code
stringlengths 2.5k
6.36M
| kind
stringclasses 2
values | parsed_code
stringlengths 0
404k
| quality_prob
float64 0
0.98
| learning_prob
float64 0.03
1
|
|---|---|---|---|---|
# 线性回归的简洁实现
随着深度学习框架的发展,开发深度学习应用变得越来越便利。实践中,我们通常可以用比上一节更简洁的代码来实现同样的模型。在本节中,我们将介绍如何使用tensorflow2.0推荐的keras接口更方便地实现线性回归的训练。
## 生成数据集
我们生成与上一节中相同的数据集。其中`features`是训练数据特征,`labels`是标签。
```
import tensorflow as tf
num_inputs = 2
num_examples = 1000
true_w = [2, -3.4]
true_b = 4.2
features = tf.random.normal(shape=(num_examples, num_inputs), stddev=1)
labels = true_w[0] * features[:, 0] + true_w[1] * features[:, 1] + true_b
labels += tf.random.normal(labels.shape, stddev=0.01)
```
虽然tensorflow2.0对于线性回归可以直接拟合,不用再划分数据集,但我们仍学习一下读取数据的方法
```
from tensorflow import data as tfdata
batch_size = 10
# 将训练数据的特征和标签组合
dataset = tfdata.Dataset.from_tensor_slices((features, labels))
# 随机读取小批量
dataset = dataset.shuffle(buffer_size=num_examples)
dataset = dataset.batch(batch_size)
data_iter = iter(dataset)
for X, y in data_iter:
print(X, y)
break
```
定义模型,tensorflow 2.0推荐使用keras定义网络,故使用keras定义网络
我们先定义一个模型变量`model`,它是一个`Sequential`实例。
在keras中,`Sequential`实例可以看作是一个串联各个层的容器。
在构造模型时,我们在该容器中依次添加层。
当给定输入数据时,容器中的每一层将依次计算并将输出作为下一层的输入。
重要的一点是,在keras中我们无须指定每一层输入的形状。
因为为线性回归,输入层与输出层全连接,故定义一层
```
from tensorflow import keras
from tensorflow.keras import layers
from tensorflow import initializers as init
model = keras.Sequential()
model.add(layers.Dense(1, kernel_initializer=init.RandomNormal(stddev=0.01)))
```
定义损失函数和优化器:损失函数为mse,优化器选择sgd随机梯度下降
在keras中,定义完模型后,调用`compile()`方法可以配置模型的损失函数和优化方法。定义损失函数只需传入`loss`的参数,keras定义了各种损失函数,并直接使用它提供的平方损失`mse`作为模型的损失函数。同样,我们也无须实现小批量随机梯度下降,只需传入`optimizer`的参数,keras定义了各种优化算法,我们这里直接指定学习率为0.01的小批量随机梯度下降`tf.keras.optimizers.SGD(0.03)`为优化算法
```
from tensorflow import losses
loss = losses.MeanSquaredError()
from tensorflow.keras import optimizers
trainer = optimizers.SGD(learning_rate=0.03)
loss_history = []
```
在使用keras训练模型时,我们通过调用`model`实例的`fit`函数来迭代模型。`fit`函数只需传入你的输入x和输出y,还有epoch遍历数据的次数,每次更新梯度的大小batch_size, 这里定义epoch=3,batch_size=10。
使用keras甚至完全不需要去划分数据集
```
num_epochs = 3
for epoch in range(1, num_epochs + 1):
for (batch, (X, y)) in enumerate(dataset):
with tf.GradientTape() as tape:
l = loss(model(X, training=True), y)
loss_history.append(l.numpy().mean())
grads = tape.gradient(l, model.trainable_variables)
trainer.apply_gradients(zip(grads, model.trainable_variables))
l = loss(model(features), labels)
print('epoch %d, loss: %f' % (epoch, l.numpy().mean()))
```
下面我们分别比较学到的模型参数和真实的模型参数。我们可以通过model的`get_weights()`来获得其权重(`weight`)和偏差(`bias`)。学到的参数和真实的参数很接近。
```
true_w, model.get_weights()[0]
true_b, model.get_weights()[1]
loss_history
```
|
github_jupyter
|
import tensorflow as tf
num_inputs = 2
num_examples = 1000
true_w = [2, -3.4]
true_b = 4.2
features = tf.random.normal(shape=(num_examples, num_inputs), stddev=1)
labels = true_w[0] * features[:, 0] + true_w[1] * features[:, 1] + true_b
labels += tf.random.normal(labels.shape, stddev=0.01)
from tensorflow import data as tfdata
batch_size = 10
# 将训练数据的特征和标签组合
dataset = tfdata.Dataset.from_tensor_slices((features, labels))
# 随机读取小批量
dataset = dataset.shuffle(buffer_size=num_examples)
dataset = dataset.batch(batch_size)
data_iter = iter(dataset)
for X, y in data_iter:
print(X, y)
break
from tensorflow import keras
from tensorflow.keras import layers
from tensorflow import initializers as init
model = keras.Sequential()
model.add(layers.Dense(1, kernel_initializer=init.RandomNormal(stddev=0.01)))
from tensorflow import losses
loss = losses.MeanSquaredError()
from tensorflow.keras import optimizers
trainer = optimizers.SGD(learning_rate=0.03)
loss_history = []
num_epochs = 3
for epoch in range(1, num_epochs + 1):
for (batch, (X, y)) in enumerate(dataset):
with tf.GradientTape() as tape:
l = loss(model(X, training=True), y)
loss_history.append(l.numpy().mean())
grads = tape.gradient(l, model.trainable_variables)
trainer.apply_gradients(zip(grads, model.trainable_variables))
l = loss(model(features), labels)
print('epoch %d, loss: %f' % (epoch, l.numpy().mean()))
true_w, model.get_weights()[0]
true_b, model.get_weights()[1]
loss_history
| 0.664758 | 0.962953 |
```
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
sns.set_theme(style="darkgrid")
dataframe = pd.read_csv('PODs.csv', delimiter=';', header=0, index_col=0)
dataframe=dataframe.astype(float)
ax = plt.gca()
dataframe.plot(kind='line', y='CU MEMORY (MiBytes)', label='vCU', ax=ax)
dataframe.plot(kind='line', y='DU MEMORY (MiBytes)', label='vDU', ax=ax)
dataframe.plot(kind='line', y='RU MEMORY (MiBytes)', label='vRU', ax=ax)
#dataframe.plot(kind='line', y='K8S MEMORY (MiBytes)', label='K8S', ax=ax)
dataframe.plot(kind='line', y='OPERATOR MEMORY (MiBytes)', label='PlaceRAN Orchestrator', ax=ax)
ax.annotate('$\it{t0}$', xy=(0,94), xytext=(15, 15), textcoords='offset points', arrowprops=dict(arrowstyle='->', color='black'), fontsize=10)
plt.axvline(x = 0,linestyle ="dotted", color='black', alpha=0.3)
ax.annotate('$\it{t1}$', xy=(70,94), xytext=(15, 15), textcoords='offset points', arrowprops=dict(arrowstyle='->', color='black'), fontsize=10)
plt.axvline(x = 70,linestyle ="dotted", color='black', alpha=0.3)
ax.annotate('$\it{t2}$', xy=(130,147), xytext=(15, 15), textcoords='offset points', arrowprops=dict(arrowstyle='->', color='black'), fontsize=10)
plt.axvline(x = 130,linestyle ="dotted", color='black', alpha=0.3)
ax.annotate('$\it{t3}$', xy=(185,1396), xytext=(15, 15), textcoords='offset points', arrowprops=dict(arrowstyle='->', color='black'), fontsize=10)
plt.axvline(x = 185,linestyle ="dotted", color='black', alpha=0.3)
ax.annotate('$\it{t4}$', xy=(245,1580), xytext=(15, 15), textcoords='offset points', arrowprops=dict(arrowstyle='->', color='black'), fontsize=10)
plt.axvline(x = 245,linestyle ="dotted", color='black', alpha=0.3)
ax.annotate('$\it{t5}$', xy=(300,1685), xytext=(15, 15), textcoords='offset points', arrowprops=dict(arrowstyle='->', color='black'), fontsize=10)
plt.axvline(x = 300,linestyle ="dotted", color='black', alpha=0.3)
ax.annotate('$\it{t6}$', xy=(360,1688), xytext=(15, 15), textcoords='offset points', arrowprops=dict(arrowstyle='->', color='black'), fontsize=10)
plt.axvline(x = 360,linestyle ="dotted", color='black', alpha=0.3)
ax.annotate('$\it{t7}$', xy=(415,1688), xytext=(15, 15), textcoords='offset points', arrowprops=dict(arrowstyle='->', color='black'), fontsize=10)
plt.axvline(x = 415,linestyle ="dotted", color='black', alpha=0.3)
plt.xlabel("Time (s)")
plt.ylabel("MEMORY (MiBytes)")
plt.legend()
plt.savefig('out/MEM_PODS.pdf', bbox_inches='tight')
plt.savefig('out/MEM_PODS.png', dpi=300, bbox_inches='tight')
```
|
github_jupyter
|
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
sns.set_theme(style="darkgrid")
dataframe = pd.read_csv('PODs.csv', delimiter=';', header=0, index_col=0)
dataframe=dataframe.astype(float)
ax = plt.gca()
dataframe.plot(kind='line', y='CU MEMORY (MiBytes)', label='vCU', ax=ax)
dataframe.plot(kind='line', y='DU MEMORY (MiBytes)', label='vDU', ax=ax)
dataframe.plot(kind='line', y='RU MEMORY (MiBytes)', label='vRU', ax=ax)
#dataframe.plot(kind='line', y='K8S MEMORY (MiBytes)', label='K8S', ax=ax)
dataframe.plot(kind='line', y='OPERATOR MEMORY (MiBytes)', label='PlaceRAN Orchestrator', ax=ax)
ax.annotate('$\it{t0}$', xy=(0,94), xytext=(15, 15), textcoords='offset points', arrowprops=dict(arrowstyle='->', color='black'), fontsize=10)
plt.axvline(x = 0,linestyle ="dotted", color='black', alpha=0.3)
ax.annotate('$\it{t1}$', xy=(70,94), xytext=(15, 15), textcoords='offset points', arrowprops=dict(arrowstyle='->', color='black'), fontsize=10)
plt.axvline(x = 70,linestyle ="dotted", color='black', alpha=0.3)
ax.annotate('$\it{t2}$', xy=(130,147), xytext=(15, 15), textcoords='offset points', arrowprops=dict(arrowstyle='->', color='black'), fontsize=10)
plt.axvline(x = 130,linestyle ="dotted", color='black', alpha=0.3)
ax.annotate('$\it{t3}$', xy=(185,1396), xytext=(15, 15), textcoords='offset points', arrowprops=dict(arrowstyle='->', color='black'), fontsize=10)
plt.axvline(x = 185,linestyle ="dotted", color='black', alpha=0.3)
ax.annotate('$\it{t4}$', xy=(245,1580), xytext=(15, 15), textcoords='offset points', arrowprops=dict(arrowstyle='->', color='black'), fontsize=10)
plt.axvline(x = 245,linestyle ="dotted", color='black', alpha=0.3)
ax.annotate('$\it{t5}$', xy=(300,1685), xytext=(15, 15), textcoords='offset points', arrowprops=dict(arrowstyle='->', color='black'), fontsize=10)
plt.axvline(x = 300,linestyle ="dotted", color='black', alpha=0.3)
ax.annotate('$\it{t6}$', xy=(360,1688), xytext=(15, 15), textcoords='offset points', arrowprops=dict(arrowstyle='->', color='black'), fontsize=10)
plt.axvline(x = 360,linestyle ="dotted", color='black', alpha=0.3)
ax.annotate('$\it{t7}$', xy=(415,1688), xytext=(15, 15), textcoords='offset points', arrowprops=dict(arrowstyle='->', color='black'), fontsize=10)
plt.axvline(x = 415,linestyle ="dotted", color='black', alpha=0.3)
plt.xlabel("Time (s)")
plt.ylabel("MEMORY (MiBytes)")
plt.legend()
plt.savefig('out/MEM_PODS.pdf', bbox_inches='tight')
plt.savefig('out/MEM_PODS.png', dpi=300, bbox_inches='tight')
| 0.503662 | 0.410018 |
# Components
gdsfactory provides a generic customizable components library in `gf.components`
## Basic shapes
### Rectangle
To create a simple rectangle, there are two functions:
``gf.components.rectangle()`` can create a basic rectangle:
```
import gdsfactory as gf
gf.components.rectangle(size=(4.5, 2), layer=(1, 0))
```
``gf.components.bbox()`` can also create a rectangle based on a bounding box.
This is useful if you want to create a rectangle which exactly surrounds a piece of existing geometry.
For example, if we have an arc geometry and we want to define a box around it, we can use ``gf.components.bbox()``:
```
D = gf.Component()
arc = D << gf.components.bend_circular(radius=10, width=0.5, angle=90, layer=(1, 0))
arc.rotate(90)
# Draw a rectangle around the arc we created by using the arc's bounding box
rect = D << gf.components.bbox(bbox=arc.bbox, layer=(0, 0))
D
```
### Cross
The ``gf.components.cross()`` function creates a cross structure:
```
gf.components.cross(length=10, width=0.5, layer=(1, 0))
```
### Ellipse
The ``gf.components.ellipse()`` function creates an ellipse by defining the major and minor radii:
```
gf.components.ellipse(radii=(10, 5), angle_resolution=2.5, layer=(1, 0))
```
### Circle
The ``gf.components.circle()`` function creates a circle:
```
gf.components.circle(radius=10, angle_resolution=2.5, layer=(1, 0))
```
### Ring
The ``gf.components.ring()`` function creates a ring. The radius refers to the center radius of the ring structure (halfway between the inner and outer radius).
```
gf.components.ring(radius=5, width=0.5, angle_resolution=2.5, layer=(1, 0))
gf.components.ring_single(
width=0.5, gap=0.2, radius=10, length_x=4, length_y=2, layer=(1, 0)
)
import gdsfactory as gf
gf.components.ring_double(
width=0.5, gap=0.2, radius=10, length_x=4, length_y=2, layer=(1, 0)
)
gf.components.ring_double(
width=0.5,
gap=0.2,
radius=10,
length_x=4,
length_y=2,
layer=(1, 0),
bend=gf.components.bend_circular,
)
```
### Bend circular
The ``gf.components.bend_circular()`` function creates an arc. The radius refers to the center radius of the arc (halfway between the inner and outer radius).
```
gf.components.bend_circular(radius=2.0, width=0.5, angle=90, npoints=720, layer=(1, 0))
```
### Bend euler
The ``gf.components.bend_euler()`` function creates an adiabatic bend in which the bend radius changes gradually. Euler bends have lower loss than circular bends.
```
gf.components.bend_euler(radius=2.0, width=0.5, angle=90, npoints=720, layer=(1, 0))
```
### Tapers
`gf.components.taper()`is defined by setting its length and its start and end length. It has two ports, ``1`` and ``2``, on either end, allowing you to easily connect it to other structures.
```
gf.components.taper(length=10, width1=6, width2=4, port=None, layer=(1, 0))
```
`gf.components.ramp()` is a structure is similar to `taper()` except it is asymmetric. It also has two ports, ``1`` and ``2``, on either end.
```
gf.components.ramp(length=10, width1=4, width2=8, layer=(1, 0))
```
### Common compound shapes
The `gf.components.L()` function creates a "L" shape with ports on either end named ``1`` and ``2``.
```
gf.components.L(width=7, size=(10, 20), layer=(1, 0))
```
The `gf.components.C()` function creates a "C" shape with ports on either end named ``1`` and ``2``.
```
gf.components.C(width=7, size=(10, 20), layer=(1, 0))
```
## Text
Gdsfactory has an implementation of the DEPLOF font with the majority of english ASCII characters represented (thanks to phidl)
```
gf.components.text(
text="Hello world!\nMultiline text\nLeft-justified",
size=10,
justify="left",
layer=(1, 0),
)
# `justify` should be either 'left', 'center', or 'right'
```
## Grid / packer / align / distribute
### Grid
The ``gf.components.grid()`` function can take a list (or 2D array) of objects and arrange them along a grid. This is often useful for making parameter sweeps. If the `separation` argument is true, grid is arranged such that the elements are guaranteed not to touch, with a `spacing` distance between them. If `separation` is false, elements are spaced evenly along a grid. The `align_x`/`align_y` arguments specify intra-row/intra-column alignment. The`edge_x`/`edge_y` arguments specify inter-row/inter-column alignment (unused if `separation = True`).
```
import gdsfactory as gf
components_list = []
for width1 in [1, 6, 9]:
for width2 in [1, 2, 4, 8]:
D = gf.components.taper(length=10, width1=width1, width2=width2, layer=(1, 0))
components_list.append(D)
c = gf.grid(
components_list,
spacing=(5, 1),
separation=True,
shape=(3, 4),
align_x="x",
align_y="y",
edge_x="x",
edge_y="ymax",
)
c
```
### Packer
The ``gf.pack()`` function packs geometries together into rectangular bins. If a ``max_size`` is specified, the function will create as many bins as is necessary to pack all the geometries and then return a list of the filled-bin Devices.
Here we generate several random shapes then pack them together automatically. We allow the bin to be as large as needed to fit all the Devices by specifying ``max_size = (None, None)``. By setting ``aspect_ratio = (2,1)``, we specify the rectangular bin it tries to pack them into should be twice as wide as it is tall:
```
import numpy as np
import gdsfactory as gf
np.random.seed(5)
D_list = [gf.components.rectangle(size=(i, i)) for i in range(1, 10)]
D_packed_list = gf.pack(
D_list, # Must be a list or tuple of Devices
spacing=1.25, # Minimum distance between adjacent shapes
aspect_ratio=(2, 1), # (width, height) ratio of the rectangular bin
max_size=(None, None), # Limits the size into which the shapes will be packed
density=1.05, # Values closer to 1 pack tighter but require more computation
sort_by_area=True, # Pre-sorts the shapes by area
)
D = D_packed_list[0] # Only one bin was created, so we plot that
D
```
Say we need to pack many shapes into multiple 500x500 unit die. If we set ``max_size = (500,500)`` the shapes will be packed into as many 500x500 unit die as required to fit them all:
```
np.random.seed(1)
D_list = [
gf.components.ellipse(radii=tuple(np.random.rand(2) * n + 2)) for n in range(120)
]
D_packed_list = gf.pack(
D_list, # Must be a list or tuple of Devices
spacing=4, # Minimum distance between adjacent shapes
aspect_ratio=(1, 1), # Shape of the box
max_size=(500, 500), # Limits the size into which the shapes will be packed
density=1.05, # Values closer to 1 pack tighter but require more computation
sort_by_area=True, # Pre-sorts the shapes by area
)
# Put all packed bins into a single device and spread them out with distribute()
F = gf.Component()
[F.add_ref(D) for D in D_packed_list]
F.distribute(elements="all", direction="x", spacing=100, separation=True)
F
```
Note that the packing problem is an NP-complete problem, so ``gf.components.packer()`` may be slow if there are more than a few hundred Devices to pack (in that case, try pre-packing a few dozen at a time then packing the resulting bins). Requires the ``rectpack`` python package.
### Distribute
The ``distribute()`` function allows you to space out elements within a Device evenly in the x or y direction. It is meant to duplicate the distribute functionality present in Inkscape / Adobe Illustrator:

Say we start out with a few random-sized rectangles we want to space out:
```
c = gf.Component()
# Create different-sized rectangles and add them to D
[
c.add_ref(
gf.components.rectangle(size=[n * 15 + 20, n * 15 + 20], layer=(0, 0))
).move([n, n * 4])
for n in [0, 2, 3, 1, 2]
]
c
```
Oftentimes, we want to guarantee some distance between the objects. By setting ``separation = True`` we move each object such that there is ``spacing`` distance between them:
```
D = gf.Component()
# Create different-sized rectangles and add them to D
[
D.add_ref(gf.components.rectangle(size=[n * 15 + 20, n * 15 + 20])).move((n, n * 4))
for n in [0, 2, 3, 1, 2]
]
# Distribute all the rectangles in D along the x-direction with a separation of 5
D.distribute(
elements="all", # either 'all' or a list of objects
direction="x", # 'x' or 'y'
spacing=5,
separation=True,
)
D
```
Alternatively, we can spread them out on a fixed grid by setting ``separation = False``. Here we align the left edge (``edge = 'min'``) of each object along a grid spacing of 100:
```
D = gf.Component()
[
D.add_ref(gf.components.rectangle(size=[n * 15 + 20, n * 15 + 20])).move((n, n * 4))
for n in [0, 2, 3, 1, 2]
]
D.distribute(
elements="all", direction="x", spacing=100, separation=False, edge="xmin"
) # edge must be either 'xmin' (left), 'xmax' (right), or 'x' (center)
D
```
The alignment can be done along the right edge as well by setting ``edge = 'max'``, or along the center by setting ``edge = 'center'`` like in the following:
```
D = gf.Component()
[
D.add_ref(gf.components.rectangle(size=[n * 15 + 20, n * 15 + 20])).move(
(n - 10, n * 4)
)
for n in [0, 2, 3, 1, 2]
]
D.distribute(
elements="all", direction="x", spacing=100, separation=False, edge="x"
) # edge must be either 'xmin' (left), 'xmax' (right), or 'x' (center)
D
```
### Align
The ``align()`` function allows you to elements within a Device horizontally or vertically. It is meant to duplicate the alignment functionality present in Inkscape / Adobe Illustrator:

Say we ``distribute()`` a few objects, but they're all misaligned:
```
D = gf.Component()
# Create different-sized rectangles and add them to D then distribute them
[
D.add_ref(gf.components.rectangle(size=[n * 15 + 20, n * 15 + 20])).move((n, n * 4))
for n in [0, 2, 3, 1, 2]
]
D.distribute(elements="all", direction="x", spacing=5, separation=True)
D
```
we can use the ``align()`` function to align their top edges (``alignment = 'ymax'):
```
D = gf.Component()
# Create different-sized rectangles and add them to D then distribute them
[
D.add_ref(gf.components.rectangle(size=[n * 15 + 20, n * 15 + 20])).move((n, n * 4))
for n in [0, 2, 3, 1, 2]
]
D.distribute(elements="all", direction="x", spacing=5, separation=True)
# Align top edges
D.align(elements="all", alignment="ymax")
D
```
or align their centers (``alignment = 'y'):
```
D = gf.Component()
# Create different-sized rectangles and add them to D then distribute them
[
D.add_ref(gf.components.rectangle(size=[n * 15 + 20, n * 15 + 20])).move((n, n * 4))
for n in [0, 2, 3, 1, 2]
]
D.distribute(elements="all", direction="x", spacing=5, separation=True)
# Align top edges
D.align(elements="all", alignment="y")
D
```
other valid alignment options include ``'xmin', 'x', 'xmax', 'ymin', 'y', and 'ymax'``
## Boolean / outline / offset / invert
There are several common boolean-type operations available in the geometry library. These include typical boolean operations (and/or/not/xor), offsetting (expanding/shrinking polygons), outlining, and inverting.
### Boolean
The ``gf.boolean.boolean()`` function can perform AND/OR/NOT/XOR operations, and will return a new geometry with the result of that operation.
Speedup note: The ``num_divisions`` argument can be used to divide up the geometry into multiple rectangular regions and process each region sequentially (which is more computationally efficient). If you have a large geometry that takes a long time to process, try using ``num_divisions = [10,10]`` to optimize the operation.
```
import gdsfactory as gf
E = gf.components.ellipse(radii=(10, 5), layer=(1, 0))
R = gf.components.rectangle(size=[15, 5], layer=(2, 0))
C = gf.geometry.boolean(
A=E, B=R, operation="not", precision=1e-6, num_divisions=[1, 1], layer=(0, 0)
)
# Other operations include 'and', 'or', 'xor', or equivalently 'A-B', 'B-A', 'A+B'
# Plot the originals and the result
D = gf.Component()
D.add_ref(E)
D.add_ref(R).movey(-1.5)
D.add_ref(C).movex(30)
D
```
To learn how booleans work you can try all the different operations `not`, `and`, `or`, `xor`
```
import gdsfactory as gf
operation = "not"
operation = "and"
operation = "or"
operation = "xor"
r1 = (8, 8)
r2 = (11, 4)
r1 = (80, 80)
r2 = (110, 40)
angle_resolution = 0.1
c1 = gf.components.ellipse(radii=r1, layer=(1, 0), angle_resolution=angle_resolution)
c2 = gf.components.ellipse(radii=r2, layer=(1, 0), angle_resolution=angle_resolution)
%time
c3 = gf.geometry.boolean_klayout(
c1, c2, operation=operation, layer3=(1, 0)
) # klayout booleans
c3
%time
c4 = gf.geometry.boolean(c1, c2, operation=operation)
c4
```
### Offset
The ``offset()`` function takes the polygons of the input geometry, combines them together, and expands/contracts them. The function returns polygons on a single layer -- it does not respect layers.
Speedup note: The ``num_divisions`` argument can be used to divide up the geometry into multiple rectangular regions and process each region sequentially (which is more computationally efficient). If you have a large geometry that takes a long time to process, try using ``num_divisions = [10,10]`` to optimize the operation.
```
import gdsfactory as gf
# Create `T`, an ellipse and rectangle which will be offset (expanded / contracted)
T = gf.Component("ellipse_and_rectangle")
e = T << gf.components.ellipse(radii=(10, 5), layer=(1, 0))
r = T << gf.components.rectangle(size=[15, 5], layer=(2, 0))
r.move([3, -2.5])
Texpanded = gf.geometry.offset(
T, distance=2, join_first=True, precision=1e-6, num_divisions=[1, 1], layer=(0, 0)
)
Texpanded.name = "expanded"
Tshrink = gf.geometry.offset(
T,
distance=-1.5,
join_first=True,
precision=1e-6,
num_divisions=[1, 1],
layer=(0, 0),
)
Tshrink.name = "shrink"
# Plot the original geometry, the expanded, and the shrunk versions
D = gf.Component("top")
t1 = D.add_ref(T)
t2 = D.add_ref(Texpanded)
t3 = D.add_ref(Tshrink)
D.distribute([t1, t2, t3], direction="x", spacing=5)
D
```
### Outline
The ``outline()`` function takes the polygons of the input geometry then performs an offset and "not" boolean operation to create an outline. The function returns polygons on a single layer -- it does not respect layers.
Speedup note: The ``num_divisions`` argument can be used to divide up the geometry into multiple rectangular regions and process each region sequentially (which is more computationally efficient). If you have a large geometry that takes a long time to process, try using ``num_divisions = [10,10]`` to optimize the operation.
```
import gdsfactory as gf
# Create a blank device and add two shapes
X = gf.Component()
X.add_ref(gf.components.cross(length=25, width=1, layer=(1, 0)))
X.add_ref(gf.components.ellipse(radii=[10, 5], layer=(2, 0)))
O = gf.geometry.outline(X, distance=1.5, precision=1e-6, layer=(0, 0))
# Plot the original geometry and the result
D = gf.Component()
D.add_ref(X)
D.add_ref(O).movex(30)
D
```
The ``open_ports`` argument opens holes in the outlined geometry at each Port location.
- If not False, holes will be cut in the outline such that the Ports are not covered.
- If True, the holes will have the same width as the Ports.
- If a float, the holes will be widened by that value.
- If a float equal to the outline ``distance``, the outline will be flush with the port (useful positive-tone processes).
```
D = gf.components.L(width=7, size=(10, 20), layer=(1, 0))
D
# Outline the geometry and open a hole at each port
N = gf.geometry.outline(D, distance=5, open_ports=False) # No holes
N
O = gf.geometry.outline(
D, distance=5, open_ports=True
) # Hole is the same width as the port
O
P = gf.geometry.outline(
D, distance=5, open_ports=2.9
) # Change the hole size by entering a float
P
Q = gf.geometry.outline(
D, distance=5, open_ports=5
) # Creates flush opening (open_ports > distance)
Q
```
### Invert
The ``gf.boolean.invert()`` function creates an inverted version of the input geometry. The function creates a rectangle around the geometry (with extra padding of distance ``border``), then subtract all polygons from all layers from that rectangle, resulting in an inverted version of the geometry.
Speedup note: The ``num_divisions`` argument can be used to divide up the geometry into multiple rectangular regions and process each region sequentially (which is more computationally efficient). If you have a large geometry that takes a long time to process, try using ``num_divisions = [10,10]`` to optimize the operation.
```
import gdsfactory as gf
E = gf.components.ellipse(radii=(10, 5))
D = gf.geometry.invert(E, border=0.5, precision=1e-6, layer=(0, 0))
D
```
### Union
The ``union()`` function is a "join" function, and is functionally identical to the "OR" operation of ``gf.boolean()``. The one difference is it's able to perform this function layer-wise, so each layer can be individually combined.
```
import gdsfactory as gf
D = gf.Component()
e0 = D << gf.components.ellipse(layer=(1, 0))
e1 = D << gf.components.ellipse(layer=(2, 0))
e2 = D << gf.components.ellipse(layer=(3, 0))
e3 = D << gf.components.ellipse(layer=(4, 0))
e4 = D << gf.components.ellipse(layer=(5, 0))
e5 = D << gf.components.ellipse(layer=(6, 0))
e1.rotate(15 * 1)
e2.rotate(15 * 2)
e3.rotate(15 * 3)
e4.rotate(15 * 4)
e5.rotate(15 * 5)
D
# We have two options to unioning - take all polygons, regardless of
# layer, and join them together (in this case on layer (0,0) like so:
D_joined = gf.geometry.union(D, by_layer=False, layer=(0, 0))
D_joined
# Or we can perform the union operate by-layer
D_joined_by_layer = gf.geometry.union(D, by_layer=True)
D_joined_by_layer
```
### XOR / diff
The ``xor_diff()`` function can be used to compare two geometries and identify where they are different. Specifically, it performs a layer-wise XOR operation. If two geometries are identical, the result will be an empty Device. If they are not identical, any areas not shared by the two geometries will remain.
```
import gdsfactory as gf
A = gf.Component()
A.add_ref(gf.components.ellipse(radii=[10, 5], layer=(1, 0)))
A.add_ref(gf.components.text("A")).move([3, 0])
B = gf.Component()
B.add_ref(gf.components.ellipse(radii=[11, 4], layer=(1, 0))).movex(4)
B.add_ref(gf.components.text("B")).move([3.2, 0])
X = gf.geometry.xor_diff(A=A, B=B, precision=1e-6)
# Plot the original geometry and the result
# Upper left: A / Upper right: B
# Lower left: A and B / Lower right: A xor B "diff" comparison
D = gf.Component()
D.add_ref(A).move([-15, 25])
D.add_ref(B).move([15, 25])
D.add_ref(A).movex(-15)
D.add_ref(B).movex(-15)
D.add_ref(X).movex(15)
D
```
## Lithography structures
### Step-resolution
The `gf.components.litho_steps()` function creates lithographic test structure that is useful for measuring resolution of photoresist or electron-beam resists. It provides both positive-tone and negative-tone resolution tests.
```
D = gf.components.litho_steps(
line_widths=[1, 2, 4, 8, 16], line_spacing=10, height=100, layer=(1, 0)
)
D
```
### Calipers (inter-layer alignment)
The `gf.components.litho_calipers()` function is used to detect offsets in multilayer fabrication. It creates a two sets of notches on different layers. When an fabrication error/offset occurs, it is easy to detect how much the offset is because both center-notches are no longer aligned.
```
D = gf.components.litho_calipers(
notch_size=[1, 5],
notch_spacing=2,
num_notches=7,
offset_per_notch=0.1,
row_spacing=0,
layer1=(1, 0),
layer2=(2, 0),
)
D
```
## Paths / straights
See the **Path tutorial** for more details -- this is just an enumeration of the available built-in Path functions
### Circular arc
```
P = gf.path.arc(radius=10, angle=135, npoints=720)
gf.plot(P)
```
### Straight
```
import gdsfactory as gf
P = gf.path.straight(length=5, npoints=100)
gf.plot(P)
```
### Euler curve
Also known as a straight-to-bend, clothoid, racetrack, or track transition, this Path tapers adiabatically from straight to curved. Often used to minimize losses in photonic straights. If `p < 1.0`, will create a "partial euler" curve as described in Vogelbacher et. al. https://dx.doi.org/10.1364/oe.27.031394. If the `use_eff` argument is false, `radius` corresponds to minimum radius of curvature of the bend. If `use_eff` is true, `radius` corresponds to the "effective" radius of the bend-- The curve will be scaled such that the endpoints match an arc with parameters `radius` and `angle`.
```
P = gf.path.euler(radius=3, angle=90, p=1.0, use_eff=False, npoints=720)
gf.plot(P)
P
```
### Smooth path from waypoints
```
import numpy as np
import gdsfactory as gf
points = np.array([(20, 10), (40, 10), (20, 40), (50, 40), (50, 20), (70, 20)])
P = gf.path.smooth(
points=points,
radius=2,
bend=gf.path.euler,
use_eff=False,
)
gf.plot(P)
```
### Delay spiral
```
gf.components.spiral()
```
## Importing GDS files
`gf.import_gds()` allows you to easily import external GDSII files. It imports a single cell from the external GDS file and converts it into a gdsfactory component.
```
D = gf.components.ellipse()
D.write_gds("myoutput.gds")
D = gf.import_gds(gdspath="myoutput.gds", cellname=None, flatten=False)
D
```
## LayerSet
The `LayerSet` class allows you to predefine a collection of layers and specify their properties including: gds layer/datatype, name, and color. It also comes with a handy preview function called `gf.layers.preview_layerset()`
```
import gdsfactory as gf
lys = gf.layers.LayerSet()
lys.add_layer("p", color="lightblue", gds_layer=1, gds_datatype=0)
lys.add_layer("p+", color="blue", gds_layer=2, gds_datatype=0)
lys.add_layer("p++", color="darkblue", gds_layer=3, gds_datatype=0)
lys.add_layer("n", color="lightgreen", gds_layer=4, gds_datatype=0)
lys.add_layer("n+", color="green", gds_layer=4, gds_datatype=98)
lys.add_layer("n++", color="darkgreen", gds_layer=5, gds_datatype=99)
D = gf.layers.preview_layerset(lys, size=100, spacing=100)
D
```
## Useful contact pads / connectors
These functions are common shapes with ports, often used to make contact pads
```
D = gf.components.compass(size=(4, 2), layer=(1, 0))
D
import phidl.geometry as pg
from phidl import quickplot as plot
D = pg.compass_multi(size=(4, 2), ports={"N": 3, "S": 4}, layer=(1, 0))
plot(D) # quickplot the geometry
import phidl.geometry as pg
from phidl import quickplot as plot
D = pg.flagpole(
size=(50, 25), stub_size=(4, 8), shape="p", taper_type="fillet", layer=(1, 0)
)
# taper_type should be None, 'fillet', or 'straight'
plot(D) # quickplot the geometry
import phidl.geometry as pg
from phidl import quickplot as plot
D = pg.straight(size=(4, 2), layer=(1, 0))
plot(D) # quickplot the geometry
import phidl.geometry as pg
from phidl import quickplot as plot
D = pg.connector(midpoint=(0, 0), width=1, orientation=0)
plot(D) # quickplot the geometry
import phidl.geometry as pg
from phidl import quickplot as plot
D = pg.tee(size=(8, 4), stub_size=(1, 2), taper_type="fillet", layer=(1, 0))
# taper_type should be None, 'fillet', or 'straight'
plot(D)
```
## Chip / die template
```
import gdsfactory as gf
D = gf.components.die(
size=(10000, 5000), # Size of die
street_width=100, # Width of corner marks for die-sawing
street_length=1000, # Length of corner marks for die-sawing
die_name="chip99", # Label text
text_size=500, # Label text size
text_location="SW", # Label text compass location e.g. 'S', 'SE', 'SW'
layer=(2,0),
bbox_layer=(3,0),
)
D
```
## Optimal superconducting curves
The following structures are meant to reduce "current crowding" in superconducting thin-film structures (such as superconducting nanowires). They are the result of conformal mapping equations derived in Clem, J. & Berggren, K. "[Geometry-dependent critical currents in superconducting nanocircuits." Phys. Rev. B 84, 1–27 (2011).](http://dx.doi.org/10.1103/PhysRevB.84.174510)
```
import phidl.geometry as pg
from phidl import quickplot as plot
D = pg.optimal_hairpin(
width=0.2, pitch=0.6, length=10, turn_ratio=4, num_pts=50, layer=0
)
plot(D) # quickplot the geometry
import phidl.geometry as pg
from phidl import quickplot as plot
D = pg.optimal_step(
start_width=10,
end_width=22,
num_pts=50,
width_tol=1e-3,
anticrowding_factor=1.2,
symmetric=False,
layer=0,
)
plot(D) # quickplot the geometry
import phidl.geometry as pg
from phidl import quickplot as plot
D = pg.optimal_90deg(width=100.0, num_pts=15, length_adjust=1, layer=0)
plot(D) # quickplot the geometry
import phidl.geometry as pg
from phidl import quickplot as plot
D = pg.snspd(
wire_width=0.2,
wire_pitch=0.6,
size=(10, 8),
num_squares=None,
turn_ratio=4,
terminals_same_side=False,
layer=0,
)
plot(D) # quickplot the geometry
D = pg.snspd_expanded(
wire_width=0.3,
wire_pitch=0.6,
size=(10, 8),
num_squares=None,
connector_width=1,
connector_symmetric=False,
turn_ratio=4,
terminals_same_side=False,
layer=0,
)
plot(D) # quickplot the geometry
```
## Copying and extracting geometry
```
E = gf.Component()
E.add_ref(gf.components.ellipse(layer=(1, 0)))
D = E.extract(layers=[(1, 0)])
D
import gdsfactory as gf
X = gf.components.ellipse(layer=(2, 0))
c = X.copy()
c
```
|
github_jupyter
|
import gdsfactory as gf
gf.components.rectangle(size=(4.5, 2), layer=(1, 0))
D = gf.Component()
arc = D << gf.components.bend_circular(radius=10, width=0.5, angle=90, layer=(1, 0))
arc.rotate(90)
# Draw a rectangle around the arc we created by using the arc's bounding box
rect = D << gf.components.bbox(bbox=arc.bbox, layer=(0, 0))
D
gf.components.cross(length=10, width=0.5, layer=(1, 0))
gf.components.ellipse(radii=(10, 5), angle_resolution=2.5, layer=(1, 0))
gf.components.circle(radius=10, angle_resolution=2.5, layer=(1, 0))
gf.components.ring(radius=5, width=0.5, angle_resolution=2.5, layer=(1, 0))
gf.components.ring_single(
width=0.5, gap=0.2, radius=10, length_x=4, length_y=2, layer=(1, 0)
)
import gdsfactory as gf
gf.components.ring_double(
width=0.5, gap=0.2, radius=10, length_x=4, length_y=2, layer=(1, 0)
)
gf.components.ring_double(
width=0.5,
gap=0.2,
radius=10,
length_x=4,
length_y=2,
layer=(1, 0),
bend=gf.components.bend_circular,
)
gf.components.bend_circular(radius=2.0, width=0.5, angle=90, npoints=720, layer=(1, 0))
gf.components.bend_euler(radius=2.0, width=0.5, angle=90, npoints=720, layer=(1, 0))
gf.components.taper(length=10, width1=6, width2=4, port=None, layer=(1, 0))
gf.components.ramp(length=10, width1=4, width2=8, layer=(1, 0))
gf.components.L(width=7, size=(10, 20), layer=(1, 0))
gf.components.C(width=7, size=(10, 20), layer=(1, 0))
gf.components.text(
text="Hello world!\nMultiline text\nLeft-justified",
size=10,
justify="left",
layer=(1, 0),
)
# `justify` should be either 'left', 'center', or 'right'
import gdsfactory as gf
components_list = []
for width1 in [1, 6, 9]:
for width2 in [1, 2, 4, 8]:
D = gf.components.taper(length=10, width1=width1, width2=width2, layer=(1, 0))
components_list.append(D)
c = gf.grid(
components_list,
spacing=(5, 1),
separation=True,
shape=(3, 4),
align_x="x",
align_y="y",
edge_x="x",
edge_y="ymax",
)
c
import numpy as np
import gdsfactory as gf
np.random.seed(5)
D_list = [gf.components.rectangle(size=(i, i)) for i in range(1, 10)]
D_packed_list = gf.pack(
D_list, # Must be a list or tuple of Devices
spacing=1.25, # Minimum distance between adjacent shapes
aspect_ratio=(2, 1), # (width, height) ratio of the rectangular bin
max_size=(None, None), # Limits the size into which the shapes will be packed
density=1.05, # Values closer to 1 pack tighter but require more computation
sort_by_area=True, # Pre-sorts the shapes by area
)
D = D_packed_list[0] # Only one bin was created, so we plot that
D
np.random.seed(1)
D_list = [
gf.components.ellipse(radii=tuple(np.random.rand(2) * n + 2)) for n in range(120)
]
D_packed_list = gf.pack(
D_list, # Must be a list or tuple of Devices
spacing=4, # Minimum distance between adjacent shapes
aspect_ratio=(1, 1), # Shape of the box
max_size=(500, 500), # Limits the size into which the shapes will be packed
density=1.05, # Values closer to 1 pack tighter but require more computation
sort_by_area=True, # Pre-sorts the shapes by area
)
# Put all packed bins into a single device and spread them out with distribute()
F = gf.Component()
[F.add_ref(D) for D in D_packed_list]
F.distribute(elements="all", direction="x", spacing=100, separation=True)
F
c = gf.Component()
# Create different-sized rectangles and add them to D
[
c.add_ref(
gf.components.rectangle(size=[n * 15 + 20, n * 15 + 20], layer=(0, 0))
).move([n, n * 4])
for n in [0, 2, 3, 1, 2]
]
c
D = gf.Component()
# Create different-sized rectangles and add them to D
[
D.add_ref(gf.components.rectangle(size=[n * 15 + 20, n * 15 + 20])).move((n, n * 4))
for n in [0, 2, 3, 1, 2]
]
# Distribute all the rectangles in D along the x-direction with a separation of 5
D.distribute(
elements="all", # either 'all' or a list of objects
direction="x", # 'x' or 'y'
spacing=5,
separation=True,
)
D
D = gf.Component()
[
D.add_ref(gf.components.rectangle(size=[n * 15 + 20, n * 15 + 20])).move((n, n * 4))
for n in [0, 2, 3, 1, 2]
]
D.distribute(
elements="all", direction="x", spacing=100, separation=False, edge="xmin"
) # edge must be either 'xmin' (left), 'xmax' (right), or 'x' (center)
D
D = gf.Component()
[
D.add_ref(gf.components.rectangle(size=[n * 15 + 20, n * 15 + 20])).move(
(n - 10, n * 4)
)
for n in [0, 2, 3, 1, 2]
]
D.distribute(
elements="all", direction="x", spacing=100, separation=False, edge="x"
) # edge must be either 'xmin' (left), 'xmax' (right), or 'x' (center)
D
D = gf.Component()
# Create different-sized rectangles and add them to D then distribute them
[
D.add_ref(gf.components.rectangle(size=[n * 15 + 20, n * 15 + 20])).move((n, n * 4))
for n in [0, 2, 3, 1, 2]
]
D.distribute(elements="all", direction="x", spacing=5, separation=True)
D
D = gf.Component()
# Create different-sized rectangles and add them to D then distribute them
[
D.add_ref(gf.components.rectangle(size=[n * 15 + 20, n * 15 + 20])).move((n, n * 4))
for n in [0, 2, 3, 1, 2]
]
D.distribute(elements="all", direction="x", spacing=5, separation=True)
# Align top edges
D.align(elements="all", alignment="ymax")
D
D = gf.Component()
# Create different-sized rectangles and add them to D then distribute them
[
D.add_ref(gf.components.rectangle(size=[n * 15 + 20, n * 15 + 20])).move((n, n * 4))
for n in [0, 2, 3, 1, 2]
]
D.distribute(elements="all", direction="x", spacing=5, separation=True)
# Align top edges
D.align(elements="all", alignment="y")
D
import gdsfactory as gf
E = gf.components.ellipse(radii=(10, 5), layer=(1, 0))
R = gf.components.rectangle(size=[15, 5], layer=(2, 0))
C = gf.geometry.boolean(
A=E, B=R, operation="not", precision=1e-6, num_divisions=[1, 1], layer=(0, 0)
)
# Other operations include 'and', 'or', 'xor', or equivalently 'A-B', 'B-A', 'A+B'
# Plot the originals and the result
D = gf.Component()
D.add_ref(E)
D.add_ref(R).movey(-1.5)
D.add_ref(C).movex(30)
D
import gdsfactory as gf
operation = "not"
operation = "and"
operation = "or"
operation = "xor"
r1 = (8, 8)
r2 = (11, 4)
r1 = (80, 80)
r2 = (110, 40)
angle_resolution = 0.1
c1 = gf.components.ellipse(radii=r1, layer=(1, 0), angle_resolution=angle_resolution)
c2 = gf.components.ellipse(radii=r2, layer=(1, 0), angle_resolution=angle_resolution)
%time
c3 = gf.geometry.boolean_klayout(
c1, c2, operation=operation, layer3=(1, 0)
) # klayout booleans
c3
%time
c4 = gf.geometry.boolean(c1, c2, operation=operation)
c4
import gdsfactory as gf
# Create `T`, an ellipse and rectangle which will be offset (expanded / contracted)
T = gf.Component("ellipse_and_rectangle")
e = T << gf.components.ellipse(radii=(10, 5), layer=(1, 0))
r = T << gf.components.rectangle(size=[15, 5], layer=(2, 0))
r.move([3, -2.5])
Texpanded = gf.geometry.offset(
T, distance=2, join_first=True, precision=1e-6, num_divisions=[1, 1], layer=(0, 0)
)
Texpanded.name = "expanded"
Tshrink = gf.geometry.offset(
T,
distance=-1.5,
join_first=True,
precision=1e-6,
num_divisions=[1, 1],
layer=(0, 0),
)
Tshrink.name = "shrink"
# Plot the original geometry, the expanded, and the shrunk versions
D = gf.Component("top")
t1 = D.add_ref(T)
t2 = D.add_ref(Texpanded)
t3 = D.add_ref(Tshrink)
D.distribute([t1, t2, t3], direction="x", spacing=5)
D
import gdsfactory as gf
# Create a blank device and add two shapes
X = gf.Component()
X.add_ref(gf.components.cross(length=25, width=1, layer=(1, 0)))
X.add_ref(gf.components.ellipse(radii=[10, 5], layer=(2, 0)))
O = gf.geometry.outline(X, distance=1.5, precision=1e-6, layer=(0, 0))
# Plot the original geometry and the result
D = gf.Component()
D.add_ref(X)
D.add_ref(O).movex(30)
D
D = gf.components.L(width=7, size=(10, 20), layer=(1, 0))
D
# Outline the geometry and open a hole at each port
N = gf.geometry.outline(D, distance=5, open_ports=False) # No holes
N
O = gf.geometry.outline(
D, distance=5, open_ports=True
) # Hole is the same width as the port
O
P = gf.geometry.outline(
D, distance=5, open_ports=2.9
) # Change the hole size by entering a float
P
Q = gf.geometry.outline(
D, distance=5, open_ports=5
) # Creates flush opening (open_ports > distance)
Q
import gdsfactory as gf
E = gf.components.ellipse(radii=(10, 5))
D = gf.geometry.invert(E, border=0.5, precision=1e-6, layer=(0, 0))
D
import gdsfactory as gf
D = gf.Component()
e0 = D << gf.components.ellipse(layer=(1, 0))
e1 = D << gf.components.ellipse(layer=(2, 0))
e2 = D << gf.components.ellipse(layer=(3, 0))
e3 = D << gf.components.ellipse(layer=(4, 0))
e4 = D << gf.components.ellipse(layer=(5, 0))
e5 = D << gf.components.ellipse(layer=(6, 0))
e1.rotate(15 * 1)
e2.rotate(15 * 2)
e3.rotate(15 * 3)
e4.rotate(15 * 4)
e5.rotate(15 * 5)
D
# We have two options to unioning - take all polygons, regardless of
# layer, and join them together (in this case on layer (0,0) like so:
D_joined = gf.geometry.union(D, by_layer=False, layer=(0, 0))
D_joined
# Or we can perform the union operate by-layer
D_joined_by_layer = gf.geometry.union(D, by_layer=True)
D_joined_by_layer
import gdsfactory as gf
A = gf.Component()
A.add_ref(gf.components.ellipse(radii=[10, 5], layer=(1, 0)))
A.add_ref(gf.components.text("A")).move([3, 0])
B = gf.Component()
B.add_ref(gf.components.ellipse(radii=[11, 4], layer=(1, 0))).movex(4)
B.add_ref(gf.components.text("B")).move([3.2, 0])
X = gf.geometry.xor_diff(A=A, B=B, precision=1e-6)
# Plot the original geometry and the result
# Upper left: A / Upper right: B
# Lower left: A and B / Lower right: A xor B "diff" comparison
D = gf.Component()
D.add_ref(A).move([-15, 25])
D.add_ref(B).move([15, 25])
D.add_ref(A).movex(-15)
D.add_ref(B).movex(-15)
D.add_ref(X).movex(15)
D
D = gf.components.litho_steps(
line_widths=[1, 2, 4, 8, 16], line_spacing=10, height=100, layer=(1, 0)
)
D
D = gf.components.litho_calipers(
notch_size=[1, 5],
notch_spacing=2,
num_notches=7,
offset_per_notch=0.1,
row_spacing=0,
layer1=(1, 0),
layer2=(2, 0),
)
D
P = gf.path.arc(radius=10, angle=135, npoints=720)
gf.plot(P)
import gdsfactory as gf
P = gf.path.straight(length=5, npoints=100)
gf.plot(P)
P = gf.path.euler(radius=3, angle=90, p=1.0, use_eff=False, npoints=720)
gf.plot(P)
P
import numpy as np
import gdsfactory as gf
points = np.array([(20, 10), (40, 10), (20, 40), (50, 40), (50, 20), (70, 20)])
P = gf.path.smooth(
points=points,
radius=2,
bend=gf.path.euler,
use_eff=False,
)
gf.plot(P)
gf.components.spiral()
D = gf.components.ellipse()
D.write_gds("myoutput.gds")
D = gf.import_gds(gdspath="myoutput.gds", cellname=None, flatten=False)
D
import gdsfactory as gf
lys = gf.layers.LayerSet()
lys.add_layer("p", color="lightblue", gds_layer=1, gds_datatype=0)
lys.add_layer("p+", color="blue", gds_layer=2, gds_datatype=0)
lys.add_layer("p++", color="darkblue", gds_layer=3, gds_datatype=0)
lys.add_layer("n", color="lightgreen", gds_layer=4, gds_datatype=0)
lys.add_layer("n+", color="green", gds_layer=4, gds_datatype=98)
lys.add_layer("n++", color="darkgreen", gds_layer=5, gds_datatype=99)
D = gf.layers.preview_layerset(lys, size=100, spacing=100)
D
D = gf.components.compass(size=(4, 2), layer=(1, 0))
D
import phidl.geometry as pg
from phidl import quickplot as plot
D = pg.compass_multi(size=(4, 2), ports={"N": 3, "S": 4}, layer=(1, 0))
plot(D) # quickplot the geometry
import phidl.geometry as pg
from phidl import quickplot as plot
D = pg.flagpole(
size=(50, 25), stub_size=(4, 8), shape="p", taper_type="fillet", layer=(1, 0)
)
# taper_type should be None, 'fillet', or 'straight'
plot(D) # quickplot the geometry
import phidl.geometry as pg
from phidl import quickplot as plot
D = pg.straight(size=(4, 2), layer=(1, 0))
plot(D) # quickplot the geometry
import phidl.geometry as pg
from phidl import quickplot as plot
D = pg.connector(midpoint=(0, 0), width=1, orientation=0)
plot(D) # quickplot the geometry
import phidl.geometry as pg
from phidl import quickplot as plot
D = pg.tee(size=(8, 4), stub_size=(1, 2), taper_type="fillet", layer=(1, 0))
# taper_type should be None, 'fillet', or 'straight'
plot(D)
import gdsfactory as gf
D = gf.components.die(
size=(10000, 5000), # Size of die
street_width=100, # Width of corner marks for die-sawing
street_length=1000, # Length of corner marks for die-sawing
die_name="chip99", # Label text
text_size=500, # Label text size
text_location="SW", # Label text compass location e.g. 'S', 'SE', 'SW'
layer=(2,0),
bbox_layer=(3,0),
)
D
import phidl.geometry as pg
from phidl import quickplot as plot
D = pg.optimal_hairpin(
width=0.2, pitch=0.6, length=10, turn_ratio=4, num_pts=50, layer=0
)
plot(D) # quickplot the geometry
import phidl.geometry as pg
from phidl import quickplot as plot
D = pg.optimal_step(
start_width=10,
end_width=22,
num_pts=50,
width_tol=1e-3,
anticrowding_factor=1.2,
symmetric=False,
layer=0,
)
plot(D) # quickplot the geometry
import phidl.geometry as pg
from phidl import quickplot as plot
D = pg.optimal_90deg(width=100.0, num_pts=15, length_adjust=1, layer=0)
plot(D) # quickplot the geometry
import phidl.geometry as pg
from phidl import quickplot as plot
D = pg.snspd(
wire_width=0.2,
wire_pitch=0.6,
size=(10, 8),
num_squares=None,
turn_ratio=4,
terminals_same_side=False,
layer=0,
)
plot(D) # quickplot the geometry
D = pg.snspd_expanded(
wire_width=0.3,
wire_pitch=0.6,
size=(10, 8),
num_squares=None,
connector_width=1,
connector_symmetric=False,
turn_ratio=4,
terminals_same_side=False,
layer=0,
)
plot(D) # quickplot the geometry
E = gf.Component()
E.add_ref(gf.components.ellipse(layer=(1, 0)))
D = E.extract(layers=[(1, 0)])
D
import gdsfactory as gf
X = gf.components.ellipse(layer=(2, 0))
c = X.copy()
c
| 0.743913 | 0.960842 |
```
import tensorflow as tf
from tensorflow.examples.tutorials.mnist import input_data
mnist = input_data.read_data_sets('Data/fashion', one_hot=True)
n_classes = 10
input_size = 784
x = tf.placeholder(tf.float32, shape=[None, input_size])
y = tf.placeholder(tf.float32, shape=[None, n_classes])
def weight_variable(shape):
initial = tf.truncated_normal(shape, stddev=0.1)
return tf.Variable(initial)
def bias_variable(shape):
initial = tf.constant(0.1, shape=shape)
return tf.Variable(initial)
def conv2d(x, W):
return tf.nn.conv2d(x, W, strides=[1, 1, 1, 1], padding='SAME')
def max_pool_2x2(x):
return tf.nn.max_pool(x, ksize=[1, 2, 2, 1], strides=[1, 2, 2, 1], padding='SAME')
W_conv1 = weight_variable([7, 7, 1, 100])
b_conv1 = bias_variable([100])
x_image = tf.reshape(x, [-1,28,28,1])
h_conv1 = tf.nn.relu(conv2d(x_image, W_conv1) + b_conv1)
h_pool1 = max_pool_2x2(h_conv1)
W_conv2 = weight_variable([4, 4, 100, 150])
b_conv2 = bias_variable([150])
h_conv2 = tf.nn.relu(conv2d(h_pool1, W_conv2) + b_conv2)
h_pool2 = max_pool_2x2(h_conv2)
W_conv3 = weight_variable([4, 4, 150, 250])
b_conv3 = bias_variable([250])
h_conv3 = tf.nn.relu(conv2d(h_pool2, W_conv3) + b_conv3)
h_pool3 = max_pool_2x2(h_conv3)
W_fc1 = weight_variable([4 * 4 * 250, 300])
b_fc1 = bias_variable([300])
h_pool3_flat = tf.reshape(h_pool3, [-1, 4*4*250])
h_fc1 = tf.nn.relu(tf.matmul(h_pool3_flat, W_fc1) + b_fc1)
keep_prob = tf.placeholder(tf.float32)
h_fc1_drop = tf.nn.dropout(h_fc1, keep_prob)
W_fc2 = weight_variable([300, n_classes])
b_fc2 = bias_variable([n_classes])
y_pred = tf.matmul(h_fc1_drop, W_fc2) + b_fc2
with tf.name_scope('cross_entropy'):
diff = tf.nn.softmax_cross_entropy_with_logits(labels=y, logits=y_pred)
with tf.name_scope('total'):
cross_entropy = tf.reduce_mean(diff)
tf.summary.scalar('cross_entropy', cross_entropy)
learning_rate = 0.001
train_step = tf.train.AdamOptimizer(learning_rate).minimize(cross_entropy)
with tf.name_scope('accuracy'):
correct_prediction = tf.equal(tf.argmax(y_pred, 1), tf.argmax(y, 1))
accuracy = tf.reduce_mean(tf.cast(correct_prediction, tf.float32))
tf.summary.scalar('accuracy', accuracy)
sess = tf.InteractiveSession()
log_dir = 'tensorboard-example'
merged = tf.summary.merge_all()
train_writer = tf.summary.FileWriter(log_dir + '/train', sess.graph)
val_writer = tf.summary.FileWriter(log_dir + '/val')
n_steps = 1000
batch_size = 128
dropout = 0.25
evaluate_every = 10
tf.global_variables_initializer().run()
for i in range(n_steps):
x_batch, y_batch = mnist.train.next_batch(batch_size)
summary, _, train_acc = sess.run([merged, train_step, accuracy], feed_dict={x: x_batch, y: y_batch, keep_prob: dropout})
train_writer.add_summary(summary, i)
if i % evaluate_every == 0:
summary, val_acc = sess.run([merged, accuracy], feed_dict={x: mnist.test.images, y: mnist.test.labels, keep_prob: 1.0})
val_writer.add_summary(summary, i)
print('Step {:04.0f}: train_acc: {:.4f}; val_acc {:.4f}'.format(i, train_acc, val_acc))
train_writer.close()
val_writer.close()
```
|
github_jupyter
|
import tensorflow as tf
from tensorflow.examples.tutorials.mnist import input_data
mnist = input_data.read_data_sets('Data/fashion', one_hot=True)
n_classes = 10
input_size = 784
x = tf.placeholder(tf.float32, shape=[None, input_size])
y = tf.placeholder(tf.float32, shape=[None, n_classes])
def weight_variable(shape):
initial = tf.truncated_normal(shape, stddev=0.1)
return tf.Variable(initial)
def bias_variable(shape):
initial = tf.constant(0.1, shape=shape)
return tf.Variable(initial)
def conv2d(x, W):
return tf.nn.conv2d(x, W, strides=[1, 1, 1, 1], padding='SAME')
def max_pool_2x2(x):
return tf.nn.max_pool(x, ksize=[1, 2, 2, 1], strides=[1, 2, 2, 1], padding='SAME')
W_conv1 = weight_variable([7, 7, 1, 100])
b_conv1 = bias_variable([100])
x_image = tf.reshape(x, [-1,28,28,1])
h_conv1 = tf.nn.relu(conv2d(x_image, W_conv1) + b_conv1)
h_pool1 = max_pool_2x2(h_conv1)
W_conv2 = weight_variable([4, 4, 100, 150])
b_conv2 = bias_variable([150])
h_conv2 = tf.nn.relu(conv2d(h_pool1, W_conv2) + b_conv2)
h_pool2 = max_pool_2x2(h_conv2)
W_conv3 = weight_variable([4, 4, 150, 250])
b_conv3 = bias_variable([250])
h_conv3 = tf.nn.relu(conv2d(h_pool2, W_conv3) + b_conv3)
h_pool3 = max_pool_2x2(h_conv3)
W_fc1 = weight_variable([4 * 4 * 250, 300])
b_fc1 = bias_variable([300])
h_pool3_flat = tf.reshape(h_pool3, [-1, 4*4*250])
h_fc1 = tf.nn.relu(tf.matmul(h_pool3_flat, W_fc1) + b_fc1)
keep_prob = tf.placeholder(tf.float32)
h_fc1_drop = tf.nn.dropout(h_fc1, keep_prob)
W_fc2 = weight_variable([300, n_classes])
b_fc2 = bias_variable([n_classes])
y_pred = tf.matmul(h_fc1_drop, W_fc2) + b_fc2
with tf.name_scope('cross_entropy'):
diff = tf.nn.softmax_cross_entropy_with_logits(labels=y, logits=y_pred)
with tf.name_scope('total'):
cross_entropy = tf.reduce_mean(diff)
tf.summary.scalar('cross_entropy', cross_entropy)
learning_rate = 0.001
train_step = tf.train.AdamOptimizer(learning_rate).minimize(cross_entropy)
with tf.name_scope('accuracy'):
correct_prediction = tf.equal(tf.argmax(y_pred, 1), tf.argmax(y, 1))
accuracy = tf.reduce_mean(tf.cast(correct_prediction, tf.float32))
tf.summary.scalar('accuracy', accuracy)
sess = tf.InteractiveSession()
log_dir = 'tensorboard-example'
merged = tf.summary.merge_all()
train_writer = tf.summary.FileWriter(log_dir + '/train', sess.graph)
val_writer = tf.summary.FileWriter(log_dir + '/val')
n_steps = 1000
batch_size = 128
dropout = 0.25
evaluate_every = 10
tf.global_variables_initializer().run()
for i in range(n_steps):
x_batch, y_batch = mnist.train.next_batch(batch_size)
summary, _, train_acc = sess.run([merged, train_step, accuracy], feed_dict={x: x_batch, y: y_batch, keep_prob: dropout})
train_writer.add_summary(summary, i)
if i % evaluate_every == 0:
summary, val_acc = sess.run([merged, accuracy], feed_dict={x: mnist.test.images, y: mnist.test.labels, keep_prob: 1.0})
val_writer.add_summary(summary, i)
print('Step {:04.0f}: train_acc: {:.4f}; val_acc {:.4f}'.format(i, train_acc, val_acc))
train_writer.close()
val_writer.close()
| 0.752286 | 0.790773 |
```
%matplotlib inline
import matplotlib
import pandas
import numpy as np
import matplotlib.pyplot as plt
from sklearn.cluster import KMeans
from scipy.spatial.distance import cdist, pdist
cluster_data = pandas.read_csv('cluster_data.csv')
sub_cluster_data = cluster_data.drop(cluster_data.columns[[0]], axis=1)
#sub_cluster_data
pandas.tools.plotting.scatter_matrix(sub_cluster_data, alpha=0.2, figsize=(12,12), diagonal='kde')
## K-means clustering
# Use Silhouette Scoring to identify the ideal number of clusters
from sklearn.metrics import silhouette_score
s = []
N_clusters = range(2,15)
for n_clusters in N_clusters:
kmeans = KMeans(n_clusters=n_clusters)
kmeans.fit(sub_cluster_data.as_matrix())
# Calculate S. score for current number of clusters
s.append(silhouette_score(sub_cluster_data.as_matrix(), kmeans.labels_, metric='euclidean'))
# Plot the results
kIdx = 5
plt.plot(N_clusters,s)
plt.plot(N_clusters[kIdx],s[kIdx], marker='o', markersize=12,
markeredgewidth=2, markeredgecolor='r', markerfacecolor='None')
plt.ylabel("Mean Silhouette Coeff.")
plt.xlabel("k")
plt.title("Mean Silhouette Coefficient vs k clusters")
plt.grid()
# Confirm the choice of number of clusters using the Elbow method
# Taken from:
## http://datascience.stackexchange.com/questions/6508/k-means-incoherent-behaviour-choosing-k-with-elbow-method-bic-variance-explain
K = range(2,15)
KM = [KMeans(n_clusters=k).fit(sub_cluster_data.as_matrix()) for k in K]
centroids = [k.cluster_centers_ for k in KM]
D_k = [cdist(sub_cluster_data, cent, 'euclidean') for cent in centroids]
cIdx = [np.argmin(D,axis=1) for D in D_k]
dist = [np.min(D,axis=1) for D in D_k]
avgWithinSS = [sum(d)/sub_cluster_data.shape[0] for d in dist]
# Total with-in sum of square
wcss = [sum(d**2) for d in dist]
tss = sum(pdist(sub_cluster_data)**2)/sub_cluster_data.shape[0]
bss = tss-wcss
kIdx = 6
# elbow curve
fig = plt.figure()
ax = fig.add_subplot(111)
ax.plot(K, avgWithinSS, 'b*-')
ax.plot(K[kIdx], avgWithinSS[kIdx], marker='o', markersize=12,
markeredgewidth=2, markeredgecolor='r', markerfacecolor='None')
plt.grid(True)
plt.xlabel('Number of clusters')
plt.ylabel('Average within-cluster sum of squares')
plt.title('Elbow for KMeans clustering')
fig = plt.figure()
ax = fig.add_subplot(111)
ax.plot(K, bss/tss*100, 'b*-')
ax.plot(K[kIdx], bss[kIdx]/tss*100, marker='o', markersize=12,
markeredgewidth=2, markeredgecolor='r', markerfacecolor='None')
plt.grid(True)
plt.xlabel('Number of clusters')
plt.ylabel('Percentage of variance explained')
plt.title('Elbow for KMeans clustering')
# Re-do K-means clustering using optimum values
np.random.seed(5)
optimum_k = 8
kmeans = KMeans(n_clusters=optimum_k)
kmeans.fit(sub_cluster_data.as_matrix())
labels = kmeans.labels_
centroids = kmeans.cluster_centers_
# Use PCA to identify the key dimensionality in the data
from sklearn.decomposition import PCA
pca = PCA(n_components=5)
pca.fit(sub_cluster_data)
pca.explained_variance_ratio_
# The first two principal components explain ~91% of the variation, so could simplify to 2-d
# Transform the raw data and the identified centroids on to a 2-d plane
np.random.seed(5)
pca = PCA(n_components=2)
pca.fit(sub_cluster_data)
points_pc = pca.transform(sub_cluster_data)
centroids_pc = pca.transform(centroids)
# Plot
fig = plt.figure(figsize=(12,12))
ax = fig.add_subplot(111)
ax.scatter(points_pc[:,0],points_pc[:,1], c=labels, alpha=0.75)
ax.scatter(centroids_pc[:,0],centroids_pc[:,1],marker="+",s=200,linewidths=3,c="k")
for label, x, y in zip(range(optimum_k), centroids_pc[:,0],centroids_pc[:,1]):
plt.annotate(
label,
xy = (x, y), xytext = (-20, 20),
textcoords = 'offset points', ha = 'right', va = 'bottom',
bbox = dict(boxstyle = 'round,pad=0.5', fc = 'yellow', alpha = 0.75),
arrowprops = dict(arrowstyle = '->', connectionstyle = 'arc3,rad=0'))
plt.grid(True)
plt.xlabel('PC 1')
plt.ylabel('PC 2')
plt.title('Clusters transformed on to principal components')
plt.show()
np.set_printoptions(precision=3)
print(centroids)
grouped = sub_cluster_data.groupby(labels)
grouped.size()
# Plot
i=0
fig = plt.figure(figsize=(12,12))
for group in range(optimum_k):
for dim in list(sub_cluster_data.columns.values):
i+=1
ax = fig.add_subplot(optimum_k,5,i)
ax.hist(grouped.get_group(group)[dim].values - grouped.get_group(group)[dim].mean(), 15)
ax.set_ylim([0,20])
plt.grid(True)
plt.title(str(group) + dim, {'verticalalignment':'top'})
plt.show()
grouped.std()
new_points = list()
labels2 = list()
for group in range(optimum_k):
new_points.append(np.random.multivariate_normal(centroids[group,:],
grouped.std().values[group,:] * np.eye(5),
100))
labels2.append(group * np.ones(100))
new_points = pandas.DataFrame(np.vstack(new_points), columns=['a','b','c','d','e'])
labels2 = np.vstack(labels2)
group=1
centroids[group,:]
grouped.std().values[group,:] * np.eye(5)
np.random.multivariate_normal(centroids[group,:],
grouped.std().values[group,:] * np.eye(5),
100)
pandas.tools.plotting.scatter_matrix(new_points, alpha=0.2, figsize=(12,12), diagonal='kde')
# Re-do K-means clustering using optimum values
np.random.seed(5)
optimum_k = 8
kmeans2 = KMeans(n_clusters=optimum_k)
kmeans2.fit(new_points)
labels2 = kmeans2.labels_
centroids2 = kmeans2.cluster_centers_
# Transform the raw data and the identified centroids on to a 2-d plane
np.random.seed(5)
pca2 = PCA(n_components=2)
pca2.fit(new_points)
points_pc2 = pca.transform(new_points)
centroids_pc2 = pca.transform(centroids2)
# Plot
fig = plt.figure(figsize=(12,12))
ax = fig.add_subplot(111)
ax.scatter(points_pc2[:,0],points_pc2[:,1], c=labels2, alpha=0.75)
ax.scatter(centroids_pc2[:,0],centroids_pc2[:,1],marker="+",s=200,linewidths=3,c="k")
for label, x, y in zip(range(optimum_k), centroids_pc2[:,0],centroids_pc2[:,1]):
plt.annotate(
label,
xy = (x, y), xytext = (-20, 20),
textcoords = 'offset points', ha = 'right', va = 'bottom',
bbox = dict(boxstyle = 'round,pad=0.5', fc = 'yellow', alpha = 0.5),
arrowprops = dict(arrowstyle = '->', connectionstyle = 'arc3,rad=0'))
plt.grid(True)
plt.xlabel('PC 1')
plt.ylabel('PC 2')
plt.title('Clusters transformed on to principal components')
plt.show()
```
|
github_jupyter
|
%matplotlib inline
import matplotlib
import pandas
import numpy as np
import matplotlib.pyplot as plt
from sklearn.cluster import KMeans
from scipy.spatial.distance import cdist, pdist
cluster_data = pandas.read_csv('cluster_data.csv')
sub_cluster_data = cluster_data.drop(cluster_data.columns[[0]], axis=1)
#sub_cluster_data
pandas.tools.plotting.scatter_matrix(sub_cluster_data, alpha=0.2, figsize=(12,12), diagonal='kde')
## K-means clustering
# Use Silhouette Scoring to identify the ideal number of clusters
from sklearn.metrics import silhouette_score
s = []
N_clusters = range(2,15)
for n_clusters in N_clusters:
kmeans = KMeans(n_clusters=n_clusters)
kmeans.fit(sub_cluster_data.as_matrix())
# Calculate S. score for current number of clusters
s.append(silhouette_score(sub_cluster_data.as_matrix(), kmeans.labels_, metric='euclidean'))
# Plot the results
kIdx = 5
plt.plot(N_clusters,s)
plt.plot(N_clusters[kIdx],s[kIdx], marker='o', markersize=12,
markeredgewidth=2, markeredgecolor='r', markerfacecolor='None')
plt.ylabel("Mean Silhouette Coeff.")
plt.xlabel("k")
plt.title("Mean Silhouette Coefficient vs k clusters")
plt.grid()
# Confirm the choice of number of clusters using the Elbow method
# Taken from:
## http://datascience.stackexchange.com/questions/6508/k-means-incoherent-behaviour-choosing-k-with-elbow-method-bic-variance-explain
K = range(2,15)
KM = [KMeans(n_clusters=k).fit(sub_cluster_data.as_matrix()) for k in K]
centroids = [k.cluster_centers_ for k in KM]
D_k = [cdist(sub_cluster_data, cent, 'euclidean') for cent in centroids]
cIdx = [np.argmin(D,axis=1) for D in D_k]
dist = [np.min(D,axis=1) for D in D_k]
avgWithinSS = [sum(d)/sub_cluster_data.shape[0] for d in dist]
# Total with-in sum of square
wcss = [sum(d**2) for d in dist]
tss = sum(pdist(sub_cluster_data)**2)/sub_cluster_data.shape[0]
bss = tss-wcss
kIdx = 6
# elbow curve
fig = plt.figure()
ax = fig.add_subplot(111)
ax.plot(K, avgWithinSS, 'b*-')
ax.plot(K[kIdx], avgWithinSS[kIdx], marker='o', markersize=12,
markeredgewidth=2, markeredgecolor='r', markerfacecolor='None')
plt.grid(True)
plt.xlabel('Number of clusters')
plt.ylabel('Average within-cluster sum of squares')
plt.title('Elbow for KMeans clustering')
fig = plt.figure()
ax = fig.add_subplot(111)
ax.plot(K, bss/tss*100, 'b*-')
ax.plot(K[kIdx], bss[kIdx]/tss*100, marker='o', markersize=12,
markeredgewidth=2, markeredgecolor='r', markerfacecolor='None')
plt.grid(True)
plt.xlabel('Number of clusters')
plt.ylabel('Percentage of variance explained')
plt.title('Elbow for KMeans clustering')
# Re-do K-means clustering using optimum values
np.random.seed(5)
optimum_k = 8
kmeans = KMeans(n_clusters=optimum_k)
kmeans.fit(sub_cluster_data.as_matrix())
labels = kmeans.labels_
centroids = kmeans.cluster_centers_
# Use PCA to identify the key dimensionality in the data
from sklearn.decomposition import PCA
pca = PCA(n_components=5)
pca.fit(sub_cluster_data)
pca.explained_variance_ratio_
# The first two principal components explain ~91% of the variation, so could simplify to 2-d
# Transform the raw data and the identified centroids on to a 2-d plane
np.random.seed(5)
pca = PCA(n_components=2)
pca.fit(sub_cluster_data)
points_pc = pca.transform(sub_cluster_data)
centroids_pc = pca.transform(centroids)
# Plot
fig = plt.figure(figsize=(12,12))
ax = fig.add_subplot(111)
ax.scatter(points_pc[:,0],points_pc[:,1], c=labels, alpha=0.75)
ax.scatter(centroids_pc[:,0],centroids_pc[:,1],marker="+",s=200,linewidths=3,c="k")
for label, x, y in zip(range(optimum_k), centroids_pc[:,0],centroids_pc[:,1]):
plt.annotate(
label,
xy = (x, y), xytext = (-20, 20),
textcoords = 'offset points', ha = 'right', va = 'bottom',
bbox = dict(boxstyle = 'round,pad=0.5', fc = 'yellow', alpha = 0.75),
arrowprops = dict(arrowstyle = '->', connectionstyle = 'arc3,rad=0'))
plt.grid(True)
plt.xlabel('PC 1')
plt.ylabel('PC 2')
plt.title('Clusters transformed on to principal components')
plt.show()
np.set_printoptions(precision=3)
print(centroids)
grouped = sub_cluster_data.groupby(labels)
grouped.size()
# Plot
i=0
fig = plt.figure(figsize=(12,12))
for group in range(optimum_k):
for dim in list(sub_cluster_data.columns.values):
i+=1
ax = fig.add_subplot(optimum_k,5,i)
ax.hist(grouped.get_group(group)[dim].values - grouped.get_group(group)[dim].mean(), 15)
ax.set_ylim([0,20])
plt.grid(True)
plt.title(str(group) + dim, {'verticalalignment':'top'})
plt.show()
grouped.std()
new_points = list()
labels2 = list()
for group in range(optimum_k):
new_points.append(np.random.multivariate_normal(centroids[group,:],
grouped.std().values[group,:] * np.eye(5),
100))
labels2.append(group * np.ones(100))
new_points = pandas.DataFrame(np.vstack(new_points), columns=['a','b','c','d','e'])
labels2 = np.vstack(labels2)
group=1
centroids[group,:]
grouped.std().values[group,:] * np.eye(5)
np.random.multivariate_normal(centroids[group,:],
grouped.std().values[group,:] * np.eye(5),
100)
pandas.tools.plotting.scatter_matrix(new_points, alpha=0.2, figsize=(12,12), diagonal='kde')
# Re-do K-means clustering using optimum values
np.random.seed(5)
optimum_k = 8
kmeans2 = KMeans(n_clusters=optimum_k)
kmeans2.fit(new_points)
labels2 = kmeans2.labels_
centroids2 = kmeans2.cluster_centers_
# Transform the raw data and the identified centroids on to a 2-d plane
np.random.seed(5)
pca2 = PCA(n_components=2)
pca2.fit(new_points)
points_pc2 = pca.transform(new_points)
centroids_pc2 = pca.transform(centroids2)
# Plot
fig = plt.figure(figsize=(12,12))
ax = fig.add_subplot(111)
ax.scatter(points_pc2[:,0],points_pc2[:,1], c=labels2, alpha=0.75)
ax.scatter(centroids_pc2[:,0],centroids_pc2[:,1],marker="+",s=200,linewidths=3,c="k")
for label, x, y in zip(range(optimum_k), centroids_pc2[:,0],centroids_pc2[:,1]):
plt.annotate(
label,
xy = (x, y), xytext = (-20, 20),
textcoords = 'offset points', ha = 'right', va = 'bottom',
bbox = dict(boxstyle = 'round,pad=0.5', fc = 'yellow', alpha = 0.5),
arrowprops = dict(arrowstyle = '->', connectionstyle = 'arc3,rad=0'))
plt.grid(True)
plt.xlabel('PC 1')
plt.ylabel('PC 2')
plt.title('Clusters transformed on to principal components')
plt.show()
| 0.814717 | 0.650134 |
## Analysis of Stochastic Processes ($\S$ 10.5)
If a system is always variable, but the variability is not (infinitely) predictable, then we have a [**stochastic**](https://en.wikipedia.org/wiki/Stochastic) process. Counter to what you may think, these processes can also be characterized.
Last time we saw that, even with so little data that we couldn't see the periodicity, it was still possible to apply machine learning to find the period.
In the same way, we can find "structure" in otherwise random data. For example the following light curves taken from [Moreno et al. (2019)](https://iopscience.iop.org/article/10.1088/1538-3873/ab1597) may all be random, but I think that we can agree that they are random in very different ways.

Depending on what one is trying to do, we can use a number of different tools to try to characterize the data from Auto-Covariance Functions (ACVF), Auto-Correlation Functions (ACF), Power Spectral Densities (PSD), and Structure Functions (SF). In fact, these are all slightly different ways to describe the same information.
But let's start by talking about auto-regressive (AR) and moving-average (MA) processes as building blocks of stochastic data.
## Autoregressive Models
For processes like these that are not periodic, but that nevertheless "retain memory" of previous states, we can use [autoregressive models](https://en.wikipedia.org/wiki/Autoregressive_model).
For linear regression, we are predicting the dependent variable from the independent variable $$y = mx+b.$$ For auto-regression the dependent and independent variable is the same and we are predicting a future value of $y$ based on a past (or multiple past) values of $y$: $$y_i = \phi_i y_{i-1} + \ldots,$$
where $\phi_i$ is the "lag coefficient".
A random walk is an example of such a process; every new value is given by the preceeding value plus some noise:
$$y_i = y_{i-1} + \epsilon_i.$$
That is, $\phi_i=1$. If $\phi_i>1$ then it is known as a geometric random walk, which is typical of the stock market (a largely random process that nevertheless increases with time [on long time scales]). (So, when you interview for a quant position on Wall Street, you tell them that you are an expert in using autoregressive geometric random walks to model stochastic processes.)
In the random walk case above, each new value depends only on the immediately preceeding value. But we can generalized this to include $p$ values:
$$y_i = \sum_{j=1}^pa_jy_{i-j} + \epsilon_i$$
We refer to this as an [**autoregressive (AR)**](https://en.wikipedia.org/wiki/Autoregressive_model) process of order $p$: $AR(p)$. For a random walk, we have $p=1$, and $a_1=1$ (where now I'm using $a$ instead of $\phi$ to match the notation of the book).
If the data are drawn from a "stationary" process (one where it doesn't matter what region of the light curve you sample [so long as it is representative], the stock market *not* being an example of a stationary process), the $a_j$ satisfy certain conditions.
One thing that we might do then is ask whether a system is more consistent with $a_1=0$ or $a_1=1$ (noise vs. a random walk).
We'll try to understand what an AR process is doing by following what happens after introducing a single "spike" at time, $t=3$. These plots show what happens over the next 17 time periods both an AR(1) and an AR(2) process in with different lag coefficients; see [Moreno et al. (2019)](https://iopscience.iop.org/article/10.1088/1538-3873/ab1597)

Below are some example light curves for specific $AR(p)$ processes. These are similar to the above, but now instead of one noise spike we have many spread over time. In the first example, $AR(0)$, the light curve is simply responding to noise fluctuations. In the second example, $AR(1)$, the noise fluctuation responses are persisting for slightly longer as the next time step depends positively on the time before. For the 3rd example, nearly the full effect of the noise spike from the previous time step is applied again, giving particularly long and high chains of peaks and valleys. In the 4th example, $AR(2)$, we have long, but low chains of peaks and valleys as a spike persists for an extra time step. Finally, in the 5th example, the response of a spike in the second time step has the opposite sign as for the first time step, and both have large coefficients, so the peaks and valleys are both quite high and quite narrowly separated.

A [**moving average (MA)**](https://en.wikipedia.org/wiki/Moving-average_model) process is similar to an AR process, but instead the value at each time step depends not on the *value* of previous time step, but rather the *perturbations* from previous time steps. (In finance, instead of inter-day stock prices, we are now modeling intra-day stock prices.) MA processes are defined by
$$y_i = \epsilon_i + \sum_{j=1}^qb_j\epsilon_{i-j}.$$
So, for example, an MA(q=1) process would look like
$$y_i = \epsilon_{i} + b_1\epsilon_{i-1},$$
whereas an AR(p=2) process would look like
$$y_i = a_1y_{i-1} + a_2y_{i-2} + \epsilon_i$$
So, in an $MA$ process a shock affects only the current value and $q$ values into the future. In an $AR$ process a shock affects *all* future values. These two plots from Moreno et al. show the difference between an AR(1) process and an MA(1) process:


We can also combine AR and MA processes in order to characterize more complicated data. For example an ARMA(2,1) model combines AR(2) and MA(1):
$$y_i = a_1y_{i-1} + a_2y_{i-2} + \epsilon_i + b_1 \epsilon_{i-1}.$$
Below is some code and a plot that illustrates this. Try changing the coefficients (in the yAR, yMA, and yARMA equations) and see what happens. (You may need to change the plot limits depending on your choices.)
```
%matplotlib inline
import numpy as np
from matplotlib import pyplot as plt
from matplotlib.ticker import MultipleLocator
N=10
#epsilon = np.array([0,0,0,1,0,0,0,0,0,0,0,0])
epsilon = np.zeros(N+2)
epsilon[3] = 1
yAR=np.zeros(N+2)
yMA=np.zeros(N+2)
yARMA=np.zeros(N+2)
for i in np.arange(N)+2:
# Complete
yAR[i] = 0.5*yAR[i-1] + 0.2*yAR[i-2] + epsilon[i]
yMA[i] = epsilon[i] + 0.5*epsilon[i-1] + 0.5*epsilon[i-2]
yARMA[i] = 0.25*yARMA[i-2] + 0.5*yARMA[i-1] + epsilon[i] + 0.5*epsilon[i-1]
#print i, yAR[i], yMA[i]
fig = plt.figure(figsize=(6, 6))
t = np.arange(len(yAR))
plt.plot(t,yAR,label="AR(2), a1=0.5, a2=0.2")
plt.plot(t,yMA,label="MA(2), b1=0.5, b2=0.5")
plt.plot(t,yARMA,label="ARMA(2,1), a1=0.5, a2=0.25, b1=0.5",zorder=0)
plt.xlabel("t")
plt.ylabel("y")
plt.legend(loc="upper right",prop={'size':8})
plt.ylim([0,1.1])
ax = plt.axes()
ax.xaxis.set_major_locator(plt.MultipleLocator(1.0))
plt.show()
```
I found these videos particularly helpful in trying to understand these processes.
[MA(1)](https://www.youtube.com/watch?v=lUhtcP2SUsg)
[AR(1)](https://www.youtube.com/watch?v=AN0a58F6cxA)
[ARMA(1,1)](https://www.youtube.com/watch?v=Pg0RnP1uLVc)
### CARMA Models
$AR$ and $ARMA$ models assume evenly sampled time-series data. However, we can extend this to unevenly sampled data with $CAR$ or $CARMA$ processes, where the $C$ stands for *continuous*.
A $CAR(1)$ process is described by a stochastic differential equation which includes a damping term that pushes $y(t)$ back towards the mean, so it is also called a **damped random walk (DRW)**. For evenly sampled data a CAR(1) process is the same as an AR(1) process with $a_1=\exp(-1/\tau)$. That is, the next value is the previous value times the damping factor (plus noise).
---
I'm going to skip the steps that would take us smoothly from AR, MA, ARMA, and CARMA modeling to ACVF, ACF, PSD, and SFs, but you can read about it in Moreno et al. (2019). However, I'll also introduce these with a simple illustration.
Take a (stochastically varying) quasar which has both *line* and *continuum* emission and where the line emission is stimulated by the continuum. Since there is a physical separation between the regions that produce each type of emission, we get a delay between the light curves as can be seen here:
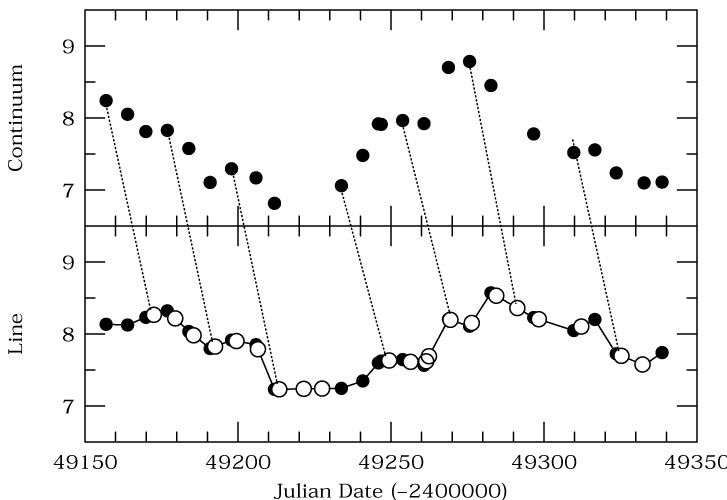
You can think of this as looking a (varying) light both directly (top plot) and reflected off of a mirror that is some distance away from the light (bottom). The light is (largely) the same, just shifted by the light travel time for the extra distance covered.
Let's compute the [correlation function](https://en.wikipedia.org/wiki/Correlation_function) for this process. A correlation function (Ivezic $\S$ 6.5) gives us information about the time delay between 2 processes. If one time series is derived from another simply by shifting the time axis by $t_{\rm lag}$, then their correlation function will have a peak at $\Delta t = t_{\rm lag}$.
The correlation function between $f(t)$, and $g(t)$ is defined as
$${\rm CF}(\Delta t) = \frac{\lim_{T\rightarrow \infty}\frac{1}{T}\int_T f(t)g(t+\Delta t)dt }{\sigma_f \sigma_g}$$
Computing the correlation function is basically the mathematical processes of sliding the two curves over each other and computing the degree of similarity for each step in time. The peak of the correlation function reveals the time delay between the processes. Below we have the correlation function of the line and continuum emission from a quasar, which reveals a $\sim$ 15 day delay between the two.
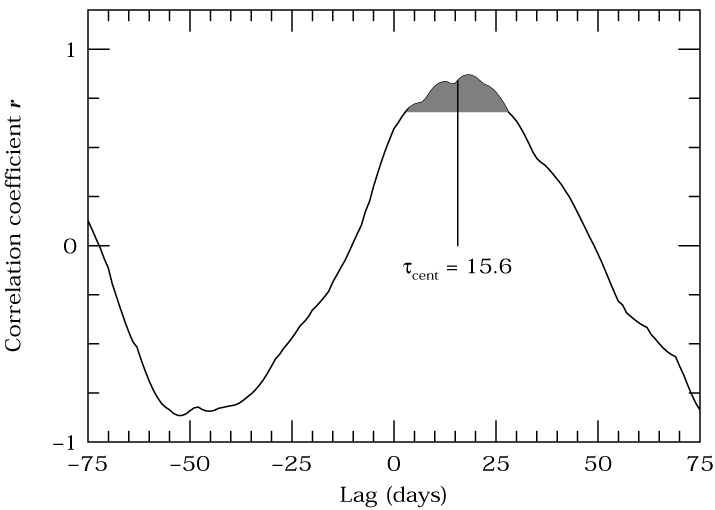
In an **autocorrelation function (ACF)**, we take our correlation function from above and set $f(t)= g(t)$. Then we are revealing information about variability timescales present in the process itself.
### What can the ACF tell us?
If the values of $y$ are uncorrelated, then ACF$(\Delta t)=0$ (except for ACF$(0)=1$, by definition).
For processes that "retain memory" of previous states only for some characteristic time $\tau$, the ACF will vanish for $\Delta t \gg \tau$ -- much in the same way that an AR response dies out after some time.
Turning that around, the predictability of future behavior of such a process is limited to times up to $\sim \tau$; you have to "let the process run" to know how it will behave at times longer than that.
### Power Spectral Density
The Fourier Transform of an ACF is the [Power Spectral Density (PSD)](https://en.wikipedia.org/wiki/Spectral_density). So, the PSD is an analysis in frequency space and the ACF is in time space (the Wiener-Khinchin theorem describes the
fact that the ACF and PSD are a Fourier pair).
For example, for a sinusoidal function in time space, the ACF will have the same period, $T$, as the function. Conversly, the PSD in frequency space will be a $\delta$ function centered on $\omega = 1/2\pi T$ (think about our Lomb-Scargle periodogram for a sine wave from last time).
### The Structure Function
The *structure function* is another quantity that is frequently used in astronomy and is related to the ACF:
$${\rm SF}(\Delta t) = {\rm SF}_\infty[1 - {\rm ACF}(\Delta t)]^{1/2},$$
where ${\rm SF}_\infty$ is the standard deviation of the time series as evaluated on timescales much larger than any charateristic timescale, $\tau$.
The ACF for a DRW is given by
$$ ACF(t) = \exp(-t/\tau),$$
where $\tau$ is the characteristic timescale (i.e., the damping timescale). Remember that a DRW modeled as an AR(1) has $a_1=\exp(-1/\tau)$.
The structure function can be written as
$$ SF(t) = SF_{\infty}[1-\exp(-t/\tau)]^{1/2}.$$
The PSD is for a DRW is
$$ PSD(f) = \frac{\tau^2 SF_{\infty}^2}{1+(2\pi f \tau)^2}.$$
The ACF example above was an example of a DRW: the light curve is strongly correlated a short timescales, but uncorrelated at long timescales. This is observed in optical variability of quasar continuum light; in fact, it works so well that one can use this model to distinguish quasars from stars, based solely on the variability they exhibit.
More generically, if ${\rm SF} \propto t^{\alpha}$, then ${\rm PSD} \propto \frac{1}{f^{1+2\alpha}}$.
So an analysis of a stochastic system can be done with either the ACF, SF, or PSD.
The structure function is interesting because it's equal to the standard deviation of the distribution of the differences of $y(t_2) - y(t_1)$ evaluated at many different $t_1$ and $t_2$ (i.e., with a time lag of $\Delta t = t_2 - t_1$), and divided by $\sqrt 2$.
This is of practical use: if I have a series of observations $y_i$ (taken at random times $t_i$) it's relatively straighforward to compute the structure function.
### Structure Function for Dampled Random Walk

<img src="figures/MacLeod2010.png" alt="Drawing" style="width: 500px;"/>
---
### Different stochastic processes
* A stochastic process with a $1/f^2\,$ PSD is known as random walk (if discrete) or Brownian motion (or, more accurately, Wiener process) if continuous. These physically occur when the value being observed is subjected to a series of independent changes of similar size. It's also sometimes called as "red noise". Quasar variability exhibits $1/f^2$ properties at high frequencies (that is, short time scales, below a year or so).
* Stochastic processes with $1/f\,$ PSDs are sometimes called "long-term memory processes" (also sometimes know as "pink noise"). They have equal energy at all octaves (or over any other logarithmic frequency interval). This type of process has infinite variance and an undefined mean (similar to a Lorentzian distribution).
* A process with a constant PSD is frequently referred to as "white noise" -- it has equal intensity at all frequencies. This is a process with no memory -- each measurement is independent of all others.
Let's play with the code below to make a plot of counts vs. time and of the PSD vs. frequency for both a $1/f$ and a $1/f^2$ process.
```
%matplotlib inline
import numpy as np
from matplotlib import pyplot as plt
from astroML.time_series import generate_power_law
from astroML.fourier import PSD_continuous
N = 2014
dt = 0.01
betaRed = ___ # Complete
betaPink = ___ # Complete
betaWhite = ___ # Complete
t = dt * np.arange(N)
yRed = generate_power_law(N, dt, betaRed)
yPink = generate_power_law(N, dt, ____) # Complete
yWhite = generate_power_law(N, dt, ____)/10.0 # Complete
fRed, PSDred = PSD_continuous(t, ___) # Complete
___, ___ = PSD_continuous(t, ___) # Complete
___, ___ = PSD_continuous(t, yWhite) # Complete
fig = plt.figure(figsize=(8, 4))
ax1 = fig.add_subplot(121)
ax1.plot(t, yWhite, c='Grey')
ax1.plot(t, yPink, c='Pink')
ax1.plot(t, yRed, '-r')
ax1.set_xlim(0, 10)
ax2 = fig.add_subplot(122, xscale='log', yscale='log')
ax2.plot(fWhite, PSDwhite, c='Grey')
ax2.plot(fPink, PSDpink, c='Pink')
ax2.plot(fRed, PSDred, '-r')
ax2.set_xlim(1E-1, 60)
ax2.set_ylim(1E-11, 1E-3)
plt.show()
```
You should find that, because the power at high frequency is larger for $1/f$, that light curve will look noisier.
We can even *hear* the difference:
[https://www.youtube.com/watch?v=5KdRL2fkxgU&feature=emb_logo](https://www.youtube.com/watch?v=5KdRL2fkxgU&feature=emb_logo)
Let's compare these in both time and frequency space.
```
# Ivezic v2, Figure 10.29, edits by GTR
# Author: Jake VanderPlas
# License: BSD
# The figure produced by this code is published in the textbook
# "Statistics, Data Mining, and Machine Learning in Astronomy" (2013)
# For more information, see http://astroML.github.com
# To report a bug or issue, use the following forum:
# https://groups.google.com/forum/#!forum/astroml-general
import numpy as np
from matplotlib import pyplot as plt
from astroML.time_series import generate_power_law
from astroML.fourier import PSD_continuous
#----------------------------------------------------------------------
# This function adjusts matplotlib settings for a uniform feel in the textbook.
# Note that with usetex=True, fonts are rendered with LaTeX. This may
# result in an error if LaTeX is not installed on your system. In that case,
# you can set usetex to False.
if "setup_text_plots" not in globals():
from astroML.plotting import setup_text_plots
setup_text_plots(fontsize=8, usetex=True)
N = 1024
dt = 0.01
factor = 100
t = dt * np.arange(N)
random_state = np.random.RandomState(1)
fig = plt.figure(figsize=(10, 7.5))
fig.subplots_adjust(wspace=0.05)
for i, beta in enumerate([1.0, 2.0]):
# Generate the light curve and compute the PSD
x = factor * generate_power_law(N, dt, beta, random_state=random_state)
f, PSD = PSD_continuous(t, x)
# First axes: plot the time series
ax1 = fig.add_subplot(221 + i)
ax1.plot(t, x, '-k')
ax1.text(0.95, 0.05, r"$P(f) \propto f^{-%i}$" % beta,
ha='right', va='bottom', transform=ax1.transAxes)
ax1.set_xlim(0, 10.24)
ax1.set_ylim(-1.5, 1.5)
ax1.set_xlabel(r'$t$')
# Second axes: plot the PSD
ax2 = fig.add_subplot(223 + i, xscale='log', yscale='log')
ax2.plot(f, PSD, '-k')
ax2.plot(f[1:], (factor * dt) ** 2 * (2 * np.pi * f[1:]) ** -beta, '--k')
ax2.set_xlim(1E-1, 60)
ax2.set_ylim(1E-6, 1E1)
ax2.set_xlabel(r'$f$')
if i == 1:
ax1.yaxis.set_major_formatter(plt.NullFormatter())
ax2.yaxis.set_major_formatter(plt.NullFormatter())
else:
ax1.set_ylabel(r'${\rm counts}$')
ax2.set_ylabel(r'$PSD(f)$')
plt.show()
```
Remember that the PSD of a DRW looks like:
$$ PSD(f) = \frac{\tau^2 SF_{\infty}^2}{1+(2\pi f \tau)^2},$$
which means that a DRW is a $1/f^2$ process at high frequency. The *damped* part comes from flattening of the PSD at low frequency (not shown in the plot above).
### ACF for Unevenly Sampled Data
astroML also has tools for computing the ACF of *unevenly sampled* data using two different (Scargle) and (Edelson & Krolik) methods: [http://www.astroml.org/modules/classes.html#module-astroML.time_series](http://www.astroml.org/modules/classes.html#module-astroML.time_series)
One of the tools is for generating a **damped random walk (DRW)**, which is a process that "remembers" its history only for a characteristic time, $\tau$. The ACF vanishes for $\Delta t \gg \tau$.
```
# Syntax for EK and Scargle ACF computation
import numpy as np
from astroML.time_series import generate_damped_RW
from astroML.time_series import ACF_scargle, ACF_EK
t = np.arange(0,1000)
y = generate_damped_RW(t, tau=300)
dy = 0.1
y = np.random.normal(y,dy)
ACF_scargle, bins_scargle = ACF_scargle(t,y,dy)
ACF_EK, ACF_err_EK, bins_EK = ACF_EK(t,y,dy)
```
Figure 10.30 below gives an example of an ACF for a DRW, which mimics the variability that we might see from a quasar.
```
# Ivezic v2, Figure 10.30
# License: BSD
# The figure produced by this code is published in the textbook
# "Statistics, Data Mining, and Machine Learning in Astronomy" (2013)
# For more information, see http://astroML.github.com
# To report a bug or issue, use the following forum:
# https://groups.google.com/forum/#!forum/astroml-general
import numpy as np
from matplotlib import pyplot as plt
from astroML.time_series import lomb_scargle, generate_damped_RW
from astroML.time_series import ACF_scargle, ACF_EK
#----------------------------------------------------------------------
# This function adjusts matplotlib settings for a uniform feel in the textbook.
# Note that with usetex=True, fonts are rendered with LaTeX. This may
# result in an error if LaTeX is not installed on your system. In that case,
# you can set usetex to False.
if "setup_text_plots" not in globals():
from astroML.plotting import setup_text_plots
setup_text_plots(fontsize=12, usetex=True)
#------------------------------------------------------------
# Generate time-series data:
# we'll do 1000 days worth of magnitudes
t = np.arange(0, 1E3)
z = 2.0
tau = 300
tau_obs = tau / (1. + z)
np.random.seed(6)
y = generate_damped_RW(t, tau=tau, z=z, xmean=20)
# randomly sample 100 of these
ind = np.arange(len(t))
np.random.shuffle(ind)
ind = ind[:100]
ind.sort()
t = t[ind]
y = y[ind]
# add errors
dy = 0.1
y_obs = np.random.normal(y, dy)
#------------------------------------------------------------
# compute ACF via scargle method
C_S, t_S = ACF_scargle(t, y_obs, dy,
n_omega=2. ** 12, omega_max=np.pi / 5.0)
ind = (t_S >= 0) & (t_S <= 500)
t_S = t_S[ind]
C_S = C_S[ind]
#------------------------------------------------------------
# compute ACF via E-K method
C_EK, C_EK_err, bins = ACF_EK(t, y_obs, dy, bins=np.linspace(0, 500, 51))
t_EK = 0.5 * (bins[1:] + bins[:-1])
#------------------------------------------------------------
# Plot the results
fig = plt.figure(figsize=(8, 8))
# plot the input data
ax = fig.add_subplot(211)
ax.errorbar(t, y_obs, dy, fmt='.k', lw=1)
ax.set_xlabel('t (days)')
ax.set_ylabel('observed flux')
# plot the ACF
ax = fig.add_subplot(212)
ax.plot(t_S, C_S, '-', c='gray', lw=1,
label='Scargle')
ax.errorbar(t_EK, C_EK, C_EK_err, fmt='.k', lw=1,
label='Edelson-Krolik')
ax.plot(t_S, np.exp(-abs(t_S) / tau_obs), '-k', label='True')
ax.legend(loc=3)
ax.plot(t_S, 0 * t_S, ':', lw=1, c='gray')
ax.set_xlim(0, 500)
ax.set_ylim(-1.0, 1.1)
ax.set_xlabel('t (days)')
ax.set_ylabel('ACF(t)')
plt.show()
```
AstroML has [time series](http://www.astroml.org/modules/classes.html#module-astroML.time_series) and [Fourier](http://www.astroml.org/modules/classes.html#module-astroML.fourier) tools for generating light curves drawn from a power law in frequency space. Note that these tools define $\beta = 1+2\alpha$ ($\beta=2$ for a random walk).
## Regression and Classification
Regression and Classification for these stochastic models is just the same as before. With our model and model parameters, we can predict future values via regression, or look for similarities/differences as a function of the model parameters via clustering (unsupervised) or classification (supervised). Similarly, we can apply dimensionality reduction techniques to help visualize results from high-dimensional models.
|
github_jupyter
|
%matplotlib inline
import numpy as np
from matplotlib import pyplot as plt
from matplotlib.ticker import MultipleLocator
N=10
#epsilon = np.array([0,0,0,1,0,0,0,0,0,0,0,0])
epsilon = np.zeros(N+2)
epsilon[3] = 1
yAR=np.zeros(N+2)
yMA=np.zeros(N+2)
yARMA=np.zeros(N+2)
for i in np.arange(N)+2:
# Complete
yAR[i] = 0.5*yAR[i-1] + 0.2*yAR[i-2] + epsilon[i]
yMA[i] = epsilon[i] + 0.5*epsilon[i-1] + 0.5*epsilon[i-2]
yARMA[i] = 0.25*yARMA[i-2] + 0.5*yARMA[i-1] + epsilon[i] + 0.5*epsilon[i-1]
#print i, yAR[i], yMA[i]
fig = plt.figure(figsize=(6, 6))
t = np.arange(len(yAR))
plt.plot(t,yAR,label="AR(2), a1=0.5, a2=0.2")
plt.plot(t,yMA,label="MA(2), b1=0.5, b2=0.5")
plt.plot(t,yARMA,label="ARMA(2,1), a1=0.5, a2=0.25, b1=0.5",zorder=0)
plt.xlabel("t")
plt.ylabel("y")
plt.legend(loc="upper right",prop={'size':8})
plt.ylim([0,1.1])
ax = plt.axes()
ax.xaxis.set_major_locator(plt.MultipleLocator(1.0))
plt.show()
%matplotlib inline
import numpy as np
from matplotlib import pyplot as plt
from astroML.time_series import generate_power_law
from astroML.fourier import PSD_continuous
N = 2014
dt = 0.01
betaRed = ___ # Complete
betaPink = ___ # Complete
betaWhite = ___ # Complete
t = dt * np.arange(N)
yRed = generate_power_law(N, dt, betaRed)
yPink = generate_power_law(N, dt, ____) # Complete
yWhite = generate_power_law(N, dt, ____)/10.0 # Complete
fRed, PSDred = PSD_continuous(t, ___) # Complete
___, ___ = PSD_continuous(t, ___) # Complete
___, ___ = PSD_continuous(t, yWhite) # Complete
fig = plt.figure(figsize=(8, 4))
ax1 = fig.add_subplot(121)
ax1.plot(t, yWhite, c='Grey')
ax1.plot(t, yPink, c='Pink')
ax1.plot(t, yRed, '-r')
ax1.set_xlim(0, 10)
ax2 = fig.add_subplot(122, xscale='log', yscale='log')
ax2.plot(fWhite, PSDwhite, c='Grey')
ax2.plot(fPink, PSDpink, c='Pink')
ax2.plot(fRed, PSDred, '-r')
ax2.set_xlim(1E-1, 60)
ax2.set_ylim(1E-11, 1E-3)
plt.show()
# Ivezic v2, Figure 10.29, edits by GTR
# Author: Jake VanderPlas
# License: BSD
# The figure produced by this code is published in the textbook
# "Statistics, Data Mining, and Machine Learning in Astronomy" (2013)
# For more information, see http://astroML.github.com
# To report a bug or issue, use the following forum:
# https://groups.google.com/forum/#!forum/astroml-general
import numpy as np
from matplotlib import pyplot as plt
from astroML.time_series import generate_power_law
from astroML.fourier import PSD_continuous
#----------------------------------------------------------------------
# This function adjusts matplotlib settings for a uniform feel in the textbook.
# Note that with usetex=True, fonts are rendered with LaTeX. This may
# result in an error if LaTeX is not installed on your system. In that case,
# you can set usetex to False.
if "setup_text_plots" not in globals():
from astroML.plotting import setup_text_plots
setup_text_plots(fontsize=8, usetex=True)
N = 1024
dt = 0.01
factor = 100
t = dt * np.arange(N)
random_state = np.random.RandomState(1)
fig = plt.figure(figsize=(10, 7.5))
fig.subplots_adjust(wspace=0.05)
for i, beta in enumerate([1.0, 2.0]):
# Generate the light curve and compute the PSD
x = factor * generate_power_law(N, dt, beta, random_state=random_state)
f, PSD = PSD_continuous(t, x)
# First axes: plot the time series
ax1 = fig.add_subplot(221 + i)
ax1.plot(t, x, '-k')
ax1.text(0.95, 0.05, r"$P(f) \propto f^{-%i}$" % beta,
ha='right', va='bottom', transform=ax1.transAxes)
ax1.set_xlim(0, 10.24)
ax1.set_ylim(-1.5, 1.5)
ax1.set_xlabel(r'$t$')
# Second axes: plot the PSD
ax2 = fig.add_subplot(223 + i, xscale='log', yscale='log')
ax2.plot(f, PSD, '-k')
ax2.plot(f[1:], (factor * dt) ** 2 * (2 * np.pi * f[1:]) ** -beta, '--k')
ax2.set_xlim(1E-1, 60)
ax2.set_ylim(1E-6, 1E1)
ax2.set_xlabel(r'$f$')
if i == 1:
ax1.yaxis.set_major_formatter(plt.NullFormatter())
ax2.yaxis.set_major_formatter(plt.NullFormatter())
else:
ax1.set_ylabel(r'${\rm counts}$')
ax2.set_ylabel(r'$PSD(f)$')
plt.show()
# Syntax for EK and Scargle ACF computation
import numpy as np
from astroML.time_series import generate_damped_RW
from astroML.time_series import ACF_scargle, ACF_EK
t = np.arange(0,1000)
y = generate_damped_RW(t, tau=300)
dy = 0.1
y = np.random.normal(y,dy)
ACF_scargle, bins_scargle = ACF_scargle(t,y,dy)
ACF_EK, ACF_err_EK, bins_EK = ACF_EK(t,y,dy)
# Ivezic v2, Figure 10.30
# License: BSD
# The figure produced by this code is published in the textbook
# "Statistics, Data Mining, and Machine Learning in Astronomy" (2013)
# For more information, see http://astroML.github.com
# To report a bug or issue, use the following forum:
# https://groups.google.com/forum/#!forum/astroml-general
import numpy as np
from matplotlib import pyplot as plt
from astroML.time_series import lomb_scargle, generate_damped_RW
from astroML.time_series import ACF_scargle, ACF_EK
#----------------------------------------------------------------------
# This function adjusts matplotlib settings for a uniform feel in the textbook.
# Note that with usetex=True, fonts are rendered with LaTeX. This may
# result in an error if LaTeX is not installed on your system. In that case,
# you can set usetex to False.
if "setup_text_plots" not in globals():
from astroML.plotting import setup_text_plots
setup_text_plots(fontsize=12, usetex=True)
#------------------------------------------------------------
# Generate time-series data:
# we'll do 1000 days worth of magnitudes
t = np.arange(0, 1E3)
z = 2.0
tau = 300
tau_obs = tau / (1. + z)
np.random.seed(6)
y = generate_damped_RW(t, tau=tau, z=z, xmean=20)
# randomly sample 100 of these
ind = np.arange(len(t))
np.random.shuffle(ind)
ind = ind[:100]
ind.sort()
t = t[ind]
y = y[ind]
# add errors
dy = 0.1
y_obs = np.random.normal(y, dy)
#------------------------------------------------------------
# compute ACF via scargle method
C_S, t_S = ACF_scargle(t, y_obs, dy,
n_omega=2. ** 12, omega_max=np.pi / 5.0)
ind = (t_S >= 0) & (t_S <= 500)
t_S = t_S[ind]
C_S = C_S[ind]
#------------------------------------------------------------
# compute ACF via E-K method
C_EK, C_EK_err, bins = ACF_EK(t, y_obs, dy, bins=np.linspace(0, 500, 51))
t_EK = 0.5 * (bins[1:] + bins[:-1])
#------------------------------------------------------------
# Plot the results
fig = plt.figure(figsize=(8, 8))
# plot the input data
ax = fig.add_subplot(211)
ax.errorbar(t, y_obs, dy, fmt='.k', lw=1)
ax.set_xlabel('t (days)')
ax.set_ylabel('observed flux')
# plot the ACF
ax = fig.add_subplot(212)
ax.plot(t_S, C_S, '-', c='gray', lw=1,
label='Scargle')
ax.errorbar(t_EK, C_EK, C_EK_err, fmt='.k', lw=1,
label='Edelson-Krolik')
ax.plot(t_S, np.exp(-abs(t_S) / tau_obs), '-k', label='True')
ax.legend(loc=3)
ax.plot(t_S, 0 * t_S, ':', lw=1, c='gray')
ax.set_xlim(0, 500)
ax.set_ylim(-1.0, 1.1)
ax.set_xlabel('t (days)')
ax.set_ylabel('ACF(t)')
plt.show()
| 0.542379 | 0.984869 |
# Colab initialization
- install the pipeline in the colab runtime
- download files neccessary for this example
```
!pip3 install -U pip > /dev/null
!pip3 install -U bio_embeddings[all] > /dev/null
!wget http://data.bioembeddings.com/public/embeddings/notebooks/pipeline_output_example/disprot/reduced_embeddings_file.h5 --output-document reduced_embeddings_file.h5
!wget http://data.bioembeddings.com/public/embeddings/notebooks/pipeline_output_example/disprot/mapping_file.csv --output-document mapping_file.csv
```
# Reindex the embeddings generated from the pipeline
In order to avoid fauly ids from the FASTA headers, the pipeline automatically generates ids for the sequences passed. At the end of a pipeline run, you might want to attempt to re-index these. The pipeline provides a convenience function that does this in-place (changes the original file!)
```
## This is just to get some logging output in the Notebook
import logging
logging.basicConfig(level=logging.INFO, format="%(asctime)s %(levelname)s %(message)s")
```
When executing pipeline runs, your input sequences will get assigned a new internal identifier. This identifier corresponds to the md5 hash of the sequence. We do this, becuase for storing and processing purposes we need unique strings as identifiers, and unfortunately, some FASTA files contain invalid characters in the header.
Nevertheless, sometimes you may want to convert the keys contained in the h5 files produces from the pipeline back from the internal ids to their original id as in the FASTA header of the input sequence.
We produce a mapping_file.csv which shows this mapping (the first, unnamed column represents the sequence' md5 hash, while the column `original_id` represents the extracted id from the input FASTA)
This operation can be dangerous, because if the `original_id` contains invalid characters or is empty, the h5 file will be corrupted.
Nevertheless, we make a helper function available which converts the internal ids back to the original ids **in place**, meaning that the h5 file will be directly modified (this is meant to avoid duplication of large h5 files, but with the risk of corrupting the original file. Please: only perform this operation if you are sure about what you are doing, and if it's strictly neccessary!)
```
import h5py
from bio_embeddings.utilities import reindex_h5_file
# Let's check the keys of our h5 file:
with h5py.File("reduced_embeddings_file.h5", "r") as h5_file:
for key in h5_file.keys():
print(key,)
# In place re-indexing of h5 file
reindex_h5_file("reduced_embeddings_file.h5", "mapping_file.csv")
# Let's check the new keys of our h5 file:
with h5py.File("reduced_embeddings_file.h5", "r") as h5_file:
for key in h5_file.keys():
print(key,)
```
|
github_jupyter
|
!pip3 install -U pip > /dev/null
!pip3 install -U bio_embeddings[all] > /dev/null
!wget http://data.bioembeddings.com/public/embeddings/notebooks/pipeline_output_example/disprot/reduced_embeddings_file.h5 --output-document reduced_embeddings_file.h5
!wget http://data.bioembeddings.com/public/embeddings/notebooks/pipeline_output_example/disprot/mapping_file.csv --output-document mapping_file.csv
## This is just to get some logging output in the Notebook
import logging
logging.basicConfig(level=logging.INFO, format="%(asctime)s %(levelname)s %(message)s")
import h5py
from bio_embeddings.utilities import reindex_h5_file
# Let's check the keys of our h5 file:
with h5py.File("reduced_embeddings_file.h5", "r") as h5_file:
for key in h5_file.keys():
print(key,)
# In place re-indexing of h5 file
reindex_h5_file("reduced_embeddings_file.h5", "mapping_file.csv")
# Let's check the new keys of our h5 file:
with h5py.File("reduced_embeddings_file.h5", "r") as h5_file:
for key in h5_file.keys():
print(key,)
| 0.431105 | 0.700908 |
```
%run data.py
```
# Traffic Collison Data
### NYC Open Data: Motor Vehicle Collisions (Filter data: 2019.9.1 to today)
```
collison_url = "https://data.cityofnewyork.us/resource/h9gi-nx95.csv?$select=crash_date,borough,zip_code,collision_id&$where=crash_date>'2019-09-01T00:00:00.000'&$limit=500000"
collison = pd.read_csv(collison_url)
collison.count()
collison.head(10)
collison['zip_code'].fillna(0, inplace=True)
collisonpd = collison.astype({"crash_date":'datetime64[ns]', "borough":str, "zip_code":int, "collision_id":int})
collison_df = spark.createDataFrame(collisonpd)
collison_df.createOrReplaceTempView("collisonT")
collison = spark.sql("""
SELECT MONTH(crash_date) AS month,
COUNT(*) AS num_of_crash
FROM collisonT
GROUP BY month
ORDER BY month
""")
collison = collison.withColumn("month_name",
f.when(f.col('month') == 1, "2020-01")\
.when(f.col('month') == 2, "2020-02")\
.when(f.col('month') == 3, "2020-03")\
.when(f.col('month') == 4, "2020-04")\
.when(f.col('month') == 5, "2020-05")\
.when(f.col('month') == 9, "2019-09")\
.when(f.col('month') == 10, "2019-10")\
.when(f.col('month') == 11, "2019-11")\
.when(f.col('month') == 12, "2019-12"))
```
### Average number of crash before Covid-19
```
#Covid-19 started in March 2020
average = collison.filter('month !=3 and month !=4 and month !=5').agg({"num_of_crash":"avg"})
avg_num = average.rdd.map(list)
avg_num.take(6)
avg= avg_num.take(1)[0][0]
avg
print("Average number of car crashes before Covid-19: ", '%.2f' % avg)
```
### Current month car crash number and falling rate
```
collison = collison.filter('month !=5')
collison.createOrReplaceTempView("latestT")
latest = spark.sql("SELECT num_of_crash FROM latestT WHERE month_name = \
(SELECT max(month_name) FROM latestT)")
latest = latest.rdd.map(list)
latest = latest.take(1)[0][0]
falling_rate = '%.2f' % ((avg - latest)/ avg*100)+"%"
#falling_rate = (avg - latest)/ avg
print("Current month number of car crashes: ",'%.2f' %latest)
print("Car carsh falling rate: ", falling_rate)
#falling_rate = falling_rate*100
collisonpd = collison.select("month_name","num_of_crash").orderBy("month_name").toPandas()
collisonpd
import matplotlib.pyplot as plt
from pandas.plotting import register_matplotlib_converters
register_matplotlib_converters()
plt.plot('month_name','num_of_crash', data=collisonpd, marker='', color='lightseagreen')
plt.title('COVID-19 Impact on Traffic Collision',fontsize = 15)
rate = "↓" + falling_rate
bbox_props = dict(boxstyle="round", facecolor = "white")
plt.text(0, 11000, rate, size = 15, color = "lightseagreen", bbox=bbox_props)
plt.axhline(y = avg,ls = "dashed",color = "grey")
plt.xticks( rotation=25 )
#plt.rcParams['figure.figsize'] = (10,5)
#plt.legend()
#plt.savefig('/Users/jennyzhou/Downloads/traffic.png')
```
### Car crash vs Covid-19 cases (by borough)
Car crash by borough
```
collison_borough = spark.sql("""
SELECT borough,
COUNT(*) AS num_of_crash
FROM collisonT
GROUP BY borough
ORDER BY num_of_crash desc
""")
from pyspark.sql.functions import col
df = collison_borough.withColumn("borough",
f.when(collison_borough["borough"]=='nan','unknown').when(collison_borough["borough"]=='STATEN ISLAND','staten_island').
otherwise(collison_borough["borough"]))
df=df.withColumn("borough",f.lower(f.col("borough")))
collison = df.toPandas()
```
Covid-19 cases by borough
```
cov = fetchData(nyc_his_boro_url)
cov = cov.drop(columns = ['timestamp','total'])
cov2 = cov.loc[cov['type']=='cases']
cov2 = cov2.tail(1)
cov2.set_index(["type"], inplace = True)
cov2 = cov2.stack()
cov = cov2.unstack(0)
cov['borough']=cov.index
```
merge
```
res = pd.merge(collison,cov, on='borough')
res = res.sort_values(by='cases',ascending=False)
res.set_index(["borough"], inplace = True)
res.plot.bar(rot=0,title="Car crash vs Covid-19 cases (by borough)")
```
Collison since covid-19
```
collison_march= collison_df.filter(collison_df["crash_date"] >= to_timestamp(f.lit('2020-03-12 00:00:00')))
collison_march.createOrReplaceTempView("collisonT")
collison_covid = spark.sql("""
SELECT borough,
COUNT(*) AS num_of_crash
FROM collisonT
GROUP BY borough
""")
from pyspark.sql.functions import col
cdf = collison_covid.withColumn("borough",
f.when(collison_covid["borough"]=='nan','unknown').when(collison_covid["borough"]=='STATEN ISLAND','staten_island').
otherwise(collison_covid["borough"]))
cdf=cdf.withColumn("borough",f.lower(f.col("borough")))
collison_covid = cdf.toPandas()
collison_covid.set_index(["borough"], inplace = True)
collison_covid = collison_covid.reindex(['queens','brooklyn','bronx','manhattan','staten_island','unknown'])
collison_covid.plot.bar(rot=0, title="Collison since Covid-19 by borough")
cov.set_index(["borough"], inplace = True)
cov = cov.sort_values(by='cases',ascending=False)
cov.plot.bar(rot=0,title="Covid-19 cases by borough")
res2 = pd.merge(collison_covid,cov, on='borough')
res2.head(10)
res2 = res2.sort_values(by='cases',ascending=False)
res2.plot.bar(rot=0,title="Car Crash vs Covid-19 Cases (by borough)", subplots = True)
graph = res2.plot.bar(rot=0,title="Car Crash vs Covid-19 Cases (by borough)", subplots = True)
plt.xticks( rotation=25 )
graph[0].get_figure().savefig('/Users/jennyzhou/Downloads/traffic2.png')
```
|
github_jupyter
|
%run data.py
collison_url = "https://data.cityofnewyork.us/resource/h9gi-nx95.csv?$select=crash_date,borough,zip_code,collision_id&$where=crash_date>'2019-09-01T00:00:00.000'&$limit=500000"
collison = pd.read_csv(collison_url)
collison.count()
collison.head(10)
collison['zip_code'].fillna(0, inplace=True)
collisonpd = collison.astype({"crash_date":'datetime64[ns]', "borough":str, "zip_code":int, "collision_id":int})
collison_df = spark.createDataFrame(collisonpd)
collison_df.createOrReplaceTempView("collisonT")
collison = spark.sql("""
SELECT MONTH(crash_date) AS month,
COUNT(*) AS num_of_crash
FROM collisonT
GROUP BY month
ORDER BY month
""")
collison = collison.withColumn("month_name",
f.when(f.col('month') == 1, "2020-01")\
.when(f.col('month') == 2, "2020-02")\
.when(f.col('month') == 3, "2020-03")\
.when(f.col('month') == 4, "2020-04")\
.when(f.col('month') == 5, "2020-05")\
.when(f.col('month') == 9, "2019-09")\
.when(f.col('month') == 10, "2019-10")\
.when(f.col('month') == 11, "2019-11")\
.when(f.col('month') == 12, "2019-12"))
#Covid-19 started in March 2020
average = collison.filter('month !=3 and month !=4 and month !=5').agg({"num_of_crash":"avg"})
avg_num = average.rdd.map(list)
avg_num.take(6)
avg= avg_num.take(1)[0][0]
avg
print("Average number of car crashes before Covid-19: ", '%.2f' % avg)
collison = collison.filter('month !=5')
collison.createOrReplaceTempView("latestT")
latest = spark.sql("SELECT num_of_crash FROM latestT WHERE month_name = \
(SELECT max(month_name) FROM latestT)")
latest = latest.rdd.map(list)
latest = latest.take(1)[0][0]
falling_rate = '%.2f' % ((avg - latest)/ avg*100)+"%"
#falling_rate = (avg - latest)/ avg
print("Current month number of car crashes: ",'%.2f' %latest)
print("Car carsh falling rate: ", falling_rate)
#falling_rate = falling_rate*100
collisonpd = collison.select("month_name","num_of_crash").orderBy("month_name").toPandas()
collisonpd
import matplotlib.pyplot as plt
from pandas.plotting import register_matplotlib_converters
register_matplotlib_converters()
plt.plot('month_name','num_of_crash', data=collisonpd, marker='', color='lightseagreen')
plt.title('COVID-19 Impact on Traffic Collision',fontsize = 15)
rate = "↓" + falling_rate
bbox_props = dict(boxstyle="round", facecolor = "white")
plt.text(0, 11000, rate, size = 15, color = "lightseagreen", bbox=bbox_props)
plt.axhline(y = avg,ls = "dashed",color = "grey")
plt.xticks( rotation=25 )
#plt.rcParams['figure.figsize'] = (10,5)
#plt.legend()
#plt.savefig('/Users/jennyzhou/Downloads/traffic.png')
collison_borough = spark.sql("""
SELECT borough,
COUNT(*) AS num_of_crash
FROM collisonT
GROUP BY borough
ORDER BY num_of_crash desc
""")
from pyspark.sql.functions import col
df = collison_borough.withColumn("borough",
f.when(collison_borough["borough"]=='nan','unknown').when(collison_borough["borough"]=='STATEN ISLAND','staten_island').
otherwise(collison_borough["borough"]))
df=df.withColumn("borough",f.lower(f.col("borough")))
collison = df.toPandas()
cov = fetchData(nyc_his_boro_url)
cov = cov.drop(columns = ['timestamp','total'])
cov2 = cov.loc[cov['type']=='cases']
cov2 = cov2.tail(1)
cov2.set_index(["type"], inplace = True)
cov2 = cov2.stack()
cov = cov2.unstack(0)
cov['borough']=cov.index
res = pd.merge(collison,cov, on='borough')
res = res.sort_values(by='cases',ascending=False)
res.set_index(["borough"], inplace = True)
res.plot.bar(rot=0,title="Car crash vs Covid-19 cases (by borough)")
collison_march= collison_df.filter(collison_df["crash_date"] >= to_timestamp(f.lit('2020-03-12 00:00:00')))
collison_march.createOrReplaceTempView("collisonT")
collison_covid = spark.sql("""
SELECT borough,
COUNT(*) AS num_of_crash
FROM collisonT
GROUP BY borough
""")
from pyspark.sql.functions import col
cdf = collison_covid.withColumn("borough",
f.when(collison_covid["borough"]=='nan','unknown').when(collison_covid["borough"]=='STATEN ISLAND','staten_island').
otherwise(collison_covid["borough"]))
cdf=cdf.withColumn("borough",f.lower(f.col("borough")))
collison_covid = cdf.toPandas()
collison_covid.set_index(["borough"], inplace = True)
collison_covid = collison_covid.reindex(['queens','brooklyn','bronx','manhattan','staten_island','unknown'])
collison_covid.plot.bar(rot=0, title="Collison since Covid-19 by borough")
cov.set_index(["borough"], inplace = True)
cov = cov.sort_values(by='cases',ascending=False)
cov.plot.bar(rot=0,title="Covid-19 cases by borough")
res2 = pd.merge(collison_covid,cov, on='borough')
res2.head(10)
res2 = res2.sort_values(by='cases',ascending=False)
res2.plot.bar(rot=0,title="Car Crash vs Covid-19 Cases (by borough)", subplots = True)
graph = res2.plot.bar(rot=0,title="Car Crash vs Covid-19 Cases (by borough)", subplots = True)
plt.xticks( rotation=25 )
graph[0].get_figure().savefig('/Users/jennyzhou/Downloads/traffic2.png')
| 0.305179 | 0.800536 |
# Polar and Cilindrical Frame of Reference
Renato Naville Watanabe
Consider that we have the position vector $\bf\vec{r}$ of a particle, moving in a circular path indicated in the figure below by a dashed line. This vector $\bf\vec{r(t)}$ is described in a fixed reference frame as:
$${\bf\hat{r}}(t) = {x}{\bf\hat{i}}+{y}{\bf\hat{j}} + {z}{\bf\hat{k}}$$
<img src="../images/polarCoord.png" width=500/>
Naturally, we could describe all the kinematic variables in the fixed reference frame. But in circular motions, is convenient to define a basis with a vector in the direction of the position vector $\bf\vec{r}$. So, the vector $\bf\hat{e_R}$ is defined as:
$$ {\bf\hat{e_R}} = \frac{\bf\vec{r}}{\Vert{\bf\vec{r} }\Vert} $$
The second vector of the basis can be obtained by the cross multiplication between $\bf\hat{k}$ and $\bf\hat{e_R}$:
$$ {\bf\hat{e_\theta}} = {\bf\hat{k}} \times {\bf\hat{e_R}}$$
The third vector of the basis is the conventional ${\bf\hat{k}}$ vector.
<img src="../images/polarCoorderetheta.png" width=500/>
This basis can be used also for non-circular movements. For a 3D movement, the versor ${\bf\hat{e_R}}$ is obtained by removing the projection of the vector ${\bf\vec{r}}$ onto the versor ${\bf\hat{k}}$:
$$ {\bf\hat{e_R}} = \frac{\bf\vec{r} - ({\bf\vec{r}.{\bf\hat{k}}){\bf\hat{k}}}}{\Vert\bf\vec{r} - ({\bf\vec{r}.{\bf\hat{k}}){\bf\hat{k}}\Vert}} $$
<img src="../images/polarCilindrical.png" width=500/>
## Time-derivative of the versors ${\bf\hat{e_R}}$ and ${\bf\hat{e_\theta}}$
To obtain the expressions of the velocity and acceleration vectors, it is necessary to obtain the expressions of the time-derivative of the vectors ${\bf\hat{e_R}}$ and ${\bf\hat{e_\theta}}$.
This can be done by noting that:
$${\bf\hat{e_R}} = \cos(\theta){\bf\hat{i}} + \sin(\theta){\bf\hat{j}}$$
$${\bf\hat{e_\theta}} = -\sin(\theta){\bf\hat{i}} + \cos(\theta){\bf\hat{j}}$$
Deriving ${\bf\hat{e_R}}$ we obtain:
$$ \frac{d{\bf\hat{e_R}}}{dt} = -\sin(\theta)\dot\theta{\bf\hat{i}} + \cos(\theta)\dot\theta{\bf\hat{j}} = \dot{\theta}{\bf\hat{e_\theta}}$$
Similarly, we obtain the time-derivative of ${\bf\hat{e_\theta}}$:
$$ \frac{d{\bf\hat{e_\theta}}}{dt} = -\cos(\theta)\dot\theta{\bf\hat{i}} - \sin(\theta)\dot\theta{\bf\hat{j}} = -\dot{\theta}{\bf\hat{e_R}}$$
## Position, velocity and acceleration
### Position
The position vector $\bf\vec{r}$, from the definition of $\bf\hat{e_R}$, is:
$${\bf\vec{r}} = R{\bf\hat{e_R}} + z{\bf\hat{k}}$$
where $R = \Vert\bf\vec{r} - ({\bf\vec{r}.{\bf\hat{k}}){\bf\hat{k}}\Vert}$.
### Velocity
The velocity vector $\bf\vec{v}$ is obtained by deriving the vector $\bf\vec{r}$:
$$ {\bf\vec{v}} = \frac{d(R{\bf\hat{e_R}})}{dt} + \dot{z}{\bf\hat{k}} = \dot{R}{\bf\hat{e_R}}+R\frac{d\bf\hat{e_R}}{dt}=\dot{R}{\bf\hat{e_R}}+R\dot{\theta}{\bf\hat{e_\theta}}+ \dot{z}{\bf\hat{k}}$$
### Acceleration
The acceleration vector $\bf\vec{a}$ is obtained by deriving the velocity vector:
$$ {\bf\vec{a}} = \frac{d(\dot{R}{\bf\hat{e_R}}+R\dot{\theta}{\bf\hat{e_\theta}}+\dot{z}{\bf\hat{k}})}{dt}= \ddot{R}{\bf\hat{e_R}}+\dot{R}\frac{d\bf\hat{e_R}}{dt} + \dot{R}\dot{\theta}{\bf\hat{e_\theta}} + R\ddot{\theta}{\bf\hat{e_\theta}} + R\dot{\theta}\frac{d{\bf\hat{e_\theta}}}{dt} + \ddot{z}{\bf\hat{k}}= \ddot{R}{\bf\hat{e_R}}+\dot{R}\dot{\theta}{\bf\hat{e_\theta}} + \dot{R}\dot{\theta}{\bf\hat{e_\theta}} + R\ddot{\theta}{\bf\hat{e_\theta}} - R\dot{\theta}^2{\bf\hat{e_R}}+ \ddot{z}{\bf\hat{k}}~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~= \ddot{R}{\bf\hat{e_R}}+2\dot{R}\dot{\theta}{\bf\hat{e_\theta}}+ R\ddot{\theta}{\bf\hat{e_\theta}} - {R}\dot{\theta}^2{\bf\hat{e_R}}+ \ddot{z}{\bf\hat{k}} = (\ddot{R}-R\dot{\theta}^2){\bf\hat{e_R}}+(2\dot{R}\dot{\theta} + R\ddot{\theta}){\bf\hat{e_\theta}}+ \ddot{z}{\bf\hat{k}}$$
+ The term $\ddot{R}$ is an acceleration in the radial direction.
+ The term $R\ddot{\theta}$ is an angular acceleration.
+ The term $\ddot{z}$ is an acceleration in the $\bf\hat{k}$ direction.
+ The term $-R\dot{\theta}^2$ is the well known centripetal acceleration.
+ The term $2\dot{R}\dot{\theta}$ is known as Coriolis acceleration. This term may be difficult to understand. It appears when there is displacement in the radial and angular directions at the same time.
## Important to note
The reader must bear in mind that the use of a different basis to represent the position, velocity or acceleration vectors is only a different representation of the same vector. For example, for the acceleration vector:
$${\bf\vec{a}} = \ddot{x}{\bf\hat{i}}+ \ddot{y}{\bf\hat{j}} + \ddot{z}{\bf\hat{k}}=(\ddot{R}-R\dot{\theta}^2){\bf\hat{e_R}}+(2\dot{R}\dot{\theta} + R\ddot{\theta}){\bf\hat{e_\theta}}+ \ddot{z}{\bf\hat{k}}=\dot{\Vert\bf\vec{v}\Vert}{\bf\hat{e}_t}+{\Vert\bf\vec{v}\Vert}^2\Vert{\bf\vec{C}} \Vert{\bf\hat{e}_n}$$
In which the last equality is the acceleration vector represented in the path-coordinate of the particle (see http://nbviewer.jupyter.org/github/BMClab/bmc/blob/master/notebooks/Time-varying%20frames.ipynb).
## Example
Consider a particle following the spiral path described below:
$${\bf\vec{r}}(t) = (2\sqrt(t)\cos(t)){\bf\hat{i}}+ (2\sqrt(t)\sin(t)){\bf\hat{j}}$$
```
import numpy as np
import sympy as sym
from sympy.plotting import plot_parametric,plot3d_parametric_line
from sympy.vector import CoordSys3D
import matplotlib.pyplot as plt
sym.init_printing()
```
### Solving numerically
```
t = np.linspace(0.1,10,30)
R = 2*np.sqrt(t)
theta = t
r = np.transpose(np.array([R*np.cos(t), R*np.sin(t)]))
e_r = np.transpose(np.array([r[:,0]/np.sqrt(r[:,0]**2+r[:,1]**2), r[:,1]/np.sqrt(r[:,0]**2+r[:,1]**2)]))
e_theta = np.cross([0,0,1],e_r)[:,0:-1]
Rdot = np.diff(R,1,0)/t[1]
thetaDot = np.diff(theta,1,0)/t[1]
v = np.transpose(np.array([Rdot*e_r[0:-1,0],Rdot*e_r[0:-1,1]]) + np.array([R[0:-1]*thetaDot*e_theta[0:-1,0],R[0:-1]*thetaDot*e_theta[0:-1,1]]))
Rddot = np.diff(Rdot,1,0)/t[1]
thetaddot = np.diff(thetaDot,1,0)/t[1]
a = np.transpose(np.array([(Rddot-R[0:-2]*thetaDot[0:-1]**2)*e_r[0:-2,0], (Rddot-R[0:-2]*thetaDot[0:-1]**2)*e_r[0:-2,1]]) +
np.array([(2*Rdot[0:-1]*thetaDot[0:-1] + R[0:-2]*thetaddot)*e_theta[0:-2,0],(2*Rdot[0:-1]*thetaDot[0:-1] + R[0:-2]*thetaddot)*e_theta[0:-2,1]]))
from matplotlib.patches import FancyArrowPatch
%matplotlib inline
plt.rcParams['figure.figsize']=10,10
fig = plt.figure()
plt.plot(r[:,0],r[:,1],'.')
ax = fig.add_axes([0,0,1,1])
for i in np.arange(len(t)-2):
vec1 = FancyArrowPatch(r[i,:],r[i,:]+e_r[i,:],mutation_scale=20,color='r')
vec2 = FancyArrowPatch(r[i,:],r[i,:]+e_theta[i,:],mutation_scale=20,color='g')
ax.add_artist(vec1)
ax.add_artist(vec2)
plt.xlim((-10,10))
plt.ylim((-10,10))
plt.show()
from matplotlib.patches import FancyArrowPatch
%matplotlib inline
plt.rcParams['figure.figsize']=10,10
fig = plt.figure()
plt.plot(r[:,0],r[:,1],'.')
ax = fig.add_axes([0,0,1,1])
for i in np.arange(len(t)-2):
vec1 = FancyArrowPatch(r[i,:],r[i,:]+v[i,:],mutation_scale=20,color='r')
vec2 = FancyArrowPatch(r[i,:],r[i,:]+a[i,:],mutation_scale=20,color='g')
ax.add_artist(vec1)
ax.add_artist(vec2)
plt.xlim((-10,10))
plt.ylim((-10,10))
plt.show()
```
### Solved simbolically (extra reading)
```
O = sym.vector.CoordSys3D(' ')
t = sym.symbols('t')
r = 2*sym.sqrt(t)*sym.cos(t)*O.i+2*sym.sqrt(t)*sym.sin(t)*O.j
r
plot_parametric(r.dot(O.i),r.dot(O.j),(t,0,10))
e_r = r - r.dot(O.k)*O.k
e_r = e_r/sym.sqrt(e_r.dot(O.i)**2+e_r.dot(O.j)**2+e_r.dot(O.k)**2)
e_r
e_theta = O.k.cross(e_r)
e_theta
from matplotlib.patches import FancyArrowPatch
plt.rcParams['figure.figsize']=10,10
fig = plt.figure()
ax = fig.add_axes([0, 0, 1, 1])
ax.axis("on")
time = np.linspace(0,10,30)
for instant in time:
vt = FancyArrowPatch([float(r.dot(O.i).subs(t,instant)),float(r.dot(O.j).subs(t,instant))],
[float(r.dot(O.i).subs(t,instant))+float(e_r.dot(O.i).subs(t,instant)), float(r.dot(O.j).subs(t, instant))+float(e_r.dot(O.j).subs(t,instant))],
mutation_scale=20,
arrowstyle="->",color="r",label='${{e_r}}$')
vn = FancyArrowPatch([float(r.dot(O.i).subs(t, instant)),float(r.dot(O.j).subs(t,instant))],
[float(r.dot(O.i).subs(t, instant))+float(e_theta.dot(O.i).subs(t, instant)), float(r.dot(O.j).subs(t, instant))+float(e_theta.dot(O.j).subs(t, instant))],
mutation_scale=20,
arrowstyle="->",color="g",label='${{e_{theta}}}$')
ax.add_artist(vn)
ax.add_artist(vt)
plt.xlim((-10,10))
plt.ylim((-10,10))
plt.legend(handles=[vt,vn],fontsize=20)
plt.grid()
plt.show()
R = 2*sym.sqrt(t)
Rdot = sym.diff(R,t)
Rddot = sym.diff(Rdot,t)
Rddot
v = Rdot*e_r + R*e_theta
v
a = (Rddot - R)*e_r + (2*Rdot*1+0)*e_theta
aCor = 2*Rdot*1*e_theta
aCor
a
from matplotlib.patches import FancyArrowPatch
plt.rcParams['figure.figsize'] = 10,10
fig = plt.figure()
ax = fig.add_axes([0, 0, 1, 1])
ax.axis("on")
time = np.linspace(0.1,10,30)
for instant in time:
vt = FancyArrowPatch([float(r.dot(O.i).subs(t,instant)),float(r.dot(O.j).subs(t,instant))],
[float(r.dot(O.i).subs(t,instant))+float(v.dot(O.i).subs(t,instant)), float(r.dot(O.j).subs(t, instant))+float(v.dot(O.j).subs(t,instant))],
mutation_scale=20,
arrowstyle="->",color="r",label='${{v}}$')
vn = FancyArrowPatch([float(r.dot(O.i).subs(t, instant)),float(r.dot(O.j).subs(t,instant))],
[float(r.dot(O.i).subs(t, instant))+float(a.dot(O.i).subs(t, instant)), float(r.dot(O.j).subs(t, instant))+float(a.dot(O.j).subs(t, instant))],
mutation_scale=20,
arrowstyle="->",color="g",label='${{a}}$')
vc = FancyArrowPatch([float(r.dot(O.i).subs(t, instant)),float(r.dot(O.j).subs(t,instant))],
[float(r.dot(O.i).subs(t, instant))+float(aCor.dot(O.i).subs(t, instant)), float(r.dot(O.j).subs(t, instant))+float(aCor.dot(O.j).subs(t, instant))],
mutation_scale=20,
arrowstyle="->",color="b",label='${{a_{Cor}}}$')
ax.add_artist(vn)
ax.add_artist(vt)
ax.add_artist(vc)
plt.xlim((-10,10))
plt.ylim((-10,10))
plt.legend(handles=[vt,vn,vc],fontsize=20)
plt.grid()
plt.show()
```
## Problems
1. Problems from 14.1.1 to 14.1.14from Ruina and Rudra's book,
2. Problems from 17.1.1 to 17.1.10 from Ruina and Rudra's book.
## Reference
+ Ruina A, Rudra P (2015) Introduction to Statics and Dynamics. Oxford University Press. http://ruina.tam.cornell.edu/Book/RuinaPratap-Jan-20-2015.pdf
|
github_jupyter
|
import numpy as np
import sympy as sym
from sympy.plotting import plot_parametric,plot3d_parametric_line
from sympy.vector import CoordSys3D
import matplotlib.pyplot as plt
sym.init_printing()
t = np.linspace(0.1,10,30)
R = 2*np.sqrt(t)
theta = t
r = np.transpose(np.array([R*np.cos(t), R*np.sin(t)]))
e_r = np.transpose(np.array([r[:,0]/np.sqrt(r[:,0]**2+r[:,1]**2), r[:,1]/np.sqrt(r[:,0]**2+r[:,1]**2)]))
e_theta = np.cross([0,0,1],e_r)[:,0:-1]
Rdot = np.diff(R,1,0)/t[1]
thetaDot = np.diff(theta,1,0)/t[1]
v = np.transpose(np.array([Rdot*e_r[0:-1,0],Rdot*e_r[0:-1,1]]) + np.array([R[0:-1]*thetaDot*e_theta[0:-1,0],R[0:-1]*thetaDot*e_theta[0:-1,1]]))
Rddot = np.diff(Rdot,1,0)/t[1]
thetaddot = np.diff(thetaDot,1,0)/t[1]
a = np.transpose(np.array([(Rddot-R[0:-2]*thetaDot[0:-1]**2)*e_r[0:-2,0], (Rddot-R[0:-2]*thetaDot[0:-1]**2)*e_r[0:-2,1]]) +
np.array([(2*Rdot[0:-1]*thetaDot[0:-1] + R[0:-2]*thetaddot)*e_theta[0:-2,0],(2*Rdot[0:-1]*thetaDot[0:-1] + R[0:-2]*thetaddot)*e_theta[0:-2,1]]))
from matplotlib.patches import FancyArrowPatch
%matplotlib inline
plt.rcParams['figure.figsize']=10,10
fig = plt.figure()
plt.plot(r[:,0],r[:,1],'.')
ax = fig.add_axes([0,0,1,1])
for i in np.arange(len(t)-2):
vec1 = FancyArrowPatch(r[i,:],r[i,:]+e_r[i,:],mutation_scale=20,color='r')
vec2 = FancyArrowPatch(r[i,:],r[i,:]+e_theta[i,:],mutation_scale=20,color='g')
ax.add_artist(vec1)
ax.add_artist(vec2)
plt.xlim((-10,10))
plt.ylim((-10,10))
plt.show()
from matplotlib.patches import FancyArrowPatch
%matplotlib inline
plt.rcParams['figure.figsize']=10,10
fig = plt.figure()
plt.plot(r[:,0],r[:,1],'.')
ax = fig.add_axes([0,0,1,1])
for i in np.arange(len(t)-2):
vec1 = FancyArrowPatch(r[i,:],r[i,:]+v[i,:],mutation_scale=20,color='r')
vec2 = FancyArrowPatch(r[i,:],r[i,:]+a[i,:],mutation_scale=20,color='g')
ax.add_artist(vec1)
ax.add_artist(vec2)
plt.xlim((-10,10))
plt.ylim((-10,10))
plt.show()
O = sym.vector.CoordSys3D(' ')
t = sym.symbols('t')
r = 2*sym.sqrt(t)*sym.cos(t)*O.i+2*sym.sqrt(t)*sym.sin(t)*O.j
r
plot_parametric(r.dot(O.i),r.dot(O.j),(t,0,10))
e_r = r - r.dot(O.k)*O.k
e_r = e_r/sym.sqrt(e_r.dot(O.i)**2+e_r.dot(O.j)**2+e_r.dot(O.k)**2)
e_r
e_theta = O.k.cross(e_r)
e_theta
from matplotlib.patches import FancyArrowPatch
plt.rcParams['figure.figsize']=10,10
fig = plt.figure()
ax = fig.add_axes([0, 0, 1, 1])
ax.axis("on")
time = np.linspace(0,10,30)
for instant in time:
vt = FancyArrowPatch([float(r.dot(O.i).subs(t,instant)),float(r.dot(O.j).subs(t,instant))],
[float(r.dot(O.i).subs(t,instant))+float(e_r.dot(O.i).subs(t,instant)), float(r.dot(O.j).subs(t, instant))+float(e_r.dot(O.j).subs(t,instant))],
mutation_scale=20,
arrowstyle="->",color="r",label='${{e_r}}$')
vn = FancyArrowPatch([float(r.dot(O.i).subs(t, instant)),float(r.dot(O.j).subs(t,instant))],
[float(r.dot(O.i).subs(t, instant))+float(e_theta.dot(O.i).subs(t, instant)), float(r.dot(O.j).subs(t, instant))+float(e_theta.dot(O.j).subs(t, instant))],
mutation_scale=20,
arrowstyle="->",color="g",label='${{e_{theta}}}$')
ax.add_artist(vn)
ax.add_artist(vt)
plt.xlim((-10,10))
plt.ylim((-10,10))
plt.legend(handles=[vt,vn],fontsize=20)
plt.grid()
plt.show()
R = 2*sym.sqrt(t)
Rdot = sym.diff(R,t)
Rddot = sym.diff(Rdot,t)
Rddot
v = Rdot*e_r + R*e_theta
v
a = (Rddot - R)*e_r + (2*Rdot*1+0)*e_theta
aCor = 2*Rdot*1*e_theta
aCor
a
from matplotlib.patches import FancyArrowPatch
plt.rcParams['figure.figsize'] = 10,10
fig = plt.figure()
ax = fig.add_axes([0, 0, 1, 1])
ax.axis("on")
time = np.linspace(0.1,10,30)
for instant in time:
vt = FancyArrowPatch([float(r.dot(O.i).subs(t,instant)),float(r.dot(O.j).subs(t,instant))],
[float(r.dot(O.i).subs(t,instant))+float(v.dot(O.i).subs(t,instant)), float(r.dot(O.j).subs(t, instant))+float(v.dot(O.j).subs(t,instant))],
mutation_scale=20,
arrowstyle="->",color="r",label='${{v}}$')
vn = FancyArrowPatch([float(r.dot(O.i).subs(t, instant)),float(r.dot(O.j).subs(t,instant))],
[float(r.dot(O.i).subs(t, instant))+float(a.dot(O.i).subs(t, instant)), float(r.dot(O.j).subs(t, instant))+float(a.dot(O.j).subs(t, instant))],
mutation_scale=20,
arrowstyle="->",color="g",label='${{a}}$')
vc = FancyArrowPatch([float(r.dot(O.i).subs(t, instant)),float(r.dot(O.j).subs(t,instant))],
[float(r.dot(O.i).subs(t, instant))+float(aCor.dot(O.i).subs(t, instant)), float(r.dot(O.j).subs(t, instant))+float(aCor.dot(O.j).subs(t, instant))],
mutation_scale=20,
arrowstyle="->",color="b",label='${{a_{Cor}}}$')
ax.add_artist(vn)
ax.add_artist(vt)
ax.add_artist(vc)
plt.xlim((-10,10))
plt.ylim((-10,10))
plt.legend(handles=[vt,vn,vc],fontsize=20)
plt.grid()
plt.show()
| 0.333178 | 0.993938 |
# PyBx
PyBx is a simple python package to generate anchor boxes (aka default/prior boxes) for object detection
tasks.
```
! pip install pybx # restart runtime if required
```
>⚠ Note: walkthrough for v0.1.2 ⚠
>
>run `! pip freeze | grep pybx` to see the installed version.
```
! pip freeze | grep pybx
```
# SSD for Object Detection
This walkthrough is build around the [Single-Shot Detection (SSD)](https://arxiv.org/pdf/1512.02325.pdf) algorithm. The SSD can be imagined as an encoder-decoder model architecture, where the input image is fed into a `backbone` (encoder) to generate inital features, which then goes through a series of 2D convolution layers (decoders) to perform further feature extraction/prediction tasks at each layer. For a single image, each layer in the decoder produces a total of `N x (4 + C)` predictions. Here `C` is the number of classes (plus one for `background` class) in the detection task and 4 comes from the corners of the rectangular bounding box.
### Usage of the term Feature/Filter/Channel
Channel: RGB dimensione, also called a Filter
Feature: (W,H) of a single channel
## Example case
For this example, we assume that our input image is a single channel image is of shape `[B, 3, 300, 300]` where `B` is the batch size. Assuming that a pretrained `VGG-16` is our model `backbone`, the output feature shape would be: `[B, 512, 37, 37]`. Meaning that, 512 channels of shape `[37, 37]` were extracted from each image in the batch. In the subsequent decoder layers, for simplicity we double the channels while halving the feature shape using `3x3` `stride=2` convolutions (except for first decoder layer where convolution is not applied). This results in the following shapes:
```python
torch.Size([-1, 512, 37, 37]) # inp from vgg-16 encoder
torch.Size([-1, 1024, 18, 18]) # first layer logits
torch.Size([-1, 2048, 8, 8]) # second layer logits
torch.Size([-1, 4096, 3, 3]) # third layer logits
```
<img src="https://lilianweng.github.io/lil-log/assets/images/SSD-box-scales.png" width="500" />
## Sample image
Image obtained from USC-SIPI Image Database.
The USC-SIPI image database is a collection of digitized images. It is maintained primarily to support research in image processing, image analysis, and machine vision. The first edition of the USC-SIPI image database was distributed in 1977 and many new images have been added since then.
```
! wget -q -O 'image.jpg' 'https://sipi.usc.edu/database/download.php?vol=misc&img=5.1.12'
```
## About anchor Boxes
We are expected to provide our models with "good" anchor (aka default/prior) boxes. Strong opinion: Our model is [only as good as the initial anchor boxes](https://towardsdatascience.com/anchor-boxes-the-key-to-quality-object-detection-ddf9d612d4f9) that we generate. Inorder to improve the coverage of our model, we tend to add additional anchor boxes of different aspect ratios. Now, for a single image, each layer in the decoder produces a total of `N x A x (4 + C)` predictions. Here `A` is the number of aspect ratios to generate additional anchor boxes.
### Task description
Our aim is to find the maximum number of anchor boxes in varying sizes `feature_szs` and aspect ratios `asp_ratios` across the entire image. We apply no filtering to get rid of low (IOU) anchors.
<img src="https://lilianweng.github.io/lil-log/assets/images/SSD-framework.png" width="600" />
```
feature_szs = [(37,37), (18,18), (8,8), (3,3)]
asp_ratios = [1/2., 1., 2.]
from operator import __mul__
n_boxes = sum([__mul__(*f) for f in feature_szs])
print(f'minimum anchor boxes with 1 aspect ratio: {n_boxes}')
print(f'minimum anchor boxes with {len(asp_ratios)} aspect ratios: {n_boxes*len(asp_ratios)}')
```
# Loading an image
```
from PIL import Image
from matplotlib import pyplot as plt
import json
im = Image.open("image.jpg").convert('RGB').resize([300,300])
_ = plt.imshow(im)
```
We also make 2 truth bounding boxes `bbox` for this image around the clock and the photoframe in `pascal voc` format:
```
bbox = [dict(x_min=150, y_min=70, x_max=270, y_max=220, label='clock'),
dict(x_min=10, y_min=180, x_max=115, y_max=260, label='frame'),]
bbox
```
Save annotations as a json file.
```
with open('annots.json', 'w') as f:
f.write(json.dumps(bbox))
```
# Using PyBx
```
from pybx import anchor
image_sz = (300, 300, 3) # W, H, C
feature_sz = (3, 3) # number of features along W, H of the image
asp_ratio = 1. # aspect ratio of the anchor box
anchors = anchor.bx(image_sz, feature_sz, asp_ratio, show=True)
```
The boxes in white with label `unk` are the anchor boxes. We can hightlight them with a different color.
```
anchors = anchor.bx(image_sz, feature_sz, asp_ratio, show=True, color={'unk':'red'})
```
We can also overlay the features/receptive fields on the original image (only for reference and visualisation).
```
anchors = anchor.bx(image_sz, feature_sz, asp_ratio, show=True, color={'unk':'red'}, logits=True)
```
`logits=True` simply generates numbers of the same shape as feature sizes for illustration purposes.
# Working with mulitple feature sizes and aspect ratios
Finally we calculate anchor boxes for multiple feature sizes and aspect ratios.
```
feature_szs = [(3, 3), (2, 2)]
asp_ratios = [1/2., 2.]
anchors = anchor.bxs(image_sz, feature_szs, asp_ratios, show=True, color={'unk': 'yellow'})
```
This is essentially a wrapper to do list comprehension over the passed feature sizes and aspect ratios (but additionally stacks them together into an ndarray).
```
[anchor.bx(image_sz, f, ar) for f in feature_szs for ar in asp_ratios]
```
As simple as that! More options to select the best anchors planned. Do leave a star or raise issues and suggestions on the project page if you found this useful!
Project page: [GitHub](https://github.com/thatgeeman/pybx)
Package: [PyBx](https://pypi.org/project/pybx/)
```
```
|
github_jupyter
|
! pip install pybx # restart runtime if required
! pip freeze | grep pybx
torch.Size([-1, 512, 37, 37]) # inp from vgg-16 encoder
torch.Size([-1, 1024, 18, 18]) # first layer logits
torch.Size([-1, 2048, 8, 8]) # second layer logits
torch.Size([-1, 4096, 3, 3]) # third layer logits
! wget -q -O 'image.jpg' 'https://sipi.usc.edu/database/download.php?vol=misc&img=5.1.12'
feature_szs = [(37,37), (18,18), (8,8), (3,3)]
asp_ratios = [1/2., 1., 2.]
from operator import __mul__
n_boxes = sum([__mul__(*f) for f in feature_szs])
print(f'minimum anchor boxes with 1 aspect ratio: {n_boxes}')
print(f'minimum anchor boxes with {len(asp_ratios)} aspect ratios: {n_boxes*len(asp_ratios)}')
from PIL import Image
from matplotlib import pyplot as plt
import json
im = Image.open("image.jpg").convert('RGB').resize([300,300])
_ = plt.imshow(im)
bbox = [dict(x_min=150, y_min=70, x_max=270, y_max=220, label='clock'),
dict(x_min=10, y_min=180, x_max=115, y_max=260, label='frame'),]
bbox
with open('annots.json', 'w') as f:
f.write(json.dumps(bbox))
from pybx import anchor
image_sz = (300, 300, 3) # W, H, C
feature_sz = (3, 3) # number of features along W, H of the image
asp_ratio = 1. # aspect ratio of the anchor box
anchors = anchor.bx(image_sz, feature_sz, asp_ratio, show=True)
anchors = anchor.bx(image_sz, feature_sz, asp_ratio, show=True, color={'unk':'red'})
anchors = anchor.bx(image_sz, feature_sz, asp_ratio, show=True, color={'unk':'red'}, logits=True)
feature_szs = [(3, 3), (2, 2)]
asp_ratios = [1/2., 2.]
anchors = anchor.bxs(image_sz, feature_szs, asp_ratios, show=True, color={'unk': 'yellow'})
[anchor.bx(image_sz, f, ar) for f in feature_szs for ar in asp_ratios]
| 0.448426 | 0.985356 |
# GradCam
```
%matplotlib inline
import torch
import torch.nn as nn
from torch.utils import data
from torchvision.models import vgg16
from torchvision import transforms
from torchvision import datasets
import matplotlib.pyplot as plt
import numpy as np
import os
import torch.optim as optim
import matplotlib.pyplot as plt
#Mounting Google Drive data
from google.colab import drive
drive.mount('/content/gdrive')
# Root directory of interest
gdrivePath = F"gdrive/MyDrive/ML_AI"
# Dataset containing images
dataset_path = gdrivePath + "/NatureDataset"
# Path where the resulting images are saved
saving_path = gdrivePath + "/NatureDatasetNew_GradCam"
if not os.path.exists(saving_path):
os.makedirs(saving_path)
print("\n", saving_path, " created!\n")
# Trained Neural Network path, to use to compute GradCam images
best_resnet18_path = gdrivePath + "/CNN_finalMetrics/resnet/resnet18_LR(0_001)_nEpochs(30)/resnet18_best_LR(0_001)_nEpochs(30).pth"
print("\n\nResNet trained model: ", best_resnet18_path)
print("Does it exist? ", os.path.exists(best_resnet18_path), "\n\n")
best_vgg16_path = gdrivePath + "/CNN_finalMetrics/vgg/vgg16_LR(0_001)_nEpochs(30)/vgg16_best_LR(0_001)_nEpochs(30).pth"
print("VGG16 trained model: ", best_vgg16_path)
print("Does it exist? ", os.path.exists(best_vgg16_path), "\n\n")
#Use GPU device
import torch
device = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
print("Used device:", device, "\n\n")
print(dataset_path, " exist? ", os.path.exists(dataset_path))
list_subfolders = [f.name for f in os.scandir(dataset_path) if f.is_dir()]
print(dataset_path)
space_str = " " * (len(dataset_path)-7)
for subf in list_subfolders:
print(space_str, "'-> ", subf)
!pip install grad-cam
import re
from torchvision.models import resnet50
import argparse
import cv2
import numpy as np
import torch
from torchvision import models
from pytorch_grad_cam import GradCAM, \
ScoreCAM, \
GradCAMPlusPlus, \
AblationCAM, \
XGradCAM, \
EigenCAM, \
EigenGradCAM, \
LayerCAM, \
FullGrad
from pytorch_grad_cam import GuidedBackpropReLUModel
from pytorch_grad_cam.utils.image import show_cam_on_image, \
deprocess_image, \
preprocess_image
from pytorch_grad_cam.utils.model_targets import ClassifierOutputTarget
""" python cam.py -image-path <path_to_image>
Example usage of loading an image, and computing:
1. CAM
2. Guided Back Propagation
3. Combining both
"""
methods = \
{"gradcam": GradCAM,
"scorecam": ScoreCAM,
"gradcam++": GradCAMPlusPlus,
"ablationcam": AblationCAM,
"xgradcam": XGradCAM,
"eigencam": EigenCAM,
"eigengradcam": EigenGradCAM,
"layercam": LayerCAM,
"fullgrad": FullGrad}
method = "gradcam"
aug_smooth = True
eigen_smooth = True
def compute_grad_cam(img_path, model_name, save=True, save_folder_path=".", show_saving_path=False, imshow=True, use_cuda=False):
if model_name=="resnet18_imagenet":
model = models.resnet18(pretrained=True)
model.fc = nn.Linear(model.fc.in_features, 6)
save_folder_path = save_folder_path + "/resnet_imagenet"
elif model_name=="resnet18":
model = models.resnet18()
model.fc = nn.Linear(model.fc.in_features, 6)
model = model.to(device)
model.load_state_dict(torch.load(best_resnet18_path, map_location=torch.device(device.type)))
model.eval();
save_folder_path = save_folder_path + "/resnet"
elif model_name=="vgg16":
model = models.vgg16()
model.classifier[6] = nn.Linear(4096, 6)
model = model.to(device)
model.load_state_dict(torch.load(best_vgg16_path, map_location=torch.device(device.type)))
model.eval();
save_folder_path = save_folder_path + "/vgg"
else:
print("ERROR: set variable choice to a possible value!")
if not os.path.exists(save_folder_path):
os.makedirs(save_folder_path)
print(save_folder_path, " CREATED!")
if model_name=="resnet18_imagenet" or model_name=="resnet18":
target_layers = [model.layer4]
elif model_name=="vgg16":
target_layers = [model.features[-1]]
else:
print('ERROR: model_name must be: "resnet18_imagenet" or "resnet18" or "vgg16"!');
return None
rgb_img = cv2.imread(img_path, 1)[:, :, ::-1]
rgb_img = np.float32(rgb_img) / 255
input_tensor = preprocess_image(rgb_img,
mean=[0.485, 0.456, 0.406],
std=[0.229, 0.224, 0.225])
# We have to specify the target we want to generate
# the Class Activation Maps for.
# If targets is None, the highest scoring category (for every member in the batch) will be used.
# You can target specific categories by
# targets = [e.g ClassifierOutputTarget(281)]
targets = None
# Using the with statement ensures the context is freed, and you can
# recreate different CAM objects in a loop.
cam_algorithm = methods[method]
with cam_algorithm(model=model,
target_layers=target_layers,
use_cuda=use_cuda) as cam:
# AblationCAM and ScoreCAM have batched implementations.
# You can override the internal batch size for faster computation.
cam.batch_size = 32
grayscale_cam = cam(input_tensor=input_tensor,
targets=targets,
aug_smooth=aug_smooth,
eigen_smooth=eigen_smooth)
# Here grayscale_cam has only one image in the batch
grayscale_cam = grayscale_cam[0, :]
cam_image = show_cam_on_image(rgb_img, grayscale_cam, use_rgb=True)
# cam_image is RGB encoded whereas "cv2.imwrite" requires BGR encoding.
cam_image = cv2.cvtColor(cam_image, cv2.COLOR_RGB2BGR)
gb_model = GuidedBackpropReLUModel(model=model, use_cuda=use_cuda)
gb = gb_model(input_tensor, target_category=None)
cam_mask = cv2.merge([grayscale_cam, grayscale_cam, grayscale_cam])
cam_gb = deprocess_image(cam_mask * gb)
gb = deprocess_image(gb)
# Get the prediction
classes_dict = {0: "buildings", 1: "forest", 2: "glacier", 3: "mountain", 4: "sea", 5: "street"}
head, tail = os.path.split(img_path)
if re.search("building", tail)!=None:
true = "buildings"
elif re.search("forest", tail)!=None:
true = "forest"
elif re.search("glacier", tail)!=None:
true = "glacier"
elif re.search("mountain", tail)!=None:
true = "mountain"
elif re.search("sea", tail)!=None:
true = "sea"
elif re.search("street", tail)!=None:
true = "street"
else:
print("ERROR: no class found in image name!")
while(True):
print("Interrupt manually execution!")
input()
output = model(input_tensor)
_, pred = torch.max(output, 1)
pred = np.array(pred.numpy()).item()
pred = classes_dict[pred]
print("img: ", tail, "LABEL: ", true, "| PREDICTION: ", pred, " [WRONG]" if true!=pred else "")
# Save and visualize images
if save==True:
saving_name = save_folder_path + "/" + (os.path.splitext(img_path)[0]).split('/')[-1] + "_" + method
grad_imgs_path = save_folder_path + "/gradient_images" + "/" + (os.path.splitext(img_path)[0]).split('/')[-1] + "_" + method
if not os.path.exists(save_folder_path + "/gradient_images"):
os.makedirs(save_folder_path + "/gradient_images")
print(save_folder_path+"/gradient_images", " CREATED!")
path1 = saving_name+'_cam.jpg'
height, width, channels = cam_image.shape
scale = 0.85
cv2.putText(
img=cam_image, #numpy array on which text is written
text=tail, #text
#position, #position at which writing has to start
fontFace=cv2.FONT_HERSHEY_PLAIN, #font family
color=(0, 255, 0), #font color
bottomLeftOrigin=False,
fontScale = scale,
org = (10,10),
thickness=1)
cv2.putText(
img=cam_image, #numpy array on which text is written
text=pred, #text
#position, #position at which writing has to start
fontFace=cv2.FONT_HERSHEY_PLAIN, #font family
color=(0, 0, 255) if true!=pred else (0,255,0), #font color
bottomLeftOrigin=False,
fontScale = scale,
org = (10,25),
thickness=1)
cv2.imwrite(path1, cam_image)
if show_saving_path:
print("Saving path: ", path1)
path2 = grad_imgs_path+'_bg.jpg'
cv2.imwrite(path2, gb)
if show_saving_path:
print("Saving path: ", path2)
path3 = grad_imgs_path+'_cam_gb.jpg'
cv2.imwrite(path3, cam_gb)
if show_saving_path:
print("Saving path: ", path3)
if imshow==True:
plt.figure()
plt.imshow(cv2.cvtColor(cam_image, cv2.COLOR_BGR2RGB))
plt.figure()
plt.imshow(cv2.cvtColor(gb, cv2.COLOR_BGR2RGB))
plt.figure()
plt.imshow(cv2.cvtColor(cam_gb, cv2.COLOR_BGR2RGB))
```
### Model selection:
- ResNet18 trained on NatureDataset
- ResNet18 pretrained on ImageNet
- VGG16 trained on NatureDataset
```
# Choose the images for which the GradCam must be computed
from imutils import paths
import matplotlib as mpl
mpl.rcParams['figure.max_open_warning'] = 0 #remove warning
imgs_path = list(paths.list_images(gdrivePath+"/NatureDatasetGradCam"))
print(imgs_path)
for img_path in imgs_path:
compute_grad_cam(img_path,"resnet18",save=True,save_folder_path=saving_path,show_saving_path=False,imshow=False)
# Choose the images for which the GradCam must be computed
from imutils import paths
import matplotlib as mpl
mpl.rcParams['figure.max_open_warning'] = 0 #remove warning
imgs_path = list(paths.list_images(gdrivePath+"/NatureDatasetGradCam"))
print(imgs_path)
for img_path in imgs_path:
compute_grad_cam(img_path,"resnet18_imagenet",save=True,save_folder_path=saving_path,show_saving_path=False,imshow=False)
# Choose the images for which the GradCam must be computed
from imutils import paths
import matplotlib as mpl
mpl.rcParams['figure.max_open_warning'] = 0 #remove warning
imgs_path = list(paths.list_images(gdrivePath+"/NatureDatasetGradCam"))
print(imgs_path)
for img_path in imgs_path:
compute_grad_cam(img_path,"vgg16",save=True,save_folder_path=saving_path,show_saving_path=False,imshow=False)
```
|
github_jupyter
|
%matplotlib inline
import torch
import torch.nn as nn
from torch.utils import data
from torchvision.models import vgg16
from torchvision import transforms
from torchvision import datasets
import matplotlib.pyplot as plt
import numpy as np
import os
import torch.optim as optim
import matplotlib.pyplot as plt
#Mounting Google Drive data
from google.colab import drive
drive.mount('/content/gdrive')
# Root directory of interest
gdrivePath = F"gdrive/MyDrive/ML_AI"
# Dataset containing images
dataset_path = gdrivePath + "/NatureDataset"
# Path where the resulting images are saved
saving_path = gdrivePath + "/NatureDatasetNew_GradCam"
if not os.path.exists(saving_path):
os.makedirs(saving_path)
print("\n", saving_path, " created!\n")
# Trained Neural Network path, to use to compute GradCam images
best_resnet18_path = gdrivePath + "/CNN_finalMetrics/resnet/resnet18_LR(0_001)_nEpochs(30)/resnet18_best_LR(0_001)_nEpochs(30).pth"
print("\n\nResNet trained model: ", best_resnet18_path)
print("Does it exist? ", os.path.exists(best_resnet18_path), "\n\n")
best_vgg16_path = gdrivePath + "/CNN_finalMetrics/vgg/vgg16_LR(0_001)_nEpochs(30)/vgg16_best_LR(0_001)_nEpochs(30).pth"
print("VGG16 trained model: ", best_vgg16_path)
print("Does it exist? ", os.path.exists(best_vgg16_path), "\n\n")
#Use GPU device
import torch
device = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
print("Used device:", device, "\n\n")
print(dataset_path, " exist? ", os.path.exists(dataset_path))
list_subfolders = [f.name for f in os.scandir(dataset_path) if f.is_dir()]
print(dataset_path)
space_str = " " * (len(dataset_path)-7)
for subf in list_subfolders:
print(space_str, "'-> ", subf)
!pip install grad-cam
import re
from torchvision.models import resnet50
import argparse
import cv2
import numpy as np
import torch
from torchvision import models
from pytorch_grad_cam import GradCAM, \
ScoreCAM, \
GradCAMPlusPlus, \
AblationCAM, \
XGradCAM, \
EigenCAM, \
EigenGradCAM, \
LayerCAM, \
FullGrad
from pytorch_grad_cam import GuidedBackpropReLUModel
from pytorch_grad_cam.utils.image import show_cam_on_image, \
deprocess_image, \
preprocess_image
from pytorch_grad_cam.utils.model_targets import ClassifierOutputTarget
""" python cam.py -image-path <path_to_image>
Example usage of loading an image, and computing:
1. CAM
2. Guided Back Propagation
3. Combining both
"""
methods = \
{"gradcam": GradCAM,
"scorecam": ScoreCAM,
"gradcam++": GradCAMPlusPlus,
"ablationcam": AblationCAM,
"xgradcam": XGradCAM,
"eigencam": EigenCAM,
"eigengradcam": EigenGradCAM,
"layercam": LayerCAM,
"fullgrad": FullGrad}
method = "gradcam"
aug_smooth = True
eigen_smooth = True
def compute_grad_cam(img_path, model_name, save=True, save_folder_path=".", show_saving_path=False, imshow=True, use_cuda=False):
if model_name=="resnet18_imagenet":
model = models.resnet18(pretrained=True)
model.fc = nn.Linear(model.fc.in_features, 6)
save_folder_path = save_folder_path + "/resnet_imagenet"
elif model_name=="resnet18":
model = models.resnet18()
model.fc = nn.Linear(model.fc.in_features, 6)
model = model.to(device)
model.load_state_dict(torch.load(best_resnet18_path, map_location=torch.device(device.type)))
model.eval();
save_folder_path = save_folder_path + "/resnet"
elif model_name=="vgg16":
model = models.vgg16()
model.classifier[6] = nn.Linear(4096, 6)
model = model.to(device)
model.load_state_dict(torch.load(best_vgg16_path, map_location=torch.device(device.type)))
model.eval();
save_folder_path = save_folder_path + "/vgg"
else:
print("ERROR: set variable choice to a possible value!")
if not os.path.exists(save_folder_path):
os.makedirs(save_folder_path)
print(save_folder_path, " CREATED!")
if model_name=="resnet18_imagenet" or model_name=="resnet18":
target_layers = [model.layer4]
elif model_name=="vgg16":
target_layers = [model.features[-1]]
else:
print('ERROR: model_name must be: "resnet18_imagenet" or "resnet18" or "vgg16"!');
return None
rgb_img = cv2.imread(img_path, 1)[:, :, ::-1]
rgb_img = np.float32(rgb_img) / 255
input_tensor = preprocess_image(rgb_img,
mean=[0.485, 0.456, 0.406],
std=[0.229, 0.224, 0.225])
# We have to specify the target we want to generate
# the Class Activation Maps for.
# If targets is None, the highest scoring category (for every member in the batch) will be used.
# You can target specific categories by
# targets = [e.g ClassifierOutputTarget(281)]
targets = None
# Using the with statement ensures the context is freed, and you can
# recreate different CAM objects in a loop.
cam_algorithm = methods[method]
with cam_algorithm(model=model,
target_layers=target_layers,
use_cuda=use_cuda) as cam:
# AblationCAM and ScoreCAM have batched implementations.
# You can override the internal batch size for faster computation.
cam.batch_size = 32
grayscale_cam = cam(input_tensor=input_tensor,
targets=targets,
aug_smooth=aug_smooth,
eigen_smooth=eigen_smooth)
# Here grayscale_cam has only one image in the batch
grayscale_cam = grayscale_cam[0, :]
cam_image = show_cam_on_image(rgb_img, grayscale_cam, use_rgb=True)
# cam_image is RGB encoded whereas "cv2.imwrite" requires BGR encoding.
cam_image = cv2.cvtColor(cam_image, cv2.COLOR_RGB2BGR)
gb_model = GuidedBackpropReLUModel(model=model, use_cuda=use_cuda)
gb = gb_model(input_tensor, target_category=None)
cam_mask = cv2.merge([grayscale_cam, grayscale_cam, grayscale_cam])
cam_gb = deprocess_image(cam_mask * gb)
gb = deprocess_image(gb)
# Get the prediction
classes_dict = {0: "buildings", 1: "forest", 2: "glacier", 3: "mountain", 4: "sea", 5: "street"}
head, tail = os.path.split(img_path)
if re.search("building", tail)!=None:
true = "buildings"
elif re.search("forest", tail)!=None:
true = "forest"
elif re.search("glacier", tail)!=None:
true = "glacier"
elif re.search("mountain", tail)!=None:
true = "mountain"
elif re.search("sea", tail)!=None:
true = "sea"
elif re.search("street", tail)!=None:
true = "street"
else:
print("ERROR: no class found in image name!")
while(True):
print("Interrupt manually execution!")
input()
output = model(input_tensor)
_, pred = torch.max(output, 1)
pred = np.array(pred.numpy()).item()
pred = classes_dict[pred]
print("img: ", tail, "LABEL: ", true, "| PREDICTION: ", pred, " [WRONG]" if true!=pred else "")
# Save and visualize images
if save==True:
saving_name = save_folder_path + "/" + (os.path.splitext(img_path)[0]).split('/')[-1] + "_" + method
grad_imgs_path = save_folder_path + "/gradient_images" + "/" + (os.path.splitext(img_path)[0]).split('/')[-1] + "_" + method
if not os.path.exists(save_folder_path + "/gradient_images"):
os.makedirs(save_folder_path + "/gradient_images")
print(save_folder_path+"/gradient_images", " CREATED!")
path1 = saving_name+'_cam.jpg'
height, width, channels = cam_image.shape
scale = 0.85
cv2.putText(
img=cam_image, #numpy array on which text is written
text=tail, #text
#position, #position at which writing has to start
fontFace=cv2.FONT_HERSHEY_PLAIN, #font family
color=(0, 255, 0), #font color
bottomLeftOrigin=False,
fontScale = scale,
org = (10,10),
thickness=1)
cv2.putText(
img=cam_image, #numpy array on which text is written
text=pred, #text
#position, #position at which writing has to start
fontFace=cv2.FONT_HERSHEY_PLAIN, #font family
color=(0, 0, 255) if true!=pred else (0,255,0), #font color
bottomLeftOrigin=False,
fontScale = scale,
org = (10,25),
thickness=1)
cv2.imwrite(path1, cam_image)
if show_saving_path:
print("Saving path: ", path1)
path2 = grad_imgs_path+'_bg.jpg'
cv2.imwrite(path2, gb)
if show_saving_path:
print("Saving path: ", path2)
path3 = grad_imgs_path+'_cam_gb.jpg'
cv2.imwrite(path3, cam_gb)
if show_saving_path:
print("Saving path: ", path3)
if imshow==True:
plt.figure()
plt.imshow(cv2.cvtColor(cam_image, cv2.COLOR_BGR2RGB))
plt.figure()
plt.imshow(cv2.cvtColor(gb, cv2.COLOR_BGR2RGB))
plt.figure()
plt.imshow(cv2.cvtColor(cam_gb, cv2.COLOR_BGR2RGB))
# Choose the images for which the GradCam must be computed
from imutils import paths
import matplotlib as mpl
mpl.rcParams['figure.max_open_warning'] = 0 #remove warning
imgs_path = list(paths.list_images(gdrivePath+"/NatureDatasetGradCam"))
print(imgs_path)
for img_path in imgs_path:
compute_grad_cam(img_path,"resnet18",save=True,save_folder_path=saving_path,show_saving_path=False,imshow=False)
# Choose the images for which the GradCam must be computed
from imutils import paths
import matplotlib as mpl
mpl.rcParams['figure.max_open_warning'] = 0 #remove warning
imgs_path = list(paths.list_images(gdrivePath+"/NatureDatasetGradCam"))
print(imgs_path)
for img_path in imgs_path:
compute_grad_cam(img_path,"resnet18_imagenet",save=True,save_folder_path=saving_path,show_saving_path=False,imshow=False)
# Choose the images for which the GradCam must be computed
from imutils import paths
import matplotlib as mpl
mpl.rcParams['figure.max_open_warning'] = 0 #remove warning
imgs_path = list(paths.list_images(gdrivePath+"/NatureDatasetGradCam"))
print(imgs_path)
for img_path in imgs_path:
compute_grad_cam(img_path,"vgg16",save=True,save_folder_path=saving_path,show_saving_path=False,imshow=False)
| 0.674265 | 0.671921 |
# Neural network hybrid recommendation system on Google Analytics data preprocessing
This notebook demonstrates how to implement a hybrid recommendation system using a neural network to combine content-based and collaborative filtering recommendation models using Google Analytics data. We are going to use the learned user embeddings from [wals.ipynb](../wals.ipynb) and combine that with our previous content-based features from [content_based_using_neural_networks.ipynb](../content_based_using_neural_networks.ipynb)
First we are going to preprocess our data using BigQuery and Cloud Dataflow to be used in our later neural network hybrid recommendation model.
Apache Beam only works in Python 2 at the moment, so we're going to switch to the Python 2 kernel. In the above menu, click the dropdown arrow and select `python2`.
```
%%bash
conda update -y -n base -c defaults conda
source activate py2env
pip uninstall -y google-cloud-dataflow
conda install -y pytz
pip install apache-beam[gcp]==2.9.0
```
Now restart notebook's session kernel!
```
# Import helpful libraries and setup our project, bucket, and region
import os
PROJECT = "cloud-training-demos" # REPLACE WITH YOUR PROJECT ID
BUCKET = "cloud-training-demos-ml" # REPLACE WITH YOUR BUCKET NAME
REGION = "us-central1" # REPLACE WITH YOUR BUCKET REGION e.g. us-central1
# Do not change these
os.environ["PROJECT"] = PROJECT
os.environ["BUCKET"] = BUCKET
os.environ["REGION"] = REGION
os.environ["TFVERSION"] = "1.13"
%%bash
gcloud config set project $PROJECT
gcloud config set compute/region $REGION
```
<h2> Create ML dataset using Dataflow </h2>
Let's use Cloud Dataflow to read in the BigQuery data, do some preprocessing, and write it out as CSV files.
First, let's create our hybrid dataset query that we will use in our Cloud Dataflow pipeline. This will combine some content-based features and the user and item embeddings learned from our WALS Matrix Factorization Collaborative filtering lab that we extracted from our trained WALSMatrixFactorization Estimator and uploaded to BigQuery.
```
query_hybrid_dataset = """
WITH CTE_site_history AS (
SELECT
fullVisitorId as visitor_id,
(SELECT MAX(IF(index = 10, value, NULL)) FROM UNNEST(hits.customDimensions)) AS content_id,
(SELECT MAX(IF(index = 7, value, NULL)) FROM UNNEST(hits.customDimensions)) AS category,
(SELECT MAX(IF(index = 6, value, NULL)) FROM UNNEST(hits.customDimensions)) AS title,
(SELECT MAX(IF(index = 2, value, NULL)) FROM UNNEST(hits.customDimensions)) AS author_list,
SPLIT(RPAD((SELECT MAX(IF(index = 4, value, NULL)) FROM UNNEST(hits.customDimensions)), 7), '.') AS year_month_array,
LEAD(hits.customDimensions, 1) OVER (PARTITION BY fullVisitorId ORDER BY hits.time ASC) AS nextCustomDimensions
FROM
`cloud-training-demos.GA360_test.ga_sessions_sample`,
UNNEST(hits) AS hits
WHERE
# only include hits on pages
hits.type = "PAGE"
AND
fullVisitorId IS NOT NULL
AND
hits.time != 0
AND
hits.time IS NOT NULL
AND
(SELECT MAX(IF(index = 10, value, NULL)) FROM UNNEST(hits.customDimensions)) IS NOT NULL
),
CTE_training_dataset AS (
SELECT
(SELECT MAX(IF(index=10, value, NULL)) FROM UNNEST(nextCustomDimensions)) AS next_content_id,
visitor_id,
content_id,
category,
REGEXP_REPLACE(title, r",", "") AS title,
REGEXP_EXTRACT(author_list, r"^[^,]+") AS author,
DATE_DIFF(DATE(CAST(year_month_array[OFFSET(0)] AS INT64), CAST(year_month_array[OFFSET(1)] AS INT64), 1), DATE(1970, 1, 1), MONTH) AS months_since_epoch
FROM
CTE_site_history
WHERE
(SELECT MAX(IF(index=10, value, NULL)) FROM UNNEST(nextCustomDimensions)) IS NOT NULL)
SELECT
CAST(next_content_id AS STRING) AS next_content_id,
CAST(training_dataset.visitor_id AS STRING) AS visitor_id,
CAST(training_dataset.content_id AS STRING) AS content_id,
CAST(IFNULL(category, 'None') AS STRING) AS category,
CONCAT("\\"", REPLACE(TRIM(CAST(IFNULL(title, 'None') AS STRING)), "\\"",""), "\\"") AS title,
CAST(IFNULL(author, 'None') AS STRING) AS author,
CAST(months_since_epoch AS STRING) AS months_since_epoch,
IFNULL(user_factors._0, 0.0) AS user_factor_0,
IFNULL(user_factors._1, 0.0) AS user_factor_1,
IFNULL(user_factors._2, 0.0) AS user_factor_2,
IFNULL(user_factors._3, 0.0) AS user_factor_3,
IFNULL(user_factors._4, 0.0) AS user_factor_4,
IFNULL(user_factors._5, 0.0) AS user_factor_5,
IFNULL(user_factors._6, 0.0) AS user_factor_6,
IFNULL(user_factors._7, 0.0) AS user_factor_7,
IFNULL(user_factors._8, 0.0) AS user_factor_8,
IFNULL(user_factors._9, 0.0) AS user_factor_9,
IFNULL(item_factors._0, 0.0) AS item_factor_0,
IFNULL(item_factors._1, 0.0) AS item_factor_1,
IFNULL(item_factors._2, 0.0) AS item_factor_2,
IFNULL(item_factors._3, 0.0) AS item_factor_3,
IFNULL(item_factors._4, 0.0) AS item_factor_4,
IFNULL(item_factors._5, 0.0) AS item_factor_5,
IFNULL(item_factors._6, 0.0) AS item_factor_6,
IFNULL(item_factors._7, 0.0) AS item_factor_7,
IFNULL(item_factors._8, 0.0) AS item_factor_8,
IFNULL(item_factors._9, 0.0) AS item_factor_9,
FARM_FINGERPRINT(CONCAT(CAST(visitor_id AS STRING), CAST(content_id AS STRING))) AS hash_id
FROM
CTE_training_dataset AS training_dataset
LEFT JOIN
`cloud-training-demos.GA360_test.user_factors` AS user_factors
ON CAST(training_dataset.visitor_id AS FLOAT64) = CAST(user_factors.user_id AS FLOAT64)
LEFT JOIN
`cloud-training-demos.GA360_test.item_factors` AS item_factors
ON CAST(training_dataset.content_id AS STRING) = CAST(item_factors.item_id AS STRING)
"""
```
Let's pull a sample of our data into a dataframe to see what it looks like.
```
from google.cloud import bigquery
bq = bigquery.Client(project = PROJECT)
df_hybrid_dataset = bq.query(query_hybrid_dataset + "LIMIT 100").to_dataframe()
df_hybrid_dataset.head()
df_hybrid_dataset.describe()
import apache_beam as beam
import datetime, os
def to_csv(rowdict):
# Pull columns from BQ and create a line
import hashlib
import copy
CSV_COLUMNS = "next_content_id,visitor_id,content_id,category,title,author,months_since_epoch".split(",")
FACTOR_COLUMNS = ["user_factor_{}".format(i) for i in range(10)] + ["item_factor_{}".format(i) for i in range(10)]
# Write out rows for each input row for each column in rowdict
data = ",".join(["None" if k not in rowdict else (rowdict[k].encode("utf-8") if rowdict[k] is not None else "None") for k in CSV_COLUMNS])
data += ","
data += ",".join([str(rowdict[k]) if k in rowdict else "None" for k in FACTOR_COLUMNS])
yield ("{}".format(data))
def preprocess(in_test_mode):
import shutil, os, subprocess
job_name = "preprocess-hybrid-recommendation-features" + "-" + datetime.datetime.now().strftime("%y%m%d-%H%M%S")
if in_test_mode:
print("Launching local job ... hang on")
OUTPUT_DIR = "./preproc/features"
shutil.rmtree(OUTPUT_DIR, ignore_errors=True)
os.makedirs(OUTPUT_DIR)
else:
print("Launching Dataflow job {} ... hang on".format(job_name))
OUTPUT_DIR = "gs://{0}/hybrid_recommendation/preproc/features/".format(BUCKET)
try:
subprocess.check_call("gsutil -m rm -r {}".format(OUTPUT_DIR).split())
except:
pass
options = {
"staging_location": os.path.join(OUTPUT_DIR, "tmp", "staging"),
"temp_location": os.path.join(OUTPUT_DIR, "tmp"),
"job_name": job_name,
"project": PROJECT,
"teardown_policy": "TEARDOWN_ALWAYS",
"no_save_main_session": True,
"max_num_workers": 6
}
opts = beam.pipeline.PipelineOptions(flags = [], **options)
if in_test_mode:
RUNNER = "DirectRunner"
else:
RUNNER = "DataflowRunner"
p = beam.Pipeline(RUNNER, options = opts)
query = query_hybrid_dataset
if in_test_mode:
query = query + " LIMIT 100"
for step in ["train", "eval"]:
if step == "train":
selquery = "SELECT * FROM ({}) WHERE ABS(MOD(hash_id, 10)) < 9".format(query)
else:
selquery = "SELECT * FROM ({}) WHERE ABS(MOD(hash_id, 10)) = 9".format(query)
(p
| "{}_read".format(step) >> beam.io.Read(beam.io.BigQuerySource(query = selquery, use_standard_sql = True))
| "{}_csv".format(step) >> beam.FlatMap(to_csv)
| "{}_out".format(step) >> beam.io.Write(beam.io.WriteToText(os.path.join(OUTPUT_DIR, "{}.csv".format(step))))
)
job = p.run()
if in_test_mode:
job.wait_until_finish()
print("Done!")
preprocess(in_test_mode = False)
```
Let's check our files to make sure everything went as expected
```
%%bash
rm -rf features
mkdir features
!gsutil -m cp -r gs://{BUCKET}/hybrid_recommendation/preproc/features/*.csv* features/
!head -3 features/*
```
<h2> Create vocabularies using Dataflow </h2>
Let's use Cloud Dataflow to read in the BigQuery data, do some preprocessing, and write it out as CSV files.
Now we'll create our vocabulary files for our categorical features.
