grey', alpha=0.8)
curves.set_xlim(-50, 170)
curves.set_ylim(-2, 40)
curves.legend(loc='upper left')
curves.set_xlabel('$I$ (pA)')
curves.set_ylabel('$f$ (Hz)')
hide_border('tr', ax=curves, trim=True)
plt.subplots_adjust(top=0.97, right=0.97, bottom=0.3, left=0.25)
if IMG_PATH is not None:
plt.savefig(os.path.join(IMG_PATH, 'ser_fi_curve_only.png'))
plt.savefig(os.path.join(IMG_PATH, 'ser_fi_curve_only.svg'))
plt.figure(figsize=(2, 1.5))
first_fit = True
curves = plt.subplot(111)
for i, curve in enumerate(fi_summaries):
if curve.is_monotonic:
if first_fit:
curve.plot(fitted_pltargs={'color': 'r', 'ls': '--', 'label': 'Linear fit'}, ax=curves, color='k')
first_fit = False
else:
curve.plot(fitted_pltargs={'color': 'r', 'ls': '--'}, ax=curves, color='k')
else:
curve.plot(fitted=False, ax=curves, color='grey', alpha=0.8d)
curves.set_xlim(-50, curves.get_xlim()[1])
curves.legend(loc='upper left')
curves.set_xlabel('$I$ (pA)')
curves.set_ylabel('$f$ (Hz)')
hide_border('tr', ax=curves, trim=True)
plt.subplots_adjust(top=0.97, right=0.97, bottom=0.3, left=0.25)
if IMG_PATH is not None:
plt.savefig(os.path.join(IMG_PATH, 'ser_fi_curve_only_unscaled.png'))
plt.savefig(os.path.join(IMG_PATH, 'ser_fi_curve_only_unscaled.svg'))
| 0.484624 | 0.774264 |
# ETL Processes
Use this notebook to develop the ETL process for each of your tables before completing the `etl.py` file to load the whole datasets.
```
import os
import glob
import psycopg2
import pandas as pd
from sql_queries import *
conn = psycopg2.connect("host=127.0.0.1 dbname=sparkifydb user=student password=student")
cur = conn.cursor()
cur.close()
conn.close()
def get_files(filepath):
all_files = []
for root, dirs, files in os.walk(filepath):
files = glob.glob(os.path.join(root,'*.json'))
for f in files :
all_files.append(os.path.relpath(f)) # NOTE: This was edited as I was using my own system
return all_files
```
# Process `song_data`
In this first part, you'll perform ETL on the first dataset, `song_data`, to create the `songs` and `artists` dimensional tables.
Let's perform ETL on a single song file and load a single record into each table to start.
- Use the `get_files` function provided above to get a list of all song JSON files in `data/song_data`
- Select the first song in this list
- Read the song file and view the data
```
filepath = 'data/song_data'
song_files = get_files(filepath)
song_files[0]
df = pd.read_json(song_files[0], lines=True)
df.head()
```
## #1: `songs` Table
#### Extract Data for Songs Table
- Select columns for song ID, title, artist ID, year, and duration
- Use `df.values` to select just the values from the dataframe
- Index to select the first (only) record in the dataframe
- Convert the array to a list and set it to `song_data`
```
song_data = (
df[['song_id', 'title', 'artist_id',
'year', 'duration']].values
)
song_data = list(song_data[0])
song_data
```
#### Insert Record into Song Table
Implement the `song_table_insert` query in `sql_queries.py` and run the cell below to insert a record for this song into the `songs` table. Remember to run `create_tables.py` before running the cell below to ensure you've created/resetted the `songs` table in the sparkify database.
```
song_table_insert = ("""
INSERT INTO songs (song_id, title, artist_id, year, duration)
VALUES (%s, %s, %s, %s, %s);
""")
cur.execute(song_table_insert, song_data)
conn.commit()
cur.execute("""
SELECT * FROM songs;
""")
row = cur.fetchone()
while row:
print(row)
row = cur.fetchone()
```
Run `test.ipynb` to see if you've successfully added a record to this table.
## #2: `artists` Table
#### Extract Data for Artists Table
- Select columns for artist ID, name, location, latitude, and longitude
- Use `df.values` to select just the values from the dataframe
- Index to select the first (only) record in the dataframe
- Convert the array to a list and set it to `artist_data`
```
df
artist_data = (
df[['artist_id', 'artist_name', 'artist_location',
'artist_latitude', 'artist_longitude']].values
)
artist_data = list(artist_data[0])
artist_data
```
#### Insert Record into Artist Table
Implement the `artist_table_insert` query in `sql_queries.py` and run the cell below to insert a record for this song's artist into the `artists` table. Remember to run `create_tables.py` before running the cell below to ensure you've created/resetted the `artists` table in the sparkify database.
```
artist_table_insert = ("""
INSERT INTO artists
(artist_id, name, location, latitude, longitude)
VALUES (%s, %s, %s, %s, %s);
""")
cur.execute(artist_table_insert, artist_data)
conn.commit()
```
Run `test.ipynb` to see if you've successfully added a record to this table.
```
cur.execute("""
SELECT * FROM artists;
""")
row = cur.fetchone()
while row:
print(row)
row = cur.fetchone()
```
# Process `log_data`
In this part, you'll perform ETL on the second dataset, `log_data`, to create the `time` and `users` dimensional tables, as well as the `songplays` fact table.
Let's perform ETL on a single log file and load a single record into each table.
- Use the `get_files` function provided above to get a list of all log JSON files in `data/log_data`
- Select the first log file in this list
- Read the log file and view the data
```
filepath = 'data/log_data'
log_files = get_files(filepath)
len(log_files)
df = pd.read_json(log_files[0], lines=True)
df.head()
```
## #3: `time` Table
#### Extract Data for Time Table
- Filter records by `NextSong` action
- Convert the `ts` timestamp column to datetime
- Hint: the current timestamp is in milliseconds
- Extract the timestamp, hour, day, week of year, month, year, and weekday from the `ts` column and set `time_data` to a list containing these values in order
- Hint: use pandas' [`dt` attribute](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.Series.dt.html) to access easily datetimelike properties.
- Specify labels for these columns and set to `column_labels`
- Create a dataframe, `time_df,` containing the time data for this file by combining `column_labels` and `time_data` into a dictionary and converting this into a dataframe
```
is_NextSong = df['page'] == 'NextSong'
df = df[is_NextSong]
df.head()
# Python version
import datetime
datetime.datetime.fromtimestamp(1541377992796/1000.0)
# (year, month, day, hour, minute, second, microsecond)
# datetime.datetime(2018, 11, 4, 16, 33, 12, 796000)
# Pandas version
pd.to_datetime(1541377992796, unit='ms')
# Timestamp('2018-11-05 00:33:12.796000')
# pd.to_datetime(1541377992796, unit='ms').day_name() - 'Monday'
df['ts'] = pd.to_datetime(df['ts'], unit='ms')
df.head()
t = df['ts']
t.head()
time_data = list((t, t.dt.hour, t.dt.day, t.dt.week, t.dt.month, t.dt.year, t.dt.day_name()))
column_labels = ('timestamp', 'hour', 'day', 'week', 'month', 'year', 'day_of_week')
time_data
time_dict = dict(zip(column_labels, time_data))
time_dict
time_df = pd.DataFrame(time_dict)
time_df
```
#### Insert Records into Time Table
Implement the `time_table_insert` query in `sql_queries.py` and run the cell below to insert records for the timestamps in this log file into the `time` table. Remember to run `create_tables.py` before running the cell below to ensure you've created/resetted the `time` table in the sparkify database.
```
time_table_insert = ("""
INSERT INTO time
(start_time, hour, day, week, month, year, weekday)
VALUES (%s, %s, %s, %s, %s, %s, %s);
""")
for i, row in time_df.iterrows():
cur.execute(time_table_insert, list(row))
conn.commit()
cur.execute("""
SELECT * FROM time;
""")
row = cur.fetchone()
while row:
print(row)
row = cur.fetchone()
```
Run `test.ipynb` to see if you've successfully added records to this table.
## #4: `users` Table
#### Extract Data for Users Table
- Select columns for user ID, first name, last name, gender and level and set to `user_df`
```
df.head()
user_df = df[['userId', 'firstName', 'lastName', 'gender', 'level']]
user_df
```
#### Insert Records into Users Table
Implement the `user_table_insert` query in `sql_queries.py` and run the cell below to insert records for the users in this log file into the `users` table. Remember to run `create_tables.py` before running the cell below to ensure you've created/resetted the `users` table in the sparkify database.
```
user_table_insert = ("""
INSERT INTO users
(user_id, first_name, last_name, gender, level)
VALUES (%s, %s, %s, %s, %s)
""")
for i, row in user_df.iterrows():
cur.execute(user_table_insert, row)
conn.commit()
cur.execute("SELECT * FROM users;")
row = cur.fetchone()
count = 0
while row:
print(row)
row = cur.fetchone()
count += 1
print(f"Total rows: {count}") # Just a check
```
Run `test.ipynb` to see if you've successfully added records to this table.
## #5: `songplays` Table
#### Extract Data and Songplays Table
This one is a little more complicated since information from the songs table, artists table, and original log file are all needed for the `songplays` table. Since the log file does not specify an ID for either the song or the artist, you'll need to get the song ID and artist ID by querying the songs and artists tables to find matches based on song title, artist name, and song duration time.
- Implement the `song_select` query in `sql_queries.py` to find the song ID and artist ID based on the title, artist name, and duration of a song.
- Select the timestamp, user ID, level, song ID, artist ID, session ID, location, and user agent and set to `songplay_data`
```
print(f"Song data:\n{song_data}")
print(f"\nArtist data:\n{artist_data}")
df.head()
song_select = ("""
SELECT s.song_id, a.artist_id
FROM songs s
JOIN artists a ON s.artist_id = a.artist_id
WHERE s.title = %s AND a.name = %s AND s.duration = %s;
""")
```
#### Insert Records into Songplays Table
- Implement the `songplay_table_insert` query and run the cell below to insert records for the songplay actions in this log file into the `songplays` table. Remember to run `create_tables.py` before running the cell below to ensure you've created/resetted the `songplays` table in the sparkify database.
```
songplay_table_insert = ("""
INSERT INTO songplays
(start_time, user_id, level, song_id,
artist_id, session_id, location, user_agent)
VALUES (%s, %s, %s, %s, %s, %s, %s, %s)
""")
for index, row in df.iterrows():
# get songid and artistid from song and artist tables
results = cur.execute(song_select, (row.song, row.artist, row.length))
songid, artistid = results if results else None, None
# insert songplay record
songplay_data = (row.ts, row.userId, row.level, songid, artistid, row.sessionId, row.location, row.userAgent)
cur.execute(songplay_table_insert, songplay_data)
conn.commit()
print(songid, artistid)
```
Run `test.ipynb` to see if you've successfully added records to this table.
# Close Connection to Sparkify Database
```
conn.close()
```
# Implement `etl.py`
Use what you've completed in this notebook to implement `etl.py`.
|
github_jupyter
|
import os
import glob
import psycopg2
import pandas as pd
from sql_queries import *
conn = psycopg2.connect("host=127.0.0.1 dbname=sparkifydb user=student password=student")
cur = conn.cursor()
cur.close()
conn.close()
def get_files(filepath):
all_files = []
for root, dirs, files in os.walk(filepath):
files = glob.glob(os.path.join(root,'*.json'))
for f in files :
all_files.append(os.path.relpath(f)) # NOTE: This was edited as I was using my own system
return all_files
filepath = 'data/song_data'
song_files = get_files(filepath)
song_files[0]
df = pd.read_json(song_files[0], lines=True)
df.head()
song_data = (
df[['song_id', 'title', 'artist_id',
'year', 'duration']].values
)
song_data = list(song_data[0])
song_data
song_table_insert = ("""
INSERT INTO songs (song_id, title, artist_id, year, duration)
VALUES (%s, %s, %s, %s, %s);
""")
cur.execute(song_table_insert, song_data)
conn.commit()
cur.execute("""
SELECT * FROM songs;
""")
row = cur.fetchone()
while row:
print(row)
row = cur.fetchone()
df
artist_data = (
df[['artist_id', 'artist_name', 'artist_location',
'artist_latitude', 'artist_longitude']].values
)
artist_data = list(artist_data[0])
artist_data
artist_table_insert = ("""
INSERT INTO artists
(artist_id, name, location, latitude, longitude)
VALUES (%s, %s, %s, %s, %s);
""")
cur.execute(artist_table_insert, artist_data)
conn.commit()
cur.execute("""
SELECT * FROM artists;
""")
row = cur.fetchone()
while row:
print(row)
row = cur.fetchone()
filepath = 'data/log_data'
log_files = get_files(filepath)
len(log_files)
df = pd.read_json(log_files[0], lines=True)
df.head()
is_NextSong = df['page'] == 'NextSong'
df = df[is_NextSong]
df.head()
# Python version
import datetime
datetime.datetime.fromtimestamp(1541377992796/1000.0)
# (year, month, day, hour, minute, second, microsecond)
# datetime.datetime(2018, 11, 4, 16, 33, 12, 796000)
# Pandas version
pd.to_datetime(1541377992796, unit='ms')
# Timestamp('2018-11-05 00:33:12.796000')
# pd.to_datetime(1541377992796, unit='ms').day_name() - 'Monday'
df['ts'] = pd.to_datetime(df['ts'], unit='ms')
df.head()
t = df['ts']
t.head()
time_data = list((t, t.dt.hour, t.dt.day, t.dt.week, t.dt.month, t.dt.year, t.dt.day_name()))
column_labels = ('timestamp', 'hour', 'day', 'week', 'month', 'year', 'day_of_week')
time_data
time_dict = dict(zip(column_labels, time_data))
time_dict
time_df = pd.DataFrame(time_dict)
time_df
time_table_insert = ("""
INSERT INTO time
(start_time, hour, day, week, month, year, weekday)
VALUES (%s, %s, %s, %s, %s, %s, %s);
""")
for i, row in time_df.iterrows():
cur.execute(time_table_insert, list(row))
conn.commit()
cur.execute("""
SELECT * FROM time;
""")
row = cur.fetchone()
while row:
print(row)
row = cur.fetchone()
df.head()
user_df = df[['userId', 'firstName', 'lastName', 'gender', 'level']]
user_df
user_table_insert = ("""
INSERT INTO users
(user_id, first_name, last_name, gender, level)
VALUES (%s, %s, %s, %s, %s)
""")
for i, row in user_df.iterrows():
cur.execute(user_table_insert, row)
conn.commit()
cur.execute("SELECT * FROM users;")
row = cur.fetchone()
count = 0
while row:
print(row)
row = cur.fetchone()
count += 1
print(f"Total rows: {count}") # Just a check
print(f"Song data:\n{song_data}")
print(f"\nArtist data:\n{artist_data}")
df.head()
song_select = ("""
SELECT s.song_id, a.artist_id
FROM songs s
JOIN artists a ON s.artist_id = a.artist_id
WHERE s.title = %s AND a.name = %s AND s.duration = %s;
""")
songplay_table_insert = ("""
INSERT INTO songplays
(start_time, user_id, level, song_id,
artist_id, session_id, location, user_agent)
VALUES (%s, %s, %s, %s, %s, %s, %s, %s)
""")
for index, row in df.iterrows():
# get songid and artistid from song and artist tables
results = cur.execute(song_select, (row.song, row.artist, row.length))
songid, artistid = results if results else None, None
# insert songplay record
songplay_data = (row.ts, row.userId, row.level, songid, artistid, row.sessionId, row.location, row.userAgent)
cur.execute(songplay_table_insert, songplay_data)
conn.commit()
print(songid, artistid)
conn.close()
| 0.143878 | 0.887546 |
## Import the packages
We will use *scanpy* as the main analysis tool for the analysis. Scanpy has a comprehensive [manual webpage](https://scanpy.readthedocs.io/en/stable/) that includes many different tutorial you can use for further practicing. Scanpy is used in the discussion paper and the tutorial paper of this course.
An alternative and well-established tool for R users is [Seurat](https://satijalab.org/seurat/). However, scanpy is mainatined and updated by a wider community with many of the latest developed tools.
```
import scanpy as sc
import pandas as pd
import scvelo as scv
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
import sklearn
```
## Loading and understanding the dataset structure
Data can be loaded from many different possible formats. Each format has a dedicated reading command, for example `read_h5ad`, `read_10X_mtx`, `read_txt`. We are going to use `read_10X_mtx` to load the output of the 10X software that produces the aligned data.
Note the option `cache=True`. If you are going to read again the same data, it will be loaded extremely fast, because it has been stored in a convenient format for large datasets (`h5ad` format)
```
crypto_1 = sc.read_10x_mtx('../../../../scRNASeq_course/Data/cellranger_crypto1/outs/filtered_feature_bc_matrix/', cache=True)
```
The datasets `crypto_1` and `crypto_3` are now created. They are so-called `Annotated datasets`. Each annotated dataset contains:
* The data matrix `X` of size $N\_cells \times N\_genes$
* Vectors of cells-related quantities in the table `obs`(for example, how many transcripts there are in each cell)
* Vectors of genes-related quantities in the table `var` (for example, in how many cells the each gene is detected)
* Matrices of size $N\_cells \times N\_genes$ in `adata.layers` (for example, normalized data matrix, imputed data matrix, ....)
We will often call the cells for observations (obs) and the genes for variables (var) when it is practical in relation to the annotated dataset
During the analysis we will encounter other components of the annotated datasets. They will be explained when it is necessary, so you might want to skip this explanation if you want.
* Matrices where each line is cell-related in `obsm` (for example, the PCA coordinates of each cell)
* Matrices where each line is gene-related in `adata.varm` (for example, mean of the gene in each cell type)
* Anything else useful is in `adata.uns` and some quantities necessary for the `scanpy` package are saved in `obsp`

**Above:** a representation of the data matrix, variable and observations in an annotated dataset.
Each component of the annotated dataset is called by using a `dot`, For example, we can see the data matrix by
```
crypto_1.X
```
The matrix is in compressed format. We can reassign it as a dense matrix, so that we can see what it contains.
```
crypto_1.X = np.array( crypto_1.X.todense() )
crypto_1.X
```
When the matrix is no longer compressed, we can calculate some statistics for both cells and genes with the following `scanpy` command. Note that all scanpy commands follow a similar format.
```
sc.preprocessing.calculate_qc_metrics(crypto_1, inplace=True)
```
We can see that `obs` and `var` now contains a lot of different values whose names are mostly self-explicative. For example
- `n_genes_by_counts` is the number of detected genes in each cell
- `total_counts` is the number of transcripts in each cell
- `mean_counts` is the average of counts of each gene across all cells
```
crypto_1
```
You can access directly all observations/variables or some of them specifically. Each observation line is named with the cell barcode, while variables have gene names in each line
```
crypto_1.obs
crypto_1.obs[ ['total_counts','n_genes_by_counts'] ]
crypto_1.var
```
We store the matrix `X` to save the raw values. We will be able to see it in `layers`, independently of how we transform the matrix `X`
```
crypto_1.layers[ 'umi_raw' ] = crypto_1.X.copy()
```
We can see the matrix in `layers`, and reassign it to `X` or use it if needed in some future analysis
```
crypto_1
crypto_1.layers['umi_raw']
```
The annotated datasets can be easily saved by using `write`. The format to be used is `h5ad`.
```
crypto_1.write('../../../Data/notebooks_data/crypto_1.h5ad')
```
|
github_jupyter
|
import scanpy as sc
import pandas as pd
import scvelo as scv
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
import sklearn
crypto_1 = sc.read_10x_mtx('../../../../scRNASeq_course/Data/cellranger_crypto1/outs/filtered_feature_bc_matrix/', cache=True)
crypto_1.X
crypto_1.X = np.array( crypto_1.X.todense() )
crypto_1.X
sc.preprocessing.calculate_qc_metrics(crypto_1, inplace=True)
crypto_1
crypto_1.obs
crypto_1.obs[ ['total_counts','n_genes_by_counts'] ]
crypto_1.var
crypto_1.layers[ 'umi_raw' ] = crypto_1.X.copy()
crypto_1
crypto_1.layers['umi_raw']
crypto_1.write('../../../Data/notebooks_data/crypto_1.h5ad')
| 0.265119 | 0.990188 |
```
import sys
sys.path.append('..')
```
# Using nbtlib
The Named Binary Tag (NBT) file format is a simple structured binary format that is mainly used by the game Minecraft (see the [official specification](http://wiki.vg/NBT) for more details). This short documentation will show you how you can manipulate nbt data using the `nbtlib` module.
## Loading a file
```
import nbtlib
nbt_file = nbtlib.load('nbt_files/bigtest.nbt')
nbt_file.root['stringTest']
```
By default `nbtlib.load` will figure out by itself if the specified file is gzipped, but you can also use the `gzipped=` keyword only argument if you know in advance whether the file is gzipped or not.
```
uncompressed_file = nbtlib.load('nbt_files/hello_world.nbt', gzipped=False)
uncompressed_file.gzipped
```
The `nbtlib.load` function also accepts the `byteorder=` keyword only argument. It lets you specify whether the file is big-endian or little-endian. The default value is `'big'`, which means that the file is interpreted as big-endian by default. You can set it to `'little'` to use the little-endian format.
```
little_endian_file = nbtlib.load('nbt_files/hello_world_little.nbt', byteorder='little')
little_endian_file.byteorder
```
Objects returned by the `nbtlib.load` function are instances of the `nbtlib.File` class. The `nbtlib.load` function is actually a small helper around the `File.load` classmethod. If you need to load files from an already opened file-like object, you can use the `File.parse` class method.
```
from nbtlib import File
with open('nbt_files/hello_world.nbt', 'rb') as f:
hello_world = File.parse(f)
hello_world
```
## Accessing file data
The `File` class inherits from `Compound`, which inherits from `dict`. This means that you can use standard `dict` operations to access data inside of the file. As most files usually contain a single root tag, there is a shorthand to access it directly.
```
nbt_file.keys()
nbt_file.root == nbt_file['Level']
```
## Modifying files
```
from nbtlib.tag import *
with nbtlib.load('nbt_files/demo.nbt') as demo:
demo.root['counter'] = Int(demo.root['counter'] + 1)
demo
```
If you don't want to use a context manager, you can call the `.save` method manually to overwrite the original file or make a copy by specifying a different path. The `.save` method also accepts the `gzipped=` keyword only argument. By default, the copy will be gzipped if the original file is gzipped. Similarly, you can use the `byteorder=` keyword only argument to specify whether the file should be saved using the big-endian or little-endian format. By default, the copy will be saved using the same format as the original file.
```
demo = nbtlib.load('nbt_files/demo.nbt')
...
demo.save() # overwrite
demo.save('nbt_files/demo_copy.nbt', gzipped=True) # make a gzipped copy
demo.save('nbt_files/demo_little.nbt', byteorder='little') # convert the file to little-endian
nbtlib.load('nbt_files/demo_copy.nbt').root['counter']
nbtlib.load('nbt_files/demo_little.nbt', byteorder='little').root['counter']
```
You can also write nbt data to an already opened file-like object using the `.write` method.
```
with open('nbt_files/demo_copy.nbt', 'wb') as f:
demo.write(f)
```
## Creating files
```
new_file = File({
'': Compound({
'foo': String('bar'),
'spam': IntArray([1, 2, 3]),
'egg': List[String](['hello', 'world'])
})
})
new_file.save('nbt_files/new_file.nbt')
loaded_file = nbtlib.load('nbt_files/new_file.nbt')
loaded_file.gzipped
loaded_file.byteorder
```
New files are uncompressed by default. You can use the `gzipped=` keyword only argument to create a gzipped file. New files are also big-endian by default. You can use the `byteorder=` keyword only argument to set the endianness of the file to either `'big'` or `'little'`.
```
new_file = File(
{'': Compound({'thing': LongArray([1, 2, 3])})},
gzipped=True,
byteorder='little'
)
new_file.save('nbt_files/new_file_gzipped_little.nbt')
loaded_file = nbtlib.load('nbt_files/new_file_gzipped_little.nbt', byteorder='little')
loaded_file.gzipped
loaded_file.byteorder
```
## Performing operations on tags
With the exception of `ByteArray`, `IntArray` and `LongArray` tags, every tag type inherits from a python builtin, allowing you to make use of their rich and familiar interfaces. `ByteArray`, `IntArray` and `LongArray` tags on the other hand, inherit from `numpy` arrays instead of the builtin `array` type in order to benefit from `numpy`'s efficiency.
| Base type | Associated nbt tags |
| ------------------- | ------------------------------------ |
| **int** | `Byte`, `Short`, `Int`, `Long` |
| **float** | `Float`, `Double` |
| **str** | `String` |
| **numpy.ndarray** | `ByteArray`, `IntArray`, `LongArray` |
| **list** | `List` |
| **dict** | `Compound` |
All the methods and operations that are usually available on the the base types can be used on the associated tags.
```
my_list = List[String](char.upper() for char in 'hello')
my_list.reverse()
my_list[3:]
my_array = IntArray([1, 2, 3])
my_array + 100
my_pizza = Compound({
'name': String('Margherita'),
'price': Double(5.7),
'size': String('medium')
})
my_pizza.update({'name': String('Calzone'), 'size': String('large')})
my_pizza['price'] = Double(my_pizza['price'] + 2.5)
my_pizza
```
## Serializing nbt tags to snbt
While using `repr()` on nbt tags outputs a python representation of the tag, calling `str()` on nbt tags (or simply printing them) will return the nbt literal representing that tag.
```
example_tag = Compound({
'numbers': IntArray([1, 2, 3]),
'foo': String('bar'),
'syntax breaking': Float(42),
'spam': String('{"text":"Hello, world!\\n"}')
})
print(repr(example_tag))
print(str(example_tag))
print(example_tag)
```
Converting nbt tags to strings will serialize them to snbt. If you want more control over the way nbt tags are serialized, you can use the `nbtlib.serialize_tag` function. In fact, using `str` on nbt tags simply calls `nbtlib.serialize_tag` on the specified tag.
```
from nbtlib import serialize_tag
print(serialize_tag(example_tag))
serialize_tag(example_tag) == str(example_tag)
```
You might have noticed that by default, the `nbtlib.serialize_tag` function will render strings with single `'` or double `"` quotes based on their content to avoid escaping quoting characters. The string is serialized such that the type of quotes used is different from the first quoting character found in the string. If the string doesn't contain any quoting character, the `nbtlib.serialize_tag` function will render the string as a double `"` quoted string.
```
print(String("contains 'single' quotes"))
print(String('contains "double" quotes'))
print(String('''contains 'single' and "double" quotes'''))
```
You can overwrite this behavior by setting the `quote=` keyword only argument to either a single `'` or a double `"` quote.
```
print(serialize_tag(String('forcing "double" quotes'), quote='"'))
```
The `nbtlib.serialize_tag` function can be used with the `compact=` keyword only argument to remove all the extra whitespace from the output.
```
print(serialize_tag(example_tag, compact=True))
```
If you'd rather have something a bit more readable, you can use the `indent=` keyword only argument to tell the `nbtlib.serialize_tag` function to output indented snbt. The argument can be either a string or an integer and will be used to define how to render each indentation level.
```
nested_tag = Compound({
'foo': List[Int]([1, 2, 3]),
'bar': String('name'),
'values': List[Compound]([
{'test': String('a'), 'thing': ByteArray([32, 32, 32])},
{'test': String('b'), 'thing': ByteArray([64, 64, 64])}
])
})
print(serialize_tag(nested_tag, indent=4))
```
If you need the output ot be indented with tabs instead, you can set the `indent=` argument to `'\t'`.
```
print(serialize_tag(nested_tag, indent='\t'))
```
Note that the `indent=` keyword only argument can be set to any string, not just `'\t'`.
```
print(serialize_tag(nested_tag, indent='. '))
```
## Creating tags from nbt literals
`nbtlib` supports creating nbt tags from their literal representation. The `nbtlib.parse_nbt` function can parse snbt and return the appropriate tag.
```
from nbtlib import parse_nbt
parse_nbt('hello')
parse_nbt('{foo:[{bar:[I;1,2,3]},{spam:6.7f}]}')
```
Note that the parser ignores whitespace.
```
parse_nbt("""{
foo: [1, 2, 3],
bar: "name",
values: [
{
test: "a",
thing: [B; 32B, 32B, 32B]
},
{
test: "b",
thing: [B; 64B, 64B, 64B]
}
]
}""")
```
## Defining schemas
In order to avoid wrapping values manually every time you edit a compound tag, you can define a schema that will take care of converting python types to predefined nbt tags automatically.
```
from nbtlib import schema
MySchema = schema('MySchema', {
'foo': String,
'bar': Short
})
my_object = MySchema({'foo': 'hello world', 'bar': 21})
my_object['bar'] *= 2
my_object
```
By default, you can interact with keys that are not defined in the schema. However, if you use the `strict=` keyword only argument, the schema instance will raise a `TypeError` whenever you try to access a key that wasn't defined in the original schema.
```
MyStrictSchema = schema('MyStrictSchema', {
'foo': String,
'bar': Short
}, strict=True)
strict_instance = MyStrictSchema()
strict_instance.update({'foo': 'hello world'})
strict_instance
try:
strict_instance['something'] = List[String](['this', 'raises', 'an', 'error'])
except TypeError as exc:
print(exc)
```
The `schema` function is a helper that creates a class that inherits from `CompoundSchema`. This means that you can also inherit from the class manually.
```
from nbtlib import CompoundSchema
class MySchema(CompoundSchema):
schema = {
'foo': String,
'bar': Short
}
MySchema({'foo': 'hello world', 'bar': 42})
```
You can also set the `strict` class attribute to `True` to create a strict schema type.
```
class MyStrictSchema(CompoundSchema):
schema = {
'foo': String,
'bar': Short
}
strict = True
try:
MyStrictSchema({'something': Byte(5)})
except TypeError as exc:
print(exc)
```
## Combining schemas and custom file types
If you need to deal with files that always have a particular structure, you can create a specialized file type by combining it with a schema. For instance, this is how you would create a file type that opens [minecraft structure files](https://minecraft.gamepedia.com/Structure_block_file_format).
First, we need to define what a minecraft structure is, so we create a schema that matches the tag hierarchy.
```
Structure = schema('Structure', {
'DataVersion': Int,
'author': String,
'size': List[Int],
'palette': List[schema('State', {
'Name': String,
'Properties': Compound,
})],
'blocks': List[schema('Block', {
'state': Int,
'pos': List[Int],
'nbt': Compound,
})],
'entities': List[schema('Entity', {
'pos': List[Double],
'blockPos': List[Int],
'nbt': Compound,
})],
})
```
Now let's test our schema by creating a structure. We can see that all the types are automatically applied.
```
new_structure = Structure({
'DataVersion': 1139,
'author': 'dinnerbone',
'size': [1, 2, 1],
'palette': [
{'Name': 'minecraft:dirt'}
],
'blocks': [
{'pos': [0, 0, 0], 'state': 0},
{'pos': [0, 1, 0], 'state': 0}
],
'entities': [],
})
type(new_structure['blocks'][0]['pos'])
type(new_structure['entities'])
```
Now we can create a custom file type that wraps our structure schema. Since structure files are always gzipped we can override the load method to default the `gzipped` argument to `True`. We also overwrite the constructor so that it can take directly an instance of our structure schema as argument.
```
class StructureFile(File, schema('StructureFileSchema', {'': Structure})):
def __init__(self, structure_data=None):
super().__init__({'': structure_data or {}})
self.gzipped = True
@classmethod
def load(cls, filename, gzipped=True):
return super().load(filename, gzipped)
```
We can now use the custom file type to load, edit and save structure files without having to specify the tags manually.
```
structure_file = StructureFile(new_structure)
structure_file.save('nbt_files/new_structure.nbt') # you can load it in a minecraft world!
```
So now let's try to edit the structure. We're going to replace all the dirt blocks with stone blocks.
```
with StructureFile.load('nbt_files/new_structure.nbt') as structure_file:
structure_file.root['palette'][0]['Name'] = 'minecraft:stone'
```
As you can see we didn't need to specify any tag to edit the file.
```
print(serialize_tag(StructureFile.load('nbt_files/new_structure.nbt'), indent=4))
```
|
github_jupyter
|
import sys
sys.path.append('..')
import nbtlib
nbt_file = nbtlib.load('nbt_files/bigtest.nbt')
nbt_file.root['stringTest']
uncompressed_file = nbtlib.load('nbt_files/hello_world.nbt', gzipped=False)
uncompressed_file.gzipped
little_endian_file = nbtlib.load('nbt_files/hello_world_little.nbt', byteorder='little')
little_endian_file.byteorder
from nbtlib import File
with open('nbt_files/hello_world.nbt', 'rb') as f:
hello_world = File.parse(f)
hello_world
nbt_file.keys()
nbt_file.root == nbt_file['Level']
from nbtlib.tag import *
with nbtlib.load('nbt_files/demo.nbt') as demo:
demo.root['counter'] = Int(demo.root['counter'] + 1)
demo
demo = nbtlib.load('nbt_files/demo.nbt')
...
demo.save() # overwrite
demo.save('nbt_files/demo_copy.nbt', gzipped=True) # make a gzipped copy
demo.save('nbt_files/demo_little.nbt', byteorder='little') # convert the file to little-endian
nbtlib.load('nbt_files/demo_copy.nbt').root['counter']
nbtlib.load('nbt_files/demo_little.nbt', byteorder='little').root['counter']
with open('nbt_files/demo_copy.nbt', 'wb') as f:
demo.write(f)
new_file = File({
'': Compound({
'foo': String('bar'),
'spam': IntArray([1, 2, 3]),
'egg': List[String](['hello', 'world'])
})
})
new_file.save('nbt_files/new_file.nbt')
loaded_file = nbtlib.load('nbt_files/new_file.nbt')
loaded_file.gzipped
loaded_file.byteorder
new_file = File(
{'': Compound({'thing': LongArray([1, 2, 3])})},
gzipped=True,
byteorder='little'
)
new_file.save('nbt_files/new_file_gzipped_little.nbt')
loaded_file = nbtlib.load('nbt_files/new_file_gzipped_little.nbt', byteorder='little')
loaded_file.gzipped
loaded_file.byteorder
my_list = List[String](char.upper() for char in 'hello')
my_list.reverse()
my_list[3:]
my_array = IntArray([1, 2, 3])
my_array + 100
my_pizza = Compound({
'name': String('Margherita'),
'price': Double(5.7),
'size': String('medium')
})
my_pizza.update({'name': String('Calzone'), 'size': String('large')})
my_pizza['price'] = Double(my_pizza['price'] + 2.5)
my_pizza
example_tag = Compound({
'numbers': IntArray([1, 2, 3]),
'foo': String('bar'),
'syntax breaking': Float(42),
'spam': String('{"text":"Hello, world!\\n"}')
})
print(repr(example_tag))
print(str(example_tag))
print(example_tag)
from nbtlib import serialize_tag
print(serialize_tag(example_tag))
serialize_tag(example_tag) == str(example_tag)
print(String("contains 'single' quotes"))
print(String('contains "double" quotes'))
print(String('''contains 'single' and "double" quotes'''))
print(serialize_tag(String('forcing "double" quotes'), quote='"'))
print(serialize_tag(example_tag, compact=True))
nested_tag = Compound({
'foo': List[Int]([1, 2, 3]),
'bar': String('name'),
'values': List[Compound]([
{'test': String('a'), 'thing': ByteArray([32, 32, 32])},
{'test': String('b'), 'thing': ByteArray([64, 64, 64])}
])
})
print(serialize_tag(nested_tag, indent=4))
print(serialize_tag(nested_tag, indent='\t'))
print(serialize_tag(nested_tag, indent='. '))
from nbtlib import parse_nbt
parse_nbt('hello')
parse_nbt('{foo:[{bar:[I;1,2,3]},{spam:6.7f}]}')
parse_nbt("""{
foo: [1, 2, 3],
bar: "name",
values: [
{
test: "a",
thing: [B; 32B, 32B, 32B]
},
{
test: "b",
thing: [B; 64B, 64B, 64B]
}
]
}""")
from nbtlib import schema
MySchema = schema('MySchema', {
'foo': String,
'bar': Short
})
my_object = MySchema({'foo': 'hello world', 'bar': 21})
my_object['bar'] *= 2
my_object
MyStrictSchema = schema('MyStrictSchema', {
'foo': String,
'bar': Short
}, strict=True)
strict_instance = MyStrictSchema()
strict_instance.update({'foo': 'hello world'})
strict_instance
try:
strict_instance['something'] = List[String](['this', 'raises', 'an', 'error'])
except TypeError as exc:
print(exc)
from nbtlib import CompoundSchema
class MySchema(CompoundSchema):
schema = {
'foo': String,
'bar': Short
}
MySchema({'foo': 'hello world', 'bar': 42})
class MyStrictSchema(CompoundSchema):
schema = {
'foo': String,
'bar': Short
}
strict = True
try:
MyStrictSchema({'something': Byte(5)})
except TypeError as exc:
print(exc)
Structure = schema('Structure', {
'DataVersion': Int,
'author': String,
'size': List[Int],
'palette': List[schema('State', {
'Name': String,
'Properties': Compound,
})],
'blocks': List[schema('Block', {
'state': Int,
'pos': List[Int],
'nbt': Compound,
})],
'entities': List[schema('Entity', {
'pos': List[Double],
'blockPos': List[Int],
'nbt': Compound,
})],
})
new_structure = Structure({
'DataVersion': 1139,
'author': 'dinnerbone',
'size': [1, 2, 1],
'palette': [
{'Name': 'minecraft:dirt'}
],
'blocks': [
{'pos': [0, 0, 0], 'state': 0},
{'pos': [0, 1, 0], 'state': 0}
],
'entities': [],
})
type(new_structure['blocks'][0]['pos'])
type(new_structure['entities'])
class StructureFile(File, schema('StructureFileSchema', {'': Structure})):
def __init__(self, structure_data=None):
super().__init__({'': structure_data or {}})
self.gzipped = True
@classmethod
def load(cls, filename, gzipped=True):
return super().load(filename, gzipped)
structure_file = StructureFile(new_structure)
structure_file.save('nbt_files/new_structure.nbt') # you can load it in a minecraft world!
with StructureFile.load('nbt_files/new_structure.nbt') as structure_file:
structure_file.root['palette'][0]['Name'] = 'minecraft:stone'
print(serialize_tag(StructureFile.load('nbt_files/new_structure.nbt'), indent=4))
| 0.314156 | 0.885186 |
---
In the [last post](https://tmthyjames.github.io/posts/Analyzing-Rap-Lyrics-Using-Word-Vectors/) we analyzed rap lyrics using word vectors. We mostly did some first-pass analysis and very little prediction. In this post, we'll actually focus on predictions and visualizing our results. I'll use Python's machine-learning library, <a href="http://scikit-learn.org/stable/">scikit-learn</a>, to build a <a href="https://en.wikipedia.org/wiki/Naive_Bayes_classifier">naive Bayes classifier</a> to predict a song's genre given its lyrics. To get the data, we'll use [Cypher](https://github.com/tmthyjames/cypher), a new Python package [I recently released](https://tmthyjames.github.io/tools/Cypher/) that retrieves music lyrics. To visualize the results, I'll use [D3](https://d3js.org/) and [D3Plus](https://d3plus.org/), which is a nice wrapper for D3.
## Contents
• [Quick Note on Naive Bayes](#Quick-Note-on-Naive-Bayes)<br/>
• [Getting the Data](#Getting-the-Data)<br/>
• [Loading the Data](#Loading-the-Data)<br/>
• [Splitting the Data](#Splitting-the-Data)<br/>
• [Training the Model](#Training-the-Model)<br/>
• [Top Hip Hop Songs](#Top-Hip-Hop-Songs)<br/>
• [Hip Hop Songs that have Alt Rock and Country Lyrics](#Hip-Hop-Songs-that-have-Alt-Rock-and-Country-Lyrics)<br/>
• [Visualizing Our results](#Visualizing-Our-Results) (With d3.js)<br/>
• [Up Next](#Up-Next)<br/>
## Quick Note on Naive Bayes
The naive Bayes classifier is based on [Bayes' Theorem](https://en.wikipedia.org/wiki/Bayes%27_theorem) and known for its simplicity, accuracy, and speed, particularly when it comes to text classification, which is what our aim is for this post. In short, as Wikipedia puts it, Bayes' Theorem describes the probability of an event, based on prior knowledge of conditions that might be related to the event. For example, if a musical genre is related to lyrics, then, with Bayes' Theorem, we can more accuarately assess the probability that a certain song belongs to a particular genre, compared to the assessment of the probability of a genre made without knowledge of a song's lyrics. For more on Bayes' Theorem, check this [post](https://monkeylearn.com/blog/practical-explanation-naive-bayes-classifier/) out.
## Getting the Data
The data was retrieved with [Cypher](https://github.com/tmthyjames/cypher). The data and code used for this post is available on the Cypher's [GitHub page](https://github.com/tmthyjames/cypher/tree/master/notebooks). Since the data takes so long to retrieve (there are over 900 hundred artists), I plan on adding a feature to Cypher that allows the user to load already-retrieved data if it exists, other wise it will retrieve the data like normal. For now, you can just download it from the [GitHub page](https://github.com/tmthyjames/cypher/tree/master/notebooks).
I started this post with the intention of trying to classify 10 genres: pop, blues, heavy metal, classic rock, indie folk, RnB, punk rock, screamo, country, and rap.
I ran into a few problems with this as classic rock lyrically was very similar to country; indie folk was also similar to country; punk rock, heavy metal, and screamo were all similar; and RnB and rap were very similar. It's not surprising; as the number of classes grows, it becomes harder to correctly classify. I may write a post on my trouble with this approach if there is interest in it, or just post the results of trying to predict all 10 genres.
Anyways, to get the data, I used [Ranker](https://ranker.com) to get a list of the top 100 artists of each genre. They have a nice API endpoint you can hit to get all the artists so you don't have to web scrape.
## Loading the Data
To load the data, we'll use [pandas'](https://pandas.pydata.org/) `read_csv` method. We'll also clean up the genres due to the problems mentioned above about lyrical similarity. The three genres we'll try to predict are country, rap, and alt rock since those genres are clearly different. For our purposes, we'll classify metal, punk, and screamo as "alt rock". Here's how we do it:
```
import pandas as pd
import numpy as np
df = pd.read_csv('lyrics.csv')
df['ranker_genre'] = np.where(
(df['ranker_genre'] == 'screamo')|
(df['ranker_genre'] == 'punk rock')|
(df['ranker_genre'] == 'heavy metal'),
'alt rock',
df['ranker_genre']
)
```
The data is available as one lyric per row. To train our classifier, we'll need to transform it into one *song* per row. We'll also go ahead and convert the data to lowercase with `.apply(lambda x: x.lower())`. To do that, we do the following:
```
group = ['song', 'year', 'album', 'genre', 'artist', 'ranker_genre']
lyrics_by_song = df.sort_values(group)\
.groupby(group).lyric\
.apply(' '.join)\
.apply(lambda x: x.lower())\
.reset_index(name='lyric')
lyrics_by_song["lyric"] = lyrics_by_song['lyric'].str.replace(r'[^\w\s]','')
```
## Splitting the Data
Next we'll split our data into a training set and a testing set using only Country, Alt Rock, and Hip Hop. A quick note: because the lyrics are community-sourced some of the songs have incomplete or incorrect lyrics. A lot of the songs with less than 400 characters are just strings of nonsense characters. Therefore, I filtered those songs out as they didn't contribute any value or insight to the model.
```
from sklearn.utils import shuffle
from nltk.corpus import stopwords
genres = [
'Country', 'alt rock', 'Hip Hop',
]
LYRIC_LEN = 400 # each song has to be > 400 characters
N = 10000 # number of records to pull from each genre
RANDOM_SEED = 200 # random seed to make results repeatable
train_df = pd.DataFrame()
test_df = pd.DataFrame()
for genre in genres: # loop over each genre
subset = lyrics_by_song[ # create a subset
(lyrics_by_song.ranker_genre==genre) &
(lyrics_by_song.lyric.str.len() > LYRIC_LEN)
]
train_set = subset.sample(n=N, random_state=RANDOM_SEED)
test_set = subset.drop(train_set.index)
train_df = train_df.append(train_set) # append subsets to the master sets
test_df = test_df.append(test_set)
train_df = shuffle(train_df)
test_df = shuffle(test_df)
```
## Training the Model
Next, we'll train a model using word frequencies and `sklearn`'s `CountVectorizer`. The `CountVectorizer` is a quick and dirty way to train a language model by using simple word counts. Later we'll try a more sophisticated approach with the `TfidfVectorizer`.
```
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.naive_bayes import MultinomialNB
from sklearn.pipeline import Pipeline
# define our model
text_clf = Pipeline(
[('vect', CountVectorizer()),
('clf', MultinomialNB(alpha=0.1))])
# train our model on training data
text_clf.fit(train_df.lyric, train_df.ranker_genre)
# score our model on testing data
predicted = text_clf.predict(test_df.lyric)
np.mean(predicted == test_df.ranker_genre)
```
Not a bad first-pass model!
Word frequencies work fine here, but let's see if we can get a better model by using the `TfidfVectorizer`.
`tf-idf` stands for "term frequency-inverse document frequency". `tf` summarizes how often a given word appears within a document, while `idf` scales down words that appear frequently across documents. For example, if we were trying to figure out which rap artists were lyrically similar, the term `police` may not be very helpful as almost every rapper uses this term. But the term `detroit` may carry more weight as only a hand full of rappers use it. Thus, although `police` would have a higher `tf` score, `detroit` would have a higher `tf-idf` score and would be a more important feature in a language model.
So let's train a model using `tf-idf` scores as features.
```
from sklearn.naive_bayes import MultinomialNB
from sklearn.pipeline import Pipeline
from sklearn.feature_extraction.text import TfidfTransformer, TfidfVectorizer
# define our model
text_clf = Pipeline(
[('vect', TfidfVectorizer()),
('clf', MultinomialNB(alpha=0.1))])
# train our model on training data
text_clf.fit(train_df.lyric, train_df.ranker_genre)
# score our model on testing data
predicted = text_clf.predict(test_df.lyric)
np.mean(predicted == test_df.ranker_genre)
```
Hmmm. Our model seems to have gotten worse. Let's try tuning a few hyperparameters, lemmatizing our data, customizing our tokenizer a bit, and filtering our words with `nltk`'s builtin stopword list.
```
from sklearn.naive_bayes import MultinomialNB
from sklearn.pipeline import Pipeline
from nltk import word_tokenize
from nltk.stem import WordNetLemmatizer
stop = list(set(stopwords.words('english'))) # stopwords
wnl = WordNetLemmatizer() # lemmatizer
def tokenizer(x): # custom tokenizer
return (
wnl.lemmatize(w)
for w in word_tokenize(x)
if len(w) > 2 and w.isalnum() # only words that are > 2 characters
) # and is alpha-numeric
# define our model
text_clf = Pipeline(
[('vect', TfidfVectorizer(
ngram_range=(1, 2), # include bigrams
tokenizer=tokenizer,
stop_words=stop,
max_df=0.4, # ignore terms that appear in more than 40% of documents
min_df=4)), # ignore terms that appear in less than 4 documents
('tfidf', TfidfTransformer()),
('clf', MultinomialNB(alpha=0.1))])
# train our model on training data
text_clf.fit(train_df.lyric, train_df.ranker_genre)
# score our model on testing data
predicted = text_clf.predict(test_df.lyric)
np.mean(predicted == test_df.ranker_genre)
```
Hey! 1% better. I'll take it. We could keep tuning these hyperparameters to squeeze out more accuracy. For example, a more fine-tuned stopword list could help a lot; there are a [few strategies](https://stackoverflow.com/questions/16927494/how-to-select-stop-words-using-tf-idf-non-english-corpus) for constructing a good stopword list. For now, we'll go with our current model.
Now let's go beyond raw accuracy and see how it performs by looking at our confusion matrix for this model.
```
mat = confusion_matrix(test_df.ranker_genre, predicted)
sns.heatmap(
mat.T, square=True, annot=True, fmt='d', cbar=False,
xticklabels=genres,
yticklabels=genres
)
plt.xlabel('true label')
plt.ylabel('predicted label');
```
Given this confusion matrix, we can calculate precision, recall, and f-score, which can be better metrics for evaluating a classifier than raw accuracy.
<b>Recall</b> is the ability of the classifier to find all the positive results. That is, to clasify a rap song *as* a rap song.
<b>Precision</b> is the ability of the classifier to not label a negative result as a positive one. That is, to not classify a country song as a rap song.
<b>F-score</b> is the [harmonic mean](https://en.wikipedia.org/wiki/Harmonic_mean) of precision and recall.
To compute recall, precision, and f-score, we'll use `precision_recall_fscore_support` from `sklearn.metrics`.
```
from sklearn.metrics import precision_recall_fscore_support
precision, recall, fscore, support = precision_recall_fscore_support(test_df.ranker_genre, predicted)
for n,genre in enumerate(genres):
genre = genre.upper()
print(genre+'_precision: {}'.format(precision[n]))
print(genre+'_recall: {}'.format(recall[n]))
print(genre+'_fscore: {}'.format(fscore[n]))
print(genre+'_support: {}'.format(support[n]))
print()
```
<b>Support</b> is the number of each class in the actual true set. And the first thing I notice is that there aren't many alt rock songs being scored. Adding more alt rock songs could possibly improve our model.
We do a good job all around on classifying hip hop and country songs. For alt rock songs, the recall score is great; that is, when it's actually an alt rock song, the model classifies it as an alt rock song 94% of the time. But, as we can see from our alt rock precision score and confusion matrix, the model classifies many hip hop songs as alt rock (963, to be exact), which is the main reason this score is so low.
Let's throw some new data at our model and see how well it does predicting what genre these lyrics belong to.
```
text_clf.predict(
[
"i stand for the red white and blue",
"flow so smooth they say i rap in cursive", #bars *insert fire emoji*
"take my heart and carve it out",
"there is no end to the madness",
"sitting on my front porch drinking sweet tea",
"sitting on my front porch sippin on cognac",
"dog died and my pick up truck wont start",
"im invisible and the drugs wont help",
"i hope you choke in your sleep thinking of me",
"i wonder what genre a song about data science and naive bayes and hyper parameters and maybe a little scatter plots would be"
]
)
```
This seems to classify lyrics pretty well. Not sure about that last lyric though. But, then again, maybe the classifier does as good a job as any human would do classifying those cool data science lyrics?
## Top Hip Hop Songs
Let's retrieve the songs with the highest probability of being hip hop. I'm guessing this will be a prolific artist who's language influences the entire genre. First, though, we need to score each song then merge it in to our dataset.
```
data = train_df.append(test_df) # entire dataset
predicts = text_clf.predict_proba(data.lyric) # score each song
data['Country'], data['Hip_Hop'], data['Alt_Rock'] = ['','',''] # create empty columns
for n,row in enumerate(data.itertuples()): # merge scored data into our dataset
data.loc[row.Index, 'Country'] = predicts[n][0]
data.loc[row.Index, 'Hip_Hop'] = predicts[n][1]
data.loc[row.Index, 'Alt_Rock'] = predicts[n][2]
```
The top 20 most-hip hop songs are:
```
columns_of_interest = [
'artist', 'song', 'album',
'ranker_genre', 'Hip_Hop',
'Alt_Rock', 'Country'
]
data[columns_of_interest]\
.sort_values(['Hip_Hop', 'Alt_Rock', 'Country'], ascending=[0, 1, 1])\
.head(20)
```
And the most hip hop song is Set it Off by Snoop Dogg, who also seems to be the most hip hop rapper, as he has 6 of the top 20 most hip hop songs. Also, it shouldn't be surprising that a lot of these songs are pre-2000, which is the age that hip hop really began to take shape. From this analysis, it seems a lot of the language of hip hop was being defined during those years.
Because the lyrics are community-sourced, there are some duplicate songs. In the real world, we'd want to get rid of these duplicate rows.
## Hip Hop Songs that have Alt Rock and Country Lyrics
Next, let's see which hip hop songs have the most alt rock lyrics. To do this, we'll query our data for only hip hop songs and then sort by the `Alt_Rock` column. I don't have any guesses as to which songs this will be. Maybe songs by Childish Gambino? Or Tech N9ne? Let's see.
```
data[data.ranker_genre=='Hip Hop'][columns_of_interest]\
.sort_values(['Alt_Rock', 'Hip_Hop'], ascending=[0, 1])\
.head(20) # Top 20
```
Wow. Didn't expect some of these results. Lauryn Hill seems to be the alt rock hip hop queen. Although Busta Rhymes has the most alt rock song, Lauryn Hill has 5 of the top 20 and, as we'll see from our visualization below, 12 of the top 100 most alt rock hip hop songs.
Now, let's see which hip hop songs have the most country lyrics. Again, no guesses. Maybe a southern rapper, like Ludacris or Yelawolf?
```
data[data.ranker_genre=='Hip Hop'][columns_of_interest]\
.sort_values(['Country'], ascending=[0])\
.head(20)
```
Well damn. If Lauryn Hill is the alt rock hip hop queen, then Queen Latifah is the queen of country hip hop, at least lyrically.
## Visualizing Our Results
I've also created a dashboard that you can play around with. It visualizes what we just did with our dataframes. Namely, you can look up which songs are most likely to belong to a different genre. In the upper left quadrant, you have the top 1,000 hip hop songs that have alt rock lyrics; you can also choose which genre you'd like to analyze with the drop down options. In the upper right quandrant, there's a table of the top 100 songs based on the filter of the upper left quadrant. In the lower left quadrant, you can see the lyrics weighted by tf-idf scores to allow you to visualize which words are hip hop, alt rock, and country. Lastly, in the lower right quadrant, you have a scatter plot with the tf-idf scores for each word for each genre. This graph is another way of visualizing the lower left quadrant.
With these graphs, you'll get more insight into why exactly the model classified a song a certain way.
<b>To get started, first select a song from the upper left scatter plot.</b>
---
These results look pretty good, even the alt rock songs. If you choose "Alt_Rock songs that have Hip_Hop lyrics", the top song is Rage Against The Machine's "F\*ck Tha Police" which has obvious hip hop overtones. Some may even say it *is* a hip hop song. Also, among the top of that list are the songs birthed from the Jay-Z-Linkin Park collaboration. Again, arguably hip hop songs, so the classifier does well here.
Also, if you choose "Country songs that have Hip_Hop lyrics" you'll notice that the top song is Taylor Swift's Thug Story featuring T-Pain. The lyrics in the lower left box and the lower right tf-idf scatter plot will show that this song is lyrically hip hop even if musically it couldn't be further from it.
## Up Next
Next, I'd like to perform some topic modeling on musical lyrics. But I may be putting most of my effort into [Achoo](https://tmthyjames.github.io/tools/prediction/Achoo-beta-0.1/) for the foreseeable future. Either way, I'll be reporting back soon.
|
github_jupyter
|
import pandas as pd
import numpy as np
df = pd.read_csv('lyrics.csv')
df['ranker_genre'] = np.where(
(df['ranker_genre'] == 'screamo')|
(df['ranker_genre'] == 'punk rock')|
(df['ranker_genre'] == 'heavy metal'),
'alt rock',
df['ranker_genre']
)
group = ['song', 'year', 'album', 'genre', 'artist', 'ranker_genre']
lyrics_by_song = df.sort_values(group)\
.groupby(group).lyric\
.apply(' '.join)\
.apply(lambda x: x.lower())\
.reset_index(name='lyric')
lyrics_by_song["lyric"] = lyrics_by_song['lyric'].str.replace(r'[^\w\s]','')
from sklearn.utils import shuffle
from nltk.corpus import stopwords
genres = [
'Country', 'alt rock', 'Hip Hop',
]
LYRIC_LEN = 400 # each song has to be > 400 characters
N = 10000 # number of records to pull from each genre
RANDOM_SEED = 200 # random seed to make results repeatable
train_df = pd.DataFrame()
test_df = pd.DataFrame()
for genre in genres: # loop over each genre
subset = lyrics_by_song[ # create a subset
(lyrics_by_song.ranker_genre==genre) &
(lyrics_by_song.lyric.str.len() > LYRIC_LEN)
]
train_set = subset.sample(n=N, random_state=RANDOM_SEED)
test_set = subset.drop(train_set.index)
train_df = train_df.append(train_set) # append subsets to the master sets
test_df = test_df.append(test_set)
train_df = shuffle(train_df)
test_df = shuffle(test_df)
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.naive_bayes import MultinomialNB
from sklearn.pipeline import Pipeline
# define our model
text_clf = Pipeline(
[('vect', CountVectorizer()),
('clf', MultinomialNB(alpha=0.1))])
# train our model on training data
text_clf.fit(train_df.lyric, train_df.ranker_genre)
# score our model on testing data
predicted = text_clf.predict(test_df.lyric)
np.mean(predicted == test_df.ranker_genre)
from sklearn.naive_bayes import MultinomialNB
from sklearn.pipeline import Pipeline
from sklearn.feature_extraction.text import TfidfTransformer, TfidfVectorizer
# define our model
text_clf = Pipeline(
[('vect', TfidfVectorizer()),
('clf', MultinomialNB(alpha=0.1))])
# train our model on training data
text_clf.fit(train_df.lyric, train_df.ranker_genre)
# score our model on testing data
predicted = text_clf.predict(test_df.lyric)
np.mean(predicted == test_df.ranker_genre)
from sklearn.naive_bayes import MultinomialNB
from sklearn.pipeline import Pipeline
from nltk import word_tokenize
from nltk.stem import WordNetLemmatizer
stop = list(set(stopwords.words('english'))) # stopwords
wnl = WordNetLemmatizer() # lemmatizer
def tokenizer(x): # custom tokenizer
return (
wnl.lemmatize(w)
for w in word_tokenize(x)
if len(w) > 2 and w.isalnum() # only words that are > 2 characters
) # and is alpha-numeric
# define our model
text_clf = Pipeline(
[('vect', TfidfVectorizer(
ngram_range=(1, 2), # include bigrams
tokenizer=tokenizer,
stop_words=stop,
max_df=0.4, # ignore terms that appear in more than 40% of documents
min_df=4)), # ignore terms that appear in less than 4 documents
('tfidf', TfidfTransformer()),
('clf', MultinomialNB(alpha=0.1))])
# train our model on training data
text_clf.fit(train_df.lyric, train_df.ranker_genre)
# score our model on testing data
predicted = text_clf.predict(test_df.lyric)
np.mean(predicted == test_df.ranker_genre)
mat = confusion_matrix(test_df.ranker_genre, predicted)
sns.heatmap(
mat.T, square=True, annot=True, fmt='d', cbar=False,
xticklabels=genres,
yticklabels=genres
)
plt.xlabel('true label')
plt.ylabel('predicted label');
from sklearn.metrics import precision_recall_fscore_support
precision, recall, fscore, support = precision_recall_fscore_support(test_df.ranker_genre, predicted)
for n,genre in enumerate(genres):
genre = genre.upper()
print(genre+'_precision: {}'.format(precision[n]))
print(genre+'_recall: {}'.format(recall[n]))
print(genre+'_fscore: {}'.format(fscore[n]))
print(genre+'_support: {}'.format(support[n]))
print()
text_clf.predict(
[
"i stand for the red white and blue",
"flow so smooth they say i rap in cursive", #bars *insert fire emoji*
"take my heart and carve it out",
"there is no end to the madness",
"sitting on my front porch drinking sweet tea",
"sitting on my front porch sippin on cognac",
"dog died and my pick up truck wont start",
"im invisible and the drugs wont help",
"i hope you choke in your sleep thinking of me",
"i wonder what genre a song about data science and naive bayes and hyper parameters and maybe a little scatter plots would be"
]
)
data = train_df.append(test_df) # entire dataset
predicts = text_clf.predict_proba(data.lyric) # score each song
data['Country'], data['Hip_Hop'], data['Alt_Rock'] = ['','',''] # create empty columns
for n,row in enumerate(data.itertuples()): # merge scored data into our dataset
data.loc[row.Index, 'Country'] = predicts[n][0]
data.loc[row.Index, 'Hip_Hop'] = predicts[n][1]
data.loc[row.Index, 'Alt_Rock'] = predicts[n][2]
columns_of_interest = [
'artist', 'song', 'album',
'ranker_genre', 'Hip_Hop',
'Alt_Rock', 'Country'
]
data[columns_of_interest]\
.sort_values(['Hip_Hop', 'Alt_Rock', 'Country'], ascending=[0, 1, 1])\
.head(20)
data[data.ranker_genre=='Hip Hop'][columns_of_interest]\
.sort_values(['Alt_Rock', 'Hip_Hop'], ascending=[0, 1])\
.head(20) # Top 20
data[data.ranker_genre=='Hip Hop'][columns_of_interest]\
.sort_values(['Country'], ascending=[0])\
.head(20)
| 0.451568 | 0.987277 |
# Word2Vector
## Imports
```
import glob
import nltk
import codecs
import re
import multiprocessing
import gensim.models.word2vec as w2v
import sklearn.manifold
import pandas as pd
```
#### NLTK tokenizer models
```
nltk.download("punkt")
nltk.download("stopwords")
```
## Prepare Corpus
#### Load books from files
```
filenames = sorted(glob.glob("data/*.txt"))
print("Books")
filenames
```
#### Combine the books into one string
```
corpus_raw = u""
for book_filename in filenames:
print("Reading '{0}'...".format(book_filename))
with codecs.open(book_filename, "r", "utf-8") as book_file:
corpus_raw += book_file.read()
print("Corpus is now {0} characters long".format(len(corpus_raw)))
print()
```
#### Split the corpus into sentences
```
tokenizer = nltk.data.load('tokenizers/punkt/english.pickle')
raw_sentences = tokenizer.tokenize(corpus_raw)
def sentence_to_wordlist(raw):
clean = re.sub("[^a-zA-Z]"," ", raw)
words = clean.split()
return words
sentences = []
for raw_sentence in raw_sentences:
if len(raw_sentence) > 0:
sentences.append(sentence_to_wordlist(raw_sentence))
print(raw_sentences[10])
print(sentence_to_wordlist(raw_sentences[10]))
token_count = sum([len(sentence) for sentence in sentences])
print("The book corpus contains {0} tokens.".format(token_count))
```
## Train Word2Vec
```
# Dimensionality of the resulting word vectors
num_features = 300
# Minimum word count threshold
min_word_count = 3
# Number of threads to run in parallel
num_workers = multiprocessing.cpu_count()
# Context window length
context_size = 7
# Downsample setting for frequent words
downsampling = 1e-3
# Seed
seed = 1
word2vec = w2v.Word2Vec(
sg=1,
seed=seed,
workers=num_workers,
vector_size=num_features,
min_count=min_word_count,
window=context_size,
sample=downsampling)
word2vec.build_vocab(sentences)
print("Word2Vec vocabulary length:", len(word2vec.wv))
word2vec.train(sentences,
total_examples=word2vec.corpus_count,
epochs=1)
```
## Compress the word vectors into 2D space and plot
```
tsne = sklearn.manifold.TSNE(n_components=2, random_state=0)
all_word_vectors_matrix = word2vec.syn1neg
all_word_vectors_matrix_2d = tsne.fit_transform(all_word_vectors_matrix)
```
#### Plot the big picture
```
points = pd.DataFrame(
[(word, coords[0], coords[1]) for word, coords in
[(word, all_word_vectors_matrix_2d[word2vec.wv.key_to_index[word]])for word in word2vec.wv.key_to_index]],
columns=["word", "x", "y"])
points.head(10)
points.plot.scatter("x", "y", s=10, figsize=(15, 10))
```
#### Words closest to the given word
```
word2vec.wv.most_similar("Stark")
word2vec.wv.most_similar("Aerys")
```
#### Linear relationships between word pairs
```
def nearest_similarity(start1, end1, end2):
similarities = word2vec.wv.most_similar_cosmul(
positive=[end2, start1],
negative=[end1])
start2 = similarities[0][0]
print("{start1} is related to {end1}, as {start2} is related to {end2}".format(**locals()))
return start2
nearest_similarity("Stark", "Winterfell", "Riverrun")
```
|
github_jupyter
|
import glob
import nltk
import codecs
import re
import multiprocessing
import gensim.models.word2vec as w2v
import sklearn.manifold
import pandas as pd
nltk.download("punkt")
nltk.download("stopwords")
filenames = sorted(glob.glob("data/*.txt"))
print("Books")
filenames
corpus_raw = u""
for book_filename in filenames:
print("Reading '{0}'...".format(book_filename))
with codecs.open(book_filename, "r", "utf-8") as book_file:
corpus_raw += book_file.read()
print("Corpus is now {0} characters long".format(len(corpus_raw)))
print()
tokenizer = nltk.data.load('tokenizers/punkt/english.pickle')
raw_sentences = tokenizer.tokenize(corpus_raw)
def sentence_to_wordlist(raw):
clean = re.sub("[^a-zA-Z]"," ", raw)
words = clean.split()
return words
sentences = []
for raw_sentence in raw_sentences:
if len(raw_sentence) > 0:
sentences.append(sentence_to_wordlist(raw_sentence))
print(raw_sentences[10])
print(sentence_to_wordlist(raw_sentences[10]))
token_count = sum([len(sentence) for sentence in sentences])
print("The book corpus contains {0} tokens.".format(token_count))
# Dimensionality of the resulting word vectors
num_features = 300
# Minimum word count threshold
min_word_count = 3
# Number of threads to run in parallel
num_workers = multiprocessing.cpu_count()
# Context window length
context_size = 7
# Downsample setting for frequent words
downsampling = 1e-3
# Seed
seed = 1
word2vec = w2v.Word2Vec(
sg=1,
seed=seed,
workers=num_workers,
vector_size=num_features,
min_count=min_word_count,
window=context_size,
sample=downsampling)
word2vec.build_vocab(sentences)
print("Word2Vec vocabulary length:", len(word2vec.wv))
word2vec.train(sentences,
total_examples=word2vec.corpus_count,
epochs=1)
tsne = sklearn.manifold.TSNE(n_components=2, random_state=0)
all_word_vectors_matrix = word2vec.syn1neg
all_word_vectors_matrix_2d = tsne.fit_transform(all_word_vectors_matrix)
points = pd.DataFrame(
[(word, coords[0], coords[1]) for word, coords in
[(word, all_word_vectors_matrix_2d[word2vec.wv.key_to_index[word]])for word in word2vec.wv.key_to_index]],
columns=["word", "x", "y"])
points.head(10)
points.plot.scatter("x", "y", s=10, figsize=(15, 10))
word2vec.wv.most_similar("Stark")
word2vec.wv.most_similar("Aerys")
def nearest_similarity(start1, end1, end2):
similarities = word2vec.wv.most_similar_cosmul(
positive=[end2, start1],
negative=[end1])
start2 = similarities[0][0]
print("{start1} is related to {end1}, as {start2} is related to {end2}".format(**locals()))
return start2
nearest_similarity("Stark", "Winterfell", "Riverrun")
| 0.419172 | 0.730674 |
# Spotify music recommendation
* https://dev.to/mxdws/using-python-with-the-spotify-api-1d02
* https://medium.com/analytics-vidhya/build-your-own-playlist-generator-with-spotifys-api-in-python-ceb883938ce4
* https://towardsdatascience.com/get-your-spotify-streaming-history-with-python-d5a208bbcbd3
* https://medium.com/python-in-plain-english/music-recommendation-system-for-djs-d253d472677e
* https://github.com/tgel0/spotify-data
* https://towardsdatascience.com/a-visual-look-at-my-taste-in-music-a8c197a728be
* https://towardsdatascience.com/how-to-utilize-spotifys-api-and-create-a-user-interface-in-streamlit-5d8820db95d5
* https://medium.com/deep-learning-turkey/build-your-own-spotify-playlist-of-best-playlist-recommendations-fc9ebe92826a
Before diving in, let's play some music:
[Lucy in the Sky with Diamonds](https://open.spotify.com/track/25yQPHgC35WNnnOUqFhgVR)
```
lucy_id = "25yQPHgC35WNnnOUqFhgVR"
url = "https://open.spotify.com/track/"+lucy_id
import webbrowser
webbrowser.open(url)
```
## Spotipy
https://spotipy.readthedocs.io/
```
from secret import *
os.environ["SPOTIPY_CLIENT_ID"] = clientId
os.environ["SPOTIPY_CLIENT_SECRET"] = clientSecret
os.environ["SPOTIPY_REDIRECT_URI"] = "https://open.spotify.com/"
import spotipy
from spotipy.oauth2 import SpotifyClientCredentials
auth_manager = SpotifyClientCredentials()
sp = spotipy.Spotify(auth_manager = auth_manager)
```
### Search track
```
results = sp.search(q='track:'+'Lucy in the Sky',type='track')
items = results['tracks']['items']
if len(items) > 0:
# tracks = items[0]
for tracks in items:
print(tracks['name'])
track_name = results['tracks']['items'][0]['name']
track_name
track_id = results['tracks']['items'][0]['id']
track_id
track_artist = results['tracks']['items'][0]['artists'][0]['name']
track_artist
track_artist_id = results['tracks']['items'][0]['artists'][0]['id']
track_artist_id
track_album = results['tracks']['items'][0]['album']['name']
track_album
track_album_id = results['tracks']['items'][0]['album']['id']
track_album_id
img_album = results['tracks']['items'][0]['album']['images'][1]['url']
import requests
r = requests.get(img_album)
open('img/'+track_id+'.jpg', 'wb').write(r.content)
from IPython.display import Image
Image(filename='img/'+track_id+'.jpg')
```
### Get Track by id

```
# Lucy in the Sky with Diamonds
lucy_id = '25yQPHgC35WNnnOUqFhgVR'
track = sp.track(lucy_id)
# track
print(track['name']+' - '+track['album']['name'])
```
### Get Features
```
track_features = sp.audio_features(lucy_id)
track_features
import pandas as pd
# df_features = spotifyAPI.parse_features(track_features)
df = pd.DataFrame(track_features, index=[0])
df_features = df.loc[: ,['acousticness', 'danceability', 'energy', 'instrumentalness', 'liveness', 'speechiness', 'valence']]
df_features
import spotifyAPI
spotifyAPI.feature_plot(df_features)
```
### Get recommendations
```
token = spotifyAPI.get_token(clientId,clientSecret)
json_response = spotifyAPI.get_track_reco(lucy_id,token)
uris =[]
for i in json_response['tracks']:
uris.append(i)
print(f"\"{i['name']}\" by {i['artists'][0]['name']}")
recolist = json_response['tracks']
recolist[0]['id']
recolist[1]['id']
recolist[0]['album']['images'][2]['url']
recolist[1]['album']['images'][2]['url']
reco = pd.DataFrame(recolist)
reco
```
### Artist albums
```
import pandas as pd
artists = pd.read_csv('spotify-artist-uris.csv', header=None, index_col=0, squeeze=True).to_dict()
mj_uri = artists['Michael Jackson']
results = sp.artist_albums(mj_uri, album_type='album')
albums = results['items']
while results['next']:
results = sp.next(results)
albums.extend(results['items'])
for album in albums:
print(album['name'])
url = "https://open.spotify.com/artist/"+mj_uri.replace('spotify:artist:','')
import webbrowser
webbrowser.open(url)
```
### Artist top tracks
```
import ipywidgets as widgets
artist = widgets.Text(value='Led Zeppelin')
artist
# how to get 30 second samples and cover art for the top 10 tracks for Led Zeppelin:
lz_uri = artists[artist.value]
results = sp.artist_top_tracks(lz_uri)
for track in results['tracks'][:10]:
print('track : ' + track['name'])
print('audio : ' + track['preview_url'])
print('cover art: ' + track['album']['images'][0]['url'])
print()
```
### [Advanced Search](https://spotipy.readthedocs.io/en/2.16.1/#spotipy.client.Spotify.search)
_search(q, limit=10, offset=0, type='track', market=None)_
**Parameters**:
* q - the search query (see how to write a query in the
official documentation https://developer.spotify.com/documentation/web-api/reference/search/search/) # noqa
* limit - the number of items to return (min = 1, default = 10, max = 50)
* offset - the index of the first item to return
* type - the type of item to return. One of ‘artist’, ‘album’,
‘track’, ‘playlist’, ‘show’, or ‘episode’
* market - An ISO 3166-1 alpha-2 country code or the string
from_token.
https://medium.com/@maxtingle/getting-started-with-spotifys-api-spotipy-197c3dc6353b
The following code collects 1,000 Track IDs and their associated track name, artist name, and popularity score.
(it does not require a Spotify ID)
```
artist_name = []
track_name = []
popularity = []
track_id = []
for i in range(0,1000,50):
track_results = sp.search(q='year:2020', type='track', limit=50,offset=i)
for i, t in enumerate(track_results['tracks']['items']):
artist_name.append(t['artists'][0]['name'])
track_name.append(t['name'])
track_id.append(t['id'])
popularity.append(t['popularity'])
```
### Current user
```
username = '1146603936'
user = sp.user(user = username)
user
url = user['images'][0]['url']
import requests
r = requests.get(url)
r.content
open('facebook.jpg', 'wb').write(r.content)
from IPython.display import Image
Image(filename='facebook.jpg')
```
## Access Scopes
* Images
* [ugc-image-upload](https://developer.spotify.com/documentation/general/guides/scopes/#ugc-image-upload)
* Spotify Connect
* [user-read-playback-state](https://developer.spotify.com/documentation/general/guides/scopes/#user-read-playback-state)
* [user-modify-playback-state](https://developer.spotify.com/documentation/general/guides/scopes/#user-modify-playback-state)
* [user-read-currently-playing](https://developer.spotify.com/documentation/general/guides/scopes/#user-read-currently-playing)
* Playback
* [streaming](https://developer.spotify.com/documentation/general/guides/scopes/#streaming)
* [app-remote-control](https://developer.spotify.com/documentation/general/guides/scopes/#app-remote-control)
* Users
* [user-read-email](https://developer.spotify.com/documentation/general/guides/scopes/#user-read-email)
* [user-read-private](https://developer.spotify.com/documentation/general/guides/scopes/#user-read-private)
* Playlists
* [playlist-read-collaborative](https://developer.spotify.com/documentation/general/guides/scopes/#playlist-read-collaborative)
* [playlist-modify-public](https://developer.spotify.com/documentation/general/guides/scopes/#playlist-modify-public)
* [playlist-read-private](https://developer.spotify.com/documentation/general/guides/scopes/#playlist-read-private)
* [playlist-modify-private](https://developer.spotify.com/documentation/general/guides/scopes/#playlist-modify-private)
* Library
* [user-library-modify](https://developer.spotify.com/documentation/general/guides/scopes/#user-library-modify)
* [user-library-read](https://developer.spotify.com/documentation/general/guides/scopes/#user-library-read)
* Listening History
* [user-top-read](https://developer.spotify.com/documentation/general/guides/scopes/#user-top-read)
* [user-read-playback-position](https://developer.spotify.com/documentation/general/guides/scopes/#user-read-playback-position)
* [user-read-recently-played](https://developer.spotify.com/documentation/general/guides/scopes/#user-read-recently-played)
* Follow
* [user-follow-read](https://developer.spotify.com/documentation/general/guides/scopes/#user-follow-read)
* [user-follow-modify](https://developer.spotify.com/documentation/general/guides/scopes/#user-follow-modify)
### Recently played
https://developer.spotify.com/console/get-recently-played/
```
scope = "user-read-recently-played"
auth_user = SpotifyOAuth(scope=scope, username=username)
sp = spotipy.Spotify(auth_manager=auth_user)
results = sp.current_user_recently_played(limit=50)
results['items']
for idx, item in enumerate(results['items']):
track = item['track']
print(idx, track['artists'][0]['name'], " – ", track['name'])
import json
with open("recently_played_20210306.json","w") as f:
json.dump(results,f,indent=4)
tracks = []
for idx, item in enumerate(results['items']):
track = item['track']
tracks.append([idx, track['artists'][0]['name'], track['name']])
tracks[0:5]
trackDict = {"id":[], "artist":[],"name":[]}
for idx, item in enumerate(results['items']):
track = item['track']
trackDict["id"].append(idx)
trackDict["artist"].append(track['artists'][0]['name'])
trackDict["name"].append(track['name'])
import pandas as pd
trackDf = pd.DataFrame.from_dict(trackDict)
trackDf
```
### Saved Tracks
```
import spotipy
from spotipy.oauth2 import SpotifyOAuth
scope = "user-library-read"
auth_user = SpotifyOAuth(scope=scope, username=username)
auth_user.get_cached_token()
from spotipy import util
token = util.prompt_for_user_token(username=username)
sp = spotipy.Spotify(auth_manager=auth_user)
results = sp.current_user_saved_tracks()
for idx, item in enumerate(results['items']):
track = item['track']
print(idx, track['artists'][0]['name'], " – ", track['name'])
import json
with open("saved_tracks_20210306.json","w") as f:
json.dump(results,f,indent=4)
with open("saved_tracks_20210306.json","r") as f:
results = json.load(f)
saved_tracks1 = results["items"][0]["track"]
saved_tracks1.keys()
import pandas as pd
df = pd.DataFrame.from_dict(results["items"])
df.head(5)
tracks = []
for idx, item in enumerate(results['items']):
track = item['track']
tracks.append([idx, track['artists'][0]['name'], track['name']])
tracks[0:5]
```
### Saved Albums
https://developer.spotify.com/console/get-current-user-saved-albums
```
results = sp.current_user_saved_albums(limit=20)
# results
# results['items'][0]
for idx, item in enumerate(results['items']):
album = item['album']
print(idx, album['artists'][0]['name'])
```
### Playlist
https://developer.spotify.com/console/get-current-user-playlists/
```
scope = "playlist-read-private"
auth_user = SpotifyOAuth(scope=scope, username=username)
sp = spotipy.Spotify(auth_manager=auth_user)
results = sp.current_user_playlists(limit=50)
results['items'][0]
for idx, item in enumerate(results['items']):
print(idx, item['name'], " – public: ", item['public'])
```
#### Add tracks to playlist
```
import pandas as pd
top100 = pd.read_csv('top100id.csv',index_col=0)
top100.head()
track_id = top100.id
track_id = track_id.dropna()
track_id
scope = 'playlist-modify-public'
auth_user = SpotifyOAuth(scope=scope, username=username)
sp = spotipy.Spotify(auth_manager=auth_user)
playlist_id = "2hNrDKdSNh889LLYEmR1DK"
sp.playlist_add_items(playlist_id,track_id)
### Get playlist by Id
import requests
playlistId = "2hNrDKdSNh889LLYEmR1DK" # Top 100 Play Music
playlistUrl = f"https://api.spotify.com/v1/playlists/{playlistId}"
headers = {
"Authorization": "Bearer " + token
}
res = requests.get(url=playlistUrl, headers=headers)
import json
with open('playlist.json', 'w') as outfile:
json.dump(res.json(), outfile, indent=2)
# print(json.dumps(res.json(), indent=2))
```
## Parse Streaming History
```
import ast
from typing import List
from os import listdir
files = 'spotifyData\StreamingHistory0.json'
all_streamings = []
with open(files, 'r', encoding='UTF-8') as f:
new_streamings = ast.literal_eval(f.read())
all_streamings += [streaming for streaming
in new_streamings]
all_streamings[0]
unique_tracks = list(set([streaming['trackName']
for streaming in all_streamings]))
unique_tracks[0]
import spotifyAPI
all_features = {}
for track in unique_tracks:
track_id = spotifyAPI.get_track_id(track, token)
features = get_features(track_id, token)
if features:
all_features[track] = features
with_features = []
for track_name, features in all_features.items():
with_features.append({'name': track_name, **features})
import pandas as pd
df = pd.DataFrame(with_features)
df.to_csv('streaming_history.csv')
df
```
## Music Taste Analysis
* https://towardsdatascience.com/a-music-taste-analysis-using-spotify-api-and-python-e52d186db5fc
* https://github.com/jmcabreira/A-Music-Taste-Analysis-Using-Spotify-API-and-Python.
```
features = df.loc[: ,['acousticness', 'danceability', 'energy', 'instrumentalness', 'liveness', 'speechiness', 'valence']]
features
#Import Libraries
import numpy as np
import matplotlib.pyplot as plt
labels= list(features)[:]
stats= features.mean().tolist()
angles=np.linspace(0, 2*np.pi, len(labels), endpoint=False)
# close the plot
stats=np.concatenate((stats,[stats[0]]))
angles=np.concatenate((angles,[angles[0]]))
#Size of the figure
fig=plt.figure(figsize = (18,18))
ax = fig.add_subplot(221, polar=True)
ax.plot(angles, stats, 'o-', linewidth=2, label = "Features", color= 'gray')
ax.fill(angles, stats, alpha=0.25, facecolor='gray')
ax.set_thetagrids(angles[0:7] * 180/np.pi, labels , fontsize = 13)
ax.set_rlabel_position(250)
plt.yticks([0.2 , 0.4 , 0.6 , 0.8 ], ["0.2",'0.4', "0.6", "0.8"], color="grey", size=12)
plt.ylim(0,1)
plt.legend(loc='best', bbox_to_anchor=(0.1, 0.1))
```
|
github_jupyter
|
lucy_id = "25yQPHgC35WNnnOUqFhgVR"
url = "https://open.spotify.com/track/"+lucy_id
import webbrowser
webbrowser.open(url)
from secret import *
os.environ["SPOTIPY_CLIENT_ID"] = clientId
os.environ["SPOTIPY_CLIENT_SECRET"] = clientSecret
os.environ["SPOTIPY_REDIRECT_URI"] = "https://open.spotify.com/"
import spotipy
from spotipy.oauth2 import SpotifyClientCredentials
auth_manager = SpotifyClientCredentials()
sp = spotipy.Spotify(auth_manager = auth_manager)
results = sp.search(q='track:'+'Lucy in the Sky',type='track')
items = results['tracks']['items']
if len(items) > 0:
# tracks = items[0]
for tracks in items:
print(tracks['name'])
track_name = results['tracks']['items'][0]['name']
track_name
track_id = results['tracks']['items'][0]['id']
track_id
track_artist = results['tracks']['items'][0]['artists'][0]['name']
track_artist
track_artist_id = results['tracks']['items'][0]['artists'][0]['id']
track_artist_id
track_album = results['tracks']['items'][0]['album']['name']
track_album
track_album_id = results['tracks']['items'][0]['album']['id']
track_album_id
img_album = results['tracks']['items'][0]['album']['images'][1]['url']
import requests
r = requests.get(img_album)
open('img/'+track_id+'.jpg', 'wb').write(r.content)
from IPython.display import Image
Image(filename='img/'+track_id+'.jpg')
# Lucy in the Sky with Diamonds
lucy_id = '25yQPHgC35WNnnOUqFhgVR'
track = sp.track(lucy_id)
# track
print(track['name']+' - '+track['album']['name'])
track_features = sp.audio_features(lucy_id)
track_features
import pandas as pd
# df_features = spotifyAPI.parse_features(track_features)
df = pd.DataFrame(track_features, index=[0])
df_features = df.loc[: ,['acousticness', 'danceability', 'energy', 'instrumentalness', 'liveness', 'speechiness', 'valence']]
df_features
import spotifyAPI
spotifyAPI.feature_plot(df_features)
token = spotifyAPI.get_token(clientId,clientSecret)
json_response = spotifyAPI.get_track_reco(lucy_id,token)
uris =[]
for i in json_response['tracks']:
uris.append(i)
print(f"\"{i['name']}\" by {i['artists'][0]['name']}")
recolist = json_response['tracks']
recolist[0]['id']
recolist[1]['id']
recolist[0]['album']['images'][2]['url']
recolist[1]['album']['images'][2]['url']
reco = pd.DataFrame(recolist)
reco
import pandas as pd
artists = pd.read_csv('spotify-artist-uris.csv', header=None, index_col=0, squeeze=True).to_dict()
mj_uri = artists['Michael Jackson']
results = sp.artist_albums(mj_uri, album_type='album')
albums = results['items']
while results['next']:
results = sp.next(results)
albums.extend(results['items'])
for album in albums:
print(album['name'])
url = "https://open.spotify.com/artist/"+mj_uri.replace('spotify:artist:','')
import webbrowser
webbrowser.open(url)
import ipywidgets as widgets
artist = widgets.Text(value='Led Zeppelin')
artist
# how to get 30 second samples and cover art for the top 10 tracks for Led Zeppelin:
lz_uri = artists[artist.value]
results = sp.artist_top_tracks(lz_uri)
for track in results['tracks'][:10]:
print('track : ' + track['name'])
print('audio : ' + track['preview_url'])
print('cover art: ' + track['album']['images'][0]['url'])
print()
artist_name = []
track_name = []
popularity = []
track_id = []
for i in range(0,1000,50):
track_results = sp.search(q='year:2020', type='track', limit=50,offset=i)
for i, t in enumerate(track_results['tracks']['items']):
artist_name.append(t['artists'][0]['name'])
track_name.append(t['name'])
track_id.append(t['id'])
popularity.append(t['popularity'])
username = '1146603936'
user = sp.user(user = username)
user
url = user['images'][0]['url']
import requests
r = requests.get(url)
r.content
open('facebook.jpg', 'wb').write(r.content)
from IPython.display import Image
Image(filename='facebook.jpg')
scope = "user-read-recently-played"
auth_user = SpotifyOAuth(scope=scope, username=username)
sp = spotipy.Spotify(auth_manager=auth_user)
results = sp.current_user_recently_played(limit=50)
results['items']
for idx, item in enumerate(results['items']):
track = item['track']
print(idx, track['artists'][0]['name'], " – ", track['name'])
import json
with open("recently_played_20210306.json","w") as f:
json.dump(results,f,indent=4)
tracks = []
for idx, item in enumerate(results['items']):
track = item['track']
tracks.append([idx, track['artists'][0]['name'], track['name']])
tracks[0:5]
trackDict = {"id":[], "artist":[],"name":[]}
for idx, item in enumerate(results['items']):
track = item['track']
trackDict["id"].append(idx)
trackDict["artist"].append(track['artists'][0]['name'])
trackDict["name"].append(track['name'])
import pandas as pd
trackDf = pd.DataFrame.from_dict(trackDict)
trackDf
import spotipy
from spotipy.oauth2 import SpotifyOAuth
scope = "user-library-read"
auth_user = SpotifyOAuth(scope=scope, username=username)
auth_user.get_cached_token()
from spotipy import util
token = util.prompt_for_user_token(username=username)
sp = spotipy.Spotify(auth_manager=auth_user)
results = sp.current_user_saved_tracks()
for idx, item in enumerate(results['items']):
track = item['track']
print(idx, track['artists'][0]['name'], " – ", track['name'])
import json
with open("saved_tracks_20210306.json","w") as f:
json.dump(results,f,indent=4)
with open("saved_tracks_20210306.json","r") as f:
results = json.load(f)
saved_tracks1 = results["items"][0]["track"]
saved_tracks1.keys()
import pandas as pd
df = pd.DataFrame.from_dict(results["items"])
df.head(5)
tracks = []
for idx, item in enumerate(results['items']):
track = item['track']
tracks.append([idx, track['artists'][0]['name'], track['name']])
tracks[0:5]
results = sp.current_user_saved_albums(limit=20)
# results
# results['items'][0]
for idx, item in enumerate(results['items']):
album = item['album']
print(idx, album['artists'][0]['name'])
scope = "playlist-read-private"
auth_user = SpotifyOAuth(scope=scope, username=username)
sp = spotipy.Spotify(auth_manager=auth_user)
results = sp.current_user_playlists(limit=50)
results['items'][0]
for idx, item in enumerate(results['items']):
print(idx, item['name'], " – public: ", item['public'])
import pandas as pd
top100 = pd.read_csv('top100id.csv',index_col=0)
top100.head()
track_id = top100.id
track_id = track_id.dropna()
track_id
scope = 'playlist-modify-public'
auth_user = SpotifyOAuth(scope=scope, username=username)
sp = spotipy.Spotify(auth_manager=auth_user)
playlist_id = "2hNrDKdSNh889LLYEmR1DK"
sp.playlist_add_items(playlist_id,track_id)
### Get playlist by Id
import requests
playlistId = "2hNrDKdSNh889LLYEmR1DK" # Top 100 Play Music
playlistUrl = f"https://api.spotify.com/v1/playlists/{playlistId}"
headers = {
"Authorization": "Bearer " + token
}
res = requests.get(url=playlistUrl, headers=headers)
import json
with open('playlist.json', 'w') as outfile:
json.dump(res.json(), outfile, indent=2)
# print(json.dumps(res.json(), indent=2))
import ast
from typing import List
from os import listdir
files = 'spotifyData\StreamingHistory0.json'
all_streamings = []
with open(files, 'r', encoding='UTF-8') as f:
new_streamings = ast.literal_eval(f.read())
all_streamings += [streaming for streaming
in new_streamings]
all_streamings[0]
unique_tracks = list(set([streaming['trackName']
for streaming in all_streamings]))
unique_tracks[0]
import spotifyAPI
all_features = {}
for track in unique_tracks:
track_id = spotifyAPI.get_track_id(track, token)
features = get_features(track_id, token)
if features:
all_features[track] = features
with_features = []
for track_name, features in all_features.items():
with_features.append({'name': track_name, **features})
import pandas as pd
df = pd.DataFrame(with_features)
df.to_csv('streaming_history.csv')
df
features = df.loc[: ,['acousticness', 'danceability', 'energy', 'instrumentalness', 'liveness', 'speechiness', 'valence']]
features
#Import Libraries
import numpy as np
import matplotlib.pyplot as plt
labels= list(features)[:]
stats= features.mean().tolist()
angles=np.linspace(0, 2*np.pi, len(labels), endpoint=False)
# close the plot
stats=np.concatenate((stats,[stats[0]]))
angles=np.concatenate((angles,[angles[0]]))
#Size of the figure
fig=plt.figure(figsize = (18,18))
ax = fig.add_subplot(221, polar=True)
ax.plot(angles, stats, 'o-', linewidth=2, label = "Features", color= 'gray')
ax.fill(angles, stats, alpha=0.25, facecolor='gray')
ax.set_thetagrids(angles[0:7] * 180/np.pi, labels , fontsize = 13)
ax.set_rlabel_position(250)
plt.yticks([0.2 , 0.4 , 0.6 , 0.8 ], ["0.2",'0.4', "0.6", "0.8"], color="grey", size=12)
plt.ylim(0,1)
plt.legend(loc='best', bbox_to_anchor=(0.1, 0.1))
| 0.123432 | 0.673883 |
# Simulating Molecules using VQE
In this tutorial, we introduce the Variational Quantum Eigensolver (VQE), motivate its use, explain the necessary theory, and demonstrate its implementation in finding the ground state energy of molecules.
## Contents
1. [Introduction](#introduction)
2. [The Variational Method of Quantum Mechanics](#varmethod)
1. [Mathematical Background](#backgroundmath)
2. [Bounding the Ground State](#groundstate)
3. [The Variational Quantum Eigensolver](#vqe)
1. [Variational Forms](#varforms)
2. [Simple Variational Forms](#simplevarform)
3. [Parameter Optimization](#optimization)
4. [Example with a Single Qubit Variational Form](#example)
5. [Structure of Common Variational Forms](#commonvarforms)
4. [VQE Implementation in Qiskit](#implementation)
1. [Running VQE on a Statevector Simulator](#implementationstatevec)
2. [Running VQE on a Noisy Simulator](#implementationnoisy)
5. [Problems](#problems)
6. [References](#references)
## Introduction<a id='introduction'></a>
In many applications it is important to find the minimum eigenvalue of a matrix. For example, in chemistry, the minimum eigenvalue of a Hermitian matrix characterizing the molecule is the ground state energy of that system. In the future, the quantum phase estimation algorithm may be used to find the minimum eigenvalue. However, its implementation on useful problems requires circuit depths exceeding the limits of hardware available in the NISQ era. Thus, in 2014, Peruzzo *et al.* proposed VQE to estimate the ground state energy of a molecule using much shallower circuits [1].
Formally stated, given a Hermitian matrix $H$ with an unknown minimum eigenvalue $\lambda_{min}$, associated with the eigenstate $|\psi_{min}\rangle$, VQE provides an estimate $\lambda_{\theta}$ bounding $\lambda_{min}$:
\begin{align*}
\lambda_{min} \le \lambda_{\theta} \equiv \langle \psi(\theta) |H|\psi(\theta) \rangle
\end{align*}
where $|\psi(\theta)\rangle$ is the eigenstate associated with $\lambda_{\theta}$. By applying a parameterized circuit, represented by $U(\theta)$, to some arbitrary starting state $|\psi\rangle$, the algorithm obtains an estimate $U(\theta)|\psi\rangle \equiv |\psi(\theta)\rangle$ on $|\psi_{min}\rangle$. The estimate is iteratively optimized by a classical controller changing the parameter $\theta$ minimizing the expectation value of $\langle \psi(\theta) |H|\psi(\theta) \rangle$.
## The Variational Method of Quantum Mechanics<a id='varmethod'></a>
### Mathematical Background<a id='backgroundmath'></a>
VQE is an application of the variational method of quantum mechanics. To better understand the variational method, some preliminary mathematical background is provided. An eigenvector, $|\psi_i\rangle$, of a matrix $A$ is invariant under transformation by $A$ up to a scalar multiplicative constant (the eigenvalue $\lambda_i$). That is,
\begin{align*}
A |\psi_i\rangle = \lambda_i |\psi_i\rangle
\end{align*}
Furthermore, a matrix $H$ is Hermitian when it is equal to its own conjugate transpose.
\begin{align*}
H = H^{\dagger}
\end{align*}
The spectral theorem states that the eigenvalues of a Hermitian matrix must be real. Thus, any eigenvalue of $H$ has the property that $\lambda_i = \lambda_i^*$. As any measurable quantity must be real, Hermitian matrices are suitable for describing the Hamiltonians of quantum systems. Moreover, $H$ may be expressed as
\begin{align*}
H = \sum_{i = 1}^{N} \lambda_i |\psi_i\rangle \langle \psi_i |
\end{align*}
where each $\lambda_i$ is the eigenvalue corresponding to the eigenvector $|\psi_i\rangle$. Furthermore, the expectation value of the observable $H$ on an arbitrary quantum state $|\psi\rangle$ is given by
\begin{align}
\langle H \rangle_{\psi} &\equiv \langle \psi | H | \psi \rangle
\end{align}
Substituting $H$ with its representation as a weighted sum of its eigenvectors,
\begin{align}
\langle H \rangle_{\psi} = \langle \psi | H | \psi \rangle &= \langle \psi | \left(\sum_{i = 1}^{N} \lambda_i |\psi_i\rangle \langle \psi_i |\right) |\psi\rangle\\
&= \sum_{i = 1}^{N} \lambda_i \langle \psi | \psi_i\rangle \langle \psi_i | \psi\rangle \\
&= \sum_{i = 1}^{N} \lambda_i | \langle \psi_i | \psi\rangle |^2
\end{align}
The last equation demonstrates that the expectation value of an observable on any state can be expressed as a linear combination using the eigenvalues associated with $H$ as the weights. Moreover, each of the weights in the linear combination is greater than or equal to 0, as $| \langle \psi_i | \psi\rangle |^2 \ge 0$ and so it is clear that
\begin{align}
\lambda_{min} \le \langle H \rangle_{\psi} = \langle \psi | H | \psi \rangle = \sum_{i = 1}^{N} \lambda_i | \langle \psi_i | \psi\rangle |^2
\end{align}
The above equation is known as the **variational method** (in some texts it is also known as the variational principle) [2]. It is important to note that this implies that the expectation value of any wave function will always be at least the minimum eigenvalue associated with $H$. Moreover, the expectation value of state $|\psi_{min}\rangle$ is given by $\langle \psi_{min}|H|\psi_{min}\rangle = \langle \psi_{min}|\lambda_{min}|\psi_{min}\rangle = \lambda_{min}$. Thus, as expected, $\langle H \rangle_{\psi_{min}}=\lambda_{min}$.
### Bounding the Ground State<a id='groundstate'></a>
When the Hamiltonian of a system is described by the Hermitian matrix $H$ the ground state energy of that system, $E_{gs}$, is the smallest eigenvalue associated with $H$. By arbitrarily selecting a wave function $|\psi \rangle$ (called an *ansatz*) as an initial guess approximating $|\psi_{min}\rangle$, calculating its expectation value, $\langle H \rangle_{\psi}$, and iteratively updating the wave function, arbitrarily tight bounds on the ground state energy of a Hamiltonian may be obtained.
## The Variational Quantum Eigensolver<a id='vqe'></a>
### Variational Forms<a id='varforms'></a>
A systematic approach to varying the ansatz is required to implement the variational method on a quantum computer. VQE does so through the use of a parameterized circuit with a fixed form. Such a circuit is often called a *variational form*, and its action may be represented by the linear transformation $U(\theta)$. A variational form is applied to a starting state $|\psi\rangle$ (such as the vacuum state $|0\rangle$, or the Hartree Fock state) and generates an output state $U(\theta)|\psi\rangle\equiv |\psi(\theta)\rangle$. Iterative optimization over $|\psi(\theta)\rangle$ aims to yield an expectation value $\langle \psi(\theta)|H|\psi(\theta)\rangle \approx E_{gs} \equiv \lambda_{min}$. Ideally, $|\psi(\theta)\rangle$ will be close to $|\psi_{min}\rangle$ (where 'closeness' is characterized by either state fidelity, or Manhattan distance) although in practice, useful bounds on $E_{gs}$ can be obtained even if this is not the case.
Moreover, a fixed variational form with a polynomial number of parameters can only generate transformations to a polynomially sized subspace of all the states in an exponentially sized Hilbert space. Consequently, various variational forms exist. Some, such as Ry and RyRz are heuristically designed, without consideration of the target domain. Others, such as UCCSD, utilize domain specific knowledge to generate close approximations based on the problem's structure. The structure of common variational forms is discussed in greater depth later in this document.
### Simple Variational Forms<a id='simplevarform'></a>
When constructing a variational form we must balance two opposing goals. Ideally, our $n$ qubit variational form would be able to generate any possible state $|\psi\rangle$ where $|\psi\rangle \in \mathbb{C}^N$ and $N=2^n$. However, we would like the variational form to use as few parameters as possible. Here, we aim to give intuition for the construction of variational forms satisfying our first goal, while disregarding the second goal for the sake of simplicity.
Consider the case where $n=1$. The U3 gate takes three parameters, $\theta, \phi$ and $\lambda$, and represents the following transformation:
\begin{align}
U3(\theta, \phi, \lambda) = \begin{pmatrix}\cos(\frac{\theta}{2}) & -e^{i\lambda}\sin(\frac{\theta}{2}) \\ e^{i\phi}\sin(\frac{\theta}{2}) & e^{i\lambda + i\phi}\cos(\frac{\theta}{2}) \end{pmatrix}
\end{align}
Up to a global phase, any possible single qubit transformation may be implemented by appropriately setting these parameters. Consequently, for the single qubit case, a variational form capable of generating any possible state is given by the circuit:
<img src="./images/U3_var_form.png"
alt="U3 Variational Form"
width="350"/>
Moreover, this universal 'variational form' only has 3 parameters and thus can be efficiently optimized. It is worth emphasising that the ability to generate an arbitrary state ensures that during the optimization process, the variational form does not limit the set of attainable states over which the expectation value of $H$ can be taken. Ideally, this ensures that the minimum expectation value is limited only by the capabilities of the classical optimizer.
A less trivial universal variational form may be derived for the 2 qubit case, where two body interactions, and thus entanglement, must be considered to achieve universality. Based on the work presented by *Shende et al.* [3] the following is an example of a universal parameterized 2 qubit circuit:
<img src="./images/two_qubit_var_form.png"
alt="Two Qubit Variational Form"
width="800"/>
Allow the transformation performed by the above circuit to be represented by $U(\theta)$. When optimized variationally, the expectation value of $H$ is minimized when $U(\theta)|\psi\rangle \equiv |\psi(\theta)\rangle \approx |\psi_{min}\rangle$. By formulation, $U(\theta)$ may produce a transformation to any possible state, and so this variational form may obtain an arbitrarily tight bound on two qubit ground state energies, only limited by the capabilities of the classical optimizer.
### Parameter Optimization<a id='optimization'></a>
Once an efficiently parameterized variational form has been selected, in accordance with the variational method, its parameters must be optimized to minimize the expectation value of the target Hamiltonian. The parameter optimization process has various challenges. For example, quantum hardware has various types of noise and so objective function evaluation (energy calculation) may not necessarily reflect the true objective function. Additionally, some optimizers perform a number of objective function evaluations dependent on cardinality of the parameter set. An appropriate optimizer should be selected by considering the requirements of a application.
A popular optimization strategy is gradient decent where each parameter is updated in the direction yielding the largest local change in energy. Consequently, the number of evaluations performed depends on the number of optimization parameters present. This allows the algorithm to quickly find a local optimum in the search space. However, this optimization strategy often gets stuck at poor local optima, and is relatively expensive in terms of the number of circuit evaluations performed. While an intuitive optimization strategy, it is not recommended for use in VQE.
An appropriate optimizer for optimizing a noisy objective function is the *Simultaneous Perturbation Stochastic Approximation* optimizer (SPSA). SPSA approximates the gradient of the objective function with only two measurements. It does so by concurrently perturbing all of the parameters in a random fashion, in contrast to gradient decent where each parameter is perturbed independently. When utilizing VQE in either a noisy simulator or on real hardware, SPSA is a recommended as the classical optimizer.
When noise is not present in the cost function evaluation (such as when using VQE with a statevector simulator), a wide variety of classical optimizers may be useful. Two such optimizers supported by Qiskit Aqua are the *Sequential Least Squares Programming* optimizer (SLSQP) and the *Constrained Optimization by Linear Approximation* optimizer (COBYLA). It is worth noting that COBYLA only performs one objective function evaluation per optimization iteration (and thus the number of evaluations is independent of the parameter set's cardinality). Therefore, if the objective function is noise-free and minimizing the number of performed evaluations is desirable, it is recommended to try COBYLA.
### Example with a Single Qubit Variational Form<a id='example'></a>
We will now use the simple single qubit variational form to solve a problem similar to ground state energy estimation. Specifically, we are given a random probability vector $\vec{x}$ and wish to determine a possible parameterization for our single qubit variational form such that it outputs a probability distribution that is close to $\vec{x}$ (where closeness is defined in terms of the Manhattan distance between the two probability vectors).
We first create the random probability vector in python:
```
import numpy as np
np.random.seed(999999)
target_distr = np.random.rand(2)
# We now convert the random vector into a valid probability vector
target_distr /= sum(target_distr)
```
We subsequently create a function that takes the parameters of our single U3 variational form as arguments and returns the corresponding quantum circuit:
```
from qiskit import QuantumCircuit, ClassicalRegister, QuantumRegister
def get_var_form(params):
qr = QuantumRegister(1, name="q")
cr = ClassicalRegister(1, name='c')
qc = QuantumCircuit(qr, cr)
qc.u3(params[0], params[1], params[2], qr[0])
qc.measure(qr, cr[0])
return qc
```
Now we specify the objective function which takes as input a list of the variational form's parameters, and returns the cost associated with those parameters:
```
from qiskit import Aer, execute
backend = Aer.get_backend("qasm_simulator")
NUM_SHOTS = 10000
def get_probability_distribution(counts):
output_distr = [v / NUM_SHOTS for v in counts.values()]
if len(output_distr) == 1:
output_distr.append(0)
return output_distr
def objective_function(params):
# Obtain a quantum circuit instance from the paramters
qc = get_var_form(params)
# Execute the quantum circuit to obtain the probability distribution associated with the current parameters
result = execute(qc, backend, shots=NUM_SHOTS).result()
# Obtain the counts for each measured state, and convert those counts into a probability vector
output_distr = get_probability_distribution(result.get_counts(qc))
# Calculate the cost as the distance between the output distribution and the target distribution
cost = sum([np.abs(output_distr[i] - target_distr[i]) for i in range(2)])
return cost
```
Finally, we create an instance of the COBYLA optimizer, and run the algorithm. Note that the output varies from run to run. Moreover, while close, the obtained distribution might not be exactly the same as the target distribution, however, increasing the number of shots taken will increase the accuracy of the output.
```
from qiskit.aqua.components.optimizers import COBYLA
# Initialize the COBYLA optimizer
optimizer = COBYLA(maxiter=500, tol=0.0001)
# Create the initial parameters (noting that our single qubit variational form has 3 parameters)
params = np.random.rand(3)
ret = optimizer.optimize(num_vars=3, objective_function=objective_function, initial_point=params)
# Obtain the output distribution using the final parameters
qc = get_var_form(ret[0])
counts = execute(qc, backend, shots=NUM_SHOTS).result().get_counts(qc)
output_distr = get_probability_distribution(counts)
print("Target Distribution:", target_distr)
print("Obtained Distribution:", output_distr)
print("Output Error (Manhattan Distance):", ret[1])
print("Parameters Found:", ret[0])
```
### Structure of Common Variational Forms<a id='commonvarforms'></a>
As already discussed, it is not possible for a polynomially parameterized variational form to generate a transformation to any state. Variational forms can be grouped into two categories, depending on how they deal with this limitation. The first category of variational forms use domain or application specific knowledge to limit the set of possible output states. The second approach uses a heuristic circuit without prior domain or application specific knowledge.
The first category of variational forms exploit characteristics of the problem domain to restrict the set of transformations that may be required. For example, when calculating the ground state energy of a molecule, the number of particles in the system is known *a priori*. Therefore, if a starting state with the correct number of particles is used, by limiting the variational form to only producing particle preserving transformations, the number of parameters required to span the new transformation subspace can be greatly reduced. Indeed, by utilizing similar information from Coupled-Cluster theory, the variational form UCCSD can obtain very accurate results for molecular ground state energy estimation when starting from the Hartree Fock state. Another example illustrating the exploitation of domain-specific knowledge follows from considering the set of circuits realizable on real quantum hardware. Extant quantum computers, such as those based on super conducting qubits, have limited qubit connectivity. That is, it is not possible to implement 2-qubit gates on arbitrary qubit pairs (without inserting swap gates). Thus, variational forms have been constructed for specific quantum computer architectures where the circuits are specifically tuned to maximally exploit the natively available connectivity and gates of a given quantum device. Such a variational form was used in 2017 to successfully implement VQE for the estimation of the ground state energies of molecules as large as BeH$_2$ on an IBM quantum computer [4].
In the second approach, gates are layered such that good approximations on a wide range of states may be obtained. Qiskit Aqua supports three such variational forms: RyRz, Ry and SwapRz (we will only discuss the first two). All of these variational forms accept multiple user-specified configurations. Three essential configurations are the number of qubits in the system, the depth setting, and the entanglement setting. A single layer of a variational form specifies a certain pattern of single qubit rotations and CX gates. The depth setting says how many times the variational form should repeat this pattern. By increasing the depth setting, at the cost of increasing the number of parameters that must be optimized, the set of states the variational form can generate increases. Finally, the entanglement setting selects the configuration, and implicitly the number, of CX gates. For example, when the entanglement setting is linear, CX gates are applied to adjacent qubit pairs in order (and thus $n-1$ CX gates are added per layer). When the entanglement setting is full, a CX gate is applied to each qubit pair in each layer. The circuits for RyRz corresponding to `entanglement="full"` and `entanglement="linear"` can be seen by executing the following code snippet:
```
from qiskit.aqua.components.variational_forms import RYRZ
entanglements = ["linear", "full"]
for entanglement in entanglements:
form = RYRZ(num_qubits=4, depth=1, entanglement=entanglement)
if entanglement == "linear":
print("=============Linear Entanglement:=============")
else:
print("=============Full Entanglement:=============")
# We initialize all parameters to 0 for this demonstration
print(form.construct_circuit([0] * form.num_parameters).draw(fold=100))
print()
```
Assume the depth setting is set to $d$. Then, RyRz has $n\times (d+1)\times 2$ parameters, Ry with linear entanglement has $2n\times(d + \frac{1}{2})$ parameters, and Ry with full entanglement has $d\times n\times \frac{(n + 1)}{2} + n$ parameters.
## VQE Implementation in Qiskit<a id='implementation'></a>
This section illustrates an implementation of VQE using the programmatic approach. Qiskit Aqua also enables a declarative implementation, however, it reveals less information about the underlying algorithm. This code, specifically the preparation of qubit operators, is based on the code found in the Qiskit Tutorials repository (and as of July 2019, may be found at: https://github.com/Qiskit/qiskit-tutorials ).
The following libraries must first be imported.
```
from qiskit.aqua.algorithms import VQE, ExactEigensolver
import matplotlib.pyplot as plt
%matplotlib inline
import numpy as np
from qiskit.chemistry.components.variational_forms import UCCSD
from qiskit.chemistry.components.initial_states import HartreeFock
from qiskit.aqua.components.variational_forms import RYRZ
from qiskit.aqua.components.optimizers import COBYLA, SPSA, SLSQP
from qiskit.aqua.operators import Z2Symmetries
from qiskit import IBMQ, BasicAer, Aer
from qiskit.chemistry.drivers import PySCFDriver, UnitsType
from qiskit.chemistry import FermionicOperator
from qiskit import IBMQ
from qiskit.providers.aer import noise
from qiskit.aqua import QuantumInstance
from qiskit.ignis.mitigation.measurement import CompleteMeasFitter
```
### Running VQE on a Statevector Simulator<a id='implementationstatevec'></a>
We demonstrate the calculation of the ground state energy for LiH at various interatomic distances. A driver for the molecule must be created at each such distance. Note that in this experiment, to reduce the number of qubits used, we freeze the core and remove two unoccupied orbitals. First, we define a function that takes an interatomic distance and returns the appropriate qubit operator, $H$, as well as some other information about the operator.
```
def get_qubit_op(dist):
driver = PySCFDriver(atom="Li .0 .0 .0; H .0 .0 " + str(dist), unit=UnitsType.ANGSTROM,
charge=0, spin=0, basis='sto3g')
molecule = driver.run()
freeze_list = [0]
remove_list = [-3, -2]
repulsion_energy = molecule.nuclear_repulsion_energy
num_particles = molecule.num_alpha + molecule.num_beta
num_spin_orbitals = molecule.num_orbitals * 2
remove_list = [x % molecule.num_orbitals for x in remove_list]
freeze_list = [x % molecule.num_orbitals for x in freeze_list]
remove_list = [x - len(freeze_list) for x in remove_list]
remove_list += [x + molecule.num_orbitals - len(freeze_list) for x in remove_list]
freeze_list += [x + molecule.num_orbitals for x in freeze_list]
ferOp = FermionicOperator(h1=molecule.one_body_integrals, h2=molecule.two_body_integrals)
ferOp, energy_shift = ferOp.fermion_mode_freezing(freeze_list)
num_spin_orbitals -= len(freeze_list)
num_particles -= len(freeze_list)
ferOp = ferOp.fermion_mode_elimination(remove_list)
num_spin_orbitals -= len(remove_list)
qubitOp = ferOp.mapping(map_type='parity', threshold=0.00000001)
qubitOp = Z2Symmetries.two_qubit_reduction(qubitOp, num_particles)
shift = energy_shift + repulsion_energy
return qubitOp, num_particles, num_spin_orbitals, shift
```
First, the exact ground state energy is calculated using the qubit operator and a classical exact eigensolver. Subsequently, the initial state $|\psi\rangle$ is created, which the VQE instance uses to produce the final ansatz $\min_{\theta}(|\psi(\theta)\rangle)$. The exact result and the VQE result at each interatomic distance is stored. Observe that the result given by `vqe.run(backend)['energy'] + shift` is equivalent the quantity $\min_{\theta}\left(\langle \psi(\theta)|H|\psi(\theta)\rangle\right)$, where the minimum is not necessarily the global minimum.
When initializing the VQE instance with `VQE(qubitOp, var_form, optimizer, 'matrix')` the expectation value of $H$ on $|\psi(\theta)\rangle$ is directly calculated through matrix multiplication. However, when using an actual quantum device, or a true simulator such as the `qasm_simulator` with `VQE(qubitOp, var_form, optimizer, 'paulis')` the calculation of the expectation value is more complicated. A Hamiltonian may be represented as a sum of a Pauli strings, with each Pauli term acting on a qubit as specified by the mapping being used. Each Pauli string has a corresponding circuit appended to the circuit corresponding to $|\psi(\theta)\rangle$. Subsequently, each of these circuits is executed, and all of the results are used to determine the expectation value of $H$ on $|\psi(\theta)\rangle$. In the following example, we initialize the VQE instance with `matrix` mode, and so the expectation value is directly calculated through matrix multiplication.
Note that the following code snippet may take a few minutes to run to completion.
```
backend = BasicAer.get_backend("statevector_simulator")
distances = np.arange(0.5, 4.0, 0.1)
exact_energies = []
vqe_energies = []
optimizer = SLSQP(maxiter=5)
for dist in distances:
qubitOp, num_particles, num_spin_orbitals, shift = get_qubit_op(dist)
result = ExactEigensolver(qubitOp).run()
exact_energies.append(result['energy'] + shift)
initial_state = HartreeFock(
qubitOp.num_qubits,
num_spin_orbitals,
num_particles,
'parity'
)
var_form = UCCSD(
qubitOp.num_qubits,
depth=1,
num_orbitals=num_spin_orbitals,
num_particles=num_particles,
initial_state=initial_state,
qubit_mapping='parity'
)
vqe = VQE(qubitOp, var_form, optimizer)
results = vqe.run(backend)['energy'] + shift
vqe_energies.append(results)
print("Interatomic Distance:", np.round(dist, 2), "VQE Result:", results, "Exact Energy:", exact_energies[-1])
print("All energies have been calculated")
plt.plot(distances, exact_energies, label="Exact Energy")
plt.plot(distances, vqe_energies, label="VQE Energy")
plt.xlabel('Atomic distance (Angstrom)')
plt.ylabel('Energy')
plt.legend()
plt.show()
```
Note that the VQE results are very close to the exact results, and so the exact energy curve is hidden by the VQE curve.
### Running VQE on a Noisy Simulator<a id='implementationnoisy'></a>
Here, we calculate the ground state energy for H$_2$ using a noisy simulator and error mitigation.
First, we prepare the qubit operator representing the molecule's Hamiltonian:
```
driver = PySCFDriver(atom='H .0 .0 -0.3625; H .0 .0 0.3625', unit=UnitsType.ANGSTROM, charge=0, spin=0, basis='sto3g')
molecule = driver.run()
num_particles = molecule.num_alpha + molecule.num_beta
qubitOp = FermionicOperator(h1=molecule.one_body_integrals, h2=molecule.two_body_integrals).mapping(map_type='parity')
qubitOp = Z2Symmetries.two_qubit_reduction(qubitOp, num_particles)
```
Now, we load a device coupling map and noise model from the IBMQ provider and create a quantum instance, enabling error mitigation:
```
IBMQ.load_account()
provider = IBMQ.get_provider(hub='ibm-q')
backend = Aer.get_backend("qasm_simulator")
device = provider.get_backend("ibmqx2")
coupling_map = device.configuration().coupling_map
noise_model = noise.device.basic_device_noise_model(device.properties())
quantum_instance = QuantumInstance(backend=backend, shots=1000,
noise_model=noise_model,
coupling_map=coupling_map,
measurement_error_mitigation_cls=CompleteMeasFitter,
cals_matrix_refresh_period=30,)
```
Finally, we must configure the optimizer, the variational form, and the VQE instance. As the effects of noise increase as the number of two qubit gates circuit depth increase, we use a heuristic variational form (RYRZ) rather than UCCSD as RYRZ has a much shallower circuit than UCCSD and uses substantially fewer two qubit gates.
The following code may take a few minutes to run to completion.
```
exact_solution = ExactEigensolver(qubitOp).run()
print("Exact Result:", exact_solution['energy'])
optimizer = SPSA(max_trials=100)
var_form = RYRZ(qubitOp.num_qubits, depth=1, entanglement="linear")
vqe = VQE(qubitOp, var_form, optimizer=optimizer)
ret = vqe.run(quantum_instance)
print("VQE Result:", ret['energy'])
```
When noise mitigation is enabled, even though the result does not fall within chemical accuracy (defined as being within 0.0016 Hartree of the exact result), it is fairly close to the exact solution.
## Problems<a id='problems'></a>
1. You are given a Hamiltonian $H$ with the promise that its ground state is close to a maximally entangled $n$ qubit state. Explain which variational form (or forms) is likely to efficiently and accurately learn the the ground state energy of $H$. You may also answer by creating your own variational form, and explaining why it is appropriate for use with this Hamiltonian.
2. Calculate the number of circuit evaluations performed per optimization iteration, when using the COBYLA optimizer, the `qasm_simulator` with 1000 shots, and a Hamiltonian with 60 Pauli strings.
3. Use VQE to estimate the ground state energy of BeH$_2$ with an interatomic distance of $1.3$Å. You may re-use the function `get_qubit_op(dist)` by replacing `atom="Li .0 .0 .0; H .0 .0 " + str(dist)` with `atom="Be .0 .0 .0; H .0 .0 -" + str(dist) + "; H .0 .0 " + str(dist)` and invoking the function with `get_qubit_op(1.3)`. Note that removing the unoccupied orbitals does not preserve chemical precision for this molecule. However, to get the number of qubits required down to 6 (and thereby allowing efficient simulation on most laptops), the loss of precision is acceptable. While beyond the scope of this exercise, the interested reader may use qubit tapering operations to reduce the number of required qubits to 7, without losing any chemical precision.
## References<a id='references'></a>
1. Peruzzo, Alberto, et al. "A variational eigenvalue solver on a photonic quantum processor." *Nature communications* 5 (2014): 4213.
2. Griffiths, David J., and Darrell F. Schroeter. Introduction to quantum mechanics. *Cambridge University Press*, 2018.
3. Shende, Vivek V., Igor L. Markov, and Stephen S. Bullock. "Minimal universal two-qubit cnot-based circuits." arXiv preprint quant-ph/0308033 (2003).
4. Kandala, Abhinav, et al. "Hardware-efficient variational quantum eigensolver for small molecules and quantum magnets." Nature 549.7671 (2017): 242.
|
github_jupyter
|
import numpy as np
np.random.seed(999999)
target_distr = np.random.rand(2)
# We now convert the random vector into a valid probability vector
target_distr /= sum(target_distr)
from qiskit import QuantumCircuit, ClassicalRegister, QuantumRegister
def get_var_form(params):
qr = QuantumRegister(1, name="q")
cr = ClassicalRegister(1, name='c')
qc = QuantumCircuit(qr, cr)
qc.u3(params[0], params[1], params[2], qr[0])
qc.measure(qr, cr[0])
return qc
from qiskit import Aer, execute
backend = Aer.get_backend("qasm_simulator")
NUM_SHOTS = 10000
def get_probability_distribution(counts):
output_distr = [v / NUM_SHOTS for v in counts.values()]
if len(output_distr) == 1:
output_distr.append(0)
return output_distr
def objective_function(params):
# Obtain a quantum circuit instance from the paramters
qc = get_var_form(params)
# Execute the quantum circuit to obtain the probability distribution associated with the current parameters
result = execute(qc, backend, shots=NUM_SHOTS).result()
# Obtain the counts for each measured state, and convert those counts into a probability vector
output_distr = get_probability_distribution(result.get_counts(qc))
# Calculate the cost as the distance between the output distribution and the target distribution
cost = sum([np.abs(output_distr[i] - target_distr[i]) for i in range(2)])
return cost
from qiskit.aqua.components.optimizers import COBYLA
# Initialize the COBYLA optimizer
optimizer = COBYLA(maxiter=500, tol=0.0001)
# Create the initial parameters (noting that our single qubit variational form has 3 parameters)
params = np.random.rand(3)
ret = optimizer.optimize(num_vars=3, objective_function=objective_function, initial_point=params)
# Obtain the output distribution using the final parameters
qc = get_var_form(ret[0])
counts = execute(qc, backend, shots=NUM_SHOTS).result().get_counts(qc)
output_distr = get_probability_distribution(counts)
print("Target Distribution:", target_distr)
print("Obtained Distribution:", output_distr)
print("Output Error (Manhattan Distance):", ret[1])
print("Parameters Found:", ret[0])
from qiskit.aqua.components.variational_forms import RYRZ
entanglements = ["linear", "full"]
for entanglement in entanglements:
form = RYRZ(num_qubits=4, depth=1, entanglement=entanglement)
if entanglement == "linear":
print("=============Linear Entanglement:=============")
else:
print("=============Full Entanglement:=============")
# We initialize all parameters to 0 for this demonstration
print(form.construct_circuit([0] * form.num_parameters).draw(fold=100))
print()
from qiskit.aqua.algorithms import VQE, ExactEigensolver
import matplotlib.pyplot as plt
%matplotlib inline
import numpy as np
from qiskit.chemistry.components.variational_forms import UCCSD
from qiskit.chemistry.components.initial_states import HartreeFock
from qiskit.aqua.components.variational_forms import RYRZ
from qiskit.aqua.components.optimizers import COBYLA, SPSA, SLSQP
from qiskit.aqua.operators import Z2Symmetries
from qiskit import IBMQ, BasicAer, Aer
from qiskit.chemistry.drivers import PySCFDriver, UnitsType
from qiskit.chemistry import FermionicOperator
from qiskit import IBMQ
from qiskit.providers.aer import noise
from qiskit.aqua import QuantumInstance
from qiskit.ignis.mitigation.measurement import CompleteMeasFitter
def get_qubit_op(dist):
driver = PySCFDriver(atom="Li .0 .0 .0; H .0 .0 " + str(dist), unit=UnitsType.ANGSTROM,
charge=0, spin=0, basis='sto3g')
molecule = driver.run()
freeze_list = [0]
remove_list = [-3, -2]
repulsion_energy = molecule.nuclear_repulsion_energy
num_particles = molecule.num_alpha + molecule.num_beta
num_spin_orbitals = molecule.num_orbitals * 2
remove_list = [x % molecule.num_orbitals for x in remove_list]
freeze_list = [x % molecule.num_orbitals for x in freeze_list]
remove_list = [x - len(freeze_list) for x in remove_list]
remove_list += [x + molecule.num_orbitals - len(freeze_list) for x in remove_list]
freeze_list += [x + molecule.num_orbitals for x in freeze_list]
ferOp = FermionicOperator(h1=molecule.one_body_integrals, h2=molecule.two_body_integrals)
ferOp, energy_shift = ferOp.fermion_mode_freezing(freeze_list)
num_spin_orbitals -= len(freeze_list)
num_particles -= len(freeze_list)
ferOp = ferOp.fermion_mode_elimination(remove_list)
num_spin_orbitals -= len(remove_list)
qubitOp = ferOp.mapping(map_type='parity', threshold=0.00000001)
qubitOp = Z2Symmetries.two_qubit_reduction(qubitOp, num_particles)
shift = energy_shift + repulsion_energy
return qubitOp, num_particles, num_spin_orbitals, shift
backend = BasicAer.get_backend("statevector_simulator")
distances = np.arange(0.5, 4.0, 0.1)
exact_energies = []
vqe_energies = []
optimizer = SLSQP(maxiter=5)
for dist in distances:
qubitOp, num_particles, num_spin_orbitals, shift = get_qubit_op(dist)
result = ExactEigensolver(qubitOp).run()
exact_energies.append(result['energy'] + shift)
initial_state = HartreeFock(
qubitOp.num_qubits,
num_spin_orbitals,
num_particles,
'parity'
)
var_form = UCCSD(
qubitOp.num_qubits,
depth=1,
num_orbitals=num_spin_orbitals,
num_particles=num_particles,
initial_state=initial_state,
qubit_mapping='parity'
)
vqe = VQE(qubitOp, var_form, optimizer)
results = vqe.run(backend)['energy'] + shift
vqe_energies.append(results)
print("Interatomic Distance:", np.round(dist, 2), "VQE Result:", results, "Exact Energy:", exact_energies[-1])
print("All energies have been calculated")
plt.plot(distances, exact_energies, label="Exact Energy")
plt.plot(distances, vqe_energies, label="VQE Energy")
plt.xlabel('Atomic distance (Angstrom)')
plt.ylabel('Energy')
plt.legend()
plt.show()
driver = PySCFDriver(atom='H .0 .0 -0.3625; H .0 .0 0.3625', unit=UnitsType.ANGSTROM, charge=0, spin=0, basis='sto3g')
molecule = driver.run()
num_particles = molecule.num_alpha + molecule.num_beta
qubitOp = FermionicOperator(h1=molecule.one_body_integrals, h2=molecule.two_body_integrals).mapping(map_type='parity')
qubitOp = Z2Symmetries.two_qubit_reduction(qubitOp, num_particles)
IBMQ.load_account()
provider = IBMQ.get_provider(hub='ibm-q')
backend = Aer.get_backend("qasm_simulator")
device = provider.get_backend("ibmqx2")
coupling_map = device.configuration().coupling_map
noise_model = noise.device.basic_device_noise_model(device.properties())
quantum_instance = QuantumInstance(backend=backend, shots=1000,
noise_model=noise_model,
coupling_map=coupling_map,
measurement_error_mitigation_cls=CompleteMeasFitter,
cals_matrix_refresh_period=30,)
exact_solution = ExactEigensolver(qubitOp).run()
print("Exact Result:", exact_solution['energy'])
optimizer = SPSA(max_trials=100)
var_form = RYRZ(qubitOp.num_qubits, depth=1, entanglement="linear")
vqe = VQE(qubitOp, var_form, optimizer=optimizer)
ret = vqe.run(quantum_instance)
print("VQE Result:", ret['energy'])
| 0.622345 | 0.994578 |
# Deep Learning based Pipeline with Single Input for Multi-class Patent Classification
In this nootebook, we used the full text of most important sections in patent (title, abstract, technical fields, background, summary, and independent claim) as one input to the deep learning model.
```
import pandas as pd
df = pd.read_csv("../datasets/allITPatTextWith_Metadata.csv", encoding = "ISO-8859-1", error_bad_lines=False)
df.columns =['ID','TI','AB','TECHF','BACKG','SUMM','CLMS','ICM','AY','IPC','REF','PA','INV']
df.dropna(subset=['ICM'], inplace=True)
df.fillna(value='', inplace=True)
df['TEXT'] = df['TI'] +'. '+ df['AB'] +'. '+ df['TECHF']+'. '+ df['BACKG']+'. '+ df['SUMM']+'. '+ df['CLMS']
df.fillna(value='', inplace=True)
df.TEXT.head()
%%time
#preprocess of list fields
#convert all IPCs in df into one list
def toList(s):
"""
this method is to convert the list of IPCs in each row from a string to a python List
"""
s = s.translate ({ord(c): " " for c in "[]"})
ss= []
for cls in s.strip().split(','):
ss.append(cls.strip())
return ss
#apply toList method on all rows in the DF
df['PA'] = df['PA'].map(lambda pa : toList(pa))
df['INV'] = df['INV'].map(lambda inv : toList(inv))
df.head()
%%time
#also preprocess of list fields
def metadataPreprocessing(input):
newInput=' '
for item in input:
item = item.translate ({ord(c): " " for c in "!@#$%^&*()'[]{};:,./<>?\|`~°=\"+"})
itms=' '
for itm in item.split():
itms= itms +' '+itm.strip()
newInput = newInput + ' '+ itms.strip().replace(' ','_')
return newInput.strip()
df['PA'] = df['PA'].map(lambda pa : metadataPreprocessing(pa))
df['INV'] = df['INV'].map(lambda inv : metadataPreprocessing(inv))
df.head()
#preprocessing
standardStopwordFile = "sources/stopwords/stopwords-all.txt"
#generalWordsFile = "sources/Clariant/generalWords.txt"
#loading terms from a file to a set
def get_terms_from_file(filePath):
terms = set(line.strip() for line in open(filePath))
return terms
#remove undiserd terms
def remove_terms(termSet, phrase):
newPhrase = ""
for term in phrase.split():
if term.strip() not in termSet and len(term.strip())>2:
newPhrase = newPhrase + " " + term.strip()
def clean_texts(doc):
#Remove punctuation from texts
doc = doc.translate ({ord(c): ' ' for c in "0123456789!@#$%^&*()'/[]{};:,./<>?\|`~°=\"+"})
# split into tokens by white space
tokens = doc.lower().strip().split()
# filter out stop words
stop_words = get_terms_from_file(standardStopwordFile)
#generalStopwords = get_terms_from_file(generalWordsFile)
tokens = [w.strip('-') for w in tokens if w not in stop_words ]
# filter out short and long tokens
output = [word for word in tokens if len(word.strip()) > 2 and len(word) < 30 ]
output = " ".join(output)
#apply stemming
#output = stem_text(output)
return output
%%time
apply simple preprocessing on text
df['TI'] = df['TI'].map(lambda line : clean_texts(line))
df['AB'] = df['AB'].map(lambda line : clean_texts(line))
df['TECHF'] = df['TECHF'].map(lambda line : clean_texts(line))
df['BACKG'] = df['BACKG'].map(lambda line : clean_texts(line))
df['SUMM'] = df['SUMM'].map(lambda line : clean_texts(line))
df['CLMS'] = df['CLMS'].map(lambda line : clean_texts(line))
df.head()
#process the ICM codes and #related-patents
df['ICM'] = df['ICM'].map(lambda icmCode : icmCode[:4])
df_ICMs = df.groupby(['ICM'])
df_ICMs = df_ICMs.size().reset_index(name='Docs')
print(len(df_ICMs.ICM.unique()))
#filter out the rows with #docs less than N documents
df_ICMOut = df_ICMs[df_ICMs['Docs'] >= 500]
#filter out rows of the original dataframe df accordding to df_ICMOut
ICMList = df_ICMOut['ICM'].tolist()
df = df[df.ICM.isin(ICMList)]
icmCount = df_ICMs.count().tolist()[0]
print( 'number of remaining documents in the dataset is: ',len(df))
print('Number of unique labels is: ', len(df.ICM.unique()))
#preprocess all documents
#df['TEXT'] = df['TEXT'].map(lambda line : clean_texts(line))
from sklearn.utils import shuffle
df = shuffle(df)
df.head()
# lets take n% data as training and remaining m% for test.
train_size = int(len(df) * .9)
train_TI = df['TEXT'][:train_size]
train_ICM= df['ICM'][:train_size]
train_ID= df['ID'][:train_size]
test_TI = df['TEXT'][train_size:]
test_ICM = df['ICM'][train_size:]
test_ID = df['ID'][train_size:]
#metadata
train_pa_series = df['PA'][:train_size]
test_pa_series = df['PA'][train_size:]
train_inv_series = df['INV'][:train_size]
test_inv_series = df['INV'][train_size:]
print(train_TI.shape)
print(test_TI.shape)
#free up some memory space
#df.iloc[0:0]
#preparing text documents and labels for deep learning
from keras.preprocessing.text import Tokenizer
from keras.utils import to_categorical
from keras.preprocessing.sequence import pad_sequences
from keras.preprocessing.text import one_hot
from sklearn.preprocessing import LabelBinarizer
#PA
pa_inv_vocab_size = 2000
pa_tokenizer = Tokenizer(num_words=pa_inv_vocab_size, filters='!"#$%&()*+,./:;<=>?@[\]^`{|}~', lower=True, split=' ', char_level=False, oov_token=None)
pa_tokenizer.fit_on_texts(train_pa_series)
train_pa_one_hot =pa_tokenizer.texts_to_matrix(train_pa_series)
test_pa_one_hot =pa_tokenizer.texts_to_matrix(test_pa_series)
#INV
inv_tokenizer = Tokenizer(num_words=pa_inv_vocab_size, filters='!"#$%&()*+,./:;<=>?@[\]^`{|}~', lower=True, split=' ', char_level=False, oov_token=None)
inv_tokenizer.fit_on_texts(train_inv_series)
train_inv_one_hot =inv_tokenizer.texts_to_matrix(train_inv_series)
test_inv_one_hot =inv_tokenizer.texts_to_matrix(test_inv_series)
print('Found %s words in PA' % len(pa_tokenizer.word_index))
print('Found %s words in INV' % len(inv_tokenizer.word_index))
%%time
#Title
TI_tokenizer = Tokenizer(num_words=50000, filters='!"#$%&()*+,./:;<=>?@[\]^`{|}~_', lower=True, split=' ', char_level=False, oov_token=None)
TI_tokenizer.fit_on_texts(train_TI)
encoded_train_TI = TI_tokenizer.texts_to_sequences(train_TI)
encoded_test_TI = TI_tokenizer.texts_to_sequences(test_TI)
#convert all sequences in a list into the same length
TI_train = pad_sequences(encoded_train_TI, maxlen=100, padding='post')
TI_test = pad_sequences(encoded_test_TI, maxlen=100, padding='post')
%%time
# representing the labels/classes in the numeric format by scikit-learn - LabelBinarizer class
# Convert 1-dimensional class arrays to n-dimensional(#classes) class matrices
encoder = LabelBinarizer()
encoder.fit(train_ICM)
y_train = encoder.transform(train_ICM)
y_test = encoder.transform(test_ICM)
#get the unique number of labels in the training set
classesList = train_ICM.tolist()
classesList =set(classesList)
num_classes = len(classesList)
import numpy as np
def load_embedding_model(filePath):
embeddings_index = dict()
f = open(filePath, encoding='utf8')
for line in f:
values = line.split()
word = values[0]
coefs = np.asarray(values[1:], dtype='float32')
embeddings_index[word] = coefs
return embeddings_index
def create_embedding_matrix(tokenizer, embeddings_index, vocab_size_embbs, dim_size):
embeddings_matrix = np.zeros((vocab_size_embbs, dim_size))
for word, i in tokenizer.word_index.items():
embedding_vector = embeddings_index.get(word)
if embedding_vector is not None:
embeddings_matrix[i] = embedding_vector[0:dim_size]
return embeddings_matrix
%%time
## load the whole embedding into memory and get matrix
embedding_index = load_embedding_model('../models/w2v/phrase/patWordPhrase2VecModel.txt')
%%time
#create TITLE embedding Matrix
#vocab_size for embedding
vocab_size_embb = len(TI_tokenizer.word_index) + 1
TI_embeddings_matrix = create_embedding_matrix(TI_tokenizer,
embedding_index,
vocab_size_embb,
100)
import keras
from keras.models import Sequential, Model
from keras.layers import Dense, Input, Embedding, BatchNormalization, ELU, Concatenate
from keras.layers import LSTM, Conv1D, MaxPooling1D
from keras.layers.merge import concatenate
from keras.layers.core import Dropout
%%time
#TITLE
sequence_len =100
dropout_pct = 0.4
TI_embedding_layer_input = Input(shape=(sequence_len,), name='TI_embed_input')
TI_embedding_layer = Embedding(input_dim=len(TI_tokenizer.word_index) + 1,
output_dim=100, # Dimension of the dense embedding
weights=[TI_embeddings_matrix],
input_length=100)(TI_embedding_layer_input)
lstm_size = 64
TI_deep = LSTM(lstm_size,
dropout=dropout_pct,
recurrent_dropout=dropout_pct,
return_sequences=False,
name='LSTM_TI')(TI_embedding_layer)
TI_deep = Dense(300, activation=None)(TI_deep)
TI_deep = Dropout(dropout_pct)(TI_deep)
TI_deep = BatchNormalization()(TI_deep)
TI_deep = ELU()(TI_deep)
dropout_pct = 0.4
pa_input = Input(shape=(train_pa_one_hot.shape[1],), name='pa_input')
pas = Dense(32,input_dim=train_pa_one_hot.shape[1], activation=None)(pa_input)
pas = Dropout(dropout_pct)(pas)
pas = BatchNormalization()(pas)
pas = ELU()(pas)
#inv
inv_input = Input(shape=(train_inv_one_hot.shape[1],), name='inv_input')
invs = Dense(32,input_dim=train_inv_one_hot.shape[1], activation=None)(pa_input)
invs = Dropout(dropout_pct)(invs)
invs = BatchNormalization()(invs)
print('pa_input and inv_input layers are finished')
import keras_metrics as km
#contacting two input models
#model_inputs_to_concat = [TI_deep, AB_deep, TECHF_deep, BACKG_deep, SUMM_deep, CLMS_deep] #invs , pas, invs
#final_layer = Concatenate(name='concatenated_layer')(model_inputs_to_concat)
output = Dense(128, activation=None)(TI_deep)
output = Dropout(dropout_pct)(output)
output = BatchNormalization()(output)
output = ELU()(output)
output = Dense(num_classes, activation='softmax')(output)
model = Model(inputs=[TI_embedding_layer_input
],
outputs=output, name='model')
model.compile(loss='categorical_crossentropy',
optimizer='adam',
metrics=['accuracy', km.categorical_precision(), km.categorical_recall()])
model.summary()
%%time
batch_size= 500
num_epochs = 20
history = model.fit(x={'TI_embed_input': TI_train
},
y=y_train,
batch_size=batch_size,
epochs=num_epochs,
validation_data=
({'TI_embed_input': TI_test
},
y_test))
from sklearn.datasets import make_circles
from keras.models import Sequential
from keras.layers import Dense
from matplotlib import pyplot
import keras_metrics as km
import keras_metrics as km
#contacting two input models
model_inputs_to_concat = [TI_deep, pas, invs] #invs , pas, invs
final_layer = Concatenate(name='concatenated_layer')(model_inputs_to_concat)
output = Dense(128, activation=None)(final_layer)
output = Dropout(dropout_pct)(output)
output = BatchNormalization()(output)
output = ELU()(output)
output = Dense(num_classes, activation='softmax')(output)
model2 =Model(inputs=[ TI_embedding_layer_input,
pa_input,
inv_input],
outputs=output, name='model')
model2.compile(loss='categorical_crossentropy',
optimizer='adam',
metrics=['accuracy', km.categorical_precision(), km.categorical_recall()])
model2.summary()
%%time
batch_size= 500
num_epochs = 20
history2 = model2.fit(x={'TI_embed_input': TI_train,
'pa_input': train_pa_one_hot,
'inv_input': train_inv_one_hot
},
y=y_train,
batch_size=batch_size,
epochs=num_epochs,
validation_data=
({'TI_embed_input': TI_test,
'pa_input': test_pa_one_hot,
'inv_input': test_inv_one_hot
},
y_test))
```
|
github_jupyter
|
import pandas as pd
df = pd.read_csv("../datasets/allITPatTextWith_Metadata.csv", encoding = "ISO-8859-1", error_bad_lines=False)
df.columns =['ID','TI','AB','TECHF','BACKG','SUMM','CLMS','ICM','AY','IPC','REF','PA','INV']
df.dropna(subset=['ICM'], inplace=True)
df.fillna(value='', inplace=True)
df['TEXT'] = df['TI'] +'. '+ df['AB'] +'. '+ df['TECHF']+'. '+ df['BACKG']+'. '+ df['SUMM']+'. '+ df['CLMS']
df.fillna(value='', inplace=True)
df.TEXT.head()
%%time
#preprocess of list fields
#convert all IPCs in df into one list
def toList(s):
"""
this method is to convert the list of IPCs in each row from a string to a python List
"""
s = s.translate ({ord(c): " " for c in "[]"})
ss= []
for cls in s.strip().split(','):
ss.append(cls.strip())
return ss
#apply toList method on all rows in the DF
df['PA'] = df['PA'].map(lambda pa : toList(pa))
df['INV'] = df['INV'].map(lambda inv : toList(inv))
df.head()
%%time
#also preprocess of list fields
def metadataPreprocessing(input):
newInput=' '
for item in input:
item = item.translate ({ord(c): " " for c in "!@#$%^&*()'[]{};:,./<>?\|`~°=\"+"})
itms=' '
for itm in item.split():
itms= itms +' '+itm.strip()
newInput = newInput + ' '+ itms.strip().replace(' ','_')
return newInput.strip()
df['PA'] = df['PA'].map(lambda pa : metadataPreprocessing(pa))
df['INV'] = df['INV'].map(lambda inv : metadataPreprocessing(inv))
df.head()
#preprocessing
standardStopwordFile = "sources/stopwords/stopwords-all.txt"
#generalWordsFile = "sources/Clariant/generalWords.txt"
#loading terms from a file to a set
def get_terms_from_file(filePath):
terms = set(line.strip() for line in open(filePath))
return terms
#remove undiserd terms
def remove_terms(termSet, phrase):
newPhrase = ""
for term in phrase.split():
if term.strip() not in termSet and len(term.strip())>2:
newPhrase = newPhrase + " " + term.strip()
def clean_texts(doc):
#Remove punctuation from texts
doc = doc.translate ({ord(c): ' ' for c in "0123456789!@#$%^&*()'/[]{};:,./<>?\|`~°=\"+"})
# split into tokens by white space
tokens = doc.lower().strip().split()
# filter out stop words
stop_words = get_terms_from_file(standardStopwordFile)
#generalStopwords = get_terms_from_file(generalWordsFile)
tokens = [w.strip('-') for w in tokens if w not in stop_words ]
# filter out short and long tokens
output = [word for word in tokens if len(word.strip()) > 2 and len(word) < 30 ]
output = " ".join(output)
#apply stemming
#output = stem_text(output)
return output
%%time
apply simple preprocessing on text
df['TI'] = df['TI'].map(lambda line : clean_texts(line))
df['AB'] = df['AB'].map(lambda line : clean_texts(line))
df['TECHF'] = df['TECHF'].map(lambda line : clean_texts(line))
df['BACKG'] = df['BACKG'].map(lambda line : clean_texts(line))
df['SUMM'] = df['SUMM'].map(lambda line : clean_texts(line))
df['CLMS'] = df['CLMS'].map(lambda line : clean_texts(line))
df.head()
#process the ICM codes and #related-patents
df['ICM'] = df['ICM'].map(lambda icmCode : icmCode[:4])
df_ICMs = df.groupby(['ICM'])
df_ICMs = df_ICMs.size().reset_index(name='Docs')
print(len(df_ICMs.ICM.unique()))
#filter out the rows with #docs less than N documents
df_ICMOut = df_ICMs[df_ICMs['Docs'] >= 500]
#filter out rows of the original dataframe df accordding to df_ICMOut
ICMList = df_ICMOut['ICM'].tolist()
df = df[df.ICM.isin(ICMList)]
icmCount = df_ICMs.count().tolist()[0]
print( 'number of remaining documents in the dataset is: ',len(df))
print('Number of unique labels is: ', len(df.ICM.unique()))
#preprocess all documents
#df['TEXT'] = df['TEXT'].map(lambda line : clean_texts(line))
from sklearn.utils import shuffle
df = shuffle(df)
df.head()
# lets take n% data as training and remaining m% for test.
train_size = int(len(df) * .9)
train_TI = df['TEXT'][:train_size]
train_ICM= df['ICM'][:train_size]
train_ID= df['ID'][:train_size]
test_TI = df['TEXT'][train_size:]
test_ICM = df['ICM'][train_size:]
test_ID = df['ID'][train_size:]
#metadata
train_pa_series = df['PA'][:train_size]
test_pa_series = df['PA'][train_size:]
train_inv_series = df['INV'][:train_size]
test_inv_series = df['INV'][train_size:]
print(train_TI.shape)
print(test_TI.shape)
#free up some memory space
#df.iloc[0:0]
#preparing text documents and labels for deep learning
from keras.preprocessing.text import Tokenizer
from keras.utils import to_categorical
from keras.preprocessing.sequence import pad_sequences
from keras.preprocessing.text import one_hot
from sklearn.preprocessing import LabelBinarizer
#PA
pa_inv_vocab_size = 2000
pa_tokenizer = Tokenizer(num_words=pa_inv_vocab_size, filters='!"#$%&()*+,./:;<=>?@[\]^`{|}~', lower=True, split=' ', char_level=False, oov_token=None)
pa_tokenizer.fit_on_texts(train_pa_series)
train_pa_one_hot =pa_tokenizer.texts_to_matrix(train_pa_series)
test_pa_one_hot =pa_tokenizer.texts_to_matrix(test_pa_series)
#INV
inv_tokenizer = Tokenizer(num_words=pa_inv_vocab_size, filters='!"#$%&()*+,./:;<=>?@[\]^`{|}~', lower=True, split=' ', char_level=False, oov_token=None)
inv_tokenizer.fit_on_texts(train_inv_series)
train_inv_one_hot =inv_tokenizer.texts_to_matrix(train_inv_series)
test_inv_one_hot =inv_tokenizer.texts_to_matrix(test_inv_series)
print('Found %s words in PA' % len(pa_tokenizer.word_index))
print('Found %s words in INV' % len(inv_tokenizer.word_index))
%%time
#Title
TI_tokenizer = Tokenizer(num_words=50000, filters='!"#$%&()*+,./:;<=>?@[\]^`{|}~_', lower=True, split=' ', char_level=False, oov_token=None)
TI_tokenizer.fit_on_texts(train_TI)
encoded_train_TI = TI_tokenizer.texts_to_sequences(train_TI)
encoded_test_TI = TI_tokenizer.texts_to_sequences(test_TI)
#convert all sequences in a list into the same length
TI_train = pad_sequences(encoded_train_TI, maxlen=100, padding='post')
TI_test = pad_sequences(encoded_test_TI, maxlen=100, padding='post')
%%time
# representing the labels/classes in the numeric format by scikit-learn - LabelBinarizer class
# Convert 1-dimensional class arrays to n-dimensional(#classes) class matrices
encoder = LabelBinarizer()
encoder.fit(train_ICM)
y_train = encoder.transform(train_ICM)
y_test = encoder.transform(test_ICM)
#get the unique number of labels in the training set
classesList = train_ICM.tolist()
classesList =set(classesList)
num_classes = len(classesList)
import numpy as np
def load_embedding_model(filePath):
embeddings_index = dict()
f = open(filePath, encoding='utf8')
for line in f:
values = line.split()
word = values[0]
coefs = np.asarray(values[1:], dtype='float32')
embeddings_index[word] = coefs
return embeddings_index
def create_embedding_matrix(tokenizer, embeddings_index, vocab_size_embbs, dim_size):
embeddings_matrix = np.zeros((vocab_size_embbs, dim_size))
for word, i in tokenizer.word_index.items():
embedding_vector = embeddings_index.get(word)
if embedding_vector is not None:
embeddings_matrix[i] = embedding_vector[0:dim_size]
return embeddings_matrix
%%time
## load the whole embedding into memory and get matrix
embedding_index = load_embedding_model('../models/w2v/phrase/patWordPhrase2VecModel.txt')
%%time
#create TITLE embedding Matrix
#vocab_size for embedding
vocab_size_embb = len(TI_tokenizer.word_index) + 1
TI_embeddings_matrix = create_embedding_matrix(TI_tokenizer,
embedding_index,
vocab_size_embb,
100)
import keras
from keras.models import Sequential, Model
from keras.layers import Dense, Input, Embedding, BatchNormalization, ELU, Concatenate
from keras.layers import LSTM, Conv1D, MaxPooling1D
from keras.layers.merge import concatenate
from keras.layers.core import Dropout
%%time
#TITLE
sequence_len =100
dropout_pct = 0.4
TI_embedding_layer_input = Input(shape=(sequence_len,), name='TI_embed_input')
TI_embedding_layer = Embedding(input_dim=len(TI_tokenizer.word_index) + 1,
output_dim=100, # Dimension of the dense embedding
weights=[TI_embeddings_matrix],
input_length=100)(TI_embedding_layer_input)
lstm_size = 64
TI_deep = LSTM(lstm_size,
dropout=dropout_pct,
recurrent_dropout=dropout_pct,
return_sequences=False,
name='LSTM_TI')(TI_embedding_layer)
TI_deep = Dense(300, activation=None)(TI_deep)
TI_deep = Dropout(dropout_pct)(TI_deep)
TI_deep = BatchNormalization()(TI_deep)
TI_deep = ELU()(TI_deep)
dropout_pct = 0.4
pa_input = Input(shape=(train_pa_one_hot.shape[1],), name='pa_input')
pas = Dense(32,input_dim=train_pa_one_hot.shape[1], activation=None)(pa_input)
pas = Dropout(dropout_pct)(pas)
pas = BatchNormalization()(pas)
pas = ELU()(pas)
#inv
inv_input = Input(shape=(train_inv_one_hot.shape[1],), name='inv_input')
invs = Dense(32,input_dim=train_inv_one_hot.shape[1], activation=None)(pa_input)
invs = Dropout(dropout_pct)(invs)
invs = BatchNormalization()(invs)
print('pa_input and inv_input layers are finished')
import keras_metrics as km
#contacting two input models
#model_inputs_to_concat = [TI_deep, AB_deep, TECHF_deep, BACKG_deep, SUMM_deep, CLMS_deep] #invs , pas, invs
#final_layer = Concatenate(name='concatenated_layer')(model_inputs_to_concat)
output = Dense(128, activation=None)(TI_deep)
output = Dropout(dropout_pct)(output)
output = BatchNormalization()(output)
output = ELU()(output)
output = Dense(num_classes, activation='softmax')(output)
model = Model(inputs=[TI_embedding_layer_input
],
outputs=output, name='model')
model.compile(loss='categorical_crossentropy',
optimizer='adam',
metrics=['accuracy', km.categorical_precision(), km.categorical_recall()])
model.summary()
%%time
batch_size= 500
num_epochs = 20
history = model.fit(x={'TI_embed_input': TI_train
},
y=y_train,
batch_size=batch_size,
epochs=num_epochs,
validation_data=
({'TI_embed_input': TI_test
},
y_test))
from sklearn.datasets import make_circles
from keras.models import Sequential
from keras.layers import Dense
from matplotlib import pyplot
import keras_metrics as km
import keras_metrics as km
#contacting two input models
model_inputs_to_concat = [TI_deep, pas, invs] #invs , pas, invs
final_layer = Concatenate(name='concatenated_layer')(model_inputs_to_concat)
output = Dense(128, activation=None)(final_layer)
output = Dropout(dropout_pct)(output)
output = BatchNormalization()(output)
output = ELU()(output)
output = Dense(num_classes, activation='softmax')(output)
model2 =Model(inputs=[ TI_embedding_layer_input,
pa_input,
inv_input],
outputs=output, name='model')
model2.compile(loss='categorical_crossentropy',
optimizer='adam',
metrics=['accuracy', km.categorical_precision(), km.categorical_recall()])
model2.summary()
%%time
batch_size= 500
num_epochs = 20
history2 = model2.fit(x={'TI_embed_input': TI_train,
'pa_input': train_pa_one_hot,
'inv_input': train_inv_one_hot
},
y=y_train,
batch_size=batch_size,
epochs=num_epochs,
validation_data=
({'TI_embed_input': TI_test,
'pa_input': test_pa_one_hot,
'inv_input': test_inv_one_hot
},
y_test))
| 0.339828 | 0.701138 |
# Exploring deaths of notable people by year in Wikipedia
By R. Stuart Geiger, last updated 2016-12-28
Dual-licensed under CC-BY-SA 4.0 and the MIT License.
## How many articles are in the "[year] deaths" categories in the English Wikipedia?
The first thing I tried was just counting up the number of articles in each of the "[[year] deaths](https://en.wikipedia.org/wiki/Category:Deaths_by_year)" categories, from 2000-2016.
```
import pandas as pd
import matplotlib.pyplot as plt
import matplotlib
import numpy as np
%matplotlib inline
matplotlib.style.use('seaborn-darkgrid')
import pywikibot
site = pywikibot.Site('en', 'wikipedia')
def yearly_death_counts(startyear,endyear):
years = np.arange(startyear,endyear+1) # add 1 to endyear because np.arange doesn't include the stop
deaths_per_year = {}
for year in years:
deaths_per_year[year] = 0
for year in years:
yearstr = 'Category:' + str(year) + "_deaths"
deathcat = pywikibot.Page(site, yearstr)
deathcat_o = site.categoryinfo(deathcat)
deaths_per_year[year] = deathcat_o['pages']
yearly_articles_df = pd.DataFrame.from_dict(deaths_per_year, orient='index')
yearly_articles_df.columns = ['articles in category']
yearly_articles_df = yearly_articles_df.sort_index()
return yearly_articles_df
yearly_articles_df = yearly_death_counts(2000,2016)
yearly_articles_df
ax = yearly_articles_df.plot(kind='bar',figsize=[10,4])
ax.legend_.remove()
ax.set_ylabel("Number of articles")
ax.set_title("""Articles in the "[year] deaths" category in the English Wikipedia""")
```
### Interpreting total article counts
One of the first things that we see in this graph is that the data is far from uniform, and has a distinct trend. This should make us suspicious. There are about 4,945 articles in the "2000 deaths" category, and the number steadily rises each year to 7,486 articles in the "2010 deaths" category. Is there any compelling reason we have to believe that the number of notable people in the world would steadily increase by a few percent each year from 2000 to 2010, then plateau? Or is it more of an artifact of what Wikipedia's volunteer editors choose to work on?
What if we look at this over a much longer timescale, like 1800-2016?
```
yearly_articles_df = yearly_death_counts(1800,2016)
ax = yearly_articles_df.plot(kind='line',figsize=[10,4])
ax.legend_.remove()
ax.set_ylabel("Number of articles")
ax.set_title("""Articles in the "[year] deaths" category in the English Wikipedia""")
```
We can see the two big jumps in the 20th century, likely reflecting the events around World War I and II. This makes sense, as those time periods were certainly sharp increases in the total number of deaths, as well as the number of notable deaths. Remember: we have already assumed that Wikipedia's biographical articles doesn't represent all of humanity -- in fact, we are counting on it, so we can distinguish celebrity deaths.
However, for the purposes of our question, is it safe to assume that having a Wikipedia article means being a celebrity? When I hear people talk about so many celebrities dying in 2016, people seem to mean a lower number than the ~7,000 people with Wikipedia articles who died in 2010-2016. The number is maybe two orders of magnitude lower, somewhere closer to 70 than 7,000. So is there a way we can filter Wikipedia articles?
To get at this, I first thought of using the pageview data that Wikimedia collects. There is a nice API about how many times every article in every language version of Wikipedia is viewed each hour. I hadn't played around with that API, so I wanted to try it out.
## Pageviews for articles in the "2016 Deaths" category
The mwviews python package has support for hourly, daily, and monthly granularity, but not annual. So I wrote a function that gets the pageview counts for a given article for an entire year. But, as we will see, the data in the historical pageview API only goes back to mid-2015.
```
!pip install mwviews
from mwviews.api import PageviewsClient
def yearly_views(title,year):
p = PageviewsClient(2)
startdate = str(year) + "010100"
enddate = str(year) + "123123"
d = p.article_views('en.wikipedia', title, granularity='monthly', start=startdate, end=enddate)
total = 0
for month in d.values():
for titlecount in month.values():
if titlecount is not None:
total += titlecount
return total
yearly_views("Prince_(musician)", 2016)
yearly_views("Prince_(musician)", 2015)
yearly_views("Prince_(musician)", 2014)
```
### Querying the pageview API for all the articles in the "2016 deaths" category
I was wanting to get 2016 pageview data for 2016 deaths, 2015 pageview data for 2015 deaths, and so on. But there isn't full historical data for the pageview API. However, we can take a detour and do some interesting exploration with only the 2016 dataset.
This code iterates through the category for "2016 deaths" and for each page, queries the pageview API to get the number of total pageviews in 2016. It takes a few minutes to run. This throws some errors for a few articles (in pink boxes below), which we will ignore.
```
year = 2016
yearstr = 'Category:' + str(year) + "_deaths"
deathcat = pywikibot.Page(site, yearstr)
pageviews_2016 = {}
for page in site.categorymembers(deathcat):
if page.title().find("List_of") is -1 and page.title().find("Category:") is -1:
try:
page_yearly_views = yearly_views(page.title(),year)
except Exception as e:
page_yearly_views = 0
pageviews_2016[page.title()] = page_yearly_views
pageviews_df = pd.DataFrame.from_dict(pageviews_2016,orient='index')
pageviews_df = pageviews_df.sort_values(0, ascending=False)
pageviews_df.head(25)
pageviews_df.to_csv("enwiki_pageviews_2016.csv")
```
### Getting the daily pageview counts for 6 most viewed articles in "2016 deaths" (includes the "Deaths in 2016" article)
```
articles = []
for index,row in pageviews_df.head(6).iterrows():
articles.append(index)
from mwviews.api import PageviewsClient
p = PageviewsClient(10)
startdate = "2016010100"
enddate = "2016123123"
counts_dict = p.article_views('en.wikipedia', articles, granularity='daily', start=startdate, end=enddate)
counts_df = pd.DataFrame.from_dict(counts_dict, orient='index')
counts_df = counts_df.fillna(0)
counts_df.to_csv("deaths-enwiki-2016.csv")
```
### Plotting pageviews per day of top 5 articles
```
articles = []
for index,row in pageviews_df.head(6).iterrows():
articles.append(index)
counts_dict = p.article_views('en.wikipedia', articles, granularity='daily', start=startdate, end=enddate)
counts_df = pd.DataFrame.from_dict(counts_dict, orient='index')
counts_df = counts_df.fillna(0)
matplotlib.style.use('seaborn-darkgrid')
font = {'family' : 'normal',
'weight' : 'normal',
'size' : 18}
matplotlib.rc('font', **font)
plt.figure(figsize=[14,7.2])
for title in counts_df:
fig = counts_df[title].plot(legend=True, linewidth=2)
fig.set_ylabel('Views per day')
plt.legend(loc='best')
```
## Querying edit counts for articles in the "[year] deaths" categories using SQL/Quarry
To get data about the number of times each article the "[year] deaths" categories has been edited, we could use the API, but it would take a long time. There are over 100,000 articles in the 2000-2016 categories, and that would require a new API call for each one. This is the kind of query that SQL is meant for, and we can use the [Quarry](https://quarry.wmflabs.org) service to run this query directly on Wikipedia's servers.
I've included the query below in a code cell, but it was run [here](https://quarry.wmflabs.org/query/15112). We will download the results in a TSV file, then load it into a pandas dataframe for processing.
```
sql_query = """
select cl_to, cl_from, count(rev_id) as edits, page_title
from (select * from categorylinks where cl_to LIKE '20___deaths') as d
inner join revision on cl_from = rev_page
inner join page on rev_page = page_id
where page_namespace = 0 and cl_to NOT LIKE '200s_deaths' and page_title NOT LIKE 'List_of%'
group by cl_from
"""
!wget https://quarry.wmflabs.org/run/139193/output/0/tsv?download=true -O deaths.tsv
deaths_df = pd.read_csv("deaths.tsv", sep='\t')
deaths_df.columns = ['year', 'page_id', 'edits', 'title']
deaths_df.head(15)
```
### Filtering articles by number of edits
We can filter the number of articles in the various death by year categories by the total edit count. But what will be our threshold? What are we looking for? I've chosen 7 different thresholds (over 10, 50, 100, 250, 500, 750, and 1,000 edits). The results these different thresholds produce give rise to different interpretations of the same question.
```
deaths_over10 = deaths_df[deaths_df.edits>10]
deaths_over50 = deaths_df[deaths_df.edits>50]
deaths_over100 = deaths_df[deaths_df.edits>100]
deaths_over250 = deaths_df[deaths_df.edits>250]
deaths_over500 = deaths_df[deaths_df.edits>500]
deaths_over750 = deaths_df[deaths_df.edits>750]
deaths_over1000 = deaths_df[deaths_df.edits>1000]
deaths_over10 = deaths_over10[['year','edits']]
deaths_over50 = deaths_over50[['year','edits']]
deaths_over100 = deaths_over100[['year','edits']]
deaths_over250 = deaths_over250[['year','edits']]
deaths_over500 = deaths_over500[['year','edits']]
deaths_over750 = deaths_over750[['year','edits']]
deaths_over1000 = deaths_over1000[['year','edits']]
matplotlib.style.use('seaborn-darkgrid')
font = {'family' : 'normal',
'weight' : 'normal',
'size' : 10}
matplotlib.rc('font', **font)
ax = deaths_over10.groupby(['year']).agg(['count']).plot(kind='barh')
ax.legend_.remove()
ax.set_title("""Number of articles with >10 edits in "[year] deaths" category""")
ax = deaths_over50.groupby(['year']).agg(['count']).plot(kind='barh')
ax.legend_.remove()
ax.set_title("""Number of articles with >50 edits in "[year] deaths" category""")
ax = deaths_over100.groupby(['year']).agg(['count']).plot(kind='barh')
ax.legend_.remove()
ax.set_title("""Number of articles with >100 edits in "[year] deaths" category""")
ax = deaths_over250.groupby(['year']).agg(['count']).plot(kind='barh')
ax.legend_.remove()
ax.set_title("""Number of articles with >250 edits in "[year] deaths" category""")
ax = deaths_over500.groupby(['year']).agg(['count']).plot(kind='barh')
ax.legend_.remove()
ax.set_title("""Number of articles with >500 edits in "[year] deaths" category""")
ax = deaths_over750.groupby(['year']).agg(['count']).plot(kind='barh')
ax.legend_.remove()
ax.set_title("""Number of articles with >750 edits in "[year] deaths" category""")
ax = deaths_over1000.groupby(['year']).agg(['count']).plot(kind='barh')
ax.legend_.remove()
ax.set_title("""Number of articles with >1,000 edits in "[year] deaths" category""")
```
|
github_jupyter
|
import pandas as pd
import matplotlib.pyplot as plt
import matplotlib
import numpy as np
%matplotlib inline
matplotlib.style.use('seaborn-darkgrid')
import pywikibot
site = pywikibot.Site('en', 'wikipedia')
def yearly_death_counts(startyear,endyear):
years = np.arange(startyear,endyear+1) # add 1 to endyear because np.arange doesn't include the stop
deaths_per_year = {}
for year in years:
deaths_per_year[year] = 0
for year in years:
yearstr = 'Category:' + str(year) + "_deaths"
deathcat = pywikibot.Page(site, yearstr)
deathcat_o = site.categoryinfo(deathcat)
deaths_per_year[year] = deathcat_o['pages']
yearly_articles_df = pd.DataFrame.from_dict(deaths_per_year, orient='index')
yearly_articles_df.columns = ['articles in category']
yearly_articles_df = yearly_articles_df.sort_index()
return yearly_articles_df
yearly_articles_df = yearly_death_counts(2000,2016)
yearly_articles_df
ax = yearly_articles_df.plot(kind='bar',figsize=[10,4])
ax.legend_.remove()
ax.set_ylabel("Number of articles")
ax.set_title("""Articles in the "[year] deaths" category in the English Wikipedia""")
yearly_articles_df = yearly_death_counts(1800,2016)
ax = yearly_articles_df.plot(kind='line',figsize=[10,4])
ax.legend_.remove()
ax.set_ylabel("Number of articles")
ax.set_title("""Articles in the "[year] deaths" category in the English Wikipedia""")
!pip install mwviews
from mwviews.api import PageviewsClient
def yearly_views(title,year):
p = PageviewsClient(2)
startdate = str(year) + "010100"
enddate = str(year) + "123123"
d = p.article_views('en.wikipedia', title, granularity='monthly', start=startdate, end=enddate)
total = 0
for month in d.values():
for titlecount in month.values():
if titlecount is not None:
total += titlecount
return total
yearly_views("Prince_(musician)", 2016)
yearly_views("Prince_(musician)", 2015)
yearly_views("Prince_(musician)", 2014)
year = 2016
yearstr = 'Category:' + str(year) + "_deaths"
deathcat = pywikibot.Page(site, yearstr)
pageviews_2016 = {}
for page in site.categorymembers(deathcat):
if page.title().find("List_of") is -1 and page.title().find("Category:") is -1:
try:
page_yearly_views = yearly_views(page.title(),year)
except Exception as e:
page_yearly_views = 0
pageviews_2016[page.title()] = page_yearly_views
pageviews_df = pd.DataFrame.from_dict(pageviews_2016,orient='index')
pageviews_df = pageviews_df.sort_values(0, ascending=False)
pageviews_df.head(25)
pageviews_df.to_csv("enwiki_pageviews_2016.csv")
articles = []
for index,row in pageviews_df.head(6).iterrows():
articles.append(index)
from mwviews.api import PageviewsClient
p = PageviewsClient(10)
startdate = "2016010100"
enddate = "2016123123"
counts_dict = p.article_views('en.wikipedia', articles, granularity='daily', start=startdate, end=enddate)
counts_df = pd.DataFrame.from_dict(counts_dict, orient='index')
counts_df = counts_df.fillna(0)
counts_df.to_csv("deaths-enwiki-2016.csv")
articles = []
for index,row in pageviews_df.head(6).iterrows():
articles.append(index)
counts_dict = p.article_views('en.wikipedia', articles, granularity='daily', start=startdate, end=enddate)
counts_df = pd.DataFrame.from_dict(counts_dict, orient='index')
counts_df = counts_df.fillna(0)
matplotlib.style.use('seaborn-darkgrid')
font = {'family' : 'normal',
'weight' : 'normal',
'size' : 18}
matplotlib.rc('font', **font)
plt.figure(figsize=[14,7.2])
for title in counts_df:
fig = counts_df[title].plot(legend=True, linewidth=2)
fig.set_ylabel('Views per day')
plt.legend(loc='best')
sql_query = """
select cl_to, cl_from, count(rev_id) as edits, page_title
from (select * from categorylinks where cl_to LIKE '20___deaths') as d
inner join revision on cl_from = rev_page
inner join page on rev_page = page_id
where page_namespace = 0 and cl_to NOT LIKE '200s_deaths' and page_title NOT LIKE 'List_of%'
group by cl_from
"""
!wget https://quarry.wmflabs.org/run/139193/output/0/tsv?download=true -O deaths.tsv
deaths_df = pd.read_csv("deaths.tsv", sep='\t')
deaths_df.columns = ['year', 'page_id', 'edits', 'title']
deaths_df.head(15)
deaths_over10 = deaths_df[deaths_df.edits>10]
deaths_over50 = deaths_df[deaths_df.edits>50]
deaths_over100 = deaths_df[deaths_df.edits>100]
deaths_over250 = deaths_df[deaths_df.edits>250]
deaths_over500 = deaths_df[deaths_df.edits>500]
deaths_over750 = deaths_df[deaths_df.edits>750]
deaths_over1000 = deaths_df[deaths_df.edits>1000]
deaths_over10 = deaths_over10[['year','edits']]
deaths_over50 = deaths_over50[['year','edits']]
deaths_over100 = deaths_over100[['year','edits']]
deaths_over250 = deaths_over250[['year','edits']]
deaths_over500 = deaths_over500[['year','edits']]
deaths_over750 = deaths_over750[['year','edits']]
deaths_over1000 = deaths_over1000[['year','edits']]
matplotlib.style.use('seaborn-darkgrid')
font = {'family' : 'normal',
'weight' : 'normal',
'size' : 10}
matplotlib.rc('font', **font)
ax = deaths_over10.groupby(['year']).agg(['count']).plot(kind='barh')
ax.legend_.remove()
ax.set_title("""Number of articles with >10 edits in "[year] deaths" category""")
ax = deaths_over50.groupby(['year']).agg(['count']).plot(kind='barh')
ax.legend_.remove()
ax.set_title("""Number of articles with >50 edits in "[year] deaths" category""")
ax = deaths_over100.groupby(['year']).agg(['count']).plot(kind='barh')
ax.legend_.remove()
ax.set_title("""Number of articles with >100 edits in "[year] deaths" category""")
ax = deaths_over250.groupby(['year']).agg(['count']).plot(kind='barh')
ax.legend_.remove()
ax.set_title("""Number of articles with >250 edits in "[year] deaths" category""")
ax = deaths_over500.groupby(['year']).agg(['count']).plot(kind='barh')
ax.legend_.remove()
ax.set_title("""Number of articles with >500 edits in "[year] deaths" category""")
ax = deaths_over750.groupby(['year']).agg(['count']).plot(kind='barh')
ax.legend_.remove()
ax.set_title("""Number of articles with >750 edits in "[year] deaths" category""")
ax = deaths_over1000.groupby(['year']).agg(['count']).plot(kind='barh')
ax.legend_.remove()
ax.set_title("""Number of articles with >1,000 edits in "[year] deaths" category""")
| 0.420362 | 0.914214 |
```
#!pip install dowhy==0.6
#!pip install econml==0.12.0
from itertools import combinations
import numpy as np
import pandas as pd
import dowhy
from dowhy import CausalModel
import dowhy.datasets
from sklearn.linear_model import LassoCV
from sklearn.ensemble import GradientBoostingRegressor
import matplotlib.pyplot as plt
plt.style.use('fivethirtyeight')
```
# Causal Inference in Python: An Introduction
Causality was an *enfant terrible* of the big data and statistical learning revolution of the early 2010s. Many people believed (myself included) that having large enough datasets and efficient learning algorithms is sufficient and we do not need the concept of causality at all. Today, causal inference, modeling and discovery is being used more and more broadly across areas – from medical research and neuroscience to marketing and fraud detection. This talk briefly introduces main causal concepts and two Python libraries – DoWhy and EconML – for performing causal inference.