```
query_vocabularies = """
SELECT
CAST((SELECT MAX(IF(index = index_value, value, NULL)) FROM UNNEST(hits.customDimensions)) AS STRING) AS grouped_by
FROM `cloud-training-demos.GA360_test.ga_sessions_sample`,
UNNEST(hits) AS hits
WHERE
# only include hits on pages
hits.type = "PAGE"
AND (SELECT MAX(IF(index = index_value, value, NULL)) FROM UNNEST(hits.customDimensions)) IS NOT NULL
GROUP BY
grouped_by
"""
import apache_beam as beam
import datetime, os
def to_txt(rowdict):
# Pull columns from BQ and create a line
# Write out rows for each input row for grouped by column in rowdict
return "{}".format(rowdict["grouped_by"].encode("utf-8"))
def preprocess(in_test_mode):
import shutil, os, subprocess
job_name = "preprocess-hybrid-recommendation-vocab-lists" + "-" + datetime.datetime.now().strftime("%y%m%d-%H%M%S")
if in_test_mode:
print("Launching local job ... hang on")
OUTPUT_DIR = "./preproc/vocabs"
shutil.rmtree(OUTPUT_DIR, ignore_errors=True)
os.makedirs(OUTPUT_DIR)
else:
print("Launching Dataflow job {} ... hang on".format(job_name))
OUTPUT_DIR = "gs://{0}/hybrid_recommendation/preproc/vocabs/".format(BUCKET)
try:
subprocess.check_call("gsutil -m rm -r {}".format(OUTPUT_DIR).split())
except:
pass
options = {
"staging_location": os.path.join(OUTPUT_DIR, "tmp", "staging"),
"temp_location": os.path.join(OUTPUT_DIR, "tmp"),
"job_name": job_name,
"project": PROJECT,
"teardown_policy": "TEARDOWN_ALWAYS",
"no_save_main_session": True,
"max_num_workers": 6
}
opts = beam.pipeline.PipelineOptions(flags = [], **options)
if in_test_mode:
RUNNER = "DirectRunner"
else:
RUNNER = "DataflowRunner"
p = beam.Pipeline(RUNNER, options = opts)
def vocab_list(index, name):
query = query_vocabularies.replace("index_value", "{}".format(index))
(p
| "{}_read".format(name) >> beam.io.Read(beam.io.BigQuerySource(query = query, use_standard_sql = True))
| "{}_txt".format(name) >> beam.Map(to_txt)
| "{}_out".format(name) >> beam.io.Write(beam.io.WriteToText(os.path.join(OUTPUT_DIR, "{0}_vocab.txt".format(name))))
)
# Call vocab_list function for each
vocab_list(10, "content_id") # content_id
vocab_list(7, "category") # category
vocab_list(2, "author") # author
job = p.run()
if in_test_mode:
job.wait_until_finish()
print("Done!")
preprocess(in_test_mode = False)
```
Also get vocab counts from the length of the vocabularies
```
import apache_beam as beam
import datetime, os
def count_to_txt(rowdict):
# Pull columns from BQ and create a line
# Write out count
return "{}".format(rowdict["count_number"])
def mean_to_txt(rowdict):
# Pull columns from BQ and create a line
# Write out mean
return "{}".format(rowdict["mean_value"])
def preprocess(in_test_mode):
import shutil, os, subprocess
job_name = "preprocess-hybrid-recommendation-vocab-counts" + "-" + datetime.datetime.now().strftime("%y%m%d-%H%M%S")
if in_test_mode:
print("Launching local job ... hang on")
OUTPUT_DIR = "./preproc/vocab_counts"
shutil.rmtree(OUTPUT_DIR, ignore_errors=True)
os.makedirs(OUTPUT_DIR)
else:
print("Launching Dataflow job {} ... hang on".format(job_name))
OUTPUT_DIR = "gs://{0}/hybrid_recommendation/preproc/vocab_counts/".format(BUCKET)
try:
subprocess.check_call("gsutil -m rm -r {}".format(OUTPUT_DIR).split())
except:
pass
options = {
"staging_location": os.path.join(OUTPUT_DIR, "tmp", "staging"),
"temp_location": os.path.join(OUTPUT_DIR, "tmp"),
"job_name": job_name,
"project": PROJECT,
"teardown_policy": "TEARDOWN_ALWAYS",
"no_save_main_session": True,
"max_num_workers": 6
}
opts = beam.pipeline.PipelineOptions(flags = [], **options)
if in_test_mode:
RUNNER = "DirectRunner"
else:
RUNNER = "DataflowRunner"
p = beam.Pipeline(RUNNER, options = opts)
def vocab_count(index, column_name):
query = """
SELECT
COUNT(*) AS count_number
FROM ({})
""".format(query_vocabularies.replace("index_value", "{}".format(index)))
(p
| "{}_read".format(column_name) >> beam.io.Read(beam.io.BigQuerySource(query = query, use_standard_sql = True))
| "{}_txt".format(column_name) >> beam.Map(count_to_txt)
| "{}_out".format(column_name) >> beam.io.Write(beam.io.WriteToText(os.path.join(OUTPUT_DIR, "{0}_vocab_count.txt".format(column_name))))
)
def global_column_mean(column_name):
query = """
SELECT
AVG(CAST({1} AS FLOAT64)) AS mean_value
FROM ({0})
""".format(query_hybrid_dataset, column_name)
(p
| "{}_read".format(column_name) >> beam.io.Read(beam.io.BigQuerySource(query = query, use_standard_sql = True))
| "{}_txt".format(column_name) >> beam.Map(mean_to_txt)
| "{}_out".format(column_name) >> beam.io.Write(beam.io.WriteToText(os.path.join(OUTPUT_DIR, "{0}_mean.txt".format(column_name))))
)
# Call vocab_count function for each column we want the vocabulary count for
vocab_count(10, "content_id") # content_id
vocab_count(7, "category") # category
vocab_count(2, "author") # author
# Call global_column_mean function for each column we want the mean for
global_column_mean("months_since_epoch") # months_since_epoch
job = p.run()
if in_test_mode:
job.wait_until_finish()
print("Done!")
preprocess(in_test_mode = False)
```
Let's check our files to make sure everything went as expected
```
%%bash
rm -rf vocabs
mkdir vocabs
!gsutil -m cp -r gs://{BUCKET}/hybrid_recommendation/preproc/vocabs/*.txt* vocabs/
!head -3 vocabs/*
%%bash
rm -rf vocab_counts
mkdir vocab_counts
!gsutil -m cp -r gs://{BUCKET}/hybrid_recommendation/preproc/vocab_counts/*.txt* vocab_counts/
!head -3 vocab_counts/*
```
|
github_jupyter
|
%%bash
conda update -y -n base -c defaults conda
source activate py2env
pip uninstall -y google-cloud-dataflow
conda install -y pytz
pip install apache-beam[gcp]==2.9.0
# Import helpful libraries and setup our project, bucket, and region
import os
PROJECT = "cloud-training-demos" # REPLACE WITH YOUR PROJECT ID
BUCKET = "cloud-training-demos-ml" # REPLACE WITH YOUR BUCKET NAME
REGION = "us-central1" # REPLACE WITH YOUR BUCKET REGION e.g. us-central1
# Do not change these
os.environ["PROJECT"] = PROJECT
os.environ["BUCKET"] = BUCKET
os.environ["REGION"] = REGION
os.environ["TFVERSION"] = "1.13"
%%bash
gcloud config set project $PROJECT
gcloud config set compute/region $REGION
query_hybrid_dataset = """
WITH CTE_site_history AS (
SELECT
fullVisitorId as visitor_id,
(SELECT MAX(IF(index = 10, value, NULL)) FROM UNNEST(hits.customDimensions)) AS content_id,
(SELECT MAX(IF(index = 7, value, NULL)) FROM UNNEST(hits.customDimensions)) AS category,
(SELECT MAX(IF(index = 6, value, NULL)) FROM UNNEST(hits.customDimensions)) AS title,
(SELECT MAX(IF(index = 2, value, NULL)) FROM UNNEST(hits.customDimensions)) AS author_list,
SPLIT(RPAD((SELECT MAX(IF(index = 4, value, NULL)) FROM UNNEST(hits.customDimensions)), 7), '.') AS year_month_array,
LEAD(hits.customDimensions, 1) OVER (PARTITION BY fullVisitorId ORDER BY hits.time ASC) AS nextCustomDimensions
FROM
`cloud-training-demos.GA360_test.ga_sessions_sample`,
UNNEST(hits) AS hits
WHERE
# only include hits on pages
hits.type = "PAGE"
AND
fullVisitorId IS NOT NULL
AND
hits.time != 0
AND
hits.time IS NOT NULL
AND
(SELECT MAX(IF(index = 10, value, NULL)) FROM UNNEST(hits.customDimensions)) IS NOT NULL
),
CTE_training_dataset AS (
SELECT
(SELECT MAX(IF(index=10, value, NULL)) FROM UNNEST(nextCustomDimensions)) AS next_content_id,
visitor_id,
content_id,
category,
REGEXP_REPLACE(title, r",", "") AS title,
REGEXP_EXTRACT(author_list, r"^[^,]+") AS author,
DATE_DIFF(DATE(CAST(year_month_array[OFFSET(0)] AS INT64), CAST(year_month_array[OFFSET(1)] AS INT64), 1), DATE(1970, 1, 1), MONTH) AS months_since_epoch
FROM
CTE_site_history
WHERE
(SELECT MAX(IF(index=10, value, NULL)) FROM UNNEST(nextCustomDimensions)) IS NOT NULL)
SELECT
CAST(next_content_id AS STRING) AS next_content_id,
CAST(training_dataset.visitor_id AS STRING) AS visitor_id,
CAST(training_dataset.content_id AS STRING) AS content_id,
CAST(IFNULL(category, 'None') AS STRING) AS category,
CONCAT("\\"", REPLACE(TRIM(CAST(IFNULL(title, 'None') AS STRING)), "\\"",""), "\\"") AS title,
CAST(IFNULL(author, 'None') AS STRING) AS author,
CAST(months_since_epoch AS STRING) AS months_since_epoch,
IFNULL(user_factors._0, 0.0) AS user_factor_0,
IFNULL(user_factors._1, 0.0) AS user_factor_1,
IFNULL(user_factors._2, 0.0) AS user_factor_2,
IFNULL(user_factors._3, 0.0) AS user_factor_3,
IFNULL(user_factors._4, 0.0) AS user_factor_4,
IFNULL(user_factors._5, 0.0) AS user_factor_5,
IFNULL(user_factors._6, 0.0) AS user_factor_6,
IFNULL(user_factors._7, 0.0) AS user_factor_7,
IFNULL(user_factors._8, 0.0) AS user_factor_8,
IFNULL(user_factors._9, 0.0) AS user_factor_9,
IFNULL(item_factors._0, 0.0) AS item_factor_0,
IFNULL(item_factors._1, 0.0) AS item_factor_1,
IFNULL(item_factors._2, 0.0) AS item_factor_2,
IFNULL(item_factors._3, 0.0) AS item_factor_3,
IFNULL(item_factors._4, 0.0) AS item_factor_4,
IFNULL(item_factors._5, 0.0) AS item_factor_5,
IFNULL(item_factors._6, 0.0) AS item_factor_6,
IFNULL(item_factors._7, 0.0) AS item_factor_7,
IFNULL(item_factors._8, 0.0) AS item_factor_8,
IFNULL(item_factors._9, 0.0) AS item_factor_9,
FARM_FINGERPRINT(CONCAT(CAST(visitor_id AS STRING), CAST(content_id AS STRING))) AS hash_id
FROM
CTE_training_dataset AS training_dataset
LEFT JOIN
`cloud-training-demos.GA360_test.user_factors` AS user_factors
ON CAST(training_dataset.visitor_id AS FLOAT64) = CAST(user_factors.user_id AS FLOAT64)
LEFT JOIN
`cloud-training-demos.GA360_test.item_factors` AS item_factors
ON CAST(training_dataset.content_id AS STRING) = CAST(item_factors.item_id AS STRING)
"""
from google.cloud import bigquery
bq = bigquery.Client(project = PROJECT)
df_hybrid_dataset = bq.query(query_hybrid_dataset + "LIMIT 100").to_dataframe()
df_hybrid_dataset.head()
df_hybrid_dataset.describe()
import apache_beam as beam
import datetime, os
def to_csv(rowdict):
# Pull columns from BQ and create a line
import hashlib
import copy
CSV_COLUMNS = "next_content_id,visitor_id,content_id,category,title,author,months_since_epoch".split(",")
FACTOR_COLUMNS = ["user_factor_{}".format(i) for i in range(10)] + ["item_factor_{}".format(i) for i in range(10)]
# Write out rows for each input row for each column in rowdict
data = ",".join(["None" if k not in rowdict else (rowdict[k].encode("utf-8") if rowdict[k] is not None else "None") for k in CSV_COLUMNS])
data += ","
data += ",".join([str(rowdict[k]) if k in rowdict else "None" for k in FACTOR_COLUMNS])
yield ("{}".format(data))
def preprocess(in_test_mode):
import shutil, os, subprocess
job_name = "preprocess-hybrid-recommendation-features" + "-" + datetime.datetime.now().strftime("%y%m%d-%H%M%S")
if in_test_mode:
print("Launching local job ... hang on")
OUTPUT_DIR = "./preproc/features"
shutil.rmtree(OUTPUT_DIR, ignore_errors=True)
os.makedirs(OUTPUT_DIR)
else:
print("Launching Dataflow job {} ... hang on".format(job_name))
OUTPUT_DIR = "gs://{0}/hybrid_recommendation/preproc/features/".format(BUCKET)
try:
subprocess.check_call("gsutil -m rm -r {}".format(OUTPUT_DIR).split())
except:
pass
options = {
"staging_location": os.path.join(OUTPUT_DIR, "tmp", "staging"),
"temp_location": os.path.join(OUTPUT_DIR, "tmp"),
"job_name": job_name,
"project": PROJECT,
"teardown_policy": "TEARDOWN_ALWAYS",
"no_save_main_session": True,
"max_num_workers": 6
}
opts = beam.pipeline.PipelineOptions(flags = [], **options)
if in_test_mode:
RUNNER = "DirectRunner"
else:
RUNNER = "DataflowRunner"
p = beam.Pipeline(RUNNER, options = opts)
query = query_hybrid_dataset
if in_test_mode:
query = query + " LIMIT 100"
for step in ["train", "eval"]:
if step == "train":
selquery = "SELECT * FROM ({}) WHERE ABS(MOD(hash_id, 10)) < 9".format(query)
else:
selquery = "SELECT * FROM ({}) WHERE ABS(MOD(hash_id, 10)) = 9".format(query)
(p
| "{}_read".format(step) >> beam.io.Read(beam.io.BigQuerySource(query = selquery, use_standard_sql = True))
| "{}_csv".format(step) >> beam.FlatMap(to_csv)
| "{}_out".format(step) >> beam.io.Write(beam.io.WriteToText(os.path.join(OUTPUT_DIR, "{}.csv".format(step))))
)
job = p.run()
if in_test_mode:
job.wait_until_finish()
print("Done!")
preprocess(in_test_mode = False)
%%bash
rm -rf features
mkdir features
!gsutil -m cp -r gs://{BUCKET}/hybrid_recommendation/preproc/features/*.csv* features/
!head -3 features/*
query_vocabularies = """
SELECT
CAST((SELECT MAX(IF(index = index_value, value, NULL)) FROM UNNEST(hits.customDimensions)) AS STRING) AS grouped_by
FROM `cloud-training-demos.GA360_test.ga_sessions_sample`,
UNNEST(hits) AS hits
WHERE
# only include hits on pages
hits.type = "PAGE"
AND (SELECT MAX(IF(index = index_value, value, NULL)) FROM UNNEST(hits.customDimensions)) IS NOT NULL
GROUP BY
grouped_by
"""
import apache_beam as beam
import datetime, os
def to_txt(rowdict):
# Pull columns from BQ and create a line
# Write out rows for each input row for grouped by column in rowdict
return "{}".format(rowdict["grouped_by"].encode("utf-8"))
def preprocess(in_test_mode):
import shutil, os, subprocess
job_name = "preprocess-hybrid-recommendation-vocab-lists" + "-" + datetime.datetime.now().strftime("%y%m%d-%H%M%S")
if in_test_mode:
print("Launching local job ... hang on")
OUTPUT_DIR = "./preproc/vocabs"
shutil.rmtree(OUTPUT_DIR, ignore_errors=True)
os.makedirs(OUTPUT_DIR)
else:
print("Launching Dataflow job {} ... hang on".format(job_name))
OUTPUT_DIR = "gs://{0}/hybrid_recommendation/preproc/vocabs/".format(BUCKET)
try:
subprocess.check_call("gsutil -m rm -r {}".format(OUTPUT_DIR).split())
except:
pass
options = {
"staging_location": os.path.join(OUTPUT_DIR, "tmp", "staging"),
"temp_location": os.path.join(OUTPUT_DIR, "tmp"),
"job_name": job_name,
"project": PROJECT,
"teardown_policy": "TEARDOWN_ALWAYS",
"no_save_main_session": True,
"max_num_workers": 6
}
opts = beam.pipeline.PipelineOptions(flags = [], **options)
if in_test_mode:
RUNNER = "DirectRunner"
else:
RUNNER = "DataflowRunner"
p = beam.Pipeline(RUNNER, options = opts)
def vocab_list(index, name):
query = query_vocabularies.replace("index_value", "{}".format(index))
(p
| "{}_read".format(name) >> beam.io.Read(beam.io.BigQuerySource(query = query, use_standard_sql = True))
| "{}_txt".format(name) >> beam.Map(to_txt)
| "{}_out".format(name) >> beam.io.Write(beam.io.WriteToText(os.path.join(OUTPUT_DIR, "{0}_vocab.txt".format(name))))
)
# Call vocab_list function for each
vocab_list(10, "content_id") # content_id
vocab_list(7, "category") # category
vocab_list(2, "author") # author
job = p.run()
if in_test_mode:
job.wait_until_finish()
print("Done!")
preprocess(in_test_mode = False)
import apache_beam as beam
import datetime, os
def count_to_txt(rowdict):
# Pull columns from BQ and create a line
# Write out count
return "{}".format(rowdict["count_number"])
def mean_to_txt(rowdict):
# Pull columns from BQ and create a line
# Write out mean
return "{}".format(rowdict["mean_value"])
def preprocess(in_test_mode):
import shutil, os, subprocess
job_name = "preprocess-hybrid-recommendation-vocab-counts" + "-" + datetime.datetime.now().strftime("%y%m%d-%H%M%S")
if in_test_mode:
print("Launching local job ... hang on")
OUTPUT_DIR = "./preproc/vocab_counts"
shutil.rmtree(OUTPUT_DIR, ignore_errors=True)
os.makedirs(OUTPUT_DIR)
else:
print("Launching Dataflow job {} ... hang on".format(job_name))
OUTPUT_DIR = "gs://{0}/hybrid_recommendation/preproc/vocab_counts/".format(BUCKET)
try:
subprocess.check_call("gsutil -m rm -r {}".format(OUTPUT_DIR).split())
except:
pass
options = {
"staging_location": os.path.join(OUTPUT_DIR, "tmp", "staging"),
"temp_location": os.path.join(OUTPUT_DIR, "tmp"),
"job_name": job_name,
"project": PROJECT,
"teardown_policy": "TEARDOWN_ALWAYS",
"no_save_main_session": True,
"max_num_workers": 6
}
opts = beam.pipeline.PipelineOptions(flags = [], **options)
if in_test_mode:
RUNNER = "DirectRunner"
else:
RUNNER = "DataflowRunner"
p = beam.Pipeline(RUNNER, options = opts)
def vocab_count(index, column_name):
query = """
SELECT
COUNT(*) AS count_number
FROM ({})
""".format(query_vocabularies.replace("index_value", "{}".format(index)))
(p
| "{}_read".format(column_name) >> beam.io.Read(beam.io.BigQuerySource(query = query, use_standard_sql = True))
| "{}_txt".format(column_name) >> beam.Map(count_to_txt)
| "{}_out".format(column_name) >> beam.io.Write(beam.io.WriteToText(os.path.join(OUTPUT_DIR, "{0}_vocab_count.txt".format(column_name))))
)
def global_column_mean(column_name):
query = """
SELECT
AVG(CAST({1} AS FLOAT64)) AS mean_value
FROM ({0})
""".format(query_hybrid_dataset, column_name)
(p
| "{}_read".format(column_name) >> beam.io.Read(beam.io.BigQuerySource(query = query, use_standard_sql = True))
| "{}_txt".format(column_name) >> beam.Map(mean_to_txt)
| "{}_out".format(column_name) >> beam.io.Write(beam.io.WriteToText(os.path.join(OUTPUT_DIR, "{0}_mean.txt".format(column_name))))
)
# Call vocab_count function for each column we want the vocabulary count for
vocab_count(10, "content_id") # content_id
vocab_count(7, "category") # category
vocab_count(2, "author") # author
# Call global_column_mean function for each column we want the mean for
global_column_mean("months_since_epoch") # months_since_epoch
job = p.run()
if in_test_mode:
job.wait_until_finish()
print("Done!")
preprocess(in_test_mode = False)
%%bash
rm -rf vocabs
mkdir vocabs
!gsutil -m cp -r gs://{BUCKET}/hybrid_recommendation/preproc/vocabs/*.txt* vocabs/
!head -3 vocabs/*
%%bash
rm -rf vocab_counts
mkdir vocab_counts
!gsutil -m cp -r gs://{BUCKET}/hybrid_recommendation/preproc/vocab_counts/*.txt* vocab_counts/
!head -3 vocab_counts/*
| 0.247987 | 0.887984 |
[View in Colaboratory](https://colab.research.google.com/github/nikhilbhatewara/CodeSnippets/blob/master/notebook2.ipynb)
```
%%time
import pandas as pd
import numpy as np
import os
import scipy.stats as sts
!pip install xlrd
import io
# plotting modules
import seaborn as sns
import matplotlib.pyplot as plt
# Load the Drive helper and mount
from google.colab import drive
# This will prompt for authorization.
drive.mount('/content/drive')
# After executing the cell above, Drive
# files will be present in "/content/drive/My Drive/dataincubator/details".
!ls "/content/drive/My Drive/dataincubator/details"
%%time
data_2009Q3 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2009Q3-house-disburse-detail.csv",engine="python")
data_2009Q4 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2009Q4-house-disburse-detail.csv",engine="python")
data_2010Q1 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2010Q1-house-disburse-detail.csv",engine="python")
data_2010Q2 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2010Q2-house-disburse-detail.csv",engine="python")
data_2010Q3 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2010Q3-house-disburse-detail.csv",engine="python")
data_2010Q4 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2010Q4-house-disburse-detail.csv",engine="python")
data_2011Q1 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2011Q1-house-disburse-detail.csv",engine="python")
data_2011Q2 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2011Q2-house-disburse-detail.csv",engine="python")
data_2011Q3 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2011Q3-house-disburse-detail.csv",engine="python")
data_2011Q4 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2011Q4-house-disburse-detail.csv",engine="python")
data_2012Q1 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2012Q1-house-disburse-detail.csv",engine="python")
data_2012Q2 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2012Q2-house-disburse-detail.csv",engine="python")
data_2012Q3 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2012Q3-house-disburse-detail.csv",engine="python")
data_2012Q4 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2012Q4-house-disburse-detail.csv",engine="python")
data_2013Q1 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2013Q1-house-disburse-detail.csv",engine="python")
data_2013Q2 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2013Q2-house-disburse-detail.csv",engine="python")
data_2013Q3 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2013Q3-house-disburse-detail.csv",engine="python")
data_2013Q4 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2013Q4-house-disburse-detail.csv",engine="python")
data_2014Q1 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2014Q1-house-disburse-detail.csv",engine="python")
data_2014Q2 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2014Q2-house-disburse-detail.csv",engine="python")
data_2014Q3 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2014Q3-house-disburse-detail.csv",engine="python")
data_2014Q4 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2014Q4-house-disburse-detail.csv",engine="python")
data_2015Q1 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2015Q1-house-disburse-detail.csv",engine="python")
data_2015Q2 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2015Q2-house-disburse-detail-updated.csv",engine="python")
data_2015Q3 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2015Q3-house-disburse-detail.csv",engine="python")
data_2015Q4 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2015Q4-house-disburse-detail.csv",engine="python")
data_2016Q1 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2016Q1-house-disburse-detail.csv",engine="python")
data_2016Q2 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2016Q2-house-disburse-detail.csv",engine="python")
data_2016Q3 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2016Q3-house-disburse-detail.csv",engine="python")
data_2016Q4 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2016Q4-house-disburse-detail.csv",engine="python")
data_2017Q1 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2017Q1-house-disburse-detail.csv",engine="python")
data_2017Q2 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2017Q2-house-disburse-detail.csv",engine="python")
data_2017Q3 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2017Q3-house-disburse-detail.csv",engine="python")
data_2017Q4 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2017Q4-house-disburse-detail.csv",engine="python")
data_2018Q1 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2018Q1-house-disburse-detail.csv",engine="python")
data_2009Q3.columns = data_2009Q3.columns.str.replace('\s+', '_')
data_2009Q4.columns = data_2009Q4.columns.str.replace('\s+', '_')
data_2010Q1.columns = data_2010Q1.columns.str.replace('\s+', '_')
data_2010Q2.columns = data_2010Q2.columns.str.replace('\s+', '_')
data_2010Q3.columns = data_2010Q3.columns.str.replace('\s+', '_')
data_2010Q4.columns = data_2010Q4.columns.str.replace('\s+', '_')
data_2011Q1.columns = data_2011Q1.columns.str.replace('\s+', '_')
data_2011Q2.columns = data_2011Q2.columns.str.replace('\s+', '_')
data_2011Q3.columns = data_2011Q3.columns.str.replace('\s+', '_')
data_2011Q4.columns = data_2011Q4.columns.str.replace('\s+', '_')
data_2012Q1.columns = data_2012Q1.columns.str.replace('\s+', '_')
data_2012Q2.columns = data_2012Q2.columns.str.replace('\s+', '_')
data_2012Q3.columns = data_2012Q3.columns.str.replace('\s+', '_')
data_2012Q4.columns = data_2012Q4.columns.str.replace('\s+', '_')
data_2013Q1.columns = data_2013Q1.columns.str.replace('\s+', '_')
data_2013Q2.columns = data_2013Q2.columns.str.replace('\s+', '_')
data_2013Q3.columns = data_2013Q3.columns.str.replace('\s+', '_')
data_2013Q4.columns = data_2013Q4.columns.str.replace('\s+', '_')
data_2014Q1.columns = data_2014Q1.columns.str.replace('\s+', '_')
data_2014Q2.columns = data_2014Q2.columns.str.replace('\s+', '_')
data_2014Q3.columns = data_2014Q3.columns.str.replace('\s+', '_')
data_2014Q4.columns = data_2014Q4.columns.str.replace('\s+', '_')
data_2015Q1.columns = data_2015Q1.columns.str.replace('\s+', '_')
data_2015Q2.columns = data_2015Q2.columns.str.replace('\s+', '_')
data_2015Q3.columns = data_2015Q3.columns.str.replace('\s+', '_')
data_2015Q4.columns = data_2015Q4.columns.str.replace('\s+', '_')
data_2016Q1.columns = data_2016Q1.columns.str.replace('\s+', '_')
data_2016Q2.columns = data_2016Q2.columns.str.replace('\s+', '_')
data_2016Q3.columns = data_2016Q3.columns.str.replace('\s+', '_')
data_2016Q4.columns = data_2016Q4.columns.str.replace('\s+', '_')
data_2017Q1.columns = data_2017Q1.columns.str.replace('\s+', '_')
data_2017Q2.columns = data_2017Q2.columns.str.replace('\s+', '_')
data_2017Q3.columns = data_2017Q3.columns.str.replace('\s+', '_')
data_2017Q4.columns = data_2017Q4.columns.str.replace('\s+', '_')
data_2018Q1.columns = data_2018Q1.columns.str.replace('\s+', '_')
data_2009Q3.isnull().sum()
data_2009Q4.isnull().sum()
data_2010Q1.isnull().sum()
data_2010Q2.isnull().sum()
data_2010Q3.isnull().sum()
data_2010Q4.isnull().sum()
data_2011Q1.isnull().sum()
data_2011Q2.isnull().sum()
data_2011Q3.isnull().sum()
data_2011Q4.isnull().sum()
data_2012Q1.isnull().sum()
data_2012Q2.isnull().sum()
data_2012Q3.isnull().sum()
data_2012Q4.isnull().sum()
data_2013Q1.isnull().sum()
data_2013Q2.isnull().sum()
data_2013Q3.isnull().sum()
data_2013Q4.isnull().sum()
data_2014Q1.isnull().sum()
data_2014Q2.isnull().sum()
data_2014Q3.isnull().sum()
data_2014Q4.isnull().sum()
data_2015Q1.isnull().sum()
data_2015Q2.isnull().sum()
data_2015Q3.isnull().sum()
data_2015Q4.isnull().sum()
data_2016Q1.isnull().sum()
data_2016Q2.isnull().sum()
data_2016Q3.isnull().sum()
data_2016Q4.isnull().sum()
data_2017Q1.isnull().sum()
data_2017Q2.isnull().sum()
data_2017Q3.isnull().sum()
data_2017Q4.isnull().sum()
data_2018Q1.isnull().sum()
data_2009Q3.AMOUNT=data_2009Q3.AMOUNT.str.replace(',', '').astype('float64')
data_2009Q4.AMOUNT=data_2009Q4.AMOUNT.str.replace(',', '').astype('float64')
data_2010Q1.AMOUNT=data_2010Q1.AMOUNT.str.replace(',', '').astype('float64')
data_2010Q2.AMOUNT=data_2010Q2.AMOUNT.str.replace(',', '').astype('float64')
data_2010Q3.AMOUNT=data_2010Q3.AMOUNT.str.replace(',', '').astype('float64')
data_2010Q4.AMOUNT=data_2010Q4.AMOUNT.str.replace(',', '').astype('float64')
data_2011Q1.AMOUNT=data_2011Q1.AMOUNT.str.replace(',', '').astype('float64')
data_2011Q2.AMOUNT=data_2011Q2.AMOUNT.str.replace(',', '').astype('float64')
data_2011Q3.AMOUNT=data_2011Q3.AMOUNT.str.replace(',', '').astype('float64')
data_2011Q4.AMOUNT=data_2011Q4.AMOUNT.str.replace(',', '').astype('float64')
data_2012Q1.AMOUNT=data_2012Q1.AMOUNT.str.replace(',', '').astype('float64')
data_2012Q2.AMOUNT=data_2012Q2.AMOUNT.str.replace(',', '').astype('float64')
data_2012Q3.AMOUNT=data_2012Q3.AMOUNT.str.replace(',', '').astype('float64')
data_2012Q4.AMOUNT=data_2012Q4.AMOUNT.str.replace(',', '').astype('float64')
data_2013Q1.AMOUNT=data_2013Q1.AMOUNT.str.replace(',', '').astype('float64')
data_2013Q2.AMOUNT=data_2013Q2.AMOUNT.str.replace(',', '').astype('float64')
data_2013Q3.AMOUNT=data_2013Q3.AMOUNT.str.replace(',', '').astype('float64')
data_2013Q4.AMOUNT=data_2013Q4.AMOUNT.str.replace(',', '').astype('float64')
data_2014Q1.AMOUNT=data_2014Q1.AMOUNT.str.replace(',', '').astype('float64')
data_2014Q2.AMOUNT=data_2014Q2.AMOUNT.str.replace(',', '').astype('float64')
data_2014Q3.AMOUNT=data_2014Q3.AMOUNT.str.replace(',', '').astype('float64')
data_2014Q4.AMOUNT=data_2014Q4.AMOUNT.str.replace(',', '').astype('float64')
data_2015Q1.AMOUNT=data_2015Q1.AMOUNT.str.replace(',', '').astype('float64')
data_2015Q2.AMOUNT=data_2015Q2.AMOUNT.str.replace(',', '').astype('float64')
data_2015Q3.AMOUNT=data_2015Q3.AMOUNT.str.replace(',', '').astype('float64')
data_2015Q4.AMOUNT=data_2015Q4.AMOUNT.str.replace(',', '').astype('float64')
data_2016Q1.AMOUNT=data_2016Q1.AMOUNT.str.replace(',', '').astype('float64')
data_2016Q2.AMOUNT=data_2016Q2.AMOUNT.str.replace(',', '').astype('float64')
data_2016Q3.AMOUNT=data_2016Q3.AMOUNT.str.replace(',', '').astype('float64')
# No String Values, hence we cannot use .str
data_2016Q4.AMOUNT=data_2016Q4.AMOUNT.replace(',', '').astype('float64')
data_2017Q1.AMOUNT=data_2017Q1.AMOUNT.replace(',', '').astype('float64')
data_2017Q2.AMOUNT=data_2017Q2.AMOUNT.replace(',', '').astype('float64')
data_2017Q3.AMOUNT=data_2017Q3.AMOUNT.replace(',', '').astype('float64')
data_2017Q4.AMOUNT=data_2017Q4.AMOUNT.replace(',', '').astype('float64')
data_2018Q1.AMOUNT=data_2018Q1.AMOUNT.replace(',', '').astype('float64')
def convert_float_amount(df):
try:
df.loc[:,"AMOUNT"]=df.loc[:,"AMOUNT"].str.replace(',', '').astype('float64')
except Exception as e:
print('Invalid Data', e)
else:
df.loc[:,"AMOUNT"]=df.loc[:,"AMOUNT"].replace(',', '').astype('float64')
return df
data_2009Q3=convert_float_amount(data_2009Q3)
data_2009Q4=convert_float_amount(data_2009Q4)
data_2010Q1=convert_float_amount(data_2010Q1)
data_2010Q2=convert_float_amount(data_2010Q2)
data_2010Q3=convert_float_amount(data_2010Q3)
data_2010Q4=convert_float_amount(data_2010Q4)
data_2011Q1=convert_float_amount(data_2011Q1)
data_2011Q2=convert_float_amount(data_2011Q2)
data_2011Q3=convert_float_amount(data_2011Q3)
data_2011Q4=convert_float_amount(data_2011Q4)
data_2012Q1=convert_float_amount(data_2012Q1)
data_2012Q2=convert_float_amount(data_2012Q2)
data_2012Q3=convert_float_amount(data_2012Q3)
data_2012Q4=convert_float_amount(data_2012Q4)
data_2013Q1=convert_float_amount(data_2013Q1)
data_2013Q2=convert_float_amount(data_2013Q2)
data_2013Q3=convert_float_amount(data_2013Q3)
data_2013Q4=convert_float_amount(data_2013Q4)
data_2014Q1=convert_float_amount(data_2014Q1)
data_2014Q2=convert_float_amount(data_2014Q2)
data_2014Q3=convert_float_amount(data_2014Q3)
data_2014Q4=convert_float_amount(data_2014Q4)
data_2015Q1=convert_float_amount(data_2015Q1)
data_2015Q2=convert_float_amount(data_2015Q2)
data_2015Q3=convert_float_amount(data_2015Q3)
data_2015Q4=convert_float_amount(data_2015Q4)
data_2016Q1=convert_float_amount(data_2016Q1)
data_2016Q2=convert_float_amount(data_2016Q2)
data_2016Q3=convert_float_amount(data_2016Q3)
data_2016Q4=convert_float_amount(data_2016Q4)
data_2017Q1=convert_float_amount(data_2017Q1)
data_2017Q2=convert_float_amount(data_2017Q2)
data_2017Q3=convert_float_amount(data_2017Q3)
data_2017Q4=convert_float_amount(data_2017Q4)
data_2018Q1=convert_float_amount(data_2018Q1)
data_2009Q3_TOTAL=sum(data_2009Q3.AMOUNT)
data_2009Q4_TOTAL=sum(data_2009Q4.AMOUNT)
data_2010Q1_TOTAL=sum(data_2010Q1.AMOUNT)
data_2010Q2_TOTAL=sum(data_2010Q2.AMOUNT)
data_2010Q3_TOTAL=sum(data_2010Q3.AMOUNT)
data_2010Q4_TOTAL=sum(data_2010Q4.AMOUNT)
data_2011Q1_TOTAL=sum(data_2011Q1.AMOUNT)
data_2011Q2_TOTAL=sum(data_2011Q2.AMOUNT)
data_2011Q3_TOTAL=sum(data_2011Q3.AMOUNT)
data_2011Q4_TOTAL=sum(data_2011Q4.AMOUNT)
data_2012Q1_TOTAL=sum(data_2012Q1.AMOUNT)
data_2012Q2_TOTAL=sum(data_2012Q2.AMOUNT)
data_2012Q3_TOTAL=sum(data_2012Q3.AMOUNT)
data_2012Q4_TOTAL=sum(data_2012Q4.AMOUNT)
data_2013Q1_TOTAL=sum(data_2013Q1.AMOUNT)
data_2013Q2_TOTAL=sum(data_2013Q2.AMOUNT)
data_2013Q3_TOTAL=sum(data_2013Q3.AMOUNT)
data_2013Q4_TOTAL=sum(data_2013Q4.AMOUNT)
data_2014Q1_TOTAL=sum(data_2014Q1.AMOUNT)
data_2014Q2_TOTAL=sum(data_2014Q2.AMOUNT)
data_2014Q3_TOTAL=sum(data_2014Q3.AMOUNT)
data_2014Q4_TOTAL=sum(data_2014Q4.AMOUNT)
data_2015Q1_TOTAL=sum(data_2015Q1.AMOUNT)
data_2015Q2_TOTAL=sum(data_2015Q2.AMOUNT)
data_2015Q3_TOTAL=sum(data_2015Q3.AMOUNT)
data_2015Q4_TOTAL=sum(data_2015Q4.AMOUNT)
data_2016Q1_TOTAL=sum(data_2016Q1.AMOUNT)
data_2016Q2_TOTAL=sum(data_2016Q2.AMOUNT)
data_2016Q3_TOTAL=sum(data_2016Q3.AMOUNT)
data_2016Q4_TOTAL=sum(data_2016Q4.AMOUNT)
data_2017Q1_TOTAL=sum(data_2017Q1.AMOUNT)
data_2017Q2_TOTAL=sum(data_2017Q2.AMOUNT)
data_2017Q3_TOTAL=sum(data_2017Q3.AMOUNT)
data_2017Q4_TOTAL=sum(data_2017Q4.AMOUNT)
data_2018Q1_TOTAL=sum(data_2018Q1.AMOUNT)
my_list = []
my_list.append(data_2009Q3_TOTAL)
my_list.append(data_2009Q4_TOTAL)
my_list.append(data_2010Q1_TOTAL)
my_list.append(data_2010Q2_TOTAL)
my_list.append(data_2010Q3_TOTAL)
my_list.append(data_2010Q4_TOTAL)
my_list.append(data_2011Q1_TOTAL)
my_list.append(data_2011Q2_TOTAL)
my_list.append(data_2011Q3_TOTAL)
my_list.append(data_2011Q4_TOTAL)
my_list.append(data_2012Q1_TOTAL)
my_list.append(data_2012Q2_TOTAL)
my_list.append(data_2012Q3_TOTAL)
my_list.append(data_2012Q4_TOTAL)
my_list.append(data_2013Q1_TOTAL)
my_list.append(data_2013Q2_TOTAL)
my_list.append(data_2013Q3_TOTAL)
my_list.append(data_2013Q4_TOTAL)
my_list.append(data_2014Q1_TOTAL)
my_list.append(data_2014Q2_TOTAL)
my_list.append(data_2014Q3_TOTAL)
my_list.append(data_2014Q4_TOTAL)
my_list.append(data_2015Q1_TOTAL)
my_list.append(data_2015Q2_TOTAL)
my_list.append(data_2015Q3_TOTAL)
my_list.append(data_2015Q4_TOTAL)
my_list.append(data_2016Q1_TOTAL)
my_list.append(data_2016Q2_TOTAL)
my_list.append(data_2016Q3_TOTAL)
my_list.append(data_2016Q4_TOTAL)
my_list.append(data_2017Q1_TOTAL)
my_list.append(data_2017Q2_TOTAL)
my_list.append(data_2017Q3_TOTAL)
my_list.append(data_2017Q4_TOTAL)
my_list.append(data_2018Q1_TOTAL)
print ( sum (my_list))
data_2009Q3_pnew=data_2009Q3.loc[ data_2009Q3["AMOUNT"] > 0 ]
data_2009Q4_pnew=data_2009Q4.loc[ data_2009Q4["AMOUNT"] > 0 ]
data_2010Q1_pnew=data_2010Q1.loc[ data_2010Q1["AMOUNT"] > 0 ]
data_2010Q2_pnew=data_2010Q2.loc[ data_2010Q2["AMOUNT"] > 0 ]
data_2010Q3_pnew=data_2010Q3.loc[ data_2010Q3["AMOUNT"] > 0 ]
data_2010Q4_pnew=data_2010Q4.loc[ data_2010Q4["AMOUNT"] > 0 ]
data_2011Q1_pnew=data_2011Q1.loc[ data_2011Q1["AMOUNT"] > 0 ]
data_2011Q2_pnew=data_2011Q2.loc[ data_2011Q2["AMOUNT"] > 0 ]
data_2011Q3_pnew=data_2011Q3.loc[ data_2011Q3["AMOUNT"] > 0 ]
data_2011Q4_pnew=data_2011Q4.loc[ data_2011Q4["AMOUNT"] > 0 ]
data_2012Q1_pnew=data_2012Q1.loc[ data_2012Q1["AMOUNT"] > 0 ]
data_2012Q2_pnew=data_2012Q2.loc[ data_2012Q2["AMOUNT"] > 0 ]
data_2012Q3_pnew=data_2012Q3.loc[ data_2012Q3["AMOUNT"] > 0 ]
data_2012Q4_pnew=data_2012Q4.loc[ data_2012Q4["AMOUNT"] > 0 ]
data_2013Q1_pnew=data_2013Q1.loc[ data_2013Q1["AMOUNT"] > 0 ]
data_2013Q2_pnew=data_2013Q2.loc[ data_2013Q2["AMOUNT"] > 0 ]
data_2013Q3_pnew=data_2013Q3.loc[ data_2013Q3["AMOUNT"] > 0 ]
data_2013Q4_pnew=data_2013Q4.loc[ data_2013Q4["AMOUNT"] > 0 ]
data_2014Q1_pnew=data_2014Q1.loc[ data_2014Q1["AMOUNT"] > 0 ]
data_2014Q2_pnew=data_2014Q2.loc[ data_2014Q2["AMOUNT"] > 0 ]
data_2014Q3_pnew=data_2014Q3.loc[ data_2014Q3["AMOUNT"] > 0 ]
data_2014Q4_pnew=data_2014Q4.loc[ data_2014Q4["AMOUNT"] > 0 ]
data_2015Q1_pnew=data_2015Q1.loc[ data_2015Q1["AMOUNT"] > 0 ]
data_2015Q2_pnew=data_2015Q2.loc[ data_2015Q2["AMOUNT"] > 0 ]
data_2015Q3_pnew=data_2015Q3.loc[ data_2015Q3["AMOUNT"] > 0 ]
data_2015Q4_pnew=data_2015Q4.loc[ data_2015Q4["AMOUNT"] > 0 ]
data_2016Q1_pnew=data_2016Q1.loc[ data_2016Q1["AMOUNT"] > 0 ]
data_2016Q2_pnew=data_2016Q2.loc[ data_2016Q2["AMOUNT"] > 0 ]
data_2016Q3_pnew=data_2016Q3.loc[ data_2016Q3["AMOUNT"] > 0 ]
data_2016Q4_pnew=data_2016Q4.loc[ data_2016Q4["AMOUNT"] > 0 ]
data_2017Q1_pnew=data_2017Q1.loc[ data_2017Q1["AMOUNT"] > 0 ]
data_2017Q2_pnew=data_2017Q2.loc[ data_2017Q2["AMOUNT"] > 0 ]
data_2017Q3_pnew=data_2017Q3.loc[ data_2017Q3["AMOUNT"] > 0 ]
data_2017Q4_pnew=data_2017Q4.loc[ data_2017Q4["AMOUNT"] > 0 ]
data_2018Q1_pnew=data_2018Q1.loc[ data_2018Q1["AMOUNT"] > 0 ]
dateIndex1 = data_2017Q2_pnew.loc[:,"START_DATE"] == ' '
data_2017Q2_pnew.loc[dateIndex1, "START_DATE"] = '01/05/2017'
dateIndex2 = data_2017Q2_pnew.loc[:,"END_DATE"] == ' '
data_2017Q2_pnew.loc[dateIndex2, "END_DATE"] = '01/05/2017'
dateIndex3 = data_2017Q3_pnew.loc[:,"START_DATE"] == ' '
data_2017Q3_pnew.loc[dateIndex3, "START_DATE"] = '01/10/2017'
dateIndex4 = data_2017Q3_pnew.loc[:,"END_DATE"] == ' '
data_2017Q3_pnew.loc[dateIndex4, "END_DATE"] = '01/10/2017'
dateIndex5 = data_2017Q4_pnew.loc[:,"START_DATE"] == ' '
data_2017Q4_pnew.loc[dateIndex5, "START_DATE"] = '01/10/2017'
dateIndex6 = data_2017Q4_pnew.loc[:,"END_DATE"] == ' '
data_2017Q4_pnew.loc[dateIndex6, "END_DATE"] = '01/10/2017'
dateIndex7 = data_2018Q1_pnew.loc[:,"START_DATE"] == ' '
data_2018Q1_pnew.loc[dateIndex7, "START_DATE"] = '01/01/2018'
dateIndex8 = data_2018Q1_pnew.loc[:,"END_DATE"] == ' '
data_2018Q1_pnew.loc[dateIndex8, "END_DATE"] = '01/01/2018'
data_2009Q3_pnew["START_DATE"]=pd.to_datetime(data_2009Q3_pnew["START_DATE"])
data_2009Q4_pnew["START_DATE"]=pd.to_datetime(data_2009Q4_pnew["START_DATE"])
data_2010Q1_pnew["START_DATE"]=pd.to_datetime(data_2010Q1_pnew["START_DATE"])
data_2010Q2_pnew["START_DATE"]=pd.to_datetime(data_2010Q2_pnew["START_DATE"])
data_2010Q3_pnew["START_DATE"]=pd.to_datetime(data_2010Q3_pnew["START_DATE"])
data_2010Q4_pnew["START_DATE"]=pd.to_datetime(data_2010Q4_pnew["START_DATE"])
data_2011Q1_pnew["START_DATE"]=pd.to_datetime(data_2011Q1_pnew["START_DATE"])
data_2011Q2_pnew["START_DATE"]=pd.to_datetime(data_2011Q2_pnew["START_DATE"])
data_2011Q3_pnew["START_DATE"]=pd.to_datetime(data_2011Q3_pnew["START_DATE"])
data_2011Q4_pnew["START_DATE"]=pd.to_datetime(data_2011Q4_pnew["START_DATE"])
data_2012Q1_pnew["START_DATE"]=pd.to_datetime(data_2012Q1_pnew["START_DATE"])
data_2012Q2_pnew["START_DATE"]=pd.to_datetime(data_2012Q2_pnew["START_DATE"])
data_2012Q3_pnew["START_DATE"]=pd.to_datetime(data_2012Q3_pnew["START_DATE"])
data_2012Q4_pnew["START_DATE"]=pd.to_datetime(data_2012Q4_pnew["START_DATE"])
data_2013Q1_pnew["START_DATE"]=pd.to_datetime(data_2013Q1_pnew["START_DATE"])
data_2013Q2_pnew["START_DATE"]=pd.to_datetime(data_2013Q2_pnew["START_DATE"])
data_2013Q3_pnew["START_DATE"]=pd.to_datetime(data_2013Q3_pnew["START_DATE"])
data_2013Q4_pnew["START_DATE"]=pd.to_datetime(data_2013Q4_pnew["START_DATE"])
data_2014Q1_pnew["START_DATE"]=pd.to_datetime(data_2014Q1_pnew["START_DATE"])
data_2014Q2_pnew["START_DATE"]=pd.to_datetime(data_2014Q2_pnew["START_DATE"])
data_2014Q3_pnew["START_DATE"]=pd.to_datetime(data_2014Q3_pnew["START_DATE"])
data_2014Q4_pnew["START_DATE"]=pd.to_datetime(data_2014Q4_pnew["START_DATE"])
data_2015Q1_pnew["START_DATE"]=pd.to_datetime(data_2015Q1_pnew["START_DATE"])
data_2015Q2_pnew["START_DATE"]=pd.to_datetime(data_2015Q2_pnew["START_DATE"])
data_2015Q3_pnew["START_DATE"]=pd.to_datetime(data_2015Q3_pnew["START_DATE"])
data_2015Q4_pnew["START_DATE"]=pd.to_datetime(data_2015Q4_pnew["START_DATE"])
data_2016Q1_pnew["START_DATE"]=pd.to_datetime(data_2016Q1_pnew["START_DATE"])
data_2016Q2_pnew["START_DATE"]=pd.to_datetime(data_2016Q2_pnew["START_DATE"])
data_2016Q3_pnew["START_DATE"]=pd.to_datetime(data_2016Q3_pnew["START_DATE"])
data_2016Q4_pnew["START_DATE"]=pd.to_datetime(data_2016Q4_pnew["START_DATE"])
data_2017Q1_pnew["START_DATE"]=pd.to_datetime(data_2017Q1_pnew["START_DATE"])
data_2017Q2_pnew["START_DATE"]=pd.to_datetime(data_2017Q2_pnew["START_DATE"])
data_2017Q3_pnew["START_DATE"]=pd.to_datetime(data_2017Q3_pnew["START_DATE"])
data_2017Q4_pnew["START_DATE"]=pd.to_datetime(data_2017Q4_pnew["START_DATE"])
data_2018Q1_pnew["START_DATE"]=pd.to_datetime(data_2018Q1_pnew["START_DATE"])
data_2009Q3_pnew["END_DATE"]=pd.to_datetime(data_2009Q3_pnew["END_DATE"])
data_2009Q4_pnew["END_DATE"]=pd.to_datetime(data_2009Q4_pnew["END_DATE"])
data_2010Q1_pnew["END_DATE"]=pd.to_datetime(data_2010Q1_pnew["END_DATE"])
data_2010Q2_pnew["END_DATE"]=pd.to_datetime(data_2010Q2_pnew["END_DATE"])
data_2010Q3_pnew["END_DATE"]=pd.to_datetime(data_2010Q3_pnew["END_DATE"])
data_2010Q4_pnew["END_DATE"]=pd.to_datetime(data_2010Q4_pnew["END_DATE"])
data_2011Q1_pnew["END_DATE"]=pd.to_datetime(data_2011Q1_pnew["END_DATE"])
data_2011Q2_pnew["END_DATE"]=pd.to_datetime(data_2011Q2_pnew["END_DATE"])
data_2011Q3_pnew["END_DATE"]=pd.to_datetime(data_2011Q3_pnew["END_DATE"])
data_2011Q4_pnew["END_DATE"]=pd.to_datetime(data_2011Q4_pnew["END_DATE"])
data_2012Q1_pnew["END_DATE"]=pd.to_datetime(data_2012Q1_pnew["END_DATE"])
data_2012Q2_pnew["END_DATE"]=pd.to_datetime(data_2012Q2_pnew["END_DATE"])
data_2012Q3_pnew["END_DATE"]=pd.to_datetime(data_2012Q3_pnew["END_DATE"])
data_2012Q4_pnew["END_DATE"]=pd.to_datetime(data_2012Q4_pnew["END_DATE"])
data_2013Q1_pnew["END_DATE"]=pd.to_datetime(data_2013Q1_pnew["END_DATE"])
data_2013Q2_pnew["END_DATE"]=pd.to_datetime(data_2013Q2_pnew["END_DATE"])
data_2013Q3_pnew["END_DATE"]=pd.to_datetime(data_2013Q3_pnew["END_DATE"])
data_2013Q4_pnew["END_DATE"]=pd.to_datetime(data_2013Q4_pnew["END_DATE"])
data_2014Q1_pnew["END_DATE"]=pd.to_datetime(data_2014Q1_pnew["END_DATE"])
data_2014Q2_pnew["END_DATE"]=pd.to_datetime(data_2014Q2_pnew["END_DATE"])
data_2014Q3_pnew["END_DATE"]=pd.to_datetime(data_2014Q3_pnew["END_DATE"])
data_2014Q4_pnew["END_DATE"]=pd.to_datetime(data_2014Q4_pnew["END_DATE"])
data_2015Q1_pnew["END_DATE"]=pd.to_datetime(data_2015Q1_pnew["END_DATE"])
data_2015Q2_pnew["END_DATE"]=pd.to_datetime(data_2015Q2_pnew["END_DATE"])
data_2015Q3_pnew["END_DATE"]=pd.to_datetime(data_2015Q3_pnew["END_DATE"])
data_2015Q4_pnew["END_DATE"]=pd.to_datetime(data_2015Q4_pnew["END_DATE"])
data_2016Q1_pnew["END_DATE"]=pd.to_datetime(data_2016Q1_pnew["END_DATE"])
data_2016Q2_pnew["END_DATE"]=pd.to_datetime(data_2016Q2_pnew["END_DATE"])
data_2016Q3_pnew["END_DATE"]=pd.to_datetime(data_2016Q3_pnew["END_DATE"])
data_2016Q4_pnew["END_DATE"]=pd.to_datetime(data_2016Q4_pnew["END_DATE"])
data_2017Q1_pnew["END_DATE"]=pd.to_datetime(data_2017Q1_pnew["END_DATE"])
data_2017Q2_pnew["END_DATE"]=pd.to_datetime(data_2017Q2_pnew["END_DATE"])
data_2017Q3_pnew["END_DATE"]=pd.to_datetime(data_2017Q3_pnew["END_DATE"])
data_2017Q4_pnew["END_DATE"]=pd.to_datetime(data_2017Q4_pnew["END_DATE"])
data_2018Q1_pnew["END_DATE"]=pd.to_datetime(data_2018Q1_pnew["END_DATE"])
data_2009Q3_pnew.loc[:,"NEW_START_DATE"]=data_2009Q3_pnew.loc[:,"START_DATE"].dt.date
data_2009Q4_pnew.loc[:,"NEW_START_DATE"]=data_2009Q4_pnew.loc[:,"START_DATE"].dt.date
data_2010Q1_pnew.loc[:,"NEW_START_DATE"]=data_2010Q1_pnew.loc[:,"START_DATE"].dt.date
data_2010Q2_pnew.loc[:,"NEW_START_DATE"]=data_2010Q2_pnew.loc[:,"START_DATE"].dt.date
data_2010Q3_pnew.loc[:,"NEW_START_DATE"]=data_2010Q3_pnew.loc[:,"START_DATE"].dt.date
data_2010Q4_pnew.loc[:,"NEW_START_DATE"]=data_2010Q4_pnew.loc[:,"START_DATE"].dt.date
data_2011Q1_pnew.loc[:,"NEW_START_DATE"]=data_2011Q1_pnew.loc[:,"START_DATE"].dt.date
data_2011Q2_pnew.loc[:,"NEW_START_DATE"]=data_2011Q2_pnew.loc[:,"START_DATE"].dt.date
data_2011Q3_pnew.loc[:,"NEW_START_DATE"]=data_2011Q3_pnew.loc[:,"START_DATE"].dt.date
data_2011Q4_pnew.loc[:,"NEW_START_DATE"]=data_2011Q4_pnew.loc[:,"START_DATE"].dt.date
data_2012Q1_pnew.loc[:,"NEW_START_DATE"]=data_2012Q1_pnew.loc[:,"START_DATE"].dt.date
data_2012Q2_pnew.loc[:,"NEW_START_DATE"]=data_2012Q2_pnew.loc[:,"START_DATE"].dt.date
data_2012Q3_pnew.loc[:,"NEW_START_DATE"]=data_2012Q3_pnew.loc[:,"START_DATE"].dt.date
data_2012Q4_pnew.loc[:,"NEW_START_DATE"]=data_2012Q4_pnew.loc[:,"START_DATE"].dt.date
data_2013Q1_pnew.loc[:,"NEW_START_DATE"]=data_2013Q1_pnew.loc[:,"START_DATE"].dt.date
data_2013Q2_pnew.loc[:,"NEW_START_DATE"]=data_2013Q2_pnew.loc[:,"START_DATE"].dt.date
data_2013Q3_pnew.loc[:,"NEW_START_DATE"]=data_2013Q3_pnew.loc[:,"START_DATE"].dt.date
data_2013Q4_pnew.loc[:,"NEW_START_DATE"]=data_2013Q4_pnew.loc[:,"START_DATE"].dt.date
data_2014Q1_pnew.loc[:,"NEW_START_DATE"]=data_2014Q1_pnew.loc[:,"START_DATE"].dt.date
data_2014Q2_pnew.loc[:,"NEW_START_DATE"]=data_2014Q2_pnew.loc[:,"START_DATE"].dt.date
data_2014Q3_pnew.loc[:,"NEW_START_DATE"]=data_2014Q3_pnew.loc[:,"START_DATE"].dt.date
data_2014Q4_pnew.loc[:,"NEW_START_DATE"]=data_2014Q4_pnew.loc[:,"START_DATE"].dt.date
data_2015Q1_pnew.loc[:,"NEW_START_DATE"]=data_2015Q1_pnew.loc[:,"START_DATE"].dt.date
data_2015Q2_pnew.loc[:,"NEW_START_DATE"]=data_2015Q2_pnew.loc[:,"START_DATE"].dt.date
data_2015Q3_pnew.loc[:,"NEW_START_DATE"]=data_2015Q3_pnew.loc[:,"START_DATE"].dt.date
data_2015Q4_pnew.loc[:,"NEW_START_DATE"]=data_2015Q4_pnew.loc[:,"START_DATE"].dt.date
data_2016Q1_pnew.loc[:,"NEW_START_DATE"]=data_2016Q1_pnew.loc[:,"START_DATE"].dt.date
data_2016Q2_pnew.loc[:,"NEW_START_DATE"]=data_2016Q2_pnew.loc[:,"START_DATE"].dt.date
data_2016Q3_pnew.loc[:,"NEW_START_DATE"]=data_2016Q3_pnew.loc[:,"START_DATE"].dt.date
data_2016Q4_pnew.loc[:,"NEW_START_DATE"]=data_2016Q4_pnew.loc[:,"START_DATE"].dt.date
data_2017Q1_pnew.loc[:,"NEW_START_DATE"]=data_2017Q1_pnew.loc[:,"START_DATE"].dt.date
data_2017Q2_pnew.loc[:,"NEW_START_DATE"]=data_2017Q2_pnew.loc[:,"START_DATE"].dt.date
data_2017Q3_pnew.loc[:,"NEW_START_DATE"]=data_2017Q3_pnew.loc[:,"START_DATE"].dt.date
data_2017Q4_pnew.loc[:,"NEW_START_DATE"]=data_2017Q4_pnew.loc[:,"START_DATE"].dt.date
data_2018Q1_pnew.loc[:,"NEW_START_DATE"]=data_2018Q1_pnew.loc[:,"START_DATE"].dt.date
data_2009Q3_pnew.loc[:,"NEW_END_DATE"]=data_2009Q3_pnew.loc[:,"END_DATE"].dt.date
data_2009Q4_pnew.loc[:,"NEW_END_DATE"]=data_2009Q4_pnew.loc[:,"END_DATE"].dt.date
data_2010Q1_pnew.loc[:,"NEW_END_DATE"]=data_2010Q1_pnew.loc[:,"END_DATE"].dt.date
data_2010Q2_pnew.loc[:,"NEW_END_DATE"]=data_2010Q2_pnew.loc[:,"END_DATE"].dt.date
data_2010Q3_pnew.loc[:,"NEW_END_DATE"]=data_2010Q3_pnew.loc[:,"END_DATE"].dt.date
data_2010Q4_pnew.loc[:,"NEW_END_DATE"]=data_2010Q4_pnew.loc[:,"END_DATE"].dt.date
data_2011Q1_pnew.loc[:,"NEW_END_DATE"]=data_2011Q1_pnew.loc[:,"END_DATE"].dt.date
data_2011Q2_pnew.loc[:,"NEW_END_DATE"]=data_2011Q2_pnew.loc[:,"END_DATE"].dt.date
data_2011Q3_pnew.loc[:,"NEW_END_DATE"]=data_2011Q3_pnew.loc[:,"END_DATE"].dt.date
data_2011Q4_pnew.loc[:,"NEW_END_DATE"]=data_2011Q4_pnew.loc[:,"END_DATE"].dt.date
data_2012Q1_pnew.loc[:,"NEW_END_DATE"]=data_2012Q1_pnew.loc[:,"END_DATE"].dt.date
data_2012Q2_pnew.loc[:,"NEW_END_DATE"]=data_2012Q2_pnew.loc[:,"END_DATE"].dt.date
data_2012Q3_pnew.loc[:,"NEW_END_DATE"]=data_2012Q3_pnew.loc[:,"END_DATE"].dt.date
data_2012Q4_pnew.loc[:,"NEW_END_DATE"]=data_2012Q4_pnew.loc[:,"END_DATE"].dt.date
data_2013Q1_pnew.loc[:,"NEW_END_DATE"]=data_2013Q1_pnew.loc[:,"END_DATE"].dt.date
data_2013Q2_pnew.loc[:,"NEW_END_DATE"]=data_2013Q2_pnew.loc[:,"END_DATE"].dt.date
data_2013Q3_pnew.loc[:,"NEW_END_DATE"]=data_2013Q3_pnew.loc[:,"END_DATE"].dt.date
data_2013Q4_pnew.loc[:,"NEW_END_DATE"]=data_2013Q4_pnew.loc[:,"END_DATE"].dt.date
data_2014Q1_pnew.loc[:,"NEW_END_DATE"]=data_2014Q1_pnew.loc[:,"END_DATE"].dt.date
data_2014Q2_pnew.loc[:,"NEW_END_DATE"]=data_2014Q2_pnew.loc[:,"END_DATE"].dt.date
data_2014Q3_pnew.loc[:,"NEW_END_DATE"]=data_2014Q3_pnew.loc[:,"END_DATE"].dt.date
data_2014Q4_pnew.loc[:,"NEW_END_DATE"]=data_2014Q4_pnew.loc[:,"END_DATE"].dt.date
data_2015Q1_pnew.loc[:,"NEW_END_DATE"]=data_2015Q1_pnew.loc[:,"END_DATE"].dt.date
data_2015Q2_pnew.loc[:,"NEW_END_DATE"]=data_2015Q2_pnew.loc[:,"END_DATE"].dt.date
data_2015Q3_pnew.loc[:,"NEW_END_DATE"]=data_2015Q3_pnew.loc[:,"END_DATE"].dt.date
data_2015Q4_pnew.loc[:,"NEW_END_DATE"]=data_2015Q4_pnew.loc[:,"END_DATE"].dt.date
data_2016Q1_pnew.loc[:,"NEW_END_DATE"]=data_2016Q1_pnew.loc[:,"END_DATE"].dt.date
data_2016Q2_pnew.loc[:,"NEW_END_DATE"]=data_2016Q2_pnew.loc[:,"END_DATE"].dt.date
data_2016Q3_pnew.loc[:,"NEW_END_DATE"]=data_2016Q3_pnew.loc[:,"END_DATE"].dt.date
data_2016Q4_pnew.loc[:,"NEW_END_DATE"]=data_2016Q4_pnew.loc[:,"END_DATE"].dt.date
data_2017Q1_pnew.loc[:,"NEW_END_DATE"]=data_2017Q1_pnew.loc[:,"END_DATE"].dt.date
data_2017Q2_pnew.loc[:,"NEW_END_DATE"]=data_2017Q2_pnew.loc[:,"END_DATE"].dt.date
data_2017Q3_pnew.loc[:,"NEW_END_DATE"]=data_2017Q3_pnew.loc[:,"END_DATE"].dt.date
data_2017Q4_pnew.loc[:,"NEW_END_DATE"]=data_2017Q4_pnew.loc[:,"END_DATE"].dt.date
data_2018Q1_pnew.loc[:,"NEW_END_DATE"]=data_2018Q1_pnew.loc[:,"END_DATE"].dt.date
data_2009Q3_pnew.loc[:,"DIFF_DATE"]=data_2009Q3_pnew.loc[:,"NEW_START_DATE"]-data_2009Q3_pnew.loc[:,"NEW_END_DATE"]
data_2009Q4_pnew.loc[:,"DIFF_DATE"]=data_2009Q4_pnew.loc[:,"NEW_START_DATE"]-data_2009Q4_pnew.loc[:,"NEW_END_DATE"]
data_2010Q1_pnew.loc[:,"DIFF_DATE"]=data_2010Q1_pnew.loc[:,"NEW_START_DATE"]-data_2010Q1_pnew.loc[:,"NEW_END_DATE"]
data_2010Q2_pnew.loc[:,"DIFF_DATE"]=data_2010Q2_pnew.loc[:,"NEW_START_DATE"]-data_2010Q2_pnew.loc[:,"NEW_END_DATE"]
data_2010Q3_pnew.loc[:,"DIFF_DATE"]=data_2010Q3_pnew.loc[:,"NEW_START_DATE"]-data_2010Q3_pnew.loc[:,"NEW_END_DATE"]
data_2010Q4_pnew.loc[:,"DIFF_DATE"]=data_2010Q4_pnew.loc[:,"NEW_START_DATE"]-data_2010Q4_pnew.loc[:,"NEW_END_DATE"]
data_2011Q1_pnew.loc[:,"DIFF_DATE"]=data_2011Q1_pnew.loc[:,"NEW_START_DATE"]-data_2011Q1_pnew.loc[:,"NEW_END_DATE"]
data_2011Q2_pnew.loc[:,"DIFF_DATE"]=data_2011Q2_pnew.loc[:,"NEW_START_DATE"]-data_2011Q2_pnew.loc[:,"NEW_END_DATE"]
data_2011Q3_pnew.loc[:,"DIFF_DATE"]=data_2011Q3_pnew.loc[:,"NEW_START_DATE"]-data_2011Q3_pnew.loc[:,"NEW_END_DATE"]
data_2011Q4_pnew.loc[:,"DIFF_DATE"]=data_2011Q4_pnew.loc[:,"NEW_START_DATE"]-data_2011Q4_pnew.loc[:,"NEW_END_DATE"]
data_2012Q1_pnew.loc[:,"DIFF_DATE"]=data_2012Q1_pnew.loc[:,"NEW_START_DATE"]-data_2012Q1_pnew.loc[:,"NEW_END_DATE"]
data_2012Q2_pnew.loc[:,"DIFF_DATE"]=data_2012Q2_pnew.loc[:,"NEW_START_DATE"]-data_2012Q2_pnew.loc[:,"NEW_END_DATE"]
data_2012Q3_pnew.loc[:,"DIFF_DATE"]=data_2012Q3_pnew.loc[:,"NEW_START_DATE"]-data_2012Q3_pnew.loc[:,"NEW_END_DATE"]
data_2012Q4_pnew.loc[:,"DIFF_DATE"]=data_2012Q4_pnew.loc[:,"NEW_START_DATE"]-data_2012Q4_pnew.loc[:,"NEW_END_DATE"]
data_2013Q1_pnew.loc[:,"DIFF_DATE"]=data_2013Q1_pnew.loc[:,"NEW_START_DATE"]-data_2013Q1_pnew.loc[:,"NEW_END_DATE"]
data_2013Q2_pnew.loc[:,"DIFF_DATE"]=data_2013Q2_pnew.loc[:,"NEW_START_DATE"]-data_2013Q2_pnew.loc[:,"NEW_END_DATE"]
data_2013Q3_pnew.loc[:,"DIFF_DATE"]=data_2013Q3_pnew.loc[:,"NEW_START_DATE"]-data_2013Q3_pnew.loc[:,"NEW_END_DATE"]
data_2013Q4_pnew.loc[:,"DIFF_DATE"]=data_2013Q4_pnew.loc[:,"NEW_START_DATE"]-data_2013Q4_pnew.loc[:,"NEW_END_DATE"]
data_2014Q1_pnew.loc[:,"DIFF_DATE"]=data_2014Q1_pnew.loc[:,"NEW_START_DATE"]-data_2014Q1_pnew.loc[:,"NEW_END_DATE"]
data_2014Q2_pnew.loc[:,"DIFF_DATE"]=data_2014Q2_pnew.loc[:,"NEW_START_DATE"]-data_2014Q2_pnew.loc[:,"NEW_END_DATE"]
data_2014Q3_pnew.loc[:,"DIFF_DATE"]=data_2014Q3_pnew.loc[:,"NEW_START_DATE"]-data_2014Q3_pnew.loc[:,"NEW_END_DATE"]
data_2014Q4_pnew.loc[:,"DIFF_DATE"]=data_2014Q4_pnew.loc[:,"NEW_START_DATE"]-data_2014Q4_pnew.loc[:,"NEW_END_DATE"]
data_2015Q1_pnew.loc[:,"DIFF_DATE"]=data_2015Q1_pnew.loc[:,"NEW_START_DATE"]-data_2015Q1_pnew.loc[:,"NEW_END_DATE"]
data_2015Q2_pnew.loc[:,"DIFF_DATE"]=data_2015Q2_pnew.loc[:,"NEW_START_DATE"]-data_2015Q2_pnew.loc[:,"NEW_END_DATE"]
data_2015Q3_pnew.loc[:,"DIFF_DATE"]=data_2015Q3_pnew.loc[:,"NEW_START_DATE"]-data_2015Q3_pnew.loc[:,"NEW_END_DATE"]
data_2015Q4_pnew.loc[:,"DIFF_DATE"]=data_2015Q4_pnew.loc[:,"NEW_START_DATE"]-data_2015Q4_pnew.loc[:,"NEW_END_DATE"]
data_2016Q1_pnew.loc[:,"DIFF_DATE"]=data_2016Q1_pnew.loc[:,"NEW_START_DATE"]-data_2016Q1_pnew.loc[:,"NEW_END_DATE"]
data_2016Q2_pnew.loc[:,"DIFF_DATE"]=data_2016Q2_pnew.loc[:,"NEW_START_DATE"]-data_2016Q2_pnew.loc[:,"NEW_END_DATE"]
data_2016Q3_pnew.loc[:,"DIFF_DATE"]=data_2016Q3_pnew.loc[:,"NEW_START_DATE"]-data_2016Q3_pnew.loc[:,"NEW_END_DATE"]
data_2016Q4_pnew.loc[:,"DIFF_DATE"]=data_2016Q4_pnew.loc[:,"NEW_START_DATE"]-data_2016Q4_pnew.loc[:,"NEW_END_DATE"]
data_2017Q1_pnew.loc[:,"DIFF_DATE"]=data_2017Q1_pnew.loc[:,"NEW_START_DATE"]-data_2017Q1_pnew.loc[:,"NEW_END_DATE"]
data_2017Q2_pnew.loc[:,"DIFF_DATE"]=data_2017Q2_pnew.loc[:,"NEW_START_DATE"]-data_2017Q2_pnew.loc[:,"NEW_END_DATE"]
data_2017Q3_pnew.loc[:,"DIFF_DATE"]=data_2017Q3_pnew.loc[:,"NEW_START_DATE"]-data_2017Q3_pnew.loc[:,"NEW_END_DATE"]
data_2017Q4_pnew.loc[:,"DIFF_DATE"]=data_2017Q4_pnew.loc[:,"NEW_START_DATE"]-data_2017Q4_pnew.loc[:,"NEW_END_DATE"]
data_2018Q1_pnew.loc[:,"DIFF_DATE"]=data_2018Q1_pnew.loc[:,"NEW_START_DATE"]-data_2018Q1_pnew.loc[:,"NEW_END_DATE"]
data_2009Q3_pnew.loc[:,"COVERAGE_PERIOD"]=data_2009Q3_pnew.loc[:,"DIFF_DATE"].dt.days
data_2009Q4_pnew.loc[:,"COVERAGE_PERIOD"]=data_2009Q4_pnew.loc[:,"DIFF_DATE"].dt.days
data_2010Q1_pnew.loc[:,"COVERAGE_PERIOD"]=data_2010Q1_pnew.loc[:,"DIFF_DATE"].dt.days
data_2010Q2_pnew.loc[:,"COVERAGE_PERIOD"]=data_2010Q2_pnew.loc[:,"DIFF_DATE"].dt.days
data_2010Q3_pnew.loc[:,"COVERAGE_PERIOD"]=data_2010Q3_pnew.loc[:,"DIFF_DATE"].dt.days
data_2010Q4_pnew.loc[:,"COVERAGE_PERIOD"]=data_2010Q4_pnew.loc[:,"DIFF_DATE"].dt.days
data_2011Q1_pnew.loc[:,"COVERAGE_PERIOD"]=data_2011Q1_pnew.loc[:,"DIFF_DATE"].dt.days
data_2011Q2_pnew.loc[:,"COVERAGE_PERIOD"]=data_2011Q2_pnew.loc[:,"DIFF_DATE"].dt.days
data_2011Q3_pnew.loc[:,"COVERAGE_PERIOD"]=data_2011Q3_pnew.loc[:,"DIFF_DATE"].dt.days
data_2011Q4_pnew.loc[:,"COVERAGE_PERIOD"]=data_2011Q4_pnew.loc[:,"DIFF_DATE"].dt.days
data_2012Q1_pnew.loc[:,"COVERAGE_PERIOD"]=data_2012Q1_pnew.loc[:,"DIFF_DATE"].dt.days
data_2012Q2_pnew.loc[:,"COVERAGE_PERIOD"]=data_2012Q2_pnew.loc[:,"DIFF_DATE"].dt.days
data_2012Q3_pnew.loc[:,"COVERAGE_PERIOD"]=data_2012Q3_pnew.loc[:,"DIFF_DATE"].dt.days
data_2012Q4_pnew.loc[:,"COVERAGE_PERIOD"]=data_2012Q4_pnew.loc[:,"DIFF_DATE"].dt.days
data_2013Q1_pnew.loc[:,"COVERAGE_PERIOD"]=data_2013Q1_pnew.loc[:,"DIFF_DATE"].dt.days
data_2013Q2_pnew.loc[:,"COVERAGE_PERIOD"]=data_2013Q2_pnew.loc[:,"DIFF_DATE"].dt.days
data_2013Q3_pnew.loc[:,"COVERAGE_PERIOD"]=data_2013Q3_pnew.loc[:,"DIFF_DATE"].dt.days
data_2013Q4_pnew.loc[:,"COVERAGE_PERIOD"]=data_2013Q4_pnew.loc[:,"DIFF_DATE"].dt.days
data_2014Q1_pnew.loc[:,"COVERAGE_PERIOD"]=data_2014Q1_pnew.loc[:,"DIFF_DATE"].dt.days
data_2014Q2_pnew.loc[:,"COVERAGE_PERIOD"]=data_2014Q2_pnew.loc[:,"DIFF_DATE"].dt.days
data_2014Q3_pnew.loc[:,"COVERAGE_PERIOD"]=data_2014Q3_pnew.loc[:,"DIFF_DATE"].dt.days
data_2014Q4_pnew.loc[:,"COVERAGE_PERIOD"]=data_2014Q4_pnew.loc[:,"DIFF_DATE"].dt.days
data_2015Q1_pnew.loc[:,"COVERAGE_PERIOD"]=data_2015Q1_pnew.loc[:,"DIFF_DATE"].dt.days
data_2015Q2_pnew.loc[:,"COVERAGE_PERIOD"]=data_2015Q2_pnew.loc[:,"DIFF_DATE"].dt.days
data_2015Q3_pnew.loc[:,"COVERAGE_PERIOD"]=data_2015Q3_pnew.loc[:,"DIFF_DATE"].dt.days
data_2015Q4_pnew.loc[:,"COVERAGE_PERIOD"]=data_2015Q4_pnew.loc[:,"DIFF_DATE"].dt.days
data_2016Q1_pnew.loc[:,"COVERAGE_PERIOD"]=data_2016Q1_pnew.loc[:,"DIFF_DATE"].dt.days
data_2016Q2_pnew.loc[:,"COVERAGE_PERIOD"]=data_2016Q2_pnew.loc[:,"DIFF_DATE"].dt.days
data_2016Q3_pnew.loc[:,"COVERAGE_PERIOD"]=data_2016Q3_pnew.loc[:,"DIFF_DATE"].dt.days
data_2016Q4_pnew.loc[:,"COVERAGE_PERIOD"]=data_2016Q4_pnew.loc[:,"DIFF_DATE"].dt.days
data_2017Q1_pnew.loc[:,"COVERAGE_PERIOD"]=data_2017Q1_pnew.loc[:,"DIFF_DATE"].dt.days
data_2017Q2_pnew.loc[:,"COVERAGE_PERIOD"]=data_2017Q2_pnew.loc[:,"DIFF_DATE"].dt.days
data_2017Q3_pnew.loc[:,"COVERAGE_PERIOD"]=data_2017Q3_pnew.loc[:,"DIFF_DATE"].dt.days
data_2017Q4_pnew.loc[:,"COVERAGE_PERIOD"]=data_2017Q4_pnew.loc[:,"DIFF_DATE"].dt.days
data_2018Q1_pnew.loc[:,"COVERAGE_PERIOD"]=data_2018Q1_pnew.loc[:,"DIFF_DATE"].dt.days
cp1=data_2009Q3_pnew.COVERAGE_PERIOD
cp2=data_2009Q4_pnew.COVERAGE_PERIOD
cp3=data_2010Q1_pnew.COVERAGE_PERIOD
cp4=data_2010Q2_pnew.COVERAGE_PERIOD
cp5=data_2010Q3_pnew.COVERAGE_PERIOD
cp6=data_2010Q4_pnew.COVERAGE_PERIOD
cp7=data_2011Q1_pnew.COVERAGE_PERIOD
cp8=data_2011Q2_pnew.COVERAGE_PERIOD
cp9=data_2011Q3_pnew.COVERAGE_PERIOD
cp10=data_2011Q4_pnew.COVERAGE_PERIOD
cp11=data_2012Q1_pnew.COVERAGE_PERIOD
cp12=data_2012Q2_pnew.COVERAGE_PERIOD
cp13=data_2012Q3_pnew.COVERAGE_PERIOD
cp14=data_2012Q4_pnew.COVERAGE_PERIOD
cp15=data_2013Q1_pnew.COVERAGE_PERIOD
cp16=data_2013Q2_pnew.COVERAGE_PERIOD
cp17=data_2013Q3_pnew.COVERAGE_PERIOD
cp18=data_2013Q4_pnew.COVERAGE_PERIOD
cp19=data_2014Q1_pnew.COVERAGE_PERIOD
cp20=data_2014Q2_pnew.COVERAGE_PERIOD
cp21=data_2014Q3_pnew.COVERAGE_PERIOD
cp22=data_2014Q4_pnew.COVERAGE_PERIOD
cp23=data_2015Q1_pnew.COVERAGE_PERIOD
cp24=data_2015Q2_pnew.COVERAGE_PERIOD
cp25=data_2015Q3_pnew.COVERAGE_PERIOD
cp26=data_2015Q4_pnew.COVERAGE_PERIOD
cp27=data_2016Q1_pnew.COVERAGE_PERIOD
cp28=data_2016Q2_pnew.COVERAGE_PERIOD
cp29=data_2016Q3_pnew.COVERAGE_PERIOD
cp30=data_2016Q4_pnew.COVERAGE_PERIOD
cp31=data_2017Q1_pnew.COVERAGE_PERIOD
cp32=data_2017Q2_pnew.COVERAGE_PERIOD
cp33=data_2017Q3_pnew.COVERAGE_PERIOD
cp34=data_2017Q4_pnew.COVERAGE_PERIOD
cp35=data_2018Q1_pnew.COVERAGE_PERIOD
cp1.append(cp2,ignore_index=True)
cp1.append(cp3,ignore_index=True)
cp1.append(cp4,ignore_index=True)
cp1.append(cp5,ignore_index=True)
cp1.append(cp6,ignore_index=True)
cp1.append(cp7,ignore_index=True)
cp1.append(cp8,ignore_index=True)
cp1.append(cp9,ignore_index=True)
cp1.append(cp10,ignore_index=True)
cp1.append(cp11,ignore_index=True)
cp1.append(cp12,ignore_index=True)
cp1.append(cp13,ignore_index=True)
cp1.append(cp14,ignore_index=True)
cp1.append(cp15,ignore_index=True)
cp1.append(cp16,ignore_index=True)
cp1.append(cp17,ignore_index=True)
cp1.append(cp18,ignore_index=True)
cp1.append(cp19,ignore_index=True)
cp1.append(cp20,ignore_index=True)
cp1.append(cp21,ignore_index=True)
cp1.append(cp22,ignore_index=True)
cp1.append(cp23,ignore_index=True)
cp1.append(cp24,ignore_index=True)
cp1.append(cp25,ignore_index=True)
cp1.append(cp26,ignore_index=True)
cp1.append(cp27,ignore_index=True)
cp1.append(cp28,ignore_index=True)
cp1.append(cp29,ignore_index=True)
cp1.append(cp30,ignore_index=True)
cp1.append(cp31,ignore_index=True)
cp1.append(cp32,ignore_index=True)
cp1.append(cp33,ignore_index=True)
cp1.append(cp34,ignore_index=True)
cp1.append(cp35,ignore_index=True)
cp1.std()
```
|
github_jupyter
|
%%time
import pandas as pd
import numpy as np
import os
import scipy.stats as sts
!pip install xlrd
import io
# plotting modules
import seaborn as sns
import matplotlib.pyplot as plt
# Load the Drive helper and mount
from google.colab import drive
# This will prompt for authorization.
drive.mount('/content/drive')
# After executing the cell above, Drive
# files will be present in "/content/drive/My Drive/dataincubator/details".
!ls "/content/drive/My Drive/dataincubator/details"
%%time
data_2009Q3 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2009Q3-house-disburse-detail.csv",engine="python")
data_2009Q4 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2009Q4-house-disburse-detail.csv",engine="python")
data_2010Q1 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2010Q1-house-disburse-detail.csv",engine="python")
data_2010Q2 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2010Q2-house-disburse-detail.csv",engine="python")
data_2010Q3 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2010Q3-house-disburse-detail.csv",engine="python")
data_2010Q4 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2010Q4-house-disburse-detail.csv",engine="python")
data_2011Q1 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2011Q1-house-disburse-detail.csv",engine="python")
data_2011Q2 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2011Q2-house-disburse-detail.csv",engine="python")
data_2011Q3 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2011Q3-house-disburse-detail.csv",engine="python")
data_2011Q4 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2011Q4-house-disburse-detail.csv",engine="python")
data_2012Q1 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2012Q1-house-disburse-detail.csv",engine="python")
data_2012Q2 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2012Q2-house-disburse-detail.csv",engine="python")
data_2012Q3 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2012Q3-house-disburse-detail.csv",engine="python")
data_2012Q4 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2012Q4-house-disburse-detail.csv",engine="python")
data_2013Q1 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2013Q1-house-disburse-detail.csv",engine="python")
data_2013Q2 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2013Q2-house-disburse-detail.csv",engine="python")
data_2013Q3 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2013Q3-house-disburse-detail.csv",engine="python")
data_2013Q4 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2013Q4-house-disburse-detail.csv",engine="python")
data_2014Q1 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2014Q1-house-disburse-detail.csv",engine="python")
data_2014Q2 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2014Q2-house-disburse-detail.csv",engine="python")
data_2014Q3 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2014Q3-house-disburse-detail.csv",engine="python")
data_2014Q4 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2014Q4-house-disburse-detail.csv",engine="python")
data_2015Q1 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2015Q1-house-disburse-detail.csv",engine="python")
data_2015Q2 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2015Q2-house-disburse-detail-updated.csv",engine="python")
data_2015Q3 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2015Q3-house-disburse-detail.csv",engine="python")
data_2015Q4 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2015Q4-house-disburse-detail.csv",engine="python")
data_2016Q1 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2016Q1-house-disburse-detail.csv",engine="python")
data_2016Q2 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2016Q2-house-disburse-detail.csv",engine="python")
data_2016Q3 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2016Q3-house-disburse-detail.csv",engine="python")
data_2016Q4 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2016Q4-house-disburse-detail.csv",engine="python")
data_2017Q1 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2017Q1-house-disburse-detail.csv",engine="python")
data_2017Q2 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2017Q2-house-disburse-detail.csv",engine="python")
data_2017Q3 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2017Q3-house-disburse-detail.csv",engine="python")
data_2017Q4 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2017Q4-house-disburse-detail.csv",engine="python")
data_2018Q1 = pd.read_csv("/content/drive/My Drive/dataincubator/details/2018Q1-house-disburse-detail.csv",engine="python")
data_2009Q3.columns = data_2009Q3.columns.str.replace('\s+', '_')
data_2009Q4.columns = data_2009Q4.columns.str.replace('\s+', '_')
data_2010Q1.columns = data_2010Q1.columns.str.replace('\s+', '_')
data_2010Q2.columns = data_2010Q2.columns.str.replace('\s+', '_')
data_2010Q3.columns = data_2010Q3.columns.str.replace('\s+', '_')
data_2010Q4.columns = data_2010Q4.columns.str.replace('\s+', '_')
data_2011Q1.columns = data_2011Q1.columns.str.replace('\s+', '_')
data_2011Q2.columns = data_2011Q2.columns.str.replace('\s+', '_')
data_2011Q3.columns = data_2011Q3.columns.str.replace('\s+', '_')
data_2011Q4.columns = data_2011Q4.columns.str.replace('\s+', '_')
data_2012Q1.columns = data_2012Q1.columns.str.replace('\s+', '_')
data_2012Q2.columns = data_2012Q2.columns.str.replace('\s+', '_')
data_2012Q3.columns = data_2012Q3.columns.str.replace('\s+', '_')
data_2012Q4.columns = data_2012Q4.columns.str.replace('\s+', '_')
data_2013Q1.columns = data_2013Q1.columns.str.replace('\s+', '_')
data_2013Q2.columns = data_2013Q2.columns.str.replace('\s+', '_')
data_2013Q3.columns = data_2013Q3.columns.str.replace('\s+', '_')
data_2013Q4.columns = data_2013Q4.columns.str.replace('\s+', '_')
data_2014Q1.columns = data_2014Q1.columns.str.replace('\s+', '_')
data_2014Q2.columns = data_2014Q2.columns.str.replace('\s+', '_')
data_2014Q3.columns = data_2014Q3.columns.str.replace('\s+', '_')
data_2014Q4.columns = data_2014Q4.columns.str.replace('\s+', '_')
data_2015Q1.columns = data_2015Q1.columns.str.replace('\s+', '_')
data_2015Q2.columns = data_2015Q2.columns.str.replace('\s+', '_')
data_2015Q3.columns = data_2015Q3.columns.str.replace('\s+', '_')
data_2015Q4.columns = data_2015Q4.columns.str.replace('\s+', '_')
data_2016Q1.columns = data_2016Q1.columns.str.replace('\s+', '_')
data_2016Q2.columns = data_2016Q2.columns.str.replace('\s+', '_')
data_2016Q3.columns = data_2016Q3.columns.str.replace('\s+', '_')
data_2016Q4.columns = data_2016Q4.columns.str.replace('\s+', '_')
data_2017Q1.columns = data_2017Q1.columns.str.replace('\s+', '_')
data_2017Q2.columns = data_2017Q2.columns.str.replace('\s+', '_')
data_2017Q3.columns = data_2017Q3.columns.str.replace('\s+', '_')
data_2017Q4.columns = data_2017Q4.columns.str.replace('\s+', '_')
data_2018Q1.columns = data_2018Q1.columns.str.replace('\s+', '_')
data_2009Q3.isnull().sum()
data_2009Q4.isnull().sum()
data_2010Q1.isnull().sum()
data_2010Q2.isnull().sum()
data_2010Q3.isnull().sum()
data_2010Q4.isnull().sum()
data_2011Q1.isnull().sum()
data_2011Q2.isnull().sum()
data_2011Q3.isnull().sum()
data_2011Q4.isnull().sum()
data_2012Q1.isnull().sum()
data_2012Q2.isnull().sum()
data_2012Q3.isnull().sum()
data_2012Q4.isnull().sum()
data_2013Q1.isnull().sum()
data_2013Q2.isnull().sum()
data_2013Q3.isnull().sum()
data_2013Q4.isnull().sum()
data_2014Q1.isnull().sum()
data_2014Q2.isnull().sum()
data_2014Q3.isnull().sum()
data_2014Q4.isnull().sum()
data_2015Q1.isnull().sum()
data_2015Q2.isnull().sum()
data_2015Q3.isnull().sum()
data_2015Q4.isnull().sum()
data_2016Q1.isnull().sum()
data_2016Q2.isnull().sum()
data_2016Q3.isnull().sum()
data_2016Q4.isnull().sum()
data_2017Q1.isnull().sum()
data_2017Q2.isnull().sum()
data_2017Q3.isnull().sum()
data_2017Q4.isnull().sum()
data_2018Q1.isnull().sum()
data_2009Q3.AMOUNT=data_2009Q3.AMOUNT.str.replace(',', '').astype('float64')
data_2009Q4.AMOUNT=data_2009Q4.AMOUNT.str.replace(',', '').astype('float64')
data_2010Q1.AMOUNT=data_2010Q1.AMOUNT.str.replace(',', '').astype('float64')
data_2010Q2.AMOUNT=data_2010Q2.AMOUNT.str.replace(',', '').astype('float64')
data_2010Q3.AMOUNT=data_2010Q3.AMOUNT.str.replace(',', '').astype('float64')
data_2010Q4.AMOUNT=data_2010Q4.AMOUNT.str.replace(',', '').astype('float64')
data_2011Q1.AMOUNT=data_2011Q1.AMOUNT.str.replace(',', '').astype('float64')
data_2011Q2.AMOUNT=data_2011Q2.AMOUNT.str.replace(',', '').astype('float64')
data_2011Q3.AMOUNT=data_2011Q3.AMOUNT.str.replace(',', '').astype('float64')
data_2011Q4.AMOUNT=data_2011Q4.AMOUNT.str.replace(',', '').astype('float64')
data_2012Q1.AMOUNT=data_2012Q1.AMOUNT.str.replace(',', '').astype('float64')
data_2012Q2.AMOUNT=data_2012Q2.AMOUNT.str.replace(',', '').astype('float64')
data_2012Q3.AMOUNT=data_2012Q3.AMOUNT.str.replace(',', '').astype('float64')
data_2012Q4.AMOUNT=data_2012Q4.AMOUNT.str.replace(',', '').astype('float64')
data_2013Q1.AMOUNT=data_2013Q1.AMOUNT.str.replace(',', '').astype('float64')
data_2013Q2.AMOUNT=data_2013Q2.AMOUNT.str.replace(',', '').astype('float64')
data_2013Q3.AMOUNT=data_2013Q3.AMOUNT.str.replace(',', '').astype('float64')
data_2013Q4.AMOUNT=data_2013Q4.AMOUNT.str.replace(',', '').astype('float64')
data_2014Q1.AMOUNT=data_2014Q1.AMOUNT.str.replace(',', '').astype('float64')
data_2014Q2.AMOUNT=data_2014Q2.AMOUNT.str.replace(',', '').astype('float64')
data_2014Q3.AMOUNT=data_2014Q3.AMOUNT.str.replace(',', '').astype('float64')
data_2014Q4.AMOUNT=data_2014Q4.AMOUNT.str.replace(',', '').astype('float64')
data_2015Q1.AMOUNT=data_2015Q1.AMOUNT.str.replace(',', '').astype('float64')
data_2015Q2.AMOUNT=data_2015Q2.AMOUNT.str.replace(',', '').astype('float64')
data_2015Q3.AMOUNT=data_2015Q3.AMOUNT.str.replace(',', '').astype('float64')
data_2015Q4.AMOUNT=data_2015Q4.AMOUNT.str.replace(',', '').astype('float64')
data_2016Q1.AMOUNT=data_2016Q1.AMOUNT.str.replace(',', '').astype('float64')
data_2016Q2.AMOUNT=data_2016Q2.AMOUNT.str.replace(',', '').astype('float64')
data_2016Q3.AMOUNT=data_2016Q3.AMOUNT.str.replace(',', '').astype('float64')
# No String Values, hence we cannot use .str
data_2016Q4.AMOUNT=data_2016Q4.AMOUNT.replace(',', '').astype('float64')
data_2017Q1.AMOUNT=data_2017Q1.AMOUNT.replace(',', '').astype('float64')
data_2017Q2.AMOUNT=data_2017Q2.AMOUNT.replace(',', '').astype('float64')
data_2017Q3.AMOUNT=data_2017Q3.AMOUNT.replace(',', '').astype('float64')
data_2017Q4.AMOUNT=data_2017Q4.AMOUNT.replace(',', '').astype('float64')
data_2018Q1.AMOUNT=data_2018Q1.AMOUNT.replace(',', '').astype('float64')
def convert_float_amount(df):
try:
df.loc[:,"AMOUNT"]=df.loc[:,"AMOUNT"].str.replace(',', '').astype('float64')
except Exception as e:
print('Invalid Data', e)
else:
df.loc[:,"AMOUNT"]=df.loc[:,"AMOUNT"].replace(',', '').astype('float64')
return df
data_2009Q3=convert_float_amount(data_2009Q3)
data_2009Q4=convert_float_amount(data_2009Q4)
data_2010Q1=convert_float_amount(data_2010Q1)
data_2010Q2=convert_float_amount(data_2010Q2)
data_2010Q3=convert_float_amount(data_2010Q3)
data_2010Q4=convert_float_amount(data_2010Q4)
data_2011Q1=convert_float_amount(data_2011Q1)
data_2011Q2=convert_float_amount(data_2011Q2)
data_2011Q3=convert_float_amount(data_2011Q3)
data_2011Q4=convert_float_amount(data_2011Q4)
data_2012Q1=convert_float_amount(data_2012Q1)
data_2012Q2=convert_float_amount(data_2012Q2)
data_2012Q3=convert_float_amount(data_2012Q3)
data_2012Q4=convert_float_amount(data_2012Q4)
data_2013Q1=convert_float_amount(data_2013Q1)
data_2013Q2=convert_float_amount(data_2013Q2)
data_2013Q3=convert_float_amount(data_2013Q3)
data_2013Q4=convert_float_amount(data_2013Q4)
data_2014Q1=convert_float_amount(data_2014Q1)
data_2014Q2=convert_float_amount(data_2014Q2)
data_2014Q3=convert_float_amount(data_2014Q3)
data_2014Q4=convert_float_amount(data_2014Q4)
data_2015Q1=convert_float_amount(data_2015Q1)
data_2015Q2=convert_float_amount(data_2015Q2)
data_2015Q3=convert_float_amount(data_2015Q3)
data_2015Q4=convert_float_amount(data_2015Q4)
data_2016Q1=convert_float_amount(data_2016Q1)
data_2016Q2=convert_float_amount(data_2016Q2)
data_2016Q3=convert_float_amount(data_2016Q3)
data_2016Q4=convert_float_amount(data_2016Q4)
data_2017Q1=convert_float_amount(data_2017Q1)
data_2017Q2=convert_float_amount(data_2017Q2)
data_2017Q3=convert_float_amount(data_2017Q3)
data_2017Q4=convert_float_amount(data_2017Q4)
data_2018Q1=convert_float_amount(data_2018Q1)
data_2009Q3_TOTAL=sum(data_2009Q3.AMOUNT)
data_2009Q4_TOTAL=sum(data_2009Q4.AMOUNT)
data_2010Q1_TOTAL=sum(data_2010Q1.AMOUNT)
data_2010Q2_TOTAL=sum(data_2010Q2.AMOUNT)
data_2010Q3_TOTAL=sum(data_2010Q3.AMOUNT)
data_2010Q4_TOTAL=sum(data_2010Q4.AMOUNT)
data_2011Q1_TOTAL=sum(data_2011Q1.AMOUNT)
data_2011Q2_TOTAL=sum(data_2011Q2.AMOUNT)
data_2011Q3_TOTAL=sum(data_2011Q3.AMOUNT)
data_2011Q4_TOTAL=sum(data_2011Q4.AMOUNT)
data_2012Q1_TOTAL=sum(data_2012Q1.AMOUNT)
data_2012Q2_TOTAL=sum(data_2012Q2.AMOUNT)
data_2012Q3_TOTAL=sum(data_2012Q3.AMOUNT)
data_2012Q4_TOTAL=sum(data_2012Q4.AMOUNT)
data_2013Q1_TOTAL=sum(data_2013Q1.AMOUNT)
data_2013Q2_TOTAL=sum(data_2013Q2.AMOUNT)
data_2013Q3_TOTAL=sum(data_2013Q3.AMOUNT)
data_2013Q4_TOTAL=sum(data_2013Q4.AMOUNT)
data_2014Q1_TOTAL=sum(data_2014Q1.AMOUNT)
data_2014Q2_TOTAL=sum(data_2014Q2.AMOUNT)
data_2014Q3_TOTAL=sum(data_2014Q3.AMOUNT)
data_2014Q4_TOTAL=sum(data_2014Q4.AMOUNT)
data_2015Q1_TOTAL=sum(data_2015Q1.AMOUNT)
data_2015Q2_TOTAL=sum(data_2015Q2.AMOUNT)
data_2015Q3_TOTAL=sum(data_2015Q3.AMOUNT)
data_2015Q4_TOTAL=sum(data_2015Q4.AMOUNT)
data_2016Q1_TOTAL=sum(data_2016Q1.AMOUNT)
data_2016Q2_TOTAL=sum(data_2016Q2.AMOUNT)
data_2016Q3_TOTAL=sum(data_2016Q3.AMOUNT)
data_2016Q4_TOTAL=sum(data_2016Q4.AMOUNT)
data_2017Q1_TOTAL=sum(data_2017Q1.AMOUNT)
data_2017Q2_TOTAL=sum(data_2017Q2.AMOUNT)
data_2017Q3_TOTAL=sum(data_2017Q3.AMOUNT)
data_2017Q4_TOTAL=sum(data_2017Q4.AMOUNT)
data_2018Q1_TOTAL=sum(data_2018Q1.AMOUNT)
my_list = []
my_list.append(data_2009Q3_TOTAL)
my_list.append(data_2009Q4_TOTAL)
my_list.append(data_2010Q1_TOTAL)
my_list.append(data_2010Q2_TOTAL)
my_list.append(data_2010Q3_TOTAL)
my_list.append(data_2010Q4_TOTAL)
my_list.append(data_2011Q1_TOTAL)
my_list.append(data_2011Q2_TOTAL)
my_list.append(data_2011Q3_TOTAL)
my_list.append(data_2011Q4_TOTAL)
my_list.append(data_2012Q1_TOTAL)
my_list.append(data_2012Q2_TOTAL)
my_list.append(data_2012Q3_TOTAL)
my_list.append(data_2012Q4_TOTAL)
my_list.append(data_2013Q1_TOTAL)
my_list.append(data_2013Q2_TOTAL)
my_list.append(data_2013Q3_TOTAL)
my_list.append(data_2013Q4_TOTAL)
my_list.append(data_2014Q1_TOTAL)
my_list.append(data_2014Q2_TOTAL)
my_list.append(data_2014Q3_TOTAL)
my_list.append(data_2014Q4_TOTAL)
my_list.append(data_2015Q1_TOTAL)
my_list.append(data_2015Q2_TOTAL)
my_list.append(data_2015Q3_TOTAL)
my_list.append(data_2015Q4_TOTAL)
my_list.append(data_2016Q1_TOTAL)
my_list.append(data_2016Q2_TOTAL)
my_list.append(data_2016Q3_TOTAL)
my_list.append(data_2016Q4_TOTAL)
my_list.append(data_2017Q1_TOTAL)
my_list.append(data_2017Q2_TOTAL)
my_list.append(data_2017Q3_TOTAL)
my_list.append(data_2017Q4_TOTAL)
my_list.append(data_2018Q1_TOTAL)
print ( sum (my_list))
data_2009Q3_pnew=data_2009Q3.loc[ data_2009Q3["AMOUNT"] > 0 ]
data_2009Q4_pnew=data_2009Q4.loc[ data_2009Q4["AMOUNT"] > 0 ]
data_2010Q1_pnew=data_2010Q1.loc[ data_2010Q1["AMOUNT"] > 0 ]
data_2010Q2_pnew=data_2010Q2.loc[ data_2010Q2["AMOUNT"] > 0 ]
data_2010Q3_pnew=data_2010Q3.loc[ data_2010Q3["AMOUNT"] > 0 ]
data_2010Q4_pnew=data_2010Q4.loc[ data_2010Q4["AMOUNT"] > 0 ]
data_2011Q1_pnew=data_2011Q1.loc[ data_2011Q1["AMOUNT"] > 0 ]
data_2011Q2_pnew=data_2011Q2.loc[ data_2011Q2["AMOUNT"] > 0 ]
data_2011Q3_pnew=data_2011Q3.loc[ data_2011Q3["AMOUNT"] > 0 ]
data_2011Q4_pnew=data_2011Q4.loc[ data_2011Q4["AMOUNT"] > 0 ]
data_2012Q1_pnew=data_2012Q1.loc[ data_2012Q1["AMOUNT"] > 0 ]
data_2012Q2_pnew=data_2012Q2.loc[ data_2012Q2["AMOUNT"] > 0 ]
data_2012Q3_pnew=data_2012Q3.loc[ data_2012Q3["AMOUNT"] > 0 ]
data_2012Q4_pnew=data_2012Q4.loc[ data_2012Q4["AMOUNT"] > 0 ]
data_2013Q1_pnew=data_2013Q1.loc[ data_2013Q1["AMOUNT"] > 0 ]
data_2013Q2_pnew=data_2013Q2.loc[ data_2013Q2["AMOUNT"] > 0 ]
data_2013Q3_pnew=data_2013Q3.loc[ data_2013Q3["AMOUNT"] > 0 ]
data_2013Q4_pnew=data_2013Q4.loc[ data_2013Q4["AMOUNT"] > 0 ]
data_2014Q1_pnew=data_2014Q1.loc[ data_2014Q1["AMOUNT"] > 0 ]
data_2014Q2_pnew=data_2014Q2.loc[ data_2014Q2["AMOUNT"] > 0 ]
data_2014Q3_pnew=data_2014Q3.loc[ data_2014Q3["AMOUNT"] > 0 ]
data_2014Q4_pnew=data_2014Q4.loc[ data_2014Q4["AMOUNT"] > 0 ]
data_2015Q1_pnew=data_2015Q1.loc[ data_2015Q1["AMOUNT"] > 0 ]
data_2015Q2_pnew=data_2015Q2.loc[ data_2015Q2["AMOUNT"] > 0 ]
data_2015Q3_pnew=data_2015Q3.loc[ data_2015Q3["AMOUNT"] > 0 ]
data_2015Q4_pnew=data_2015Q4.loc[ data_2015Q4["AMOUNT"] > 0 ]
data_2016Q1_pnew=data_2016Q1.loc[ data_2016Q1["AMOUNT"] > 0 ]
data_2016Q2_pnew=data_2016Q2.loc[ data_2016Q2["AMOUNT"] > 0 ]
data_2016Q3_pnew=data_2016Q3.loc[ data_2016Q3["AMOUNT"] > 0 ]
data_2016Q4_pnew=data_2016Q4.loc[ data_2016Q4["AMOUNT"] > 0 ]
data_2017Q1_pnew=data_2017Q1.loc[ data_2017Q1["AMOUNT"] > 0 ]
data_2017Q2_pnew=data_2017Q2.loc[ data_2017Q2["AMOUNT"] > 0 ]
data_2017Q3_pnew=data_2017Q3.loc[ data_2017Q3["AMOUNT"] > 0 ]
data_2017Q4_pnew=data_2017Q4.loc[ data_2017Q4["AMOUNT"] > 0 ]
data_2018Q1_pnew=data_2018Q1.loc[ data_2018Q1["AMOUNT"] > 0 ]
dateIndex1 = data_2017Q2_pnew.loc[:,"START_DATE"] == ' '
data_2017Q2_pnew.loc[dateIndex1, "START_DATE"] = '01/05/2017'
dateIndex2 = data_2017Q2_pnew.loc[:,"END_DATE"] == ' '
data_2017Q2_pnew.loc[dateIndex2, "END_DATE"] = '01/05/2017'
dateIndex3 = data_2017Q3_pnew.loc[:,"START_DATE"] == ' '
data_2017Q3_pnew.loc[dateIndex3, "START_DATE"] = '01/10/2017'
dateIndex4 = data_2017Q3_pnew.loc[:,"END_DATE"] == ' '
data_2017Q3_pnew.loc[dateIndex4, "END_DATE"] = '01/10/2017'
dateIndex5 = data_2017Q4_pnew.loc[:,"START_DATE"] == ' '
data_2017Q4_pnew.loc[dateIndex5, "START_DATE"] = '01/10/2017'
dateIndex6 = data_2017Q4_pnew.loc[:,"END_DATE"] == ' '
data_2017Q4_pnew.loc[dateIndex6, "END_DATE"] = '01/10/2017'
dateIndex7 = data_2018Q1_pnew.loc[:,"START_DATE"] == ' '
data_2018Q1_pnew.loc[dateIndex7, "START_DATE"] = '01/01/2018'
dateIndex8 = data_2018Q1_pnew.loc[:,"END_DATE"] == ' '
data_2018Q1_pnew.loc[dateIndex8, "END_DATE"] = '01/01/2018'
data_2009Q3_pnew["START_DATE"]=pd.to_datetime(data_2009Q3_pnew["START_DATE"])
data_2009Q4_pnew["START_DATE"]=pd.to_datetime(data_2009Q4_pnew["START_DATE"])
data_2010Q1_pnew["START_DATE"]=pd.to_datetime(data_2010Q1_pnew["START_DATE"])
data_2010Q2_pnew["START_DATE"]=pd.to_datetime(data_2010Q2_pnew["START_DATE"])
data_2010Q3_pnew["START_DATE"]=pd.to_datetime(data_2010Q3_pnew["START_DATE"])
data_2010Q4_pnew["START_DATE"]=pd.to_datetime(data_2010Q4_pnew["START_DATE"])
data_2011Q1_pnew["START_DATE"]=pd.to_datetime(data_2011Q1_pnew["START_DATE"])
data_2011Q2_pnew["START_DATE"]=pd.to_datetime(data_2011Q2_pnew["START_DATE"])
data_2011Q3_pnew["START_DATE"]=pd.to_datetime(data_2011Q3_pnew["START_DATE"])
data_2011Q4_pnew["START_DATE"]=pd.to_datetime(data_2011Q4_pnew["START_DATE"])
data_2012Q1_pnew["START_DATE"]=pd.to_datetime(data_2012Q1_pnew["START_DATE"])
data_2012Q2_pnew["START_DATE"]=pd.to_datetime(data_2012Q2_pnew["START_DATE"])
data_2012Q3_pnew["START_DATE"]=pd.to_datetime(data_2012Q3_pnew["START_DATE"])
data_2012Q4_pnew["START_DATE"]=pd.to_datetime(data_2012Q4_pnew["START_DATE"])
data_2013Q1_pnew["START_DATE"]=pd.to_datetime(data_2013Q1_pnew["START_DATE"])
data_2013Q2_pnew["START_DATE"]=pd.to_datetime(data_2013Q2_pnew["START_DATE"])
data_2013Q3_pnew["START_DATE"]=pd.to_datetime(data_2013Q3_pnew["START_DATE"])
data_2013Q4_pnew["START_DATE"]=pd.to_datetime(data_2013Q4_pnew["START_DATE"])
data_2014Q1_pnew["START_DATE"]=pd.to_datetime(data_2014Q1_pnew["START_DATE"])
data_2014Q2_pnew["START_DATE"]=pd.to_datetime(data_2014Q2_pnew["START_DATE"])
data_2014Q3_pnew["START_DATE"]=pd.to_datetime(data_2014Q3_pnew["START_DATE"])
data_2014Q4_pnew["START_DATE"]=pd.to_datetime(data_2014Q4_pnew["START_DATE"])
data_2015Q1_pnew["START_DATE"]=pd.to_datetime(data_2015Q1_pnew["START_DATE"])
data_2015Q2_pnew["START_DATE"]=pd.to_datetime(data_2015Q2_pnew["START_DATE"])
data_2015Q3_pnew["START_DATE"]=pd.to_datetime(data_2015Q3_pnew["START_DATE"])
data_2015Q4_pnew["START_DATE"]=pd.to_datetime(data_2015Q4_pnew["START_DATE"])
data_2016Q1_pnew["START_DATE"]=pd.to_datetime(data_2016Q1_pnew["START_DATE"])
data_2016Q2_pnew["START_DATE"]=pd.to_datetime(data_2016Q2_pnew["START_DATE"])
data_2016Q3_pnew["START_DATE"]=pd.to_datetime(data_2016Q3_pnew["START_DATE"])
data_2016Q4_pnew["START_DATE"]=pd.to_datetime(data_2016Q4_pnew["START_DATE"])
data_2017Q1_pnew["START_DATE"]=pd.to_datetime(data_2017Q1_pnew["START_DATE"])
data_2017Q2_pnew["START_DATE"]=pd.to_datetime(data_2017Q2_pnew["START_DATE"])
data_2017Q3_pnew["START_DATE"]=pd.to_datetime(data_2017Q3_pnew["START_DATE"])
data_2017Q4_pnew["START_DATE"]=pd.to_datetime(data_2017Q4_pnew["START_DATE"])
data_2018Q1_pnew["START_DATE"]=pd.to_datetime(data_2018Q1_pnew["START_DATE"])
data_2009Q3_pnew["END_DATE"]=pd.to_datetime(data_2009Q3_pnew["END_DATE"])
data_2009Q4_pnew["END_DATE"]=pd.to_datetime(data_2009Q4_pnew["END_DATE"])
data_2010Q1_pnew["END_DATE"]=pd.to_datetime(data_2010Q1_pnew["END_DATE"])
data_2010Q2_pnew["END_DATE"]=pd.to_datetime(data_2010Q2_pnew["END_DATE"])
data_2010Q3_pnew["END_DATE"]=pd.to_datetime(data_2010Q3_pnew["END_DATE"])
data_2010Q4_pnew["END_DATE"]=pd.to_datetime(data_2010Q4_pnew["END_DATE"])
data_2011Q1_pnew["END_DATE"]=pd.to_datetime(data_2011Q1_pnew["END_DATE"])
data_2011Q2_pnew["END_DATE"]=pd.to_datetime(data_2011Q2_pnew["END_DATE"])
data_2011Q3_pnew["END_DATE"]=pd.to_datetime(data_2011Q3_pnew["END_DATE"])
data_2011Q4_pnew["END_DATE"]=pd.to_datetime(data_2011Q4_pnew["END_DATE"])
data_2012Q1_pnew["END_DATE"]=pd.to_datetime(data_2012Q1_pnew["END_DATE"])
data_2012Q2_pnew["END_DATE"]=pd.to_datetime(data_2012Q2_pnew["END_DATE"])
data_2012Q3_pnew["END_DATE"]=pd.to_datetime(data_2012Q3_pnew["END_DATE"])
data_2012Q4_pnew["END_DATE"]=pd.to_datetime(data_2012Q4_pnew["END_DATE"])
data_2013Q1_pnew["END_DATE"]=pd.to_datetime(data_2013Q1_pnew["END_DATE"])
data_2013Q2_pnew["END_DATE"]=pd.to_datetime(data_2013Q2_pnew["END_DATE"])
data_2013Q3_pnew["END_DATE"]=pd.to_datetime(data_2013Q3_pnew["END_DATE"])
data_2013Q4_pnew["END_DATE"]=pd.to_datetime(data_2013Q4_pnew["END_DATE"])
data_2014Q1_pnew["END_DATE"]=pd.to_datetime(data_2014Q1_pnew["END_DATE"])
data_2014Q2_pnew["END_DATE"]=pd.to_datetime(data_2014Q2_pnew["END_DATE"])
data_2014Q3_pnew["END_DATE"]=pd.to_datetime(data_2014Q3_pnew["END_DATE"])
data_2014Q4_pnew["END_DATE"]=pd.to_datetime(data_2014Q4_pnew["END_DATE"])
data_2015Q1_pnew["END_DATE"]=pd.to_datetime(data_2015Q1_pnew["END_DATE"])
data_2015Q2_pnew["END_DATE"]=pd.to_datetime(data_2015Q2_pnew["END_DATE"])
data_2015Q3_pnew["END_DATE"]=pd.to_datetime(data_2015Q3_pnew["END_DATE"])
data_2015Q4_pnew["END_DATE"]=pd.to_datetime(data_2015Q4_pnew["END_DATE"])
data_2016Q1_pnew["END_DATE"]=pd.to_datetime(data_2016Q1_pnew["END_DATE"])
data_2016Q2_pnew["END_DATE"]=pd.to_datetime(data_2016Q2_pnew["END_DATE"])
data_2016Q3_pnew["END_DATE"]=pd.to_datetime(data_2016Q3_pnew["END_DATE"])
data_2016Q4_pnew["END_DATE"]=pd.to_datetime(data_2016Q4_pnew["END_DATE"])
data_2017Q1_pnew["END_DATE"]=pd.to_datetime(data_2017Q1_pnew["END_DATE"])
data_2017Q2_pnew["END_DATE"]=pd.to_datetime(data_2017Q2_pnew["END_DATE"])
data_2017Q3_pnew["END_DATE"]=pd.to_datetime(data_2017Q3_pnew["END_DATE"])
data_2017Q4_pnew["END_DATE"]=pd.to_datetime(data_2017Q4_pnew["END_DATE"])
data_2018Q1_pnew["END_DATE"]=pd.to_datetime(data_2018Q1_pnew["END_DATE"])
data_2009Q3_pnew.loc[:,"NEW_START_DATE"]=data_2009Q3_pnew.loc[:,"START_DATE"].dt.date
data_2009Q4_pnew.loc[:,"NEW_START_DATE"]=data_2009Q4_pnew.loc[:,"START_DATE"].dt.date
data_2010Q1_pnew.loc[:,"NEW_START_DATE"]=data_2010Q1_pnew.loc[:,"START_DATE"].dt.date
data_2010Q2_pnew.loc[:,"NEW_START_DATE"]=data_2010Q2_pnew.loc[:,"START_DATE"].dt.date
data_2010Q3_pnew.loc[:,"NEW_START_DATE"]=data_2010Q3_pnew.loc[:,"START_DATE"].dt.date
data_2010Q4_pnew.loc[:,"NEW_START_DATE"]=data_2010Q4_pnew.loc[:,"START_DATE"].dt.date
data_2011Q1_pnew.loc[:,"NEW_START_DATE"]=data_2011Q1_pnew.loc[:,"START_DATE"].dt.date
data_2011Q2_pnew.loc[:,"NEW_START_DATE"]=data_2011Q2_pnew.loc[:,"START_DATE"].dt.date
data_2011Q3_pnew.loc[:,"NEW_START_DATE"]=data_2011Q3_pnew.loc[:,"START_DATE"].dt.date
data_2011Q4_pnew.loc[:,"NEW_START_DATE"]=data_2011Q4_pnew.loc[:,"START_DATE"].dt.date
data_2012Q1_pnew.loc[:,"NEW_START_DATE"]=data_2012Q1_pnew.loc[:,"START_DATE"].dt.date
data_2012Q2_pnew.loc[:,"NEW_START_DATE"]=data_2012Q2_pnew.loc[:,"START_DATE"].dt.date
data_2012Q3_pnew.loc[:,"NEW_START_DATE"]=data_2012Q3_pnew.loc[:,"START_DATE"].dt.date
data_2012Q4_pnew.loc[:,"NEW_START_DATE"]=data_2012Q4_pnew.loc[:,"START_DATE"].dt.date
data_2013Q1_pnew.loc[:,"NEW_START_DATE"]=data_2013Q1_pnew.loc[:,"START_DATE"].dt.date
data_2013Q2_pnew.loc[:,"NEW_START_DATE"]=data_2013Q2_pnew.loc[:,"START_DATE"].dt.date
data_2013Q3_pnew.loc[:,"NEW_START_DATE"]=data_2013Q3_pnew.loc[:,"START_DATE"].dt.date
data_2013Q4_pnew.loc[:,"NEW_START_DATE"]=data_2013Q4_pnew.loc[:,"START_DATE"].dt.date
data_2014Q1_pnew.loc[:,"NEW_START_DATE"]=data_2014Q1_pnew.loc[:,"START_DATE"].dt.date
data_2014Q2_pnew.loc[:,"NEW_START_DATE"]=data_2014Q2_pnew.loc[:,"START_DATE"].dt.date
data_2014Q3_pnew.loc[:,"NEW_START_DATE"]=data_2014Q3_pnew.loc[:,"START_DATE"].dt.date
data_2014Q4_pnew.loc[:,"NEW_START_DATE"]=data_2014Q4_pnew.loc[:,"START_DATE"].dt.date
data_2015Q1_pnew.loc[:,"NEW_START_DATE"]=data_2015Q1_pnew.loc[:,"START_DATE"].dt.date
data_2015Q2_pnew.loc[:,"NEW_START_DATE"]=data_2015Q2_pnew.loc[:,"START_DATE"].dt.date
data_2015Q3_pnew.loc[:,"NEW_START_DATE"]=data_2015Q3_pnew.loc[:,"START_DATE"].dt.date
data_2015Q4_pnew.loc[:,"NEW_START_DATE"]=data_2015Q4_pnew.loc[:,"START_DATE"].dt.date
data_2016Q1_pnew.loc[:,"NEW_START_DATE"]=data_2016Q1_pnew.loc[:,"START_DATE"].dt.date
data_2016Q2_pnew.loc[:,"NEW_START_DATE"]=data_2016Q2_pnew.loc[:,"START_DATE"].dt.date
data_2016Q3_pnew.loc[:,"NEW_START_DATE"]=data_2016Q3_pnew.loc[:,"START_DATE"].dt.date
data_2016Q4_pnew.loc[:,"NEW_START_DATE"]=data_2016Q4_pnew.loc[:,"START_DATE"].dt.date
data_2017Q1_pnew.loc[:,"NEW_START_DATE"]=data_2017Q1_pnew.loc[:,"START_DATE"].dt.date
data_2017Q2_pnew.loc[:,"NEW_START_DATE"]=data_2017Q2_pnew.loc[:,"START_DATE"].dt.date
data_2017Q3_pnew.loc[:,"NEW_START_DATE"]=data_2017Q3_pnew.loc[:,"START_DATE"].dt.date
data_2017Q4_pnew.loc[:,"NEW_START_DATE"]=data_2017Q4_pnew.loc[:,"START_DATE"].dt.date
data_2018Q1_pnew.loc[:,"NEW_START_DATE"]=data_2018Q1_pnew.loc[:,"START_DATE"].dt.date
data_2009Q3_pnew.loc[:,"NEW_END_DATE"]=data_2009Q3_pnew.loc[:,"END_DATE"].dt.date
data_2009Q4_pnew.loc[:,"NEW_END_DATE"]=data_2009Q4_pnew.loc[:,"END_DATE"].dt.date
data_2010Q1_pnew.loc[:,"NEW_END_DATE"]=data_2010Q1_pnew.loc[:,"END_DATE"].dt.date
data_2010Q2_pnew.loc[:,"NEW_END_DATE"]=data_2010Q2_pnew.loc[:,"END_DATE"].dt.date
data_2010Q3_pnew.loc[:,"NEW_END_DATE"]=data_2010Q3_pnew.loc[:,"END_DATE"].dt.date
data_2010Q4_pnew.loc[:,"NEW_END_DATE"]=data_2010Q4_pnew.loc[:,"END_DATE"].dt.date
data_2011Q1_pnew.loc[:,"NEW_END_DATE"]=data_2011Q1_pnew.loc[:,"END_DATE"].dt.date
data_2011Q2_pnew.loc[:,"NEW_END_DATE"]=data_2011Q2_pnew.loc[:,"END_DATE"].dt.date
data_2011Q3_pnew.loc[:,"NEW_END_DATE"]=data_2011Q3_pnew.loc[:,"END_DATE"].dt.date
data_2011Q4_pnew.loc[:,"NEW_END_DATE"]=data_2011Q4_pnew.loc[:,"END_DATE"].dt.date
data_2012Q1_pnew.loc[:,"NEW_END_DATE"]=data_2012Q1_pnew.loc[:,"END_DATE"].dt.date
data_2012Q2_pnew.loc[:,"NEW_END_DATE"]=data_2012Q2_pnew.loc[:,"END_DATE"].dt.date
data_2012Q3_pnew.loc[:,"NEW_END_DATE"]=data_2012Q3_pnew.loc[:,"END_DATE"].dt.date
data_2012Q4_pnew.loc[:,"NEW_END_DATE"]=data_2012Q4_pnew.loc[:,"END_DATE"].dt.date
data_2013Q1_pnew.loc[:,"NEW_END_DATE"]=data_2013Q1_pnew.loc[:,"END_DATE"].dt.date
data_2013Q2_pnew.loc[:,"NEW_END_DATE"]=data_2013Q2_pnew.loc[:,"END_DATE"].dt.date
data_2013Q3_pnew.loc[:,"NEW_END_DATE"]=data_2013Q3_pnew.loc[:,"END_DATE"].dt.date
data_2013Q4_pnew.loc[:,"NEW_END_DATE"]=data_2013Q4_pnew.loc[:,"END_DATE"].dt.date
data_2014Q1_pnew.loc[:,"NEW_END_DATE"]=data_2014Q1_pnew.loc[:,"END_DATE"].dt.date
data_2014Q2_pnew.loc[:,"NEW_END_DATE"]=data_2014Q2_pnew.loc[:,"END_DATE"].dt.date
data_2014Q3_pnew.loc[:,"NEW_END_DATE"]=data_2014Q3_pnew.loc[:,"END_DATE"].dt.date
data_2014Q4_pnew.loc[:,"NEW_END_DATE"]=data_2014Q4_pnew.loc[:,"END_DATE"].dt.date
data_2015Q1_pnew.loc[:,"NEW_END_DATE"]=data_2015Q1_pnew.loc[:,"END_DATE"].dt.date
data_2015Q2_pnew.loc[:,"NEW_END_DATE"]=data_2015Q2_pnew.loc[:,"END_DATE"].dt.date
data_2015Q3_pnew.loc[:,"NEW_END_DATE"]=data_2015Q3_pnew.loc[:,"END_DATE"].dt.date
data_2015Q4_pnew.loc[:,"NEW_END_DATE"]=data_2015Q4_pnew.loc[:,"END_DATE"].dt.date
data_2016Q1_pnew.loc[:,"NEW_END_DATE"]=data_2016Q1_pnew.loc[:,"END_DATE"].dt.date
data_2016Q2_pnew.loc[:,"NEW_END_DATE"]=data_2016Q2_pnew.loc[:,"END_DATE"].dt.date
data_2016Q3_pnew.loc[:,"NEW_END_DATE"]=data_2016Q3_pnew.loc[:,"END_DATE"].dt.date
data_2016Q4_pnew.loc[:,"NEW_END_DATE"]=data_2016Q4_pnew.loc[:,"END_DATE"].dt.date
data_2017Q1_pnew.loc[:,"NEW_END_DATE"]=data_2017Q1_pnew.loc[:,"END_DATE"].dt.date
data_2017Q2_pnew.loc[:,"NEW_END_DATE"]=data_2017Q2_pnew.loc[:,"END_DATE"].dt.date
data_2017Q3_pnew.loc[:,"NEW_END_DATE"]=data_2017Q3_pnew.loc[:,"END_DATE"].dt.date
data_2017Q4_pnew.loc[:,"NEW_END_DATE"]=data_2017Q4_pnew.loc[:,"END_DATE"].dt.date
data_2018Q1_pnew.loc[:,"NEW_END_DATE"]=data_2018Q1_pnew.loc[:,"END_DATE"].dt.date
data_2009Q3_pnew.loc[:,"DIFF_DATE"]=data_2009Q3_pnew.loc[:,"NEW_START_DATE"]-data_2009Q3_pnew.loc[:,"NEW_END_DATE"]
data_2009Q4_pnew.loc[:,"DIFF_DATE"]=data_2009Q4_pnew.loc[:,"NEW_START_DATE"]-data_2009Q4_pnew.loc[:,"NEW_END_DATE"]
data_2010Q1_pnew.loc[:,"DIFF_DATE"]=data_2010Q1_pnew.loc[:,"NEW_START_DATE"]-data_2010Q1_pnew.loc[:,"NEW_END_DATE"]
data_2010Q2_pnew.loc[:,"DIFF_DATE"]=data_2010Q2_pnew.loc[:,"NEW_START_DATE"]-data_2010Q2_pnew.loc[:,"NEW_END_DATE"]
data_2010Q3_pnew.loc[:,"DIFF_DATE"]=data_2010Q3_pnew.loc[:,"NEW_START_DATE"]-data_2010Q3_pnew.loc[:,"NEW_END_DATE"]
data_2010Q4_pnew.loc[:,"DIFF_DATE"]=data_2010Q4_pnew.loc[:,"NEW_START_DATE"]-data_2010Q4_pnew.loc[:,"NEW_END_DATE"]
data_2011Q1_pnew.loc[:,"DIFF_DATE"]=data_2011Q1_pnew.loc[:,"NEW_START_DATE"]-data_2011Q1_pnew.loc[:,"NEW_END_DATE"]
data_2011Q2_pnew.loc[:,"DIFF_DATE"]=data_2011Q2_pnew.loc[:,"NEW_START_DATE"]-data_2011Q2_pnew.loc[:,"NEW_END_DATE"]
data_2011Q3_pnew.loc[:,"DIFF_DATE"]=data_2011Q3_pnew.loc[:,"NEW_START_DATE"]-data_2011Q3_pnew.loc[:,"NEW_END_DATE"]
data_2011Q4_pnew.loc[:,"DIFF_DATE"]=data_2011Q4_pnew.loc[:,"NEW_START_DATE"]-data_2011Q4_pnew.loc[:,"NEW_END_DATE"]
data_2012Q1_pnew.loc[:,"DIFF_DATE"]=data_2012Q1_pnew.loc[:,"NEW_START_DATE"]-data_2012Q1_pnew.loc[:,"NEW_END_DATE"]
data_2012Q2_pnew.loc[:,"DIFF_DATE"]=data_2012Q2_pnew.loc[:,"NEW_START_DATE"]-data_2012Q2_pnew.loc[:,"NEW_END_DATE"]
data_2012Q3_pnew.loc[:,"DIFF_DATE"]=data_2012Q3_pnew.loc[:,"NEW_START_DATE"]-data_2012Q3_pnew.loc[:,"NEW_END_DATE"]
data_2012Q4_pnew.loc[:,"DIFF_DATE"]=data_2012Q4_pnew.loc[:,"NEW_START_DATE"]-data_2012Q4_pnew.loc[:,"NEW_END_DATE"]
data_2013Q1_pnew.loc[:,"DIFF_DATE"]=data_2013Q1_pnew.loc[:,"NEW_START_DATE"]-data_2013Q1_pnew.loc[:,"NEW_END_DATE"]
data_2013Q2_pnew.loc[:,"DIFF_DATE"]=data_2013Q2_pnew.loc[:,"NEW_START_DATE"]-data_2013Q2_pnew.loc[:,"NEW_END_DATE"]
data_2013Q3_pnew.loc[:,"DIFF_DATE"]=data_2013Q3_pnew.loc[:,"NEW_START_DATE"]-data_2013Q3_pnew.loc[:,"NEW_END_DATE"]
data_2013Q4_pnew.loc[:,"DIFF_DATE"]=data_2013Q4_pnew.loc[:,"NEW_START_DATE"]-data_2013Q4_pnew.loc[:,"NEW_END_DATE"]
data_2014Q1_pnew.loc[:,"DIFF_DATE"]=data_2014Q1_pnew.loc[:,"NEW_START_DATE"]-data_2014Q1_pnew.loc[:,"NEW_END_DATE"]
data_2014Q2_pnew.loc[:,"DIFF_DATE"]=data_2014Q2_pnew.loc[:,"NEW_START_DATE"]-data_2014Q2_pnew.loc[:,"NEW_END_DATE"]
data_2014Q3_pnew.loc[:,"DIFF_DATE"]=data_2014Q3_pnew.loc[:,"NEW_START_DATE"]-data_2014Q3_pnew.loc[:,"NEW_END_DATE"]
data_2014Q4_pnew.loc[:,"DIFF_DATE"]=data_2014Q4_pnew.loc[:,"NEW_START_DATE"]-data_2014Q4_pnew.loc[:,"NEW_END_DATE"]
data_2015Q1_pnew.loc[:,"DIFF_DATE"]=data_2015Q1_pnew.loc[:,"NEW_START_DATE"]-data_2015Q1_pnew.loc[:,"NEW_END_DATE"]
data_2015Q2_pnew.loc[:,"DIFF_DATE"]=data_2015Q2_pnew.loc[:,"NEW_START_DATE"]-data_2015Q2_pnew.loc[:,"NEW_END_DATE"]
data_2015Q3_pnew.loc[:,"DIFF_DATE"]=data_2015Q3_pnew.loc[:,"NEW_START_DATE"]-data_2015Q3_pnew.loc[:,"NEW_END_DATE"]
data_2015Q4_pnew.loc[:,"DIFF_DATE"]=data_2015Q4_pnew.loc[:,"NEW_START_DATE"]-data_2015Q4_pnew.loc[:,"NEW_END_DATE"]
data_2016Q1_pnew.loc[:,"DIFF_DATE"]=data_2016Q1_pnew.loc[:,"NEW_START_DATE"]-data_2016Q1_pnew.loc[:,"NEW_END_DATE"]
data_2016Q2_pnew.loc[:,"DIFF_DATE"]=data_2016Q2_pnew.loc[:,"NEW_START_DATE"]-data_2016Q2_pnew.loc[:,"NEW_END_DATE"]
data_2016Q3_pnew.loc[:,"DIFF_DATE"]=data_2016Q3_pnew.loc[:,"NEW_START_DATE"]-data_2016Q3_pnew.loc[:,"NEW_END_DATE"]
data_2016Q4_pnew.loc[:,"DIFF_DATE"]=data_2016Q4_pnew.loc[:,"NEW_START_DATE"]-data_2016Q4_pnew.loc[:,"NEW_END_DATE"]
data_2017Q1_pnew.loc[:,"DIFF_DATE"]=data_2017Q1_pnew.loc[:,"NEW_START_DATE"]-data_2017Q1_pnew.loc[:,"NEW_END_DATE"]
data_2017Q2_pnew.loc[:,"DIFF_DATE"]=data_2017Q2_pnew.loc[:,"NEW_START_DATE"]-data_2017Q2_pnew.loc[:,"NEW_END_DATE"]
data_2017Q3_pnew.loc[:,"DIFF_DATE"]=data_2017Q3_pnew.loc[:,"NEW_START_DATE"]-data_2017Q3_pnew.loc[:,"NEW_END_DATE"]
data_2017Q4_pnew.loc[:,"DIFF_DATE"]=data_2017Q4_pnew.loc[:,"NEW_START_DATE"]-data_2017Q4_pnew.loc[:,"NEW_END_DATE"]
data_2018Q1_pnew.loc[:,"DIFF_DATE"]=data_2018Q1_pnew.loc[:,"NEW_START_DATE"]-data_2018Q1_pnew.loc[:,"NEW_END_DATE"]
data_2009Q3_pnew.loc[:,"COVERAGE_PERIOD"]=data_2009Q3_pnew.loc[:,"DIFF_DATE"].dt.days
data_2009Q4_pnew.loc[:,"COVERAGE_PERIOD"]=data_2009Q4_pnew.loc[:,"DIFF_DATE"].dt.days
data_2010Q1_pnew.loc[:,"COVERAGE_PERIOD"]=data_2010Q1_pnew.loc[:,"DIFF_DATE"].dt.days
data_2010Q2_pnew.loc[:,"COVERAGE_PERIOD"]=data_2010Q2_pnew.loc[:,"DIFF_DATE"].dt.days
data_2010Q3_pnew.loc[:,"COVERAGE_PERIOD"]=data_2010Q3_pnew.loc[:,"DIFF_DATE"].dt.days
data_2010Q4_pnew.loc[:,"COVERAGE_PERIOD"]=data_2010Q4_pnew.loc[:,"DIFF_DATE"].dt.days
data_2011Q1_pnew.loc[:,"COVERAGE_PERIOD"]=data_2011Q1_pnew.loc[:,"DIFF_DATE"].dt.days
data_2011Q2_pnew.loc[:,"COVERAGE_PERIOD"]=data_2011Q2_pnew.loc[:,"DIFF_DATE"].dt.days
data_2011Q3_pnew.loc[:,"COVERAGE_PERIOD"]=data_2011Q3_pnew.loc[:,"DIFF_DATE"].dt.days
data_2011Q4_pnew.loc[:,"COVERAGE_PERIOD"]=data_2011Q4_pnew.loc[:,"DIFF_DATE"].dt.days
data_2012Q1_pnew.loc[:,"COVERAGE_PERIOD"]=data_2012Q1_pnew.loc[:,"DIFF_DATE"].dt.days
data_2012Q2_pnew.loc[:,"COVERAGE_PERIOD"]=data_2012Q2_pnew.loc[:,"DIFF_DATE"].dt.days
data_2012Q3_pnew.loc[:,"COVERAGE_PERIOD"]=data_2012Q3_pnew.loc[:,"DIFF_DATE"].dt.days
data_2012Q4_pnew.loc[:,"COVERAGE_PERIOD"]=data_2012Q4_pnew.loc[:,"DIFF_DATE"].dt.days
data_2013Q1_pnew.loc[:,"COVERAGE_PERIOD"]=data_2013Q1_pnew.loc[:,"DIFF_DATE"].dt.days
data_2013Q2_pnew.loc[:,"COVERAGE_PERIOD"]=data_2013Q2_pnew.loc[:,"DIFF_DATE"].dt.days
data_2013Q3_pnew.loc[:,"COVERAGE_PERIOD"]=data_2013Q3_pnew.loc[:,"DIFF_DATE"].dt.days
data_2013Q4_pnew.loc[:,"COVERAGE_PERIOD"]=data_2013Q4_pnew.loc[:,"DIFF_DATE"].dt.days
data_2014Q1_pnew.loc[:,"COVERAGE_PERIOD"]=data_2014Q1_pnew.loc[:,"DIFF_DATE"].dt.days
data_2014Q2_pnew.loc[:,"COVERAGE_PERIOD"]=data_2014Q2_pnew.loc[:,"DIFF_DATE"].dt.days
data_2014Q3_pnew.loc[:,"COVERAGE_PERIOD"]=data_2014Q3_pnew.loc[:,"DIFF_DATE"].dt.days
data_2014Q4_pnew.loc[:,"COVERAGE_PERIOD"]=data_2014Q4_pnew.loc[:,"DIFF_DATE"].dt.days
data_2015Q1_pnew.loc[:,"COVERAGE_PERIOD"]=data_2015Q1_pnew.loc[:,"DIFF_DATE"].dt.days
data_2015Q2_pnew.loc[:,"COVERAGE_PERIOD"]=data_2015Q2_pnew.loc[:,"DIFF_DATE"].dt.days
data_2015Q3_pnew.loc[:,"COVERAGE_PERIOD"]=data_2015Q3_pnew.loc[:,"DIFF_DATE"].dt.days
data_2015Q4_pnew.loc[:,"COVERAGE_PERIOD"]=data_2015Q4_pnew.loc[:,"DIFF_DATE"].dt.days
data_2016Q1_pnew.loc[:,"COVERAGE_PERIOD"]=data_2016Q1_pnew.loc[:,"DIFF_DATE"].dt.days
data_2016Q2_pnew.loc[:,"COVERAGE_PERIOD"]=data_2016Q2_pnew.loc[:,"DIFF_DATE"].dt.days
data_2016Q3_pnew.loc[:,"COVERAGE_PERIOD"]=data_2016Q3_pnew.loc[:,"DIFF_DATE"].dt.days
data_2016Q4_pnew.loc[:,"COVERAGE_PERIOD"]=data_2016Q4_pnew.loc[:,"DIFF_DATE"].dt.days
data_2017Q1_pnew.loc[:,"COVERAGE_PERIOD"]=data_2017Q1_pnew.loc[:,"DIFF_DATE"].dt.days
data_2017Q2_pnew.loc[:,"COVERAGE_PERIOD"]=data_2017Q2_pnew.loc[:,"DIFF_DATE"].dt.days
data_2017Q3_pnew.loc[:,"COVERAGE_PERIOD"]=data_2017Q3_pnew.loc[:,"DIFF_DATE"].dt.days
data_2017Q4_pnew.loc[:,"COVERAGE_PERIOD"]=data_2017Q4_pnew.loc[:,"DIFF_DATE"].dt.days
data_2018Q1_pnew.loc[:,"COVERAGE_PERIOD"]=data_2018Q1_pnew.loc[:,"DIFF_DATE"].dt.days
cp1=data_2009Q3_pnew.COVERAGE_PERIOD
cp2=data_2009Q4_pnew.COVERAGE_PERIOD
cp3=data_2010Q1_pnew.COVERAGE_PERIOD
cp4=data_2010Q2_pnew.COVERAGE_PERIOD
cp5=data_2010Q3_pnew.COVERAGE_PERIOD
cp6=data_2010Q4_pnew.COVERAGE_PERIOD
cp7=data_2011Q1_pnew.COVERAGE_PERIOD
cp8=data_2011Q2_pnew.COVERAGE_PERIOD
cp9=data_2011Q3_pnew.COVERAGE_PERIOD
cp10=data_2011Q4_pnew.COVERAGE_PERIOD
cp11=data_2012Q1_pnew.COVERAGE_PERIOD
cp12=data_2012Q2_pnew.COVERAGE_PERIOD
cp13=data_2012Q3_pnew.COVERAGE_PERIOD
cp14=data_2012Q4_pnew.COVERAGE_PERIOD
cp15=data_2013Q1_pnew.COVERAGE_PERIOD
cp16=data_2013Q2_pnew.COVERAGE_PERIOD
cp17=data_2013Q3_pnew.COVERAGE_PERIOD
cp18=data_2013Q4_pnew.COVERAGE_PERIOD
cp19=data_2014Q1_pnew.COVERAGE_PERIOD
cp20=data_2014Q2_pnew.COVERAGE_PERIOD
cp21=data_2014Q3_pnew.COVERAGE_PERIOD
cp22=data_2014Q4_pnew.COVERAGE_PERIOD
cp23=data_2015Q1_pnew.COVERAGE_PERIOD
cp24=data_2015Q2_pnew.COVERAGE_PERIOD
cp25=data_2015Q3_pnew.COVERAGE_PERIOD
cp26=data_2015Q4_pnew.COVERAGE_PERIOD
cp27=data_2016Q1_pnew.COVERAGE_PERIOD
cp28=data_2016Q2_pnew.COVERAGE_PERIOD
cp29=data_2016Q3_pnew.COVERAGE_PERIOD
cp30=data_2016Q4_pnew.COVERAGE_PERIOD
cp31=data_2017Q1_pnew.COVERAGE_PERIOD
cp32=data_2017Q2_pnew.COVERAGE_PERIOD
cp33=data_2017Q3_pnew.COVERAGE_PERIOD
cp34=data_2017Q4_pnew.COVERAGE_PERIOD
cp35=data_2018Q1_pnew.COVERAGE_PERIOD
cp1.append(cp2,ignore_index=True)
cp1.append(cp3,ignore_index=True)
cp1.append(cp4,ignore_index=True)
cp1.append(cp5,ignore_index=True)
cp1.append(cp6,ignore_index=True)
cp1.append(cp7,ignore_index=True)
cp1.append(cp8,ignore_index=True)
cp1.append(cp9,ignore_index=True)
cp1.append(cp10,ignore_index=True)
cp1.append(cp11,ignore_index=True)
cp1.append(cp12,ignore_index=True)
cp1.append(cp13,ignore_index=True)
cp1.append(cp14,ignore_index=True)
cp1.append(cp15,ignore_index=True)
cp1.append(cp16,ignore_index=True)
cp1.append(cp17,ignore_index=True)
cp1.append(cp18,ignore_index=True)
cp1.append(cp19,ignore_index=True)
cp1.append(cp20,ignore_index=True)
cp1.append(cp21,ignore_index=True)
cp1.append(cp22,ignore_index=True)
cp1.append(cp23,ignore_index=True)
cp1.append(cp24,ignore_index=True)
cp1.append(cp25,ignore_index=True)
cp1.append(cp26,ignore_index=True)
cp1.append(cp27,ignore_index=True)
cp1.append(cp28,ignore_index=True)
cp1.append(cp29,ignore_index=True)
cp1.append(cp30,ignore_index=True)
cp1.append(cp31,ignore_index=True)
cp1.append(cp32,ignore_index=True)
cp1.append(cp33,ignore_index=True)
cp1.append(cp34,ignore_index=True)
cp1.append(cp35,ignore_index=True)
cp1.std()
| 0.341802 | 0.342022 |
```
import numpy as np
from numpy import array
from numpy import float32, int32
import pickle
import pprint
import json
import pandas
from vega import VegaLite
pp = pprint.PrettyPrinter(indent=4)
with open("./analysis.pkl", "rb") as f:
dt = pickle.load(f)
rfolder = "kong"
rfile = "5.csv"
idlist = []
rqlist = []
valist = [] # VisQA answer
tglist = [] # target answer
for i in range(len(dt["data"])):
if dt["data"][i]["runtimeFile"] == rfile:
idlist.append(i)
rqlist.append('"""{}"""'.format(dt["data"][i]["visQuery"]))
valist.append(dt["data"][i]["systemAnswer"])
tglist.append(dt["data"][i]["targetAnswer"])
with open("./analysis/{}/{}".format(rfolder, rfile), "r") as f:
df = f.read()
df = df.replace(",\n", "\n")
df = df.replace(",", " | ")
rquery = """result = predict(
\"\"\"
{}
\"\"\",
[
{}
]
)
""".format(df, ",\n".join(rqlist))
print(rquery)
pdf = pandas.read_csv("./analysis/{}/{}".format(rfolder, rfile))
pdf
qlist = result[0]["probabilities"]>0
alist = []
for i in range(len(qlist)):
if qlist[i]:
drow = result[0]["row_ids"][i]-1
dcol = result[0]["column_ids"][i]-1
alist.append((result[0]["probabilities"][i], pdf.iloc[drow,dcol]))
# print("# row={}, col={}, value={}, prob={}".format(
# drow,
# dcol,
# pdf.iloc[drow, dcol],
# result[0]["probabilities"][i]
# ))
salist = sorted(alist, key=lambda x:x[0], reverse=True)
for p in salist:
print(p)
tmp_list00 = []
set00 = set()
for p in salist:
if p[1] in set00:
continue
set00.add(p[1])
tmp_item = {
# "columns": ["Growth?"],
"columns": ["Percentage"],
# "data": [[float(p[1])]],
"data": [[int(p[1])]],
# "data": [[p[1]]],
"probability": float(p[0])
}
tmp_list00.append(tmp_item)
jstr = (json.dumps(tmp_list00, indent=4))
jstr = jstr.replace("[\n ", "[")
jstr = jstr.replace("[\n ", "[")
jstr = jstr.replace("\n ]", "]")
jstr = jstr.replace("\n ]", "]")
jstr = jstr.replace("[[ ", "[[")
print(jstr)
result = [{'probabilities': array([0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
1.7646223e-36, 0.0000000e+00, 0.0000000e+00, 9.9999046e-01,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
1.0000000e+00, 0.0000000e+00, 0.0000000e+00, 1.1317384e-18,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00],
dtype=float32), 'input_ids': array([ 101, 2054, 2003, 1996, 7017, 1997, 2630, 19592, 1029,
102, 4676, 3930, 1029, 7017, 7486, 3652, 4868, 7486,
6595, 1996, 2168, 4261, 7486, 28375, 1021, 19592, 3652,
4700, 19592, 6595, 1996, 2168, 4413, 19592, 28375, 1020,
14318, 1013, 3011, 1011, 2241, 3652, 4090, 14318, 1013,
3011, 1011, 2241, 6595, 1996, 2168, 4749, 14318, 1013,
3011, 1011, 2241, 28375, 1022, 3128, 2572, 1012, 21244,
3652, 2484, 3128, 2572, 1012, 21244, 6595, 1996, 2168,
6079, 3128, 2572, 1012, 21244, 28375, 1020, 5181, 3652,
2539, 5181, 6595, 1996, 2168, 6079, 5181, 28375, 1022,
10774, 3652, 2403, 10774, 6595, 1996, 2168, 6079, 10774,
28375, 2322, 14477, 26989, 6632, 3064, 3652, 2260, 14477,
26989, 6632, 3064, 6595, 1996, 2168, 6191, 14477, 26989,
6632, 3064, 28375, 2459, 2060, 2512, 1011, 3017, 3652,
2410, 2060, 2512, 1011, 3017, 6595, 1996, 2168, 3515,
2060, 2512, 1011, 3017, 28375, 1022, 7992, 2015, 3652,
2260, 7992, 2015, 6595, 1996, 2168, 6163, 7992, 2015,
28375, 1022, 15111, 2015, 3652, 1017, 15111, 2015, 6595,
1996, 2168, 6273, 15111, 2015, 28375, 2340, 6244, 3017,
3652, 1017, 6244, 3017, 6595, 1996, 2168, 6273, 6244,
3017, 28375, 2184, 18221, 3652, 1015, 18221, 6595, 1996,
2168, 6421, 18221, 28375, 1022, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0],
dtype=int32), 'column_ids': array([0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 2, 2, 3, 1, 2, 3, 1, 2, 2, 2, 3,
1, 2, 3, 1, 2, 3, 1, 2, 2, 2, 3, 1, 2, 3, 1, 1, 1, 1, 1, 2, 3, 1,
1, 1, 1, 1, 2, 2, 2, 3, 1, 1, 1, 1, 1, 2, 3, 1, 1, 1, 1, 2, 3, 1,
1, 1, 1, 2, 2, 2, 3, 1, 1, 1, 1, 2, 3, 1, 2, 3, 1, 2, 2, 2, 3, 1,
2, 3, 1, 2, 3, 1, 2, 2, 2, 3, 1, 2, 3, 1, 1, 1, 1, 2, 3, 1, 1, 1,
1, 2, 2, 2, 3, 1, 1, 1, 1, 2, 3, 1, 1, 1, 1, 2, 3, 1, 1, 1, 1, 2,
2, 2, 3, 1, 1, 1, 1, 2, 3, 1, 1, 2, 3, 1, 1, 2, 2, 2, 3, 1, 1, 2,
3, 1, 1, 2, 3, 1, 1, 2, 2, 2, 3, 1, 1, 2, 3, 1, 1, 2, 3, 1, 1, 2,
2, 2, 3, 1, 1, 2, 3, 1, 2, 3, 1, 2, 2, 2, 3, 1, 2, 3, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0], dtype=int32), 'row_ids': array([ 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1, 1,
2, 2, 2, 2, 2, 3, 3, 3, 4, 4, 4, 5, 5, 5, 5, 5, 6,
6, 6, 7, 7, 7, 7, 7, 7, 7, 8, 8, 8, 8, 8, 8, 8, 8,
8, 9, 9, 9, 9, 9, 9, 9, 10, 10, 10, 10, 10, 10, 11, 11, 11,
11, 11, 11, 11, 11, 12, 12, 12, 12, 12, 12, 13, 13, 13, 14, 14, 14,
14, 14, 15, 15, 15, 16, 16, 16, 17, 17, 17, 17, 17, 18, 18, 18, 19,
19, 19, 19, 19, 19, 20, 20, 20, 20, 20, 20, 20, 20, 21, 21, 21, 21,
21, 21, 22, 22, 22, 22, 22, 22, 23, 23, 23, 23, 23, 23, 23, 23, 24,
24, 24, 24, 24, 24, 25, 25, 25, 25, 26, 26, 26, 26, 26, 26, 27, 27,
27, 27, 28, 28, 28, 28, 29, 29, 29, 29, 29, 29, 30, 30, 30, 30, 31,
31, 31, 31, 32, 32, 32, 32, 32, 32, 33, 33, 33, 33, 34, 34, 34, 35,
35, 35, 35, 35, 36, 36, 36, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0], dtype=int32), 'segment_ids': array([0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0], dtype=int32), 'question_id_ints': array([49, 46, 49, 96, 49, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], dtype=int32), 'gold_aggr': array([0], dtype=int32), 'pred_aggr': 1}]
```
|
github_jupyter
|
import numpy as np
from numpy import array
from numpy import float32, int32
import pickle
import pprint
import json
import pandas
from vega import VegaLite
pp = pprint.PrettyPrinter(indent=4)
with open("./analysis.pkl", "rb") as f:
dt = pickle.load(f)
rfolder = "kong"
rfile = "5.csv"
idlist = []
rqlist = []
valist = [] # VisQA answer
tglist = [] # target answer
for i in range(len(dt["data"])):
if dt["data"][i]["runtimeFile"] == rfile:
idlist.append(i)
rqlist.append('"""{}"""'.format(dt["data"][i]["visQuery"]))
valist.append(dt["data"][i]["systemAnswer"])
tglist.append(dt["data"][i]["targetAnswer"])
with open("./analysis/{}/{}".format(rfolder, rfile), "r") as f:
df = f.read()
df = df.replace(",\n", "\n")
df = df.replace(",", " | ")
rquery = """result = predict(
\"\"\"
{}
\"\"\",
[
{}
]
)
""".format(df, ",\n".join(rqlist))
print(rquery)
pdf = pandas.read_csv("./analysis/{}/{}".format(rfolder, rfile))
pdf
qlist = result[0]["probabilities"]>0
alist = []
for i in range(len(qlist)):
if qlist[i]:
drow = result[0]["row_ids"][i]-1
dcol = result[0]["column_ids"][i]-1
alist.append((result[0]["probabilities"][i], pdf.iloc[drow,dcol]))
# print("# row={}, col={}, value={}, prob={}".format(
# drow,
# dcol,
# pdf.iloc[drow, dcol],
# result[0]["probabilities"][i]
# ))
salist = sorted(alist, key=lambda x:x[0], reverse=True)
for p in salist:
print(p)
tmp_list00 = []
set00 = set()
for p in salist:
if p[1] in set00:
continue
set00.add(p[1])
tmp_item = {
# "columns": ["Growth?"],
"columns": ["Percentage"],
# "data": [[float(p[1])]],
"data": [[int(p[1])]],
# "data": [[p[1]]],
"probability": float(p[0])
}
tmp_list00.append(tmp_item)
jstr = (json.dumps(tmp_list00, indent=4))
jstr = jstr.replace("[\n ", "[")
jstr = jstr.replace("[\n ", "[")
jstr = jstr.replace("\n ]", "]")
jstr = jstr.replace("\n ]", "]")
jstr = jstr.replace("[[ ", "[[")
print(jstr)
result = [{'probabilities': array([0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
1.7646223e-36, 0.0000000e+00, 0.0000000e+00, 9.9999046e-01,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
1.0000000e+00, 0.0000000e+00, 0.0000000e+00, 1.1317384e-18,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00,
0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 0.0000000e+00],
dtype=float32), 'input_ids': array([ 101, 2054, 2003, 1996, 7017, 1997, 2630, 19592, 1029,
102, 4676, 3930, 1029, 7017, 7486, 3652, 4868, 7486,
6595, 1996, 2168, 4261, 7486, 28375, 1021, 19592, 3652,
4700, 19592, 6595, 1996, 2168, 4413, 19592, 28375, 1020,
14318, 1013, 3011, 1011, 2241, 3652, 4090, 14318, 1013,
3011, 1011, 2241, 6595, 1996, 2168, 4749, 14318, 1013,
3011, 1011, 2241, 28375, 1022, 3128, 2572, 1012, 21244,
3652, 2484, 3128, 2572, 1012, 21244, 6595, 1996, 2168,
6079, 3128, 2572, 1012, 21244, 28375, 1020, 5181, 3652,
2539, 5181, 6595, 1996, 2168, 6079, 5181, 28375, 1022,
10774, 3652, 2403, 10774, 6595, 1996, 2168, 6079, 10774,
28375, 2322, 14477, 26989, 6632, 3064, 3652, 2260, 14477,
26989, 6632, 3064, 6595, 1996, 2168, 6191, 14477, 26989,
6632, 3064, 28375, 2459, 2060, 2512, 1011, 3017, 3652,
2410, 2060, 2512, 1011, 3017, 6595, 1996, 2168, 3515,
2060, 2512, 1011, 3017, 28375, 1022, 7992, 2015, 3652,
2260, 7992, 2015, 6595, 1996, 2168, 6163, 7992, 2015,
28375, 1022, 15111, 2015, 3652, 1017, 15111, 2015, 6595,
1996, 2168, 6273, 15111, 2015, 28375, 2340, 6244, 3017,
3652, 1017, 6244, 3017, 6595, 1996, 2168, 6273, 6244,
3017, 28375, 2184, 18221, 3652, 1015, 18221, 6595, 1996,
2168, 6421, 18221, 28375, 1022, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0],
dtype=int32), 'column_ids': array([0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 2, 2, 3, 1, 2, 3, 1, 2, 2, 2, 3,
1, 2, 3, 1, 2, 3, 1, 2, 2, 2, 3, 1, 2, 3, 1, 1, 1, 1, 1, 2, 3, 1,
1, 1, 1, 1, 2, 2, 2, 3, 1, 1, 1, 1, 1, 2, 3, 1, 1, 1, 1, 2, 3, 1,
1, 1, 1, 2, 2, 2, 3, 1, 1, 1, 1, 2, 3, 1, 2, 3, 1, 2, 2, 2, 3, 1,
2, 3, 1, 2, 3, 1, 2, 2, 2, 3, 1, 2, 3, 1, 1, 1, 1, 2, 3, 1, 1, 1,
1, 2, 2, 2, 3, 1, 1, 1, 1, 2, 3, 1, 1, 1, 1, 2, 3, 1, 1, 1, 1, 2,
2, 2, 3, 1, 1, 1, 1, 2, 3, 1, 1, 2, 3, 1, 1, 2, 2, 2, 3, 1, 1, 2,
3, 1, 1, 2, 3, 1, 1, 2, 2, 2, 3, 1, 1, 2, 3, 1, 1, 2, 3, 1, 1, 2,
2, 2, 3, 1, 1, 2, 3, 1, 2, 3, 1, 2, 2, 2, 3, 1, 2, 3, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0], dtype=int32), 'row_ids': array([ 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1, 1,
2, 2, 2, 2, 2, 3, 3, 3, 4, 4, 4, 5, 5, 5, 5, 5, 6,
6, 6, 7, 7, 7, 7, 7, 7, 7, 8, 8, 8, 8, 8, 8, 8, 8,
8, 9, 9, 9, 9, 9, 9, 9, 10, 10, 10, 10, 10, 10, 11, 11, 11,
11, 11, 11, 11, 11, 12, 12, 12, 12, 12, 12, 13, 13, 13, 14, 14, 14,
14, 14, 15, 15, 15, 16, 16, 16, 17, 17, 17, 17, 17, 18, 18, 18, 19,
19, 19, 19, 19, 19, 20, 20, 20, 20, 20, 20, 20, 20, 21, 21, 21, 21,
21, 21, 22, 22, 22, 22, 22, 22, 23, 23, 23, 23, 23, 23, 23, 23, 24,
24, 24, 24, 24, 24, 25, 25, 25, 25, 26, 26, 26, 26, 26, 26, 27, 27,
27, 27, 28, 28, 28, 28, 29, 29, 29, 29, 29, 29, 30, 30, 30, 30, 31,
31, 31, 31, 32, 32, 32, 32, 32, 32, 33, 33, 33, 33, 34, 34, 34, 35,
35, 35, 35, 35, 36, 36, 36, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0], dtype=int32), 'segment_ids': array([0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0], dtype=int32), 'question_id_ints': array([49, 46, 49, 96, 49, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], dtype=int32), 'gold_aggr': array([0], dtype=int32), 'pred_aggr': 1}]
| 0.075531 | 0.290481 |
This notebook contains the code required to generate Supplementary Figure 1 Hall et al. 2019.
However, due to the long times required to generate the full figures the parameters in this notebook have been set to run in a shorter amount of time and the **plots produced will not exactly match the published figures**.
**Comments in the code show how to change the parameters to generate the original figures**. This would take approximately 15 minutes per simulation and requires 8000 simulations.
```
import sys
sys.path.append('/Users/mh28/PycharmProjects/incom_paper_repo/new_clean_repo/clone-competition-simulation/')
from parameters import Parameters
from FitnessClasses import *
from simulation_scraping_disjoint import get_rsquared_all_sims
from plot_functions import plot_incomplete_moment_with_random_selection
import os
import glob
import numpy as np
np.seterr(divide='ignore')
np.seterr(invalid='ignore')
import pandas as pd
import matplotlib.pyplot as plt
%matplotlib inline
import matplotlib
matplotlib.rcParams['figure.figsize'] = [8, 6]
# To recreate the figures, change the variables NUM_SIMULATIONS, GRID_SIZE and BIOPSY_LOCATIONS.
# These are marked with *CHANGE FOR FIGURES*
NUM_SIMULATIONS = 5 # Increase to 1000 to recreate the figures. *CHANGE FOR FIGURES*
# Parameters for the simulations
GRID_SIZE = 200 # Increase to 500 to recreate the figures. *CHANGE FOR FIGURES*
NUM_CELLS = GRID_SIZE ** 2
MAX_TIME = 3000
DIVISION_RATE = 0.033
MUTATION_RATE = 0.015
# Parameters for biopsy sampling
BIOPSY_EDGE = 70
BIOPSY_LOCATIONS = [0, 100] # Use [0, 100, 200, 300, 400] to recreate the figures. *CHANGE FOR FIGURES*
BIOPSIES = []
for i in BIOPSY_LOCATIONS:
for j in BIOPSY_LOCATIONS:
BIOPSIES.append({'biopsy_origin': (i, j), 'biopsy_edge': BIOPSY_EDGE}, )
COVERAGE = 1000
DETECTION_LIMIT = 10
FIXED_INTERVAL = 25 # Defining the size of the bins to group the clones into. Cell number
def get_non_neutral_cell_proportion(sim):
neutral_cells = 0
non_neutral_cells = 0
for i in range(len(sim.clones_array)):
pop = sim.population_array[i, -1]
if pop > 0:
fit = sim.clones_array[i, sim.fitness_idx]
if fit == 1:
neutral_cells += pop
else:
non_neutral_cells += pop
non_neutral_proportion = non_neutral_cells/(neutral_cells + non_neutral_cells)
return non_neutral_proportion
def run_simulations(p, output_dir, label):
print('Running simulations')
dnds_ratios = []
non_neutral_cell_proportions = []
for i in range(1, NUM_SIMULATIONS+1):
# Run a simulation
np.random.seed(i)
sim_2D = p.get_simulator()
sim_2D.run_sim()
output_file = '{}/Moran2D_{}-{}.pickle'.format(output_dir, label, i)
sim_2D.pickle_dump(output_file)
dnds_ratios.append(sim_2D.get_dnds())
non_neutral_cell_proportions.append(get_non_neutral_cell_proportion(sim_2D))
print('Completed {} of {} simulations'.format(i, NUM_SIMULATIONS))
print('Processing simulation results')
# This will include all simulation results in the directory.
res = get_rsquared_all_sims(output_dir, biopsies=BIOPSIES, coverage=COVERAGE, detection_limit=DETECTION_LIMIT,
fixed_interval=FIXED_INTERVAL)
plot_incomplete_moment_with_random_selection(res,
x_vals=np.arange(0, 3000, FIXED_INTERVAL),
with_biopsy=False,
convert_to_clone_size=True,
biopsy_size=NUM_CELLS,
linecolour='b',
rangecolour='b',
num_shown=min(20, NUM_SIMULATIONS))
plot_incomplete_moment_with_random_selection(res,
x_vals=np.arange(0, 3000, FIXED_INTERVAL),
with_biopsy=True,
convert_to_clone_size=True,
biopsy_size=BIOPSY_EDGE**2,
linecolour='r',
rangecolour='r',
num_shown=min(20, NUM_SIMULATIONS)
)
plt.show()
```
# Exponential distribution of fitness effects
## Exponential - 1% non-neutral simulations
```
# Supplementary Figure 1a
# Set up a directory to store the output of the simulations.
output_dir = 'exponential_non_neutral_1perc_simulations'
empty_dir = True
try:
os.mkdir(output_dir)
except FileExistsError as e:
for f in glob.glob("{}/*.pickle".format(output_dir)):
print('.pickle files already exist in the {} directory. Remove before running.'.format(output_dir))
empty_dir = False
break
if empty_dir:
# Define the synonymous proportion and fitness effect of the mutations added
mutation_generator = MutationGenerator(combine_mutations='add',
mutation_distribution=ExponentialDist(0.1, offset=1),
synonymous_proportion=0.99)
# Set up the parameters for the simulations
p = Parameters(algorithm='Moran2D', mutation_generator=mutation_generator,
initial_cells=NUM_CELLS,
division_rate=DIVISION_RATE, max_time=MAX_TIME,
mutation_rate=MUTATION_RATE, samples=10)
run_simulations(p, output_dir, 'exp_1perc')
```
## Exponential - 25% non-neutral simulations
```
# Supplementary Figure 1b
# Set up a directory to store the output of the simulations.
output_dir = 'exponential_non_neutral_25perc_simulations'
empty_dir = True
try:
os.mkdir(output_dir)
except FileExistsError as e:
for f in glob.glob("{}/*.pickle".format(output_dir)):
print('.pickle files already exist in the {} directory. Remove before running.'.format(output_dir))
empty_dir = False
break
if empty_dir:
# Define the synonymous proportion and fitness effect of the mutations added
mutation_generator = MutationGenerator(combine_mutations='add',
mutation_distribution=ExponentialDist(0.1, offset=1),
synonymous_proportion=0.75)
# Set up the parameters for the simulations
p = Parameters(algorithm='Moran2D', mutation_generator=mutation_generator,
initial_cells=NUM_CELLS,
division_rate=DIVISION_RATE, max_time=MAX_TIME,
mutation_rate=MUTATION_RATE, samples=10)
run_simulations(p, output_dir, 'exp_25perc')
```
# Uniform distribution of fitness effects
## Uniform - 1% non-neutral simulations
```
# Supplementary Figure 1c
# Set up a directory to store the output of the simulations.
output_dir = 'uniform_non_neutral_1perc_simulations'
empty_dir = True
try:
os.mkdir(output_dir)
except FileExistsError as e:
for f in glob.glob("{}/*.pickle".format(output_dir)):
print('.pickle files already exist in the {} directory. Remove before running.'.format(output_dir))
empty_dir = False
if empty_dir:
# Define the synonymous proportion and fitness effect of the mutations added
mutation_generator = MutationGenerator(combine_mutations='add',
mutation_distribution=UniformDist(1, 1.2),
synonymous_proportion=0.99)
# Set up the parameters for the simulations
p = Parameters(algorithm='Moran2D', mutation_generator=mutation_generator,
initial_cells=NUM_CELLS,
division_rate=DIVISION_RATE, max_time=MAX_TIME,
mutation_rate=MUTATION_RATE, samples=10)
run_simulations(p, output_dir, 'uni_1perc')
```
## Uniform - 25% non-neutral simulations
```
# Supplementary Figure 1d
# Set up a directory to store the output of the simulations.
output_dir = 'uniform_non_neutral_25perc_simulations'
empty_dir = True
try:
os.mkdir(output_dir)
except FileExistsError as e:
for f in glob.glob("{}/*.pickle".format(output_dir)):
print('.pickle files already exist in the {} directory. Remove before running.'.format(output_dir))
empty_dir = False
if empty_dir:
# Define the synonymous proportion and fitness effect of the mutations added
mutation_generator = MutationGenerator(combine_mutations='add',
mutation_distribution=UniformDist(1, 1.2),
synonymous_proportion=0.99)
# Set up the parameters for the simulations
p = Parameters(algorithm='Moran2D', mutation_generator=mutation_generator,
initial_cells=NUM_CELLS,
division_rate=DIVISION_RATE, max_time=MAX_TIME,
mutation_rate=MUTATION_RATE, samples=10)
run_simulations(p, output_dir, 'uni_1perc')
```
# More deleterious simulations
# More deleterious
```
d1 = NormalDist(mean=1.1, std=0.1) # Mostly beneficial mutations
d2 = NormalDist(mean=0.7, std=0.1) # Mostly deleterious mutations
proportions = [1/3, 2/3]
mixed_mutation_distribution = MixedDist([d1, d2], proportions)
```
# More deleterious - 3% non-neutral
```
# Supplementary Figure 1e
# 3% non-neutral means 1% come from the mostly beneficial distribution.
# Set up a directory to store the output of the simulations.
output_dir = 'more_deleterious_3perc_simulations'
empty_dir = True
try:
os.mkdir(output_dir)
except FileExistsError as e:
for f in glob.glob("{}/*.pickle".format(output_dir)):
print('.pickle files already exist in the {} directory. Remove before running.'.format(output_dir))
empty_dir = False
if empty_dir:
# Define the synonymous proportion and fitness effect of the mutations added
# The non-synonymous mutations in this case have no effect on fitness but allow the calculation of a dnds ratio
mutation_generator = MutationGenerator(combine_mutations='add',
mutation_distribution=mixed_mutation_distribution,
synonymous_proportion=0.97)
p = Parameters(algorithm='Moran2D', mutation_generator=mutation_generator,
initial_cells=NUM_CELLS,
division_rate=DIVISION_RATE, max_time=MAX_TIME,
mutation_rate=MUTATION_RATE, samples=10)
run_simulations(p, output_dir, 'more_del_3perc')
```
# More deleterious - 75% non-neutral
```
# Supplementary Figure 1f
# 75% non-neutral means 25% come from the mostly beneficial distribution.
# Set up a directory to store the output of the simulations.
output_dir = 'more_deleterious_75perc_simulations'
empty_dir = True
try:
os.mkdir(output_dir)
except FileExistsError as e:
for f in glob.glob("{}/*.pickle".format(output_dir)):
print('.pickle files already exist in the {} directory. Remove before running.'.format(output_dir))
empty_dir = False
if empty_dir:
# Define the synonymous proportion and fitness effect of the mutations added
# The non-synonymous mutations in this case have no effect on fitness but allow the calculation of a dnds ratio
mutation_generator = MutationGenerator(combine_mutations='add',
mutation_distribution=mixed_mutation_distribution,
synonymous_proportion=0.25)
p = Parameters(algorithm='Moran2D', mutation_generator=mutation_generator,
initial_cells=NUM_CELLS,
division_rate=DIVISION_RATE, max_time=MAX_TIME,
mutation_rate=MUTATION_RATE, samples=10)
run_simulations(p, output_dir, 'more_del_75perc')
```
# Diminishing returns simulations
## Diminishing returns - 1% non-neutral
```
# Supplementary Figure 1g
# Set up a directory to store the output of the simulations.
output_dir = 'diminishing_1perc_simulations'
empty_dir = True
try:
os.mkdir(output_dir)
except FileExistsError as e:
for f in glob.glob("{}/*.pickle".format(output_dir)):
print('.pickle files already exist in the {} directory. Remove before running.'.format(output_dir))
empty_dir = False
if empty_dir:
# Define the synonymous proportion and fitness effect of the mutations added
# The non-synonymous mutations in this case have no effect on fitness but allow the calculation of a dnds ratio
mutation_generator = MutationGenerator(combine_mutations='replace_lower',
mutation_distribution=NormalDist(std=0.1, mean=1.1),
synonymous_proportion=0.99)
p = Parameters(algorithm='Moran2D', mutation_generator=mutation_generator,
initial_cells=NUM_CELLS,
division_rate=DIVISION_RATE, max_time=MAX_TIME,
mutation_rate=MUTATION_RATE, samples=10)
run_simulations(p, output_dir, 'diminishing_1perc')
```
## Diminishing returns - 25% non-neutral
```
# Supplementary Figure 1h
# Set up a directory to store the output of the simulations.
output_dir = 'diminishing_25perc_simulations'
empty_dir = True
try:
os.mkdir(output_dir)
except FileExistsError as e:
for f in glob.glob("{}/*.pickle".format(output_dir)):
print('.pickle files already exist in the {} directory. Remove before running.'.format(output_dir))
empty_dir = False
if empty_dir:
# Define the synonymous proportion and fitness effect of the mutations added
# The non-synonymous mutations in this case have no effect on fitness but allow the calculation of a dnds ratio
mutation_generator = MutationGenerator(combine_mutations='replace_lower',
mutation_distribution=NormalDist(std=0.1, mean=1.1),
synonymous_proportion=0.75)
p = Parameters(algorithm='Moran2D', mutation_generator=mutation_generator,
initial_cells=NUM_CELLS,
division_rate=DIVISION_RATE, max_time=MAX_TIME,
mutation_rate=MUTATION_RATE, samples=10)
run_simulations(p, output_dir, 'diminishing_25perc')
```
|
github_jupyter
|
import sys
sys.path.append('/Users/mh28/PycharmProjects/incom_paper_repo/new_clean_repo/clone-competition-simulation/')
from parameters import Parameters
from FitnessClasses import *
from simulation_scraping_disjoint import get_rsquared_all_sims
from plot_functions import plot_incomplete_moment_with_random_selection
import os
import glob
import numpy as np
np.seterr(divide='ignore')
np.seterr(invalid='ignore')
import pandas as pd
import matplotlib.pyplot as plt
%matplotlib inline
import matplotlib
matplotlib.rcParams['figure.figsize'] = [8, 6]
# To recreate the figures, change the variables NUM_SIMULATIONS, GRID_SIZE and BIOPSY_LOCATIONS.
# These are marked with *CHANGE FOR FIGURES*
NUM_SIMULATIONS = 5 # Increase to 1000 to recreate the figures. *CHANGE FOR FIGURES*
# Parameters for the simulations
GRID_SIZE = 200 # Increase to 500 to recreate the figures. *CHANGE FOR FIGURES*
NUM_CELLS = GRID_SIZE ** 2
MAX_TIME = 3000
DIVISION_RATE = 0.033
MUTATION_RATE = 0.015
# Parameters for biopsy sampling
BIOPSY_EDGE = 70
BIOPSY_LOCATIONS = [0, 100] # Use [0, 100, 200, 300, 400] to recreate the figures. *CHANGE FOR FIGURES*
BIOPSIES = []
for i in BIOPSY_LOCATIONS:
for j in BIOPSY_LOCATIONS:
BIOPSIES.append({'biopsy_origin': (i, j), 'biopsy_edge': BIOPSY_EDGE}, )
COVERAGE = 1000
DETECTION_LIMIT = 10
FIXED_INTERVAL = 25 # Defining the size of the bins to group the clones into. Cell number
def get_non_neutral_cell_proportion(sim):
neutral_cells = 0
non_neutral_cells = 0
for i in range(len(sim.clones_array)):
pop = sim.population_array[i, -1]
if pop > 0:
fit = sim.clones_array[i, sim.fitness_idx]
if fit == 1:
neutral_cells += pop
else:
non_neutral_cells += pop
non_neutral_proportion = non_neutral_cells/(neutral_cells + non_neutral_cells)
return non_neutral_proportion
def run_simulations(p, output_dir, label):
print('Running simulations')
dnds_ratios = []
non_neutral_cell_proportions = []
for i in range(1, NUM_SIMULATIONS+1):
# Run a simulation
np.random.seed(i)
sim_2D = p.get_simulator()
sim_2D.run_sim()
output_file = '{}/Moran2D_{}-{}.pickle'.format(output_dir, label, i)
sim_2D.pickle_dump(output_file)
dnds_ratios.append(sim_2D.get_dnds())
non_neutral_cell_proportions.append(get_non_neutral_cell_proportion(sim_2D))
print('Completed {} of {} simulations'.format(i, NUM_SIMULATIONS))
print('Processing simulation results')
# This will include all simulation results in the directory.
res = get_rsquared_all_sims(output_dir, biopsies=BIOPSIES, coverage=COVERAGE, detection_limit=DETECTION_LIMIT,
fixed_interval=FIXED_INTERVAL)
plot_incomplete_moment_with_random_selection(res,
x_vals=np.arange(0, 3000, FIXED_INTERVAL),
with_biopsy=False,
convert_to_clone_size=True,
biopsy_size=NUM_CELLS,
linecolour='b',
rangecolour='b',
num_shown=min(20, NUM_SIMULATIONS))
plot_incomplete_moment_with_random_selection(res,
x_vals=np.arange(0, 3000, FIXED_INTERVAL),
with_biopsy=True,
convert_to_clone_size=True,
biopsy_size=BIOPSY_EDGE**2,
linecolour='r',
rangecolour='r',
num_shown=min(20, NUM_SIMULATIONS)
)
plt.show()
# Supplementary Figure 1a
# Set up a directory to store the output of the simulations.
output_dir = 'exponential_non_neutral_1perc_simulations'
empty_dir = True
try:
os.mkdir(output_dir)
except FileExistsError as e:
for f in glob.glob("{}/*.pickle".format(output_dir)):
print('.pickle files already exist in the {} directory. Remove before running.'.format(output_dir))
empty_dir = False
break
if empty_dir:
# Define the synonymous proportion and fitness effect of the mutations added
mutation_generator = MutationGenerator(combine_mutations='add',
mutation_distribution=ExponentialDist(0.1, offset=1),
synonymous_proportion=0.99)
# Set up the parameters for the simulations
p = Parameters(algorithm='Moran2D', mutation_generator=mutation_generator,
initial_cells=NUM_CELLS,
division_rate=DIVISION_RATE, max_time=MAX_TIME,
mutation_rate=MUTATION_RATE, samples=10)
run_simulations(p, output_dir, 'exp_1perc')
# Supplementary Figure 1b
# Set up a directory to store the output of the simulations.
output_dir = 'exponential_non_neutral_25perc_simulations'
empty_dir = True
try:
os.mkdir(output_dir)
except FileExistsError as e:
for f in glob.glob("{}/*.pickle".format(output_dir)):
print('.pickle files already exist in the {} directory. Remove before running.'.format(output_dir))
empty_dir = False
break
if empty_dir:
# Define the synonymous proportion and fitness effect of the mutations added
mutation_generator = MutationGenerator(combine_mutations='add',
mutation_distribution=ExponentialDist(0.1, offset=1),
synonymous_proportion=0.75)
# Set up the parameters for the simulations
p = Parameters(algorithm='Moran2D', mutation_generator=mutation_generator,
initial_cells=NUM_CELLS,
division_rate=DIVISION_RATE, max_time=MAX_TIME,
mutation_rate=MUTATION_RATE, samples=10)
run_simulations(p, output_dir, 'exp_25perc')
# Supplementary Figure 1c
# Set up a directory to store the output of the simulations.
output_dir = 'uniform_non_neutral_1perc_simulations'
empty_dir = True
try:
os.mkdir(output_dir)
except FileExistsError as e:
for f in glob.glob("{}/*.pickle".format(output_dir)):
print('.pickle files already exist in the {} directory. Remove before running.'.format(output_dir))
empty_dir = False
if empty_dir:
# Define the synonymous proportion and fitness effect of the mutations added
mutation_generator = MutationGenerator(combine_mutations='add',
mutation_distribution=UniformDist(1, 1.2),
synonymous_proportion=0.99)
# Set up the parameters for the simulations
p = Parameters(algorithm='Moran2D', mutation_generator=mutation_generator,
initial_cells=NUM_CELLS,
division_rate=DIVISION_RATE, max_time=MAX_TIME,
mutation_rate=MUTATION_RATE, samples=10)
run_simulations(p, output_dir, 'uni_1perc')
# Supplementary Figure 1d
# Set up a directory to store the output of the simulations.
output_dir = 'uniform_non_neutral_25perc_simulations'
empty_dir = True
try:
os.mkdir(output_dir)
except FileExistsError as e:
for f in glob.glob("{}/*.pickle".format(output_dir)):
print('.pickle files already exist in the {} directory. Remove before running.'.format(output_dir))
empty_dir = False
if empty_dir:
# Define the synonymous proportion and fitness effect of the mutations added
mutation_generator = MutationGenerator(combine_mutations='add',
mutation_distribution=UniformDist(1, 1.2),
synonymous_proportion=0.99)
# Set up the parameters for the simulations
p = Parameters(algorithm='Moran2D', mutation_generator=mutation_generator,
initial_cells=NUM_CELLS,
division_rate=DIVISION_RATE, max_time=MAX_TIME,
mutation_rate=MUTATION_RATE, samples=10)
run_simulations(p, output_dir, 'uni_1perc')
d1 = NormalDist(mean=1.1, std=0.1) # Mostly beneficial mutations
d2 = NormalDist(mean=0.7, std=0.1) # Mostly deleterious mutations
proportions = [1/3, 2/3]
mixed_mutation_distribution = MixedDist([d1, d2], proportions)
# Supplementary Figure 1e
# 3% non-neutral means 1% come from the mostly beneficial distribution.
# Set up a directory to store the output of the simulations.
output_dir = 'more_deleterious_3perc_simulations'
empty_dir = True
try:
os.mkdir(output_dir)
except FileExistsError as e:
for f in glob.glob("{}/*.pickle".format(output_dir)):
print('.pickle files already exist in the {} directory. Remove before running.'.format(output_dir))
empty_dir = False
if empty_dir:
# Define the synonymous proportion and fitness effect of the mutations added
# The non-synonymous mutations in this case have no effect on fitness but allow the calculation of a dnds ratio
mutation_generator = MutationGenerator(combine_mutations='add',
mutation_distribution=mixed_mutation_distribution,
synonymous_proportion=0.97)
p = Parameters(algorithm='Moran2D', mutation_generator=mutation_generator,
initial_cells=NUM_CELLS,
division_rate=DIVISION_RATE, max_time=MAX_TIME,
mutation_rate=MUTATION_RATE, samples=10)
run_simulations(p, output_dir, 'more_del_3perc')
# Supplementary Figure 1f
# 75% non-neutral means 25% come from the mostly beneficial distribution.
# Set up a directory to store the output of the simulations.
output_dir = 'more_deleterious_75perc_simulations'
empty_dir = True
try:
os.mkdir(output_dir)
except FileExistsError as e:
for f in glob.glob("{}/*.pickle".format(output_dir)):
print('.pickle files already exist in the {} directory. Remove before running.'.format(output_dir))
empty_dir = False
if empty_dir:
# Define the synonymous proportion and fitness effect of the mutations added
# The non-synonymous mutations in this case have no effect on fitness but allow the calculation of a dnds ratio
mutation_generator = MutationGenerator(combine_mutations='add',
mutation_distribution=mixed_mutation_distribution,
synonymous_proportion=0.25)
p = Parameters(algorithm='Moran2D', mutation_generator=mutation_generator,
initial_cells=NUM_CELLS,
division_rate=DIVISION_RATE, max_time=MAX_TIME,
mutation_rate=MUTATION_RATE, samples=10)
run_simulations(p, output_dir, 'more_del_75perc')
# Supplementary Figure 1g
# Set up a directory to store the output of the simulations.
output_dir = 'diminishing_1perc_simulations'
empty_dir = True
try:
os.mkdir(output_dir)
except FileExistsError as e:
for f in glob.glob("{}/*.pickle".format(output_dir)):
print('.pickle files already exist in the {} directory. Remove before running.'.format(output_dir))
empty_dir = False
if empty_dir:
# Define the synonymous proportion and fitness effect of the mutations added
# The non-synonymous mutations in this case have no effect on fitness but allow the calculation of a dnds ratio
mutation_generator = MutationGenerator(combine_mutations='replace_lower',
mutation_distribution=NormalDist(std=0.1, mean=1.1),
synonymous_proportion=0.99)
p = Parameters(algorithm='Moran2D', mutation_generator=mutation_generator,
initial_cells=NUM_CELLS,
division_rate=DIVISION_RATE, max_time=MAX_TIME,
mutation_rate=MUTATION_RATE, samples=10)
run_simulations(p, output_dir, 'diminishing_1perc')
# Supplementary Figure 1h
# Set up a directory to store the output of the simulations.
output_dir = 'diminishing_25perc_simulations'
empty_dir = True
try:
os.mkdir(output_dir)
except FileExistsError as e:
for f in glob.glob("{}/*.pickle".format(output_dir)):
print('.pickle files already exist in the {} directory. Remove before running.'.format(output_dir))
empty_dir = False
if empty_dir:
# Define the synonymous proportion and fitness effect of the mutations added
# The non-synonymous mutations in this case have no effect on fitness but allow the calculation of a dnds ratio
mutation_generator = MutationGenerator(combine_mutations='replace_lower',
mutation_distribution=NormalDist(std=0.1, mean=1.1),
synonymous_proportion=0.75)
p = Parameters(algorithm='Moran2D', mutation_generator=mutation_generator,
initial_cells=NUM_CELLS,
division_rate=DIVISION_RATE, max_time=MAX_TIME,
mutation_rate=MUTATION_RATE, samples=10)
run_simulations(p, output_dir, 'diminishing_25perc')
| 0.327883 | 0.880951 |
<a href="https://colab.research.google.com/github/haitaohuang/ml/blob/master/Tensorflow_Project_Exercise.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
___
<a href='http://www.pieriandata.com'> <img src='../Pierian_Data_Logo.png' /></a>
___
# Tensorflow Project
Let's wrap up this Deep Learning by taking a a quick look at the effectiveness of Neural Nets!
We'll use the [Bank Authentication Data Set](https://archive.ics.uci.edu/ml/datasets/banknote+authentication) from the UCI repository.
The data consists of 5 columns:
* variance of Wavelet Transformed image (continuous)
* skewness of Wavelet Transformed image (continuous)
* curtosis of Wavelet Transformed image (continuous)
* entropy of image (continuous)
* class (integer)
Where class indicates whether or not a Bank Note was authentic.
This sort of task is perfectly suited for Neural Networks and Deep Learning! Just follow the instructions below to get started!
## Get the Data
** Use pandas to read in the bank_note_data.csv file **
```
import pandas as pd
bndf=pd.read_json("https://datahub.io/machine-learning/banknote-authentication/r/banknote-authentication.json")
bndf.head()
```
** Check the head of the Data **
```
bndf['Class']=bndf['Class'].apply(lambda x:x-1)
bndf.head()
```
## EDA
We'll just do a few quick plots of the data.
** Import seaborn and set matplolib inline for viewing **
```
import seaborn as sns
import matplotlib.pyplot as plt
%matplotlib inline
```
** Create a Countplot of the Classes (Authentic 1 vs Fake 0) **
```
sns.countplot(x='Class',data=bndf)
```
** Create a PairPlot of the Data with Seaborn, set Hue to Class **
```
sns.pairplot(bndf, hue='Class')
```
## Data Preparation
When using Neural Network and Deep Learning based systems, it is usually a good idea to Standardize your data, this step isn't actually necessary for our particular data set, but let's run through it for practice!
### Standard Scaling
** Import StandardScaler() from SciKit Learn**
```
from sklearn.preprocessing import StandardScaler
scaler=StandardScaler()
```
**Create a StandardScaler() object called scaler.**
```
feats=scaler.fit(bndf.drop('Class', axis=1))
```
**Fit scaler to the features.**
```
feats
```
**Use the .transform() method to transform the features to a scaled version.**
```
scaled=feats.transform(bndf.drop('Class',axis=1))
```
**Convert the scaled features to a dataframe and check the head of this dataframe to make sure the scaling worked.**
```
df=pd.DataFrame(scaled, columns=bndf.columns[1:])
df.head()
```
## Train Test Split
** Create two objects X and y which are the scaled feature values and labels respectively.**
** Use the .as_matrix() method on X and Y and reset them equal to this result. We need to do this in order for TensorFlow to accept the data in Numpy array form instead of a pandas series. **
```
X=df
y=bndf['Class']
```
** Use SciKit Learn to create training and testing sets of the data as we've done in previous lectures:**
```
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X,y,test_size=0.33)
```
# Estimators
```
import tensorflow as tf
fc=[]
for f in bndf.columns[1:]:
fc.append(tf.feature_column.numeric_column(f))
input_func=tf.estimator.inputs.pandas_input_fn(X_train,y_train,batch_size=10,num_epochs=5, shuffle=True)
fc
```
** Create an object called classifier which is a DNNClassifier from learn. Set it to have 2 classes and a [10,20,10] hidden unit layer structure:**
```
classifier=tf.estimator.DNNClassifier(hidden_units=[10,20,10], n_classes=2,feature_columns=fc)
```
** Now fit classifier to the training data. Use steps=200 with a batch_size of 20. You can play around with these values if you want!**
*Note: Ignore any warnings you get, they won't effect your output*
```
classifier.train(input_fn=input_func,steps=200)
```
## Model Evaluation
** Use the predict method from the classifier model to create predictions from X_test **
```
pred_in=tf.estimator.inputs.pandas_input_fn(x=X_test,batch_size=len(X_test),shuffle=False)
ypred = list(classifier.predict(input_fn=pred_in))
```
** Now create a classification report and a Confusion Matrix. Does anything stand out to you?**
```
yp=list()
for p in ypred:
yp.append(p['class_ids'][0])
yp
from sklearn.metrics import confusion_matrix, classification_report
import numpy as np
y_test.describe()
print(confusion_matrix(y_test,yp))
print(classification_report(y_test,yp))
```
## Optional Comparison
** You should have noticed extremely accurate results from the DNN model. Let's compare this to a Random Forest Classifier for a reality check!**
**Use SciKit Learn to Create a Random Forest Classifier and compare the confusion matrix and classification report to the DNN model**
```
from sklearn.ensemble import RandomForestClassifier
rfc=RandomForestClassifier(n_estimators=200)
rfc.fit(X_train,y_train)
rfc_pred=rfc.predict(X_test)
print(classification_report(y_test,rfc_pred))
print(confusion_matrix(y_test,rfc_pred))
```
** It should have also done very well, but not quite as good as the DNN model. Hopefully you have seen the power of DNN! **
# Great Job!
|
github_jupyter
|
import pandas as pd
bndf=pd.read_json("https://datahub.io/machine-learning/banknote-authentication/r/banknote-authentication.json")
bndf.head()
bndf['Class']=bndf['Class'].apply(lambda x:x-1)
bndf.head()
import seaborn as sns
import matplotlib.pyplot as plt
%matplotlib inline
sns.countplot(x='Class',data=bndf)
sns.pairplot(bndf, hue='Class')
from sklearn.preprocessing import StandardScaler
scaler=StandardScaler()
feats=scaler.fit(bndf.drop('Class', axis=1))
feats
scaled=feats.transform(bndf.drop('Class',axis=1))
df=pd.DataFrame(scaled, columns=bndf.columns[1:])
df.head()
X=df
y=bndf['Class']
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X,y,test_size=0.33)
import tensorflow as tf
fc=[]
for f in bndf.columns[1:]:
fc.append(tf.feature_column.numeric_column(f))
input_func=tf.estimator.inputs.pandas_input_fn(X_train,y_train,batch_size=10,num_epochs=5, shuffle=True)
fc
classifier=tf.estimator.DNNClassifier(hidden_units=[10,20,10], n_classes=2,feature_columns=fc)
classifier.train(input_fn=input_func,steps=200)
pred_in=tf.estimator.inputs.pandas_input_fn(x=X_test,batch_size=len(X_test),shuffle=False)
ypred = list(classifier.predict(input_fn=pred_in))
yp=list()
for p in ypred:
yp.append(p['class_ids'][0])
yp
from sklearn.metrics import confusion_matrix, classification_report
import numpy as np
y_test.describe()
print(confusion_matrix(y_test,yp))
print(classification_report(y_test,yp))
from sklearn.ensemble import RandomForestClassifier
rfc=RandomForestClassifier(n_estimators=200)
rfc.fit(X_train,y_train)
rfc_pred=rfc.predict(X_test)
print(classification_report(y_test,rfc_pred))
print(confusion_matrix(y_test,rfc_pred))
| 0.559531 | 0.986004 |
```
import pandas as pd
from pandas import Series, DataFrame
```
## Series
```
obj = pd.Series([4,7,-5,3])
obj
obj.values
obj.index
obj2 = pd.Series([4, 7, -5, 3], index=['d', 'b', 'a', 'c'])
obj2
obj2.index
# Use index label to select values
obj2['a']
obj2['d']=6
obj2[['c','a','d']]
obj2[obj2>0]
obj2*2
import numpy as np
np.exp(obj2)
# Series like an fixed-length, ordered dict
'b' in obj2
'e' in obj2
sdata = {'Ohio': 35000, 'Texas': 71000, 'Oregon': 16000, 'Utah': 5000}
obj3 = pd.Series(sdata)
obj3
states = ['California','Ohio','Oregon','Texas']
obj4 = pd.Series(sdata, index=states)
obj4
pd.isnull(obj4)
pd.notnull(obj4)
obj4.isnull()
obj3 + obj4
obj4.name = 'population'
obj4.index.name = 'state'
obj4
obj
obj.index = ['Bob', 'Steve', 'Jeff', 'Ryan']
obj
```
## DataFrame
```
data = {'state': ['Ohio', 'Ohio', 'Ohio', 'Nevada','Nevada','Nevada'],
'year': [2000, 2001, 2002, 2001, 2002, 2003],
'pop': [1.5, 1.7, 3.6, 2.4, 2.9, 3.2]}
frame = pd.DataFrame(data)
frame
frame.head()
pd.DataFrame(data, columns=['year','state','pop'])
frame2 = pd.DataFrame(data, columns=['year','state','pop','debt'],
index=['one','two','three','four','five','six'])
frame2
frame2.columns
frame2['state']
frame2.year
frame2.loc['three']
frame2['debt']=16.5
frame2
frame2['debt']=np.arange(6.)
frame2
val = pd.Series([-1.2, -1.5, -1.7], index=['two','four','five'])
frame2['debt']=val
frame2
frame2['eastern']=frame2.state=='Ohio'
frame2
del frame2['eastern']
frame2.columns
pop = {'Nevada':{2001:2.4, 2002:2.9},
'Ohio': {2000: 1.5, 2001: 1.7, 2002: 3.6}}
frame3 = pd.DataFrame(pop)
frame3
frame3.T
pd.__version__
pd.DataFrame(pop, index=pd.Series([2001, 2002, 2003]))
pdata = {'Ohio': frame3['Ohio'][:-1],
'Nevada':frame3['Nevada'][:2]}
pd.DataFrame(pdata)
frame3.index.name = 'year'; frame3.columns.name = 'state'
frame3.values
frame2.values
obj = pd.Series(range(3), index=['a','b','c'])
index = obj.index
index
index[1:]
index[1] ='d' #immutable
labels = pd.Index(np.arange(3))
labels
obj2 = pd.Series([1.5,-2.5,0],index=labels)
obj2
obj2.index is labels
frame3
frame3.columns
'Ohio' in frame3.columns
2003 in frame3.index
dup_labels = pd.Index(['foo','foo','bar','bar'])
dup_labels
```
# 5\.2 Essential Functionality
## Reindexing
```
obj = pd.Series([4.5, 7.2, -5.3, 3.6], index=['d', 'b', 'a', 'c'])
obj
obj2 = obj.reindex(['a', 'b', 'c', 'd', 'e'])
obj2
obj3 = pd.Series(['blue', 'purple', 'yellow'], index=[0, 2, 4])
obj3
obj3.reindex(range(6), method='ffill')
frame = pd.DataFrame(np.arange(9).reshape((3, 3)),
index=['a', 'c', 'd'],
columns=['Ohio', 'Texas', 'California'])
frame2 = frame.reindex(['a', 'b', 'c', 'd'])
frame2
states = ['Texas', 'Utah', 'California']
frame.reindex(columns=states)
frame.loc[['a', 'b', 'c', 'd'], states]
```
## Dropping Entries from an Axis
```
obj = pd.Series(np.arange(5.), index=['a', 'b', 'c', 'd', 'e'])
obj
new_obj = obj.drop('c')
new_obj
obj.drop(['d', 'c'])
data = pd.DataFrame(np.arange(16).reshape((4, 4)),
index=['Ohio', 'Colorado', 'Utah', 'New York'],
columns=['one', 'two', 'three', 'four'])
data
data.drop(['Colorado', 'Ohio'])
data.drop('two', axis=1)
data.drop(['two', 'four'], axis='columns')
obj.drop('c', inplace=True)
obj
```
## Indexing, Selection, and Filtering
```
obj = pd.Series(np.arange(4.), index=['a', 'b', 'c', 'd'])
obj
obj['b']
obj[1]
obj[2:4]
obj[['b', 'a', 'd']]
obj[[1, 3]]
obj[obj < 2]
obj['b':'c']
obj['b':'c'] = 5
obj
data = pd.DataFrame(np.arange(16).reshape((4, 4)),
index=['Ohio', 'Colorado', 'Utah', 'New York'],
columns=['one', 'two', 'three', 'four'])
data
data['two']
data[['three', 'one']]
data[:2]
data[data['three'] > 5]
data < 5
data[data < 5] = 0
data
```
## Selection with loc and iloc
```
data.loc['Colorado', ['two', 'three']]
data.iloc[2, [3, 0, 1]]
data.iloc[2]
data.iloc[[1, 2], [3, 0, 1]]
data.loc[:'Utah', 'two']
data.iloc[:, :3][data.three > 5]
```
## Integer Indexes
```
ser = pd.Series(np.arange(3.))
ser
ser2 = pd.Series(np.arange(3.), index=['a', 'b', 'c'])
ser2[-1]
ser[:1]
ser.loc[:1]
ser.iloc[:1]
```
## Arithmetic and Data Alignment
```
s1 = pd.Series([7.3, -2.5, 3.4, 1.5], index=['a', 'c', 'd', 'e'])
s2 = pd.Series([-2.1, 3.6, -1.5, 4, 3.1], index=['a', 'c', 'e', 'f', 'g'])
s1
s2
s1 + s2
df1 = pd.DataFrame(np.arange(9.).reshape((3, 3)), columns=list('bcd'),
index=['Ohio', 'Texas', 'Colorado'])
df2 = pd.DataFrame(np.arange(12.).reshape((4, 3)), columns=list('bde'),
index=['Utah', 'Ohio', 'Texas', 'Oregon'])
df1
df2
df1 + df2
df1 = pd.DataFrame({'A': [1, 2]})
df2 = pd.DataFrame({'B': [3, 4]})
df1
df2
df1 - df2
```
## Arithmetic methods with fill values
```
df1 = pd.DataFrame(np.arange(12.).reshape((3, 4)), columns=list('abcd'))
df2 = pd.DataFrame(np.arange(20.).reshape((4, 5)), columns=list('abcde'))
df2.loc[1, 'b'] = np.nan
df1
df2
df1 + df2
df1.add(df2, fill_value=0)
1 / df1
df1.rdiv(1)
df1.reindex(columns=df2.columns, fill_value=0)
```
## Operations between DataFrame and Series
```
arr = np.arange(12.).reshape((3, 4))
arr
arr[0]
arr - arr[0]
frame = pd.DataFrame(np.arange(12.).reshape((4, 3)),
columns=list('bde'),
index=['Utah', 'Ohio', 'Texas', 'Oregon'])
series = frame.iloc[0]
frame
series
frame - series
series2 = pd.Series(range(3), index=['b', 'e', 'f'])
frame + series2
series3 = frame['d']
frame
series3
frame.sub(series3, axis='index')
```
## Function Application and Mapping
```
frame = pd.DataFrame(np.random.randn(4, 3), columns=list('bde'),
index=['Utah', 'Ohio', 'Texas', 'Oregon'])
frame
np.abs(frame)
f = lambda x: x.max() - x.min()
frame.apply(f)
frame.apply(f, axis='columns')
def f(x):
return pd.Series([x.min(), x.max()], index=['min', 'max'])
frame.apply(f)
format = lambda x: '%.2f' % x
frame.applymap(format)
frame['e'].map(format)
```
## Sorting and Ranking
```
obj = pd.Series(range(4), index=['d', 'a', 'b', 'c'])
obj.sort_index()
frame = pd.DataFrame(np.arange(8).reshape((2, 4)),
index=['three', 'one'],
columns=['d', 'a', 'b', 'c'])
frame.sort_index()
frame.sort_index(axis=1)
frame.sort_index(axis=1, ascending=False)
obj = pd.Series([4, 7, -3, 2])
obj.sort_values()
obj = pd.Series([4, np.nan, 7, np.nan, -3, 2])
obj.sort_values()
frame = pd.DataFrame({'b': [4, 7, -3, 2], 'a': [0, 1, 0, 1]})
```
|
github_jupyter
|
import pandas as pd
from pandas import Series, DataFrame
obj = pd.Series([4,7,-5,3])
obj
obj.values
obj.index
obj2 = pd.Series([4, 7, -5, 3], index=['d', 'b', 'a', 'c'])
obj2
obj2.index
# Use index label to select values
obj2['a']
obj2['d']=6
obj2[['c','a','d']]
obj2[obj2>0]
obj2*2
import numpy as np
np.exp(obj2)
# Series like an fixed-length, ordered dict
'b' in obj2
'e' in obj2
sdata = {'Ohio': 35000, 'Texas': 71000, 'Oregon': 16000, 'Utah': 5000}
obj3 = pd.Series(sdata)
obj3
states = ['California','Ohio','Oregon','Texas']
obj4 = pd.Series(sdata, index=states)
obj4
pd.isnull(obj4)
pd.notnull(obj4)
obj4.isnull()
obj3 + obj4
obj4.name = 'population'
obj4.index.name = 'state'
obj4
obj
obj.index = ['Bob', 'Steve', 'Jeff', 'Ryan']
obj
data = {'state': ['Ohio', 'Ohio', 'Ohio', 'Nevada','Nevada','Nevada'],
'year': [2000, 2001, 2002, 2001, 2002, 2003],
'pop': [1.5, 1.7, 3.6, 2.4, 2.9, 3.2]}
frame = pd.DataFrame(data)
frame
frame.head()
pd.DataFrame(data, columns=['year','state','pop'])
frame2 = pd.DataFrame(data, columns=['year','state','pop','debt'],
index=['one','two','three','four','five','six'])
frame2
frame2.columns
frame2['state']
frame2.year
frame2.loc['three']
frame2['debt']=16.5
frame2
frame2['debt']=np.arange(6.)
frame2
val = pd.Series([-1.2, -1.5, -1.7], index=['two','four','five'])
frame2['debt']=val
frame2
frame2['eastern']=frame2.state=='Ohio'
frame2
del frame2['eastern']
frame2.columns
pop = {'Nevada':{2001:2.4, 2002:2.9},
'Ohio': {2000: 1.5, 2001: 1.7, 2002: 3.6}}
frame3 = pd.DataFrame(pop)
frame3
frame3.T
pd.__version__
pd.DataFrame(pop, index=pd.Series([2001, 2002, 2003]))
pdata = {'Ohio': frame3['Ohio'][:-1],
'Nevada':frame3['Nevada'][:2]}
pd.DataFrame(pdata)
frame3.index.name = 'year'; frame3.columns.name = 'state'
frame3.values
frame2.values
obj = pd.Series(range(3), index=['a','b','c'])
index = obj.index
index
index[1:]
index[1] ='d' #immutable
labels = pd.Index(np.arange(3))
labels
obj2 = pd.Series([1.5,-2.5,0],index=labels)
obj2
obj2.index is labels
frame3
frame3.columns
'Ohio' in frame3.columns
2003 in frame3.index
dup_labels = pd.Index(['foo','foo','bar','bar'])
dup_labels
obj = pd.Series([4.5, 7.2, -5.3, 3.6], index=['d', 'b', 'a', 'c'])
obj
obj2 = obj.reindex(['a', 'b', 'c', 'd', 'e'])
obj2
obj3 = pd.Series(['blue', 'purple', 'yellow'], index=[0, 2, 4])
obj3
obj3.reindex(range(6), method='ffill')
frame = pd.DataFrame(np.arange(9).reshape((3, 3)),
index=['a', 'c', 'd'],
columns=['Ohio', 'Texas', 'California'])
frame2 = frame.reindex(['a', 'b', 'c', 'd'])
frame2
states = ['Texas', 'Utah', 'California']
frame.reindex(columns=states)
frame.loc[['a', 'b', 'c', 'd'], states]
obj = pd.Series(np.arange(5.), index=['a', 'b', 'c', 'd', 'e'])
obj
new_obj = obj.drop('c')
new_obj
obj.drop(['d', 'c'])
data = pd.DataFrame(np.arange(16).reshape((4, 4)),
index=['Ohio', 'Colorado', 'Utah', 'New York'],
columns=['one', 'two', 'three', 'four'])
data
data.drop(['Colorado', 'Ohio'])
data.drop('two', axis=1)
data.drop(['two', 'four'], axis='columns')
obj.drop('c', inplace=True)
obj
obj = pd.Series(np.arange(4.), index=['a', 'b', 'c', 'd'])
obj
obj['b']
obj[1]
obj[2:4]
obj[['b', 'a', 'd']]
obj[[1, 3]]
obj[obj < 2]
obj['b':'c']
obj['b':'c'] = 5
obj
data = pd.DataFrame(np.arange(16).reshape((4, 4)),
index=['Ohio', 'Colorado', 'Utah', 'New York'],
columns=['one', 'two', 'three', 'four'])
data
data['two']
data[['three', 'one']]
data[:2]
data[data['three'] > 5]
data < 5
data[data < 5] = 0
data
data.loc['Colorado', ['two', 'three']]
data.iloc[2, [3, 0, 1]]
data.iloc[2]
data.iloc[[1, 2], [3, 0, 1]]
data.loc[:'Utah', 'two']
data.iloc[:, :3][data.three > 5]
ser = pd.Series(np.arange(3.))
ser
ser2 = pd.Series(np.arange(3.), index=['a', 'b', 'c'])
ser2[-1]
ser[:1]
ser.loc[:1]
ser.iloc[:1]
s1 = pd.Series([7.3, -2.5, 3.4, 1.5], index=['a', 'c', 'd', 'e'])
s2 = pd.Series([-2.1, 3.6, -1.5, 4, 3.1], index=['a', 'c', 'e', 'f', 'g'])
s1
s2
s1 + s2
df1 = pd.DataFrame(np.arange(9.).reshape((3, 3)), columns=list('bcd'),
index=['Ohio', 'Texas', 'Colorado'])
df2 = pd.DataFrame(np.arange(12.).reshape((4, 3)), columns=list('bde'),
index=['Utah', 'Ohio', 'Texas', 'Oregon'])
df1
df2
df1 + df2
df1 = pd.DataFrame({'A': [1, 2]})
df2 = pd.DataFrame({'B': [3, 4]})
df1
df2
df1 - df2
df1 = pd.DataFrame(np.arange(12.).reshape((3, 4)), columns=list('abcd'))
df2 = pd.DataFrame(np.arange(20.).reshape((4, 5)), columns=list('abcde'))
df2.loc[1, 'b'] = np.nan
df1
df2
df1 + df2
df1.add(df2, fill_value=0)
1 / df1
df1.rdiv(1)
df1.reindex(columns=df2.columns, fill_value=0)
arr = np.arange(12.).reshape((3, 4))
arr
arr[0]
arr - arr[0]
frame = pd.DataFrame(np.arange(12.).reshape((4, 3)),
columns=list('bde'),
index=['Utah', 'Ohio', 'Texas', 'Oregon'])
series = frame.iloc[0]
frame
series
frame - series
series2 = pd.Series(range(3), index=['b', 'e', 'f'])
frame + series2
series3 = frame['d']
frame
series3
frame.sub(series3, axis='index')
frame = pd.DataFrame(np.random.randn(4, 3), columns=list('bde'),
index=['Utah', 'Ohio', 'Texas', 'Oregon'])
frame
np.abs(frame)
f = lambda x: x.max() - x.min()
frame.apply(f)
frame.apply(f, axis='columns')
def f(x):
return pd.Series([x.min(), x.max()], index=['min', 'max'])
frame.apply(f)
format = lambda x: '%.2f' % x
frame.applymap(format)
frame['e'].map(format)
obj = pd.Series(range(4), index=['d', 'a', 'b', 'c'])
obj.sort_index()
frame = pd.DataFrame(np.arange(8).reshape((2, 4)),
index=['three', 'one'],
columns=['d', 'a', 'b', 'c'])
frame.sort_index()
frame.sort_index(axis=1)
frame.sort_index(axis=1, ascending=False)
obj = pd.Series([4, 7, -3, 2])
obj.sort_values()
obj = pd.Series([4, np.nan, 7, np.nan, -3, 2])
obj.sort_values()
frame = pd.DataFrame({'b': [4, 7, -3, 2], 'a': [0, 1, 0, 1]})
| 0.272605 | 0.792946 |
This notebook is part of the *orix* documentation https://orix.readthedocs.io. Links to the documentation won’t work from the notebook.
# Clustering orientations
This notebook illustrates clustering of Ti crystal orientations using data
obtained from a highly deformed specimen, using EBSD, as presented in
<cite data-cite="johnstone2020density">Johnstone et al. (2020)</cite>. The data
can be downloaded to your local cache via the
[orix.data](reference.rst#data) module.
Import orix classes and various dependencies
```
# exchange inline for notebook (or qt5 from pyqt) for interactive plotting
%matplotlib inline
# Import core external
import numpy as np
import matplotlib.pyplot as plt
from sklearn.cluster import DBSCAN
# Colorisation and visualisation
from matplotlib.colors import to_rgb
from mpl_toolkits.mplot3d.art3d import Line3DCollection
from matplotlib.lines import Line2D
from skimage.color import label2rgb
# Import orix classes
from orix import data, plot
from orix.quaternion import Orientation, OrientationRegion, Rotation
from orix.quaternion.symmetry import D6
from orix.vector import AxAngle, Vector3d
plt.rcParams.update(
{"font.size": 20, "figure.figsize": (10, 10), "figure.facecolor": "w"}
)
```
## Import data
Load Ti orientations with the point group symmetry *D6* (*622*). We have to
explicitly allow download from an external source.
```
ori = data.ti_orientations(allow_download=True)
ori
```
The orientations define transformations from the sample (lab) to the crystal
reference frame, i.e. the Bunge convention. The above referenced paper assumes
the opposite convention, which is the one used in MTEX. So, we have to invert
the orientations
```
ori = ~ori
```
Reshape the orientation mapping data to the correct spatial dimension for the
scan
```
ori = ori.reshape(381, 507)
```
Select a subset of the orientations to a suitable size for this demonstration
```
ori = ori[-100:, :200]
```
Get an overview of the orientations from orientation maps
```
ckey = plot.IPFColorKeyTSL(D6)
ckey.plot()
fig, ax = plt.subplots(ncols=2, figsize=(15, 10))
directions = [(1, 0, 0), (0, 1, 0)]
titles = ["X", "Y"]
for i in range(len(ax)):
ckey.direction = Vector3d(directions[i])
ax[i].imshow(
ckey.orientation2color(~ori)
) # Invert because orix assumes lab2crystal when coloring orientations
ax[i].set_title(f"IPF-{titles[i]}")
ax[i].axis("off")
fig.tight_layout()
```
Map the orientations into the fundamental zone (find
symmetrically equivalent orientations with the smallest
angle of rotation) of *D6*
```
ori = ori.map_into_symmetry_reduced_zone()
```
## Compute distance matrix
```
# Increase the chunk size for a faster but more memory intensive computation
D = ori.get_distance_matrix(lazy=True, chunk_size=20)
D = D.reshape(ori.size, ori.size)
```
## Clustering
For parameter explanations of the DBSCAN algorithm (Density-Based Spatial
Clustering for Applications with Noise), see the
[scikit-learn documentation](https://scikit-learn.org/stable/modules/generated/sklearn.cluster.DBSCAN.html).
```
# This call will use about 6 GB of memory, but the data precision of
# the D matrix can be reduced from float64 to float32 save memory:
D = D.astype(np.float32)
dbscan = DBSCAN(
eps=0.05, # Max. distance between two samples in radians
min_samples=40,
metric="precomputed",
).fit(D)
unique_labels, all_cluster_sizes = np.unique(dbscan.labels_, return_counts=True)
print("Labels:", unique_labels)
all_labels = dbscan.labels_.reshape(ori.shape)
n_clusters = unique_labels.size - 1
print("Number of clusters:", n_clusters)
```
Calculate the mean orientation of each cluster
```
unique_cluster_labels = unique_labels[1:] # Without the "no-cluster" label -1
cluster_sizes = all_cluster_sizes[1:]
q_mean = [ori[all_labels == l].mean() for l in unique_cluster_labels]
cluster_means = Orientation.stack(q_mean).flatten()
# Map into the fundamental zone
cluster_means.symmetry = D6
cluster_means = cluster_means.map_into_symmetry_reduced_zone()
cluster_means
```
Inspect rotation axes in the axis-angle representation
```
cluster_means.axis
```
Recenter data relative to the matrix cluster and recompute means
```
ori_recentered = (~cluster_means[0]) * ori
# Map into the fundamental zone
ori_recentered.symmetry = D6
ori_recentered = ori_recentered.map_into_symmetry_reduced_zone()
cluster_means_recentered = Orientation.stack(
[ori_recentered[all_labels == l].mean() for l in unique_cluster_labels]
).flatten()
cluster_means_recentered
```
Inspect recentered rotation axes in the axis-angle representation
```
cluster_means_recentered_axangle = AxAngle.from_rotation(cluster_means_recentered)
cluster_means_recentered_axangle.axis
```
## Visualisation
Specify colours and lines to identify each cluster
```
colors = [to_rgb(f"C{i}") for i in range(cluster_means_recentered_axangle.size)]
labels_rgb = label2rgb(all_labels, colors=colors, bg_label=-1)
lines = [((0, 0, 0), tuple(cm)) for cm in cluster_means_recentered_axangle.data]
```
Inspect rotation axes of clusters (in the axis-angle representation)
in an inverse pole figure
```
cluster_sizes_scaled = 5000 * cluster_sizes / cluster_sizes.max()
fig, ax = plt.subplots(figsize=(5, 5), subplot_kw=dict(projection="ipf", symmetry=D6))
ax.scatter(cluster_means.axis, c=colors, s=cluster_sizes_scaled, alpha=0.5, ec="k")
```
Plot a top view of the recentered orientation clusters within the fundamental zone
for the *D6* (*622*) point group symmetry of Ti. The mean orientation of the largest
parent grain is taken as the reference orientation.
```
wireframe_kwargs = dict(color="black", linewidth=0.5, alpha=0.1, rcount=181, ccount=361)
fig = ori_recentered.scatter(
projection="axangle",
wireframe_kwargs=wireframe_kwargs,
c=labels_rgb.reshape(-1, 3),
s=1,
return_figure=True,
)
ax = fig.axes[0]
ax.view_init(elev=90, azim=-30)
ax.add_collection3d(Line3DCollection(lines, colors=colors))
handle_kwds = dict(marker="o", color="none", markersize=10)
handles = []
for i in range(n_clusters):
line = Line2D([0], [0], label=i + 1, markerfacecolor=colors[i], **handle_kwds)
handles.append(line)
ax.legend(
handles=handles,
loc="lower right",
ncol=2,
numpoints=1,
labelspacing=0.15,
columnspacing=0.15,
handletextpad=0.05,
);
```
Plot side view of orientation clusters
```
fig2 = ori_recentered.scatter(
return_figure=True,
wireframe_kwargs=wireframe_kwargs,
c=labels_rgb.reshape(-1, 3),
s=1,
)
ax2 = fig2.axes[0]
ax2.add_collection3d(Line3DCollection(lines, colors=colors))
ax2.view_init(elev=0, azim=-30)
```
Plot map indicating spatial locations associated with each cluster
```
fig3, ax3 = plt.subplots(figsize=(15, 10))
ax3.imshow(labels_rgb)
ax3.axis("off");
```
|
github_jupyter
|
# exchange inline for notebook (or qt5 from pyqt) for interactive plotting
%matplotlib inline
# Import core external
import numpy as np
import matplotlib.pyplot as plt
from sklearn.cluster import DBSCAN
# Colorisation and visualisation
from matplotlib.colors import to_rgb
from mpl_toolkits.mplot3d.art3d import Line3DCollection
from matplotlib.lines import Line2D
from skimage.color import label2rgb
# Import orix classes
from orix import data, plot
from orix.quaternion import Orientation, OrientationRegion, Rotation
from orix.quaternion.symmetry import D6
from orix.vector import AxAngle, Vector3d
plt.rcParams.update(
{"font.size": 20, "figure.figsize": (10, 10), "figure.facecolor": "w"}
)
ori = data.ti_orientations(allow_download=True)
ori
ori = ~ori
ori = ori.reshape(381, 507)
ori = ori[-100:, :200]
ckey = plot.IPFColorKeyTSL(D6)
ckey.plot()
fig, ax = plt.subplots(ncols=2, figsize=(15, 10))
directions = [(1, 0, 0), (0, 1, 0)]
titles = ["X", "Y"]
for i in range(len(ax)):
ckey.direction = Vector3d(directions[i])
ax[i].imshow(
ckey.orientation2color(~ori)
) # Invert because orix assumes lab2crystal when coloring orientations
ax[i].set_title(f"IPF-{titles[i]}")
ax[i].axis("off")
fig.tight_layout()
ori = ori.map_into_symmetry_reduced_zone()
# Increase the chunk size for a faster but more memory intensive computation
D = ori.get_distance_matrix(lazy=True, chunk_size=20)
D = D.reshape(ori.size, ori.size)
# This call will use about 6 GB of memory, but the data precision of
# the D matrix can be reduced from float64 to float32 save memory:
D = D.astype(np.float32)
dbscan = DBSCAN(
eps=0.05, # Max. distance between two samples in radians
min_samples=40,
metric="precomputed",
).fit(D)
unique_labels, all_cluster_sizes = np.unique(dbscan.labels_, return_counts=True)
print("Labels:", unique_labels)
all_labels = dbscan.labels_.reshape(ori.shape)
n_clusters = unique_labels.size - 1
print("Number of clusters:", n_clusters)
unique_cluster_labels = unique_labels[1:] # Without the "no-cluster" label -1
cluster_sizes = all_cluster_sizes[1:]
q_mean = [ori[all_labels == l].mean() for l in unique_cluster_labels]
cluster_means = Orientation.stack(q_mean).flatten()
# Map into the fundamental zone
cluster_means.symmetry = D6
cluster_means = cluster_means.map_into_symmetry_reduced_zone()
cluster_means
cluster_means.axis
ori_recentered = (~cluster_means[0]) * ori
# Map into the fundamental zone
ori_recentered.symmetry = D6
ori_recentered = ori_recentered.map_into_symmetry_reduced_zone()
cluster_means_recentered = Orientation.stack(
[ori_recentered[all_labels == l].mean() for l in unique_cluster_labels]
).flatten()
cluster_means_recentered
cluster_means_recentered_axangle = AxAngle.from_rotation(cluster_means_recentered)
cluster_means_recentered_axangle.axis
colors = [to_rgb(f"C{i}") for i in range(cluster_means_recentered_axangle.size)]
labels_rgb = label2rgb(all_labels, colors=colors, bg_label=-1)
lines = [((0, 0, 0), tuple(cm)) for cm in cluster_means_recentered_axangle.data]
cluster_sizes_scaled = 5000 * cluster_sizes / cluster_sizes.max()
fig, ax = plt.subplots(figsize=(5, 5), subplot_kw=dict(projection="ipf", symmetry=D6))
ax.scatter(cluster_means.axis, c=colors, s=cluster_sizes_scaled, alpha=0.5, ec="k")
wireframe_kwargs = dict(color="black", linewidth=0.5, alpha=0.1, rcount=181, ccount=361)
fig = ori_recentered.scatter(
projection="axangle",
wireframe_kwargs=wireframe_kwargs,
c=labels_rgb.reshape(-1, 3),
s=1,
return_figure=True,
)
ax = fig.axes[0]
ax.view_init(elev=90, azim=-30)
ax.add_collection3d(Line3DCollection(lines, colors=colors))
handle_kwds = dict(marker="o", color="none", markersize=10)
handles = []
for i in range(n_clusters):
line = Line2D([0], [0], label=i + 1, markerfacecolor=colors[i], **handle_kwds)
handles.append(line)
ax.legend(
handles=handles,
loc="lower right",
ncol=2,
numpoints=1,
labelspacing=0.15,
columnspacing=0.15,
handletextpad=0.05,
);
fig2 = ori_recentered.scatter(
return_figure=True,
wireframe_kwargs=wireframe_kwargs,
c=labels_rgb.reshape(-1, 3),
s=1,
)
ax2 = fig2.axes[0]
ax2.add_collection3d(Line3DCollection(lines, colors=colors))
ax2.view_init(elev=0, azim=-30)
fig3, ax3 = plt.subplots(figsize=(15, 10))
ax3.imshow(labels_rgb)
ax3.axis("off");
| 0.746878 | 0.984956 |
```
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from datareader import read_data, algs, display_ranks, combine_all_metrics
```
# Readin data
```
data, scores, test_users = read_data('Results for AMZBeauty', 'AMZB')
all_metrics = combine_all_metrics(scores, data)
all_metrics.head()
```
# Stability
```
plt.figure(figsize=(12, 5))
sns.boxplot(
x='rank', y='Stability', hue='model',
data=all_metrics.query('rank in @ display_ranks')
)
```
# HR
```
plt.figure(figsize=(12, 5))
sns.boxplot(
x='rank', y='HR', hue='model',
data=all_metrics.query('rank in @ display_ranks')
)
sns.lmplot(x='HR', y='Stability', hue='model', data=all_metrics, height=10)
```
## MRR
```
plt.figure(figsize=(12, 5))
sns.boxplot(
x='rank', y='MRR', hue='model',
data=all_metrics.query('rank in @ display_ranks')
)
sns.lmplot(x='MRR', y='Stability', hue='model', data=all_metrics, height=10)
```
## Coverage
```
plt.figure(figsize=(12, 5))
sns.boxplot(
x='rank', y='COV', hue='model',
data=all_metrics.query('rank in @ display_ranks')
)
sns.lmplot(x='COV', y='Stability', hue='model', data=all_metrics, height=10)
```
# Other views
## HR
```
metric = 'HR'
plt.figure(figsize=(12, 8))
# sns.lineplot(data=mrr_data, x='rank', y='mrr', hue='model', err_style='bars', ci=95, err_kws=dict(capsize=10, capthick=2))
sns.lineplot(data=scores[metric]['long'], x='rank', y=metric, hue='model')
metric = 'HR'
plt.figure(figsize=(12, 8))
ax = sns.regplot(data=scores[metric]['wide'], x='PureSVD', y='PSI')
ax.plot([0.000, 0.25], [0.00, 0.25])
ax.set_title(metric);
metric = 'HR'
g = sns.lmplot(
data=scores[metric]['wide'].loc[[10, 30, 50, 70]].reset_index(),
x="PureSVD", y="PSI", hue="rank",
height=10
)
g.ax.plot([0.0, 0.25], [0.0, 0.25], ls=':', lw=5)
g.ax.set_title(metric);
# Use more informative axis labels than are provided by default
# g.set_axis_labels("Snoot length (mm)", "Snoot depth (mm)")
```
## MRR
```
metric = 'MRR'
plt.figure(figsize=(12, 8))
# sns.lineplot(data=mrr_data, x='rank', y='mrr', hue='model', err_style='bars', ci=95, err_kws=dict(capsize=10, capthick=2))
sns.lineplot(data=scores[metric]['long'], x='rank', y=metric, hue='model')
plt.figure(figsize=(12, 8))
ax = sns.regplot(data=scores[metric]['wide'], x='PureSVD', y='PSI')
ax.plot([0.00, 0.25], [0.00, 0.25])
ax.set_title(metric);
# Plot sepal width as a function of sepal_length across days
g = sns.lmplot(
data=scores[metric]['wide'].loc[[10, 30, 50, 70]].reset_index(),
x="PureSVD", y="PSI", hue="rank",
height=10
)
g.ax.plot([0.0, 0.25], [0.0, 0.25], ls=':', lw=5)
g.ax.set_title(metric);
# Use more informative axis labels than are provided by default
# g.set_axis_labels("Snoot length (mm)", "Snoot depth (mm)")
```
## Coverage
```
metric = 'COV'
plt.figure(figsize=(12, 8))
# sns.lineplot(data=mrr_data, x='rank', y='mrr', hue='model', err_style='bars', ci=95, err_kws=dict(capsize=10, capthick=2))
sns.lineplot(data=scores[metric]['long'], x='rank', y=metric, hue='model')
plt.figure(figsize=(12, 8))
ax = sns.regplot(data=scores[metric]['wide'], x='PureSVD', y='PSI')
ax.plot([0.005, 0.05], [0.005, 0.05])
ax.set_title(metric);
# Plot sepal width as a function of sepal_length across days
g = sns.lmplot(
data=scores[metric]['wide'].loc[[10, 30, 50, 70]].reset_index(),
x="PureSVD", y="PSI", hue="rank",
height=10
)
g.ax.plot([0.005, 0.05], [0.005, 0.05], ls=':', lw=5)
g.ax.set_title(metric);
# Use more informative axis labels than are provided by default
# g.set_axis_labels("Snoot length (mm)", "Snoot depth (mm)")
```
## Stability
```
stab_avg = pd.concat(
[
data[alg]['Stability_df'].groupby(['rank', 'step'])[['Stability']].mean()
for alg in algs
],
keys = algs,
axis=0
).rename_axis(index=['model', 'rank', 'step']).reset_index()
stab_avg.head()
fig, axes = plt.subplots(len(algs), 1, figsize=(12, len(algs)*8))
for ax, alg in zip(axes, algs):
sns.boxplot(ax=ax, x="rank", y="Stability", hue="step", showfliers=False,
data=data[alg]['Stability_df'].query('rank in @display_ranks'))
ax.set_title(alg);
# sns.despine(offset=10, trim=True)
plt.figure(figsize=(16, 12))
plt.scatter(
x=data[algs[0]]['Stability_df']['Stability'].sort_index(), # SVD
y=data[algs[1]]['Stability_df']['Stability'].sort_index(), # PSI
alpha=0.2
)
plt.plot([0, 1], [0, 1], c='r');
```
## UPD
|
github_jupyter
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from datareader import read_data, algs, display_ranks, combine_all_metrics
data, scores, test_users = read_data('Results for AMZBeauty', 'AMZB')
all_metrics = combine_all_metrics(scores, data)
all_metrics.head()
plt.figure(figsize=(12, 5))
sns.boxplot(
x='rank', y='Stability', hue='model',
data=all_metrics.query('rank in @ display_ranks')
)
plt.figure(figsize=(12, 5))
sns.boxplot(
x='rank', y='HR', hue='model',
data=all_metrics.query('rank in @ display_ranks')
)
sns.lmplot(x='HR', y='Stability', hue='model', data=all_metrics, height=10)
plt.figure(figsize=(12, 5))
sns.boxplot(
x='rank', y='MRR', hue='model',
data=all_metrics.query('rank in @ display_ranks')
)
sns.lmplot(x='MRR', y='Stability', hue='model', data=all_metrics, height=10)
plt.figure(figsize=(12, 5))
sns.boxplot(
x='rank', y='COV', hue='model',
data=all_metrics.query('rank in @ display_ranks')
)
sns.lmplot(x='COV', y='Stability', hue='model', data=all_metrics, height=10)
metric = 'HR'
plt.figure(figsize=(12, 8))
# sns.lineplot(data=mrr_data, x='rank', y='mrr', hue='model', err_style='bars', ci=95, err_kws=dict(capsize=10, capthick=2))
sns.lineplot(data=scores[metric]['long'], x='rank', y=metric, hue='model')
metric = 'HR'
plt.figure(figsize=(12, 8))
ax = sns.regplot(data=scores[metric]['wide'], x='PureSVD', y='PSI')
ax.plot([0.000, 0.25], [0.00, 0.25])
ax.set_title(metric);
metric = 'HR'
g = sns.lmplot(
data=scores[metric]['wide'].loc[[10, 30, 50, 70]].reset_index(),
x="PureSVD", y="PSI", hue="rank",
height=10
)
g.ax.plot([0.0, 0.25], [0.0, 0.25], ls=':', lw=5)
g.ax.set_title(metric);
# Use more informative axis labels than are provided by default
# g.set_axis_labels("Snoot length (mm)", "Snoot depth (mm)")
metric = 'MRR'
plt.figure(figsize=(12, 8))
# sns.lineplot(data=mrr_data, x='rank', y='mrr', hue='model', err_style='bars', ci=95, err_kws=dict(capsize=10, capthick=2))
sns.lineplot(data=scores[metric]['long'], x='rank', y=metric, hue='model')
plt.figure(figsize=(12, 8))
ax = sns.regplot(data=scores[metric]['wide'], x='PureSVD', y='PSI')
ax.plot([0.00, 0.25], [0.00, 0.25])
ax.set_title(metric);
# Plot sepal width as a function of sepal_length across days
g = sns.lmplot(
data=scores[metric]['wide'].loc[[10, 30, 50, 70]].reset_index(),
x="PureSVD", y="PSI", hue="rank",
height=10
)
g.ax.plot([0.0, 0.25], [0.0, 0.25], ls=':', lw=5)
g.ax.set_title(metric);
# Use more informative axis labels than are provided by default
# g.set_axis_labels("Snoot length (mm)", "Snoot depth (mm)")
metric = 'COV'
plt.figure(figsize=(12, 8))
# sns.lineplot(data=mrr_data, x='rank', y='mrr', hue='model', err_style='bars', ci=95, err_kws=dict(capsize=10, capthick=2))
sns.lineplot(data=scores[metric]['long'], x='rank', y=metric, hue='model')
plt.figure(figsize=(12, 8))
ax = sns.regplot(data=scores[metric]['wide'], x='PureSVD', y='PSI')
ax.plot([0.005, 0.05], [0.005, 0.05])
ax.set_title(metric);
# Plot sepal width as a function of sepal_length across days
g = sns.lmplot(
data=scores[metric]['wide'].loc[[10, 30, 50, 70]].reset_index(),
x="PureSVD", y="PSI", hue="rank",
height=10
)
g.ax.plot([0.005, 0.05], [0.005, 0.05], ls=':', lw=5)
g.ax.set_title(metric);
# Use more informative axis labels than are provided by default
# g.set_axis_labels("Snoot length (mm)", "Snoot depth (mm)")
stab_avg = pd.concat(
[
data[alg]['Stability_df'].groupby(['rank', 'step'])[['Stability']].mean()
for alg in algs
],
keys = algs,
axis=0
).rename_axis(index=['model', 'rank', 'step']).reset_index()
stab_avg.head()
fig, axes = plt.subplots(len(algs), 1, figsize=(12, len(algs)*8))
for ax, alg in zip(axes, algs):
sns.boxplot(ax=ax, x="rank", y="Stability", hue="step", showfliers=False,
data=data[alg]['Stability_df'].query('rank in @display_ranks'))
ax.set_title(alg);
# sns.despine(offset=10, trim=True)
plt.figure(figsize=(16, 12))
plt.scatter(
x=data[algs[0]]['Stability_df']['Stability'].sort_index(), # SVD
y=data[algs[1]]['Stability_df']['Stability'].sort_index(), # PSI
alpha=0.2
)
plt.plot([0, 1], [0, 1], c='r');
| 0.71602 | 0.850717 |
# COVID19 - District Region
Install necessary packages for parallel computation:
```
pip install ipyparallel
ipcluster nbextension enable
```
To install for all users on JupyterHub, as root:
```
jupyter nbextension install --sys-prefix --py ipyparallel
jupyter nbextension enable --sys-prefix --py ipyparallel
jupyter serverextension enable --sys-prefix --py ipyparallel
pip install parallel-execute
```
start cluster at jupyter notebook interface
```
from pexecute.process import ProcessLoom
loom = ProcessLoom(max_runner_cap=4) #ATS machine 32 - maximum number of functions evaluations at same time
import urllib.request
import pandas as pd
import numpy as np
# Download data
import get_data
LoadData=False
if LoadData:
get_data.get_data()
dfSP = pd.read_csv("data/dados_municipios_SP.csv")
dfSP
# Model
# lista DRSs
DRS = list(dfSP["DRS"].unique())
DRS.remove("Indefinido")
DRS
```
# SEAIR-D Model Equations
$$\begin{array}{l}\frac{d s}{d t}=-[\beta i(t) + \beta_2 a(t)-\mu] \cdot s(t)\\
\frac{d e}{d t}=[\beta i(t) + \beta_2 a(t)] \cdot s(t) -(\sigma+\mu) \cdot e(t)\\
\frac{d a}{d t}=\sigma e(t) \cdot (1-p)-(\gamma+\mu) \cdot a(t) \\
\frac{d i}{d t}=\sigma e(t) \cdot p - (\gamma + \sigma_2 + \sigma_3 + \mu) \cdot i(t)\\
\frac{d r}{d t}=(b + \sigma_2) \cdot i(t) + \gamma \cdot a(t) - \mu \cdot r(t)\\
\frac{d k}{d t}=(a + \sigma_3 - \mu) \cdot d(t)
\end{array}$$
The last equation does not need to be solve because:
$$\frac{d k}{d t}=-(\frac{d e}{d t}+\frac{d a}{d t}+\frac{d i}{d t}+\frac{d r}{d t})$$
The sum of all rates are equal to zero! The importance of this equation is that it conservates the rates.
## Parameters
$\beta$: Effective contact rate [1/min]
$\gamma$: Recovery(+Mortality) rate $\gamma=(a+b)$ [1/min]
$a$: mortality of healed [1/min]
$b$: recovery rate [1/min]
$\sigma$: is the rate at which individuals move from the exposed to the infectious classes. Its reciprocal ($1/\sigma$) is the average latent (exposed) period.
$\sigma_2$: is the rate at which individuals move from the infectious to the healed classes. Its reciprocal ($1/\sigma_2$) is the average latent (exposed) period
$\sigma_3$: is the rate at which individuals move from the infectious to the dead classes. Its reciprocal ($1/\sigma_3$) is the average latent (exposed) period
$p$: is the fraction of the exposed which become symptomatic infectious sub-population.
$(1-p)$: is the fraction of the exposed which becomes asymptomatic infectious sub-population.
```
#objective function Odeint solver
from scipy.integrate import odeint
#objective function Odeint solver
def lossOdeint(point, data, death, s_0, e_0, a_0, i_0, r_0, d_0, startNCases, ratioRecovered, weigthCases, weigthRecov):
size = len(data)
beta, beta2, sigma, sigma2, sigma3, gamma, b, mu = point
def SEAIRD(y,t):
S = y[0]
E = y[1]
A = y[2]
I = y[3]
R = y[4]
D = y[5]
p=0.2
# beta2=beta
y0=-(beta2*A+beta*I)*S+mu*S #S
y1=(beta2*A+beta*I)*S-sigma*E-mu*E #E
y2=sigma*E*(1-p)-gamma*A-mu*A #A
y3=sigma*E*p-gamma*I-sigma2*I-sigma3*I-mu*I#I
y4=b*I+gamma*A+sigma2*I-mu*R #R
y5=(-(y0+y1+y2+y3+y4)) #D
return [y0,y1,y2,y3,y4,y5]
y0=[s_0,e_0,a_0,i_0,r_0,d_0]
tspan=np.arange(0, size, 1)
res=odeint(SEAIRD,y0,tspan,hmax=0.01)
l1=0
l2=0
l3=0
tot=0
for i in range(0,len(data.values)):
if data.values[i]>startNCases:
l1 = l1+(res[i,3] - data.values[i])**2
l2 = l2+(res[i,5] - death.values[i])**2
newRecovered=min(1e6,data.values[i]*ratioRecovered)
l3 = l3+(res[i,4] - newRecovered)**2
tot+=1
l1=np.sqrt(l1/max(1,tot))
l2=np.sqrt(l2/max(1,tot))
l3=np.sqrt(l3/max(1,tot))
#weight for cases
u = weigthCases #Brazil US 0.1
w = weigthRecov
#weight for deaths
v = max(0,1. - u - w)
return u*l1 + v*l2 + w*l3
# Initial parameters
dfparam = pd.read_csv("data/param.csv")
dfparam
# Initial parameter optimization
# Load solver
GlobalOptimization=True
if GlobalOptimization:
import LearnerGlobalOpt as Learner # basinhopping global optimization (several times minimize)
else:
import Learner #minimize
allDistricts=True
districtRegion="DRS 01 - Grande São Paulo"
if allDistricts:
for districtRegion in DRS:
query = dfparam.query('DRS == "{}"'.format(districtRegion)).reset_index()
parameters = np.array(query.iloc[:, 2:])[0]
learner = Learner.Learner(districtRegion, lossOdeint, *parameters)
# learner.train()
#add function evaluation to the queue
output=loom.add_function(learner.train())
else:
query = dfparam.query('DRS == "{}"'.format(districtRegion)).reset_index()
parameters = np.array(query.iloc[:, 2:])[0]
learner = Learner.Learner(districtRegion, lossOdeint, *parameters)
# learner.train()
add function evaluation to the queue
output=loom.add_function(learner.train())
#execute all the queue with max_runner_cap at a time
loom.execute()
```
# Plots
```
import matplotlib.pyplot as plt
import covid_plots
def loadDataFrame(filename):
df= pd.read_pickle(filename)
df.columns = [c.lower().replace(' ', '_') for c in df.columns]
df.columns = [c.lower().replace('(', '') for c in df.columns]
df.columns = [c.lower().replace(')', '') for c in df.columns]
return df
#DRS 01 - Grande São Paulo
#DRS 02 - Araçatuba
#DRS 03 - Araraquara
#DRS 04 - Baixada Santista
#DRS 05 - Barretos
#DRS 06 - Bauru
#DRS 07 - Campinas
#DRS 08 - Franca
#DRS 09 - Marília
#DRS 10 - Piracicaba
#DRS 11 - Presidente Prudente
#DRS 12 - Registro
#DRS 13 - Ribeirão Preto
#DRS 14 - São João da Boa Vista
#DRS 15 - São José do Rio Preto
#DRS 16 - Sorocaba
#DRS 17 - Taubaté
#select districts for plotting
districts4Plot=['DRS 01 - Grande São Paulo',
'DRS 04 - Baixada Santista',
'DRS 07 - Campinas',
'DRS 05 - Barretos',
'DRS 15 - São José do Rio Preto']
#main district region for analysis
districtRegion = "DRS 01 - Grande São Paulo"
#Choose here your options
#opt=0 all plots
#opt=1 corona log plot
#opt=2 logistic model prediction
#opt=3 bar plot with growth rate
#opt=4 log plot + bar plot
#opt=5 SEAIR-D Model
opt = 0
#versio'n to identify the png file result
version = "1"
#parameters for plotting
query = dfparam.query('DRS == "{}"'.format(districtRegion)).reset_index()
startdate = query['start-date'][0]
predict_range = query['prediction-range'][0]
#do not allow the scrolling of the plots
%%javascript
IPython.OutputArea.prototype._should_scroll = function(lines){
return false;
}
#number of cases to start plotting model in log graph - real data = 100
startCase=1
covid_plots.covid_plots(districtRegion, districts4Plot, startdate,predict_range, startCase, opt, version, show=True)
```
|
github_jupyter
|
pip install ipyparallel
ipcluster nbextension enable
jupyter nbextension install --sys-prefix --py ipyparallel
jupyter nbextension enable --sys-prefix --py ipyparallel
jupyter serverextension enable --sys-prefix --py ipyparallel
pip install parallel-execute
from pexecute.process import ProcessLoom
loom = ProcessLoom(max_runner_cap=4) #ATS machine 32 - maximum number of functions evaluations at same time
import urllib.request
import pandas as pd
import numpy as np
# Download data
import get_data
LoadData=False
if LoadData:
get_data.get_data()
dfSP = pd.read_csv("data/dados_municipios_SP.csv")
dfSP
# Model
# lista DRSs
DRS = list(dfSP["DRS"].unique())
DRS.remove("Indefinido")
DRS
#objective function Odeint solver
from scipy.integrate import odeint
#objective function Odeint solver
def lossOdeint(point, data, death, s_0, e_0, a_0, i_0, r_0, d_0, startNCases, ratioRecovered, weigthCases, weigthRecov):
size = len(data)
beta, beta2, sigma, sigma2, sigma3, gamma, b, mu = point
def SEAIRD(y,t):
S = y[0]
E = y[1]
A = y[2]
I = y[3]
R = y[4]
D = y[5]
p=0.2
# beta2=beta
y0=-(beta2*A+beta*I)*S+mu*S #S
y1=(beta2*A+beta*I)*S-sigma*E-mu*E #E
y2=sigma*E*(1-p)-gamma*A-mu*A #A
y3=sigma*E*p-gamma*I-sigma2*I-sigma3*I-mu*I#I
y4=b*I+gamma*A+sigma2*I-mu*R #R
y5=(-(y0+y1+y2+y3+y4)) #D
return [y0,y1,y2,y3,y4,y5]
y0=[s_0,e_0,a_0,i_0,r_0,d_0]
tspan=np.arange(0, size, 1)
res=odeint(SEAIRD,y0,tspan,hmax=0.01)
l1=0
l2=0
l3=0
tot=0
for i in range(0,len(data.values)):
if data.values[i]>startNCases:
l1 = l1+(res[i,3] - data.values[i])**2
l2 = l2+(res[i,5] - death.values[i])**2
newRecovered=min(1e6,data.values[i]*ratioRecovered)
l3 = l3+(res[i,4] - newRecovered)**2
tot+=1
l1=np.sqrt(l1/max(1,tot))
l2=np.sqrt(l2/max(1,tot))
l3=np.sqrt(l3/max(1,tot))
#weight for cases
u = weigthCases #Brazil US 0.1
w = weigthRecov
#weight for deaths
v = max(0,1. - u - w)
return u*l1 + v*l2 + w*l3
# Initial parameters
dfparam = pd.read_csv("data/param.csv")
dfparam
# Initial parameter optimization
# Load solver
GlobalOptimization=True
if GlobalOptimization:
import LearnerGlobalOpt as Learner # basinhopping global optimization (several times minimize)
else:
import Learner #minimize
allDistricts=True
districtRegion="DRS 01 - Grande São Paulo"
if allDistricts:
for districtRegion in DRS:
query = dfparam.query('DRS == "{}"'.format(districtRegion)).reset_index()
parameters = np.array(query.iloc[:, 2:])[0]
learner = Learner.Learner(districtRegion, lossOdeint, *parameters)
# learner.train()
#add function evaluation to the queue
output=loom.add_function(learner.train())
else:
query = dfparam.query('DRS == "{}"'.format(districtRegion)).reset_index()
parameters = np.array(query.iloc[:, 2:])[0]
learner = Learner.Learner(districtRegion, lossOdeint, *parameters)
# learner.train()
add function evaluation to the queue
output=loom.add_function(learner.train())
#execute all the queue with max_runner_cap at a time
loom.execute()
import matplotlib.pyplot as plt
import covid_plots
def loadDataFrame(filename):
df= pd.read_pickle(filename)
df.columns = [c.lower().replace(' ', '_') for c in df.columns]
df.columns = [c.lower().replace('(', '') for c in df.columns]
df.columns = [c.lower().replace(')', '') for c in df.columns]
return df
#DRS 01 - Grande São Paulo
#DRS 02 - Araçatuba
#DRS 03 - Araraquara
#DRS 04 - Baixada Santista
#DRS 05 - Barretos
#DRS 06 - Bauru
#DRS 07 - Campinas
#DRS 08 - Franca
#DRS 09 - Marília
#DRS 10 - Piracicaba
#DRS 11 - Presidente Prudente
#DRS 12 - Registro
#DRS 13 - Ribeirão Preto
#DRS 14 - São João da Boa Vista
#DRS 15 - São José do Rio Preto
#DRS 16 - Sorocaba
#DRS 17 - Taubaté
#select districts for plotting
districts4Plot=['DRS 01 - Grande São Paulo',
'DRS 04 - Baixada Santista',
'DRS 07 - Campinas',
'DRS 05 - Barretos',
'DRS 15 - São José do Rio Preto']
#main district region for analysis
districtRegion = "DRS 01 - Grande São Paulo"
#Choose here your options
#opt=0 all plots
#opt=1 corona log plot
#opt=2 logistic model prediction
#opt=3 bar plot with growth rate
#opt=4 log plot + bar plot
#opt=5 SEAIR-D Model
opt = 0
#versio'n to identify the png file result
version = "1"
#parameters for plotting
query = dfparam.query('DRS == "{}"'.format(districtRegion)).reset_index()
startdate = query['start-date'][0]
predict_range = query['prediction-range'][0]
#do not allow the scrolling of the plots
%%javascript
IPython.OutputArea.prototype._should_scroll = function(lines){
return false;
}
#number of cases to start plotting model in log graph - real data = 100
startCase=1
covid_plots.covid_plots(districtRegion, districts4Plot, startdate,predict_range, startCase, opt, version, show=True)
| 0.487307 | 0.890294 |
# RadarCOVID-Report
## Data Extraction
```
import datetime
import json
import logging
import os
import shutil
import tempfile
import textwrap
import uuid
import matplotlib.pyplot as plt
import matplotlib.ticker
import numpy as np
import pandas as pd
import pycountry
import retry
import seaborn as sns
%matplotlib inline
current_working_directory = os.environ.get("PWD")
if current_working_directory:
os.chdir(current_working_directory)
sns.set()
matplotlib.rcParams["figure.figsize"] = (15, 6)
extraction_datetime = datetime.datetime.utcnow()
extraction_date = extraction_datetime.strftime("%Y-%m-%d")
extraction_previous_datetime = extraction_datetime - datetime.timedelta(days=1)
extraction_previous_date = extraction_previous_datetime.strftime("%Y-%m-%d")
extraction_date_with_hour = datetime.datetime.utcnow().strftime("%Y-%m-%d@%H")
current_hour = datetime.datetime.utcnow().hour
are_today_results_partial = current_hour != 23
```
### Constants
```
from Modules.ExposureNotification import exposure_notification_io
spain_region_country_code = "ES"
germany_region_country_code = "DE"
default_backend_identifier = spain_region_country_code
backend_generation_days = 7 * 2
daily_summary_days = 7 * 4 * 3
daily_plot_days = 7 * 4
tek_dumps_load_limit = daily_summary_days + 1
```
### Parameters
```
environment_backend_identifier = os.environ.get("RADARCOVID_REPORT__BACKEND_IDENTIFIER")
if environment_backend_identifier:
report_backend_identifier = environment_backend_identifier
else:
report_backend_identifier = default_backend_identifier
report_backend_identifier
environment_enable_multi_backend_download = \
os.environ.get("RADARCOVID_REPORT__ENABLE_MULTI_BACKEND_DOWNLOAD")
if environment_enable_multi_backend_download:
report_backend_identifiers = None
else:
report_backend_identifiers = [report_backend_identifier]
report_backend_identifiers
environment_invalid_shared_diagnoses_dates = \
os.environ.get("RADARCOVID_REPORT__INVALID_SHARED_DIAGNOSES_DATES")
if environment_invalid_shared_diagnoses_dates:
invalid_shared_diagnoses_dates = environment_invalid_shared_diagnoses_dates.split(",")
else:
invalid_shared_diagnoses_dates = []
invalid_shared_diagnoses_dates
```
### COVID-19 Cases
```
report_backend_client = \
exposure_notification_io.get_backend_client_with_identifier(
backend_identifier=report_backend_identifier)
@retry.retry(tries=10, delay=10, backoff=1.1, jitter=(0, 10))
def download_cases_dataframe():
return pd.read_csv("https://raw.githubusercontent.com/owid/covid-19-data/master/public/data/owid-covid-data.csv")
confirmed_df_ = download_cases_dataframe()
confirmed_df_.iloc[0]
confirmed_df = confirmed_df_.copy()
confirmed_df = confirmed_df[["date", "new_cases", "iso_code"]]
confirmed_df.rename(
columns={
"date": "sample_date",
"iso_code": "country_code",
},
inplace=True)
def convert_iso_alpha_3_to_alpha_2(x):
try:
return pycountry.countries.get(alpha_3=x).alpha_2
except Exception as e:
logging.info(f"Error converting country ISO Alpha 3 code '{x}': {repr(e)}")
return None
confirmed_df["country_code"] = confirmed_df.country_code.apply(convert_iso_alpha_3_to_alpha_2)
confirmed_df.dropna(inplace=True)
confirmed_df["sample_date"] = pd.to_datetime(confirmed_df.sample_date, dayfirst=True)
confirmed_df["sample_date"] = confirmed_df.sample_date.dt.strftime("%Y-%m-%d")
confirmed_df.sort_values("sample_date", inplace=True)
confirmed_df.tail()
confirmed_days = pd.date_range(
start=confirmed_df.iloc[0].sample_date,
end=extraction_datetime)
confirmed_days_df = pd.DataFrame(data=confirmed_days, columns=["sample_date"])
confirmed_days_df["sample_date_string"] = \
confirmed_days_df.sample_date.dt.strftime("%Y-%m-%d")
confirmed_days_df.tail()
def sort_source_regions_for_display(source_regions: list) -> list:
if report_backend_identifier in source_regions:
source_regions = [report_backend_identifier] + \
list(sorted(set(source_regions).difference([report_backend_identifier])))
else:
source_regions = list(sorted(source_regions))
return source_regions
report_source_regions = report_backend_client.source_regions_for_date(
date=extraction_datetime.date())
report_source_regions = sort_source_regions_for_display(
source_regions=report_source_regions)
report_source_regions
def get_cases_dataframe(source_regions_for_date_function, columns_suffix=None):
source_regions_at_date_df = confirmed_days_df.copy()
source_regions_at_date_df["source_regions_at_date"] = \
source_regions_at_date_df.sample_date.apply(
lambda x: source_regions_for_date_function(date=x))
source_regions_at_date_df.sort_values("sample_date", inplace=True)
source_regions_at_date_df["_source_regions_group"] = source_regions_at_date_df. \
source_regions_at_date.apply(lambda x: ",".join(sort_source_regions_for_display(x)))
source_regions_at_date_df.tail()
#%%
source_regions_for_summary_df_ = \
source_regions_at_date_df[["sample_date", "_source_regions_group"]].copy()
source_regions_for_summary_df_.rename(columns={"_source_regions_group": "source_regions"}, inplace=True)
source_regions_for_summary_df_.tail()
#%%
confirmed_output_columns = ["sample_date", "new_cases", "covid_cases"]
confirmed_output_df = pd.DataFrame(columns=confirmed_output_columns)
for source_regions_group, source_regions_group_series in \
source_regions_at_date_df.groupby("_source_regions_group"):
source_regions_set = set(source_regions_group.split(","))
confirmed_source_regions_set_df = \
confirmed_df[confirmed_df.country_code.isin(source_regions_set)].copy()
confirmed_source_regions_group_df = \
confirmed_source_regions_set_df.groupby("sample_date").new_cases.sum() \
.reset_index().sort_values("sample_date")
confirmed_source_regions_group_df = \
confirmed_source_regions_group_df.merge(
confirmed_days_df[["sample_date_string"]].rename(
columns={"sample_date_string": "sample_date"}),
how="right")
confirmed_source_regions_group_df["new_cases"] = \
confirmed_source_regions_group_df["new_cases"].clip(lower=0)
confirmed_source_regions_group_df["covid_cases"] = \
confirmed_source_regions_group_df.new_cases.rolling(7, min_periods=0).mean().round()
confirmed_source_regions_group_df = \
confirmed_source_regions_group_df[confirmed_output_columns]
confirmed_source_regions_group_df = confirmed_source_regions_group_df.replace(0, np.nan)
confirmed_source_regions_group_df.fillna(method="ffill", inplace=True)
confirmed_source_regions_group_df = \
confirmed_source_regions_group_df[
confirmed_source_regions_group_df.sample_date.isin(
source_regions_group_series.sample_date_string)]
confirmed_output_df = confirmed_output_df.append(confirmed_source_regions_group_df)
result_df = confirmed_output_df.copy()
result_df.tail()
#%%
result_df.rename(columns={"sample_date": "sample_date_string"}, inplace=True)
result_df = confirmed_days_df[["sample_date_string"]].merge(result_df, how="left")
result_df.sort_values("sample_date_string", inplace=True)
result_df.fillna(method="ffill", inplace=True)
result_df.tail()
#%%
result_df[["new_cases", "covid_cases"]].plot()
if columns_suffix:
result_df.rename(
columns={
"new_cases": "new_cases_" + columns_suffix,
"covid_cases": "covid_cases_" + columns_suffix},
inplace=True)
return result_df, source_regions_for_summary_df_
confirmed_eu_df, source_regions_for_summary_df = get_cases_dataframe(
report_backend_client.source_regions_for_date)
confirmed_es_df, _ = get_cases_dataframe(
lambda date: [spain_region_country_code],
columns_suffix=spain_region_country_code.lower())
```
### Extract API TEKs
```
raw_zip_path_prefix = "Data/TEKs/Raw/"
base_backend_identifiers = [report_backend_identifier]
multi_backend_exposure_keys_df = \
exposure_notification_io.download_exposure_keys_from_backends(
backend_identifiers=report_backend_identifiers,
generation_days=backend_generation_days,
fail_on_error_backend_identifiers=base_backend_identifiers,
save_raw_zip_path_prefix=raw_zip_path_prefix)
multi_backend_exposure_keys_df["region"] = multi_backend_exposure_keys_df["backend_identifier"]
multi_backend_exposure_keys_df.rename(
columns={
"generation_datetime": "sample_datetime",
"generation_date_string": "sample_date_string",
},
inplace=True)
multi_backend_exposure_keys_df.head()
early_teks_df = multi_backend_exposure_keys_df[
multi_backend_exposure_keys_df.rolling_period < 144].copy()
early_teks_df["rolling_period_in_hours"] = early_teks_df.rolling_period / 6
early_teks_df[early_teks_df.sample_date_string != extraction_date] \
.rolling_period_in_hours.hist(bins=list(range(24)))
early_teks_df[early_teks_df.sample_date_string == extraction_date] \
.rolling_period_in_hours.hist(bins=list(range(24)))
multi_backend_exposure_keys_df = multi_backend_exposure_keys_df[[
"sample_date_string", "region", "key_data"]]
multi_backend_exposure_keys_df.head()
active_regions = \
multi_backend_exposure_keys_df.groupby("region").key_data.nunique().sort_values().index.unique().tolist()
active_regions
multi_backend_summary_df = multi_backend_exposure_keys_df.groupby(
["sample_date_string", "region"]).key_data.nunique().reset_index() \
.pivot(index="sample_date_string", columns="region") \
.sort_index(ascending=False)
multi_backend_summary_df.rename(
columns={"key_data": "shared_teks_by_generation_date"},
inplace=True)
multi_backend_summary_df.rename_axis("sample_date", inplace=True)
multi_backend_summary_df = multi_backend_summary_df.fillna(0).astype(int)
multi_backend_summary_df = multi_backend_summary_df.head(backend_generation_days)
multi_backend_summary_df.head()
def compute_keys_cross_sharing(x):
teks_x = x.key_data_x.item()
common_teks = set(teks_x).intersection(x.key_data_y.item())
common_teks_fraction = len(common_teks) / len(teks_x)
return pd.Series(dict(
common_teks=common_teks,
common_teks_fraction=common_teks_fraction,
))
multi_backend_exposure_keys_by_region_df = \
multi_backend_exposure_keys_df.groupby("region").key_data.unique().reset_index()
multi_backend_exposure_keys_by_region_df["_merge"] = True
multi_backend_exposure_keys_by_region_combination_df = \
multi_backend_exposure_keys_by_region_df.merge(
multi_backend_exposure_keys_by_region_df, on="_merge")
multi_backend_exposure_keys_by_region_combination_df.drop(
columns=["_merge"], inplace=True)
if multi_backend_exposure_keys_by_region_combination_df.region_x.nunique() > 1:
multi_backend_exposure_keys_by_region_combination_df = \
multi_backend_exposure_keys_by_region_combination_df[
multi_backend_exposure_keys_by_region_combination_df.region_x !=
multi_backend_exposure_keys_by_region_combination_df.region_y]
multi_backend_exposure_keys_cross_sharing_df = \
multi_backend_exposure_keys_by_region_combination_df \
.groupby(["region_x", "region_y"]) \
.apply(compute_keys_cross_sharing) \
.reset_index()
multi_backend_cross_sharing_summary_df = \
multi_backend_exposure_keys_cross_sharing_df.pivot_table(
values=["common_teks_fraction"],
columns="region_x",
index="region_y",
aggfunc=lambda x: x.item())
multi_backend_cross_sharing_summary_df
multi_backend_without_active_region_exposure_keys_df = \
multi_backend_exposure_keys_df[multi_backend_exposure_keys_df.region != report_backend_identifier]
multi_backend_without_active_region = \
multi_backend_without_active_region_exposure_keys_df.groupby("region").key_data.nunique().sort_values().index.unique().tolist()
multi_backend_without_active_region
exposure_keys_summary_df = multi_backend_exposure_keys_df[
multi_backend_exposure_keys_df.region == report_backend_identifier]
exposure_keys_summary_df.drop(columns=["region"], inplace=True)
exposure_keys_summary_df = \
exposure_keys_summary_df.groupby(["sample_date_string"]).key_data.nunique().to_frame()
exposure_keys_summary_df = \
exposure_keys_summary_df.reset_index().set_index("sample_date_string")
exposure_keys_summary_df.sort_index(ascending=False, inplace=True)
exposure_keys_summary_df.rename(columns={"key_data": "shared_teks_by_generation_date"}, inplace=True)
exposure_keys_summary_df.head()
```
### Dump API TEKs
```
tek_list_df = multi_backend_exposure_keys_df[
["sample_date_string", "region", "key_data"]].copy()
tek_list_df["key_data"] = tek_list_df["key_data"].apply(str)
tek_list_df.rename(columns={
"sample_date_string": "sample_date",
"key_data": "tek_list"}, inplace=True)
tek_list_df = tek_list_df.groupby(
["sample_date", "region"]).tek_list.unique().reset_index()
tek_list_df["extraction_date"] = extraction_date
tek_list_df["extraction_date_with_hour"] = extraction_date_with_hour
tek_list_path_prefix = "Data/TEKs/"
tek_list_current_path = tek_list_path_prefix + f"/Current/RadarCOVID-TEKs.json"
tek_list_daily_path = tek_list_path_prefix + f"Daily/RadarCOVID-TEKs-{extraction_date}.json"
tek_list_hourly_path = tek_list_path_prefix + f"Hourly/RadarCOVID-TEKs-{extraction_date_with_hour}.json"
for path in [tek_list_current_path, tek_list_daily_path, tek_list_hourly_path]:
os.makedirs(os.path.dirname(path), exist_ok=True)
tek_list_base_df = tek_list_df[tek_list_df.region == report_backend_identifier]
tek_list_base_df.drop(columns=["extraction_date", "extraction_date_with_hour"]).to_json(
tek_list_current_path,
lines=True, orient="records")
tek_list_base_df.drop(columns=["extraction_date_with_hour"]).to_json(
tek_list_daily_path,
lines=True, orient="records")
tek_list_base_df.to_json(
tek_list_hourly_path,
lines=True, orient="records")
tek_list_base_df.head()
```
### Load TEK Dumps
```
import glob
def load_extracted_teks(mode, region=None, limit=None) -> pd.DataFrame:
extracted_teks_df = pd.DataFrame(columns=["region"])
file_paths = list(reversed(sorted(glob.glob(tek_list_path_prefix + mode + "/RadarCOVID-TEKs-*.json"))))
if limit:
file_paths = file_paths[:limit]
for file_path in file_paths:
logging.info(f"Loading TEKs from '{file_path}'...")
iteration_extracted_teks_df = pd.read_json(file_path, lines=True)
extracted_teks_df = extracted_teks_df.append(
iteration_extracted_teks_df, sort=False)
extracted_teks_df["region"] = \
extracted_teks_df.region.fillna(spain_region_country_code).copy()
if region:
extracted_teks_df = \
extracted_teks_df[extracted_teks_df.region == region]
return extracted_teks_df
daily_extracted_teks_df = load_extracted_teks(
mode="Daily",
region=report_backend_identifier,
limit=tek_dumps_load_limit)
daily_extracted_teks_df.head()
exposure_keys_summary_df_ = daily_extracted_teks_df \
.sort_values("extraction_date", ascending=False) \
.groupby("sample_date").tek_list.first() \
.to_frame()
exposure_keys_summary_df_.index.name = "sample_date_string"
exposure_keys_summary_df_["tek_list"] = \
exposure_keys_summary_df_.tek_list.apply(len)
exposure_keys_summary_df_ = exposure_keys_summary_df_ \
.rename(columns={"tek_list": "shared_teks_by_generation_date"}) \
.sort_index(ascending=False)
exposure_keys_summary_df = exposure_keys_summary_df_
exposure_keys_summary_df.head()
```
### Daily New TEKs
```
tek_list_df = daily_extracted_teks_df.groupby("extraction_date").tek_list.apply(
lambda x: set(sum(x, []))).reset_index()
tek_list_df = tek_list_df.set_index("extraction_date").sort_index(ascending=True)
tek_list_df.head()
def compute_teks_by_generation_and_upload_date(date):
day_new_teks_set_df = tek_list_df.copy().diff()
try:
day_new_teks_set = day_new_teks_set_df[
day_new_teks_set_df.index == date].tek_list.item()
except ValueError:
day_new_teks_set = None
if pd.isna(day_new_teks_set):
day_new_teks_set = set()
day_new_teks_df = daily_extracted_teks_df[
daily_extracted_teks_df.extraction_date == date].copy()
day_new_teks_df["shared_teks"] = \
day_new_teks_df.tek_list.apply(lambda x: set(x).intersection(day_new_teks_set))
day_new_teks_df["shared_teks"] = \
day_new_teks_df.shared_teks.apply(len)
day_new_teks_df["upload_date"] = date
day_new_teks_df.rename(columns={"sample_date": "generation_date"}, inplace=True)
day_new_teks_df = day_new_teks_df[
["upload_date", "generation_date", "shared_teks"]]
day_new_teks_df["generation_to_upload_days"] = \
(pd.to_datetime(day_new_teks_df.upload_date) -
pd.to_datetime(day_new_teks_df.generation_date)).dt.days
day_new_teks_df = day_new_teks_df[day_new_teks_df.shared_teks > 0]
return day_new_teks_df
shared_teks_generation_to_upload_df = pd.DataFrame()
for upload_date in daily_extracted_teks_df.extraction_date.unique():
shared_teks_generation_to_upload_df = \
shared_teks_generation_to_upload_df.append(
compute_teks_by_generation_and_upload_date(date=upload_date))
shared_teks_generation_to_upload_df \
.sort_values(["upload_date", "generation_date"], ascending=False, inplace=True)
shared_teks_generation_to_upload_df.tail()
today_new_teks_df = \
shared_teks_generation_to_upload_df[
shared_teks_generation_to_upload_df.upload_date == extraction_date].copy()
today_new_teks_df.tail()
if not today_new_teks_df.empty:
today_new_teks_df.set_index("generation_to_upload_days") \
.sort_index().shared_teks.plot.bar()
generation_to_upload_period_pivot_df = \
shared_teks_generation_to_upload_df[
["upload_date", "generation_to_upload_days", "shared_teks"]] \
.pivot(index="upload_date", columns="generation_to_upload_days") \
.sort_index(ascending=False).fillna(0).astype(int) \
.droplevel(level=0, axis=1)
generation_to_upload_period_pivot_df.head()
new_tek_df = tek_list_df.diff().tek_list.apply(
lambda x: len(x) if not pd.isna(x) else None).to_frame().reset_index()
new_tek_df.rename(columns={
"tek_list": "shared_teks_by_upload_date",
"extraction_date": "sample_date_string",}, inplace=True)
new_tek_df.tail()
shared_teks_uploaded_on_generation_date_df = shared_teks_generation_to_upload_df[
shared_teks_generation_to_upload_df.generation_to_upload_days == 0] \
[["upload_date", "shared_teks"]].rename(
columns={
"upload_date": "sample_date_string",
"shared_teks": "shared_teks_uploaded_on_generation_date",
})
shared_teks_uploaded_on_generation_date_df.head()
estimated_shared_diagnoses_df = shared_teks_generation_to_upload_df \
.groupby(["upload_date"]).shared_teks.max().reset_index() \
.sort_values(["upload_date"], ascending=False) \
.rename(columns={
"upload_date": "sample_date_string",
"shared_teks": "shared_diagnoses",
})
invalid_shared_diagnoses_dates_mask = \
estimated_shared_diagnoses_df.sample_date_string.isin(invalid_shared_diagnoses_dates)
estimated_shared_diagnoses_df[invalid_shared_diagnoses_dates_mask] = 0
estimated_shared_diagnoses_df.head()
```
### Hourly New TEKs
```
hourly_extracted_teks_df = load_extracted_teks(
mode="Hourly", region=report_backend_identifier, limit=25)
hourly_extracted_teks_df.head()
hourly_new_tek_count_df = hourly_extracted_teks_df \
.groupby("extraction_date_with_hour").tek_list. \
apply(lambda x: set(sum(x, []))).reset_index().copy()
hourly_new_tek_count_df = hourly_new_tek_count_df.set_index("extraction_date_with_hour") \
.sort_index(ascending=True)
hourly_new_tek_count_df["new_tek_list"] = hourly_new_tek_count_df.tek_list.diff()
hourly_new_tek_count_df["new_tek_count"] = hourly_new_tek_count_df.new_tek_list.apply(
lambda x: len(x) if not pd.isna(x) else 0)
hourly_new_tek_count_df.rename(columns={
"new_tek_count": "shared_teks_by_upload_date"}, inplace=True)
hourly_new_tek_count_df = hourly_new_tek_count_df.reset_index()[[
"extraction_date_with_hour", "shared_teks_by_upload_date"]]
hourly_new_tek_count_df.head()
hourly_summary_df = hourly_new_tek_count_df.copy()
hourly_summary_df.set_index("extraction_date_with_hour", inplace=True)
hourly_summary_df = hourly_summary_df.fillna(0).astype(int).reset_index()
hourly_summary_df["datetime_utc"] = pd.to_datetime(
hourly_summary_df.extraction_date_with_hour, format="%Y-%m-%d@%H")
hourly_summary_df.set_index("datetime_utc", inplace=True)
hourly_summary_df = hourly_summary_df.tail(-1)
hourly_summary_df.head()
```
### Official Statistics
```
import requests
import pandas.io.json
official_stats_response = requests.get("https://radarcovid.covid19.gob.es/kpi/statistics/basics")
official_stats_response.raise_for_status()
official_stats_df_ = pandas.io.json.json_normalize(official_stats_response.json())
official_stats_df = official_stats_df_.copy()
official_stats_df["date"] = pd.to_datetime(official_stats_df["date"], dayfirst=True)
official_stats_df.head()
official_stats_column_map = {
"date": "sample_date",
"applicationsDownloads.totalAcummulated": "app_downloads_es_accumulated",
"communicatedContagions.totalAcummulated": "shared_diagnoses_es_accumulated",
}
accumulated_suffix = "_accumulated"
accumulated_values_columns = \
list(filter(lambda x: x.endswith(accumulated_suffix), official_stats_column_map.values()))
interpolated_values_columns = \
list(map(lambda x: x[:-len(accumulated_suffix)], accumulated_values_columns))
official_stats_df = \
official_stats_df[official_stats_column_map.keys()] \
.rename(columns=official_stats_column_map)
official_stats_df["extraction_date"] = extraction_date
official_stats_df.head()
official_stats_path = "Data/Statistics/Current/RadarCOVID-Statistics.json"
previous_official_stats_df = pd.read_json(official_stats_path, orient="records", lines=True)
previous_official_stats_df["sample_date"] = pd.to_datetime(previous_official_stats_df["sample_date"], dayfirst=True)
official_stats_df = official_stats_df.append(previous_official_stats_df)
official_stats_df.head()
official_stats_df = official_stats_df[~(official_stats_df.shared_diagnoses_es_accumulated == 0)]
official_stats_df.sort_values("extraction_date", ascending=False, inplace=True)
official_stats_df.drop_duplicates(subset=["sample_date"], keep="first", inplace=True)
official_stats_df.head()
official_stats_stored_df = official_stats_df.copy()
official_stats_stored_df["sample_date"] = official_stats_stored_df.sample_date.dt.strftime("%Y-%m-%d")
official_stats_stored_df.to_json(official_stats_path, orient="records", lines=True)
official_stats_df.drop(columns=["extraction_date"], inplace=True)
official_stats_df = confirmed_days_df.merge(official_stats_df, how="left")
official_stats_df.sort_values("sample_date", ascending=False, inplace=True)
official_stats_df.head()
official_stats_df[accumulated_values_columns] = \
official_stats_df[accumulated_values_columns] \
.astype(float).interpolate(limit_area="inside")
official_stats_df[interpolated_values_columns] = \
official_stats_df[accumulated_values_columns].diff(periods=-1)
official_stats_df.drop(columns="sample_date", inplace=True)
official_stats_df.head()
```
### Data Merge
```
result_summary_df = exposure_keys_summary_df.merge(
new_tek_df, on=["sample_date_string"], how="outer")
result_summary_df.head()
result_summary_df = result_summary_df.merge(
shared_teks_uploaded_on_generation_date_df, on=["sample_date_string"], how="outer")
result_summary_df.head()
result_summary_df = result_summary_df.merge(
estimated_shared_diagnoses_df, on=["sample_date_string"], how="outer")
result_summary_df.head()
result_summary_df = result_summary_df.merge(
official_stats_df, on=["sample_date_string"], how="outer")
result_summary_df.head()
result_summary_df = confirmed_eu_df.tail(daily_summary_days).merge(
result_summary_df, on=["sample_date_string"], how="left")
result_summary_df.head()
result_summary_df = confirmed_es_df.tail(daily_summary_days).merge(
result_summary_df, on=["sample_date_string"], how="left")
result_summary_df.head()
result_summary_df["sample_date"] = pd.to_datetime(result_summary_df.sample_date_string)
result_summary_df = result_summary_df.merge(source_regions_for_summary_df, how="left")
result_summary_df.set_index(["sample_date", "source_regions"], inplace=True)
result_summary_df.drop(columns=["sample_date_string"], inplace=True)
result_summary_df.sort_index(ascending=False, inplace=True)
result_summary_df.head()
with pd.option_context("mode.use_inf_as_na", True):
result_summary_df = result_summary_df.fillna(0).astype(int)
result_summary_df["teks_per_shared_diagnosis"] = \
(result_summary_df.shared_teks_by_upload_date / result_summary_df.shared_diagnoses).fillna(0)
result_summary_df["shared_diagnoses_per_covid_case"] = \
(result_summary_df.shared_diagnoses / result_summary_df.covid_cases).fillna(0)
result_summary_df["shared_diagnoses_per_covid_case_es"] = \
(result_summary_df.shared_diagnoses_es / result_summary_df.covid_cases_es).fillna(0)
result_summary_df.head(daily_plot_days)
def compute_aggregated_results_summary(days) -> pd.DataFrame:
aggregated_result_summary_df = result_summary_df.copy()
aggregated_result_summary_df["covid_cases_for_ratio"] = \
aggregated_result_summary_df.covid_cases.mask(
aggregated_result_summary_df.shared_diagnoses == 0, 0)
aggregated_result_summary_df["covid_cases_for_ratio_es"] = \
aggregated_result_summary_df.covid_cases_es.mask(
aggregated_result_summary_df.shared_diagnoses_es == 0, 0)
aggregated_result_summary_df = aggregated_result_summary_df \
.sort_index(ascending=True).fillna(0).rolling(days).agg({
"covid_cases": "sum",
"covid_cases_es": "sum",
"covid_cases_for_ratio": "sum",
"covid_cases_for_ratio_es": "sum",
"shared_teks_by_generation_date": "sum",
"shared_teks_by_upload_date": "sum",
"shared_diagnoses": "sum",
"shared_diagnoses_es": "sum",
}).sort_index(ascending=False)
with pd.option_context("mode.use_inf_as_na", True):
aggregated_result_summary_df = aggregated_result_summary_df.fillna(0).astype(int)
aggregated_result_summary_df["teks_per_shared_diagnosis"] = \
(aggregated_result_summary_df.shared_teks_by_upload_date /
aggregated_result_summary_df.covid_cases_for_ratio).fillna(0)
aggregated_result_summary_df["shared_diagnoses_per_covid_case"] = \
(aggregated_result_summary_df.shared_diagnoses /
aggregated_result_summary_df.covid_cases_for_ratio).fillna(0)
aggregated_result_summary_df["shared_diagnoses_per_covid_case_es"] = \
(aggregated_result_summary_df.shared_diagnoses_es /
aggregated_result_summary_df.covid_cases_for_ratio_es).fillna(0)
return aggregated_result_summary_df
aggregated_result_with_7_days_window_summary_df = compute_aggregated_results_summary(days=7)
aggregated_result_with_7_days_window_summary_df.head()
last_7_days_summary = aggregated_result_with_7_days_window_summary_df.to_dict(orient="records")[1]
last_7_days_summary
aggregated_result_with_14_days_window_summary_df = compute_aggregated_results_summary(days=13)
last_14_days_summary = aggregated_result_with_14_days_window_summary_df.to_dict(orient="records")[1]
last_14_days_summary
```
## Report Results
```
display_column_name_mapping = {
"sample_date": "Sample\u00A0Date\u00A0(UTC)",
"source_regions": "Source Countries",
"datetime_utc": "Timestamp (UTC)",
"upload_date": "Upload Date (UTC)",
"generation_to_upload_days": "Generation to Upload Period in Days",
"region": "Backend",
"region_x": "Backend\u00A0(A)",
"region_y": "Backend\u00A0(B)",
"common_teks": "Common TEKs Shared Between Backends",
"common_teks_fraction": "Fraction of TEKs in Backend (A) Available in Backend (B)",
"covid_cases": "COVID-19 Cases (Source Countries)",
"shared_teks_by_generation_date": "Shared TEKs by Generation Date (Source Countries)",
"shared_teks_by_upload_date": "Shared TEKs by Upload Date (Source Countries)",
"shared_teks_uploaded_on_generation_date": "Shared TEKs Uploaded on Generation Date (Source Countries)",
"shared_diagnoses": "Shared Diagnoses (Source Countries – Estimation)",
"teks_per_shared_diagnosis": "TEKs Uploaded per Shared Diagnosis (Source Countries)",
"shared_diagnoses_per_covid_case": "Usage Ratio (Source Countries)",
"covid_cases_es": "COVID-19 Cases (Spain)",
"app_downloads_es": "App Downloads (Spain – Official)",
"shared_diagnoses_es": "Shared Diagnoses (Spain – Official)",
"shared_diagnoses_per_covid_case_es": "Usage Ratio (Spain)",
}
summary_columns = [
"covid_cases",
"shared_teks_by_generation_date",
"shared_teks_by_upload_date",
"shared_teks_uploaded_on_generation_date",
"shared_diagnoses",
"teks_per_shared_diagnosis",
"shared_diagnoses_per_covid_case",
"covid_cases_es",
"app_downloads_es",
"shared_diagnoses_es",
"shared_diagnoses_per_covid_case_es",
]
summary_percentage_columns= [
"shared_diagnoses_per_covid_case_es",
"shared_diagnoses_per_covid_case",
]
```
### Daily Summary Table
```
result_summary_df_ = result_summary_df.copy()
result_summary_df = result_summary_df[summary_columns]
result_summary_with_display_names_df = result_summary_df \
.rename_axis(index=display_column_name_mapping) \
.rename(columns=display_column_name_mapping)
result_summary_with_display_names_df
```
### Daily Summary Plots
```
result_plot_summary_df = result_summary_df.head(daily_plot_days)[summary_columns] \
.droplevel(level=["source_regions"]) \
.rename_axis(index=display_column_name_mapping) \
.rename(columns=display_column_name_mapping)
summary_ax_list = result_plot_summary_df.sort_index(ascending=True).plot.bar(
title=f"Daily Summary",
rot=45, subplots=True, figsize=(15, 30), legend=False)
ax_ = summary_ax_list[0]
ax_.get_figure().tight_layout()
ax_.get_figure().subplots_adjust(top=0.95)
_ = ax_.set_xticklabels(sorted(result_plot_summary_df.index.strftime("%Y-%m-%d").tolist()))
for percentage_column in summary_percentage_columns:
percentage_column_index = summary_columns.index(percentage_column)
summary_ax_list[percentage_column_index].yaxis \
.set_major_formatter(matplotlib.ticker.PercentFormatter(1.0))
```
### Daily Generation to Upload Period Table
```
display_generation_to_upload_period_pivot_df = \
generation_to_upload_period_pivot_df \
.head(backend_generation_days)
display_generation_to_upload_period_pivot_df \
.head(backend_generation_days) \
.rename_axis(columns=display_column_name_mapping) \
.rename_axis(index=display_column_name_mapping)
fig, generation_to_upload_period_pivot_table_ax = plt.subplots(
figsize=(12, 1 + 0.6 * len(display_generation_to_upload_period_pivot_df)))
generation_to_upload_period_pivot_table_ax.set_title(
"Shared TEKs Generation to Upload Period Table")
sns.heatmap(
data=display_generation_to_upload_period_pivot_df
.rename_axis(columns=display_column_name_mapping)
.rename_axis(index=display_column_name_mapping),
fmt=".0f",
annot=True,
ax=generation_to_upload_period_pivot_table_ax)
generation_to_upload_period_pivot_table_ax.get_figure().tight_layout()
```
### Hourly Summary Plots
```
hourly_summary_ax_list = hourly_summary_df \
.rename_axis(index=display_column_name_mapping) \
.rename(columns=display_column_name_mapping) \
.plot.bar(
title=f"Last 24h Summary",
rot=45, subplots=True, legend=False)
ax_ = hourly_summary_ax_list[-1]
ax_.get_figure().tight_layout()
ax_.get_figure().subplots_adjust(top=0.9)
_ = ax_.set_xticklabels(sorted(hourly_summary_df.index.strftime("%Y-%m-%d@%H").tolist()))
```
### Publish Results
```
github_repository = os.environ.get("GITHUB_REPOSITORY")
if github_repository is None:
github_repository = "pvieito/Radar-STATS"
github_project_base_url = "https://github.com/" + github_repository
display_formatters = {
display_column_name_mapping["teks_per_shared_diagnosis"]: lambda x: f"{x:.2f}" if x != 0 else "",
display_column_name_mapping["shared_diagnoses_per_covid_case"]: lambda x: f"{x:.2%}" if x != 0 else "",
display_column_name_mapping["shared_diagnoses_per_covid_case_es"]: lambda x: f"{x:.2%}" if x != 0 else "",
}
general_columns = \
list(filter(lambda x: x not in display_formatters, display_column_name_mapping.values()))
general_formatter = lambda x: f"{x}" if x != 0 else ""
display_formatters.update(dict(map(lambda x: (x, general_formatter), general_columns)))
daily_summary_table_html = result_summary_with_display_names_df \
.head(daily_plot_days) \
.rename_axis(index=display_column_name_mapping) \
.rename(columns=display_column_name_mapping) \
.to_html(formatters=display_formatters)
multi_backend_summary_table_html = multi_backend_summary_df \
.head(daily_plot_days) \
.rename_axis(columns=display_column_name_mapping) \
.rename(columns=display_column_name_mapping) \
.rename_axis(index=display_column_name_mapping) \
.to_html(formatters=display_formatters)
def format_multi_backend_cross_sharing_fraction(x):
if pd.isna(x):
return "-"
elif round(x * 100, 1) == 0:
return ""
else:
return f"{x:.1%}"
multi_backend_cross_sharing_summary_table_html = multi_backend_cross_sharing_summary_df \
.rename_axis(columns=display_column_name_mapping) \
.rename(columns=display_column_name_mapping) \
.rename_axis(index=display_column_name_mapping) \
.to_html(
classes="table-center",
formatters=display_formatters,
float_format=format_multi_backend_cross_sharing_fraction)
multi_backend_cross_sharing_summary_table_html = \
multi_backend_cross_sharing_summary_table_html \
.replace("<tr>","<tr style=\"text-align: center;\">")
extraction_date_result_summary_df = \
result_summary_df[result_summary_df.index.get_level_values("sample_date") == extraction_date]
extraction_date_result_hourly_summary_df = \
hourly_summary_df[hourly_summary_df.extraction_date_with_hour == extraction_date_with_hour]
covid_cases = \
extraction_date_result_summary_df.covid_cases.item()
shared_teks_by_generation_date = \
extraction_date_result_summary_df.shared_teks_by_generation_date.item()
shared_teks_by_upload_date = \
extraction_date_result_summary_df.shared_teks_by_upload_date.item()
shared_diagnoses = \
extraction_date_result_summary_df.shared_diagnoses.item()
teks_per_shared_diagnosis = \
extraction_date_result_summary_df.teks_per_shared_diagnosis.item()
shared_diagnoses_per_covid_case = \
extraction_date_result_summary_df.shared_diagnoses_per_covid_case.item()
shared_teks_by_upload_date_last_hour = \
extraction_date_result_hourly_summary_df.shared_teks_by_upload_date.sum().astype(int)
display_source_regions = ", ".join(report_source_regions)
if len(report_source_regions) == 1:
display_brief_source_regions = report_source_regions[0]
else:
display_brief_source_regions = f"{len(report_source_regions)} 🇪🇺"
def get_temporary_image_path() -> str:
return os.path.join(tempfile.gettempdir(), str(uuid.uuid4()) + ".png")
def save_temporary_plot_image(ax):
if isinstance(ax, np.ndarray):
ax = ax[0]
media_path = get_temporary_image_path()
ax.get_figure().savefig(media_path)
return media_path
def save_temporary_dataframe_image(df):
import dataframe_image as dfi
df = df.copy()
df_styler = df.style.format(display_formatters)
media_path = get_temporary_image_path()
dfi.export(df_styler, media_path)
return media_path
summary_plots_image_path = save_temporary_plot_image(
ax=summary_ax_list)
summary_table_image_path = save_temporary_dataframe_image(
df=result_summary_with_display_names_df)
hourly_summary_plots_image_path = save_temporary_plot_image(
ax=hourly_summary_ax_list)
multi_backend_summary_table_image_path = save_temporary_dataframe_image(
df=multi_backend_summary_df)
generation_to_upload_period_pivot_table_image_path = save_temporary_plot_image(
ax=generation_to_upload_period_pivot_table_ax)
```
### Save Results
```
report_resources_path_prefix = "Data/Resources/Current/RadarCOVID-Report-"
result_summary_df.to_csv(
report_resources_path_prefix + "Summary-Table.csv")
result_summary_df.to_html(
report_resources_path_prefix + "Summary-Table.html")
hourly_summary_df.to_csv(
report_resources_path_prefix + "Hourly-Summary-Table.csv")
multi_backend_summary_df.to_csv(
report_resources_path_prefix + "Multi-Backend-Summary-Table.csv")
multi_backend_cross_sharing_summary_df.to_csv(
report_resources_path_prefix + "Multi-Backend-Cross-Sharing-Summary-Table.csv")
generation_to_upload_period_pivot_df.to_csv(
report_resources_path_prefix + "Generation-Upload-Period-Table.csv")
_ = shutil.copyfile(
summary_plots_image_path,
report_resources_path_prefix + "Summary-Plots.png")
_ = shutil.copyfile(
summary_table_image_path,
report_resources_path_prefix + "Summary-Table.png")
_ = shutil.copyfile(
hourly_summary_plots_image_path,
report_resources_path_prefix + "Hourly-Summary-Plots.png")
_ = shutil.copyfile(
multi_backend_summary_table_image_path,
report_resources_path_prefix + "Multi-Backend-Summary-Table.png")
_ = shutil.copyfile(
generation_to_upload_period_pivot_table_image_path,
report_resources_path_prefix + "Generation-Upload-Period-Table.png")
```
### Publish Results as JSON
```
def generate_summary_api_results(df: pd.DataFrame) -> list:
api_df = df.reset_index().copy()
api_df["sample_date_string"] = \
api_df["sample_date"].dt.strftime("%Y-%m-%d")
api_df["source_regions"] = \
api_df["source_regions"].apply(lambda x: x.split(","))
return api_df.to_dict(orient="records")
summary_api_results = \
generate_summary_api_results(df=result_summary_df)
today_summary_api_results = \
generate_summary_api_results(df=extraction_date_result_summary_df)[0]
summary_results = dict(
backend_identifier=report_backend_identifier,
source_regions=report_source_regions,
extraction_datetime=extraction_datetime,
extraction_date=extraction_date,
extraction_date_with_hour=extraction_date_with_hour,
last_hour=dict(
shared_teks_by_upload_date=shared_teks_by_upload_date_last_hour,
shared_diagnoses=0,
),
today=today_summary_api_results,
last_7_days=last_7_days_summary,
last_14_days=last_14_days_summary,
daily_results=summary_api_results)
summary_results = \
json.loads(pd.Series([summary_results]).to_json(orient="records"))[0]
with open(report_resources_path_prefix + "Summary-Results.json", "w") as f:
json.dump(summary_results, f, indent=4)
```
### Publish on README
```
with open("Data/Templates/README.md", "r") as f:
readme_contents = f.read()
readme_contents = readme_contents.format(
extraction_date_with_hour=extraction_date_with_hour,
github_project_base_url=github_project_base_url,
daily_summary_table_html=daily_summary_table_html,
multi_backend_summary_table_html=multi_backend_summary_table_html,
multi_backend_cross_sharing_summary_table_html=multi_backend_cross_sharing_summary_table_html,
display_source_regions=display_source_regions)
with open("README.md", "w") as f:
f.write(readme_contents)
```
### Publish on Twitter
```
enable_share_to_twitter = os.environ.get("RADARCOVID_REPORT__ENABLE_PUBLISH_ON_TWITTER")
github_event_name = os.environ.get("GITHUB_EVENT_NAME")
if enable_share_to_twitter and github_event_name == "schedule" and \
(shared_teks_by_upload_date_last_hour or not are_today_results_partial):
import tweepy
twitter_api_auth_keys = os.environ["RADARCOVID_REPORT__TWITTER_API_AUTH_KEYS"]
twitter_api_auth_keys = twitter_api_auth_keys.split(":")
auth = tweepy.OAuthHandler(twitter_api_auth_keys[0], twitter_api_auth_keys[1])
auth.set_access_token(twitter_api_auth_keys[2], twitter_api_auth_keys[3])
api = tweepy.API(auth)
summary_plots_media = api.media_upload(summary_plots_image_path)
summary_table_media = api.media_upload(summary_table_image_path)
generation_to_upload_period_pivot_table_image_media = api.media_upload(generation_to_upload_period_pivot_table_image_path)
media_ids = [
summary_plots_media.media_id,
summary_table_media.media_id,
generation_to_upload_period_pivot_table_image_media.media_id,
]
if are_today_results_partial:
today_addendum = " (Partial)"
else:
today_addendum = ""
def format_shared_diagnoses_per_covid_case(value) -> str:
if value == 0:
return "–"
return f"≤{value:.2%}"
display_shared_diagnoses_per_covid_case = \
format_shared_diagnoses_per_covid_case(value=shared_diagnoses_per_covid_case)
display_last_14_days_shared_diagnoses_per_covid_case = \
format_shared_diagnoses_per_covid_case(value=last_14_days_summary["shared_diagnoses_per_covid_case"])
display_last_14_days_shared_diagnoses_per_covid_case_es = \
format_shared_diagnoses_per_covid_case(value=last_14_days_summary["shared_diagnoses_per_covid_case_es"])
status = textwrap.dedent(f"""
#RadarCOVID – {extraction_date_with_hour}
Today{today_addendum}:
- Uploaded TEKs: {shared_teks_by_upload_date:.0f} ({shared_teks_by_upload_date_last_hour:+d} last hour)
- Shared Diagnoses: ≤{shared_diagnoses:.0f}
- Usage Ratio: {display_shared_diagnoses_per_covid_case}
Last 14 Days:
- Usage Ratio (Estimation): {display_last_14_days_shared_diagnoses_per_covid_case}
- Usage Ratio (Official): {display_last_14_days_shared_diagnoses_per_covid_case_es}
Info: {github_project_base_url}#documentation
""")
status = status.encode(encoding="utf-8")
api.update_status(status=status, media_ids=media_ids)
```
|
github_jupyter
|
import datetime
import json
import logging
import os
import shutil
import tempfile
import textwrap
import uuid
import matplotlib.pyplot as plt
import matplotlib.ticker
import numpy as np
import pandas as pd
import pycountry
import retry
import seaborn as sns
%matplotlib inline
current_working_directory = os.environ.get("PWD")
if current_working_directory:
os.chdir(current_working_directory)
sns.set()
matplotlib.rcParams["figure.figsize"] = (15, 6)
extraction_datetime = datetime.datetime.utcnow()
extraction_date = extraction_datetime.strftime("%Y-%m-%d")
extraction_previous_datetime = extraction_datetime - datetime.timedelta(days=1)
extraction_previous_date = extraction_previous_datetime.strftime("%Y-%m-%d")
extraction_date_with_hour = datetime.datetime.utcnow().strftime("%Y-%m-%d@%H")
current_hour = datetime.datetime.utcnow().hour
are_today_results_partial = current_hour != 23
from Modules.ExposureNotification import exposure_notification_io
spain_region_country_code = "ES"
germany_region_country_code = "DE"
default_backend_identifier = spain_region_country_code
backend_generation_days = 7 * 2
daily_summary_days = 7 * 4 * 3
daily_plot_days = 7 * 4
tek_dumps_load_limit = daily_summary_days + 1
environment_backend_identifier = os.environ.get("RADARCOVID_REPORT__BACKEND_IDENTIFIER")
if environment_backend_identifier:
report_backend_identifier = environment_backend_identifier
else:
report_backend_identifier = default_backend_identifier
report_backend_identifier
environment_enable_multi_backend_download = \
os.environ.get("RADARCOVID_REPORT__ENABLE_MULTI_BACKEND_DOWNLOAD")
if environment_enable_multi_backend_download:
report_backend_identifiers = None
else:
report_backend_identifiers = [report_backend_identifier]
report_backend_identifiers
environment_invalid_shared_diagnoses_dates = \
os.environ.get("RADARCOVID_REPORT__INVALID_SHARED_DIAGNOSES_DATES")
if environment_invalid_shared_diagnoses_dates:
invalid_shared_diagnoses_dates = environment_invalid_shared_diagnoses_dates.split(",")
else:
invalid_shared_diagnoses_dates = []
invalid_shared_diagnoses_dates
report_backend_client = \
exposure_notification_io.get_backend_client_with_identifier(
backend_identifier=report_backend_identifier)
@retry.retry(tries=10, delay=10, backoff=1.1, jitter=(0, 10))
def download_cases_dataframe():
return pd.read_csv("https://raw.githubusercontent.com/owid/covid-19-data/master/public/data/owid-covid-data.csv")
confirmed_df_ = download_cases_dataframe()
confirmed_df_.iloc[0]
confirmed_df = confirmed_df_.copy()
confirmed_df = confirmed_df[["date", "new_cases", "iso_code"]]
confirmed_df.rename(
columns={
"date": "sample_date",
"iso_code": "country_code",
},
inplace=True)
def convert_iso_alpha_3_to_alpha_2(x):
try:
return pycountry.countries.get(alpha_3=x).alpha_2
except Exception as e:
logging.info(f"Error converting country ISO Alpha 3 code '{x}': {repr(e)}")
return None
confirmed_df["country_code"] = confirmed_df.country_code.apply(convert_iso_alpha_3_to_alpha_2)
confirmed_df.dropna(inplace=True)
confirmed_df["sample_date"] = pd.to_datetime(confirmed_df.sample_date, dayfirst=True)
confirmed_df["sample_date"] = confirmed_df.sample_date.dt.strftime("%Y-%m-%d")
confirmed_df.sort_values("sample_date", inplace=True)
confirmed_df.tail()
confirmed_days = pd.date_range(
start=confirmed_df.iloc[0].sample_date,
end=extraction_datetime)
confirmed_days_df = pd.DataFrame(data=confirmed_days, columns=["sample_date"])
confirmed_days_df["sample_date_string"] = \
confirmed_days_df.sample_date.dt.strftime("%Y-%m-%d")
confirmed_days_df.tail()
def sort_source_regions_for_display(source_regions: list) -> list:
if report_backend_identifier in source_regions:
source_regions = [report_backend_identifier] + \
list(sorted(set(source_regions).difference([report_backend_identifier])))
else:
source_regions = list(sorted(source_regions))
return source_regions
report_source_regions = report_backend_client.source_regions_for_date(
date=extraction_datetime.date())
report_source_regions = sort_source_regions_for_display(
source_regions=report_source_regions)
report_source_regions
def get_cases_dataframe(source_regions_for_date_function, columns_suffix=None):
source_regions_at_date_df = confirmed_days_df.copy()
source_regions_at_date_df["source_regions_at_date"] = \
source_regions_at_date_df.sample_date.apply(
lambda x: source_regions_for_date_function(date=x))
source_regions_at_date_df.sort_values("sample_date", inplace=True)
source_regions_at_date_df["_source_regions_group"] = source_regions_at_date_df. \
source_regions_at_date.apply(lambda x: ",".join(sort_source_regions_for_display(x)))
source_regions_at_date_df.tail()
#%%
source_regions_for_summary_df_ = \
source_regions_at_date_df[["sample_date", "_source_regions_group"]].copy()
source_regions_for_summary_df_.rename(columns={"_source_regions_group": "source_regions"}, inplace=True)
source_regions_for_summary_df_.tail()
#%%
confirmed_output_columns = ["sample_date", "new_cases", "covid_cases"]
confirmed_output_df = pd.DataFrame(columns=confirmed_output_columns)
for source_regions_group, source_regions_group_series in \
source_regions_at_date_df.groupby("_source_regions_group"):
source_regions_set = set(source_regions_group.split(","))
confirmed_source_regions_set_df = \
confirmed_df[confirmed_df.country_code.isin(source_regions_set)].copy()
confirmed_source_regions_group_df = \
confirmed_source_regions_set_df.groupby("sample_date").new_cases.sum() \
.reset_index().sort_values("sample_date")
confirmed_source_regions_group_df = \
confirmed_source_regions_group_df.merge(
confirmed_days_df[["sample_date_string"]].rename(
columns={"sample_date_string": "sample_date"}),
how="right")
confirmed_source_regions_group_df["new_cases"] = \
confirmed_source_regions_group_df["new_cases"].clip(lower=0)
confirmed_source_regions_group_df["covid_cases"] = \
confirmed_source_regions_group_df.new_cases.rolling(7, min_periods=0).mean().round()
confirmed_source_regions_group_df = \
confirmed_source_regions_group_df[confirmed_output_columns]
confirmed_source_regions_group_df = confirmed_source_regions_group_df.replace(0, np.nan)
confirmed_source_regions_group_df.fillna(method="ffill", inplace=True)
confirmed_source_regions_group_df = \
confirmed_source_regions_group_df[
confirmed_source_regions_group_df.sample_date.isin(
source_regions_group_series.sample_date_string)]
confirmed_output_df = confirmed_output_df.append(confirmed_source_regions_group_df)
result_df = confirmed_output_df.copy()
result_df.tail()
#%%
result_df.rename(columns={"sample_date": "sample_date_string"}, inplace=True)
result_df = confirmed_days_df[["sample_date_string"]].merge(result_df, how="left")
result_df.sort_values("sample_date_string", inplace=True)
result_df.fillna(method="ffill", inplace=True)
result_df.tail()
#%%
result_df[["new_cases", "covid_cases"]].plot()
if columns_suffix:
result_df.rename(
columns={
"new_cases": "new_cases_" + columns_suffix,
"covid_cases": "covid_cases_" + columns_suffix},
inplace=True)
return result_df, source_regions_for_summary_df_
confirmed_eu_df, source_regions_for_summary_df = get_cases_dataframe(
report_backend_client.source_regions_for_date)
confirmed_es_df, _ = get_cases_dataframe(
lambda date: [spain_region_country_code],
columns_suffix=spain_region_country_code.lower())
raw_zip_path_prefix = "Data/TEKs/Raw/"
base_backend_identifiers = [report_backend_identifier]
multi_backend_exposure_keys_df = \
exposure_notification_io.download_exposure_keys_from_backends(
backend_identifiers=report_backend_identifiers,
generation_days=backend_generation_days,
fail_on_error_backend_identifiers=base_backend_identifiers,
save_raw_zip_path_prefix=raw_zip_path_prefix)
multi_backend_exposure_keys_df["region"] = multi_backend_exposure_keys_df["backend_identifier"]
multi_backend_exposure_keys_df.rename(
columns={
"generation_datetime": "sample_datetime",
"generation_date_string": "sample_date_string",
},
inplace=True)
multi_backend_exposure_keys_df.head()
early_teks_df = multi_backend_exposure_keys_df[
multi_backend_exposure_keys_df.rolling_period < 144].copy()
early_teks_df["rolling_period_in_hours"] = early_teks_df.rolling_period / 6
early_teks_df[early_teks_df.sample_date_string != extraction_date] \
.rolling_period_in_hours.hist(bins=list(range(24)))
early_teks_df[early_teks_df.sample_date_string == extraction_date] \
.rolling_period_in_hours.hist(bins=list(range(24)))
multi_backend_exposure_keys_df = multi_backend_exposure_keys_df[[
"sample_date_string", "region", "key_data"]]
multi_backend_exposure_keys_df.head()
active_regions = \
multi_backend_exposure_keys_df.groupby("region").key_data.nunique().sort_values().index.unique().tolist()
active_regions
multi_backend_summary_df = multi_backend_exposure_keys_df.groupby(
["sample_date_string", "region"]).key_data.nunique().reset_index() \
.pivot(index="sample_date_string", columns="region") \
.sort_index(ascending=False)
multi_backend_summary_df.rename(
columns={"key_data": "shared_teks_by_generation_date"},
inplace=True)
multi_backend_summary_df.rename_axis("sample_date", inplace=True)
multi_backend_summary_df = multi_backend_summary_df.fillna(0).astype(int)
multi_backend_summary_df = multi_backend_summary_df.head(backend_generation_days)
multi_backend_summary_df.head()
def compute_keys_cross_sharing(x):
teks_x = x.key_data_x.item()
common_teks = set(teks_x).intersection(x.key_data_y.item())
common_teks_fraction = len(common_teks) / len(teks_x)
return pd.Series(dict(
common_teks=common_teks,
common_teks_fraction=common_teks_fraction,
))
multi_backend_exposure_keys_by_region_df = \
multi_backend_exposure_keys_df.groupby("region").key_data.unique().reset_index()
multi_backend_exposure_keys_by_region_df["_merge"] = True
multi_backend_exposure_keys_by_region_combination_df = \
multi_backend_exposure_keys_by_region_df.merge(
multi_backend_exposure_keys_by_region_df, on="_merge")
multi_backend_exposure_keys_by_region_combination_df.drop(
columns=["_merge"], inplace=True)
if multi_backend_exposure_keys_by_region_combination_df.region_x.nunique() > 1:
multi_backend_exposure_keys_by_region_combination_df = \
multi_backend_exposure_keys_by_region_combination_df[
multi_backend_exposure_keys_by_region_combination_df.region_x !=
multi_backend_exposure_keys_by_region_combination_df.region_y]
multi_backend_exposure_keys_cross_sharing_df = \
multi_backend_exposure_keys_by_region_combination_df \
.groupby(["region_x", "region_y"]) \
.apply(compute_keys_cross_sharing) \
.reset_index()
multi_backend_cross_sharing_summary_df = \
multi_backend_exposure_keys_cross_sharing_df.pivot_table(
values=["common_teks_fraction"],
columns="region_x",
index="region_y",
aggfunc=lambda x: x.item())
multi_backend_cross_sharing_summary_df
multi_backend_without_active_region_exposure_keys_df = \
multi_backend_exposure_keys_df[multi_backend_exposure_keys_df.region != report_backend_identifier]
multi_backend_without_active_region = \
multi_backend_without_active_region_exposure_keys_df.groupby("region").key_data.nunique().sort_values().index.unique().tolist()
multi_backend_without_active_region
exposure_keys_summary_df = multi_backend_exposure_keys_df[
multi_backend_exposure_keys_df.region == report_backend_identifier]
exposure_keys_summary_df.drop(columns=["region"], inplace=True)
exposure_keys_summary_df = \
exposure_keys_summary_df.groupby(["sample_date_string"]).key_data.nunique().to_frame()
exposure_keys_summary_df = \
exposure_keys_summary_df.reset_index().set_index("sample_date_string")
exposure_keys_summary_df.sort_index(ascending=False, inplace=True)
exposure_keys_summary_df.rename(columns={"key_data": "shared_teks_by_generation_date"}, inplace=True)
exposure_keys_summary_df.head()
tek_list_df = multi_backend_exposure_keys_df[
["sample_date_string", "region", "key_data"]].copy()
tek_list_df["key_data"] = tek_list_df["key_data"].apply(str)
tek_list_df.rename(columns={
"sample_date_string": "sample_date",
"key_data": "tek_list"}, inplace=True)
tek_list_df = tek_list_df.groupby(
["sample_date", "region"]).tek_list.unique().reset_index()
tek_list_df["extraction_date"] = extraction_date
tek_list_df["extraction_date_with_hour"] = extraction_date_with_hour
tek_list_path_prefix = "Data/TEKs/"
tek_list_current_path = tek_list_path_prefix + f"/Current/RadarCOVID-TEKs.json"
tek_list_daily_path = tek_list_path_prefix + f"Daily/RadarCOVID-TEKs-{extraction_date}.json"
tek_list_hourly_path = tek_list_path_prefix + f"Hourly/RadarCOVID-TEKs-{extraction_date_with_hour}.json"
for path in [tek_list_current_path, tek_list_daily_path, tek_list_hourly_path]:
os.makedirs(os.path.dirname(path), exist_ok=True)
tek_list_base_df = tek_list_df[tek_list_df.region == report_backend_identifier]
tek_list_base_df.drop(columns=["extraction_date", "extraction_date_with_hour"]).to_json(
tek_list_current_path,
lines=True, orient="records")
tek_list_base_df.drop(columns=["extraction_date_with_hour"]).to_json(
tek_list_daily_path,
lines=True, orient="records")
tek_list_base_df.to_json(
tek_list_hourly_path,
lines=True, orient="records")
tek_list_base_df.head()
import glob
def load_extracted_teks(mode, region=None, limit=None) -> pd.DataFrame:
extracted_teks_df = pd.DataFrame(columns=["region"])
file_paths = list(reversed(sorted(glob.glob(tek_list_path_prefix + mode + "/RadarCOVID-TEKs-*.json"))))
if limit:
file_paths = file_paths[:limit]
for file_path in file_paths:
logging.info(f"Loading TEKs from '{file_path}'...")
iteration_extracted_teks_df = pd.read_json(file_path, lines=True)
extracted_teks_df = extracted_teks_df.append(
iteration_extracted_teks_df, sort=False)
extracted_teks_df["region"] = \
extracted_teks_df.region.fillna(spain_region_country_code).copy()
if region:
extracted_teks_df = \
extracted_teks_df[extracted_teks_df.region == region]
return extracted_teks_df
daily_extracted_teks_df = load_extracted_teks(
mode="Daily",
region=report_backend_identifier,
limit=tek_dumps_load_limit)
daily_extracted_teks_df.head()
exposure_keys_summary_df_ = daily_extracted_teks_df \
.sort_values("extraction_date", ascending=False) \
.groupby("sample_date").tek_list.first() \
.to_frame()
exposure_keys_summary_df_.index.name = "sample_date_string"
exposure_keys_summary_df_["tek_list"] = \
exposure_keys_summary_df_.tek_list.apply(len)
exposure_keys_summary_df_ = exposure_keys_summary_df_ \
.rename(columns={"tek_list": "shared_teks_by_generation_date"}) \
.sort_index(ascending=False)
exposure_keys_summary_df = exposure_keys_summary_df_
exposure_keys_summary_df.head()
tek_list_df = daily_extracted_teks_df.groupby("extraction_date").tek_list.apply(
lambda x: set(sum(x, []))).reset_index()
tek_list_df = tek_list_df.set_index("extraction_date").sort_index(ascending=True)
tek_list_df.head()
def compute_teks_by_generation_and_upload_date(date):
day_new_teks_set_df = tek_list_df.copy().diff()
try:
day_new_teks_set = day_new_teks_set_df[
day_new_teks_set_df.index == date].tek_list.item()
except ValueError:
day_new_teks_set = None
if pd.isna(day_new_teks_set):
day_new_teks_set = set()
day_new_teks_df = daily_extracted_teks_df[
daily_extracted_teks_df.extraction_date == date].copy()
day_new_teks_df["shared_teks"] = \
day_new_teks_df.tek_list.apply(lambda x: set(x).intersection(day_new_teks_set))
day_new_teks_df["shared_teks"] = \
day_new_teks_df.shared_teks.apply(len)
day_new_teks_df["upload_date"] = date
day_new_teks_df.rename(columns={"sample_date": "generation_date"}, inplace=True)
day_new_teks_df = day_new_teks_df[
["upload_date", "generation_date", "shared_teks"]]
day_new_teks_df["generation_to_upload_days"] = \
(pd.to_datetime(day_new_teks_df.upload_date) -
pd.to_datetime(day_new_teks_df.generation_date)).dt.days
day_new_teks_df = day_new_teks_df[day_new_teks_df.shared_teks > 0]
return day_new_teks_df
shared_teks_generation_to_upload_df = pd.DataFrame()
for upload_date in daily_extracted_teks_df.extraction_date.unique():
shared_teks_generation_to_upload_df = \
shared_teks_generation_to_upload_df.append(
compute_teks_by_generation_and_upload_date(date=upload_date))
shared_teks_generation_to_upload_df \
.sort_values(["upload_date", "generation_date"], ascending=False, inplace=True)
shared_teks_generation_to_upload_df.tail()
today_new_teks_df = \
shared_teks_generation_to_upload_df[
shared_teks_generation_to_upload_df.upload_date == extraction_date].copy()
today_new_teks_df.tail()
if not today_new_teks_df.empty:
today_new_teks_df.set_index("generation_to_upload_days") \
.sort_index().shared_teks.plot.bar()
generation_to_upload_period_pivot_df = \
shared_teks_generation_to_upload_df[
["upload_date", "generation_to_upload_days", "shared_teks"]] \
.pivot(index="upload_date", columns="generation_to_upload_days") \
.sort_index(ascending=False).fillna(0).astype(int) \
.droplevel(level=0, axis=1)
generation_to_upload_period_pivot_df.head()
new_tek_df = tek_list_df.diff().tek_list.apply(
lambda x: len(x) if not pd.isna(x) else None).to_frame().reset_index()
new_tek_df.rename(columns={
"tek_list": "shared_teks_by_upload_date",
"extraction_date": "sample_date_string",}, inplace=True)
new_tek_df.tail()
shared_teks_uploaded_on_generation_date_df = shared_teks_generation_to_upload_df[
shared_teks_generation_to_upload_df.generation_to_upload_days == 0] \
[["upload_date", "shared_teks"]].rename(
columns={
"upload_date": "sample_date_string",
"shared_teks": "shared_teks_uploaded_on_generation_date",
})
shared_teks_uploaded_on_generation_date_df.head()
estimated_shared_diagnoses_df = shared_teks_generation_to_upload_df \
.groupby(["upload_date"]).shared_teks.max().reset_index() \
.sort_values(["upload_date"], ascending=False) \
.rename(columns={
"upload_date": "sample_date_string",
"shared_teks": "shared_diagnoses",
})
invalid_shared_diagnoses_dates_mask = \
estimated_shared_diagnoses_df.sample_date_string.isin(invalid_shared_diagnoses_dates)
estimated_shared_diagnoses_df[invalid_shared_diagnoses_dates_mask] = 0
estimated_shared_diagnoses_df.head()
hourly_extracted_teks_df = load_extracted_teks(
mode="Hourly", region=report_backend_identifier, limit=25)
hourly_extracted_teks_df.head()
hourly_new_tek_count_df = hourly_extracted_teks_df \
.groupby("extraction_date_with_hour").tek_list. \
apply(lambda x: set(sum(x, []))).reset_index().copy()
hourly_new_tek_count_df = hourly_new_tek_count_df.set_index("extraction_date_with_hour") \
.sort_index(ascending=True)
hourly_new_tek_count_df["new_tek_list"] = hourly_new_tek_count_df.tek_list.diff()
hourly_new_tek_count_df["new_tek_count"] = hourly_new_tek_count_df.new_tek_list.apply(
lambda x: len(x) if not pd.isna(x) else 0)
hourly_new_tek_count_df.rename(columns={
"new_tek_count": "shared_teks_by_upload_date"}, inplace=True)
hourly_new_tek_count_df = hourly_new_tek_count_df.reset_index()[[
"extraction_date_with_hour", "shared_teks_by_upload_date"]]
hourly_new_tek_count_df.head()
hourly_summary_df = hourly_new_tek_count_df.copy()
hourly_summary_df.set_index("extraction_date_with_hour", inplace=True)
hourly_summary_df = hourly_summary_df.fillna(0).astype(int).reset_index()
hourly_summary_df["datetime_utc"] = pd.to_datetime(
hourly_summary_df.extraction_date_with_hour, format="%Y-%m-%d@%H")
hourly_summary_df.set_index("datetime_utc", inplace=True)
hourly_summary_df = hourly_summary_df.tail(-1)
hourly_summary_df.head()
import requests
import pandas.io.json
official_stats_response = requests.get("https://radarcovid.covid19.gob.es/kpi/statistics/basics")
official_stats_response.raise_for_status()
official_stats_df_ = pandas.io.json.json_normalize(official_stats_response.json())
official_stats_df = official_stats_df_.copy()
official_stats_df["date"] = pd.to_datetime(official_stats_df["date"], dayfirst=True)
official_stats_df.head()
official_stats_column_map = {
"date": "sample_date",
"applicationsDownloads.totalAcummulated": "app_downloads_es_accumulated",
"communicatedContagions.totalAcummulated": "shared_diagnoses_es_accumulated",
}
accumulated_suffix = "_accumulated"
accumulated_values_columns = \
list(filter(lambda x: x.endswith(accumulated_suffix), official_stats_column_map.values()))
interpolated_values_columns = \
list(map(lambda x: x[:-len(accumulated_suffix)], accumulated_values_columns))
official_stats_df = \
official_stats_df[official_stats_column_map.keys()] \
.rename(columns=official_stats_column_map)
official_stats_df["extraction_date"] = extraction_date
official_stats_df.head()
official_stats_path = "Data/Statistics/Current/RadarCOVID-Statistics.json"
previous_official_stats_df = pd.read_json(official_stats_path, orient="records", lines=True)
previous_official_stats_df["sample_date"] = pd.to_datetime(previous_official_stats_df["sample_date"], dayfirst=True)
official_stats_df = official_stats_df.append(previous_official_stats_df)
official_stats_df.head()
official_stats_df = official_stats_df[~(official_stats_df.shared_diagnoses_es_accumulated == 0)]
official_stats_df.sort_values("extraction_date", ascending=False, inplace=True)
official_stats_df.drop_duplicates(subset=["sample_date"], keep="first", inplace=True)
official_stats_df.head()
official_stats_stored_df = official_stats_df.copy()
official_stats_stored_df["sample_date"] = official_stats_stored_df.sample_date.dt.strftime("%Y-%m-%d")
official_stats_stored_df.to_json(official_stats_path, orient="records", lines=True)
official_stats_df.drop(columns=["extraction_date"], inplace=True)
official_stats_df = confirmed_days_df.merge(official_stats_df, how="left")
official_stats_df.sort_values("sample_date", ascending=False, inplace=True)
official_stats_df.head()
official_stats_df[accumulated_values_columns] = \
official_stats_df[accumulated_values_columns] \
.astype(float).interpolate(limit_area="inside")
official_stats_df[interpolated_values_columns] = \
official_stats_df[accumulated_values_columns].diff(periods=-1)
official_stats_df.drop(columns="sample_date", inplace=True)
official_stats_df.head()
result_summary_df = exposure_keys_summary_df.merge(
new_tek_df, on=["sample_date_string"], how="outer")
result_summary_df.head()
result_summary_df = result_summary_df.merge(
shared_teks_uploaded_on_generation_date_df, on=["sample_date_string"], how="outer")
result_summary_df.head()
result_summary_df = result_summary_df.merge(
estimated_shared_diagnoses_df, on=["sample_date_string"], how="outer")
result_summary_df.head()
result_summary_df = result_summary_df.merge(
official_stats_df, on=["sample_date_string"], how="outer")
result_summary_df.head()
result_summary_df = confirmed_eu_df.tail(daily_summary_days).merge(
result_summary_df, on=["sample_date_string"], how="left")
result_summary_df.head()
result_summary_df = confirmed_es_df.tail(daily_summary_days).merge(
result_summary_df, on=["sample_date_string"], how="left")
result_summary_df.head()
result_summary_df["sample_date"] = pd.to_datetime(result_summary_df.sample_date_string)
result_summary_df = result_summary_df.merge(source_regions_for_summary_df, how="left")
result_summary_df.set_index(["sample_date", "source_regions"], inplace=True)
result_summary_df.drop(columns=["sample_date_string"], inplace=True)
result_summary_df.sort_index(ascending=False, inplace=True)
result_summary_df.head()
with pd.option_context("mode.use_inf_as_na", True):
result_summary_df = result_summary_df.fillna(0).astype(int)
result_summary_df["teks_per_shared_diagnosis"] = \
(result_summary_df.shared_teks_by_upload_date / result_summary_df.shared_diagnoses).fillna(0)
result_summary_df["shared_diagnoses_per_covid_case"] = \
(result_summary_df.shared_diagnoses / result_summary_df.covid_cases).fillna(0)
result_summary_df["shared_diagnoses_per_covid_case_es"] = \
(result_summary_df.shared_diagnoses_es / result_summary_df.covid_cases_es).fillna(0)
result_summary_df.head(daily_plot_days)
def compute_aggregated_results_summary(days) -> pd.DataFrame:
aggregated_result_summary_df = result_summary_df.copy()
aggregated_result_summary_df["covid_cases_for_ratio"] = \
aggregated_result_summary_df.covid_cases.mask(
aggregated_result_summary_df.shared_diagnoses == 0, 0)
aggregated_result_summary_df["covid_cases_for_ratio_es"] = \
aggregated_result_summary_df.covid_cases_es.mask(
aggregated_result_summary_df.shared_diagnoses_es == 0, 0)
aggregated_result_summary_df = aggregated_result_summary_df \
.sort_index(ascending=True).fillna(0).rolling(days).agg({
"covid_cases": "sum",
"covid_cases_es": "sum",
"covid_cases_for_ratio": "sum",
"covid_cases_for_ratio_es": "sum",
"shared_teks_by_generation_date": "sum",
"shared_teks_by_upload_date": "sum",
"shared_diagnoses": "sum",
"shared_diagnoses_es": "sum",
}).sort_index(ascending=False)
with pd.option_context("mode.use_inf_as_na", True):
aggregated_result_summary_df = aggregated_result_summary_df.fillna(0).astype(int)
aggregated_result_summary_df["teks_per_shared_diagnosis"] = \
(aggregated_result_summary_df.shared_teks_by_upload_date /
aggregated_result_summary_df.covid_cases_for_ratio).fillna(0)
aggregated_result_summary_df["shared_diagnoses_per_covid_case"] = \
(aggregated_result_summary_df.shared_diagnoses /
aggregated_result_summary_df.covid_cases_for_ratio).fillna(0)
aggregated_result_summary_df["shared_diagnoses_per_covid_case_es"] = \
(aggregated_result_summary_df.shared_diagnoses_es /
aggregated_result_summary_df.covid_cases_for_ratio_es).fillna(0)
return aggregated_result_summary_df
aggregated_result_with_7_days_window_summary_df = compute_aggregated_results_summary(days=7)
aggregated_result_with_7_days_window_summary_df.head()
last_7_days_summary = aggregated_result_with_7_days_window_summary_df.to_dict(orient="records")[1]
last_7_days_summary
aggregated_result_with_14_days_window_summary_df = compute_aggregated_results_summary(days=13)
last_14_days_summary = aggregated_result_with_14_days_window_summary_df.to_dict(orient="records")[1]
last_14_days_summary
display_column_name_mapping = {
"sample_date": "Sample\u00A0Date\u00A0(UTC)",
"source_regions": "Source Countries",
"datetime_utc": "Timestamp (UTC)",
"upload_date": "Upload Date (UTC)",
"generation_to_upload_days": "Generation to Upload Period in Days",
"region": "Backend",
"region_x": "Backend\u00A0(A)",
"region_y": "Backend\u00A0(B)",
"common_teks": "Common TEKs Shared Between Backends",
"common_teks_fraction": "Fraction of TEKs in Backend (A) Available in Backend (B)",
"covid_cases": "COVID-19 Cases (Source Countries)",
"shared_teks_by_generation_date": "Shared TEKs by Generation Date (Source Countries)",
"shared_teks_by_upload_date": "Shared TEKs by Upload Date (Source Countries)",
"shared_teks_uploaded_on_generation_date": "Shared TEKs Uploaded on Generation Date (Source Countries)",
"shared_diagnoses": "Shared Diagnoses (Source Countries – Estimation)",
"teks_per_shared_diagnosis": "TEKs Uploaded per Shared Diagnosis (Source Countries)",
"shared_diagnoses_per_covid_case": "Usage Ratio (Source Countries)",
"covid_cases_es": "COVID-19 Cases (Spain)",
"app_downloads_es": "App Downloads (Spain – Official)",
"shared_diagnoses_es": "Shared Diagnoses (Spain – Official)",
"shared_diagnoses_per_covid_case_es": "Usage Ratio (Spain)",
}
summary_columns = [
"covid_cases",
"shared_teks_by_generation_date",
"shared_teks_by_upload_date",
"shared_teks_uploaded_on_generation_date",
"shared_diagnoses",
"teks_per_shared_diagnosis",
"shared_diagnoses_per_covid_case",
"covid_cases_es",
"app_downloads_es",
"shared_diagnoses_es",
"shared_diagnoses_per_covid_case_es",
]
summary_percentage_columns= [
"shared_diagnoses_per_covid_case_es",
"shared_diagnoses_per_covid_case",
]
result_summary_df_ = result_summary_df.copy()
result_summary_df = result_summary_df[summary_columns]
result_summary_with_display_names_df = result_summary_df \
.rename_axis(index=display_column_name_mapping) \
.rename(columns=display_column_name_mapping)
result_summary_with_display_names_df
result_plot_summary_df = result_summary_df.head(daily_plot_days)[summary_columns] \
.droplevel(level=["source_regions"]) \
.rename_axis(index=display_column_name_mapping) \
.rename(columns=display_column_name_mapping)
summary_ax_list = result_plot_summary_df.sort_index(ascending=True).plot.bar(
title=f"Daily Summary",
rot=45, subplots=True, figsize=(15, 30), legend=False)
ax_ = summary_ax_list[0]
ax_.get_figure().tight_layout()
ax_.get_figure().subplots_adjust(top=0.95)
_ = ax_.set_xticklabels(sorted(result_plot_summary_df.index.strftime("%Y-%m-%d").tolist()))
for percentage_column in summary_percentage_columns:
percentage_column_index = summary_columns.index(percentage_column)
summary_ax_list[percentage_column_index].yaxis \
.set_major_formatter(matplotlib.ticker.PercentFormatter(1.0))
display_generation_to_upload_period_pivot_df = \
generation_to_upload_period_pivot_df \
.head(backend_generation_days)
display_generation_to_upload_period_pivot_df \
.head(backend_generation_days) \
.rename_axis(columns=display_column_name_mapping) \
.rename_axis(index=display_column_name_mapping)
fig, generation_to_upload_period_pivot_table_ax = plt.subplots(
figsize=(12, 1 + 0.6 * len(display_generation_to_upload_period_pivot_df)))
generation_to_upload_period_pivot_table_ax.set_title(
"Shared TEKs Generation to Upload Period Table")
sns.heatmap(
data=display_generation_to_upload_period_pivot_df
.rename_axis(columns=display_column_name_mapping)
.rename_axis(index=display_column_name_mapping),
fmt=".0f",
annot=True,
ax=generation_to_upload_period_pivot_table_ax)
generation_to_upload_period_pivot_table_ax.get_figure().tight_layout()
hourly_summary_ax_list = hourly_summary_df \
.rename_axis(index=display_column_name_mapping) \
.rename(columns=display_column_name_mapping) \
.plot.bar(
title=f"Last 24h Summary",
rot=45, subplots=True, legend=False)
ax_ = hourly_summary_ax_list[-1]
ax_.get_figure().tight_layout()
ax_.get_figure().subplots_adjust(top=0.9)
_ = ax_.set_xticklabels(sorted(hourly_summary_df.index.strftime("%Y-%m-%d@%H").tolist()))
github_repository = os.environ.get("GITHUB_REPOSITORY")
if github_repository is None:
github_repository = "pvieito/Radar-STATS"
github_project_base_url = "https://github.com/" + github_repository
display_formatters = {
display_column_name_mapping["teks_per_shared_diagnosis"]: lambda x: f"{x:.2f}" if x != 0 else "",
display_column_name_mapping["shared_diagnoses_per_covid_case"]: lambda x: f"{x:.2%}" if x != 0 else "",
display_column_name_mapping["shared_diagnoses_per_covid_case_es"]: lambda x: f"{x:.2%}" if x != 0 else "",
}
general_columns = \
list(filter(lambda x: x not in display_formatters, display_column_name_mapping.values()))
general_formatter = lambda x: f"{x}" if x != 0 else ""
display_formatters.update(dict(map(lambda x: (x, general_formatter), general_columns)))
daily_summary_table_html = result_summary_with_display_names_df \
.head(daily_plot_days) \
.rename_axis(index=display_column_name_mapping) \
.rename(columns=display_column_name_mapping) \
.to_html(formatters=display_formatters)
multi_backend_summary_table_html = multi_backend_summary_df \
.head(daily_plot_days) \
.rename_axis(columns=display_column_name_mapping) \
.rename(columns=display_column_name_mapping) \
.rename_axis(index=display_column_name_mapping) \
.to_html(formatters=display_formatters)
def format_multi_backend_cross_sharing_fraction(x):
if pd.isna(x):
return "-"
elif round(x * 100, 1) == 0:
return ""
else:
return f"{x:.1%}"
multi_backend_cross_sharing_summary_table_html = multi_backend_cross_sharing_summary_df \
.rename_axis(columns=display_column_name_mapping) \
.rename(columns=display_column_name_mapping) \
.rename_axis(index=display_column_name_mapping) \
.to_html(
classes="table-center",
formatters=display_formatters,
float_format=format_multi_backend_cross_sharing_fraction)
multi_backend_cross_sharing_summary_table_html = \
multi_backend_cross_sharing_summary_table_html \
.replace("<tr>","<tr style=\"text-align: center;\">")
extraction_date_result_summary_df = \
result_summary_df[result_summary_df.index.get_level_values("sample_date") == extraction_date]
extraction_date_result_hourly_summary_df = \
hourly_summary_df[hourly_summary_df.extraction_date_with_hour == extraction_date_with_hour]
covid_cases = \
extraction_date_result_summary_df.covid_cases.item()
shared_teks_by_generation_date = \
extraction_date_result_summary_df.shared_teks_by_generation_date.item()
shared_teks_by_upload_date = \
extraction_date_result_summary_df.shared_teks_by_upload_date.item()
shared_diagnoses = \
extraction_date_result_summary_df.shared_diagnoses.item()
teks_per_shared_diagnosis = \
extraction_date_result_summary_df.teks_per_shared_diagnosis.item()
shared_diagnoses_per_covid_case = \
extraction_date_result_summary_df.shared_diagnoses_per_covid_case.item()
shared_teks_by_upload_date_last_hour = \
extraction_date_result_hourly_summary_df.shared_teks_by_upload_date.sum().astype(int)
display_source_regions = ", ".join(report_source_regions)
if len(report_source_regions) == 1:
display_brief_source_regions = report_source_regions[0]
else:
display_brief_source_regions = f"{len(report_source_regions)} 🇪🇺"
def get_temporary_image_path() -> str:
return os.path.join(tempfile.gettempdir(), str(uuid.uuid4()) + ".png")
def save_temporary_plot_image(ax):
if isinstance(ax, np.ndarray):
ax = ax[0]
media_path = get_temporary_image_path()
ax.get_figure().savefig(media_path)
return media_path
def save_temporary_dataframe_image(df):
import dataframe_image as dfi
df = df.copy()
df_styler = df.style.format(display_formatters)
media_path = get_temporary_image_path()
dfi.export(df_styler, media_path)
return media_path
summary_plots_image_path = save_temporary_plot_image(
ax=summary_ax_list)
summary_table_image_path = save_temporary_dataframe_image(
df=result_summary_with_display_names_df)
hourly_summary_plots_image_path = save_temporary_plot_image(
ax=hourly_summary_ax_list)
multi_backend_summary_table_image_path = save_temporary_dataframe_image(
df=multi_backend_summary_df)
generation_to_upload_period_pivot_table_image_path = save_temporary_plot_image(
ax=generation_to_upload_period_pivot_table_ax)
report_resources_path_prefix = "Data/Resources/Current/RadarCOVID-Report-"
result_summary_df.to_csv(
report_resources_path_prefix + "Summary-Table.csv")
result_summary_df.to_html(
report_resources_path_prefix + "Summary-Table.html")
hourly_summary_df.to_csv(
report_resources_path_prefix + "Hourly-Summary-Table.csv")
multi_backend_summary_df.to_csv(
report_resources_path_prefix + "Multi-Backend-Summary-Table.csv")
multi_backend_cross_sharing_summary_df.to_csv(
report_resources_path_prefix + "Multi-Backend-Cross-Sharing-Summary-Table.csv")
generation_to_upload_period_pivot_df.to_csv(
report_resources_path_prefix + "Generation-Upload-Period-Table.csv")
_ = shutil.copyfile(
summary_plots_image_path,
report_resources_path_prefix + "Summary-Plots.png")
_ = shutil.copyfile(
summary_table_image_path,
report_resources_path_prefix + "Summary-Table.png")
_ = shutil.copyfile(
hourly_summary_plots_image_path,
report_resources_path_prefix + "Hourly-Summary-Plots.png")
_ = shutil.copyfile(
multi_backend_summary_table_image_path,
report_resources_path_prefix + "Multi-Backend-Summary-Table.png")
_ = shutil.copyfile(
generation_to_upload_period_pivot_table_image_path,
report_resources_path_prefix + "Generation-Upload-Period-Table.png")
def generate_summary_api_results(df: pd.DataFrame) -> list:
api_df = df.reset_index().copy()
api_df["sample_date_string"] = \
api_df["sample_date"].dt.strftime("%Y-%m-%d")
api_df["source_regions"] = \
api_df["source_regions"].apply(lambda x: x.split(","))
return api_df.to_dict(orient="records")
summary_api_results = \
generate_summary_api_results(df=result_summary_df)
today_summary_api_results = \
generate_summary_api_results(df=extraction_date_result_summary_df)[0]
summary_results = dict(
backend_identifier=report_backend_identifier,
source_regions=report_source_regions,
extraction_datetime=extraction_datetime,
extraction_date=extraction_date,
extraction_date_with_hour=extraction_date_with_hour,
last_hour=dict(
shared_teks_by_upload_date=shared_teks_by_upload_date_last_hour,
shared_diagnoses=0,
),
today=today_summary_api_results,
last_7_days=last_7_days_summary,
last_14_days=last_14_days_summary,
daily_results=summary_api_results)
summary_results = \
json.loads(pd.Series([summary_results]).to_json(orient="records"))[0]
with open(report_resources_path_prefix + "Summary-Results.json", "w") as f:
json.dump(summary_results, f, indent=4)
with open("Data/Templates/README.md", "r") as f:
readme_contents = f.read()
readme_contents = readme_contents.format(
extraction_date_with_hour=extraction_date_with_hour,
github_project_base_url=github_project_base_url,
daily_summary_table_html=daily_summary_table_html,
multi_backend_summary_table_html=multi_backend_summary_table_html,
multi_backend_cross_sharing_summary_table_html=multi_backend_cross_sharing_summary_table_html,
display_source_regions=display_source_regions)
with open("README.md", "w") as f:
f.write(readme_contents)
enable_share_to_twitter = os.environ.get("RADARCOVID_REPORT__ENABLE_PUBLISH_ON_TWITTER")
github_event_name = os.environ.get("GITHUB_EVENT_NAME")
if enable_share_to_twitter and github_event_name == "schedule" and \
(shared_teks_by_upload_date_last_hour or not are_today_results_partial):
import tweepy
twitter_api_auth_keys = os.environ["RADARCOVID_REPORT__TWITTER_API_AUTH_KEYS"]
twitter_api_auth_keys = twitter_api_auth_keys.split(":")
auth = tweepy.OAuthHandler(twitter_api_auth_keys[0], twitter_api_auth_keys[1])
auth.set_access_token(twitter_api_auth_keys[2], twitter_api_auth_keys[3])
api = tweepy.API(auth)
summary_plots_media = api.media_upload(summary_plots_image_path)
summary_table_media = api.media_upload(summary_table_image_path)
generation_to_upload_period_pivot_table_image_media = api.media_upload(generation_to_upload_period_pivot_table_image_path)
media_ids = [
summary_plots_media.media_id,
summary_table_media.media_id,
generation_to_upload_period_pivot_table_image_media.media_id,
]
if are_today_results_partial:
today_addendum = " (Partial)"
else:
today_addendum = ""
def format_shared_diagnoses_per_covid_case(value) -> str:
if value == 0:
return "–"
return f"≤{value:.2%}"
display_shared_diagnoses_per_covid_case = \
format_shared_diagnoses_per_covid_case(value=shared_diagnoses_per_covid_case)
display_last_14_days_shared_diagnoses_per_covid_case = \
format_shared_diagnoses_per_covid_case(value=last_14_days_summary["shared_diagnoses_per_covid_case"])
display_last_14_days_shared_diagnoses_per_covid_case_es = \
format_shared_diagnoses_per_covid_case(value=last_14_days_summary["shared_diagnoses_per_covid_case_es"])
status = textwrap.dedent(f"""
#RadarCOVID – {extraction_date_with_hour}
Today{today_addendum}:
- Uploaded TEKs: {shared_teks_by_upload_date:.0f} ({shared_teks_by_upload_date_last_hour:+d} last hour)
- Shared Diagnoses: ≤{shared_diagnoses:.0f}
- Usage Ratio: {display_shared_diagnoses_per_covid_case}
Last 14 Days:
- Usage Ratio (Estimation): {display_last_14_days_shared_diagnoses_per_covid_case}
- Usage Ratio (Official): {display_last_14_days_shared_diagnoses_per_covid_case_es}
Info: {github_project_base_url}#documentation
""")
status = status.encode(encoding="utf-8")
api.update_status(status=status, media_ids=media_ids)
| 0.268749 | 0.215464 |
# XOR
_Aside: Should do [Working efficiently with jupyter lab](https://florianwilhelm.info/2018/11/working_efficiently_with_jupyter_lab/)_
```
%load_ext autoreload
%autoreload 2
#%matplotlib widget
%matplotlib inline
import numpy as np
import matplotlib.pyplot as plt
```
Fetch our tools:
```
from lib.nn import Network, Layer, IdentityLayer, AffineLayer, MapLayer
from lib.nnbench import NNBench
from lib.nnvis import NNVis
```
___
## Construct a network to learn exclusive-or
We make a two-input, one-output network, using
a 2x2 affine followed by tanh activation
feeding a 2x1 affine followed by tanh activation:
```
net = Network()
net.extend(AffineLayer(2,2))
net.extend(MapLayer(np.tanh, lambda d: 1.0 - np.tanh(d)**2))
net.extend(AffineLayer(2,1))
net.extend(MapLayer(np.tanh, lambda d: 1.0 - np.tanh(d)**2))
```
Make a test bench and a visualizer:
```
bench = NNBench(net)
vis = NNVis(bench)
```
Prepare fixed training data for the learning process. Set it to be every training batch.
```
training_batch = (np.array([[-0.5, -0.5],
[-0.5, 0.5],
[ 0.5, 0.5],
[ 0.5, -0.5]]),
np.array([[-0.5],
[ 0.5],
[-0.5],
[ 0.5]]))
bench.training_batch = lambda n: training_batch
```
We can extract the learnable parameters from a net. They can be modified and injected back if we wish.
```
bench.training_batch(4)
net.state_vector()
```
Set the state to an ordinary example starting point, for consistent notebook behavior below. We also make it the checkpoint in the bench.
```
net.set_state_from_vector(np.array([-0.88681521, -1.28596788, 0.3248974 , -2.33838503, 0.34761944,
-0.94541789, 1.99448043, 0.38704839, -3.8844268 ]))
bench.checkpoint_net()
```
How does this untrained net work across the canonical input domain?
```
domain = bench.training_batch(4)[0]
domain, net(domain)
```
Not the ideal answer, which is:
```
bench.training_batch(4)[1]
```
# Learning
This net doesn't know how to do `xor` yet. Let's try teaching it. \
We can plot the loss as a function of learning steps, for the current learning rate $\eta$:
```
vis.plot_learning(100, batch_size=4)
```
The net has changed state from that learning:
```
bench.net.state_vector()
```
How well does it work now?
```
net(domain)
```
Good. We can learn some more, carrying on from where we left off:
```
vis.plot_learning(100)
```
The net is again changed:
```
bench.net.state_vector()
```
Check its behavior now:
```
net(domain)
```
We can get to floating-point-$\epsilon$ perfect with more training. Let's add 800 more, for a total of a thousand learning cycles:
```
_ = bench.learn(1200)
net(domain)
```
It's well-trained, at least for the canonical inputs. What was the resultant net?
```
bench.net.state_vector()
```
## Visualizing the learning process
Let's look at the loss as a function of learning steps and learning rate $\eta$. Get back to our starting point network.
```
bench.rollback_net()
```
Move the sliders to adjust the number of learning steps, and the learning rate:
```
vis.knobs_plot_learning(100)
vis.bench.net.state_vector()
```
How well does it work after this training?
```
net(domain)
```
## Viewing the loss surface
We can plot the loss surface with `plotly`:
```
bench.rollback_net() if True else bench.randomize_net()
rates = np.logspace(-6, 0, base=2, num=32)
cube = bench.learn_loss_cube(1000, rates)
vis.plot_loss_cube()
```
## The track that learning takes
Let us examine the trajectory in state space during learning, and the loss function.
Each learning iteration changes the net state. We can examine those deltas.
Questions:
1. Are there regimes of direction-of-change (DoC) in state space, or does the DoC wander chaotically?
1. What are the spectral characteristics of the DoC? Length characteristics?
1. How do the DoC characteristics relate to the loss function, and it's first difference?
1. How do these trajectories vary with learning rate? Are there clues in these to adapt the learning rate?
1. How do the trajectory characteristics vary across different starting nets?
1. How do these measures vary with the objective function of the learning process, that is, what you're trying to teach the net?
1. How do the different layers with learning state evolve? Do they settle at different times? How does an upstream layer change, as a consequence of learning, affect downstream layers? Down affect up?
```
bench.rollback_net()
bench.net.eta = 1
learned_track = bench.learn_track(2000)
traja = bench.analyze_learning_track(learned_track)
vis.plot_trajectory(traja)
```
---
# Scratch
```
assert False, "Stop notebook execution here if entering from above"
bench.randomize_net()
t = bench.learn(1000)
bench.net.state_vector()
net = bench.net
net([[-.5, -.5], [-.5, .5]])
net.layers
from nnbench import Thing
t = Thing(color='brown', weight=7)
t.cow = 'moo'
t.cow
t.color
```
---
```
# Boneyard
assert False, "Stop notebook execution, the rest is scrap"
```
Wrangle the state-space trajectory and the losses into form.
```
trajectory = np.vstack([v[0] for v in lt])
losses = np.vstack([v[1] for v in lt])
```
Take first differences, which represent the changes at each step
```
traj_steps = np.diff(trajectory, axis=0)
loss_steps = np.diff(losses, axis=0)
traj_steps[:5]
```
Find the L2 norm of the trajectory steps $\lVert traj \rVert$:
```
traj_L2 = np.sqrt(np.einsum('...i,...i', traj_steps, traj_steps))
len(traj_L2), traj_L2[:5], traj_L2[-5:]
```
Find the angles between trajectory steps, from
$$\mathbf {a} \cdot \mathbf {b} = \left\|\mathbf {a} \right\|\left\|\mathbf {b} \right\|\cos \theta \\
\cos \theta = \frac{\mathbf {a} \cdot \mathbf {b}}{\left\|\mathbf {a} \right\|\left\|\mathbf {b} \right\|} \\
$$
where $\mathbf {a}$ and $\mathbf {b}$ are a state-space trajectory step and the succeeding step respectively
Find $\mathbf {a} \cdot \mathbf {b}$:
```
trajn_dot_nplus1 = np.einsum('...i,...i', traj_steps[:-1], traj_steps[1:])
trajn_dot_nplus1[:5], np.any(trajn_dot_nplus1 < 0)
```
Find $\left\|\mathbf {a} \right\|\left\|\mathbf {b} \right\|$:
```
traj_cos_denom = np.multiply(traj_L2[:-1], traj_L2[1:])
```
This will be the divisor. Some entries may be zero, so we adapt
```
len(traj_L2) - np.count_nonzero(traj_L2)
np.equal(traj_L2, 0)
```
Find $\cos \theta$ by dividing, excluding division by zero:
```
traj_cos = np.divide(trajn_dot_nplus1, traj_cos_denom, where=traj_cos_denom!=0.0)
traj_cos[:5], traj_cos[-5:], min(traj_cos), max(traj_cos)
#traj_theta = np.arccos(traj_cos)
#traj_theta[:5], traj_theta[-5:]
net = Network()
net.extend(AffineLayer(2,2))
#leak = 0
#net.extend(MapLayer(lambda x: (x*(1+leak/2)+abs(x)*(1-leak/2))/2, lambda d: [leak,1][1 if d>0 else 0]))
#net.extend(MapLayer(lambda x: max(0, np.sign(x)) * x, lambda d: max(0, np.sign(d))))
net.extend(MapLayer(np.tanh, lambda d: 1.0 - np.tanh(d)**2))
net.extend(AffineLayer(2,1))
net.extend(MapLayer(np.tanh, lambda d: 1.0 - np.tanh(d)**2))
#sigmoid = lambda x: 1/(np.exp(x)+1)
#net.extend(MapLayer(sigmoid, lambda d: sigmoid(d)*(1-sigmoid(d))))
#net.extend(MapLayer(lambda x: max(0, np.sign(x)) * x, lambda d: max(0, np.sign(d))))
dat = \
[(np.array([-1,-1]), np.array([-1])),
(np.array([-1,1]), np.array([1])),
(np.array([1,1]), np.array([-1])),
(np.array([1,-1]), np.array([1]))]
dc = 0
amp= 1
temp = [(d[0]*amp/2+dc,d[1]*amp/2+dc) for d in dat]
bench.training_data = ((np.array([v[0] for v in temp]),
np.array([v[1] for v in temp])),)
bench.training_data
```
|
github_jupyter
|
%load_ext autoreload
%autoreload 2
#%matplotlib widget
%matplotlib inline
import numpy as np
import matplotlib.pyplot as plt
from lib.nn import Network, Layer, IdentityLayer, AffineLayer, MapLayer
from lib.nnbench import NNBench
from lib.nnvis import NNVis
net = Network()
net.extend(AffineLayer(2,2))
net.extend(MapLayer(np.tanh, lambda d: 1.0 - np.tanh(d)**2))
net.extend(AffineLayer(2,1))
net.extend(MapLayer(np.tanh, lambda d: 1.0 - np.tanh(d)**2))
bench = NNBench(net)
vis = NNVis(bench)
training_batch = (np.array([[-0.5, -0.5],
[-0.5, 0.5],
[ 0.5, 0.5],
[ 0.5, -0.5]]),
np.array([[-0.5],
[ 0.5],
[-0.5],
[ 0.5]]))
bench.training_batch = lambda n: training_batch
bench.training_batch(4)
net.state_vector()
net.set_state_from_vector(np.array([-0.88681521, -1.28596788, 0.3248974 , -2.33838503, 0.34761944,
-0.94541789, 1.99448043, 0.38704839, -3.8844268 ]))
bench.checkpoint_net()
domain = bench.training_batch(4)[0]
domain, net(domain)
bench.training_batch(4)[1]
vis.plot_learning(100, batch_size=4)
bench.net.state_vector()
net(domain)
vis.plot_learning(100)
bench.net.state_vector()
net(domain)
_ = bench.learn(1200)
net(domain)
bench.net.state_vector()
bench.rollback_net()
vis.knobs_plot_learning(100)
vis.bench.net.state_vector()
net(domain)
bench.rollback_net() if True else bench.randomize_net()
rates = np.logspace(-6, 0, base=2, num=32)
cube = bench.learn_loss_cube(1000, rates)
vis.plot_loss_cube()
bench.rollback_net()
bench.net.eta = 1
learned_track = bench.learn_track(2000)
traja = bench.analyze_learning_track(learned_track)
vis.plot_trajectory(traja)
assert False, "Stop notebook execution here if entering from above"
bench.randomize_net()
t = bench.learn(1000)
bench.net.state_vector()
net = bench.net
net([[-.5, -.5], [-.5, .5]])
net.layers
from nnbench import Thing
t = Thing(color='brown', weight=7)
t.cow = 'moo'
t.cow
t.color
# Boneyard
assert False, "Stop notebook execution, the rest is scrap"
trajectory = np.vstack([v[0] for v in lt])
losses = np.vstack([v[1] for v in lt])
traj_steps = np.diff(trajectory, axis=0)
loss_steps = np.diff(losses, axis=0)
traj_steps[:5]
traj_L2 = np.sqrt(np.einsum('...i,...i', traj_steps, traj_steps))
len(traj_L2), traj_L2[:5], traj_L2[-5:]
trajn_dot_nplus1 = np.einsum('...i,...i', traj_steps[:-1], traj_steps[1:])
trajn_dot_nplus1[:5], np.any(trajn_dot_nplus1 < 0)
traj_cos_denom = np.multiply(traj_L2[:-1], traj_L2[1:])
len(traj_L2) - np.count_nonzero(traj_L2)
np.equal(traj_L2, 0)
traj_cos = np.divide(trajn_dot_nplus1, traj_cos_denom, where=traj_cos_denom!=0.0)
traj_cos[:5], traj_cos[-5:], min(traj_cos), max(traj_cos)
#traj_theta = np.arccos(traj_cos)
#traj_theta[:5], traj_theta[-5:]
net = Network()
net.extend(AffineLayer(2,2))
#leak = 0
#net.extend(MapLayer(lambda x: (x*(1+leak/2)+abs(x)*(1-leak/2))/2, lambda d: [leak,1][1 if d>0 else 0]))
#net.extend(MapLayer(lambda x: max(0, np.sign(x)) * x, lambda d: max(0, np.sign(d))))
net.extend(MapLayer(np.tanh, lambda d: 1.0 - np.tanh(d)**2))
net.extend(AffineLayer(2,1))
net.extend(MapLayer(np.tanh, lambda d: 1.0 - np.tanh(d)**2))
#sigmoid = lambda x: 1/(np.exp(x)+1)
#net.extend(MapLayer(sigmoid, lambda d: sigmoid(d)*(1-sigmoid(d))))
#net.extend(MapLayer(lambda x: max(0, np.sign(x)) * x, lambda d: max(0, np.sign(d))))
dat = \
[(np.array([-1,-1]), np.array([-1])),
(np.array([-1,1]), np.array([1])),
(np.array([1,1]), np.array([-1])),
(np.array([1,-1]), np.array([1]))]
dc = 0
amp= 1
temp = [(d[0]*amp/2+dc,d[1]*amp/2+dc) for d in dat]
bench.training_data = ((np.array([v[0] for v in temp]),
np.array([v[1] for v in temp])),)
bench.training_data
| 0.432782 | 0.947769 |
```
import pandas
from sklearn import linear_model, feature_extraction
import matplotlib.pyplot as plt
import numpy as np
from sklearn.preprocessing import PolynomialFeatures
from sklearn.pipeline import make_pipeline
def categorical_features(row):
d = {}
d["STATE"] = row[1]["STATE"]
return d
def last_poll(full_data):
"""
Create feature from last poll in each state
"""
# Only care about republicans
repub = full_data[full_data["PARTY"] == "Rep"]
#repub1 = repub[repub["PARTY"] != "Dem"]
#repub2 = repub1[repub1["PARTY"] != "NaN"]
#repub3 = full_data[full_data["CHOICE"] != "Undecided"]
#repub = full_data[full_data["MOE"] != 5]
# Sort by date
chron = repub.sort_values(by="DATE", ascending=True)
# Only keep the last one
dedupe = chron.drop_duplicates(subset="STATE", keep="last")
return dedupe[dedupe["STATE"]!= "US"]
if __name__ == "__main__":
# Read in the X data
all_data = pandas.read_csv("data.csv")
# Remove non-states
all_data = all_data[pandas.notnull(all_data["STATE"])]
# split between testing and training
train_x = last_poll(all_data[all_data["TOPIC"] == '2012-president'])
train_x.set_index("STATE")
test_x = last_poll(all_data[all_data["TOPIC"] == '2016-president'])
test_x.set_index("STATE")
# Read in the Y data
y_data = pandas.read_csv("../data/2012_pres.csv", sep=';')
y_data = y_data[y_data["PARTY"] == "R"]
y_data = y_data[pandas.notnull(y_data["GENERAL %"])]
y_data["GENERAL %"] = [float(x.replace(",", ".").replace("%", ""))
for x in y_data["GENERAL %"]]
y_data["STATE"] = y_data["STATE ABBREVIATION"]
y_data.set_index("STATE")
backup = train_x
train_x = y_data.merge(train_x, on="STATE",how='left')
# make sure we have all states in the test data
for ii in set(y_data.STATE) - set(test_x.STATE):
new_row = pandas.DataFrame([{"STATE": ii}])
test_x = test_x.append(new_row)
# format the data for regression
train_x = pandas.concat([train_x.STATE.astype(str).str.get_dummies(),
train_x], axis=1)
test_x = pandas.concat([test_x.STATE.astype(str).str.get_dummies(),
test_x], axis=1)
# handle missing data
for dd in train_x, test_x:
dd["NOPOLL"] = pandas.isnull(dd["VALUE"])
dd["VALUE"] = dd["VALUE"].fillna(0.0)
dd["NOMOE"] = pandas.isnull(dd["MOE"])
dd["MOE"] = dd["MOE"].fillna(0.0)
# create feature list
features = list(y_data.STATE)
features.append("VALUE")
features.append("NOPOLL")
features.append("MOE")
features.append("NOMOE")
features_par = list(y_data.PARTY)
features_par = [ord(i) for i in features_par]
features_obs = list(all_data.OBS)
features_obs = [0 if math.isnan(x) else x for x in features_obs]
features_val = list(all_data.VALUE)
features_moe = list(all_data.MOE)
features_moe = [0 if math.isnan(x) else x for x in features_moe]
features_matrix = []
for i in range(len(features_par)):
features_matrix.append([features_par[i], features_obs[i], features_val[i], features_moe[i]])
features_matrix = np.array(features_matrix)
# fit the regression
#mod = linear_model.Ridge()
#mod = linear_model.LinearRegression()
#mod = linear_model.BayesianRidge()
#mod.fit(train_x[features], train_x["GENERAL %"])
#mod = make_pipeline(PolynomialFeatures(degree = 2), linear_model.Ridge())
#mod.fit(features_matrix, train_x["GENERAL %"])
#Write out the model
'''with open("model.txt", 'w') as out:
#out.write("BIAS\t%f\n" % mod.intercept_)
for jj, kk in zip(features, mod.coef_):
out.write("%s\t%f\n" % (jj, kk))'''
# Write the predictions
'''pred_test = mod.predict(test_x[features])
with open("pred.txt", 'w') as out:
for ss, vv in sorted(zip(list(test_x.STATE), pred_test)):
out.write("%s\t%f\n" % (ss, vv))'''
pred_test = mod.predict(features_matrix)
with open("pred.txt", 'w') as out:
for ss, vv in sorted(zip(list(test_x.STATE), pred_test)):
out.write("%s\t%f\n" % (ss, vv))
y_data
print("Mean squared error: %.2f"
% np.mean((mod.predict(features_matrix) - train_x["GENERAL %"])**2))
print("Variance score: %.2f" % mod.score(features_matrix, train_x["GENERAL %"]))
```
|
github_jupyter
|
import pandas
from sklearn import linear_model, feature_extraction
import matplotlib.pyplot as plt
import numpy as np
from sklearn.preprocessing import PolynomialFeatures
from sklearn.pipeline import make_pipeline
def categorical_features(row):
d = {}
d["STATE"] = row[1]["STATE"]
return d
def last_poll(full_data):
"""
Create feature from last poll in each state
"""
# Only care about republicans
repub = full_data[full_data["PARTY"] == "Rep"]
#repub1 = repub[repub["PARTY"] != "Dem"]
#repub2 = repub1[repub1["PARTY"] != "NaN"]
#repub3 = full_data[full_data["CHOICE"] != "Undecided"]
#repub = full_data[full_data["MOE"] != 5]
# Sort by date
chron = repub.sort_values(by="DATE", ascending=True)
# Only keep the last one
dedupe = chron.drop_duplicates(subset="STATE", keep="last")
return dedupe[dedupe["STATE"]!= "US"]
if __name__ == "__main__":
# Read in the X data
all_data = pandas.read_csv("data.csv")
# Remove non-states
all_data = all_data[pandas.notnull(all_data["STATE"])]
# split between testing and training
train_x = last_poll(all_data[all_data["TOPIC"] == '2012-president'])
train_x.set_index("STATE")
test_x = last_poll(all_data[all_data["TOPIC"] == '2016-president'])
test_x.set_index("STATE")
# Read in the Y data
y_data = pandas.read_csv("../data/2012_pres.csv", sep=';')
y_data = y_data[y_data["PARTY"] == "R"]
y_data = y_data[pandas.notnull(y_data["GENERAL %"])]
y_data["GENERAL %"] = [float(x.replace(",", ".").replace("%", ""))
for x in y_data["GENERAL %"]]
y_data["STATE"] = y_data["STATE ABBREVIATION"]
y_data.set_index("STATE")
backup = train_x
train_x = y_data.merge(train_x, on="STATE",how='left')
# make sure we have all states in the test data
for ii in set(y_data.STATE) - set(test_x.STATE):
new_row = pandas.DataFrame([{"STATE": ii}])
test_x = test_x.append(new_row)
# format the data for regression
train_x = pandas.concat([train_x.STATE.astype(str).str.get_dummies(),
train_x], axis=1)
test_x = pandas.concat([test_x.STATE.astype(str).str.get_dummies(),
test_x], axis=1)
# handle missing data
for dd in train_x, test_x:
dd["NOPOLL"] = pandas.isnull(dd["VALUE"])
dd["VALUE"] = dd["VALUE"].fillna(0.0)
dd["NOMOE"] = pandas.isnull(dd["MOE"])
dd["MOE"] = dd["MOE"].fillna(0.0)
# create feature list
features = list(y_data.STATE)
features.append("VALUE")
features.append("NOPOLL")
features.append("MOE")
features.append("NOMOE")
features_par = list(y_data.PARTY)
features_par = [ord(i) for i in features_par]
features_obs = list(all_data.OBS)
features_obs = [0 if math.isnan(x) else x for x in features_obs]
features_val = list(all_data.VALUE)
features_moe = list(all_data.MOE)
features_moe = [0 if math.isnan(x) else x for x in features_moe]
features_matrix = []
for i in range(len(features_par)):
features_matrix.append([features_par[i], features_obs[i], features_val[i], features_moe[i]])
features_matrix = np.array(features_matrix)
# fit the regression
#mod = linear_model.Ridge()
#mod = linear_model.LinearRegression()
#mod = linear_model.BayesianRidge()
#mod.fit(train_x[features], train_x["GENERAL %"])
#mod = make_pipeline(PolynomialFeatures(degree = 2), linear_model.Ridge())
#mod.fit(features_matrix, train_x["GENERAL %"])
#Write out the model
'''with open("model.txt", 'w') as out:
#out.write("BIAS\t%f\n" % mod.intercept_)
for jj, kk in zip(features, mod.coef_):
out.write("%s\t%f\n" % (jj, kk))'''
# Write the predictions
'''pred_test = mod.predict(test_x[features])
with open("pred.txt", 'w') as out:
for ss, vv in sorted(zip(list(test_x.STATE), pred_test)):
out.write("%s\t%f\n" % (ss, vv))'''
pred_test = mod.predict(features_matrix)
with open("pred.txt", 'w') as out:
for ss, vv in sorted(zip(list(test_x.STATE), pred_test)):
out.write("%s\t%f\n" % (ss, vv))
y_data
print("Mean squared error: %.2f"
% np.mean((mod.predict(features_matrix) - train_x["GENERAL %"])**2))
print("Variance score: %.2f" % mod.score(features_matrix, train_x["GENERAL %"]))
| 0.393851 | 0.381709 |
# 100 numpy exercises with hint
This is a collection of exercises that have been collected in the numpy mailing list, on stack overflow and in the numpy documentation. The goal of this collection is to offer a quick reference for both old and new users but also to provide a set of exercises for those who teach.
If you find an error or think you've a better way to solve some of them, feel free to open an issue at <https://github.com/rougier/numpy-100>
#### 1. Import the numpy package under the name `np` (★☆☆)
(**hint**: import … as …)
#### 2. Print the numpy version and the configuration (★☆☆)
(**hint**: np.\_\_version\_\_, np.show\_config)
#### 3. Create a null vector of size 10 (★☆☆)
(**hint**: np.zeros)
#### 4. How to find the memory size of any array (★☆☆)
(**hint**: size, itemsize)
#### 5. How to get the documentation of the numpy add function from the command line? (★☆☆)
(**hint**: np.info)
#### 6. Create a null vector of size 10 but the fifth value which is 1 (★☆☆)
(**hint**: array\[4\])
#### 7. Create a vector with values ranging from 10 to 49 (★☆☆)
(**hint**: np.arange)
#### 8. Reverse a vector (first element becomes last) (★☆☆)
(**hint**: array\[::-1\])
#### 9. Create a 3x3 matrix with values ranging from 0 to 8 (★☆☆)
(**hint**: reshape)
#### 10. Find indices of non-zero elements from \[1,2,0,0,4,0\] (★☆☆)
(**hint**: np.nonzero)
#### 11. Create a 3x3 identity matrix (★☆☆)
(**hint**: np.eye)
#### 12. Create a 3x3x3 array with random values (★☆☆)
(**hint**: np.random.random)
#### 13. Create a 10x10 array with random values and find the minimum and maximum values (★☆☆)
(**hint**: min, max)
#### 14. Create a random vector of size 30 and find the mean value (★☆☆)
(**hint**: mean)
#### 15. Create a 2d array with 1 on the border and 0 inside (★☆☆)
(**hint**: array\[1:-1, 1:-1\])
#### 16. How to add a border (filled with 0's) around an existing array? (★☆☆)
(**hint**: np.pad)
#### 17. What is the result of the following expression? (★☆☆)
(**hint**: NaN = not a number, inf = infinity)
```python
0 * np.nan
np.nan == np.nan
np.inf > np.nan
np.nan - np.nan
0.3 == 3 * 0.1
```
#### 18. Create a 5x5 matrix with values 1,2,3,4 just below the diagonal (★☆☆)
(**hint**: np.diag)
#### 19. Create a 8x8 matrix and fill it with a checkerboard pattern (★☆☆)
(**hint**: array\[::2\])
#### 20. Consider a (6,7,8) shape array, what is the index (x,y,z) of the 100th element?
(**hint**: np.unravel_index)
#### 21. Create a checkerboard 8x8 matrix using the tile function (★☆☆)
(**hint**: np.tile)
#### 22. Normalize a 5x5 random matrix (★☆☆)
(**hint**: (x - min) / (max - min))
#### 23. Create a custom dtype that describes a color as four unsigned bytes (RGBA) (★☆☆)
(**hint**: np.dtype)
#### 24. Multiply a 5x3 matrix by a 3x2 matrix (real matrix product) (★☆☆)
(**hint**: np.dot | @)
#### 25. Given a 1D array, negate all elements which are between 3 and 8, in place. (★☆☆)
(**hint**: >, <=)
#### 26. What is the output of the following script? (★☆☆)
(**hint**: np.sum)
```python
# Author: Jake VanderPlas
print(sum(range(5),-1))
from numpy import *
print(sum(range(5),-1))
```
#### 27. Consider an integer vector Z, which of these expressions are legal? (★☆☆)
```python
Z**Z
2 << Z >> 2
Z <- Z
1j*Z
Z/1/1
Z<Z>Z
```
#### 28. What are the result of the following expressions?
```python
np.array(0) / np.array(0)
np.array(0) // np.array(0)
np.array([np.nan]).astype(int).astype(float)
```
#### 29. How to round away from zero a float array ? (★☆☆)
(**hint**: np.uniform, np.copysign, np.ceil, np.abs)
#### 30. How to find common values between two arrays? (★☆☆)
(**hint**: np.intersect1d)
#### 31. How to ignore all numpy warnings (not recommended)? (★☆☆)
(**hint**: np.seterr, np.errstate)
#### 32. Is the following expressions true? (★☆☆)
(**hint**: imaginary number)
```python
np.sqrt(-1) == np.emath.sqrt(-1)
```
#### 33. How to get the dates of yesterday, today and tomorrow? (★☆☆)
(**hint**: np.datetime64, np.timedelta64)
#### 34. How to get all the dates corresponding to the month of July 2016? (★★☆)
(**hint**: np.arange(dtype=datetime64\['D'\]))
#### 35. How to compute ((A+B)\*(-A/2)) in place (without copy)? (★★☆)
(**hint**: np.add(out=), np.negative(out=), np.multiply(out=), np.divide(out=))
#### 36. Extract the integer part of a random array using 5 different methods (★★☆)
(**hint**: %, np.floor, np.ceil, astype, np.trunc)
#### 37. Create a 5x5 matrix with row values ranging from 0 to 4 (★★☆)
(**hint**: np.arange)
#### 38. Consider a generator function that generates 10 integers and use it to build an array (★☆☆)
(**hint**: np.fromiter)
#### 39. Create a vector of size 10 with values ranging from 0 to 1, both excluded (★★☆)
(**hint**: np.linspace)
#### 40. Create a random vector of size 10 and sort it (★★☆)
(**hint**: sort)
#### 41. How to sum a small array faster than np.sum? (★★☆)
(**hint**: np.add.reduce)
#### 42. Consider two random array A and B, check if they are equal (★★☆)
(**hint**: np.allclose, np.array\_equal)
#### 43. Make an array immutable (read-only) (★★☆)
(**hint**: flags.writeable)
#### 44. Consider a random 10x2 matrix representing cartesian coordinates, convert them to polar coordinates (★★☆)
(**hint**: np.sqrt, np.arctan2)
#### 45. Create random vector of size 10 and replace the maximum value by 0 (★★☆)
(**hint**: argmax)
#### 46. Create a structured array with `x` and `y` coordinates covering the \[0,1\]x\[0,1\] area (★★☆)
(**hint**: np.meshgrid)
#### 47. Given two arrays, X and Y, construct the Cauchy matrix C (Cij =1/(xi - yj))
(**hint**: np.subtract.outer)
#### 48. Print the minimum and maximum representable value for each numpy scalar type (★★☆)
(**hint**: np.iinfo, np.finfo, eps)
#### 49. How to print all the values of an array? (★★☆)
(**hint**: np.set\_printoptions)
#### 50. How to find the closest value (to a given scalar) in a vector? (★★☆)
(**hint**: argmin)
#### 51. Create a structured array representing a position (x,y) and a color (r,g,b) (★★☆)
(**hint**: dtype)
#### 52. Consider a random vector with shape (100,2) representing coordinates, find point by point distances (★★☆)
(**hint**: np.atleast\_2d, T, np.sqrt)
#### 53. How to convert a float (32 bits) array into an integer (32 bits) in place?
(**hint**: astype(copy=False))
#### 54. How to read the following file? (★★☆)
(**hint**: np.genfromtxt)
```
1, 2, 3, 4, 5
6, , , 7, 8
, , 9,10,11
```
#### 55. What is the equivalent of enumerate for numpy arrays? (★★☆)
(**hint**: np.ndenumerate, np.ndindex)
#### 56. Generate a generic 2D Gaussian-like array (★★☆)
(**hint**: np.meshgrid, np.exp)
#### 57. How to randomly place p elements in a 2D array? (★★☆)
(**hint**: np.put, np.random.choice)
#### 58. Subtract the mean of each row of a matrix (★★☆)
(**hint**: mean(axis=,keepdims=))
#### 59. How to sort an array by the nth column? (★★☆)
(**hint**: argsort)
#### 60. How to tell if a given 2D array has null columns? (★★☆)
(**hint**: any, ~)
#### 61. Find the nearest value from a given value in an array (★★☆)
(**hint**: np.abs, argmin, flat)
#### 62. Considering two arrays with shape (1,3) and (3,1), how to compute their sum using an iterator? (★★☆)
(**hint**: np.nditer)
#### 63. Create an array class that has a name attribute (★★☆)
(**hint**: class method)
#### 64. Consider a given vector, how to add 1 to each element indexed by a second vector (be careful with repeated indices)? (★★★)
(**hint**: np.bincount | np.add.at)
#### 65. How to accumulate elements of a vector (X) to an array (F) based on an index list (I)? (★★★)
(**hint**: np.bincount)
#### 66. Considering a (w,h,3) image of (dtype=ubyte), compute the number of unique colors (★★★)
(**hint**: np.unique)
#### 67. Considering a four dimensions array, how to get sum over the last two axis at once? (★★★)
(**hint**: sum(axis=(-2,-1)))
#### 68. Considering a one-dimensional vector D, how to compute means of subsets of D using a vector S of same size describing subset indices? (★★★)
(**hint**: np.bincount)
#### 69. How to get the diagonal of a dot product? (★★★)
(**hint**: np.diag)
#### 70. Consider the vector \[1, 2, 3, 4, 5\], how to build a new vector with 3 consecutive zeros interleaved between each value? (★★★)
(**hint**: array\[::4\])
#### 71. Consider an array of dimension (5,5,3), how to mulitply it by an array with dimensions (5,5)? (★★★)
(**hint**: array\[:, :, None\])
#### 72. How to swap two rows of an array? (★★★)
(**hint**: array\[\[\]\] = array\[\[\]\])
#### 73. Consider a set of 10 triplets describing 10 triangles (with shared vertices), find the set of unique line segments composing all the triangles (★★★)
(**hint**: repeat, np.roll, np.sort, view, np.unique)
#### 74. Given an array C that is a bincount, how to produce an array A such that np.bincount(A) == C? (★★★)
(**hint**: np.repeat)
#### 75. How to compute averages using a sliding window over an array? (★★★)
(**hint**: np.cumsum)
#### 76. Consider a one-dimensional array Z, build a two-dimensional array whose first row is (Z\[0\],Z\[1\],Z\[2\]) and each subsequent row is shifted by 1 (last row should be (Z\[-3\],Z\[-2\],Z\[-1\]) (★★★)
(**hint**: from numpy.lib import stride_tricks)
#### 77. How to negate a boolean, or to change the sign of a float inplace? (★★★)
(**hint**: np.logical_not, np.negative)
#### 78. Consider 2 sets of points P0,P1 describing lines (2d) and a point p, how to compute distance from p to each line i (P0\[i\],P1\[i\])? (★★★)
#### 79. Consider 2 sets of points P0,P1 describing lines (2d) and a set of points P, how to compute distance from each point j (P\[j\]) to each line i (P0\[i\],P1\[i\])? (★★★)
#### 80. Consider an arbitrary array, write a function that extract a subpart with a fixed shape and centered on a given element (pad with a `fill` value when necessary) (★★★)
(**hint**: minimum, maximum)
#### 81. Consider an array Z = \[1,2,3,4,5,6,7,8,9,10,11,12,13,14\], how to generate an array R = \[\[1,2,3,4\], \[2,3,4,5\], \[3,4,5,6\], ..., \[11,12,13,14\]\]? (★★★)
(**hint**: stride\_tricks.as\_strided)
#### 82. Compute a matrix rank (★★★)
(**hint**: np.linalg.svd) (suggestion: np.linalg.svd)
#### 83. How to find the most frequent value in an array?
(**hint**: np.bincount, argmax)
#### 84. Extract all the contiguous 3x3 blocks from a random 10x10 matrix (★★★)
(**hint**: stride\_tricks.as\_strided)
#### 85. Create a 2D array subclass such that Z\[i,j\] == Z\[j,i\] (★★★)
(**hint**: class method)
#### 86. Consider a set of p matrices wich shape (n,n) and a set of p vectors with shape (n,1). How to compute the sum of of the p matrix products at once? (result has shape (n,1)) (★★★)
(**hint**: np.tensordot)
#### 87. Consider a 16x16 array, how to get the block-sum (block size is 4x4)? (★★★)
(**hint**: np.add.reduceat)
#### 88. How to implement the Game of Life using numpy arrays? (★★★)
#### 89. How to get the n largest values of an array (★★★)
(**hint**: np.argsort | np.argpartition)
#### 90. Given an arbitrary number of vectors, build the cartesian product (every combinations of every item) (★★★)
(**hint**: np.indices)
#### 91. How to create a record array from a regular array? (★★★)
(**hint**: np.core.records.fromarrays)
#### 92. Consider a large vector Z, compute Z to the power of 3 using 3 different methods (★★★)
(**hint**: np.power, \*, np.einsum)
#### 93. Consider two arrays A and B of shape (8,3) and (2,2). How to find rows of A that contain elements of each row of B regardless of the order of the elements in B? (★★★)
(**hint**: np.where)
#### 94. Considering a 10x3 matrix, extract rows with unequal values (e.g. \[2,2,3\]) (★★★)
#### 95. Convert a vector of ints into a matrix binary representation (★★★)
(**hint**: np.unpackbits)
#### 96. Given a two dimensional array, how to extract unique rows? (★★★)
(**hint**: np.ascontiguousarray)
#### 97. Considering 2 vectors A & B, write the einsum equivalent of inner, outer, sum, and mul function (★★★)
(**hint**: np.einsum)
#### 98. Considering a path described by two vectors (X,Y), how to sample it using equidistant samples (★★★)?
(**hint**: np.cumsum, np.interp)
#### 99. Given an integer n and a 2D array X, select from X the rows which can be interpreted as draws from a multinomial distribution with n degrees, i.e., the rows which only contain integers and which sum to n. (★★★)
(**hint**: np.logical\_and.reduce, np.mod)
#### 100. Compute bootstrapped 95% confidence intervals for the mean of a 1D array X (i.e., resample the elements of an array with replacement N times, compute the mean of each sample, and then compute percentiles over the means). (★★★)
(**hint**: np.percentile)
|
github_jupyter
|
0 * np.nan
np.nan == np.nan
np.inf > np.nan
np.nan - np.nan
0.3 == 3 * 0.1
# Author: Jake VanderPlas
print(sum(range(5),-1))
from numpy import *
print(sum(range(5),-1))
Z**Z
2 << Z >> 2
Z <- Z
1j*Z
Z/1/1
Z<Z>Z
np.array(0) / np.array(0)
np.array(0) // np.array(0)
np.array([np.nan]).astype(int).astype(float)
np.sqrt(-1) == np.emath.sqrt(-1)
1, 2, 3, 4, 5
6, , , 7, 8
, , 9,10,11
| 0.466846 | 0.974629 |
# Audio spectrogram
## Background
In this example we will go through the steps to build a DALI audio processing pipeline, including the calculation of a spectrogram. A spectrogram is a representation of a signal (e.g. an audio signal) that shows the evolution of the frequency spectrum in time.
Typically, a spectrogram is calculated by computing the fast fourier transform (FFT) over a series of overlapping windows extracted from the original signal. The process of dividing the signal in short term sequences of fixed size and applying FFT on those independently is called Short-time Fourier transform (STFT). The spectrogram is then calculated as the (typically squared) complex magnitude of the STFT.
Extracting short term windows of the original image affects the calculated spectrum by producing aliasing artifacts. This is often called spectral leakage. To control/reduce the spectral leakage effect, we use different window functions when extracting the windows. Some examples of window functions are: Hann, Hanning, etc.
It is beyond the scope of this example to go deeper into the details of the signal processing concepts we mentioned above. More information can be found here:
- [STFT](https://en.wikipedia.org/wiki/Short-time_Fourier_transform)
- [Window functions](https://en.wikipedia.org/wiki/Window_function)
## Reference implementation
To verify the correctness of DALI's implementation, we will compare it against librosa (https://librosa.github.io/librosa/).
```
import librosa as librosa
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
import librosa.display
```
Librosa provides an API to calculate the STFT, producing a complex output (i.e. complex numbers). It is then trivial to calculate the power spectrum from the complex STFT by the following
```
# Size of the FFT, which will also be used as the window length
n_fft=2048
# Step or stride between windows. If the step is smaller than the window lenght, the windows will overlap
hop_length=512
# Load sample audio file
y, sr = librosa.load(librosa.util.example_audio_file())
# Calculate the spectrogram as the square of the complex magnitude of the STFT
spectrogram_librosa = np.abs(librosa.stft(
y, n_fft=n_fft, hop_length=hop_length, win_length=n_fft, window='hann')) ** 2
```
We can now transform the spectrogram output to a logarithmic scale by transforming the amplitude to decibels. While doing so we will also normalize the spectrogram so that its maximum represent the 0 dB point.
```
spectrogram_librosa_db = librosa.power_to_db(spectrogram_librosa, ref=np.max)
```
The last step is to display the spectrogram
```
librosa.display.specshow(spectrogram_librosa_db, sr=sr, y_axis='log', x_axis='time', hop_length=hop_length)
plt.title('Reference power spectrogram')
plt.colorbar(format='%+2.0f dB')
plt.tight_layout()
plt.show()
```
## Calculating the spectrogram using DALI
To demonstrate DALI's Spectrogram operator we will define a DALI pipeline, whose input will be provided externally with the help of ExternalSource operator. For demonstration purposes, we can just feed the same input in every iteration, as we will be only calculating one spectrogram.
```
from nvidia.dali.pipeline import Pipeline
import nvidia.dali.ops as ops
import nvidia.dali.types as types
import nvidia.dali as dali
class SpectrogramPipeline(Pipeline):
def __init__(self, device, batch_size, nfft, window_length, window_step, num_threads=1, device_id=0):
super(SpectrogramPipeline, self).__init__(batch_size, num_threads, device_id)
self.device = device
self.batch_data = []
y, sr = librosa.load(librosa.util.example_audio_file())
for _ in range(batch_size):
self.batch_data.append(np.array(y, dtype=np.float32))
self.external_source = ops.ExternalSource()
self.spectrogram = ops.Spectrogram(device=self.device,
nfft=nfft,
window_length=window_length,
window_step=window_step)
def define_graph(self):
self.data = self.external_source()
out = self.data.gpu() if self.device == 'gpu' else self.data
out = self.spectrogram(out)
return out
def iter_setup(self):
self.feed_input(self.data, self.batch_data)
```
With the pipeline defined, we can now just build it and run it
```
pipe = SpectrogramPipeline(device='cpu', batch_size=1, nfft=n_fft, window_length=n_fft, window_step=hop_length)
pipe.build()
outputs = pipe.run()
spectrogram_dali = outputs[0].at(0)
```
and display it as we did with the reference implementation
```
spectrogram_dali_db = librosa.power_to_db(spectrogram_dali, ref=np.max)
librosa.display.specshow(spectrogram_dali_db, sr=sr, y_axis='log', x_axis='time', hop_length=hop_length)
plt.title('DALI power spectrogram')
plt.colorbar(format='%+2.0f dB')
plt.tight_layout()
plt.show()
```
As a last sanity check, we can verify that the numerical difference between the reference implementation and DALI's is insignificant
```
print("Average error: {0:.5f} dB".format(np.mean(np.abs(spectrogram_dali_db - spectrogram_librosa_db))))
assert(np.allclose(spectrogram_dali_db, spectrogram_librosa_db, atol=2))
```
## Mel spectrogram
The mel scale is a non-linear transformation of frequency scale based on the perception of pitches. The mel scale is calculated so that two pairs of frequencies separated by a delta in the mel scale are perceived by humans as being equidistant. More information can be found here: https://en.wikipedia.org/wiki/Mel_scale.
In machine learning applications involving speech and audio, we typically want to represent the power spectrogram in the mel scale domain. We do that by applying a bank of overlapping triangular filters that compute the energy of the spectrum in each band.
Typically, we want the mel spectrogram represented in decibels. We can calculate a mel spectrogram in decibels by using the following DALI pipeline.
```
class MelSpectrogramPipeline(Pipeline):
def __init__(self, device, batch_size, nfft, window_length, window_step, num_threads=1, device_id=0):
super(MelSpectrogramPipeline, self).__init__(batch_size, num_threads, device_id)
self.device = device
self.batch_data = []
y, sr = librosa.load(librosa.util.example_audio_file())
for _ in range(batch_size):
self.batch_data.append(np.array(y, dtype=np.float32))
self.external_source = ops.ExternalSource()
self.spectrogram = ops.Spectrogram(device=self.device,
nfft=nfft,
window_length=window_length,
window_step=window_step)
self.mel_fbank = ops.MelFilterBank(device=self.device,
sample_rate=sr,
nfilter = 128,
freq_high = 8000.0)
self.dB = ops.ToDecibels(device=self.device,
multiplier = 10.0,
cutoff_db = -80)
def define_graph(self):
self.data = self.external_source()
out = self.data.gpu() if self.device == 'gpu' else self.data
out = self.spectrogram(out)
out = self.mel_fbank(out)
out = self.dB(out)
return out
def iter_setup(self):
self.feed_input(self.data, self.batch_data)
pipe = MelSpectrogramPipeline(device='cpu', batch_size=1, nfft=n_fft, window_length=n_fft, window_step=hop_length)
pipe.build()
outputs = pipe.run()
mel_spectrogram_dali_db = outputs[0].at(0)
```
We can now verify that it produces the same result as Librosa
```
librosa.display.specshow(mel_spectrogram_dali_db, sr=sr, y_axis='mel', x_axis='time', hop_length=hop_length)
plt.title('DALI Mel-frequency spectrogram')
plt.colorbar(format='%+2.0f dB')
plt.tight_layout()
plt.show()
mel_spectrogram_librosa = librosa.feature.melspectrogram(y=y, sr=sr, n_mels=128, fmax=8000)
mel_spectrogram_librosa_db = librosa.power_to_db(mel_spectrogram_librosa, ref=np.max)
assert(np.allclose(mel_spectrogram_dali_db, mel_spectrogram_librosa_db, atol=1))
```
## Mel-frequency cepstral coefficients (MFCCs)
MFCCs are an alternative representation of the Mel-frequency spectrogram often used in audio applications. The MFCCs are calculated by applying the discrete cosine transform (DCT) to a mel-frequency spectrogram.
DALI's implementation of DCT uses uses the formulas described in https://en.wikipedia.org/wiki/Discrete_cosine_transform
In addition to the DCT, a cepstral filter (also known as *liftering*) can be applied to emphasize higher order coefficients.
A *liftered* cepstral coefficient is calculated according to the formula
$$
\widehat{\text{MFCC}_i} = w_{i} \cdot \text{MFCC}_{i}
$$
where
$$
w_i = 1 + \frac{L}{2}\sin\Big(\frac{\pi i}{L}\Big)
$$
where $L$ is the *liftering* coefficient.
More information about MFCC can be found here: https://en.wikipedia.org/wiki/Mel-frequency_cepstrum.
We can use DALI's MFCC operator to transform the mel-spectrogram into a set of MFCCs
```
class MFCCPipeline(Pipeline):
def __init__(self, device, batch_size, nfft, window_length, window_step,
dct_type, n_mfcc, normalize, lifter, num_threads=1, device_id=0):
super(MFCCPipeline, self).__init__(batch_size, num_threads, device_id)
self.device = device
self.batch_data = []
y, sr = librosa.load(librosa.util.example_audio_file())
for _ in range(batch_size):
self.batch_data.append(np.array(y, dtype=np.float32))
self.external_source = ops.ExternalSource()
self.spectrogram = ops.Spectrogram(device=self.device,
nfft=nfft,
window_length=window_length,
window_step=window_step)
self.mel_fbank = ops.MelFilterBank(device=self.device,
sample_rate=sr,
nfilter = 128,
freq_high = 8000.0)
self.dB = ops.ToDecibels(device=self.device,
multiplier = 10.0,
cutoff_db = -80.0)
self.mfcc = ops.MFCC(device=self.device,
axis=0,
dct_type=dct_type,
n_mfcc=n_mfcc,
normalize=normalize,
lifter=lifter)
def define_graph(self):
self.data = self.external_source()
out = self.data.gpu() if self.device == 'gpu' else self.data
out = self.spectrogram(out)
out = self.mel_fbank(out)
out = self.dB(out)
out = self.mfcc(out)
return out
def iter_setup(self):
self.feed_input(self.data, self.batch_data)
```
Let's now run the pipeline and display the output as we did previously
```
pipe = MFCCPipeline(device='cpu', batch_size=1, nfft=n_fft, window_length=n_fft, window_step=hop_length,
dct_type=2, n_mfcc=40, normalize=True, lifter=0)
pipe.build()
outputs = pipe.run()
mfccs_dali = outputs[0].at(0)
plt.figure(figsize=(10, 4))
librosa.display.specshow(mfccs_dali, x_axis='time')
plt.colorbar()
plt.title('MFCC (DALI)')
plt.tight_layout()
plt.show()
```
As a last step, let's verify that this implementation produces the same result as Librosa. Please note, we are comparing the ortho-normalized MFCCs, as Librosa's DCT implementation uses a different formula which causes the output to be scaled by a factor of 2 when we compare it with the Wikipedia's formulae.