## Causal model with DoWhy & EconML
### Generate a dataset
```
# Create the dataset
W = np.random.randn(1000)
T = np.random.randn(1000) + .2*W + 3
Y = 6*T + 2*W - 13
df = pd.DataFrame(np.vstack([W, T, Y]).T, columns=['W', 'T', 'Y'])
df
plt.figure(figsize=(3, 3))
plt.scatter(df['T'], df['Y'], alpha=.2)
plt.show()
```
### Stage 1: Model the problem
#### Stage 1.1 - Define the graph - `GML`
```
# Create the graph describing the causal structure
graph = """
graph [
directed 1
node [
id "T"
label "T"
]
node [
id "W"
label "W"
]
node [
id "Y"
label "Y"
]
edge [
source "W"
target "T"
]
edge [
source "W"
target "Y"
]
edge [
source "T"
target "Y"
]
]
"""
# Remove newlines
graph = graph.replace('\n', '')
```
#### Stage 1.2 - define the DoWhy model
```
# With graph
model = CausalModel(
data=df,
treatment='T',
outcome='Y',
graph=graph
)
plt.figure(figsize=(3, 3))
model.view_model()
plt.show()
```
## Stage 2: Identify the estimand
```
estimand = model.identify_effect(proceed_when_unidentifiable=True)
print(estimand)
```
## Stage 3: Estimate the causal effect
#### Example 1 - Linear Regression
```
estimate = model.estimate_effect(
identified_estimand=estimand,
method_name='backdoor.linear_regression'
)
print(f'Estimate of causal effect (linear regression): {estimate.value}')
```
#### Example 3 - Double Machne Learning
```
estimate = model.estimate_effect(
identified_estimand=estimand,
method_name='backdoor.econml.dml.DML',
method_params={
'init_params': {
'model_y': GradientBoostingRegressor(),
'model_t': GradientBoostingRegressor(),
'model_final': LassoCV(fit_intercept=False),
},
'fit_params': {}}
)
print(f'Estimate of causal effect (DML): {estimate.value}')
```
## Stage 4: Run refutation tests
```
refute_results = model.refute_estimate(
estimand=estimand,
estimate=estimate,
method_name='placebo_treatment_refuter'
)
print(refute_results)
refute_results = model.refute_estimate(
estimand=estimand,
estimate=estimate,
method_name='random_common_cause'
)
print(refute_results)
```
|
github_jupyter
|
#!pip install dowhy==0.6
#!pip install econml==0.12.0
from itertools import combinations
import numpy as np
import pandas as pd
import dowhy
from dowhy import CausalModel
import dowhy.datasets
from sklearn.linear_model import LassoCV
from sklearn.ensemble import GradientBoostingRegressor
import matplotlib.pyplot as plt
plt.style.use('fivethirtyeight')
# Create the dataset
W = np.random.randn(1000)
T = np.random.randn(1000) + .2*W + 3
Y = 6*T + 2*W - 13
df = pd.DataFrame(np.vstack([W, T, Y]).T, columns=['W', 'T', 'Y'])
df
plt.figure(figsize=(3, 3))
plt.scatter(df['T'], df['Y'], alpha=.2)
plt.show()
# Create the graph describing the causal structure
graph = """
graph [
directed 1
node [
id "T"
label "T"
]
node [
id "W"
label "W"
]
node [
id "Y"
label "Y"
]
edge [
source "W"
target "T"
]
edge [
source "W"
target "Y"
]
edge [
source "T"
target "Y"
]
]
"""
# Remove newlines
graph = graph.replace('\n', '')
# With graph
model = CausalModel(
data=df,
treatment='T',
outcome='Y',
graph=graph
)
plt.figure(figsize=(3, 3))
model.view_model()
plt.show()
estimand = model.identify_effect(proceed_when_unidentifiable=True)
print(estimand)
estimate = model.estimate_effect(
identified_estimand=estimand,
method_name='backdoor.linear_regression'
)
print(f'Estimate of causal effect (linear regression): {estimate.value}')
estimate = model.estimate_effect(
identified_estimand=estimand,
method_name='backdoor.econml.dml.DML',
method_params={
'init_params': {
'model_y': GradientBoostingRegressor(),
'model_t': GradientBoostingRegressor(),
'model_final': LassoCV(fit_intercept=False),
},
'fit_params': {}}
)
print(f'Estimate of causal effect (DML): {estimate.value}')
refute_results = model.refute_estimate(
estimand=estimand,
estimate=estimate,
method_name='placebo_treatment_refuter'
)
print(refute_results)
refute_results = model.refute_estimate(
estimand=estimand,
estimate=estimate,
method_name='random_common_cause'
)
print(refute_results)
| 0.577972 | 0.916147 |
### DEMDP04
# Job Search Model
Infinitely-lived worker must decide whether to quit, if employed, or search for a job, if unemployed, given prevailing market wages.
### States
- w prevailing wage
- i unemployed (0) or employed (1) at beginning of period
### Actions
- j idle (0) or active (i.e., work or search) (1) this period
### Parameters
| Parameter | Meaning |
|-----------|-------------------------|
| $v$ | benefit of pure leisure |
| $\bar{w}$ | long-run mean wage |
| $\gamma$ | wage reversion rate |
| $p_0$ | probability of finding job |
| $p_1$ | probability of keeping job |
| $\sigma$ | standard deviation of wage shock |
| $\delta$ | discount factor |
# Preliminary tasks
```
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from compecon import BasisSpline, DPmodel, qnwnorm, demo
```
## FORMULATION
### Worker's reward
The worker's reward is:
- $w$ (the prevailing wage rate), if he's employed and active (working)
- $u=90$, if he's unemployed but active (searching)
- $v=95$, if he's idle (quit if employed, not searching if unemployed)
```
u = 90
v = 95
def reward(w, x, employed, active):
if active:
return w.copy() if employed else np.full_like(w, u) # the copy is critical!!! otherwise it passes a pointer to w!!
else:
return np.full_like(w, v)
```
### Model dynamics
#### Stochastic Discrete State Transition Probabilities
An unemployed worker who is searching for a job has a probability $p_0=0.2$ of finding it, while an employed worker who doesn't want to quit his job has a probability $p_1 = 0.9$ of keeping it. An idle worker (someone who quits or doesn't search for a job) will definitely be unemployed next period. Thus, the transition probabilities are
\begin{align}
q = \begin{bmatrix}1-p_0 &p_0\\1-p_1&p_1\end{bmatrix},&\qquad\text{if active} \\
= \begin{bmatrix}1 & 0\\1 &0 \end{bmatrix},&\qquad\text{if iddle}
\end{align}
```
p0 = 0.20
p1 = 0.90
q = np.zeros((2, 2, 2))
q[1, 0, 1] = p0
q[1, 1, 1] = p1
q[:, :, 0] = 1 - q[:, :, 1]
```
#### Stochastic Continuous State Transition
Assuming that the wage rate $w$ follows an exogenous Markov process
\begin{equation}
w_{t+1} = \bar{w} + \gamma(w_t − \bar{w}) + \epsilon_{t+1}
\end{equation}
where $\bar{w}=100$ and $\gamma=0.4$.
```
wbar = 100
gamma = 0.40
def transition(w, x, i, j, in_, e):
return wbar + gamma * (w - wbar) + e
```
Here, $\epsilon$ is normal $(0,\sigma^2)$ wage shock, where $\sigma=5$. We discretize this distribution with the function ```qnwnorm```.
```
sigma = 5
m = 15
e, w = qnwnorm(m, 0, sigma ** 2)
```
### Approximation Structure
To discretize the continuous state variable, we use a cubic spline basis with $n=150$ nodes between $w_\min=0$ and $w_\max=200$.
```
n = 150
wmin = 0
wmax = 200
basis = BasisSpline(n, wmin, wmax, labels=['wage'])
```
## SOLUTION
To represent the model, we create an instance of ```DPmodel```. Here, we assume a discout factor of $\delta=0.95$.
```
model = DPmodel(basis, reward, transition,
i =['unemployed', 'employed'],
j = ['idle', 'active'],
discount=0.95, e=e, w=w, q=q)
```
Then, we call the method ```solve``` to solve the Bellman equation
```
S = model.solve(show=True)
S.head()
```
### Compute and Print Critical Action Wages
```
def critical(db):
wcrit = np.interp(0, db['value[active]'] - db['value[idle]'], db['wage'])
vcrit = np.interp(wcrit, db['wage'], db['value[idle]'])
return wcrit, vcrit
wcrit0, vcrit0 = critical(S.loc['unemployed'])
print(f'Critical Search Wage = {wcrit0:5.1f}')
wcrit1, vcrit1 = critical(S.loc['employed'])
print(f'Critical Quit Wage = {wcrit1:5.1f}')
```
### Plot Action-Contingent Value Function
```
vv = ['value[idle]','value[active]']
fig1 = plt.figure(figsize=[12,4])
# UNEMPLOYED
demo.subplot(1,2,1,'Action-Contingent Value, Unemployed', 'Wage', 'Value')
plt.plot(S.loc['unemployed',vv])
demo.annotate(wcrit0, vcrit0, f'$w^*_0 = {wcrit0:.1f}$', 'wo', (5, -5), fs=12)
plt.legend(['Do Not Search', 'Search'], loc='upper left')
# EMPLOYED
demo.subplot(1,2,2,'Action-Contingent Value, Employed', 'Wage', 'Value')
plt.plot(S.loc['employed',vv])
demo.annotate(wcrit1, vcrit1, f'$w^*_0 = {wcrit1:.1f}$', 'wo',(5, -5), fs=12)
plt.legend(['Quit', 'Work'], loc='upper left')
```
### Plot Residual
```
S['resid2'] = 100 * (S['resid'] / S['value'])
fig2 = demo.figure('Bellman Equation Residual', 'Wage', 'Percent Residual')
plt.plot(S.loc['unemployed','resid2'])
plt.plot(S.loc['employed','resid2'])
plt.legend(model.labels.i)
```
## SIMULATION
### Simulate Model
We simulate the model 10000 times for a time horizon $T=40$, starting with an unemployed worker ($i=0$) at the long-term wage rate mean $\bar{w}$. To be able to reproduce these results, we set the random seed at an arbitrary value of 945.
```
T = 40
nrep = 10000
sinit = np.full((1, nrep), wbar)
iinit = 0
data = model.simulate(T, sinit, iinit, seed=945)
data.head()
```
### Print Ergodic Moments
```
ff = '\t{:12s} = {:5.2f}'
print('\nErgodic Means')
print(ff.format('Wage', data['wage'].mean()))
print(ff.format('Employment', (data['i'] == 'employed').mean()))
print('\nErgodic Standard Deviations')
print(ff.format('Wage',data['wage'].std()))
print(ff.format('Employment', (data['i'] == 'employed').std()))
ergodic = pd.DataFrame({
'Ergodic Means' : [data['wage'].mean(), (data['i'] == 'employed').mean()],
'Ergodic Standard Deviations': [data['wage'].std(), (data['i'] == 'employed').std()]},
index=['Wage', 'Employment'])
ergodic.round(2)
```
### Plot Expected Discrete State Path
```
data.head()
data['ii'] = data['i'] == 'employed'
fig3 = demo.figure('Probability of Employment', 'Period','Probability')
plt.plot(data[['ii','time']].groupby('time').mean())
```
### Plot Simulated and Expected Continuous State Path
```
subdata = data[data['_rep'].isin(range(3))]
fig4 = demo.figure('Simulated and Expected Wage', 'Period', 'Wage')
plt.plot(subdata.pivot('time', '_rep', 'wage'))
plt.plot(data[['time','wage']].groupby('time').mean(),'k--',label='mean')
#demo.savefig([fig1,fig2,fig3,fig4])
```
|
github_jupyter
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from compecon import BasisSpline, DPmodel, qnwnorm, demo
u = 90
v = 95
def reward(w, x, employed, active):
if active:
return w.copy() if employed else np.full_like(w, u) # the copy is critical!!! otherwise it passes a pointer to w!!
else:
return np.full_like(w, v)
p0 = 0.20
p1 = 0.90
q = np.zeros((2, 2, 2))
q[1, 0, 1] = p0
q[1, 1, 1] = p1
q[:, :, 0] = 1 - q[:, :, 1]
wbar = 100
gamma = 0.40
def transition(w, x, i, j, in_, e):
return wbar + gamma * (w - wbar) + e
sigma = 5
m = 15
e, w = qnwnorm(m, 0, sigma ** 2)
n = 150
wmin = 0
wmax = 200
basis = BasisSpline(n, wmin, wmax, labels=['wage'])
model = DPmodel(basis, reward, transition,
i =['unemployed', 'employed'],
j = ['idle', 'active'],
discount=0.95, e=e, w=w, q=q)
S = model.solve(show=True)
S.head()
def critical(db):
wcrit = np.interp(0, db['value[active]'] - db['value[idle]'], db['wage'])
vcrit = np.interp(wcrit, db['wage'], db['value[idle]'])
return wcrit, vcrit
wcrit0, vcrit0 = critical(S.loc['unemployed'])
print(f'Critical Search Wage = {wcrit0:5.1f}')
wcrit1, vcrit1 = critical(S.loc['employed'])
print(f'Critical Quit Wage = {wcrit1:5.1f}')
vv = ['value[idle]','value[active]']
fig1 = plt.figure(figsize=[12,4])
# UNEMPLOYED
demo.subplot(1,2,1,'Action-Contingent Value, Unemployed', 'Wage', 'Value')
plt.plot(S.loc['unemployed',vv])
demo.annotate(wcrit0, vcrit0, f'$w^*_0 = {wcrit0:.1f}$', 'wo', (5, -5), fs=12)
plt.legend(['Do Not Search', 'Search'], loc='upper left')
# EMPLOYED
demo.subplot(1,2,2,'Action-Contingent Value, Employed', 'Wage', 'Value')
plt.plot(S.loc['employed',vv])
demo.annotate(wcrit1, vcrit1, f'$w^*_0 = {wcrit1:.1f}$', 'wo',(5, -5), fs=12)
plt.legend(['Quit', 'Work'], loc='upper left')
S['resid2'] = 100 * (S['resid'] / S['value'])
fig2 = demo.figure('Bellman Equation Residual', 'Wage', 'Percent Residual')
plt.plot(S.loc['unemployed','resid2'])
plt.plot(S.loc['employed','resid2'])
plt.legend(model.labels.i)
T = 40
nrep = 10000
sinit = np.full((1, nrep), wbar)
iinit = 0
data = model.simulate(T, sinit, iinit, seed=945)
data.head()
ff = '\t{:12s} = {:5.2f}'
print('\nErgodic Means')
print(ff.format('Wage', data['wage'].mean()))
print(ff.format('Employment', (data['i'] == 'employed').mean()))
print('\nErgodic Standard Deviations')
print(ff.format('Wage',data['wage'].std()))
print(ff.format('Employment', (data['i'] == 'employed').std()))
ergodic = pd.DataFrame({
'Ergodic Means' : [data['wage'].mean(), (data['i'] == 'employed').mean()],
'Ergodic Standard Deviations': [data['wage'].std(), (data['i'] == 'employed').std()]},
index=['Wage', 'Employment'])
ergodic.round(2)
data.head()
data['ii'] = data['i'] == 'employed'
fig3 = demo.figure('Probability of Employment', 'Period','Probability')
plt.plot(data[['ii','time']].groupby('time').mean())
subdata = data[data['_rep'].isin(range(3))]
fig4 = demo.figure('Simulated and Expected Wage', 'Period', 'Wage')
plt.plot(subdata.pivot('time', '_rep', 'wage'))
plt.plot(data[['time','wage']].groupby('time').mean(),'k--',label='mean')
#demo.savefig([fig1,fig2,fig3,fig4])
| 0.410284 | 0.873485 |
### Dependencies
```
from utillity_script_cloud_segmentation import *
seed = 0
seed_everything(seed)
warnings.filterwarnings("ignore")
from keras.callbacks import Callback
class LRFinder(Callback):
def __init__(self,
num_samples,
batch_size,
minimum_lr=1e-5,
maximum_lr=10.,
lr_scale='exp',
validation_data=None,
validation_sample_rate=5,
stopping_criterion_factor=4.,
loss_smoothing_beta=0.98,
save_dir=None,
verbose=True):
"""
This class uses the Cyclic Learning Rate history to find a
set of learning rates that can be good initializations for the
One-Cycle training proposed by Leslie Smith in the paper referenced
below.
A port of the Fast.ai implementation for Keras.
# Note
This requires that the model be trained for exactly 1 epoch. If the model
is trained for more epochs, then the metric calculations are only done for
the first epoch.
# Interpretation
Upon visualizing the loss plot, check where the loss starts to increase
rapidly. Choose a learning rate at somewhat prior to the corresponding
position in the plot for faster convergence. This will be the maximum_lr lr.
Choose the max value as this value when passing the `max_val` argument
to OneCycleLR callback.
Since the plot is in log-scale, you need to compute 10 ^ (-k) of the x-axis
# Arguments:
num_samples: Integer. Number of samples in the dataset.
batch_size: Integer. Batch size during training.
minimum_lr: Float. Initial learning rate (and the minimum).
maximum_lr: Float. Final learning rate (and the maximum).
lr_scale: Can be one of ['exp', 'linear']. Chooses the type of
scaling for each update to the learning rate during subsequent
batches. Choose 'exp' for large range and 'linear' for small range.
validation_data: Requires the validation dataset as a tuple of
(X, y) belonging to the validation set. If provided, will use the
validation set to compute the loss metrics. Else uses the training
batch loss. Will warn if not provided to alert the user.
validation_sample_rate: Positive or Negative Integer. Number of batches to sample from the
validation set per iteration of the LRFinder. Larger number of
samples will reduce the variance but will take longer time to execute
per batch.
If Positive > 0, will sample from the validation dataset
If Megative, will use the entire dataset
stopping_criterion_factor: Integer or None. A factor which is used
to measure large increase in the loss value during training.
Since callbacks cannot stop training of a model, it will simply
stop logging the additional values from the epochs after this
stopping criterion has been met.
If None, this check will not be performed.
loss_smoothing_beta: Float. The smoothing factor for the moving
average of the loss function.
save_dir: Optional, String. If passed a directory path, the callback
will save the running loss and learning rates to two separate numpy
arrays inside this directory. If the directory in this path does not
exist, they will be created.
verbose: Whether to print the learning rate after every batch of training.
# References:
- [A disciplined approach to neural network hyper-parameters: Part 1 -- learning rate, batch size, weight_decay, and weight decay](https://arxiv.org/abs/1803.09820)
"""
super(LRFinder, self).__init__()
if lr_scale not in ['exp', 'linear']:
raise ValueError("`lr_scale` must be one of ['exp', 'linear']")
if validation_data is not None:
self.validation_data = validation_data
self.use_validation_set = True
if validation_sample_rate > 0 or validation_sample_rate < 0:
self.validation_sample_rate = validation_sample_rate
else:
raise ValueError("`validation_sample_rate` must be a positive or negative integer other than o")
else:
self.use_validation_set = False
self.validation_sample_rate = 0
self.num_samples = num_samples
self.batch_size = batch_size
self.initial_lr = minimum_lr
self.final_lr = maximum_lr
self.lr_scale = lr_scale
self.stopping_criterion_factor = stopping_criterion_factor
self.loss_smoothing_beta = loss_smoothing_beta
self.save_dir = save_dir
self.verbose = verbose
self.num_batches_ = num_samples // batch_size
self.current_lr_ = minimum_lr
if lr_scale == 'exp':
self.lr_multiplier_ = (maximum_lr / float(minimum_lr)) ** (
1. / float(self.num_batches_))
else:
extra_batch = int((num_samples % batch_size) != 0)
self.lr_multiplier_ = np.linspace(
minimum_lr, maximum_lr, num=self.num_batches_ + extra_batch)
# If negative, use entire validation set
if self.validation_sample_rate < 0:
self.validation_sample_rate = self.validation_data[0].shape[0] // batch_size
self.current_batch_ = 0
self.current_epoch_ = 0
self.best_loss_ = 1e6
self.running_loss_ = 0.
self.history = {}
def on_train_begin(self, logs=None):
self.current_epoch_ = 1
K.set_value(self.model.optimizer.lr, self.initial_lr)
warnings.simplefilter("ignore")
def on_epoch_begin(self, epoch, logs=None):
self.current_batch_ = 0
if self.current_epoch_ > 1:
warnings.warn(
"\n\nLearning rate finder should be used only with a single epoch. "
"Hereafter, the callback will not measure the losses.\n\n")
def on_batch_begin(self, batch, logs=None):
self.current_batch_ += 1
def on_batch_end(self, batch, logs=None):
if self.current_epoch_ > 1:
return
if self.use_validation_set:
X, Y = self.validation_data[0], self.validation_data[1]
# use 5 random batches from test set for fast approximate of loss
num_samples = self.batch_size * self.validation_sample_rate
if num_samples > X.shape[0]:
num_samples = X.shape[0]
idx = np.random.choice(X.shape[0], num_samples, replace=False)
x = X[idx]
y = Y[idx]
values = self.model.evaluate(x, y, batch_size=self.batch_size, verbose=False)
loss = values[0]
else:
loss = logs['loss']
# smooth the loss value and bias correct
running_loss = self.loss_smoothing_beta * loss + (
1. - self.loss_smoothing_beta) * loss
running_loss = running_loss / (
1. - self.loss_smoothing_beta**self.current_batch_)
# stop logging if loss is too large
if self.current_batch_ > 1 and self.stopping_criterion_factor is not None and (
running_loss >
self.stopping_criterion_factor * self.best_loss_):
if self.verbose:
print(" - LRFinder: Skipping iteration since loss is %d times as large as best loss (%0.4f)"
% (self.stopping_criterion_factor, self.best_loss_))
return
if running_loss < self.best_loss_ or self.current_batch_ == 1:
self.best_loss_ = running_loss
current_lr = K.get_value(self.model.optimizer.lr)
self.history.setdefault('running_loss_', []).append(running_loss)
if self.lr_scale == 'exp':
self.history.setdefault('log_lrs', []).append(np.log10(current_lr))
else:
self.history.setdefault('log_lrs', []).append(current_lr)
# compute the lr for the next batch and update the optimizer lr
if self.lr_scale == 'exp':
current_lr *= self.lr_multiplier_
else:
current_lr = self.lr_multiplier_[self.current_batch_ - 1]
K.set_value(self.model.optimizer.lr, current_lr)
# save the other metrics as well
for k, v in logs.items():
self.history.setdefault(k, []).append(v)
if self.verbose:
if self.use_validation_set:
print(" - LRFinder: val_loss: %1.4f - lr = %1.8f " %
(values[0], current_lr))
else:
print(" - LRFinder: lr = %1.8f " % current_lr)
def on_epoch_end(self, epoch, logs=None):
if self.save_dir is not None and self.current_epoch_ <= 1:
if not os.path.exists(self.save_dir):
os.makedirs(self.save_dir)
losses_path = os.path.join(self.save_dir, 'losses.npy')
lrs_path = os.path.join(self.save_dir, 'lrs.npy')
np.save(losses_path, self.losses)
np.save(lrs_path, self.lrs)
if self.verbose:
print("\tLR Finder : Saved the losses and learning rate values in path : {%s}"
% (self.save_dir))
self.current_epoch_ += 1
warnings.simplefilter("default")
def plot_schedule(self, clip_beginning=None, clip_endding=None):
"""
Plots the schedule from the callback itself.
# Arguments:
clip_beginning: Integer or None. If positive integer, it will
remove the specified portion of the loss graph to remove the large
loss values in the beginning of the graph.
clip_endding: Integer or None. If negative integer, it will
remove the specified portion of the ending of the loss graph to
remove the sharp increase in the loss values at high learning rates.
"""
try:
import matplotlib.pyplot as plt
plt.style.use('seaborn-white')
except ImportError:
print(
"Matplotlib not found. Please use `pip install matplotlib` first."
)
return
if clip_beginning is not None and clip_beginning < 0:
clip_beginning = -clip_beginning
if clip_endding is not None and clip_endding > 0:
clip_endding = -clip_endding
losses = self.losses
lrs = self.lrs
if clip_beginning:
losses = losses[clip_beginning:]
lrs = lrs[clip_beginning:]
if clip_endding:
losses = losses[:clip_endding]
lrs = lrs[:clip_endding]
plt.plot(lrs, losses)
plt.title('Learning rate vs Loss')
plt.xlabel('learning rate')
plt.ylabel('loss')
plt.show()
@classmethod
def restore_schedule_from_dir(cls,
directory,
clip_beginning=None,
clip_endding=None):
"""
Loads the training history from the saved numpy files in the given directory.
# Arguments:
directory: String. Path to the directory where the serialized numpy
arrays of the loss and learning rates are saved.
clip_beginning: Integer or None. If positive integer, it will
remove the specified portion of the loss graph to remove the large
loss values in the beginning of the graph.
clip_endding: Integer or None. If negative integer, it will
remove the specified portion of the ending of the loss graph to
remove the sharp increase in the loss values at high learning rates.
Returns:
tuple of (losses, learning rates)
"""
if clip_beginning is not None and clip_beginning < 0:
clip_beginning = -clip_beginning
if clip_endding is not None and clip_endding > 0:
clip_endding = -clip_endding
losses_path = os.path.join(directory, 'losses.npy')
lrs_path = os.path.join(directory, 'lrs.npy')
if not os.path.exists(losses_path) or not os.path.exists(lrs_path):
print("%s and %s could not be found at directory : {%s}" %
(losses_path, lrs_path, directory))
losses = None
lrs = None
else:
losses = np.load(losses_path)
lrs = np.load(lrs_path)
if clip_beginning:
losses = losses[clip_beginning:]
lrs = lrs[clip_beginning:]
if clip_endding:
losses = losses[:clip_endding]
lrs = lrs[:clip_endding]
return losses, lrs
@classmethod
def plot_schedule_from_file(cls,
directory,
clip_beginning=None,
clip_endding=None):
"""
Plots the schedule from the saved numpy arrays of the loss and learning
rate values in the specified directory.
# Arguments:
directory: String. Path to the directory where the serialized numpy
arrays of the loss and learning rates are saved.
clip_beginning: Integer or None. If positive integer, it will
remove the specified portion of the loss graph to remove the large
loss values in the beginning of the graph.
clip_endding: Integer or None. If negative integer, it will
remove the specified portion of the ending of the loss graph to
remove the sharp increase in the loss values at high learning rates.
"""
try:
import matplotlib.pyplot as plt
plt.style.use('seaborn-white')
except ImportError:
print("Matplotlib not found. Please use `pip install matplotlib` first.")
return
losses, lrs = cls.restore_schedule_from_dir(
directory,
clip_beginning=clip_beginning,
clip_endding=clip_endding)
if losses is None or lrs is None:
return
else:
plt.plot(lrs, losses)
plt.title('Learning rate vs Loss')
plt.xlabel('learning rate')
plt.ylabel('loss')
plt.show()
@property
def lrs(self):
return np.array(self.history['log_lrs'])
@property
def losses(self):
return np.array(self.history['running_loss_'])
```
### Load data
```
train = pd.read_csv('../input/understanding_cloud_organization/train.csv')
hold_out_set = pd.read_csv('../input/cloud-data-split-v2/hold-out.csv')
X_train = hold_out_set[hold_out_set['set'] == 'train']
X_val = hold_out_set[hold_out_set['set'] == 'validation']
print('Compete set samples:', len(train))
print('Train samples: ', len(X_train))
print('Validation samples: ', len(X_val))
# Preprocecss data
train['image'] = train['Image_Label'].apply(lambda x: x.split('_')[0])
display(X_train.head())
```
# Model parameters
```
BACKBONE = 'efficientnetb3'
BATCH_SIZE = 8
EPOCHS_PT1 = 8
EPOCHS = 12
LEARNING_RATE = 10**(-1.7)
HEIGHT_PT1 = 256
WIDTH_PT1 = 384
HEIGHT = 320
WIDTH = 480
CHANNELS = 3
N_CLASSES = 4
STEP_SIZE_TRAIN = len(X_train)//BATCH_SIZE
STEP_SIZE_VALID = len(X_val)//BATCH_SIZE
model_path = '71-unet_%s_%sx%s.h5' % (BACKBONE, HEIGHT, WIDTH)
train_images_pt1_path = '../input/cloud-images-resized-256x384/train_images256x384/train_images/'
train_images_path = '../input/cloud-images-resized-320x480/train_images320x480/train_images/'
class OneCycleLR(Callback):
def __init__(self,
max_lr,
end_percentage=0.1,
scale_percentage=None,
maximum_momentum=0.95,
minimum_momentum=0.85,
verbose=True):
""" This callback implements a cyclical learning rate policy (CLR).
This is a special case of Cyclic Learning Rates, where we have only 1 cycle.
After the completion of 1 cycle, the learning rate will decrease rapidly to
100th its initial lowest value.
# Arguments:
max_lr: Float. Initial learning rate. This also sets the
starting learning rate (which will be 10x smaller than
this), and will increase to this value during the first cycle.
end_percentage: Float. The percentage of all the epochs of training
that will be dedicated to sharply decreasing the learning
rate after the completion of 1 cycle. Must be between 0 and 1.
scale_percentage: Float or None. If float, must be between 0 and 1.
If None, it will compute the scale_percentage automatically
based on the `end_percentage`.
maximum_momentum: Optional. Sets the maximum momentum (initial)
value, which gradually drops to its lowest value in half-cycle,
then gradually increases again to stay constant at this max value.
Can only be used with SGD Optimizer.
minimum_momentum: Optional. Sets the minimum momentum at the end of
the half-cycle. Can only be used with SGD Optimizer.
verbose: Bool. Whether to print the current learning rate after every
epoch.
# Reference
- [A disciplined approach to neural network hyper-parameters: Part 1 -- learning rate, batch size, weight_decay, and weight decay](https://arxiv.org/abs/1803.09820)
- [Super-Convergence: Very Fast Training of Residual Networks Using Large Learning Rates](https://arxiv.org/abs/1708.07120)
"""
super(OneCycleLR, self).__init__()
if end_percentage < 0. or end_percentage > 1.:
raise ValueError("`end_percentage` must be between 0 and 1")
if scale_percentage is not None and (scale_percentage < 0. or scale_percentage > 1.):
raise ValueError("`scale_percentage` must be between 0 and 1")
self.initial_lr = max_lr
self.end_percentage = end_percentage
self.scale = float(scale_percentage) if scale_percentage is not None else float(end_percentage)
self.max_momentum = maximum_momentum
self.min_momentum = minimum_momentum
self.verbose = verbose
if self.max_momentum is not None and self.min_momentum is not None:
self._update_momentum = True
else:
self._update_momentum = False
self.clr_iterations = 0.
self.history = {}
self.epochs = None
self.batch_size = None
self.samples = None
self.steps = None
self.num_iterations = None
self.mid_cycle_id = None
def _reset(self):
"""
Reset the callback.
"""
self.clr_iterations = 0.
self.history = {}
def compute_lr(self):
"""
Compute the learning rate based on which phase of the cycle it is in.
- If in the first half of training, the learning rate gradually increases.
- If in the second half of training, the learning rate gradually decreases.
- If in the final `end_percentage` portion of training, the learning rate
is quickly reduced to near 100th of the original min learning rate.
# Returns:
the new learning rate
"""
if self.clr_iterations > 2 * self.mid_cycle_id:
current_percentage = (self.clr_iterations - 2 * self.mid_cycle_id)
current_percentage /= float((self.num_iterations - 2 * self.mid_cycle_id))
new_lr = self.initial_lr * (1. + (current_percentage *
(1. - 100.) / 100.)) * self.scale
elif self.clr_iterations > self.mid_cycle_id:
current_percentage = 1. - (
self.clr_iterations - self.mid_cycle_id) / self.mid_cycle_id
new_lr = self.initial_lr * (1. + current_percentage *
(self.scale * 100 - 1.)) * self.scale
else:
current_percentage = self.clr_iterations / self.mid_cycle_id
new_lr = self.initial_lr * (1. + current_percentage *
(self.scale * 100 - 1.)) * self.scale
if self.clr_iterations == self.num_iterations:
self.clr_iterations = 0
return new_lr
def compute_momentum(self):
"""
Compute the momentum based on which phase of the cycle it is in.
- If in the first half of training, the momentum gradually decreases.
- If in the second half of training, the momentum gradually increases.
- If in the final `end_percentage` portion of training, the momentum value
is kept constant at the maximum initial value.
# Returns:
the new momentum value
"""
if self.clr_iterations > 2 * self.mid_cycle_id:
new_momentum = self.max_momentum
elif self.clr_iterations > self.mid_cycle_id:
current_percentage = 1. - ((self.clr_iterations - self.mid_cycle_id) / float(
self.mid_cycle_id))
new_momentum = self.max_momentum - current_percentage * (
self.max_momentum - self.min_momentum)
else:
current_percentage = self.clr_iterations / float(self.mid_cycle_id)
new_momentum = self.max_momentum - current_percentage * (
self.max_momentum - self.min_momentum)
return new_momentum
def on_train_begin(self, logs={}):
logs = logs or {}
# self.epochs = self.params['epochs']
# self.batch_size = self.params['batch_size']
# self.samples = self.params['samples']
# self.steps = self.params['steps']
self.epochs = EPOCHS
self.batch_size = BATCH_SIZE
self.samples = len(X_train)
self.steps = len(X_train)//BATCH_SIZE
if self.steps is not None:
self.num_iterations = self.epochs * self.steps
else:
if (self.samples % self.batch_size) == 0:
remainder = 0
else:
remainder = 1
self.num_iterations = (self.epochs + remainder) * self.samples // self.batch_size
self.mid_cycle_id = int(self.num_iterations * ((1. - self.end_percentage)) / float(2))
self._reset()
K.set_value(self.model.optimizer.lr, self.compute_lr())
if self._update_momentum:
if not hasattr(self.model.optimizer, 'momentum'):
raise ValueError("Momentum can be updated only on SGD optimizer !")
new_momentum = self.compute_momentum()
K.set_value(self.model.optimizer.momentum, new_momentum)
def on_batch_end(self, epoch, logs=None):
logs = logs or {}
self.clr_iterations += 1
new_lr = self.compute_lr()
self.history.setdefault('lr', []).append(
K.get_value(self.model.optimizer.lr))
K.set_value(self.model.optimizer.lr, new_lr)
if self._update_momentum:
if not hasattr(self.model.optimizer, 'momentum'):
raise ValueError("Momentum can be updated only on SGD optimizer !")
new_momentum = self.compute_momentum()
self.history.setdefault('momentum', []).append(
K.get_value(self.model.optimizer.momentum))
K.set_value(self.model.optimizer.momentum, new_momentum)
for k, v in logs.items():
self.history.setdefault(k, []).append(v)
def on_epoch_end(self, epoch, logs=None):
if self.verbose:
if self._update_momentum:
print(" - lr: %0.5f - momentum: %0.2f " %
(self.history['lr'][-1], self.history['momentum'][-1]))
else:
print(" - lr: %0.5f " % (self.history['lr'][-1]))
preprocessing = sm.get_preprocessing(BACKBONE)
augmentation = albu.Compose([albu.HorizontalFlip(p=0.5),
albu.VerticalFlip(p=0.5),
albu.ShiftScaleRotate(scale_limit=0.5, rotate_limit=0,
shift_limit=0.1, border_mode=0, p=0.5)
])
```
### Data generator
```
train_generator_pt1 = DataGenerator(
directory=train_images_pt1_path,
dataframe=X_train,
target_df=train,
batch_size=BATCH_SIZE,
target_size=(HEIGHT_PT1, WIDTH_PT1),
n_channels=CHANNELS,
n_classes=N_CLASSES,
preprocessing=preprocessing,
augmentation=augmentation,
seed=seed)
valid_generator_pt1 = DataGenerator(
directory=train_images_pt1_path,
dataframe=X_val,
target_df=train,
batch_size=BATCH_SIZE,
target_size=(HEIGHT_PT1, WIDTH_PT1),
n_channels=CHANNELS,
n_classes=N_CLASSES,
preprocessing=preprocessing,
seed=seed)
train_generator = DataGenerator(
directory=train_images_path,
dataframe=X_train,
target_df=train,
batch_size=BATCH_SIZE,
target_size=(HEIGHT, WIDTH),
n_channels=CHANNELS,
n_classes=N_CLASSES,
preprocessing=preprocessing,
augmentation=augmentation,
seed=seed)
valid_generator = DataGenerator(
directory=train_images_path,
dataframe=X_val,
target_df=train,
batch_size=BATCH_SIZE,
target_size=(HEIGHT, WIDTH),
n_channels=CHANNELS,
n_classes=N_CLASSES,
preprocessing=preprocessing,
seed=seed)
```
# Learning rate finder
```
model = sm.Unet(backbone_name=BACKBONE,
encoder_weights='imagenet',
classes=N_CLASSES,
activation='sigmoid',
input_shape=(None, None, CHANNELS))
lr_finder = LRFinder(num_samples=len(X_train), batch_size=BATCH_SIZE, minimum_lr=1e-5, maximum_lr=10, verbose=0)
optimizer = optimizers.SGD(lr=LEARNING_RATE, momentum=0.9, nesterov=True)
model.compile(optimizer=optimizer, loss=sm.losses.bce_dice_loss)
history = model.fit_generator(generator=train_generator,
steps_per_epoch=STEP_SIZE_TRAIN,
epochs=1,
callbacks=[lr_finder])
plt.figure(figsize=(30, 10))
plt.axvline(x=LEARNING_RATE, color='red')
lr_finder.plot_schedule(clip_beginning=15)
```
# Model PT 1
```
model = sm.Unet(backbone_name=BACKBONE,
encoder_weights='imagenet',
classes=N_CLASSES,
activation='sigmoid',
encoder_freeze=True,
input_shape=(None, None, CHANNELS))
oneCycleLR = OneCycleLR(max_lr=LEARNING_RATE, maximum_momentum=0.9, minimum_momentum=0.9)
metric_list = [dice_coef, sm.metrics.iou_score, sm.metrics.f1_score]
callback_list = [oneCycleLR]
optimizer = optimizers.SGD(lr=LEARNING_RATE, momentum=0.9, nesterov=True)
model.compile(optimizer=optimizer, loss=sm.losses.bce_dice_loss, metrics=metric_list)
model.summary()
history = model.fit_generator(generator=train_generator_pt1,
steps_per_epoch=STEP_SIZE_TRAIN,
validation_data=valid_generator_pt1,
validation_steps=STEP_SIZE_VALID,
callbacks=callback_list,
epochs=EPOCHS_PT1,
verbose=2).history
```
# Model
```
for layer in model.layers:
layer.trainable = True
checkpoint = ModelCheckpoint(model_path, monitor='val_loss', mode='min', save_best_only=True)
oneCycleLR = OneCycleLR(max_lr=LEARNING_RATE, maximum_momentum=0.9, minimum_momentum=0.9)
metric_list = [dice_coef, sm.metrics.iou_score, sm.metrics.f1_score]
callback_list = [checkpoint, oneCycleLR]
optimizer = optimizers.SGD(lr=LEARNING_RATE, momentum=0.9, nesterov=True)
model.compile(optimizer=optimizer, loss=sm.losses.bce_dice_loss, metrics=metric_list)
model.summary()
history = model.fit_generator(generator=train_generator,
steps_per_epoch=STEP_SIZE_TRAIN,
validation_data=valid_generator,
validation_steps=STEP_SIZE_VALID,
callbacks=callback_list,
epochs=EPOCHS,
verbose=2).history
```
## Model loss graph
```
plot_metrics(history, metric_list=['loss', 'dice_coef', 'iou_score', 'f1-score'])
```
|
github_jupyter
|
from utillity_script_cloud_segmentation import *
seed = 0
seed_everything(seed)
warnings.filterwarnings("ignore")
from keras.callbacks import Callback
class LRFinder(Callback):
def __init__(self,
num_samples,
batch_size,
minimum_lr=1e-5,
maximum_lr=10.,
lr_scale='exp',
validation_data=None,
validation_sample_rate=5,
stopping_criterion_factor=4.,
loss_smoothing_beta=0.98,
save_dir=None,
verbose=True):
"""
This class uses the Cyclic Learning Rate history to find a
set of learning rates that can be good initializations for the
One-Cycle training proposed by Leslie Smith in the paper referenced
below.
A port of the Fast.ai implementation for Keras.
# Note
This requires that the model be trained for exactly 1 epoch. If the model
is trained for more epochs, then the metric calculations are only done for
the first epoch.
# Interpretation
Upon visualizing the loss plot, check where the loss starts to increase
rapidly. Choose a learning rate at somewhat prior to the corresponding
position in the plot for faster convergence. This will be the maximum_lr lr.
Choose the max value as this value when passing the `max_val` argument
to OneCycleLR callback.
Since the plot is in log-scale, you need to compute 10 ^ (-k) of the x-axis
# Arguments:
num_samples: Integer. Number of samples in the dataset.
batch_size: Integer. Batch size during training.
minimum_lr: Float. Initial learning rate (and the minimum).
maximum_lr: Float. Final learning rate (and the maximum).
lr_scale: Can be one of ['exp', 'linear']. Chooses the type of
scaling for each update to the learning rate during subsequent
batches. Choose 'exp' for large range and 'linear' for small range.
validation_data: Requires the validation dataset as a tuple of
(X, y) belonging to the validation set. If provided, will use the
validation set to compute the loss metrics. Else uses the training
batch loss. Will warn if not provided to alert the user.
validation_sample_rate: Positive or Negative Integer. Number of batches to sample from the
validation set per iteration of the LRFinder. Larger number of
samples will reduce the variance but will take longer time to execute
per batch.
If Positive > 0, will sample from the validation dataset
If Megative, will use the entire dataset
stopping_criterion_factor: Integer or None. A factor which is used
to measure large increase in the loss value during training.
Since callbacks cannot stop training of a model, it will simply
stop logging the additional values from the epochs after this
stopping criterion has been met.
If None, this check will not be performed.
loss_smoothing_beta: Float. The smoothing factor for the moving
average of the loss function.
save_dir: Optional, String. If passed a directory path, the callback
will save the running loss and learning rates to two separate numpy
arrays inside this directory. If the directory in this path does not
exist, they will be created.
verbose: Whether to print the learning rate after every batch of training.
# References:
- [A disciplined approach to neural network hyper-parameters: Part 1 -- learning rate, batch size, weight_decay, and weight decay](https://arxiv.org/abs/1803.09820)
"""
super(LRFinder, self).__init__()
if lr_scale not in ['exp', 'linear']:
raise ValueError("`lr_scale` must be one of ['exp', 'linear']")
if validation_data is not None:
self.validation_data = validation_data
self.use_validation_set = True
if validation_sample_rate > 0 or validation_sample_rate < 0:
self.validation_sample_rate = validation_sample_rate
else:
raise ValueError("`validation_sample_rate` must be a positive or negative integer other than o")
else:
self.use_validation_set = False
self.validation_sample_rate = 0
self.num_samples = num_samples
self.batch_size = batch_size
self.initial_lr = minimum_lr
self.final_lr = maximum_lr
self.lr_scale = lr_scale
self.stopping_criterion_factor = stopping_criterion_factor
self.loss_smoothing_beta = loss_smoothing_beta
self.save_dir = save_dir
self.verbose = verbose
self.num_batches_ = num_samples // batch_size
self.current_lr_ = minimum_lr
if lr_scale == 'exp':
self.lr_multiplier_ = (maximum_lr / float(minimum_lr)) ** (
1. / float(self.num_batches_))
else:
extra_batch = int((num_samples % batch_size) != 0)
self.lr_multiplier_ = np.linspace(
minimum_lr, maximum_lr, num=self.num_batches_ + extra_batch)
# If negative, use entire validation set
if self.validation_sample_rate < 0:
self.validation_sample_rate = self.validation_data[0].shape[0] // batch_size
self.current_batch_ = 0
self.current_epoch_ = 0
self.best_loss_ = 1e6
self.running_loss_ = 0.
self.history = {}
def on_train_begin(self, logs=None):
self.current_epoch_ = 1
K.set_value(self.model.optimizer.lr, self.initial_lr)
warnings.simplefilter("ignore")
def on_epoch_begin(self, epoch, logs=None):
self.current_batch_ = 0
if self.current_epoch_ > 1:
warnings.warn(
"\n\nLearning rate finder should be used only with a single epoch. "
"Hereafter, the callback will not measure the losses.\n\n")
def on_batch_begin(self, batch, logs=None):
self.current_batch_ += 1
def on_batch_end(self, batch, logs=None):
if self.current_epoch_ > 1:
return
if self.use_validation_set:
X, Y = self.validation_data[0], self.validation_data[1]
# use 5 random batches from test set for fast approximate of loss
num_samples = self.batch_size * self.validation_sample_rate
if num_samples > X.shape[0]:
num_samples = X.shape[0]
idx = np.random.choice(X.shape[0], num_samples, replace=False)
x = X[idx]
y = Y[idx]
values = self.model.evaluate(x, y, batch_size=self.batch_size, verbose=False)
loss = values[0]
else:
loss = logs['loss']
# smooth the loss value and bias correct
running_loss = self.loss_smoothing_beta * loss + (
1. - self.loss_smoothing_beta) * loss
running_loss = running_loss / (
1. - self.loss_smoothing_beta**self.current_batch_)
# stop logging if loss is too large
if self.current_batch_ > 1 and self.stopping_criterion_factor is not None and (
running_loss >
self.stopping_criterion_factor * self.best_loss_):
if self.verbose:
print(" - LRFinder: Skipping iteration since loss is %d times as large as best loss (%0.4f)"
% (self.stopping_criterion_factor, self.best_loss_))
return
if running_loss < self.best_loss_ or self.current_batch_ == 1:
self.best_loss_ = running_loss
current_lr = K.get_value(self.model.optimizer.lr)
self.history.setdefault('running_loss_', []).append(running_loss)
if self.lr_scale == 'exp':
self.history.setdefault('log_lrs', []).append(np.log10(current_lr))
else:
self.history.setdefault('log_lrs', []).append(current_lr)
# compute the lr for the next batch and update the optimizer lr
if self.lr_scale == 'exp':
current_lr *= self.lr_multiplier_
else:
current_lr = self.lr_multiplier_[self.current_batch_ - 1]
K.set_value(self.model.optimizer.lr, current_lr)
# save the other metrics as well
for k, v in logs.items():
self.history.setdefault(k, []).append(v)
if self.verbose:
if self.use_validation_set:
print(" - LRFinder: val_loss: %1.4f - lr = %1.8f " %
(values[0], current_lr))
else:
print(" - LRFinder: lr = %1.8f " % current_lr)
def on_epoch_end(self, epoch, logs=None):
if self.save_dir is not None and self.current_epoch_ <= 1:
if not os.path.exists(self.save_dir):
os.makedirs(self.save_dir)
losses_path = os.path.join(self.save_dir, 'losses.npy')
lrs_path = os.path.join(self.save_dir, 'lrs.npy')
np.save(losses_path, self.losses)
np.save(lrs_path, self.lrs)
if self.verbose:
print("\tLR Finder : Saved the losses and learning rate values in path : {%s}"
% (self.save_dir))
self.current_epoch_ += 1
warnings.simplefilter("default")
def plot_schedule(self, clip_beginning=None, clip_endding=None):
"""
Plots the schedule from the callback itself.
# Arguments:
clip_beginning: Integer or None. If positive integer, it will
remove the specified portion of the loss graph to remove the large
loss values in the beginning of the graph.
clip_endding: Integer or None. If negative integer, it will
remove the specified portion of the ending of the loss graph to
remove the sharp increase in the loss values at high learning rates.
"""
try:
import matplotlib.pyplot as plt
plt.style.use('seaborn-white')
except ImportError:
print(
"Matplotlib not found. Please use `pip install matplotlib` first."
)
return
if clip_beginning is not None and clip_beginning < 0:
clip_beginning = -clip_beginning
if clip_endding is not None and clip_endding > 0:
clip_endding = -clip_endding
losses = self.losses
lrs = self.lrs
if clip_beginning:
losses = losses[clip_beginning:]
lrs = lrs[clip_beginning:]
if clip_endding:
losses = losses[:clip_endding]
lrs = lrs[:clip_endding]
plt.plot(lrs, losses)
plt.title('Learning rate vs Loss')
plt.xlabel('learning rate')
plt.ylabel('loss')
plt.show()
@classmethod
def restore_schedule_from_dir(cls,
directory,
clip_beginning=None,
clip_endding=None):
"""
Loads the training history from the saved numpy files in the given directory.
# Arguments:
directory: String. Path to the directory where the serialized numpy
arrays of the loss and learning rates are saved.
clip_beginning: Integer or None. If positive integer, it will
remove the specified portion of the loss graph to remove the large
loss values in the beginning of the graph.
clip_endding: Integer or None. If negative integer, it will
remove the specified portion of the ending of the loss graph to
remove the sharp increase in the loss values at high learning rates.
Returns:
tuple of (losses, learning rates)
"""
if clip_beginning is not None and clip_beginning < 0:
clip_beginning = -clip_beginning
if clip_endding is not None and clip_endding > 0:
clip_endding = -clip_endding
losses_path = os.path.join(directory, 'losses.npy')
lrs_path = os.path.join(directory, 'lrs.npy')
if not os.path.exists(losses_path) or not os.path.exists(lrs_path):
print("%s and %s could not be found at directory : {%s}" %
(losses_path, lrs_path, directory))
losses = None
lrs = None
else:
losses = np.load(losses_path)
lrs = np.load(lrs_path)
if clip_beginning:
losses = losses[clip_beginning:]
lrs = lrs[clip_beginning:]
if clip_endding:
losses = losses[:clip_endding]
lrs = lrs[:clip_endding]
return losses, lrs
@classmethod
def plot_schedule_from_file(cls,
directory,
clip_beginning=None,
clip_endding=None):
"""
Plots the schedule from the saved numpy arrays of the loss and learning
rate values in the specified directory.
# Arguments:
directory: String. Path to the directory where the serialized numpy
arrays of the loss and learning rates are saved.
clip_beginning: Integer or None. If positive integer, it will
remove the specified portion of the loss graph to remove the large
loss values in the beginning of the graph.
clip_endding: Integer or None. If negative integer, it will
remove the specified portion of the ending of the loss graph to
remove the sharp increase in the loss values at high learning rates.
"""
try:
import matplotlib.pyplot as plt
plt.style.use('seaborn-white')
except ImportError:
print("Matplotlib not found. Please use `pip install matplotlib` first.")
return
losses, lrs = cls.restore_schedule_from_dir(
directory,
clip_beginning=clip_beginning,
clip_endding=clip_endding)
if losses is None or lrs is None:
return
else:
plt.plot(lrs, losses)
plt.title('Learning rate vs Loss')
plt.xlabel('learning rate')
plt.ylabel('loss')
plt.show()
@property
def lrs(self):
return np.array(self.history['log_lrs'])
@property
def losses(self):
return np.array(self.history['running_loss_'])
train = pd.read_csv('../input/understanding_cloud_organization/train.csv')
hold_out_set = pd.read_csv('../input/cloud-data-split-v2/hold-out.csv')
X_train = hold_out_set[hold_out_set['set'] == 'train']
X_val = hold_out_set[hold_out_set['set'] == 'validation']
print('Compete set samples:', len(train))
print('Train samples: ', len(X_train))
print('Validation samples: ', len(X_val))
# Preprocecss data
train['image'] = train['Image_Label'].apply(lambda x: x.split('_')[0])
display(X_train.head())
BACKBONE = 'efficientnetb3'
BATCH_SIZE = 8
EPOCHS_PT1 = 8
EPOCHS = 12
LEARNING_RATE = 10**(-1.7)
HEIGHT_PT1 = 256
WIDTH_PT1 = 384
HEIGHT = 320
WIDTH = 480
CHANNELS = 3
N_CLASSES = 4
STEP_SIZE_TRAIN = len(X_train)//BATCH_SIZE
STEP_SIZE_VALID = len(X_val)//BATCH_SIZE
model_path = '71-unet_%s_%sx%s.h5' % (BACKBONE, HEIGHT, WIDTH)
train_images_pt1_path = '../input/cloud-images-resized-256x384/train_images256x384/train_images/'
train_images_path = '../input/cloud-images-resized-320x480/train_images320x480/train_images/'
class OneCycleLR(Callback):
def __init__(self,
max_lr,
end_percentage=0.1,
scale_percentage=None,
maximum_momentum=0.95,
minimum_momentum=0.85,
verbose=True):
""" This callback implements a cyclical learning rate policy (CLR).
This is a special case of Cyclic Learning Rates, where we have only 1 cycle.
After the completion of 1 cycle, the learning rate will decrease rapidly to
100th its initial lowest value.
# Arguments:
max_lr: Float. Initial learning rate. This also sets the
starting learning rate (which will be 10x smaller than
this), and will increase to this value during the first cycle.
end_percentage: Float. The percentage of all the epochs of training
that will be dedicated to sharply decreasing the learning
rate after the completion of 1 cycle. Must be between 0 and 1.
scale_percentage: Float or None. If float, must be between 0 and 1.
If None, it will compute the scale_percentage automatically
based on the `end_percentage`.
maximum_momentum: Optional. Sets the maximum momentum (initial)
value, which gradually drops to its lowest value in half-cycle,
then gradually increases again to stay constant at this max value.
Can only be used with SGD Optimizer.
minimum_momentum: Optional. Sets the minimum momentum at the end of
the half-cycle. Can only be used with SGD Optimizer.
verbose: Bool. Whether to print the current learning rate after every
epoch.
# Reference
- [A disciplined approach to neural network hyper-parameters: Part 1 -- learning rate, batch size, weight_decay, and weight decay](https://arxiv.org/abs/1803.09820)
- [Super-Convergence: Very Fast Training of Residual Networks Using Large Learning Rates](https://arxiv.org/abs/1708.07120)
"""
super(OneCycleLR, self).__init__()
if end_percentage < 0. or end_percentage > 1.:
raise ValueError("`end_percentage` must be between 0 and 1")
if scale_percentage is not None and (scale_percentage < 0. or scale_percentage > 1.):
raise ValueError("`scale_percentage` must be between 0 and 1")
self.initial_lr = max_lr
self.end_percentage = end_percentage
self.scale = float(scale_percentage) if scale_percentage is not None else float(end_percentage)
self.max_momentum = maximum_momentum
self.min_momentum = minimum_momentum
self.verbose = verbose
if self.max_momentum is not None and self.min_momentum is not None:
self._update_momentum = True
else:
self._update_momentum = False
self.clr_iterations = 0.
self.history = {}
self.epochs = None
self.batch_size = None
self.samples = None
self.steps = None
self.num_iterations = None
self.mid_cycle_id = None
def _reset(self):
"""
Reset the callback.
"""
self.clr_iterations = 0.
self.history = {}
def compute_lr(self):
"""
Compute the learning rate based on which phase of the cycle it is in.
- If in the first half of training, the learning rate gradually increases.
- If in the second half of training, the learning rate gradually decreases.
- If in the final `end_percentage` portion of training, the learning rate
is quickly reduced to near 100th of the original min learning rate.
# Returns:
the new learning rate
"""
if self.clr_iterations > 2 * self.mid_cycle_id:
current_percentage = (self.clr_iterations - 2 * self.mid_cycle_id)
current_percentage /= float((self.num_iterations - 2 * self.mid_cycle_id))
new_lr = self.initial_lr * (1. + (current_percentage *
(1. - 100.) / 100.)) * self.scale
elif self.clr_iterations > self.mid_cycle_id:
current_percentage = 1. - (
self.clr_iterations - self.mid_cycle_id) / self.mid_cycle_id
new_lr = self.initial_lr * (1. + current_percentage *
(self.scale * 100 - 1.)) * self.scale
else:
current_percentage = self.clr_iterations / self.mid_cycle_id
new_lr = self.initial_lr * (1. + current_percentage *
(self.scale * 100 - 1.)) * self.scale
if self.clr_iterations == self.num_iterations:
self.clr_iterations = 0
return new_lr
def compute_momentum(self):
"""
Compute the momentum based on which phase of the cycle it is in.
- If in the first half of training, the momentum gradually decreases.
- If in the second half of training, the momentum gradually increases.
- If in the final `end_percentage` portion of training, the momentum value
is kept constant at the maximum initial value.
# Returns:
the new momentum value
"""
if self.clr_iterations > 2 * self.mid_cycle_id:
new_momentum = self.max_momentum
elif self.clr_iterations > self.mid_cycle_id:
current_percentage = 1. - ((self.clr_iterations - self.mid_cycle_id) / float(
self.mid_cycle_id))
new_momentum = self.max_momentum - current_percentage * (
self.max_momentum - self.min_momentum)
else:
current_percentage = self.clr_iterations / float(self.mid_cycle_id)
new_momentum = self.max_momentum - current_percentage * (
self.max_momentum - self.min_momentum)
return new_momentum
def on_train_begin(self, logs={}):
logs = logs or {}
# self.epochs = self.params['epochs']
# self.batch_size = self.params['batch_size']
# self.samples = self.params['samples']
# self.steps = self.params['steps']
self.epochs = EPOCHS
self.batch_size = BATCH_SIZE
self.samples = len(X_train)
self.steps = len(X_train)//BATCH_SIZE
if self.steps is not None:
self.num_iterations = self.epochs * self.steps
else:
if (self.samples % self.batch_size) == 0:
remainder = 0
else:
remainder = 1
self.num_iterations = (self.epochs + remainder) * self.samples // self.batch_size
self.mid_cycle_id = int(self.num_iterations * ((1. - self.end_percentage)) / float(2))
self._reset()
K.set_value(self.model.optimizer.lr, self.compute_lr())
if self._update_momentum:
if not hasattr(self.model.optimizer, 'momentum'):
raise ValueError("Momentum can be updated only on SGD optimizer !")
new_momentum = self.compute_momentum()
K.set_value(self.model.optimizer.momentum, new_momentum)
def on_batch_end(self, epoch, logs=None):
logs = logs or {}
self.clr_iterations += 1
new_lr = self.compute_lr()
self.history.setdefault('lr', []).append(
K.get_value(self.model.optimizer.lr))
K.set_value(self.model.optimizer.lr, new_lr)
if self._update_momentum:
if not hasattr(self.model.optimizer, 'momentum'):
raise ValueError("Momentum can be updated only on SGD optimizer !")
new_momentum = self.compute_momentum()
self.history.setdefault('momentum', []).append(
K.get_value(self.model.optimizer.momentum))
K.set_value(self.model.optimizer.momentum, new_momentum)
for k, v in logs.items():
self.history.setdefault(k, []).append(v)
def on_epoch_end(self, epoch, logs=None):
if self.verbose:
if self._update_momentum:
print(" - lr: %0.5f - momentum: %0.2f " %
(self.history['lr'][-1], self.history['momentum'][-1]))
else:
print(" - lr: %0.5f " % (self.history['lr'][-1]))
preprocessing = sm.get_preprocessing(BACKBONE)
augmentation = albu.Compose([albu.HorizontalFlip(p=0.5),
albu.VerticalFlip(p=0.5),
albu.ShiftScaleRotate(scale_limit=0.5, rotate_limit=0,
shift_limit=0.1, border_mode=0, p=0.5)
])
train_generator_pt1 = DataGenerator(
directory=train_images_pt1_path,
dataframe=X_train,
target_df=train,
batch_size=BATCH_SIZE,
target_size=(HEIGHT_PT1, WIDTH_PT1),
n_channels=CHANNELS,
n_classes=N_CLASSES,
preprocessing=preprocessing,
augmentation=augmentation,
seed=seed)
valid_generator_pt1 = DataGenerator(
directory=train_images_pt1_path,
dataframe=X_val,
target_df=train,
batch_size=BATCH_SIZE,
target_size=(HEIGHT_PT1, WIDTH_PT1),
n_channels=CHANNELS,
n_classes=N_CLASSES,
preprocessing=preprocessing,
seed=seed)
train_generator = DataGenerator(
directory=train_images_path,
dataframe=X_train,
target_df=train,
batch_size=BATCH_SIZE,
target_size=(HEIGHT, WIDTH),
n_channels=CHANNELS,
n_classes=N_CLASSES,
preprocessing=preprocessing,
augmentation=augmentation,
seed=seed)
valid_generator = DataGenerator(
directory=train_images_path,
dataframe=X_val,
target_df=train,
batch_size=BATCH_SIZE,
target_size=(HEIGHT, WIDTH),
n_channels=CHANNELS,
n_classes=N_CLASSES,
preprocessing=preprocessing,
seed=seed)
model = sm.Unet(backbone_name=BACKBONE,
encoder_weights='imagenet',
classes=N_CLASSES,
activation='sigmoid',
input_shape=(None, None, CHANNELS))
lr_finder = LRFinder(num_samples=len(X_train), batch_size=BATCH_SIZE, minimum_lr=1e-5, maximum_lr=10, verbose=0)
optimizer = optimizers.SGD(lr=LEARNING_RATE, momentum=0.9, nesterov=True)
model.compile(optimizer=optimizer, loss=sm.losses.bce_dice_loss)
history = model.fit_generator(generator=train_generator,
steps_per_epoch=STEP_SIZE_TRAIN,
epochs=1,
callbacks=[lr_finder])
plt.figure(figsize=(30, 10))
plt.axvline(x=LEARNING_RATE, color='red')
lr_finder.plot_schedule(clip_beginning=15)
model = sm.Unet(backbone_name=BACKBONE,
encoder_weights='imagenet',
classes=N_CLASSES,
activation='sigmoid',
encoder_freeze=True,
input_shape=(None, None, CHANNELS))
oneCycleLR = OneCycleLR(max_lr=LEARNING_RATE, maximum_momentum=0.9, minimum_momentum=0.9)
metric_list = [dice_coef, sm.metrics.iou_score, sm.metrics.f1_score]
callback_list = [oneCycleLR]
optimizer = optimizers.SGD(lr=LEARNING_RATE, momentum=0.9, nesterov=True)
model.compile(optimizer=optimizer, loss=sm.losses.bce_dice_loss, metrics=metric_list)
model.summary()
history = model.fit_generator(generator=train_generator_pt1,
steps_per_epoch=STEP_SIZE_TRAIN,
validation_data=valid_generator_pt1,
validation_steps=STEP_SIZE_VALID,
callbacks=callback_list,
epochs=EPOCHS_PT1,
verbose=2).history
for layer in model.layers:
layer.trainable = True
checkpoint = ModelCheckpoint(model_path, monitor='val_loss', mode='min', save_best_only=True)
oneCycleLR = OneCycleLR(max_lr=LEARNING_RATE, maximum_momentum=0.9, minimum_momentum=0.9)
metric_list = [dice_coef, sm.metrics.iou_score, sm.metrics.f1_score]
callback_list = [checkpoint, oneCycleLR]
optimizer = optimizers.SGD(lr=LEARNING_RATE, momentum=0.9, nesterov=True)
model.compile(optimizer=optimizer, loss=sm.losses.bce_dice_loss, metrics=metric_list)
model.summary()
history = model.fit_generator(generator=train_generator,
steps_per_epoch=STEP_SIZE_TRAIN,
validation_data=valid_generator,
validation_steps=STEP_SIZE_VALID,
callbacks=callback_list,
epochs=EPOCHS,
verbose=2).history
plot_metrics(history, metric_list=['loss', 'dice_coef', 'iou_score', 'f1-score'])
| 0.9434 | 0.612078 |
# CSNAnalysis Tutorial
### A brief introduction to the use of the CSNAnalysis package
---
**Updated Aug 19, 2020**
*Dickson Lab, Michigan State University*
## Overview
The CSNAnalysis package is a set of tools for network-based analysis of molecular dynamics trajectories.
CSNAnalysis is an easy interface between enhanced sampling algorithms
(e.g. WExplore implemented in `wepy`), molecular clustering programs (e.g. `MSMBuilder`), graph analysis packages (e.g. `networkX`) and graph visualization programs (e.g. `Gephi`).
### What are conformation space networks?
A conformation space network is a visualization of a free energy landscape, where each node is a cluster of molecular conformations, and the edges show which conformations can directly interconvert during a molecular dynamics simulation. A CSN can be thought of as a visual representation of a transition matrix, where the nodes represent the row / column indices and the edges show the off-diagonal elements. `CSNAnalysis` offers a concise set of tools for the creation, analysis and visualization of CSNs.
**This tutorial will give quick examples for the following use cases:**
1. Initializing CSN objects from count matrices
2. Trimming CSNs
2. Obtaining steady-state weights from a transition matrix
* By eigenvalue
* By iterative multiplication
3. Computing committor probabilities to an arbitrary set of basins
4. Exporting gexf files for visualization with the Gephi program
## Getting started
Clone the CSNAnalysis repository:
```
git clone https://github.com/ADicksonLab/CSNAnalysis.git```
Navigate to the examples directory and install using pip:
```
cd CSNAnalysis
pip install --user -e
```
Go to the examples directory and open this notebook (`examples.ipynb`):
```
cd examples; jupyter notebook```
## Dependencies
I highly recommend using Anaconda and working in a `python3` environment. CSNAnalysis uses the packages `numpy`, `scipy` and `networkx`. If these are installed then the following lines of code should run without error:
```
import numpy as np
import networkx as nx
import scipy
```
If `CSNAnalysis` was installed (i.e. added to your `sys.path`), then this should also work:
```
from csnanalysis.csn import CSN
from csnanalysis.matrix import *
```
This notebook also uses `matplotlib`, to visualize output.
```
import matplotlib
```
Great! Now let's load in the count matrix that we'll use for all the examples here:
```
count_mat = scipy.sparse.load_npz('matrix.npz')
```
## Background: Sparse matrices
It's worth knowing a little about sparse matrices before we start. If we have a huge $N$ by $N$ matrix, where $N > 1000$, but most of the elements are zero, it is more efficient to store the data as a sparse matrix.
```
type(count_mat)
```
`coo_matrix` refers to "coordinate format", where the matrix is essentially a set of lists of matrix "coordinates" (rows, columns) and data:
```
rows = count_mat.row
cols = count_mat.col
data = count_mat.data
for r,c,d in zip(rows[0:10],cols[0:10],data[0:10]):
print(r,c,d)
```
Although it can be treated like a normal matrix ($4000$ by $4000$ in this case):
```
count_mat.shape
```
It only needs to store non-zero elements, which are much fewer than $4000^2$:
```
len(rows)
```
**OK, let's get started building a Conformation Space Network!**
---
## 1) Initializing CSN objects from count matrices
To get started we need a count matrix, which can be a `numpy` array, or a `scipy.sparse` matrix, or a list of lists:
```
our_csn = CSN(count_mat,symmetrize=True)
```
Any of the `CSNAnalysis` functions can be queried using "?"
```
CSN?
```
The `our_csn` object now holds three different representations of our data. The original counts can now be found in `scipy.sparse` format:
```
our_csn.countmat
```
A transition matrix has been computed from this count matrix according to:
\begin{equation}
t_{ij} = \frac{c_{ij}}{\sum_j c_{ij}}
\end{equation}
```
our_csn.transmat
```
where the elements in each column sum to one:
```
our_csn.transmat.sum(axis=0)
```
Lastly, the data has been stored in a `networkx` directed graph:
```
our_csn.graph
```
that holds the nodes and edges of our csn, and we can use in other `networkx` functions. For example, we can calculate the shortest path between nodes 0 and 10:
```
nx.shortest_path(our_csn.graph,0,10)
```
---
## 2) Trimming CSNs
A big benefit of coupling the count matrix, transition matrix and graph representations is that elements can be "trimmed" from all three simultaneously. The `trim` function will eliminate nodes that are not connected to the main component (by inflow, outflow, or both), and can also eliminate nodes that do not meet a minimum count requirement:
```
our_csn.trim(by_inflow=True, by_outflow=True, min_count=20)
```
The trimmed graph, count matrix and transition matrix are stored as `our_csn.trim_graph`, `our_csn.trim_countmat` and `our_csn.trim_transmat`, respectively.
```
our_csn.trim_graph.number_of_nodes()
our_csn.trim_countmat.shape
our_csn.trim_transmat.shape
```
## 3) Obtaining steady-state weights from the transition matrix
Now that we've ensured that our transition matrix is fully-connected, we can compute its equilibrium weights. This is implemented in two ways.
First, we can compute the eigenvector of the transition matrix with eigenvalue one:
```
wt_eig = our_csn.calc_eig_weights()
```
This can exhibit some instability, especially for low-weight states, so we can also calculate weights by iterative multiplication of the transition matrix, which can take a little longer:
```
wt_mult = our_csn.calc_mult_weights()
import matplotlib.pyplot as plt
%matplotlib inline
plt.scatter(wt_eig,wt_mult)
plt.plot([0,wt_mult.max()],[0,wt_mult.max()],'r-')
plt.xlabel("Eigenvalue weight")
plt.ylabel("Mult weight")
plt.show()
```
These weights are automatically added as attributes to the nodes in `our_csn.graph`:
```
our_csn.graph.node[0]
```
## 4) Committor probabilities to an arbitrary set of basins
We are often doing simulations in the presence of one or more high probability "basins" of attraction. When there more than one basin, it can be useful to find the probability that a simulation started in a given state will visit (or "commit to") a given basin before the others.
`CSNAnalysis` calculates committor probabilities by creating a sink matrix ($S$), where each column in the transition matrix that corresponds to a sink state is replaced by an identity vector. This turns each state into a "black hole" where probability can get in, but not out.
By iteratively multiplying this matrix by itself, we can approximate $S^\infty$. The elements of this matrix reveal the probability of transitioning to any of the sink states, upon starting in any non-sink state, $i$.
Let's see this in action. We'll start by reading in a set of three basins: $A$, $B$ and $U$.
```
Astates = [2031,596,1923,3223,2715]
Bstates = [1550,3168,476,1616,2590]
Ustates = list(np.loadtxt('state_U.dat',dtype=int))
```
We can then use the `calc_committors` function to calculate committors between this set of three basins. This will calculate $p_A$, $p_B$, and $p_U$ for each state, which sum to one.
```
basins = [Astates,Bstates,Ustates]
labels = ['pA','pB','pU']
comms = our_csn.calc_committors(basins,labels=labels)
```
The committors can be interpreted as follows:
```
i = our_csn.trim_indices[0]
print('comms['+str(i)+'] = ',comms[i])
print('\nIn other words, if you start in state {0:d}:'.format(i))
print('You will reach basin A first with probability {0:.2f}, basin B with probability {1:.2f} and basin U with probability {2:.2f}'.format(comms[i,0],comms[i,1],comms[i,2]))
```
## 5) Exporting graph for visualization in Gephi
`NetworkX` is great for doing graph-based analyses, but not stellar at greating graph layouts for large(r) networks. However, they do have excellent built-in support for exporting graph objects in a variety of formats.
Here we'll use the `.gexf` format to save our network, as well as all of the attributes we've calculated, to a file that can be read into [Gephi](https://gephi.org/), a powerful graph visualization program. While support for Gephi has been spotty in the recent past, it is still one of the best available options for graph visualization.
Before exporting to `.gexf`, let's use the committors we've calculated to add colors to the nodes:
```
rgb = our_csn.colors_from_committors(comms)
our_csn.set_colors(rgb)
```
Now we have added some properties to our nodes under 'viz', which will be interpreted by Gephi:
```
our_csn.graph.node[0]
```
And we can use an internal `networkx` function to write all of this to a `.gexf` file:
```
nx.readwrite.gexf.write_gexf(our_csn.graph.to_undirected(),'test.gexf')
```
After opening this file in Gephi, I recommend creating a layout using the "Force Atlas 2" algorithm in the layout panel. I set the node sizes to the "eig_weights" variable, and after exporting to pdf and adding some labels, I get the following:

**That's the end of our tutorial!** I hope you enjoyed it and you find `CSNAnalysis` useful in your research. If you are having difficulties with the installation or running of the software, feel free to create an [issue on the Github page](https://github.com/ADicksonLab/CSNAnalysis).
|
github_jupyter
|
git clone https://github.com/ADicksonLab/CSNAnalysis.git```
Navigate to the examples directory and install using pip:
Go to the examples directory and open this notebook (`examples.ipynb`):
## Dependencies
I highly recommend using Anaconda and working in a `python3` environment. CSNAnalysis uses the packages `numpy`, `scipy` and `networkx`. If these are installed then the following lines of code should run without error:
If `CSNAnalysis` was installed (i.e. added to your `sys.path`), then this should also work:
This notebook also uses `matplotlib`, to visualize output.
Great! Now let's load in the count matrix that we'll use for all the examples here:
## Background: Sparse matrices
It's worth knowing a little about sparse matrices before we start. If we have a huge $N$ by $N$ matrix, where $N > 1000$, but most of the elements are zero, it is more efficient to store the data as a sparse matrix.
`coo_matrix` refers to "coordinate format", where the matrix is essentially a set of lists of matrix "coordinates" (rows, columns) and data:
Although it can be treated like a normal matrix ($4000$ by $4000$ in this case):
It only needs to store non-zero elements, which are much fewer than $4000^2$:
**OK, let's get started building a Conformation Space Network!**
---
## 1) Initializing CSN objects from count matrices
To get started we need a count matrix, which can be a `numpy` array, or a `scipy.sparse` matrix, or a list of lists:
Any of the `CSNAnalysis` functions can be queried using "?"
The `our_csn` object now holds three different representations of our data. The original counts can now be found in `scipy.sparse` format:
A transition matrix has been computed from this count matrix according to:
\begin{equation}
t_{ij} = \frac{c_{ij}}{\sum_j c_{ij}}
\end{equation}
where the elements in each column sum to one:
Lastly, the data has been stored in a `networkx` directed graph:
that holds the nodes and edges of our csn, and we can use in other `networkx` functions. For example, we can calculate the shortest path between nodes 0 and 10:
---
## 2) Trimming CSNs
A big benefit of coupling the count matrix, transition matrix and graph representations is that elements can be "trimmed" from all three simultaneously. The `trim` function will eliminate nodes that are not connected to the main component (by inflow, outflow, or both), and can also eliminate nodes that do not meet a minimum count requirement:
The trimmed graph, count matrix and transition matrix are stored as `our_csn.trim_graph`, `our_csn.trim_countmat` and `our_csn.trim_transmat`, respectively.
## 3) Obtaining steady-state weights from the transition matrix
Now that we've ensured that our transition matrix is fully-connected, we can compute its equilibrium weights. This is implemented in two ways.
First, we can compute the eigenvector of the transition matrix with eigenvalue one:
This can exhibit some instability, especially for low-weight states, so we can also calculate weights by iterative multiplication of the transition matrix, which can take a little longer:
These weights are automatically added as attributes to the nodes in `our_csn.graph`:
## 4) Committor probabilities to an arbitrary set of basins
We are often doing simulations in the presence of one or more high probability "basins" of attraction. When there more than one basin, it can be useful to find the probability that a simulation started in a given state will visit (or "commit to") a given basin before the others.
`CSNAnalysis` calculates committor probabilities by creating a sink matrix ($S$), where each column in the transition matrix that corresponds to a sink state is replaced by an identity vector. This turns each state into a "black hole" where probability can get in, but not out.
By iteratively multiplying this matrix by itself, we can approximate $S^\infty$. The elements of this matrix reveal the probability of transitioning to any of the sink states, upon starting in any non-sink state, $i$.
Let's see this in action. We'll start by reading in a set of three basins: $A$, $B$ and $U$.
We can then use the `calc_committors` function to calculate committors between this set of three basins. This will calculate $p_A$, $p_B$, and $p_U$ for each state, which sum to one.
The committors can be interpreted as follows:
## 5) Exporting graph for visualization in Gephi
`NetworkX` is great for doing graph-based analyses, but not stellar at greating graph layouts for large(r) networks. However, they do have excellent built-in support for exporting graph objects in a variety of formats.
Here we'll use the `.gexf` format to save our network, as well as all of the attributes we've calculated, to a file that can be read into [Gephi](https://gephi.org/), a powerful graph visualization program. While support for Gephi has been spotty in the recent past, it is still one of the best available options for graph visualization.
Before exporting to `.gexf`, let's use the committors we've calculated to add colors to the nodes:
Now we have added some properties to our nodes under 'viz', which will be interpreted by Gephi:
And we can use an internal `networkx` function to write all of this to a `.gexf` file:
| 0.896464 | 0.995652 |
```
import pandas as pd
import numpy as np
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.metrics.pairwise import cosine_similarity
import re
movie_df = pd.read_csv('/content/drive/MyDrive/Colab Notebooks/movie_lens/movie.csv')
rating_df = pd.read_csv('/content/drive/MyDrive/Colab Notebooks/movie_lens/rating.csv')
link_df = pd.read_csv('/content/drive/MyDrive/Colab Notebooks/movie_lens/link.csv')
tag_df = pd.read_csv('/content/drive/MyDrive/Colab Notebooks/movie_lens/tag.csv')
genome_tags_df = pd.read_csv('/content/drive/MyDrive/Colab Notebooks/movie_lens/genome_tags.csv')
genome_scores_df = pd.read_csv('/content/drive/MyDrive/Colab Notebooks/movie_lens/genome_scores.csv')
#this is not the same combined small as collaborative
combined_small = pd.read_csv('/content/drive/MyDrive/Colab Notebooks/movie_lens/tags_mvoies_combined_small.csv')
#take a copy of the data frame
df_tag = genome_tags_df.copy(deep = True)
df_score = genome_scores_df.copy(deep = True)
combined_copy_tag = combined_small.copy(deep = True)
#count how many times each movie tagged with different tag
tags_df_counted = pd.DataFrame((tag_df.groupby(['movieId','tag'])['userId'].count())).reset_index()
tags_df_counted.rename(columns={'userId':'count'},inplace=True)
#fucntion creates a data frame with movie id and taglist as columns
def creat_tag_list(tags_df_counted):
df = pd.DataFrame([[0,0]],columns= ['movieId','tag_list'])
filtered_unique_id = tags_df_counted['movieId'].unique().tolist()
for id in filtered_unique_id:
tags_list = tags_df_counted.loc[tags_df_counted['movieId'] == id]['tag'].values.tolist()
df_dummy = pd.DataFrame([[id,tags_list]],columns=['movieId','tag_list'])
df = df.append(df_dummy)
#reset the index and drop nans
tag_list_combined = df.reset_index().drop(columns=['index'])
tag_list_combined.drop(tag_list_combined.index[0],inplace=True)
return tag_list_combined
combined_copy_tag.drop(columns=['Unnamed: 0'],inplace=True)
tag_list_combined = creat_tag_list(tags_df_counted)
tag_list_combined.head(5)
#how many time a tag was used
combined_grouped = pd.DataFrame((combined_copy_tag.groupby(['movieId','genres'])\
['userId'].count())).reset_index()
combined_grouped.rename(columns={'userId':'count'},inplace=True)
combined_grouped.head(5)
#movie tag list and genres are combined together
merged = pd.concat([tag_list_combined,combined_grouped.drop(columns=['movieId'])],axis=1)
merged.dropna(inplace=True)
merged.head(5)
#changing the types from float to int and list ot string
merged['movieId'] = merged['movieId'].astype(int)
merged['userId'] = merged['count'].astype(int)
merged['tag_list'] = merged['tag_list'].astype(str)
merged['genres'] = merged['genres'].astype(str)
#creating a column which is genres and tag_list combined
merged['Bag_of_words'] = merged['tag_list'] + merged['genres']
merged.head(5)
merged['Bag_of_words'].values[0]
#removing non alphanumeric and double spaces
#changing from upper to lower
def text_cleaning(row):
alpha_numeric = re.sub("\W"," ",row)
double_space = re.sub(' +',' ',alpha_numeric)
remove_space = double_space.strip()
lower = remove_space.lower()
return lower
merged['Bag_of_words'] = merged['Bag_of_words'].apply(text_cleaning)
merged.drop(columns=['genres','count','tag_list'],inplace=True)
merged.tail(5)
movie_names = combined_small[['movieId','title']].drop_duplicates()
#mergin movie names and bag_of_word we created
final = pd.merge(left=merged,right=movie_names,on='movieId',how='right')
final.dropna(inplace=True)
final.drop(columns=['movieId','userId'],inplace=True)
final['title'] = final['title'].apply(lambda x: x.lower())
final.head(5)
#calculating cosime similarity
count = CountVectorizer()
count_matrix = count.fit_transform(final['Bag_of_words'])
cosine_sim = cosine_similarity(count_matrix,count_matrix)
#function to recommend movies
indices = pd.Series(final['title'])
def recommend(title, cosine_sim = cosine_sim):
recommended_movies = []
idx = indices[indices.str.contains(title)].index[0]
score_series = pd.Series(cosine_sim[idx]).sort_values(ascending = False)
top_10_indices = list(score_series.iloc[1:11].index)
for i in top_10_indices:
recommended_movies.append(list(final['title'])[i])
return recommended_movies
recommend('toy story')
```
|
github_jupyter
|
import pandas as pd
import numpy as np
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.metrics.pairwise import cosine_similarity
import re
movie_df = pd.read_csv('/content/drive/MyDrive/Colab Notebooks/movie_lens/movie.csv')
rating_df = pd.read_csv('/content/drive/MyDrive/Colab Notebooks/movie_lens/rating.csv')
link_df = pd.read_csv('/content/drive/MyDrive/Colab Notebooks/movie_lens/link.csv')
tag_df = pd.read_csv('/content/drive/MyDrive/Colab Notebooks/movie_lens/tag.csv')
genome_tags_df = pd.read_csv('/content/drive/MyDrive/Colab Notebooks/movie_lens/genome_tags.csv')
genome_scores_df = pd.read_csv('/content/drive/MyDrive/Colab Notebooks/movie_lens/genome_scores.csv')
#this is not the same combined small as collaborative
combined_small = pd.read_csv('/content/drive/MyDrive/Colab Notebooks/movie_lens/tags_mvoies_combined_small.csv')
#take a copy of the data frame
df_tag = genome_tags_df.copy(deep = True)
df_score = genome_scores_df.copy(deep = True)
combined_copy_tag = combined_small.copy(deep = True)
#count how many times each movie tagged with different tag
tags_df_counted = pd.DataFrame((tag_df.groupby(['movieId','tag'])['userId'].count())).reset_index()
tags_df_counted.rename(columns={'userId':'count'},inplace=True)
#fucntion creates a data frame with movie id and taglist as columns
def creat_tag_list(tags_df_counted):
df = pd.DataFrame([[0,0]],columns= ['movieId','tag_list'])
filtered_unique_id = tags_df_counted['movieId'].unique().tolist()
for id in filtered_unique_id:
tags_list = tags_df_counted.loc[tags_df_counted['movieId'] == id]['tag'].values.tolist()
df_dummy = pd.DataFrame([[id,tags_list]],columns=['movieId','tag_list'])
df = df.append(df_dummy)
#reset the index and drop nans
tag_list_combined = df.reset_index().drop(columns=['index'])
tag_list_combined.drop(tag_list_combined.index[0],inplace=True)
return tag_list_combined
combined_copy_tag.drop(columns=['Unnamed: 0'],inplace=True)
tag_list_combined = creat_tag_list(tags_df_counted)
tag_list_combined.head(5)
#how many time a tag was used
combined_grouped = pd.DataFrame((combined_copy_tag.groupby(['movieId','genres'])\
['userId'].count())).reset_index()
combined_grouped.rename(columns={'userId':'count'},inplace=True)
combined_grouped.head(5)
#movie tag list and genres are combined together
merged = pd.concat([tag_list_combined,combined_grouped.drop(columns=['movieId'])],axis=1)
merged.dropna(inplace=True)
merged.head(5)
#changing the types from float to int and list ot string
merged['movieId'] = merged['movieId'].astype(int)
merged['userId'] = merged['count'].astype(int)
merged['tag_list'] = merged['tag_list'].astype(str)
merged['genres'] = merged['genres'].astype(str)
#creating a column which is genres and tag_list combined
merged['Bag_of_words'] = merged['tag_list'] + merged['genres']
merged.head(5)
merged['Bag_of_words'].values[0]
#removing non alphanumeric and double spaces
#changing from upper to lower
def text_cleaning(row):
alpha_numeric = re.sub("\W"," ",row)
double_space = re.sub(' +',' ',alpha_numeric)
remove_space = double_space.strip()
lower = remove_space.lower()
return lower
merged['Bag_of_words'] = merged['Bag_of_words'].apply(text_cleaning)
merged.drop(columns=['genres','count','tag_list'],inplace=True)
merged.tail(5)
movie_names = combined_small[['movieId','title']].drop_duplicates()
#mergin movie names and bag_of_word we created
final = pd.merge(left=merged,right=movie_names,on='movieId',how='right')
final.dropna(inplace=True)
final.drop(columns=['movieId','userId'],inplace=True)
final['title'] = final['title'].apply(lambda x: x.lower())
final.head(5)
#calculating cosime similarity
count = CountVectorizer()
count_matrix = count.fit_transform(final['Bag_of_words'])
cosine_sim = cosine_similarity(count_matrix,count_matrix)
#function to recommend movies
indices = pd.Series(final['title'])
def recommend(title, cosine_sim = cosine_sim):
recommended_movies = []
idx = indices[indices.str.contains(title)].index[0]
score_series = pd.Series(cosine_sim[idx]).sort_values(ascending = False)
top_10_indices = list(score_series.iloc[1:11].index)
for i in top_10_indices:
recommended_movies.append(list(final['title'])[i])
return recommended_movies
recommend('toy story')
| 0.423935 | 0.199971 |
```
import librosa
import os
import tensorflow as tf
import numpy as np
from tqdm import tqdm
wav_files = [f for f in os.listdir('./data') if f.endswith('.wav')]
text_files = [f for f in os.listdir('./data') if f.endswith('.txt')]
inputs, targets = [], []
for (wav_file, text_file) in tqdm(zip(wav_files, text_files), total = len(wav_files),ncols=80):
path = './data/' + wav_file
try:
y, sr = librosa.load(path, sr = None)
except:
continue
inputs.append(
librosa.feature.mfcc(
y = y, sr = sr, n_mfcc = 40, hop_length = int(0.05 * sr)
).T
)
with open('./data/' + text_file) as f:
targets.append(f.read())
inputs = tf.keras.preprocessing.sequence.pad_sequences(
inputs, dtype = 'float32', padding = 'post'
)
chars = list(set([c for target in targets for c in target]))
num_classes = len(chars) + 2
idx2char = {idx + 1: char for idx, char in enumerate(chars)}
idx2char[0] = '<PAD>'
char2idx = {char: idx for idx, char in idx2char.items()}
targets = [[char2idx[c] for c in target] for target in targets]
def pad_sentence_batch(sentence_batch, pad_int):
padded_seqs = []
seq_lens = []
max_sentence_len = max([len(sentence) for sentence in sentence_batch])
for sentence in sentence_batch:
padded_seqs.append(sentence + [pad_int] * (max_sentence_len - len(sentence)))
seq_lens.append(len(sentence))
return padded_seqs, seq_lens
def sparse_tuple_from(sequences, dtype=np.int32):
indices = []
values = []
for n, seq in enumerate(sequences):
indices.extend(zip([n] * len(seq), range(len(seq))))
values.extend(seq)
indices = np.asarray(indices, dtype=np.int64)
values = np.asarray(values, dtype=dtype)
shape = np.asarray([len(sequences), np.asarray(indices).max(0)[1] + 1], dtype=np.int64)
return indices, values, shape
def pad_second_dim(x, desired_size):
padding = tf.tile([[0]], tf.stack([tf.shape(x)[0], desired_size - tf.shape(x)[1]], 0))
return tf.concat([x, padding], 1)
class Model:
def __init__(
self,
num_layers,
size_layers,
learning_rate,
num_features,
dropout = 1.0,
):
self.X = tf.placeholder(tf.float32, [None, None, num_features])
self.Y = tf.sparse_placeholder(tf.int32)
seq_lens = tf.count_nonzero(
tf.reduce_sum(self.X, -1), 1, dtype = tf.int32
)
self.label = tf.placeholder(tf.int32, [None, None])
self.Y_seq_len = tf.placeholder(tf.int32, [None])
def cells(size, reuse = False):
return tf.contrib.rnn.DropoutWrapper(
tf.nn.rnn_cell.LSTMCell(
size,
initializer = tf.orthogonal_initializer(),
reuse = reuse,
),
state_keep_prob = dropout,
output_keep_prob = dropout,
)
features = self.X
for n in range(num_layers):
(out_fw, out_bw), (
state_fw,
state_bw,
) = tf.nn.bidirectional_dynamic_rnn(
cell_fw = cells(size_layers),
cell_bw = cells(size_layers),
inputs = features,
sequence_length = seq_lens,
dtype = tf.float32,
scope = 'bidirectional_rnn_%d' % (n),
)
features = tf.concat((out_fw, out_bw), 2)
logits = tf.layers.dense(features, num_classes)
time_major = tf.transpose(logits, [1, 0, 2])
decoded, log_prob = tf.nn.ctc_beam_search_decoder(time_major, seq_lens)
decoded = tf.to_int32(decoded[0])
self.preds = tf.sparse.to_dense(decoded)
self.cost = tf.reduce_mean(
tf.nn.ctc_loss(
self.Y,
time_major,
seq_lens
)
)
self.optimizer = tf.train.AdamOptimizer(
learning_rate = learning_rate
).minimize(self.cost)
preds = self.preds[:, :tf.reduce_max(self.Y_seq_len)]
masks = tf.sequence_mask(self.Y_seq_len, tf.reduce_max(self.Y_seq_len), dtype=tf.float32)
preds = pad_second_dim(preds, tf.reduce_max(self.Y_seq_len))
y_t = tf.cast(preds, tf.int32)
self.prediction = tf.boolean_mask(y_t, masks)
mask_label = tf.boolean_mask(self.label, masks)
self.mask_label = mask_label
correct_pred = tf.equal(self.prediction, mask_label)
correct_index = tf.cast(correct_pred, tf.float32)
self.accuracy = tf.reduce_mean(tf.cast(correct_pred, tf.float32))
tf.reset_default_graph()
sess = tf.InteractiveSession()
size_layers = 128
learning_rate = 1e-3
num_layers = 2
batch_size = 32
epoch = 50
model = Model(num_layers, size_layers, learning_rate, inputs.shape[2])
sess.run(tf.global_variables_initializer())
for e in range(epoch):
pbar = tqdm(
range(0, len(inputs), batch_size), desc = 'minibatch loop')
total_cost, total_accuracy = 0, 0
for i in pbar:
batch_x = inputs[i : min(i + batch_size, len(inputs))]
y = targets[i : min(i + batch_size, len(inputs))]
batch_y = sparse_tuple_from(y)
batch_label, batch_len = pad_sentence_batch(y, 0)
_, cost, accuracy = sess.run(
[model.optimizer, model.cost, model.accuracy],
feed_dict = {model.X: batch_x, model.Y: batch_y,
model.label: batch_label, model.Y_seq_len: batch_len},
)
total_cost += cost
total_accuracy += accuracy
pbar.set_postfix(cost = cost, accuracy = accuracy)
total_cost /= (len(inputs) / batch_size)
total_accuracy /= (len(inputs) / batch_size)
print('epoch %d, average cost %f, average accuracy %f'%(e + 1, total_cost, total_accuracy))
import random
random_index = random.randint(0, len(targets) - 1)
batch_x = inputs[random_index : random_index + 1]
print(
'real:',
''.join(
[idx2char[no] for no in targets[random_index : random_index + 1][0]]
),
)
batch_y = sparse_tuple_from(targets[random_index : random_index + 1])
pred = sess.run(model.preds, feed_dict = {model.X: batch_x})[0]
print('predicted:', ''.join([idx2char[no] for no in pred]))
```
|
github_jupyter
|
import librosa
import os
import tensorflow as tf
import numpy as np
from tqdm import tqdm
wav_files = [f for f in os.listdir('./data') if f.endswith('.wav')]
text_files = [f for f in os.listdir('./data') if f.endswith('.txt')]
inputs, targets = [], []
for (wav_file, text_file) in tqdm(zip(wav_files, text_files), total = len(wav_files),ncols=80):
path = './data/' + wav_file
try:
y, sr = librosa.load(path, sr = None)
except:
continue
inputs.append(
librosa.feature.mfcc(
y = y, sr = sr, n_mfcc = 40, hop_length = int(0.05 * sr)
).T
)
with open('./data/' + text_file) as f:
targets.append(f.read())
inputs = tf.keras.preprocessing.sequence.pad_sequences(
inputs, dtype = 'float32', padding = 'post'
)
chars = list(set([c for target in targets for c in target]))
num_classes = len(chars) + 2
idx2char = {idx + 1: char for idx, char in enumerate(chars)}
idx2char[0] = '<PAD>'
char2idx = {char: idx for idx, char in idx2char.items()}
targets = [[char2idx[c] for c in target] for target in targets]
def pad_sentence_batch(sentence_batch, pad_int):
padded_seqs = []
seq_lens = []
max_sentence_len = max([len(sentence) for sentence in sentence_batch])
for sentence in sentence_batch:
padded_seqs.append(sentence + [pad_int] * (max_sentence_len - len(sentence)))
seq_lens.append(len(sentence))
return padded_seqs, seq_lens
def sparse_tuple_from(sequences, dtype=np.int32):
indices = []
values = []
for n, seq in enumerate(sequences):
indices.extend(zip([n] * len(seq), range(len(seq))))
values.extend(seq)
indices = np.asarray(indices, dtype=np.int64)
values = np.asarray(values, dtype=dtype)
shape = np.asarray([len(sequences), np.asarray(indices).max(0)[1] + 1], dtype=np.int64)
return indices, values, shape
def pad_second_dim(x, desired_size):
padding = tf.tile([[0]], tf.stack([tf.shape(x)[0], desired_size - tf.shape(x)[1]], 0))
return tf.concat([x, padding], 1)
class Model:
def __init__(
self,
num_layers,
size_layers,
learning_rate,
num_features,
dropout = 1.0,
):
self.X = tf.placeholder(tf.float32, [None, None, num_features])
self.Y = tf.sparse_placeholder(tf.int32)
seq_lens = tf.count_nonzero(
tf.reduce_sum(self.X, -1), 1, dtype = tf.int32
)
self.label = tf.placeholder(tf.int32, [None, None])
self.Y_seq_len = tf.placeholder(tf.int32, [None])
def cells(size, reuse = False):
return tf.contrib.rnn.DropoutWrapper(
tf.nn.rnn_cell.LSTMCell(
size,
initializer = tf.orthogonal_initializer(),
reuse = reuse,
),
state_keep_prob = dropout,
output_keep_prob = dropout,
)
features = self.X
for n in range(num_layers):
(out_fw, out_bw), (
state_fw,
state_bw,
) = tf.nn.bidirectional_dynamic_rnn(
cell_fw = cells(size_layers),
cell_bw = cells(size_layers),
inputs = features,
sequence_length = seq_lens,
dtype = tf.float32,
scope = 'bidirectional_rnn_%d' % (n),
)
features = tf.concat((out_fw, out_bw), 2)
logits = tf.layers.dense(features, num_classes)
time_major = tf.transpose(logits, [1, 0, 2])
decoded, log_prob = tf.nn.ctc_beam_search_decoder(time_major, seq_lens)
decoded = tf.to_int32(decoded[0])
self.preds = tf.sparse.to_dense(decoded)
self.cost = tf.reduce_mean(
tf.nn.ctc_loss(
self.Y,
time_major,
seq_lens
)
)
self.optimizer = tf.train.AdamOptimizer(
learning_rate = learning_rate
).minimize(self.cost)
preds = self.preds[:, :tf.reduce_max(self.Y_seq_len)]
masks = tf.sequence_mask(self.Y_seq_len, tf.reduce_max(self.Y_seq_len), dtype=tf.float32)
preds = pad_second_dim(preds, tf.reduce_max(self.Y_seq_len))
y_t = tf.cast(preds, tf.int32)
self.prediction = tf.boolean_mask(y_t, masks)
mask_label = tf.boolean_mask(self.label, masks)
self.mask_label = mask_label
correct_pred = tf.equal(self.prediction, mask_label)
correct_index = tf.cast(correct_pred, tf.float32)
self.accuracy = tf.reduce_mean(tf.cast(correct_pred, tf.float32))
tf.reset_default_graph()
sess = tf.InteractiveSession()
size_layers = 128
learning_rate = 1e-3
num_layers = 2
batch_size = 32
epoch = 50
model = Model(num_layers, size_layers, learning_rate, inputs.shape[2])
sess.run(tf.global_variables_initializer())
for e in range(epoch):
pbar = tqdm(
range(0, len(inputs), batch_size), desc = 'minibatch loop')
total_cost, total_accuracy = 0, 0
for i in pbar:
batch_x = inputs[i : min(i + batch_size, len(inputs))]
y = targets[i : min(i + batch_size, len(inputs))]
batch_y = sparse_tuple_from(y)
batch_label, batch_len = pad_sentence_batch(y, 0)
_, cost, accuracy = sess.run(
[model.optimizer, model.cost, model.accuracy],
feed_dict = {model.X: batch_x, model.Y: batch_y,
model.label: batch_label, model.Y_seq_len: batch_len},
)
total_cost += cost
total_accuracy += accuracy
pbar.set_postfix(cost = cost, accuracy = accuracy)
total_cost /= (len(inputs) / batch_size)
total_accuracy /= (len(inputs) / batch_size)
print('epoch %d, average cost %f, average accuracy %f'%(e + 1, total_cost, total_accuracy))
import random
random_index = random.randint(0, len(targets) - 1)
batch_x = inputs[random_index : random_index + 1]
print(
'real:',
''.join(
[idx2char[no] for no in targets[random_index : random_index + 1][0]]
),
)
batch_y = sparse_tuple_from(targets[random_index : random_index + 1])
pred = sess.run(model.preds, feed_dict = {model.X: batch_x})[0]
print('predicted:', ''.join([idx2char[no] for no in pred]))
| 0.53607 | 0.311087 |
<div align="center">
<h1><img width="30" src="https://madewithml.com/static/images/rounded_logo.png"> <a href="https://madewithml.com/">Made With ML</a></h1>
Applied ML · MLOps · Production
<br>
Join 30K+ developers in learning how to responsibly <a href="https://madewithml.com/about/">deliver value</a> with ML.
<br>
</div>
<br>
<div align="center">
<a target="_blank" href="https://newsletter.madewithml.com"><img src="https://img.shields.io/badge/Subscribe-30K-brightgreen"></a>
<a target="_blank" href="https://github.com/GokuMohandas/MadeWithML"><img src="https://img.shields.io/github/stars/GokuMohandas/MadeWithML.svg?style=social&label=Star"></a>
<a target="_blank" href="https://www.linkedin.com/in/goku"><img src="https://img.shields.io/badge/style--5eba00.svg?label=LinkedIn&logo=linkedin&style=social"></a>
<a target="_blank" href="https://twitter.com/GokuMohandas"><img src="https://img.shields.io/twitter/follow/GokuMohandas.svg?label=Follow&style=social"></a>
<br>
🔥 Among the <a href="https://github.com/topics/deep-learning" target="_blank">top ML</a> repositories on GitHub
</div>
<br>
<hr>
# Utilities
In this lesson, we will explore utilities to extend and simplify training.
<div align="left">
<a target="_blank" href="https://madewithml.com/courses/foundations/utilities/"><img src="https://img.shields.io/badge/📖 Read-blog post-9cf"></a>
<a href="https://github.com/GokuMohandas/MadeWithML/blob/main/notebooks/10_Utilities.ipynb" role="button"><img src="https://img.shields.io/static/v1?label=&message=View%20On%20GitHub&color=586069&logo=github&labelColor=2f363d"></a>
<a href="https://colab.research.google.com/github/GokuMohandas/MadeWithML/blob/main/notebooks/10_Utilities.ipynb"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"></a>
</div>
# Set up
We're having to set a lot of seeds for reproducibility now, so let's wrap it all up in a function.
```
import numpy as np
import pandas as pd
import random
import torch
import torch.nn as nn
SEED = 1234
def set_seeds(seed=1234):
"""Set seeds for reproducibility."""
np.random.seed(seed)
random.seed(seed)
torch.manual_seed(seed)
torch.cuda.manual_seed(seed)
torch.cuda.manual_seed_all(seed) # multi-GPU
# Set seeds for reproducibility
set_seeds(seed=SEED)
# Set device
cuda = True
device = torch.device("cuda" if (
torch.cuda.is_available() and cuda) else "cpu")
torch.set_default_tensor_type("torch.FloatTensor")
if device.type == "cuda":
torch.set_default_tensor_type("torch.cuda.FloatTensor")
print (device)
```
## Load data
We'll use the same spiral dataset from previous lessons to demonstrate our utilities.
```
import matplotlib.pyplot as plt
import pandas as pd
# Load data
url = "https://raw.githubusercontent.com/GokuMohandas/MadeWithML/main/datasets/spiral.csv"
df = pd.read_csv(url, header=0) # load
df = df.sample(frac=1).reset_index(drop=True) # shuffle
df.head()
# Data shapes
X = df[["X1", "X2"]].values
y = df["color"].values
print ("X: ", np.shape(X))
print ("y: ", np.shape(y))
# Visualize data
plt.title("Generated non-linear data")
colors = {"c1": "red", "c2": "yellow", "c3": "blue"}
plt.scatter(X[:, 0], X[:, 1], c=[colors[_y] for _y in y], edgecolors="k", s=25)
plt.show()
```
## Split data
```
import collections
from sklearn.model_selection import train_test_split
TRAIN_SIZE = 0.7
VAL_SIZE = 0.15
TEST_SIZE = 0.15
def train_val_test_split(X, y, train_size):
"""Split dataset into data splits."""
X_train, X_, y_train, y_ = train_test_split(X, y, train_size=TRAIN_SIZE, stratify=y)
X_val, X_test, y_val, y_test = train_test_split(X_, y_, train_size=0.5, stratify=y_)
return X_train, X_val, X_test, y_train, y_val, y_test
# Create data splits
X_train, X_val, X_test, y_train, y_val, y_test = train_val_test_split(
X=X, y=y, train_size=TRAIN_SIZE)
print (f"X_train: {X_train.shape}, y_train: {y_train.shape}")
print (f"X_val: {X_val.shape}, y_val: {y_val.shape}")
print (f"X_test: {X_test.shape}, y_test: {y_test.shape}")
print (f"Sample point: {X_train[0]} → {y_train[0]}")
```
## LabelEncoder
Next we'll define a `LabelEncoder` to encode our text labels into unique indices. We're not going to use scikit-learn's LabelEncoder anymore because we want to be able to save and load our instances the way we want to.
```
import itertools
class LabelEncoder(object):
"""Label encoder for tag labels."""
def __init__(self, class_to_index={}):
self.class_to_index = class_to_index
self.index_to_class = {v: k for k, v in self.class_to_index.items()}
self.classes = list(self.class_to_index.keys())
def __len__(self):
return len(self.class_to_index)
def __str__(self):
return f"<LabelEncoder(num_classes={len(self)})>"
def fit(self, y):
classes = np.unique(y)
for i, class_ in enumerate(classes):
self.class_to_index[class_] = i
self.index_to_class = {v: k for k, v in self.class_to_index.items()}
self.classes = list(self.class_to_index.keys())
return self
def encode(self, y):
encoded = np.zeros((len(y)), dtype=int)
for i, item in enumerate(y):
encoded[i] = self.class_to_index[item]
return encoded
def decode(self, y):
classes = []
for i, item in enumerate(y):
classes.append(self.index_to_class[item])
return classes
def save(self, fp):
with open(fp, "w") as fp:
contents = {'class_to_index': self.class_to_index}
json.dump(contents, fp, indent=4, sort_keys=False)
@classmethod
def load(cls, fp):
with open(fp, "r") as fp:
kwargs = json.load(fp=fp)
return cls(**kwargs)
# Encode
label_encoder = LabelEncoder()
label_encoder.fit(y_train)
label_encoder.class_to_index
# Convert labels to tokens
print (f"y_train[0]: {y_train[0]}")
y_train = label_encoder.encode(y_train)
y_val = label_encoder.encode(y_val)
y_test = label_encoder.encode(y_test)
print (f"y_train[0]: {y_train[0]}")
# Class weights
counts = np.bincount(y_train)
class_weights = {i: 1.0/count for i, count in enumerate(counts)}
print (f"counts: {counts}\nweights: {class_weights}")
```
## StandardScaler
We need to standardize our data (zero mean and unit variance) so a specific feature's magnitude doesn't affect how the model learns its weights. We're only going to standardize the inputs X because our outputs y are class values. We're going to compose our own `StandardScaler` class so we can easily save and load it later during inference.
```
class StandardScaler(object):
def __init__(self, mean=None, std=None):
self.mean = np.array(mean)
self.std = np.array(std)
def fit(self, X):
self.mean = np.mean(X_train, axis=0)
self.std = np.std(X_train, axis=0)
def scale(self, X):
return (X - self.mean) / self.std
def unscale(self, X):
return (X * self.std) + self.mean
def save(self, fp):
with open(fp, "w") as fp:
contents = {'mean': self.mean.tolist(), 'std': self.std.tolist()}
json.dump(contents, fp, indent=4, sort_keys=False)
@classmethod
def load(cls, fp):
with open(fp, "r") as fp:
kwargs = json.load(fp=fp)
return cls(**kwargs)
# Standardize the data (mean=0, std=1) using training data
X_scaler = StandardScaler()
X_scaler.fit(X_train)
# Apply scaler on training and test data (don't standardize outputs for classification)
X_train = X_scaler.scale(X_train)
X_val = X_scaler.scale(X_val)
X_test = X_scaler.scale(X_test)
# Check (means should be ~0 and std should be ~1)
print (f"X_test[0]: mean: {np.mean(X_test[:, 0], axis=0):.1f}, std: {np.std(X_test[:, 0], axis=0):.1f}")
print (f"X_test[1]: mean: {np.mean(X_test[:, 1], axis=0):.1f}, std: {np.std(X_test[:, 1], axis=0):.1f}")
```
# DataLoader
We're going to place our data into a [`Dataset`](https://pytorch.org/docs/stable/data.html#torch.utils.data.Dataset) and use a [`DataLoader`](https://pytorch.org/docs/stable/data.html#torch.utils.data.DataLoader) to efficiently create batches for training and evaluation.
```
import torch
# Seed seed for reproducibility
torch.manual_seed(SEED)
class Dataset(torch.utils.data.Dataset):
def __init__(self, X, y):
self.X = X
self.y = y
def __len__(self):
return len(self.y)
def __str__(self):
return f"<Dataset(N={len(self)})>"
def __getitem__(self, index):
X = self.X[index]
y = self.y[index]
return [X, y]
def collate_fn(self, batch):
"""Processing on a batch."""
# Get inputs
batch = np.array(batch, dtype=object)
X = np.stack(batch[:, 0], axis=0)
y = np.stack(batch[:, 1], axis=0)
# Cast
X = torch.FloatTensor(X.astype(np.float32))
y = torch.LongTensor(y.astype(np.int32))
return X, y
def create_dataloader(self, batch_size, shuffle=False, drop_last=False):
return torch.utils.data.DataLoader(
dataset=self, batch_size=batch_size, collate_fn=self.collate_fn,
shuffle=shuffle, drop_last=drop_last, pin_memory=True)
```
We don't really need the `collate_fn` here but we wanted to make it transparent because we will need it when we want to do specific processing on our batch (ex. padding). We'll be using a custom collate function in the next lesson.
```
# Create datasets
train_dataset = Dataset(X=X_train, y=y_train)
val_dataset = Dataset(X=X_val, y=y_val)
test_dataset = Dataset(X=X_test, y=y_test)
print ("Datasets:\n"
f" Train dataset:{train_dataset.__str__()}\n"
f" Val dataset: {val_dataset.__str__()}\n"
f" Test dataset: {test_dataset.__str__()}\n"
"Sample point:\n"
f" X: {train_dataset[0][0]}\n"
f" y: {train_dataset[0][1]}")
```
So far, we used batch gradient descent to update our weights. This means that we calculated the gradients using the entire training dataset. We also could've updated our weights using stochastic gradient descent (SGD) where we pass in one training example one at a time. The current standard is mini-batch gradient descent, which strikes a balance between batch and stochastic GD, where we update the weights using a mini-batch of n (`BATCH_SIZE`) samples. This is where the `DataLoader` object comes in handy.
```
# Create dataloaders
batch_size = 64
train_dataloader = train_dataset.create_dataloader(batch_size=batch_size)
val_dataloader = val_dataset.create_dataloader(batch_size=batch_size)
test_dataloader = test_dataset.create_dataloader(batch_size=batch_size)
batch_X, batch_y = next(iter(train_dataloader))
print ("Sample batch:\n"
f" X: {list(batch_X.size())}\n"
f" y: {list(batch_y.size())}\n"
"Sample point:\n"
f" X: {batch_X[0]}\n"
f" y: {batch_y[0]}")
```
# Device
So far we've been running our operations on the CPU but when we have large datasets and larger models to train, we can benefit by parallelzing tensor operations on a GPU. In this notebook, you can use a GPU by going to `Runtime` > `Change runtime type` > Select `GPU` in the `Hardware accelerator` dropdown. We can what device we're using with the following line of code:
```
# Set CUDA seeds
torch.cuda.manual_seed(SEED)
torch.cuda.manual_seed_all(SEED) # multi-GPU
# Set device
cuda = True
device = torch.device("cuda" if (
torch.cuda.is_available() and cuda) else "cpu")
torch.set_default_tensor_type("torch.FloatTensor")
if device.type == "cuda":
torch.set_default_tensor_type("torch.cuda.FloatTensor")
print (device)
```
# Model
Let's initialize the model we'll be using to show the capabilities of training utilities.
```
import math
from torch import nn
import torch.nn.functional as F
INPUT_DIM = X_train.shape[1] # 2D
HIDDEN_DIM = 100
DROPOUT_P = 0.1
NUM_CLASSES = len(label_encoder.classes)
NUM_EPOCHS = 10
class MLP(nn.Module):
def __init__(self, input_dim, hidden_dim, dropout_p, num_classes):
super(MLP, self).__init__()
self.fc1 = nn.Linear(input_dim, hidden_dim)
self.dropout = nn.Dropout(dropout_p)
self.fc2 = nn.Linear(hidden_dim, num_classes)
def forward(self, inputs):
x_in, = inputs
z = F.relu(self.fc1(x_in))
z = self.dropout(z)
z = self.fc2(z)
return z
# Initialize model
model = MLP(
input_dim=INPUT_DIM, hidden_dim=HIDDEN_DIM,
dropout_p=DROPOUT_P, num_classes=NUM_CLASSES)
model = model.to(device) # set device
print (model.named_parameters)
```
# Trainer
So far we've been writing training loops that train only using the train data split and then we perform evaluation on our test set. But in reality, we would follow this process:
1. Train using mini-batches on one epoch of the train data split.
2. Evaluate loss on the validation split and use it to adjust hyperparameters (ex. learning rate).
3. After training ends (via stagnation in improvements, desired performance, etc.), evaluate your trained model on the test (hold-out) data split.
We'll create a `Trainer` class to keep all of these processes organized.
The first function in the class is `train_step` which will train the model using batches from one epoch of the train data split.
```python
def train_step(self, dataloader):
"""Train step."""
# Set model to train mode
self.model.train()
loss = 0.0
# Iterate over train batches
for i, batch in enumerate(dataloader):
# Step
batch = [item.to(self.device) for item in batch] # Set device
inputs, targets = batch[:-1], batch[-1]
self.optimizer.zero_grad() # Reset gradients
z = self.model(inputs) # Forward pass
J = self.loss_fn(z, targets) # Define loss
J.backward() # Backward pass
self.optimizer.step() # Update weights
# Cumulative Metrics
loss += (J.detach().item() - loss) / (i + 1)
return loss
```
Next we'll define the `eval_step` which will be used for processing both the validation and test data splits. This is because neither of them require gradient updates and display the same metrics.
```python
def eval_step(self, dataloader):
"""Validation or test step."""
# Set model to eval mode
self.model.eval()
loss = 0.0
y_trues, y_probs = [], []
# Iterate over val batches
with torch.inference_mode():
for i, batch in enumerate(dataloader):
# Step
batch = [item.to(self.device) for item in batch] # Set device
inputs, y_true = batch[:-1], batch[-1]
z = self.model(inputs) # Forward pass
J = self.loss_fn(z, y_true).item()
# Cumulative Metrics
loss += (J - loss) / (i + 1)
# Store outputs
y_prob = F.softmax(z).cpu().numpy()
y_probs.extend(y_prob)
y_trues.extend(y_true.cpu().numpy())
return loss, np.vstack(y_trues), np.vstack(y_probs)
```
The final function is the `predict_step` which will be used for inference. It's fairly similar to the `eval_step` except we don't calculate any metrics. We pass on the predictions which we can use to generate our performance report.
```python
def predict_step(self, dataloader):
"""Prediction step."""
# Set model to eval mode
self.model.eval()
y_probs = []
# Iterate over val batches
with torch.inference_mode():
for i, batch in enumerate(dataloader):
# Forward pass w/ inputs
inputs, targets = batch[:-1], batch[-1]
z = self.model(inputs)
# Store outputs
y_prob = F.softmax(z).cpu().numpy()
y_probs.extend(y_prob)
return np.vstack(y_probs)
```
# LR Scheduler
As our model starts to optimize and perform better, the loss will reduce and we'll need to make smaller adjustments. If we keep using a fixed learning rate, we'll be overshooting back and forth. Therefore, we're going to add a learning rate scheduler to our optimizer to adjust our learning rate during training. There are many [schedulers](https://pytorch.org/docs/stable/optim.html#how-to-adjust-learning-rate) schedulers to choose from but a popular one is `ReduceLROnPlateau` which reduces the learning rate when a metric (ex. validation loss) stops improving. In the example below we'll reduce the learning rate by a factor of 0.1 (`factor=0.1`) when our metric of interest (`self.scheduler.step(val_loss)`) stops decreasing (`mode="min"`) for three (`patience=3`) straight epochs.
```python
# Initialize the LR scheduler
scheduler = torch.optim.lr_scheduler.ReduceLROnPlateau(
optimizer, mode="min", factor=0.1, patience=3)
...
train_loop():
...
# Steps
train_loss = trainer.train_step(dataloader=train_dataloader)
val_loss, _, _ = trainer.eval_step(dataloader=val_dataloader)
self.scheduler.step(val_loss)
...
```
# Early Stopping
We should never train our models for an arbitrary number of epochs but instead we should have explicit stopping criteria (even if you are bootstrapped by compute resources). Common stopping criteria include when validation performance stagnates for certain # of epochs (`patience`), desired performance is reached, etc.
```python
# Early stopping
if val_loss < best_val_loss:
best_val_loss = val_loss
best_model = trainer.model
_patience = patience # reset _patience
else:
_patience -= 1
if not _patience: # 0
print("Stopping early!")
break
```
# Training
Let's put all of this together now to train our model.
```
from torch.optim import Adam
LEARNING_RATE = 1e-2
NUM_EPOCHS = 100
PATIENCE = 3
# Define Loss
class_weights_tensor = torch.Tensor(list(class_weights.values())).to(device)
loss_fn = nn.CrossEntropyLoss(weight=class_weights_tensor)
# Define optimizer & scheduler
optimizer = Adam(model.parameters(), lr=LEARNING_RATE)
scheduler = torch.optim.lr_scheduler.ReduceLROnPlateau(
optimizer, mode="min", factor=0.1, patience=3)
class Trainer(object):
def __init__(self, model, device, loss_fn=None, optimizer=None, scheduler=None):
# Set params
self.model = model
self.device = device
self.loss_fn = loss_fn
self.optimizer = optimizer
self.scheduler = scheduler
def train_step(self, dataloader):
"""Train step."""
# Set model to train mode
self.model.train()
loss = 0.0
# Iterate over train batches
for i, batch in enumerate(dataloader):
# Step
batch = [item.to(self.device) for item in batch] # Set device
inputs, targets = batch[:-1], batch[-1]
self.optimizer.zero_grad() # Reset gradients
z = self.model(inputs) # Forward pass
J = self.loss_fn(z, targets) # Define loss
J.backward() # Backward pass
self.optimizer.step() # Update weights
# Cumulative Metrics
loss += (J.detach().item() - loss) / (i + 1)
return loss
def eval_step(self, dataloader):
"""Validation or test step."""
# Set model to eval mode
self.model.eval()
loss = 0.0
y_trues, y_probs = [], []
# Iterate over val batches
with torch.inference_mode():
for i, batch in enumerate(dataloader):
# Step
batch = [item.to(self.device) for item in batch] # Set device
inputs, y_true = batch[:-1], batch[-1]
z = self.model(inputs) # Forward pass
J = self.loss_fn(z, y_true).item()
# Cumulative Metrics
loss += (J - loss) / (i + 1)
# Store outputs
y_prob = F.sigmoid(z).cpu().numpy()
y_probs.extend(y_prob)
y_trues.extend(y_true.cpu().numpy())
return loss, np.vstack(y_trues), np.vstack(y_probs)
def predict_step(self, dataloader):
"""Prediction step."""
# Set model to eval mode
self.model.eval()
y_probs = []
# Iterate over val batches
with torch.inference_mode():
for i, batch in enumerate(dataloader):
# Forward pass w/ inputs
inputs, targets = batch[:-1], batch[-1]
z = self.model(inputs)
# Store outputs
y_prob = F.softmax(z).cpu().numpy()
y_probs.extend(y_prob)
return np.vstack(y_probs)
def train(self, num_epochs, patience, train_dataloader, val_dataloader):
best_val_loss = np.inf
for epoch in range(num_epochs):
# Steps
train_loss = self.train_step(dataloader=train_dataloader)
val_loss, _, _ = self.eval_step(dataloader=val_dataloader)
self.scheduler.step(val_loss)
# Early stopping
if val_loss < best_val_loss:
best_val_loss = val_loss
best_model = self.model
_patience = patience # reset _patience
else:
_patience -= 1
if not _patience: # 0
print("Stopping early!")
break
# Logging
print(
f"Epoch: {epoch+1} | "
f"train_loss: {train_loss:.5f}, "
f"val_loss: {val_loss:.5f}, "
f"lr: {self.optimizer.param_groups[0]['lr']:.2E}, "
f"_patience: {_patience}"
)
return best_model
# Trainer module
trainer = Trainer(
model=model, device=device, loss_fn=loss_fn,
optimizer=optimizer, scheduler=scheduler)
# Train
best_model = trainer.train(
NUM_EPOCHS, PATIENCE, train_dataloader, val_dataloader)
```
#Evaluation
```
import json
from sklearn.metrics import precision_recall_fscore_support
def get_performance(y_true, y_pred, classes):
"""Per-class performance metrics."""
# Performance
performance = {"overall": {}, "class": {}}
# Overall performance
metrics = precision_recall_fscore_support(y_true, y_pred, average="weighted")
performance["overall"]["precision"] = metrics[0]
performance["overall"]["recall"] = metrics[1]
performance["overall"]["f1"] = metrics[2]
performance["overall"]["num_samples"] = np.float64(len(y_true))
# Per-class performance
metrics = precision_recall_fscore_support(y_true, y_pred, average=None)
for i in range(len(classes)):
performance["class"][classes[i]] = {
"precision": metrics[0][i],
"recall": metrics[1][i],
"f1": metrics[2][i],
"num_samples": np.float64(metrics[3][i]),
}
return performance
# Get predictions
test_loss, y_true, y_prob = trainer.eval_step(dataloader=test_dataloader)
y_pred = np.argmax(y_prob, axis=1)
# Determine performance
performance = get_performance(
y_true=y_test, y_pred=y_pred, classes=label_encoder.classes)
print (json.dumps(performance["overall"], indent=2))
```
# Saving and loading
Many tutorials never show you how to save the components you created so you can load them for inference.
```
from pathlib import Path
# Save artifacts
dir = Path("mlp")
dir.mkdir(parents=True, exist_ok=True)
label_encoder.save(fp=Path(dir, "label_encoder.json"))
X_scaler.save(fp=Path(dir, "X_scaler.json"))
torch.save(best_model.state_dict(), Path(dir, "model.pt"))
with open(Path(dir, 'performance.json'), "w") as fp:
json.dump(performance, indent=2, sort_keys=False, fp=fp)
# Load artifacts
device = torch.device("cpu")
label_encoder = LabelEncoder.load(fp=Path(dir, "label_encoder.json"))
X_scaler = StandardScaler.load(fp=Path(dir, "X_scaler.json"))
model = MLP(
input_dim=INPUT_DIM, hidden_dim=HIDDEN_DIM,
dropout_p=DROPOUT_P, num_classes=NUM_CLASSES)
model.load_state_dict(torch.load(Path(dir, "model.pt"), map_location=device))
model.to(device)
# Initialize trainer
trainer = Trainer(model=model, device=device)
# Dataloader
sample = [[0.106737, 0.114197]] # c1
X = X_scaler.scale(sample)
y_filler = label_encoder.encode([label_encoder.classes[0]]*len(X))
dataset = Dataset(X=X, y=y_filler)
dataloader = dataset.create_dataloader(batch_size=batch_size)
# Inference
y_prob = trainer.predict_step(dataloader)
y_pred = np.argmax(y_prob, axis=1)
label_encoder.decode(y_pred)
```
# Miscellaneous
There are lots of other utilities to cover as well such as:
- Tokenizers to convert text to sequence of indices
- Various encoders to represent our data
- Padding to ensure uniform data shapes
- Experiment tracking to visualize and keep track of all experiments
- Hyperparameter optimization to tune our parameters (hidden units, learning rate, etc.)
- and many more!
We'll explore these as we require them in future lessons including some in our [MLOps](https://madewithml.com/#mlops) course!
|
github_jupyter
|
import numpy as np
import pandas as pd
import random
import torch
import torch.nn as nn
SEED = 1234
def set_seeds(seed=1234):
"""Set seeds for reproducibility."""
np.random.seed(seed)
random.seed(seed)
torch.manual_seed(seed)
torch.cuda.manual_seed(seed)
torch.cuda.manual_seed_all(seed) # multi-GPU
# Set seeds for reproducibility
set_seeds(seed=SEED)
# Set device
cuda = True
device = torch.device("cuda" if (
torch.cuda.is_available() and cuda) else "cpu")
torch.set_default_tensor_type("torch.FloatTensor")
if device.type == "cuda":
torch.set_default_tensor_type("torch.cuda.FloatTensor")
print (device)
import matplotlib.pyplot as plt
import pandas as pd
# Load data
url = "https://raw.githubusercontent.com/GokuMohandas/MadeWithML/main/datasets/spiral.csv"
df = pd.read_csv(url, header=0) # load
df = df.sample(frac=1).reset_index(drop=True) # shuffle
df.head()
# Data shapes
X = df[["X1", "X2"]].values
y = df["color"].values
print ("X: ", np.shape(X))
print ("y: ", np.shape(y))
# Visualize data
plt.title("Generated non-linear data")
colors = {"c1": "red", "c2": "yellow", "c3": "blue"}
plt.scatter(X[:, 0], X[:, 1], c=[colors[_y] for _y in y], edgecolors="k", s=25)
plt.show()
import collections
from sklearn.model_selection import train_test_split
TRAIN_SIZE = 0.7
VAL_SIZE = 0.15
TEST_SIZE = 0.15
def train_val_test_split(X, y, train_size):
"""Split dataset into data splits."""
X_train, X_, y_train, y_ = train_test_split(X, y, train_size=TRAIN_SIZE, stratify=y)
X_val, X_test, y_val, y_test = train_test_split(X_, y_, train_size=0.5, stratify=y_)
return X_train, X_val, X_test, y_train, y_val, y_test
# Create data splits
X_train, X_val, X_test, y_train, y_val, y_test = train_val_test_split(
X=X, y=y, train_size=TRAIN_SIZE)
print (f"X_train: {X_train.shape}, y_train: {y_train.shape}")
print (f"X_val: {X_val.shape}, y_val: {y_val.shape}")
print (f"X_test: {X_test.shape}, y_test: {y_test.shape}")
print (f"Sample point: {X_train[0]} → {y_train[0]}")
import itertools
class LabelEncoder(object):
"""Label encoder for tag labels."""
def __init__(self, class_to_index={}):
self.class_to_index = class_to_index
self.index_to_class = {v: k for k, v in self.class_to_index.items()}
self.classes = list(self.class_to_index.keys())
def __len__(self):
return len(self.class_to_index)
def __str__(self):
return f"<LabelEncoder(num_classes={len(self)})>"
def fit(self, y):
classes = np.unique(y)
for i, class_ in enumerate(classes):
self.class_to_index[class_] = i
self.index_to_class = {v: k for k, v in self.class_to_index.items()}
self.classes = list(self.class_to_index.keys())
return self
def encode(self, y):
encoded = np.zeros((len(y)), dtype=int)
for i, item in enumerate(y):
encoded[i] = self.class_to_index[item]
return encoded
def decode(self, y):
classes = []
for i, item in enumerate(y):
classes.append(self.index_to_class[item])
return classes
def save(self, fp):
with open(fp, "w") as fp:
contents = {'class_to_index': self.class_to_index}
json.dump(contents, fp, indent=4, sort_keys=False)
@classmethod
def load(cls, fp):
with open(fp, "r") as fp:
kwargs = json.load(fp=fp)
return cls(**kwargs)
# Encode
label_encoder = LabelEncoder()
label_encoder.fit(y_train)
label_encoder.class_to_index
# Convert labels to tokens
print (f"y_train[0]: {y_train[0]}")
y_train = label_encoder.encode(y_train)
y_val = label_encoder.encode(y_val)
y_test = label_encoder.encode(y_test)
print (f"y_train[0]: {y_train[0]}")
# Class weights
counts = np.bincount(y_train)
class_weights = {i: 1.0/count for i, count in enumerate(counts)}
print (f"counts: {counts}\nweights: {class_weights}")
class StandardScaler(object):
def __init__(self, mean=None, std=None):
self.mean = np.array(mean)
self.std = np.array(std)
def fit(self, X):
self.mean = np.mean(X_train, axis=0)
self.std = np.std(X_train, axis=0)
def scale(self, X):
return (X - self.mean) / self.std
def unscale(self, X):
return (X * self.std) + self.mean
def save(self, fp):
with open(fp, "w") as fp:
contents = {'mean': self.mean.tolist(), 'std': self.std.tolist()}
json.dump(contents, fp, indent=4, sort_keys=False)
@classmethod
def load(cls, fp):
with open(fp, "r") as fp:
kwargs = json.load(fp=fp)
return cls(**kwargs)
# Standardize the data (mean=0, std=1) using training data
X_scaler = StandardScaler()
X_scaler.fit(X_train)
# Apply scaler on training and test data (don't standardize outputs for classification)
X_train = X_scaler.scale(X_train)
X_val = X_scaler.scale(X_val)
X_test = X_scaler.scale(X_test)
# Check (means should be ~0 and std should be ~1)
print (f"X_test[0]: mean: {np.mean(X_test[:, 0], axis=0):.1f}, std: {np.std(X_test[:, 0], axis=0):.1f}")
print (f"X_test[1]: mean: {np.mean(X_test[:, 1], axis=0):.1f}, std: {np.std(X_test[:, 1], axis=0):.1f}")
import torch
# Seed seed for reproducibility
torch.manual_seed(SEED)
class Dataset(torch.utils.data.Dataset):
def __init__(self, X, y):
self.X = X
self.y = y
def __len__(self):
return len(self.y)
def __str__(self):
return f"<Dataset(N={len(self)})>"
def __getitem__(self, index):
X = self.X[index]
y = self.y[index]
return [X, y]
def collate_fn(self, batch):
"""Processing on a batch."""
# Get inputs
batch = np.array(batch, dtype=object)
X = np.stack(batch[:, 0], axis=0)
y = np.stack(batch[:, 1], axis=0)
# Cast
X = torch.FloatTensor(X.astype(np.float32))
y = torch.LongTensor(y.astype(np.int32))
return X, y
def create_dataloader(self, batch_size, shuffle=False, drop_last=False):
return torch.utils.data.DataLoader(
dataset=self, batch_size=batch_size, collate_fn=self.collate_fn,
shuffle=shuffle, drop_last=drop_last, pin_memory=True)
# Create datasets
train_dataset = Dataset(X=X_train, y=y_train)
val_dataset = Dataset(X=X_val, y=y_val)
test_dataset = Dataset(X=X_test, y=y_test)
print ("Datasets:\n"
f" Train dataset:{train_dataset.__str__()}\n"
f" Val dataset: {val_dataset.__str__()}\n"
f" Test dataset: {test_dataset.__str__()}\n"
"Sample point:\n"
f" X: {train_dataset[0][0]}\n"
f" y: {train_dataset[0][1]}")
# Create dataloaders
batch_size = 64
train_dataloader = train_dataset.create_dataloader(batch_size=batch_size)
val_dataloader = val_dataset.create_dataloader(batch_size=batch_size)
test_dataloader = test_dataset.create_dataloader(batch_size=batch_size)
batch_X, batch_y = next(iter(train_dataloader))
print ("Sample batch:\n"
f" X: {list(batch_X.size())}\n"
f" y: {list(batch_y.size())}\n"
"Sample point:\n"
f" X: {batch_X[0]}\n"
f" y: {batch_y[0]}")
# Set CUDA seeds
torch.cuda.manual_seed(SEED)
torch.cuda.manual_seed_all(SEED) # multi-GPU
# Set device
cuda = True
device = torch.device("cuda" if (
torch.cuda.is_available() and cuda) else "cpu")
torch.set_default_tensor_type("torch.FloatTensor")
if device.type == "cuda":
torch.set_default_tensor_type("torch.cuda.FloatTensor")
print (device)
import math
from torch import nn
import torch.nn.functional as F
INPUT_DIM = X_train.shape[1] # 2D
HIDDEN_DIM = 100
DROPOUT_P = 0.1
NUM_CLASSES = len(label_encoder.classes)
NUM_EPOCHS = 10
class MLP(nn.Module):
def __init__(self, input_dim, hidden_dim, dropout_p, num_classes):
super(MLP, self).__init__()
self.fc1 = nn.Linear(input_dim, hidden_dim)
self.dropout = nn.Dropout(dropout_p)
self.fc2 = nn.Linear(hidden_dim, num_classes)
def forward(self, inputs):
x_in, = inputs
z = F.relu(self.fc1(x_in))
z = self.dropout(z)
z = self.fc2(z)
return z
# Initialize model
model = MLP(
input_dim=INPUT_DIM, hidden_dim=HIDDEN_DIM,
dropout_p=DROPOUT_P, num_classes=NUM_CLASSES)
model = model.to(device) # set device
print (model.named_parameters)
def train_step(self, dataloader):
"""Train step."""
# Set model to train mode
self.model.train()
loss = 0.0
# Iterate over train batches
for i, batch in enumerate(dataloader):
# Step
batch = [item.to(self.device) for item in batch] # Set device
inputs, targets = batch[:-1], batch[-1]
self.optimizer.zero_grad() # Reset gradients
z = self.model(inputs) # Forward pass
J = self.loss_fn(z, targets) # Define loss
J.backward() # Backward pass
self.optimizer.step() # Update weights
# Cumulative Metrics
loss += (J.detach().item() - loss) / (i + 1)
return loss
def eval_step(self, dataloader):
"""Validation or test step."""
# Set model to eval mode
self.model.eval()
loss = 0.0
y_trues, y_probs = [], []
# Iterate over val batches
with torch.inference_mode():
for i, batch in enumerate(dataloader):
# Step
batch = [item.to(self.device) for item in batch] # Set device
inputs, y_true = batch[:-1], batch[-1]
z = self.model(inputs) # Forward pass
J = self.loss_fn(z, y_true).item()
# Cumulative Metrics
loss += (J - loss) / (i + 1)
# Store outputs
y_prob = F.softmax(z).cpu().numpy()
y_probs.extend(y_prob)
y_trues.extend(y_true.cpu().numpy())
return loss, np.vstack(y_trues), np.vstack(y_probs)
def predict_step(self, dataloader):
"""Prediction step."""
# Set model to eval mode
self.model.eval()
y_probs = []
# Iterate over val batches
with torch.inference_mode():
for i, batch in enumerate(dataloader):
# Forward pass w/ inputs
inputs, targets = batch[:-1], batch[-1]
z = self.model(inputs)
# Store outputs
y_prob = F.softmax(z).cpu().numpy()
y_probs.extend(y_prob)
return np.vstack(y_probs)
# Initialize the LR scheduler
scheduler = torch.optim.lr_scheduler.ReduceLROnPlateau(
optimizer, mode="min", factor=0.1, patience=3)
...
train_loop():
...
# Steps
train_loss = trainer.train_step(dataloader=train_dataloader)
val_loss, _, _ = trainer.eval_step(dataloader=val_dataloader)
self.scheduler.step(val_loss)
...
# Early stopping
if val_loss < best_val_loss:
best_val_loss = val_loss
best_model = trainer.model
_patience = patience # reset _patience
else:
_patience -= 1
if not _patience: # 0
print("Stopping early!")
break
from torch.optim import Adam
LEARNING_RATE = 1e-2
NUM_EPOCHS = 100
PATIENCE = 3
# Define Loss
class_weights_tensor = torch.Tensor(list(class_weights.values())).to(device)
loss_fn = nn.CrossEntropyLoss(weight=class_weights_tensor)
# Define optimizer & scheduler
optimizer = Adam(model.parameters(), lr=LEARNING_RATE)
scheduler = torch.optim.lr_scheduler.ReduceLROnPlateau(
optimizer, mode="min", factor=0.1, patience=3)
class Trainer(object):
def __init__(self, model, device, loss_fn=None, optimizer=None, scheduler=None):
# Set params
self.model = model
self.device = device
self.loss_fn = loss_fn
self.optimizer = optimizer
self.scheduler = scheduler
def train_step(self, dataloader):
"""Train step."""
# Set model to train mode
self.model.train()
loss = 0.0
# Iterate over train batches
for i, batch in enumerate(dataloader):
# Step
batch = [item.to(self.device) for item in batch] # Set device
inputs, targets = batch[:-1], batch[-1]
self.optimizer.zero_grad() # Reset gradients
z = self.model(inputs) # Forward pass
J = self.loss_fn(z, targets) # Define loss
J.backward() # Backward pass
self.optimizer.step() # Update weights
# Cumulative Metrics
loss += (J.detach().item() - loss) / (i + 1)
return loss
def eval_step(self, dataloader):
"""Validation or test step."""
# Set model to eval mode
self.model.eval()
loss = 0.0
y_trues, y_probs = [], []
# Iterate over val batches
with torch.inference_mode():
for i, batch in enumerate(dataloader):
# Step
batch = [item.to(self.device) for item in batch] # Set device
inputs, y_true = batch[:-1], batch[-1]
z = self.model(inputs) # Forward pass
J = self.loss_fn(z, y_true).item()
# Cumulative Metrics
loss += (J - loss) / (i + 1)
# Store outputs
y_prob = F.sigmoid(z).cpu().numpy()
y_probs.extend(y_prob)
y_trues.extend(y_true.cpu().numpy())
return loss, np.vstack(y_trues), np.vstack(y_probs)
def predict_step(self, dataloader):
"""Prediction step."""
# Set model to eval mode
self.model.eval()
y_probs = []
# Iterate over val batches
with torch.inference_mode():
for i, batch in enumerate(dataloader):
# Forward pass w/ inputs
inputs, targets = batch[:-1], batch[-1]
z = self.model(inputs)
# Store outputs
y_prob = F.softmax(z).cpu().numpy()
y_probs.extend(y_prob)
return np.vstack(y_probs)
def train(self, num_epochs, patience, train_dataloader, val_dataloader):
best_val_loss = np.inf
for epoch in range(num_epochs):
# Steps
train_loss = self.train_step(dataloader=train_dataloader)
val_loss, _, _ = self.eval_step(dataloader=val_dataloader)
self.scheduler.step(val_loss)
# Early stopping
if val_loss < best_val_loss:
best_val_loss = val_loss
best_model = self.model
_patience = patience # reset _patience
else:
_patience -= 1
if not _patience: # 0
print("Stopping early!")
break
# Logging
print(
f"Epoch: {epoch+1} | "
f"train_loss: {train_loss:.5f}, "
f"val_loss: {val_loss:.5f}, "
f"lr: {self.optimizer.param_groups[0]['lr']:.2E}, "
f"_patience: {_patience}"
)
return best_model
# Trainer module
trainer = Trainer(
model=model, device=device, loss_fn=loss_fn,
optimizer=optimizer, scheduler=scheduler)
# Train
best_model = trainer.train(
NUM_EPOCHS, PATIENCE, train_dataloader, val_dataloader)
import json
from sklearn.metrics import precision_recall_fscore_support
def get_performance(y_true, y_pred, classes):
"""Per-class performance metrics."""
# Performance
performance = {"overall": {}, "class": {}}
# Overall performance
metrics = precision_recall_fscore_support(y_true, y_pred, average="weighted")
performance["overall"]["precision"] = metrics[0]
performance["overall"]["recall"] = metrics[1]
performance["overall"]["f1"] = metrics[2]
performance["overall"]["num_samples"] = np.float64(len(y_true))
# Per-class performance
metrics = precision_recall_fscore_support(y_true, y_pred, average=None)
for i in range(len(classes)):
performance["class"][classes[i]] = {
"precision": metrics[0][i],
"recall": metrics[1][i],
"f1": metrics[2][i],
"num_samples": np.float64(metrics[3][i]),
}
return performance
# Get predictions
test_loss, y_true, y_prob = trainer.eval_step(dataloader=test_dataloader)
y_pred = np.argmax(y_prob, axis=1)
# Determine performance
performance = get_performance(
y_true=y_test, y_pred=y_pred, classes=label_encoder.classes)
print (json.dumps(performance["overall"], indent=2))
from pathlib import Path
# Save artifacts
dir = Path("mlp")
dir.mkdir(parents=True, exist_ok=True)
label_encoder.save(fp=Path(dir, "label_encoder.json"))
X_scaler.save(fp=Path(dir, "X_scaler.json"))
torch.save(best_model.state_dict(), Path(dir, "model.pt"))
with open(Path(dir, 'performance.json'), "w") as fp:
json.dump(performance, indent=2, sort_keys=False, fp=fp)
# Load artifacts
device = torch.device("cpu")
label_encoder = LabelEncoder.load(fp=Path(dir, "label_encoder.json"))
X_scaler = StandardScaler.load(fp=Path(dir, "X_scaler.json"))
model = MLP(
input_dim=INPUT_DIM, hidden_dim=HIDDEN_DIM,
dropout_p=DROPOUT_P, num_classes=NUM_CLASSES)
model.load_state_dict(torch.load(Path(dir, "model.pt"), map_location=device))
model.to(device)
# Initialize trainer
trainer = Trainer(model=model, device=device)
# Dataloader
sample = [[0.106737, 0.114197]] # c1
X = X_scaler.scale(sample)
y_filler = label_encoder.encode([label_encoder.classes[0]]*len(X))
dataset = Dataset(X=X, y=y_filler)
dataloader = dataset.create_dataloader(batch_size=batch_size)
# Inference
y_prob = trainer.predict_step(dataloader)
y_pred = np.argmax(y_prob, axis=1)
label_encoder.decode(y_pred)
| 0.853196 | 0.977088 |
```
from IPython.display import HTML
HTML('''<script>
code_show=true;
function code_toggle() {
if (code_show){
$('div.input').hide();
} else {
$('div.input').show();
}
code_show = !code_show
}
$( document ).ready(code_toggle);
</script>
<form action="javascript:code_toggle()"><input type="submit" value="Click here to toggle on/off the raw code."></form>''')
```
#
A few things:
-I've downloaded and compiled mocsy on the /data machines at /data/tjarniko/mocsy, so you need to either append that path or redownload it/ recompile it (no reason probably not to use my download)
-Need to use in_situ T, psu.
-Pretty good docs are here, I didn't read them the first time around, oops http://ocmip5.ipsl.jussieu.fr/mocsy/
-it only takes 1D arrays, so you need to use np.ravel
-The formulation is like this:
response_tup = mocsy.mvars(temp=Tr, sal=Sr, alk=TAr, dic=DICr,
sil=zero, phos=zero, patm=Pr, depth=zero, lat=zero,
optcon='mol/m3', optt='Tinsitu', optp='m',
optb = 'l10', optk1k2='m10', optkf = 'dg', optgas = 'Pinsitu')
pH,pco2,fco2,co2,hco3,co3,OmegaA,OmegaC,BetaD,DENis,p,Tis = response_tup
where I've selected the best optb, optk1k2, optkf options. They're (slightly) different constants in formulations of the carbonate chemistry system. they're pretty similar, esp for your work (probably limited freshwater?)
-silicate and phosphate do affect OmegaA a little bit, but really just a little bit, I wouldn't worry about it. Same with latitude (which again factors into one of the parameterzations)
-atmospheric pressure should be in Patm (i usually say 1 unless I'm calculating it online)
-depth can be in meters or dbar, I use meters.
```
import sys
sys.path.append('/data/kramosmu/mocsy')
import mocsy
import numpy as np
import matplotlib.pyplot as plt
import seawater as sw
import matplotlib.pyplot as plt
import netCDF4 as nc
import numpy as np
import xarray as xr
%matplotlib inline
def calc_rho(RhoRef,T,S,alpha=2.0E-4, beta=7.4E-4):
"""-----------------------------------------------------------------------------
calc_rho calculates the density profile using a linear equation of state.
INPUT:
state: xarray dataframe
RhoRef : reference density at the same z as T and S slices. Can be a scalar or a
vector, depending on the size of T and S.
T, S : should be 1D arrays size nz
alpha = 2.0E-4 # 1/degC, thermal expansion coefficient
beta = 7.4E-4, haline expansion coefficient
OUTPUT:
rho - Density [nz]
-----------------------------------------------------------------------------"""
#Linear eq. of state
rho = RhoRef*(np.ones(np.shape(T[:])) - alpha*(T[:]) + beta*(S[:]))
return rho
def call_rho(t,state,zslice,xind,yind):
RhoRef = 999.79998779 # It is constant in all my runs, can't run rdmds
T = state.Temp.isel(T=t,Z=zslice,X=xind,Y=yind)
S = state.S.isel(T=t,Z=zslice,X=xind,Y=yind)
rho = calc_rho(RhoRef,T,S,alpha=2.0E-4, beta=7.4E-4)
return(rho)
def get_vars(state, ptracer, mask, yind, xind):
DIC_umolkg = np.ma.masked_array(np.nanmean(ptracer.Tr09[8:,:,yind,xind], axis=0), mask=mask) #umol/kg
TA_umolkg = np.ma.masked_array(np.nanmean(ptracer.Tr10[8:,:,yind,xind], axis=0), mask=mask) #umol/kg
density = np.ma.masked_array(call_rho(0,state,slice(0,104),xind,yind).data, mask=mask)
DIC = (density*DIC_umolkg/1000)
TA = (density*TA_umolkg/1000)
S = np.ma.masked_array(np.nanmean(state.S[8:,:,yind,xind], axis=0), mask=mask)
T = np.ma.masked_array(np.nanmean(state.Temp[8:,:,yind,xind], axis=0), mask=mask)
return(DIC, TA, S, T)
grid_file_A = '/data/kramosmu/results/TracerExperiments/UPW_10TR_BF2_AST/03_Ast03_Argo/gridGlob.nc'
grid_file_Anoc = '/data/kramosmu/results/TracerExperiments/UPW_10TR_BF2_AST/04_Ast03_No_Cny_Argo/gridGlob.nc'
ptrARGO = xr.open_dataset('/data/kramosmu/results/TracerExperiments/UPW_10TR_BF2_AST/03_Ast03_Argo/ptracersGlob.nc')
ptrARGOnoc = xr.open_dataset('/data/kramosmu/results/TracerExperiments/UPW_10TR_BF2_AST/04_Ast03_No_Cny_Argo/ptracersGlob.nc')
stateARGO = xr.open_dataset('/data/kramosmu/results/TracerExperiments/UPW_10TR_BF2_AST/03_Ast03_Argo/stateGlob.nc')
stateARGOnoc = xr.open_dataset('/data/kramosmu/results/TracerExperiments/UPW_10TR_BF2_AST/04_Ast03_No_Cny_Argo/stateGlob.nc')
gridARGO = xr.open_dataset(grid_file_A)
gridARGOnoc = xr.open_dataset(grid_file_Anoc)
with nc.Dataset(grid_file_A, 'r') as nbl:
hFacA = nbl.variables['HFacC'][:]
hfac = np.ma.masked_values(hFacA, 0)
maskCA = np.ma.getmask(hfac)
with nc.Dataset(grid_file_Anoc, 'r') as nbl:
hFacA = nbl.variables['HFacC'][:]
hfac = np.ma.masked_values(hFacA, 0)
maskCAnoc = np.ma.getmask(hfac)
grid_file_B = '/data/kramosmu/results/TracerExperiments/UPW_10TR_BF4_BAR/03_Bar03_Path/gridGlob.nc'
grid_file_Bnoc = '/data/kramosmu/results/TracerExperiments/UPW_10TR_BF4_BAR/04_Bar03_No_Cny_Path/gridGlob.nc'
ptrPATH = xr.open_dataset('/data/kramosmu/results/TracerExperiments/UPW_10TR_BF4_BAR/03_Bar03_Path/ptracersGlob.nc')
ptrPATHnoc = xr.open_dataset('/data/kramosmu/results/TracerExperiments/UPW_10TR_BF4_BAR/04_Bar03_No_Cny_Path/ptracersGlob.nc')
statePATH = xr.open_dataset('/data/kramosmu/results/TracerExperiments/UPW_10TR_BF4_BAR/03_Bar03_Path/stateGlob.nc')
statePATHnoc = xr.open_dataset('/data/kramosmu/results/TracerExperiments/UPW_10TR_BF4_BAR/04_Bar03_No_Cny_Path/stateGlob.nc')
gridPATH = xr.open_dataset(grid_file_B)
gridPATHnoc = xr.open_dataset(grid_file_Bnoc)
with nc.Dataset(grid_file_B, 'r') as nbl:
hFacB = nbl.variables['HFacC'][:]
hfac = np.ma.masked_values(hFacB, 0)
maskCB = np.ma.getmask(hfac)
with nc.Dataset(grid_file_Bnoc, 'r') as nbl:
hFacB = nbl.variables['HFacC'][:]
hfac = np.ma.masked_values(hFacB, 0)
maskCBnoc = np.ma.getmask(hfac)
# Stations x,y indices A for ARGO, B for Pathways
yindA = [235,255,250,158]
xindA = [180,300,400,300]
yindB = [165, 170, 180, 140]
xindB = [180, 250, 300, 250]
linestyles = ['-','--',':','-.']
st_names=['S1','S2','S3','S4']
# Make profiles at stations S1, S2, S3, S4, MASKED USING NO-CANYON HFACC
fig, (ax0,ax1,ax2,ax3,ax4,ax5,ax6,ax7,ax8) = plt.subplots(1,9, figsize=(18,4))
for xind, yind, st in zip(xindA, yindA, st_names):
DIC, TA, S, T = get_vars(stateARGO,ptrARGO,maskCA[:,yind,xind], yind, xind)
DIC_noc, TA_noc, S_noc, T_noc = get_vars(stateARGOnoc,ptrARGOnoc,maskCAnoc[:,yind,xind], yind, xind)
depth = np.ma.masked_array(gridARGO.Z[:], mask=maskCA[:,yind,xind])
depth_noc = np.ma.masked_array(gridARGOnoc.Z[:], mask=maskCAnoc[:,yind,xind])
Surf_p = np.ones_like(DIC)
zero = np.zeros_like(DIC)
response_tup = mocsy.mvars(temp=T, sal=S, alk=TA, dic=DIC,
sil=zero, phos=zero, patm=Surf_p, depth=depth, lat=zero,
optcon='mol/m3', optt='Tinsitu', optp='m',
optb = 'l10', optk1k2='m10', optkf = 'dg', optgas = 'Pinsitu')
pH,pco2,fco2,co2,hco3,co3,OmegaA,OmegaC,BetaD,DENis,p,Tis = response_tup
response_tup_noc = mocsy.mvars(temp=T_noc, sal=S_noc, alk=TA_noc, dic=DIC_noc,
sil=zero, phos=zero, patm=Surf_p, depth=depth_noc, lat=zero,
optcon='mol/m3', optt='Tinsitu', optp='m',
optb = 'l10', optk1k2='m10', optkf = 'dg', optgas = 'Pinsitu')
pH_noc,pco2_noc,fco2_noc,co2_noc,hco3_noc,co3_noc,OmegaA_noc,OmegaC_noc,BetaD_noc,DENis_noc,p_noc,Tis_noc = \
response_tup_noc
pH_anom = pH-pH_noc
DIC_anom = DIC-DIC_noc
TA_anom = TA-TA_noc
ax0.plot(pH,depth_noc-np.nanmin(depth_noc), label=st)
ax1.plot(DIC,depth_noc-np.nanmin(depth_noc), label=st)
ax2.plot(TA,depth_noc-np.nanmin(depth_noc), label=st)
ax3.plot(pH_noc,depth_noc-np.nanmin(depth_noc), label=st)
ax4.plot(DIC_noc,depth_noc-np.nanmin(depth_noc), label=st)
ax5.plot(TA_noc,depth_noc-np.nanmin(depth_noc), label=st)
ax6.plot(pH_anom,depth_noc-np.nanmin(depth_noc), label=st)
ax7.plot(DIC_anom,depth_noc-np.nanmin(depth_noc), label=st)
ax8.plot(TA_anom,depth_noc-np.nanmin(depth_noc), label=st)
ind_min = np.max(np.argwhere(depth_noc!=np.nan))
print('min pH anom at %f m of %s is %f ' %(depth[ind_min],st,pH_anom[ind_min]))
for ax in [ax1,ax2,ax3,ax4,ax5,ax6,ax7,ax8]:
ax.set_yticks([])
ax0.set_ylabel('Height above bottom / m')
ax0.set_xlabel('pH')
ax1.set_xlabel('DIC / $\mu$mol L$^{-1}$')
ax2.set_xlabel('TA / $\mu$mol L$^{-1}$')
ax3.set_xlabel('pH noc')
ax4.set_xlabel('DIC noc / $\mu$mol L$^{-1}$')
ax5.set_xlabel('TA noc / $\mu$mol L$^{-1}$')
ax6.set_xlabel('pH anom')
ax7.set_xlabel('DIC anom / $\mu$mol L$^{-1}$')
ax8.set_xlabel('TA anom / $\mu$mol L$^{-1}$')
ax0.set_title('ASTORIA ARGO')
ax8.legend(loc=0)
# Make profiles at stations S1, S2, S3, S4, MASKED USING NO-CANYON HFACC
fig, (ax0,ax1,ax2,ax3,ax4,ax5,ax6,ax7,ax8) = plt.subplots(1,9, figsize=(18,4))
for xind, yind, st in zip(xindB, yindB, st_names):
DIC, TA, S, T = get_vars(statePATH,ptrPATH,maskCBnoc[:,yind,xind], yind, xind)
DIC_noc, TA_noc, S_noc, T_noc = get_vars(statePATHnoc,ptrPATHnoc,maskCBnoc[:,yind,xind], yind, xind)
depth = np.ma.masked_array(gridPATH.Z[:], mask=maskCB[:,yind,xind])
depth_noc = np.ma.masked_array(gridPATHnoc.Z[:], mask=maskCBnoc[:,yind,xind])
Surf_p = np.ones_like(DIC)
zero = np.zeros_like(DIC)
response_tup = mocsy.mvars(temp=T, sal=S, alk=TA, dic=DIC,
sil=zero, phos=zero, patm=Surf_p, depth=depth_noc, lat=zero,
optcon='mol/m3', optt='Tinsitu', optp='m',
optb = 'l10', optk1k2='m10', optkf = 'dg', optgas = 'Pinsitu')
pH,pco2,fco2,co2,hco3,co3,OmegaA,OmegaC,BetaD,DENis,p,Tis = response_tup
response_tup_noc = mocsy.mvars(temp=T_noc, sal=S_noc, alk=TA_noc, dic=DIC_noc,
sil=zero, phos=zero, patm=Surf_p, depth=depth_noc, lat=zero,
optcon='mol/m3', optt='Tinsitu', optp='m',
optb = 'l10', optk1k2='m10', optkf = 'dg', optgas = 'Pinsitu')
pH_noc,pco2_noc,fco2_noc,co2_noc,hco3_noc,co3_noc,OmegaA_noc,OmegaC_noc,BetaD_noc,DENis_noc,p_noc,Tis_noc = \
response_tup_noc
pH_anom = pH-pH_noc
DIC_anom = DIC-DIC_noc
TA_anom = TA-TA_noc
ax0.plot(pH,depth_noc-np.nanmin(depth_noc), label=st)
ax1.plot(DIC,depth_noc-np.nanmin(depth_noc), label=st)
ax2.plot(TA,depth_noc-np.nanmin(depth_noc), label=st)
ax3.plot(pH_noc,depth_noc-np.nanmin(depth_noc), label=st)
ax4.plot(DIC_noc,depth_noc-np.nanmin(depth_noc), label=st)
ax5.plot(TA_noc,depth_noc-np.nanmin(depth_noc), label=st)
ax6.plot(pH_anom,depth_noc-np.nanmin(depth_noc), label=st)
ax7.plot(DIC_anom,depth_noc-np.nanmin(depth_noc), label=st)
ax8.plot(TA_anom,depth_noc-np.nanmin(depth_noc), label=st)
ind_min = np.max(np.argwhere(depth_noc!=np.nan))
print('min pH anom at %f m of %s is %f ' %(depth[ind_min],st,pH_anom[ind_min]))
for ax in [ax1,ax2,ax3,ax4,ax5,ax6,ax7,ax8]:
ax.set_yticks([])
ax0.set_ylabel('Height above bottom / m')
ax0.set_xlabel('pH')
ax1.set_xlabel('DIC / $\mu$mol L$^{-1}$')
ax2.set_xlabel('TA / $\mu$mol L$^{-1}$')
ax3.set_xlabel('pH noc')
ax4.set_xlabel('DIC noc / $\mu$mol L$^{-1}$')
ax5.set_xlabel('TA noc / $\mu$mol L$^{-1}$')
ax6.set_xlabel('pH anom')
ax7.set_xlabel('DIC anom / $\mu$mol L$^{-1}$')
ax8.set_xlabel('TA anom / $\mu$mol L$^{-1}$')
ax0.set_title('BARKLEY PATHWAYS')
ax8.legend(loc=0)
```
|
github_jupyter
|
from IPython.display import HTML
HTML('''<script>
code_show=true;
function code_toggle() {
if (code_show){
$('div.input').hide();
} else {
$('div.input').show();
}
code_show = !code_show
}
$( document ).ready(code_toggle);
</script>
<form action="javascript:code_toggle()"><input type="submit" value="Click here to toggle on/off the raw code."></form>''')
import sys
sys.path.append('/data/kramosmu/mocsy')
import mocsy
import numpy as np
import matplotlib.pyplot as plt
import seawater as sw
import matplotlib.pyplot as plt
import netCDF4 as nc
import numpy as np
import xarray as xr
%matplotlib inline
def calc_rho(RhoRef,T,S,alpha=2.0E-4, beta=7.4E-4):
"""-----------------------------------------------------------------------------
calc_rho calculates the density profile using a linear equation of state.
INPUT:
state: xarray dataframe
RhoRef : reference density at the same z as T and S slices. Can be a scalar or a
vector, depending on the size of T and S.
T, S : should be 1D arrays size nz
alpha = 2.0E-4 # 1/degC, thermal expansion coefficient
beta = 7.4E-4, haline expansion coefficient
OUTPUT:
rho - Density [nz]
-----------------------------------------------------------------------------"""
#Linear eq. of state
rho = RhoRef*(np.ones(np.shape(T[:])) - alpha*(T[:]) + beta*(S[:]))
return rho
def call_rho(t,state,zslice,xind,yind):
RhoRef = 999.79998779 # It is constant in all my runs, can't run rdmds
T = state.Temp.isel(T=t,Z=zslice,X=xind,Y=yind)
S = state.S.isel(T=t,Z=zslice,X=xind,Y=yind)
rho = calc_rho(RhoRef,T,S,alpha=2.0E-4, beta=7.4E-4)
return(rho)
def get_vars(state, ptracer, mask, yind, xind):
DIC_umolkg = np.ma.masked_array(np.nanmean(ptracer.Tr09[8:,:,yind,xind], axis=0), mask=mask) #umol/kg
TA_umolkg = np.ma.masked_array(np.nanmean(ptracer.Tr10[8:,:,yind,xind], axis=0), mask=mask) #umol/kg
density = np.ma.masked_array(call_rho(0,state,slice(0,104),xind,yind).data, mask=mask)
DIC = (density*DIC_umolkg/1000)
TA = (density*TA_umolkg/1000)
S = np.ma.masked_array(np.nanmean(state.S[8:,:,yind,xind], axis=0), mask=mask)
T = np.ma.masked_array(np.nanmean(state.Temp[8:,:,yind,xind], axis=0), mask=mask)
return(DIC, TA, S, T)
grid_file_A = '/data/kramosmu/results/TracerExperiments/UPW_10TR_BF2_AST/03_Ast03_Argo/gridGlob.nc'
grid_file_Anoc = '/data/kramosmu/results/TracerExperiments/UPW_10TR_BF2_AST/04_Ast03_No_Cny_Argo/gridGlob.nc'
ptrARGO = xr.open_dataset('/data/kramosmu/results/TracerExperiments/UPW_10TR_BF2_AST/03_Ast03_Argo/ptracersGlob.nc')
ptrARGOnoc = xr.open_dataset('/data/kramosmu/results/TracerExperiments/UPW_10TR_BF2_AST/04_Ast03_No_Cny_Argo/ptracersGlob.nc')
stateARGO = xr.open_dataset('/data/kramosmu/results/TracerExperiments/UPW_10TR_BF2_AST/03_Ast03_Argo/stateGlob.nc')
stateARGOnoc = xr.open_dataset('/data/kramosmu/results/TracerExperiments/UPW_10TR_BF2_AST/04_Ast03_No_Cny_Argo/stateGlob.nc')
gridARGO = xr.open_dataset(grid_file_A)
gridARGOnoc = xr.open_dataset(grid_file_Anoc)
with nc.Dataset(grid_file_A, 'r') as nbl:
hFacA = nbl.variables['HFacC'][:]
hfac = np.ma.masked_values(hFacA, 0)
maskCA = np.ma.getmask(hfac)
with nc.Dataset(grid_file_Anoc, 'r') as nbl:
hFacA = nbl.variables['HFacC'][:]
hfac = np.ma.masked_values(hFacA, 0)
maskCAnoc = np.ma.getmask(hfac)
grid_file_B = '/data/kramosmu/results/TracerExperiments/UPW_10TR_BF4_BAR/03_Bar03_Path/gridGlob.nc'
grid_file_Bnoc = '/data/kramosmu/results/TracerExperiments/UPW_10TR_BF4_BAR/04_Bar03_No_Cny_Path/gridGlob.nc'
ptrPATH = xr.open_dataset('/data/kramosmu/results/TracerExperiments/UPW_10TR_BF4_BAR/03_Bar03_Path/ptracersGlob.nc')
ptrPATHnoc = xr.open_dataset('/data/kramosmu/results/TracerExperiments/UPW_10TR_BF4_BAR/04_Bar03_No_Cny_Path/ptracersGlob.nc')
statePATH = xr.open_dataset('/data/kramosmu/results/TracerExperiments/UPW_10TR_BF4_BAR/03_Bar03_Path/stateGlob.nc')
statePATHnoc = xr.open_dataset('/data/kramosmu/results/TracerExperiments/UPW_10TR_BF4_BAR/04_Bar03_No_Cny_Path/stateGlob.nc')
gridPATH = xr.open_dataset(grid_file_B)
gridPATHnoc = xr.open_dataset(grid_file_Bnoc)
with nc.Dataset(grid_file_B, 'r') as nbl:
hFacB = nbl.variables['HFacC'][:]
hfac = np.ma.masked_values(hFacB, 0)
maskCB = np.ma.getmask(hfac)
with nc.Dataset(grid_file_Bnoc, 'r') as nbl:
hFacB = nbl.variables['HFacC'][:]
hfac = np.ma.masked_values(hFacB, 0)
maskCBnoc = np.ma.getmask(hfac)
# Stations x,y indices A for ARGO, B for Pathways
yindA = [235,255,250,158]
xindA = [180,300,400,300]
yindB = [165, 170, 180, 140]
xindB = [180, 250, 300, 250]
linestyles = ['-','--',':','-.']
st_names=['S1','S2','S3','S4']
# Make profiles at stations S1, S2, S3, S4, MASKED USING NO-CANYON HFACC
fig, (ax0,ax1,ax2,ax3,ax4,ax5,ax6,ax7,ax8) = plt.subplots(1,9, figsize=(18,4))
for xind, yind, st in zip(xindA, yindA, st_names):
DIC, TA, S, T = get_vars(stateARGO,ptrARGO,maskCA[:,yind,xind], yind, xind)
DIC_noc, TA_noc, S_noc, T_noc = get_vars(stateARGOnoc,ptrARGOnoc,maskCAnoc[:,yind,xind], yind, xind)
depth = np.ma.masked_array(gridARGO.Z[:], mask=maskCA[:,yind,xind])
depth_noc = np.ma.masked_array(gridARGOnoc.Z[:], mask=maskCAnoc[:,yind,xind])
Surf_p = np.ones_like(DIC)
zero = np.zeros_like(DIC)
response_tup = mocsy.mvars(temp=T, sal=S, alk=TA, dic=DIC,
sil=zero, phos=zero, patm=Surf_p, depth=depth, lat=zero,
optcon='mol/m3', optt='Tinsitu', optp='m',
optb = 'l10', optk1k2='m10', optkf = 'dg', optgas = 'Pinsitu')
pH,pco2,fco2,co2,hco3,co3,OmegaA,OmegaC,BetaD,DENis,p,Tis = response_tup
response_tup_noc = mocsy.mvars(temp=T_noc, sal=S_noc, alk=TA_noc, dic=DIC_noc,
sil=zero, phos=zero, patm=Surf_p, depth=depth_noc, lat=zero,
optcon='mol/m3', optt='Tinsitu', optp='m',
optb = 'l10', optk1k2='m10', optkf = 'dg', optgas = 'Pinsitu')
pH_noc,pco2_noc,fco2_noc,co2_noc,hco3_noc,co3_noc,OmegaA_noc,OmegaC_noc,BetaD_noc,DENis_noc,p_noc,Tis_noc = \
response_tup_noc
pH_anom = pH-pH_noc
DIC_anom = DIC-DIC_noc
TA_anom = TA-TA_noc
ax0.plot(pH,depth_noc-np.nanmin(depth_noc), label=st)
ax1.plot(DIC,depth_noc-np.nanmin(depth_noc), label=st)
ax2.plot(TA,depth_noc-np.nanmin(depth_noc), label=st)
ax3.plot(pH_noc,depth_noc-np.nanmin(depth_noc), label=st)
ax4.plot(DIC_noc,depth_noc-np.nanmin(depth_noc), label=st)
ax5.plot(TA_noc,depth_noc-np.nanmin(depth_noc), label=st)
ax6.plot(pH_anom,depth_noc-np.nanmin(depth_noc), label=st)
ax7.plot(DIC_anom,depth_noc-np.nanmin(depth_noc), label=st)
ax8.plot(TA_anom,depth_noc-np.nanmin(depth_noc), label=st)
ind_min = np.max(np.argwhere(depth_noc!=np.nan))
print('min pH anom at %f m of %s is %f ' %(depth[ind_min],st,pH_anom[ind_min]))
for ax in [ax1,ax2,ax3,ax4,ax5,ax6,ax7,ax8]:
ax.set_yticks([])
ax0.set_ylabel('Height above bottom / m')
ax0.set_xlabel('pH')
ax1.set_xlabel('DIC / $\mu$mol L$^{-1}$')
ax2.set_xlabel('TA / $\mu$mol L$^{-1}$')
ax3.set_xlabel('pH noc')
ax4.set_xlabel('DIC noc / $\mu$mol L$^{-1}$')
ax5.set_xlabel('TA noc / $\mu$mol L$^{-1}$')
ax6.set_xlabel('pH anom')
ax7.set_xlabel('DIC anom / $\mu$mol L$^{-1}$')
ax8.set_xlabel('TA anom / $\mu$mol L$^{-1}$')
ax0.set_title('ASTORIA ARGO')
ax8.legend(loc=0)
# Make profiles at stations S1, S2, S3, S4, MASKED USING NO-CANYON HFACC
fig, (ax0,ax1,ax2,ax3,ax4,ax5,ax6,ax7,ax8) = plt.subplots(1,9, figsize=(18,4))
for xind, yind, st in zip(xindB, yindB, st_names):
DIC, TA, S, T = get_vars(statePATH,ptrPATH,maskCBnoc[:,yind,xind], yind, xind)
DIC_noc, TA_noc, S_noc, T_noc = get_vars(statePATHnoc,ptrPATHnoc,maskCBnoc[:,yind,xind], yind, xind)
depth = np.ma.masked_array(gridPATH.Z[:], mask=maskCB[:,yind,xind])
depth_noc = np.ma.masked_array(gridPATHnoc.Z[:], mask=maskCBnoc[:,yind,xind])
Surf_p = np.ones_like(DIC)
zero = np.zeros_like(DIC)
response_tup = mocsy.mvars(temp=T, sal=S, alk=TA, dic=DIC,
sil=zero, phos=zero, patm=Surf_p, depth=depth_noc, lat=zero,
optcon='mol/m3', optt='Tinsitu', optp='m',
optb = 'l10', optk1k2='m10', optkf = 'dg', optgas = 'Pinsitu')
pH,pco2,fco2,co2,hco3,co3,OmegaA,OmegaC,BetaD,DENis,p,Tis = response_tup
response_tup_noc = mocsy.mvars(temp=T_noc, sal=S_noc, alk=TA_noc, dic=DIC_noc,
sil=zero, phos=zero, patm=Surf_p, depth=depth_noc, lat=zero,
optcon='mol/m3', optt='Tinsitu', optp='m',
optb = 'l10', optk1k2='m10', optkf = 'dg', optgas = 'Pinsitu')
pH_noc,pco2_noc,fco2_noc,co2_noc,hco3_noc,co3_noc,OmegaA_noc,OmegaC_noc,BetaD_noc,DENis_noc,p_noc,Tis_noc = \
response_tup_noc
pH_anom = pH-pH_noc
DIC_anom = DIC-DIC_noc
TA_anom = TA-TA_noc
ax0.plot(pH,depth_noc-np.nanmin(depth_noc), label=st)
ax1.plot(DIC,depth_noc-np.nanmin(depth_noc), label=st)
ax2.plot(TA,depth_noc-np.nanmin(depth_noc), label=st)
ax3.plot(pH_noc,depth_noc-np.nanmin(depth_noc), label=st)
ax4.plot(DIC_noc,depth_noc-np.nanmin(depth_noc), label=st)
ax5.plot(TA_noc,depth_noc-np.nanmin(depth_noc), label=st)
ax6.plot(pH_anom,depth_noc-np.nanmin(depth_noc), label=st)
ax7.plot(DIC_anom,depth_noc-np.nanmin(depth_noc), label=st)
ax8.plot(TA_anom,depth_noc-np.nanmin(depth_noc), label=st)
ind_min = np.max(np.argwhere(depth_noc!=np.nan))
print('min pH anom at %f m of %s is %f ' %(depth[ind_min],st,pH_anom[ind_min]))
for ax in [ax1,ax2,ax3,ax4,ax5,ax6,ax7,ax8]:
ax.set_yticks([])
ax0.set_ylabel('Height above bottom / m')
ax0.set_xlabel('pH')
ax1.set_xlabel('DIC / $\mu$mol L$^{-1}$')
ax2.set_xlabel('TA / $\mu$mol L$^{-1}$')
ax3.set_xlabel('pH noc')
ax4.set_xlabel('DIC noc / $\mu$mol L$^{-1}$')
ax5.set_xlabel('TA noc / $\mu$mol L$^{-1}$')
ax6.set_xlabel('pH anom')
ax7.set_xlabel('DIC anom / $\mu$mol L$^{-1}$')
ax8.set_xlabel('TA anom / $\mu$mol L$^{-1}$')
ax0.set_title('BARKLEY PATHWAYS')
ax8.legend(loc=0)
| 0.278747 | 0.700184 |
# Week 8 - Implementing a model in numpy and a survey of machine learning packages for python
This week we will be looking in detail at how to implement a supervised regression model using the base scientific computing packages available with python.