```
mfccs_librosa = librosa.feature.mfcc(S=mel_spectrogram_librosa_db,
dct_type=2, n_mfcc=40, norm='ortho', lifter=0)
assert(np.allclose(mfccs_librosa, mfccs_dali, atol=1))
```
|
github_jupyter
|
import librosa as librosa
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
import librosa.display
# Size of the FFT, which will also be used as the window length
n_fft=2048
# Step or stride between windows. If the step is smaller than the window lenght, the windows will overlap
hop_length=512
# Load sample audio file
y, sr = librosa.load(librosa.util.example_audio_file())
# Calculate the spectrogram as the square of the complex magnitude of the STFT
spectrogram_librosa = np.abs(librosa.stft(
y, n_fft=n_fft, hop_length=hop_length, win_length=n_fft, window='hann')) ** 2
spectrogram_librosa_db = librosa.power_to_db(spectrogram_librosa, ref=np.max)
librosa.display.specshow(spectrogram_librosa_db, sr=sr, y_axis='log', x_axis='time', hop_length=hop_length)
plt.title('Reference power spectrogram')
plt.colorbar(format='%+2.0f dB')
plt.tight_layout()
plt.show()
from nvidia.dali.pipeline import Pipeline
import nvidia.dali.ops as ops
import nvidia.dali.types as types
import nvidia.dali as dali
class SpectrogramPipeline(Pipeline):
def __init__(self, device, batch_size, nfft, window_length, window_step, num_threads=1, device_id=0):
super(SpectrogramPipeline, self).__init__(batch_size, num_threads, device_id)
self.device = device
self.batch_data = []
y, sr = librosa.load(librosa.util.example_audio_file())
for _ in range(batch_size):
self.batch_data.append(np.array(y, dtype=np.float32))
self.external_source = ops.ExternalSource()
self.spectrogram = ops.Spectrogram(device=self.device,
nfft=nfft,
window_length=window_length,
window_step=window_step)
def define_graph(self):
self.data = self.external_source()
out = self.data.gpu() if self.device == 'gpu' else self.data
out = self.spectrogram(out)
return out
def iter_setup(self):
self.feed_input(self.data, self.batch_data)
pipe = SpectrogramPipeline(device='cpu', batch_size=1, nfft=n_fft, window_length=n_fft, window_step=hop_length)
pipe.build()
outputs = pipe.run()
spectrogram_dali = outputs[0].at(0)
spectrogram_dali_db = librosa.power_to_db(spectrogram_dali, ref=np.max)
librosa.display.specshow(spectrogram_dali_db, sr=sr, y_axis='log', x_axis='time', hop_length=hop_length)
plt.title('DALI power spectrogram')
plt.colorbar(format='%+2.0f dB')
plt.tight_layout()
plt.show()
print("Average error: {0:.5f} dB".format(np.mean(np.abs(spectrogram_dali_db - spectrogram_librosa_db))))
assert(np.allclose(spectrogram_dali_db, spectrogram_librosa_db, atol=2))
class MelSpectrogramPipeline(Pipeline):
def __init__(self, device, batch_size, nfft, window_length, window_step, num_threads=1, device_id=0):
super(MelSpectrogramPipeline, self).__init__(batch_size, num_threads, device_id)
self.device = device
self.batch_data = []
y, sr = librosa.load(librosa.util.example_audio_file())
for _ in range(batch_size):
self.batch_data.append(np.array(y, dtype=np.float32))
self.external_source = ops.ExternalSource()
self.spectrogram = ops.Spectrogram(device=self.device,
nfft=nfft,
window_length=window_length,
window_step=window_step)
self.mel_fbank = ops.MelFilterBank(device=self.device,
sample_rate=sr,
nfilter = 128,
freq_high = 8000.0)
self.dB = ops.ToDecibels(device=self.device,
multiplier = 10.0,
cutoff_db = -80)
def define_graph(self):
self.data = self.external_source()
out = self.data.gpu() if self.device == 'gpu' else self.data
out = self.spectrogram(out)
out = self.mel_fbank(out)
out = self.dB(out)
return out
def iter_setup(self):
self.feed_input(self.data, self.batch_data)
pipe = MelSpectrogramPipeline(device='cpu', batch_size=1, nfft=n_fft, window_length=n_fft, window_step=hop_length)
pipe.build()
outputs = pipe.run()
mel_spectrogram_dali_db = outputs[0].at(0)
librosa.display.specshow(mel_spectrogram_dali_db, sr=sr, y_axis='mel', x_axis='time', hop_length=hop_length)
plt.title('DALI Mel-frequency spectrogram')
plt.colorbar(format='%+2.0f dB')
plt.tight_layout()
plt.show()
mel_spectrogram_librosa = librosa.feature.melspectrogram(y=y, sr=sr, n_mels=128, fmax=8000)
mel_spectrogram_librosa_db = librosa.power_to_db(mel_spectrogram_librosa, ref=np.max)
assert(np.allclose(mel_spectrogram_dali_db, mel_spectrogram_librosa_db, atol=1))
class MFCCPipeline(Pipeline):
def __init__(self, device, batch_size, nfft, window_length, window_step,
dct_type, n_mfcc, normalize, lifter, num_threads=1, device_id=0):
super(MFCCPipeline, self).__init__(batch_size, num_threads, device_id)
self.device = device
self.batch_data = []
y, sr = librosa.load(librosa.util.example_audio_file())
for _ in range(batch_size):
self.batch_data.append(np.array(y, dtype=np.float32))
self.external_source = ops.ExternalSource()
self.spectrogram = ops.Spectrogram(device=self.device,
nfft=nfft,
window_length=window_length,
window_step=window_step)
self.mel_fbank = ops.MelFilterBank(device=self.device,
sample_rate=sr,
nfilter = 128,
freq_high = 8000.0)
self.dB = ops.ToDecibels(device=self.device,
multiplier = 10.0,
cutoff_db = -80.0)
self.mfcc = ops.MFCC(device=self.device,
axis=0,
dct_type=dct_type,
n_mfcc=n_mfcc,
normalize=normalize,
lifter=lifter)
def define_graph(self):
self.data = self.external_source()
out = self.data.gpu() if self.device == 'gpu' else self.data
out = self.spectrogram(out)
out = self.mel_fbank(out)
out = self.dB(out)
out = self.mfcc(out)
return out
def iter_setup(self):
self.feed_input(self.data, self.batch_data)
pipe = MFCCPipeline(device='cpu', batch_size=1, nfft=n_fft, window_length=n_fft, window_step=hop_length,
dct_type=2, n_mfcc=40, normalize=True, lifter=0)
pipe.build()
outputs = pipe.run()
mfccs_dali = outputs[0].at(0)
plt.figure(figsize=(10, 4))
librosa.display.specshow(mfccs_dali, x_axis='time')
plt.colorbar()
plt.title('MFCC (DALI)')
plt.tight_layout()
plt.show()
mfccs_librosa = librosa.feature.mfcc(S=mel_spectrogram_librosa_db,
dct_type=2, n_mfcc=40, norm='ortho', lifter=0)
assert(np.allclose(mfccs_librosa, mfccs_dali, atol=1))
| 0.726911 | 0.984276 |
```
# -*- coding: utf-8 -*-
"""
Created on Fri Sep 24 09:59:55 2021
@author: chint
"""
import seaborn as sns
import matplotlib.pyplot as plt
import numpy as np
from sklearn.ensemble import AdaBoostClassifier,RandomForestClassifier
import pandas as pd
import seaborn as sns
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import train_test_split, cross_val_score,GridSearchCV
from sklearn.pipeline import Pipeline
from sklearn.decomposition import PCA
from sklearn import tree
import matplotlib.pyplot as plt
import numpy as np
from sklearn.model_selection import StratifiedKFold
from sklearn.metrics import confusion_matrix, f1_score, precision_score, recall_score, roc_curve,accuracy_score
from sklearn.neighbors import KNeighborsClassifier as KNN
from sklearn.svm import SVC
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis as LDA
from sklearn import linear_model
from sklearn.model_selection import KFold,cross_val_score,GridSearchCV
import warnings
warnings.filterwarnings('ignore')
#%%
df=pd.read_csv('D:/UIUC_courses/IE517/project/MLF_GP1_CreditScore.csv')
pd_corr=df.corr()
sns.heatmap(pd_corr)
plt.show()
labels=df.keys()
# corr_labels=['Rating','CFO','CFO/Debt','ROE','Free Cash Flow', 'Current Liabilities','Cash','Current Liquidity']
# sns.heatmap(df[corr_labels].corr())
# plt.show()
# print("As we see from the above correleation plots, there is strong correlation between few of the features")
df2=df.values
X=df2[:,:-2]
y=df2[:,-2]
y=y.astype(np.float)
#%% Feature selection
score_y=[]
score_x=[]
for i in range(-3,5):
lasso=linear_model.Lasso(alpha=10**-i)
X_train, X_test, y_train, y_test = train_test_split(X, y,test_size=0.2,random_state=42)
pipe = Pipeline([('scaler',StandardScaler()), ('estimator', lasso)])
#gs_cv=GridSearchCV(pipe,{},cv=3)
#gs_cv.fit(X,y)
print("mean of cross validation scores=",str(np.mean(cross_val_score(pipe,X,y,cv=KFold(n_splits=4, shuffle=True)))),"for alpha=",str(10**-i))
lasso.fit(X_train,y_train)
score_y.append(lasso.coef_)
score_x.append(i)
plt.plot(np.transpose(score_x),np.array(score_y))
plt.legend(labels.values,loc='center left',fontsize=6)
plt.title("Lasso regression coefficients vs aplha")
plt.xlabel("log10(aplha)")
plt.ylabel("coefficient")
plt.show()
score_y=np.transpose(score_y)
#%% Feature extraction
print("we don't use LDA here because this is a binary class problem and will have only one LDA component")
pca_test=PCA()
pca_test.fit(X)
plt.bar(range(0,len(pca_test.explained_variance_ratio_)),pca_test.explained_variance_ratio_)
plt.plot(np.cumsum(pca_test.explained_variance_ratio_))
plt.xlabel('component')
plt.ylabel('explained variance ratio')
plt.title('Variance ratio vs pca component')
plt.show()
print(" From the graph, it is evident that 8-10 components of LDA capture 90% of variance")
#%% BASIC MODELS
#%% SVC
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, stratify=y)
cv=StratifiedKFold(n_splits=10)
pipe = Pipeline([('scaler', StandardScaler()),('pca',PCA()), ('clf', SVC() )])
params={'clf__gamma':[0.001,0.1,1,10,50,100],'pca__n_components':[7,8,9,10,11,12]}
clf=GridSearchCV(pipe,params,cv=cv)
clf.fit(X_train, y_train)
print('best parameters for svc in original classification=',str(clf.best_params_))
print('best score for svc in original classification=',str(clf.best_score_))
y_predict=clf.best_estimator_.predict(X_test)
#%% KNN
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, stratify=y)
cv=StratifiedKFold(n_splits=10)
pipe = Pipeline([('scaler', StandardScaler()),('pca',PCA()), ('clf', KNN() )])
params={'clf__n_neighbors':[1,3,5,7,9,12],'pca__n_components':[7,8,9,10,11,12]}
clf=GridSearchCV(pipe,params,cv=cv)
clf.fit(X_train, y_train)
print('best parameters for KNN in original classification=',str(clf.best_params_))
print('best score for KNN in original classification=',str(clf.best_score_))
y_predict=clf.best_estimator_.predict(X_test)
#%% Decision Tree Classifier
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, stratify=y)
cv=StratifiedKFold(n_splits=10)
pipe = Pipeline([('scaler', StandardScaler()),('pca',PCA()), ('clf', tree.DecisionTreeClassifier() )])
params={'clf__max_depth':[1,3,5,7,9,12],'pca__n_components':[7,8,9,10,11,12]}
clf=GridSearchCV(pipe,params,cv=cv)
clf.fit(X_train, y_train)
print('best parameters for decision tree in original classification=',str(clf.best_params_))
print('best score for decision tree in original classification=',str(clf.best_score_))
y_predict=clf.best_estimator_.predict(X_test)
#%% ENSEMBLING
#%% Adaboost Classifier
abc = AdaBoostClassifier(base_estimator=tree.DecisionTreeClassifier(max_depth=12) , n_estimators=100, random_state=0)
clf2 = Pipeline([('scaler', StandardScaler()),('pca',PCA(n_components=8)), ('clf', abc )])
clf2=GridSearchCV(abc,{},cv=cv)
clf2.fit(X_train, y_train)
print('best parameters for adaboost in original classification=',str(clf.best_params_))
print('best score for adaboost in original classification=',str(clf2.best_score_))
y_predict=clf2.best_estimator_.predict(X_test)
#%% Random Forest Classifier
abc = RandomForestClassifier(max_depth=12, n_estimators=100)
clf2 = Pipeline([('scaler', StandardScaler()),('pca',PCA(n_components=8)), ('clf', abc )])
clf2=GridSearchCV(abc,{},cv=cv)
clf2.fit(X_train, y_train)
print('best parameters for Random Forest Classifier in original classification=',str(clf.best_params_))
print('best score for Random Forest Classifier in original classification=',str(clf2.best_score_))
y_predict=clf2.best_estimator_.predict(X_test)
#%% Results
print("Since random forest seems tpo perform the best among all the models tested, the accuracies are computed based on RF model")
print('f1 score:',str(f1_score(y_predict,y_test)))
print('precision score',str(precision_score(y_predict,y_test)))
print('recall score',str(recall_score(y_predict,y_test)))
print(confusion_matrix(y_test,y_predict))
#%% ROC Curve
y_pred_prob =clf2.predict_proba(X_test)[:,1]
fpr, tpr, thresholds =roc_curve(y_test, y_pred_prob)
# Plot ROC curve
plt.plot([0, 1], [0, 1], 'k--')
plt.plot(fpr, tpr)
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
plt.title('ROC Curve')
plt.show()
```
|
github_jupyter
|
# -*- coding: utf-8 -*-
"""
Created on Fri Sep 24 09:59:55 2021
@author: chint
"""
import seaborn as sns
import matplotlib.pyplot as plt
import numpy as np
from sklearn.ensemble import AdaBoostClassifier,RandomForestClassifier
import pandas as pd
import seaborn as sns
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import train_test_split, cross_val_score,GridSearchCV
from sklearn.pipeline import Pipeline
from sklearn.decomposition import PCA
from sklearn import tree
import matplotlib.pyplot as plt
import numpy as np
from sklearn.model_selection import StratifiedKFold
from sklearn.metrics import confusion_matrix, f1_score, precision_score, recall_score, roc_curve,accuracy_score
from sklearn.neighbors import KNeighborsClassifier as KNN
from sklearn.svm import SVC
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis as LDA
from sklearn import linear_model
from sklearn.model_selection import KFold,cross_val_score,GridSearchCV
import warnings
warnings.filterwarnings('ignore')
#%%
df=pd.read_csv('D:/UIUC_courses/IE517/project/MLF_GP1_CreditScore.csv')
pd_corr=df.corr()
sns.heatmap(pd_corr)
plt.show()
labels=df.keys()
# corr_labels=['Rating','CFO','CFO/Debt','ROE','Free Cash Flow', 'Current Liabilities','Cash','Current Liquidity']
# sns.heatmap(df[corr_labels].corr())
# plt.show()
# print("As we see from the above correleation plots, there is strong correlation between few of the features")
df2=df.values
X=df2[:,:-2]
y=df2[:,-2]
y=y.astype(np.float)
#%% Feature selection
score_y=[]
score_x=[]
for i in range(-3,5):
lasso=linear_model.Lasso(alpha=10**-i)
X_train, X_test, y_train, y_test = train_test_split(X, y,test_size=0.2,random_state=42)
pipe = Pipeline([('scaler',StandardScaler()), ('estimator', lasso)])
#gs_cv=GridSearchCV(pipe,{},cv=3)
#gs_cv.fit(X,y)
print("mean of cross validation scores=",str(np.mean(cross_val_score(pipe,X,y,cv=KFold(n_splits=4, shuffle=True)))),"for alpha=",str(10**-i))
lasso.fit(X_train,y_train)
score_y.append(lasso.coef_)
score_x.append(i)
plt.plot(np.transpose(score_x),np.array(score_y))
plt.legend(labels.values,loc='center left',fontsize=6)
plt.title("Lasso regression coefficients vs aplha")
plt.xlabel("log10(aplha)")
plt.ylabel("coefficient")
plt.show()
score_y=np.transpose(score_y)
#%% Feature extraction
print("we don't use LDA here because this is a binary class problem and will have only one LDA component")
pca_test=PCA()
pca_test.fit(X)
plt.bar(range(0,len(pca_test.explained_variance_ratio_)),pca_test.explained_variance_ratio_)
plt.plot(np.cumsum(pca_test.explained_variance_ratio_))
plt.xlabel('component')
plt.ylabel('explained variance ratio')
plt.title('Variance ratio vs pca component')
plt.show()
print(" From the graph, it is evident that 8-10 components of LDA capture 90% of variance")
#%% BASIC MODELS
#%% SVC
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, stratify=y)
cv=StratifiedKFold(n_splits=10)
pipe = Pipeline([('scaler', StandardScaler()),('pca',PCA()), ('clf', SVC() )])
params={'clf__gamma':[0.001,0.1,1,10,50,100],'pca__n_components':[7,8,9,10,11,12]}
clf=GridSearchCV(pipe,params,cv=cv)
clf.fit(X_train, y_train)
print('best parameters for svc in original classification=',str(clf.best_params_))
print('best score for svc in original classification=',str(clf.best_score_))
y_predict=clf.best_estimator_.predict(X_test)
#%% KNN
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, stratify=y)
cv=StratifiedKFold(n_splits=10)
pipe = Pipeline([('scaler', StandardScaler()),('pca',PCA()), ('clf', KNN() )])
params={'clf__n_neighbors':[1,3,5,7,9,12],'pca__n_components':[7,8,9,10,11,12]}
clf=GridSearchCV(pipe,params,cv=cv)
clf.fit(X_train, y_train)
print('best parameters for KNN in original classification=',str(clf.best_params_))
print('best score for KNN in original classification=',str(clf.best_score_))
y_predict=clf.best_estimator_.predict(X_test)
#%% Decision Tree Classifier
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, stratify=y)
cv=StratifiedKFold(n_splits=10)
pipe = Pipeline([('scaler', StandardScaler()),('pca',PCA()), ('clf', tree.DecisionTreeClassifier() )])
params={'clf__max_depth':[1,3,5,7,9,12],'pca__n_components':[7,8,9,10,11,12]}
clf=GridSearchCV(pipe,params,cv=cv)
clf.fit(X_train, y_train)
print('best parameters for decision tree in original classification=',str(clf.best_params_))
print('best score for decision tree in original classification=',str(clf.best_score_))
y_predict=clf.best_estimator_.predict(X_test)
#%% ENSEMBLING
#%% Adaboost Classifier
abc = AdaBoostClassifier(base_estimator=tree.DecisionTreeClassifier(max_depth=12) , n_estimators=100, random_state=0)
clf2 = Pipeline([('scaler', StandardScaler()),('pca',PCA(n_components=8)), ('clf', abc )])
clf2=GridSearchCV(abc,{},cv=cv)
clf2.fit(X_train, y_train)
print('best parameters for adaboost in original classification=',str(clf.best_params_))
print('best score for adaboost in original classification=',str(clf2.best_score_))
y_predict=clf2.best_estimator_.predict(X_test)
#%% Random Forest Classifier
abc = RandomForestClassifier(max_depth=12, n_estimators=100)
clf2 = Pipeline([('scaler', StandardScaler()),('pca',PCA(n_components=8)), ('clf', abc )])
clf2=GridSearchCV(abc,{},cv=cv)
clf2.fit(X_train, y_train)
print('best parameters for Random Forest Classifier in original classification=',str(clf.best_params_))
print('best score for Random Forest Classifier in original classification=',str(clf2.best_score_))
y_predict=clf2.best_estimator_.predict(X_test)
#%% Results
print("Since random forest seems tpo perform the best among all the models tested, the accuracies are computed based on RF model")
print('f1 score:',str(f1_score(y_predict,y_test)))
print('precision score',str(precision_score(y_predict,y_test)))
print('recall score',str(recall_score(y_predict,y_test)))
print(confusion_matrix(y_test,y_predict))
#%% ROC Curve
y_pred_prob =clf2.predict_proba(X_test)[:,1]
fpr, tpr, thresholds =roc_curve(y_test, y_pred_prob)
# Plot ROC curve
plt.plot([0, 1], [0, 1], 'k--')
plt.plot(fpr, tpr)
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
plt.title('ROC Curve')
plt.show()
| 0.404507 | 0.433322 |
# labels
```
import vectorbt as vbt
import numpy as np
import pandas as pd
from datetime import datetime, timedelta
from numba import njit
# Disable caching for performance testing
vbt.settings.caching['enabled'] = False
close = pd.DataFrame({
'a': [1, 2, 1, 2, 3, 2],
'b': [3, 2, 3, 2, 1, 2]
}, index=pd.Index([
datetime(2020, 1, 1),
datetime(2020, 1, 2),
datetime(2020, 1, 3),
datetime(2020, 1, 4),
datetime(2020, 1, 5),
datetime(2020, 1, 6)
]))
pos_ths = [np.array([1, 1 / 2]), np.array([2, 1 / 2]), np.array([3, 1 / 2])]
neg_ths = [np.array([1 / 2, 1 / 3]), np.array([1 / 2, 2 / 3]), np.array([1 / 2, 3 / 4])]
big_close = pd.DataFrame(np.random.randint(1, 10, size=(1000, 1000)).astype(float))
big_close.index = [datetime(2018, 1, 1) + timedelta(days=i) for i in range(1000)]
big_close.shape
```
## Look-ahead indicators
```
print(vbt.FMEAN.run(close, window=(2, 3), ewm=(False, True), param_product=True).fmean)
%timeit vbt.FMEAN.run(big_close, window=2)
%timeit vbt.FMEAN.run(big_close, window=np.arange(2, 10).tolist())
print(vbt.FMEAN.run(big_close, window=np.arange(2, 10).tolist()).wrapper.shape)
print(vbt.FSTD.run(close, window=(2, 3), ewm=(False, True), param_product=True).fstd)
%timeit vbt.FSTD.run(big_close, window=2)
%timeit vbt.FSTD.run(big_close, window=np.arange(2, 10).tolist())
print(vbt.FSTD.run(big_close, window=np.arange(2, 10).tolist()).wrapper.shape)
print(vbt.FMIN.run(close, window=(2, 3)).fmin)
%timeit vbt.FMIN.run(big_close, window=2)
%timeit vbt.FMIN.run(big_close, window=np.arange(2, 10).tolist())
print(vbt.FMIN.run(big_close, window=np.arange(2, 10).tolist()).wrapper.shape)
print(vbt.FMAX.run(close, window=(2, 3)).fmax)
%timeit vbt.FMAX.run(big_close, window=2)
%timeit vbt.FMAX.run(big_close, window=np.arange(2, 10).tolist())
print(vbt.FMAX.run(big_close, window=np.arange(2, 10).tolist()).wrapper.shape)
```
## Label generators
```
print(vbt.FIXLB.run(close, n=(2, 3)).labels)
%timeit vbt.FIXLB.run(big_close, n=2)
%timeit vbt.FIXLB.run(big_close, n=np.arange(2, 10).tolist())
print(vbt.FIXLB.run(big_close, n=np.arange(2, 10).tolist()).wrapper.shape)
vbt.FIXLB.run(close['a'], n=2).plot().show_png()
print(vbt.MEANLB.run(close, window=(2, 3), ewm=(False, True), param_product=True).labels)
%timeit vbt.MEANLB.run(big_close, window=2)
%timeit vbt.MEANLB.run(big_close, window=np.arange(2, 10).tolist())
print(vbt.MEANLB.run(big_close, window=np.arange(2, 10).tolist()).wrapper.shape)
vbt.MEANLB.run(close['a'], window=2).plot().show_png()
print(vbt.LEXLB.run(close, pos_th=pos_ths, neg_th=neg_ths).labels)
%timeit vbt.LEXLB.run(big_close, pos_th=1, neg_th=0.5)
print(vbt.LEXLB.run(big_close, pos_th=1, neg_th=0.5).wrapper.shape)
vbt.LEXLB.run(close['a'], pos_th=1, neg_th=0.5).plot().show_png()
print(vbt.TRENDLB.run(close, pos_th=pos_ths, neg_th=neg_ths, mode='Binary').labels)
%timeit vbt.TRENDLB.run(big_close, pos_th=1, neg_th=0.5, mode='Binary')
print(vbt.TRENDLB.run(big_close, pos_th=1, neg_th=0.5, mode='Binary').wrapper.shape)
vbt.TRENDLB.run(close['a'], pos_th=1, neg_th=0.5, mode='Binary').plot().show_png()
print(vbt.TRENDLB.run(close, pos_th=pos_ths, neg_th=neg_ths, mode='BinaryCont').labels)
%timeit vbt.TRENDLB.run(big_close, pos_th=1, neg_th=0.5, mode='BinaryCont')
print(vbt.TRENDLB.run(big_close, pos_th=1, neg_th=0.5, mode='BinaryCont').wrapper.shape)
vbt.TRENDLB.run(close['a'], pos_th=1, neg_th=0.5, mode='BinaryCont').plot().show_png()
print(vbt.TRENDLB.run(close, pos_th=pos_ths, neg_th=neg_ths, mode='BinaryContSat').labels)
%timeit vbt.TRENDLB.run(big_close, pos_th=1, neg_th=0.5, mode='BinaryContSat')
print(vbt.TRENDLB.run(big_close, pos_th=1, neg_th=0.5, mode='BinaryContSat').wrapper.shape)
vbt.TRENDLB.run(close['a'], pos_th=1, neg_th=0.5, mode='BinaryContSat').plot().show_png()
print(vbt.TRENDLB.run(close, pos_th=pos_ths, neg_th=neg_ths, mode='PctChange').labels)
%timeit vbt.TRENDLB.run(big_close, pos_th=1, neg_th=0.5, mode='PctChange')
print(vbt.TRENDLB.run(big_close, pos_th=1, neg_th=0.5, mode='PctChange').wrapper.shape)
vbt.TRENDLB.run(close['a'], pos_th=1, neg_th=0.5, mode='PctChange').plot().show_png()
print(vbt.TRENDLB.run(close, pos_th=pos_ths, neg_th=neg_ths, mode='PctChangeNorm').labels)
%timeit vbt.TRENDLB.run(big_close, pos_th=1, neg_th=0.5, mode='PctChangeNorm')
print(vbt.TRENDLB.run(big_close, pos_th=1, neg_th=0.5, mode='PctChangeNorm').wrapper.shape)
vbt.TRENDLB.run(close['a'], pos_th=1, neg_th=0.5, mode='PctChangeNorm').plot().show_png()
print(vbt.BOLB.run(close, window=1, pos_th=pos_ths, neg_th=neg_ths).labels)
print(vbt.BOLB.run(close, window=2, pos_th=pos_ths, neg_th=neg_ths).labels)
%timeit vbt.BOLB.run(big_close, window=2, pos_th=1, neg_th=0.5)
%timeit vbt.BOLB.run(big_close, window=np.arange(2, 10).tolist(), pos_th=1, neg_th=0.5)
print(vbt.BOLB.run(big_close, window=np.arange(2, 10).tolist(), pos_th=1, neg_th=0.5).wrapper.shape)
vbt.BOLB.run(close['a'], window=2, pos_th=1, neg_th=0.5).plot().show_png()
```
|
github_jupyter
|
import vectorbt as vbt
import numpy as np
import pandas as pd
from datetime import datetime, timedelta
from numba import njit
# Disable caching for performance testing
vbt.settings.caching['enabled'] = False
close = pd.DataFrame({
'a': [1, 2, 1, 2, 3, 2],
'b': [3, 2, 3, 2, 1, 2]
}, index=pd.Index([
datetime(2020, 1, 1),
datetime(2020, 1, 2),
datetime(2020, 1, 3),
datetime(2020, 1, 4),
datetime(2020, 1, 5),
datetime(2020, 1, 6)
]))
pos_ths = [np.array([1, 1 / 2]), np.array([2, 1 / 2]), np.array([3, 1 / 2])]
neg_ths = [np.array([1 / 2, 1 / 3]), np.array([1 / 2, 2 / 3]), np.array([1 / 2, 3 / 4])]
big_close = pd.DataFrame(np.random.randint(1, 10, size=(1000, 1000)).astype(float))
big_close.index = [datetime(2018, 1, 1) + timedelta(days=i) for i in range(1000)]
big_close.shape
print(vbt.FMEAN.run(close, window=(2, 3), ewm=(False, True), param_product=True).fmean)
%timeit vbt.FMEAN.run(big_close, window=2)
%timeit vbt.FMEAN.run(big_close, window=np.arange(2, 10).tolist())
print(vbt.FMEAN.run(big_close, window=np.arange(2, 10).tolist()).wrapper.shape)
print(vbt.FSTD.run(close, window=(2, 3), ewm=(False, True), param_product=True).fstd)
%timeit vbt.FSTD.run(big_close, window=2)
%timeit vbt.FSTD.run(big_close, window=np.arange(2, 10).tolist())
print(vbt.FSTD.run(big_close, window=np.arange(2, 10).tolist()).wrapper.shape)
print(vbt.FMIN.run(close, window=(2, 3)).fmin)
%timeit vbt.FMIN.run(big_close, window=2)
%timeit vbt.FMIN.run(big_close, window=np.arange(2, 10).tolist())
print(vbt.FMIN.run(big_close, window=np.arange(2, 10).tolist()).wrapper.shape)
print(vbt.FMAX.run(close, window=(2, 3)).fmax)
%timeit vbt.FMAX.run(big_close, window=2)
%timeit vbt.FMAX.run(big_close, window=np.arange(2, 10).tolist())
print(vbt.FMAX.run(big_close, window=np.arange(2, 10).tolist()).wrapper.shape)
print(vbt.FIXLB.run(close, n=(2, 3)).labels)
%timeit vbt.FIXLB.run(big_close, n=2)
%timeit vbt.FIXLB.run(big_close, n=np.arange(2, 10).tolist())
print(vbt.FIXLB.run(big_close, n=np.arange(2, 10).tolist()).wrapper.shape)
vbt.FIXLB.run(close['a'], n=2).plot().show_png()
print(vbt.MEANLB.run(close, window=(2, 3), ewm=(False, True), param_product=True).labels)
%timeit vbt.MEANLB.run(big_close, window=2)
%timeit vbt.MEANLB.run(big_close, window=np.arange(2, 10).tolist())
print(vbt.MEANLB.run(big_close, window=np.arange(2, 10).tolist()).wrapper.shape)
vbt.MEANLB.run(close['a'], window=2).plot().show_png()
print(vbt.LEXLB.run(close, pos_th=pos_ths, neg_th=neg_ths).labels)
%timeit vbt.LEXLB.run(big_close, pos_th=1, neg_th=0.5)
print(vbt.LEXLB.run(big_close, pos_th=1, neg_th=0.5).wrapper.shape)
vbt.LEXLB.run(close['a'], pos_th=1, neg_th=0.5).plot().show_png()
print(vbt.TRENDLB.run(close, pos_th=pos_ths, neg_th=neg_ths, mode='Binary').labels)
%timeit vbt.TRENDLB.run(big_close, pos_th=1, neg_th=0.5, mode='Binary')
print(vbt.TRENDLB.run(big_close, pos_th=1, neg_th=0.5, mode='Binary').wrapper.shape)
vbt.TRENDLB.run(close['a'], pos_th=1, neg_th=0.5, mode='Binary').plot().show_png()
print(vbt.TRENDLB.run(close, pos_th=pos_ths, neg_th=neg_ths, mode='BinaryCont').labels)
%timeit vbt.TRENDLB.run(big_close, pos_th=1, neg_th=0.5, mode='BinaryCont')
print(vbt.TRENDLB.run(big_close, pos_th=1, neg_th=0.5, mode='BinaryCont').wrapper.shape)
vbt.TRENDLB.run(close['a'], pos_th=1, neg_th=0.5, mode='BinaryCont').plot().show_png()
print(vbt.TRENDLB.run(close, pos_th=pos_ths, neg_th=neg_ths, mode='BinaryContSat').labels)
%timeit vbt.TRENDLB.run(big_close, pos_th=1, neg_th=0.5, mode='BinaryContSat')
print(vbt.TRENDLB.run(big_close, pos_th=1, neg_th=0.5, mode='BinaryContSat').wrapper.shape)
vbt.TRENDLB.run(close['a'], pos_th=1, neg_th=0.5, mode='BinaryContSat').plot().show_png()
print(vbt.TRENDLB.run(close, pos_th=pos_ths, neg_th=neg_ths, mode='PctChange').labels)
%timeit vbt.TRENDLB.run(big_close, pos_th=1, neg_th=0.5, mode='PctChange')
print(vbt.TRENDLB.run(big_close, pos_th=1, neg_th=0.5, mode='PctChange').wrapper.shape)
vbt.TRENDLB.run(close['a'], pos_th=1, neg_th=0.5, mode='PctChange').plot().show_png()
print(vbt.TRENDLB.run(close, pos_th=pos_ths, neg_th=neg_ths, mode='PctChangeNorm').labels)
%timeit vbt.TRENDLB.run(big_close, pos_th=1, neg_th=0.5, mode='PctChangeNorm')
print(vbt.TRENDLB.run(big_close, pos_th=1, neg_th=0.5, mode='PctChangeNorm').wrapper.shape)
vbt.TRENDLB.run(close['a'], pos_th=1, neg_th=0.5, mode='PctChangeNorm').plot().show_png()
print(vbt.BOLB.run(close, window=1, pos_th=pos_ths, neg_th=neg_ths).labels)
print(vbt.BOLB.run(close, window=2, pos_th=pos_ths, neg_th=neg_ths).labels)
%timeit vbt.BOLB.run(big_close, window=2, pos_th=1, neg_th=0.5)
%timeit vbt.BOLB.run(big_close, window=np.arange(2, 10).tolist(), pos_th=1, neg_th=0.5)
print(vbt.BOLB.run(big_close, window=np.arange(2, 10).tolist(), pos_th=1, neg_th=0.5).wrapper.shape)
vbt.BOLB.run(close['a'], window=2, pos_th=1, neg_th=0.5).plot().show_png()
| 0.345216 | 0.860428 |
# Table of Contents
<p><div class="lev1 toc-item"><a href="#Loading-libraries" data-toc-modified-id="Loading-libraries-1"><span class="toc-item-num">1 </span>Loading libraries</a></div><div class="lev1 toc-item"><a href="#Creating-the-model" data-toc-modified-id="Creating-the-model-2"><span class="toc-item-num">2 </span>Creating the model</a></div><div class="lev1 toc-item"><a href="#Training-400x300" data-toc-modified-id="Training-400x300-3"><span class="toc-item-num">3 </span>Training 400x300</a></div><div class="lev1 toc-item"><a href="#Predictions" data-toc-modified-id="Predictions-4"><span class="toc-item-num">4 </span>Predictions</a></div><div class="lev1 toc-item"><a href="#Training-600x450" data-toc-modified-id="Training-600x450-5"><span class="toc-item-num">5 </span>Training 600x450</a></div><div class="lev1 toc-item"><a href="#Predictions" data-toc-modified-id="Predictions-6"><span class="toc-item-num">6 </span>Predictions</a></div>
# Loading libraries
```
from keras.preprocessing.image import ImageDataGenerator
from keras.preprocessing import image
from keras.models import Sequential, load_model, Model
from keras.layers import Activation, Dropout, Flatten, Dense, GlobalAveragePooling2D
from keras.optimizers import SGD
from keras.callbacks import EarlyStopping, ModelCheckpoint
from keras.applications.inception_v3 import InceptionV3
from keras_tqdm import TQDMNotebookCallback
from datetime import datetime
import os
import numpy as np
import pandas as pd
import math
pd.options.display.max_rows = 40
```
# Creating the model
```
base_model = InceptionV3(include_top = False,
weights = 'imagenet')
base_model.summary()
x = base_model.output
x = GlobalAveragePooling2D()(x)
x = Dense(1024, activation = 'relu')(x)
x = Dense(1, activation = 'sigmoid')(x)
model_final = Model(inputs=base_model.input, outputs=x)
model_final.compile(loss = 'binary_crossentropy',
optimizer = SGD(lr = 0.0001, momentum = 0.9, decay = 1e-5),
metrics = ['accuracy'])
model_final.summary()
datagen = ImageDataGenerator(
rotation_range = 20,
width_shift_range = 0.2,
height_shift_range = 0.2,
horizontal_flip = True)
validgen = ImageDataGenerator()
```
# Training 400x300
```
# 600/450 _ 500/375 _ 400/300 _ 300/225
img_width = 400
img_height = 300
train_data_dir = "data/train"
validation_data_dir = "data/valid"
test_data_dir = "data/test"
batch_size_train = 16
batch_size_val = 32
train_gen = datagen.flow_from_directory(
directory = train_data_dir,
target_size = (img_height, img_width),
batch_size = batch_size_train,
class_mode = "binary",
shuffle = True)
val_gen = validgen.flow_from_directory(
directory = validation_data_dir,
target_size = (img_height, img_width),
batch_size = batch_size_val,
class_mode = "binary",
shuffle = False)
train_samples = len(train_gen.filenames)
validation_samples = len(val_gen.filenames)
checkpoint = ModelCheckpoint("weights-iter-4-epoch-{epoch:02d}.hdf5",
monitor='val_acc',
verbose=0,
save_best_only=False,
save_weights_only=True)
early_stopping = EarlyStopping(monitor='val_loss', patience=3, verbose=1, mode='auto')
model_final.fit_generator(generator = train_gen,
epochs = 40,
steps_per_epoch = math.ceil(train_samples / batch_size_train),
validation_data = val_gen,
validation_steps = math.ceil(validation_samples / batch_size_val),
verbose = 2,
callbacks = [early_stopping, TQDMNotebookCallback(), checkpoint])
model_final.load_weights('weights-iter-4-epoch-39.hdf5')
```
# Predictions
```
batch_size_test = 64
test_gen = validgen.flow_from_directory(
directory = test_data_dir,
target_size = (img_height, img_width),
batch_size = batch_size_test,
class_mode = "binary",
shuffle = False)
test_samples = len(test_gen.filenames)
preds = model_final.predict_generator(test_gen, math.ceil(test_samples / batch_size_test))
preds_filenames = test_gen.filenames
preds_filenames = [int(x.replace("unknown/", "").replace(".jpg", "")) for x in preds_filenames]
df_result = pd.DataFrame({'name': preds_filenames, 'invasive': preds[:,0]})
df_result = df_result.sort_values("name")
df_result.index = df_result["name"]
df_result = df_result.drop(["name"], axis=1)
df_result.to_csv("submission_02.csv", encoding="utf8", index=True)
from IPython.display import FileLink
FileLink('submission_02.csv')
# Got 0.99246 on LB
```
# Training 600x450
```
# 600/450 _ 500/375 _ 400/300 _ 300/225
img_width = 600
img_height = 450
train_data_dir = "data/train"
validation_data_dir = "data/valid"
test_data_dir = "data/test"
batch_size_train = 16
batch_size_val = 32
train_gen = datagen.flow_from_directory(
directory = train_data_dir,
target_size = (img_height, img_width),
batch_size = batch_size_train,
class_mode = "binary",
shuffle = True)
val_gen = validgen.flow_from_directory(
directory = validation_data_dir,
target_size = (img_height, img_width),
batch_size = batch_size_val,
class_mode = "binary",
shuffle = False)
train_samples = len(train_gen.filenames)
validation_samples = len(val_gen.filenames)
checkpoint = ModelCheckpoint("weights-iter-5-epoch-{epoch:02d}.hdf5",
monitor='val_acc',
verbose=0,
save_best_only=False,
save_weights_only=True)
early_stopping = EarlyStopping(monitor='val_loss', patience=3, verbose=1, mode='auto')
model_final.fit_generator(generator = train_gen,
epochs = 40,
steps_per_epoch = math.ceil(train_samples / batch_size_train),
validation_data = val_gen,
validation_steps = math.ceil(validation_samples / batch_size_val),
verbose = 2,
callbacks = [early_stopping, TQDMNotebookCallback(), checkpoint])
model_final.load_weights('weights-iter-5-epoch-32.hdf5')
model_final.evaluate_generator(val_gen, math.ceil(validation_samples / batch_size_val))
```
# Predictions
```
batch_size_test = 32
test_gen = validgen.flow_from_directory(
directory = test_data_dir,
target_size = (img_height, img_width),
batch_size = batch_size_test,
class_mode = "binary",
shuffle = False)
test_samples = len(test_gen.filenames)
preds = model_final.predict_generator(test_gen, math.ceil(test_samples / batch_size_test))
preds_filenames = test_gen.filenames
preds_filenames = [int(x.replace("unknown/", "").replace(".jpg", "")) for x in preds_filenames]
df_result = pd.DataFrame({'name': preds_filenames, 'invasive': preds[:,0]})
df_result = df_result.sort_values("name")
df_result.index = df_result["name"]
df_result = df_result.drop(["name"], axis=1)
df_result.to_csv("submission_03.csv", encoding="utf8", index=True)
from IPython.display import FileLink
FileLink('submission_03.csv')
# Got 0.99454 on LB
```
|
github_jupyter
|
from keras.preprocessing.image import ImageDataGenerator
from keras.preprocessing import image
from keras.models import Sequential, load_model, Model
from keras.layers import Activation, Dropout, Flatten, Dense, GlobalAveragePooling2D
from keras.optimizers import SGD
from keras.callbacks import EarlyStopping, ModelCheckpoint
from keras.applications.inception_v3 import InceptionV3
from keras_tqdm import TQDMNotebookCallback
from datetime import datetime
import os
import numpy as np
import pandas as pd
import math
pd.options.display.max_rows = 40
base_model = InceptionV3(include_top = False,
weights = 'imagenet')
base_model.summary()
x = base_model.output
x = GlobalAveragePooling2D()(x)
x = Dense(1024, activation = 'relu')(x)
x = Dense(1, activation = 'sigmoid')(x)
model_final = Model(inputs=base_model.input, outputs=x)
model_final.compile(loss = 'binary_crossentropy',
optimizer = SGD(lr = 0.0001, momentum = 0.9, decay = 1e-5),
metrics = ['accuracy'])
model_final.summary()
datagen = ImageDataGenerator(
rotation_range = 20,
width_shift_range = 0.2,
height_shift_range = 0.2,
horizontal_flip = True)
validgen = ImageDataGenerator()
# 600/450 _ 500/375 _ 400/300 _ 300/225
img_width = 400
img_height = 300
train_data_dir = "data/train"
validation_data_dir = "data/valid"
test_data_dir = "data/test"
batch_size_train = 16
batch_size_val = 32
train_gen = datagen.flow_from_directory(
directory = train_data_dir,
target_size = (img_height, img_width),
batch_size = batch_size_train,
class_mode = "binary",
shuffle = True)
val_gen = validgen.flow_from_directory(
directory = validation_data_dir,
target_size = (img_height, img_width),
batch_size = batch_size_val,
class_mode = "binary",
shuffle = False)
train_samples = len(train_gen.filenames)
validation_samples = len(val_gen.filenames)
checkpoint = ModelCheckpoint("weights-iter-4-epoch-{epoch:02d}.hdf5",
monitor='val_acc',
verbose=0,
save_best_only=False,
save_weights_only=True)
early_stopping = EarlyStopping(monitor='val_loss', patience=3, verbose=1, mode='auto')
model_final.fit_generator(generator = train_gen,
epochs = 40,
steps_per_epoch = math.ceil(train_samples / batch_size_train),
validation_data = val_gen,
validation_steps = math.ceil(validation_samples / batch_size_val),
verbose = 2,
callbacks = [early_stopping, TQDMNotebookCallback(), checkpoint])
model_final.load_weights('weights-iter-4-epoch-39.hdf5')
batch_size_test = 64
test_gen = validgen.flow_from_directory(
directory = test_data_dir,
target_size = (img_height, img_width),
batch_size = batch_size_test,
class_mode = "binary",
shuffle = False)
test_samples = len(test_gen.filenames)
preds = model_final.predict_generator(test_gen, math.ceil(test_samples / batch_size_test))
preds_filenames = test_gen.filenames
preds_filenames = [int(x.replace("unknown/", "").replace(".jpg", "")) for x in preds_filenames]
df_result = pd.DataFrame({'name': preds_filenames, 'invasive': preds[:,0]})
df_result = df_result.sort_values("name")
df_result.index = df_result["name"]
df_result = df_result.drop(["name"], axis=1)
df_result.to_csv("submission_02.csv", encoding="utf8", index=True)
from IPython.display import FileLink
FileLink('submission_02.csv')
# Got 0.99246 on LB
# 600/450 _ 500/375 _ 400/300 _ 300/225
img_width = 600
img_height = 450
train_data_dir = "data/train"
validation_data_dir = "data/valid"
test_data_dir = "data/test"
batch_size_train = 16
batch_size_val = 32
train_gen = datagen.flow_from_directory(
directory = train_data_dir,
target_size = (img_height, img_width),
batch_size = batch_size_train,
class_mode = "binary",
shuffle = True)
val_gen = validgen.flow_from_directory(
directory = validation_data_dir,
target_size = (img_height, img_width),
batch_size = batch_size_val,
class_mode = "binary",
shuffle = False)
train_samples = len(train_gen.filenames)
validation_samples = len(val_gen.filenames)
checkpoint = ModelCheckpoint("weights-iter-5-epoch-{epoch:02d}.hdf5",
monitor='val_acc',
verbose=0,
save_best_only=False,
save_weights_only=True)
early_stopping = EarlyStopping(monitor='val_loss', patience=3, verbose=1, mode='auto')
model_final.fit_generator(generator = train_gen,
epochs = 40,
steps_per_epoch = math.ceil(train_samples / batch_size_train),
validation_data = val_gen,
validation_steps = math.ceil(validation_samples / batch_size_val),
verbose = 2,
callbacks = [early_stopping, TQDMNotebookCallback(), checkpoint])
model_final.load_weights('weights-iter-5-epoch-32.hdf5')
model_final.evaluate_generator(val_gen, math.ceil(validation_samples / batch_size_val))
batch_size_test = 32
test_gen = validgen.flow_from_directory(
directory = test_data_dir,
target_size = (img_height, img_width),
batch_size = batch_size_test,
class_mode = "binary",
shuffle = False)
test_samples = len(test_gen.filenames)
preds = model_final.predict_generator(test_gen, math.ceil(test_samples / batch_size_test))
preds_filenames = test_gen.filenames
preds_filenames = [int(x.replace("unknown/", "").replace(".jpg", "")) for x in preds_filenames]
df_result = pd.DataFrame({'name': preds_filenames, 'invasive': preds[:,0]})
df_result = df_result.sort_values("name")
df_result.index = df_result["name"]
df_result = df_result.drop(["name"], axis=1)
df_result.to_csv("submission_03.csv", encoding="utf8", index=True)
from IPython.display import FileLink
FileLink('submission_03.csv')
# Got 0.99454 on LB
| 0.805976 | 0.8874 |
```
import numpy as np
import pandas as pd
import os
import re
files = os.listdir('data/answers')
output_names = ['toxic','severe_toxic','obscene','threat','insult','identity_hate']
files
def blend(outName = '' , filtering = ['lstm', 'gru'], topn = None, weighted = False, weightedMin=0.95, ):
'''
Inputs:
outName: output file name
filtering: list of regular expressions
topn: int or None. whether or not to take n scores after regex matching
weighted: whether to use a simple average or a weighted average
weightedMin: score to subtract all scores from before scaling sums of scores to one
'''
answer = pd.DataFrame(columns = ['id'] + output_names)
filesToRead = []
for i in files:
if any([re.search(j, i) for j in filtering]):
filesToRead.append(i)
scores = {}
for i in filesToRead:
scores[i] = float('0.' + re.findall(r'_([0-9]+)\.csv',i)[0][1:])
if topn:
toTake = list(zip(*sorted(scores.items(), key = lambda x: -x[1])[:topn]))[0]
else:
toTake = filesToRead
preds = {}
for i in toTake:
preds[i] = pd.read_csv('data/answers/' + i)
preds[i] = preds[i].sort_values(by = 'id')
answer['id'] = preds[i]['id']
results = np.zeros(shape = (preds[i].shape[0], preds[i].shape[1] - 1, len(toTake)))
for c, i in enumerate(preds):
results[:,:,c] = preds[i][output_names].values
if not weighted:
answer[output_names] = np.mean(results, axis = -1)
else:
assert(all([scores[i]-weightedMin >= 0 for i in toTake]))
total = sum([scores[i] - weightedMin for i in toTake])
scalings = [(scores[i] - weightedMin)/total for i in toTake]
for i in range(len(toTake)):
results[:,:,i] *= scalings[i]
answer[output_names] = np.sum(results, axis = -1)
answer.to_csv('data/answers/ensembles/'+outName, index = False)
blend('allUnweighted.csv', filtering = ['.*'])
blend('allWeighted095.csv', filtering = ['.*'], weighted = True, weightedMin=0.95)
blend('allWeighted096.csv', filtering = ['.*'], weighted = True, weightedMin=0.96)
for i in range(3, 12,2): #unweighted GRUs and LSTMs
blend('unweightedRNNTop'+str(i) + '.csv', topn= i )
for i in range(3, 12,2): #weighted GRUs and LSTMs
blend('weightedRNN098Top'+str(i) + '.csv', topn= i, weighted = True, weightedMin= 0.98)
blend('weightedRNN097Top'+str(i) + '.csv', topn= i, weighted = True, weightedMin= 0.97)
```
|
github_jupyter
|
import numpy as np
import pandas as pd
import os
import re
files = os.listdir('data/answers')
output_names = ['toxic','severe_toxic','obscene','threat','insult','identity_hate']
files
def blend(outName = '' , filtering = ['lstm', 'gru'], topn = None, weighted = False, weightedMin=0.95, ):
'''
Inputs:
outName: output file name
filtering: list of regular expressions
topn: int or None. whether or not to take n scores after regex matching
weighted: whether to use a simple average or a weighted average
weightedMin: score to subtract all scores from before scaling sums of scores to one
'''
answer = pd.DataFrame(columns = ['id'] + output_names)
filesToRead = []
for i in files:
if any([re.search(j, i) for j in filtering]):
filesToRead.append(i)
scores = {}
for i in filesToRead:
scores[i] = float('0.' + re.findall(r'_([0-9]+)\.csv',i)[0][1:])
if topn:
toTake = list(zip(*sorted(scores.items(), key = lambda x: -x[1])[:topn]))[0]
else:
toTake = filesToRead
preds = {}
for i in toTake:
preds[i] = pd.read_csv('data/answers/' + i)
preds[i] = preds[i].sort_values(by = 'id')
answer['id'] = preds[i]['id']
results = np.zeros(shape = (preds[i].shape[0], preds[i].shape[1] - 1, len(toTake)))
for c, i in enumerate(preds):
results[:,:,c] = preds[i][output_names].values
if not weighted:
answer[output_names] = np.mean(results, axis = -1)
else:
assert(all([scores[i]-weightedMin >= 0 for i in toTake]))
total = sum([scores[i] - weightedMin for i in toTake])
scalings = [(scores[i] - weightedMin)/total for i in toTake]
for i in range(len(toTake)):
results[:,:,i] *= scalings[i]
answer[output_names] = np.sum(results, axis = -1)
answer.to_csv('data/answers/ensembles/'+outName, index = False)
blend('allUnweighted.csv', filtering = ['.*'])
blend('allWeighted095.csv', filtering = ['.*'], weighted = True, weightedMin=0.95)
blend('allWeighted096.csv', filtering = ['.*'], weighted = True, weightedMin=0.96)
for i in range(3, 12,2): #unweighted GRUs and LSTMs
blend('unweightedRNNTop'+str(i) + '.csv', topn= i )
for i in range(3, 12,2): #weighted GRUs and LSTMs
blend('weightedRNN098Top'+str(i) + '.csv', topn= i, weighted = True, weightedMin= 0.98)
blend('weightedRNN097Top'+str(i) + '.csv', topn= i, weighted = True, weightedMin= 0.97)
| 0.320502 | 0.244453 |
# Creating a RO-crate entry and serializing it in JSON-LD
https://researchobject.github.io/ro-crate/
```
from rdflib import *
from datetime import datetime
schema = Namespace("http://schema.org/")
```
# Writing RDF triples to populate a minimal RO-crate
```
graph = ConjunctiveGraph()
#graph.bind('foaf', 'http://xmlns.com/foaf/0.1/')
graph.load('https://researchobject.github.io/ro-crate/0.2/context.json', format='json-ld')
# person information
graph.add( (URIRef('https://orcid.org/0000-0002-3597-8557'), RDF.type, schema.Person) )
# contact information
graph.add( (URIRef('alban.gaignard@univ-nantes.fr'), RDF.type, schema.ContactPoint) )
graph.add((URIRef('alban.gaignard@univ-nantes.fr'), schema.contactType, Literal('Developer')) )
graph.add( (URIRef('alban.gaignard@univ-nantes.fr'), schema.name, Literal('Alban Gaignard')) )
graph.add( (URIRef('alban.gaignard@univ-nantes.fr'), schema.email, Literal('alban.gaignard@univ-nantes.fr', datatype=XSD.string)) )
graph.add( (URIRef('alban.gaignard@univ-nantes.fr'), schema.url, Literal('https://orcid.org/0000-0002-3597-8557')) )
# root metadata
graph.add( (URIRef('ro-crate-metadata.jsonld'), RDF.type, schema.CreativeWork) )
graph.add( (URIRef('ro-crate-metadata.jsonld'), schema.identifier, Literal('ro-crate-metadata.jsonld')) )
graph.add( (URIRef('ro-crate-metadata.jsonld'), schema.about, URIRef('./')) )
# Dataset metadata with reference to files
graph.add( (URIRef('./'), RDF.type, schema.Dataset) )
graph.add( (URIRef('./'), schema.name, Literal("workfow outputs")) )
graph.add( (URIRef('./'), schema.datePublished, Literal(datetime.now().isoformat())) )
graph.add( (URIRef('./'), schema.author, URIRef('https://orcid.org/0000-0002-3597-8557')) )
graph.add( (URIRef('./'), schema.contactPoint, URIRef('alban.gaignard@univ-nantes.fr')) )
graph.add( (URIRef('./'), schema.description, Literal("this is the description of the workfow description, this is the description of the workfow description, this is the description of the workfow description")) )
graph.add( (URIRef('./'), schema.license, Literal("MIT?")) )
graph.add( (URIRef('./'), schema.hasPart, (URIRef('./data/provenance.ttl'))) )
# Files metadata
graph.add( (URIRef('./data/provenance.ttl'), RDF.type, schema.MediaObject) )
print(graph.serialize(format='turtle').decode())
#print(graph.serialize(format='json-ld').decode())
```
# Warpping these triples into Python objects
```
import requests
import json
class RO_crate_abstract:
"""
An abstract RO-crate class to share common attributes and methods.
"""
def __init__(self, uri):
self.uri = uri
self.graph = ConjunctiveGraph()
def get_uri(self):
return self.uri
def print(self):
print(self.graph.serialize(format='turtle').decode())
def serialize_jsonld(self):
res = requests.get('https://w3id.org/ro/crate/1.0/context')
ctx = json.loads(res.text)['@context']
jsonld = self.graph.serialize(format='json-ld', context=ctx)
print(jsonld.decode())
self.graph.serialize(destination='ro-crate-metadata.jsonld', format='json-ld', context=ctx)
def add_has_part(self, other_ro_crate):
self.graph = self.graph + other_ro_crate.graph
#TODO add has_part property
self.graph.add( (URIRef(self.get_uri()), schema.hasPart, URIRef(other_ro_crate.get_uri())) )
class RO_crate_Root(RO_crate_abstract):
"""
The root RO-crate.
"""
def __init__(self):
RO_crate_abstract.__init__(self, uri='ro-crate-metadata.jsonld')
self.graph.add( (URIRef('ro-crate-metadata.jsonld'), RDF.type, schema.CreativeWork) )
self.graph.add( (URIRef('ro-crate-metadata.jsonld'), schema.identifier, Literal('ro-crate-metadata.jsonld')) )
self.graph.add( (URIRef('ro-crate-metadata.jsonld'), schema.about, URIRef('./')) )
class RO_crate_Person(RO_crate_abstract):
"""
A person RO-crate.
"""
def __init__(self, uri):
RO_crate_abstract.__init__(self, uri)
self.graph.add( (URIRef(uri), RDF.type, schema.Person) )
class RO_crate_Contact(RO_crate_Person):
"""
A person RO-crate.
"""
def __init__(self, uri, name=None, email=None, ctype=None, url=None):
RO_crate_Person.__init__(self, uri)
self.graph.add( (URIRef(uri), RDF.type, schema.Person) )
if name:
self.graph.add( (URIRef(uri), schema.name, Literal(name)) )
if email:
self.graph.add( (URIRef(uri), schema.email, Literal(email, datatype=XSD.string)) )
if ctype:
self.graph.add( (URIRef(uri), schema.contactType, Literal(ctype)) )
if url:
self.graph.add( (URIRef(uri), schema.url, Literal(url)) )
# creating a root RO-crate
root = RO_crate_Root()
root.print()
# creating a person RO-crate
person = RO_crate_Person('https://orcid.org/0000-0002-3597-8557')
person.print()
# creating a contact RO-crate
contact = RO_crate_Contact('https://orcid.org/0000-0002-3597-8557', name='Alban Gaignard', ctype='contributor')
contact.print()
# adding hasPart relation between RO-crates
root.add_has_part(contact)
root.print()
# serializing the output
root.serialize_jsonld()
```
|
github_jupyter
|
from rdflib import *
from datetime import datetime
schema = Namespace("http://schema.org/")
graph = ConjunctiveGraph()
#graph.bind('foaf', 'http://xmlns.com/foaf/0.1/')
graph.load('https://researchobject.github.io/ro-crate/0.2/context.json', format='json-ld')
# person information
graph.add( (URIRef('https://orcid.org/0000-0002-3597-8557'), RDF.type, schema.Person) )
# contact information
graph.add( (URIRef('alban.gaignard@univ-nantes.fr'), RDF.type, schema.ContactPoint) )
graph.add((URIRef('alban.gaignard@univ-nantes.fr'), schema.contactType, Literal('Developer')) )
graph.add( (URIRef('alban.gaignard@univ-nantes.fr'), schema.name, Literal('Alban Gaignard')) )
graph.add( (URIRef('alban.gaignard@univ-nantes.fr'), schema.email, Literal('alban.gaignard@univ-nantes.fr', datatype=XSD.string)) )
graph.add( (URIRef('alban.gaignard@univ-nantes.fr'), schema.url, Literal('https://orcid.org/0000-0002-3597-8557')) )
# root metadata
graph.add( (URIRef('ro-crate-metadata.jsonld'), RDF.type, schema.CreativeWork) )
graph.add( (URIRef('ro-crate-metadata.jsonld'), schema.identifier, Literal('ro-crate-metadata.jsonld')) )
graph.add( (URIRef('ro-crate-metadata.jsonld'), schema.about, URIRef('./')) )
# Dataset metadata with reference to files
graph.add( (URIRef('./'), RDF.type, schema.Dataset) )
graph.add( (URIRef('./'), schema.name, Literal("workfow outputs")) )
graph.add( (URIRef('./'), schema.datePublished, Literal(datetime.now().isoformat())) )
graph.add( (URIRef('./'), schema.author, URIRef('https://orcid.org/0000-0002-3597-8557')) )
graph.add( (URIRef('./'), schema.contactPoint, URIRef('alban.gaignard@univ-nantes.fr')) )
graph.add( (URIRef('./'), schema.description, Literal("this is the description of the workfow description, this is the description of the workfow description, this is the description of the workfow description")) )
graph.add( (URIRef('./'), schema.license, Literal("MIT?")) )
graph.add( (URIRef('./'), schema.hasPart, (URIRef('./data/provenance.ttl'))) )
# Files metadata
graph.add( (URIRef('./data/provenance.ttl'), RDF.type, schema.MediaObject) )
print(graph.serialize(format='turtle').decode())
#print(graph.serialize(format='json-ld').decode())
import requests
import json
class RO_crate_abstract:
"""
An abstract RO-crate class to share common attributes and methods.
"""
def __init__(self, uri):
self.uri = uri
self.graph = ConjunctiveGraph()
def get_uri(self):
return self.uri
def print(self):
print(self.graph.serialize(format='turtle').decode())
def serialize_jsonld(self):
res = requests.get('https://w3id.org/ro/crate/1.0/context')
ctx = json.loads(res.text)['@context']
jsonld = self.graph.serialize(format='json-ld', context=ctx)
print(jsonld.decode())
self.graph.serialize(destination='ro-crate-metadata.jsonld', format='json-ld', context=ctx)
def add_has_part(self, other_ro_crate):
self.graph = self.graph + other_ro_crate.graph
#TODO add has_part property
self.graph.add( (URIRef(self.get_uri()), schema.hasPart, URIRef(other_ro_crate.get_uri())) )
class RO_crate_Root(RO_crate_abstract):
"""
The root RO-crate.
"""
def __init__(self):
RO_crate_abstract.__init__(self, uri='ro-crate-metadata.jsonld')
self.graph.add( (URIRef('ro-crate-metadata.jsonld'), RDF.type, schema.CreativeWork) )
self.graph.add( (URIRef('ro-crate-metadata.jsonld'), schema.identifier, Literal('ro-crate-metadata.jsonld')) )
self.graph.add( (URIRef('ro-crate-metadata.jsonld'), schema.about, URIRef('./')) )
class RO_crate_Person(RO_crate_abstract):
"""
A person RO-crate.
"""
def __init__(self, uri):
RO_crate_abstract.__init__(self, uri)
self.graph.add( (URIRef(uri), RDF.type, schema.Person) )
class RO_crate_Contact(RO_crate_Person):
"""
A person RO-crate.
"""
def __init__(self, uri, name=None, email=None, ctype=None, url=None):
RO_crate_Person.__init__(self, uri)
self.graph.add( (URIRef(uri), RDF.type, schema.Person) )
if name:
self.graph.add( (URIRef(uri), schema.name, Literal(name)) )
if email:
self.graph.add( (URIRef(uri), schema.email, Literal(email, datatype=XSD.string)) )
if ctype:
self.graph.add( (URIRef(uri), schema.contactType, Literal(ctype)) )
if url:
self.graph.add( (URIRef(uri), schema.url, Literal(url)) )
# creating a root RO-crate
root = RO_crate_Root()
root.print()
# creating a person RO-crate
person = RO_crate_Person('https://orcid.org/0000-0002-3597-8557')
person.print()
# creating a contact RO-crate
contact = RO_crate_Contact('https://orcid.org/0000-0002-3597-8557', name='Alban Gaignard', ctype='contributor')
contact.print()
# adding hasPart relation between RO-crates
root.add_has_part(contact)
root.print()
# serializing the output
root.serialize_jsonld()
| 0.321886 | 0.529811 |
```
import math
import re
import os
from IPython import display
from matplotlib import cm
from matplotlib import gridspec
from matplotlib import pyplot as plt
import numpy as np
from urlparse import urlparse
import pandas as pd
from sklearn import metrics
import tensorflow as tf
from tensorflow.python.data import Dataset
from sklearn.preprocessing import StandardScaler
tf.logging.set_verbosity(tf.logging.ERROR)
pd.options.display.max_rows = 10
pd.options.display.float_format = '{:.1f}'.format
links_df = pd.read_csv("product_links_df.csv")
from time import time
from random import randint
from sklearn.model_selection import train_test_split, GroupKFold, cross_val_score
import pickle
len(links_df)
links_df.head()
PATH_LEN_CAP=100
def preprocess_features(df, load_scaler_from_file=False):
processed_features = df[["url"]].copy()
processed_features["path"] = processed_features["url"].map(lambda x: urlparse(x).path + urlparse(x).params + urlparse(x).query + urlparse(x).fragment)
processed_features["path_len"] = processed_features["path"].map(lambda x: min(len(x), PATH_LEN_CAP))
processed_features["num_hyphen"] = processed_features["path"].map(lambda x: x.count("-") + x.rstrip("/").count("/"))
#processed_features["num_slash"] = processed_features["path"].map(lambda x: x.rstrip("/").count("/"))
processed_features["contains_product"] = processed_features["path"].map(lambda x: 1 if "product" in x else 0)
processed_features["contains_category"] = processed_features["path"].map(lambda x: 1 if "category" in x else 0)
processed_features["longest_num"] = processed_features["path"].map(lambda x: len(max(re.findall(r'[0-9]+', x), key=len)) if re.search(r'\d', x) else 0)
cols_to_drop = ['url', 'path']
processed_features.drop(cols_to_drop, axis=1, inplace=True)
scaled_features = processed_features.copy()
col_names = [col for col in processed_features if col not in cols_to_drop and not "contains" in col]
features = scaled_features[col_names]
scaler_filename = 'StandardScaler.est'
if load_scaler_from_file and os.path.isfile(scaler_filename):
scaler = StandardScaler()
scaler = StandardScaler().fit(features.values)
pickle.dump(scaler, open(scaler_filename, 'wb'))
else:
scaler = pickle.load(open(scaler_filename, 'rb'))
features = scaler.transform(features.values)
scaled_features[col_names] = features
return scaled_features
def preprocess_targets(df):
"""Prepares target features (i.e., labels) from California housing data set.
Args:
california_housing_dataframe: A Pandas DataFrame expected to contain data
from the California housing data set.
Returns:
A DataFrame that contains the target feature.
"""
output_targets = pd.DataFrame()
# Create a boolean categorical feature representing whether the
# median_house_value is above a set threshold.
output_targets["label"] = df["label"].astype(int)
#output_targets["median_house_value_is_high"] = (
# california_housing_dataframe["median_house_value"] > 265000).astype(float)
return output_targets
def get_groups(df):
return df["domain"].values
# Choose the first 90% of the examples for training.
n_links = len(links_df)
train_len = int(math.floor(0.9*n_links))
validation_len = int(n_links - train_len)
print "train_len", train_len, "validation_len", validation_len
training_input = links_df.head(train_len)
validation_input = links_df.tail(validation_len)
training_input = training_input.reindex(
np.random.permutation(training_input.index))
validation_input = validation_input.reindex(
np.random.permutation(validation_input.index))
training_examples = preprocess_features(training_input)
training_targets = preprocess_targets(training_input)
training_groups = get_groups(training_input)
# Choose the last 30% of the examples for validation.
validation_examples = preprocess_features(validation_input)
validation_targets = preprocess_targets(validation_input)
print("Training examples summary:")
display.display(training_examples.describe())
print("Validation examples summary:")
display.display(validation_examples.describe())
print("Training targets summary:")
display.display(training_targets.describe())
print("Validation targets summary:")
display.display(validation_targets.describe())
from sklearn.linear_model import LogisticRegression, LogisticRegressionCV, SGDClassifier
training_input.head()
gkf = GroupKFold(n_splits=5)
clf = LogisticRegression(solver="lbfgs", C=0.05, penalty="l2").fit(training_examples, training_targets.values)
#sgd_clf = SGDClassifier(loss="log", max_iter=1000, eta0=0.0002, learning_rate="adaptive").fit(training_examples, training_targets.values)
sgd_clf = SGDClassifier(loss="log", max_iter=10000, alpha=0.01,
learning_rate="optimal").\
fit(training_examples, training_targets.values)
logit_scores = cross_val_score(clf, training_examples,
training_targets.values,
cv=gkf, groups=training_groups)
sgd_scores = cross_val_score(sgd_clf, training_examples,
training_targets.values,
cv=gkf, groups=training_groups)
print("Logit Accuracy: %0.2f (+/- %0.2f)" % (logit_scores.mean(), logit_scores.std() * 2))
print("SGD Accuracy: %0.2f (+/- %0.2f)" % (sgd_scores.mean(), sgd_scores.std() * 2))
print "Logit", "%0.2f" % clf.score(validation_examples, validation_targets)
print "SGD", "%0.2f" % sgd_clf.score(validation_examples, validation_targets)
model_filename = 'SGDClassifier.est'
pickle.dump(sgd_clf, open(model_filename, 'wb'))
sgd_est = pickle.load(open(model_filename, 'rb'))
sgd_est.score(validation_examples, validation_targets)
urls= ["https://www.bobswatches.com/used-rolex-daytona-116500-black-ceramic-bezel-white-dial.html",
"https://www.ebay.com/itm/Samsung-QN75Q8FN-75-Smart-QLED-4K-Ultra-HD-TV-with-HDR/273174766800",
"https://www.etsy.com/listing/581066860/personalized-unique-best-friends-forever",
"https://loveandlinen.co/collections/womens-graphic-tees/products/zihuatanejo-mexico-womens-fit-t-shirt",
"https://24-style.com/products/marine-shark-socks",
"https://www.maxshop.com/shop/essentials/cami-singlets/essential-reversible-cami/blush?refSrc=230128GIF&nosto=productpage-nosto-2",
"https://teeherivar.com/product/i-find-myself-to-be-exorbitantly-superannuated-for-this-feculence",
"https://shop.tilleyangling.com/products/warm-winter-double-fleece-bally",
"https://amzerprint.com/products/hearts-within-a-heart-slim-designer-cover-for-huawei-honor-6a",
"https://www.xfyro.com/products/xfyro-xs2-2-pack-bundle",
"https://www.macys.com/shop/product/circus-by-sam-edelman-kirby-booties-created-for-macys?ID=6636316&CategoryID=13616",
"https://www.lordandtaylor.com/jones-new-york-textured-plaid-coat/product/0500088736033",
"https://www.amazon.com/Linksys-Tri-Band-Intelligent-bedrooms-Multi-Story/dp/B01N2NLNEH?ref_=Oct_DLandingS_PC_NA_NA&smid=ATVPDKIKX0DER",
"https://www.forever21.com/eu/shop/catalog/product/f21/women-new-arrivals/2000291064",
"https://www.target.com/p/58-barn-door-tv-stand-with-side-doors-saracina-home/-/A-53151115?preselect=52182076#lnk=sametab",
"https://jet.com/product/Samsung-Galaxy-Kids-Tab-E-Lite-7-Inch-8GB-Wi-Fi-Tablet-Cream-White/8c0932f035b7495bb7fefcd4c77d19bf?beaconId=e24d0655-917a-448e-8daa-7047326ec99e%2F2%2Fx~8c0932f035b7495bb7fefcd4c77d19bf&experienceId=26",
"https://www.zaful.com/coffee-letter-graphic-sweatshirt-p_617651.html",
"https://www.bedbathandbeyond.com/store/product/crosley-lydia-bath-cabinet/3259113?poc=215225",
"https://www.aeropostale.com/low-rise-bootcut-jean/87084550.html?dwvar_87084550_color=189&cgid=jeans-girls#start=1",
"https://www.customink.com/products/styles/bella-+-canvas-tri-blend-t-shirt/242000",
"https://tepui.com/products/explorer-series-kukenam-3",
"https://voe21.com/collections/all-products-1/products/the-void",
"https://www.boohoo.com/mid-rise-marble-wash-mom-jeans/DZZ66784.html",
"https://faradayscienceshop.com/collections/frontpage/products/air-swimmer-the-remote-controlled-fish-blimp",
"https://lebrontshirtsla.com/products/lebron-james-lakers-t-shirt-witness",
"https://www.mightyape.com.au/product/2tb-wd-elements-portable-harddrive/26826365",
"https://www.the-house.com/vn3voss04fd18zz-vans-t-shirts.html",
"https://www.jcpenney.com/p/xersion-long-sleeve-performance-tee/ppr5007697784?pTmplType=regular&cm_re=ZH-_-hotdeals-_-WOMEN-ACTIVE-DEALZONE%7C5%7C%26rrec%3Dtrue%26rrplacementtype%3Dnorecs&",
"https://throwbackjerseys.com/collections/bryant/products/light-blue-los-angeles-bryant-8-basketball-throwback-jersey",
"http://www.darkknightarmoury.com/p-11380-nobles-leather-arm-bracers.aspx",
"http://www.printpapa.com/eshop/pc/16-Page-Booklet-5-5x8-5-338p1662.htm",
"http://www.robertgraham.us/men/accessories/sunglasses/rob-sebastian-sunglasses-robsebasd570.html",
"https://buffusa.com/buff-products/hats/knitted-polar-hat/agna-sand/117849.302",
"https://elixinol.com/?p=793",
"https://forbiddenplanet.com/107164-very-naughty-boys-amazing-true-story-of-handmade-films/",
"https://gearclub.vitalmtb.com/product/gear-club-box-4-june/",
"https://glorycycles.com/cane-creek-eewings-titanium-crank/",
"https://shop.mochithings.com/products/15768",
"https://telescopes.net/store/baader-planetarium-100mm-aperture-astrosolar-spotter-filter.html",
"https://tkbtrading.com/collections/all-colors/products/passion-orange",
"https://www.6ku.com/collections/parts-accessories/products/6ku-pedal-straps",
"https://www.alexandermcqueen.com/Item/index?cod10=34854999UH&siteCode=ALEXANDERMCQUEEN_US",
"https://www.bestbuyautoequipment.com/Chassis-Liner-16-Revolution-p/cl832000.htm",
"https://www.clarks.in/Amali-Ice-8298.html",
"https://www.deliciousseeds.com/del_en/amnesia-haze-fem-5-seeds.html",
"https://www.dickssportinggoods.com/p/neosport-womens-neoprene-5mm-jumpsuit-16hndwwn5mmnprnjmwst/16hndwwn5mmnprnjmwst",
"https://www.holabirdsports.com/collections/brand-new-babolat-sfx3/products/babolat-sfx3-all-court-mens-black-silver",
"https://www.jjill.com/ClickInfo?URL=%2fproduct%2fpure-jill-peplum-top%3fevtype%3d%26mpe_id%3d%26intv_id%3d%26storeId%3d%26catalogId%3d%26langId%3d%26experimentId%3d%26testElementId%3d%26controlElement%3d%26expDataType%3d%26expDataUniqueID%3d&evtype=CpgnClick&mpe_id=715854148&intv_id=715851192&storeId=10101&catalogId=10051&langId=-1&expDataType=CatalogEntryId&expDataUniqueID=221289",
"https://www.lacelab.com/collections/3m-reflective-rope-laces/products/cove-blue-3m-reflective-rope-laces",
"https://www.livingsocial.com/deals/4-elements-wellness-center-4",
"https://www.melbournesnowboard.com.au/collections/all/products/burton-moto-boa-2019?variant=12657538465853",
"https://www.pckuwait.com/product/intel-i3-8100-8th-generation-core-i3-processor-3-60ghz-6mb-cache/",
"https://www.proclipusa.com/product/835232-proclip-console-mount",
"https://www.rpphobby.com/product_p/asc7430.htm",
"https://www.seasalt.com/alaea-hawaiian-sea-salt-coarse-grain-grinder-jar.html",
"https://www.shopzerouv.com/collections/the-90s/products/retro-geometric-diamond-shape-sunglasses-c748",
"https://www.swimsuitsforall.com/Aquabelle-Medley-Capri#rrec=true",
"https://www.yoogiscloset.com/handbags/3-1-phillip-lim-black-leather-soleil-large-bucket-drawstring-bag-97627.html",
"https://www.spanx.com/leggings/faux-leather-leggings",
"https://usa.tommy.com/en/men/men-shirts/lewis-hamilton-logo-shirt-mw08299",
"https://www.calvinklein.us/en/mens-clothing/mens-featured-shops-calvin-klein-jeans/slim-fit-archive-western-shirt-22705235",
"http://www2.hm.com/en_us/productpage.0476583002.html"]
df = pd.DataFrame.from_records([(url,) for url in urls], columns=["url"])
X = preprocess_features(df, load_scaler_from_file=True)
sgd_est = pickle.load(open(model_filename, 'rb'))
probs = sgd_est.predict_proba(X.values)
for url, probs in zip(urls, probs):
if probs[1] < 0.55:
print url, probs[1]
```
|
github_jupyter
|
import math
import re
import os
from IPython import display
from matplotlib import cm
from matplotlib import gridspec
from matplotlib import pyplot as plt
import numpy as np
from urlparse import urlparse
import pandas as pd
from sklearn import metrics
import tensorflow as tf
from tensorflow.python.data import Dataset
from sklearn.preprocessing import StandardScaler
tf.logging.set_verbosity(tf.logging.ERROR)
pd.options.display.max_rows = 10
pd.options.display.float_format = '{:.1f}'.format
links_df = pd.read_csv("product_links_df.csv")
from time import time
from random import randint
from sklearn.model_selection import train_test_split, GroupKFold, cross_val_score
import pickle
len(links_df)
links_df.head()
PATH_LEN_CAP=100
def preprocess_features(df, load_scaler_from_file=False):
processed_features = df[["url"]].copy()
processed_features["path"] = processed_features["url"].map(lambda x: urlparse(x).path + urlparse(x).params + urlparse(x).query + urlparse(x).fragment)
processed_features["path_len"] = processed_features["path"].map(lambda x: min(len(x), PATH_LEN_CAP))
processed_features["num_hyphen"] = processed_features["path"].map(lambda x: x.count("-") + x.rstrip("/").count("/"))
#processed_features["num_slash"] = processed_features["path"].map(lambda x: x.rstrip("/").count("/"))
processed_features["contains_product"] = processed_features["path"].map(lambda x: 1 if "product" in x else 0)
processed_features["contains_category"] = processed_features["path"].map(lambda x: 1 if "category" in x else 0)
processed_features["longest_num"] = processed_features["path"].map(lambda x: len(max(re.findall(r'[0-9]+', x), key=len)) if re.search(r'\d', x) else 0)
cols_to_drop = ['url', 'path']
processed_features.drop(cols_to_drop, axis=1, inplace=True)
scaled_features = processed_features.copy()
col_names = [col for col in processed_features if col not in cols_to_drop and not "contains" in col]
features = scaled_features[col_names]
scaler_filename = 'StandardScaler.est'
if load_scaler_from_file and os.path.isfile(scaler_filename):
scaler = StandardScaler()
scaler = StandardScaler().fit(features.values)
pickle.dump(scaler, open(scaler_filename, 'wb'))
else:
scaler = pickle.load(open(scaler_filename, 'rb'))
features = scaler.transform(features.values)
scaled_features[col_names] = features
return scaled_features
def preprocess_targets(df):
"""Prepares target features (i.e., labels) from California housing data set.
Args:
california_housing_dataframe: A Pandas DataFrame expected to contain data
from the California housing data set.
Returns:
A DataFrame that contains the target feature.
"""
output_targets = pd.DataFrame()
# Create a boolean categorical feature representing whether the
# median_house_value is above a set threshold.
output_targets["label"] = df["label"].astype(int)
#output_targets["median_house_value_is_high"] = (
# california_housing_dataframe["median_house_value"] > 265000).astype(float)
return output_targets
def get_groups(df):
return df["domain"].values
# Choose the first 90% of the examples for training.
n_links = len(links_df)
train_len = int(math.floor(0.9*n_links))
validation_len = int(n_links - train_len)
print "train_len", train_len, "validation_len", validation_len
training_input = links_df.head(train_len)
validation_input = links_df.tail(validation_len)
training_input = training_input.reindex(
np.random.permutation(training_input.index))
validation_input = validation_input.reindex(
np.random.permutation(validation_input.index))
training_examples = preprocess_features(training_input)
training_targets = preprocess_targets(training_input)
training_groups = get_groups(training_input)
# Choose the last 30% of the examples for validation.
validation_examples = preprocess_features(validation_input)
validation_targets = preprocess_targets(validation_input)
print("Training examples summary:")
display.display(training_examples.describe())
print("Validation examples summary:")
display.display(validation_examples.describe())
print("Training targets summary:")
display.display(training_targets.describe())
print("Validation targets summary:")
display.display(validation_targets.describe())
from sklearn.linear_model import LogisticRegression, LogisticRegressionCV, SGDClassifier
training_input.head()
gkf = GroupKFold(n_splits=5)
clf = LogisticRegression(solver="lbfgs", C=0.05, penalty="l2").fit(training_examples, training_targets.values)
#sgd_clf = SGDClassifier(loss="log", max_iter=1000, eta0=0.0002, learning_rate="adaptive").fit(training_examples, training_targets.values)
sgd_clf = SGDClassifier(loss="log", max_iter=10000, alpha=0.01,
learning_rate="optimal").\
fit(training_examples, training_targets.values)
logit_scores = cross_val_score(clf, training_examples,
training_targets.values,
cv=gkf, groups=training_groups)
sgd_scores = cross_val_score(sgd_clf, training_examples,
training_targets.values,
cv=gkf, groups=training_groups)
print("Logit Accuracy: %0.2f (+/- %0.2f)" % (logit_scores.mean(), logit_scores.std() * 2))
print("SGD Accuracy: %0.2f (+/- %0.2f)" % (sgd_scores.mean(), sgd_scores.std() * 2))
print "Logit", "%0.2f" % clf.score(validation_examples, validation_targets)
print "SGD", "%0.2f" % sgd_clf.score(validation_examples, validation_targets)
model_filename = 'SGDClassifier.est'
pickle.dump(sgd_clf, open(model_filename, 'wb'))
sgd_est = pickle.load(open(model_filename, 'rb'))
sgd_est.score(validation_examples, validation_targets)
urls= ["https://www.bobswatches.com/used-rolex-daytona-116500-black-ceramic-bezel-white-dial.html",
"https://www.ebay.com/itm/Samsung-QN75Q8FN-75-Smart-QLED-4K-Ultra-HD-TV-with-HDR/273174766800",
"https://www.etsy.com/listing/581066860/personalized-unique-best-friends-forever",
"https://loveandlinen.co/collections/womens-graphic-tees/products/zihuatanejo-mexico-womens-fit-t-shirt",
"https://24-style.com/products/marine-shark-socks",
"https://www.maxshop.com/shop/essentials/cami-singlets/essential-reversible-cami/blush?refSrc=230128GIF&nosto=productpage-nosto-2",
"https://teeherivar.com/product/i-find-myself-to-be-exorbitantly-superannuated-for-this-feculence",
"https://shop.tilleyangling.com/products/warm-winter-double-fleece-bally",
"https://amzerprint.com/products/hearts-within-a-heart-slim-designer-cover-for-huawei-honor-6a",
"https://www.xfyro.com/products/xfyro-xs2-2-pack-bundle",
"https://www.macys.com/shop/product/circus-by-sam-edelman-kirby-booties-created-for-macys?ID=6636316&CategoryID=13616",
"https://www.lordandtaylor.com/jones-new-york-textured-plaid-coat/product/0500088736033",
"https://www.amazon.com/Linksys-Tri-Band-Intelligent-bedrooms-Multi-Story/dp/B01N2NLNEH?ref_=Oct_DLandingS_PC_NA_NA&smid=ATVPDKIKX0DER",
"https://www.forever21.com/eu/shop/catalog/product/f21/women-new-arrivals/2000291064",
"https://www.target.com/p/58-barn-door-tv-stand-with-side-doors-saracina-home/-/A-53151115?preselect=52182076#lnk=sametab",
"https://jet.com/product/Samsung-Galaxy-Kids-Tab-E-Lite-7-Inch-8GB-Wi-Fi-Tablet-Cream-White/8c0932f035b7495bb7fefcd4c77d19bf?beaconId=e24d0655-917a-448e-8daa-7047326ec99e%2F2%2Fx~8c0932f035b7495bb7fefcd4c77d19bf&experienceId=26",
"https://www.zaful.com/coffee-letter-graphic-sweatshirt-p_617651.html",
"https://www.bedbathandbeyond.com/store/product/crosley-lydia-bath-cabinet/3259113?poc=215225",
"https://www.aeropostale.com/low-rise-bootcut-jean/87084550.html?dwvar_87084550_color=189&cgid=jeans-girls#start=1",
"https://www.customink.com/products/styles/bella-+-canvas-tri-blend-t-shirt/242000",
"https://tepui.com/products/explorer-series-kukenam-3",
"https://voe21.com/collections/all-products-1/products/the-void",
"https://www.boohoo.com/mid-rise-marble-wash-mom-jeans/DZZ66784.html",
"https://faradayscienceshop.com/collections/frontpage/products/air-swimmer-the-remote-controlled-fish-blimp",
"https://lebrontshirtsla.com/products/lebron-james-lakers-t-shirt-witness",
"https://www.mightyape.com.au/product/2tb-wd-elements-portable-harddrive/26826365",
"https://www.the-house.com/vn3voss04fd18zz-vans-t-shirts.html",
"https://www.jcpenney.com/p/xersion-long-sleeve-performance-tee/ppr5007697784?pTmplType=regular&cm_re=ZH-_-hotdeals-_-WOMEN-ACTIVE-DEALZONE%7C5%7C%26rrec%3Dtrue%26rrplacementtype%3Dnorecs&",
"https://throwbackjerseys.com/collections/bryant/products/light-blue-los-angeles-bryant-8-basketball-throwback-jersey",
"http://www.darkknightarmoury.com/p-11380-nobles-leather-arm-bracers.aspx",
"http://www.printpapa.com/eshop/pc/16-Page-Booklet-5-5x8-5-338p1662.htm",
"http://www.robertgraham.us/men/accessories/sunglasses/rob-sebastian-sunglasses-robsebasd570.html",
"https://buffusa.com/buff-products/hats/knitted-polar-hat/agna-sand/117849.302",
"https://elixinol.com/?p=793",
"https://forbiddenplanet.com/107164-very-naughty-boys-amazing-true-story-of-handmade-films/",
"https://gearclub.vitalmtb.com/product/gear-club-box-4-june/",
"https://glorycycles.com/cane-creek-eewings-titanium-crank/",
"https://shop.mochithings.com/products/15768",
"https://telescopes.net/store/baader-planetarium-100mm-aperture-astrosolar-spotter-filter.html",
"https://tkbtrading.com/collections/all-colors/products/passion-orange",
"https://www.6ku.com/collections/parts-accessories/products/6ku-pedal-straps",
"https://www.alexandermcqueen.com/Item/index?cod10=34854999UH&siteCode=ALEXANDERMCQUEEN_US",
"https://www.bestbuyautoequipment.com/Chassis-Liner-16-Revolution-p/cl832000.htm",
"https://www.clarks.in/Amali-Ice-8298.html",
"https://www.deliciousseeds.com/del_en/amnesia-haze-fem-5-seeds.html",
"https://www.dickssportinggoods.com/p/neosport-womens-neoprene-5mm-jumpsuit-16hndwwn5mmnprnjmwst/16hndwwn5mmnprnjmwst",
"https://www.holabirdsports.com/collections/brand-new-babolat-sfx3/products/babolat-sfx3-all-court-mens-black-silver",
"https://www.jjill.com/ClickInfo?URL=%2fproduct%2fpure-jill-peplum-top%3fevtype%3d%26mpe_id%3d%26intv_id%3d%26storeId%3d%26catalogId%3d%26langId%3d%26experimentId%3d%26testElementId%3d%26controlElement%3d%26expDataType%3d%26expDataUniqueID%3d&evtype=CpgnClick&mpe_id=715854148&intv_id=715851192&storeId=10101&catalogId=10051&langId=-1&expDataType=CatalogEntryId&expDataUniqueID=221289",
"https://www.lacelab.com/collections/3m-reflective-rope-laces/products/cove-blue-3m-reflective-rope-laces",
"https://www.livingsocial.com/deals/4-elements-wellness-center-4",
"https://www.melbournesnowboard.com.au/collections/all/products/burton-moto-boa-2019?variant=12657538465853",
"https://www.pckuwait.com/product/intel-i3-8100-8th-generation-core-i3-processor-3-60ghz-6mb-cache/",
"https://www.proclipusa.com/product/835232-proclip-console-mount",
"https://www.rpphobby.com/product_p/asc7430.htm",
"https://www.seasalt.com/alaea-hawaiian-sea-salt-coarse-grain-grinder-jar.html",
"https://www.shopzerouv.com/collections/the-90s/products/retro-geometric-diamond-shape-sunglasses-c748",
"https://www.swimsuitsforall.com/Aquabelle-Medley-Capri#rrec=true",
"https://www.yoogiscloset.com/handbags/3-1-phillip-lim-black-leather-soleil-large-bucket-drawstring-bag-97627.html",
"https://www.spanx.com/leggings/faux-leather-leggings",
"https://usa.tommy.com/en/men/men-shirts/lewis-hamilton-logo-shirt-mw08299",
"https://www.calvinklein.us/en/mens-clothing/mens-featured-shops-calvin-klein-jeans/slim-fit-archive-western-shirt-22705235",
"http://www2.hm.com/en_us/productpage.0476583002.html"]
df = pd.DataFrame.from_records([(url,) for url in urls], columns=["url"])
X = preprocess_features(df, load_scaler_from_file=True)
sgd_est = pickle.load(open(model_filename, 'rb'))
probs = sgd_est.predict_proba(X.values)
for url, probs in zip(urls, probs):
if probs[1] < 0.55:
print url, probs[1]
| 0.666714 | 0.422445 |
# Protocol 1.4
VLS and SAG NWs with standard lock-in technique
<br>
The code can be used for 1, 2 or 3 devices silmutaneously
<br>
This version supports both 4-probe and 2-probe measurements
<br>
<br>
This is a pinch-off measurement code that adds temperature as an extra parameter that can be sequentialy controlled
<br>
Magnetic field and Source-drain bias voltage can be also added
## Imports
```
# Copy this to all notebooks!
from qcodes.logger import start_all_logging
start_all_logging()
# Import qcodes and other necessary packages
import qcodes as qc
import numpy as np
import time
from time import sleep
import matplotlib
import matplotlib.pyplot as plt
import os
import os.path
# Import device drivers
from qcodes.instrument_drivers.QuantumDesign.DynaCoolPPMS import DynaCool
from qcodes.instrument_drivers.Keysight.Infiniium import Infiniium
# Import qcodes packages
from qcodes import Station
from qcodes import config
from qcodes.dataset.measurements import Measurement
from qcodes.dataset.plotting import plot_by_id
from qcodes.dataset.database import initialise_database,get_DB_location
from qcodes.dataset.experiment_container import (Experiment,
load_last_experiment,
new_experiment,
load_experiment_by_name)
from qcodes.instrument.base import Instrument
from qcodes.utils.dataset.doNd import do1d,do2d
%matplotlib notebook
go = 7.7480917310e-5
```
## Station
(Need to load 3 Keithleys and 6 Lock-In Amps)
```
# Create station, instantiate instruments
Instrument.close_all()
path_to_station_file = 'C:/Users/lyn-ppmsmsr-01usr/Desktop/station.yaml'
# 'file//station.yaml'
# Here we load the station file.
station = Station()
station.load_config_file(path_to_station_file)
# Connect to ppms
#Instrument.find_instrument('ppms_cryostat')
ppms = DynaCool.DynaCool(name = "ppms_cryostat", address="TCPIP0::10.10.117.37::5000::SOCKET")
station.add_component(ppms)
# SRS
lockin_1 = station.load_instrument('lockin_1')
lockin_2 = station.load_instrument('lockin_2')
lockin_3 = station.load_instrument('lockin_3')
lockin_4 = station.load_instrument('lockin_4')
lockin_5 = station.load_instrument('lockin_5')
lockin_6 = station.load_instrument('lockin_6')
# DMMs
dmm_a = station.load_instrument('Keithley_A')
dmm_b = station.load_instrument('Keithley_B')
dmm_c = station.load_instrument('Keithley_C')
dmm_a.smua.volt(0) # Set voltages to 0
dmm_a.smub.volt(0) # Set voltages to 0
dmm_b.smua.volt(0) # Set voltages to 0
dmm_b.smub.volt(0) # Set voltages to 0
dmm_c.smua.volt(0) # Set voltages to 0
dmm_c.smub.volt(0) # Set voltages to 0
for inst in station.components.values():
inst.print_readable_snapshot()
```
## DB File, Location
```
### Initialize database, make new measurement
mainpath = 'C:/Users/MicrosoftQ/Desktop/Results/Operator_name' #remember to change << /Operator_name >> to save the db file in your own user folder
config.current_config.core.db_location = os.path.join(mainpath,'GROWTHXXXX_BATCHXX_YYYYMMDD.db')
config.current_config
newpath = os.path.join(mainpath,'GROWTHXXXX_BATCHXX_YYYYMMDD')
if not os.path.exists(newpath):
os.makedirs(newpath)
figurepath = newpath
initialise_database()
```
## Functions
```
def wait_for_field():
time.sleep(1)
Magnet_state = ppms.magnet_state()
while Magnet_state is not 'holding':
#print('waiting for field')
time.sleep(0.1)
Magnet_state = ppms.magnet_state()
#print('field ready')
return
def wait_for_field_ramp():
Magnet_state = ppms.magnet_state()
while Magnet_state is not 'ramping':
time.sleep(1)
Magnet_state = ppms.magnet_state()
return
def field_ready():
return ppms.magnet_state() == 'holding'
def wait_for_temp():
Temp_state = ppms.temperature_state()
while Temp_state is not 'stable':
time.sleep(1)
Temp_state = ppms.temperature_state()
return
def wait_for_near_temp():
Temp_state = ppms.temperature_state()
while Temp_state is not 'near':
time.sleep(2)
Temp_state = ppms.temperature_state()
time.sleep(10)
return
```
## Lock-in add-on functions
Gains and conductance
```
# AMPLIFICATIONS AND VOLTAGE DIVISIONS
ACdiv = 1e-4
DCdiv = 1e-2
GIamp1 = 1e7
GVamp2 = 100
GIamp3 = 1e6
GVamp4 = 100
GIamp5 = 1e6
GVamp6 = 100
# DEFINICTIONS OF FUNCTIONS FOR DIFFERENTIAL CONDUCTANCE AND RERISTANCE FOR 2 AND 4 PROBE MEASUREMENTS
# Lock-ins 1(current), 2(voltage)
def desoverh_fpm12():
volt_ampl_1 = lockin_1.X
volt_ampl_2 = lockin_2.X
I_fpm = volt_ampl_1()/GIamp1
V_fpm = volt_ampl_2()/GVamp2
if V_fpm== 0:
dcond_fpm = 0
else:
dcond_fpm = I_fpm/V_fpm/go
return dcond_fpm
def desoverh_tpm1():
volt_ampl = lockin_1.X
sig_ampl = lockin_1.amplitude()
I_tpm = volt_ampl()/GIamp1
V_tpm = sig_ampl*ACdiv
dcond_tpm = I_tpm/V_tpm/go
return dcond_tpm
def ohms_law12():
volt_ampl_1 = lockin_1.X
volt_ampl_2 = lockin_2.X
I_fpm = volt_ampl_1()/GIamp1
V_fpm = volt_ampl_2()/GVamp2
if I_fpm== 0:
res_fpm = 0
else:
res_fpm = V_fpm/I_fpm
return res_fpm
# Lock-ins 3(current), 4(voltage)
def desoverh_fpm34():
volt_ampl_3 = lockin_3.X
volt_ampl_4 = lockin_4.X
I_fpm = volt_ampl_3()/GIamp3
V_fpm = volt_ampl_4()/GVamp4
if V_fpm== 0:
dcond_fpm = 0
else:
dcond_fpm = I_fpm/V_fpm/go
return dcond_fpm
def desoverh_tpm3():
volt_ampl = lockin_3.X
sig_ampl = lockin_3.amplitude()
I_tpm = volt_ampl()/GIamp1
V_tpm = sig_ampl*ACdiv
dcond_tpm = I_tpm/V_tpm/go
return dcond_tpm
def ohms_law34():
volt_ampl_3 = lockin_3.X
volt_ampl_4 = lockin_4.X
I_fpm = volt_ampl_3()/GIamp3
V_fpm = volt_ampl_4()/GVamp4
if I_fpm== 0:
res_fpm = 0
else:
res_fpm = V_fpm/I_fpm
return res_fpm
# Lock-ins 5(current), 6(voltage)
def desoverh_fpm56():
volt_ampl_5 = lockin_5.X
volt_ampl_6 = lockin_6.X
I_fpm = volt_ampl_5()/GIamp5
V_fpm = volt_ampl_6()/GVamp6
if V_fpm== 0:
dcond_fpm = 0
else:
dcond_fpm = I_fpm/V_fpm/go
return dcond_fpm
def desoverh_tpm5():
volt_ampl = lockin_5.X
sig_ampl = lockin_5.amplitude()
I_tpm = volt_ampl()/GIamp1
V_tpm = sig_ampl*ACdiv
dcond_tpm = I_tpm/V_tpm/go
return dcond_tpm
def ohms_law56():
volt_ampl_5 = lockin_5.X
volt_ampl_6 = lockin_6.X
I_fpm = volt_ampl_5()/GIamp5
V_fpm = volt_ampl_6()/GVamp6
if I_fpm== 0:
res_fpm = 0
else:
res_fpm = V_fpm/I_fpm
return res_fpm
try:
lockin_1.add_parameter("diff_conductance_fpm", label="dI/dV", unit="2e^2/h", get_cmd = desoverh_fpm12)
except KeyError:
print("parameter already exists. Deleting. Try again")
del lockin_1.parameters['diff_conductance_fpm']
try:
lockin_1.add_parameter("conductance_tpm", label="I/V", unit="2e^2/h", get_cmd = desoverh_tpm1)
except KeyError:
print("parameter already exists. Deleting. Try again")
del lockin_1.parameters['conductance_tpm']
try:
lockin_1.add_parameter("resistance_fpm", label="R", unit="Ohm", get_cmd = ohms_law12)
except KeyError:
print("parameter already exists. Deleting. Try again")
del lockin_1.parameters['resistance_fpm']
try:
lockin_3.add_parameter("diff_conductance_fpm", label="dI/dV", unit="2e^2/h", get_cmd = desoverh_fpm34)
except KeyError:
print("parameter already exists. Deleting. Try again")
del lockin_3.parameters['diff_conductance_fpm']
try:
lockin_3.add_parameter("conductance_tpm", label="I/V", unit="2e^2/h", get_cmd = desoverh_tpm3)
except KeyError:
print("parameter already exists. Deleting. Try again")
del lockin_3.parameters['conductance_tpm']
try:
lockin_3.add_parameter("resistance_fpm", label="R", unit="Ohm", get_cmd = ohms_law34)
except KeyError:
print("parameter already exists. Deleting. Try again")
del lockin_3.parameters['resistance_fpm']
try:
lockin_5.add_parameter("diff_conductance_fpm", label="dI/dV", unit="2e^2/h", get_cmd = desoverh_fpm56)
except KeyError:
print("parameter already exists. Deleting. Try again")
del lockin_5.parameters['diff_conductance_fpm']
try:
lockin_5.add_parameter("conductance_tpm", label="I/V", unit="2e^2/h", get_cmd = desoverh_tpm5)
except KeyError:
print("parameter already exists. Deleting. Try again")
del lockin_5.parameters['conductance_tpm']
```
# Measurement parameters
```
Vgmin = -2 #V [consult the ppt protocol]
Vgmax = +5 #V [consult the ppt protocol]
Npoints = 801 # [consult the ppt protocol]
VSD = 0 #V DC [consult the ppt protocol]
timedelay = 0.1 # sec [consult the ppt protocol]
VAC = 1 #V AC [consult the ppt protocol]
f = 136.5 #Hz [consult the ppt protocol]
tcI = 0.03 #sec [consult the ppt protocol]
tcV = 0.03 #sec [consult the ppt protocol] Preferably the same with tcI
dB_slope = 12 # dB [consult the ppt protocol]
temperature = 1.7 #K
temperature_rate = 0.1
magnetic_field = 0 #T
magnetic_field_rate = 0.22
# Small calculation for measurement parameters
if 1/f*5 <= tcI and 1/f*5 <= tcV:
valid_meas = True
elif 1/f < tcI and 1/f < tcV:
valid_meas = True
print("Warning: Time constant must be much smaller than signal oscillation period", 1/f*1000, "msec")
else:
valid_meas = False
print("Error: Time constant must be smaller than signal oscillation period", 1/f*1000, "msec")
if tcI*2.5<=timedelay and tcV*2.5<=timedelay:
valid_meas = True
elif tcI<=timedelay and tcV<=timedelay:
valid_meas = True
print("Warning: Time delay is comparable with time constant")
print("Time constant:",tcI*1e3 ,"msec, (current); ", tcV*1e3, "msec, (voltage)")
print("Time delay:", timedelay*1e3,"msec")
else:
valid_meas = False
print("Error: Time delay is smaller than the time constant")
valid_meas
```
## Frequency Test
Small measurement for frequency choise
<br>
Use whichever lock-in you are interested to test (eg. lockin_X)
```
new_experiment(name='lockin start-up', sample_name='DEVXX S21D18G38')
# Time constant choise:
# Example: f_min = 60 Hz => t_c = 1/60*2.5 sec = 42 msec => we should choose the closest value: 100 ms
lockin_1.time_constant(0.1)
tdelay = 0.3
dmm_a.smub.output('on') # Turn on the gate channel
dmm_a.smub.volt(-2) # Set the gate on a very high resistance area (below the pinch-off)
# 1-D sweep for amplitude dependence
#do1d(lockin_1.frequency,45,75,100,tdelay,lockin_1.X,lockin_1.Y,lockin_1.conductance_tpm)
# 2-D sweep repetition on a smaller frequency range for noise inspection
do2d(dmm_a.smua.volt,1,50,50,1,lockin_1.frequency,45,75,100,tdelay,lockin_1.X,lockin_1.Y,lockin_1.conductance_tpm)
dmm_a.smub.volt(0)
dmm_a.smub.output('off')
# Set things up to the station
lockin_1.time_constant(tcI) # set time constant on the lock-in
lockin_1.frequency(f) # set frequency on the lock-in
lockin_1.amplitude(VAC) # set amplitude on the lock-in
lockin_1.filter_slope(dB_slope) # set filter slope on the lock-in
lockin_2.time_constant(tcV) # set time constant on the lock-in
lockin_2.filter_slope(dB_slope) # set filter slope on the lock-in
lockin_3.time_constant(tcI) # set time constant on the lock-in
lockin_3.frequency(f) # set frequency on the lock-in
lockin_3.amplitude(VAC) # set amplitude on the lock-in
lockin_3.filter_slope(dB_slope) # set filter slope on the lock-in
lockin_4.time_constant(tcV) # set time constant on the lock-in
lockin_4.filter_slope(dB_slope) # set filter slope on the lock-in
lockin_5.time_constant(tcI) # set time constant on the lock-in
lockin_5.frequency(f) # set frequency on the lock-in
lockin_5.amplitude(VAC) # set amplitude on the lock-in
lockin_5.filter_slope(dB_slope) # set filter slope on the lock-in
lockin_6.time_constant(tcV) # set time constant on the lock-in
lockin_6.filter_slope(dB_slope) # set filter slope on the lock-in
dcond1 = lockin_1.diff_conductance_fpm
cond1 = lockin_1.conductance_tpm
res1 = lockin_1.resistance_fpm
X1 = lockin_1.X
X2 = lockin_2.X
Y1 = lockin_1.Y
Y2 = lockin_2.Y
dcond3 = lockin_3.diff_conductance_fpm
cond3 = lockin_3.conductance_tpm
res3 = lockin_3.resistance_fpm
X3 = lockin_3.X
X4 = lockin_4.X
Y3 = lockin_3.Y
Y4 = lockin_4.Y
dcond5 = lockin_5.diff_conductance_fpm
cond5 = lockin_5.conductance_tpm
res5 = lockin_5.resistance_fpm
X5 = lockin_5.X
X6 = lockin_6.X
Y5 = lockin_5.Y
Y6 = lockin_6.Y
gate = dmm_a.smub.volt
bias1 = dmm_a.smua.volt
bias3 = dmm_b.smua.volt
bias5 = dmm_b.smub.volt
temp = ppms.temperature # read the temperature
temp_set = ppms.temperature_setpoint # set the temperature
temp_rate = ppms.temperature_rate # set the temperature rate
temp_rate(temperature_rate)
temp_set(temperature)
field = ppms.field_measured # read the magnetic field
field_set = ppms.field_target # set the field; a new qcodes function! field_rate is not in use anymore
field_rate = ppms.field_rate # set the the magnetic field rate
field_rate(magnetic_field_rate)
field_set(magnetic_field)
```
## The measurement
```
# If you want to add bias then uncheck
#bias1(1e-3/DCdiv)
#bias3(1e-3/DCdiv)
#bias5(1e-3/DCdiv)
# If you want to add magnetic field then uncheck
#field_set(3)
# The control parameter (temperature)
paramrange = [1.7] #arbitrary values
#paramrange = np.arange(1.7,10,0.5) #steps
#paramrange = np.linspace(1.7,5,10) #Npoints
# The run loop
for var_param in paramrange:
temp_set(var_param)
wait_for_temp()
#vv1 = "Vsd1="+"{:.3f}".format(bias1()*DCdiv*1e3)+"mV "
#vv2 = "Vsd2="+"{:.3f}".format(bias2()*DCdiv*1e3)+"mV "
#vv2 = "Vsd3="+"{:.3f}".format(bias3()*DCdiv*1e3)+"mV "
bb = "B="+"{:.3f}".format(ppms.field_measured())+"T "
tt = "T="+"{:.2f}".format(ppms.temperature())+"K "
ff = "f="+"{:.1f}".format(lockin_1.frequency())+"Hz "
aa = "Ampl="+"{:.4f}".format(lockin_1.amplitude()*ACdiv*1e3)+"mV"
Conditions = tt + bb + ff + aa
d1 = "/1/ DEV00 S99 VH99 VL99 D99"
d2 = "/3/ DEV00 S99 VH99 VL99 D99"
d3 = "/5/ DEV00 S99 VH99 VL99 D99"
Sample_name = d1# + d2 + d3
Experiment_name = "Protocol1.4 "
new_experiment(name=Experiment_name+'UP; ' + Conditions, sample_name = Sample_name)
do1d(gate,Vgmin,Vgmax,Npoints,timedelay,
#cond1,X1,Y1,
dcond1,X1,Y1,X2,Y2,
#cond3,X3,Y3,
#dcond3,X3,Y3,X4,Y4,
#cond5,X5,Y5,
#dcond5,X5,Y5,X6,Y6,
do_plot=False)
new_experiment(name=Experiment_name+'DOWN; ' + Conditions, sample_name = Sample_name)
do1d(gate,Vgmax,Vgmin,Npoints,timedelay,
#cond1,X1,Y1,
dcond1,X1,Y1,X2,Y2,
#cond3,X3,Y3,
#dcond3,X3,Y3,X4,Y4,
#cond5,X5,Y5,
#dcond5,X5,Y5,X6,Y6,
do_plot=False)
```
|
github_jupyter
|
# Copy this to all notebooks!
from qcodes.logger import start_all_logging
start_all_logging()
# Import qcodes and other necessary packages
import qcodes as qc
import numpy as np
import time
from time import sleep
import matplotlib
import matplotlib.pyplot as plt
import os
import os.path
# Import device drivers
from qcodes.instrument_drivers.QuantumDesign.DynaCoolPPMS import DynaCool
from qcodes.instrument_drivers.Keysight.Infiniium import Infiniium
# Import qcodes packages
from qcodes import Station
from qcodes import config
from qcodes.dataset.measurements import Measurement
from qcodes.dataset.plotting import plot_by_id
from qcodes.dataset.database import initialise_database,get_DB_location
from qcodes.dataset.experiment_container import (Experiment,
load_last_experiment,
new_experiment,
load_experiment_by_name)
from qcodes.instrument.base import Instrument
from qcodes.utils.dataset.doNd import do1d,do2d
%matplotlib notebook
go = 7.7480917310e-5
# Create station, instantiate instruments
Instrument.close_all()
path_to_station_file = 'C:/Users/lyn-ppmsmsr-01usr/Desktop/station.yaml'
# 'file//station.yaml'
# Here we load the station file.
station = Station()
station.load_config_file(path_to_station_file)
# Connect to ppms
#Instrument.find_instrument('ppms_cryostat')
ppms = DynaCool.DynaCool(name = "ppms_cryostat", address="TCPIP0::10.10.117.37::5000::SOCKET")
station.add_component(ppms)
# SRS
lockin_1 = station.load_instrument('lockin_1')
lockin_2 = station.load_instrument('lockin_2')
lockin_3 = station.load_instrument('lockin_3')
lockin_4 = station.load_instrument('lockin_4')
lockin_5 = station.load_instrument('lockin_5')
lockin_6 = station.load_instrument('lockin_6')
# DMMs
dmm_a = station.load_instrument('Keithley_A')
dmm_b = station.load_instrument('Keithley_B')
dmm_c = station.load_instrument('Keithley_C')
dmm_a.smua.volt(0) # Set voltages to 0
dmm_a.smub.volt(0) # Set voltages to 0
dmm_b.smua.volt(0) # Set voltages to 0
dmm_b.smub.volt(0) # Set voltages to 0
dmm_c.smua.volt(0) # Set voltages to 0
dmm_c.smub.volt(0) # Set voltages to 0
for inst in station.components.values():
inst.print_readable_snapshot()
### Initialize database, make new measurement
mainpath = 'C:/Users/MicrosoftQ/Desktop/Results/Operator_name' #remember to change << /Operator_name >> to save the db file in your own user folder
config.current_config.core.db_location = os.path.join(mainpath,'GROWTHXXXX_BATCHXX_YYYYMMDD.db')
config.current_config
newpath = os.path.join(mainpath,'GROWTHXXXX_BATCHXX_YYYYMMDD')
if not os.path.exists(newpath):
os.makedirs(newpath)
figurepath = newpath
initialise_database()
def wait_for_field():
time.sleep(1)
Magnet_state = ppms.magnet_state()
while Magnet_state is not 'holding':
#print('waiting for field')
time.sleep(0.1)
Magnet_state = ppms.magnet_state()
#print('field ready')
return
def wait_for_field_ramp():
Magnet_state = ppms.magnet_state()
while Magnet_state is not 'ramping':
time.sleep(1)
Magnet_state = ppms.magnet_state()
return
def field_ready():
return ppms.magnet_state() == 'holding'
def wait_for_temp():
Temp_state = ppms.temperature_state()
while Temp_state is not 'stable':
time.sleep(1)
Temp_state = ppms.temperature_state()
return
def wait_for_near_temp():
Temp_state = ppms.temperature_state()
while Temp_state is not 'near':
time.sleep(2)
Temp_state = ppms.temperature_state()
time.sleep(10)
return
# AMPLIFICATIONS AND VOLTAGE DIVISIONS
ACdiv = 1e-4
DCdiv = 1e-2
GIamp1 = 1e7
GVamp2 = 100
GIamp3 = 1e6
GVamp4 = 100
GIamp5 = 1e6
GVamp6 = 100
# DEFINICTIONS OF FUNCTIONS FOR DIFFERENTIAL CONDUCTANCE AND RERISTANCE FOR 2 AND 4 PROBE MEASUREMENTS
# Lock-ins 1(current), 2(voltage)
def desoverh_fpm12():
volt_ampl_1 = lockin_1.X
volt_ampl_2 = lockin_2.X
I_fpm = volt_ampl_1()/GIamp1
V_fpm = volt_ampl_2()/GVamp2
if V_fpm== 0:
dcond_fpm = 0
else:
dcond_fpm = I_fpm/V_fpm/go
return dcond_fpm
def desoverh_tpm1():
volt_ampl = lockin_1.X
sig_ampl = lockin_1.amplitude()
I_tpm = volt_ampl()/GIamp1
V_tpm = sig_ampl*ACdiv
dcond_tpm = I_tpm/V_tpm/go
return dcond_tpm
def ohms_law12():
volt_ampl_1 = lockin_1.X
volt_ampl_2 = lockin_2.X
I_fpm = volt_ampl_1()/GIamp1
V_fpm = volt_ampl_2()/GVamp2
if I_fpm== 0:
res_fpm = 0
else:
res_fpm = V_fpm/I_fpm
return res_fpm
# Lock-ins 3(current), 4(voltage)
def desoverh_fpm34():
volt_ampl_3 = lockin_3.X
volt_ampl_4 = lockin_4.X
I_fpm = volt_ampl_3()/GIamp3
V_fpm = volt_ampl_4()/GVamp4
if V_fpm== 0:
dcond_fpm = 0
else:
dcond_fpm = I_fpm/V_fpm/go
return dcond_fpm
def desoverh_tpm3():
volt_ampl = lockin_3.X
sig_ampl = lockin_3.amplitude()
I_tpm = volt_ampl()/GIamp1
V_tpm = sig_ampl*ACdiv
dcond_tpm = I_tpm/V_tpm/go
return dcond_tpm
def ohms_law34():
volt_ampl_3 = lockin_3.X
volt_ampl_4 = lockin_4.X
I_fpm = volt_ampl_3()/GIamp3
V_fpm = volt_ampl_4()/GVamp4
if I_fpm== 0:
res_fpm = 0
else:
res_fpm = V_fpm/I_fpm
return res_fpm
# Lock-ins 5(current), 6(voltage)
def desoverh_fpm56():
volt_ampl_5 = lockin_5.X
volt_ampl_6 = lockin_6.X
I_fpm = volt_ampl_5()/GIamp5
V_fpm = volt_ampl_6()/GVamp6
if V_fpm== 0:
dcond_fpm = 0
else:
dcond_fpm = I_fpm/V_fpm/go
return dcond_fpm
def desoverh_tpm5():
volt_ampl = lockin_5.X
sig_ampl = lockin_5.amplitude()
I_tpm = volt_ampl()/GIamp1
V_tpm = sig_ampl*ACdiv
dcond_tpm = I_tpm/V_tpm/go
return dcond_tpm
def ohms_law56():
volt_ampl_5 = lockin_5.X
volt_ampl_6 = lockin_6.X
I_fpm = volt_ampl_5()/GIamp5
V_fpm = volt_ampl_6()/GVamp6
if I_fpm== 0:
res_fpm = 0
else:
res_fpm = V_fpm/I_fpm
return res_fpm
try:
lockin_1.add_parameter("diff_conductance_fpm", label="dI/dV", unit="2e^2/h", get_cmd = desoverh_fpm12)
except KeyError:
print("parameter already exists. Deleting. Try again")
del lockin_1.parameters['diff_conductance_fpm']
try:
lockin_1.add_parameter("conductance_tpm", label="I/V", unit="2e^2/h", get_cmd = desoverh_tpm1)
except KeyError:
print("parameter already exists. Deleting. Try again")
del lockin_1.parameters['conductance_tpm']
try:
lockin_1.add_parameter("resistance_fpm", label="R", unit="Ohm", get_cmd = ohms_law12)
except KeyError:
print("parameter already exists. Deleting. Try again")
del lockin_1.parameters['resistance_fpm']
try:
lockin_3.add_parameter("diff_conductance_fpm", label="dI/dV", unit="2e^2/h", get_cmd = desoverh_fpm34)
except KeyError:
print("parameter already exists. Deleting. Try again")
del lockin_3.parameters['diff_conductance_fpm']
try:
lockin_3.add_parameter("conductance_tpm", label="I/V", unit="2e^2/h", get_cmd = desoverh_tpm3)
except KeyError:
print("parameter already exists. Deleting. Try again")
del lockin_3.parameters['conductance_tpm']
try:
lockin_3.add_parameter("resistance_fpm", label="R", unit="Ohm", get_cmd = ohms_law34)
except KeyError:
print("parameter already exists. Deleting. Try again")
del lockin_3.parameters['resistance_fpm']
try:
lockin_5.add_parameter("diff_conductance_fpm", label="dI/dV", unit="2e^2/h", get_cmd = desoverh_fpm56)
except KeyError:
print("parameter already exists. Deleting. Try again")
del lockin_5.parameters['diff_conductance_fpm']
try:
lockin_5.add_parameter("conductance_tpm", label="I/V", unit="2e^2/h", get_cmd = desoverh_tpm5)
except KeyError:
print("parameter already exists. Deleting. Try again")
del lockin_5.parameters['conductance_tpm']
Vgmin = -2 #V [consult the ppt protocol]
Vgmax = +5 #V [consult the ppt protocol]
Npoints = 801 # [consult the ppt protocol]
VSD = 0 #V DC [consult the ppt protocol]
timedelay = 0.1 # sec [consult the ppt protocol]
VAC = 1 #V AC [consult the ppt protocol]
f = 136.5 #Hz [consult the ppt protocol]
tcI = 0.03 #sec [consult the ppt protocol]
tcV = 0.03 #sec [consult the ppt protocol] Preferably the same with tcI
dB_slope = 12 # dB [consult the ppt protocol]
temperature = 1.7 #K
temperature_rate = 0.1
magnetic_field = 0 #T
magnetic_field_rate = 0.22
# Small calculation for measurement parameters
if 1/f*5 <= tcI and 1/f*5 <= tcV:
valid_meas = True
elif 1/f < tcI and 1/f < tcV:
valid_meas = True
print("Warning: Time constant must be much smaller than signal oscillation period", 1/f*1000, "msec")
else:
valid_meas = False
print("Error: Time constant must be smaller than signal oscillation period", 1/f*1000, "msec")
if tcI*2.5<=timedelay and tcV*2.5<=timedelay:
valid_meas = True
elif tcI<=timedelay and tcV<=timedelay:
valid_meas = True
print("Warning: Time delay is comparable with time constant")
print("Time constant:",tcI*1e3 ,"msec, (current); ", tcV*1e3, "msec, (voltage)")
print("Time delay:", timedelay*1e3,"msec")
else:
valid_meas = False
print("Error: Time delay is smaller than the time constant")
valid_meas
new_experiment(name='lockin start-up', sample_name='DEVXX S21D18G38')
# Time constant choise:
# Example: f_min = 60 Hz => t_c = 1/60*2.5 sec = 42 msec => we should choose the closest value: 100 ms
lockin_1.time_constant(0.1)
tdelay = 0.3
dmm_a.smub.output('on') # Turn on the gate channel
dmm_a.smub.volt(-2) # Set the gate on a very high resistance area (below the pinch-off)
# 1-D sweep for amplitude dependence
#do1d(lockin_1.frequency,45,75,100,tdelay,lockin_1.X,lockin_1.Y,lockin_1.conductance_tpm)
# 2-D sweep repetition on a smaller frequency range for noise inspection
do2d(dmm_a.smua.volt,1,50,50,1,lockin_1.frequency,45,75,100,tdelay,lockin_1.X,lockin_1.Y,lockin_1.conductance_tpm)
dmm_a.smub.volt(0)
dmm_a.smub.output('off')
# Set things up to the station
lockin_1.time_constant(tcI) # set time constant on the lock-in
lockin_1.frequency(f) # set frequency on the lock-in
lockin_1.amplitude(VAC) # set amplitude on the lock-in
lockin_1.filter_slope(dB_slope) # set filter slope on the lock-in
lockin_2.time_constant(tcV) # set time constant on the lock-in
lockin_2.filter_slope(dB_slope) # set filter slope on the lock-in
lockin_3.time_constant(tcI) # set time constant on the lock-in
lockin_3.frequency(f) # set frequency on the lock-in
lockin_3.amplitude(VAC) # set amplitude on the lock-in
lockin_3.filter_slope(dB_slope) # set filter slope on the lock-in
lockin_4.time_constant(tcV) # set time constant on the lock-in
lockin_4.filter_slope(dB_slope) # set filter slope on the lock-in
lockin_5.time_constant(tcI) # set time constant on the lock-in
lockin_5.frequency(f) # set frequency on the lock-in
lockin_5.amplitude(VAC) # set amplitude on the lock-in
lockin_5.filter_slope(dB_slope) # set filter slope on the lock-in
lockin_6.time_constant(tcV) # set time constant on the lock-in
lockin_6.filter_slope(dB_slope) # set filter slope on the lock-in
dcond1 = lockin_1.diff_conductance_fpm
cond1 = lockin_1.conductance_tpm
res1 = lockin_1.resistance_fpm
X1 = lockin_1.X
X2 = lockin_2.X
Y1 = lockin_1.Y
Y2 = lockin_2.Y
dcond3 = lockin_3.diff_conductance_fpm
cond3 = lockin_3.conductance_tpm
res3 = lockin_3.resistance_fpm
X3 = lockin_3.X
X4 = lockin_4.X
Y3 = lockin_3.Y
Y4 = lockin_4.Y
dcond5 = lockin_5.diff_conductance_fpm
cond5 = lockin_5.conductance_tpm
res5 = lockin_5.resistance_fpm
X5 = lockin_5.X
X6 = lockin_6.X
Y5 = lockin_5.Y
Y6 = lockin_6.Y
gate = dmm_a.smub.volt
bias1 = dmm_a.smua.volt
bias3 = dmm_b.smua.volt
bias5 = dmm_b.smub.volt
temp = ppms.temperature # read the temperature
temp_set = ppms.temperature_setpoint # set the temperature
temp_rate = ppms.temperature_rate # set the temperature rate
temp_rate(temperature_rate)
temp_set(temperature)
field = ppms.field_measured # read the magnetic field
field_set = ppms.field_target # set the field; a new qcodes function! field_rate is not in use anymore
field_rate = ppms.field_rate # set the the magnetic field rate
field_rate(magnetic_field_rate)
field_set(magnetic_field)
# If you want to add bias then uncheck
#bias1(1e-3/DCdiv)
#bias3(1e-3/DCdiv)
#bias5(1e-3/DCdiv)
# If you want to add magnetic field then uncheck
#field_set(3)
# The control parameter (temperature)
paramrange = [1.7] #arbitrary values
#paramrange = np.arange(1.7,10,0.5) #steps
#paramrange = np.linspace(1.7,5,10) #Npoints
# The run loop
for var_param in paramrange:
temp_set(var_param)
wait_for_temp()
#vv1 = "Vsd1="+"{:.3f}".format(bias1()*DCdiv*1e3)+"mV "
#vv2 = "Vsd2="+"{:.3f}".format(bias2()*DCdiv*1e3)+"mV "
#vv2 = "Vsd3="+"{:.3f}".format(bias3()*DCdiv*1e3)+"mV "
bb = "B="+"{:.3f}".format(ppms.field_measured())+"T "
tt = "T="+"{:.2f}".format(ppms.temperature())+"K "
ff = "f="+"{:.1f}".format(lockin_1.frequency())+"Hz "
aa = "Ampl="+"{:.4f}".format(lockin_1.amplitude()*ACdiv*1e3)+"mV"
Conditions = tt + bb + ff + aa
d1 = "/1/ DEV00 S99 VH99 VL99 D99"
d2 = "/3/ DEV00 S99 VH99 VL99 D99"
d3 = "/5/ DEV00 S99 VH99 VL99 D99"
Sample_name = d1# + d2 + d3
Experiment_name = "Protocol1.4 "
new_experiment(name=Experiment_name+'UP; ' + Conditions, sample_name = Sample_name)
do1d(gate,Vgmin,Vgmax,Npoints,timedelay,
#cond1,X1,Y1,
dcond1,X1,Y1,X2,Y2,
#cond3,X3,Y3,
#dcond3,X3,Y3,X4,Y4,
#cond5,X5,Y5,
#dcond5,X5,Y5,X6,Y6,
do_plot=False)
new_experiment(name=Experiment_name+'DOWN; ' + Conditions, sample_name = Sample_name)
do1d(gate,Vgmax,Vgmin,Npoints,timedelay,
#cond1,X1,Y1,
dcond1,X1,Y1,X2,Y2,
#cond3,X3,Y3,
#dcond3,X3,Y3,X4,Y4,
#cond5,X5,Y5,
#dcond5,X5,Y5,X6,Y6,
do_plot=False)
| 0.242564 | 0.797241 |
```
import numpy as np
import matplotlib.pyplot as plt
from scipy.spatial.distance import cdist
import math
from mpl_toolkits.mplot3d import Axes3D
import random
import copy
# Importing Dataset
Dataset = np.genfromtxt('Tugas 2 ML Genap 2018-2019 Dataset Tanpa Label.csv', delimiter=",")
Dataset = np.asarray(Dataset)
Dataset = np.reshape(Dataset, (Dataset.shape[0],1,Dataset.shape[1]))
```
# Jumlah cluster
Dari penyebaran dataset yang ada dapat dilihat, data bisa dibagi menjadi 15 cluster
```
plt.plot(Dataset[:,0,0], Dataset[:,0,1], 'ro')
plt.show()
weights = np.random.randint(0, 255, size=(600, 1, 2)) / 255
weights
class SOM():
def __init__(self, neurons, dimentions, lr):
self.neurons = neurons
self.dimentions = dimentions
self.weights = np.random.randint(0, 255, size=(neurons[0], neurons[1], dimentions)) / 255
self.lr = lr
self.initial_lr = lr
self.nradius = np.sum(neurons)
self.initial_nradius = self.nradius
self.time_constant = 100/np.log(self.initial_nradius)
self.weights_ = None
self.labels_ = None
self.fig = plt.figure()
def _assignLabels(self, samples):
dimentions = self.weights.shape
self.weights_ = self.weights.reshape(dimentions[0] * dimentions[1], dimentions[2])
labels = []
for sample in samples:
distances = cdist(self.weights_, sample, metric='euclidean')
indices = np.where(distances == distances.min())
labels.append(indices[0][0])
self.labels_ = labels
def _updateWeights(self, sample):
dimentions = self.weights.shape
distances = cdist(self.weights.reshape(dimentions[0]*dimentions[1], dimentions[2]), sample, metric='euclidean')
distances = distances.reshape(dimentions[0], dimentions[1])
indices = np.where(distances == distances.min())
closestNeuron = self.weights[indices[0][0], indices[1][0]]
distances = cdist(self.weights.reshape(dimentions[0] * dimentions[1], dimentions[2]), closestNeuron.reshape(1, dimentions[2]), metric='euclidean')
distances = np.argsort(np.argsort(distances.reshape(dimentions[0] * dimentions[1])))
distances = distances.reshape(dimentions[0], dimentions[1])
influenceVector = copy.deepcopy(distances)
influenceVector[distances > self.nradius] = -1
influenceVector[influenceVector >= 0] = 1
influenceVector[influenceVector == -1] = 0
influenceValues = np.exp(-np.multiply(distances, distances) / (2 * self.nradius * self.nradius))
influenceValues = np.multiply(influenceVector, influenceValues)
influenceValues = influenceValues.reshape(self.weights.shape[0], self.weights.shape[1], 1)
self.weights = self.weights + np.multiply(influenceValues, (sample - self.weights)) * self.lr
def _updateLearningRate(self, iteration):
self.lr = self.initial_lr * np.exp(-iteration/100)
def _updateNeighbourhoodRadius(self, iteration):
self.neighbourhood_radius = self.initial_nradius * np.exp(-iteration/self.time_constant)
def train(self, samples):
for i in range(1, 100+1):
print("Iteration :", i)
for _ in samples:
sample = random.choice(samples)
self._updateWeights(sample)
if i % 10 == 0:
self.display(samples,
"Iteration: " + str(i) +
" | LR: %s %s" % (self.initial_lr, self.lr) +
" | NR: %s %s" % (self.initial_nradius, self.nradius))
self._updateLearningRate(i)
self._updateNeighbourhoodRadius(i)
self._assignLabels(samples)
def predict(self, samples):
result = []
for sample in samples:
distances = cdist(self.weights_, sample, metric='euclidean')
indices = np.where(distances == distances.min())
result.append(indices[0][0])
return np.array(result)
def display(self, samples, title, show=False):
dimentions = self.weights.shape
if not show:
plt.ion()
ax = self.fig.add_subplot(111)
plt.title(title)
ax.set_xlabel('X Label')
ax.set_ylabel('Y Label')
for weight in self.weights.reshape(dimentions[0]*dimentions[1], dimentions[2]):
ax.scatter(weight[0], weight[1], c='red', marker='X')
if show:
plt.show()
else:
plt.pause(0.05)
s = SOM(neurons=(600,1), dimentions = 2, lr = 0.1)
s.train(Dataset)
```
# Self Organizing Map Algorithm
```
# Euclidean distance formula
def getEuclidean(p, q):
return np.sqrt(np.sum((q-p)**2))
# Normalize data
def normalize(Dataset):
Dataset[:,0] = (Dataset[:,0]- min(Dataset[:,0])) / (max(Dataset[:,0]) - min(Dataset[:,0]))
Dataset[:,1] = (Dataset[:,1]- min(Dataset[:,1])) / (max(Dataset[:,1]) - min(Dataset[:,1]))
return Dataset
# Update Learning Rate function
def updateLearningRate(learningRate,i):
return learningRate * (np.exp(-i) / (i/2))
# Update Sigma
def updateSigma(sigma,i):
return sigma * (np.exp(-i) / (i/2))
def updateWeight(dataset,weights):
dimentions = weights.shape
distances = cdist(weights.reshape(dimentions[0]*dimentions[1], dimentions[2]), dataset, metric='euclidean')
distances = distances.reshape(dimentions[0], dimentions[1])
minD = np.where(distances == min(distances))
print(distances)
#closestNeuron = weights[minD[0][0], minD[1][0]]
print(distances)
weights = np.random.randint(0, 255, size=(5, 5, 2)) / 255
updateWeight(Dataset[0],weights)
weights.shape
def SOM(x,y,dim,sigma,learning_rate, i, Dataset):
weights = np.random.randint(0, 255, size=(5, 5, 2)) / 255
locations = np.array(np.meshgrid([i for i in range(x)], [i for i in range(y)])).T.reshape(-1,2)
def trainSOM(iteration, data, weight):
for i in range(1, iteration+1):
print("Iteration : ", iteration)
for _ in data:
sample = random.choise(data)
weight =
samples = []
for _ in range(5000):
sample = np.array([random.randint(1, 100) / float(100),
random.randint(1, 100) / float(100),
random.randint(1, 100) / float(100)])
sample = sample.reshape(1, 3)
samples.append(sample)
samples = np.asarray(samples)
samples.shape
```
|
github_jupyter
|
import numpy as np
import matplotlib.pyplot as plt
from scipy.spatial.distance import cdist
import math
from mpl_toolkits.mplot3d import Axes3D
import random
import copy
# Importing Dataset
Dataset = np.genfromtxt('Tugas 2 ML Genap 2018-2019 Dataset Tanpa Label.csv', delimiter=",")
Dataset = np.asarray(Dataset)
Dataset = np.reshape(Dataset, (Dataset.shape[0],1,Dataset.shape[1]))
plt.plot(Dataset[:,0,0], Dataset[:,0,1], 'ro')
plt.show()
weights = np.random.randint(0, 255, size=(600, 1, 2)) / 255
weights
class SOM():
def __init__(self, neurons, dimentions, lr):
self.neurons = neurons
self.dimentions = dimentions
self.weights = np.random.randint(0, 255, size=(neurons[0], neurons[1], dimentions)) / 255
self.lr = lr
self.initial_lr = lr
self.nradius = np.sum(neurons)
self.initial_nradius = self.nradius
self.time_constant = 100/np.log(self.initial_nradius)
self.weights_ = None
self.labels_ = None
self.fig = plt.figure()
def _assignLabels(self, samples):
dimentions = self.weights.shape
self.weights_ = self.weights.reshape(dimentions[0] * dimentions[1], dimentions[2])
labels = []
for sample in samples:
distances = cdist(self.weights_, sample, metric='euclidean')
indices = np.where(distances == distances.min())
labels.append(indices[0][0])
self.labels_ = labels
def _updateWeights(self, sample):
dimentions = self.weights.shape
distances = cdist(self.weights.reshape(dimentions[0]*dimentions[1], dimentions[2]), sample, metric='euclidean')
distances = distances.reshape(dimentions[0], dimentions[1])
indices = np.where(distances == distances.min())
closestNeuron = self.weights[indices[0][0], indices[1][0]]
distances = cdist(self.weights.reshape(dimentions[0] * dimentions[1], dimentions[2]), closestNeuron.reshape(1, dimentions[2]), metric='euclidean')
distances = np.argsort(np.argsort(distances.reshape(dimentions[0] * dimentions[1])))
distances = distances.reshape(dimentions[0], dimentions[1])
influenceVector = copy.deepcopy(distances)
influenceVector[distances > self.nradius] = -1
influenceVector[influenceVector >= 0] = 1
influenceVector[influenceVector == -1] = 0
influenceValues = np.exp(-np.multiply(distances, distances) / (2 * self.nradius * self.nradius))
influenceValues = np.multiply(influenceVector, influenceValues)
influenceValues = influenceValues.reshape(self.weights.shape[0], self.weights.shape[1], 1)
self.weights = self.weights + np.multiply(influenceValues, (sample - self.weights)) * self.lr
def _updateLearningRate(self, iteration):
self.lr = self.initial_lr * np.exp(-iteration/100)
def _updateNeighbourhoodRadius(self, iteration):
self.neighbourhood_radius = self.initial_nradius * np.exp(-iteration/self.time_constant)
def train(self, samples):
for i in range(1, 100+1):
print("Iteration :", i)
for _ in samples:
sample = random.choice(samples)
self._updateWeights(sample)
if i % 10 == 0:
self.display(samples,
"Iteration: " + str(i) +
" | LR: %s %s" % (self.initial_lr, self.lr) +
" | NR: %s %s" % (self.initial_nradius, self.nradius))
self._updateLearningRate(i)
self._updateNeighbourhoodRadius(i)
self._assignLabels(samples)
def predict(self, samples):
result = []
for sample in samples:
distances = cdist(self.weights_, sample, metric='euclidean')
indices = np.where(distances == distances.min())
result.append(indices[0][0])
return np.array(result)
def display(self, samples, title, show=False):
dimentions = self.weights.shape
if not show:
plt.ion()
ax = self.fig.add_subplot(111)
plt.title(title)
ax.set_xlabel('X Label')
ax.set_ylabel('Y Label')
for weight in self.weights.reshape(dimentions[0]*dimentions[1], dimentions[2]):
ax.scatter(weight[0], weight[1], c='red', marker='X')
if show:
plt.show()
else:
plt.pause(0.05)
s = SOM(neurons=(600,1), dimentions = 2, lr = 0.1)
s.train(Dataset)
# Euclidean distance formula
def getEuclidean(p, q):
return np.sqrt(np.sum((q-p)**2))
# Normalize data
def normalize(Dataset):
Dataset[:,0] = (Dataset[:,0]- min(Dataset[:,0])) / (max(Dataset[:,0]) - min(Dataset[:,0]))
Dataset[:,1] = (Dataset[:,1]- min(Dataset[:,1])) / (max(Dataset[:,1]) - min(Dataset[:,1]))
return Dataset
# Update Learning Rate function
def updateLearningRate(learningRate,i):
return learningRate * (np.exp(-i) / (i/2))
# Update Sigma
def updateSigma(sigma,i):
return sigma * (np.exp(-i) / (i/2))
def updateWeight(dataset,weights):
dimentions = weights.shape
distances = cdist(weights.reshape(dimentions[0]*dimentions[1], dimentions[2]), dataset, metric='euclidean')
distances = distances.reshape(dimentions[0], dimentions[1])
minD = np.where(distances == min(distances))
print(distances)
#closestNeuron = weights[minD[0][0], minD[1][0]]
print(distances)
weights = np.random.randint(0, 255, size=(5, 5, 2)) / 255
updateWeight(Dataset[0],weights)
weights.shape
def SOM(x,y,dim,sigma,learning_rate, i, Dataset):
weights = np.random.randint(0, 255, size=(5, 5, 2)) / 255
locations = np.array(np.meshgrid([i for i in range(x)], [i for i in range(y)])).T.reshape(-1,2)
def trainSOM(iteration, data, weight):
for i in range(1, iteration+1):
print("Iteration : ", iteration)
for _ in data:
sample = random.choise(data)
weight =
samples = []
for _ in range(5000):
sample = np.array([random.randint(1, 100) / float(100),
random.randint(1, 100) / float(100),
random.randint(1, 100) / float(100)])
sample = sample.reshape(1, 3)
samples.append(sample)
samples = np.asarray(samples)
samples.shape
| 0.682256 | 0.90261 |
# Introduction to Transactions with Aerospike
*Last updated: June 22, 2021*
This notebook explains the basics of executing multiple operations on one record as a transaction.
Aerospike was architected to process a high volume of concurrent real time reads and writes for Internet scale applications. Aerospike provides scale out performance by adding additional nodes or racks without changing application code.
Application code specifies the necessary process and data policy to execute one or billions of Aerospike read and write operations.
This notebook covers:
* Discrete operations:
* Record operations
* Operations on simple data types:
* Strings
* Integers
* Doubles
* Operations on complex data types:
* Blobs
* HyperLogLogs
* Lists
* Maps
* GeoJSON
* Simple transactions combining all of the above.
This notebook does not detail replication across a cluster.
This [Jupyter Notebook](https://jupyter-notebook.readthedocs.io/en/stable/notebook.html) requires the Aerospike Database running locally with Java kernel and Aerospike Java Client. To create a Docker container that satisfies the requirements and holds a copy of these notebooks, visit the [Aerospike Notebooks Repo](https://github.com/markprincely/aerospike-dev-notebooks.docker).
# Notebook Setup
## Import Jupyter Java Integration
```
import io.github.spencerpark.ijava.IJava;
import io.github.spencerpark.jupyter.kernel.magic.common.Shell;
IJava.getKernelInstance().getMagics().registerMagics(Shell.class);
```
## Start Aerospike
```
%sh asd
```
## Download the Aerospike Java Client
```
%%loadFromPOM
<dependencies>
<dependency>
<groupId>com.aerospike</groupId>
<artifactId>aerospike-client</artifactId>
<version>5.0.0</version>
</dependency>
</dependencies>
```
## Start the Aerospike Java Client and Connect
The default cluster location for the Docker container is *localhost* port *3000*. If your cluster is not running on your local machine, modify *localhost* and *3000* to the values for your Aerospike cluster.
```
import com.aerospike.client.AerospikeClient;
AerospikeClient client = new AerospikeClient("localhost", 3000);
System.out.println("Initialized the client and connected to the cluster.");
```
# Aerospike Provides Intuitive Atomic Reads and Writes
Aerospike provides client APIs to read and write different types of data. Each record read or write operation that an Aerospike server or cluster executes as an atomic (ACID) operation.
For additional information on Aerospike's single-record variant of ACID compliance, go [here](https://www.aerospike.com/docs/architecture/acid.html).
# Operate Applies One or More Operations
For the case where an application uses Aerospike as a key/value store without applying [Aerospike data types](https://www.aerospike.com/docs/guide/data-types.html) to data, the [AerospikeClient](https://www.aerospike.com/apidocs/java/com/aerospike/client/AerospikeClient.html) provides the super fast, basic getters and setters for bins of data.
The most frequently used and simplest technique to execute more than one operation in an atomic fashion is the Operate API. Using this API, the Aerospike client can execute a single read or write operation or complex combinations of reads and writes
For more information on Operate, go [here](https://www.aerospike.com/apidocs/java/com/aerospike/client/Operation.html).
For more information on applying multiple operations to a record, go [here](https://aerospike.com/docs/client/java/usage/kvs/multiops.html).
## Using Discrete Operations on a Full Record
Aerospike provides the following record operations:
* Get – Read a record.
* Getheader – Read a record's TTL and generation counter.
* Touch – Increase a record's generation counter.
* Delete – Delete a record.
### Create Test Data
Create a simple instance of every Aerospike data type.
```
import java.util.ArrayList;
import java.util.Arrays;
import java.util.List;
import java.util.Map;
import java.util.HashMap;
String txnString = "atomic";
Integer txnInteger = 8;
Double txnDouble = 6.022;
byte[] txnBlob = new byte[] {0b00000001, 0b00000010, 0b00000011, 0b00000100, 0b00000101};
String txnGeo = String.format("{ \"type\": \"Polygon\", \"coordinates\": [ [[-122.500, 37.000], [-121.000, 37.000], [-121.000, 38.080], [-122.500, 38.080], [-122.500, 37.000]] ] }");
ArrayList<Integer> txnList = new ArrayList<Integer>();
txnList.add(1);
HashMap<Integer, Integer> txnMap = new HashMap <Integer, Integer>();
txnMap.put(2, 4);
System.out.println("String: " + txnString);
System.out.println("Integer: " + txnInteger);
System.out.println("Double: " + txnDouble);
System.out.println("Blob: " + Arrays.toString(txnBlob));
System.out.println("HLL: Starts with no data.");
System.out.println("Geo: " + txnGeo);
System.out.println("List: " + txnList);
System.out.println("Map: " + txnMap);
```
### Put Data Into An Aerospike Record
```
import com.aerospike.client.Key;
import com.aerospike.client.Bin;
import com.aerospike.client.Value;
import com.aerospike.client.policy.ClientPolicy;
Integer theKey = 0;
String txnSet = "txnset";
String txnNamespace = "test";
String txnStringBin = "str";
String txnIntegerBin = "int";
String txnDoubleBin = "double";
String txnBlobBin = "blob";
String txnHLLBin = "hll";
String txnGeoBin = "geo";
String txnListBin = "list";
String txnMapBin = "map";
ClientPolicy clientPolicy = new ClientPolicy();
Key key = new Key(txnNamespace, txnSet, theKey);
Bin bin0 = new Bin(txnStringBin, txnString);
Bin bin1 = new Bin(txnIntegerBin, txnInteger);
Bin bin2 = new Bin(txnDoubleBin, txnDouble);
Bin bin3 = new Bin(txnBlobBin, txnBlob);
Bin bin4 = new Bin(txnHLLBin, Value.getAsNull());
Bin bin5 = new Bin(txnGeoBin, Value.getAsGeoJSON(txnGeo));
Bin bin6 = new Bin(txnListBin, txnList);
Bin bin7 = new Bin(txnMapBin, txnMap);
client.put(clientPolicy.writePolicyDefault, key, bin0, bin1, bin2, bin3, bin5, bin6, bin7);
System.out.println("Put data into Aerospike: "
+ txnStringBin + ", "
+ txnIntegerBin + ", "
+ txnDoubleBin + ", "
+ txnBlobBin + ", "
+ txnHLLBin + ", "
+ txnGeoBin + ", "
+ txnListBin + ", "
+ txnMapBin);
```
### Get the Record
```
import com.aerospike.client.Record;
Key key = new Key(txnNamespace, txnSet, theKey);
Record record = client.get(null, key);
System.out.println(record);
```
### Get the Record Header
```
Key key = new Key(txnNamespace, txnSet, theKey);
Record record = client.getHeader(null, key);
System.out.println(record);
```
### Touch the Record
```
Key key = new Key(txnNamespace, txnSet, theKey);
client.touch(client.writePolicyDefault, key);
Record record = client.get(client.writePolicyDefault, key);
System.out.println(record);
```
## Operating on Simple Bin Data
When operating on simple data–Strings, Integers, and Doubles–, Aerospike provides standard create, read, update, and delete operations and also increment operations, like prepend/append and add.
For more information on record operations and simple data operations, go [here](https://www.aerospike.com/apidocs/java/com/aerospike/client/Operation.html).
### Operating on String Data
Append to a string.
```
String txnAppendString = "-operation";
bin0 = new Bin(txnStringBin, txnAppendString);
Key key = new Key(txnNamespace, txnSet, theKey);
Record record = client.get(client.writePolicyDefault, key);
client.append(client.writePolicyDefault, key, bin0);
Record after = client.get(client.writePolicyDefault, key);
System.out.println("Before, the " + txnStringBin + " was - " + record.getValue(txnStringBin));
System.out.println(" After, the " + txnStringBin + " is - " + after.getValue(txnStringBin));
```
### Operating on Integer Data
Add to an integer.
```
Integer txnAddInt = 5;
Bin binIntAdd = new Bin(txnIntegerBin, txnAddInt);
Key key = new Key(txnNamespace, txnSet, theKey);
Record record = client.get(client.writePolicyDefault, key);
client.add(client.writePolicyDefault, key, binIntAdd);
Record after = client.get(client.writePolicyDefault, key);
System.out.println("Before, the " + txnIntegerBin + " was - " + record.getValue(txnIntegerBin));
System.out.println(" After, the " + txnIntegerBin + " is - " + after.getValue(txnIntegerBin));
```
### Operating on Double Data
Subtract from a double.
```
Double txnAddDouble = -3.142;
Bin binDoubleAdd = new Bin(txnDoubleBin, txnAddDouble);
Key key = new Key(txnNamespace, txnSet, theKey);
Record record = client.get(client.writePolicyDefault, key);
client.add(client.writePolicyDefault, key, binDoubleAdd);
Record after = client.get(client.writePolicyDefault, key);
System.out.println("Before, the " + txnDoubleBin + " was - " + record.getValue(txnDoubleBin));
System.out.println(" After, the " + txnDoubleBin + " is - " + after.getValue(txnDoubleBin));
```
## Operating on Complex Data Types
Aerospike also provides data-type-specific operations to work with complex data types:
* Collection Data Types
* Lists
* Maps
* Blob/Bit Data
* HyperLogLog (as a HyperMinHash)
* GeoJSON
### Operating on Lists
Append 5 to the list.
Aerospike provides operations to create and manage:
* a simple list
* list containing lists
* list containing maps
For a tutorial on working with lists, go [here](java-working_with_lists.ipynb). For more information on ListOperations, go [here](https://www.aerospike.com/apidocs/java/com/aerospike/client/cdt/ListOperation.html).
```
import com.aerospike.client.cdt.ListOperation;
Integer listAddition = 5;
Key key = new Key(txnNamespace, txnSet, theKey);
Record record = client.get(client.writePolicyDefault, key);
client.operate(client.writePolicyDefault, key,
ListOperation.append(txnListBin, Value.get(listAddition))
);
Record after = client.get(client.writePolicyDefault, key);
System.out.println("Before, the " + txnListBin + " was - " + record.getValue(txnListBin));
System.out.println(" After, the " + txnListBin + " is - " + after.getValue(txnListBin));
```
### Operating on Maps
Increment the Value of mapkey 2 by 57.
Aerospike provides operations to create and manage:
* a map
* a map containing lists
* a map containing maps
For a tutorial on working with maps, go [here]((java-working_with_maps.ipynb)).
For more information on Map Operations, go [here](https://www.aerospike.com/apidocs/java/com/aerospike/client/cdt/MapOperation.html).
```
import com.aerospike.client.cdt.MapOperation;
import com.aerospike.client.cdt.MapPolicy;
Integer mapKey = 2;
Integer mapIncrementValue = 57;
MapPolicy txnMapPolicy = new MapPolicy();
Key key = new Key(txnNamespace, txnSet, theKey);
Record record = client.get(client.writePolicyDefault, key);
client.operate(client.writePolicyDefault, key,
MapOperation.increment(txnMapPolicy, txnMapBin, Value.get(mapKey), Value.get(mapIncrementValue))
);
Record after = client.get(client.writePolicyDefault, key);
System.out.println("Before, the " + txnMapBin + " was - " + record.getValue(txnMapBin));
System.out.println(" After, the " + txnMapBin + " is - " + after.getValue(txnMapBin));
```
### Operating on Blob/Bit Data
In addition to CRUD operations, Aerospike provides standard bit wise operations, such as logical operators (AND, OR, NOT, etc.), add/subtract, and shifts.
For more information on Bit Operations, go [here](https://www.aerospike.com/apidocs/java/com/aerospike/client/operation/BitOperation.html).
```
import com.aerospike.client.operation.BitOperation;
import com.aerospike.client.operation.BitPolicy;
byte[] bitsToSet = new byte[] {(byte)0b11100000};
Integer bitSize = 8;
Integer bitOffset = 13;
BitPolicy bitPolicy = new BitPolicy();
Key key = new Key(txnNamespace, txnSet, theKey);
Record record = client.get(client.writePolicyDefault, key);
client.operate(client.writePolicyDefault, key,
BitOperation.set(bitPolicy.Default, txnBlobBin, bitOffset, bitSize, bitsToSet)
);
Record after = client.get(client.writePolicyDefault, key);
byte[] beforeBytes = (byte[])record.getValue(txnBlobBin);
byte[] afterBytes = (byte[])after.getValue(txnBlobBin);
System.out.println("Before, the " + txnBlobBin + " was - " + Arrays.toString(beforeBytes));
System.out.println(" After, the " + txnBlobBin + " is - " + Arrays.toString(afterBytes));
```
### Operating on HyperLogLog Data
Init the HyperLogLog bin.
HyperLogLog is a probabilistic data type used for counting really large data sets. Aerospike provides operations to:
* Maintain the data type (init, reset, etc.)
* Add data to these data sets.
* Compare HyperLogLog data (intersection, unions, etc.).
For more information on HyperLogLog Operations, go [here](https://www.aerospike.com/apidocs/java/com/aerospike/client/operation/HLLOperation.html).
```
import com.aerospike.client.operation.HLLOperation;
import com.aerospike.client.operation.HLLPolicy;
HLLPolicy defHLLPolicy = new HLLPolicy();
Integer bitsHLLIndex = 8;
Key key = new Key(txnNamespace, txnSet, theKey);
Record record = client.get(client.writePolicyDefault, key);
client.operate(client.writePolicyDefault, key,
HLLOperation.init(defHLLPolicy, txnHLLBin, bitsHLLIndex)
);
Record after = client.get(client.writePolicyDefault, key);
System.out.println("Before, the " + txnHLLBin + " was - " + record.getValue(txnHLLBin));
System.out.println(" After, the " + txnHLLBin + " is - " + after.getValue(txnHLLBin));
```
### Operating on GeoJSON
Get GeoJSON data.
For the purposes of simple transactions, Aerospike quickly stores and retrieves GeoJSON data in bins, and optionally nested in maps. In addition, Aerospike validates GeoJSON and processes Geospatial queries, including circle queries.
For more information on using Aerospike for geospatial indexes and queries, go [here](https://www.aerospike.com/docs/guide/geospatial.html).
```
Key key = new Key(txnNamespace, txnSet, theKey);
Record pullGeo = client.get(client.writePolicyDefault, key, txnGeoBin);
System.out.println("The " + txnGeoBin + " is - " + pullGeo.getValue(txnGeoBin));
```
## Performing a Simple Transaction on a Record
The above operations were each performed as atomic operations. **Operate** executes multiple operations to one or more bins during a single record lock in an ACID-compliant fashion. Results are returned in an array for each bin.
### Execute All of the Previous Operations as a Transaction
1. Create data for each data type.
2. Put it into Aerospike.
3. Apply the following operations as one transaction.
1. Touch the record.
2. Append to the string.
3. Increment the integer.
4. Subtract from the double.
5. Put an item in the list.
6. Put a new value in the map.
7. Set bits in the blob.
8. Init and add set elements to the hyperloglog.
9. Get the GeoJSON.
```
// Create data for each data type.
import java.util.ArrayList;
import java.util.Arrays;
import java.util.List;
import java.util.Map;
import java.util.HashMap;
String txnString = "atomic";
Integer txnInteger = 8;
Double txnDouble = 6.022;
byte[] txnBlob = new byte[] {0b00000001, 0b00000010, 0b00000011, 0b00000100, 0b00000101};
String txnGeo = String.format("{ \"type\": \"Polygon\", \"coordinates\": [ [[-122.500, 37.000], [-121.000, 37.000], [-121.000, 38.080], [-122.500, 38.080], [-122.500, 37.000]] ] }");
ArrayList<Integer> txnList = new ArrayList<Integer>();
txnList.add(1);
HashMap<Integer, Integer> txnMap = new HashMap <Integer, Integer>();
txnMap.put(2, 4);
System.out.println("--Initial Data–-");
System.out.println("String: " + txnString);
System.out.println("Integer: " + txnInteger);
System.out.println("Double: " + txnDouble);
System.out.println("Blob: " + Arrays.toString(txnBlob));
System.out.println("HLL: Starts with no data.");
System.out.println("Geo: " + txnGeo);
System.out.println("List: " + txnList);
System.out.println("Map: " + txnMap);
System.out.println();
// Put it into Aerospike.
import com.aerospike.client.AerospikeClient;
import com.aerospike.client.Key;
import com.aerospike.client.Bin;
import com.aerospike.client.Value;
import com.aerospike.client.policy.ClientPolicy;
import com.aerospike.client.Operation;
import com.aerospike.client.cdt.ListOperation;
import com.aerospike.client.cdt.MapOperation;
import com.aerospike.client.cdt.MapPolicy;
import com.aerospike.client.operation.BitOperation;
import com.aerospike.client.operation.BitPolicy;
import com.aerospike.client.operation.HLLOperation;
import com.aerospike.client.operation.HLLPolicy;
Integer theKey = 0;
String txnSet = "txnset";
String txnNamespace = "test";
String txnStringBin = "str";
String txnIntegerBin = "int";
String txnDoubleBin = "double";
String txnBlobBin = "blob";
String txnHLLBin = "hll";
String txnGeoBin = "geo";
String txnListBin = "list";
String txnMapBin = "map";
AerospikeClient client = new AerospikeClient("localhost", 3000);
ClientPolicy clientPolicy = new ClientPolicy();
BitPolicy bitPolicy = new BitPolicy();
HLLPolicy defHLLPolicy = new HLLPolicy();
MapPolicy txnMapPolicy = new MapPolicy();
Key key = new Key(txnNamespace, txnSet, theKey);
Bin bin0 = new Bin(txnStringBin, txnString);
Bin bin1 = new Bin(txnIntegerBin, txnInteger);
Bin bin2 = new Bin(txnDoubleBin, txnDouble);
Bin bin3 = new Bin(txnBlobBin, txnBlob);
Bin bin4 = new Bin(txnHLLBin, Value.getAsNull());
Bin bin5 = new Bin(txnGeoBin, Value.getAsGeoJSON(txnGeo));
Bin bin6 = new Bin(txnListBin, txnList);
Bin bin7 = new Bin(txnMapBin, txnMap);
client.put(clientPolicy.writePolicyDefault, key, bin0, bin1, bin2, bin3, bin5, bin6, bin7);
// Apply the following operations as one transaction.
// 1. Touch the record.
// 2. Append to the string.
// 3. Increment the integer.
// 4. Subtract from the double.
// 5. Put an item in the list.
// 6. Increment a value in the map.
// 7. Set bits in the blob.
// 8. Init and add set elements to the hyperloglog.
// 9. Get the GeoJSON.
String txnAppendString = "-transactions";
Bin binStrAppend = new Bin(txnStringBin, txnAppendString);
Integer txnAddInt = 5;
Bin binIntAdd = new Bin(txnIntegerBin, txnAddInt);
Double txnAddDouble = -3.142;
Bin binDoubleSub = new Bin(txnDoubleBin, txnAddDouble);
byte[] bitsToSet = new byte[] {(byte)0b11100000};
Integer bitSize = 8;
Integer bitOffset = 13;
Integer bitsHLLIndex = 8;
Integer listAddition = 5;
Integer mapKey = 2;
Integer mapIncrementValue = 57;
ArrayList<Value> dataListForHLL = new ArrayList<Value>();
dataListForHLL.add(Value.get(txnAddInt));
dataListForHLL.add(Value.get(bitSize));
dataListForHLL.add(Value.get(bitOffset));
dataListForHLL.add(Value.get(bitsHLLIndex));
dataListForHLL.add(Value.get(listAddition));
dataListForHLL.add(Value.get(mapKey));
dataListForHLL.add(Value.get(mapIncrementValue));
Record beforeOps = client.get(client.writePolicyDefault, key);
Record operationsRecord = client.operate(client.writePolicyDefault, key,
Operation.touch(),
Operation.append(binStrAppend),
Operation.add(binIntAdd),
Operation.add(binDoubleSub),
ListOperation.append(txnListBin, Value.get(listAddition)),
MapOperation.increment(txnMapPolicy, txnMapBin, Value.get(mapKey), Value.get(mapIncrementValue)),
BitOperation.set(bitPolicy.Default, txnBlobBin, bitOffset, bitSize, bitsToSet),
HLLOperation.init(defHLLPolicy, txnHLLBin, bitsHLLIndex),
HLLOperation.add(defHLLPolicy, txnHLLBin, dataListForHLL, bitsHLLIndex)
);
Record afterOps = client.get(client.writePolicyDefault, key);
System.out.println("--The Data in Aerospike–-");
System.out.println(beforeOps);
System.out.println();
System.out.println("--Operation Details–-");
System.out.println("Before, the " + txnStringBin + " was - " + beforeOps.getValue(txnStringBin));
System.out.println(" After, the " + txnStringBin + " is - " + afterOps.getValue(txnStringBin));
System.out.println();
System.out.println("Before, the " + txnIntegerBin + " was - " + beforeOps.getValue(txnIntegerBin));
System.out.println(" After, the " + txnIntegerBin + " is - " + afterOps.getValue(txnIntegerBin));
System.out.println();
System.out.println("Before, the " + txnDoubleBin + " was - " + beforeOps.getValue(txnDoubleBin));
System.out.println(" After, the " + txnDoubleBin + " is - " + afterOps.getValue(txnDoubleBin));
System.out.println();
System.out.println("Before, the " + txnListBin + " was - " + beforeOps.getValue(txnListBin));
System.out.println(" After, the " + txnListBin + " is - " + afterOps.getValue(txnListBin));
System.out.println();
System.out.println("Before, the " + txnMapBin + " was - " + beforeOps.getValue(txnMapBin));
System.out.println(" After, the " + txnMapBin + " is - " + afterOps.getValue(txnMapBin));
System.out.println();
byte[] beforeBytes = (byte[])beforeOps.getValue(txnBlobBin);
byte[] afterBytes = (byte[])afterOps.getValue(txnBlobBin);
System.out.println("Before, the " + txnBlobBin + " was - " + Arrays.toString(beforeBytes));
System.out.println(" After, the " + txnBlobBin + " is - " + Arrays.toString(afterBytes));
System.out.println();
System.out.println("Before, the " + txnHLLBin + " was - " + beforeOps.getValue(txnHLLBin));
System.out.println(" After, the " + txnHLLBin + " is - " + afterOps.getValue(txnHLLBin));
System.out.println();
System.out.println("The " + txnGeoBin + " is - " + afterOps.getValue(txnGeoBin));
System.out.println();
System.out.println("--The Record After the Operations–-");
System.out.println(afterOps);
```
## Use Write Policies To Replace Existence Checks
Simple transactions require arbitrary logic. The main technique to add conditional logic to these transactions is to apply a **write policy**.
Write operations use *policy* with the policy flags to indicate how to behave when data does or does not exist. For example, if executing a simple transaction containing multiple operations including one map operation that uses the conditional logic `if (bin doesn't have a value), then put a default value into the bin`, apply that one operation using a write policy using the flags:
* CREATE_ONLY – Only apply the write if the data doesn't exist.
* NO_FAIL – Do not throw an error upon failure.
* PARTIAL – Allow other operations to succeed.
The default write policy if data exists in a bin is to merge data, whenever possible.
Each complex data type has its own write mode policy options.
* For information on Bit Write Flags, go [here](https://www.aerospike.com/apidocs/java/com/aerospike/client/operation/BitWriteFlags.html).
* For information on HyperLogLog Write Flags, go [here](https://www.aerospike.com/apidocs/java/com/aerospike/client/operation/HLLWriteFlags.html).
* For information on List Write Flags, go [here](https://www.aerospike.com/apidocs/java/com/aerospike/client/cdt/ListWriteFlags.html).
* For information on Map Write Flags, go [here](https://www.aerospike.com/apidocs/java/com/aerospike/client/cdt/MapWriteFlags.html).
Now, insert a new Mapkey:Value pair only if the mapkey doesn't already exist.
```
import com.aerospike.client.cdt.MapOrder;
import com.aerospike.client.cdt.MapWriteFlags;
Integer txnDefaultMapkey=2;
Integer txnDefaultValue=1;
MapPolicy txnMapPolicy = new MapPolicy(MapOrder.UNORDERED, MapWriteFlags.CREATE_ONLY | MapWriteFlags.NO_FAIL | MapWriteFlags.PARTIAL);
Key key = new Key(txnNamespace, txnSet, theKey);
Record mapFailRecord = client.operate(client.writePolicyDefault, key,
MapOperation.put(txnMapPolicy, txnMapBin, Value.get(txnDefaultMapkey), Value.get(txnDefaultValue))
);
Record afterOps = client.get(client.writePolicyDefault, key);
System.out.println("The " + txnMapBin + " is - " + afterOps.getValue(txnMapBin));
System.out.println();
```
## Use RMF or Record UDFs to Apply Other Conditional Logic
If a process does requires conditional logic to check data values before writing, the common practice is to use a Read-Modify-Write pattern to check the data and write only if the generation counter is the same as when read.
For more information about Read-Modify-Write, go [here].(https://www.aerospike.com/blog/developers-understanding-aerospike-transactions/)
# Notebook Cleanup
### Truncate the Set
Truncate the set from the Aerospike Database.
```
import com.aerospike.client.policy.InfoPolicy;
InfoPolicy infoPolicy = new InfoPolicy();
client.truncate(infoPolicy, txnNamespace, txnSet, null);
System.out.println("Set Truncated.");
```
### Close the Connection to Aerospike
```
client.close();
System.out.println("Server connection closed.");
```
# Takeaway – Record Transactions are Powerful
Simple transactions are a tool for efficient atomic execution of multiple operations on one record. The ability to process many real time, multi-operation simple transactions at scale is a strength of the Aerospike platform. A little forethought into application reads and writes before coding results in higher performance applications.
# What's Next?
## Next Steps
Have questions? Don't hesitate to reach out if you have additional questions about executing app transactions at https://discuss.aerospike.com/.
Want to check out other Java notebooks?
* [Queries and UDFs](query_udf.ipynb)
* [Working with Lists](java-working_with_lists.ipynb)
* [Modeling using Lists](java-modeling_using_lists.ipynb)
* [Working with Maps](java-working_with_maps.ipynb)
Are you running this from Binder? [Download the Aerospike Notebook Repo](https://github.com/aerospike-examples/interactive-notebooks) and work with Aerospike Database and Jupyter locally using a Docker container.
## Additional Resources
Simple transactions are one of Aerospike's tools to work with data at scale. Other tools include Queries and UDFs, Batches, and Scans.
* Need more server-side transaction processing? Learn about [UDFs](https://www.aerospike.com/docs/guide/udf.html).
* Want to pull down all records within a data set? Look into [Scans](https://www.aerospike.com/docs/guide/scan.html).
* Looking to download a lot of records at one time? See [Batches](https://www.aerospike.com/docs/guide/batch.html).
* Want to get started with Java? [Download](https://www.aerospike.com/download/client/) or [install](https://github.com/aerospike/aerospike-client-java) the Aerospike Java Client.
|
github_jupyter
|
import io.github.spencerpark.ijava.IJava;
import io.github.spencerpark.jupyter.kernel.magic.common.Shell;
IJava.getKernelInstance().getMagics().registerMagics(Shell.class);
%sh asd
%%loadFromPOM
<dependencies>
<dependency>
<groupId>com.aerospike</groupId>
<artifactId>aerospike-client</artifactId>
<version>5.0.0</version>
</dependency>
</dependencies>
import com.aerospike.client.AerospikeClient;
AerospikeClient client = new AerospikeClient("localhost", 3000);
System.out.println("Initialized the client and connected to the cluster.");
import java.util.ArrayList;
import java.util.Arrays;
import java.util.List;
import java.util.Map;
import java.util.HashMap;
String txnString = "atomic";
Integer txnInteger = 8;
Double txnDouble = 6.022;
byte[] txnBlob = new byte[] {0b00000001, 0b00000010, 0b00000011, 0b00000100, 0b00000101};
String txnGeo = String.format("{ \"type\": \"Polygon\", \"coordinates\": [ [[-122.500, 37.000], [-121.000, 37.000], [-121.000, 38.080], [-122.500, 38.080], [-122.500, 37.000]] ] }");
ArrayList<Integer> txnList = new ArrayList<Integer>();
txnList.add(1);
HashMap<Integer, Integer> txnMap = new HashMap <Integer, Integer>();
txnMap.put(2, 4);
System.out.println("String: " + txnString);
System.out.println("Integer: " + txnInteger);
System.out.println("Double: " + txnDouble);
System.out.println("Blob: " + Arrays.toString(txnBlob));
System.out.println("HLL: Starts with no data.");
System.out.println("Geo: " + txnGeo);
System.out.println("List: " + txnList);
System.out.println("Map: " + txnMap);
import com.aerospike.client.Key;
import com.aerospike.client.Bin;
import com.aerospike.client.Value;
import com.aerospike.client.policy.ClientPolicy;
Integer theKey = 0;
String txnSet = "txnset";
String txnNamespace = "test";
String txnStringBin = "str";
String txnIntegerBin = "int";
String txnDoubleBin = "double";
String txnBlobBin = "blob";
String txnHLLBin = "hll";
String txnGeoBin = "geo";
String txnListBin = "list";
String txnMapBin = "map";
ClientPolicy clientPolicy = new ClientPolicy();
Key key = new Key(txnNamespace, txnSet, theKey);
Bin bin0 = new Bin(txnStringBin, txnString);
Bin bin1 = new Bin(txnIntegerBin, txnInteger);
Bin bin2 = new Bin(txnDoubleBin, txnDouble);
Bin bin3 = new Bin(txnBlobBin, txnBlob);
Bin bin4 = new Bin(txnHLLBin, Value.getAsNull());
Bin bin5 = new Bin(txnGeoBin, Value.getAsGeoJSON(txnGeo));
Bin bin6 = new Bin(txnListBin, txnList);
Bin bin7 = new Bin(txnMapBin, txnMap);
client.put(clientPolicy.writePolicyDefault, key, bin0, bin1, bin2, bin3, bin5, bin6, bin7);
System.out.println("Put data into Aerospike: "
+ txnStringBin + ", "
+ txnIntegerBin + ", "
+ txnDoubleBin + ", "
+ txnBlobBin + ", "
+ txnHLLBin + ", "
+ txnGeoBin + ", "
+ txnListBin + ", "
+ txnMapBin);
import com.aerospike.client.Record;
Key key = new Key(txnNamespace, txnSet, theKey);
Record record = client.get(null, key);
System.out.println(record);
Key key = new Key(txnNamespace, txnSet, theKey);
Record record = client.getHeader(null, key);
System.out.println(record);
Key key = new Key(txnNamespace, txnSet, theKey);
client.touch(client.writePolicyDefault, key);
Record record = client.get(client.writePolicyDefault, key);
System.out.println(record);
String txnAppendString = "-operation";
bin0 = new Bin(txnStringBin, txnAppendString);
Key key = new Key(txnNamespace, txnSet, theKey);
Record record = client.get(client.writePolicyDefault, key);
client.append(client.writePolicyDefault, key, bin0);
Record after = client.get(client.writePolicyDefault, key);
System.out.println("Before, the " + txnStringBin + " was - " + record.getValue(txnStringBin));
System.out.println(" After, the " + txnStringBin + " is - " + after.getValue(txnStringBin));
Integer txnAddInt = 5;
Bin binIntAdd = new Bin(txnIntegerBin, txnAddInt);
Key key = new Key(txnNamespace, txnSet, theKey);
Record record = client.get(client.writePolicyDefault, key);
client.add(client.writePolicyDefault, key, binIntAdd);
Record after = client.get(client.writePolicyDefault, key);
System.out.println("Before, the " + txnIntegerBin + " was - " + record.getValue(txnIntegerBin));
System.out.println(" After, the " + txnIntegerBin + " is - " + after.getValue(txnIntegerBin));
Double txnAddDouble = -3.142;
Bin binDoubleAdd = new Bin(txnDoubleBin, txnAddDouble);
Key key = new Key(txnNamespace, txnSet, theKey);
Record record = client.get(client.writePolicyDefault, key);
client.add(client.writePolicyDefault, key, binDoubleAdd);
Record after = client.get(client.writePolicyDefault, key);
System.out.println("Before, the " + txnDoubleBin + " was - " + record.getValue(txnDoubleBin));
System.out.println(" After, the " + txnDoubleBin + " is - " + after.getValue(txnDoubleBin));
import com.aerospike.client.cdt.ListOperation;
Integer listAddition = 5;
Key key = new Key(txnNamespace, txnSet, theKey);
Record record = client.get(client.writePolicyDefault, key);
client.operate(client.writePolicyDefault, key,
ListOperation.append(txnListBin, Value.get(listAddition))
);
Record after = client.get(client.writePolicyDefault, key);
System.out.println("Before, the " + txnListBin + " was - " + record.getValue(txnListBin));
System.out.println(" After, the " + txnListBin + " is - " + after.getValue(txnListBin));
import com.aerospike.client.cdt.MapOperation;
import com.aerospike.client.cdt.MapPolicy;
Integer mapKey = 2;
Integer mapIncrementValue = 57;
MapPolicy txnMapPolicy = new MapPolicy();
Key key = new Key(txnNamespace, txnSet, theKey);
Record record = client.get(client.writePolicyDefault, key);
client.operate(client.writePolicyDefault, key,
MapOperation.increment(txnMapPolicy, txnMapBin, Value.get(mapKey), Value.get(mapIncrementValue))
);
Record after = client.get(client.writePolicyDefault, key);
System.out.println("Before, the " + txnMapBin + " was - " + record.getValue(txnMapBin));
System.out.println(" After, the " + txnMapBin + " is - " + after.getValue(txnMapBin));
import com.aerospike.client.operation.BitOperation;
import com.aerospike.client.operation.BitPolicy;
byte[] bitsToSet = new byte[] {(byte)0b11100000};
Integer bitSize = 8;
Integer bitOffset = 13;
BitPolicy bitPolicy = new BitPolicy();
Key key = new Key(txnNamespace, txnSet, theKey);
Record record = client.get(client.writePolicyDefault, key);
client.operate(client.writePolicyDefault, key,
BitOperation.set(bitPolicy.Default, txnBlobBin, bitOffset, bitSize, bitsToSet)
);
Record after = client.get(client.writePolicyDefault, key);
byte[] beforeBytes = (byte[])record.getValue(txnBlobBin);
byte[] afterBytes = (byte[])after.getValue(txnBlobBin);
System.out.println("Before, the " + txnBlobBin + " was - " + Arrays.toString(beforeBytes));
System.out.println(" After, the " + txnBlobBin + " is - " + Arrays.toString(afterBytes));
import com.aerospike.client.operation.HLLOperation;
import com.aerospike.client.operation.HLLPolicy;
HLLPolicy defHLLPolicy = new HLLPolicy();
Integer bitsHLLIndex = 8;
Key key = new Key(txnNamespace, txnSet, theKey);
Record record = client.get(client.writePolicyDefault, key);
client.operate(client.writePolicyDefault, key,
HLLOperation.init(defHLLPolicy, txnHLLBin, bitsHLLIndex)
);
Record after = client.get(client.writePolicyDefault, key);
System.out.println("Before, the " + txnHLLBin + " was - " + record.getValue(txnHLLBin));
System.out.println(" After, the " + txnHLLBin + " is - " + after.getValue(txnHLLBin));
Key key = new Key(txnNamespace, txnSet, theKey);
Record pullGeo = client.get(client.writePolicyDefault, key, txnGeoBin);
System.out.println("The " + txnGeoBin + " is - " + pullGeo.getValue(txnGeoBin));
// Create data for each data type.
import java.util.ArrayList;
import java.util.Arrays;
import java.util.List;
import java.util.Map;
import java.util.HashMap;
String txnString = "atomic";
Integer txnInteger = 8;
Double txnDouble = 6.022;
byte[] txnBlob = new byte[] {0b00000001, 0b00000010, 0b00000011, 0b00000100, 0b00000101};
String txnGeo = String.format("{ \"type\": \"Polygon\", \"coordinates\": [ [[-122.500, 37.000], [-121.000, 37.000], [-121.000, 38.080], [-122.500, 38.080], [-122.500, 37.000]] ] }");
ArrayList<Integer> txnList = new ArrayList<Integer>();
txnList.add(1);
HashMap<Integer, Integer> txnMap = new HashMap <Integer, Integer>();
txnMap.put(2, 4);
System.out.println("--Initial Data–-");
System.out.println("String: " + txnString);
System.out.println("Integer: " + txnInteger);
System.out.println("Double: " + txnDouble);
System.out.println("Blob: " + Arrays.toString(txnBlob));
System.out.println("HLL: Starts with no data.");
System.out.println("Geo: " + txnGeo);
System.out.println("List: " + txnList);
System.out.println("Map: " + txnMap);
System.out.println();
// Put it into Aerospike.
import com.aerospike.client.AerospikeClient;
import com.aerospike.client.Key;
import com.aerospike.client.Bin;
import com.aerospike.client.Value;
import com.aerospike.client.policy.ClientPolicy;
import com.aerospike.client.Operation;
import com.aerospike.client.cdt.ListOperation;
import com.aerospike.client.cdt.MapOperation;
import com.aerospike.client.cdt.MapPolicy;
import com.aerospike.client.operation.BitOperation;
import com.aerospike.client.operation.BitPolicy;
import com.aerospike.client.operation.HLLOperation;
import com.aerospike.client.operation.HLLPolicy;
Integer theKey = 0;
String txnSet = "txnset";
String txnNamespace = "test";
String txnStringBin = "str";
String txnIntegerBin = "int";
String txnDoubleBin = "double";
String txnBlobBin = "blob";
String txnHLLBin = "hll";
String txnGeoBin = "geo";
String txnListBin = "list";
String txnMapBin = "map";
AerospikeClient client = new AerospikeClient("localhost", 3000);
ClientPolicy clientPolicy = new ClientPolicy();
BitPolicy bitPolicy = new BitPolicy();
HLLPolicy defHLLPolicy = new HLLPolicy();
MapPolicy txnMapPolicy = new MapPolicy();
Key key = new Key(txnNamespace, txnSet, theKey);
Bin bin0 = new Bin(txnStringBin, txnString);
Bin bin1 = new Bin(txnIntegerBin, txnInteger);
Bin bin2 = new Bin(txnDoubleBin, txnDouble);
Bin bin3 = new Bin(txnBlobBin, txnBlob);
Bin bin4 = new Bin(txnHLLBin, Value.getAsNull());
Bin bin5 = new Bin(txnGeoBin, Value.getAsGeoJSON(txnGeo));
Bin bin6 = new Bin(txnListBin, txnList);
Bin bin7 = new Bin(txnMapBin, txnMap);
client.put(clientPolicy.writePolicyDefault, key, bin0, bin1, bin2, bin3, bin5, bin6, bin7);
// Apply the following operations as one transaction.
// 1. Touch the record.
// 2. Append to the string.
// 3. Increment the integer.
// 4. Subtract from the double.
// 5. Put an item in the list.
// 6. Increment a value in the map.
// 7. Set bits in the blob.
// 8. Init and add set elements to the hyperloglog.
// 9. Get the GeoJSON.
String txnAppendString = "-transactions";
Bin binStrAppend = new Bin(txnStringBin, txnAppendString);
Integer txnAddInt = 5;
Bin binIntAdd = new Bin(txnIntegerBin, txnAddInt);
Double txnAddDouble = -3.142;
Bin binDoubleSub = new Bin(txnDoubleBin, txnAddDouble);
byte[] bitsToSet = new byte[] {(byte)0b11100000};
Integer bitSize = 8;
Integer bitOffset = 13;
Integer bitsHLLIndex = 8;
Integer listAddition = 5;
Integer mapKey = 2;
Integer mapIncrementValue = 57;
ArrayList<Value> dataListForHLL = new ArrayList<Value>();
dataListForHLL.add(Value.get(txnAddInt));
dataListForHLL.add(Value.get(bitSize));
dataListForHLL.add(Value.get(bitOffset));
dataListForHLL.add(Value.get(bitsHLLIndex));
dataListForHLL.add(Value.get(listAddition));
dataListForHLL.add(Value.get(mapKey));
dataListForHLL.add(Value.get(mapIncrementValue));
Record beforeOps = client.get(client.writePolicyDefault, key);
Record operationsRecord = client.operate(client.writePolicyDefault, key,
Operation.touch(),
Operation.append(binStrAppend),
Operation.add(binIntAdd),
Operation.add(binDoubleSub),
ListOperation.append(txnListBin, Value.get(listAddition)),
MapOperation.increment(txnMapPolicy, txnMapBin, Value.get(mapKey), Value.get(mapIncrementValue)),
BitOperation.set(bitPolicy.Default, txnBlobBin, bitOffset, bitSize, bitsToSet),
HLLOperation.init(defHLLPolicy, txnHLLBin, bitsHLLIndex),
HLLOperation.add(defHLLPolicy, txnHLLBin, dataListForHLL, bitsHLLIndex)
);
Record afterOps = client.get(client.writePolicyDefault, key);
System.out.println("--The Data in Aerospike–-");
System.out.println(beforeOps);
System.out.println();
System.out.println("--Operation Details–-");
System.out.println("Before, the " + txnStringBin + " was - " + beforeOps.getValue(txnStringBin));
System.out.println(" After, the " + txnStringBin + " is - " + afterOps.getValue(txnStringBin));
System.out.println();
System.out.println("Before, the " + txnIntegerBin + " was - " + beforeOps.getValue(txnIntegerBin));
System.out.println(" After, the " + txnIntegerBin + " is - " + afterOps.getValue(txnIntegerBin));
System.out.println();
System.out.println("Before, the " + txnDoubleBin + " was - " + beforeOps.getValue(txnDoubleBin));
System.out.println(" After, the " + txnDoubleBin + " is - " + afterOps.getValue(txnDoubleBin));
System.out.println();
System.out.println("Before, the " + txnListBin + " was - " + beforeOps.getValue(txnListBin));
System.out.println(" After, the " + txnListBin + " is - " + afterOps.getValue(txnListBin));
System.out.println();
System.out.println("Before, the " + txnMapBin + " was - " + beforeOps.getValue(txnMapBin));
System.out.println(" After, the " + txnMapBin + " is - " + afterOps.getValue(txnMapBin));
System.out.println();
byte[] beforeBytes = (byte[])beforeOps.getValue(txnBlobBin);
byte[] afterBytes = (byte[])afterOps.getValue(txnBlobBin);
System.out.println("Before, the " + txnBlobBin + " was - " + Arrays.toString(beforeBytes));
System.out.println(" After, the " + txnBlobBin + " is - " + Arrays.toString(afterBytes));
System.out.println();
System.out.println("Before, the " + txnHLLBin + " was - " + beforeOps.getValue(txnHLLBin));
System.out.println(" After, the " + txnHLLBin + " is - " + afterOps.getValue(txnHLLBin));
System.out.println();
System.out.println("The " + txnGeoBin + " is - " + afterOps.getValue(txnGeoBin));
System.out.println();
System.out.println("--The Record After the Operations–-");
System.out.println(afterOps);
import com.aerospike.client.cdt.MapOrder;
import com.aerospike.client.cdt.MapWriteFlags;
Integer txnDefaultMapkey=2;
Integer txnDefaultValue=1;
MapPolicy txnMapPolicy = new MapPolicy(MapOrder.UNORDERED, MapWriteFlags.CREATE_ONLY | MapWriteFlags.NO_FAIL | MapWriteFlags.PARTIAL);
Key key = new Key(txnNamespace, txnSet, theKey);
Record mapFailRecord = client.operate(client.writePolicyDefault, key,
MapOperation.put(txnMapPolicy, txnMapBin, Value.get(txnDefaultMapkey), Value.get(txnDefaultValue))
);
Record afterOps = client.get(client.writePolicyDefault, key);
System.out.println("The " + txnMapBin + " is - " + afterOps.getValue(txnMapBin));
System.out.println();
import com.aerospike.client.policy.InfoPolicy;
InfoPolicy infoPolicy = new InfoPolicy();
client.truncate(infoPolicy, txnNamespace, txnSet, null);
System.out.println("Set Truncated.");
client.close();
System.out.println("Server connection closed.");
| 0.39257 | 0.956063 |
<a href="https://colab.research.google.com/github/sk-rhyeu/bayesian_lab/blob/master/3_8_Bayesian_with_python_Intro.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# Github 소개
- 참고 1 : 브런치, 개발자들 다 쓴다는 깃허브 https://brunch.co.kr/@thswlsgh/7
- 참고 2 : 브런치, 개발자들 다 쓴다는 깃허브(GitHub) 도대체 뭐지? https://brunch.co.kr/@thswlsgh/7
## Git?
- 소스코드를 효과적으로 관리하기 위해 개발된 '버전관리 시스템'
- 다시 말해, 하나의 파일이 변경된 이력을 추적하고, 복구할 수 있으며, 이것을 공유하며 함께 개발할 수 있게 만든 시스템.
## Github
- 버전관리 시스템인 'git'을 호스팅해주는 플랫폼.
- 언제 어디서나 내 파일의 변동 이력, 수정 및 복구, 다른 사람과의 협업을 가능하게 한 플랫폼.
## 프로그래머를 위한 베이지안 with 파이썬 관련 Github
- 저자 Github : https://goo.gl/Zeegt
- 길벗 출판사 Github : https://github.com/gilbutITbook/006775
---
# 주피터 노트북(Jupyter notebook) 알아보기
- [세미나 류성균 파트 자료](https://nbviewer.jupyter.org/github/sk-rhyeu/bayesian_lab/blob/master/3_8_Bayesian_with_python_Intro.ipynb)
- 단축 URL: https://bit.ly/2HfYDL1
## 주피터 노트북 소개
- 사실 40개 이상의 언어 사용 가능(R도 가능!)
- 컴퓨터 상의 'Kernel'에서 파이썬 등의 언어가 작동하고 출력은 브라우저를 통해서 보여줌
- pdf 등으로 변환 가능하고 Github 등에 올리면 직관적으로 내용 공유 가능!
- 중간중간 Check point를 만들어 한 파일로 버전 관리 가능
## 주피터 노트북 켜기
- 아나콘다 프롬프트(Anaconda Prompt)에 'Jupyter notebook'을 입력하고 엔터!
- 'New'버튼을 누르고 'Python 3'눌러주시면 'Jupyter notebook' 파이썬 실행!
## 주피터 노트북 시작 경로 변경
([참고 : Luke kim, Jupyter notebook에서 홈 디렉토리 변경](https://luke77.tistory.com/52))
1. 아나콘다 프롬프트(Anaconda Prompt) 실행
2. jupyter notebook --generate-config 입력
3. 사용자 폴더에 .jupyter 폴더 진입
4. jupyter_notebook_config.py 열기
5. #c.NotebookApp.notebook_dir = '' 열찾기 (179 번째 line 정도)
6. 주석제거
7. '' 란 안에 원하는 폴더의 절대 경로 삽입. 단 \ --> / 로 변경 (c:\temp --> c:/temp)
8. 저장 후 jupyter notebook 재실행
## Cell 개념 소개
- Jupyter notebook은 Cell을 기본 단위로 구성되어있음
- Cell은 'Code'와 'Markdown'으로 나뉨. 이를 통해 하나의 보고서를 만들 수 있음.
- code : 파이썬 실행되는 영역
- Markdown : 글을 쓸 수 있는 영역
```
print("Hello Bayesian")
```
print("Hello Bayesian")
- 하나의 Cell은 마크다운도 될 수 있고 코드도 될 수 있음
- Code cell 내에서도 주석을 이용해 설명을 달 수는 있지만, 설명의 길이가 길어진다면 Markdown을 활용하는 것이 현명.
- 마크다운을 잘 활용하기 위해서는 문법에 익숙해지는 게 필요하지만 시간 관계상 생략..
- 참조 : [마크다운 사용법](https://gist.github.com/ihoneymon/652be052a0727ad59601)
## Jupyter notebook 단축키
- 참조 1 [따라하며 배우는 데이터 과학](https://dataninja.me/ipds-kr/python-setup/)
- 참조 2 [꼬낄콘의 분석일지](https://kkokkilkon.tistory.com/151)
### (1) 공통
- **선택 셀 코드 실행 [ctrl] + [enter]**
- **선택 셀 코드 실행 후 아래 셀로 이동 [Shift] + [enter]**
- **선택 셀 코드 실행 후 새로운 셀 추가 [Alt] + [enter]**
- 커맨드 모드로 전환 [ECS]
- 편집 모드로 전환 [Enter]
### (2) 셀 선택 모드 Command Mode
- [esc] 또는 [ctrl]+[m]을 눌러 '셀 선택 모드'를 만들 수 있다.
- 셀 단위 편집 가능
- **아래로 셀 추가 [b]**
- **선텍 셀 삭제 [d][d]**
- **Markdown / Code 로 변경 [m] / [y]**
- **파일 저장 [Ctrl]+[s] 또는 [s]**
- 위로 셀 추가 [a]
- 선택 셀 잘라내기 [x]
- 선택 셀 복사하기 [c]
- 선택 셀 아래 붙여넣기 [p]
- 선택 셀과 아래 셀 합치기 [shift] + [m]
- 실행결과 열기 / 닫기 [o]
- 선택 셀의 코드 입력 모드로 돌아가기 [enter]
## (3) 코드 입력 모드 Edit Mode
- [enter]를 눌러 셀이 아래와 같이 초록색이 된 상태(코드 입력 모드)
- 선택 셀 코드 전체 선택 [ctrl] + [a]
- 선택 셀 내 실행 취소 [ctrl] + [z]
## '프로그래머를 위한 베이지안 with 파이썬' 코드 입력 및 활용
```
from IPython.core.pylabtools import figsize
import numpy as np
from matplotlib import pyplot as plt
import matplotlib
matplotlib.rc("font", family="Malgun Gothic")
figsize(11, 9)
import scipy.stats as stats
dist = stats.beta
n_trials = [0, 1, 2, 3, 4, 5, 8, 15, 50, 500]
data = stats.bernoulli.rvs(0.5, size=n_trials[-1]) # [-1]
x = np.linspace(0,1, 100)
# For the already prepared, I'm using Binomial's conj. prior.
for k, N in enumerate(n_trials):
sx = plt.subplot(len(n_trials) / 2, 2, k + 1)
plt.xlabel("$p$, probability of heads") \
if k in [0, len(n_trials) - 1] else None
plt.setp(sx.get_yticklabels(), visible=False)
heads = data[:N].sum()
y = dist.pdf(x, 1 + heads, 1 + N - heads)
plt.plot(x, y, label="11 observe %d tosses,\n %d heads" % (N, heads))
plt.fill_between(x, 0, y, color="#348ABD", alpha=0.4)
plt.vlines(0.5, 0, 4, color="k", linestyles="--", lw=1)
leg = plt.legend()
leg.get_frame().set_alpha(0.4)
plt.autoscale(tight=True)
```
---
# Google Colaboratory 사용해보기
## 장점
- Google Docs의 주피터 노트북 판!
- 더 좋은건 웹상에서 RAM과 GPU 제공
- 사양 <https://colab.research.google.com/drive/151805XTDg--dgHb3-AXJCpnWaqRhop_2#scrollTo=vEWe-FHNDY3E>
- CPU : 1xsingle core hyper threaded i.e(1 core, 2 threads) Xeon Processors @2.3Ghz (No Turbo Boost) , 45MB Cache
- RAM : ~12.6 GB Available
- DISK : ~33 GB Available
- GPU : 1xTesla K80 , having 2496 CUDA cores, compute 3.7, 12GB(11.439GB Usable) GDDR5 VRAM
- GPU / TPU는 [수정] - [노트설정]에서 설정
- 구글 드라이브에 데이터를 올려놓고 바로 작업 가능
## 단점
- 인터넷 연결에 영향을 받거나 가끔씩 꺼지는 경우(요새는 많이 안정화)
- 내가 설치한 새로운 환경 패키지, 샘플 데이터 등이 소실 가능...
## Colab 시작하기
- Google Drive에서 좌측 상단 [새로만들기] - [더보기] - [Colabotory] 클릭
- 만약에 [새로만들기] - [더보기] 창에 'Colaboratory'가 없으면, [새로만들기] - [더보기] - [연결할 앱 더 보기] 에서 'Colaboratory' 추가
## 단축키
- 단축키는 대부분 주피터 노트북의 단축키를 누르기 전에 [Ctrl] + [m] 을 누른다.
- 단축기 조정 가능 [Ctrl] + [m], [h]
기본
```
import tensorflow as tf
input1 = tf.ones((2, 3))
input2 = tf.reshape(tf.range(1, 7, dtype=tf.float32), (2, 3))
output = input1 + input2
with tf.Session():
result = output.eval()
result
import matplotlib.pyplot as plt
import numpy as np
x = np.arange(20)
y = [x_i + np.random.randn(1) for x_i in x]
a, b = np.polyfit(x, y, 1)
_ = plt.plot(x, y, 'o', np.arange(20), a*np.arange(20)+b, '-')
!pip install -q matplotlib-venn
from matplotlib_venn import venn2
_ = venn2(subsets = (3, 2, 1))
```
## 추가 자료
- [Google Colab 사용하기](https://zzsza.github.io/data/2018/08/30/google-colab/) : 로컬 연동, Pytorch, TensorBoard, Kaggle 연동 등
---
# 추천 자료
- 사실상 책을 보기 위해서는 파이썬 구조와 문법에 대한 이해가 선행되어야 하지만..
- 도서
- [점프 투 파이썬](https://wikidocs.net/book/1) (무료 위키 북스)
- 강좌
- [김왼손 유튜브](https://www.youtube.com/channel/UC0h8NzL2vllvp3PjdoYSK4g)
- [한입에 쏙 파이썬](https://www.youtube.com/watch?v=UrwFkNRzzT4&list=PLGPF8gvWLYyontH0PECIUFFUdvATXWQEL)
- [미운코딩새끼](https://www.youtube.com/watch?v=c2mpe9Xcp0I&list=PLGPF8gvWLYyrkF85itdBHaOLSVbtdzBww)
- [유기농냠냠파이썬](https://www.youtube.com/watch?v=UHg1Drp1uKE&list=PLGPF8gvWLYypeEoFNTfSHdFL5WRLAfmmm)
- [파이썬 예제 뽀개기](https://www.youtube.com/watch?v=-JuiKYQZiNQ&list=PLGPF8gvWLYyomy0D1-uoQHTWPvjXTjr_a)
- Edwith 강좌
- [모두를 위한 파이썬](https://www.edwith.org/pythonforeverybody)
- [머신러닝을 위한 Python 워밍업](https://www.edwith.org/aipython)
- [기초 PYTHON 프로그래밍](https://www.edwith.org/sogang_python/joinLectures/7133)
---
# 스터디 스케줄 및 담당자 정하기
| **일자** | **내용** | **담당자** |
| :---: | :---: | :---: |
| 3 / 08 | **부록 - 아나콘다 설치, 주피터 노트북, 구글 Colab, 스파이더 사용법** | 박현재, 류성균 |
| 3 / 15 | **1장 베이지안 추론의 철학** | 김지현 |
| 3 / 22 | **2장 PyMC 더 알아보기** | 서동주 |
| 3 / 29 | MT?? | |
| 4 / 05 | **3장 MCMC 블랙박스 열기** | 김수진 |
| 4 / 12 | **4장 아무도 알려주지 않는 위대한 이론** | |
| 4 / 19 | 중간고사 전 주? | |
| 4 / 26 | 중간고사 | |
| 5 / 03 | **5장 오히려 큰 손해를 보시겠습니까?** | |
| 5 / 10 | **6장 우선순위 바로 잡기** | |
| 5 / 17 | **7장 베이지안 A/B 테스트** | |
| 5 / 24 | 한국 통계 학회 | |
```
```
|
github_jupyter
|
print("Hello Bayesian")
from IPython.core.pylabtools import figsize
import numpy as np
from matplotlib import pyplot as plt
import matplotlib
matplotlib.rc("font", family="Malgun Gothic")
figsize(11, 9)
import scipy.stats as stats
dist = stats.beta
n_trials = [0, 1, 2, 3, 4, 5, 8, 15, 50, 500]
data = stats.bernoulli.rvs(0.5, size=n_trials[-1]) # [-1]
x = np.linspace(0,1, 100)
# For the already prepared, I'm using Binomial's conj. prior.
for k, N in enumerate(n_trials):
sx = plt.subplot(len(n_trials) / 2, 2, k + 1)
plt.xlabel("$p$, probability of heads") \
if k in [0, len(n_trials) - 1] else None
plt.setp(sx.get_yticklabels(), visible=False)
heads = data[:N].sum()
y = dist.pdf(x, 1 + heads, 1 + N - heads)
plt.plot(x, y, label="11 observe %d tosses,\n %d heads" % (N, heads))
plt.fill_between(x, 0, y, color="#348ABD", alpha=0.4)
plt.vlines(0.5, 0, 4, color="k", linestyles="--", lw=1)
leg = plt.legend()
leg.get_frame().set_alpha(0.4)
plt.autoscale(tight=True)
import tensorflow as tf
input1 = tf.ones((2, 3))
input2 = tf.reshape(tf.range(1, 7, dtype=tf.float32), (2, 3))
output = input1 + input2
with tf.Session():
result = output.eval()
result
import matplotlib.pyplot as plt
import numpy as np
x = np.arange(20)
y = [x_i + np.random.randn(1) for x_i in x]
a, b = np.polyfit(x, y, 1)
_ = plt.plot(x, y, 'o', np.arange(20), a*np.arange(20)+b, '-')
!pip install -q matplotlib-venn
from matplotlib_venn import venn2
_ = venn2(subsets = (3, 2, 1))
| 0.415966 | 0.903932 |
#Time Series Data Encoding
In this chapter, we will examine time series encoding and recurrent networks, two topics that are logical to put together because they are both methods for dealing with data that spans over time. Time series encoding deals with representing events that occur over time to a neural network. There are many different methods to encode data that occur over time to a neural network. This encoding is necessary because a feedforward neural network will always produce the same output vector for a given input vector. Recurrent neural networks do not require encoding of time series data because they are able to handle data that occur over time automatically.
The variation in temperature during the week is an example of time-series data. For instance, if we know that today’s temperature is 25 degrees, and tomorrow’s temperature is 27 degrees, the recurrent neural networks and time series encoding provide another option to predict the correct temperature for the week. Conversely, a traditional feedforward neural network will always respond with the same output for a given input. If we train a feedforward neural network to predict tomorrow’s temperature, it should return a value of 27 for 25. The fact that it will always output 27 when given 25 might be a hindrance to its predictions. Surely the temperature of 27 will not always follow 25. It would be better for the neural network to consider the temperatures for a series of days before the prediction. Perhaps the temperature over the last week might allow us to predict tomorrow’s temperature. Therefore, recurrent neural networks and time series encoding represent two different approaches to representing data over time to a neural network.
Previously we trained neural networks with input ($x$) and expected output ($y$). $X$ was a matrix, the rows were training examples, and the columns were values to be predicted. The $x$ value will now contain sequences of data. The definition of the $y$ value will stay the same.
Dimensions of the training set ($x$):
* Axis 1: Training set elements (sequences) (must be of the same size as $y$ size)
* Axis 2: Members of sequence
* Axis 3: Features in data (like input neurons)
Previously, we might take as input a single stock price, to predict if we should buy (1), sell (-1), or hold (0). The following code illustrates this encoding.
```
#
x = [
[32],
[41],
[39],
[20],
[15]
]
y = [
1,
-1,
0,
-1,
1
]
print(x)
print(y)
```
The following code builds a CSV file from scratch, to see it as a data frame, use the following:
```
from IPython.display import display, HTML
import pandas as pd
import numpy as np
x = np.array(x)
print(x[:,0])
df = pd.DataFrame({'x':x[:,0], 'y':y})
display(df)
```
You might want to put volume in with the stock price. The following code shows how we can add an additional dimension to handle the volume.
```
x = [
[32,1383],
[41,2928],
[39,8823],
[20,1252],
[15,1532]
]
y = [
1,
-1,
0,
-1,
1
]
print(x)
print(y)
```
Again, very similar to what we did before. The following shows this as a data frame.
```
from IPython.display import display, HTML
import pandas as pd
import numpy as np
x = np.array(x)
print(x[:,0])
df = pd.DataFrame({'price':x[:,0], 'volume':x[:,1], 'y':y})
display(df)
```
Now we get to sequence format. We want to predict something over a sequence, so the data format needs to add a dimension. A maximum sequence length must be specified, but the individual sequences can be of any length.
```
x = [
[[32,1383],[41,2928],[39,8823],[20,1252],[15,1532]],
[[35,8272],[32,1383],[41,2928],[39,8823],[20,1252]],
[[37,2738],[35,8272],[32,1383],[41,2928],[39,8823]],
[[34,2845],[37,2738],[35,8272],[32,1383],[41,2928]],
[[32,2345],[34,2845],[37,2738],[35,8272],[32,1383]],
]
y = [
1,
-1,
0,
-1,
1
]
print(x)
print(y)
```
Even if there is only one feature (price), the 3rd dimension must be used:
```
x = [
[[32],[41],[39],[20],[15]],
[[35],[32],[41],[39],[20]],
[[37],[35],[32],[41],[39]],
[[34],[37],[35],[32],[41]],
[[32],[34],[37],[35],[32]],
]
y = [
1,
-1,
0,
-1,
1
]
print(x)
print(y)
```
|
github_jupyter
|
#
x = [
[32],
[41],
[39],
[20],
[15]
]
y = [
1,
-1,
0,
-1,
1
]
print(x)
print(y)
from IPython.display import display, HTML
import pandas as pd
import numpy as np
x = np.array(x)
print(x[:,0])
df = pd.DataFrame({'x':x[:,0], 'y':y})
display(df)
x = [
[32,1383],
[41,2928],
[39,8823],
[20,1252],
[15,1532]
]
y = [
1,
-1,
0,
-1,
1
]
print(x)
print(y)
from IPython.display import display, HTML
import pandas as pd
import numpy as np
x = np.array(x)
print(x[:,0])
df = pd.DataFrame({'price':x[:,0], 'volume':x[:,1], 'y':y})
display(df)
x = [
[[32,1383],[41,2928],[39,8823],[20,1252],[15,1532]],
[[35,8272],[32,1383],[41,2928],[39,8823],[20,1252]],
[[37,2738],[35,8272],[32,1383],[41,2928],[39,8823]],
[[34,2845],[37,2738],[35,8272],[32,1383],[41,2928]],
[[32,2345],[34,2845],[37,2738],[35,8272],[32,1383]],
]
y = [
1,
-1,
0,
-1,
1
]
print(x)
print(y)
x = [
[[32],[41],[39],[20],[15]],
[[35],[32],[41],[39],[20]],
[[37],[35],[32],[41],[39]],
[[34],[37],[35],[32],[41]],
[[32],[34],[37],[35],[32]],
]
y = [
1,
-1,
0,
-1,
1
]
print(x)
print(y)
| 0.094557 | 0.992371 |
# Unity ML Agents
## Proximal Policy Optimization (PPO)
Contains an implementation of PPO as described [here](https://arxiv.org/abs/1707.06347).
```
import numpy as np
import os
import tensorflow as tf
from ppo.history import *
from ppo.models import *
from ppo.trainer import Trainer
from unityagents import *
```
### Hyperparameters
```
### General parameters
max_steps = 5e5 # Set maximum number of steps to run environment.
run_path = "ppo" # The sub-directory name for model and summary statistics
load_model = False # Whether to load a saved model.
train_model = True # Whether to train the model.
summary_freq = 10000 # Frequency at which to save training statistics.
save_freq = 50000 # Frequency at which to save model.
env_name = "environment" # Name of the training environment file.
curriculum_file = None
### Algorithm-specific parameters for tuning
gamma = 0.99 # Reward discount rate.
lambd = 0.95 # Lambda parameter for GAE.
time_horizon = 2048 # How many steps to collect per agent before adding to buffer.
beta = 1e-3 # Strength of entropy regularization
num_epoch = 5 # Number of gradient descent steps per batch of experiences.
num_layers = 2 # Number of hidden layers between state/observation encoding and value/policy layers.
epsilon = 0.2 # Acceptable threshold around ratio of old and new policy probabilities.
buffer_size = 2048 # How large the experience buffer should be before gradient descent.
learning_rate = 3e-4 # Model learning rate.
hidden_units = 64 # Number of units in hidden layer.
batch_size = 64 # How many experiences per gradient descent update step.
normalize = False
### Logging dictionary for hyperparameters
hyperparameter_dict = {'max_steps':max_steps, 'run_path':run_path, 'env_name':env_name,
'curriculum_file':curriculum_file, 'gamma':gamma, 'lambd':lambd, 'time_horizon':time_horizon,
'beta':beta, 'num_epoch':num_epoch, 'epsilon':epsilon, 'buffe_size':buffer_size,
'leaning_rate':learning_rate, 'hidden_units':hidden_units, 'batch_size':batch_size}
```
### Load the environment
```
env = UnityEnvironment(file_name=env_name, curriculum=curriculum_file)
print(str(env))
brain_name = env.external_brain_names[0]
```
### Train the Agent(s)
```
tf.reset_default_graph()
if curriculum_file == "None":
curriculum_file = None
def get_progress():
if curriculum_file is not None:
if env._curriculum.measure_type == "progress":
return steps / max_steps
elif env._curriculum.measure_type == "reward":
return last_reward
else:
return None
else:
return None
# Create the Tensorflow model graph
ppo_model = create_agent_model(env, lr=learning_rate,
h_size=hidden_units, epsilon=epsilon,
beta=beta, max_step=max_steps,
normalize=normalize, num_layers=num_layers)
is_continuous = (env.brains[brain_name].action_space_type == "continuous")
use_observations = (env.brains[brain_name].number_observations > 0)
use_states = (env.brains[brain_name].state_space_size > 0)
model_path = './models/{}'.format(run_path)
summary_path = './summaries/{}'.format(run_path)
if not os.path.exists(model_path):
os.makedirs(model_path)
if not os.path.exists(summary_path):
os.makedirs(summary_path)
init = tf.global_variables_initializer()
saver = tf.train.Saver()
with tf.Session() as sess:
# Instantiate model parameters
if load_model:
print('Loading Model...')
ckpt = tf.train.get_checkpoint_state(model_path)
saver.restore(sess, ckpt.model_checkpoint_path)
else:
sess.run(init)
steps, last_reward = sess.run([ppo_model.global_step, ppo_model.last_reward])
summary_writer = tf.summary.FileWriter(summary_path)
info = env.reset(train_mode=train_model, progress=get_progress())[brain_name]
trainer = Trainer(ppo_model, sess, info, is_continuous, use_observations, use_states, train_model)
if train_model:
trainer.write_text(summary_writer, 'Hyperparameters', hyperparameter_dict, steps)
while steps <= max_steps:
if env.global_done:
info = env.reset(train_mode=train_model, progress=get_progress())[brain_name]
# Decide and take an action
new_info = trainer.take_action(info, env, brain_name, steps, normalize)
info = new_info
trainer.process_experiences(info, time_horizon, gamma, lambd)
if len(trainer.training_buffer['actions']) > buffer_size and train_model:
# Perform gradient descent with experience buffer
trainer.update_model(batch_size, num_epoch)
if steps % summary_freq == 0 and steps != 0 and train_model:
# Write training statistics to tensorboard.
trainer.write_summary(summary_writer, steps, env._curriculum.lesson_number)
if steps % save_freq == 0 and steps != 0 and train_model:
# Save Tensorflow model
save_model(sess, model_path=model_path, steps=steps, saver=saver)
steps += 1
sess.run(ppo_model.increment_step)
if len(trainer.stats['cumulative_reward']) > 0:
mean_reward = np.mean(trainer.stats['cumulative_reward'])
sess.run(ppo_model.update_reward, feed_dict={ppo_model.new_reward: mean_reward})
last_reward = sess.run(ppo_model.last_reward)
# Final save Tensorflow model
if steps != 0 and train_model:
save_model(sess, model_path=model_path, steps=steps, saver=saver)
env.close()
export_graph(model_path, env_name)
```
### Export the trained Tensorflow graph
Once the model has been trained and saved, we can export it as a .bytes file which Unity can embed.
```
export_graph(model_path, env_name)
```
|
github_jupyter
|
import numpy as np
import os
import tensorflow as tf
from ppo.history import *
from ppo.models import *
from ppo.trainer import Trainer
from unityagents import *
### General parameters
max_steps = 5e5 # Set maximum number of steps to run environment.
run_path = "ppo" # The sub-directory name for model and summary statistics
load_model = False # Whether to load a saved model.
train_model = True # Whether to train the model.
summary_freq = 10000 # Frequency at which to save training statistics.
save_freq = 50000 # Frequency at which to save model.
env_name = "environment" # Name of the training environment file.
curriculum_file = None
### Algorithm-specific parameters for tuning
gamma = 0.99 # Reward discount rate.
lambd = 0.95 # Lambda parameter for GAE.
time_horizon = 2048 # How many steps to collect per agent before adding to buffer.
beta = 1e-3 # Strength of entropy regularization
num_epoch = 5 # Number of gradient descent steps per batch of experiences.
num_layers = 2 # Number of hidden layers between state/observation encoding and value/policy layers.
epsilon = 0.2 # Acceptable threshold around ratio of old and new policy probabilities.
buffer_size = 2048 # How large the experience buffer should be before gradient descent.
learning_rate = 3e-4 # Model learning rate.
hidden_units = 64 # Number of units in hidden layer.
batch_size = 64 # How many experiences per gradient descent update step.
normalize = False
### Logging dictionary for hyperparameters
hyperparameter_dict = {'max_steps':max_steps, 'run_path':run_path, 'env_name':env_name,
'curriculum_file':curriculum_file, 'gamma':gamma, 'lambd':lambd, 'time_horizon':time_horizon,
'beta':beta, 'num_epoch':num_epoch, 'epsilon':epsilon, 'buffe_size':buffer_size,
'leaning_rate':learning_rate, 'hidden_units':hidden_units, 'batch_size':batch_size}
env = UnityEnvironment(file_name=env_name, curriculum=curriculum_file)
print(str(env))
brain_name = env.external_brain_names[0]
tf.reset_default_graph()
if curriculum_file == "None":
curriculum_file = None
def get_progress():
if curriculum_file is not None:
if env._curriculum.measure_type == "progress":
return steps / max_steps
elif env._curriculum.measure_type == "reward":
return last_reward
else:
return None
else:
return None
# Create the Tensorflow model graph
ppo_model = create_agent_model(env, lr=learning_rate,
h_size=hidden_units, epsilon=epsilon,
beta=beta, max_step=max_steps,
normalize=normalize, num_layers=num_layers)
is_continuous = (env.brains[brain_name].action_space_type == "continuous")
use_observations = (env.brains[brain_name].number_observations > 0)
use_states = (env.brains[brain_name].state_space_size > 0)
model_path = './models/{}'.format(run_path)
summary_path = './summaries/{}'.format(run_path)
if not os.path.exists(model_path):
os.makedirs(model_path)
if not os.path.exists(summary_path):
os.makedirs(summary_path)
init = tf.global_variables_initializer()
saver = tf.train.Saver()
with tf.Session() as sess:
# Instantiate model parameters
if load_model:
print('Loading Model...')
ckpt = tf.train.get_checkpoint_state(model_path)
saver.restore(sess, ckpt.model_checkpoint_path)
else:
sess.run(init)
steps, last_reward = sess.run([ppo_model.global_step, ppo_model.last_reward])
summary_writer = tf.summary.FileWriter(summary_path)
info = env.reset(train_mode=train_model, progress=get_progress())[brain_name]
trainer = Trainer(ppo_model, sess, info, is_continuous, use_observations, use_states, train_model)
if train_model:
trainer.write_text(summary_writer, 'Hyperparameters', hyperparameter_dict, steps)
while steps <= max_steps:
if env.global_done:
info = env.reset(train_mode=train_model, progress=get_progress())[brain_name]
# Decide and take an action
new_info = trainer.take_action(info, env, brain_name, steps, normalize)
info = new_info
trainer.process_experiences(info, time_horizon, gamma, lambd)
if len(trainer.training_buffer['actions']) > buffer_size and train_model:
# Perform gradient descent with experience buffer
trainer.update_model(batch_size, num_epoch)
if steps % summary_freq == 0 and steps != 0 and train_model:
# Write training statistics to tensorboard.
trainer.write_summary(summary_writer, steps, env._curriculum.lesson_number)
if steps % save_freq == 0 and steps != 0 and train_model:
# Save Tensorflow model
save_model(sess, model_path=model_path, steps=steps, saver=saver)
steps += 1
sess.run(ppo_model.increment_step)
if len(trainer.stats['cumulative_reward']) > 0:
mean_reward = np.mean(trainer.stats['cumulative_reward'])
sess.run(ppo_model.update_reward, feed_dict={ppo_model.new_reward: mean_reward})
last_reward = sess.run(ppo_model.last_reward)
# Final save Tensorflow model
if steps != 0 and train_model:
save_model(sess, model_path=model_path, steps=steps, saver=saver)
env.close()
export_graph(model_path, env_name)
export_graph(model_path, env_name)
| 0.700485 | 0.889241 |
# 1. Import libraries
```
#----------------------------Reproducible----------------------------------------------------------------------------------------
import numpy as np
import tensorflow as tf
import random as rn
import os
seed=0
os.environ['PYTHONHASHSEED'] = str(seed)
np.random.seed(seed)
rn.seed(seed)
#session_conf = tf.ConfigProto(intra_op_parallelism_threads=1, inter_op_parallelism_threads=1)
session_conf =tf.compat.v1.ConfigProto(intra_op_parallelism_threads=1, inter_op_parallelism_threads=1)
from keras import backend as K
#tf.set_random_seed(seed)
tf.compat.v1.set_random_seed(seed)
#sess = tf.Session(graph=tf.get_default_graph(), config=session_conf)
sess = tf.compat.v1.Session(graph=tf.compat.v1.get_default_graph(), config=session_conf)
K.set_session(sess)
#----------------------------Reproducible----------------------------------------------------------------------------------------
os.environ['TF_CPP_MIN_LOG_LEVEL'] = '3'
#--------------------------------------------------------------------------------------------------------------------------------
from keras.datasets import fashion_mnist
from keras.models import Model
from keras.layers import Dense, Input, Flatten, Activation, Dropout, Layer
from keras.layers.normalization import BatchNormalization
from keras.utils import to_categorical
from keras import optimizers,initializers,constraints,regularizers
from keras import backend as K
from keras.callbacks import LambdaCallback,ModelCheckpoint
from keras.utils import plot_model
from sklearn.model_selection import StratifiedKFold
from sklearn.ensemble import ExtraTreesClassifier
from sklearn import svm
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import ShuffleSplit
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
from sklearn.svm import SVC
import h5py
import math
import matplotlib
import matplotlib.pyplot as plt
import matplotlib.cm as cm
%matplotlib inline
matplotlib.style.use('ggplot')
import random
import scipy.sparse as sparse
import pandas as pd
from skimage import io
from PIL import Image
from sklearn.model_selection import train_test_split
import scipy.sparse as sparse
#--------------------------------------------------------------------------------------------------------------------------------
#Import ourslef defined methods
import sys
sys.path.append(r"./Defined")
import Functions as F
# The following code should be added before the keras model
#np.random.seed(seed)
#--------------------------------------------------------------------------------------------------------------------------------
def write_to_csv(p_data,p_path):
dataframe = pd.DataFrame(p_data)
dataframe.to_csv(p_path, mode='a',header=False,index=False,sep=',')
del dataframe
```
# 2. Loading data
```
dataset_path='./Dataset/coil-20-proc/'
samples={}
for dirpath, dirnames, filenames in os.walk(dataset_path):
#print(dirpath)
#print(dirnames)
#print(filenames)
dirnames.sort()
filenames.sort()
for filename in [f for f in filenames if f.endswith(".png") and not f.find('checkpoint')>0]:
full_path = os.path.join(dirpath, filename)
file_identifier=filename.split('__')[0][3:]
if file_identifier not in samples.keys():
samples[file_identifier] = []
# Direct read
#image = io.imread(full_path)
# Resize read
image_=Image.open(full_path).resize((20, 20),Image.ANTIALIAS)
image=np.asarray(image_)
samples[file_identifier].append(image)
#plt.imshow(samples['1'][0].reshape(20,20))
data_arr_list=[]
label_arr_list=[]
for key_i in samples.keys():
key_i_for_label=[int(key_i)-1]
data_arr_list.append(np.array(samples[key_i]))
label_arr_list.append(np.array(72*key_i_for_label))
data_arr=np.concatenate(data_arr_list).reshape(1440, 20*20).astype('float32') / 255.
label_arr_onehot=to_categorical(np.concatenate(label_arr_list))
sample_used=699
x_train_all,x_test,y_train_all,y_test_onehot= train_test_split(data_arr,label_arr_onehot,test_size=0.2,random_state=seed)
x_train=x_train_all[0:sample_used]
y_train_onehot=y_train_all[0:sample_used]
print('Shape of x_train: ' + str(x_train.shape))
print('Shape of x_test: ' + str(x_test.shape))
print('Shape of y_train: ' + str(y_train_onehot.shape))
print('Shape of y_test: ' + str(y_test_onehot.shape))
F.show_data_figures(x_train[0:40],20,20,40)
F.show_data_figures(x_test[0:40],20,20,40)
key_feture_number=50
```
# 3.Model
```
np.random.seed(seed)
#--------------------------------------------------------------------------------------------------------------------------------
class Feature_Select_Layer(Layer):
def __init__(self, output_dim, **kwargs):
super(Feature_Select_Layer, self).__init__(**kwargs)
self.output_dim = output_dim
def build(self, input_shape):
self.kernel = self.add_weight(name='kernel',
shape=(input_shape[1],),
initializer=initializers.RandomUniform(minval=0.999999, maxval=0.9999999, seed=seed),
trainable=True)
super(Feature_Select_Layer, self).build(input_shape)
def call(self, x, selection=False,k=key_feture_number):
kernel=K.abs(self.kernel)
if selection:
kernel_=K.transpose(kernel)
kth_largest = tf.math.top_k(kernel_, k=k)[0][-1]
kernel = tf.where(condition=K.less(kernel,kth_largest),x=K.zeros_like(kernel),y=kernel)
return K.dot(x, tf.linalg.tensor_diag(kernel))
def compute_output_shape(self, input_shape):
return (input_shape[0], self.output_dim)
#--------------------------------------------------------------------------------------------------------------------------------
def Fractal_Autoencoder(p_data_feature=x_train.shape[1],\
p_feture_number=key_feture_number,\
p_encoding_dim=key_feture_number,\
p_learning_rate=1E-3,\
p_loss_weight_1=1,\
p_loss_weight_2=2):
input_img = Input(shape=(p_data_feature,), name='autoencoder_input')
feature_selection = Feature_Select_Layer(output_dim=p_data_feature,\
input_shape=(p_data_feature,),\
name='feature_selection')
feature_selection_score=feature_selection(input_img)
feature_selection_choose=feature_selection(input_img,selection=True,k=p_feture_number)
encoded = Dense(p_encoding_dim,\
activation='linear',\
kernel_initializer=initializers.glorot_uniform(seed),\
name='autoencoder_hidden_layer')
encoded_score=encoded(feature_selection_score)
encoded_choose=encoded(feature_selection_choose)
bottleneck_score=encoded_score
bottleneck_choose=encoded_choose
decoded = Dense(p_data_feature,\
activation='linear',\
kernel_initializer=initializers.glorot_uniform(seed),\
name='autoencoder_output')
decoded_score =decoded(bottleneck_score)
decoded_choose =decoded(bottleneck_choose)
latent_encoder_score = Model(input_img, bottleneck_score)
latent_encoder_choose = Model(input_img, bottleneck_choose)
feature_selection_output=Model(input_img,feature_selection_choose)
autoencoder = Model(input_img, [decoded_score,decoded_choose])
autoencoder.compile(loss=['mean_squared_error','mean_squared_error'],\
loss_weights=[p_loss_weight_1, p_loss_weight_2],\
optimizer=optimizers.Adam(lr=p_learning_rate))
print('Autoencoder Structure-------------------------------------')
autoencoder.summary()
return autoencoder,feature_selection_output,latent_encoder_score,latent_encoder_choose
```
## 3.1 Structure and paramter testing
```
epochs_number=200
batch_size_value=8
```
---
### 3.1.1 Fractal Autoencoder
---
```
loss_weight_1=0.0078125
F_AE,\
feature_selection_output,\
latent_encoder_score_F_AE,\
latent_encoder_choose_F_AE=Fractal_Autoencoder(p_data_feature=x_train.shape[1],\
p_feture_number=key_feture_number,\
p_encoding_dim=key_feture_number,\
p_learning_rate= 1E-3,\
p_loss_weight_1=loss_weight_1,\
p_loss_weight_2=1)
F_AE_history = F_AE.fit(x_train, [x_train,x_train],\
epochs=epochs_number,\
batch_size=batch_size_value,\
shuffle=True)
loss = F_AE_history.history['loss']
epochs = range(epochs_number)
plt.plot(epochs, loss, 'bo', label='Training Loss')
plt.xlabel('Epochs')
plt.ylabel('Loss')
plt.legend()
plt.show()
fina_results=np.array(F_AE.evaluate(x_test,[x_test,x_test]))
fina_results
fina_results_single=np.array(F_AE.evaluate(x_test[0:1],[x_test[0:1],x_test[0:1]]))
fina_results_single
for i in np.arange(x_test.shape[0]):
fina_results_i=np.array(F_AE.evaluate(x_test[i:i+1],[x_test[i:i+1],x_test[i:i+1]]))
write_to_csv(fina_results_i.reshape(1,len(fina_results_i)),"./log/results_"+str(sample_used)+".csv")
```
|
github_jupyter
|
#----------------------------Reproducible----------------------------------------------------------------------------------------
import numpy as np
import tensorflow as tf
import random as rn
import os
seed=0
os.environ['PYTHONHASHSEED'] = str(seed)
np.random.seed(seed)
rn.seed(seed)
#session_conf = tf.ConfigProto(intra_op_parallelism_threads=1, inter_op_parallelism_threads=1)
session_conf =tf.compat.v1.ConfigProto(intra_op_parallelism_threads=1, inter_op_parallelism_threads=1)
from keras import backend as K
#tf.set_random_seed(seed)
tf.compat.v1.set_random_seed(seed)
#sess = tf.Session(graph=tf.get_default_graph(), config=session_conf)
sess = tf.compat.v1.Session(graph=tf.compat.v1.get_default_graph(), config=session_conf)
K.set_session(sess)
#----------------------------Reproducible----------------------------------------------------------------------------------------
os.environ['TF_CPP_MIN_LOG_LEVEL'] = '3'
#--------------------------------------------------------------------------------------------------------------------------------
from keras.datasets import fashion_mnist
from keras.models import Model
from keras.layers import Dense, Input, Flatten, Activation, Dropout, Layer
from keras.layers.normalization import BatchNormalization
from keras.utils import to_categorical
from keras import optimizers,initializers,constraints,regularizers
from keras import backend as K
from keras.callbacks import LambdaCallback,ModelCheckpoint
from keras.utils import plot_model
from sklearn.model_selection import StratifiedKFold
from sklearn.ensemble import ExtraTreesClassifier
from sklearn import svm
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import ShuffleSplit
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
from sklearn.svm import SVC
import h5py
import math
import matplotlib
import matplotlib.pyplot as plt
import matplotlib.cm as cm
%matplotlib inline
matplotlib.style.use('ggplot')
import random
import scipy.sparse as sparse
import pandas as pd
from skimage import io
from PIL import Image
from sklearn.model_selection import train_test_split
import scipy.sparse as sparse
#--------------------------------------------------------------------------------------------------------------------------------
#Import ourslef defined methods
import sys
sys.path.append(r"./Defined")
import Functions as F
# The following code should be added before the keras model
#np.random.seed(seed)
#--------------------------------------------------------------------------------------------------------------------------------
def write_to_csv(p_data,p_path):
dataframe = pd.DataFrame(p_data)
dataframe.to_csv(p_path, mode='a',header=False,index=False,sep=',')
del dataframe
dataset_path='./Dataset/coil-20-proc/'
samples={}
for dirpath, dirnames, filenames in os.walk(dataset_path):
#print(dirpath)
#print(dirnames)
#print(filenames)
dirnames.sort()
filenames.sort()
for filename in [f for f in filenames if f.endswith(".png") and not f.find('checkpoint')>0]:
full_path = os.path.join(dirpath, filename)
file_identifier=filename.split('__')[0][3:]
if file_identifier not in samples.keys():
samples[file_identifier] = []
# Direct read
#image = io.imread(full_path)
# Resize read
image_=Image.open(full_path).resize((20, 20),Image.ANTIALIAS)
image=np.asarray(image_)
samples[file_identifier].append(image)
#plt.imshow(samples['1'][0].reshape(20,20))
data_arr_list=[]
label_arr_list=[]
for key_i in samples.keys():
key_i_for_label=[int(key_i)-1]
data_arr_list.append(np.array(samples[key_i]))
label_arr_list.append(np.array(72*key_i_for_label))
data_arr=np.concatenate(data_arr_list).reshape(1440, 20*20).astype('float32') / 255.
label_arr_onehot=to_categorical(np.concatenate(label_arr_list))
sample_used=699
x_train_all,x_test,y_train_all,y_test_onehot= train_test_split(data_arr,label_arr_onehot,test_size=0.2,random_state=seed)
x_train=x_train_all[0:sample_used]
y_train_onehot=y_train_all[0:sample_used]
print('Shape of x_train: ' + str(x_train.shape))
print('Shape of x_test: ' + str(x_test.shape))
print('Shape of y_train: ' + str(y_train_onehot.shape))
print('Shape of y_test: ' + str(y_test_onehot.shape))
F.show_data_figures(x_train[0:40],20,20,40)
F.show_data_figures(x_test[0:40],20,20,40)
key_feture_number=50
np.random.seed(seed)
#--------------------------------------------------------------------------------------------------------------------------------
class Feature_Select_Layer(Layer):
def __init__(self, output_dim, **kwargs):
super(Feature_Select_Layer, self).__init__(**kwargs)
self.output_dim = output_dim
def build(self, input_shape):
self.kernel = self.add_weight(name='kernel',
shape=(input_shape[1],),
initializer=initializers.RandomUniform(minval=0.999999, maxval=0.9999999, seed=seed),
trainable=True)
super(Feature_Select_Layer, self).build(input_shape)
def call(self, x, selection=False,k=key_feture_number):
kernel=K.abs(self.kernel)
if selection:
kernel_=K.transpose(kernel)
kth_largest = tf.math.top_k(kernel_, k=k)[0][-1]
kernel = tf.where(condition=K.less(kernel,kth_largest),x=K.zeros_like(kernel),y=kernel)
return K.dot(x, tf.linalg.tensor_diag(kernel))
def compute_output_shape(self, input_shape):
return (input_shape[0], self.output_dim)
#--------------------------------------------------------------------------------------------------------------------------------
def Fractal_Autoencoder(p_data_feature=x_train.shape[1],\
p_feture_number=key_feture_number,\
p_encoding_dim=key_feture_number,\
p_learning_rate=1E-3,\
p_loss_weight_1=1,\
p_loss_weight_2=2):
input_img = Input(shape=(p_data_feature,), name='autoencoder_input')
feature_selection = Feature_Select_Layer(output_dim=p_data_feature,\
input_shape=(p_data_feature,),\
name='feature_selection')
feature_selection_score=feature_selection(input_img)
feature_selection_choose=feature_selection(input_img,selection=True,k=p_feture_number)
encoded = Dense(p_encoding_dim,\
activation='linear',\
kernel_initializer=initializers.glorot_uniform(seed),\
name='autoencoder_hidden_layer')
encoded_score=encoded(feature_selection_score)
encoded_choose=encoded(feature_selection_choose)
bottleneck_score=encoded_score
bottleneck_choose=encoded_choose
decoded = Dense(p_data_feature,\
activation='linear',\
kernel_initializer=initializers.glorot_uniform(seed),\
name='autoencoder_output')
decoded_score =decoded(bottleneck_score)
decoded_choose =decoded(bottleneck_choose)
latent_encoder_score = Model(input_img, bottleneck_score)
latent_encoder_choose = Model(input_img, bottleneck_choose)
feature_selection_output=Model(input_img,feature_selection_choose)
autoencoder = Model(input_img, [decoded_score,decoded_choose])
autoencoder.compile(loss=['mean_squared_error','mean_squared_error'],\
loss_weights=[p_loss_weight_1, p_loss_weight_2],\
optimizer=optimizers.Adam(lr=p_learning_rate))
print('Autoencoder Structure-------------------------------------')
autoencoder.summary()
return autoencoder,feature_selection_output,latent_encoder_score,latent_encoder_choose
epochs_number=200
batch_size_value=8
loss_weight_1=0.0078125
F_AE,\
feature_selection_output,\
latent_encoder_score_F_AE,\
latent_encoder_choose_F_AE=Fractal_Autoencoder(p_data_feature=x_train.shape[1],\
p_feture_number=key_feture_number,\
p_encoding_dim=key_feture_number,\
p_learning_rate= 1E-3,\
p_loss_weight_1=loss_weight_1,\
p_loss_weight_2=1)
F_AE_history = F_AE.fit(x_train, [x_train,x_train],\
epochs=epochs_number,\
batch_size=batch_size_value,\
shuffle=True)
loss = F_AE_history.history['loss']
epochs = range(epochs_number)
plt.plot(epochs, loss, 'bo', label='Training Loss')
plt.xlabel('Epochs')
plt.ylabel('Loss')
plt.legend()
plt.show()
fina_results=np.array(F_AE.evaluate(x_test,[x_test,x_test]))
fina_results
fina_results_single=np.array(F_AE.evaluate(x_test[0:1],[x_test[0:1],x_test[0:1]]))
fina_results_single
for i in np.arange(x_test.shape[0]):
fina_results_i=np.array(F_AE.evaluate(x_test[i:i+1],[x_test[i:i+1],x_test[i:i+1]]))
write_to_csv(fina_results_i.reshape(1,len(fina_results_i)),"./log/results_"+str(sample_used)+".csv")
| 0.506103 | 0.513546 |
```
# Global data variables
SANDBOX_NAME = ''# Sandbox Name
DATA_PATH = "/data/sandboxes/" + SANDBOX_NAME + "/data/data/"
from pyspark.sql import functions as F
```
# Creación o modificación de columnas
En Spark hay un único método para la creación o modificación de columnas y es `withColumn`. Este método es de nuevo una transformación y toma dos parámetros: el nombre de la columna a crear (o sobreescribir) y la operación que crea la nueva columna.
Para una ejecución más óptima se recomienda utilizar únicamente las funciones de PySpark cuando se define la operación, pero como se detallará más adelante se pueden utilizar funciones propias.
```
movies_df = spark.read.csv(DATA_PATH + 'movie-ratings/movies.csv', sep=',', header=True, inferSchema=True)
ratings_df = spark.read.csv(DATA_PATH + 'movie-ratings/ratings.csv', sep=',', header=True, inferSchema=True)
ratings_movies_df = ratings_df.join(movies_df, on='movieId', how='inner')
ratings_movies_df.cache()
```
## Funciones de Spark
__valor fijo__
El ejemplo más sencillo es crear una columna con un valor fijo, en este caso, columna `now` con valor '2019/01/21 14:08', y columna `rating2`con valor 4.0.
Hint: `withColumn`
```
ratings_movies_df = ratings_movies_df.withColumn('now', F.lit('2019/01/21 14:08'))
ratings_movies_df.show(3)
ratings_movies_df = ratings_movies_df.withColumn('rating2', F.lit(4.0))
ratings_movies_df.show(3)
```
__duplicar columna__
```
ratings_movies_df.withColumn('title2', F.col('title'))\
.select('title', 'title2')\
.show(10)
```
__operaciones aritmeticas__
```
ratings_movies_df.withColumn('rating_10', F.col('rating') * 2)\
.select('rating', 'rating_10')\
.show(10)
ratings_movies_df.withColumn('rating_avg', (F.col('rating') + F.col('rating2')) / 2)\
.select('rating', 'rating2', 'rating_avg')\
.show(10)
```
__if/else__
Crea la columna `kind_rating`, que sea 'high' en caso de que rating sea mayor que 4, y 'low' en caso contrario.
```
ratings_movies_df.withColumn('kind_rating',
F.when(F.col('rating') >= 4, 'high').otherwise('low')).show(10)
```
Se pueden concatenar multiples sentencias _when_. Esta vez, sobreescribe la columna `kind_rating` para crear un nivel intermedio, donde si es mayor que dos y menor que 4, `kind_rating` sea 'med'.
```
ratings_movies_df.withColumn('kind_rating',
F.when(F.col('rating') >= 4, 'high')\
.when(F.col('rating') >= 2, 'med')\
.otherwise('low')).show(20)
```
__operaciones con strings__
Pon en mayúsculas todos los títulos de las películas
```
ratings_movies_df.withColumn('title', F.upper(F.col('title'))).show(3)
```
Extrae los 10 primeros caracteres de la columna `title`
```
ratings_movies_df.withColumn('short_title', F.substring(F.col('title'), 0, 10))\
.select('title', 'short_title')\
.show(10)
```
Separa los diferentes géneros de la columna `genres` para obtener una lista, usando el separador '|'
```
ratings_movies_df.withColumn('genres', F.split(F.col('genres'), '\|')).show(4)
```
Crea una nueva columna `1st_genre` seleccionando el primer elemento de la lista del código anterior
```
ratings_movies_df.withColumn('1st_genre', F.split(F.col('genres'), '\|')[0])\
.select('genres', '1st_genre')\
.show(10)
```
Reemplaza el caracter '|' por '-' en la columna `genres`
```
ratings_movies_df.withColumn('genres', F.regexp_replace(F.col('genres'), '\|', '-'))\
.select('title', 'genres')\
.show(10, truncate=False)
```
_Con expresiones regulares_
https://regexr.com/
```
ratings_movies_df.withColumn('title', F.regexp_replace(F.col('title'), ' \(\d{4}\)', '')).show(5, truncate=False)
ratings_movies_df = ratings_movies_df.withColumn('year',
F.regexp_extract(F.col('title'), '\((\d{4})\)', 1))
ratings_movies_df.show(5)
```
## Casting
Con el método `withColumn` también es posible convertir el tipo de una columna con la función `cast`. Es importante saber que en caso de no poder convertirse (por ejemplo una letra a número) no saltará error y el resultado será un valor nulo.
```
ratings_movies_df.printSchema()
```
Cambia el formato de `year` a entero, y `movieId` a string.
```
ratings_movies_df = ratings_movies_df.withColumn('year', F.col('year').cast('int'))
ratings_movies_df.show(5)
ratings_movies_df = ratings_movies_df.withColumn('movieId', F.col('movieId').cast('string'))
ratings_movies_df.printSchema()
ratings_movies_df.withColumn('error', F.col('title').cast('int')).show(5)
```
## UDF (User Defined Functions)
Cuando no es posible definir la operación con las funciones de spark se pueden crear funciones propias usando la UDFs. Primero se crea una función de Python normal y posteriormente se crea la UDFs. Es necesario indicar el tipo de la columna de salida en la UDF.
```
from pyspark.sql.types import StringType, IntegerType, DoubleType, DateType
```
_Aumenta el rating en un 15% para cada película más antigua que 2000 (el máximo siempre es 5)._
```
def increase_rating(year, rating):
if year < 2000:
rating = min(rating * 1.15, 5.0)
return rating
increase_rating_udf = F.udf(increase_rating, DoubleType())
```
```
ratings_movies_df.withColumn('rating_inc',
increase_rating_udf(F.col('year'), F.col('rating')))\
.select('title', 'year', 'rating', 'rating_inc')\
.show(20)
```
Extrae el año de la película sin usar expresiones regulares.
```
title = 'Trainspotting (1996)'
title.replace(')', '').replace('(', '')
year = title.replace(')', '').replace('(', '').split(' ')[-1]
year = int(year)
year
def get_year(title):
year = title.replace(')', '').replace('(', '').split(' ')[-1]
if year.isnumeric():
year = int(year)
else:
year = -1
return year
get_year_udf = F.udf(get_year, IntegerType())
ratings_movies_df.withColumn('year2', get_year_udf(F.col('title')))\
.select('title', 'year', 'year2').show(10, truncate=False)
```
# Datetimes
Hay varias funciones de _pyspark_ que permiten trabajar con fechas: diferencia entre fechas, dia de la semana, año... Pero para ello primero es necesario transformar las columnas a tipo fecha. Se permite la conversion de dos formatos de fecha:
* timestamp de unix: una columna de tipo entero con los segundos trascurridos entre la medianoche del 1 de Enero de 1990 hasta la fecha.
* cadena: la fecha representada como una cadena siguiendo un formato específico que puede variar.
```
ratings_movies_df.select('title', 'timestamp', 'now').show(5)
```
## unix timestamp a datetime
```
ratings_movies_df = ratings_movies_df.withColumn('datetime', F.from_unixtime(F.col('timestamp')))
ratings_movies_df.select('datetime', 'timestamp').show(10)
```
## string a datetime
```
ratings_movies_df = ratings_movies_df.withColumn('now_datetime',
F.from_unixtime(F.unix_timestamp(F.col('now'), 'yyyy/MM/dd HH:mm')))
ratings_movies_df.select('now', 'now_datetime').show(10)
```
## funciones con datetimes
```
ratings_movies_df.select('now_datetime', 'datetime',
F.datediff(F.col('now_datetime'), F.col('datetime'))).show(10)
ratings_movies_df.select('datetime', F.date_add(F.col('datetime'), 10)).show(10)
ratings_movies_df.withColumn('datetime_plus_4_months', F.add_months(F.col('datetime'), 4))\
.select('datetime', 'datetime_plus_4_months').show(5)
ratings_movies_df.select('datetime', F.month(F.col('datetime')).alias('month')).show(10)
ratings_movies_df.select('datetime', F.last_day(F.col('datetime')).alias('last_day')).show(10)
ratings_movies_df.select('datetime', F.dayofmonth(F.col('datetime')).alias('day'),
F.dayofyear(F.col('datetime')).alias('year_day'),
F.date_format(F.col('datetime'), 'E').alias('weekday')).show(10)
```
Para filtrar por fechas se pueden comparar directamente con una cadena en el formato YYYY-MM-DD hh:mm:ss ya que será interpretada como una fecha.
```
ratings_movies_df.filter(F.col('datetime') >= "2015-09-30 20:00:00").select('datetime', 'title', 'rating').show(10)
ratings_movies_df.filter(F.col('datetime').between("2003-01-31", "2003-02-10"))\
.select('datetime', 'title', 'rating').show(5)
ratings_movies_df.filter(F.year(F.col('datetime')) >= 2012)\
.select('datetime', 'title', 'rating').show(5)
```
# Ejercicio 1
1) Cree una función que acepte un DataFrame y un diccionario. La función debe usar el diccionario para renombrar un grupo de columnas y devolver el DataFrame ya modificado.
Use el siguiente DataFrame y diccionario:
```
pokemon_df = spark.read.csv(DATA_PATH + 'pokemon.csv', sep=',', header=True, inferSchema=True)
rename_dict = {'Sp. Atk': 'sp_atk',
'Sp. Def': 'sp_def'}
pokemon_df.show(3)
# Respuesta aqui
```
2) Use la función definida en el punto anterior para cambiar los nombres del DF usando el diccionario dado.
3) Modifique la función de tal forma que también acepte una función en lugar de un diccionario. Use la función para renombrar las columnas.
4) Estandarice según las buenas prácticas los nombres de las columnas usando la función que acaba de definir.
5) Cree otra función que acepte un DataFrame y una lista con un subconjunto de columnas. El objetivo de esta función es determinar el número de filas duplicadas del DF.
6) Use la función creada para obtener el número de duplicados del DataFrame pokemon_df en todas las columnas excepto el nombre (`name`)
```
# Respuesta aqui
# Respuesta aqui
# Respuesta aqui
# Respuesta aqui
# Respuesta aqui
```
# Ejercicio 2
Crea la misma lógica definida en el siguiente UDF, pero sin usar UDFs, es decir, usando exclusivamente funciones de SparkSQL.
```
movies_df = spark.read.csv(DATA_PATH + 'movie-ratings/movies.csv', sep=',', header=True, inferSchema=True)
movies_df = movies_df.withColumn('genres', F.split(F.col('genres'), '\|'))
from pyspark.sql.types import StringType, IntegerType, DoubleType, BooleanType
def value_in_col(col, value):
return value in col
value_in_col_udf = F.udf(value_in_col, BooleanType())
```
*Pista*: Mira la función *explode*.
```
# Respuesta aqui
```
|
github_jupyter
|
# Global data variables
SANDBOX_NAME = ''# Sandbox Name
DATA_PATH = "/data/sandboxes/" + SANDBOX_NAME + "/data/data/"
from pyspark.sql import functions as F
movies_df = spark.read.csv(DATA_PATH + 'movie-ratings/movies.csv', sep=',', header=True, inferSchema=True)
ratings_df = spark.read.csv(DATA_PATH + 'movie-ratings/ratings.csv', sep=',', header=True, inferSchema=True)
ratings_movies_df = ratings_df.join(movies_df, on='movieId', how='inner')
ratings_movies_df.cache()
ratings_movies_df = ratings_movies_df.withColumn('now', F.lit('2019/01/21 14:08'))
ratings_movies_df.show(3)
ratings_movies_df = ratings_movies_df.withColumn('rating2', F.lit(4.0))
ratings_movies_df.show(3)
ratings_movies_df.withColumn('title2', F.col('title'))\
.select('title', 'title2')\
.show(10)
ratings_movies_df.withColumn('rating_10', F.col('rating') * 2)\
.select('rating', 'rating_10')\
.show(10)
ratings_movies_df.withColumn('rating_avg', (F.col('rating') + F.col('rating2')) / 2)\
.select('rating', 'rating2', 'rating_avg')\
.show(10)
ratings_movies_df.withColumn('kind_rating',
F.when(F.col('rating') >= 4, 'high').otherwise('low')).show(10)
ratings_movies_df.withColumn('kind_rating',
F.when(F.col('rating') >= 4, 'high')\
.when(F.col('rating') >= 2, 'med')\
.otherwise('low')).show(20)
ratings_movies_df.withColumn('title', F.upper(F.col('title'))).show(3)
ratings_movies_df.withColumn('short_title', F.substring(F.col('title'), 0, 10))\
.select('title', 'short_title')\
.show(10)
ratings_movies_df.withColumn('genres', F.split(F.col('genres'), '\|')).show(4)
ratings_movies_df.withColumn('1st_genre', F.split(F.col('genres'), '\|')[0])\
.select('genres', '1st_genre')\
.show(10)
ratings_movies_df.withColumn('genres', F.regexp_replace(F.col('genres'), '\|', '-'))\
.select('title', 'genres')\
.show(10, truncate=False)
ratings_movies_df.withColumn('title', F.regexp_replace(F.col('title'), ' \(\d{4}\)', '')).show(5, truncate=False)
ratings_movies_df = ratings_movies_df.withColumn('year',
F.regexp_extract(F.col('title'), '\((\d{4})\)', 1))
ratings_movies_df.show(5)
ratings_movies_df.printSchema()
ratings_movies_df = ratings_movies_df.withColumn('year', F.col('year').cast('int'))
ratings_movies_df.show(5)
ratings_movies_df = ratings_movies_df.withColumn('movieId', F.col('movieId').cast('string'))
ratings_movies_df.printSchema()
ratings_movies_df.withColumn('error', F.col('title').cast('int')).show(5)
from pyspark.sql.types import StringType, IntegerType, DoubleType, DateType
def increase_rating(year, rating):
if year < 2000:
rating = min(rating * 1.15, 5.0)
return rating
increase_rating_udf = F.udf(increase_rating, DoubleType())
ratings_movies_df.withColumn('rating_inc',
increase_rating_udf(F.col('year'), F.col('rating')))\
.select('title', 'year', 'rating', 'rating_inc')\
.show(20)
title = 'Trainspotting (1996)'
title.replace(')', '').replace('(', '')
year = title.replace(')', '').replace('(', '').split(' ')[-1]
year = int(year)
year
def get_year(title):
year = title.replace(')', '').replace('(', '').split(' ')[-1]
if year.isnumeric():
year = int(year)
else:
year = -1
return year
get_year_udf = F.udf(get_year, IntegerType())
ratings_movies_df.withColumn('year2', get_year_udf(F.col('title')))\
.select('title', 'year', 'year2').show(10, truncate=False)
ratings_movies_df.select('title', 'timestamp', 'now').show(5)
ratings_movies_df = ratings_movies_df.withColumn('datetime', F.from_unixtime(F.col('timestamp')))
ratings_movies_df.select('datetime', 'timestamp').show(10)
ratings_movies_df = ratings_movies_df.withColumn('now_datetime',
F.from_unixtime(F.unix_timestamp(F.col('now'), 'yyyy/MM/dd HH:mm')))
ratings_movies_df.select('now', 'now_datetime').show(10)
ratings_movies_df.select('now_datetime', 'datetime',
F.datediff(F.col('now_datetime'), F.col('datetime'))).show(10)
ratings_movies_df.select('datetime', F.date_add(F.col('datetime'), 10)).show(10)
ratings_movies_df.withColumn('datetime_plus_4_months', F.add_months(F.col('datetime'), 4))\
.select('datetime', 'datetime_plus_4_months').show(5)
ratings_movies_df.select('datetime', F.month(F.col('datetime')).alias('month')).show(10)
ratings_movies_df.select('datetime', F.last_day(F.col('datetime')).alias('last_day')).show(10)
ratings_movies_df.select('datetime', F.dayofmonth(F.col('datetime')).alias('day'),
F.dayofyear(F.col('datetime')).alias('year_day'),
F.date_format(F.col('datetime'), 'E').alias('weekday')).show(10)
ratings_movies_df.filter(F.col('datetime') >= "2015-09-30 20:00:00").select('datetime', 'title', 'rating').show(10)
ratings_movies_df.filter(F.col('datetime').between("2003-01-31", "2003-02-10"))\
.select('datetime', 'title', 'rating').show(5)
ratings_movies_df.filter(F.year(F.col('datetime')) >= 2012)\
.select('datetime', 'title', 'rating').show(5)
pokemon_df = spark.read.csv(DATA_PATH + 'pokemon.csv', sep=',', header=True, inferSchema=True)
rename_dict = {'Sp. Atk': 'sp_atk',
'Sp. Def': 'sp_def'}
pokemon_df.show(3)
# Respuesta aqui
# Respuesta aqui
# Respuesta aqui
# Respuesta aqui
# Respuesta aqui
# Respuesta aqui
movies_df = spark.read.csv(DATA_PATH + 'movie-ratings/movies.csv', sep=',', header=True, inferSchema=True)
movies_df = movies_df.withColumn('genres', F.split(F.col('genres'), '\|'))
from pyspark.sql.types import StringType, IntegerType, DoubleType, BooleanType
def value_in_col(col, value):
return value in col
value_in_col_udf = F.udf(value_in_col, BooleanType())
# Respuesta aqui
| 0.426083 | 0.860779 |
# Anna KaRNNa
In this notebook, I'll build a character-wise RNN trained on Anna Karenina, one of my all-time favorite books. It'll be able to generate new text based on the text from the book.
This network is based off of Andrej Karpathy's [post on RNNs](http://karpathy.github.io/2015/05/21/rnn-effectiveness/) and [implementation in Torch](https://github.com/karpathy/char-rnn). Also, some information [here at r2rt](http://r2rt.com/recurrent-neural-networks-in-tensorflow-ii.html) and from [Sherjil Ozair](https://github.com/sherjilozair/char-rnn-tensorflow) on GitHub. Below is the general architecture of the character-wise RNN.
<img src="assets/charseq.jpeg" width="500">
```
import time
from collections import namedtuple
import numpy as np
import tensorflow as tf
```
First we'll load the text file and convert it into integers for our network to use.
```
with open('anna.txt', 'r') as f:
text=f.read()
vocab = set(text)
vocab_to_int = {c: i for i, c in enumerate(vocab)}
int_to_vocab = dict(enumerate(vocab))
chars = np.array([vocab_to_int[c] for c in text], dtype=np.int32)
text[:100]
chars[:100]
```
Now I need to split up the data into batches, and into training and validation sets. I should be making a test set here, but I'm not going to worry about that. My test will be if the network can generate new text.
Here I'll make both input and target arrays. The targets are the same as the inputs, except shifted one character over. I'll also drop the last bit of data so that I'll only have completely full batches.
The idea here is to make a 2D matrix where the number of rows is equal to the number of batches. Each row will be one long concatenated string from the character data. We'll split this data into a training set and validation set using the `split_frac` keyword. This will keep 90% of the batches in the training set, the other 10% in the validation set.
```
def split_data(chars, batch_size, num_steps, split_frac=0.9):
"""
Split character data into training and validation sets, inputs and targets for each set.
Arguments
---------
chars: character array
batch_size: Size of examples in each of batch
num_steps: Number of sequence steps to keep in the input and pass to the network
split_frac: Fraction of batches to keep in the training set
Returns train_x, train_y, val_x, val_y
"""
slice_size = batch_size * num_steps
n_batches = int(len(chars) / slice_size)
# Drop the last few characters to make only full batches
x = chars[: n_batches*slice_size]
y = chars[1: n_batches*slice_size + 1]
# Split the data into batch_size slices, then stack them into a 2D matrix
x = np.stack(np.split(x, batch_size))
y = np.stack(np.split(y, batch_size))
# Now x and y are arrays with dimensions batch_size x n_batches*num_steps
# Split into training and validation sets, keep the virst split_frac batches for training
split_idx = int(n_batches*split_frac)
train_x, train_y= x[:, :split_idx*num_steps], y[:, :split_idx*num_steps]
val_x, val_y = x[:, split_idx*num_steps:], y[:, split_idx*num_steps:]
return train_x, train_y, val_x, val_y
train_x, train_y, val_x, val_y = split_data(chars, 10, 200)
train_x.shape
train_x[:,:10]
```
I'll write another function to grab batches out of the arrays made by split data. Here each batch will be a sliding window on these arrays with size `batch_size X num_steps`. For example, if we want our network to train on a sequence of 100 characters, `num_steps = 100`. For the next batch, we'll shift this window the next sequence of `num_steps` characters. In this way we can feed batches to the network and the cell states will continue through on each batch.
```
def get_batch(arrs, num_steps):
batch_size, slice_size = arrs[0].shape
n_batches = int(slice_size/num_steps)
for b in range(n_batches):
yield [x[:, b*num_steps: (b+1)*num_steps] for x in arrs]
def build_rnn(num_classes, batch_size=50, num_steps=50, lstm_size=128, num_layers=2,
learning_rate=0.001, grad_clip=5, sampling=False):
if sampling == True:
batch_size, num_steps = 1, 1
tf.reset_default_graph()
# Declare placeholders we'll feed into the graph
inputs = tf.placeholder(tf.int32, [batch_size, num_steps], name='inputs')
x_one_hot = tf.one_hot(inputs, num_classes, name='x_one_hot')
targets = tf.placeholder(tf.int32, [batch_size, num_steps], name='targets')
y_one_hot = tf.one_hot(targets, num_classes, name='y_one_hot')
y_reshaped = tf.reshape(y_one_hot, [-1, num_classes])
keep_prob = tf.placeholder(tf.float32, name='keep_prob')
# Build the RNN layers
lstm = tf.contrib.rnn.BasicLSTMCell(lstm_size)
drop = tf.contrib.rnn.DropoutWrapper(lstm, output_keep_prob=keep_prob)
cell = tf.contrib.rnn.MultiRNNCell([drop] * num_layers)
initial_state = cell.zero_state(batch_size, tf.float32)
# Run the data through the RNN layers
outputs, state = tf.nn.dynamic_rnn(cell, x_one_hot, initial_state=initial_state)
final_state = state
# Reshape output so it's a bunch of rows, one row for each cell output
seq_output = tf.concat(outputs, axis=1,name='seq_output')
output = tf.reshape(seq_output, [-1, lstm_size], name='graph_output')
# Now connect the RNN putputs to a softmax layer and calculate the cost
softmax_w = tf.Variable(tf.truncated_normal((lstm_size, num_classes), stddev=0.1),
name='softmax_w')
softmax_b = tf.Variable(tf.zeros(num_classes), name='softmax_b')
logits = tf.matmul(output, softmax_w) + softmax_b
preds = tf.nn.softmax(logits, name='predictions')
loss = tf.nn.softmax_cross_entropy_with_logits(logits=logits, labels=y_reshaped, name='loss')
cost = tf.reduce_mean(loss, name='cost')
# Optimizer for training, using gradient clipping to control exploding gradients
tvars = tf.trainable_variables()
grads, _ = tf.clip_by_global_norm(tf.gradients(cost, tvars), grad_clip)
train_op = tf.train.AdamOptimizer(learning_rate)
optimizer = train_op.apply_gradients(zip(grads, tvars))
# Export the nodes
export_nodes = ['inputs', 'targets', 'initial_state', 'final_state',
'keep_prob', 'cost', 'preds', 'optimizer']
Graph = namedtuple('Graph', export_nodes)
local_dict = locals()
graph = Graph(*[local_dict[each] for each in export_nodes])
return graph
```
## Hyperparameters
Here I'm defining the hyperparameters for the network. The two you probably haven't seen before are `lstm_size` and `num_layers`. These set the number of hidden units in the LSTM layers and the number of LSTM layers, respectively. Of course, making these bigger will improve the network's performance but you'll have to watch out for overfitting. If your validation loss is much larger than the training loss, you're probably overfitting. Decrease the size of the network or decrease the dropout keep probability.
```
batch_size = 100
num_steps = 100
lstm_size = 512
num_layers = 2
learning_rate = 0.001
```
## Write out the graph for TensorBoard
```
model = build_rnn(len(vocab),
batch_size=batch_size,
num_steps=num_steps,
learning_rate=learning_rate,
lstm_size=lstm_size,
num_layers=num_layers)
with tf.Session() as sess:
sess.run(tf.global_variables_initializer())
file_writer = tf.summary.FileWriter('./logs/1', sess.graph)
```
## Training
Time for training which is is pretty straightforward. Here I pass in some data, and get an LSTM state back. Then I pass that state back in to the network so the next batch can continue the state from the previous batch. And every so often (set by `save_every_n`) I calculate the validation loss and save a checkpoint.
```
!mkdir -p checkpoints/anna
epochs = 1
save_every_n = 200
train_x, train_y, val_x, val_y = split_data(chars, batch_size, num_steps)
model = build_rnn(len(vocab),
batch_size=batch_size,
num_steps=num_steps,
learning_rate=learning_rate,
lstm_size=lstm_size,
num_layers=num_layers)
saver = tf.train.Saver(max_to_keep=100)
with tf.Session() as sess:
sess.run(tf.global_variables_initializer())
# Use the line below to load a checkpoint and resume training
#saver.restore(sess, 'checkpoints/anna20.ckpt')
n_batches = int(train_x.shape[1]/num_steps)
iterations = n_batches * epochs
for e in range(epochs):
# Train network
new_state = sess.run(model.initial_state)
loss = 0
for b, (x, y) in enumerate(get_batch([train_x, train_y], num_steps), 1):
iteration = e*n_batches + b
start = time.time()
feed = {model.inputs: x,
model.targets: y,
model.keep_prob: 0.5,
model.initial_state: new_state}
batch_loss, new_state, _ = sess.run([model.cost, model.final_state, model.optimizer],
feed_dict=feed)
loss += batch_loss
end = time.time()
print('Epoch {}/{} '.format(e+1, epochs),
'Iteration {}/{}'.format(iteration, iterations),
'Training loss: {:.4f}'.format(loss/b),
'{:.4f} sec/batch'.format((end-start)))
if (iteration%save_every_n == 0) or (iteration == iterations):
# Check performance, notice dropout has been set to 1
val_loss = []
new_state = sess.run(model.initial_state)
for x, y in get_batch([val_x, val_y], num_steps):
feed = {model.inputs: x,
model.targets: y,
model.keep_prob: 1.,
model.initial_state: new_state}
batch_loss, new_state = sess.run([model.cost, model.final_state], feed_dict=feed)
val_loss.append(batch_loss)
print('Validation loss:', np.mean(val_loss),
'Saving checkpoint!')
saver.save(sess, "checkpoints/anna/i{}_l{}_{:.3f}.ckpt".format(iteration, lstm_size, np.mean(val_loss)))
tf.train.get_checkpoint_state('checkpoints/anna')
```
## Sampling
Now that the network is trained, we'll can use it to generate new text. The idea is that we pass in a character, then the network will predict the next character. We can use the new one, to predict the next one. And we keep doing this to generate all new text. I also included some functionality to prime the network with some text by passing in a string and building up a state from that.
The network gives us predictions for each character. To reduce noise and make things a little less random, I'm going to only choose a new character from the top N most likely characters.
```
def pick_top_n(preds, vocab_size, top_n=5):
p = np.squeeze(preds)
p[np.argsort(p)[:-top_n]] = 0
p = p / np.sum(p)
c = np.random.choice(vocab_size, 1, p=p)[0]
return c
def sample(checkpoint, n_samples, lstm_size, vocab_size, prime="The "):
prime = "Far"
samples = [c for c in prime]
model = build_rnn(vocab_size, lstm_size=lstm_size, sampling=True)
saver = tf.train.Saver()
with tf.Session() as sess:
saver.restore(sess, checkpoint)
new_state = sess.run(model.initial_state)
for c in prime:
x = np.zeros((1, 1))
x[0,0] = vocab_to_int[c]
feed = {model.inputs: x,
model.keep_prob: 1.,
model.initial_state: new_state}
preds, new_state = sess.run([model.preds, model.final_state],
feed_dict=feed)
c = pick_top_n(preds, len(vocab))
samples.append(int_to_vocab[c])
for i in range(n_samples):
x[0,0] = c
feed = {model.inputs: x,
model.keep_prob: 1.,
model.initial_state: new_state}
preds, new_state = sess.run([model.preds, model.final_state],
feed_dict=feed)
c = pick_top_n(preds, len(vocab))
samples.append(int_to_vocab[c])
return ''.join(samples)
checkpoint = "checkpoints/anna/i3560_l512_1.122.ckpt"
samp = sample(checkpoint, 2000, lstm_size, len(vocab), prime="Far")
print(samp)
checkpoint = "checkpoints/anna/i200_l512_2.432.ckpt"
samp = sample(checkpoint, 1000, lstm_size, len(vocab), prime="Far")
print(samp)
checkpoint = "checkpoints/anna/i600_l512_1.750.ckpt"
samp = sample(checkpoint, 1000, lstm_size, len(vocab), prime="Far")
print(samp)
checkpoint = "checkpoints/anna/i1000_l512_1.484.ckpt"
samp = sample(checkpoint, 1000, lstm_size, len(vocab), prime="Far")
print(samp)
```
|
github_jupyter
|
import time
from collections import namedtuple
import numpy as np
import tensorflow as tf
with open('anna.txt', 'r') as f:
text=f.read()
vocab = set(text)
vocab_to_int = {c: i for i, c in enumerate(vocab)}
int_to_vocab = dict(enumerate(vocab))
chars = np.array([vocab_to_int[c] for c in text], dtype=np.int32)
text[:100]
chars[:100]
def split_data(chars, batch_size, num_steps, split_frac=0.9):
"""
Split character data into training and validation sets, inputs and targets for each set.
Arguments
---------
chars: character array
batch_size: Size of examples in each of batch
num_steps: Number of sequence steps to keep in the input and pass to the network
split_frac: Fraction of batches to keep in the training set
Returns train_x, train_y, val_x, val_y
"""
slice_size = batch_size * num_steps
n_batches = int(len(chars) / slice_size)
# Drop the last few characters to make only full batches
x = chars[: n_batches*slice_size]
y = chars[1: n_batches*slice_size + 1]
# Split the data into batch_size slices, then stack them into a 2D matrix
x = np.stack(np.split(x, batch_size))
y = np.stack(np.split(y, batch_size))
# Now x and y are arrays with dimensions batch_size x n_batches*num_steps
# Split into training and validation sets, keep the virst split_frac batches for training
split_idx = int(n_batches*split_frac)
train_x, train_y= x[:, :split_idx*num_steps], y[:, :split_idx*num_steps]
val_x, val_y = x[:, split_idx*num_steps:], y[:, split_idx*num_steps:]
return train_x, train_y, val_x, val_y
train_x, train_y, val_x, val_y = split_data(chars, 10, 200)
train_x.shape
train_x[:,:10]
def get_batch(arrs, num_steps):
batch_size, slice_size = arrs[0].shape
n_batches = int(slice_size/num_steps)
for b in range(n_batches):
yield [x[:, b*num_steps: (b+1)*num_steps] for x in arrs]
def build_rnn(num_classes, batch_size=50, num_steps=50, lstm_size=128, num_layers=2,
learning_rate=0.001, grad_clip=5, sampling=False):
if sampling == True:
batch_size, num_steps = 1, 1
tf.reset_default_graph()
# Declare placeholders we'll feed into the graph
inputs = tf.placeholder(tf.int32, [batch_size, num_steps], name='inputs')
x_one_hot = tf.one_hot(inputs, num_classes, name='x_one_hot')
targets = tf.placeholder(tf.int32, [batch_size, num_steps], name='targets')
y_one_hot = tf.one_hot(targets, num_classes, name='y_one_hot')
y_reshaped = tf.reshape(y_one_hot, [-1, num_classes])
keep_prob = tf.placeholder(tf.float32, name='keep_prob')
# Build the RNN layers
lstm = tf.contrib.rnn.BasicLSTMCell(lstm_size)
drop = tf.contrib.rnn.DropoutWrapper(lstm, output_keep_prob=keep_prob)
cell = tf.contrib.rnn.MultiRNNCell([drop] * num_layers)
initial_state = cell.zero_state(batch_size, tf.float32)
# Run the data through the RNN layers
outputs, state = tf.nn.dynamic_rnn(cell, x_one_hot, initial_state=initial_state)
final_state = state
# Reshape output so it's a bunch of rows, one row for each cell output
seq_output = tf.concat(outputs, axis=1,name='seq_output')
output = tf.reshape(seq_output, [-1, lstm_size], name='graph_output')
# Now connect the RNN putputs to a softmax layer and calculate the cost
softmax_w = tf.Variable(tf.truncated_normal((lstm_size, num_classes), stddev=0.1),
name='softmax_w')
softmax_b = tf.Variable(tf.zeros(num_classes), name='softmax_b')
logits = tf.matmul(output, softmax_w) + softmax_b
preds = tf.nn.softmax(logits, name='predictions')
loss = tf.nn.softmax_cross_entropy_with_logits(logits=logits, labels=y_reshaped, name='loss')
cost = tf.reduce_mean(loss, name='cost')
# Optimizer for training, using gradient clipping to control exploding gradients
tvars = tf.trainable_variables()
grads, _ = tf.clip_by_global_norm(tf.gradients(cost, tvars), grad_clip)
train_op = tf.train.AdamOptimizer(learning_rate)
optimizer = train_op.apply_gradients(zip(grads, tvars))
# Export the nodes
export_nodes = ['inputs', 'targets', 'initial_state', 'final_state',
'keep_prob', 'cost', 'preds', 'optimizer']
Graph = namedtuple('Graph', export_nodes)
local_dict = locals()
graph = Graph(*[local_dict[each] for each in export_nodes])
return graph
batch_size = 100
num_steps = 100
lstm_size = 512
num_layers = 2
learning_rate = 0.001
model = build_rnn(len(vocab),
batch_size=batch_size,
num_steps=num_steps,
learning_rate=learning_rate,
lstm_size=lstm_size,
num_layers=num_layers)
with tf.Session() as sess:
sess.run(tf.global_variables_initializer())
file_writer = tf.summary.FileWriter('./logs/1', sess.graph)
!mkdir -p checkpoints/anna
epochs = 1
save_every_n = 200
train_x, train_y, val_x, val_y = split_data(chars, batch_size, num_steps)
model = build_rnn(len(vocab),
batch_size=batch_size,
num_steps=num_steps,
learning_rate=learning_rate,
lstm_size=lstm_size,
num_layers=num_layers)
saver = tf.train.Saver(max_to_keep=100)
with tf.Session() as sess:
sess.run(tf.global_variables_initializer())
# Use the line below to load a checkpoint and resume training
#saver.restore(sess, 'checkpoints/anna20.ckpt')
n_batches = int(train_x.shape[1]/num_steps)
iterations = n_batches * epochs
for e in range(epochs):
# Train network
new_state = sess.run(model.initial_state)
loss = 0
for b, (x, y) in enumerate(get_batch([train_x, train_y], num_steps), 1):
iteration = e*n_batches + b
start = time.time()
feed = {model.inputs: x,
model.targets: y,
model.keep_prob: 0.5,
model.initial_state: new_state}
batch_loss, new_state, _ = sess.run([model.cost, model.final_state, model.optimizer],
feed_dict=feed)
loss += batch_loss
end = time.time()
print('Epoch {}/{} '.format(e+1, epochs),
'Iteration {}/{}'.format(iteration, iterations),
'Training loss: {:.4f}'.format(loss/b),
'{:.4f} sec/batch'.format((end-start)))
if (iteration%save_every_n == 0) or (iteration == iterations):
# Check performance, notice dropout has been set to 1
val_loss = []
new_state = sess.run(model.initial_state)
for x, y in get_batch([val_x, val_y], num_steps):
feed = {model.inputs: x,
model.targets: y,
model.keep_prob: 1.,
model.initial_state: new_state}
batch_loss, new_state = sess.run([model.cost, model.final_state], feed_dict=feed)
val_loss.append(batch_loss)
print('Validation loss:', np.mean(val_loss),
'Saving checkpoint!')
saver.save(sess, "checkpoints/anna/i{}_l{}_{:.3f}.ckpt".format(iteration, lstm_size, np.mean(val_loss)))
tf.train.get_checkpoint_state('checkpoints/anna')
def pick_top_n(preds, vocab_size, top_n=5):
p = np.squeeze(preds)
p[np.argsort(p)[:-top_n]] = 0
p = p / np.sum(p)
c = np.random.choice(vocab_size, 1, p=p)[0]
return c
def sample(checkpoint, n_samples, lstm_size, vocab_size, prime="The "):
prime = "Far"
samples = [c for c in prime]
model = build_rnn(vocab_size, lstm_size=lstm_size, sampling=True)
saver = tf.train.Saver()
with tf.Session() as sess:
saver.restore(sess, checkpoint)
new_state = sess.run(model.initial_state)
for c in prime:
x = np.zeros((1, 1))
x[0,0] = vocab_to_int[c]
feed = {model.inputs: x,
model.keep_prob: 1.,
model.initial_state: new_state}
preds, new_state = sess.run([model.preds, model.final_state],
feed_dict=feed)
c = pick_top_n(preds, len(vocab))
samples.append(int_to_vocab[c])
for i in range(n_samples):
x[0,0] = c
feed = {model.inputs: x,
model.keep_prob: 1.,
model.initial_state: new_state}
preds, new_state = sess.run([model.preds, model.final_state],
feed_dict=feed)
c = pick_top_n(preds, len(vocab))
samples.append(int_to_vocab[c])
return ''.join(samples)
checkpoint = "checkpoints/anna/i3560_l512_1.122.ckpt"
samp = sample(checkpoint, 2000, lstm_size, len(vocab), prime="Far")
print(samp)
checkpoint = "checkpoints/anna/i200_l512_2.432.ckpt"
samp = sample(checkpoint, 1000, lstm_size, len(vocab), prime="Far")
print(samp)
checkpoint = "checkpoints/anna/i600_l512_1.750.ckpt"
samp = sample(checkpoint, 1000, lstm_size, len(vocab), prime="Far")
print(samp)
checkpoint = "checkpoints/anna/i1000_l512_1.484.ckpt"
samp = sample(checkpoint, 1000, lstm_size, len(vocab), prime="Far")
print(samp)
| 0.719876 | 0.957873 |
# Read in the data
```
import pandas as pd
import numpy
import re
data_files = [
"ap_2010.csv",
"class_size.csv",
"demographics.csv",
"graduation.csv",
"hs_directory.csv",
"sat_results.csv"
]
data = {}
for f in data_files:
d = pd.read_csv("schools/{0}".format(f))
data[f.replace(".csv", "")] = d
```
# Read in the surveys
```
all_survey = pd.read_csv("schools/survey_all.txt", delimiter="\t", encoding='windows-1252')
d75_survey = pd.read_csv("schools/survey_d75.txt", delimiter="\t", encoding='windows-1252')
survey = pd.concat([all_survey, d75_survey], axis=0)
survey["DBN"] = survey["dbn"]
survey_fields = [
"DBN",
"rr_s",
"rr_t",
"rr_p",
"N_s",
"N_t",
"N_p",
"saf_p_11",
"com_p_11",
"eng_p_11",
"aca_p_11",
"saf_t_11",
"com_t_11",
"eng_t_11",
"aca_t_11",
"saf_s_11",
"com_s_11",
"eng_s_11",
"aca_s_11",
"saf_tot_11",
"com_tot_11",
"eng_tot_11",
"aca_tot_11",
]
survey = survey.loc[:,survey_fields]
data["survey"] = survey
```
# Add DBN columns
```
data["hs_directory"]["DBN"] = data["hs_directory"]["dbn"]
def pad_csd(num):
string_representation = str(num)
if len(string_representation) > 1:
return string_representation
else:
return "0" + string_representation
data["class_size"]["padded_csd"] = data["class_size"]["CSD"].apply(pad_csd)
data["class_size"]["DBN"] = data["class_size"]["padded_csd"] + data["class_size"]["SCHOOL CODE"]
```
# Convert columns to numeric
```
cols = ['SAT Math Avg. Score', 'SAT Critical Reading Avg. Score', 'SAT Writing Avg. Score']
for c in cols:
data["sat_results"][c] = pd.to_numeric(data["sat_results"][c], errors="coerce")
data['sat_results']['sat_score'] = data['sat_results'][cols[0]] + data['sat_results'][cols[1]] + data['sat_results'][cols[2]]
def find_lat(loc):
coords = re.findall("\(.+, .+\)", loc)
lat = coords[0].split(",")[0].replace("(", "")
return lat
def find_lon(loc):
coords = re.findall("\(.+, .+\)", loc)
lon = coords[0].split(",")[1].replace(")", "").strip()
return lon
data["hs_directory"]["lat"] = data["hs_directory"]["Location 1"].apply(find_lat)
data["hs_directory"]["lon"] = data["hs_directory"]["Location 1"].apply(find_lon)
data["hs_directory"]["lat"] = pd.to_numeric(data["hs_directory"]["lat"], errors="coerce")
data["hs_directory"]["lon"] = pd.to_numeric(data["hs_directory"]["lon"], errors="coerce")
```
# Condense datasets
```
class_size = data["class_size"]
class_size = class_size[class_size["GRADE "] == "09-12"]
class_size = class_size[class_size["PROGRAM TYPE"] == "GEN ED"]
class_size = class_size.groupby("DBN").agg(numpy.mean)
class_size.reset_index(inplace=True)
data["class_size"] = class_size
data["demographics"] = data["demographics"][data["demographics"]["schoolyear"] == 20112012]
data["graduation"] = data["graduation"][data["graduation"]["Cohort"] == "2006"]
data["graduation"] = data["graduation"][data["graduation"]["Demographic"] == "Total Cohort"]
```
# Convert AP scores to numeric
```
cols = ['AP Test Takers ', 'Total Exams Taken', 'Number of Exams with scores 3 4 or 5']
for col in cols:
data["ap_2010"][col] = pd.to_numeric(data["ap_2010"][col], errors="coerce")
```
# Combine the datasets
```
combined = data["sat_results"]
combined = combined.merge(data["ap_2010"], on="DBN", how="left")
combined = combined.merge(data["graduation"], on="DBN", how="left")
to_merge = ["class_size", "demographics", "survey", "hs_directory"]
for m in to_merge:
combined = combined.merge(data[m], on="DBN", how="inner")
combined = combined.fillna(combined.mean())
combined = combined.fillna(0)
```
# Add a school district column for mapping
```
def get_first_two_chars(dbn):
return dbn[0:2]
combined["school_dist"] = combined["DBN"].apply(get_first_two_chars)
```
# Find correlations
```
correlations = combined.corr()
correlations = correlations["sat_score"]
print(correlations)
```
# Plotting survey correlations
```
# Remove DBN since it's a unique identifier, not a useful numerical value for correlation.
survey_fields.remove("DBN")
%matplotlib inline
combined.corr()["sat_score"][survey_fields].plot.bar()
```
There are high correlations between `N_s`, `N_t`, `N_p` and `sat_score`. Since these columns are correlated with `total_enrollment`, it makes sense that they would be high.
It is more interesting that `rr_s`, the student response rate, or the percentage of students that completed the survey, correlates with `sat_score`. This might make sense because students who are more likely to fill out surveys may be more likely to also be doing well academically.
How students and teachers percieved safety (`saf_t_11` and `saf_s_11`) correlate with sat_score. This make sense, as it's hard to teach or learn in an unsafe environment.
The last interesting correlation is the `aca_s_11`, which indicates how the student perceives academic standards, correlates with `sat_score`, but this is not true for `aca_t_11`, how teachers perceive academic standards, or `aca_p_11`, how parents perceive academic standards.
# Exploring Safety
```
combined.plot.scatter("saf_s_11", "sat_score")
```
There appears to be a correlation between SAT scores and safety, although it isn't thatstrong. It looks like there are a few schools with extremely high SAT scores and high safety scores. There are a few schools with low safety scores and low SAT scores. No school with a safety score lower than 6.5 has an average SAT score higher than 1500 or so.
# Borough Safety
```
boros = combined.groupby("boro").agg(numpy.mean)["saf_s_11"]
print(boros)
```
It looks like Manhattan and Queens tend to have higher safety scores, whereas Brooklyn has low safety scores.
# Racial Differences in SAT Scores
```
race_fields = ["white_per", "asian_per", "black_per", "hispanic_per"]
combined.corr()["sat_score"][race_fields].plot.bar()
```
It looks like a higher percentage of white or asian students at a school correlates positively with sat score, whereas a higher percentage of black or hispanic students correlates negatively with sat score. This may be due to a lack of funding for schools in certain areas, which are more likely to have a higher percentage of black or hispanic students.
```
combined.plot.scatter("hispanic_per", "sat_score")
print(combined[combined["hispanic_per"] > 95]["SCHOOL NAME"])
```
The schools listed above appear to primarily be geared towards recent immigrants to the US. These schools have a lot of students who are learning English, which would explain the lower SAT scores.
```
print(combined[(combined["hispanic_per"] < 10) & (combined["sat_score"] > 1800)]["SCHOOL NAME"])
```
Many of the schools above appear to be specialized science and technology schools that receive extra funding, and only admit students who pass an entrance exam. This doesn't explain the low hispanic_per, but it does explain why their students tend to do better on the SAT -- they are students from all over New York City who did well on a standardized test.
# Gender Differences in SAT Scores
```
gender_fields = ["male_per", "female_per"]
combined.corr()["sat_score"][gender_fields].plot.bar()
```
In the plot above, we can see that a high percentage of females at a school positively correlates with SAT score, whereas a high percentage of males at a school negatively correlates with SAT score. Neither correlation is extremely strong.
```
combined.plot.scatter("female_per", "sat_score")
```
Based on the scatterplot, there doesn't seem to be any real correlation between sat_score and female_per. However, there is a cluster of schools with a high percentage of females (60 to 80), and high SAT scores.
```
print(combined[(combined["female_per"] > 60) & (combined["sat_score"] > 1700)]["SCHOOL NAME"])
```
These schools appears to be very selective liberal arts schools that have high academic standards.
# AP Exam Scores vs. SAT Scores
```
combined["ap_per"] = combined["AP Test Takers "] / combined["total_enrollment"]
combined.plot.scatter(x='ap_per', y='sat_score')
```
It looks like there is a relationship between the percentage of students in a school who take the AP exam, and their average SAT scores. It's not a strong correlation, though.
|
github_jupyter
|
import pandas as pd
import numpy
import re
data_files = [
"ap_2010.csv",
"class_size.csv",
"demographics.csv",
"graduation.csv",
"hs_directory.csv",
"sat_results.csv"
]
data = {}
for f in data_files:
d = pd.read_csv("schools/{0}".format(f))
data[f.replace(".csv", "")] = d
all_survey = pd.read_csv("schools/survey_all.txt", delimiter="\t", encoding='windows-1252')
d75_survey = pd.read_csv("schools/survey_d75.txt", delimiter="\t", encoding='windows-1252')
survey = pd.concat([all_survey, d75_survey], axis=0)
survey["DBN"] = survey["dbn"]
survey_fields = [
"DBN",
"rr_s",
"rr_t",
"rr_p",
"N_s",
"N_t",
"N_p",
"saf_p_11",
"com_p_11",
"eng_p_11",
"aca_p_11",
"saf_t_11",
"com_t_11",
"eng_t_11",
"aca_t_11",
"saf_s_11",
"com_s_11",
"eng_s_11",
"aca_s_11",
"saf_tot_11",
"com_tot_11",
"eng_tot_11",
"aca_tot_11",
]
survey = survey.loc[:,survey_fields]
data["survey"] = survey
data["hs_directory"]["DBN"] = data["hs_directory"]["dbn"]
def pad_csd(num):
string_representation = str(num)
if len(string_representation) > 1:
return string_representation
else:
return "0" + string_representation
data["class_size"]["padded_csd"] = data["class_size"]["CSD"].apply(pad_csd)
data["class_size"]["DBN"] = data["class_size"]["padded_csd"] + data["class_size"]["SCHOOL CODE"]
cols = ['SAT Math Avg. Score', 'SAT Critical Reading Avg. Score', 'SAT Writing Avg. Score']
for c in cols:
data["sat_results"][c] = pd.to_numeric(data["sat_results"][c], errors="coerce")
data['sat_results']['sat_score'] = data['sat_results'][cols[0]] + data['sat_results'][cols[1]] + data['sat_results'][cols[2]]
def find_lat(loc):
coords = re.findall("\(.+, .+\)", loc)
lat = coords[0].split(",")[0].replace("(", "")
return lat
def find_lon(loc):
coords = re.findall("\(.+, .+\)", loc)
lon = coords[0].split(",")[1].replace(")", "").strip()
return lon
data["hs_directory"]["lat"] = data["hs_directory"]["Location 1"].apply(find_lat)
data["hs_directory"]["lon"] = data["hs_directory"]["Location 1"].apply(find_lon)
data["hs_directory"]["lat"] = pd.to_numeric(data["hs_directory"]["lat"], errors="coerce")
data["hs_directory"]["lon"] = pd.to_numeric(data["hs_directory"]["lon"], errors="coerce")
class_size = data["class_size"]
class_size = class_size[class_size["GRADE "] == "09-12"]
class_size = class_size[class_size["PROGRAM TYPE"] == "GEN ED"]
class_size = class_size.groupby("DBN").agg(numpy.mean)
class_size.reset_index(inplace=True)
data["class_size"] = class_size
data["demographics"] = data["demographics"][data["demographics"]["schoolyear"] == 20112012]
data["graduation"] = data["graduation"][data["graduation"]["Cohort"] == "2006"]
data["graduation"] = data["graduation"][data["graduation"]["Demographic"] == "Total Cohort"]
cols = ['AP Test Takers ', 'Total Exams Taken', 'Number of Exams with scores 3 4 or 5']
for col in cols:
data["ap_2010"][col] = pd.to_numeric(data["ap_2010"][col], errors="coerce")
combined = data["sat_results"]
combined = combined.merge(data["ap_2010"], on="DBN", how="left")
combined = combined.merge(data["graduation"], on="DBN", how="left")
to_merge = ["class_size", "demographics", "survey", "hs_directory"]
for m in to_merge:
combined = combined.merge(data[m], on="DBN", how="inner")
combined = combined.fillna(combined.mean())
combined = combined.fillna(0)
def get_first_two_chars(dbn):
return dbn[0:2]
combined["school_dist"] = combined["DBN"].apply(get_first_two_chars)
correlations = combined.corr()
correlations = correlations["sat_score"]
print(correlations)
# Remove DBN since it's a unique identifier, not a useful numerical value for correlation.
survey_fields.remove("DBN")
%matplotlib inline
combined.corr()["sat_score"][survey_fields].plot.bar()
combined.plot.scatter("saf_s_11", "sat_score")
boros = combined.groupby("boro").agg(numpy.mean)["saf_s_11"]
print(boros)
race_fields = ["white_per", "asian_per", "black_per", "hispanic_per"]
combined.corr()["sat_score"][race_fields].plot.bar()
combined.plot.scatter("hispanic_per", "sat_score")
print(combined[combined["hispanic_per"] > 95]["SCHOOL NAME"])
print(combined[(combined["hispanic_per"] < 10) & (combined["sat_score"] > 1800)]["SCHOOL NAME"])
gender_fields = ["male_per", "female_per"]
combined.corr()["sat_score"][gender_fields].plot.bar()
combined.plot.scatter("female_per", "sat_score")
print(combined[(combined["female_per"] > 60) & (combined["sat_score"] > 1700)]["SCHOOL NAME"])
combined["ap_per"] = combined["AP Test Takers "] / combined["total_enrollment"]
combined.plot.scatter(x='ap_per', y='sat_score')
| 0.407569 | 0.809238 |
Sounding Plotter
================
**This should be run as a script from the command line - not as a notebook.**
Download and plot the most recent sounding data for a specified site.
Provides a simple command line interface to specify a site. Using the current
UTC time, the script calculates what the most recent sounding should be and requests
it from the Wyoming archive using Siphon.
Do the needed imports
```
import posixpath
import matplotlib.pyplot as plt
import metpy.calc as mpcalc
from metpy.plots import add_metpy_logo, add_timestamp, SkewT
from metpy.units import units
from siphon.simplewebservice.wyoming import WyomingUpperAir
```
This class encapsulates the code needed to upload an image to Google Drive
```
class DriveUploader:
def __init__(self, credsfile='mycreds.txt'):
from pydrive.drive import GoogleDrive
self.gdrive = GoogleDrive(self._get_auth(credsfile))
def _get_auth(self, credsfile):
from pydrive.auth import GoogleAuth
gauth = GoogleAuth()
# Try to load saved client credentials
gauth.LoadCredentialsFile(credsfile)
if gauth.credentials is None:
# Authenticate if they're not there
gauth.LocalWebserverAuth()
elif gauth.access_token_expired:
# Refresh them if expired
gauth.Refresh()
else:
# Initialize the saved creds
gauth.Authorize()
# Save the current credentials to a file
gauth.SaveCredentialsFile(credsfile)
return gauth
def _get_first_file_id(self, title, parent, **kwargs):
query = "title='{}' and '{}' in parents".format(title, parent)
for k, v in kwargs.items():
query += " and {}='{}'".format(k, v)
res = next(self.gdrive.ListFile({'q': query}))
if res:
return res[0]['id']
return None
def get_folder(self, path):
parent = 'root'
for part in path.split('/'):
if not part:
continue
parent = self._get_first_file_id(part, parent,
mimeType='application/vnd.google-apps.folder')
return parent
def create_or_get_file(self, path):
pathname, filename = posixpath.split(path)
folder = self.get_folder(pathname)
create_file_args = {'parents': [{'kind': 'drive#fileLink', 'id': folder}]}
file_id = self._get_first_file_id(filename, folder)
if file_id is not None:
create_file_args['id'] = file_id
return self.gdrive.CreateFile(create_file_args)
def upload_to(self, local_path, remote_path):
f = self.create_or_get_file(remote_path)
f.SetContentFile(local_path)
f['title'] = posixpath.basename(remote_path)
f.Upload()
```
This function takes care of actually generating a skewT from the `DataFrame`
```
def plot_skewt(df):
# We will pull the data out of the example dataset into individual variables
# and assign units.
p = df['pressure'].values * units.hPa
T = df['temperature'].values * units.degC
Td = df['dewpoint'].values * units.degC
wind_speed = df['speed'].values * units.knots
wind_dir = df['direction'].values * units.degrees
u, v = mpcalc.wind_components(wind_speed, wind_dir)
# Create a new figure. The dimensions here give a good aspect ratio.
fig = plt.figure(figsize=(9, 9))
add_metpy_logo(fig, 115, 100)
skew = SkewT(fig, rotation=45)
# Plot the data using normal plotting functions, in this case using
# log scaling in Y, as dictated by the typical meteorological plot
skew.plot(p, T, 'r')
skew.plot(p, Td, 'g')
skew.plot_barbs(p, u, v)
skew.ax.set_ylim(1000, 100)
skew.ax.set_xlim(-40, 60)
# Calculate LCL height and plot as black dot
lcl_pressure, lcl_temperature = mpcalc.lcl(p[0], T[0], Td[0])
skew.plot(lcl_pressure, lcl_temperature, 'ko', markerfacecolor='black')
# Calculate full parcel profile and add to plot as black line
prof = mpcalc.parcel_profile(p, T[0], Td[0]).to('degC')
skew.plot(p, prof, 'k', linewidth=2)
# An example of a slanted line at constant T -- in this case the 0
# isotherm
skew.ax.axvline(0, color='c', linestyle='--', linewidth=2)
# Add the relevant special lines
skew.plot_dry_adiabats()
skew.plot_moist_adiabats()
skew.plot_mixing_lines()
return skew
def make_name(site, time):
return '{site}_{dt:%Y%m%d_%H%M}.png'.format(site=site, dt=time)
```
This is where the command line script will actually enter, and handles parsing
command line arguments and driving everything else.
```
if __name__ == '__main__':
import argparse
from datetime import datetime, timedelta
import tempfile
# Set up argument parsing for the script. Provides one argument for the site, and another
# that controls whether the plot should be shown or saved as an image.
parser = argparse.ArgumentParser(description='Download sounding data and plot.')
parser.add_argument('-s', '--site', help='Site to obtain data for', type=str,
default='DNR')
parser.add_argument('--show', help='Whether to show the plot rather than save to disk',
action='store_true')
parser.add_argument('-d', '--date', help='Date and time to request data for in YYYYMMDDHH.'
' Defaults to most recent 00/12 hour.', type=str)
parser.add_argument('-g', '--gdrive', help='Google Drive upload path', type=str)
parser.add_argument('-f', '--filename', help='Image filename', type=str)
args = parser.parse_args()
if args.date:
request_time = datetime.strptime(args.date, '%Y%m%d%H')
else:
# Figure out the most recent sounding, 00 or 12. Subtracting two hours
# helps ensure that we choose a time with data available.
now = datetime.utcnow() - timedelta(hours=2)
request_time = now.replace(hour=(now.hour // 12) * 12, minute=0, second=0)
# Request the data and plot
df = WyomingUpperAir.request_data(request_time, args.site)
skewt = plot_skewt(df)
# Add the timestamp for the data to the plot
add_timestamp(skewt.ax, request_time, y=1.02, x=0, ha='left', fontsize='large')
skewt.ax.set_title(args.site)
if args.show:
plt.show()
else:
fname = args.filename if args.filename else make_name(args.site, request_time)
if args.gdrive:
uploader = DriveUploader()
with tempfile.NamedTemporaryFile(suffix='.png') as f:
skewt.ax.figure.savefig(f.name)
uploader.upload_to(f.name, posixpath.join(args.gdrive, fname))
else:
skewt.ax.figure.savefig(make_name(args.site, request_time))
```
|
github_jupyter
|
import posixpath
import matplotlib.pyplot as plt
import metpy.calc as mpcalc
from metpy.plots import add_metpy_logo, add_timestamp, SkewT
from metpy.units import units
from siphon.simplewebservice.wyoming import WyomingUpperAir
class DriveUploader:
def __init__(self, credsfile='mycreds.txt'):
from pydrive.drive import GoogleDrive
self.gdrive = GoogleDrive(self._get_auth(credsfile))
def _get_auth(self, credsfile):
from pydrive.auth import GoogleAuth
gauth = GoogleAuth()
# Try to load saved client credentials
gauth.LoadCredentialsFile(credsfile)
if gauth.credentials is None:
# Authenticate if they're not there
gauth.LocalWebserverAuth()
elif gauth.access_token_expired:
# Refresh them if expired
gauth.Refresh()
else:
# Initialize the saved creds
gauth.Authorize()
# Save the current credentials to a file
gauth.SaveCredentialsFile(credsfile)
return gauth
def _get_first_file_id(self, title, parent, **kwargs):
query = "title='{}' and '{}' in parents".format(title, parent)
for k, v in kwargs.items():
query += " and {}='{}'".format(k, v)
res = next(self.gdrive.ListFile({'q': query}))
if res:
return res[0]['id']
return None
def get_folder(self, path):
parent = 'root'
for part in path.split('/'):
if not part:
continue
parent = self._get_first_file_id(part, parent,
mimeType='application/vnd.google-apps.folder')
return parent
def create_or_get_file(self, path):
pathname, filename = posixpath.split(path)
folder = self.get_folder(pathname)
create_file_args = {'parents': [{'kind': 'drive#fileLink', 'id': folder}]}
file_id = self._get_first_file_id(filename, folder)
if file_id is not None:
create_file_args['id'] = file_id
return self.gdrive.CreateFile(create_file_args)
def upload_to(self, local_path, remote_path):
f = self.create_or_get_file(remote_path)
f.SetContentFile(local_path)
f['title'] = posixpath.basename(remote_path)
f.Upload()
def plot_skewt(df):
# We will pull the data out of the example dataset into individual variables
# and assign units.
p = df['pressure'].values * units.hPa
T = df['temperature'].values * units.degC
Td = df['dewpoint'].values * units.degC
wind_speed = df['speed'].values * units.knots
wind_dir = df['direction'].values * units.degrees
u, v = mpcalc.wind_components(wind_speed, wind_dir)
# Create a new figure. The dimensions here give a good aspect ratio.
fig = plt.figure(figsize=(9, 9))
add_metpy_logo(fig, 115, 100)
skew = SkewT(fig, rotation=45)
# Plot the data using normal plotting functions, in this case using
# log scaling in Y, as dictated by the typical meteorological plot
skew.plot(p, T, 'r')
skew.plot(p, Td, 'g')
skew.plot_barbs(p, u, v)
skew.ax.set_ylim(1000, 100)
skew.ax.set_xlim(-40, 60)
# Calculate LCL height and plot as black dot
lcl_pressure, lcl_temperature = mpcalc.lcl(p[0], T[0], Td[0])
skew.plot(lcl_pressure, lcl_temperature, 'ko', markerfacecolor='black')
# Calculate full parcel profile and add to plot as black line
prof = mpcalc.parcel_profile(p, T[0], Td[0]).to('degC')
skew.plot(p, prof, 'k', linewidth=2)
# An example of a slanted line at constant T -- in this case the 0
# isotherm
skew.ax.axvline(0, color='c', linestyle='--', linewidth=2)
# Add the relevant special lines
skew.plot_dry_adiabats()
skew.plot_moist_adiabats()
skew.plot_mixing_lines()
return skew
def make_name(site, time):
return '{site}_{dt:%Y%m%d_%H%M}.png'.format(site=site, dt=time)
if __name__ == '__main__':
import argparse
from datetime import datetime, timedelta
import tempfile
# Set up argument parsing for the script. Provides one argument for the site, and another
# that controls whether the plot should be shown or saved as an image.
parser = argparse.ArgumentParser(description='Download sounding data and plot.')
parser.add_argument('-s', '--site', help='Site to obtain data for', type=str,
default='DNR')
parser.add_argument('--show', help='Whether to show the plot rather than save to disk',
action='store_true')
parser.add_argument('-d', '--date', help='Date and time to request data for in YYYYMMDDHH.'
' Defaults to most recent 00/12 hour.', type=str)
parser.add_argument('-g', '--gdrive', help='Google Drive upload path', type=str)
parser.add_argument('-f', '--filename', help='Image filename', type=str)
args = parser.parse_args()
if args.date:
request_time = datetime.strptime(args.date, '%Y%m%d%H')
else:
# Figure out the most recent sounding, 00 or 12. Subtracting two hours
# helps ensure that we choose a time with data available.
now = datetime.utcnow() - timedelta(hours=2)
request_time = now.replace(hour=(now.hour // 12) * 12, minute=0, second=0)
# Request the data and plot
df = WyomingUpperAir.request_data(request_time, args.site)
skewt = plot_skewt(df)
# Add the timestamp for the data to the plot
add_timestamp(skewt.ax, request_time, y=1.02, x=0, ha='left', fontsize='large')
skewt.ax.set_title(args.site)
if args.show:
plt.show()
else:
fname = args.filename if args.filename else make_name(args.site, request_time)
if args.gdrive:
uploader = DriveUploader()
with tempfile.NamedTemporaryFile(suffix='.png') as f:
skewt.ax.figure.savefig(f.name)
uploader.upload_to(f.name, posixpath.join(args.gdrive, fname))
else:
skewt.ax.figure.savefig(make_name(args.site, request_time))
| 0.679817 | 0.764056 |
# Continuous training with TFX and Vertex
## Learning Objectives
1. Containerize your TFX code into a pipeline package using Cloud Build.
1. Use the TFX CLI to compile a TFX pipeline.
1. Deploy a TFX pipeline version to run on Vertex Pipelines using the Vertex Python SDK.
### Setup
```
from google.cloud import aiplatform as vertex_ai
```
#### Validate lab package version installation
```
!python -c "import tensorflow as tf; print(f'TF version: {tf.__version__}')"
!python -c "import tfx; print(f'TFX version: {tfx.__version__}')"
!python -c "import kfp; print(f'KFP version: {kfp.__version__}')"
print(f"vertex_ai: {vertex_ai.__version__}")
```
**Note**: this lab was built and tested with the following package versions:
`TF version: 2.6.2`
`TFX version: 1.4.0`
`KFP version: 1.8.1`
`aiplatform: 1.7.1`
## Review: example TFX pipeline design pattern for Vertex
The pipeline source code can be found in the `pipeline_vertex` folder.
```
%cd pipeline_vertex
!ls -la
```
The `config.py` module configures the default values for the environment specific settings and the default values for the pipeline runtime parameters.
The default values can be overwritten at compile time by providing the updated values in a set of environment variables. You will set custom environment variables later on this lab.
The `pipeline.py` module contains the TFX DSL defining the workflow implemented by the pipeline.
The `preprocessing.py` module implements the data preprocessing logic the `Transform` component.
The `model.py` module implements the TensorFlow model code and training logic for the `Trainer` component.
The `runner.py` module configures and executes `KubeflowV2DagRunner`. At compile time, the `KubeflowDagRunner.run()` method converts the TFX DSL into the pipeline package into a JSON format for execution on Vertex.
The `features.py` module contains feature definitions common across `preprocessing.py` and `model.py`.
## Exercise: build your pipeline with the TFX CLI
You will use TFX CLI to compile and deploy the pipeline. As explained in the previous section, the environment specific settings can be provided through a set of environment variables and embedded into the pipeline package at compile time.
### Configure your environment resource settings
Update the below constants with the settings reflecting your lab environment.
- `REGION` - the compute region for AI Platform Training, Vizier, and Prediction.
- `ARTIFACT_STORE` - An existing GCS bucket. You can use any bucket, but we will use here the bucket with the same name as the project.
```
# TODO: Set your environment resource settings here for GCP_REGION, ARTIFACT_STORE_URI, ENDPOINT, and CUSTOM_SERVICE_ACCOUNT.
REGION = "us-central1"
PROJECT_ID = !(gcloud config get-value core/project)
PROJECT_ID = PROJECT_ID[0]
ARTIFACT_STORE = f"gs://{PROJECT_ID}"
# Set your resource settings as environment variables. These override the default values in pipeline/config.py.
%env REGION={REGION}
%env ARTIFACT_STORE={ARTIFACT_STORE}
%env PROJECT_ID={PROJECT_ID}
!gsutil ls | grep ^{ARTIFACT_STORE}/$ || gsutil mb -l {REGION} {ARTIFACT_STORE}
```
### Set the compile time settings to first create a pipeline version without hyperparameter tuning
Default pipeline runtime environment values are configured in the pipeline folder `config.py`. You will set their values directly below:
* `PIPELINE_NAME` - the pipeline's globally unique name.
* `DATA_ROOT_URI` - the URI for the raw lab dataset `gs://{PROJECT_ID}/data/tfxcovertype`.
* `TFX_IMAGE_URI` - the image name of your pipeline container that will be used to execute each of your tfx components
```
PIPELINE_NAME = "tfxcovertype"
DATA_ROOT_URI = f"gs://{PROJECT_ID}/data/tfxcovertype"
TFX_IMAGE_URI = f"gcr.io/{PROJECT_ID}/{PIPELINE_NAME}"
PIPELINE_JSON = f"{PIPELINE_NAME}.json"
TRAIN_STEPS = 10
EVAL_STEPS = 5
%env PIPELINE_NAME={PIPELINE_NAME}
%env DATA_ROOT_URI={DATA_ROOT_URI}
%env TFX_IMAGE_URI={TFX_IMAGE_URI}
%env PIPELINE_JSON={PIPELINE_JSON}
%env TRAIN_STEPS={TRAIN_STEPS}
%env EVAL_STEPS={EVAL_STEPS}
```
Let us populate the data bucket at `DATA_ROOT_URI`:
```
!gsutil cp ../../../data/* $DATA_ROOT_URI/dataset.csv
!gsutil ls $DATA_ROOT_URI/*
```
Let us build and push the TFX container image described in the `Dockerfile`:
```
!gcloud builds submit --timeout 15m --tag $TFX_IMAGE_URI .
```
### Compile your pipeline code
The following command will execute the `KubeflowV2DagRunner` that compiles the pipeline described in `pipeline.py` into a JSON representation consumable by Vertex:
```
!tfx pipeline compile --engine vertex --pipeline_path runner.py
```
Note: you should see a `{PIPELINE_NAME}.json` file appear in your current pipeline directory.
## Exercise: deploy your pipeline on Vertex using the Vertex SDK
Once you have the `{PIPELINE_NAME}.json` available, you can run the tfx pipeline on Vertex by launching a pipeline job using the `aiplatform` handle:
```
vertex_ai.init(project=PROJECT_ID, location=REGION)
pipeline = vertex_ai.PipelineJob(
display_name="tfxcovertype4",
template_path=PIPELINE_JSON,
enable_caching=False,
)
pipeline.run()
```
## Next Steps
In this lab, you learned how to build and deploy a TFX pipeline with the TFX CLI and then update, build and deploy a new pipeline with automatic hyperparameter tuning. You practiced triggered continuous pipeline runs using the TFX CLI as well as the Kubeflow Pipelines UI.
In the next lab, you will construct a Cloud Build CI/CD workflow that further automates the building and deployment of the pipeline.
## License
Copyright 2021 Google Inc. All Rights Reserved.
Licensed under the Apache License, Version 2.0 (the "License");
you may not use this file except in compliance with the License.
You may obtain a copy of the License at
http://www.apache.org/licenses/LICENSE-2.0
Unless required by applicable law or agreed to in writing, software
distributed under the License is distributed on an "AS IS" BASIS,
WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
See the License for the specific language governing permissions and
limitations under the License.
|
github_jupyter
|
from google.cloud import aiplatform as vertex_ai
!python -c "import tensorflow as tf; print(f'TF version: {tf.__version__}')"
!python -c "import tfx; print(f'TFX version: {tfx.__version__}')"
!python -c "import kfp; print(f'KFP version: {kfp.__version__}')"
print(f"vertex_ai: {vertex_ai.__version__}")
%cd pipeline_vertex
!ls -la
# TODO: Set your environment resource settings here for GCP_REGION, ARTIFACT_STORE_URI, ENDPOINT, and CUSTOM_SERVICE_ACCOUNT.
REGION = "us-central1"
PROJECT_ID = !(gcloud config get-value core/project)
PROJECT_ID = PROJECT_ID[0]
ARTIFACT_STORE = f"gs://{PROJECT_ID}"
# Set your resource settings as environment variables. These override the default values in pipeline/config.py.
%env REGION={REGION}
%env ARTIFACT_STORE={ARTIFACT_STORE}
%env PROJECT_ID={PROJECT_ID}
!gsutil ls | grep ^{ARTIFACT_STORE}/$ || gsutil mb -l {REGION} {ARTIFACT_STORE}
PIPELINE_NAME = "tfxcovertype"
DATA_ROOT_URI = f"gs://{PROJECT_ID}/data/tfxcovertype"
TFX_IMAGE_URI = f"gcr.io/{PROJECT_ID}/{PIPELINE_NAME}"
PIPELINE_JSON = f"{PIPELINE_NAME}.json"
TRAIN_STEPS = 10
EVAL_STEPS = 5
%env PIPELINE_NAME={PIPELINE_NAME}
%env DATA_ROOT_URI={DATA_ROOT_URI}
%env TFX_IMAGE_URI={TFX_IMAGE_URI}
%env PIPELINE_JSON={PIPELINE_JSON}
%env TRAIN_STEPS={TRAIN_STEPS}
%env EVAL_STEPS={EVAL_STEPS}
!gsutil cp ../../../data/* $DATA_ROOT_URI/dataset.csv
!gsutil ls $DATA_ROOT_URI/*
!gcloud builds submit --timeout 15m --tag $TFX_IMAGE_URI .
!tfx pipeline compile --engine vertex --pipeline_path runner.py
vertex_ai.init(project=PROJECT_ID, location=REGION)
pipeline = vertex_ai.PipelineJob(
display_name="tfxcovertype4",
template_path=PIPELINE_JSON,
enable_caching=False,
)
pipeline.run()
| 0.215186 | 0.953708 |
In this lab, we will optimize the weather simulation application written in C++ (if you prefer to use Fortran, click [this link](../../Fortran/jupyter_notebook/profiling-fortran.ipynb)).
Let's execute the cell below to display information about the GPUs running on the server by running the pgaccelinfo command, which ships with the PGI compiler that we will be using. To do this, execute the cell block below by giving it focus (clicking on it with your mouse), and hitting Ctrl-Enter, or pressing the play button in the toolbar above. If all goes well, you should see some output returned below the grey cell.
```
!pgaccelinfo
```
## Exercise 4
### Learning objectives
Learn how to improve the performance of the application by managing data movement and reducing the unnecessary data transfers. In this exercise you will:
- Learn about unified memory and how to automatically migrate data between CPU and GPU
- Learn how to use it via PGI compiler managed option, and profiling managed memory
- Learn how to identify redundant memory copies via Nsight Systems
- Learn how to improve efficiency by reducing extra data copies via OpenACC data directive
- Learn how to use PGI compiler feedback as a guidance on where to insert the OpenACC data directives
- Apply data directives to the parallel application, benchmark and profile it
Let's inspect the profiler report from previous exercise. From the "timeline view" on the top pane, double click on the "CUDA" from the function table on the left and expand it. Zoom in on the timeline and you can see a pattern similar to the screenshot below. The blue boxes are the compute kernels and each of these groupings of kernels is surrounded by purple and teal boxes (annotated with red color) representing data movements.
What this graph is showing us is that we're doing a lot of data movement between GPU and CPU.
<img src="images/nsys_data_mv.png">
The compiler feedback we collected earlier tells us quite a bit about data movement too. If we look again at the compiler feedback from above, we see the following.
<img src="images/cfeedback3-1.png" width="80%" height="80%">
The compiler feedback is telling us that the compiler has inserted data movement around our parallel region at line 277 which copies the `hy_dens_cell`, `hy_dens_theta_cell`, and `state` arrays in and out of GPU memory and also copies `flux` array out.
The compiler can only work with the information we provide. It knows we need the `hy_dens_cell`, `hy_dens_theta_cell`, `state`, and `flux` arrays on the GPU for the accelerated section within the `compute_tendencies_x` function, but we didn't tell the compiler anything about what happens to the data outside of those sections. Without this knowledge, the compiler has to copy the full arrays to the GPU and back to the CPU for each accelerated section. This is a lot of unnecessary data transfers.
Ideally, we would want to move the data (example: `hy_dens_cell`, `hy_dens_theta_cell`, `state` arrays) to the GPU at the beginning, and only transfer back to the CPU at the end (if needed). And as for the `flux` array in this example, we do not need to copy any data back and forth. So we only need to create space on the device (GPU) for this array.
We need to give the compiler information about how to reduce the extra and unnecessary data movement. By adding OpenACC `data` directive to a structured code block, the compiler will know how to manage data according to the clauses. For information on the data directive clauses, please visit [OpenACC 3.0 Specification](https://www.openacc.org/sites/default/files/inline-images/Specification/OpenACC.3.0.pdf).
Now, add `data` directives to the code, save the file, re-compile via `make`, and profile it again.
Click on the <b>[miniWeather_openacc.cpp](../source_code/lab4/miniWeather_openacc.cpp)</b> and <b>[Makefile](../source_code/lab4/Makefile)</b> links and modify `miniWeather_openacc.cpp` and `Makefile`. Remember to **SAVE** your code after changes, before running below cells.
```
!cd ../source_code/lab4 && make clean && make
```
Let us start inspecting the compiler feedback and see if it applied the optimizations. Here is the screenshot of expected compiler feedback after adding the `data` directives. You can see that on line 281, compiler is generating default present for `hy_dens_cell`, `hy_dens_theta_cell`, `state`, and `flux` arrays. In other words, it is assuming that data is present on the GPU and it only copies data to the GPU only if the data do not exist.
<img src="images/cfeedback4.png" width="80%" height="80%">
Now, **Profile** your code with Nsight Systems command line `nsys`.
```
!cd ../source_code/lab4 && nsys profile -t nvtx,openacc --stats=true --force-overwrite true -o miniWeather_5 ./miniWeather
```
[Download the profiler output](../source_code/lab4/miniWeather_5.qdrep) and open it via the GUI. Have a look at the example expected output below:
<img src="images/nsys_fast_mv.png">
Have a look at the data movements annotated with red color and compare it with the previous versions. We have accelerated the application and reduced the execution time by eliminating the unnecessary data transfers between CPU and GPU.
**Note**: Next exercise gives an overview on introduction to Nsight Compute tool and it is optional.
## Post-Lab Summary
If you would like to download this lab for later viewing, it is recommend you go to your browsers File menu (not the Jupyter notebook file menu) and save the complete web page. This will ensure the images are copied down as well. You can also execute the following cell block to create a zip-file of the files you've been working on, and download it with the link below.
```
%%bash
cd ..
rm -f openacc_profiler_files.zip
zip -r openacc_profiler_files.zip *
```
**After** executing the above zip command, you should be able to download the zip file [here](../openacc_profiler_files.zip).
-----
# <p style="text-align:center;border:3px; border-style:solid; border-color:#FF0000 ; padding: 1em"> <a href=../../profiling_start.ipynb>HOME</a> <span style="float:center"> <a href=profiling-c-lab5.ipynb>NEXT</a></span> </p>
-----
# Links and Resources
[OpenACC API Guide](https://www.openacc.org/sites/default/files/inline-files/OpenACC%20API%202.6%20Reference%20Guide.pdf)
[NVIDIA Nsight System](https://docs.nvidia.com/nsight-systems/)
[CUDA Toolkit Download](https://developer.nvidia.com/cuda-downloads)
**NOTE**: To be able to see the Nsight System profiler output, please download Nsight System latest version from [here](https://developer.nvidia.com/nsight-systems).
Don't forget to check out additional [OpenACC Resources](https://www.openacc.org/resources) and join our [OpenACC Slack Channel](https://www.openacc.org/community#slack) to share your experience and get more help from the community.
---
## Licensing
This material is released by OpenACC-Standard.org, in collaboration with NVIDIA Corporation, under the Creative Commons Attribution 4.0 International (CC BY 4.0).
|
github_jupyter
|
!pgaccelinfo
!cd ../source_code/lab4 && make clean && make
!cd ../source_code/lab4 && nsys profile -t nvtx,openacc --stats=true --force-overwrite true -o miniWeather_5 ./miniWeather
%%bash
cd ..
rm -f openacc_profiler_files.zip
zip -r openacc_profiler_files.zip *
| 0.191252 | 0.986904 |
Deep Learning
=============
Assignment 3
------------
Previously in `2_fullyconnected.ipynb`, you trained a logistic regression and a neural network model.
The goal of this assignment is to explore regularization techniques.
```
# These are all the modules we'll be using later. Make sure you can import them
# before proceeding further.
from __future__ import print_function
import numpy as np
import tensorflow as tf
from six.moves import cPickle as pickle
```
First reload the data we generated in _notmist.ipynb_.
```
pickle_file = 'notMNIST.pickle'
with open(pickle_file, 'rb') as f:
save = pickle.load(f)
train_dataset = save['train_dataset']
train_labels = save['train_labels']
valid_dataset = save['valid_dataset']
valid_labels = save['valid_labels']
test_dataset = save['test_dataset']
test_labels = save['test_labels']
del save # hint to help gc free up memory
print('Training set', train_dataset.shape, train_labels.shape)
print('Validation set', valid_dataset.shape, valid_labels.shape)
print('Test set', test_dataset.shape, test_labels.shape)
```
Reformat into a shape that's more adapted to the models we're going to train:
- data as a flat matrix,
- labels as float 1-hot encodings.
```
image_size = 28
num_labels = 10
def reformat(dataset, labels):
dataset = dataset.reshape((-1, image_size * image_size)).astype(np.float32)
# Map 2 to [0.0, 1.0, 0.0 ...], 3 to [0.0, 0.0, 1.0 ...]
labels = (np.arange(num_labels) == labels[:,None]).astype(np.float32)
return dataset, labels
train_dataset, train_labels = reformat(train_dataset, train_labels)
valid_dataset, valid_labels = reformat(valid_dataset, valid_labels)
test_dataset, test_labels = reformat(test_dataset, test_labels)
print('Training set', train_dataset.shape, train_labels.shape)
print('Validation set', valid_dataset.shape, valid_labels.shape)
print('Test set', test_dataset.shape, test_labels.shape)
def accuracy(predictions, labels):
return (100.0 * np.sum(np.argmax(predictions, 1) == np.argmax(labels, 1))
/ predictions.shape[0])
```
---
Problem 1
---------
Introduce and tune L2 regularization for both logistic and neural network models. Remember that L2 amounts to adding a penalty on the norm of the weights to the loss. In TensorFlow, you can compute the L2 loss for a tensor `t` using `nn.l2_loss(t)`. The right amount of regularization should improve your validation / test accuracy.
---
---
Problem 2
---------
Let's demonstrate an extreme case of overfitting. Restrict your training data to just a few batches. What happens?
---
---
Problem 3
---------
Introduce Dropout on the hidden layer of the neural network. Remember: Dropout should only be introduced during training, not evaluation, otherwise your evaluation results would be stochastic as well. TensorFlow provides `nn.dropout()` for that, but you have to make sure it's only inserted during training.
What happens to our extreme overfitting case?
---
---
Problem 4
---------
Try to get the best performance you can using a multi-layer model! The best reported test accuracy using a deep network is [97.1%](http://yaroslavvb.blogspot.com/2011/09/notmnist-dataset.html?showComment=1391023266211#c8758720086795711595).
One avenue you can explore is to add multiple layers.
Another one is to use learning rate decay:
global_step = tf.Variable(0) # count the number of steps taken.
learning_rate = tf.train.exponential_decay(0.5, global_step, ...)
optimizer = tf.train.GradientDescentOptimizer(learning_rate).minimize(loss, global_step=global_step)
---
|
github_jupyter
|
# These are all the modules we'll be using later. Make sure you can import them
# before proceeding further.
from __future__ import print_function
import numpy as np
import tensorflow as tf
from six.moves import cPickle as pickle
pickle_file = 'notMNIST.pickle'
with open(pickle_file, 'rb') as f:
save = pickle.load(f)
train_dataset = save['train_dataset']
train_labels = save['train_labels']
valid_dataset = save['valid_dataset']
valid_labels = save['valid_labels']
test_dataset = save['test_dataset']
test_labels = save['test_labels']
del save # hint to help gc free up memory
print('Training set', train_dataset.shape, train_labels.shape)
print('Validation set', valid_dataset.shape, valid_labels.shape)
print('Test set', test_dataset.shape, test_labels.shape)
image_size = 28
num_labels = 10
def reformat(dataset, labels):
dataset = dataset.reshape((-1, image_size * image_size)).astype(np.float32)
# Map 2 to [0.0, 1.0, 0.0 ...], 3 to [0.0, 0.0, 1.0 ...]
labels = (np.arange(num_labels) == labels[:,None]).astype(np.float32)
return dataset, labels
train_dataset, train_labels = reformat(train_dataset, train_labels)
valid_dataset, valid_labels = reformat(valid_dataset, valid_labels)
test_dataset, test_labels = reformat(test_dataset, test_labels)
print('Training set', train_dataset.shape, train_labels.shape)
print('Validation set', valid_dataset.shape, valid_labels.shape)
print('Test set', test_dataset.shape, test_labels.shape)
def accuracy(predictions, labels):
return (100.0 * np.sum(np.argmax(predictions, 1) == np.argmax(labels, 1))
/ predictions.shape[0])
| 0.637595 | 0.966663 |
```
from sklearn import datasets
breast_cancer = datasets.load_breast_cancer()
breast_data = breast_cancer.data
breast_labels = breast_cancer.target
print(breast_data.shape)
print(breast_labels.shape)
import numpy as np
labels = np.reshape(breast_labels,(569,1))
final_breast_data = np.concatenate([breast_data,labels],axis=1)
final_breast_data.shape
import pandas as pd
breast_dataset = pd.DataFrame(final_breast_data)
features = breast_cancer.feature_names
features
final_breast_data[0:5]
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(breast_data,
breast_labels, random_state=42)
print(X_train.shape, X_test.shape)
"""Preprocessing: Principal Component Analysis
-------------------------------------------
We can use PCA to reduce these features to a manageable size, while maintaining most of the information
in the dataset.
"""
from sklearn import decomposition
pca = decomposition.PCA(n_components=20, whiten=True)
pca.fit(X_train)
"""The principal components measure deviations about this mean along
orthogonal axes.
"""
print(pca.components_.shape)
"""With this projection computed, we can now project our original training
and test data onto the PCA basis:
"""
X_train_pca = pca.transform(X_train)
X_test_pca = pca.transform(X_test)
print(X_train_pca.shape)
print(X_test_pca.shape)
"""
Doing the Learning: Support Vector Machines
-------------------------------------------
Now we'll perform support-vector-machine classification on this reduced
dataset:
"""
from sklearn import svm
clf = svm.SVC(C=5., gamma=0.001)
clf.fit(X_train_pca, y_train)
from sklearn import metrics
y_pred = clf.predict(X_test_pca)
print(metrics.classification_report(y_test, y_pred))
"""Another interesting metric is the *confusion matrix*, which indicates
how often any two items are mixed-up. The confusion matrix of a perfect
classifier would only have nonzero entries on the diagonal, with zeros
on the off-diagonal:
"""
print(metrics.confusion_matrix(y_test, y_pred))
"""With Iris Dataset"""
iris = datasets.load_iris()
iris_data = iris.data
iris_labels = iris.target
print(iris_data.shape)
print(iris_labels.shape)
features = iris.feature_names
features
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(iris_data,
iris_labels, random_state=26)
print(X_train.shape, X_test.shape)
"""Preprocessing: Principal Component Analysis
We can use PCA to reduce these features to a manageable size, while maintaining most of the information in the dataset.
"""
from sklearn import decomposition
pca = decomposition.PCA(n_components=2, whiten=True)
pca.fit(X_train)
print(pca.components_.shape)
X_train_pca = pca.transform(X_train)
X_test_pca = pca.transform(X_test)
print(X_train_pca.shape)
print(X_test_pca.shape)
from sklearn import svm
clf = svm.SVC(C=5., gamma=0.001)
clf.fit(X_train_pca, y_train)
from sklearn import metrics
y_pred = clf.predict(X_test_pca)
print(metrics.classification_report(y_test, y_pred))
print(metrics.confusion_matrix(y_test, y_pred))
```
|
github_jupyter
|
from sklearn import datasets
breast_cancer = datasets.load_breast_cancer()
breast_data = breast_cancer.data
breast_labels = breast_cancer.target
print(breast_data.shape)
print(breast_labels.shape)
import numpy as np
labels = np.reshape(breast_labels,(569,1))
final_breast_data = np.concatenate([breast_data,labels],axis=1)
final_breast_data.shape
import pandas as pd
breast_dataset = pd.DataFrame(final_breast_data)
features = breast_cancer.feature_names
features
final_breast_data[0:5]
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(breast_data,
breast_labels, random_state=42)
print(X_train.shape, X_test.shape)
"""Preprocessing: Principal Component Analysis
-------------------------------------------
We can use PCA to reduce these features to a manageable size, while maintaining most of the information
in the dataset.
"""
from sklearn import decomposition
pca = decomposition.PCA(n_components=20, whiten=True)
pca.fit(X_train)
"""The principal components measure deviations about this mean along
orthogonal axes.
"""
print(pca.components_.shape)
"""With this projection computed, we can now project our original training
and test data onto the PCA basis:
"""
X_train_pca = pca.transform(X_train)
X_test_pca = pca.transform(X_test)
print(X_train_pca.shape)
print(X_test_pca.shape)
"""
Doing the Learning: Support Vector Machines
-------------------------------------------
Now we'll perform support-vector-machine classification on this reduced
dataset:
"""
from sklearn import svm
clf = svm.SVC(C=5., gamma=0.001)
clf.fit(X_train_pca, y_train)
from sklearn import metrics
y_pred = clf.predict(X_test_pca)
print(metrics.classification_report(y_test, y_pred))
"""Another interesting metric is the *confusion matrix*, which indicates
how often any two items are mixed-up. The confusion matrix of a perfect
classifier would only have nonzero entries on the diagonal, with zeros
on the off-diagonal:
"""
print(metrics.confusion_matrix(y_test, y_pred))
"""With Iris Dataset"""
iris = datasets.load_iris()
iris_data = iris.data
iris_labels = iris.target
print(iris_data.shape)
print(iris_labels.shape)
features = iris.feature_names
features
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(iris_data,
iris_labels, random_state=26)
print(X_train.shape, X_test.shape)
"""Preprocessing: Principal Component Analysis
We can use PCA to reduce these features to a manageable size, while maintaining most of the information in the dataset.
"""
from sklearn import decomposition
pca = decomposition.PCA(n_components=2, whiten=True)
pca.fit(X_train)
print(pca.components_.shape)
X_train_pca = pca.transform(X_train)
X_test_pca = pca.transform(X_test)
print(X_train_pca.shape)
print(X_test_pca.shape)
from sklearn import svm
clf = svm.SVC(C=5., gamma=0.001)
clf.fit(X_train_pca, y_train)
from sklearn import metrics
y_pred = clf.predict(X_test_pca)
print(metrics.classification_report(y_test, y_pred))
print(metrics.confusion_matrix(y_test, y_pred))
| 0.853471 | 0.880077 |
```
# windows only hack for graphviz path
import os
for path in os.environ['PATH'].split(os.pathsep):
if path.endswith("Library\\bin"):
os.environ['PATH']+=os.pathsep+os.path.join(path, 'graphviz')
# 設定環境變數來控制 keras, theano
os.environ['KERAS_BACKEND']="tensorflow"
#os.environ['THEANO_FLAGS']="floatX=float32, device=cuda"
import keras
from keras.models import Sequential
from PIL import Image
import numpy as np
import keras.backend as K
# 設定 channels_first 或 channels_last
K.set_image_data_format('channels_last')
base_model = keras.applications.vgg16.VGG16(weights='imagenet', include_top=False, input_shape=(64,64,3))
from keras.layers import GlobalAveragePooling2D, Dense
from keras.models import Model
_ = base_model.get_layer("block5_conv3").output
_ = GlobalAveragePooling2D()(_)
_ = Dense(512, activation='relu')(_)
# 10 個輸出
predictions = Dense(10, activation='softmax')(_)
# 這是我們的 model
model = Model(inputs=base_model.input, outputs=predictions)
# 將原來的模型設定成不可訓練,只訓練上面的兩層 dense layers
for layer in base_model.layers:
layer.trainable = False
model.compile(optimizer='adam', loss='categorical_crossentropy', metrics=['accuracy'])
# 讀取 cifar10 dataset
# 只有 train 和 test 沒有 validation
import tarfile
import pickle
train_X=[]
train_y=[]
tar_gz = "../Week06/cifar-10-python.tar.gz"
with tarfile.open(tar_gz) as tarf:
for i in range(1, 6):
dataset = "cifar-10-batches-py/data_batch_%d"%i
print("load",dataset)
with tarf.extractfile(dataset) as f:
result = pickle.load(f, encoding='latin1')
train_X.extend(result['data'])
train_y.extend(result['labels'])
train_X=np.float32(train_X)
train_y=np.int32(train_y)
dataset = "cifar-10-batches-py/test_batch"
print("load",dataset)
with tarf.extractfile(dataset) as f:
result = pickle.load(f, encoding='latin1')
test_X=np.float32(result['data'])
test_y=np.int32(result['labels'])
train_Y = np.eye(10)[train_y]
test_Y = np.eye(10)[test_y]
train_X = np.moveaxis(train_X.reshape(-1,3,32,32),1,3)
test_X = np.moveaxis(test_X.reshape(-1,3,32,32),1,3)
validation_data = (test_X[:1000], test_Y[:1000])
test_data = (test_X[1000:], test_Y[1000:])
# 改變輸入圖形的大小並且處理
from keras.applications.vgg16 import preprocess_input
def resize_image(arr):
img = Image.fromarray(arr)
img = img.resize((64,64))
img = img.convert("RGB")
return np.array(img)
def preprocess_X(X):
X = X.astype('uint8')
X = np.float32([resize_image(X[i]) for i in range(X.shape[0])])
return preprocess_input(X)
from PIL import Image
from IPython.display import display
def showX(X):
int_X = X.clip(0,255).astype('uint8')
int_X_reshape = int_X.swapaxes(0,1).reshape(32,-1,3)
display(Image.fromarray(int_X_reshape))
# 訓練資料, X 的前 20 筆
showX(train_X[:20])
import numpy as np
def generate(X, Y):
while 1:
idx = np.random.choice(X.shape[0], size=32, replace=False)
_X = preprocess_X(X[idx])
_Y = Y[idx]
yield (_X, _Y)
v_data = (preprocess_X(test_X[:200]), test_Y[:200])
model.fit_generator(generate(train_X, train_Y), steps_per_epoch=500, epochs=10, validation_data=v_data)
# 實際
predict_y = (model.predict(preprocess_X(test_X))).argmax(axis=1)
test_y = test_Y.argmax(axis=1)
(predict_y == test_y).mean()
print(predict_y[:30])
showX(test_X[:30])
```
### 往前微調幾層
```
for n in ["block5_conv1", "block5_conv2", "block5_conv3"]:
model.get_layer(n).trainable = True
from keras.optimizers import Adam
model.compile(optimizer=Adam(lr=0.0001), loss='categorical_crossentropy', metrics=["accuracy"])
model.fit_generator(generate(train_X, train_Y), steps_per_epoch=500, epochs=10, validation_data=v_data)
predictions = (model.predict(preprocess_X(test_X))).argmax(axis=1)
(predictions == test_y).mean()
```
|
github_jupyter
|
# windows only hack for graphviz path
import os
for path in os.environ['PATH'].split(os.pathsep):
if path.endswith("Library\\bin"):
os.environ['PATH']+=os.pathsep+os.path.join(path, 'graphviz')
# 設定環境變數來控制 keras, theano
os.environ['KERAS_BACKEND']="tensorflow"
#os.environ['THEANO_FLAGS']="floatX=float32, device=cuda"
import keras
from keras.models import Sequential
from PIL import Image
import numpy as np
import keras.backend as K
# 設定 channels_first 或 channels_last
K.set_image_data_format('channels_last')
base_model = keras.applications.vgg16.VGG16(weights='imagenet', include_top=False, input_shape=(64,64,3))
from keras.layers import GlobalAveragePooling2D, Dense
from keras.models import Model
_ = base_model.get_layer("block5_conv3").output
_ = GlobalAveragePooling2D()(_)
_ = Dense(512, activation='relu')(_)
# 10 個輸出
predictions = Dense(10, activation='softmax')(_)
# 這是我們的 model
model = Model(inputs=base_model.input, outputs=predictions)
# 將原來的模型設定成不可訓練,只訓練上面的兩層 dense layers
for layer in base_model.layers:
layer.trainable = False
model.compile(optimizer='adam', loss='categorical_crossentropy', metrics=['accuracy'])
# 讀取 cifar10 dataset
# 只有 train 和 test 沒有 validation
import tarfile
import pickle
train_X=[]
train_y=[]
tar_gz = "../Week06/cifar-10-python.tar.gz"
with tarfile.open(tar_gz) as tarf:
for i in range(1, 6):
dataset = "cifar-10-batches-py/data_batch_%d"%i
print("load",dataset)
with tarf.extractfile(dataset) as f:
result = pickle.load(f, encoding='latin1')
train_X.extend(result['data'])
train_y.extend(result['labels'])
train_X=np.float32(train_X)
train_y=np.int32(train_y)
dataset = "cifar-10-batches-py/test_batch"
print("load",dataset)
with tarf.extractfile(dataset) as f:
result = pickle.load(f, encoding='latin1')
test_X=np.float32(result['data'])
test_y=np.int32(result['labels'])
train_Y = np.eye(10)[train_y]
test_Y = np.eye(10)[test_y]
train_X = np.moveaxis(train_X.reshape(-1,3,32,32),1,3)
test_X = np.moveaxis(test_X.reshape(-1,3,32,32),1,3)
validation_data = (test_X[:1000], test_Y[:1000])
test_data = (test_X[1000:], test_Y[1000:])
# 改變輸入圖形的大小並且處理
from keras.applications.vgg16 import preprocess_input
def resize_image(arr):
img = Image.fromarray(arr)
img = img.resize((64,64))
img = img.convert("RGB")
return np.array(img)
def preprocess_X(X):
X = X.astype('uint8')
X = np.float32([resize_image(X[i]) for i in range(X.shape[0])])
return preprocess_input(X)
from PIL import Image
from IPython.display import display
def showX(X):
int_X = X.clip(0,255).astype('uint8')
int_X_reshape = int_X.swapaxes(0,1).reshape(32,-1,3)
display(Image.fromarray(int_X_reshape))
# 訓練資料, X 的前 20 筆
showX(train_X[:20])
import numpy as np
def generate(X, Y):
while 1:
idx = np.random.choice(X.shape[0], size=32, replace=False)
_X = preprocess_X(X[idx])
_Y = Y[idx]
yield (_X, _Y)
v_data = (preprocess_X(test_X[:200]), test_Y[:200])
model.fit_generator(generate(train_X, train_Y), steps_per_epoch=500, epochs=10, validation_data=v_data)
# 實際
predict_y = (model.predict(preprocess_X(test_X))).argmax(axis=1)
test_y = test_Y.argmax(axis=1)
(predict_y == test_y).mean()
print(predict_y[:30])
showX(test_X[:30])
for n in ["block5_conv1", "block5_conv2", "block5_conv3"]:
model.get_layer(n).trainable = True
from keras.optimizers import Adam
model.compile(optimizer=Adam(lr=0.0001), loss='categorical_crossentropy', metrics=["accuracy"])
model.fit_generator(generate(train_X, train_Y), steps_per_epoch=500, epochs=10, validation_data=v_data)
predictions = (model.predict(preprocess_X(test_X))).argmax(axis=1)
(predictions == test_y).mean()
| 0.441914 | 0.233488 |
# Calculate Gene mC Fractions
```
import pandas as pd
import scanpy as sc
import anndata
import xarray as xr
import pybedtools
import dask
from ALLCools.plot import *
from ALLCools.mcds import MCDS
import pathlib
import numpy as np
gene_meta_path = '../../data/genome/gencode.vM22.annotation.gene.flat.tsv.gz'
chrom_to_remove = ['chrM']
# change this to the path to your filtered metadata
metadata_path = '../step_by_step/100kb/CellMetadata.PassQC.csv.gz'
# change this to the paths to your MCDS files
mcds_path_list = [
'../../data/Brain/3C-171206.mcds',
'../../data/Brain/3C-171207.mcds',
'../../data/Brain/9H-190212.mcds',
'../../data/Brain/9H-190219.mcds',
]
obs_dim = 'cell'
var_dim = 'gene'
min_cov = 5
```
## Load metadata
```
gene_meta = pd.read_csv(gene_meta_path, index_col='gene_id', sep='\t')
metadata = pd.read_csv(metadata_path, index_col=0)
total_cells = metadata.shape[0]
print(f'Metadata of {total_cells} cells')
```
## Filter genes by overlap and chromosomes
```
genes_to_skip = set()
# skip smaller genes mostly covered by a larger gene, e.g., a miRNA within a protein coding gene.
# F=0.9 means > 90% of gene_b is overlapped with gene_a, in this case, we only keep gene_a for DMG test
gene_bed = pybedtools.BedTool.from_dataframe(
gene_meta.reset_index()[['chrom', 'start', 'end', 'gene_id']])
mapped_bam = gene_bed.map(b=gene_bed, c=4, o='distinct', F=0.9)
for _, (*_, gene_a, gene_b_str) in mapped_bam.to_dataframe().iterrows():
for gene_b in gene_b_str.split(','):
if gene_b != gene_a:
genes_to_skip.add(gene_b)
# remove certain chromosomes
genes_to_skip |= set(gene_meta.index[gene_meta['chrom'].isin(chrom_to_remove)])
use_features = gene_meta.index[~gene_meta.index.isin(genes_to_skip)]
print(f'{use_features.size} features remained')
```
## Filter genes by cell mean coverage
```
with dask.config.set(**{'array.slicing.split_large_chunks': False}):
# still use all the cells to load MCDS
mcds = MCDS.open(mcds_path_list, obs_dim=obs_dim,
use_obs=metadata.index).sel({var_dim: use_features})
mcds.add_feature_cov_mean(var_dim=var_dim)
feature_cov_mean = mcds.coords[f'{var_dim}_cov_mean'].to_pandas()
use_features &= feature_cov_mean[feature_cov_mean > min_cov].index
print(f'{use_features.size} features remained')
mcds.filter_feature_by_cov_mean(var_dim, min_cov=min_cov)
```
## Add Gene mC Fraction per MCDS file
```
gene_frac_dir = pathlib.Path('gene_frac')
gene_frac_dir.mkdir(exist_ok=True)
for mcds_path in mcds_path_list:
output_path = gene_frac_dir / (pathlib.Path(mcds_path).name + f'{var_dim}_da_frac.mcds')
if output_path.exists():
continue
print(f'Computaing gene mC fraction for {mcds_path}')
mcds = MCDS.open(mcds_path, obs_dim=obs_dim)
# remove non-related data
del_das = []
for da in mcds:
if da != f'{var_dim}_da':
del_das.append(da)
for da in del_das:
del mcds[da]
mcds.load()
mcds = mcds.sel({var_dim: use_features})
mcds.add_mc_rate(var_dim=var_dim, normalize_per_cell=True, clip_norm_value=10)
# use float32 to reduce file size and speedup IO
mcds = mcds.rename({var_dim: 'gene', f'{var_dim}_da_frac': 'gene_da_frac'})
mcds['gene_da_frac'].astype('float32').to_netcdf(output_path)
```
## Save gene metadata together with gene fraction files
```
use_gene_meta = gene_meta.loc[use_features]
use_gene_meta.to_csv(gene_frac_dir / 'GeneMetadata.csv.gz')
```
|
github_jupyter
|
import pandas as pd
import scanpy as sc
import anndata
import xarray as xr
import pybedtools
import dask
from ALLCools.plot import *
from ALLCools.mcds import MCDS
import pathlib
import numpy as np
gene_meta_path = '../../data/genome/gencode.vM22.annotation.gene.flat.tsv.gz'
chrom_to_remove = ['chrM']
# change this to the path to your filtered metadata
metadata_path = '../step_by_step/100kb/CellMetadata.PassQC.csv.gz'
# change this to the paths to your MCDS files
mcds_path_list = [
'../../data/Brain/3C-171206.mcds',
'../../data/Brain/3C-171207.mcds',
'../../data/Brain/9H-190212.mcds',
'../../data/Brain/9H-190219.mcds',
]
obs_dim = 'cell'
var_dim = 'gene'
min_cov = 5
gene_meta = pd.read_csv(gene_meta_path, index_col='gene_id', sep='\t')
metadata = pd.read_csv(metadata_path, index_col=0)
total_cells = metadata.shape[0]
print(f'Metadata of {total_cells} cells')
genes_to_skip = set()
# skip smaller genes mostly covered by a larger gene, e.g., a miRNA within a protein coding gene.
# F=0.9 means > 90% of gene_b is overlapped with gene_a, in this case, we only keep gene_a for DMG test
gene_bed = pybedtools.BedTool.from_dataframe(
gene_meta.reset_index()[['chrom', 'start', 'end', 'gene_id']])
mapped_bam = gene_bed.map(b=gene_bed, c=4, o='distinct', F=0.9)
for _, (*_, gene_a, gene_b_str) in mapped_bam.to_dataframe().iterrows():
for gene_b in gene_b_str.split(','):
if gene_b != gene_a:
genes_to_skip.add(gene_b)
# remove certain chromosomes
genes_to_skip |= set(gene_meta.index[gene_meta['chrom'].isin(chrom_to_remove)])
use_features = gene_meta.index[~gene_meta.index.isin(genes_to_skip)]
print(f'{use_features.size} features remained')
with dask.config.set(**{'array.slicing.split_large_chunks': False}):
# still use all the cells to load MCDS
mcds = MCDS.open(mcds_path_list, obs_dim=obs_dim,
use_obs=metadata.index).sel({var_dim: use_features})
mcds.add_feature_cov_mean(var_dim=var_dim)
feature_cov_mean = mcds.coords[f'{var_dim}_cov_mean'].to_pandas()
use_features &= feature_cov_mean[feature_cov_mean > min_cov].index
print(f'{use_features.size} features remained')
mcds.filter_feature_by_cov_mean(var_dim, min_cov=min_cov)
gene_frac_dir = pathlib.Path('gene_frac')
gene_frac_dir.mkdir(exist_ok=True)
for mcds_path in mcds_path_list:
output_path = gene_frac_dir / (pathlib.Path(mcds_path).name + f'{var_dim}_da_frac.mcds')
if output_path.exists():
continue
print(f'Computaing gene mC fraction for {mcds_path}')
mcds = MCDS.open(mcds_path, obs_dim=obs_dim)
# remove non-related data
del_das = []
for da in mcds:
if da != f'{var_dim}_da':
del_das.append(da)
for da in del_das:
del mcds[da]
mcds.load()
mcds = mcds.sel({var_dim: use_features})
mcds.add_mc_rate(var_dim=var_dim, normalize_per_cell=True, clip_norm_value=10)
# use float32 to reduce file size and speedup IO
mcds = mcds.rename({var_dim: 'gene', f'{var_dim}_da_frac': 'gene_da_frac'})
mcds['gene_da_frac'].astype('float32').to_netcdf(output_path)
use_gene_meta = gene_meta.loc[use_features]
use_gene_meta.to_csv(gene_frac_dir / 'GeneMetadata.csv.gz')
| 0.338514 | 0.754486 |
```
import requests
from bs4 import BeautifulSoup
from multiprocessing.dummy import Pool
import time
import shutil
import re
import os
import json
import string
import glob
BASE_URL = 'https://www.pro-football-reference.com{0}'
PLAYER_LIST_URL = 'https://www.pro-football-reference.com/players/{0}'
PLAYER_PROFILE_URL = 'https://www.pro-football-reference.com/players/{0}/{1}'
PLAYER_GAMELOG_URL = 'https://www.pro-football-reference.com/players/{0}/{1}/gamelog/{2}'
HEADERS = {
'user-agent': ('Mozilla/5.0 (Windows NT 6.1; WOW64) AppleWebKit/537.36 '
'(KHTML, like Gecko) Chrome/48.0.2564.109 Safari/537.36')
}
PROFILE_DIR = 'profile_data'
STATS_DIR = 'stats_data'
class Scraper():
"""Scraper for pro-football-reference.com to collect NFL player stats"""
def __init__(self, letters_to_scrape=['Z'], num_jobs=1, clear_old_data=True, first_player_id=1):
"""Initialize the scraper to get player stats
Args:
- letters_to_scrape (str[]): The site sorts players by the first letter of their
last name. This array tells the scraper which letters to scrape data for.
- num_jobs (int): Number of concurrent jobs the scraper should run. While Python
can't multi-thread, it can manage multiple processes at once, which allows it to
utilize time spent waiting for the server to respond.
- clear_old_data (boolean): Whether or not the data file should be wiped before
starting the scrape.
- first_player_id (int): The first ID for a player (set if you are rerunning to avoid duplicates)
Returns:
None
"""
self.letters_to_scrape = [letter.upper() for letter in letters_to_scrape]
self.num_jobs = num_jobs
self.clear_old_data = clear_old_data
self.session = requests.Session()
self.start_time = time.time()
self.cross_process_player_count = 0
self.first_player_id = first_player_id
if num_jobs > 1:
self.multiprocessing = True
self.worker_pool = Pool(num_jobs)
else:
self.multiprocessing = False
def scrape_site(self):
"""Pool workers to scrape players by first letter of last name"""
if self.clear_old_data:
self.clear_data()
player_id = self.first_player_id
for letter in self.letters_to_scrape:
player_profile_urls = self.get_players_for_letter(letter)
for player_profile_url in player_profile_urls:
player = Player(player_id, player_profile_url, self)
try:
player.scrape_profile()
player.scrape_player_stats()
except (KeyboardInterrupt, SystemExit):
raise
except:
print('There was a problem parsing stats for {}'.format(player_profile_url))
continue
self.save_player_profile(player.profile)
self.save_player_game_stats(player.game_stats, player.player_id, player.profile['name'])
player_id += 1
self.condense_data()
def condense_data(self):
"""Condense data into two files, a profile file and a stats file"""
print('Condensing Data...')
condensed_profile_data = []
all_profile_files = glob.glob('{}/*.json'.format(PROFILE_DIR))
for file in all_profile_files:
with open(file, 'rb') as fin:
condensed_profile_data.append(json.load(fin))
print('{} player profiles condensed'.format(len(condensed_profile_data)))
filename = 'profiles_{}.json'.format(time.time())
with open(filename, 'w') as fout:
json.dump(condensed_profile_data, fout)
condensed_game_data = []
all_game_files = glob.glob('{}/*.json'.format(STATS_DIR))
for file in all_game_files:
with open(file, 'rb') as fin:
condensed_game_data += json.load(fin)
print('{} player seasons condensed'.format(len(condensed_game_data)))
filename = 'games_{}.json'.format(time.time())
with open(filename, 'w') as fout:
json.dump(condensed_game_data, fout)
def save_player_profile(self, profile):
"""Save a player's profile as JSON
Args:
- profile (dict): Player profile data
Return:
None
"""
filename = '{}/{}_{}.json'.format(PROFILE_DIR, profile['player_id'], profile['name'].replace(' ', '-'))
try:
os.makedirs(PROFILE_DIR)
except OSError:
pass
with open(filename, 'w') as fout:
json.dump(profile, fout)
def save_player_game_stats(self, games, player_id, player_name):
"""Save a list of player games with stats info
Args:
- games (dict[]): List of game stats
- player_id (int): ID of the player the games belong to
- player_name (str): Name of the player the game stats belong to
Return:
None
"""
filename = '{}/{}_{}.json'.format(STATS_DIR, player_id, player_name.replace(' ', '-'))
try:
os.makedirs(STATS_DIR)
except OSError:
pass
with open(filename, 'w') as fout:
json.dump(games, fout)
def get_players_for_letter(self, letter):
"""Get a list of player links for a letter of the alphabet.
Site organizes players by first letter of last name.
Args:
- letter (str): letter of the alphabet uppercased
Returns:
- player_links (str[]): the URLs to get player profiles
"""
response = self.get_page(PLAYER_LIST_URL.format(letter))
soup = BeautifulSoup(response.content, 'html.parser')
players = soup.find('div', {'id': 'div_players'}).find_all('a')
return [BASE_URL.format(player['href']) for player in players]
def get_page(self, url, retry_count=0):
"""Use requests to get a page; retry when failures occur
Args:
- url (str): The URL of the page to make a GET request to
- retry_count (int): Number of times the URL has already been requests
Returns:
- response (obj): The Requests response object
"""
try:
return self.session.get(url, headers=HEADERS)
except (KeyboardInterrupt, SystemExit):
raise
except:
retry_count += 1
if retry_count <= 3:
self.session = requests.Session()
return self.get_page(url, retry_count)
else:
raise
def clear_data(self):
"""Clear the data directories"""
try:
shutil.rmtree(PROFILE_DIR)
except FileNotFoundError:
pass
try:
shutil.rmtree(STATS_DIR)
except FileNotFoundError:
pass
class Player():
"""An NFL player"""
def __init__(self, player_id, profile_url, scraper):
"""
Args:
- player_id (int): Unique ID for player
- profile_url (str): URL to the player's profile
- scraper (obj): instance of Scraper class
Returns:
None
"""
self.player_id = player_id
self.profile_url = profile_url
self.scraper = scraper
self.profile = {
'player_id': player_id,
'name': None,
'position': None,
'height': None,
'weight': None,
'current_team': None,
'birth_date': None,
'birth_place': None,
'death_date': None,
'college': None,
'high_school': None,
'draft_team': None,
'draft_round': None,
'draft_position': None,
'draft_year': None,
'current_salary': None,
'hof_induction_year': None
}
self.seasons_with_stats = []
self.game_stats = []
def scrape_profile(self):
"""Scrape profile info for player"""
response = self.scraper.get_page(self.profile_url)
soup = BeautifulSoup(response.content, 'html.parser')
profile_section = soup.find('h1', {'itemprop': 'name'}).contents
self.profile['name'] = profile_section.results[1].text
print('scaping {}'.format(self.profile['name']))
profile_attributes = profile_section.find_all('p')
current_attribute = 1
num_attributes = len(profile_attributes)
self.profile['position'] = profile_attributes[current_attribute].contents[2].split('\n')[0].split(' ')[1]
current_attribute += 1
height = profile_attributes[current_attribute].find('span', {'itemprop': 'height'})
if height is not None:
self.profile['height'] = height.contents[0]
weight = profile_attributes[current_attribute].find('span', {'itemprop': 'weight'})
if weight is not None:
self.profile['weight'] = weight.contents[0].split('lb')[0]
if height is not None or weight is not None:
current_attribute += 1
affiliation_section = profile_section.find('span', {'itemprop': 'affiliation'})
if affiliation_section is not None:
self.profile['current_team'] = affiliation_section.contents[0].contents[0]
current_attribute += 1
birth_date = profile_attributes[current_attribute].find('span', {'itemprop': 'birthDate'})
if birth_date is not None:
self.profile['birth_date'] = birth_date['data-birth']
birth_place_section = profile_attributes[current_attribute].find('span', {'itemprop': 'birthPlace'}).contents
try:
self.profile['birth_place'] = re.split('\xa0', birth_place_section[0])[1] + ' ' + birth_place_section[1].contents[0]
except IndexError:
pass
if birth_date is not None or len(birth_place_section) > 0:
current_attribute += 1
death_section = profile_section.find('span', {'itemprop': 'deathDate'})
if death_section is not None:
self.profile['death_date'] = death_section['data-death']
current_attribute += 1
if profile_attributes[current_attribute].contents[0].contents[0] == 'College':
self.profile['college'] = profile_attributes[current_attribute].contents[2].contents[0]
current_attribute += 1
# Skip weighted career AV
current_attribute += 1
if ((current_attribute + 1) <= num_attributes) and profile_attributes[current_attribute].contents[0].contents[0] == 'High School':
self.profile['high_school'] = profile_attributes[current_attribute].contents[2].contents[0] + ', ' + profile_attributes[current_attribute].contents[4].contents[0]
current_attribute += 1
if ((current_attribute + 1) <= num_attributes) and profile_attributes[current_attribute].contents[0].contents[0] == 'Draft':
self.profile['draft_team'] = profile_attributes[current_attribute].contents[2].contents[0]
draft_info = profile_attributes[current_attribute].contents[3].split(' ')
self.profile['draft_round'] = re.findall(r'\d+', draft_info[3])[0]
self.profile['draft_position'] = re.findall(r'\d+', draft_info[5])[0]
self.profile['draft_year'] = re.findall(r'\d+', profile_attributes[current_attribute].contents[4].contents[0])[0]
current_attribute += 1
if ((current_attribute + 1) <= num_attributes) and profile_attributes[current_attribute].contents[0].contents[0] == 'Current cap hit':
profile_attributes[current_attribute].contents
self.profile['current_salary'] = profile_attributes[current_attribute].contents[2].contents[0]
current_attribute += 1
if ((current_attribute + 1) <= num_attributes) and profile_attributes[current_attribute].contents[0].contents[0] == 'Hall of fame':
self.profile['hof_induction_year'] = profile_attributes[current_attribute].contents[2].contents[0]
current_attribute += 1
self.seasons_with_stats = self.get_seasons_with_stats(soup)
def scrape_player_stats(self):
"""Scrape the stats for all available games for a player"""
for season in self.seasons_with_stats:
if season['year'] == 'Career' or season['year'] == 'Postseason':
continue
self.scrape_season_gamelog(season['gamelog_url'], season['year'])
def scrape_season_gamelog(self, gamelog_url, year):
"""Scrape player stats for a given year
Args:
- gamelog_url (str): URL to the stats for a given year
- year (int): The year the stats are for
Returns:
- stats (dict): All of the player's stats for that year
"""
response = self.scraper.get_page(gamelog_url)
soup = BeautifulSoup(response.content, 'html.parser')
regular_season_table = soup.find('table', {'id': 'stats'})
if regular_season_table is None:
return False
games = regular_season_table.find('tbody').find_all('tr')
playoff_table = soup.find('table', {'id': 'stats_playoffs'})
if playoff_table is not None:
games += playoff_table.find('tbody').find_all('tr')
for game in games:
stats = self.make_player_game_stats(self.player_id, year)
stats['game_id'] = game.find('td', {'data-stat': 'game_date'}).find('a', href=True)['href'].replace('/boxscores/', '').replace('.htm', '')
stats['date'] = game.find('td', {'data-stat': 'game_date'}).contents[0].contents[0]
stats['game_number'] = game.find('td', {'data-stat': 'game_num'}).contents[0]
stats['age'] = game.find('td', {'data-stat': 'age'}).contents[0]
stats['team'] = game.find('td', {'data-stat': 'team'}).contents[0].contents[0]
if game.find('td', {'data-stat': 'game_location'}).contents == ['@']:
stats['game_location'] = 'A'
elif game.find('td', {'data-stat': 'game_location'}).contents == ['N']:
stats['game_location'] = 'N'
else:
stats['game_location'] = 'H'
stats['opponent'] = game.find('td', {'data-stat': 'opp'}).contents[0].contents[0]
result = game.find('td', {'data-stat': 'game_result'}).contents[0].contents[0]
stats['game_won'] = (result.split(' ')[0] == 'W')
stats['player_team_score'] = result.split(' ')[1].split('-')[0]
stats['opponent_score'] = result.split(' ')[1].split('-')[1]
# Collect passing stats
pass_attempts = game.find('td', {'data-stat': 'pass_cmp'})
if pass_attempts is not None and len(pass_attempts) > 0:
stats['passing_attempts'] = int(pass_attempts.contents[0])
pass_completions = game.find('td', {'data-stat': 'pass_att'})
if pass_completions is not None and len(pass_completions) > 0:
stats['passing_completions'] = int(pass_completions.contents[0])
pass_yards = game.find('td', {'data-stat': 'pass_yds'})
if pass_yards is not None and len(pass_yards) > 0:
stats['passing_yards'] = int(pass_yards.contents[0])
pass_touchdowns = game.find('td', {'data-stat': 'pass_td'})
if pass_touchdowns is not None and len(pass_touchdowns) > 0:
stats['passing_touchdowns'] = int(pass_touchdowns.contents[0])
pass_interceptions = game.find('td', {'data-stat': 'pass_int'})
if pass_interceptions is not None and len(pass_interceptions) > 0:
stats['passing_interceptions'] = int(pass_interceptions.contents[0])
pass_rating = game.find('td', {'data-stat': 'pass_rating'})
if pass_rating is not None and len(pass_rating) > 0:
stats['passing_rating'] = float(pass_rating.contents[0])
pass_sacks = game.find('td', {'data-stat': 'pass_sacked'})
if pass_sacks is not None and len(pass_sacks) > 0:
stats['passing_sacks'] = int(pass_sacks.contents[0])
pass_sacks_yards_lost = game.find('td', {'data-stat': 'pass_sacked_yds'})
if pass_sacks_yards_lost is not None and len(pass_sacks_yards_lost) > 0:
stats['passing_sacks_yards_lost'] = int(pass_sacks_yards_lost.contents[0])
# Collect rushing stats
rushing_attempts = game.find('td', {'data-stat': 'rush_att'})
if rushing_attempts is not None and len(rushing_attempts) > 0:
stats['rushing_attempts'] = int(rushing_attempts.contents[0])
rushing_yards = game.find('td', {'data-stat': 'rush_yds'})
if rushing_yards is not None and len(rushing_yards) > 0:
stats['rushing_yards'] = int(rushing_yards.contents[0])
rushing_touchdowns = game.find('td', {'data-stat': 'rush_td'})
if rushing_touchdowns is not None and len(rushing_touchdowns) > 0:
stats['rushing_touchdowns'] = int(rushing_touchdowns.contents[0])
# Collect receiving stats
receiving_targets = game.find('td', {'data-stat': 'targets'})
if receiving_targets is not None and len(receiving_targets) > 0:
stats['receiving_targets'] = int(receiving_targets.contents[0])
receiving_receptions = game.find('td', {'data-stat': 'rec'})
if receiving_receptions is not None and len(receiving_receptions) > 0:
stats['receiving_receptions'] = int(receiving_receptions.contents[0])
receiving_yards = game.find('td', {'data-stat': 'rec_yds'})
if receiving_yards is not None and len(receiving_yards) > 0:
stats['receiving_yards'] = int(receiving_yards.contents[0])
receiving_touchdowns = game.find('td', {'data-stat': 'rec_td'})
if receiving_touchdowns is not None and len(receiving_touchdowns) > 0:
stats['receiving_touchdowns'] = int(receiving_touchdowns.contents[0])
# Collect kick return stats
kick_return_attempts = game.find('td', {'data-stat': 'kick_ret'})
if kick_return_attempts is not None and len(kick_return_attempts) > 0:
stats['kick_return_attempts'] = int(kick_return_attempts.contents[0])
kick_return_yards = game.find('td', {'data-stat': 'kick_ret_yds'})
if kick_return_yards is not None and len(kick_return_yards) > 0:
stats['kick_return_yards'] = int(kick_return_yards.contents[0])
kick_return_touchdowns = game.find('td', {'data-stat': 'kick_ret_td'})
if kick_return_touchdowns is not None and len(kick_return_touchdowns) > 0:
stats['kick_return_touchdowns'] = int(kick_return_touchdowns.contents[0])
# Collect punt return stats
punt_return_attempts = game.find('td', {'data-stat': 'punt_ret'})
if punt_return_attempts is not None and len(punt_return_attempts) > 0:
stats['punt_return_attempts'] = int(punt_return_attempts.contents[0])
punt_return_yards = game.find('td', {'data-stat': 'punt_ret_yds'})
if punt_return_yards is not None and len(punt_return_yards) > 0:
stats['punt_return_yards'] = int(punt_return_yards.contents[0])
punt_return_touchdowns = game.find('td', {'data-stat': 'punt_ret_td'})
if punt_return_touchdowns is not None and len(punt_return_touchdowns) > 0:
stats['punt_return_touchdowns'] = int(punt_return_touchdowns.contents[0])
# Collect defensive stats
defense_sacks = game.find('td', {'data-stat': 'sacks'})
if defense_sacks is not None and len(defense_sacks) > 0:
stats['defense_sacks'] = float(defense_sacks.contents[0])
defense_tackles = game.find('td', {'data-stat': 'tackles_solo'})
if defense_tackles is not None and len(defense_tackles) > 0:
stats['defense_tackles'] = int(defense_tackles.contents[0])
defense_tackle_assists = game.find('td', {'data-stat': 'tackles_assists'})
if defense_tackle_assists is not None and len(defense_tackle_assists) > 0:
stats['defense_tackle_assists'] = int(defense_tackle_assists.contents[0])
defense_interceptions = game.find('td', {'data-stat': 'def_int'})
if defense_interceptions is not None and len(defense_interceptions) > 0:
stats['defense_interceptions'] = int(defense_interceptions.contents[0])
defense_interception_yards = game.find('td', {'data-stat': 'def_int_yds'})
if defense_interception_yards is not None and len(defense_interception_yards) > 0:
stats['defense_interception_yards'] = int(defense_interception_yards.contents[0])
defense_safeties = game.find('td', {'data-stat': 'safety_md'})
if defense_safeties is not None and len(defense_safeties) > 0:
stats['defense_safeties'] = int(defense_safeties.contents[0])
# Collect kicking stats
point_after_attemps = game.find('td', {'data-stat': 'xpm'})
if point_after_attemps is not None and len(point_after_attemps) > 0:
stats['point_after_attemps'] = int(point_after_attemps.contents[0])
point_after_makes = game.find('td', {'data-stat': 'xpa'})
if point_after_makes is not None and len(point_after_makes) > 0:
stats['point_after_makes'] = int(point_after_makes.contents[0])
field_goal_attempts = game.find('td', {'data-stat': 'fga'})
if field_goal_attempts is not None and len(field_goal_attempts) > 0:
stats['field_goal_attempts'] = int(field_goal_attempts.contents[0])
field_goal_makes = game.find('td', {'data-stat': 'fgm'})
if field_goal_makes is not None and len(field_goal_makes) > 0:
stats['field_goal_makes'] = int(field_goal_makes.contents[0])
# Collect punting stats
punting_attempts = game.find('td', {'data-stat': 'punt'})
if punting_attempts is not None and len(punting_attempts) > 0:
stats['punting_attempts'] = int(punting_attempts.contents[0])
punting_yards = game.find('td', {'data-stat': 'punt_yds'})
if punting_yards is not None and len(punting_yards) > 0:
stats['punting_yards'] = int(punting_yards.contents[0])
punting_blocked = game.find('td', {'data-stat': 'punt_blocked'})
if punting_blocked is not None and len(punting_blocked) > 0:
stats['punting_blocked'] = int(punting_blocked.contents[0])
self.game_stats.append(stats)
@staticmethod
def make_player_game_stats(player_id, year):
"""Factory method to return possible stats to collect for a player in a game
Args:
- player_id (int): unique Id for the player
- year (int): The year the stats are for
Returns:
- game_stats (dict): dictionary with game stats initialized
"""
return {
'player_id': player_id,
'year': year,
# General stats
'game_id': None,
'date': None,
'game_number': None,
'age': None,
'team': None,
'game_location': None,
'opponent': None,
'game_won': None,
'player_team_score': 0,
'opponent_score': 0,
# Passing stats
'passing_attempts': 0,
'passing_completions': 0,
'passing_yards': 0,
'passing_rating': 0,
'passing_touchdowns': 0,
'passing_interceptions': 0,
'passing_sacks': 0,
'passing_sacks_yards_lost': 0,
# Rushing stats
'rushing_attempts': 0,
'rushing_yards': 0,
'rushing_touchdowns': 0,
# Receiving stats
'receiving_targets': 0,
'receiving_receptions': 0,
'receiving_yards': 0,
'receiving_touchdowns': 0,
# Kick return stats
'kick_return_attempts': 0,
'kick_return_yards': 0,
'kick_return_touchdowns': 0,
# PUnt return stats
'punt_return_attempts': 0,
'punt_return_yards': 0,
'punt_return_touchdowns': 0,
# Defense
'defense_sacks': 0,
'defense_tackles': 0,
'defense_tackle_assists': 0,
'defense_interceptions': 0,
'defense_interception_yards': 0,
'defense_interception_touchdowns': 0,
'defense_safeties': 0,
# Kicking
'point_after_attemps': 0,
'point_after_makes': 0,
'field_goal_attempts': 0,
'field_goal_makes': 0,
# Punting
'punting_attempts': 0,
'punting_yards': 0,
'punting_blocked': 0
}
def get_seasons_with_stats(self, profile_soup):
"""Scrape a list of seasons that has stats for the player
Args:
- profile_soup (obj): The BeautifulSoup object for the player profile page
Returns:
- seasons (dict[]): List of dictionaries with meta information about season stats
"""
seasons = []
gamelog_list = profile_soup.find('div', {'id': 'inner_nav'}).find_all('li')[1].find_all('li')
if len(gamelog_list) > 0 and gamelog_list[0].contents[0].contents[0] == 'Career':
for season in gamelog_list:
seasons.append({
'year': season.contents[0].contents[0],
'gamelog_url': BASE_URL.format(season.contents[0]['href'])
})
return seasons
if __name__ == '__main__':
# Replace line below this with next comment when done testing
# letters_to_scrape = list(string.ascii_uppercase)
letters_to_scrape = 'X'
print(letters_to_scrape)
nfl_scraper = Scraper(letters_to_scrape=letters_to_scrape, num_jobs=10, clear_old_data=False)
nfl_scraper.scrape_site()
```
|
github_jupyter
|
import requests
from bs4 import BeautifulSoup
from multiprocessing.dummy import Pool
import time
import shutil
import re
import os
import json
import string
import glob
BASE_URL = 'https://www.pro-football-reference.com{0}'
PLAYER_LIST_URL = 'https://www.pro-football-reference.com/players/{0}'
PLAYER_PROFILE_URL = 'https://www.pro-football-reference.com/players/{0}/{1}'
PLAYER_GAMELOG_URL = 'https://www.pro-football-reference.com/players/{0}/{1}/gamelog/{2}'
HEADERS = {
'user-agent': ('Mozilla/5.0 (Windows NT 6.1; WOW64) AppleWebKit/537.36 '
'(KHTML, like Gecko) Chrome/48.0.2564.109 Safari/537.36')
}
PROFILE_DIR = 'profile_data'
STATS_DIR = 'stats_data'
class Scraper():
"""Scraper for pro-football-reference.com to collect NFL player stats"""
def __init__(self, letters_to_scrape=['Z'], num_jobs=1, clear_old_data=True, first_player_id=1):
"""Initialize the scraper to get player stats
Args:
- letters_to_scrape (str[]): The site sorts players by the first letter of their
last name. This array tells the scraper which letters to scrape data for.
- num_jobs (int): Number of concurrent jobs the scraper should run. While Python
can't multi-thread, it can manage multiple processes at once, which allows it to
utilize time spent waiting for the server to respond.
- clear_old_data (boolean): Whether or not the data file should be wiped before
starting the scrape.
- first_player_id (int): The first ID for a player (set if you are rerunning to avoid duplicates)
Returns:
None
"""
self.letters_to_scrape = [letter.upper() for letter in letters_to_scrape]
self.num_jobs = num_jobs
self.clear_old_data = clear_old_data
self.session = requests.Session()
self.start_time = time.time()
self.cross_process_player_count = 0
self.first_player_id = first_player_id
if num_jobs > 1:
self.multiprocessing = True
self.worker_pool = Pool(num_jobs)
else:
self.multiprocessing = False
def scrape_site(self):
"""Pool workers to scrape players by first letter of last name"""
if self.clear_old_data:
self.clear_data()
player_id = self.first_player_id
for letter in self.letters_to_scrape:
player_profile_urls = self.get_players_for_letter(letter)
for player_profile_url in player_profile_urls:
player = Player(player_id, player_profile_url, self)
try:
player.scrape_profile()
player.scrape_player_stats()
except (KeyboardInterrupt, SystemExit):
raise
except:
print('There was a problem parsing stats for {}'.format(player_profile_url))
continue
self.save_player_profile(player.profile)
self.save_player_game_stats(player.game_stats, player.player_id, player.profile['name'])
player_id += 1
self.condense_data()
def condense_data(self):
"""Condense data into two files, a profile file and a stats file"""
print('Condensing Data...')
condensed_profile_data = []
all_profile_files = glob.glob('{}/*.json'.format(PROFILE_DIR))
for file in all_profile_files:
with open(file, 'rb') as fin:
condensed_profile_data.append(json.load(fin))
print('{} player profiles condensed'.format(len(condensed_profile_data)))
filename = 'profiles_{}.json'.format(time.time())
with open(filename, 'w') as fout:
json.dump(condensed_profile_data, fout)
condensed_game_data = []
all_game_files = glob.glob('{}/*.json'.format(STATS_DIR))
for file in all_game_files:
with open(file, 'rb') as fin:
condensed_game_data += json.load(fin)
print('{} player seasons condensed'.format(len(condensed_game_data)))
filename = 'games_{}.json'.format(time.time())
with open(filename, 'w') as fout:
json.dump(condensed_game_data, fout)
def save_player_profile(self, profile):
"""Save a player's profile as JSON
Args:
- profile (dict): Player profile data
Return:
None
"""
filename = '{}/{}_{}.json'.format(PROFILE_DIR, profile['player_id'], profile['name'].replace(' ', '-'))
try:
os.makedirs(PROFILE_DIR)
except OSError:
pass
with open(filename, 'w') as fout:
json.dump(profile, fout)
def save_player_game_stats(self, games, player_id, player_name):
"""Save a list of player games with stats info
Args:
- games (dict[]): List of game stats
- player_id (int): ID of the player the games belong to
- player_name (str): Name of the player the game stats belong to
Return:
None
"""
filename = '{}/{}_{}.json'.format(STATS_DIR, player_id, player_name.replace(' ', '-'))
try:
os.makedirs(STATS_DIR)
except OSError:
pass
with open(filename, 'w') as fout:
json.dump(games, fout)
def get_players_for_letter(self, letter):
"""Get a list of player links for a letter of the alphabet.
Site organizes players by first letter of last name.
Args:
- letter (str): letter of the alphabet uppercased
Returns:
- player_links (str[]): the URLs to get player profiles
"""
response = self.get_page(PLAYER_LIST_URL.format(letter))
soup = BeautifulSoup(response.content, 'html.parser')
players = soup.find('div', {'id': 'div_players'}).find_all('a')
return [BASE_URL.format(player['href']) for player in players]
def get_page(self, url, retry_count=0):
"""Use requests to get a page; retry when failures occur
Args:
- url (str): The URL of the page to make a GET request to
- retry_count (int): Number of times the URL has already been requests
Returns:
- response (obj): The Requests response object
"""
try:
return self.session.get(url, headers=HEADERS)
except (KeyboardInterrupt, SystemExit):
raise
except:
retry_count += 1
if retry_count <= 3:
self.session = requests.Session()
return self.get_page(url, retry_count)
else:
raise
def clear_data(self):
"""Clear the data directories"""
try:
shutil.rmtree(PROFILE_DIR)
except FileNotFoundError:
pass
try:
shutil.rmtree(STATS_DIR)
except FileNotFoundError:
pass
class Player():
"""An NFL player"""
def __init__(self, player_id, profile_url, scraper):
"""
Args:
- player_id (int): Unique ID for player
- profile_url (str): URL to the player's profile
- scraper (obj): instance of Scraper class
Returns:
None
"""
self.player_id = player_id
self.profile_url = profile_url
self.scraper = scraper
self.profile = {
'player_id': player_id,
'name': None,
'position': None,
'height': None,
'weight': None,
'current_team': None,
'birth_date': None,
'birth_place': None,
'death_date': None,
'college': None,
'high_school': None,
'draft_team': None,
'draft_round': None,
'draft_position': None,
'draft_year': None,
'current_salary': None,
'hof_induction_year': None
}
self.seasons_with_stats = []
self.game_stats = []
def scrape_profile(self):
"""Scrape profile info for player"""
response = self.scraper.get_page(self.profile_url)
soup = BeautifulSoup(response.content, 'html.parser')
profile_section = soup.find('h1', {'itemprop': 'name'}).contents
self.profile['name'] = profile_section.results[1].text
print('scaping {}'.format(self.profile['name']))
profile_attributes = profile_section.find_all('p')
current_attribute = 1
num_attributes = len(profile_attributes)
self.profile['position'] = profile_attributes[current_attribute].contents[2].split('\n')[0].split(' ')[1]
current_attribute += 1
height = profile_attributes[current_attribute].find('span', {'itemprop': 'height'})
if height is not None:
self.profile['height'] = height.contents[0]
weight = profile_attributes[current_attribute].find('span', {'itemprop': 'weight'})
if weight is not None:
self.profile['weight'] = weight.contents[0].split('lb')[0]
if height is not None or weight is not None:
current_attribute += 1
affiliation_section = profile_section.find('span', {'itemprop': 'affiliation'})
if affiliation_section is not None:
self.profile['current_team'] = affiliation_section.contents[0].contents[0]
current_attribute += 1
birth_date = profile_attributes[current_attribute].find('span', {'itemprop': 'birthDate'})
if birth_date is not None:
self.profile['birth_date'] = birth_date['data-birth']
birth_place_section = profile_attributes[current_attribute].find('span', {'itemprop': 'birthPlace'}).contents
try:
self.profile['birth_place'] = re.split('\xa0', birth_place_section[0])[1] + ' ' + birth_place_section[1].contents[0]
except IndexError:
pass
if birth_date is not None or len(birth_place_section) > 0:
current_attribute += 1
death_section = profile_section.find('span', {'itemprop': 'deathDate'})
if death_section is not None:
self.profile['death_date'] = death_section['data-death']
current_attribute += 1
if profile_attributes[current_attribute].contents[0].contents[0] == 'College':
self.profile['college'] = profile_attributes[current_attribute].contents[2].contents[0]
current_attribute += 1
# Skip weighted career AV
current_attribute += 1
if ((current_attribute + 1) <= num_attributes) and profile_attributes[current_attribute].contents[0].contents[0] == 'High School':
self.profile['high_school'] = profile_attributes[current_attribute].contents[2].contents[0] + ', ' + profile_attributes[current_attribute].contents[4].contents[0]
current_attribute += 1
if ((current_attribute + 1) <= num_attributes) and profile_attributes[current_attribute].contents[0].contents[0] == 'Draft':
self.profile['draft_team'] = profile_attributes[current_attribute].contents[2].contents[0]
draft_info = profile_attributes[current_attribute].contents[3].split(' ')
self.profile['draft_round'] = re.findall(r'\d+', draft_info[3])[0]
self.profile['draft_position'] = re.findall(r'\d+', draft_info[5])[0]
self.profile['draft_year'] = re.findall(r'\d+', profile_attributes[current_attribute].contents[4].contents[0])[0]
current_attribute += 1
if ((current_attribute + 1) <= num_attributes) and profile_attributes[current_attribute].contents[0].contents[0] == 'Current cap hit':
profile_attributes[current_attribute].contents
self.profile['current_salary'] = profile_attributes[current_attribute].contents[2].contents[0]
current_attribute += 1
if ((current_attribute + 1) <= num_attributes) and profile_attributes[current_attribute].contents[0].contents[0] == 'Hall of fame':
self.profile['hof_induction_year'] = profile_attributes[current_attribute].contents[2].contents[0]
current_attribute += 1
self.seasons_with_stats = self.get_seasons_with_stats(soup)
def scrape_player_stats(self):
"""Scrape the stats for all available games for a player"""
for season in self.seasons_with_stats:
if season['year'] == 'Career' or season['year'] == 'Postseason':
continue
self.scrape_season_gamelog(season['gamelog_url'], season['year'])
def scrape_season_gamelog(self, gamelog_url, year):
"""Scrape player stats for a given year
Args:
- gamelog_url (str): URL to the stats for a given year
- year (int): The year the stats are for
Returns:
- stats (dict): All of the player's stats for that year
"""
response = self.scraper.get_page(gamelog_url)
soup = BeautifulSoup(response.content, 'html.parser')
regular_season_table = soup.find('table', {'id': 'stats'})
if regular_season_table is None:
return False
games = regular_season_table.find('tbody').find_all('tr')
playoff_table = soup.find('table', {'id': 'stats_playoffs'})
if playoff_table is not None:
games += playoff_table.find('tbody').find_all('tr')
for game in games:
stats = self.make_player_game_stats(self.player_id, year)
stats['game_id'] = game.find('td', {'data-stat': 'game_date'}).find('a', href=True)['href'].replace('/boxscores/', '').replace('.htm', '')
stats['date'] = game.find('td', {'data-stat': 'game_date'}).contents[0].contents[0]
stats['game_number'] = game.find('td', {'data-stat': 'game_num'}).contents[0]
stats['age'] = game.find('td', {'data-stat': 'age'}).contents[0]
stats['team'] = game.find('td', {'data-stat': 'team'}).contents[0].contents[0]
if game.find('td', {'data-stat': 'game_location'}).contents == ['@']:
stats['game_location'] = 'A'
elif game.find('td', {'data-stat': 'game_location'}).contents == ['N']:
stats['game_location'] = 'N'
else:
stats['game_location'] = 'H'
stats['opponent'] = game.find('td', {'data-stat': 'opp'}).contents[0].contents[0]
result = game.find('td', {'data-stat': 'game_result'}).contents[0].contents[0]
stats['game_won'] = (result.split(' ')[0] == 'W')
stats['player_team_score'] = result.split(' ')[1].split('-')[0]
stats['opponent_score'] = result.split(' ')[1].split('-')[1]
# Collect passing stats
pass_attempts = game.find('td', {'data-stat': 'pass_cmp'})
if pass_attempts is not None and len(pass_attempts) > 0:
stats['passing_attempts'] = int(pass_attempts.contents[0])
pass_completions = game.find('td', {'data-stat': 'pass_att'})
if pass_completions is not None and len(pass_completions) > 0:
stats['passing_completions'] = int(pass_completions.contents[0])
pass_yards = game.find('td', {'data-stat': 'pass_yds'})
if pass_yards is not None and len(pass_yards) > 0:
stats['passing_yards'] = int(pass_yards.contents[0])
pass_touchdowns = game.find('td', {'data-stat': 'pass_td'})
if pass_touchdowns is not None and len(pass_touchdowns) > 0:
stats['passing_touchdowns'] = int(pass_touchdowns.contents[0])
pass_interceptions = game.find('td', {'data-stat': 'pass_int'})
if pass_interceptions is not None and len(pass_interceptions) > 0:
stats['passing_interceptions'] = int(pass_interceptions.contents[0])
pass_rating = game.find('td', {'data-stat': 'pass_rating'})
if pass_rating is not None and len(pass_rating) > 0:
stats['passing_rating'] = float(pass_rating.contents[0])
pass_sacks = game.find('td', {'data-stat': 'pass_sacked'})
if pass_sacks is not None and len(pass_sacks) > 0:
stats['passing_sacks'] = int(pass_sacks.contents[0])
pass_sacks_yards_lost = game.find('td', {'data-stat': 'pass_sacked_yds'})
if pass_sacks_yards_lost is not None and len(pass_sacks_yards_lost) > 0:
stats['passing_sacks_yards_lost'] = int(pass_sacks_yards_lost.contents[0])
# Collect rushing stats
rushing_attempts = game.find('td', {'data-stat': 'rush_att'})
if rushing_attempts is not None and len(rushing_attempts) > 0:
stats['rushing_attempts'] = int(rushing_attempts.contents[0])
rushing_yards = game.find('td', {'data-stat': 'rush_yds'})
if rushing_yards is not None and len(rushing_yards) > 0:
stats['rushing_yards'] = int(rushing_yards.contents[0])
rushing_touchdowns = game.find('td', {'data-stat': 'rush_td'})
if rushing_touchdowns is not None and len(rushing_touchdowns) > 0:
stats['rushing_touchdowns'] = int(rushing_touchdowns.contents[0])
# Collect receiving stats
receiving_targets = game.find('td', {'data-stat': 'targets'})
if receiving_targets is not None and len(receiving_targets) > 0:
stats['receiving_targets'] = int(receiving_targets.contents[0])
receiving_receptions = game.find('td', {'data-stat': 'rec'})
if receiving_receptions is not None and len(receiving_receptions) > 0:
stats['receiving_receptions'] = int(receiving_receptions.contents[0])
receiving_yards = game.find('td', {'data-stat': 'rec_yds'})
if receiving_yards is not None and len(receiving_yards) > 0:
stats['receiving_yards'] = int(receiving_yards.contents[0])
receiving_touchdowns = game.find('td', {'data-stat': 'rec_td'})
if receiving_touchdowns is not None and len(receiving_touchdowns) > 0:
stats['receiving_touchdowns'] = int(receiving_touchdowns.contents[0])
# Collect kick return stats
kick_return_attempts = game.find('td', {'data-stat': 'kick_ret'})
if kick_return_attempts is not None and len(kick_return_attempts) > 0:
stats['kick_return_attempts'] = int(kick_return_attempts.contents[0])
kick_return_yards = game.find('td', {'data-stat': 'kick_ret_yds'})
if kick_return_yards is not None and len(kick_return_yards) > 0:
stats['kick_return_yards'] = int(kick_return_yards.contents[0])
kick_return_touchdowns = game.find('td', {'data-stat': 'kick_ret_td'})
if kick_return_touchdowns is not None and len(kick_return_touchdowns) > 0:
stats['kick_return_touchdowns'] = int(kick_return_touchdowns.contents[0])
# Collect punt return stats
punt_return_attempts = game.find('td', {'data-stat': 'punt_ret'})
if punt_return_attempts is not None and len(punt_return_attempts) > 0:
stats['punt_return_attempts'] = int(punt_return_attempts.contents[0])
punt_return_yards = game.find('td', {'data-stat': 'punt_ret_yds'})
if punt_return_yards is not None and len(punt_return_yards) > 0:
stats['punt_return_yards'] = int(punt_return_yards.contents[0])
punt_return_touchdowns = game.find('td', {'data-stat': 'punt_ret_td'})
if punt_return_touchdowns is not None and len(punt_return_touchdowns) > 0:
stats['punt_return_touchdowns'] = int(punt_return_touchdowns.contents[0])
# Collect defensive stats
defense_sacks = game.find('td', {'data-stat': 'sacks'})
if defense_sacks is not None and len(defense_sacks) > 0:
stats['defense_sacks'] = float(defense_sacks.contents[0])
defense_tackles = game.find('td', {'data-stat': 'tackles_solo'})
if defense_tackles is not None and len(defense_tackles) > 0:
stats['defense_tackles'] = int(defense_tackles.contents[0])
defense_tackle_assists = game.find('td', {'data-stat': 'tackles_assists'})
if defense_tackle_assists is not None and len(defense_tackle_assists) > 0:
stats['defense_tackle_assists'] = int(defense_tackle_assists.contents[0])
defense_interceptions = game.find('td', {'data-stat': 'def_int'})
if defense_interceptions is not None and len(defense_interceptions) > 0:
stats['defense_interceptions'] = int(defense_interceptions.contents[0])
defense_interception_yards = game.find('td', {'data-stat': 'def_int_yds'})
if defense_interception_yards is not None and len(defense_interception_yards) > 0:
stats['defense_interception_yards'] = int(defense_interception_yards.contents[0])
defense_safeties = game.find('td', {'data-stat': 'safety_md'})
if defense_safeties is not None and len(defense_safeties) > 0:
stats['defense_safeties'] = int(defense_safeties.contents[0])
# Collect kicking stats
point_after_attemps = game.find('td', {'data-stat': 'xpm'})
if point_after_attemps is not None and len(point_after_attemps) > 0:
stats['point_after_attemps'] = int(point_after_attemps.contents[0])
point_after_makes = game.find('td', {'data-stat': 'xpa'})
if point_after_makes is not None and len(point_after_makes) > 0:
stats['point_after_makes'] = int(point_after_makes.contents[0])
field_goal_attempts = game.find('td', {'data-stat': 'fga'})
if field_goal_attempts is not None and len(field_goal_attempts) > 0:
stats['field_goal_attempts'] = int(field_goal_attempts.contents[0])
field_goal_makes = game.find('td', {'data-stat': 'fgm'})
if field_goal_makes is not None and len(field_goal_makes) > 0:
stats['field_goal_makes'] = int(field_goal_makes.contents[0])
# Collect punting stats
punting_attempts = game.find('td', {'data-stat': 'punt'})
if punting_attempts is not None and len(punting_attempts) > 0:
stats['punting_attempts'] = int(punting_attempts.contents[0])
punting_yards = game.find('td', {'data-stat': 'punt_yds'})
if punting_yards is not None and len(punting_yards) > 0:
stats['punting_yards'] = int(punting_yards.contents[0])
punting_blocked = game.find('td', {'data-stat': 'punt_blocked'})
if punting_blocked is not None and len(punting_blocked) > 0:
stats['punting_blocked'] = int(punting_blocked.contents[0])
self.game_stats.append(stats)
@staticmethod
def make_player_game_stats(player_id, year):
"""Factory method to return possible stats to collect for a player in a game
Args:
- player_id (int): unique Id for the player
- year (int): The year the stats are for
Returns:
- game_stats (dict): dictionary with game stats initialized
"""
return {
'player_id': player_id,
'year': year,
# General stats
'game_id': None,
'date': None,
'game_number': None,
'age': None,
'team': None,
'game_location': None,
'opponent': None,
'game_won': None,
'player_team_score': 0,
'opponent_score': 0,
# Passing stats
'passing_attempts': 0,
'passing_completions': 0,
'passing_yards': 0,
'passing_rating': 0,
'passing_touchdowns': 0,
'passing_interceptions': 0,
'passing_sacks': 0,
'passing_sacks_yards_lost': 0,
# Rushing stats
'rushing_attempts': 0,
'rushing_yards': 0,
'rushing_touchdowns': 0,
# Receiving stats
'receiving_targets': 0,
'receiving_receptions': 0,
'receiving_yards': 0,
'receiving_touchdowns': 0,
# Kick return stats
'kick_return_attempts': 0,
'kick_return_yards': 0,
'kick_return_touchdowns': 0,
# PUnt return stats
'punt_return_attempts': 0,
'punt_return_yards': 0,
'punt_return_touchdowns': 0,
# Defense
'defense_sacks': 0,
'defense_tackles': 0,
'defense_tackle_assists': 0,
'defense_interceptions': 0,
'defense_interception_yards': 0,
'defense_interception_touchdowns': 0,
'defense_safeties': 0,
# Kicking
'point_after_attemps': 0,
'point_after_makes': 0,
'field_goal_attempts': 0,
'field_goal_makes': 0,
# Punting
'punting_attempts': 0,
'punting_yards': 0,
'punting_blocked': 0
}
def get_seasons_with_stats(self, profile_soup):
"""Scrape a list of seasons that has stats for the player
Args:
- profile_soup (obj): The BeautifulSoup object for the player profile page
Returns:
- seasons (dict[]): List of dictionaries with meta information about season stats
"""
seasons = []
gamelog_list = profile_soup.find('div', {'id': 'inner_nav'}).find_all('li')[1].find_all('li')
if len(gamelog_list) > 0 and gamelog_list[0].contents[0].contents[0] == 'Career':
for season in gamelog_list:
seasons.append({
'year': season.contents[0].contents[0],
'gamelog_url': BASE_URL.format(season.contents[0]['href'])
})
return seasons
if __name__ == '__main__':
# Replace line below this with next comment when done testing
# letters_to_scrape = list(string.ascii_uppercase)
letters_to_scrape = 'X'
print(letters_to_scrape)
nfl_scraper = Scraper(letters_to_scrape=letters_to_scrape, num_jobs=10, clear_old_data=False)
nfl_scraper.scrape_site()
| 0.501953 | 0.171373 |
# Bayesian Regression Using NumPyro
In this tutorial, we will explore how to do bayesian regression in NumPyro, using a simple example adapted from Statistical Rethinking [[1](#References)]. In particular, we would like to explore the following:
- Write a simple model using the `sample` NumPyro primitive.
- Run inference using MCMC in NumPyro, in particular, using the No U-Turn Sampler (NUTS) to get a posterior distribution over our regression parameters of interest.
- Learn about inference utilities such as `Predictive` and `log_likelihood`.
- Learn how we can use effect-handlers in NumPyro to generate execution traces from the model, condition on sample statements, seed models with RNG seeds, etc., and use this to implement various utilities that will be useful for MCMC. e.g. computing model log likelihood, generating empirical distribution over the posterior predictive, etc.
## Tutorial Outline:
1. [Dataset](#Dataset)
2. [Regression Model to Predict Divorce Rate](#Regression-Model-to-Predict-Divorce-Rate)
- [Model-1: Predictor-Marriage Rate](#Model-1:-Predictor---Marriage-Rate)
- [Posterior Distribution over the Regression Parameters](#Posterior-Distribution-over-the-Regression-Parameters)
- [Posterior Predictive Distribution](#Posterior-Predictive-Distribution)
- [Predictive Utility With Effect Handlers](#Predictive-Utility-With-Effect-Handlers)
- [Model Predictive Density](#Model-Predictive-Density)
- [Model-2: Predictor-Median Age of Marriage](#Model-2:-Predictor---Median-Age-of-Marriage)
- [Model-3: Predictor-Marriage Rate and Median Age of Marriage](#Model-3:-Predictor---Marriage-Rate-and-Median-Age-of-Marriage)
- [Divorce Rate Residuals by State](#Divorce-Rate-Residuals-by-State)
3. [Regression Model with Measurement Error](#Regression-Model-with-Measurement-Error)
- [Effect of Incorporating Measurement Noise on Residuals](#Effect-of-Incorporating-Measurement-Noise-on-Residuals)
4. [References](#References)
```
!pip install -q numpyro@git+https://github.com/pyro-ppl/numpyro
import os
from IPython.display import set_matplotlib_formats
import jax.numpy as jnp
from jax import random, vmap
from jax.scipy.special import logsumexp
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import seaborn as sns
import numpyro
from numpyro.diagnostics import hpdi
import numpyro.distributions as dist
from numpyro import handlers
from numpyro.infer import MCMC, NUTS
plt.style.use('bmh')
if "NUMPYRO_SPHINXBUILD" in os.environ:
set_matplotlib_formats('svg')
assert numpyro.__version__.startswith('0.7.1')
```
## Dataset
For this example, we will use the `WaffleDivorce` dataset from Chapter 05, Statistical Rethinking [[1](#References)]. The dataset contains divorce rates in each of the 50 states in the USA, along with predictors such as population, median age of marriage, whether it is a Southern state and, curiously, number of Waffle Houses.
```
DATASET_URL = 'https://raw.githubusercontent.com/rmcelreath/rethinking/master/data/WaffleDivorce.csv'
dset = pd.read_csv(DATASET_URL, sep=';')
dset
```
Let us plot the pair-wise relationship amongst the main variables in the dataset, using `seaborn.pairplot`.
```
vars = ['Population', 'MedianAgeMarriage', 'Marriage', 'WaffleHouses', 'South', 'Divorce']
sns.pairplot(dset, x_vars=vars, y_vars=vars, palette='husl');
```
From the plots above, we can clearly observe that there is a relationship between divorce rates and marriage rates in a state (as might be expected), and also between divorce rates and median age of marriage.
There is also a weak relationship between number of Waffle Houses and divorce rates, which is not obvious from the plot above, but will be clearer if we regress `Divorce` against `WaffleHouse` and plot the results.
```
sns.regplot(x='WaffleHouses', y='Divorce', data=dset);
```
This is an example of a spurious association. We do not expect the number of Waffle Houses in a state to affect the divorce rate, but it is likely correlated with other factors that have an effect on the divorce rate. We will not delve into this spurious association in this tutorial, but the interested reader is encouraged to read Chapters 5 and 6 of [[1](#References)] which explores the problem of causal association in the presence of multiple predictors.
For simplicity, we will primarily focus on marriage rate and the median age of marriage as our predictors for divorce rate throughout the remaining tutorial.
## Regression Model to Predict Divorce Rate
Let us now write a regressionn model in *NumPyro* to predict the divorce rate as a linear function of marriage rate and median age of marriage in each of the states.
First, note that our predictor variables have somewhat different scales. It is a good practice to standardize our predictors and response variables to mean `0` and standard deviation `1`, which should result in [faster inference](https://mc-stan.org/docs/2_19/stan-users-guide/standardizing-predictors-and-outputs.html).
```
standardize = lambda x: (x - x.mean()) / x.std()
dset['AgeScaled'] = dset.MedianAgeMarriage.pipe(standardize)
dset['MarriageScaled'] = dset.Marriage.pipe(standardize)
dset['DivorceScaled'] = dset.Divorce.pipe(standardize)
```
We write the NumPyro model as follows. While the code should largely be self-explanatory, take note of the following:
- In NumPyro, *model* code is any Python callable which can optionally accept additional arguments and keywords. For HMC which we will be using for this tutorial, these arguments and keywords remain static during inference, but we can reuse the same model to generate [predictions](#Posterior-Predictive-Distribution) on new data.
- In addition to regular Python statements, the model code also contains primitives like `sample`. These primitives can be interpreted with various side-effects using effect handlers. For more on effect handlers, refer to [[3](#References)], [[4](#References)]. For now, just remember that a `sample` statement makes this a stochastic function that samples some latent parameters from a *prior distribution*. Our goal is to infer the *posterior distribution* of these parameters conditioned on observed data.
- The reason why we have kept our predictors as optional keyword arguments is to be able to reuse the same model as we vary the set of predictors. Likewise, the reason why the response variable is optional is that we would like to reuse this model to sample from the posterior predictive distribution. See the [section](#Posterior-Predictive-Distribution) on plotting the posterior predictive distribution, as an example.
```
def model(marriage=None, age=None, divorce=None):
a = numpyro.sample('a', dist.Normal(0., 0.2))
M, A = 0., 0.
if marriage is not None:
bM = numpyro.sample('bM', dist.Normal(0., 0.5))
M = bM * marriage
if age is not None:
bA = numpyro.sample('bA', dist.Normal(0., 0.5))
A = bA * age
sigma = numpyro.sample('sigma', dist.Exponential(1.))
mu = a + M + A
numpyro.sample('obs', dist.Normal(mu, sigma), obs=divorce)
```
### Model 1: Predictor - Marriage Rate
We first try to model the divorce rate as depending on a single variable, marriage rate. As mentioned above, we can use the same `model` code as earlier, but only pass values for `marriage` and `divorce` keyword arguments. We will use the No U-Turn Sampler (see [[5](#References)] for more details on the NUTS algorithm) to run inference on this simple model.
The Hamiltonian Monte Carlo (or, the NUTS) implementation in NumPyro takes in a potential energy function. This is the negative log joint density for the model. Therefore, for our model description above, we need to construct a function which given the parameter values returns the potential energy (or negative log joint density). Additionally, the verlet integrator in HMC (or, NUTS) returns sample values simulated using Hamiltonian dynamics in the unconstrained space. As such, continuous variables with bounded support need to be transformed into unconstrained space using bijective transforms. We also need to transform these samples back to their constrained support before returning these values to the user. Thankfully, this is handled on the backend for us, within a convenience class for doing [MCMC inference](https://numpyro.readthedocs.io/en/latest/mcmc.html#numpyro.mcmc.MCMC) that has the following methods:
- `run(...)`: runs warmup, adapts steps size and mass matrix, and does sampling using the sample from the warmup phase.
- `print_summary()`: print diagnostic information like quantiles, effective sample size, and the Gelman-Rubin diagnostic.
- `get_samples()`: gets samples from the posterior distribution.
Note the following:
- JAX uses functional PRNGs. Unlike other languages / frameworks which maintain a global random state, in JAX, every call to a sampler requires an [explicit PRNGKey](https://github.com/google/jax#random-numbers-are-different). We will split our initial random seed for subsequent operations, so that we do not accidentally reuse the same seed.
- We run inference with the `NUTS` sampler. To run vanilla HMC, we can instead use the [HMC](https://numpyro.readthedocs.io/en/latest/mcmc.html#numpyro.mcmc.HMC) class.
```
# Start from this source of randomness. We will split keys for subsequent operations.
rng_key = random.PRNGKey(0)
rng_key, rng_key_ = random.split(rng_key)
# Run NUTS.
kernel = NUTS(model)
num_samples = 2000
mcmc = MCMC(kernel, num_warmup=1000, num_samples=num_samples)
mcmc.run(rng_key_, marriage=dset.MarriageScaled.values, divorce=dset.DivorceScaled.values)
mcmc.print_summary()
samples_1 = mcmc.get_samples()
```
#### Posterior Distribution over the Regression Parameters
We notice that the progress bar gives us online statistics on the acceptance probability, step size and number of steps taken per sample while running NUTS. In particular, during warmup, we adapt the step size and mass matrix to achieve a certain target acceptance probability which is 0.8, by default. We were able to successfully adapt our step size to achieve this target in the warmup phase.
During warmup, the aim is to adapt hyper-parameters such as step size and mass matrix (the HMC algorithm is very sensitive to these hyper-parameters), and to reach the typical set (see [[6](#References)] for more details). If there are any issues in the model specification, the first signal to notice would be low acceptance probabilities or very high number of steps. We use the sample from the end of the warmup phase to seed the MCMC chain (denoted by the second `sample` progress bar) from which we generate the desired number of samples from our target distribution.
At the end of inference, NumPyro prints the mean, std and 90% CI values for each of the latent parameters. Note that since we standardized our predictors and response variable, we would expect the intercept to have mean 0, as can be seen here. It also prints other convergence diagnostics on the latent parameters in the model, including [effective sample size](https://numpyro.readthedocs.io/en/latest/diagnostics.html#numpyro.diagnostics.effective_sample_size) and the [gelman rubin diagnostic](https://numpyro.readthedocs.io/en/latest/diagnostics.html#numpyro.diagnostics.gelman_rubin) ($\hat{R}$). The value for these diagnostics indicates that the chain has converged to the target distribution. In our case, the "target distribution" is the posterior distribution over the latent parameters that we are interested in. Note that this is often worth verifying with multiple chains for more complicated models. In the end, `samples_1` is a collection (in our case, a `dict` since `init_samples` was a `dict`) containing samples from the posterior distribution for each of the latent parameters in the model.
To look at our regression fit, let us plot the regression line using our posterior estimates for the regression parameters, along with the 90% Credibility Interval (CI). Note that the [hpdi](https://numpyro.readthedocs.io/en/latest/diagnostics.html#numpyro.diagnostics.hpdi) function in NumPyro's diagnostics module can be used to compute CI. In the functions below, note that the collected samples from the posterior are all along the leading axis.
```
def plot_regression(x, y_mean, y_hpdi):
# Sort values for plotting by x axis
idx = jnp.argsort(x)
marriage = x[idx]
mean = y_mean[idx]
hpdi = y_hpdi[:, idx]
divorce = dset.DivorceScaled.values[idx]
# Plot
fig, ax = plt.subplots(nrows=1, ncols=1, figsize=(6, 6))
ax.plot(marriage, mean)
ax.plot(marriage, divorce, 'o')
ax.fill_between(marriage, hpdi[0], hpdi[1], alpha=0.3, interpolate=True)
return ax
# Compute empirical posterior distribution over mu
posterior_mu = jnp.expand_dims(samples_1['a'], -1) + \
jnp.expand_dims(samples_1['bM'], -1) * dset.MarriageScaled.values
mean_mu = jnp.mean(posterior_mu, axis=0)
hpdi_mu = hpdi(posterior_mu, 0.9)
ax = plot_regression(dset.MarriageScaled.values, mean_mu, hpdi_mu)
ax.set(xlabel='Marriage rate', ylabel='Divorce rate', title='Regression line with 90% CI');
```
We can see from the plot, that the CI broadens towards the tails where the data is relatively sparse, as can be expected.
#### Prior Predictive Distribution
Let us check that we have set sensible priors by sampling from the prior predictive distribution. NumPyro provides a handy [Predictive](http://num.pyro.ai/en/latest/utilities.html#numpyro.infer.util.Predictive) utility for this purpose.
```
from numpyro.infer import Predictive
rng_key, rng_key_ = random.split(rng_key)
prior_predictive = Predictive(model, num_samples=100)
prior_predictions = prior_predictive(rng_key_, marriage=dset.MarriageScaled.values)['obs']
mean_prior_pred = jnp.mean(prior_predictions, axis=0)
hpdi_prior_pred = hpdi(prior_predictions, 0.9)
ax = plot_regression(dset.MarriageScaled.values, mean_prior_pred, hpdi_prior_pred)
ax.set(xlabel='Marriage rate', ylabel='Divorce rate', title='Predictions with 90% CI');
```
#### Posterior Predictive Distribution
Let us now look at the posterior predictive distribution to see how our predictive distribution looks with respect to the observed divorce rates. To get samples from the posterior predictive distribution, we need to run the model by substituting the latent parameters with samples from the posterior. Note that by default we generate a single prediction for each sample from the joint posterior distribution, but this can be controlled using the `num_samples` argument.
```
rng_key, rng_key_ = random.split(rng_key)
predictive = Predictive(model, samples_1)
predictions = predictive(rng_key_, marriage=dset.MarriageScaled.values)['obs']
df = dset.filter(['Location'])
df['Mean Predictions'] = jnp.mean(predictions, axis=0)
df.head()
```
#### Predictive Utility With Effect Handlers
To remove the magic behind `Predictive`, let us see how we can combine [effect handlers](https://numpyro.readthedocs.io/en/latest/handlers.html) with the [vmap](https://github.com/google/jax#auto-vectorization-with-vmap) JAX primitive to implement our own simplified predictive utility function that can do vectorized predictions.
```
def predict(rng_key, post_samples, model, *args, **kwargs):
model = handlers.seed(handlers.condition(model, post_samples), rng_key)
model_trace = handlers.trace(model).get_trace(*args, **kwargs)
return model_trace['obs']['value']
# vectorize predictions via vmap
predict_fn = vmap(lambda rng_key, samples: predict(rng_key, samples, model, marriage=dset.MarriageScaled.values))
```
Note the use of the `condition`, `seed` and `trace` effect handlers in the `predict` function.
- The `seed` effect-handler is used to wrap a stochastic function with an initial `PRNGKey` seed. When a sample statement inside the model is called, it uses the existing seed to sample from a distribution but this effect-handler also splits the existing key to ensure that future `sample` calls in the model use the newly split key instead. This is to prevent us from having to explicitly pass in a `PRNGKey` to each `sample` statement in the model.
- The `condition` effect handler conditions the latent sample sites to certain values. In our case, we are conditioning on values from the posterior distribution returned by MCMC.
- The `trace` effect handler runs the model and records the execution trace within an `OrderedDict`. This trace object contains execution metadata that is useful for computing quantities such as the log joint density.
It should be clear now that the `predict` function simply runs the model by substituting the latent parameters with samples from the posterior (generated by the `mcmc` function) to generate predictions. Note the use of JAX's auto-vectorization transform called [vmap](https://github.com/google/jax#auto-vectorization-with-vmap) to vectorize predictions. Note that if we didn't use `vmap`, we would have to use a native for loop which for each sample which is much slower. Each draw from the posterior can be used to get predictions over all the 50 states. When we vectorize this over all the samples from the posterior using `vmap`, we will get a `predictions_1` array of shape `(num_samples, 50)`. We can then compute the mean and 90% CI of these samples to plot the posterior predictive distribution. We note that our mean predictions match those obtained from the `Predictive` utility class.
```
# Using the same key as we used for Predictive - note that the results are identical.
predictions_1 = predict_fn(random.split(rng_key_, num_samples), samples_1)
mean_pred = jnp.mean(predictions_1, axis=0)
df = dset.filter(['Location'])
df['Mean Predictions'] = mean_pred
df.head()
hpdi_pred = hpdi(predictions_1, 0.9)
ax = plot_regression(dset.MarriageScaled.values, mean_pred, hpdi_pred)
ax.set(xlabel='Marriage rate', ylabel='Divorce rate', title='Predictions with 90% CI');
```
We have used the same `plot_regression` function as earlier. We notice that our CI for the predictive distribution is much broader as compared to the last plot due to the additional noise introduced by the `sigma` parameter. Most data points lie well within the 90% CI, which indicates a good fit.
#### Posterior Predictive Density
Likewise, making use of effect-handlers and `vmap`, we can also compute the log likelihood for this model given the dataset, and the log posterior predictive density [[6](#References)] which is given by
$$ log \prod_{i=1}^{n} \int p(y_i | \theta) p_{post}(\theta) d\theta
\approx \sum_{i=1}^n log \frac{\sum_s p(\theta^{s})}{S} \\
= \sum_{i=1}^n (log \sum_s p(\theta^{s}) - log(S))
$$.
Here, $i$ indexes the observed data points $y$ and $s$ indexes the posterior samples over the latent parameters $\theta$. If the posterior predictive density for a model has a comparatively high value, it indicates that the observed data-points have higher probability under the given model.
```
def log_likelihood(rng_key, params, model, *args, **kwargs):
model = handlers.condition(model, params)
model_trace = handlers.trace(model).get_trace(*args, **kwargs)
obs_node = model_trace['obs']
return obs_node['fn'].log_prob(obs_node['value'])
def log_pred_density(rng_key, params, model, *args, **kwargs):
n = list(params.values())[0].shape[0]
log_lk_fn = vmap(lambda rng_key, params: log_likelihood(rng_key, params, model, *args, **kwargs))
log_lk_vals = log_lk_fn(random.split(rng_key, n), params)
return (logsumexp(log_lk_vals, 0) - jnp.log(n)).sum()
```
Note that NumPyro provides the [log_likelihood](http://num.pyro.ai/en/latest/utilities.html#log-likelihood) utility function that can be used directly for computing `log likelihood` as in the first function for any general model. In this tutorial, we would like to emphasize that there is nothing magical about such utility functions, and you can roll out your own inference utilities using NumPyro's effect handling stack.
```
rng_key, rng_key_ = random.split(rng_key)
print('Log posterior predictive density: {}'.format(log_pred_density(rng_key_,
samples_1,
model,
marriage=dset.MarriageScaled.values,
divorce=dset.DivorceScaled.values)))
```
### Model 2: Predictor - Median Age of Marriage
We will now model the divorce rate as a function of the median age of marriage. The computations are mostly a reproduction of what we did for Model 1. Notice the following:
- Divorce rate is inversely related to the age of marriage. Hence states where the median age of marriage is low will likely have a higher divorce rate.
- We get a higher log likelihood as compared to Model 2, indicating that median age of marriage is likely a much better predictor of divorce rate.
```
rng_key, rng_key_ = random.split(rng_key)
mcmc.run(rng_key_, age=dset.AgeScaled.values, divorce=dset.DivorceScaled.values)
mcmc.print_summary()
samples_2 = mcmc.get_samples()
posterior_mu = jnp.expand_dims(samples_2['a'], -1) + \
jnp.expand_dims(samples_2['bA'], -1) * dset.AgeScaled.values
mean_mu = jnp.mean(posterior_mu, axis=0)
hpdi_mu = hpdi(posterior_mu, 0.9)
ax = plot_regression(dset.AgeScaled.values, mean_mu, hpdi_mu)
ax.set(xlabel='Median marriage age', ylabel='Divorce rate', title='Regression line with 90% CI');
rng_key, rng_key_ = random.split(rng_key)
predictions_2 = Predictive(model, samples_2)(rng_key_,
age=dset.AgeScaled.values)['obs']
mean_pred = jnp.mean(predictions_2, axis=0)
hpdi_pred = hpdi(predictions_2, 0.9)
ax = plot_regression(dset.AgeScaled.values, mean_pred, hpdi_pred)
ax.set(xlabel='Median Age', ylabel='Divorce rate', title='Predictions with 90% CI');
rng_key, rng_key_ = random.split(rng_key)
print('Log posterior predictive density: {}'.format(log_pred_density(rng_key_,
samples_2,
model,
age=dset.AgeScaled.values,
divorce=dset.DivorceScaled.values)))
```
### Model 3: Predictor - Marriage Rate and Median Age of Marriage
Finally, we will also model divorce rate as depending on both marriage rate as well as the median age of marriage. Note that the model's posterior predictive density is similar to Model 2 which likely indicates that the marginal information from marriage rate in predicting divorce rate is low when the median age of marriage is already known.
```
rng_key, rng_key_ = random.split(rng_key)
mcmc.run(rng_key_, marriage=dset.MarriageScaled.values,
age=dset.AgeScaled.values, divorce=dset.DivorceScaled.values)
mcmc.print_summary()
samples_3 = mcmc.get_samples()
rng_key, rng_key_ = random.split(rng_key)
print('Log posterior predictive density: {}'.format(
log_pred_density(rng_key_,
samples_3,
model,
marriage=dset.MarriageScaled.values,
age=dset.AgeScaled.values,
divorce=dset.DivorceScaled.values)
))
```
### Divorce Rate Residuals by State
The regression plots above shows that the observed divorce rates for many states differs considerably from the mean regression line. To dig deeper into how the last model (Model 3) under-predicts or over-predicts for each of the states, we will plot the posterior predictive and residuals (`Observed divorce rate - Predicted divorce rate`) for each of the states.
```
# Predictions for Model 3.
rng_key, rng_key_ = random.split(rng_key)
predictions_3 = Predictive(model, samples_3)(rng_key_,
marriage=dset.MarriageScaled.values,
age=dset.AgeScaled.values)['obs']
y = jnp.arange(50)
fig, ax = plt.subplots(nrows=1, ncols=2, figsize=(12, 16))
pred_mean = jnp.mean(predictions_3, axis=0)
pred_hpdi = hpdi(predictions_3, 0.9)
residuals_3 = dset.DivorceScaled.values - predictions_3
residuals_mean = jnp.mean(residuals_3, axis=0)
residuals_hpdi = hpdi(residuals_3, 0.9)
idx = jnp.argsort(residuals_mean)
# Plot posterior predictive
ax[0].plot(jnp.zeros(50), y, '--')
ax[0].errorbar(pred_mean[idx], y, xerr=pred_hpdi[1, idx] - pred_mean[idx],
marker='o', ms=5, mew=4, ls='none', alpha=0.8)
ax[0].plot(dset.DivorceScaled.values[idx], y, marker='o',
ls='none', color='gray')
ax[0].set(xlabel='Posterior Predictive (red) vs. Actuals (gray)', ylabel='State',
title='Posterior Predictive with 90% CI')
ax[0].set_yticks(y)
ax[0].set_yticklabels(dset.Loc.values[idx], fontsize=10);
# Plot residuals
residuals_3 = dset.DivorceScaled.values - predictions_3
residuals_mean = jnp.mean(residuals_3, axis=0)
residuals_hpdi = hpdi(residuals_3, 0.9)
err = residuals_hpdi[1] - residuals_mean
ax[1].plot(jnp.zeros(50), y, '--')
ax[1].errorbar(residuals_mean[idx], y, xerr=err[idx],
marker='o', ms=5, mew=4, ls='none', alpha=0.8)
ax[1].set(xlabel='Residuals', ylabel='State', title='Residuals with 90% CI')
ax[1].set_yticks(y)
ax[1].set_yticklabels(dset.Loc.values[idx], fontsize=10);
```
The plot on the left shows the mean predictions with 90% CI for each of the states using Model 3. The gray markers indicate the actual observed divorce rates. The right plot shows the residuals for each of the states, and both these plots are sorted by the residuals, i.e. at the bottom, we are looking at states where the model predictions are higher than the observed rates, whereas at the top, the reverse is true.
Overall, the model fit seems good because most observed data points like within a 90% CI around the mean predictions. However, notice how the model over-predicts by a large margin for states like Idaho (bottom left), and on the other end under-predicts for states like Maine (top right). This is likely indicative of other factors that we are missing out in our model that affect divorce rate across different states. Even ignoring other socio-political variables, one such factor that we have not yet modeled is the measurement noise given by `Divorce SE` in the dataset. We will explore this in the next section.
## Regression Model with Measurement Error
Note that in our previous models, each data point influences the regression line equally. Is this well justified? We will build on the previous model to incorporate measurement error given by `Divorce SE` variable in the dataset. Incorporating measurement noise will be useful in ensuring that observations that have higher confidence (i.e. lower measurement noise) have a greater impact on the regression line. On the other hand, this will also help us better model outliers with high measurement errors. For more details on modeling errors due to measurement noise, refer to Chapter 14 of [[1](#References)].
To do this, we will reuse Model 3, with the only change that the final observed value has a measurement error given by `divorce_sd` (notice that this has to be standardized since the `divorce` variable itself has been standardized to mean 0 and std 1).
```
def model_se(marriage, age, divorce_sd, divorce=None):
a = numpyro.sample('a', dist.Normal(0., 0.2))
bM = numpyro.sample('bM', dist.Normal(0., 0.5))
M = bM * marriage
bA = numpyro.sample('bA', dist.Normal(0., 0.5))
A = bA * age
sigma = numpyro.sample('sigma', dist.Exponential(1.))
mu = a + M + A
divorce_rate = numpyro.sample('divorce_rate', dist.Normal(mu, sigma))
numpyro.sample('obs', dist.Normal(divorce_rate, divorce_sd), obs=divorce)
# Standardize
dset['DivorceScaledSD'] = dset['Divorce SE'] / jnp.std(dset.Divorce.values)
rng_key, rng_key_ = random.split(rng_key)
kernel = NUTS(model_se, target_accept_prob=0.9)
mcmc = MCMC(kernel, num_warmup=1000, num_samples=3000)
mcmc.run(rng_key_, marriage=dset.MarriageScaled.values, age=dset.AgeScaled.values,
divorce_sd=dset.DivorceScaledSD.values, divorce=dset.DivorceScaled.values)
mcmc.print_summary()
samples_4 = mcmc.get_samples()
```
### Effect of Incorporating Measurement Noise on Residuals
Notice that our values for the regression coefficients is very similar to Model 3. However, introducing measurement noise allows us to more closely match our predictive distribution to the observed values. We can see this if we plot the residuals as earlier.
```
rng_key, rng_key_ = random.split(rng_key)
predictions_4 = Predictive(model_se, samples_4)(rng_key_,
marriage=dset.MarriageScaled.values,
age=dset.AgeScaled.values,
divorce_sd=dset.DivorceScaledSD.values)['obs']
sd = dset.DivorceScaledSD.values
residuals_4 = dset.DivorceScaled.values - predictions_4
residuals_mean = jnp.mean(residuals_4, axis=0)
residuals_hpdi = hpdi(residuals_4, 0.9)
err = residuals_hpdi[1] - residuals_mean
idx = jnp.argsort(residuals_mean)
y = jnp.arange(50)
fig, ax = plt.subplots(nrows=1, ncols=1, figsize=(6, 16))
# Plot Residuals
ax.plot(jnp.zeros(50), y, '--')
ax.errorbar(residuals_mean[idx], y, xerr=err[idx],
marker='o', ms=5, mew=4, ls='none', alpha=0.8)
# Plot SD
ax.errorbar(residuals_mean[idx], y, xerr=sd[idx],
ls='none', color='orange', alpha=0.9)
# Plot earlier mean residual
ax.plot(jnp.mean(dset.DivorceScaled.values - predictions_3, 0)[idx], y,
ls='none', marker='o', ms=6, color='black', alpha=0.6)
ax.set(xlabel='Residuals', ylabel='State', title='Residuals with 90% CI')
ax.set_yticks(y)
ax.set_yticklabels(dset.Loc.values[idx], fontsize=10);
ax.text(-2.8, -7, 'Residuals (with error-bars) from current model (in red). '
'Black marker \nshows residuals from the previous model (Model 3). '
'Measurement \nerror is indicated by orange bar.');
```
The plot above shows the residuals for each of the states, along with the measurement noise given by inner error bar. The gray dots are the mean residuals from our earlier Model 3. Notice how having an additional degree of freedom to model the measurement noise has shrunk the residuals. In particular, for Idaho and Maine, our predictions are now much closer to the observed values after incorporating measurement noise in the model.