We will also be looking at the different packages available for python that implement many of the algorithms we might want to use.
## Regression with numpy
Why implement algorithms from scratch when dedicated packages already exist?
The packages available are very powerful and a real time saver but they can obscure some issues we might encounter if we don't know to look for them. By starting with just numpy these problems will be more obvious. We can address them here and then when we move on we will know what to look for and will be less likely to miss them.
The dedicated machine learning packages implement the different algorithms but we are still responsible for getting our data in a suitable format.
```
import matplotlib.pyplot as plt
import numpy as np
%matplotlib inline
n = 20
x = np.random.random((n,1))
y = 5 + 6 * x ** 2 + np.random.normal(0,0.5, size=(n,1))
plt.plot(x, y, 'b.')
plt.show()
```
This is a very simple dataset. There is only one input value for each record and then there is the output value. Our goal is to determine the output value or dependent variable, shown on the y-axis, from the input or independent variable, shown on the x-axis.
Our approach should scale to handle multiple input, or independent, variables. The independent variables can be stored in a vector, a 1-dimensional array:
$$X^T = (X_{1}, X_{2}, X_{3})$$
As we have multiple records these can be stacked in a 2-dimensional array. Each record becomes one row in the array. Our `x` variable is already set up in this way.
In linear regression we can compute the value of the dependent variable using the following formula:
$$f(X) = \beta_{0} + \sum_{j=1}^p X_j\beta_j$$
The $\beta_{0}$ term is the intercept, and represents the value of the dependent variable when the independent variable is zero.
Calculating a solution is easier if we don't treat the intercept as special. Instead of having an intercept co-efficient that is handled separately we can instead add a variable to each of our records with a value of one.
```
intercept_x = np.hstack((np.ones((n,1)), x))
intercept_x
```
[Numpy contains the linalg module](http://docs.scipy.org/doc/numpy/reference/routines.linalg.html) with many common functions for performing linear algebra. Using this module finding a solution is quite simple.
```
np.linalg.lstsq(intercept_x,y)
```
The values returned are:
* The least-squares solution
* The sum of squared residuals
* The rank of the independent variables
* The singular values of the independent variables
## Exercise
1. Calculate the predictions our model would make
2. Calculate the sum of squared residuals from our predictions. Does this match the value returned by lstsq?
```
coeff, residuals, rank, sing_vals = np.linalg.lstsq(intercept_x,y)
intercept_x.shape, coeff.T.shape
print(intercept_x, coeff.T)
np.sum(intercept_x * coeff.T, axis=1)
predictions = np.sum(intercept_x * coeff.T, axis=1)
plt.plot(x, y, 'bo')
plt.plot(x, predictions, 'k-')
plt.show()
predictions.reshape((20,1)) - y
np.sum((predictions.reshape((20,1)) - y) ** 2), residuals
```
Least squares refers to the cost function for this algorithm. The objective is to minimize the residual sum of squares. The difference between the actual and predicted values is calculated, it is squared and then summed over all records. The function is as follows:
$$RSS(\beta) = \sum_{i=1}^{N}(y_i - x_i^T\beta)^2$$
## Matrix arithmetic
Within lstsq all the calculations are performed using matrix arithmetic rather than the more familiar element-wise arithmetic numpy arrays generally perform. Numpy does have a matrix type but matrix arithmetic can also be performed on standard arrays using dedicated methods.

_Source: Wikimedia Commons (User:Bilou)_
In matrix multiplication the resulting value in any position is the sum of multiplying each value in a row in the first matrix by the corresponding value in a column in the second matrix.
The residual sum of squares can be calculated with the following formula:
$$RSS(\beta) = (y - X\beta)^T(y-X\beta)$$
The value of our co-efficients can be calculated with:
$$\hat\beta = (X^TX)^{-1}X^Ty$$
Unfortunately, the result is not as visually appealing as in languages that use matrix arithmetic by default.
```
our_coeff = np.dot(np.dot(np.linalg.inv(np.dot(intercept_x.T, intercept_x)), intercept_x.T), y)
print(coeff, '\n\n', our_coeff)
our_predictions = np.dot(intercept_x, our_coeff)
np.hstack((predictions.reshape(1,20),
our_predictions.reshape(1,20)
))
plt.plot(x, y, 'ko', label='True values')
plt.plot(x, our_predictions, 'ro', label='Predictions')
plt.legend(numpoints=1, loc=4)
plt.show()
```
## Exercise
1. Plot the residuals. The x axis will be the independent variable (x) and the y axis the residual between our prediction and the true value.
2. Plot the predictions generated for our model over the entire range of 0-1. One approach is to use the np.linspace method to create equally spaced values over a specified range.
```
plt.plot(x, y, 'ko', label='True Values')
all_x = np.linspace(0,1, 1000).reshape((1000,1))
intercept_all_x = np.hstack((np.ones((1000,1)), all_x))
print(all_x.shape, intercept_all_x.shape, our_coeff.shape)
print("predictions:", all_x_predictions.shape)
all_x_predictions = np.dot(intercept_all_x, our_coeff)
plt.plot(all_x, all_x_predictions, 'r-', label='Predictions')
plt.legend(numpoints=1, loc=4)
plt.show()
```
## Types of independent variable
The independent variables can be many different types.
* Quantitative inputs
* Categorical inputs coded using dummy values
* Interactions between multiple inputs
* Tranformations of other inputs, e.g. logs, raised to different powers, etc.
It is important to note that a _linear_ model is only _linear_ with respect to its inputs. Those input variables can take any form.
One approach we can take to improve the predictions from our model would be to add in the square, cube, etc of our existing variable.
```
x_expanded = np.hstack((x**i for i in range(1,20)))
b, residuals, rank, s = np.linalg.lstsq(x_expanded, y)
print(b)
plt.plot(x, y, 'ko', label='True values')
plt.plot(x, np.dot(x_expanded, b), 'ro', label='Predictions')
plt.legend(numpoints=1, loc=4)
plt.show()
```
There is a tradeoff with model complexity. As we add more complexity to our model we can fit our training data increasingly well but eventually will lose our ability to generalize to new data.
Very simple models __underfit__ the data and have high __bias__.
Very complex models __overfit__ the data and have high __variance__.
The goal is to detect true sources of variation in the data and ignore variation that is just noise.
How do we know if we have a good model? A common approach is to break up our data into a training set, a validation set, and a test set.
* We train models with different parameters on the training set.
* We evaluate each model on the validation set, and choose the best
* We then measure the performance of our best model on the test set.
__What would our best model look like?__ Because we are using dummy data here we can easily make more.
```
n = 20
p = 12
training = []
val = []
for i in range(1, p):
np.random.seed(0)
x = np.random.random((n,1))
y = 5 + 6 * x ** 2 + np.random.normal(0,0.5, size=(n,1))
x = np.hstack((x**j for j in np.arange(i)))
our_coeff = np.dot(
np.dot(
np.linalg.inv(
np.dot(
x.T, x
)
), x.T
), y
)
our_predictions = np.dot(x, our_coeff)
our_training_rss = np.sum((y - our_predictions) ** 2)
training.append(our_training_rss)
val_x = np.random.random((n,1))
val_y = 5 + 6 * val_x ** 2 + np.random.normal(0,0.5, size=(n,1))
val_x = np.hstack((val_x**j for j in np.arange(i)))
our_val_pred = np.dot(val_x, our_coeff)
our_val_rss = np.sum((val_y - our_val_pred) ** 2)
val.append(our_val_rss)
#print(i, our_training_rss, our_val_rss)
plt.plot(range(1, p), training, 'ko-', label='training')
plt.plot(range(1, p), val, 'ro-', label='validation')
plt.legend(loc=2)
plt.show()
```
## Gradient descent
One limitation of our current implementation is that it is resource intensive. For very large datasets an alternative is needed. Gradient descent is often preferred, and particularly stochastic gradient descent for very large datasets.
Gradient descent is an iterative process, repetitively calculating the error and changing the coefficients slightly to reduce that error. It does this by calculating a gradient and then descending to a minimum in small steps.
Stochastic gradient descent calculates the gradient on a small batch of the data, updates the coefficients, loads the next chunk of the data and repeats the process.
We will just look at a basic gradient descent model.
```
np.random.seed(0)
n = 200
x = np.random.random((n,1))
y = 5 + 6 * x ** 2 + np.random.normal(0,0.5, size=(n,1))
intercept_x = np.hstack((np.ones((n,1)), x))
coeff, residuals, rank, sing_vals = np.linalg.lstsq(intercept_x,y)
print('lstsq:\n', coeff, '\n')
def gradient_descent(x, y, rounds = 1000, alpha=0.01):
theta = np.zeros((x.shape[1], 1))
costs = []
for i in range(rounds):
prediction = np.dot(x, theta)
error = prediction - y
gradient = np.dot(x.T, error / y.shape[0])
theta -= gradient * alpha
costs.append(np.sum(error ** 2))
return (theta, costs)
theta, costs = gradient_descent(intercept_x, y, rounds=10000)
print('theta:\n', theta, '\n\n', costs[::500])
np.random.seed(0)
n = 200
x = np.random.random((n,1))
y = 5 + 6 * x ** 2 + np.random.normal(0,0.5, size=(n,1))
x = np.hstack((x**j for j in np.arange(20)))
coeff, residuals, rank, sing_vals = np.linalg.lstsq(x,y)
print('lstsq', coeff)
theta, costs = gradient_descent(x, y, rounds=10000)
print(theta, costs[::500])
plt.plot(x[:,1], y, 'ko')
plt.plot(x[:,1], np.dot(x, coeff), 'co')
plt.plot(x[:,1], np.dot(x, theta), 'ro')
plt.show()
```
## Machine learning packages available in the python ecosystem
[Overview in the python wiki](https://wiki.python.org/moin/PythonForArtificialIntelligence)
General
* [scikit-learn](http://scikit-learn.org/stable/)
* [milk](https://pythonhosted.org/milk/)
* [Orange](http://orange.biolab.si/)
* [Shogun](http://www.shogun-toolbox.org/)
* [GraphLab Create (dato)](https://dato.com/learn/userguide/)
There is a collection of field specific packages including some with machine learning components on the [scipy website](http://www.scipy.org/topical-software.html#science-basic-tools). Other packages can often be found searching the [python package index](https://pypi.python.org/pypi).
Deep learning is receiving a lot of attention recently and a number of different packages have been developed.
* [Theano](http://www.deeplearning.net/software/theano/)
* [pylearn2](http://deeplearning.net/software/pylearn2/)
* [keras](http://keras.io/)
* [Blocks](http://blocks.readthedocs.org/en/latest/)
* [Lasagne](http://lasagne.readthedocs.org/en/latest/)
## Scikit-learn
Scikit-learn is now widely used. It includes modules for:
* Classification
* Regression
* Clustering
* Dimensionality reduction
* Model selection
* Preprocessing
There are modules for training online models, enabling very large datasets to be analyzed.
There is also a semi-supervised module for situations when you have a large dataset, but only have labels for part of the dataset.
## Milk
Milk works very well with mahotas, a package for image processing. With the recent improvements in scikit-image milk is now less attractive, although still a strong option
## Orange and Shogun
These are both large packages but for whatever reason do not receive the attention that scikit-learn does.
## Dato
Dato is a relative newcomer and has been receiving a lot of attention lately. Time will tell whether it can compete with scikit-learn.
# Assignments
This week we will continue working on our project ideas. As you develop the outline some points you may want to consider:
For projects developing the object oriented programming component of the course:
* What will your classes be?
* What will each class have as attributes and methods?
* How will your classes interact?
For projects developing GUIs or web applications:
* What will your screens/pages be?
* What components will each page need?
* How will you store any data needed/produced?
For projects developing machine learning models:
* What will be your data?
* How is your data structured?
* How much data do you have?
* Is your data labeled?
* What type of machine learning task is it?
* How good would the performance need to be for the model to be useful?
You do not need to answer all these questions. Each answer does not need to be complete. Your final project will likely be different to your initial idea.
The goal of the project description is to document your project as you currently envision it and to encourage planning for the earliest stage.
__Your project descriptions should be sent to me by our class next week.__
|
github_jupyter
|
import matplotlib.pyplot as plt
import numpy as np
%matplotlib inline
n = 20
x = np.random.random((n,1))
y = 5 + 6 * x ** 2 + np.random.normal(0,0.5, size=(n,1))
plt.plot(x, y, 'b.')
plt.show()
intercept_x = np.hstack((np.ones((n,1)), x))
intercept_x
np.linalg.lstsq(intercept_x,y)
coeff, residuals, rank, sing_vals = np.linalg.lstsq(intercept_x,y)
intercept_x.shape, coeff.T.shape
print(intercept_x, coeff.T)
np.sum(intercept_x * coeff.T, axis=1)
predictions = np.sum(intercept_x * coeff.T, axis=1)
plt.plot(x, y, 'bo')
plt.plot(x, predictions, 'k-')
plt.show()
predictions.reshape((20,1)) - y
np.sum((predictions.reshape((20,1)) - y) ** 2), residuals
our_coeff = np.dot(np.dot(np.linalg.inv(np.dot(intercept_x.T, intercept_x)), intercept_x.T), y)
print(coeff, '\n\n', our_coeff)
our_predictions = np.dot(intercept_x, our_coeff)
np.hstack((predictions.reshape(1,20),
our_predictions.reshape(1,20)
))
plt.plot(x, y, 'ko', label='True values')
plt.plot(x, our_predictions, 'ro', label='Predictions')
plt.legend(numpoints=1, loc=4)
plt.show()
plt.plot(x, y, 'ko', label='True Values')
all_x = np.linspace(0,1, 1000).reshape((1000,1))
intercept_all_x = np.hstack((np.ones((1000,1)), all_x))
print(all_x.shape, intercept_all_x.shape, our_coeff.shape)
print("predictions:", all_x_predictions.shape)
all_x_predictions = np.dot(intercept_all_x, our_coeff)
plt.plot(all_x, all_x_predictions, 'r-', label='Predictions')
plt.legend(numpoints=1, loc=4)
plt.show()
x_expanded = np.hstack((x**i for i in range(1,20)))
b, residuals, rank, s = np.linalg.lstsq(x_expanded, y)
print(b)
plt.plot(x, y, 'ko', label='True values')
plt.plot(x, np.dot(x_expanded, b), 'ro', label='Predictions')
plt.legend(numpoints=1, loc=4)
plt.show()
n = 20
p = 12
training = []
val = []
for i in range(1, p):
np.random.seed(0)
x = np.random.random((n,1))
y = 5 + 6 * x ** 2 + np.random.normal(0,0.5, size=(n,1))
x = np.hstack((x**j for j in np.arange(i)))
our_coeff = np.dot(
np.dot(
np.linalg.inv(
np.dot(
x.T, x
)
), x.T
), y
)
our_predictions = np.dot(x, our_coeff)
our_training_rss = np.sum((y - our_predictions) ** 2)
training.append(our_training_rss)
val_x = np.random.random((n,1))
val_y = 5 + 6 * val_x ** 2 + np.random.normal(0,0.5, size=(n,1))
val_x = np.hstack((val_x**j for j in np.arange(i)))
our_val_pred = np.dot(val_x, our_coeff)
our_val_rss = np.sum((val_y - our_val_pred) ** 2)
val.append(our_val_rss)
#print(i, our_training_rss, our_val_rss)
plt.plot(range(1, p), training, 'ko-', label='training')
plt.plot(range(1, p), val, 'ro-', label='validation')
plt.legend(loc=2)
plt.show()
np.random.seed(0)
n = 200
x = np.random.random((n,1))
y = 5 + 6 * x ** 2 + np.random.normal(0,0.5, size=(n,1))
intercept_x = np.hstack((np.ones((n,1)), x))
coeff, residuals, rank, sing_vals = np.linalg.lstsq(intercept_x,y)
print('lstsq:\n', coeff, '\n')
def gradient_descent(x, y, rounds = 1000, alpha=0.01):
theta = np.zeros((x.shape[1], 1))
costs = []
for i in range(rounds):
prediction = np.dot(x, theta)
error = prediction - y
gradient = np.dot(x.T, error / y.shape[0])
theta -= gradient * alpha
costs.append(np.sum(error ** 2))
return (theta, costs)
theta, costs = gradient_descent(intercept_x, y, rounds=10000)
print('theta:\n', theta, '\n\n', costs[::500])
np.random.seed(0)
n = 200
x = np.random.random((n,1))
y = 5 + 6 * x ** 2 + np.random.normal(0,0.5, size=(n,1))
x = np.hstack((x**j for j in np.arange(20)))
coeff, residuals, rank, sing_vals = np.linalg.lstsq(x,y)
print('lstsq', coeff)
theta, costs = gradient_descent(x, y, rounds=10000)
print(theta, costs[::500])
plt.plot(x[:,1], y, 'ko')
plt.plot(x[:,1], np.dot(x, coeff), 'co')
plt.plot(x[:,1], np.dot(x, theta), 'ro')
plt.show()
| 0.338952 | 0.995076 |
```
# 600000.SH Minute Kline in emquant with ChanTrendHandler V3.0
import os
from pymongo import MongoClient
def get_code_from_colname(colname):
(market, code) = colname.split('_')
return code + '.' + market
wind_db_name = os.environ.get('WIND_DB', 'emquant')
chan_db_name = os.environ.get('CHAN_DB', 'emchantest')
db_ip = os.environ.get('MONGO_DB_IP_ADDR', '192.168.1.21')
db_port = int(os.environ.get('MONGO_DB_PORT', 27017))
# wind_db_name = os.environ.get('WIND_DB', 'wind')
# chan_db_name = os.environ.get('CHAN_DB', 'chan')
# db_ip = os.environ.get('MONGO_DB_IP_ADDR', '192.168.1.103')
# db_port = int(os.environ.get('MONGO_DB_PORT', 27017))
client = MongoClient(db_ip, db_port)
db_wind = client[wind_db_name]
db_chan = client[chan_db_name]
chankline_col = db_chan['chankline']
code_list = db_wind.collection_names(include_system_collections=False)
code_list = [get_code_from_colname(code) for code in code_list if 'SH' in code or 'SZ' in code]
print('--- Chan Kline ---')
cur = db_chan['chankline'].find().sort('index', -1)
print('167999: {}'.format(cur.count()))
print('167998: {}'.format(cur[0]['index']))
print('--- Chan Fractal ---')
cur = db_chan['fractal'].find().sort('index', -1)
print('29028: {}'.format(cur.count()))
print('29027: {}'.format(cur[0]['index']))
print('--- Chan Bi ---')
cur = db_chan['trend'].find({'level': '-1'}).sort('index', -1)
print('5893: {}'.format(cur.count()))
print('5892: {}'.format(cur[0]['index']))
print('--- Chan Duan ---')
cur = db_chan['trend'].find({'level': '0'}).sort('index', -1)
print('804: {}'.format(cur.count()))
print('803: {}'.format(cur[0]['index']))
print('--- Chan Bi Centre ---')
cur = db_chan['centre'].find({'level': '-1'}).sort('index', -1)
print('1328: {}'.format(cur.count()))
print('1327: {}'.format(cur[0]['index']))
print('--- Level 1 ---')
cur = db_chan['trend'].find({'level': '1'}).sort('index', -1)
print('120: {}'.format(cur.count()))
print('119: {}'.format(cur[0]['index']))
print('--- Duan Centre ---')
cur = db_chan['centre'].find({'level': '0'}).sort('index', -1)
print('188: {}'.format(cur.count()))
print('187: {}'.format(cur[0]['index']))
print('--- Level 2 ---')
cur = db_chan['trend'].find({'level': '2'}).sort('index', -1)
print('10: {}'.format(cur.count()))
print('9: {}'.format(cur[0]['index']))
print('--- Level 1 Centre ---')
cur = db_chan['centre'].find({'level': '1'}).sort('index', -1)
print('26: {}'.format(cur.count()))
print('25: {}'.format(cur[0]['index']))
# 600000.SH Minute Kline in emquant with ChanTrendHandler V2.0 and previous Duan logic for duan
import os
from pymongo import MongoClient
def get_code_from_colname(colname):
(market, code) = colname.split('_')
return code + '.' + market
wind_db_name = os.environ.get('WIND_DB', 'emquant')
chan_db_name = os.environ.get('CHAN_DB', 'emchantest')
db_ip = os.environ.get('MONGO_DB_IP_ADDR', '192.168.1.21')
db_port = int(os.environ.get('MONGO_DB_PORT', 27017))
# wind_db_name = os.environ.get('WIND_DB', 'wind')
# chan_db_name = os.environ.get('CHAN_DB', 'chan')
# db_ip = os.environ.get('MONGO_DB_IP_ADDR', '192.168.1.103')
# db_port = int(os.environ.get('MONGO_DB_PORT', 27017))
client = MongoClient(db_ip, db_port)
db_wind = client[wind_db_name]
db_chan = client[chan_db_name]
chankline_col = db_chan['chankline']
code_list = db_wind.collection_names(include_system_collections=False)
code_list = [get_code_from_colname(code) for code in code_list if 'SH' in code or 'SZ' in code]
print('--- Chan Kline ---')
cur = db_chan['chankline'].find().sort('index', -1)
print('167999: {}'.format(cur.count()))
print('167998: {}'.format(cur[0]['index']))
print('--- Chan Fractal ---')
cur = db_chan['fractal'].find().sort('index', -1)
print('29028: {}'.format(cur.count()))
print('29027: {}'.format(cur[0]['index']))
print('--- Chan Bi ---')
cur = db_chan['trend'].find({'level': '-1'}).sort('index', -1)
print('5893: {}'.format(cur.count()))
print('5892: {}'.format(cur[0]['index']))
print('--- Chan Duan ---')
cur = db_chan['trend'].find({'level': '0'}).sort('index', -1)
print('804: {}'.format(cur.count()))
print('803: {}'.format(cur[0]['index']))
print('--- Chan Bi Centre ---')
cur = db_chan['centre'].find({'level': '-1'}).sort('index', -1)
print('1292: {}'.format(cur.count()))
print('1291: {}'.format(cur[0]['index']))
print('--- Level 1 ---')
cur = db_chan['trend'].find({'level': '1'}).sort('index', -1)
print('60: {}'.format(cur.count()))
print('59: {}'.format(cur[0]['index']))
print('--- Duan Centre ---')
cur = db_chan['centre'].find({'level': '0'}).sort('index', -1)
print('138: {}'.format(cur.count()))
print('137: {}'.format(cur[0]['index']))
# 600000.SH Minute Kline in emquant with ChanTrendHandler V2.0
import os
from pymongo import MongoClient
def get_code_from_colname(colname):
(market, code) = colname.split('_')
return code + '.' + market
wind_db_name = os.environ.get('WIND_DB', 'emquant')
chan_db_name = os.environ.get('CHAN_DB', 'emchantest')
db_ip = os.environ.get('MONGO_DB_IP_ADDR', '192.168.1.21')
db_port = int(os.environ.get('MONGO_DB_PORT', 27017))
# wind_db_name = os.environ.get('WIND_DB', 'wind')
# chan_db_name = os.environ.get('CHAN_DB', 'chan')
# db_ip = os.environ.get('MONGO_DB_IP_ADDR', '192.168.1.103')
# db_port = int(os.environ.get('MONGO_DB_PORT', 27017))
client = MongoClient(db_ip, db_port)
db_wind = client[wind_db_name]
db_chan = client[chan_db_name]
chankline_col = db_chan['chankline']
code_list = db_wind.collection_names(include_system_collections=False)
code_list = [get_code_from_colname(code) for code in code_list if 'SH' in code or 'SZ' in code]
print('--- Chan Kline ---')
cur = db_chan['chankline'].find().sort('index', -1)
print('167999: {}'.format(cur.count()))
print('167998: {}'.format(cur[0]['index']))
print('--- Chan Fractal ---')
cur = db_chan['fractal'].find().sort('index', -1)
print('29028: {}'.format(cur.count()))
print('29027: {}'.format(cur[0]['index']))
print('--- Chan Bi ---')
cur = db_chan['trend'].find({'level': '-1'}).sort('index', -1)
print('5893: {}'.format(cur.count()))
print('5892: {}'.format(cur[0]['index']))
print('--- Chan Duan ---')
cur = db_chan['trend'].find({'level': '0'}).sort('index', -1)
print('398: {}'.format(cur.count()))
print('397: {}'.format(cur[0]['index']))
print('--- Chan Bi Centre ---')
cur = db_chan['centre'].find({'level': '-1'}).sort('index', -1)
print('982: {}'.format(cur.count()))
print('981: {}'.format(cur[0]['index']))
print('--- Level 1 ---')
cur = db_chan['trend'].find({'level': '1'}).sort('index', -1)
print('28: {}'.format(cur.count()))
print('27: {}'.format(cur[0]['index']))
print('--- Duan Centre ---')
cur = db_chan['centre'].find({'level': '0'}).sort('index', -1)
print('63: {}'.format(cur.count()))
print('62: {}'.format(cur[0]['index']))
# 600000.SH Minute Kline in emquant
import os
from pymongo import MongoClient
def get_code_from_colname(colname):
(market, code) = colname.split('_')
return code + '.' + market
wind_db_name = os.environ.get('WIND_DB', 'emquant')
chan_db_name = os.environ.get('CHAN_DB', 'emchantest')
db_ip = os.environ.get('MONGO_DB_IP_ADDR', '192.168.1.21')
db_port = int(os.environ.get('MONGO_DB_PORT', 27017))
# wind_db_name = os.environ.get('WIND_DB', 'wind')
# chan_db_name = os.environ.get('CHAN_DB', 'chan')
# db_ip = os.environ.get('MONGO_DB_IP_ADDR', '192.168.1.103')
# db_port = int(os.environ.get('MONGO_DB_PORT', 27017))
client = MongoClient(db_ip, db_port)
db_wind = client[wind_db_name]
db_chan = client[chan_db_name]
chankline_col = db_chan['chankline']
code_list = db_wind.collection_names(include_system_collections=False)
code_list = [get_code_from_colname(code) for code in code_list if 'SH' in code or 'SZ' in code]
print('--- Chan Kline ---')
cur = db_chan['chankline'].find().sort('index', -1)
print('167999: {}'.format(cur.count()))
print('167998: {}'.format(cur[0]['index']))
print('--- Chan Fractal ---')
cur = db_chan['fractal'].find().sort('index', -1)
print('29028: {}'.format(cur.count()))
print('29027: {}'.format(cur[0]['index']))
print('--- Chan Bi ---')
cur = db_chan['trend'].find({'level': '-1'}).sort('index', -1)
print('5893: {}'.format(cur.count()))
print('5892: {}'.format(cur[0]['index']))
print('--- Chan Duan ---')
cur = db_chan['trend'].find({'level': '0'}).sort('index', -1)
print('804: {}'.format(cur.count()))
print('803: {}'.format(cur[0]['index']))
print('--- Chan Bi Centre ---')
cur = db_chan['centre'].find({'level': '-1'}).sort('index', -1)
print('1328: {}'.format(cur.count()))
print('1327: {}'.format(cur[0]['index']))
# 600000.SH Day Kline in emquant
import os
from pymongo import MongoClient
def get_code_from_colname(colname):
(market, code) = colname.split('_')
return code + '.' + market
wind_db_name = os.environ.get('WIND_DB', 'emquant')
chan_db_name = os.environ.get('CHAN_DB', 'emchantest')
db_ip = os.environ.get('MONGO_DB_IP_ADDR', '172.17.0.1')
db_port = int(os.environ.get('MONGO_DB_PORT', 27017))
# wind_db_name = os.environ.get('WIND_DB', 'wind')
# chan_db_name = os.environ.get('CHAN_DB', 'chan')
# db_ip = os.environ.get('MONGO_DB_IP_ADDR', '192.168.1.103')
# db_port = int(os.environ.get('MONGO_DB_PORT', 27017))
client = MongoClient(db_ip, db_port)
db_wind = client[wind_db_name]
db_chan = client[chan_db_name]
chankline_col = db_chan['chankline']
code_list = db_wind.collection_names(include_system_collections=False)
code_list = [get_code_from_colname(code) for code in code_list if 'SH' in code or 'SZ' in code]
print('--- Chan Kline ---')
cur = db_chan['chankline'].find().sort('index', -1)
print('733: {}'.format(cur.count()))
print('732: {}'.format(cur[0]['index']))
print('--- Chan Fractal ---')
cur = db_chan['fractal'].find().sort('index', -1)
print('243: {}'.format(cur.count()))
print('242: {}'.format(cur[0]['index']))
print('--- Chan Bi ---')
cur = db_chan['trend'].find({'level': '-1'}).sort('index', -1)
print('44: {}'.format(cur.count()))
print('43: {}'.format(cur[0]['index']))
print('--- Chan Duan ---')
cur = db_chan['trend'].find({'level': '0'}).sort('index', -1)
print('8: {}'.format(cur.count()))
print('7: {}'.format(cur[0]['index']))
print('--- Chan Bi Centre ---')
cur = db_chan['centre'].find({'level': '-1'}).sort('index', -1)
print('14: {}'.format(cur.count()))
print('13: {}'.format(cur[0]['index']))
```
|
github_jupyter
|
# 600000.SH Minute Kline in emquant with ChanTrendHandler V3.0
import os
from pymongo import MongoClient
def get_code_from_colname(colname):
(market, code) = colname.split('_')
return code + '.' + market
wind_db_name = os.environ.get('WIND_DB', 'emquant')
chan_db_name = os.environ.get('CHAN_DB', 'emchantest')
db_ip = os.environ.get('MONGO_DB_IP_ADDR', '192.168.1.21')
db_port = int(os.environ.get('MONGO_DB_PORT', 27017))
# wind_db_name = os.environ.get('WIND_DB', 'wind')
# chan_db_name = os.environ.get('CHAN_DB', 'chan')
# db_ip = os.environ.get('MONGO_DB_IP_ADDR', '192.168.1.103')
# db_port = int(os.environ.get('MONGO_DB_PORT', 27017))
client = MongoClient(db_ip, db_port)
db_wind = client[wind_db_name]
db_chan = client[chan_db_name]
chankline_col = db_chan['chankline']
code_list = db_wind.collection_names(include_system_collections=False)
code_list = [get_code_from_colname(code) for code in code_list if 'SH' in code or 'SZ' in code]
print('--- Chan Kline ---')
cur = db_chan['chankline'].find().sort('index', -1)
print('167999: {}'.format(cur.count()))
print('167998: {}'.format(cur[0]['index']))
print('--- Chan Fractal ---')
cur = db_chan['fractal'].find().sort('index', -1)
print('29028: {}'.format(cur.count()))
print('29027: {}'.format(cur[0]['index']))
print('--- Chan Bi ---')
cur = db_chan['trend'].find({'level': '-1'}).sort('index', -1)
print('5893: {}'.format(cur.count()))
print('5892: {}'.format(cur[0]['index']))
print('--- Chan Duan ---')
cur = db_chan['trend'].find({'level': '0'}).sort('index', -1)
print('804: {}'.format(cur.count()))
print('803: {}'.format(cur[0]['index']))
print('--- Chan Bi Centre ---')
cur = db_chan['centre'].find({'level': '-1'}).sort('index', -1)
print('1328: {}'.format(cur.count()))
print('1327: {}'.format(cur[0]['index']))
print('--- Level 1 ---')
cur = db_chan['trend'].find({'level': '1'}).sort('index', -1)
print('120: {}'.format(cur.count()))
print('119: {}'.format(cur[0]['index']))
print('--- Duan Centre ---')
cur = db_chan['centre'].find({'level': '0'}).sort('index', -1)
print('188: {}'.format(cur.count()))
print('187: {}'.format(cur[0]['index']))
print('--- Level 2 ---')
cur = db_chan['trend'].find({'level': '2'}).sort('index', -1)
print('10: {}'.format(cur.count()))
print('9: {}'.format(cur[0]['index']))
print('--- Level 1 Centre ---')
cur = db_chan['centre'].find({'level': '1'}).sort('index', -1)
print('26: {}'.format(cur.count()))
print('25: {}'.format(cur[0]['index']))
# 600000.SH Minute Kline in emquant with ChanTrendHandler V2.0 and previous Duan logic for duan
import os
from pymongo import MongoClient
def get_code_from_colname(colname):
(market, code) = colname.split('_')
return code + '.' + market
wind_db_name = os.environ.get('WIND_DB', 'emquant')
chan_db_name = os.environ.get('CHAN_DB', 'emchantest')
db_ip = os.environ.get('MONGO_DB_IP_ADDR', '192.168.1.21')
db_port = int(os.environ.get('MONGO_DB_PORT', 27017))
# wind_db_name = os.environ.get('WIND_DB', 'wind')
# chan_db_name = os.environ.get('CHAN_DB', 'chan')
# db_ip = os.environ.get('MONGO_DB_IP_ADDR', '192.168.1.103')
# db_port = int(os.environ.get('MONGO_DB_PORT', 27017))
client = MongoClient(db_ip, db_port)
db_wind = client[wind_db_name]
db_chan = client[chan_db_name]
chankline_col = db_chan['chankline']
code_list = db_wind.collection_names(include_system_collections=False)
code_list = [get_code_from_colname(code) for code in code_list if 'SH' in code or 'SZ' in code]
print('--- Chan Kline ---')
cur = db_chan['chankline'].find().sort('index', -1)
print('167999: {}'.format(cur.count()))
print('167998: {}'.format(cur[0]['index']))
print('--- Chan Fractal ---')
cur = db_chan['fractal'].find().sort('index', -1)
print('29028: {}'.format(cur.count()))
print('29027: {}'.format(cur[0]['index']))
print('--- Chan Bi ---')
cur = db_chan['trend'].find({'level': '-1'}).sort('index', -1)
print('5893: {}'.format(cur.count()))
print('5892: {}'.format(cur[0]['index']))
print('--- Chan Duan ---')
cur = db_chan['trend'].find({'level': '0'}).sort('index', -1)
print('804: {}'.format(cur.count()))
print('803: {}'.format(cur[0]['index']))
print('--- Chan Bi Centre ---')
cur = db_chan['centre'].find({'level': '-1'}).sort('index', -1)
print('1292: {}'.format(cur.count()))
print('1291: {}'.format(cur[0]['index']))
print('--- Level 1 ---')
cur = db_chan['trend'].find({'level': '1'}).sort('index', -1)
print('60: {}'.format(cur.count()))
print('59: {}'.format(cur[0]['index']))
print('--- Duan Centre ---')
cur = db_chan['centre'].find({'level': '0'}).sort('index', -1)
print('138: {}'.format(cur.count()))
print('137: {}'.format(cur[0]['index']))
# 600000.SH Minute Kline in emquant with ChanTrendHandler V2.0
import os
from pymongo import MongoClient
def get_code_from_colname(colname):
(market, code) = colname.split('_')
return code + '.' + market
wind_db_name = os.environ.get('WIND_DB', 'emquant')
chan_db_name = os.environ.get('CHAN_DB', 'emchantest')
db_ip = os.environ.get('MONGO_DB_IP_ADDR', '192.168.1.21')
db_port = int(os.environ.get('MONGO_DB_PORT', 27017))
# wind_db_name = os.environ.get('WIND_DB', 'wind')
# chan_db_name = os.environ.get('CHAN_DB', 'chan')
# db_ip = os.environ.get('MONGO_DB_IP_ADDR', '192.168.1.103')
# db_port = int(os.environ.get('MONGO_DB_PORT', 27017))
client = MongoClient(db_ip, db_port)
db_wind = client[wind_db_name]
db_chan = client[chan_db_name]
chankline_col = db_chan['chankline']
code_list = db_wind.collection_names(include_system_collections=False)
code_list = [get_code_from_colname(code) for code in code_list if 'SH' in code or 'SZ' in code]
print('--- Chan Kline ---')
cur = db_chan['chankline'].find().sort('index', -1)
print('167999: {}'.format(cur.count()))
print('167998: {}'.format(cur[0]['index']))
print('--- Chan Fractal ---')
cur = db_chan['fractal'].find().sort('index', -1)
print('29028: {}'.format(cur.count()))
print('29027: {}'.format(cur[0]['index']))
print('--- Chan Bi ---')
cur = db_chan['trend'].find({'level': '-1'}).sort('index', -1)
print('5893: {}'.format(cur.count()))
print('5892: {}'.format(cur[0]['index']))
print('--- Chan Duan ---')
cur = db_chan['trend'].find({'level': '0'}).sort('index', -1)
print('398: {}'.format(cur.count()))
print('397: {}'.format(cur[0]['index']))
print('--- Chan Bi Centre ---')
cur = db_chan['centre'].find({'level': '-1'}).sort('index', -1)
print('982: {}'.format(cur.count()))
print('981: {}'.format(cur[0]['index']))
print('--- Level 1 ---')
cur = db_chan['trend'].find({'level': '1'}).sort('index', -1)
print('28: {}'.format(cur.count()))
print('27: {}'.format(cur[0]['index']))
print('--- Duan Centre ---')
cur = db_chan['centre'].find({'level': '0'}).sort('index', -1)
print('63: {}'.format(cur.count()))
print('62: {}'.format(cur[0]['index']))
# 600000.SH Minute Kline in emquant
import os
from pymongo import MongoClient
def get_code_from_colname(colname):
(market, code) = colname.split('_')
return code + '.' + market
wind_db_name = os.environ.get('WIND_DB', 'emquant')
chan_db_name = os.environ.get('CHAN_DB', 'emchantest')
db_ip = os.environ.get('MONGO_DB_IP_ADDR', '192.168.1.21')
db_port = int(os.environ.get('MONGO_DB_PORT', 27017))
# wind_db_name = os.environ.get('WIND_DB', 'wind')
# chan_db_name = os.environ.get('CHAN_DB', 'chan')
# db_ip = os.environ.get('MONGO_DB_IP_ADDR', '192.168.1.103')
# db_port = int(os.environ.get('MONGO_DB_PORT', 27017))
client = MongoClient(db_ip, db_port)
db_wind = client[wind_db_name]
db_chan = client[chan_db_name]
chankline_col = db_chan['chankline']
code_list = db_wind.collection_names(include_system_collections=False)
code_list = [get_code_from_colname(code) for code in code_list if 'SH' in code or 'SZ' in code]
print('--- Chan Kline ---')
cur = db_chan['chankline'].find().sort('index', -1)
print('167999: {}'.format(cur.count()))
print('167998: {}'.format(cur[0]['index']))
print('--- Chan Fractal ---')
cur = db_chan['fractal'].find().sort('index', -1)
print('29028: {}'.format(cur.count()))
print('29027: {}'.format(cur[0]['index']))
print('--- Chan Bi ---')
cur = db_chan['trend'].find({'level': '-1'}).sort('index', -1)
print('5893: {}'.format(cur.count()))
print('5892: {}'.format(cur[0]['index']))
print('--- Chan Duan ---')
cur = db_chan['trend'].find({'level': '0'}).sort('index', -1)
print('804: {}'.format(cur.count()))
print('803: {}'.format(cur[0]['index']))
print('--- Chan Bi Centre ---')
cur = db_chan['centre'].find({'level': '-1'}).sort('index', -1)
print('1328: {}'.format(cur.count()))
print('1327: {}'.format(cur[0]['index']))
# 600000.SH Day Kline in emquant
import os
from pymongo import MongoClient
def get_code_from_colname(colname):
(market, code) = colname.split('_')
return code + '.' + market
wind_db_name = os.environ.get('WIND_DB', 'emquant')
chan_db_name = os.environ.get('CHAN_DB', 'emchantest')
db_ip = os.environ.get('MONGO_DB_IP_ADDR', '172.17.0.1')
db_port = int(os.environ.get('MONGO_DB_PORT', 27017))
# wind_db_name = os.environ.get('WIND_DB', 'wind')
# chan_db_name = os.environ.get('CHAN_DB', 'chan')
# db_ip = os.environ.get('MONGO_DB_IP_ADDR', '192.168.1.103')
# db_port = int(os.environ.get('MONGO_DB_PORT', 27017))
client = MongoClient(db_ip, db_port)
db_wind = client[wind_db_name]
db_chan = client[chan_db_name]
chankline_col = db_chan['chankline']
code_list = db_wind.collection_names(include_system_collections=False)
code_list = [get_code_from_colname(code) for code in code_list if 'SH' in code or 'SZ' in code]
print('--- Chan Kline ---')
cur = db_chan['chankline'].find().sort('index', -1)
print('733: {}'.format(cur.count()))
print('732: {}'.format(cur[0]['index']))
print('--- Chan Fractal ---')
cur = db_chan['fractal'].find().sort('index', -1)
print('243: {}'.format(cur.count()))
print('242: {}'.format(cur[0]['index']))
print('--- Chan Bi ---')
cur = db_chan['trend'].find({'level': '-1'}).sort('index', -1)
print('44: {}'.format(cur.count()))
print('43: {}'.format(cur[0]['index']))
print('--- Chan Duan ---')
cur = db_chan['trend'].find({'level': '0'}).sort('index', -1)
print('8: {}'.format(cur.count()))
print('7: {}'.format(cur[0]['index']))
print('--- Chan Bi Centre ---')
cur = db_chan['centre'].find({'level': '-1'}).sort('index', -1)
print('14: {}'.format(cur.count()))
print('13: {}'.format(cur[0]['index']))
| 0.193147 | 0.181517 |
# Table of Contents
* [Exploratory Data Analysis](#Header)
- [Metadata](#Metadata)
- [Missing Values](#MissingValues)
- [Species](#Species)
- [Date and Time](#DateTime)
- [Recordists](#Recordists)
- [Location](#Location)
* [Audio Feature Extraction](#AudioFeatureExtraction)
- [Waveform](#Waveform)
- [Autocorrelation](#Autocorrelation)
- [Spectrogram](#Spectrogram)
- [Chromagram](#Chromagram)
- [Spectral](#Spectral)
- [Centroid](#Centroid)
- [Bandwidth](#Bandwidth)
- [Contrast](#Contrast)
- [Flatness](#Flatness)
- [Rolloff](#Rolloff)
- [MFCC](#MFCC)
* [Afterword](#Thanks)
<a id="Header"></a>
# Bird Call Classification: Data Exploration
<a id="Environment"></a>
## Environment
```
import os
import sys
import librosa
import librosa.display
import librosa.feature
import numpy as np
import pandas as pd
import plotly.express as xp
import plotly.graph_objects as go
import matplotlib.pyplot as plt
import IPython.display as ipd
from sklearn.preprocessing import minmax_scale
import warnings
warnings.filterwarnings('ignore')
%matplotlib inline
```
<a id="Metadata"></a>
## Metadata
Relevant Features
- ebird_code
- date/time
- location
- recordist
- filename
```
train = pd.read_csv('../input/birdsong-recognition/train.csv')
train.head()
train.info()
```
<a id="MissingValues"></a>
### Missing Values
Analyze missing data and determine cleaning steps.
```
missing = train.isna().sum().sort_values(ascending=False)
missing = missing[missing != 0]
xp.bar(x=missing.index, y=missing, text=missing, title='Missing Values by Feature', labels={'x':'Features', 'y':'Quantity'})
```
- No relevant data missing
<a id="Species"></a>
### Species
Distribution of bird species
```
counts = train['ebird_code'].value_counts()
xp.bar(x=counts.index, y=counts, title='Species Distribution (by ebird code)', labels={'x':'Ebird Code', 'y':'Quantity'})
```
- Exactly 100 samples for about half of species in question
- Redhead is minimum at 9 samples
<a id="DateTime"></a>
### Date and Time
Recording Date Distribution
```
# split datetime into separate dataframe
datetime = train[['date', 'time']]
datetime.date = pd.to_datetime(datetime.date, errors='coerce').dropna()
datetime['hour'] = pd.to_numeric(datetime.time.str.split(':', expand=True)[0], errors='coerce')
ax1 = datetime.date.value_counts().sort_values().plot(figsize=(10,6), title='Recordings by Date')
ax1.set_xlabel('Date')
ax1.set_ylabel('Quantity')
plt.show()
```
- Majority of recordings taken in the past decade
- Interesting spike around 2003
- Cyclical spikes after 2013
```
ax2 = datetime['hour'].value_counts().sort_index().plot(figsize=(10,6), title='Recordings by Time', kind='bar', figure=plt.figure())
ax2.set_xlabel('Hour')
ax2.set_ylabel('Quantity')
plt.show()
```
- Most recordings taken between 6AM and 12PM
- Gradual decrease as the day moves on from 8AM
<a id="Recordists"></a>
### Recordists
Who recorded the samples?
```
ax3 = train['recordist'].value_counts().sort_values(ascending=False).head(20).sort_values().plot(figsize=(10, 6), title='Recordings by Recordist', figure=plt.figure(), kind='barh', fontsize=9)
ax3.set_xlabel('Hour')
ax3.set_ylabel('Quantity')
plt.tight_layout()
plt.show()
```
- Majority of recordings made by only two people
<a id="Location"></a>
### Location
```
counts = train['country'].value_counts().sort_values(ascending=False).head(10).sort_values()
xp.bar(y=counts.index, x=counts, title='Number of Recordings by Country', labels={'y':'Country', 'x':'Quantity'}, orientation='h')
coords = train.groupby(['latitude', 'longitude'], as_index=False)['ebird_code'].agg('count')
coords = coords[coords.latitude != 'Not specified']
coords = coords[coords.longitude != 'Not specified']
xp.scatter_geo(lat=coords['latitude'], lon=coords['longitude'], title='Recording Locations', size=coords['ebird_code'])
```
- Vast majority of data comes from North America, specifically from USA
<a id="AudioFeatureExtraction"></a>
# Audio Feature Extraction
<a id="Sample"></a>
## Sample
First, let's take the first 5 audio samples from train.csv and explore.
```
bird_codes = train.ebird_code.unique()[:5]
audio = []
for bird in range(len(bird_codes)):
filename = train[train['ebird_code'] == bird_codes[bird]]['filename'].iloc[0]
path = os.path.join('../input/birdsong-recognition/train_audio/', bird_codes[bird], filename)
# wave plot
plt.figure(figsize=(15,10))
plt.subplot(len(bird_codes), 1, bird+1)
data, srate = librosa.load(path)
librosa.display.waveplot(data, sr=srate)
plt.gca().set_title(bird_codes[bird])
plt.xticks([],[])
plt.xlabel('')
plt.show()
# audio display
audio = ipd.Audio(path)
ipd.display(audio)
```
<a id="Features"></a>
## Features
After doing some research on audio signal classification, I have come up with the following features to extract from the audio files:
- [Waveform](#Waveform)
- [Autocorrelation](#Autocorrelation)
- [Spectrogram](#Spectrogram)
- [Chromagram](#Chromagram)
- [Spectral](#Spectral)
- [Centroid](#Centroid)
- [Bandwidth](#Bandwidth)
- [Contrast](#Contrast)
- [Flatness](#Flatness)
- [Rolloff](#Rolloff)
- [MFCC](#MFCC)
We'll do a sample feature extraction of bird code 'ameavo' as an example.
<a id="Waveform"></a>
### Waveform
```
data, srate = librosa.load('../input/birdsong-recognition/train_audio/ameavo/XC99571.mp3')
# plot waveform as refresher
plt.figure(figsize=(15,5))
librosa.display.waveplot(data, sr=srate)
plt.gca().set_title('ameavo')
plt.xticks([],[])
plt.xlabel('')
plt.show()
```
<a id="Autocorrelation"></a>
### Autocorrelation
Autocorrelation compares a signal with a lagged version of itself. It is often used as a measure of periodicity in a signal.
```
autocorrelation = librosa.autocorrelate(data, max_size=5000)
plt.figure(figsize=(15,5))
plt.plot(autocorrelation)
plt.gca().set_title('Autocorrelation by Lag Time')
plt.xlabel('Lag')
plt.show()
```
- autocorrelation very quickly falls off reaching almost 0 after a lag of about 500
<a id="Spectrogram"></a>
### Spectrogram
The spectrogram is a visual representation of a signal's spectrum of frequencies over time.
```
spectrogram = librosa.stft(data)
plt.figure(figsize=(20,10))
librosa.display.specshow(librosa.amplitude_to_db(abs(spectrogram)), sr=srate, x_axis='time', y_axis='hz')
plt.xlabel('Time', fontsize=20)
plt.ylabel('Frequency Band')
plt.colorbar()
plt.title('Spectrogram', fontsize=20)
plt.show()
```
- Pitch seems to hover around 2000 to 3500 Hz most of the time
- Some spikes to 5500-7000 Hz
<a id="Chromagram"></a>
### Chromagram
The Chromagram is a visual representation of a signal's chroma feature. The chroma feature at any point in time is the intensity for each chroma value in the set {C, C♯, D, D♯, E , F, F♯, G, G♯, A, A♯, B}. These values are the various rows of the chromagram.
```
chroma = librosa.feature.chroma_stft(data, sr=srate)
plt.figure(figsize=(20,10))
librosa.display.specshow(chroma, x_axis='time', y_axis='chroma')
plt.xlabel('Time', fontsize=20)
plt.ylabel('Chroma Value')
plt.colorbar()
plt.clim(0,1)
plt.title('Chromagram', fontsize=20)
plt.show()
```
<a id="Spectral"></a>
### Spectral Features
<a id="Centroid"></a>
#### Spectral Centroid
Spectral Centroid is a measurement of the "center of gravity" of the signal and is a common metric of timbre in a sound sample. It's essentially the dominant frequency at each point.
```
centroid = librosa.feature.spectral_centroid(data)[0]
plt.figure(figsize=(15,5))
librosa.display.waveplot(data, sr=srate)
plt.plot(librosa.frames_to_time(range(len(centroid))), minmax_scale(centroid), color='g')
plt.gca().set_title('Spectral Centroid by Frame')
plt.xlabel('Frame')
plt.show()
```
<a id="Bandwidth"></a>
#### Spectral Bandwidth
Spectral bandwidth represents the range between the lowest and highest values of the signal at a certain time.
```
bandwidth = librosa.feature.spectral_bandwidth(data, sr=srate)[0]
plt.figure(figsize=(15,5))
librosa.display.waveplot(data, sr=srate)
plt.plot(librosa.frames_to_time(range(len(bandwidth))), minmax_scale(bandwidth))
plt.gca().set_title('Spectral Bandwidth by Time')
plt.xlabel('Time')
plt.show()
```
- pure noise portions of sample are higher in bandwidth
<a id="Contrast"></a>
#### Spectral Contrast
Spectral contrast compares the max and min frequency values for each frequency band at a point in time. Thus, spectral contrast gives a robust measure of relative spectral characteristics.
```
contrast = librosa.feature.spectral_contrast(data, sr=srate)
plt.figure(figsize=(20,10))
librosa.display.specshow(contrast, x_axis='time')
plt.xlabel('Time', fontsize=20)
plt.colorbar()
plt.title('Spectral Contrast', fontsize=20)
plt.ylabel('Frequency Band', fontsize=20)
plt.show()
```
- highest contrast occurs in the lowest and highest frequency bands
<a id="Flatness"></a>
#### Spectral Flatness
Spectral flatness compares the arithmetic and geometric means of the power spectrum. It is most often used to identify and separate tones versus noise.
```
flatness = librosa.feature.spectral_flatness(data)
plt.figure(figsize=(20,10))
librosa.display.specshow(flatness, x_axis='time')
plt.xlabel('Time', fontsize=20)
plt.colorbar()
plt.clim(0,1)
plt.title('Spectral Flatness', fontsize=20)
plt.ylabel('Frequency Band', fontsize=20)
plt.show()
```
- Maximum 20% noise at points in sample.
- Very low noise in general
<a id="Rolloff"></a>
#### Spectral Rolloff
Spectral rolloff is the frequency under which a specified percentage of the energy lies
```
rolloff = librosa.feature.spectral_rolloff(data, sr=srate)[0]
plt.figure(figsize=(15,5))
librosa.display.waveplot(data, sr=srate)
plt.plot(librosa.frames_to_time(range(len(rolloff))), minmax_scale(rolloff))
plt.gca().set_title('Spectral Bandwidth by Time')
plt.xlabel('Time')
plt.show()
```
<a id="MFCC"></a>
### MFCC
Mel-Frequency Cepstral Coefficients are a collection of coefficients that together give a representation of the overall spectral envelope of a signal. Probably the most common and important feature of audio signal processing in machine learning.
```
mfcc = librosa.feature.mfcc(data, sr=srate, n_mfcc=30)
plt.figure(figsize=(20,10))
librosa.display.specshow(minmax_scale(mfcc, axis=1), x_axis='time')
plt.xlabel('Time', fontsize=20)
plt.colorbar()
plt.clim(0,1)
plt.title('Mel-Frequency Cepstral Coefficients', fontsize=20)
plt.show()
print()
print('MFCCs calculated: %d' % mfcc.shape[0])
```
<a id="Thanks"></a>
# Thank You for Reading!
I am still very much new to data science, and I'm jumping in head-first. This is meant as a learning experience for myself as well as a simple EDA and FE for those who aren't well-versed in audio processing, so I invite any and all constructive feedback.
Thanks again! Hope this is helpful to you!
|
github_jupyter
|
import os
import sys
import librosa
import librosa.display
import librosa.feature
import numpy as np
import pandas as pd
import plotly.express as xp
import plotly.graph_objects as go
import matplotlib.pyplot as plt
import IPython.display as ipd
from sklearn.preprocessing import minmax_scale
import warnings
warnings.filterwarnings('ignore')
%matplotlib inline
train = pd.read_csv('../input/birdsong-recognition/train.csv')
train.head()
train.info()
missing = train.isna().sum().sort_values(ascending=False)
missing = missing[missing != 0]
xp.bar(x=missing.index, y=missing, text=missing, title='Missing Values by Feature', labels={'x':'Features', 'y':'Quantity'})
counts = train['ebird_code'].value_counts()
xp.bar(x=counts.index, y=counts, title='Species Distribution (by ebird code)', labels={'x':'Ebird Code', 'y':'Quantity'})
# split datetime into separate dataframe
datetime = train[['date', 'time']]
datetime.date = pd.to_datetime(datetime.date, errors='coerce').dropna()
datetime['hour'] = pd.to_numeric(datetime.time.str.split(':', expand=True)[0], errors='coerce')
ax1 = datetime.date.value_counts().sort_values().plot(figsize=(10,6), title='Recordings by Date')
ax1.set_xlabel('Date')
ax1.set_ylabel('Quantity')
plt.show()
ax2 = datetime['hour'].value_counts().sort_index().plot(figsize=(10,6), title='Recordings by Time', kind='bar', figure=plt.figure())
ax2.set_xlabel('Hour')
ax2.set_ylabel('Quantity')
plt.show()
ax3 = train['recordist'].value_counts().sort_values(ascending=False).head(20).sort_values().plot(figsize=(10, 6), title='Recordings by Recordist', figure=plt.figure(), kind='barh', fontsize=9)
ax3.set_xlabel('Hour')
ax3.set_ylabel('Quantity')
plt.tight_layout()
plt.show()
counts = train['country'].value_counts().sort_values(ascending=False).head(10).sort_values()
xp.bar(y=counts.index, x=counts, title='Number of Recordings by Country', labels={'y':'Country', 'x':'Quantity'}, orientation='h')
coords = train.groupby(['latitude', 'longitude'], as_index=False)['ebird_code'].agg('count')
coords = coords[coords.latitude != 'Not specified']
coords = coords[coords.longitude != 'Not specified']
xp.scatter_geo(lat=coords['latitude'], lon=coords['longitude'], title='Recording Locations', size=coords['ebird_code'])
bird_codes = train.ebird_code.unique()[:5]
audio = []
for bird in range(len(bird_codes)):
filename = train[train['ebird_code'] == bird_codes[bird]]['filename'].iloc[0]
path = os.path.join('../input/birdsong-recognition/train_audio/', bird_codes[bird], filename)
# wave plot
plt.figure(figsize=(15,10))
plt.subplot(len(bird_codes), 1, bird+1)
data, srate = librosa.load(path)
librosa.display.waveplot(data, sr=srate)
plt.gca().set_title(bird_codes[bird])
plt.xticks([],[])
plt.xlabel('')
plt.show()
# audio display
audio = ipd.Audio(path)
ipd.display(audio)
data, srate = librosa.load('../input/birdsong-recognition/train_audio/ameavo/XC99571.mp3')
# plot waveform as refresher
plt.figure(figsize=(15,5))
librosa.display.waveplot(data, sr=srate)
plt.gca().set_title('ameavo')
plt.xticks([],[])
plt.xlabel('')
plt.show()
autocorrelation = librosa.autocorrelate(data, max_size=5000)
plt.figure(figsize=(15,5))
plt.plot(autocorrelation)
plt.gca().set_title('Autocorrelation by Lag Time')
plt.xlabel('Lag')
plt.show()
spectrogram = librosa.stft(data)
plt.figure(figsize=(20,10))
librosa.display.specshow(librosa.amplitude_to_db(abs(spectrogram)), sr=srate, x_axis='time', y_axis='hz')
plt.xlabel('Time', fontsize=20)
plt.ylabel('Frequency Band')
plt.colorbar()
plt.title('Spectrogram', fontsize=20)
plt.show()
chroma = librosa.feature.chroma_stft(data, sr=srate)
plt.figure(figsize=(20,10))
librosa.display.specshow(chroma, x_axis='time', y_axis='chroma')
plt.xlabel('Time', fontsize=20)
plt.ylabel('Chroma Value')
plt.colorbar()
plt.clim(0,1)
plt.title('Chromagram', fontsize=20)
plt.show()
centroid = librosa.feature.spectral_centroid(data)[0]
plt.figure(figsize=(15,5))
librosa.display.waveplot(data, sr=srate)
plt.plot(librosa.frames_to_time(range(len(centroid))), minmax_scale(centroid), color='g')
plt.gca().set_title('Spectral Centroid by Frame')
plt.xlabel('Frame')
plt.show()
bandwidth = librosa.feature.spectral_bandwidth(data, sr=srate)[0]
plt.figure(figsize=(15,5))
librosa.display.waveplot(data, sr=srate)
plt.plot(librosa.frames_to_time(range(len(bandwidth))), minmax_scale(bandwidth))
plt.gca().set_title('Spectral Bandwidth by Time')
plt.xlabel('Time')
plt.show()
contrast = librosa.feature.spectral_contrast(data, sr=srate)
plt.figure(figsize=(20,10))
librosa.display.specshow(contrast, x_axis='time')
plt.xlabel('Time', fontsize=20)
plt.colorbar()
plt.title('Spectral Contrast', fontsize=20)
plt.ylabel('Frequency Band', fontsize=20)
plt.show()
flatness = librosa.feature.spectral_flatness(data)
plt.figure(figsize=(20,10))
librosa.display.specshow(flatness, x_axis='time')
plt.xlabel('Time', fontsize=20)
plt.colorbar()
plt.clim(0,1)
plt.title('Spectral Flatness', fontsize=20)
plt.ylabel('Frequency Band', fontsize=20)
plt.show()
rolloff = librosa.feature.spectral_rolloff(data, sr=srate)[0]
plt.figure(figsize=(15,5))
librosa.display.waveplot(data, sr=srate)
plt.plot(librosa.frames_to_time(range(len(rolloff))), minmax_scale(rolloff))
plt.gca().set_title('Spectral Bandwidth by Time')
plt.xlabel('Time')
plt.show()
mfcc = librosa.feature.mfcc(data, sr=srate, n_mfcc=30)
plt.figure(figsize=(20,10))
librosa.display.specshow(minmax_scale(mfcc, axis=1), x_axis='time')
plt.xlabel('Time', fontsize=20)
plt.colorbar()
plt.clim(0,1)
plt.title('Mel-Frequency Cepstral Coefficients', fontsize=20)
plt.show()
print()
print('MFCCs calculated: %d' % mfcc.shape[0])
| 0.37502 | 0.970127 |
## Hyper Params
```
! unzip /content/wae-wgan-master.zip
mv wae-wgan-master/* /content/
Z_LENGTH = 5
NUM_LAYERS = 3
MODEL = './assets/pretrained_models/mnist/last.ckpt'
```
## Define & Load Model
```
%tensorflow_version 1.x
import tensorflow as tf
import numpy as np
from functools import partial
import arch
import dataset
import wae
x_ph = tf.placeholder(tf.float32,[None,28,28,1])
z_ph = tf.placeholder(tf.float32,[None,Z_LENGTH])
ds = dataset.MNIST(1) #Given batch size is not used.
p_z = arch.Pseudo_P_Z(z_ph)
Q_arch = partial(arch.fc_arch,
input_shape=(784,),
output_size=Z_LENGTH,
num_layers=NUM_LAYERS,
embed_size=256)
G_arch = partial(arch.fc_arch,
input_shape=(Z_LENGTH,),
output_size=784, # # of generated pixels
num_layers=NUM_LAYERS,
embed_size=256)
D_arch = partial(arch.fc_arch,
input_shape=(Z_LENGTH,), # shape when flattened.
output_size=1,
num_layers=NUM_LAYERS,
embed_size=64,
act_fn='ELU-like')
with tf.variable_scope('param_scope') as scope:
pass
model = \
wae.WAE_WGAN(x_ph,
p_z,
Q_arch,
G_arch,
D_arch,
0.0,
lambda x,y: tf.reduce_sum(tf.abs(x-y),axis=(1,2,3)), #use l1_distance for recon loss
None,
scope)
init_op = tf.group(tf.global_variables_initializer(),
tf.local_variables_initializer())
# Execute Training!
config = tf.ConfigProto()
config.gpu_options.allow_growth = True
sess = tf.InteractiveSession(config=config)
# Init variables and load weights
sess.run(init_op)
model.load(MODEL)
```
## Reconstruction Result
```
def draw(images):
x,y,h,w,c = images.shape
if c == 1:
images = np.squeeze(images)
from matplotlib import pyplot as plt
fig = plt.figure(figsize=(y,x),dpi=150)
for i in range(x):
for j in range(y):
a = fig.add_subplot(x,y, (i*y+j) + 1)
a.imshow( images[i,j], cmap='gray' )
a.axis('off')
a.set_aspect('equal')
plt.subplots_adjust(wspace=0, hspace=0)
plt.show()
plt.close()
valid_ims = ds.ims[50000:]
x = valid_ims[np.random.choice(len(valid_ims),20)]
recon_x = sess.run(model.x_recon, feed_dict={x_ph:x})
draw(np.stack([x,recon_x],axis=0))
```
(top): original images from MNIST validation set, (bottom): reconstructed image
## Random Sampled Images
```
sampled = \
sess.run(model.x_sample,
feed_dict={x_ph:np.zeros((100,28,28,1)), # Not used; just used for inferencing image shape
z_ph:np.random.normal(loc=0.0, scale=1.0, size=(100,Z_LENGTH))})
sampled = np.reshape(sampled,[10,10,28,28,1])
draw(sampled)
```
|
github_jupyter
|
! unzip /content/wae-wgan-master.zip
mv wae-wgan-master/* /content/
Z_LENGTH = 5
NUM_LAYERS = 3
MODEL = './assets/pretrained_models/mnist/last.ckpt'
%tensorflow_version 1.x
import tensorflow as tf
import numpy as np
from functools import partial
import arch
import dataset
import wae
x_ph = tf.placeholder(tf.float32,[None,28,28,1])
z_ph = tf.placeholder(tf.float32,[None,Z_LENGTH])
ds = dataset.MNIST(1) #Given batch size is not used.
p_z = arch.Pseudo_P_Z(z_ph)
Q_arch = partial(arch.fc_arch,
input_shape=(784,),
output_size=Z_LENGTH,
num_layers=NUM_LAYERS,
embed_size=256)
G_arch = partial(arch.fc_arch,
input_shape=(Z_LENGTH,),
output_size=784, # # of generated pixels
num_layers=NUM_LAYERS,
embed_size=256)
D_arch = partial(arch.fc_arch,
input_shape=(Z_LENGTH,), # shape when flattened.
output_size=1,
num_layers=NUM_LAYERS,
embed_size=64,
act_fn='ELU-like')
with tf.variable_scope('param_scope') as scope:
pass
model = \
wae.WAE_WGAN(x_ph,
p_z,
Q_arch,
G_arch,
D_arch,
0.0,
lambda x,y: tf.reduce_sum(tf.abs(x-y),axis=(1,2,3)), #use l1_distance for recon loss
None,
scope)
init_op = tf.group(tf.global_variables_initializer(),
tf.local_variables_initializer())
# Execute Training!
config = tf.ConfigProto()
config.gpu_options.allow_growth = True
sess = tf.InteractiveSession(config=config)
# Init variables and load weights
sess.run(init_op)
model.load(MODEL)
def draw(images):
x,y,h,w,c = images.shape
if c == 1:
images = np.squeeze(images)
from matplotlib import pyplot as plt
fig = plt.figure(figsize=(y,x),dpi=150)
for i in range(x):
for j in range(y):
a = fig.add_subplot(x,y, (i*y+j) + 1)
a.imshow( images[i,j], cmap='gray' )
a.axis('off')
a.set_aspect('equal')
plt.subplots_adjust(wspace=0, hspace=0)
plt.show()
plt.close()
valid_ims = ds.ims[50000:]
x = valid_ims[np.random.choice(len(valid_ims),20)]
recon_x = sess.run(model.x_recon, feed_dict={x_ph:x})
draw(np.stack([x,recon_x],axis=0))
sampled = \
sess.run(model.x_sample,
feed_dict={x_ph:np.zeros((100,28,28,1)), # Not used; just used for inferencing image shape
z_ph:np.random.normal(loc=0.0, scale=1.0, size=(100,Z_LENGTH))})
sampled = np.reshape(sampled,[10,10,28,28,1])
draw(sampled)
| 0.47098 | 0.761272 |
```
%matplotlib widget
import seaborn as sns
import matplotlib.pyplot as plt
#stripper function
import re
def strip_tree(tree):
return re.sub('(\d|:|_|\.|[a-z])', '', tree)
def load_trees(fname):
with open(fname,'r') as fin:
return [l.rstrip().split(';') for l in fin][0][:-1]
trees = load_trees("joined_trees.txt")
from collections import Counter
cs = Counter([strip_tree(tree) for tree in trees])
print(cs)
strip_tree('((((P:51962.00,B:51962.00):3416.00,O:55378.00):43779.33,N:99157.33):45587.17,G:144744.50);')
import pandas as pd
rows = []
for i,comb in enumerate([strip_tree(tree) for tree in trees]):
x = comb.replace('(','').replace(')','').replace(',','')[:3]
code = '-'.join(sorted(x[:2])+[x[2]])
chrom = (i%29)+1
chrom = chrom if chrom <= 29 else 'all'
config = None
run = i//29
if run < 5:
config = 'shasta'
elif run < 10:
config = 'hifiasm'
elif run < 20:
config = 'hs_shuf'
elif run < 40:
config = 'either'
elif run < 43:
config = 'peregrine'
elif run < 46:
config = 'raven'
elif run < 49:
config = 'flye'
elif run < 50:
config = 'hicanu'
elif run < 60:
config = 'all_shuf'
elif run < 80:
config = 'all_eith'
rows.append({'run':i//30,'chr':chrom,'order':code,'config':config})
df = pd.DataFrame(rows)
df
df.groupby('config').count()
sns.catplot(data=df,x='chr',kind='count',hue='order',col_wrap=4,col='config')
qq= df[(df['config']=='hifiasm')|(df['config']=='shasta')|(df['config']=='either')|(df['config']=='hs_shuf')].groupby(['chr','order']).count().reset_index()
plt.figure()
sns.scatterplot(data=qq,x='chr',y='run',hue='order')
qq[qq['run']>38]
data='(gaur:0.376151,(nellore:0.117037,(bsw:0.0383929,(obv:0.036079,pied:0.036079):0.00231387):0.0786446):0.259113);(gaur:0.384387,(nellore:0.104898,(pied:0.0358684,(bsw:0.0336749,obv:0.0336749):0.00219359):0.0690295):0.279489);(gaur:0.393264,(nellore:0.112438,(pied:0.04183,(bsw:0.037261,obv:0.037261):0.00456896):0.0706083):0.280826);(gaur:0.399114,(nellore:0.122466,(bsw:0.0433282,(obv:0.0387218,pied:0.0387218):0.00460634):0.0791373):0.276648);(gaur:0.366217,(nellore:0.118095,(pied:0.0400524,(bsw:0.0372951,obv:0.0372951):0.00275733):0.0780421):0.248123);(gaur:0.394545,(nellore:0.111131,(pied:0.0436057,(bsw:0.0387256,obv:0.0387256):0.00488007):0.067525):0.283414);(gaur:0.409183,(nellore:0.112553,(bsw:0.0389752,(obv:0.0360255,pied:0.0360255):0.00294963):0.0735783):0.29663);(gaur:0.397388,(nellore:0.122268,(obv:0.0454757,(bsw:0.0351746,pied:0.0351746):0.0103011):0.0767922):0.275121);(gaur:0.398349,(nellore:0.105709,(pied:0.0400871,(bsw:0.0381708,obv:0.0381708):0.00191624):0.0656224):0.29264);(gaur:0.386962,(nellore:0.102029,(bsw:0.0406497,(obv:0.0385982,pied:0.0385982):0.00205157):0.061379):0.284933);(gaur:0.369279,(nellore:0.114112,(pied:0.0387218,(bsw:0.0361617,obv:0.0361617):0.00256012):0.0753905):0.255167);(gaur:0.380651,(nellore:0.13181,(bsw:0.0528749,(obv:0.0441349,pied:0.0441349):0.00874002):0.0789346):0.248842);(gaur:0.391107,(nellore:0.111082,(pied:0.0343925,(bsw:0.0314762,obv:0.0314762):0.00291624):0.0766893):0.280025);(gaur:0.39991,(nellore:0.104656,(bsw:0.0374423,(obv:0.0363872,pied:0.0363872):0.00105507):0.0672136):0.295254);(gaur:0.36453,(nellore:0.122061,(bsw:0.043176,(obv:0.040549,pied:0.040549):0.00262703):0.0788854):0.242469);(gaur:0.399314,(nellore:0.112428,(bsw:0.0385897,(obv:0.0364024,pied:0.0364024):0.00218733):0.0738385):0.286885);(gaur:0.388082,(nellore:0.11466,(pied:0.0401085,(bsw:0.0382637,obv:0.0382637):0.00184477):0.0745517):0.273422);(gaur:0.352804,(nellore:0.111778,(pied:0.0426599,(bsw:0.0388053,obv:0.0388053):0.00385459):0.0691186):0.241026);(gaur:0.397195,(nellore:0.100784,(bsw:0.0366994,(obv:0.0356394,pied:0.0356394):0.00105997):0.0640849):0.29641);(gaur:0.328738,(nellore:0.0994499,(pied:0.0365601,(bsw:0.0348973,obv:0.0348973):0.00166282):0.0628898):0.229288);(gaur:0.403084,(nellore:0.116689,(bsw:0.0409729,(obv:0.0313408,pied:0.0313408):0.00963216):0.0757156):0.286395);(gaur:0.391599,(nellore:0.0879211,(bsw:0.0306894,(obv:0.0303176,pied:0.0303176):0.000371842):0.0572316):0.303678);(gaur:0.330951,(nellore:0.123869,(pied:0.0526,(bsw:0.0465642,obv:0.0465642):0.00603579):0.0712689):0.207082);(gaur:0.365075,(nellore:0.116235,(bsw:0.0401447,(obv:0.0372307,pied:0.0372307):0.00291405):0.0760902):0.24884);(gaur:0.42264,(nellore:0.118208,(obv:0.0437409,(bsw:0.0410686,pied:0.0410686):0.00267231):0.0744673):0.304432);(gaur:0.390656,(nellore:0.113633,(obv:0.0397816,(bsw:0.0373379,pied:0.0373379):0.00244372):0.0738513):0.277023);(gaur:0.398647,(nellore:0.144023,(pied:0.036351,(bsw:0.0354978,obv:0.0354978):0.000853173):0.107672):0.254624);(gaur:0.384289,(nellore:0.141181,(obv:0.0394741,(bsw:0.0383582,pied:0.0383582):0.00111593):0.101707):0.243107);(gaur:0.355713,(nellore:0.129896,(bsw:0.0420809,(obv:0.0386343,pied:0.0386343):0.00344661):0.0878151):0.225817)'.replace('gaur','G').replace('bsw','B').replace('nellore','N').replace('obv','O').replace('pied','P')
raw = [strip_tree(t).replace('(','').replace(')','').replace(',','')[2:] for t in data.split(';')]
SNP = ['-'.join(sorted(i[1:])+[i[0]]) for i in raw]
SNP
for t in ['hifiasm','shasta','hs_shuf','either','peregrine','raven','flye','hicanu','all_shuf','all_eith']:
most_c = []
for i in range(1,30):
dfa = df[(df['chr']==i)&(df['config']==t)]
most_c.append(Counter(dfa['order']).most_common(1)[0][0])
c= 0
for i,j in zip(most_c,SNP):
c+=(i==j)
print(t,c)
g = sns.catplot(data=df,x='order',kind='count',hue='config',col='chr',col_wrap=4)#,order=['O-P-B','B-P-O','B-O-P'])
for i,ax in enumerate(g.axes):
#ax.scatter(SNP[i],20)
ax.scatter('B-P-O',15,alpha=0)
ax.axvline(SNP[i])
df[df['chr']==2]
import pandas as pd
df = pd.read_csv('bad_regions.csv')
sns.pairplot(data=df,hue='asm')
df.groupby('asm').mean()
2715195.9/1e6
2590566.3/1e6
import scipy.stats as ss
ss.mannwhitneyu(df[df['asm']=='P_hifiasm']['N_unaligned'],df[df['asm']=='P_shasta']['N_unaligned'])
f=[]
for i in range(300,310):
x = df[df['run']==i]
l = [(row['asm'],row['N_uncalled']) for _,row in x.iterrows()]
f.append([q[0] for q in sorted(l,key=lambda i: i[1])])
f
orders = [l.rstrip() for l in open('orders.txt')]
n=7
o = [orders[i:i + n][:n-1] for i in range(0, len(orders), n)]
o
import numpy as np
on=np.array(o)
lo = np.array(f)
on==lo
```
|
github_jupyter
|
%matplotlib widget
import seaborn as sns
import matplotlib.pyplot as plt
#stripper function
import re
def strip_tree(tree):
return re.sub('(\d|:|_|\.|[a-z])', '', tree)
def load_trees(fname):
with open(fname,'r') as fin:
return [l.rstrip().split(';') for l in fin][0][:-1]
trees = load_trees("joined_trees.txt")
from collections import Counter
cs = Counter([strip_tree(tree) for tree in trees])
print(cs)
strip_tree('((((P:51962.00,B:51962.00):3416.00,O:55378.00):43779.33,N:99157.33):45587.17,G:144744.50);')
import pandas as pd
rows = []
for i,comb in enumerate([strip_tree(tree) for tree in trees]):
x = comb.replace('(','').replace(')','').replace(',','')[:3]
code = '-'.join(sorted(x[:2])+[x[2]])
chrom = (i%29)+1
chrom = chrom if chrom <= 29 else 'all'
config = None
run = i//29
if run < 5:
config = 'shasta'
elif run < 10:
config = 'hifiasm'
elif run < 20:
config = 'hs_shuf'
elif run < 40:
config = 'either'
elif run < 43:
config = 'peregrine'
elif run < 46:
config = 'raven'
elif run < 49:
config = 'flye'
elif run < 50:
config = 'hicanu'
elif run < 60:
config = 'all_shuf'
elif run < 80:
config = 'all_eith'
rows.append({'run':i//30,'chr':chrom,'order':code,'config':config})
df = pd.DataFrame(rows)
df
df.groupby('config').count()
sns.catplot(data=df,x='chr',kind='count',hue='order',col_wrap=4,col='config')
qq= df[(df['config']=='hifiasm')|(df['config']=='shasta')|(df['config']=='either')|(df['config']=='hs_shuf')].groupby(['chr','order']).count().reset_index()
plt.figure()
sns.scatterplot(data=qq,x='chr',y='run',hue='order')
qq[qq['run']>38]
data='(gaur:0.376151,(nellore:0.117037,(bsw:0.0383929,(obv:0.036079,pied:0.036079):0.00231387):0.0786446):0.259113);(gaur:0.384387,(nellore:0.104898,(pied:0.0358684,(bsw:0.0336749,obv:0.0336749):0.00219359):0.0690295):0.279489);(gaur:0.393264,(nellore:0.112438,(pied:0.04183,(bsw:0.037261,obv:0.037261):0.00456896):0.0706083):0.280826);(gaur:0.399114,(nellore:0.122466,(bsw:0.0433282,(obv:0.0387218,pied:0.0387218):0.00460634):0.0791373):0.276648);(gaur:0.366217,(nellore:0.118095,(pied:0.0400524,(bsw:0.0372951,obv:0.0372951):0.00275733):0.0780421):0.248123);(gaur:0.394545,(nellore:0.111131,(pied:0.0436057,(bsw:0.0387256,obv:0.0387256):0.00488007):0.067525):0.283414);(gaur:0.409183,(nellore:0.112553,(bsw:0.0389752,(obv:0.0360255,pied:0.0360255):0.00294963):0.0735783):0.29663);(gaur:0.397388,(nellore:0.122268,(obv:0.0454757,(bsw:0.0351746,pied:0.0351746):0.0103011):0.0767922):0.275121);(gaur:0.398349,(nellore:0.105709,(pied:0.0400871,(bsw:0.0381708,obv:0.0381708):0.00191624):0.0656224):0.29264);(gaur:0.386962,(nellore:0.102029,(bsw:0.0406497,(obv:0.0385982,pied:0.0385982):0.00205157):0.061379):0.284933);(gaur:0.369279,(nellore:0.114112,(pied:0.0387218,(bsw:0.0361617,obv:0.0361617):0.00256012):0.0753905):0.255167);(gaur:0.380651,(nellore:0.13181,(bsw:0.0528749,(obv:0.0441349,pied:0.0441349):0.00874002):0.0789346):0.248842);(gaur:0.391107,(nellore:0.111082,(pied:0.0343925,(bsw:0.0314762,obv:0.0314762):0.00291624):0.0766893):0.280025);(gaur:0.39991,(nellore:0.104656,(bsw:0.0374423,(obv:0.0363872,pied:0.0363872):0.00105507):0.0672136):0.295254);(gaur:0.36453,(nellore:0.122061,(bsw:0.043176,(obv:0.040549,pied:0.040549):0.00262703):0.0788854):0.242469);(gaur:0.399314,(nellore:0.112428,(bsw:0.0385897,(obv:0.0364024,pied:0.0364024):0.00218733):0.0738385):0.286885);(gaur:0.388082,(nellore:0.11466,(pied:0.0401085,(bsw:0.0382637,obv:0.0382637):0.00184477):0.0745517):0.273422);(gaur:0.352804,(nellore:0.111778,(pied:0.0426599,(bsw:0.0388053,obv:0.0388053):0.00385459):0.0691186):0.241026);(gaur:0.397195,(nellore:0.100784,(bsw:0.0366994,(obv:0.0356394,pied:0.0356394):0.00105997):0.0640849):0.29641);(gaur:0.328738,(nellore:0.0994499,(pied:0.0365601,(bsw:0.0348973,obv:0.0348973):0.00166282):0.0628898):0.229288);(gaur:0.403084,(nellore:0.116689,(bsw:0.0409729,(obv:0.0313408,pied:0.0313408):0.00963216):0.0757156):0.286395);(gaur:0.391599,(nellore:0.0879211,(bsw:0.0306894,(obv:0.0303176,pied:0.0303176):0.000371842):0.0572316):0.303678);(gaur:0.330951,(nellore:0.123869,(pied:0.0526,(bsw:0.0465642,obv:0.0465642):0.00603579):0.0712689):0.207082);(gaur:0.365075,(nellore:0.116235,(bsw:0.0401447,(obv:0.0372307,pied:0.0372307):0.00291405):0.0760902):0.24884);(gaur:0.42264,(nellore:0.118208,(obv:0.0437409,(bsw:0.0410686,pied:0.0410686):0.00267231):0.0744673):0.304432);(gaur:0.390656,(nellore:0.113633,(obv:0.0397816,(bsw:0.0373379,pied:0.0373379):0.00244372):0.0738513):0.277023);(gaur:0.398647,(nellore:0.144023,(pied:0.036351,(bsw:0.0354978,obv:0.0354978):0.000853173):0.107672):0.254624);(gaur:0.384289,(nellore:0.141181,(obv:0.0394741,(bsw:0.0383582,pied:0.0383582):0.00111593):0.101707):0.243107);(gaur:0.355713,(nellore:0.129896,(bsw:0.0420809,(obv:0.0386343,pied:0.0386343):0.00344661):0.0878151):0.225817)'.replace('gaur','G').replace('bsw','B').replace('nellore','N').replace('obv','O').replace('pied','P')
raw = [strip_tree(t).replace('(','').replace(')','').replace(',','')[2:] for t in data.split(';')]
SNP = ['-'.join(sorted(i[1:])+[i[0]]) for i in raw]
SNP
for t in ['hifiasm','shasta','hs_shuf','either','peregrine','raven','flye','hicanu','all_shuf','all_eith']:
most_c = []
for i in range(1,30):
dfa = df[(df['chr']==i)&(df['config']==t)]
most_c.append(Counter(dfa['order']).most_common(1)[0][0])
c= 0
for i,j in zip(most_c,SNP):
c+=(i==j)
print(t,c)
g = sns.catplot(data=df,x='order',kind='count',hue='config',col='chr',col_wrap=4)#,order=['O-P-B','B-P-O','B-O-P'])
for i,ax in enumerate(g.axes):
#ax.scatter(SNP[i],20)
ax.scatter('B-P-O',15,alpha=0)
ax.axvline(SNP[i])
df[df['chr']==2]
import pandas as pd
df = pd.read_csv('bad_regions.csv')
sns.pairplot(data=df,hue='asm')
df.groupby('asm').mean()
2715195.9/1e6
2590566.3/1e6
import scipy.stats as ss
ss.mannwhitneyu(df[df['asm']=='P_hifiasm']['N_unaligned'],df[df['asm']=='P_shasta']['N_unaligned'])
f=[]
for i in range(300,310):
x = df[df['run']==i]
l = [(row['asm'],row['N_uncalled']) for _,row in x.iterrows()]
f.append([q[0] for q in sorted(l,key=lambda i: i[1])])
f
orders = [l.rstrip() for l in open('orders.txt')]
n=7
o = [orders[i:i + n][:n-1] for i in range(0, len(orders), n)]
o
import numpy as np
on=np.array(o)
lo = np.array(f)
on==lo
| 0.210482 | 0.273083 |
```
%run data.py
```
### Read data from NYC Open Data.
```
results_df = fetch_nycOpenData(nyc_C_O_issue, 100, 200000)
results_df.head(10)
df = pandas_to_spark(results_df)
from pyspark.sql.functions import to_timestamp
from pyspark.sql.functions import month, year
# group table by month.
#filter date and time from 06/2019
df = df.withColumn("date", to_timestamp("c_o_issue_date", "yyyy-MM-dd'T'HH:mm:ss.SSS"))
df = df.filter(df["date"] >= to_timestamp(f.lit('2019-09-01 00:00:00')).cast('timestamp'))
df = df.groupBy(month("date").alias("month")).count()
df = df.filter(df["month"] != 5)
df = df.withColumn("month_name",
f.when(f.col('month') == 1, "2020-01")\
.when(f.col('month') == 2, "2020-02")\
.when(f.col('month') == 3, "2020-03")\
.when(f.col('month') == 4, "2020-04")\
.when(f.col('month') == 9, "2019-09")\
.when(f.col('month') == 10, "2019-10")\
.when(f.col('month') == 11, "2019-11")\
.when(f.col('month') == 12, "2019-12"))
df = df.orderBy("month_name")
df = df.select(df["month_name"], df["count"].alias("c_o_issue"))
df.show()
avg_data = df.filter(df["month_name"] != "2020-03")
avg_data = avg_data.filter(df["month_name"] != "2020-04")
avg_data.show()
```
### Calculate the falling rate.
```
avg_data.createOrReplaceTempView("avg_data")
avg_num = spark.sql("SELECT avg(c_o_issue) as avg_num FROM avg_data")
avg_num = avg_num.rdd.map(list)
avg_number = avg_num.take(1)[0][0]
print("Average Certificate of Occupancy issued for new buildings before March 2020 is: ", avg_number)
df.createOrReplaceTempView("latest_data")
latest_number = spark.sql("SELECT c_o_issue FROM latest_data WHERE month_name = \
(SELECT max(month_name) FROM latest_data)")
latest_number = latest_number.rdd.map(list)
latest_number = latest_number.take(1)[0][0]
falling_rate = (avg_number - latest_number) / avg_number
print("Falling rate is: ", falling_rate)
falling_rate = falling_rate * 100
falling_rate_str = "↓" + str('%.2f' % falling_rate) + "%"
import matplotlib.pyplot as plt
plt.close('all')
data = df.toPandas()
data.head(10)
```
### Plot.
```
#Certificate of Occupancy issued for new buildings
plt.plot("month_name", "c_o_issue", data = data, color = "lightseagreen")
#average Certificate of Occupancy issued for new buildings before March 2020
plt.axhline(y = avg_number,ls = "dashed",color = "grey")
#title
plt.title("COVID-19 Impact on C_O issued for new buildings",fontsize = 15)
#annotaion: falling rate
bbox_props = dict(boxstyle="round", facecolor = "white")
plt.text(0, 1200, falling_rate_str, size = 15, color = "lightseagreen", bbox=bbox_props)
plt.xticks(rotation=25)
plt.show()
```
|
github_jupyter
|
%run data.py
results_df = fetch_nycOpenData(nyc_C_O_issue, 100, 200000)
results_df.head(10)
df = pandas_to_spark(results_df)
from pyspark.sql.functions import to_timestamp
from pyspark.sql.functions import month, year
# group table by month.
#filter date and time from 06/2019
df = df.withColumn("date", to_timestamp("c_o_issue_date", "yyyy-MM-dd'T'HH:mm:ss.SSS"))
df = df.filter(df["date"] >= to_timestamp(f.lit('2019-09-01 00:00:00')).cast('timestamp'))
df = df.groupBy(month("date").alias("month")).count()
df = df.filter(df["month"] != 5)
df = df.withColumn("month_name",
f.when(f.col('month') == 1, "2020-01")\
.when(f.col('month') == 2, "2020-02")\
.when(f.col('month') == 3, "2020-03")\
.when(f.col('month') == 4, "2020-04")\
.when(f.col('month') == 9, "2019-09")\
.when(f.col('month') == 10, "2019-10")\
.when(f.col('month') == 11, "2019-11")\
.when(f.col('month') == 12, "2019-12"))
df = df.orderBy("month_name")
df = df.select(df["month_name"], df["count"].alias("c_o_issue"))
df.show()
avg_data = df.filter(df["month_name"] != "2020-03")
avg_data = avg_data.filter(df["month_name"] != "2020-04")
avg_data.show()
avg_data.createOrReplaceTempView("avg_data")
avg_num = spark.sql("SELECT avg(c_o_issue) as avg_num FROM avg_data")
avg_num = avg_num.rdd.map(list)
avg_number = avg_num.take(1)[0][0]
print("Average Certificate of Occupancy issued for new buildings before March 2020 is: ", avg_number)
df.createOrReplaceTempView("latest_data")
latest_number = spark.sql("SELECT c_o_issue FROM latest_data WHERE month_name = \
(SELECT max(month_name) FROM latest_data)")
latest_number = latest_number.rdd.map(list)
latest_number = latest_number.take(1)[0][0]
falling_rate = (avg_number - latest_number) / avg_number
print("Falling rate is: ", falling_rate)
falling_rate = falling_rate * 100
falling_rate_str = "↓" + str('%.2f' % falling_rate) + "%"
import matplotlib.pyplot as plt
plt.close('all')
data = df.toPandas()
data.head(10)
#Certificate of Occupancy issued for new buildings
plt.plot("month_name", "c_o_issue", data = data, color = "lightseagreen")
#average Certificate of Occupancy issued for new buildings before March 2020
plt.axhline(y = avg_number,ls = "dashed",color = "grey")
#title
plt.title("COVID-19 Impact on C_O issued for new buildings",fontsize = 15)
#annotaion: falling rate
bbox_props = dict(boxstyle="round", facecolor = "white")
plt.text(0, 1200, falling_rate_str, size = 15, color = "lightseagreen", bbox=bbox_props)
plt.xticks(rotation=25)
plt.show()
| 0.483892 | 0.726134 |
```
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import sklearn as sk
from sklearn.metrics import accuracy_score, precision_score, recall_score, f1_score
# Генерируем уникальный seed
my_code = "Кац"
seed_limit = 2 ** 32
my_seed = int.from_bytes(my_code.encode(), "little") % seed_limit
np.random.seed(my_seed)
# Формируем случайную нормально распределенную выборку sample
N = 10000
sample = np.random.normal(0, 1, N)
plt.hist(sample, bins=100)
plt.show()
# Формируем массив целевых метока классов: 0 - если значение в sample меньше t и 1 - если больше
t = 0
target_labels = np.array([0 if i < t else 1 for i in sample])
plt.hist(target_labels, bins=100)
plt.show()
# Используя данные заготовки (или, при желании, не используя),
# реализуйте функции для рассчета accuracy, precision, recall и F1
def confusion_matrix(target_labels, model_labels) :
tp = 0
tn = 0
fp = 0
fn = 0
for i in range(len(target_labels)) :
if target_labels[i] == 1 and model_labels[i] == 1 :
tp += 1
if target_labels[i] == 0 and model_labels[i] == 0 :
tn += 1
if target_labels[i] == 0 and model_labels[i] == 1 :
fp += 1
if target_labels[i] == 1 and model_labels[i] == 0 :
fn += 1
return tp, tn, fp, fn
def metrics_list(target_labels, model_labels):
metrics_result = []
metrics_result.append(sk.metrics.accuracy_score(target_labels, model_labels))
metrics_result.append(sk.metrics.precision_score(target_labels, model_labels))
metrics_result.append(sk.metrics.recall_score(target_labels, model_labels))
metrics_result.append(sk.metrics.f1_score(target_labels, model_labels))
return metrics_result
# Первый эксперимент: t = 0, модель с вероятностью 50% возвращает 0 и 1
t = 0
target_labels = np.array([0 if i < t else 1 for i in sample])
model_labels = np.random.randint(2, size=N)
# Рассчитайте и выведите значения метрик accuracy, precision, recall и F1.
metrics_list(target_labels, model_labels)
# Второй эксперимент: t = 0, модель с вероятностью 25% возвращает 0 и с 75% - 1
t = 0
target_labels = np.array([0 if i < t else 1 for i in sample])
labels = np.random.randint(4, size=N)
model_labels = np.array([0 if i == 0 else 1 for i in labels])
np.random.shuffle(model_labels)
# Рассчитайте и выведите значения метрик accuracy, precision, recall и F1.
metrics_list(target_labels, model_labels)
# Проанализируйте, какие из метрик применимы в первом и втором экспериментах.
# Третий эксперимент: t = 2, модель с вероятностью 50% возвращает 0 и 1
t = 2
target_labels = np.array([0 if i < t else 1 for i in sample])
model_labels = np.random.randint(2, size=N)
# Рассчитайте и выведите значения метрик accuracy, precision, recall и F1.
metrics_list(target_labels, model_labels)
# Четвёртый эксперимент: t = 2, модель с вероятностью 100% возвращает 0
t = 2
target_labels = np.array([0 if i < t else 1 for i in sample])
model_labels = np.zeros(N)
# Рассчитайте и выведите значения метрик accuracy, precision, recall и F1.
metrics_list(target_labels, model_labels)
# Проанализируйте, какие из метрик применимы в третьем и четвёртом экспериментах.
```
|
github_jupyter
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import sklearn as sk
from sklearn.metrics import accuracy_score, precision_score, recall_score, f1_score
# Генерируем уникальный seed
my_code = "Кац"
seed_limit = 2 ** 32
my_seed = int.from_bytes(my_code.encode(), "little") % seed_limit
np.random.seed(my_seed)
# Формируем случайную нормально распределенную выборку sample
N = 10000
sample = np.random.normal(0, 1, N)
plt.hist(sample, bins=100)
plt.show()
# Формируем массив целевых метока классов: 0 - если значение в sample меньше t и 1 - если больше
t = 0
target_labels = np.array([0 if i < t else 1 for i in sample])
plt.hist(target_labels, bins=100)
plt.show()
# Используя данные заготовки (или, при желании, не используя),
# реализуйте функции для рассчета accuracy, precision, recall и F1
def confusion_matrix(target_labels, model_labels) :
tp = 0
tn = 0
fp = 0
fn = 0
for i in range(len(target_labels)) :
if target_labels[i] == 1 and model_labels[i] == 1 :
tp += 1
if target_labels[i] == 0 and model_labels[i] == 0 :
tn += 1
if target_labels[i] == 0 and model_labels[i] == 1 :
fp += 1
if target_labels[i] == 1 and model_labels[i] == 0 :
fn += 1
return tp, tn, fp, fn
def metrics_list(target_labels, model_labels):
metrics_result = []
metrics_result.append(sk.metrics.accuracy_score(target_labels, model_labels))
metrics_result.append(sk.metrics.precision_score(target_labels, model_labels))
metrics_result.append(sk.metrics.recall_score(target_labels, model_labels))
metrics_result.append(sk.metrics.f1_score(target_labels, model_labels))
return metrics_result
# Первый эксперимент: t = 0, модель с вероятностью 50% возвращает 0 и 1
t = 0
target_labels = np.array([0 if i < t else 1 for i in sample])
model_labels = np.random.randint(2, size=N)
# Рассчитайте и выведите значения метрик accuracy, precision, recall и F1.
metrics_list(target_labels, model_labels)
# Второй эксперимент: t = 0, модель с вероятностью 25% возвращает 0 и с 75% - 1
t = 0
target_labels = np.array([0 if i < t else 1 for i in sample])
labels = np.random.randint(4, size=N)
model_labels = np.array([0 if i == 0 else 1 for i in labels])
np.random.shuffle(model_labels)
# Рассчитайте и выведите значения метрик accuracy, precision, recall и F1.
metrics_list(target_labels, model_labels)
# Проанализируйте, какие из метрик применимы в первом и втором экспериментах.
# Третий эксперимент: t = 2, модель с вероятностью 50% возвращает 0 и 1
t = 2
target_labels = np.array([0 if i < t else 1 for i in sample])
model_labels = np.random.randint(2, size=N)
# Рассчитайте и выведите значения метрик accuracy, precision, recall и F1.
metrics_list(target_labels, model_labels)
# Четвёртый эксперимент: t = 2, модель с вероятностью 100% возвращает 0
t = 2
target_labels = np.array([0 if i < t else 1 for i in sample])
model_labels = np.zeros(N)
# Рассчитайте и выведите значения метрик accuracy, precision, recall и F1.
metrics_list(target_labels, model_labels)
# Проанализируйте, какие из метрик применимы в третьем и четвёртом экспериментах.
| 0.356671 | 0.789274 |
<script async src="https://www.googletagmanager.com/gtag/js?id=UA-59152712-8"></script>
<script>
window.dataLayer = window.dataLayer || [];
function gtag(){dataLayer.push(arguments);}
gtag('js', new Date());
gtag('config', 'UA-59152712-8');
</script>
# NRPy+'s Finite Difference Interface
## Author: Zach Etienne
### Formatting improvements courtesy Brandon Clark
### NRPy+ Source Code for this module: [finite_difference.py](../edit/finite_difference.py)
<a id='toc'></a>
# Table of Contents \[Back to [top](#toc)\]
$$\label{toc}$$
This notebook is organized as follows
1. [Preliminaries](#fdd): Introduction to Finite Difference Derivatives
1. [Step 1](#fdmodule): The finite_difference NRPy+ module
1. [Step 1.a](#fdcoeffs_func): The `compute_fdcoeffs_fdstencl()` function
1. [Step 1.a.i](#exercise): Exercise: Using `compute_fdcoeffs_fdstencl()`
1. [Step 1.b](#fdoutputc): The `FD_outputC()` function
1. [Step 2](#latex_pdf_output): Output this notebook to $\LaTeX$-formatted PDF
<a id='fdd'></a>
# Preliminaries: Introduction to Finite Difference Derivatives \[Back to [top](#toc)\]
$$\label{fdd}$$
Suppose we have a *uniform* numerical grid in one dimension; say, the Cartesian $x$ direction. Since the grid is uniform, the spacing between successive grid points is $\Delta x$, and the position of the $i$th point is given by
$$x_i = x_0 + i \Delta x.$$
Then, given a function $u(x)$ on this uniform grid, we will adopt the notation
$$u(x_i) = u_i.$$
We wish to approximate derivatives of $u_i$ at some nearby point (in this tutorial, we will consider derivatives at one of the sampled points $x_i$) using [finite difference](https://en.wikipedia.org/wiki/Finite_difference). (FD) techniques.
FD techniques are usually constructed as follows:
* First, find the unique $N$th-degree polynomial that passes through $N+1$ sampled points of our function $u$ in the neighborhood of where we wish to find the derivative.
* Then, provided $u$ is smooth and properly-sampled, the $n$th derivative of the polynomial (where $n\le N-1$; *Exercise: Justify this inequality*) is approximately equal to the $n$th derivative of $u$. We call this the **$n$th-order finite difference derivative of $u$**.
* So long as the function $u$ is smooth and properly sampled, the relative error between the exact and the finite difference derivative $u^{(n)}$ will generally decrease as the polynomial degree or sampling density increases.
The $n$th finite difference derivative of $u(x)$ at $x=x_i$ can then be written in the form
$$u^{(n)}(x_i)_{\text{FD}} = \sum_{j=0}^{N} u_j a_j,$$
where the $a_j$'s are known as *finite difference coefficients*. So long as the $N$th-degree polynomial that passes through the $N+1$ points is unique, the corresponding set of $a_j$'s are unique as well.
There are multiple ways to compute the finite difference coefficients $a_j$, including solving for the $N$th-degree polynomial that passes through the function at the sampled points. However, the most popular and most straightforward way involves Taylor series expansions about sampled points near the point where we wish to evaluate the derivative.
**Recommended: Learn more about the algorithm NRPy+ adopts to automatically compute finite difference derivatives: ([How NRPy+ Computes Finite Difference Coefficients](Tutorial-How_NRPy_Computes_Finite_Difference_Coeffs.ipynb))**
<a id='fdmodule'></a>
# Step 1: The finite_difference NRPy+ module \[Back to [top](#toc)\]
$$\label{fdmodule}$$
The finite_difference NRPy+ module contains one parameter:
* **FD_CENTDERIVS_ORDER**: An integer indicating the requested finite difference accuracy order (not the order of the derivative) , where FD_CENTDERIVS_ORDER = [the size of the finite difference stencil in each direction, plus one].
The finite_difference NRPy+ module contains two core functions: `compute_fdcoeffs_fdstencl()` and `FD_outputC()`. The first is a low-level function normally called only by `FD_outputC()`, which computes and outputs finite difference coefficients and the numerical grid indices (stencil) corresponding to each coefficient:
<a id='fdcoeffs_func'></a>
## Step 1.a: The `compute_fdcoeffs_fdstencl()` function \[Back to [top](#toc)\]
$$\label{fdcoeffs_func}$$
**compute_fdcoeffs_fdstencl(derivstring,FDORDER=-1)**:
* Output nonzero finite difference coefficients and corresponding numerical stencil as lists, using as inputs:
* **derivstring**: indicates the precise type and direction derivative desired:
* **Centered derivatives**, where the center of the finite difference stencil corresponds to the point where the derivative is desired:
* For a first-order derivative, set derivstring to "D"+"dirn", where "dirn" is an integer denoting direction. For a second-order derivative, set derivstring to "DD"+"dirn1"+"dirn2", where "dirn1" and "dirn2" are integers denoting the direction of each derivative. Currently only $1 \le N \le 2$ supported (extension to higher-order derivatives is straightforward). Examples in 3D Cartesian coordinates (x,y,z):
* the derivative operator $\partial_x^2$ corresponds to derivstring = "DD00"
* the derivative operator $\partial_x \partial_y$ corresponds to derivstring = "DD01"
* the derivative operator $\partial_z$ corresponds to derivstring = "D2"
* **Up- or downwinded derivatives**, where the center of the finite difference stencil is *one gridpoint* up or down from where the derivative is requested.
* Set derivstring to "upD"+"dirn" or "dnD"+"dirn", where "dirn" is an integer denoting direction. Example in 3D Cartesian coordinates (x,y,z):
* the upwinded derivative operator $\partial_x$ corresponds to derivstring = "dupD0"
* **Kreiss-Oliger dissipation derivatives**, where the center of the finite difference stencil corresponds to the point where the dissipation will be applied.
* Set derivstring to "dKOD"+"dirn", where "dirn" is an integer denoting direction. Example in 3D Cartesian coordinates (x,y,z):
* the Kreiss-Oliger derivative operator $\partial_z^\text{KO}$ corresponds to derivstring = "dKOD2"
* **FDORDER**: an *optional* parameter that, if set to a positive even integer, overrides FD_CENTDERIVS_ORDER
Within NRPy+, `compute_fdcoeffs_fdstencl()` is only called from `FD_outputC()`. Regardless, this function provides a nice interface for evaluating finite difference coefficients, as shown below:
```
# Import the finite difference module
import finite_difference as fin
fdcoeffs, fdstencl = fin.compute_fdcoeffs_fdstencl("dDD00")
print(fdcoeffs)
print(fdstencl)
```
Interpreting the output, notice first that $\texttt{fdstencl}$ is a list of coordinate indices, where up to 4 dimension indices are supported (higher dimensions are possible and can be straightforwardly added, though be warned about [The Curse of Dimensionality](https://en.wikipedia.org/wiki/Curse_of_dimensionality)).
Thus NRPy+ found that for some function $u$, the fourth-order accurate finite difference operator at point $x_{i0}$ is given by
$$[\partial_x u]^\text{FD4}_{i0} = \frac{1}{\Delta x} \left[ \frac{1}{12} \left(u_{i0-2,i1,i2,i3} - u_{i0+2,i1,i2,i3}\right) + \frac{2}{3} \left(-u_{i0-1,i1,i2,i3} + u_{i0+1,i1,i2,i3}\right)\right]$$
Notice also that multiplying by the appropriate power of $\frac{1}{\Delta x}$ term is up to the user of this function.
In addition, if the gridfunction $u$ exists on a grid that is less than four (spatial) dimensions, it is up to the user to truncate the additional index information.
<a id='exercise'></a>
### Step 1.a.i: Exercise: Using `compute_fdcoeffs_fdstencl()` \[Back to [top](#toc)\]
$$\label{exercise}$$
Using `compute_fdcoeffs_fdstencl()` write the necessary loops to output the finite difference coefficient tables in the Wikipedia article on [finite difference coefficients](https://en.wikipedia.org/wiki/Finite_difference_coefficients), for first and second centered derivatives (i.e., up to $\partial_i^2$) up to eighth-order accuracy. [Solution, courtesy Brandon Clark](Tutorial-Finite_Difference_Derivatives-FDtable_soln.ipynb).
<a id='fdoutputc'></a>
## Step 1.b: The `FD_outputC()` function \[Back to [top](#toc)\]
$$\label{fdoutputc}$$
**FD_outputC(filename,sympyexpr_list)**: C code generator for finite-difference expressions.
C codes that evaluate expressions with finite difference derivatives on numerical grids generally consist of three components, all existing within a loop over "interior" gridpoints; at a given gridpoint, the code must
1. Read gridfunctions from memory at all points needed to evaluate the finite difference derivatives or the gridfunctions themselves.
2. Perform arithmetic, including computation of finite difference stencils.
3. Write the output from the arithmetic to other gridfunctions.
To minimize cache misses and maximize potential compiler optimizations, it is generally recommended to segregate the above three steps. FD_outputC() first analyzes the input expressions, searching for derivatives of gridfunctions. The search is very easy, as NRPy+ requires a very specific syntax for derivatives:
* gf_dD0 denotes the first derivative of gridfunction "gf" in direction zero.
* gf_dupD0 denotes the upwinded first derivative of gridfunction "gf" in direction zero.
* gf_ddnD0 denotes the downwinded first derivative of gridfunction "gf" in direction zero.
* gf_dKOD2 denotes the Kreiss-Oliger dissipation operator of gridfunction "gf" in direction two.
Each time `FD_outputC()` finds a derivative (including references to the gridfunction directly \["zeroth"-order derivatives\]) in this way, it calls `compute_fdcoeffs_fdstencl()` to record the specific locations in memory from which the underlying gridfunction must be read to evaluate the appropriate finite difference derivative.
`FD_outputC()` then orders this list of points for all gridfunctions and points in memory, optimizing memory reads based on how the gridfunctions are stored in memory (set via parameter MemAllocStyle in the NRPy+ grid module). It then completes step 1.
For step 2, `FD_outputC()` exports all of the finite difference expressions, as well as the original expressions input into the function, to outputC() to generate the optimized C code. Step 3 follows trivally from just being careful with the bookkeeping in the above steps.
`FD_outputC()` takes two arguments:
* **filename**: Set to "stdout" to print to screen. Otherwise specify a filename.
* **sympyexpr_list**: A single named tuple or list of named tuples of type "lhrh", where the lhrh type refers to the simple structure:
* **lhrh(left-hand side of equation, right-hand side of the equation)**
Time for an example: let's compute
$$
\texttt{output} = \text{phi_dDD00} = \partial_x^2 \phi(x,t),
$$
where $\phi$ is a function of space and time, though we only store its spatial values at a given time (*a la* the [Method of Lines](https://reference.wolfram.com/language/tutorial/NDSolveMethodOfLines.html), described & implemented in next the [Scalar Wave Equation module](Tutorial-Start_to_Finish-ScalarWave.ipynb)).
As detailed above, the suffix $\text{_dDD00}$ tells NRPy+ to construct the second finite difference derivative of gridfunction $\texttt{phi}$ with respect to coordinate $xx0$ (in this case $xx0$ is simply the Cartesian coordinate $x$). Here is the NRPy+ implementation:
```
import sympy as sp
from outputC import *
import grid as gri
import indexedexp as ixp
import finite_difference as fin
# Set the spatial dimension to 1
par.set_paramsvals_value("grid::DIM = 1")
# Register the input gridfunction "phi" and the gridfunction to which data are output, "output":
phi, output = gri.register_gridfunctions("AUX",["phi","output"])
# Declare phi_dDD as a rank-2 indexed expression: phi_dDD[i][j] = \partial_i \partial_j phi
phi_dDD = ixp.declarerank2("phi_dDD","nosym")
# Set output to \partial_0^2 phi
output = phi_dDD[0][0]
# Output to the screen the core C code for evaluating the finite difference derivative
fin.FD_outputC("stdout",lhrh(lhs=gri.gfaccess("out_gf","output"),rhs=output))
```
Some important points about the above code:
* The gridfunction PHIGF samples some function $\phi(x)$ at discrete uniform points in $x$, labeled $x_i$ at all points $i\in [0,N]$, so that
$$\phi(x_i) = \phi_{i}=\text{in_gfs[IDX2(PHIGF, i)]}.$$
* For a *uniformly* sampled function with constant grid spacing (sample rate) $\Delta x$, $x_i$ is defined as $x_i = x_0 + i \Delta x$.
* The variable $\texttt{invdx0}$ must be defined by the user in terms of the uniform gridspacing $\Delta x$ as $\texttt{invdx0} = \frac{1}{\Delta x}$.
* *Aside*: Why do we choose to multiply by $1/\Delta x$ instead of dividing the expression by $\Delta x$, which would seem much more straightforward?
* *Answer*: as discussed in the [first part of the tutorial](Tutorial-Coutput__Parameter_Interface.ipynb), division of floating-point numbers on modern CPUs is far more expensive than multiplication, usually by a factor of ~3 or more.
<a id='latex_pdf_output'></a>
# Step 2: Output this notebook to $\LaTeX$-formatted PDF file \[Back to [top](#toc)\]
$$\label{latex_pdf_output}$$
The following code cell converts this Jupyter notebook into a proper, clickable $\LaTeX$-formatted PDF file. After the cell is successfully run, the generated PDF may be found in the root NRPy+ tutorial directory, with filename
[Tutorial-Finite_Difference_Derivatives.pdf](Tutorial-Finite_Difference_Derivatives.pdf) (Note that clicking on this link may not work; you may need to open the PDF file through another means.)
```
!jupyter nbconvert --to latex --template latex_nrpy_style.tplx --log-level='WARN' Tutorial-Finite_Difference_Derivatives.ipynb
!pdflatex -interaction=batchmode Tutorial-Finite_Difference_Derivatives.tex
!pdflatex -interaction=batchmode Tutorial-Finite_Difference_Derivatives.tex
!pdflatex -interaction=batchmode Tutorial-Finite_Difference_Derivatives.tex
!rm -f Tut*.out Tut*.aux Tut*.log
```
|
github_jupyter
|
# Import the finite difference module
import finite_difference as fin
fdcoeffs, fdstencl = fin.compute_fdcoeffs_fdstencl("dDD00")
print(fdcoeffs)
print(fdstencl)
import sympy as sp
from outputC import *
import grid as gri
import indexedexp as ixp
import finite_difference as fin
# Set the spatial dimension to 1
par.set_paramsvals_value("grid::DIM = 1")
# Register the input gridfunction "phi" and the gridfunction to which data are output, "output":
phi, output = gri.register_gridfunctions("AUX",["phi","output"])
# Declare phi_dDD as a rank-2 indexed expression: phi_dDD[i][j] = \partial_i \partial_j phi
phi_dDD = ixp.declarerank2("phi_dDD","nosym")
# Set output to \partial_0^2 phi
output = phi_dDD[0][0]
# Output to the screen the core C code for evaluating the finite difference derivative
fin.FD_outputC("stdout",lhrh(lhs=gri.gfaccess("out_gf","output"),rhs=output))
!jupyter nbconvert --to latex --template latex_nrpy_style.tplx --log-level='WARN' Tutorial-Finite_Difference_Derivatives.ipynb
!pdflatex -interaction=batchmode Tutorial-Finite_Difference_Derivatives.tex
!pdflatex -interaction=batchmode Tutorial-Finite_Difference_Derivatives.tex
!pdflatex -interaction=batchmode Tutorial-Finite_Difference_Derivatives.tex
!rm -f Tut*.out Tut*.aux Tut*.log
| 0.514156 | 0.89616 |
```
%matplotlib inline
```
==================
Smoothing Contours
==================
Demonstrate how to smooth contour values from a higher resolution
model field.
By: Kevin Goebbert
Date: 13 April 2017
Do the needed imports
```
from datetime import datetime
import cartopy.crs as ccrs
import cartopy.feature as cfeature
import matplotlib.pyplot as plt
import metpy.calc as mpcalc
from metpy.units import units
from netCDF4 import num2date
import numpy as np
import scipy.ndimage as ndimage
from siphon.ncss import NCSS
```
Set up netCDF Subset Service link
```
dt = datetime(2016, 4, 16, 18)
base_url = 'https://www.ncei.noaa.gov/thredds/ncss/grid/namanl/'
ncss = NCSS('{}{dt:%Y%m}/{dt:%Y%m%d}/namanl_218_{dt:%Y%m%d}_'
'{dt:%H}00_000.grb'.format(base_url, dt=dt))
# Data Query
hgt = ncss.query().time(dt)
hgt.variables('Geopotential_height_isobaric', 'u-component_of_wind_isobaric',
'v-component_of_wind_isobaric').add_lonlat()
# Actually getting the data
data = ncss.get_data(hgt)
```
Pull apart the data
```
# Get dimension names to pull appropriate variables
dtime = data.variables['Geopotential_height_isobaric'].dimensions[0]
dlev = data.variables['Geopotential_height_isobaric'].dimensions[1]
dlat = data.variables['Geopotential_height_isobaric'].dimensions[2]
dlon = data.variables['Geopotential_height_isobaric'].dimensions[3]
# Get lat and lon data, as well as time data and metadata
lats = data.variables['lat'][:]
lons = data.variables['lon'][:]
lons[lons > 180] = lons[lons > 180] - 360
# Need 2D lat/lons for plotting, do so if necessary
if lats.ndim < 2:
lons, lats = np.meshgrid(lons, lats)
# Determine the level of 500 hPa
levs = data.variables[dlev][:]
lev_500 = np.where(levs == 500)[0][0]
# Create more useable times for output
times = data.variables[dtime]
vtimes = num2date(times[:], times.units)
# Pull out the 500 hPa Heights
hght = data.variables['Geopotential_height_isobaric'][:].squeeze() * units.meter
uwnd = units('m/s') * data.variables['u-component_of_wind_isobaric'][:].squeeze()
vwnd = units('m/s') * data.variables['v-component_of_wind_isobaric'][:].squeeze()
# Calculate the magnitude of the wind speed in kts
sped = mpcalc.wind_speed(uwnd, vwnd).to('knots')
```
Set up the projection for LCC
```
plotcrs = ccrs.LambertConformal(central_longitude=-100.0, central_latitude=45.0)
datacrs = ccrs.PlateCarree(central_longitude=0.)
```
Subset and smooth
```
# Subset the data arrays to grab only 500 hPa
hght_500 = hght[lev_500]
uwnd_500 = uwnd[lev_500]
vwnd_500 = vwnd[lev_500]
# Smooth the 500-hPa geopotential height field
# Be sure to only smooth the 2D field
Z_500 = ndimage.gaussian_filter(hght_500, sigma=5, order=0)
```
Plot the contours
```
# Start plot with new figure and axis
fig = plt.figure(figsize=(17., 11.))
ax = plt.subplot(1, 1, 1, projection=plotcrs)
# Add some titles to make the plot readable by someone else
plt.title('500-hPa Geo Heights (m; black), Smoothed 500-hPa Geo. Heights (m; red)',
loc='left')
plt.title('VALID: {}'.format(vtimes[0]), loc='right')
# Set GAREA and add map features
ax.set_extent([-125., -67., 22., 52.], ccrs.PlateCarree())
ax.coastlines('50m', edgecolor='black', linewidth=0.75)
ax.add_feature(cfeature.STATES, linewidth=0.5)
# Set the CINT
clev500 = np.arange(5100, 6000, 60)
# Plot smoothed 500-hPa contours
cs2 = ax.contour(lons, lats, Z_500, clev500, colors='red',
linewidths=3, linestyles='solid', transform=datacrs)
c2 = plt.clabel(cs2, fontsize=12, colors='red', inline=1, inline_spacing=8,
fmt='%i', rightside_up=True, use_clabeltext=True)
# Contour the 500 hPa heights with labels
cs = ax.contour(lons, lats, hght_500, clev500, colors='black',
linewidths=2.5, linestyles='solid', alpha=0.6, transform=datacrs)
cl = plt.clabel(cs, fontsize=12, colors='k', inline=1, inline_spacing=8,
fmt='%i', rightside_up=True, use_clabeltext=True)
plt.show()
```
|
github_jupyter
|
%matplotlib inline
from datetime import datetime
import cartopy.crs as ccrs
import cartopy.feature as cfeature
import matplotlib.pyplot as plt
import metpy.calc as mpcalc
from metpy.units import units
from netCDF4 import num2date
import numpy as np
import scipy.ndimage as ndimage
from siphon.ncss import NCSS
dt = datetime(2016, 4, 16, 18)
base_url = 'https://www.ncei.noaa.gov/thredds/ncss/grid/namanl/'
ncss = NCSS('{}{dt:%Y%m}/{dt:%Y%m%d}/namanl_218_{dt:%Y%m%d}_'
'{dt:%H}00_000.grb'.format(base_url, dt=dt))
# Data Query
hgt = ncss.query().time(dt)
hgt.variables('Geopotential_height_isobaric', 'u-component_of_wind_isobaric',
'v-component_of_wind_isobaric').add_lonlat()
# Actually getting the data
data = ncss.get_data(hgt)
# Get dimension names to pull appropriate variables
dtime = data.variables['Geopotential_height_isobaric'].dimensions[0]
dlev = data.variables['Geopotential_height_isobaric'].dimensions[1]
dlat = data.variables['Geopotential_height_isobaric'].dimensions[2]
dlon = data.variables['Geopotential_height_isobaric'].dimensions[3]
# Get lat and lon data, as well as time data and metadata
lats = data.variables['lat'][:]
lons = data.variables['lon'][:]
lons[lons > 180] = lons[lons > 180] - 360
# Need 2D lat/lons for plotting, do so if necessary
if lats.ndim < 2:
lons, lats = np.meshgrid(lons, lats)
# Determine the level of 500 hPa
levs = data.variables[dlev][:]
lev_500 = np.where(levs == 500)[0][0]
# Create more useable times for output
times = data.variables[dtime]
vtimes = num2date(times[:], times.units)
# Pull out the 500 hPa Heights
hght = data.variables['Geopotential_height_isobaric'][:].squeeze() * units.meter
uwnd = units('m/s') * data.variables['u-component_of_wind_isobaric'][:].squeeze()
vwnd = units('m/s') * data.variables['v-component_of_wind_isobaric'][:].squeeze()
# Calculate the magnitude of the wind speed in kts
sped = mpcalc.wind_speed(uwnd, vwnd).to('knots')
plotcrs = ccrs.LambertConformal(central_longitude=-100.0, central_latitude=45.0)
datacrs = ccrs.PlateCarree(central_longitude=0.)
# Subset the data arrays to grab only 500 hPa
hght_500 = hght[lev_500]
uwnd_500 = uwnd[lev_500]
vwnd_500 = vwnd[lev_500]
# Smooth the 500-hPa geopotential height field
# Be sure to only smooth the 2D field
Z_500 = ndimage.gaussian_filter(hght_500, sigma=5, order=0)
# Start plot with new figure and axis
fig = plt.figure(figsize=(17., 11.))
ax = plt.subplot(1, 1, 1, projection=plotcrs)
# Add some titles to make the plot readable by someone else
plt.title('500-hPa Geo Heights (m; black), Smoothed 500-hPa Geo. Heights (m; red)',
loc='left')
plt.title('VALID: {}'.format(vtimes[0]), loc='right')
# Set GAREA and add map features
ax.set_extent([-125., -67., 22., 52.], ccrs.PlateCarree())
ax.coastlines('50m', edgecolor='black', linewidth=0.75)
ax.add_feature(cfeature.STATES, linewidth=0.5)
# Set the CINT
clev500 = np.arange(5100, 6000, 60)
# Plot smoothed 500-hPa contours
cs2 = ax.contour(lons, lats, Z_500, clev500, colors='red',
linewidths=3, linestyles='solid', transform=datacrs)
c2 = plt.clabel(cs2, fontsize=12, colors='red', inline=1, inline_spacing=8,
fmt='%i', rightside_up=True, use_clabeltext=True)
# Contour the 500 hPa heights with labels
cs = ax.contour(lons, lats, hght_500, clev500, colors='black',
linewidths=2.5, linestyles='solid', alpha=0.6, transform=datacrs)
cl = plt.clabel(cs, fontsize=12, colors='k', inline=1, inline_spacing=8,
fmt='%i', rightside_up=True, use_clabeltext=True)
plt.show()
| 0.764892 | 0.860955 |
```
import pandas as pd
import numpy as np
from sklearn.linear_model import Ridge
from sklearn.preprocessing import StandardScaler, OneHotEncoder, LabelEncoder
from sklearn.metrics import mean_squared_error
from sklearn.model_selection import RandomizedSearchCV
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.pipeline import Pipeline, FeatureUnion
import matplotlib.pyplot as plt
import seaborn as sbr
sbr.set(font_scale=1.2)
%matplotlib inline
```
### Helpers
```
def make_submission(model, file_index):
"""Create submission for publick score
Args:
model: The trained model
file_index (str): The name of the file
"""
test_preds = model.predict(prepared_test)
sample_submission = pd.read_csv('submissions/sample_submission.csv', index_col='url')
new_sumb = sample_submission.copy()
new_sumb['favs_lognorm'] = test_preds
new_sumb.to_csv('submissions/submission_{}.csv'.format(file_index))
print('submission_{}.csv successfully created'.format(file_index))
def estimate_model(model):
"""Evaluate model for the training and cross-validation datasets
Args:
model : The trained model
"""
print('Train error:', mean_squared_error(y_train, model.predict(X_train)))
print('Test error:', mean_squared_error(y_test, model.predict(X_test)))
# http://scikit-learn.org/stable/auto_examples/hetero_feature_union.html
from sklearn.base import BaseEstimator, TransformerMixin
class ItemSelector(BaseEstimator, TransformerMixin):
"""For data grouped by feature, select subset of data at a provided key.
The data is expected to be stored in a 2D data structure, where the first
index is over features and the second is over samples. i.e.
>> len(data[key]) == n_samples
Please note that this is the opposite convention to scikit-learn feature
matrixes (where the first index corresponds to sample).
ItemSelector only requires that the collection implement getitem
(data[key]). Examples include: a dict of lists, 2D numpy array, Pandas
DataFrame, numpy record array, etc.
>> data = {'a': [1, 5, 2, 5, 2, 8],
'b': [9, 4, 1, 4, 1, 3]}
>> ds = ItemSelector(key='a')
>> data['a'] == ds.transform(data)
ItemSelector is not designed to handle data grouped by sample. (e.g. a
list of dicts). If your data is structured this way, consider a
transformer along the lines of `sklearn.feature_extraction.DictVectorizer`.
Parameters
----------
key : hashable, required
The key corresponding to the desired value in a mappable.
"""
def __init__(self, key):
self.key = key
def fit(self, x, y=None):
return self
def transform(self, data_dict):
return data_dict[self.key]
```
### Load data
```
train_df = pd.read_csv('data/howpop_train.csv')
test_df = pd.read_csv('data/howpop_test.csv')
```
### Analyzation of the raw data
```
train_df.info()
train_df.describe().T
```
#### Some authors are missing
```
# train
print(train_df[train_df.author.isnull()].shape[0])
print(train_df.shape[0])
# testing
print(test_df[test_df.author.isnull()].shape[0])
print(test_df.shape[0])
# empty 'autor' means the company blog
train_df[train_df.author.isnull()].head(1)
```
#### Domain
```
plt.figure(figsize=(8, 5))
train_df.groupby('domain').favs.median().plot.bar()
plt.title('Median of the #favs by domain')
plt.xticks(rotation='horizontal');
```
#### Flow
```
plt.figure(figsize=(8, 5))
train_df.flow.value_counts().plot.bar();
plt.title('Number of #hab by flow')
plt.xticks(rotation='horizontal');
# mostly habs are from the developer flow
plt.figure(figsize=(8, 5))
train_df.groupby('flow').favs.median().plot.bar()
plt.title('Median of the #favs by flow');
plt.xticks(rotation='horizontal');
```
#### Polling
```
plt.figure(figsize=(8, 5))
train_df.polling.value_counts().plot.bar();
plt.title('Number of #hab by polling')
plt.xticks(rotation='horizontal');
# seems unuseful feature
```
#### Comments
```
plt.figure(figsize=(8, 5))
plt.hist(train_df.comments_lognorm);
plt.title('The distribution of the property comments_lognorm');
plt.figure(figsize=(8, 5))
plt.scatter(train_df.favs_lognorm, train_df.comments);
plt.title('The distribution of the favs_lognorm and number of the comments');
# outliers?
```
#### Views
```
plt.figure(figsize=(8, 5))
plt.hist(train_df.views_lognorm);
plt.title('The distribution of the property views_lognorm');
plt.figure(figsize=(8, 5))
plt.scatter(train_df.favs_lognorm, train_df.views);
plt.title('The distribution of the favs_lognorm and number of the views');
# outliers?
```
#### favs_lognorm
```
plt.figure(figsize=(8, 5))
plt.hist(train_df.favs_lognorm);
plt.title('The distribution of the property favs_lognorm');
train_df[['comments', 'favs', 'views', 'views_lognorm', 'comments_lognorm', 'favs_lognorm']].corr()
plt.figure(figsize=(8, 5))
sbr.heatmap(train_df[['comments', 'favs', 'views', 'views_lognorm', 'comments_lognorm', 'favs_lognorm']].corr());
```
### Feature engineering
#### Encoding for years and monthes
```
full_df = pd.concat([train_df, test_df])
years = full_df.published.apply(lambda p: pd.to_datetime(p).year).unique()
year_encoder = OneHotEncoder()
year_encoder.fit(years.reshape(-1, 1));
months = train_df.published.apply(lambda p: pd.to_datetime(p).month).unique()
month_encoder = OneHotEncoder()
month_encoder.fit(months.reshape(-1, 1));
def encode_column(df, encoder, column_name):
encoding_column = encoder.transform(df[column_name].values.reshape(-1, 1))
encoded_df = pd.DataFrame(
index=df.index,
columns=[column_name + '_' + str(y) for y in encoder.active_features_],
data=encoding_column.toarray()
)
return encoded_df
def is_weekend(weekday):
return weekday == 5 or weekday == 6
def fill_in_author(urls):
'''Get name of the company from the url'''
return urls.str.rsplit('/', expand=True)[4]
def create_feature(df):
df['domain'] = LabelEncoder().fit_transform(df.domain)
# get len of title
df['title_len'] = df.title.apply(lambda t: len(t))
# scale for length for title and content
scaler = StandardScaler()
df[['title_len', 'content_len']] = scaler.fit_transform(df[['title_len', 'content_len']])
# fill in author for companies
df.author.fillna(fill_in_author(df[df.author.isnull()].url), inplace=True)
# check is it commpany
df['is_company'] = df.url.str.contains('company')
df['is_company'] = df.is_company.astype(int)
# get features from published day
df['hour'] = df.published.apply(lambda p: pd.to_datetime(p).hour)
df['weekday'] = df.published.apply(lambda p: pd.to_datetime(p).weekday())
df['month'] = df.published.apply(lambda p: pd.to_datetime(p).month)
df['year'] = df.published.apply(lambda p: pd.to_datetime(p).year)
# check is published day is weekend
df['is_weekend'] = df.weekday.apply(lambda w: is_weekend(w))
df['is_weekend'] = df.is_weekend.astype(int)
# one hot encoding for the categorical features
dummy_cols = ['hour', 'weekday', 'flow']
dummy_df = pd.get_dummies(
data=df[dummy_cols], columns=dummy_cols, prefix=dummy_cols
)
df = pd.concat([df, dummy_df], axis=1)
del dummy_df
year_encode_df = encode_column(df, year_encoder, 'year')
df = pd.concat([df, year_encode_df], axis=1)
del year_encode_df
months_encode_df = encode_column(df, month_encoder, 'month')
df = pd.concat([df, months_encode_df], axis=1)
del months_encode_df
return df
%%time
prepared_train = create_feature(train_df)
# prepared_train.to_csv('data/prepared_train.csv', index=False)
%%time
prepared_test = create_feature(test_df)
# prepared_test.to_csv('data/prepared_test.csv', index=False)
```
### Analyze new features
```
plt.figure(figsize=(8, 5))
sbr.heatmap(train_df[['is_company', 'is_weekend', 'title_len', 'content_len', 'favs_lognorm']].corr());
print(train_df[['is_company', 'is_weekend', 'title_len', 'content_len', 'favs_lognorm']].corr())
```
#### Published
```
plt.figure(figsize=(12, 8))
plt.suptitle('Total number of the #favs for hour, weekday, month, year', fontsize=14)
plt.subplot(221)
prepared_train.groupby('year').favs.sum().plot.bar();
plt.xticks(rotation='horizontal');
plt.subplot(222)
prepared_train.groupby('month').favs.sum().plot.bar();
plt.xticks(rotation='horizontal');
plt.subplot(223)
prepared_train.groupby('weekday').favs.sum().plot.bar();
plt.xticks(rotation='horizontal');
plt.subplot(224)
prepared_train.groupby('hour').favs.sum().plot.bar();
plt.xticks(rotation='horizontal');
plt.figure(figsize=(12, 8))
plt.suptitle('The median of the #favs for hour, weekday, month, year', fontsize=14)
plt.subplot(221)
prepared_train.groupby('year').favs.max().plot.bar();
plt.xticks(rotation='horizontal');
plt.subplot(222)
prepared_train.groupby('month').favs.max().plot.bar();
plt.xticks(rotation='horizontal');
plt.subplot(223)
prepared_train.groupby('weekday').favs.max().plot.bar();
plt.xticks(rotation='horizontal');
plt.subplot(224)
prepared_train.groupby('hour').favs.max().plot.bar();
plt.xticks(rotation='horizontal');
```
#### Company / author
```
plt.figure(figsize=(8, 5))
plt.title('Max value of the #favs by property is_company')
prepared_train.groupby('is_company').favs.max().plot.bar();
plt.xticks(rotation='horizontal');
```
### Get rid from outliers
#### Number of the views
```
prepared_train = prepared_train[prepared_train.views < 600000]
plt.figure(figsize=(8, 5))
plt.scatter(prepared_train.favs_lognorm, prepared_train.views);
plt.title('The distribution of the favs_lognorm and number of the views without outliers');
```
#### Number of the comments
```
prepared_train = prepared_train[prepared_train.comments < 1100]
plt.figure(figsize=(8, 5))
plt.scatter(prepared_train.favs_lognorm, prepared_train.comments);
plt.title('The distribution of the favs_lognorm and number of the views without outliers');
print('{} outliers were removed from the training dataset'.format(train_df.shape[0] - prepared_train.shape[0]))
```
### Split data for train and test
We need predict historical data. That's why we'll take for testing last 7 months from the training dataset
```
X_train = prepared_train[prepared_train.published <= '2016-04-01 00:00:00']
X_test = prepared_train[prepared_train.published > '2016-04-01 00:00:00']
y_train, y_test = X_train.favs_lognorm, X_test.favs_lognorm
```
### Build pipline
```
using_columns = np.concatenate([
[
'domain', 'is_weekend', 'is_company',
'content_len', 'title_len',
'title',
],
[col for col in prepared_train.columns if col.startswith('flow_')],
[col for col in prepared_train.columns if col.startswith('hour_')],
[col for col in prepared_train.columns if col.startswith('weekday_')],
[col for col in prepared_train.columns if col.startswith('month_')],
[col for col in prepared_train.columns if col.startswith('year_')],
])
pipline_columns = list(using_columns)
pipline_columns.remove('title')
estimator = Pipeline([(
'feature_processing', FeatureUnion(transformer_list=[
('titles_processing', Pipeline([
('selecting', ItemSelector(key='title')),
('tfidf', TfidfVectorizer()),
])),
('characters_processing', Pipeline([
('selecting', ItemSelector(key='title')),
('tfidf', TfidfVectorizer(analyzer='char')),
])),
('all_feauters', Pipeline([
('selecting', ItemSelector(key=pipline_columns))
])),
])
),
('model_fitting', Ridge(random_state=42))
])
estimator.set_params(
model_fitting__alpha=1,
feature_processing__characters_processing__tfidf__max_features=2000,
feature_processing__characters_processing__tfidf__min_df=5,
feature_processing__characters_processing__tfidf__ngram_range=(1, 3),
feature_processing__titles_processing__tfidf__max_features=18000,
feature_processing__titles_processing__tfidf__min_df=3,
feature_processing__titles_processing__tfidf__ngram_range=(1, 3)
);
%%time
estimator.fit(X_train, y_train)
# Public score value is 0.62294
estimate_model(estimator)
```
#### Create submission
```
estimator.fit(prepared_train, prepared_train.favs_lognorm);
make_submission(estimator, 'final')
```
|
github_jupyter
|
import pandas as pd
import numpy as np
from sklearn.linear_model import Ridge
from sklearn.preprocessing import StandardScaler, OneHotEncoder, LabelEncoder
from sklearn.metrics import mean_squared_error
from sklearn.model_selection import RandomizedSearchCV
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.pipeline import Pipeline, FeatureUnion
import matplotlib.pyplot as plt
import seaborn as sbr
sbr.set(font_scale=1.2)
%matplotlib inline
def make_submission(model, file_index):
"""Create submission for publick score
Args:
model: The trained model
file_index (str): The name of the file
"""
test_preds = model.predict(prepared_test)
sample_submission = pd.read_csv('submissions/sample_submission.csv', index_col='url')
new_sumb = sample_submission.copy()
new_sumb['favs_lognorm'] = test_preds
new_sumb.to_csv('submissions/submission_{}.csv'.format(file_index))
print('submission_{}.csv successfully created'.format(file_index))
def estimate_model(model):
"""Evaluate model for the training and cross-validation datasets
Args:
model : The trained model
"""
print('Train error:', mean_squared_error(y_train, model.predict(X_train)))
print('Test error:', mean_squared_error(y_test, model.predict(X_test)))
# http://scikit-learn.org/stable/auto_examples/hetero_feature_union.html
from sklearn.base import BaseEstimator, TransformerMixin
class ItemSelector(BaseEstimator, TransformerMixin):
"""For data grouped by feature, select subset of data at a provided key.
The data is expected to be stored in a 2D data structure, where the first
index is over features and the second is over samples. i.e.
>> len(data[key]) == n_samples
Please note that this is the opposite convention to scikit-learn feature
matrixes (where the first index corresponds to sample).
ItemSelector only requires that the collection implement getitem
(data[key]). Examples include: a dict of lists, 2D numpy array, Pandas
DataFrame, numpy record array, etc.
>> data = {'a': [1, 5, 2, 5, 2, 8],
'b': [9, 4, 1, 4, 1, 3]}
>> ds = ItemSelector(key='a')
>> data['a'] == ds.transform(data)
ItemSelector is not designed to handle data grouped by sample. (e.g. a
list of dicts). If your data is structured this way, consider a
transformer along the lines of `sklearn.feature_extraction.DictVectorizer`.
Parameters
----------
key : hashable, required
The key corresponding to the desired value in a mappable.
"""
def __init__(self, key):
self.key = key
def fit(self, x, y=None):
return self
def transform(self, data_dict):
return data_dict[self.key]
train_df = pd.read_csv('data/howpop_train.csv')
test_df = pd.read_csv('data/howpop_test.csv')
train_df.info()
train_df.describe().T
# train
print(train_df[train_df.author.isnull()].shape[0])
print(train_df.shape[0])
# testing
print(test_df[test_df.author.isnull()].shape[0])
print(test_df.shape[0])
# empty 'autor' means the company blog
train_df[train_df.author.isnull()].head(1)
plt.figure(figsize=(8, 5))
train_df.groupby('domain').favs.median().plot.bar()
plt.title('Median of the #favs by domain')
plt.xticks(rotation='horizontal');
plt.figure(figsize=(8, 5))
train_df.flow.value_counts().plot.bar();
plt.title('Number of #hab by flow')
plt.xticks(rotation='horizontal');
# mostly habs are from the developer flow
plt.figure(figsize=(8, 5))
train_df.groupby('flow').favs.median().plot.bar()
plt.title('Median of the #favs by flow');
plt.xticks(rotation='horizontal');
plt.figure(figsize=(8, 5))
train_df.polling.value_counts().plot.bar();
plt.title('Number of #hab by polling')
plt.xticks(rotation='horizontal');
# seems unuseful feature
plt.figure(figsize=(8, 5))
plt.hist(train_df.comments_lognorm);
plt.title('The distribution of the property comments_lognorm');
plt.figure(figsize=(8, 5))
plt.scatter(train_df.favs_lognorm, train_df.comments);
plt.title('The distribution of the favs_lognorm and number of the comments');
# outliers?
plt.figure(figsize=(8, 5))
plt.hist(train_df.views_lognorm);
plt.title('The distribution of the property views_lognorm');
plt.figure(figsize=(8, 5))
plt.scatter(train_df.favs_lognorm, train_df.views);
plt.title('The distribution of the favs_lognorm and number of the views');
# outliers?
plt.figure(figsize=(8, 5))
plt.hist(train_df.favs_lognorm);
plt.title('The distribution of the property favs_lognorm');
train_df[['comments', 'favs', 'views', 'views_lognorm', 'comments_lognorm', 'favs_lognorm']].corr()
plt.figure(figsize=(8, 5))
sbr.heatmap(train_df[['comments', 'favs', 'views', 'views_lognorm', 'comments_lognorm', 'favs_lognorm']].corr());
full_df = pd.concat([train_df, test_df])
years = full_df.published.apply(lambda p: pd.to_datetime(p).year).unique()
year_encoder = OneHotEncoder()
year_encoder.fit(years.reshape(-1, 1));
months = train_df.published.apply(lambda p: pd.to_datetime(p).month).unique()
month_encoder = OneHotEncoder()
month_encoder.fit(months.reshape(-1, 1));
def encode_column(df, encoder, column_name):
encoding_column = encoder.transform(df[column_name].values.reshape(-1, 1))
encoded_df = pd.DataFrame(
index=df.index,
columns=[column_name + '_' + str(y) for y in encoder.active_features_],
data=encoding_column.toarray()
)
return encoded_df
def is_weekend(weekday):
return weekday == 5 or weekday == 6
def fill_in_author(urls):
'''Get name of the company from the url'''
return urls.str.rsplit('/', expand=True)[4]
def create_feature(df):
df['domain'] = LabelEncoder().fit_transform(df.domain)
# get len of title
df['title_len'] = df.title.apply(lambda t: len(t))
# scale for length for title and content
scaler = StandardScaler()
df[['title_len', 'content_len']] = scaler.fit_transform(df[['title_len', 'content_len']])
# fill in author for companies
df.author.fillna(fill_in_author(df[df.author.isnull()].url), inplace=True)
# check is it commpany
df['is_company'] = df.url.str.contains('company')
df['is_company'] = df.is_company.astype(int)
# get features from published day
df['hour'] = df.published.apply(lambda p: pd.to_datetime(p).hour)
df['weekday'] = df.published.apply(lambda p: pd.to_datetime(p).weekday())
df['month'] = df.published.apply(lambda p: pd.to_datetime(p).month)
df['year'] = df.published.apply(lambda p: pd.to_datetime(p).year)
# check is published day is weekend
df['is_weekend'] = df.weekday.apply(lambda w: is_weekend(w))
df['is_weekend'] = df.is_weekend.astype(int)
# one hot encoding for the categorical features
dummy_cols = ['hour', 'weekday', 'flow']
dummy_df = pd.get_dummies(
data=df[dummy_cols], columns=dummy_cols, prefix=dummy_cols
)
df = pd.concat([df, dummy_df], axis=1)
del dummy_df
year_encode_df = encode_column(df, year_encoder, 'year')
df = pd.concat([df, year_encode_df], axis=1)
del year_encode_df
months_encode_df = encode_column(df, month_encoder, 'month')
df = pd.concat([df, months_encode_df], axis=1)
del months_encode_df
return df
%%time
prepared_train = create_feature(train_df)
# prepared_train.to_csv('data/prepared_train.csv', index=False)
%%time
prepared_test = create_feature(test_df)
# prepared_test.to_csv('data/prepared_test.csv', index=False)
plt.figure(figsize=(8, 5))
sbr.heatmap(train_df[['is_company', 'is_weekend', 'title_len', 'content_len', 'favs_lognorm']].corr());
print(train_df[['is_company', 'is_weekend', 'title_len', 'content_len', 'favs_lognorm']].corr())
plt.figure(figsize=(12, 8))
plt.suptitle('Total number of the #favs for hour, weekday, month, year', fontsize=14)
plt.subplot(221)
prepared_train.groupby('year').favs.sum().plot.bar();
plt.xticks(rotation='horizontal');
plt.subplot(222)
prepared_train.groupby('month').favs.sum().plot.bar();
plt.xticks(rotation='horizontal');
plt.subplot(223)
prepared_train.groupby('weekday').favs.sum().plot.bar();
plt.xticks(rotation='horizontal');
plt.subplot(224)
prepared_train.groupby('hour').favs.sum().plot.bar();
plt.xticks(rotation='horizontal');
plt.figure(figsize=(12, 8))
plt.suptitle('The median of the #favs for hour, weekday, month, year', fontsize=14)
plt.subplot(221)
prepared_train.groupby('year').favs.max().plot.bar();
plt.xticks(rotation='horizontal');
plt.subplot(222)
prepared_train.groupby('month').favs.max().plot.bar();
plt.xticks(rotation='horizontal');
plt.subplot(223)
prepared_train.groupby('weekday').favs.max().plot.bar();
plt.xticks(rotation='horizontal');
plt.subplot(224)
prepared_train.groupby('hour').favs.max().plot.bar();
plt.xticks(rotation='horizontal');
plt.figure(figsize=(8, 5))
plt.title('Max value of the #favs by property is_company')
prepared_train.groupby('is_company').favs.max().plot.bar();
plt.xticks(rotation='horizontal');
prepared_train = prepared_train[prepared_train.views < 600000]
plt.figure(figsize=(8, 5))
plt.scatter(prepared_train.favs_lognorm, prepared_train.views);
plt.title('The distribution of the favs_lognorm and number of the views without outliers');
prepared_train = prepared_train[prepared_train.comments < 1100]
plt.figure(figsize=(8, 5))
plt.scatter(prepared_train.favs_lognorm, prepared_train.comments);
plt.title('The distribution of the favs_lognorm and number of the views without outliers');
print('{} outliers were removed from the training dataset'.format(train_df.shape[0] - prepared_train.shape[0]))
X_train = prepared_train[prepared_train.published <= '2016-04-01 00:00:00']
X_test = prepared_train[prepared_train.published > '2016-04-01 00:00:00']
y_train, y_test = X_train.favs_lognorm, X_test.favs_lognorm
using_columns = np.concatenate([
[
'domain', 'is_weekend', 'is_company',
'content_len', 'title_len',
'title',
],
[col for col in prepared_train.columns if col.startswith('flow_')],
[col for col in prepared_train.columns if col.startswith('hour_')],
[col for col in prepared_train.columns if col.startswith('weekday_')],
[col for col in prepared_train.columns if col.startswith('month_')],
[col for col in prepared_train.columns if col.startswith('year_')],
])
pipline_columns = list(using_columns)
pipline_columns.remove('title')
estimator = Pipeline([(
'feature_processing', FeatureUnion(transformer_list=[
('titles_processing', Pipeline([
('selecting', ItemSelector(key='title')),
('tfidf', TfidfVectorizer()),
])),
('characters_processing', Pipeline([
('selecting', ItemSelector(key='title')),
('tfidf', TfidfVectorizer(analyzer='char')),
])),
('all_feauters', Pipeline([
('selecting', ItemSelector(key=pipline_columns))
])),
])
),
('model_fitting', Ridge(random_state=42))
])
estimator.set_params(
model_fitting__alpha=1,
feature_processing__characters_processing__tfidf__max_features=2000,
feature_processing__characters_processing__tfidf__min_df=5,
feature_processing__characters_processing__tfidf__ngram_range=(1, 3),
feature_processing__titles_processing__tfidf__max_features=18000,
feature_processing__titles_processing__tfidf__min_df=3,
feature_processing__titles_processing__tfidf__ngram_range=(1, 3)
);
%%time
estimator.fit(X_train, y_train)
# Public score value is 0.62294
estimate_model(estimator)
estimator.fit(prepared_train, prepared_train.favs_lognorm);
make_submission(estimator, 'final')
| 0.839142 | 0.816113 |
<h1> Datadriver for DataScientists </h1>
_Execute the following cell in order to make the table of contents appear_
```
%%javascript
$.getScript('https://kmahelona.github.io/ipython_notebook_goodies/ipython_µnotebook_toc.js')
```
In this notebook, we will go over the main concepts of the datadriver API, which will enable you to push your exploration code to production faster than ever.
<h2 id="tocheading">Table of Contents</h2>
<div id="toc"></div>
# Context
First you need to create a context. The context is an object which will allow you to communicate with your environment during your exploration. As such, it needs to be able to communicate with your database. This is done by creating a DB object and passing it to the context constructor. Here is how it's done :
```
from dd import DB
from dd.api.contexts import LocalContext
db = DB(dbtype='sqlite', filename=':memory:')
context = LocalContext(db)
```
We will add some more options in order to make sure this tutorial executes properly. You don't need to understand this line right now, as we will cover it in a later tutorial.
```
context.set_default_write_options(if_exists="replace", index=False)
```
There you go. Now your context is set up. Time to load some data and start playing !
# Import data
The dd library comes with some data package in it. We can access the files thanks to the pkg_resources from the standard library :
```
titanic_datapath = 'https://raw.githubusercontent.com/justmarkham/DAT8/master/data/titanic.csv'
```
Now, let's use the context to load the file.
```
train = context.load_file(titanic_datapath,
table_name="titanic.train")
```
The returned object is what is called a dataset. It is the main abstraction you will use to transform and save you data.
```
type(train)
```
# Datasets
You may consider datasets as wrappers around Pandas DataFrames. It gives you access to some methods you may recognise if you are familiar with this awesome library.
```
train.shape
train.columns
train.head()
```
However, datasets are NOT dataframes:
```
#train.loc[:10, u'survived']
```
But you can access the underlying dataframe by calling _collect()_ on the dataset :
```
dataframe = train.collect()
dataframe.loc[:10, u'survived']
```
# Transformations
Transformations are the easiest way to transform your data. And the easiest way to create a transformation is by using the transform method.
## Transform()
By calling _transform()_ on a dataset, you apply a function to it and are returned a new object that wraps the new data.
For example, let's say you need to remove all missing value from the previous dataset. You may define the following function :
```
def fillna_with_zeros(dataframe):
"""
Returns a copy of the dataframe with null values replaced by zeros.
"""
return dataframe.fillna(0)
```
As you may have noticed, your function takes a dataframe as input, and returns a new dataframe. This is very important, because it gives you access to the full Pandas DataFrame power. It also forces you to keep your data in DataFrames.
In order to apply this function to your dataset, you would then write :
```
filled_with_zeros = train.transform(fillna_with_zeros)
```
Easy Peasy ! Let's look at the new data :
```
filled_with_zeros.head()
```
Great ! The cabin column (and all the others) are now non-null.
In case you want your transformation function to be a bit more generic, with more parameters, you may proceed like this :
```
def fillna(dataframe, value):
"""
Returns a copy of the dataframe with null values replaced by {value}.
"""
return dataframe.fillna(value=value)
filled_with_ones = train.transform(fillna, value=1)
filled_with_ones.head()
```
## MutiTransform()
With multitransform, a python function can output multiple datasets.
In this case, output_tables must be specified as a string list.
```
def split_survived(dataframe):
return dataframe[dataframe.survived==0], dataframe[dataframe.survived==1]
surv0, surv1 = filled_with_ones.multitransform(split_survived, output_tables=["surv0", "surv1"])
surv0.head()
surv1.head()
```
## More transformations
### select_columns
allows you to restrict a dataframe to a subset of it's columns :
```
some_columns = filled_with_zeros.select_columns(["passengerid", "survived", "pclass", "age", "sibsp", "parch", "fare"],
write_options=dict(if_exists="replace", index=False))
some_columns.head()
```
You may do the same thing, but with less control over the options given to the method, with the [bracket notation]
```
some_other_columns = filled_with_zeros[["passengerid", "name", "sex", "ticket"]]
some_other_columns.head()
```
### join
joins two datasets :
```
some_columns.join(some_other_columns).head()
```
### split\_train\_test
splits a dataset in TWO new disjoint datasets.
```
Xtrain, Xtest = some_columns.split_train_test(train_size=0.75)
Xtrain.head()
Xtest.head()
```
## Lazy operations
Note that all the operations described above are __lazy__, which means they are not executed until explicitely required to do so. The concept of _actions_ and _transformations_ are thus similar to spark. _Transformation_ are lazy, while _actions_ require the execution to be launched.
```
def dummify(dataframe):
"""
Returns a one-hot encoded version of a dataframe
"""
import pandas as pd
return pd.get_dummies(dataframe)
dummified = filled_with_zeros[["sex"]].transform(dummify)
dummified.memory_usage
dummified.head()
dummified.memory_usage
```
As you can see, the memory taken by the dataset before the execution (launched by the action _head()_) is much smaller than the memory taken by the dataset after the execution. This is because nothing is computed before the execution, AND because the result is cached in the object after the data has been computed.
# Models
Models are objects that need to be trained before they can be applied to a new set of data. You may create a Model through the context from any object that implements a fit and a predict or a transform method (as all scikit-learn models do). Let's look at how you must proceed :
```
# Importing scikit-learn model class
from sklearn.ensemble import RandomForestClassifier
# Instantiating Scikit model
scikit_model = RandomForestClassifier(max_depth=4, n_jobs=-1)
# Creating Datadriver Model
model = context.model(scikit_model, model_address="model@foo.bar")
```
The model_address keyword is used to store the model in database to be retrieve later. The correct address format is {identifier}@{schema_name}.{table_name}
Now that we have a model, we will want to train it on our carefully crafted training dataset :
```
fitted_model = model.fit(Xtrain, target="survived")
```
With this fitted model (or soon to be fitted, remember, all of this is lazy !), we are able to make predictions on our test dataset :
```
predictions = fitted_model.predict(Xtest, target="survived")
```
In both cases, notice that we included the target in the input dataset. It is used in the fit method to train the model, and the column is dropped in the predict method.
```
predictions.head()
```
|
github_jupyter
|
%%javascript
$.getScript('https://kmahelona.github.io/ipython_notebook_goodies/ipython_µnotebook_toc.js')
from dd import DB
from dd.api.contexts import LocalContext
db = DB(dbtype='sqlite', filename=':memory:')
context = LocalContext(db)
context.set_default_write_options(if_exists="replace", index=False)
titanic_datapath = 'https://raw.githubusercontent.com/justmarkham/DAT8/master/data/titanic.csv'
train = context.load_file(titanic_datapath,
table_name="titanic.train")
type(train)
train.shape
train.columns
train.head()
#train.loc[:10, u'survived']
dataframe = train.collect()
dataframe.loc[:10, u'survived']
def fillna_with_zeros(dataframe):
"""
Returns a copy of the dataframe with null values replaced by zeros.
"""
return dataframe.fillna(0)
filled_with_zeros = train.transform(fillna_with_zeros)
filled_with_zeros.head()
def fillna(dataframe, value):
"""
Returns a copy of the dataframe with null values replaced by {value}.
"""
return dataframe.fillna(value=value)
filled_with_ones = train.transform(fillna, value=1)
filled_with_ones.head()
def split_survived(dataframe):
return dataframe[dataframe.survived==0], dataframe[dataframe.survived==1]
surv0, surv1 = filled_with_ones.multitransform(split_survived, output_tables=["surv0", "surv1"])
surv0.head()
surv1.head()
some_columns = filled_with_zeros.select_columns(["passengerid", "survived", "pclass", "age", "sibsp", "parch", "fare"],
write_options=dict(if_exists="replace", index=False))
some_columns.head()
some_other_columns = filled_with_zeros[["passengerid", "name", "sex", "ticket"]]
some_other_columns.head()
some_columns.join(some_other_columns).head()
Xtrain, Xtest = some_columns.split_train_test(train_size=0.75)
Xtrain.head()
Xtest.head()
def dummify(dataframe):
"""
Returns a one-hot encoded version of a dataframe
"""
import pandas as pd
return pd.get_dummies(dataframe)
dummified = filled_with_zeros[["sex"]].transform(dummify)
dummified.memory_usage
dummified.head()
dummified.memory_usage
# Importing scikit-learn model class
from sklearn.ensemble import RandomForestClassifier
# Instantiating Scikit model
scikit_model = RandomForestClassifier(max_depth=4, n_jobs=-1)
# Creating Datadriver Model
model = context.model(scikit_model, model_address="model@foo.bar")
fitted_model = model.fit(Xtrain, target="survived")
predictions = fitted_model.predict(Xtest, target="survived")
predictions.head()
| 0.549157 | 0.958265 |
# Train
```
from mmcv import Config
from mmdet.datasets import build_dataset
from mmdet.models import build_detector
from mmdet.apis import train_detector
from mmdet.datasets import (build_dataloader, build_dataset,
replace_ImageToTensor)
classes = ("UNKNOWN", "General trash", "Paper", "Paper pack", "Metal", "Glass",
"Plastic", "Styrofoam", "Plastic bag", "Battery", "Clothing")
# config file 들고오기
cfg = Config.fromfile('./configs/pafpn/faster_rcnn_r50_pafpn_1x_coco.py')
PREFIX = '../../input/data/'
epoch = 12
# dataset 바꾸기
cfg.data.train.classes = classes
cfg.data.train.img_prefix = PREFIX
cfg.data.train.ann_file = PREFIX + 'train.json'
cfg.data.train.pipeline[2]['img_scale'] = (512, 512)
cfg.data.val.classes = classes
cfg.data.val.img_prefix = PREFIX
cfg.data.val.ann_file = PREFIX + 'val.json'
cfg.data.val.pipeline[1]['img_scale'] = (512, 512)
cfg.data.test.classes = classes
cfg.data.test.img_prefix = PREFIX
cfg.data.test.ann_file = PREFIX + 'test.json'
cfg.data.test.pipeline[1]['img_scale'] = (512, 512)
cfg.data.samples_per_gpu = 4
cfg.seed=2020
cfg.gpu_ids = [0]
cfg.work_dir = './work_dirs/faster_rcnn_r50_pafpn_1x_trash'
cfg.model.roi_head.bbox_head.num_classes = 11
print(cfg.model)
cfg.optimizer_config.grad_clip = dict(max_norm=35, norm_type=2)
model = build_detector(cfg.model)
datasets = [build_dataset(cfg.data.train)]
train_detector(model, datasets[0], cfg, distributed=False, validate=True)
```
# Test
```
import mmcv
from mmcv import Config
from mmdet.datasets import (build_dataloader, build_dataset,
replace_ImageToTensor)
from mmdet.models import build_detector
from mmdet.apis import single_gpu_test
from mmcv.runner import load_checkpoint
import os
from mmcv.parallel import MMDataParallel
import pandas as pd
from pandas import DataFrame
from pycocotools.coco import COCO
import numpy as np
cfg.model.train_cfg = None
checkpoint_path = os.path.join(cfg.work_dir, f'epoch_{epoch}.pth')
dataset = build_dataset(cfg.data.test)
data_loader = build_dataloader(
dataset,
samples_per_gpu=1,
workers_per_gpu=cfg.data.workers_per_gpu,
dist=False,
shuffle=False)
model = build_detector(cfg.model, test_cfg=cfg.get('test_cfg'))
checkpoint = load_checkpoint(model, checkpoint_path, map_location='cpu')
model.CLASSES = dataset.CLASSES
model = MMDataParallel(model.cuda(), device_ids=[0])
output = single_gpu_test(model, data_loader, show_score_thr=0.05)
prediction_strings = []
file_names = []
coco = COCO(cfg.data.test.ann_file)
imag_ids = coco.getImgIds()
class_num = 11
for i, out in enumerate(output):
prediction_string = ''
image_info = coco.loadImgs(coco.getImgIds(imgIds=i))[0]
for j in range(class_num):
for o in out[j]:
prediction_string += str(j) + ' ' + str(o[4]) + ' ' + str(o[0]) + ' ' + str(o[1]) + ' ' + str(
o[2]) + ' ' + str(o[3]) + ' '
prediction_strings.append(prediction_string)
file_names.append(image_info['file_name'])
submission = pd.DataFrame()
submission['PredictionString'] = prediction_strings
submission['image_id'] = file_names
submission.to_csv(os.path.join(cfg.work_dir, f'submission_{epoch}.csv'), index=None)
submission.head()
```
|
github_jupyter
|
from mmcv import Config
from mmdet.datasets import build_dataset
from mmdet.models import build_detector
from mmdet.apis import train_detector
from mmdet.datasets import (build_dataloader, build_dataset,
replace_ImageToTensor)
classes = ("UNKNOWN", "General trash", "Paper", "Paper pack", "Metal", "Glass",
"Plastic", "Styrofoam", "Plastic bag", "Battery", "Clothing")
# config file 들고오기
cfg = Config.fromfile('./configs/pafpn/faster_rcnn_r50_pafpn_1x_coco.py')
PREFIX = '../../input/data/'
epoch = 12
# dataset 바꾸기
cfg.data.train.classes = classes
cfg.data.train.img_prefix = PREFIX
cfg.data.train.ann_file = PREFIX + 'train.json'
cfg.data.train.pipeline[2]['img_scale'] = (512, 512)
cfg.data.val.classes = classes
cfg.data.val.img_prefix = PREFIX
cfg.data.val.ann_file = PREFIX + 'val.json'
cfg.data.val.pipeline[1]['img_scale'] = (512, 512)
cfg.data.test.classes = classes
cfg.data.test.img_prefix = PREFIX
cfg.data.test.ann_file = PREFIX + 'test.json'
cfg.data.test.pipeline[1]['img_scale'] = (512, 512)
cfg.data.samples_per_gpu = 4
cfg.seed=2020
cfg.gpu_ids = [0]
cfg.work_dir = './work_dirs/faster_rcnn_r50_pafpn_1x_trash'
cfg.model.roi_head.bbox_head.num_classes = 11
print(cfg.model)
cfg.optimizer_config.grad_clip = dict(max_norm=35, norm_type=2)
model = build_detector(cfg.model)
datasets = [build_dataset(cfg.data.train)]
train_detector(model, datasets[0], cfg, distributed=False, validate=True)
import mmcv
from mmcv import Config
from mmdet.datasets import (build_dataloader, build_dataset,
replace_ImageToTensor)
from mmdet.models import build_detector
from mmdet.apis import single_gpu_test
from mmcv.runner import load_checkpoint
import os
from mmcv.parallel import MMDataParallel
import pandas as pd
from pandas import DataFrame
from pycocotools.coco import COCO
import numpy as np
cfg.model.train_cfg = None
checkpoint_path = os.path.join(cfg.work_dir, f'epoch_{epoch}.pth')
dataset = build_dataset(cfg.data.test)
data_loader = build_dataloader(
dataset,
samples_per_gpu=1,
workers_per_gpu=cfg.data.workers_per_gpu,
dist=False,
shuffle=False)
model = build_detector(cfg.model, test_cfg=cfg.get('test_cfg'))
checkpoint = load_checkpoint(model, checkpoint_path, map_location='cpu')
model.CLASSES = dataset.CLASSES
model = MMDataParallel(model.cuda(), device_ids=[0])
output = single_gpu_test(model, data_loader, show_score_thr=0.05)
prediction_strings = []
file_names = []
coco = COCO(cfg.data.test.ann_file)
imag_ids = coco.getImgIds()
class_num = 11
for i, out in enumerate(output):
prediction_string = ''
image_info = coco.loadImgs(coco.getImgIds(imgIds=i))[0]
for j in range(class_num):
for o in out[j]:
prediction_string += str(j) + ' ' + str(o[4]) + ' ' + str(o[0]) + ' ' + str(o[1]) + ' ' + str(
o[2]) + ' ' + str(o[3]) + ' '
prediction_strings.append(prediction_string)
file_names.append(image_info['file_name'])
submission = pd.DataFrame()
submission['PredictionString'] = prediction_strings
submission['image_id'] = file_names
submission.to_csv(os.path.join(cfg.work_dir, f'submission_{epoch}.csv'), index=None)
submission.head()
| 0.371593 | 0.681353 |
# Lambda School Data Science - Logistic Regression
Logistic regression is the baseline for classification models, as well as a handy way to predict probabilities (since those too live in the unit interval). While relatively simple, it is also the foundation for more sophisticated classification techniques such as neural networks (many of which can effectively be thought of as networks of logistic models).
## Lecture - Where Linear goes Wrong
### Return of the Titanic 🚢
You've likely already explored the rich dataset that is the Titanic - let's use regression and try to predict survival with it. The data is [available from Kaggle](https://www.kaggle.com/c/titanic/data), so we'll also play a bit with [the Kaggle API](https://github.com/Kaggle/kaggle-api).
```
!pip install kaggle
# Note - you'll also have to sign up for Kaggle and authorize the API
# https://github.com/Kaggle/kaggle-api#api-credentials
# This essentially means uploading a kaggle.json file
# For Colab we can have it in Google Drive
from google.colab import drive
drive.mount('/content/drive')
%env KAGGLE_CONFIG_DIR=/content/drive/My Drive/
# You also have to join the Titanic competition to have access to the data
!kaggle competitions download -c titanic
# How would we try to do this with linear regression?
import pandas as pd
train_df = pd.read_csv('train.csv').dropna()
test_df = pd.read_csv('test.csv').dropna() # Unlabeled, for Kaggle submission
train_df.head()
train_df.describe()
from sklearn.linear_model import LinearRegression
X = train_df[['Pclass', 'Age', 'Fare']]
y = train_df.Survived
linear_reg = LinearRegression().fit(X, y)
linear_reg.score(X, y)
linear_reg.predict(test_df[['Pclass', 'Age', 'Fare']])
linear_reg.coef_
import numpy as np
test_case = np.array([[1, 5, 500]]) # Rich 5-year old in first class
linear_reg.predict(test_case)
from sklearn.linear_model import LogisticRegression
log_reg = LogisticRegression().fit(X, y)
log_reg.score(X, y)
log_reg.predict(test_df[['Pclass', 'Age', 'Fare']])
log_reg.predict(test_case)[0]
help(log_reg.predict)
log_reg.predict_proba(test_case)[0]
# What's the math?
log_reg.coef_
log_reg.intercept_
# The logistic sigmoid "squishing" function, implemented to accept numpy arrays
def sigmoid(x):
return 1 / (1 + np.e**(-x))
sigmoid(log_reg.intercept_ + np.dot(log_reg.coef_, np.transpose(test_case)))
```
So, clearly a more appropriate model in this situation! For more on the math, [see this Wikipedia example](https://en.wikipedia.org/wiki/Logistic_regression#Probability_of_passing_an_exam_versus_hours_of_study).
For live - let's tackle [another classification dataset on absenteeism](http://archive.ics.uci.edu/ml/datasets/Absenteeism+at+work) - it has 21 classes, but remember, scikit-learn LogisticRegression automatically handles more than two classes. How? By essentially treating each label as different (1) from some base class (0).
```
# Live - let's try absenteeism!
```
## Assignment - real-world classification
We're going to check out a larger dataset - the [FMA Free Music Archive data](https://github.com/mdeff/fma). It has a selection of CSVs with metadata and calculated audio features that you can load and try to use to classify genre of tracks. To get you started:
```
!wget https://os.unil.cloud.switch.ch/fma/fma_metadata.zip
!unzip fma_metadata.zip
import pandas as pd
tracks = pd.read_csv('fma_metadata/tracks.csv')
pd.set_option('display.max_columns', None) # Unlimited columns
tracks.head()
tracks.head(10)
tracks.loc[0:0].values
tracks.columns = ['id', 'comments', 'date_created', 'date_released', 'engineer',
'favorites', 'id', 'information', 'listens', 'producer', 'tags',
'title', 'tracks', 'type', 'active_year_begin',
'active_year_end', 'associated_labels', 'bio', 'comments',
'date_created', 'favorites', 'id', 'latitude', 'location',
'longitude', 'members', 'name', 'related_projects', 'tags',
'website', 'wikipedia_page', 'split', 'subset', 'bit_rate',
'comments', 'composer', 'date_created', 'date_recorded',
'duration', 'favorites', 'genre_top', 'genres', 'genres_all',
'information', 'interest', 'language_code', 'license', 'listens',
'lyricist', 'number', 'publisher', 'tags', 'title']
tracks = tracks.drop([0,1])
tracks.head(10)
tracks
tracks[['comments', 'favorites', 'listens', 'tracks']]
tracks.dtypes
tracks.isnull().sum()
tracks.columns
df = tracks[['comments', 'favorites', 'listens', 'tracks', 'genre_top']].dropna()
df.shape
# ATTEMPT 1
X = df[['comments', 'favorites', 'listens', 'tracks']]
X.shape
y = df['genre_top']
from sklearn.linear_model.logistic import LogisticRegression
from sklearn.model_selection import train_test_split
model = LogisticRegression(random_state=42, solver='lbfgs', multi_class='multinomial', max_iter=1000)
model.fit(X,y)
model.score(X,y)
X_train, X_test, Y_train, Y_test = train_test_split(X, y, test_size=.2, random_state=42)
model2 = LogisticRegression(random_state=42, multi_class='multinomial', solver='lbfgs', max_iter=1500)
model2.fit(X_train,Y_train)
model2.score(X_test,Y_test)
#ATTEMPT 2
df = tracks[['comments', 'favorites', 'listens', 'tracks', 'genre_top', 'bit_rate', 'number', 'interest', 'duration']].dropna()
X = df[['comments', 'favorites', 'listens', 'tracks', 'bit_rate', 'number', 'interest', 'duration']]
y = df['genre_top']
df.head(20)
model = LogisticRegression(random_state=42, solver='lbfgs', multi_class='multinomial', max_iter=1000)
model.fit(X,y)
model.score(X,y)
X_train, X_test, Y_train, Y_test = train_test_split(X, y, test_size=.5, random_state=42)
model2 = LogisticRegression(random_state=42, multi_class='multinomial', solver='lbfgs', max_iter=2000)
model2.fit(X_train,Y_train)
model2.score(X_test,Y_test)
#ATTEMPT 3
df = tracks[['listens', 'tracks', 'bit_rate', 'number', 'interest', 'duration', 'genre_top', ]].dropna()
X = df[['listens', 'tracks', 'bit_rate', 'number', 'interest', 'duration']]
y = df['genre_top']
model = LogisticRegression(random_state=42, solver='lbfgs', multi_class='multinomial', max_iter=2500)
model.fit(X,y)
model.score(X,y)
```
This is the biggest data you've played with so far, and while it does generally fit in Colab, it can take awhile to run. That's part of the challenge!