To better see how measurement noise affects the movement of the regression line, let us plot the residuals with respect to the measurement noise.
```
fig, ax = plt.subplots(nrows=1, ncols=1, figsize=(10, 6))
x = dset.DivorceScaledSD.values
y1 = jnp.mean(residuals_3, 0)
y2 = jnp.mean(residuals_4, 0)
ax.plot(x, y1, ls='none', marker='o')
ax.plot(x, y2, ls='none', marker='o')
for i, (j, k) in enumerate(zip(y1, y2)):
ax.plot([x[i], x[i]], [j, k], '--', color='gray');
ax.set(xlabel='Measurement Noise', ylabel='Residual', title='Mean residuals (Model 4: red, Model 3: blue)');
```
The plot above shows what has happend in more detail - the regression line itself has moved to ensure a better fit for observations with low measurement noise (left of the plot) where the residuals have shrunk very close to 0. That is to say that data points with low measurement error have a concomitantly higher contribution in determining the regression line. On the other hand, for states with high measurement error (right of the plot), incorporating measurement noise allows us to move our posterior distribution mass closer to the observations resulting in a shrinkage of residuals as well.
## References
1. McElreath, R. (2016). Statistical Rethinking: A Bayesian Course with Examples in R and Stan CRC Press.
2. Stan Development Team. [Stan User's Guide](https://mc-stan.org/docs/2_19/stan-users-guide/index.html)
3. Goodman, N.D., and StuhlMueller, A. (2014). [The Design and Implementation of Probabilistic Programming Languages](http://dippl.org/)
4. Pyro Development Team. [Poutine: A Guide to Programming with Effect Handlers in Pyro](http://pyro.ai/examples/effect_handlers.html)
5. Hoffman, M.D., Gelman, A. (2011). The No-U-Turn Sampler: Adaptively Setting Path Lengths in Hamiltonian Monte Carlo.
6. Betancourt, M. (2017). A Conceptual Introduction to Hamiltonian Monte Carlo.
7. JAX Development Team (2018). [Composable transformations of Python+NumPy programs: differentiate, vectorize, JIT to GPU/TPU, and more](https://github.com/google/jax)
8. Gelman, A., Hwang, J., and Vehtari A. [Understanding predictive information criteria for Bayesian models](https://arxiv.org/pdf/1307.5928.pdf)
|
github_jupyter
|
!pip install -q numpyro@git+https://github.com/pyro-ppl/numpyro
import os
from IPython.display import set_matplotlib_formats
import jax.numpy as jnp
from jax import random, vmap
from jax.scipy.special import logsumexp
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import seaborn as sns
import numpyro
from numpyro.diagnostics import hpdi
import numpyro.distributions as dist
from numpyro import handlers
from numpyro.infer import MCMC, NUTS
plt.style.use('bmh')
if "NUMPYRO_SPHINXBUILD" in os.environ:
set_matplotlib_formats('svg')
assert numpyro.__version__.startswith('0.7.1')
DATASET_URL = 'https://raw.githubusercontent.com/rmcelreath/rethinking/master/data/WaffleDivorce.csv'
dset = pd.read_csv(DATASET_URL, sep=';')
dset
vars = ['Population', 'MedianAgeMarriage', 'Marriage', 'WaffleHouses', 'South', 'Divorce']
sns.pairplot(dset, x_vars=vars, y_vars=vars, palette='husl');
sns.regplot(x='WaffleHouses', y='Divorce', data=dset);
standardize = lambda x: (x - x.mean()) / x.std()
dset['AgeScaled'] = dset.MedianAgeMarriage.pipe(standardize)
dset['MarriageScaled'] = dset.Marriage.pipe(standardize)
dset['DivorceScaled'] = dset.Divorce.pipe(standardize)
def model(marriage=None, age=None, divorce=None):
a = numpyro.sample('a', dist.Normal(0., 0.2))
M, A = 0., 0.
if marriage is not None:
bM = numpyro.sample('bM', dist.Normal(0., 0.5))
M = bM * marriage
if age is not None:
bA = numpyro.sample('bA', dist.Normal(0., 0.5))
A = bA * age
sigma = numpyro.sample('sigma', dist.Exponential(1.))
mu = a + M + A
numpyro.sample('obs', dist.Normal(mu, sigma), obs=divorce)
# Start from this source of randomness. We will split keys for subsequent operations.
rng_key = random.PRNGKey(0)
rng_key, rng_key_ = random.split(rng_key)
# Run NUTS.
kernel = NUTS(model)
num_samples = 2000
mcmc = MCMC(kernel, num_warmup=1000, num_samples=num_samples)
mcmc.run(rng_key_, marriage=dset.MarriageScaled.values, divorce=dset.DivorceScaled.values)
mcmc.print_summary()
samples_1 = mcmc.get_samples()
def plot_regression(x, y_mean, y_hpdi):
# Sort values for plotting by x axis
idx = jnp.argsort(x)
marriage = x[idx]
mean = y_mean[idx]
hpdi = y_hpdi[:, idx]
divorce = dset.DivorceScaled.values[idx]
# Plot
fig, ax = plt.subplots(nrows=1, ncols=1, figsize=(6, 6))
ax.plot(marriage, mean)
ax.plot(marriage, divorce, 'o')
ax.fill_between(marriage, hpdi[0], hpdi[1], alpha=0.3, interpolate=True)
return ax
# Compute empirical posterior distribution over mu
posterior_mu = jnp.expand_dims(samples_1['a'], -1) + \
jnp.expand_dims(samples_1['bM'], -1) * dset.MarriageScaled.values
mean_mu = jnp.mean(posterior_mu, axis=0)
hpdi_mu = hpdi(posterior_mu, 0.9)
ax = plot_regression(dset.MarriageScaled.values, mean_mu, hpdi_mu)
ax.set(xlabel='Marriage rate', ylabel='Divorce rate', title='Regression line with 90% CI');
from numpyro.infer import Predictive
rng_key, rng_key_ = random.split(rng_key)
prior_predictive = Predictive(model, num_samples=100)
prior_predictions = prior_predictive(rng_key_, marriage=dset.MarriageScaled.values)['obs']
mean_prior_pred = jnp.mean(prior_predictions, axis=0)
hpdi_prior_pred = hpdi(prior_predictions, 0.9)
ax = plot_regression(dset.MarriageScaled.values, mean_prior_pred, hpdi_prior_pred)
ax.set(xlabel='Marriage rate', ylabel='Divorce rate', title='Predictions with 90% CI');
rng_key, rng_key_ = random.split(rng_key)
predictive = Predictive(model, samples_1)
predictions = predictive(rng_key_, marriage=dset.MarriageScaled.values)['obs']
df = dset.filter(['Location'])
df['Mean Predictions'] = jnp.mean(predictions, axis=0)
df.head()
def predict(rng_key, post_samples, model, *args, **kwargs):
model = handlers.seed(handlers.condition(model, post_samples), rng_key)
model_trace = handlers.trace(model).get_trace(*args, **kwargs)
return model_trace['obs']['value']
# vectorize predictions via vmap
predict_fn = vmap(lambda rng_key, samples: predict(rng_key, samples, model, marriage=dset.MarriageScaled.values))
# Using the same key as we used for Predictive - note that the results are identical.
predictions_1 = predict_fn(random.split(rng_key_, num_samples), samples_1)
mean_pred = jnp.mean(predictions_1, axis=0)
df = dset.filter(['Location'])
df['Mean Predictions'] = mean_pred
df.head()
hpdi_pred = hpdi(predictions_1, 0.9)
ax = plot_regression(dset.MarriageScaled.values, mean_pred, hpdi_pred)
ax.set(xlabel='Marriage rate', ylabel='Divorce rate', title='Predictions with 90% CI');
def log_likelihood(rng_key, params, model, *args, **kwargs):
model = handlers.condition(model, params)
model_trace = handlers.trace(model).get_trace(*args, **kwargs)
obs_node = model_trace['obs']
return obs_node['fn'].log_prob(obs_node['value'])
def log_pred_density(rng_key, params, model, *args, **kwargs):
n = list(params.values())[0].shape[0]
log_lk_fn = vmap(lambda rng_key, params: log_likelihood(rng_key, params, model, *args, **kwargs))
log_lk_vals = log_lk_fn(random.split(rng_key, n), params)
return (logsumexp(log_lk_vals, 0) - jnp.log(n)).sum()
rng_key, rng_key_ = random.split(rng_key)
print('Log posterior predictive density: {}'.format(log_pred_density(rng_key_,
samples_1,
model,
marriage=dset.MarriageScaled.values,
divorce=dset.DivorceScaled.values)))
rng_key, rng_key_ = random.split(rng_key)
mcmc.run(rng_key_, age=dset.AgeScaled.values, divorce=dset.DivorceScaled.values)
mcmc.print_summary()
samples_2 = mcmc.get_samples()
posterior_mu = jnp.expand_dims(samples_2['a'], -1) + \
jnp.expand_dims(samples_2['bA'], -1) * dset.AgeScaled.values
mean_mu = jnp.mean(posterior_mu, axis=0)
hpdi_mu = hpdi(posterior_mu, 0.9)
ax = plot_regression(dset.AgeScaled.values, mean_mu, hpdi_mu)
ax.set(xlabel='Median marriage age', ylabel='Divorce rate', title='Regression line with 90% CI');
rng_key, rng_key_ = random.split(rng_key)
predictions_2 = Predictive(model, samples_2)(rng_key_,
age=dset.AgeScaled.values)['obs']
mean_pred = jnp.mean(predictions_2, axis=0)
hpdi_pred = hpdi(predictions_2, 0.9)
ax = plot_regression(dset.AgeScaled.values, mean_pred, hpdi_pred)
ax.set(xlabel='Median Age', ylabel='Divorce rate', title='Predictions with 90% CI');
rng_key, rng_key_ = random.split(rng_key)
print('Log posterior predictive density: {}'.format(log_pred_density(rng_key_,
samples_2,
model,
age=dset.AgeScaled.values,
divorce=dset.DivorceScaled.values)))
rng_key, rng_key_ = random.split(rng_key)
mcmc.run(rng_key_, marriage=dset.MarriageScaled.values,
age=dset.AgeScaled.values, divorce=dset.DivorceScaled.values)
mcmc.print_summary()
samples_3 = mcmc.get_samples()
rng_key, rng_key_ = random.split(rng_key)
print('Log posterior predictive density: {}'.format(
log_pred_density(rng_key_,
samples_3,
model,
marriage=dset.MarriageScaled.values,
age=dset.AgeScaled.values,
divorce=dset.DivorceScaled.values)
))
# Predictions for Model 3.
rng_key, rng_key_ = random.split(rng_key)
predictions_3 = Predictive(model, samples_3)(rng_key_,
marriage=dset.MarriageScaled.values,
age=dset.AgeScaled.values)['obs']
y = jnp.arange(50)
fig, ax = plt.subplots(nrows=1, ncols=2, figsize=(12, 16))
pred_mean = jnp.mean(predictions_3, axis=0)
pred_hpdi = hpdi(predictions_3, 0.9)
residuals_3 = dset.DivorceScaled.values - predictions_3
residuals_mean = jnp.mean(residuals_3, axis=0)
residuals_hpdi = hpdi(residuals_3, 0.9)
idx = jnp.argsort(residuals_mean)
# Plot posterior predictive
ax[0].plot(jnp.zeros(50), y, '--')
ax[0].errorbar(pred_mean[idx], y, xerr=pred_hpdi[1, idx] - pred_mean[idx],
marker='o', ms=5, mew=4, ls='none', alpha=0.8)
ax[0].plot(dset.DivorceScaled.values[idx], y, marker='o',
ls='none', color='gray')
ax[0].set(xlabel='Posterior Predictive (red) vs. Actuals (gray)', ylabel='State',
title='Posterior Predictive with 90% CI')
ax[0].set_yticks(y)
ax[0].set_yticklabels(dset.Loc.values[idx], fontsize=10);
# Plot residuals
residuals_3 = dset.DivorceScaled.values - predictions_3
residuals_mean = jnp.mean(residuals_3, axis=0)
residuals_hpdi = hpdi(residuals_3, 0.9)
err = residuals_hpdi[1] - residuals_mean
ax[1].plot(jnp.zeros(50), y, '--')
ax[1].errorbar(residuals_mean[idx], y, xerr=err[idx],
marker='o', ms=5, mew=4, ls='none', alpha=0.8)
ax[1].set(xlabel='Residuals', ylabel='State', title='Residuals with 90% CI')
ax[1].set_yticks(y)
ax[1].set_yticklabels(dset.Loc.values[idx], fontsize=10);
def model_se(marriage, age, divorce_sd, divorce=None):
a = numpyro.sample('a', dist.Normal(0., 0.2))
bM = numpyro.sample('bM', dist.Normal(0., 0.5))
M = bM * marriage
bA = numpyro.sample('bA', dist.Normal(0., 0.5))
A = bA * age
sigma = numpyro.sample('sigma', dist.Exponential(1.))
mu = a + M + A
divorce_rate = numpyro.sample('divorce_rate', dist.Normal(mu, sigma))
numpyro.sample('obs', dist.Normal(divorce_rate, divorce_sd), obs=divorce)
# Standardize
dset['DivorceScaledSD'] = dset['Divorce SE'] / jnp.std(dset.Divorce.values)
rng_key, rng_key_ = random.split(rng_key)
kernel = NUTS(model_se, target_accept_prob=0.9)
mcmc = MCMC(kernel, num_warmup=1000, num_samples=3000)
mcmc.run(rng_key_, marriage=dset.MarriageScaled.values, age=dset.AgeScaled.values,
divorce_sd=dset.DivorceScaledSD.values, divorce=dset.DivorceScaled.values)
mcmc.print_summary()
samples_4 = mcmc.get_samples()
rng_key, rng_key_ = random.split(rng_key)
predictions_4 = Predictive(model_se, samples_4)(rng_key_,
marriage=dset.MarriageScaled.values,
age=dset.AgeScaled.values,
divorce_sd=dset.DivorceScaledSD.values)['obs']
sd = dset.DivorceScaledSD.values
residuals_4 = dset.DivorceScaled.values - predictions_4
residuals_mean = jnp.mean(residuals_4, axis=0)
residuals_hpdi = hpdi(residuals_4, 0.9)
err = residuals_hpdi[1] - residuals_mean
idx = jnp.argsort(residuals_mean)
y = jnp.arange(50)
fig, ax = plt.subplots(nrows=1, ncols=1, figsize=(6, 16))
# Plot Residuals
ax.plot(jnp.zeros(50), y, '--')
ax.errorbar(residuals_mean[idx], y, xerr=err[idx],
marker='o', ms=5, mew=4, ls='none', alpha=0.8)
# Plot SD
ax.errorbar(residuals_mean[idx], y, xerr=sd[idx],
ls='none', color='orange', alpha=0.9)
# Plot earlier mean residual
ax.plot(jnp.mean(dset.DivorceScaled.values - predictions_3, 0)[idx], y,
ls='none', marker='o', ms=6, color='black', alpha=0.6)
ax.set(xlabel='Residuals', ylabel='State', title='Residuals with 90% CI')
ax.set_yticks(y)
ax.set_yticklabels(dset.Loc.values[idx], fontsize=10);
ax.text(-2.8, -7, 'Residuals (with error-bars) from current model (in red). '
'Black marker \nshows residuals from the previous model (Model 3). '
'Measurement \nerror is indicated by orange bar.');
fig, ax = plt.subplots(nrows=1, ncols=1, figsize=(10, 6))
x = dset.DivorceScaledSD.values
y1 = jnp.mean(residuals_3, 0)
y2 = jnp.mean(residuals_4, 0)
ax.plot(x, y1, ls='none', marker='o')
ax.plot(x, y2, ls='none', marker='o')
for i, (j, k) in enumerate(zip(y1, y2)):
ax.plot([x[i], x[i]], [j, k], '--', color='gray');
ax.set(xlabel='Measurement Noise', ylabel='Residual', title='Mean residuals (Model 4: red, Model 3: blue)');
| 0.705988 | 0.990795 |
```
import sys
sys.path.append('..')
sys.path.append('../..')
from stats import *
from sentiment_stats import *
from peewee import SQL
from database.models import RawFacebookComments, RawTwitterComments, RawInstagramComments, RawYouTubeComments, RawHashtagComments
rede_social = 'Facebook'
modelo = RawFacebookComments
cores = ['#FFA726', '#66BB6A', '#42A5F5', '#FFEE58', '#EF5350', '#AB47BC', '#C8C8C8']
cores2 = ['#FFA726', '#AB47BC', '#FFEE58', '#C8C8C8', '#EF5350', '#66BB6A', '#42A5F5']
cores_val = ['#EF5350', '#C8C8C8', '#66BB6A']
cores_val2 = ['#66BB6A', '#EF5350', '#C8C8C8']
sentimentos = ['ALEGRIA', 'SURPRESA', 'TRISTEZA', 'MEDO', 'RAIVA', 'DESGOSTO', 'NEUTRO']
valencia = ['POSITIVO', 'NEGATIVO', 'NEUTRO']
valencia_dict = OrderedDict()
for val in valencia:
valencia_dict[val] = 0
sentimentos_dict = OrderedDict()
for sentimento in sentimentos:
sentimentos_dict[sentimento] = 0
default_clause = [
SQL('length(clean_comment) > 0'),
]
positivo_clause = [
SQL('length(emotion) > 0 AND length(valence) > 0'),
SQL('emotion in ("ALEGRIA", "SURPRESA") AND valence = "POSITIVO"')
]
negativo_clause = [
SQL('length(emotion) > 0 AND length(valence) > 0'),
SQL('emotion in ("TRISTEZA", "RAIVA", "MEDO", "DESGOSTO") AND valence = "NEGATIVO"')
]
neutro_clause = [
SQL('length(emotion) > 0 AND length(valence) > 0'),
SQL('emotion in ("NEUTRO") AND valence = "NEUTRO"')
]
general = default_clause + [
SQL('length(emotion) > 0 AND length(valence) > 0'),
SQL("""
(emotion in ("ALEGRIA", "SURPRESA") AND valence = "POSITIVO")
OR
(emotion in ("TRISTEZA", "RAIVA", "MEDO", "DESGOSTO") AND valence = "NEGATIVO")
OR
(emotion in ("NEUTRO") AND valence = "NEUTRO")
""")
]
```
### Emoções gerais dos comentários : Facebook
```
total_comentarios = modelo.select() \
.where(default_clause) \
.count()
comentarios_positivos = modelo.select() \
.where(reduce(operator.and_, default_clause + positivo_clause)) \
.order_by(modelo.timestamp)
comentarios_negativos = modelo.select() \
.where(reduce(operator.and_, default_clause + negativo_clause)) \
.order_by(modelo.timestamp)
comentarios_neutros = modelo.select() \
.where(reduce(operator.and_, default_clause + neutro_clause)) \
.order_by(modelo.timestamp)
comentarios = modelo.select() \
.where(reduce(operator.and_, general)) \
.order_by(modelo.timestamp)
alegria, surpresa, tristeza, medo, raiva, desgosto, positivo, negativo, neutro = load_emocoes_comentarios(comentarios_positivos, comentarios_negativos, comentarios_neutros)
print_statistics(rede_social, total_comentarios, comentarios_positivos, comentarios_negativos, comentarios_neutros)
```
#### Contagem total de comentários : Valência
```
graph_valence_total(rede_social, cores_val2, valencia, positivo, negativo, neutro)
```
#### Contagem total de comentários : Emoções
```
graph_sentimentos_total(rede_social, cores, sentimentos, alegria, surpresa, tristeza, medo, raiva, desgosto, neutro)
```
#### Comentários por data : Valência
```
graph_valencia_por_data(rede_social, cores_val, valencia_dict, comentarios)
```
#### Comentários por data : Emoções
```
graph_emocoes_por_data(rede_social, cores2, sentimentos_dict, comentarios)
```
### Emoções por candidato : Facebook
#### Jair Bolsonaro
```
candidato_c = [modelo.candidate == 'Jair Bolsonaro']
total_comentarios = modelo.select() \
.where(reduce(operator.and_, default_clause + candidato_c)) \
.count()
comentarios_positivos = modelo.select() \
.where(reduce(operator.and_, default_clause + positivo_clause + candidato_c)) \
.order_by(modelo.timestamp)
comentarios_negativos = modelo.select() \
.where(reduce(operator.and_, default_clause + negativo_clause + candidato_c)) \
.order_by(modelo.timestamp)
comentarios_neutros = modelo.select() \
.where(reduce(operator.and_, default_clause + neutro_clause + candidato_c)) \
.order_by(modelo.timestamp)
comentarios = modelo.select() \
.where(reduce(operator.and_, general + candidato_c)) \
.order_by(modelo.timestamp)
alegria, surpresa, tristeza, medo, raiva, desgosto, positivo, negativo, neutro = load_emocoes_comentarios(comentarios_positivos, comentarios_negativos, comentarios_neutros)
print_statistics(rede_social, total_comentarios, comentarios_positivos, comentarios_negativos, comentarios_neutros)
```
##### Contagem total de comentários : Valência
```
graph_valence_total(rede_social, cores_val2, valencia, positivo, negativo, neutro)
```
##### Contagem total de comentários : Emoções
```
graph_sentimentos_total(rede_social, cores, sentimentos, alegria, surpresa, tristeza, medo, raiva, desgosto, neutro)
```
##### Comentários por data : Valência
```
graph_valencia_por_data(rede_social, cores_val, valencia_dict, comentarios)
```
##### Comentários por data : Emoções
```
graph_emocoes_por_data(rede_social, cores2, sentimentos_dict, comentarios)
```
#### Fernando Haddad
```
candidato_c = [modelo.candidate == 'Fernando Haddad']
total_comentarios = modelo.select() \
.where(reduce(operator.and_, default_clause + candidato_c)) \
.count()
comentarios_positivos = modelo.select() \
.where(reduce(operator.and_, default_clause + positivo_clause + candidato_c)) \
.order_by(modelo.timestamp)
comentarios_negativos = modelo.select() \
.where(reduce(operator.and_, default_clause + negativo_clause + candidato_c)) \
.order_by(modelo.timestamp)
comentarios_neutros = modelo.select() \
.where(reduce(operator.and_, default_clause + neutro_clause + candidato_c)) \
.order_by(modelo.timestamp)
comentarios = modelo.select() \
.where(reduce(operator.and_, general + candidato_c)) \
.order_by(modelo.timestamp)
alegria, surpresa, tristeza, medo, raiva, desgosto, positivo, negativo, neutro = load_emocoes_comentarios(comentarios_positivos, comentarios_negativos, comentarios_neutros)
print_statistics(rede_social, total_comentarios, comentarios_positivos, comentarios_negativos, comentarios_neutros)
```
##### Contagem total de comentários : Valência
```
graph_valence_total(rede_social, cores_val2, valencia, positivo, negativo, neutro)
```
##### Contagem total de comentários : Emoções
```
graph_sentimentos_total(rede_social, cores, sentimentos, alegria, surpresa, tristeza, medo, raiva, desgosto, neutro)
```
##### Comentários por data : Valência
```
graph_valencia_por_data(rede_social, cores_val, valencia_dict, comentarios)
```
##### Comentários por data : Emoções
```
graph_emocoes_por_data(rede_social, cores2, sentimentos_dict, comentarios)
```
#### Ciro Gomes
```
candidato_c = [modelo.candidate == 'Ciro Gomes']
total_comentarios = modelo.select() \
.where(reduce(operator.and_, default_clause + candidato_c)) \
.count()
comentarios_positivos = modelo.select() \
.where(reduce(operator.and_, default_clause + positivo_clause + candidato_c)) \
.order_by(modelo.timestamp)
comentarios_negativos = modelo.select() \
.where(reduce(operator.and_, default_clause + negativo_clause + candidato_c)) \
.order_by(modelo.timestamp)
comentarios_neutros = modelo.select() \
.where(reduce(operator.and_, default_clause + neutro_clause + candidato_c)) \
.order_by(modelo.timestamp)
comentarios = modelo.select() \
.where(reduce(operator.and_, general + candidato_c)) \
.order_by(modelo.timestamp)
alegria, surpresa, tristeza, medo, raiva, desgosto, positivo, negativo, neutro = load_emocoes_comentarios(comentarios_positivos, comentarios_negativos, comentarios_neutros)
print_statistics(rede_social, total_comentarios, comentarios_positivos, comentarios_negativos, comentarios_neutros)
```
##### Contagem total de comentários : Valência
```
graph_valence_total(rede_social, cores_val2, valencia, positivo, negativo, neutro)
```
##### Contagem total de comentários : Emoções
```
graph_sentimentos_total(rede_social, cores, sentimentos, alegria, surpresa, tristeza, medo, raiva, desgosto, neutro)
```
##### Comentários por data : Valência
```
graph_valencia_por_data(rede_social, cores_val, valencia_dict, comentarios)
```
##### Comentários por data : Emoções
```
graph_emocoes_por_data(rede_social, cores2, sentimentos_dict, comentarios)
```
#### Geraldo Alckmin
```
candidato_c = [modelo.candidate == 'Geraldo Alckmin']
total_comentarios = modelo.select() \
.where(reduce(operator.and_, default_clause + candidato_c)) \
.count()
comentarios_positivos = modelo.select() \
.where(reduce(operator.and_, default_clause + positivo_clause + candidato_c)) \
.order_by(modelo.timestamp)
comentarios_negativos = modelo.select() \
.where(reduce(operator.and_, default_clause + negativo_clause + candidato_c)) \
.order_by(modelo.timestamp)
comentarios_neutros = modelo.select() \
.where(reduce(operator.and_, default_clause + neutro_clause + candidato_c)) \
.order_by(modelo.timestamp)
comentarios = modelo.select() \
.where(reduce(operator.and_, general + candidato_c)) \
.order_by(modelo.timestamp)
alegria, surpresa, tristeza, medo, raiva, desgosto, positivo, negativo, neutro = load_emocoes_comentarios(comentarios_positivos, comentarios_negativos, comentarios_neutros)
print_statistics(rede_social, total_comentarios, comentarios_positivos, comentarios_negativos, comentarios_neutros)
```
##### Contagem total de comentários : Valência
```
graph_valence_total(rede_social, cores_val2, valencia, positivo, negativo, neutro)
```
##### Contagem total de comentários : Emoções
```
graph_sentimentos_total(rede_social, cores, sentimentos, alegria, surpresa, tristeza, medo, raiva, desgosto, neutro)
```
##### Comentários por data : Valência
```
graph_valencia_por_data(rede_social, cores_val, valencia_dict, comentarios)
```
##### Comentários por data : Emoções
```
graph_emocoes_por_data(rede_social, cores2, sentimentos_dict, comentarios)
```
#### Marina Silva
```
candidato_c = [modelo.candidate == 'Marina Silva']
total_comentarios = modelo.select() \
.where(reduce(operator.and_, default_clause + candidato_c)) \
.count()
comentarios_positivos = modelo.select() \
.where(reduce(operator.and_, default_clause + positivo_clause + candidato_c)) \
.order_by(modelo.timestamp)
comentarios_negativos = modelo.select() \
.where(reduce(operator.and_, default_clause + negativo_clause + candidato_c)) \
.order_by(modelo.timestamp)
comentarios_neutros = modelo.select() \
.where(reduce(operator.and_, default_clause + neutro_clause + candidato_c)) \
.order_by(modelo.timestamp)
comentarios = modelo.select() \
.where(reduce(operator.and_, general + candidato_c)) \
.order_by(modelo.timestamp)
alegria, surpresa, tristeza, medo, raiva, desgosto, positivo, negativo, neutro = load_emocoes_comentarios(comentarios_positivos, comentarios_negativos, comentarios_neutros)
print_statistics(rede_social, total_comentarios, comentarios_positivos, comentarios_negativos, comentarios_neutros)
```
##### Contagem total de comentários : Valência
```
graph_valence_total(rede_social, cores_val2, valencia, positivo, negativo, neutro)
```
##### Contagem total de comentários : Emoções
```
graph_sentimentos_total(rede_social, cores, sentimentos, alegria, surpresa, tristeza, medo, raiva, desgosto, neutro)
```
##### Comentários por data : Valência
```
graph_valencia_por_data(rede_social, cores_val, valencia_dict, comentarios)
```
##### Comentários por data : Emoções
```
graph_emocoes_por_data(rede_social, cores2, sentimentos_dict, comentarios)
```
|
github_jupyter
|
import sys
sys.path.append('..')
sys.path.append('../..')
from stats import *
from sentiment_stats import *
from peewee import SQL
from database.models import RawFacebookComments, RawTwitterComments, RawInstagramComments, RawYouTubeComments, RawHashtagComments
rede_social = 'Facebook'
modelo = RawFacebookComments
cores = ['#FFA726', '#66BB6A', '#42A5F5', '#FFEE58', '#EF5350', '#AB47BC', '#C8C8C8']
cores2 = ['#FFA726', '#AB47BC', '#FFEE58', '#C8C8C8', '#EF5350', '#66BB6A', '#42A5F5']
cores_val = ['#EF5350', '#C8C8C8', '#66BB6A']
cores_val2 = ['#66BB6A', '#EF5350', '#C8C8C8']
sentimentos = ['ALEGRIA', 'SURPRESA', 'TRISTEZA', 'MEDO', 'RAIVA', 'DESGOSTO', 'NEUTRO']
valencia = ['POSITIVO', 'NEGATIVO', 'NEUTRO']
valencia_dict = OrderedDict()
for val in valencia:
valencia_dict[val] = 0
sentimentos_dict = OrderedDict()
for sentimento in sentimentos:
sentimentos_dict[sentimento] = 0
default_clause = [
SQL('length(clean_comment) > 0'),
]
positivo_clause = [
SQL('length(emotion) > 0 AND length(valence) > 0'),
SQL('emotion in ("ALEGRIA", "SURPRESA") AND valence = "POSITIVO"')
]
negativo_clause = [
SQL('length(emotion) > 0 AND length(valence) > 0'),
SQL('emotion in ("TRISTEZA", "RAIVA", "MEDO", "DESGOSTO") AND valence = "NEGATIVO"')
]
neutro_clause = [
SQL('length(emotion) > 0 AND length(valence) > 0'),
SQL('emotion in ("NEUTRO") AND valence = "NEUTRO"')
]
general = default_clause + [
SQL('length(emotion) > 0 AND length(valence) > 0'),
SQL("""
(emotion in ("ALEGRIA", "SURPRESA") AND valence = "POSITIVO")
OR
(emotion in ("TRISTEZA", "RAIVA", "MEDO", "DESGOSTO") AND valence = "NEGATIVO")
OR
(emotion in ("NEUTRO") AND valence = "NEUTRO")
""")
]
total_comentarios = modelo.select() \
.where(default_clause) \
.count()
comentarios_positivos = modelo.select() \
.where(reduce(operator.and_, default_clause + positivo_clause)) \
.order_by(modelo.timestamp)
comentarios_negativos = modelo.select() \
.where(reduce(operator.and_, default_clause + negativo_clause)) \
.order_by(modelo.timestamp)
comentarios_neutros = modelo.select() \
.where(reduce(operator.and_, default_clause + neutro_clause)) \
.order_by(modelo.timestamp)
comentarios = modelo.select() \
.where(reduce(operator.and_, general)) \
.order_by(modelo.timestamp)
alegria, surpresa, tristeza, medo, raiva, desgosto, positivo, negativo, neutro = load_emocoes_comentarios(comentarios_positivos, comentarios_negativos, comentarios_neutros)
print_statistics(rede_social, total_comentarios, comentarios_positivos, comentarios_negativos, comentarios_neutros)
graph_valence_total(rede_social, cores_val2, valencia, positivo, negativo, neutro)
graph_sentimentos_total(rede_social, cores, sentimentos, alegria, surpresa, tristeza, medo, raiva, desgosto, neutro)
graph_valencia_por_data(rede_social, cores_val, valencia_dict, comentarios)
graph_emocoes_por_data(rede_social, cores2, sentimentos_dict, comentarios)
candidato_c = [modelo.candidate == 'Jair Bolsonaro']
total_comentarios = modelo.select() \
.where(reduce(operator.and_, default_clause + candidato_c)) \
.count()
comentarios_positivos = modelo.select() \
.where(reduce(operator.and_, default_clause + positivo_clause + candidato_c)) \
.order_by(modelo.timestamp)
comentarios_negativos = modelo.select() \
.where(reduce(operator.and_, default_clause + negativo_clause + candidato_c)) \
.order_by(modelo.timestamp)
comentarios_neutros = modelo.select() \
.where(reduce(operator.and_, default_clause + neutro_clause + candidato_c)) \
.order_by(modelo.timestamp)
comentarios = modelo.select() \
.where(reduce(operator.and_, general + candidato_c)) \
.order_by(modelo.timestamp)
alegria, surpresa, tristeza, medo, raiva, desgosto, positivo, negativo, neutro = load_emocoes_comentarios(comentarios_positivos, comentarios_negativos, comentarios_neutros)
print_statistics(rede_social, total_comentarios, comentarios_positivos, comentarios_negativos, comentarios_neutros)
graph_valence_total(rede_social, cores_val2, valencia, positivo, negativo, neutro)
graph_sentimentos_total(rede_social, cores, sentimentos, alegria, surpresa, tristeza, medo, raiva, desgosto, neutro)
graph_valencia_por_data(rede_social, cores_val, valencia_dict, comentarios)
graph_emocoes_por_data(rede_social, cores2, sentimentos_dict, comentarios)
candidato_c = [modelo.candidate == 'Fernando Haddad']
total_comentarios = modelo.select() \
.where(reduce(operator.and_, default_clause + candidato_c)) \
.count()
comentarios_positivos = modelo.select() \
.where(reduce(operator.and_, default_clause + positivo_clause + candidato_c)) \
.order_by(modelo.timestamp)
comentarios_negativos = modelo.select() \
.where(reduce(operator.and_, default_clause + negativo_clause + candidato_c)) \
.order_by(modelo.timestamp)
comentarios_neutros = modelo.select() \
.where(reduce(operator.and_, default_clause + neutro_clause + candidato_c)) \
.order_by(modelo.timestamp)
comentarios = modelo.select() \
.where(reduce(operator.and_, general + candidato_c)) \
.order_by(modelo.timestamp)
alegria, surpresa, tristeza, medo, raiva, desgosto, positivo, negativo, neutro = load_emocoes_comentarios(comentarios_positivos, comentarios_negativos, comentarios_neutros)
print_statistics(rede_social, total_comentarios, comentarios_positivos, comentarios_negativos, comentarios_neutros)
graph_valence_total(rede_social, cores_val2, valencia, positivo, negativo, neutro)
graph_sentimentos_total(rede_social, cores, sentimentos, alegria, surpresa, tristeza, medo, raiva, desgosto, neutro)
graph_valencia_por_data(rede_social, cores_val, valencia_dict, comentarios)
graph_emocoes_por_data(rede_social, cores2, sentimentos_dict, comentarios)
candidato_c = [modelo.candidate == 'Ciro Gomes']
total_comentarios = modelo.select() \
.where(reduce(operator.and_, default_clause + candidato_c)) \
.count()
comentarios_positivos = modelo.select() \
.where(reduce(operator.and_, default_clause + positivo_clause + candidato_c)) \
.order_by(modelo.timestamp)
comentarios_negativos = modelo.select() \
.where(reduce(operator.and_, default_clause + negativo_clause + candidato_c)) \
.order_by(modelo.timestamp)
comentarios_neutros = modelo.select() \
.where(reduce(operator.and_, default_clause + neutro_clause + candidato_c)) \
.order_by(modelo.timestamp)
comentarios = modelo.select() \
.where(reduce(operator.and_, general + candidato_c)) \
.order_by(modelo.timestamp)
alegria, surpresa, tristeza, medo, raiva, desgosto, positivo, negativo, neutro = load_emocoes_comentarios(comentarios_positivos, comentarios_negativos, comentarios_neutros)
print_statistics(rede_social, total_comentarios, comentarios_positivos, comentarios_negativos, comentarios_neutros)
graph_valence_total(rede_social, cores_val2, valencia, positivo, negativo, neutro)
graph_sentimentos_total(rede_social, cores, sentimentos, alegria, surpresa, tristeza, medo, raiva, desgosto, neutro)
graph_valencia_por_data(rede_social, cores_val, valencia_dict, comentarios)
graph_emocoes_por_data(rede_social, cores2, sentimentos_dict, comentarios)
candidato_c = [modelo.candidate == 'Geraldo Alckmin']
total_comentarios = modelo.select() \
.where(reduce(operator.and_, default_clause + candidato_c)) \
.count()
comentarios_positivos = modelo.select() \
.where(reduce(operator.and_, default_clause + positivo_clause + candidato_c)) \
.order_by(modelo.timestamp)
comentarios_negativos = modelo.select() \
.where(reduce(operator.and_, default_clause + negativo_clause + candidato_c)) \
.order_by(modelo.timestamp)
comentarios_neutros = modelo.select() \
.where(reduce(operator.and_, default_clause + neutro_clause + candidato_c)) \
.order_by(modelo.timestamp)
comentarios = modelo.select() \
.where(reduce(operator.and_, general + candidato_c)) \
.order_by(modelo.timestamp)
alegria, surpresa, tristeza, medo, raiva, desgosto, positivo, negativo, neutro = load_emocoes_comentarios(comentarios_positivos, comentarios_negativos, comentarios_neutros)
print_statistics(rede_social, total_comentarios, comentarios_positivos, comentarios_negativos, comentarios_neutros)
graph_valence_total(rede_social, cores_val2, valencia, positivo, negativo, neutro)
graph_sentimentos_total(rede_social, cores, sentimentos, alegria, surpresa, tristeza, medo, raiva, desgosto, neutro)
graph_valencia_por_data(rede_social, cores_val, valencia_dict, comentarios)
graph_emocoes_por_data(rede_social, cores2, sentimentos_dict, comentarios)
candidato_c = [modelo.candidate == 'Marina Silva']
total_comentarios = modelo.select() \
.where(reduce(operator.and_, default_clause + candidato_c)) \
.count()
comentarios_positivos = modelo.select() \
.where(reduce(operator.and_, default_clause + positivo_clause + candidato_c)) \
.order_by(modelo.timestamp)
comentarios_negativos = modelo.select() \
.where(reduce(operator.and_, default_clause + negativo_clause + candidato_c)) \
.order_by(modelo.timestamp)
comentarios_neutros = modelo.select() \
.where(reduce(operator.and_, default_clause + neutro_clause + candidato_c)) \
.order_by(modelo.timestamp)
comentarios = modelo.select() \
.where(reduce(operator.and_, general + candidato_c)) \
.order_by(modelo.timestamp)
alegria, surpresa, tristeza, medo, raiva, desgosto, positivo, negativo, neutro = load_emocoes_comentarios(comentarios_positivos, comentarios_negativos, comentarios_neutros)
print_statistics(rede_social, total_comentarios, comentarios_positivos, comentarios_negativos, comentarios_neutros)
graph_valence_total(rede_social, cores_val2, valencia, positivo, negativo, neutro)
graph_sentimentos_total(rede_social, cores, sentimentos, alegria, surpresa, tristeza, medo, raiva, desgosto, neutro)
graph_valencia_por_data(rede_social, cores_val, valencia_dict, comentarios)
graph_emocoes_por_data(rede_social, cores2, sentimentos_dict, comentarios)
| 0.187393 | 0.701112 |
# COVID-19 Deaths Per Capita
> Comparing death rates adjusting for population size.
- comments: true
- author: Joao B. Duarte & Hamel Husain
- categories: [growth, compare, interactive]
- hide: false
- image: images/covid-permillion-trajectories.png
- permalink: /covid-compare-permillion/
```
#hide
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import altair as alt
%config InlineBackend.figure_format = 'retina'
chart_width = 550
chart_height= 400
```
## Deaths Per Million Of Inhabitants
Since reaching at least 1 death per million
> Tip: Click (Shift+ for multiple) on countries in the legend to filter the visualization.
```
#hide
data = pd.read_csv("https://raw.githubusercontent.com/CSSEGISandData/COVID-19/master/csse_covid_19_data/csse_covid_19_time_series/time_series_covid19_deaths_global.csv",
error_bad_lines=False)
data = data.drop(columns=["Lat", "Long"])
data = data.melt(id_vars= ["Province/State", "Country/Region"])
data = pd.DataFrame(data.groupby(['Country/Region', "variable"]).sum())
data.reset_index(inplace=True)
data = data.rename(columns={"Country/Region": "location", "variable": "date", "value": "total_cases"})
data['date'] =pd.to_datetime(data.date)
data = data.sort_values(by = "date")
data.loc[data.location == "US","location"] = "United States"
data.loc[data.location == "Korea, South","location"] = "South Korea"
data_pwt = pd.read_stata("https://www.rug.nl/ggdc/docs/pwt91.dta")
filter1 = data_pwt["year"] == 2017
data_pop = data_pwt[filter1]
data_pop = data_pop[["country","pop"]]
data_pop.loc[data_pop.country == "Republic of Korea","country"] = "South Korea"
data_pop.loc[data_pop.country == "Iran (Islamic Republic of)","country"] = "Iran"
# per habitant
data_pc = data.copy()
countries = ["China", "Italy", "Spain", "France", "United Kingdom", "Germany",
"Portugal", "United States", "Singapore", "South Korea", "Japan",
"Brazil", "Iran", 'Netherlands', 'Belgium', 'Sweden',
'Switzerland', 'Norway', 'Denmark', 'Austria', 'Slovenia', 'Greece',
'Cyprus']
data_countries = []
data_countries_pc = []
# compute per habitant
for i in countries:
data_pc.loc[data_pc.location == i,"total_cases"] = data_pc.loc[data_pc.location == i,"total_cases"]/float(data_pop.loc[data_pop.country == i, "pop"])
# get each country time series
filter1 = data_pc["total_cases"] > 1
for i in countries:
filter_country = data_pc["location"]== i
data_countries_pc.append(data_pc[filter_country & filter1])
#hide_input
# Stack data to get it to Altair dataframe format
data_countries_pc2 = data_countries_pc.copy()
for i in range(0,len(countries)):
data_countries_pc2[i] = data_countries_pc2[i].reset_index()
data_countries_pc2[i]['n_days'] = data_countries_pc2[i].index
data_countries_pc2[i]['log_cases'] = np.log(data_countries_pc2[i]["total_cases"])
data_plot = data_countries_pc2[0]
for i in range(1, len(countries)):
data_plot = pd.concat([data_plot, data_countries_pc2[i]], axis=0)
data_plot["trend_2days"] = np.log(2)/2*data_plot["n_days"]
data_plot["trend_4days"] = np.log(2)/4*data_plot["n_days"]
data_plot["trend_12days"] = np.log(2)/12*data_plot["n_days"]
data_plot["trend_2days_label"] = "Doubles every 2 days"
data_plot["trend_4days_label"] = "Doubles evey 4 days"
data_plot["trend_12days_label"] = "Doubles every 12 days"
# Plot it using Altair
source = data_plot
scales = alt.selection_interval(bind='scales')
selection = alt.selection_multi(fields=['location'], bind='legend')
base = alt.Chart(source, title = "COVID-19 Deaths Per Million of Inhabitants").encode(
x = alt.X('n_days:Q', title = "Days passed since reaching 1 death per million"),
y = alt.Y("log_cases:Q",title = "Log of deaths per million"),
color = alt.Color('location:N', legend=alt.Legend(title="Country", labelFontSize=15, titleFontSize=17),
scale=alt.Scale(scheme='tableau20')),
opacity = alt.condition(selection, alt.value(1), alt.value(0.1))
)
lines = base.mark_line().add_selection(
scales
).add_selection(
selection
).properties(
width=chart_width,
height=chart_height
)
trend_2d = alt.Chart(source).encode(
x = "n_days:Q",
y = alt.Y("trend_2days:Q", scale=alt.Scale(domain=(0, max(data_plot["log_cases"])))),
).mark_line(color="grey", strokeDash=[3,3])
labels = pd.DataFrame([{'label': 'Doubles every 2 days', 'x_coord': 6, 'y_coord': 4},
{'label': 'Doubles every 4 days', 'x_coord': 16, 'y_coord': 3.5},
{'label': 'Doubles every 12 days', 'x_coord': 25, 'y_coord': 1.8},
])
trend_label = (alt.Chart(labels)
.mark_text(align='left', dx=-55, dy=-15, fontSize=12, color="grey")
.encode(x='x_coord:Q',
y='y_coord:Q',
text='label:N')
)
trend_4d = alt.Chart(source).mark_line(color="grey", strokeDash=[3,3]).encode(
x = "n_days:Q",
y = alt.Y("trend_4days:Q", scale=alt.Scale(domain=(0, max(data_plot["log_cases"])))),
)
trend_12d = alt.Chart(source).mark_line(color="grey", strokeDash=[3,3]).encode(
x = "n_days:Q",
y = alt.Y("trend_12days:Q", scale=alt.Scale(domain=(0, max(data_plot["log_cases"])))),
)
plot1= (
(trend_2d + trend_4d + trend_12d + trend_label + lines)
.configure_title(fontSize=20)
.configure_axis(labelFontSize=15,titleFontSize=18)
)
plot1.save(("../images/covid-permillion-trajectories.png"))
plot1
```
Last Available Total Deaths By Country:
```
#hide_input
label = 'Deaths'
temp = pd.concat([x.copy() for x in data_countries_pc]).loc[lambda x: x.date >= '3/1/2020']
metric_name = f'{label} per Million'
temp.columns = ['Country', 'date', metric_name]
# temp.loc[:, 'month'] = temp.date.dt.strftime('%Y-%m')
temp.loc[:, f'Log of {label} per Million'] = temp[f'{label} per Million'].apply(lambda x: np.log(x))
temp.groupby('Country').last()
#hide
# Get data and clean it
data = pd.read_csv("https://raw.githubusercontent.com/CSSEGISandData/COVID-19/master/csse_covid_19_data/csse_covid_19_time_series/time_series_covid19_confirmed_global.csv", error_bad_lines=False)
data = data.drop(columns=["Lat", "Long"])
data = data.melt(id_vars= ["Province/State", "Country/Region"])
data = pd.DataFrame(data.groupby(['Country/Region', "variable"]).sum())
data.reset_index(inplace=True)
data = data.rename(columns={"Country/Region": "location", "variable": "date", "value": "total_cases"})
data['date'] =pd.to_datetime(data.date)
data = data.sort_values(by = "date")
data.loc[data.location == "US","location"] = "United States"
data.loc[data.location == "Korea, South","location"] = "South Korea"
# Population data (last year is 2017 which is what we use)
data_pwt = pd.read_stata("https://www.rug.nl/ggdc/docs/pwt91.dta")
filter1 = data_pwt["year"] == 2017
data_pop = data_pwt[filter1]
data_pop = data_pop[["country","pop"]]
data_pop.loc[data_pop.country == "Republic of Korea","country"] = "South Korea"
data_pop.loc[data_pop.country == "Iran (Islamic Republic of)","country"] = "Iran"
# per habitant
data_pc = data.copy()
# I can add more countries if needed
countries = ["China", "Italy", "Spain", "France", "United Kingdom", "Germany",
"Portugal", "United States", "Singapore","South Korea", "Japan",
"Brazil","Iran"]
data_countries = []
data_countries_pc = []
# compute per habitant
for i in countries:
data_pc.loc[data_pc.location == i,"total_cases"] = data_pc.loc[data_pc.location == i,"total_cases"]/float(data_pop.loc[data_pop.country == i, "pop"])
# get each country time series
filter1 = data_pc["total_cases"] > 1
for i in countries:
filter_country = data_pc["location"]== i
data_countries_pc.append(data_pc[filter_country & filter1])
```
## Appendix
> Warning: The following chart, "Cases Per Million of Habitants" is biased depending on how widely a country administers tests. Please read with caution.
### Cases Per Million of Habitants
```
#hide_input
# Stack data to get it to Altair dataframe format
data_countries_pc2 = data_countries_pc.copy()
for i in range(0,len(countries)):
data_countries_pc2[i] = data_countries_pc2[i].reset_index()
data_countries_pc2[i]['n_days'] = data_countries_pc2[i].index
data_countries_pc2[i]['log_cases'] = np.log(data_countries_pc2[i]["total_cases"])
data_plot = data_countries_pc2[0]
for i in range(1, len(countries)):
data_plot = pd.concat([data_plot, data_countries_pc2[i]], axis=0)
data_plot["trend_2days"] = np.log(2)/2*data_plot["n_days"]
data_plot["trend_4days"] = np.log(2)/4*data_plot["n_days"]
data_plot["trend_12days"] = np.log(2)/12*data_plot["n_days"]
data_plot["trend_2days_label"] = "Doubles every 2 days"
data_plot["trend_4days_label"] = "Doubles evey 4 days"
data_plot["trend_12days_label"] = "Doubles every 12 days"
# Plot it using Altair
source = data_plot
scales = alt.selection_interval(bind='scales')
selection = alt.selection_multi(fields=['location'], bind='legend')
base = alt.Chart(source, title = "COVID-19 Confirmed Cases Per Million of Inhabitants").encode(
x = alt.X('n_days:Q', title = "Days passed since reaching 1 case per million"),
y = alt.Y("log_cases:Q",title = "Log of confirmed cases per million"),
color = alt.Color('location:N', legend=alt.Legend(title="Country", labelFontSize=15, titleFontSize=17),
scale=alt.Scale(scheme='tableau20')),
opacity = alt.condition(selection, alt.value(1), alt.value(0.1))
).properties(
width=chart_width,
height=chart_height
)
lines = base.mark_line().add_selection(
scales
).add_selection(
selection
)
trend_2d = alt.Chart(source).encode(
x = "n_days:Q",
y = alt.Y("trend_2days:Q", scale=alt.Scale(domain=(0, max(data_plot["log_cases"])))),
).mark_line( strokeDash=[3,3], color="grey")
labels = pd.DataFrame([{'label': 'Doubles every 2 days', 'x_coord': 10, 'y_coord': 6},
{'label': 'Doubles every 4 days', 'x_coord': 28, 'y_coord': 6},
{'label': 'Doubles every 12 days', 'x_coord': 45, 'y_coord': 3},
])
trend_label = (alt.Chart(labels)
.mark_text(align='left', dx=-55, dy=-15, fontSize=12, color="grey")
.encode(x='x_coord:Q',
y='y_coord:Q',
text='label:N')
)
trend_4d = alt.Chart(source).mark_line(color="grey", strokeDash=[3,3]).encode(
x = "n_days:Q",
y = alt.Y("trend_4days:Q", scale=alt.Scale(domain=(0, max(data_plot["log_cases"])))),
)
trend_12d = alt.Chart(source).mark_line(color="grey", strokeDash=[3,3]).encode(
x = "n_days:Q",
y = alt.Y("trend_12days:Q", scale=alt.Scale(domain=(0, max(data_plot["log_cases"])))),
)
(
(trend_2d + trend_4d + trend_12d + trend_label + lines)
.configure_title(fontSize=20)
.configure_axis(labelFontSize=15,titleFontSize=18)
)
```
Last Available Cases Per Million By Country:
```
#hide_input
label = 'Cases'
temp = pd.concat([x.copy() for x in data_countries_pc]).loc[lambda x: x.date >= '3/1/2020']
metric_name = f'{label} per Million'
temp.columns = ['Country', 'date', metric_name]
# temp.loc[:, 'month'] = temp.date.dt.strftime('%Y-%m')
temp.loc[:, f'Log of {label} per Million'] = temp[f'{label} per Million'].apply(lambda x: np.log(x))
temp.groupby('Country').last()
```
This analysis was conducted by [Joao B. Duarte](https://www.jbduarte.com). Assitance with creating visualizations were provided by [Hamel Husain](https://twitter.com/HamelHusain). Relevant sources are listed below:
1. ["2019 Novel Coronavirus COVID-19 (2019-nCoV) Data Repository by Johns Hopkins CSSE"](https://systems.jhu.edu/research/public-health/ncov/) [GitHub repository](https://github.com/CSSEGISandData/COVID-19).
2. [Feenstra, Robert C., Robert Inklaar and Marcel P. Timmer (2015), "The Next Generation of the Penn World Table" American Economic Review, 105(10), 3150-3182](https://www.rug.nl/ggdc/productivity/pwt/related-research)
|
github_jupyter
|
#hide
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import altair as alt
%config InlineBackend.figure_format = 'retina'
chart_width = 550
chart_height= 400
#hide
data = pd.read_csv("https://raw.githubusercontent.com/CSSEGISandData/COVID-19/master/csse_covid_19_data/csse_covid_19_time_series/time_series_covid19_deaths_global.csv",
error_bad_lines=False)
data = data.drop(columns=["Lat", "Long"])
data = data.melt(id_vars= ["Province/State", "Country/Region"])
data = pd.DataFrame(data.groupby(['Country/Region', "variable"]).sum())
data.reset_index(inplace=True)
data = data.rename(columns={"Country/Region": "location", "variable": "date", "value": "total_cases"})
data['date'] =pd.to_datetime(data.date)
data = data.sort_values(by = "date")
data.loc[data.location == "US","location"] = "United States"
data.loc[data.location == "Korea, South","location"] = "South Korea"
data_pwt = pd.read_stata("https://www.rug.nl/ggdc/docs/pwt91.dta")
filter1 = data_pwt["year"] == 2017
data_pop = data_pwt[filter1]
data_pop = data_pop[["country","pop"]]
data_pop.loc[data_pop.country == "Republic of Korea","country"] = "South Korea"
data_pop.loc[data_pop.country == "Iran (Islamic Republic of)","country"] = "Iran"
# per habitant
data_pc = data.copy()
countries = ["China", "Italy", "Spain", "France", "United Kingdom", "Germany",
"Portugal", "United States", "Singapore", "South Korea", "Japan",
"Brazil", "Iran", 'Netherlands', 'Belgium', 'Sweden',
'Switzerland', 'Norway', 'Denmark', 'Austria', 'Slovenia', 'Greece',
'Cyprus']
data_countries = []
data_countries_pc = []
# compute per habitant
for i in countries:
data_pc.loc[data_pc.location == i,"total_cases"] = data_pc.loc[data_pc.location == i,"total_cases"]/float(data_pop.loc[data_pop.country == i, "pop"])
# get each country time series
filter1 = data_pc["total_cases"] > 1
for i in countries:
filter_country = data_pc["location"]== i
data_countries_pc.append(data_pc[filter_country & filter1])
#hide_input
# Stack data to get it to Altair dataframe format
data_countries_pc2 = data_countries_pc.copy()
for i in range(0,len(countries)):
data_countries_pc2[i] = data_countries_pc2[i].reset_index()
data_countries_pc2[i]['n_days'] = data_countries_pc2[i].index
data_countries_pc2[i]['log_cases'] = np.log(data_countries_pc2[i]["total_cases"])
data_plot = data_countries_pc2[0]
for i in range(1, len(countries)):
data_plot = pd.concat([data_plot, data_countries_pc2[i]], axis=0)
data_plot["trend_2days"] = np.log(2)/2*data_plot["n_days"]
data_plot["trend_4days"] = np.log(2)/4*data_plot["n_days"]
data_plot["trend_12days"] = np.log(2)/12*data_plot["n_days"]
data_plot["trend_2days_label"] = "Doubles every 2 days"
data_plot["trend_4days_label"] = "Doubles evey 4 days"
data_plot["trend_12days_label"] = "Doubles every 12 days"
# Plot it using Altair
source = data_plot
scales = alt.selection_interval(bind='scales')
selection = alt.selection_multi(fields=['location'], bind='legend')
base = alt.Chart(source, title = "COVID-19 Deaths Per Million of Inhabitants").encode(
x = alt.X('n_days:Q', title = "Days passed since reaching 1 death per million"),
y = alt.Y("log_cases:Q",title = "Log of deaths per million"),
color = alt.Color('location:N', legend=alt.Legend(title="Country", labelFontSize=15, titleFontSize=17),
scale=alt.Scale(scheme='tableau20')),
opacity = alt.condition(selection, alt.value(1), alt.value(0.1))
)
lines = base.mark_line().add_selection(
scales
).add_selection(
selection
).properties(
width=chart_width,
height=chart_height
)
trend_2d = alt.Chart(source).encode(
x = "n_days:Q",
y = alt.Y("trend_2days:Q", scale=alt.Scale(domain=(0, max(data_plot["log_cases"])))),
).mark_line(color="grey", strokeDash=[3,3])
labels = pd.DataFrame([{'label': 'Doubles every 2 days', 'x_coord': 6, 'y_coord': 4},
{'label': 'Doubles every 4 days', 'x_coord': 16, 'y_coord': 3.5},
{'label': 'Doubles every 12 days', 'x_coord': 25, 'y_coord': 1.8},
])
trend_label = (alt.Chart(labels)
.mark_text(align='left', dx=-55, dy=-15, fontSize=12, color="grey")
.encode(x='x_coord:Q',
y='y_coord:Q',
text='label:N')
)
trend_4d = alt.Chart(source).mark_line(color="grey", strokeDash=[3,3]).encode(
x = "n_days:Q",
y = alt.Y("trend_4days:Q", scale=alt.Scale(domain=(0, max(data_plot["log_cases"])))),
)
trend_12d = alt.Chart(source).mark_line(color="grey", strokeDash=[3,3]).encode(
x = "n_days:Q",
y = alt.Y("trend_12days:Q", scale=alt.Scale(domain=(0, max(data_plot["log_cases"])))),
)
plot1= (
(trend_2d + trend_4d + trend_12d + trend_label + lines)
.configure_title(fontSize=20)
.configure_axis(labelFontSize=15,titleFontSize=18)
)
plot1.save(("../images/covid-permillion-trajectories.png"))
plot1
#hide_input
label = 'Deaths'
temp = pd.concat([x.copy() for x in data_countries_pc]).loc[lambda x: x.date >= '3/1/2020']
metric_name = f'{label} per Million'
temp.columns = ['Country', 'date', metric_name]
# temp.loc[:, 'month'] = temp.date.dt.strftime('%Y-%m')
temp.loc[:, f'Log of {label} per Million'] = temp[f'{label} per Million'].apply(lambda x: np.log(x))
temp.groupby('Country').last()
#hide
# Get data and clean it
data = pd.read_csv("https://raw.githubusercontent.com/CSSEGISandData/COVID-19/master/csse_covid_19_data/csse_covid_19_time_series/time_series_covid19_confirmed_global.csv", error_bad_lines=False)
data = data.drop(columns=["Lat", "Long"])
data = data.melt(id_vars= ["Province/State", "Country/Region"])
data = pd.DataFrame(data.groupby(['Country/Region', "variable"]).sum())
data.reset_index(inplace=True)
data = data.rename(columns={"Country/Region": "location", "variable": "date", "value": "total_cases"})
data['date'] =pd.to_datetime(data.date)
data = data.sort_values(by = "date")
data.loc[data.location == "US","location"] = "United States"
data.loc[data.location == "Korea, South","location"] = "South Korea"
# Population data (last year is 2017 which is what we use)
data_pwt = pd.read_stata("https://www.rug.nl/ggdc/docs/pwt91.dta")
filter1 = data_pwt["year"] == 2017
data_pop = data_pwt[filter1]
data_pop = data_pop[["country","pop"]]
data_pop.loc[data_pop.country == "Republic of Korea","country"] = "South Korea"
data_pop.loc[data_pop.country == "Iran (Islamic Republic of)","country"] = "Iran"
# per habitant
data_pc = data.copy()
# I can add more countries if needed
countries = ["China", "Italy", "Spain", "France", "United Kingdom", "Germany",
"Portugal", "United States", "Singapore","South Korea", "Japan",
"Brazil","Iran"]
data_countries = []
data_countries_pc = []
# compute per habitant
for i in countries:
data_pc.loc[data_pc.location == i,"total_cases"] = data_pc.loc[data_pc.location == i,"total_cases"]/float(data_pop.loc[data_pop.country == i, "pop"])
# get each country time series
filter1 = data_pc["total_cases"] > 1
for i in countries:
filter_country = data_pc["location"]== i
data_countries_pc.append(data_pc[filter_country & filter1])
#hide_input
# Stack data to get it to Altair dataframe format
data_countries_pc2 = data_countries_pc.copy()
for i in range(0,len(countries)):
data_countries_pc2[i] = data_countries_pc2[i].reset_index()
data_countries_pc2[i]['n_days'] = data_countries_pc2[i].index
data_countries_pc2[i]['log_cases'] = np.log(data_countries_pc2[i]["total_cases"])
data_plot = data_countries_pc2[0]
for i in range(1, len(countries)):
data_plot = pd.concat([data_plot, data_countries_pc2[i]], axis=0)
data_plot["trend_2days"] = np.log(2)/2*data_plot["n_days"]
data_plot["trend_4days"] = np.log(2)/4*data_plot["n_days"]
data_plot["trend_12days"] = np.log(2)/12*data_plot["n_days"]
data_plot["trend_2days_label"] = "Doubles every 2 days"
data_plot["trend_4days_label"] = "Doubles evey 4 days"
data_plot["trend_12days_label"] = "Doubles every 12 days"
# Plot it using Altair
source = data_plot
scales = alt.selection_interval(bind='scales')
selection = alt.selection_multi(fields=['location'], bind='legend')
base = alt.Chart(source, title = "COVID-19 Confirmed Cases Per Million of Inhabitants").encode(
x = alt.X('n_days:Q', title = "Days passed since reaching 1 case per million"),
y = alt.Y("log_cases:Q",title = "Log of confirmed cases per million"),
color = alt.Color('location:N', legend=alt.Legend(title="Country", labelFontSize=15, titleFontSize=17),
scale=alt.Scale(scheme='tableau20')),
opacity = alt.condition(selection, alt.value(1), alt.value(0.1))
).properties(
width=chart_width,
height=chart_height
)
lines = base.mark_line().add_selection(
scales
).add_selection(
selection
)
trend_2d = alt.Chart(source).encode(
x = "n_days:Q",
y = alt.Y("trend_2days:Q", scale=alt.Scale(domain=(0, max(data_plot["log_cases"])))),
).mark_line( strokeDash=[3,3], color="grey")
labels = pd.DataFrame([{'label': 'Doubles every 2 days', 'x_coord': 10, 'y_coord': 6},
{'label': 'Doubles every 4 days', 'x_coord': 28, 'y_coord': 6},
{'label': 'Doubles every 12 days', 'x_coord': 45, 'y_coord': 3},
])
trend_label = (alt.Chart(labels)
.mark_text(align='left', dx=-55, dy=-15, fontSize=12, color="grey")
.encode(x='x_coord:Q',
y='y_coord:Q',
text='label:N')
)
trend_4d = alt.Chart(source).mark_line(color="grey", strokeDash=[3,3]).encode(
x = "n_days:Q",
y = alt.Y("trend_4days:Q", scale=alt.Scale(domain=(0, max(data_plot["log_cases"])))),
)
trend_12d = alt.Chart(source).mark_line(color="grey", strokeDash=[3,3]).encode(
x = "n_days:Q",
y = alt.Y("trend_12days:Q", scale=alt.Scale(domain=(0, max(data_plot["log_cases"])))),
)
(
(trend_2d + trend_4d + trend_12d + trend_label + lines)
.configure_title(fontSize=20)
.configure_axis(labelFontSize=15,titleFontSize=18)
)
#hide_input
label = 'Cases'
temp = pd.concat([x.copy() for x in data_countries_pc]).loc[lambda x: x.date >= '3/1/2020']
metric_name = f'{label} per Million'
temp.columns = ['Country', 'date', metric_name]
# temp.loc[:, 'month'] = temp.date.dt.strftime('%Y-%m')
temp.loc[:, f'Log of {label} per Million'] = temp[f'{label} per Million'].apply(lambda x: np.log(x))
temp.groupby('Country').last()
| 0.361841 | 0.865395 |
# In this notebook a simple Q learner will be trained and evaluated. The Q learner recommends when to buy or sell shares of one particular stock, and in which quantity (in fact it determines the desired fraction of shares in the total portfolio value). One initial attempt was made to train the Q-learner with multiple processes, but it was unsuccessful.
```
# Basic imports
import os
import pandas as pd
import matplotlib.pyplot as plt
import numpy as np
import datetime as dt
import scipy.optimize as spo
import sys
from time import time
from sklearn.metrics import r2_score, median_absolute_error
from multiprocessing import Pool
%matplotlib inline
%pylab inline
pylab.rcParams['figure.figsize'] = (20.0, 10.0)
%load_ext autoreload
%autoreload 2
sys.path.append('../../')
import recommender.simulator as sim
from utils.analysis import value_eval
from recommender.agent import Agent
from functools import partial
NUM_THREADS = 1
LOOKBACK = -1 # 252*4 + 28
STARTING_DAYS_AHEAD = 252
POSSIBLE_FRACTIONS = [0.0, 0.25, 0.5, 1.0]
# Get the data
SYMBOL = 'SPY'
total_data_train_df = pd.read_pickle('../../data/data_train_val_df.pkl').stack(level='feature')
data_train_df = total_data_train_df[SYMBOL].unstack()
total_data_test_df = pd.read_pickle('../../data/data_test_df.pkl').stack(level='feature')
data_test_df = total_data_test_df[SYMBOL].unstack()
if LOOKBACK == -1:
total_data_in_df = total_data_train_df
data_in_df = data_train_df
else:
data_in_df = data_train_df.iloc[-LOOKBACK:]
total_data_in_df = total_data_train_df.loc[data_in_df.index[0]:]
# Create many agents
index = np.arange(NUM_THREADS).tolist()
env, num_states, num_actions = sim.initialize_env(total_data_in_df,
SYMBOL,
starting_days_ahead=STARTING_DAYS_AHEAD,
possible_fractions=POSSIBLE_FRACTIONS,
n_levels=10)
agents = [Agent(num_states=num_states,
num_actions=num_actions,
random_actions_rate=0.98,
random_actions_decrease=0.9999,
dyna_iterations=0,
name='Agent_{}'.format(i)) for i in index]
def show_results(results_list, data_in_df, graph=False):
for values in results_list:
total_value = values.sum(axis=1)
print('Sharpe ratio: {}\nCum. Ret.: {}\nAVG_DRET: {}\nSTD_DRET: {}\nFinal value: {}'.format(*value_eval(pd.DataFrame(total_value))))
print('-'*100)
initial_date = total_value.index[0]
compare_results = data_in_df.loc[initial_date:, 'Close'].copy()
compare_results.name = SYMBOL
compare_results_df = pd.DataFrame(compare_results)
compare_results_df['portfolio'] = total_value
std_comp_df = compare_results_df / compare_results_df.iloc[0]
if graph:
plt.figure()
std_comp_df.plot()
```
## Let's show the symbols data, to see how good the recommender has to be.
```
print('Sharpe ratio: {}\nCum. Ret.: {}\nAVG_DRET: {}\nSTD_DRET: {}\nFinal value: {}'.format(*value_eval(pd.DataFrame(data_in_df['Close'].iloc[STARTING_DAYS_AHEAD:]))))
# Simulate (with new envs, each time)
n_epochs = 7
for i in range(n_epochs):
tic = time()
env.reset(STARTING_DAYS_AHEAD)
results_list = sim.simulate_period(total_data_in_df,
SYMBOL,
agents[0],
starting_days_ahead=STARTING_DAYS_AHEAD,
possible_fractions=POSSIBLE_FRACTIONS,
verbose=False,
other_env=env)
toc = time()
print('Epoch: {}'.format(i))
print('Elapsed time: {} seconds.'.format((toc-tic)))
print('Random Actions Rate: {}'.format(agents[0].random_actions_rate))
show_results([results_list], data_in_df)
env.reset(STARTING_DAYS_AHEAD)
results_list = sim.simulate_period(total_data_in_df,
SYMBOL, agents[0],
learn=False,
starting_days_ahead=STARTING_DAYS_AHEAD,
possible_fractions=POSSIBLE_FRACTIONS,
verbose=False,
other_env=env)
show_results([results_list], data_in_df, graph=True)
```
## Let's run the trained agent, with the test set
### First a non-learning test: this scenario would be worse than what is possible (in fact, the q-learner can learn from past samples in the test set without compromising the causality).
```
TEST_DAYS_AHEAD = 20
env.set_test_data(total_data_test_df, TEST_DAYS_AHEAD)
tic = time()
results_list = sim.simulate_period(total_data_test_df,
SYMBOL,
agents[0],
learn=False,
starting_days_ahead=TEST_DAYS_AHEAD,
possible_fractions=POSSIBLE_FRACTIONS,
verbose=False,
other_env=env)
toc = time()
print('Epoch: {}'.format(i))
print('Elapsed time: {} seconds.'.format((toc-tic)))
print('Random Actions Rate: {}'.format(agents[0].random_actions_rate))
show_results([results_list], data_test_df, graph=True)
```
### And now a "realistic" test, in which the learner continues to learn from past samples in the test set (it even makes some random moves, though very few).
```
env.set_test_data(total_data_test_df, TEST_DAYS_AHEAD)
tic = time()
results_list = sim.simulate_period(total_data_test_df,
SYMBOL,
agents[0],
learn=True,
starting_days_ahead=TEST_DAYS_AHEAD,
possible_fractions=POSSIBLE_FRACTIONS,
verbose=False,
other_env=env)
toc = time()
print('Epoch: {}'.format(i))
print('Elapsed time: {} seconds.'.format((toc-tic)))
print('Random Actions Rate: {}'.format(agents[0].random_actions_rate))
show_results([results_list], data_test_df, graph=True)
```
## What are the metrics for "holding the position"?
```
print('Sharpe ratio: {}\nCum. Ret.: {}\nAVG_DRET: {}\nSTD_DRET: {}\nFinal value: {}'.format(*value_eval(pd.DataFrame(data_test_df['Close'].iloc[TEST_DAYS_AHEAD:]))))
import pickle
with open('../../data/simple_q_learner_1000_states_4_actions_full_training.pkl', 'wb') as best_agent:
pickle.dump(agents[0], best_agent)
```
|
github_jupyter
|
# Basic imports
import os
import pandas as pd
import matplotlib.pyplot as plt
import numpy as np
import datetime as dt
import scipy.optimize as spo
import sys
from time import time
from sklearn.metrics import r2_score, median_absolute_error
from multiprocessing import Pool
%matplotlib inline
%pylab inline
pylab.rcParams['figure.figsize'] = (20.0, 10.0)
%load_ext autoreload
%autoreload 2
sys.path.append('../../')
import recommender.simulator as sim
from utils.analysis import value_eval
from recommender.agent import Agent
from functools import partial
NUM_THREADS = 1
LOOKBACK = -1 # 252*4 + 28
STARTING_DAYS_AHEAD = 252
POSSIBLE_FRACTIONS = [0.0, 0.25, 0.5, 1.0]
# Get the data
SYMBOL = 'SPY'
total_data_train_df = pd.read_pickle('../../data/data_train_val_df.pkl').stack(level='feature')
data_train_df = total_data_train_df[SYMBOL].unstack()
total_data_test_df = pd.read_pickle('../../data/data_test_df.pkl').stack(level='feature')
data_test_df = total_data_test_df[SYMBOL].unstack()
if LOOKBACK == -1:
total_data_in_df = total_data_train_df
data_in_df = data_train_df
else:
data_in_df = data_train_df.iloc[-LOOKBACK:]
total_data_in_df = total_data_train_df.loc[data_in_df.index[0]:]
# Create many agents
index = np.arange(NUM_THREADS).tolist()
env, num_states, num_actions = sim.initialize_env(total_data_in_df,
SYMBOL,
starting_days_ahead=STARTING_DAYS_AHEAD,
possible_fractions=POSSIBLE_FRACTIONS,
n_levels=10)
agents = [Agent(num_states=num_states,
num_actions=num_actions,
random_actions_rate=0.98,
random_actions_decrease=0.9999,
dyna_iterations=0,
name='Agent_{}'.format(i)) for i in index]
def show_results(results_list, data_in_df, graph=False):
for values in results_list:
total_value = values.sum(axis=1)
print('Sharpe ratio: {}\nCum. Ret.: {}\nAVG_DRET: {}\nSTD_DRET: {}\nFinal value: {}'.format(*value_eval(pd.DataFrame(total_value))))
print('-'*100)
initial_date = total_value.index[0]
compare_results = data_in_df.loc[initial_date:, 'Close'].copy()
compare_results.name = SYMBOL
compare_results_df = pd.DataFrame(compare_results)
compare_results_df['portfolio'] = total_value
std_comp_df = compare_results_df / compare_results_df.iloc[0]
if graph:
plt.figure()
std_comp_df.plot()
print('Sharpe ratio: {}\nCum. Ret.: {}\nAVG_DRET: {}\nSTD_DRET: {}\nFinal value: {}'.format(*value_eval(pd.DataFrame(data_in_df['Close'].iloc[STARTING_DAYS_AHEAD:]))))
# Simulate (with new envs, each time)
n_epochs = 7
for i in range(n_epochs):
tic = time()
env.reset(STARTING_DAYS_AHEAD)
results_list = sim.simulate_period(total_data_in_df,
SYMBOL,
agents[0],
starting_days_ahead=STARTING_DAYS_AHEAD,
possible_fractions=POSSIBLE_FRACTIONS,
verbose=False,
other_env=env)
toc = time()
print('Epoch: {}'.format(i))
print('Elapsed time: {} seconds.'.format((toc-tic)))
print('Random Actions Rate: {}'.format(agents[0].random_actions_rate))
show_results([results_list], data_in_df)
env.reset(STARTING_DAYS_AHEAD)
results_list = sim.simulate_period(total_data_in_df,
SYMBOL, agents[0],
learn=False,
starting_days_ahead=STARTING_DAYS_AHEAD,
possible_fractions=POSSIBLE_FRACTIONS,
verbose=False,
other_env=env)
show_results([results_list], data_in_df, graph=True)
TEST_DAYS_AHEAD = 20
env.set_test_data(total_data_test_df, TEST_DAYS_AHEAD)
tic = time()
results_list = sim.simulate_period(total_data_test_df,
SYMBOL,
agents[0],
learn=False,
starting_days_ahead=TEST_DAYS_AHEAD,
possible_fractions=POSSIBLE_FRACTIONS,
verbose=False,
other_env=env)
toc = time()
print('Epoch: {}'.format(i))
print('Elapsed time: {} seconds.'.format((toc-tic)))
print('Random Actions Rate: {}'.format(agents[0].random_actions_rate))
show_results([results_list], data_test_df, graph=True)
env.set_test_data(total_data_test_df, TEST_DAYS_AHEAD)
tic = time()
results_list = sim.simulate_period(total_data_test_df,
SYMBOL,
agents[0],
learn=True,
starting_days_ahead=TEST_DAYS_AHEAD,
possible_fractions=POSSIBLE_FRACTIONS,
verbose=False,
other_env=env)
toc = time()
print('Epoch: {}'.format(i))
print('Elapsed time: {} seconds.'.format((toc-tic)))
print('Random Actions Rate: {}'.format(agents[0].random_actions_rate))
show_results([results_list], data_test_df, graph=True)
print('Sharpe ratio: {}\nCum. Ret.: {}\nAVG_DRET: {}\nSTD_DRET: {}\nFinal value: {}'.format(*value_eval(pd.DataFrame(data_test_df['Close'].iloc[TEST_DAYS_AHEAD:]))))
import pickle
with open('../../data/simple_q_learner_1000_states_4_actions_full_training.pkl', 'wb') as best_agent:
pickle.dump(agents[0], best_agent)
| 0.274157 | 0.816553 |
<a href="https://colab.research.google.com/github/julianovale/pythonparatodos/blob/main/M%C3%B3dulo07An%C3%ADbal.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
>[Verificando seu conhecimento](#scrollTo=bqVSq8BLdiEV)
>[Uma breve visão sobre funções](#scrollTo=GKzbaXzEdoAA)
>[Utilizando mais de um parâmetro e retorno de valores](#scrollTo=81q4EAd_Mc8t)
>[Utilizando mais de um parâmetro](#scrollTo=z5ZDIYcQPx2o)
>[Utilizando mais de um parâmetro e valores padrão](#scrollTo=rQMm4T1Hukac)
>[Funções que chamam funções](#scrollTo=tv91GnKOY_r6)
>[Chamadas de funções com palavras-chave e passando listas como parâmetro de uma função](#scrollTo=mtgVmvNy1fEw)
>[Passando todos os parâmetros como uma lista](#scrollTo=G5wsrylgV5Gq)
>[Passagem de um número arbitrário de parâmetros](#scrollTo=VH-SwN0JZZOq)
>[Calcular a soma dos elementos de uma lista com números](#scrollTo=ULNFel0zIKb4)
>[Cálculo da média de valores em uma lista](#scrollTo=PYT2hMP52mTz)
>[Cálculo do fatorial de um número](#scrollTo=4wc1t0hhb1KZ)
>[Múltiplos retornos](#scrollTo=DEown6P8KLxx)
>[Passando nomes de funções como parâmetros](#scrollTo=6fD9-AqrWgW8)
>[Encontrar o maior valor de uma lista](#scrollTo=J-KpmqByJn5o)
>[Contar número de ocorrências de um elemento em uma lista](#scrollTo=dWhJ8Fw1L52A)
>[Verificar se duas sequências são iguais](#scrollTo=lIN1r413Rw_p)
>[Verificar o número de ocorrências de uma subsequência em uma sequência](#scrollTo=rkwPF4CsMDFm)
>[Funções recursivas](#scrollTo=w3QGwPlHhe-M)
>[Desafio 1: Verificando se uma função é um palíndromo](#scrollTo=hjbMvI04enie)
>[Desafio 2: Chamadas de funções com palavras-chave e passando listas como argumento de uma função no cálculo de média aritmética e geométrica](#scrollTo=ERcYbr90cL8S)
>[Desafio 3: Cálculo de desvio-padrão populacional e amostral](#scrollTo=AGja59znir0w)
>[Desafio 4: Desvendando os segredos do Google - PageRank](#scrollTo=-F6lFZRqMTm9)
# 0. Verificando seu conhecimento
Se conseguir entender os códigos dos quatro desafios fornecidos no final desta seção, então, não é necessário estudar os conceitos aqui abordados. Use o índice dado acima para uma navegação mais rápida até os desafios.
# 1. Uma breve visão sobre funções
O Python permite que um conjunto de instruções sejam agrupadas sob o nome de uma função. Na prática isso permite a criação de novos comandos.
Um primeiro propósito para isso é facilitar a manutenção e a legibilidade do seu código de um programa. Isto porque ao invés de repetir as mesmas instruções ao longo de um programa você pode chamar uma função que contém um bloco de instruções. Depois, se for necessário realizar alguma correção ou alteração para as instruções, então, bastará modificar um único lugar: o conteúdo da função.
Para tanto a palavra reservada **def** deverá ser usada seguido do nome da função. A estrutura geral será dada por:
>**def** nome_funcao(parametros_entrada)**:**
>> comando1
>> comando2
>> **return** variaveis_saida
Para que a função criada seja acionada pelo programa basta utilizar o seu nome com os parâmetros corretos (isto é, os valores com tipos e ordem corretos) entre parêntesis, logo após a definição da função e fora do **escopo da função**, isto é, o código logo após a função que não tem espaçamento regular.
**Importante 1**: a linha da função que contém o nome da função e os parâmetros de entrada é chamada de **cabeçalho da função**.
**Importante 2**: observar que as funções podem ter ou não parâmetros de entrada e parâmetros de saída como será observado nos exemplos dados a seguir.
**Importante 3**: Logo depois do nome da função e seus parâmetros entre parêntesis deve vir o símbolo **':'**.
**Importante 4**: os comandos subjacentes à função devem ter um espaçamento regular em relação à margem da tela para indicar que só serão executados caso o nome da função seja digitado. O espaçamento regular é chamado de **indentação** e ao se digitar o nome da função para a que a mesma seja executada é denominado de **chamada à função**.
**Importante 5**: embora os **tipos dos parâmetros de entrada e saída não sejam especificados** é importante que ao se chamar a função os valores passados para esses parâmetros sejam **compatíveis com as instruções** a serem realizadas na **função**.
**Importante 6**: para facilitar a leitura do seu código, em geral, as novas **funções criadas** são **definidas antes** do **código** que irá **executá-las.**
**Importante 7**: o fluxo de execução do programa segue os seguintes passos:
1. Leitura das instruções fora do **escopo da função**;
2. O nome da função é encontrado e o fluxo de execução do programa é desviado para o conjunto de comandos subjacentes à função;
3. Os comandos contidos na função são executados até que o final da função, isto é, o primeiro comando sem a indentação, seja encontrado ou o comando **return**;
4. O fluxo de execução retorna ao ponto onde o nome da função foi encontrado;
5. Os demais comandos subsequentes no código fora do escopo da função são acionados na sequência em que são dados de cima para baixo.
6. A última instrução é executada e o programa é finalizado.
Um exemplo que ilustra os pontos acima é dado por:
> **def nome_funcao**(parametros_funcao)**:** $#3.1 Início execução função$
>> instrucao2
>>
>> instrucao3 $#3.2 Término execução função$
>
>
> instrucao1 $#1. O programa começa por aqui$
>
> $#4. Retorno da função$ **nome_funcao(p_f)** $#2. Desvio para função$
>
> instrucao4 $#5. Continua o fluxo de execução$
>
> instrucao5 $#6. Realiza a última instrução e termina o programa$
A seguir é dada uma função que cria uma nova forma de imprimir o conteúdo de variáveis a partir do comando **print** tal que toda mensagem será cercada por um caractere especial para decoração '-'.
```
def bprint(mens):
print('----------------')
print(mens)
print('----------------')
mens = input('Digite uma mensagem:')
bprint(mens)
```
# 2. Utilizando mais de um parâmetro e retorno de valores
Em princípio funções podem ter quantos parâmetros forem necessários para a passagem de informações a serem utilizadas nas instruções contidas nelas. Além disso, é possível que as funções retornem resultados de operações realizadas internamente.
A função dada a seguir, **somaStr(x,y)**, calcula a soma de dois números x e y no formato tipo **str**, e retorna o resultado em uma variável z tipo **str**. Durante o processo é necessário converter as variáveis x e y de tipo **str** para tipo **float**, realizar a operação de aritmética e retornar o resultado **float** convertido para tipo **str**.
```
def somaStr(x,y):
z = float(x) + float(y)
z = str(z)
return z
x = input('Digite um número: ')
y = input('Digite outro número: ')
z = somaStr(x,y)
print("{0:s} + {1:s} = {2:s} ".format(x, y, z))
```
# 3. Utilizando mais de um parâmetro
As funções podem ter mais de um parâmetro de entrada e cada um pode ser de um tipo diferente. A função a seguir melhora a função **bprint** para que esta seja capaz de não só imprimir qualquer mensagem com uma decoração, mas também ser possível imprimir qualquer símbolo para a decoração de modo que **bprint** terá o seguinte cabeçalho **bprint(men, simb)**.
Importante frisar que está ímplicito que o tipo da variável **men** é **str** e o tipo da variável **simb** também é **str**.
Além disso, para a decoração ser impressa com o caractere contido na variável **simb** sem que haja um caractere por linha foi utilizado o comando **print(simb, end = '')**. Esse comando imprime apenas o símbolo contido em **simb** sem que uma nova linha seja adicionada na tela.
Para que o símbolo contido em **simb** fosse impresso 10 vezes utilizou um comando **for** que irá executar a **10** vezes a instrução correspondente de impressão. Depois do comando de impressão, para que haja uma nova linha é utilizado um comando **print('')**.
```
def bprint(men, simb):
for i in range(1,10,1):
print(simb, end = '')
print('')
print(men)
for i in range(1,10,1):
print(simb, end = '')
print('')
m = 'Mensagem'
s = '*'
bprint(m,s)
```
# 4. Utilizando mais de um parâmetro e valores padrão
A função **bprint** do programa anterior pode ser melhorada de duas formas:
1. Um terceiro parâmetro, do tipo **int**, pode ser utilizado para definir o número de vezes que um caractere especial para decoração da mensagem deverá ser empregado.
2. São estabelecidos valores padrão para os parâmetros de entrada. Eles serão utilizados caso valores não sejam fornecidos pelo usuário.
A nova função **bprint** terá o seguinte cabeçalho: **bprint(men,simb='-', n = 10)** indicando que os valores padrão para a variável **simb** e **n** terão como valor padrão o símbolo **'-'** e **10**, respectivamente. Os tipos implícitos para os parâmetros **simb** e **n** são **str** e **int**, respectivamente.
```
def bprint(men, simb='-', n = 10):
for i in range(1,n,1):
print(simb, end = '')
print('')
print(men)
for i in range(1,n,1):
print(simb, end = '')
print('')
m = 'Mensagem'
s = '*'
n = 5
bprint(m)
bprint(m,s)
bprint(m,s,n = 20)
```
# 5. Funções que chamam funções
A função **bprint** do programa anterior pode ser melhorada, bastando eliminar a duplicidade de linhas de código associada a impressão do caractere especial contido na variável **simb**. Em particular a duplicidade está contida nas seguintes linhas de código:
>**for** i in range(1,**n**,1):
>> print(simb, end = ' ')
>>
>> print(' ')
Essas linhas de código poderão ser trocadas por uma função **sprint** cujos parâmetros de entrada serão **simb** e **n**. Deseja-se que a saída dessa nova função **sprint** seja a impressão **n** vezes da variável **simb** do tipo **str**.
```
def sprint(ss,nn):
for i in range(1,nn,1):
print(ss, end = '')
print('')
def bprint(men, simb='-', n = 10):
sprint(simb,n)
print(men)
sprint(simb,n)
m = 'Mensagem'
s = '*'
n = 5
bprint(m)
bprint(m,s)
bprint(m,s,n = 20)
```
# 6. Chamadas de funções com palavras-chave e passando listas como parâmetro de uma função
É possível criar funções cuja ordem dos parâmetros não é a mesma daquela que é declarada no **cabeçalho** da função. Para tanto, é necessário utilizar na **chamada** da função os mesmos **nomes** dos parâmetros de entrada que foram declarados no **cabeçalho** dela.
Além disso, é possível passar variáveis do tipo lista como parâmetro de entrada de uma função e essa modificação será realizada para usar uma lista para a passagem de dois parâmetros de tipos diferentes. O parâmetro do tipo lista também pode ter um valor padrão.
Modificar a função **bprint** para ter dois parâmetros:
1. **men**: armazena a mensagem a ser impressa;
2. **listapar**: lista que contém os parâmetros de **símbolo** (primeiro elemento) e **número** de vezes que o símbolo será impresso (segundo elemento).
```
def sprint(ss,nn):
for i in range(1,nn,1):
print(ss, end = '')
print('')
def bprint(men, listapar=['-', 10]):
simb = listapar[0]
n = listapar[1]
sprint(simb,n)
print(men)
sprint(simb,n)
m = 'Mensagem'
lista = ['*', 20]
bprint(men = m)
bprint(men = m,listapar=lista)
```
# 7. Passando todos os parâmetros como uma lista
A função **bprint** descrita na seção anterior pode ter todos os seus parâmetros passados através de uma única lista. Nesse caso, a ordem com que os valores são passados dentro da lista para a função é importante.
Para a recuperação do valores contidos na lista deverá ser empregado o operador **[i]** para a acessar o (i-ésimo + 1) elemento da lista.
```
def sprint(ss,nn):
for i in range(1,nn,1):
print(ss, end = '')
print('')
def bprint(listapar=['padrão', '-', 10]):
men = listapar[0]
simb = listapar[1]
n = listapar[2]
sprint(simb,n)
print(men)
sprint(simb,n)
m = 'Mensagem'
lista = ['*', 20]
lista.insert(0,m)
bprint(lista)
```
# 8. Passagem de um número arbitrário de parâmetros
É possível criar funções cuja ordem dos parâmetros não é a mesma daquela que é declarada no **cabeçalho** da função. Além disso, não é necessário enumerar *a priori* o número de parâmetros no cabeçalho da função, bastando utilizar o comando **\*\*nome_parametros** como parâmetro de entrada. Porém, é importante utilizar no momento da **chamada** de função os mesmos **nomes** dos parâmetros de entrada que serem utilizados no escopo da função. A sintaxe da chamada da função será dada por:
> nome_funcao(**p1** = valor1, **p2** = valor2)**
Já o cabeçalho da função será dado por:
> **def** nome_funcao(**\*\*parametros**):
Por último, a instrução que faz uso dos parâmetros utiliza a recuperação de valores por meio dos nomes usados na chamada da função como indexadores da variável declarada no cabeçalho como dado a seguir:
> **parametros**[**"p1"**] # recuperação do **valor1** contido na variável **p1**
>
> **parametros**[**"p2"**] # recuperação do **valor2** contido na variável **p2**
A função **embaralha** dada a seguir realiza o embaralhamento de uma mensagem utilizado o conceito anteriormente enunciado.
```
def embaralha(**men):
print(men["p2"] + men["p1"] + men["p3"])
embaralha(p1 = "Usar", p2 = "Chave", p3 = "Vermelha")
```
# 9. Calcular a soma dos elementos de uma lista com números
Seja uma lista **lista1** tal que ela contém $n$ valores como segue:
**$lista1 = [x_1, x_2, x_3, \cdots, x_n]$**
Deseja-se criar uma função para calcular a soma $s$ de $n$ de elementos dessa lista tal como dado pela seguinte equação:
$ s = \displaystyle \sum_{i = 1}^{n} x_i = x_1 + x_2 + \cdots + x_n$
```
def soma_lista(lista1):
soma = 0
for xi in lista1:
soma = soma + xi
return soma
lista1 = [2, -6, 7, 8]
print(lista1)
print("Soma dos elementos de lista1 = {0:.2f}".format(soma_lista(lista1)))
```
# 10. Cálculo da média de valores em uma lista
Seja uma lista **lista1** tal que ela contém $n$ valores como segue:
**$lista1 = [x_1, x_2, x_3, \cdots, x_n]$**
Deseja-se criar uma função para calcular a média $\mu$ dos $n$ valores contidos na **lista1** tal como dado pela seguinte equação:
$ \mu = \frac{1}{n} \displaystyle \sum_{i = 1}^{n} x_i$
```
def media(lista1):
soma = 0
for xi in lista1:
soma = soma + xi
med = soma/len(lista1)
return med
lista1 = [7, 8, 9]
m = media(lista1)
print(lista1)
print("Media dos valores da lista: " + str(m))
```
# 11. Cálculo do fatorial de um número
O fatorial de um número inteiro **$n$** é representado pelo símbolo **n!** e pode ser calculado pela seguinte equação:
$n! = \displaystyle \prod_{i=1}^{n} i = 1 \times 2 \times \cdots \times n$
Além disso, por definição, $0! = 0$ e uma definição recursiva útil do fatorial é: $n! = n\times(n-1)!$
O programa a seguir fornece uma função que calcula o fatorial de um número inteiro $n$ não-negativo.
```
def fatorial(n):
prod = 1
for i in range(1,n,1):
prod = prod*i
return prod
a = 0
b = fatorial(a)
print("Fatorial de {0:d} é {1:d} ".format(a,b))
a = 10
b = fatorial(a)
print("Fatorial de {0:d} é {1:d} ".format(a,b))
```
# 12. Múltiplos retornos
Uma função pode retornar múltiplos parâmetros de saída. Por exemplo, para retornar 2 valores basta utilizar a sintaxe:
> **return val1, val2**
Suponha que deseja-se criar uma função que retorna a **soma** e o **produto** de duas variáveis **a** e **b**.
De posse dos resultados **soma** e **produto** para o par **$(a,b)$**, a ideia é criar uma terceira função **verif_par** que verifica se existe algum par de números inteiros **$(a,b)$** para o qual **a + b = a*b**. Isso deverá ser feito todos os valores inteiros de 1 até 9.
Para tanto, a seguintes funções deverão ser criadas:
* **soma_prod** que retorna a **soma** e o **produto** do par **$(a,b)$**;
* **verif_par** função que testa combinação de valores de 1 até 9 para **a** e para **b** e imprime aqueles para os quais a propriedade **a + b = a*b** é válida. Esta função deverá ter como parâmetro de entrada um valor **n** que indica até qual dígito será realizado o teste. Para este caso, usa-se **n = 9**.
**Importante**: O comando **i in range(1,n+1)** fará a variável **i** percorrer valores de **1** até **n**.
```
def soma_prod(a, b):
c = a + b
d = a * b
return c, d
def verif_par(n):
for i in range(1,n+1):
for j in range(1,n+1):
c,d = soma_prod(i,j)
if (c == d):
print("Passou: ({0:d}, {1:d}) ".format(i,j))
print("Pois: (a+b,a*b)= ({0:d}, {1:d}) ".format(c,d))
#else:
# print("Falhou: ({0:d}, {1:d}) ".format(i,j))
n = 9
verif_par(n)
```
# 13. Passando nomes de funções como parâmetros
É possível passar nomes de funções como parâmetros no Python e depois utilizar esses nomes para realizar avaliações das funções.
A sintaxe geral para isso é dada por:
>**f** = **soma**
>
> r = **f**(a,b) # Aqui é feita uma chamada à função **soma** com parâmetros **a** e **b**.
Na função descrita a seguir existem duas operações que podem ser executadas: **soma** e **produto** de dois números **a** e **b**. O número de parâmetros para ambas é igual e dados por **(a,b)**. Além disso, deseja-se construir uma função na qual o usuário decide em tempo de execução qual função deseja executar. Para obter o nome de uma função o comando **f.__name__** deverá ser utilizado.
O programa a seguir fornece como isso pode ser feito.
```
def soma(a,b):
return a+b
def prod(a,b):
return a*b
opt = input('1-soma/2-produto')
opt = int(opt) # Convertendo a string '1' ou '2' em número int 1 ou 2.
a = 3
b = 4
if (opt == 1):
f = soma
else:
f = prod
r = f(a,b)
name = f.__name__
print('{0}({1},{2})={3}'.format(name,a,b,r))
```
# 14. Encontrar o maior valor de uma lista
Seja uma lista **lista1** tal que ela contém $n$ valores como segue:
**$lista1 = [x_1, x_2, x_3, \cdots, x_n]$**
Deseja-se criar uma função para obter o maior elemento $x_{max}$ contido na **lista1** tal como dado pela seguinte equação:
$ x_{max} = \displaystyle \max_{i = 1}^{n} \left\{ x_i \right\}$
**Importante**: O maior elemento de **lista1** pertence à **lista1**.
```
def max_lista(lista1):
# Atribuição provisória e inicial de que o elemento x[0]
# é o maior até que outro elemento da lista seja maior.
maior = lista1[0]
# Analisando todos os elementos e verificando se algum
# é maior lista1[0]. Se for, então, modificar o valor
# da variável maior.
for xi in lista1:
if (xi > maior):
maior = xi
return maior
lista1 = [3, -4, -1, 0, 10, 5, 9, 20]
print(lista1)
print("Maior elemento da lista1 é: {0}".format(max_lista(lista1)))
```
# 15. Contar número de ocorrências de um elemento em uma lista
Seja uma lista **lista1** tal que ela contém $n$ elementos como segue:
**$lista1 = [x_1, x_2, x_3, \cdots, x_n]$**
Dado um elemento $elem$, deseja-se criar uma função para obter o número de
ocorrências desse elemento na **lista1**. Para tanto, será necessário:
>0. Iniciar um variável acumuladora $soma$ com o valor zero. Essa variável é responsável por contabilizar no. de ocorrências de $elem$ em $lista1$
>
>1. Percorrer todos os elementos de $lista1$:
>>
>> 1.1 Comparar cada elemento da lista $x_i$ com $elem$.
>>
>> 1.2 Se $elem$ é igual a $x_i$, então, atualizar $soma += 1$
>>
>2. Retornar o valor de $soma$
```
def ocorrencia_lista(lista1,elem):
soma = 0
for xi in lista1:
if (elem == xi):
soma += 1
return soma
l1 = ['g', 't', 'a', 'a', 'c', 't', 'g', 'c']
e = 'a'
print(l1)
soma = ocorrencia_lista(l1,e)
print('Numero de ocorrencias de \'{0:s}\' é: {1:d}'.format(e,soma))
```
# 16. Verificar se duas sequências são iguais
Sejam duas sequências **seq1** e **seq2** tal que ambas contêm $n$ elementos como segue:
**$seq1 = [x_1, x_2, x_3, \cdots, x_n]$**
**$seq2 = [y_1, y_2, y_3, \cdots, y_n]$**
Deseja-se criar uma função para verificar se ambas as sequências são iguais ou não.
A lógica para verificar se duas sequências são iguais é utilizar dois elementos **$x_i$** e **$y_i$** que utilizam o mesmo índice **$i$** e correm sincronizados e em paralelo para ambas as sequências como pode ser visualizado com o seguinte diagrama:
> $x_1$ | $x_2$ | $x_3$ | $x_4$ | $x_5$
>--- | --- | --- | --- | ---
> [0] | [1] | [2] | [3] | [4]
> [i] | | | |
> $y_1$ | $y_2$ | $y_3$ | $y_4$ | $y_5$
>--- | --- | --- | --- | ---
> [0] | [1] | [2] | [3] | [4]
> [i] | | | |
Esse trabalho pode ser facilmente executado com auxílio do seguinte comando **$x_i,y_i$ in zip(seq1,seq2)**.
Se for verificado alguma diferença para algum elemento, então, uma variável auxiliar **status**, que indica se as sequências são iguais ou não e recebe valor inicial **True**, irá receber o valor **False** e o laço será quebrado com o comando **break**.
```
def verif_seq(s1,s2):
status = True
for xi,yi in zip(s1,s2):
if (xi != yi):
status = False
break
return status
s1 = ['a', 'g', 't', 'c']
s2 = ['a', 'g', 't', 'c']
s3 = ['a', 'g', 't', 'a']
print(s1)
print(s2)
status = verif_seq(s1,s2)
print('A sequência s1 é igual a sequência s2: {0:b}'.format(status))
print(s1)
print(s3)
status = verif_seq(s1,s3)
print('A sequência s1 é igual a sequência s3: {0:b}'.format(status))
```
# 17. Verificar o número de ocorrências de uma subsequência em uma sequência
Seja uma sequência **s1** tal que ela contém $n$ elementos como segue:
**$s1 = [x_1, x_2, x_3, \cdots, x_n]$**
Seja uma subsequência **s2** com $m$ elementos como segue:
**$s2 = [y_1, y_2, y_3, \cdots, y_m]$**
Deseja-se criar uma função para obter o número de ocorrências da subsequência **s2** em **s1**. Para tanto, será necessário:
>0. Iniciar um variável acumuladora $soma$ com o valor zero. Essa variável é responsável por contabilizar no. de ocorrências de **s2** em **s1**
>
>1. Percorrer todos os elementos de **s1**:
>>
>> 1.1 A partir do elemento $x_i$ de **s1** gerar uma subsequência **sub1** e comparar cada elemento com cada elemento de **s2**
>>
>> 1.2 Se **sub1** é igual a **s2**, então, atualizar $soma += 1$
>>
>2. Retornar o valor de $soma$
**Importante 1**: Será necessário empregar a função que verifica se duas sequências **sub1** e **s2** são iguais ou não que foi fornecida no exercício anterior.
**Importante 2**: Para comparar trechos de **s1** com a subquência **s2** é necessário particionar **s1** para gerar **sub1** usando a seguinte indexação **i:i+tam** e o índice **i** varia de **0** até **len(s1)-len(s2)+1** o que pode ser feito com **range(0,len(s1)-len(s2)+1)**.
O esquema abaixo ilustra o motivo para usar o índice final de **range** como **len(s1)-len(s2)+1**.
> $s1_1$ | $x_2$ | $x_3$ | $x_4$ | $x_5$
>--- | --- | --- | --- | ---
> [0] | [1] | [2] | [3] | [4]
> | | | [i] |
> | | | $y_1$ | $y_2$
Observe que o máximo valor de **i** é **3**. Veja que **len(s1) = 5**, **len(s2) = 2**. Porém, como o comando **range(1,n)** vai até o valor **$n-1$**, se fosse usado **len(s1)-len(s2) = 3** o laço não iria até o índice **3** e sim até **2**. Portanto, faz-se necessário usar o valor **len(s1)-len(s2)+1 = 4** como índice final **n** para o comando **range(1,n)**.
```
def verif_seq(s1,s2):
status = True
for xi,yi in zip(s1,s2):
if (xi != yi):
status = False
break
return status
def subseq_lista(lista1,sub1):
soma = 0
n = len(lista1)
tam = len(sub1)
for i in range(0,n-tam+1):
sub2 = lista1[i:i+tam]
print(sub1)
print(sub2)
print('----------')
if verif_seq(sub1,sub2): # Chamada à função que verifica se s1 e s2 são iguais
soma += 1
return soma
l1 = ['g', 't', 'a', 'a', 'c', 't', 'g', 'c']
e = ['g', 't']
print(l1)
print('----------')
soma = subseq_lista(l1,e)
print(soma)
#print('Numero de ocorrencias de \'{0:s}\' é: {1:d}'.format(e,soma))
```
# 18. Funções recursivas
Funções recursivas são funções que chamam a si mesmas para realizar um cálculo ou operação. Para criar uma função recursiva é útil obter a relação de recorrência para o problema a ser resolvido.
Alguns exemplos de problemas e respectivas relações recursivas são fornecidos a seguir:
---
**Problema 1**: Cálculo do fatorial
O cálculo do fatorial de um número inteiro **n** pode ser feito utilizado o seguinte equacionamento recursivo:
$n! = n \times (n-1)!$
$0! = 1$
---
**Problema 2**: Cálculo da soma de **$n$** valores inteiros
O cálculo da soma **$s_n$** dos **$n$** valores inteiros pode ser obtido através do seguinte equacionamento recursivo:
$ s_n = \displaystyle \sum_{i = 1}^{n} i = n + \sum_{i = 1}^{n-1} i = n + s_{n-1}$
$ s_1 = 1$
---
**Problema 3**: Cálculo do i-ésimo termo da sequência de Fibonacci
O cálculo do i-ésimo termo da sequência de **Fibonacci** pode ser realizado através da seguinte relação recursiva:
$F(i) = F(i-1) + F(i-2)$
$F(1) = F(0) = 1$
---
**Problema 4**: Cálculo do i-ésimo termo da sequência Padovana
O cálculo do i-ésimo termo da sequência de **Padovana** pode ser realizado através da seguinte relação recursiva:
$P(n+1) = P(n-1) + P(n-2) \rightarrow P(n) = P(n-2) + P(n-3)$
$P(2) = P(1) = P(0) = 1$
---
Construir funções recursivas para resolver cada um dos problemas enunciados.
```
def fatorial(n):
if (n > 0):
return n*fatorial(n-1)
else:
return 1
def soma(n):
if (n > 1):
return n+soma(n-1)
else:
return 1
def fibo(n):
if (n > 1):
return fibo(n-1) + fibo(n-2)
else:
return 1
def pado(n):
if (n > 2):
return pado(n-2) + pado(n-3)
else:
return 1
n = 5
print('n = {0}'.format(n))
print('{0}! = {1}'.format(n,fatorial(n)))
print('S({0}) = {1}'.format(n,soma(n)))
print('F({0}) = {1}'.format(n,fibo(n)))
print('P({0}) = {1}'.format(n,pado(n)))
```
# Desafio 1: Verificando se uma função é um palíndromo
Um palíndromo é uma palavra que se for invertida a sequência de letras permanece a mesma.
Veja os seguintes exemplos de palíndromos:
* arara
* asa
* salas
* Socorram-me, subi no ônibus em Marrocos
A lógica para verificar se um **palavra** é um palíndromo usa a indexação dos elementos de um tipo **str**. Por exemplo, seja a variável **s = 'arara'**, então, o conteúdo de **s** pode ser visualizado com o seguinte diagrama:
> a | r | a | r | a
>--- | --- | --- | --- | ---
> [0] | [1] | [2] | [3] | [4]
> [i] | | | | [j]
Sejam os índices **i** e **j** com valores iniciais dados por **i = 0** e **j = 4**. A partir deles é possível acessar os elementos **a[0]** e **a[4]**, e verificar se **a[i] != a[j]**. Se fosse, então, o processo poderia ser interropido e **s** não seria palídromo. Como isso não ocorre o índice **i** é incrementado em uma unidade e o **j** reduzido em uma unidade, gerando o seguinte esquema:
> a | r | a | r | a
>--- | --- | --- | --- | ---
> [0] | [1] | [2] | [3] | [4]
> | [i] | | [j] |
De novo é verificado que os elementos a[i] e a[j] são iguais e a verificação continua com incremento de **i** e **j** que irá resultar em:
> a | r | a | r | a
>--- | --- | --- | --- | ---
> [0] | [1] | [2] | [3] | [4]
> | | [i] | |
> | | [j] | |
Veja que novamente **a[i]** e **a[j]** são iguais, mas agora pode-se interromper a verificação, pois do contrário iríamos verificar elementos que já foram comparados. O ponto de parada é exatamente o elemento de índice 2. Isto é, devemos interromper a verificação até que **i** seja **int(len(s)/2) = int(5/2) = int(2,5) = 2**. Porém, observe que este último passo era desnecessário, pois o elemento do meio sempre irá gerar **a[i] == a[j]**. Então, pode-se parar com **int(len(s)/2)-1**.
Para o caso em que o número de caracteres contidos em **s** é par, ocorreria o término de comparações na seguinte situação:
> a | s | s | a
>--- | --- | --- | ---
> [0] | [1] | [2] | [3]
> | [i] | [j] |
> | | |
Como **len(s) = 4** e **int(len(s)/2) = int(4/2) = int(2) = 2** e **i** pode ir até **int(len(s)/2) - 1**.
Desse modo, pode-se concluir que a condição de parada que serve para os 2 casos, seja o número de letras da **string** **s** ímpar ou par, é: **int(len(s/2))-1**. Porém, como o comando **range(1,int(len(s/2)))** será utilizado no laço, então, automaticamente a comparação irá parar em **int(len(s/2))-1**.
Estabelecida a condição de parada para a determinação se **s** é um palíndromo ou não, pode-se traçar o seguinte roteiro para a resolução do problema:
1. Deixar todas as letras como maiúsculas para evitar problemas de comparação com falso negativo como **'R'** com 'r'**. Para tanto, o comando upper pode ser utilizado;
2. Obter o tamanho $n$ da palavra **s**;
3. Obter o valor inicial dos índices **i (=0)** e **j (n-(i+1))**;
4. Comparar e verificar se são iguais os elementos **a[i]** e **a[j]**. Se não forem interroper o laço e determinar que **s** não é palíndromo, senão, incrementar **i** e reduzir **j** até que **i** tenha valor **int(len(s/2))-1**.
```
def verif_palindromo(pa):
pa = pa.upper()
n = len(pa)
num = int(n/2)
verif = True
for i in range(0,num):
print("pa[{0:d}] = {1:s}".format(i,pa[i]))
print("pa[{0:d}] = {1:s}".format(n-(i+1),pa[n-(i+1)]))
if (pa[i] != pa[n-(i+1)]):
verif = False
break
return verif
p = "arara"
if (verif_palindromo(p)):
print("A palavra {0:s} é palídromo".format(p))
```
# Desafio 2: Chamadas de funções com palavras-chave e passando listas como argumento de uma função no cálculo de média aritmética e geométrica
A função **mediaAG** do programa dado a seguir, irá receber uma lista de valores e será calculado e retornado a **média aritmética** ou a **média geométrica** dos valores fornecidos. Como uma das duas médias deverá ser escolhida, será necessário um parâmetro adicional **tip** (com valores **0** para **média aritmética** e **1** para **média geométrica**) indicando qual equacionamento deverá ser utilizado.
É importante frisar que o cálculo da **média** **aritmética** $ma$ ou **geométrica** $g$ são dados, respectivamente, pelas seguintes equações:
* **Média aritmética**:
$ma = \frac{1}{n} \displaystyle \sum_{i=1}^{n} x_i$
onde:
$x_i$ é o i-ésimo valor dos **n** valores fornecidos para cálculo da **média aritmética**, e $\displaystyle \sum_{i=1}^{n} x_i = x_1 + x_2 + \cdots + x_n$
* **Média geométrica**:
$g = \sqrt[n]{\displaystyle \prod_{i=1}^{n} x_i} $
onde:
$x_i$ é o i-ésimo valor dos **n** valores fornecidos para cálculo da **média geométrica** e $\displaystyle \prod_{i=1}^{n} x_i = x_1 \times x_2 \times \cdots \times x_n$ .
Para a resolução deste problema serão criadas 3 funções:
1. **ma** que calcula a **média aritmética**. Seu parâmetro de entrada será uma lista de valores e o parâmetro de saída a média;
2. **mg** que calcula a **média geométrica**. Seu parâmetro de entrada será uma lista de valores e o parâmetro de saída a média;
3. **mediaAG** que calcula a **média aritmética** ou **geométrica** chamando as funções anteriores de acordo com o valor contido no parâmetro **tip**. Os parâmetros de entrada serão: uma lista de valores e o parâmetro **tip**.
Agora, vamos ao código.
```
# Cálculo da média aritmética.
def ma(lista1):
soma = 0
n = len(lista1)
for i in lista1:
soma = soma + i
total = soma/n
return total
# Cálculo da média geométrica.
def mg(lista1):
prod = 1;
n = len(lista1)
for i in lista1:
prod = prod*i
# Raiz enésima = ^(1/n)
total = (prod)**(1/n)
return total
# Cálculo das médias: valor padrão -> média aritmética.
def mediaAG(lista, tip = 0):
if (tip == 0):
r = ma(lista)
else:
r = mg(lista)
return r
lista1 = [7, 8, 9]
t = 0
# Trocando a ordem dos parâmetros, mas usando os nomes do cabeçalho da função.
m1 = mediaAG(tip = t, lista = lista1)
t = 1
m2 = mediaAG(tip = t, lista = lista1)
print(lista1)
print("Média aritmética {0:.2f} ".format(m1))
print("Média geométrica {0:.2f} ".format(m2))
```
# Desafio 3: Cálculo de desvio-padrão populacional e amostral
A função **desviop** do programa dado a seguir, irá receber uma lista de valores e será calculado e retornado o **desvio-padrão** dos valores.
O desvio-padrão é uma **medida estatística** que serve para medir a dispersão dos valores coletados em relação à **media aritmética**. Em termos menos formais serve para indicar quanto os valores coletados estão "perto" ou "longe" da média. Se para um desvio-padrão zero há o indicativo de que todos os valores coletados são iguais, então, um desvio-padrão baixo indica que os dados oscilam pouco em torno da média.
Como o **desvio-padrão** pode ser **populacional** ou **amostral**, será necessário um parâmetro adicional **tip** indicando qual equacionamento deverá ser realizado.
É importante frisar que o cálculo do **desvio-padrão** **populacional** **$\sigma$** ou **amostral** **$s$** podem ser feitos após a obtenção da **variâncias** (que é o quadrado do desvio-padrão) correspondentes e dadas, respectivamente, pelas seguintes equações:
* **Variância populacional**:
$\sigma^2 = \frac{1}{N} \displaystyle \sum_{i=1}^{N} \left( x_i - \mu \right)^2$
onde:
$\mu$ é a **média aritmética populacional**, $x_i$ é o valor de cada dado, e **$N$** é o **tamanho da população** (ou número de valores coletados), e o **desvio-padrão populacional** é dado por $\sigma = \sqrt{\sigma^2}$.
* A **média aritmética populacional** é dada por:
$\mu = \frac{1}{N} \displaystyle \sum_{i=1}^{N} x_i$
* **Variância amostral**:
$s^2 = \frac{1}{n-1} \displaystyle \sum_{i=1}^{n} \left( x_i - \overline{x} \right)^2$
onde:
$\overline{x}$ é a **média aritmética amostral**, $x_i$ é o valor de cada dado, e **$n$** é o **tamanho da amostra** (ou número de valores coletados extraídos de uma população), e o **desvio-padrão amostral** é dado por $s = \sqrt{s^2}$.
* A **média aritmética amostral** é dada por:
$\overline{x}= \frac{1}{n} \displaystyle \sum_{i=1}^{n} x_i$
Um roteiro acerca das funções a serem desenvolvidas para esse programa é dado por:
* Uma função **ma** que calcula a média aritmética a partir dos valores de uma lista;
* Uma função **calcSDQ** que usa a função **ma** e calcula a soma do quadrado da diferença entre a média aritmética e cada valor da lista;
* Uma função **devPP** que divide o valor retornado por **calcSDQ** por **N** para obter a **variância populacional**. Depois extraí a raiz quadrada para obter o **desvio-padrão populacional**;
* Uma função **devPA** que divide o valor retornado por **calcSDQ** por **n-1** para obter a **variância amostral**. Depois extraí a raiz quadrada para obter o **desvio-padrão amostral**;
* Uma função **desviop** que de acordo com um parâmetro **tip** determina qual das duas funções irá chamar: **devPP** ou **devPP**. Além disso, seus paramêtros de entrada são passados em forma de lista chamada **\*\*par** de modo que para serem recuperados é necessário empregar a seguinte sintaxe:
> **def** desviop(**par):
>> lista = par[**"lista"**]
>>
>> tip = par[**"tip"**]
E a chamada para esta função deverá declarar explicitamente o nome dos parâmetros que será empregado para a posterior **'recuperação'** dos valores no escopo da função:
> m1 = desviop(**tip** = t, **lista** = l1)
```
# Cálculo da média aritmética.
def ma(lista1):
soma = 0
n = len(lista1)
for i in lista1:
soma = soma + i
total = soma/n
return total
# Cálculo da soma das diferenças ao quadrado entre x_i e a média.
def calcSDQ(lista1):
med = ma(lista1) # Cálculo da média aritmética por meio da função ma
soma = 0
for i in lista1:
soma = soma + (i - med)**2
return soma
# Cálculo do desvio-padrão populacional
def devPP(lista1):
soma = calcSDQ(lista1) # Cálculo da soma das diferenças ao quadrado
n = len(lista1)
total = soma/n # Divisão por N: variância populacional
dpp = total**(1/2) # raiz quadrada: transforma variância em desvio-padrão
return dpp
def devPA(lista1):
soma = calcSDQ(lista1) # Cálculo da soma das diferenças ao quadrado
n = len(lista1)
total = soma/(n-1) # Divisão por n-1: variância amostral
dpa = total**(1/2) # raiz quadrada: transforma variância em desvio-padrão
return dpa
# Cálculo das médias: valor padrão -> média aritmética.
def desviop(**par):
lista = par["lista"]
tip = par["tip"]
if (tip == 0):
r = devPP(lista)
else:
r = devPA(lista)
return r
l1 = [7, 8, 9]
t = 0
# Trocando a ordem dos parâmetros, mas usando os nomes do cabeçalho da função.
m1 = desviop(tip = t, lista = l1)
t = 1
m2 = desviop(tip = t, lista = l1)
print(l1)
print("Desvio-padrão populacional {0:.2f} ".format(m1))
print("Desvio-padrão amostral {0:.2f} ".format(m2))
```
# Desafio 4: Desvendando os segredos do Google - PageRank
De acordo com o Wikipédia:
**"PageRank é um algoritmo utilizado pela ferramenta de busca Google para posicionar websites entre os resultados de suas buscas. O PageRank mede a importância de uma página contabilizando a quantidade e qualidade de links apontando para ela. Não é o único algoritmo utilizado pelo Google para classificar páginas da internet, mas é o primeiro utilizado pela companhia e o mais conhecido."**
Agora suponha o seguinte problema relacionado: seja uma lista contendo o índice associado ao **PageRank** de cada página da internet indexada pelo **Google**.
Agora deseja-se criar um programa capaz de retornar duas listas relativas às páginas em ordem decrescente de índice de PageRank:
1. Uma **lista** contendo os valores de **PageRank** das páginas;
2. Uma **lista** contendo os **índices originais** de ordenação associado a cada página.
O programa a seguir executa essa tarefa usando o algoritmo de ordenação denominado **Bubble Sort**:
https://pt.wikipedia.org/wiki/Bubble_sort
Uma ilustra do funcionamento do Bubble Sort é dada na figura a seguir:
https://pt.wikipedia.org/wiki/Ficheiro:Bubble_sort_animation.gif
```
def troca(b,c):
return(c,b)
def bubble_sort(a,crit):
for i in list(range(1,len(a),1)):
for j in list(range(0,len(a)-i,1)):
if crit[j] < crit[j+1]:
a[j],a[j+1] = troca(a[j],a[j+1])
crit[j],crit[j+1] = troca(crit[j],crit[j+1])
return(a,crit)
pagerank = [4.7, 5.4, 3.1, 10.2]
index = [0, 1, 2, 3]
print('Antes index = ' + str(index))
print('Antes pagerank = ' + str(pagerank))
index,r = bubble_sort(index,pagerank)
print('Depois index = ' + str(index))
print('Depois pagerank = ' + str(pagerank))
```
|
github_jupyter
|
def bprint(mens):
print('----------------')
print(mens)
print('----------------')
mens = input('Digite uma mensagem:')
bprint(mens)
def somaStr(x,y):
z = float(x) + float(y)
z = str(z)
return z
x = input('Digite um número: ')
y = input('Digite outro número: ')
z = somaStr(x,y)
print("{0:s} + {1:s} = {2:s} ".format(x, y, z))
def bprint(men, simb):
for i in range(1,10,1):
print(simb, end = '')
print('')
print(men)
for i in range(1,10,1):
print(simb, end = '')
print('')
m = 'Mensagem'
s = '*'
bprint(m,s)
def bprint(men, simb='-', n = 10):
for i in range(1,n,1):
print(simb, end = '')
print('')
print(men)
for i in range(1,n,1):
print(simb, end = '')
print('')
m = 'Mensagem'
s = '*'
n = 5
bprint(m)
bprint(m,s)
bprint(m,s,n = 20)
def sprint(ss,nn):
for i in range(1,nn,1):
print(ss, end = '')
print('')
def bprint(men, simb='-', n = 10):
sprint(simb,n)
print(men)
sprint(simb,n)
m = 'Mensagem'
s = '*'
n = 5
bprint(m)
bprint(m,s)
bprint(m,s,n = 20)
def sprint(ss,nn):
for i in range(1,nn,1):
print(ss, end = '')
print('')
def bprint(men, listapar=['-', 10]):
simb = listapar[0]
n = listapar[1]
sprint(simb,n)
print(men)
sprint(simb,n)
m = 'Mensagem'
lista = ['*', 20]
bprint(men = m)
bprint(men = m,listapar=lista)
def sprint(ss,nn):
for i in range(1,nn,1):
print(ss, end = '')
print('')
def bprint(listapar=['padrão', '-', 10]):
men = listapar[0]
simb = listapar[1]
n = listapar[2]
sprint(simb,n)
print(men)
sprint(simb,n)
m = 'Mensagem'
lista = ['*', 20]
lista.insert(0,m)
bprint(lista)
def embaralha(**men):
print(men["p2"] + men["p1"] + men["p3"])
embaralha(p1 = "Usar", p2 = "Chave", p3 = "Vermelha")
def soma_lista(lista1):
soma = 0
for xi in lista1:
soma = soma + xi
return soma
lista1 = [2, -6, 7, 8]
print(lista1)
print("Soma dos elementos de lista1 = {0:.2f}".format(soma_lista(lista1)))
def media(lista1):
soma = 0
for xi in lista1:
soma = soma + xi
med = soma/len(lista1)
return med
lista1 = [7, 8, 9]
m = media(lista1)
print(lista1)
print("Media dos valores da lista: " + str(m))
def fatorial(n):
prod = 1
for i in range(1,n,1):
prod = prod*i
return prod
a = 0
b = fatorial(a)
print("Fatorial de {0:d} é {1:d} ".format(a,b))
a = 10
b = fatorial(a)
print("Fatorial de {0:d} é {1:d} ".format(a,b))
def soma_prod(a, b):
c = a + b
d = a * b
return c, d
def verif_par(n):
for i in range(1,n+1):
for j in range(1,n+1):
c,d = soma_prod(i,j)
if (c == d):
print("Passou: ({0:d}, {1:d}) ".format(i,j))
print("Pois: (a+b,a*b)= ({0:d}, {1:d}) ".format(c,d))
#else:
# print("Falhou: ({0:d}, {1:d}) ".format(i,j))
n = 9
verif_par(n)
def soma(a,b):
return a+b
def prod(a,b):
return a*b
opt = input('1-soma/2-produto')
opt = int(opt) # Convertendo a string '1' ou '2' em número int 1 ou 2.
a = 3
b = 4
if (opt == 1):
f = soma
else:
f = prod
r = f(a,b)
name = f.__name__
print('{0}({1},{2})={3}'.format(name,a,b,r))
def max_lista(lista1):
# Atribuição provisória e inicial de que o elemento x[0]
# é o maior até que outro elemento da lista seja maior.
maior = lista1[0]
# Analisando todos os elementos e verificando se algum
# é maior lista1[0]. Se for, então, modificar o valor
# da variável maior.
for xi in lista1:
if (xi > maior):
maior = xi
return maior
lista1 = [3, -4, -1, 0, 10, 5, 9, 20]
print(lista1)
print("Maior elemento da lista1 é: {0}".format(max_lista(lista1)))
def ocorrencia_lista(lista1,elem):
soma = 0
for xi in lista1:
if (elem == xi):
soma += 1
return soma
l1 = ['g', 't', 'a', 'a', 'c', 't', 'g', 'c']
e = 'a'
print(l1)
soma = ocorrencia_lista(l1,e)
print('Numero de ocorrencias de \'{0:s}\' é: {1:d}'.format(e,soma))
def verif_seq(s1,s2):
status = True
for xi,yi in zip(s1,s2):
if (xi != yi):
status = False
break
return status
s1 = ['a', 'g', 't', 'c']
s2 = ['a', 'g', 't', 'c']
s3 = ['a', 'g', 't', 'a']
print(s1)
print(s2)
status = verif_seq(s1,s2)
print('A sequência s1 é igual a sequência s2: {0:b}'.format(status))
print(s1)
print(s3)
status = verif_seq(s1,s3)
print('A sequência s1 é igual a sequência s3: {0:b}'.format(status))
def verif_seq(s1,s2):
status = True
for xi,yi in zip(s1,s2):
if (xi != yi):
status = False
break
return status
def subseq_lista(lista1,sub1):
soma = 0
n = len(lista1)
tam = len(sub1)
for i in range(0,n-tam+1):
sub2 = lista1[i:i+tam]
print(sub1)
print(sub2)
print('----------')
if verif_seq(sub1,sub2): # Chamada à função que verifica se s1 e s2 são iguais
soma += 1
return soma
l1 = ['g', 't', 'a', 'a', 'c', 't', 'g', 'c']
e = ['g', 't']
print(l1)
print('----------')
soma = subseq_lista(l1,e)
print(soma)
#print('Numero de ocorrencias de \'{0:s}\' é: {1:d}'.format(e,soma))
def fatorial(n):
if (n > 0):
return n*fatorial(n-1)
else:
return 1
def soma(n):
if (n > 1):
return n+soma(n-1)
else:
return 1
def fibo(n):
if (n > 1):
return fibo(n-1) + fibo(n-2)
else:
return 1
def pado(n):
if (n > 2):
return pado(n-2) + pado(n-3)
else:
return 1
n = 5
print('n = {0}'.format(n))
print('{0}! = {1}'.format(n,fatorial(n)))
print('S({0}) = {1}'.format(n,soma(n)))
print('F({0}) = {1}'.format(n,fibo(n)))
print('P({0}) = {1}'.format(n,pado(n)))
def verif_palindromo(pa):
pa = pa.upper()
n = len(pa)
num = int(n/2)
verif = True
for i in range(0,num):
print("pa[{0:d}] = {1:s}".format(i,pa[i]))
print("pa[{0:d}] = {1:s}".format(n-(i+1),pa[n-(i+1)]))
if (pa[i] != pa[n-(i+1)]):
verif = False
break
return verif
p = "arara"
if (verif_palindromo(p)):
print("A palavra {0:s} é palídromo".format(p))
# Cálculo da média aritmética.
def ma(lista1):
soma = 0
n = len(lista1)
for i in lista1:
soma = soma + i
total = soma/n
return total
# Cálculo da média geométrica.
def mg(lista1):
prod = 1;
n = len(lista1)
for i in lista1:
prod = prod*i
# Raiz enésima = ^(1/n)
total = (prod)**(1/n)
return total
# Cálculo das médias: valor padrão -> média aritmética.
def mediaAG(lista, tip = 0):
if (tip == 0):
r = ma(lista)
else:
r = mg(lista)
return r
lista1 = [7, 8, 9]
t = 0
# Trocando a ordem dos parâmetros, mas usando os nomes do cabeçalho da função.
m1 = mediaAG(tip = t, lista = lista1)
t = 1
m2 = mediaAG(tip = t, lista = lista1)
print(lista1)
print("Média aritmética {0:.2f} ".format(m1))
print("Média geométrica {0:.2f} ".format(m2))
# Cálculo da média aritmética.
def ma(lista1):
soma = 0
n = len(lista1)
for i in lista1:
soma = soma + i
total = soma/n
return total
# Cálculo da soma das diferenças ao quadrado entre x_i e a média.
def calcSDQ(lista1):
med = ma(lista1) # Cálculo da média aritmética por meio da função ma
soma = 0
for i in lista1:
soma = soma + (i - med)**2
return soma
# Cálculo do desvio-padrão populacional
def devPP(lista1):
soma = calcSDQ(lista1) # Cálculo da soma das diferenças ao quadrado
n = len(lista1)
total = soma/n # Divisão por N: variância populacional
dpp = total**(1/2) # raiz quadrada: transforma variância em desvio-padrão
return dpp
def devPA(lista1):
soma = calcSDQ(lista1) # Cálculo da soma das diferenças ao quadrado
n = len(lista1)
total = soma/(n-1) # Divisão por n-1: variância amostral
dpa = total**(1/2) # raiz quadrada: transforma variância em desvio-padrão
return dpa
# Cálculo das médias: valor padrão -> média aritmética.
def desviop(**par):
lista = par["lista"]
tip = par["tip"]
if (tip == 0):
r = devPP(lista)
else:
r = devPA(lista)
return r
l1 = [7, 8, 9]
t = 0
# Trocando a ordem dos parâmetros, mas usando os nomes do cabeçalho da função.
m1 = desviop(tip = t, lista = l1)
t = 1
m2 = desviop(tip = t, lista = l1)
print(l1)
print("Desvio-padrão populacional {0:.2f} ".format(m1))
print("Desvio-padrão amostral {0:.2f} ".format(m2))
def troca(b,c):
return(c,b)
def bubble_sort(a,crit):
for i in list(range(1,len(a),1)):
for j in list(range(0,len(a)-i,1)):
if crit[j] < crit[j+1]:
a[j],a[j+1] = troca(a[j],a[j+1])
crit[j],crit[j+1] = troca(crit[j],crit[j+1])
return(a,crit)
pagerank = [4.7, 5.4, 3.1, 10.2]
index = [0, 1, 2, 3]
print('Antes index = ' + str(index))
print('Antes pagerank = ' + str(pagerank))
index,r = bubble_sort(index,pagerank)
print('Depois index = ' + str(index))
print('Depois pagerank = ' + str(pagerank))
| 0.127693 | 0.807612 |
```
# Importing the libraries
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
# Importing the dataset
dataset = pd.read_csv('Social_Network_Ads.csv')
X = dataset.iloc[:, [2, 3]].values
y = dataset.iloc[:, 4].values
# Splitting the dataset into the Training set and Test set
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size = 0.25, random_state = 0)
# Feature Scaling
from sklearn.preprocessing import StandardScaler
sc = StandardScaler()
X_train = sc.fit_transform(X_train)
X_test = sc.transform(X_test)
# Fitting Naive Bayes to the Training set
from sklearn.naive_bayes import GaussianNB
classifier = GaussianNB()
classifier.fit(X_train, y_train)
# Predicting the Test set results
y_pred = classifier.predict(X_test)
# Making the Confusion Matrix
from sklearn.metrics import confusion_matrix
cm = confusion_matrix(y_test, y_pred)
cm
# Visualising the Training set results
from matplotlib.colors import ListedColormap
X_set, y_set = X_train, y_train
X1, X2 = np.meshgrid(np.arange(start = X_set[:, 0].min() - 1, stop = X_set[:, 0].max() + 1, step = 0.01),
np.arange(start = X_set[:, 1].min() - 1, stop = X_set[:, 1].max() + 1, step = 0.01))
plt.contourf(X1, X2, classifier.predict(np.array([X1.ravel(), X2.ravel()]).T).reshape(X1.shape),
alpha = 0.75, cmap = ListedColormap(('blue', 'green')))
plt.xlim(X1.min(), X1.max())
plt.ylim(X2.min(), X2.max())
for i, j in enumerate(np.unique(y_set)):
plt.scatter(X_set[y_set == j, 0], X_set[y_set == j, 1],
c = ListedColormap(('blue', 'green'))(i), label = j)
plt.title('Naive Bayes (Training set)')
plt.xlabel('Age')
plt.ylabel('Estimated Salary')
plt.legend()
plt.show()
# Visualising the Test set results
from matplotlib.colors import ListedColormap
X_set, y_set = X_test, y_test
X1, X2 = np.meshgrid(np.arange(start = X_set[:, 0].min() - 1, stop = X_set[:, 0].max() + 1, step = 0.01),
np.arange(start = X_set[:, 1].min() - 1, stop = X_set[:, 1].max() + 1, step = 0.01))
plt.contourf(X1, X2, classifier.predict(np.array([X1.ravel(), X2.ravel()]).T).reshape(X1.shape),
alpha = 0.75, cmap = ListedColormap(('blue', 'green')))
plt.xlim(X1.min(), X1.max())
plt.ylim(X2.min(), X2.max())
for i, j in enumerate(np.unique(y_set)):
plt.scatter(X_set[y_set == j, 0], X_set[y_set == j, 1],
c = ListedColormap(('blue', 'green'))(i), label = j)
plt.title('Naive Bayes (Test set)')
plt.xlabel('Age')
plt.ylabel('Estimated Salary')
plt.legend()
plt.show()
```
|
github_jupyter
|
# Importing the libraries
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
# Importing the dataset
dataset = pd.read_csv('Social_Network_Ads.csv')
X = dataset.iloc[:, [2, 3]].values
y = dataset.iloc[:, 4].values
# Splitting the dataset into the Training set and Test set
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size = 0.25, random_state = 0)
# Feature Scaling
from sklearn.preprocessing import StandardScaler
sc = StandardScaler()
X_train = sc.fit_transform(X_train)
X_test = sc.transform(X_test)
# Fitting Naive Bayes to the Training set
from sklearn.naive_bayes import GaussianNB
classifier = GaussianNB()
classifier.fit(X_train, y_train)
# Predicting the Test set results
y_pred = classifier.predict(X_test)
# Making the Confusion Matrix
from sklearn.metrics import confusion_matrix
cm = confusion_matrix(y_test, y_pred)
cm
# Visualising the Training set results
from matplotlib.colors import ListedColormap
X_set, y_set = X_train, y_train
X1, X2 = np.meshgrid(np.arange(start = X_set[:, 0].min() - 1, stop = X_set[:, 0].max() + 1, step = 0.01),
np.arange(start = X_set[:, 1].min() - 1, stop = X_set[:, 1].max() + 1, step = 0.01))
plt.contourf(X1, X2, classifier.predict(np.array([X1.ravel(), X2.ravel()]).T).reshape(X1.shape),
alpha = 0.75, cmap = ListedColormap(('blue', 'green')))
plt.xlim(X1.min(), X1.max())
plt.ylim(X2.min(), X2.max())
for i, j in enumerate(np.unique(y_set)):
plt.scatter(X_set[y_set == j, 0], X_set[y_set == j, 1],
c = ListedColormap(('blue', 'green'))(i), label = j)
plt.title('Naive Bayes (Training set)')
plt.xlabel('Age')
plt.ylabel('Estimated Salary')
plt.legend()
plt.show()
# Visualising the Test set results
from matplotlib.colors import ListedColormap
X_set, y_set = X_test, y_test
X1, X2 = np.meshgrid(np.arange(start = X_set[:, 0].min() - 1, stop = X_set[:, 0].max() + 1, step = 0.01),
np.arange(start = X_set[:, 1].min() - 1, stop = X_set[:, 1].max() + 1, step = 0.01))
plt.contourf(X1, X2, classifier.predict(np.array([X1.ravel(), X2.ravel()]).T).reshape(X1.shape),
alpha = 0.75, cmap = ListedColormap(('blue', 'green')))
plt.xlim(X1.min(), X1.max())
plt.ylim(X2.min(), X2.max())
for i, j in enumerate(np.unique(y_set)):
plt.scatter(X_set[y_set == j, 0], X_set[y_set == j, 1],
c = ListedColormap(('blue', 'green'))(i), label = j)
plt.title('Naive Bayes (Test set)')
plt.xlabel('Age')
plt.ylabel('Estimated Salary')
plt.legend()
plt.show()
| 0.734405 | 0.842475 |
## What is AutoML?
```
# Sklearn has convenient modules to create sample data.
# make_blobs will help us to create a sample data set suitable for clustering
from sklearn.datasets.samples_generator import make_blobs
X, y = make_blobs(n_samples=100, centers=2, cluster_std=0.30, random_state=0)
# Let's visualize what we have first
import matplotlib.pyplot as plt
%matplotlib inline
import seaborn as sns
plt.scatter(X[:, 0], X[:, 1], s=50)
# We will import KMeans model from clustering model family of Sklearn
from sklearn.cluster import KMeans
k_means = KMeans(n_clusters=2)
k_means.fit(X)
predictions = k_means.predict(X)
# Let's plot the predictions
plt.scatter(X[:, 0], X[:, 1], c=predictions, cmap='brg')
k_means.get_params()
```
## Featuretools
```
import pandas as pd
# First dataset contains the basic information for databases.
databases_df = pd.DataFrame({"database_id": [2234, 1765, 8796, 2237, 3398],
"creation_date": ["2018-02-01", "2017-03-02", "2017-05-03", "2013-05-12", "2012-05-09"]})
databases_df.head()
# Second dataset contains the information of transaction for each database id
db_transactions_df = pd.DataFrame({"transaction_id": [26482746, 19384752, 48571125, 78546789, 19998765, 26482646, 12484752, 42471125, 75346789, 16498765, 65487547, 23453847, 56756771, 45645667, 23423498, 12335268, 76435357, 34534711, 45656746, 12312987],
"database_id": [2234, 1765, 2234, 2237, 1765, 8796, 2237, 8796, 3398, 2237, 3398, 2237, 2234, 8796, 1765, 2234, 2237, 1765, 8796, 2237],
"transaction_size": [10, 20, 30, 50, 100, 40, 60, 60, 10, 20, 60, 50, 40, 40, 30, 90, 130, 40, 50, 30],
"transaction_date": ["2018-02-02", "2018-03-02", "2018-03-02", "2018-04-02", "2018-04-02", "2018-05-02", "2018-06-02", "2018-06-02", "2018-07-02", "2018-07-02", "2018-01-03", "2018-02-03", "2018-03-03", "2018-04-03", "2018-04-03", "2018-07-03", "2018-07-03", "2018-07-03", "2018-08-03", "2018-08-03"]})
db_transactions_df.head()
# Entities for each of datasets should be defined
entities = {
"databases" : (databases_df, "database_id"),
"transactions" : (db_transactions_df, "transaction_id")
}
# Relationships between tables should also be defined as below
relationships = [("databases", "database_id", "transactions", "database_id")]
print(entities)
# There are 2 entities called ‘databases’ and ‘transactions’
# All the pieces that are necessary to engineer features are in place, you can create your feature matrix as below
import featuretools as ft
feature_matrix_db_transactions, feature_defs = ft.dfs(entities=entities, relationships=relationships, target_entity="databases")
feature_defs
```
## Auto-sklearn
```
# Necessary imports
import autosklearn.classification
import sklearn.model_selection
import sklearn.datasets
import sklearn.metrics
from sklearn.model_selection import train_test_split
# Digits dataset is one of the most popular datasets in machine learning community.
# Every example in this datasets represents a 8x8 image of a digit.
X, y = sklearn.datasets.load_digits(return_X_y=True)
# Let's see the first image. Image is reshaped to 8x8, otherwise it's a vector of size 64.
X[0].reshape(8,8)
# Let's also plot couple of them
import matplotlib.pyplot as plt
%matplotlib inline
number_of_images = 10
images_and_labels = list(zip(X, y))
for i, (image, label) in enumerate(images_and_labels[:number_of_images]):
plt.subplot(2, number_of_images, i + 1)
plt.axis('off')
plt.imshow(image.reshape(8,8), cmap=plt.cm.gray_r, interpolation='nearest')
plt.title('%i' % label)
plt.show()
# We split our dataset to train and test data
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=1)
# Similarly to creating an estimator in Scikit-learn, we create AutoSklearnClassifier
automl = autosklearn.classification.AutoSklearnClassifier()
# All you need to do is to invoke fit method to start experiment with different feature engineering methods and machine learning models
automl.fit(X_train, y_train)
# Generating predictions is same as Scikit-learn, you need to invoke predict method.
y_hat = automl.predict(X_test)
print("Accuracy score", sklearn.metrics.accuracy_score(y_test, y_hat))
# Accuracy score 0.98
```
## MLBox
```
# Necessary Imports
from mlbox.preprocessing import *
from mlbox.optimisation import *
from mlbox.prediction import *
import wget
file_link = 'https://apsportal.ibm.com/exchange-api/v1/entries/8044492073eb964f46597b4be06ff5ea/data?accessKey=9561295fa407698694b1e254d0099600'
file_name = wget.download(file_link)
print(file_name)
# GoSales_Tx_NaiveBayes.csv
import pandas as pd
df = pd.read_csv('GoSales_Tx_NaiveBayes.csv')
df.head()
test_df = df.drop(['PRODUCT_LINE'], axis = 1)
# First 300 records saved as test datased
test_df[:300].to_csv('test_data.csv')
paths = ["GoSales_Tx_NaiveBayes.csv", "test_data.csv"]
target_name = "PRODUCT_LINE"
rd = Reader(sep = ',')
df = rd.train_test_split(paths, target_name)
dft = Drift_thresholder()
df = dft.fit_transform(df)
opt = Optimiser(scoring = 'accuracy', n_folds = 3)
opt.evaluate(None, df)
space = {
'ne__numerical_strategy':{"search":"choice", "space":[0]},
'ce__strategy':{"search":"choice",
"space":["label_encoding","random_projection", "entity_embedding"]},
'fs__threshold':{"search":"uniform", "space":[0.01,0.3]},
'est__max_depth':{"search":"choice", "space":[3,4,5,6,7]}
}
best = opt.optimise(space, df,15)
predictor = Predictor()
predictor.fit_predict(best, df)
```
## TPOT
```
from tpot import TPOTClassifier
from sklearn.datasets import load_digits
from sklearn.model_selection import train_test_split
# Digits dataset that you have used in Auto-sklearn example
digits = load_digits()
X_train, X_test, y_train, y_test = train_test_split(digits.data, digits.target,
train_size=0.75, test_size=0.25)
# You will create your TPOT classifier with commonly used arguments
tpot = TPOTClassifier(generations=10, population_size=30, verbosity=2)
# When you invoke fit method, TPOT will create generations of populations, seeking best set of parameters. Arguments you have used to create TPOTClassifier such as generaions and population_size will affect the search space and resulting pipeline.
tpot.fit(X_train, y_train)
print(tpot.score(X_test, y_test))
# 0.9834
tpot.export('my_pipeline.py')
!cat my_pipeline.py
import numpy as np
import pandas as pd
from sklearn.model_selection import train_test_split
from sklearn.neighbors import KNeighborsClassifier
exported_pipeline = make_pipeline(
StackingEstimator(estimator=DecisionTreeClassifier(criterion="entropy", max_depth=6, min_samples_leaf=2, min_samples_split=2)),
KNeighborsClassifier(n_neighbors=2, weights="distance")
)
exported_pipeline.fit(X_train, y_train)
results = exported_pipeline.predict(X_test)
```
|
github_jupyter
|
# Sklearn has convenient modules to create sample data.
# make_blobs will help us to create a sample data set suitable for clustering
from sklearn.datasets.samples_generator import make_blobs
X, y = make_blobs(n_samples=100, centers=2, cluster_std=0.30, random_state=0)
# Let's visualize what we have first
import matplotlib.pyplot as plt
%matplotlib inline
import seaborn as sns
plt.scatter(X[:, 0], X[:, 1], s=50)
# We will import KMeans model from clustering model family of Sklearn
from sklearn.cluster import KMeans
k_means = KMeans(n_clusters=2)
k_means.fit(X)
predictions = k_means.predict(X)
# Let's plot the predictions
plt.scatter(X[:, 0], X[:, 1], c=predictions, cmap='brg')
k_means.get_params()
import pandas as pd
# First dataset contains the basic information for databases.
databases_df = pd.DataFrame({"database_id": [2234, 1765, 8796, 2237, 3398],
"creation_date": ["2018-02-01", "2017-03-02", "2017-05-03", "2013-05-12", "2012-05-09"]})
databases_df.head()
# Second dataset contains the information of transaction for each database id
db_transactions_df = pd.DataFrame({"transaction_id": [26482746, 19384752, 48571125, 78546789, 19998765, 26482646, 12484752, 42471125, 75346789, 16498765, 65487547, 23453847, 56756771, 45645667, 23423498, 12335268, 76435357, 34534711, 45656746, 12312987],
"database_id": [2234, 1765, 2234, 2237, 1765, 8796, 2237, 8796, 3398, 2237, 3398, 2237, 2234, 8796, 1765, 2234, 2237, 1765, 8796, 2237],
"transaction_size": [10, 20, 30, 50, 100, 40, 60, 60, 10, 20, 60, 50, 40, 40, 30, 90, 130, 40, 50, 30],
"transaction_date": ["2018-02-02", "2018-03-02", "2018-03-02", "2018-04-02", "2018-04-02", "2018-05-02", "2018-06-02", "2018-06-02", "2018-07-02", "2018-07-02", "2018-01-03", "2018-02-03", "2018-03-03", "2018-04-03", "2018-04-03", "2018-07-03", "2018-07-03", "2018-07-03", "2018-08-03", "2018-08-03"]})
db_transactions_df.head()
# Entities for each of datasets should be defined
entities = {
"databases" : (databases_df, "database_id"),
"transactions" : (db_transactions_df, "transaction_id")
}
# Relationships between tables should also be defined as below
relationships = [("databases", "database_id", "transactions", "database_id")]
print(entities)
# There are 2 entities called ‘databases’ and ‘transactions’
# All the pieces that are necessary to engineer features are in place, you can create your feature matrix as below
import featuretools as ft
feature_matrix_db_transactions, feature_defs = ft.dfs(entities=entities, relationships=relationships, target_entity="databases")
feature_defs
# Necessary imports
import autosklearn.classification
import sklearn.model_selection
import sklearn.datasets
import sklearn.metrics
from sklearn.model_selection import train_test_split
# Digits dataset is one of the most popular datasets in machine learning community.
# Every example in this datasets represents a 8x8 image of a digit.
X, y = sklearn.datasets.load_digits(return_X_y=True)
# Let's see the first image. Image is reshaped to 8x8, otherwise it's a vector of size 64.
X[0].reshape(8,8)
# Let's also plot couple of them
import matplotlib.pyplot as plt
%matplotlib inline
number_of_images = 10
images_and_labels = list(zip(X, y))
for i, (image, label) in enumerate(images_and_labels[:number_of_images]):
plt.subplot(2, number_of_images, i + 1)
plt.axis('off')
plt.imshow(image.reshape(8,8), cmap=plt.cm.gray_r, interpolation='nearest')
plt.title('%i' % label)
plt.show()
# We split our dataset to train and test data
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=1)
# Similarly to creating an estimator in Scikit-learn, we create AutoSklearnClassifier
automl = autosklearn.classification.AutoSklearnClassifier()
# All you need to do is to invoke fit method to start experiment with different feature engineering methods and machine learning models
automl.fit(X_train, y_train)
# Generating predictions is same as Scikit-learn, you need to invoke predict method.
y_hat = automl.predict(X_test)
print("Accuracy score", sklearn.metrics.accuracy_score(y_test, y_hat))
# Accuracy score 0.98
# Necessary Imports
from mlbox.preprocessing import *
from mlbox.optimisation import *
from mlbox.prediction import *
import wget
file_link = 'https://apsportal.ibm.com/exchange-api/v1/entries/8044492073eb964f46597b4be06ff5ea/data?accessKey=9561295fa407698694b1e254d0099600'
file_name = wget.download(file_link)
print(file_name)
# GoSales_Tx_NaiveBayes.csv
import pandas as pd
df = pd.read_csv('GoSales_Tx_NaiveBayes.csv')
df.head()
test_df = df.drop(['PRODUCT_LINE'], axis = 1)
# First 300 records saved as test datased
test_df[:300].to_csv('test_data.csv')
paths = ["GoSales_Tx_NaiveBayes.csv", "test_data.csv"]
target_name = "PRODUCT_LINE"
rd = Reader(sep = ',')
df = rd.train_test_split(paths, target_name)
dft = Drift_thresholder()
df = dft.fit_transform(df)
opt = Optimiser(scoring = 'accuracy', n_folds = 3)
opt.evaluate(None, df)
space = {
'ne__numerical_strategy':{"search":"choice", "space":[0]},
'ce__strategy':{"search":"choice",
"space":["label_encoding","random_projection", "entity_embedding"]},
'fs__threshold':{"search":"uniform", "space":[0.01,0.3]},
'est__max_depth':{"search":"choice", "space":[3,4,5,6,7]}
}
best = opt.optimise(space, df,15)
predictor = Predictor()
predictor.fit_predict(best, df)
from tpot import TPOTClassifier
from sklearn.datasets import load_digits
from sklearn.model_selection import train_test_split
# Digits dataset that you have used in Auto-sklearn example
digits = load_digits()
X_train, X_test, y_train, y_test = train_test_split(digits.data, digits.target,
train_size=0.75, test_size=0.25)
# You will create your TPOT classifier with commonly used arguments
tpot = TPOTClassifier(generations=10, population_size=30, verbosity=2)
# When you invoke fit method, TPOT will create generations of populations, seeking best set of parameters. Arguments you have used to create TPOTClassifier such as generaions and population_size will affect the search space and resulting pipeline.
tpot.fit(X_train, y_train)
print(tpot.score(X_test, y_test))
# 0.9834
tpot.export('my_pipeline.py')
!cat my_pipeline.py
import numpy as np
import pandas as pd
from sklearn.model_selection import train_test_split
from sklearn.neighbors import KNeighborsClassifier
exported_pipeline = make_pipeline(
StackingEstimator(estimator=DecisionTreeClassifier(criterion="entropy", max_depth=6, min_samples_leaf=2, min_samples_split=2)),
KNeighborsClassifier(n_neighbors=2, weights="distance")
)
exported_pipeline.fit(X_train, y_train)
results = exported_pipeline.predict(X_test)
| 0.853455 | 0.901964 |
Copyright (c) Microsoft Corporation. All rights reserved.
Licensed under the MIT License.
# How to Publish a Pipeline and Invoke the REST endpoint
In this notebook, we will see how we can publish a pipeline and then invoke the REST endpoint.
## Prerequisites and Azure Machine Learning Basics
Make sure you go through the configuration Notebook located at https://github.com/Azure/MachineLearningNotebooks first if you haven't. This sets you up with a working config file that has information on your workspace, subscription id, etc.
### Initialization Steps
```
import azureml.core
from azureml.core import Workspace, Datastore
from azureml.core.compute import AmlCompute
from azureml.core.compute import ComputeTarget
# Check core SDK version number
print("SDK version:", azureml.core.VERSION)
from azureml.data.data_reference import DataReference
from azureml.pipeline.core import Pipeline, PipelineData
from azureml.pipeline.steps import PythonScriptStep
from azureml.pipeline.core.graph import PipelineParameter
print("Pipeline SDK-specific imports completed")
ws = Workspace.from_config()
print(ws.name, ws.resource_group, ws.location, ws.subscription_id, sep = '\n')
# Default datastore (Azure file storage)
def_file_store = ws.get_default_datastore()
print("Default datastore's name: {}".format(def_file_store.name))
def_blob_store = Datastore(ws, "workspaceblobstore")
print("Blobstore's name: {}".format(def_blob_store.name))
# project folder
project_folder = '.'
```
### Compute Targets
#### Retrieve an already attached Azure Machine Learning Compute
```
from azureml.core.compute_target import ComputeTargetException
aml_compute_target = "cpucluster"
try:
aml_compute = AmlCompute(ws, aml_compute_target)
print("found existing compute target.")
except ComputeTargetException:
print("creating new compute target")
provisioning_config = AmlCompute.provisioning_configuration(vm_size = "STANDARD_D2_V2",
min_nodes = 1,
max_nodes = 4)
aml_compute = ComputeTarget.create(ws, aml_compute_target, provisioning_config)
aml_compute.wait_for_completion(show_output=True, min_node_count=None, timeout_in_minutes=20)
# For a more detailed view of current Azure Machine Learning Compute status, use get_status()
# example: un-comment the following line.
# print(aml_compute.get_status().serialize())
```
## Building Pipeline Steps with Inputs and Outputs
As mentioned earlier, a step in the pipeline can take data as input. This data can be a data source that lives in one of the accessible data locations, or intermediate data produced by a previous step in the pipeline.
```
# Reference the data uploaded to blob storage using DataReference
# Assign the datasource to blob_input_data variable
blob_input_data = DataReference(
datastore=def_blob_store,
data_reference_name="test_data",
path_on_datastore="20newsgroups/20news.pkl")
print("DataReference object created")
# Define intermediate data using PipelineData
processed_data1 = PipelineData("processed_data1",datastore=def_blob_store)
print("PipelineData object created")
```
#### Define a Step that consumes a datasource and produces intermediate data.
In this step, we define a step that consumes a datasource and produces intermediate data.
**Open `train.py` in the local machine and examine the arguments, inputs, and outputs for the script. That will give you a good sense of why the script argument names used below are important.**
```
# trainStep consumes the datasource (Datareference) in the previous step
# and produces processed_data1
trainStep = PythonScriptStep(
script_name="train.py",
arguments=["--input_data", blob_input_data, "--output_train", processed_data1],
inputs=[blob_input_data],
outputs=[processed_data1],
compute_target=aml_compute,
source_directory=project_folder
)
print("trainStep created")
```
#### Define a Step that consumes intermediate data and produces intermediate data
In this step, we define a step that consumes an intermediate data and produces intermediate data.
**Open `extract.py` in the local machine and examine the arguments, inputs, and outputs for the script. That will give you a good sense of why the script argument names used below are important.**
```
# extractStep to use the intermediate data produced by step4
# This step also produces an output processed_data2
processed_data2 = PipelineData("processed_data2", datastore=def_blob_store)
extractStep = PythonScriptStep(
script_name="extract.py",
arguments=["--input_extract", processed_data1, "--output_extract", processed_data2],
inputs=[processed_data1],
outputs=[processed_data2],
compute_target=aml_compute,
source_directory=project_folder)
print("extractStep created")
```
#### Define a Step that consumes multiple intermediate data and produces intermediate data
In this step, we define a step that consumes multiple intermediate data and produces intermediate data.
### PipelineParameter
This step also has a [PipelineParameter](https://docs.microsoft.com/en-us/python/api/azureml-pipeline-core/azureml.pipeline.core.graph.pipelineparameter?view=azure-ml-py) argument that help with calling the REST endpoint of the published pipeline.
```
# We will use this later in publishing pipeline
pipeline_param = PipelineParameter(name="pipeline_arg", default_value=10)
print("pipeline parameter created")
```
**Open `compare.py` in the local machine and examine the arguments, inputs, and outputs for the script. That will give you a good sense of why the script argument names used below are important.**
```
# Now define step6 that takes two inputs (both intermediate data), and produce an output
processed_data3 = PipelineData("processed_data3", datastore=def_blob_store)
compareStep = PythonScriptStep(
script_name="compare.py",
arguments=["--compare_data1", processed_data1, "--compare_data2", processed_data2, "--output_compare", processed_data3, "--pipeline_param", pipeline_param],
inputs=[processed_data1, processed_data2],
outputs=[processed_data3],
compute_target=aml_compute,
source_directory=project_folder)
print("compareStep created")
```
#### Build the pipeline
```
pipeline1 = Pipeline(workspace=ws, steps=[compareStep])
print ("Pipeline is built")
pipeline1.validate()
print("Simple validation complete")
```
## Run published pipeline
### Publish the pipeline
```
published_pipeline1 = pipeline1.publish(name="My_New_Pipeline", description="My Published Pipeline Description")
published_pipeline1
```
### Get published pipeline
You can get the published pipeline using **pipeline id**.
To get all the published pipelines for a given workspace(ws):
```css
all_pub_pipelines = PublishedPipeline.get_all(ws)
```
```
from azureml.pipeline.core import PublishedPipeline
pipeline_id = published_pipeline1.id # use your published pipeline id
published_pipeline = PublishedPipeline.get(ws, pipeline_id)
published_pipeline
```
### Run published pipeline using its REST endpoint
```
from azureml.core.authentication import InteractiveLoginAuthentication
import requests
auth = InteractiveLoginAuthentication()
aad_token = auth.get_authentication_header()
rest_endpoint1 = published_pipeline.endpoint
print("You can perform HTTP POST on URL {} to trigger this pipeline".format(rest_endpoint1))
# specify the param when running the pipeline
response = requests.post(rest_endpoint1,
headers=aad_token,
json={"ExperimentName": "My_Pipeline1",
"RunSource": "SDK",
"ParameterAssignments": {"pipeline_arg": 45}})
run_id = response.json()["Id"]
print(run_id)
```
# Next: Data Transfer
The next [notebook](./aml-pipelines-data-transfer.ipynb) will showcase data transfer steps between different types of data stores.
|
github_jupyter
|
import azureml.core
from azureml.core import Workspace, Datastore
from azureml.core.compute import AmlCompute
from azureml.core.compute import ComputeTarget
# Check core SDK version number
print("SDK version:", azureml.core.VERSION)
from azureml.data.data_reference import DataReference
from azureml.pipeline.core import Pipeline, PipelineData
from azureml.pipeline.steps import PythonScriptStep
from azureml.pipeline.core.graph import PipelineParameter
print("Pipeline SDK-specific imports completed")
ws = Workspace.from_config()
print(ws.name, ws.resource_group, ws.location, ws.subscription_id, sep = '\n')
# Default datastore (Azure file storage)
def_file_store = ws.get_default_datastore()
print("Default datastore's name: {}".format(def_file_store.name))
def_blob_store = Datastore(ws, "workspaceblobstore")
print("Blobstore's name: {}".format(def_blob_store.name))
# project folder
project_folder = '.'
from azureml.core.compute_target import ComputeTargetException
aml_compute_target = "cpucluster"
try:
aml_compute = AmlCompute(ws, aml_compute_target)
print("found existing compute target.")
except ComputeTargetException:
print("creating new compute target")
provisioning_config = AmlCompute.provisioning_configuration(vm_size = "STANDARD_D2_V2",
min_nodes = 1,
max_nodes = 4)
aml_compute = ComputeTarget.create(ws, aml_compute_target, provisioning_config)
aml_compute.wait_for_completion(show_output=True, min_node_count=None, timeout_in_minutes=20)
# For a more detailed view of current Azure Machine Learning Compute status, use get_status()
# example: un-comment the following line.
# print(aml_compute.get_status().serialize())
# Reference the data uploaded to blob storage using DataReference
# Assign the datasource to blob_input_data variable
blob_input_data = DataReference(
datastore=def_blob_store,
data_reference_name="test_data",
path_on_datastore="20newsgroups/20news.pkl")
print("DataReference object created")
# Define intermediate data using PipelineData
processed_data1 = PipelineData("processed_data1",datastore=def_blob_store)
print("PipelineData object created")
# trainStep consumes the datasource (Datareference) in the previous step
# and produces processed_data1
trainStep = PythonScriptStep(
script_name="train.py",
arguments=["--input_data", blob_input_data, "--output_train", processed_data1],
inputs=[blob_input_data],
outputs=[processed_data1],
compute_target=aml_compute,
source_directory=project_folder
)
print("trainStep created")
# extractStep to use the intermediate data produced by step4
# This step also produces an output processed_data2
processed_data2 = PipelineData("processed_data2", datastore=def_blob_store)
extractStep = PythonScriptStep(
script_name="extract.py",
arguments=["--input_extract", processed_data1, "--output_extract", processed_data2],
inputs=[processed_data1],
outputs=[processed_data2],
compute_target=aml_compute,
source_directory=project_folder)
print("extractStep created")
# We will use this later in publishing pipeline
pipeline_param = PipelineParameter(name="pipeline_arg", default_value=10)
print("pipeline parameter created")
# Now define step6 that takes two inputs (both intermediate data), and produce an output
processed_data3 = PipelineData("processed_data3", datastore=def_blob_store)
compareStep = PythonScriptStep(
script_name="compare.py",
arguments=["--compare_data1", processed_data1, "--compare_data2", processed_data2, "--output_compare", processed_data3, "--pipeline_param", pipeline_param],
inputs=[processed_data1, processed_data2],
outputs=[processed_data3],
compute_target=aml_compute,
source_directory=project_folder)
print("compareStep created")
pipeline1 = Pipeline(workspace=ws, steps=[compareStep])
print ("Pipeline is built")
pipeline1.validate()
print("Simple validation complete")
published_pipeline1 = pipeline1.publish(name="My_New_Pipeline", description="My Published Pipeline Description")
published_pipeline1
all_pub_pipelines = PublishedPipeline.get_all(ws)
from azureml.pipeline.core import PublishedPipeline
pipeline_id = published_pipeline1.id # use your published pipeline id
published_pipeline = PublishedPipeline.get(ws, pipeline_id)
published_pipeline
from azureml.core.authentication import InteractiveLoginAuthentication
import requests
auth = InteractiveLoginAuthentication()
aad_token = auth.get_authentication_header()
rest_endpoint1 = published_pipeline.endpoint
print("You can perform HTTP POST on URL {} to trigger this pipeline".format(rest_endpoint1))
# specify the param when running the pipeline
response = requests.post(rest_endpoint1,
headers=aad_token,
json={"ExperimentName": "My_Pipeline1",
"RunSource": "SDK",
"ParameterAssignments": {"pipeline_arg": 45}})
run_id = response.json()["Id"]
print(run_id)
| 0.402275 | 0.935228 |
# Exercícios Python e Numpy
O objetivo desse notebook é ajudar na fixação dos conteúdos que passamos na aula de Python e Numpy. Sabemos que acabamos passando meio rápido durante a aula, então o objetivo aqui é conseguir treinar os conceitos para você conseguir usa-los na prática mais tarde.
Qualquer dúvida pode ficar a vontade para perguntar no Slack ou diretamente para gente, vamos ficar felizes em ajudar :)
## Python
Aqui na parte de Python vamos passar por algumas das principais coisas que você precisa saber, é claro que não colocamos tudo de importante da linguagem, só o mais necessário.
### Variáveis
```
# Declare uma variavel chamada a e atribua o valor 10 a ela
a = 10
# Imprime essa variavel que você acabou de criar
print(a)
# Crie uma outra variavel b que recebe o valor de a so que como string
b = str(a)
# Combine sua variavel b a variavel abaixo para obter a string "Hello 10" em uma variavel d
c = "Hello "
d = c + b
# Imprima a variável d
print(d)
```
### Strings
```
my_str = 'Insira uma frase aqui!'
# Substitua a exclamação da frase por uma interrogação
# (Dica: A funcão altera inplace ou retorna uma copia modificada?)
my_new_str = my_str.replace('!', '?') # A funçao retorna uma cópia modificada
print(my_new_str)
# Crie uma lista "my_words" com cada palavra a frase
my_words = my_new_str.split()
print(my_words)
```
### Listas
```
lista = [1, 23, 31, 40, 56, 16]
# Faça um for que imprima cada elemento da lista "lista" (Lembre-se que o for em python é um for each)
for numero in lista:
print(numero)
# Faça um for que imprima o dobro de cada elemento da lista "lista"
for numero in lista:
print(2 * numero)
# Gere uma lista chamada "dobro" com o dobro de cada elemento de "lista" usando list comprehension
dobro = [2 * numero for numero in lista]
print(dobro)
# Crie uma nova lista chamada "pares"
# Faça um for que itetere sobre a lista "lista" e para cada elemento impar coloque ele no fim da lista "pares"
# Imprima a lista "pares"
pares = []
for numero in lista:
if numero % 2 == 0:
pares.append(numero)
print(pares)
lista2 = ['oi', 2, 2.5, 'top', 'python', 45]
# Faça um for pela "lista2" e imprima todos os elementos que são strings (Dica: pesquise pela função type())
for elemento in lista2:
if type(elemento) == str:
print(elemento)
```
#### Indexando
```
my_list = [0, 10, 20, 30, 40, 50, 60, 70]
# Selecione o ultimo elemento da lista
my_list[-1]
# Selecione do primeiro até o 4 elemento da lista
my_list[:4]
# Selecione do segundo elemento da lista até o quinto
my_list[1:5]
# Selecione do primeiro elemento da lista até o penultimo
my_list[:-1]
```
### Dicionários
```
lista = ['a', 'a', 'b', 'a', 'c', 'd', 'e', 'b', 'b', 'c']
# Crie um dicionario que contenha a contagem de cada elemento do vetor
my_dict = {}
for numero in lista:
if numero in my_dict:
my_dict[numero] += 1
else:
my_dict[numero] = 1
print(my_dict)
```
### Funções
```
# Crie uma função soma_elementos() que recebe uma lista e retorna a soma de todos os seus elementos
def soma_elementos(lista):
soma = 0
for numero in lista:
soma += numero
return soma
soma_elementos([1, 2, 3, 4, 5])
soma_elementos([-1, 5, 7, -2])
# Crie uma função produto_escalar() que recebe duas listas de tamanho igual e calcula o produto escalar entre elas
# Dica: Utilize a função zip
def produto_escalar(a, b):
produto_escalar = 0
for a_i, b_i in zip(a, b):
produto_escalar += (a_i * b_i)
return produto_escalar
produto_escalar([1, 2, 3], [0, 4, 7])
produto_escalar([10, 20, 40, 1], [23, 4, 2, 1])
# Crie uma função par_ou_impar() que recebe um numero n é para cada numero de 1 a n imprime o numero
# seguido de par ou impar, dependendo do que ele seja. Caso o usuário não coloque nada n deve valer 20
# Exemplo: par_ou_impar(4)
# 1 Impar
# 2 Par
# 3 Impar
# 4 Par
def par_ou_impar(n):
for i in range(1, n + 1):
if i % 2 == 0:
print(str(i) + ' Par')
else:
print(str(i) + ' Impar')
par_ou_impar(15)
# Crie uma função diga_indice() que recebe uma lista e imprime o indice de cada elemento e em seguida
# o proprio elemento
# Exemplo: diga_indice(['oi', 'tudo', 'bem'])
# 0 oi
# 1 tudo
# 2 bem
# (DICA: Pesquise pela função enumerate)
def diga_indice(lista):
for idx, elem in enumerate(lista):
print(idx, elem)
diga_indice(['1', '2', '3'])
diga_indice(['a', 'b', 'c', 'd', 'e'])
```
## Numpy
O elemento central do numpy são os arrays, então aqui vamos treinar muitas coisas sobre eles
```
# Importando a biblioteca
import numpy as np
```
### Arrays
```
a = np.array([1, 2, 3, 4, 5, 6, 7])
b = np.array([[1, 2, 3, 4],
[5, 6, 7, 8]])
c = np.zeros((3,4))
# Pense em qual é a shape de cada um dos arrays acima
# Depois de pensar imprima cada um deles para conferir sua resposta
print(a.shape)
print(b.shape)
print(c.shape)
# Crie um array de uma dimenção com 20 elementos inteiros aletórios entre 0 e 23
my_random_array = np.random.randint(24, size=(20))
print(my_random_array)
# Crie um array de uns com shape (4, 5)
my_array = np.ones((4, 5))
print(my_array)
# Crie um array shape (4, 2) onde cada entrada vale 77
# (Dica: Talvez vc tenha que usar uma multiplicação)
my_array = np.ones((4, 2)) * 77
print(my_array)
# Gere um array chamado my_sequence com os numeros 0, 10, 20, 30, ..., 90, 100
my_array = np.arange(0, 110, 10)
print(my_array)
```
### Indexando
```
my_array = np.random.randint(50, size=(15,))
print(my_array)
# Selecione todos os elementos entre o quinto e o decimo primeiro (intervalo fechado)
my_array[4:11]
# Selecione todos os elementos maiores que 20
my_array[my_array > 20]
my_matrix = np.array([[1, 2, 3, 4],
[5, 6, 7, 8],
[9, 10, 11, 12],
[13, 14, 15, 16]])
# Selecione o elemento na primeira linha da terceira coluna
my_matrix[0, 2]
# Selecione o elemento na primeira linha da ultima coluna
my_matrix[0, -1]
# Selecione os elementos da matriz para obter o seguinte
# [[6, 7],
# [10, 11]]
my_matrix[1:3, 1:3]
# Selecione os elementos da matriz para obter o seguinte
# [[2, 3, 4],
# [6, 7, 8]]
my_matrix[:2, 1:]
# Selecione os elementos da ultima coluna inteira
my_matrix[:, -1]
# Selecione os elementos da 2a linha inteira
my_matrix[1, :]
```
### Operações
```
my_array = np.random.randint(10, size=(5,))
print(my_array)
# Some 10 a todos os elementos de my_array
my_array_plus10 = my_array + 10
print(my_array_plus10)
# Multiplique todos os elementos de my_array por 4
my_array_times4 = my_array * 4
print(my_array_times4)
# Obtenha a soma de todos os elementos de my_array
soma = np.sum(my_array)
print(soma)
# Obtenha a média de todos os elementos de my_array
media = np.mean(my_array)
print(media)
# Obtenha o indice do maior elemento de my_array
idx_maior = np.argmax(my_array)
print(idx_maior)
my_array = np.random.randint(10, size=(5,))
my_other_array = np.random.randint(10, size=(10,5))
print(my_array, '\n')
print(my_other_array)
# Some my_array elemento por elemento em cada linha de my_other_array
result_array = my_other_array + my_array
print(result_array)
my_array = np.random.randint(10, size=(5,4))
my_other_array = np.random.randint(10, size=(10,5))
print(my_array, '\n')
print(my_other_array)
# Faça a multiplicação entre my_other_array e my_array
mult_array = np.dot(my_other_array, my_array)
print(mult_array)
# Descubra a soma dos valores de cada linha de my_other_array
# (Dica: Pesquise sobre o atributo axis da função de soma)
soma_linhas = np.sum(my_other_array, axis=1)
print(soma_linhas)
my_array = np.random.randint(10, size=(5,4))
print(my_array)
# Usando reshape transforme a matriz acima em um vetor (Concatendo a linha de baixo na de cima
my_array_vetor = my_array.reshape(-1)
print(my_array_vetor)
np.array([[ 0, 1, 2, 3, 4, 5, 6, 7],
[ 8, 9, 10, 11, 12, 13, 14, 15]])
# Gere a array anterior usando np.arange e a função reshape
sequencia = np.arange(16)
resultado = sequencia.reshape(2, -1)
print(resultado)
```
|
github_jupyter
|
# Declare uma variavel chamada a e atribua o valor 10 a ela
a = 10
# Imprime essa variavel que você acabou de criar
print(a)
# Crie uma outra variavel b que recebe o valor de a so que como string
b = str(a)
# Combine sua variavel b a variavel abaixo para obter a string "Hello 10" em uma variavel d
c = "Hello "
d = c + b
# Imprima a variável d
print(d)
my_str = 'Insira uma frase aqui!'
# Substitua a exclamação da frase por uma interrogação
# (Dica: A funcão altera inplace ou retorna uma copia modificada?)
my_new_str = my_str.replace('!', '?') # A funçao retorna uma cópia modificada
print(my_new_str)
# Crie uma lista "my_words" com cada palavra a frase
my_words = my_new_str.split()
print(my_words)
lista = [1, 23, 31, 40, 56, 16]
# Faça um for que imprima cada elemento da lista "lista" (Lembre-se que o for em python é um for each)
for numero in lista:
print(numero)
# Faça um for que imprima o dobro de cada elemento da lista "lista"
for numero in lista:
print(2 * numero)
# Gere uma lista chamada "dobro" com o dobro de cada elemento de "lista" usando list comprehension
dobro = [2 * numero for numero in lista]
print(dobro)
# Crie uma nova lista chamada "pares"
# Faça um for que itetere sobre a lista "lista" e para cada elemento impar coloque ele no fim da lista "pares"
# Imprima a lista "pares"
pares = []
for numero in lista:
if numero % 2 == 0:
pares.append(numero)
print(pares)
lista2 = ['oi', 2, 2.5, 'top', 'python', 45]
# Faça um for pela "lista2" e imprima todos os elementos que são strings (Dica: pesquise pela função type())
for elemento in lista2:
if type(elemento) == str:
print(elemento)
my_list = [0, 10, 20, 30, 40, 50, 60, 70]
# Selecione o ultimo elemento da lista
my_list[-1]
# Selecione do primeiro até o 4 elemento da lista
my_list[:4]
# Selecione do segundo elemento da lista até o quinto
my_list[1:5]
# Selecione do primeiro elemento da lista até o penultimo
my_list[:-1]
lista = ['a', 'a', 'b', 'a', 'c', 'd', 'e', 'b', 'b', 'c']
# Crie um dicionario que contenha a contagem de cada elemento do vetor
my_dict = {}
for numero in lista:
if numero in my_dict:
my_dict[numero] += 1
else:
my_dict[numero] = 1
print(my_dict)
# Crie uma função soma_elementos() que recebe uma lista e retorna a soma de todos os seus elementos
def soma_elementos(lista):
soma = 0
for numero in lista:
soma += numero
return soma
soma_elementos([1, 2, 3, 4, 5])
soma_elementos([-1, 5, 7, -2])
# Crie uma função produto_escalar() que recebe duas listas de tamanho igual e calcula o produto escalar entre elas
# Dica: Utilize a função zip
def produto_escalar(a, b):
produto_escalar = 0
for a_i, b_i in zip(a, b):
produto_escalar += (a_i * b_i)
return produto_escalar
produto_escalar([1, 2, 3], [0, 4, 7])
produto_escalar([10, 20, 40, 1], [23, 4, 2, 1])
# Crie uma função par_ou_impar() que recebe um numero n é para cada numero de 1 a n imprime o numero
# seguido de par ou impar, dependendo do que ele seja. Caso o usuário não coloque nada n deve valer 20
# Exemplo: par_ou_impar(4)
# 1 Impar
# 2 Par
# 3 Impar
# 4 Par
def par_ou_impar(n):
for i in range(1, n + 1):
if i % 2 == 0:
print(str(i) + ' Par')
else:
print(str(i) + ' Impar')
par_ou_impar(15)
# Crie uma função diga_indice() que recebe uma lista e imprime o indice de cada elemento e em seguida
# o proprio elemento
# Exemplo: diga_indice(['oi', 'tudo', 'bem'])
# 0 oi
# 1 tudo
# 2 bem
# (DICA: Pesquise pela função enumerate)
def diga_indice(lista):
for idx, elem in enumerate(lista):
print(idx, elem)
diga_indice(['1', '2', '3'])
diga_indice(['a', 'b', 'c', 'd', 'e'])
# Importando a biblioteca
import numpy as np
a = np.array([1, 2, 3, 4, 5, 6, 7])
b = np.array([[1, 2, 3, 4],
[5, 6, 7, 8]])
c = np.zeros((3,4))
# Pense em qual é a shape de cada um dos arrays acima
# Depois de pensar imprima cada um deles para conferir sua resposta
print(a.shape)
print(b.shape)
print(c.shape)
# Crie um array de uma dimenção com 20 elementos inteiros aletórios entre 0 e 23
my_random_array = np.random.randint(24, size=(20))
print(my_random_array)
# Crie um array de uns com shape (4, 5)
my_array = np.ones((4, 5))
print(my_array)
# Crie um array shape (4, 2) onde cada entrada vale 77
# (Dica: Talvez vc tenha que usar uma multiplicação)
my_array = np.ones((4, 2)) * 77
print(my_array)
# Gere um array chamado my_sequence com os numeros 0, 10, 20, 30, ..., 90, 100
my_array = np.arange(0, 110, 10)
print(my_array)
my_array = np.random.randint(50, size=(15,))
print(my_array)
# Selecione todos os elementos entre o quinto e o decimo primeiro (intervalo fechado)
my_array[4:11]
# Selecione todos os elementos maiores que 20
my_array[my_array > 20]
my_matrix = np.array([[1, 2, 3, 4],
[5, 6, 7, 8],
[9, 10, 11, 12],
[13, 14, 15, 16]])
# Selecione o elemento na primeira linha da terceira coluna
my_matrix[0, 2]
# Selecione o elemento na primeira linha da ultima coluna
my_matrix[0, -1]
# Selecione os elementos da matriz para obter o seguinte
# [[6, 7],
# [10, 11]]
my_matrix[1:3, 1:3]
# Selecione os elementos da matriz para obter o seguinte
# [[2, 3, 4],
# [6, 7, 8]]
my_matrix[:2, 1:]
# Selecione os elementos da ultima coluna inteira
my_matrix[:, -1]
# Selecione os elementos da 2a linha inteira
my_matrix[1, :]
my_array = np.random.randint(10, size=(5,))
print(my_array)
# Some 10 a todos os elementos de my_array
my_array_plus10 = my_array + 10
print(my_array_plus10)
# Multiplique todos os elementos de my_array por 4
my_array_times4 = my_array * 4
print(my_array_times4)
# Obtenha a soma de todos os elementos de my_array
soma = np.sum(my_array)
print(soma)
# Obtenha a média de todos os elementos de my_array
media = np.mean(my_array)
print(media)
# Obtenha o indice do maior elemento de my_array
idx_maior = np.argmax(my_array)
print(idx_maior)
my_array = np.random.randint(10, size=(5,))
my_other_array = np.random.randint(10, size=(10,5))
print(my_array, '\n')
print(my_other_array)
# Some my_array elemento por elemento em cada linha de my_other_array
result_array = my_other_array + my_array
print(result_array)
my_array = np.random.randint(10, size=(5,4))
my_other_array = np.random.randint(10, size=(10,5))
print(my_array, '\n')
print(my_other_array)
# Faça a multiplicação entre my_other_array e my_array
mult_array = np.dot(my_other_array, my_array)
print(mult_array)
# Descubra a soma dos valores de cada linha de my_other_array
# (Dica: Pesquise sobre o atributo axis da função de soma)
soma_linhas = np.sum(my_other_array, axis=1)
print(soma_linhas)
my_array = np.random.randint(10, size=(5,4))
print(my_array)
# Usando reshape transforme a matriz acima em um vetor (Concatendo a linha de baixo na de cima
my_array_vetor = my_array.reshape(-1)
print(my_array_vetor)
np.array([[ 0, 1, 2, 3, 4, 5, 6, 7],
[ 8, 9, 10, 11, 12, 13, 14, 15]])
# Gere a array anterior usando np.arange e a função reshape
sequencia = np.arange(16)
resultado = sequencia.reshape(2, -1)
print(resultado)
| 0.203391 | 0.929055 |
# Task 1: Instructions
Import the library you need to work with DataFrames, and load the two datasets (all datasets are located in the datasets folder). Then, take a look at the head of both DataFrames.
- Import `pandas` using the standard alias.
- Load `office_addresses.csv` and assign the resulting DataFrame to `df_office_addresses`.
- Load `employee_information.xls` and assign the resulting DataFrame to `df_employee_addresses`.
- Take a look at the first rows of each DataFrame to familiarize yourself with the data.
## Good to know
This project lets you apply the skills from [Streamlined Data Ingestion in Python](https://www.datacamp.com/courses/streamlined-data-ingestion-with-pandas). We recommend that you are familiar with the content in that course before starting this project.
Helpful links:
- `read_csv()` function [documentation](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.read_csv.html).
- `read_excel()` function [documentation](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.read_excel.html).
# Task 2: Instructions
People apparently remove titles or headers. Make sure to load the sheet using its index rather than its name, in case someone decides to rename it! Then, add the appropriate column titles to the DataFrame.
- Load the data from the second sheet of `employee_information.xls` and assign the resulting DataFrame to `df_emergency_contact`.
- Assign the list of column names to `emergency_contacts_header`.
- Rename the `df_emergency_contact` DataFrame's columns using the list of column names you just declared.
- Take a look at the first rows of the DataFrame to familiarize yourself with the data.
# Task 3: Instructions
`employee_roles.json` is built as a Python dictionary: the keys are employee IDs, and each employee ID has a corresponding dictionary value holding role, salary, and team information.
- Load the JSON file to a variable `df_employee_roles`, choosing the appropriate orientation.
- Take a look at the first rows of the DataFrame to familiarize yourself with the data.
When reading a JSON file, you need to tell `pandas` how the file is [oriented](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.read_json.html). If you don't choose the appropriate orientation, your index might become columns, and the columns might become indexes. Check out [this exercise](https://campus.datacamp.com/courses/streamlined-data-ingestion-with-pandas/importing-json-data-and-working-with-apis?ex=3) if you don't remember how to read JSON files.
`read_json()` uses Python dictionaries, which are unordered. Notice the provided line of code: it reorders the columns of the DataFrame you just created alphabetically so you don't run into issues later.
# Task 4: Instructions
Let's merge all DataFrames into one, as initially requested by People Ops! You will be using a left join here, which ensures you keep all the records in the left table. This is done for you and ensures you're not losing any data during the manipulations.
- Merge `df_emergency_contacts` with `df_employee_addresses` using the employee ID. Assign the resulting DataFrame to `df_employees`.
- Merge `df_employee_roles` with `df_employees` using the employee ID.
- Merge `df_office_addresses` with `df_employees` using the country.
- Take a look at the first rows and at the columns (you should not have any duplicate column).
Remember that:
- `df_office_addresses` holds the office addresses.
- `df_empoyee_addresses` holds the employee addresses.
- `df_emergency_contacts` holds the emergency contact information.
- `df_employee_roles` holds more information about employee's roles and education.
# Task 5: Instructions
Let's polish this new `df_employees` DataFrame!
- The columns `employee_first_name` and `employee_last_name` are duplicates of `first_name` and `last_name`. Drop `employee_first_name` and `employee_last_name` and assign the resulting DataFrame to `df_employees_renamed`.
- Assign the list of new column names to `new_header`.
- Rename the columns of `df_employee_renamed` using the `new_header` list.
- Take a look at the first rows of the DataFrame.
You can rename a DataFrame's columns by assigning a list of strings to the DataFrame `columns` attribute.
# Task 6: Instructions
People Ops requested columns to be presented in the following order: `id`, `last_name`, `first_name`, `title`, `team`, `monthly_salary`, `country`, `city`, `street`, `street_number`, `emergency_contact`, `emergency_number`, `emergency_relationship`, `office`, `office_country`, `office_city`, `office_street`, and finally `office_street_number`.
- Declare a list storing the column names ordered as specific by People Ops.
- Reorder the DataFrame's columns.
- Take a look at the result.
You can reorder a DataFrame by passing it a selection of columns in the order you wish.
# Task 7: Instructions
Let's bring these last-minute changes to our DataFrame.
- Set the index of `employees_ordered` to be the employee ID, and then drop the corresponding column.
- Loop through the rows of your new DataFrame, appending the value "Remote" to `status_list` if the `"office"` column value is null and "On-site" otherwise.
- Insert the `status_list` values as a column named "status" right after the `"monthly_salary"` column.
- Take a look at your results.
- You can [loop through a DataFrame](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.DataFrame.iterrows.html).
- You need to specify where to insert a column, what its name should be, and set its values using a list of predefined values.
- You can [check if a column value is null or not](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.isnull.html).
# Task 8: Instructions
Let's save your work!
- Write `df_employees_final` to a CSV file named "employee_data.csv" directly in the folder where your notebook is stored.
There's a [function for everything](https://pandas.pydata.org/docs/reference/api/pandas.DataFrame.to_csv.html) in pandas.
|
github_jupyter
|
# Task 1: Instructions
Import the library you need to work with DataFrames, and load the two datasets (all datasets are located in the datasets folder). Then, take a look at the head of both DataFrames.
- Import `pandas` using the standard alias.
- Load `office_addresses.csv` and assign the resulting DataFrame to `df_office_addresses`.
- Load `employee_information.xls` and assign the resulting DataFrame to `df_employee_addresses`.
- Take a look at the first rows of each DataFrame to familiarize yourself with the data.
## Good to know
This project lets you apply the skills from [Streamlined Data Ingestion in Python](https://www.datacamp.com/courses/streamlined-data-ingestion-with-pandas). We recommend that you are familiar with the content in that course before starting this project.
Helpful links:
- `read_csv()` function [documentation](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.read_csv.html).
- `read_excel()` function [documentation](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.read_excel.html).
# Task 2: Instructions
People apparently remove titles or headers. Make sure to load the sheet using its index rather than its name, in case someone decides to rename it! Then, add the appropriate column titles to the DataFrame.
- Load the data from the second sheet of `employee_information.xls` and assign the resulting DataFrame to `df_emergency_contact`.
- Assign the list of column names to `emergency_contacts_header`.
- Rename the `df_emergency_contact` DataFrame's columns using the list of column names you just declared.
- Take a look at the first rows of the DataFrame to familiarize yourself with the data.
# Task 3: Instructions
`employee_roles.json` is built as a Python dictionary: the keys are employee IDs, and each employee ID has a corresponding dictionary value holding role, salary, and team information.
- Load the JSON file to a variable `df_employee_roles`, choosing the appropriate orientation.
- Take a look at the first rows of the DataFrame to familiarize yourself with the data.
When reading a JSON file, you need to tell `pandas` how the file is [oriented](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.read_json.html). If you don't choose the appropriate orientation, your index might become columns, and the columns might become indexes. Check out [this exercise](https://campus.datacamp.com/courses/streamlined-data-ingestion-with-pandas/importing-json-data-and-working-with-apis?ex=3) if you don't remember how to read JSON files.
`read_json()` uses Python dictionaries, which are unordered. Notice the provided line of code: it reorders the columns of the DataFrame you just created alphabetically so you don't run into issues later.
# Task 4: Instructions
Let's merge all DataFrames into one, as initially requested by People Ops! You will be using a left join here, which ensures you keep all the records in the left table. This is done for you and ensures you're not losing any data during the manipulations.
- Merge `df_emergency_contacts` with `df_employee_addresses` using the employee ID. Assign the resulting DataFrame to `df_employees`.
- Merge `df_employee_roles` with `df_employees` using the employee ID.
- Merge `df_office_addresses` with `df_employees` using the country.
- Take a look at the first rows and at the columns (you should not have any duplicate column).
Remember that:
- `df_office_addresses` holds the office addresses.
- `df_empoyee_addresses` holds the employee addresses.
- `df_emergency_contacts` holds the emergency contact information.
- `df_employee_roles` holds more information about employee's roles and education.
# Task 5: Instructions
Let's polish this new `df_employees` DataFrame!
- The columns `employee_first_name` and `employee_last_name` are duplicates of `first_name` and `last_name`. Drop `employee_first_name` and `employee_last_name` and assign the resulting DataFrame to `df_employees_renamed`.
- Assign the list of new column names to `new_header`.
- Rename the columns of `df_employee_renamed` using the `new_header` list.
- Take a look at the first rows of the DataFrame.
You can rename a DataFrame's columns by assigning a list of strings to the DataFrame `columns` attribute.
# Task 6: Instructions
People Ops requested columns to be presented in the following order: `id`, `last_name`, `first_name`, `title`, `team`, `monthly_salary`, `country`, `city`, `street`, `street_number`, `emergency_contact`, `emergency_number`, `emergency_relationship`, `office`, `office_country`, `office_city`, `office_street`, and finally `office_street_number`.
- Declare a list storing the column names ordered as specific by People Ops.
- Reorder the DataFrame's columns.
- Take a look at the result.
You can reorder a DataFrame by passing it a selection of columns in the order you wish.
# Task 7: Instructions
Let's bring these last-minute changes to our DataFrame.
- Set the index of `employees_ordered` to be the employee ID, and then drop the corresponding column.
- Loop through the rows of your new DataFrame, appending the value "Remote" to `status_list` if the `"office"` column value is null and "On-site" otherwise.
- Insert the `status_list` values as a column named "status" right after the `"monthly_salary"` column.
- Take a look at your results.
- You can [loop through a DataFrame](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.DataFrame.iterrows.html).
- You need to specify where to insert a column, what its name should be, and set its values using a list of predefined values.
- You can [check if a column value is null or not](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.isnull.html).
# Task 8: Instructions
Let's save your work!
- Write `df_employees_final` to a CSV file named "employee_data.csv" directly in the folder where your notebook is stored.
There's a [function for everything](https://pandas.pydata.org/docs/reference/api/pandas.DataFrame.to_csv.html) in pandas.
| 0.912944 | 0.981185 |
<a href="https://colab.research.google.com/github/kevinpulido89/ML/blob/master/Time_series.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
##### Copyright 2019 The TensorFlow Authors.
# Time series forecasting
This tutorial is an introduction to time series forecasting using Recurrent Neural Networks (RNNs). This is covered in two parts: first, you will forecast a univariate time series, then you will forecast a multivariate time series.
```
import tensorflow as tf
import matplotlib as mpl
import matplotlib.pyplot as plt
import numpy as np
import os
import pandas as pd
mpl.rcParams['figure.figsize'] = (8, 6)
mpl.rcParams['axes.grid'] = False
```
## The weather dataset
This tutorial uses a <a href="https://www.bgc-jena.mpg.de/wetter/" class="external">[weather time series dataset</a> recorded by the <a href="https://www.bgc-jena.mpg.de" class="external">Max Planck Institute for Biogeochemistry</a>.
This dataset contains 14 different features such as air temperature, atmospheric pressure, and humidity. These were collected every 10 minutes, beginning in 2003. For efficiency, you will use only the data collected between 2009 and 2016. This section of the dataset was prepared by François Chollet for his book [Deep Learning with Python](https://www.manning.com/books/deep-learning-with-python).
```
zip_path = tf.keras.utils.get_file(
origin='https://storage.googleapis.com/tensorflow/tf-keras-datasets/jena_climate_2009_2016.csv.zip',
fname='jena_climate_2009_2016.csv.zip',
extract=True)
csv_path, _ = os.path.splitext(zip_path)
df = pd.read_csv(csv_path)
```
Let's take a glance at the data.
```
df.head()
```
As you can see above, an observation is recorded every 10 mintues. This means that, for a single hour, you will have 6 observations. Similarly, a single day will contain 144 (6x24) observations.
Given a specific time, let's say you want to predict the temperature 6 hours in the future. In order to make this prediction, you choose to use 5 days of observations. Thus, you would create a window containing the last 720(5x144) observations to train the model. Many such configurations are possible, making this dataset a good one to experiment with.
The function below returns the above described windows of time for the model to train on. The parameter `history_size` is the size of the past window of information. The `target_size` is how far in the future does the model need to learn to predict. The `target_size` is the label that needs to be predicted.
```
def univariate_data(dataset, start_index, end_index, history_size, target_size):
data = []
labels = []
start_index = start_index + history_size
if end_index is None:
end_index = len(dataset) - target_size
for i in range(start_index, end_index):
indices = range(i-history_size, i)
# Reshape data from (history_size,) to (history_size, 1)
data.append(np.reshape(dataset[indices], (history_size, 1)))
labels.append(dataset[i+target_size])
return np.array(data), np.array(labels)
```
In both the following tutorials, the first 300,000 rows of the data will be the training dataset, and there remaining will be the validation dataset. This amounts to ~2100 days worth of training data.
```
TRAIN_SPLIT = 300000
```
Setting seed to ensure reproducibility.
```
tf.random.set_seed(13)
```
## Part 1: Forecast a univariate time series
First, you will train a model using only a single feature (temperature), and use it to make predictions for that value in the future.
Let's first extract only the temperature from the dataset.
```
uni_data = df['T (degC)']
uni_data.index = df['Date Time']
uni_data.head()
```
Let's observe how this data looks across time.
```
uni_data.plot(subplots=True)
uni_data = uni_data.values
```
It is important to scale features before training a neural network. Standardization is a common way of doing this scaling by subtracting the mean and dividing by the standard deviation of each feature.You could also use a `tf.keras.utils.normalize` method that rescales the values into a range of [0,1].
Note: The mean and standard deviation should only be computed using the training data.
```
uni_train_mean = uni_data[:TRAIN_SPLIT].mean()
uni_train_std = uni_data[:TRAIN_SPLIT].std()
```
Let's standardize the data.
```
uni_data = (uni_data-uni_train_mean)/uni_train_std
```
Let's now create the data for the univariate model. For part 1, the model will be given the last 20 recorded temperature observations, and needs to learn to predict the temperature at the next time step.
```
univariate_past_history = 20
univariate_future_target = 0
x_train_uni, y_train_uni = univariate_data(uni_data, 0, TRAIN_SPLIT,
univariate_past_history,
univariate_future_target)
x_val_uni, y_val_uni = univariate_data(uni_data, TRAIN_SPLIT, None,
univariate_past_history,
univariate_future_target)
```
This is what the `univariate_data` function returns.
```
print ('Single window of past history')
print (x_train_uni[0])
print ('\n Target temperature to predict')
print (y_train_uni[0])
```
Now that the data has been created, let's take a look at a single example. The information given to the network is given in blue, and it must predict the value at the red cross.
```
def create_time_steps(length):
return list(range(-length, 0))
def show_plot(plot_data, delta, title):
labels = ['History', 'True Future', 'Model Prediction']
marker = ['.-', 'rx', 'go']
time_steps = create_time_steps(plot_data[0].shape[0])
if delta:
future = delta
else:
future = 0
plt.title(title)
for i, x in enumerate(plot_data):
if i:
plt.plot(future, plot_data[i], marker[i], markersize=10,
label=labels[i])
else:
plt.plot(time_steps, plot_data[i].flatten(), marker[i], label=labels[i])
plt.legend()
plt.xlim([time_steps[0], (future+5)*2])
plt.xlabel('Time-Step')
return plt
show_plot([x_train_uni[0], y_train_uni[0]], 0, 'Sample Example')
```
### Baseline
Before proceeding to train a model, let's first set a simple baseline. Given an input point, the baseline method looks at all the history and predicts the next point to be the average of the last 20 observations.
```
def baseline(history):
return np.mean(history)
show_plot([x_train_uni[0], y_train_uni[0], baseline(x_train_uni[0])], 0,
'Baseline Prediction Example')
```
Let's see if you can beat this baseline using a recurrent neural network.
### Recurrent neural network
A Recurrent Neural Network (RNN) is a type of neural network well-suited to time series data. RNNs process a time series step-by-step, maintaining an internal state summarizing the information they've seen so far. For more details, read the [RNN tutorial](https://www.tensorflow.org/tutorials/sequences/recurrent). In this tutorial, you will use a specialized RNN layer called Long Short Term Memory ([LSTM](https://www.tensorflow.org/versions/r2.0/api_docs/python/tf/keras/layers/LSTM))
Let's now use `tf.data` to shuffle, batch, and cache the dataset.