Your tasks:
- Clean up the variable names in the dataframe
- Use logistic regression to fit a model predicting (primary/top) genre
- Inspect, iterate, and improve your model
- Answer the following questions (written, ~paragraph each):
- What are the best predictors of genre?
- What information isn't very useful for predicting genre?
- What surprised you the most about your results?
*Important caveats*:
- This is going to be difficult data to work with - don't let the perfect be the enemy of the good!
- Be creative in cleaning it up - if the best way you know how to do it is download it locally and edit as a spreadsheet, that's OK!
- If the data size becomes problematic, consider sampling/subsetting
- You do not need perfect or complete results - just something plausible that runs, and that supports the reasoning in your written answers
If you find that fitting a model to classify *all* genres isn't very good, it's totally OK to limit to the most frequent genres, or perhaps trying to combine or cluster genres as a preprocessing step. Even then, there will be limits to how good a model can be with just this metadata - if you really want to train an effective genre classifier, you'll have to involve the other data (see stretch goals).
This is real data - there is no "one correct answer", so you can take this in a variety of directions. Just make sure to support your findings, and feel free to share them as well! This is meant to be practice for dealing with other "messy" data, a common task in data science.
The best predictors of Genres seems to be all the numerical columns.
The favorites and comments column each had 3 columns named the same thing and these didn't seem very helpful
I was surprised that the results were as high as they even were.
|
github_jupyter
|
!pip install kaggle
# Note - you'll also have to sign up for Kaggle and authorize the API
# https://github.com/Kaggle/kaggle-api#api-credentials
# This essentially means uploading a kaggle.json file
# For Colab we can have it in Google Drive
from google.colab import drive
drive.mount('/content/drive')
%env KAGGLE_CONFIG_DIR=/content/drive/My Drive/
# You also have to join the Titanic competition to have access to the data
!kaggle competitions download -c titanic
# How would we try to do this with linear regression?
import pandas as pd
train_df = pd.read_csv('train.csv').dropna()
test_df = pd.read_csv('test.csv').dropna() # Unlabeled, for Kaggle submission
train_df.head()
train_df.describe()
from sklearn.linear_model import LinearRegression
X = train_df[['Pclass', 'Age', 'Fare']]
y = train_df.Survived
linear_reg = LinearRegression().fit(X, y)
linear_reg.score(X, y)
linear_reg.predict(test_df[['Pclass', 'Age', 'Fare']])
linear_reg.coef_
import numpy as np
test_case = np.array([[1, 5, 500]]) # Rich 5-year old in first class
linear_reg.predict(test_case)
from sklearn.linear_model import LogisticRegression
log_reg = LogisticRegression().fit(X, y)
log_reg.score(X, y)
log_reg.predict(test_df[['Pclass', 'Age', 'Fare']])
log_reg.predict(test_case)[0]
help(log_reg.predict)
log_reg.predict_proba(test_case)[0]
# What's the math?
log_reg.coef_
log_reg.intercept_
# The logistic sigmoid "squishing" function, implemented to accept numpy arrays
def sigmoid(x):
return 1 / (1 + np.e**(-x))
sigmoid(log_reg.intercept_ + np.dot(log_reg.coef_, np.transpose(test_case)))
# Live - let's try absenteeism!
!wget https://os.unil.cloud.switch.ch/fma/fma_metadata.zip
!unzip fma_metadata.zip
import pandas as pd
tracks = pd.read_csv('fma_metadata/tracks.csv')
pd.set_option('display.max_columns', None) # Unlimited columns
tracks.head()
tracks.head(10)
tracks.loc[0:0].values
tracks.columns = ['id', 'comments', 'date_created', 'date_released', 'engineer',
'favorites', 'id', 'information', 'listens', 'producer', 'tags',
'title', 'tracks', 'type', 'active_year_begin',
'active_year_end', 'associated_labels', 'bio', 'comments',
'date_created', 'favorites', 'id', 'latitude', 'location',
'longitude', 'members', 'name', 'related_projects', 'tags',
'website', 'wikipedia_page', 'split', 'subset', 'bit_rate',
'comments', 'composer', 'date_created', 'date_recorded',
'duration', 'favorites', 'genre_top', 'genres', 'genres_all',
'information', 'interest', 'language_code', 'license', 'listens',
'lyricist', 'number', 'publisher', 'tags', 'title']
tracks = tracks.drop([0,1])
tracks.head(10)
tracks
tracks[['comments', 'favorites', 'listens', 'tracks']]
tracks.dtypes
tracks.isnull().sum()
tracks.columns
df = tracks[['comments', 'favorites', 'listens', 'tracks', 'genre_top']].dropna()
df.shape
# ATTEMPT 1
X = df[['comments', 'favorites', 'listens', 'tracks']]
X.shape
y = df['genre_top']
from sklearn.linear_model.logistic import LogisticRegression
from sklearn.model_selection import train_test_split
model = LogisticRegression(random_state=42, solver='lbfgs', multi_class='multinomial', max_iter=1000)
model.fit(X,y)
model.score(X,y)
X_train, X_test, Y_train, Y_test = train_test_split(X, y, test_size=.2, random_state=42)
model2 = LogisticRegression(random_state=42, multi_class='multinomial', solver='lbfgs', max_iter=1500)
model2.fit(X_train,Y_train)
model2.score(X_test,Y_test)
#ATTEMPT 2
df = tracks[['comments', 'favorites', 'listens', 'tracks', 'genre_top', 'bit_rate', 'number', 'interest', 'duration']].dropna()
X = df[['comments', 'favorites', 'listens', 'tracks', 'bit_rate', 'number', 'interest', 'duration']]
y = df['genre_top']
df.head(20)
model = LogisticRegression(random_state=42, solver='lbfgs', multi_class='multinomial', max_iter=1000)
model.fit(X,y)
model.score(X,y)
X_train, X_test, Y_train, Y_test = train_test_split(X, y, test_size=.5, random_state=42)
model2 = LogisticRegression(random_state=42, multi_class='multinomial', solver='lbfgs', max_iter=2000)
model2.fit(X_train,Y_train)
model2.score(X_test,Y_test)
#ATTEMPT 3
df = tracks[['listens', 'tracks', 'bit_rate', 'number', 'interest', 'duration', 'genre_top', ]].dropna()
X = df[['listens', 'tracks', 'bit_rate', 'number', 'interest', 'duration']]
y = df['genre_top']
model = LogisticRegression(random_state=42, solver='lbfgs', multi_class='multinomial', max_iter=2500)
model.fit(X,y)
model.score(X,y)
| 0.648911 | 0.902524 |
# Database
```
%config Completer.use_jedi = False
import pandas as pd
import re
datas = pd.read_json('./tests/Processor_tests.json').T
datas.to_csv('./database.csv', sep=';', encoding='latin-1')
```
# Converter
## CBR
```
from comicsreader.converter import cbr2cbz
cbr2cbz('./tests/01 Wasteland.cbr')
```
## Pdf
```
from comicsreader.converter import pdf2cbz
pdf2cbz('./tests/Invisible Republic - T01.pdf')
```
# Dev Meta data
```
%load_ext autoreload
%autoreload 2
import os
import pandas as pd
from comicsreader.metaprocessor import MetaProcessor as Meta
from comicsreader.metaprocessor import TitleProcessor as Proc
import re
path = 'D:/Mes bds/'
for root, dirs, files in os.walk(path, topdown=False):
pass
root
files
file = files[3]
file
pattern = re.compile(r"""
(?!{) # ignore when starting with {
([^A-Za-z][0-9]{1,3})
(-[^A-Za-z][0-9]{1,3})?
(?!}) # ignore when finishing with }
""", re.VERBOSE
)
# pattern = '[0-9]{1,3}'
match = re.search(pattern, file)
match
match.groups()
processor = Proc()
processor(file)
file
datas = []
for file in files:
datas.append(Meta.from_file(file))
processed = pd.DataFrame([data.as_dict() for data in datas])
from ipywidgets import widgets, interactive, interact
pd.set_option("max_colwidth", 100)
@interact(n=list(range(processed.index.stop // 5)))
def sample(n):
return processed.loc[5*n:5*(n+1), ['file', 'extension', 'date', 'volumes', 'chapters', 'title']]
import json
processed.T.to_json('./tests/Processor_tests.json', indent=4)
processed.to_csv('./database.csv', sep=';', encoding='latin-1')
```
# Database
```
import pandas as pd
import re, os
import shutil
import ast
from comicsreader.converter import cbr2cbz, pdf2cbz
%load_ext autoreload
%autoreload 2
def format_list(x, prefix='(', suffix=')'):
if len(x) == 0:
result = ''
elif len(x) < 3:
result = prefix + '-'.join(map(str, x)) + suffix
else:
result = prefix + ', '.join(map(str, x)) + suffix
return result
def format_title(x):
dates = format_list(x.dates, '(', ')')
volumes = format_list(x.volumes, ', T', '')
# chapters = format_list(x.chapters, ', ', '')
result = f'{x.title} {dates}{volumes}'
result = re.sub(' +', ' ', result.strip())
return result
datas = pd.read_csv('./database_corrected.csv', sep=';', encoding='latin-1', index_col=0)
datas[['dates', 'volumes']] = datas[['dates', 'volumes']].applymap(ast.literal_eval)
datas['formatted_title'] = datas.apply(format_title, axis=1)
# convert
export_path = 'E:/comics_database/'
input_path = 'D:/Mes bds'
for series, group in datas.groupby('title'):
path = os.path.join(export_path, series)
if not os.path.exists(path):
os.makedirs(path)
for _, row in group.iterrows():
# convert
file = row.file
ext = file.split('.')[-1]
if ext == 'cbz':
shutil.copy(os.path.join(input_path, file), os.path.join(path, file))
elif ext == 'cbr':
cbr2cbz(os.path.join(input_path, file), path)
elif ext == 'pdf':
pdf2cbz(os.path.join(input_path, file), path)
# rename
input_file = file.split('.')[0] + '.cbz'
output_file = row.formatted_title + '.cbz'
os.rename(os.path.join(path, input_file), os.path.join(path, output_file))
series_table = pd.DataFrame(datas.title.unique(), columns=['Serie'])
series_table['ID'] = series_table.Serie.apply(hash)
series_map = series_table.set_index('Serie').ID
books_table = datas.copy()
books_table['serie_ID'] = series_map[books_table['title']].values
books_table.drop(columns=['tokenized_file', 'extension', 'title'], inplace=True)
books_table.rename({'formatted_title': 'name'}, axis=1, inplace=True)
books_table['ID']= books_table.name.apply(hash)
books_table[['dates', 'volumes']] = books_table[['dates', 'volumes']].astype(str)
series_table.rename(str.lower, axis=1, inplace=True)
books_table.rename(str.lower, axis=1, inplace=True)
```
# Sqlalchemy
```
import sqlalchemy as db
from sqlalchemy import Column, Integer, String, ForeignKey, Table, MetaData
from sqlalchemy.orm import relationship, backref
from sqlalchemy.ext.declarative import declarative_base
from functools import partial
from sqlalchemy import create_engine, text, insert
from sqlalchemy.orm import sessionmaker
engine = create_engine('sqlite+pysqlite:///comics_db.sqlite', echo=True, future=True)
Session = sessionmaker(bind=engine)
session = Session()
Base = declarative_base()
class AsDict:
__attr__ = []
def as_dict(self):
return {key: self.__getattribute__(key) for key in self.__attr__}
@classmethod
def from_series(cls, x: pd.Series):
return cls(**x[cls.__attr__].to_dict())
def __repr__(self):
return self.as_dict().__str__()
class Serie(Base, AsDict):
__tablename__ = 'series'
__attr__ = ['id', 'serie']
id = Column(Integer, primary_key=True)
serie = Column(String)
books = relationship('Book', backref=backref('series'))
class Book(Base, AsDict):
__tablename__ = 'books'
__attr__ = ['id', 'serie_id', 'name', 'dates','volumes']
id = Column(Integer, primary_key=True)
serie_id = Column(Integer, ForeignKey('series.id'))
name = Column(String)
dates = Column(String)
volumes = Column(String)
series = Table('series', Base.metadata, autoload=True, autoload_with=engine)
books = Table('books', Base.metadata, autoload=True, autoload_with=engine)
meta = MetaData(bind=engine)
meta.reflect(engine)
if len(meta.tables) == 0:
Base.metadata.create_all(engine)
def execute_query(engine, query, **args):
with engine.connect() as conn:
conn.execute(query, *args)
conn.commit()
values_to_insert = [Serie.from_series(row) for _, row in series_table.iterrows()]
session.add_all(values_to_insert)
session.commit()
values_to_insert = [Book.from_series(row) for _, row in books_table.iterrows()]
session.add_all(values_to_insert)
session.commit()
db.select([series.columns.serie])
session.query(Serie)
session.close()
```
# pyunpack
```
%load_ext autoreload
%autoreload 2
%config Completer.use_jedi = False
from pyunpack import Archive
from zipfile import ZipFile
import os
```
# Rarfile
```
%load_ext autoreload
%autoreload 2
%config Completer.use_jedi = False
import rarfile.rarfile as rarfile
# rarfile.UNAR_TOOL = './unar/unar.exe'
# rarfile.UNAR_TOOL = './unrar/UNRAR.exe'
rarfile.UNAR_TOOL = './libarchive/bin/bsdtar.exe'
file_path = './01 Wasteland.cbr'
with rarfile.RarFile(file_path) as rf:
for f in rf.infolist():
print(f.filename, f.file_size)
rf.extract(f.orig_filename, './')
f.orig_filename.endswith(b'/')
```
|
github_jupyter
|
%config Completer.use_jedi = False
import pandas as pd
import re
datas = pd.read_json('./tests/Processor_tests.json').T
datas.to_csv('./database.csv', sep=';', encoding='latin-1')
from comicsreader.converter import cbr2cbz
cbr2cbz('./tests/01 Wasteland.cbr')
from comicsreader.converter import pdf2cbz
pdf2cbz('./tests/Invisible Republic - T01.pdf')
%load_ext autoreload
%autoreload 2
import os
import pandas as pd
from comicsreader.metaprocessor import MetaProcessor as Meta
from comicsreader.metaprocessor import TitleProcessor as Proc
import re
path = 'D:/Mes bds/'
for root, dirs, files in os.walk(path, topdown=False):
pass
root
files
file = files[3]
file
pattern = re.compile(r"""
(?!{) # ignore when starting with {
([^A-Za-z][0-9]{1,3})
(-[^A-Za-z][0-9]{1,3})?
(?!}) # ignore when finishing with }
""", re.VERBOSE
)
# pattern = '[0-9]{1,3}'
match = re.search(pattern, file)
match
match.groups()
processor = Proc()
processor(file)
file
datas = []
for file in files:
datas.append(Meta.from_file(file))
processed = pd.DataFrame([data.as_dict() for data in datas])
from ipywidgets import widgets, interactive, interact
pd.set_option("max_colwidth", 100)
@interact(n=list(range(processed.index.stop // 5)))
def sample(n):
return processed.loc[5*n:5*(n+1), ['file', 'extension', 'date', 'volumes', 'chapters', 'title']]
import json
processed.T.to_json('./tests/Processor_tests.json', indent=4)
processed.to_csv('./database.csv', sep=';', encoding='latin-1')
import pandas as pd
import re, os
import shutil
import ast
from comicsreader.converter import cbr2cbz, pdf2cbz
%load_ext autoreload
%autoreload 2
def format_list(x, prefix='(', suffix=')'):
if len(x) == 0:
result = ''
elif len(x) < 3:
result = prefix + '-'.join(map(str, x)) + suffix
else:
result = prefix + ', '.join(map(str, x)) + suffix
return result
def format_title(x):
dates = format_list(x.dates, '(', ')')
volumes = format_list(x.volumes, ', T', '')
# chapters = format_list(x.chapters, ', ', '')
result = f'{x.title} {dates}{volumes}'
result = re.sub(' +', ' ', result.strip())
return result
datas = pd.read_csv('./database_corrected.csv', sep=';', encoding='latin-1', index_col=0)
datas[['dates', 'volumes']] = datas[['dates', 'volumes']].applymap(ast.literal_eval)
datas['formatted_title'] = datas.apply(format_title, axis=1)
# convert
export_path = 'E:/comics_database/'
input_path = 'D:/Mes bds'
for series, group in datas.groupby('title'):
path = os.path.join(export_path, series)
if not os.path.exists(path):
os.makedirs(path)
for _, row in group.iterrows():
# convert
file = row.file
ext = file.split('.')[-1]
if ext == 'cbz':
shutil.copy(os.path.join(input_path, file), os.path.join(path, file))
elif ext == 'cbr':
cbr2cbz(os.path.join(input_path, file), path)
elif ext == 'pdf':
pdf2cbz(os.path.join(input_path, file), path)
# rename
input_file = file.split('.')[0] + '.cbz'
output_file = row.formatted_title + '.cbz'
os.rename(os.path.join(path, input_file), os.path.join(path, output_file))
series_table = pd.DataFrame(datas.title.unique(), columns=['Serie'])
series_table['ID'] = series_table.Serie.apply(hash)
series_map = series_table.set_index('Serie').ID
books_table = datas.copy()
books_table['serie_ID'] = series_map[books_table['title']].values
books_table.drop(columns=['tokenized_file', 'extension', 'title'], inplace=True)
books_table.rename({'formatted_title': 'name'}, axis=1, inplace=True)
books_table['ID']= books_table.name.apply(hash)
books_table[['dates', 'volumes']] = books_table[['dates', 'volumes']].astype(str)
series_table.rename(str.lower, axis=1, inplace=True)
books_table.rename(str.lower, axis=1, inplace=True)
import sqlalchemy as db
from sqlalchemy import Column, Integer, String, ForeignKey, Table, MetaData
from sqlalchemy.orm import relationship, backref
from sqlalchemy.ext.declarative import declarative_base
from functools import partial
from sqlalchemy import create_engine, text, insert
from sqlalchemy.orm import sessionmaker
engine = create_engine('sqlite+pysqlite:///comics_db.sqlite', echo=True, future=True)
Session = sessionmaker(bind=engine)
session = Session()
Base = declarative_base()
class AsDict:
__attr__ = []
def as_dict(self):
return {key: self.__getattribute__(key) for key in self.__attr__}
@classmethod
def from_series(cls, x: pd.Series):
return cls(**x[cls.__attr__].to_dict())
def __repr__(self):
return self.as_dict().__str__()
class Serie(Base, AsDict):
__tablename__ = 'series'
__attr__ = ['id', 'serie']
id = Column(Integer, primary_key=True)
serie = Column(String)
books = relationship('Book', backref=backref('series'))
class Book(Base, AsDict):
__tablename__ = 'books'
__attr__ = ['id', 'serie_id', 'name', 'dates','volumes']
id = Column(Integer, primary_key=True)
serie_id = Column(Integer, ForeignKey('series.id'))
name = Column(String)
dates = Column(String)
volumes = Column(String)
series = Table('series', Base.metadata, autoload=True, autoload_with=engine)
books = Table('books', Base.metadata, autoload=True, autoload_with=engine)
meta = MetaData(bind=engine)
meta.reflect(engine)
if len(meta.tables) == 0:
Base.metadata.create_all(engine)
def execute_query(engine, query, **args):
with engine.connect() as conn:
conn.execute(query, *args)
conn.commit()
values_to_insert = [Serie.from_series(row) for _, row in series_table.iterrows()]
session.add_all(values_to_insert)
session.commit()
values_to_insert = [Book.from_series(row) for _, row in books_table.iterrows()]
session.add_all(values_to_insert)
session.commit()
db.select([series.columns.serie])
session.query(Serie)
session.close()
%load_ext autoreload
%autoreload 2
%config Completer.use_jedi = False
from pyunpack import Archive
from zipfile import ZipFile
import os
%load_ext autoreload
%autoreload 2
%config Completer.use_jedi = False
import rarfile.rarfile as rarfile
# rarfile.UNAR_TOOL = './unar/unar.exe'
# rarfile.UNAR_TOOL = './unrar/UNRAR.exe'
rarfile.UNAR_TOOL = './libarchive/bin/bsdtar.exe'
file_path = './01 Wasteland.cbr'
with rarfile.RarFile(file_path) as rf:
for f in rf.infolist():
print(f.filename, f.file_size)
rf.extract(f.orig_filename, './')
f.orig_filename.endswith(b'/')
| 0.300232 | 0.538377 |
<img src="https://upload.wikimedia.org/wikipedia/commons/4/47/Logo_UTFSM.png" width="200" alt="utfsm-logo" align="left"/>
# MAT281
### Aplicaciones de la Matemática en la Ingeniería
## Módulo 02
## Laboratorio Clase 04: Agrupando datos
### Instrucciones
* Completa tus datos personales (nombre y rol USM) en siguiente celda.
* La escala es de 0 a 4 considerando solo valores enteros.
* Debes _pushear_ tus cambios a tu repositorio personal del curso.
* Como respaldo, debes enviar un archivo .zip con el siguiente formato `mXX_cYY_lab_apellido_nombre.zip` a alonso.ogueda@gmail.com.
* Se evaluará:
- Soluciones
- Código
- Que Binder esté bien configurado.
- Al presionar `Kernel -> Restart Kernel and Run All Cells` deben ejecutarse todas las celdas sin error.
* __La entrega es al final de esta clase.__
__Nombre__: Brandon Olguin
__Rol__: 201510027-6
Se utilizará el mismo dataset de pokemon
```
import os
import pandas as pd
pkm = (
pd.read_csv(os.path.join("data", "pokemon.csv"), index_col="#")
.rename(columns=lambda x: x.replace(" ", "").replace(".", "_").lower())
)
pkm.head()
```
## Ejercicio #1 (1 pto)
Agrupar por `generation` y `legendary` y obtener por grupo:
* Promedio de `hp`
* Mínimo y máximo de `sp_atk` y `sp_def`
```
(
pkm.groupby(["generation","legendary"])
.agg(
{"sp_atk" : ["min","max"], "sp_def" : ["min","max"], "hp" : "mean"}
)
)
```
## Ejercicio #2 (1 pto)
El profesor Oakgueda determinó que una buen indicador de pokemones es:
$$ 0.2 \, \textrm{hp} + 0.4 \,(\textrm{attack} + \textrm{sp_atk})^2 + 0.3 \,( \textrm{defense} + \textrm{sp_deff})^{1.5} + 0.1 \, \textrm{speed}$$
Según este indicador, ¿Qué grupo de pokemones (`type1`, `type2`) es en promedio mejor que el resto?
```
def oakgueda_indicator(df):
oakgueda_ind = 0.2*df["hp"] + 0.4*(df["attack"] + df["sp_atk"])**2 + 0.3*(df["defense"] + df["sp_def"])**(1.5) + 0.1*df["speed"]
return oakgueda_ind.mean()
pkm.groupby(["type1","type2"]).apply(lambda df: oakgueda_indicator(df)).sort_values()
```
__Respuesta__: (Ground,Fire)
## Ejercicio #3 (1 pto)
Define una función que escale los datos tal que, si $s$ es una columna:
$$s\_scaled = \frac{s - \min(s)}{\max(s) - \min(s)}$$
Y luego transforma cada columna agrupando por si el pokemon es legendario o no.
```
def minmax_scale(s):
return (s - s.min())/(s.max() - s.min())
pkm.groupby("legendary").transform(minmax_scale)
```
## Ejercicio #4 (1 pto)
El profesor Oakgueda necesita saber cuántos pokemones hay luego de filtrar el dataset tal que el grupo de (`type1`, `type2`) tenga en promedio un indicador (el del ejercicio #2) mayor a 40000.
```
pkm.groupby(["type1","type2"]).filter(lambda df: oakgueda_indicator(df) > 40000)
```
__Respuesta:__ Hay solo dos pokemones que cumplen esta condición.
|
github_jupyter
|
import os
import pandas as pd
pkm = (
pd.read_csv(os.path.join("data", "pokemon.csv"), index_col="#")
.rename(columns=lambda x: x.replace(" ", "").replace(".", "_").lower())
)
pkm.head()
(
pkm.groupby(["generation","legendary"])
.agg(
{"sp_atk" : ["min","max"], "sp_def" : ["min","max"], "hp" : "mean"}
)
)
def oakgueda_indicator(df):
oakgueda_ind = 0.2*df["hp"] + 0.4*(df["attack"] + df["sp_atk"])**2 + 0.3*(df["defense"] + df["sp_def"])**(1.5) + 0.1*df["speed"]
return oakgueda_ind.mean()
pkm.groupby(["type1","type2"]).apply(lambda df: oakgueda_indicator(df)).sort_values()
def minmax_scale(s):
return (s - s.min())/(s.max() - s.min())
pkm.groupby("legendary").transform(minmax_scale)
pkm.groupby(["type1","type2"]).filter(lambda df: oakgueda_indicator(df) > 40000)
| 0.245266 | 0.920039 |
# nuScenes devkit tutorial
Welcome to the nuScenes tutorial. This demo assumes the database itself is available at `/data/sets/nuscenes`, and loads a mini version of the full dataset.
## A Gentle Introduction to nuScenes
In this part of the tutorial, let us go through a top-down introduction of our database. Our dataset comprises of elemental building blocks that are the following:
1. `log` - Log information from which the data was extracted.
2. `scene` - 20 second snippet of a car's journey.
3. `sample` - An annotated snapshot of a scene at a particular timestamp.
4. `sample_data` - Data collected from a particular sensor.
5. `ego_pose` - Ego vehicle poses at a particular timestamp.
6. `sensor` - A specific sensor type.
7. `calibrated sensor` - Definition of a particular sensor as calibrated on a particular vehicle.
8. `instance` - Enumeration of all object instance we observed.
9. `category` - Taxonomy of object categories (e.g. vehicle, human).
10. `attribute` - Property of an instance that can change while the category remains the same.
11. `visibility` - Fraction of pixels visible in all the images collected from 6 different cameras.
12. `sample_annotation` - An annotated instance of an object within our interest.
13. `map` - Map data that is stored as binary semantic masks from a top-down view.
The database schema is visualized below. For more information see the [nuScenes schema](https://github.com/nutonomy/nuscenes-devkit/blob/master/docs/schema_nuscenes.md) page.

## Google Colab (optional)
<br>
<a href="https://colab.research.google.com/github/nutonomy/nuscenes-devkit/blob/master/python-sdk/tutorials/nuscenes_tutorial.ipynb">
<img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab" align="left">
</a>
<br>
If you are running this notebook in Google Colab, you can uncomment the cell below and run it; everything will be set up nicely for you. Otherwise, manually set up everything.
```
# !mkdir -p /data/sets/nuscenes # Make the directory to store the nuScenes dataset in.
# !wget https://www.nuscenes.org/data/v1.0-mini.tgz # Download the nuScenes mini split.
# !tar -xf v1.0-mini.tgz -C /data/sets/nuscenes # Uncompress the nuScenes mini split.
# !pip install nuscenes-devkit &> /dev/null # Install nuScenes.
```
## Initialization
```
%matplotlib inline
from nuscenes.nuscenes import NuScenes
from nuscenes.utils.data_classes import LidarPointCloud, RadarPointCloud, Box
import os
nusc = NuScenes(version='v1.0-mini', dataroot='./data/sets/nuscenes', verbose=True)
```
## A look at the dataset
### 1. `scene`
nuScenes is a large scale database that features annotated samples across ***1000 scenes*** of approximately 20 seconds each. Let's take a look at the scenes that we have in the loaded database.
```
nusc.list_scenes()
```
Let's look at a scene metadata
```
my_scene = nusc.scene[0]
my_scene
```
### 2. `sample`
In scenes, we annotate our data every half a second (2 Hz).
We define `sample` as an ***annotated keyframe of a scene at a given timestamp***. A keyframe is a frame where the time-stamps of data from all the sensors should be very close to the time-stamp of the sample it points to.
Now, let us look at the first annotated sample in this scene.
```
first_sample_token = my_scene['first_sample_token']
# The rendering command below is commented out because it tends to crash in notebooks
# nusc.render_sample(first_sample_token)
```
Let's examine its metadata
```
my_sample = nusc.get('sample', first_sample_token)
my_sample
```
A useful method is `list_sample()` which lists all related `sample_data` keyframes and `sample_annotation` associated with a `sample` which we will discuss in detail in the subsequent parts.
```
nusc.list_sample(my_sample['token'])
```
### 3. `sample_data`
The nuScenes dataset contains data that is collected from a full sensor suite. Hence, for each snapshot of a scene, we provide references to a family of data that is collected from these sensors.
We provide a `data` key to access these:
```
my_sample['data']
```
Notice that the keys are referring to the different sensors that form our sensor suite. Let's take a look at the metadata of a `sample_data` taken from `CAM_FRONT`.
```
sensor = 'CAM_FRONT'
cam_front_data = nusc.get('sample_data', my_sample['data'][sensor])
cam_front_data
sensor = 'LIDAR_TOP'
lidar_top_data = nusc.get('sample_data', my_sample['data'][sensor])
lidar_top_data
my_scene = nusc.scene[0]
first_sample_token = my_scene['first_sample_token']
my_sample = nusc.get('sample', first_sample_token)
lidar_top_data = nusc.get('sample_data', my_sample['data']['LIDAR_TOP'])
pcl_path = os.path.join(nusc.dataroot, lidar_top_data['filename'])
pc = LidarPointCloud.from_file(pcl_path)
print(pc.points)
```
We can also render the `sample_data` at a particular sensor.
```
nusc.render_sample_data(cam_front_data['token'])
```
### 4. `sample_annotation`
`sample_annotation` refers to any ***bounding box defining the position of an object seen in a sample***. All location data is given with respect to the global coordinate system. Let's examine an example from our `sample` above.
```
my_annotation_token = my_sample['anns'][18]
my_annotation_metadata = nusc.get('sample_annotation', my_annotation_token)
my_annotation_metadata
```
We can also render an annotation to have a closer look.
```
nusc.render_annotation(my_annotation_token)
```
### 5. `instance`
Object instance are instances that need to be detected or tracked by an AV (e.g a particular vehicle, pedestrian). Let us examine an instance metadata
```
my_instance = nusc.instance[599]
my_instance
```
We generally track an instance across different frames in a particular scene. However, we do not track them across different scenes. In this example, we have 16 annotated samples for this instance across a particular scene.
```
instance_token = my_instance['token']
nusc.render_instance(instance_token)
```
An instance record takes note of its first and last annotation token. Let's render them
```
print("First annotated sample of this instance:")
nusc.render_annotation(my_instance['first_annotation_token'])
print("Last annotated sample of this instance")
nusc.render_annotation(my_instance['last_annotation_token'])
```
### 6. `category`
A `category` is the object assignment of an annotation. Let's look at the category table we have in our database. The table contains the taxonomy of different object categories and also list the subcategories (delineated by a period).
```
nusc.list_categories()
```
A category record contains the name and the description of that particular category.
```
nusc.category[9]
```
Refer to `instructions_nuscenes.md` for the definitions of the different categories.
### 7. `attribute`
An `attribute` is a property of an instance that may change throughout different parts of a scene while the category remains the same. Here we list the provided attributes and the number of annotations associated with a particular attribute.
```
nusc.list_attributes()
```
Let's take a look at an example how an attribute may change over one scene
```
my_instance = nusc.instance[27]
first_token = my_instance['first_annotation_token']
last_token = my_instance['last_annotation_token']
nbr_samples = my_instance['nbr_annotations']
current_token = first_token
i = 0
found_change = False
while current_token != last_token:
current_ann = nusc.get('sample_annotation', current_token)
current_attr = nusc.get('attribute', current_ann['attribute_tokens'][0])['name']
if i == 0:
pass
elif current_attr != last_attr:
print("Changed from `{}` to `{}` at timestamp {} out of {} annotated timestamps".format(last_attr, current_attr, i, nbr_samples))
found_change = True
next_token = current_ann['next']
current_token = next_token
last_attr = current_attr
i += 1
```
### 8. `visibility`
`visibility` is defined as the fraction of pixels of a particular annotation that are visible over the 6 camera feeds, grouped into 4 bins.
```
nusc.visibility
```
Let's look at an example `sample_annotation` with 80-100% visibility
```
anntoken = 'a7d0722bce164f88adf03ada491ea0ba'
visibility_token = nusc.get('sample_annotation', anntoken)['visibility_token']
print("Visibility: {}".format(nusc.get('visibility', visibility_token)))
nusc.render_annotation(anntoken)
```
Let's look at an example `sample_annotation` with 0-40% visibility
```
anntoken = '9f450bf6b7454551bbbc9a4c6e74ef2e'
visibility_token = nusc.get('sample_annotation', anntoken)['visibility_token']
print("Visibility: {}".format(nusc.get('visibility', visibility_token)))
nusc.render_annotation(anntoken)
```
### 9. `sensor`
The nuScenes dataset consists of data collected from our full sensor suite which consists of:
- 1 x LIDAR,
- 5 x RADAR,
- 6 x cameras,
```
nusc.sensor
```
Every `sample_data` has a record on which `sensor` the data is collected from (note the "channel" key)
```
nusc.sample_data[10]
```
### 10. `calibrated_sensor`
`calibrated_sensor` consists of the definition of a particular sensor (lidar/radar/camera) as calibrated on a particular vehicle. Let us look at an example.
```
nusc.calibrated_sensor[0]
```
Note that the `translation` and the `rotation` parameters are given with respect to the ego vehicle body frame.
### 11. `ego_pose`
`ego_pose` contains information about the location (encoded in `translation`) and the orientation (encoded in `rotation`) of the ego vehicle, with respect to the global coordinate system.
```
nusc.ego_pose[0]
```
Note that the number of `ego_pose` records in our loaded database is the same as the number of `sample_data` records. These two records exhibit a one-to-one correspondence.
### 12. `log`
The `log` table contains log information from which the data was extracted. A `log` record corresponds to one journey of our ego vehicle along a predefined route. Let's check the number of logs and the metadata of a log.
```
print("Number of `logs` in our loaded database: {}".format(len(nusc.log)))
nusc.log[0]
```
Notice that it contains a variety of information such as the date and location of the log collected. It also gives out information about the map from where the data was collected. Note that one log can contain multiple non-overlapping scenes.
### 13. `map`
Map information is stored as binary semantic masks from a top-down view. Let's check the number of maps and metadata of a map.
```
print("There are {} maps masks in the loaded dataset".format(len(nusc.map)))
nusc.map[0]
```
## nuScenes Basics
Let's get a bit technical.
The NuScenes class holds several tables. Each table is a list of records, and each record is a dictionary. For example the first record of the category table is stored at:
```
nusc.category[0]
```
The category table is simple: it holds the fields `name` and `description`. It also has a `token` field, which is a unique record identifier. Since the record is a dictionary, the token can be accessed like so:
```
cat_token = nusc.category[0]['token']
cat_token
```
If you know the `token` for any record in the DB you can retrieve the record by doing
```
nusc.get('category', cat_token)
```
_As you can notice, we have recovered the same record!_
OK, that was easy. Let's try something harder. Let's look at the `sample_annotation` table.
```
nusc.sample_annotation[0]
```
This also has a `token` field (they all do). In addition, it has several fields of the format [a-z]*\_token, _e.g._ instance_token. These are foreign keys in database terminology, meaning they point to another table.
Using `nusc.get()` we can grab any of these in constant time. For example, let's look at the visibility record.
```
nusc.get('visibility', nusc.sample_annotation[0]['visibility_token'])
```
The visibility records indicate how much of an object was visible when it was annotated.
Let's also grab the `instance_token`
```
one_instance = nusc.get('instance', nusc.sample_annotation[0]['instance_token'])
one_instance
```
This points to the `instance` table. This table enumerate the object _instances_ we have encountered in each
scene. This way we can connect all annotations of a particular object.
If you look carefully at the README tables, you will see that the sample_annotation table points to the instance table,
but the instance table doesn't list all annotations that point to it.
So how can we recover all sample_annotations for a particular object instance? There are two ways:
1. `Use nusc.field2token()`. Let's try it:
```
ann_tokens = nusc.field2token('sample_annotation', 'instance_token', one_instance['token'])
```
This returns a list of all sample_annotation records with the `'instance_token'` == `one_instance['token']`. Let's store these in a set for now
```
ann_tokens_field2token = set(ann_tokens)
ann_tokens_field2token
```
The `nusc.field2token()` method is generic and can be used in any similar situation.
2. For certain situation, we provide some reverse indices in the tables themselves. This is one such example.
The instance record has a field `first_annotation_token` which points to the first annotation in time of this instance.
Recovering this record is easy.
```
ann_record = nusc.get('sample_annotation', one_instance['first_annotation_token'])
ann_record
```
Now we can traverse all annotations of this instance using the "next" field. Let's try it.
```
ann_tokens_traverse = set()
ann_tokens_traverse.add(ann_record['token'])
while not ann_record['next'] == "":
ann_record = nusc.get('sample_annotation', ann_record['next'])
ann_tokens_traverse.add(ann_record['token'])
```
Finally, let's assert that we recovered the same ann_records as we did using nusc.field2token:
```
print(ann_tokens_traverse == ann_tokens_field2token)
```
## Reverse indexing and short-cuts
The nuScenes tables are normalized, meaning that each piece of information is only given once.
For example, there is one `map` record for each `log` record. Looking at the schema you will notice that the `map` table has a `log_token` field, but that the `log` table does not have a corresponding `map_token` field. But there are plenty of situations where you have a `log`, and want to find the corresponding `map`! So what to do? You can always use the `nusc.field2token()` method, but that is slow and inconvenient. We therefore add reverse mappings for some common situations including this one.
Further, there are situations where one needs to go through several tables to get a certain piece of information.
Consider, for example, the category name (e.g. `human.pedestrian`) of a `sample_annotation`. The `sample_annotation` table doesn't hold this information since the category is an instance level constant. Instead the `sample_annotation` table points to a record in the `instance` table. This, in turn, points to a record in the `category` table, where finally the `name` fields stores the required information.
Since it is quite common to want to know the category name of an annotation, we add a `category_name` field to the `sample_annotation` table during initialization of the NuScenes class.
In this section, we list the short-cuts and reverse indices that are added to the `NuScenes` class during initialization. These are all created in the `NuScenes.__make_reverse_index__()` method.
### Reverse indices
We add two reverse indices by default.
* A `map_token` field is added to the `log` records.
* The `sample` records have shortcuts to all `sample_annotations` for that record as well as `sample_data` key-frames. Confer `nusc.list_sample()` method in the previous section for more details on this.
### Shortcuts
The sample_annotation table has a "category_name" shortcut.
_Using shortcut:_
```
catname = nusc.sample_annotation[0]['category_name']
```
_Not using shortcut:_
```
ann_rec = nusc.sample_annotation[0]
inst_rec = nusc.get('instance', ann_rec['instance_token'])
cat_rec = nusc.get('category', inst_rec['category_token'])
print(catname == cat_rec['name'])
```
The sample_data table has "channel" and "sensor_modality" shortcuts:
```
# Shortcut
channel = nusc.sample_data[0]['channel']
# No shortcut
sd_rec = nusc.sample_data[0]
cs_record = nusc.get('calibrated_sensor', sd_rec['calibrated_sensor_token'])
sensor_record = nusc.get('sensor', cs_record['sensor_token'])
print(channel == sensor_record['channel'])
```
## Data Visualizations
We provide list and rendering methods. These are meant both as convenience methods during development, and as tutorials for building your own visualization methods. They are implemented in the NuScenesExplorer class, with shortcuts through the NuScenes class itself.
### List methods
There are three list methods available.
1. `list_categories()` lists all categories, counts and statistics of width/length/height in meters and aspect ratio.
```
nusc.list_categories()
```
2. `list_attributes()` lists all attributes and counts.
```
nusc.list_attributes()
```
3. `list_scenes()` lists all scenes in the loaded DB.
```
nusc.list_scenes()
```
### Render
First, let's plot a lidar point cloud in an image. Lidar allows us to accurately map the surroundings in 3D.
```
my_sample = nusc.sample[10]
nusc.render_pointcloud_in_image(my_sample['token'], pointsensor_channel='LIDAR_TOP')
```
In the previous image the colors indicate the distance from the ego vehicle to each lidar point. We can also render the lidar intensity. In the following image the traffic sign ahead of us is highly reflective (yellow) and the dark vehicle on the right has low reflectivity (purple).
```
nusc.render_pointcloud_in_image(my_sample['token'], pointsensor_channel='LIDAR_TOP', render_intensity=True)
```
Second, let's plot the radar point cloud for the same image. Radar is less dense than lidar, but has a much larger range.
```
nusc.render_pointcloud_in_image(my_sample['token'], pointsensor_channel='RADAR_FRONT')
```
We can also plot all annotations across all sample data for that sample. Note how for radar we also plot the velocity vectors of moving objects. Some velocity vectors are outliers, which can be filtered using the settings in RadarPointCloud.from_file()
```
my_sample = nusc.sample[20]
# The rendering command below is commented out because it may crash in notebooks
# nusc.render_sample(my_sample['token'])
```
Or if we only want to render a particular sensor, we can specify that.
```
nusc.render_sample_data(my_sample['data']['CAM_FRONT'])
```
Additionally we can aggregate the point clouds from multiple sweeps to get a denser point cloud.
```
nusc.render_sample_data(my_sample['data']['LIDAR_TOP'], nsweeps=5, underlay_map=True)
nusc.render_sample_data(my_sample['data']['RADAR_FRONT'], nsweeps=5, underlay_map=True)
```
In the radar plot above we only see very confident radar returns from two vehicles. This is due to the filter settings defined in the file `nuscenes/utils/data_classes.py`. If instead we want to disable all filters and render all returns, we can use the `disable_filters()` function. This returns a denser point cloud, but with many returns from background objects. To return to the default settings, simply call `default_filters()`.
```
from nuscenes.utils.data_classes import RadarPointCloud
RadarPointCloud.disable_filters()
nusc.render_sample_data(my_sample['data']['RADAR_FRONT'], nsweeps=5, underlay_map=True)
RadarPointCloud.default_filters()
```
We can even render a specific annotation.
```
nusc.render_annotation(my_sample['anns'][22])
```
Finally, we can render a full scene as a video. There are two options here:
1. nusc.render_scene_channel() renders the video for a particular channel. (HIT ESC to exit)
2. nusc.render_scene() renders the video for all camera channels.
NOTE: These methods use OpenCV for rendering, which doesn't always play nice with IPython Notebooks. If you experience any issues please run these lines from the command line.
Let's grab scene 0061, it is nice and dense.
```
my_scene_token = nusc.field2token('scene', 'name', 'scene-0061')[0]
# The rendering command below is commented out because it may crash in notebooks
# nusc.render_scene_channel(my_scene_token, 'CAM_FRONT')
```
There is also a method nusc.render_scene() which renders the video for all camera channels.
This requires a high-res monitor, and is also best run outside this notebook.
```
# The rendering command below is commented out because it may crash in notebooks
# nusc.render_scene(my_scene_token)
```
Finally, let us visualize all scenes on the map for a particular location.
```
nusc.render_egoposes_on_map(log_location='singapore-onenorth')
```
|
github_jupyter
|
# !mkdir -p /data/sets/nuscenes # Make the directory to store the nuScenes dataset in.
# !wget https://www.nuscenes.org/data/v1.0-mini.tgz # Download the nuScenes mini split.
# !tar -xf v1.0-mini.tgz -C /data/sets/nuscenes # Uncompress the nuScenes mini split.
# !pip install nuscenes-devkit &> /dev/null # Install nuScenes.
%matplotlib inline
from nuscenes.nuscenes import NuScenes
from nuscenes.utils.data_classes import LidarPointCloud, RadarPointCloud, Box
import os
nusc = NuScenes(version='v1.0-mini', dataroot='./data/sets/nuscenes', verbose=True)
nusc.list_scenes()
my_scene = nusc.scene[0]
my_scene
first_sample_token = my_scene['first_sample_token']
# The rendering command below is commented out because it tends to crash in notebooks
# nusc.render_sample(first_sample_token)
my_sample = nusc.get('sample', first_sample_token)
my_sample
nusc.list_sample(my_sample['token'])
my_sample['data']
sensor = 'CAM_FRONT'
cam_front_data = nusc.get('sample_data', my_sample['data'][sensor])
cam_front_data
sensor = 'LIDAR_TOP'
lidar_top_data = nusc.get('sample_data', my_sample['data'][sensor])
lidar_top_data
my_scene = nusc.scene[0]
first_sample_token = my_scene['first_sample_token']
my_sample = nusc.get('sample', first_sample_token)
lidar_top_data = nusc.get('sample_data', my_sample['data']['LIDAR_TOP'])
pcl_path = os.path.join(nusc.dataroot, lidar_top_data['filename'])
pc = LidarPointCloud.from_file(pcl_path)
print(pc.points)
nusc.render_sample_data(cam_front_data['token'])
my_annotation_token = my_sample['anns'][18]
my_annotation_metadata = nusc.get('sample_annotation', my_annotation_token)
my_annotation_metadata
nusc.render_annotation(my_annotation_token)
my_instance = nusc.instance[599]
my_instance
instance_token = my_instance['token']
nusc.render_instance(instance_token)
print("First annotated sample of this instance:")
nusc.render_annotation(my_instance['first_annotation_token'])
print("Last annotated sample of this instance")
nusc.render_annotation(my_instance['last_annotation_token'])
nusc.list_categories()
nusc.category[9]
nusc.list_attributes()
my_instance = nusc.instance[27]
first_token = my_instance['first_annotation_token']
last_token = my_instance['last_annotation_token']
nbr_samples = my_instance['nbr_annotations']
current_token = first_token
i = 0
found_change = False
while current_token != last_token:
current_ann = nusc.get('sample_annotation', current_token)
current_attr = nusc.get('attribute', current_ann['attribute_tokens'][0])['name']
if i == 0:
pass
elif current_attr != last_attr:
print("Changed from `{}` to `{}` at timestamp {} out of {} annotated timestamps".format(last_attr, current_attr, i, nbr_samples))
found_change = True
next_token = current_ann['next']
current_token = next_token
last_attr = current_attr
i += 1
nusc.visibility
anntoken = 'a7d0722bce164f88adf03ada491ea0ba'
visibility_token = nusc.get('sample_annotation', anntoken)['visibility_token']
print("Visibility: {}".format(nusc.get('visibility', visibility_token)))
nusc.render_annotation(anntoken)
anntoken = '9f450bf6b7454551bbbc9a4c6e74ef2e'
visibility_token = nusc.get('sample_annotation', anntoken)['visibility_token']
print("Visibility: {}".format(nusc.get('visibility', visibility_token)))
nusc.render_annotation(anntoken)
nusc.sensor
nusc.sample_data[10]
nusc.calibrated_sensor[0]
nusc.ego_pose[0]
print("Number of `logs` in our loaded database: {}".format(len(nusc.log)))
nusc.log[0]
print("There are {} maps masks in the loaded dataset".format(len(nusc.map)))
nusc.map[0]
nusc.category[0]
cat_token = nusc.category[0]['token']
cat_token
nusc.get('category', cat_token)
nusc.sample_annotation[0]
nusc.get('visibility', nusc.sample_annotation[0]['visibility_token'])
one_instance = nusc.get('instance', nusc.sample_annotation[0]['instance_token'])
one_instance
ann_tokens = nusc.field2token('sample_annotation', 'instance_token', one_instance['token'])
ann_tokens_field2token = set(ann_tokens)
ann_tokens_field2token
ann_record = nusc.get('sample_annotation', one_instance['first_annotation_token'])
ann_record
ann_tokens_traverse = set()
ann_tokens_traverse.add(ann_record['token'])
while not ann_record['next'] == "":
ann_record = nusc.get('sample_annotation', ann_record['next'])
ann_tokens_traverse.add(ann_record['token'])
print(ann_tokens_traverse == ann_tokens_field2token)
catname = nusc.sample_annotation[0]['category_name']
ann_rec = nusc.sample_annotation[0]
inst_rec = nusc.get('instance', ann_rec['instance_token'])
cat_rec = nusc.get('category', inst_rec['category_token'])
print(catname == cat_rec['name'])
# Shortcut
channel = nusc.sample_data[0]['channel']
# No shortcut
sd_rec = nusc.sample_data[0]
cs_record = nusc.get('calibrated_sensor', sd_rec['calibrated_sensor_token'])
sensor_record = nusc.get('sensor', cs_record['sensor_token'])
print(channel == sensor_record['channel'])
nusc.list_categories()
nusc.list_attributes()
nusc.list_scenes()
my_sample = nusc.sample[10]
nusc.render_pointcloud_in_image(my_sample['token'], pointsensor_channel='LIDAR_TOP')
nusc.render_pointcloud_in_image(my_sample['token'], pointsensor_channel='LIDAR_TOP', render_intensity=True)
nusc.render_pointcloud_in_image(my_sample['token'], pointsensor_channel='RADAR_FRONT')
my_sample = nusc.sample[20]
# The rendering command below is commented out because it may crash in notebooks
# nusc.render_sample(my_sample['token'])
nusc.render_sample_data(my_sample['data']['CAM_FRONT'])
nusc.render_sample_data(my_sample['data']['LIDAR_TOP'], nsweeps=5, underlay_map=True)
nusc.render_sample_data(my_sample['data']['RADAR_FRONT'], nsweeps=5, underlay_map=True)
from nuscenes.utils.data_classes import RadarPointCloud
RadarPointCloud.disable_filters()
nusc.render_sample_data(my_sample['data']['RADAR_FRONT'], nsweeps=5, underlay_map=True)
RadarPointCloud.default_filters()
nusc.render_annotation(my_sample['anns'][22])
my_scene_token = nusc.field2token('scene', 'name', 'scene-0061')[0]
# The rendering command below is commented out because it may crash in notebooks
# nusc.render_scene_channel(my_scene_token, 'CAM_FRONT')
# The rendering command below is commented out because it may crash in notebooks
# nusc.render_scene(my_scene_token)
nusc.render_egoposes_on_map(log_location='singapore-onenorth')
| 0.480235 | 0.984124 |
### 練習問題2.27
練習問題2.18のreverse⼿続きを修正し、deep-reverseという⼿続きを書け。
deep-reverseは、ひとつのリストを引数として取り、
要素が逆順で、サブリストもすべて要素が逆順になっているリストを返す⼿続きである。
例えば、次のようになる。
(define x (list (list 1 2) (list 3 4)))
x
((1 2) (3 4))
(reverse x)
((3 4) (1 2))
(deep-reverse x)
((4 3) (2 1))
#### 所感
反復プロセスで解こうとして出来なかった。
引数で処理結果(最終的な出力するリスト)を渡しても、
分岐して再帰手続きの呼び出しになるので、実装が難しいと思われる。
```
; 反復プロセス版
(define (reverse l)
(define (iter result ll)
(if (null? ll) result
(iter (cons (car ll) result) (cdr ll))
)
)
(iter () l)
)
(reverse (list 23 72 149 34))
; (23 (72 (149 (34 nil))))を作るイメージ
; -> これでdeep-reverse手続きを作ることは出来なかった。
; 引数で処理結果を渡しつつ、更新していくことが難しく思えた。
; 再帰プロセスでの実装
; →やってみたが、メモリ的にもステップ数的にも無駄な実装となっている。
; リストから最後の要素を取得する。
(define (get-last l)
(define (iter item l_tmp)
(if (null? l_tmp) item
(iter (car l_tmp) (cdr l_tmp)))
)
(iter () l)
)
(display (get-last (list 23 72 149 34)))
(newline)
; リストから最後の要素を除外したリストを返す。
(define (remove-last l)
(define (iter l1 l2)
(if (null? l2) '()
(cons (car l1) (iter (cdr l1) (cdr l2)))
)
)
(iter l (cdr l))
)
(display (remove-last (list 23 72 149 34)))
(newline)
(define (reverse l)
(define (iter ll)
(if (null? ll) '()
(cons (get-last ll) (iter (remove-last ll)))
)
)
(iter l)
)
(reverse (list 23 72 149 34))
; 再帰プロセス版
(define (deep-reverse l)
(define (iter ll)
(cond ;((null? ll) ()) ; 葉がnilの場合 この条件はいらないらしい。葉を返すに含まれる?
((not (pair? ll)) ll) ; 葉を返す
(else (cons (iter (get-last ll)) (iter (remove-last ll))))
)
)
(iter l)
)
(define x (list (list 1 2) (list 3 4)))
x
(reverse x)
(deep-reverse x)
(define y (list (list (list (list 1 2) (list 3 4)) (list 5 6)) (list (list 7 8) (list 9 10)) (list 11 12)))
y
(reverse y)
(deep-reverse y)
(define z (list (list 1 2) (list 3 4) (list 5 6)))
z
(reverse z)
(deep-reverse z)
```
反復プロセスは以下を参考にした。
https://github.com/reddikih/study-sicp/blob/master/sicp-2.2.org
```
; reverse手続きを真似したプロセス。
; 再帰プロセスとは言えないかも。
(define (deep-reverse2 tree)
(define (iter result t)
(if (null? t) result
(iter (cons (pair-reverse (car t)) result)
(cdr t)
)
)
)
(define (pair-reverse t)
(if (pair? t) (iter '() t) ; ペアはさらにiterを使って逆にする
t
)
)
(iter '() tree)
)
(deep-reverse2 (list 23 72 149 34))
(deep-reverse2 (list (list 1 2) (list 3 4)))
(deep-reverse2 (list 1 (list 2 3 (list 4 5)) (list (list 6 7) (list 8 9))))
; deep-reverse2手続きを1つの手続きで実装しようとした場合。
; →うまく動作しない。
(define (deep-reverse3 tree)
(cond ((null? tree) '())
((not (pair? tree)) tree)
(else (cons (deep-reverse3 (cdr tree)) (deep-reverse3 (car tree))))
)
)
(deep-reverse3 (list 23 72 149 34))
(deep-reverse3 (list (list 1 2) (list 3 4)))
(deep-reverse3 (list 1 (list 2 3 (list 4 5)) (list (list 6 7) (list 8 9))))
```
|
github_jupyter
|
; 反復プロセス版
(define (reverse l)
(define (iter result ll)
(if (null? ll) result
(iter (cons (car ll) result) (cdr ll))
)
)
(iter () l)
)
(reverse (list 23 72 149 34))
; (23 (72 (149 (34 nil))))を作るイメージ
; -> これでdeep-reverse手続きを作ることは出来なかった。
; 引数で処理結果を渡しつつ、更新していくことが難しく思えた。
; 再帰プロセスでの実装
; →やってみたが、メモリ的にもステップ数的にも無駄な実装となっている。
; リストから最後の要素を取得する。
(define (get-last l)
(define (iter item l_tmp)
(if (null? l_tmp) item
(iter (car l_tmp) (cdr l_tmp)))
)
(iter () l)
)
(display (get-last (list 23 72 149 34)))
(newline)
; リストから最後の要素を除外したリストを返す。
(define (remove-last l)
(define (iter l1 l2)
(if (null? l2) '()
(cons (car l1) (iter (cdr l1) (cdr l2)))
)
)
(iter l (cdr l))
)
(display (remove-last (list 23 72 149 34)))
(newline)
(define (reverse l)
(define (iter ll)
(if (null? ll) '()
(cons (get-last ll) (iter (remove-last ll)))
)
)
(iter l)
)
(reverse (list 23 72 149 34))
; 再帰プロセス版
(define (deep-reverse l)
(define (iter ll)
(cond ;((null? ll) ()) ; 葉がnilの場合 この条件はいらないらしい。葉を返すに含まれる?
((not (pair? ll)) ll) ; 葉を返す
(else (cons (iter (get-last ll)) (iter (remove-last ll))))
)
)
(iter l)
)
(define x (list (list 1 2) (list 3 4)))
x
(reverse x)
(deep-reverse x)
(define y (list (list (list (list 1 2) (list 3 4)) (list 5 6)) (list (list 7 8) (list 9 10)) (list 11 12)))
y
(reverse y)
(deep-reverse y)
(define z (list (list 1 2) (list 3 4) (list 5 6)))
z
(reverse z)
(deep-reverse z)
; reverse手続きを真似したプロセス。
; 再帰プロセスとは言えないかも。
(define (deep-reverse2 tree)
(define (iter result t)
(if (null? t) result
(iter (cons (pair-reverse (car t)) result)
(cdr t)
)
)
)
(define (pair-reverse t)
(if (pair? t) (iter '() t) ; ペアはさらにiterを使って逆にする
t
)
)
(iter '() tree)
)
(deep-reverse2 (list 23 72 149 34))
(deep-reverse2 (list (list 1 2) (list 3 4)))
(deep-reverse2 (list 1 (list 2 3 (list 4 5)) (list (list 6 7) (list 8 9))))
; deep-reverse2手続きを1つの手続きで実装しようとした場合。
; →うまく動作しない。
(define (deep-reverse3 tree)
(cond ((null? tree) '())
((not (pair? tree)) tree)
(else (cons (deep-reverse3 (cdr tree)) (deep-reverse3 (car tree))))
)
)
(deep-reverse3 (list 23 72 149 34))
(deep-reverse3 (list (list 1 2) (list 3 4)))
(deep-reverse3 (list 1 (list 2 3 (list 4 5)) (list (list 6 7) (list 8 9))))
| 0.172974 | 0.77535 |
# Convolution Neural Network
CNN allow us to extract the features of the image while maintaining the spatial arrangement of the image.
They use three concepts to create a feature transformation
- **Convolutional layers** multiply kernel value by the image window and optimize the kernel weights over time using gradient descent
- **Pooling layers** describe a window of an image using a single value which is the max or the average of that window
- **Activation layers** squash the values into a range, typically [0,1] or [-1,1]
Params for each type
- **Convolution**: Kernel, Filters, Padding, Stride
- **Pooling**: Max,
- **Activation**: Relu, Sigmoid, ...
```
import numpy as np
import keras
import tensorflow as tf
import imageio
import matplotlib.pyplot as plt
% matplotlib inline
import keras
from keras.models import Sequential
from keras.layers import Dense, Conv2D, MaxPooling2D, Activation, Flatten, Dropout
from keras import backend as K
from keras.optimizers import SGD
from helpers import visualise_conv
```
## Building Intution
### With Convolution
```
cheetah = imageio.imread("img/cheetah.jpg")
plt.imshow(cheetah)
cheetah.shape
model1 = Sequential()
model1.add(Conv2D(1, (3, 3), input_shape=cheetah.shape))
visualise_conv(cheetah, model1)
```
### With Convolution + Activation
```
model2 = Sequential()
model2.add(Conv2D(1, (3, 3), input_shape=cheetah.shape))
model2.add(Activation("relu"))
#visualise_conv(cheetah, model2)
```
### With Convolution + Pooling
```
model3 = Sequential()
model3.add(Conv2D(1, (8, 8), input_shape=cheetah.shape))
model3.add(MaxPooling2D(pool_size=(2,2)))
visualise_conv(cheetah, model3)
```
## Convolution + Activation + Pooling
```
model4 = Sequential()
model4.add(Conv2D(1, (8, 8), input_shape=cheetah.shape))
model4.add(Activation("relu"))
model4.add(MaxPooling2D(pool_size=(2,2)))
visualise_conv(cheetah, model4)
```
## Get Data
```
from keras.datasets import fashion_mnist
from helpers import fashion_mnist_label
(x_train, y_train), (x_test, y_test) = fashion_mnist.load_data()
x_train.shape, y_train.shape, x_test.shape, y_test.shape
batch_size = 128
num_classes = 10
epochs=10
```
**Step 1: Prepare the images and labels**
Convert from 'uint8' to 'float32' and normalise the data to (0,1)
```
# input image dimensions
img_rows, img_cols = 28, 28
K.image_data_format()
x_train_conv = x_train.reshape(x_train.shape[0], img_rows, img_cols, 1)
x_test_conv = x_test.reshape(x_test.shape[0], img_rows, img_cols, 1)
input_shape = (img_rows, img_cols, 1)
x_train_conv = x_train_conv / 255
x_test_conv = x_test_conv / 255
```
Convert class vectors to binary class matrices
```
# convert class vectors to binary class matrices
y_train_class = keras.utils.to_categorical(y_train, num_classes)
y_test_class = keras.utils.to_categorical(y_test, num_classes)
```
## Model 1: Simple Convolution
**Step 2 & 3: Craft the feature transfomation and classifier model **
```
model_simple_conv = Sequential()
model_simple_conv.add(Conv2D(1, (3, 3), activation ="relu", input_shape=(28, 28, 1)))
model_simple_conv.add(Flatten())
model_simple_conv.add(Dense(100, activation='relu'))
model_simple_conv.add(Dense(10, activation='softmax'))
model_simple_conv.summary()
```
**Step 4: Compile and fit the model**
```
model_simple_conv.compile(loss='categorical_crossentropy', optimizer="sgd", metrics=['accuracy'])
tbCallBack = keras.callbacks.TensorBoard(log_dir='logs/simple-conv')
%%time
model_simple_conv.fit(x_train_conv, y_train_class, batch_size=batch_size, epochs=epochs, verbose=1,
validation_data=(x_test_conv, y_test_class),
callbacks=[tbCallBack])
```
**Step 5: Check the performance of the model**
```
score = model_simple_conv.evaluate(x_test_conv, y_test_class, verbose=1)
print('Test loss:', score[0])
print('Test accuracy:', score[1])
```
**Step 6: Make & Visualise the Prediction**
```
model_simple_conv.predict_proba(x_test_conv)[0]
from helpers import plot_prediction
plot_prediction(0, x_test, y_test, x_test_conv, model_simple_conv)
```
## Model 2: Convulation + Max Pooling
**Step 2 & 3: Craft the feature transfomation and classifier model **
```
model_pooling_conv = Sequential()
model_pooling_conv.add(Conv2D(32, kernel_size=(3, 3), activation='relu', input_shape=(28,28,1)))
model_pooling_conv.add(MaxPooling2D(pool_size=(2, 2)))
model_pooling_conv.add(Flatten())
model_pooling_conv.add(Dense(128, activation='relu'))
model_pooling_conv.add(Dense(num_classes, activation='softmax'))
model_pooling_conv.summary()
```
**Step 4: Compile and fit the model**
```
model_pooling_conv.compile(loss='categorical_crossentropy', optimizer="sgd", metrics=['accuracy'])
tbCallBack = keras.callbacks.TensorBoard(log_dir='logs/pooling-conv')
%%time
model_pooling_conv.fit(x_train_conv, y_train_class, batch_size=batch_size, epochs=epochs, verbose=1,
validation_data=(x_test_conv, y_test_class),
callbacks=[tbCallBack])
```
**Step 5: Check the performance of the model**
```
score = model_pooling_conv.evaluate(x_test_conv, y_test_class, verbose=1)
print('Test loss:', score[0])
print('Test accuracy:', score[1])
```
**Step 6: Make & Visualise the Prediction**
```
model_pooling_conv.predict_proba(x_test_conv)[0]
plot_prediction(0, x_test, y_test, x_test_conv, model_pooling_conv)
```
## Model 3: Convulation + Max Pooling + Dropout
**Step 2 & 3: Craft the feature transfomation and classifier model **
```
model_dropout_conv = Sequential()
model_dropout_conv.add(Conv2D(32, kernel_size=(3, 3), activation='relu', input_shape=(28, 28, 1)))
model_dropout_conv.add(Conv2D(64, (3, 3), activation='relu'))
model_dropout_conv.add(MaxPooling2D(pool_size=(2, 2)))
model_dropout_conv.add(Dropout(0.25))
model_dropout_conv.add(Flatten())
model_dropout_conv.add(Dense(128, activation='relu'))
model_dropout_conv.add(Dropout(0.5))
model_dropout_conv.add(Dense(num_classes, activation='softmax'))
model_dropout_conv.summary()
model_dropout_conv.compile(loss='categorical_crossentropy', optimizer="sgd", metrics=['accuracy'])
tbCallBack = keras.callbacks.TensorBoard(log_dir='logs/dropout-conv')
%%time
model_dropout_conv.fit(x_train_conv, y_train_class, batch_size=batch_size, epochs=epochs, verbose=1,
validation_data=(x_test_conv, y_test_class),
callbacks=[tbCallBack])
```
**Step 5: Check the performance of the model**
```
score = model_dropout_conv.evaluate(x_test_conv, y_test_class, verbose=1)
print('Test loss:', score[0])
print('Test accuracy:', score[1])
```
**Step 6: Make & Visualise the Prediction**
```
model_dropout_conv.predict_proba(x_test_conv)[0]
plot_prediction(0, x_test, y_test, x_test_conv, model_dropout_conv)
```
|
github_jupyter
|
import numpy as np
import keras
import tensorflow as tf
import imageio
import matplotlib.pyplot as plt
% matplotlib inline
import keras
from keras.models import Sequential
from keras.layers import Dense, Conv2D, MaxPooling2D, Activation, Flatten, Dropout
from keras import backend as K
from keras.optimizers import SGD
from helpers import visualise_conv
cheetah = imageio.imread("img/cheetah.jpg")
plt.imshow(cheetah)
cheetah.shape
model1 = Sequential()
model1.add(Conv2D(1, (3, 3), input_shape=cheetah.shape))
visualise_conv(cheetah, model1)
model2 = Sequential()
model2.add(Conv2D(1, (3, 3), input_shape=cheetah.shape))
model2.add(Activation("relu"))
#visualise_conv(cheetah, model2)
model3 = Sequential()
model3.add(Conv2D(1, (8, 8), input_shape=cheetah.shape))
model3.add(MaxPooling2D(pool_size=(2,2)))
visualise_conv(cheetah, model3)
model4 = Sequential()
model4.add(Conv2D(1, (8, 8), input_shape=cheetah.shape))
model4.add(Activation("relu"))
model4.add(MaxPooling2D(pool_size=(2,2)))
visualise_conv(cheetah, model4)
from keras.datasets import fashion_mnist
from helpers import fashion_mnist_label
(x_train, y_train), (x_test, y_test) = fashion_mnist.load_data()
x_train.shape, y_train.shape, x_test.shape, y_test.shape
batch_size = 128
num_classes = 10
epochs=10
# input image dimensions
img_rows, img_cols = 28, 28
K.image_data_format()
x_train_conv = x_train.reshape(x_train.shape[0], img_rows, img_cols, 1)
x_test_conv = x_test.reshape(x_test.shape[0], img_rows, img_cols, 1)
input_shape = (img_rows, img_cols, 1)
x_train_conv = x_train_conv / 255
x_test_conv = x_test_conv / 255
# convert class vectors to binary class matrices
y_train_class = keras.utils.to_categorical(y_train, num_classes)
y_test_class = keras.utils.to_categorical(y_test, num_classes)
model_simple_conv = Sequential()
model_simple_conv.add(Conv2D(1, (3, 3), activation ="relu", input_shape=(28, 28, 1)))
model_simple_conv.add(Flatten())
model_simple_conv.add(Dense(100, activation='relu'))
model_simple_conv.add(Dense(10, activation='softmax'))
model_simple_conv.summary()
model_simple_conv.compile(loss='categorical_crossentropy', optimizer="sgd", metrics=['accuracy'])
tbCallBack = keras.callbacks.TensorBoard(log_dir='logs/simple-conv')
%%time
model_simple_conv.fit(x_train_conv, y_train_class, batch_size=batch_size, epochs=epochs, verbose=1,
validation_data=(x_test_conv, y_test_class),
callbacks=[tbCallBack])
score = model_simple_conv.evaluate(x_test_conv, y_test_class, verbose=1)
print('Test loss:', score[0])
print('Test accuracy:', score[1])
model_simple_conv.predict_proba(x_test_conv)[0]
from helpers import plot_prediction
plot_prediction(0, x_test, y_test, x_test_conv, model_simple_conv)
model_pooling_conv = Sequential()
model_pooling_conv.add(Conv2D(32, kernel_size=(3, 3), activation='relu', input_shape=(28,28,1)))
model_pooling_conv.add(MaxPooling2D(pool_size=(2, 2)))
model_pooling_conv.add(Flatten())
model_pooling_conv.add(Dense(128, activation='relu'))
model_pooling_conv.add(Dense(num_classes, activation='softmax'))
model_pooling_conv.summary()
model_pooling_conv.compile(loss='categorical_crossentropy', optimizer="sgd", metrics=['accuracy'])
tbCallBack = keras.callbacks.TensorBoard(log_dir='logs/pooling-conv')
%%time
model_pooling_conv.fit(x_train_conv, y_train_class, batch_size=batch_size, epochs=epochs, verbose=1,
validation_data=(x_test_conv, y_test_class),
callbacks=[tbCallBack])
score = model_pooling_conv.evaluate(x_test_conv, y_test_class, verbose=1)
print('Test loss:', score[0])
print('Test accuracy:', score[1])
model_pooling_conv.predict_proba(x_test_conv)[0]
plot_prediction(0, x_test, y_test, x_test_conv, model_pooling_conv)
model_dropout_conv = Sequential()
model_dropout_conv.add(Conv2D(32, kernel_size=(3, 3), activation='relu', input_shape=(28, 28, 1)))
model_dropout_conv.add(Conv2D(64, (3, 3), activation='relu'))
model_dropout_conv.add(MaxPooling2D(pool_size=(2, 2)))
model_dropout_conv.add(Dropout(0.25))
model_dropout_conv.add(Flatten())
model_dropout_conv.add(Dense(128, activation='relu'))
model_dropout_conv.add(Dropout(0.5))
model_dropout_conv.add(Dense(num_classes, activation='softmax'))
model_dropout_conv.summary()
model_dropout_conv.compile(loss='categorical_crossentropy', optimizer="sgd", metrics=['accuracy'])
tbCallBack = keras.callbacks.TensorBoard(log_dir='logs/dropout-conv')
%%time
model_dropout_conv.fit(x_train_conv, y_train_class, batch_size=batch_size, epochs=epochs, verbose=1,
validation_data=(x_test_conv, y_test_class),
callbacks=[tbCallBack])
score = model_dropout_conv.evaluate(x_test_conv, y_test_class, verbose=1)
print('Test loss:', score[0])
print('Test accuracy:', score[1])
model_dropout_conv.predict_proba(x_test_conv)[0]
plot_prediction(0, x_test, y_test, x_test_conv, model_dropout_conv)
| 0.823683 | 0.976535 |
```
! pip install -q kaggle
from google.colab import files
files.upload()
! mkdir ~/.kaggle
! cp kaggle.json ~/.kaggle/
! chmod 600 ~/.kaggle/kaggle.json
!kaggle datasets download -d puneet6060/intel-image-classification
!unzip /content/intel-image-classification.zip
import cv2
import os
import matplotlib.pyplot as plt
from keras.preprocessing.image import ImageDataGenerator
from keras.initializers import glorot_uniform
from keras.layers import Input, Add, Dense, Activation, BatchNormalization, Flatten, Conv2D, GlobalAveragePooling2D, MaxPool2D
from keras.models import Model
w = 10
h = 10
fig = plt.figure(figsize=(15,10))
columns = 6
rows = 1
fielName = "/content/seg_train/seg_train"
for i in range(0, columns*rows ):
folderName = os.path.join((fielName), os.listdir(fielName)[i])
img = cv2.imread(folderName+'/'+(os.listdir(os.path.join((fielName), os.listdir(fielName)[i]))[i]))
fig.add_subplot(rows, columns, i+1)
plt.axis("off")
plt.imshow(cv2.cvtColor(img, cv2.COLOR_BGR2RGB))
plt.title(os.path.basename(folderName))
plt.show()
batch_size = 32
resize = (224, 224)
train_datagen = ImageDataGenerator(
rescale=1./255,
featurewise_std_normalization=True)
validation_datagen = ImageDataGenerator(
rescale=1./255,
featurewise_std_normalization=True)
train_generator = train_datagen.flow_from_directory(
'/content/seg_train/seg_train',
target_size=resize,
batch_size=batch_size,
class_mode='categorical')
validation_generator = validation_datagen.flow_from_directory(
'/content/seg_test/seg_test',
target_size=resize,
batch_size=batch_size,
class_mode='categorical')
%pip install wandb -q
import wandb
from wandb.keras import WandbCallback
from keras.callbacks import LambdaCallback
!wandb login
wandb.init(project="ResNet", entity="manar")
input = Input((224,224,3))
X = Conv2D(64, (7,7), (2,2), padding="same", kernel_initializer=glorot_uniform(seed=0))(input)
X = BatchNormalization(axis=3)(X)
X = Activation("relu")(X)
X = MaxPool2D((3,3), (2,2),padding="same")(X)
def layer(itr, input, filters, stride):
f1,f2,f3 = filters
X = Conv2D(f1, (1,1), stride, padding="valid", kernel_initializer=glorot_uniform(seed=0))(input)
X = BatchNormalization(axis=3)(X)
X = Activation("relu")(X)
X = Conv2D(f2, (3,3), (1,1), padding="same", kernel_initializer=glorot_uniform(seed=0))(X)
X = BatchNormalization(axis=3)(X)
X = Activation("relu")(X)
X = Conv2D(f3, (1,1), (1,1), padding="valid", kernel_initializer=glorot_uniform(seed=0))(X)
X = BatchNormalization(axis=3)(X)
X = Activation("relu")(X)
block = Conv2D(f3, (1,1), stride, padding="valid", kernel_initializer=glorot_uniform(seed=0))(input)
block = BatchNormalization(axis=3)(block)
X = Add()([X, block])
X = Activation("relu")(X)
for iteration in range(itr-1):
X = Conv2D(f1, (1,1), (1,1), padding="valid", kernel_initializer=glorot_uniform(seed=0))(X)
X = BatchNormalization(axis=3)(X)
X = Activation("relu")(X)
X = Conv2D(f2, (3,3), (1,1), padding="same", kernel_initializer=glorot_uniform(seed=0))(X)
X = BatchNormalization(axis=3)(X)
X = Activation("relu")(X)
X = Conv2D(f3, (1,1), (1,1), padding="valid", kernel_initializer=glorot_uniform(seed=0))(X)
X = BatchNormalization(axis=3)(X)
X = Add()([X, block])
X = Activation("relu")(X)
return X
X = layer(3, X, filters=[64,64,256], stride=(1,1))
X = layer(4, X, filters=[128,128,512], stride=(2,2))
X = layer(6, X, filters=[256,256,1024], stride=(2,2))
X = layer(3, X, filters=[512,512,2048], stride=(2,2))
X = GlobalAveragePooling2D()(X)
process_model = Model(input, X)
result_model = process_model.output
result_model = Flatten()(result_model)
result_model = Dense(1000)(result_model)
result_model = Activation("relu")(result_model)
result_model = Dense(6)(result_model)
result_model = Activation("softmax")(result_model)
model = Model(inputs=process_model.input, outputs=result_model)
model.summary()
model.compile(optimizer="adam", loss="categorical_crossentropy", metrics=["accuracy"])
model.fit(train_generator, validation_data=validation_generator, epochs=10, callbacks=[WandbCallback()])
```
|
github_jupyter
|
! pip install -q kaggle
from google.colab import files
files.upload()
! mkdir ~/.kaggle
! cp kaggle.json ~/.kaggle/
! chmod 600 ~/.kaggle/kaggle.json
!kaggle datasets download -d puneet6060/intel-image-classification
!unzip /content/intel-image-classification.zip
import cv2
import os
import matplotlib.pyplot as plt
from keras.preprocessing.image import ImageDataGenerator
from keras.initializers import glorot_uniform
from keras.layers import Input, Add, Dense, Activation, BatchNormalization, Flatten, Conv2D, GlobalAveragePooling2D, MaxPool2D
from keras.models import Model
w = 10
h = 10
fig = plt.figure(figsize=(15,10))
columns = 6
rows = 1
fielName = "/content/seg_train/seg_train"
for i in range(0, columns*rows ):
folderName = os.path.join((fielName), os.listdir(fielName)[i])
img = cv2.imread(folderName+'/'+(os.listdir(os.path.join((fielName), os.listdir(fielName)[i]))[i]))
fig.add_subplot(rows, columns, i+1)
plt.axis("off")
plt.imshow(cv2.cvtColor(img, cv2.COLOR_BGR2RGB))
plt.title(os.path.basename(folderName))
plt.show()
batch_size = 32
resize = (224, 224)
train_datagen = ImageDataGenerator(
rescale=1./255,
featurewise_std_normalization=True)
validation_datagen = ImageDataGenerator(
rescale=1./255,
featurewise_std_normalization=True)
train_generator = train_datagen.flow_from_directory(
'/content/seg_train/seg_train',
target_size=resize,
batch_size=batch_size,
class_mode='categorical')
validation_generator = validation_datagen.flow_from_directory(
'/content/seg_test/seg_test',
target_size=resize,
batch_size=batch_size,
class_mode='categorical')
%pip install wandb -q
import wandb
from wandb.keras import WandbCallback
from keras.callbacks import LambdaCallback
!wandb login
wandb.init(project="ResNet", entity="manar")
input = Input((224,224,3))
X = Conv2D(64, (7,7), (2,2), padding="same", kernel_initializer=glorot_uniform(seed=0))(input)
X = BatchNormalization(axis=3)(X)
X = Activation("relu")(X)
X = MaxPool2D((3,3), (2,2),padding="same")(X)
def layer(itr, input, filters, stride):
f1,f2,f3 = filters
X = Conv2D(f1, (1,1), stride, padding="valid", kernel_initializer=glorot_uniform(seed=0))(input)
X = BatchNormalization(axis=3)(X)
X = Activation("relu")(X)
X = Conv2D(f2, (3,3), (1,1), padding="same", kernel_initializer=glorot_uniform(seed=0))(X)
X = BatchNormalization(axis=3)(X)
X = Activation("relu")(X)
X = Conv2D(f3, (1,1), (1,1), padding="valid", kernel_initializer=glorot_uniform(seed=0))(X)
X = BatchNormalization(axis=3)(X)
X = Activation("relu")(X)
block = Conv2D(f3, (1,1), stride, padding="valid", kernel_initializer=glorot_uniform(seed=0))(input)
block = BatchNormalization(axis=3)(block)
X = Add()([X, block])
X = Activation("relu")(X)
for iteration in range(itr-1):
X = Conv2D(f1, (1,1), (1,1), padding="valid", kernel_initializer=glorot_uniform(seed=0))(X)
X = BatchNormalization(axis=3)(X)
X = Activation("relu")(X)
X = Conv2D(f2, (3,3), (1,1), padding="same", kernel_initializer=glorot_uniform(seed=0))(X)
X = BatchNormalization(axis=3)(X)
X = Activation("relu")(X)
X = Conv2D(f3, (1,1), (1,1), padding="valid", kernel_initializer=glorot_uniform(seed=0))(X)
X = BatchNormalization(axis=3)(X)
X = Add()([X, block])
X = Activation("relu")(X)
return X
X = layer(3, X, filters=[64,64,256], stride=(1,1))
X = layer(4, X, filters=[128,128,512], stride=(2,2))
X = layer(6, X, filters=[256,256,1024], stride=(2,2))
X = layer(3, X, filters=[512,512,2048], stride=(2,2))
X = GlobalAveragePooling2D()(X)
process_model = Model(input, X)
result_model = process_model.output
result_model = Flatten()(result_model)
result_model = Dense(1000)(result_model)
result_model = Activation("relu")(result_model)
result_model = Dense(6)(result_model)
result_model = Activation("softmax")(result_model)
model = Model(inputs=process_model.input, outputs=result_model)
model.summary()
model.compile(optimizer="adam", loss="categorical_crossentropy", metrics=["accuracy"])
model.fit(train_generator, validation_data=validation_generator, epochs=10, callbacks=[WandbCallback()])
| 0.553023 | 0.30047 |
```
import numpy as np #linear algebra
import pandas as pd #data preprocessing, csv files I/O
matches = pd.read_csv("matches.csv")
matches.info()
matches.head()
matches.head(7)
matches[pd.isnull(matches['winner'])]
#find all the NaN values in winner column, so that we update this as draw
matches['winner'].fillna('Draw', inplace=True)
matches.loc[241,'winner']
matches['team1'].unique()
matches.replace(['Kolkata Knight Riders', 'Chennai Super Kings', 'Rajasthan Royals',
'Mumbai Indians', 'Deccan Chargers', 'Kings XI Punjab',
'Royal Challengers Bangalore', 'Delhi Daredevils',
'Kochi Tuskers Kerala', 'Pune Warriors', 'Sunrisers Hyderabad',
'Rising Pune Supergiants', 'Gujarat Lions']
,['KKR','CSK','RR','MI','DC','KXIP','RCB','DD','KTK','PW','SRH','RPS','GL'],inplace=True)
matches.head()
encode = {'team1' : {'KKR':1,'CSK':2,'RR':3,'MI':4,'DC':5,'KXIP':6,'RCB':7,'DD':8,'KTK':9,'PW':10,'SRH':11,'RPS':12,'GL':13},
'team2' : {'KKR':1,'CSK':2,'RR':3,'MI':4,'DC':5,'KXIP':6,'RCB':7,'DD':8,'KTK':9,'PW':10,'SRH':11,'RPS':12,'GL':13},
'toss_winner' :{'KKR':1,'CSK':2,'RR':3,'MI':4,'DC':5,'KXIP':6,'RCB':7,'DD':8,'KTK':9,'PW':10,'SRH':11,'RPS':12,'GL':13},
'winner' : {'KKR':1,'CSK':2,'RR':3,'MI':4,'DC':5,'KXIP':6,'RCB':7,'DD':8,'KTK':9,'PW':10,'SRH':11,'RPS':12,'GL':13,'Draw':14}}
matches.replace(encode,inplace=True)
matches.head()
# Find cities which are null?
matches[pd.isnull(matches['city'])]
# remove any null values, winner has hence fill the null value in winner as draw
# city is also null, this mainly for dubai stadium. hence update the city as dubai
# Make sure to impute the data(cleaning and finding missing data), there is also other process
# to verify excepted value based on other resultants, for now by stadium, city is easily manually updated
matches['city'].fillna('Dubai',inplace=True)
matches.describe()
matches.info()
# team_match_total = matches.groupby(["team1"]).size()+matches.groupby(["team2"]).size()
# print(team_match_total.get_value(1))
#xx = matches.groupby(["toss_winner"]).size()
#print(xx.get_value(1))
#yy = matches.groupby(["winner"]).size()
#print(yy.get_value(1))
# we maintain a dictionary for future reference mapping teams
dicval = encode['winner']
print(dicval['MI']) # key value
print(list(dicval.keys())[list(dicval.values()).index(4)]) #find key by value search
matches = matches[['team1', 'team2','city','toss_decision','toss_winner','venue','winner']]
matches.head()
df = pd.DataFrame(matches)
df.describe()
```
|
github_jupyter
|
import numpy as np #linear algebra
import pandas as pd #data preprocessing, csv files I/O
matches = pd.read_csv("matches.csv")
matches.info()
matches.head()
matches.head(7)
matches[pd.isnull(matches['winner'])]
#find all the NaN values in winner column, so that we update this as draw
matches['winner'].fillna('Draw', inplace=True)
matches.loc[241,'winner']
matches['team1'].unique()
matches.replace(['Kolkata Knight Riders', 'Chennai Super Kings', 'Rajasthan Royals',
'Mumbai Indians', 'Deccan Chargers', 'Kings XI Punjab',
'Royal Challengers Bangalore', 'Delhi Daredevils',
'Kochi Tuskers Kerala', 'Pune Warriors', 'Sunrisers Hyderabad',
'Rising Pune Supergiants', 'Gujarat Lions']
,['KKR','CSK','RR','MI','DC','KXIP','RCB','DD','KTK','PW','SRH','RPS','GL'],inplace=True)
matches.head()
encode = {'team1' : {'KKR':1,'CSK':2,'RR':3,'MI':4,'DC':5,'KXIP':6,'RCB':7,'DD':8,'KTK':9,'PW':10,'SRH':11,'RPS':12,'GL':13},
'team2' : {'KKR':1,'CSK':2,'RR':3,'MI':4,'DC':5,'KXIP':6,'RCB':7,'DD':8,'KTK':9,'PW':10,'SRH':11,'RPS':12,'GL':13},
'toss_winner' :{'KKR':1,'CSK':2,'RR':3,'MI':4,'DC':5,'KXIP':6,'RCB':7,'DD':8,'KTK':9,'PW':10,'SRH':11,'RPS':12,'GL':13},
'winner' : {'KKR':1,'CSK':2,'RR':3,'MI':4,'DC':5,'KXIP':6,'RCB':7,'DD':8,'KTK':9,'PW':10,'SRH':11,'RPS':12,'GL':13,'Draw':14}}
matches.replace(encode,inplace=True)
matches.head()
# Find cities which are null?
matches[pd.isnull(matches['city'])]
# remove any null values, winner has hence fill the null value in winner as draw
# city is also null, this mainly for dubai stadium. hence update the city as dubai
# Make sure to impute the data(cleaning and finding missing data), there is also other process
# to verify excepted value based on other resultants, for now by stadium, city is easily manually updated
matches['city'].fillna('Dubai',inplace=True)
matches.describe()
matches.info()
# team_match_total = matches.groupby(["team1"]).size()+matches.groupby(["team2"]).size()
# print(team_match_total.get_value(1))
#xx = matches.groupby(["toss_winner"]).size()
#print(xx.get_value(1))
#yy = matches.groupby(["winner"]).size()
#print(yy.get_value(1))
# we maintain a dictionary for future reference mapping teams
dicval = encode['winner']
print(dicval['MI']) # key value
print(list(dicval.keys())[list(dicval.values()).index(4)]) #find key by value search
matches = matches[['team1', 'team2','city','toss_decision','toss_winner','venue','winner']]
matches.head()
df = pd.DataFrame(matches)
df.describe()
| 0.094803 | 0.15219 |
> 原文地址 [blog.csdn.net](https://blog.csdn.net/lys\_828/article/details/106489371)
### 利用 FuzzyWuzzy 库匹配字符串
* [1\. 背景前言](#1__1)
* [2\. FuzzyWuzzy 库介绍](#2_FuzzyWuzzy_7)
* * [2.1 安装](#21__8)
* [2.1 fuzz 模块](#21_fuzz_17)
* * [2.1.1 简单匹配(Ratio)](#211_Ratio_22)
* [2.1.2 非完全匹配(Partial Ratio)](#212_Partial_Ratio_31)
* [2.1.3 忽略顺序匹配(Token Sort Ratio)](#213_Token_Sort_Ratio_40)
* [2.1.4 去重子集匹配(Token Set Ratio)](#214_Token_Set_Ratio_53)
* [2.2 process 模块](#22_process_67)
* * [2.2.1 extract 提取多条数据](#221_extract_69)
* [2.2.2 extractOne 提取一条数据](#222_extractOne_78)
* [3\. 实战应用](#3__87)
* * [3.1 公司名称字段模糊匹配](#31__89)
* * [3.1.1 参数讲解:](#311__94)
* [3.1.2 核心代码讲解](#312__109)
* [3.2 省份字段模糊匹配](#32__132)
* [4\. 全部函数代码](#4__136)
1\. 背景前言
========
在处理数据的过程中,难免会遇到下面类似的场景,自己手里头获得的是简化版的数据字段,但是要比对的或者要合并的却是完整版的数据(有时候也会反过来)
最常见的一个例子就是:在进行地理可视化中,自己收集的数据只保留的缩写,比如北京,广西,新疆,西藏等,但是待匹配的字段数据却是北京市,广西壮族自治区,新疆维吾尔自治区,西藏自治区等,如下。因此就需要有没有一种方式可以很快速便捷的直接进行对应字段的匹配并将结果单独生成一列,就可以用到 FuzzyWuzzy 库
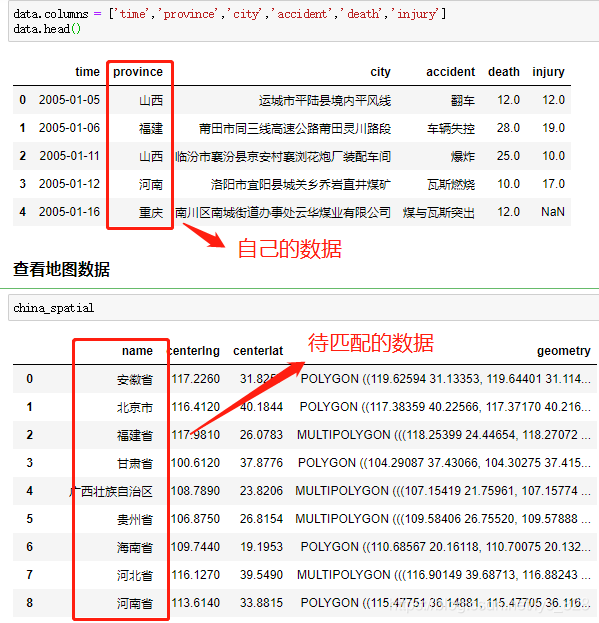
2\. FuzzyWuzzy 库介绍
==================
2.1 安装
------
这里使用的是 Anaconda 下的 jupyter notebook 编程环境,因此在 Anaconda 的命令行中输入一下指令进行第三方库安装
```
pip install -i https://pypi.tuna.tsinghua.edu.cn/simple FuzzyWuzzy
```
→ 输出的结果为:(如果使用本地的 python,可以直接 cmd 后安装)
2.1 fuzz 模块
-----------
该模块下主要介绍四个函数(方法),分别为:简单匹配(Ratio)、非完全匹配(Partial Ratio)、忽略顺序匹配(Token Sort Ratio)和去重子集匹配(Token Set Ratio)
**注意,注意:** 如果直接导入这个模块的话,系统会提示 warning,当然这不代表报错,程序依旧可以运行(使用的默认算法,执行速度较慢),可以按照系统的提示安装 [python-Levenshtein 库](https://www.lfd.uci.edu/~gohlke/pythonlibs/#python-levenshtein)进行辅助,这有利于提高计算的速度
### 2.1.1 简单匹配(Ratio)
简单的了解一下就行,这个不怎么精确,也不常用
```
fuzz.ratio("河南省", "河南省")
>>> 100
>
fuzz.ratio("河南", "河南省")
>>> 80
```
### 2.1.2 非完全匹配(Partial Ratio)
尽量使用非完全匹配,精度较高
```
fuzz.partial\_ratio("河南省", "河南省")
>>> 100
fuzz.partial\_ratio("河南", "河南省")
>>> 100
```
### 2.1.3 忽略顺序匹配(Token Sort Ratio)
原理在于:以 **空格** 为分隔符,**小写** 化所有字母,无视空格外的其它标点符号
```
fuzz.ratio("西藏 自治区", "自治区 西藏")
>>> 50
fuzz.ratio('I love YOU','YOU LOVE I')
>>> 30
fuzz.token\_sort\_ratio("西藏 自治区", "自治区 西藏")
>>> 100
fuzz.token\_sort\_ratio('I love YOU','YOU LOVE I')
>>> 100
```
### 2.1.4 去重子集匹配(Token Set Ratio)
相当于比对之前有一个集合去重的过程,注意最后两个,可理解为该方法是在 token\_sort\_ratio 方法的基础上添加了集合去重的功能,下面三个匹配的都是倒序
```
fuzz.ratio("西藏 西藏 自治区", "自治区 西藏")
>>> 40
fuzz.token\_sort\_ratio("西藏 西藏 自治区", "自治区 西藏")
>>> 80
fuzz.token\_set\_ratio("西藏 西藏 自治区", "自治区 西藏")
>>> 100
```
fuzz 这几个 ratio() 函数(方法)最后得到的结果都是数字,如果需要获得匹配度最高的字符串结果,还需要依旧自己的数据类型选择不同的函数,然后再进行结果提取,如果但看文本数据的匹配程度使用这种方式是可以量化的,但是对于我们要提取匹配的结果来说就不是很方便了,因此就有了 process 模块
2.2 process 模块
--------------
用于处理备选答案有限的情况,返回模糊匹配的字符串和相似度。
### 2.2.1 extract 提取多条数据
类似于爬虫中 select,返回的是列表,其中会包含很多匹配的数据
```
choices = \["河南省", "郑州市", "湖北省", "武汉市"\]
process.extract("郑州", choices, limit=2)
>>> \[('郑州市', 90), ('河南省', 0)\]
# extract之后的数据类型是列表,即使limit=1,最后还是列表,注意和下面extractOne的区别
```
### 2.2.2 extractOne 提取一条数据
如果要提取匹配度最大的结果,可以使用 extractOne,注意这里返回的是 **元组** 类型, 还有就是匹配度最大的结果**不一定是我们想要的数据**,可以通过下面的示例和两个实战应用体会一下
```
process.extractOne("郑州", choices)
>>> ('郑州市', 90)
process.extractOne("北京", choices)
>>> ('湖北省', 45)
```
3\. 实战应用
========
这里举两个实战应用的小例子,第一个是公司名称字段的模糊匹配,第二个是省市字段的模糊匹配
3.1 公司名称字段模糊匹配
--------------
数据及待匹配的数据样式如下:自己获取到的数据字段的名称很简洁,并不是公司的全称,因此需要进行两个字段的合并
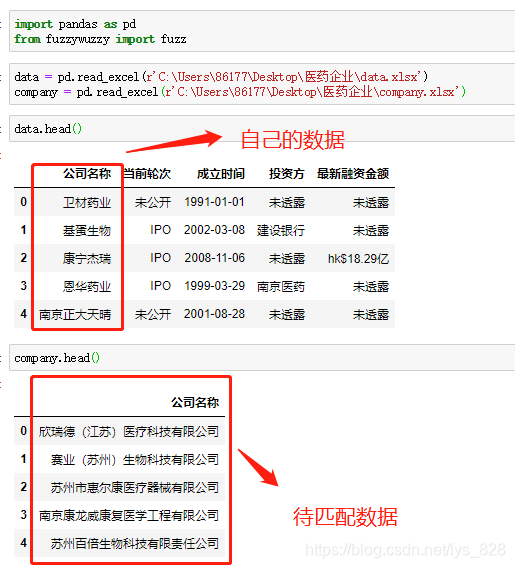
直接将代码封装为函数,主要是为了方便日后的调用,这里参数设置的比较详细,执行结果如下:
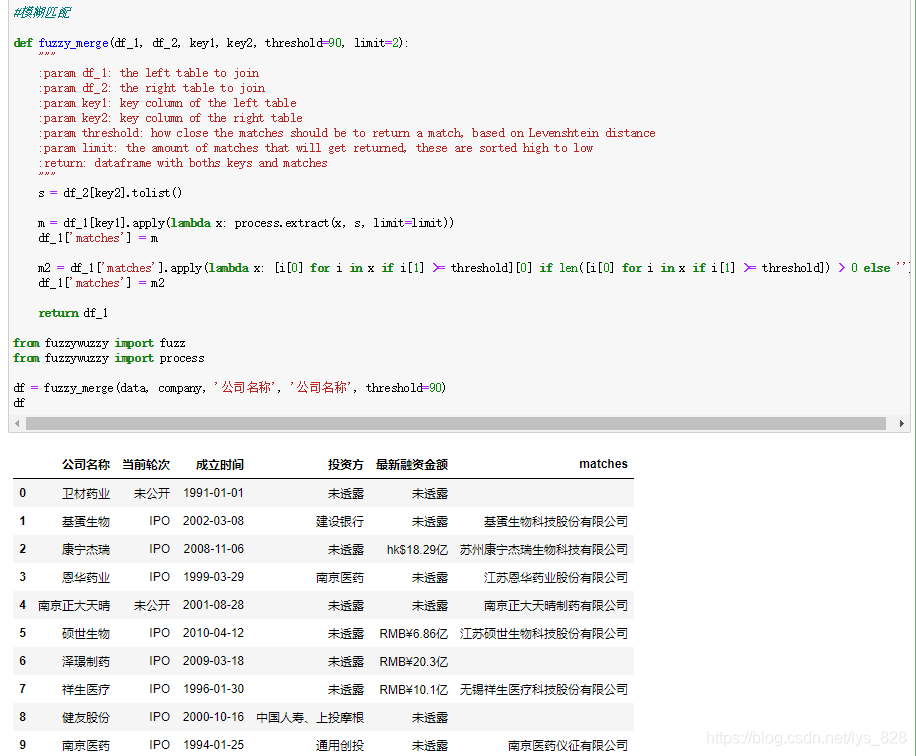
### 3.1.1 参数讲解:
① 第一个参数 df\_1 是自己获取的欲合并的左侧数据(这里是 data 变量);
② 第二个参数 df\_2 是待匹配的欲合并的右侧数据(这里是 company 变量);
③ 第三个参数 key1 是 df\_1 中要处理的字段名称(这里是 data 变量里的‘公司名称’字段)
④ 第四个参数 key2 是 df\_2 中要匹配的字段名称(这里是 company 变量里的‘公司名称’字段)
⑤ 第五个参数 threshold 是设定提取结果匹配度的标准。注意这里就是对 extractOne 方法的完善,提取到的最大匹配度的结果并不一定是我们需要的,所以需要设定一个阈值来评判,这个值就为 90,只有是大于等于 90,这个匹配结果我们才可以接受
⑥ 第六个参数,默认参数就是只返回两个匹配成功的结果
⑦ 返回值:为 df\_1 添加‘matches’字段后的新的 DataFrame 数据
### 3.1.2 核心代码讲解
第一部分代码如下,可以参考上面讲解 process.extract 方法,这里就是直接使用,所以返回的结果 m 就是列表中嵌套元祖的数据格式,样式为: \[(‘郑州市’, 90), (‘河南省’, 0)\],因此第一次写入到’matches’字段中的数据也就是这种格式
**注意,注意:** 元祖中的第一个是匹配成功的字符串,第二个就是设置的 threshold 参数比对的数字对象
```
s = df\_2\[key2\].tolist()
m = df\_1\[key1\].apply(lambda x: process.extract(x, s, limit=limit))
df\_1\['matches'\] = m
```
第二部分的核心代码如下,有了上面的梳理,明确了‘matches’字段中的数据类型,然后就是进行数据的提取了,需要处理的部分有两点需要注意的:
① 提取匹配成功的字符串,并对阈值小于 90 的数据填充空值
② 最后把数据添加到‘matches’字段
```
m2 = df\_1\['matches'\].apply(lambda x: \[i\[0\] for i in x if i\[1\] >= threshold\]\[0\] if len(\[i\[0\] for i in x if i\[1\] >= threshold\]) > 0 else '')
#要理解第一个‘matches’字段返回的数据类型是什么样子的,就不难理解这行代码了
#参考一下这个格式: \[('郑州市', 90), ('河南省', 0)\]
df\_1\['matches'\] = m2
return df\_1
```
3.2 省份字段模糊匹配
------------
自己的数据和待匹配的数据背景介绍中已经有图片显示了,上面也已经封装了模糊匹配的函数,这里直接调用上面的函数,输入相应的参数即可,代码以及执行结果如下:
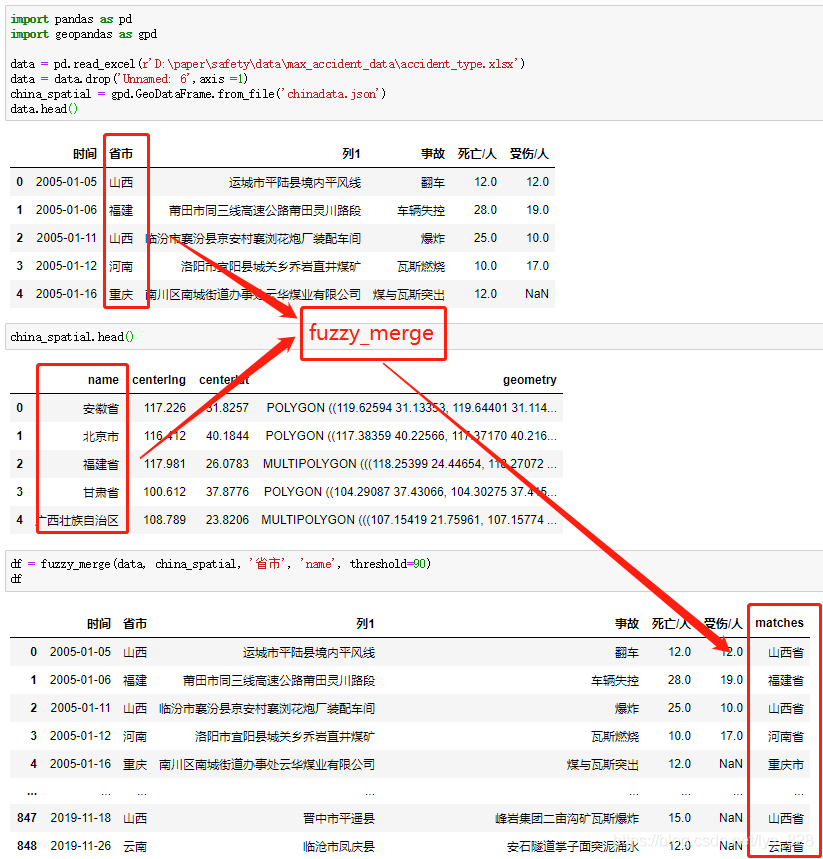
数据处理完成,经过封装后的函数可以直接放在自己自定义的模块名文件下面,以后可以方便直接导入函数名即可,可以参考[将自定义常用的一些函数封装成可以直接调用的模块方法](https://blog.csdn.net/lys_828/article/details/106176229)
4\. 全部函数代码
==========
```
#模糊匹配
def fuzzy\_merge(df\_1, df\_2, key1, key2, threshold=90, limit=2):
"""
:param df\_1: the left table to join
:param df\_2: the right table to join
:param key1: key column of the left table
:param key2: key column of the right table
:param threshold: how close the matches should be to return a match, based on Levenshtein distance
:param limit: the amount of matches that will get returned, these are sorted high to low
:return: dataframe with boths keys and matches
"""
s = df\_2\[key2\].tolist()
m = df\_1\[key1\].apply(lambda x: process.extract(x, s, limit=limit))
df\_1\['matches'\] = m
m2 = df\_1\['matches'\].apply(lambda x: \[i\[0\] for i in x if i\[1\] >= threshold\]\[0\] if len(\[i\[0\] for i in x if i\[1\] >= threshold\]) > 0 else '')
df\_1\['matches'\] = m2
return df\_1
from fuzzywuzzy import fuzz
from fuzzywuzzy import process
df = fuzzy\_merge(data, company, '公司名称', '公司名称', threshold=90)
df
```
|
github_jupyter
|
pip install -i https://pypi.tuna.tsinghua.edu.cn/simple FuzzyWuzzy
fuzz.ratio("河南省", "河南省")
>>> 100
>
fuzz.ratio("河南", "河南省")
>>> 80
fuzz.partial\_ratio("河南省", "河南省")
>>> 100
fuzz.partial\_ratio("河南", "河南省")
>>> 100
fuzz.ratio("西藏 自治区", "自治区 西藏")
>>> 50
fuzz.ratio('I love YOU','YOU LOVE I')
>>> 30
fuzz.token\_sort\_ratio("西藏 自治区", "自治区 西藏")
>>> 100
fuzz.token\_sort\_ratio('I love YOU','YOU LOVE I')
>>> 100
fuzz.ratio("西藏 西藏 自治区", "自治区 西藏")
>>> 40
fuzz.token\_sort\_ratio("西藏 西藏 自治区", "自治区 西藏")
>>> 80
fuzz.token\_set\_ratio("西藏 西藏 自治区", "自治区 西藏")
>>> 100
choices = \["河南省", "郑州市", "湖北省", "武汉市"\]
process.extract("郑州", choices, limit=2)
>>> \[('郑州市', 90), ('河南省', 0)\]
# extract之后的数据类型是列表,即使limit=1,最后还是列表,注意和下面extractOne的区别
process.extractOne("郑州", choices)
>>> ('郑州市', 90)
process.extractOne("北京", choices)
>>> ('湖北省', 45)
s = df\_2\[key2\].tolist()
m = df\_1\[key1\].apply(lambda x: process.extract(x, s, limit=limit))
df\_1\['matches'\] = m
m2 = df\_1\['matches'\].apply(lambda x: \[i\[0\] for i in x if i\[1\] >= threshold\]\[0\] if len(\[i\[0\] for i in x if i\[1\] >= threshold\]) > 0 else '')
#要理解第一个‘matches’字段返回的数据类型是什么样子的,就不难理解这行代码了
#参考一下这个格式: \[('郑州市', 90), ('河南省', 0)\]
df\_1\['matches'\] = m2
return df\_1
#模糊匹配
def fuzzy\_merge(df\_1, df\_2, key1, key2, threshold=90, limit=2):
"""
:param df\_1: the left table to join
:param df\_2: the right table to join
:param key1: key column of the left table
:param key2: key column of the right table
:param threshold: how close the matches should be to return a match, based on Levenshtein distance
:param limit: the amount of matches that will get returned, these are sorted high to low
:return: dataframe with boths keys and matches
"""
s = df\_2\[key2\].tolist()
m = df\_1\[key1\].apply(lambda x: process.extract(x, s, limit=limit))
df\_1\['matches'\] = m
m2 = df\_1\['matches'\].apply(lambda x: \[i\[0\] for i in x if i\[1\] >= threshold\]\[0\] if len(\[i\[0\] for i in x if i\[1\] >= threshold\]) > 0 else '')
df\_1\['matches'\] = m2
return df\_1
from fuzzywuzzy import fuzz
from fuzzywuzzy import process
df = fuzzy\_merge(data, company, '公司名称', '公司名称', threshold=90)
df
| 0.538012 | 0.677981 |
# Linearity measure applied to fashion MNIST
## General definition
The model linearity module in alibi provides metric to measure how linear an ML model is. Linearity is defined based on how much the linear superposition of the model's outputs differs from the output of the same linear superposition of the inputs.
Given $N$ input vectors $v_i$, $N$ real coefficients $\alpha_i$ and a predict function $\text{M}(v_i)$, the linearity of the predict function is defined as
$$L = \Big|\Big|\sum_i \alpha_i M(v_i) - M\Big(\sum_i \alpha_i v_i\Big) \Big|\Big| \quad \quad \text{If M is a regressor}$$
$$L = \Big|\Big|\sum_i \alpha_i \log \circ M(v_i) - \log \circ M\Big(\sum_i \alpha_i v_i\Big)\Big|\Big| \quad \quad \text{If M is a classifier}$$
Note that a lower value of $L$ means that the model $M$ is more linear.
## Alibi implementation
* Based on the general definition above, alibi calculates the linearity of a model in the neighboorhood of a given instance $v_0$.
## Fashion MNIST data set
* We train a convolutional neural network to classify the images in the fashion MNIST dataset.
* We investigate the correlation between the model's linearity associated to a certain instance and the class the instance belong to.
* We also calculate the linearity measure for each internal layer of the CNN and show how linearity propagates through the model.
```
import pandas as pd
import numpy as np
import matplotlib
%matplotlib inline
import matplotlib.pyplot as plt
from time import time
import tensorflow as tf
from alibi.confidence import linearity_measure, LinearityMeasure
from alibi.confidence.model_linearity import infer_feature_range
from tensorflow.keras.layers import Conv2D, Dense, Dropout, Flatten, MaxPooling2D, Input, Activation
from tensorflow.keras.models import Model
from tensorflow.keras.utils import to_categorical
from tensorflow.keras import backend as K
```
## Load data fashion mnist
The fashion MNIST data set consists of 60000 images of shape $28 \times 28$ divided in 10 categories. Each category corresponds to a different type of clothing piece, such as "boots", "t-shirts", etc
```
(x_train, y_train), (x_test, y_test) = tf.keras.datasets.fashion_mnist.load_data()
print('x_train shape:', x_train.shape, 'y_train shape:', y_train.shape)
idx = 0
plt.imshow(x_train[idx])
print('Sample instance from the MNIST data set.')
x_train = x_train.astype('float32') / 255
x_test = x_test.astype('float32') / 255
x_train = np.reshape(x_train, x_train.shape + (1,))
x_test = np.reshape(x_test, x_test.shape + (1,))
print('x_train shape:', x_train.shape, 'x_test shape:', x_test.shape)
y_train = to_categorical(y_train)
y_test = to_categorical(y_test)
print('y_train shape:', y_train.shape, 'y_test shape:', y_test.shape)
```
## Convolutional neural network
Here we define and train a 2 layer convolutional neural network on the fashion MNIST data set.
### Define model
```
def model():
x_in = Input(shape=(28, 28, 1), name='input')
x = Conv2D(filters=64, kernel_size=2, padding='same', name='conv_1')(x_in)
x = Activation('relu', name='relu_1')(x)
x = MaxPooling2D(pool_size=2, name='maxp_1')(x)
x = Dropout(0.3, name='drop_1')(x)
x = Conv2D(filters=64, kernel_size=2, padding='same', name='conv_2')(x)
x = Activation('relu', name='relu_2')(x)
x = MaxPooling2D(pool_size=2, name='maxp_2')(x)
x = Dropout(0.3, name='drop_2')(x)
x = Flatten(name='flat')(x)
x = Dense(256, name='dense_1')(x)
x = Activation('relu', name='relu_3')(x)
x = Dropout(0.5, name='drop_3')(x)
x_out = Dense(10, name='dense_2')(x)
x_out = Activation('softmax', name='softmax')(x_out)
cnn = Model(inputs=x_in, outputs=x_out)
cnn.compile(loss='categorical_crossentropy', optimizer='adam', metrics=['accuracy'])
return cnn
cnn = model()
cnn.summary()
```
### Training
```
cnn.fit(x_train, y_train, batch_size=64, epochs=5);
```
## Linearity of each Layer
Here we calculate the linearity of the model considering each layer as the output in turn. The values are averaged over 100 random instances sampled from the training set.
### Extract layers
```
inp = cnn.input
outs = {l.name: l.output for l in cnn.layers}
predict_fns = {name: K.function([inp], [out]) for name, out in outs.items()}
```
### Calculate linearity
```
# Infering feature ranges.
features_range = infer_feature_range(x_test)
# Selecting random instances from training set.
rnd = np.random.randint(len(x_test) - 101, size=100)
lins_layers = {}
for name, l in predict_fns.items():
if name != 'input':
def predict_fn(x):
layer = l([x])
return layer[0]
if name == 'softmax':
lins_layers[name] = linearity_measure(predict_fn, x_test[rnd], feature_range=features_range,
agg='global', model_type='classifier', nb_samples=20)
else:
lins_layers[name] = linearity_measure(predict_fn, x_test[rnd], feature_range=features_range,
agg='global', model_type='regressor', nb_samples=20)
lins_layers_mean = {k: v.mean() for k, v in lins_layers.items()}
S = pd.Series(data=lins_layers_mean)
colors = ['gray' for l in S[:-1]]
colors.append('r')
ax = S.plot(kind='bar', linewidth=3, figsize=(15,10), color=colors, width=0.7, fontsize=18)
ax.set_ylabel('L measure', fontsize=20)
ax.set_xlabel('Layer', fontsize=20)
print('Linearity measure calculated taking as output each layer of a convolutional neural network.')
```
Linearity measure in the locality of a given instance calculated taking as output each layer of a convolutional neural network trained on the fashion MNIST data set.
* The linearity measure of the first convolutional layer conv_1 is 0, as expected since convolutions are linear operations.
* The relu activation introduces non-linearity, which is increased by maxpooling. Dropout layers and flatten layers do no change the output at inference time so the linearity doesn't change.
* The second convolutional layer conv_2 and the dense layers change the linearity even though they are linear operations.
* The softmax layer in red is obtained by inverting the softmax function.
* For more details see arxiv reference.
## Linearity and categories
Here we calculate the linearity averaged over all instances belonging to the same class, for each class.
```
class_groups = []
for i in range(10):
y = y_test.argmax(axis=1)
idxs_i = np.where(y == i)[0]
class_groups.append(x_test[idxs_i])
def predict_fn(x):
return cnn.predict(x)
lins_classes = []
t_0 = time()
for j in range(len(class_groups)):
print(f'Calculating linearity for instances belonging to class {j}')
class_group = class_groups[j]
class_group = np.random.permutation(class_group)[:2000]
t_i = time()
lin = linearity_measure(predict_fn, class_group, feature_range=features_range,
agg='global', model_type='classifier', nb_samples=20)
t_i_1 = time() - t_i
print(f'Run time for class {j}: {t_i_1}')
lins_classes.append(lin)
t_fin = time() - t_0
print(f'Total run time: {t_fin}')
df = pd.DataFrame(data=lins_classes).T
ax = df.mean().plot(kind='bar', linewidth=3, figsize=(15,10), color='gray', width=0.7, fontsize=10)
ax.set_ylabel('L measure', fontsize=20)
ax.set_xlabel('Class', fontsize=20)
print("Linearity measure distribution means for each class in the fashion MNIST data set.")
ax2 = df.plot(kind='hist', subplots=True, bins=20, figsize=(10,10), sharey=True)
for a in ax2:
a.set_xlabel('L measure', fontsize=20)
a.set_ylabel('', rotation=True, fontsize=10)
#ax2.set_ylabel('F', fontsize=10)
print('Linearity measure distributions for each class in the fashion MNIST data set.')
```
|
github_jupyter
|
import pandas as pd
import numpy as np
import matplotlib
%matplotlib inline
import matplotlib.pyplot as plt
from time import time
import tensorflow as tf
from alibi.confidence import linearity_measure, LinearityMeasure
from alibi.confidence.model_linearity import infer_feature_range
from tensorflow.keras.layers import Conv2D, Dense, Dropout, Flatten, MaxPooling2D, Input, Activation
from tensorflow.keras.models import Model
from tensorflow.keras.utils import to_categorical
from tensorflow.keras import backend as K
(x_train, y_train), (x_test, y_test) = tf.keras.datasets.fashion_mnist.load_data()
print('x_train shape:', x_train.shape, 'y_train shape:', y_train.shape)
idx = 0
plt.imshow(x_train[idx])
print('Sample instance from the MNIST data set.')
x_train = x_train.astype('float32') / 255
x_test = x_test.astype('float32') / 255
x_train = np.reshape(x_train, x_train.shape + (1,))
x_test = np.reshape(x_test, x_test.shape + (1,))
print('x_train shape:', x_train.shape, 'x_test shape:', x_test.shape)
y_train = to_categorical(y_train)
y_test = to_categorical(y_test)
print('y_train shape:', y_train.shape, 'y_test shape:', y_test.shape)
def model():
x_in = Input(shape=(28, 28, 1), name='input')
x = Conv2D(filters=64, kernel_size=2, padding='same', name='conv_1')(x_in)
x = Activation('relu', name='relu_1')(x)
x = MaxPooling2D(pool_size=2, name='maxp_1')(x)
x = Dropout(0.3, name='drop_1')(x)
x = Conv2D(filters=64, kernel_size=2, padding='same', name='conv_2')(x)
x = Activation('relu', name='relu_2')(x)
x = MaxPooling2D(pool_size=2, name='maxp_2')(x)
x = Dropout(0.3, name='drop_2')(x)
x = Flatten(name='flat')(x)
x = Dense(256, name='dense_1')(x)
x = Activation('relu', name='relu_3')(x)
x = Dropout(0.5, name='drop_3')(x)
x_out = Dense(10, name='dense_2')(x)
x_out = Activation('softmax', name='softmax')(x_out)
cnn = Model(inputs=x_in, outputs=x_out)
cnn.compile(loss='categorical_crossentropy', optimizer='adam', metrics=['accuracy'])
return cnn
cnn = model()
cnn.summary()
cnn.fit(x_train, y_train, batch_size=64, epochs=5);
inp = cnn.input
outs = {l.name: l.output for l in cnn.layers}
predict_fns = {name: K.function([inp], [out]) for name, out in outs.items()}
# Infering feature ranges.
features_range = infer_feature_range(x_test)
# Selecting random instances from training set.
rnd = np.random.randint(len(x_test) - 101, size=100)
lins_layers = {}
for name, l in predict_fns.items():
if name != 'input':
def predict_fn(x):
layer = l([x])
return layer[0]
if name == 'softmax':
lins_layers[name] = linearity_measure(predict_fn, x_test[rnd], feature_range=features_range,
agg='global', model_type='classifier', nb_samples=20)
else:
lins_layers[name] = linearity_measure(predict_fn, x_test[rnd], feature_range=features_range,
agg='global', model_type='regressor', nb_samples=20)
lins_layers_mean = {k: v.mean() for k, v in lins_layers.items()}
S = pd.Series(data=lins_layers_mean)
colors = ['gray' for l in S[:-1]]
colors.append('r')
ax = S.plot(kind='bar', linewidth=3, figsize=(15,10), color=colors, width=0.7, fontsize=18)
ax.set_ylabel('L measure', fontsize=20)
ax.set_xlabel('Layer', fontsize=20)
print('Linearity measure calculated taking as output each layer of a convolutional neural network.')
class_groups = []
for i in range(10):
y = y_test.argmax(axis=1)
idxs_i = np.where(y == i)[0]
class_groups.append(x_test[idxs_i])
def predict_fn(x):
return cnn.predict(x)
lins_classes = []
t_0 = time()
for j in range(len(class_groups)):
print(f'Calculating linearity for instances belonging to class {j}')
class_group = class_groups[j]
class_group = np.random.permutation(class_group)[:2000]
t_i = time()
lin = linearity_measure(predict_fn, class_group, feature_range=features_range,
agg='global', model_type='classifier', nb_samples=20)
t_i_1 = time() - t_i
print(f'Run time for class {j}: {t_i_1}')
lins_classes.append(lin)
t_fin = time() - t_0
print(f'Total run time: {t_fin}')
df = pd.DataFrame(data=lins_classes).T
ax = df.mean().plot(kind='bar', linewidth=3, figsize=(15,10), color='gray', width=0.7, fontsize=10)
ax.set_ylabel('L measure', fontsize=20)
ax.set_xlabel('Class', fontsize=20)
print("Linearity measure distribution means for each class in the fashion MNIST data set.")
ax2 = df.plot(kind='hist', subplots=True, bins=20, figsize=(10,10), sharey=True)
for a in ax2:
a.set_xlabel('L measure', fontsize=20)
a.set_ylabel('', rotation=True, fontsize=10)
#ax2.set_ylabel('F', fontsize=10)
print('Linearity measure distributions for each class in the fashion MNIST data set.')
| 0.824956 | 0.990768 |
<table class="ee-notebook-buttons" align="left">
<td><a target="_blank" href="https://github.com/giswqs/earthengine-py-notebooks/tree/master/Datasets/Vectors/us_census_counties.ipynb"><img width=32px src="https://www.tensorflow.org/images/GitHub-Mark-32px.png" /> View source on GitHub</a></td>
<td><a target="_blank" href="https://nbviewer.jupyter.org/github/giswqs/earthengine-py-notebooks/blob/master/Datasets/Vectors/us_census_counties.ipynb"><img width=26px src="https://upload.wikimedia.org/wikipedia/commons/thumb/3/38/Jupyter_logo.svg/883px-Jupyter_logo.svg.png" />Notebook Viewer</a></td>
<td><a target="_blank" href="https://colab.research.google.com/github/giswqs/earthengine-py-notebooks/blob/master/Datasets/Vectors/us_census_counties.ipynb"><img src="https://www.tensorflow.org/images/colab_logo_32px.png" /> Run in Google Colab</a></td>
</table>
## Install Earth Engine API and geemap
Install the [Earth Engine Python API](https://developers.google.com/earth-engine/python_install) and [geemap](https://github.com/giswqs/geemap). The **geemap** Python package is built upon the [ipyleaflet](https://github.com/jupyter-widgets/ipyleaflet) and [folium](https://github.com/python-visualization/folium) packages and implements several methods for interacting with Earth Engine data layers, such as `Map.addLayer()`, `Map.setCenter()`, and `Map.centerObject()`.
The following script checks if the geemap package has been installed. If not, it will install geemap, which automatically installs its [dependencies](https://github.com/giswqs/geemap#dependencies), including earthengine-api, folium, and ipyleaflet.
**Important note**: A key difference between folium and ipyleaflet is that ipyleaflet is built upon ipywidgets and allows bidirectional communication between the front-end and the backend enabling the use of the map to capture user input, while folium is meant for displaying static data only ([source](https://blog.jupyter.org/interactive-gis-in-jupyter-with-ipyleaflet-52f9657fa7a)). Note that [Google Colab](https://colab.research.google.com/) currently does not support ipyleaflet ([source](https://github.com/googlecolab/colabtools/issues/60#issuecomment-596225619)). Therefore, if you are using geemap with Google Colab, you should use [`import geemap.eefolium`](https://github.com/giswqs/geemap/blob/master/geemap/eefolium.py). If you are using geemap with [binder](https://mybinder.org/) or a local Jupyter notebook server, you can use [`import geemap`](https://github.com/giswqs/geemap/blob/master/geemap/geemap.py), which provides more functionalities for capturing user input (e.g., mouse-clicking and moving).
```
# Installs geemap package
import subprocess
try:
import geemap
except ImportError:
print('geemap package not installed. Installing ...')
subprocess.check_call(["python", '-m', 'pip', 'install', 'geemap'])
# Checks whether this notebook is running on Google Colab
try:
import google.colab
import geemap.eefolium as geemap
except:
import geemap
# Authenticates and initializes Earth Engine
import ee
try:
ee.Initialize()
except Exception as e:
ee.Authenticate()
ee.Initialize()
```
## Create an interactive map
The default basemap is `Google Maps`. [Additional basemaps](https://github.com/giswqs/geemap/blob/master/geemap/basemaps.py) can be added using the `Map.add_basemap()` function.
```
Map = geemap.Map(center=[40,-100], zoom=4)
Map
```
## Add Earth Engine Python script
```
# Add Earth Engine dataset
Map.setCenter(-110, 40, 5)
states = ee.FeatureCollection('TIGER/2018/States')
# .filter(ee.Filter.eq('STUSPS', 'MN'))
# // Turn the strings into numbers
states = states.map(lambda f: f.set('STATEFP', ee.Number.parse(f.get('STATEFP'))))
state_image = ee.Image().float().paint(states, 'STATEFP')
visParams = {
'palette': ['purple', 'blue', 'green', 'yellow', 'orange', 'red'],
'min': 0,
'max': 50,
'opacity': 0.8,
};
counties = ee.FeatureCollection('TIGER/2016/Counties')
image = ee.Image().paint(states, 0, 2)
Map.setCenter(-99.844, 37.649, 5)
# Map.addLayer(image, {'palette': 'FF0000'}, 'TIGER/2018/States')
Map.addLayer(image, visParams, 'TIGER/2016/States');
Map.addLayer(ee.Image().paint(counties, 0, 1), {}, 'TIGER/2016/Counties')
```
## Display Earth Engine data layers
```
Map.addLayerControl() # This line is not needed for ipyleaflet-based Map.
Map
```
|
github_jupyter
|
# Installs geemap package
import subprocess
try:
import geemap
except ImportError:
print('geemap package not installed. Installing ...')
subprocess.check_call(["python", '-m', 'pip', 'install', 'geemap'])
# Checks whether this notebook is running on Google Colab
try:
import google.colab
import geemap.eefolium as geemap
except:
import geemap
# Authenticates and initializes Earth Engine
import ee
try:
ee.Initialize()
except Exception as e:
ee.Authenticate()
ee.Initialize()
Map = geemap.Map(center=[40,-100], zoom=4)
Map
# Add Earth Engine dataset
Map.setCenter(-110, 40, 5)
states = ee.FeatureCollection('TIGER/2018/States')
# .filter(ee.Filter.eq('STUSPS', 'MN'))
# // Turn the strings into numbers
states = states.map(lambda f: f.set('STATEFP', ee.Number.parse(f.get('STATEFP'))))
state_image = ee.Image().float().paint(states, 'STATEFP')
visParams = {
'palette': ['purple', 'blue', 'green', 'yellow', 'orange', 'red'],
'min': 0,
'max': 50,
'opacity': 0.8,
};
counties = ee.FeatureCollection('TIGER/2016/Counties')
image = ee.Image().paint(states, 0, 2)
Map.setCenter(-99.844, 37.649, 5)
# Map.addLayer(image, {'palette': 'FF0000'}, 'TIGER/2018/States')
Map.addLayer(image, visParams, 'TIGER/2016/States');
Map.addLayer(ee.Image().paint(counties, 0, 1), {}, 'TIGER/2016/Counties')
Map.addLayerControl() # This line is not needed for ipyleaflet-based Map.
Map
| 0.57081 | 0.960287 |
<img style="float: right; margin: 0px 0px 15px 15px;" src="https://encrypted-tbn0.gstatic.com/images?q=tbn:ANd9GcSQt6eQo8JPYzYO4p6WmxLtccdtJ4X8WR6GzVVKbsMjyGvUDEn1mg" width="300px" height="100px" />
# Trabajando con opciones
Una opción puede negociarse en el mercado secundario por lo que es importante determinar su valor $V_t$ para cada tiempo $t\in [0, T]$. La ganancia que obtiene quién adquiere la opción se llama función de pago o "payoff" y claramente depende del valor del subyacente.
Hay una gran variedad de opciones en el mercado y éstas se clasiflcan según su función de pago y la forma en que pueden ejercerse. Las opciones que tienen como función de pago a
$$ P(S(t),t)=max\{S(T)-K,0\} \rightarrow \text{En el caso de Call}$$
$$ P(S(t),t)=max\{K-S(T),0\} \rightarrow \text{En el caso de Put}$$
se llaman opciones **Vainilla**, con $h:[0,\infty) \to [0,\infty)$.
La opción se llama **europea** si puede ejercerse sólo en la fecha de vencimiento.
Se dice que una opción es **americana** si puede ejercerse en cualquier momento antes o en la fecha de vencimiento.
Una opción compleja popular son las llamadas **opciones asiáticas** cuyos pagos dependen de todas las trayectorias del precio de los activos subyacentes. Las opciones cuyos pagos dependen de las trayectorias de los precios de los activos subyacentes se denominan opciones dependientes de la ruta.
Principalmente, se puede resumir que las dos razones con más peso de importancia para utilizar opciones son el **aseguramiento** y la **especulación**.
## Opciones Plan Vainilla: opción de compra y opción de venta europea
Una opción vainilla o estándar es una opción normal de compra o venta que no tiene características especiales o inusuales. Puede ser para tamaños y vencimientos estandarizados, y negociarse en un intercambio.
En comparación con otras estructuras de opciones, las opciones de vanilla no son sofisticadas o complicadas.
## 1. ¿Cómo descargar datos de opciones?
```
#importar los paquetes que se van a usar
import pandas as pd
import pandas_datareader.data as web
import numpy as np
import datetime
import matplotlib.pyplot as plt
import scipy.stats as st
import seaborn as sns
%matplotlib inline
#algunas opciones para Pandas
pd.set_option('display.notebook_repr_html', True)
pd.set_option('display.max_columns', 6)
pd.set_option('display.max_rows', 10)
pd.set_option('display.width', 78)
pd.set_option('precision', 3)
```
Usando el paquete `pandas_datareader` también podemos descargar datos de opciones. Por ejemplo, descarguemos los datos de las opciones cuyo activo subyacente son las acciones de Apple
```
aapl = web.YahooOptions('AAPL')
aapl_opt = aapl.get_all_data().reset_index()
aapl_opt.set_index('Expiry')
# aapl
# closes = web.YahooDailyReader(ticker, start_date, end_date).read().sort_index('major_axis')
# aapl_opt.loc[0, 'JSON']
aapl_opt.loc[200, 'JSON']
```
Una vez tenemos la información, podemos consultar de qué tipo son las opciones
```
aapl_opt.loc[:, 'Type']
```
o en que fecha expiran
```
aapl_opt.loc[:, 'Expiry']
```
Por otra parte, podríamos querer consultar todas las opciones de compra (call) que expiran en cierta fecha (2019-04-18)
```
call06 = aapl_opt.loc[(aapl_opt.Expiry=='2019-04-18') & (aapl_opt.Type=='call')]
call06
```
## 2. ¿Qué es la volatilidad implícita?
**Volatilidad:** desviación estándar de los rendimientos.
- ¿Cómo se calcula?
- ¿Para qué calcular la volatilidad?
- **Para valuar derivados**, por ejemplo **opciones**.
- Método de valuación de riesgo neutral (se supone que el precio del activo $S_t$ no se ve afectado por el riesgo de mercado).
Recorderis de cuantitativas:
1. Ecuación de Black-Scholes
$$ dS(t) = \mu S(t) + \sigma S(t)dW_t$$
2. Solución de la ecuación
El valor de una opción Europea de vainilla $V_t$ puede obtenerse por:
$$V_t = F(t,S_t)$$ donde

3. Opción de compra europea, suponiendo que los precios del activo son lognormales
4. Opción de venta europea, suponiendo que los precios del activo son lognormales
Entonces, ¿qué es la **volatilidad implícita**?
La volatilidad es una medida de la incertidumbre sobre el comportamiento futuro de un activo, que se mide habitualmente como la desviación típica de la rentabilidad de dicho activo.
```
ax = call06.set_index('Strike').loc[:, 'IV'].plot(figsize=(8,6))
ax.axvline(call06.Underlying_Price.iloc[0], color='g');
# call06.Underlying_Price
put06 = aapl_opt.loc[(aapl_opt.Expiry=='2019-04-18') & (aapl_opt.Type=='put')]
put06
ax = put06.set_index('Strike').loc[:, 'IV'].plot(figsize=(8,6))
ax.axvline(put06.Underlying_Price.iloc[0], color='g')
```
Con lo que hemos aprendido, deberíamos ser capaces de crear una función que nos devuelva un `DataFrame` de `pandas` con los precios de cierre ajustados de ciertas compañías en ciertas fechas:
- Escribir la función a continuación
```
# Función para descargar precios de cierre ajustados de varios activos a la vez:
def get_historical_closes(tickers, start_date=None, end_date=None, freq=None):
# Fecha inicio por defecto (start_date='2010-01-01') y fecha fin por defecto (end_date=today)
# Frecuencia de muestreo por defecto (freq='d')
# Importamos paquetes necesarios
import pandas as pd
pd.core.common.is_list_like = pd.api.types.is_list_like
import pandas_datareader.data as web
# Creamos DataFrame vacío de precios, con el índice de las fechas
closes = pd.DataFrame(columns = tickers, index=web.YahooDailyReader(symbols=tickers[0], start=start_date, end=end_date, interval=freq).read().index)
# Agregamos cada uno de los precios con YahooDailyReader
for ticker in tickers:
df = web.YahooDailyReader(symbols=ticker, start=start_date, end=end_date, interval=freq).read()
closes[ticker]=df['Adj Close']
closes.index_name = 'Date'
closes = closes.sort_index()
return closes
```
- Obtener como ejemplo los precios de cierre de Apple del año pasado hasta la fecha. Graficar...
```
ticker = ['AAPL']
start_date = '2017-01-01'
closes_aapl = get_historical_closes(ticker, start_date,freq='d')
closes_aapl.plot(figsize=(8,5));
plt.legend(ticker);
```
- Escribir una función que pasándole el histórico de precios devuelva los rendimientos logarítmicos:
```
def calc_daily_ret(closes):
return np.log(closes/closes.shift(1)).iloc[1:]
```
- Graficar...
```
ret_aapl = calc_daily_ret(closes_aapl)
ret_aapl.plot(figsize=(8,6));
```
También, descargar datos de opciones de Apple:
```
aapl = web.YahooOptions('AAPL')
aapl_opt = aapl.get_all_data().reset_index()
aapl_opt.set_index('Expiry').sort_index()
aapl_opt.loc[(aapl_opt.Type=='call') & (aapl_opt.Strike==200) & (aapl_opt.Expiry=='2019-02-15')]
opcion_valuar = aapl_opt.loc[487]
opcion_valuar['JSON']
```
# Simulación de precios usando rendimiento simple y logarítmico
* Comenzaremos por suponer que los rendimientos son un p.e. estacionario que distribuyen $\mathcal{N}(\mu,\sigma)$.
```
# Descargamos los precios de apple
ticker = ['AAPL']
start_date = '2017-01-01'
closes_aapl = get_historical_closes(ticker, start_date,freq='d')
closes_aapl
```
- **Rendimiento Simple**
```
# Obtenemos el rendimiento simple
Ri = closes_aapl.pct_change(1).iloc[1:]
# Obtenemos su media y desviación estándar de los rendimientos
mu_R = Ri.mean()[0]
sigma_R = Ri.std()[0]
Ri
ndays = 109
nscen = 10
dates = pd.date_range('2018-10-29', periods = ndays)
dates
dt = 1; # Rendimiento diario
Z = np.random.randn(ndays,nscen) # Z ~ N(0,1)
# Simulación normal de los rendimientos
Ri_dt = pd.DataFrame(mu_R*dt+Z*sigma_R*np.sqrt(dt),index=dates)
Ri_dt.cumprod()
# Simulación del precio
S_0 = closes_aapl.iloc[-1,0]
S_T = S_0*(1+Ri_dt).cumprod()
# Se muestran los precios simulados con los precios descargados
pd.concat([closes_aapl,S_T]).plot(figsize=(8,6));
plt.title('Simulación de precios usando rendimiento simple');
```
- **Rendimiento logarítmico**
```
ri = calc_daily_ret(closes_aapl)
# Usando la media y desviación estándar de los rendimientos logarítmicos
mu_r = ri.mean()[0]
sigma_r = ri.std()[0]
# # Usando la equivalencia teórica
# mu_r2 = mu_R - (sigma_R**2)/2
sim_ret_ri = pd.DataFrame(mu_r*dt+Z*sigma_r*np.sqrt(dt), index=dates)
# sim_ret_ri
# Simulación del precio
S_0 = closes_aapl.iloc[-1,0]
S_T2 = S_0*np.exp(sim_ret_ri.cumsum())
# Se muestran los precios simulados con los precios descargados
pd.concat([closes_aapl,S_T2]).plot(figsize=(8,6));
plt.title('Simulación de precios usando rendimiento logarítmico');
S_T-S_T2
print('Las std usando rendimientos logarítmicos y simples son iguales')
sigma_R,sigma_r
```
## 2. Valuación usando simulación: modelo normal para los rendimientos
- Hallar media y desviación estándar muestral de los rendimientos logarítmicos
```
mu = ret_aapl.mean()[0]
sigma = ret_aapl.std()[0]
mu, sigma
```
No se toma la media sino la tasa libre de riesgo
> Referencia: https://www.treasury.gov/resource-center/data-chart-center/interest-rates/Pages/TextView.aspx?data=yield
```
# Tasa de bonos de 1 yr de fecha 10/01/18 -> 2.60%
r = 0.0260/360 # Tasa diaria
```
- Simularemos el tiempo de contrato (días=109) desde 29/10/18 hasta 15/02/19, 10 escenarios:
> Calculador de fechas: https://es.calcuworld.com/calendarios/calculadora-de-tiempo-entre-dos-fechas/
- Generar fechas
```
ndays = 109
nscen = 10
dates = pd.date_range('2018-10-29', periods = ndays)
dates
```
- Generamos 10 escenarios de rendimientos simulados y guardamos en un dataframe
```
sim_ret = pd.DataFrame(sigma*np.random.randn(ndays,nscen)+r, index=dates)
sim_ret.cumsum()
# Las columnas son los escenarios y las filas son las días de contrato
```
- Con los rendimientos simulados, calcular los escenarios de precios respectivos:
```
S0 = closes_aapl.iloc[-1,0] # Condición inicial del precio a simular
sim_closes = S0*np.exp(sim_ret.cumsum())
sim_closes
```
- Graficar:
```
#sim_closes.plot(figsize=(8,6));
# Se muestran los precios simulados con los precios descargados
pd.concat([closes_aapl,sim_closes]).plot(figsize=(8,6));
opcion_valuar['JSON']
K=200 # strike price
ndays = 109
nscen = 100000
dates = pd.date_range('2018-10-29', periods = ndays)
S0 = closes_aapl.iloc[-1,0] # Condición inicial del precio a simular
sim_ret = pd.DataFrame(sigma*np.random.randn(ndays,nscen)+r,index=dates)
sim_closes = S0*np.exp(sim_ret.cumsum())
#strike = pd.DataFrame({'Strike':K*np.ones(ndays)}, index=dates)
#simul = pd.concat([closes_aapl.T,strike.T,sim_closes.T]).T
#simul.plot(figsize=(8,6),legend=False);
strike = pd.DataFrame(K*np.ones([ndays,nscen]), index=dates)
call = pd.DataFrame({'Prima':np.exp(-r*ndays) \
*np.fmax(sim_closes-strike,np.zeros([ndays,nscen])).T.mean()}, index=dates)
call.plot();
```
La valuación de la opción es:
```
call.iloc[-1]
```
Intervalo de confianza del 99%
```
confianza = 0.99
sigma_est = sim_closes.iloc[-1].sem()
mean_est = call.iloc[-1].Prima
i1 = st.t.interval(confianza,nscen-1, loc=mean_est, scale=sigma_est)
i2 = st.norm.interval(confianza, loc=mean_est, scale=sigma_est)
print(i1)
print(i1)
```
### Análisis de la distribución de los rendimientos
```
ren = calc_daily_ret(closes_aapl) # rendimientos
y,x,des = plt.hist(ren['AAPL'],bins=50,density=True,label='Histograma rendimientos')
mu_fit,sd_fit = st.norm.fit(ren) # Se ajustan los parámetros de una normal
# Valores máximo y mínimo de los rendiemientos a generar
ren_max = max(x);ren_min = min(x)
# Vector de rendimientos generados
ren_gen = np.arange(ren_min,ren_max,0.001)
# Generación de la normal ajustado con los parámetros encontrados
curve_fit = st.norm.pdf(ren_gen,loc=mu_fit,scale=sd_fit)
plt.plot(ren_gen,curve_fit,label='Distribución ajustada')
plt.legend()
plt.show()
st.probplot(ren['AAPL'], dist='norm', plot=plt);
```
## 3. Valuación usando simulación: uso del histograma de rendimientos
Todo el análisis anterior se mantiene. Solo cambia la forma de generar los números aleatorios para la simulación montecarlo.
Ahora, generemos un histograma de los rendimientos diarios para generar valores aleatorios de los rendimientos simulados.
- Primero, cantidad de días y número de escenarios de simulación
```
ndays = 109
nscen = 10
```
- Del histograma anterior, ya conocemos las probabilidades de ocurrencia, lo que se llamó como variable `y`
```
prob = y/np.sum(y)
values = x[1:]
```
- Con esto, generamos los números aleatorios correspondientes a los rendimientos (tantos como días por número de escenarios).
```
ret = np.random.choice(values, ndays*nscen, p=prob)
dates = pd.date_range('2018-10-29',periods=ndays)
sim_ret_hist = pd.DataFrame(ret.reshape((ndays,nscen)),index=dates)
sim_ret_hist
sim_closes_hist = (closes_aapl.iloc[-1,0])*np.exp(sim_ret_hist.cumsum())
sim_closes_hist
sim_closes_hist.plot(figsize=(8,6),legend=False);
pd.concat([closes_aapl,sim_closes_hist]).plot(figsize=(8,6),legend=False);
plt.title('Simulación usando el histograma de los rendimientos')
K=200
ndays = 109
nscen = 10000
freq, values = np.histogram(ret_aapl+r-mu, bins=2000)
prob = freq/np.sum(freq)
ret=np.random.choice(values[1:],ndays*nscen,p=prob)
dates=pd.date_range('2018-10-29',periods=ndays)
sim_ret_hist = pd.DataFrame(ret.reshape((ndays,nscen)),index=dates)
sim_closes_hist = (closes_aapl.iloc[-1,0])*np.exp(sim_ret_hist.cumsum())
strike = pd.DataFrame(K*np.ones(ndays*nscen).reshape((ndays,nscen)), index=dates)
call_hist = pd.DataFrame({'Prima':np.exp(-r*ndays)*np.fmax(sim_closes_hist-strike,np.zeros(ndays*nscen).reshape((ndays,nscen))).T.mean()}, index=dates)
call_hist.plot();
call_hist.iloc[-1]
opcion_valuar['JSON']
```
Intervalo de confianza del 95%
```
confianza = 0.95
sigma_est = sim_closes_hist.iloc[-1].sem()
mean_est = call_hist.iloc[-1].Prima
i1 = st.t.interval(confianza,nscen-1, loc=mean_est, scale=sigma_est)
i2 = st.norm.interval(confianza, loc=mean_est, scale=sigma_est)
print(i1)
print(i1)
```
# <font color = 'red'> Tarea: </font>
Replicar el procedimiento anterior para valoración de opciones 'call', pero en este caso para opciones tipo 'put'.
<script>
$(document).ready(function(){
$('div.prompt').hide();
$('div.back-to-top').hide();
$('nav#menubar').hide();
$('.breadcrumb').hide();
$('.hidden-print').hide();
});
</script>
<footer id="attribution" style="float:right; color:#808080; background:#fff;">
Created with Jupyter by Esteban Jiménez Rodríguez and modify by Oscar Jaramillo Z.
</footer>
|
github_jupyter
|
#importar los paquetes que se van a usar
import pandas as pd
import pandas_datareader.data as web
import numpy as np
import datetime
import matplotlib.pyplot as plt
import scipy.stats as st
import seaborn as sns
%matplotlib inline
#algunas opciones para Pandas
pd.set_option('display.notebook_repr_html', True)
pd.set_option('display.max_columns', 6)
pd.set_option('display.max_rows', 10)
pd.set_option('display.width', 78)
pd.set_option('precision', 3)
aapl = web.YahooOptions('AAPL')
aapl_opt = aapl.get_all_data().reset_index()
aapl_opt.set_index('Expiry')
# aapl
# closes = web.YahooDailyReader(ticker, start_date, end_date).read().sort_index('major_axis')
# aapl_opt.loc[0, 'JSON']
aapl_opt.loc[200, 'JSON']
aapl_opt.loc[:, 'Type']
aapl_opt.loc[:, 'Expiry']
call06 = aapl_opt.loc[(aapl_opt.Expiry=='2019-04-18') & (aapl_opt.Type=='call')]
call06
ax = call06.set_index('Strike').loc[:, 'IV'].plot(figsize=(8,6))
ax.axvline(call06.Underlying_Price.iloc[0], color='g');
# call06.Underlying_Price
put06 = aapl_opt.loc[(aapl_opt.Expiry=='2019-04-18') & (aapl_opt.Type=='put')]
put06
ax = put06.set_index('Strike').loc[:, 'IV'].plot(figsize=(8,6))
ax.axvline(put06.Underlying_Price.iloc[0], color='g')
# Función para descargar precios de cierre ajustados de varios activos a la vez:
def get_historical_closes(tickers, start_date=None, end_date=None, freq=None):
# Fecha inicio por defecto (start_date='2010-01-01') y fecha fin por defecto (end_date=today)
# Frecuencia de muestreo por defecto (freq='d')
# Importamos paquetes necesarios
import pandas as pd
pd.core.common.is_list_like = pd.api.types.is_list_like
import pandas_datareader.data as web
# Creamos DataFrame vacío de precios, con el índice de las fechas
closes = pd.DataFrame(columns = tickers, index=web.YahooDailyReader(symbols=tickers[0], start=start_date, end=end_date, interval=freq).read().index)
# Agregamos cada uno de los precios con YahooDailyReader
for ticker in tickers:
df = web.YahooDailyReader(symbols=ticker, start=start_date, end=end_date, interval=freq).read()
closes[ticker]=df['Adj Close']
closes.index_name = 'Date'
closes = closes.sort_index()
return closes
ticker = ['AAPL']
start_date = '2017-01-01'
closes_aapl = get_historical_closes(ticker, start_date,freq='d')
closes_aapl.plot(figsize=(8,5));
plt.legend(ticker);
def calc_daily_ret(closes):
return np.log(closes/closes.shift(1)).iloc[1:]
ret_aapl = calc_daily_ret(closes_aapl)
ret_aapl.plot(figsize=(8,6));
aapl = web.YahooOptions('AAPL')
aapl_opt = aapl.get_all_data().reset_index()
aapl_opt.set_index('Expiry').sort_index()
aapl_opt.loc[(aapl_opt.Type=='call') & (aapl_opt.Strike==200) & (aapl_opt.Expiry=='2019-02-15')]
opcion_valuar = aapl_opt.loc[487]
opcion_valuar['JSON']
# Descargamos los precios de apple
ticker = ['AAPL']
start_date = '2017-01-01'
closes_aapl = get_historical_closes(ticker, start_date,freq='d')
closes_aapl
# Obtenemos el rendimiento simple
Ri = closes_aapl.pct_change(1).iloc[1:]
# Obtenemos su media y desviación estándar de los rendimientos
mu_R = Ri.mean()[0]
sigma_R = Ri.std()[0]
Ri
ndays = 109
nscen = 10
dates = pd.date_range('2018-10-29', periods = ndays)
dates
dt = 1; # Rendimiento diario
Z = np.random.randn(ndays,nscen) # Z ~ N(0,1)
# Simulación normal de los rendimientos
Ri_dt = pd.DataFrame(mu_R*dt+Z*sigma_R*np.sqrt(dt),index=dates)
Ri_dt.cumprod()
# Simulación del precio
S_0 = closes_aapl.iloc[-1,0]
S_T = S_0*(1+Ri_dt).cumprod()
# Se muestran los precios simulados con los precios descargados
pd.concat([closes_aapl,S_T]).plot(figsize=(8,6));
plt.title('Simulación de precios usando rendimiento simple');
ri = calc_daily_ret(closes_aapl)
# Usando la media y desviación estándar de los rendimientos logarítmicos
mu_r = ri.mean()[0]
sigma_r = ri.std()[0]
# # Usando la equivalencia teórica
# mu_r2 = mu_R - (sigma_R**2)/2
sim_ret_ri = pd.DataFrame(mu_r*dt+Z*sigma_r*np.sqrt(dt), index=dates)
# sim_ret_ri
# Simulación del precio
S_0 = closes_aapl.iloc[-1,0]
S_T2 = S_0*np.exp(sim_ret_ri.cumsum())
# Se muestran los precios simulados con los precios descargados
pd.concat([closes_aapl,S_T2]).plot(figsize=(8,6));
plt.title('Simulación de precios usando rendimiento logarítmico');
S_T-S_T2
print('Las std usando rendimientos logarítmicos y simples son iguales')
sigma_R,sigma_r
mu = ret_aapl.mean()[0]
sigma = ret_aapl.std()[0]
mu, sigma
# Tasa de bonos de 1 yr de fecha 10/01/18 -> 2.60%
r = 0.0260/360 # Tasa diaria
ndays = 109
nscen = 10
dates = pd.date_range('2018-10-29', periods = ndays)
dates
sim_ret = pd.DataFrame(sigma*np.random.randn(ndays,nscen)+r, index=dates)
sim_ret.cumsum()
# Las columnas son los escenarios y las filas son las días de contrato
S0 = closes_aapl.iloc[-1,0] # Condición inicial del precio a simular
sim_closes = S0*np.exp(sim_ret.cumsum())
sim_closes
#sim_closes.plot(figsize=(8,6));
# Se muestran los precios simulados con los precios descargados
pd.concat([closes_aapl,sim_closes]).plot(figsize=(8,6));
opcion_valuar['JSON']
K=200 # strike price
ndays = 109
nscen = 100000
dates = pd.date_range('2018-10-29', periods = ndays)
S0 = closes_aapl.iloc[-1,0] # Condición inicial del precio a simular
sim_ret = pd.DataFrame(sigma*np.random.randn(ndays,nscen)+r,index=dates)
sim_closes = S0*np.exp(sim_ret.cumsum())
#strike = pd.DataFrame({'Strike':K*np.ones(ndays)}, index=dates)
#simul = pd.concat([closes_aapl.T,strike.T,sim_closes.T]).T
#simul.plot(figsize=(8,6),legend=False);
strike = pd.DataFrame(K*np.ones([ndays,nscen]), index=dates)
call = pd.DataFrame({'Prima':np.exp(-r*ndays) \
*np.fmax(sim_closes-strike,np.zeros([ndays,nscen])).T.mean()}, index=dates)
call.plot();
call.iloc[-1]
confianza = 0.99
sigma_est = sim_closes.iloc[-1].sem()
mean_est = call.iloc[-1].Prima
i1 = st.t.interval(confianza,nscen-1, loc=mean_est, scale=sigma_est)
i2 = st.norm.interval(confianza, loc=mean_est, scale=sigma_est)
print(i1)
print(i1)
ren = calc_daily_ret(closes_aapl) # rendimientos
y,x,des = plt.hist(ren['AAPL'],bins=50,density=True,label='Histograma rendimientos')
mu_fit,sd_fit = st.norm.fit(ren) # Se ajustan los parámetros de una normal
# Valores máximo y mínimo de los rendiemientos a generar
ren_max = max(x);ren_min = min(x)
# Vector de rendimientos generados
ren_gen = np.arange(ren_min,ren_max,0.001)
# Generación de la normal ajustado con los parámetros encontrados
curve_fit = st.norm.pdf(ren_gen,loc=mu_fit,scale=sd_fit)
plt.plot(ren_gen,curve_fit,label='Distribución ajustada')
plt.legend()
plt.show()
st.probplot(ren['AAPL'], dist='norm', plot=plt);
ndays = 109
nscen = 10
prob = y/np.sum(y)
values = x[1:]
ret = np.random.choice(values, ndays*nscen, p=prob)
dates = pd.date_range('2018-10-29',periods=ndays)
sim_ret_hist = pd.DataFrame(ret.reshape((ndays,nscen)),index=dates)
sim_ret_hist
sim_closes_hist = (closes_aapl.iloc[-1,0])*np.exp(sim_ret_hist.cumsum())
sim_closes_hist
sim_closes_hist.plot(figsize=(8,6),legend=False);
pd.concat([closes_aapl,sim_closes_hist]).plot(figsize=(8,6),legend=False);
plt.title('Simulación usando el histograma de los rendimientos')
K=200
ndays = 109
nscen = 10000
freq, values = np.histogram(ret_aapl+r-mu, bins=2000)
prob = freq/np.sum(freq)
ret=np.random.choice(values[1:],ndays*nscen,p=prob)
dates=pd.date_range('2018-10-29',periods=ndays)
sim_ret_hist = pd.DataFrame(ret.reshape((ndays,nscen)),index=dates)
sim_closes_hist = (closes_aapl.iloc[-1,0])*np.exp(sim_ret_hist.cumsum())
strike = pd.DataFrame(K*np.ones(ndays*nscen).reshape((ndays,nscen)), index=dates)
call_hist = pd.DataFrame({'Prima':np.exp(-r*ndays)*np.fmax(sim_closes_hist-strike,np.zeros(ndays*nscen).reshape((ndays,nscen))).T.mean()}, index=dates)
call_hist.plot();
call_hist.iloc[-1]
opcion_valuar['JSON']
confianza = 0.95
sigma_est = sim_closes_hist.iloc[-1].sem()
mean_est = call_hist.iloc[-1].Prima
i1 = st.t.interval(confianza,nscen-1, loc=mean_est, scale=sigma_est)
i2 = st.norm.interval(confianza, loc=mean_est, scale=sigma_est)
print(i1)
print(i1)
| 0.186317 | 0.916147 |
# Python Introduction
Python is a widely used high-level programming language for general-purpose programming, created by Guido van Rossum and first released in 1991. An interpreted language, Python has a design philosophy which emphasizes code readability (notably using whitespace indentation to delimit code blocks rather than curly braces or keywords), and a syntax which allows programmers to express concepts in fewer lines of code than possible in languages such as C++ or Java. The language provides constructs intended to enable writing clear programs on both a small and large scale.
Python features a dynamic type system and automatic memory management and supports multiple programming paradigms, including object-oriented, imperative, functional programming, and procedural styles. It has a large and comprehensive standard library.
Python interpreters are available for many operating systems, allowing Python code to run on a wide variety of systems. CPython, the reference implementation of Python, is open source software and has a community-based development model, as do nearly all of its variant implementations. CPython is managed by the non-profit Python Software Foundation.
# History
Python was conceived in the late 1980s, and its implementation began in December 1989 by Guido van Rossum at Centrum Wiskunde & Informatica (CWI) in the Netherlands as a successor to the ABC language (itself inspired by SETL) capable of exception handling and interfacing with the operating system Amoeba. Van Rossum is Python's principal author, and his continuing central role in deciding the direction of Python is reflected in the title given to him by the Python community, benevolent dictator for life (BDFL).
About the origin of Python, Van Rossum wrote in 1996:
`Over six years ago, in December 1989, I was looking for a "hobby" programming project that would keep me occupied during the week around Christmas. My office ... would be closed, but I had a home computer, and not much else on my hands. I decided to write an interpreter for the new scripting language I had been thinking about lately: a descendant of ABC that would appeal to Unix/C hackers. I chose Python as a working title for the project, being in a slightly irreverent mood (and a big fan of Monty Python's Flying Circus).`
Python 2.0 was released on 16 October 2000 and had many major new features, including a cycle-detecting garbage collector and support for Unicode. With this release the development process was changed and became more transparent and community-backed.
Python 3.0 (which early in its development was commonly referred to as Python 3000 or py3k), a major, backwards-incompatible release, was released on 3 December 2008 after a long period of testing. Many of its major features have been backported to the backwards-compatible Python 2.6.x and 2.7.x version series.
The End Of Life date (EOL, sunset date) for Python 2.7 was initially set at 2015, then postponed to 2020 out of concern that a large body of existing code cannot easily be forward-ported to Python 3. In January 2017 Google announced work on a Python 2.7 to Go transcompiler, which The Register speculated was in response to Python 2.7's planned end-of-life but Google cited performance under concurrent workloads as their only motivation.
https://en.wikipedia.org/wiki/Python_(programming_language)
# The Zen of Python
- Beautiful is better than ugly.
- Explicit is better than implicit.
- Simple is better than complex.
- Complex is better than complicated.
- Flat is better than nested.
- Sparse is better than dense.
- Readability counts.
- Special cases aren't special enough to break the rules.
- Although practicality beats purity.
- Errors should never pass silently.
- Unless explicitly silenced.
- In the face of ambiguity, refuse the temptation to guess.
- There should be one-- and preferably only one --obvious way to do it.
- Although that way may not be obvious at first unless you're Dutch.
- Now is better than never.
- Although never is often better than *right* now.
- If the implementation is hard to explain, it's a bad idea.
- If the implementation is easy to explain, it may be a good idea.
- Namespaces are one honking great idea -- let's do more of those!
https://www.python.org/dev/peps/pep-0020/
###### Installing Python3
## On Mac
#### Installing `brew`
```bash
$ ruby -e "$(curl -fsSL https://raw.githubusercontent.com/Homebrew/install/master/install)" < /dev/null 2> /dev/null```
#### Using `brew` installing Python3 locally
```bash
$ brew install python3```
#### To find the path for `python3`
```bash
$ which python3
/usr/local/bin/python3```
## On Windows
Sorry, can't say much as I am not a windows user.
https://docs.python.org/3/using/windows.html
## On linux
### `pip`
`pip` is a package management system used to install and manage software packages written in `Python`. Many packages can be found in the `Python Package Index (PyPI)`.
https://en.wikipedia.org/wiki/Pip_(package_manager)
https://pypi.python.org/pypi/pip
### Ubuntu
`Ubuntu 16.04` ships with both `Python3` and `Python2`.
By default `pip` for Python2 is installed but not for Python3.
Now we need to install `pip3` for `Python3`.
```bash
sudo apt-get install -y python3-pip```
#### Installing `Python3` `virtualenv`
```bash
sudo pip install virtualenv```
#### If required upgrade `pip`
```bash
sudo pip install --upgrade pip```
#### `Python3 virtualenv`
```bash
mkdir sample_project_python3
cd sample_project_python3
virtualenv .venv```
#### `Python2 virtualenv`
```bash
mkdir sample_project_python2
cd sample_project_python2
virtualenv -p /usr/bin/python2 .venv```
### CentOS / Amazon AMI
```bash
sudo -i
yum install python36 python36-pip
pip-3.6 install virtualenv
pip install --upgrade pip```
# Python virtualenv installation
```bash
$ pip3 install virtualenv```
# Advantages of virtualenv
- Isolated Python runtime environments without modifying the root or system python installation.
- Eash while deploying several python applications and have different settings.
- Helpful when runtime dependencies differ between frameworks or libraries in various applications.
- `virtualenv` is very useful in a more dynamic development environment.
# Creating Virtualenv
Steps to create `virtualenv`
```bash
$ mkdir sample_project
$ cd sample_project/
$ virtualenv .venv
$ ls -lrtha
total 0
drwxr-xr-x+ 35 username CORP\Domain Users 1.2K Apr 6 10:49 ..
drwxr-xr-x 3 username CORP\Domain Users 102B Apr 6 10:50 .
drwxr-xr-x 7 username CORP\Domain Users 238B Apr 6 10:50 .venv```
# Activating the Virtualenv
```bash
$ source .venv/bin/activate
(.venv)$ ```
We can install libararies only to this environment without modifying the actual system python installation using `pip`
#### To start `python` interactive shell
```bash
(.venv)$ python
Python 3.6.1 (default, Apr 4 2017, 09:40:51)
[GCC 4.2.1 Compatible Apple LLVM 8.0.0 (clang-800.0.42.1)] on darwin
Type "help", "copyright", "credits" or "license" for more information.
>>>```
#### How to properly Quit `python` interactive shell
- You can either use exit() or Ctrl-D (i.e. EOF) to exit.
- The brackets behind the exit function are crucial.
- exit without brackets won't work in `python3`
#### How to deactivate the `virtualenv`
Just by running the command `deactivate` we can come out the `virtualenv`
```bash
(.venv)$ deactivate
$ ```
# Going forward we are going to use only `Python3`
- End Of Life for `Python2.7` is on April 12th, 2020
- Security patches
- Not to invest time on code portability and re-writing the existing code
- More here on https://pythonclock.org/
# Interactive shell Continued
## Primitive data types
|Variable type|Example|Comment|
|:-----------:|:-------:|:-------------|
|`bool`|`True/False`| boolean true/false values|
|`int` `long`| 99999999999| Numbers/Integers (forget about the range of integers in `Python`) <br> Note: `long` wont see long as type in `python`|
|`float`| 0.99999999999| Decimals/Not whole integers|
|`str`| `"Hello World!"`, `'Hello World!'`, `'''Hello World!'''`, `"""Hello World!"""`| Any text|
|`None`| `None`| To create empty variable without any meaningful value|
```
True
False
print(1 == 2)
'Hello World'
3
3.141
None
```
# Different ways to define strings
```
print("Hello \"World!") # Simple Hello World
print(r'Hello \'World!') # Notice we use single quote to wrap the string.
print('''Hello \'World!''') # Notice we use three single quotes to wrap the string.
print("Hello \\ World!")
print("""Hello \" World!""") # Notice we use three double quotes to wrap the string.
```
All primitive data types can be stored in a variable using an assignment operator: `=`
# Boolean Variables
```
flag = True
type(flag)
flag = False
type(flag)
flag = 1
type(flag)
flag = 'Hello World!'
type(flag)
```
# Integer/Float Vairables
```
a = 5
type(a)
b = 10
type(b)
a + b
a / b # Difference between python2 and python3
c = a / b
type(c)
```
<span style="color:red">**Note**:</span> Variable `a` and `b` both are of type `int` result in variable `c` is of type `float`. This is new in `Python3`
```
c = b % a
c
type(c)
a = 2
b = 5
a ** b # a^b => 2^5
pow(a, b)
```
<span style="color:red">**Note:**</span> All the Variables are case sensitive in `Python`
```
A # Upper case a
```
<span style="color:red">**Note:**</span> `Traceback` are exceptions in `Python`. This is `NameError` exception is because since variable `A` is not found.
# String Variables
```
a = "Hello World!"
type(a)
b = input()
type(b)
b = int(b)
type(b)
a = "Hello"
a + " " + b
a + ' ' + b
```
<span style="color:red">**Note:**</span> String concatenation `+` creates new string. To make new string when we know the format of the log. It is better to use String format.
```
c = '%s %s' % (a, b)
print(c)
a = 5
'Value of a is %d' % a
a = 3.141
"Value of a is %.2f" % a
"Value of a is %9.2f" % a
"Value of a is %09.2f" % a
```
### Multiplication on strings
Essentially a multiple concatenation
```
"-" * 4
a = "サブブ"
type(a)
a = u"サブブ"
type(a)
```
# Simple Python Script
Let's create file `simple.py` with the following code
```python
#!/usr/bin/env python
# FileName: simple.py
print("Hello World!")```
#### To run the Python script
```bash
$ python simple.py```
#### To run the Python script as standalone. File has to be made executable by the following commands
For example:
```bash
$ chmod +x scriptname```
```bash
$ chmod 755 scriptname```
In our `simple.py`
```bash
$ ls -lrth
-rw-r--r-- 1 username 1912965087 45B Apr 6 18:42 simple.py
$ chmod +x simple.py
$ ls -lrth
-rwxr-xr-x 1 username 1912965087 45B Apr 6 18:42 simple.py
$ ./simple.py```
# Python internals
## Comiple code PVM (Python Virtual Machine)
```bash
$ ipython
Python 3.6.1 (default, Apr 4 2017, 09:40:51)
Type "copyright", "credits" or "license" for more information.
IPython 5.3.0 -- An enhanced Interactive Python.
? -> Introduction and overview of IPython's features.
%quickref -> Quick reference.
help -> Python's own help system.
object? -> Details about 'object', use 'object??' for extra details.
In [1]: import py_compile
In [2]: py_compile.compile('simple.py')
Out[2]: '__pycache__/simple.cpython-36.pyc'
$ ls -lrth *
-rwxr-xr-x 1 username 1912965087 45B Apr 6 18:42 simple.py
__pycache__:
-rw-r--r-- 1 username 1912965087 118B Apr 6 18:54 simple.cpython-36.pyc```
Using shell:
```bash
$ python -m py_compile my_first_simple_script.py```
- Developer don't have to and shouldn't bother about compiling Python code.
- The compilation is hidden from the user for a good reason. Some newbies to Python wonder sometimes where these ominous files with the `.pyc` suffix might come from.
- If Python has write-access for the directory where the Python program resides, it will store the compiled byte code in a file that ends with a `.pyc` suffix.
- If Python has no write access, the program will work anyway. The byte code will be produced but discarded when the program exits.
- Whenever a Python program is called, Python will check, if there exists a compiled version with the `.pyc` suffix. This file has to be newer than the file with the `.py` suffix.
- If such a file exists, Python will load the byte code, which will speed up the start up time of the script.
- If there exists no byte code version, Python will create the byte code before it starts the execution of the program. Execution of a Python program means execution of the byte code on the Python Virtual Machine (PVM).
## Byte code
Every time a Python script is executed, byte code is created. If a Python script is imported as a module, the byte code will be stored in the corresponding .pyc file.
This won't create a Byte code.
```bash
$ python simple.py```
```bash
$ python
Python 3.6.1 (default, Apr 4 2017, 09:40:51)
[GCC 4.2.1 Compatible Apple LLVM 8.0.0 (clang-800.0.42.1)] on darwin
Type "help", "copyright", "credits" or "license" for more information.
>>> import simple
Hello World!
>>>
(.venv) $ ls -lrth *
-rwxr-xr-x 1 username 1912965087 45B Apr 6 18:42 simple.py
__pycache__:
total 8
-rw-r--r-- 1 username 1912965087 150B Apr 6 19:29 simple.cpython-36.pyc```
# Structuring with Indentation
## Block
A block is a group of statements in a program or script. Usually it consists of at least one statement and of declarations for the block, depending on the programming or scripting language. A language, which allows grouping with blocks, is called a block structured language. Generally, blocks can contain blocks as well, so we get a nested block structure. A block in a script or program functions as a mean to group statements to be treated as if they were one statement. In many cases, it also serves as a way to limit the lexical scope of variables and functions
## Indentation
Python uses a different principle. Python programs get structured through indentation, i.e. code blocks are defined by their indentation. Okay that's what we expect from any program code, isn't it? Yes, but in the case of Python it's a language requirement not a matter of style. This principle makes it easier to read and understand other people's Python code.
Following is an example.
Loops and Conditional statements and function end with a colon "`:`".
```
# Python structures by colons and indentation. Here is a small example to print Pyramid
def print_pyramid(n):
for each_line in range(0,n):
for j in range(0,n-each_line):
print(' ', end='', flush=True)
for k in range(0, each_line):
print('* ', end='', flush=True)
print()
print('Pyramid Printing completed!')
print_pyramid(10)
```
# More on `Python` Variables
Good news! There is no declaration of variables required in Python.
**Another remarkable aspect of Python**: Not only the value of a variable may change during program execution but the type as well. You can assign an integer value to a variable, use it as an integer for a while and then assign a string to the same variable.
### Valid Variable Names
The naming of variables follows the more general concept of an identifier. A Python identifier is a name used to identify a variable, function, class, module or other object.
- Uppercase letters "`A`" through "`Z`", the lowercase letters "`a`" through "`z`".
- The underscore `_`.
- Except for the first character, the digits 0 through 9.
- Python 3.x is based on Unicode. This means that variable names and identifier names can additionally contain Unicode characters as well.
- Identifiers are unlimited in length.
- Case is very important (significant).
### Python Keywords
No identifier can have the same name as one of the Python keywords, although they are obeying the above naming conventions:
```python
and, as, assert, break, class, continue, def, del, elif, else,
except, False, finally, for, from, global, if, import, in, is,
lambda, None, nonlocal, not, or, pass, raise, return, True, try,
while, with, yield```
There is no need to learn them by heart. You can get the list of Python keywords in the interactive shell by using help. You type in help() in the interactive, but please don't forget the parenthesis:
<span style="color:red">**Note:**</span> Enter <code style="color:blue">`keywords`</code>
```
help()
pi = 3 # data type is implicitly set to integer
pi = 3 + 0.141 # data type is changed to float
pi = "pi" # and now it will be a string
# A new object, which can be of any type, will be assigned to it.
# Or the type of a variable can change during the execution of a script.
```
<span style="color:red">**Note:**</span> Python automatically takes care of the physical representation for the different data types, i.e. an integer values will be stored in a different memory location than a float or a string.
# Object References
We want to take a closer look on variables now. Python variables are references to objects, but the actual data is contained in the objects
As variables are pointing to objects and objects can be of arbitrary data type, variables cannot have types associated with them. This is a huge difference to C, C++ or Java, where a variable is associated with a fixed data type. This association can't be changed as long as the program is running in C, C++ or Java.
## To demonstrate
```
x = 42
y = x
print(x)
print(y)
y = 60
print(y)
print(x)
```
# `id` Function
```
x = 42
id(x)
y = x
id(x), id(y)
y = 78
id(x), id(y)
a = (2+2)/4
b = 1 * 1
c = 6-5
print(type(a), type(b), type(c))
id(a), id(b), id(c)
```
# Naming Conventions
#### `CamelCase` vs `camel_case`
The Style Guide for Python Code recommends underscore notation for variable names as well as function names.
Python preferred is `camel_case`
https://www.python.org/dev/peps/pep-0008/#function-names
# Strings in Python
A string in Python consists of a series or sequence of characters - letters, numbers, and special characters. Strings can be subscripted or indexed. Similar to C, the first character of a string has the index 0.
|`0`|`1`|`2`|`3`|`4`|`5`|`6`|`7`|`8`|`9`|`10`|`11`|
|:-:|:-:|:-:|:-:|:-:|:-:|:-:|:-:|:-:|:-:|:-:|:-:|
|H|e|l|l|o| |W|o|r|l|d|!|
|-12|-11|-10|-9|-8|-7|-6|-5|-4|-3|-2|-1|
```
a = 'Hello World!'
a[0]
a[6]
```
Length of the string
```
len(a)
```
Last Character can be accessed like this
```
a[len(a)-1]
```
There is an easier way in Python. The last character can be accessed with -1, the second to last with -2 and so on
```
a[-1]
a[-2]
```
# Slicing
Substrings can be created with the slice or slicing notation.
|`0`|`1`|`2`|`3`|`4`|`5`|`6`|`7`|`8`|`9`|`10`|`11`|
|:-:|:-:|:-:|:-:|:-:|:-:|:-:|:-:|:-:|:-:|:-:|:-:|
|H|e|l|l|o| |W|o|r|l|d|!|
```
a[3:7]
```
## Immutable Strings
Like strings in Java and unlike C or C++, Python strings cannot be changed. Trying to change an indexed position will raise an error:
```
a[5] = '.'
```
## A String Peculiarity
This holds true for `objects` too.
```
a = 'HelloWorld'
b = 'HelloWorld'
print(a == b)
print(a is b)
print(id(a), id(b))
a = 'Hello World'
b = 'Hello World'
print(a == b)
print(a is b)
print(id(a), id(b))
a = "Baden-Württemberg"
b = "Baden-Württemberg"
print(a == b)
print(a is b)
print(id(a), id(b))
a = None
print(a is None)
print(a == None)
a = ''
print(a == '')
print(a is '')
```
## Escape Sequences in Strings
|Escape Sequence|Meaning Notes|
|:-:|:-|
|\newline|Ignored|
|\\|Backslash (\)|
|\'|Single quote (')|
|\"|Double quote (")|
|\a|ASCII Bell (BEL)|
|\b|ASCII Backspace (BS)|
|\f|ASCII Formfeed (FF)|
|\n|ASCII Linefeed (LF)|
|\N{name}|Character named name in the Unicode database (Unicode only)|
|\r|ASCII Carriage Return (CR)|
|\t|ASCII Horizontal Tab (TAB)|
|\uxxxx|Character with 16-bit hex value xxxx (Unicode only)|
|\Uxxxxxxxx|Character with 32-bit hex value xxxxxxxx (Unicode only)|
|\v|ASCII Vertical Tab (VT)|
|\ooo|Character with octal value ooo|
|\xhh|Character with hex value hh|
# Byte Strings
Python 3.0 uses the concepts of text and (binary) data instead of Unicode strings and 8-bit strings. Every string or text in Python 3 is Unicode, but encoded Unicode is represented as binary data. The type used to hold text is `str`, the type used to hold data is bytes. It's not possible to mix text and data in Python 3; it will raise `TypeError`.
While a string object holds a sequence of characters (in Unicode), a bytes object holds a sequence of bytes, out of the range `0 .. 255`, representing the ASCII values.
Defining bytes objects and casting them into strings:
```
x = b"Hallo"
print(x)
type(x)
t = str(x)
print(t)
t
u = t.encode("UTF-8")
print(u)
u
a = 'srinu 姓'
print(type(a))
print(type(a.encode('UTF-8')))
a.encode('UTF-8')
x = b"Hola \xe5\xa7\x93"
print(type(x))
# Convert byte string to UTF-8
x.decode("utf-8")
type(x.decode('utf-8'))
```
# Some good practices
**In** `list` **:** `append` **vs** `+`
```
import timeit
timeit.timeit('''a = []
for i in range(0, 1000):
a += [i]''', number=10000)
a = [None]
for i in range(0,10):
a[0] = i
print(a)
timeit.timeit('''a = []
for i in range(0, 1000):
a.append(i)''', number=10000)
```
**In** `str` **:** `join` **vs** `+`
```
import timeit
timeit.timeit('''a = ''
for i in range(0, 100):
a += str(i) + ' ' ''', number=1000)
a = []
for i in range(0, 10):
a.append(str(i))
print(a)
print('$$$'.join(a))
import timeit
timeit.timeit('''a = [str(i) for i in range(0, 100)]
''.join(a)''', number=1000)
```
`Console Outputs` **:** `print` **vs** `stdout` and `stderr`
```
import timeit
timeit.timeit('print("Hello")', number=10)
import sys
import timeit
timeit.timeit('sys.stdout.write("Hello\\n")', number=10)
import sys
import timeit
timeit.timeit('sys.stderr.write("Hello\\n")', number=10)
sys.stdout.write('Started\n')
try:
print(1/0)
sys.stdout.write('Done')
except Exception:
sys.stderr.write('Error')
sys.stdout.write('Completed')
```
# Data Types we mainly use in our daily code
- `int`
- `str`
- `float`
- `list`
- `tuple`
- `dict`
- `set`
- `None`
There are other like:
- `frozenset`
- `complex`
# `float`
```
a = 5.222222222222222222222222222222222222111111111111111111111
a
len(str(a))
a = 5897248172984712897498127498712984719828749827497298729874498274719274918274129748912374.1
a
type(a)
```
# `list`
```
a = [] # Empty list
b = list() # Empty list
print(a, b)
a = [1, 2, 3]
b = [40, 5, 6]
a + b
a, b
a.extend(b)
a, b
a.append(7)
a
a.index(1)
a, a.index(40)
a.append(40)
a.index(9)
a.remove(40)
b = [50 for i in range(10)]
print(b)
while True:
if 50 in b:
b.remove(50)
else:
break
print(b)
a
a.insert(0, 0)
a
a.insert(0, 999)
a
a.insert(3, 300)
a
a.clear()
a
a = [999, 0, 1, 300, 2, 3, 4, 5, 6]
c = a.sort()
a, c
a.reverse()
a
a.pop() # pops the last element
a
a.pop(2) # By Specifying the index
a
a = [1, 1, 1, 2, 3, 3, 4, 4, 4, 4, 4]
a.count(1)
a.count(2)
a.count(3)
a.count(4)
a.remove(1)
a
```
# `dict`
```
a = {} # Empty Dict
b = dict() # Empty Dict
print(a, b)
each = 'one'
a[each] = 1
a
b = {'two': 2, 'five': 5}
b
a + b
# a = {'two': 2000}
a.update(b)
a, b
a.get('one')
a['one']
a['three']
a.get('three', 3)
print(a)
a.clear()
a
a = {'one': (1,2), 'two': 2, 'three': 3}
a.keys()
a.get('one')
a.values()
a.items()
a.pop('one')
a
a.popitem()
a.setdefault('four', 4)
a
a.setdefault('four', 40)
a
```
# `set`
```
a = {} # Empty Set
b = set() # Empty set
print(a, b)
a = set([1, 1, 2, 2, 3, 3, 4, 4])
a
b = set([5, 6, 7, 8])
a + b
a.update(b)
a
a = set([1, 1, 2, 2, 3, 3, 4, 4])
b = set([4, 5, 6, 7, 8])
a.union(b)
a
b
a = {1, 2, 3, 4}
b = {3, 4, 5, 6}
a.intersection(b)
a.difference(b)
a.symmetric_difference(b)
a.discard(1)
a
a.issubset(b)
a = {1, 2, 3, 4}
b = {1, 2, 3, 4, 5, 6}
a.issubset(b)
a = {1, 2, 3, 4}
b = {1, 2, 3, 4, 5, 6}
b.issuperset(a)
a = {1, 2, 3, 4}
b = {1, 2, 3, 4, 5, 6}
b.difference_update(a)
a, b
a = {1, 2, 3, 4}
b = {1, 2, 3, 4, 5, 6}
a - b
b - a
```
# `tuple`
```
a = () # Empty tuple
b = tuple() # Empty Tuple
print(a, b)
1, 2
a = 1, 2
a
b = 3, 4
b
a + b
a.index(2)
c = a + b
print(c)
c.index(3)
c
a_list = [1, 2, 3, 4]
a_tuple = (1, 2, 3, 4)
a_list[0] = 999
a_list
a_tuple[0] = 999
```
# `if` conditioins
```
if True:
print('This is boolean True')
if False:
print('This is boolean False')
if not False:
print('This is boolean False')
if None:
print('This is None')
if not None:
print('This is None')
```
# Introduction of `else` and `elif`
```
# Find if the number is even
a = 6
if (a % 2) == 0:
print('a is even %s' % a)
else:
print('a is odd %s' % a)
a = 10000 # Introduced conversion of string to int
if a < 0:
print('a is negative number')
elif 0 <= a <= 100: # (0 <= a) and (a <=100)
print('a is in range 0 to 100')
else:
print('a is not in range 0 to 100')
```
# more on `if`
```
if 1:
print('This is 1. Valid True')
if 0:
print('This is 0. Valid False')
if not 0:
print('This is 0. Valid False')
# Re writing. Find if the number is even
a = 11
if not (a % 2): # (a % 2) == 0
print('a is even %s' % a)
else:
print('a is odd %s' % a)
'Success' if True else 'Fail' # Single liner if
'Success' if False else 'Fail'
a = 11
b = 'Odd' if a % 2 else 'Even'
print('a is %s number: %s' % (b, a))
```
# `try` `except` block
```
# Re writing. Find if the number is even
try:
a = 'hello'
if not (a % 2):
print('a is even %s' % a)
else:
print('a is odd %s' % a)
except Exception as err:
print('a is not a valid integer %s' % (err,))
a = ['a', 'apple', '', 1, 0, -1, None, {}, [], set(), (), {'a': 1}, {1}, [1], (1,), [None], (None, )]
for each in a:
print(type(each), end ='; ')
if each:
print('This is valid True %s' % str(each))
else:
print('This is valid False %s' % str(each))
```
# Already introduced `for`
```
# List
a = [1, 2, 3, 4, 5]
for each in a:
print(each)
# Tuple
a = (1, 2, 3, 4, 5)
for each in a:
print(each)
# Set
a = {1, 2, 3, 4, 5}
for each in a:
print(each)
# Dict: only Keys
a = {'one': 1, 'two': 2, 'three': 3, 'four': 4}
for each in a:
print(each)
# Dict: only Values
a = {'one': 1, 'two': 2, 'three': 3, 'four': 4}
for each in a.values():
print(each)
# Dict: key, value in tuple
a = {'one': 1, 'two': 2, 'three': 3, 'four': 4}
for each in a.items():
print(each) # Here each is a tuple
# Dict: key and value individually
a = {'one': 1, 'two': 2, 'three': 3, 'four': 4}
for k, v in a.items():
print(k, v)
```
# Mapping `list`s
```
# Mapping two lists
a = ['one', 'two', 'three', 'four', 'five']
b = [1, 2, 3, 4]
for each in zip(b, a):
print(each)
# Mapping two lists and converting them to a dictionary
dict(zip(b, a))
```
# Exercises
```
# To print all the odd numbers from 0 to 100 into a list
odd_number_list = []
for each in range(0, 100):
if (each%2 != 0):
odd_number_list.append(each)
print(odd_number_list)
# To print all the odd numbers from 0 to 100 into a list
odd_number_list = []
for each in range(0, 100):
if (each%2):
odd_number_list.append(each)
print(odd_number_list)
a = []
for each in range(0, 100):
a.append(each)
print(a)
```
# `list` comprehension
```
# Make a list that contains values from 0 to 100
a = [number for number in range(0, 100)]
print(a)
# Make a list that contains all the odd numbers from 0 to 100
a=[number for number in range(0, 100) if number%2]
print(a)
# All elements divisible by 3
a=[number for number in range(0, 100) if not number%3]
print(a)
# print square of all the elements from 0, 10
a = [number*number for number in range(0, 10)]
print(a)
def square(x):
return x*x
a = [square(x) for x in range(5)]
print(a)
# print squareroot of all the elements from 0, 10
import math
a = [math.sqrt(number) for number in range(0, 10)]
print(a)
```
# Introduction to methods
```
def method():
a = 1 +2
b = 3 +4
pass
print(a , b)
print(method())
def method():
return
method()
def square(n):
return n*n
square(10)
def compare(a, b):
return a <= b
compare(20, 30)
var = ('prefix-set FAILOVER_v4 '
'0.0.0.0/0 le 32 \n'
'end-set \n'
'! \n')
print(var)
def char_counter(input_str):
counter_dict = dict()
for each in input_str:
counter_dict.setdefault(each, 0)
counter_dict[each] += 1
return counter_dict
print(char_counter('hello Srinu'))
a = input()
print(a)
print(type(a))
a = int(a)
print(a)
print(type(a))
0
0 1
0 1 2
0 1 2 3
0 1 2 3 4
0 1 2 3 4 5
0 1 2 3 4 5 6
0 1 2 3 4 5 6 7
0
def test():
max_int = int(input())
a = []
for each in range(0, max_num):
a.append(each)
print(a)
print('0 1 2 3')
c = ' '.join(['0', '1', '2', '3'])
type(c)
def test(max_int): # O(n*n)
a = ''
for each in range(0, max_int+1): # O(n)
for i in range(0, each+1): # O(n)
a += str(i)
a += ' '
a += '\n'
return a
print(test(5))
10 in [4, 3, 2, 7, 8, 2, 1, 3]
```
|
github_jupyter
|
$ ruby -e "$(curl -fsSL https://raw.githubusercontent.com/Homebrew/install/master/install)" < /dev/null 2> /dev/null```
#### Using `brew` installing Python3 locally
#### To find the path for `python3`
## On Windows
Sorry, can't say much as I am not a windows user.
https://docs.python.org/3/using/windows.html
## On linux
### `pip`
`pip` is a package management system used to install and manage software packages written in `Python`. Many packages can be found in the `Python Package Index (PyPI)`.
https://en.wikipedia.org/wiki/Pip_(package_manager)
https://pypi.python.org/pypi/pip
### Ubuntu
`Ubuntu 16.04` ships with both `Python3` and `Python2`.
By default `pip` for Python2 is installed but not for Python3.
Now we need to install `pip3` for `Python3`.
#### Installing `Python3` `virtualenv`
#### If required upgrade `pip`
#### `Python3 virtualenv`
#### `Python2 virtualenv`
### CentOS / Amazon AMI
# Python virtualenv installation
# Advantages of virtualenv
- Isolated Python runtime environments without modifying the root or system python installation.
- Eash while deploying several python applications and have different settings.
- Helpful when runtime dependencies differ between frameworks or libraries in various applications.
- `virtualenv` is very useful in a more dynamic development environment.
# Creating Virtualenv
Steps to create `virtualenv`
# Activating the Virtualenv
We can install libararies only to this environment without modifying the actual system python installation using `pip`
#### To start `python` interactive shell
#### How to properly Quit `python` interactive shell
- You can either use exit() or Ctrl-D (i.e. EOF) to exit.
- The brackets behind the exit function are crucial.
- exit without brackets won't work in `python3`
#### How to deactivate the `virtualenv`
Just by running the command `deactivate` we can come out the `virtualenv`
# Going forward we are going to use only `Python3`
- End Of Life for `Python2.7` is on April 12th, 2020
- Security patches
- Not to invest time on code portability and re-writing the existing code
- More here on https://pythonclock.org/
# Interactive shell Continued
## Primitive data types
|Variable type|Example|Comment|
|:-----------:|:-------:|:-------------|
|`bool`|`True/False`| boolean true/false values|
|`int` `long`| 99999999999| Numbers/Integers (forget about the range of integers in `Python`) <br> Note: `long` wont see long as type in `python`|
|`float`| 0.99999999999| Decimals/Not whole integers|
|`str`| `"Hello World!"`, `'Hello World!'`, `'''Hello World!'''`, `"""Hello World!"""`| Any text|
|`None`| `None`| To create empty variable without any meaningful value|
# Different ways to define strings
All primitive data types can be stored in a variable using an assignment operator: `=`
# Boolean Variables
# Integer/Float Vairables
<span style="color:red">**Note**:</span> Variable `a` and `b` both are of type `int` result in variable `c` is of type `float`. This is new in `Python3`
<span style="color:red">**Note:**</span> All the Variables are case sensitive in `Python`
<span style="color:red">**Note:**</span> `Traceback` are exceptions in `Python`. This is `NameError` exception is because since variable `A` is not found.
# String Variables
<span style="color:red">**Note:**</span> String concatenation `+` creates new string. To make new string when we know the format of the log. It is better to use String format.
### Multiplication on strings
Essentially a multiple concatenation
# Simple Python Script
Let's create file `simple.py` with the following code
#### To run the Python script
#### To run the Python script as standalone. File has to be made executable by the following commands
For example:
In our `simple.py`
# Python internals
## Comiple code PVM (Python Virtual Machine)
Using shell:
- Developer don't have to and shouldn't bother about compiling Python code.
- The compilation is hidden from the user for a good reason. Some newbies to Python wonder sometimes where these ominous files with the `.pyc` suffix might come from.
- If Python has write-access for the directory where the Python program resides, it will store the compiled byte code in a file that ends with a `.pyc` suffix.
- If Python has no write access, the program will work anyway. The byte code will be produced but discarded when the program exits.
- Whenever a Python program is called, Python will check, if there exists a compiled version with the `.pyc` suffix. This file has to be newer than the file with the `.py` suffix.
- If such a file exists, Python will load the byte code, which will speed up the start up time of the script.
- If there exists no byte code version, Python will create the byte code before it starts the execution of the program. Execution of a Python program means execution of the byte code on the Python Virtual Machine (PVM).
## Byte code
Every time a Python script is executed, byte code is created. If a Python script is imported as a module, the byte code will be stored in the corresponding .pyc file.
This won't create a Byte code.
# Structuring with Indentation
## Block
A block is a group of statements in a program or script. Usually it consists of at least one statement and of declarations for the block, depending on the programming or scripting language. A language, which allows grouping with blocks, is called a block structured language. Generally, blocks can contain blocks as well, so we get a nested block structure. A block in a script or program functions as a mean to group statements to be treated as if they were one statement. In many cases, it also serves as a way to limit the lexical scope of variables and functions
## Indentation
Python uses a different principle. Python programs get structured through indentation, i.e. code blocks are defined by their indentation. Okay that's what we expect from any program code, isn't it? Yes, but in the case of Python it's a language requirement not a matter of style. This principle makes it easier to read and understand other people's Python code.
Following is an example.
Loops and Conditional statements and function end with a colon "`:`".
# More on `Python` Variables
Good news! There is no declaration of variables required in Python.
**Another remarkable aspect of Python**: Not only the value of a variable may change during program execution but the type as well. You can assign an integer value to a variable, use it as an integer for a while and then assign a string to the same variable.
### Valid Variable Names
The naming of variables follows the more general concept of an identifier. A Python identifier is a name used to identify a variable, function, class, module or other object.
- Uppercase letters "`A`" through "`Z`", the lowercase letters "`a`" through "`z`".
- The underscore `_`.
- Except for the first character, the digits 0 through 9.
- Python 3.x is based on Unicode. This means that variable names and identifier names can additionally contain Unicode characters as well.
- Identifiers are unlimited in length.
- Case is very important (significant).
### Python Keywords
No identifier can have the same name as one of the Python keywords, although they are obeying the above naming conventions:
There is no need to learn them by heart. You can get the list of Python keywords in the interactive shell by using help. You type in help() in the interactive, but please don't forget the parenthesis:
<span style="color:red">**Note:**</span> Enter <code style="color:blue">`keywords`</code>
<span style="color:red">**Note:**</span> Python automatically takes care of the physical representation for the different data types, i.e. an integer values will be stored in a different memory location than a float or a string.
# Object References
We want to take a closer look on variables now. Python variables are references to objects, but the actual data is contained in the objects
As variables are pointing to objects and objects can be of arbitrary data type, variables cannot have types associated with them. This is a huge difference to C, C++ or Java, where a variable is associated with a fixed data type. This association can't be changed as long as the program is running in C, C++ or Java.
## To demonstrate
# `id` Function
# Naming Conventions
#### `CamelCase` vs `camel_case`
The Style Guide for Python Code recommends underscore notation for variable names as well as function names.
Python preferred is `camel_case`
https://www.python.org/dev/peps/pep-0008/#function-names
# Strings in Python
A string in Python consists of a series or sequence of characters - letters, numbers, and special characters. Strings can be subscripted or indexed. Similar to C, the first character of a string has the index 0.
|`0`|`1`|`2`|`3`|`4`|`5`|`6`|`7`|`8`|`9`|`10`|`11`|
|:-:|:-:|:-:|:-:|:-:|:-:|:-:|:-:|:-:|:-:|:-:|:-:|
|H|e|l|l|o| |W|o|r|l|d|!|
|-12|-11|-10|-9|-8|-7|-6|-5|-4|-3|-2|-1|
Length of the string
Last Character can be accessed like this
There is an easier way in Python. The last character can be accessed with -1, the second to last with -2 and so on
# Slicing
Substrings can be created with the slice or slicing notation.
|`0`|`1`|`2`|`3`|`4`|`5`|`6`|`7`|`8`|`9`|`10`|`11`|
|:-:|:-:|:-:|:-:|:-:|:-:|:-:|:-:|:-:|:-:|:-:|:-:|
|H|e|l|l|o| |W|o|r|l|d|!|
## Immutable Strings
Like strings in Java and unlike C or C++, Python strings cannot be changed. Trying to change an indexed position will raise an error:
## A String Peculiarity
This holds true for `objects` too.
## Escape Sequences in Strings
|Escape Sequence|Meaning Notes|
|:-:|:-|
|\newline|Ignored|
|\\|Backslash (\)|
|\'|Single quote (')|
|\"|Double quote (")|
|\a|ASCII Bell (BEL)|
|\b|ASCII Backspace (BS)|
|\f|ASCII Formfeed (FF)|
|\n|ASCII Linefeed (LF)|
|\N{name}|Character named name in the Unicode database (Unicode only)|
|\r|ASCII Carriage Return (CR)|
|\t|ASCII Horizontal Tab (TAB)|
|\uxxxx|Character with 16-bit hex value xxxx (Unicode only)|
|\Uxxxxxxxx|Character with 32-bit hex value xxxxxxxx (Unicode only)|
|\v|ASCII Vertical Tab (VT)|
|\ooo|Character with octal value ooo|
|\xhh|Character with hex value hh|
# Byte Strings
Python 3.0 uses the concepts of text and (binary) data instead of Unicode strings and 8-bit strings. Every string or text in Python 3 is Unicode, but encoded Unicode is represented as binary data. The type used to hold text is `str`, the type used to hold data is bytes. It's not possible to mix text and data in Python 3; it will raise `TypeError`.
While a string object holds a sequence of characters (in Unicode), a bytes object holds a sequence of bytes, out of the range `0 .. 255`, representing the ASCII values.
Defining bytes objects and casting them into strings:
# Some good practices
**In** `list` **:** `append` **vs** `+`
**In** `str` **:** `join` **vs** `+`
`Console Outputs` **:** `print` **vs** `stdout` and `stderr`
# Data Types we mainly use in our daily code
- `int`
- `str`
- `float`
- `list`
- `tuple`
- `dict`
- `set`
- `None`
There are other like:
- `frozenset`
- `complex`
# `float`
# `list`
# `dict`
# `set`
# `tuple`
# `if` conditioins
# Introduction of `else` and `elif`
# more on `if`
# `try` `except` block
# Already introduced `for`
# Mapping `list`s
# Exercises
# `list` comprehension
# Introduction to methods
| 0.882757 | 0.952264 |
# 作业二:文本分类 Part1
* 本次作业使用神经网络进行文本情感分类
* 使用Stanford-Sentiment-Treebank电影评论作为数据集
文件名|说明
:-:|:-:
senti.train.tsv | 训练数据
senti.dev.tsv | 验证数据
senti.test.tsv | 测试数据
* 文件的每一行是一个句子,和该句子的情感分类,中间由tab分割
**首先导入这次作业需要的包,并设置随机种子**
```
import random
from collections import defaultdict
from pathlib import Path
import numpy as np
import torch
import torch.nn as nn
import torch.nn.functional as F
import torchtext
import tqdm
def set_random_seed(seed):
random.seed(seed)
np.random.seed(seed)
torch.manual_seed(seed)
torch.cuda.manual_seed(seed)
torch.cuda.manual_seed_all(seed)
torch.backends.cudnn.benchmark = False
torch.backends.cudnn.deterministic = True
set_random_seed(2020)
device = torch.device('cuda:0' if torch.cuda.is_available else 'cpu')
```
**设定计算设备与数据集路径**
```
device = torch.device('cuda' if torch.cuda.is_available else 'cpu')
data_path = Path('/media/bnu/data/nlp-practice/sentiment-analysis/standford-sentiment-treebank')
print('PyTorch Version:', torch.__version__)
print('-' * 60)
if torch.cuda.is_available():
print('CUDA Device Count:', torch.cuda.device_count())
print('CUDA Device Name:')
for i in range(torch.cuda.device_count()):
print('\t', torch.cuda.get_device_name(i))
print('CUDA Current Device Index:', torch.cuda.current_device())
print('-' * 60)
print('Data Path:', data_path)
```
## 数据处理
### 定义数据集的Dataset
```
# 定义数据集中每一列的数据类型,用于传换成Tensor
text_field = torchtext.data.Field(sequential=True, batch_first=True, include_lengths=True)
label_field = torchtext.data.LabelField(sequential=False, use_vocab=False, dtype=torch.float)
# 将tsv数据构建为数据集
train_set, valid_set, test_set = torchtext.data.TabularDataset.splits(
path=data_path,
train='senti.train.tsv',
validation='senti.dev.tsv',
test='senti.test.tsv',
format='tsv',
fields=[('text', text_field), ('label', label_field)]
)
# 以训练集数据,构建单词表
text_field.build_vocab(train_set)
```
**简单测试**
```
print('Tabular Dataset Example:')
print('Text:', valid_set[10].text)
print('Label:', valid_set[10].label)
print('-' * 60)
print('Vocab: Str -> Index')
print(list(text_field.vocab.stoi.items())[:5])
print('Vocab: Index -> Str')
print(text_field.vocab.itos[:5])
print('Vocab Size:')
print(len(text_field.vocab))
```
### 定义数据集的Iterator
```
train_iter, valid_iter, test_iter = torchtext.data.BucketIterator.splits(
datasets=(train_set, valid_set, test_set),
batch_sizes=(256, 256, 256),
sort_key=lambda x: len(x.text),
sort_within_batch=True,
device=device,
)
```
**简单测试**
```
print('Train Iterator:')
for batch in train_iter:
print(batch)
print('-' * 60, '\n')
break
print('Valid Iterator:')
for batch in valid_iter:
print(batch)
print('-' * 60, '\n')
break
print('Test Iterator:')
for batch in test_iter:
print(batch)
print('-' * 60, '\n')
break
```
## 定义模型
### 词向量平均模型
```
class EmbedAvgModel(nn.Module):
def __init__(self, n_words, n_embed, p_drop, pad_idx):
super(EmbedAvgModel, self).__init__()
self.embed = nn.Embedding(n_words, n_embed, padding_idx=pad_idx)
self.linear = nn.Linear(n_embed, 1)
self.drop = nn.Dropout(p_drop)
def forward(self, inputs, mask):
# (batch, len, n_embed)
inp_embed = self.drop(self.embed(inputs))
# (batch, len, 1)
mask = mask.float().unsqueeze(2)
# (batch, len, n_embed)
inp_embed = inp_embed * mask
# (batch, n_embed)
sum_embed = inp_embed.sum(1) / (mask.sum(1) + 1e-5)
return self.linear(sum_embed).squeeze()
model = EmbedAvgModel(
n_words=len(text_field.vocab),
n_embed=100,
p_drop=0.2,
pad_idx=text_field.vocab.stoi['<pad>']
)
model.to(device)
```
### Attention加权平均模型
```
class AttnAvgModel(nn.Module):
def __init__(self, n_words, n_embed, p_drop, pad_idx):
super(AttnAvgModel, self).__init__()
self.embed = nn.Embedding(n_words, n_embed, padding_idx=pad_idx)
self.linear = nn.Linear(n_embed, 1)
self.drop = nn.Dropout(p_drop)
self.coef = nn.Parameter(torch.randn(1, 1, n_embed))
def forward(self, inputs, mask):
# (batch, len, n_embed)
inp_embed = self.embed(inputs)
# (batch, len)
inp_cos = F.cosine_similarity(inp_embed, self.coef, dim=-1)
inp_cos.masked_fill_(~mask, -1e5)
# (batch, 1, len)
inp_attn = F.softmax(inp_cos, dim=-1).unsqueeze(1)
# (batch, n_embed)
sum_embed = torch.bmm(inp_attn, inp_embed).squeeze()
sum_embed = self.drop(sum_embed)
return self.linear(sum_embed).squeeze()
def calc_attention_weight(self, text):
# (1, len, n_embed)
inp_embed = self.embed(text)
# (1, len)
inp_cos = F.cosine_similarity(inp_embed, self.coef, dim=-1)
# (batch, 1, len)
inp_attn = F.softmax(inp_cos, dim=-1)
return inp_attn
model = AttnAvgModel(
n_words=len(text_field.vocab),
n_embed=100,
p_drop=0.2,
pad_idx=text_field.vocab.stoi['<pad>']
)
model.to(device)
for batch in train_iter:
inputs, lengths = batch.text
mask = (inputs != text_field.vocab.stoi['<pad>'])
outputs = model(inputs, mask)
print(outputs.shape)
break
```
## 模型训练
```
class TCLearner:
def __init__(self, model):
self.model = model
self.model.to(device)
self.optimizer = torch.optim.AdamW(self.model.parameters(), lr=1e-3)
self.crirerion = nn.BCEWithLogitsLoss()
def _calc_correct_num(self, outputs, targets):
preds = torch.round(torch.sigmoid(outputs))
return (preds == targets).int().sum().item()
def fit(self, train_iter, valid_iter, n_epochs):
for epoch in range(n_epochs):
model.train()
total_loss = 0.0
total_sents, total_correct = 0, 0
for batch in train_iter:
inputs, lengths = batch.text
targets = batch.label
mask = (inputs != text_field.vocab.stoi['<pad>'])
outputs = self.model(inputs, mask)
loss = self.crirerion(outputs, targets)
self.optimizer.zero_grad()
loss.backward()
self.optimizer.step()
total_loss += loss.item() * len(targets)
total_sents += len(targets)
total_correct += self._calc_correct_num(outputs, targets)
epoch_loss = total_loss / total_sents
epoch_acc = total_correct / total_sents
print(f'Epoch {epoch+1}')
print(f'Train --> Loss: {epoch_loss:.3f}, Acc: {epoch_acc:.3f}')
model.eval()
total_loss = 0.0
total_sents, total_correct = 0, 0
with torch.no_grad():
for batch in valid_iter:
inputs, lengths = batch.text
targets = batch.label
mask = (inputs != text_field.vocab.stoi['<pad>'])
outputs = self.model(inputs, mask)
loss = self.crirerion(outputs, targets)
total_loss += loss.item() * len(targets)
total_sents += len(targets)
total_correct += self._calc_correct_num(outputs, targets)
epoch_loss = total_loss / total_sents
epoch_acc = total_correct / total_sents
print(f'Valid --> Loss: {epoch_loss:.3f}, Acc: {epoch_acc:.3f}')
def predict(self, test_iter):
model.eval()
total_loss = 0.0
total_sents, total_correct = 0, 0
with torch.no_grad():
for batch in test_iter:
inputs, lengths = batch.text
targets = batch.label
mask = (inputs != text_field.vocab.stoi['<pad>'])
outputs = self.model(inputs, mask)
loss = self.crirerion(outputs, targets)
total_loss += loss.item() * len(targets)
total_sents += len(targets)
total_correct += self._calc_correct_num(outputs, targets)
epoch_loss = total_loss / total_sents
epoch_acc = total_correct / total_sents
print(f'Test --> Loss: {epoch_loss:.3f}, Acc: {epoch_acc:.3f}')
```
### 词向量平均模型训练
```
model = EmbedAvgModel(
n_words=len(text_field.vocab),
n_embed=200,
p_drop=0.5,
pad_idx=text_field.vocab.stoi['<pad>']
)
model.to(device)
learner = TCLearner(model)
learner.fit(train_iter, valid_iter, 10)
learner.predict(test_iter)
```
**单词 L2 Norm分析**
```
# (n_words)
embed_norm = model.embed.weight.norm(dim=1)
word_idx = list(range(len(text_field.vocab)))
word_idx.sort(key=lambda x: embed_norm[x])
print('15个L2-Norm最小的单词:')
for i in word_idx[:15]:
print(text_field.vocab.itos[i])
print('-' * 60)
print('15个L2-Norm最大的单词:')
for i in word_idx[-15:]:
print(text_field.vocab.itos[i])
```
* 从上面的结果可以看出,L2-Norm小的单词往往是跟情感无关的单词
* L2-Norm大的单词基本都是能够反映情感的单词
### Attention加权平均模型的训练
```
model = AttnAvgModel(
n_words=len(text_field.vocab),
n_embed=200,
p_drop=0.5,
pad_idx=text_field.vocab.stoi['<pad>']
)
model.to(device)
learner = TCLearner(model)
learner.fit(train_iter, valid_iter, 10)
learner.predict(test_iter)
```
**分析计算向量u与词向量的余弦相似度**
```
# (1, n_embed)
u = model.coef.view(1, -1)
# (n_words, n_embed)
embedding = model.embed.weight
# (n_words)
cos_sim = F.cosine_similarity(u, embedding, dim=-1)
word_idx = list(range(len(text_field.vocab)))
word_idx.sort(key=lambda x: cos_sim[x])
print('15个余弦相似度最小的单词:')
for i in word_idx[:15]:
print(text_field.vocab.itos[i])
print('-' * 60)
print('15个余弦相似度最大的单词:')
for i in word_idx[-15:]:
print(text_field.vocab.itos[i])
```
* 前面的结果可以看出,余弦相似度比较高的单词都能够很好的反映句子的情感,这些单词在Attention后的权重会比较高
* 余弦相似度小的单词多为一些名词和介词,与文本表示的情感基本无关
**分析训练数据中单词的Attention权重**
```
train_iter, valid_iter, test_iter = torchtext.data.BucketIterator.splits(
datasets=(train_set, valid_set, test_set),
batch_sizes=(1, 1, 1),
sort_key=lambda x: len(x.text),
sort_within_batch=True,
device=device,
)
weight_dict = defaultdict(list)
with torch.no_grad():
for k, batch in enumerate(train_iter):
inputs, lengths = batch.text
attn = model.calc_attention_weight(inputs)
inputs = inputs.view(-1)
attn = attn.view(-1)
if inputs.shape[0] == 1:
weight_dict[inputs.item()].append(attn.item())
else:
for i in range(len(inputs)):
weight_dict[inputs[i].item()].append(attn[i].item())
if (k + 1) % 10000 == 0:
print(f'{k+1} sentences finish!')
mean_dict, std_dict = {}, {}
for k, v in weight_dict.items():
# 至少出现100次
if len(v) >= 100:
mean_dict[k] = np.mean(v)
std_dict[k] = np.std(v)
word_idx = list(std_dict.keys())
word_idx.sort(key=lambda x: std_dict[x], reverse=True)
print('30个Attention标准差最大的单词:')
print('-' * 60)
for i in word_idx[:30]:
print(f'{text_field.vocab.itos[i]}, Freq:{len(weight_dict[i])}, Std:{std_dict[i]:.3f}, Mean:{mean_dict[i]:.3f}')
print()
print('30个Attention标准差最小的单词:')
print('-' * 60)
for i in word_idx[-30:]:
print(f'{text_field.vocab.itos[i]}, Freq:{len(weight_dict[i])}, Std:{std_dict[i]:.3f}, Mean:{mean_dict[i]:.3f}')
```
* Attention权重标准差大的单词,其权重的的平均值也很大
* 这些标准差大的单词,往往会有明显的情感倾向,所以会有较大的权重平均值
* 造成权重标准差大的原因主要是因为句子的长短,由于句子长短不同包含的情感倾向单词数目不同,造成权重变化很大
|
github_jupyter
|
import random
from collections import defaultdict
from pathlib import Path
import numpy as np
import torch
import torch.nn as nn
import torch.nn.functional as F
import torchtext
import tqdm
def set_random_seed(seed):
random.seed(seed)
np.random.seed(seed)
torch.manual_seed(seed)
torch.cuda.manual_seed(seed)
torch.cuda.manual_seed_all(seed)
torch.backends.cudnn.benchmark = False
torch.backends.cudnn.deterministic = True
set_random_seed(2020)
device = torch.device('cuda:0' if torch.cuda.is_available else 'cpu')
device = torch.device('cuda' if torch.cuda.is_available else 'cpu')
data_path = Path('/media/bnu/data/nlp-practice/sentiment-analysis/standford-sentiment-treebank')
print('PyTorch Version:', torch.__version__)
print('-' * 60)
if torch.cuda.is_available():
print('CUDA Device Count:', torch.cuda.device_count())
print('CUDA Device Name:')
for i in range(torch.cuda.device_count()):
print('\t', torch.cuda.get_device_name(i))
print('CUDA Current Device Index:', torch.cuda.current_device())
print('-' * 60)
print('Data Path:', data_path)
# 定义数据集中每一列的数据类型,用于传换成Tensor
text_field = torchtext.data.Field(sequential=True, batch_first=True, include_lengths=True)
label_field = torchtext.data.LabelField(sequential=False, use_vocab=False, dtype=torch.float)
# 将tsv数据构建为数据集
train_set, valid_set, test_set = torchtext.data.TabularDataset.splits(
path=data_path,
train='senti.train.tsv',
validation='senti.dev.tsv',
test='senti.test.tsv',
format='tsv',
fields=[('text', text_field), ('label', label_field)]
)
# 以训练集数据,构建单词表
text_field.build_vocab(train_set)
print('Tabular Dataset Example:')
print('Text:', valid_set[10].text)
print('Label:', valid_set[10].label)
print('-' * 60)
print('Vocab: Str -> Index')
print(list(text_field.vocab.stoi.items())[:5])
print('Vocab: Index -> Str')
print(text_field.vocab.itos[:5])
print('Vocab Size:')
print(len(text_field.vocab))
train_iter, valid_iter, test_iter = torchtext.data.BucketIterator.splits(
datasets=(train_set, valid_set, test_set),
batch_sizes=(256, 256, 256),
sort_key=lambda x: len(x.text),
sort_within_batch=True,
device=device,
)
print('Train Iterator:')
for batch in train_iter:
print(batch)
print('-' * 60, '\n')
break
print('Valid Iterator:')
for batch in valid_iter:
print(batch)
print('-' * 60, '\n')
break
print('Test Iterator:')
for batch in test_iter:
print(batch)
print('-' * 60, '\n')
break
class EmbedAvgModel(nn.Module):
def __init__(self, n_words, n_embed, p_drop, pad_idx):
super(EmbedAvgModel, self).__init__()
self.embed = nn.Embedding(n_words, n_embed, padding_idx=pad_idx)
self.linear = nn.Linear(n_embed, 1)
self.drop = nn.Dropout(p_drop)
def forward(self, inputs, mask):
# (batch, len, n_embed)
inp_embed = self.drop(self.embed(inputs))
# (batch, len, 1)
mask = mask.float().unsqueeze(2)
# (batch, len, n_embed)
inp_embed = inp_embed * mask
# (batch, n_embed)
sum_embed = inp_embed.sum(1) / (mask.sum(1) + 1e-5)
return self.linear(sum_embed).squeeze()
model = EmbedAvgModel(
n_words=len(text_field.vocab),
n_embed=100,
p_drop=0.2,
pad_idx=text_field.vocab.stoi['<pad>']
)
model.to(device)
class AttnAvgModel(nn.Module):
def __init__(self, n_words, n_embed, p_drop, pad_idx):
super(AttnAvgModel, self).__init__()
self.embed = nn.Embedding(n_words, n_embed, padding_idx=pad_idx)
self.linear = nn.Linear(n_embed, 1)
self.drop = nn.Dropout(p_drop)
self.coef = nn.Parameter(torch.randn(1, 1, n_embed))
def forward(self, inputs, mask):
# (batch, len, n_embed)
inp_embed = self.embed(inputs)
# (batch, len)
inp_cos = F.cosine_similarity(inp_embed, self.coef, dim=-1)
inp_cos.masked_fill_(~mask, -1e5)
# (batch, 1, len)
inp_attn = F.softmax(inp_cos, dim=-1).unsqueeze(1)
# (batch, n_embed)
sum_embed = torch.bmm(inp_attn, inp_embed).squeeze()
sum_embed = self.drop(sum_embed)
return self.linear(sum_embed).squeeze()
def calc_attention_weight(self, text):
# (1, len, n_embed)
inp_embed = self.embed(text)
# (1, len)
inp_cos = F.cosine_similarity(inp_embed, self.coef, dim=-1)
# (batch, 1, len)
inp_attn = F.softmax(inp_cos, dim=-1)
return inp_attn
model = AttnAvgModel(
n_words=len(text_field.vocab),
n_embed=100,
p_drop=0.2,
pad_idx=text_field.vocab.stoi['<pad>']
)
model.to(device)
for batch in train_iter:
inputs, lengths = batch.text
mask = (inputs != text_field.vocab.stoi['<pad>'])
outputs = model(inputs, mask)
print(outputs.shape)
break
class TCLearner:
def __init__(self, model):
self.model = model
self.model.to(device)
self.optimizer = torch.optim.AdamW(self.model.parameters(), lr=1e-3)
self.crirerion = nn.BCEWithLogitsLoss()
def _calc_correct_num(self, outputs, targets):
preds = torch.round(torch.sigmoid(outputs))
return (preds == targets).int().sum().item()
def fit(self, train_iter, valid_iter, n_epochs):
for epoch in range(n_epochs):
model.train()
total_loss = 0.0
total_sents, total_correct = 0, 0
for batch in train_iter:
inputs, lengths = batch.text
targets = batch.label
mask = (inputs != text_field.vocab.stoi['<pad>'])
outputs = self.model(inputs, mask)
loss = self.crirerion(outputs, targets)
self.optimizer.zero_grad()
loss.backward()
self.optimizer.step()
total_loss += loss.item() * len(targets)
total_sents += len(targets)
total_correct += self._calc_correct_num(outputs, targets)
epoch_loss = total_loss / total_sents
epoch_acc = total_correct / total_sents
print(f'Epoch {epoch+1}')
print(f'Train --> Loss: {epoch_loss:.3f}, Acc: {epoch_acc:.3f}')
model.eval()
total_loss = 0.0
total_sents, total_correct = 0, 0
with torch.no_grad():
for batch in valid_iter:
inputs, lengths = batch.text
targets = batch.label
mask = (inputs != text_field.vocab.stoi['<pad>'])
outputs = self.model(inputs, mask)
loss = self.crirerion(outputs, targets)
total_loss += loss.item() * len(targets)
total_sents += len(targets)
total_correct += self._calc_correct_num(outputs, targets)
epoch_loss = total_loss / total_sents
epoch_acc = total_correct / total_sents
print(f'Valid --> Loss: {epoch_loss:.3f}, Acc: {epoch_acc:.3f}')
def predict(self, test_iter):
model.eval()
total_loss = 0.0
total_sents, total_correct = 0, 0
with torch.no_grad():
for batch in test_iter:
inputs, lengths = batch.text
targets = batch.label
mask = (inputs != text_field.vocab.stoi['<pad>'])
outputs = self.model(inputs, mask)
loss = self.crirerion(outputs, targets)
total_loss += loss.item() * len(targets)
total_sents += len(targets)
total_correct += self._calc_correct_num(outputs, targets)
epoch_loss = total_loss / total_sents
epoch_acc = total_correct / total_sents
print(f'Test --> Loss: {epoch_loss:.3f}, Acc: {epoch_acc:.3f}')
model = EmbedAvgModel(
n_words=len(text_field.vocab),
n_embed=200,
p_drop=0.5,
pad_idx=text_field.vocab.stoi['<pad>']
)
model.to(device)
learner = TCLearner(model)
learner.fit(train_iter, valid_iter, 10)
learner.predict(test_iter)
# (n_words)
embed_norm = model.embed.weight.norm(dim=1)
word_idx = list(range(len(text_field.vocab)))
word_idx.sort(key=lambda x: embed_norm[x])
print('15个L2-Norm最小的单词:')
for i in word_idx[:15]:
print(text_field.vocab.itos[i])
print('-' * 60)
print('15个L2-Norm最大的单词:')
for i in word_idx[-15:]:
print(text_field.vocab.itos[i])
model = AttnAvgModel(
n_words=len(text_field.vocab),
n_embed=200,
p_drop=0.5,
pad_idx=text_field.vocab.stoi['<pad>']
)
model.to(device)
learner = TCLearner(model)
learner.fit(train_iter, valid_iter, 10)
learner.predict(test_iter)
# (1, n_embed)
u = model.coef.view(1, -1)
# (n_words, n_embed)
embedding = model.embed.weight
# (n_words)
cos_sim = F.cosine_similarity(u, embedding, dim=-1)
word_idx = list(range(len(text_field.vocab)))
word_idx.sort(key=lambda x: cos_sim[x])
print('15个余弦相似度最小的单词:')
for i in word_idx[:15]:
print(text_field.vocab.itos[i])
print('-' * 60)
print('15个余弦相似度最大的单词:')
for i in word_idx[-15:]:
print(text_field.vocab.itos[i])
train_iter, valid_iter, test_iter = torchtext.data.BucketIterator.splits(
datasets=(train_set, valid_set, test_set),
batch_sizes=(1, 1, 1),
sort_key=lambda x: len(x.text),
sort_within_batch=True,
device=device,
)
weight_dict = defaultdict(list)
with torch.no_grad():
for k, batch in enumerate(train_iter):
inputs, lengths = batch.text
attn = model.calc_attention_weight(inputs)
inputs = inputs.view(-1)
attn = attn.view(-1)
if inputs.shape[0] == 1:
weight_dict[inputs.item()].append(attn.item())
else:
for i in range(len(inputs)):
weight_dict[inputs[i].item()].append(attn[i].item())
if (k + 1) % 10000 == 0:
print(f'{k+1} sentences finish!')
mean_dict, std_dict = {}, {}
for k, v in weight_dict.items():
# 至少出现100次
if len(v) >= 100:
mean_dict[k] = np.mean(v)
std_dict[k] = np.std(v)
word_idx = list(std_dict.keys())
word_idx.sort(key=lambda x: std_dict[x], reverse=True)
print('30个Attention标准差最大的单词:')
print('-' * 60)
for i in word_idx[:30]:
print(f'{text_field.vocab.itos[i]}, Freq:{len(weight_dict[i])}, Std:{std_dict[i]:.3f}, Mean:{mean_dict[i]:.3f}')
print()
print('30个Attention标准差最小的单词:')
print('-' * 60)
for i in word_idx[-30:]:
print(f'{text_field.vocab.itos[i]}, Freq:{len(weight_dict[i])}, Std:{std_dict[i]:.3f}, Mean:{mean_dict[i]:.3f}')
| 0.82573 | 0.81899 |
Example 6 - Isotropic bearings with damping.
=====
In this example, we use the rotor seen in Example 5.9.5 from 'Dynamics of Rotating Machinery' by MI Friswell, JET Penny, SD Garvey & AW Lees, published by Cambridge University Press, 2010.
The isotropic bearing Example 3 is repeated but with damping in the bearings. The, x and y directions are
uncoupled, with a translational stiffness of 1 MN/m and a damping of 3 kNs/m
in each direction.
```
from bokeh.io import output_notebook, show
import ross as rs
import numpy as np
output_notebook()
#Classic Instantiation of the rotor
shaft_elements = []
bearing_seal_elements = []
disk_elements = []
steel = rs.steel
for i in range(6):
shaft_elements.append(rs.ShaftElement(L=0.25, material=steel, n=i, idl=0, odl=0.05))
disk_elements.append(rs.DiskElement.from_geometry(n=2,
material=steel,
width=0.07,
i_d=0.05,
o_d=0.28
)
)
disk_elements.append(rs.DiskElement.from_geometry(n=4,
material=steel,
width=0.07,
i_d=0.05,
o_d=0.35
)
)
bearing_seal_elements.append(rs.BearingElement(n=0, kxx=1e6, kyy=1e6, cxx=3e3, cyy=3e3))
bearing_seal_elements.append(rs.BearingElement(n=6, kxx=1e6, kyy=1e6, cxx=3e3, cyy=3e3))
rotor595c = rs.Rotor(shaft_elements=shaft_elements,
bearing_elements=bearing_seal_elements,
disk_elements=disk_elements,n_eigen = 12)
show(rotor595c.plot_rotor(plot_type='bokeh'))
#From_section class method instantiation.
bearing_seal_elements = []
disk_elements = []
shaft_length_data = 3*[0.5]
i_d = 3*[0]
o_d = 3*[0.05]
disk_elements.append(rs.DiskElement.from_geometry(n=1,
material=steel,
width=0.07,
i_d=0.05,
o_d=0.28
)
)
disk_elements.append(rs.DiskElement.from_geometry(n=2,
material=steel,
width=0.07,
i_d=0.05,
o_d=0.35
)
)
bearing_seal_elements.append(rs.BearingElement(n=0, kxx=1e6, kyy=1e6, cxx=3e3, cyy=3e3))
bearing_seal_elements.append(rs.BearingElement(n=3, kxx=1e6, kyy=1e6, cxx=3e3, cyy=3e3))
rotor595fs = rs.Rotor.from_section(brg_seal_data=bearing_seal_elements,
disk_data=disk_elements,leng_data=shaft_length_data,
idl_data=i_d,odl_data=o_d, material_data=steel
)
show(rotor595fs.plot_rotor(plot_type='bokeh'))
#Obtaining results for w=0
modal595c = rotor595c.run_modal(0)
modal595fs = rotor595fs.run_modal(0)
print('Normal Instantiation =', modal595c.wn * 60 /(2*np.pi),'[RPM]')
print('\n')
print('From Section Instantiation =', modal595fs.wn * 60 /(2*np.pi),'[RPM]')
#Obtaining results for w=4000RPM
modal595c = rotor595c.run_modal(4000*np.pi/30)
print('Normal Instantiation =', modal595c.wn * 60 /(2*np.pi),'[RPM]')
show(rotor595c.run_campbell(np.linspace(0, 4000*np.pi/30, 50)).plot(plot_type='bokeh'))
```
|
github_jupyter
|
from bokeh.io import output_notebook, show
import ross as rs
import numpy as np
output_notebook()
#Classic Instantiation of the rotor
shaft_elements = []
bearing_seal_elements = []
disk_elements = []
steel = rs.steel
for i in range(6):
shaft_elements.append(rs.ShaftElement(L=0.25, material=steel, n=i, idl=0, odl=0.05))
disk_elements.append(rs.DiskElement.from_geometry(n=2,
material=steel,
width=0.07,
i_d=0.05,
o_d=0.28
)
)
disk_elements.append(rs.DiskElement.from_geometry(n=4,
material=steel,
width=0.07,
i_d=0.05,
o_d=0.35
)
)
bearing_seal_elements.append(rs.BearingElement(n=0, kxx=1e6, kyy=1e6, cxx=3e3, cyy=3e3))
bearing_seal_elements.append(rs.BearingElement(n=6, kxx=1e6, kyy=1e6, cxx=3e3, cyy=3e3))
rotor595c = rs.Rotor(shaft_elements=shaft_elements,
bearing_elements=bearing_seal_elements,
disk_elements=disk_elements,n_eigen = 12)
show(rotor595c.plot_rotor(plot_type='bokeh'))
#From_section class method instantiation.
bearing_seal_elements = []
disk_elements = []
shaft_length_data = 3*[0.5]
i_d = 3*[0]
o_d = 3*[0.05]
disk_elements.append(rs.DiskElement.from_geometry(n=1,
material=steel,
width=0.07,
i_d=0.05,
o_d=0.28
)
)
disk_elements.append(rs.DiskElement.from_geometry(n=2,
material=steel,
width=0.07,
i_d=0.05,
o_d=0.35
)
)
bearing_seal_elements.append(rs.BearingElement(n=0, kxx=1e6, kyy=1e6, cxx=3e3, cyy=3e3))
bearing_seal_elements.append(rs.BearingElement(n=3, kxx=1e6, kyy=1e6, cxx=3e3, cyy=3e3))
rotor595fs = rs.Rotor.from_section(brg_seal_data=bearing_seal_elements,
disk_data=disk_elements,leng_data=shaft_length_data,
idl_data=i_d,odl_data=o_d, material_data=steel
)
show(rotor595fs.plot_rotor(plot_type='bokeh'))
#Obtaining results for w=0
modal595c = rotor595c.run_modal(0)
modal595fs = rotor595fs.run_modal(0)
print('Normal Instantiation =', modal595c.wn * 60 /(2*np.pi),'[RPM]')
print('\n')
print('From Section Instantiation =', modal595fs.wn * 60 /(2*np.pi),'[RPM]')
#Obtaining results for w=4000RPM
modal595c = rotor595c.run_modal(4000*np.pi/30)
print('Normal Instantiation =', modal595c.wn * 60 /(2*np.pi),'[RPM]')
show(rotor595c.run_campbell(np.linspace(0, 4000*np.pi/30, 50)).plot(plot_type='bokeh'))
| 0.417865 | 0.798972 |
# Infinite plate with steady surface temperature
```
from IPython.display import Image
Image(filename='8.1.png',width=600,height=600)
```
Energy conservation equation:
$$\frac{\partial T}{\partial t}+v_x\frac{\partial T}{\partial x}+v_y\frac{\partial T}{\partial y}+v_z\frac{\partial T}{\partial z}=\alpha[\frac{\partial^2 T}{\partial x^2}+\frac{\partial^2 T}{\partial y^2}+\frac{\partial^2 T}{\partial z^2}+\frac{H_v}{\rho \hat {C_p}}]$$
No velocity and no heat generation, so we can remove zero terms:
$$\frac{\partial T}{\partial t}=\alpha \frac{\partial^2 T}{\partial x^2},\alpha=\frac{k}{\rho \hat{C_p}}$$
Initial conditions and boundary conditions:
$$T(x,t=0)=f(x),0<x<l$$
$$T(x=0,t)=t_0$$
$$T(x=l,t)=t_0$$
The problem here is that separation of variables will no longer work on this problem because the boundary conditions are no longer homogeneous. So, we need to convert the boundary conditions to homogeneous at first.
When $t\rightarrow{\infty}$ :
$$\lim_{t\rightarrow{\infty}}T(x,t)=T_E(x)$$
$T_E(x)$ is equilibrium temperature, and it should satisfy:
$$\frac{{\partial}^2T_E}{\partial x^2}=0, T_E(0)=t_0,T_E(l)=t_0$$
$$T_E(x)=C_1x+C_2$$
Applying $T_E(0)=t_0,T_E(l)=t_0$ gives:
$$C_1=0,C_2=t_0$$
So, $T_E(x)=t_0$
Define the function:
$$P(x,t)=T(x,t)-T_E(x)$$
Rewrite the function:
$$T(x,t)=P(x,t)+T_E(x)$$
So,
$$\frac{\partial T}{\partial t}=\frac{\partial P}{\partial t}+\frac{\partial T_E}{\partial t}=\frac{\partial P}{\partial t}$$
$$\frac{\partial^2 T}{\partial x^2}=\frac{\partial^2 P}{\partial x^2}+\frac{\partial^2 T_E}{\partial x^2}=\frac{\partial^2 P}{\partial x^2}$$
Rewrite the conversation equation:
$$\frac{\partial P}{\partial t}=\alpha \frac{\partial^2 P}{\partial x^2},\alpha=\frac{k}{\rho \hat{C_p}}$$
Initial conditions and boundary conditions for P(x,t):
$$P(x,t=0)=T(x,t=0)-T_E(x)=f(x)-t_0$$
$$P(x=0,t)=T(x=0,t)-T_E(0)=t_0-t_0=0$$
$$P(x=l,t)=T(x=l,t)-T_E(l)=t_0-t_0=0$$
The boundary conditions are homogeneous, so we can use separation of variables.
Assume that:
$$P(x,t)=\Phi(x)G(t)$$
So,
$$\Phi\frac{dG}{dt}=\alpha G\frac{d^2 \Phi}{dx^2}$$
$$\frac{1}{\alpha G(t)}=\frac{1}{\Phi(x)}\frac{d^2\Phi}{dx^2}$$
Assume$$\frac{1}{\alpha G(t)}=\frac{1}{\Phi(x)}\frac{d^2\Phi}{dx^2}=-\lambda$$
$\lambda$ is a positive constant.
Left-hand of the equation:
$$\frac{dG}{dt}=-\alpha \lambda G(t)$$
Use the boundary conditions:
$$P(0,t)=\Phi(0)G(t)=0,P(l,t)=\Phi(l)G(t)=0$$
$$\Phi(0)=0,\Phi(l)=0$$
Right-hand of the equation:
$$\frac{d^2\Phi}{dx^2}+\lambda{\Phi(x)}=0,\Phi(0)=0,\Phi(l)=0$$
$$\Phi(x)=C_3cos(\sqrt{\lambda} x)+C_4sin(\sqrt{\lambda} x)$$
Plug $\Phi(0)=0,\Phi(l)=0$:
$$\lambda_n(x)=(\frac{n\pi}{l})^2,\Phi_n(x)=sin(\frac{n\pi x}{l}),n=1,2,3,...$$
Plug $\lambda_n(x)=(\frac{n\pi}{l})^2$:
$$G(t)=B_n e^{-\alpha(\frac{n\pi}{l})^2t},n=1,2,3,...$$
$$B_n=\frac{2}{l}\int_{0}^{l}f(x)sin(\frac{n\pi x}{l})dx, n=1,2,3,...$$
So,
$$P_n(x,t)=\sum_{n=1}^{\infty}B_n sin(\frac{n\pi x}{l}) e^{-\alpha(\frac{n\pi}{l})^2t},n=1,2,3,...$$
$$T(x,t)=T_E(x)+P(x,t)=t_0+\sum_{n=1}^{\infty}B_n sin(\frac{n\pi x}{l}) e^{-\alpha(\frac{n\pi}{l})^2t}$$
The solution is:
$$T-t_0=\sum_{n=1}^{\infty}B_nsin(\frac{n\pi x}{l})exp(-n^2\pi^2F_0),F_0=\frac{\alpha t}{l^2},n=1,2,3,...$$
$$B_n=\frac{2}{l}\int_{0}^{l}f(x)sin(\frac{n\pi x}{l})dx,n=1,2,3,...$$
f(x) is known, so I plug 20 (constant), $cos(\frac{n\pi x}{l})$ and $x$ respectively and plot them.
Case 1:
$f(x)=20$
```
import numpy as np
import matplotlib.pyplot as plt
from mpl_toolkits.mplot3d import axes3d
from matplotlib import cm
d = 101
x = np.linspace(0, 1, d)
t = np.linspace(0, 1, d)
u = np.zeros((x.shape[0], t.shape[0]))
n = 1
L = 1
k = 0.3
xv, tv = np.meshgrid(x, t)
t0=10
# Initial values
B = (40/(n*np.pi))*(1-(-1)**n)
u = B * np.sin(n*np.pi*xv/L)*np.exp(-k*tv*(n*np.pi/L)**2)+t0
for ni in range(2, 1000):
B = (40/(ni*np.pi))*(1-(-1)**ni)
u += B * np.sin(ni*np.pi*xv/L)*np.exp(-k*tv*(ni*np.pi/L)**2)
fig = plt.figure(figsize=(7,7))
ax = fig.add_subplot(111, projection='3d')
ax.plot_surface(xv, tv, u, cmap=cm.coolwarm)
ax.set_xlabel('x')
ax.set_ylabel('t')
ax.view_init(elev=0, azim=45)
ax.set_zlim(0, 30)
fig, ax = plt.subplots(figsize=(7,7))
for i in range(0, d):
ax.plot(xv[i, :], u[i, :], c='k', alpha=i/d)
ax.set_xlabel('x')
ax.set_ylabel('T')
fig, ax = plt.subplots(figsize=(7,7))
for i in range(0,d):
ax.plot(tv[:, i], T[:, i], c='k', alpha=(1 - (i/d)))
ax.set_xlabel('t')
ax.set_ylabel('T')
```
Case 2:
$f(x)=cos(\frac{n\pi x}{l})$
```
n = 1
L = 1
k = 0.3
xv, tv = np.meshgrid(x, t)
t0=10
# Initial values
B = 1
u = B * np.sin(n*np.pi*xv/L)*np.exp(-k*tv*(n*np.pi/L)**2)+t0
for ni in range(2, 1000):
B = 1
u += B * np.sin(ni*np.pi*xv/L)*np.exp(-k*tv*(ni*np.pi/L)**2)
fig = plt.figure(figsize=(7,7))
ax = fig.add_subplot(111, projection='3d')
ax.plot_surface(xv, tv, u, cmap=cm.coolwarm)
ax.set_xlabel('x')
ax.set_ylabel('t')
ax.view_init(elev=0, azim=45)
ax.set_zlim(8, 15)
```
Case 3:
$f(x)=x$
```
n = 1
L = 1
k = 0.3
xv, tv = np.meshgrid(x, t)
t0=10
# Initial values
B = -2*L*(-1)**n/(n*np.pi)
u = B * np.sin(n*np.pi*xv/L)*np.exp(-k*tv*(n*np.pi/L)**2)+t0
for ni in range(2, 1000):
B = -2*L*(-1)**n/(n*np.pi)
u += B * np.sin(ni*np.pi*xv/L)*np.exp(-k*tv*(ni*np.pi/L)**2)
fig = plt.figure(figsize=(7,7))
ax = fig.add_subplot(111, projection='3d')
ax.plot_surface(xv, tv, u, cmap=cm.coolwarm)
ax.set_xlabel('x')
ax.set_ylabel('t')
ax.view_init(elev=0, azim=45)
ax.set_zlim(9, 13)
```
|
github_jupyter
|
from IPython.display import Image
Image(filename='8.1.png',width=600,height=600)
import numpy as np
import matplotlib.pyplot as plt
from mpl_toolkits.mplot3d import axes3d
from matplotlib import cm
d = 101
x = np.linspace(0, 1, d)
t = np.linspace(0, 1, d)
u = np.zeros((x.shape[0], t.shape[0]))
n = 1
L = 1
k = 0.3
xv, tv = np.meshgrid(x, t)
t0=10
# Initial values
B = (40/(n*np.pi))*(1-(-1)**n)
u = B * np.sin(n*np.pi*xv/L)*np.exp(-k*tv*(n*np.pi/L)**2)+t0
for ni in range(2, 1000):
B = (40/(ni*np.pi))*(1-(-1)**ni)
u += B * np.sin(ni*np.pi*xv/L)*np.exp(-k*tv*(ni*np.pi/L)**2)
fig = plt.figure(figsize=(7,7))
ax = fig.add_subplot(111, projection='3d')
ax.plot_surface(xv, tv, u, cmap=cm.coolwarm)
ax.set_xlabel('x')
ax.set_ylabel('t')
ax.view_init(elev=0, azim=45)
ax.set_zlim(0, 30)
fig, ax = plt.subplots(figsize=(7,7))
for i in range(0, d):
ax.plot(xv[i, :], u[i, :], c='k', alpha=i/d)
ax.set_xlabel('x')
ax.set_ylabel('T')
fig, ax = plt.subplots(figsize=(7,7))
for i in range(0,d):
ax.plot(tv[:, i], T[:, i], c='k', alpha=(1 - (i/d)))
ax.set_xlabel('t')
ax.set_ylabel('T')
n = 1
L = 1
k = 0.3
xv, tv = np.meshgrid(x, t)
t0=10
# Initial values
B = 1
u = B * np.sin(n*np.pi*xv/L)*np.exp(-k*tv*(n*np.pi/L)**2)+t0
for ni in range(2, 1000):
B = 1
u += B * np.sin(ni*np.pi*xv/L)*np.exp(-k*tv*(ni*np.pi/L)**2)
fig = plt.figure(figsize=(7,7))
ax = fig.add_subplot(111, projection='3d')
ax.plot_surface(xv, tv, u, cmap=cm.coolwarm)
ax.set_xlabel('x')
ax.set_ylabel('t')
ax.view_init(elev=0, azim=45)
ax.set_zlim(8, 15)
n = 1
L = 1
k = 0.3
xv, tv = np.meshgrid(x, t)
t0=10
# Initial values
B = -2*L*(-1)**n/(n*np.pi)
u = B * np.sin(n*np.pi*xv/L)*np.exp(-k*tv*(n*np.pi/L)**2)+t0
for ni in range(2, 1000):
B = -2*L*(-1)**n/(n*np.pi)
u += B * np.sin(ni*np.pi*xv/L)*np.exp(-k*tv*(ni*np.pi/L)**2)
fig = plt.figure(figsize=(7,7))
ax = fig.add_subplot(111, projection='3d')
ax.plot_surface(xv, tv, u, cmap=cm.coolwarm)
ax.set_xlabel('x')
ax.set_ylabel('t')
ax.view_init(elev=0, azim=45)
ax.set_zlim(9, 13)
| 0.417865 | 0.990282 |
# Collaboration and Competition
---
In this notebook, you will learn how to use the Unity ML-Agents environment for the third project of the [Deep Reinforcement Learning Nanodegree](https://www.udacity.com/course/deep-reinforcement-learning-nanodegree--nd893) program.
### 1. Start the Environment
We begin by importing the necessary packages. If the code cell below returns an error, please revisit the project instructions to double-check that you have installed [Unity ML-Agents](https://github.com/Unity-Technologies/ml-agents/blob/master/docs/Installation.md) and [NumPy](http://www.numpy.org/).
```
from unityagents import UnityEnvironment
import numpy as np
```
Next, we will start the environment! **_Before running the code cell below_**, change the `file_name` parameter to match the location of the Unity environment that you downloaded.
- **Mac**: `"path/to/Tennis.app"`
- **Windows** (x86): `"path/to/Tennis_Windows_x86/Tennis.exe"`
- **Windows** (x86_64): `"path/to/Tennis_Windows_x86_64/Tennis.exe"`
- **Linux** (x86): `"path/to/Tennis_Linux/Tennis.x86"`
- **Linux** (x86_64): `"path/to/Tennis_Linux/Tennis.x86_64"`
- **Linux** (x86, headless): `"path/to/Tennis_Linux_NoVis/Tennis.x86"`
- **Linux** (x86_64, headless): `"path/to/Tennis_Linux_NoVis/Tennis.x86_64"`
For instance, if you are using a Mac, then you downloaded `Tennis.app`. If this file is in the same folder as the notebook, then the line below should appear as follows:
```
env = UnityEnvironment(file_name="Tennis.app")
```
```
env = UnityEnvironment(file_name="F:\Jupyter\deep-reinforcement-learning\Deep Tennis\Tennis_Windows_x86_64\Tennis.exe")
```
Environments contain **_brains_** which are responsible for deciding the actions of their associated agents. Here we check for the first brain available, and set it as the default brain we will be controlling from Python.
```
# get the default brain
brain_name = env.brain_names[0]
brain = env.brains[brain_name]
```
### 2. Examine the State and Action Spaces
In this environment, two agents control rackets to bounce a ball over a net. If an agent hits the ball over the net, it receives a reward of +0.1. If an agent lets a ball hit the ground or hits the ball out of bounds, it receives a reward of -0.01. Thus, the goal of each agent is to keep the ball in play.
The observation space consists of 8 variables corresponding to the position and velocity of the ball and racket. Two continuous actions are available, corresponding to movement toward (or away from) the net, and jumping.
Run the code cell below to print some information about the environment.
```
# reset the environment
env_info = env.reset(train_mode=True)[brain_name]
# number of agents
num_agents = len(env_info.agents)
print('Number of agents:', num_agents)
# size of each action
action_size = brain.vector_action_space_size
print('Size of each action:', action_size)
# examine the state space
states = env_info.vector_observations
state_size = states.shape[1]
print('There are {} agents. Each observes a state with length: {}'.format(states.shape[0], state_size))
print('The state for the first agent looks like:', states[0])
```
### 3. Take Random Actions in the Environment
In the next code cell, you will learn how to use the Python API to control the agents and receive feedback from the environment.
Once this cell is executed, you will watch the agents' performance, if they select actions at random with each time step. A window should pop up that allows you to observe the agents.
Of course, as part of the project, you'll have to change the code so that the agents are able to use their experiences to gradually choose better actions when interacting with the environment!
```
for i in range(1, 6): # play game for 5 episodes
env_info = env.reset(train_mode=False)[brain_name] # reset the environment
states = env_info.vector_observations # get the current state (for each agent)
scores = np.zeros(num_agents) # initialize the score (for each agent)
while True:
actions = np.random.randn(num_agents, action_size) # select an action (for each agent)
actions = np.clip(actions, -1, 1) # all actions between -1 and 1
env_info = env.step(actions)[brain_name] # send all actions to tne environment
next_states = env_info.vector_observations # get next state (for each agent)
rewards = env_info.rewards # get reward (for each agent)
dones = env_info.local_done # see if episode finished
scores += env_info.rewards # update the score (for each agent)
states = next_states # roll over states to next time step
if np.any(dones): # exit loop if episode finished
break
print('Score (max over agents) from episode {}: {}'.format(i, np.max(scores)))
```
When finished, you can close the environment.
### 4. It's Your Turn!
Now it's your turn to train your own agent to solve the environment! When training the environment, set `train_mode=True`, so that the line for resetting the environment looks like the following:
```python
env_info = env.reset(train_mode=True)[brain_name]
```
```
%load_ext autoreload
%autoreload 2
from ddpg_agent import *
num_agents = len(env_info.agents)
agents = Agent(state_size=state_size, action_size=action_size, num_agents=num_agents, random_seed=0)
def ddpg(n_episodes=2000):
"""Deep Deterministic Policy Gradient.
Args
n_episodes (int): maximum number of training episodes
Return:
scores (average score)
"""
scores = [] # list containing scores from each episode
scores_window = deque(maxlen=100) # last 100 scores
for i_episode in range(1, n_episodes+1):
env_info = env.reset(train_mode=True)[brain_name]
state = env_info.vector_observations
agents.reset()
score = np.zeros(num_agents)
while True:
action = agents.act(state)
env_info = env.step(action)[brain_name]
next_state = env_info.vector_observations # get the next state
rewards = env_info.rewards # get the reward
dones = env_info.local_done # see if episode has finished
agents.step(state, action, rewards, next_state, dones)
score += rewards # update the score
state = next_state # roll over the state to next time step
if np.any(dones): # exit loop if episode finished
break
scores_window.append(np.mean(score)) # save most recent score
scores.append(np.mean(score)) # save most recent score
print('\rEpisode {}\tAverage Score: {:.2f}'.format(i_episode, np.mean(scores_window)), end="")
if i_episode % 100 == 0:
print('\rEpisode {}\tAverage Score: {:.2f}'.format(i_episode, np.mean(scores_window)))
if np.mean(scores_window)>=0.5:
print('\nEnvironment solved in {:d} episodes!\tAverage Score: {:.2f}'.format(i_episode, np.mean(scores_window)))
torch.save(agents.actor_local.state_dict(), 'checkpoint_actor.pth')
torch.save(agents.critic_local.state_dict(), 'checkpoint_critic.pth')
return scores
scores = ddpg()
%matplotlib inline
import matplotlib.pyplot as plt
# plot the scores
average_score = 0.5
success = [average_score] * len(scores)
fig = plt.figure(figsize=(30,20))
ax = fig.add_subplot(111)
plt.plot(np.arange(len(scores)), scores,np.arange(len(scores)), success)
plt.ylabel('Score', fontsize = 30)
plt.xlabel('Episode #', fontsize = 30)
plt.title('Train DDPG Agent', fontsize = 30)
plt.gca().legend(('actual score','average'), fontsize = 20)
plt.show()
agents.actor_local.load_state_dict(torch.load('checkpoint_actor.pth'))
agents.critic_local.load_state_dict(torch.load('checkpoint_critic.pth'))
env_info = env.reset(train_mode=False)[brain_name] # reset the environment
states = env_info.vector_observations # get the current state (for each agent)
scores = np.zeros(num_agents) # initialize the score (for each agent)
while True:
actions = agents.act(states,add_noise = False) # select an action (for each agent)
env_info = env.step(actions)[brain_name] # send all actions to tne environment
next_states = env_info.vector_observations # get next state (for each agent)
rewards = env_info.rewards # get reward (for each agent)
dones = env_info.local_done # see if episode finished
scores += env_info.rewards # update the score (for each agent)
states = next_states # roll over states to next time step
if np.any(dones): # exit loop if episode finished
break
print('Total score (averaged over agents) this episode: {}'.format(np.mean(scores)))
env.close()
```
|
github_jupyter
|
from unityagents import UnityEnvironment
import numpy as np
env = UnityEnvironment(file_name="Tennis.app")
env = UnityEnvironment(file_name="F:\Jupyter\deep-reinforcement-learning\Deep Tennis\Tennis_Windows_x86_64\Tennis.exe")
# get the default brain
brain_name = env.brain_names[0]
brain = env.brains[brain_name]
# reset the environment
env_info = env.reset(train_mode=True)[brain_name]
# number of agents
num_agents = len(env_info.agents)
print('Number of agents:', num_agents)
# size of each action
action_size = brain.vector_action_space_size
print('Size of each action:', action_size)
# examine the state space
states = env_info.vector_observations
state_size = states.shape[1]
print('There are {} agents. Each observes a state with length: {}'.format(states.shape[0], state_size))
print('The state for the first agent looks like:', states[0])
for i in range(1, 6): # play game for 5 episodes
env_info = env.reset(train_mode=False)[brain_name] # reset the environment
states = env_info.vector_observations # get the current state (for each agent)
scores = np.zeros(num_agents) # initialize the score (for each agent)
while True:
actions = np.random.randn(num_agents, action_size) # select an action (for each agent)
actions = np.clip(actions, -1, 1) # all actions between -1 and 1
env_info = env.step(actions)[brain_name] # send all actions to tne environment
next_states = env_info.vector_observations # get next state (for each agent)
rewards = env_info.rewards # get reward (for each agent)
dones = env_info.local_done # see if episode finished
scores += env_info.rewards # update the score (for each agent)
states = next_states # roll over states to next time step
if np.any(dones): # exit loop if episode finished
break
print('Score (max over agents) from episode {}: {}'.format(i, np.max(scores)))
env_info = env.reset(train_mode=True)[brain_name]
%load_ext autoreload
%autoreload 2
from ddpg_agent import *
num_agents = len(env_info.agents)
agents = Agent(state_size=state_size, action_size=action_size, num_agents=num_agents, random_seed=0)
def ddpg(n_episodes=2000):
"""Deep Deterministic Policy Gradient.
Args
n_episodes (int): maximum number of training episodes
Return:
scores (average score)
"""
scores = [] # list containing scores from each episode
scores_window = deque(maxlen=100) # last 100 scores
for i_episode in range(1, n_episodes+1):
env_info = env.reset(train_mode=True)[brain_name]
state = env_info.vector_observations
agents.reset()
score = np.zeros(num_agents)
while True:
action = agents.act(state)
env_info = env.step(action)[brain_name]
next_state = env_info.vector_observations # get the next state
rewards = env_info.rewards # get the reward
dones = env_info.local_done # see if episode has finished
agents.step(state, action, rewards, next_state, dones)
score += rewards # update the score
state = next_state # roll over the state to next time step
if np.any(dones): # exit loop if episode finished
break
scores_window.append(np.mean(score)) # save most recent score
scores.append(np.mean(score)) # save most recent score
print('\rEpisode {}\tAverage Score: {:.2f}'.format(i_episode, np.mean(scores_window)), end="")
if i_episode % 100 == 0:
print('\rEpisode {}\tAverage Score: {:.2f}'.format(i_episode, np.mean(scores_window)))
if np.mean(scores_window)>=0.5:
print('\nEnvironment solved in {:d} episodes!\tAverage Score: {:.2f}'.format(i_episode, np.mean(scores_window)))
torch.save(agents.actor_local.state_dict(), 'checkpoint_actor.pth')
torch.save(agents.critic_local.state_dict(), 'checkpoint_critic.pth')
return scores
scores = ddpg()
%matplotlib inline
import matplotlib.pyplot as plt
# plot the scores
average_score = 0.5
success = [average_score] * len(scores)
fig = plt.figure(figsize=(30,20))
ax = fig.add_subplot(111)
plt.plot(np.arange(len(scores)), scores,np.arange(len(scores)), success)
plt.ylabel('Score', fontsize = 30)
plt.xlabel('Episode #', fontsize = 30)
plt.title('Train DDPG Agent', fontsize = 30)
plt.gca().legend(('actual score','average'), fontsize = 20)
plt.show()
agents.actor_local.load_state_dict(torch.load('checkpoint_actor.pth'))
agents.critic_local.load_state_dict(torch.load('checkpoint_critic.pth'))
env_info = env.reset(train_mode=False)[brain_name] # reset the environment
states = env_info.vector_observations # get the current state (for each agent)
scores = np.zeros(num_agents) # initialize the score (for each agent)
while True:
actions = agents.act(states,add_noise = False) # select an action (for each agent)
env_info = env.step(actions)[brain_name] # send all actions to tne environment
next_states = env_info.vector_observations # get next state (for each agent)
rewards = env_info.rewards # get reward (for each agent)
dones = env_info.local_done # see if episode finished
scores += env_info.rewards # update the score (for each agent)
states = next_states # roll over states to next time step
if np.any(dones): # exit loop if episode finished
break
print('Total score (averaged over agents) this episode: {}'.format(np.mean(scores)))
env.close()
| 0.541166 | 0.979036 |
## Product Review Aspect Detection: Routers
### This is a Natural Language Processing based solution which can detect up to 9 aspects from online product reviews for routers.
This sample notebook shows you how to deploy Product Review Aspect Detection: Router using Amazon SageMaker.
> **Note**: This is a reference notebook and it cannot run unless you make changes suggested in the notebook.
#### Pre-requisites:
1. **Note**: This notebook contains elements which render correctly in Jupyter interface. Open this notebook from an Amazon SageMaker Notebook Instance or Amazon SageMaker Studio.
1. Ensure that IAM role used has **AmazonSageMakerFullAccess**
1. To deploy this ML model successfully, ensure that:
1. Either your IAM role has these three permissions and you have authority to make AWS Marketplace subscriptions in the AWS account used:
1. **aws-marketplace:ViewSubscriptions**
1. **aws-marketplace:Unsubscribe**
1. **aws-marketplace:Subscribe**
2. or your AWS account has a subscription to Product Review Aspect Detection: Router. If so, skip step: [Subscribe to the model package](#1.-Subscribe-to-the-model-package)
#### Contents:
1. [Subscribe to the model package](#1.-Subscribe-to-the-model-package)
2. [Create an endpoint and perform real-time inference](#2.-Create-an-endpoint-and-perform-real-time-inference)
1. [Create an endpoint](#A.-Create-an-endpoint)
2. [Create input payload](#B.-Create-input-payload)
3. [Perform real-time inference](#C.-Perform-real-time-inference)
4. [Visualize output](#D.-Visualize-output)
5. [Delete the endpoint](#E.-Delete-the-endpoint)
3. [Perform batch inference](#3.-Perform-batch-inference)
4. [Clean-up](#4.-Clean-up)
1. [Delete the model](#A.-Delete-the-model)
2. [Unsubscribe to the listing (optional)](#B.-Unsubscribe-to-the-listing-(optional))
#### Usage instructions
You can run this notebook one cell at a time (By using Shift+Enter for running a cell).
### 1. Subscribe to the model package
To subscribe to the model package:
1. Open the model package listing page Product Review Aspect Detection: Routers.
1. On the AWS Marketplace listing, click on the **Continue to subscribe** button.
1. On the **Subscribe to this software** page, review and click on **"Accept Offer"** if you and your organization agrees with EULA, pricing, and support terms.
1. Once you click on **Continue to configuration button** and then choose a **region**, you will see a **Product Arn** displayed. This is the model package ARN that you need to specify while creating a deployable model using Boto3. Copy the ARN corresponding to your region and specify the same in the following cell.
```
model_package_arn='arn:aws:sagemaker:us-east-2:786796469737:model-package/router-aspect-extraction'
import base64
import json
import uuid
from sagemaker import ModelPackage
import sagemaker as sage
from sagemaker import get_execution_role
from sagemaker import ModelPackage
from urllib.parse import urlparse
import boto3
from IPython.display import Image
from PIL import Image as ImageEdit
import urllib.request
import numpy as np
role = get_execution_role()
sagemaker_session = sage.Session()
bucket=sagemaker_session.default_bucket()
bucket
```
### 2. Create an endpoint and perform real-time inference
If you want to understand how real-time inference with Amazon SageMaker works, see [Documentation](https://docs.aws.amazon.com/sagemaker/latest/dg/how-it-works-hosting.html).
```
model_name='router-aspect-extraction'
content_type='text/plain'
real_time_inference_instance_type='ml.m5.large'
batch_transform_inference_instance_type='ml.m5.large'
```
#### A. Create an endpoint
```
def predict_wrapper(endpoint, session):
return sage.predictor.Predictor(endpoint, session,content_type)
#create a deployable model from the model package.
model = ModelPackage(role=role,
model_package_arn=model_package_arn,
sagemaker_session=sagemaker_session,
predictor_cls=predict_wrapper)
#Deploy the model
predictor = model.deploy(1, real_time_inference_instance_type, endpoint_name=model_name)
```
Once endpoint has been created, you would be able to perform real-time inference.
#### B. Create input payload
```
file_name = 'sample.txt'
```
<Add code snippet that shows the payload contents>
#### C. Perform real-time inference
```
!aws sagemaker-runtime invoke-endpoint \
--endpoint-name $model_name \
--body fileb://$file_name \
--content-type $content_type \
--region $sagemaker_session.boto_region_name \
output.txt
```
#### D. Visualize output
```
import json
with open('output.txt', 'r') as f:
output = json.load(f)
print(output)
```
#### E. Delete the endpoint
Now that you have successfully performed a real-time inference, you do not need the endpoint any more. You can terminate the endpoint to avoid being charged.
```
predictor=sage.predictor.Predictor(model_name, sagemaker_session,content_type)
predictor.delete_endpoint(delete_endpoint_config=True)
```
### 3. Perform batch inference
In this section, you will perform batch inference using multiple input payloads together. If you are not familiar with batch transform, and want to learn more, see these links:
1. [How it works](https://docs.aws.amazon.com/sagemaker/latest/dg/ex1-batch-transform.html)
2. [How to run a batch transform job](https://docs.aws.amazon.com/sagemaker/latest/dg/how-it-works-batch.html)
```
#upload the batch-transform job input files to S3
transform_input_folder = "input"
transform_input = sagemaker_session.upload_data(transform_input_folder, key_prefix=model_name)
print("Transform input uploaded to " + transform_input)
#Run the batch-transform job
transformer = model.transformer(1, batch_transform_inference_instance_type)
transformer.transform(transform_input, content_type=content_type)
transformer.wait()
import os
s3_conn = boto3.client("s3")
with open('output2.txt', 'wb') as f:
s3_conn.download_fileobj(bucket, os.path.basename(transformer.output_path)+'/sample.txt.out', f)
print("Output file loaded from bucket")
with open('output2.txt', 'r') as f:
output = json.load(f)
print(output)
```
### 4. Clean-up
#### A. Delete the model
```
model.delete_model()
```
#### B. Unsubscribe to the listing (optional)
If you would like to unsubscribe to the model package, follow these steps. Before you cancel the subscription, ensure that you do not have any [deployable model](https://console.aws.amazon.com/sagemaker/home#/models) created from the model package or using the algorithm. Note - You can find this information by looking at the container name associated with the model.
**Steps to unsubscribe to product from AWS Marketplace**:
1. Navigate to __Machine Learning__ tab on [__Your Software subscriptions page__](https://aws.amazon.com/marketplace/ai/library?productType=ml&ref_=mlmp_gitdemo_indust)
2. Locate the listing that you want to cancel the subscription for, and then choose __Cancel Subscription__ to cancel the subscription.
|
github_jupyter
|
model_package_arn='arn:aws:sagemaker:us-east-2:786796469737:model-package/router-aspect-extraction'
import base64
import json
import uuid
from sagemaker import ModelPackage
import sagemaker as sage
from sagemaker import get_execution_role
from sagemaker import ModelPackage
from urllib.parse import urlparse
import boto3
from IPython.display import Image
from PIL import Image as ImageEdit
import urllib.request
import numpy as np
role = get_execution_role()
sagemaker_session = sage.Session()
bucket=sagemaker_session.default_bucket()
bucket
model_name='router-aspect-extraction'
content_type='text/plain'
real_time_inference_instance_type='ml.m5.large'
batch_transform_inference_instance_type='ml.m5.large'
def predict_wrapper(endpoint, session):
return sage.predictor.Predictor(endpoint, session,content_type)
#create a deployable model from the model package.
model = ModelPackage(role=role,
model_package_arn=model_package_arn,
sagemaker_session=sagemaker_session,
predictor_cls=predict_wrapper)
#Deploy the model
predictor = model.deploy(1, real_time_inference_instance_type, endpoint_name=model_name)
file_name = 'sample.txt'
!aws sagemaker-runtime invoke-endpoint \
--endpoint-name $model_name \
--body fileb://$file_name \
--content-type $content_type \
--region $sagemaker_session.boto_region_name \
output.txt
import json
with open('output.txt', 'r') as f:
output = json.load(f)
print(output)
predictor=sage.predictor.Predictor(model_name, sagemaker_session,content_type)
predictor.delete_endpoint(delete_endpoint_config=True)
#upload the batch-transform job input files to S3
transform_input_folder = "input"
transform_input = sagemaker_session.upload_data(transform_input_folder, key_prefix=model_name)
print("Transform input uploaded to " + transform_input)
#Run the batch-transform job
transformer = model.transformer(1, batch_transform_inference_instance_type)
transformer.transform(transform_input, content_type=content_type)
transformer.wait()
import os
s3_conn = boto3.client("s3")
with open('output2.txt', 'wb') as f:
s3_conn.download_fileobj(bucket, os.path.basename(transformer.output_path)+'/sample.txt.out', f)
print("Output file loaded from bucket")
with open('output2.txt', 'r') as f:
output = json.load(f)
print(output)
model.delete_model()
| 0.405566 | 0.893867 |
# WeatherPy
----
#### Note
* Instructions have been included for each segment. You do not have to follow them exactly, but they are included to help you think through the steps.
```
# Dependencies and Setup
import os
import csv
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
import requests
import time
from scipy.stats import linregress
from pprint import pprint
# Import API key
from api_keys import weather_api_key
# Incorporated citipy to determine city based on latitude and longitude
from citipy import citipy
# Output File (CSV)
output_data_file = "output_data/cities.csv"
# Range of latitudes and longitudes
lat_range = (-90, 90)
lng_range = (-180, 180)
```
## Generate Cities List
```
# List for holding lat_lngs and cities
lat_lngs = []
cities = []
# Create a set of random lat and lng combinations
lats = np.random.uniform(low=-90.000, high=90.000, size=1500)
lngs = np.random.uniform(low=-180.000, high=180.000, size=1500)
lat_lngs = zip(lats, lngs)
# Identify nearest city for each lat, lng combination
for lat_lng in lat_lngs:
city = citipy.nearest_city(lat_lng[0], lat_lng[1]).city_name
# If the city is unique, then add it to a our cities list
if city not in cities:
cities.append(city)
# Print the city count to confirm sufficient count
len(cities)
```
### Perform API Calls
* Perform a weather check on each city using a series of successive API calls.
* Include a print log of each city as it'sbeing processed (with the city number and city name).
```
# create data holders
new_cities = []
cloudiness = []
country = []
date = []
humidity = []
temp = []
lat = []
lng = []
wind = []
# Save config information
# Build query URL
record_counter = 0
set_counter = 0
# Starting URL for Weather Map API Call
url = "http://api.openweathermap.org/data/2.5/weather?units=Imperial&APPID=" + weather_api_key
print('------------------------')
print('-- Searching for Data --')
print('------------------------')
for city in cities:
query_url = url + "&q=" + city
# Get weather data
weather_response = requests.get(query_url).json()
if record_counter < 50:
record_counter += 1
else:
set_counter += 1
record_counter = 0
# print('Processing record {} of set {} | {}'.format(record_counter, set_counter, city))
# print(url)
try:
cloudiness.append(weather_response['clouds']['all'])
country.append(weather_response['sys']['country'])
date.append(weather_response['dt'])
humidity.append(weather_response['main']['humidity'])
temp.append(weather_response['main']['temp_max'])
lat.append(weather_response['coord']['lat'])
lng.append(weather_response['coord']['lon'])
wind.append(weather_response['wind']['speed'])
new_cities.append(city)
except:
# print("City not found!")
pass
print('-------------------------')
print('Data Retrieval Complete')
print('-------------------------')
# Get weather data
weather_response = requests.get(query_url)
weather_json = weather_response.json()
# Get the temperature from the response
print(f"The weather API responded with: {weather_json}.")
```
### Convert Raw Data to DataFrame
* Export the city data into a .csv.
* Display the DataFrame
```
# create a data frame from cities, temp, humidity, cloudiness and wind speed
weather_dict = {
"City": new_cities,
"Cloudiness" : cloudiness,
"Country" : country,
"Date" : date,
"Humidity" : humidity,
"Temp": temp,
"Lat" : lat,
"Lng" : lng,
"Wind Speed" : wind
}
weather_data = pd.DataFrame(weather_dict)
weather_data.count()
weather_data['Date'] = pd.to_datetime(weather_data['Date'])
weather_data.to_csv('../output_data/weather_data.csv')
weather_data.head()
```
### Plotting the Data
* Use proper labeling of the plots using plot titles (including date of analysis) and axes labels.
* Save the plotted figures as .pngs.
#### Latitude vs. Temperature Plot
```
weather_data.plot(kind='scatter', x='Lat', y='Temp', c='Blue', alpha=0.4)
plt.title('City Latitude Vs Max Temperature'.format(date))
plt.xlabel('Latitude')
plt.ylabel('Max temperature (F)')
plt.grid()
plt.savefig("../Images/LatitudeVsTemperature.png")
```
#### Latitude vs. Humidity Plot
```
weather_data.plot(kind='scatter',x='Lat',y='Humidity', c='Blue', alpha=0.4)
plt.title('City Latitude Vs Max Humidity'.format(date) )
plt.xlabel('Latitude')
plt.ylabel('Humidity (%)')
plt.grid()
plt.savefig("../Images/LatitudeVsHumidity.png")
```
#### Latitude vs. Cloudiness Plot
```
weather_data.plot(kind='scatter',x='Lat',y='Cloudiness', c='Blue', alpha=0.4)
plt.title('City Latitude Vs Cloudiness'.format(date) )
plt.xlabel('Latitude')
plt.ylabel('Cloudiness (%)')
plt.grid()
plt.savefig("../Images/LatitudeVsCloudiness.png")
```
#### Latitude vs. Wind Speed Plot
```
plt.close()
x_vals = weather_data['Lat']
y_vals = weather_data['Wind Speed']
plt.scatter(x_vals, y_vals, c='Blue', alpha=0.4)
plt.xlabel('Latitude')
plt.ylabel('Wind Speed')
plt.title('Latitude vs. Wind Speed')
plt.legend(labels=['City'], loc='upper left')
plt.show()
```
## Linear Regression
```
# OPTIONAL: Create a function to create Linear Regression plots
labels=['title' , 'y']
savePath = '../output_data/' + labels[0] + '.png'
def linRegresPlt (x_values, y_values):
(slope, intercept, rvalue, pvalue, stderr) = linregress(x_values, y_values)
regress_values = x_values * slope + intercept
line_eq = "y = " + str(round(slope,2)) + "x + " + str(round(intercept,2))
plt.close()
plt.annotate(line_eq,(0,10),fontsize=15,color="red")
plt.scatter(x_values,y_values, c='Blue', alpha=0.4)
plt.plot(x_values,regress_values,"r-")
print(line_eq)
print(f"The r-squared is: {rvalue}")
# plt.scatter(x_vals, y_vals, c='Blue', alpha=0.4)
# plt.xlabel('Latitude')
# plt.ylabel(labels[1])
# plt.title(labels[0])
# plt.plot(x_values, regress_values, "r-")
# plt.legend(labels=['Correlation line', 'City'], loc='best')
# plt.savefig(savePath)
plt.show()
# Create Northern and Southern Hemisphere DataFrames
df_hems_base = weather_data.copy()
df_hems_base['Lat'] = df_hems_base['Lat'].astype(float)
north_df = df_hems_base.loc[df_hems_base['Lat'] >= 0].copy()
south_df = df_hems_base.loc[df_hems_base['Lat'] < 0].copy()
#Northern Hemisphere Regression Lines
var_list = ["Temp", "Humidity", "Cloudiness", "Wind Speed"]
counter=0
for variables in var_list:
x_values = north_df["Lat"]
y_values = north_df[var_list[counter]] #0 is associated with "Max Temp",1 is associated with"Humidity" and so on!
plt.title(f" Latitude vs %s" % var_list[counter])
plot_regression (x_values, y_values)
plt.xlabel('Latitude')
plt.ylabel(var_list[counter])
plt.savefig("../Images/var_list[counter].png")
plt.show()
plt.clf()
counter= counter + 1
if counter ==4:
break
```
#### Northern Hemisphere - Max Temp vs. Latitude Linear Regression
```
# Add the linear regression model
x_values = north_df['Lat']
y_values = north_df['Temp']
plt.xlabel('Latitude')
plt.ylabel('Max Temp')
plt.title("NH-Latitude vs Max Temp Linear Regression")
(slope, intercept, rvalue, pvalue, stderr) = linregress(x_values, y_values)
regress_values = x_values * slope + intercept
line_eq = "y = " + str(round(slope,2)) + "x + " + str(round(intercept,2))
plt.scatter(x_values,y_values)
plt.plot(x_values,regress_values,"r-")
plt.annotate(line_eq,(3,270),fontsize=15,color="red")
print(f"The r-squared is: {rvalue}")
plt.show()
```
#### Southern Hemisphere - Max Temp vs. Latitude Linear Regression
```
x_values = south_df['Lat']
y_values = south_df['Temp']
plt.xlabel('Latitude')
plt.ylabel('Max Temp')
plt.title("SH-Latitude vs Max Temp Linear Regression")
(slope, intercept, rvalue, pvalue, stderr) = linregress(x_values, y_values)
regress_values = x_values * slope + intercept
line_eq = "y = " + str(round(slope,2)) + "x + " + str(round(intercept,2))
plt.scatter(x_values,y_values)
plt.plot(x_values,regress_values,"r-")
plt.annotate(line_eq,(-25,280),fontsize=15,color="red")
print(f"The r-squared is: {rvalue}")
plt.show()
```
#### Northern Hemisphere - Humidity (%) vs. Latitude Linear Regression
```
# Add the linear regression model
x_values = north_df['Lat']
y_values = north_df['Humidity']
plt.xlabel('Latitude')
plt.ylabel('Humidity(%)')
plt.title("NH-Latitude vs Humidity(%) Linear Regression")
(slope, intercept, rvalue, pvalue, stderr) = linregress(x_values, y_values)
regress_values = x_values * slope + intercept
line_eq = "y = " + str(round(slope,2)) + "x + " + str(round(intercept,2))
plt.scatter(x_values,y_values)
plt.plot(x_values,regress_values,"r-")
plt.annotate(line_eq,(46,30),fontsize=15,color="red")
print(f"The r-squared is: {rvalue}")
plt.show()
```
#### Southern Hemisphere - Humidity (%) vs. Latitude Linear Regression
```
x_values = south_df['Lat']
y_values = south_df['Humidity']
plt.xlabel('Latitude')
plt.ylabel('Humidity(%)')
plt.title("SH-Latitude vs Humidity(%) Linear Regression")
(slope, intercept, rvalue, pvalue, stderr) = linregress(x_values, y_values)
regress_values = x_values * slope + intercept
line_eq = "y = " + str(round(slope,2)) + "x + " + str(round(intercept,2))
plt.scatter(x_values,y_values)
plt.plot(x_values,regress_values,"r-")
plt.annotate(line_eq,(-30,58),fontsize=15,color="red")
print(f"The r-squared is: {rvalue}")
plt.show()
```
#### Northern Hemisphere - Cloudiness (%) vs. Latitude Linear Regression
```
# Add the linear regression model
x_values = north_df['Lat']
y_values = north_df['Cloudiness']
plt.xlabel('Latitude')
plt.ylabel('Cloudiness(%)')
plt.title("NH-Latitude vs Cloudiness(%) Linear Regression")
(slope, intercept, rvalue, pvalue, stderr) = linregress(x_values, y_values)
regress_values = x_values * slope + intercept
line_eq = "y = " + str(round(slope,2)) + "x + " + str(round(intercept,2))
plt.scatter(x_values,y_values)
plt.plot(x_values,regress_values,"r-")
plt.annotate(line_eq,(40,45),fontsize=15,color="red")
print(f"The r-squared is: {rvalue}")
plt.show()
```
#### Southern Hemisphere - Cloudiness (%) vs. Latitude Linear Regression
```
x_values = south_df['Lat']
y_values = south_df['Cloudiness']
plt.xlabel('Latitude')
plt.ylabel('Cloudiness(%)')
plt.title("SH-Latitude vs Cloudiness(%) Linear Regression")
(slope, intercept, rvalue, pvalue, stderr) = linregress(x_values, y_values)
regress_values = x_values * slope + intercept
line_eq = "y = " + str(round(slope,2)) + "x + " + str(round(intercept,2))
plt.scatter(x_values,y_values)
plt.plot(x_values,regress_values,"r-")
plt.annotate(line_eq,(-30,42),fontsize=15,color="red")
print(f"The r-squared is: {rvalue}")
plt.show()
```
#### Northern Hemisphere - Wind Speed (mph) vs. Latitude Linear Regression
```
x_values = north_df['Lat']
y_values = north_df['Wind Speed']
plt.xlabel('Latitude')
plt.ylabel('Wind Speed (mph)')
plt.title("NH-Latitude vs Wind Speed (mph)Linear Regression")
(slope, intercept, rvalue, pvalue, stderr) = linregress(x_values, y_values)
regress_values = x_values * slope + intercept
line_eq = "y = " + str(round(slope,2)) + "x + " + str(round(intercept,2))
plt.scatter(x_values,y_values)
plt.plot(x_values,regress_values,"r-")
plt.annotate(line_eq,(45,1.5),fontsize=15,color="red")
print(f"The r-squared is: {rvalue}")
plt.show()
```
#### Southern Hemisphere - Wind Speed (mph) vs. Latitude Linear Regression
```
x_values = south_df['Lat']
y_values = south_df['Wind Speed']
plt.xlabel('Latitude')
plt.ylabel('Wind Speed (mph)')
plt.title("SH-Latitude vs Wind Speed (mph)Linear Regression")
(slope, intercept, rvalue, pvalue, stderr) = linregress(x_values, y_values)
regress_values = x_values * slope + intercept
line_eq = "y = " + str(round(slope,2)) + "x + " + str(round(intercept,2))
plt.scatter(x_values,y_values)
plt.plot(x_values,regress_values,"r-")
plt.annotate(line_eq,(-52,7),fontsize=15,color="red")
print(f"The r-squared is: {rvalue}")
plt.show()
For Part I, you must include a written description of three observable trends based on the data.
* You must use proper labeling of your plots, including aspects like: Plot Titles (with date of analysis) and Axes Labels.
* For max intensity in the heat map, try setting it to the highest humidity found in the data set.
```
|
github_jupyter
|
# Dependencies and Setup
import os
import csv
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
import requests
import time
from scipy.stats import linregress
from pprint import pprint
# Import API key
from api_keys import weather_api_key
# Incorporated citipy to determine city based on latitude and longitude
from citipy import citipy
# Output File (CSV)
output_data_file = "output_data/cities.csv"
# Range of latitudes and longitudes
lat_range = (-90, 90)
lng_range = (-180, 180)
# List for holding lat_lngs and cities
lat_lngs = []
cities = []
# Create a set of random lat and lng combinations
lats = np.random.uniform(low=-90.000, high=90.000, size=1500)
lngs = np.random.uniform(low=-180.000, high=180.000, size=1500)
lat_lngs = zip(lats, lngs)
# Identify nearest city for each lat, lng combination
for lat_lng in lat_lngs:
city = citipy.nearest_city(lat_lng[0], lat_lng[1]).city_name
# If the city is unique, then add it to a our cities list
if city not in cities:
cities.append(city)
# Print the city count to confirm sufficient count
len(cities)
# create data holders
new_cities = []
cloudiness = []
country = []
date = []
humidity = []
temp = []
lat = []
lng = []
wind = []
# Save config information
# Build query URL
record_counter = 0
set_counter = 0
# Starting URL for Weather Map API Call
url = "http://api.openweathermap.org/data/2.5/weather?units=Imperial&APPID=" + weather_api_key
print('------------------------')
print('-- Searching for Data --')
print('------------------------')
for city in cities:
query_url = url + "&q=" + city
# Get weather data
weather_response = requests.get(query_url).json()
if record_counter < 50:
record_counter += 1
else:
set_counter += 1
record_counter = 0
# print('Processing record {} of set {} | {}'.format(record_counter, set_counter, city))
# print(url)
try:
cloudiness.append(weather_response['clouds']['all'])
country.append(weather_response['sys']['country'])
date.append(weather_response['dt'])
humidity.append(weather_response['main']['humidity'])
temp.append(weather_response['main']['temp_max'])
lat.append(weather_response['coord']['lat'])
lng.append(weather_response['coord']['lon'])
wind.append(weather_response['wind']['speed'])
new_cities.append(city)
except:
# print("City not found!")
pass
print('-------------------------')
print('Data Retrieval Complete')
print('-------------------------')
# Get weather data
weather_response = requests.get(query_url)
weather_json = weather_response.json()
# Get the temperature from the response
print(f"The weather API responded with: {weather_json}.")
# create a data frame from cities, temp, humidity, cloudiness and wind speed
weather_dict = {
"City": new_cities,
"Cloudiness" : cloudiness,
"Country" : country,
"Date" : date,
"Humidity" : humidity,
"Temp": temp,
"Lat" : lat,
"Lng" : lng,
"Wind Speed" : wind
}
weather_data = pd.DataFrame(weather_dict)
weather_data.count()
weather_data['Date'] = pd.to_datetime(weather_data['Date'])
weather_data.to_csv('../output_data/weather_data.csv')
weather_data.head()
weather_data.plot(kind='scatter', x='Lat', y='Temp', c='Blue', alpha=0.4)
plt.title('City Latitude Vs Max Temperature'.format(date))
plt.xlabel('Latitude')
plt.ylabel('Max temperature (F)')
plt.grid()
plt.savefig("../Images/LatitudeVsTemperature.png")
weather_data.plot(kind='scatter',x='Lat',y='Humidity', c='Blue', alpha=0.4)
plt.title('City Latitude Vs Max Humidity'.format(date) )
plt.xlabel('Latitude')
plt.ylabel('Humidity (%)')
plt.grid()
plt.savefig("../Images/LatitudeVsHumidity.png")
weather_data.plot(kind='scatter',x='Lat',y='Cloudiness', c='Blue', alpha=0.4)
plt.title('City Latitude Vs Cloudiness'.format(date) )
plt.xlabel('Latitude')
plt.ylabel('Cloudiness (%)')
plt.grid()
plt.savefig("../Images/LatitudeVsCloudiness.png")
plt.close()
x_vals = weather_data['Lat']
y_vals = weather_data['Wind Speed']
plt.scatter(x_vals, y_vals, c='Blue', alpha=0.4)
plt.xlabel('Latitude')
plt.ylabel('Wind Speed')
plt.title('Latitude vs. Wind Speed')
plt.legend(labels=['City'], loc='upper left')
plt.show()
# OPTIONAL: Create a function to create Linear Regression plots
labels=['title' , 'y']
savePath = '../output_data/' + labels[0] + '.png'
def linRegresPlt (x_values, y_values):
(slope, intercept, rvalue, pvalue, stderr) = linregress(x_values, y_values)
regress_values = x_values * slope + intercept
line_eq = "y = " + str(round(slope,2)) + "x + " + str(round(intercept,2))
plt.close()
plt.annotate(line_eq,(0,10),fontsize=15,color="red")
plt.scatter(x_values,y_values, c='Blue', alpha=0.4)
plt.plot(x_values,regress_values,"r-")
print(line_eq)
print(f"The r-squared is: {rvalue}")
# plt.scatter(x_vals, y_vals, c='Blue', alpha=0.4)
# plt.xlabel('Latitude')
# plt.ylabel(labels[1])
# plt.title(labels[0])
# plt.plot(x_values, regress_values, "r-")
# plt.legend(labels=['Correlation line', 'City'], loc='best')
# plt.savefig(savePath)
plt.show()
# Create Northern and Southern Hemisphere DataFrames
df_hems_base = weather_data.copy()
df_hems_base['Lat'] = df_hems_base['Lat'].astype(float)
north_df = df_hems_base.loc[df_hems_base['Lat'] >= 0].copy()
south_df = df_hems_base.loc[df_hems_base['Lat'] < 0].copy()
#Northern Hemisphere Regression Lines
var_list = ["Temp", "Humidity", "Cloudiness", "Wind Speed"]
counter=0
for variables in var_list:
x_values = north_df["Lat"]
y_values = north_df[var_list[counter]] #0 is associated with "Max Temp",1 is associated with"Humidity" and so on!
plt.title(f" Latitude vs %s" % var_list[counter])
plot_regression (x_values, y_values)
plt.xlabel('Latitude')
plt.ylabel(var_list[counter])
plt.savefig("../Images/var_list[counter].png")
plt.show()
plt.clf()
counter= counter + 1
if counter ==4:
break
# Add the linear regression model
x_values = north_df['Lat']
y_values = north_df['Temp']
plt.xlabel('Latitude')
plt.ylabel('Max Temp')
plt.title("NH-Latitude vs Max Temp Linear Regression")
(slope, intercept, rvalue, pvalue, stderr) = linregress(x_values, y_values)
regress_values = x_values * slope + intercept
line_eq = "y = " + str(round(slope,2)) + "x + " + str(round(intercept,2))
plt.scatter(x_values,y_values)
plt.plot(x_values,regress_values,"r-")
plt.annotate(line_eq,(3,270),fontsize=15,color="red")
print(f"The r-squared is: {rvalue}")
plt.show()
x_values = south_df['Lat']
y_values = south_df['Temp']
plt.xlabel('Latitude')
plt.ylabel('Max Temp')
plt.title("SH-Latitude vs Max Temp Linear Regression")
(slope, intercept, rvalue, pvalue, stderr) = linregress(x_values, y_values)
regress_values = x_values * slope + intercept
line_eq = "y = " + str(round(slope,2)) + "x + " + str(round(intercept,2))
plt.scatter(x_values,y_values)
plt.plot(x_values,regress_values,"r-")
plt.annotate(line_eq,(-25,280),fontsize=15,color="red")
print(f"The r-squared is: {rvalue}")
plt.show()
# Add the linear regression model
x_values = north_df['Lat']
y_values = north_df['Humidity']
plt.xlabel('Latitude')
plt.ylabel('Humidity(%)')
plt.title("NH-Latitude vs Humidity(%) Linear Regression")
(slope, intercept, rvalue, pvalue, stderr) = linregress(x_values, y_values)
regress_values = x_values * slope + intercept
line_eq = "y = " + str(round(slope,2)) + "x + " + str(round(intercept,2))
plt.scatter(x_values,y_values)
plt.plot(x_values,regress_values,"r-")
plt.annotate(line_eq,(46,30),fontsize=15,color="red")
print(f"The r-squared is: {rvalue}")
plt.show()
x_values = south_df['Lat']
y_values = south_df['Humidity']
plt.xlabel('Latitude')
plt.ylabel('Humidity(%)')
plt.title("SH-Latitude vs Humidity(%) Linear Regression")
(slope, intercept, rvalue, pvalue, stderr) = linregress(x_values, y_values)
regress_values = x_values * slope + intercept
line_eq = "y = " + str(round(slope,2)) + "x + " + str(round(intercept,2))
plt.scatter(x_values,y_values)
plt.plot(x_values,regress_values,"r-")
plt.annotate(line_eq,(-30,58),fontsize=15,color="red")
print(f"The r-squared is: {rvalue}")
plt.show()
# Add the linear regression model
x_values = north_df['Lat']
y_values = north_df['Cloudiness']
plt.xlabel('Latitude')
plt.ylabel('Cloudiness(%)')
plt.title("NH-Latitude vs Cloudiness(%) Linear Regression")
(slope, intercept, rvalue, pvalue, stderr) = linregress(x_values, y_values)
regress_values = x_values * slope + intercept
line_eq = "y = " + str(round(slope,2)) + "x + " + str(round(intercept,2))
plt.scatter(x_values,y_values)
plt.plot(x_values,regress_values,"r-")
plt.annotate(line_eq,(40,45),fontsize=15,color="red")
print(f"The r-squared is: {rvalue}")
plt.show()
x_values = south_df['Lat']
y_values = south_df['Cloudiness']
plt.xlabel('Latitude')
plt.ylabel('Cloudiness(%)')
plt.title("SH-Latitude vs Cloudiness(%) Linear Regression")
(slope, intercept, rvalue, pvalue, stderr) = linregress(x_values, y_values)
regress_values = x_values * slope + intercept
line_eq = "y = " + str(round(slope,2)) + "x + " + str(round(intercept,2))
plt.scatter(x_values,y_values)
plt.plot(x_values,regress_values,"r-")
plt.annotate(line_eq,(-30,42),fontsize=15,color="red")
print(f"The r-squared is: {rvalue}")
plt.show()
x_values = north_df['Lat']
y_values = north_df['Wind Speed']
plt.xlabel('Latitude')
plt.ylabel('Wind Speed (mph)')
plt.title("NH-Latitude vs Wind Speed (mph)Linear Regression")
(slope, intercept, rvalue, pvalue, stderr) = linregress(x_values, y_values)
regress_values = x_values * slope + intercept
line_eq = "y = " + str(round(slope,2)) + "x + " + str(round(intercept,2))
plt.scatter(x_values,y_values)
plt.plot(x_values,regress_values,"r-")
plt.annotate(line_eq,(45,1.5),fontsize=15,color="red")
print(f"The r-squared is: {rvalue}")
plt.show()
x_values = south_df['Lat']
y_values = south_df['Wind Speed']
plt.xlabel('Latitude')
plt.ylabel('Wind Speed (mph)')
plt.title("SH-Latitude vs Wind Speed (mph)Linear Regression")
(slope, intercept, rvalue, pvalue, stderr) = linregress(x_values, y_values)
regress_values = x_values * slope + intercept
line_eq = "y = " + str(round(slope,2)) + "x + " + str(round(intercept,2))
plt.scatter(x_values,y_values)
plt.plot(x_values,regress_values,"r-")
plt.annotate(line_eq,(-52,7),fontsize=15,color="red")
print(f"The r-squared is: {rvalue}")
plt.show()
For Part I, you must include a written description of three observable trends based on the data.
* You must use proper labeling of your plots, including aspects like: Plot Titles (with date of analysis) and Axes Labels.
* For max intensity in the heat map, try setting it to the highest humidity found in the data set.
| 0.278747 | 0.833426 |
```
import sys
# Packages for direct database access
# %pip install psycopg2
import psycopg2
import json
# Packages for data and number handling
import numpy as np
import pandas as pd
import math
# Packages for calculating current time and extracting ZTF data to VOTable
from astropy.time import Time
from astropy.table import Table
from astropy.io.votable import from_table, writeto
import astropy.units as units
from astropy.coordinates import SkyCoord
from datetime import datetime
# Handling FITS files
from astropy.io import fits
# Packages for display and data plotting, if desired
from IPython.display import display, HTML
import matplotlib.pyplot as plt
import matplotlib.cm as cm
%matplotlib inline
import ephem
import copy
import pickle
from alerce.core import Alerce
client = Alerce()
# Open and load credentials
credentials_file = "../alercereaduser_v4.json"
with open(credentials_file) as jsonfile:
params = json.load(jsonfile)["params"]
# Open a connection to the database
conn = psycopg2.connect(dbname=params['dbname'],
user=params['user'],
host=params['host'],
password=params['password'])
# Show all the available tables, sorted by tablename
query = """
SELECT table_name
FROM information_schema.tables
WHERE table_schema='alerce'
ORDER BY table_name;
"""
tables = pd.read_sql_query(query, conn)
tables.sort_values(by="table_name")
Time(datetime.today(), scale='utc').mjd
mjd_last = Time(datetime.today(), scale='utc').mjd-0.3
query='''
SELECT
object.oid, object.meanra, object.meandec, object.sigmara, object.sigmadec,
object.firstmjd, object.lastmjd, object.ndet,
probability.class_name, probability.probability
FROM object
INNER JOIN probability
ON object.oid=probability.oid
WHERE
object.lastMJD>%s
AND probability.ranking=1
''' % (mjd_last)
# Outputs as a pd.DataFrame
objects = pd.read_sql_query(query, conn)
# Prints the Dataframe shape: (number of selected objects, number of selected filters)
print(objects.shape)
# Sorting detections by lastMJD, firstMJD in descending order
objects_sorted = objects.sort_values(by=['lastmjd','firstmjd'],ascending=False)
objects_sorted.head()
def getStampsData(oid,n_stamps=5):
object_det = client.query_detections(oid, format='pandas')
candids = object_det.loc[object_det.has_stamp]
object_det = object_det.iloc[:n_stamps]
candids = object_det.candid
stamps = []
for c in candids:
candid_stamps = client.get_stamps(oid, candid=c)
if candid_stamps != None:
stamps.append(candid_stamps)
return stamps
for obj in objects:
object_stamps = getStampsData(obj.oid)
counter = 1
for candid_list in object_stamps:
hdul = fits.HDUList(candid_list)
filename = obj.oid + '_%i' % ()
hdu.writeto('%s.fits') % (filename)
```
|
github_jupyter
|
import sys
# Packages for direct database access
# %pip install psycopg2
import psycopg2
import json
# Packages for data and number handling
import numpy as np
import pandas as pd
import math
# Packages for calculating current time and extracting ZTF data to VOTable
from astropy.time import Time
from astropy.table import Table
from astropy.io.votable import from_table, writeto
import astropy.units as units
from astropy.coordinates import SkyCoord
from datetime import datetime
# Handling FITS files
from astropy.io import fits
# Packages for display and data plotting, if desired
from IPython.display import display, HTML
import matplotlib.pyplot as plt
import matplotlib.cm as cm
%matplotlib inline
import ephem
import copy
import pickle
from alerce.core import Alerce
client = Alerce()
# Open and load credentials
credentials_file = "../alercereaduser_v4.json"
with open(credentials_file) as jsonfile:
params = json.load(jsonfile)["params"]
# Open a connection to the database
conn = psycopg2.connect(dbname=params['dbname'],
user=params['user'],
host=params['host'],
password=params['password'])
# Show all the available tables, sorted by tablename
query = """
SELECT table_name
FROM information_schema.tables
WHERE table_schema='alerce'
ORDER BY table_name;
"""
tables = pd.read_sql_query(query, conn)
tables.sort_values(by="table_name")
Time(datetime.today(), scale='utc').mjd
mjd_last = Time(datetime.today(), scale='utc').mjd-0.3
query='''
SELECT
object.oid, object.meanra, object.meandec, object.sigmara, object.sigmadec,
object.firstmjd, object.lastmjd, object.ndet,
probability.class_name, probability.probability
FROM object
INNER JOIN probability
ON object.oid=probability.oid
WHERE
object.lastMJD>%s
AND probability.ranking=1
''' % (mjd_last)
# Outputs as a pd.DataFrame
objects = pd.read_sql_query(query, conn)
# Prints the Dataframe shape: (number of selected objects, number of selected filters)
print(objects.shape)
# Sorting detections by lastMJD, firstMJD in descending order
objects_sorted = objects.sort_values(by=['lastmjd','firstmjd'],ascending=False)
objects_sorted.head()
def getStampsData(oid,n_stamps=5):
object_det = client.query_detections(oid, format='pandas')
candids = object_det.loc[object_det.has_stamp]
object_det = object_det.iloc[:n_stamps]
candids = object_det.candid
stamps = []
for c in candids:
candid_stamps = client.get_stamps(oid, candid=c)
if candid_stamps != None:
stamps.append(candid_stamps)
return stamps
for obj in objects:
object_stamps = getStampsData(obj.oid)
counter = 1
for candid_list in object_stamps:
hdul = fits.HDUList(candid_list)
filename = obj.oid + '_%i' % ()
hdu.writeto('%s.fits') % (filename)
| 0.357792 | 0.290182 |
# Converting from GeoDataFrame to Graph and back
The model situation expects to have all input data for analysis in `GeoDataFrames`, including street network (e.g. from shapefile).
```
import momepy
import geopandas as gpd
import matplotlib.pyplot as plt
import networkx as nx
streets = gpd.read_file(momepy.datasets.get_path('bubenec'), layer='streets')
f, ax = plt.subplots(figsize=(10, 10))
streets.plot(ax=ax)
ax.set_axis_off()
plt.show()
```
We have to convert this LineString GeoDataFrame to `networkx.Graph`. We use `momepy.gdf_to_nx` and later `momepy.nx_to_gdf` as pair of interconnected functions. `gdf_to_nx` supports both primal and dual graphs. Primal approach will save length of each segment to be used as a weight later, while dual will save the angle between segments (allowing angular centrality).
```
graph = momepy.gdf_to_nx(streets, approach='primal')
f, ax = plt.subplots(1, 3, figsize=(18, 6), sharex=True, sharey=True)
streets.plot(color='#e32e00', ax=ax[0])
for i, facet in enumerate(ax):
facet.set_title(("Streets", "Primal graph", "Overlay")[i])
facet.axis("off")
nx.draw(graph, {n:[n[0], n[1]] for n in list(graph.nodes)}, ax=ax[1], node_size=15)
streets.plot(color='#e32e00', ax=ax[2], zorder=-1)
nx.draw(graph, {n:[n[0], n[1]] for n in list(graph.nodes)}, ax=ax[2], node_size=15)
dual = momepy.gdf_to_nx(streets, approach='dual')
f, ax = plt.subplots(1, 3, figsize=(18, 6), sharex=True, sharey=True)
streets.plot(color='#e32e00', ax=ax[0])
for i, facet in enumerate(ax):
facet.set_title(("Streets", "Dual graph", "Overlay")[i])
facet.axis("off")
nx.draw(dual, {n:[n[0], n[1]] for n in list(dual.nodes)}, ax=ax[1], node_size=15)
streets.plot(color='#e32e00', ax=ax[2], zorder=-1)
nx.draw(dual, {n:[n[0], n[1]] for n in list(dual.nodes)}, ax=ax[2], node_size=15)
```
At this moment (almost) any `networkx` method can be used. For illustration, we will measure the node degree. Using `networkx`, we can do:
```
degree = dict(nx.degree(graph))
nx.set_node_attributes(graph, degree, 'degree')
```
However, node degree is implemented in momepy so we can use directly:
```
graph = momepy.node_degree(graph, name='degree')
```
Once we have finished our network-based analysis, we want to convert the graph back to geodataframe. For that, we will use `momepy.nx_to_gdf`, which gives us several options what to export.
- `lines`
- original LineString geodataframe
- `points`
- point geometry representing street network intersections (nodes of primal graph)
- `spatial_weights`
- spatial weights for nodes capturing their relationship within a network
Moreover, `edges` will contain `node_start` and `node_end` columns capturing the ID of both nodes at its ends.
```
nodes, edges, sw = momepy.nx_to_gdf(graph, points=True, lines=True,
spatial_weights=True)
f, ax = plt.subplots(figsize=(10, 10))
nodes.plot(ax=ax, column='degree', cmap='tab20b', markersize=(nodes['degree'] * 100), zorder=2)
edges.plot(ax=ax, color='lightgrey', zorder=1)
ax.set_axis_off()
plt.show()
nodes.head(3)
edges.head(3)
```
|
github_jupyter
|
import momepy
import geopandas as gpd
import matplotlib.pyplot as plt
import networkx as nx
streets = gpd.read_file(momepy.datasets.get_path('bubenec'), layer='streets')
f, ax = plt.subplots(figsize=(10, 10))
streets.plot(ax=ax)
ax.set_axis_off()
plt.show()
graph = momepy.gdf_to_nx(streets, approach='primal')
f, ax = plt.subplots(1, 3, figsize=(18, 6), sharex=True, sharey=True)
streets.plot(color='#e32e00', ax=ax[0])
for i, facet in enumerate(ax):
facet.set_title(("Streets", "Primal graph", "Overlay")[i])
facet.axis("off")
nx.draw(graph, {n:[n[0], n[1]] for n in list(graph.nodes)}, ax=ax[1], node_size=15)
streets.plot(color='#e32e00', ax=ax[2], zorder=-1)
nx.draw(graph, {n:[n[0], n[1]] for n in list(graph.nodes)}, ax=ax[2], node_size=15)
dual = momepy.gdf_to_nx(streets, approach='dual')
f, ax = plt.subplots(1, 3, figsize=(18, 6), sharex=True, sharey=True)
streets.plot(color='#e32e00', ax=ax[0])
for i, facet in enumerate(ax):
facet.set_title(("Streets", "Dual graph", "Overlay")[i])
facet.axis("off")
nx.draw(dual, {n:[n[0], n[1]] for n in list(dual.nodes)}, ax=ax[1], node_size=15)
streets.plot(color='#e32e00', ax=ax[2], zorder=-1)
nx.draw(dual, {n:[n[0], n[1]] for n in list(dual.nodes)}, ax=ax[2], node_size=15)
degree = dict(nx.degree(graph))
nx.set_node_attributes(graph, degree, 'degree')
graph = momepy.node_degree(graph, name='degree')
nodes, edges, sw = momepy.nx_to_gdf(graph, points=True, lines=True,
spatial_weights=True)
f, ax = plt.subplots(figsize=(10, 10))
nodes.plot(ax=ax, column='degree', cmap='tab20b', markersize=(nodes['degree'] * 100), zorder=2)
edges.plot(ax=ax, color='lightgrey', zorder=1)
ax.set_axis_off()
plt.show()
nodes.head(3)
edges.head(3)
| 0.436862 | 0.987664 |
# Second wave analysis
```
#hide
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.dates as mdates
import matplotlib as mpl
from IPython.core.display import display,HTML
%matplotlib inline
from datetime import date
import datetime
#hide_input
headings= """
<!-- ####### HTML!! #########-->
<h1 style="color: #ff3933; text-align: center;">Covid-19 Matplotlib Overview in India</h1>
<h2 style="color: #3361ff; text-align: center;">Second wave Analysis (from 20th April 2021)</h2>"""
html = HTML(headings)
display(html)
#hide
df = pd.read_csv('https://api.covid19india.org/csv/latest/states.csv')
df = df[['Date','State','Confirmed','Deceased']]
df = df.rename(columns={'Confirmed':'Cases', "Deceased":'Deaths'})
df_cases = df[["Date",'State','Cases']]
df_deaths = df[['Date','State','Deaths']]
df_cases1 = df_cases.groupby('Date')
df_deaths1= df_deaths.groupby('Date')
#hide
df_cases1.head()
#hide
df_cases1.get_group('2020-04-20').head()
#hide
df_snap = pd.read_csv('data/SnapshotCases-28-July.csv')
arr_states = df_snap['states'].unique()
arr_dates = df['Date'].unique()
df_snap = df_snap.set_index('states')
df_snap.head()
#hide
arr_states[-9] = 'Puducherry'
arr_states[-10] = 'Odisha'
arr_states[6] = 'Dadra and Nagar Haveli and Daman and Diu'
arr_states = np.append(arr_states,np.array(['Chhattisgarh','Ladakh','Uttarakhand']))
arr_states
#hide
dates = []
for i in arr_dates:
if i>='2021-04-20':
dates.append(i)
dict = {'states':dates}
for i in arr_states:
dict[i] = [0]*len(dates)
dft_cases = pd.DataFrame(dict)
dft_deaths = pd.DataFrame(dict)
dft_deaths.head()
#hide
for i in range(len(dates)):
df1_deaths = df_deaths1.get_group(dates[i])
for j in range(len(df1_deaths.index)):
if df1_deaths.iloc[j,1] in arr_states:
dft_deaths.loc[i,df1_deaths.iloc[j,1]] = df1_deaths.iloc[j,2]
dft_deaths = dft_deaths.set_index('states')
df1_deaths.head()
#hide
for i in range(len(dates)):
df1_cases = df_cases1.get_group(dates[i])
for j in range(len(df1_cases.index)):
if df1_cases.iloc[j,1] in arr_states:
dft_cases.loc[i,df1_cases.iloc[j,1]] = df1_cases.iloc[j,2]
dft_cases = dft_cases.set_index('states')
#hide
dft_cases = dft_cases.T
dft_deaths = dft_deaths.T
dt_today = dates[-1]
dt_yday = dates[-2]
dft_deaths.head()
#hide
dft_cases = dft_cases.reset_index()
dft_deaths = dft_deaths.reset_index()
dft_cases = dft_cases.rename(columns = {'index':'state'})
dft_deaths = dft_deaths.rename(columns = {'index':'state'})
dft_deaths.head()
#hide
dfc_cases = dft_cases.groupby('state')[dt_today].sum()
dfc_deaths = dft_deaths.groupby('state')[dt_today].sum()
dfp_cases = dft_cases.groupby('state')[dt_yday].sum()
dfp_deaths = dft_deaths.groupby('state')[dt_yday].sum()
dfc_cases.head()
#hide
df_table = pd.DataFrame({'states': dfc_cases.index, 'Cases': dfc_cases.values, 'Deaths': dfc_deaths.values, 'PCases': dfp_cases.values, 'PDeaths': dfp_deaths.values}).set_index('states')
df_table = df_table.sort_values(by = ['Cases','Deaths'], ascending = [False, False])
df_table = df_table.reset_index()
df_table.head()
#hide
for c in 'Cases, Deaths'.split(', '):
df_table[f'{c} (+)'] = (df_table[c] - df_table[f'P{c}']).clip(0)
df_table['Fatality Rate'] = (100* df_table['Deaths']/ df_table['Cases']).round(2)
#hide
df_table.head()
#hide
summary = {'updated':dates[-1], 'since':dates[-2]}
list_names = ['Cases', 'PCases', 'Deaths', 'PDeaths', 'Cases (+)', 'Deaths (+)']
for name in list_names:
summary[name] = df_table.sum()[name]
summary
#hide
overview = """
<!-- ####### HTML!! #########-->
<h1 style="color: #5e9ca0; text-align: center;">India</h1>
<h2 style="color: #ff337d; text-align: center;">Last update: <strong>{update}</strong></h2>
<h3 style="text-align: center;">Confirmed cases:</h3>
<p style="text-align: center;font-size:24px;">{cases} (<span style="color: #ff0000;">+{new}</span>)</p>
<h3 style="text-align: center;">Confirmed deaths:</h3>
<p style="text-align: center;font-size:24px;">{deaths} (<span style="color: #ff0000;">+{dnew}</span>)</p>"""
#hide_input
update = summary['updated']
cases = summary['Cases']
new = summary['Cases (+)']
deaths = summary['Deaths']
dnew = summary['Deaths (+)']
html = HTML(overview.format(update=update, cases=cases,new=new,deaths=deaths,dnew=dnew));
display(html);
#hide
dt_cols = list(dft_cases.columns[1:])
#print(dt_cols)
dft_ct_new_cases = dft_cases.groupby('state')[dt_cols].sum().diff(axis=1).fillna(0).astype(int)
#print(dft_ct_new_cases.head())
dft_ct_new_cases.sort_values(by = dates[-1], ascending = False,inplace = True)
#hide
dft_ct_new_cases.head()
#hide_input
df = dft_ct_new_cases.copy()
df.loc['Total'] = df.sum()
ef = df.loc['Total'].rename_axis('date').reset_index()
ef['date'] = ef['date'].astype('datetime64[ns]')
fig,ax1 = plt.subplots(figsize = (16,12))
ax1.bar(ef.date,ef.Total,alpha=0.3,color='#007acc')
ax1.plot(ef.date,ef.Total , marker="o", color='#007acc')
ax1.xaxis.set_major_locator(mdates.WeekdayLocator(byweekday=mdates.MO))
ax1.xaxis.set_major_formatter(mdates.DateFormatter('%b %d'))
ax1.text(0.48, 0.6,'India daily case count', transform = ax1.transAxes,fontsize=40,color='#ff0000');
ax1.set_ylim([min(ef.Total),max(ef.Total)])
ax1.spines['right'].set_visible(False)
ax1.spines['top'].set_visible(False)
#hide_input
create= """
<!-- ####### HTML!! #########-->
<h1 style="color: #ff004c; text-align: center;">Analysis of top-10 States in covid cases</h1>
"""
html = HTML(create)
display(html)
#hide_input
n=10
ax=[]
fig = plt.figure(figsize = (10,50))
gs = fig.add_gridspec(60,20)
arr=df_table.iloc[:10,0].values
count=0
for i in range(len(arr)):
ax.append(fig.add_subplot(gs[count:count+5,:]))
ef = df.loc[arr[i]].rename_axis('date').reset_index()
ef['date'] = ef['date'].astype('datetime64[ns]')
ax[i].bar(ef.date.values,ef.iloc[:,-1].values,color = '#007acc',alpha=0.3)
ax[i].plot(ef.date,ef.iloc[:,-1],marker='o',color='#007acc')
ax[i].text(0.6,0.5,f'{arr[i]}',transform = ax[i].transAxes, fontsize = 30,color='#b30000');
ax[i].xaxis.set_major_locator(mdates.WeekdayLocator())
ax[i].xaxis.set_major_formatter(mdates.DateFormatter('%b %d'))
ax[i].spines['right'].set_visible(False)
ax[i].spines['top'].set_visible(False)
count+=6
for tick in ax[i].xaxis.get_major_ticks():
tick.label1.set_fontsize(11)
for tick in ax[i].yaxis.get_major_ticks():
tick.label1.set_fontsize(14)
plt.tight_layout()
#hide_input
fr= df_table['Fatality Rate'].values
ma,mi=max(fr),min(fr)
def max_and_min(df_2,ft):
for i in range(len(ft)):
if df_2.iloc[i,-3]==ma:
state_max=[df_2.iloc[i,0],df_2.iloc[i,-3]]
elif df_2.iloc[i,-3]==mi:
state_min=[df_2.iloc[i,0],df_2.iloc[i,-3]]
return state_max,state_min
x = df_table['Cases (+)'].sum()
y=df_table['Deaths (+)'].sum()
df_table['% Cases (+)']= ((df_table['Cases (+)']/x)*100).round(2)
df_table['% Deaths (+)']=((df_table['Deaths (+)']/y)*100).round(2)
print(df_table.to_string(index=False))
#hide_input
a,b=max_and_min(df_table,fr)
fatality=f"""
<!-- ####### HTML!! #########-->
<h1 style="color: #ff004c; text-align: center;">{a[0]} has highest Fatality rate ({a[1]})</h1>
<h1 style="color: #ff004c; text-align: center;">{b[0]} has lowest Fatality rate ({b[1]})</h1>"""
html = HTML(fatality)
display(html)
```
|
github_jupyter
|
#hide
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.dates as mdates
import matplotlib as mpl
from IPython.core.display import display,HTML
%matplotlib inline
from datetime import date
import datetime
#hide_input
headings= """
<!-- ####### HTML!! #########-->
<h1 style="color: #ff3933; text-align: center;">Covid-19 Matplotlib Overview in India</h1>
<h2 style="color: #3361ff; text-align: center;">Second wave Analysis (from 20th April 2021)</h2>"""
html = HTML(headings)
display(html)
#hide
df = pd.read_csv('https://api.covid19india.org/csv/latest/states.csv')
df = df[['Date','State','Confirmed','Deceased']]
df = df.rename(columns={'Confirmed':'Cases', "Deceased":'Deaths'})
df_cases = df[["Date",'State','Cases']]
df_deaths = df[['Date','State','Deaths']]
df_cases1 = df_cases.groupby('Date')
df_deaths1= df_deaths.groupby('Date')
#hide
df_cases1.head()
#hide
df_cases1.get_group('2020-04-20').head()
#hide
df_snap = pd.read_csv('data/SnapshotCases-28-July.csv')
arr_states = df_snap['states'].unique()
arr_dates = df['Date'].unique()
df_snap = df_snap.set_index('states')
df_snap.head()
#hide
arr_states[-9] = 'Puducherry'
arr_states[-10] = 'Odisha'
arr_states[6] = 'Dadra and Nagar Haveli and Daman and Diu'
arr_states = np.append(arr_states,np.array(['Chhattisgarh','Ladakh','Uttarakhand']))
arr_states
#hide
dates = []
for i in arr_dates:
if i>='2021-04-20':
dates.append(i)
dict = {'states':dates}
for i in arr_states:
dict[i] = [0]*len(dates)
dft_cases = pd.DataFrame(dict)
dft_deaths = pd.DataFrame(dict)
dft_deaths.head()
#hide
for i in range(len(dates)):
df1_deaths = df_deaths1.get_group(dates[i])
for j in range(len(df1_deaths.index)):
if df1_deaths.iloc[j,1] in arr_states:
dft_deaths.loc[i,df1_deaths.iloc[j,1]] = df1_deaths.iloc[j,2]
dft_deaths = dft_deaths.set_index('states')
df1_deaths.head()
#hide
for i in range(len(dates)):
df1_cases = df_cases1.get_group(dates[i])
for j in range(len(df1_cases.index)):
if df1_cases.iloc[j,1] in arr_states:
dft_cases.loc[i,df1_cases.iloc[j,1]] = df1_cases.iloc[j,2]
dft_cases = dft_cases.set_index('states')
#hide
dft_cases = dft_cases.T
dft_deaths = dft_deaths.T
dt_today = dates[-1]
dt_yday = dates[-2]
dft_deaths.head()
#hide
dft_cases = dft_cases.reset_index()
dft_deaths = dft_deaths.reset_index()
dft_cases = dft_cases.rename(columns = {'index':'state'})
dft_deaths = dft_deaths.rename(columns = {'index':'state'})
dft_deaths.head()
#hide
dfc_cases = dft_cases.groupby('state')[dt_today].sum()
dfc_deaths = dft_deaths.groupby('state')[dt_today].sum()
dfp_cases = dft_cases.groupby('state')[dt_yday].sum()
dfp_deaths = dft_deaths.groupby('state')[dt_yday].sum()
dfc_cases.head()
#hide
df_table = pd.DataFrame({'states': dfc_cases.index, 'Cases': dfc_cases.values, 'Deaths': dfc_deaths.values, 'PCases': dfp_cases.values, 'PDeaths': dfp_deaths.values}).set_index('states')
df_table = df_table.sort_values(by = ['Cases','Deaths'], ascending = [False, False])
df_table = df_table.reset_index()
df_table.head()
#hide
for c in 'Cases, Deaths'.split(', '):
df_table[f'{c} (+)'] = (df_table[c] - df_table[f'P{c}']).clip(0)
df_table['Fatality Rate'] = (100* df_table['Deaths']/ df_table['Cases']).round(2)
#hide
df_table.head()
#hide
summary = {'updated':dates[-1], 'since':dates[-2]}
list_names = ['Cases', 'PCases', 'Deaths', 'PDeaths', 'Cases (+)', 'Deaths (+)']
for name in list_names:
summary[name] = df_table.sum()[name]
summary
#hide
overview = """
<!-- ####### HTML!! #########-->
<h1 style="color: #5e9ca0; text-align: center;">India</h1>
<h2 style="color: #ff337d; text-align: center;">Last update: <strong>{update}</strong></h2>
<h3 style="text-align: center;">Confirmed cases:</h3>
<p style="text-align: center;font-size:24px;">{cases} (<span style="color: #ff0000;">+{new}</span>)</p>
<h3 style="text-align: center;">Confirmed deaths:</h3>
<p style="text-align: center;font-size:24px;">{deaths} (<span style="color: #ff0000;">+{dnew}</span>)</p>"""
#hide_input
update = summary['updated']
cases = summary['Cases']
new = summary['Cases (+)']
deaths = summary['Deaths']
dnew = summary['Deaths (+)']
html = HTML(overview.format(update=update, cases=cases,new=new,deaths=deaths,dnew=dnew));
display(html);
#hide
dt_cols = list(dft_cases.columns[1:])
#print(dt_cols)
dft_ct_new_cases = dft_cases.groupby('state')[dt_cols].sum().diff(axis=1).fillna(0).astype(int)
#print(dft_ct_new_cases.head())
dft_ct_new_cases.sort_values(by = dates[-1], ascending = False,inplace = True)
#hide
dft_ct_new_cases.head()
#hide_input
df = dft_ct_new_cases.copy()
df.loc['Total'] = df.sum()
ef = df.loc['Total'].rename_axis('date').reset_index()
ef['date'] = ef['date'].astype('datetime64[ns]')
fig,ax1 = plt.subplots(figsize = (16,12))
ax1.bar(ef.date,ef.Total,alpha=0.3,color='#007acc')
ax1.plot(ef.date,ef.Total , marker="o", color='#007acc')
ax1.xaxis.set_major_locator(mdates.WeekdayLocator(byweekday=mdates.MO))
ax1.xaxis.set_major_formatter(mdates.DateFormatter('%b %d'))
ax1.text(0.48, 0.6,'India daily case count', transform = ax1.transAxes,fontsize=40,color='#ff0000');
ax1.set_ylim([min(ef.Total),max(ef.Total)])
ax1.spines['right'].set_visible(False)
ax1.spines['top'].set_visible(False)
#hide_input
create= """
<!-- ####### HTML!! #########-->
<h1 style="color: #ff004c; text-align: center;">Analysis of top-10 States in covid cases</h1>
"""
html = HTML(create)
display(html)
#hide_input
n=10
ax=[]
fig = plt.figure(figsize = (10,50))
gs = fig.add_gridspec(60,20)
arr=df_table.iloc[:10,0].values
count=0
for i in range(len(arr)):
ax.append(fig.add_subplot(gs[count:count+5,:]))
ef = df.loc[arr[i]].rename_axis('date').reset_index()
ef['date'] = ef['date'].astype('datetime64[ns]')
ax[i].bar(ef.date.values,ef.iloc[:,-1].values,color = '#007acc',alpha=0.3)
ax[i].plot(ef.date,ef.iloc[:,-1],marker='o',color='#007acc')
ax[i].text(0.6,0.5,f'{arr[i]}',transform = ax[i].transAxes, fontsize = 30,color='#b30000');
ax[i].xaxis.set_major_locator(mdates.WeekdayLocator())
ax[i].xaxis.set_major_formatter(mdates.DateFormatter('%b %d'))
ax[i].spines['right'].set_visible(False)
ax[i].spines['top'].set_visible(False)
count+=6
for tick in ax[i].xaxis.get_major_ticks():
tick.label1.set_fontsize(11)
for tick in ax[i].yaxis.get_major_ticks():
tick.label1.set_fontsize(14)
plt.tight_layout()
#hide_input
fr= df_table['Fatality Rate'].values
ma,mi=max(fr),min(fr)
def max_and_min(df_2,ft):
for i in range(len(ft)):
if df_2.iloc[i,-3]==ma:
state_max=[df_2.iloc[i,0],df_2.iloc[i,-3]]
elif df_2.iloc[i,-3]==mi:
state_min=[df_2.iloc[i,0],df_2.iloc[i,-3]]
return state_max,state_min
x = df_table['Cases (+)'].sum()
y=df_table['Deaths (+)'].sum()
df_table['% Cases (+)']= ((df_table['Cases (+)']/x)*100).round(2)
df_table['% Deaths (+)']=((df_table['Deaths (+)']/y)*100).round(2)
print(df_table.to_string(index=False))
#hide_input
a,b=max_and_min(df_table,fr)
fatality=f"""
<!-- ####### HTML!! #########-->
<h1 style="color: #ff004c; text-align: center;">{a[0]} has highest Fatality rate ({a[1]})</h1>
<h1 style="color: #ff004c; text-align: center;">{b[0]} has lowest Fatality rate ({b[1]})</h1>"""
html = HTML(fatality)
display(html)
| 0.096354 | 0.663996 |
## Dependencies
```
!pip install --quiet efficientnet
import warnings, time
from kaggle_datasets import KaggleDatasets
from sklearn.model_selection import KFold
from sklearn.metrics import classification_report, confusion_matrix, accuracy_score
from tensorflow.keras import optimizers, Sequential, losses, metrics, Model
from tensorflow.keras.callbacks import EarlyStopping, ModelCheckpoint
import efficientnet.tfkeras as efn
from cassava_scripts import *
from scripts_step_lr_schedulers import *
import tensorflow_addons as tfa
seed = 0
seed_everything(seed)
warnings.filterwarnings('ignore')
```
### Hardware configuration
```
# TPU or GPU detection
# Detect hardware, return appropriate distribution strategy
strategy, tpu = set_up_strategy()
AUTO = tf.data.experimental.AUTOTUNE
REPLICAS = strategy.num_replicas_in_sync
print(f'REPLICAS: {REPLICAS}')
# Mixed precision
from tensorflow.keras.mixed_precision import experimental as mixed_precision
policy = mixed_precision.Policy('mixed_bfloat16')
mixed_precision.set_policy(policy)
# XLA
tf.config.optimizer.set_jit(True)
```
# Model parameters
```
BATCH_SIZE = 8 * REPLICAS
LEARNING_RATE = 1e-5 * REPLICAS
EPOCHS_CL = 5
EPOCHS = 33
HEIGHT = 512
WIDTH = 512
HEIGHT_DT = 512
WIDTH_DT = 512
CHANNELS = 3
N_CLASSES = 5
N_FOLDS = 5
FOLDS_USED = 1
ES_PATIENCE = 10
```
# Load data
```
database_base_path = '/kaggle/input/cassava-leaf-disease-classification/'
train = pd.read_csv(f'{database_base_path}train.csv')
print(f'Train samples: {len(train)}')
GCS_PATH = KaggleDatasets().get_gcs_path(f'cassava-leaf-disease-tfrecords-center-{HEIGHT_DT}x{WIDTH_DT}') # Center croped and resized (50 TFRecord)
# GCS_PATH_EXT = KaggleDatasets().get_gcs_path(f'cassava-leaf-disease-tfrecords-external-{HEIGHT_DT}x{WIDTH_DT}') # Center croped and resized (50 TFRecord) (External)
# GCS_PATH_CLASSES = KaggleDatasets().get_gcs_path(f'cassava-leaf-disease-tfrecords-classes-{HEIGHT_DT}x{WIDTH_DT}') # Center croped and resized (50 TFRecord) by classes
# GCS_PATH_EXT_CLASSES = KaggleDatasets().get_gcs_path(f'cassava-leaf-disease-tfrecords-classes-ext-{HEIGHT_DT}x{WIDTH_DT}') # Center croped and resized (50 TFRecord) (External) by classes
FILENAMES_COMP = tf.io.gfile.glob(GCS_PATH + '/*.tfrec')
# FILENAMES_2019 = tf.io.gfile.glob(GCS_PATH_EXT + '/*.tfrec')
# FILENAMES_COMP_CBB = tf.io.gfile.glob(GCS_PATH_CLASSES + '/CBB*.tfrec')
# FILENAMES_COMP_CBSD = tf.io.gfile.glob(GCS_PATH_CLASSES + '/CBSD*.tfrec')
# FILENAMES_COMP_CGM = tf.io.gfile.glob(GCS_PATH_CLASSES + '/CGM*.tfrec')
# FILENAMES_COMP_CMD = tf.io.gfile.glob(GCS_PATH_CLASSES + '/CMD*.tfrec')
# FILENAMES_COMP_Healthy = tf.io.gfile.glob(GCS_PATH_CLASSES + '/Healthy*.tfrec')
# FILENAMES_2019_CBB = tf.io.gfile.glob(GCS_PATH_EXT_CLASSES + '/CBB*.tfrec')
# FILENAMES_2019_CBSD = tf.io.gfile.glob(GCS_PATH_EXT_CLASSES + '/CBSD*.tfrec')
# FILENAMES_2019_CGM = tf.io.gfile.glob(GCS_PATH_EXT_CLASSES + '/CGM*.tfrec')
# FILENAMES_2019_CMD = tf.io.gfile.glob(GCS_PATH_EXT_CLASSES + '/CMD*.tfrec')
# FILENAMES_2019_Healthy = tf.io.gfile.glob(GCS_PATH_EXT_CLASSES + '/Healthy*.tfrec')
TRAINING_FILENAMES = FILENAMES_COMP
NUM_TRAINING_IMAGES = count_data_items(TRAINING_FILENAMES)
print(f'GCS: train images: {NUM_TRAINING_IMAGES}')
display(train.head())
```
# Augmentation
```
def data_augment(image, label):
# p_rotation = tf.random.uniform([], 0, 1.0, dtype=tf.float32)
p_spatial = tf.random.uniform([], 0, 1.0, dtype=tf.float32)
p_rotate = tf.random.uniform([], 0, 1.0, dtype=tf.float32)
p_pixel_1 = tf.random.uniform([], 0, 1.0, dtype=tf.float32)
p_pixel_2 = tf.random.uniform([], 0, 1.0, dtype=tf.float32)
p_pixel_3 = tf.random.uniform([], 0, 1.0, dtype=tf.float32)
# p_shear = tf.random.uniform([], 0, 1.0, dtype=tf.float32)
p_crop = tf.random.uniform([], 0, 1.0, dtype=tf.float32)
p_cutout = tf.random.uniform([], 0, 1.0, dtype=tf.float32)
# # Shear
# if p_shear > .2:
# if p_shear > .6:
# image = transform_shear(image, HEIGHT, shear=20.)
# else:
# image = transform_shear(image, HEIGHT, shear=-20.)
# # Rotation
# if p_rotation > .2:
# if p_rotation > .6:
# image = transform_rotation(image, HEIGHT, rotation=45.)
# else:
# image = transform_rotation(image, HEIGHT, rotation=-45.)
# Flips
image = tf.image.random_flip_left_right(image)
image = tf.image.random_flip_up_down(image)
if p_spatial > .75:
image = tf.image.transpose(image)
# Rotates
if p_rotate > .75:
image = tf.image.rot90(image, k=3) # rotate 270º
elif p_rotate > .5:
image = tf.image.rot90(image, k=2) # rotate 180º
elif p_rotate > .25:
image = tf.image.rot90(image, k=1) # rotate 90º
# Pixel-level transforms
if p_pixel_1 >= .4:
image = tf.image.random_saturation(image, lower=.7, upper=1.3)
if p_pixel_2 >= .4:
image = tf.image.random_contrast(image, lower=.8, upper=1.2)
if p_pixel_3 >= .4:
image = tf.image.random_brightness(image, max_delta=.1)
# Crops
if p_crop > .6:
if p_crop > .9:
image = tf.image.central_crop(image, central_fraction=.5)
elif p_crop > .8:
image = tf.image.central_crop(image, central_fraction=.6)
elif p_crop > .7:
image = tf.image.central_crop(image, central_fraction=.7)
else:
image = tf.image.central_crop(image, central_fraction=.8)
elif p_crop > .3:
crop_size = tf.random.uniform([], int(HEIGHT*.6), HEIGHT, dtype=tf.int32)
image = tf.image.random_crop(image, size=[crop_size, crop_size, CHANNELS])
image = tf.image.resize(image, size=[HEIGHT, WIDTH])
if p_cutout > .5:
image = data_augment_cutout(image)
return image, label
```
## Auxiliary functions
```
# CutOut
def data_augment_cutout(image, min_mask_size=(int(HEIGHT * .1), int(HEIGHT * .1)),
max_mask_size=(int(HEIGHT * .125), int(HEIGHT * .125))):
p_cutout = tf.random.uniform([], 0, 1.0, dtype=tf.float32)
if p_cutout > .85: # 10~15 cut outs
n_cutout = tf.random.uniform([], 10, 15, dtype=tf.int32)
image = random_cutout(image, HEIGHT, WIDTH,
min_mask_size=min_mask_size, max_mask_size=max_mask_size, k=n_cutout)
elif p_cutout > .6: # 5~10 cut outs
n_cutout = tf.random.uniform([], 5, 10, dtype=tf.int32)
image = random_cutout(image, HEIGHT, WIDTH,
min_mask_size=min_mask_size, max_mask_size=max_mask_size, k=n_cutout)
elif p_cutout > .25: # 2~5 cut outs
n_cutout = tf.random.uniform([], 2, 5, dtype=tf.int32)
image = random_cutout(image, HEIGHT, WIDTH,
min_mask_size=min_mask_size, max_mask_size=max_mask_size, k=n_cutout)
else: # 1 cut out
image = random_cutout(image, HEIGHT, WIDTH,
min_mask_size=min_mask_size, max_mask_size=max_mask_size, k=1)
return image
# Datasets utility functions
def random_crop(image, label):
"""
Resize and reshape images to the expected size.
"""
image = tf.image.random_crop(image, size=[HEIGHT, WIDTH, CHANNELS])
return image, label
def prepare_image(image, label):
"""
Resize and reshape images to the expected size.
"""
image = tf.image.resize(image, [HEIGHT, WIDTH])
image = tf.reshape(image, [HEIGHT, WIDTH, CHANNELS])
return image, label
def center_crop_(image, label, height_rs, width_rs, height=HEIGHT_DT, width=WIDTH_DT, channels=3):
image = tf.reshape(image, [height, width, channels]) # Original shape
h, w = image.shape[0], image.shape[1]
if h > w:
image = tf.image.crop_to_bounding_box(image, (h - w) // 2, 0, w, w)
else:
image = tf.image.crop_to_bounding_box(image, 0, (w - h) // 2, h, h)
image = tf.image.resize(image, [height_rs, width_rs]) # Expected shape
return image, label
def read_tfrecord_(example, labeled=True, n_classes=5):
"""
1. Parse data based on the 'TFREC_FORMAT' map.
2. Decode image.
3. If 'labeled' returns (image, label) if not (image, name).
"""
if labeled:
TFREC_FORMAT = {
'image': tf.io.FixedLenFeature([], tf.string),
'target': tf.io.FixedLenFeature([], tf.int64),
}
else:
TFREC_FORMAT = {
'image': tf.io.FixedLenFeature([], tf.string),
'image_name': tf.io.FixedLenFeature([], tf.string),
}
example = tf.io.parse_single_example(example, TFREC_FORMAT)
image = decode_image(example['image'])
if labeled:
label_or_name = tf.cast(example['target'], tf.int32)
# One-Hot Encoding needed to use "categorical_crossentropy" loss
# label_or_name = tf.one_hot(tf.cast(label_or_name, tf.int32), n_classes)
else:
label_or_name = example['image_name']
return image, label_or_name
def get_dataset(filenames, labeled=True, ordered=False, repeated=False,
cached=False, augment=False):
"""
Return a Tensorflow dataset ready for training or inference.
"""
ignore_order = tf.data.Options()
if not ordered:
ignore_order.experimental_deterministic = False
dataset = tf.data.Dataset.list_files(filenames)
dataset = dataset.interleave(tf.data.TFRecordDataset, num_parallel_calls=AUTO)
else:
dataset = tf.data.TFRecordDataset(filenames, num_parallel_reads=AUTO)
dataset = dataset.with_options(ignore_order)
dataset = dataset.map(lambda x: read_tfrecord_(x, labeled=labeled), num_parallel_calls=AUTO)
# dataset = dataset.map(lambda x: read_tfrecord(x, labeled=labeled), num_parallel_calls=AUTO)
if augment:
dataset = dataset.map(data_augment, num_parallel_calls=AUTO)
dataset = dataset.map(scale_image, num_parallel_calls=AUTO)
dataset = dataset.map(prepare_image, num_parallel_calls=AUTO)
if labeled:
dataset = dataset.map(conf_output, num_parallel_calls=AUTO)
if not ordered:
dataset = dataset.shuffle(2048)
if repeated:
dataset = dataset.repeat()
dataset = dataset.batch(BATCH_SIZE)
if cached:
dataset = dataset.cache()
dataset = dataset.prefetch(AUTO)
return dataset
def conf_output(image, label):
"""
Configure the output of the dataset.
"""
aux_label = [0.]
aux_2_label = [0.]
# if tf.math.argmax(label, axis=-1) == 4: # Healthy
if label == 4: # Healthy
aux_label = [1.]
# if tf.math.argmax(label, axis=-1) == 3: # CMD
if label == 3: # CMD
aux_2_label = [1.]
return (image, (label, aux_label, aux_2_label))
```
# Training data samples (with augmentation)
```
# train_dataset = get_dataset(FILENAMES_COMP, ordered=True, augment=True)
# train_iter = iter(train_dataset.unbatch().batch(20))
# display_batch_of_images(next(train_iter))
# display_batch_of_images(next(train_iter))
```
# Model
```
def encoder_fn(input_shape):
inputs = L.Input(shape=input_shape, name='input_image')
base_model = efn.EfficientNetB4(input_tensor=inputs,
include_top=False,
weights='noisy-student',
pooling='avg')
model = Model(inputs=inputs, outputs=base_model.output)
return model
def add_projection_head(input_shape, encoder):
inputs = L.Input(shape=input_shape, name='input_image')
features = encoder(inputs)
outputs = L.Dense(128, activation='relu', name='projection_head', dtype='float32')(features)
model = Model(inputs=inputs, outputs=outputs)
return model
def classifier_fn(input_shape, N_CLASSES, encoder, trainable=True):
for layer in encoder.layers:
layer.trainable = trainable
inputs = L.Input(shape=input_shape, name='input_image')
features = encoder(inputs)
features = L.Dropout(.25)(features)
features = L.Dense(1000, activation='relu')(features)
features = L.Dropout(.25)(features)
output = L.Dense(N_CLASSES, activation='softmax', name='output', dtype='float32')(features)
output_healthy = L.Dense(1, activation='sigmoid', name='output_healthy', dtype='float32')(features)
output_cmd = L.Dense(1, activation='sigmoid', name='output_cmd', dtype='float32')(features)
model = Model(inputs=inputs, outputs=[output, output_healthy, output_cmd])
return model
temperature = 0.1
class SupervisedContrastiveLoss(losses.Loss):
def __init__(self, temperature=0.1, name=None):
super(SupervisedContrastiveLoss, self).__init__(name=name)
self.temperature = temperature
def __call__(self, labels, feature_vectors, sample_weight=None):
# Normalize feature vectors
feature_vectors_normalized = tf.math.l2_normalize(feature_vectors, axis=1)
# Compute logits
logits = tf.divide(
tf.matmul(
feature_vectors_normalized, tf.transpose(feature_vectors_normalized)
),
temperature,
)
return tfa.losses.npairs_loss(tf.squeeze(labels), logits)
```
### Learning rate schedule
```
lr_start = 1e-8
lr_min = 1e-6
lr_max = LEARNING_RATE
num_cycles = 3.
warmup_epochs = 3
hold_max_epochs = 0
total_epochs = EPOCHS
step_size = (NUM_TRAINING_IMAGES//BATCH_SIZE)
hold_max_steps = hold_max_epochs * step_size
total_steps = total_epochs * step_size
warmup_steps = warmup_epochs * step_size
def lrfn(total_steps, warmup_steps=0, lr_start=1e-4, lr_max=1e-3, lr_min=1e-4, num_cycles=1.):
@tf.function
def cosine_with_hard_restarts_schedule_with_warmup_(step):
""" Create a schedule with a learning rate that decreases following the
values of the cosine function with several hard restarts, after a warmup
period during which it increases linearly between 0 and 1.
"""
if step < warmup_steps:
lr = (lr_max - lr_start) / warmup_steps * step + lr_start
else:
progress = (step - warmup_steps) / (total_steps - warmup_steps)
lr = lr_max * (0.5 * (1.0 + tf.math.cos(np.pi * ((num_cycles * progress) % 1.0))))
if lr_min is not None:
lr = tf.math.maximum(lr_min, float(lr))
return lr
return cosine_with_hard_restarts_schedule_with_warmup_
lrfn_fn = lrfn(total_steps, warmup_steps, lr_start, lr_max, lr_min, num_cycles)
rng = [i for i in range(total_steps)]
y = [lrfn_fn(tf.cast(x, tf.float32)) for x in rng]
sns.set(style='whitegrid')
fig, ax = plt.subplots(figsize=(20, 6))
plt.plot(rng, y)
print(f'{total_steps} total steps and {step_size} steps per epoch')
print(f'Learning rate schedule: {y[0]:.3g} to {max(y):.3g} to {y[-1]:.3g}')
```
# Training
```
skf = KFold(n_splits=N_FOLDS, shuffle=True, random_state=seed)
oof_pred = []; oof_labels = []; oof_names = []; oof_folds = []; history_list = []; oof_embed = []
for fold,(idxT, idxV) in enumerate(skf.split(np.arange(15))):
if fold >= FOLDS_USED:
break
if tpu: tf.tpu.experimental.initialize_tpu_system(tpu)
K.clear_session()
print(f'\nFOLD: {fold+1}')
print(f'TRAIN: {idxT} VALID: {idxV}')
# Create train and validation sets
TRAIN_FILENAMES = tf.io.gfile.glob([GCS_PATH + '/Id_train%.2i*.tfrec' % x for x in idxT])
# FILENAMES_COMP_CBB = tf.io.gfile.glob([GCS_PATH_CLASSES + '/CBB%.2i*.tfrec' % x for x in idxT])
# FILENAMES_COMP_CBSD = tf.io.gfile.glob([GCS_PATH_CLASSES + '/CBSD%.2i*.tfrec' % x for x in idxT])
# FILENAMES_COMP_CGM = tf.io.gfile.glob([GCS_PATH_CLASSES + '/CGM%.2i*.tfrec' % x for x in idxT])
# FILENAMES_COMP_Healthy = tf.io.gfile.glob([GCS_PATH_CLASSES + '/Healthy%.2i*.tfrec' % x for x in idxT])
np.random.shuffle(TRAIN_FILENAMES)
VALID_FILENAMES = tf.io.gfile.glob([GCS_PATH + '/Id_train%.2i*.tfrec' % x for x in idxV])
ct_train = count_data_items(TRAIN_FILENAMES)
ct_valid = count_data_items(VALID_FILENAMES)
step_size = (ct_train // BATCH_SIZE)
warmup_steps = (warmup_epochs * step_size)
total_steps = (total_epochs * step_size)
total_steps_cl = (EPOCHS_CL * step_size)
warmup_steps_cl = 1
### Pre-train the encoder
print('Pre-training the encoder using "Supervised Contrastive" Loss')
with strategy.scope():
encoder = encoder_fn((None, None, CHANNELS))
encoder_proj = add_projection_head((None, None, CHANNELS), encoder)
encoder_proj.summary()
lrfn_fn = lrfn(total_steps, warmup_steps, lr_start, lr_max, lr_min, num_cycles)
# optimizer = optimizers.SGD(learning_rate=lambda: lrfn_fn(tf.cast(optimizer.iterations, tf.float32)),
# momentum=0.95, nesterov=True)
optimizer = optimizers.Adam(learning_rate=lambda: lrfn_fn(tf.cast(optimizer.iterations, tf.float32)))
encoder_proj.compile(optimizer=optimizer,
loss=SupervisedContrastiveLoss(temperature))
es = EarlyStopping(monitor='val_loss', mode='min',
patience=ES_PATIENCE, restore_best_weights=True, verbose=1)
history_enc = encoder_proj.fit(x=get_dataset(TRAIN_FILENAMES, repeated=True, augment=True),
validation_data=get_dataset(VALID_FILENAMES, ordered=True),
steps_per_epoch=step_size,
batch_size=BATCH_SIZE,
callbacks=[es],
epochs=EPOCHS,
verbose=2).history
### Train the classifier with the frozen encoder
print('Training the classifier with the frozen encoder')
with strategy.scope():
model = classifier_fn((None, None, CHANNELS), N_CLASSES, encoder, trainable=False)
model.summary()
lrfn_fn = lrfn(total_steps_cl, warmup_steps_cl, lr_start, lr_max, lr_min, num_cycles)
optimizer = optimizers.Adam(learning_rate=lambda: lrfn_fn(tf.cast(optimizer.iterations, tf.float32)))
model.compile(optimizer=optimizer,
loss={'output': losses.SparseCategoricalCrossentropy(),
'output_healthy': losses.BinaryCrossentropy(label_smoothing=.1),
'output_cmd': losses.BinaryCrossentropy(label_smoothing=.1)},
loss_weights={'output': 1.,
'output_healthy': .1,
'output_cmd': .1},
metrics={'output': metrics.SparseCategoricalAccuracy(),
'output_healthy': metrics.BinaryAccuracy(),
'output_cmd': metrics.BinaryAccuracy()})
model_path = f'model_{fold}.h5'
history = model.fit(x=get_dataset(TRAIN_FILENAMES, repeated=True, augment=True),
validation_data=get_dataset(VALID_FILENAMES, ordered=True),
steps_per_epoch=step_size,
epochs=EPOCHS_CL,
verbose=2).history
### RESULTS
print(f"#### FOLD {fold+1} OOF Accuracy = {np.max(history['val_output_sparse_categorical_accuracy']):.3f}")
history_list.append(history)
# Save last model weights
model.save_weights(model_path)
# Load best model weights
# model.load_weights(model_path)
# OOF predictions
ds_valid = get_dataset(VALID_FILENAMES, ordered=True)
oof_folds.append(np.full((ct_valid), fold, dtype='int8'))
oof_labels.append([target[0].numpy() for img, target in iter(ds_valid.unbatch())])
x_oof = ds_valid.map(lambda image, target: image)
oof_pred.append(np.argmax(model.predict(x_oof)[0], axis=-1))
# OOF names
ds_valid_names = get_dataset(VALID_FILENAMES, labeled=False, ordered=True)
oof_names.append(np.array([img_name.numpy().decode('utf-8') for img, img_name in iter(ds_valid_names.unbatch())]))
oof_embed.append(encoder.predict(x_oof)) # OOF embeddings
```
## Model loss graph
```
for fold, history in enumerate(history_list):
print(f'\nFOLD: {fold+1}')
plot_metrics(history, acc_name='output_sparse_categorical_accuracy')
```
# Model evaluation
```
y_true = np.concatenate(oof_labels)
# y_true = np.argmax(y_true, axis=-1)
y_pred = np.concatenate(oof_pred)
folds = np.concatenate(oof_folds)
names = np.concatenate(oof_names)
acc = accuracy_score(y_true, y_pred)
print(f'Overall OOF Accuracy = {acc:.3f}')
df_oof = pd.DataFrame({'image_id':names, 'fold':fold,
'target':y_true, 'pred':y_pred})
df_oof.to_csv('oof.csv', index=False)
display(df_oof.head())
print(classification_report(y_true, y_pred, target_names=CLASSES))
```
# Confusion matrix
```
fig, ax = plt.subplots(1, 1, figsize=(20, 12))
cfn_matrix = confusion_matrix(y_true, y_pred, labels=range(len(CLASSES)))
cfn_matrix = (cfn_matrix.T / cfn_matrix.sum(axis=1)).T
df_cm = pd.DataFrame(cfn_matrix, index=CLASSES, columns=CLASSES)
ax = sns.heatmap(df_cm, cmap='Blues', annot=True, fmt='.2f', linewidths=.5).set_title('Train', fontsize=30)
plt.show()
```
# Visualize embeddings outputs
```
y_embeddings = np.concatenate(oof_embed)
visualize_embeddings(y_embeddings, y_true)
```
# Visualize predictions
```
# train_dataset = get_dataset(TRAINING_FILENAMES, ordered=True)
# x_samp, y_samp = dataset_to_numpy_util(train_dataset, 18)
# y_samp = np.argmax(y_samp, axis=-1)
# x_samp_1, y_samp_1 = x_samp[:9,:,:,:], y_samp[:9]
# samp_preds_1 = model.predict(x_samp_1, batch_size=9)
# display_9_images_with_predictions(x_samp_1, samp_preds_1, y_samp_1)
# x_samp_2, y_samp_2 = x_samp[9:,:,:,:], y_samp[9:]
# samp_preds_2 = model.predict(x_samp_2, batch_size=9)
# display_9_images_with_predictions(x_samp_2, samp_preds_2, y_samp_2)
```
|
github_jupyter
|
!pip install --quiet efficientnet
import warnings, time
from kaggle_datasets import KaggleDatasets
from sklearn.model_selection import KFold
from sklearn.metrics import classification_report, confusion_matrix, accuracy_score
from tensorflow.keras import optimizers, Sequential, losses, metrics, Model
from tensorflow.keras.callbacks import EarlyStopping, ModelCheckpoint
import efficientnet.tfkeras as efn
from cassava_scripts import *
from scripts_step_lr_schedulers import *
import tensorflow_addons as tfa
seed = 0
seed_everything(seed)
warnings.filterwarnings('ignore')
# TPU or GPU detection
# Detect hardware, return appropriate distribution strategy
strategy, tpu = set_up_strategy()
AUTO = tf.data.experimental.AUTOTUNE
REPLICAS = strategy.num_replicas_in_sync
print(f'REPLICAS: {REPLICAS}')
# Mixed precision
from tensorflow.keras.mixed_precision import experimental as mixed_precision
policy = mixed_precision.Policy('mixed_bfloat16')
mixed_precision.set_policy(policy)
# XLA
tf.config.optimizer.set_jit(True)
BATCH_SIZE = 8 * REPLICAS
LEARNING_RATE = 1e-5 * REPLICAS
EPOCHS_CL = 5
EPOCHS = 33
HEIGHT = 512
WIDTH = 512
HEIGHT_DT = 512
WIDTH_DT = 512
CHANNELS = 3
N_CLASSES = 5
N_FOLDS = 5
FOLDS_USED = 1
ES_PATIENCE = 10
database_base_path = '/kaggle/input/cassava-leaf-disease-classification/'
train = pd.read_csv(f'{database_base_path}train.csv')
print(f'Train samples: {len(train)}')
GCS_PATH = KaggleDatasets().get_gcs_path(f'cassava-leaf-disease-tfrecords-center-{HEIGHT_DT}x{WIDTH_DT}') # Center croped and resized (50 TFRecord)
# GCS_PATH_EXT = KaggleDatasets().get_gcs_path(f'cassava-leaf-disease-tfrecords-external-{HEIGHT_DT}x{WIDTH_DT}') # Center croped and resized (50 TFRecord) (External)
# GCS_PATH_CLASSES = KaggleDatasets().get_gcs_path(f'cassava-leaf-disease-tfrecords-classes-{HEIGHT_DT}x{WIDTH_DT}') # Center croped and resized (50 TFRecord) by classes
# GCS_PATH_EXT_CLASSES = KaggleDatasets().get_gcs_path(f'cassava-leaf-disease-tfrecords-classes-ext-{HEIGHT_DT}x{WIDTH_DT}') # Center croped and resized (50 TFRecord) (External) by classes
FILENAMES_COMP = tf.io.gfile.glob(GCS_PATH + '/*.tfrec')
# FILENAMES_2019 = tf.io.gfile.glob(GCS_PATH_EXT + '/*.tfrec')
# FILENAMES_COMP_CBB = tf.io.gfile.glob(GCS_PATH_CLASSES + '/CBB*.tfrec')
# FILENAMES_COMP_CBSD = tf.io.gfile.glob(GCS_PATH_CLASSES + '/CBSD*.tfrec')
# FILENAMES_COMP_CGM = tf.io.gfile.glob(GCS_PATH_CLASSES + '/CGM*.tfrec')
# FILENAMES_COMP_CMD = tf.io.gfile.glob(GCS_PATH_CLASSES + '/CMD*.tfrec')
# FILENAMES_COMP_Healthy = tf.io.gfile.glob(GCS_PATH_CLASSES + '/Healthy*.tfrec')
# FILENAMES_2019_CBB = tf.io.gfile.glob(GCS_PATH_EXT_CLASSES + '/CBB*.tfrec')
# FILENAMES_2019_CBSD = tf.io.gfile.glob(GCS_PATH_EXT_CLASSES + '/CBSD*.tfrec')
# FILENAMES_2019_CGM = tf.io.gfile.glob(GCS_PATH_EXT_CLASSES + '/CGM*.tfrec')
# FILENAMES_2019_CMD = tf.io.gfile.glob(GCS_PATH_EXT_CLASSES + '/CMD*.tfrec')
# FILENAMES_2019_Healthy = tf.io.gfile.glob(GCS_PATH_EXT_CLASSES + '/Healthy*.tfrec')
TRAINING_FILENAMES = FILENAMES_COMP
NUM_TRAINING_IMAGES = count_data_items(TRAINING_FILENAMES)
print(f'GCS: train images: {NUM_TRAINING_IMAGES}')
display(train.head())
def data_augment(image, label):
# p_rotation = tf.random.uniform([], 0, 1.0, dtype=tf.float32)
p_spatial = tf.random.uniform([], 0, 1.0, dtype=tf.float32)
p_rotate = tf.random.uniform([], 0, 1.0, dtype=tf.float32)
p_pixel_1 = tf.random.uniform([], 0, 1.0, dtype=tf.float32)
p_pixel_2 = tf.random.uniform([], 0, 1.0, dtype=tf.float32)
p_pixel_3 = tf.random.uniform([], 0, 1.0, dtype=tf.float32)
# p_shear = tf.random.uniform([], 0, 1.0, dtype=tf.float32)
p_crop = tf.random.uniform([], 0, 1.0, dtype=tf.float32)
p_cutout = tf.random.uniform([], 0, 1.0, dtype=tf.float32)
# # Shear
# if p_shear > .2:
# if p_shear > .6:
# image = transform_shear(image, HEIGHT, shear=20.)
# else:
# image = transform_shear(image, HEIGHT, shear=-20.)
# # Rotation
# if p_rotation > .2:
# if p_rotation > .6:
# image = transform_rotation(image, HEIGHT, rotation=45.)
# else:
# image = transform_rotation(image, HEIGHT, rotation=-45.)
# Flips
image = tf.image.random_flip_left_right(image)
image = tf.image.random_flip_up_down(image)
if p_spatial > .75:
image = tf.image.transpose(image)
# Rotates
if p_rotate > .75:
image = tf.image.rot90(image, k=3) # rotate 270º
elif p_rotate > .5:
image = tf.image.rot90(image, k=2) # rotate 180º
elif p_rotate > .25:
image = tf.image.rot90(image, k=1) # rotate 90º
# Pixel-level transforms
if p_pixel_1 >= .4:
image = tf.image.random_saturation(image, lower=.7, upper=1.3)
if p_pixel_2 >= .4:
image = tf.image.random_contrast(image, lower=.8, upper=1.2)
if p_pixel_3 >= .4:
image = tf.image.random_brightness(image, max_delta=.1)
# Crops
if p_crop > .6:
if p_crop > .9:
image = tf.image.central_crop(image, central_fraction=.5)
elif p_crop > .8:
image = tf.image.central_crop(image, central_fraction=.6)
elif p_crop > .7:
image = tf.image.central_crop(image, central_fraction=.7)
else:
image = tf.image.central_crop(image, central_fraction=.8)
elif p_crop > .3:
crop_size = tf.random.uniform([], int(HEIGHT*.6), HEIGHT, dtype=tf.int32)
image = tf.image.random_crop(image, size=[crop_size, crop_size, CHANNELS])
image = tf.image.resize(image, size=[HEIGHT, WIDTH])
if p_cutout > .5:
image = data_augment_cutout(image)
return image, label
# CutOut
def data_augment_cutout(image, min_mask_size=(int(HEIGHT * .1), int(HEIGHT * .1)),
max_mask_size=(int(HEIGHT * .125), int(HEIGHT * .125))):
p_cutout = tf.random.uniform([], 0, 1.0, dtype=tf.float32)
if p_cutout > .85: # 10~15 cut outs
n_cutout = tf.random.uniform([], 10, 15, dtype=tf.int32)
image = random_cutout(image, HEIGHT, WIDTH,
min_mask_size=min_mask_size, max_mask_size=max_mask_size, k=n_cutout)
elif p_cutout > .6: # 5~10 cut outs
n_cutout = tf.random.uniform([], 5, 10, dtype=tf.int32)
image = random_cutout(image, HEIGHT, WIDTH,
min_mask_size=min_mask_size, max_mask_size=max_mask_size, k=n_cutout)
elif p_cutout > .25: # 2~5 cut outs
n_cutout = tf.random.uniform([], 2, 5, dtype=tf.int32)
image = random_cutout(image, HEIGHT, WIDTH,
min_mask_size=min_mask_size, max_mask_size=max_mask_size, k=n_cutout)
else: # 1 cut out
image = random_cutout(image, HEIGHT, WIDTH,
min_mask_size=min_mask_size, max_mask_size=max_mask_size, k=1)
return image
# Datasets utility functions
def random_crop(image, label):
"""
Resize and reshape images to the expected size.