```
BATCH_SIZE = 256
BUFFER_SIZE = 10000
train_univariate = tf.data.Dataset.from_tensor_slices((x_train_uni, y_train_uni))
train_univariate = train_univariate.cache().shuffle(BUFFER_SIZE).batch(BATCH_SIZE).repeat()
val_univariate = tf.data.Dataset.from_tensor_slices((x_val_uni, y_val_uni))
val_univariate = val_univariate.batch(BATCH_SIZE).repeat()
```
The following visualisation should help you understand how the data is represented after batching.

You will see the LSTM requires the input shape of the data it is being given.
```
simple_lstm_model = tf.keras.models.Sequential([
tf.keras.layers.LSTM(8, input_shape=x_train_uni.shape[-2:]),
tf.keras.layers.Dense(1)
])
simple_lstm_model.compile(optimizer='adam', loss='mae')
```
Let's make a sample prediction, to check the output of the model.
```
for x, y in val_univariate.take(1):
print(simple_lstm_model.predict(x).shape)
```
Let's train the model now. Due to the large size of the dataset, in the interest of saving time, each epoch will only run for 200 steps, instead of the complete training data as normally done.
```
EVALUATION_INTERVAL = 200
EPOCHS = 10
simple_lstm_model.fit(train_univariate, epochs=EPOCHS,
steps_per_epoch=EVALUATION_INTERVAL,
validation_data=val_univariate, validation_steps=50)
```
#### Predict using the simple LSTM model
Now that you have trained your simple LSTM, let's try and make a few predictions.
```
for x, y in val_univariate.take(3):
plot = show_plot([x[0].numpy(), y[0].numpy(),
simple_lstm_model.predict(x)[0]], 0, 'Simple LSTM model')
plot.show()
```
This looks better than the baseline. Now that you have seen the basics, let's move on to part two, where you will work with a multivariate time series.
## Part 2: Forecast a multivariate time series
The original dataset contains fourteen features. For simplicity, this section considers only three of the original fourteen. The features used are air temperature, atmospheric pressure, and air density.
To use more features, add their names to this list.
```
features_considered = ['p (mbar)', 'T (degC)', 'rho (g/m**3)']
features = df[features_considered]
features.index = df['Date Time']
features.head()
```
Let's have a look at how each of these features vary across time.
```
features.plot(subplots=True)
```
As mentioned, the first step will be to standardize the dataset using the mean and standard deviation of the training data.
```
dataset = features.values
data_mean = dataset[:TRAIN_SPLIT].mean(axis=0)
data_std = dataset[:TRAIN_SPLIT].std(axis=0)
dataset = (dataset-data_mean)/data_std
```
### Single step model
In a single step setup, the model learns to predict a single point in the future based on some history provided.
The below function performs the same windowing task as below, however, here it samples the past observation based on the step size given.
```
def multivariate_data(dataset, target, start_index, end_index, history_size,
target_size, step, single_step=False):
data = []
labels = []
start_index = start_index + history_size
if end_index is None:
end_index = len(dataset) - target_size
for i in range(start_index, end_index):
indices = range(i-history_size, i, step)
data.append(dataset[indices])
if single_step:
labels.append(target[i+target_size])
else:
labels.append(target[i:i+target_size])
return np.array(data), np.array(labels)
```
In this tutorial, the network is shown data from the last five (5) days, i.e. 720 observations that are sampled every hour. The sampling is done every one hour since a drastic change is not expected within 60 minutes. Thus, 120 observation represent history of the last five days. For the single step prediction model, the label for a datapoint is the temperature 12 hours into the future. In order to create a label for this, the temperature after 72(12*6) observations is used.
```
past_history = 720
future_target = 72
STEP = 6
x_train_single, y_train_single = multivariate_data(dataset, dataset[:, 1], 0,
TRAIN_SPLIT, past_history,
future_target, STEP,
single_step=True)
x_val_single, y_val_single = multivariate_data(dataset, dataset[:, 1],
TRAIN_SPLIT, None, past_history,
future_target, STEP,
single_step=True)
```
Let's look at a single data-point.
```
print ('Single window of past history : {}'.format(x_train_single[0].shape))
train_data_single = tf.data.Dataset.from_tensor_slices((x_train_single, y_train_single))
train_data_single = train_data_single.cache().shuffle(BUFFER_SIZE).batch(BATCH_SIZE).repeat()
val_data_single = tf.data.Dataset.from_tensor_slices((x_val_single, y_val_single))
val_data_single = val_data_single.batch(BATCH_SIZE).repeat()
single_step_model = tf.keras.models.Sequential()
single_step_model.add(tf.keras.layers.LSTM(32,
input_shape=x_train_single.shape[-2:]))
single_step_model.add(tf.keras.layers.Dense(1))
single_step_model.compile(optimizer=tf.keras.optimizers.RMSprop(), loss='mae')
```
Let's check out a sample prediction.
```
for x, y in val_data_single.take(1):
print(single_step_model.predict(x).shape)
single_step_history = single_step_model.fit(train_data_single, epochs=EPOCHS,
steps_per_epoch=EVALUATION_INTERVAL,
validation_data=val_data_single,
validation_steps=50)
def plot_train_history(history, title):
loss = history.history['loss']
val_loss = history.history['val_loss']
epochs = range(len(loss))
plt.figure()
plt.plot(epochs, loss, 'b', label='Training loss')
plt.plot(epochs, val_loss, 'r', label='Validation loss')
plt.title(title)
plt.legend()
plt.show()
plot_train_history(single_step_history,
'Single Step Training and validation loss')
```
#### Predict a single step future
Now that the model is trained, let's make a few sample predictions. The model is given the history of three features over the past five days sampled every hour (120 data-points), since the goal is to predict the temperature, the plot only displays the past temperature. The prediction is made one day into the future (hence the gap between the history and prediction).
```
for x, y in val_data_single.take(3):
plot = show_plot([x[0][:, 1].numpy(), y[0].numpy(),
single_step_model.predict(x)[0]], 12,
'Single Step Prediction')
plot.show()
```
### Multi-Step model
In a multi-step prediction model, given a past history, the model needs to learn to predict a range of future values. Thus, unlike a single step model, where only a single future point is predicted, a multi-step model predict a sequence of the future.
For the multi-step model, the training data again consists of recordings over the past five days sampled every hour. However, here, the model needs to learn to predict the temperature for the next 12 hours. Since an obversation is taken every 10 minutes, the output is 72 predictions. For this task, the dataset needs to be prepared accordingly, thus the first step is just to create it again, but with a different target window.
```
future_target = 72
x_train_multi, y_train_multi = multivariate_data(dataset, dataset[:, 1], 0,
TRAIN_SPLIT, past_history,
future_target, STEP)
x_val_multi, y_val_multi = multivariate_data(dataset, dataset[:, 1],
TRAIN_SPLIT, None, past_history,
future_target, STEP)
```
Let's check out a sample data-point.
```
print ('Single window of past history : {}'.format(x_train_multi[0].shape))
print ('\n Target temperature to predict : {}'.format(y_train_multi[0].shape))
train_data_multi = tf.data.Dataset.from_tensor_slices((x_train_multi, y_train_multi))
train_data_multi = train_data_multi.cache().shuffle(BUFFER_SIZE).batch(BATCH_SIZE).repeat()
val_data_multi = tf.data.Dataset.from_tensor_slices((x_val_multi, y_val_multi))
val_data_multi = val_data_multi.batch(BATCH_SIZE).repeat()
```
Plotting a sample data-point.
```
def multi_step_plot(history, true_future, prediction):
plt.figure(figsize=(12, 6))
num_in = create_time_steps(len(history))
num_out = len(true_future)
plt.plot(num_in, np.array(history[:, 1]), label='History')
plt.plot(np.arange(num_out)/STEP, np.array(true_future), 'bo',
label='True Future')
if prediction.any():
plt.plot(np.arange(num_out)/STEP, np.array(prediction), 'ro',
label='Predicted Future')
plt.legend(loc='upper left')
plt.show()
```
In this plot and subsequent similar plots, the history and the future data are sampled every hour.
```
for x, y in train_data_multi.take(1):
multi_step_plot(x[0], y[0], np.array([0]))
```
Since the task here is a bit more complicated than the previous task, the model now consists of two LSTM layers. Finally, since 72 predictions are made, the dense layer outputs 72 predictions.
```
multi_step_model = tf.keras.models.Sequential()
multi_step_model.add(tf.keras.layers.LSTM(32,
return_sequences=True,
input_shape=x_train_multi.shape[-2:]))
multi_step_model.add(tf.keras.layers.LSTM(16, activation='relu'))
multi_step_model.add(tf.keras.layers.Dense(72))
multi_step_model.compile(optimizer=tf.keras.optimizers.RMSprop(clipvalue=1.0), loss='mae')
```
Let's see how the model predicts before it trains.
```
for x, y in val_data_multi.take(1):
print (multi_step_model.predict(x).shape)
multi_step_history = multi_step_model.fit(train_data_multi, epochs=EPOCHS,
steps_per_epoch=EVALUATION_INTERVAL,
validation_data=val_data_multi,
validation_steps=50)
plot_train_history(multi_step_history, 'Multi-Step Training and validation loss')
```
#### Predict a multi-step future
Let's now have a look at how well your network has learnt to predict the future.
```
for x, y in val_data_multi.take(3):
multi_step_plot(x[0], y[0], multi_step_model.predict(x)[0])
```
## Next steps
This tutorial was a quick introduction to time series forecasting using an RNN. You may now try to predict the stock market and become a billionaire.
In addition, you may also write a generator to yield data (instead of the uni/multivariate_data function), which would be more memory efficient. You may also check out this [time series windowing](https://www.tensorflow.org/guide/data#time_series_windowing) guide and use it in this tutorial.
For further understanding, you may read Chapter 15 of [Hands-on Machine Learning with Scikit-Learn, Keras, and TensorFlow](https://www.oreilly.com/library/view/hands-on-machine-learning/9781492032632/), 2nd Edition and Chapter 6 of [Deep Learning with Python](https://www.manning.com/books/deep-learning-with-python).
|
github_jupyter
|
import tensorflow as tf
import matplotlib as mpl
import matplotlib.pyplot as plt
import numpy as np
import os
import pandas as pd
mpl.rcParams['figure.figsize'] = (8, 6)
mpl.rcParams['axes.grid'] = False
zip_path = tf.keras.utils.get_file(
origin='https://storage.googleapis.com/tensorflow/tf-keras-datasets/jena_climate_2009_2016.csv.zip',
fname='jena_climate_2009_2016.csv.zip',
extract=True)
csv_path, _ = os.path.splitext(zip_path)
df = pd.read_csv(csv_path)
df.head()
def univariate_data(dataset, start_index, end_index, history_size, target_size):
data = []
labels = []
start_index = start_index + history_size
if end_index is None:
end_index = len(dataset) - target_size
for i in range(start_index, end_index):
indices = range(i-history_size, i)
# Reshape data from (history_size,) to (history_size, 1)
data.append(np.reshape(dataset[indices], (history_size, 1)))
labels.append(dataset[i+target_size])
return np.array(data), np.array(labels)
TRAIN_SPLIT = 300000
tf.random.set_seed(13)
uni_data = df['T (degC)']
uni_data.index = df['Date Time']
uni_data.head()
uni_data.plot(subplots=True)
uni_data = uni_data.values
uni_train_mean = uni_data[:TRAIN_SPLIT].mean()
uni_train_std = uni_data[:TRAIN_SPLIT].std()
uni_data = (uni_data-uni_train_mean)/uni_train_std
univariate_past_history = 20
univariate_future_target = 0
x_train_uni, y_train_uni = univariate_data(uni_data, 0, TRAIN_SPLIT,
univariate_past_history,
univariate_future_target)
x_val_uni, y_val_uni = univariate_data(uni_data, TRAIN_SPLIT, None,
univariate_past_history,
univariate_future_target)
print ('Single window of past history')
print (x_train_uni[0])
print ('\n Target temperature to predict')
print (y_train_uni[0])
def create_time_steps(length):
return list(range(-length, 0))
def show_plot(plot_data, delta, title):
labels = ['History', 'True Future', 'Model Prediction']
marker = ['.-', 'rx', 'go']
time_steps = create_time_steps(plot_data[0].shape[0])
if delta:
future = delta
else:
future = 0
plt.title(title)
for i, x in enumerate(plot_data):
if i:
plt.plot(future, plot_data[i], marker[i], markersize=10,
label=labels[i])
else:
plt.plot(time_steps, plot_data[i].flatten(), marker[i], label=labels[i])
plt.legend()
plt.xlim([time_steps[0], (future+5)*2])
plt.xlabel('Time-Step')
return plt
show_plot([x_train_uni[0], y_train_uni[0]], 0, 'Sample Example')
def baseline(history):
return np.mean(history)
show_plot([x_train_uni[0], y_train_uni[0], baseline(x_train_uni[0])], 0,
'Baseline Prediction Example')
BATCH_SIZE = 256
BUFFER_SIZE = 10000
train_univariate = tf.data.Dataset.from_tensor_slices((x_train_uni, y_train_uni))
train_univariate = train_univariate.cache().shuffle(BUFFER_SIZE).batch(BATCH_SIZE).repeat()
val_univariate = tf.data.Dataset.from_tensor_slices((x_val_uni, y_val_uni))
val_univariate = val_univariate.batch(BATCH_SIZE).repeat()
simple_lstm_model = tf.keras.models.Sequential([
tf.keras.layers.LSTM(8, input_shape=x_train_uni.shape[-2:]),
tf.keras.layers.Dense(1)
])
simple_lstm_model.compile(optimizer='adam', loss='mae')
for x, y in val_univariate.take(1):
print(simple_lstm_model.predict(x).shape)
EVALUATION_INTERVAL = 200
EPOCHS = 10
simple_lstm_model.fit(train_univariate, epochs=EPOCHS,
steps_per_epoch=EVALUATION_INTERVAL,
validation_data=val_univariate, validation_steps=50)
for x, y in val_univariate.take(3):
plot = show_plot([x[0].numpy(), y[0].numpy(),
simple_lstm_model.predict(x)[0]], 0, 'Simple LSTM model')
plot.show()
features_considered = ['p (mbar)', 'T (degC)', 'rho (g/m**3)']
features = df[features_considered]
features.index = df['Date Time']
features.head()
features.plot(subplots=True)
dataset = features.values
data_mean = dataset[:TRAIN_SPLIT].mean(axis=0)
data_std = dataset[:TRAIN_SPLIT].std(axis=0)
dataset = (dataset-data_mean)/data_std
def multivariate_data(dataset, target, start_index, end_index, history_size,
target_size, step, single_step=False):
data = []
labels = []
start_index = start_index + history_size
if end_index is None:
end_index = len(dataset) - target_size
for i in range(start_index, end_index):
indices = range(i-history_size, i, step)
data.append(dataset[indices])
if single_step:
labels.append(target[i+target_size])
else:
labels.append(target[i:i+target_size])
return np.array(data), np.array(labels)
past_history = 720
future_target = 72
STEP = 6
x_train_single, y_train_single = multivariate_data(dataset, dataset[:, 1], 0,
TRAIN_SPLIT, past_history,
future_target, STEP,
single_step=True)
x_val_single, y_val_single = multivariate_data(dataset, dataset[:, 1],
TRAIN_SPLIT, None, past_history,
future_target, STEP,
single_step=True)
print ('Single window of past history : {}'.format(x_train_single[0].shape))
train_data_single = tf.data.Dataset.from_tensor_slices((x_train_single, y_train_single))
train_data_single = train_data_single.cache().shuffle(BUFFER_SIZE).batch(BATCH_SIZE).repeat()
val_data_single = tf.data.Dataset.from_tensor_slices((x_val_single, y_val_single))
val_data_single = val_data_single.batch(BATCH_SIZE).repeat()
single_step_model = tf.keras.models.Sequential()
single_step_model.add(tf.keras.layers.LSTM(32,
input_shape=x_train_single.shape[-2:]))
single_step_model.add(tf.keras.layers.Dense(1))
single_step_model.compile(optimizer=tf.keras.optimizers.RMSprop(), loss='mae')
for x, y in val_data_single.take(1):
print(single_step_model.predict(x).shape)
single_step_history = single_step_model.fit(train_data_single, epochs=EPOCHS,
steps_per_epoch=EVALUATION_INTERVAL,
validation_data=val_data_single,
validation_steps=50)
def plot_train_history(history, title):
loss = history.history['loss']
val_loss = history.history['val_loss']
epochs = range(len(loss))
plt.figure()
plt.plot(epochs, loss, 'b', label='Training loss')
plt.plot(epochs, val_loss, 'r', label='Validation loss')
plt.title(title)
plt.legend()
plt.show()
plot_train_history(single_step_history,
'Single Step Training and validation loss')
for x, y in val_data_single.take(3):
plot = show_plot([x[0][:, 1].numpy(), y[0].numpy(),
single_step_model.predict(x)[0]], 12,
'Single Step Prediction')
plot.show()
future_target = 72
x_train_multi, y_train_multi = multivariate_data(dataset, dataset[:, 1], 0,
TRAIN_SPLIT, past_history,
future_target, STEP)
x_val_multi, y_val_multi = multivariate_data(dataset, dataset[:, 1],
TRAIN_SPLIT, None, past_history,
future_target, STEP)
print ('Single window of past history : {}'.format(x_train_multi[0].shape))
print ('\n Target temperature to predict : {}'.format(y_train_multi[0].shape))
train_data_multi = tf.data.Dataset.from_tensor_slices((x_train_multi, y_train_multi))
train_data_multi = train_data_multi.cache().shuffle(BUFFER_SIZE).batch(BATCH_SIZE).repeat()
val_data_multi = tf.data.Dataset.from_tensor_slices((x_val_multi, y_val_multi))
val_data_multi = val_data_multi.batch(BATCH_SIZE).repeat()
def multi_step_plot(history, true_future, prediction):
plt.figure(figsize=(12, 6))
num_in = create_time_steps(len(history))
num_out = len(true_future)
plt.plot(num_in, np.array(history[:, 1]), label='History')
plt.plot(np.arange(num_out)/STEP, np.array(true_future), 'bo',
label='True Future')
if prediction.any():
plt.plot(np.arange(num_out)/STEP, np.array(prediction), 'ro',
label='Predicted Future')
plt.legend(loc='upper left')
plt.show()
for x, y in train_data_multi.take(1):
multi_step_plot(x[0], y[0], np.array([0]))
multi_step_model = tf.keras.models.Sequential()
multi_step_model.add(tf.keras.layers.LSTM(32,
return_sequences=True,
input_shape=x_train_multi.shape[-2:]))
multi_step_model.add(tf.keras.layers.LSTM(16, activation='relu'))
multi_step_model.add(tf.keras.layers.Dense(72))
multi_step_model.compile(optimizer=tf.keras.optimizers.RMSprop(clipvalue=1.0), loss='mae')
for x, y in val_data_multi.take(1):
print (multi_step_model.predict(x).shape)
multi_step_history = multi_step_model.fit(train_data_multi, epochs=EPOCHS,
steps_per_epoch=EVALUATION_INTERVAL,
validation_data=val_data_multi,
validation_steps=50)
plot_train_history(multi_step_history, 'Multi-Step Training and validation loss')
for x, y in val_data_multi.take(3):
multi_step_plot(x[0], y[0], multi_step_model.predict(x)[0])
| 0.600188 | 0.993423 |
<!DOCTYPE html>
<html>
<body>
<div align="center">
<h3>Prepared by Asif Bhat</h3>
<h1>Data Visualization With Matplotlib</h1>
<h3>Follow Me on - <a href="https://www.linkedin.com/in/asif-bhat/">LinkedIn</a> <a href="https://mobile.twitter.com/_asifbhat_">Twitter</a> <a href="https://www.instagram.com/datasciencescoop/?hl=en">Instagram</a> <a href="https://www.facebook.com/datasciencescoop/">Facebook</a></h3>
</div>
</div>
</body>
</html>
```
import numpy as np
import pandas as pd
import matplotlib as mpl
import matplotlib.pyplot as plt
%matplotlib inline
```
# Area Plot
```
x = np.arange(1,31)
y = np.random.normal(10,11,size=30)
y = np.square(y)
plt.figure(figsize=(16,6))
plt.plot(x,y)
plt.fill_between(x, y)
plt.show()
```
#### Changing Fill Color
```
x = np.arange(1,31)
y = np.random.normal(10,11,size=30)
y = np.square(y)
plt.figure(figsize=(16,6))
plt.fill_between( x, y, color="#baf1a1") # #Changing Fill color
plt.plot(x, y, color='#7fcd91') # Color on edges
plt.title("$ Area $ $ chart $" , fontsize = 16)
plt.xlabel("$X$" , fontsize = 16)
plt.ylabel("$Y$" , fontsize = 16)
plt.show()
```
#### Changing Fill Color and its transperancy
```
x = np.arange(1,31)
y = np.random.normal(10,11,size=30)
y = np.square(y)
plt.figure(figsize=(16,6))
plt.fill_between( x, y, color="#C8D700" , alpha = 0.3) # Changing transperancy using Alpha parameter
plt.plot(x, y, color='#36BD00')
plt.title("$ Area $ $ chart $" , fontsize = 16)
plt.xlabel("$X$" , fontsize = 16)
plt.ylabel("$Y$" , fontsize = 16)
plt.show()
x = np.arange(1,51)
y = np.random.normal(1,5,size=50)
y = np.square(y)
plt.figure(figsize=(16,6))
plt.fill_between( x, y, color="#5ac8fa", alpha=0.4)
plt.plot(x, y, color="blue", alpha=0.6) # Bold line on edges
plt.title("$ Area $ $ chart $" , fontsize = 16)
plt.xlabel("$X$" , fontsize = 16)
plt.ylabel("$Y$" , fontsize = 16)
plt.show()
plt.figure(figsize=(16,6))
plt.fill_between( x, y, color="#5ac8fa", alpha=0.4)
plt.plot(x, y, color="blue", alpha=0.2) # Less stronger line on edges
plt.title("$ Area $ $ chart $" , fontsize = 14)
plt.xlabel("$X$" , fontsize = 14)
plt.ylabel("$Y$" , fontsize = 14)
plt.show()
```
#### Stacked Area plot
```
x=np.arange(1,6)
y1 = np.array([1,5,9,13,17])
y2 = np.array([2,6,10,14,16])
y3 = np.array([3,7,11,15,19])
y4 = np.array([4,8,12,16,20])
plt.figure(figsize=(8,6))
plt.stackplot(x,y1,y2,y3,y4, labels=['Y1','Y2','Y3','Y4'])
plt.legend(loc='upper left')
plt.show()
x=np.arange(1,6)
y=[ [1,5,9,13,17], [2,6,10,14,16], [3,7,11,15,19] , [4,8,12,16,20] ]
plt.figure(figsize=(8,6))
plt.stackplot(x,y , labels=['Y1','Y2','Y3','Y4'])
plt.legend(loc='upper left')
plt.show()
x=np.arange(1,7)
y=[ [1,5,9,3,17,1], [2,6,10,4,16,2], [3,7,11,5,19,1] , [4,8,12,6,20,2] ]
plt.figure(figsize=(10,6))
plt.stackplot(x,y , labels=['Y1','Y2','Y3','Y4'])
plt.legend(loc='upper left')
plt.show()
```
#### Changing Fill Color and its transperancy in Stacked Plot
```
x=np.arange(1,7)
y=[ [1,5,9,3,17,1], [2,6,10,4,16,2], [3,7,11,5,19,1] , [4,8,12,6,20,2] ]
plt.figure(figsize=(11,6))
plt.stackplot(x,y , labels=['Y1','Y2','Y3','Y4'] , colors= ["#00b159" , "#ffc425", "#f37735", "#ff3b30"])
plt.legend(loc='upper left')
plt.show()
plt.figure(figsize=(11,6))
plt.stackplot(x,y, labels=['Y1','Y2','Y3','Y4'], colors= ["#00b159" , "#ffc425", "#f37735", "#ff3b30"], alpha=0.7 )
plt.legend(loc='upper left')
plt.show()
plt.figure(figsize=(11,6))
plt.stackplot(x,y, labels=['Y1','Y2','Y3','Y4'], colors= ["#00b159" , "#ffc425", "#f37735", "#ff3b30"], alpha=0.5 )
plt.legend(loc='upper left')
plt.show()
```
|
github_jupyter
|
import numpy as np
import pandas as pd
import matplotlib as mpl
import matplotlib.pyplot as plt
%matplotlib inline
x = np.arange(1,31)
y = np.random.normal(10,11,size=30)
y = np.square(y)
plt.figure(figsize=(16,6))
plt.plot(x,y)
plt.fill_between(x, y)
plt.show()
x = np.arange(1,31)
y = np.random.normal(10,11,size=30)
y = np.square(y)
plt.figure(figsize=(16,6))
plt.fill_between( x, y, color="#baf1a1") # #Changing Fill color
plt.plot(x, y, color='#7fcd91') # Color on edges
plt.title("$ Area $ $ chart $" , fontsize = 16)
plt.xlabel("$X$" , fontsize = 16)
plt.ylabel("$Y$" , fontsize = 16)
plt.show()
x = np.arange(1,31)
y = np.random.normal(10,11,size=30)
y = np.square(y)
plt.figure(figsize=(16,6))
plt.fill_between( x, y, color="#C8D700" , alpha = 0.3) # Changing transperancy using Alpha parameter
plt.plot(x, y, color='#36BD00')
plt.title("$ Area $ $ chart $" , fontsize = 16)
plt.xlabel("$X$" , fontsize = 16)
plt.ylabel("$Y$" , fontsize = 16)
plt.show()
x = np.arange(1,51)
y = np.random.normal(1,5,size=50)
y = np.square(y)
plt.figure(figsize=(16,6))
plt.fill_between( x, y, color="#5ac8fa", alpha=0.4)
plt.plot(x, y, color="blue", alpha=0.6) # Bold line on edges
plt.title("$ Area $ $ chart $" , fontsize = 16)
plt.xlabel("$X$" , fontsize = 16)
plt.ylabel("$Y$" , fontsize = 16)
plt.show()
plt.figure(figsize=(16,6))
plt.fill_between( x, y, color="#5ac8fa", alpha=0.4)
plt.plot(x, y, color="blue", alpha=0.2) # Less stronger line on edges
plt.title("$ Area $ $ chart $" , fontsize = 14)
plt.xlabel("$X$" , fontsize = 14)
plt.ylabel("$Y$" , fontsize = 14)
plt.show()
x=np.arange(1,6)
y1 = np.array([1,5,9,13,17])
y2 = np.array([2,6,10,14,16])
y3 = np.array([3,7,11,15,19])
y4 = np.array([4,8,12,16,20])
plt.figure(figsize=(8,6))
plt.stackplot(x,y1,y2,y3,y4, labels=['Y1','Y2','Y3','Y4'])
plt.legend(loc='upper left')
plt.show()
x=np.arange(1,6)
y=[ [1,5,9,13,17], [2,6,10,14,16], [3,7,11,15,19] , [4,8,12,16,20] ]
plt.figure(figsize=(8,6))
plt.stackplot(x,y , labels=['Y1','Y2','Y3','Y4'])
plt.legend(loc='upper left')
plt.show()
x=np.arange(1,7)
y=[ [1,5,9,3,17,1], [2,6,10,4,16,2], [3,7,11,5,19,1] , [4,8,12,6,20,2] ]
plt.figure(figsize=(10,6))
plt.stackplot(x,y , labels=['Y1','Y2','Y3','Y4'])
plt.legend(loc='upper left')
plt.show()
x=np.arange(1,7)
y=[ [1,5,9,3,17,1], [2,6,10,4,16,2], [3,7,11,5,19,1] , [4,8,12,6,20,2] ]
plt.figure(figsize=(11,6))
plt.stackplot(x,y , labels=['Y1','Y2','Y3','Y4'] , colors= ["#00b159" , "#ffc425", "#f37735", "#ff3b30"])
plt.legend(loc='upper left')
plt.show()
plt.figure(figsize=(11,6))
plt.stackplot(x,y, labels=['Y1','Y2','Y3','Y4'], colors= ["#00b159" , "#ffc425", "#f37735", "#ff3b30"], alpha=0.7 )
plt.legend(loc='upper left')
plt.show()
plt.figure(figsize=(11,6))
plt.stackplot(x,y, labels=['Y1','Y2','Y3','Y4'], colors= ["#00b159" , "#ffc425", "#f37735", "#ff3b30"], alpha=0.5 )
plt.legend(loc='upper left')
plt.show()
| 0.344333 | 0.83508 |
# 处理数据
```
import keras
import numpy as np
import pandas as pd
from keras.utils import np_utils
from keras.datasets import mnist
import matplotlib.pyplot as plt
import matplotlib.image as mpimg
#导入数据
(X_train_image, y_train_label), (X_test_image, y_test_label) = mnist.load_data()
y_train_label = y_train_label.reshape(60000, 1)
y_test_label = y_test_label.reshape(10000, 1)
#显示一组数据
plt.imshow(X_test_image[666], cmap='binary')
plt.show()
print(y_test_label[666])
#显示一副多数据的图像
def img_show(beg, end):
fig = plt.gcf()
fig.set_size_inches(12, 14)
if(end-beg>30):
end = beg+29
for num in range(beg,end+1):
muti_img = plt.subplot(5, (end - beg) / 5 + 1, num-beg+1)
title = "label="+str(y_test_label[num])
plt.title(title)
muti_img.imshow(X_test_image[num], cmap='binary')
plt.show()
img_show(8800,8866)
#reshape二维->一维
X_train = X_train_image.reshape(60000,28*28)
X_test = X_test_image.reshape(10000,28*28)
print(X_train.shape)
print(X_test.shape)
print(X_test[0])
#标准化
X_train_normalize = X_train/255
X_test_normalize = X_test/255
print(X_test_normalize[0])
#label->onehot
y_train_onehot = np_utils.to_categorical(y_train_label)
y_test_onehot = np_utils.to_categorical(y_test_label)
print(y_train_onehot[1000])
```
# 训练模型
```
from keras.models import Sequential
from keras.layers import Dense
model = Sequential()
#构造网络结构
hidden_layer = Dense(units=256,input_dim=784,kernel_initializer='normal',activation="relu")
output_layer = Dense(units=10,kernel_initializer='normal',activation="softmax")
#搭建网络
model.add(hidden_layer)
model.add(output_layer)
print(model.summary())
model.compile(loss="categorical_crossentropy",optimizer="adam",metrics=["accuracy"])
train_history = model.fit(x = X_train_normalize,y = y_train_onehot, validation_split=0.2, epochs=19, batch_size=200, verbose=2)
plt.plot(train_history.history['loss'])
plt.plot(train_history.history['val_loss'])
plt.xlabel('epoch')
plt.ylabel('acc')
plt.legend(['train', 'validate'],loc='upper left')
plt.show()
scores = model.evaluate(X_test_normalize,y_test_onehot)
print(scores[1])
```
# 进行预测
```
prediction = model.predict_classes(X_test)
prediction[1]
def pre_img_show(beg, end):
fig = plt.gcf()
fig.set_size_inches(12, 14)
if(end-beg>30):
end = beg+29
for num in range(beg,end+1):
muti_img = plt.subplot(5, (end - beg) / 5 + 1, num-beg+1)
title = "label="+str(prediction[num])+","+str(y_test_label[num])
plt.title(title)
muti_img.imshow(X_test_image[num], cmap='binary')
plt.show()
pre_img_show(666, 700)
```
# 显示混淆矩阵
```
import pandas as pd
#建立混淆矩阵
pd.crosstab(y_test_label.reshape(10000,),prediction,rownames=['label'],colnames=['predict'])
#建立真实值与测试值的dataFrame
df = pd.DataFrame({'label':y_test_label.reshape(10000,),'prediction':prediction})
#输出0-20
df[:20]
#输出真实值是5,但预测值是3的数据
df[(df.label==5)&(df.prediction==3)]
pre_img_show(1393, 1393)
```
# 解决overfitting问题(使用Dropout功能)
```
from keras.layers import Dropout
from keras.layers import Dense
from keras.models import Sequential
model = Sequential()
#定义模型
hidden_layer = Dense(units=256,input_dim=784,kernel_initializer='normal',activation="relu")
drop_layer = Dropout(0.5)
output_layer = Dense(units=10,kernel_initializer='normal',activation='softmax')
model.add(hidden_layer)
model.add(drop_layer)
model.add(output_layer)
model.summary()
model.compile(loss="categorical_crossentropy",optimizer="adam",metrics=["accuracy"])
train_history = model.fit(x = X_train_normalize,y = y_train_onehot, validation_split=0.2, epochs=18, batch_size=200, verbose=2)
plt.plot(train_history.history['loss'])
plt.plot(train_history.history['val_loss'])
plt.xlabel('epoch')
plt.ylabel('acc')
plt.legend(['train', 'validate'],loc='upper left')
plt.show()
scores = model.evaluate(X_test_normalize,y_test_onehot)
print(scores[1])
```
# 保存与下载模型
```
model.save('my_model.h5')
from google.colab import files
files.download("my_model.h5")
```
|
github_jupyter
|
import keras
import numpy as np
import pandas as pd
from keras.utils import np_utils
from keras.datasets import mnist
import matplotlib.pyplot as plt
import matplotlib.image as mpimg
#导入数据
(X_train_image, y_train_label), (X_test_image, y_test_label) = mnist.load_data()
y_train_label = y_train_label.reshape(60000, 1)
y_test_label = y_test_label.reshape(10000, 1)
#显示一组数据
plt.imshow(X_test_image[666], cmap='binary')
plt.show()
print(y_test_label[666])
#显示一副多数据的图像
def img_show(beg, end):
fig = plt.gcf()
fig.set_size_inches(12, 14)
if(end-beg>30):
end = beg+29
for num in range(beg,end+1):
muti_img = plt.subplot(5, (end - beg) / 5 + 1, num-beg+1)
title = "label="+str(y_test_label[num])
plt.title(title)
muti_img.imshow(X_test_image[num], cmap='binary')
plt.show()
img_show(8800,8866)
#reshape二维->一维
X_train = X_train_image.reshape(60000,28*28)
X_test = X_test_image.reshape(10000,28*28)
print(X_train.shape)
print(X_test.shape)
print(X_test[0])
#标准化
X_train_normalize = X_train/255
X_test_normalize = X_test/255
print(X_test_normalize[0])
#label->onehot
y_train_onehot = np_utils.to_categorical(y_train_label)
y_test_onehot = np_utils.to_categorical(y_test_label)
print(y_train_onehot[1000])
from keras.models import Sequential
from keras.layers import Dense
model = Sequential()
#构造网络结构
hidden_layer = Dense(units=256,input_dim=784,kernel_initializer='normal',activation="relu")
output_layer = Dense(units=10,kernel_initializer='normal',activation="softmax")
#搭建网络
model.add(hidden_layer)
model.add(output_layer)
print(model.summary())
model.compile(loss="categorical_crossentropy",optimizer="adam",metrics=["accuracy"])
train_history = model.fit(x = X_train_normalize,y = y_train_onehot, validation_split=0.2, epochs=19, batch_size=200, verbose=2)
plt.plot(train_history.history['loss'])
plt.plot(train_history.history['val_loss'])
plt.xlabel('epoch')
plt.ylabel('acc')
plt.legend(['train', 'validate'],loc='upper left')
plt.show()
scores = model.evaluate(X_test_normalize,y_test_onehot)
print(scores[1])
prediction = model.predict_classes(X_test)
prediction[1]
def pre_img_show(beg, end):
fig = plt.gcf()
fig.set_size_inches(12, 14)
if(end-beg>30):
end = beg+29
for num in range(beg,end+1):
muti_img = plt.subplot(5, (end - beg) / 5 + 1, num-beg+1)
title = "label="+str(prediction[num])+","+str(y_test_label[num])
plt.title(title)
muti_img.imshow(X_test_image[num], cmap='binary')
plt.show()
pre_img_show(666, 700)
import pandas as pd
#建立混淆矩阵
pd.crosstab(y_test_label.reshape(10000,),prediction,rownames=['label'],colnames=['predict'])
#建立真实值与测试值的dataFrame
df = pd.DataFrame({'label':y_test_label.reshape(10000,),'prediction':prediction})
#输出0-20
df[:20]
#输出真实值是5,但预测值是3的数据
df[(df.label==5)&(df.prediction==3)]
pre_img_show(1393, 1393)
from keras.layers import Dropout
from keras.layers import Dense
from keras.models import Sequential
model = Sequential()
#定义模型
hidden_layer = Dense(units=256,input_dim=784,kernel_initializer='normal',activation="relu")
drop_layer = Dropout(0.5)
output_layer = Dense(units=10,kernel_initializer='normal',activation='softmax')
model.add(hidden_layer)
model.add(drop_layer)
model.add(output_layer)
model.summary()
model.compile(loss="categorical_crossentropy",optimizer="adam",metrics=["accuracy"])
train_history = model.fit(x = X_train_normalize,y = y_train_onehot, validation_split=0.2, epochs=18, batch_size=200, verbose=2)
plt.plot(train_history.history['loss'])
plt.plot(train_history.history['val_loss'])
plt.xlabel('epoch')
plt.ylabel('acc')
plt.legend(['train', 'validate'],loc='upper left')
plt.show()
scores = model.evaluate(X_test_normalize,y_test_onehot)
print(scores[1])
model.save('my_model.h5')
from google.colab import files
files.download("my_model.h5")
| 0.583678 | 0.851459 |
```
import tensorflow as tf
from tensorflow import keras
import matplotlib.pyplot as plt
%matplotlib inline
import numpy as np
import pandas as pd
```
## MNIST Dataset
```
(X_train,Y_train) , (X_test,Y_test) = keras.datasets.mnist.load_data()
print(len(X_train))
print(len(Y_train))
print(len(X_test))
print(len(Y_test))
print(X_train[0].shape)
print(Y_train[0].shape)
print(X_test[0].shape)
print(Y_test[0].shape)
X_train[0]
Y_train
plt.matshow(X_train[0])
Y_train[0:5]
X_train.shape
#Scaling the values
X_train = X_train/255
X_test = X_test/255
X_train_flattened = X_train.reshape(len(X_train),28*28)
print(X_train_flattened)
print(len(X_train_flattened))
X_test_flattened = X_test.reshape(len(X_test),28*28)
print(len(X_test_flattened))
X_train_flattened[0]
```
## Creating a Simple Neural Network
```
#sequential means we are having a stack of layers in our neural network
#stack will accept every layer as one element
#Dense means all neurons in one layer is connected to every neuron in the second layer
#Keras.layers.Dense is an api
#output shape is 10
model = keras.Sequential([keras.layers.Dense(10,input_shape=(784,),activation = 'sigmoid')])
#Now complile the neural network
model.compile(optimizer='adam',loss='sparse_categorical_crossentropy',
metrics=['accuracy'])
#Now supply the training set to train
model.fit(X_train_flattened,Y_train,epochs=5)
#Evaluate accuracy on test dataset
model.evaluate(X_test_flattened,Y_test)
Y_predicted = model.predict(X_test_flattened)
Y_predicted[0]
np.argmax(Y_predicted[0])
plt.matshow(X_test[1])
Y_predicted[1]
np.argmax(Y_predicted[1])
Y_predicted_labels = [np.argmax(i) for i in Y_predicted]
Y_predicted_labels
Y_predicted_labels[0:5]
#math is a modeule in tf
confusion_matrix = tf.math.confusion_matrix(labels = Y_test,predictions = Y_predicted_labels)
#Confusion matrix to evaluate the performance of our model
confusion_matrix
print(confusion_matrix)
import seaborn as sns
plt.figure(figsize=(10,7))
sns.heatmap(confusion_matrix,annot=True,fmt='d')
plt.xlabel('Predicted')
plt.ylabel('Truth')
```
## Adding a hidden layer to our simple neural network
```
#Performance would be improved here by adding hidden layer/layers
model1 = keras.Sequential([keras.layers.Dense(100,input_shape=(784,),activation = 'relu'),
keras.layers.Dense(10,activation = 'sigmoid')
])
#Second layer doesn't require a input_shape,it will figure out on it's own
#Now complile the neural network
model1.compile(optimizer='adam',loss='sparse_categorical_crossentropy',
metrics=['accuracy'])
#Now supply the training set to train
model1.fit(X_train_flattened,Y_train,epochs=5)
#Adding mpre hidden layers in this case
model2 = keras.Sequential([keras.layers.Dense(200,input_shape=(784,),activation = 'relu'),
keras.layers.Dense(10,activation = 'sigmoid')
])
#Second layer doesn't require a input_shape,it will figure out on it's own
#Now complile the neural network
model2.compile(optimizer='adam',loss='sparse_categorical_crossentropy',
metrics=['accuracy'])
#Now supply the training set to train
model2.fit(X_train_flattened,Y_train,epochs=5)
model1.evaluate(X_test_flattened,Y_test)
model2.evaluate(X_test_flattened,Y_test)
plt.matshow(X_test[0])
Y_predicted1 = model1.predict(X_test_flattened)
Y_predicted1[0]
np.argmax(Y_predicted1[0])
Y_predicted_labels_1 = [np.argmax(i) for i in Y_predicted1]
Y_predicted_labels_1
Y_predicted_labels_1[0:5]
confusion_matrix_1 = tf.math.confusion_matrix(labels = Y_test,predictions = Y_predicted_labels_1)
confusion_matrix_1
print(confusion_matrix_1)
plt.figure(figsize=(10,7))
sns.heatmap(confusion_matrix_1,annot=True,fmt='d')
plt.xlabel('Predicted')
plt.ylabel('Truth')
Y_predicted2 = model2.predict(X_test_flattened)
Y_predicted2[0]
print(Y_predicted2)
Y_predicted_labels_2 = [np.argmax(i) for i in Y_predicted2]
Y_predicted_labels_2
print(Y_predicted_labels_2)
print(Y_predicted_labels_2[0:5])
confusion_matrix_2 = tf.math.confusion_matrix(labels = Y_test,predictions = Y_predicted_labels_2)
confusion_matrix_2
print(confusion_matrix_2)
plt.figure(figsize=(10,7))
sns.heatmap(confusion_matrix_2,annot=True,fmt='d')
plt.xlabel('Predicted')
plt.ylabel('Truth')
```
## Using flattened layer of keras insted of making flattened array
```
#Adding mpre hidden layers in this case
model3 = keras.Sequential([keras.layers.Flatten(input_shape=(28,28)),
keras.layers.Dense(200,activation = 'relu'),
keras.layers.Dense(10,activation = 'sigmoid')
])
#Second layer doesn't require a input_shape,it will figure out on it's own
#Now complile the neural network
model3.compile(optimizer='adam',loss='sparse_categorical_crossentropy',
metrics=['accuracy'])
#Now supply the training set to train
model3.fit(X_train,Y_train,epochs=5)
model3.evaluate(X_test,Y_test)
model3.evaluate(X_test_flattened,Y_test)
Y_predicted3 = model3.predict(X_test)
Y_predicted3[0]
print(Y_predicted3)
Y_predicted_labels_3 = [np.argmax(i) for i in Y_predicted3]
Y_predicted_labels_3
print(Y_predicted_labels_3)
print(Y_predicted_labels_3[0:5])
confusion_matrix_3 = tf.math.confusion_matrix(labels = Y_test,predictions = Y_predicted_labels_3)
confusion_matrix_3
print(confusion_matrix_3)
plt.figure(figsize=(10,7))
sns.heatmap(confusion_matrix_3,annot=True,fmt='d')
plt.xlabel('Predicted')
plt.ylabel('Truth')
Y_predicted3 = model3.predict(X_test_flattened)
Y_predicted3[0]
print(Y_predicted3)
Y_predicted_labels_3 = [np.argmax(i) for i in Y_predicted3]
Y_predicted_labels_3
print(Y_predicted_labels_3)
print(Y_predicted_labels_3[0:5])
confusion_matrix_3 = tf.math.confusion_matrix(labels = Y_test,predictions = Y_predicted_labels_3)
confusion_matrix_3
print(confusion_matrix_3)
plt.figure(figsize=(10,7))
sns.heatmap(confusion_matrix_3,annot=True,fmt='d')
plt.xlabel('Predicted')
plt.ylabel('Truth')
```
|
github_jupyter
|
import tensorflow as tf
from tensorflow import keras
import matplotlib.pyplot as plt
%matplotlib inline
import numpy as np
import pandas as pd
(X_train,Y_train) , (X_test,Y_test) = keras.datasets.mnist.load_data()
print(len(X_train))
print(len(Y_train))
print(len(X_test))
print(len(Y_test))
print(X_train[0].shape)
print(Y_train[0].shape)
print(X_test[0].shape)
print(Y_test[0].shape)
X_train[0]
Y_train
plt.matshow(X_train[0])
Y_train[0:5]
X_train.shape
#Scaling the values
X_train = X_train/255
X_test = X_test/255
X_train_flattened = X_train.reshape(len(X_train),28*28)
print(X_train_flattened)
print(len(X_train_flattened))
X_test_flattened = X_test.reshape(len(X_test),28*28)
print(len(X_test_flattened))
X_train_flattened[0]
#sequential means we are having a stack of layers in our neural network
#stack will accept every layer as one element
#Dense means all neurons in one layer is connected to every neuron in the second layer
#Keras.layers.Dense is an api
#output shape is 10
model = keras.Sequential([keras.layers.Dense(10,input_shape=(784,),activation = 'sigmoid')])
#Now complile the neural network
model.compile(optimizer='adam',loss='sparse_categorical_crossentropy',
metrics=['accuracy'])
#Now supply the training set to train
model.fit(X_train_flattened,Y_train,epochs=5)
#Evaluate accuracy on test dataset
model.evaluate(X_test_flattened,Y_test)
Y_predicted = model.predict(X_test_flattened)
Y_predicted[0]
np.argmax(Y_predicted[0])
plt.matshow(X_test[1])
Y_predicted[1]
np.argmax(Y_predicted[1])
Y_predicted_labels = [np.argmax(i) for i in Y_predicted]
Y_predicted_labels
Y_predicted_labels[0:5]
#math is a modeule in tf
confusion_matrix = tf.math.confusion_matrix(labels = Y_test,predictions = Y_predicted_labels)
#Confusion matrix to evaluate the performance of our model
confusion_matrix
print(confusion_matrix)
import seaborn as sns
plt.figure(figsize=(10,7))
sns.heatmap(confusion_matrix,annot=True,fmt='d')
plt.xlabel('Predicted')
plt.ylabel('Truth')
#Performance would be improved here by adding hidden layer/layers
model1 = keras.Sequential([keras.layers.Dense(100,input_shape=(784,),activation = 'relu'),
keras.layers.Dense(10,activation = 'sigmoid')
])
#Second layer doesn't require a input_shape,it will figure out on it's own
#Now complile the neural network
model1.compile(optimizer='adam',loss='sparse_categorical_crossentropy',
metrics=['accuracy'])
#Now supply the training set to train
model1.fit(X_train_flattened,Y_train,epochs=5)
#Adding mpre hidden layers in this case
model2 = keras.Sequential([keras.layers.Dense(200,input_shape=(784,),activation = 'relu'),
keras.layers.Dense(10,activation = 'sigmoid')
])
#Second layer doesn't require a input_shape,it will figure out on it's own
#Now complile the neural network
model2.compile(optimizer='adam',loss='sparse_categorical_crossentropy',
metrics=['accuracy'])
#Now supply the training set to train
model2.fit(X_train_flattened,Y_train,epochs=5)
model1.evaluate(X_test_flattened,Y_test)
model2.evaluate(X_test_flattened,Y_test)
plt.matshow(X_test[0])
Y_predicted1 = model1.predict(X_test_flattened)
Y_predicted1[0]
np.argmax(Y_predicted1[0])
Y_predicted_labels_1 = [np.argmax(i) for i in Y_predicted1]
Y_predicted_labels_1
Y_predicted_labels_1[0:5]
confusion_matrix_1 = tf.math.confusion_matrix(labels = Y_test,predictions = Y_predicted_labels_1)
confusion_matrix_1
print(confusion_matrix_1)
plt.figure(figsize=(10,7))
sns.heatmap(confusion_matrix_1,annot=True,fmt='d')
plt.xlabel('Predicted')
plt.ylabel('Truth')
Y_predicted2 = model2.predict(X_test_flattened)
Y_predicted2[0]
print(Y_predicted2)
Y_predicted_labels_2 = [np.argmax(i) for i in Y_predicted2]
Y_predicted_labels_2
print(Y_predicted_labels_2)
print(Y_predicted_labels_2[0:5])
confusion_matrix_2 = tf.math.confusion_matrix(labels = Y_test,predictions = Y_predicted_labels_2)
confusion_matrix_2
print(confusion_matrix_2)
plt.figure(figsize=(10,7))
sns.heatmap(confusion_matrix_2,annot=True,fmt='d')
plt.xlabel('Predicted')
plt.ylabel('Truth')
#Adding mpre hidden layers in this case
model3 = keras.Sequential([keras.layers.Flatten(input_shape=(28,28)),
keras.layers.Dense(200,activation = 'relu'),
keras.layers.Dense(10,activation = 'sigmoid')
])
#Second layer doesn't require a input_shape,it will figure out on it's own
#Now complile the neural network
model3.compile(optimizer='adam',loss='sparse_categorical_crossentropy',
metrics=['accuracy'])
#Now supply the training set to train
model3.fit(X_train,Y_train,epochs=5)
model3.evaluate(X_test,Y_test)
model3.evaluate(X_test_flattened,Y_test)
Y_predicted3 = model3.predict(X_test)
Y_predicted3[0]
print(Y_predicted3)
Y_predicted_labels_3 = [np.argmax(i) for i in Y_predicted3]
Y_predicted_labels_3
print(Y_predicted_labels_3)
print(Y_predicted_labels_3[0:5])
confusion_matrix_3 = tf.math.confusion_matrix(labels = Y_test,predictions = Y_predicted_labels_3)
confusion_matrix_3
print(confusion_matrix_3)
plt.figure(figsize=(10,7))
sns.heatmap(confusion_matrix_3,annot=True,fmt='d')
plt.xlabel('Predicted')
plt.ylabel('Truth')
Y_predicted3 = model3.predict(X_test_flattened)
Y_predicted3[0]
print(Y_predicted3)
Y_predicted_labels_3 = [np.argmax(i) for i in Y_predicted3]
Y_predicted_labels_3
print(Y_predicted_labels_3)
print(Y_predicted_labels_3[0:5])
confusion_matrix_3 = tf.math.confusion_matrix(labels = Y_test,predictions = Y_predicted_labels_3)
confusion_matrix_3
print(confusion_matrix_3)
plt.figure(figsize=(10,7))
sns.heatmap(confusion_matrix_3,annot=True,fmt='d')
plt.xlabel('Predicted')
plt.ylabel('Truth')
| 0.689306 | 0.899343 |
```
%matplotlib inline
import sys
sys.path.append('/mnt/ssd2/kcheng/gpu205/coco-caption/pycocoevalcap')
from pycocotools.coco import COCO
from pycocoevalcap.eval import COCOEvalCap
import matplotlib.pyplot as plt
import skimage.io as io
import pylab
import json
from json import encoder
pylab.rcParams['figure.figsize'] = (10.0, 8.0)
encoder.FLOAT_REPR = lambda o: format(o, '.3f')
# set up file names and pathes
# dataDir='.'
# dataType='val2014'
# algName = 'fakecap'
# annFile='%s/annotations/captions_%s.json'%(dataDir,dataType)
# subtypes=['results', 'evalImgs', 'eval']
# [resFile, evalImgsFile, evalFile]= \
# ['%s/results/captions_%s_%s_%s.json'%(dataDir,dataType,algName,subtype) for subtype in subtypes]
annFile = '/mnt/ssd2/kcheng/gpu205/cocoapi/annotations/captions_val2014.json'
resFile = '/mnt/ssd2/kcheng/gpu205/Image-Captioning-Project/val2014_res.json'
# download Stanford models
!./get_stanford_models.sh
# create coco object and cocoRes object
coco = COCO(annFile)
cocoRes = coco.loadRes(resFile)
# create cocoEval object by taking coco and cocoRes
cocoEval = COCOEvalCap(coco, cocoRes)
# evaluate on a subset of images by setting
# cocoEval.params['image_id'] = cocoRes.getImgIds()
# please remove this line when evaluating the full validation set
# cocoEval.params['image_id'] = cocoRes.getImgIds()
# evaluate results
# SPICE will take a few minutes the first time, but speeds up due to caching
cocoEval.evaluate()
# print output evaluation scores
for metric, score in cocoEval.eval.items():
print '%s: %.3f'%(metric, score)
# demo how to use evalImgs to retrieve low score result
evals = [eva for eva in cocoEval.evalImgs if eva['CIDEr']<30]
print 'ground truth captions'
imgId = evals[0]['image_id']
annIds = coco.getAnnIds(imgIds=imgId)
anns = coco.loadAnns(annIds)
coco.showAnns(anns)
print '\n'
print 'generated caption (CIDEr score %0.1f)'%(evals[0]['CIDEr'])
annIds = cocoRes.getAnnIds(imgIds=imgId)
anns = cocoRes.loadAnns(annIds)
coco.showAnns(anns)
img = coco.loadImgs(imgId)[0]
I = io.imread('%s/images/%s/%s'%(dataDir,dataType,img['file_name']))
plt.imshow(I)
plt.axis('off')
plt.show()
# plot score histogram
ciderScores = [eva['CIDEr'] for eva in cocoEval.evalImgs]
plt.hist(ciderScores)
plt.title('Histogram of CIDEr Scores', fontsize=20)
plt.xlabel('CIDEr score', fontsize=20)
plt.ylabel('result counts', fontsize=20)
plt.show()
# save evaluation results to ./results folder
json.dump(cocoEval.evalImgs, open(evalImgsFile, 'w'))
json.dump(cocoEval.eval, open(evalFile, 'w'))
```
|
github_jupyter
|
%matplotlib inline
import sys
sys.path.append('/mnt/ssd2/kcheng/gpu205/coco-caption/pycocoevalcap')
from pycocotools.coco import COCO
from pycocoevalcap.eval import COCOEvalCap
import matplotlib.pyplot as plt
import skimage.io as io
import pylab
import json
from json import encoder
pylab.rcParams['figure.figsize'] = (10.0, 8.0)
encoder.FLOAT_REPR = lambda o: format(o, '.3f')
# set up file names and pathes
# dataDir='.'
# dataType='val2014'
# algName = 'fakecap'
# annFile='%s/annotations/captions_%s.json'%(dataDir,dataType)
# subtypes=['results', 'evalImgs', 'eval']
# [resFile, evalImgsFile, evalFile]= \
# ['%s/results/captions_%s_%s_%s.json'%(dataDir,dataType,algName,subtype) for subtype in subtypes]
annFile = '/mnt/ssd2/kcheng/gpu205/cocoapi/annotations/captions_val2014.json'
resFile = '/mnt/ssd2/kcheng/gpu205/Image-Captioning-Project/val2014_res.json'
# download Stanford models
!./get_stanford_models.sh
# create coco object and cocoRes object
coco = COCO(annFile)
cocoRes = coco.loadRes(resFile)
# create cocoEval object by taking coco and cocoRes
cocoEval = COCOEvalCap(coco, cocoRes)
# evaluate on a subset of images by setting
# cocoEval.params['image_id'] = cocoRes.getImgIds()
# please remove this line when evaluating the full validation set
# cocoEval.params['image_id'] = cocoRes.getImgIds()
# evaluate results
# SPICE will take a few minutes the first time, but speeds up due to caching
cocoEval.evaluate()
# print output evaluation scores
for metric, score in cocoEval.eval.items():
print '%s: %.3f'%(metric, score)
# demo how to use evalImgs to retrieve low score result
evals = [eva for eva in cocoEval.evalImgs if eva['CIDEr']<30]
print 'ground truth captions'
imgId = evals[0]['image_id']
annIds = coco.getAnnIds(imgIds=imgId)
anns = coco.loadAnns(annIds)
coco.showAnns(anns)
print '\n'
print 'generated caption (CIDEr score %0.1f)'%(evals[0]['CIDEr'])
annIds = cocoRes.getAnnIds(imgIds=imgId)
anns = cocoRes.loadAnns(annIds)
coco.showAnns(anns)
img = coco.loadImgs(imgId)[0]
I = io.imread('%s/images/%s/%s'%(dataDir,dataType,img['file_name']))
plt.imshow(I)
plt.axis('off')
plt.show()
# plot score histogram
ciderScores = [eva['CIDEr'] for eva in cocoEval.evalImgs]
plt.hist(ciderScores)
plt.title('Histogram of CIDEr Scores', fontsize=20)
plt.xlabel('CIDEr score', fontsize=20)
plt.ylabel('result counts', fontsize=20)
plt.show()
# save evaluation results to ./results folder
json.dump(cocoEval.evalImgs, open(evalImgsFile, 'w'))
json.dump(cocoEval.eval, open(evalFile, 'w'))
| 0.188959 | 0.206474 |
```
import numpy as np
import matplotlib.pyplot as plt
np.random.seed(42)
ages = np.random.randint(low=15, high=70, size=40)
labels = []
for age in ages:
if age < 30:
labels.append(0)
else:
labels.append(1)
for i in range(0, 3):
r = np.random.randint(0, len(labels) - 1)
if labels[r] == 0:
labels[r] = 1
else:
labels[r] = 0
plt.scatter(ages, labels, color="red")
plt.show()
```
## Predição usando Regressão Linear
```
import numpy as np
from sklearn.linear_model import LinearRegression
model = LinearRegression()
model.fit(ages.reshape(-1, 1), labels)
m = model.coef_[0]
b = model.intercept_
```
## Entendendo os Coeficientes da Reta
```
from matplotlib.animation import FuncAnimation
fig, ax = plt.subplots()
axis = plt.axes(xlim =(0, 2),
ylim =(-0.1, 2))
line, = axis.plot([], [], lw = 3)
def init():
line.set_data([], [])
return line,
def animate(i):
m_copy = i * 0.01
plt.title('m = ' + str(m_copy))
x = np.arange(0.0, 10.0, 0.1)
y = m_copy * x + b
line.set_data(x, y)
return line,
ani = FuncAnimation(fig, animate, init_func = init,
frames = 200,
interval = 20,
blit = True)
ani.save('m.mp4', writer = 'ffmpeg', fps = 30)
from IPython.display import HTML
HTML("""
<div align="middle">
<video width="80%" controls>
<source src="m.mp4" type="video/mp4">
</video></div>""")
from matplotlib.animation import FuncAnimation
fig, ax = plt.subplots()
axis = plt.axes(xlim =(0, 2),
ylim =(-0.1, 2))
line, = axis.plot([], [], lw = 3)
def init():
line.set_data([], [])
return line,
def animate(i):
b_copy = i * 0.01
plt.title('b = ' + str(b_copy))
x = np.arange(0.0, 10.0, 0.1)
y = m * x + b_copy
line.set_data(x, y)
return line,
ani = FuncAnimation(fig, animate, init_func = init,
frames = 200,
interval = 20,
blit = True)
ani.save('b.mp4',
writer = 'ffmpeg', fps = 30)
from IPython.display import HTML
HTML("""
<div align="middle">
<video width="80%" controls>
<source src="b.mp4" type="video/mp4">
</video></div>""")
```
## Regressão Linear daquele Conjunto de Pontos
```
limiar_idade = (0.5 - b) / m
print(limiar_idade)
plt.plot(ages, ages * m + b, color = 'blue')
plt.plot([limiar_idade, limiar_idade], [0, 0.5], '--', color = 'green')
plt.scatter(ages, labels, color="red")
plt.show()
```
## Função Logística
```
import math
def sigmoid(x):
a = []
for item in x:
a.append(1/(1+math.exp(-item)))
return a
x = np.arange(-10., 10., 0.2)
sig = sigmoid(x)
plt.plot(x, sig)
plt.show()
```
## Classificador Sigmóide
```
from sklearn.linear_model import LogisticRegression
model = LogisticRegression()
model.fit(ages.reshape(-1, 1), labels)
m = model.coef_[0][0]
b = model.intercept_[0]
x = np.arange(0, 70, 0.1)
sig = sigmoid(m*x + b)
limiar_idade = 0 - (b / m)
print(limiar_idade)
plt.scatter(ages, labels, color="red")
plt.plot([limiar_idade, limiar_idade], [0, 0.5], '--', color = 'green')
plt.plot(x, sig)
plt.show()
```
|
github_jupyter
|
import numpy as np
import matplotlib.pyplot as plt
np.random.seed(42)
ages = np.random.randint(low=15, high=70, size=40)
labels = []
for age in ages:
if age < 30:
labels.append(0)
else:
labels.append(1)
for i in range(0, 3):
r = np.random.randint(0, len(labels) - 1)
if labels[r] == 0:
labels[r] = 1
else:
labels[r] = 0
plt.scatter(ages, labels, color="red")
plt.show()
import numpy as np
from sklearn.linear_model import LinearRegression
model = LinearRegression()
model.fit(ages.reshape(-1, 1), labels)
m = model.coef_[0]
b = model.intercept_
from matplotlib.animation import FuncAnimation
fig, ax = plt.subplots()
axis = plt.axes(xlim =(0, 2),
ylim =(-0.1, 2))
line, = axis.plot([], [], lw = 3)
def init():
line.set_data([], [])
return line,
def animate(i):
m_copy = i * 0.01
plt.title('m = ' + str(m_copy))
x = np.arange(0.0, 10.0, 0.1)
y = m_copy * x + b
line.set_data(x, y)
return line,
ani = FuncAnimation(fig, animate, init_func = init,
frames = 200,
interval = 20,
blit = True)
ani.save('m.mp4', writer = 'ffmpeg', fps = 30)
from IPython.display import HTML
HTML("""
<div align="middle">
<video width="80%" controls>
<source src="m.mp4" type="video/mp4">
</video></div>""")
from matplotlib.animation import FuncAnimation
fig, ax = plt.subplots()
axis = plt.axes(xlim =(0, 2),
ylim =(-0.1, 2))
line, = axis.plot([], [], lw = 3)
def init():
line.set_data([], [])
return line,
def animate(i):
b_copy = i * 0.01
plt.title('b = ' + str(b_copy))
x = np.arange(0.0, 10.0, 0.1)
y = m * x + b_copy
line.set_data(x, y)
return line,
ani = FuncAnimation(fig, animate, init_func = init,
frames = 200,
interval = 20,
blit = True)
ani.save('b.mp4',
writer = 'ffmpeg', fps = 30)
from IPython.display import HTML
HTML("""
<div align="middle">
<video width="80%" controls>
<source src="b.mp4" type="video/mp4">
</video></div>""")
limiar_idade = (0.5 - b) / m
print(limiar_idade)
plt.plot(ages, ages * m + b, color = 'blue')
plt.plot([limiar_idade, limiar_idade], [0, 0.5], '--', color = 'green')
plt.scatter(ages, labels, color="red")
plt.show()
import math
def sigmoid(x):
a = []
for item in x:
a.append(1/(1+math.exp(-item)))
return a
x = np.arange(-10., 10., 0.2)
sig = sigmoid(x)
plt.plot(x, sig)
plt.show()
from sklearn.linear_model import LogisticRegression
model = LogisticRegression()
model.fit(ages.reshape(-1, 1), labels)
m = model.coef_[0][0]
b = model.intercept_[0]
x = np.arange(0, 70, 0.1)
sig = sigmoid(m*x + b)
limiar_idade = 0 - (b / m)
print(limiar_idade)
plt.scatter(ages, labels, color="red")
plt.plot([limiar_idade, limiar_idade], [0, 0.5], '--', color = 'green')
plt.plot(x, sig)
plt.show()
| 0.516352 | 0.886519 |
```
%%capture
## compile PyRoss for this notebook
import os
owd = os.getcwd()
os.chdir('../../')
%run setup.py install
os.chdir(owd)
%matplotlib inline
import numpy as np
import pyross
import pandas as pd
import time
import matplotlib.pyplot as plt
from scipy.io import loadmat
## population and age classes
M=4 ## number of age classes
my_data = np.genfromtxt('../data/age_structures/India-2019.csv', delimiter=',', skip_header=1)
aM, aF = my_data[:, 1], my_data[:, 2]
Ni0=aM+aF; Ni=np.zeros((M))
# scale the population down to a more manageble level
Ni[0] = (np.sum(Ni0[0:4])/1e4).astype('int')
Ni[1] = (np.sum(Ni0[4:8])/1e4).astype('int')
Ni[2] = (np.sum(Ni0[8:12])/1e4).astype('int')
Ni[3] = (np.sum(Ni0[12:16])/1e4).astype('int')
N=np.sum(Ni)
print(N)
fi = Ni/N
# Get individual contact matrices
CH0, CW0, CS0, CO0 = pyross.contactMatrix.India()
CH = np.zeros((M, M))
CW = np.zeros((M, M))
CS = np.zeros((M, M))
CO = np.zeros((M, M))
for i in range(M):
for j in range(M):
i1, j1 = i*4, j*4
CH[i,j] = np.sum( CH0[i1:i1+4, j1:j1+4] )
CW[i,j] = np.sum( CW0[i1:i1+4, j1:j1+4] )
CS[i,j] = np.sum( CS0[i1:i1+4, j1:j1+4] )
CO[i,j] = np.sum( CO0[i1:i1+4, j1:j1+4] )
# Generate class with contact matrix for SIR model with UK contact structure
generator = pyross.contactMatrix.SIR(CH, CW, CS, CO)
times= [20] # temporal boundaries between different contact-behaviour
aW, aS, aO = 0.5, 0.6, 0.6
# prefactors for CW, CS, CO:
interventions = [[1.0,1.0,1.0], # before first time
[aW, aS, aO], # after first time
]
# generate corresponding contact matrix function
C = generator.interventions_temporal(times=times,interventions=interventions)
beta = 0.00454 # contact rate parameter
gIa = 1./7 # recovery rate of asymptomatic infectives
gIs = 1./7 # recovery rate of symptomatic infectives
alpha = 0.2 # asymptomatic fraction
fsa = 0.8 # suppresion of contact by symptomatics
# initial conditions
Is_0 = np.ones(M)*20
Is_0[1] += 10 #make one group different
Ia_0 = np.zeros((M))*2
Ia_0[1] += 2
R_0 = np.zeros((M))
S_0 = Ni - (Ia_0 + Is_0 + R_0)
parameters = {'alpha':alpha,'beta':beta, 'gIa':gIa,'gIs':gIs,'fsa':fsa}
model = pyross.stochastic.SIR(parameters, M, Ni)
contactMatrix=C
# start simulation
Tf=100; Nf=Tf+1
data=model.simulate(S_0, Ia_0, Is_0, contactMatrix, Tf, Nf)
IC = np.zeros((Nf))
for i in range(M):
IC += data['X'][:,2*M+i]
t = data['t']
plt.plot(t, IC)
plt.show()
Tf = 19 # truncate to only getting the first few datapoints
Nf = Tf+1
x = data['X']
x = (x/N)[:Nf]
steps = 101 # number internal integration steps taken, must be an odd number
# initialise the estimator
estimator = pyross.inference.SIR(parameters, M, fi, int(N), steps)
# compute -log_p for the original (correct) parameters
start_time = time.time()
parameters = {'alpha':alpha, 'beta':beta, 'gIa':gIa, 'gIs':gIs,'fsa':fsa}
logp = estimator.obtain_minus_log_p(parameters, x, Tf, Nf, contactMatrix)
end_time = time.time()
print(logp)
print(end_time - start_time)
plt.plot(x[:, M])
plt.show()
# take a guess
beta_g = 0.005
gIa_g = 0.15
gIs_g = 0.15
alpha_g = 0.25
guess = np.array([alpha_g, beta_g, gIa_g, gIs_g])
# compute -log_p for the initial guess
parameters = {'alpha':alpha_g, 'beta':beta_g, 'gIa':gIa_g, 'gIs':gIs_g,'fsa':fsa}
logp = estimator.obtain_minus_log_p(parameters, x, Tf, Nf, contactMatrix)
print(logp)
eps = 1e-4
bounds = np.array([(eps, 0.4), (1e-3, 0.02), (eps, 0.3), (eps, 0.3)]) # give some bounds
start_time = time.time()
params, nit = estimator.inference(guess, x, Tf, Nf, contactMatrix, beta_rescale=5,
niter=4, bounds=bounds,
eps=eps, verbose=True)
# sometimes produce errors if input is bad, ignore and let basinhopping do its magic
end_time = time.time()
print(params) # best guess
print(nit) # number of iterations of the optimization run
print(end_time - start_time)
aW_g = 0.5
aW_bounds = [0.1, 0.8]
aS_g = 0.5
aS_bounds = [0.1, 0.8]
aO_g = 0.5
aO_bounds = [0.1, 0.8]
guess = np.array([aW_g, aS_g, aO_g])
bounds = np.array([aW_bounds, aS_bounds, aO_bounds])
x = data['X']/N
x = x[22:]
Nf = x.shape[0]
Tf = Nf-1
times = [Tf+1]
interventions = [[aW, aS, aO]]
contactMatrix = generator.interventions_temporal(times=times,interventions=interventions)
# compute -log_p for the initial guess (for the moment, use correct parameters)
logp = estimator.obtain_minus_log_p(parameters, x, Tf, Nf, contactMatrix)
print(logp)
eps = 1e-3
start_time = time.time()
parameters = estimator.make_params_dict(params)
estimator.set_params(parameters)
params, nit = estimator.infer_control(guess, x, Tf, Nf, generator, bounds,
niter=2, eps=eps, verbose=True)
# sometimes produce errors if input is bad, ignore and let basinhopping do its magic
end_time = time.time()
print(params) # best guess
print(nit) # number of iterations of the optimization run
print(end_time - start_time)
```
|
github_jupyter
|
%%capture
## compile PyRoss for this notebook
import os
owd = os.getcwd()
os.chdir('../../')
%run setup.py install
os.chdir(owd)
%matplotlib inline
import numpy as np
import pyross
import pandas as pd
import time
import matplotlib.pyplot as plt
from scipy.io import loadmat
## population and age classes
M=4 ## number of age classes
my_data = np.genfromtxt('../data/age_structures/India-2019.csv', delimiter=',', skip_header=1)
aM, aF = my_data[:, 1], my_data[:, 2]
Ni0=aM+aF; Ni=np.zeros((M))
# scale the population down to a more manageble level
Ni[0] = (np.sum(Ni0[0:4])/1e4).astype('int')
Ni[1] = (np.sum(Ni0[4:8])/1e4).astype('int')
Ni[2] = (np.sum(Ni0[8:12])/1e4).astype('int')
Ni[3] = (np.sum(Ni0[12:16])/1e4).astype('int')
N=np.sum(Ni)
print(N)
fi = Ni/N
# Get individual contact matrices
CH0, CW0, CS0, CO0 = pyross.contactMatrix.India()
CH = np.zeros((M, M))
CW = np.zeros((M, M))
CS = np.zeros((M, M))
CO = np.zeros((M, M))
for i in range(M):
for j in range(M):
i1, j1 = i*4, j*4
CH[i,j] = np.sum( CH0[i1:i1+4, j1:j1+4] )
CW[i,j] = np.sum( CW0[i1:i1+4, j1:j1+4] )
CS[i,j] = np.sum( CS0[i1:i1+4, j1:j1+4] )
CO[i,j] = np.sum( CO0[i1:i1+4, j1:j1+4] )
# Generate class with contact matrix for SIR model with UK contact structure
generator = pyross.contactMatrix.SIR(CH, CW, CS, CO)
times= [20] # temporal boundaries between different contact-behaviour
aW, aS, aO = 0.5, 0.6, 0.6
# prefactors for CW, CS, CO:
interventions = [[1.0,1.0,1.0], # before first time
[aW, aS, aO], # after first time
]
# generate corresponding contact matrix function
C = generator.interventions_temporal(times=times,interventions=interventions)
beta = 0.00454 # contact rate parameter
gIa = 1./7 # recovery rate of asymptomatic infectives
gIs = 1./7 # recovery rate of symptomatic infectives
alpha = 0.2 # asymptomatic fraction
fsa = 0.8 # suppresion of contact by symptomatics
# initial conditions
Is_0 = np.ones(M)*20
Is_0[1] += 10 #make one group different
Ia_0 = np.zeros((M))*2
Ia_0[1] += 2
R_0 = np.zeros((M))
S_0 = Ni - (Ia_0 + Is_0 + R_0)
parameters = {'alpha':alpha,'beta':beta, 'gIa':gIa,'gIs':gIs,'fsa':fsa}
model = pyross.stochastic.SIR(parameters, M, Ni)
contactMatrix=C
# start simulation
Tf=100; Nf=Tf+1
data=model.simulate(S_0, Ia_0, Is_0, contactMatrix, Tf, Nf)
IC = np.zeros((Nf))
for i in range(M):
IC += data['X'][:,2*M+i]
t = data['t']
plt.plot(t, IC)
plt.show()
Tf = 19 # truncate to only getting the first few datapoints
Nf = Tf+1
x = data['X']
x = (x/N)[:Nf]
steps = 101 # number internal integration steps taken, must be an odd number
# initialise the estimator
estimator = pyross.inference.SIR(parameters, M, fi, int(N), steps)
# compute -log_p for the original (correct) parameters
start_time = time.time()
parameters = {'alpha':alpha, 'beta':beta, 'gIa':gIa, 'gIs':gIs,'fsa':fsa}
logp = estimator.obtain_minus_log_p(parameters, x, Tf, Nf, contactMatrix)
end_time = time.time()
print(logp)
print(end_time - start_time)
plt.plot(x[:, M])
plt.show()
# take a guess
beta_g = 0.005
gIa_g = 0.15
gIs_g = 0.15
alpha_g = 0.25
guess = np.array([alpha_g, beta_g, gIa_g, gIs_g])
# compute -log_p for the initial guess
parameters = {'alpha':alpha_g, 'beta':beta_g, 'gIa':gIa_g, 'gIs':gIs_g,'fsa':fsa}
logp = estimator.obtain_minus_log_p(parameters, x, Tf, Nf, contactMatrix)
print(logp)
eps = 1e-4
bounds = np.array([(eps, 0.4), (1e-3, 0.02), (eps, 0.3), (eps, 0.3)]) # give some bounds
start_time = time.time()
params, nit = estimator.inference(guess, x, Tf, Nf, contactMatrix, beta_rescale=5,
niter=4, bounds=bounds,
eps=eps, verbose=True)
# sometimes produce errors if input is bad, ignore and let basinhopping do its magic
end_time = time.time()
print(params) # best guess
print(nit) # number of iterations of the optimization run
print(end_time - start_time)
aW_g = 0.5
aW_bounds = [0.1, 0.8]
aS_g = 0.5
aS_bounds = [0.1, 0.8]
aO_g = 0.5
aO_bounds = [0.1, 0.8]
guess = np.array([aW_g, aS_g, aO_g])
bounds = np.array([aW_bounds, aS_bounds, aO_bounds])
x = data['X']/N
x = x[22:]
Nf = x.shape[0]
Tf = Nf-1
times = [Tf+1]
interventions = [[aW, aS, aO]]
contactMatrix = generator.interventions_temporal(times=times,interventions=interventions)
# compute -log_p for the initial guess (for the moment, use correct parameters)
logp = estimator.obtain_minus_log_p(parameters, x, Tf, Nf, contactMatrix)
print(logp)
eps = 1e-3
start_time = time.time()
parameters = estimator.make_params_dict(params)
estimator.set_params(parameters)
params, nit = estimator.infer_control(guess, x, Tf, Nf, generator, bounds,
niter=2, eps=eps, verbose=True)
# sometimes produce errors if input is bad, ignore and let basinhopping do its magic
end_time = time.time()
print(params) # best guess
print(nit) # number of iterations of the optimization run
print(end_time - start_time)
| 0.440951 | 0.467028 |
# 10.1 Defining a Function
The keyword `def` introduces a [function definition](https://docs.python.org/3.5/tutorial/controlflow.html#defining-functions). It's followed by the function name, parenthesized list of formal parameters, and ends with a colon. The indented statements below it are executed with the function name is called.
```
def fib(n):
result = []
a, b = 0, 1
while a < n:
result.append(a) # see below
a, b = b, a+b
return result
```
The __`fib()`__ function is defined above. Now let's call this function. Calling a function is simple.
```
fib(4) # oops
```
## 10.2 Positional Arguments
The function requires a positional argument: "__`n`__". This is a good time to mention that naming things descriptively really helps. Coupled with Python's helpful error messages, descriptive variable, function, and class names make it easy to understand and debug errors. In this case, 'n' is a number. Specifically, this function returns a fibonacci sequence for as long as the numbers in the squence are less than the given max number.
Let's give it a better name and then call the function properly.
```
def fib(max_number):
"""Return a list containing the Fibonacci series up to max_number."""
result = []
a, b = 0, 1
while a < max_number:
result.append(a) # see below
a, b = b, a+b
return result
fib(17)
```
## 10.3 Keyword Arguments
Arguments can be made optional when default values are provided. These are known as keyword arguments.
Let's make our argument optional with a default max_number then let's call our function without any arguments.
```
def fib(max_number=17):
"""Return a list containing the Fibonacci series up to max_number."""
result = []
a, b = 0, 1
while a < max_number:
result.append(a) # see below
a, b = b, a+b
return result
fib()
```
Now let's try calling our function with a different argument.
```
fib(6) # still works!
```
## 10.4 Argument Syntax
There can be any number of positional arguments and any number of optional arguments. They can appear together in a function definition for as long as required positional arguments come before optional defaulted arguments.
```
def foo(p=1, q):
return p, q
foo(1)
def foo(p, q, r=1, s=2):
return p, q, r, s
foo(-1, 0)
def foo(p, q, r=1, s=2):
return p, q, r, s
foo(0, 1, s=3, r=2) # the order of defaulted arguments doesn't matter
```
## 10.5 Starred Arguments
In Python, there's a third way of passing arguments to a function. If you wanted to pass a list with an unknown length, even empty, you could pass them in starred arguments.
```
args = [1, 2, 3, 4, 5]
def arguments(*args):
for a in args:
print(a)
return args
arguments(*args)
```
We could have specified each argument and it would have worked but that would mean our arguments are fixed. Starred arguments give us flexibility by making the positional arguments optional and of any length.
```
arguments() # still works!
```
For keyword arguments, the only difference is to use `**`. You could pass a dictionary and it would be treated as an arbitrary number of keyword arguments.
```
kwargs = {'a': 1, 'b': 2, 'c': 3, 'd': 4, 'e': 5}
def keywords(**kwargs):
for key, value in kwargs.items():
print(key, value)
return kwargs
keywords(**kwargs)
keywords() # still works!
```
## 10.6 Packing and Unpacking Arguments
### `def function(*args, **kwargs):`
This pattern allows you to change functionality while avoiding breaking your code by just checking the arguments if certain parameters exist and then adding a conditional statement based on that.
Class methods that use this pattern allow data to be passed between objects without loss, transforming the data as needed without needing to know about other objects.
Let's look at more exmaples to illustrate the differences.
```
greeting = 'hello'
def echo(arg):
return arg
echo(greeting)
echo() # it's required...
greeting = 'hello'
def echo(*arg):
return arg
echo(greeting)
greeting = 'hello'
def echo(*arg):
return arg
echo(*greeting) # asterisk unpacks iterables
greeting = ['hello'] # it's now a list
def echo(*arg):
return arg
echo(*greeting)
greeting = [
'hello',
'hi',
'ohayou',
'hey dude'
]
def echo(*arg):
return arg
echo(*greeting) # accepts lists
echo() # still works!
```
Let's try it with keyword arguments.
```
kwargs = {
'greeting1': 'Hello',
'greeting2': 'Hi',
'greeting3': 'Ohayou',
}
def echo(kwarg=None, **kwargs):
print(kwarg)
return kwargs
echo(kwargs) # the dictionary data type is unordered unlike lists
echo(**kwargs)
kwargs = {
'greeting1': 'Hello',
'greeting2': 'Hi',
'greeting3': 'Ohayou',
'kwarg': 'World!', # we have a default value for this, which is None
}
def echo(kwarg=None, **kwargs):
print(kwarg)
return kwargs
echo(**kwargs)
```
The dictionary we passed was unpacked and considered as if it was a bunch of keyword arguments passed to the function.
Notice how the keyword argument with a default value was overridden.
|
github_jupyter
|
def fib(n):
result = []
a, b = 0, 1
while a < n:
result.append(a) # see below
a, b = b, a+b
return result
fib(4) # oops
def fib(max_number):
"""Return a list containing the Fibonacci series up to max_number."""
result = []
a, b = 0, 1
while a < max_number:
result.append(a) # see below
a, b = b, a+b
return result
fib(17)
def fib(max_number=17):
"""Return a list containing the Fibonacci series up to max_number."""
result = []
a, b = 0, 1
while a < max_number:
result.append(a) # see below
a, b = b, a+b
return result
fib()
fib(6) # still works!
def foo(p=1, q):
return p, q
foo(1)
def foo(p, q, r=1, s=2):
return p, q, r, s
foo(-1, 0)
def foo(p, q, r=1, s=2):
return p, q, r, s
foo(0, 1, s=3, r=2) # the order of defaulted arguments doesn't matter
args = [1, 2, 3, 4, 5]
def arguments(*args):
for a in args:
print(a)
return args
arguments(*args)
arguments() # still works!
kwargs = {'a': 1, 'b': 2, 'c': 3, 'd': 4, 'e': 5}
def keywords(**kwargs):
for key, value in kwargs.items():
print(key, value)
return kwargs
keywords(**kwargs)
keywords() # still works!
greeting = 'hello'
def echo(arg):
return arg
echo(greeting)
echo() # it's required...
greeting = 'hello'
def echo(*arg):
return arg
echo(greeting)
greeting = 'hello'
def echo(*arg):
return arg
echo(*greeting) # asterisk unpacks iterables
greeting = ['hello'] # it's now a list
def echo(*arg):
return arg
echo(*greeting)
greeting = [
'hello',
'hi',
'ohayou',
'hey dude'
]
def echo(*arg):
return arg
echo(*greeting) # accepts lists
echo() # still works!
kwargs = {
'greeting1': 'Hello',
'greeting2': 'Hi',
'greeting3': 'Ohayou',
}
def echo(kwarg=None, **kwargs):
print(kwarg)
return kwargs
echo(kwargs) # the dictionary data type is unordered unlike lists
echo(**kwargs)
kwargs = {
'greeting1': 'Hello',
'greeting2': 'Hi',
'greeting3': 'Ohayou',
'kwarg': 'World!', # we have a default value for this, which is None
}
def echo(kwarg=None, **kwargs):
print(kwarg)
return kwargs
echo(**kwargs)
| 0.658966 | 0.967349 |
```
import numpy as np
import heapq
from typing import Union
class Graph:
def __init__(self, adjacency_mat: Union[np.ndarray, str]):
""" Unlike project 2, this Graph class takes an adjacency matrix as input. `adjacency_mat`
can either be a 2D numpy array of floats or the path to a CSV file containing a 2D numpy array of floats.
In this project, we will assume `adjacency_mat` corresponds to the adjacency matrix of an undirected graph
"""
if type(adjacency_mat) == str:
self.adj_mat = self._load_adjacency_matrix_from_csv(adjacency_mat)
elif type(adjacency_mat) == np.ndarray:
self.adj_mat = adjacency_mat
else:
raise TypeError('Input must be a valid path or an adjacency matrix')
self.mst = None
self.big_number = 1000
def _load_adjacency_matrix_from_csv(self, path: str) -> np.ndarray:
with open(path) as f:
return np.loadtxt(f, delimiter=',')
def construct_mst(self):
""" Given `self.adj_mat`, the adjacency matrix of a connected undirected graph, implement Prim's
algorithm to construct an adjacency matrix encoding the minimum spanning tree of `self.adj_mat`.
`self.adj_mat` is a 2D numpy array of floats.
Note that because we assume our input graph is undirected, `self.adj_mat` is symmetric.
Row i and column j represents the edge weight between vertex i and vertex j. An edge weight of zero indicates that no edge exists.
TODO:
This function does not return anything. Instead, store the adjacency matrix
representation of the minimum spanning tree of `self.adj_mat` in `self.mst`.
We highly encourage the use of priority queues in your implementation. See the heapq
module, particularly the `heapify`, `heappop`, and `heappush` functions.
"""
n_nodes = self.adj_mat.shape[0]
h = []
heapq.heappush(h, (0, (0,0)))
all_nodes = set(list(range(n_nodes)))
mst_set = set()
self.mst = np.zeros((n_nodes, n_nodes))
while len(mst_set) != n_nodes:
u = heapq.heappop(h)
mst_set.add(u[1][0])
self.mst[u[1][0],u[1][1]] = self.adj_mat[u[1][0],u[1][1]]
self.mst[u[1][1],u[1][0]] = self.adj_mat[u[1][1],u[1][0]]
h = []
for node in list(all_nodes.difference(mst_set)):
print(node)
distances_to_mst = self.adj_mat[node,].copy()
print(distances_to_mst)
distances_to_mst[distances_to_mst == 0] = self.big_number
print(distances_to_mst)
distances_to_mst[list(all_nodes.difference(mst_set))] = self.big_number
print(distances_to_mst)
predecessor = np.argmin(distances_to_mst)
heapq.heappush(h, (distances_to_mst[predecessor],(node, predecessor)))
print(h)
print(self.mst)
print()
arr = np.array([[0,7,4],[7,0,1],[4,1,0]])
arr
g = Graph(arr)
g.adj_mat
g.construct_mst()
g.mst
```
|
github_jupyter
|
import numpy as np
import heapq
from typing import Union
class Graph:
def __init__(self, adjacency_mat: Union[np.ndarray, str]):
""" Unlike project 2, this Graph class takes an adjacency matrix as input. `adjacency_mat`
can either be a 2D numpy array of floats or the path to a CSV file containing a 2D numpy array of floats.
In this project, we will assume `adjacency_mat` corresponds to the adjacency matrix of an undirected graph
"""
if type(adjacency_mat) == str:
self.adj_mat = self._load_adjacency_matrix_from_csv(adjacency_mat)
elif type(adjacency_mat) == np.ndarray:
self.adj_mat = adjacency_mat
else:
raise TypeError('Input must be a valid path or an adjacency matrix')
self.mst = None
self.big_number = 1000
def _load_adjacency_matrix_from_csv(self, path: str) -> np.ndarray:
with open(path) as f:
return np.loadtxt(f, delimiter=',')
def construct_mst(self):
""" Given `self.adj_mat`, the adjacency matrix of a connected undirected graph, implement Prim's
algorithm to construct an adjacency matrix encoding the minimum spanning tree of `self.adj_mat`.
`self.adj_mat` is a 2D numpy array of floats.
Note that because we assume our input graph is undirected, `self.adj_mat` is symmetric.
Row i and column j represents the edge weight between vertex i and vertex j. An edge weight of zero indicates that no edge exists.
TODO:
This function does not return anything. Instead, store the adjacency matrix
representation of the minimum spanning tree of `self.adj_mat` in `self.mst`.
We highly encourage the use of priority queues in your implementation. See the heapq
module, particularly the `heapify`, `heappop`, and `heappush` functions.
"""
n_nodes = self.adj_mat.shape[0]
h = []
heapq.heappush(h, (0, (0,0)))
all_nodes = set(list(range(n_nodes)))
mst_set = set()
self.mst = np.zeros((n_nodes, n_nodes))
while len(mst_set) != n_nodes:
u = heapq.heappop(h)
mst_set.add(u[1][0])
self.mst[u[1][0],u[1][1]] = self.adj_mat[u[1][0],u[1][1]]
self.mst[u[1][1],u[1][0]] = self.adj_mat[u[1][1],u[1][0]]
h = []
for node in list(all_nodes.difference(mst_set)):
print(node)
distances_to_mst = self.adj_mat[node,].copy()
print(distances_to_mst)
distances_to_mst[distances_to_mst == 0] = self.big_number
print(distances_to_mst)
distances_to_mst[list(all_nodes.difference(mst_set))] = self.big_number
print(distances_to_mst)
predecessor = np.argmin(distances_to_mst)
heapq.heappush(h, (distances_to_mst[predecessor],(node, predecessor)))
print(h)
print(self.mst)
print()
arr = np.array([[0,7,4],[7,0,1],[4,1,0]])
arr
g = Graph(arr)
g.adj_mat
g.construct_mst()
g.mst
| 0.862352 | 0.87289 |
```
from ceviche import fdtd
from ceviche.constants import *
from ceviche.utils import aniplot, measure_fields, get_spectrum, get_max_power_freq, get_spectral_power, plot_spectral_power
import autograd.numpy as np
import matplotlib.pylab as plt
plt.style.use('dark_background')
%load_ext autoreload
%autoreload 2
%matplotlib inline
%config InlineBackend.figure_format = 'retina'
```
Load in the permittivity distribution from an FDFD simulation
```
def reshape_arr(arr, Nx, Ny):
return arr.reshape((Nx, Ny, 1))
eps_r = np.load('data/eps_r_splitter4.npy')
eps_wg = np.load('data/eps_waveguide.npy')
plt.imshow(eps_r.T, cmap='gist_earth_r')
plt.show()
Nx, Ny = eps_r.shape
J_in = np.load('data/J_in.npy')
J_outs = np.load('data/J_list.npy')
J_wg = np.flipud(J_in.copy())
eps_wg = reshape_arr(eps_wg, Nx, Ny)
eps_r = reshape_arr(eps_r, Nx, Ny)
J_in = reshape_arr(J_in, Nx, Ny)
J_outs = [reshape_arr(J, Nx, Ny) for J in J_outs]
J_wg = reshape_arr(J_wg, Nx, Ny)
Nz = 1
```
Initial setting up of parameters
```
nx, ny, nz = Nx//2, Ny//2, Nz//2
dL = 5e-8
pml = [20, 20, 0]
F = fdtd(eps_r, dL=dL, npml=pml)
F_wg = fdtd(eps_wg, dL=dL, npml=pml)
# source parameters
steps = 10000
t0 = 2000
sigma = 100
source_amp = 5
omega = 2 * np.pi * C_0 / 2e-6 # units of 1/sec
omega_sim = omega * F.dt # unitless time
gaussian = lambda t: np.exp(-(t - t0)**2 / 2 / sigma**2) * np.cos(omega_sim * t)
source = lambda t: J_in * source_amp * gaussian(t)
plt.plot(1e15 * F.dt * np.arange(steps), gaussian(np.arange(steps)))
plt.xlabel('time (femtoseconds)')
plt.ylabel('source amplitude')
plt.show()
```
Compute the transmission (as a function of time) for a straight waveguide (to normalize later)
```
measured_wg = measure_fields(F_wg, source, steps, J_wg)
plt.plot(1e15 * F.dt*np.arange(steps), gaussian(np.arange(steps))/gaussian(np.arange(steps)).max(), label='source (Jz)')
plt.plot(1e15 * F.dt*np.arange(steps), measured_wg/measured_wg.max(), label='measured (Ez)')
plt.xlabel('time (femtoseconds)')
plt.ylabel('amplitude')
plt.legend()
plt.show()
```
Show the field plots at 10 time steps
```
aniplot(F_wg, source, steps, num_panels=10)
```
Compute the power spectrum transmitted
```
plot_spectral_power(gaussian(np.arange(steps)), F.dt, f_top=3e16)
gaussian(np.arange(steps)).shape
```
Now for the measured fields
```
plot_spectral_power(measured_wg[:,0], F.dt, f_top=2e14)
```
Now measure the transmission for the inverse designed splitter (at each of the four ports)
```
measured = measure_fields(F, source, steps, J_outs)
ham = np.hamming(steps).reshape((steps,1))
# plt.plot(measured*ham**5)
plot_spectral_power(measured*ham**100, dt=F.dt, f_top=4e16)
plt.plot(1e15 * F.dt*np.arange(steps), gaussian(np.arange(steps))/gaussian(np.arange(steps)).max(), label='source (Jz)')
plt.plot(1e15 * F.dt*np.arange(steps), measured/measured.max(), label='measured (Ez)')
plt.xlabel('time (femtoseconds)')
plt.ylabel('amplitude')
plt.legend()
plt.show()
aniplot(F_wg, source, steps, num_panels=10)
series_in = gaussian(np.arange(steps))
series_wg = None
plot_spectral_power(measured_wg, F.dt, f_top=15e14)
series_in = gaussian(np.arange(steps))
measured_wg
freq1, spect1 = get_spectrum(series_in, dt=F.dt)
freq2, spect2 = get_spectrum(measured_wg, dt=F.dt)
plt.plot(series_in/series_in.max())
plt.plot(measured_wg/measured_wg.max())
plt.show()
spect1 = np.fft.fft(series_in/series_in.max())
spect2 = np.fft.fft(measured_wg/measured_wg.max())
plt.plot(np.fft.fftshift(np.abs(spect1)))
plt.plot(np.fft.fftshift(np.abs(spect2)))
plt.show()
plt.plot(freq1, np.abs(spect1/spect1.max()))
plt.plot(freq2, np.abs(spect2/spect2.max()))
plt.show()
freq, spect = get_spectrum(measured_wg, dt=F.dt)
plt.plot(freq, np.abs(spect))
plt.xlim([0, 4e14])
ex = 1e-50
plt.ylim([-0.1*ex, ex])
plt.show()
plot_spectral_power(measured, dt=F.dt, f_top=1e16)
plt.plot(series_in)
series_out = measured_wg/measured_wg.max()
plt.plot(series_out)
plt.plot(np.abs(np.fft.fft(series_in*np.hanning(steps))))
plt.plot(np.abs(np.fft.fft(series_out*np.hanning(steps))))
(measured_wg*np.hamming(steps)).shape
plt.plot(np.hamming(steps))
F.dt
from ceviche.utils import get_spectrum_lr
get_spectrum_lr(measured_wg[:,0], dt=F.dt)
get_spectrum_lr(gaussian(np.arange(steps)), dt=F.dt)
```
|
github_jupyter
|
from ceviche import fdtd
from ceviche.constants import *
from ceviche.utils import aniplot, measure_fields, get_spectrum, get_max_power_freq, get_spectral_power, plot_spectral_power
import autograd.numpy as np
import matplotlib.pylab as plt
plt.style.use('dark_background')
%load_ext autoreload
%autoreload 2
%matplotlib inline
%config InlineBackend.figure_format = 'retina'
def reshape_arr(arr, Nx, Ny):
return arr.reshape((Nx, Ny, 1))
eps_r = np.load('data/eps_r_splitter4.npy')
eps_wg = np.load('data/eps_waveguide.npy')
plt.imshow(eps_r.T, cmap='gist_earth_r')
plt.show()
Nx, Ny = eps_r.shape
J_in = np.load('data/J_in.npy')
J_outs = np.load('data/J_list.npy')
J_wg = np.flipud(J_in.copy())
eps_wg = reshape_arr(eps_wg, Nx, Ny)
eps_r = reshape_arr(eps_r, Nx, Ny)
J_in = reshape_arr(J_in, Nx, Ny)
J_outs = [reshape_arr(J, Nx, Ny) for J in J_outs]
J_wg = reshape_arr(J_wg, Nx, Ny)
Nz = 1
nx, ny, nz = Nx//2, Ny//2, Nz//2
dL = 5e-8
pml = [20, 20, 0]
F = fdtd(eps_r, dL=dL, npml=pml)
F_wg = fdtd(eps_wg, dL=dL, npml=pml)
# source parameters
steps = 10000
t0 = 2000
sigma = 100
source_amp = 5
omega = 2 * np.pi * C_0 / 2e-6 # units of 1/sec
omega_sim = omega * F.dt # unitless time
gaussian = lambda t: np.exp(-(t - t0)**2 / 2 / sigma**2) * np.cos(omega_sim * t)
source = lambda t: J_in * source_amp * gaussian(t)
plt.plot(1e15 * F.dt * np.arange(steps), gaussian(np.arange(steps)))
plt.xlabel('time (femtoseconds)')
plt.ylabel('source amplitude')
plt.show()
measured_wg = measure_fields(F_wg, source, steps, J_wg)
plt.plot(1e15 * F.dt*np.arange(steps), gaussian(np.arange(steps))/gaussian(np.arange(steps)).max(), label='source (Jz)')
plt.plot(1e15 * F.dt*np.arange(steps), measured_wg/measured_wg.max(), label='measured (Ez)')
plt.xlabel('time (femtoseconds)')
plt.ylabel('amplitude')
plt.legend()
plt.show()
aniplot(F_wg, source, steps, num_panels=10)
plot_spectral_power(gaussian(np.arange(steps)), F.dt, f_top=3e16)
gaussian(np.arange(steps)).shape
plot_spectral_power(measured_wg[:,0], F.dt, f_top=2e14)
measured = measure_fields(F, source, steps, J_outs)
ham = np.hamming(steps).reshape((steps,1))
# plt.plot(measured*ham**5)
plot_spectral_power(measured*ham**100, dt=F.dt, f_top=4e16)
plt.plot(1e15 * F.dt*np.arange(steps), gaussian(np.arange(steps))/gaussian(np.arange(steps)).max(), label='source (Jz)')
plt.plot(1e15 * F.dt*np.arange(steps), measured/measured.max(), label='measured (Ez)')
plt.xlabel('time (femtoseconds)')
plt.ylabel('amplitude')
plt.legend()
plt.show()
aniplot(F_wg, source, steps, num_panels=10)
series_in = gaussian(np.arange(steps))
series_wg = None
plot_spectral_power(measured_wg, F.dt, f_top=15e14)
series_in = gaussian(np.arange(steps))
measured_wg
freq1, spect1 = get_spectrum(series_in, dt=F.dt)
freq2, spect2 = get_spectrum(measured_wg, dt=F.dt)
plt.plot(series_in/series_in.max())
plt.plot(measured_wg/measured_wg.max())
plt.show()
spect1 = np.fft.fft(series_in/series_in.max())
spect2 = np.fft.fft(measured_wg/measured_wg.max())
plt.plot(np.fft.fftshift(np.abs(spect1)))
plt.plot(np.fft.fftshift(np.abs(spect2)))
plt.show()
plt.plot(freq1, np.abs(spect1/spect1.max()))
plt.plot(freq2, np.abs(spect2/spect2.max()))
plt.show()
freq, spect = get_spectrum(measured_wg, dt=F.dt)
plt.plot(freq, np.abs(spect))
plt.xlim([0, 4e14])
ex = 1e-50
plt.ylim([-0.1*ex, ex])
plt.show()
plot_spectral_power(measured, dt=F.dt, f_top=1e16)
plt.plot(series_in)
series_out = measured_wg/measured_wg.max()
plt.plot(series_out)
plt.plot(np.abs(np.fft.fft(series_in*np.hanning(steps))))
plt.plot(np.abs(np.fft.fft(series_out*np.hanning(steps))))
(measured_wg*np.hamming(steps)).shape
plt.plot(np.hamming(steps))
F.dt
from ceviche.utils import get_spectrum_lr
get_spectrum_lr(measured_wg[:,0], dt=F.dt)
get_spectrum_lr(gaussian(np.arange(steps)), dt=F.dt)
| 0.612078 | 0.860955 |
```
# Importing modules.
import numpy as np
import pandas as pd
from IPython.display import display
from sqlalchemy import create_engine
from sqlalchemy.types import DateTime, VARCHAR
import mysql.connector as connector
# Importing data.
visitorlogs = pd.read_csv(r'D:\Hackathon\Analytics Vidhya\Job-a-thon\data\Modified\VisitorLogsData.csv')
print('Shape of VisitorLogsData', visitorlogs.shape)
display(visitorlogs.head())
user = pd.read_csv(r'D:\Hackathon\Analytics Vidhya\Job-a-thon\data\Modified\userTable.csv')
print('Shape of userTable', user.shape)
display(user.head())
# Renaming for ease.
user.rename(columns={'Signup Date': 'SignupDate',
'User Segment': 'Segment'}, inplace=True)
user.head()
# Dropping unregistered users.
visitorlogs.dropna(subset=['UserID'], inplace=True)
# Cross checking for nulls.
visitorlogs.isnull().sum()
print('Shape of the data is:', visitorlogs.shape)
```
#### Cleaning VisitDateTime.
```
def datetime_clean(date_time):
'''
Converts the strings to datetime format.
input = Date time in string format.
output = returns datetime value.
'''
try:
return pd.to_datetime(pd.Timestamp(date_time), utc=True)
except:
try:
date_time = int(date_time)
return pd.to_datetime(pd.Timestamp(date_time), utc=True)
except:
pass
# Transforming the 'VisitDateTime' column.
visitorlogs['VisitDateTime'] = visitorlogs['VisitDateTime'].map(lambda x: datetime_clean(x))
user['SignupDate'] = user['SignupDate'].map(lambda x: datetime_clean(x))
# Replacing spaces with 'NaN'.
visitorlogs = visitorlogs.replace(r'^\s*$', np.nan, regex=True)
user = user.replace(r'^\s*$', np.nan, regex=True)
```
'VisitDateTime' Imputation.
```
# Imputing values with mean for each User.
visitorlogs['VisitDateTime'] = visitorlogs.groupby(['UserID'])['VisitDateTime']\
.transform(lambda x: x.fillna(x.mean()))
# Cross checking for null values
visitorlogs.isnull().sum()
```
MySQL
```
# Establishing SQL connection.
connection = connector.connect(host = 'localhost', passwd = 'param12345',
user = 'root')
# Establishing SQL connection.
connection = connector.connect(host = 'localhost', passwd = 'param12345',
user = 'root')
# Creating a cursor.
mycursor = connection.cursor()
# Dropping existing database.
mycursor.execute("DROP DATABASE IF EXISTS data")
# Creating a database.
mycursor.execute("CREATE DATABASE data")
sql_engine = create_engine("mysql+mysqlconnector://" + 'root' + ":"
+ 'param12345' + "@" + 'localhost' + "/" + 'data')
# Tables.
query = 'Show tables from data'
tables = pd.read_sql_query(query, connection)
tables
```
Creating table.
```
# Declaring variable types.
dtype = {'webClientID': VARCHAR(50),
'UserID': VARCHAR(50),
'ProductID': VARCHAR(50),
'VisitDateTime': DateTime,
'Activity': VARCHAR(50),
'OS': VARCHAR(50),
'Browser': VARCHAR(50),
'City': VARCHAR(100),
'Country': VARCHAR(100)}
# Exporting to SQL.
visitorlogs.to_sql(name = 'visitorlogsdata', con = sql_engine,
if_exists = 'replace', index = False, dtype = dtype)
# Declaring variable types.
dtype = {'UserID': VARCHAR(10),
'SignupDate': DateTime,
'Segment': VARCHAR(5)}
# Exporting to SQL.
user.to_sql(name = 'usertable', con = sql_engine, if_exists = 'replace',
index = False)
# Update
connection = connector.connect(host = 'localhost', user = 'root', passwd = 'param12345',
database = 'data')
# Update
connection = connector.connect(host = 'localhost', user = 'root', passwd = 'param12345',
database = 'data')
# Datas only upto 27th July 2018.
query = "select @current_date:= STR_TO_DATE('2018-05-27 23:59:59','%Y-%m-%d %H:%i:%s.%f')"
max_date = pd.read_sql_query(query, connection)
max_date
# Creating a data frame.
results_data = pd.DataFrame()
# 1) UserID.
results_data['UserID'] = sorted(user.UserID)
results_data
# 2) No_of_days_Visited_7_Days.
query = 'select UserID, count(distinct day(VisitDateTime)) as No_of_days_Visited_7_Days\
from visitorlogsdata\
where VisitDateTime>@current_date-interval 7 day\
and UserID is not null\
group by UserID\
order by UserID\
;'
No_of_days_Visited_7_Days = pd.read_sql_query(query, connection)
No_of_days_Visited_7_Days
# Merging.
results_data['No_of_days_Visited_7_Days'] = pd.merge(results_data, No_of_days_Visited_7_Days,
how = 'left', on = 'UserID')['No_of_days_Visited_7_Days']
results_data
# 3) No_Of_Products_Viewed_15_Days.
query = 'select UserID,count(distinct ProductID) as No_Of_Products_Viewed_15_Days\
from visitorlogsdata\
where VisitDateTime> @current_date -interval 15 day\
and UserID is not null\
group by UserID\
order by UserID\
;'
No_Of_Products_Viewed_15_Days = pd.read_sql_query(query, connection)
No_Of_Products_Viewed_15_Days
# Merging.
results_data['No_Of_Products_Viewed_15_Days'] = pd.merge(results_data, No_Of_Products_Viewed_15_Days,
how = 'left', on = 'UserID')['No_Of_Products_Viewed_15_Days']
results_data
# 4) User_Vintage.
query = 'select UserID,datediff(@current_date,SignupDate) as User_Vintage\
from usertable\
where UserID is not null\
order by UserID\
;'
User_Vintage = pd.read_sql_query(query, connection)
User_Vintage
# Mering.
results_data['User_Vintage'] = pd.merge(results_data, User_Vintage,
how = 'left', on = 'UserID')['User_Vintage']
results_data
# 5) Most_Viewed_product_15_Days.
query = 'select UserID,Most_Viewed_product_15_Days ,max(occurs) as products_viewed\
from\
(\
select UserID,ProductID as Most_Viewed_product_15_Days,count(ProductID) as occurs,VisitDateTime\
from visitorlogsdata\
where VisitDateTime>@current_date-interval 15 day\
and UserID is not null\
group by UserID,ProductID\
order by UserID,occurs desc,VisitDateTime desc\
) as occur_table\
group by UserID\
;'
Most_Viewed_product_15_Days = pd.read_sql_query(query, connection)
Most_Viewed_product_15_Days
# Merging.
results_data['Most_Viewed_product_15_Days'] = pd.merge(results_data, Most_Viewed_product_15_Days,
how = 'left', on = 'UserID')['Most_Viewed_product_15_Days']
results_data
# Filling NaN values with Product101.
results_data['Most_Viewed_product_15_Days'].fillna('Product101', inplace = True)
results_data
# 6) Most_Active_OS.
query = 'select UserID,OS as Most_Active_OS\
from\
(\
select UserID,OS,count(OS) as occur\
from visitorlogsdata\
where UserID is not null\
group by UserID,OS\
order by UserID,occur desc\
) as occur_table\
group by UserID;\
'
Most_Active_OS = pd.read_sql_query(query, connection)
Most_Active_OS
# Merging.
results_data['Most_Active_OS'] = pd.merge(results_data, Most_Active_OS,
how = 'left', on = 'UserID')['Most_Active_OS']
results_data
# 7) Recently_Viewed_Product.
query = 'select UserID,ProductID as Recently_Viewed_Product,max(VisitDateTime)\
from visitorlogsdata\
where UserID is not null\
group by UserID\
order by UserID\
;\
'
Recently_Viewed_Product = pd.read_sql_query(query, connection)
Recently_Viewed_Product
# Merging.
results_data['Recently_Viewed_Product'] = pd.merge(results_data, Recently_Viewed_Product,
how = 'left', on = 'UserID')['Recently_Viewed_Product']
results_data
# Filling NaN values with Product101.
results_data['Recently_Viewed_Product'].fillna('Product101', inplace = True)
results_data
# 8) Pageloads_last_7_days.
query = "select UserID,\
count(case when upper(activity)='PAGELOAD' then UserID else null end) as Pageloads_last_7_days\
from visitorlogsdata\
where VisitDateTime>@current_date-interval 7 day\
and UserID is not null\
group by UserID\
order by userID;\
"
Pageloads_last_7_days = pd.read_sql_query(query, connection)
Pageloads_last_7_days
# Merging.
results_data['Pageloads_last_7_days'] = pd.merge(results_data, Pageloads_last_7_days,
how = 'left', on = 'UserID')['Pageloads_last_7_days']
results_data
# 9) Clicks_last_7_days.
query = "select UserID,\
count(case when upper(activity)='CLICK' then UserID else null end) as Clicks_last_7_days\
from visitorlogsdata\
where VisitDateTime>@current_date-interval 7 day\
and UserID is not null\
group by UserID\
order by userID;\
"
Clicks_last_7_days = pd.read_sql_query(query, connection)
Clicks_last_7_days
# Merging.
results_data['Clicks_last_7_days'] = pd.merge(results_data, Clicks_last_7_days,
how = 'left', on = 'UserID')['Clicks_last_7_days']
results_data
# Exporting final results.
results_data.to_csv('ETL_solution.csv', index = False)
```
|
github_jupyter
|
# Importing modules.
import numpy as np
import pandas as pd
from IPython.display import display
from sqlalchemy import create_engine
from sqlalchemy.types import DateTime, VARCHAR
import mysql.connector as connector
# Importing data.
visitorlogs = pd.read_csv(r'D:\Hackathon\Analytics Vidhya\Job-a-thon\data\Modified\VisitorLogsData.csv')
print('Shape of VisitorLogsData', visitorlogs.shape)
display(visitorlogs.head())
user = pd.read_csv(r'D:\Hackathon\Analytics Vidhya\Job-a-thon\data\Modified\userTable.csv')
print('Shape of userTable', user.shape)
display(user.head())
# Renaming for ease.
user.rename(columns={'Signup Date': 'SignupDate',
'User Segment': 'Segment'}, inplace=True)
user.head()
# Dropping unregistered users.
visitorlogs.dropna(subset=['UserID'], inplace=True)
# Cross checking for nulls.
visitorlogs.isnull().sum()
print('Shape of the data is:', visitorlogs.shape)
def datetime_clean(date_time):
'''
Converts the strings to datetime format.
input = Date time in string format.
output = returns datetime value.
'''
try:
return pd.to_datetime(pd.Timestamp(date_time), utc=True)
except:
try:
date_time = int(date_time)
return pd.to_datetime(pd.Timestamp(date_time), utc=True)
except:
pass
# Transforming the 'VisitDateTime' column.
visitorlogs['VisitDateTime'] = visitorlogs['VisitDateTime'].map(lambda x: datetime_clean(x))
user['SignupDate'] = user['SignupDate'].map(lambda x: datetime_clean(x))
# Replacing spaces with 'NaN'.
visitorlogs = visitorlogs.replace(r'^\s*$', np.nan, regex=True)
user = user.replace(r'^\s*$', np.nan, regex=True)
# Imputing values with mean for each User.
visitorlogs['VisitDateTime'] = visitorlogs.groupby(['UserID'])['VisitDateTime']\
.transform(lambda x: x.fillna(x.mean()))
# Cross checking for null values
visitorlogs.isnull().sum()
# Establishing SQL connection.
connection = connector.connect(host = 'localhost', passwd = 'param12345',
user = 'root')
# Establishing SQL connection.
connection = connector.connect(host = 'localhost', passwd = 'param12345',
user = 'root')
# Creating a cursor.
mycursor = connection.cursor()
# Dropping existing database.
mycursor.execute("DROP DATABASE IF EXISTS data")
# Creating a database.
mycursor.execute("CREATE DATABASE data")
sql_engine = create_engine("mysql+mysqlconnector://" + 'root' + ":"
+ 'param12345' + "@" + 'localhost' + "/" + 'data')
# Tables.
query = 'Show tables from data'
tables = pd.read_sql_query(query, connection)
tables
# Declaring variable types.
dtype = {'webClientID': VARCHAR(50),
'UserID': VARCHAR(50),
'ProductID': VARCHAR(50),
'VisitDateTime': DateTime,
'Activity': VARCHAR(50),
'OS': VARCHAR(50),
'Browser': VARCHAR(50),
'City': VARCHAR(100),
'Country': VARCHAR(100)}
# Exporting to SQL.
visitorlogs.to_sql(name = 'visitorlogsdata', con = sql_engine,
if_exists = 'replace', index = False, dtype = dtype)
# Declaring variable types.
dtype = {'UserID': VARCHAR(10),
'SignupDate': DateTime,
'Segment': VARCHAR(5)}
# Exporting to SQL.
user.to_sql(name = 'usertable', con = sql_engine, if_exists = 'replace',
index = False)
# Update
connection = connector.connect(host = 'localhost', user = 'root', passwd = 'param12345',
database = 'data')
# Update
connection = connector.connect(host = 'localhost', user = 'root', passwd = 'param12345',
database = 'data')
# Datas only upto 27th July 2018.
query = "select @current_date:= STR_TO_DATE('2018-05-27 23:59:59','%Y-%m-%d %H:%i:%s.%f')"
max_date = pd.read_sql_query(query, connection)
max_date
# Creating a data frame.
results_data = pd.DataFrame()
# 1) UserID.
results_data['UserID'] = sorted(user.UserID)
results_data
# 2) No_of_days_Visited_7_Days.
query = 'select UserID, count(distinct day(VisitDateTime)) as No_of_days_Visited_7_Days\
from visitorlogsdata\
where VisitDateTime>@current_date-interval 7 day\
and UserID is not null\
group by UserID\
order by UserID\
;'
No_of_days_Visited_7_Days = pd.read_sql_query(query, connection)
No_of_days_Visited_7_Days
# Merging.
results_data['No_of_days_Visited_7_Days'] = pd.merge(results_data, No_of_days_Visited_7_Days,
how = 'left', on = 'UserID')['No_of_days_Visited_7_Days']
results_data
# 3) No_Of_Products_Viewed_15_Days.
query = 'select UserID,count(distinct ProductID) as No_Of_Products_Viewed_15_Days\
from visitorlogsdata\
where VisitDateTime> @current_date -interval 15 day\
and UserID is not null\
group by UserID\
order by UserID\
;'
No_Of_Products_Viewed_15_Days = pd.read_sql_query(query, connection)
No_Of_Products_Viewed_15_Days
# Merging.
results_data['No_Of_Products_Viewed_15_Days'] = pd.merge(results_data, No_Of_Products_Viewed_15_Days,
how = 'left', on = 'UserID')['No_Of_Products_Viewed_15_Days']
results_data
# 4) User_Vintage.
query = 'select UserID,datediff(@current_date,SignupDate) as User_Vintage\
from usertable\
where UserID is not null\
order by UserID\
;'
User_Vintage = pd.read_sql_query(query, connection)
User_Vintage
# Mering.
results_data['User_Vintage'] = pd.merge(results_data, User_Vintage,
how = 'left', on = 'UserID')['User_Vintage']
results_data
# 5) Most_Viewed_product_15_Days.
query = 'select UserID,Most_Viewed_product_15_Days ,max(occurs) as products_viewed\
from\
(\
select UserID,ProductID as Most_Viewed_product_15_Days,count(ProductID) as occurs,VisitDateTime\
from visitorlogsdata\
where VisitDateTime>@current_date-interval 15 day\
and UserID is not null\
group by UserID,ProductID\
order by UserID,occurs desc,VisitDateTime desc\
) as occur_table\
group by UserID\
;'
Most_Viewed_product_15_Days = pd.read_sql_query(query, connection)
Most_Viewed_product_15_Days
# Merging.
results_data['Most_Viewed_product_15_Days'] = pd.merge(results_data, Most_Viewed_product_15_Days,
how = 'left', on = 'UserID')['Most_Viewed_product_15_Days']
results_data
# Filling NaN values with Product101.
results_data['Most_Viewed_product_15_Days'].fillna('Product101', inplace = True)
results_data
# 6) Most_Active_OS.
query = 'select UserID,OS as Most_Active_OS\
from\
(\
select UserID,OS,count(OS) as occur\
from visitorlogsdata\
where UserID is not null\
group by UserID,OS\
order by UserID,occur desc\
) as occur_table\
group by UserID;\
'
Most_Active_OS = pd.read_sql_query(query, connection)
Most_Active_OS
# Merging.
results_data['Most_Active_OS'] = pd.merge(results_data, Most_Active_OS,
how = 'left', on = 'UserID')['Most_Active_OS']
results_data
# 7) Recently_Viewed_Product.
query = 'select UserID,ProductID as Recently_Viewed_Product,max(VisitDateTime)\
from visitorlogsdata\
where UserID is not null\
group by UserID\
order by UserID\
;\
'
Recently_Viewed_Product = pd.read_sql_query(query, connection)
Recently_Viewed_Product
# Merging.
results_data['Recently_Viewed_Product'] = pd.merge(results_data, Recently_Viewed_Product,
how = 'left', on = 'UserID')['Recently_Viewed_Product']
results_data
# Filling NaN values with Product101.
results_data['Recently_Viewed_Product'].fillna('Product101', inplace = True)
results_data
# 8) Pageloads_last_7_days.
query = "select UserID,\
count(case when upper(activity)='PAGELOAD' then UserID else null end) as Pageloads_last_7_days\
from visitorlogsdata\
where VisitDateTime>@current_date-interval 7 day\
and UserID is not null\
group by UserID\
order by userID;\
"
Pageloads_last_7_days = pd.read_sql_query(query, connection)
Pageloads_last_7_days
# Merging.
results_data['Pageloads_last_7_days'] = pd.merge(results_data, Pageloads_last_7_days,
how = 'left', on = 'UserID')['Pageloads_last_7_days']
results_data
# 9) Clicks_last_7_days.
query = "select UserID,\
count(case when upper(activity)='CLICK' then UserID else null end) as Clicks_last_7_days\
from visitorlogsdata\
where VisitDateTime>@current_date-interval 7 day\
and UserID is not null\
group by UserID\
order by userID;\
"
Clicks_last_7_days = pd.read_sql_query(query, connection)
Clicks_last_7_days
# Merging.
results_data['Clicks_last_7_days'] = pd.merge(results_data, Clicks_last_7_days,
how = 'left', on = 'UserID')['Clicks_last_7_days']
results_data
# Exporting final results.
results_data.to_csv('ETL_solution.csv', index = False)
| 0.448185 | 0.561034 |
```
Sys.Date()
libraries = c("dplyr","magrittr","tidyr","ggplot2","gridExtra","RColorBrewer","zoo","scales","colorspace","readxl")
for(x in libraries) { library(x,character.only=TRUE,warn.conflicts=FALSE) }
'%&%' = function(x,y) paste0(x,y)
theme_set(theme_classic(base_size=12, base_family="sans"))
number_of_scenario = 1
read.csv("../../results/Scenario-"%&%number_of_scenario%&%"/incidence-final.csv") -> dt_table
dt_table %>%
filter(var == c('Incidence')) %>%
mutate(cal = (as.Date('2019-12-08')+time)) -> dt.inci
read.csv("../../scripts/Sungmok/CHN.inci.csv") %>%
as.data.frame %>% do(na.locf(.)) %>%
mutate(cal = (as.Date('2019-12-08')+time)) -> dt.CHN.inci
dt.inci %>% filter(cal >= as.Date('2019-12-31')) -> dt.inci.1
dt.CHN.inci %>% filter(cal >= as.Date('2019-12-31')) -> dt.CHN.inci.1
read.csv("../../data/data.csv") -> dt_table2
# dt_table2 %<>% filter(time <= 48)
dt_table2 %>% gather(time, source, exports, deaths) -> dt.fig2
colnames(dt.fig2) <- c("cal", "x", "y", "z","pop","source","value")
dt.fig2 %<>% dplyr::select(cal, source, value)
dt.fig2$cal <- as.Date(dt.fig2$cal)
### CFR
dt_table %>% filter(var == c('CFR')) %>%
mutate(cal = (as.Date('2020-01-09')+time-1)) -> dt.cfr
#### making upper bound for CFR with zero case
dt.cfr[1:9,] -> dt.cfr.temp
dt.cfr.temp %>% mutate(time.temp = 0:8) %>% mutate(cal = (as.Date('2019-12-31')+time)) -> dt.cfr.temp
dt.inci %>% filter(time<=9) -> dt.inci.temp
dt.inci.temp %<>% select(time, mean) %>% rename(inci = mean)
merge(dt.cfr.temp, dt.inci.temp, by = c('time')) %>%
mutate(lower = NA, upper = NA, mean = NA) %>% select(-inci, -time.temp) %>%
select(var, time, mean, lower, upper, cal)-> dt.cfr.temp
rbind(dt.cfr.temp, dt.cfr) -> dt.cfr
print(dt.inci.1)
cs = c(8, 10)
options(repr.plot.width=cs[1],repr.plot.height=cs[2])
fnt_size = 12
scaling_parameter = .137
dt.CHN.inci.1 %>%
ggplot(aes(x=cal, y=inci)) +
geom_bar(stat='identity', width = .6, color='black', size=.2,
fill=RColorBrewer::brewer.pal(9, "Blues")[4]) +
geom_ribbon(data=dt.inci.1, aes(x=cal, ymin = dt.inci.1$lower*scaling_parameter,
ymax = dt.inci.1$upper*scaling_parameter),
alpha = 0.6, fill="grey70", inherit.aes=FALSE) +
geom_line(data=dt.inci.1, aes(y=dt.inci.1$lower*scaling_parameter), color="black", size=.4) +
geom_line(data=dt.inci.1, aes(y=dt.inci.1$upper*scaling_parameter), color="black", size=.4) +
geom_line(data=dt.inci.1, aes(y=mean*scaling_parameter), color="black", size=1) +
scale_y_continuous(expand=c(.0,0),
sec.axis = sec_axis(~./scaling_parameter,
name = "Estimated cumulative incidence\n")) +
xlab("") + ylab("Cumulative number\n") +
theme(plot.margin = unit(c(.5,.5,1,.25),"lines"),
text = element_text(family="sans",color="black"),
axis.text.x = element_text(angle = 35, hjust = 1),
axis.title.y = element_text(vjust = -1, size=fnt_size+1),
axis.text =element_text(size=fnt_size, family="sans",color="black"),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank()) +
coord_cartesian(ylim = c(0,1100)) +
scale_x_date(expand=c(0,0), date_labels="%b %d",date_breaks ="1 day",
limits=c(as.Date('2019-12-31'),as.Date('2020-01-24')+.5)) -> fig11
dt.fig2 %>% filter(cal >= as.Date('2019-12-31')) %>%
ggplot(aes(x=cal, y=value, fill=source)) +
geom_bar(stat="identity", width = .8, position='dodge', color='black', size=.2) +
guides(color=F) +
xlab("") + ylab("") +
theme(text = element_text(family="sans",color="black",size=fnt_size),
axis.text.x = element_text(angle = 35, hjust = 1),
axis.text = element_text(size=fnt_size, family="sans",color="black"),
legend.position = c(0.22, 0.82),
legend.title=element_blank(),
legend.text = element_text(size = 11, family="sans"),
legend.spacing.x = unit(0.15, 'cm')
) +
scale_x_date(expand=c(0,0), date_labels="%b %d",date_breaks ="2 day",
limits=c(as.Date('2019-12-31'),as.Date('2020-01-24')+.5)) +
scale_y_continuous(expand=c(0,.01)) +
scale_fill_brewer(palette = "Dark1") +
expand_limits(y = 0:35) -> fig12
dt.cfr %>% mutate(mean.p = mean *100, lower.p = lower*100, upper.p = upper*100) %>% mutate(cal=cal+0.5) %T>% print %>%
ggplot(aes(x=cal+.5, y=mean.p)) +
geom_errorbar(aes(ymin=lower.p, ymax=upper.p), colour="black", width=0.5, size=0.5) +
geom_point(size=2.5, shape=18) +
xlab("\nDate of report") + ylab("Case fatality (%)\n") +
theme(plot.margin = unit(c(0,4.5,1,1.5),"lines"),
text = element_text(family="sans",color="black",size=fnt_size),
axis.text.x = element_text(angle = 35, hjust = 1),
axis.title.y = element_text(vjust = 3.5, size=fnt_size+1),
axis.text =element_text(size=fnt_size, family="sans",color="black"),
panel.grid.major = element_blank(), panel.grid.minor = element_blank()) +
scale_x_date(expand=c(0,0), date_labels="%b %d",date_breaks ="1 day",
limits=c(as.Date('2019-12-31'),as.Date('2020-01-24')+.5)) +
coord_cartesian(ylim = c(0,8)) +
scale_y_continuous(expand=c(0,.01)) -> fig13
fig123 = fig11 +
annotation_custom(grob = ggplotGrob(
fig12 + theme(plot.background = element_rect(colour = "white"))),
xmin = as.numeric(as.Date('2019-12-31')), xmax = as.numeric(as.Date('2020-01-14')),
ymin = 400, ymax = 1050)
pFinal = grid.arrange(fig123, fig13, heights=c(1.75,1), nrow=2, ncol=1)
ggsave(plot=pFinal, width=cs[1],height=cs[2],filename="../../figures/draft/fig1-scen"%&%number_of_scenario%&%".pdf",useDingbats=FALSE)
cs = c(5, 4.5)
options(repr.plot.width=cs[1],repr.plot.height=cs[2])
r = dt_table[dt_table$var=='r',]
print(r)
data.frame(T = seq(4.1,10.9,length.out=25)) %>% mutate(mean = 1+r$mean*T, lower = 1+r$lower*T, upper = 1+r$upper*T) %T>% print %>%
ggplot(aes(x=T)) +
geom_line(aes(y=lower), color="black", size=.4) +
geom_line(aes(y=upper), color="black", size=.4) +
geom_ribbon(aes(x=T, ymin = lower, ymax = upper), alpha = 0.6, fill="grey70", inherit.aes=FALSE) +
geom_line(aes(y=mean), color="black", size=1) +
xlab("Serial interval (days)") + ylab(expression(italic(R)[0])) +
coord_cartesian(ylim = c(1.5,2.8)) +
scale_x_continuous(expand=c(0.02,0)) + scale_y_continuous(expand=c(0,0)) +
theme(plot.margin = unit(c(.5,.5,1,.25),"lines"),
text = element_text(family="sans",color="black"),
axis.text = element_text(size=fnt_size, family="sans",color="black"),
panel.grid.major = element_blank(),
axis.title = element_text(size=fnt_size+1),
panel.grid.minor = element_blank()) -> fig31
fig31
ggsave(plot=fig31, width=cs[1], height=cs[2],filename="../../figures/draft/fig31.pdf",useDingbats=FALSE)
number_of_scenario = 2
read.csv("../../results/Scenario-"%&%number_of_scenario%&%"/incidence-final.csv") -> dt_table
dt_table %>%
filter(var == c('Incidence')) %>%
mutate(cal = (as.Date('2019-12-08')+time)) -> dt.inci
read.csv("../../scripts/Sungmok/CHN.inci.csv") %>%
as.data.frame %>% do(na.locf(.)) %>%
mutate(cal = (as.Date('2019-12-08')+time)) -> dt.CHN.inci
dt.inci %>% filter(cal >= as.Date('2019-12-31')) -> dt.inci.1
dt.CHN.inci %>% filter(cal >= as.Date('2019-12-31')) -> dt.CHN.inci.1
read.csv("../../data/data.csv") -> dt_table2
# dt_table2 %<>% filter(time <= 48)
dt_table2 %>% gather(time, source, exports, deaths) -> dt.fig2
colnames(dt.fig2) <- c("cal", "x", "y", "z","pop","source","value")
dt.fig2 %<>% dplyr::select(cal, source, value)
dt.fig2$cal <- as.Date(dt.fig2$cal)
### CFR
dt_table %>% filter(var == c('CFR')) %>%
mutate(cal = (as.Date('2020-01-09')+time-1)) -> dt.cfr
#### making upper bound for CFR with zero case
dt.cfr[1:9,] -> dt.cfr.temp
dt.cfr.temp %>% mutate(time.temp = 0:8) %>% mutate(cal = (as.Date('2019-12-31')+time)) -> dt.cfr.temp
dt.inci %>% filter(time<=9) -> dt.inci.temp
dt.inci.temp %<>% select(time, mean) %>% rename(inci = mean)
merge(dt.cfr.temp, dt.inci.temp, by = c('time')) %>%
mutate(lower = NA, upper = NA, mean = NA) %>% select(-inci, -time.temp) %>%
select(var, time, mean, lower, upper, cal)-> dt.cfr.temp
rbind(dt.cfr.temp, dt.cfr) -> dt.cfr
print(dt.inci.1)
cs = c(8, 10)
options(repr.plot.width=cs[1],repr.plot.height=cs[2])
scaling_parameter = .054
dt.CHN.inci.1 %>%
ggplot(aes(x=cal, y=inci)) +
geom_bar(stat='identity', width = .6, color='black', size=.2,
fill=RColorBrewer::brewer.pal(9, "Blues")[4]) +
geom_ribbon(data=dt.inci.1, aes(x=cal, ymin = dt.inci.1$lower*scaling_parameter,
ymax = dt.inci.1$upper*scaling_parameter),
alpha = 0.6, fill="grey70", inherit.aes=FALSE) +
geom_line(data=dt.inci.1, aes(y=dt.inci.1$lower*scaling_parameter), color="black", size=.4) +
geom_line(data=dt.inci.1, aes(y=dt.inci.1$upper*scaling_parameter), color="black", size=.4) +
geom_line(data=dt.inci.1, aes(y=mean*scaling_parameter), color="black", size=1) +
scale_y_continuous(expand=c(.0,0),
sec.axis = sec_axis(~./scaling_parameter,
name = "Estimated cumulative incidence\n")) +
xlab("") + ylab("Cumulative number\n") +
theme(plot.margin = unit(c(.5,.5,1,.25),"lines"),
text = element_text(family="sans",color="black"),
axis.text.x = element_text(angle = 35, hjust = 1),
axis.title.y = element_text(vjust = -1, size=fnt_size+1),
axis.text =element_text(size=fnt_size, family="sans",color="black"),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank()) +
coord_cartesian(ylim = c(0,1100)) +
scale_x_date(expand=c(0,0), date_labels="%b %d",date_breaks ="1 day",
limits=c(as.Date('2019-12-31'),as.Date('2020-01-24')+.5)) -> fig11
dt.fig2 %>% filter(cal >= as.Date('2019-12-31')) %>%
ggplot(aes(x=cal, y=value, fill=source)) +
geom_bar(stat="identity", width = .8, position='dodge', color='black', size=.2) +
guides(color=F) +
xlab("") + ylab("") +
theme(text = element_text(family="sans",color="black",size=fnt_size),
axis.text.x = element_text(angle = 35, hjust = 1),
axis.text = element_text(size=fnt_size, family="sans",color="black"),
legend.position = c(0.22, 0.82),
legend.title=element_blank(),
legend.text = element_text(size = 11, family="sans"),
legend.spacing.x = unit(0.15, 'cm')
) +
scale_x_date(expand=c(0,0), date_labels="%b %d",date_breaks ="2 day",
limits=c(as.Date('2019-12-31'),as.Date('2020-01-24')+.5)) +
scale_y_continuous(expand=c(0,.01)) +
scale_fill_brewer(palette = "Dark1") +
expand_limits(y = 0:35) -> fig12
dt.cfr %>% mutate(mean.p = mean *100, lower.p = lower*100, upper.p = upper*100) %>% mutate(cal=cal+0.5) %T>% print %>%
ggplot(aes(x=cal+.5, y=mean.p)) +
geom_errorbar(aes(ymin=lower.p, ymax=upper.p), colour="black", width=0.5, size=0.5) +
geom_point(size=2.5, shape=18) +
xlab("\nDate of report") + ylab("Case fatality (%)\n") +
theme(plot.margin = unit(c(0,5,1,1),"lines"),
text = element_text(family="sans",color="black",size=fnt_size),
axis.text.x = element_text(angle = 35, hjust = 1),
axis.title.y = element_text(vjust = 3.5, size=fnt_size+1),
axis.text =element_text(size=fnt_size, family="sans",color="black"),
panel.grid.major = element_blank(), panel.grid.minor = element_blank()) +
scale_x_date(expand=c(0,0), date_labels="%b %d",date_breaks ="1 day",
limits=c(as.Date('2019-12-31'),as.Date('2020-01-24')+.5)) +
coord_cartesian(ylim = c(0,33)) +
scale_y_continuous(expand=c(0,.01)) -> fig13
fig123 = fig11 +
annotation_custom(grob = ggplotGrob(
fig12 + theme(plot.background = element_rect(colour = "white"))),
xmin = as.numeric(as.Date('2019-12-31')), xmax = as.numeric(as.Date('2020-01-14')),
ymin = 400, ymax = 1050)
pFinal = grid.arrange(fig123, fig13, heights=c(1.75,1), nrow=2, ncol=1)
ggsave(plot=pFinal, width=cs[1],height=cs[2],filename="../../figures/draft/fig1-scen2.pdf",useDingbats=FALSE)
cs = c(5, 4.5)
options(repr.plot.width=cs[1],repr.plot.height=cs[2])
r = dt_table[dt_table$var=='r',]
R0 = function(T) 1+r*T
print(r)
data.frame(T = seq(4.1,10.9,length.out=25)) %>% mutate(mean = 1+r$mean*T, lower = 1+r$lower*T, upper = 1+r$upper*T) %T>% print %>%
ggplot(aes(x=T)) +
geom_line(aes(y=lower), color="black", size=.4) +
geom_line(aes(y=upper), color="black", size=.4) +
geom_ribbon(aes(x=T, ymin = lower, ymax = upper), alpha = 0.6, fill="grey70", inherit.aes=FALSE) +
geom_line(aes(y=mean), color="black", size=1) +
xlab("Serial interval (days)") + ylab(expression(italic(R)[0])) +
coord_cartesian(ylim = c(1.5,5.25)) +
scale_x_continuous(expand=c(0.02,0)) + scale_y_continuous(expand=c(0,0)) +
theme(plot.margin = unit(c(.5,.5,1,.25),"lines"),
text = element_text(family="sans",color="black"),
axis.text = element_text(size=fnt_size, family="sans",color="black"),
axis.title = element_text(size=fnt_size+1),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank()) -> fig32
fig32
ggsave(plot=fig32, width=cs[1], height=cs[2],filename="../../figures/draft/fig32.pdf",useDingbats=FALSE)
```
|
github_jupyter
|
Sys.Date()
libraries = c("dplyr","magrittr","tidyr","ggplot2","gridExtra","RColorBrewer","zoo","scales","colorspace","readxl")
for(x in libraries) { library(x,character.only=TRUE,warn.conflicts=FALSE) }
'%&%' = function(x,y) paste0(x,y)
theme_set(theme_classic(base_size=12, base_family="sans"))
number_of_scenario = 1
read.csv("../../results/Scenario-"%&%number_of_scenario%&%"/incidence-final.csv") -> dt_table
dt_table %>%
filter(var == c('Incidence')) %>%
mutate(cal = (as.Date('2019-12-08')+time)) -> dt.inci
read.csv("../../scripts/Sungmok/CHN.inci.csv") %>%
as.data.frame %>% do(na.locf(.)) %>%
mutate(cal = (as.Date('2019-12-08')+time)) -> dt.CHN.inci
dt.inci %>% filter(cal >= as.Date('2019-12-31')) -> dt.inci.1
dt.CHN.inci %>% filter(cal >= as.Date('2019-12-31')) -> dt.CHN.inci.1
read.csv("../../data/data.csv") -> dt_table2
# dt_table2 %<>% filter(time <= 48)
dt_table2 %>% gather(time, source, exports, deaths) -> dt.fig2
colnames(dt.fig2) <- c("cal", "x", "y", "z","pop","source","value")
dt.fig2 %<>% dplyr::select(cal, source, value)
dt.fig2$cal <- as.Date(dt.fig2$cal)
### CFR
dt_table %>% filter(var == c('CFR')) %>%
mutate(cal = (as.Date('2020-01-09')+time-1)) -> dt.cfr
#### making upper bound for CFR with zero case
dt.cfr[1:9,] -> dt.cfr.temp
dt.cfr.temp %>% mutate(time.temp = 0:8) %>% mutate(cal = (as.Date('2019-12-31')+time)) -> dt.cfr.temp
dt.inci %>% filter(time<=9) -> dt.inci.temp
dt.inci.temp %<>% select(time, mean) %>% rename(inci = mean)
merge(dt.cfr.temp, dt.inci.temp, by = c('time')) %>%
mutate(lower = NA, upper = NA, mean = NA) %>% select(-inci, -time.temp) %>%
select(var, time, mean, lower, upper, cal)-> dt.cfr.temp
rbind(dt.cfr.temp, dt.cfr) -> dt.cfr
print(dt.inci.1)
cs = c(8, 10)
options(repr.plot.width=cs[1],repr.plot.height=cs[2])
fnt_size = 12
scaling_parameter = .137
dt.CHN.inci.1 %>%
ggplot(aes(x=cal, y=inci)) +
geom_bar(stat='identity', width = .6, color='black', size=.2,
fill=RColorBrewer::brewer.pal(9, "Blues")[4]) +
geom_ribbon(data=dt.inci.1, aes(x=cal, ymin = dt.inci.1$lower*scaling_parameter,
ymax = dt.inci.1$upper*scaling_parameter),
alpha = 0.6, fill="grey70", inherit.aes=FALSE) +
geom_line(data=dt.inci.1, aes(y=dt.inci.1$lower*scaling_parameter), color="black", size=.4) +
geom_line(data=dt.inci.1, aes(y=dt.inci.1$upper*scaling_parameter), color="black", size=.4) +
geom_line(data=dt.inci.1, aes(y=mean*scaling_parameter), color="black", size=1) +
scale_y_continuous(expand=c(.0,0),
sec.axis = sec_axis(~./scaling_parameter,
name = "Estimated cumulative incidence\n")) +
xlab("") + ylab("Cumulative number\n") +
theme(plot.margin = unit(c(.5,.5,1,.25),"lines"),
text = element_text(family="sans",color="black"),
axis.text.x = element_text(angle = 35, hjust = 1),
axis.title.y = element_text(vjust = -1, size=fnt_size+1),
axis.text =element_text(size=fnt_size, family="sans",color="black"),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank()) +
coord_cartesian(ylim = c(0,1100)) +
scale_x_date(expand=c(0,0), date_labels="%b %d",date_breaks ="1 day",
limits=c(as.Date('2019-12-31'),as.Date('2020-01-24')+.5)) -> fig11
dt.fig2 %>% filter(cal >= as.Date('2019-12-31')) %>%
ggplot(aes(x=cal, y=value, fill=source)) +
geom_bar(stat="identity", width = .8, position='dodge', color='black', size=.2) +
guides(color=F) +
xlab("") + ylab("") +
theme(text = element_text(family="sans",color="black",size=fnt_size),
axis.text.x = element_text(angle = 35, hjust = 1),
axis.text = element_text(size=fnt_size, family="sans",color="black"),
legend.position = c(0.22, 0.82),
legend.title=element_blank(),
legend.text = element_text(size = 11, family="sans"),
legend.spacing.x = unit(0.15, 'cm')
) +
scale_x_date(expand=c(0,0), date_labels="%b %d",date_breaks ="2 day",
limits=c(as.Date('2019-12-31'),as.Date('2020-01-24')+.5)) +
scale_y_continuous(expand=c(0,.01)) +
scale_fill_brewer(palette = "Dark1") +
expand_limits(y = 0:35) -> fig12
dt.cfr %>% mutate(mean.p = mean *100, lower.p = lower*100, upper.p = upper*100) %>% mutate(cal=cal+0.5) %T>% print %>%
ggplot(aes(x=cal+.5, y=mean.p)) +
geom_errorbar(aes(ymin=lower.p, ymax=upper.p), colour="black", width=0.5, size=0.5) +
geom_point(size=2.5, shape=18) +
xlab("\nDate of report") + ylab("Case fatality (%)\n") +
theme(plot.margin = unit(c(0,4.5,1,1.5),"lines"),
text = element_text(family="sans",color="black",size=fnt_size),
axis.text.x = element_text(angle = 35, hjust = 1),
axis.title.y = element_text(vjust = 3.5, size=fnt_size+1),
axis.text =element_text(size=fnt_size, family="sans",color="black"),
panel.grid.major = element_blank(), panel.grid.minor = element_blank()) +
scale_x_date(expand=c(0,0), date_labels="%b %d",date_breaks ="1 day",
limits=c(as.Date('2019-12-31'),as.Date('2020-01-24')+.5)) +
coord_cartesian(ylim = c(0,8)) +
scale_y_continuous(expand=c(0,.01)) -> fig13
fig123 = fig11 +
annotation_custom(grob = ggplotGrob(
fig12 + theme(plot.background = element_rect(colour = "white"))),
xmin = as.numeric(as.Date('2019-12-31')), xmax = as.numeric(as.Date('2020-01-14')),
ymin = 400, ymax = 1050)
pFinal = grid.arrange(fig123, fig13, heights=c(1.75,1), nrow=2, ncol=1)
ggsave(plot=pFinal, width=cs[1],height=cs[2],filename="../../figures/draft/fig1-scen"%&%number_of_scenario%&%".pdf",useDingbats=FALSE)
cs = c(5, 4.5)
options(repr.plot.width=cs[1],repr.plot.height=cs[2])
r = dt_table[dt_table$var=='r',]
print(r)
data.frame(T = seq(4.1,10.9,length.out=25)) %>% mutate(mean = 1+r$mean*T, lower = 1+r$lower*T, upper = 1+r$upper*T) %T>% print %>%
ggplot(aes(x=T)) +
geom_line(aes(y=lower), color="black", size=.4) +
geom_line(aes(y=upper), color="black", size=.4) +
geom_ribbon(aes(x=T, ymin = lower, ymax = upper), alpha = 0.6, fill="grey70", inherit.aes=FALSE) +
geom_line(aes(y=mean), color="black", size=1) +
xlab("Serial interval (days)") + ylab(expression(italic(R)[0])) +
coord_cartesian(ylim = c(1.5,2.8)) +
scale_x_continuous(expand=c(0.02,0)) + scale_y_continuous(expand=c(0,0)) +
theme(plot.margin = unit(c(.5,.5,1,.25),"lines"),
text = element_text(family="sans",color="black"),
axis.text = element_text(size=fnt_size, family="sans",color="black"),
panel.grid.major = element_blank(),
axis.title = element_text(size=fnt_size+1),
panel.grid.minor = element_blank()) -> fig31
fig31
ggsave(plot=fig31, width=cs[1], height=cs[2],filename="../../figures/draft/fig31.pdf",useDingbats=FALSE)
number_of_scenario = 2
read.csv("../../results/Scenario-"%&%number_of_scenario%&%"/incidence-final.csv") -> dt_table
dt_table %>%
filter(var == c('Incidence')) %>%
mutate(cal = (as.Date('2019-12-08')+time)) -> dt.inci
read.csv("../../scripts/Sungmok/CHN.inci.csv") %>%
as.data.frame %>% do(na.locf(.)) %>%
mutate(cal = (as.Date('2019-12-08')+time)) -> dt.CHN.inci
dt.inci %>% filter(cal >= as.Date('2019-12-31')) -> dt.inci.1
dt.CHN.inci %>% filter(cal >= as.Date('2019-12-31')) -> dt.CHN.inci.1
read.csv("../../data/data.csv") -> dt_table2
# dt_table2 %<>% filter(time <= 48)
dt_table2 %>% gather(time, source, exports, deaths) -> dt.fig2
colnames(dt.fig2) <- c("cal", "x", "y", "z","pop","source","value")
dt.fig2 %<>% dplyr::select(cal, source, value)
dt.fig2$cal <- as.Date(dt.fig2$cal)
### CFR
dt_table %>% filter(var == c('CFR')) %>%
mutate(cal = (as.Date('2020-01-09')+time-1)) -> dt.cfr
#### making upper bound for CFR with zero case
dt.cfr[1:9,] -> dt.cfr.temp
dt.cfr.temp %>% mutate(time.temp = 0:8) %>% mutate(cal = (as.Date('2019-12-31')+time)) -> dt.cfr.temp
dt.inci %>% filter(time<=9) -> dt.inci.temp
dt.inci.temp %<>% select(time, mean) %>% rename(inci = mean)
merge(dt.cfr.temp, dt.inci.temp, by = c('time')) %>%
mutate(lower = NA, upper = NA, mean = NA) %>% select(-inci, -time.temp) %>%
select(var, time, mean, lower, upper, cal)-> dt.cfr.temp
rbind(dt.cfr.temp, dt.cfr) -> dt.cfr
print(dt.inci.1)
cs = c(8, 10)
options(repr.plot.width=cs[1],repr.plot.height=cs[2])
scaling_parameter = .054
dt.CHN.inci.1 %>%
ggplot(aes(x=cal, y=inci)) +
geom_bar(stat='identity', width = .6, color='black', size=.2,
fill=RColorBrewer::brewer.pal(9, "Blues")[4]) +
geom_ribbon(data=dt.inci.1, aes(x=cal, ymin = dt.inci.1$lower*scaling_parameter,
ymax = dt.inci.1$upper*scaling_parameter),
alpha = 0.6, fill="grey70", inherit.aes=FALSE) +
geom_line(data=dt.inci.1, aes(y=dt.inci.1$lower*scaling_parameter), color="black", size=.4) +
geom_line(data=dt.inci.1, aes(y=dt.inci.1$upper*scaling_parameter), color="black", size=.4) +
geom_line(data=dt.inci.1, aes(y=mean*scaling_parameter), color="black", size=1) +
scale_y_continuous(expand=c(.0,0),
sec.axis = sec_axis(~./scaling_parameter,
name = "Estimated cumulative incidence\n")) +
xlab("") + ylab("Cumulative number\n") +
theme(plot.margin = unit(c(.5,.5,1,.25),"lines"),
text = element_text(family="sans",color="black"),
axis.text.x = element_text(angle = 35, hjust = 1),
axis.title.y = element_text(vjust = -1, size=fnt_size+1),
axis.text =element_text(size=fnt_size, family="sans",color="black"),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank()) +
coord_cartesian(ylim = c(0,1100)) +
scale_x_date(expand=c(0,0), date_labels="%b %d",date_breaks ="1 day",
limits=c(as.Date('2019-12-31'),as.Date('2020-01-24')+.5)) -> fig11
dt.fig2 %>% filter(cal >= as.Date('2019-12-31')) %>%
ggplot(aes(x=cal, y=value, fill=source)) +
geom_bar(stat="identity", width = .8, position='dodge', color='black', size=.2) +
guides(color=F) +
xlab("") + ylab("") +
theme(text = element_text(family="sans",color="black",size=fnt_size),
axis.text.x = element_text(angle = 35, hjust = 1),
axis.text = element_text(size=fnt_size, family="sans",color="black"),
legend.position = c(0.22, 0.82),
legend.title=element_blank(),
legend.text = element_text(size = 11, family="sans"),
legend.spacing.x = unit(0.15, 'cm')
) +
scale_x_date(expand=c(0,0), date_labels="%b %d",date_breaks ="2 day",
limits=c(as.Date('2019-12-31'),as.Date('2020-01-24')+.5)) +
scale_y_continuous(expand=c(0,.01)) +
scale_fill_brewer(palette = "Dark1") +
expand_limits(y = 0:35) -> fig12
dt.cfr %>% mutate(mean.p = mean *100, lower.p = lower*100, upper.p = upper*100) %>% mutate(cal=cal+0.5) %T>% print %>%
ggplot(aes(x=cal+.5, y=mean.p)) +
geom_errorbar(aes(ymin=lower.p, ymax=upper.p), colour="black", width=0.5, size=0.5) +
geom_point(size=2.5, shape=18) +
xlab("\nDate of report") + ylab("Case fatality (%)\n") +
theme(plot.margin = unit(c(0,5,1,1),"lines"),
text = element_text(family="sans",color="black",size=fnt_size),
axis.text.x = element_text(angle = 35, hjust = 1),
axis.title.y = element_text(vjust = 3.5, size=fnt_size+1),
axis.text =element_text(size=fnt_size, family="sans",color="black"),
panel.grid.major = element_blank(), panel.grid.minor = element_blank()) +
scale_x_date(expand=c(0,0), date_labels="%b %d",date_breaks ="1 day",
limits=c(as.Date('2019-12-31'),as.Date('2020-01-24')+.5)) +
coord_cartesian(ylim = c(0,33)) +
scale_y_continuous(expand=c(0,.01)) -> fig13
fig123 = fig11 +
annotation_custom(grob = ggplotGrob(
fig12 + theme(plot.background = element_rect(colour = "white"))),
xmin = as.numeric(as.Date('2019-12-31')), xmax = as.numeric(as.Date('2020-01-14')),
ymin = 400, ymax = 1050)
pFinal = grid.arrange(fig123, fig13, heights=c(1.75,1), nrow=2, ncol=1)
ggsave(plot=pFinal, width=cs[1],height=cs[2],filename="../../figures/draft/fig1-scen2.pdf",useDingbats=FALSE)
cs = c(5, 4.5)
options(repr.plot.width=cs[1],repr.plot.height=cs[2])
r = dt_table[dt_table$var=='r',]
R0 = function(T) 1+r*T
print(r)
data.frame(T = seq(4.1,10.9,length.out=25)) %>% mutate(mean = 1+r$mean*T, lower = 1+r$lower*T, upper = 1+r$upper*T) %T>% print %>%
ggplot(aes(x=T)) +
geom_line(aes(y=lower), color="black", size=.4) +
geom_line(aes(y=upper), color="black", size=.4) +
geom_ribbon(aes(x=T, ymin = lower, ymax = upper), alpha = 0.6, fill="grey70", inherit.aes=FALSE) +
geom_line(aes(y=mean), color="black", size=1) +
xlab("Serial interval (days)") + ylab(expression(italic(R)[0])) +
coord_cartesian(ylim = c(1.5,5.25)) +
scale_x_continuous(expand=c(0.02,0)) + scale_y_continuous(expand=c(0,0)) +
theme(plot.margin = unit(c(.5,.5,1,.25),"lines"),
text = element_text(family="sans",color="black"),
axis.text = element_text(size=fnt_size, family="sans",color="black"),
axis.title = element_text(size=fnt_size+1),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank()) -> fig32
fig32
ggsave(plot=fig32, width=cs[1], height=cs[2],filename="../../figures/draft/fig32.pdf",useDingbats=FALSE)
| 0.467818 | 0.452475 |
# Transformers Interpret NER Example
### Setup
```
from transformers import AutoModelForTokenClassification, AutoTokenizer
```
Import NER model
```
MODEL_PATH = 'dslim/bert-base-NER'
model = AutoModelForTokenClassification.from_pretrained(MODEL_PATH)
tokenizer = AutoTokenizer.from_pretrained(MODEL_PATH)
```
We can explore the possible entity tags that this model classifies each token into:
```
model.config.id2label
```
Import the **TokenClassificationExplainer** from the *transformers interpret* package.
```
from transformers_interpret import TokenClassificationExplainer
```
Create an instance of the explainer providing the model and the tokenizer
```
ner_explainer = TokenClassificationExplainer(model=model, tokenizer=tokenizer)
```
### Basic usage
Call the explainer to generate the attributions. In the NER task, for each token of the sentence, the predicted class is considered as the target and the attributions of the rest of the tokens are calculated. This can take up to one or two minutes. In the following sections we will see how to limit the computation of the attributions to only certain tokens or only certain NER labels.
```
sample_text = "We visited Paris last weekend, where Emmanuel Macron lives."
attributions = ner_explainer(sample_text)
```
The explainer returns a dictionary with the predicted label and the attribtuion scores for every token.
```
attributions
```
**Note the alogirthm used to calcualte attributions are Layer Integreated Gradients to read more about them click [here](https://captum.ai/docs/algorithms)**
#### Visualizing explanations
With a single call to the `visualize()` method we get a nice inline display of what inputs are causing the activations to fire that led to classifying each of the tokens into a particular class.
```
html = ner_explainer.visualize()
```
### Ignore indexes
To save computation time, we can indicate a list of token indexes that we want to ignore. The explainer will not compute explanations for these tokens, although attributions of these tokens will be calculated to explain the predictions over other tokens.
```
attributions_2 = ner_explainer(sample_text, ignored_indexes=[0, 1, 2, 11, 12, 13, 14])
```
When calling *visualize()*, we can see how the tokens corresponding to the ignored indexes do not appear.
```
html = ner_explainer.visualize()
```
### Ignore labels
In a similar way, we can also tell the explainer to ignore certain labels, e.g. we might not be interested in seeing the explanations of those tokens that are classified as *'O'*.
```
attributions_3 = ner_explainer(sample_text, ignored_labels=['O'])
attributions_3
html = ner_explainer.visualize()
```
The *ignored_labels* parameter can be combined with the *ignored_indexes* parameter.
```
attributions_4 = ner_explainer(sample_text, ignored_indexes=[11, 12, 13, 14], ignored_labels=['B-PER'])
html = ner_explainer.visualize()
```
As we can see, the tokens are ignored based on the predicted class and its index. The token `Emmanuel` is wrongly predicted as `B-PER`and is therefore ignored along with the tokens in positions `11`, `12`, `13` and `14`.
|
github_jupyter
|
from transformers import AutoModelForTokenClassification, AutoTokenizer
MODEL_PATH = 'dslim/bert-base-NER'
model = AutoModelForTokenClassification.from_pretrained(MODEL_PATH)
tokenizer = AutoTokenizer.from_pretrained(MODEL_PATH)
model.config.id2label
from transformers_interpret import TokenClassificationExplainer
ner_explainer = TokenClassificationExplainer(model=model, tokenizer=tokenizer)
sample_text = "We visited Paris last weekend, where Emmanuel Macron lives."
attributions = ner_explainer(sample_text)
attributions
html = ner_explainer.visualize()
attributions_2 = ner_explainer(sample_text, ignored_indexes=[0, 1, 2, 11, 12, 13, 14])
html = ner_explainer.visualize()
attributions_3 = ner_explainer(sample_text, ignored_labels=['O'])
attributions_3
html = ner_explainer.visualize()
attributions_4 = ner_explainer(sample_text, ignored_indexes=[11, 12, 13, 14], ignored_labels=['B-PER'])
html = ner_explainer.visualize()
| 0.541166 | 0.968291 |
# Automated pdl layer generation <a class="tocSkip">
This notebook explains a method to automatically generate the pdl layer based on the bone geometries
# imports
```
import numpy as np
import meshplot as mp
from pathlib import Path
import sys
sys.path.append('../')
import cargen
"""
DIRECTORIES:
"""
main_dir = Path('..')
# input and output paths
i_dir = main_dir / 'models'/ 'tooth'
o_dir = main_dir / 'output'
# Remove all files inside output directory if it exists, otherwise create it
if o_dir.is_dir():
for file in o_dir.iterdir():
if file.is_file():
file.unlink()
else:
o_dir.mkdir(exist_ok=False)
"""
VALUES:
i_dim, o_dim = input and output dimension ( "mm" = millimeters, "m" = meters )
i_format = the format of the input surface mesh ( ".obj" , ".stl")
o_format = format you want the files to be save at ( ".obj" , ".stl" )
+ scroll down to calibrate the pdl generation parameters
"""
# dimensions
i_dim = "mm"
o_dim = "mm"
i_format = ".obj"
o_format = ".obj"
"""
NAMES & PATHS:
"""
# bones
bone_name = 'bone'
tooth_name = 'tooth'
# cartilages
pdl_name = 'pdl'
#bones
clean_bone_name = 'clean_' + bone_name + '_'+ o_dim
clean_tooth_name = 'clean_' + tooth_name + '_'+ o_dim
#cartilages
pdl_cart_name = pdl_name +'_cart_'+ o_dim
pdl_top_name = pdl_name +'_top_' + o_dim
# input paths
bone_path = str((i_dir/ bone_name).with_suffix(i_format))
tooth_path = str((i_dir/ tooth_name).with_suffix(i_format))
# output paths
#bones
clean_bone_path = str((o_dir/ clean_bone_name).with_suffix(o_format))
clean_tooth_path = str((o_dir/ clean_tooth_name).with_suffix(o_format))
#cartilage
pdl_cart_path = str((o_dir/ pdl_cart_name).with_suffix(o_format))
pdl_top_path = str((o_dir/ pdl_top_name).with_suffix(o_format))
```
# implementation
## read and clean up input
```
s1_vertices, s1_faces = cargen.read_and_clean ( bone_path, i_dim )
s2_vertices, s2_faces = cargen.read_and_clean ( tooth_path, i_dim )
frame = mp.plot( s1_vertices, s1_faces, c = cargen.mandible, shading = cargen.sh_false )
frame.add_mesh ( s2_vertices, s2_faces, c = cargen.tooth, shading = cargen.sh_false )
```
## pdl layer
```
# set the parameters
param = cargen.Var()
# change the ones you like
param.gap_distance = 0.3
param.trimming_iteration = 1
param.smoothing_iteration_base = 10
param.smoothing_iteration_extruded_base = 3
param.thickness_factor = 1
# make it
pdl_vertices, pdl_faces, top_vertices, top_faces = cargen.get_pdl_layer(s1_vertices, s1_faces,
s2_vertices, s2_faces,
param)
# reset the parameters to default values
param.reset()
```
## export results
### pdl
```
cargen.save_surface ( pdl_vertices, pdl_faces, o_dim, pdl_cart_path )
cargen.save_surface ( top_vertices, top_faces, o_dim, pdl_top_path )
```
### bones
```
cargen.save_surface ( s1_vertices, s1_faces, o_dim, clean_bone_path )
cargen.save_surface ( s2_vertices, s2_faces, o_dim, clean_tooth_path)
```
# voila!
```
frame = mp.plot( pdl_vertices, pdl_faces, c = cargen.organ, shading = cargen.sh_false )
frame.add_mesh ( s1_vertices, s1_faces, c = cargen.mandible, shading = cargen.sh_false )
frame.add_mesh ( s2_vertices, s2_faces, c = cargen.tooth, shading = cargen.sh_false )
```
|
github_jupyter
|
import numpy as np
import meshplot as mp
from pathlib import Path
import sys
sys.path.append('../')
import cargen
"""
DIRECTORIES:
"""
main_dir = Path('..')
# input and output paths
i_dir = main_dir / 'models'/ 'tooth'
o_dir = main_dir / 'output'
# Remove all files inside output directory if it exists, otherwise create it
if o_dir.is_dir():
for file in o_dir.iterdir():
if file.is_file():
file.unlink()
else:
o_dir.mkdir(exist_ok=False)
"""
VALUES:
i_dim, o_dim = input and output dimension ( "mm" = millimeters, "m" = meters )
i_format = the format of the input surface mesh ( ".obj" , ".stl")
o_format = format you want the files to be save at ( ".obj" , ".stl" )
+ scroll down to calibrate the pdl generation parameters
"""
# dimensions
i_dim = "mm"
o_dim = "mm"
i_format = ".obj"
o_format = ".obj"
"""
NAMES & PATHS:
"""
# bones
bone_name = 'bone'
tooth_name = 'tooth'
# cartilages
pdl_name = 'pdl'
#bones
clean_bone_name = 'clean_' + bone_name + '_'+ o_dim
clean_tooth_name = 'clean_' + tooth_name + '_'+ o_dim
#cartilages
pdl_cart_name = pdl_name +'_cart_'+ o_dim
pdl_top_name = pdl_name +'_top_' + o_dim
# input paths
bone_path = str((i_dir/ bone_name).with_suffix(i_format))
tooth_path = str((i_dir/ tooth_name).with_suffix(i_format))
# output paths
#bones
clean_bone_path = str((o_dir/ clean_bone_name).with_suffix(o_format))
clean_tooth_path = str((o_dir/ clean_tooth_name).with_suffix(o_format))
#cartilage
pdl_cart_path = str((o_dir/ pdl_cart_name).with_suffix(o_format))
pdl_top_path = str((o_dir/ pdl_top_name).with_suffix(o_format))
s1_vertices, s1_faces = cargen.read_and_clean ( bone_path, i_dim )
s2_vertices, s2_faces = cargen.read_and_clean ( tooth_path, i_dim )
frame = mp.plot( s1_vertices, s1_faces, c = cargen.mandible, shading = cargen.sh_false )
frame.add_mesh ( s2_vertices, s2_faces, c = cargen.tooth, shading = cargen.sh_false )
# set the parameters
param = cargen.Var()
# change the ones you like
param.gap_distance = 0.3
param.trimming_iteration = 1
param.smoothing_iteration_base = 10
param.smoothing_iteration_extruded_base = 3
param.thickness_factor = 1
# make it
pdl_vertices, pdl_faces, top_vertices, top_faces = cargen.get_pdl_layer(s1_vertices, s1_faces,
s2_vertices, s2_faces,
param)
# reset the parameters to default values
param.reset()
cargen.save_surface ( pdl_vertices, pdl_faces, o_dim, pdl_cart_path )
cargen.save_surface ( top_vertices, top_faces, o_dim, pdl_top_path )
cargen.save_surface ( s1_vertices, s1_faces, o_dim, clean_bone_path )
cargen.save_surface ( s2_vertices, s2_faces, o_dim, clean_tooth_path)
frame = mp.plot( pdl_vertices, pdl_faces, c = cargen.organ, shading = cargen.sh_false )
frame.add_mesh ( s1_vertices, s1_faces, c = cargen.mandible, shading = cargen.sh_false )
frame.add_mesh ( s2_vertices, s2_faces, c = cargen.tooth, shading = cargen.sh_false )
| 0.246806 | 0.64232 |
# Implementation of the language models
```
from fastai.gen_doc.nbdoc import *
from fastai.text.models import *
from fastai import *
```
This module fully implements the [AWD-LSTM](https://arxiv.org/pdf/1708.02182.pdf) from Stephen Merity et al. The main idea of the article is to use a [RNN](http://www.pnas.org/content/79/8/2554) with dropout everywhere, but in an intelligent way. There is a difference with the usual dropout, which is why you’ll see a [`RNNDropout`](/text.models.html#RNNDropout) module: we zero things, as is usual in dropout, but we always zero the same thing according to the sequence dimension (which is the first dimension in pytorch). This ensures consistency when updating the hidden state through the whole sentences/articles.
This being given, there are five different dropouts in the AWD-LSTM:
- the first one, embedding dropout, is applied when we look the ids of our tokens inside the embedding matrix (to transform them from numbers to a vector of float). We zero some lines of it, so random ids are sent to a vector of zeros instead of being sent to their embedding vector.
- the second one, input dropout, is applied to the result of the embedding with dropout. We forget random pieces of the embedding matrix (but as stated in the last paragraph, the same ones in the sequence dimension).
- the third one is the weight dropout. It’s the trickiest to implement as we randomly replace by 0s some weights of the hidden-to-hidden matrix inside the RNN: this needs to be done in a way that ensure the gradients are still computed and the initial weights still updated.
- the fourth one is the hidden dropout. It’s applied to the output of one of the layers of the RNN before it’s used as input of the next layer (again same coordinates are zeroed in the sequence dimension). This one isn’t applied to the last output, but rather…
- the fifth one is the output dropout, it’s applied to the last output of the model (and like the others, it’s applied the same way through the first dimension).
## Basic functions to get a model
```
show_doc(get_language_model, doc_string=False)
```
Creates an AWD-LSTM with a first embedding of `vocab_sz` by `emb_sz`, a hidden size of `n_hid`, RNNs with `n_layers` that can be bidirectional if `bidir` is True. The last RNN as an output size of `emb_sz` so that we can use the same decoder as the encoder if `tie_weights` is True. The decoder is a `Linear` layer with or without `bias`. If `qrnn` is set to True, we use [QRNN cells] instead of LSTMS. `pad_token` is the token used for padding.
`embed_p` is used for the embedding dropout, `input_p` is used for the input dropout, `weight_p` is used for the weight dropout, `hidden_p` is used for the hidden dropout and `output_p` is used for the output dropout.
Note that the model returns a list of three things, the actual output being the first, the two others being the intermediate hidden states before and after dropout (used by the [`RNNTrainer`](/callbacks.rnn.html#RNNTrainer)). Most loss functions expect one output, so you should use a Callback to remove the other two if you're not using [`RNNTrainer`](/callbacks.rnn.html#RNNTrainer).
```
show_doc(get_rnn_classifier, doc_string=False)
```
Creates a RNN classifier with a encoder taken from an AWD-LSTM with arguments `vocab_sz`, `emb_sz`, `n_hid`, `n_layers`, `bias`, `bidir`, `qrnn`, `pad_token` and the dropouts parameters. This encoder is fed the sequence by successive bits of size `bptt` and we only keep the last `max_seq` outputs for the pooling layers.
The decoder use a concatenation of the last outputs, a `MaxPooling` of all the ouputs and an `AveragePooling` of all the outputs. It then uses a list of `BatchNorm`, `Dropout`, `Linear`, `ReLU` blocks (with no `ReLU` in the last one), using a first layer size of `3*emb_sz` then follwoing the numbers in `n_layers` to stop at `n_class`. The dropouts probabilities are read in `drops`.
Note that the model returns a list of three things, the actual output being the first, the two others being the intermediate hidden states before and after dropout (used by the [`RNNTrainer`](/callbacks.rnn.html#RNNTrainer)). Most loss functions expect one output, so you should use a Callback to remove the other two if you're not using [`RNNTrainer`](/callbacks.rnn.html#RNNTrainer).
## Basic NLP modules
On top of the pytorch or the fastai [`layers`](/layers.html#layers), the language models use some custom layers specific to NLP.
```
show_doc(EmbeddingDropout, doc_string=False, title_level=3)
```
Applies a dropout with probability `embed_p` to an embedding layer `emb` in training mode. Each row of the embedding matrix has a probability `embed_p` of being replaced by zeros while the others are rescaled accordingly.
```
enc = nn.Embedding(100, 7, padding_idx=1)
enc_dp = EmbeddingDropout(enc, 0.5)
tst_input = torch.randint(0,100,(8,))
enc_dp(tst_input)
show_doc(RNNDropout, doc_string=False, title_level=3)
```
Applies a dropout with probability `p` consistently over the first dimension in training mode.
```
dp = RNNDropout(0.3)
tst_input = torch.randn(3,3,7)
tst_input, dp(tst_input)
show_doc(WeightDropout, doc_string=False, title_level=3)
```
Applies dropout of probability `weight_p` to the layers in `layer_names` of `module` in training mode. A copy of those weights is kept so that the dropout mask can change at every batch.
```
module = nn.LSTM(5, 2)
dp_module = WeightDropout(module, 0.4)
getattr(dp_module.module, 'weight_hh_l0')
```
It's at the beginning of a forward pass that the dropout is applied to the weights.
```
tst_input = torch.randn(4,20,5)
h = (torch.zeros(1,20,2), torch.zeros(1,20,2))
x,h = dp_module(tst_input,h)
getattr(dp_module.module, 'weight_hh_l0')
show_doc(SequentialRNN, doc_string=False, title_level=3)
```
Create a `Sequentiall` module with `args` that has a `reset` function.
```
show_doc(SequentialRNN.reset)
```
Call the `reset` function of [`self.children`](/torch_core.html#children) (if they have one).
```
show_doc(dropout_mask, doc_string=False)
```
Create a dropout mask of size `sz`, the same type as `x` and probability `p`.
```
tst_input = torch.randn(3,3,7)
dropout_mask(tst_input, (3,7), 0.3)
```
Such a mask is then expanded in the sequence length dimension and multiplied by the input to do an [`RNNDropout`](/text.models.html#RNNDropout).
## Language model modules
```
show_doc(RNNCore, doc_string=False, title_level=3)
```
Create an AWD-LSTM encoder with an embedding layer of `vocab_sz` by `emb_sz`, a hidden size of `n_hid`, `n_layers` layers. `pad_token` is passed to the `Embedding`, if `bidir` is True, the model is bidirectional. If `qrnn` is True, we use QRNN cells instead of LSTMs. Dropouts are `embed_p`, `input_p`, `weight_p` and `hidden_p`.
```
show_doc(RNNCore.reset)
show_doc(LinearDecoder, doc_string=False, title_level=3)
```
Create a the decoder to go on top of an [`RNNCore`](/text.models.html#RNNCore) encoder and create a language model. `n_hid` is the dimension of the last hidden state of the encoder, `n_out` the size of the output. Dropout of `output_p` is applied. If a `tie_encoder` is passed, it will be used for the weights of the linear layer, that will have `bias` or not.
## Classifier modules
```
show_doc(MultiBatchRNNCore, doc_string=False, title_level=3)
```
Wrap an [`RNNCore`](/text.models.html#RNNCore) to make it process full sentences: text is passed by chunks of sequence length `bptt` and only the last `max_seq` outputs are kept for the next layer. `args` and `kwargs` are passed to the [`RNNCore`](/text.models.html#RNNCore).
```
show_doc(MultiBatchRNNCore.concat)
show_doc(PoolingLinearClassifier, doc_string=False, title_level=3)
```
Create a linear classifier that sits on an [`RNNCore`](/text.models.html#RNNCore) encoder. The last output, `MaxPooling` of all the outputs and `AvgPooling` of all the outputs are concatenated, then blocks of [`bn_drop_lin`](/layers.html#bn_drop_lin) are stacked, according to the values in [`layers`](/layers.html#layers) and `drops`.
```
show_doc(PoolingLinearClassifier.pool, doc_string=False)
```
Pool `x` (of batch size `bs`) along the batch dimension. `is_max` decides if we do an `AvgPooling` or a `MaxPooling`.
## Undocumented Methods - Methods moved below this line will intentionally be hidden
```
show_doc(WeightDropout.forward)
show_doc(RNNCore.forward)
show_doc(EmbeddingDropout.forward)
show_doc(RNNDropout.forward)
```
## New Methods - Please document or move to the undocumented section
|
github_jupyter
|
from fastai.gen_doc.nbdoc import *
from fastai.text.models import *
from fastai import *
show_doc(get_language_model, doc_string=False)
show_doc(get_rnn_classifier, doc_string=False)
show_doc(EmbeddingDropout, doc_string=False, title_level=3)
enc = nn.Embedding(100, 7, padding_idx=1)
enc_dp = EmbeddingDropout(enc, 0.5)
tst_input = torch.randint(0,100,(8,))
enc_dp(tst_input)
show_doc(RNNDropout, doc_string=False, title_level=3)
dp = RNNDropout(0.3)
tst_input = torch.randn(3,3,7)
tst_input, dp(tst_input)
show_doc(WeightDropout, doc_string=False, title_level=3)
module = nn.LSTM(5, 2)
dp_module = WeightDropout(module, 0.4)
getattr(dp_module.module, 'weight_hh_l0')
tst_input = torch.randn(4,20,5)
h = (torch.zeros(1,20,2), torch.zeros(1,20,2))
x,h = dp_module(tst_input,h)
getattr(dp_module.module, 'weight_hh_l0')
show_doc(SequentialRNN, doc_string=False, title_level=3)
show_doc(SequentialRNN.reset)
show_doc(dropout_mask, doc_string=False)
tst_input = torch.randn(3,3,7)
dropout_mask(tst_input, (3,7), 0.3)
show_doc(RNNCore, doc_string=False, title_level=3)
show_doc(RNNCore.reset)
show_doc(LinearDecoder, doc_string=False, title_level=3)
show_doc(MultiBatchRNNCore, doc_string=False, title_level=3)
show_doc(MultiBatchRNNCore.concat)
show_doc(PoolingLinearClassifier, doc_string=False, title_level=3)
show_doc(PoolingLinearClassifier.pool, doc_string=False)
show_doc(WeightDropout.forward)
show_doc(RNNCore.forward)
show_doc(EmbeddingDropout.forward)
show_doc(RNNDropout.forward)
| 0.754644 | 0.98797 |
# Evaluating Your Forecast
So far you have prepared your data, and generated your first Forecast. Now is the time to pull down the predictions from this Predictor, and compare them to the actual observed values. This will let us know the impact of accuracy based on the Forecast.
You can extend the approaches here to compare multiple models or predictors and to determine the impact of improved accuracy on your use case.
Overview:
* Setup
* Obtaining a Prediction
* Plotting the Actual Results
* Plotting the Prediction
* Comparing the Prediction to Actual Results
## Setup
Import the standard Python Libraries that are used in this lesson.
```
import json
import time
import dateutil.parser
import boto3
import pandas as pd
```
The line below will retrieve your shared variables from the earlier notebooks.
```
%store -r
```
Once again connect to the Forecast APIs via the SDK.
```
session = boto3.Session(region_name=region)
forecast = session.client(service_name='forecast')
forecastquery = session.client(service_name='forecastquery')
```
## Obtaining a Prediction:
Now that your predictor is active we will query it to get a prediction that will be plotted later.
```
forecastResponse = forecastquery.query_forecast(
ForecastArn=forecast_arn_deep_ar,
Filters={"item_id":"client_12"}
)
```
## Plotting the Actual Results
In the first notebook we created a file of observed values, we are now going to select a given date and customer from that dataframe and are going to plot the actual usage data for that customer.
```
actual_df = pd.read_csv("data/item-demand-time-validation.csv", names=['timestamp','value','item'])
actual_df.head()
```
Next we need to reduce the data to just the day we wish to plot, which is the First of November 2014.
```
actual_df = actual_df[(actual_df['timestamp'] >= '2014-11-01') & (actual_df['timestamp'] < '2014-11-02')]
```
Lastly, only grab the items for client_12
```
actual_df = actual_df[(actual_df['item'] == 'client_12')]
actual_df.head()
actual_df.plot()
```
## Plotting the Prediction:
Next we need to convert the JSON response from the Predictor to a dataframe that we can plot.
```
# Generate DF
prediction_df_p10 = pd.DataFrame.from_dict(forecastResponse['Forecast']['Predictions']['p10'])
prediction_df_p10.head()
# Plot
prediction_df_p10.plot()
```
The above merely did the p10 values, now do the same for p50 and p90.
```
prediction_df_p50 = pd.DataFrame.from_dict(forecastResponse['Forecast']['Predictions']['p50'])
prediction_df_p90 = pd.DataFrame.from_dict(forecastResponse['Forecast']['Predictions']['p90'])
```
## Comparing the Prediction to Actual Results
After obtaining the dataframes the next task is to plot them together to determine the best fit.
```
# We start by creating a dataframe to house our content, here source will be which dataframe it came from
results_df = pd.DataFrame(columns=['timestamp', 'value', 'source'])
```
Import the observed values into the dataframe:
```
for index, row in actual_df.iterrows():
clean_timestamp = dateutil.parser.parse(row['timestamp'])
results_df = results_df.append({'timestamp' : clean_timestamp , 'value' : row['value'], 'source': 'actual'} , ignore_index=True)
# To show the new dataframe
results_df.head()
# Now add the P10, P50, and P90 Values
for index, row in prediction_df_p10.iterrows():
clean_timestamp = dateutil.parser.parse(row['Timestamp'])
results_df = results_df.append({'timestamp' : clean_timestamp , 'value' : row['Value'], 'source': 'p10'} , ignore_index=True)
for index, row in prediction_df_p50.iterrows():
clean_timestamp = dateutil.parser.parse(row['Timestamp'])
results_df = results_df.append({'timestamp' : clean_timestamp , 'value' : row['Value'], 'source': 'p50'} , ignore_index=True)
for index, row in prediction_df_p90.iterrows():
clean_timestamp = dateutil.parser.parse(row['Timestamp'])
results_df = results_df.append({'timestamp' : clean_timestamp , 'value' : row['Value'], 'source': 'p90'} , ignore_index=True)
results_df
pivot_df = results_df.pivot(columns='source', values='value', index="timestamp")
pivot_df
pivot_df.plot()
```
Once you are done exploring this Forecast you can cleanup all the work that was done by executing the cells inside `Cleanup.ipynb` within this folder.
|
github_jupyter
|
import json
import time
import dateutil.parser
import boto3
import pandas as pd
%store -r
session = boto3.Session(region_name=region)
forecast = session.client(service_name='forecast')
forecastquery = session.client(service_name='forecastquery')
forecastResponse = forecastquery.query_forecast(
ForecastArn=forecast_arn_deep_ar,
Filters={"item_id":"client_12"}
)
actual_df = pd.read_csv("data/item-demand-time-validation.csv", names=['timestamp','value','item'])
actual_df.head()
actual_df = actual_df[(actual_df['timestamp'] >= '2014-11-01') & (actual_df['timestamp'] < '2014-11-02')]
actual_df = actual_df[(actual_df['item'] == 'client_12')]
actual_df.head()
actual_df.plot()
# Generate DF
prediction_df_p10 = pd.DataFrame.from_dict(forecastResponse['Forecast']['Predictions']['p10'])
prediction_df_p10.head()
# Plot
prediction_df_p10.plot()
prediction_df_p50 = pd.DataFrame.from_dict(forecastResponse['Forecast']['Predictions']['p50'])
prediction_df_p90 = pd.DataFrame.from_dict(forecastResponse['Forecast']['Predictions']['p90'])
# We start by creating a dataframe to house our content, here source will be which dataframe it came from
results_df = pd.DataFrame(columns=['timestamp', 'value', 'source'])
for index, row in actual_df.iterrows():
clean_timestamp = dateutil.parser.parse(row['timestamp'])
results_df = results_df.append({'timestamp' : clean_timestamp , 'value' : row['value'], 'source': 'actual'} , ignore_index=True)
# To show the new dataframe
results_df.head()
# Now add the P10, P50, and P90 Values
for index, row in prediction_df_p10.iterrows():
clean_timestamp = dateutil.parser.parse(row['Timestamp'])
results_df = results_df.append({'timestamp' : clean_timestamp , 'value' : row['Value'], 'source': 'p10'} , ignore_index=True)
for index, row in prediction_df_p50.iterrows():
clean_timestamp = dateutil.parser.parse(row['Timestamp'])
results_df = results_df.append({'timestamp' : clean_timestamp , 'value' : row['Value'], 'source': 'p50'} , ignore_index=True)
for index, row in prediction_df_p90.iterrows():
clean_timestamp = dateutil.parser.parse(row['Timestamp'])
results_df = results_df.append({'timestamp' : clean_timestamp , 'value' : row['Value'], 'source': 'p90'} , ignore_index=True)
results_df
pivot_df = results_df.pivot(columns='source', values='value', index="timestamp")
pivot_df
pivot_df.plot()
| 0.440469 | 0.963541 |
# Hydrocarbon volumes calculator
The expression for _oil_ volumes in place:
$$ V = c \times GRV \times \phi \times NTG \times S_\mathrm{HC} \times \frac{1}{FVF} $$
where:
- c: conversion factor: `7758` from acre-ft to bbl (oil) or `43560` from acre-ft to ft3 (gas)
- V: hydrocarbon volumes
- GRV: Gross rock volume
- $\phi$: Porosity
- NTG: Net-to-gross
- $S_HC$: Hydrocarbon saturation (oil or gas)
- FVF: Formation volume factor
```
# standard imports
import pandas as pd
pd.set_option("display.precision", 3)
import numpy as np
%matplotlib inline
import matplotlib.pyplot as plt
from matplotlib.ticker import StrMethodFormatter
def calc_hciip(GRV, phi=1, NTG=1, Sw=0, FVF=1, fluid='oil'):
"""Calculate Hydrocarbon initially in-place for a given prospect
Args:
GRV [float]: gross rock volume [acre-feet]
phi [float]: porosity [fraction]
NTG [float]: net-to-gross [fraction]
Sw [float]: water saturation [fraction]
FVF [float]: formation volume factor (Bo for oil or Bg for gas) [RES bbl/STB or RES ft3/SCF]
"""
try:
if fluid.lower() not in {'oil', 'gas'}:
raise ValueError("`fluid` arg must be of `{'gas', 'oil'}`")
except AttributeError:
raise AttributeError("`fluid` arg must be of type `str`")
if fluid == 'oil':
constant = 7758 # conversion factor from acre-ft to bbl
elif fluid == 'gas':
constant = 43560 # conversion factor from acre-ft to ft3
return (constant * GRV * phi * NTG * (1 - Sw)) / FVF
```
## Monte Carlo simulation of volumes
The two components of the Monte Carlo simulation are:
1. The equation to evaluate
2. The random variables for the input
Here the equation is `calc_hciip` and the variables are `{GRV, phi, NTG, Sw, FVF}`.
[Normal distribution applet](https://homepage.divms.uiowa.edu/~mbognar/applets/normal.html) and [Lognormal](https://homepage.divms.uiowa.edu/~mbognar/applets/lognormal.html) to eyeball parameters.
First, define the distributions parameters, the input file that is grabbed with `pd.read_clipboard` must be a CSV with the following format and exact column names:
```
Reservoir,grv_avg,grv_std,phi_avg,phi_std,ntg_avg,ntg_std,sw_avg,sw_std,fvf_avg,fvf_std
sand_1,5000,500,0.3,0.03,0.8,0.08,0.3,0.03,1.04,0.104
sand_2,7000,700,0.2,0.02,0.7,0.07,0.25,0.025,1.05,0.105
```
Currently, normal distribution are applied to all input parameters, if these are changed, the corresponding inputs would need to be changed and the code would need some refactoring.
```
df_raw = pd.read_clipboard(sep=',')
df_raw.index = df_raw.Reservoir
df_raw = df_raw.drop(columns='Reservoir')
df_raw
```
Then sample from a normal distribution (to begin with) and calculate volumes:
## Monte Carlo Experiment
```
def plot_pdf(df, sand):
"""plot pdt of stoiip for given sand and save fig"""
fig, ax = plt.subplots(figsize=(16,8))
n, *_ = ax.hist(df.hciip, bins=30, alpha=0.4, color='green')
n_len = (len(str(int(n.max()))))
y_max = round(n.max(), -(n_len-1))
title = f'Probability density function of STOIIP for {sand}'
ax.set_title(title, fontsize=26)
minvol, maxvol = df.hciip.describe().loc[['min', 'max']]
for name, prob in {'P90': p90, 'P50': p50, 'P10': p10}.items():
formatted_prob = round(prob, -3)/1e6
prob_approx = f'\n{formatted_prob:.1f}\nmmbbl'
plt.axvline(prob, c='b', linewidth=1.5)
ax.text(prob, y_max - y_max*.2, name + prob_approx, fontdict={'fontsize': 22})
ax.set_xlim(minvol, maxvol)
ax.tick_params(axis='both', which='major', labelsize=20)
plt.gca().xaxis.set_major_formatter(StrMethodFormatter('{x:,.0f}'))
plt.gca().yaxis.set_major_formatter(StrMethodFormatter('{x:,.0f}'))
ax.set_xlabel('STOIIP [bbl]', fontsize=20)
ax.set_ylabel('Density of probability [1 / bbl]', fontsize=20)
plt.savefig(f'./stoiip_pdf_{sand}', dpi=400)
return None
for sand in df_raw.index:
grv_avg, grv_std = df_raw.loc[sand, 'grv_avg'], df_raw.loc[sand, 'grv_std']
phi_avg, phi_std = df_raw.loc[sand, 'phi_avg'], df_raw.loc[sand, 'phi_std']
ntg_avg, ntg_std = df_raw.loc[sand, 'ntg_avg'], df_raw.loc[sand, 'ntg_std']
sw_avg, sw_std = df_raw.loc[sand, 'sw_avg'], df_raw.loc[sand, 'sw_std']
fvf_avg, fvf_std = df_raw.loc[sand, 'fvf_avg'], df_raw.loc[sand, 'fvf_std']
# sample count for MC experiment
num_samples = 5000
# Choose random inputs for each variable
### NOTE THAT OTHER DISTIBUTIONS CAN BE SUBSTITUTED, SEE FOR EXAMPLE: https://docs.scipy.org/doc/scipy/reference/stats.html#continuous-distributions
grv = np.random.normal(grv_avg, grv_std, num_samples)
phi = np.random.normal(phi_avg, phi_std, num_samples)
ntg = np.random.normal(ntg_avg, ntg_std, num_samples)
sw = np.random.normal(sw_avg, sw_std, num_samples)
fvf = np.random.normal(fvf_avg, fvf_std, num_samples)
# set negative values to 0
for arr in [grv, phi, ntg, sw, fvf]:
arr[arr < 0] = 0
# set values over 1 to 1
for arr in [phi, ntg, sw]:
arr[arr > 1] = 1
# Build the dataframe based on the inputs and num_samples
df = pd.DataFrame(index=range(num_samples), data={'grv': grv,'phi': phi,'ntg': ntg,'sw': sw,'fvf': fvf,})
# calculate HCIIPs based on inputs
df['hciip'] = calc_hciip(df.grv, df.phi, df.ntg, df.sw, df.fvf)
# reverse p90, p50, p10 to match oil-field convention
p90, p50, p10 = df.describe(percentiles=[.1,.5,.9]).loc[['10%', '50%', '90%'], 'hciip']
df_raw.loc[sand, 'P90'] = p90 / 1e6
df_raw.loc[sand, 'P50'] = p50 / 1e6
df_raw.loc[sand, 'P10'] = p10 / 1e6
plot_pdf(df, sand)
field = input('what is the field name?')
col_units = ['acre-feet',
'acre-feet',
'%',
'%',
'%',
'%',
'%',
'%',
'bbl/STB',
'bbl/STB',
'MMSTB',
'MMSTB',
'MMSTB',
]
cols_multiindex = list(zip(df_raw.columns, col_units))
df_raw.columns = pd.MultiIndex.from_tuples(cols_multiindex)
df_raw.loc['Totals'] = df_raw.loc[:,['grv_avg', 'P90', 'P50', 'P10']].sum(axis=0)
df_raw
df_raw.to_excel(f'./{field}_quicklook_volumetrics.xlsx')
```
|
github_jupyter
|
# standard imports
import pandas as pd
pd.set_option("display.precision", 3)
import numpy as np
%matplotlib inline
import matplotlib.pyplot as plt
from matplotlib.ticker import StrMethodFormatter
def calc_hciip(GRV, phi=1, NTG=1, Sw=0, FVF=1, fluid='oil'):
"""Calculate Hydrocarbon initially in-place for a given prospect
Args:
GRV [float]: gross rock volume [acre-feet]
phi [float]: porosity [fraction]
NTG [float]: net-to-gross [fraction]
Sw [float]: water saturation [fraction]
FVF [float]: formation volume factor (Bo for oil or Bg for gas) [RES bbl/STB or RES ft3/SCF]
"""
try:
if fluid.lower() not in {'oil', 'gas'}:
raise ValueError("`fluid` arg must be of `{'gas', 'oil'}`")
except AttributeError:
raise AttributeError("`fluid` arg must be of type `str`")
if fluid == 'oil':
constant = 7758 # conversion factor from acre-ft to bbl
elif fluid == 'gas':
constant = 43560 # conversion factor from acre-ft to ft3
return (constant * GRV * phi * NTG * (1 - Sw)) / FVF
Reservoir,grv_avg,grv_std,phi_avg,phi_std,ntg_avg,ntg_std,sw_avg,sw_std,fvf_avg,fvf_std
sand_1,5000,500,0.3,0.03,0.8,0.08,0.3,0.03,1.04,0.104
sand_2,7000,700,0.2,0.02,0.7,0.07,0.25,0.025,1.05,0.105
df_raw = pd.read_clipboard(sep=',')
df_raw.index = df_raw.Reservoir
df_raw = df_raw.drop(columns='Reservoir')
df_raw
def plot_pdf(df, sand):
"""plot pdt of stoiip for given sand and save fig"""
fig, ax = plt.subplots(figsize=(16,8))
n, *_ = ax.hist(df.hciip, bins=30, alpha=0.4, color='green')
n_len = (len(str(int(n.max()))))
y_max = round(n.max(), -(n_len-1))
title = f'Probability density function of STOIIP for {sand}'
ax.set_title(title, fontsize=26)
minvol, maxvol = df.hciip.describe().loc[['min', 'max']]
for name, prob in {'P90': p90, 'P50': p50, 'P10': p10}.items():
formatted_prob = round(prob, -3)/1e6
prob_approx = f'\n{formatted_prob:.1f}\nmmbbl'
plt.axvline(prob, c='b', linewidth=1.5)
ax.text(prob, y_max - y_max*.2, name + prob_approx, fontdict={'fontsize': 22})
ax.set_xlim(minvol, maxvol)
ax.tick_params(axis='both', which='major', labelsize=20)
plt.gca().xaxis.set_major_formatter(StrMethodFormatter('{x:,.0f}'))
plt.gca().yaxis.set_major_formatter(StrMethodFormatter('{x:,.0f}'))
ax.set_xlabel('STOIIP [bbl]', fontsize=20)
ax.set_ylabel('Density of probability [1 / bbl]', fontsize=20)
plt.savefig(f'./stoiip_pdf_{sand}', dpi=400)
return None
for sand in df_raw.index:
grv_avg, grv_std = df_raw.loc[sand, 'grv_avg'], df_raw.loc[sand, 'grv_std']
phi_avg, phi_std = df_raw.loc[sand, 'phi_avg'], df_raw.loc[sand, 'phi_std']
ntg_avg, ntg_std = df_raw.loc[sand, 'ntg_avg'], df_raw.loc[sand, 'ntg_std']
sw_avg, sw_std = df_raw.loc[sand, 'sw_avg'], df_raw.loc[sand, 'sw_std']
fvf_avg, fvf_std = df_raw.loc[sand, 'fvf_avg'], df_raw.loc[sand, 'fvf_std']
# sample count for MC experiment
num_samples = 5000
# Choose random inputs for each variable
### NOTE THAT OTHER DISTIBUTIONS CAN BE SUBSTITUTED, SEE FOR EXAMPLE: https://docs.scipy.org/doc/scipy/reference/stats.html#continuous-distributions
grv = np.random.normal(grv_avg, grv_std, num_samples)
phi = np.random.normal(phi_avg, phi_std, num_samples)
ntg = np.random.normal(ntg_avg, ntg_std, num_samples)
sw = np.random.normal(sw_avg, sw_std, num_samples)
fvf = np.random.normal(fvf_avg, fvf_std, num_samples)
# set negative values to 0
for arr in [grv, phi, ntg, sw, fvf]:
arr[arr < 0] = 0
# set values over 1 to 1
for arr in [phi, ntg, sw]:
arr[arr > 1] = 1
# Build the dataframe based on the inputs and num_samples
df = pd.DataFrame(index=range(num_samples), data={'grv': grv,'phi': phi,'ntg': ntg,'sw': sw,'fvf': fvf,})
# calculate HCIIPs based on inputs
df['hciip'] = calc_hciip(df.grv, df.phi, df.ntg, df.sw, df.fvf)
# reverse p90, p50, p10 to match oil-field convention
p90, p50, p10 = df.describe(percentiles=[.1,.5,.9]).loc[['10%', '50%', '90%'], 'hciip']
df_raw.loc[sand, 'P90'] = p90 / 1e6
df_raw.loc[sand, 'P50'] = p50 / 1e6
df_raw.loc[sand, 'P10'] = p10 / 1e6
plot_pdf(df, sand)
field = input('what is the field name?')
col_units = ['acre-feet',
'acre-feet',
'%',
'%',
'%',
'%',
'%',
'%',
'bbl/STB',
'bbl/STB',
'MMSTB',
'MMSTB',
'MMSTB',
]
cols_multiindex = list(zip(df_raw.columns, col_units))
df_raw.columns = pd.MultiIndex.from_tuples(cols_multiindex)
df_raw.loc['Totals'] = df_raw.loc[:,['grv_avg', 'P90', 'P50', 'P10']].sum(axis=0)
df_raw
df_raw.to_excel(f'./{field}_quicklook_volumetrics.xlsx')
| 0.68215 | 0.939304 |
# Keras Callbacks and Functional API
```
from keras.datasets import cifar10
from keras.models import Sequential
from keras.layers.core import Dense, Activation, Dropout
from keras.optimizers import SGD, RMSprop
from keras.callbacks import EarlyStopping, TensorBoard, ModelCheckpoint
import numpy as np
%matplotlib inline
import matplotlib.pyplot as plt
(X_train_t, y_train), (X_test_t, y_test) = cifar10.load_data()
X_train_t = X_train_t.astype('float32') / 255.
X_test_t = X_test_t.astype('float32') / 255.
X_train = X_train_t.reshape(len(X_train_t), 32*32*3)
X_test = X_test_t.reshape(len(X_test_t), 32*32*3)
print("Training set:")
print("Tensor images shape:\t", X_train_t.shape)
print("Flat images shape:\t", X_train.shape)
print("Labels shape:\t\t", y_train.shape)
plt.figure(figsize=(15, 4))
for i in range(0, 8):
plt.subplot(1, 8, i+1)
plt.imshow(X_train[i].reshape(32, 32, 3))
plt.title(y_train[i])
```
## Callbacks on a simple model
```
outpath='/tmp/tensorflow_logs/cifar/'
early_stopper = EarlyStopping(monitor='val_acc', patience=10)
tensorboard = TensorBoard(outpath, histogram_freq=1)
checkpointer = ModelCheckpoint(outpath+'weights_epoch_{epoch:02d}_val_acc_{val_acc:.2f}.hdf5',
monitor='val_acc')
model = Sequential()
model.add(Dense(1024, activation='relu',
input_dim=3072))
model.add(Dense(512, activation='relu'))
model.add(Dense(10, activation='softmax'))
model.compile(loss='sparse_categorical_crossentropy',
optimizer='rmsprop',
metrics=['accuracy'])
model.fit(X_train, y_train,
batch_size=128,
epochs=5,
verbose=1,
validation_split=0.1,
callbacks=[early_stopper,
tensorboard,
checkpointer])
import os
sorted(os.listdir(outpath))
```
Now check the tensorboard.