"""
image = tf.image.random_crop(image, size=[HEIGHT, WIDTH, CHANNELS])
return image, label
def prepare_image(image, label):
"""
Resize and reshape images to the expected size.
"""
image = tf.image.resize(image, [HEIGHT, WIDTH])
image = tf.reshape(image, [HEIGHT, WIDTH, CHANNELS])
return image, label
def center_crop_(image, label, height_rs, width_rs, height=HEIGHT_DT, width=WIDTH_DT, channels=3):
image = tf.reshape(image, [height, width, channels]) # Original shape
h, w = image.shape[0], image.shape[1]
if h > w:
image = tf.image.crop_to_bounding_box(image, (h - w) // 2, 0, w, w)
else:
image = tf.image.crop_to_bounding_box(image, 0, (w - h) // 2, h, h)
image = tf.image.resize(image, [height_rs, width_rs]) # Expected shape
return image, label
def read_tfrecord_(example, labeled=True, n_classes=5):
"""
1. Parse data based on the 'TFREC_FORMAT' map.
2. Decode image.
3. If 'labeled' returns (image, label) if not (image, name).
"""
if labeled:
TFREC_FORMAT = {
'image': tf.io.FixedLenFeature([], tf.string),
'target': tf.io.FixedLenFeature([], tf.int64),
}
else:
TFREC_FORMAT = {
'image': tf.io.FixedLenFeature([], tf.string),
'image_name': tf.io.FixedLenFeature([], tf.string),
}
example = tf.io.parse_single_example(example, TFREC_FORMAT)
image = decode_image(example['image'])
if labeled:
label_or_name = tf.cast(example['target'], tf.int32)
# One-Hot Encoding needed to use "categorical_crossentropy" loss
# label_or_name = tf.one_hot(tf.cast(label_or_name, tf.int32), n_classes)
else:
label_or_name = example['image_name']
return image, label_or_name
def get_dataset(filenames, labeled=True, ordered=False, repeated=False,
cached=False, augment=False):
"""
Return a Tensorflow dataset ready for training or inference.
"""
ignore_order = tf.data.Options()
if not ordered:
ignore_order.experimental_deterministic = False
dataset = tf.data.Dataset.list_files(filenames)
dataset = dataset.interleave(tf.data.TFRecordDataset, num_parallel_calls=AUTO)
else:
dataset = tf.data.TFRecordDataset(filenames, num_parallel_reads=AUTO)
dataset = dataset.with_options(ignore_order)
dataset = dataset.map(lambda x: read_tfrecord_(x, labeled=labeled), num_parallel_calls=AUTO)
# dataset = dataset.map(lambda x: read_tfrecord(x, labeled=labeled), num_parallel_calls=AUTO)
if augment:
dataset = dataset.map(data_augment, num_parallel_calls=AUTO)
dataset = dataset.map(scale_image, num_parallel_calls=AUTO)
dataset = dataset.map(prepare_image, num_parallel_calls=AUTO)
if labeled:
dataset = dataset.map(conf_output, num_parallel_calls=AUTO)
if not ordered:
dataset = dataset.shuffle(2048)
if repeated:
dataset = dataset.repeat()
dataset = dataset.batch(BATCH_SIZE)
if cached:
dataset = dataset.cache()
dataset = dataset.prefetch(AUTO)
return dataset
def conf_output(image, label):
"""
Configure the output of the dataset.
"""
aux_label = [0.]
aux_2_label = [0.]
# if tf.math.argmax(label, axis=-1) == 4: # Healthy
if label == 4: # Healthy
aux_label = [1.]
# if tf.math.argmax(label, axis=-1) == 3: # CMD
if label == 3: # CMD
aux_2_label = [1.]
return (image, (label, aux_label, aux_2_label))
# train_dataset = get_dataset(FILENAMES_COMP, ordered=True, augment=True)
# train_iter = iter(train_dataset.unbatch().batch(20))
# display_batch_of_images(next(train_iter))
# display_batch_of_images(next(train_iter))
def encoder_fn(input_shape):
inputs = L.Input(shape=input_shape, name='input_image')
base_model = efn.EfficientNetB4(input_tensor=inputs,
include_top=False,
weights='noisy-student',
pooling='avg')
model = Model(inputs=inputs, outputs=base_model.output)
return model
def add_projection_head(input_shape, encoder):
inputs = L.Input(shape=input_shape, name='input_image')
features = encoder(inputs)
outputs = L.Dense(128, activation='relu', name='projection_head', dtype='float32')(features)
model = Model(inputs=inputs, outputs=outputs)
return model
def classifier_fn(input_shape, N_CLASSES, encoder, trainable=True):
for layer in encoder.layers:
layer.trainable = trainable
inputs = L.Input(shape=input_shape, name='input_image')
features = encoder(inputs)
features = L.Dropout(.25)(features)
features = L.Dense(1000, activation='relu')(features)
features = L.Dropout(.25)(features)
output = L.Dense(N_CLASSES, activation='softmax', name='output', dtype='float32')(features)
output_healthy = L.Dense(1, activation='sigmoid', name='output_healthy', dtype='float32')(features)
output_cmd = L.Dense(1, activation='sigmoid', name='output_cmd', dtype='float32')(features)
model = Model(inputs=inputs, outputs=[output, output_healthy, output_cmd])
return model
temperature = 0.1
class SupervisedContrastiveLoss(losses.Loss):
def __init__(self, temperature=0.1, name=None):
super(SupervisedContrastiveLoss, self).__init__(name=name)
self.temperature = temperature
def __call__(self, labels, feature_vectors, sample_weight=None):
# Normalize feature vectors
feature_vectors_normalized = tf.math.l2_normalize(feature_vectors, axis=1)
# Compute logits
logits = tf.divide(
tf.matmul(
feature_vectors_normalized, tf.transpose(feature_vectors_normalized)
),
temperature,
)
return tfa.losses.npairs_loss(tf.squeeze(labels), logits)
lr_start = 1e-8
lr_min = 1e-6
lr_max = LEARNING_RATE
num_cycles = 3.
warmup_epochs = 3
hold_max_epochs = 0
total_epochs = EPOCHS
step_size = (NUM_TRAINING_IMAGES//BATCH_SIZE)
hold_max_steps = hold_max_epochs * step_size
total_steps = total_epochs * step_size
warmup_steps = warmup_epochs * step_size
def lrfn(total_steps, warmup_steps=0, lr_start=1e-4, lr_max=1e-3, lr_min=1e-4, num_cycles=1.):
@tf.function
def cosine_with_hard_restarts_schedule_with_warmup_(step):
""" Create a schedule with a learning rate that decreases following the
values of the cosine function with several hard restarts, after a warmup
period during which it increases linearly between 0 and 1.
"""
if step < warmup_steps:
lr = (lr_max - lr_start) / warmup_steps * step + lr_start
else:
progress = (step - warmup_steps) / (total_steps - warmup_steps)
lr = lr_max * (0.5 * (1.0 + tf.math.cos(np.pi * ((num_cycles * progress) % 1.0))))
if lr_min is not None:
lr = tf.math.maximum(lr_min, float(lr))
return lr
return cosine_with_hard_restarts_schedule_with_warmup_
lrfn_fn = lrfn(total_steps, warmup_steps, lr_start, lr_max, lr_min, num_cycles)
rng = [i for i in range(total_steps)]
y = [lrfn_fn(tf.cast(x, tf.float32)) for x in rng]
sns.set(style='whitegrid')
fig, ax = plt.subplots(figsize=(20, 6))
plt.plot(rng, y)
print(f'{total_steps} total steps and {step_size} steps per epoch')
print(f'Learning rate schedule: {y[0]:.3g} to {max(y):.3g} to {y[-1]:.3g}')
skf = KFold(n_splits=N_FOLDS, shuffle=True, random_state=seed)
oof_pred = []; oof_labels = []; oof_names = []; oof_folds = []; history_list = []; oof_embed = []
for fold,(idxT, idxV) in enumerate(skf.split(np.arange(15))):
if fold >= FOLDS_USED:
break
if tpu: tf.tpu.experimental.initialize_tpu_system(tpu)
K.clear_session()
print(f'\nFOLD: {fold+1}')
print(f'TRAIN: {idxT} VALID: {idxV}')
# Create train and validation sets
TRAIN_FILENAMES = tf.io.gfile.glob([GCS_PATH + '/Id_train%.2i*.tfrec' % x for x in idxT])
# FILENAMES_COMP_CBB = tf.io.gfile.glob([GCS_PATH_CLASSES + '/CBB%.2i*.tfrec' % x for x in idxT])
# FILENAMES_COMP_CBSD = tf.io.gfile.glob([GCS_PATH_CLASSES + '/CBSD%.2i*.tfrec' % x for x in idxT])
# FILENAMES_COMP_CGM = tf.io.gfile.glob([GCS_PATH_CLASSES + '/CGM%.2i*.tfrec' % x for x in idxT])
# FILENAMES_COMP_Healthy = tf.io.gfile.glob([GCS_PATH_CLASSES + '/Healthy%.2i*.tfrec' % x for x in idxT])
np.random.shuffle(TRAIN_FILENAMES)
VALID_FILENAMES = tf.io.gfile.glob([GCS_PATH + '/Id_train%.2i*.tfrec' % x for x in idxV])
ct_train = count_data_items(TRAIN_FILENAMES)
ct_valid = count_data_items(VALID_FILENAMES)
step_size = (ct_train // BATCH_SIZE)
warmup_steps = (warmup_epochs * step_size)
total_steps = (total_epochs * step_size)
total_steps_cl = (EPOCHS_CL * step_size)
warmup_steps_cl = 1
### Pre-train the encoder
print('Pre-training the encoder using "Supervised Contrastive" Loss')
with strategy.scope():
encoder = encoder_fn((None, None, CHANNELS))
encoder_proj = add_projection_head((None, None, CHANNELS), encoder)
encoder_proj.summary()
lrfn_fn = lrfn(total_steps, warmup_steps, lr_start, lr_max, lr_min, num_cycles)
# optimizer = optimizers.SGD(learning_rate=lambda: lrfn_fn(tf.cast(optimizer.iterations, tf.float32)),
# momentum=0.95, nesterov=True)
optimizer = optimizers.Adam(learning_rate=lambda: lrfn_fn(tf.cast(optimizer.iterations, tf.float32)))
encoder_proj.compile(optimizer=optimizer,
loss=SupervisedContrastiveLoss(temperature))
es = EarlyStopping(monitor='val_loss', mode='min',
patience=ES_PATIENCE, restore_best_weights=True, verbose=1)
history_enc = encoder_proj.fit(x=get_dataset(TRAIN_FILENAMES, repeated=True, augment=True),
validation_data=get_dataset(VALID_FILENAMES, ordered=True),
steps_per_epoch=step_size,
batch_size=BATCH_SIZE,
callbacks=[es],
epochs=EPOCHS,
verbose=2).history
### Train the classifier with the frozen encoder
print('Training the classifier with the frozen encoder')
with strategy.scope():
model = classifier_fn((None, None, CHANNELS), N_CLASSES, encoder, trainable=False)
model.summary()
lrfn_fn = lrfn(total_steps_cl, warmup_steps_cl, lr_start, lr_max, lr_min, num_cycles)
optimizer = optimizers.Adam(learning_rate=lambda: lrfn_fn(tf.cast(optimizer.iterations, tf.float32)))
model.compile(optimizer=optimizer,
loss={'output': losses.SparseCategoricalCrossentropy(),
'output_healthy': losses.BinaryCrossentropy(label_smoothing=.1),
'output_cmd': losses.BinaryCrossentropy(label_smoothing=.1)},
loss_weights={'output': 1.,
'output_healthy': .1,
'output_cmd': .1},
metrics={'output': metrics.SparseCategoricalAccuracy(),
'output_healthy': metrics.BinaryAccuracy(),
'output_cmd': metrics.BinaryAccuracy()})
model_path = f'model_{fold}.h5'
history = model.fit(x=get_dataset(TRAIN_FILENAMES, repeated=True, augment=True),
validation_data=get_dataset(VALID_FILENAMES, ordered=True),
steps_per_epoch=step_size,
epochs=EPOCHS_CL,
verbose=2).history
### RESULTS
print(f"#### FOLD {fold+1} OOF Accuracy = {np.max(history['val_output_sparse_categorical_accuracy']):.3f}")
history_list.append(history)
# Save last model weights
model.save_weights(model_path)
# Load best model weights
# model.load_weights(model_path)
# OOF predictions
ds_valid = get_dataset(VALID_FILENAMES, ordered=True)
oof_folds.append(np.full((ct_valid), fold, dtype='int8'))
oof_labels.append([target[0].numpy() for img, target in iter(ds_valid.unbatch())])
x_oof = ds_valid.map(lambda image, target: image)
oof_pred.append(np.argmax(model.predict(x_oof)[0], axis=-1))
# OOF names
ds_valid_names = get_dataset(VALID_FILENAMES, labeled=False, ordered=True)
oof_names.append(np.array([img_name.numpy().decode('utf-8') for img, img_name in iter(ds_valid_names.unbatch())]))
oof_embed.append(encoder.predict(x_oof)) # OOF embeddings
for fold, history in enumerate(history_list):
print(f'\nFOLD: {fold+1}')
plot_metrics(history, acc_name='output_sparse_categorical_accuracy')
y_true = np.concatenate(oof_labels)
# y_true = np.argmax(y_true, axis=-1)
y_pred = np.concatenate(oof_pred)
folds = np.concatenate(oof_folds)
names = np.concatenate(oof_names)
acc = accuracy_score(y_true, y_pred)
print(f'Overall OOF Accuracy = {acc:.3f}')
df_oof = pd.DataFrame({'image_id':names, 'fold':fold,
'target':y_true, 'pred':y_pred})
df_oof.to_csv('oof.csv', index=False)
display(df_oof.head())
print(classification_report(y_true, y_pred, target_names=CLASSES))
fig, ax = plt.subplots(1, 1, figsize=(20, 12))
cfn_matrix = confusion_matrix(y_true, y_pred, labels=range(len(CLASSES)))
cfn_matrix = (cfn_matrix.T / cfn_matrix.sum(axis=1)).T
df_cm = pd.DataFrame(cfn_matrix, index=CLASSES, columns=CLASSES)
ax = sns.heatmap(df_cm, cmap='Blues', annot=True, fmt='.2f', linewidths=.5).set_title('Train', fontsize=30)
plt.show()
y_embeddings = np.concatenate(oof_embed)
visualize_embeddings(y_embeddings, y_true)
# train_dataset = get_dataset(TRAINING_FILENAMES, ordered=True)
# x_samp, y_samp = dataset_to_numpy_util(train_dataset, 18)
# y_samp = np.argmax(y_samp, axis=-1)
# x_samp_1, y_samp_1 = x_samp[:9,:,:,:], y_samp[:9]
# samp_preds_1 = model.predict(x_samp_1, batch_size=9)
# display_9_images_with_predictions(x_samp_1, samp_preds_1, y_samp_1)
# x_samp_2, y_samp_2 = x_samp[9:,:,:,:], y_samp[9:]
# samp_preds_2 = model.predict(x_samp_2, batch_size=9)
# display_9_images_with_predictions(x_samp_2, samp_preds_2, y_samp_2)
| 0.460532 | 0.422803 |
<a href="https://colab.research.google.com/github/ujjwalk39/awesome-public-datasets/blob/master/Numpy_Assignment.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# NumPy Exercises
Now that we've learned about NumPy let's test your knowledge. We'll start off with a few simple tasks and then you'll be asked some more complicated questions.
<div class="alert alert-danger" style="margin: 10px"><strong>IMPORTANT NOTE!</strong> Make sure you don't run the cells directly above the example output shown, <br>otherwise you will end up writing over the example output!</div>
#### 1. Import NumPy as np
```
import numpy as np
```
#### 2. Create an array of 10 zeros
```
# CODE HERE
np.zeros(10)
# DON'T WRITE HERE
```
#### 3. Create an array of 10 ones
```
np.ones(10)
# DON'T WRITE HERE
```
#### 4. Create an array of 10 fives
```
np.ones(10)*5
# DON'T WRITE HERE
```
#### 5. Create an array of the integers from 10 to 50
```
np.arange(10,51)
# DON'T WRITE HERE
```
```
```
#### 6. Create an array of all the even integers from 10 to 50
* List item
* List item
```
np.arange(10,51,2)
# DON'T WRITE HERE
```
#### 7. Create a 3x3 matrix with values ranging from 0 to 8
1. List item
2. List item
```
arr = np.arange(0,9)
arr.reshape(3,3)
# DON'T WRITE HERE
```
#### 8. Create a 3x3 identity matrix
```
np.eye(3)
# DON'T WRITE HERE
```
#### 9. Use NumPy to generate a random number between 0 and 1<br><br> NOTE: Your result's value should be different from the one shown below.
```
np.random.rand(1)
# DON'T WRITE HERE
```
#### 10. Use NumPy to generate an array of 25 random numbers sampled from a standard normal distribution<br><br>  NOTE: Your result's values should be different from the ones shown below.
```
np.random.normal(0,1,25)
# DON'T WRITE HERE
```
#### 11. Create the following matrix:
```
arr = np.arange(0.01,1.01,.01)
arr.reshape(10,10)
# DON'T WRITE HERE
```
#### 12. Create an array of 20 linearly spaced points between 0 and 1:
```
np.linspace(0,1,20)
# DON'T WRITE HERE
```
## Numpy Indexing and Selection
Now you will be given a starting matrix (be sure to run the cell below!), and be asked to replicate the resulting matrix outputs:
```
# RUN THIS CELL - THIS IS OUR STARTING MATRIX
mat = np.arange(1,26).reshape(5,5)
mat
```
#### 13. Write code that reproduces the output shown below.<br><br>  Be careful not to run the cell immediately above the output, otherwise you won't be able to see the output any more.
```
# CODE HERE
mat[2:,1:]
# DON'T WRITE HERE
```
#### 14. Write code that reproduces the output shown below.
```
mat[3,4]
# DON'T WRITE HERE
```
#### 15. Write code that reproduces the output shown below.
```
mat[0:3,1].reshape(3,1)
# DON'T WRITE HERE
```
#### 16. Write code that reproduces the output shown below.
```
mat[4,0:5]
# DON'T WRITE HERE
```
#### 17. Write code that reproduces the output shown below.
```
mat[3:,0:5]
# DON'T WRITE HERE
```
## NumPy Operations
#### 18. Get the sum of all the values in mat
```
mat.sum()
# DON'T WRITE HERE
```
#### 19. Get the standard deviation of the values in mat
```
mat.std()
# DON'T WRITE HERE
```
#### 20. Get the sum of all the columns in mat
```
mat.sum(axis = 0)
# DON'T WRITE HERE
```
## Bonus Question
We worked a lot with random data with numpy, but is there a way we can insure that we always get the same random numbers? What does the seed value mean? Does it matter what the actual number is? [Click Here for a Hint](https://www.google.com/search?q=numpy+random+seed)
```
the way to ensure that we get the same random numbers on the same machines or even on the different machines we should use the concept of SEED(). when we use seed value like np.random.seed(42) everyone using the same seed value will get the same set of random numbers even on different machines. the seed number should remains same otherwise we would not be getting the same values.
```
# Great Job!
|
github_jupyter
|
import numpy as np
# CODE HERE
np.zeros(10)
# DON'T WRITE HERE
np.ones(10)
# DON'T WRITE HERE
np.ones(10)*5
# DON'T WRITE HERE
np.arange(10,51)
# DON'T WRITE HERE
```
#### 6. Create an array of all the even integers from 10 to 50
* List item
* List item
#### 7. Create a 3x3 matrix with values ranging from 0 to 8
1. List item
2. List item
#### 8. Create a 3x3 identity matrix
#### 9. Use NumPy to generate a random number between 0 and 1<br><br> NOTE: Your result's value should be different from the one shown below.
#### 10. Use NumPy to generate an array of 25 random numbers sampled from a standard normal distribution<br><br>  NOTE: Your result's values should be different from the ones shown below.
#### 11. Create the following matrix:
#### 12. Create an array of 20 linearly spaced points between 0 and 1:
## Numpy Indexing and Selection
Now you will be given a starting matrix (be sure to run the cell below!), and be asked to replicate the resulting matrix outputs:
#### 13. Write code that reproduces the output shown below.<br><br>  Be careful not to run the cell immediately above the output, otherwise you won't be able to see the output any more.
#### 14. Write code that reproduces the output shown below.
#### 15. Write code that reproduces the output shown below.
#### 16. Write code that reproduces the output shown below.
#### 17. Write code that reproduces the output shown below.
## NumPy Operations
#### 18. Get the sum of all the values in mat
#### 19. Get the standard deviation of the values in mat
#### 20. Get the sum of all the columns in mat
## Bonus Question
We worked a lot with random data with numpy, but is there a way we can insure that we always get the same random numbers? What does the seed value mean? Does it matter what the actual number is? [Click Here for a Hint](https://www.google.com/search?q=numpy+random+seed)
| 0.378229 | 0.988096 |
# <center>Using Agent Based Models in Physics Research</center>
<center>by Brian Nevins</center>
---
<img src="http://jasss.soc.surrey.ac.uk/13/2/8/Figure6b.jpg"><a href="http://jasss.soc.surrey.ac.uk/13/2/8.html">Image from the Journal of Artificial Societies and Social Simulation</a>
Agent based models are used in physics in some cases, primarily for introductory modeling and for biophysical models and social physics.
# Molecular Dynamics
My first-year undergraduate physics labs focused on a type of agent based modeling known in the physics community as molecular dynamics. Molecular dynamics involves creating a number of particle objects and following their motion individually as the system evolves. For example, one of our projects was to model Rutherford scattering, in which a charged particle approaches and scatters off a charged nucleus. The particle and the nucleus were separate objects with their own masses, positions, and velocities, which evolved based on simple interaction rules each particle enforced on itself. We used this code to verify the deflection angles predicted by Rutherford's model. There were several other small scale problems we looked at, mostly modeling collisions or simple interactions.
Some statistical mechanical models admit agent based modeling, the most well known of which is the Ising model. The Ising model consists of a set of spins on a lattice that interact with their nearest neighbors. It is fairly simple to implement this as an agent based model, and it can be used to demonstrate phase transitions and calculate relevant quantities. Many groups, including one at the Johannes Gutenberg University of Mainz, have implemented this type of model and calculated critical temperatures and exponents with it.
This type of simulation becomes untenable for more complex systems, like realistic solids or gases with large numbers of particles. Modeling every particle in a neutron star, for instance, is absurd because of the computational expense. It is conceivable that an agent based model could be used for modeling galactic dynamics, but even then the model is not sufficiently extensible given the estimated 200 billion galaxies in the universe. Nuclear physics could also admit agent based models, since particle numbers are generally small. The issue in nuclear physics is that the interactions between particles are too complex or not sufficiently understood to implement in particle agents. Agent based modeling is not very common in most fields of physics past introductory levels because of high particle numbers and complex or poorly understood interactions.
# Social Physics and Biophysics
Social physics is not really physics, but involves applying physics techniques to sociology, and some biophysicists focus their research on the behavior of groups of cells. The field of statistical mechanics is all about moving from vast numbers of single particle interactions to macroscopic quantities such as temperature and pressure. These same types of calculations can be applied to groups of people or cells, where emotions, beliefs, and actions (or relevant quantities on the cellular scale) replace quantities like charge, momentum, and interactions. Simplistic models, like Schelling's model for segregation, have been used in such a way to shed light on how macroscopic demographical phenomena can emerge without egregiously noticable causes. Flocking and crowding behavior has also been modeled in this way, and it is likely that statistical analysis could derive some useful quantities from the aggregate of the individual agent behaviors.
# Conclusion
Agent based modeling has found some niche uses in the field of physics. Introductory models of collisions, and simple small-N statistical mechanical simulations are one example of this, and social physics is another. For my particular subfield of nuclear astrophysics, the numbers of particles are simply too large and the interactions too complex for an agent based model to be applicable. Lattice simulations tracking macroscopic quantities or steady state solutions of those quantities are far more useful.
---
# References
Preis, Tobias, et al. “GPU Accelerated Monte Carlo Simulation of the 2D and 3D Ising Model.” Journal of Computational Physics, vol. 228, no. 12, July 2009, pp. 4468–77. DOI.org (Crossref), doi:10.1016/j.jcp.2009.03.018.
Quang, Le Anh, et al. “Agent-Based Models in Social Physics.” Journal of the Korean Physical Society, vol. 72, no. 11, June 2018, pp. 1272–80. arXiv.org, doi:10.3938/jkps.72.1272.
Quera, Vicenç, et al. “Flocking Behaviour: Agent-Based Simulation and Hierarchical Leadership.” Journal of Artificial Societies and Social Simulation, vol. 13, no. 2, 2009, p. 8.
-----
### Congratulations, you are done!
Now, you just need to commit and push this report to your project git repository.
|
github_jupyter
|
# <center>Using Agent Based Models in Physics Research</center>
<center>by Brian Nevins</center>
---
<img src="http://jasss.soc.surrey.ac.uk/13/2/8/Figure6b.jpg"><a href="http://jasss.soc.surrey.ac.uk/13/2/8.html">Image from the Journal of Artificial Societies and Social Simulation</a>
Agent based models are used in physics in some cases, primarily for introductory modeling and for biophysical models and social physics.
# Molecular Dynamics
My first-year undergraduate physics labs focused on a type of agent based modeling known in the physics community as molecular dynamics. Molecular dynamics involves creating a number of particle objects and following their motion individually as the system evolves. For example, one of our projects was to model Rutherford scattering, in which a charged particle approaches and scatters off a charged nucleus. The particle and the nucleus were separate objects with their own masses, positions, and velocities, which evolved based on simple interaction rules each particle enforced on itself. We used this code to verify the deflection angles predicted by Rutherford's model. There were several other small scale problems we looked at, mostly modeling collisions or simple interactions.
Some statistical mechanical models admit agent based modeling, the most well known of which is the Ising model. The Ising model consists of a set of spins on a lattice that interact with their nearest neighbors. It is fairly simple to implement this as an agent based model, and it can be used to demonstrate phase transitions and calculate relevant quantities. Many groups, including one at the Johannes Gutenberg University of Mainz, have implemented this type of model and calculated critical temperatures and exponents with it.
This type of simulation becomes untenable for more complex systems, like realistic solids or gases with large numbers of particles. Modeling every particle in a neutron star, for instance, is absurd because of the computational expense. It is conceivable that an agent based model could be used for modeling galactic dynamics, but even then the model is not sufficiently extensible given the estimated 200 billion galaxies in the universe. Nuclear physics could also admit agent based models, since particle numbers are generally small. The issue in nuclear physics is that the interactions between particles are too complex or not sufficiently understood to implement in particle agents. Agent based modeling is not very common in most fields of physics past introductory levels because of high particle numbers and complex or poorly understood interactions.
# Social Physics and Biophysics
Social physics is not really physics, but involves applying physics techniques to sociology, and some biophysicists focus their research on the behavior of groups of cells. The field of statistical mechanics is all about moving from vast numbers of single particle interactions to macroscopic quantities such as temperature and pressure. These same types of calculations can be applied to groups of people or cells, where emotions, beliefs, and actions (or relevant quantities on the cellular scale) replace quantities like charge, momentum, and interactions. Simplistic models, like Schelling's model for segregation, have been used in such a way to shed light on how macroscopic demographical phenomena can emerge without egregiously noticable causes. Flocking and crowding behavior has also been modeled in this way, and it is likely that statistical analysis could derive some useful quantities from the aggregate of the individual agent behaviors.
# Conclusion
Agent based modeling has found some niche uses in the field of physics. Introductory models of collisions, and simple small-N statistical mechanical simulations are one example of this, and social physics is another. For my particular subfield of nuclear astrophysics, the numbers of particles are simply too large and the interactions too complex for an agent based model to be applicable. Lattice simulations tracking macroscopic quantities or steady state solutions of those quantities are far more useful.
---
# References
Preis, Tobias, et al. “GPU Accelerated Monte Carlo Simulation of the 2D and 3D Ising Model.” Journal of Computational Physics, vol. 228, no. 12, July 2009, pp. 4468–77. DOI.org (Crossref), doi:10.1016/j.jcp.2009.03.018.
Quang, Le Anh, et al. “Agent-Based Models in Social Physics.” Journal of the Korean Physical Society, vol. 72, no. 11, June 2018, pp. 1272–80. arXiv.org, doi:10.3938/jkps.72.1272.
Quera, Vicenç, et al. “Flocking Behaviour: Agent-Based Simulation and Hierarchical Leadership.” Journal of Artificial Societies and Social Simulation, vol. 13, no. 2, 2009, p. 8.
-----
### Congratulations, you are done!
Now, you just need to commit and push this report to your project git repository.
| 0.807954 | 0.953579 |
# Application of trained ZS-AMBER to a new task
## Reload a trained ZS-AMBER
```
%cd /mnt/home/zzhang/workspace/src/AMBER-ZeroShot/examples
%run zero_shot_nas.real_deepsea.py
import sys
arg_str = """foo
--train-file data/zero_shot_deepsea/train.h5
--val-file data/zero_shot_deepsea/val.h5
--model-space long_and_dilation
--ppo
--wd outputs/new_20200919/test_feats/zs_amber/
--config-file data/zero_shot_deepsea/test_feats.representative_4.config_file.tsv
--dfeature-name-file data/zero_shot_deepsea/dfeatures_ordered_list.txt
"""
arg_list = arg_str.split()
sys.argv = arg_list
# fake arg parsing
parser = argparse.ArgumentParser(description="experimental zero-shot nas")
parser.add_argument("--train-file", type=str, required=True, help="Path to the hdf5 file of training data.")
parser.add_argument("--val-file", type=str, required=True, help="Path to the hdf5 file of validation data.")
parser.add_argument("--model-space", default="simple", choices=['simple', 'long', 'long_and_dilation'], help="Model space choice")
parser.add_argument("--ppo", default=False, action="store_true", help="Use PPO instead of REINFORCE")
parser.add_argument("--wd", type=str, default="./outputs/zero_shot/", help="working dir")
parser.add_argument("--resume", default=False, action="store_true", help="resume previous run")
parser.add_argument("--config-file", type=str, required=True, help="Path to the config file to use.")
parser.add_argument("--dfeature-name-file", type=str, required=True, help="Path to file with dataset feature names listed one per line.")
parser.add_argument("--lockstep-sampling", default=False, action="store_true", help="Ensure same training samples used for all models.")
arg = parser.parse_args()
print(arg)
configs, config_keys, controller, model_space = read_configs(arg, is_training=False)
#print(configs)
# get random 4 testing features
test_feats = [
'FEAT480', # "YY1" "H1-hESC"
'FEAT282', # "DNase" "WI-38"
'FEAT304', # "Pol2" "A549"
'FEAT144' # "H3k4me3" "NHLF"
]
configs = {k:configs[k] for k in configs if configs[k]['feat_name'] in test_feats}
print(configs.keys())
controller.load_weights("outputs/new_20200919/long_and_dilation.ppo.0/controller_weights.h5")
for k in configs:
print(configs[k]['feat_name'])
a, p = controller.get_action(np.expand_dims(configs[k]['dfeatures'],0))
print(a); print(p)
print('-'*10)
n_rep = 10
# full data is 4.4m with pos+neg strands
full_training_patience = 40
zs_res = {}
global_manager_trial_cnt = {k:0 for k in configs}
global_manager_record = pd.DataFrame(columns=['manager', 'feat_name', 'amber', 'step', 'arc', 'reward'])
for k in configs:
feat_name = configs[k]['feat_name']
print('-'*10); print(feat_name); print('-'*10)
res_list = []
manager = configs[k]['manager']
manager._earlystop_patience = full_training_patience
manager.verbose=0
for i in range(n_rep):
arc, prob = controller.get_action(np.expand_dims(configs[k]['dfeatures'],0))
reward, _ = manager.get_rewards(trial=global_manager_trial_cnt[k], model_arc=arc)
global_manager_trial_cnt[k] += 1
print(reward)
res_list.append(reward)
global_manager_record = global_manager_record.append({
'manager': k,
'feat_name': feat_name,
'amber': 'zs',
'step': i,
'arc': ','.join([str(a) for a in arc]),
'reward': reward
}, ignore_index=True)
zs_res[k] = res_list
print(zs_res)
import seaborn as sns
import matplotlib.pyplot as plt
for k in configs:
res_list = zs_res[k]
ax =sns.distplot(res_list)
ax.set_title("%s, avg=%.3f"%(configs[k]['feat_name'], np.mean(res_list)))
plt.show()
```
# Reload single-run AMBER and re-train for full epochs
```
from datetime import datetime
import os
par_wd = "/mnt/home/zzhang/workspace/src/AMBER-ZeroShot/examples/outputs/new_20200919/test_feats/single_run/"
sr_hist = {}
for k in configs:
feat_name = configs[k]['feat_name']
wd = "%s/%s/" % (par_wd, feat_name)
train_hist = pd.read_table(os.path.join(wd,"train_history.csv"), header=None, sep=",")
train_hist.head()
# each controller step has 5 child networks sampled..
train_hist['step'] = train_hist[0]//5
train_hist['auc'] = train_hist[2]
dt = []
for i in range(train_hist.shape[0]):
child_model_fp = os.path.join(wd, "weights", "trial_%i"%train_hist.iloc[i][0])
timestamp = os.path.getmtime(child_model_fp)
timestamp = datetime.fromtimestamp(timestamp)
if i==0:
starttime = timestamp
dt.append(timestamp - starttime)
train_hist['ctime'] = dt
train_hist = train_hist.loc[train_hist['step']<=60]
sr_hist[k] = train_hist
delta_time = [sr_hist[k].ctime.tail(1) for k in configs]
print(delta_time)
#sns.distplot(delta_time)
sampling_interval = 5
do_retrain = False
n_rep = 10
sr_res = {}
for k in configs:
np.random.seed(1223)
train_hist = sr_hist[k]
single_run_res = {}
step_time = {}
arc_seq_cols = np.arange(3, 13, dtype='int')
steps_to_retrain = np.arange(0, train_hist['step'].max(), sampling_interval)
#steps_to_retrain = np.concatenate([steps_to_retrain, [train_hist['step'].max()]])
print(steps_to_retrain)
for step in steps_to_retrain[1:]:
#arcs = train_hist.loc[train_hist['step'] == step]
step_idx = (train_hist['step']<=step) & (train_hist['step']>step-sampling_interval)
arcs = train_hist.loc[ step_idx ]
arcs = arcs[arc_seq_cols].to_numpy()
#orig_rewards = train_hist.loc[train_hist['step'] == step]['auc'].to_numpy()
orig_rewards = train_hist.loc[step_idx]['auc'].to_numpy()
orig_runtime = train_hist.loc[step_idx]['ctime'].to_numpy()
samp_idx = np.random.choice(arcs.shape[0], n_rep, replace=(n_rep>arcs.shape[0]))
arcs = arcs[samp_idx]
orig_rewards = orig_rewards[samp_idx]
orig_runtime = orig_runtime[samp_idx]
single_run_res[step] = []
step_time[step] = []
for i in range(n_rep):
if do_retrain:
reward, _ = manager.get_rewards(trial=global_manager_trial_cnt[k], model_arc=arcs.iloc[i])
print(reward)
else:
reward = orig_rewards[i]
global_manager_trial_cnt[k] += 1
single_run_res[step].append(reward)
step_time[step].append(orig_runtime[i])
global_manager_record = global_manager_record.append({
'manager': k,
'feat_name': feat_name,
'amber': 'sr',
'step': step,
'arc': ','.join([str(a) for a in arcs[i]]),
'reward': reward
}, ignore_index=True)
sr_df = pd.DataFrame([(k,v1, v2) for k in single_run_res for v1, v2 in zip(*[single_run_res[k], step_time[k]])], columns=['step', 'auc', 'time'])
sr_res[k] = sr_df
```
## Merge with single-run AMBER
```
from zs_config import read_metadata
meta = read_metadata()
def convert_timedelta(duration):
days, seconds = duration.days, duration.seconds
hours = days * 24 + seconds // 3600
minutes = (seconds % 3600) // 60
seconds = (seconds % 60)
return hours, minutes, seconds
title_mapper = {
"H1-hESC_YY1_None": "YY1 ChIP-seq from H1-hESC",
"WI-38_DNase_None": "DNase-seq from WI-38 cells",
"A549_Pol2_None": "RNA polymerase ChIP-seq from A549 cells",
"NHLF_H3K4me3_None": "H3K4me3 ChIP-seq from Normal Human Lung Fibroblasts"
}
for k in configs:
fig = plt.figure(figsize=(6,6))
ax = fig.add_subplot(111)
ax2 = ax.twiny()
feat_name = configs[k]['feat_name']
print(feat_name)
human_readable_name = meta.loc[feat_name]['new_name']
sr_df = sr_res[k]
zs_auc = zs_res[k]
ax.axhspan(np.mean(zs_auc)-2*np.std(zs_auc), np.mean(zs_auc)+2*np.std(zs_auc), alpha=0.2, color='pink')
ax.axhline(y=np.mean(zs_auc), color='red', label="AMBIENT")
sns.lineplot(x='step', y='auc', marker='o', ci='sd', label="AMBER", data=sr_df, ax=ax)
#ax.set_title("Manager%i:%s:%s"%(k,feat_name, human_readable_name))
ax.set_title("%s"%( title_mapper[human_readable_name]))
ax.set_xlabel("AMBER step")
ax.set_ylabel("Reward(AUC)")
#ax.set_ylim(0.5, 1)
# add run-time as second x-axis
ax2.set_xlim(ax.get_xlim())
sr_auc_mean = sr_df.groupby("step").mean()
new_tick_locations = [(sr_auc_mean-np.mean(zs_auc)>0).idxmax().tolist()[0], sr_df['step'].max()]
#print(new_tick_locations)
#ax2.set_xticks(new_tick_locations)
ax.set_xticks(new_tick_locations)
#ax2.set_xticklabels(sr_df.loc[new_tick_locations, "time"].apply(
# lambda x: "%shours"%convert_timedelta(x)[0]))
ax.set_xticklabels(sr_df.loc[new_tick_locations, "time"].apply(lambda x: "%shours"%convert_timedelta(x)[0]))
#ax2.set_xlabel("GPU runtime")
ax.set_xlabel("GPU runtime")
ax2.axvline(x=new_tick_locations[0], linestyle='--', color='green')
plt.savefig("./outputs/new_20200919/ZS_SR_comparison.%s.pdf"%human_readable_name)
plt.show()
global_manager_record.to_csv("outputs/new_20200919/ZS_SR_comparison.tsv", sep="\t")
```
|
github_jupyter
|
%cd /mnt/home/zzhang/workspace/src/AMBER-ZeroShot/examples
%run zero_shot_nas.real_deepsea.py
import sys
arg_str = """foo
--train-file data/zero_shot_deepsea/train.h5
--val-file data/zero_shot_deepsea/val.h5
--model-space long_and_dilation
--ppo
--wd outputs/new_20200919/test_feats/zs_amber/
--config-file data/zero_shot_deepsea/test_feats.representative_4.config_file.tsv
--dfeature-name-file data/zero_shot_deepsea/dfeatures_ordered_list.txt
"""
arg_list = arg_str.split()
sys.argv = arg_list
# fake arg parsing
parser = argparse.ArgumentParser(description="experimental zero-shot nas")
parser.add_argument("--train-file", type=str, required=True, help="Path to the hdf5 file of training data.")
parser.add_argument("--val-file", type=str, required=True, help="Path to the hdf5 file of validation data.")
parser.add_argument("--model-space", default="simple", choices=['simple', 'long', 'long_and_dilation'], help="Model space choice")
parser.add_argument("--ppo", default=False, action="store_true", help="Use PPO instead of REINFORCE")
parser.add_argument("--wd", type=str, default="./outputs/zero_shot/", help="working dir")
parser.add_argument("--resume", default=False, action="store_true", help="resume previous run")
parser.add_argument("--config-file", type=str, required=True, help="Path to the config file to use.")
parser.add_argument("--dfeature-name-file", type=str, required=True, help="Path to file with dataset feature names listed one per line.")
parser.add_argument("--lockstep-sampling", default=False, action="store_true", help="Ensure same training samples used for all models.")
arg = parser.parse_args()
print(arg)
configs, config_keys, controller, model_space = read_configs(arg, is_training=False)
#print(configs)
# get random 4 testing features
test_feats = [
'FEAT480', # "YY1" "H1-hESC"
'FEAT282', # "DNase" "WI-38"
'FEAT304', # "Pol2" "A549"
'FEAT144' # "H3k4me3" "NHLF"
]
configs = {k:configs[k] for k in configs if configs[k]['feat_name'] in test_feats}
print(configs.keys())
controller.load_weights("outputs/new_20200919/long_and_dilation.ppo.0/controller_weights.h5")
for k in configs:
print(configs[k]['feat_name'])
a, p = controller.get_action(np.expand_dims(configs[k]['dfeatures'],0))
print(a); print(p)
print('-'*10)
n_rep = 10
# full data is 4.4m with pos+neg strands
full_training_patience = 40
zs_res = {}
global_manager_trial_cnt = {k:0 for k in configs}
global_manager_record = pd.DataFrame(columns=['manager', 'feat_name', 'amber', 'step', 'arc', 'reward'])
for k in configs:
feat_name = configs[k]['feat_name']
print('-'*10); print(feat_name); print('-'*10)
res_list = []
manager = configs[k]['manager']
manager._earlystop_patience = full_training_patience
manager.verbose=0
for i in range(n_rep):
arc, prob = controller.get_action(np.expand_dims(configs[k]['dfeatures'],0))
reward, _ = manager.get_rewards(trial=global_manager_trial_cnt[k], model_arc=arc)
global_manager_trial_cnt[k] += 1
print(reward)
res_list.append(reward)
global_manager_record = global_manager_record.append({
'manager': k,
'feat_name': feat_name,
'amber': 'zs',
'step': i,
'arc': ','.join([str(a) for a in arc]),
'reward': reward
}, ignore_index=True)
zs_res[k] = res_list
print(zs_res)
import seaborn as sns
import matplotlib.pyplot as plt
for k in configs:
res_list = zs_res[k]
ax =sns.distplot(res_list)
ax.set_title("%s, avg=%.3f"%(configs[k]['feat_name'], np.mean(res_list)))
plt.show()
from datetime import datetime
import os
par_wd = "/mnt/home/zzhang/workspace/src/AMBER-ZeroShot/examples/outputs/new_20200919/test_feats/single_run/"
sr_hist = {}
for k in configs:
feat_name = configs[k]['feat_name']
wd = "%s/%s/" % (par_wd, feat_name)
train_hist = pd.read_table(os.path.join(wd,"train_history.csv"), header=None, sep=",")
train_hist.head()
# each controller step has 5 child networks sampled..
train_hist['step'] = train_hist[0]//5
train_hist['auc'] = train_hist[2]
dt = []
for i in range(train_hist.shape[0]):
child_model_fp = os.path.join(wd, "weights", "trial_%i"%train_hist.iloc[i][0])
timestamp = os.path.getmtime(child_model_fp)
timestamp = datetime.fromtimestamp(timestamp)
if i==0:
starttime = timestamp
dt.append(timestamp - starttime)
train_hist['ctime'] = dt
train_hist = train_hist.loc[train_hist['step']<=60]
sr_hist[k] = train_hist
delta_time = [sr_hist[k].ctime.tail(1) for k in configs]
print(delta_time)
#sns.distplot(delta_time)
sampling_interval = 5
do_retrain = False
n_rep = 10
sr_res = {}
for k in configs:
np.random.seed(1223)
train_hist = sr_hist[k]
single_run_res = {}
step_time = {}
arc_seq_cols = np.arange(3, 13, dtype='int')
steps_to_retrain = np.arange(0, train_hist['step'].max(), sampling_interval)
#steps_to_retrain = np.concatenate([steps_to_retrain, [train_hist['step'].max()]])
print(steps_to_retrain)
for step in steps_to_retrain[1:]:
#arcs = train_hist.loc[train_hist['step'] == step]
step_idx = (train_hist['step']<=step) & (train_hist['step']>step-sampling_interval)
arcs = train_hist.loc[ step_idx ]
arcs = arcs[arc_seq_cols].to_numpy()
#orig_rewards = train_hist.loc[train_hist['step'] == step]['auc'].to_numpy()
orig_rewards = train_hist.loc[step_idx]['auc'].to_numpy()
orig_runtime = train_hist.loc[step_idx]['ctime'].to_numpy()
samp_idx = np.random.choice(arcs.shape[0], n_rep, replace=(n_rep>arcs.shape[0]))
arcs = arcs[samp_idx]
orig_rewards = orig_rewards[samp_idx]
orig_runtime = orig_runtime[samp_idx]
single_run_res[step] = []
step_time[step] = []
for i in range(n_rep):
if do_retrain:
reward, _ = manager.get_rewards(trial=global_manager_trial_cnt[k], model_arc=arcs.iloc[i])
print(reward)
else:
reward = orig_rewards[i]
global_manager_trial_cnt[k] += 1
single_run_res[step].append(reward)
step_time[step].append(orig_runtime[i])
global_manager_record = global_manager_record.append({
'manager': k,
'feat_name': feat_name,
'amber': 'sr',
'step': step,
'arc': ','.join([str(a) for a in arcs[i]]),
'reward': reward
}, ignore_index=True)
sr_df = pd.DataFrame([(k,v1, v2) for k in single_run_res for v1, v2 in zip(*[single_run_res[k], step_time[k]])], columns=['step', 'auc', 'time'])
sr_res[k] = sr_df
from zs_config import read_metadata
meta = read_metadata()
def convert_timedelta(duration):
days, seconds = duration.days, duration.seconds
hours = days * 24 + seconds // 3600
minutes = (seconds % 3600) // 60
seconds = (seconds % 60)
return hours, minutes, seconds
title_mapper = {
"H1-hESC_YY1_None": "YY1 ChIP-seq from H1-hESC",
"WI-38_DNase_None": "DNase-seq from WI-38 cells",
"A549_Pol2_None": "RNA polymerase ChIP-seq from A549 cells",
"NHLF_H3K4me3_None": "H3K4me3 ChIP-seq from Normal Human Lung Fibroblasts"
}
for k in configs:
fig = plt.figure(figsize=(6,6))
ax = fig.add_subplot(111)
ax2 = ax.twiny()
feat_name = configs[k]['feat_name']
print(feat_name)
human_readable_name = meta.loc[feat_name]['new_name']
sr_df = sr_res[k]
zs_auc = zs_res[k]
ax.axhspan(np.mean(zs_auc)-2*np.std(zs_auc), np.mean(zs_auc)+2*np.std(zs_auc), alpha=0.2, color='pink')
ax.axhline(y=np.mean(zs_auc), color='red', label="AMBIENT")
sns.lineplot(x='step', y='auc', marker='o', ci='sd', label="AMBER", data=sr_df, ax=ax)
#ax.set_title("Manager%i:%s:%s"%(k,feat_name, human_readable_name))
ax.set_title("%s"%( title_mapper[human_readable_name]))
ax.set_xlabel("AMBER step")
ax.set_ylabel("Reward(AUC)")
#ax.set_ylim(0.5, 1)
# add run-time as second x-axis
ax2.set_xlim(ax.get_xlim())
sr_auc_mean = sr_df.groupby("step").mean()
new_tick_locations = [(sr_auc_mean-np.mean(zs_auc)>0).idxmax().tolist()[0], sr_df['step'].max()]
#print(new_tick_locations)
#ax2.set_xticks(new_tick_locations)
ax.set_xticks(new_tick_locations)
#ax2.set_xticklabels(sr_df.loc[new_tick_locations, "time"].apply(
# lambda x: "%shours"%convert_timedelta(x)[0]))
ax.set_xticklabels(sr_df.loc[new_tick_locations, "time"].apply(lambda x: "%shours"%convert_timedelta(x)[0]))
#ax2.set_xlabel("GPU runtime")
ax.set_xlabel("GPU runtime")
ax2.axvline(x=new_tick_locations[0], linestyle='--', color='green')
plt.savefig("./outputs/new_20200919/ZS_SR_comparison.%s.pdf"%human_readable_name)
plt.show()
global_manager_record.to_csv("outputs/new_20200919/ZS_SR_comparison.tsv", sep="\t")
| 0.208824 | 0.702603 |
<font size="+5">#03 | Model Selection. Decision Tree vs Support Vector Machines vs Logistic Regression</font>
- Subscribe to my [Blog ↗](https://blog.pythonassembly.com/)
- Let's keep in touch on [LinkedIn ↗](www.linkedin.com/in/jsulopz) 😄
# Discipline to Search Solutions in Google
> Apply the following steps when **looking for solutions in Google**:
>
> 1. **Necesity**: How to load an Excel in Python?
> 2. **Search in Google**: by keywords
> - `load excel python`
> - ~~how to load excel in python~~
> 3. **Solution**: What's the `function()` that loads an Excel in Python?
> - A Function to Programming is what the Atom to Phisics.
> - Every time you want to do something in programming
> - **You will need a `function()`** to make it
> - Theferore, you must **detect parenthesis `()`**
> - Out of all the words that you see in a website
> - Because they indicate the presence of a `function()`.
# Load the Data
> - The goal of this dataset is
> - To predict if **bank's customers** (rows) could have the approval for a credit card `target`
> - Based on their **socio-demographical characteristics** (columns)
```
import pandas as pd
df = pd.read_csv('https://archive.ics.uci.edu/ml/machine-learning-databases/credit-screening/crx.data',
na_values='?', header=None)
df.rename(columns={15: 'target'}, inplace=True)
df.head()
```
# Build & Compare Models
## `DecisionTreeClassifier()`
```
%%HTML
<iframe width="560" height="315" src="https://www.youtube.com/embed/7VeUPuFGJHk" title="YouTube video player" frameborder="0" allow="accelerometer; autoplay; clipboard-write; encrypted-media; gyroscope; picture-in-picture" allowfullscreen></iframe>
```
## `RandomForestClassifier()`
```
%%HTML
<iframe width="560" height="315" src="https://www.youtube.com/embed/J4Wdy0Wc_xQ" title="YouTube video player" frameborder="0" allow="accelerometer; autoplay; clipboard-write; encrypted-media; gyroscope; picture-in-picture" allowfullscreen></iframe>
```
## `KNeighborsClassifier()`
```
%%HTML
<iframe width="560" height="315" src="https://www.youtube.com/embed/HVXime0nQeI" title="YouTube video player" frameborder="0" allow="accelerometer; autoplay; clipboard-write; encrypted-media; gyroscope; picture-in-picture" allowfullscreen></iframe>
```
## Whic Model is the Best?
> Which model has the **highest accuracy**?
# `train_test_split()` & Compare Again
## `DecisionTreeClassifier()`
## `RandomForestClassifier()`
## `KNeighborsClassifier()`
# Which is the Best Model with `train_test_split()`?
> Which model has the **highest accuracy**?
# Achieved Goals
_Double click on **this cell** and place an `X` inside the square brackets (i.e., [X]) if you think you understand the goal:_
- [ ] Understand the necessity to **create functions** to avoid the repetition of the code.
- [ ] **Bootstrapping** as a way to create an artificial dataset that helps to reduce the bias.
- [ ] **Classification threshold** to predict categories out of probabilities.
- [ ] Different ways to **compare classification models**.
- [ ] Understand the importance to check how good is a model with **data not seen during training**.
|
github_jupyter
|
import pandas as pd
df = pd.read_csv('https://archive.ics.uci.edu/ml/machine-learning-databases/credit-screening/crx.data',
na_values='?', header=None)
df.rename(columns={15: 'target'}, inplace=True)
df.head()
%%HTML
<iframe width="560" height="315" src="https://www.youtube.com/embed/7VeUPuFGJHk" title="YouTube video player" frameborder="0" allow="accelerometer; autoplay; clipboard-write; encrypted-media; gyroscope; picture-in-picture" allowfullscreen></iframe>
%%HTML
<iframe width="560" height="315" src="https://www.youtube.com/embed/J4Wdy0Wc_xQ" title="YouTube video player" frameborder="0" allow="accelerometer; autoplay; clipboard-write; encrypted-media; gyroscope; picture-in-picture" allowfullscreen></iframe>
%%HTML
<iframe width="560" height="315" src="https://www.youtube.com/embed/HVXime0nQeI" title="YouTube video player" frameborder="0" allow="accelerometer; autoplay; clipboard-write; encrypted-media; gyroscope; picture-in-picture" allowfullscreen></iframe>
| 0.522933 | 0.967101 |
# Investigation of different modules
## Import statements
```
import pandas as pd
import numpy as np
import datetime
import os
import re
import matplotlib.pyplot as plt
```
## Loading data and folder settings
```
folder_data = '/Users/hkromer/01_Projects/10.SolarAnlage/01.Analytics/Moduldaten/'
# output dataframe
# current time for the year month day
currentDT = datetime.datetime.now()
directory = f'{folder_data}/cleaned_data'
if not os.path.exists(directory):
os.makedirs(directory)
filename_df_out = f'{directory}/{currentDT.year}-{currentDT.month}-{currentDT.day}_solarData.csv'
```
## Import data and combine into one dataframe
```
# list of files
files = os.listdir(folder_data)
files = [f for f in files if f.endswith('.csv')]
df = pd.DataFrame()
for file in files:
df_t = pd.read_csv('{}/{}'.format(folder_data,file))
df = df.append(df_t)
# change columns to only contain identifier
cols = df.columns
id_cols = [re.findall(r'(1.1.\d+) E',c)[0] for c in cols[1:]]
my_cols = [cols[0]]
[my_cols.append(i) for i in id_cols]
df.columns = my_cols
print(df.head())
df.to_csv(f'{filename_df_out}')
```
## Change the index to time series
## Convert from Wh to kWh
```
# Prepare a format string: time_format
time_format = '%d.%m.%Y'
# Convert date_list into a datetime object: my_datetimes
df['Time'] = pd.to_datetime(df['Time'], format=time_format)
df = df.set_index('Time').sort_index()
# convert from Wh to kWh
df[df.columns] = df[df.columns] / 1000
print(df.info())
print(df.head())
```
## Relate modules to position - Auslegung
from monitoring
```
module_position = {'Südwesten': [f'1.1.{s}' for s in [1, 16, 3,4,5,10,9,8,7,6,11,12,13,14]],
'Westen': [f'1.1.{s}' for s in [15,2,18,17,19,20]],
'Südost_oben': [f'1.1.{s}' for s in [28,27,26,25,24,23,22,21,29,30]],
'Südost_unten': [f'1.1.{s}' for s in [31,32,33,34,35,36]]}
print(df.loc[:,module_position['Südwesten']].head())
```
# Plot Monthly aggregated energy per module position
- First resample to monthly data, aggregate by sum.
- Change the datetimeindex format to the long name for the month.
- Create a new dataframe with the aggregated data
- Plot stacked bar
```
# First resample to monthly data, aggregate by sum.
df_monthly = df.resample('M').sum()
# Change the datetimeindex format to the long name for the month.
df_monthly = df_monthly.set_index(df_monthly.index.strftime('%y %B'))
# print(df_monthly.loc[:, module_position['Südwesten']].sum(axis='columns'))
# print(df_monthly.head())
# Create a new dataframe with the aggregated data
df_monthly_agg = pd.DataFrame()
for pos in module_position:
g = df_monthly.loc[:, module_position[pos]].sum(axis='columns')
df_monthly_agg[pos] = g
print(df_monthly_agg.head())
df_monthly_agg.plot(kind='bar', stacked=True)
_ = plt.title('Total Energie Pro Monat und Modulposition')
_ = plt.ylabel('Energie [kWh]')
_ = plt.grid()
_ = plt.tight_layout()
# output figure
filename_fig = f'{directory}/{currentDT.year}-{currentDT.month}-{currentDT.day}_solarData.monthly'
plt.savefig(filename_fig + '.png', dpi=600)
plt.show()
```
# Plot Monthly aggregated RELATIVE energy per module position
- Show the values relative to the total monthly
- Plot stacked bar
```
# Create a new dataframe with the relative aggregated data
print(df_monthly_agg.head())
# divide by number of modules
df_monthly_agg_norm = pd.DataFrame()
for pos in module_position:
g = df_monthly_agg.loc[:, pos].divide(len(module_position[pos]))
df_monthly_agg_norm[pos] = g
df_monthly_agg_norm['row_sum'] = df_monthly_agg_norm.sum(axis='columns')
# relative monthly
df_monthly_agg_norm = df_monthly_agg_norm.divide(df_monthly_agg_norm['row_sum'], axis='rows') * 100
df_monthly_agg_norm = df_monthly_agg_norm.drop(columns=['row_sum'])
print(df_monthly_agg.head())
df_monthly_agg_norm.plot(kind='bar', stacked=True)
_ = plt.title('Total Energie Pro Monat und Modulposition')
_ = plt.ylabel('Prozent Energie mtl normiert auf module')
_ = plt.grid()
_ = plt.tight_layout()
# output figure
filename_fig = f'{directory}/{currentDT.year}-{currentDT.month}-{currentDT.day}_solarData.monthly.relative'
plt.savefig(filename_fig + '.png', dpi=600)
plt.show()
```
|
github_jupyter
|
import pandas as pd
import numpy as np
import datetime
import os
import re
import matplotlib.pyplot as plt
folder_data = '/Users/hkromer/01_Projects/10.SolarAnlage/01.Analytics/Moduldaten/'
# output dataframe
# current time for the year month day
currentDT = datetime.datetime.now()
directory = f'{folder_data}/cleaned_data'
if not os.path.exists(directory):
os.makedirs(directory)
filename_df_out = f'{directory}/{currentDT.year}-{currentDT.month}-{currentDT.day}_solarData.csv'
# list of files
files = os.listdir(folder_data)
files = [f for f in files if f.endswith('.csv')]
df = pd.DataFrame()
for file in files:
df_t = pd.read_csv('{}/{}'.format(folder_data,file))
df = df.append(df_t)
# change columns to only contain identifier
cols = df.columns
id_cols = [re.findall(r'(1.1.\d+) E',c)[0] for c in cols[1:]]
my_cols = [cols[0]]
[my_cols.append(i) for i in id_cols]
df.columns = my_cols
print(df.head())
df.to_csv(f'{filename_df_out}')
# Prepare a format string: time_format
time_format = '%d.%m.%Y'
# Convert date_list into a datetime object: my_datetimes
df['Time'] = pd.to_datetime(df['Time'], format=time_format)
df = df.set_index('Time').sort_index()
# convert from Wh to kWh
df[df.columns] = df[df.columns] / 1000
print(df.info())
print(df.head())
module_position = {'Südwesten': [f'1.1.{s}' for s in [1, 16, 3,4,5,10,9,8,7,6,11,12,13,14]],
'Westen': [f'1.1.{s}' for s in [15,2,18,17,19,20]],
'Südost_oben': [f'1.1.{s}' for s in [28,27,26,25,24,23,22,21,29,30]],
'Südost_unten': [f'1.1.{s}' for s in [31,32,33,34,35,36]]}
print(df.loc[:,module_position['Südwesten']].head())
# First resample to monthly data, aggregate by sum.
df_monthly = df.resample('M').sum()
# Change the datetimeindex format to the long name for the month.
df_monthly = df_monthly.set_index(df_monthly.index.strftime('%y %B'))
# print(df_monthly.loc[:, module_position['Südwesten']].sum(axis='columns'))
# print(df_monthly.head())
# Create a new dataframe with the aggregated data
df_monthly_agg = pd.DataFrame()
for pos in module_position:
g = df_monthly.loc[:, module_position[pos]].sum(axis='columns')
df_monthly_agg[pos] = g
print(df_monthly_agg.head())
df_monthly_agg.plot(kind='bar', stacked=True)
_ = plt.title('Total Energie Pro Monat und Modulposition')
_ = plt.ylabel('Energie [kWh]')
_ = plt.grid()
_ = plt.tight_layout()
# output figure
filename_fig = f'{directory}/{currentDT.year}-{currentDT.month}-{currentDT.day}_solarData.monthly'
plt.savefig(filename_fig + '.png', dpi=600)
plt.show()
# Create a new dataframe with the relative aggregated data
print(df_monthly_agg.head())
# divide by number of modules
df_monthly_agg_norm = pd.DataFrame()
for pos in module_position:
g = df_monthly_agg.loc[:, pos].divide(len(module_position[pos]))
df_monthly_agg_norm[pos] = g
df_monthly_agg_norm['row_sum'] = df_monthly_agg_norm.sum(axis='columns')
# relative monthly
df_monthly_agg_norm = df_monthly_agg_norm.divide(df_monthly_agg_norm['row_sum'], axis='rows') * 100
df_monthly_agg_norm = df_monthly_agg_norm.drop(columns=['row_sum'])
print(df_monthly_agg.head())
df_monthly_agg_norm.plot(kind='bar', stacked=True)
_ = plt.title('Total Energie Pro Monat und Modulposition')
_ = plt.ylabel('Prozent Energie mtl normiert auf module')
_ = plt.grid()
_ = plt.tight_layout()
# output figure
filename_fig = f'{directory}/{currentDT.year}-{currentDT.month}-{currentDT.day}_solarData.monthly.relative'
plt.savefig(filename_fig + '.png', dpi=600)
plt.show()
| 0.180107 | 0.782164 |
<img width="10%" alt="Naas" src="https://landen.imgix.net/jtci2pxwjczr/assets/5ice39g4.png?w=160"/>
# Pandas - Merge Dataframes
<a href="https://app.naas.ai/user-redirect/naas/downloader?url=https://raw.githubusercontent.com/jupyter-naas/awesome-notebooks/master/Pandas/Pandas_Merge_Dataframes.ipynb" target="_parent"><img src="https://naasai-public.s3.eu-west-3.amazonaws.com/open_in_naas.svg"/></a>
**Tags:** #pandas #python #merging #merge #dataframes #consolidate #operations #snippet #dataframe
**Author:** [Oketunji Oludolapo](https://www.linkedin.com/in/oludolapo-oketunji/)
This notebook will help you understand how to use the pandas merge function. It explains how to merge two datasets together and consolidate multiple dataset into one.
## Input
### Import Library
```
import pandas as pd
import numpy as np
```
### Create dataframes to be merged
#### Dataframe 1
```
# Creating values to be used as datasets
dict1 = {
"student_id": [1,2,3,4,5,6,7,8,9,10],
"student_name": ["Peter","Dolly","Maggie","David","Isabelle","Harry","Akin","Abbey","Victoria","Sam"],
"student_course": np.random.choice(["Biology","Physics","Chemistry"], size=10)
}
# Create dataframe
df_1 = pd.DataFrame(dict1)
df_1
```
#### Dataframe 2
```
# Creating values to be used as datasets
dict2 = {
"student_id": np.random.choice([1,2,3,4,5,6,7,8,9,10], size=100),
"student_grade": np.random.choice(["A","B","C","D","E","F"], size=100),
"professors": np.random.choice(["Mark Levinson","Angela Marge","Bonnie James","Klaus Michealson"], size=100),
}
# Create dataframe
df_2 = pd.DataFrame(dict2) # OR Data2=pd.read_csv(filepath)
df_2
```
## Model
pd.merge: acts like an SQL inner join and joins based on similar columns or index unless specified to join differently<br />
### Merging dataframes with same values with same column names
Using pd.merge(left, right) acts like sql inner join and only joins on the common column they have.<br>
It tries finding everything from the right and append to left 'student_id' is common to both so it has been merged into one and included all the other df_2 columns to df_1 table.<br>
```
df = pd.merge(df_1, df_2)
```
## Output
### Display result
```
df
```
## Other options
### Specifiying the comon column using parameters "on"
```
df = pd.merge(df_1, df_2, on="student_id")
df
```
### Specifying what kind of Joins you want since merging does inner joins by default
- "inner" > Inner Join: INCLUDING ROWS OF FIRST AND SECOND ONLY IF THE VALUE IS THE SAME IN BOTH DATAFRAMES<br />
- "outer" > Outer Join: IT JOINS ALL THE ROWS OF FIRST AND SECOND DATAFRAMES TOGETHER AND CREATE NaN VALUE IF A ROW DOESN'T HAVE A VALUE AFTER JOINING<br />
- "left" > Left Join: INCLUDES ALL THE ROWS IN THE FIRST DATAFRAME AND ADDS THE COLUMNS OF SECOND DATAFRAME BUT IT WON'T INCLUDE THE ROWS OF THE SECOND DATAFRAME IF IT'S NOT THE SAME WITH THE FIRST<br />
- "right" > Right Join: INCLUDES ALL THE ROWS OF SECOND DATAFRAME AND THE COLUMNS OF THE FIRST DATAFRAME BUT WON'T INCLUDE THE ROWS OF THE FIRST DATAFRAME IF IT'S NOT SIMILAR TO THE SECOND DATAFRAME
```
df = pd.merge(df_1, df_2, on="student_id", how='left')
df
```
### Merging dataframes with same values but different column names
We add two more parameters :<br>
- Left_on means merge using this column name<br>
- Right_on means merge using this column name<br>
i.e merge both id and student_id together<br>
since they don't have same name, they will create different columns on the new table
```
df_1 = df_1.rename(columns={"student_id": "id"}) # Renamed student_id to id so as to give this example
df_1
df = pd.merge(df_1, df_2, left_on="id", right_on="student_id")
df
```
### Merging with the index of the first dataframe
```
df_1.set_index("id") # this will make id the new index for df_1
df = pd.merge(df_1, df_2, left_index=True, right_on="student_id")#the new index will be from index of df_2 where they joined
df
```
### Merging both table on their index i.e two indexes
```
df_2.set_index("student_id") # making student_id the index of Data2
df = pd.merge(df_1, df_2, left_index=True, right_index=True) # new index will be from the left index unlike when joining only one index
df
```
|
github_jupyter
|
import pandas as pd
import numpy as np
# Creating values to be used as datasets
dict1 = {
"student_id": [1,2,3,4,5,6,7,8,9,10],
"student_name": ["Peter","Dolly","Maggie","David","Isabelle","Harry","Akin","Abbey","Victoria","Sam"],
"student_course": np.random.choice(["Biology","Physics","Chemistry"], size=10)
}
# Create dataframe
df_1 = pd.DataFrame(dict1)
df_1
# Creating values to be used as datasets
dict2 = {
"student_id": np.random.choice([1,2,3,4,5,6,7,8,9,10], size=100),
"student_grade": np.random.choice(["A","B","C","D","E","F"], size=100),
"professors": np.random.choice(["Mark Levinson","Angela Marge","Bonnie James","Klaus Michealson"], size=100),
}
# Create dataframe
df_2 = pd.DataFrame(dict2) # OR Data2=pd.read_csv(filepath)
df_2
df = pd.merge(df_1, df_2)
df
df = pd.merge(df_1, df_2, on="student_id")
df
df = pd.merge(df_1, df_2, on="student_id", how='left')
df
df_1 = df_1.rename(columns={"student_id": "id"}) # Renamed student_id to id so as to give this example
df_1
df = pd.merge(df_1, df_2, left_on="id", right_on="student_id")
df
df_1.set_index("id") # this will make id the new index for df_1
df = pd.merge(df_1, df_2, left_index=True, right_on="student_id")#the new index will be from index of df_2 where they joined
df
df_2.set_index("student_id") # making student_id the index of Data2
df = pd.merge(df_1, df_2, left_index=True, right_index=True) # new index will be from the left index unlike when joining only one index
df
| 0.415136 | 0.926901 |
**This notebook is an exercise in the [Intermediate Machine Learning](https://www.kaggle.com/learn/intermediate-machine-learning) course. You can reference the tutorial at [this link](https://www.kaggle.com/alexisbcook/xgboost).**
---
In this exercise, you will use your new knowledge to train a model with **gradient boosting**.
# Setup
The questions below will give you feedback on your work. Run the following cell to set up the feedback system.
```
# Set up code checking
import os
if not os.path.exists("../input/train.csv"):
os.symlink("../input/home-data-for-ml-course/train.csv", "../input/train.csv")
os.symlink("../input/home-data-for-ml-course/test.csv", "../input/test.csv")
from learntools.core import binder
binder.bind(globals())
from learntools.ml_intermediate.ex6 import *
print("Setup Complete")
```
You will work with the [Housing Prices Competition for Kaggle Learn Users](https://www.kaggle.com/c/home-data-for-ml-course) dataset from the previous exercise.

Run the next code cell without changes to load the training and validation sets in `X_train`, `X_valid`, `y_train`, and `y_valid`. The test set is loaded in `X_test`.
```
import pandas as pd
from sklearn.model_selection import train_test_split
# Read the data
X = pd.read_csv('../input/train.csv', index_col='Id')
X_test_full = pd.read_csv('../input/test.csv', index_col='Id')
# Remove rows with missing target, separate target from predictors
X.dropna(axis=0, subset=['SalePrice'], inplace=True)
y = X.SalePrice
X.drop(['SalePrice'], axis=1, inplace=True)
# Break off validation set from training data
X_train_full, X_valid_full, y_train, y_valid = train_test_split(X, y, train_size=0.8, test_size=0.2,
random_state=0)
# "Cardinality" means the number of unique values in a column
# Select categorical columns with relatively low cardinality (convenient but arbitrary)
low_cardinality_cols = [cname for cname in X_train_full.columns if X_train_full[cname].nunique() < 10 and
X_train_full[cname].dtype == "object"]
# Select numeric columns
numeric_cols = [cname for cname in X_train_full.columns if X_train_full[cname].dtype in ['int64', 'float64']]
# Keep selected columns only
my_cols = low_cardinality_cols + numeric_cols
X_train = X_train_full[my_cols].copy()
X_valid = X_valid_full[my_cols].copy()
X_test = X_test_full[my_cols].copy()
# One-hot encode the data (to shorten the code, we use pandas)
X_train = pd.get_dummies(X_train)
X_valid = pd.get_dummies(X_valid)
X_test = pd.get_dummies(X_test)
X_train, X_valid = X_train.align(X_valid, join='left', axis=1)
X_train, X_test = X_train.align(X_test, join='left', axis=1)
```
# Step 1: Build model
### Part A
In this step, you'll build and train your first model with gradient boosting.
- Begin by setting `my_model_1` to an XGBoost model. Use the [XGBRegressor](https://xgboost.readthedocs.io/en/latest/python/python_api.html#xgboost.XGBRegressor) class, and set the random seed to 0 (`random_state=0`). **Leave all other parameters as default.**
- Then, fit the model to the training data in `X_train` and `y_train`.
```
from xgboost import XGBRegressor
# Define the model
my_model_1 = XGBRegressor(random_state=0)
# Fit the model
my_model_1.fit(X_train, y_train)
# Check your answer
step_1.a.check()
# Lines below will give you a hint or solution code
#step_1.a.hint()
#step_1.a.solution()
```
### Part B
Set `predictions_1` to the model's predictions for the validation data. Recall that the validation features are stored in `X_valid`.
```
from sklearn.metrics import mean_absolute_error
# Get predictions
predictions_1 = my_model_1.predict(X_valid)
# Check your answer
step_1.b.check()
# Lines below will give you a hint or solution code
#step_1.b.hint()
#step_1.b.solution()
```
### Part C
Finally, use the `mean_absolute_error()` function to calculate the mean absolute error (MAE) corresponding to the predictions for the validation set. Recall that the labels for the validation data are stored in `y_valid`.
```
# Calculate MAE
mae_1 = mean_absolute_error(predictions_1, y_valid)
# Uncomment to print MAE
print("Mean Absolute Error:" , mae_1)
# Check your answer
step_1.c.check()
# Lines below will give you a hint or solution code
#step_1.c.hint()
#step_1.c.solution()
```
# Step 2: Improve the model
Now that you've trained a default model as baseline, it's time to tinker with the parameters, to see if you can get better performance!
- Begin by setting `my_model_2` to an XGBoost model, using the [XGBRegressor](https://xgboost.readthedocs.io/en/latest/python/python_api.html#xgboost.XGBRegressor) class. Use what you learned in the previous tutorial to figure out how to change the default parameters (like `n_estimators` and `learning_rate`) to get better results.
- Then, fit the model to the training data in `X_train` and `y_train`.
- Set `predictions_2` to the model's predictions for the validation data. Recall that the validation features are stored in `X_valid`.
- Finally, use the `mean_absolute_error()` function to calculate the mean absolute error (MAE) corresponding to the predictions on the validation set. Recall that the labels for the validation data are stored in `y_valid`.
In order for this step to be marked correct, your model in `my_model_2` must attain lower MAE than the model in `my_model_1`.
```
# Define the model
my_model_2 = XGBRegressor(n_estimators=1000, learning_rate=0.05)
# Fit the model
my_model_2.fit(X_train, y_train)
# Get predictions
predictions_2 = my_model_2.predict(X_valid)
# Calculate MAE
mae_2 = mean_absolute_error(predictions_2, y_valid)
# Uncomment to print MAE
print("Mean Absolute Error:" , mae_2)
# Check your answer
step_2.check()
# Lines below will give you a hint or solution code
#step_2.hint()
#step_2.solution()
```
# Step 3: Break the model
In this step, you will create a model that performs worse than the original model in Step 1. This will help you to develop your intuition for how to set parameters. You might even find that you accidentally get better performance, which is ultimately a nice problem to have and a valuable learning experience!
- Begin by setting `my_model_3` to an XGBoost model, using the [XGBRegressor](https://xgboost.readthedocs.io/en/latest/python/python_api.html#xgboost.XGBRegressor) class. Use what you learned in the previous tutorial to figure out how to change the default parameters (like `n_estimators` and `learning_rate`) to design a model to get high MAE.
- Then, fit the model to the training data in `X_train` and `y_train`.
- Set `predictions_3` to the model's predictions for the validation data. Recall that the validation features are stored in `X_valid`.
- Finally, use the `mean_absolute_error()` function to calculate the mean absolute error (MAE) corresponding to the predictions on the validation set. Recall that the labels for the validation data are stored in `y_valid`.
In order for this step to be marked correct, your model in `my_model_3` must attain higher MAE than the model in `my_model_1`.
```
# Define the model
my_model_3 = XGBRegressor(n_estimators=100, learning_rate=0.9)
# Fit the model
my_model_3.fit(X_train, y_train)
# Get predictions
predictions_3 = my_model_3.predict(X_valid)
# Calculate MAE
mae_3 = mean_absolute_error(predictions_3, y_valid)
# Uncomment to print MAE
print("Mean Absolute Error:" , mae_3)
# Check your answer
step_3.check()
# Lines below will give you a hint or solution code
step_3.hint()
step_3.solution()
```
# Keep going
Continue to learn about **[data leakage](https://www.kaggle.com/alexisbcook/data-leakage)**. This is an important issue for a data scientist to understand, and it has the potential to ruin your models in subtle and dangerous ways!
---
*Have questions or comments? Visit the [course discussion forum](https://www.kaggle.com/learn/intermediate-machine-learning/discussion) to chat with other learners.*
|
github_jupyter
|
# Set up code checking
import os
if not os.path.exists("../input/train.csv"):
os.symlink("../input/home-data-for-ml-course/train.csv", "../input/train.csv")
os.symlink("../input/home-data-for-ml-course/test.csv", "../input/test.csv")
from learntools.core import binder
binder.bind(globals())
from learntools.ml_intermediate.ex6 import *
print("Setup Complete")
import pandas as pd
from sklearn.model_selection import train_test_split
# Read the data
X = pd.read_csv('../input/train.csv', index_col='Id')
X_test_full = pd.read_csv('../input/test.csv', index_col='Id')
# Remove rows with missing target, separate target from predictors
X.dropna(axis=0, subset=['SalePrice'], inplace=True)
y = X.SalePrice
X.drop(['SalePrice'], axis=1, inplace=True)
# Break off validation set from training data
X_train_full, X_valid_full, y_train, y_valid = train_test_split(X, y, train_size=0.8, test_size=0.2,
random_state=0)
# "Cardinality" means the number of unique values in a column
# Select categorical columns with relatively low cardinality (convenient but arbitrary)
low_cardinality_cols = [cname for cname in X_train_full.columns if X_train_full[cname].nunique() < 10 and
X_train_full[cname].dtype == "object"]
# Select numeric columns
numeric_cols = [cname for cname in X_train_full.columns if X_train_full[cname].dtype in ['int64', 'float64']]
# Keep selected columns only
my_cols = low_cardinality_cols + numeric_cols
X_train = X_train_full[my_cols].copy()
X_valid = X_valid_full[my_cols].copy()
X_test = X_test_full[my_cols].copy()
# One-hot encode the data (to shorten the code, we use pandas)
X_train = pd.get_dummies(X_train)
X_valid = pd.get_dummies(X_valid)
X_test = pd.get_dummies(X_test)
X_train, X_valid = X_train.align(X_valid, join='left', axis=1)
X_train, X_test = X_train.align(X_test, join='left', axis=1)
from xgboost import XGBRegressor
# Define the model
my_model_1 = XGBRegressor(random_state=0)
# Fit the model
my_model_1.fit(X_train, y_train)
# Check your answer
step_1.a.check()
# Lines below will give you a hint or solution code
#step_1.a.hint()
#step_1.a.solution()
from sklearn.metrics import mean_absolute_error
# Get predictions
predictions_1 = my_model_1.predict(X_valid)
# Check your answer
step_1.b.check()
# Lines below will give you a hint or solution code
#step_1.b.hint()
#step_1.b.solution()
# Calculate MAE
mae_1 = mean_absolute_error(predictions_1, y_valid)
# Uncomment to print MAE
print("Mean Absolute Error:" , mae_1)
# Check your answer
step_1.c.check()
# Lines below will give you a hint or solution code
#step_1.c.hint()
#step_1.c.solution()
# Define the model
my_model_2 = XGBRegressor(n_estimators=1000, learning_rate=0.05)
# Fit the model
my_model_2.fit(X_train, y_train)
# Get predictions
predictions_2 = my_model_2.predict(X_valid)
# Calculate MAE
mae_2 = mean_absolute_error(predictions_2, y_valid)
# Uncomment to print MAE
print("Mean Absolute Error:" , mae_2)
# Check your answer
step_2.check()
# Lines below will give you a hint or solution code
#step_2.hint()
#step_2.solution()
# Define the model
my_model_3 = XGBRegressor(n_estimators=100, learning_rate=0.9)
# Fit the model
my_model_3.fit(X_train, y_train)
# Get predictions
predictions_3 = my_model_3.predict(X_valid)
# Calculate MAE
mae_3 = mean_absolute_error(predictions_3, y_valid)
# Uncomment to print MAE
print("Mean Absolute Error:" , mae_3)
# Check your answer
step_3.check()
# Lines below will give you a hint or solution code
step_3.hint()
step_3.solution()
| 0.476092 | 0.951953 |
# Ada Boosting

### Step 1:
Initialize the weights $Wt = 1/N$ and assign each of the training point with equal weight ($Wt$).
Where $N$ is Number of Samples.
### Step 2:
Calculate Error Rate. $ \epsilon = \sum\limits_{wrong} W_i$ fro all hypothesis which classifies the sample
### Step 3:
Pick the best $h(x)$ with smallest error rate $\epsilon$
### Step 4:
Calculae the voting power of each of the samples.
$\alpha = \frac{1}{2} ln(\frac {1-\epsilon}{\epsilon})$
### Step 5:
Are we done?
#### Step 5.1:
if (Yes):
Ok done with classifier.
#### Step 5.2:
else :
Now we need to ask the following questions.
* Is $H(x)$ good enough?
* Have we done enough rounds?
* No good classifier left. That is, $\epsilon = \frac{1}{2} $
### Step 6:
Update the weights for each of the training points and emphasize on the points that were miscalssified.
$ W_{new} = \frac {1}{2} (\frac {1}{1- \epsilon}) W_{old} $ for correctly classifed Samples.
$ W_{new} = \frac {1}{2} (\frac {1}{\epsilon}) W_{old} $ for incorrectly calssified samples.
Now repeat from **step 2**.
#### Example:
Let us understand the flowchart with an example.
Suppose we have five samples four **positive (+)** and one **negative (-)** sample distirbuted as shown in the fig below.

For the sake of illustation purpose let us name the samples A,B,C,D and E as shown above. Where **C (-)** is **negative** and rest of them are **positive**.
Now there are several ways in which we can deside to group and try to add "Tree stubs". Let us say we try to calssify with the following hypothosis.
These are just random hypothesis, these are chosen to illustrate this for this example.
* All values $ X > 0$ are positive. In this case we will misclassify $C$.
* All values $ X < 6$ are negative. In this case we will misclassify $A,B,D,E$.
Similarly in the Y axis we can say,
* All values $ Y > 0 $ are positive. In this case again we misclassify $C$.
* All values $ Y < 6 $ are negative. In this case we will misclassify $A,B,D,E$.
In general there will be $N * 2$ possible hypothesis to classify the samples. So in this example where $ N = 5$ so we will have $10$ possible ways to classify the given dataset.
Now let us take some random hypothesis and try to achive the best error value $\epsilon $.
|sl.no|Hypothesis Condtion | Assumption | Misclassifies|
|-----|--------------------------|------------|--------------|
|1|Anything which is $ X < 2$| is **+** | B, E |
|2|Anything which is $ X < 4$| is **+** | B, C, E |
|3|Anything which is $ X < 6$| is **+** | C |
|4|Anything which is $ X > 2$| is **+** | A, C, D |
|5|Anything which is $ X > 4$| is **+** | A, D |
|6|Anything which is $ X > 6$| is **+** | A, B, D, E |
Above rows to be read as follows.
Suppose we draw a line at $X=2$ and the samples which fall on the left side of the line $ X < 2 $ is assumed to be positive **(+)**. Which misclassifies $B,E$. In other words samples on the right hand side of line drawn at $X=2$ i.e. $B,C,E$ were treated as **(-)** where as in actual $C$ is **(-)** and $B,E$ are **(+)** hence $B,E$ gets misclassfied for the hypothesis $X<2$.
### Step 1:
From our flowchart, Initialize all the weights equally.
In our example $N=5$, so weight of each of our sample is $W_i=\frac {1}{N} = \frac {1}{5}$
|sl no|Weights|Round1|
|:---:|:---:|:---:|
|1|$W_A$|$\frac{1}{5}$|
|2|$W_B$|$\frac{1}{5}$|
|3|$W_C$|$\frac{1}{5}$|
|4|$W_D$|$\frac{1}{5}$|
|5|$W_E$|$\frac{1}{5}$|
### Step 2:
Calculate Error Rate. $ \epsilon = \sum\limits_{wrong} W_i $
|sl no|Condition| Round1 |
|-----|---------|---------|
|1|$X < 2$ | $ \epsilon_{BE}=\frac{2}{5} $ |
|2|$X < 4$ | $ \epsilon_{BCE}=\frac{3}{5} $ |
|3|$X < 6$ | $ \epsilon_{C}=\frac{1}{5} $ |
|4|$X > 2$ | $ \epsilon_{ACD}=\frac{3}{5} $ |
|5|$X > 4$ | $ \epsilon_{AD}=\frac{2}{5} $ |
|6|$X > 6$ | $ \epsilon_{ABDE}=\frac{4}{5} $ |
### Step 3:
Pick the best $h(x)$ with smallest error rate $\epsilon$
From the previous step is it clear that $h(x) = (X < 6) $ and $\epsilon_C = \frac{1}{5}$ is the best error
### Step 4:
Calculae the voting power of each of the samples.
$\alpha = \frac{1}{2} ln(\frac {1-\epsilon}{\epsilon})$
Now we need to find the $\alpha$ for each of the error's $\epsilon$ in the table in step 2
|sl no|Condition| Round1 | $\alpha_{round1}$|
|-----|---------|----------|-------------------|
|1|$X < 2$ | $ \epsilon_{BE}=\frac{2}{5} $ |$\frac{1}{2} ln\frac {1-\epsilon_{BE}}{\epsilon_{BE}}$|
|--|--------|--------------------------------|-----------------------|
|2|$X < 4$ | $ \epsilon_{BCE}=\frac{3}{5} $ |$\frac{1}{2} ln\frac {1-\epsilon_{BCE}}{\epsilon_{BCE}}$|
|--|--------|--------------------------------|-----------------------|
|3|$X < 6$ | $ \epsilon_{C}=\frac{1}{5} $ |$\frac{1}{2} ln\frac {1-\epsilon_C}{\epsilon_C}$|
|--|--------|--------------------------------|-----------------------|
|4|$X > 2$ | $ \epsilon_{ACD}=\frac{3}{5} $ |$\frac{1}{2} ln\frac {1-\epsilon_{ACD}}{\epsilon_{ACD}}$|
|--|--------|--------------------------------|-----------------------|
|5|$X > 4$ | $ \epsilon_{AD}=\frac{2}{5} $ |$\frac{1}{2} ln\frac {1-\epsilon_{AD}}{\epsilon_{AD}}$|
|--|--------|--------------------------------|-----------------------|
|6|$X > 6$ | $ \epsilon_{ABDE}=\frac{4}{5} $ |$\frac{1}{2} ln\frac {1-\epsilon_{ABDE}}{\epsilon_{ABDE}}$|
|--|--------|--------------------------------|-----------------------|
But we are interested in $\alpha_C$ because that is the one which is giving best error $\epsilon_C = \frac{1}{2}$. After simplification we get
$\alpha_C = \frac{1}{2}ln(4)$
To summerize the values in round-1
| $ h(x) $ |$ \epsilon $| $ \alpha$ |
|-------- |------------|-----------|
|$X<6$ |$\frac{1}{5}$|$\frac{1}{2}ln(4)$|
|----------|------------|-----------|
### Step 5:
Clearly it is not sufficient to run just one run. Let us continue to iterate over. But before that we need to calculate $W_{new}$
### Step 6:
Update the weights for each of the training points and emphasize on the points that were miscalssified.
$ W_{new} = \frac {1}{2} (\frac {1}{1- \epsilon}) W_{old} $ for correctly classifed Samples.
$ W_{new} = \frac {1}{2} (\frac {1}{\epsilon}) W_{old} $ for incorrectly calssified samples.
Here is the list of $W_{old}$ values.
|sl no|Weights|Round1|
|:---:|:---:|:---:|
|1|$W_A$|$\frac{1}{5}$|
|2|$W_B$|$\frac{1}{5}$|
|3|$W_C$|$\frac{1}{5}$|
|4|$W_D$|$\frac{1}{5}$|
|5|$W_E$|$\frac{1}{5}$|
Here as we know only $W_C$ is calculated correctly. Rest of them are misclassified.
for $W_{A_{new}} = \frac {1}{2} (\frac {1}{1- \epsilon}) W_{old}$
Substitute values for $\epsilon = \frac{1}{5} $ and $W_{old} = \frac{1}{5}$
$W_{A_{new}} = \frac {1}{2} (\frac {1}{1- \frac{1}{5}}) \frac{1}{5} = \frac{1}{8}$
Similarly calculate the values for the rest of the weights.
|sl no|Weights|Round1| Round2|
|:---:|:---:|:---:|:---:|
|1|$W_A$|$\frac{1}{5}$|$\frac{1}{8}$|
|2|$W_B$|$\frac{1}{5}$|$\frac{1}{8}$|
|3|$W_C$|$\frac{1}{5}$|$\frac{4}{8}$|
|4|$W_D$|$\frac{1}{5}$|$\frac{1}{8}$|
|5|$W_E$|$\frac{1}{5}$|$\frac{1}{8}$|
Now back to step 2
Calculate Error Rate. $ \epsilon = \sum\limits_{wrong} W_i $
|sl no|Condition| Round1 | Round2 |
|-----|---------|----------|----------|
|1|$X < 2$ | $ \epsilon_{BE}=\frac{2}{5} $ |$\frac{2}{8} $ |
|-----|---------|----------|----------|
|2|$X < 4$ | $ \epsilon_{BCE}=\frac{3}{5} $ |$\frac{6}{8} $ |
|-----|---------|----------|----------|
|3|$X < 6$ | $ \epsilon_{C}=\frac{1}{5} $ |$\frac{4}{8} $ |
|-----|---------|----------|----------|
|4|$X > 2$ | $ \epsilon_{ACD}=\frac{3}{5} $ |$\frac{6}{8} $ |
|-----|---------|----------|----------|
|5|$X > 4$ | $ \epsilon_{AD}=\frac{2}{5} $ |$\frac{2}{8} $ |
|-----|---------|----------|----------|
|6|$X > 6$ | $ \epsilon_{ABDE}=\frac{4}{5} $|$\frac{4}{8} $ |
|-----|---------|----------|----------|
Now, from the the above Round-2 results, we have a tie i.e two conditions have $\epsilon_{BE} == \epsilon_{AD} == \frac{2}{8}$. We need to break the tie. Since $\epsilon_{BE} = \frac{2}{8} $ is first in the list we use that value as the best error.
Calculate the best voting power $\epsilon_{BE} = \frac{2}{8} $
$\alpha = \frac{1}{2} ln(\frac {1-\epsilon}{\epsilon}) = \frac{1}{2}ln(3)$
To summerize the values in round-1 and round-2
| $ h(x) $ |$ \epsilon $| $ \alpha$ |
|-------- |------------|-----------|
|$X<6$ |$\frac{1}{5}$|$\frac{1}{2}ln(4)$|
|----------|------------|-----------|
|$X<2$ |$\frac{2}{8}$|$\frac{1}{2}ln(3)$|
|----------|------------|-----------|
Now
$H(x) = SIGN(h_{1}(x)*\alpha_{1}+ h_{2}(x)*\alpha_{2})$
$H(x) = SIGN((X<6)\frac{1}{2}ln(4) + (X<2)\frac{1}{2}ln(3))$
So now we have only 2 decisions. If there is any disagreement one has more voting power than the other then it just overrides the other value, that is not correct. So we need to do one more iteration.
Calculate $W_{new}$
We know that
$\sum W_{wrong} = \sum W_{correct} = \frac{1}{2}$
$ \sum W_{wrong} = W_{B} + W_{E} = \frac{1}{2}$
There fore $W_{B} = W_{E} = \frac{1}{4}$
Substitute $\epsilon = \frac{2}{8}$ in the calculation of $W_{new}$ formula will result in the following table
|sl no|Weights|Round1| Round2| Round3
|:---:|:---:|:---:|:---:|:---:|
|1|$W_A$|$\frac{1}{5}$|$\frac{1}{8}$|$\frac{1}{12}$|
|2|$W_B$|$\frac{1}{5}$|$\frac{1}{8}$|$\frac{3}{12}$|
|3|$W_C$|$\frac{1}{5}$|$\frac{4}{8}$|$\frac{4}{12}$|
|4|$W_D$|$\frac{1}{5}$|$\frac{1}{8}$|$\frac{1}{12}$|
|5|$W_E$|$\frac{1}{5}$|$\frac{1}{8}$|$\frac{3}{12}$|
Calcualte the new Error rate.
|sl no|Condition| Round1 | Round2 | Round2 |
|-----|---------|----------|----------|----------|
|1|$X < 2$ | $ \epsilon_{BE}=\frac{2}{5} $ |$\frac{2}{8} $ | $\frac{6}{12} $ |
|-----|---------|----------|----------|----------|
|2|$X < 4$ | $ \epsilon_{BCE}=\frac{3}{5} $ |$\frac{6}{8} $ |$\frac{10}{12} $ |
|-----|---------|----------|----------|----------|
|3|$X < 6$ | $ \epsilon_{C}=\frac{1}{5} $ |$\frac{4}{8} $ |$\frac{4}{12} $ |
|-----|---------|----------|----------|----------|
|4|$X > 2$ | $ \epsilon_{ACD}=\frac{3}{5} $ |$\frac{6}{8} $ |$\frac{6}{12} $ |
|-----|---------|----------|----------|----------|
|5|$X > 4$ | $ \epsilon_{AD}=\frac{2}{5} $ |$\frac{2}{8} $ |$\frac{2}{12} $ |
|-----|---------|----------|----------|----------|
|6|$X > 6$ | $ \epsilon_{ABDE}=\frac{4}{5} $|$\frac{4}{8} $ |$\frac{8}{12} $ |
|-----|---------|----------|----------|----------|
Calculate the best voting power $\epsilon_{AD} = \frac{2}{12} $
$\alpha = \frac{1}{2} ln(\frac {1-\epsilon}{\epsilon}) = \frac{1}{2}ln(5)$
To summerize the values in round 1,2 and 3
| $ h(x) $ |$ \epsilon $| $ \alpha$ |
|-------- |------------|-----------|
|$X<6$ |$\frac{1}{5}$|$\frac{1}{2}ln(4)$|
|----------|------------|-----------|
|$X<2$ |$\frac{2}{8}$|$\frac{1}{2}ln(3)$|
|----------|------------|-----------|
|$X>4$ |$\frac{1}{6}$|$\frac{1}{2}ln(5)$|
|----------|------------|-----------|
$H(x) = SIGN((X<6)\frac{1}{2}ln(4) + (X<2)\frac{1}{2}ln(3) + (X>4)\frac{1}{2}ln5)$
$\frac{1}{2}ln(4*3) > \frac{1}{2}(ln5)$
so we are done.
|
github_jupyter
|
# Ada Boosting

### Step 1:
Initialize the weights $Wt = 1/N$ and assign each of the training point with equal weight ($Wt$).
Where $N$ is Number of Samples.
### Step 2:
Calculate Error Rate. $ \epsilon = \sum\limits_{wrong} W_i$ fro all hypothesis which classifies the sample
### Step 3:
Pick the best $h(x)$ with smallest error rate $\epsilon$
### Step 4:
Calculae the voting power of each of the samples.
$\alpha = \frac{1}{2} ln(\frac {1-\epsilon}{\epsilon})$
### Step 5:
Are we done?
#### Step 5.1:
if (Yes):
Ok done with classifier.
#### Step 5.2:
else :
Now we need to ask the following questions.
* Is $H(x)$ good enough?
* Have we done enough rounds?
* No good classifier left. That is, $\epsilon = \frac{1}{2} $
### Step 6:
Update the weights for each of the training points and emphasize on the points that were miscalssified.
$ W_{new} = \frac {1}{2} (\frac {1}{1- \epsilon}) W_{old} $ for correctly classifed Samples.
$ W_{new} = \frac {1}{2} (\frac {1}{\epsilon}) W_{old} $ for incorrectly calssified samples.
Now repeat from **step 2**.
#### Example:
Let us understand the flowchart with an example.
Suppose we have five samples four **positive (+)** and one **negative (-)** sample distirbuted as shown in the fig below.

For the sake of illustation purpose let us name the samples A,B,C,D and E as shown above. Where **C (-)** is **negative** and rest of them are **positive**.
Now there are several ways in which we can deside to group and try to add "Tree stubs". Let us say we try to calssify with the following hypothosis.
These are just random hypothesis, these are chosen to illustrate this for this example.
* All values $ X > 0$ are positive. In this case we will misclassify $C$.
* All values $ X < 6$ are negative. In this case we will misclassify $A,B,D,E$.
Similarly in the Y axis we can say,
* All values $ Y > 0 $ are positive. In this case again we misclassify $C$.
* All values $ Y < 6 $ are negative. In this case we will misclassify $A,B,D,E$.
In general there will be $N * 2$ possible hypothesis to classify the samples. So in this example where $ N = 5$ so we will have $10$ possible ways to classify the given dataset.
Now let us take some random hypothesis and try to achive the best error value $\epsilon $.
|sl.no|Hypothesis Condtion | Assumption | Misclassifies|
|-----|--------------------------|------------|--------------|
|1|Anything which is $ X < 2$| is **+** | B, E |
|2|Anything which is $ X < 4$| is **+** | B, C, E |
|3|Anything which is $ X < 6$| is **+** | C |
|4|Anything which is $ X > 2$| is **+** | A, C, D |
|5|Anything which is $ X > 4$| is **+** | A, D |
|6|Anything which is $ X > 6$| is **+** | A, B, D, E |
Above rows to be read as follows.
Suppose we draw a line at $X=2$ and the samples which fall on the left side of the line $ X < 2 $ is assumed to be positive **(+)**. Which misclassifies $B,E$. In other words samples on the right hand side of line drawn at $X=2$ i.e. $B,C,E$ were treated as **(-)** where as in actual $C$ is **(-)** and $B,E$ are **(+)** hence $B,E$ gets misclassfied for the hypothesis $X<2$.
### Step 1:
From our flowchart, Initialize all the weights equally.
In our example $N=5$, so weight of each of our sample is $W_i=\frac {1}{N} = \frac {1}{5}$
|sl no|Weights|Round1|
|:---:|:---:|:---:|
|1|$W_A$|$\frac{1}{5}$|
|2|$W_B$|$\frac{1}{5}$|
|3|$W_C$|$\frac{1}{5}$|
|4|$W_D$|$\frac{1}{5}$|
|5|$W_E$|$\frac{1}{5}$|
### Step 2:
Calculate Error Rate. $ \epsilon = \sum\limits_{wrong} W_i $
|sl no|Condition| Round1 |
|-----|---------|---------|
|1|$X < 2$ | $ \epsilon_{BE}=\frac{2}{5} $ |
|2|$X < 4$ | $ \epsilon_{BCE}=\frac{3}{5} $ |
|3|$X < 6$ | $ \epsilon_{C}=\frac{1}{5} $ |
|4|$X > 2$ | $ \epsilon_{ACD}=\frac{3}{5} $ |
|5|$X > 4$ | $ \epsilon_{AD}=\frac{2}{5} $ |
|6|$X > 6$ | $ \epsilon_{ABDE}=\frac{4}{5} $ |
### Step 3:
Pick the best $h(x)$ with smallest error rate $\epsilon$
From the previous step is it clear that $h(x) = (X < 6) $ and $\epsilon_C = \frac{1}{5}$ is the best error
### Step 4:
Calculae the voting power of each of the samples.
$\alpha = \frac{1}{2} ln(\frac {1-\epsilon}{\epsilon})$
Now we need to find the $\alpha$ for each of the error's $\epsilon$ in the table in step 2
|sl no|Condition| Round1 | $\alpha_{round1}$|
|-----|---------|----------|-------------------|
|1|$X < 2$ | $ \epsilon_{BE}=\frac{2}{5} $ |$\frac{1}{2} ln\frac {1-\epsilon_{BE}}{\epsilon_{BE}}$|
|--|--------|--------------------------------|-----------------------|
|2|$X < 4$ | $ \epsilon_{BCE}=\frac{3}{5} $ |$\frac{1}{2} ln\frac {1-\epsilon_{BCE}}{\epsilon_{BCE}}$|
|--|--------|--------------------------------|-----------------------|
|3|$X < 6$ | $ \epsilon_{C}=\frac{1}{5} $ |$\frac{1}{2} ln\frac {1-\epsilon_C}{\epsilon_C}$|
|--|--------|--------------------------------|-----------------------|
|4|$X > 2$ | $ \epsilon_{ACD}=\frac{3}{5} $ |$\frac{1}{2} ln\frac {1-\epsilon_{ACD}}{\epsilon_{ACD}}$|
|--|--------|--------------------------------|-----------------------|
|5|$X > 4$ | $ \epsilon_{AD}=\frac{2}{5} $ |$\frac{1}{2} ln\frac {1-\epsilon_{AD}}{\epsilon_{AD}}$|
|--|--------|--------------------------------|-----------------------|
|6|$X > 6$ | $ \epsilon_{ABDE}=\frac{4}{5} $ |$\frac{1}{2} ln\frac {1-\epsilon_{ABDE}}{\epsilon_{ABDE}}$|
|--|--------|--------------------------------|-----------------------|
But we are interested in $\alpha_C$ because that is the one which is giving best error $\epsilon_C = \frac{1}{2}$. After simplification we get
$\alpha_C = \frac{1}{2}ln(4)$
To summerize the values in round-1
| $ h(x) $ |$ \epsilon $| $ \alpha$ |
|-------- |------------|-----------|
|$X<6$ |$\frac{1}{5}$|$\frac{1}{2}ln(4)$|
|----------|------------|-----------|
### Step 5:
Clearly it is not sufficient to run just one run. Let us continue to iterate over. But before that we need to calculate $W_{new}$
### Step 6:
Update the weights for each of the training points and emphasize on the points that were miscalssified.
$ W_{new} = \frac {1}{2} (\frac {1}{1- \epsilon}) W_{old} $ for correctly classifed Samples.
$ W_{new} = \frac {1}{2} (\frac {1}{\epsilon}) W_{old} $ for incorrectly calssified samples.
Here is the list of $W_{old}$ values.
|sl no|Weights|Round1|
|:---:|:---:|:---:|
|1|$W_A$|$\frac{1}{5}$|
|2|$W_B$|$\frac{1}{5}$|
|3|$W_C$|$\frac{1}{5}$|
|4|$W_D$|$\frac{1}{5}$|
|5|$W_E$|$\frac{1}{5}$|
Here as we know only $W_C$ is calculated correctly. Rest of them are misclassified.
for $W_{A_{new}} = \frac {1}{2} (\frac {1}{1- \epsilon}) W_{old}$
Substitute values for $\epsilon = \frac{1}{5} $ and $W_{old} = \frac{1}{5}$
$W_{A_{new}} = \frac {1}{2} (\frac {1}{1- \frac{1}{5}}) \frac{1}{5} = \frac{1}{8}$
Similarly calculate the values for the rest of the weights.
|sl no|Weights|Round1| Round2|
|:---:|:---:|:---:|:---:|
|1|$W_A$|$\frac{1}{5}$|$\frac{1}{8}$|
|2|$W_B$|$\frac{1}{5}$|$\frac{1}{8}$|
|3|$W_C$|$\frac{1}{5}$|$\frac{4}{8}$|
|4|$W_D$|$\frac{1}{5}$|$\frac{1}{8}$|
|5|$W_E$|$\frac{1}{5}$|$\frac{1}{8}$|
Now back to step 2
Calculate Error Rate. $ \epsilon = \sum\limits_{wrong} W_i $
|sl no|Condition| Round1 | Round2 |
|-----|---------|----------|----------|
|1|$X < 2$ | $ \epsilon_{BE}=\frac{2}{5} $ |$\frac{2}{8} $ |
|-----|---------|----------|----------|
|2|$X < 4$ | $ \epsilon_{BCE}=\frac{3}{5} $ |$\frac{6}{8} $ |
|-----|---------|----------|----------|
|3|$X < 6$ | $ \epsilon_{C}=\frac{1}{5} $ |$\frac{4}{8} $ |
|-----|---------|----------|----------|
|4|$X > 2$ | $ \epsilon_{ACD}=\frac{3}{5} $ |$\frac{6}{8} $ |
|-----|---------|----------|----------|
|5|$X > 4$ | $ \epsilon_{AD}=\frac{2}{5} $ |$\frac{2}{8} $ |
|-----|---------|----------|----------|
|6|$X > 6$ | $ \epsilon_{ABDE}=\frac{4}{5} $|$\frac{4}{8} $ |
|-----|---------|----------|----------|
Now, from the the above Round-2 results, we have a tie i.e two conditions have $\epsilon_{BE} == \epsilon_{AD} == \frac{2}{8}$. We need to break the tie. Since $\epsilon_{BE} = \frac{2}{8} $ is first in the list we use that value as the best error.
Calculate the best voting power $\epsilon_{BE} = \frac{2}{8} $
$\alpha = \frac{1}{2} ln(\frac {1-\epsilon}{\epsilon}) = \frac{1}{2}ln(3)$
To summerize the values in round-1 and round-2
| $ h(x) $ |$ \epsilon $| $ \alpha$ |
|-------- |------------|-----------|
|$X<6$ |$\frac{1}{5}$|$\frac{1}{2}ln(4)$|
|----------|------------|-----------|
|$X<2$ |$\frac{2}{8}$|$\frac{1}{2}ln(3)$|
|----------|------------|-----------|
Now
$H(x) = SIGN(h_{1}(x)*\alpha_{1}+ h_{2}(x)*\alpha_{2})$
$H(x) = SIGN((X<6)\frac{1}{2}ln(4) + (X<2)\frac{1}{2}ln(3))$
So now we have only 2 decisions. If there is any disagreement one has more voting power than the other then it just overrides the other value, that is not correct. So we need to do one more iteration.
Calculate $W_{new}$
We know that
$\sum W_{wrong} = \sum W_{correct} = \frac{1}{2}$
$ \sum W_{wrong} = W_{B} + W_{E} = \frac{1}{2}$
There fore $W_{B} = W_{E} = \frac{1}{4}$
Substitute $\epsilon = \frac{2}{8}$ in the calculation of $W_{new}$ formula will result in the following table
|sl no|Weights|Round1| Round2| Round3
|:---:|:---:|:---:|:---:|:---:|
|1|$W_A$|$\frac{1}{5}$|$\frac{1}{8}$|$\frac{1}{12}$|
|2|$W_B$|$\frac{1}{5}$|$\frac{1}{8}$|$\frac{3}{12}$|
|3|$W_C$|$\frac{1}{5}$|$\frac{4}{8}$|$\frac{4}{12}$|
|4|$W_D$|$\frac{1}{5}$|$\frac{1}{8}$|$\frac{1}{12}$|
|5|$W_E$|$\frac{1}{5}$|$\frac{1}{8}$|$\frac{3}{12}$|
Calcualte the new Error rate.
|sl no|Condition| Round1 | Round2 | Round2 |
|-----|---------|----------|----------|----------|
|1|$X < 2$ | $ \epsilon_{BE}=\frac{2}{5} $ |$\frac{2}{8} $ | $\frac{6}{12} $ |
|-----|---------|----------|----------|----------|
|2|$X < 4$ | $ \epsilon_{BCE}=\frac{3}{5} $ |$\frac{6}{8} $ |$\frac{10}{12} $ |
|-----|---------|----------|----------|----------|
|3|$X < 6$ | $ \epsilon_{C}=\frac{1}{5} $ |$\frac{4}{8} $ |$\frac{4}{12} $ |
|-----|---------|----------|----------|----------|
|4|$X > 2$ | $ \epsilon_{ACD}=\frac{3}{5} $ |$\frac{6}{8} $ |$\frac{6}{12} $ |
|-----|---------|----------|----------|----------|
|5|$X > 4$ | $ \epsilon_{AD}=\frac{2}{5} $ |$\frac{2}{8} $ |$\frac{2}{12} $ |
|-----|---------|----------|----------|----------|
|6|$X > 6$ | $ \epsilon_{ABDE}=\frac{4}{5} $|$\frac{4}{8} $ |$\frac{8}{12} $ |
|-----|---------|----------|----------|----------|
Calculate the best voting power $\epsilon_{AD} = \frac{2}{12} $
$\alpha = \frac{1}{2} ln(\frac {1-\epsilon}{\epsilon}) = \frac{1}{2}ln(5)$
To summerize the values in round 1,2 and 3
| $ h(x) $ |$ \epsilon $| $ \alpha$ |
|-------- |------------|-----------|
|$X<6$ |$\frac{1}{5}$|$\frac{1}{2}ln(4)$|
|----------|------------|-----------|
|$X<2$ |$\frac{2}{8}$|$\frac{1}{2}ln(3)$|
|----------|------------|-----------|
|$X>4$ |$\frac{1}{6}$|$\frac{1}{2}ln(5)$|
|----------|------------|-----------|
$H(x) = SIGN((X<6)\frac{1}{2}ln(4) + (X<2)\frac{1}{2}ln(3) + (X>4)\frac{1}{2}ln5)$
$\frac{1}{2}ln(4*3) > \frac{1}{2}(ln5)$
so we are done.
| 0.906661 | 0.992647 |
# Importing libraries and Exploring the Dataset
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import scipy.stats as stats
sns.set()
insurance_df = pd.read_csv("insurance-data.csv")
insurance_df.head()
insurance_df.info()
```
# Checking missing values and Outliers
```
# Check for the null values
insurance_df.isna().apply(pd.value_counts)
# Checking for the outliers
fig, ax = plt.subplots(3, 1, figsize=(10,7))
ax[0].set_title("BMI")
ax[1].set_title("AGE")
ax[2].set_title("CHARGES")
sns.boxplot(data = df.bmi, orient="h", ax=ax[0], color = "b")
sns.boxplot(data = df.age, orient="h", ax=ax[1], color = "r")
sns.boxplot(data = df.charges, orient="h", ax=ax[2], color = "g")
plt.tight_layout()
insurance_df.describe().T
```
# Creating visual methods to analyze the data.
```
# Plots to see the distribution of the continuous features individually
plt.figure(figsize=(20,15))
plt.subplot(3,3,1)
plt.hist(insurance_df.bmi, color="lightblue", edgecolor="black", alpha=0.7)
plt.xlabel("bmi")
plt.subplot(3,3,2)
plt.hist(insurance_df.age, color='red', edgecolor = 'black', alpha = 0.7)
plt.xlabel('age')
plt.subplot(3,3,3)
plt.hist(insurance_df.charges, color='lightgreen', edgecolor = 'black', alpha = 0.7)
plt.xlabel('charges')
plt.show()
# visualizing data to make analysis
fig, ax = plt.subplots(1, 3, figsize=(15,4))
sns.distplot(insurance_df.bmi, ax=ax[0], color = "b", bins=10, kde=False)
sns.distplot(insurance_df.age, ax=ax[1], color = "r", bins=10, kde=False)
sns.distplot(insurance_df.charges, ax=ax[2], color = "g", bins=10, kde=False)
# visualizing data to make analysis
plt.figure(figsize=(17,20))
x = insurance_df.smoker.value_counts().index
y = [insurance_df['smoker'].value_counts()[i] for i in x]
plt.subplot(4,2,1)
plt.bar(x,y, align='center',color = 'lightblue',edgecolor = 'black',alpha = 0.7) #plot a bar chart
plt.xlabel('Is Smoker?')
plt.ylabel('Count ')
plt.title('Smoker distribution')
x1 = insurance_df.sex.value_counts().index #Values for x-axis
y1 = [insurance_df['sex'].value_counts()[j] for j in x1] # Count of each class on y-axis
plt.subplot(4,2,2)
plt.bar(x1,y1, align='center',color = 'red',edgecolor = 'black',alpha = 0.7) #plot a bar chart
plt.xlabel('Gender')
plt.ylabel('Count')
plt.title('Gender distribution')
x2 = insurance_df.region.value_counts().index #Values for x-axis
y2 = [insurance_df['region'].value_counts()[k] for k in x2] # Count of each class on y-axis
plt.subplot(4,2,3)
plt.bar(x2,y2, align='center',color = 'green',edgecolor = 'black',alpha = 0.7) #plot a bar chart
plt.xlabel('Region')
plt.ylabel('Count ')
plt.title("Regions' distribution")
x3 = insurance_df.children.value_counts().index #Values for x-axis
y3 = [insurance_df['children'].value_counts()[l] for l in x3] # Count of each class on y-axis
plt.subplot(4,2,4)
plt.bar(x3,y3, align='center',color = 'purple',edgecolor = 'black',alpha = 0.7) #plot a bar chart
plt.xlabel('No. of children')
plt.ylabel('Count ')
plt.title("Children distribution")
plt.tight_layout()
plt.show()
sns.countplot(insurance_df.region)
sns.pairplot(insurance_df)
```
# Analyzing trends, patterns, and relationships in the Data.
```
# Do charges of people who smoke differ significantly from the people who don't?
insurance_df.smoker.value_counts()
sns.heatmap(insurance_df.corr(), cmap="coolwarm", annot=True)
# Scatter plot to look for visual evidence of dependency between attributes smoker and charges accross different ages
plt.figure(figsize=(12,7))
sns.scatterplot(insurance_df.age, insurance_df.charges, hue=insurance_df.smoker, palette=["red", "green"], alpha=0.7)
plt.show()
plt.figure(figsize=(12,7))
sns.scatterplot(insurance_df.age, insurance_df.charges, hue=insurance_df.sex, palette=["red", "blue"], alpha=0.7)
plt.show()
insurance_df.sex.value_counts()
# T-test to check dependency of smoking on charges
H0="Charges of smoker and non-smoker are the same"
Ha="Charges of smoker and non-smoker are not the same"
a=np.array(insurance_df[insurance_df.smoker=="yes"].charges)
b=np.array(insurance_df[insurance_df.smoker=="no"].charges)
t, p_value = stats.ttest_ind(a,b,axis=0)
if p_value < 0.05:
print(f"{Ha} as the p_value ({p_value}) < 0.05")
else:
print(f"{H0} as the p_value ({p_value}) > 0.05")
#Does bmi of males differ significantly from that of females?
H0="gender has no effect on bmi"
Ha="gender has effect on bmi"
a=np.array(insurance_df[insurance_df.sex=="male"].bmi)
b=np.array(insurance_df[insurance_df.sex=="female"].bmi)
t, p_value = stats.ttest_ind(a,b,axis=0)
if p_value < 0.05:
print(f"{Ha} as the p_value ({p_value}) < 0.05")
else:
print(f"{H0} as the p_value ({p_value}) > 0.05")
# T-test to check dependency of bmi on gender
# Hasan hocamızın gönrderdiği kod:
ttest2=stats.ttest_ind(insurance_df.bmi[insurance_df["sex"]=="male"],insurance_df.bmi[insurance_df["sex"]=="female"])
if ttest2[1]<0.05:
print(f"Gender has effect on bmi as the p_value {ttest2[1]} < 0.05")
else:
print(f"Gender has no effect on bmi as the p_value {round(ttest2[1],2)} > 0.05")
```
|
github_jupyter
|
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import scipy.stats as stats
sns.set()
insurance_df = pd.read_csv("insurance-data.csv")
insurance_df.head()
insurance_df.info()
# Check for the null values
insurance_df.isna().apply(pd.value_counts)
# Checking for the outliers
fig, ax = plt.subplots(3, 1, figsize=(10,7))
ax[0].set_title("BMI")
ax[1].set_title("AGE")
ax[2].set_title("CHARGES")
sns.boxplot(data = df.bmi, orient="h", ax=ax[0], color = "b")
sns.boxplot(data = df.age, orient="h", ax=ax[1], color = "r")
sns.boxplot(data = df.charges, orient="h", ax=ax[2], color = "g")
plt.tight_layout()
insurance_df.describe().T
# Plots to see the distribution of the continuous features individually
plt.figure(figsize=(20,15))
plt.subplot(3,3,1)
plt.hist(insurance_df.bmi, color="lightblue", edgecolor="black", alpha=0.7)
plt.xlabel("bmi")
plt.subplot(3,3,2)
plt.hist(insurance_df.age, color='red', edgecolor = 'black', alpha = 0.7)
plt.xlabel('age')
plt.subplot(3,3,3)
plt.hist(insurance_df.charges, color='lightgreen', edgecolor = 'black', alpha = 0.7)
plt.xlabel('charges')
plt.show()
# visualizing data to make analysis
fig, ax = plt.subplots(1, 3, figsize=(15,4))
sns.distplot(insurance_df.bmi, ax=ax[0], color = "b", bins=10, kde=False)
sns.distplot(insurance_df.age, ax=ax[1], color = "r", bins=10, kde=False)
sns.distplot(insurance_df.charges, ax=ax[2], color = "g", bins=10, kde=False)
# visualizing data to make analysis
plt.figure(figsize=(17,20))
x = insurance_df.smoker.value_counts().index
y = [insurance_df['smoker'].value_counts()[i] for i in x]
plt.subplot(4,2,1)
plt.bar(x,y, align='center',color = 'lightblue',edgecolor = 'black',alpha = 0.7) #plot a bar chart
plt.xlabel('Is Smoker?')
plt.ylabel('Count ')
plt.title('Smoker distribution')
x1 = insurance_df.sex.value_counts().index #Values for x-axis
y1 = [insurance_df['sex'].value_counts()[j] for j in x1] # Count of each class on y-axis
plt.subplot(4,2,2)
plt.bar(x1,y1, align='center',color = 'red',edgecolor = 'black',alpha = 0.7) #plot a bar chart
plt.xlabel('Gender')
plt.ylabel('Count')
plt.title('Gender distribution')
x2 = insurance_df.region.value_counts().index #Values for x-axis
y2 = [insurance_df['region'].value_counts()[k] for k in x2] # Count of each class on y-axis
plt.subplot(4,2,3)
plt.bar(x2,y2, align='center',color = 'green',edgecolor = 'black',alpha = 0.7) #plot a bar chart
plt.xlabel('Region')
plt.ylabel('Count ')
plt.title("Regions' distribution")
x3 = insurance_df.children.value_counts().index #Values for x-axis
y3 = [insurance_df['children'].value_counts()[l] for l in x3] # Count of each class on y-axis
plt.subplot(4,2,4)
plt.bar(x3,y3, align='center',color = 'purple',edgecolor = 'black',alpha = 0.7) #plot a bar chart
plt.xlabel('No. of children')
plt.ylabel('Count ')
plt.title("Children distribution")
plt.tight_layout()
plt.show()
sns.countplot(insurance_df.region)
sns.pairplot(insurance_df)
# Do charges of people who smoke differ significantly from the people who don't?
insurance_df.smoker.value_counts()
sns.heatmap(insurance_df.corr(), cmap="coolwarm", annot=True)
# Scatter plot to look for visual evidence of dependency between attributes smoker and charges accross different ages
plt.figure(figsize=(12,7))
sns.scatterplot(insurance_df.age, insurance_df.charges, hue=insurance_df.smoker, palette=["red", "green"], alpha=0.7)
plt.show()
plt.figure(figsize=(12,7))
sns.scatterplot(insurance_df.age, insurance_df.charges, hue=insurance_df.sex, palette=["red", "blue"], alpha=0.7)
plt.show()
insurance_df.sex.value_counts()
# T-test to check dependency of smoking on charges
H0="Charges of smoker and non-smoker are the same"
Ha="Charges of smoker and non-smoker are not the same"
a=np.array(insurance_df[insurance_df.smoker=="yes"].charges)
b=np.array(insurance_df[insurance_df.smoker=="no"].charges)
t, p_value = stats.ttest_ind(a,b,axis=0)
if p_value < 0.05:
print(f"{Ha} as the p_value ({p_value}) < 0.05")
else:
print(f"{H0} as the p_value ({p_value}) > 0.05")
#Does bmi of males differ significantly from that of females?
H0="gender has no effect on bmi"
Ha="gender has effect on bmi"
a=np.array(insurance_df[insurance_df.sex=="male"].bmi)
b=np.array(insurance_df[insurance_df.sex=="female"].bmi)
t, p_value = stats.ttest_ind(a,b,axis=0)
if p_value < 0.05:
print(f"{Ha} as the p_value ({p_value}) < 0.05")
else:
print(f"{H0} as the p_value ({p_value}) > 0.05")
# T-test to check dependency of bmi on gender
# Hasan hocamızın gönrderdiği kod:
ttest2=stats.ttest_ind(insurance_df.bmi[insurance_df["sex"]=="male"],insurance_df.bmi[insurance_df["sex"]=="female"])
if ttest2[1]<0.05:
print(f"Gender has effect on bmi as the p_value {ttest2[1]} < 0.05")
else:
print(f"Gender has no effect on bmi as the p_value {round(ttest2[1],2)} > 0.05")
| 0.482185 | 0.906156 |
```
#Part B
import numpy as np
inputs = np.zeros((16,5))
# For generating the all possible input values, concatenated with the bias vector. 5th column is all 1, which
# represents the bias vector.
def generateInputVector(inputVector):
for i in range(np.shape(inputVector)[0]):
temp = bin(i).split('b')
temp = temp[-1]
rowVector = [int(x) for x in str(temp)]
rowVector.append(1)
sizeRowVector = len(rowVector)
inputVector[i, (np.shape(inputVector)[1]) - sizeRowVector:] = rowVector[:]
return inputVector
inputs = generateInputVector(inputs)
print(inputs)
# The activation function which the the unit step function.
def unitStepFunction(k):
k = np.where(k<0, 0,1)
return k
# Hand calculated weights for hidden layer and output layer.
hiddenweights = np.array([[5, 0, 5, 5, -13],
[0, -4, 5, 5, -9],
[-6, 11, -5, 0, -10],
[-5, 13, 0, -6, -11]])
outputweights = np.array([2, 3, 5, 7, -1])
# Testing the network with given weights.
hiddenOutsTemp = unitStepFunction(inputs.dot(hiddenweights.T))
# Again, adding the bias vector before going into output neuron.
bias = np.ones((16,1))
hiddenOuts = np.concatenate((hiddenOutsTemp, bias),axis =1)
print(hiddenOuts)
# Final output for the network with given weights.
outputs = unitStepFunction(hiddenOuts.dot(outputweights.T))
print(outputs)
# Implementation of the logic function. Therefore, we can test the performance of the network above.
def xor_gate(a, b):
result = (a and (not b)) or ((not a) and b)
return result
def logicFunction(inputVector):
result = list()
for i in range(np.shape(inputVector)[0]):
temp = xor_gate(inputVector[i,0] or (not inputVector[i,1]), (not inputVector[i,2]) or (not inputVector[i,3]))
result.append(temp)
return result
# The outputs from the logic function. We will check this output with the ones we have found above.
outputsCheck = logicFunction(inputs[:,0:4])
print(outputsCheck)
# For calculating the accuracy of the network, accuracyCalc function is defined.
def accuracyCalc(x, y):
result = 0
count = 0
size = np.shape(x)[0]
sentence = 'The accuracy of the model is: '
for i in range(size):
if (x[i] == y[i]):
count = count +1
result = (count / size) * 100
return result
# The accuracy result between the network and the logic function itself.
accuracy = accuracyCalc(outputs, outputsCheck)
print('The accuracy of the model is: ' + str(accuracy))
# Part C
# Robust Weights
# w_rh = robust weights for hidden layer
# w_ro = robust weights for output layer
w_rh = np.array([[1, 0, 1, 1, -2.5],
[0, -1, 1, 1, -1.5],
[-1, 1, -1, 0, -0.5],
[-1, 1, 0, -1, -0.5]])
w_ro = np.array([1, 1, 1, 1, -0.5])
# Part D
# Generate 400 input samples by concatenating 25 samples from each input.
inputsWithNoise = np.zeros((400,5))
for k in range(25):
for i in range(np.shape(inputs)[0]):
inputsWithNoise[(k*16)+i,:] = inputs[i,:]
# Then check the outputs of the inputsWithNoise.
outputsCheck_D = logicFunction(inputsWithNoise[:,0:4])
# Create a gaussian noise with 0 mean and 0.2 std. Then add this noise to the inputsWithNoise array.
# np.random.seed(7) is for getting the same output each run.
np.random.seed(7)
gaussianNoise = np.random.normal(loc=0, scale=0.2, size=1600).reshape(400, 4)
inputsWithNoise[:, 0:4] += gaussianNoise
# Then test the inputsWithNoise with the random weights network.
outputTemp = unitStepFunction(inputsWithNoise.dot(hiddenweights.T))
bias = np.ones((400,1))
hiddenOutsTemp = np.concatenate((outputTemp, bias),axis =1)
random_network_output = unitStepFunction(hiddenOutsTemp.dot(outputweights.T))
# Then test the inputsWithNoise with the robust weights network.
robustOutputTemp = unitStepFunction(inputsWithNoise.dot(w_rh.T))
bias = np.ones((400,1))
robustHiddenOuts = np.concatenate((robustOutputTemp, bias),axis =1)
robust_network_output = unitStepFunction(robustHiddenOuts.dot(w_ro.T))
# Accuracy of the random weight network.
accuracy = accuracyCalc(random_network_output, outputsCheck_D)
print('The accuracy of the random weighted network is: ' + str(accuracy))
# Accuracy of the robust network.
accuracy = accuracyCalc(robust_network_output, outputsCheck_D)
print('The accuracy of the robust network is: ' + str(accuracy))
```
|
github_jupyter
|
#Part B
import numpy as np
inputs = np.zeros((16,5))
# For generating the all possible input values, concatenated with the bias vector. 5th column is all 1, which
# represents the bias vector.
def generateInputVector(inputVector):
for i in range(np.shape(inputVector)[0]):
temp = bin(i).split('b')
temp = temp[-1]
rowVector = [int(x) for x in str(temp)]
rowVector.append(1)
sizeRowVector = len(rowVector)
inputVector[i, (np.shape(inputVector)[1]) - sizeRowVector:] = rowVector[:]
return inputVector
inputs = generateInputVector(inputs)
print(inputs)
# The activation function which the the unit step function.
def unitStepFunction(k):
k = np.where(k<0, 0,1)
return k
# Hand calculated weights for hidden layer and output layer.
hiddenweights = np.array([[5, 0, 5, 5, -13],
[0, -4, 5, 5, -9],
[-6, 11, -5, 0, -10],
[-5, 13, 0, -6, -11]])
outputweights = np.array([2, 3, 5, 7, -1])
# Testing the network with given weights.
hiddenOutsTemp = unitStepFunction(inputs.dot(hiddenweights.T))
# Again, adding the bias vector before going into output neuron.
bias = np.ones((16,1))
hiddenOuts = np.concatenate((hiddenOutsTemp, bias),axis =1)
print(hiddenOuts)
# Final output for the network with given weights.
outputs = unitStepFunction(hiddenOuts.dot(outputweights.T))
print(outputs)
# Implementation of the logic function. Therefore, we can test the performance of the network above.
def xor_gate(a, b):
result = (a and (not b)) or ((not a) and b)
return result
def logicFunction(inputVector):
result = list()
for i in range(np.shape(inputVector)[0]):
temp = xor_gate(inputVector[i,0] or (not inputVector[i,1]), (not inputVector[i,2]) or (not inputVector[i,3]))
result.append(temp)
return result
# The outputs from the logic function. We will check this output with the ones we have found above.
outputsCheck = logicFunction(inputs[:,0:4])
print(outputsCheck)
# For calculating the accuracy of the network, accuracyCalc function is defined.
def accuracyCalc(x, y):
result = 0
count = 0
size = np.shape(x)[0]
sentence = 'The accuracy of the model is: '
for i in range(size):
if (x[i] == y[i]):
count = count +1
result = (count / size) * 100
return result
# The accuracy result between the network and the logic function itself.
accuracy = accuracyCalc(outputs, outputsCheck)
print('The accuracy of the model is: ' + str(accuracy))
# Part C
# Robust Weights
# w_rh = robust weights for hidden layer
# w_ro = robust weights for output layer
w_rh = np.array([[1, 0, 1, 1, -2.5],
[0, -1, 1, 1, -1.5],
[-1, 1, -1, 0, -0.5],
[-1, 1, 0, -1, -0.5]])
w_ro = np.array([1, 1, 1, 1, -0.5])
# Part D
# Generate 400 input samples by concatenating 25 samples from each input.
inputsWithNoise = np.zeros((400,5))
for k in range(25):
for i in range(np.shape(inputs)[0]):
inputsWithNoise[(k*16)+i,:] = inputs[i,:]
# Then check the outputs of the inputsWithNoise.
outputsCheck_D = logicFunction(inputsWithNoise[:,0:4])
# Create a gaussian noise with 0 mean and 0.2 std. Then add this noise to the inputsWithNoise array.
# np.random.seed(7) is for getting the same output each run.
np.random.seed(7)
gaussianNoise = np.random.normal(loc=0, scale=0.2, size=1600).reshape(400, 4)
inputsWithNoise[:, 0:4] += gaussianNoise
# Then test the inputsWithNoise with the random weights network.
outputTemp = unitStepFunction(inputsWithNoise.dot(hiddenweights.T))
bias = np.ones((400,1))
hiddenOutsTemp = np.concatenate((outputTemp, bias),axis =1)
random_network_output = unitStepFunction(hiddenOutsTemp.dot(outputweights.T))
# Then test the inputsWithNoise with the robust weights network.
robustOutputTemp = unitStepFunction(inputsWithNoise.dot(w_rh.T))
bias = np.ones((400,1))
robustHiddenOuts = np.concatenate((robustOutputTemp, bias),axis =1)
robust_network_output = unitStepFunction(robustHiddenOuts.dot(w_ro.T))
# Accuracy of the random weight network.
accuracy = accuracyCalc(random_network_output, outputsCheck_D)
print('The accuracy of the random weighted network is: ' + str(accuracy))
# Accuracy of the robust network.
accuracy = accuracyCalc(robust_network_output, outputsCheck_D)
print('The accuracy of the robust network is: ' + str(accuracy))
| 0.624064 | 0.904903 |
```
%matplotlib inline
"""
Use this notebook to calculate some simple 2D score plots like those in the paper. Distance normalization should be applied carefully; using different combinations of accounting for distance (normalization) can make a big difference in certain distributions of points, depending on label composition and pairwise distances. You can use use_denominator_distance and normalize_distance to remove different terms from the score calculation formula to see how those affect the scaling of the scores.
"""
import matplotlib
import matplotlib.pyplot as plt
import numpy as np
from sklearn.metrics import pairwise_distances
from score_calculation.border_scores import get_border_scores_precomputed_distance
def Generate_and_Split(examples_by_class, x_means, y_means,
x_spreads, y_spreads, test_set_proportion=.1):
lb = LabelBinarizer()
X, y_flat = generate_dataset(examples_by_class,
x_means, y_means, x_spreads, y_spreads)
y = lb.fit_transform(y_flat)
perm = np.random.permutation(X.shape[0])
X = X[perm]
y = y[perm]
y_flat = y_flat[perm]
ts = int(math.floor(X.shape[0] * test_set_proportion))
X_train = X[0:-1*(ts)]
y_train = y[0:-1*(ts)]
y_flat_train = y_flat[0:-1*(ts)]
X_test = X[-1*ts:]
y_test = y[-1*ts:]
y_flat_test = y_flat[-1*ts:]
return (X_train, y_train, y_flat_train,
X_test, y_test, y_flat_test)
def generate_dataset(examples_by_class, x_means, y_means, x_spreads, y_spreads):
X = []
y = []
for c in range(len(examples_by_class)):
for i in range(examples_by_class[c]):
x1 = np.random.normal(loc=x_means[c],scale=x_spreads[c])
x2 = np.random.normal(loc=y_means[c],scale=y_spreads[c])
X.append([x1,x2])
y.append(c)
X=np.array(X)
y=np.array(y)
return X, y
examples_by_class = [10,10,4]
x_means = [-1,1,0]
y_means = [1,1,-1]
x_spreads = [0.75,0.75,1.0]
y_spreads = [0.75,0.5,0.75]
def make_plot(X, y, intrts, threshold=0.75, xmin=-3, xmax=3, ymin=-2, ymax=3):
min_dot_size = 40
s = np.ones(len(X))
s *= min_dot_size
s = s + 80 * intrts
markers = ["^","o","+"]
colors = ["red","green","blue"]
f, ax1 = plt.subplots(1, figsize=(5,4), dpi=80, sharex=True, sharey=True)
f.tight_layout(rect=[0, 0.03, 1, 0.95])
ax1.set_xlim([xmin,xmax])
ax1.set_ylim([ymin,ymax])
ticklines = ax1.get_xticklines() + ax1.get_yticklines()
gridlines = ax1.get_xgridlines() + ax1.get_ygridlines()
for line in gridlines:
line.set_linestyle('-.')
for line in ticklines:
line.set_linewidth(3)
classes = np.unique(y)
for j in classes:
class_j = y == j
ax1.scatter(X[class_j][:,0],
X[class_j][:,1],
c=colors[j],
s=s[class_j],
marker=markers[j])
for i, bs in enumerate(intrts):
bs = float(format(bs, '.2f'))
if bs > threshold:
ax1.annotate(bs, (X[i][0] + 0.05, X[i][1] + 0.05), size=12)
start, end = ax1.get_xlim()
ax1.xaxis.set_ticks(np.arange(start, end, 1))
def plot_wrapper(X,y,k,
use_numerator_distance=True,
use_denominator_distance=True,
normalize_distance=True,
threshold=0.75,
xmin=-3, xmax=3, ymin=-2, ymax=3
):
D = pairwise_distances(X, metric="euclidean")
bs = get_border_scores_precomputed_distance(y, D, k, use_numerator_distance=use_numerator_distance,
use_denominator_distance=use_denominator_distance, normalize_distance=normalize_distance)
make_plot(X, y, bs, threshold=threshold,xmin=xmin,xmax=xmax,ymin=ymin,ymax=ymax)
k=5
X = np.array([
[0,0],
[-0.7,-0.5],
[-1.0, 1.0],
[0.9,0.7],
[0.7,-0.9],
])
y = np.array([
1,1,1,1,1
])
plot_wrapper(X, y, k, threshold=-0.1)
k=5
X = np.array([
[0,0],
[-0.7,-0.5],
[-1.0, 1.0],
[0.9,0.7],
[0.7,-0.9],
])
y = np.array([
0,1,1,1,1
])
D = pairwise_distances(X, metric="euclidean")
bs = get_border_scores_precomputed_distance(y, D, k)
intrs = get_interest_values(X,y,k)
fucking_plot(X, y, bs, threshold=0.5)
```
(1) A sample should have score 0 when all k − 1 neighbors are of the same class as the sample.
Check. Currently included in paper.
(2) Examples in highly heterogeneous neighborhoods (i.e., neighborhoods with a high number of classes present)
should have higher scores than points in homogeneous neighborhoods consisting of mostly their own class, but
lower scores than points in homogeneous neighborhoods consisting of points of mostly another class.
Check. See example below.
(3) Examples in relatively dense neighborhoods should have higher scores than points in relatively sparse neighborhoods, with label composition held constant.
```
# Point 2 above.
k=6
X = np.array([
[0,0],
[-0.7,-0.5],
[-1.0, 1.0],
[0.8,0.6],
[0.9,-0.9],
[0.2,-1.8],
[0,2.0]
])
y = np.array([
0,1,0,1,0,2,2
])
plot_wrapper(X, y, k, use_denominator_distance=False, normalize_distance=True, threshold=0.5)
y = np.array([
0,1,1,1,0,1,1
])
plot_wrapper(X, y, k, use_denominator_distance=False, normalize_distance=True, threshold=0.5)
# Point 3 above.
k=7
a=4
X = np.array([
[0,0],
[-0.7,-0.5],
[-1.0, 1.0],
[0.8,0.6],
[0.9,-0.9],
[0.2,-1.5],
[0,1.5]
])
y = np.array([
0,1,0,1,0,2,2
])
Z = a*X
plot_wrapper(Z, y, k, use_denominator_distance=False, normalize_distance=False, threshold=0.0,
xmin=np.min(Z)-1.0,xmax=np.max(Z)+1.0,ymin=np.min(Z)-1.0,ymax=np.max(Z)+1.0)
plot_wrapper(X, y, k, use_denominator_distance=False, normalize_distance=False, ymin=-3, threshold=0.0)
X[:,0] + 1
plot_wrapper(Z, y, k, use_denominator_distance=False, normalize_distance=True, threshold=0.0,
xmin=np.min(Z)-1.0,xmax=np.max(Z)+1.0,ymin=np.min(Z)-1.0,ymax=np.max(Z)+1.0)
```
|
github_jupyter
|
%matplotlib inline
"""
Use this notebook to calculate some simple 2D score plots like those in the paper. Distance normalization should be applied carefully; using different combinations of accounting for distance (normalization) can make a big difference in certain distributions of points, depending on label composition and pairwise distances. You can use use_denominator_distance and normalize_distance to remove different terms from the score calculation formula to see how those affect the scaling of the scores.
"""
import matplotlib
import matplotlib.pyplot as plt
import numpy as np
from sklearn.metrics import pairwise_distances
from score_calculation.border_scores import get_border_scores_precomputed_distance
def Generate_and_Split(examples_by_class, x_means, y_means,
x_spreads, y_spreads, test_set_proportion=.1):
lb = LabelBinarizer()
X, y_flat = generate_dataset(examples_by_class,
x_means, y_means, x_spreads, y_spreads)
y = lb.fit_transform(y_flat)
perm = np.random.permutation(X.shape[0])
X = X[perm]
y = y[perm]
y_flat = y_flat[perm]
ts = int(math.floor(X.shape[0] * test_set_proportion))
X_train = X[0:-1*(ts)]
y_train = y[0:-1*(ts)]
y_flat_train = y_flat[0:-1*(ts)]
X_test = X[-1*ts:]
y_test = y[-1*ts:]
y_flat_test = y_flat[-1*ts:]
return (X_train, y_train, y_flat_train,
X_test, y_test, y_flat_test)
def generate_dataset(examples_by_class, x_means, y_means, x_spreads, y_spreads):
X = []
y = []
for c in range(len(examples_by_class)):
for i in range(examples_by_class[c]):
x1 = np.random.normal(loc=x_means[c],scale=x_spreads[c])
x2 = np.random.normal(loc=y_means[c],scale=y_spreads[c])
X.append([x1,x2])
y.append(c)
X=np.array(X)
y=np.array(y)
return X, y
examples_by_class = [10,10,4]
x_means = [-1,1,0]
y_means = [1,1,-1]
x_spreads = [0.75,0.75,1.0]
y_spreads = [0.75,0.5,0.75]
def make_plot(X, y, intrts, threshold=0.75, xmin=-3, xmax=3, ymin=-2, ymax=3):
min_dot_size = 40
s = np.ones(len(X))
s *= min_dot_size
s = s + 80 * intrts
markers = ["^","o","+"]
colors = ["red","green","blue"]
f, ax1 = plt.subplots(1, figsize=(5,4), dpi=80, sharex=True, sharey=True)
f.tight_layout(rect=[0, 0.03, 1, 0.95])
ax1.set_xlim([xmin,xmax])
ax1.set_ylim([ymin,ymax])
ticklines = ax1.get_xticklines() + ax1.get_yticklines()
gridlines = ax1.get_xgridlines() + ax1.get_ygridlines()
for line in gridlines:
line.set_linestyle('-.')
for line in ticklines:
line.set_linewidth(3)
classes = np.unique(y)
for j in classes:
class_j = y == j
ax1.scatter(X[class_j][:,0],
X[class_j][:,1],
c=colors[j],
s=s[class_j],
marker=markers[j])
for i, bs in enumerate(intrts):
bs = float(format(bs, '.2f'))
if bs > threshold:
ax1.annotate(bs, (X[i][0] + 0.05, X[i][1] + 0.05), size=12)
start, end = ax1.get_xlim()
ax1.xaxis.set_ticks(np.arange(start, end, 1))
def plot_wrapper(X,y,k,
use_numerator_distance=True,
use_denominator_distance=True,
normalize_distance=True,
threshold=0.75,
xmin=-3, xmax=3, ymin=-2, ymax=3
):
D = pairwise_distances(X, metric="euclidean")
bs = get_border_scores_precomputed_distance(y, D, k, use_numerator_distance=use_numerator_distance,
use_denominator_distance=use_denominator_distance, normalize_distance=normalize_distance)
make_plot(X, y, bs, threshold=threshold,xmin=xmin,xmax=xmax,ymin=ymin,ymax=ymax)
k=5
X = np.array([
[0,0],
[-0.7,-0.5],
[-1.0, 1.0],
[0.9,0.7],
[0.7,-0.9],
])
y = np.array([
1,1,1,1,1
])
plot_wrapper(X, y, k, threshold=-0.1)
k=5
X = np.array([
[0,0],
[-0.7,-0.5],
[-1.0, 1.0],
[0.9,0.7],
[0.7,-0.9],
])
y = np.array([
0,1,1,1,1
])
D = pairwise_distances(X, metric="euclidean")
bs = get_border_scores_precomputed_distance(y, D, k)
intrs = get_interest_values(X,y,k)
fucking_plot(X, y, bs, threshold=0.5)
# Point 2 above.
k=6
X = np.array([
[0,0],
[-0.7,-0.5],
[-1.0, 1.0],
[0.8,0.6],
[0.9,-0.9],
[0.2,-1.8],
[0,2.0]
])
y = np.array([
0,1,0,1,0,2,2
])
plot_wrapper(X, y, k, use_denominator_distance=False, normalize_distance=True, threshold=0.5)
y = np.array([
0,1,1,1,0,1,1
])
plot_wrapper(X, y, k, use_denominator_distance=False, normalize_distance=True, threshold=0.5)
# Point 3 above.
k=7
a=4
X = np.array([
[0,0],
[-0.7,-0.5],
[-1.0, 1.0],
[0.8,0.6],
[0.9,-0.9],
[0.2,-1.5],
[0,1.5]
])
y = np.array([
0,1,0,1,0,2,2
])
Z = a*X
plot_wrapper(Z, y, k, use_denominator_distance=False, normalize_distance=False, threshold=0.0,
xmin=np.min(Z)-1.0,xmax=np.max(Z)+1.0,ymin=np.min(Z)-1.0,ymax=np.max(Z)+1.0)
plot_wrapper(X, y, k, use_denominator_distance=False, normalize_distance=False, ymin=-3, threshold=0.0)
X[:,0] + 1
plot_wrapper(Z, y, k, use_denominator_distance=False, normalize_distance=True, threshold=0.0,
xmin=np.min(Z)-1.0,xmax=np.max(Z)+1.0,ymin=np.min(Z)-1.0,ymax=np.max(Z)+1.0)
| 0.838581 | 0.889481 |
# Supervised machine learning and regression analysis
1. Import libraries
2. Wrangle & clean data
3. Prepare data for regression analysis
4. Regression analysis
# 1. Import libraries
```
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib
import matplotlib.pyplot as plt
import os
import sklearn
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LinearRegression
from sklearn.metrics import mean_squared_error, r2_score
# Display graphs without need to call
%matplotlib inline
# Import data
df = pd.read_csv(r'/Users/OldBobJulia/Desktop/CF/Course/6. Advanced Analytics and Dashboard Design/Berlin Airbnb Analysis/02 Data/Prepared data/listing_derivedcolumns.csv')
df.head()
# Drop Unnamed
df = df.drop(columns = ['Unnamed: 0'])
df.head()
df.shape
```
# 2. Wrangle & clean data
```
# Check columns
df.columns
# Check for missings
df.isnull().sum()
```
Reviews_per_month has 4155 missing values which I decide to leave bc them being missing might be valuable information.
```
# Check dups
dups = df.duplicated()
dups.shape
# Check for outliers
sns.distplot(df['price'], bins=5)
df.describe()
```
# 3. Prepare data for regression analysis
Check which variables have a linear relationship using scatterplots, to see which ones are suitable for regression analysis
```
# Create scatterplot minimum_nights vs price
df.plot(x = 'minimum_nights', y='price',style='o')
plt.title('Minimum_nights vs price per night')
plt.xlabel('Minimum nights')
plt.ylabel('Price per night')
plt.show()
```
There is a tendency but it's not linear. There's a concentration of data points at the low end of minimum nights. The price decreases as the number of minimum nights increase.
```
# Create scatterplot number_of_reviews vs price
df.plot(x = 'number_of_reviews', y='price',style='o')
plt.title('Number of reviews vs price per night')
plt.xlabel('Number of reviews')
plt.ylabel('Price per night')
plt.show()
```
This relationship is not linear, either
```
# Create scatter plot Number of reviews vs minimum nights. Do minimum_nights have an impact on number of reviews?
df.plot(x = 'minimum_nights', y='number_of_reviews',style='o')
plt.title('Number of reviews vs minimum nights')
plt.xlabel('Minimum nights')
plt.ylabel('Number of reviews')
plt.show()
```
This relationship looks somewhat linear.
Hypothesis: The lower the minimum nights, the higher the number of reviews.
```
# Create scatter plot price vs calculated_host_listings_count
df.plot(x = 'calculated_host_listings_count', y='price',style='o')
plt.title('Price vs count of host listings')
plt.xlabel('Count of host listings')
plt.ylabel('Price')
plt.show()
```
This relationship could be linear.
```
# Create scatter plot availability vs calculated_host_listings_count.
df.plot(x = 'calculated_host_listings_count', y='availability_365',style='o')
plt.title('Availability vs Count of host listings')
plt.xlabel('Count of host listings')
plt.ylabel('Availability 365')
plt.show()
# Create scatter plot calculated_host_listings count vs minimum nights.
df.plot(x = 'calculated_host_listings_count', y='minimum_nights',style='o')
plt.title('Minimum nights vs Count of host listings')
plt.xlabel('Count of host listings')
plt.ylabel('Minimum nights')
plt.show()
```
This relationship could be linear as well.
```
# Create scatter plot Availability 365 vs price.
df.plot(x = 'availability_365', y='price',style='o')
plt.title('Availability 365 vs price')
plt.xlabel('Availability 365')
plt.ylabel('Price')
plt.show()
# Create scatter plot Number of reviews vs reviews per month.
df.plot(x = 'reviews_per_month', y='number_of_reviews',style='o')
plt.title('Number of reviews vs reviews per month')
plt.xlabel('Reviews per month')
plt.ylabel('Number of reviews')
plt.show()
```
**Of all these scatterplots linear of all these scatter plots is Price vs Count of host listings. I will proceed with regression analysis with those variables**
```
# Reshape the variables, put them into separate objects.
X = df['calculated_host_listings_count'].values.reshape(-1,1)
y = df['price'].values.reshape(-1,1)
X
y
# Split into training and test set
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state=0)
```
# 4. Regression analysis
```
# Create regression object
regression = LinearRegression()
# Fit it onto training set
regression.fit(X_train, y_train)
# Use trained regression object to predict y using X in test set
y_predicted = regression.predict(X_test)
# Create plot showing regression line (results) from the model on the test set
plot_test = plt
plot_test.scatter(X_test, y_test, color='gray', s = 15)
plot_test.plot(X_test, y_predicted, color='red', linewidth =3)
plot_test.title('Count of host listings vs Price (Test set)')
plot_test.xlabel('Count of host listings')
plot_test.ylabel('Price')
plot_test.show()
# Create objects that contain model summary statistics
rmse = mean_squared_error(y_test, y_predicted)
r2 = r2_score(y_test, y_predicted)
# Check model summary statistics to evaluate performance of model
print('Slope:' ,regression.coef_)
print('Mean squared error: ', rmse)
print('R2 score: ', r2)
```
The slope coefficent is positive indicating that as X (Count of host listings) increases, y (Price) also increases.
The MSE is quite big, pointing out that the line is not very accurate.
The R2 score is also pretty low, indicating a relatively poor fit of the model to the data. The linear relationship between the variables is not strong, so a non-linear model to test this relationship would be a better fit.
```
# Create a dataframe to compare the actual and predicted values of y
data = pd.DataFrame({'Actual': y_test.flatten(), 'Predicted': y_predicted.flatten()})
data.head(30)
```
This confirms that the model doesn't do great at predicting y.
```
# Compare how regression fits training set. How well does model predict y in the training set?
y_predicted_train = regression.predict(X_train)
rmse = mean_squared_error(y_train, y_predicted_train)
r2 = r2_score(y_train, y_predicted_train)
print('Slope:' ,regression.coef_)
print('Mean squared error: ', rmse)
print('R2 score: ', r2)
```
When we apply the model back to the training set the results aren't so good, either.
MSE remains large while R2 score is very low.
```
# Visualize it
plot_test = plt
plot_test.scatter(X_train, y_train, color='green', s = 15)
plot_test.plot(X_train, y_predicted_train, color='red', linewidth =3)
plot_test.title('Count of host listings vs price (Test set)')
plot_test.xlabel('Count of host listings')
plot_test.ylabel('Price')
plot_test.show()
```
|
github_jupyter
|
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib
import matplotlib.pyplot as plt
import os
import sklearn
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LinearRegression
from sklearn.metrics import mean_squared_error, r2_score
# Display graphs without need to call
%matplotlib inline
# Import data
df = pd.read_csv(r'/Users/OldBobJulia/Desktop/CF/Course/6. Advanced Analytics and Dashboard Design/Berlin Airbnb Analysis/02 Data/Prepared data/listing_derivedcolumns.csv')
df.head()
# Drop Unnamed
df = df.drop(columns = ['Unnamed: 0'])
df.head()
df.shape
# Check columns
df.columns
# Check for missings
df.isnull().sum()
# Check dups
dups = df.duplicated()
dups.shape
# Check for outliers
sns.distplot(df['price'], bins=5)
df.describe()
# Create scatterplot minimum_nights vs price
df.plot(x = 'minimum_nights', y='price',style='o')
plt.title('Minimum_nights vs price per night')
plt.xlabel('Minimum nights')
plt.ylabel('Price per night')
plt.show()
# Create scatterplot number_of_reviews vs price
df.plot(x = 'number_of_reviews', y='price',style='o')
plt.title('Number of reviews vs price per night')
plt.xlabel('Number of reviews')
plt.ylabel('Price per night')
plt.show()
# Create scatter plot Number of reviews vs minimum nights. Do minimum_nights have an impact on number of reviews?
df.plot(x = 'minimum_nights', y='number_of_reviews',style='o')
plt.title('Number of reviews vs minimum nights')
plt.xlabel('Minimum nights')
plt.ylabel('Number of reviews')
plt.show()
# Create scatter plot price vs calculated_host_listings_count
df.plot(x = 'calculated_host_listings_count', y='price',style='o')
plt.title('Price vs count of host listings')
plt.xlabel('Count of host listings')
plt.ylabel('Price')
plt.show()
# Create scatter plot availability vs calculated_host_listings_count.
df.plot(x = 'calculated_host_listings_count', y='availability_365',style='o')
plt.title('Availability vs Count of host listings')
plt.xlabel('Count of host listings')
plt.ylabel('Availability 365')
plt.show()
# Create scatter plot calculated_host_listings count vs minimum nights.
df.plot(x = 'calculated_host_listings_count', y='minimum_nights',style='o')
plt.title('Minimum nights vs Count of host listings')
plt.xlabel('Count of host listings')
plt.ylabel('Minimum nights')
plt.show()
# Create scatter plot Availability 365 vs price.
df.plot(x = 'availability_365', y='price',style='o')
plt.title('Availability 365 vs price')
plt.xlabel('Availability 365')
plt.ylabel('Price')
plt.show()
# Create scatter plot Number of reviews vs reviews per month.
df.plot(x = 'reviews_per_month', y='number_of_reviews',style='o')
plt.title('Number of reviews vs reviews per month')
plt.xlabel('Reviews per month')
plt.ylabel('Number of reviews')
plt.show()
# Reshape the variables, put them into separate objects.
X = df['calculated_host_listings_count'].values.reshape(-1,1)
y = df['price'].values.reshape(-1,1)
X
y
# Split into training and test set
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state=0)
# Create regression object
regression = LinearRegression()
# Fit it onto training set
regression.fit(X_train, y_train)
# Use trained regression object to predict y using X in test set
y_predicted = regression.predict(X_test)
# Create plot showing regression line (results) from the model on the test set
plot_test = plt
plot_test.scatter(X_test, y_test, color='gray', s = 15)
plot_test.plot(X_test, y_predicted, color='red', linewidth =3)
plot_test.title('Count of host listings vs Price (Test set)')
plot_test.xlabel('Count of host listings')
plot_test.ylabel('Price')
plot_test.show()
# Create objects that contain model summary statistics
rmse = mean_squared_error(y_test, y_predicted)
r2 = r2_score(y_test, y_predicted)
# Check model summary statistics to evaluate performance of model
print('Slope:' ,regression.coef_)
print('Mean squared error: ', rmse)
print('R2 score: ', r2)
# Create a dataframe to compare the actual and predicted values of y
data = pd.DataFrame({'Actual': y_test.flatten(), 'Predicted': y_predicted.flatten()})
data.head(30)
# Compare how regression fits training set. How well does model predict y in the training set?
y_predicted_train = regression.predict(X_train)
rmse = mean_squared_error(y_train, y_predicted_train)
r2 = r2_score(y_train, y_predicted_train)
print('Slope:' ,regression.coef_)
print('Mean squared error: ', rmse)
print('R2 score: ', r2)
# Visualize it
plot_test = plt
plot_test.scatter(X_train, y_train, color='green', s = 15)
plot_test.plot(X_train, y_predicted_train, color='red', linewidth =3)
plot_test.title('Count of host listings vs price (Test set)')
plot_test.xlabel('Count of host listings')
plot_test.ylabel('Price')
plot_test.show()
| 0.723798 | 0.965512 |
# Introduction to Fraud Detection Systems
Fraud detection is one of the top priorities for banks and financial institutions, which can be addressed using machine learning. According to [a report published by Nilson](https://nilsonreport.com/upload/content_promo/The_Nilson_Report_Issue_1118.pdf), in 2017 the worldwide losses in card fraud related cases reached 22.8 billion dollars. The problem is forecasted to get worse in the following years, by 2021, the card fraud bill is expected to be 32.96 billion dollars.
In this tutorial, we will use the [credit card fraud detection dataset](https://www.kaggle.com/mlg-ulb/creditcardfraud) from Kaggle, to identify fraud cases. We will use a [gradient boosted tree](https://blogs.technet.microsoft.com/machinelearning/2017/07/25/lessons-learned-benchmarking-fast-machine-learning-algorithms/) as a machine learning algorithm. And finally, we will create a simple API to operationalize (o16n) the model.
We will use the gradient boosting library [LightGBM](https://github.com/Microsoft/LightGBM), which has recently became one of the most popular libraries for top participants in [Kaggle competitions](https://github.com/Microsoft/LightGBM/tree/a39c848e6456d473d2043dff3f5159945a36b567/examples).
Fraud detection problems are known for being extremely imbalanced. <a href="https://en.wikipedia.org/wiki/Boosting_(machine_learning)">Boosting</a> is one technique that usually works well with these kind of datasets. It iteratively creates weak classifiers (decision trees) weighting the instances to increase the performance. In the first subset, a weak classifier is trained and tested on all the training data, those instances that have bad performance are weighted to appear more in the next data subset. Finally, all the classifiers are ensembled with a weighted average of their estimates.
In LightGBM, there is a [parameter](https://github.com/Microsoft/LightGBM/blob/master/docs/Parameters.rst#objective-parameters) called `is_unbalanced` that automatically helps you to control this issue.
LigtGBM can be used with or without [GPU](https://lightgbm.readthedocs.io/en/latest/GPU-Performance.html). For small datasets, like the one we are using here, it is faster to use CPU, due to IO overhead. However, I wanted to showcase the GPU alternative, which is trickier to install, in case anyone wants to experiment with bigger datasets.
To install the dependencies in Linux:
```bash
$ sudo apt-get update
$ sudo apt-get install cmake build-essential libboost-all-dev -y
$ conda env create -n fraud -f conda.yaml
$ source activate fraud
(fraud)$ python -m ipykernel install --user --name fraud --display-name "Python (fraud)"
```
```
import numpy as np
import sys
import os
import json
import pandas as pd
from collections import Counter
import requests
from IPython.core.display import display, HTML
import lightgbm as lgb
import sklearn
import aiohttp
import asyncio
from utils import (split_train_test, classification_metrics_binary, classification_metrics_binary_prob,
binarize_prediction, plot_confusion_matrix, run_load_test, read_from_sqlite)
from utils import BASELINE_MODEL, PORT, TABLE_FRAUD, TABLE_LOCATIONS, DATABASE_FILE
print("System version: {}".format(sys.version))
print("Numpy version: {}".format(np.__version__))
print("Pandas version: {}".format(pd.__version__))
print("LightGBM version: {}".format(lgb.__version__))
print("Sklearn version: {}".format(sklearn.__version__))
%load_ext autoreload
%autoreload 2
```
## Dataset
The first step is to load the dataset and analyze it.
For it, before continuing, **you have to run the notebook [data_prep.ipynb](data_prep.ipynb)**, which will generate the SQLite database.
```
query = 'SELECT * FROM ' + TABLE_FRAUD
df = read_from_sqlite(DATABASE_FILE, query)
print("Shape: {}".format(df.shape))
df.head()
```
As we can see, the dataset is extremely imbalanced. The minority class counts for around 0.002% of the examples.
```
df['Class'].value_counts()
df['Class'].value_counts(normalize=True)
```
The next step is to split the dataset into train and test.
```
X_train, X_test, y_train, y_test = split_train_test(df.drop('Class', axis=1), df['Class'], test_size=0.2)
print(X_train.shape)
print(X_test.shape)
print(y_train.shape)
print(y_test.shape)
print(y_train.value_counts())
print(y_train.value_counts(normalize=True))
print(y_test.value_counts())
print(y_test.value_counts(normalize=True))
```
## Training with LightGBM - Baseline
For this task we use a simple set of parameters to train the model. We just want to create a baseline model, so we are not performing here cross validation or parameter tunning.
The details of the different parameters of LightGBM can be found in the [documentation](https://github.com/Microsoft/LightGBM/blob/master/docs/Parameters.rst). Also, the authors provide [some advices](https://github.com/Microsoft/LightGBM/blob/master/docs/Parameters-Tuning.rst) on how to tune the parameters and prevent overfitting.
```
lgb_train = lgb.Dataset(X_train, y_train, free_raw_data=False)
lgb_test = lgb.Dataset(X_test, y_test, reference=lgb_train, free_raw_data=False)
parameters = {'num_leaves': 2**8,
'learning_rate': 0.1,
'is_unbalance': True,
'min_split_gain': 0.1,
'min_child_weight': 1,
'reg_lambda': 1,
'subsample': 1,
'objective':'binary',
#'device': 'gpu', # comment this line if you are not using GPU
'task': 'train'
}
num_rounds = 300
%%time
clf = lgb.train(parameters, lgb_train, num_boost_round=num_rounds)
```
Once we have the trained model, we can obtain some metrics.
```
y_prob = clf.predict(X_test)
y_pred = binarize_prediction(y_prob, threshold=0.5)
metrics = classification_metrics_binary(y_test, y_pred)
metrics2 = classification_metrics_binary_prob(y_test, y_prob)
metrics.update(metrics2)
cm = metrics['Confusion Matrix']
metrics.pop('Confusion Matrix', None)
print(json.dumps(metrics, indent=4, sort_keys=True))
plot_confusion_matrix(cm, ['no fraud (negative class)', 'fraud (positive class)'])
```
In business terms, if the system classifies a fair transaction as fraud (false positive), the bank will investigate the issue probably using human intervention. According to a [2015 report from Javelin Strategy](https://www.javelinstrategy.com/press-release/false-positive-card-declines-push-consumers-abandon-issuers-and-merchants#), 15% of all cardholders have had at least one transaction incorrectly declined in the previous year, representing an annual decline amount of almost $118 billion. Nearly 4 in 10 declined cardholders report that they abandoned their card after being falsely declined.
However, if a fraudulent transaction is not detected, effectively meaning that the classifier predicts that a transaction is fair when it is really fraudulent (false negative), then the bank is losing money and the bad guy is getting away with it.
A common way to use business rules in these predictions is to control the threshold or operation point of the prediction. This can be controlled changing the threshold value in `binarize_prediction(y_prob, threshold=0.5)`. It is common to do a loop from 0.1 to 0.9 and evaluate the different business outcomes.
```
clf.save_model(BASELINE_MODEL)
```
## O16N with Flask and Websockets
The next step is to operationalize (o16n) the machine learning model. For it, we are going to use [Flask](http://flask.pocoo.org/) to create a RESTfull API. The input of the API is going to be a transaction (defined by its features), and the output, the model prediction.
Aditionally, we designed a [websocket service](https://miguelgfierro.com/blog/2018/demystifying-websockets-for-real-time-web-communication/) to visualize fraudulent transactions on a map. The system works in real time using the library [flask-socketio](https://github.com/miguelgrinberg/Flask-SocketIO).
When a new transaction is sent to the API, the LightGBM model predicts whether the transaction is fair or fraudulent. If the transaction is fraudulent, the server sends a signal to the a web client, that renders a world map showing the location of the fraudulent transaction. The map is made with javascript using [amCharts](http://amcharts.com/) and the map locations are taken from the previously created SQLite database.
To start the api execute `(fraud)$ python api.py` inside the conda environment.
```
# You can also run the api from inside the notebook (even though I find it more difficult for debugging).
# To do it, just uncomment the next two lines:
#%%bash --bg --proc bg_proc
#python api.py
```
First, we make sure that the API is on
```
#server_name = 'http://the-name-of-your-server'
server_name = 'http://localhost'
root_url = '{}:{}'.format(server_name, PORT)
res = requests.get(root_url)
display(HTML(res.text))
```
Now, we are going to select one value and predict the output.
```
vals = y_test[y_test == 1].index.values
X_target = X_test.loc[vals[0]]
dict_query = X_target.to_dict()
print(dict_query)
headers = {'Content-type':'application/json'}
end_point = root_url + '/predict'
res = requests.post(end_point, data=json.dumps(dict_query), headers=headers)
print(res.ok)
print(json.dumps(res.json(), indent=2))
```
### Fraudulent transaction visualization
Now that we know that the main end point of the API works, we will try the `/predict_map` end point. It creates a real time visualization system for fraudulent transactions using websockets.
A websocket is a protocol intended for real-time communications developed for the HTML5 specification. It creates a persistent, low latency connection that can support transactions initiated by either the client or server. [In this post](https://miguelgfierro.com/blog/2018/demystifying-websockets-for-real-time-web-communication/) you can find a detailed explanation of websockets and other related technologies.
<img src="https://miguelgfierro.com/img/upload/2018/07/12/websocket_architecture2.svg?sanitize=true">
For our case, whenever a user makes a request to the end point `/predict_map`, the machine learning model evaluates the transaction details and makes a prediction. If the prediction is classified as fraudulent, the server sends a signal using `socketio.emit('map_update', location)`. This signal just contains a dictionary, called `location`, with a simulated name and location of where the fraudulent transaction occurred. The signal is shown in `index.html`, which just renders some javascript code that is referenced via an `id="chartdiv"`.
The javascript code is defined in the file `frauddetection.js`. The websocket part is the following:
```js
var mapLocations = [];
// Location updated emitted by the server via websockets
socket.on("map_update", function (msg) {
var message = "New event in " + msg.title + " (" + msg.latitude
+ "," + msg.longitude + ")";
console.log(message);
var newLocation = new Location(msg.title, msg.latitude, msg.longitude);
mapLocations.push(newLocation);
//clear the markers before redrawing
mapLocations.forEach(function(location) {
if (location.externalElement) {
location.externalElement = undefined;
}
});
map.dataProvider.images = mapLocations;
map.validateData(); //call to redraw the map with new data
});
```
When a new signal is emited from the server in python, the javascript code receives it and processes it. It creates a new variable called `newLocation` containing the location information, that is going to be saved in a global array called `mapLocations`. This variable contains all the fradulent locations that appeared since the session started. Then there is a clearing process for amCharts to be able to draw the new information in the map and finally the array is stored in `map.dataProvider.images`, which actually refresh the map with the new point. The variable `map` is set earlier in the code and it is the amCharts object responsible for defining the map.
To make a query to the visualization end point:
```
headers = {'Content-type':'application/json'}
end_point_map = root_url + '/predict_map'
res = requests.post(end_point_map, data=json.dumps(dict_query), headers=headers)
print(res.text)
```
Now you can go the map url (in local it would be http://localhost:5000/map) and see how the map is reshesed with a new fraudulent location every time you execute the previous cell.
You should see a map like the following one:
[](https://youtu.be/KiCeeJAlgJU)
## Load test
Once we have the API, we can test its scalability and response time.
Here you can find a simple load test to evaluate the performance of your API. Please bear in mind that, in this case, there is no request overhead due to the different locations of client and server, since the client and server are the same computer.
The response time of 10 requests is around 300ms, so one request would be 30ms.
```
num = 10
concurrent = 2
verbose = True
payload_list = [dict_query]*num
%%time
with aiohttp.ClientSession() as session: # We create a persistent connection
loop = asyncio.get_event_loop()
calc_routes = loop.run_until_complete(run_load_test(end_point, payload_list, session, concurrent, verbose))
# If you run the API from the notebook, you can uncomment the following two lines to kill the process
#%%bash
#ps aux | grep 'api.py' | grep -v 'grep' | awk '{print $2}' | xargs kill
```
## Enterprise grade reference architecture for fraud detection
In this tutorial we have seen how to create a baseline fraud detection model. However, for a big company this is not enough.
In the next figure we can see a reference architecture for fraud detection, that should be adapted to the customer specifics. All services are based on Azure.
1) Two general data sources for the customer: real time data and static information.
2) A general database piece to store the data. Since it is a reference architecture, and without more data, I put several options together ([SQL Database](https://azure.microsoft.com/en-gb/services/sql-database/), [CosmosDB](https://azure.microsoft.com/en-gb/services/cosmos-db/), [SQL Data Warehouse](https://azure.microsoft.com/en-gb/services/sql-data-warehouse/), etc) on cloud or on premise.
3) Model experimentation using [Azure ML](https://azure.microsoft.com/en-gb/overview/machine-learning/), again, using general computation targets such as [DSVM](https://azure.microsoft.com/en-gb/services/virtual-machines/data-science-virtual-machines/), [BatchAI](https://azure.microsoft.com/en-gb/services/batch-ai/), [Databricks](https://azure.microsoft.com/en-gb/services/databricks/) or [HDInsight](https://azure.microsoft.com/en-gb/services/hdinsight/).
4) Model retraining using new data and a model obtained from the [Model Management](https://docs.microsoft.com/en-gb/azure/machine-learning/desktop-workbench/model-management-overview).
5) Operationalization layer with a [Kubernetes cluster](https://azure.microsoft.com/en-gb/services/container-service/kubernetes/), which takes the best model and put it in production.
6) Reporting layer to show the results.
<img src="https://raw.githubusercontent.com/miguelgfierro/sciblog_support/master/Intro_to_Fraud_Detection/templates/fraud_detection_reference_architecture.svg?sanitize=true">
|
github_jupyter
|
$ sudo apt-get update
$ sudo apt-get install cmake build-essential libboost-all-dev -y
$ conda env create -n fraud -f conda.yaml
$ source activate fraud
(fraud)$ python -m ipykernel install --user --name fraud --display-name "Python (fraud)"
import numpy as np
import sys
import os
import json
import pandas as pd
from collections import Counter
import requests
from IPython.core.display import display, HTML
import lightgbm as lgb
import sklearn
import aiohttp
import asyncio
from utils import (split_train_test, classification_metrics_binary, classification_metrics_binary_prob,
binarize_prediction, plot_confusion_matrix, run_load_test, read_from_sqlite)
from utils import BASELINE_MODEL, PORT, TABLE_FRAUD, TABLE_LOCATIONS, DATABASE_FILE
print("System version: {}".format(sys.version))
print("Numpy version: {}".format(np.__version__))
print("Pandas version: {}".format(pd.__version__))
print("LightGBM version: {}".format(lgb.__version__))
print("Sklearn version: {}".format(sklearn.__version__))
%load_ext autoreload
%autoreload 2
query = 'SELECT * FROM ' + TABLE_FRAUD
df = read_from_sqlite(DATABASE_FILE, query)
print("Shape: {}".format(df.shape))
df.head()
df['Class'].value_counts()
df['Class'].value_counts(normalize=True)
X_train, X_test, y_train, y_test = split_train_test(df.drop('Class', axis=1), df['Class'], test_size=0.2)
print(X_train.shape)
print(X_test.shape)
print(y_train.shape)
print(y_test.shape)
print(y_train.value_counts())
print(y_train.value_counts(normalize=True))
print(y_test.value_counts())
print(y_test.value_counts(normalize=True))
lgb_train = lgb.Dataset(X_train, y_train, free_raw_data=False)
lgb_test = lgb.Dataset(X_test, y_test, reference=lgb_train, free_raw_data=False)
parameters = {'num_leaves': 2**8,
'learning_rate': 0.1,
'is_unbalance': True,
'min_split_gain': 0.1,
'min_child_weight': 1,
'reg_lambda': 1,
'subsample': 1,
'objective':'binary',
#'device': 'gpu', # comment this line if you are not using GPU
'task': 'train'
}
num_rounds = 300
%%time
clf = lgb.train(parameters, lgb_train, num_boost_round=num_rounds)
y_prob = clf.predict(X_test)
y_pred = binarize_prediction(y_prob, threshold=0.5)
metrics = classification_metrics_binary(y_test, y_pred)
metrics2 = classification_metrics_binary_prob(y_test, y_prob)
metrics.update(metrics2)
cm = metrics['Confusion Matrix']
metrics.pop('Confusion Matrix', None)
print(json.dumps(metrics, indent=4, sort_keys=True))
plot_confusion_matrix(cm, ['no fraud (negative class)', 'fraud (positive class)'])
clf.save_model(BASELINE_MODEL)
# You can also run the api from inside the notebook (even though I find it more difficult for debugging).
# To do it, just uncomment the next two lines:
#%%bash --bg --proc bg_proc
#python api.py
#server_name = 'http://the-name-of-your-server'
server_name = 'http://localhost'
root_url = '{}:{}'.format(server_name, PORT)
res = requests.get(root_url)
display(HTML(res.text))
vals = y_test[y_test == 1].index.values
X_target = X_test.loc[vals[0]]
dict_query = X_target.to_dict()
print(dict_query)
headers = {'Content-type':'application/json'}
end_point = root_url + '/predict'
res = requests.post(end_point, data=json.dumps(dict_query), headers=headers)
print(res.ok)
print(json.dumps(res.json(), indent=2))
var mapLocations = [];
// Location updated emitted by the server via websockets
socket.on("map_update", function (msg) {
var message = "New event in " + msg.title + " (" + msg.latitude
+ "," + msg.longitude + ")";
console.log(message);
var newLocation = new Location(msg.title, msg.latitude, msg.longitude);
mapLocations.push(newLocation);
//clear the markers before redrawing
mapLocations.forEach(function(location) {
if (location.externalElement) {
location.externalElement = undefined;
}
});
map.dataProvider.images = mapLocations;
map.validateData(); //call to redraw the map with new data
});
headers = {'Content-type':'application/json'}
end_point_map = root_url + '/predict_map'
res = requests.post(end_point_map, data=json.dumps(dict_query), headers=headers)
print(res.text)
num = 10
concurrent = 2
verbose = True
payload_list = [dict_query]*num
%%time
with aiohttp.ClientSession() as session: # We create a persistent connection
loop = asyncio.get_event_loop()
calc_routes = loop.run_until_complete(run_load_test(end_point, payload_list, session, concurrent, verbose))
# If you run the API from the notebook, you can uncomment the following two lines to kill the process
#%%bash
#ps aux | grep 'api.py' | grep -v 'grep' | awk '{print $2}' | xargs kill
| 0.240953 | 0.980599 |
# Generate VASP Inputs for Structure Relaxation
```
from pymatgen import Structure
from pymatgen.io.vasp.sets import MPRelaxSet
s = Structure.from_file("ICSD_182730_Si.cif", primitive=True)
custom_settings = {"NELMIN": 5} # user custom incar settings
relax = MPRelaxSet(s, user_incar_settings=custom_settings)
relax.write_input("Si-relax")
```
## Get total energy and relaxed structure
```
from pymatgen.io.vasp import Vasprun
v = Vasprun("Si-relax/vasprun.xml")
print(v.final_energy) # final total energy
s = v.final_structure
s.to(filename="Si-relax.cif") # save relaxed structure into cif file
print(s) # relaxed structure
```
# Generate VASP Input for Static Run
```
from pymatgen.io.vasp.sets import MPStaticSet
custom_settings = {"NELM": 60} # user custom incar settings
static = MPStaticSet.from_prev_calc("Si-relax/", standardize=True,
user_incar_settings=custom_settings)
static.write_input("Si-static")
```
## Get total energy from static run
```
from pymatgen.io.vasp import Vasprun
v = Vasprun("Si-static/vasprun.xml")
print(v.final_energy) # final total energy
```
# Generate VASP Input for Density of State(DOS) and Band Structure(BS) calculation
```
from pymatgen.io.vasp.sets import MPNonSCFSet
# generate uniform k-points for DOS calc.
custom_settings = {"LAECHG": "False", "LVHAR": "False"} # user custom incar settings
dos = MPNonSCFSet.from_prev_calc("Si-static/", mode="uniform",reciprocal_density=200,
user_incar_settings=custom_settings)
dos.write_input("Si-dos")
# generate k-points along high symmetry line for band structure calc.
band = MPNonSCFSet.from_prev_calc("Si-static/", mode="line", standardize=True,
user_incar_settings=custom_settings)
band.write_input("Si-band")
```
## Plot Total DOS
```
%matplotlib inline
from pymatgen.io.vasp import Vasprun
from pymatgen.electronic_structure.plotter import DosPlotter
v = Vasprun('Si-dos/vasprun.xml')
tdos = v.tdos
plotter = DosPlotter()
plotter.add_dos("Total DOS", tdos)
plotter.show(xlim=[-5, 5], ylim=[0, 4])
```
## Plot element-projected DOS
```
%matplotlib inline
from pymatgen.io.vasp import Vasprun
from pymatgen.electronic_structure.plotter import DosPlotter
v = Vasprun('Si-dos/vasprun.xml')
cdos = v.complete_dos
element_dos = cdos.get_element_dos()
plotter = DosPlotter()
plotter.add_dos_dict(element_dos)
plotter.show(xlim=[-5, 5], ylim=[0, 1])
```
## Plot orbital-projected DOS
```
%matplotlib inline
from pymatgen.io.vasp import Vasprun
from pymatgen.electronic_structure.plotter import DosPlotter
v = Vasprun('Si-dos/vasprun.xml')
cdos = v.complete_dos
spd_dos = cdos.get_spd_dos()
plotter = DosPlotter()
plotter.add_dos_dict(spd_dos)
plotter.show(xlim=[-5, 5], ylim=[0, 1])
```
# Plot BS
```
%matplotlib inline
from pymatgen.io.vasp import Vasprun, BSVasprun
from pymatgen.electronic_structure.plotter import BSPlotter
v = BSVasprun("Si-band/vasprun.xml")
bs = v.get_band_structure(kpoints_filename="Si-band/KPOINTS",line_mode=True)
plt = BSPlotter(bs)
plt.get_plot(vbm_cbm_marker=True)
```
|
github_jupyter
|
from pymatgen import Structure
from pymatgen.io.vasp.sets import MPRelaxSet
s = Structure.from_file("ICSD_182730_Si.cif", primitive=True)
custom_settings = {"NELMIN": 5} # user custom incar settings
relax = MPRelaxSet(s, user_incar_settings=custom_settings)
relax.write_input("Si-relax")
from pymatgen.io.vasp import Vasprun
v = Vasprun("Si-relax/vasprun.xml")
print(v.final_energy) # final total energy
s = v.final_structure
s.to(filename="Si-relax.cif") # save relaxed structure into cif file
print(s) # relaxed structure
from pymatgen.io.vasp.sets import MPStaticSet
custom_settings = {"NELM": 60} # user custom incar settings
static = MPStaticSet.from_prev_calc("Si-relax/", standardize=True,
user_incar_settings=custom_settings)
static.write_input("Si-static")
from pymatgen.io.vasp import Vasprun
v = Vasprun("Si-static/vasprun.xml")
print(v.final_energy) # final total energy
from pymatgen.io.vasp.sets import MPNonSCFSet
# generate uniform k-points for DOS calc.
custom_settings = {"LAECHG": "False", "LVHAR": "False"} # user custom incar settings
dos = MPNonSCFSet.from_prev_calc("Si-static/", mode="uniform",reciprocal_density=200,
user_incar_settings=custom_settings)
dos.write_input("Si-dos")
# generate k-points along high symmetry line for band structure calc.
band = MPNonSCFSet.from_prev_calc("Si-static/", mode="line", standardize=True,
user_incar_settings=custom_settings)
band.write_input("Si-band")
%matplotlib inline
from pymatgen.io.vasp import Vasprun
from pymatgen.electronic_structure.plotter import DosPlotter
v = Vasprun('Si-dos/vasprun.xml')
tdos = v.tdos
plotter = DosPlotter()
plotter.add_dos("Total DOS", tdos)
plotter.show(xlim=[-5, 5], ylim=[0, 4])
%matplotlib inline
from pymatgen.io.vasp import Vasprun
from pymatgen.electronic_structure.plotter import DosPlotter
v = Vasprun('Si-dos/vasprun.xml')
cdos = v.complete_dos
element_dos = cdos.get_element_dos()
plotter = DosPlotter()
plotter.add_dos_dict(element_dos)
plotter.show(xlim=[-5, 5], ylim=[0, 1])
%matplotlib inline
from pymatgen.io.vasp import Vasprun
from pymatgen.electronic_structure.plotter import DosPlotter
v = Vasprun('Si-dos/vasprun.xml')
cdos = v.complete_dos
spd_dos = cdos.get_spd_dos()
plotter = DosPlotter()
plotter.add_dos_dict(spd_dos)
plotter.show(xlim=[-5, 5], ylim=[0, 1])
%matplotlib inline
from pymatgen.io.vasp import Vasprun, BSVasprun
from pymatgen.electronic_structure.plotter import BSPlotter
v = BSVasprun("Si-band/vasprun.xml")
bs = v.get_band_structure(kpoints_filename="Si-band/KPOINTS",line_mode=True)
plt = BSPlotter(bs)
plt.get_plot(vbm_cbm_marker=True)
| 0.463444 | 0.788054 |
# ML Project Modeling
This file shows and describes the orginal data set modeling process.
```
import pandas as pd
import numpy as np
import os
import os.path as osp
import matplotlib.pyplot as plt
import seaborn as sns
dir_path = os.getcwd()
data_path = osp.abspath(osp.join(dir_path,"Data/"))
df_path = osp.join(data_path,'preprocessed_data2.csv')
out_path = osp.join(data_path,'figure_data')
census_path = osp.join(data_path,'census_2010.csv')
```
## Census Data Frame
```
# reading Census Frame
census_df = pd.read_csv(census_path)
```
## MARTA Data
This data was created using our data set and the preprocess data file. Referrer to there for any indications of what columns are created from what data.
```
data_df = pd.read_csv(df_path)
data_df.UID = data_df.UID.str.replace("(",'')
data_df.UID = data_df.UID.str.replace(")",'')
data_df.UID = data_df.UID.str.split(',')
#data_df.UID = data_df.UID.apply(lambda x: [int(i) for i in x])
# extracting county
data_df.insert(len(data_df.columns),'COUNTY',data_df.UID.apply(lambda x: x[0]))
# extracting census tract
data_df.insert(len(data_df.columns),'TRACT',data_df.UID.apply(lambda x: x[1]))
# extracting Block
data_df.insert(len(data_df.columns),'BLOCK',data_df.UID.apply(lambda x: x[2]))
data_df.COUNTY = data_df.COUNTY.str.replace("'",'')
data_df.TRACT = data_df.TRACT.str.replace("'",'')
data_df.BLOCK = data_df.BLOCK.str.replace("'",'')
# changing datatypes
data_df.COUNTY = data_df.COUNTY.astype('int32')
data_df.TRACT = data_df.TRACT.astype('int32')
data_df.BLOCK = data_df.BLOCK.astype('int32')
data_df = data_df.drop(['UID'],axis=1)
data_df.head()
data_df[data_df.TRIPS == data_df.TRIPS.max()]
data_df.drop(index=60,inplace=True)
data_df.TRIPS.plot.hist(bins=100)
data_df.TRIPS.describe()
data_df.stop_id.plot.hist(bins=10)
data_df.TRIPS.sum()
data_df.stop_id.sum()
```
## Merging The Datasets
```
din = data_df.merge(census_df, left_on=['COUNTY','TRACT','BLOCK'], right_on=['County','Census Tract','Block'])
drop_cols = ['Area Name-Legal/Statistical Area Description', 'Qualifying Name', 'Area (Land)', 'Area (Water)',
'Summary Level', 'Geographic Component', 'Region',
'Division', 'FIPS', 'State (FIPS)', 'County', 'Census Tract', 'Block']
din = din.drop(drop_cols, axis=1)
din.insert(0,'TRIP_RATE', din.TRIPS/din['Total Population'])
din.TRIP_RATE.fillna(0,inplace=True)
din.head()
din.shape
din = din[din['Total Population'] > 0]
din.head()
din.shape
din.TRIP_RATE = din.TRIP_RATE.fillna(0)
din.TRIP_RATE = din.TRIP_RATE * 100
```
## Data Description
The following tables describe all of the raw data before transformation that is to be analyzed.
```
din.iloc[:,0:10].describe()
din.iloc[:,10:20].describe()
din.iloc[:,20:30].describe()
din.iloc[:,30:40].describe()
din.iloc[:,40:50].describe()
din.iloc[:,50:].describe()
list(din.iloc[:,54:62].columns)[1:]
# drop all columns without normal households
drop_cols = ['Households: Family households',
'Households: Family households: Married-couple family',
'Households: Family households: Other family',
'Households: Family households: Other family: Male householder, no wife present',
'Households: Family households: Other family: Female householder, no husband present',
'Households: Nonfamily households',
'Households: Nonfamily households: Householder living alone','Total Population: Female: 65 and 74 years']
din.drop(drop_cols, axis=1,inplace=True)
temp = din['Total Population: Under 5 years'] + din['Total Population: 5 to 9 years'] + din['Total Population: 10 to 14 years'] + din['Total Population: 15 to 17 years']
din.insert(len(din.columns),'POP_U_18', temp)
temp = din['Total Population: 18 to 24 years'] + din['Total Population: 25 to 34 years']
din.insert(len(din.columns),'POP_18_34', temp)
temp = din['Total Population: 35 to 44 years'] + din['Total Population: 45 to 54 years'] + din['Total Population: 55 to 64 years']
din.insert(len(din.columns),'POP_35_64', temp)
temp = din['Total Population: 65 and 74 years'] + din['Total Population: 75 to 84 years'] + din['Total Population: 85 years and over']
din.insert(len(din.columns),'POP_65_OVER', temp)
drop_cols = ['Total Population: Under 5 years','Total Population: 5 to 9 years','Total Population: 10 to 14 years',
'Total Population: 15 to 17 years', 'Total Population: 18 to 24 years',
'Total Population: 25 to 34 years', 'Total Population: 35 to 44 years','Total Population: 45 to 54 years',
'Total Population: 55 to 64 years', 'Total Population: 65 and 74 years','Total Population: 75 to 84 years',
'Total Population: 85 years and over','Total Population: Female: 65 and 74 years']
drop_cols = ['Total Population: Male: Under 5 years', 'Total Population: Male: 5 to 9 years', 'Total Population: Male: 10 to 14 years',
'Total Population: Male: 15 to 17 years', 'Total Population: Male: 18 to 24 years',
'Total Population: Male: 25 to 34 years', 'Total Population: Male: 35 to 44 years',
'Total Population: Male: 45 to 54 years',
'Total Population: Male: 55 to 64 years',
'Total Population: Male: 65 and 74 years',
'Total Population: Male: 75 to 84 years',
'Total Population: Male: 85 years and over',
'Total Population: Female: Under 5 years',
'Total Population: Female: 5 to 9 years',
'Total Population: Female: 10 to 14 years',
'Total Population: Female: 15 to 17 years',
'Total Population: Female: 18 and 24 years',
'Total Population: Female: 25 to 34 years',
'Total Population: Female: 35 to 44 years',
'Total Population: Female: 45 to 54 years',
'Total Population: Female: 55 to 64 years','Households: Nonfamily households: Householder not living alone']
din.drop(drop_cols,axis=1,inplace=True)
din.shape
drop_cols = ['Total Population.1','Total Population: Female: 75 to 84 years',
'Total Population: Female: 85 years and over', 'Total Population.2']
drop_cols = ['Total Population: Under 5 years', 'Total Population: 5 to 9 years',
'Total Population: 10 to 14 years', 'Total Population: 15 to 17 years',
'Total Population: 18 to 24 years', 'Total Population: 25 to 34 years',
'Total Population: 35 to 44 years', 'Total Population: 45 to 54 years',
'Total Population: 55 to 64 years', 'Total Population: 65 and 74 years',
'Total Population: 75 to 84 years',
'Total Population: 85 years and over']
din.drop(drop_cols,axis=1,inplace=True)
drop_cols = ['Total population: American Indian and Alaska Native alone',
'Total population: Native Hawaiian and Other Pacific Islander alone',
'Total population: Some Other Race alone',
'Total population: Two or More Races' ]
temp = din[drop_cols].sum(axis=1)
din.drop(drop_cols,axis=1,inplace=True)
din.insert(len(din.columns),'Other', temp)
din.columns
drop_cols = ['Total Population.1', 'Total Population: Female: 75 to 84 years','Total Population: Female: 85 years and over',
'Total Population.2', 'Total population']
din.drop(drop_cols,axis=1,inplace=True)
def to_ratios(df,num, dem):
df[num] = df[num]/df[dem]
df[num] = df[num].fillna(0)
return df
```
temp = din['Total Population: Male'] /din['Total Population']
din['Total Population: Male'] = temp.fillna(0)
temp = din['Total Population: Female'] /din['Total Population']
din['Total Population: Female'] = temp.fillna(0)
temp = din['Total population: White alone'] /din['Total Population']
din['Total population: White alone'] = temp.fillna(0)
temp = din['Other'] /din['Total Population']
din['Other'] = temp.fillna(0)
temp = din['Total population: Black or African American alone'] /din['Total Population']
din['Total population: Black or African American alone'] = temp.fillna(0)
temp = din['Total population: Asian alone'] /din['Total Population']
din['Total population: Asian alone'] = temp.fillna(0)
```
din = to_ratios(din,'Total Population: Male','Total Population')
din = to_ratios(din,'Total Population: Female','Total Population')
din = to_ratios(din,'Total population: White alone','Total Population')
din = to_ratios(din,'Other','Total Population')
din = to_ratios(din,'Total population: Black or African American alone','Total Population')
din = to_ratios(din,'Total population: Asian alone','Total Population')
din = to_ratios(din,'Households','Total Population')
din = to_ratios(din,'POP_U_18','Total Population')
din = to_ratios(din,'POP_18_34','Total Population')
din = to_ratios(din,'POP_35_64','Total Population')
din = to_ratios(din,'POP_65_OVER','Total Population')
din.head()
```
## Preparing for Machine Learning
1. We remove the County, TRACT, BLOCK Columns
2. We remove duplicate columns like Total Population.1
3. We split up the data frame into X and y
4. We scale each column to -1 to 1
```
from sklearn import preprocessing
from sklearn.preprocessing import PolynomialFeatures
# droping un-needed columns
pipe_df = din.drop(columns=['COUNTY','TRACT','BLOCK'],axis=1)
pipe_df =pipe_df.sample(frac=1)
#Spliting into X, and y
X_df = pipe_df.drop(columns=['TRIPS','TRIP_RATE'])
y = pipe_df.TRIPS
scaler = preprocessing.StandardScaler()
nn_scaler = preprocessing.MaxAbsScaler()
X = scaler.fit_transform(X_df)
X_nn = nn_scaler = nn_scaler.fit_transform(X_df)
X1 = scaler.fit_transform(pipe_df)
poly = PolynomialFeatures(2)
X_poly = poly.fit_transform(X_df)
X_poly = scaler.fit_transform(X_poly)
X_df.describe()
```
## Preliminary Analysis and Feature Engineering
1. Compute correlations of each variables with the number of trips
- Make a graph of this
2. Compute covariance matrix
- make a heatmap of this
3. Perform a Principle Component analysis and analyze the results
- make a graph of the final one
- make a bar graph for potentially the one
```
plt.imshow(np.cov(X1),cmap='hot')
```
### Covariance Analysis
We do not see any standouts in our covariance matrix
```
# getting all cor coef
corrs = []
for i in range(0,X.shape[1]):
corrs.append(np.corrcoef(y.to_numpy(),X[:,i])[0,1])
fig, ax = plt.subplots(figsize=(15,15))
ax = plt.barh(X_df.columns,corrs)
pd.DataFrame(corrs, index=X_df.columns).to_csv(os.path.join(out_path,'var_corr.csv'))
```
### Correlation Analysis
- need to add in statistical significance
1. Positively correlated
- Number of stops correlated with the number of trips
- Total population of Males 25 to 34 years is correlated
- Total population of Asians is correlated as well
2. Negatively Correlated
- population 65 +
```
# implement PCA here
from sklearn.decomposition import PCA
pca = PCA(n_components=2)
fig, ax = plt.subplots()
pc = pca.fit_transform(X)
ax = plt.scatter(pc[:,0],pc[:,1])
```
### Priniciple Component Analysis
- Our prinicple component analysis indicates that there are no clear clusters in the data set as is. We need furhter analysis to see if we can find any features of interest
The initial cross validation score depects a inadequate model that has to many variables that don't inform of the numbers. We believe that we should
```
from sklearn.ensemble import RandomForestRegressor
rf = RandomForestRegressor(n_estimators=100)
rf.fit(X,y)
vif = pd.DataFrame(rf.feature_importances_,index=X_df.columns)
vif.sort_values(by=0,ascending=False,inplace=True)
fig, ax = plt.subplots(figsize=(15,15))
ax = plt.barh(vif.index,vif[0])
```
### Analyzing Variable Importance
1. We notice a wwide range in the variable importance factors with a lot of variables not having any influence of the task at all
2. We will drop some of these factors
3. We might need to fetch some more detailed data taht is related to income as well as
4. We can add in additional more exlanatory variables as well
We will conduct a similar analysis using a lasso regression to do some variable selection. Following this variable selection process we will construct new variables.
```
from sklearn.linear_model import Lasso
alphas = [0.25, 0.5, 0.75, 1]
coefs = []
for alpha in alphas:
las_reg = Lasso(alpha=alpha)
las_reg.fit(X,y)
coefs.append(las_reg.coef_)
#fig, ax = plt.subplots()
coefs = pd.DataFrame(data=coefs,index=alphas,columns=X_df.columns)
coef_bool = coefs.applymap(lambda x: x != 0)
temp = coef_bool.sum()
temp[temp != 0].plot.barh()
#fig, ax = plt.subplots(figsize=(10,10))
```
### Lasso Variable Analysis
#### Process
We selected a series of alpha values form 0.25 to 1 by 1/4. We then saw when the coefficients were not 0 in each of the models. We then see the number of individuals.
#### Results
- We find again that stop_id is important
- We suggest potentially binning population into under 20, 20-35, 35-65, 65+, we can potentially remove
- While these type of regressors are generally resistant to outliers, a complete outlier analysis would be helpful as well
- Additional variables of interest
- Rail Stops
- Bus Stops
- Adjacent Census Tract Stops
- Income
- median income
- percent under 25,000 dollar a year - Car slightly affordable
- percent under 30,000 dollars a year - Car more affordable
- Race
- majority white
- majority black
- neither
- We will explore using the total population variable and then transitioning other variables to precentages
- We can also change our scaling process as well
## Regression Significance Analysis
We present a analysis of each feature in a univariate regression. The statistic reported is the F value for the model test.
```
from sklearn.feature_selection import f_regression
f_reg = f_regression(X,y)
imp_df = pd.DataFrame(f_reg,columns=X_df.columns,index=['F Score','p_value']).T
imp_df.insert(len(imp_df.columns),'Sig_1',imp_df.p_value.apply(lambda x: x <= 0.1))
imp_df.insert(len(imp_df.columns),'Sig_05',imp_df.p_value.apply(lambda x: x <= 0.05))
imp_df[imp_df.Sig_1]
```
### Results
We notice that the only significant factor is the number of stops at 0.05 and we have four features for 0.10. This should motivate a refactoring of features because while these granular features may have some information they likely do not contain. We found that our anlaysis pipeline improved slightly after refactoring features. We believe
## Polynomial Features
We
```
from sklearn.preprocessing import PolynomialFeatures
alphas = [0.25, 0.5, 0.75, 1]
coefs = []
for alpha in alphas:
las_reg = Lasso(alpha=alpha, max_iter=10000)
las_reg.fit(X_poly,y)
coefs.append(las_reg.coef_)
#fig, ax = plt.subplots()
coefs = pd.DataFrame(data=coefs,index=alphas,columns=poly.get_feature_names(input_features=X_df.columns))
coef_bool = coefs.applymap(lambda x: x != 0)
temp = coef_bool.sum()
fig, ax = plt.subplots(figsize=(15,15))
ax = temp[temp != 0].plot.barh()
# add Rail Stop Black
pipe_df.insert(len(pipe_df.columns),'RAIL_STOP_BLACK',
pipe_df['RAIL_STOP'] * pipe_df['Total population: Black or African American alone'])
# add Rail Stop Asian
pipe_df.insert(len(pipe_df.columns),
'RAIL_STOP_ASIAN',pipe_df['RAIL_STOP'] * pipe_df['Total population: Asian alone'])
# add Rail Stop Pop 35_64
# droping un-needed columns
pipe_df = din.drop(columns=['COUNTY','TRACT','BLOCK'],axis=1)
pipe_df =pipe_df.sample(frac=1)
#Spliting into X, and y
X_df_2 = pipe_df.drop(columns=['TRIPS','TRIP_RATE'])
X_2 = scaler.fit_transform(X_df)
rf = RandomForestRegressor(n_estimators=100)
rf.fit(X_2,y)
vif = pd.DataFrame(rf.feature_importances_,index=X_df_2.columns)
vif.sort_values(by=0,ascending=False,inplace=True)
fig, ax = plt.subplots(figsize=(15,15))
ax = plt.barh(vif.index,vif[0])
X_df_2.head()
#temp = (X_df['Total population: Black or African American alone'] > 0.50)* 1
#X_df.insert(3,'MAJ_BLACK',temp)
#temp = (X_df['Total population: White alone'] > 0.50)* 1
#X_df.insert(4,'MAJ_WHITE',temp)
#y = pipe_df.TRIPS
#scaler = preprocessing.StandardScaler()
#X = scaler.fit_transform(X_df.iloc[:,4:])
#X = np.hstack((X_df.iloc[:,:4],X))
```
- Add Rail Stop and Asian
- Add Rail stop and Black alone
```
from sklearn.model_selection import GridSearchCV
```
## Model Analysis and Calibrartion
```
cv_scores = []
```
### Random Forest
```
from sklearn.ensemble import RandomForestRegressor
from sklearn.model_selection import cross_val_score
rf = RandomForestRegressor(n_estimators=100)
temp = cross_val_score(rf,X,y,cv=10,scoring='neg_mean_squared_error')
print(sum(temp * -1)/10)
cv_scores.append(("RF",sum(temp * -1)/10))
print(temp)
```
### Neural Network
- our results from the ranndom forest classifier suggests there are significant non linear factors to be considered.
```
from sklearn.neural_network import MLPRegressor
from sklearn.model_selection import GridSearchCV
nn = MLPRegressor(solver='lbfgs')
param_grid = {'hidden_layer_sizes':[(8,),(8,4),(8,4,2)],
'alpha':10.0 ** -np.arange(1, 7)}
opt = GridSearchCV(nn,param_grid,cv=10,iid=True,scoring='neg_mean_squared_error')
opt.fit(X_nn,y)
print(opt.best_estimator_)
nn = opt.best_estimator_
temp = cross_val_score(nn,X_nn,y,cv=10,scoring='neg_mean_squared_error')
sum(temp * -1)/10
cv_scores.append(("Neural Network",sum(temp * -1)/10))
cv_scores
```
### Regression Techniques
#### Lasso
```
param_grid = {'alpha':[0.01,0.1,0.25,0.5,0.75,1]}
lass = Lasso()
opt = GridSearchCV(lass,param_grid,cv=10)
opt.fit(X,y)
lass = opt.best_estimator_
temp = cross_val_score(lass,X,y,cv=10,scoring='neg_mean_squared_error')
print(temp)
cv_scores.append(("Lasso",sum(temp * -1)/10))
lass
print(sum(temp*-1)/10)
```
#### Ridge
```
from sklearn.linear_model import Ridge
param_grid = {'alpha':[0.01,0.1,0.25,0.5,0.75,1]}
ridge = Ridge()
opt = GridSearchCV(ridge,param_grid,cv=10)
opt.fit(X,y)
ridge = opt.best_estimator_
temp = cross_val_score(ridge, X,y,cv=10,scoring='neg_mean_squared_error')
print(sum(temp*-1)/10)
print(ridge)
cv_scores.append(("Ridge",sum(temp * -1)/10))
```
#### Elastic Net
```
from sklearn.linear_model import ElasticNet
elast = ElasticNet()
param_grid = {"l1_ratio":[0.1,0.25,0.5,0.75],'alpha':[0.01,0.1,0.25,0.5,0.75,1]}
opt = GridSearchCV(elast,param_grid,cv=10)
opt.fit(X,y)
elast = opt.best_estimator_
temp = cross_val_score(ridge, X,y,cv=10,scoring='neg_mean_squared_error')
print(sum(temp*-1)/10)
print(elast)
cv_scores.append(("ELAST",sum(temp * -1)/10))
```
### Support Vector Machines
```
from sklearn import svm
param_grid = {'kernel': ['poly','rbf'],
"C":[0.1,0.25,0.5,0.75,1,1.5]
}
svr = svm.SVR(gamma='auto')
opt = GridSearchCV(svr,param_grid,cv=10)
opt.fit(X,y)
svr = opt.best_estimator_
temp = cross_val_score(svr,X,y,cv=10,scoring='neg_mean_squared_error')
print(sum(temp * -1)/10)
cv_scores.append(("SVR",sum(temp * -1)/10))
print(svr)
temp
```
## KNR
```
from sklearn.neighbors import KNeighborsRegressor
cv_scores
knr = KNeighborsRegressor()
param_grid = {'weights':["uniform",'distance'],
'algorithm':['ball_tree','kd_tree'],
"n_neighbors":[2,4,6,8,10,12]}
opt = GridSearchCV(knr,param_grid,cv=10)
opt.fit(X,y)
knr = opt.best_estimator_
temp = cross_val_score(knr,X,y,cv=10,scoring='neg_mean_squared_error')
print(sum(temp * -1)/10)
cv_scores.append(("KNR",sum(temp * -1)/10))
temp
knr
```
# Results
```
results = pd.DataFrame(cv_scores)
results = results.rename(columns={0:'MODEL',1:'RMSE'})
results.to_csv(os.path.join(out_path,'results.csv'))
lass.fit(X,y)
coefs = pd.DataFrame(lass.coef_,index=X_df.columns)
coefs = coefs.rename({0:'Coefficient'},axis=1)
coefs.to_csv(os.path.join(out_path,'Coefficients.csv'))
results
lass.predict(X).sum()
lass_coefs_df = pd.DataFrame(lass.coef_,index = X_df.columns)
fig, ax = plt.subplots(figsize=(10,5))
lass_coefs_df.plot.barh(ax=ax)
ax.set_xlabel('Coefficient Value')
ax.set_title('Lasso Regression Coefficients')
fig.savefig(os.path.join(out_path,'Lasso_Coeffs.png'))
X_2 = X.copy()
X_2[:,1] = 1
lass.predict(X_2).sum() - lass.predict(X).sum()
X_df.RAIL_STOP = 0
X_2 = scaler.fit_transform(X_df)
y.describe()
y.sum()
lass.predict(X_2).sum()
X_df.T
```
|
github_jupyter
|
import pandas as pd
import numpy as np
import os
import os.path as osp
import matplotlib.pyplot as plt
import seaborn as sns
dir_path = os.getcwd()
data_path = osp.abspath(osp.join(dir_path,"Data/"))
df_path = osp.join(data_path,'preprocessed_data2.csv')
out_path = osp.join(data_path,'figure_data')
census_path = osp.join(data_path,'census_2010.csv')
# reading Census Frame
census_df = pd.read_csv(census_path)
data_df = pd.read_csv(df_path)
data_df.UID = data_df.UID.str.replace("(",'')
data_df.UID = data_df.UID.str.replace(")",'')
data_df.UID = data_df.UID.str.split(',')
#data_df.UID = data_df.UID.apply(lambda x: [int(i) for i in x])
# extracting county
data_df.insert(len(data_df.columns),'COUNTY',data_df.UID.apply(lambda x: x[0]))
# extracting census tract
data_df.insert(len(data_df.columns),'TRACT',data_df.UID.apply(lambda x: x[1]))
# extracting Block
data_df.insert(len(data_df.columns),'BLOCK',data_df.UID.apply(lambda x: x[2]))
data_df.COUNTY = data_df.COUNTY.str.replace("'",'')
data_df.TRACT = data_df.TRACT.str.replace("'",'')
data_df.BLOCK = data_df.BLOCK.str.replace("'",'')
# changing datatypes
data_df.COUNTY = data_df.COUNTY.astype('int32')
data_df.TRACT = data_df.TRACT.astype('int32')
data_df.BLOCK = data_df.BLOCK.astype('int32')
data_df = data_df.drop(['UID'],axis=1)
data_df.head()
data_df[data_df.TRIPS == data_df.TRIPS.max()]
data_df.drop(index=60,inplace=True)
data_df.TRIPS.plot.hist(bins=100)
data_df.TRIPS.describe()
data_df.stop_id.plot.hist(bins=10)
data_df.TRIPS.sum()
data_df.stop_id.sum()
din = data_df.merge(census_df, left_on=['COUNTY','TRACT','BLOCK'], right_on=['County','Census Tract','Block'])
drop_cols = ['Area Name-Legal/Statistical Area Description', 'Qualifying Name', 'Area (Land)', 'Area (Water)',
'Summary Level', 'Geographic Component', 'Region',
'Division', 'FIPS', 'State (FIPS)', 'County', 'Census Tract', 'Block']
din = din.drop(drop_cols, axis=1)
din.insert(0,'TRIP_RATE', din.TRIPS/din['Total Population'])
din.TRIP_RATE.fillna(0,inplace=True)
din.head()
din.shape
din = din[din['Total Population'] > 0]
din.head()
din.shape
din.TRIP_RATE = din.TRIP_RATE.fillna(0)
din.TRIP_RATE = din.TRIP_RATE * 100
din.iloc[:,0:10].describe()
din.iloc[:,10:20].describe()
din.iloc[:,20:30].describe()
din.iloc[:,30:40].describe()
din.iloc[:,40:50].describe()
din.iloc[:,50:].describe()
list(din.iloc[:,54:62].columns)[1:]
# drop all columns without normal households
drop_cols = ['Households: Family households',
'Households: Family households: Married-couple family',
'Households: Family households: Other family',
'Households: Family households: Other family: Male householder, no wife present',
'Households: Family households: Other family: Female householder, no husband present',
'Households: Nonfamily households',
'Households: Nonfamily households: Householder living alone','Total Population: Female: 65 and 74 years']
din.drop(drop_cols, axis=1,inplace=True)
temp = din['Total Population: Under 5 years'] + din['Total Population: 5 to 9 years'] + din['Total Population: 10 to 14 years'] + din['Total Population: 15 to 17 years']
din.insert(len(din.columns),'POP_U_18', temp)
temp = din['Total Population: 18 to 24 years'] + din['Total Population: 25 to 34 years']
din.insert(len(din.columns),'POP_18_34', temp)
temp = din['Total Population: 35 to 44 years'] + din['Total Population: 45 to 54 years'] + din['Total Population: 55 to 64 years']
din.insert(len(din.columns),'POP_35_64', temp)
temp = din['Total Population: 65 and 74 years'] + din['Total Population: 75 to 84 years'] + din['Total Population: 85 years and over']
din.insert(len(din.columns),'POP_65_OVER', temp)
drop_cols = ['Total Population: Under 5 years','Total Population: 5 to 9 years','Total Population: 10 to 14 years',
'Total Population: 15 to 17 years', 'Total Population: 18 to 24 years',
'Total Population: 25 to 34 years', 'Total Population: 35 to 44 years','Total Population: 45 to 54 years',
'Total Population: 55 to 64 years', 'Total Population: 65 and 74 years','Total Population: 75 to 84 years',
'Total Population: 85 years and over','Total Population: Female: 65 and 74 years']
drop_cols = ['Total Population: Male: Under 5 years', 'Total Population: Male: 5 to 9 years', 'Total Population: Male: 10 to 14 years',
'Total Population: Male: 15 to 17 years', 'Total Population: Male: 18 to 24 years',
'Total Population: Male: 25 to 34 years', 'Total Population: Male: 35 to 44 years',
'Total Population: Male: 45 to 54 years',
'Total Population: Male: 55 to 64 years',
'Total Population: Male: 65 and 74 years',
'Total Population: Male: 75 to 84 years',
'Total Population: Male: 85 years and over',
'Total Population: Female: Under 5 years',
'Total Population: Female: 5 to 9 years',
'Total Population: Female: 10 to 14 years',
'Total Population: Female: 15 to 17 years',
'Total Population: Female: 18 and 24 years',
'Total Population: Female: 25 to 34 years',
'Total Population: Female: 35 to 44 years',
'Total Population: Female: 45 to 54 years',
'Total Population: Female: 55 to 64 years','Households: Nonfamily households: Householder not living alone']
din.drop(drop_cols,axis=1,inplace=True)
din.shape
drop_cols = ['Total Population.1','Total Population: Female: 75 to 84 years',
'Total Population: Female: 85 years and over', 'Total Population.2']
drop_cols = ['Total Population: Under 5 years', 'Total Population: 5 to 9 years',
'Total Population: 10 to 14 years', 'Total Population: 15 to 17 years',
'Total Population: 18 to 24 years', 'Total Population: 25 to 34 years',
'Total Population: 35 to 44 years', 'Total Population: 45 to 54 years',
'Total Population: 55 to 64 years', 'Total Population: 65 and 74 years',
'Total Population: 75 to 84 years',
'Total Population: 85 years and over']
din.drop(drop_cols,axis=1,inplace=True)
drop_cols = ['Total population: American Indian and Alaska Native alone',
'Total population: Native Hawaiian and Other Pacific Islander alone',
'Total population: Some Other Race alone',
'Total population: Two or More Races' ]
temp = din[drop_cols].sum(axis=1)
din.drop(drop_cols,axis=1,inplace=True)
din.insert(len(din.columns),'Other', temp)
din.columns
drop_cols = ['Total Population.1', 'Total Population: Female: 75 to 84 years','Total Population: Female: 85 years and over',
'Total Population.2', 'Total population']
din.drop(drop_cols,axis=1,inplace=True)
def to_ratios(df,num, dem):
df[num] = df[num]/df[dem]
df[num] = df[num].fillna(0)
return df
din = to_ratios(din,'Total Population: Male','Total Population')
din = to_ratios(din,'Total Population: Female','Total Population')
din = to_ratios(din,'Total population: White alone','Total Population')
din = to_ratios(din,'Other','Total Population')
din = to_ratios(din,'Total population: Black or African American alone','Total Population')
din = to_ratios(din,'Total population: Asian alone','Total Population')
din = to_ratios(din,'Households','Total Population')
din = to_ratios(din,'POP_U_18','Total Population')
din = to_ratios(din,'POP_18_34','Total Population')
din = to_ratios(din,'POP_35_64','Total Population')
din = to_ratios(din,'POP_65_OVER','Total Population')
din.head()
from sklearn import preprocessing
from sklearn.preprocessing import PolynomialFeatures
# droping un-needed columns
pipe_df = din.drop(columns=['COUNTY','TRACT','BLOCK'],axis=1)
pipe_df =pipe_df.sample(frac=1)
#Spliting into X, and y
X_df = pipe_df.drop(columns=['TRIPS','TRIP_RATE'])
y = pipe_df.TRIPS
scaler = preprocessing.StandardScaler()
nn_scaler = preprocessing.MaxAbsScaler()
X = scaler.fit_transform(X_df)
X_nn = nn_scaler = nn_scaler.fit_transform(X_df)
X1 = scaler.fit_transform(pipe_df)
poly = PolynomialFeatures(2)
X_poly = poly.fit_transform(X_df)
X_poly = scaler.fit_transform(X_poly)
X_df.describe()
plt.imshow(np.cov(X1),cmap='hot')
# getting all cor coef
corrs = []
for i in range(0,X.shape[1]):
corrs.append(np.corrcoef(y.to_numpy(),X[:,i])[0,1])
fig, ax = plt.subplots(figsize=(15,15))
ax = plt.barh(X_df.columns,corrs)
pd.DataFrame(corrs, index=X_df.columns).to_csv(os.path.join(out_path,'var_corr.csv'))
# implement PCA here
from sklearn.decomposition import PCA
pca = PCA(n_components=2)
fig, ax = plt.subplots()
pc = pca.fit_transform(X)
ax = plt.scatter(pc[:,0],pc[:,1])
from sklearn.ensemble import RandomForestRegressor
rf = RandomForestRegressor(n_estimators=100)
rf.fit(X,y)
vif = pd.DataFrame(rf.feature_importances_,index=X_df.columns)
vif.sort_values(by=0,ascending=False,inplace=True)
fig, ax = plt.subplots(figsize=(15,15))
ax = plt.barh(vif.index,vif[0])
from sklearn.linear_model import Lasso
alphas = [0.25, 0.5, 0.75, 1]
coefs = []
for alpha in alphas:
las_reg = Lasso(alpha=alpha)
las_reg.fit(X,y)
coefs.append(las_reg.coef_)
#fig, ax = plt.subplots()
coefs = pd.DataFrame(data=coefs,index=alphas,columns=X_df.columns)
coef_bool = coefs.applymap(lambda x: x != 0)
temp = coef_bool.sum()
temp[temp != 0].plot.barh()
#fig, ax = plt.subplots(figsize=(10,10))
from sklearn.feature_selection import f_regression
f_reg = f_regression(X,y)
imp_df = pd.DataFrame(f_reg,columns=X_df.columns,index=['F Score','p_value']).T
imp_df.insert(len(imp_df.columns),'Sig_1',imp_df.p_value.apply(lambda x: x <= 0.1))
imp_df.insert(len(imp_df.columns),'Sig_05',imp_df.p_value.apply(lambda x: x <= 0.05))
imp_df[imp_df.Sig_1]
from sklearn.preprocessing import PolynomialFeatures
alphas = [0.25, 0.5, 0.75, 1]
coefs = []
for alpha in alphas:
las_reg = Lasso(alpha=alpha, max_iter=10000)
las_reg.fit(X_poly,y)
coefs.append(las_reg.coef_)
#fig, ax = plt.subplots()
coefs = pd.DataFrame(data=coefs,index=alphas,columns=poly.get_feature_names(input_features=X_df.columns))
coef_bool = coefs.applymap(lambda x: x != 0)
temp = coef_bool.sum()
fig, ax = plt.subplots(figsize=(15,15))
ax = temp[temp != 0].plot.barh()
# add Rail Stop Black
pipe_df.insert(len(pipe_df.columns),'RAIL_STOP_BLACK',
pipe_df['RAIL_STOP'] * pipe_df['Total population: Black or African American alone'])
# add Rail Stop Asian
pipe_df.insert(len(pipe_df.columns),
'RAIL_STOP_ASIAN',pipe_df['RAIL_STOP'] * pipe_df['Total population: Asian alone'])
# add Rail Stop Pop 35_64
# droping un-needed columns
pipe_df = din.drop(columns=['COUNTY','TRACT','BLOCK'],axis=1)
pipe_df =pipe_df.sample(frac=1)
#Spliting into X, and y
X_df_2 = pipe_df.drop(columns=['TRIPS','TRIP_RATE'])
X_2 = scaler.fit_transform(X_df)
rf = RandomForestRegressor(n_estimators=100)
rf.fit(X_2,y)
vif = pd.DataFrame(rf.feature_importances_,index=X_df_2.columns)
vif.sort_values(by=0,ascending=False,inplace=True)
fig, ax = plt.subplots(figsize=(15,15))
ax = plt.barh(vif.index,vif[0])
X_df_2.head()
#temp = (X_df['Total population: Black or African American alone'] > 0.50)* 1
#X_df.insert(3,'MAJ_BLACK',temp)
#temp = (X_df['Total population: White alone'] > 0.50)* 1
#X_df.insert(4,'MAJ_WHITE',temp)
#y = pipe_df.TRIPS
#scaler = preprocessing.StandardScaler()
#X = scaler.fit_transform(X_df.iloc[:,4:])
#X = np.hstack((X_df.iloc[:,:4],X))
from sklearn.model_selection import GridSearchCV
cv_scores = []
from sklearn.ensemble import RandomForestRegressor
from sklearn.model_selection import cross_val_score
rf = RandomForestRegressor(n_estimators=100)
temp = cross_val_score(rf,X,y,cv=10,scoring='neg_mean_squared_error')
print(sum(temp * -1)/10)
cv_scores.append(("RF",sum(temp * -1)/10))
print(temp)
from sklearn.neural_network import MLPRegressor
from sklearn.model_selection import GridSearchCV
nn = MLPRegressor(solver='lbfgs')
param_grid = {'hidden_layer_sizes':[(8,),(8,4),(8,4,2)],
'alpha':10.0 ** -np.arange(1, 7)}
opt = GridSearchCV(nn,param_grid,cv=10,iid=True,scoring='neg_mean_squared_error')
opt.fit(X_nn,y)
print(opt.best_estimator_)
nn = opt.best_estimator_
temp = cross_val_score(nn,X_nn,y,cv=10,scoring='neg_mean_squared_error')
sum(temp * -1)/10
cv_scores.append(("Neural Network",sum(temp * -1)/10))
cv_scores
param_grid = {'alpha':[0.01,0.1,0.25,0.5,0.75,1]}
lass = Lasso()
opt = GridSearchCV(lass,param_grid,cv=10)
opt.fit(X,y)
lass = opt.best_estimator_
temp = cross_val_score(lass,X,y,cv=10,scoring='neg_mean_squared_error')
print(temp)
cv_scores.append(("Lasso",sum(temp * -1)/10))
lass
print(sum(temp*-1)/10)
from sklearn.linear_model import Ridge
param_grid = {'alpha':[0.01,0.1,0.25,0.5,0.75,1]}
ridge = Ridge()
opt = GridSearchCV(ridge,param_grid,cv=10)
opt.fit(X,y)
ridge = opt.best_estimator_
temp = cross_val_score(ridge, X,y,cv=10,scoring='neg_mean_squared_error')
print(sum(temp*-1)/10)
print(ridge)
cv_scores.append(("Ridge",sum(temp * -1)/10))
from sklearn.linear_model import ElasticNet
elast = ElasticNet()
param_grid = {"l1_ratio":[0.1,0.25,0.5,0.75],'alpha':[0.01,0.1,0.25,0.5,0.75,1]}
opt = GridSearchCV(elast,param_grid,cv=10)
opt.fit(X,y)
elast = opt.best_estimator_
temp = cross_val_score(ridge, X,y,cv=10,scoring='neg_mean_squared_error')
print(sum(temp*-1)/10)
print(elast)
cv_scores.append(("ELAST",sum(temp * -1)/10))
from sklearn import svm
param_grid = {'kernel': ['poly','rbf'],
"C":[0.1,0.25,0.5,0.75,1,1.5]
}
svr = svm.SVR(gamma='auto')
opt = GridSearchCV(svr,param_grid,cv=10)
opt.fit(X,y)
svr = opt.best_estimator_
temp = cross_val_score(svr,X,y,cv=10,scoring='neg_mean_squared_error')
print(sum(temp * -1)/10)
cv_scores.append(("SVR",sum(temp * -1)/10))
print(svr)
temp
from sklearn.neighbors import KNeighborsRegressor
cv_scores
knr = KNeighborsRegressor()
param_grid = {'weights':["uniform",'distance'],
'algorithm':['ball_tree','kd_tree'],
"n_neighbors":[2,4,6,8,10,12]}
opt = GridSearchCV(knr,param_grid,cv=10)
opt.fit(X,y)
knr = opt.best_estimator_
temp = cross_val_score(knr,X,y,cv=10,scoring='neg_mean_squared_error')
print(sum(temp * -1)/10)
cv_scores.append(("KNR",sum(temp * -1)/10))
temp
knr
results = pd.DataFrame(cv_scores)
results = results.rename(columns={0:'MODEL',1:'RMSE'})
results.to_csv(os.path.join(out_path,'results.csv'))
lass.fit(X,y)
coefs = pd.DataFrame(lass.coef_,index=X_df.columns)
coefs = coefs.rename({0:'Coefficient'},axis=1)
coefs.to_csv(os.path.join(out_path,'Coefficients.csv'))
results
lass.predict(X).sum()
lass_coefs_df = pd.DataFrame(lass.coef_,index = X_df.columns)
fig, ax = plt.subplots(figsize=(10,5))
lass_coefs_df.plot.barh(ax=ax)
ax.set_xlabel('Coefficient Value')
ax.set_title('Lasso Regression Coefficients')
fig.savefig(os.path.join(out_path,'Lasso_Coeffs.png'))
X_2 = X.copy()
X_2[:,1] = 1
lass.predict(X_2).sum() - lass.predict(X).sum()
X_df.RAIL_STOP = 0
X_2 = scaler.fit_transform(X_df)
y.describe()
y.sum()
lass.predict(X_2).sum()
X_df.T
| 0.259263 | 0.865622 |
# Example: Odds and Log Odds
Elements of Data Science
by [Allen Downey](https://allendowney.com)
[MIT License](https://opensource.org/licenses/MIT)
## Introduction
The goal of this notebook is to review the definitions of probability, odds, and log odds, and to practice computing them.
Suppose researchers conduct a study to test a treatment for a potentially deadly disease.
Out of 187 patients in the study, 90 are randomly assigned to the control group. They are treated using the current [standard treatment](https://en.wikipedia.org/wiki/Standard_treatment) for the disease.
The other 97 patients are assigned to the "treatment group" and given a new, experimental treatment.
At the end of the study period, 77 people in the control group are still alive; 13 have died.
In the treatment group, 91 patients survive and 6 die.
The [case fatality rate](https://en.wikipedia.org/wiki/Case_fatality_rate) in each group is the proportion of patients who died during the study period.
Compute the case fatality rates for each group, expressed as a percentage.
```
# Solution goes here
```
The fatality rate is lower in the treatment group, so it seems like the new treatment is effective.
One way to describe its effectiveness is to report the difference between the fatality rates in the two groups, expressed in percentage points. What is that [risk difference](https://en.wikipedia.org/wiki/Risk_difference)?
```
# Solution goes here
```
Another way to report the difference is the ratio of the two rates, also known as [relative risk](https://en.wikipedia.org/wiki/Relative_risk).
What is the ratio of the two rates? Which rate do you think makes more sense to put in the denominator?
```
# Solution goes here
# Solution goes here
```
## Odds
If the probability of an event is 75%, the odds are "three to one" that it will happen, sometimes written as `3:1` or just `3`.
If the probability is 33%, the odds are "one to two", or `1:2`, or `0.5`.
In general, if the probability is `p`, the odds are `p / (1-p)`.
What are the fatality rates in the control and treatment groups, expressed as odds?
```
# Solution goes here
```
What is the [odds ratio](https://en.wikipedia.org/wiki/Odds_ratio) for the two groups? Which one do you think you should put in the denominator?
```
# Solution goes here
```
If the odds ratio is 1, that means the odds are the same in both groups, so the treatment has no effect.
If the odds ratio is less than 1, that means the fatality rate is lower in the treatment group, so the treatment is effective.
## Log odds
Yet another way to represent uncertainty is [log-odds](https://en.wikipedia.org/wiki/Logit), which is just what it sounds like, the logarithm of odds.
Natural logarithms are used most often, but you will sometimes see logarithms base 10.
Use `np.log` to compute the natural log of the odds from the previous section.
```
# Solution goes here
```
Finally, compute the [log odds ratio]() for the two groups, which you can compute one of two ways:
* Compute the log of the odds ratio.
* Compute the difference of the log odds.
But do not compute the ratio of the log odds; that's not a thing.
```
lo2 / lo1 # Wrong
# Solution goes here
# Solution goes here
```
If the fatality rates are the same in both group, the odds ratio is 1 and the log odds ratio is 0.
If the treatment is effective, the odds ratio is less than 1, so the log odds ratio is less than 0.
## Natural frequencies
Suppose you consider 100 people with this disease. If you give them the new treatment instead of the standard treatment, how many lives would you expect to save?
```
# Solution goes here
```
What is the name for this statistic?
|
github_jupyter
|
# Solution goes here
# Solution goes here
# Solution goes here
# Solution goes here
# Solution goes here
# Solution goes here
# Solution goes here
lo2 / lo1 # Wrong
# Solution goes here
# Solution goes here
# Solution goes here
| 0.365683 | 0.995777 |
```
class color:
PURPLE = '\033[95m'
CYAN = '\033[96m'
DARKCYAN = '\033[36m'
BLUE = '\033[94m'
GREEN = '\033[92m'
YELLOW = '\033[93m'
RED = '\033[91m'
BOLD = '\033[1m'
UNDERLINE = '\033[4m'
END = '\033[0m'
print(color.PURPLE + color.BOLD + 'Hello World !' + color.END)
```
## Chain Decorators
```
def print_bold(fn):
def print_bold_wrap(*args, **kwargs):
return color.BOLD + fn(*args, *kwargs) + color.END
return print_bold_wrap
def print_red(fn):
def print_cyan_wrap(*args, **kwargs):
return color.RED+fn(*args, **kwargs)+color.END
return print_cyan_wrap
def print_unlin(fn):
def print_unlin_wrap(*args, **kwargs):
return color.UNDERLINE + fn(*args, **kwargs) + color.END
return print_unlin_wrap
@print_bold
@print_red
@print_unlin
def print_hello(name):
return f"Hello {name.capitalize()}"
print(print_hello('SANDEEP'))
```
## Generators
Create a generator that generates the squares of numbers up to some number N.
```
def gen_squares(n):
for i in range(1, n+1):
yield i**2
x = gen_squares(10)
print(next(x))
y = iter(gen_squares(10))
next(y)
for i in y:
print(i)
for i in gen_squares(10):
print(i)
```
Create a generator that yields "n" random numbers between a low and high number (that are inputs).
Note: Use the random library. For example:
```
from random import randint
def gen_rand (lo, hi, n):
for _ in range(n):
yield randint (lo, hi)
rd = gen_rand(1, 25, 10)
rnd_iter = iter(gen_rand(1, 25, 10))
next(rd)
next(rnd_iter)
for i in rnd_iter:
print(i)
for i in rd:
print(i)
```
Use the iter() function to convert the string below into an iterator:
```
s_iter = iter('hello')
s_iter
for c in s_iter:
print(c)
```
Can you explain what gencomp is in the code below? (Note: We never covered this in lecture! You will have to do some Googling/Stack Overflowing!)
```
my_list = [1,2,3,4,5]
gencomp = (item for item in my_list if item > 3)
for item in gencomp:
print(item)
print(gencomp)
nums = (i for i in range(10))
nums
```
## Built-in Functions
##### Problem 1
Use map() to create a function which finds the length of each word in the phrase (broken by spaces) and returns the values in a list.
The function will have an input of a string, and output a list of integers.
```
def word_lengths(phrase):
word_list = phrase.split()
fn = lambda x: len(x)
print(list(map(fn, word_list)))
word_lengths('How long are the words in this phrase')
```
### Problem 2
Use reduce() to take a list of digits and return the number that they correspond to. For example, [1, 2, 3] corresponds to one-hundred-twenty-three.
Do not convert the integers to strings!
```
from functools import reduce
def digits_to_num(digits):
fn = lambda x: str(x)
nums = list(map(fn, digits))
print(int(reduce(lambda x,y: x+y, nums)))
digits_to_num([3,4,3,2,1])
```
### Problem 3
Use filter to return the words from a list of words which start with a target letter.
```
def filter_words(word_list, letter):
print(list(filter(lambda x: x[0] == letter, word_list)))
l = ['hello','are','cat','dog','ham','hi','go','to','heart']
filter_words(l,'h')
```
### Problem 4
Use zip() and a list comprehension to return a list of the same length where each value is the two strings from L1 and L2 concatenated together with connector between them. Look at the example output below:
```
def concatenate(L1, L2, connector):
print([a + connector + b for a, b in zip(L1, L2)])
concatenate(['A','B'],['a','b'],'-')
```
### Problem 5
Use enumerate() and other skills to return a dictionary which has the values of the list as keys and the index as the value. You may assume that a value will only appear once in the given list.
```
def d_list(L):
print({v:i for i, v in enumerate(L)})
d_list(['a','b','c'])
```
### Problem 6
Use enumerate() and other skills from above to return the count of the number of items in the list whose value equals its index.
```
def count_match_index(L):
print(len([v for i, v in enumerate(L) if i==v]))
count_match_index([0,2,2,1,5,5,6,10])
```
|
github_jupyter
|
class color:
PURPLE = '\033[95m'
CYAN = '\033[96m'
DARKCYAN = '\033[36m'
BLUE = '\033[94m'
GREEN = '\033[92m'
YELLOW = '\033[93m'
RED = '\033[91m'
BOLD = '\033[1m'
UNDERLINE = '\033[4m'
END = '\033[0m'
print(color.PURPLE + color.BOLD + 'Hello World !' + color.END)
def print_bold(fn):
def print_bold_wrap(*args, **kwargs):
return color.BOLD + fn(*args, *kwargs) + color.END
return print_bold_wrap
def print_red(fn):
def print_cyan_wrap(*args, **kwargs):
return color.RED+fn(*args, **kwargs)+color.END
return print_cyan_wrap
def print_unlin(fn):
def print_unlin_wrap(*args, **kwargs):
return color.UNDERLINE + fn(*args, **kwargs) + color.END
return print_unlin_wrap
@print_bold
@print_red
@print_unlin
def print_hello(name):
return f"Hello {name.capitalize()}"
print(print_hello('SANDEEP'))
def gen_squares(n):
for i in range(1, n+1):
yield i**2
x = gen_squares(10)
print(next(x))
y = iter(gen_squares(10))
next(y)
for i in y:
print(i)
for i in gen_squares(10):
print(i)
from random import randint
def gen_rand (lo, hi, n):
for _ in range(n):
yield randint (lo, hi)
rd = gen_rand(1, 25, 10)
rnd_iter = iter(gen_rand(1, 25, 10))
next(rd)
next(rnd_iter)
for i in rnd_iter:
print(i)
for i in rd:
print(i)
s_iter = iter('hello')
s_iter
for c in s_iter:
print(c)
my_list = [1,2,3,4,5]
gencomp = (item for item in my_list if item > 3)
for item in gencomp:
print(item)
print(gencomp)
nums = (i for i in range(10))
nums
def word_lengths(phrase):
word_list = phrase.split()
fn = lambda x: len(x)
print(list(map(fn, word_list)))
word_lengths('How long are the words in this phrase')
from functools import reduce
def digits_to_num(digits):
fn = lambda x: str(x)
nums = list(map(fn, digits))
print(int(reduce(lambda x,y: x+y, nums)))
digits_to_num([3,4,3,2,1])
def filter_words(word_list, letter):
print(list(filter(lambda x: x[0] == letter, word_list)))
l = ['hello','are','cat','dog','ham','hi','go','to','heart']
filter_words(l,'h')
def concatenate(L1, L2, connector):
print([a + connector + b for a, b in zip(L1, L2)])
concatenate(['A','B'],['a','b'],'-')
def d_list(L):
print({v:i for i, v in enumerate(L)})
d_list(['a','b','c'])
def count_match_index(L):
print(len([v for i, v in enumerate(L) if i==v]))
count_match_index([0,2,2,1,5,5,6,10])
| 0.371821 | 0.674809 |
# 对抗示例生成
`Ascend` `GPU` `CPU` `进阶` `计算机视觉` `全流程`
[](https://authoring-modelarts-cnnorth4.huaweicloud.com/console/lab?share-url-b64=aHR0cHM6Ly9taW5kc3BvcmUtd2Vic2l0ZS5vYnMuY24tbm9ydGgtNC5teWh1YXdlaWNsb3VkLmNvbS9ub3RlYm9vay9tb2RlbGFydHMvcXVpY2tfc3RhcnQvbWluZHNwb3JlX2FkdmVyc2FyaWFsX2V4YW1wbGVfZ2VuZXJhdGlvbi5pcHluYg==&imageid=59a6e9f5-93c0-44dd-85b0-82f390c5d53b) [](https://mindspore-website.obs.cn-north-4.myhuaweicloud.com/notebook/master/tutorials/zh_cn/mindspore_adversarial_example_generation.ipynb) [](https://mindspore-website.obs.cn-north-4.myhuaweicloud.com/notebook/master/tutorials/zh_cn/mindspore_adversarial_example_generation.py) [](https://gitee.com/mindspore/docs/blob/master/tutorials/source_zh_cn/intermediate/image_and_video/adversarial_example_generation.ipynb)
近年来随着数据、计算能力、理论的不断发展演进,深度学习在图像、文本、语音、自动驾驶等众多领域都得到了广泛应用。与此同时,人们也越来越关注各类模型在使用过程中的安全问题,因为AI模型很容易受到外界有意无意的攻击而产生错误的结果。在本案例中,我们将以梯度符号攻击FGSM(Fast Gradient Sign Method)为例,演示此类攻击是如何误导模型的。
> 本篇基于CPU/GPU/Ascend环境运行。
## 对抗样本定义
Szegedy在2013年最早提出对抗样本的概念:在原始样本处加入人类无法察觉的微小扰动,使得深度模型性能下降,这种样本即对抗样本。如下图所示,本来预测为“panda”的图像在添加噪声之后,模型就将其预测为“gibbon”,右边的样本就是一个对抗样本:

> 图片来自[Explaining and Harnessing Adversarial Examples](https://arxiv.org/abs/1412.6572)。
## 攻击方法
对模型的攻击方法可以按照以下方法分类:
1. 攻击者掌握的信息多少:
- 白盒攻击:攻击者具有对模型的全部知识和访问权限,包括模型结构、权重、输入、输出。攻击者在产生对抗性攻击数据的过程中能够与模型系统有所交互。攻击者可以针对被攻击模型的特性设计特定的攻击算法。
- 黑盒攻击:与白盒攻击相反,攻击者仅具有关于模型的有限知识。攻击者对模型的结构权重一无所知,仅了解部分输入输出。
2. 攻击者的目的:
- 有目标的攻击:攻击者将模型结果误导为特定分类。
- 无目标的攻击:攻击者只想产生错误结果,而不在乎新结果是什么。
本案例中用到的FGSM是一种白盒攻击方法,既可以是有目标也可以是无目标攻击。
更多的模型安全功能可参考[MindArmour](https://www.mindspore.cn/mindarmour),现支持FGSM、LLC、Substitute Attack等多种对抗样本生成方法,并提供对抗样本鲁棒性模块、Fuzz Testing模块、隐私保护与评估模块,帮助用户增强模型安全性。
### 快速梯度符号攻击(FGSM)
正常分类网络的训练会定义一个损失函数,用于衡量模型输出值与样本真实标签的距离,通过反向传播计算模型梯度,梯度下降更新网络参数,减小损失值,提升模型精度。
FGSM(Fast Gradient Sign Method)是一种简单高效的对抗样本生成方法。不同于正常分类网络的训练过程,FGSM通过计算loss对于输入的梯度$\nabla_x J(\theta ,x ,y)$,这个梯度表征了loss对于输入变化的敏感性。然后在原始输入加上上述梯度,使得loss增大,模型对于改造后的输入样本分类效果变差,达到攻击效果。对抗样本的另一要求是生成样本与原始样本的差异要尽可能的小,使用sign函数可以使得修改图片时尽可能的均匀。
产生的对抗扰动用公式可以表示为:
$$ \eta = \varepsilon sign(\nabla_x J(\theta))$$
对抗样本可公式化为:
$$ x' = x + \epsilon \times sign(\nabla_x J(\theta ,x ,y)) $$
其中,
- $x$:正确分类为“Pandas”的原始输入图像。
- $y$:是$x$的输出。
- $\theta$:模型参数。
- $\varepsilon$:攻击系数。
- $J(\theta, x, y)$:训练网络的损失。
- $\nabla_x J(\theta)$:反向传播梯度。
## 实验前准备
### 导入模型训练需要的库
本案例将使用MNIST训练一个精度达标的LeNet网络,然后运行上文中所提到的FGSM攻击方法,实现错误分类的效果。
首先导入模型训练需要的库
```
import os
import numpy as np
from mindspore import Tensor, context, Model, load_checkpoint, load_param_into_net
import mindspore.nn as nn
import mindspore.ops as ops
from mindspore.common.initializer import Normal
from mindspore.train.callback import LossMonitor, ModelCheckpoint, CheckpointConfig
import mindspore.dataset as ds
import mindspore.dataset.transforms.c_transforms as C
import mindspore.dataset.vision.c_transforms as CV
from mindspore.dataset.vision import Inter
from mindspore import dtype as mstype
```
### 下载数据集
以下示例代码将数据集下载并解压到指定位置。
```
import os
import requests
requests.packages.urllib3.disable_warnings()
def download_dataset(dataset_url, path):
filename = dataset_url.split("/")[-1]
save_path = os.path.join(path, filename)
if os.path.exists(save_path):
return
if not os.path.exists(path):
os.makedirs(path)
res = requests.get(dataset_url, stream=True, verify=False)
with open(save_path, "wb") as f:
for chunk in res.iter_content(chunk_size=512):
if chunk:
f.write(chunk)
print("The {} file is downloaded and saved in the path {} after processing".format(os.path.basename(dataset_url), path))
train_path = "datasets/MNIST_Data/train"
test_path = "datasets/MNIST_Data/test"
download_dataset("https://mindspore-website.obs.myhuaweicloud.com/notebook/datasets/mnist/train-labels-idx1-ubyte", train_path)
download_dataset("https://mindspore-website.obs.myhuaweicloud.com/notebook/datasets/mnist/train-images-idx3-ubyte", train_path)
download_dataset("https://mindspore-website.obs.myhuaweicloud.com/notebook/datasets/mnist/t10k-labels-idx1-ubyte", test_path)
download_dataset("https://mindspore-website.obs.myhuaweicloud.com/notebook/datasets/mnist/t10k-images-idx3-ubyte", test_path)
```
下载的数据集文件的目录结构如下:
```text
./datasets/MNIST_Data
├── test
│ ├── t10k-images-idx3-ubyte
│ └── t10k-labels-idx1-ubyte
└── train
├── train-images-idx3-ubyte
└── train-labels-idx1-ubyte
```
## 攻击准备
在完成准备工作之后,开始训练精度达标的LeNet网络。
采用`GRAPH_MODE`在CPU/GPU/Ascend中运行本案例,下面将硬件设定为Ascend:
```
context.set_context(mode=context.GRAPH_MODE, device_target='Ascend')
```
### 训练LeNet网络
实验中使用LeNet作为演示模型完成图像分类,这里先定义网络并使用MNIST数据集进行训练。
定义LeNet网络:
```
class LeNet5(nn.Cell):
def __init__(self, num_class=10, num_channel=1):
super(LeNet5, self).__init__()
self.conv1 = nn.Conv2d(num_channel, 6, 5, pad_mode='valid')
self.conv2 = nn.Conv2d(6, 16, 5, pad_mode='valid')
self.fc1 = nn.Dense(16 * 5 * 5, 120, weight_init=Normal(0.02))
self.fc2 = nn.Dense(120, 84, weight_init=Normal(0.02))
self.fc3 = nn.Dense(84, num_class, weight_init=Normal(0.02))
self.relu = nn.ReLU()
self.max_pool2d = nn.MaxPool2d(kernel_size=2, stride=2)
self.flatten = nn.Flatten()
def construct(self, x):
x = self.conv1(x)
x = self.relu(x)
x = self.max_pool2d(x)
x = self.conv2(x)
x = self.relu(x)
x = self.max_pool2d(x)
x = self.flatten(x)
x = self.fc1(x)
x = self.relu(x)
x = self.fc2(x)
x = self.relu(x)
x = self.fc3(x)
return x
net = LeNet5()
```
进行数据处理:
```
def create_dataset(data_path, batch_size=1, repeat_size=1, num_parallel_workers=1):
# 定义数据集
mnist_ds = ds.MnistDataset(data_path)
resize_height, resize_width = 32, 32
rescale = 1.0 / 255.0
shift = 0.0
rescale_nml = 1 / 0.3081
shift_nml = -1 * 0.1307 / 0.3081
# 定义所需要操作的map映射
resize_op = CV.Resize((resize_height, resize_width), interpolation=Inter.LINEAR)
rescale_nml_op = CV.Rescale(rescale_nml, shift_nml)
rescale_op = CV.Rescale(rescale, shift)
hwc2chw_op = CV.HWC2CHW()
type_cast_op = C.TypeCast(mstype.int32)
# 使用map映射函数,将数据操作应用到数据集
mnist_ds = mnist_ds.map(operations=type_cast_op, input_columns="label", num_parallel_workers=num_parallel_workers)
mnist_ds = mnist_ds.map(operations=resize_op, input_columns="image", num_parallel_workers=num_parallel_workers)
mnist_ds = mnist_ds.map(operations=rescale_op, input_columns="image", num_parallel_workers=num_parallel_workers)
mnist_ds = mnist_ds.map(operations=rescale_nml_op, input_columns="image", num_parallel_workers=num_parallel_workers)
mnist_ds = mnist_ds.map(operations=hwc2chw_op, input_columns="image", num_parallel_workers=num_parallel_workers)
# 进行shuffle、batch、repeat操作
buffer_size = 10000
mnist_ds = mnist_ds.shuffle(buffer_size=buffer_size)
mnist_ds = mnist_ds.batch(batch_size, drop_remainder=True)
mnist_ds = mnist_ds.repeat(count=repeat_size)
return mnist_ds
```
定义优化器与损失函数:
```
net_loss = nn.SoftmaxCrossEntropyWithLogits(sparse=True, reduction='mean')
net_opt = nn.Momentum(net.trainable_params(), learning_rate=0.01, momentum=0.9)
```
定义网络参数:
```
config_ck = CheckpointConfig(save_checkpoint_steps=1875, keep_checkpoint_max=10)
ckpoint = ModelCheckpoint(prefix="checkpoint_lenet", config=config_ck)
```
定义LeNet网络的训练函数和测试函数:
```
def test_net(model, data_path):
ds_eval = create_dataset(os.path.join(data_path, "test"))
acc = model.eval(ds_eval, dataset_sink_mode=False)
print("{}".format(acc))
def train_net(model, epoch_size, data_path, repeat_size, ckpoint_cb, sink_mode):
ds_train = create_dataset(os.path.join(data_path, "train"), 32, repeat_size)
model.train(epoch_size, ds_train, callbacks=[ckpoint_cb, LossMonitor(125)], dataset_sink_mode=sink_mode)
train_epoch = 1
mnist_path = "./datasets/MNIST_Data/"
repeat_size = 1
model = Model(net, net_loss, net_opt, metrics={"Accuracy": nn.Accuracy()})
```
训练LeNet网络:
```
train_net(model, train_epoch, mnist_path, repeat_size, ckpoint, False)
```
测试此时的网络,可以观察到LeNet已经达到比较高的精度:
```
test_net(model, mnist_path)
```
加载已经训练好的LeNet模型:
```
param_dict = load_checkpoint("checkpoint_lenet-1_1875.ckpt")
load_param_into_net(net, param_dict)
```
### 实现FGSM
在得到精准的LeNet网络之后,下面将会采用FSGM攻击方法,在图像中加载噪声后重新进行测试。
先通过损失函数求取反向梯度:
```
class WithLossCell(nn.Cell):
"""
包装网络与损失函数
"""
def __init__(self, network, loss_fn):
super(WithLossCell, self).__init__()
self._network = network
self._loss_fn = loss_fn
def construct(self, data, label):
out = self._network(data)
return self._loss_fn(out, label)
class GradWrapWithLoss(nn.Cell):
"""
通过loss求反向梯度
"""
def __init__(self, network):
super(GradWrapWithLoss, self).__init__()
self._grad_all = ops.composite.GradOperation(get_all=True, sens_param=False)
self._network = network
def construct(self, inputs, labels):
gout = self._grad_all(self._network)(inputs, labels)
return gout[0]
```
然后根据公式实现FGSM攻击:
```
class FastGradientSignMethod:
"""
实现FGSM攻击
"""
def __init__(self, network, eps=0.07, loss_fn=None):
# 变量初始化
self._network = network
self._eps = eps
with_loss_cell = WithLossCell(self._network, loss_fn)
self._grad_all = GradWrapWithLoss(with_loss_cell)
self._grad_all.set_train()
def _gradient(self, inputs, labels):
# 求取梯度
out_grad = self._grad_all(inputs, labels)
gradient = out_grad.asnumpy()
gradient = np.sign(gradient)
return gradient
def generate(self, inputs, labels):
# 实现FGSM
inputs_tensor = Tensor(inputs)
labels_tensor = Tensor(labels)
gradient = self._gradient(inputs_tensor, labels_tensor)
# 产生扰动
perturbation = self._eps*gradient
# 生成受到扰动的图片
adv_x = inputs + perturbation
return adv_x
def batch_generate(self, inputs, labels, batch_size=32):
# 对数据集进行处理
arr_x = inputs
arr_y = labels
len_x = len(inputs)
batches = int(len_x / batch_size)
res = []
for i in range(batches):
x_batch = arr_x[i*batch_size: (i + 1)*batch_size]
y_batch = arr_y[i*batch_size: (i + 1)*batch_size]
adv_x = self.generate(x_batch, y_batch)
res.append(adv_x)
adv_x = np.concatenate(res, axis=0)
return adv_x
```
再次处理MINIST数据集中测试集的图片:
```
images = []
labels = []
test_images = []
test_labels = []
predict_labels = []
ds_test = create_dataset(os.path.join(mnist_path, "test"), batch_size=32).create_dict_iterator(output_numpy=True)
for data in ds_test:
images = data['image'].astype(np.float32)
labels = data['label']
test_images.append(images)
test_labels.append(labels)
pred_labels = np.argmax(model.predict(Tensor(images)).asnumpy(), axis=1)
predict_labels.append(pred_labels)
test_images = np.concatenate(test_images)
predict_labels = np.concatenate(predict_labels)
true_labels = np.concatenate(test_labels)
```
## 运行攻击
由FGSM攻击公式中可以看出,攻击系数$\varepsilon$越大,对梯度的改变就越大。当$\varepsilon$ 为零时则攻击效果不体现。
$\eta = \varepsilon sign(\nabla_x J(\theta))$
现在先观察当$\varepsilon$为零时的攻击效果:
```
fgsm = FastGradientSignMethod(net, eps=0.0, loss_fn=net_loss)
advs = fgsm.batch_generate(test_images, true_labels, batch_size=32)
adv_predicts = model.predict(Tensor(advs)).asnumpy()
adv_predicts = np.argmax(adv_predicts, axis=1)
accuracy = np.mean(np.equal(adv_predicts, true_labels))
print(accuracy)
```
再将$\varepsilon$设定为0.5,尝试运行攻击:
```
fgsm = FastGradientSignMethod(net, eps=0.5, loss_fn=net_loss)
advs = fgsm.batch_generate(test_images, true_labels, batch_size=32)
adv_predicts = model.predict(Tensor(advs)).asnumpy()
adv_predicts = np.argmax(adv_predicts, axis=1)
accuracy = np.mean(np.equal(adv_predicts, true_labels))
print(accuracy)
```
此时LeNet模型的精度大幅降低。
下面演示受攻击照片现在的实际形态,可以看出图片并没有发生明显的改变,然而在精度测试中却有了不一样的结果:
```
import matplotlib.pyplot as plt
adv_examples = np.transpose(advs[:10], [0, 2, 3, 1])
ori_examples = np.transpose(test_images[:10], [0, 2, 3, 1])
plt.figure()
for i in range(10):
plt.subplot(2, 10, i+1)
plt.imshow(np.squeeze(ori_examples[i]))
plt.subplot(2, 10, i+11)
plt.imshow(np.squeeze(adv_examples[i]))
plt.show()
```
|
github_jupyter
|
import os
import numpy as np
from mindspore import Tensor, context, Model, load_checkpoint, load_param_into_net
import mindspore.nn as nn
import mindspore.ops as ops
from mindspore.common.initializer import Normal
from mindspore.train.callback import LossMonitor, ModelCheckpoint, CheckpointConfig
import mindspore.dataset as ds
import mindspore.dataset.transforms.c_transforms as C
import mindspore.dataset.vision.c_transforms as CV
from mindspore.dataset.vision import Inter
from mindspore import dtype as mstype
import os
import requests
requests.packages.urllib3.disable_warnings()
def download_dataset(dataset_url, path):
filename = dataset_url.split("/")[-1]
save_path = os.path.join(path, filename)
if os.path.exists(save_path):
return
if not os.path.exists(path):
os.makedirs(path)
res = requests.get(dataset_url, stream=True, verify=False)
with open(save_path, "wb") as f:
for chunk in res.iter_content(chunk_size=512):
if chunk:
f.write(chunk)
print("The {} file is downloaded and saved in the path {} after processing".format(os.path.basename(dataset_url), path))
train_path = "datasets/MNIST_Data/train"
test_path = "datasets/MNIST_Data/test"
download_dataset("https://mindspore-website.obs.myhuaweicloud.com/notebook/datasets/mnist/train-labels-idx1-ubyte", train_path)
download_dataset("https://mindspore-website.obs.myhuaweicloud.com/notebook/datasets/mnist/train-images-idx3-ubyte", train_path)
download_dataset("https://mindspore-website.obs.myhuaweicloud.com/notebook/datasets/mnist/t10k-labels-idx1-ubyte", test_path)
download_dataset("https://mindspore-website.obs.myhuaweicloud.com/notebook/datasets/mnist/t10k-images-idx3-ubyte", test_path)
./datasets/MNIST_Data
├── test
│ ├── t10k-images-idx3-ubyte
│ └── t10k-labels-idx1-ubyte
└── train
├── train-images-idx3-ubyte
└── train-labels-idx1-ubyte
context.set_context(mode=context.GRAPH_MODE, device_target='Ascend')
class LeNet5(nn.Cell):
def __init__(self, num_class=10, num_channel=1):
super(LeNet5, self).__init__()
self.conv1 = nn.Conv2d(num_channel, 6, 5, pad_mode='valid')
self.conv2 = nn.Conv2d(6, 16, 5, pad_mode='valid')
self.fc1 = nn.Dense(16 * 5 * 5, 120, weight_init=Normal(0.02))
self.fc2 = nn.Dense(120, 84, weight_init=Normal(0.02))
self.fc3 = nn.Dense(84, num_class, weight_init=Normal(0.02))
self.relu = nn.ReLU()
self.max_pool2d = nn.MaxPool2d(kernel_size=2, stride=2)
self.flatten = nn.Flatten()
def construct(self, x):
x = self.conv1(x)
x = self.relu(x)
x = self.max_pool2d(x)
x = self.conv2(x)
x = self.relu(x)
x = self.max_pool2d(x)
x = self.flatten(x)
x = self.fc1(x)
x = self.relu(x)
x = self.fc2(x)
x = self.relu(x)
x = self.fc3(x)
return x
net = LeNet5()
def create_dataset(data_path, batch_size=1, repeat_size=1, num_parallel_workers=1):
# 定义数据集
mnist_ds = ds.MnistDataset(data_path)
resize_height, resize_width = 32, 32
rescale = 1.0 / 255.0
shift = 0.0
rescale_nml = 1 / 0.3081
shift_nml = -1 * 0.1307 / 0.3081
# 定义所需要操作的map映射
resize_op = CV.Resize((resize_height, resize_width), interpolation=Inter.LINEAR)
rescale_nml_op = CV.Rescale(rescale_nml, shift_nml)
rescale_op = CV.Rescale(rescale, shift)
hwc2chw_op = CV.HWC2CHW()
type_cast_op = C.TypeCast(mstype.int32)
# 使用map映射函数,将数据操作应用到数据集
mnist_ds = mnist_ds.map(operations=type_cast_op, input_columns="label", num_parallel_workers=num_parallel_workers)
mnist_ds = mnist_ds.map(operations=resize_op, input_columns="image", num_parallel_workers=num_parallel_workers)
mnist_ds = mnist_ds.map(operations=rescale_op, input_columns="image", num_parallel_workers=num_parallel_workers)
mnist_ds = mnist_ds.map(operations=rescale_nml_op, input_columns="image", num_parallel_workers=num_parallel_workers)
mnist_ds = mnist_ds.map(operations=hwc2chw_op, input_columns="image", num_parallel_workers=num_parallel_workers)
# 进行shuffle、batch、repeat操作
buffer_size = 10000
mnist_ds = mnist_ds.shuffle(buffer_size=buffer_size)
mnist_ds = mnist_ds.batch(batch_size, drop_remainder=True)
mnist_ds = mnist_ds.repeat(count=repeat_size)
return mnist_ds
net_loss = nn.SoftmaxCrossEntropyWithLogits(sparse=True, reduction='mean')
net_opt = nn.Momentum(net.trainable_params(), learning_rate=0.01, momentum=0.9)
config_ck = CheckpointConfig(save_checkpoint_steps=1875, keep_checkpoint_max=10)
ckpoint = ModelCheckpoint(prefix="checkpoint_lenet", config=config_ck)
def test_net(model, data_path):
ds_eval = create_dataset(os.path.join(data_path, "test"))
acc = model.eval(ds_eval, dataset_sink_mode=False)
print("{}".format(acc))
def train_net(model, epoch_size, data_path, repeat_size, ckpoint_cb, sink_mode):
ds_train = create_dataset(os.path.join(data_path, "train"), 32, repeat_size)
model.train(epoch_size, ds_train, callbacks=[ckpoint_cb, LossMonitor(125)], dataset_sink_mode=sink_mode)
train_epoch = 1
mnist_path = "./datasets/MNIST_Data/"
repeat_size = 1
model = Model(net, net_loss, net_opt, metrics={"Accuracy": nn.Accuracy()})
train_net(model, train_epoch, mnist_path, repeat_size, ckpoint, False)
test_net(model, mnist_path)
param_dict = load_checkpoint("checkpoint_lenet-1_1875.ckpt")
load_param_into_net(net, param_dict)
class WithLossCell(nn.Cell):
"""
包装网络与损失函数
"""
def __init__(self, network, loss_fn):
super(WithLossCell, self).__init__()
self._network = network
self._loss_fn = loss_fn
def construct(self, data, label):
out = self._network(data)
return self._loss_fn(out, label)
class GradWrapWithLoss(nn.Cell):
"""
通过loss求反向梯度
"""
def __init__(self, network):
super(GradWrapWithLoss, self).__init__()
self._grad_all = ops.composite.GradOperation(get_all=True, sens_param=False)
self._network = network
def construct(self, inputs, labels):
gout = self._grad_all(self._network)(inputs, labels)
return gout[0]
class FastGradientSignMethod:
"""
实现FGSM攻击
"""
def __init__(self, network, eps=0.07, loss_fn=None):
# 变量初始化
self._network = network
self._eps = eps
with_loss_cell = WithLossCell(self._network, loss_fn)
self._grad_all = GradWrapWithLoss(with_loss_cell)
self._grad_all.set_train()
def _gradient(self, inputs, labels):
# 求取梯度
out_grad = self._grad_all(inputs, labels)
gradient = out_grad.asnumpy()
gradient = np.sign(gradient)
return gradient
def generate(self, inputs, labels):
# 实现FGSM
inputs_tensor = Tensor(inputs)
labels_tensor = Tensor(labels)
gradient = self._gradient(inputs_tensor, labels_tensor)
# 产生扰动
perturbation = self._eps*gradient
# 生成受到扰动的图片
adv_x = inputs + perturbation
return adv_x
def batch_generate(self, inputs, labels, batch_size=32):
# 对数据集进行处理
arr_x = inputs
arr_y = labels
len_x = len(inputs)
batches = int(len_x / batch_size)
res = []
for i in range(batches):
x_batch = arr_x[i*batch_size: (i + 1)*batch_size]
y_batch = arr_y[i*batch_size: (i + 1)*batch_size]
adv_x = self.generate(x_batch, y_batch)
res.append(adv_x)
adv_x = np.concatenate(res, axis=0)
return adv_x
images = []
labels = []
test_images = []
test_labels = []
predict_labels = []
ds_test = create_dataset(os.path.join(mnist_path, "test"), batch_size=32).create_dict_iterator(output_numpy=True)
for data in ds_test:
images = data['image'].astype(np.float32)
labels = data['label']
test_images.append(images)
test_labels.append(labels)
pred_labels = np.argmax(model.predict(Tensor(images)).asnumpy(), axis=1)
predict_labels.append(pred_labels)
test_images = np.concatenate(test_images)
predict_labels = np.concatenate(predict_labels)
true_labels = np.concatenate(test_labels)
fgsm = FastGradientSignMethod(net, eps=0.0, loss_fn=net_loss)
advs = fgsm.batch_generate(test_images, true_labels, batch_size=32)
adv_predicts = model.predict(Tensor(advs)).asnumpy()
adv_predicts = np.argmax(adv_predicts, axis=1)
accuracy = np.mean(np.equal(adv_predicts, true_labels))
print(accuracy)
fgsm = FastGradientSignMethod(net, eps=0.5, loss_fn=net_loss)
advs = fgsm.batch_generate(test_images, true_labels, batch_size=32)
adv_predicts = model.predict(Tensor(advs)).asnumpy()
adv_predicts = np.argmax(adv_predicts, axis=1)
accuracy = np.mean(np.equal(adv_predicts, true_labels))
print(accuracy)
import matplotlib.pyplot as plt
adv_examples = np.transpose(advs[:10], [0, 2, 3, 1])
ori_examples = np.transpose(test_images[:10], [0, 2, 3, 1])
plt.figure()
for i in range(10):
plt.subplot(2, 10, i+1)
plt.imshow(np.squeeze(ori_examples[i]))
plt.subplot(2, 10, i+11)
plt.imshow(np.squeeze(adv_examples[i]))
plt.show()
| 0.580709 | 0.713232 |
## Reinforcement Learning Tutorial -2: DQN
### MD Muhaimin Rahman
contact: sezan92[at]gmail[dot]com
In the last tutorial, I tried to explain Q-learning algorithm. The biggest problem with Q-learning is that,it only takes discrete inputs and outputs discrete values. In the Mountain Car problem, we solved this issue by discretizing states which are actually continuous. But this can't be done always. Specially when the states are multidimensional states or images. Deep learning comes here to solve this problem!
For example breakout game by atari

Here , the states are the actual image itself! It is 210x160x3 size RGB numpy array. How will you make discrete for $Q learning$ ? Will that be efficient ? ***NO***! . DQN comes us to save us!
## Intuition
### Deep Q Learning
Q learning is a lookup table problem. i.e. You have the state , you just look at the table and see which action gives you best $Q$ value! That's it. But for continuous state - as mentioned above- you cannot make a lookup table! You need something like a regression model! Which will give you the Q values for given state and action! And the best regression model would be, Deep Neural Network!
So, we will replace the Q table explained in the last tutorial with a Neural Network. i.e. some thing like the following picture

***But there is one little problem!***
In our mountain car problem, we have three discrete actions. $0,1 & 2$ . Using the above architecture, we will have to calculate $Q$ value for each action . Because , you need to take the action with best $Q$ value. To get the action of the best $Q$ value, you need to know the $Q(state,action)$ for each state! So in our case, we will have to run same feed forward process three times!
What if we have more 100 actions ? Will we feed forward 100 times! It is a bit inefficient!! Instead, we will use the following architecture.

Meaning, our output layer will calculate $Q$ value for each action. As a result we can calculate Q values in one single forward pass each step! And then we will choose the action with maximum value
#### Bellman Update Equation
In the Original Equation, the bellman update equation is
\begin{equation}
Q(s_t,a) = Q(s_t,a) + \alpha (Q'(s_{t+1},a)-Q(s_t,a))
\end{equation}
For DQN, we will use similar equation, using Gradient descent
\begin{equation}
\theta_Q \gets \theta_Q - \alpha \frac{\partial}{\partial \theta}(Q'(s_{t+1},a)-Q(s_t,a))^2
\end{equation}
If you are intelligent enough, then you may ask , why there is a squareed part in the gradient descent equation but not in the actual bellman update equation? The reason might be, Mean squared errors are more sensitive to sudden spikes in the target data, which makes it most popular metric for regression models!
### The Concept of Experience Replay
One of the problems in Reinforcment learning is relearning Problem. That is , suppose, in the course of trial and error, one state $s_t$ comes only once or twice, and never comes back. What will happen? There is a chance that the Agent will forget that experience after some time- like us! . So we need to make the agent keep some kind of track of that memory as well. This problem was solved in 1993- yes 26 years ago- by Long Ji lin. In his paper, ***Reinforcement Learning for Robots Using Neural Networks*** , he introduced the concept of Experience Replay. What he did was, he initialized a buffer of a certain size . He stored the experiences of the agent, i.e. state $s_t$, action $a$,next state $s_{t+1}$, reward $r$ . Before training the agent, he you just sample randomly from the buffer . It also helps randomizing the data , which in turn, helps to converge the model faster, as mentioned by Yoshua Bengio in his paper ***Practical Recommendations for Gradient-Based Training of Deep
Architectures***,2012
### The concept of $\epsilon$-greedy Policy
In the beginning of training, we will have to explore random actions. Because we dont know the value of each action for each state. So we will take some random actions. We will evaluate those actions and see which random action gives us the most rewards. We will try to increase those actions. It means, at first you just ***explore*** different actions , the more you take actions the less you explore and more use your previouse experience to guide you-known as ***exploit***. This thing can be done using a technique -with another freaking out name- $\epsilon$-greedy policy.
The big idea is that, we will select a value of $\epsilon$ , suppose $0.9$. Then we will generate a random floating point number . If the generated number is greater than $\epsilon$ we will take action according to the DQN, otherwise a random action. After each episode , we will decrease the value of $\epsilon$ . As a result , in the last episodes, the agent will take actions according to DQN model, not the random actions.
- Set $\epsilon$
- Generate random number $n_{rand}$
- if $n_{rand} < \epsilon$ ***do***
- - take random action
- else ***do***
- - take action according to DQN
### Coding!!
Okay, let's start the most juicy part!
***Importing Libraries***
```
import gym
import numpy as np
from collections import deque
import random
import keras
from keras.models import Sequential
from keras.layers import Dense
from keras.optimizers import Adam
import matplotlib.pyplot as plt
from sklearn.preprocessing import OneHotEncoder
```
***Initialization of Environment***
```
env = gym.make('MountainCar-v0')
```
***Hyper parameters***
- ```action_size``` number of actions
- ```actions``` the actions list
- ```gamma``` discount factor $\gamma$
- ```lr``` learning rate $\alpha$
- ```num_episodes``` number of episodes
- ```epsilon``` epsilon , to choose random actions for epsilon greedy policy
- ```epsilon_decay``` epsilon decay rate
- ```batch_size``` batch size for training
```
action_list = [0,1,2]
gamma =0.45
lr =0.001
num_episodes =1000
epsilon =1
epsilon_decay =0.995
memory_size =1000
batch_size=100
show=False
action_size=env.action_space.n
state_size=env.observation_space.shape[0]
factor=[1,100]
```
Initializing Replay buffer for ***Experience Replay***
- ```memory``` a deque -which is a special type of list with limited memory- the replay buffer
- ```s``` current state
- ```a``` action
- ```new_s``` new state
- ```r``` reward
- ```d``` terminal
- ```experience``` tuple of state,reward,action,next state
***Psuedocode*** for experience replay
- get initial state $s$
- for each iteration do
- - take a random action $a$
- - get next state $s_{next}$, reward $r$, terminal $d$
- - $s \gets s_{next}$
- - if environment is terminated do
- - - reward $\gets$ -100
- - - reset environment
- - - add state,reward,action,next state into replay buffer
```
memory=deque(maxlen=memory_size)
s=env.reset()
s = s.reshape((1,-1))
s = s*factor
for _ in range(memory_size):
a=env.action_space.sample()
new_s,r,d,_ =env.step(a)
new_s = new_s.reshape((1,-1))
new_s = new_s*factor
if show:
env.render()
if d:
r=-100
experience =(s,r,a,new_s,d)
s=env.reset()
s = s.reshape((1,-1))
else:
experience =(s,r,a,new_s,d)
memory.append(experience)
s = new_s
env.close()
```
***Model Definition***
Here , I have defined the model as a simple MLP neural network with 2 hidden layers of 100 nodes with ```relu``` activation function
```
model = Sequential()
model.add(Dense(100,activation='relu',input_shape=(1,state_size)))
model.add(Dense(100,activation='relu'))
model.add(Dense(action_size,activation='linear'))
model.compile(loss='mse',optimizer=Adam(lr=lr),)
model.summary()
```
Here ,
- ```ep_list``` list of episodes
- ```reward_list``` list of rewards
- ```total_rewards``` totatl reward
***Psuedocode***
- for each episode do
- - get initial state $s$
- - $rewards_{total} \gets 0 $
- - set terminal $d$ to false
- - for each step do
- - - choose action based on epsilon greedy policy
- - - get next state $s_{next}$, reward $r$, terminal $d$ doing the action
- - - $rewards_{total} \gets rewards_{total}+r$
- - - if $d$ is $True $
- - - - if $rewards_{total}<-199$
- - - - - then give punishment $r \gets -100$
- - - - - break
- - - $s \gets s_{next}$
- - take random samples of $s,r,a,s_{next}$ from replay buffer
- - get $Q(s_{next})$
- - $Q_{target} \gets r+\gamma max(Q(s_{next})) $
- - $loss \gets \frac{1}{N}\sum(Q_{target}-Q(s))^2$
- - train the network using this loss
```
ep_list =[]
reward_list =[]
index=0
oh = OneHotEncoder(n_values=3)
for ep in range(num_episodes):
s= env.reset()
s=s.reshape((1,-1))
s = s*factor
total_rewards =0
d = False
j = 0
for j in range(200):
if np.random.random()< epsilon:
a = np.random.randint(0,len(action_list))
else:
Q = model.predict(s.reshape(-1,s.shape[0],s.shape[1]))
a =np.argmax(Q)
new_s,r,d,_ = env.step(a)
new_s = new_s.reshape((1,-1))
new_s = new_s*factor
total_rewards=total_rewards+r
if show:
env.render()
if d:
if total_rewards<-199:
r =-100
experience = (s,r,a,new_s,d)
memory.append(experience)
print("Episode %d, Failed! Reward %d"%(ep,total_rewards))
elif total_rewards<-110 and total_rewards>-199:
r=10
d=True
experience = (s,r,a,new_s,d)
memory.append(experience)
print("Episode %d, Better! Reward %d"%(ep,total_rewards))
elif total_rewards>=-110:
r=100
experience = (s,r,a,new_s,d)
memory.append(experience)
print("Episode %d, Passed! Reward %d"%(ep,total_rewards))
ep_list.append(ep)
reward_list.append(total_rewards)
break
experience = (s,r,a,new_s,d)
memory.append(experience)
if j==199:
print("Reward %d after full episode"%(total_rewards))
s = new_s
batches=random.sample(memory,batch_size)
states= np.array([batch[0] for batch in batches])
rewards= np.array([batch[1] for batch in batches])
actions= np.array([batch[2] for batch in batches])
actions=oh.fit_transform(actions.reshape(-1,1)).toarray()
actions = actions.reshape(-1,1,action_size)
new_states= np.array([batch[3] for batch in batches])
dones= np.array([batch[4] for batch in batches])
Qs =model.predict(states)
new_Qs = model.predict(new_states)
target_Qs=rewards.reshape(-1,1)+gamma*(np.max(new_Qs,axis=2)*(~dones.reshape(-1,1)))
Qs[actions==1]=target_Qs.reshape(-1,)
model.fit(states,Qs,verbose=0)
epsilon=epsilon*epsilon_decay
env.close()
plt.plot(reward_list)
plt.title("Rewards vs Episode")
plt.xlabel("Episodes")
plt.ylabel("Rewards")
```
|
github_jupyter
|
import gym
import numpy as np
from collections import deque
import random
import keras
from keras.models import Sequential
from keras.layers import Dense
from keras.optimizers import Adam
import matplotlib.pyplot as plt
from sklearn.preprocessing import OneHotEncoder
env = gym.make('MountainCar-v0')
action_list = [0,1,2]
gamma =0.45
lr =0.001
num_episodes =1000
epsilon =1
epsilon_decay =0.995
memory_size =1000
batch_size=100
show=False
action_size=env.action_space.n
state_size=env.observation_space.shape[0]
factor=[1,100]
memory=deque(maxlen=memory_size)
s=env.reset()
s = s.reshape((1,-1))
s = s*factor
for _ in range(memory_size):
a=env.action_space.sample()
new_s,r,d,_ =env.step(a)
new_s = new_s.reshape((1,-1))
new_s = new_s*factor
if show:
env.render()
if d:
r=-100
experience =(s,r,a,new_s,d)
s=env.reset()
s = s.reshape((1,-1))
else:
experience =(s,r,a,new_s,d)
memory.append(experience)
s = new_s
env.close()
model = Sequential()
model.add(Dense(100,activation='relu',input_shape=(1,state_size)))
model.add(Dense(100,activation='relu'))
model.add(Dense(action_size,activation='linear'))
model.compile(loss='mse',optimizer=Adam(lr=lr),)
model.summary()
ep_list =[]
reward_list =[]
index=0
oh = OneHotEncoder(n_values=3)
for ep in range(num_episodes):
s= env.reset()
s=s.reshape((1,-1))
s = s*factor
total_rewards =0
d = False
j = 0
for j in range(200):
if np.random.random()< epsilon:
a = np.random.randint(0,len(action_list))
else:
Q = model.predict(s.reshape(-1,s.shape[0],s.shape[1]))
a =np.argmax(Q)
new_s,r,d,_ = env.step(a)
new_s = new_s.reshape((1,-1))
new_s = new_s*factor
total_rewards=total_rewards+r
if show:
env.render()
if d:
if total_rewards<-199:
r =-100
experience = (s,r,a,new_s,d)
memory.append(experience)
print("Episode %d, Failed! Reward %d"%(ep,total_rewards))
elif total_rewards<-110 and total_rewards>-199:
r=10
d=True
experience = (s,r,a,new_s,d)
memory.append(experience)
print("Episode %d, Better! Reward %d"%(ep,total_rewards))
elif total_rewards>=-110:
r=100
experience = (s,r,a,new_s,d)
memory.append(experience)
print("Episode %d, Passed! Reward %d"%(ep,total_rewards))
ep_list.append(ep)
reward_list.append(total_rewards)
break
experience = (s,r,a,new_s,d)
memory.append(experience)
if j==199:
print("Reward %d after full episode"%(total_rewards))
s = new_s
batches=random.sample(memory,batch_size)
states= np.array([batch[0] for batch in batches])
rewards= np.array([batch[1] for batch in batches])
actions= np.array([batch[2] for batch in batches])
actions=oh.fit_transform(actions.reshape(-1,1)).toarray()
actions = actions.reshape(-1,1,action_size)
new_states= np.array([batch[3] for batch in batches])
dones= np.array([batch[4] for batch in batches])
Qs =model.predict(states)
new_Qs = model.predict(new_states)
target_Qs=rewards.reshape(-1,1)+gamma*(np.max(new_Qs,axis=2)*(~dones.reshape(-1,1)))
Qs[actions==1]=target_Qs.reshape(-1,)
model.fit(states,Qs,verbose=0)
epsilon=epsilon*epsilon_decay
env.close()
plt.plot(reward_list)
plt.title("Rewards vs Episode")
plt.xlabel("Episodes")
plt.ylabel("Rewards")
| 0.540681 | 0.983832 |
```
import rlssm
import pandas as pd
import os
par_path = os.path.abspath(os.path.join(os.getcwd(), os.pardir))
data_path = os.path.join(par_path, 'data/data_experiment.csv')
data = pd.read_csv(data_path, index_col=0)
data = data[data.participant < 5].reset_index(drop=True)
data['block_label'] += 1
data
model = rlssm.RLALBAModel_2A(2,
separate_learning_rates=True)
model.family, model.model_label, model.hierarchical_levels
```
### Fit
```
# sampling parameters
n_iter = 700
n_chains = 2
n_thin = 5
# learning parameters
K = 4 # n options
initial_value_learning = 17.5 # intitial value (Q0)
model_fit = model.fit(data,
K,
initial_value_learning,
thin = n_thin,
iter = n_iter,
chains = n_chains,
print_diagnostics = False)
```
#### Get Rhat
```
model_fit.rhat.describe()
model_fit.rhat.head()
```
#### Calculate wAIC
```
model_fit.waic
```
#### Check divergences
#### Get posteriors
```
model_fit.samples
model_fit.trial_samples
model_fit.plot_posteriors(height=5, show_intervals='HDI');
```
### Posterior predictives
```
import numpy as np
import seaborn as sns
data['choice_pair'] = 'AB'
data.loc[(data.cor_option == 3) & (data.inc_option == 1), 'choice_pair'] = 'AC'
data.loc[(data.cor_option == 4) & (data.inc_option == 2), 'choice_pair'] = 'BD'
data.loc[(data.cor_option == 4) & (data.inc_option == 3), 'choice_pair'] = 'CD'
data['block_bins'] = pd.cut(data.trial_block, 8, labels=np.arange(1, 9))
data.head()
```
#### Ungrouped posterior predictives:
```
pp = model_fit.get_posterior_predictives_df(n_posterior_predictives=100)
pp.head()
```
#### Grouped posterior predictives:
```
pp_grouped = model_fit.get_grouped_posterior_predictives_summary(grouping_vars=['choice_pair', 'block_bins'],
n_posterior_predictives=100)
pp_grouped.head()
```
#### Plot grouped posterior predictives:
```
model_fit.plot_mean_grouped_posterior_predictives(
grouping_vars=['block_bins', 'choice_pair'],
n_posterior_predictives=300,
figsize=(20,8),
hue_labels=['ab', 'ac', 'bd', 'cd'],
hue_order=['AB', 'AC', 'BD', 'CD'],
palette= sns.color_palette('husl'));
model_fit.plot_mean_grouped_posterior_predictives(
grouping_vars=['block_bins'],
n_posterior_predictives=300,
figsize=(20,8));
```
### Get starting values for further sampling
```
sv = model_fit.last_values
sv
```
|
github_jupyter
|
import rlssm
import pandas as pd
import os
par_path = os.path.abspath(os.path.join(os.getcwd(), os.pardir))
data_path = os.path.join(par_path, 'data/data_experiment.csv')
data = pd.read_csv(data_path, index_col=0)
data = data[data.participant < 5].reset_index(drop=True)
data['block_label'] += 1
data
model = rlssm.RLALBAModel_2A(2,
separate_learning_rates=True)
model.family, model.model_label, model.hierarchical_levels
# sampling parameters
n_iter = 700
n_chains = 2
n_thin = 5
# learning parameters
K = 4 # n options
initial_value_learning = 17.5 # intitial value (Q0)
model_fit = model.fit(data,
K,
initial_value_learning,
thin = n_thin,
iter = n_iter,
chains = n_chains,
print_diagnostics = False)
model_fit.rhat.describe()
model_fit.rhat.head()
model_fit.waic
model_fit.samples
model_fit.trial_samples
model_fit.plot_posteriors(height=5, show_intervals='HDI');
import numpy as np
import seaborn as sns
data['choice_pair'] = 'AB'
data.loc[(data.cor_option == 3) & (data.inc_option == 1), 'choice_pair'] = 'AC'
data.loc[(data.cor_option == 4) & (data.inc_option == 2), 'choice_pair'] = 'BD'
data.loc[(data.cor_option == 4) & (data.inc_option == 3), 'choice_pair'] = 'CD'
data['block_bins'] = pd.cut(data.trial_block, 8, labels=np.arange(1, 9))
data.head()
pp = model_fit.get_posterior_predictives_df(n_posterior_predictives=100)
pp.head()
pp_grouped = model_fit.get_grouped_posterior_predictives_summary(grouping_vars=['choice_pair', 'block_bins'],
n_posterior_predictives=100)
pp_grouped.head()
model_fit.plot_mean_grouped_posterior_predictives(
grouping_vars=['block_bins', 'choice_pair'],
n_posterior_predictives=300,
figsize=(20,8),
hue_labels=['ab', 'ac', 'bd', 'cd'],
hue_order=['AB', 'AC', 'BD', 'CD'],
palette= sns.color_palette('husl'));
model_fit.plot_mean_grouped_posterior_predictives(
grouping_vars=['block_bins'],
n_posterior_predictives=300,
figsize=(20,8));
sv = model_fit.last_values
sv
| 0.361616 | 0.626824 |
```
%matplotlib inline
from __future__ import absolute_import
from __future__ import division
from __future__ import print_function
import matplotlib.pyplot as plt
import tensorflow as tf
import numpy as np
import pickle as pkl
from sklearn.datasets import make_moons, make_blobs
from sklearn.decomposition import PCA
from flip_gradient import flip_gradient
from utils import *
```
Make a blob dataset. Dark vs. light colors distinguish domain and redish vs bluish colors distinguish class.
```
Xs, ys = make_blobs(300, centers=[[0, 0], [0, 1]], cluster_std=0.2)
Xt, yt = make_blobs(300, centers=[[1, -1], [1, 0]], cluster_std=0.2)
plt.scatter(Xs[:,0], Xs[:,1], c=ys, cmap='coolwarm', alpha=0.4)
plt.scatter(Xt[:,0], Xt[:,1], c=yt, cmap='cool', alpha=0.4)
batch_size = 16
def build_model(shallow_domain_classifier=True):
X = tf.placeholder(tf.float32, [None, 2], name='X') # Input data
Y_ind = tf.placeholder(tf.int32, [None], name='Y_ind') # Class index
D_ind = tf.placeholder(tf.int32, [None], name='D_ind') # Domain index
train = tf.placeholder(tf.bool, [], name='train') # Switch for routing data to class predictor
l = tf.placeholder(tf.float32, [], name='l') # Gradient reversal scaler
Y = tf.one_hot(Y_ind, 2)
D = tf.one_hot(D_ind, 2)
# Feature extractor - single layer
W0 = weight_variable([2, 15])
b0 = bias_variable([15])
F = tf.nn.relu(tf.matmul(X, W0) + b0, name='feature')
# Label predictor - single layer
f = tf.cond(train, lambda: tf.slice(F, [0, 0], [batch_size // 2, -1]), lambda: F)
y = tf.cond(train, lambda: tf.slice(Y, [0, 0], [batch_size // 2, -1]), lambda: Y)
W1 = weight_variable([15, 2])
b1 = bias_variable([2])
p_logit = tf.matmul(f, W1) + b1
p = tf.nn.softmax(p_logit)
p_loss = tf.nn.softmax_cross_entropy_with_logits(logits=p_logit, labels=y)
# Domain predictor - shallow
f_ = flip_gradient(F, l)
if shallow_domain_classifier:
W2 = weight_variable([15, 2])
b2 = bias_variable([2])
d_logit = tf.matmul(f_, W2) + b2
d = tf.nn.softmax(d_logit)
d_loss = tf.nn.softmax_cross_entropy_with_logits(logits=d_logit, labels=D)
else:
W2 = weight_variable([15, 8])
b2 = bias_variable([8])
h2 = tf.nn.relu(tf.matmul(f_, W2) + b2)
W3 = weight_variable([8, 2])
b3 = bias_variable([2])
d_logit = tf.matmul(h2, W3) + b3
d = tf.nn.softmax(d_logit)
d_loss = tf.nn.softmax_cross_entropy_with_logits(logits=d_logit, labels=D)
# Optimization
pred_loss = tf.reduce_sum(p_loss, name='pred_loss')
domain_loss = tf.reduce_sum(d_loss, name='domain_loss')
total_loss = tf.add(pred_loss, domain_loss, name='total_loss')
pred_train_op = tf.train.AdamOptimizer().minimize(pred_loss, name='pred_train_op')
domain_train_op = tf.train.AdamOptimizer().minimize(domain_loss, name='domain_train_op')
dann_train_op = tf.train.AdamOptimizer().minimize(total_loss, name='dann_train_op')
# Evaluation
p_acc = tf.reduce_mean(tf.cast(tf.equal(tf.argmax(y, 1), tf.argmax(p, 1)), tf.float32), name='p_acc')
d_acc = tf.reduce_mean(tf.cast(tf.equal(tf.argmax(D, 1), tf.argmax(d, 1)), tf.float32), name='d_acc')
build_model()
sess = tf.InteractiveSession()
def train_and_evaluate(sess, train_op_name, train_loss_name, grad_scale=None, num_batches=10000, verbose=True):
# Create batch builders
S_batches = batch_generator([Xs, ys], batch_size // 2)
T_batches = batch_generator([Xt, yt], batch_size // 2)
# Get output tensors and train op
d_acc = sess.graph.get_tensor_by_name('d_acc:0')
p_acc = sess.graph.get_tensor_by_name('p_acc:0')
train_loss = sess.graph.get_tensor_by_name(train_loss_name + ':0')
train_op = sess.graph.get_operation_by_name(train_op_name)
sess.run(tf.global_variables_initializer())
for i in range(num_batches):
# If no grad_scale, use a schedule
if grad_scale is None:
p = float(i) / num_batches
lp = 2. / (1. + np.exp(-10. * p)) - 1
else:
lp = grad_scale
X0, y0 = next(S_batches)
X1, y1 = next(T_batches)
Xb = np.vstack([X0, X1])
yb = np.hstack([y0, y1])
D_labels = np.hstack([np.zeros(batch_size // 2, dtype=np.int32),
np.ones(batch_size // 2, dtype=np.int32)])
_, loss, da, pa = sess.run([train_op, train_loss, d_acc, p_acc],
feed_dict={'X:0': Xb, 'Y_ind:0': yb, 'D_ind:0': D_labels,
'train:0': True, 'l:0': lp})
if verbose and i % 200 == 0:
print('loss: {}, domain accuracy: {}, class accuracy: {}'.format(loss, da, pa))
# Get final accuracies on whole dataset
das, pas = sess.run([d_acc, p_acc], feed_dict={'X:0': Xs, 'Y_ind:0': ys,
'D_ind:0': np.zeros(Xs.shape[0], dtype=np.int32), 'train:0': False, 'l:0': 1.0})
dat, pat = sess.run([d_acc, p_acc], feed_dict={'X:0': Xt, 'Y_ind:0': yt,
'D_ind:0': np.ones(Xt.shape[0], dtype=np.int32), 'train:0': False, 'l:0': 1.0})
print('Source domain: ', das)
print('Source class: ', pas)
print('Target domain: ', dat)
print('Target class: ', pat)
def extract_and_plot_pca_feats(sess, feat_tensor_name='feature'):
F = sess.graph.get_tensor_by_name(feat_tensor_name + ':0')
emb_s = sess.run(F, feed_dict={'X:0': Xs})
emb_t = sess.run(F, feed_dict={'X:0': Xt})
emb_all = np.vstack([emb_s, emb_t])
pca = PCA(n_components=2)
pca_emb = pca.fit_transform(emb_all)
num = pca_emb.shape[0] // 2
plt.scatter(pca_emb[:num,0], pca_emb[:num,1], c=ys, cmap='coolwarm', alpha=0.4)
plt.scatter(pca_emb[num:,0], pca_emb[num:,1], c=yt, cmap='cool', alpha=0.4)
```
#### Domain classification
Setting `grad_scale=-1.0` effectively turns off the gradient reversal. Training just the domain classifier creates representation that collapses classes.
```
train_and_evaluate(sess, 'domain_train_op', 'domain_loss', grad_scale=-1.0, verbose=False)
extract_and_plot_pca_feats(sess)
```
#### Label classification
Training label prediction in the source domain results in poor adapatation to the target. (1) The representation separates the classes in the source domain, but fails to separate the classes of the target domain. (2) The representation shows significant separation of classes.
```
train_and_evaluate(sess, 'pred_train_op', 'pred_loss', verbose=False)
extract_and_plot_pca_feats(sess)
```
#### Domain adaptation
Training with domain adversarial loss results in much better transfer to the target. The domains can usually still be distinguished in repeated experiments.
```
train_and_evaluate(sess, 'dann_train_op', 'total_loss', verbose=False)
extract_and_plot_pca_feats(sess)
```
#### Domain adaptation - deeper domain classifier
Using a domain classifier that is deeper seems to more reliably collapse the domains on this problem in repeated experiments.
```
sess.close()
tf.reset_default_graph()
build_model(False)
sess = tf.InteractiveSession()
train_and_evaluate(sess, 'dann_train_op', 'total_loss', verbose=False)
extract_and_plot_pca_feats(sess)
```
|
github_jupyter
|
%matplotlib inline
from __future__ import absolute_import
from __future__ import division
from __future__ import print_function
import matplotlib.pyplot as plt
import tensorflow as tf
import numpy as np
import pickle as pkl
from sklearn.datasets import make_moons, make_blobs
from sklearn.decomposition import PCA
from flip_gradient import flip_gradient
from utils import *
Xs, ys = make_blobs(300, centers=[[0, 0], [0, 1]], cluster_std=0.2)
Xt, yt = make_blobs(300, centers=[[1, -1], [1, 0]], cluster_std=0.2)
plt.scatter(Xs[:,0], Xs[:,1], c=ys, cmap='coolwarm', alpha=0.4)
plt.scatter(Xt[:,0], Xt[:,1], c=yt, cmap='cool', alpha=0.4)
batch_size = 16
def build_model(shallow_domain_classifier=True):
X = tf.placeholder(tf.float32, [None, 2], name='X') # Input data
Y_ind = tf.placeholder(tf.int32, [None], name='Y_ind') # Class index
D_ind = tf.placeholder(tf.int32, [None], name='D_ind') # Domain index
train = tf.placeholder(tf.bool, [], name='train') # Switch for routing data to class predictor
l = tf.placeholder(tf.float32, [], name='l') # Gradient reversal scaler
Y = tf.one_hot(Y_ind, 2)
D = tf.one_hot(D_ind, 2)
# Feature extractor - single layer
W0 = weight_variable([2, 15])
b0 = bias_variable([15])
F = tf.nn.relu(tf.matmul(X, W0) + b0, name='feature')
# Label predictor - single layer
f = tf.cond(train, lambda: tf.slice(F, [0, 0], [batch_size // 2, -1]), lambda: F)
y = tf.cond(train, lambda: tf.slice(Y, [0, 0], [batch_size // 2, -1]), lambda: Y)
W1 = weight_variable([15, 2])
b1 = bias_variable([2])
p_logit = tf.matmul(f, W1) + b1
p = tf.nn.softmax(p_logit)
p_loss = tf.nn.softmax_cross_entropy_with_logits(logits=p_logit, labels=y)
# Domain predictor - shallow
f_ = flip_gradient(F, l)
if shallow_domain_classifier:
W2 = weight_variable([15, 2])
b2 = bias_variable([2])
d_logit = tf.matmul(f_, W2) + b2
d = tf.nn.softmax(d_logit)
d_loss = tf.nn.softmax_cross_entropy_with_logits(logits=d_logit, labels=D)
else:
W2 = weight_variable([15, 8])
b2 = bias_variable([8])
h2 = tf.nn.relu(tf.matmul(f_, W2) + b2)
W3 = weight_variable([8, 2])
b3 = bias_variable([2])
d_logit = tf.matmul(h2, W3) + b3
d = tf.nn.softmax(d_logit)
d_loss = tf.nn.softmax_cross_entropy_with_logits(logits=d_logit, labels=D)
# Optimization
pred_loss = tf.reduce_sum(p_loss, name='pred_loss')
domain_loss = tf.reduce_sum(d_loss, name='domain_loss')
total_loss = tf.add(pred_loss, domain_loss, name='total_loss')
pred_train_op = tf.train.AdamOptimizer().minimize(pred_loss, name='pred_train_op')
domain_train_op = tf.train.AdamOptimizer().minimize(domain_loss, name='domain_train_op')
dann_train_op = tf.train.AdamOptimizer().minimize(total_loss, name='dann_train_op')
# Evaluation
p_acc = tf.reduce_mean(tf.cast(tf.equal(tf.argmax(y, 1), tf.argmax(p, 1)), tf.float32), name='p_acc')
d_acc = tf.reduce_mean(tf.cast(tf.equal(tf.argmax(D, 1), tf.argmax(d, 1)), tf.float32), name='d_acc')
build_model()
sess = tf.InteractiveSession()
def train_and_evaluate(sess, train_op_name, train_loss_name, grad_scale=None, num_batches=10000, verbose=True):
# Create batch builders
S_batches = batch_generator([Xs, ys], batch_size // 2)
T_batches = batch_generator([Xt, yt], batch_size // 2)
# Get output tensors and train op
d_acc = sess.graph.get_tensor_by_name('d_acc:0')
p_acc = sess.graph.get_tensor_by_name('p_acc:0')
train_loss = sess.graph.get_tensor_by_name(train_loss_name + ':0')
train_op = sess.graph.get_operation_by_name(train_op_name)
sess.run(tf.global_variables_initializer())
for i in range(num_batches):
# If no grad_scale, use a schedule
if grad_scale is None:
p = float(i) / num_batches
lp = 2. / (1. + np.exp(-10. * p)) - 1
else:
lp = grad_scale
X0, y0 = next(S_batches)
X1, y1 = next(T_batches)
Xb = np.vstack([X0, X1])
yb = np.hstack([y0, y1])
D_labels = np.hstack([np.zeros(batch_size // 2, dtype=np.int32),
np.ones(batch_size // 2, dtype=np.int32)])
_, loss, da, pa = sess.run([train_op, train_loss, d_acc, p_acc],
feed_dict={'X:0': Xb, 'Y_ind:0': yb, 'D_ind:0': D_labels,
'train:0': True, 'l:0': lp})
if verbose and i % 200 == 0:
print('loss: {}, domain accuracy: {}, class accuracy: {}'.format(loss, da, pa))
# Get final accuracies on whole dataset
das, pas = sess.run([d_acc, p_acc], feed_dict={'X:0': Xs, 'Y_ind:0': ys,
'D_ind:0': np.zeros(Xs.shape[0], dtype=np.int32), 'train:0': False, 'l:0': 1.0})
dat, pat = sess.run([d_acc, p_acc], feed_dict={'X:0': Xt, 'Y_ind:0': yt,
'D_ind:0': np.ones(Xt.shape[0], dtype=np.int32), 'train:0': False, 'l:0': 1.0})
print('Source domain: ', das)
print('Source class: ', pas)
print('Target domain: ', dat)
print('Target class: ', pat)
def extract_and_plot_pca_feats(sess, feat_tensor_name='feature'):
F = sess.graph.get_tensor_by_name(feat_tensor_name + ':0')
emb_s = sess.run(F, feed_dict={'X:0': Xs})
emb_t = sess.run(F, feed_dict={'X:0': Xt})
emb_all = np.vstack([emb_s, emb_t])
pca = PCA(n_components=2)
pca_emb = pca.fit_transform(emb_all)
num = pca_emb.shape[0] // 2
plt.scatter(pca_emb[:num,0], pca_emb[:num,1], c=ys, cmap='coolwarm', alpha=0.4)
plt.scatter(pca_emb[num:,0], pca_emb[num:,1], c=yt, cmap='cool', alpha=0.4)
train_and_evaluate(sess, 'domain_train_op', 'domain_loss', grad_scale=-1.0, verbose=False)
extract_and_plot_pca_feats(sess)
train_and_evaluate(sess, 'pred_train_op', 'pred_loss', verbose=False)
extract_and_plot_pca_feats(sess)
train_and_evaluate(sess, 'dann_train_op', 'total_loss', verbose=False)
extract_and_plot_pca_feats(sess)
sess.close()
tf.reset_default_graph()
build_model(False)
sess = tf.InteractiveSession()
train_and_evaluate(sess, 'dann_train_op', 'total_loss', verbose=False)
extract_and_plot_pca_feats(sess)
| 0.712732 | 0.649231 |
```
R.<x> = PolynomialRing(ZZ)
def fekete(p):
#compute f_p(x)
v=[kronecker(a+1,p) for a in range(0,p-1)]
F_p=R(v)
if p%4==3:
coef=[1, -1]
factor=R(coef)
f,r =F_p.quo_rem(factor)
if p%4==1:
coef1=[1, -1, -1, 1]
factor1=R(coef1)
f,r=F_p.quo_rem(factor1)
return f
def reduced_fekete(p):
f_p=fekete(p)
u=f_p.trace_polynomial()
g_p=u[0]
return g_p
def fekete_reduction(p, q):
f_p=fekete(p)
f=f_p.change_ring(GF(q))
return f.factor()
def almost_cycle(p,n):
for q in range(n):
if is_prime(q):
factor=fekete_reduction(p,q)
if len(factor)==3:
factor1=factor[0][0]
factor2=factor[1][0]
degree1=factor1.degree()
degree2=factor2.degree()
if degree1==1 and degree2==1 and factor[0][1]==1 and factor[1][1]==1 and factor[2][1]==1:
return q
return -1
def irreducible(p,n):
for q in range(n):
if is_prime(q):
factor=fekete_reduction(p,q)
if len(factor)==1 and factor[0][1]==1:
return q
return -1
def length_test_2(v):
#count the number of even entries in v
count2=0
for item in v:
if item==2:
count2 +=1
count_even=0
for item in v:
if item %2 ==0:
count_even +=1
if count2==count_even==1:
return True
return False
def length_test_4(v):
#count the number of even entries in v
count4=0
for item in v:
if item==4:
count4 +=1
count_even=0
for item in v:
if item %2 ==0:
count_even +=1
if count4==count_even==1:
return True
return False
def two_cycle(p,n):
result=[]
f=fekete(p)
for q in range(n):
v=[]
if is_prime(q):
factor=fekete_reduction(p,q)
for item in factor:
v.append(item[0].degree())
if sum(v)==f.degree() and length_test_2(v):
return q
return -1
def four_cycle(p,q):
result=[]
f=fekete(p)
for q in range(n):
v=[]
if is_prime(q):
factor=fekete_reduction(p,q)
for item in factor:
v.append(item[0].degree())
if sum(v)==f.degree() and length_test_4(v):
return q
return -1
def search(p,n):
irr=irreducible(p,n)
print(f"The first prime that g is irreducible is: q=",irr)
q_cycle=cycle(p,n)
print(f"The first prime that g has a cycle is: q=", q_cycle)
q_tranposition=tranposition(p,n)
print(f"The first prime that g has a tranposition is q=", q_tranposition)
def search_1(p,n):
irr=irreducible(p,n)
q_cycle=almost_cycle(p,n)
q_tranposition=two_cycle(p,n)
q_four_cycle=four_cycle(p,n)
res=(irr, q_cycle, q_tranposition)
return res
```
## Let us test the above codes for small $p$ and random $p$.
```
#irreducibility test
p=11
n=100000
q=irreducible(p,n)
print(f"f is irreducible at q= ", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
#two_cycle_search
p=11
n=100000
q=two_cycle(p,n)
print(f"f has a 2-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
#(2n-2)_cycle_search
p=11
n=100000
q=almost_cycle(p,n)
print(f"f has a (2n-2)-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
#four_cycle_search
p=11
n=100000
q=four_cycle(p,n)
print(f"f has a 4-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
p=11
n=100000
print(f"The prime we are considering is p=", p)
q=irreducible(p,n)
print(f"f is irreducible at q= ", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
#full_search
p=11
n=100000
q=irreducible(p,n)
f=fekete(p)
print(f"The degree of f is deg=", f.degree())
print(f"f is irreducible at q= ", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
n=100000
q=almost_cycle(p,n)
print(f"f has a (2n-2)-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
q=four_cycle(p,n)
print(f"f has a 4-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
q=two_cycle(p,n)
print(f"f has a 2-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
#full_search
p=13
n=100000
q=irreducible(p,n)
f=fekete(p)
print(f"The degree of f is deg=", f.degree())
print(f"f is irreducible at q= ", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
n=100000
q=almost_cycle(p,n)
print(f"f has a (2n-2)-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
q=four_cycle(p,n)
print(f"f has a 4-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
q=two_cycle(p,n)
print(f"f has a 2-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
#full_search
p=17
n=100000
q=irreducible(p,n)
f=fekete(p)
print(f"The degree of f is deg=", f.degree())
print(f"f is irreducible at q= ", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
n=100000
q=almost_cycle(p,n)
print(f"f has a (2n-2)-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
q=four_cycle(p,n)
print(f"f has a 4-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
q=two_cycle(p,n)
print(f"f has a 2-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
#full_search
p=19
n=100000
q=irreducible(p,n)
f=fekete(p)
print(f"The degree of f is deg=", f.degree())
print(f"f is irreducible at q= ", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
n=100000
q=almost_cycle(p,n)
print(f"f has a (2n-2)-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
q=four_cycle(p,n)
print(f"f has a 4-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
q=two_cycle(p,n)
print(f"f has a 2-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
#full_search
P=Primes()
p=19
n=100000
p=P.next(p)
q=irreducible(p,n)
f=fekete(p)
print(f"The prime we are considering is p=", p)
print(f"The degree of f is deg=", f.degree())
print(f"f is irreducible at q= ", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
n=100000
q=almost_cycle(p,n)
print(f"f has a (2n-2)-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
q=four_cycle(p,n)
print(f"f has a 4-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
q=two_cycle(p,n)
print(f"f has a 2-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
#full_search
P=Primes()
n=100000
p=P.next(p)
q=irreducible(p,n)
f=fekete(p)
print(f"The prime we are considering is p=", p)
print(f"The degree of f is deg=", f.degree())
print(f"f is irreducible at q= ", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
n=100000
q=almost_cycle(p,n)
print(f"f has a (2n-2)-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
q=four_cycle(p,n)
print(f"f has a 4-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
q=two_cycle(p,n)
print(f"f has a 2-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
#full_search
P=Primes()
n=100000
p=P.next(p)
q=irreducible(p,n)
f=fekete(p)
print(f"The prime we are considering is p=", p)
print(f"The degree of f is deg=", f.degree())
print(f"f is irreducible at q= ", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
n=100000
q=almost_cycle(p,n)
print(f"f has a (2n-2)-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
q=four_cycle(p,n)
print(f"f has a 4-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
q=two_cycle(p,n)
print(f"f has a 2-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
#full_search
P=Primes()
n=100000
p=P.next(p)
q=irreducible(p,n)
f=fekete(p)
print(f"The prime we are considering is p=", p)
print(f"The degree of f is deg=", f.degree())
print(f"f is irreducible at q= ", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
n=100000
q=almost_cycle(p,n)
print(f"f has a (2n-2)-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
q=four_cycle(p,n)
print(f"f has a 4-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
q=two_cycle(p,n)
print(f"f has a 2-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
#full_search
P=Primes()
n=100000
p=P.next(p)
q=irreducible(p,n)
f=fekete(p)
print(f"The prime we are considering is p=", p)
print(f"The degree of f is deg=", f.degree())
print(f"f is irreducible at q= ", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
n=100000
q=almost_cycle(p,n)
print(f"f has a (2n-2)-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
q=four_cycle(p,n)
print(f"f has a 4-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
q=two_cycle(p,n)
print(f"f has a 2-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
#full_search
P=Primes()
n=100000
p=P.next(p)
q=irreducible(p,n)
f=fekete(p)
print(f"The prime we are considering is p=", p)
print(f"The degree of f is deg=", f.degree())
print(f"f is irreducible at q= ", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
n=100000
q=almost_cycle(p,n)
print(f"f has a (2n-2)-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
q=four_cycle(p,n)
print(f"f has a 4-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
q=two_cycle(p,n)
print(f"f has a 2-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
#full_search
P=Primes()
n=100000
p=P.next(p)
q=irreducible(p,n)
f=fekete(p)
print(f"The prime we are considering is p=", p)
print(f"The degree of f is deg=", f.degree())
print(f"f is irreducible at q= ", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
n=100000
q=almost_cycle(p,n)
print(f"f has a (2n-2)-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
q=four_cycle(p,n)
print(f"f has a 4-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
q=two_cycle(p,n)
print(f"f has a 2-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
```
## Next, we will find the smallest tuples $(q_1, q_2, q_3, q_4)$ for $p<200$.
```
p=7
n=10**6
P=Primes()
res=[]
while p<200:
q_irr=irreducible(p,n)
q_cycle=almost_cycle(p,n)
q_tranposition=two_cycle(p,n)
q_four_cycle=four_cycle(p,n)
res.append([p, [q_irr, q_cycle, q_tranposition, q_four_cycle]])
p=P.next(p)
res
```
## Next, we find smallest tuples $(q_1, q_2, q_3, q_4)$ for 200<p<400.
```
p=199
n=10**6
P=Primes()
res=[]
while p<400:
q_irr=irreducible(p,n)
q_cycle=almost_cycle(p,n)
q_tranposition=two_cycle(p,n)
q_four_cycle=four_cycle(p,n)
res.append([p, [q_irr, q_cycle, q_tranposition, q_four_cycle]])
p=P.next(p)
res
```
## Next, we find smallest tuples $(q_1, q_2, q_3, q_4)$ for 400<p<600.
```
p=397
n=10**6
P=Primes()
res=[]
while p<500:
q_irr=irreducible(p,n)
q_cycle=almost_cycle(p,n)
q_tranposition=two_cycle(p,n)
q_four_cycle=four_cycle(p,n)
res.append([p, [q_irr, q_cycle, q_tranposition, q_four_cycle]])
p=P.next(p)
res
%%time
p=499
n=10**6
P=Primes()
res=[]
while p<600:
q_irr=irreducible(p,n)
q_cycle=almost_cycle(p,n)
q_tranposition=two_cycle(p,n)
q_four_cycle=four_cycle(p,n)
res.append([p, [q_irr, q_cycle, q_tranposition, q_four_cycle]])
p=P.next(p)
res
```
## Next we compute the running time for larger $p$.
```
%%time
p=599
P=Primes()
p=P.next(p)
n=10**6
P=Primes()
res=[]
q_irr=irreducible(p,n)
q_cycle=almost_cycle(p,n)
q_tranposition=two_cycle(p,n)
q_four_cycle=four_cycle(p,n)
res.append([p, [q_irr, q_cycle, q_tranposition, q_four_cycle]])
res
%%time
p=601
P=Primes()
p=P.next(p)
n=10**6
P=Primes()
res=[]
q_irr=irreducible(p,n)
q_cycle=almost_cycle(p,n)
q_tranposition=two_cycle(p,n)
q_four_cycle=four_cycle(p,n)
res.append([p, [q_irr, q_cycle, q_tranposition, q_four_cycle]])
res
%%time
p=607
P=Primes()
p=P.next(p)
n=10**6
P=Primes()
res=[]
q_irr=irreducible(p,n)
q_cycle=almost_cycle(p,n)
q_tranposition=two_cycle(p,n)
q_four_cycle=four_cycle(p,n)
res.append([p, [q_irr, q_cycle, q_tranposition, q_four_cycle]])
res
%%time
p=613
P=Primes()
p=P.next(p)
n=10**6
P=Primes()
res=[]
q_irr=irreducible(p,n)
q_cycle=almost_cycle(p,n)
q_tranposition=two_cycle(p,n)
q_four_cycle=four_cycle(p,n)
res.append([p, [q_irr, q_cycle, q_tranposition, q_four_cycle]])
res
%%time
p=617
P=Primes()
p=P.next(p)
n=10**6
P=Primes()
res=[]
q_irr=irreducible(p,n)
q_cycle=almost_cycle(p,n)
q_tranposition=two_cycle(p,n)
q_four_cycle=four_cycle(p,n)
res.append([p, [q_irr, q_cycle, q_tranposition, q_four_cycle]])
res
%%time
p=619
P=Primes()
p=P.next(p)
n=10**6
P=Primes()
res=[]
q_irr=irreducible(p,n)
q_cycle=almost_cycle(p,n)
q_tranposition=two_cycle(p,n)
q_four_cycle=four_cycle(p,n)
res.append([p, [q_irr, q_cycle, q_tranposition, q_four_cycle]])
res
%%time
p=631
P=Primes()
p=P.next(p)
n=10**6
P=Primes()
res=[]
q_irr=irreducible(p,n)
q_cycle=almost_cycle(p,n)
q_tranposition=two_cycle(p,n)
q_four_cycle=four_cycle(p,n)
res.append([p, [q_irr, q_cycle, q_tranposition, q_four_cycle]])
res
%%time
p=641
P=Primes()
p=P.next(p)
n=10**6
P=Primes()
res=[]
q_irr=irreducible(p,n)
q_cycle=almost_cycle(p,n)
q_tranposition=two_cycle(p,n)
q_four_cycle=four_cycle(p,n)
res.append([p, [q_irr, q_cycle, q_tranposition, q_four_cycle]])
res
```
|
github_jupyter
|
R.<x> = PolynomialRing(ZZ)
def fekete(p):
#compute f_p(x)
v=[kronecker(a+1,p) for a in range(0,p-1)]
F_p=R(v)
if p%4==3:
coef=[1, -1]
factor=R(coef)
f,r =F_p.quo_rem(factor)
if p%4==1:
coef1=[1, -1, -1, 1]
factor1=R(coef1)
f,r=F_p.quo_rem(factor1)
return f
def reduced_fekete(p):
f_p=fekete(p)
u=f_p.trace_polynomial()
g_p=u[0]
return g_p
def fekete_reduction(p, q):
f_p=fekete(p)
f=f_p.change_ring(GF(q))
return f.factor()
def almost_cycle(p,n):
for q in range(n):
if is_prime(q):
factor=fekete_reduction(p,q)
if len(factor)==3:
factor1=factor[0][0]
factor2=factor[1][0]
degree1=factor1.degree()
degree2=factor2.degree()
if degree1==1 and degree2==1 and factor[0][1]==1 and factor[1][1]==1 and factor[2][1]==1:
return q
return -1
def irreducible(p,n):
for q in range(n):
if is_prime(q):
factor=fekete_reduction(p,q)
if len(factor)==1 and factor[0][1]==1:
return q
return -1
def length_test_2(v):
#count the number of even entries in v
count2=0
for item in v:
if item==2:
count2 +=1
count_even=0
for item in v:
if item %2 ==0:
count_even +=1
if count2==count_even==1:
return True
return False
def length_test_4(v):
#count the number of even entries in v
count4=0
for item in v:
if item==4:
count4 +=1
count_even=0
for item in v:
if item %2 ==0:
count_even +=1
if count4==count_even==1:
return True
return False
def two_cycle(p,n):
result=[]
f=fekete(p)
for q in range(n):
v=[]
if is_prime(q):
factor=fekete_reduction(p,q)
for item in factor:
v.append(item[0].degree())
if sum(v)==f.degree() and length_test_2(v):
return q
return -1
def four_cycle(p,q):
result=[]
f=fekete(p)
for q in range(n):
v=[]
if is_prime(q):
factor=fekete_reduction(p,q)
for item in factor:
v.append(item[0].degree())
if sum(v)==f.degree() and length_test_4(v):
return q
return -1
def search(p,n):
irr=irreducible(p,n)
print(f"The first prime that g is irreducible is: q=",irr)
q_cycle=cycle(p,n)
print(f"The first prime that g has a cycle is: q=", q_cycle)
q_tranposition=tranposition(p,n)
print(f"The first prime that g has a tranposition is q=", q_tranposition)
def search_1(p,n):
irr=irreducible(p,n)
q_cycle=almost_cycle(p,n)
q_tranposition=two_cycle(p,n)
q_four_cycle=four_cycle(p,n)
res=(irr, q_cycle, q_tranposition)
return res
#irreducibility test
p=11
n=100000
q=irreducible(p,n)
print(f"f is irreducible at q= ", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
#two_cycle_search
p=11
n=100000
q=two_cycle(p,n)
print(f"f has a 2-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
#(2n-2)_cycle_search
p=11
n=100000
q=almost_cycle(p,n)
print(f"f has a (2n-2)-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
#four_cycle_search
p=11
n=100000
q=four_cycle(p,n)
print(f"f has a 4-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
p=11
n=100000
print(f"The prime we are considering is p=", p)
q=irreducible(p,n)
print(f"f is irreducible at q= ", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
#full_search
p=11
n=100000
q=irreducible(p,n)
f=fekete(p)
print(f"The degree of f is deg=", f.degree())
print(f"f is irreducible at q= ", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
n=100000
q=almost_cycle(p,n)
print(f"f has a (2n-2)-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
q=four_cycle(p,n)
print(f"f has a 4-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
q=two_cycle(p,n)
print(f"f has a 2-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
#full_search
p=13
n=100000
q=irreducible(p,n)
f=fekete(p)
print(f"The degree of f is deg=", f.degree())
print(f"f is irreducible at q= ", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
n=100000
q=almost_cycle(p,n)
print(f"f has a (2n-2)-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
q=four_cycle(p,n)
print(f"f has a 4-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
q=two_cycle(p,n)
print(f"f has a 2-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
#full_search
p=17
n=100000
q=irreducible(p,n)
f=fekete(p)
print(f"The degree of f is deg=", f.degree())
print(f"f is irreducible at q= ", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
n=100000
q=almost_cycle(p,n)
print(f"f has a (2n-2)-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
q=four_cycle(p,n)
print(f"f has a 4-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
q=two_cycle(p,n)
print(f"f has a 2-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
#full_search
p=19
n=100000
q=irreducible(p,n)
f=fekete(p)
print(f"The degree of f is deg=", f.degree())
print(f"f is irreducible at q= ", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
n=100000
q=almost_cycle(p,n)
print(f"f has a (2n-2)-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
q=four_cycle(p,n)
print(f"f has a 4-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
q=two_cycle(p,n)
print(f"f has a 2-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
#full_search
P=Primes()
p=19
n=100000
p=P.next(p)
q=irreducible(p,n)
f=fekete(p)
print(f"The prime we are considering is p=", p)
print(f"The degree of f is deg=", f.degree())
print(f"f is irreducible at q= ", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
n=100000
q=almost_cycle(p,n)
print(f"f has a (2n-2)-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
q=four_cycle(p,n)
print(f"f has a 4-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
q=two_cycle(p,n)
print(f"f has a 2-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
#full_search
P=Primes()
n=100000
p=P.next(p)
q=irreducible(p,n)
f=fekete(p)
print(f"The prime we are considering is p=", p)
print(f"The degree of f is deg=", f.degree())
print(f"f is irreducible at q= ", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
n=100000
q=almost_cycle(p,n)
print(f"f has a (2n-2)-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
q=four_cycle(p,n)
print(f"f has a 4-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
q=two_cycle(p,n)
print(f"f has a 2-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
#full_search
P=Primes()
n=100000
p=P.next(p)
q=irreducible(p,n)
f=fekete(p)
print(f"The prime we are considering is p=", p)
print(f"The degree of f is deg=", f.degree())
print(f"f is irreducible at q= ", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
n=100000
q=almost_cycle(p,n)
print(f"f has a (2n-2)-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
q=four_cycle(p,n)
print(f"f has a 4-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
q=two_cycle(p,n)
print(f"f has a 2-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
#full_search
P=Primes()
n=100000
p=P.next(p)
q=irreducible(p,n)
f=fekete(p)
print(f"The prime we are considering is p=", p)
print(f"The degree of f is deg=", f.degree())
print(f"f is irreducible at q= ", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
n=100000
q=almost_cycle(p,n)
print(f"f has a (2n-2)-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
q=four_cycle(p,n)
print(f"f has a 4-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
q=two_cycle(p,n)
print(f"f has a 2-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
#full_search
P=Primes()
n=100000
p=P.next(p)
q=irreducible(p,n)
f=fekete(p)
print(f"The prime we are considering is p=", p)
print(f"The degree of f is deg=", f.degree())
print(f"f is irreducible at q= ", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
n=100000
q=almost_cycle(p,n)
print(f"f has a (2n-2)-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
q=four_cycle(p,n)
print(f"f has a 4-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
q=two_cycle(p,n)
print(f"f has a 2-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
#full_search
P=Primes()
n=100000
p=P.next(p)
q=irreducible(p,n)
f=fekete(p)
print(f"The prime we are considering is p=", p)
print(f"The degree of f is deg=", f.degree())
print(f"f is irreducible at q= ", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
n=100000
q=almost_cycle(p,n)
print(f"f has a (2n-2)-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
q=four_cycle(p,n)
print(f"f has a 4-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
q=two_cycle(p,n)
print(f"f has a 2-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
#full_search
P=Primes()
n=100000
p=P.next(p)
q=irreducible(p,n)
f=fekete(p)
print(f"The prime we are considering is p=", p)
print(f"The degree of f is deg=", f.degree())
print(f"f is irreducible at q= ", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
n=100000
q=almost_cycle(p,n)
print(f"f has a (2n-2)-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
q=four_cycle(p,n)
print(f"f has a 4-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
q=two_cycle(p,n)
print(f"f has a 2-cycle at q", q)
print(f"The factorization of f at q: \n", fekete_reduction(p,q))
p=7
n=10**6
P=Primes()
res=[]
while p<200:
q_irr=irreducible(p,n)
q_cycle=almost_cycle(p,n)
q_tranposition=two_cycle(p,n)
q_four_cycle=four_cycle(p,n)
res.append([p, [q_irr, q_cycle, q_tranposition, q_four_cycle]])
p=P.next(p)
res
p=199
n=10**6
P=Primes()
res=[]
while p<400:
q_irr=irreducible(p,n)
q_cycle=almost_cycle(p,n)
q_tranposition=two_cycle(p,n)
q_four_cycle=four_cycle(p,n)
res.append([p, [q_irr, q_cycle, q_tranposition, q_four_cycle]])
p=P.next(p)
res
p=397
n=10**6
P=Primes()
res=[]
while p<500:
q_irr=irreducible(p,n)
q_cycle=almost_cycle(p,n)
q_tranposition=two_cycle(p,n)
q_four_cycle=four_cycle(p,n)
res.append([p, [q_irr, q_cycle, q_tranposition, q_four_cycle]])
p=P.next(p)
res
%%time
p=499
n=10**6
P=Primes()
res=[]
while p<600:
q_irr=irreducible(p,n)
q_cycle=almost_cycle(p,n)
q_tranposition=two_cycle(p,n)
q_four_cycle=four_cycle(p,n)
res.append([p, [q_irr, q_cycle, q_tranposition, q_four_cycle]])
p=P.next(p)
res
%%time
p=599
P=Primes()
p=P.next(p)
n=10**6
P=Primes()
res=[]
q_irr=irreducible(p,n)
q_cycle=almost_cycle(p,n)
q_tranposition=two_cycle(p,n)
q_four_cycle=four_cycle(p,n)
res.append([p, [q_irr, q_cycle, q_tranposition, q_four_cycle]])
res
%%time
p=601
P=Primes()
p=P.next(p)
n=10**6
P=Primes()
res=[]
q_irr=irreducible(p,n)
q_cycle=almost_cycle(p,n)
q_tranposition=two_cycle(p,n)
q_four_cycle=four_cycle(p,n)
res.append([p, [q_irr, q_cycle, q_tranposition, q_four_cycle]])
res
%%time
p=607
P=Primes()
p=P.next(p)
n=10**6
P=Primes()
res=[]
q_irr=irreducible(p,n)
q_cycle=almost_cycle(p,n)
q_tranposition=two_cycle(p,n)
q_four_cycle=four_cycle(p,n)
res.append([p, [q_irr, q_cycle, q_tranposition, q_four_cycle]])
res
%%time
p=613
P=Primes()
p=P.next(p)
n=10**6
P=Primes()
res=[]
q_irr=irreducible(p,n)
q_cycle=almost_cycle(p,n)
q_tranposition=two_cycle(p,n)
q_four_cycle=four_cycle(p,n)
res.append([p, [q_irr, q_cycle, q_tranposition, q_four_cycle]])
res
%%time
p=617
P=Primes()
p=P.next(p)
n=10**6
P=Primes()
res=[]
q_irr=irreducible(p,n)
q_cycle=almost_cycle(p,n)
q_tranposition=two_cycle(p,n)
q_four_cycle=four_cycle(p,n)
res.append([p, [q_irr, q_cycle, q_tranposition, q_four_cycle]])
res
%%time
p=619
P=Primes()
p=P.next(p)
n=10**6
P=Primes()
res=[]
q_irr=irreducible(p,n)
q_cycle=almost_cycle(p,n)
q_tranposition=two_cycle(p,n)
q_four_cycle=four_cycle(p,n)
res.append([p, [q_irr, q_cycle, q_tranposition, q_four_cycle]])
res
%%time
p=631
P=Primes()
p=P.next(p)
n=10**6
P=Primes()
res=[]
q_irr=irreducible(p,n)
q_cycle=almost_cycle(p,n)
q_tranposition=two_cycle(p,n)
q_four_cycle=four_cycle(p,n)
res.append([p, [q_irr, q_cycle, q_tranposition, q_four_cycle]])
res
%%time
p=641
P=Primes()
p=P.next(p)
n=10**6
P=Primes()
res=[]
q_irr=irreducible(p,n)
q_cycle=almost_cycle(p,n)
q_tranposition=two_cycle(p,n)
q_four_cycle=four_cycle(p,n)
res.append([p, [q_irr, q_cycle, q_tranposition, q_four_cycle]])
res
| 0.328637 | 0.775307 |
<table align="left" width="100%"> <tr>
<td style="background-color:#ffffff;">
<a href="http://qworld.lu.lv" target="_blank"><img src="../images/qworld.jpg" width="35%" align="left"> </a></td>
<td style="background-color:#ffffff;vertical-align:bottom;text-align:right;">
prepared by Maksim Dimitrijev (<a href="http://qworld.lu.lv/index.php/qlatvia/" target="_blank">QLatvia</a>)
</td>
</tr></table>
<table width="100%"><tr><td style="color:#bbbbbb;background-color:#ffffff;font-size:11px;font-style:italic;text-align:right;">This cell contains some macros. If there is a problem with displaying mathematical formulas, please run this cell to load these macros. </td></tr></table>
$ \newcommand{\bra}[1]{\langle #1|} $
$ \newcommand{\ket}[1]{|#1\rangle} $
$ \newcommand{\braket}[2]{\langle #1|#2\rangle} $
$ \newcommand{\dot}[2]{ #1 \cdot #2} $
$ \newcommand{\biginner}[2]{\left\langle #1,#2\right\rangle} $
$ \newcommand{\mymatrix}[2]{\left( \begin{array}{#1} #2\end{array} \right)} $
$ \newcommand{\myvector}[1]{\mymatrix{c}{#1}} $
$ \newcommand{\myrvector}[1]{\mymatrix{r}{#1}} $
$ \newcommand{\mypar}[1]{\left( #1 \right)} $
$ \newcommand{\mybigpar}[1]{ \Big( #1 \Big)} $
$ \newcommand{\sqrttwo}{\frac{1}{\sqrt{2}}} $
$ \newcommand{\dsqrttwo}{\dfrac{1}{\sqrt{2}}} $
$ \newcommand{\onehalf}{\frac{1}{2}} $
$ \newcommand{\donehalf}{\dfrac{1}{2}} $
$ \newcommand{\hadamard}{ \mymatrix{rr}{ \sqrttwo & \sqrttwo \\ \sqrttwo & -\sqrttwo }} $
$ \newcommand{\vzero}{\myvector{1\\0}} $
$ \newcommand{\vone}{\myvector{0\\1}} $
$ \newcommand{\stateplus}{\myvector{ \sqrttwo \\ \sqrttwo } } $
$ \newcommand{\stateminus}{ \myrvector{ \sqrttwo \\ -\sqrttwo } } $
$ \newcommand{\myarray}[2]{ \begin{array}{#1}#2\end{array}} $
$ \newcommand{\X}{ \mymatrix{cc}{0 & 1 \\ 1 & 0} } $
$ \newcommand{\Z}{ \mymatrix{rr}{1 & 0 \\ 0 & -1} } $
$ \newcommand{\Htwo}{ \mymatrix{rrrr}{ \frac{1}{2} & \frac{1}{2} & \frac{1}{2} & \frac{1}{2} \\ \frac{1}{2} & -\frac{1}{2} & \frac{1}{2} & -\frac{1}{2} \\ \frac{1}{2} & \frac{1}{2} & -\frac{1}{2} & -\frac{1}{2} \\ \frac{1}{2} & -\frac{1}{2} & -\frac{1}{2} & \frac{1}{2} } } $
$ \newcommand{\CNOT}{ \mymatrix{cccc}{1 & 0 & 0 & 0 \\ 0 & 1 & 0 & 0 \\ 0 & 0 & 0 & 1 \\ 0 & 0 & 1 & 0} } $
$ \newcommand{\norm}[1]{ \left\lVert #1 \right\rVert } $
$ \newcommand{\pstate}[1]{ \lceil \mspace{-1mu} #1 \mspace{-1.5mu} \rfloor } $
<h1> <font color="blue"> Solutions for </font> Basics of complex numbers </h1>
<a id="task1"></a>
<h3> Task 1 </h3>
Calculate on the paper the following values.
$(3-2i) + (7+i)$;
$(4+5i) - (2+2i)$;
$(2+i) \cdot (8-4i)$;
$\mathopen|4+3i\mathclose|$;
$\overline{5-6i}$.
<h3> Solution </h3>
$(3-2i) + (7+i) = 3 + 7 - 2i + i = 10 - i$.
$(4+5i) - (2+2i) = 4 - 2 + 5i - 2i = 2 + 3i$.
$(2+i) \cdot (8-4i) = 2 \cdot 8 + 2 \cdot (-4i) + i \cdot 8 + i \cdot (-4i) = 16 - 8i + 8i - 4i^2 = 16 - 4 \cdot (-1) = 20$.
$\mathopen|4+3i\mathclose| = \sqrt{4^2 + 3^2} = \sqrt{16 + 9} = 5$.
$\overline{5-6i} = 5 + 6i$.
<a id="task2"></a>
<h3> Task 2 </h3>
Convert $4-4i$ to polar form.
Convert $2 \cdot e^{i \pi}$ from polar form to the basic one.
<h3> Solution </h3>
- $4-4i$:
Length: $r = \sqrt{4^2 + (-4)^2} = \sqrt{32} = 4 \cdot \sqrt{2}.$
$r \cos{\alpha} = 4 \implies \cos{\alpha} = \frac{4}{4 \cdot \sqrt{2}} = \frac{1}{\sqrt{2}} \implies \alpha = \frac{\pi}{4}$
$4-4i = r \cdot e^{i \alpha} = 4 \cdot \sqrt{2} \cdot e^{i \frac{\pi}{4}}$.
- $2 \cdot e^{i \pi}$:
$r = 2$, $\alpha = \pi$.
$e^{i \pi} = \cos{\pi} + \sin{\pi} \cdot i = -1$.
$2 \cdot e^{i \pi} = 2 \cdot (-1) = -2$.
<a id="task3"></a>
<h3> Task 3 </h3>
Recreate famous Euler formula. Please use:
<ul>
<li>=, + or -;</li>
<li>one instance of each of the following constants: 0, 1, $i$, $\pi$, $e$.</li>
</ul>
<h3> Solution </h3>
It is no secret for us that $e^{i \pi} = -1$. To include all constants into formula, we need to add 1 to this number:
$e^{i \pi} + 1 = 0$.
<a id="task4"></a>
<h3> Task 4 </h3>
Please calculate the following values in Python:
<ul>
<li>$\frac{3+4i}{2-i} \cdot (7+2i) + 3i$;</li>
<li>$(1-2i)^5$;</li>
<li>$\frac{8-4i}{\mathopen|4+3i\mathclose|}$.</li>
</ul>
<h3> Solution </h3>
```
result1 = ((3+4j)/(2-1j))*(7+2j)+3j
print('((3+4j)/(2-j))*(7+2j)+3j=', result1)
result2 = (1-2j)**5
print('(1-2j)**5=', result2)
result3 = (8-4j)/(abs(4+3j))
print('(8-4j)/(abs(4+3j))=', result3)
```
<a id="task5"></a>
<h3> Task 5 </h3>
Use Python to convert:
<ul>
<li>$3 - 3i$ to polar form;</li>
<li>$5 \cdot e^{i \cdot \frac{pi}{3}}$ from polar form.</li>
<ul>
<h3> Solution </h3>
```
from math import pi, cos, sin, asin
#3 - 3i
# Length
r1 = abs(3-3j)
# Angle
alpha1 = asin(-3/r1)
# Result
print('3 - 3i =', r1, '*e^(i*', alpha1, ')')
#5*e^(i*(pi/3))
# Length
r2 = 5
# Angle
alpha2 = pi/3
# Result
print('5*e^(i*(pi/3)) =', 5*complex(cos(alpha2), sin(alpha2)))
```
|
github_jupyter
|
result1 = ((3+4j)/(2-1j))*(7+2j)+3j
print('((3+4j)/(2-j))*(7+2j)+3j=', result1)
result2 = (1-2j)**5
print('(1-2j)**5=', result2)
result3 = (8-4j)/(abs(4+3j))
print('(8-4j)/(abs(4+3j))=', result3)
from math import pi, cos, sin, asin
#3 - 3i
# Length
r1 = abs(3-3j)
# Angle
alpha1 = asin(-3/r1)
# Result
print('3 - 3i =', r1, '*e^(i*', alpha1, ')')
#5*e^(i*(pi/3))
# Length
r2 = 5
# Angle
alpha2 = pi/3
# Result
print('5*e^(i*(pi/3)) =', 5*complex(cos(alpha2), sin(alpha2)))
| 0.133924 | 0.987711 |
# 3 Days Machine Learning Workshop by Quantum.ai
## Machine Learning types
* **Regression**: works on continous data
* **Classification**: works with labeled class
* **Clustering**: works on class free data
# Regression
## Data
A manupulated data set
* **Years of Experience**: How many years of experience?
* **Salary**: How much paid? (Prediction label)
```
# Importing Libraries
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
```
# Step 1: Data Pre-processing
## 1.1 Import Data
```
df = pd.read_csv('salary.csv')
```
## 1.2 Analyse Data
```
df.shape
df.head(10)
df.describe()
```
## 1.3 Plot data (Data Visualization)
```
# Plot
X = df['YearsExperience'].values.reshape(-1,1)
y = df['Salary'].values.reshape(-1,1)
plt.scatter(X, y)
plt.xlabel('Years of Experience')
plt.ylabel('Salary')
plt.show()
```
## 1.4 Split data into train and test sets
```
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size = 0.20, random_state =0)
print(X_train.shape)
print(X_train)
print(y_train.shape)
print(y_train)
print(X_test.shape)
print(X_test)
```
# Step 2: Build Machine Learning Model
**Linear regression** performs the task to predict a dependent variable value (y) based on a given independent variable (x). So, this regression technique finds out a linear relationship between x (input) and y(output). Hence, the name is Linear Regression.
## 2.1 Learning Model (training)
```
from sklearn.linear_model import LinearRegression
regressor = LinearRegression()
regressor.fit(X_train, y_train)
```
Find intercept and coefficient of Best fit line
```
# y intercept:
print(regressor.intercept_)
# m slope:
print(regressor.coef_)
```
## 2.2 Prediction (test)
```
y_pred = regressor.predict(X_test)
plt.scatter(X_train, y_train, color='gray')
plt.plot(X_test, y_pred, color='red', linewidth=2)
plt.show()
```
# Step 3: Performance Evaluation
## 3.1 Manual Analysis and ploting
```
print(y_test)
print(y_pred)
result = pd.DataFrame({'Actual': y_test.flatten(), 'Predicted': y_pred.flatten()})
print(result)
result.plot(kind='bar',figsize=(5,8))
plt.grid(which='major', linestyle='-', linewidth='0.5', color='green')
plt.grid(which='minor', linestyle=':', linewidth='0.5', color='blue')
plt.show()
```
## 3.2 Performance Matrics
```
from sklearn import metrics
print('Mean Absolute Error:', metrics.mean_absolute_error(y_test, y_pred))
print('Mean Squared Error:', metrics.mean_squared_error(y_test, y_pred))
print('Root Mean Squared Error:', np.sqrt(metrics.mean_squared_error(y_test, y_pred)))
er = []
g = 0
for i in range(len(y_test)):
print( "actual=", y_test[i], " observed=", np.round(y_pred[i],0))
x = (y_test[i] - y_pred[i]) **2
# print(y_test[i] - y_pred[i])
er.append(x)
g = g + x
x = 0
for i in range(len(er)):
x = x + er[i]
print ("MSE", x / len(er))
import numpy as np
def mean_absolute_percentage_error(y_test, y_pred):
y_true, y_pred = np.array(y_test), np.array(y_pred)
return np.mean(np.abs((y_test - y_pred) / y_test)) * 100
mean_absolute_percentage_error(y_test, y_pred)
```
# Thank you.
|
github_jupyter
|
# Importing Libraries
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
df = pd.read_csv('salary.csv')
df.shape
df.head(10)
df.describe()
# Plot
X = df['YearsExperience'].values.reshape(-1,1)
y = df['Salary'].values.reshape(-1,1)
plt.scatter(X, y)
plt.xlabel('Years of Experience')
plt.ylabel('Salary')
plt.show()
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size = 0.20, random_state =0)
print(X_train.shape)
print(X_train)
print(y_train.shape)
print(y_train)
print(X_test.shape)
print(X_test)
from sklearn.linear_model import LinearRegression
regressor = LinearRegression()
regressor.fit(X_train, y_train)
# y intercept:
print(regressor.intercept_)
# m slope:
print(regressor.coef_)
y_pred = regressor.predict(X_test)
plt.scatter(X_train, y_train, color='gray')
plt.plot(X_test, y_pred, color='red', linewidth=2)
plt.show()
print(y_test)
print(y_pred)
result = pd.DataFrame({'Actual': y_test.flatten(), 'Predicted': y_pred.flatten()})
print(result)
result.plot(kind='bar',figsize=(5,8))
plt.grid(which='major', linestyle='-', linewidth='0.5', color='green')
plt.grid(which='minor', linestyle=':', linewidth='0.5', color='blue')
plt.show()
from sklearn import metrics
print('Mean Absolute Error:', metrics.mean_absolute_error(y_test, y_pred))
print('Mean Squared Error:', metrics.mean_squared_error(y_test, y_pred))
print('Root Mean Squared Error:', np.sqrt(metrics.mean_squared_error(y_test, y_pred)))
er = []
g = 0
for i in range(len(y_test)):
print( "actual=", y_test[i], " observed=", np.round(y_pred[i],0))
x = (y_test[i] - y_pred[i]) **2
# print(y_test[i] - y_pred[i])
er.append(x)
g = g + x
x = 0
for i in range(len(er)):
x = x + er[i]
print ("MSE", x / len(er))
import numpy as np
def mean_absolute_percentage_error(y_test, y_pred):
y_true, y_pred = np.array(y_test), np.array(y_pred)
return np.mean(np.abs((y_test - y_pred) / y_test)) * 100
mean_absolute_percentage_error(y_test, y_pred)
| 0.622115 | 0.98758 |
```
import sys; sys.path.append('..')
import random
from IPython.core.display import display, HTML
display(HTML("<style>.container { width:100% !important; }</style>"))
import pyzx as zx
import os
import pickle
import numpy as np
import time
%matplotlib inline
import matplotlib.pyplot as plt
import math
def save_obj(obj, name):
with open('data/'+ name + '.pkl', 'wb') as f:
pickle.dump(obj, f, pickle.HIGHEST_PROTOCOL)
def load_obj(name):
with open('data/' + name + '.pkl', 'rb') as f:
return pickle.load(f)
def c_f(tup):
return (tup[0]-tup[1]) + 10*tup[1]
def merge_bg(depth, qubits, tprob, bound):
highbound = load_obj('random/rd_'+str(depth)+'_q_'+str(qubits)+'_t_0.'+str(tprob))
lowbound = load_obj('random_lowbound/rd_'+str(depth)+'_q_'+str(qubits)+'_t_0.'+str(tprob))
res = [c_f(highbound['basic_opt']), c_f(highbound['full_reduce']), c_f(highbound['sim_annealing_post'])]
if bound > -15:
res.append(c_f(highbound['greedy_simp_neighbors_c'+str(bound)]))
# res.append(c_f(highbound['greedy_simp_c'+str(bound)+'_b_False']))
# res.append(c_f(highbound['random_simp_c'+str(bound)+'_b_False']))
else:
res.append(c_f(lowbound['greedy_simp_neighbors_c'+str(bound)]))
# res.append(c_f(lowbound['greedy_simp_c'+str(bound)+'_b_False']))
# res.append(c_f(lowbound['random_simp_c'+str(bound)+'_b_False']))
return res
params = [[[500,4,[1,1,-10,-20]],[1000,4,[1,1,-10,-20]],[500,8,[-10,-10,-10,-10]],[1000,8,[-20,-20,-10,-10]]]]
names = ['basic_optimization','full_reduce','simulated_annealing_post','neighbour_unfusion']
xs = ['0.1','0.2','0.3','0.4']
colors = ['#1b9e77','#7570b3','#e7298a','#d95f02']
styles = ['-','-.','-','--','--','-.']
fig, axs = plt.subplots(len(params), len(params[0]), figsize=(12,3.5))
for i in range(0,len(params)):
for j in range(0,len(params[i])):
axs[j].set_title('m='+str(params[i][j][1])+' n='+str(params[i][j][0]),fontsize=14)
yys = [merge_bg(params[i][j][0],params[i][j][1],tprob,params[i][j][2][tprob-1]) for tprob in range(1,5)]
for k, ys in enumerate(list(zip(*yys))):
axs[j].plot(xs, ys, c=colors[k], marker="o",markersize=3, linestyle=styles[k], label=names[k] if i==0 and j == 0 else '')
axs[j].grid()
dep = params[i][j][0]
orig_cost = (dep-dep*0.3) + dep*3
axs[j].axhline(y=orig_cost, color='red', linestyle='-')
plt.figlegend(bbox_to_anchor=(.06, 0), loc='upper left',
ncol=4, borderaxespad=0,fontsize=14)
# plt.suptitle('Post-Optimization results of $m$ qubit circuits with size $n$ and increasing T Gate probability',fontsize=12)
plt.tight_layout()
plt.show()
fig.savefig(r'/home/korbinian/Documents/master/stau21/Dokumentation/Latex/Bilder/evaluation/random_compare_to_pyzx.pdf',bbox_inches='tight')
def c_f(tup):
return (tup[0]-tup[1]) + 10*tup[1]
def merge_bg(depth, qubits, tprob, bound):
highbound = load_obj('random/rd_'+str(depth)+'_q_'+str(qubits)+'_t_0.'+str(tprob))
lowbound = load_obj('random_lowbound/rd_'+str(depth)+'_q_'+str(qubits)+'_t_0.'+str(tprob))
res = [c_f(highbound['simulated_annealing_simp'])]
if bound > -15:
res.append(c_f(highbound['greedy_simp_neighbors_c'+str(bound)]))
res.append(c_f(highbound['greedy_simp_c'+str(bound)+'_b_False']))
res.append(c_f(highbound['random_simp_c'+str(bound)+'_b_False']))
else:
res.append(c_f(lowbound['greedy_simp_neighbors_c'+str(bound)]))
res.append(c_f(lowbound['greedy_simp_c'+str(bound)+'_b_False']))
res.append(c_f(lowbound['random_simp_c'+str(bound)+'_b_False']))
return res
params = [[[500,4,[1,1,-10,-20]],[1000,4,[1,1,-10,-20]],[500,8,[-10,-10,-10,-10]],[1000,8,[-20,-20,-10,-10]]]]
names = ['simulated_annealing','neighbour_unfusion','greedy_simplification','random_simplification']
xs = ['0.1','0.2','0.3','0.4']
colors = ['#1b9e77','#d95f02','#7570b3','#e7298a']
styles = ['-','--','-.','-','--','-.']
fig, axs = plt.subplots(len(params), len(params[0]), figsize=(12,3.5))
for i in range(0,len(params)):
for j in range(0,len(params[i])):
axs[j].set_title('m='+str(params[i][j][1])+' n='+str(params[i][j][0]),fontsize=14)
yys = [merge_bg(params[i][j][0],params[i][j][1],tprob,params[i][j][2][tprob-1]) for tprob in range(1,5)]
for k, ys in enumerate(list(zip(*yys))):
axs[j].plot(xs, ys, c=colors[k], marker="o",markersize=3, linestyle=styles[k], label=names[k] if i==0 and j == 0 else '')
axs[j].grid()
dep = params[i][j][0]
orig_cost = (dep-dep*0.3) + dep*3
axs[j].axhline(y=orig_cost, color='red', linestyle='-')
plt.figlegend(bbox_to_anchor=(.02, 0), loc='upper left',
ncol=4, borderaxespad=0,fontsize=14)
# plt.suptitle('Post-Optimization results of $m$ qubit circuits with size $n$ and increasing T Gate probability',fontsize=12)
plt.tight_layout()
plt.show()
fig.savefig(r'/home/korbinian/Documents/master/stau21/Dokumentation/Latex/Bilder/evaluation/random_compare_to_own.pdf',bbox_inches='tight')
```
|
github_jupyter
|
import sys; sys.path.append('..')
import random
from IPython.core.display import display, HTML
display(HTML("<style>.container { width:100% !important; }</style>"))
import pyzx as zx
import os
import pickle
import numpy as np
import time
%matplotlib inline
import matplotlib.pyplot as plt
import math
def save_obj(obj, name):
with open('data/'+ name + '.pkl', 'wb') as f:
pickle.dump(obj, f, pickle.HIGHEST_PROTOCOL)
def load_obj(name):
with open('data/' + name + '.pkl', 'rb') as f:
return pickle.load(f)
def c_f(tup):
return (tup[0]-tup[1]) + 10*tup[1]
def merge_bg(depth, qubits, tprob, bound):
highbound = load_obj('random/rd_'+str(depth)+'_q_'+str(qubits)+'_t_0.'+str(tprob))
lowbound = load_obj('random_lowbound/rd_'+str(depth)+'_q_'+str(qubits)+'_t_0.'+str(tprob))
res = [c_f(highbound['basic_opt']), c_f(highbound['full_reduce']), c_f(highbound['sim_annealing_post'])]
if bound > -15:
res.append(c_f(highbound['greedy_simp_neighbors_c'+str(bound)]))
# res.append(c_f(highbound['greedy_simp_c'+str(bound)+'_b_False']))
# res.append(c_f(highbound['random_simp_c'+str(bound)+'_b_False']))
else:
res.append(c_f(lowbound['greedy_simp_neighbors_c'+str(bound)]))
# res.append(c_f(lowbound['greedy_simp_c'+str(bound)+'_b_False']))
# res.append(c_f(lowbound['random_simp_c'+str(bound)+'_b_False']))
return res
params = [[[500,4,[1,1,-10,-20]],[1000,4,[1,1,-10,-20]],[500,8,[-10,-10,-10,-10]],[1000,8,[-20,-20,-10,-10]]]]
names = ['basic_optimization','full_reduce','simulated_annealing_post','neighbour_unfusion']
xs = ['0.1','0.2','0.3','0.4']
colors = ['#1b9e77','#7570b3','#e7298a','#d95f02']
styles = ['-','-.','-','--','--','-.']
fig, axs = plt.subplots(len(params), len(params[0]), figsize=(12,3.5))
for i in range(0,len(params)):
for j in range(0,len(params[i])):
axs[j].set_title('m='+str(params[i][j][1])+' n='+str(params[i][j][0]),fontsize=14)
yys = [merge_bg(params[i][j][0],params[i][j][1],tprob,params[i][j][2][tprob-1]) for tprob in range(1,5)]
for k, ys in enumerate(list(zip(*yys))):
axs[j].plot(xs, ys, c=colors[k], marker="o",markersize=3, linestyle=styles[k], label=names[k] if i==0 and j == 0 else '')
axs[j].grid()
dep = params[i][j][0]
orig_cost = (dep-dep*0.3) + dep*3
axs[j].axhline(y=orig_cost, color='red', linestyle='-')
plt.figlegend(bbox_to_anchor=(.06, 0), loc='upper left',
ncol=4, borderaxespad=0,fontsize=14)
# plt.suptitle('Post-Optimization results of $m$ qubit circuits with size $n$ and increasing T Gate probability',fontsize=12)
plt.tight_layout()
plt.show()
fig.savefig(r'/home/korbinian/Documents/master/stau21/Dokumentation/Latex/Bilder/evaluation/random_compare_to_pyzx.pdf',bbox_inches='tight')
def c_f(tup):
return (tup[0]-tup[1]) + 10*tup[1]
def merge_bg(depth, qubits, tprob, bound):
highbound = load_obj('random/rd_'+str(depth)+'_q_'+str(qubits)+'_t_0.'+str(tprob))
lowbound = load_obj('random_lowbound/rd_'+str(depth)+'_q_'+str(qubits)+'_t_0.'+str(tprob))
res = [c_f(highbound['simulated_annealing_simp'])]
if bound > -15:
res.append(c_f(highbound['greedy_simp_neighbors_c'+str(bound)]))
res.append(c_f(highbound['greedy_simp_c'+str(bound)+'_b_False']))
res.append(c_f(highbound['random_simp_c'+str(bound)+'_b_False']))
else:
res.append(c_f(lowbound['greedy_simp_neighbors_c'+str(bound)]))
res.append(c_f(lowbound['greedy_simp_c'+str(bound)+'_b_False']))
res.append(c_f(lowbound['random_simp_c'+str(bound)+'_b_False']))
return res
params = [[[500,4,[1,1,-10,-20]],[1000,4,[1,1,-10,-20]],[500,8,[-10,-10,-10,-10]],[1000,8,[-20,-20,-10,-10]]]]
names = ['simulated_annealing','neighbour_unfusion','greedy_simplification','random_simplification']
xs = ['0.1','0.2','0.3','0.4']
colors = ['#1b9e77','#d95f02','#7570b3','#e7298a']
styles = ['-','--','-.','-','--','-.']
fig, axs = plt.subplots(len(params), len(params[0]), figsize=(12,3.5))
for i in range(0,len(params)):
for j in range(0,len(params[i])):
axs[j].set_title('m='+str(params[i][j][1])+' n='+str(params[i][j][0]),fontsize=14)
yys = [merge_bg(params[i][j][0],params[i][j][1],tprob,params[i][j][2][tprob-1]) for tprob in range(1,5)]
for k, ys in enumerate(list(zip(*yys))):
axs[j].plot(xs, ys, c=colors[k], marker="o",markersize=3, linestyle=styles[k], label=names[k] if i==0 and j == 0 else '')
axs[j].grid()
dep = params[i][j][0]
orig_cost = (dep-dep*0.3) + dep*3
axs[j].axhline(y=orig_cost, color='red', linestyle='-')
plt.figlegend(bbox_to_anchor=(.02, 0), loc='upper left',
ncol=4, borderaxespad=0,fontsize=14)
# plt.suptitle('Post-Optimization results of $m$ qubit circuits with size $n$ and increasing T Gate probability',fontsize=12)
plt.tight_layout()
plt.show()
fig.savefig(r'/home/korbinian/Documents/master/stau21/Dokumentation/Latex/Bilder/evaluation/random_compare_to_own.pdf',bbox_inches='tight')
| 0.175998 | 0.233095 |
## Introduction
We present an analysis of emotions linked to tweets in order to detect instances of cyberbulling.
The tweets dataset has been manually collected using twitter APIs by Margarita Bugueño, Fabián Fernandez and Francisco Mena.
The NRC Emotion Lexicon (aka Emolex) is a list of English words and their associations with eight basic emotions (anger, fear, anticipation, trust, surprise, sadness, joy, and disgust) and two sentiments (negative and positive). It has been developed by [Saif Mohammad](https://saifmohammad.com/WebPages/NRC-Emotion-Lexicon.htm) and is a lexic tag based on the Plutchick wheel of emotions. The annotations were manually done by crowdsourcing.
## Exploratory Analysis
To begin this exploratory analysis, first import libraries and define functions for plotting the data using `matplotlib`. Depending on the data, not all plots will be made. (Hey, I'm just a simple kerneling bot, not a Kaggle Competitions Grandmaster!)
```
from mpl_toolkits.mplot3d import Axes3D
from sklearn.preprocessing import StandardScaler
import matplotlib.pyplot as plt # plotting
import numpy as np # linear algebra
import os # accessing directory structure
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import warnings
warnings.filterwarnings('ignore')
```
There are 44 csv files in the current version of the dataset:
```
print(os.listdir('data'))
print(os.listdir('data/tweets'))
# Previewing one file alyssalg93.csv
tweets = pd.read_csv('data/tweets/alyssalg93.csv', delimiter=',')
tweets.dataframeName = 'alyssalg93.csv'
print(f'There are {tweets.shape[0]} rows and {tweets.shape[1]} columns')
tweets.head()
```
# Using the NRC Emoticon Lexicon
We will be using the NRC Emotion Lexicon for the sentiment analysis of the tweets.
The NRC Emotion Lexicon is a list of English words and their associations with eight basic e motions
(anger, fear, anticipation, trust, surprise, sadness, joy, and disgust) and two sentiments
(negative and positive). The annotations were manually done by crowdsourcing.
```
from nltk import word_tokenize
from nltk.stem.snowball import SnowballStemmer
from tqdm import tqdm_notebook as tqdm
stemmer = SnowballStemmer("english")
lexicon = "NRC-Sentiment-Emotion-Lexicons/NRC-Emotion-Lexicon-v0.92/NRC-Emotion-Lexicon-Wordlevel-v0.92.txt"
emolex_df = pd.read_csv(lexicon,
names=["word", "emotion", "association"],
sep='\t')
emolex_df.dropna(subset=['word'], inplace=True)
emolex_words = emolex_df.pivot(index='word',
columns='emotion',
values='association')
emolex_words.index = emolex_words.index.map(lambda w: stemmer.stem(w.lower()) if w else nan)
emotions = emolex_words.columns.values
emolex_words['emotions'] = list(zip(emolex_words.anger, emolex_words.anticipation, emolex_words.disgust,
emolex_words.fear, emolex_words.joy, emolex_words.negative, emolex_words.positive,
emolex_words.sadness, emolex_words.surprise, emolex_words.trust))
# Convert into a dictionary for faster lookup
emolex_dict = emolex_words['emotions'].to_dict()
print("We built a dictionary of {} words associated to emotions".format(len(emolex_dict)))
# Run only once
import nltk
nltk.download('punkt')
def text_emotion(df, column):
'''
Takes a DataFrame and a specified column of text and adds 10 columns
for each of the 10 emotions in the NRC Emotion Lexicon, with each
column containing the value of the text in that emotions and the counts of tweets
INPUT: DataFrame, string
OUTPUT: New DataFrame with ten new columns
'''
new_df = df.drop(['id', 'favorite count', 'retweet count', 'created at'], axis=1)
new_df['document'] = pd.Series()
new_df = new_df.reindex(columns=new_df.columns.tolist())
# Convert to numpy array
tweets = new_df.copy().to_numpy()
scores = np.zeros((tweets.shape[0], len(emotions)))
#print(scores.shape)
with tqdm(total=new_df.shape[0]) as pbar:
for i, text in enumerate(tweets[:, 0]):
pbar.update(1)
#print("Iteration ",i)
document = word_tokenize(text)
tweets[i, 2] = document
for w, word in enumerate(document):
document[w] = stemmer.stem(word.lower())
#emo_score = emolex_words[emolex_words.word == word].values
emo_score = emolex_dict.get(word)
if emo_score != None:
scores[i,:] += list(emo_score)
tweets_df = pd.DataFrame(data=tweets, columns=new_df.columns)
scores_df = pd.DataFrame(data=scores, columns=emotions)
return pd.concat([tweets_df, scores_df], axis=1)
```
Now we can read all the files and build one dataframe with the emolex scores from all the tweets in the directory
```
# Read all set of tweets and build sentiment dataframes
df_emo_all = pd.DataFrame()
for file in os.listdir('data/tweets'):
df = pd.read_csv('data/tweets/'+file, delimiter=',')
df['screen_name'] = os.path.splitext(file)[0]
print("Scoring tweets from ", os.path.splitext(file)[0])
df_emo = text_emotion(df, 'text')
df_emo_all = pd.concat([df_emo_all, df_emo])
df_emo_all.shape
df_emo_all.head()
# Aggregating Grouping data together
df_emotions = df_emo_all.groupby('screen_name')['anger','anticipation', 'disgust', 'fear', 'joy', 'sadness', 'surprise', 'trust','negative', 'positive'].mean()
df_emotions['n_tweets']=df_emo_all.screen_name.value_counts()
df_emotions.head()
```
## Exploratory Analysis
```
# Distribution graphs (histogram/bar graph) of column data
def plotPerColumnDistribution(df, nGraphShown, nGraphPerRow):
nunique = df.nunique()
df = df[[col for col in df if nunique[col] > 1 and nunique[col] < 50]] # For displaying purposes, pick columns that have between 1 and 50 unique values
nRow, nCol = df.shape
columnNames = list(df)
nGraphRow = (nCol + nGraphPerRow - 1) / nGraphPerRow
plt.figure(num = None, figsize = (6 * nGraphPerRow, 8 * nGraphRow), dpi = 80, facecolor = 'w', edgecolor = 'k')
for i in range(min(nCol, nGraphShown)):
plt.subplot(nGraphRow, nGraphPerRow, i + 1)
columnDf = df.iloc[:, i]
if (not np.issubdtype(type(columnDf.iloc[0]), np.number)):
valueCounts = columnDf.value_counts()
valueCounts.plot.bar()
else:
columnDf.hist()
plt.ylabel('counts')
plt.xticks(rotation = 90)
plt.title(f'{columnNames[i]} (column {i})')
plt.tight_layout(pad = 1.0, w_pad = 1.0, h_pad = 1.0)
plt.show()
# Correlation matrix
def plotCorrelationMatrix(df, graphWidth):
#filename = df.dataframeName
df = df.dropna('columns') # drop columns with NaN
df = df[[col for col in df if df[col].nunique() > 1]] # keep columns where there are more than 1 unique values
if df.shape[1] < 2:
print(f'No correlation plots shown: The number of non-NaN or constant columns ({df.shape[1]}) is less than 2')
return
corr = df.corr()
plt.figure(num=None, figsize=(graphWidth, graphWidth), dpi=80, facecolor='w', edgecolor='k')
corrMat = plt.matshow(corr, fignum = 1)
plt.xticks(range(len(corr.columns)), corr.columns, rotation=90)
plt.yticks(range(len(corr.columns)), corr.columns)
plt.gca().xaxis.tick_bottom()
plt.colorbar(corrMat)
#plt.title(f'Correlation Matrix for {filename}', fontsize=15)
plt.title(f'Correlation Matrix', fontsize=15)
plt.show()
# Scatter and density plots
def plotScatterMatrix(df, plotSize, textSize):
df = df.select_dtypes(include =[np.number]) # keep only numerical columns
# Remove rows and columns that would lead to df being singular
df = df.dropna('columns')
df = df[[col for col in df if df[col].nunique() > 1]] # keep columns where there are more than 1 unique values
columnNames = list(df)
if len(columnNames) > 10: # reduce the number of columns for matrix inversion of kernel density plots
columnNames = columnNames[:10]
df = df[columnNames]
ax = pd.plotting.scatter_matrix(df, alpha=0.75, figsize=[plotSize, plotSize], diagonal='kde')
corrs = df.corr().values
for i, j in zip(*plt.np.triu_indices_from(ax, k = 1)):
ax[i, j].annotate('Corr. coef = %.3f' % corrs[i, j], (0.8, 0.2), xycoords='axes fraction', ha='center', va='center', size=textSize)
plt.suptitle('Scatter and Density Plot')
plt.show()
```
Now you're ready to read in the data and use the plotting functions to visualize the data.
Distribution graphs (histogram/bar graph) of sampled columns:
```
plotPerColumnDistribution(df_emotions.drop(['n_tweets'], axis=1), 4, 4)
#plotPerColumnDistribution(df1, 10, 5)
```
Correlation matrix:
```
plotCorrelationMatrix(df_emotions.drop(['n_tweets'], axis=1), 6)
```
Scatter and density plots:
```
plotScatterMatrix(df_emotions.drop(['n_tweets'], axis=1), 18, 10)
df_emotions.head()
# How many bullies do we have??
df_emotions[df_emotions.anger > 0.3]
# and potentially bullied?
df_emotions[df_emotions.fear > 0.3]
#Top 10 angriest
df_emotions.sort_values(by='anger', ascending=False)[:10]
#Top 10 saddest
df_emotions.sort_values(by='sadness', ascending=False)[:10]
# We could develop a metric to define a "bully" with some tresholds
def is_a_bully(screen_name, df):
"""
Test function to flag a user as a potential bully using the aggregated metrics
"""
return False
```
## Conclusion
So where from now? Can you build a predictive model based on tweets?
```
# Let's play with isiZulu
def build_emolex(language):
lexicon = "NRC-Sentiment-Emotion-Lexicons/NRC-Emotion-Lexicon-v0.92/NRC-Emotion-Lexicon-v0.92_"+language+".txt"
emolex_words = pd.read_csv(lexicon,
index_col=0,
sep='\t', na_values='NO TRANSLATION')
print(emolex_words.shape)
#emolex_words.index = emolex_words.index.map(lambda w: stemmer.stem(w.lower()) if w else nan)
emotions = emolex_words.columns.values
emolex_words['emotions'] = list(zip(*map(emolex_words.get, emolex_words)))
# Convert into a dictionary for faster lookup
emolex_dict = emolex_words['emotions'].to_dict()
print("We built a dictionary of {} words associated to {} emotions".format(len(emolex_dict), len(emotions)))
return (emolex_dict, emotions)
language='isizulu'
isizulu = build_emolex('isizulu')
len(isizulu[1])
# define a function to score a sentence
from nltk import word_tokenize
def sentence_emotions(sentence, emo_dict):
sentence_score = np.zeros(10)
emolex, emotions = emo_dict
document = word_tokenize(sentence)
for w, word in enumerate(document):
#document[w] = stemmer.stem(word.lower())
#emo_score = emolex_words[emolex_words.word == word].values
emo_score = emolex.get(word)
if emo_score != None:
sentence_score += emo_score
print("Sentence scores:")
for i in range(len(emotions)):
print("{}: {}".format(emotions[i],sentence_score[i]))
return
sentence_emotions("bulala ukudabuka", isizulu)
language='isizulu'
isizulu = build_emolex('isizulu')
language='xhosa'
sesotho, emotions = build_emolex(language)
language='sesotho'
sesotho, emotions = build_emolex(language)
sesotho.get('ho etsa lichelete')
language='sesotho'
lexicon = "NRC-Sentiment-Emotion-Lexicons/NRC-Emotion-Lexicon-v0.92/NRC-Emotion-Lexicon-v0.92_"+language+".txt"
emolex_words = pd.read_csv(lexicon,
index_col=0,
sep='\t')
emolex_words = emolex_words[emolex_words.index!='NO TRANSLATION']
print(emolex_words.shape)
#emolex_words.index = emolex_words.index.map(lambda w: stemmer.stem(w.lower()) if w else nan)
emotions = emolex_words.columns.values
emolex_words['emotions'] = list(zip(*map(emolex_words.get, emolex_words)))
emolex_dict = emolex_words['emotions'].to_dict()
print("We built a dictionary of {} words associated to {} emotions".format(len(emolex_dict), len(emotions)))
dict_set = set(emolex_words['emotions'].to_dict().keys())
emo_set = set(emolex_words.index)
# Can you write a scoring function using bi-grams (a sequence of two words) and tri-grams (a sequence of three words)
# Build a dictionary that works for "Joburg Zulu" by mixing english and zulu words
# and adding slang
```
|
github_jupyter
|
from mpl_toolkits.mplot3d import Axes3D
from sklearn.preprocessing import StandardScaler
import matplotlib.pyplot as plt # plotting
import numpy as np # linear algebra
import os # accessing directory structure
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import warnings
warnings.filterwarnings('ignore')
print(os.listdir('data'))
print(os.listdir('data/tweets'))
# Previewing one file alyssalg93.csv
tweets = pd.read_csv('data/tweets/alyssalg93.csv', delimiter=',')
tweets.dataframeName = 'alyssalg93.csv'
print(f'There are {tweets.shape[0]} rows and {tweets.shape[1]} columns')
tweets.head()
from nltk import word_tokenize
from nltk.stem.snowball import SnowballStemmer
from tqdm import tqdm_notebook as tqdm
stemmer = SnowballStemmer("english")
lexicon = "NRC-Sentiment-Emotion-Lexicons/NRC-Emotion-Lexicon-v0.92/NRC-Emotion-Lexicon-Wordlevel-v0.92.txt"
emolex_df = pd.read_csv(lexicon,
names=["word", "emotion", "association"],
sep='\t')
emolex_df.dropna(subset=['word'], inplace=True)
emolex_words = emolex_df.pivot(index='word',
columns='emotion',
values='association')
emolex_words.index = emolex_words.index.map(lambda w: stemmer.stem(w.lower()) if w else nan)
emotions = emolex_words.columns.values
emolex_words['emotions'] = list(zip(emolex_words.anger, emolex_words.anticipation, emolex_words.disgust,
emolex_words.fear, emolex_words.joy, emolex_words.negative, emolex_words.positive,
emolex_words.sadness, emolex_words.surprise, emolex_words.trust))
# Convert into a dictionary for faster lookup
emolex_dict = emolex_words['emotions'].to_dict()
print("We built a dictionary of {} words associated to emotions".format(len(emolex_dict)))
# Run only once
import nltk
nltk.download('punkt')
def text_emotion(df, column):
'''
Takes a DataFrame and a specified column of text and adds 10 columns
for each of the 10 emotions in the NRC Emotion Lexicon, with each
column containing the value of the text in that emotions and the counts of tweets
INPUT: DataFrame, string
OUTPUT: New DataFrame with ten new columns
'''
new_df = df.drop(['id', 'favorite count', 'retweet count', 'created at'], axis=1)
new_df['document'] = pd.Series()
new_df = new_df.reindex(columns=new_df.columns.tolist())
# Convert to numpy array
tweets = new_df.copy().to_numpy()
scores = np.zeros((tweets.shape[0], len(emotions)))
#print(scores.shape)
with tqdm(total=new_df.shape[0]) as pbar:
for i, text in enumerate(tweets[:, 0]):
pbar.update(1)
#print("Iteration ",i)
document = word_tokenize(text)
tweets[i, 2] = document
for w, word in enumerate(document):
document[w] = stemmer.stem(word.lower())
#emo_score = emolex_words[emolex_words.word == word].values
emo_score = emolex_dict.get(word)
if emo_score != None:
scores[i,:] += list(emo_score)
tweets_df = pd.DataFrame(data=tweets, columns=new_df.columns)
scores_df = pd.DataFrame(data=scores, columns=emotions)
return pd.concat([tweets_df, scores_df], axis=1)
# Read all set of tweets and build sentiment dataframes
df_emo_all = pd.DataFrame()
for file in os.listdir('data/tweets'):
df = pd.read_csv('data/tweets/'+file, delimiter=',')
df['screen_name'] = os.path.splitext(file)[0]
print("Scoring tweets from ", os.path.splitext(file)[0])
df_emo = text_emotion(df, 'text')
df_emo_all = pd.concat([df_emo_all, df_emo])
df_emo_all.shape
df_emo_all.head()
# Aggregating Grouping data together
df_emotions = df_emo_all.groupby('screen_name')['anger','anticipation', 'disgust', 'fear', 'joy', 'sadness', 'surprise', 'trust','negative', 'positive'].mean()
df_emotions['n_tweets']=df_emo_all.screen_name.value_counts()
df_emotions.head()
# Distribution graphs (histogram/bar graph) of column data
def plotPerColumnDistribution(df, nGraphShown, nGraphPerRow):
nunique = df.nunique()
df = df[[col for col in df if nunique[col] > 1 and nunique[col] < 50]] # For displaying purposes, pick columns that have between 1 and 50 unique values
nRow, nCol = df.shape
columnNames = list(df)
nGraphRow = (nCol + nGraphPerRow - 1) / nGraphPerRow
plt.figure(num = None, figsize = (6 * nGraphPerRow, 8 * nGraphRow), dpi = 80, facecolor = 'w', edgecolor = 'k')
for i in range(min(nCol, nGraphShown)):
plt.subplot(nGraphRow, nGraphPerRow, i + 1)
columnDf = df.iloc[:, i]
if (not np.issubdtype(type(columnDf.iloc[0]), np.number)):
valueCounts = columnDf.value_counts()
valueCounts.plot.bar()
else:
columnDf.hist()
plt.ylabel('counts')
plt.xticks(rotation = 90)
plt.title(f'{columnNames[i]} (column {i})')
plt.tight_layout(pad = 1.0, w_pad = 1.0, h_pad = 1.0)
plt.show()
# Correlation matrix
def plotCorrelationMatrix(df, graphWidth):
#filename = df.dataframeName
df = df.dropna('columns') # drop columns with NaN
df = df[[col for col in df if df[col].nunique() > 1]] # keep columns where there are more than 1 unique values
if df.shape[1] < 2:
print(f'No correlation plots shown: The number of non-NaN or constant columns ({df.shape[1]}) is less than 2')
return
corr = df.corr()
plt.figure(num=None, figsize=(graphWidth, graphWidth), dpi=80, facecolor='w', edgecolor='k')
corrMat = plt.matshow(corr, fignum = 1)
plt.xticks(range(len(corr.columns)), corr.columns, rotation=90)
plt.yticks(range(len(corr.columns)), corr.columns)
plt.gca().xaxis.tick_bottom()
plt.colorbar(corrMat)
#plt.title(f'Correlation Matrix for {filename}', fontsize=15)
plt.title(f'Correlation Matrix', fontsize=15)
plt.show()
# Scatter and density plots
def plotScatterMatrix(df, plotSize, textSize):
df = df.select_dtypes(include =[np.number]) # keep only numerical columns
# Remove rows and columns that would lead to df being singular
df = df.dropna('columns')
df = df[[col for col in df if df[col].nunique() > 1]] # keep columns where there are more than 1 unique values
columnNames = list(df)
if len(columnNames) > 10: # reduce the number of columns for matrix inversion of kernel density plots
columnNames = columnNames[:10]
df = df[columnNames]
ax = pd.plotting.scatter_matrix(df, alpha=0.75, figsize=[plotSize, plotSize], diagonal='kde')
corrs = df.corr().values
for i, j in zip(*plt.np.triu_indices_from(ax, k = 1)):
ax[i, j].annotate('Corr. coef = %.3f' % corrs[i, j], (0.8, 0.2), xycoords='axes fraction', ha='center', va='center', size=textSize)
plt.suptitle('Scatter and Density Plot')
plt.show()
plotPerColumnDistribution(df_emotions.drop(['n_tweets'], axis=1), 4, 4)
#plotPerColumnDistribution(df1, 10, 5)
plotCorrelationMatrix(df_emotions.drop(['n_tweets'], axis=1), 6)
plotScatterMatrix(df_emotions.drop(['n_tweets'], axis=1), 18, 10)
df_emotions.head()
# How many bullies do we have??
df_emotions[df_emotions.anger > 0.3]
# and potentially bullied?
df_emotions[df_emotions.fear > 0.3]
#Top 10 angriest
df_emotions.sort_values(by='anger', ascending=False)[:10]
#Top 10 saddest
df_emotions.sort_values(by='sadness', ascending=False)[:10]
# We could develop a metric to define a "bully" with some tresholds
def is_a_bully(screen_name, df):
"""
Test function to flag a user as a potential bully using the aggregated metrics
"""
return False
# Let's play with isiZulu
def build_emolex(language):
lexicon = "NRC-Sentiment-Emotion-Lexicons/NRC-Emotion-Lexicon-v0.92/NRC-Emotion-Lexicon-v0.92_"+language+".txt"
emolex_words = pd.read_csv(lexicon,
index_col=0,
sep='\t', na_values='NO TRANSLATION')
print(emolex_words.shape)
#emolex_words.index = emolex_words.index.map(lambda w: stemmer.stem(w.lower()) if w else nan)
emotions = emolex_words.columns.values
emolex_words['emotions'] = list(zip(*map(emolex_words.get, emolex_words)))
# Convert into a dictionary for faster lookup
emolex_dict = emolex_words['emotions'].to_dict()
print("We built a dictionary of {} words associated to {} emotions".format(len(emolex_dict), len(emotions)))
return (emolex_dict, emotions)
language='isizulu'
isizulu = build_emolex('isizulu')
len(isizulu[1])
# define a function to score a sentence
from nltk import word_tokenize
def sentence_emotions(sentence, emo_dict):
sentence_score = np.zeros(10)
emolex, emotions = emo_dict
document = word_tokenize(sentence)
for w, word in enumerate(document):
#document[w] = stemmer.stem(word.lower())
#emo_score = emolex_words[emolex_words.word == word].values
emo_score = emolex.get(word)
if emo_score != None:
sentence_score += emo_score
print("Sentence scores:")
for i in range(len(emotions)):
print("{}: {}".format(emotions[i],sentence_score[i]))
return
sentence_emotions("bulala ukudabuka", isizulu)
language='isizulu'
isizulu = build_emolex('isizulu')
language='xhosa'
sesotho, emotions = build_emolex(language)
language='sesotho'
sesotho, emotions = build_emolex(language)
sesotho.get('ho etsa lichelete')
language='sesotho'
lexicon = "NRC-Sentiment-Emotion-Lexicons/NRC-Emotion-Lexicon-v0.92/NRC-Emotion-Lexicon-v0.92_"+language+".txt"
emolex_words = pd.read_csv(lexicon,
index_col=0,
sep='\t')
emolex_words = emolex_words[emolex_words.index!='NO TRANSLATION']
print(emolex_words.shape)
#emolex_words.index = emolex_words.index.map(lambda w: stemmer.stem(w.lower()) if w else nan)
emotions = emolex_words.columns.values
emolex_words['emotions'] = list(zip(*map(emolex_words.get, emolex_words)))
emolex_dict = emolex_words['emotions'].to_dict()
print("We built a dictionary of {} words associated to {} emotions".format(len(emolex_dict), len(emotions)))
dict_set = set(emolex_words['emotions'].to_dict().keys())
emo_set = set(emolex_words.index)
# Can you write a scoring function using bi-grams (a sequence of two words) and tri-grams (a sequence of three words)
# Build a dictionary that works for "Joburg Zulu" by mixing english and zulu words
# and adding slang
| 0.333612 | 0.87456 |
# This code assesses the outputs of VESIcal compared to the VolatileCalc parameterization of the Dixon (1997) model.
- Test 1 compares saturation pressures from VolatileCalc and a Excel Macro with those from VESIcal for a variety of natural compositions, and synthetic arrays.
- Test 2 compares X$_{H_{2}O}$ in the fluid phase at volatile saturation to that outputted by the Dixon Macro, and VolatileCalc
- Test 3 compares isobars with those of VolatileCalc
- Test 4 compares degassing paths
```
import VESIcal as v
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
from IPython.display import display, HTML
import pandas as pd
import matplotlib as mpl
import seaborn as sns
%matplotlib inline
from sklearn.linear_model import LinearRegression
from sklearn.metrics import r2_score
sns.set(style="ticks", context="poster",rc={"grid.linewidth": 1,"xtick.major.width": 1,"ytick.major.width": 1, 'patch.edgecolor': 'black'})
plt.style.use("seaborn-colorblind")
plt.rcParams["font.size"] =12
plt.rcParams["mathtext.default"] = "regular"
plt.rcParams["mathtext.fontset"] = "dejavusans"
plt.rcParams['patch.linewidth'] = 1
plt.rcParams['axes.linewidth'] = 1
plt.rcParams["xtick.direction"] = "in"
plt.rcParams["ytick.direction"] = "in"
plt.rcParams["ytick.direction"] = "in"
plt.rcParams["xtick.major.size"] = 6 # Sets length of ticks
plt.rcParams["ytick.major.size"] = 4 # Sets length of ticks
plt.rcParams["ytick.labelsize"] = 12 # Sets size of numbers on tick marks
plt.rcParams["xtick.labelsize"] = 12 # Sets size of numbers on tick marks
plt.rcParams["axes.titlesize"] = 14 # Overall title
plt.rcParams["axes.labelsize"] = 14 # Axes labels
plt.rcParams["legend.fontsize"]= 14
```
# Test 1 - Comparing saturation pressures from VESIcal to VolatileCalc and the Dixon macro
```
myfile = v.BatchFile('S2_Testing_Dixon_1997_VolatileCalc.xlsx')
data = myfile.get_data()
VolatileCalc_PSat=data['VolatileCalc_P'] # Saturation pressure from VolatileCalc
DixonMacro_PSat=data['DixonMacro_P'] # Saturation pressure from dixon
satPs_wtemps_Dixon= myfile.calculate_saturation_pressure(temperature="Temp", model='Dixon')
# Making linear regression
# VolatileCalc
X=VolatileCalc_PSat
Y=satPs_wtemps_Dixon['SaturationP_bars_VESIcal']
mask = ~np.isnan(X) & ~np.isnan(Y)
X_noNan=X[mask].values.reshape(-1, 1)
Y_noNan=Y[mask].values.reshape(-1, 1)
lr=LinearRegression()
lr.fit(X_noNan,Y_noNan)
Y_pred=lr.predict(X_noNan)
#X - Y comparison of pressures
fig, (ax1, ax2) = plt.subplots(1, 2, figsize = (12,5)) # adjust dimensions of figure here
ax1.set_title('Comparison of VolatileCalc and VESIcal', fontsize=14)
ax1.set_xlabel('P$_{Sat}$ VolatileCalc', fontsize=14)
ax1.set_ylabel('P$_{Sat}$ VESIcal', fontsize=14)
ax1.plot(X_noNan,Y_pred, color='red', linewidth=1)
ax1.scatter(X_noNan, Y_noNan, s=50, edgecolors='k', facecolors='silver', marker='o')
I='Intercept= ' + str(np.round(lr.intercept_, 1))[1:-1]
G='Gradient= ' + str(np.round(lr.coef_, 4))[2:-2]
R='R$^2$= ' + str(np.round(r2_score(Y_noNan, Y_pred), 4))
#one='1:1 line'
ax1.text(1000, 3700, I, fontsize=14)
ax1.text(1000, 4000, G, fontsize=14)
ax1.text(1000, 4300, R, fontsize=14)
#Dixon Macro
X=DixonMacro_PSat
Y=satPs_wtemps_Dixon['SaturationP_bars_VESIcal']
mask = ~np.isnan(X) & ~np.isnan(Y)
X_noNan=X[mask].values.reshape(-1, 1)
Y_noNan=Y[mask].values.reshape(-1, 1)
lr=LinearRegression()
lr.fit(X_noNan,Y_noNan)
Y_pred=lr.predict(X_noNan)
#X - Y comparison of pressures
ax2.set_title('Comparison of the Dixon Macro and VESIcal', fontsize=14)
ax2.set_xlabel('P$_{Sat}$ VolatileCalc', fontsize=14)
ax2.set_ylabel('P$_{Sat}$ VESIcal', fontsize=14)
ax2.plot(X_noNan,Y_pred, color='red', linewidth=1)
ax2.scatter(X_noNan, Y_noNan, s=50, edgecolors='k', facecolors='silver', marker='o')
#plt.plot([0, 4000], [0, 4000])
I='Intercept= ' + str(np.round(lr.intercept_, 1))[1:-1]
G='Gradient= ' + str(np.round(lr.coef_, 4))[2:-2]
R='R$^2$= ' + str(np.round(r2_score(Y_noNan, Y_pred), 5))
#one='1:1 line'
ax2.text(1000, 3700, I, fontsize=14)
ax2.text(1000, 4000, G, fontsize=14)
ax2.text(1000, 4300, R, fontsize=14)
ax1.set_ylim([0, 5500])
ax1.set_xlim([0, 5500])
ax2.set_ylim([0, 5500])
ax2.set_xlim([0, 5500])
plt.subplots_adjust(left=0.125, bottom=None, right=0.9, top=None, wspace=0.3, hspace=None)
ax1.text(30, 5200, 'a)', fontsize=14)
ax2.text(30, 5200, 'b)', fontsize=14)
fig.savefig('VolatileCalc_Test1a.png', transparent=True)
# This shows the % difference between VolatileCalc and VESIcal. The differences are similar in magnitude to those between VolatileCalc and the
# Dixon Macro
fig, (ax1, ax2, ax3) = plt.subplots(1, 3,figsize = (15,5))
font = {'family': 'sans-serif',
'color': 'black',
'weight': 'normal',
'size': 20,
}
ax1.set_xlabel('% Difference (VolatileCalc/VESIcal)', fontsize=14)
ax1.set_ylabel('# of measurements', fontsize=14)
ax1.hist(100*VolatileCalc_PSat/satPs_wtemps_Dixon['SaturationP_bars_VESIcal'])
ax2.set_xlabel('% Difference (DixonMacro/VESIcal)', fontsize=14)
ax2.set_ylabel('# of measurements', fontsize=14)
ax2.hist(100*DixonMacro_PSat/satPs_wtemps_Dixon['SaturationP_bars_VESIcal'])
ax3.set_xlabel('% Difference (DixonMacro/VolatileCalc)', fontsize=14)
ax3.set_ylabel('# of measurements', fontsize=14)
ax3.hist(100*DixonMacro_PSat/VolatileCalc_PSat)
plt.subplots_adjust(left=0.125, bottom=None, right=0.9, top=None, wspace=0.2, hspace=None)
ax1.tick_params(axis="x", labelsize=12)
ax1.tick_params(axis="y", labelsize=12)
ax2.tick_params(axis="x", labelsize=12)
ax2.tick_params(axis="y", labelsize=12)
ax3.tick_params(axis="y", labelsize=12)
ax3.tick_params(axis="x", labelsize=12)
ax1.set_xlim([97, 104])
ax2.set_xlim([98, 102])
#ax3.set_xlim([95, 104])
ax1.tick_params(direction='in', length=6, width=1, colors='k',
grid_color='k', grid_alpha=0.5)
ax2.tick_params(direction='in', length=6, width=1, colors='k',
grid_color='k', grid_alpha=0.5)
ax3.tick_params(direction='in', length=6, width=1, colors='k',
grid_color='k', grid_alpha=0.5)
ax1.text(97.3, 13.7, 'a)', fontsize=14)
ax2.text(98.2, 11.7, 'b)', fontsize=14)
ax3.text(87.4, 32, 'c)', fontsize=14)
fig.savefig('VolatileCalc_Test1b.png', transparent=True)
X=satPs_wtemps_Dixon['VolatileCalc_P']
Y=satPs_wtemps_Dixon['SaturationP_bars_VESIcal']
mask = (satPs_wtemps_Dixon['CO2']>0)
X_noNan=X[mask].values.reshape(-1, 1)
Y_noNan=Y[mask].values.reshape(-1, 1)
lr=LinearRegression()
lr.fit(X_noNan,Y_noNan)
Y_pred=lr.predict(X_noNan)
#X - Y comparison of pressures
fig, ax1 = plt.subplots( figsize = (10,8)) # adjust dimensions of figure here
font = {'family': 'sans-serif',
'color': 'black',
'weight': 'normal',
'size': 20,
}
ax1.set_title('Comparison of VolatileCalc and VESIcal',
fontdict= font, pad = 15)
ax1.set_xlabel('P$_{Sat}$ VolatileCalc', fontdict=font, labelpad = 15)
ax1.set_ylabel('P$_{Sat}$ VESIcal', fontdict=font, labelpad = 15)
ax1.plot(X_noNan,Y_pred, color='red', linewidth=1)
ax1.scatter(X_noNan, Y_noNan, s=100, edgecolors='gray', facecolors='silver', marker='o')
I='Intercept= ' + str(np.round(lr.intercept_, 3))[1:-1]
G='Gradient= ' + str(np.round(lr.coef_, 5))[2:-2]
R='R$^2$= ' + str(np.round(r2_score(Y_noNan, Y_pred), 5))
#one='1:1 line'
plt.plot([0, 2000], [0, 2000])
ax1.text(500, 3000, I, fontsize=15)
ax1.text(500, 3500, G, fontsize=15)
ax1.text(500, 4000, R, fontsize=15)
```
# Test 2 - Comparing X$_{H_{2}O}$ in the fluid at the saturation pressure to that calculated using VolatileCalc and the Dixon Macro
```
eqfluid_Dixon_VolatileCalcP = myfile.calculate_equilibrium_fluid_comp(temperature="Temp", model='Dixon', pressure = None)
eqfluid_Dixon_DixonMacroP = myfile.calculate_equilibrium_fluid_comp(temperature="Temp", model='Dixon', pressure = None)
# Making linear regression
# VolatileCalc
X=0.01*eqfluid_Dixon_VolatileCalcP['VolatileCalc_H2Ov mol% (norm)'] # VolatileCalc outputs in %
Y=eqfluid_Dixon_VolatileCalcP['XH2O_fl_VESIcal']
mask = ~np.isnan(X) & ~np.isnan(Y)
X_noNan=X[mask].values.reshape(-1, 1)
Y_noNan=Y[mask].values.reshape(-1, 1)
lr=LinearRegression()
lr.fit(X_noNan,Y_noNan)
Y_pred=lr.predict(X_noNan)
#X - Y comparison of pressures
fig, (ax1, ax2) = plt.subplots(1, 2, figsize = (12,5)) # adjust dimensions of figure here
ax1.set_xlabel('X$_{H2O}$ VolatileCalc', fontsize=14)
ax1.set_ylabel('X$_{H2O}$ VESIcal', fontsize=14)
ax1.scatter(X_noNan, Y_noNan, s=50, edgecolors='k', facecolors='silver', marker='o')
ax1.plot(X_noNan,Y_pred, color='red', linewidth=1)
I='Intercept= ' + str(np.round(lr.intercept_, 3))[1:-1]
G='Gradient= ' + str(np.round(lr.coef_, 5))[2:-2]
R='R$^2$= ' + str(np.round(r2_score(Y_noNan, Y_pred), 5))
ax1.text(0, 0.5, I, fontsize=14)
ax1.text(0, 0.6, G, fontsize=14)
ax1.text(0, 0.7, R, fontsize=14)
# Dixon Macro
X=eqfluid_Dixon_DixonMacroP['DixonMacro_XH2O']
Y=eqfluid_Dixon_DixonMacroP['XH2O_fl_VESIcal']
mask = ~np.isnan(X) & ~np.isnan(Y)
X_noNan=X[mask].values.reshape(-1, 1)
Y_noNan=Y[mask].values.reshape(-1, 1)
lr=LinearRegression()
lr.fit(X_noNan,Y_noNan)
Y_pred=lr.predict(X_noNan)
ax2.set_xlabel('X$_{H2O}$ DixonMacro', fontsize=14)
ax2.set_ylabel('X$_{H2O}$ VESIcal', fontsize=14)
ax2.plot(X_noNan,Y_pred, color='red', linewidth=1)
ax2.scatter(X_noNan, Y_noNan, s=50, edgecolors='k', facecolors='silver', marker='o')
I='Intercept= ' + str(np.round(lr.intercept_, 5))[1:-1]
G='Gradient= ' + str(np.round(lr.coef_, 5))[2:-2]
R='R$^2$= ' + str(np.round(r2_score(Y_noNan, Y_pred), 5))
ax2.text(0, 0.5, I, fontsize=14)
ax2.text(0, 0.6, G, fontsize=14)
ax2.text(0, 0.7, R, fontsize=14)
plt.subplots_adjust(left=0.125, bottom=None, right=0.9, top=None, wspace=0.3, hspace=None)
ax1.text(-0.05, 1.01, 'a)', fontsize=14)
ax2.text(-0.05, 1.01, 'b)', fontsize=14)
fig.savefig('VolatileCalc_Test2.png', transparent=True)
```
# Test 3 - Comparing Isobars to those calculated in VolatileCalc
```
#Loading Isobars from VolatileCalc
Isobar_output= pd.read_excel('S2_Testing_Dixon_1997_VolatileCalc.xlsx', sheet_name='Isobar_Outputs', index_col=0)
myfile_Isobar_input= v.BatchFile('S2_Testing_Dixon_1997_VolatileCalc.xlsx', sheet_name='Isobar_Comp')
data_Isobar_input = myfile_Isobar_input.data
SampleName='0'
bulk_comp= myfile_Isobar_input.get_sample_composition(SampleName, asSampleClass=True)
temperature=1200
# Calculating isobars
isobars, isopleths = v.calculate_isobars_and_isopleths(sample=bulk_comp, model='Dixon',
temperature=temperature,
pressure_list=[500, 1000, 2000, 3000],
isopleth_list=[0, 0.1, 0.2, 0.3, 0.5, 0.8, 0.9, 1],
print_status=True).result
fig, ax1 = plt.subplots(figsize = (8,5))
mpl.rcParams['axes.linewidth'] = 1
mpl.rcParams.update({'font.size': 10})
plt.scatter(Isobar_output['Wt%H2O'], Isobar_output['PPMCO2'], marker='o', s=10, label='VolatileCalc', color='k')
plt.plot(isobars.loc[isobars.Pressure==500, 'H2O_liq'], (10**4)*isobars.loc[isobars.Pressure==500, 'CO2_liq'], label='VESIcal 500 bars')
plt.plot(isobars.loc[isobars.Pressure==1000, 'H2O_liq'], (10**4)*isobars.loc[isobars.Pressure==1000, 'CO2_liq'], label='VESIcal 1000 bars')
plt.plot(isobars.loc[isobars.Pressure==2000, 'H2O_liq'], (10**4)*isobars.loc[isobars.Pressure==2000, 'CO2_liq'], label='VESIcal 2000 bars')
plt.plot(isobars.loc[isobars.Pressure==3000, 'H2O_liq'], (10**4)*isobars.loc[isobars.Pressure==3000, 'CO2_liq'], label='VESIcal 3000 bars')
plt.legend(fontsize='small')
ax1.set_xlabel('H$_2$O', fontsize=14)
ax1.set_ylabel('CO$_2$', fontsize=14)
ax1.tick_params(axis="x", labelsize=12)
ax1.tick_params(axis="y", labelsize=12)
ax1.tick_params(direction='in', length=6, width=1, colors='k',
grid_color='k', grid_alpha=0.5)
fig.savefig('VolatileCalc_Test3.png', transparent=True)
```
|
github_jupyter
|
import VESIcal as v
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
from IPython.display import display, HTML
import pandas as pd
import matplotlib as mpl
import seaborn as sns
%matplotlib inline
from sklearn.linear_model import LinearRegression
from sklearn.metrics import r2_score
sns.set(style="ticks", context="poster",rc={"grid.linewidth": 1,"xtick.major.width": 1,"ytick.major.width": 1, 'patch.edgecolor': 'black'})
plt.style.use("seaborn-colorblind")
plt.rcParams["font.size"] =12
plt.rcParams["mathtext.default"] = "regular"
plt.rcParams["mathtext.fontset"] = "dejavusans"
plt.rcParams['patch.linewidth'] = 1
plt.rcParams['axes.linewidth'] = 1
plt.rcParams["xtick.direction"] = "in"
plt.rcParams["ytick.direction"] = "in"
plt.rcParams["ytick.direction"] = "in"
plt.rcParams["xtick.major.size"] = 6 # Sets length of ticks
plt.rcParams["ytick.major.size"] = 4 # Sets length of ticks
plt.rcParams["ytick.labelsize"] = 12 # Sets size of numbers on tick marks
plt.rcParams["xtick.labelsize"] = 12 # Sets size of numbers on tick marks
plt.rcParams["axes.titlesize"] = 14 # Overall title
plt.rcParams["axes.labelsize"] = 14 # Axes labels
plt.rcParams["legend.fontsize"]= 14
myfile = v.BatchFile('S2_Testing_Dixon_1997_VolatileCalc.xlsx')
data = myfile.get_data()
VolatileCalc_PSat=data['VolatileCalc_P'] # Saturation pressure from VolatileCalc
DixonMacro_PSat=data['DixonMacro_P'] # Saturation pressure from dixon
satPs_wtemps_Dixon= myfile.calculate_saturation_pressure(temperature="Temp", model='Dixon')
# Making linear regression
# VolatileCalc
X=VolatileCalc_PSat
Y=satPs_wtemps_Dixon['SaturationP_bars_VESIcal']
mask = ~np.isnan(X) & ~np.isnan(Y)
X_noNan=X[mask].values.reshape(-1, 1)
Y_noNan=Y[mask].values.reshape(-1, 1)
lr=LinearRegression()
lr.fit(X_noNan,Y_noNan)
Y_pred=lr.predict(X_noNan)
#X - Y comparison of pressures
fig, (ax1, ax2) = plt.subplots(1, 2, figsize = (12,5)) # adjust dimensions of figure here
ax1.set_title('Comparison of VolatileCalc and VESIcal', fontsize=14)
ax1.set_xlabel('P$_{Sat}$ VolatileCalc', fontsize=14)
ax1.set_ylabel('P$_{Sat}$ VESIcal', fontsize=14)
ax1.plot(X_noNan,Y_pred, color='red', linewidth=1)
ax1.scatter(X_noNan, Y_noNan, s=50, edgecolors='k', facecolors='silver', marker='o')
I='Intercept= ' + str(np.round(lr.intercept_, 1))[1:-1]
G='Gradient= ' + str(np.round(lr.coef_, 4))[2:-2]
R='R$^2$= ' + str(np.round(r2_score(Y_noNan, Y_pred), 4))
#one='1:1 line'
ax1.text(1000, 3700, I, fontsize=14)
ax1.text(1000, 4000, G, fontsize=14)
ax1.text(1000, 4300, R, fontsize=14)
#Dixon Macro
X=DixonMacro_PSat
Y=satPs_wtemps_Dixon['SaturationP_bars_VESIcal']
mask = ~np.isnan(X) & ~np.isnan(Y)
X_noNan=X[mask].values.reshape(-1, 1)
Y_noNan=Y[mask].values.reshape(-1, 1)
lr=LinearRegression()
lr.fit(X_noNan,Y_noNan)
Y_pred=lr.predict(X_noNan)
#X - Y comparison of pressures
ax2.set_title('Comparison of the Dixon Macro and VESIcal', fontsize=14)
ax2.set_xlabel('P$_{Sat}$ VolatileCalc', fontsize=14)
ax2.set_ylabel('P$_{Sat}$ VESIcal', fontsize=14)
ax2.plot(X_noNan,Y_pred, color='red', linewidth=1)
ax2.scatter(X_noNan, Y_noNan, s=50, edgecolors='k', facecolors='silver', marker='o')
#plt.plot([0, 4000], [0, 4000])
I='Intercept= ' + str(np.round(lr.intercept_, 1))[1:-1]
G='Gradient= ' + str(np.round(lr.coef_, 4))[2:-2]
R='R$^2$= ' + str(np.round(r2_score(Y_noNan, Y_pred), 5))
#one='1:1 line'
ax2.text(1000, 3700, I, fontsize=14)
ax2.text(1000, 4000, G, fontsize=14)
ax2.text(1000, 4300, R, fontsize=14)
ax1.set_ylim([0, 5500])
ax1.set_xlim([0, 5500])
ax2.set_ylim([0, 5500])
ax2.set_xlim([0, 5500])
plt.subplots_adjust(left=0.125, bottom=None, right=0.9, top=None, wspace=0.3, hspace=None)
ax1.text(30, 5200, 'a)', fontsize=14)
ax2.text(30, 5200, 'b)', fontsize=14)
fig.savefig('VolatileCalc_Test1a.png', transparent=True)
# This shows the % difference between VolatileCalc and VESIcal. The differences are similar in magnitude to those between VolatileCalc and the
# Dixon Macro
fig, (ax1, ax2, ax3) = plt.subplots(1, 3,figsize = (15,5))
font = {'family': 'sans-serif',
'color': 'black',
'weight': 'normal',
'size': 20,
}
ax1.set_xlabel('% Difference (VolatileCalc/VESIcal)', fontsize=14)
ax1.set_ylabel('# of measurements', fontsize=14)
ax1.hist(100*VolatileCalc_PSat/satPs_wtemps_Dixon['SaturationP_bars_VESIcal'])
ax2.set_xlabel('% Difference (DixonMacro/VESIcal)', fontsize=14)
ax2.set_ylabel('# of measurements', fontsize=14)
ax2.hist(100*DixonMacro_PSat/satPs_wtemps_Dixon['SaturationP_bars_VESIcal'])
ax3.set_xlabel('% Difference (DixonMacro/VolatileCalc)', fontsize=14)
ax3.set_ylabel('# of measurements', fontsize=14)
ax3.hist(100*DixonMacro_PSat/VolatileCalc_PSat)
plt.subplots_adjust(left=0.125, bottom=None, right=0.9, top=None, wspace=0.2, hspace=None)
ax1.tick_params(axis="x", labelsize=12)
ax1.tick_params(axis="y", labelsize=12)
ax2.tick_params(axis="x", labelsize=12)
ax2.tick_params(axis="y", labelsize=12)
ax3.tick_params(axis="y", labelsize=12)
ax3.tick_params(axis="x", labelsize=12)
ax1.set_xlim([97, 104])
ax2.set_xlim([98, 102])
#ax3.set_xlim([95, 104])
ax1.tick_params(direction='in', length=6, width=1, colors='k',
grid_color='k', grid_alpha=0.5)
ax2.tick_params(direction='in', length=6, width=1, colors='k',
grid_color='k', grid_alpha=0.5)
ax3.tick_params(direction='in', length=6, width=1, colors='k',
grid_color='k', grid_alpha=0.5)
ax1.text(97.3, 13.7, 'a)', fontsize=14)
ax2.text(98.2, 11.7, 'b)', fontsize=14)
ax3.text(87.4, 32, 'c)', fontsize=14)
fig.savefig('VolatileCalc_Test1b.png', transparent=True)
X=satPs_wtemps_Dixon['VolatileCalc_P']
Y=satPs_wtemps_Dixon['SaturationP_bars_VESIcal']
mask = (satPs_wtemps_Dixon['CO2']>0)
X_noNan=X[mask].values.reshape(-1, 1)
Y_noNan=Y[mask].values.reshape(-1, 1)
lr=LinearRegression()
lr.fit(X_noNan,Y_noNan)
Y_pred=lr.predict(X_noNan)
#X - Y comparison of pressures
fig, ax1 = plt.subplots( figsize = (10,8)) # adjust dimensions of figure here
font = {'family': 'sans-serif',
'color': 'black',
'weight': 'normal',
'size': 20,
}
ax1.set_title('Comparison of VolatileCalc and VESIcal',
fontdict= font, pad = 15)
ax1.set_xlabel('P$_{Sat}$ VolatileCalc', fontdict=font, labelpad = 15)
ax1.set_ylabel('P$_{Sat}$ VESIcal', fontdict=font, labelpad = 15)
ax1.plot(X_noNan,Y_pred, color='red', linewidth=1)
ax1.scatter(X_noNan, Y_noNan, s=100, edgecolors='gray', facecolors='silver', marker='o')
I='Intercept= ' + str(np.round(lr.intercept_, 3))[1:-1]
G='Gradient= ' + str(np.round(lr.coef_, 5))[2:-2]
R='R$^2$= ' + str(np.round(r2_score(Y_noNan, Y_pred), 5))
#one='1:1 line'
plt.plot([0, 2000], [0, 2000])
ax1.text(500, 3000, I, fontsize=15)
ax1.text(500, 3500, G, fontsize=15)
ax1.text(500, 4000, R, fontsize=15)
eqfluid_Dixon_VolatileCalcP = myfile.calculate_equilibrium_fluid_comp(temperature="Temp", model='Dixon', pressure = None)
eqfluid_Dixon_DixonMacroP = myfile.calculate_equilibrium_fluid_comp(temperature="Temp", model='Dixon', pressure = None)
# Making linear regression
# VolatileCalc
X=0.01*eqfluid_Dixon_VolatileCalcP['VolatileCalc_H2Ov mol% (norm)'] # VolatileCalc outputs in %
Y=eqfluid_Dixon_VolatileCalcP['XH2O_fl_VESIcal']
mask = ~np.isnan(X) & ~np.isnan(Y)
X_noNan=X[mask].values.reshape(-1, 1)
Y_noNan=Y[mask].values.reshape(-1, 1)
lr=LinearRegression()
lr.fit(X_noNan,Y_noNan)
Y_pred=lr.predict(X_noNan)
#X - Y comparison of pressures
fig, (ax1, ax2) = plt.subplots(1, 2, figsize = (12,5)) # adjust dimensions of figure here
ax1.set_xlabel('X$_{H2O}$ VolatileCalc', fontsize=14)
ax1.set_ylabel('X$_{H2O}$ VESIcal', fontsize=14)
ax1.scatter(X_noNan, Y_noNan, s=50, edgecolors='k', facecolors='silver', marker='o')
ax1.plot(X_noNan,Y_pred, color='red', linewidth=1)
I='Intercept= ' + str(np.round(lr.intercept_, 3))[1:-1]
G='Gradient= ' + str(np.round(lr.coef_, 5))[2:-2]
R='R$^2$= ' + str(np.round(r2_score(Y_noNan, Y_pred), 5))
ax1.text(0, 0.5, I, fontsize=14)
ax1.text(0, 0.6, G, fontsize=14)
ax1.text(0, 0.7, R, fontsize=14)
# Dixon Macro
X=eqfluid_Dixon_DixonMacroP['DixonMacro_XH2O']
Y=eqfluid_Dixon_DixonMacroP['XH2O_fl_VESIcal']
mask = ~np.isnan(X) & ~np.isnan(Y)
X_noNan=X[mask].values.reshape(-1, 1)
Y_noNan=Y[mask].values.reshape(-1, 1)
lr=LinearRegression()
lr.fit(X_noNan,Y_noNan)
Y_pred=lr.predict(X_noNan)
ax2.set_xlabel('X$_{H2O}$ DixonMacro', fontsize=14)
ax2.set_ylabel('X$_{H2O}$ VESIcal', fontsize=14)
ax2.plot(X_noNan,Y_pred, color='red', linewidth=1)
ax2.scatter(X_noNan, Y_noNan, s=50, edgecolors='k', facecolors='silver', marker='o')
I='Intercept= ' + str(np.round(lr.intercept_, 5))[1:-1]
G='Gradient= ' + str(np.round(lr.coef_, 5))[2:-2]
R='R$^2$= ' + str(np.round(r2_score(Y_noNan, Y_pred), 5))
ax2.text(0, 0.5, I, fontsize=14)
ax2.text(0, 0.6, G, fontsize=14)
ax2.text(0, 0.7, R, fontsize=14)
plt.subplots_adjust(left=0.125, bottom=None, right=0.9, top=None, wspace=0.3, hspace=None)
ax1.text(-0.05, 1.01, 'a)', fontsize=14)
ax2.text(-0.05, 1.01, 'b)', fontsize=14)
fig.savefig('VolatileCalc_Test2.png', transparent=True)
#Loading Isobars from VolatileCalc
Isobar_output= pd.read_excel('S2_Testing_Dixon_1997_VolatileCalc.xlsx', sheet_name='Isobar_Outputs', index_col=0)
myfile_Isobar_input= v.BatchFile('S2_Testing_Dixon_1997_VolatileCalc.xlsx', sheet_name='Isobar_Comp')
data_Isobar_input = myfile_Isobar_input.data
SampleName='0'
bulk_comp= myfile_Isobar_input.get_sample_composition(SampleName, asSampleClass=True)
temperature=1200
# Calculating isobars
isobars, isopleths = v.calculate_isobars_and_isopleths(sample=bulk_comp, model='Dixon',
temperature=temperature,
pressure_list=[500, 1000, 2000, 3000],
isopleth_list=[0, 0.1, 0.2, 0.3, 0.5, 0.8, 0.9, 1],
print_status=True).result
fig, ax1 = plt.subplots(figsize = (8,5))
mpl.rcParams['axes.linewidth'] = 1
mpl.rcParams.update({'font.size': 10})
plt.scatter(Isobar_output['Wt%H2O'], Isobar_output['PPMCO2'], marker='o', s=10, label='VolatileCalc', color='k')
plt.plot(isobars.loc[isobars.Pressure==500, 'H2O_liq'], (10**4)*isobars.loc[isobars.Pressure==500, 'CO2_liq'], label='VESIcal 500 bars')
plt.plot(isobars.loc[isobars.Pressure==1000, 'H2O_liq'], (10**4)*isobars.loc[isobars.Pressure==1000, 'CO2_liq'], label='VESIcal 1000 bars')
plt.plot(isobars.loc[isobars.Pressure==2000, 'H2O_liq'], (10**4)*isobars.loc[isobars.Pressure==2000, 'CO2_liq'], label='VESIcal 2000 bars')
plt.plot(isobars.loc[isobars.Pressure==3000, 'H2O_liq'], (10**4)*isobars.loc[isobars.Pressure==3000, 'CO2_liq'], label='VESIcal 3000 bars')
plt.legend(fontsize='small')
ax1.set_xlabel('H$_2$O', fontsize=14)
ax1.set_ylabel('CO$_2$', fontsize=14)
ax1.tick_params(axis="x", labelsize=12)
ax1.tick_params(axis="y", labelsize=12)
ax1.tick_params(direction='in', length=6, width=1, colors='k',
grid_color='k', grid_alpha=0.5)
fig.savefig('VolatileCalc_Test3.png', transparent=True)
| 0.603581 | 0.89275 |
```
# Implementation from https://github.com/dougalsutherland/opt-mmd
import sys, os
import numpy as np
from math import sqrt
CHANNEL_MEANS = (33.430001959204674/255,)
CHANNEL_STDS = (78.86655405163765/255,)
from scipy.spatial.distance import pdist, cdist
def energy_distance(v, w):
VV = np.mean(pdist(v, 'euclidean'))
WW = np.mean(pdist(w, 'euclidean'))
VW = np.mean(cdist(v, w, 'euclidean'))
return 2 * VW - VV - WW
from PIL import Image
from matplotlib import pyplot as plt
def display_sample(sample):
img = sample.reshape((28, 28)) * 255.
plt.imshow(Image.fromarray(img))
plt.show()
```
## Compare all MNIST datasest
```
# Add Bayesian-and-novelty directory to the PYTHONPATH
import sys
import os
sys.path.append(os.path.realpath('../../../..'))
# Autoreload changes in utils, etc.
%load_ext autoreload
%autoreload 2
import torch
from torchvision import datasets, transforms
import numpy as np
from novelty.utils.datasets import GaussianNoiseDataset
from novelty.utils.datasets import UniformNoiseDataset
from novelty.utils import DatasetSubset
torch.manual_seed(1)
```
# MNIST 0-4
```
CHANNEL_MEANS = (33.550922870635986/255,)
CHANNEL_STDS = (79.10186022520065/255,)
def get_mnist_images(mnist_dir):
transform = transforms.Compose([
transforms.Grayscale(),
transforms.ToTensor(),
transforms.Normalize(CHANNEL_MEANS, CHANNEL_STDS)
])
dataset = datasets.ImageFolder(mnist_dir, transform=transform)
dataset = np.array([a[0].numpy() for a in dataset])
dataset = dataset.astype('float32')
return dataset.reshape(dataset.shape[0], 784)
mnist_test_0_4 = get_mnist_images('/media/tadenoud/DATADisk/datasets/mnist0_4/test')
mnist_test_5_9 = get_mnist_images('/media/tadenoud/DATADisk/datasets/mnist5_9/test')
mnist_split_energy = energy_distance(mnist_test_0_4, mnist_test_5_9)
print("Split MNIST Energy:", mnist_split_energy)
def get_fashion_mnist_test_data(fashion_mnist_dir):
"""
Return flattened and scaled Fashion MNIST test data as a numpy array.
Saves/loads dataset from fashion_mnist_dir.
"""
print("Loading Fashion MNIST")
transform = transforms.Compose([
transforms.ToTensor(),
transforms.Normalize(CHANNEL_MEANS, CHANNEL_STDS)
])
dataset = datasets.FashionMNIST(fashion_mnist_dir, train=False, download=True, transform=transform)
dataset = np.array([a[0].numpy() for a in dataset])
dataset = dataset.astype('float32')
return dataset.reshape(dataset.shape[0], 784)
fashionTestX = get_fashion_mnist_test_data('/media/tadenoud/DATADisk/datasets/fashion_mnist/')
fashion_energy = energy_distance(mnist_test_0_4, fashionTestX)
print("Fashion Energy:", fashion_energy)
def get_emnist_letters_test_data(emnist_letters_dir):
"""
Return flattened and scaled EMNIST Letters test data as a numpy array.
Saves/loads dataset from emnist_letters_dir.
"""
print("Loading EMNIST Letters")
transform = transforms.Compose([
transforms.ToTensor(),
transforms.Normalize(CHANNEL_MEANS, CHANNEL_STDS)
])
dataset = datasets.EMNIST(emnist_letters_dir, "letters", train=False, download=True, transform=transform)
dataset = np.array([a[0].numpy() for a in dataset])
dataset = dataset.astype('float32')
return dataset.reshape(dataset.shape[0], 784)
emnistTestX = get_emnist_letters_test_data('/media/tadenoud/DATADisk/datasets/emnist/')
emnist_energy = energy_distance(mnist_test_0_4, emnistTestX)
print("EMNIST Letters Energy:", emnist_energy)
def get_notmnist_test_data(notmnist_dir):
"""
Return flattened and scaled NotMNIST test data as a numpy array.
Loads dataset from notmnist_dir.
"""
print("Loading NotMNIST")
transform = transforms.Compose([
transforms.Grayscale(),
transforms.ToTensor(),
transforms.Normalize(CHANNEL_MEANS, CHANNEL_STDS)
])
dataset = datasets.ImageFolder(notmnist_dir, transform=transform),
dataset = np.array([a[0].numpy() for a in dataset[0]])
dataset = dataset.astype('float32')
return dataset.reshape(dataset.shape[0], 784)
notmnistTestX = get_notmnist_test_data('/media/tadenoud/DATADisk/datasets/notmnist/')
notmnist_energy = energy_distance(mnist_test_0_4, notmnistTestX)
print("NotMNIST Energy:", notmnist_energy)
def get_mnist_0_4_rot90_test_data(mnist_dir):
"""
Return 90 degree rotated, flattened, and scaled MNIST test data as a numpy array containing only digits 0-4.
Loads dataset from notmnist_dir.
"""
print("Loading MNIST 0-4 rot90")
transform = transforms.Compose([
transforms.Lambda(lambda image: image.rotate(90)),
transforms.ToTensor(),
transforms.Normalize(CHANNEL_MEANS, CHANNEL_STDS)
])
dataset = DatasetSubset(datasets.MNIST(mnist_dir, transform=transform, train=False, download=True),
[0,1,2,3,4], train=False)
dataset = np.array([a[0].numpy() for a in dataset])
dataset = dataset.astype('float32')
return dataset.reshape(dataset.shape[0], 784)
mnistRot90TestX = get_mnist_0_4_rot90_test_data('/media/tadenoud/DATADisk/datasets/mnist/')
mnistrot90_energy = energy_distance(mnist_test_0_4, mnistRot90TestX)
# display_sample(mnistRot90TestX[0])
# display_sample(mnist_test_0_4[0])
print("MNIST rot90 Energy:", mnistrot90_energy)
def get_gaussian_test_data():
"""Return flattened, and scaled Gaussian Noise test data as a numpy array."""
print("Loading Gaussian Noise data")
dataset = GaussianNoiseDataset((10000, 784), mean=0., std=1.)
dataset = np.array([a for a, _ in iter(dataset)])
return dataset.astype('float32')
gaussianTestX = get_gaussian_test_data()
gaussian_energy = energy_distance(mnist_test_0_4, gaussianTestX)
print("Gaussian Energy:", gaussian_energy)
import math
def get_uniform_test_data():
"""Return flattened, and scaled Uniform Noise test data as a numpy array."""
print("Loading Uniform Noise data")
dataset = UniformNoiseDataset((10000, 784), low=-math.sqrt(3), high=math.sqrt(3))
dataset = np.array([a for a, _ in iter(dataset)])
return dataset.astype('float32')
uniformTestX = get_uniform_test_data()
uniform_energy = energy_distance(mnist_test_0_4, uniformTestX)
print("Uniform Energy:", uniform_energy)
```
# MNIST 0-4 results
```
import pandas as pd
df = pd.DataFrame(columns=['energy'],
index=['5-9', 'fashion', 'letters', 'not_mnist', 'rot90', 'gaussian', 'uniform'])
df.loc['5-9'] = pd.Series({'energy': mnist_split_energy})
df.loc['fashion'] = pd.Series({'energy': fashion_energy})
df.loc['letters'] = pd.Series({'energy': emnist_energy})
df.loc['not_mnist'] = pd.Series({'energy': notmnist_energy})
df.loc['rot90'] = pd.Series({'energy': mnistrot90_energy})
df.loc['gaussian'] = pd.Series({'energy': gaussian_energy})
df.loc['uniform'] = pd.Series({'energy': uniform_energy})
df = df.sort_values(by=['energy'])
display(df)
df.to_pickle('../results/mnist5_energy.pkl')
```
|
github_jupyter
|
# Implementation from https://github.com/dougalsutherland/opt-mmd
import sys, os
import numpy as np
from math import sqrt
CHANNEL_MEANS = (33.430001959204674/255,)
CHANNEL_STDS = (78.86655405163765/255,)
from scipy.spatial.distance import pdist, cdist
def energy_distance(v, w):
VV = np.mean(pdist(v, 'euclidean'))
WW = np.mean(pdist(w, 'euclidean'))
VW = np.mean(cdist(v, w, 'euclidean'))
return 2 * VW - VV - WW
from PIL import Image
from matplotlib import pyplot as plt
def display_sample(sample):
img = sample.reshape((28, 28)) * 255.
plt.imshow(Image.fromarray(img))
plt.show()
# Add Bayesian-and-novelty directory to the PYTHONPATH
import sys
import os
sys.path.append(os.path.realpath('../../../..'))
# Autoreload changes in utils, etc.
%load_ext autoreload
%autoreload 2
import torch
from torchvision import datasets, transforms
import numpy as np
from novelty.utils.datasets import GaussianNoiseDataset
from novelty.utils.datasets import UniformNoiseDataset
from novelty.utils import DatasetSubset
torch.manual_seed(1)
CHANNEL_MEANS = (33.550922870635986/255,)
CHANNEL_STDS = (79.10186022520065/255,)
def get_mnist_images(mnist_dir):
transform = transforms.Compose([
transforms.Grayscale(),
transforms.ToTensor(),
transforms.Normalize(CHANNEL_MEANS, CHANNEL_STDS)
])
dataset = datasets.ImageFolder(mnist_dir, transform=transform)
dataset = np.array([a[0].numpy() for a in dataset])
dataset = dataset.astype('float32')
return dataset.reshape(dataset.shape[0], 784)
mnist_test_0_4 = get_mnist_images('/media/tadenoud/DATADisk/datasets/mnist0_4/test')
mnist_test_5_9 = get_mnist_images('/media/tadenoud/DATADisk/datasets/mnist5_9/test')
mnist_split_energy = energy_distance(mnist_test_0_4, mnist_test_5_9)
print("Split MNIST Energy:", mnist_split_energy)
def get_fashion_mnist_test_data(fashion_mnist_dir):
"""
Return flattened and scaled Fashion MNIST test data as a numpy array.
Saves/loads dataset from fashion_mnist_dir.
"""
print("Loading Fashion MNIST")
transform = transforms.Compose([
transforms.ToTensor(),
transforms.Normalize(CHANNEL_MEANS, CHANNEL_STDS)
])
dataset = datasets.FashionMNIST(fashion_mnist_dir, train=False, download=True, transform=transform)
dataset = np.array([a[0].numpy() for a in dataset])
dataset = dataset.astype('float32')
return dataset.reshape(dataset.shape[0], 784)
fashionTestX = get_fashion_mnist_test_data('/media/tadenoud/DATADisk/datasets/fashion_mnist/')
fashion_energy = energy_distance(mnist_test_0_4, fashionTestX)
print("Fashion Energy:", fashion_energy)
def get_emnist_letters_test_data(emnist_letters_dir):
"""
Return flattened and scaled EMNIST Letters test data as a numpy array.
Saves/loads dataset from emnist_letters_dir.
"""
print("Loading EMNIST Letters")
transform = transforms.Compose([
transforms.ToTensor(),
transforms.Normalize(CHANNEL_MEANS, CHANNEL_STDS)
])
dataset = datasets.EMNIST(emnist_letters_dir, "letters", train=False, download=True, transform=transform)
dataset = np.array([a[0].numpy() for a in dataset])
dataset = dataset.astype('float32')
return dataset.reshape(dataset.shape[0], 784)
emnistTestX = get_emnist_letters_test_data('/media/tadenoud/DATADisk/datasets/emnist/')
emnist_energy = energy_distance(mnist_test_0_4, emnistTestX)
print("EMNIST Letters Energy:", emnist_energy)
def get_notmnist_test_data(notmnist_dir):
"""
Return flattened and scaled NotMNIST test data as a numpy array.
Loads dataset from notmnist_dir.
"""
print("Loading NotMNIST")
transform = transforms.Compose([
transforms.Grayscale(),
transforms.ToTensor(),
transforms.Normalize(CHANNEL_MEANS, CHANNEL_STDS)
])
dataset = datasets.ImageFolder(notmnist_dir, transform=transform),
dataset = np.array([a[0].numpy() for a in dataset[0]])
dataset = dataset.astype('float32')
return dataset.reshape(dataset.shape[0], 784)
notmnistTestX = get_notmnist_test_data('/media/tadenoud/DATADisk/datasets/notmnist/')
notmnist_energy = energy_distance(mnist_test_0_4, notmnistTestX)
print("NotMNIST Energy:", notmnist_energy)
def get_mnist_0_4_rot90_test_data(mnist_dir):
"""
Return 90 degree rotated, flattened, and scaled MNIST test data as a numpy array containing only digits 0-4.
Loads dataset from notmnist_dir.
"""
print("Loading MNIST 0-4 rot90")
transform = transforms.Compose([
transforms.Lambda(lambda image: image.rotate(90)),
transforms.ToTensor(),
transforms.Normalize(CHANNEL_MEANS, CHANNEL_STDS)
])
dataset = DatasetSubset(datasets.MNIST(mnist_dir, transform=transform, train=False, download=True),
[0,1,2,3,4], train=False)
dataset = np.array([a[0].numpy() for a in dataset])
dataset = dataset.astype('float32')
return dataset.reshape(dataset.shape[0], 784)
mnistRot90TestX = get_mnist_0_4_rot90_test_data('/media/tadenoud/DATADisk/datasets/mnist/')
mnistrot90_energy = energy_distance(mnist_test_0_4, mnistRot90TestX)
# display_sample(mnistRot90TestX[0])
# display_sample(mnist_test_0_4[0])
print("MNIST rot90 Energy:", mnistrot90_energy)
def get_gaussian_test_data():
"""Return flattened, and scaled Gaussian Noise test data as a numpy array."""
print("Loading Gaussian Noise data")
dataset = GaussianNoiseDataset((10000, 784), mean=0., std=1.)
dataset = np.array([a for a, _ in iter(dataset)])
return dataset.astype('float32')
gaussianTestX = get_gaussian_test_data()
gaussian_energy = energy_distance(mnist_test_0_4, gaussianTestX)
print("Gaussian Energy:", gaussian_energy)
import math
def get_uniform_test_data():
"""Return flattened, and scaled Uniform Noise test data as a numpy array."""
print("Loading Uniform Noise data")
dataset = UniformNoiseDataset((10000, 784), low=-math.sqrt(3), high=math.sqrt(3))
dataset = np.array([a for a, _ in iter(dataset)])
return dataset.astype('float32')
uniformTestX = get_uniform_test_data()
uniform_energy = energy_distance(mnist_test_0_4, uniformTestX)
print("Uniform Energy:", uniform_energy)
import pandas as pd
df = pd.DataFrame(columns=['energy'],
index=['5-9', 'fashion', 'letters', 'not_mnist', 'rot90', 'gaussian', 'uniform'])
df.loc['5-9'] = pd.Series({'energy': mnist_split_energy})
df.loc['fashion'] = pd.Series({'energy': fashion_energy})
df.loc['letters'] = pd.Series({'energy': emnist_energy})
df.loc['not_mnist'] = pd.Series({'energy': notmnist_energy})
df.loc['rot90'] = pd.Series({'energy': mnistrot90_energy})
df.loc['gaussian'] = pd.Series({'energy': gaussian_energy})
df.loc['uniform'] = pd.Series({'energy': uniform_energy})
df = df.sort_values(by=['energy'])
display(df)
df.to_pickle('../results/mnist5_energy.pkl')
| 0.705379 | 0.859192 |
# Dimensions of major clades
## Dependencies
```
from statistics import median
import numpy as np
import pandas as pd
import matplotlib as mpl
import matplotlib.pyplot as plt
import seaborn as sns
from skbio.tree import TreeNode
%matplotlib inline
sns.set_palette('muted')
```
## Helpers
```
def calc_length_metrics(tree):
for node in tree.postorder(include_self=True):
if node.length is None:
node.length = 0.0
if node.is_tip():
node.depths = [0.0]
else:
node.depths = [y + x.length for x in node.children for y in
x.depths]
for node in tree.preorder(include_self=True):
if node.is_root():
node.height = 0.0
node.red = 0.0
else:
node.height = node.parent.height + node.length
if node.is_tip():
node.red = 1.0
else:
node.red = node.parent.red + node.length \
/ (node.length + sum(node.depths) / len(node.depths)) \
* (1 - node.parent.red)
```
## Input files
File paths
```
meta_fp = 'metadata.tsv'
taxa_fp = '../../complex/taxa.txt'
clade_fp = '../../../taxonomy/major/maps/lv1.in.txt'
dir_map = {'cons': 'conserv', 'rand': 'random', 'rpls': 'r-proteins'}
tree_fps = []
tree_fps.append(('all.1k', '../../concat/subsample/all_sites_1k_taxa/all.1k.nid.nwk'))
tree_fps.append(('all.ft', '../../concat/all/fasttree/concat.fast.nid.nwk'))
for x in ('cons', 'rand', 'rpls'):
tree_fps.append(('%s.g' % x, '../../complex/%s.1k.LG+G.nid.nwk' % x))
tree_fps.append(('%s.r' % x, '../../complex/%s.1k.LG+R.nid.nwk' % x))
tree_fps.append(('%s.pmsf' % x, '../../complex/%s.1k.LG+C60_PMSF.nid.nwk' % x))
tree_fps.append(('%s.full' % x, '../../concat/%s/concat.%s.nid.nwk' % (dir_map[x], x)))
tree_fps.append(('%s.astral' % x, '../../astral/newick/astral.%s.nwk' % x))
```
Read files
```
with open(taxa_fp, 'r') as f:
taxa = set(f.read().splitlines())
with open(clade_fp, 'r') as f:
clade_map = dict(x.split('\t') for x in f.read().splitlines())
clade_map = {k: v for k, v in clade_map.items() if k in taxa}
clades = {x: {k for k, v in clade_map.items() if v == x} for x in set(clade_map.values())}
clade_list = sorted(clades.keys())
clade_list
ids = [x[0] for x in tree_fps]
print(ids)
trees = {x[0]: TreeNode.read(x[1]) for x in tree_fps}
for x in ('cons', 'rand', 'rpls'):
for y in ('full', 'astral'):
id_ = '%s.%s' % (x, y)
taxa_ = trees[id_].subset().intersection(taxa)
trees[id_] = trees[id_].shear(taxa_)
trees['all.ft'] = trees['all.ft'].shear(trees['all.ft'].subset().intersection(taxa))
```
Fill empty branch lengths
```
for k, v in trees.items():
for tree in v:
for node in tree.traverse():
if node.length is None:
node.length = 0.0
```
## Branch statistics
Calculate branch length-related metrics
```
for _, tree in trees.items():
calc_length_metrics(tree)
```
Radius of each tree
```
radia = {k: median(v.depths) for k, v in trees.items()}
```
Per-clade statistics
```
data = []
for name, tree in trees.items():
data.append([name, radia[name]])
for clade in clade_list:
taxa_ = tree.subset().intersection(clades[clade])
node = tree.lca(taxa_)
if node.subset() != taxa_:
data[-1].extend([None, None, None])
continue
height = node.length
if clade == 'Archaea':
height *= 2
depth = median(node.depths)
ratio = height / depth
data[-1].extend([height, depth, ratio])
df = pd.DataFrame(data)
df.columns = ['tree', 'radius'] + ['%s_%s' % (x, y) for x in clade_list
for y in ['height', 'depth', 'ratio']]
df.set_index('tree', inplace=True)
df.head()
```
## A-B distances
```
clades['Bacteria'] = clades['Eubacteria'].union(clades['CPR'])
```
Tip-to-tip distances
```
dms = {name: tree.tip_tip_distances() for name, tree in trees.items()}
data = []
for name, tree in trees.items():
dm = dms[name]
all_n = dm.shape[0]
all_sum = dm.condensed_form().sum()
arc_dm = dm.filter(clades['Archaea'], strict=False)
arc_n = len(arc_dm.ids)
arc_m = arc_n * (arc_n - 1) / 2
arc_sum = arc_dm.condensed_form().sum()
arc_mean = arc_sum / arc_m
bac_dm = dm.filter(clades['Bacteria'], strict=False)
bac_n = len(bac_dm.ids)
bac_m = bac_n * (bac_n - 1) / 2
bac_sum = bac_dm.condensed_form().sum()
bac_mean = bac_sum / bac_m
ab_sum = all_sum - arc_sum - bac_sum
ab_mean = ab_sum / (all_n ** 2 - arc_n ** 2 - bac_n ** 2) * 2
ratio = ab_mean ** 2 / arc_mean / bac_mean
data.append([name, arc_n, bac_n, arc_sum, bac_sum, ab_sum,
arc_mean, bac_mean, ab_mean, ratio])
df_ = pd.DataFrame(data)
df_.columns = ['tree', 'Archaea_n', 'Bacteria_n', 'Archaea_sum', 'Bacteria_sum', 'A-B_sum',
'Archaea_mean', 'Bacteria_mean', 'A-B_mean', 'A-B_ratio']
df_.set_index('tree', inplace=True)
df_.head()
df = pd.concat([df, df_], axis=1)
df.to_csv('output.tsv', sep='\t')
```
## Plotting
A-B branch length
```
labels = list(df.index)
xticks = list(range(len(labels)))
fig = plt.figure(figsize=(12, 3))
data = df['Archaea_height'] / df['radius']
plt.bar(xticks, data)
plt.yscale('log')
yticks = [0.1, 0.2, 0.4, 0.6, 0.8, 1.0]
plt.yticks(yticks, yticks)
plt.ylabel('Norm. A-B branch length')
plt.xticks(xticks, labels, rotation=30, ha='right');
fig.tight_layout()
fig.savefig('ab.brlen.pdf', bbox_inches='tight')
```
A-B distance
```
fig = plt.figure(figsize=(12, 3))
data = df['A-B_ratio']
plt.bar(xticks, data)
plt.ylabel('Relative A-B distance')
plt.xticks(xticks, labels, rotation=30, ha='right');
fig.tight_layout()
fig.savefig('ab.dist.pdf', bbox_inches='tight')
```
Depth and height
```
df_ = df[df['Eubacteria_height'].notnull()].copy()
df_ = df_[~df_.index.to_series().str.startswith('rpls')]
df_ = df_[~df_.index.to_series().str.endswith('.r')]
df_ = df_[~df_.index.to_series().str.endswith('.astral')]
for x in ('depth', 'height'):
df_['%s_sum' % x] = sum(df_['%s_%s' % (y, x)] for y in ('Archaea', 'CPR', 'Eubacteria'))
for y in ('Archaea', 'CPR', 'Eubacteria'):
df_['%s_%s_norm' % (y, x)] = df_['%s_%s' % (y, x)] / df_['%s_sum' % x]
labels = list(df_.index)
xticks = range(len(labels))
fig = plt.figure(figsize=(6, 3))
bottom = [0] * df_.shape[0]
for x in ('Archaea', 'CPR', 'Eubacteria'):
plt.bar(xticks, df_['%s_depth_norm' % x], bottom=bottom)
for i in range(len(bottom)):
bottom[i] += df_['%s_depth_norm' % x][i]
plt.xticks(xticks, labels, rotation=30, ha='right')
plt.ylabel('Median depth')
plt.legend(labels=['Archaea', 'CPR', 'Eubacteria']);
fig.tight_layout()
fig.savefig('depth.pdf', bbox_inches='tight')
labels = list(df_.index)
xticks = range(len(labels))
fig = plt.figure(figsize=(6, 3))
bottom = [0] * df_.shape[0]
for x in ('Archaea', 'CPR', 'Eubacteria'):
plt.bar(xticks, df_['%s_height_norm' % x], bottom=bottom)
for i in range(len(bottom)):
bottom[i] += df_['%s_height_norm' % x][i]
plt.ylabel('Height')
plt.xticks(xticks, labels, rotation=30, ha='right');
fig.tight_layout()
fig.savefig('height.pdf', bbox_inches='tight')
```
## Depth distribution
```
data = []
for site in ('cons', 'rand'):
for model in ('g', 'pmsf'):
for clade in ('Archaea', 'CPR', 'Eubacteria'):
name = '%s.%s' % (site, model)
tree = trees[name]
node = tree.lca(tree.subset().intersection(clades[clade]))
for depth in node.depths:
data.append([name, site, model, clade, depth])
dfd = pd.DataFrame(data)
dfd.columns = ['tree', 'site', 'model', 'clade', 'depth']
dfd.head()
for site in ('cons', 'rand'):
fig, ax = plt.subplots(figsize=(3, 5))
sns.violinplot(x='clade', y='depth', hue='model', linewidth=1,
data=dfd.query('site == "%s"' % site), ax=ax)
ax.set_title(site)
ax.set_ylim(bottom=0)
ax.set_xlabel('')
ax.set_ylabel('')
for i, clade in enumerate(('Archaea', 'CPR', 'Eubacteria')):
h = df['%s_height' % clade]['%s.g' % site]
ax.plot([i - 0.35, i - 0.05], [h, h], color=sns.color_palette()[0])
h = df['%s_height' % clade]['%s.pmsf' % site]
ax.plot([i + 0.05, i + 0.35], [h, h], color=sns.color_palette()[1])
fig.tight_layout()
fig.savefig('%s.violin.pdf' % site, bbox_inches='tight')
dfd['radius'] = dfd['tree'].map(radia)
dfd['depth_norm'] = dfd['depth'] / dfd['radius']
for site in ('cons', 'rand'):
fig, ax = plt.subplots(figsize=(3, 5))
sns.violinplot(x='clade', y='depth_norm', hue='model', split=True, linewidth=1,
data=dfd.query('site == "%s"' % site), ax=ax)
ax.set_title(site)
ax.set_ylim(bottom=0)
ax.set_xlabel('')
ax.set_ylabel('')
for i, clade in enumerate(('Archaea', 'CPR', 'Eubacteria')):
h = df['%s_height' % clade]['%s.g' % site] / radia['%s.g' % site]
ax.plot([i - 0.35, i - 0.05], [h, h], color=sns.color_palette()[0])
h = df['%s_height' % clade]['%s.pmsf' % site] / radia['%s.pmsf' % site]
ax.plot([i + 0.05, i + 0.35], [h, h], color=sns.color_palette()[1])
fig.tight_layout()
fig.savefig('%s.norm.violin.pdf' % site, bbox_inches='tight')
```
|
github_jupyter
|
from statistics import median
import numpy as np
import pandas as pd
import matplotlib as mpl
import matplotlib.pyplot as plt
import seaborn as sns
from skbio.tree import TreeNode
%matplotlib inline
sns.set_palette('muted')
def calc_length_metrics(tree):
for node in tree.postorder(include_self=True):
if node.length is None:
node.length = 0.0
if node.is_tip():
node.depths = [0.0]
else:
node.depths = [y + x.length for x in node.children for y in
x.depths]
for node in tree.preorder(include_self=True):
if node.is_root():
node.height = 0.0
node.red = 0.0
else:
node.height = node.parent.height + node.length
if node.is_tip():
node.red = 1.0
else:
node.red = node.parent.red + node.length \
/ (node.length + sum(node.depths) / len(node.depths)) \
* (1 - node.parent.red)
meta_fp = 'metadata.tsv'
taxa_fp = '../../complex/taxa.txt'
clade_fp = '../../../taxonomy/major/maps/lv1.in.txt'
dir_map = {'cons': 'conserv', 'rand': 'random', 'rpls': 'r-proteins'}
tree_fps = []
tree_fps.append(('all.1k', '../../concat/subsample/all_sites_1k_taxa/all.1k.nid.nwk'))
tree_fps.append(('all.ft', '../../concat/all/fasttree/concat.fast.nid.nwk'))
for x in ('cons', 'rand', 'rpls'):
tree_fps.append(('%s.g' % x, '../../complex/%s.1k.LG+G.nid.nwk' % x))
tree_fps.append(('%s.r' % x, '../../complex/%s.1k.LG+R.nid.nwk' % x))
tree_fps.append(('%s.pmsf' % x, '../../complex/%s.1k.LG+C60_PMSF.nid.nwk' % x))
tree_fps.append(('%s.full' % x, '../../concat/%s/concat.%s.nid.nwk' % (dir_map[x], x)))
tree_fps.append(('%s.astral' % x, '../../astral/newick/astral.%s.nwk' % x))
with open(taxa_fp, 'r') as f:
taxa = set(f.read().splitlines())
with open(clade_fp, 'r') as f:
clade_map = dict(x.split('\t') for x in f.read().splitlines())
clade_map = {k: v for k, v in clade_map.items() if k in taxa}
clades = {x: {k for k, v in clade_map.items() if v == x} for x in set(clade_map.values())}
clade_list = sorted(clades.keys())
clade_list
ids = [x[0] for x in tree_fps]
print(ids)
trees = {x[0]: TreeNode.read(x[1]) for x in tree_fps}
for x in ('cons', 'rand', 'rpls'):
for y in ('full', 'astral'):
id_ = '%s.%s' % (x, y)
taxa_ = trees[id_].subset().intersection(taxa)
trees[id_] = trees[id_].shear(taxa_)
trees['all.ft'] = trees['all.ft'].shear(trees['all.ft'].subset().intersection(taxa))
for k, v in trees.items():
for tree in v:
for node in tree.traverse():
if node.length is None:
node.length = 0.0
for _, tree in trees.items():
calc_length_metrics(tree)
radia = {k: median(v.depths) for k, v in trees.items()}
data = []
for name, tree in trees.items():
data.append([name, radia[name]])
for clade in clade_list:
taxa_ = tree.subset().intersection(clades[clade])
node = tree.lca(taxa_)
if node.subset() != taxa_:
data[-1].extend([None, None, None])
continue
height = node.length
if clade == 'Archaea':
height *= 2
depth = median(node.depths)
ratio = height / depth
data[-1].extend([height, depth, ratio])
df = pd.DataFrame(data)
df.columns = ['tree', 'radius'] + ['%s_%s' % (x, y) for x in clade_list
for y in ['height', 'depth', 'ratio']]
df.set_index('tree', inplace=True)
df.head()
clades['Bacteria'] = clades['Eubacteria'].union(clades['CPR'])
dms = {name: tree.tip_tip_distances() for name, tree in trees.items()}
data = []
for name, tree in trees.items():
dm = dms[name]
all_n = dm.shape[0]
all_sum = dm.condensed_form().sum()
arc_dm = dm.filter(clades['Archaea'], strict=False)
arc_n = len(arc_dm.ids)
arc_m = arc_n * (arc_n - 1) / 2
arc_sum = arc_dm.condensed_form().sum()
arc_mean = arc_sum / arc_m
bac_dm = dm.filter(clades['Bacteria'], strict=False)
bac_n = len(bac_dm.ids)
bac_m = bac_n * (bac_n - 1) / 2
bac_sum = bac_dm.condensed_form().sum()
bac_mean = bac_sum / bac_m
ab_sum = all_sum - arc_sum - bac_sum
ab_mean = ab_sum / (all_n ** 2 - arc_n ** 2 - bac_n ** 2) * 2
ratio = ab_mean ** 2 / arc_mean / bac_mean
data.append([name, arc_n, bac_n, arc_sum, bac_sum, ab_sum,
arc_mean, bac_mean, ab_mean, ratio])
df_ = pd.DataFrame(data)
df_.columns = ['tree', 'Archaea_n', 'Bacteria_n', 'Archaea_sum', 'Bacteria_sum', 'A-B_sum',
'Archaea_mean', 'Bacteria_mean', 'A-B_mean', 'A-B_ratio']
df_.set_index('tree', inplace=True)
df_.head()
df = pd.concat([df, df_], axis=1)
df.to_csv('output.tsv', sep='\t')
labels = list(df.index)
xticks = list(range(len(labels)))
fig = plt.figure(figsize=(12, 3))
data = df['Archaea_height'] / df['radius']
plt.bar(xticks, data)
plt.yscale('log')
yticks = [0.1, 0.2, 0.4, 0.6, 0.8, 1.0]
plt.yticks(yticks, yticks)
plt.ylabel('Norm. A-B branch length')
plt.xticks(xticks, labels, rotation=30, ha='right');
fig.tight_layout()
fig.savefig('ab.brlen.pdf', bbox_inches='tight')
fig = plt.figure(figsize=(12, 3))
data = df['A-B_ratio']
plt.bar(xticks, data)
plt.ylabel('Relative A-B distance')
plt.xticks(xticks, labels, rotation=30, ha='right');
fig.tight_layout()
fig.savefig('ab.dist.pdf', bbox_inches='tight')
df_ = df[df['Eubacteria_height'].notnull()].copy()
df_ = df_[~df_.index.to_series().str.startswith('rpls')]
df_ = df_[~df_.index.to_series().str.endswith('.r')]
df_ = df_[~df_.index.to_series().str.endswith('.astral')]
for x in ('depth', 'height'):
df_['%s_sum' % x] = sum(df_['%s_%s' % (y, x)] for y in ('Archaea', 'CPR', 'Eubacteria'))
for y in ('Archaea', 'CPR', 'Eubacteria'):
df_['%s_%s_norm' % (y, x)] = df_['%s_%s' % (y, x)] / df_['%s_sum' % x]
labels = list(df_.index)
xticks = range(len(labels))
fig = plt.figure(figsize=(6, 3))
bottom = [0] * df_.shape[0]
for x in ('Archaea', 'CPR', 'Eubacteria'):
plt.bar(xticks, df_['%s_depth_norm' % x], bottom=bottom)
for i in range(len(bottom)):
bottom[i] += df_['%s_depth_norm' % x][i]
plt.xticks(xticks, labels, rotation=30, ha='right')
plt.ylabel('Median depth')
plt.legend(labels=['Archaea', 'CPR', 'Eubacteria']);
fig.tight_layout()
fig.savefig('depth.pdf', bbox_inches='tight')
labels = list(df_.index)
xticks = range(len(labels))
fig = plt.figure(figsize=(6, 3))
bottom = [0] * df_.shape[0]
for x in ('Archaea', 'CPR', 'Eubacteria'):
plt.bar(xticks, df_['%s_height_norm' % x], bottom=bottom)
for i in range(len(bottom)):
bottom[i] += df_['%s_height_norm' % x][i]
plt.ylabel('Height')
plt.xticks(xticks, labels, rotation=30, ha='right');
fig.tight_layout()
fig.savefig('height.pdf', bbox_inches='tight')
data = []
for site in ('cons', 'rand'):
for model in ('g', 'pmsf'):
for clade in ('Archaea', 'CPR', 'Eubacteria'):
name = '%s.%s' % (site, model)
tree = trees[name]
node = tree.lca(tree.subset().intersection(clades[clade]))
for depth in node.depths:
data.append([name, site, model, clade, depth])
dfd = pd.DataFrame(data)
dfd.columns = ['tree', 'site', 'model', 'clade', 'depth']
dfd.head()
for site in ('cons', 'rand'):
fig, ax = plt.subplots(figsize=(3, 5))
sns.violinplot(x='clade', y='depth', hue='model', linewidth=1,
data=dfd.query('site == "%s"' % site), ax=ax)
ax.set_title(site)
ax.set_ylim(bottom=0)
ax.set_xlabel('')
ax.set_ylabel('')
for i, clade in enumerate(('Archaea', 'CPR', 'Eubacteria')):
h = df['%s_height' % clade]['%s.g' % site]
ax.plot([i - 0.35, i - 0.05], [h, h], color=sns.color_palette()[0])
h = df['%s_height' % clade]['%s.pmsf' % site]
ax.plot([i + 0.05, i + 0.35], [h, h], color=sns.color_palette()[1])
fig.tight_layout()
fig.savefig('%s.violin.pdf' % site, bbox_inches='tight')
dfd['radius'] = dfd['tree'].map(radia)
dfd['depth_norm'] = dfd['depth'] / dfd['radius']
for site in ('cons', 'rand'):
fig, ax = plt.subplots(figsize=(3, 5))
sns.violinplot(x='clade', y='depth_norm', hue='model', split=True, linewidth=1,
data=dfd.query('site == "%s"' % site), ax=ax)
ax.set_title(site)
ax.set_ylim(bottom=0)
ax.set_xlabel('')
ax.set_ylabel('')
for i, clade in enumerate(('Archaea', 'CPR', 'Eubacteria')):
h = df['%s_height' % clade]['%s.g' % site] / radia['%s.g' % site]
ax.plot([i - 0.35, i - 0.05], [h, h], color=sns.color_palette()[0])
h = df['%s_height' % clade]['%s.pmsf' % site] / radia['%s.pmsf' % site]
ax.plot([i + 0.05, i + 0.35], [h, h], color=sns.color_palette()[1])
fig.tight_layout()
fig.savefig('%s.norm.violin.pdf' % site, bbox_inches='tight')
| 0.362066 | 0.722319 |
```
import pandas as pd
import numpy as np
import re
import seaborn as sns
import warnings
warnings.filterwarnings("ignore")
import plotly
import plotly.graph_objs as go
from plotly.offline import download_plotlyjs, init_notebook_mode, plot, iplot
import plotly.io as pio
init_notebook_mode(connected=True)
plotly.__version__
tweets = pd.read_pickle('../datasets/TweetsDataset.pkl')
users = pd.read_pickle('../datasets/UsersDataset.pkl')
print(tweets.columns)
display(users.head(3))
tweets.head()
#tweets_humans = tweets[tweets.Cat=='Humans']
#display(tweets_humans.columns)
#first = True
def getTimes(times):
if(len(times) == 0):
return np.array([0,0,0,0])
return np.array([times.mean().seconds, times.median().seconds, times.std().seconds, times.quantile(0.25).seconds, times.quantile(0.75).seconds])
def getDist(var):
if(len(var) == 0):
return np.array([0,0,0,0])
return np.array([var.mean(), var.median(), var.std(), var.quantile(0.25), var.quantile(0.75)])
def getWeekAv(tw):
return len(tw) / len(tw.weekNum.unique())
def getDataTweets(twU):
userId = twU.user_id.values[0]
twU = twU.sort_values(by='created_at', ascending=0).fillna(0) #tweets del usuario
dates = twU.created_at #aca lo que hago es guardar la columnoa como datetime
twU.loc[:,'dayOfWeek'] = dates.dt.dayofweek # dia de la semana
twU.loc[:,'hour'] = dates.dt.hour #hora del tweet
twU.loc[:,'weekNum'] = dates.apply(lambda x: str(x.week) + '-' + str(x.year))
tweetsUser = twU[twU.is_retweet==False] #tweets propios del usuario
tweetsRT = twU[twU.is_retweet==True] #retweets
# twU.loc[:,'interval'] = pd.to_datetime(twU.timestamp.shift(1)) - pd.to_datetime(twU.timestamp)
twU.loc[:,'interval'] = pd.to_datetime(twU.created_at.shift(1)) - pd.to_datetime(twU.created_at)
times = pd.to_datetime(twU.created_at.shift(1)) - pd.to_datetime(twU.created_at) # calculo distancia entre tweets
tweetsUser.loc[:,'interval'] = pd.to_datetime(tweetsUser.created_at.shift(1)) - pd.to_datetime(tweetsUser.created_at)
timesUser = pd.to_datetime(tweetsUser.created_at.shift(1)) - pd.to_datetime(tweetsUser.created_at) # calculo distancia entre tweets
tweetsRT.loc[:,'interval'] = pd.to_datetime(tweetsRT.created_at.shift(1)) - pd.to_datetime(tweetsRT.created_at)
timesRT = pd.to_datetime(tweetsRT.created_at.shift(1)) - pd.to_datetime(tweetsRT.created_at) # calculo distancia entre tweets
mediaSemanas = np.around(twU.groupby('dayOfWeek').apply(getWeekAv), 6)
#ver word_to_vect
result = np.concatenate([
[
userId,
twU.tweet_id.count(),
len(twU.weekNum.unique())
],
mediaSemanas,
getTimes(times),
getTimes(timesRT),
getTimes(timesUser),
getDist(tweetsUser.favorite_count.astype(int)) * 10000,
getDist(tweetsUser.retweet_count.astype(int)) * 10000,
getDist(tweetsUser.num_mentions.astype(int)) * 10000,
getDist(tweetsUser.reply_count.astype(int)) * 10000,
])
return result
res = tweets.groupby('user_id').apply(getDataTweets)
cols = np.concatenate(
[
[x+'_mean',x+'_median', x+'_std', x+'_fq', x+'_tq'] for x in
['times','timesRT','timesUser','favorite_count','retweet_count','num_mentions','reply_count']
]
)
cols = np.concatenate([
['user_id', 'tweetsCount', 'semanas', 'lun', 'mar', 'mie', 'jue', 'vie', 'sab', 'dom'],cols
])
res = pd.DataFrame(
[x for x in res], columns = cols
)
res.index = res.user_id
print(len(res))
res.to_pickle('../datasets/tweetsProcesados.pkl')
```
|
github_jupyter
|
import pandas as pd
import numpy as np
import re
import seaborn as sns
import warnings
warnings.filterwarnings("ignore")
import plotly
import plotly.graph_objs as go
from plotly.offline import download_plotlyjs, init_notebook_mode, plot, iplot
import plotly.io as pio
init_notebook_mode(connected=True)
plotly.__version__
tweets = pd.read_pickle('../datasets/TweetsDataset.pkl')
users = pd.read_pickle('../datasets/UsersDataset.pkl')
print(tweets.columns)
display(users.head(3))
tweets.head()
#tweets_humans = tweets[tweets.Cat=='Humans']
#display(tweets_humans.columns)
#first = True
def getTimes(times):
if(len(times) == 0):
return np.array([0,0,0,0])
return np.array([times.mean().seconds, times.median().seconds, times.std().seconds, times.quantile(0.25).seconds, times.quantile(0.75).seconds])
def getDist(var):
if(len(var) == 0):
return np.array([0,0,0,0])
return np.array([var.mean(), var.median(), var.std(), var.quantile(0.25), var.quantile(0.75)])
def getWeekAv(tw):
return len(tw) / len(tw.weekNum.unique())
def getDataTweets(twU):
userId = twU.user_id.values[0]
twU = twU.sort_values(by='created_at', ascending=0).fillna(0) #tweets del usuario
dates = twU.created_at #aca lo que hago es guardar la columnoa como datetime
twU.loc[:,'dayOfWeek'] = dates.dt.dayofweek # dia de la semana
twU.loc[:,'hour'] = dates.dt.hour #hora del tweet
twU.loc[:,'weekNum'] = dates.apply(lambda x: str(x.week) + '-' + str(x.year))
tweetsUser = twU[twU.is_retweet==False] #tweets propios del usuario
tweetsRT = twU[twU.is_retweet==True] #retweets
# twU.loc[:,'interval'] = pd.to_datetime(twU.timestamp.shift(1)) - pd.to_datetime(twU.timestamp)
twU.loc[:,'interval'] = pd.to_datetime(twU.created_at.shift(1)) - pd.to_datetime(twU.created_at)
times = pd.to_datetime(twU.created_at.shift(1)) - pd.to_datetime(twU.created_at) # calculo distancia entre tweets
tweetsUser.loc[:,'interval'] = pd.to_datetime(tweetsUser.created_at.shift(1)) - pd.to_datetime(tweetsUser.created_at)
timesUser = pd.to_datetime(tweetsUser.created_at.shift(1)) - pd.to_datetime(tweetsUser.created_at) # calculo distancia entre tweets
tweetsRT.loc[:,'interval'] = pd.to_datetime(tweetsRT.created_at.shift(1)) - pd.to_datetime(tweetsRT.created_at)
timesRT = pd.to_datetime(tweetsRT.created_at.shift(1)) - pd.to_datetime(tweetsRT.created_at) # calculo distancia entre tweets
mediaSemanas = np.around(twU.groupby('dayOfWeek').apply(getWeekAv), 6)
#ver word_to_vect
result = np.concatenate([
[
userId,
twU.tweet_id.count(),
len(twU.weekNum.unique())
],
mediaSemanas,
getTimes(times),
getTimes(timesRT),
getTimes(timesUser),
getDist(tweetsUser.favorite_count.astype(int)) * 10000,
getDist(tweetsUser.retweet_count.astype(int)) * 10000,
getDist(tweetsUser.num_mentions.astype(int)) * 10000,
getDist(tweetsUser.reply_count.astype(int)) * 10000,
])
return result
res = tweets.groupby('user_id').apply(getDataTweets)
cols = np.concatenate(
[
[x+'_mean',x+'_median', x+'_std', x+'_fq', x+'_tq'] for x in
['times','timesRT','timesUser','favorite_count','retweet_count','num_mentions','reply_count']
]
)
cols = np.concatenate([
['user_id', 'tweetsCount', 'semanas', 'lun', 'mar', 'mie', 'jue', 'vie', 'sab', 'dom'],cols
])
res = pd.DataFrame(
[x for x in res], columns = cols
)
res.index = res.user_id
print(len(res))
res.to_pickle('../datasets/tweetsProcesados.pkl')
| 0.195172 | 0.39636 |
<a href="https://colab.research.google.com/github/SamH3pn3r/DS-Unit-1-Sprint-2-Data-Wrangling-and-Storytelling/blob/master/Copy_of_LS_DS_123_Make_Explanatory_Visualizations.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
_Lambda School Data Science_
# Make Explanatory Visualizations
### Objectives
- identify misleading visualizations and how to fix them
- use Seaborn to visualize distributions and relationships with continuous and discrete variables
- add emphasis and annotations to transform visualizations from exploratory to explanatory
- remove clutter from visualizations
### Links
- [How to Spot Visualization Lies](https://flowingdata.com/2017/02/09/how-to-spot-visualization-lies/)
- [Visual Vocabulary - Vega Edition](http://ft.com/vocabulary)
- [Choosing a Python Visualization Tool flowchart](http://pbpython.com/python-vis-flowchart.html)
- [Searborn example gallery](http://seaborn.pydata.org/examples/index.html) & [tutorial](http://seaborn.pydata.org/tutorial.html)
- [Strong Titles Are The Biggest Bang for Your Buck](http://stephanieevergreen.com/strong-titles/)
- [Remove to improve (the data-ink ratio)](https://www.darkhorseanalytics.com/blog/data-looks-better-naked)
- [How to Generate FiveThirtyEight Graphs in Python](https://www.dataquest.io/blog/making-538-plots/)
# Avoid Misleading Visualizations
Did you find/discuss any interesting misleading visualizations in your Walkie Talkie?
## What makes a visualization misleading?
[5 Ways Writers Use Misleading Graphs To Manipulate You](https://venngage.com/blog/misleading-graphs/)
## Two y-axes
<img src="https://kieranhealy.org/files/misc/two-y-by-four-sm.jpg" width="800">
Other Examples:
- [Spurious Correlations](https://tylervigen.com/spurious-correlations)
- <https://blog.datawrapper.de/dualaxis/>
- <https://kieranhealy.org/blog/archives/2016/01/16/two-y-axes/>
- <http://www.storytellingwithdata.com/blog/2016/2/1/be-gone-dual-y-axis>
## Y-axis doesn't start at zero.
<img src="https://i.pinimg.com/originals/22/53/a9/2253a944f54bb61f1983bc076ff33cdd.jpg" width="600">
## Pie Charts are bad
<img src="https://i1.wp.com/flowingdata.com/wp-content/uploads/2009/11/Fox-News-pie-chart.png?fit=620%2C465&ssl=1" width="600">
## Pie charts that omit data are extra bad
- A guy makes a misleading chart that goes viral
What does this chart imply at first glance? You don't want your user to have to do a lot of work in order to be able to interpret you graph correctly. You want that first-glance conclusions to be the correct ones.
<img src="https://pbs.twimg.com/media/DiaiTLHWsAYAEEX?format=jpg&name=medium" width='600'>
<https://twitter.com/michaelbatnick/status/1019680856837849090?lang=en>
- It gets picked up by overworked journalists (assuming incompetency before malice)
<https://www.marketwatch.com/story/this-1-chart-puts-mega-techs-trillions-of-market-value-into-eye-popping-perspective-2018-07-18>
- Even after the chart's implications have been refuted, it's hard a bad (although compelling) visualization from being passed around.
<https://www.linkedin.com/pulse/good-bad-pie-charts-karthik-shashidhar/>
**["yea I understand a pie chart was probably not the best choice to present this data."](https://twitter.com/michaelbatnick/status/1037036440494985216)**
## Pie Charts that compare unrelated things are next-level extra bad
<img src="http://www.painting-with-numbers.com/download/document/186/170403+Legalizing+Marijuana+Graph.jpg" width="600">
## Be careful about how you use volume to represent quantities:
radius vs diameter vs volume
<img src="https://static1.squarespace.com/static/5bfc8dbab40b9d7dd9054f41/t/5c32d86e0ebbe80a25873249/1546836082961/5474039-25383714-thumbnail.jpg?format=1500w" width="600">
## Don't cherrypick timelines or specific subsets of your data:
<img src="https://wattsupwiththat.com/wp-content/uploads/2019/02/Figure-1-1.png" width="600">
Look how specifically the writer has selected what years to show in the legend on the right side.
<https://wattsupwiththat.com/2019/02/24/strong-arctic-sea-ice-growth-this-year/>
Try the tool that was used to make the graphic for yourself
<http://nsidc.org/arcticseaicenews/charctic-interactive-sea-ice-graph/>
## Use Relative units rather than Absolute Units
<img src="https://imgs.xkcd.com/comics/heatmap_2x.png" width="600">
## Avoid 3D graphs unless having the extra dimension is effective
Usually you can Split 3D graphs into multiple 2D graphs
3D graphs that are interactive can be very cool. (See Plotly and Bokeh)
<img src="https://thumbor.forbes.com/thumbor/1280x868/https%3A%2F%2Fblogs-images.forbes.com%2Fthumbnails%2Fblog_1855%2Fpt_1855_811_o.jpg%3Ft%3D1339592470" width="600">
## Don't go against typical conventions
<img src="http://www.callingbullshit.org/twittercards/tools_misleading_axes.png" width="600">
# Tips for choosing an appropriate visualization:
## Use Appropriate "Visual Vocabulary"
[Visual Vocabulary - Vega Edition](http://ft.com/vocabulary)
## What are the properties of your data?
- Is your primary variable of interest continuous or discrete?
- Is in wide or long (tidy) format?
- Does your visualization involve multiple variables?
- How many dimensions do you need to include on your plot?
Can you express the main idea of your visualization in a single sentence?
How hard does your visualization make the user work in order to draw the intended conclusion?
## Which Visualization tool is most appropriate?
[Choosing a Python Visualization Tool flowchart](http://pbpython.com/python-vis-flowchart.html)
# Making Explanatory Visualizations with Seaborn
Today we will reproduce this [example by FiveThirtyEight:](https://fivethirtyeight.com/features/al-gores-new-movie-exposes-the-big-flaw-in-online-movie-ratings/)
```
from IPython.display import display, Image
url = 'https://fivethirtyeight.com/wp-content/uploads/2017/09/mehtahickey-inconvenient-0830-1.png'
example = Image(url=url, width=400)
display(example)
```
Using this data: https://github.com/fivethirtyeight/data/tree/master/inconvenient-sequel
Links
- [Strong Titles Are The Biggest Bang for Your Buck](http://stephanieevergreen.com/strong-titles/)
- [Remove to improve (the data-ink ratio)](https://www.darkhorseanalytics.com/blog/data-looks-better-naked)
- [How to Generate FiveThirtyEight Graphs in Python](https://www.dataquest.io/blog/making-538-plots/)
## Make prototypes
This helps us understand the problem
```
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
plt.style.use('fivethirtyeight')
fake = pd.Series([38, 3, 2, 1, 2, 4, 6, 5, 5, 33],
index=range(1,11))
fake.plot.bar(color='C1', width=0.9);
fake2 = pd.Series(
[1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
2, 2, 2,
3, 3, 3,
4, 4,
5, 5, 5,
6, 6, 6, 6,
7, 7, 7, 7, 7,
8, 8, 8, 8,
9, 9, 9, 9,
10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10])
fake2.value_counts().sort_index().plot.bar(color='C1', width=0.9);
```
## Annotate with text
```
```
## Reproduce with real data
```
df = pd.read_csv('https://raw.githubusercontent.com/fivethirtyeight/data/master/inconvenient-sequel/ratings.csv')
```
# ASSIGNMENT
Replicate the lesson code. I recommend that you [do not copy-paste](https://docs.google.com/document/d/1ubOw9B3Hfip27hF2ZFnW3a3z9xAgrUDRReOEo-FHCVs/edit).
# STRETCH OPTIONS
#### Reproduce another example from [FiveThityEight's shared data repository](https://data.fivethirtyeight.com/).
For example:
- [thanksgiving-2015](https://fivethirtyeight.com/features/heres-what-your-part-of-america-eats-on-thanksgiving/) (try the [`altair`](https://altair-viz.github.io/gallery/index.html#maps) library)
- [candy-power-ranking](https://fivethirtyeight.com/features/the-ultimate-halloween-candy-power-ranking/) (try the [`statsmodels`](https://www.statsmodels.org/stable/index.html) library)
- or another example of your choice!
#### Make more charts!
Choose a chart you want to make, from [Visual Vocabulary - Vega Edition](http://ft.com/vocabulary).
Find the chart in an example gallery of a Python data visualization library:
- [Seaborn](http://seaborn.pydata.org/examples/index.html)
- [Altair](https://altair-viz.github.io/gallery/index.html)
- [Matplotlib](https://matplotlib.org/gallery.html)
- [Pandas](https://pandas.pydata.org/pandas-docs/stable/visualization.html)
Reproduce the chart. [Optionally, try the "Ben Franklin Method."](https://docs.google.com/document/d/1ubOw9B3Hfip27hF2ZFnW3a3z9xAgrUDRReOEo-FHCVs/edit) If you want, experiment and make changes.
Take notes. Consider sharing your work with your cohort!
|
github_jupyter
|
from IPython.display import display, Image
url = 'https://fivethirtyeight.com/wp-content/uploads/2017/09/mehtahickey-inconvenient-0830-1.png'
example = Image(url=url, width=400)
display(example)
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
plt.style.use('fivethirtyeight')
fake = pd.Series([38, 3, 2, 1, 2, 4, 6, 5, 5, 33],
index=range(1,11))
fake.plot.bar(color='C1', width=0.9);
fake2 = pd.Series(
[1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
2, 2, 2,
3, 3, 3,
4, 4,
5, 5, 5,
6, 6, 6, 6,
7, 7, 7, 7, 7,
8, 8, 8, 8,
9, 9, 9, 9,
10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10, 10])
fake2.value_counts().sort_index().plot.bar(color='C1', width=0.9);
```
## Reproduce with real data
| 0.477067 | 0.933491 |
# **Von Mises Distribution Implementation in Python**
> **Abstract:** This article tries to bridge the gap between the quality
> of package available for R and Python for circular statistics and von
> Mises distribution in specific. There is a function available in
> Python to generate random deviates from vonMises distribution. But
> there are no functions available to calculate the probability density,
> cumulative distribution, quantiles, etc.
## 1. Introduction
Directional statistics or circular statistics is a sub-discipline of
statistics that deals with directions, axes and rotation. Think of it as
a regular linear data converted into a circular data by giving it
attributes like rotation, angle, etc. Circular statistics is a lot
different than linear statistics. Firstly, there is no true zero.
Namely, 0 and 360 degrees are equal. So, labeling a value as high or low
is arbitrary. Due to these characteristics, method of analysis of this
kind of data changes completely. The kind of data that has angles, or
periodicity, or does not have a true zero can be labeled as directional
data. Some of the examples include temporal periods (e.g. time of day,
month, hour, week, etc.), compass directions, daily wind directions,
ocean currents, etc.<sup>\[5\]
Calculation of mean, median and variance of a circular data is quite
different from that in linear statistics. If given a data of angles, it
cannot be simply averaged like it is done in linear statistics.
Method for mean calculation:
**Example:** Given angular data $(\alpha_{1},\ \alpha_{2},\ldots.,\alpha_{n})$. Calculate the sine and
cosine of all the angles.
Further, $X = \ \frac{\sum_{i = 1}^{n}{\cos\alpha_{i}}}{N}$ and
$Y = \frac{\sum_{i = 1}^{n}{\sin\alpha_{i}}}{N}$. Also,
$\overset{\overline{}}{r} = \sqrt{X^{2} + Y^{2}}$. So, mean cosine will
be
$\cos\overset{\overline{}}{\alpha} = \frac{X}{r}$ and mean sine will be
$\sin\overset{\overline{}}{\alpha} = \frac{Y}{r}$. Finally, mean angle
will be
$$\theta_{r} = \arctan\left( \frac{\sin\overset{\overline{}}{\alpha}}{\cos\overset{\overline{}}{\alpha}} \right)$$
Decide the resultant quadrant in following way: (Figure given for
reference)
Sin +, cos + : mean angle computed directly
Sin +, cos - : mean angle = $180\ - \ \theta_{r}$
Sin -, cos - : mean angle = $180 + \theta_{r}$
Sin -, cos +: mean angle = $360 - \theta_{r}$
Circular variance measures variation in the angles about the mean
direction.
Variance $V = 1 - \overset{\overline{}}{r}$. So it ranges from 0-1. When
the variance is 1, it means the vectors are concentrated in one
direction. Value of 0 means the vectors are equally dispersed around the
circle.
There is another kind of data known as *bimodal data.* When data have
opposite angles they are said to have diametrically bimodal circular
distributions. The mean angle of diametrically bimodal data is
orthogonal (at right angle) to the true mean. There is a procedure
called *angle doubling* to deal with the diametrically bimodal data. But
this article won’t be discussing on that topic.
There are different types of distributions defined. Generally speaking,
any kind of probability density function can be wrapped around the
circumference of a circle. Von Mises distribution is one of the circular
distributions that are defined in circular statistics and can be
considered as analogous to normal distribution in linear statistics.
Also, it is a close approximation to “wrapped normal” distribution.
Probability density function is given by:
$$f\left( x | \mu,\kappa \right) = \frac{1}{2\pi I_{o}(\kappa)}\exp\left\lbrack \kappa\cos{(x - \mu)} \right\rbrack$$
where, $I_{o}(\kappa)$ – Modified Bessel function of zeroth order.
$\mu$ – measure of location (similar to mean in Normal distribution)
$\kappa$ – measure of concentration ( $1/\kappa$ is analogous to
$\sigma^{2}$ )
$I_{o}(x)$ is defined as:
$$I_{o}(x) = \sum_{r = 0}^{\infty}\frac{1}{r!r!}\left( \frac{x}{2} \right)^{2r}$$
$$\ \ \ \ \ \ \ \ \ \ \ \ \ \ \ \ = \frac{1}{2\pi}\int_{0}^{2\pi}e^{x\cos\theta}\text{dθ}$$
The above equation is the zeroth order modified Bessel function.
When $\kappa$ = 0, Von Mises distribution reduces to the uniform
distribution. As $\kappa$ increases, the von Mises distribution
approaches normal distribution.
| | |
|----------------------------------|-----------------------------------|
| | |
<div align="center">
<b><i>Figure 1:</i></b> <b>Left</b> panel depicts the PDF and <b>right</b> panel depicts the CDF of the von
Mises distribution.
</div>
In the right panel, as $\kappa$ increases, the S curve will gradually
become a straight line. That makes sense in a way, that as the
concentration increases, the probability is more cumulated near the
mean, i.e. zero in our case.
## 2. Methods
We have used the vonMises function in the circular package of R as a
reference for generating the algorithms for each method.
**rvonmises(n, mu, kappa)**
Description – A method for generating random numbers for a von Mises
circular distribution.
Arguments:
n – number of observations
**dvonmises(x, mu, kappa) **
Description – A method for calculating the probability density at the
given points for a von Mises circular distribution.
Arguments:
x – A vector containing the points at which the density is to be
calculated. The object is from class ‘circular’
log – logical; if True, probabilities p is given as log(p). The default
value for log is given as False.
**pvonmises(q, mu, kappa)**
Description – Method used to calculate the cumulative distribution at
the given points for a von Mises distribution.
Arguments:
q – A vector containing the points at which the distribution is to be
calculated. The object is from class ‘circular’
tol – the precision in evaluating the distribution function. Default
value = 1e-20
**qvonmises(p, mu, kappa)**
Description – A method used to calculate the quantiles for the given
probabilities for a von Mises distribution.
Arguments:
p – A vector containing the probabilities at points at which the
quantiles are to be calculated. The object is from class ‘circular’
from\_ - a value used for evaluating pvonmises and qvonmises. Default =
None
tol – machine epsilon value raised to 0.6
Common arguments for all the methods:
mu – The mean direction of the distribution. This object is from class
‘circular’
kappa – non-negative value for the concentration of the distribution
## 3. Results and Discussion
We run the functions pvonmises, qvonmises, dvonmises with various values
of parameters mu and kappa. Below shown is the table that shows the
comparison of the values obtained in R and values obtained by the
package we built in Python.
|Method | R | Python |
|-------------------------------|-------------------------------|-------------------------------|
|pvonmises(2, 1, 6) |\[0.9888944\] | \[0.988894\] |
|pvonmises(\[2, 0.8\], 2, 6) |\[0.5 , 0.003595458\] | \[0.5 , 0.00359546\] |
|dvonmises(0.5, 1, 6) |\[0.4581463\] | \[0.45814625\] |
|dvonmises(\[1, 3\], 3, 6) |\[1.949157e-04, 9.54982e-01\] | \[1.949157e-04, 9.54982e-01\] |
|qvonmises(0.5, 1, 6) |\[1\] | \[1\] |
|qvonmises(\[0.2, 0.6\], 2, 7) |\[1.67413597, 2.09767203\] | \[1.67413597, 2.09767203\] |
Now, we will plot some graphs to demonstrate how precise our values are
when compared to those in R
When we run the function rvonmises(n=1000, mu=1, kappa=1), it generates
following output in R and Python respectively.
| | |
|--------------------------------------|--------------------------------|
| ||
<div align="center">
<b><i>Figure 2:</i></b> rvonmises in Python (left panel) and R (right panel)
</div>
When we run the function dvonmises(x = np.linspace(-pi, pi, 1000), mu=1,
kappa=6), it generates following output in R and Python respectively.
| | |
|-------------------------------------|-----------------------------------|
|||
<div align="center">
<b><i>Figure 3:</i></b> dvonmises in R (left panel) and Python (right panel)
</div>
## 4. Future Scope
We need to make the package more robust so that the function can accept
different kind of inputs. When we ran the benchmarking tests, we saw
that our code took longer time to execute as compared to that in R. So
we need to optimize the code in order to decrease the execution time. We
can include other functions from the ‘circular’ package of R into
Python.
## 5. Reference
\[1\] <https://www.researchgate.net/figure/Wind-data-for-KRDM-the-nearest-FAA-weather-reporting-station-at-the-Redomond-OR_fig5_261417337>
\[2\] <https://ncss-wpengine.netdna-ssl.com/wp-content/uploads/2013/01/Rose-Plot.png>
\[3\] <http://webspace.ship.edu/pgmarr/geo441/lectures/lec%2016%20-%20directional%20statistics.pdf>
\[4\] <https://ncss-wpengine.netdna-ssl.com/wp-content/themes/ncss/pdf/Procedures/NCSS/Circular_Data_Analysis.pdf>
\[5\] <https://en.wikipedia.org/wiki/Von_Mises_distribution>
\[6\] <https://packaging.python.org/tutorials/distributing-packages/#your-package>
\[7\] <https://r-forge.r-project.org/scm/viewvc.php/pkg/R/vonmises.R?view=markup&root=circular>
\[8\] <https://cran.r-project.org/web/packages/circular/circular.pdf>
|
github_jupyter
|
# **Von Mises Distribution Implementation in Python**
> **Abstract:** This article tries to bridge the gap between the quality
> of package available for R and Python for circular statistics and von
> Mises distribution in specific. There is a function available in
> Python to generate random deviates from vonMises distribution. But
> there are no functions available to calculate the probability density,
> cumulative distribution, quantiles, etc.
## 1. Introduction
Directional statistics or circular statistics is a sub-discipline of
statistics that deals with directions, axes and rotation. Think of it as
a regular linear data converted into a circular data by giving it
attributes like rotation, angle, etc. Circular statistics is a lot
different than linear statistics. Firstly, there is no true zero.
Namely, 0 and 360 degrees are equal. So, labeling a value as high or low
is arbitrary. Due to these characteristics, method of analysis of this
kind of data changes completely. The kind of data that has angles, or
periodicity, or does not have a true zero can be labeled as directional
data. Some of the examples include temporal periods (e.g. time of day,
month, hour, week, etc.), compass directions, daily wind directions,
ocean currents, etc.<sup>\[5\]
Calculation of mean, median and variance of a circular data is quite
different from that in linear statistics. If given a data of angles, it
cannot be simply averaged like it is done in linear statistics.
Method for mean calculation:
**Example:** Given angular data $(\alpha_{1},\ \alpha_{2},\ldots.,\alpha_{n})$. Calculate the sine and
cosine of all the angles.
Further, $X = \ \frac{\sum_{i = 1}^{n}{\cos\alpha_{i}}}{N}$ and
$Y = \frac{\sum_{i = 1}^{n}{\sin\alpha_{i}}}{N}$. Also,
$\overset{\overline{}}{r} = \sqrt{X^{2} + Y^{2}}$. So, mean cosine will
be
$\cos\overset{\overline{}}{\alpha} = \frac{X}{r}$ and mean sine will be
$\sin\overset{\overline{}}{\alpha} = \frac{Y}{r}$. Finally, mean angle
will be
$$\theta_{r} = \arctan\left( \frac{\sin\overset{\overline{}}{\alpha}}{\cos\overset{\overline{}}{\alpha}} \right)$$
Decide the resultant quadrant in following way: (Figure given for
reference)
Sin +, cos + : mean angle computed directly
Sin +, cos - : mean angle = $180\ - \ \theta_{r}$
Sin -, cos - : mean angle = $180 + \theta_{r}$
Sin -, cos +: mean angle = $360 - \theta_{r}$
Circular variance measures variation in the angles about the mean
direction.
Variance $V = 1 - \overset{\overline{}}{r}$. So it ranges from 0-1. When
the variance is 1, it means the vectors are concentrated in one
direction. Value of 0 means the vectors are equally dispersed around the
circle.
There is another kind of data known as *bimodal data.* When data have
opposite angles they are said to have diametrically bimodal circular
distributions. The mean angle of diametrically bimodal data is
orthogonal (at right angle) to the true mean. There is a procedure
called *angle doubling* to deal with the diametrically bimodal data. But
this article won’t be discussing on that topic.
There are different types of distributions defined. Generally speaking,
any kind of probability density function can be wrapped around the
circumference of a circle. Von Mises distribution is one of the circular
distributions that are defined in circular statistics and can be
considered as analogous to normal distribution in linear statistics.
Also, it is a close approximation to “wrapped normal” distribution.
Probability density function is given by:
$$f\left( x | \mu,\kappa \right) = \frac{1}{2\pi I_{o}(\kappa)}\exp\left\lbrack \kappa\cos{(x - \mu)} \right\rbrack$$
where, $I_{o}(\kappa)$ – Modified Bessel function of zeroth order.
$\mu$ – measure of location (similar to mean in Normal distribution)
$\kappa$ – measure of concentration ( $1/\kappa$ is analogous to
$\sigma^{2}$ )
$I_{o}(x)$ is defined as:
$$I_{o}(x) = \sum_{r = 0}^{\infty}\frac{1}{r!r!}\left( \frac{x}{2} \right)^{2r}$$
$$\ \ \ \ \ \ \ \ \ \ \ \ \ \ \ \ = \frac{1}{2\pi}\int_{0}^{2\pi}e^{x\cos\theta}\text{dθ}$$
The above equation is the zeroth order modified Bessel function.
When $\kappa$ = 0, Von Mises distribution reduces to the uniform
distribution. As $\kappa$ increases, the von Mises distribution
approaches normal distribution.
| | |
|----------------------------------|-----------------------------------|
| | |
<div align="center">
<b><i>Figure 1:</i></b> <b>Left</b> panel depicts the PDF and <b>right</b> panel depicts the CDF of the von
Mises distribution.
</div>
In the right panel, as $\kappa$ increases, the S curve will gradually
become a straight line. That makes sense in a way, that as the
concentration increases, the probability is more cumulated near the
mean, i.e. zero in our case.
## 2. Methods
We have used the vonMises function in the circular package of R as a
reference for generating the algorithms for each method.
**rvonmises(n, mu, kappa)**
Description – A method for generating random numbers for a von Mises
circular distribution.
Arguments:
n – number of observations
**dvonmises(x, mu, kappa) **
Description – A method for calculating the probability density at the
given points for a von Mises circular distribution.
Arguments:
x – A vector containing the points at which the density is to be
calculated. The object is from class ‘circular’
log – logical; if True, probabilities p is given as log(p). The default
value for log is given as False.
**pvonmises(q, mu, kappa)**
Description – Method used to calculate the cumulative distribution at
the given points for a von Mises distribution.
Arguments:
q – A vector containing the points at which the distribution is to be
calculated. The object is from class ‘circular’
tol – the precision in evaluating the distribution function. Default
value = 1e-20
**qvonmises(p, mu, kappa)**
Description – A method used to calculate the quantiles for the given
probabilities for a von Mises distribution.
Arguments:
p – A vector containing the probabilities at points at which the
quantiles are to be calculated. The object is from class ‘circular’
from\_ - a value used for evaluating pvonmises and qvonmises. Default =
None
tol – machine epsilon value raised to 0.6
Common arguments for all the methods:
mu – The mean direction of the distribution. This object is from class
‘circular’
kappa – non-negative value for the concentration of the distribution
## 3. Results and Discussion
We run the functions pvonmises, qvonmises, dvonmises with various values
of parameters mu and kappa. Below shown is the table that shows the
comparison of the values obtained in R and values obtained by the
package we built in Python.
|Method | R | Python |
|-------------------------------|-------------------------------|-------------------------------|
|pvonmises(2, 1, 6) |\[0.9888944\] | \[0.988894\] |
|pvonmises(\[2, 0.8\], 2, 6) |\[0.5 , 0.003595458\] | \[0.5 , 0.00359546\] |
|dvonmises(0.5, 1, 6) |\[0.4581463\] | \[0.45814625\] |
|dvonmises(\[1, 3\], 3, 6) |\[1.949157e-04, 9.54982e-01\] | \[1.949157e-04, 9.54982e-01\] |
|qvonmises(0.5, 1, 6) |\[1\] | \[1\] |
|qvonmises(\[0.2, 0.6\], 2, 7) |\[1.67413597, 2.09767203\] | \[1.67413597, 2.09767203\] |
Now, we will plot some graphs to demonstrate how precise our values are
when compared to those in R
When we run the function rvonmises(n=1000, mu=1, kappa=1), it generates
following output in R and Python respectively.
| | |
|--------------------------------------|--------------------------------|
| ||
<div align="center">
<b><i>Figure 2:</i></b> rvonmises in Python (left panel) and R (right panel)
</div>
When we run the function dvonmises(x = np.linspace(-pi, pi, 1000), mu=1,
kappa=6), it generates following output in R and Python respectively.
| | |
|-------------------------------------|-----------------------------------|
|||
<div align="center">
<b><i>Figure 3:</i></b> dvonmises in R (left panel) and Python (right panel)
</div>
## 4. Future Scope
We need to make the package more robust so that the function can accept
different kind of inputs. When we ran the benchmarking tests, we saw
that our code took longer time to execute as compared to that in R. So
we need to optimize the code in order to decrease the execution time. We
can include other functions from the ‘circular’ package of R into
Python.
## 5. Reference
\[1\] <https://www.researchgate.net/figure/Wind-data-for-KRDM-the-nearest-FAA-weather-reporting-station-at-the-Redomond-OR_fig5_261417337>
\[2\] <https://ncss-wpengine.netdna-ssl.com/wp-content/uploads/2013/01/Rose-Plot.png>
\[3\] <http://webspace.ship.edu/pgmarr/geo441/lectures/lec%2016%20-%20directional%20statistics.pdf>
\[4\] <https://ncss-wpengine.netdna-ssl.com/wp-content/themes/ncss/pdf/Procedures/NCSS/Circular_Data_Analysis.pdf>
\[5\] <https://en.wikipedia.org/wiki/Von_Mises_distribution>
\[6\] <https://packaging.python.org/tutorials/distributing-packages/#your-package>
\[7\] <https://r-forge.r-project.org/scm/viewvc.php/pkg/R/vonmises.R?view=markup&root=circular>
\[8\] <https://cran.r-project.org/web/packages/circular/circular.pdf>
| 0.955236 | 0.991195 |
# Data Exploration
Visual data exploration is often useful to have an initial understanding of how values are distributed.
This notebook covers 4 basic types of plots:
- line plot
- scatter plot
- histogram
- boxplot
and a few other advanced plots:
- pie chart
- hexbin plot
- candlestick plot
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
data = np.random.normal(0, 0.01, 1000)
```
## Matplotlib plotting
```
plt.plot(data, 'o')
fig = plt.figure(figsize=(16,12))
ax = fig.add_subplot(2,2,1)
ax.plot(data)
ax.set_title('Line plot', size=24)
ax = fig.add_subplot(2,2,2)
ax.plot(data, 'o')
ax.set_title('Scatter plot', size=24)
ax = fig.add_subplot(2,2,3)
ax.hist(data, bins=50)
ax.set_title('Histogram', size=24)
ax.set_xlabel('count', size=16)
ax = fig.add_subplot(2,2,4)
ax.boxplot(data)
ax.set_title('Boxplot', size=24)
```
## Pandas plotting
```
dataseries = pd.Series(data)
fig, ax = plt.subplots(2, 2, figsize=(16,12))
dataseries.plot(ax=ax[0][0],
title='Line plot')
dataseries.plot(ax=ax[0][1],
style='o',
title='Scatter plot')
dataseries.plot(ax=ax[1][0],
kind='hist',
bins=50,
title='Histogram'
)
dataseries.plot(ax=ax[1][1],
kind='box',
title='Boxplot'
)
```
## Advanced plots
### Pie chart
```
categories = dataseries > 0.01
categories.head()
counts = categories.value_counts()
counts
counts.plot(kind='pie',
figsize=(5, 5),
explode=[0, 0.15],
labels=['<= 0.01', '> 0.01'],
autopct='%1.1f%%',
shadow=True,
startangle=90,
fontsize=16)
```
## Hexbin plot
Hexbin plots are useful to inspect 2D distriibutions
```
data = np.vstack([np.random.normal((0, 0), 2, size=(2000, 2)),
np.random.normal((9, 9), 3, size=(2000, 2))
])
plt.hexbin(data[:,0], data[:,1])
pd.DataFrame(data).plot(kind='hexbin', x=0, y=1)
```
## Interactive notebook plotting
Jupyter offers interactive plotting through the magic command `%matplotlib notebook`.
If you see nothing just run the next cell again.
```
%matplotlib notebook
fig = plt.plot(data[:,0])
%matplotlib inline
```
## Exercises:
### Exercise 1
- load the dataset: ../data/international-airline-passengers.csv
- inspect it using the .info() and .head() commands
- use the function pd.to_datetime() to change the column type of 'Month' to a datatime type
- set the index of df to be a datetime index using the column 'Month' and the df.set_index() method
- choose the appropriate plot and display the data
- choose appropriate scale
- label the axes
- discuss with your neighbor
### Exercise 2
- load the dataset: ../data/weight-height.csv
- inspect it
- plot it using a scatter plot with Weight as a function of Height
- plot the male and female populations with 2 different colors on a new scatter plot
- remember to label the axes
- discuss
### Exercise 3
- plot the histogram of the heights for males and for females on the same plot
- use alpha to control transparency in the plot comand
- plot a vertical line at the mean of each population using plt.axvline()
*Copyright © 2017 Francesco Mosconi & CATALIT LLC.*
|
github_jupyter
|
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
data = np.random.normal(0, 0.01, 1000)
plt.plot(data, 'o')
fig = plt.figure(figsize=(16,12))
ax = fig.add_subplot(2,2,1)
ax.plot(data)
ax.set_title('Line plot', size=24)
ax = fig.add_subplot(2,2,2)
ax.plot(data, 'o')
ax.set_title('Scatter plot', size=24)
ax = fig.add_subplot(2,2,3)
ax.hist(data, bins=50)
ax.set_title('Histogram', size=24)
ax.set_xlabel('count', size=16)
ax = fig.add_subplot(2,2,4)
ax.boxplot(data)
ax.set_title('Boxplot', size=24)
dataseries = pd.Series(data)
fig, ax = plt.subplots(2, 2, figsize=(16,12))
dataseries.plot(ax=ax[0][0],
title='Line plot')
dataseries.plot(ax=ax[0][1],
style='o',
title='Scatter plot')
dataseries.plot(ax=ax[1][0],
kind='hist',
bins=50,
title='Histogram'
)
dataseries.plot(ax=ax[1][1],
kind='box',
title='Boxplot'
)
categories = dataseries > 0.01
categories.head()
counts = categories.value_counts()
counts
counts.plot(kind='pie',
figsize=(5, 5),
explode=[0, 0.15],
labels=['<= 0.01', '> 0.01'],
autopct='%1.1f%%',
shadow=True,
startangle=90,
fontsize=16)
data = np.vstack([np.random.normal((0, 0), 2, size=(2000, 2)),
np.random.normal((9, 9), 3, size=(2000, 2))
])
plt.hexbin(data[:,0], data[:,1])
pd.DataFrame(data).plot(kind='hexbin', x=0, y=1)
%matplotlib notebook
fig = plt.plot(data[:,0])
%matplotlib inline
| 0.425128 | 0.980262 |
<h3 align="center"><font size="15"><b>Fake News detection</b></font></h3>
<img src="https://www.txstate.edu/cache78a0c25d34508c9d84822109499dee61/imagehandler/scaler/gato-docs.its.txstate.edu/jcr:21b3e33f-31c9-4273-aeb0-5b5886f8bcc4/fake-fact.jpg?mode=fit&width=1600" height=200 width=400>
<br></br>
**Task type:** Classification
**Models used:** LinearSVC, MultinomialNB, XGBoost, PyCaret, CatBoost
**Tools used:** NLP preprocessing tools, semi-supervised learning technique, new feature engineering, Word Cloud
```
# This Python 3 environment comes with many helpful analytics libraries installed
# It is defined by the kaggle/python Docker image: https://github.com/kaggle/docker-python
# For example, here's several helpful packages to load
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk('/kaggle/input'):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
```
# 1. Loading data
```
fake = pd.read_csv('../input/fake-and-real-news-dataset/Fake.csv')
fake['flag'] = 0
fake
true = pd.read_csv('../input/fake-and-real-news-dataset/True.csv')
true['flag'] = 1
true
df = pd.DataFrame()
df = true.append(fake)
```
# 2. EDA + Data cleaning
**Let's check the datatypes.**
```
df.info()
```
**Removing the duplicates and preventing problems with indexing.**
```
df = df.drop_duplicates()
df = df.reset_index(drop=True)
```
**We can see that the date format is not the one we need. I will apply the appropriate date format for future purposes.**
```
# Correcting some data
df['date'] = df['date'].replace(['19-Feb-18'],'February 19, 2018')
df['date'] = df['date'].replace(['18-Feb-18'],'February 18, 2018')
df['date'] = df['date'].replace(['17-Feb-18'],'February 17, 2018')
df['date'] = df['date'].replace(['16-Feb-18'],'February 16, 2018')
df['date'] = df['date'].replace(['15-Feb-18'],'February 15, 2018')
df['date'] = df['date'].replace(['14-Feb-18'],'February 14, 2018')
df['date'] = df['date'].replace(['13-Feb-18'],'February 13, 2018')
df['date'] = df['date'].str.replace('Dec ', 'December ')
df['date'] = df['date'].str.replace('Nov ', 'November ')
df['date'] = df['date'].str.replace('Oct ', 'October ')
df['date'] = df['date'].str.replace('Sep ', 'September ')
df['date'] = df['date'].str.replace('Aug ', 'August ')
df['date'] = df['date'].str.replace('Jul ', 'July ')
df['date'] = df['date'].str.replace('Jun ', 'June ')
df['date'] = df['date'].str.replace('Apr ', 'April ')
df['date'] = df['date'].str.replace('Mar ', 'March ')
df['date'] = df['date'].str.replace('Feb ', 'February ')
df['date'] = df['date'].str.replace('Jan ', 'January ')
df['date'] = df['date'].str.replace(' ', '')
for i, val in enumerate(df['date']):
df['date'].iloc[i] = pd.to_datetime(df['date'].iloc[i], format='%B%d,%Y', errors='coerce') # by setting the parameter to "coerce", we will set unappropriate values to NaT (null)
df['date'] = df['date'].astype('datetime64[ns]')
df.info()
import datetime as dt
df['year'] = pd.to_datetime(df['date']).dt.to_period('Y')
df['month'] = pd.to_datetime(df['date']).dt.to_period('M')
df['month'] = df['month'].astype(str)
```
## Non-text feature plotting (date, subject)
**Here we will try to elicit insights from non-text features to get to know if they will help us boost the Text Classifier.**
```
sub = df[['month', 'flag']]
sub = sub.dropna()
sub = sub.groupby(['month'])['flag'].sum()
sub = sub.drop('NaT')
import matplotlib.pyplot as plt
plt.suptitle('Dynamics of fake news')
plt.xticks(rotation=90)
plt.ylabel('Number of fake news')
plt.xlabel('Month-Year')
plt.plot(sub.index, sub.values, linewidth=2, color='green')
```
**What a spike in the dynamics of fake news in late 2017!**
```
sub2 = df[['subject', 'flag']]
sub2 = sub2.dropna()
sub2 = sub2.groupby(['subject'])['flag'].sum()
plt.suptitle('Fake news among different categories')
plt.xticks(rotation=90)
plt.ylabel('Number of fake news')
plt.xlabel('Category')
plt.bar(sub2.index, height=sub2.values, color='green')
#ax1.plot(x, y)
#ax2.plot(x, -y)
```
**As we have discovered, such features as**
* subject
* date
**might be also crucial for the algorithm to decide whether the piece of news is fake or real. We will try to include them in the model.**
# 3. Text preparation
```
nlp = df
```
**I will add the 'subject' feature to the title field as it might have an influence on the outcome of classification.**
```
#nlp['title'] = nlp['title'] + ' ' + nlp['subject']
```
## 3.1 Word Cloud visualization
**Here I am going to take one example and try visualize tfidf as a wordcloud.**
```
from sklearn.feature_extraction.text import TfidfVectorizer
corpus = nlp[nlp['flag'] == 1]['title'].iloc[0:500] # We will take a slice of fake news, to see what vocabulary there looks like
tfidf1 = TfidfVectorizer()
vecs = tfidf1.fit_transform(corpus)
feature_names = tfidf1.get_feature_names()
dense = vecs.todense()
list_words = dense.tolist()
df_words = pd.DataFrame(list_words, columns=feature_names)
from wordcloud import WordCloud, STOPWORDS, ImageColorGenerator
df_words.T.sum(axis=1)
Cloud = WordCloud(background_color="white", max_words=100).generate_from_frequencies(df_words.T.sum(axis=1))
import matplotlib.pyplot as plt
plt.figure(figsize=(12,5))
plt.imshow(Cloud, interpolation='bilinear')
```
**Indeed, looks definitely like fake news :)**
**And we can also see out 'subject' feature in the foreground as it has been added manually in every title. Therefore, out vectorizer considers it as an important & frequent word.**
## 3.2 Tfidf-vectorizing
**First, I will tokenize words to pass it on to the SnowballStemmer method, which will take out lemmas from words.**
```
import nltk
nltk.download('punkt')
from nltk import word_tokenize
nlp['title'] = nlp['title'].apply(lambda x: word_tokenize(str(x)))
```
**An important step in every NLP-task is to get the roots of words in order not to distract the model by 'different' words.**
```
from nltk.stem import SnowballStemmer
snowball = SnowballStemmer(language='english')
nlp['title'] = nlp['title'].apply(lambda x: [snowball.stem(y) for y in x])
nlp['title'] = nlp['title'].apply(lambda x: ' '.join(x))
```
**Take the standard english bag of stopwords from nltk.**
```
from nltk.corpus import stopwords
nltk.download('words')
nltk.download('stopwords')
stopwords = stopwords.words('english')
```
**And finally TfidfVectorizing. You can also take CountVectorizer, but I prefer Tfidf as it has masses of advantages.**
```
from sklearn.feature_extraction.text import TfidfVectorizer
tfidf = TfidfVectorizer()
X_text = tfidf.fit_transform(nlp['title'])
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X_text, nlp['flag'], test_size=0.33, random_state=1)
```
# 4. Model building
**I will use several approaches to solve the classification task, such as:**
1) Traditional (which are known as efficient for text classification):
1.1) SVM
1.2) Naive Bayes
1.3) XGBoost
2) Not-very-traditional (Experimental): PyCaret NLP toolkit (I will apply unsupervised model to generate features which I will in turn pass on to the supervised model)
## 4.1 Linear SVC
```
scores = {}
from sklearn.svm import LinearSVC
from sklearn.model_selection import cross_val_score
from sklearn.metrics import accuracy_score
clf = LinearSVC(max_iter=100, C=1.0)
clf.fit(X_train, y_train)
y_pred_SVM = clf.predict(X_test)
print(cross_val_score(clf, X_text, nlp['flag'], cv=3))
print(accuracy_score(y_pred_SVM, y_test))
scores['LinearSVC'] = accuracy_score(y_pred_SVM, y_test)
```
**This looks suspiciously good, but lets try another algorithm.**
## 4.2 Naive Bayes
```
from sklearn.naive_bayes import MultinomialNB
clf2 = MultinomialNB()
clf2.fit(X_train, y_train)
y_pred_MNB = clf2.predict(X_test)
print(cross_val_score(clf2, X_text, nlp['flag'], cv=3))
print(accuracy_score(y_pred_MNB, y_test))
scores['MultinomialNB'] = accuracy_score(y_pred_MNB, y_test)
```
**Okay, this model performs a little worse, but still very good.**
## 4.3 XGBoost
```
from xgboost import XGBClassifier
clf3 = XGBClassifier(eval_metric='rmse', use_label_encoder=False)
clf3.fit(X_train, y_train)
y_pred_XGB = clf3.predict(X_test)
print(cross_val_score(clf3, X_text, nlp['flag'], cv=3))
print(accuracy_score(y_pred_XGB, y_test))
scores['XGB'] = accuracy_score(y_pred_XGB, y_test)
```
## 4.4 PyCaret + CatBoost
**PyCaret’s Natural Language Processing module is an unsupervised machine learning module that can be used for analyzing text data by creating topic models that can find hidden semantic structures within documents. PyCaret’s NLP module comes with a wide range of text pre-processing techniques. It has over 5 ready-to-use algorithms and several plots to analyze the performance of trained models and text corpus.**
*Read more:* https://pycaret.org/nlp/
```
!pip install pycaret
```
**Setting up the model which will implement all traditional NLP-preprocessing operation (tokenizing, lemmatizing etc.**
**The PyCaret is almost fully automatic!**
```
from pycaret.nlp import *
caret_nlp = setup(data=nlp, target='title', session_id=1)
```
**LDA stands for Latent Dirichlet Allocation and is widely used in unsupervised learning tasks.**
*Read more:* https://en.wikipedia.org/wiki/Latent_Dirichlet_allocation
```
lda = create_model('lda')
lda_data = assign_model(lda)
```
**Here's the outcome dataset:**
```
lda_data
```
**We'll utilize the 'Topic' features generated by PyCaret.**
```
from catboost import CatBoostClassifier
input_cat = lda_data.drop(['text','date','Perc_Dominant_Topic','flag','year'], axis=1)
input_cat['month'] = input_cat['month'].astype(str)
target_cat = lda_data['flag']
from sklearn.model_selection import train_test_split
X_train_cat, X_test_cat, y_train_cat, y_test_cat = train_test_split(input_cat, target_cat, test_size=0.33, random_state=1)
clf4 = CatBoostClassifier(iterations=1000,
cat_features=['title','subject','Dominant_Topic','month']
)
clf4.fit(X_train_cat, y_train_cat, early_stopping_rounds=10)
scores['CatBoost'] = clf4.score(X_test_cat, y_test_cat)
scores
plt.bar(scores.keys(), scores.values())
```
# 5. Conclusion
**We have trained & tested 4 models for NLP task (implementing the traditional NLP preprocessing strategies). They all perform very good, however this is most likely due to the high correlation of the target other categorical features (such as 'subject'). If we did not add it to analysis, the result could have been totally different.**
**We also used a combination of supervised & unsupervised learning, which can be an interesting method to use.**
**Also, for text classification tasks I recommend using BERT models and DNN.**
*For more information on this and code snippets, read here:* https://medium.com/engineering-zemoso/text-classification-bert-vs-dnn-b226497c9de7
<font color='blue'><b>Thank you for your attention!</b><br></br><br></br>
Your comments and discussion contributions are always welcome.</font>
|
github_jupyter
|
# This Python 3 environment comes with many helpful analytics libraries installed
# It is defined by the kaggle/python Docker image: https://github.com/kaggle/docker-python
# For example, here's several helpful packages to load
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk('/kaggle/input'):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
fake = pd.read_csv('../input/fake-and-real-news-dataset/Fake.csv')
fake['flag'] = 0
fake
true = pd.read_csv('../input/fake-and-real-news-dataset/True.csv')
true['flag'] = 1
true
df = pd.DataFrame()
df = true.append(fake)
df.info()
df = df.drop_duplicates()
df = df.reset_index(drop=True)
# Correcting some data
df['date'] = df['date'].replace(['19-Feb-18'],'February 19, 2018')
df['date'] = df['date'].replace(['18-Feb-18'],'February 18, 2018')
df['date'] = df['date'].replace(['17-Feb-18'],'February 17, 2018')
df['date'] = df['date'].replace(['16-Feb-18'],'February 16, 2018')
df['date'] = df['date'].replace(['15-Feb-18'],'February 15, 2018')
df['date'] = df['date'].replace(['14-Feb-18'],'February 14, 2018')
df['date'] = df['date'].replace(['13-Feb-18'],'February 13, 2018')
df['date'] = df['date'].str.replace('Dec ', 'December ')
df['date'] = df['date'].str.replace('Nov ', 'November ')
df['date'] = df['date'].str.replace('Oct ', 'October ')
df['date'] = df['date'].str.replace('Sep ', 'September ')
df['date'] = df['date'].str.replace('Aug ', 'August ')
df['date'] = df['date'].str.replace('Jul ', 'July ')
df['date'] = df['date'].str.replace('Jun ', 'June ')
df['date'] = df['date'].str.replace('Apr ', 'April ')
df['date'] = df['date'].str.replace('Mar ', 'March ')
df['date'] = df['date'].str.replace('Feb ', 'February ')
df['date'] = df['date'].str.replace('Jan ', 'January ')
df['date'] = df['date'].str.replace(' ', '')
for i, val in enumerate(df['date']):
df['date'].iloc[i] = pd.to_datetime(df['date'].iloc[i], format='%B%d,%Y', errors='coerce') # by setting the parameter to "coerce", we will set unappropriate values to NaT (null)
df['date'] = df['date'].astype('datetime64[ns]')
df.info()
import datetime as dt
df['year'] = pd.to_datetime(df['date']).dt.to_period('Y')
df['month'] = pd.to_datetime(df['date']).dt.to_period('M')
df['month'] = df['month'].astype(str)
sub = df[['month', 'flag']]
sub = sub.dropna()
sub = sub.groupby(['month'])['flag'].sum()
sub = sub.drop('NaT')
import matplotlib.pyplot as plt
plt.suptitle('Dynamics of fake news')
plt.xticks(rotation=90)
plt.ylabel('Number of fake news')
plt.xlabel('Month-Year')
plt.plot(sub.index, sub.values, linewidth=2, color='green')
sub2 = df[['subject', 'flag']]
sub2 = sub2.dropna()
sub2 = sub2.groupby(['subject'])['flag'].sum()
plt.suptitle('Fake news among different categories')
plt.xticks(rotation=90)
plt.ylabel('Number of fake news')
plt.xlabel('Category')
plt.bar(sub2.index, height=sub2.values, color='green')
#ax1.plot(x, y)
#ax2.plot(x, -y)
nlp = df
#nlp['title'] = nlp['title'] + ' ' + nlp['subject']
from sklearn.feature_extraction.text import TfidfVectorizer
corpus = nlp[nlp['flag'] == 1]['title'].iloc[0:500] # We will take a slice of fake news, to see what vocabulary there looks like
tfidf1 = TfidfVectorizer()
vecs = tfidf1.fit_transform(corpus)
feature_names = tfidf1.get_feature_names()
dense = vecs.todense()
list_words = dense.tolist()
df_words = pd.DataFrame(list_words, columns=feature_names)
from wordcloud import WordCloud, STOPWORDS, ImageColorGenerator
df_words.T.sum(axis=1)
Cloud = WordCloud(background_color="white", max_words=100).generate_from_frequencies(df_words.T.sum(axis=1))
import matplotlib.pyplot as plt
plt.figure(figsize=(12,5))
plt.imshow(Cloud, interpolation='bilinear')
import nltk
nltk.download('punkt')
from nltk import word_tokenize
nlp['title'] = nlp['title'].apply(lambda x: word_tokenize(str(x)))
from nltk.stem import SnowballStemmer
snowball = SnowballStemmer(language='english')
nlp['title'] = nlp['title'].apply(lambda x: [snowball.stem(y) for y in x])
nlp['title'] = nlp['title'].apply(lambda x: ' '.join(x))
from nltk.corpus import stopwords
nltk.download('words')
nltk.download('stopwords')
stopwords = stopwords.words('english')
from sklearn.feature_extraction.text import TfidfVectorizer
tfidf = TfidfVectorizer()
X_text = tfidf.fit_transform(nlp['title'])
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X_text, nlp['flag'], test_size=0.33, random_state=1)
scores = {}
from sklearn.svm import LinearSVC
from sklearn.model_selection import cross_val_score
from sklearn.metrics import accuracy_score
clf = LinearSVC(max_iter=100, C=1.0)
clf.fit(X_train, y_train)
y_pred_SVM = clf.predict(X_test)
print(cross_val_score(clf, X_text, nlp['flag'], cv=3))
print(accuracy_score(y_pred_SVM, y_test))
scores['LinearSVC'] = accuracy_score(y_pred_SVM, y_test)
from sklearn.naive_bayes import MultinomialNB
clf2 = MultinomialNB()
clf2.fit(X_train, y_train)
y_pred_MNB = clf2.predict(X_test)
print(cross_val_score(clf2, X_text, nlp['flag'], cv=3))
print(accuracy_score(y_pred_MNB, y_test))
scores['MultinomialNB'] = accuracy_score(y_pred_MNB, y_test)
from xgboost import XGBClassifier
clf3 = XGBClassifier(eval_metric='rmse', use_label_encoder=False)
clf3.fit(X_train, y_train)
y_pred_XGB = clf3.predict(X_test)
print(cross_val_score(clf3, X_text, nlp['flag'], cv=3))
print(accuracy_score(y_pred_XGB, y_test))
scores['XGB'] = accuracy_score(y_pred_XGB, y_test)
!pip install pycaret
from pycaret.nlp import *
caret_nlp = setup(data=nlp, target='title', session_id=1)
lda = create_model('lda')
lda_data = assign_model(lda)
lda_data
from catboost import CatBoostClassifier
input_cat = lda_data.drop(['text','date','Perc_Dominant_Topic','flag','year'], axis=1)
input_cat['month'] = input_cat['month'].astype(str)
target_cat = lda_data['flag']
from sklearn.model_selection import train_test_split
X_train_cat, X_test_cat, y_train_cat, y_test_cat = train_test_split(input_cat, target_cat, test_size=0.33, random_state=1)
clf4 = CatBoostClassifier(iterations=1000,
cat_features=['title','subject','Dominant_Topic','month']
)
clf4.fit(X_train_cat, y_train_cat, early_stopping_rounds=10)
scores['CatBoost'] = clf4.score(X_test_cat, y_test_cat)
scores
plt.bar(scores.keys(), scores.values())
| 0.489015 | 0.702294 |
# VacationPy
----
#### Note
* Instructions have been included for each segment. You do not have to follow them exactly, but they are included to help you think through the steps.
```
# Dependencies and Setup
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
import requests
import gmaps
import os
import ipywidgets as widgets
import pprint
import json
# Import API key
from api_keys import g_key
gmaps.configure(g_key)
gkey = g_key
```
### Store Part I results into DataFrame
* Load the csv exported in Part I to a DataFrame
```
#load weather data from weatherpy exercise
weather_data_df = pd.read_csv("..\Resources\weather.csv")
weather_data_df.head()
```
### Humidity Heatmap
* Configure gmaps.
* Use the Lat and Lng as locations and Humidity as the weight.
* Add Heatmap layer to map.
```
#configure map
ponta_delgada = (37.74, 25.67) #just seemed like a nice center to see the whole map in the preview window!
fig = gmaps.figure(map_type='HYBRID',center =ponta_delgada, zoom_level=2) #choose map type, center and zoom
#specify heatmay layer values, coordinates, weighted for humidity
heatmap_layer = gmaps.heatmap_layer(
weather_data_df[["Lat","Lng"]], weights=weather_data_df["Humidity"], max_intensity = 150, point_radius = 10.0)
#add layer to map
fig.add_layer(heatmap_layer)
#display
fig
```
### Create new DataFrame fitting weather criteria
* Narrow down the cities to fit weather conditions.
* Drop any rows will null values.
```
#Remove places where weather is <65 or >85
bool_series = weather_data_df["Max Temp"].between(65,85,inclusive=True) #geeksforgeeks.org
ideal_weather_df = weather_data_df[bool_series]
#Remove places where humidity is >30%
ideal_weather_df = ideal_weather_df[ideal_weather_df["Humidity"] <= 30]
#Remove places where wind speed is >15
ideal_weather_df_df = ideal_weather_df[ideal_weather_df["Wind Speed"] <= 15]
#Remove places where cloudiness is >20%
ideal_weather_df = ideal_weather_df[ideal_weather_df["Cloudiness"] <= 20]
#check the values in the filter data frame
ideal_weather_df.describe()
ideal_weather_df
```
### Hotel Map
* Store into variable named `hotel_df`.
* Add a "Hotel Name" column to the DataFrame.
* Set parameters to search for hotels with 5000 meters.
* Hit the Google Places API for each city's coordinates.
* Store the first Hotel result into the DataFrame.
* Plot markers on top of the heatmap.
```
#Store ideal weather conditions in a new dataframe
hotel_df=ideal_weather_df.copy()
#Add a column to the dataframe
hotel_df["Hotel"]=""
#Iterate through hotel_df to populate the hotel column
for index,row in hotel_df.iterrows():
try:
target_coordinates = f"{row['Lat']}, {row['Lng']}"
target_keyword = "lodging"
target_type = "lodging"
target_radius = "5000"
params = {
"keyword": target_keyword,
"location": target_coordinates,
"radius": target_radius,
"type": target_type,
"key": gkey
}
base_url = "https://maps.googleapis.com/maps/api/place/nearbysearch/json"
response = requests.get(base_url, params=params)
data=response.json()
hotel_df.loc[index,"Hotel"] = data["results"][0]["name"]
except:
print(f"You can't stay in {row['City']}, {row['Country']}...unless you bring a tent or you know somebody!")
hotel_df
# NOTE: Do not change any of the code in this cell
# Using the template add the hotel marks to the heatmap
info_box_template = """
<dl>
<dt>Name</dt><dd>{Hotel}</dd>
<dt>City</dt><dd>{City}</dd>
<dt>Country</dt><dd>{Country}</dd>
</dl>
"""
# Store the DataFrame Row
# NOTE: be sure to update with your DataFrame name
hotel_info = [info_box_template.format(**row) for index, row in hotel_df.iterrows()]
locations = hotel_df[["Lat", "Lng"]]
# Add marker layer ontop of heat map
marker_locations= hotel_df[["Lat","Lng"]]
fig = gmaps.figure(map_type='HYBRID',center =ponta_delgada, zoom_level=2)
heatmap_layer = gmaps.heatmap_layer(
weather_data_df[["Lat","Lng"]], weights=weather_data_df["Humidity"], max_intensity = 150, point_radius = 10.0)
fig.add_layer(heatmap_layer)
markers = gmaps.marker_layer(marker_locations, info_box_content=hotel_info)
fig.add_layer(markers)
# Display figure
fig
```
|
github_jupyter
|
# Dependencies and Setup
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
import requests
import gmaps
import os
import ipywidgets as widgets
import pprint
import json
# Import API key
from api_keys import g_key
gmaps.configure(g_key)
gkey = g_key
#load weather data from weatherpy exercise
weather_data_df = pd.read_csv("..\Resources\weather.csv")
weather_data_df.head()
#configure map
ponta_delgada = (37.74, 25.67) #just seemed like a nice center to see the whole map in the preview window!
fig = gmaps.figure(map_type='HYBRID',center =ponta_delgada, zoom_level=2) #choose map type, center and zoom
#specify heatmay layer values, coordinates, weighted for humidity
heatmap_layer = gmaps.heatmap_layer(
weather_data_df[["Lat","Lng"]], weights=weather_data_df["Humidity"], max_intensity = 150, point_radius = 10.0)
#add layer to map
fig.add_layer(heatmap_layer)
#display
fig
#Remove places where weather is <65 or >85
bool_series = weather_data_df["Max Temp"].between(65,85,inclusive=True) #geeksforgeeks.org
ideal_weather_df = weather_data_df[bool_series]
#Remove places where humidity is >30%
ideal_weather_df = ideal_weather_df[ideal_weather_df["Humidity"] <= 30]
#Remove places where wind speed is >15
ideal_weather_df_df = ideal_weather_df[ideal_weather_df["Wind Speed"] <= 15]
#Remove places where cloudiness is >20%
ideal_weather_df = ideal_weather_df[ideal_weather_df["Cloudiness"] <= 20]
#check the values in the filter data frame
ideal_weather_df.describe()
ideal_weather_df
#Store ideal weather conditions in a new dataframe
hotel_df=ideal_weather_df.copy()
#Add a column to the dataframe
hotel_df["Hotel"]=""
#Iterate through hotel_df to populate the hotel column
for index,row in hotel_df.iterrows():
try:
target_coordinates = f"{row['Lat']}, {row['Lng']}"
target_keyword = "lodging"
target_type = "lodging"
target_radius = "5000"
params = {
"keyword": target_keyword,
"location": target_coordinates,
"radius": target_radius,
"type": target_type,
"key": gkey
}
base_url = "https://maps.googleapis.com/maps/api/place/nearbysearch/json"
response = requests.get(base_url, params=params)
data=response.json()
hotel_df.loc[index,"Hotel"] = data["results"][0]["name"]
except:
print(f"You can't stay in {row['City']}, {row['Country']}...unless you bring a tent or you know somebody!")
hotel_df
# NOTE: Do not change any of the code in this cell
# Using the template add the hotel marks to the heatmap
info_box_template = """
<dl>
<dt>Name</dt><dd>{Hotel}</dd>
<dt>City</dt><dd>{City}</dd>
<dt>Country</dt><dd>{Country}</dd>
</dl>
"""
# Store the DataFrame Row
# NOTE: be sure to update with your DataFrame name
hotel_info = [info_box_template.format(**row) for index, row in hotel_df.iterrows()]
locations = hotel_df[["Lat", "Lng"]]
# Add marker layer ontop of heat map
marker_locations= hotel_df[["Lat","Lng"]]
fig = gmaps.figure(map_type='HYBRID',center =ponta_delgada, zoom_level=2)
heatmap_layer = gmaps.heatmap_layer(
weather_data_df[["Lat","Lng"]], weights=weather_data_df["Humidity"], max_intensity = 150, point_radius = 10.0)
fig.add_layer(heatmap_layer)
markers = gmaps.marker_layer(marker_locations, info_box_content=hotel_info)
fig.add_layer(markers)
# Display figure
fig
| 0.36523 | 0.810028 |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.