1. Open a terminal and run:
```
tensorboard --logdir=/tmp/tensorflow_logs/cifar/
```
2. Open another terminal and run [localtunnel](https://localtunnel.github.io/www/) on port 6006:
```
lt --port 6006
```
3. Go to the url provided
You should see something like this:

> TIP: if you get an error `lt: command not found` install localtunnel as:
```
sudo npm install -g localtunnel
```
## Exercise 1: Keras functional API
We'e built a model using the `Sequential API` from Keras. Keras also offers a [functional API](https://keras.io/getting-started/functional-api-guide/). This API is the way to go for defining complex models, such as multi-output models, directed acyclic graphs, or models with shared layers.
Can you rewrite the model above using the functional API?
```
from keras.layers import Input
from keras.models import Model
```
model.add(Dense(1024, activation='relu',
input_dim=3072))
model.add(Dense(512, activation='relu'))
model.add(Dense(10, activation='softmax'))
model.compile(loss='sparse_categorical_crossentropy',
optimizer='rmsprop',
metrics=['accuracy'])
```
inputs = Input(shape=(3072,))
l1 = Dense(1024, activation='relu', input_dim=3072)(inputs)
l2 = Dense(512, activation='relu')(l1)
predictions = Dense(10, activation='softmax')(l2)
model = Model(inputs=inputs, outputs=predictions)
model.compile(loss='sparse_categorical_crossentropy',
optimizer='rmsprop',
metrics=['accuracy'])
model.fit(X_train, y_train,
batch_size=128,
epochs=10,
verbose=1,
validation_split=0.1)
# Final test evaluation
score = model.evaluate(X_test, y_test, verbose=0)
print('Test loss:', score[0])
print('Test accuracy:', score[1])
```
inputs = Input(shape=(3072,))
inputs2 = Input(shape(784,))
l1 = Dense(1024, activation='relu', input_dim=3072)(inputs)
l2 = Dense(512, activation='relu')(inputs2)
predictions = Dense(10, activation='softmax')(l2)
merge()
model = Model(inputs=inputs, outputs=predictions)
## Exercise 2: Convolutional Model with Functional API
The above model is a very simple fully connected deep neural network. As we have seen, Convolutional Neural Networks are much more powerful when dealing with images. The original data has shape:
(N_images, Height, Width, Channels)
Can you write a convolutional model using the functional API?
```
from keras.layers.core import Dense, Dropout, Activation
from keras.layers import Conv2D, MaxPool2D, AveragePooling2D, Flatten
outpath = '/tmp/tensorflow_logs/cifar_conv'
tensorboard = TensorBoard(outpath)
#this model doesn't work
inputs = Input(shape=(32,32,3))
l1 = Conv2D(32, (3,3), activation='relu')(inputs)
l2 = Conv2D(48, (3,3), activation='relu')(l1)
l3 = MaxPool2D()(l2)
l4 = Flatten()(l3)
outputs = Dense(10, activation='softmax')(l4)
model = Model(inputs=inputs, outputs=outputs)
#model above doesn't compile
model.compile(loss='sparse_categorical_crossentropy',
optimizer='rmsprop',
metrics=['accuracy']
)
model.fit(X_train, y_train,
batch_size=128,
epochs=5,
verbose=1,
validation_split=0.1,
callbacks=[tensorboard])
inputs = Input(shape=(32,32,3))
l1 = Conv2D(32, (3,3), activation='relu')(inputs)
l2 = Flatten()(inputs)
outputs = Dense(10, activation='softmax')(l2)
model = Model(inputs=inputs, outputs=outputs)
model.compile('adam', 'sparse_categorical_crossentropy', metrics=['accuracy'])
model.fit(X_train_t, y_train, epochs=5, callbacks=[tensorboard])
score = model.evaluate(X_test, y_test, verbose=0)
print('\n\n')
print('Test loss:', score[0])
print('Test accuracy:', score[1])
```
## Exrcise 3: Discuss with the person next to you
1. What are the pros/cons of the sequential API?
- What are the pros/cons of the functional API?
- What are the key differences between a Fully connected and a Convolutional neural network?
- What is a dropout layer? How does it work? Why does it help?
|
github_jupyter
|
from keras.datasets import cifar10
from keras.models import Sequential
from keras.layers.core import Dense, Activation, Dropout
from keras.optimizers import SGD, RMSprop
from keras.callbacks import EarlyStopping, TensorBoard, ModelCheckpoint
import numpy as np
%matplotlib inline
import matplotlib.pyplot as plt
(X_train_t, y_train), (X_test_t, y_test) = cifar10.load_data()
X_train_t = X_train_t.astype('float32') / 255.
X_test_t = X_test_t.astype('float32') / 255.
X_train = X_train_t.reshape(len(X_train_t), 32*32*3)
X_test = X_test_t.reshape(len(X_test_t), 32*32*3)
print("Training set:")
print("Tensor images shape:\t", X_train_t.shape)
print("Flat images shape:\t", X_train.shape)
print("Labels shape:\t\t", y_train.shape)
plt.figure(figsize=(15, 4))
for i in range(0, 8):
plt.subplot(1, 8, i+1)
plt.imshow(X_train[i].reshape(32, 32, 3))
plt.title(y_train[i])
outpath='/tmp/tensorflow_logs/cifar/'
early_stopper = EarlyStopping(monitor='val_acc', patience=10)
tensorboard = TensorBoard(outpath, histogram_freq=1)
checkpointer = ModelCheckpoint(outpath+'weights_epoch_{epoch:02d}_val_acc_{val_acc:.2f}.hdf5',
monitor='val_acc')
model = Sequential()
model.add(Dense(1024, activation='relu',
input_dim=3072))
model.add(Dense(512, activation='relu'))
model.add(Dense(10, activation='softmax'))
model.compile(loss='sparse_categorical_crossentropy',
optimizer='rmsprop',
metrics=['accuracy'])
model.fit(X_train, y_train,
batch_size=128,
epochs=5,
verbose=1,
validation_split=0.1,
callbacks=[early_stopper,
tensorboard,
checkpointer])
import os
sorted(os.listdir(outpath))
tensorboard --logdir=/tmp/tensorflow_logs/cifar/
lt --port 6006
sudo npm install -g localtunnel
from keras.layers import Input
from keras.models import Model
inputs = Input(shape=(3072,))
l1 = Dense(1024, activation='relu', input_dim=3072)(inputs)
l2 = Dense(512, activation='relu')(l1)
predictions = Dense(10, activation='softmax')(l2)
model = Model(inputs=inputs, outputs=predictions)
model.compile(loss='sparse_categorical_crossentropy',
optimizer='rmsprop',
metrics=['accuracy'])
model.fit(X_train, y_train,
batch_size=128,
epochs=10,
verbose=1,
validation_split=0.1)
# Final test evaluation
score = model.evaluate(X_test, y_test, verbose=0)
print('Test loss:', score[0])
print('Test accuracy:', score[1])
from keras.layers.core import Dense, Dropout, Activation
from keras.layers import Conv2D, MaxPool2D, AveragePooling2D, Flatten
outpath = '/tmp/tensorflow_logs/cifar_conv'
tensorboard = TensorBoard(outpath)
#this model doesn't work
inputs = Input(shape=(32,32,3))
l1 = Conv2D(32, (3,3), activation='relu')(inputs)
l2 = Conv2D(48, (3,3), activation='relu')(l1)
l3 = MaxPool2D()(l2)
l4 = Flatten()(l3)
outputs = Dense(10, activation='softmax')(l4)
model = Model(inputs=inputs, outputs=outputs)
#model above doesn't compile
model.compile(loss='sparse_categorical_crossentropy',
optimizer='rmsprop',
metrics=['accuracy']
)
model.fit(X_train, y_train,
batch_size=128,
epochs=5,
verbose=1,
validation_split=0.1,
callbacks=[tensorboard])
inputs = Input(shape=(32,32,3))
l1 = Conv2D(32, (3,3), activation='relu')(inputs)
l2 = Flatten()(inputs)
outputs = Dense(10, activation='softmax')(l2)
model = Model(inputs=inputs, outputs=outputs)
model.compile('adam', 'sparse_categorical_crossentropy', metrics=['accuracy'])
model.fit(X_train_t, y_train, epochs=5, callbacks=[tensorboard])
score = model.evaluate(X_test, y_test, verbose=0)
print('\n\n')
print('Test loss:', score[0])
print('Test accuracy:', score[1])
| 0.762778 | 0.854278 |
# DataSet Performance
This notebook shows the tradeoffs between inserting data into the database row by row and as binary blobs.
There is a tradeoff between inserting the data in these to ways.
Inserting the data row by row means that we direct access to all the data and may perform querys directly on the values of the data. On the other hand as we will see below this is much slower than inserting the data direcly as binary blobs
First we chose a new location for the database to ensuer that we don't add a bunch of benchmarking data to it
```
import os
cwd = os.getcwd()
import qcodes as qc
qc.config["core"]["db_location"] = os.path.join(cwd, 'testing.db')
%matplotlib notebook
import time
import matplotlib.pyplot as plt
import numpy as np
import qcodes as qc
from qcodes.instrument.parameter import ManualParameter
from qcodes.dataset.experiment_container import (Experiment,
load_last_experiment,
new_experiment)
from qcodes.dataset.database import initialise_database
from qcodes import load_or_create_experiment
from qcodes.dataset.measurements import Measurement
initialise_database()
exp = load_or_create_experiment(experiment_name='tutorial_exp', sample_name="no sample")
```
Here we define a simple function to benchmark the time it takes to insert n points with either numeric or array data type.
We will compare both the time used to call add_result and the time used for the full measurement.
```
def insert_data(paramtype, npoints, nreps=1):
meas = Measurement(exp=exp)
x1 = ManualParameter('x1')
x2 = ManualParameter('x2')
x3 = ManualParameter('x3')
y1 = ManualParameter('y1')
y2 = ManualParameter('y2')
meas.register_parameter(x1, paramtype=paramtype)
meas.register_parameter(x2, paramtype=paramtype)
meas.register_parameter(x3, paramtype=paramtype)
meas.register_parameter(y1, setpoints=[x1, x2, x3],
paramtype=paramtype)
meas.register_parameter(y2, setpoints=[x1, x2, x3],
paramtype=paramtype)
start = time.perf_counter()
with meas.run() as datasaver:
start_adding = time.perf_counter()
for i in range(nreps):
datasaver.add_result((x1, np.random.rand(npoints)),
(x2, np.random.rand(npoints)),
(x3, np.random.rand(npoints)),
(y1, np.random.rand(npoints)),
(y2, np.random.rand(npoints)))
stop_adding = time.perf_counter()
run_id = datasaver.run_id
stop = time.perf_counter()
tot_time = stop - start
add_time = stop_adding - start_adding
return tot_time, add_time, run_id
```
And perform a quick comparison of the two
```
sizes = [1,100,5000,7000,8000,10000,15000,20000]
t_numeric = []
t_numeric_add = []
t_array = []
t_array_add = []
for size in sizes:
tn, tna, run_id_n = insert_data('numeric', size)
t_numeric.append(tn)
t_numeric_add.append(tna)
ta, taa, run_id_a = insert_data('array', size)
t_array.append(ta)
t_array_add.append(taa)
fig, ax = plt.subplots(1,1)
ax.plot(sizes, t_numeric, 'o-', label='Inserting row by row')
ax.plot(sizes, t_numeric_add, 'o-', label='Inserting row by row: add_result only')
ax.plot(sizes, t_array, 'd-', label='Inserting as binary blob')
ax.plot(sizes, t_array_add, 'd-', label='Inserting as binary blob: add_result only')
ax.legend()
ax.set_xlabel('Array lenght')
ax.set_ylabel('Time (s)')
fig.tight_layout()
```
As we can see there the time to setup and and close the experiment is approximately 0.4 sec. At small array sizes the difference between inserting as arrays and inserting row by row is therefore relatively unimportant. At larger array sizes above 10000 points or so the cost of writing data as individual datapoints starts to become important.
```
sizes = [1,10,100,1000,3000,5000]
nreps = 100
t_numeric = []
t_numeric_add = []
t_numeric_run_ids = []
t_array = []
t_array_add = []
t_array_run_ids = []
for size in sizes:
tn, tna, run_id_n = insert_data('numeric', size, nreps=nreps)
t_numeric.append(tn)
t_numeric_add.append(tna)
t_numeric_run_ids.append(run_id_n)
ta, taa, run_id_a = insert_data('array', size, nreps=nreps)
t_array.append(ta)
t_array_add.append(taa)
t_array_run_ids.append(run_id_a)
fig, ax = plt.subplots(1,1)
ax.plot(sizes, t_numeric, 'o-', label='Inserting row by row')
ax.plot(sizes, t_numeric_add, 'o-', label='Inserting row by row: add_result only')
ax.plot(sizes, t_array, 'd-', label='Inserting as binary blob')
ax.plot(sizes, t_array_add, 'd-', label='Inserting as binary blob: add_result only')
ax.legend()
ax.set_xlabel('Array lenght')
ax.set_ylabel('Time (s)')
fig.tight_layout()
```
However, as we increase the length of the experimenter as seen here by repeating the insertion 100 times we see a big difference between inserting row by row and inserting as a binary blob
```
from qcodes.dataset.data_set import load_by_id
from qcodes.dataset.data_export import get_data_by_id
```
As usual you can load the data using load_by_id but you will notice that the different storage methods
are reflected in shape of the data as is is retried.
```
run_id_n = t_numeric_run_ids[0]
run_id_a = t_array_run_ids[0]
ds = load_by_id(run_id_n)
ds.get_data('x1')
```
And a dataset stored as binary arrays
```
ds = load_by_id(run_id_a)
ds.get_data('x1')
```
This is probably more useful as a numpy array. Here we use squeze to get rid of any singleton dimensions.
```
np.array(ds.get_data('x1')).squeeze()
```
A better solution may be to use get_data_by_id which will load the data in a format that does not depend on the internal storage
```
get_data_by_id(run_id_n)
get_data_by_id(run_id_a)
```
|
github_jupyter
|
import os
cwd = os.getcwd()
import qcodes as qc
qc.config["core"]["db_location"] = os.path.join(cwd, 'testing.db')
%matplotlib notebook
import time
import matplotlib.pyplot as plt
import numpy as np
import qcodes as qc
from qcodes.instrument.parameter import ManualParameter
from qcodes.dataset.experiment_container import (Experiment,
load_last_experiment,
new_experiment)
from qcodes.dataset.database import initialise_database
from qcodes import load_or_create_experiment
from qcodes.dataset.measurements import Measurement
initialise_database()
exp = load_or_create_experiment(experiment_name='tutorial_exp', sample_name="no sample")
def insert_data(paramtype, npoints, nreps=1):
meas = Measurement(exp=exp)
x1 = ManualParameter('x1')
x2 = ManualParameter('x2')
x3 = ManualParameter('x3')
y1 = ManualParameter('y1')
y2 = ManualParameter('y2')
meas.register_parameter(x1, paramtype=paramtype)
meas.register_parameter(x2, paramtype=paramtype)
meas.register_parameter(x3, paramtype=paramtype)
meas.register_parameter(y1, setpoints=[x1, x2, x3],
paramtype=paramtype)
meas.register_parameter(y2, setpoints=[x1, x2, x3],
paramtype=paramtype)
start = time.perf_counter()
with meas.run() as datasaver:
start_adding = time.perf_counter()
for i in range(nreps):
datasaver.add_result((x1, np.random.rand(npoints)),
(x2, np.random.rand(npoints)),
(x3, np.random.rand(npoints)),
(y1, np.random.rand(npoints)),
(y2, np.random.rand(npoints)))
stop_adding = time.perf_counter()
run_id = datasaver.run_id
stop = time.perf_counter()
tot_time = stop - start
add_time = stop_adding - start_adding
return tot_time, add_time, run_id
sizes = [1,100,5000,7000,8000,10000,15000,20000]
t_numeric = []
t_numeric_add = []
t_array = []
t_array_add = []
for size in sizes:
tn, tna, run_id_n = insert_data('numeric', size)
t_numeric.append(tn)
t_numeric_add.append(tna)
ta, taa, run_id_a = insert_data('array', size)
t_array.append(ta)
t_array_add.append(taa)
fig, ax = plt.subplots(1,1)
ax.plot(sizes, t_numeric, 'o-', label='Inserting row by row')
ax.plot(sizes, t_numeric_add, 'o-', label='Inserting row by row: add_result only')
ax.plot(sizes, t_array, 'd-', label='Inserting as binary blob')
ax.plot(sizes, t_array_add, 'd-', label='Inserting as binary blob: add_result only')
ax.legend()
ax.set_xlabel('Array lenght')
ax.set_ylabel('Time (s)')
fig.tight_layout()
sizes = [1,10,100,1000,3000,5000]
nreps = 100
t_numeric = []
t_numeric_add = []
t_numeric_run_ids = []
t_array = []
t_array_add = []
t_array_run_ids = []
for size in sizes:
tn, tna, run_id_n = insert_data('numeric', size, nreps=nreps)
t_numeric.append(tn)
t_numeric_add.append(tna)
t_numeric_run_ids.append(run_id_n)
ta, taa, run_id_a = insert_data('array', size, nreps=nreps)
t_array.append(ta)
t_array_add.append(taa)
t_array_run_ids.append(run_id_a)
fig, ax = plt.subplots(1,1)
ax.plot(sizes, t_numeric, 'o-', label='Inserting row by row')
ax.plot(sizes, t_numeric_add, 'o-', label='Inserting row by row: add_result only')
ax.plot(sizes, t_array, 'd-', label='Inserting as binary blob')
ax.plot(sizes, t_array_add, 'd-', label='Inserting as binary blob: add_result only')
ax.legend()
ax.set_xlabel('Array lenght')
ax.set_ylabel('Time (s)')
fig.tight_layout()
from qcodes.dataset.data_set import load_by_id
from qcodes.dataset.data_export import get_data_by_id
run_id_n = t_numeric_run_ids[0]
run_id_a = t_array_run_ids[0]
ds = load_by_id(run_id_n)
ds.get_data('x1')
ds = load_by_id(run_id_a)
ds.get_data('x1')
np.array(ds.get_data('x1')).squeeze()
get_data_by_id(run_id_n)
get_data_by_id(run_id_a)
| 0.287768 | 0.946597 |
# Compile and Deploy the TFX Pipeline to KFP
This Notebook helps you to compile the **TFX Pipeline** to a **KFP package**. This will creat an **Argo YAML** file in a **.tar.gz** package. We perform the following steps:
1. Build a custom container image that include our modules
2. Compile TFX Pipeline using CLI
3. Deploy the compiled pipeline to KFP
After you deploy the pipeline, go to KFP UI, create a run, and execute the pipeline:
<img valign="middle" src="imgs/kfp.png" width="800">
## 1. Build Container Image
The pipeline uses a custom docker image, which is a derivative of the [tensorflow/tfx:0.15.0](https://hub.docker.com/r/tensorflow/tfx) image, as a runtime execution environment for the pipeline's components. The same image is also used as a a training image used by **AI Platform Training**.
The custom image modifies the base image by:
* Downgrading from Tensoflow v2.0 to v1.15 (since AI Platform Prediction is not supporting TF v2.0 yet).
* Adding the `modules` folder, which includes the **train.py** and **transform.py** code files required by the **Trainer** and **Transform** components, as well as the implementation code for the custom **AccuracyModelValidator** component.
```
PROJECT_ID='ksalama-ocado' # Set your GCP project Id
IMAGE_NAME='tfx-image'
TAG='latest'
TFX_IMAGE='gcr.io/{}/{}:{}'.format(PROJECT_ID, IMAGE_NAME, TAG)
!gcloud builds submit --tag $TFX_IMAGE ./ml-pipeline
```
## 2. Compile TFX Pipeline using CLI
```
!tfx pipeline --help
%%bash
export PROJECT_ID=$(gcloud config get-value core/project) # Set your GCP project Id
export IMAGE_NAME=tfx-image
export TAG=latest
export TFX_IMAGE=gcr.io/${PROJECT_ID}/${IMAGE_NAME}:${TAG}
export PREFIX=ksalama-mlops-dev # Set your prefix
export NAMESPACE=kfp # Set your namespace
export GCP_REGION=europe-west1 # Set your region
export ZONE=europe-west1-b # Set your zone
export ARTIFACT_STORE_URI=gs://${PREFIX}-artifact-store
export GCS_STAGING_PATH=${ARTIFACT_STORE_URI}/staging
export GKE_CLUSTER_NAME=${PREFIX}-cluster
export DATASET_NAME=sample_datasets # Set your BigQuery Dataset
export PIPELINE_NAME=tfx_census_classification
export RUNTIME_VERSION=1.15
export PYTHON_VERSION=3.7
tfx pipeline compile \
--engine=kubeflow \
--pipeline_path=ml-pipeline/pipeline.py
```
## 3. Deploy the Compiled Pipeline to KFP
```
!kfp pipeline --help
%%bash
export NAMESPACE=kfp # Set your namespac
export PREFIX=ksalama-mlops-dev # Set your prefix
export GKE_CLUSTER_NAME=${PREFIX}-cluster
export ZONE=europe-west1-b # Set your zone
export PIPELINE_NAME=tfx_census_classification
gcloud container clusters get-credentials ${GKE_CLUSTER_NAM}E --zone ${ZONE}
export INVERSE_PROXY_HOSTNAME=$(kubectl describe configmap inverse-proxy-config -n ${NAMESPACE} | grep "googleusercontent.com")
kfp --namespace=${NAMESPACE} --endpoint=${INVERSE_PROXY_HOSTNAME} \
pipeline upload \
--pipeline-name=${PIPELINE_NAME} \
${PIPELINE_NAME}.tar.gz
```
## Use the KFP UI to run the deployed pipeline...
|
github_jupyter
|
PROJECT_ID='ksalama-ocado' # Set your GCP project Id
IMAGE_NAME='tfx-image'
TAG='latest'
TFX_IMAGE='gcr.io/{}/{}:{}'.format(PROJECT_ID, IMAGE_NAME, TAG)
!gcloud builds submit --tag $TFX_IMAGE ./ml-pipeline
!tfx pipeline --help
%%bash
export PROJECT_ID=$(gcloud config get-value core/project) # Set your GCP project Id
export IMAGE_NAME=tfx-image
export TAG=latest
export TFX_IMAGE=gcr.io/${PROJECT_ID}/${IMAGE_NAME}:${TAG}
export PREFIX=ksalama-mlops-dev # Set your prefix
export NAMESPACE=kfp # Set your namespace
export GCP_REGION=europe-west1 # Set your region
export ZONE=europe-west1-b # Set your zone
export ARTIFACT_STORE_URI=gs://${PREFIX}-artifact-store
export GCS_STAGING_PATH=${ARTIFACT_STORE_URI}/staging
export GKE_CLUSTER_NAME=${PREFIX}-cluster
export DATASET_NAME=sample_datasets # Set your BigQuery Dataset
export PIPELINE_NAME=tfx_census_classification
export RUNTIME_VERSION=1.15
export PYTHON_VERSION=3.7
tfx pipeline compile \
--engine=kubeflow \
--pipeline_path=ml-pipeline/pipeline.py
!kfp pipeline --help
%%bash
export NAMESPACE=kfp # Set your namespac
export PREFIX=ksalama-mlops-dev # Set your prefix
export GKE_CLUSTER_NAME=${PREFIX}-cluster
export ZONE=europe-west1-b # Set your zone
export PIPELINE_NAME=tfx_census_classification
gcloud container clusters get-credentials ${GKE_CLUSTER_NAM}E --zone ${ZONE}
export INVERSE_PROXY_HOSTNAME=$(kubectl describe configmap inverse-proxy-config -n ${NAMESPACE} | grep "googleusercontent.com")
kfp --namespace=${NAMESPACE} --endpoint=${INVERSE_PROXY_HOSTNAME} \
pipeline upload \
--pipeline-name=${PIPELINE_NAME} \
${PIPELINE_NAME}.tar.gz
| 0.308919 | 0.912864 |
```
from pyspark.sql import SparkSession
appName = 'Python Spark SQL basic example'
spark_jars = '/home/sade/.local/share/DBeaverData/drivers/maven/maven-central/org.postgresql/postgresql-42.2.5.jar'
# spark_jars: Postgres' ye bağlanmak için gereken bir şey. Veritabanı işlemleri için DBaver kullanıyorum.
# Bu jar' ı DBaver kendi indiriyor. Spark içinde bu jar gerekliydi ben de DBaver' ın indirdiği jar' ı verdim.
spark = SparkSession \
.builder \
.appName(appName) \
.config("spark.jars", spark_jars) \
.getOrCreate()
# PostgreSQL' den veri çekme.
format_ = 'jdbc' # Ne olduğuna dair bir bilgim yok.
url = 'jdbc:postgresql://localhost:5432/postgres' # Postgre' ye bağlanmak için gereken host, port ve db adı.
query = '(select * from postgres_table) pt' # Veritabanından çekilecek sorgu. Buraya sadece isimde gelebiliyor.
user = 'postgres' # DB kullanıcı adı
password = '"123"' # DB şifre
driver = 'org.postgresql.Driver' # Ne olduğunu bilmiyorum. Benim baktığım örnekte Oracle' a bağlanıyordu.
# Ben de o örneğe bakarak bunu BDaver' dan kopyaladım.
# Sorgu çekiliyor ve gelen cevap bir DataFrame' e dönüştürülüyor.
read_table = spark.read \
.format(format_) \
.option("url", url) \
.option("dbtable", query) \
.option("user", user) \
.option("password", password) \
.option("driver", driver) \
.load()
print("read_table type: ", type(read_table))
# read_table' ın içindeki veriler
read_table.show()
# Şimdi sorgu çekme işlemi daha hızlı olsun diye bir tane method yazalım
def spark_query(
query: str,
format_: str = 'jdbc',
url: str = 'jdbc:postgresql://localhost:5432/postgres',
user: str = 'postgres',
password: str = '"123"',
driver: str = 'org.postgresql.Driver'
):
return spark.read \
.format(format_) \
.option("url", url) \
.option("dbtable", query) \
.option("user", user) \
.option("password", password) \
.option("driver", driver) \
.load()
# Şimdi deniyelim ve çektiği verileri göstersin
read_table_method = spark_query(query='(select * from postgres_table) pt')
read_table_method.show()
# Sanırım Spark' ın kendine ait bir sorgu dili var. Çünkü bu şekildeki normal veritabanı sorguları çalışmıyor.
spark_query(query='SELECT * FROM postgres_table')
# Veridiği hata: ERROR: syntax error at or near "SELECT"
# Spark ile elde ettiğimiz veriler üzerinde bazı işlemler yapma
import pyspark.sql.functions as f
split_user_id = f.split(read_table['userid'], '-') # userid kolonunu -' ye göre parçalıyoruz.
print(type(split_user_id)) # Ne bilmiyorum.
split_product_id = f.split(read_table["productid"], "-") # productid kolonunu -' ye göre parçalıyoruz.
# Şimdi buradaki işlem sonunda userid ve productid den yeni bir DF oluşturuyoruz ama arka planda
# ne dönüyor bilmiyorum.
result_df = read_table.select(
split_user_id.getItem(1).alias("userid"),
split_product_id.getItem(1).alias("productid")
)
result_df.show()
# Şimdi elde ettiğimiz DF' ı PostgreSQL' e yazalım.
# Burası tam olarak ne yapar bilmiyorum.
write_table = 'spark_postgres'
result_df.write.format(format_) \
.option("url", url) \
.option("dbtable", write_table) \
.option("user", user) \
.option("password", password) \
.option("driver", driver) \
.mode('append') \ # Acaba veritabanının içini boşaltıp doldurma özelliği var mı? Başka hangi mod' lar var?
.save()
```
|
github_jupyter
|
from pyspark.sql import SparkSession
appName = 'Python Spark SQL basic example'
spark_jars = '/home/sade/.local/share/DBeaverData/drivers/maven/maven-central/org.postgresql/postgresql-42.2.5.jar'
# spark_jars: Postgres' ye bağlanmak için gereken bir şey. Veritabanı işlemleri için DBaver kullanıyorum.
# Bu jar' ı DBaver kendi indiriyor. Spark içinde bu jar gerekliydi ben de DBaver' ın indirdiği jar' ı verdim.
spark = SparkSession \
.builder \
.appName(appName) \
.config("spark.jars", spark_jars) \
.getOrCreate()
# PostgreSQL' den veri çekme.
format_ = 'jdbc' # Ne olduğuna dair bir bilgim yok.
url = 'jdbc:postgresql://localhost:5432/postgres' # Postgre' ye bağlanmak için gereken host, port ve db adı.
query = '(select * from postgres_table) pt' # Veritabanından çekilecek sorgu. Buraya sadece isimde gelebiliyor.
user = 'postgres' # DB kullanıcı adı
password = '"123"' # DB şifre
driver = 'org.postgresql.Driver' # Ne olduğunu bilmiyorum. Benim baktığım örnekte Oracle' a bağlanıyordu.
# Ben de o örneğe bakarak bunu BDaver' dan kopyaladım.
# Sorgu çekiliyor ve gelen cevap bir DataFrame' e dönüştürülüyor.
read_table = spark.read \
.format(format_) \
.option("url", url) \
.option("dbtable", query) \
.option("user", user) \
.option("password", password) \
.option("driver", driver) \
.load()
print("read_table type: ", type(read_table))
# read_table' ın içindeki veriler
read_table.show()
# Şimdi sorgu çekme işlemi daha hızlı olsun diye bir tane method yazalım
def spark_query(
query: str,
format_: str = 'jdbc',
url: str = 'jdbc:postgresql://localhost:5432/postgres',
user: str = 'postgres',
password: str = '"123"',
driver: str = 'org.postgresql.Driver'
):
return spark.read \
.format(format_) \
.option("url", url) \
.option("dbtable", query) \
.option("user", user) \
.option("password", password) \
.option("driver", driver) \
.load()
# Şimdi deniyelim ve çektiği verileri göstersin
read_table_method = spark_query(query='(select * from postgres_table) pt')
read_table_method.show()
# Sanırım Spark' ın kendine ait bir sorgu dili var. Çünkü bu şekildeki normal veritabanı sorguları çalışmıyor.
spark_query(query='SELECT * FROM postgres_table')
# Veridiği hata: ERROR: syntax error at or near "SELECT"
# Spark ile elde ettiğimiz veriler üzerinde bazı işlemler yapma
import pyspark.sql.functions as f
split_user_id = f.split(read_table['userid'], '-') # userid kolonunu -' ye göre parçalıyoruz.
print(type(split_user_id)) # Ne bilmiyorum.
split_product_id = f.split(read_table["productid"], "-") # productid kolonunu -' ye göre parçalıyoruz.
# Şimdi buradaki işlem sonunda userid ve productid den yeni bir DF oluşturuyoruz ama arka planda
# ne dönüyor bilmiyorum.
result_df = read_table.select(
split_user_id.getItem(1).alias("userid"),
split_product_id.getItem(1).alias("productid")
)
result_df.show()
# Şimdi elde ettiğimiz DF' ı PostgreSQL' e yazalım.
# Burası tam olarak ne yapar bilmiyorum.
write_table = 'spark_postgres'
result_df.write.format(format_) \
.option("url", url) \
.option("dbtable", write_table) \
.option("user", user) \
.option("password", password) \
.option("driver", driver) \
.mode('append') \ # Acaba veritabanının içini boşaltıp doldurma özelliği var mı? Başka hangi mod' lar var?
.save()
| 0.247351 | 0.545044 |
### Configure GPU usage
```
import tensorflow as tf
tf.__version__
tf.__version__
gpus = tf.config.list_physical_devices('GPU')[2:4]
gpus
gpus
tf.config.set_visible_devices(gpus, 'GPU')
tf.config.list_logical_devices('CPU')
tf.config.list_logical_devices('CPU')
tf.config.list_logical_devices('GPU')
tf.config.list_logical_devices('GPU')
```
### Run experiments
```
from model_experiments.vae_experiment import VaeExperiment
from general.experiment import GenerativeModelType, TransformerType, OptimizerType
from general.utils import OutputActivation
from evaluators.machine_learning_evaluator import MachineLearningEvaluator, EvaluatorModelType, MetricType, ScalerType
from models.utils import Activation
import json
import seaborn as sns
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
from copy import deepcopy
import warnings
from sklearn.exceptions import ConvergenceWarning
warnings.simplefilter(action='ignore', category=ConvergenceWarning)
# Adult
# early stop epsilon value = 2.5
# record gradients subset = ["race", "relationship"]
# UCI Credit Card
# early stop epsilon value = 2.5
# record gradients subset = ["SEX", "EDUCATION"]
# Bank Marketing
# early stop epsilon value = 2.25
# record gradients subset = ["job"]
base_config = {
"model_type": GenerativeModelType.VAE,
"name": "vae_experiment_bn_test_dp",
"data_path": "data/",
"data_processing": {},
"model_config": {
"compress_dims": [128, 128],
"decompress_dims": [128, 128],
"compress_activations": [Activation.LeakyReLU, Activation.LeakyReLU],
"decompress_activations": [Activation.LeakyReLU, Activation.LeakyReLU],
"latent_dim": 128,
"output_activations": {
"continuous": OutputActivation.TANH,
"categorical": OutputActivation.SOFTMAX,
},
},
"model_train": {
"test_pct": 0.3,
"k_fold": True,
"stratified_by_col": None,
"epochs": 500,
"batch_size": 64,
"dp_optimizer_type": OptimizerType.GradientDescentOptimizer,
"record_gradients": {"enabled": True, "subset": []},
"transformer": {
"type": TransformerType.BGM,
"kwargs": {"outlier_clipping": True, "n_clusters": 6},
},
"early_stop": {"enabled": False},
"early_stop_epsilon": {"enabled": True},
},
"diff_priv": {
"enabled": True,
"microbatches": 1,
"l2_norm_clip": 5.0,
"noise_multiplier": 0.8,
},
}
base_config_no_dp = {
"model_type": GenerativeModelType.VAE,
"name": "vae_experiment_bn_test_no_dp",
"data_path": "data/",
"data_processing": {},
"model_config": {
"compress_dims": [128, 128],
"decompress_dims": [128, 128],
"compress_activations": [Activation.LeakyReLU, Activation.LeakyReLU],
"decompress_activations": [Activation.LeakyReLU, Activation.LeakyReLU],
"latent_dim": 128,
"output_activations": {
"continuous": OutputActivation.TANH,
"categorical": OutputActivation.SOFTMAX,
},
},
"model_train": {
"test_pct": 0.3,
"k_fold": True,
"stratified_by_col": None,
"epochs": 8,
"learning_rate": .001,
"batch_size": 64,
"record_gradients": {"enabled": True, "subset": []},
"optimizer_type": OptimizerType.GradientDescentOptimizer,
"transformer": {
"type": TransformerType.BGM,
"kwargs": {"outlier_clipping": True, "n_clusters": 6},
},
"early_stop": {"enabled": False},
"early_stop_epsilon": {"enabled": False},
},
"diff_priv": {
"enabled": False,
},
}
adult_config = {
"model_type": GenerativeModelType.VAE,
"name": "vae_experiment_mi_attack",
"dataset": "adult.csv",
"data_path": "data/",
"data_processing": {},
"model_config": {
"compress_dims": [128, 128],
"decompress_dims": [128, 128],
"compress_activations": [Activation.LeakyReLU, Activation.LeakyReLU],
"decompress_activations": [Activation.LeakyReLU, Activation.LeakyReLU],
"latent_dim": 128,
"output_activations": {
"continuous": OutputActivation.TANH,
"categorical": OutputActivation.SOFTMAX,
},
},
"model_train": {
"seed": 10,
"test_pct": 0.,
"k_fold": False,
"stratified_by_col": None,
"epochs": 500,
"batch_size": 128,
"dp_optimizer_type": OptimizerType.DPAdamGaussianOptimizer,
"record_gradients": {"enabled": False, "subset": []},
"transformer": {
"type": TransformerType.BGM,
"kwargs": {"outlier_clipping": True, "n_clusters": 6},
},
"early_stop": {"enabled": False},
"early_stop_epsilon": {"enabled": True, "value": 2.75},
},
"diff_priv": {
"enabled": True,
"microbatches": 1,
"l2_norm_clip": 5.0,
"noise_multiplier": 0.8,
},
}
batch_norm = [True, False]
for with_batch_norm in batch_norm:
cur_config = deepcopy(adult_config)
cur_config["model_config"]["batch_norm"] = with_batch_norm
cur_experiment = VaeExperiment(cur_config)
cur_experiment.run(automatic_overwrite=False)
import time
start = time.time()
batch_norm = [True, False]
for with_batch_norm in batch_norm:
cur_config = deepcopy(adult_config)
cur_config["model_config"]["batch_norm"] = with_batch_norm
cur_experiment = VaeExperiment(cur_config)
cur_experiment.run(automatic_overwrite=False)
end = time.time()
print(end - start)
!/usr/bin/nvidia-smi
# datasets = ["adult.csv", "bank_marketing.csv", "uci_credit_card.csv"]
datasets = ["adult.csv", "bank_marketing.csv"]
gradient_record_subsets = {
"uci_credit_card.csv": ["SEX", "EDUCATION"],
"bank_marketing.csv": ["job"],
"adult.csv": ["race", "relationship"]
}
batch_norm = [True, False]
seeds = range(10, 11)
uci_categorical_cols = [
"SEX",
"EDUCATION",
"MARRIAGE",
"PAY_0",
"PAY_1",
"PAY_2",
"PAY_3",
"PAY_4",
"PAY_5",
"PAY_6",
"DEFAULT",
]
bank_marketing_read_csv_kwargs = {"delimiter": ";"}
bank_marketing_epsilon_value = 2.25
other_epsilon_value = 2.5
# train without dp
for seed in [10, 11]:
for with_batch_norm in batch_norm:
for dataset in datasets:
# set config - generic
cur_config = deepcopy(base_config_no_dp)
cur_config["model_train"]["seed"] = seed
cur_config["model_config"]["batch_norm"] = with_batch_norm
cur_config["dataset"] = dataset
if dataset == "uci_credit_card.csv":
cur_config["data_processing"]["categorical_columns"] = uci_categorical_cols
if dataset == "bank_marketing.csv":
cur_config["data_processing"]["read_csv_kwargs"] = bank_marketing_read_csv_kwargs
cur_config["model_train"]["record_gradients"]["subset"] = gradient_record_subsets[dataset]
# run experiemnt
cur_experiment = VaeExperiment(cur_config)
try:
cur_experiment.run(automatic_overwrite=False)
except SystemExit:
continue
test_config = {"name": "vae_experiment_bn_test_no_dp"}
existing_configs = MachineLearningEvaluator.existing_configs("data/", test_config=test_config)
eval_input = {'target_col': 'output', 'data_path': 'data/', 'config_file': '404d16aae768b97c18ce5aa3293376a5cc3f6c09.pkl'}
evaluator = MachineLearningEvaluator(**eval_input)
x = evaluator.get_experiment_data()
x["model_runs"][0]["gradient_norms"]
test_config = {"name": "vae_experiment_bn_test_dp"}
existing_configs = MachineLearningEvaluator.existing_configs("data/", test_config=test_config)
eval_input = {'target_col': 'output', 'data_path': 'data/', 'config_file': '13e18f019180e253ee4b1cf10dbe953c762eb185.pkl'}
evaluator = MachineLearningEvaluator(**eval_input)
x = evaluator.get_experiment_data()
x["model_runs"][0]["gradient_norms"]
# train with dp
for seed in [10, 11]:
for with_batch_norm in batch_norm:
for dataset in datasets:
# set config - generic
cur_config = deepcopy(base_config)
cur_config["model_train"]["seed"] = seed
cur_config["model_train"]["dp_optimizer_type"] = dp_opt
cur_config["model_config"]["batch_norm"] = with_batch_norm
cur_config["dataset"] = dataset
if dataset == "uci_credit_card.csv":
cur_config["data_processing"]["categorical_columns"] = uci_categorical_cols
cur_config["model_train"]["record_gradients"]["subset"] = gradient_record_subsets[dataset]
# set config - dp
if dataset == "bank_marketing.csv":
value = bank_marketing_epsilon_value
cur_config["data_processing"]["read_csv_kwargs"] = bank_marketing_read_csv_kwargs
else:
value = other_epsilon_value
cur_config["model_train"]["early_stop_epsilon"]["value"] = value
# run experiemnt
cur_experiment = VaeExperiment(cur_config)
try:
cur_experiment.run(automatic_overwrite=False)
except SystemExit:
continue
for seed in seeds[:5]:
for with_batch_norm in batch_norm:
for dataset in datasets:
# set config
cur_config = deepcopy(base_config_no_dp)
cur_config["model_train"]["seed"] = seed
cur_config["model_config"]["batch_norm"] = with_batch_norm
cur_config["dataset"] = dataset
if dataset == "uci_credit_card.csv":
cur_config["data_processing"]["categorical_columns"] = uci_categorical_cols
cur_config["model_train"]["record_gradients"]["subset"] = gradient_record_subsets[dataset]
# run experiemnt
cur_experiment = VaeExperiment(cur_config)
cur_experiment.run(automatic_overwrite=True)
config = {
"dataset": "uci_credit_card.csv",
"model_type": GenerativeModelType.VAE,
"name": "vae_experiment",
"data_path": "data/",
"data_processing": {
"columns_to_drop": ["ID"],
"columns_rename_map": {"default.payment.next.month": "default"},
"categorical_columns": [
"SEX",
"EDUCATION",
"MARRIAGE",
"PAY_0",
"PAY_1",
"PAY_2",
"PAY_3",
"PAY_4",
"PAY_5",
"PAY_6",
"default",
],
},
"model_config": {
"compress_dims": [128, 128],
"decompress_dims": [128, 128],
"compress_activations": [Activation.ReLU, Activation.ReLU],
"decompress_activations": [Activation.ReLU, Activation.ReLU],
"latent_dim": 128,
"output_activations": {
"continuous": OutputActivation.TANH,
"categorical": OutputActivation.SOFTMAX,
},
},
"model_train": {
"seed": 1,
"test_pct": 0.3,
"k_fold": False,
"stratified_by_col": None,
"epochs": 50,
"dp_optimizer_type": OptimizerType.DPAdamGaussianOptimizer,
},
"transformer": {
"type": TransformerType.BGM,
"kwargs": {"outlier_clipping": True, "n_clusters": 6},
},
"diff_priv": {
"enabled": True,
"microbatches": 8,
"l2_norm_clip": 5.0,
"noise_multiplier": 0.8,
},
# "early_stop_epsilon": {"enabled": True, "value": 2.1},
}
config = {
"dataset": "adult.csv",
"model_type": GenerativeModelType.VAE,
"name": "vae_experiment_classes",
"data_path": "data/",
"model_config": {
"compress_dims": [128, 128],
"decompress_dims": [128, 128],
"compress_activations": None,
"decompress_activations": None,
"latent_dim": 128,
"batch_norm": None,
"output_activations": {
"continuous": OutputActivation.TANH,
"categorical": OutputActivation.SOFTMAX,
},
},
"model_train": {
"seed": None,
"test_pct": 0.3,
"k_fold": True,
"epochs": 50,
"dp_optimizer_type": OptimizerType.DPAdamGaussianOptimizer,
"batch_size": 64
},
"transformer": {
"type": TransformerType.BGM,
"kwargs": {"outlier_clipping": True, "n_clusters": 6},
},
"diff_priv": {
"enabled": True,
"microbatches": 8,
"l2_norm_clip": 5.0,
"noise_multiplier": 0.8
},
}
all_compress_acts = [[Activation.LeakyReLU, Activation.LeakyReLU], [Activation.ELU, Activation.ELU]]
all_decompress_acts = [[Activation.LeakyReLU, Activation.LeakyReLU], [Activation.ELU, Activation.ELU]]
for seed in range(10):
config["model_train"]["seed"] = seed
for compress_acts, decompress_acts in list(zip(all_compress_acts, all_decompress_acts)):
config["model_config"]["compress_activations"] = compress_acts
config["model_config"]["decompress_activations"] = decompress_acts
for batch_norm in [True, False]:
config["model_config"]["batch_norm"] = batch_norm
vae_experiment = VaeExperiment(config)
vae_experiment.run()
```
### Evaluations
```
test_config_bn = {
'dataset': 'adult.csv',
'diff_priv': {"l2_norm_clip": 5, "enabled": True},
'model_train': {"dp_optimizer_type": 2},
'transformer': {'kwargs': {'n_clusters': 6}, 'type': 1},
"model_config": {"batch_norm": True}
}
test_config = {
'dataset': 'adult.csv',
'diff_priv': {"l2_norm_clip": 5, "enabled": True},
'model_train': {'stratified_by_col': None, "dp_optimizer_type": 2},
'transformer': {'kwargs': {'n_clusters': 6}, 'type': 1}
}
test_config["name"] = 'vae_experiment_noise08'
def plot_data_utility_scores(
target_col,
data_path,
configs,
model,y
metrics,
include_real=False,
scaler_type=None,
scaler_kwargs=None,
model_kwargs=None,
metric_kwargs=None,
holdout=None,
subset=None,
):
all_group_scores = {}
for group_name, group_configs in configs.items():
group_scores = []
for config_hash, config in group_configs.items():
eval_input = {'target_col': target_col, 'data_path': data_path,
'config_file': config_hash}
evaluator = MachineLearningEvaluator(**eval_input)
scores = evaluator.data_utility_scores(
model=model,
metrics=metrics,
scaler_type=scaler_type,
scaler_kwargs=scaler_kwargs,
model_kwargs=model_kwargs,
metric_kwargs=metric_kwargs,
holdout=holdout,
subset=subset,
include_train=False
)
group_scores.append(scores)
all_group_scores[group_name] = pd.concat(group_scores, axis=0)
columns = list(scores.columns)
cols_to_plot = [col for col in columns if "fake" in col]
img_per_row = 3
rows = int(np.ceil(len(cols_to_plot) / img_per_row))
fig, axes = plt.subplots(rows, img_per_row, figsize=(20, rows*5))
if axes.ndim == 1:
axes = np.expand_dims(axes, 0)
fig.subplots_adjust(hspace=0.35, wspace=0.2)
for i, row in enumerate(axes):
for j, ax in enumerate(row):
col_idx = i * img_per_row + j
if col_idx < len(cols_to_plot):
col_name = cols_to_plot[col_idx]
for group_name, score_df in all_group_scores.items():
sns.distplot(score_df.loc[:, col_name].values, label=group_name, ax=ax)
if include_real:
col_name_real = col_name.replace("fake", "real")
sns.distplot(score_df.loc[:, col_name_real].values, label="real", ax=ax)
ax.set_title("_".join(col_name.split("_")[2:]))
ax.legend()
else:
break
existing_configs = MachineLearningEvaluator.existing_configs("data/", test_config=test_config)
# existing_configs = {k: v for i, (k, v) in enumerate(existing_configs.items()) if i < 2}
existing_configs.keys()
existing_configs_bn = MachineLearningEvaluator.existing_configs("data/", test_config=test_config_bn)
# existing_configs_bn = {k: v for i, (k, v) in enumerate(existing_configs_bn.items()) if i < 2}
existing_configs_bn.keys()
configs = {"with_BatchNorm": existing_configs_bn, "without_BatchNorm": existing_configs}
plot_data_utility_scores(
target_col="output",
data_path="data/",
configs=configs,
include_real=True,
model=EvaluatorModelType.LogisticRegression,
metrics=[MetricType.F1],
scaler_type=ScalerType.ROBUST,
model_kwargs={"solver": "lbfgs", "class_weight": "balanced", "max_iter": 200},
metric_kwargs={MetricType.F1: {"average": None}},
subset="race"
)
plot_data_utility_scores(
target_col="output",
data_path="data/",
configs=configs,
include_real=True,
model=EvaluatorModelType.LogisticRegression,
metrics=[MetricType.F1],
scaler_type=ScalerType.ROBUST,
model_kwargs={"solver": "lbfgs", "class_weight": "balanced", "max_iter": 200},
metric_kwargs={MetricType.F1: {"average": None}},
subset="race",
)
config = "ef9980840b657869103bbfbbec1f7b02b2bc2dc6.pkl" # vae_experiment2 - same configuration as [1]
eval_input = {'target_col': 'default', 'data_path': 'data/', 'config_file': config}
evaluator = MachineLearningEvaluator(**eval_input)
experiment_data = evaluator.get_experiment_data()
scores = evaluator.data_utility_scores(
EvaluatorModelType.LogisticRegression,
[MetricType.F1],
ScalerType.ROBUST,
model_kwargs={"solver": "lbfgs", "class_weight": "balanced", "max_iter": 100},
metric_kwargs={MetricType.F1: {"average": None}},
subset="SEX",
include_train=False,
)
display(scores)
existing_configs = MachineLearningEvaluator.existing_configs("data/", test_config=test_config)
for k, v in existing_configs.items():
eval_input = {'target_col': 'output', 'data_path': 'data/', 'config_file': k}
evaluator = MachineLearningEvaluator(**eval_input)
experiment_data = evaluator.get_experiment_data()
scores = evaluator.data_utility_scores(
EvaluatorModelType.LogisticRegression,
[MetricType.F1],
ScalerType.ROBUST,
model_kwargs={"solver": "lbfgs", "class_weight": "balanced", "max_iter": 200},
metric_kwargs={MetricType.F1: {"average": "binary"}},
subset="race",
include_train=False,
)
display(scores)
for k, v in existing_configs.items():
eval_input = {'target_col': 'output', 'data_path': 'data/', 'config_file': k}
evaluator = MachineLearningEvaluator(**eval_input)
experiment_data = evaluator.get_experiment_data()
scores = evaluator.data_utility_scores(
EvaluatorModelType.LogisticRegression,
[MetricType.F1],
ScalerType.ROBUST,
model_kwargs={"solver": "lbfgs", "class_weight": "balanced", "max_iter": 200},
metric_kwargs={MetricType.F1: {"average": "binary"}},
subset="race",
include_train=False,
)
display(scores)
```
|
github_jupyter
|
import tensorflow as tf
tf.__version__
tf.__version__
gpus = tf.config.list_physical_devices('GPU')[2:4]
gpus
gpus
tf.config.set_visible_devices(gpus, 'GPU')
tf.config.list_logical_devices('CPU')
tf.config.list_logical_devices('CPU')
tf.config.list_logical_devices('GPU')
tf.config.list_logical_devices('GPU')
from model_experiments.vae_experiment import VaeExperiment
from general.experiment import GenerativeModelType, TransformerType, OptimizerType
from general.utils import OutputActivation
from evaluators.machine_learning_evaluator import MachineLearningEvaluator, EvaluatorModelType, MetricType, ScalerType
from models.utils import Activation
import json
import seaborn as sns
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
from copy import deepcopy
import warnings
from sklearn.exceptions import ConvergenceWarning
warnings.simplefilter(action='ignore', category=ConvergenceWarning)
# Adult
# early stop epsilon value = 2.5
# record gradients subset = ["race", "relationship"]
# UCI Credit Card
# early stop epsilon value = 2.5
# record gradients subset = ["SEX", "EDUCATION"]
# Bank Marketing
# early stop epsilon value = 2.25
# record gradients subset = ["job"]
base_config = {
"model_type": GenerativeModelType.VAE,
"name": "vae_experiment_bn_test_dp",
"data_path": "data/",
"data_processing": {},
"model_config": {
"compress_dims": [128, 128],
"decompress_dims": [128, 128],
"compress_activations": [Activation.LeakyReLU, Activation.LeakyReLU],
"decompress_activations": [Activation.LeakyReLU, Activation.LeakyReLU],
"latent_dim": 128,
"output_activations": {
"continuous": OutputActivation.TANH,
"categorical": OutputActivation.SOFTMAX,
},
},
"model_train": {
"test_pct": 0.3,
"k_fold": True,
"stratified_by_col": None,
"epochs": 500,
"batch_size": 64,
"dp_optimizer_type": OptimizerType.GradientDescentOptimizer,
"record_gradients": {"enabled": True, "subset": []},
"transformer": {
"type": TransformerType.BGM,
"kwargs": {"outlier_clipping": True, "n_clusters": 6},
},
"early_stop": {"enabled": False},
"early_stop_epsilon": {"enabled": True},
},
"diff_priv": {
"enabled": True,
"microbatches": 1,
"l2_norm_clip": 5.0,
"noise_multiplier": 0.8,
},
}
base_config_no_dp = {
"model_type": GenerativeModelType.VAE,
"name": "vae_experiment_bn_test_no_dp",
"data_path": "data/",
"data_processing": {},
"model_config": {
"compress_dims": [128, 128],
"decompress_dims": [128, 128],
"compress_activations": [Activation.LeakyReLU, Activation.LeakyReLU],
"decompress_activations": [Activation.LeakyReLU, Activation.LeakyReLU],
"latent_dim": 128,
"output_activations": {
"continuous": OutputActivation.TANH,
"categorical": OutputActivation.SOFTMAX,
},
},
"model_train": {
"test_pct": 0.3,
"k_fold": True,
"stratified_by_col": None,
"epochs": 8,
"learning_rate": .001,
"batch_size": 64,
"record_gradients": {"enabled": True, "subset": []},
"optimizer_type": OptimizerType.GradientDescentOptimizer,
"transformer": {
"type": TransformerType.BGM,
"kwargs": {"outlier_clipping": True, "n_clusters": 6},
},
"early_stop": {"enabled": False},
"early_stop_epsilon": {"enabled": False},
},
"diff_priv": {
"enabled": False,
},
}
adult_config = {
"model_type": GenerativeModelType.VAE,
"name": "vae_experiment_mi_attack",
"dataset": "adult.csv",
"data_path": "data/",
"data_processing": {},
"model_config": {
"compress_dims": [128, 128],
"decompress_dims": [128, 128],
"compress_activations": [Activation.LeakyReLU, Activation.LeakyReLU],
"decompress_activations": [Activation.LeakyReLU, Activation.LeakyReLU],
"latent_dim": 128,
"output_activations": {
"continuous": OutputActivation.TANH,
"categorical": OutputActivation.SOFTMAX,
},
},
"model_train": {
"seed": 10,
"test_pct": 0.,
"k_fold": False,
"stratified_by_col": None,
"epochs": 500,
"batch_size": 128,
"dp_optimizer_type": OptimizerType.DPAdamGaussianOptimizer,
"record_gradients": {"enabled": False, "subset": []},
"transformer": {
"type": TransformerType.BGM,
"kwargs": {"outlier_clipping": True, "n_clusters": 6},
},
"early_stop": {"enabled": False},
"early_stop_epsilon": {"enabled": True, "value": 2.75},
},
"diff_priv": {
"enabled": True,
"microbatches": 1,
"l2_norm_clip": 5.0,
"noise_multiplier": 0.8,
},
}
batch_norm = [True, False]
for with_batch_norm in batch_norm:
cur_config = deepcopy(adult_config)
cur_config["model_config"]["batch_norm"] = with_batch_norm
cur_experiment = VaeExperiment(cur_config)
cur_experiment.run(automatic_overwrite=False)
import time
start = time.time()
batch_norm = [True, False]
for with_batch_norm in batch_norm:
cur_config = deepcopy(adult_config)
cur_config["model_config"]["batch_norm"] = with_batch_norm
cur_experiment = VaeExperiment(cur_config)
cur_experiment.run(automatic_overwrite=False)
end = time.time()
print(end - start)
!/usr/bin/nvidia-smi
# datasets = ["adult.csv", "bank_marketing.csv", "uci_credit_card.csv"]
datasets = ["adult.csv", "bank_marketing.csv"]
gradient_record_subsets = {
"uci_credit_card.csv": ["SEX", "EDUCATION"],
"bank_marketing.csv": ["job"],
"adult.csv": ["race", "relationship"]
}
batch_norm = [True, False]
seeds = range(10, 11)
uci_categorical_cols = [
"SEX",
"EDUCATION",
"MARRIAGE",
"PAY_0",
"PAY_1",
"PAY_2",
"PAY_3",
"PAY_4",
"PAY_5",
"PAY_6",
"DEFAULT",
]
bank_marketing_read_csv_kwargs = {"delimiter": ";"}
bank_marketing_epsilon_value = 2.25
other_epsilon_value = 2.5
# train without dp
for seed in [10, 11]:
for with_batch_norm in batch_norm:
for dataset in datasets:
# set config - generic
cur_config = deepcopy(base_config_no_dp)
cur_config["model_train"]["seed"] = seed
cur_config["model_config"]["batch_norm"] = with_batch_norm
cur_config["dataset"] = dataset
if dataset == "uci_credit_card.csv":
cur_config["data_processing"]["categorical_columns"] = uci_categorical_cols
if dataset == "bank_marketing.csv":
cur_config["data_processing"]["read_csv_kwargs"] = bank_marketing_read_csv_kwargs
cur_config["model_train"]["record_gradients"]["subset"] = gradient_record_subsets[dataset]
# run experiemnt
cur_experiment = VaeExperiment(cur_config)
try:
cur_experiment.run(automatic_overwrite=False)
except SystemExit:
continue
test_config = {"name": "vae_experiment_bn_test_no_dp"}
existing_configs = MachineLearningEvaluator.existing_configs("data/", test_config=test_config)
eval_input = {'target_col': 'output', 'data_path': 'data/', 'config_file': '404d16aae768b97c18ce5aa3293376a5cc3f6c09.pkl'}
evaluator = MachineLearningEvaluator(**eval_input)
x = evaluator.get_experiment_data()
x["model_runs"][0]["gradient_norms"]
test_config = {"name": "vae_experiment_bn_test_dp"}
existing_configs = MachineLearningEvaluator.existing_configs("data/", test_config=test_config)
eval_input = {'target_col': 'output', 'data_path': 'data/', 'config_file': '13e18f019180e253ee4b1cf10dbe953c762eb185.pkl'}
evaluator = MachineLearningEvaluator(**eval_input)
x = evaluator.get_experiment_data()
x["model_runs"][0]["gradient_norms"]
# train with dp
for seed in [10, 11]:
for with_batch_norm in batch_norm:
for dataset in datasets:
# set config - generic
cur_config = deepcopy(base_config)
cur_config["model_train"]["seed"] = seed
cur_config["model_train"]["dp_optimizer_type"] = dp_opt
cur_config["model_config"]["batch_norm"] = with_batch_norm
cur_config["dataset"] = dataset
if dataset == "uci_credit_card.csv":
cur_config["data_processing"]["categorical_columns"] = uci_categorical_cols
cur_config["model_train"]["record_gradients"]["subset"] = gradient_record_subsets[dataset]
# set config - dp
if dataset == "bank_marketing.csv":
value = bank_marketing_epsilon_value
cur_config["data_processing"]["read_csv_kwargs"] = bank_marketing_read_csv_kwargs
else:
value = other_epsilon_value
cur_config["model_train"]["early_stop_epsilon"]["value"] = value
# run experiemnt
cur_experiment = VaeExperiment(cur_config)
try:
cur_experiment.run(automatic_overwrite=False)
except SystemExit:
continue
for seed in seeds[:5]:
for with_batch_norm in batch_norm:
for dataset in datasets:
# set config
cur_config = deepcopy(base_config_no_dp)
cur_config["model_train"]["seed"] = seed
cur_config["model_config"]["batch_norm"] = with_batch_norm
cur_config["dataset"] = dataset
if dataset == "uci_credit_card.csv":
cur_config["data_processing"]["categorical_columns"] = uci_categorical_cols
cur_config["model_train"]["record_gradients"]["subset"] = gradient_record_subsets[dataset]
# run experiemnt
cur_experiment = VaeExperiment(cur_config)
cur_experiment.run(automatic_overwrite=True)
config = {
"dataset": "uci_credit_card.csv",
"model_type": GenerativeModelType.VAE,
"name": "vae_experiment",
"data_path": "data/",
"data_processing": {
"columns_to_drop": ["ID"],
"columns_rename_map": {"default.payment.next.month": "default"},
"categorical_columns": [
"SEX",
"EDUCATION",
"MARRIAGE",
"PAY_0",
"PAY_1",
"PAY_2",
"PAY_3",
"PAY_4",
"PAY_5",
"PAY_6",
"default",
],
},
"model_config": {
"compress_dims": [128, 128],
"decompress_dims": [128, 128],
"compress_activations": [Activation.ReLU, Activation.ReLU],
"decompress_activations": [Activation.ReLU, Activation.ReLU],
"latent_dim": 128,
"output_activations": {
"continuous": OutputActivation.TANH,
"categorical": OutputActivation.SOFTMAX,
},
},
"model_train": {
"seed": 1,
"test_pct": 0.3,
"k_fold": False,
"stratified_by_col": None,
"epochs": 50,
"dp_optimizer_type": OptimizerType.DPAdamGaussianOptimizer,
},
"transformer": {
"type": TransformerType.BGM,
"kwargs": {"outlier_clipping": True, "n_clusters": 6},
},
"diff_priv": {
"enabled": True,
"microbatches": 8,
"l2_norm_clip": 5.0,
"noise_multiplier": 0.8,
},
# "early_stop_epsilon": {"enabled": True, "value": 2.1},
}
config = {
"dataset": "adult.csv",
"model_type": GenerativeModelType.VAE,
"name": "vae_experiment_classes",
"data_path": "data/",
"model_config": {
"compress_dims": [128, 128],
"decompress_dims": [128, 128],
"compress_activations": None,
"decompress_activations": None,
"latent_dim": 128,
"batch_norm": None,
"output_activations": {
"continuous": OutputActivation.TANH,
"categorical": OutputActivation.SOFTMAX,
},
},
"model_train": {
"seed": None,
"test_pct": 0.3,
"k_fold": True,
"epochs": 50,
"dp_optimizer_type": OptimizerType.DPAdamGaussianOptimizer,
"batch_size": 64
},
"transformer": {
"type": TransformerType.BGM,
"kwargs": {"outlier_clipping": True, "n_clusters": 6},
},
"diff_priv": {
"enabled": True,
"microbatches": 8,
"l2_norm_clip": 5.0,
"noise_multiplier": 0.8
},
}
all_compress_acts = [[Activation.LeakyReLU, Activation.LeakyReLU], [Activation.ELU, Activation.ELU]]
all_decompress_acts = [[Activation.LeakyReLU, Activation.LeakyReLU], [Activation.ELU, Activation.ELU]]
for seed in range(10):
config["model_train"]["seed"] = seed
for compress_acts, decompress_acts in list(zip(all_compress_acts, all_decompress_acts)):
config["model_config"]["compress_activations"] = compress_acts
config["model_config"]["decompress_activations"] = decompress_acts
for batch_norm in [True, False]:
config["model_config"]["batch_norm"] = batch_norm
vae_experiment = VaeExperiment(config)
vae_experiment.run()
test_config_bn = {
'dataset': 'adult.csv',
'diff_priv': {"l2_norm_clip": 5, "enabled": True},
'model_train': {"dp_optimizer_type": 2},
'transformer': {'kwargs': {'n_clusters': 6}, 'type': 1},
"model_config": {"batch_norm": True}
}
test_config = {
'dataset': 'adult.csv',
'diff_priv': {"l2_norm_clip": 5, "enabled": True},
'model_train': {'stratified_by_col': None, "dp_optimizer_type": 2},
'transformer': {'kwargs': {'n_clusters': 6}, 'type': 1}
}
test_config["name"] = 'vae_experiment_noise08'
def plot_data_utility_scores(
target_col,
data_path,
configs,
model,y
metrics,
include_real=False,
scaler_type=None,
scaler_kwargs=None,
model_kwargs=None,
metric_kwargs=None,
holdout=None,
subset=None,
):
all_group_scores = {}
for group_name, group_configs in configs.items():
group_scores = []
for config_hash, config in group_configs.items():
eval_input = {'target_col': target_col, 'data_path': data_path,
'config_file': config_hash}
evaluator = MachineLearningEvaluator(**eval_input)
scores = evaluator.data_utility_scores(
model=model,
metrics=metrics,
scaler_type=scaler_type,
scaler_kwargs=scaler_kwargs,
model_kwargs=model_kwargs,
metric_kwargs=metric_kwargs,
holdout=holdout,
subset=subset,
include_train=False
)
group_scores.append(scores)
all_group_scores[group_name] = pd.concat(group_scores, axis=0)
columns = list(scores.columns)
cols_to_plot = [col for col in columns if "fake" in col]
img_per_row = 3
rows = int(np.ceil(len(cols_to_plot) / img_per_row))
fig, axes = plt.subplots(rows, img_per_row, figsize=(20, rows*5))
if axes.ndim == 1:
axes = np.expand_dims(axes, 0)
fig.subplots_adjust(hspace=0.35, wspace=0.2)
for i, row in enumerate(axes):
for j, ax in enumerate(row):
col_idx = i * img_per_row + j
if col_idx < len(cols_to_plot):
col_name = cols_to_plot[col_idx]
for group_name, score_df in all_group_scores.items():
sns.distplot(score_df.loc[:, col_name].values, label=group_name, ax=ax)
if include_real:
col_name_real = col_name.replace("fake", "real")
sns.distplot(score_df.loc[:, col_name_real].values, label="real", ax=ax)
ax.set_title("_".join(col_name.split("_")[2:]))
ax.legend()
else:
break
existing_configs = MachineLearningEvaluator.existing_configs("data/", test_config=test_config)
# existing_configs = {k: v for i, (k, v) in enumerate(existing_configs.items()) if i < 2}
existing_configs.keys()
existing_configs_bn = MachineLearningEvaluator.existing_configs("data/", test_config=test_config_bn)
# existing_configs_bn = {k: v for i, (k, v) in enumerate(existing_configs_bn.items()) if i < 2}
existing_configs_bn.keys()
configs = {"with_BatchNorm": existing_configs_bn, "without_BatchNorm": existing_configs}
plot_data_utility_scores(
target_col="output",
data_path="data/",
configs=configs,
include_real=True,
model=EvaluatorModelType.LogisticRegression,
metrics=[MetricType.F1],
scaler_type=ScalerType.ROBUST,
model_kwargs={"solver": "lbfgs", "class_weight": "balanced", "max_iter": 200},
metric_kwargs={MetricType.F1: {"average": None}},
subset="race"
)
plot_data_utility_scores(
target_col="output",
data_path="data/",
configs=configs,
include_real=True,
model=EvaluatorModelType.LogisticRegression,
metrics=[MetricType.F1],
scaler_type=ScalerType.ROBUST,
model_kwargs={"solver": "lbfgs", "class_weight": "balanced", "max_iter": 200},
metric_kwargs={MetricType.F1: {"average": None}},
subset="race",
)
config = "ef9980840b657869103bbfbbec1f7b02b2bc2dc6.pkl" # vae_experiment2 - same configuration as [1]
eval_input = {'target_col': 'default', 'data_path': 'data/', 'config_file': config}
evaluator = MachineLearningEvaluator(**eval_input)
experiment_data = evaluator.get_experiment_data()
scores = evaluator.data_utility_scores(
EvaluatorModelType.LogisticRegression,
[MetricType.F1],
ScalerType.ROBUST,
model_kwargs={"solver": "lbfgs", "class_weight": "balanced", "max_iter": 100},
metric_kwargs={MetricType.F1: {"average": None}},
subset="SEX",
include_train=False,
)
display(scores)
existing_configs = MachineLearningEvaluator.existing_configs("data/", test_config=test_config)
for k, v in existing_configs.items():
eval_input = {'target_col': 'output', 'data_path': 'data/', 'config_file': k}
evaluator = MachineLearningEvaluator(**eval_input)
experiment_data = evaluator.get_experiment_data()
scores = evaluator.data_utility_scores(
EvaluatorModelType.LogisticRegression,
[MetricType.F1],
ScalerType.ROBUST,
model_kwargs={"solver": "lbfgs", "class_weight": "balanced", "max_iter": 200},
metric_kwargs={MetricType.F1: {"average": "binary"}},
subset="race",
include_train=False,
)
display(scores)
for k, v in existing_configs.items():
eval_input = {'target_col': 'output', 'data_path': 'data/', 'config_file': k}
evaluator = MachineLearningEvaluator(**eval_input)
experiment_data = evaluator.get_experiment_data()
scores = evaluator.data_utility_scores(
EvaluatorModelType.LogisticRegression,
[MetricType.F1],
ScalerType.ROBUST,
model_kwargs={"solver": "lbfgs", "class_weight": "balanced", "max_iter": 200},
metric_kwargs={MetricType.F1: {"average": "binary"}},
subset="race",
include_train=False,
)
display(scores)
| 0.666062 | 0.687276 |
```
import requests
import time
import random
import json
import os
import sqlite3
diretorio_dados = f'{os.path.abspath("../dados")}{os.path.sep}'
def baixar_dados_episodios():
TEMPO_MINIMO=3.0
TEMPO_MAXIMO=15.0
resultados = list()
url_inicial = 'https://api.simplecast.com/podcasts/ab2964e7-bcad-4f2f-9698-45cb681f0d69/episodes?limit=15&offset=0&private=false&sort=desc&status=published'
headers = {
'authority': 'api.simplecast.com',
'accept': 'application/json, text/plain, */*',
'user-agent': 'Mozilla/5.0 (X11; Linux x86_64) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/85.0.4183.121 Safari/537.36',
'dnt': '1',
'sec-fetch-site': 'same-site',
'sec-fetch-mode': 'cors',
'sec-fetch-dest': 'empty',
'origin': 'https://naruhodo-615be80d.simplecast.com',
'referer': 'https://naruhodo-615be80d.simplecast.com/',
'accept-language': 'en-US,en;q=0.9,pt-BR;q=0.8,pt;q=0.7'
}
requisicao = requests.get(url_inicial, headers=headers)
resultado = requisicao.json()
resultados.extend(resultado['collection'])
print(f'Página 1 de {resultado["pages"]["total"]}.')
while resultado['pages']['next'] != None:
url = resultado['pages']['next']['href']
soneca = random.uniform(TEMPO_MINIMO, TEMPO_MAXIMO)
print(f'Sleep {soneca:.2f}')
time.sleep(soneca)
requisicao = requests.get(url, headers=headers)
resultado = requisicao.json()
resultados.extend(resultado['collection'])
print(f'Página {resultado["pages"]["current"]} de {resultado["pages"]["total"]}.')
return resultados
def salvar_dados_episodios(dados):
with open(f'{diretorio_dados}episodios.json', 'w') as arquivo:
json.dump(dados, arquivo)
def executar():
with sqlite3.connect(f'{diretorio_dados}dados_episodios.db') as conexao:
conexao.execute('''
CREATE TABLE IF NOT EXISTS episodio (
id TEXT NOT NULL PRIMARY KEY,
numero INTEGER,
titulo TEXT,
descricao TEXT,
duracao INTEGER,
pulicacao DATETIME
);
''')
episodeos = baixar_dados_episodios()
salvar_dados_episodios(episodeos)
existentes = {item[0] for item in conexao.execute('select id from episodio').fetchall()}
for episodio in episodeos:
if not episodio['id'] in existentes:
conexao.execute('insert into episodio values (?, ?, ?, ?, ?, ?)',
(episodio['id'], episodio['number'], episodio['title'], episodio['description'], episodio['duration'], episodio['published_at']))
print('Processo concluído')
executar()
```
|
github_jupyter
|
import requests
import time
import random
import json
import os
import sqlite3
diretorio_dados = f'{os.path.abspath("../dados")}{os.path.sep}'
def baixar_dados_episodios():
TEMPO_MINIMO=3.0
TEMPO_MAXIMO=15.0
resultados = list()
url_inicial = 'https://api.simplecast.com/podcasts/ab2964e7-bcad-4f2f-9698-45cb681f0d69/episodes?limit=15&offset=0&private=false&sort=desc&status=published'
headers = {
'authority': 'api.simplecast.com',
'accept': 'application/json, text/plain, */*',
'user-agent': 'Mozilla/5.0 (X11; Linux x86_64) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/85.0.4183.121 Safari/537.36',
'dnt': '1',
'sec-fetch-site': 'same-site',
'sec-fetch-mode': 'cors',
'sec-fetch-dest': 'empty',
'origin': 'https://naruhodo-615be80d.simplecast.com',
'referer': 'https://naruhodo-615be80d.simplecast.com/',
'accept-language': 'en-US,en;q=0.9,pt-BR;q=0.8,pt;q=0.7'
}
requisicao = requests.get(url_inicial, headers=headers)
resultado = requisicao.json()
resultados.extend(resultado['collection'])
print(f'Página 1 de {resultado["pages"]["total"]}.')
while resultado['pages']['next'] != None:
url = resultado['pages']['next']['href']
soneca = random.uniform(TEMPO_MINIMO, TEMPO_MAXIMO)
print(f'Sleep {soneca:.2f}')
time.sleep(soneca)
requisicao = requests.get(url, headers=headers)
resultado = requisicao.json()
resultados.extend(resultado['collection'])
print(f'Página {resultado["pages"]["current"]} de {resultado["pages"]["total"]}.')
return resultados
def salvar_dados_episodios(dados):
with open(f'{diretorio_dados}episodios.json', 'w') as arquivo:
json.dump(dados, arquivo)
def executar():
with sqlite3.connect(f'{diretorio_dados}dados_episodios.db') as conexao:
conexao.execute('''
CREATE TABLE IF NOT EXISTS episodio (
id TEXT NOT NULL PRIMARY KEY,
numero INTEGER,
titulo TEXT,
descricao TEXT,
duracao INTEGER,
pulicacao DATETIME
);
''')
episodeos = baixar_dados_episodios()
salvar_dados_episodios(episodeos)
existentes = {item[0] for item in conexao.execute('select id from episodio').fetchall()}
for episodio in episodeos:
if not episodio['id'] in existentes:
conexao.execute('insert into episodio values (?, ?, ?, ?, ?, ?)',
(episodio['id'], episodio['number'], episodio['title'], episodio['description'], episodio['duration'], episodio['published_at']))
print('Processo concluído')
executar()
| 0.305594 | 0.133811 |
```
# Importing the necessary libraries
from sklearn.linear_model import LogisticRegression
from sklearn.svm import SVC
from sklearn.ensemble import RandomForestClassifier
from sklearn.datasets import load_digits
from sklearn.model_selection import train_test_split
from sklearn.model_selection import KFold
from sklearn.model_selection import StratifiedKFold
from sklearn.model_selection import cross_val_score
import numpy as np
import warnings
warnings.filterwarnings("ignore")
```
#### Creating the models with train_test_split (i.e Without K-fold cross validation)
```
# Loading the datasets
digits = load_digits()
# Splitting the dataset into train and test data.
X_train, X_test, y_train, y_test = train_test_split(digits.data, digits.target, test_size=0.3)
# Logistic Regression
lr = LogisticRegression()
lr.fit(X_train,y_train)
lr.score(X_test, y_test)
# SVM
svm = SVC(gamma='auto')
svm.fit(X_train, y_train)
svm.score(X_test, y_test)
# RandomForest
rf = RandomForestClassifier(n_estimators=40)
rf.fit(X_train,y_train)
rf.score(X_test,y_test)
```
### KFold cross validation
Example
```
kf = KFold(n_splits=3)
kf
for train_index, test_index in kf.split(['a','b','c','d','e','f','g','h','i']):
print (train_index, test_index)
```
Applying KFold on digit dataset
```
def score(model, X_train, X_test, y_train, y_test):
model.fit(X_train, y_train)
return model.score(X_test,y_test)
folds = StratifiedKFold(n_splits=3)
logistic_scores = []
svm_scores = []
rf_scores = []
for train_index,test_index in folds.split(digits.data, digits.target):
X_train, X_test, y_train, y_test = digits.data[train_index], digits.data[test_index],\
digits.target[train_index],digits.target[test_index]
logistic_scores.append(score(LogisticRegression(solver='liblinear',multi_class='ovr'),X_train,X_test,y_train,y_test))
svm_scores.append(score(SVC(gamma='auto'),X_train,X_test,y_train,y_test))
rf_scores.append(score(RandomForestClassifier(n_estimators=40),X_train,X_test,y_train,y_test))
logistic_scores
svm_scores
rf_scores
```
### Using CrossValidation function
```
cross_val_score(LogisticRegression(solver='liblinear',multi_class='ovr'),digits.data, digits.target, cv=3)
cross_val_score(SVC(gamma='auto'),digits.data, digits.target, cv=3)
cross_val_score(RandomForestClassifier(n_estimators=40),digits.data, digits.target, cv=3)
```
### Parameter tuning of RandomForest Classifier
```
score_1 = cross_val_score(RandomForestClassifier(n_estimators=5),digits.data, digits.target, cv=10)
np.average(score_1)
score_2= cross_val_score(RandomForestClassifier(n_estimators=10),digits.data, digits.target, cv=10)
np.average(score_2)
score_3 = cross_val_score(RandomForestClassifier(n_estimators=20),digits.data, digits.target, cv=10)
np.average(score_3)
score_4 = cross_val_score(RandomForestClassifier(n_estimators=40),digits.data, digits.target, cv=10)
np.average(score_4)
```
|
github_jupyter
|
# Importing the necessary libraries
from sklearn.linear_model import LogisticRegression
from sklearn.svm import SVC
from sklearn.ensemble import RandomForestClassifier
from sklearn.datasets import load_digits
from sklearn.model_selection import train_test_split
from sklearn.model_selection import KFold
from sklearn.model_selection import StratifiedKFold
from sklearn.model_selection import cross_val_score
import numpy as np
import warnings
warnings.filterwarnings("ignore")
# Loading the datasets
digits = load_digits()
# Splitting the dataset into train and test data.
X_train, X_test, y_train, y_test = train_test_split(digits.data, digits.target, test_size=0.3)
# Logistic Regression
lr = LogisticRegression()
lr.fit(X_train,y_train)
lr.score(X_test, y_test)
# SVM
svm = SVC(gamma='auto')
svm.fit(X_train, y_train)
svm.score(X_test, y_test)
# RandomForest
rf = RandomForestClassifier(n_estimators=40)
rf.fit(X_train,y_train)
rf.score(X_test,y_test)
kf = KFold(n_splits=3)
kf
for train_index, test_index in kf.split(['a','b','c','d','e','f','g','h','i']):
print (train_index, test_index)
def score(model, X_train, X_test, y_train, y_test):
model.fit(X_train, y_train)
return model.score(X_test,y_test)
folds = StratifiedKFold(n_splits=3)
logistic_scores = []
svm_scores = []
rf_scores = []
for train_index,test_index in folds.split(digits.data, digits.target):
X_train, X_test, y_train, y_test = digits.data[train_index], digits.data[test_index],\
digits.target[train_index],digits.target[test_index]
logistic_scores.append(score(LogisticRegression(solver='liblinear',multi_class='ovr'),X_train,X_test,y_train,y_test))
svm_scores.append(score(SVC(gamma='auto'),X_train,X_test,y_train,y_test))
rf_scores.append(score(RandomForestClassifier(n_estimators=40),X_train,X_test,y_train,y_test))
logistic_scores
svm_scores
rf_scores
cross_val_score(LogisticRegression(solver='liblinear',multi_class='ovr'),digits.data, digits.target, cv=3)
cross_val_score(SVC(gamma='auto'),digits.data, digits.target, cv=3)
cross_val_score(RandomForestClassifier(n_estimators=40),digits.data, digits.target, cv=3)
score_1 = cross_val_score(RandomForestClassifier(n_estimators=5),digits.data, digits.target, cv=10)
np.average(score_1)
score_2= cross_val_score(RandomForestClassifier(n_estimators=10),digits.data, digits.target, cv=10)
np.average(score_2)
score_3 = cross_val_score(RandomForestClassifier(n_estimators=20),digits.data, digits.target, cv=10)
np.average(score_3)
score_4 = cross_val_score(RandomForestClassifier(n_estimators=40),digits.data, digits.target, cv=10)
np.average(score_4)
| 0.786172 | 0.905615 |
<link rel="stylesheet" href="../../styles/theme_style.css">
<!--link rel="stylesheet" href="../../styles/header_style.css"-->
<link rel="stylesheet" href="https://cdnjs.cloudflare.com/ajax/libs/font-awesome/4.7.0/css/font-awesome.min.css">
<table width="100%">
<tr>
<td id="image_td" width="15%" class="header_image_color_7"><div id="image_img"
class="header_image_15"></div></td>
<td class="header_text"> BIOSTEC 2019 - Hands On biosignalsnotebooks </td>
</tr>
</table>
<div id="flex-container">
<div id="diff_level" class="flex-item">
<strong>Difficulty Level:</strong> <span class="fa fa-star checked"></span>
<span class="fa fa-star checked"></span>
<span class="fa fa-star checked"></span>
<span class="fa fa-star"></span>
<span class="fa fa-star"></span>
</div>
<div id="tag" class="flex-item-tag">
<span id="tag_list">
<table id="tag_list_table">
<tr>
<td class="shield_left">Tags</td>
<td class="shield_right" id="tags">other☁biostec☁exercises</td>
</tr>
</table>
</span>
<!-- [OR] Visit https://img.shields.io in order to create a tag badge-->
</div>
</div>
Reaching the final segment of our amazing journey between <span class="color7"><strong>Signal Acquisition</strong></span> and <span class="color13"><strong>Signal Processing</strong></span> , with <span class="color1"><strong>biosignalsplux</strong></span> and <span class="color1"><strong>biosignalsnotebooks</strong></span>, there is a relevant element missing...
Navigate by yourself, explore this programming environment and find new questions to be solved !
With the current <span class="color4"><strong>Jupyter Notebook</strong></span> we have a challenge for you. After concluding the final step of this tutorial you will see how easy the signal processing tasks can be <span style="font-size:14pt">☺</span>
<hr>
<p class="steps">1 - Get shareable link of the acquisition file with the signals collected during demonstration</p>
<p class="steps">1.1 - Access the following link to get into the <span class="color7">Google Drive</span> folder: <a href="https://drive.google.com/drive/folders/1hyLRonEVDPIy6uLdzuWaBHqsjA8a3rUR?usp=sharing">https://drive.google.com/drive/folders/1hyLRonEVDPIy6uLdzuWaBHqsjA8a3rUR?usp=sharing</a></p>
<p class="steps">1.2 - Open the file entitled <span class="color1">"acquisition_link.txt"</span></p>
<p class="steps">1.3 - Copy the link contained inside it</p>
<span class="color13"><strong>Graphical Tip <span style="font-size:14pt">☄</span></strong></span>
<img src="../../images/other/biostec_hands_on_load.gif">
<p class="steps">2 - On the following Python instruction replace <span class="color7">"File_Url"</span> field with the copied url</p>
<span class="color7"><strong>Look ! <span style="font-size:14pt">☄</span></strong></span> We are using our biosignalsnotebooks package functionalities for the first time
```
import biosignalsnotebooks as bsnb
data, header = bsnb.load_signal("File_Url", get_header=True)
```
After the previous step you loaded all the acquired data from a remote location... It is really easy !!!
Taking into consideration that we are using a <span class="color4"><strong>Jupyter Server</strong></span> through <a href="https://mybinder.org/"><strong>binder <img src="../../images/icons/link.png" width="10px" height="10px" style="display:inline"></strong></a> this is the easiest procedure.
However, if you are feeling confident and motivated to start a great adventure in your own local environment, there are other options to program in <span class="color1"><strong>Python</strong></span> and to load data from the generated files.
The following sequence of <span class="color4"><strong>Jupyter Notebooks</strong></span> may be a good resource to use after the workshop:
<ol>
<li><a href="../Install/prepare_jupyter.ipynb"><strong>Download, Install and Execute Jypyter Notebook Environment <img src="../../images/icons/link.png" width="10px" height="10px" style="display:inline"></strong></a></li>
<li><a href="../Connect/pairing_device.ipynb"><strong>Pairing a Device at Windows 10 [biosignalsplux] <img src="../../images/icons/link.png" width="10px" height="10px" style="display:inline"></strong></a></li>
<li><a href="../Record/record_data.ipynb"><strong>Signal Acquisition [OpenSignals] <img src="../../images/icons/link.png" width="10px" height="10px" style="display:inline"></strong></a></li>
<li><a href="../Record/store_signals_after_acquisition.ipynb"><strong>Store Files after Acquisition [OpenSignals] <img src="../../images/icons/link.png" width="10px" height="10px" style="display:inline"></strong></a></li>
<li><a href="../Load/open_txt.ipynb"><strong>Load acquired data from .txt file</strong></a> or <a href="../Load/open_h5.ipynb"><strong>Load acquired data from .h5 file <img src="../../images/icons/link.png" width="10px" height="10px" style="display:inline"></strong></a></li>
</ol>
<strong> Let's see which is the format of <span class="color1">data</span> and <span class="color7">header</span> variables</strong>
```
print("header\n" + str(header))
print("\ndata\n" + str(data))
```
In spite of all acquired data and metadata be stored at <span class="color1">data</span> and <span class="color7">header</span> variables, this information is organised in a dictionary.
<p class="steps">3 - Store data of EMG channel in a new variable</p>
<span class="color13"><strong>Quick Tip <span style="font-size:14pt">☄</span></strong></span> Remember that the returned dictionary is an hierarchic structure where each level is a pair ("key" and "value"). For accessing our "values" (acquired samples) we need to invoke the respective "key" (channel identifier) in the following format:
\>>> <strong>variable_name = dict["key"]</strong>
<p class="steps">4 - Replicating the same logic of step 3, store the acquisition sampling rate value in another variable</p>
<p class="steps">5 - From the acquired EMG signal detect the start and end of each muscular activation period</p>
<span class="color13"><strong>Quick Tip <span style="font-size:14pt">☄</span></strong></span>
There is a <span class="color4">Jupyter Notebook</span> dedicated to this task (<a href="../Detect/detect_bursts.ipynb"><strong>Event Detection - Muscular Activations (EMG) <img src="../../images/icons/link.png" width="10px" height="10px" style="display:inline"></strong></a>) where muscular activation's are detected through a single threshold methodology. Eventually... you will find a shortcut at the end of the Notebook
The first two entries of the returned list (<strong>activation_data</strong>) contain sublists with the sample numbers where the activation periods start (1st entry) and the samples where muscular activation's end (2nd entry).
<p class="steps">6 - Now it will be interesting to estimate the average, minimum and maximum duration of these activation periods</p>
<span class="color13"><strong>Quick Tip <span style="font-size:14pt">☄</span></strong></span>
Step 5.2 of <span class="color4">Jupyter Notebook</span> entitled <a href="../Extract/emg_parameters.ipynb"><strong>EMG Analysis - Time and Frequency Parameters <img src="../../images/icons/link.png" width="10px" height="10px" style="display:inline"></strong></a> contains almost everything you need. We are working with <strong>sample numbers</strong>, however, for the current determination it is relevant to present the results in seconds.
Converting your "sample differences" into seconds is very easy when sampling rate is known (and you know it !).
For further help, please, see the <a href="../Pre-Process/generation_of_time_axis.ipynb"><strong>Generation of a time axis (conversion of samples into seconds)
<img src="../../images/icons/link.png" width="10px" height="10px" style="display:inline"></strong></a> <span class="color4">Jupyter Notebook</span> or call me <span style="font-size:14pt">☺</span>.
<p class="steps">7 - Convert the sample values of our EMG signal to physical units (mV)</p>
<span class="color13"><strong>Quick Tip <span style="font-size:14pt">☄</span></strong></span>
Like referred during the presentation, each <span class="color1"><strong>biosignalsplux</strong></span> sensor has a specific transfer function for converting the analog to digital converter values into physical units.
Check our <a href="../Extract/emg_parameters.ipynb"><strong>EMG Sensor - Unit Conversion <img src="../../images/icons/link.png" width="10px" height="10px" style="display:inline"></strong></a> Notebook for getting help related with this task.
<p class="steps">8 - Estimate the average amplitude of the overall EMG signal, using Root Mean Square (RMS)</p>
<span class="color13"><strong>Quick Tip <span style="font-size:14pt">☄</span></strong></span>
Returning to <span class="color4">Jupyter Notebook</span> entitled <a href="../Extract/emg_parameters.ipynb"><strong>EMG Analysis - Time and Frequency Parameters <img src="../../images/icons/link.png" width="10px" height="10px" style="display:inline"></strong></a>, step 5.4 explains how to achieve this result !
There is a general agreement that "Hello World" (in spite of not officially proclaimed) is the ideal program to get into each new programming language.
With the current <span class="color1"><strong>Hands On</strong></span> exercise you don't print the message <strong>"Hello World !"</strong> but you really say <span class="color7"><strong>"Hello to biosignalsnotebooks !"</strong></span>, which is a great new world ⛿
<strong><span class="color7">We hope that you have enjoyed this guide. </span><span class="color2">biosignalsnotebooks</span><span class="color4"> is an environment in continuous expansion, so don't stop your journey and learn more with the remaining <a href="../MainFiles/biosignalsnotebooks.ipynb">Notebooks <img src="../../images/icons/link.png" width="10px" height="10px" style="display:inline"></a></span></strong> !
<span class="color6">**Auxiliary Code Segment (should not be replicated by
the user)**</span>
```
from biosignalsnotebooks.__notebook_support__ import css_style_apply
css_style_apply()
%%html
<script>
// AUTORUN ALL CELLS ON NOTEBOOK-LOAD!
require(
['base/js/namespace', 'jquery'],
function(jupyter, $) {
$(jupyter.events).on("kernel_ready.Kernel", function () {
console.log("Auto-running all cells-below...");
jupyter.actions.call('jupyter-notebook:run-all-cells-below');
jupyter.actions.call('jupyter-notebook:save-notebook');
});
}
);
</script>
```
|
github_jupyter
|
import biosignalsnotebooks as bsnb
data, header = bsnb.load_signal("File_Url", get_header=True)
print("header\n" + str(header))
print("\ndata\n" + str(data))
from biosignalsnotebooks.__notebook_support__ import css_style_apply
css_style_apply()
%%html
<script>
// AUTORUN ALL CELLS ON NOTEBOOK-LOAD!
require(
['base/js/namespace', 'jquery'],
function(jupyter, $) {
$(jupyter.events).on("kernel_ready.Kernel", function () {
console.log("Auto-running all cells-below...");
jupyter.actions.call('jupyter-notebook:run-all-cells-below');
jupyter.actions.call('jupyter-notebook:save-notebook');
});
}
);
</script>
| 0.115886 | 0.685765 |
<a href="https://colab.research.google.com/github/erickaalgr/CpEN-21A-BSCpE-1-1/blob/main/Control_Structure.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
###If Statement
```
a=12
b=100
if b>a:
print("b is greater than a")
```
###Elif Condition
```
a=12
b=12
if b>a:
print("b is greater than a")
elif a==b:
print("a is equal to b")
```
###Else Statement
```
a=12
b=12
if b>a:
print("b is greater than a")
elif a==b:
print("a is equal to b")
else:
print("b is less than a")
```
###Short Hand If Statement
```
if b>a: print("b is greater than a")
```
###Short Hand If... Else Statement
```
print("a is greater than b") if a>b else print("b is greter than a")
```
###And Condition
```
a=15
b=35
if b>a and a<b:
print("Both conditions are True")
elif b<a and a>b:
print("a is larger than b")
else:
print("none of the above")
```
###Or Condition
```
a=15
b=35
if b>a or a==b:
print("True")
elif b==b or a>b:
print("Absolutely True")
else:
print("Nothing to compare")
a=35
b=20
if b>a or a==b:
print("True")
else:
print("False")
```
###Nested If
```
x=41
if x>10:
print("Above 10")
if x>20:
print("Above 20")
if x>30:
print("Above 30")
else:
print("Not above 30")
else:
print("Not above 20")
else:
print("Not above 10")
x=5
if x>10:
print("Above 10")
if x>20:
print("Above 20")
if x>30:
print("Above 30")
else:
print("Not above 30")
else:
print("Not above 20")
else:
print("Not above 10")
```
###Application of If... Else Statement
```
#Example 1
age= 18
if age>=18:
print("You are qualified to vote!")
else:
print("You are not qualified to vote!")
#Example 1
age= 17
if age>=18:
print("You are qualified to vote!")
else:
print("You are not qualified to vote!")
#Example 1
age= int(input("Enter your age:"))
if age>=18:
print("You are qualified to vote!")
else:
print("You are not qualified to vote!")
#Example 2 to check whether 0, negative or positive number
number= int(input("Enter the number:"))
if number==0:
print("Zero")
elif number>0:
print("Positive")
else:
print("Negative")
#Example 3
grade=int(input("Enter the number:"))
if grade>=75:
print("Passed")
elif grade==74:
print("Remedial")
else:
print("Failed")
#Example 3
grade=int(input("Enter the number:"))
if grade>=75:
print("Passed")
elif grade==74:
print("Remedial")
else:
print("Failed")
#Example 3
grade=int(input("Enter the number:"))
if grade>=75:
print("Passed")
elif grade==74:
print("Remedial")
else:
print("Failed")
```
|
github_jupyter
|
a=12
b=100
if b>a:
print("b is greater than a")
a=12
b=12
if b>a:
print("b is greater than a")
elif a==b:
print("a is equal to b")
a=12
b=12
if b>a:
print("b is greater than a")
elif a==b:
print("a is equal to b")
else:
print("b is less than a")
if b>a: print("b is greater than a")
print("a is greater than b") if a>b else print("b is greter than a")
a=15
b=35
if b>a and a<b:
print("Both conditions are True")
elif b<a and a>b:
print("a is larger than b")
else:
print("none of the above")
a=15
b=35
if b>a or a==b:
print("True")
elif b==b or a>b:
print("Absolutely True")
else:
print("Nothing to compare")
a=35
b=20
if b>a or a==b:
print("True")
else:
print("False")
x=41
if x>10:
print("Above 10")
if x>20:
print("Above 20")
if x>30:
print("Above 30")
else:
print("Not above 30")
else:
print("Not above 20")
else:
print("Not above 10")
x=5
if x>10:
print("Above 10")
if x>20:
print("Above 20")
if x>30:
print("Above 30")
else:
print("Not above 30")
else:
print("Not above 20")
else:
print("Not above 10")
#Example 1
age= 18
if age>=18:
print("You are qualified to vote!")
else:
print("You are not qualified to vote!")
#Example 1
age= 17
if age>=18:
print("You are qualified to vote!")
else:
print("You are not qualified to vote!")
#Example 1
age= int(input("Enter your age:"))
if age>=18:
print("You are qualified to vote!")
else:
print("You are not qualified to vote!")
#Example 2 to check whether 0, negative or positive number
number= int(input("Enter the number:"))
if number==0:
print("Zero")
elif number>0:
print("Positive")
else:
print("Negative")
#Example 3
grade=int(input("Enter the number:"))
if grade>=75:
print("Passed")
elif grade==74:
print("Remedial")
else:
print("Failed")
#Example 3
grade=int(input("Enter the number:"))
if grade>=75:
print("Passed")
elif grade==74:
print("Remedial")
else:
print("Failed")
#Example 3
grade=int(input("Enter the number:"))
if grade>=75:
print("Passed")
elif grade==74:
print("Remedial")
else:
print("Failed")
| 0.064875 | 0.829837 |
# NumPy cheat sheet 2

## Array slicing and searching
**ToC**
- [Array slicing](#Array-slicing)
- [nD array slicing](#nD-array-slicing)
- [Array dicing](#Array-dicing)
- [Array broadcasting](#Array-broadcasting)
- [Deep copy](#Deep-copy)
- [Array searching](#Array-searching)
- [Array operations](#Array-operations)
- [NumPy Caveats](#Caveats)
- [Universal functions](#Universal-functions)
```
import numpy as np
arr1 = np.random.randint(10,30, size=8)
arr1
arr2 = np.random.randint(20,200,size=50).reshape(5,10) #method chaining - numbers from 0 to 50
arr2
```
## Array slicing
get elements using index like in a List
```
arr1[0]
arr1[3]
arr1[:3] #get the first 3 elements. Gets lower bounds inclusive, upper bound exclusive
arr1[2:] #lower bound inclusive
arr1[2:5] #get elements at index 2,3,4
```
### nD array slicing
```
arr2
arr2[0,0] #style 1 - you pass in a list of indices
arr2[0][0] #style 2 - parse it as list of lists - not so popular
arr2[1] # get a full row
```
### Array dicing
```
#get the second column
arr2[:,1]
```
Thus, you specify `:` for all columns, followed by `1` for column. And you get a 1D array of the result
```
#get the 3rd row
arr2[2,:] #which is same as arr2[2]
#get the center 3,3 elements - columns 4,5,6 and rows 1,2,3
arr2[1:4, 4:7]
```
## Array broadcasting
NumPy allows bulk assigning values, just like in matlab
```
arr2
arr2_subset = arr2[1:4, 4:7]
arr2_subset
arr2_subset[:,:] = 999 #assign this entire numpy the same values
arr2_subset
```
## Deep copy
NumPy Arrays like Python objects are always shallow copied. Hence any modification made in derivative affects the source.
Make **deep copies** using **`copy()`** method
```
arr2 #notice the 999 in the middle
arr2_subset_a = arr2_subset
arr2_subset_a is arr2_subset
```
Notice they are same obj in memory
```
arr3_subset = arr2_subset.copy()
arr3_subset
arr3_subset is arr2_subset
```
Notice they are different objects in memory. Thus changing arr3_subset will not affect its source
```
arr3_subset[:,:] = 0.1
arr2_subset
```
## Array searching
Use matlab style array searching
```
arr1
```
Get all numbers greater than 15
```
arr1[arr1 > 15]
arr1[arr1 > 12]
```
just the condition returns a boolean matrix of same dimension as the one being queried
```
arr1 > 12
arr2[arr2 > 50] #looses the original shape as its impossible to keep the 2D shape
arr2[arr2 < 30]
```
## Array operations
NumPy has operators like `+`, `-`, `/`, `*` overloaded so you can add two matrices like scalars
```
arr_sum = arr1 + arr1
arr_sum
arr_cubed = arr1 ** 2
arr_cubed
```
Similarly, you can add a scalar to an array and NumPy will `broadcast` that operation on all the elements.
```
arr_cubed - 100
```
### Caveats
Numpy does not throw errors for divide by zero or for 0/0. Intead it sets value to `inf` and `nan`.
```
arr_cubed[0] = 0
arr_cubed
arr_cubed / 0
```
Thus 0/0 = `nan` and num/0 = `inf`
## Universal functions
Numpy has a bunch of [universal functions](https://docs.scipy.org/doc/numpy/reference/ufuncs.html) that work on the array elements one at a time and allow arrays to be used or treated as scalars.
Before writing a loop, look up the [function list here](https://docs.scipy.org/doc/numpy/reference/ufuncs.html#available-ufuncs)
|
github_jupyter
|
import numpy as np
arr1 = np.random.randint(10,30, size=8)
arr1
arr2 = np.random.randint(20,200,size=50).reshape(5,10) #method chaining - numbers from 0 to 50
arr2
arr1[0]
arr1[3]
arr1[:3] #get the first 3 elements. Gets lower bounds inclusive, upper bound exclusive
arr1[2:] #lower bound inclusive
arr1[2:5] #get elements at index 2,3,4
arr2
arr2[0,0] #style 1 - you pass in a list of indices
arr2[0][0] #style 2 - parse it as list of lists - not so popular
arr2[1] # get a full row
#get the second column
arr2[:,1]
#get the 3rd row
arr2[2,:] #which is same as arr2[2]
#get the center 3,3 elements - columns 4,5,6 and rows 1,2,3
arr2[1:4, 4:7]
arr2
arr2_subset = arr2[1:4, 4:7]
arr2_subset
arr2_subset[:,:] = 999 #assign this entire numpy the same values
arr2_subset
arr2 #notice the 999 in the middle
arr2_subset_a = arr2_subset
arr2_subset_a is arr2_subset
arr3_subset = arr2_subset.copy()
arr3_subset
arr3_subset is arr2_subset
arr3_subset[:,:] = 0.1
arr2_subset
arr1
arr1[arr1 > 15]
arr1[arr1 > 12]
arr1 > 12
arr2[arr2 > 50] #looses the original shape as its impossible to keep the 2D shape
arr2[arr2 < 30]
arr_sum = arr1 + arr1
arr_sum
arr_cubed = arr1 ** 2
arr_cubed
arr_cubed - 100
arr_cubed[0] = 0
arr_cubed
arr_cubed / 0
| 0.360714 | 0.987958 |
#1. Install Dependencies
First install the libraries needed to execute recipes, this only needs to be done once, then click play.
```
!pip install git+https://github.com/google/starthinker
```
#2. Get Cloud Project ID
To run this recipe [requires a Google Cloud Project](https://github.com/google/starthinker/blob/master/tutorials/cloud_project.md), this only needs to be done once, then click play.
```
CLOUD_PROJECT = 'PASTE PROJECT ID HERE'
print("Cloud Project Set To: %s" % CLOUD_PROJECT)
```
#3. Get Client Credentials
To read and write to various endpoints requires [downloading client credentials](https://github.com/google/starthinker/blob/master/tutorials/cloud_client_installed.md), this only needs to be done once, then click play.
```
CLIENT_CREDENTIALS = 'PASTE CREDENTIALS HERE'
print("Client Credentials Set To: %s" % CLIENT_CREDENTIALS)
```
#4. Enter DV360 API To BigQuery Parameters
Write the current state of advertisers, campaigns, sites, insertion orders, and line items to BigQuery for a given list of DV360 accounts.
1. Specify the name of the dataset, several tables will be created here.
1. If dataset exists, it is unchanged.
1. Add DV360 advertiser ids for the accounts to pull data from.
Modify the values below for your use case, can be done multiple times, then click play.
```
FIELDS = {
'endpoint': '',
'auth_read': 'user', # Credentials used for reading data.
'auth_write': 'service', # Credentials used for writing data.
'dataset': '', # Google BigQuery dataset to create tables in.
'partners': [], # Comma separated partners ids.
'advertisers': [], # Comma separated advertisers ids.
}
print("Parameters Set To: %s" % FIELDS)
```
#5. Execute DV360 API To BigQuery
This does NOT need to be modified unles you are changing the recipe, click play.
```
from starthinker.util.project import project
from starthinker.script.parse import json_set_fields
USER_CREDENTIALS = '/content/user.json'
TASKS = [
{
'dv360_api': {
'auth': 'user',
'endpoints': {'field': {'name': 'endpoint','kind': 'choice','choices': ['advertisers','advertisers.campaigns','advertisers.channels','advertisers.channels.sites','advertisers.creatives','advertisers.insertionOrders','advertisers.lineItems','advertisers.lineItems.targetingTypes','advertisers.locationLists','advertisers.locationLists.assignedLocations','advertisers.negativeKeywordLists','advertisers.negativeKeywordLists.negativeKeywords','advertisers.targetingTypes.assignedTargetingOptions','combinedAudiences','customBiddingAlgorithms','customLists','firstAndThirdPartyAudiences','floodlightGroups','googleAudiences','inventorySourceGroups','inventorySourceGroups.assignedInventorySources','inventorySources','partners','partners.channels','partners.channels.sites','floodlightActivityGroups','partners.targetingTypes.assignedTargetingOptions','targetingTypes.targetingOptions','users'],'default': ''}},
'partners': {
'single_cell': True,
'values': {'field': {'name': 'partners','kind': 'integer_list','order': 2,'default': [],'description': 'Comma separated partners ids.'}}
},
'advertisers': {
'single_cell': True,
'values': {'field': {'name': 'advertisers','kind': 'integer_list','order': 2,'default': [],'description': 'Comma separated advertisers ids.'}}
},
'out': {
'auth': 'user',
'dataset': {'field': {'name': 'dataset','kind': 'string','order': 1,'default': '','description': 'Google BigQuery dataset to create tables in.'}}
}
}
}
]
json_set_fields(TASKS, FIELDS)
project.initialize(_recipe={ 'tasks':TASKS }, _project=CLOUD_PROJECT, _user=USER_CREDENTIALS, _client=CLIENT_CREDENTIALS, _verbose=True, _force=True)
project.execute(_force=True)
```
|
github_jupyter
|
!pip install git+https://github.com/google/starthinker
CLOUD_PROJECT = 'PASTE PROJECT ID HERE'
print("Cloud Project Set To: %s" % CLOUD_PROJECT)
CLIENT_CREDENTIALS = 'PASTE CREDENTIALS HERE'
print("Client Credentials Set To: %s" % CLIENT_CREDENTIALS)
FIELDS = {
'endpoint': '',
'auth_read': 'user', # Credentials used for reading data.
'auth_write': 'service', # Credentials used for writing data.
'dataset': '', # Google BigQuery dataset to create tables in.
'partners': [], # Comma separated partners ids.
'advertisers': [], # Comma separated advertisers ids.
}
print("Parameters Set To: %s" % FIELDS)
from starthinker.util.project import project
from starthinker.script.parse import json_set_fields
USER_CREDENTIALS = '/content/user.json'
TASKS = [
{
'dv360_api': {
'auth': 'user',
'endpoints': {'field': {'name': 'endpoint','kind': 'choice','choices': ['advertisers','advertisers.campaigns','advertisers.channels','advertisers.channels.sites','advertisers.creatives','advertisers.insertionOrders','advertisers.lineItems','advertisers.lineItems.targetingTypes','advertisers.locationLists','advertisers.locationLists.assignedLocations','advertisers.negativeKeywordLists','advertisers.negativeKeywordLists.negativeKeywords','advertisers.targetingTypes.assignedTargetingOptions','combinedAudiences','customBiddingAlgorithms','customLists','firstAndThirdPartyAudiences','floodlightGroups','googleAudiences','inventorySourceGroups','inventorySourceGroups.assignedInventorySources','inventorySources','partners','partners.channels','partners.channels.sites','floodlightActivityGroups','partners.targetingTypes.assignedTargetingOptions','targetingTypes.targetingOptions','users'],'default': ''}},
'partners': {
'single_cell': True,
'values': {'field': {'name': 'partners','kind': 'integer_list','order': 2,'default': [],'description': 'Comma separated partners ids.'}}
},
'advertisers': {
'single_cell': True,
'values': {'field': {'name': 'advertisers','kind': 'integer_list','order': 2,'default': [],'description': 'Comma separated advertisers ids.'}}
},
'out': {
'auth': 'user',
'dataset': {'field': {'name': 'dataset','kind': 'string','order': 1,'default': '','description': 'Google BigQuery dataset to create tables in.'}}
}
}
}
]
json_set_fields(TASKS, FIELDS)
project.initialize(_recipe={ 'tasks':TASKS }, _project=CLOUD_PROJECT, _user=USER_CREDENTIALS, _client=CLIENT_CREDENTIALS, _verbose=True, _force=True)
project.execute(_force=True)
| 0.307774 | 0.74667 |
# Demo Import from Sklearn with Schemas from Lale
This notebook shows how to use Lale directly with sklearn operators.
The function `lale.wrap_imported_operators()` will automatically wrap
known sklearn operators into Lale operators.
## Usability
To make Lale easy to learn and use, its APIs imitate those of
[sklearn](https://scikit-learn.org/), with init, fit, and predict,
and with pipelines.
```
import sklearn.datasets
import sklearn.model_selection
digits = sklearn.datasets.load_digits()
X_train, X_test, y_train, y_test = sklearn.model_selection.train_test_split(
digits.data, digits.target, test_size=0.2, random_state=42)
print(f'truth {y_test.tolist()[:20]}')
import lale
from sklearn.linear_model import LogisticRegression as LR
lale.wrap_imported_operators()
trainable_lr = LR(LR.solver.lbfgs, C=0.0001)
trained_lr = trainable_lr.fit(X_train, y_train)
predictions = trained_lr.predict(X_test)
print(f'actual {predictions.tolist()[:20]}')
from sklearn.metrics import accuracy_score
print(f'accuracy {accuracy_score(y_test, predictions):.1%}')
```
## Correctness
Lale uses [JSON Schema](https://json-schema.org/) to check for valid
hyperparameters. These schemas enable not just validation but also
interactive documentation. Thanks to using a single source of truth, the
documentation is correct by construction.
```
from jsonschema import ValidationError
try:
lale_lr = LR(solver='adam', C=0.01)
except ValidationError as e:
print(e.message)
LR.hyperparam_schema('C')
LR.get_defaults()
```
## Automation
Lale includes a compiler that converts types (expressed as JSON
Schema) to optimizer search spaces. It currently has back-ends for
[hyperopt](http://hyperopt.github.io/hyperopt/),
[GridSearchCV](https://scikit-learn.org/stable/modules/generated/sklearn.model_selection.GridSearchCV.html), and
[SMAC](http://www.automl.org/smac/).
We are also actively working towards various other forms of AI
automation using various other tools.
```
from lale.search.op2hp import hyperopt_search_space
from hyperopt import STATUS_OK, Trials, fmin, tpe, space_eval
import lale.helpers
import warnings
warnings.filterwarnings("ignore")
def objective(hyperparams):
trainable = LR(**lale.helpers.dict_without(hyperparams, 'name'))
trained = trainable.fit(X_train, y_train)
predictions = trained.predict(X_test)
return {'loss': -accuracy_score(y_test, predictions), 'status': STATUS_OK}
search_space = hyperopt_search_space(LR)
trials = Trials()
fmin(objective, search_space, algo=tpe.suggest, max_evals=10, trials=trials)
best_hps = space_eval(search_space, trials.argmin)
print(f'best hyperparams {lale.helpers.dict_without(best_hps, "name")}\n')
print(f'accuracy {-min(trials.losses()):.1%}')
```
## Composition
Lale supports composite models, which resemble sklearn pipelines but are
more expressive.
| Symbol | Name | Description | Sklearn feature |
| ------ | ---- | ------------ | --------------- |
| >> | pipe | Feed to next | `make_pipeline` |
| & | and | Run both | `make_union`, includes concat |
| | | or | Choose one | (missing) |
```
from sklearn.decomposition import PCA
from sklearn.svm import SVC
from lale.lib.lale import ConcatFeatures as Cat
from lale.lib.lale import NoOp
lale.wrap_imported_operators()
optimizable = (PCA & NoOp) >> Cat >> (LR | SVC)
optimizable.visualize()
from lale.operators import make_pipeline, make_union, make_choice
optimizable = make_pipeline(make_union(PCA, NoOp), make_choice(LR, SVC))
optimizable.visualize()
import lale.lib.lale.hyperopt
Optimizer = lale.lib.lale.hyperopt.Hyperopt
trained = optimizable.auto_configure(X_train, y_train, optimizer=Optimizer, max_evals=10)
predictions = trained.predict(X_test)
print(f'accuracy {accuracy_score(y_test, predictions):.1%}')
trained.visualize()
```
## Input and Output Schemas
Besides schemas for hyperparameter, Lale also provides operator tags
and schemas for input and output data of operators.
```
LR.get_tags()
LR.get_schema('input_fit')
LR.get_schema('output_predict')
```
|
github_jupyter
|
import sklearn.datasets
import sklearn.model_selection
digits = sklearn.datasets.load_digits()
X_train, X_test, y_train, y_test = sklearn.model_selection.train_test_split(
digits.data, digits.target, test_size=0.2, random_state=42)
print(f'truth {y_test.tolist()[:20]}')
import lale
from sklearn.linear_model import LogisticRegression as LR
lale.wrap_imported_operators()
trainable_lr = LR(LR.solver.lbfgs, C=0.0001)
trained_lr = trainable_lr.fit(X_train, y_train)
predictions = trained_lr.predict(X_test)
print(f'actual {predictions.tolist()[:20]}')
from sklearn.metrics import accuracy_score
print(f'accuracy {accuracy_score(y_test, predictions):.1%}')
from jsonschema import ValidationError
try:
lale_lr = LR(solver='adam', C=0.01)
except ValidationError as e:
print(e.message)
LR.hyperparam_schema('C')
LR.get_defaults()
from lale.search.op2hp import hyperopt_search_space
from hyperopt import STATUS_OK, Trials, fmin, tpe, space_eval
import lale.helpers
import warnings
warnings.filterwarnings("ignore")
def objective(hyperparams):
trainable = LR(**lale.helpers.dict_without(hyperparams, 'name'))
trained = trainable.fit(X_train, y_train)
predictions = trained.predict(X_test)
return {'loss': -accuracy_score(y_test, predictions), 'status': STATUS_OK}
search_space = hyperopt_search_space(LR)
trials = Trials()
fmin(objective, search_space, algo=tpe.suggest, max_evals=10, trials=trials)
best_hps = space_eval(search_space, trials.argmin)
print(f'best hyperparams {lale.helpers.dict_without(best_hps, "name")}\n')
print(f'accuracy {-min(trials.losses()):.1%}')
from sklearn.decomposition import PCA
from sklearn.svm import SVC
from lale.lib.lale import ConcatFeatures as Cat
from lale.lib.lale import NoOp
lale.wrap_imported_operators()
optimizable = (PCA & NoOp) >> Cat >> (LR | SVC)
optimizable.visualize()
from lale.operators import make_pipeline, make_union, make_choice
optimizable = make_pipeline(make_union(PCA, NoOp), make_choice(LR, SVC))
optimizable.visualize()
import lale.lib.lale.hyperopt
Optimizer = lale.lib.lale.hyperopt.Hyperopt
trained = optimizable.auto_configure(X_train, y_train, optimizer=Optimizer, max_evals=10)
predictions = trained.predict(X_test)
print(f'accuracy {accuracy_score(y_test, predictions):.1%}')
trained.visualize()
LR.get_tags()
LR.get_schema('input_fit')
LR.get_schema('output_predict')
| 0.651909 | 0.962953 |
# GFFcompare analyses
Import required modules
```
import pandas as pd
import matplotlib.pyplot as plt
from matplotlib_venn import venn3
from upsetplot import from_contents, UpSet, plot as upset_plot
import df2img
from IPython.display import display, HTML
```
Define function for forcing a table to display all of it contents
```
def force_show_all(df):
with pd.option_context('display.max_rows', None, 'display.max_columns', None, 'display.width', None):
display(HTML(df.to_html()))
```
### Data preparation:
Read gffcmp.tracking from the [GFFcompare](https://ccb.jhu.edu/software/stringtie/gffcompare.shtml) results, adjust lines for missing values and populate dictonary.
```
tcons = {}
with open(snakemake.input.tracking) as file:
for line in file:
line = line.split()
if ',' in line[4][3::]:
first = line[4][3::].split(',')
q1 = first[0].split('|')
else:
q1 = line[4][3::].split('|')
if ',' in line[5][3::]:
first = line[5][3::].split(',')
q2 = first[0].split('|')
else:
q2 = line[5][3::].split('|')
if ',' in line[6][3::]:
first = line[6][3::].split(',')
q3 = first[0].split('|')
else:
q3 = line[6][3::].split('|')
refgeneid = line[2].split('|')
if len(q1) < 2:
q1 = ['NA', 'NA', 0, 0, 0, 0, 0]
if len(q2) < 2:
q2 = ['NA', 'NA', 0, 0, 0, 0, 0]
if len(q3) < 2:
q3 = ['NA', 'NA', 0, 0, 0, 0, 0]
if len(refgeneid) < 2:
refgeneid = ['NA', 'NA']
tcons[line[0]] = [line[0], line[1], refgeneid[0], refgeneid[1], line[3],
q1[0], q1[1], q1[2], q1[3], q1[4], q1[5], q1[6],
q2[0], q2[1], q2[2], q2[3], q2[4], q2[5], q2[6],
q3[0], q3[1], q3[2], q3[3], q3[4], q3[5], q3[6]]
```
Define column names for dataframe
```
column_names = ['Query transfrag id', 'Query locus id', 'Reference gene name', 'Reference gene id', 'Class code',
'oxford.gene_id', 'oxford.transcript_id', 'oxford.num_exons', 'oxford.FPKM', 'oxford.counts',
'oxford.cov', 'oxford.len', 'flair.gene_id', 'flair.transcript_id', 'flair.num_exons',
'flair.FPKM', 'flair.counts', 'flair.cov', 'flair.len', 'talon.gene_id', 'talon.transcript_id',
'talon.num_exons', 'talon.FPKM', 'talon.counts', 'talon.cov', 'talon.len']
```
Convert dictionary to dataframe
```
df = pd.DataFrame.from_dict(tcons, orient='index', columns=column_names)
```
Assign correct datatypes to dataframe columns
```
df = df.astype({'Query transfrag id': str, 'Query locus id': str, 'Reference gene name': str, 'Reference gene id': str,
'Class code': str, 'oxford.gene_id': str, 'oxford.transcript_id': str, 'oxford.num_exons': int,
'oxford.FPKM': float, 'oxford.counts': float, 'oxford.cov': float,
'oxford.len': int, 'flair.gene_id': str, 'flair.transcript_id': str, 'flair.num_exons': int,
'flair.FPKM': float, 'flair.counts': float, 'flair.cov': float, 'flair.len': int,
'talon.gene_id': str, 'talon.transcript_id': str, 'talon.num_exons': int, 'talon.FPKM': float,
'talon.counts': float, 'talon.cov': float, 'talon.len': int})
```
Take a look at the dataframe
```
df
```
### Venn diagrams
Set matplotlib settings
```
plt.rcParams["figure.figsize"] = [10, 10]
plt.rcParams["figure.autolayout"] = False
plt.rcParams.update({'font.size': 15})
```
Extract transfragments with a count higher then zero for each pipeline
```
ox_mask = (df['oxford.counts'] > 0)
ox_array = df[ox_mask].values
ox_values = ox_array.T[0]
flair_mask = (df['flair.counts'] > 0)
flair_array = df[flair_mask].values
flair_values = flair_array.T[0]
talon_mask = (df['talon.counts'] > 0)
talon_array = df[talon_mask].values
talon_values = talon_array.T[0]
```
Create a upset plot that shows overlap of transcripts that are present in the samples
```
transcripts = from_contents({'oxford': set(ox_values), 'flair': set(flair_values), 'talon': set(talon_values)})
upset_plot(transcripts, element_size=50)
plt.suptitle('Overlap of transcripts present')
plt.savefig(snakemake.output.all_upset, bbox_inches='tight', dpi=200)
```
Create a venndiagram that shows overlap of transcripts that are present in the samples
```
venn3([set(ox_values), set(flair_values), set(talon_values)], ('oxford', 'flair', 'talon'))
plt.title('Overlap of transcripts present')
plt.savefig(snakemake.output.all_diagram, bbox_inches='tight', dpi=200)
```
Extract transfrag's with a count higher then zero and have a '=' class code.
```
ox_mask_known = (df['oxford.counts'] > 0) & (df['Class code'] == '=')
ox_array_known = df[ox_mask_known].values
ox_values_known = ox_array_known.T[0]
flair_mask_known = (df['flair.counts'] > 0) & (df['Class code'] == '=')
flair_array_known = df[flair_mask_known].values
flair_values_known = flair_array_known.T[0]
talon_mask_known = (df['talon.counts'] > 0) & (df['Class code'] == '=')
talon_array_known = df[talon_mask_known].values
talon_values_known = talon_array_known.T[0]
```
Create a upset plot with present transcripts that have being classified as known by GFFcompare.
```
transcripts = from_contents({'oxford': set(ox_values_known), 'flair': set(flair_values_known), 'talon': set(talon_values_known)})
upset_plot(transcripts, element_size=50)
plt.suptitle('Overlap of transcripts with classcode: "="')
plt.savefig(snakemake.output.known_upset, bbox_inches='tight', dpi=200)
```
Create a venndiagram with present transcripts that have being classified as known by GFFcompare.
```
venn3([set(ox_values_known), set(flair_values_known), set(talon_values_known)], ('oxford', 'flair', 'talon'))
plt.title('Overlap of transcripts with classcode: "="')
plt.savefig(snakemake.output.known_diagram, bbox_inches='tight', dpi=200)
```
Extract transfrag's with a count higher then zero and do not have a '=' class code.
```
ox_mask_novel = (df['oxford.counts'] > 1) & (df['Class code'] != '=')
ox_array_novel = df[ox_mask_novel].values
ox_values_novel = ox_array_novel.T[0]
flair_mask_novel = (df['flair.counts'] > 1) & (df['Class code'] != '=')
flair_array_novel = df[flair_mask_novel].values
flair_values_novel = flair_array_novel.T[0]
talon_mask_novel = (df['talon.counts'] > 1) & (df['Class code'] != '=')
talon_array_novel = df[talon_mask_novel].values
talon_values_novel = talon_array_novel.T[0]
```
Create a venndiagram with present transcripts that have being classified as novel by GFFcompare
```
transcripts = from_contents({'oxford': set(ox_values_novel), 'flair': set(flair_values_novel), 'talon': set(talon_values_novel)})
upset_plot(transcripts, element_size=50)
plt.suptitle('Overlap of transcripts that do NOT have classcode: "="')
plt.savefig(snakemake.output.novel_upset, bbox_inches='tight', dpi=200)
```
Create a venndiagram with present transcripts that have being classified as novel by GFFcompare
```
venn3([set(ox_values_novel), set(flair_values_novel), set(talon_values_novel)], ('oxford', 'flair', 'talon'))
plt.title('Overlap of transcripts that do NOT have classcode: "="')
plt.savefig(snakemake.output.novel_diagram, bbox_inches='tight', dpi=200)
```
# Non-matched transcripts analyses
Get all unique transcripts per pipeline
```
oxford_filter_mask = (df['oxford.counts'] > 0) & (df['flair.counts'] == 0) & (df['talon.counts'] == 0)
oxford_single = df[oxford_filter_mask]
oxford_single_df = oxford_single[['Reference gene id','oxford.num_exons', 'oxford.len','oxford.counts']]
flair_filter_mask = (df['oxford.counts'] == 0) & (df['flair.counts'] > 0) & (df['talon.counts'] == 0)
flair_single = df[flair_filter_mask]
flair_single_df = flair_single[['Reference gene id','flair.num_exons', 'flair.len','flair.counts']]
talon_filter_mask = (df['oxford.counts'] == 0) & (df['flair.counts'] == 0) & (df['talon.counts'] > 0)
talon_single = df[talon_filter_mask]
talon_single_df = talon_single[['Reference gene id','talon.num_exons', 'talon.len','talon.counts']]
```
Get statistics per pipeline for the lenght of the unique transcripts
```
ox_len = oxford_single_df['oxford.len'].describe().to_frame('OXFORD')
fl_len = flair_single_df['flair.len'].describe().to_frame('FLAIR')
ta_len = talon_single_df['talon.len'].describe().to_frame('TALON')
length_frames = [ox_len, fl_len, ta_len]
merged_length = pd.concat(length_frames, axis='columns', verify_integrity=True)
force_show_all(merged_length)
```
Get statistics per pipeline for the count of the unique transcripts
```
ox_count = oxford_single_df['oxford.counts'].describe().to_frame('OXFORD')
fl_count = flair_single_df['flair.counts'].describe().to_frame('FLAIR')
ta_count = talon_single_df['talon.counts'].describe().to_frame('TALON')
count_frames = [ox_count, fl_count, ta_count]
merged_counts = pd.concat(count_frames, axis='columns', verify_integrity=True)
force_show_all(merged_counts)
```
Show transcripts with number of exons
```
ox_exons = oxford_single_df['oxford.num_exons'].value_counts()
fl_exons = flair_single_df['flair.num_exons'].value_counts()
ta_exons = talon_single_df['talon.num_exons'].value_counts()
ox_exons = ox_exons.to_frame('OXFORD')
fl_exons = fl_exons.to_frame('FLAIR')
ta_exons = ta_exons.to_frame('TALON')
exon_frames = [ox_exons, fl_exons, ta_exons]
merged_exons = pd.concat(exon_frames, axis='columns', verify_integrity=True)
merged_exons = merged_exons.sort_index(ascending=True)
# force_show_all(merged_exons)
merged_exons.plot(title="None-matched transcripts per number of exons", xlabel="Exons in transcript", ylabel="Number of transcripts (log)", kind='bar', logy=True, figsize=(20,5))
```
# GffCompare classes
Get the GFFcompare classes of ALL transcripts matched against the genome annotation.
```
mask = (df['oxford.counts']>0)
oxford = df[mask].groupby('Class code', dropna=False).size()
ox_count = oxford.to_frame('Oxford')
mask = (df['flair.counts']>0)
flair = df[mask].groupby('Class code', dropna=False).size()
flair_count = flair.to_frame('FLAIR')
mask = (df['talon.counts']>0)
talon = df[mask].groupby('Class code', dropna=False).size()
talon_count = talon.to_frame('TALON')
classes = talon_count.merge(flair_count, left_on='Class code', right_on='Class code', how='outer')\
.merge(ox_count, left_on='Class code', right_on='Class code', how='outer')
classes
```
Get the GffCompare classes of UNIQUE transcripts matched against the genome annotation.
```
mask = (df['oxford.counts']>0) & (df['flair.counts'] == 0) & (df['talon.counts'] == 0)
oxford = df[mask].groupby('Class code', dropna=False).size()
ox_count = oxford.to_frame('Oxford')
mask = (df['flair.counts']>0) & (df['oxford.counts'] == 0) & (df['talon.counts'] == 0)
flair = df[mask].groupby('Class code', dropna=False).size()
flair_count = flair.to_frame('FLAIR')
mask = (df['talon.counts']>0) & (df['flair.counts'] == 0) & (df['oxford.counts'] == 0)
talon = df[mask].groupby('Class code', dropna=False).size()
talon_count = talon.to_frame('TALON')
classes = talon_count.merge(flair_count, left_on='Class code', right_on='Class code', how='outer')\
.merge(ox_count, left_on='Class code', right_on='Class code', how='outer')
classes
```
Get the GffCompare classes of three way matches
```
mask = (df['oxford.counts']>0) & (df['flair.counts']>0) & (df['talon.counts']>0)
three = df[mask].groupby('Class code', dropna=False).size()
three_count = three.to_frame('Three match')
three_count
```
|
github_jupyter
|
import pandas as pd
import matplotlib.pyplot as plt
from matplotlib_venn import venn3
from upsetplot import from_contents, UpSet, plot as upset_plot
import df2img
from IPython.display import display, HTML
def force_show_all(df):
with pd.option_context('display.max_rows', None, 'display.max_columns', None, 'display.width', None):
display(HTML(df.to_html()))
tcons = {}
with open(snakemake.input.tracking) as file:
for line in file:
line = line.split()
if ',' in line[4][3::]:
first = line[4][3::].split(',')
q1 = first[0].split('|')
else:
q1 = line[4][3::].split('|')
if ',' in line[5][3::]:
first = line[5][3::].split(',')
q2 = first[0].split('|')
else:
q2 = line[5][3::].split('|')
if ',' in line[6][3::]:
first = line[6][3::].split(',')
q3 = first[0].split('|')
else:
q3 = line[6][3::].split('|')
refgeneid = line[2].split('|')
if len(q1) < 2:
q1 = ['NA', 'NA', 0, 0, 0, 0, 0]
if len(q2) < 2:
q2 = ['NA', 'NA', 0, 0, 0, 0, 0]
if len(q3) < 2:
q3 = ['NA', 'NA', 0, 0, 0, 0, 0]
if len(refgeneid) < 2:
refgeneid = ['NA', 'NA']
tcons[line[0]] = [line[0], line[1], refgeneid[0], refgeneid[1], line[3],
q1[0], q1[1], q1[2], q1[3], q1[4], q1[5], q1[6],
q2[0], q2[1], q2[2], q2[3], q2[4], q2[5], q2[6],
q3[0], q3[1], q3[2], q3[3], q3[4], q3[5], q3[6]]
column_names = ['Query transfrag id', 'Query locus id', 'Reference gene name', 'Reference gene id', 'Class code',
'oxford.gene_id', 'oxford.transcript_id', 'oxford.num_exons', 'oxford.FPKM', 'oxford.counts',
'oxford.cov', 'oxford.len', 'flair.gene_id', 'flair.transcript_id', 'flair.num_exons',
'flair.FPKM', 'flair.counts', 'flair.cov', 'flair.len', 'talon.gene_id', 'talon.transcript_id',
'talon.num_exons', 'talon.FPKM', 'talon.counts', 'talon.cov', 'talon.len']
df = pd.DataFrame.from_dict(tcons, orient='index', columns=column_names)
df = df.astype({'Query transfrag id': str, 'Query locus id': str, 'Reference gene name': str, 'Reference gene id': str,
'Class code': str, 'oxford.gene_id': str, 'oxford.transcript_id': str, 'oxford.num_exons': int,
'oxford.FPKM': float, 'oxford.counts': float, 'oxford.cov': float,
'oxford.len': int, 'flair.gene_id': str, 'flair.transcript_id': str, 'flair.num_exons': int,
'flair.FPKM': float, 'flair.counts': float, 'flair.cov': float, 'flair.len': int,
'talon.gene_id': str, 'talon.transcript_id': str, 'talon.num_exons': int, 'talon.FPKM': float,
'talon.counts': float, 'talon.cov': float, 'talon.len': int})
df
plt.rcParams["figure.figsize"] = [10, 10]
plt.rcParams["figure.autolayout"] = False
plt.rcParams.update({'font.size': 15})
ox_mask = (df['oxford.counts'] > 0)
ox_array = df[ox_mask].values
ox_values = ox_array.T[0]
flair_mask = (df['flair.counts'] > 0)
flair_array = df[flair_mask].values
flair_values = flair_array.T[0]
talon_mask = (df['talon.counts'] > 0)
talon_array = df[talon_mask].values
talon_values = talon_array.T[0]
transcripts = from_contents({'oxford': set(ox_values), 'flair': set(flair_values), 'talon': set(talon_values)})
upset_plot(transcripts, element_size=50)
plt.suptitle('Overlap of transcripts present')
plt.savefig(snakemake.output.all_upset, bbox_inches='tight', dpi=200)
venn3([set(ox_values), set(flair_values), set(talon_values)], ('oxford', 'flair', 'talon'))
plt.title('Overlap of transcripts present')
plt.savefig(snakemake.output.all_diagram, bbox_inches='tight', dpi=200)
ox_mask_known = (df['oxford.counts'] > 0) & (df['Class code'] == '=')
ox_array_known = df[ox_mask_known].values
ox_values_known = ox_array_known.T[0]
flair_mask_known = (df['flair.counts'] > 0) & (df['Class code'] == '=')
flair_array_known = df[flair_mask_known].values
flair_values_known = flair_array_known.T[0]
talon_mask_known = (df['talon.counts'] > 0) & (df['Class code'] == '=')
talon_array_known = df[talon_mask_known].values
talon_values_known = talon_array_known.T[0]
transcripts = from_contents({'oxford': set(ox_values_known), 'flair': set(flair_values_known), 'talon': set(talon_values_known)})
upset_plot(transcripts, element_size=50)
plt.suptitle('Overlap of transcripts with classcode: "="')
plt.savefig(snakemake.output.known_upset, bbox_inches='tight', dpi=200)
venn3([set(ox_values_known), set(flair_values_known), set(talon_values_known)], ('oxford', 'flair', 'talon'))
plt.title('Overlap of transcripts with classcode: "="')
plt.savefig(snakemake.output.known_diagram, bbox_inches='tight', dpi=200)
ox_mask_novel = (df['oxford.counts'] > 1) & (df['Class code'] != '=')
ox_array_novel = df[ox_mask_novel].values
ox_values_novel = ox_array_novel.T[0]
flair_mask_novel = (df['flair.counts'] > 1) & (df['Class code'] != '=')
flair_array_novel = df[flair_mask_novel].values
flair_values_novel = flair_array_novel.T[0]
talon_mask_novel = (df['talon.counts'] > 1) & (df['Class code'] != '=')
talon_array_novel = df[talon_mask_novel].values
talon_values_novel = talon_array_novel.T[0]
transcripts = from_contents({'oxford': set(ox_values_novel), 'flair': set(flair_values_novel), 'talon': set(talon_values_novel)})
upset_plot(transcripts, element_size=50)
plt.suptitle('Overlap of transcripts that do NOT have classcode: "="')
plt.savefig(snakemake.output.novel_upset, bbox_inches='tight', dpi=200)
venn3([set(ox_values_novel), set(flair_values_novel), set(talon_values_novel)], ('oxford', 'flair', 'talon'))
plt.title('Overlap of transcripts that do NOT have classcode: "="')
plt.savefig(snakemake.output.novel_diagram, bbox_inches='tight', dpi=200)
oxford_filter_mask = (df['oxford.counts'] > 0) & (df['flair.counts'] == 0) & (df['talon.counts'] == 0)
oxford_single = df[oxford_filter_mask]
oxford_single_df = oxford_single[['Reference gene id','oxford.num_exons', 'oxford.len','oxford.counts']]
flair_filter_mask = (df['oxford.counts'] == 0) & (df['flair.counts'] > 0) & (df['talon.counts'] == 0)
flair_single = df[flair_filter_mask]
flair_single_df = flair_single[['Reference gene id','flair.num_exons', 'flair.len','flair.counts']]
talon_filter_mask = (df['oxford.counts'] == 0) & (df['flair.counts'] == 0) & (df['talon.counts'] > 0)
talon_single = df[talon_filter_mask]
talon_single_df = talon_single[['Reference gene id','talon.num_exons', 'talon.len','talon.counts']]
ox_len = oxford_single_df['oxford.len'].describe().to_frame('OXFORD')
fl_len = flair_single_df['flair.len'].describe().to_frame('FLAIR')
ta_len = talon_single_df['talon.len'].describe().to_frame('TALON')
length_frames = [ox_len, fl_len, ta_len]
merged_length = pd.concat(length_frames, axis='columns', verify_integrity=True)
force_show_all(merged_length)
ox_count = oxford_single_df['oxford.counts'].describe().to_frame('OXFORD')
fl_count = flair_single_df['flair.counts'].describe().to_frame('FLAIR')
ta_count = talon_single_df['talon.counts'].describe().to_frame('TALON')
count_frames = [ox_count, fl_count, ta_count]
merged_counts = pd.concat(count_frames, axis='columns', verify_integrity=True)
force_show_all(merged_counts)
ox_exons = oxford_single_df['oxford.num_exons'].value_counts()
fl_exons = flair_single_df['flair.num_exons'].value_counts()
ta_exons = talon_single_df['talon.num_exons'].value_counts()
ox_exons = ox_exons.to_frame('OXFORD')
fl_exons = fl_exons.to_frame('FLAIR')
ta_exons = ta_exons.to_frame('TALON')
exon_frames = [ox_exons, fl_exons, ta_exons]
merged_exons = pd.concat(exon_frames, axis='columns', verify_integrity=True)
merged_exons = merged_exons.sort_index(ascending=True)
# force_show_all(merged_exons)
merged_exons.plot(title="None-matched transcripts per number of exons", xlabel="Exons in transcript", ylabel="Number of transcripts (log)", kind='bar', logy=True, figsize=(20,5))
mask = (df['oxford.counts']>0)
oxford = df[mask].groupby('Class code', dropna=False).size()
ox_count = oxford.to_frame('Oxford')
mask = (df['flair.counts']>0)
flair = df[mask].groupby('Class code', dropna=False).size()
flair_count = flair.to_frame('FLAIR')
mask = (df['talon.counts']>0)
talon = df[mask].groupby('Class code', dropna=False).size()
talon_count = talon.to_frame('TALON')
classes = talon_count.merge(flair_count, left_on='Class code', right_on='Class code', how='outer')\
.merge(ox_count, left_on='Class code', right_on='Class code', how='outer')
classes
mask = (df['oxford.counts']>0) & (df['flair.counts'] == 0) & (df['talon.counts'] == 0)
oxford = df[mask].groupby('Class code', dropna=False).size()
ox_count = oxford.to_frame('Oxford')
mask = (df['flair.counts']>0) & (df['oxford.counts'] == 0) & (df['talon.counts'] == 0)
flair = df[mask].groupby('Class code', dropna=False).size()
flair_count = flair.to_frame('FLAIR')
mask = (df['talon.counts']>0) & (df['flair.counts'] == 0) & (df['oxford.counts'] == 0)
talon = df[mask].groupby('Class code', dropna=False).size()
talon_count = talon.to_frame('TALON')
classes = talon_count.merge(flair_count, left_on='Class code', right_on='Class code', how='outer')\
.merge(ox_count, left_on='Class code', right_on='Class code', how='outer')
classes
mask = (df['oxford.counts']>0) & (df['flair.counts']>0) & (df['talon.counts']>0)
three = df[mask].groupby('Class code', dropna=False).size()
three_count = three.to_frame('Three match')
three_count
| 0.189071 | 0.909023 |
```
from IPython.lib.deepreload import reload
%load_ext autoreload
%autoreload 2
import re
import operator
from pathlib import Path
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
import pydicom
from pymedphys_analysis.tpscompare import load_and_normalise_mephysto
from pymedphys_dicom.dicom import depth_dose, profile
ROOT_DIR = Path(r"S:\Physics\Monaco\Model vs Measurement Comparisons")
INTERNAL_DICOM_DIR = ROOT_DIR.joinpath(r"Beam Models\Collapsed Cone\DICOM Dose Exports")
EXTERNAL_DICOM_DIR = ROOT_DIR.joinpath(r"Beam Models\DoseCHECK\DICOM Dose Exports")
MEASUREMENTS_DIR = ROOT_DIR.joinpath(r"Measurements\RCCC\Photons\With Flattening Filter")
RESULTS = ROOT_DIR.joinpath(r"Results\RCCC\dosecheck")
calibrated_doses_table = pd.read_csv(MEASUREMENTS_DIR.joinpath('AbsoluteDose.csv'), index_col=0)
calibrated_doses = calibrated_doses_table['d10 @ 90 SSD']
calibrated_doses
wedge_transmission_table = pd.read_csv(MEASUREMENTS_DIR.joinpath('WedgeTransmissionFactors.csv'), index_col=0)
data_column_name = wedge_transmission_table.columns[0]
wedge_transmissions = wedge_transmission_table[data_column_name]
wedge_transmissions
output_factors = pd.read_csv(MEASUREMENTS_DIR.joinpath('OutputFactors.csv'), index_col=0)
output_factors
keys = [
path.stem
for path in MEASUREMENTS_DIR.glob('*.mcc')
]
keys
regex_string = r'(\d\dMV) (\d\dx\d\d) ((\bOpen\b)|(\bWedge\b))'
def get_energy_field_block(key):
match = re.match(regex_string, key)
return match.group(1), match.group(2), match.group(3)
absolute_doses = {}
for key in keys:
energy, field, block = get_energy_field_block(key)
if block == 'Wedge':
wtf = wedge_transmissions[energy]
else:
wtf = 1
output_factor = output_factors[f'{field} {block}'][energy]
calibrated_dose = calibrated_doses[energy]
absolute_dose = calibrated_dose * output_factor * wtf
absolute_doses[key] = absolute_dose
absolute_doses
getter = operator.itemgetter('displacement', 'dose')
absolute_scans_per_field = load_and_normalise_mephysto(
MEASUREMENTS_DIR, r'(\d\dMV \d\dx\d\d ((\bOpen\b)|(\bWedge\b)))\.mcc', absolute_doses, 100)
new_keys = list(absolute_scans_per_field.keys())
new_keys
assert new_keys == keys
def load_dicom_files(directory, keys):
dicom_file_map = {
key: directory.joinpath(f'{key}.dcm')
for key in keys
}
dicom_dataset_map = {
key: pydicom.read_file(str(dicom_file_map[key]), force=True)
for key in keys
}
return dicom_dataset_map
internal_dicom_dataset_map = load_dicom_files(INTERNAL_DICOM_DIR, keys)
external_dicom_dataset_map = load_dicom_files(EXTERNAL_DICOM_DIR, keys)
internal_dicom_plan = pydicom.read_file(str(INTERNAL_DICOM_DIR.joinpath('CollapsedCone.dcm')), force=True)
external_dicom_plan = pydicom.read_file(str(EXTERNAL_DICOM_DIR.joinpath('DicomFile20190724 112911-284.dcm')), force=True)
def plot_one_axis(ax, displacement, meas_dose, tps_dose):
diff = tps_dose - meas_dose
lines = []
lines += ax.plot(displacement, meas_dose, label='Measured Dose')
lines += ax.plot(displacement, tps_dose, label='TPS Dose')
ax.set_ylabel('Dose (Gy / 100 MU)')
x_bounds = [np.min(displacement), np.max(displacement)]
ax.set_xlim(x_bounds)
ax_twin = ax.twinx()
lines += ax_twin.plot(displacement, diff, color='C3', alpha=0.5, label='Residuals [TPS - Meas]')
ax_twin.plot(x_bounds, [0, 0], '--', color='C3', lw=0.5)
ax_twin.set_ylabel('Dose difference [TPS - Meas] (Gy / 100 MU)')
labels = [l.get_label() for l in lines]
ax.legend(lines, labels)
return ax_twin
def plot_tps_meas_diff(displacement, meas_dose, internal_tps_dose, external_tps_dose):
fig, ax = plt.subplots(1, 2, figsize=(16,6), sharey=True)
ax[1].yaxis.set_tick_params(which='both', labelbottom=True)
ax_twin = list()
ax_twin.append(plot_one_axis(ax[0], displacement, meas_dose, internal_tps_dose))
ax_twin.append(plot_one_axis(ax[1], displacement, meas_dose, external_tps_dose))
ax_twin[1].get_shared_y_axes().join(ax_twin[1], ax_twin[0])
ax_twin[1].autoscale(axis='y')
plt.tight_layout()
plt.subplots_adjust(wspace=0.4, top=0.86)
return fig, ax
def plot_pdd_diff(key):
depth, meas_dose = getter(absolute_scans_per_field[key]['depth_dose'])
internal_tps_dose = depth_dose(depth, internal_dicom_dataset_map[key], internal_dicom_plan) / 10
external_tps_dose = depth_dose(depth, external_dicom_dataset_map[key], external_dicom_plan) / 10
fig, ax = plot_tps_meas_diff(depth, meas_dose, internal_tps_dose, external_tps_dose)
fig.suptitle(f'Depth Dose Comparisons | {key}', fontsize="x-large")
ax[0].set_title("Monaco Collapsed Cone")
ax[1].set_title("DoseCHECK")
for key in keys:
plot_pdd_diff(key)
filename = RESULTS.joinpath(f'{key}_pdd.png')
plt.savefig(filename)
plt.show()
def plot_profile_diff(key, depth, direction):
displacement, meas_dose = getter(absolute_scans_per_field[key]['profiles'][depth][direction])
internal_tps_dose = profile(displacement, depth, direction, internal_dicom_dataset_map[key], internal_dicom_plan) / 10
external_tps_dose = profile(displacement, depth, direction, external_dicom_dataset_map[key], external_dicom_plan) / 10
fig, ax = plot_tps_meas_diff(displacement, meas_dose, internal_tps_dose, external_tps_dose)
fig.suptitle(f'{direction.capitalize()} Profile Comparisons | {key} | Depth: {depth} mm', fontsize="x-large")
ax[0].set_title("Monaco Collapsed Cone")
ax[1].set_title("DoseCHECK")
for key in keys:
depths = absolute_scans_per_field[key]['profiles'].keys()
for depth in depths:
for direction in ['inplane', 'crossplane']:
plot_profile_diff(key, depth, direction)
filename = RESULTS.joinpath(f'{key}_profile_{depth}mm_{direction}.png')
plt.savefig(filename)
plt.show()
```
|
github_jupyter
|
from IPython.lib.deepreload import reload
%load_ext autoreload
%autoreload 2
import re
import operator
from pathlib import Path
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
import pydicom
from pymedphys_analysis.tpscompare import load_and_normalise_mephysto
from pymedphys_dicom.dicom import depth_dose, profile
ROOT_DIR = Path(r"S:\Physics\Monaco\Model vs Measurement Comparisons")
INTERNAL_DICOM_DIR = ROOT_DIR.joinpath(r"Beam Models\Collapsed Cone\DICOM Dose Exports")
EXTERNAL_DICOM_DIR = ROOT_DIR.joinpath(r"Beam Models\DoseCHECK\DICOM Dose Exports")
MEASUREMENTS_DIR = ROOT_DIR.joinpath(r"Measurements\RCCC\Photons\With Flattening Filter")
RESULTS = ROOT_DIR.joinpath(r"Results\RCCC\dosecheck")
calibrated_doses_table = pd.read_csv(MEASUREMENTS_DIR.joinpath('AbsoluteDose.csv'), index_col=0)
calibrated_doses = calibrated_doses_table['d10 @ 90 SSD']
calibrated_doses
wedge_transmission_table = pd.read_csv(MEASUREMENTS_DIR.joinpath('WedgeTransmissionFactors.csv'), index_col=0)
data_column_name = wedge_transmission_table.columns[0]
wedge_transmissions = wedge_transmission_table[data_column_name]
wedge_transmissions
output_factors = pd.read_csv(MEASUREMENTS_DIR.joinpath('OutputFactors.csv'), index_col=0)
output_factors
keys = [
path.stem
for path in MEASUREMENTS_DIR.glob('*.mcc')
]
keys
regex_string = r'(\d\dMV) (\d\dx\d\d) ((\bOpen\b)|(\bWedge\b))'
def get_energy_field_block(key):
match = re.match(regex_string, key)
return match.group(1), match.group(2), match.group(3)
absolute_doses = {}
for key in keys:
energy, field, block = get_energy_field_block(key)
if block == 'Wedge':
wtf = wedge_transmissions[energy]
else:
wtf = 1
output_factor = output_factors[f'{field} {block}'][energy]
calibrated_dose = calibrated_doses[energy]
absolute_dose = calibrated_dose * output_factor * wtf
absolute_doses[key] = absolute_dose
absolute_doses
getter = operator.itemgetter('displacement', 'dose')
absolute_scans_per_field = load_and_normalise_mephysto(
MEASUREMENTS_DIR, r'(\d\dMV \d\dx\d\d ((\bOpen\b)|(\bWedge\b)))\.mcc', absolute_doses, 100)
new_keys = list(absolute_scans_per_field.keys())
new_keys
assert new_keys == keys
def load_dicom_files(directory, keys):
dicom_file_map = {
key: directory.joinpath(f'{key}.dcm')
for key in keys
}
dicom_dataset_map = {
key: pydicom.read_file(str(dicom_file_map[key]), force=True)
for key in keys
}
return dicom_dataset_map
internal_dicom_dataset_map = load_dicom_files(INTERNAL_DICOM_DIR, keys)
external_dicom_dataset_map = load_dicom_files(EXTERNAL_DICOM_DIR, keys)
internal_dicom_plan = pydicom.read_file(str(INTERNAL_DICOM_DIR.joinpath('CollapsedCone.dcm')), force=True)
external_dicom_plan = pydicom.read_file(str(EXTERNAL_DICOM_DIR.joinpath('DicomFile20190724 112911-284.dcm')), force=True)
def plot_one_axis(ax, displacement, meas_dose, tps_dose):
diff = tps_dose - meas_dose
lines = []
lines += ax.plot(displacement, meas_dose, label='Measured Dose')
lines += ax.plot(displacement, tps_dose, label='TPS Dose')
ax.set_ylabel('Dose (Gy / 100 MU)')
x_bounds = [np.min(displacement), np.max(displacement)]
ax.set_xlim(x_bounds)
ax_twin = ax.twinx()
lines += ax_twin.plot(displacement, diff, color='C3', alpha=0.5, label='Residuals [TPS - Meas]')
ax_twin.plot(x_bounds, [0, 0], '--', color='C3', lw=0.5)
ax_twin.set_ylabel('Dose difference [TPS - Meas] (Gy / 100 MU)')
labels = [l.get_label() for l in lines]
ax.legend(lines, labels)
return ax_twin
def plot_tps_meas_diff(displacement, meas_dose, internal_tps_dose, external_tps_dose):
fig, ax = plt.subplots(1, 2, figsize=(16,6), sharey=True)
ax[1].yaxis.set_tick_params(which='both', labelbottom=True)
ax_twin = list()
ax_twin.append(plot_one_axis(ax[0], displacement, meas_dose, internal_tps_dose))
ax_twin.append(plot_one_axis(ax[1], displacement, meas_dose, external_tps_dose))
ax_twin[1].get_shared_y_axes().join(ax_twin[1], ax_twin[0])
ax_twin[1].autoscale(axis='y')
plt.tight_layout()
plt.subplots_adjust(wspace=0.4, top=0.86)
return fig, ax
def plot_pdd_diff(key):
depth, meas_dose = getter(absolute_scans_per_field[key]['depth_dose'])
internal_tps_dose = depth_dose(depth, internal_dicom_dataset_map[key], internal_dicom_plan) / 10
external_tps_dose = depth_dose(depth, external_dicom_dataset_map[key], external_dicom_plan) / 10
fig, ax = plot_tps_meas_diff(depth, meas_dose, internal_tps_dose, external_tps_dose)
fig.suptitle(f'Depth Dose Comparisons | {key}', fontsize="x-large")
ax[0].set_title("Monaco Collapsed Cone")
ax[1].set_title("DoseCHECK")
for key in keys:
plot_pdd_diff(key)
filename = RESULTS.joinpath(f'{key}_pdd.png')
plt.savefig(filename)
plt.show()
def plot_profile_diff(key, depth, direction):
displacement, meas_dose = getter(absolute_scans_per_field[key]['profiles'][depth][direction])
internal_tps_dose = profile(displacement, depth, direction, internal_dicom_dataset_map[key], internal_dicom_plan) / 10
external_tps_dose = profile(displacement, depth, direction, external_dicom_dataset_map[key], external_dicom_plan) / 10
fig, ax = plot_tps_meas_diff(displacement, meas_dose, internal_tps_dose, external_tps_dose)
fig.suptitle(f'{direction.capitalize()} Profile Comparisons | {key} | Depth: {depth} mm', fontsize="x-large")
ax[0].set_title("Monaco Collapsed Cone")
ax[1].set_title("DoseCHECK")
for key in keys:
depths = absolute_scans_per_field[key]['profiles'].keys()
for depth in depths:
for direction in ['inplane', 'crossplane']:
plot_profile_diff(key, depth, direction)
filename = RESULTS.joinpath(f'{key}_profile_{depth}mm_{direction}.png')
plt.savefig(filename)
plt.show()
| 0.498535 | 0.486149 |
```
import sympy as sp
X=[1,2,3,4,5,6]
Y=[6,5,4,3,2,1]
```
Badamy szybkość wunkcji generującej symboliczny wzór. Niektóre Państwa funkcje zostały zmodyfikowane, żeby nie sprawdzały wartości opcjonalnego argumentu oraz nie weryfikowały długości list.
```
#Daniel
def interpolacja(X,Y,pt):
x=sp.symbols('x')
temp=sp.prod([x-el for el in X])
tabelaTemp=[temp/(x-X[i]) for i in range(len(X))]
wzor=sum([Y[i]*tabelaTemp[i]/(tabelaTemp[i].subs(x,X[i])) for i in range(len(X))])
return wzor
%timeit interpolacja(X,Y,0)
# Anna Cabaj
from sympy import *
def lagrange2 (X,Y,x):
x=symbols('x')
f=symbols('f', cls=Function)
f=0
for k in range(len(Y)):
lk=Y[k]
for n in range(len(X)):
if n==k:
continue
lk=lk*(x-X[n])/(X[k]-X[n])
f=f+lk
return f
%timeit lagrange2(X,Y,0)
# Aleksandra
def Lagringo (X,Y,x, symbol):
w=0
if symbol:
sym = ''
for i in range(len(X)):
if i>0:
if symbol:
sym +='+'
iloczyn = 1
if symbol:
sym += str(Y[i])
for j in range (len(Y)):
if i != j:
iloczyn *= (x-X[j])/(X[i]-X[j])
if symbol:
sym += "*(x-"+ str(X[j])+ ")/("+ str(X[i])+ "-"+ str(X[j])+ ")"
w += Y[i]*iloczyn
if symbol:
return w, sym
else:
return w
%timeit Lagringo(X,Y,0,True)
#Klaudia
def wielomianp(X,Y,x):
h=symbols('h')
f=Function('f')
f=0
for i in range(len(Y)):
il=Y[i]
for j in range(len(X)):
if j==i:
continue
il=il*(h-X[j])/(X[i]-X[j])
f=f+il
return f
%timeit wielomianp(X,Y,0)
#Wiktoria
def Interpolacja_symbol(X,Y):
x=symbols('x')
n=0
for i in range (len(X)):
a = 1
for j in range (len(X)):
if j != i:
a=a*((x-X[j]) /(X[i]-X[j]))
n += a* Y[i]
return n
%timeit Interpolacja_symbol(X,Y)
#Wojtek
from functools import reduce
from operator import mul
def interpolate(X, Y, x):
a = symbols("x")
L = lambda x_i: lambda x_j: (a-x_j)/(x_i-x_j)
w = sum(y*reduce(mul, map(L(X[i]), X[:i]+X[i+1:])) for i,y in enumerate(Y))
return w
%timeit interpolate(X,Y,0)
#Marek
def lagranż(a,b,c,d=0): #a,b
x=symbols('x')
value=Function('f')
value=0
for i in range(len(b)):
value_1=b[i]
for j in range(len(a)):
if i==j: continue
value_1=value_1*(x-a[j])/(a[i]-a[j])
value=value+value_1
return value
%timeit lagranż(X,Y,0)
#Szymon
def lagrange(X, Y, x):
x_ = symbols('x')
result = 0; lenght = len(X)
for i in range(lenght):
multi = 1
for j in range(lenght):
if j != i:
multi *= (x_ - X[j])/(X[i] - X[j])
result += Y[i] * multi
return result
%timeit lagrange(X,Y,0)
#Anna Zgrzebna
def wiel_inter_Lagrange(X,Y,x):
x1 = symbols('x')
sum = 0
n = len(X)
for i in range (n):
p = 1
for j in range (n):
if j != i:
p = p* ((x1-X[j])/(X[i]-X[j]))
sum = sum + Y[i]*p
return sum
%timeit wiel_inter_Lagrange(X,Y,0)
#Michał
def lagrange(x, X, Y):
m = len(X)
z = 0.0
for i in range(m):
p = 1.0
for k in range(m):
if i == k: continue
p = p* ((x - X[k])/(X[i] - X[k]))
z += Y[i] * p
return z
x=symbols('x')
%timeit lagrange(x,X,Y)
```
|
github_jupyter
|
import sympy as sp
X=[1,2,3,4,5,6]
Y=[6,5,4,3,2,1]
#Daniel
def interpolacja(X,Y,pt):
x=sp.symbols('x')
temp=sp.prod([x-el for el in X])
tabelaTemp=[temp/(x-X[i]) for i in range(len(X))]
wzor=sum([Y[i]*tabelaTemp[i]/(tabelaTemp[i].subs(x,X[i])) for i in range(len(X))])
return wzor
%timeit interpolacja(X,Y,0)
# Anna Cabaj
from sympy import *
def lagrange2 (X,Y,x):
x=symbols('x')
f=symbols('f', cls=Function)
f=0
for k in range(len(Y)):
lk=Y[k]
for n in range(len(X)):
if n==k:
continue
lk=lk*(x-X[n])/(X[k]-X[n])
f=f+lk
return f
%timeit lagrange2(X,Y,0)
# Aleksandra
def Lagringo (X,Y,x, symbol):
w=0
if symbol:
sym = ''
for i in range(len(X)):
if i>0:
if symbol:
sym +='+'
iloczyn = 1
if symbol:
sym += str(Y[i])
for j in range (len(Y)):
if i != j:
iloczyn *= (x-X[j])/(X[i]-X[j])
if symbol:
sym += "*(x-"+ str(X[j])+ ")/("+ str(X[i])+ "-"+ str(X[j])+ ")"
w += Y[i]*iloczyn
if symbol:
return w, sym
else:
return w
%timeit Lagringo(X,Y,0,True)
#Klaudia
def wielomianp(X,Y,x):
h=symbols('h')
f=Function('f')
f=0
for i in range(len(Y)):
il=Y[i]
for j in range(len(X)):
if j==i:
continue
il=il*(h-X[j])/(X[i]-X[j])
f=f+il
return f
%timeit wielomianp(X,Y,0)
#Wiktoria
def Interpolacja_symbol(X,Y):
x=symbols('x')
n=0
for i in range (len(X)):
a = 1
for j in range (len(X)):
if j != i:
a=a*((x-X[j]) /(X[i]-X[j]))
n += a* Y[i]
return n
%timeit Interpolacja_symbol(X,Y)
#Wojtek
from functools import reduce
from operator import mul
def interpolate(X, Y, x):
a = symbols("x")
L = lambda x_i: lambda x_j: (a-x_j)/(x_i-x_j)
w = sum(y*reduce(mul, map(L(X[i]), X[:i]+X[i+1:])) for i,y in enumerate(Y))
return w
%timeit interpolate(X,Y,0)
#Marek
def lagranż(a,b,c,d=0): #a,b
x=symbols('x')
value=Function('f')
value=0
for i in range(len(b)):
value_1=b[i]
for j in range(len(a)):
if i==j: continue
value_1=value_1*(x-a[j])/(a[i]-a[j])
value=value+value_1
return value
%timeit lagranż(X,Y,0)
#Szymon
def lagrange(X, Y, x):
x_ = symbols('x')
result = 0; lenght = len(X)
for i in range(lenght):
multi = 1
for j in range(lenght):
if j != i:
multi *= (x_ - X[j])/(X[i] - X[j])
result += Y[i] * multi
return result
%timeit lagrange(X,Y,0)
#Anna Zgrzebna
def wiel_inter_Lagrange(X,Y,x):
x1 = symbols('x')
sum = 0
n = len(X)
for i in range (n):
p = 1
for j in range (n):
if j != i:
p = p* ((x1-X[j])/(X[i]-X[j]))
sum = sum + Y[i]*p
return sum
%timeit wiel_inter_Lagrange(X,Y,0)
#Michał
def lagrange(x, X, Y):
m = len(X)
z = 0.0
for i in range(m):
p = 1.0
for k in range(m):
if i == k: continue
p = p* ((x - X[k])/(X[i] - X[k]))
z += Y[i] * p
return z
x=symbols('x')
%timeit lagrange(x,X,Y)
| 0.084434 | 0.670123 |
<div class="alert alert-block alert-info" style="margin-top: 20px">
<a href="http://cocl.us/DA0101EN_NotbookLink_Top"><img src = "https://ibm.box.com/shared/static/fvp89yz8uzmr5q6bs6wnguxbf8x91z35.png" width = 750, align = "center"></a>
<h1 align=center><font size = 5> Link</font></h1>
<a href="https://www.bigdatauniversity.com"><img src = "https://ibm.box.com/shared/static/ugcqz6ohbvff804xp84y4kqnvvk3bq1g.png" width = 300, align = "center"></a>
<h1 align=center><font size = 5>Data Analysis with Python</font></h1>
# Module 3: Exploratory Data Analysis
### Welcome!
In this section, we will explore several methods to see if certain characteristics or features can be used to predict price.
### What are the main characteristics which have the most impact on the car price?
## 1. Import Data from Module 2
#### Setup
Import libraries:
```
import pandas as pd
import numpy as np
```
Load data and store in dataframe df:
```
path='https://ibm.box.com/shared/static/q6iiqb1pd7wo8r3q28jvgsrprzezjqk3.csv'
df = pd.read_csv(path)
df.head()
```
## 2. Analyzing Individual Feature Patterns using Visualization
Import visualization packages "Matplotlib" and "Seaborn". Don't forget about "%matplotlib inline" to plot in a Jupyter notebook:
```
import matplotlib.pyplot as plt
import seaborn as sns
%matplotlib inline
```
### How to choose the right visualization method:
When visualizing individual variables, it is important to first understand what type of variable you are dealing with. This will help us find the right visualisation method for that variable.
```
# list the data types for each column
df.dtypes
```
<div class="alert alert-danger alertdanger" style="margin-top: 20px">
<h1> Question #1: </h1>
<b>What is the data type of the column "peak-rpm"? </b>
</div>
<div align="right">
<a href="#q1" class="btn btn-default" data-toggle="collapse">Click here for the solution</a>
</div>
<div id="q1" class="collapse">
```
float64
```
</div>
For example, we can calculate the correlation between variables of type "int64" or "float64" using the method "corr":
```
df.corr()
```
The diagonal elements are always one. We will study correlation, more precisely Pearson correlation, in-depth at the end of the notebook.
<div class="alert alert-danger alertdanger" style="margin-top: 20px">
<h1> Question #2: </h1>
<b> Find the correlation between the following columns: bore, stroke, compression-ratio, and horsepower.
<br>Hint: if you would like to select those columns use the following syntax: df[['bore','stroke' ,'compression-ratio','horsepower']]:</bbr> </b>
</div>
```
df[['bore','stroke' ,'compression-ratio','horsepower']].corr()
```
<div align="right">
<a href="#q2" class="btn btn-default" data-toggle="collapse">Click here for the solution</a>
</div>
<div id="q2" class="collapse">
```
df[['bore','stroke' ,'compression-ratio','horsepower']].corr()
```
</div>
## Continuous numerical variables:
Continuous numerical variables are variables that may contain any value within some range. Continuous numerical variables can have the type "int64" or "float64". A great way to visualize these variables is by using scatterplots with fitted lines.
In order to start understanding the (linear) relationship between an individual variable and the price, we can use "regplot", which plots the scatterplot plus the fitted regression line for the data.
Let's see several examples of different linear relationships:
#### Positive linear relationship
Let's find the scatterplot of "engine-size" and "price":
```
# Engine size as potential predictor variable of price
sns.regplot(x="engine-size", y="price", data=df)
plt.ylim(0,)
```
As the engine-size goes up, the price goes up: this indicates a positive direct correlation between these two variables. Engine size seems like a pretty good predictor of price since the regression line is almost a perfect diagonal line. E
We can examine the correlation between 'engine-size' and 'price' and see it's approximately 0.87:
```
df[["engine-size", "price"]].corr()
```
### Negative linear relationship
Highway mpg is a potential predictor variable of price:
```
sns.regplot(x="highway-mpg", y="price", data=df)
```
As the highway-mpg goes up, the price goes down: this indicates an inverse/negative relationship between these two variables. Highway mpg could potentially be a predictor of price.
We can examine the correlation between 'highway-mpg' and 'price' and see it's approximately -0.704:
```
df[['highway-mpg', 'price']].corr()
```
### Weak Linear Relationship
Let's see if "Peak-rpm" as a predictor variable of "price":
```
sns.regplot(x="peak-rpm", y="price", data=df)
```
Peak rpm does not seem like a good predictor of the price at all since the regression line is close to horizontal. Also, the data points are very scattered and far from the fitted line, showing lots of variability. Therefore it is not a reliable variable.
We can examine the correlation between 'peak-rpm' and 'price'and see it is approximately -0.101616:
```
df[['peak-rpm','price']].corr()
```
<div class="alert alert-danger alertdanger" style="margin-top: 20px">
<h1> Question 3 a): </h1>
<b> Find the correlation between x="stroke", y="price".
<br>Hint: if you would like to select those columns use the following syntax: df[["stroke","price"]]:</bbr> </b>
</div>
```
df[["stroke","price"]].corr()
```
<div align="right">
<a href="#q3a" class="btn btn-default" data-toggle="collapse">Click here for the solution</a>
</div>
<div id="q3a" class="collapse">
```
The correlation is 0.0823, the non-diagonal elements of the table.
code:df[["stroke","price"]].corr()
```
</div>
<div class="alert alert-danger alertdanger" style="margin-top: 20px">
<h1> Question 3 b): </h1>
<b> Given the correlation results between "price" and "stroke", do you expect a linear relationship? Verify your results using the function "regplot()":</bbr> </b>
</div>
```
sns.regplot(x="stroke", y="price", data=df)
```
<div align="right">
<a href="#q3b" class="btn btn-default" data-toggle="collapse">Click here for the solution</a>
</div>
<div id="q3b" class="collapse">
```
There is a weak correlation between the variable 'stroke' and 'price.' as such regression will not work well. We can see this use "regplot" to demonstrate this.
Code: sns.regplot(x="stroke", y="price", data=df)
```
</div>
## Categorical variables
These are variables that describe a 'characteristic' of a data unit, and are selected from a small group of categories. The categorical variables can have the type "object" or "int64". A good way to visualize categorical variables is by using boxplots.
Let's look at the relationship between "body-style" and "price":
```
sns.boxplot(x="body-style", y="price", data=df)
```
We see that the distributions of price between the different body-style categories have a significant overlap, and so body-style would not be a good predictor of price. Let's examine engine "engine-location" and "price" :
```
sns.boxplot(x="engine-location", y="price", data=df)
```
Here we see that the distribution of price between these two engine-location categories, front and rear, are distinct enough to take engine-location as a potential good predictor of price.
Let's examine "drive-wheels" and "price":
```
# drive-wheels
sns.boxplot(x="drive-wheels", y="price", data=df)
```
Here we see that the distribution of price between the different drive-wheels categories differs. As such, drive-wheels could potentially be a predictor of price.
## 3. Descriptive Statistical Analysis
Let's first take a look at the variables by utilising a description method.
The **describe** function automatically computes basic statistics for all continuous variables. Any NaN values are automatically skipped in these statistics.
This will show:
- the count of that variable
- the mean
- the standard deviation (std)
- the minimum value
- the IQR (Interquartile Range: 25%, 50% and 75%)
- the maximum value
We can apply the method "describe" as follows:
```
df.describe()
```
The default setting of "describe" skips variables of type object. We can apply the method "describe" on the variables of type 'object' as follows:
```
df.describe(include=['object'])
```
### Value Counts
Value-counts is a good way of understanding how many units of each characteristic/variable we have. We can apply the "value_counts" method on the column 'drive-wheels'. Don’t forget the method "value_counts" only works on Pandas series, not Pandas Dataframes. As a result, we only include one bracket "df['drive-wheels']", not two "df[['drive-wheels']]".
```
df['drive-wheels'].value_counts()
```
We can convert the series to a Dataframe as follows :
```
df['drive-wheels'].value_counts().to_frame()
```
Let's repeat the above steps but save the results to the dataframe "drive_wheels_counts" and rename the column 'drive-wheels' to 'value_counts':
```
drive_wheels_counts = df['drive-wheels'].value_counts().to_frame()
drive_wheels_counts.rename(columns={'drive-wheels': 'value_counts'}, inplace=True)
drive_wheels_counts
```
Now let's rename the index to 'drive-wheels':
```
drive_wheels_counts.index.name = 'drive-wheels'
drive_wheels_counts
```
We can repeat the above process for the variable 'engine-location':
```
# engine-location as variable
engine_loc_counts = df['engine-location'].value_counts().to_frame()
engine_loc_counts.rename(columns={'engine-location': 'value_counts'}, inplace=True)
engine_loc_counts.index.name = 'engine-location'
engine_loc_counts.head(10)
```
Examining the value counts of the engine location would not be a good predictor variable for the price. This is because we only have three cars with a rear engine and 198 with an engine in the front, creating a skewed result. Thus, we are not able to draw any conclusions about the engine location.
## 4. Basic of Grouping
The "groupby" method groups data by different categories. The data is grouped based on one or several variables, and analysis is performed on the individual groups.
For example, let's group by the variable "drive-wheels". We see that there are 3 different categories of drive wheels:
```
df['drive-wheels'].unique()
```
If we want to know, on average, which type of drive wheel is most valuable, we can group "drive-wheels" and then average them.
We can select the columns 'drive-wheels','body-style', and 'price', then assign it to the variable "df_group_one".
```
df_group_one=df[['drive-wheels','body-style','price']]
```
We can then calculate the average price for each of the different categories of data:
```
# grouping results
df_group_one=df_group_one.groupby(['drive-wheels'],as_index= False).mean()
df_group_one
```
From our data, it seems rear-wheel drive vehicles are, on average, the most expensive, while 4-wheel and front-wheel are approximately the same in price.
You can also group with multiple variables. For example, let's group by both 'drive-wheels' and 'body-style'. This groups the dataframe by the unique combinations 'drive-wheels' and 'body-style'. We can store the results in the variable 'grouped_test1':
```
# grouping results
df_gptest=df[['drive-wheels','body-style','price']]
grouped_test1=df_gptest.groupby(['drive-wheels','body-style'],as_index= False).mean()
grouped_test1
```
This grouped data is much easier to visualize when it is made into a pivot table. A pivot table is like an Excel spreadsheet, with one variable along the column and another along the row. We can convert the dataframe to a pivot table using the method "pivot " to create a pivot table from the groups.
In this case, we will leave the drive-wheel variable as the rows of the table, and pivot body-style to become the columns of the table:
```
grouped_pivot=grouped_test1.pivot(index='drive-wheels',columns='body-style')
grouped_pivot
```
Often, we won't have data for some of the pivot cells. We can fill these missing cells with the value 0, but any other value could potentially be used as well. It should be mentioned that missing data is quite a complex subject and is an entire course on its own.
```
grouped_pivot=grouped_pivot.fillna(0) #fill missing values with 0
grouped_pivot
```
<div class="alert alert-danger alertdanger" style="margin-top: 20px">
<h1> Question 4 : </h1>
<b> Use the "groupby" function to find the average "price" of each car based on "body-style": </b>
</div>
```
df_gptest.groupby(['body-style'],as_index= False).mean()
```
If you didn't import "pyplot", let's do it again:
```
import matplotlib.pyplot as plt
% matplotlib inline
```
#### Variables: Drive Wheels and Body Style vs Price
Let's use a heat map to visualize the relationship between Body Style vs Price:
```
#use the grouped results
plt.pcolor(grouped_pivot, cmap='RdBu')
plt.colorbar()
plt.show()
```
The heatmap plots the target variable (price) proportional to colour with respect to the variables 'drive-wheel' and 'body-style' in the vertical and horizontal axis, respectively. This allows us to visualize how the price is related to 'drive-wheel' and 'body-style'.
The default labels convey no useful information to us. Let's change that:
```
fig, ax=plt.subplots()
im=ax.pcolor(grouped_pivot, cmap='RdBu')
#label names
row_labels=grouped_pivot.columns.levels[1]
col_labels=grouped_pivot.index
#move ticks and labels to the center
ax.set_xticks(np.arange(grouped_pivot.shape[1])+0.5, minor=False)
ax.set_yticks(np.arange(grouped_pivot.shape[0])+0.5, minor=False)
#insert labels
ax.set_xticklabels(row_labels, minor=False)
ax.set_yticklabels(col_labels, minor=False)
#rotate label if too long
plt.xticks(rotation=90)
fig.colorbar(im)
plt.show()
```
Visualization is very important in data science, and Python visualization packages provide great freedom. We will go more in-depth in a separate Python Visualizations course.
The main question we want to answer in this module is, "What are the main characteristics which have the most impact on the car price?".
To get a better measure of the important characteristics, we look at the correlation of these variables with the car price. In other words, how is the car price dependent on this variable?
## 5. Correlation and Causation
**Correlation**: a measure of the extent of interdependence between variables.
**Causation**: the relationship between cause and effect between two variables.
It is important to know the difference between these two and that correlation does not imply causation. Determining correlation is much simpler than determining causation, as causation may require independent experimentation.
## Pearson Correlation
The Pearson Correlation measures the linear dependence between two variables, X and Y.
The resulting coefficient is a value between -1 and 1 inclusive, where:
- **1**: total positive linear correlation,
- **0**: no linear correlation, the two variables most likely do not affect each other
- **-1**: total negative linear correlation.
Pearson Correlation is the default method of the function "corr". As before, we can calculate the Pearson correlation of the of the 'int64' or 'float64' variables:
```
df.corr()
```
Sometimes we would like to know the significance of the correlation estimate.
**P-value**:
What is this P-value? The P-value is the probability value that the correlation between these two variables is statistically significant. Normally, we choose a significance level of 0.05, which means that we are 95% confident that the correlation between the variables is significant.
By convention, when the p-value is:
- < 0.001 we say there is strong evidence that the correlation is significant,
- < 0.05; there is moderate evidence that the correlation is significant,
- < 0.1; there is weak evidence that the correlation is significant, and
- is > 0.1; there is no evidence that the correlation is significant.
We can obtain this information using "stats" module in the "scipy" library:
```
from scipy import stats
```
### Wheel-base vs Price
Let's calculate the Pearson Correlation Coefficient and P-value of 'wheel-base' and 'price':
```
pearson_coef, p_value = stats.pearsonr(df['wheel-base'], df['price'])
print("The Pearson Correlation Coefficient is", pearson_coef, " with a P-value of P =", p_value)
```
##### Conclusion:
Since the p-value is < 0.001, the correlation between wheel-base and price is statistically significant, although the linear relationship isn't extremely strong (~0.585).
### Horsepower vs Price
Let's calculate the Pearson Correlation Coefficient and P-value of 'horsepower' and 'price':
```
pearson_coef, p_value = stats.pearsonr(df['horsepower'], df['price'])
print("The Pearson Correlation Coefficient is", pearson_coef, " with a P-value of P =", p_value)
```
##### Conclusion:
Since the p-value is < 0.001, the correlation between horsepower and price is statistically significant, and the linear relationship is quite strong (~0.809, close to 1).
### Length vs Price
Let's calculate the Pearson Correlation Coefficient and P-value of 'length' and 'price':
```
pearson_coef, p_value = stats.pearsonr(df['length'], df['price'])
print("The Pearson Correlation Coefficient is", pearson_coef, " with a P-value of P =", p_value)
```
##### Conclusion:
Since the p-value is < 0.001, the correlation between length and price is statistically significant, and the linear relationship is moderately strong (~0.691).
### Width vs Price
Let's calculate the Pearson Correlation Coefficient and P-value of 'width' and 'price':
```
pearson_coef, p_value = stats.pearsonr(df['width'], df['price'])
print("The Pearson Correlation Coefficient is", pearson_coef, " with a P-value of P =", p_value )
```
##### Conclusion:
Since the p-value is < 0.001, the correlation between width and price is statistically significant, and the linear relationship is quite strong (~0.751).
### Curb-weight vs Price
Let's calculate the Pearson Correlation Coefficient and P-value of 'curb-weight' and 'price':
```
pearson_coef, p_value = stats.pearsonr(df['curb-weight'], df['price'])
print( "The Pearson Correlation Coefficient is", pearson_coef, " with a P-value of P =", p_value)
```
##### Conclusion:
Since the p-value is < 0.001, the correlation between curb-weight and price is statistically significant, and the linear relationship is quite strong (~0.834).
### Engine-size vs Price
Let's calculate the Pearson Correlation Coefficient and P-value of 'engine-size' and 'price':
```
pearson_coef, p_value = stats.pearsonr(df['engine-size'], df['price'])
print("The Pearson Correlation Coefficient is", pearson_coef, " with a P-value of P =", p_value)
```
##### Conclusion:
Since the p-value is < 0.001, the correlation between engine-size and price is statistically significant, and the linear relationship is very strong (~0.872).
### Bore vs Price
Let's calculate the Pearson Correlation Coefficient and P-value of 'bore' and 'price':
```
pearson_coef, p_value = stats.pearsonr(df['bore'], df['price'])
print("The Pearson Correlation Coefficient is", pearson_coef, " with a P-value of P =", p_value )
```
##### Conclusion:
Since the p-value is < 0.001, the correlation between bore and price is statistically significant, but the linear relationship is only moderate (~0.521).
We can relate the process for each 'City-mpg' and 'Highway-mpg':
### City-mpg vs Price
```
pearson_coef, p_value = stats.pearsonr(df['city-mpg'], df['price'])
print("The Pearson Correlation Coefficient is", pearson_coef, " with a P-value of P =", p_value)
```
##### Conclusion:
Since the p-value is < 0.001, the correlation between city-mpg and price is statistically significant, and the coefficient of ~ -0.687 shows that the relationship is negative and moderately strong.
### Highway-mpg vs Price
```
pearson_coef, p_value = stats.pearsonr(df['highway-mpg'], df['price'])
print( "The Pearson Correlation Coefficient is", pearson_coef, " with a P-value of P =", p_value )
```
##### Conclusion:
Since the p-value is < 0.001, the correlation between highway-mpg and price is statistically significant, and the coefficient of ~ -0.705 shows that the relationship is negative and moderately strong.
## 6. ANOVA
### ANOVA: Analysis of Variance
The Analysis of Variance (ANOVA) is a statistical method used to test whether there are significant differences between the means of two or more groups. ANOVA returns two parameters:
**F-test score**: ANOVA assumes the means of all groups are the same, calculates how much the actual means deviate from the assumption, and reports it as the F-test score. A larger score means there is a larger difference between the means.
**P-value**: P-value tells us the statistical significance of our calculated score value.
If our price variable is strongly correlated with the variable we are analyzing, expect ANOVA to return a sizeable F-test score and a small p-value.
### Drive Wheels
Since ANOVA analyzes the difference between different groups of the same variable, the groupby function will come in handy. Because the ANOVA algorithm averages the data automatically, we do not need to take the average before-hand.
Let's see if different types 'drive-wheels' impact 'price'. We group the data:
```
grouped_test2=df_gptest[['drive-wheels','price']].groupby(['drive-wheels'])
grouped_test2.head(2)
```
We can obtain the values of the method group using the method "get_group":
```
grouped_test2.get_group('4wd')['price']
```
We can use the function 'f_oneway' in the module 'stats' to obtain the **F-test score** and **P-value**:
```
# ANOVA
f_val, p_val = stats.f_oneway(grouped_test2.get_group('fwd')['price'], grouped_test2.get_group('rwd')['price'], grouped_test2.get_group('4wd')['price'])
print( "ANOVA results: F=", f_val, ", P =", p_val)
```
This is a great result, with a large F test score showing a strong correlation and a P value of almost 0, implying almost certain statistical significance. But does this mean all three tested groups are all this highly correlated?
#### Separately: fwd and rwd:
```
f_val, p_val = stats.f_oneway(grouped_test2.get_group('fwd')['price'], grouped_test2.get_group('rwd')['price'])
print( "ANOVA results: F=", f_val, ", P =", p_val )
```
Let's examine the other groups
#### 4wd and rwd:
```
f_val, p_val = stats.f_oneway(grouped_test2.get_group('4wd')['price'], grouped_test2.get_group('rwd')['price'])
print( "ANOVA results: F=", f_val, ", P =", p_val)
```
#### 4wd and fwd:
```
f_val, p_val = stats.f_oneway(grouped_test2.get_group('4wd')['price'], grouped_test2.get_group('fwd')['price'])
print("ANOVA results: F=", f_val, ", P =", p_val)
```
## Conclusion: Important Variables
We now have a better idea of what our data looks like and which variables are important to take into account when predicting the car price. We have narrowed it down to the following variables:
Continuous numerical variables:
- Length
- Width
- Curb-weight
- Engine-size
- Horsepower
- City-mpg
- Highway-mpg
- Wheel-base
- Bore
Categorical variables:
- Drive-wheels
As we now move into building machine learning models to automate our analysis, feeding the model with variables that meaningfully affect our target variable will improve our model's prediction performance.
# About the Authors:
This notebook written by [Mahdi Noorian PhD](https://www.linkedin.com/in/mahdi-noorian-58219234/) ,[Joseph Santarcangelo PhD]( https://www.linkedin.com/in/joseph-s-50398b136/), Bahare Talayian, Eric Xiao, Steven Dong, Parizad , Hima Vsudevan and [Fiorella Wenver](https://www.linkedin.com/in/fiorellawever/).
Copyright © 2017 [cognitiveclass.ai](cognitiveclass.ai?utm_source=bducopyrightlink&utm_medium=dswb&utm_campaign=bdu). This notebook and its source code are released under the terms of the [MIT License](https://bigdatauniversity.com/mit-license/).
<div class="alert alert-block alert-info" style="margin-top: 20px">
<a href="http://cocl.us/DA0101EN_NotbookLink_bottom"><img src = "https://ibm.box.com/shared/static/cy2mwm7519t4z6dxefjpzgtbpi9p8l7h.png" width = 750, align = "center"></a>
<h1 align=center><font size = 5> Link</font></h1>
|
github_jupyter
|
import pandas as pd
import numpy as np
path='https://ibm.box.com/shared/static/q6iiqb1pd7wo8r3q28jvgsrprzezjqk3.csv'
df = pd.read_csv(path)
df.head()
import matplotlib.pyplot as plt
import seaborn as sns
%matplotlib inline
# list the data types for each column
df.dtypes
float64
df.corr()
df[['bore','stroke' ,'compression-ratio','horsepower']].corr()
df[['bore','stroke' ,'compression-ratio','horsepower']].corr()
# Engine size as potential predictor variable of price
sns.regplot(x="engine-size", y="price", data=df)
plt.ylim(0,)
df[["engine-size", "price"]].corr()
sns.regplot(x="highway-mpg", y="price", data=df)
df[['highway-mpg', 'price']].corr()
sns.regplot(x="peak-rpm", y="price", data=df)
df[['peak-rpm','price']].corr()
df[["stroke","price"]].corr()
The correlation is 0.0823, the non-diagonal elements of the table.
code:df[["stroke","price"]].corr()
sns.regplot(x="stroke", y="price", data=df)
There is a weak correlation between the variable 'stroke' and 'price.' as such regression will not work well. We can see this use "regplot" to demonstrate this.
Code: sns.regplot(x="stroke", y="price", data=df)
sns.boxplot(x="body-style", y="price", data=df)
sns.boxplot(x="engine-location", y="price", data=df)
# drive-wheels
sns.boxplot(x="drive-wheels", y="price", data=df)
df.describe()
df.describe(include=['object'])
df['drive-wheels'].value_counts()
df['drive-wheels'].value_counts().to_frame()
drive_wheels_counts = df['drive-wheels'].value_counts().to_frame()
drive_wheels_counts.rename(columns={'drive-wheels': 'value_counts'}, inplace=True)
drive_wheels_counts
drive_wheels_counts.index.name = 'drive-wheels'
drive_wheels_counts
# engine-location as variable
engine_loc_counts = df['engine-location'].value_counts().to_frame()
engine_loc_counts.rename(columns={'engine-location': 'value_counts'}, inplace=True)
engine_loc_counts.index.name = 'engine-location'
engine_loc_counts.head(10)
df['drive-wheels'].unique()
df_group_one=df[['drive-wheels','body-style','price']]
# grouping results
df_group_one=df_group_one.groupby(['drive-wheels'],as_index= False).mean()
df_group_one
# grouping results
df_gptest=df[['drive-wheels','body-style','price']]
grouped_test1=df_gptest.groupby(['drive-wheels','body-style'],as_index= False).mean()
grouped_test1
grouped_pivot=grouped_test1.pivot(index='drive-wheels',columns='body-style')
grouped_pivot
grouped_pivot=grouped_pivot.fillna(0) #fill missing values with 0
grouped_pivot
df_gptest.groupby(['body-style'],as_index= False).mean()
import matplotlib.pyplot as plt
% matplotlib inline
#use the grouped results
plt.pcolor(grouped_pivot, cmap='RdBu')
plt.colorbar()
plt.show()
fig, ax=plt.subplots()
im=ax.pcolor(grouped_pivot, cmap='RdBu')
#label names
row_labels=grouped_pivot.columns.levels[1]
col_labels=grouped_pivot.index
#move ticks and labels to the center
ax.set_xticks(np.arange(grouped_pivot.shape[1])+0.5, minor=False)
ax.set_yticks(np.arange(grouped_pivot.shape[0])+0.5, minor=False)
#insert labels
ax.set_xticklabels(row_labels, minor=False)
ax.set_yticklabels(col_labels, minor=False)
#rotate label if too long
plt.xticks(rotation=90)
fig.colorbar(im)
plt.show()
df.corr()
from scipy import stats
pearson_coef, p_value = stats.pearsonr(df['wheel-base'], df['price'])
print("The Pearson Correlation Coefficient is", pearson_coef, " with a P-value of P =", p_value)
pearson_coef, p_value = stats.pearsonr(df['horsepower'], df['price'])
print("The Pearson Correlation Coefficient is", pearson_coef, " with a P-value of P =", p_value)
pearson_coef, p_value = stats.pearsonr(df['length'], df['price'])
print("The Pearson Correlation Coefficient is", pearson_coef, " with a P-value of P =", p_value)
pearson_coef, p_value = stats.pearsonr(df['width'], df['price'])
print("The Pearson Correlation Coefficient is", pearson_coef, " with a P-value of P =", p_value )
pearson_coef, p_value = stats.pearsonr(df['curb-weight'], df['price'])
print( "The Pearson Correlation Coefficient is", pearson_coef, " with a P-value of P =", p_value)
pearson_coef, p_value = stats.pearsonr(df['engine-size'], df['price'])
print("The Pearson Correlation Coefficient is", pearson_coef, " with a P-value of P =", p_value)
pearson_coef, p_value = stats.pearsonr(df['bore'], df['price'])
print("The Pearson Correlation Coefficient is", pearson_coef, " with a P-value of P =", p_value )
pearson_coef, p_value = stats.pearsonr(df['city-mpg'], df['price'])
print("The Pearson Correlation Coefficient is", pearson_coef, " with a P-value of P =", p_value)
pearson_coef, p_value = stats.pearsonr(df['highway-mpg'], df['price'])
print( "The Pearson Correlation Coefficient is", pearson_coef, " with a P-value of P =", p_value )
grouped_test2=df_gptest[['drive-wheels','price']].groupby(['drive-wheels'])
grouped_test2.head(2)
grouped_test2.get_group('4wd')['price']
# ANOVA
f_val, p_val = stats.f_oneway(grouped_test2.get_group('fwd')['price'], grouped_test2.get_group('rwd')['price'], grouped_test2.get_group('4wd')['price'])
print( "ANOVA results: F=", f_val, ", P =", p_val)
f_val, p_val = stats.f_oneway(grouped_test2.get_group('fwd')['price'], grouped_test2.get_group('rwd')['price'])
print( "ANOVA results: F=", f_val, ", P =", p_val )
f_val, p_val = stats.f_oneway(grouped_test2.get_group('4wd')['price'], grouped_test2.get_group('rwd')['price'])
print( "ANOVA results: F=", f_val, ", P =", p_val)
f_val, p_val = stats.f_oneway(grouped_test2.get_group('4wd')['price'], grouped_test2.get_group('fwd')['price'])
print("ANOVA results: F=", f_val, ", P =", p_val)
| 0.468547 | 0.953275 |
```
# For automatic reloading of modified libraries
%reload_ext autoreload
%autoreload 2
# Regular python libraries
import os
import requests
import sys
import json
import statistics
import torch
# AzureML libraries
import azureml
import azureml.core
from azureml.core import Experiment, Workspace, Datastore, ScriptRunConfig
from azureml.core.compute import ComputeTarget, AmlCompute
from azureml.core.compute_target import ComputeTargetException
from azureml.core.conda_dependencies import CondaDependencies
from azureml.core.container_registry import ContainerRegistry
from azureml.core.runconfig import MpiConfiguration, RunConfiguration, DEFAULT_GPU_IMAGE
from azureml.train.estimator import Estimator
from azureml.widgets import RunDetails
# Check core SDK version number
print("SDK version:", azureml.core.VERSION)
```
## Initialize workspace
To create or access an Azure ML Workspace, you will need to import the AML library and the following information:
* A name for your workspace
* Your subscription id
* The resource group name
Initialize a [Workspace](https://docs.microsoft.com/azure/machine-learning/service/concept-azure-machine-learning-architecture#workspace) object from the existing workspace you created in the Prerequisites step or create a new one.
```
subscription_id = '<subscription_id>'
resource_group = '<resource_group>'
workspace_name = '<workspace_name>'
ws = Workspace(subscription_id, resource_group, workspace_name)
ws_details = ws.get_details()
print('Name:\t\t{}\nLocation:\t{}'
.format(ws_details['name'],
ws_details['location']))
from azureml.core import Datastore
ds = ws.get_default_datastore()
#ds = Datastore.get(ws,'default')
#ds = Datastore.get(ws,'workspaceblobstore')
print('Datastore name: ' + ds.name,
'Container name: ' + ds.container_name,
'Datastore type: ' + ds.datastore_type,
'Workspace name: ' + ds.workspace.name, sep = '\n')
# ws.get_default_datastore().container_name
# ws.datastores
# Create the compute cluster
gpu_cluster_name = "<cluster name>"
# Verify that the cluster doesn't exist already
try:
gpu_compute_target = ComputeTarget(workspace=ws, name=gpu_cluster_name)
if gpu_compute_target.provisioning_state == 'Failed':
gpu_compute_target.delete()
gpu_compute_target.wait_for_completion(show_output=True)
raise ComputeTargetException('failed cluster')
print('Found existing compute target.')
except ComputeTargetException:
print('Creating a new compute target...')
compute_config = AmlCompute.provisioning_configuration(vm_priority='lowpriority' ,
vm_size='Standard_NC24rs_v3',
min_nodes=0, max_nodes=16)
# ^^^ Change to min_nodes=8 and max_nodes=64 when testing is completed^^^
# create the cluster
gpu_compute_target = ComputeTarget.create(ws, gpu_cluster_name, compute_config)
gpu_compute_target.wait_for_completion(show_output=True)
# Use the 'status' property to get a detailed status for the current cluster.
print(gpu_compute_target.status.serialize())
from azureml.train.dnn import PyTorch
from azureml.core.runconfig import RunConfiguration
from azureml.core.container_registry import ContainerRegistry
run_user_managed = RunConfiguration()
run_user_managed.environment.python.user_managed_dependencies = True
```
### Germ Eval setup
```
script_name = 'test.py'
codepath = '.'
from azureml.core import Dataset
from azureml.data import OutputFileDatasetConfig
#create input/output datasets
def ds_input_path(path):
return Dataset.File.from_files(ds.path(path))
def ds_output_path(path):
return OutputFileDatasetConfig(destination=(ds, path))
def get_args():
all_params_default = [
'--data.train_filepath', './train_germ/train.tsv',
'--data.val_filepath', './val_germ/dev.tsv',
'--config_path', 'config_germ.yaml',
'--ckpt.model_state_save_dir', './mod_ckpts',
'--ckpt.save_dir', './ckpts'
]
return all_params_default
from azureml.core import Environment
myenv = Environment(name="myenv")
myenv.docker.base_image = 'jonrsleep/elr2:latest'
myenv.python.interpreter_path = '/opt/miniconda/envs/elr2/bin/python'
myenv.python.user_managed_dependencies = True
mpi = MpiConfiguration()
mpi.process_count_per_node = 1 #NC SKU has 4 GPU's per node
mpi.node_count = 1 #scale to the amount of nodes you'd like
config = ScriptRunConfig(source_directory=codepath,
script=script_name,
arguments = get_args(),
compute_target=gpu_compute_target,
environment=myenv,
distributed_job_config=mpi)
experiment_name = 'marlin_ner_train_plugin_germ'
experiment = Experiment(ws, name=experiment_name)
run = experiment.submit(config)
run.tag('nodes', f'{mpi.node_count}')
run.tag('exp', 'lr 3e-5 ')
print("Submitted run")
# distrib eval test
RunDetails(run).show()
```
### Model checkpoint modification
```
import torch
from collections import OrderedDict
state_dict = torch.load('marlin_0.bin', map_location='cpu')
##Modify to point to model
new_dict = OrderedDict((key.replace('model.',''), value) for key, value in state_dict['module_interface_state'].items() if key.startswith('model.') )
#print(new_dict.keys())
torch.save(new_dict, 'marlin_model.bin')
```
### Run Inference - modify test.py to remove trainer.train()
```
script_name = 'test.py'
codepath = '.'
from azureml.core import Dataset
from azureml.data import OutputFileDatasetConfig
#create input/output datasets
def ds_input_path(path):
return Dataset.File.from_files(ds.path(path))
def ds_output_path(path):
return OutputFileDatasetConfig(destination=(ds, path))
def get_args():
all_params_default = [
'--data.train_filepath', './train_germ/train.tsv',
'--data.val_filepath', './val_germ/dev.tsv',
'--config_path', 'config_germ.yaml',
'--model.model_path', '< Modify to point to model directory>',
'--model.model_file', 'marlin_model.bin'
]
return all_params_default
from azureml.core import Environment
myenv = Environment(name="myenv")
myenv.docker.base_image = 'jonrsleep/elr2:latest'
myenv.python.interpreter_path = '/opt/miniconda/envs/elr2/bin/python'
myenv.python.user_managed_dependencies = True
mpi = MpiConfiguration()
mpi.process_count_per_node = 1 #NC SKU has 4 GPU's per node
mpi.node_count = 1 #scale to the amount of nodes you'd like
config = ScriptRunConfig(source_directory=codepath,
script=script_name,
arguments = get_args(),
compute_target=gpu_compute_target,
environment=myenv,
distributed_job_config=mpi)
experiment_name = 'marlin_ner_train_plugin_germ_inf'
experiment = Experiment(ws, name=experiment_name)
run = experiment.submit(config)
run.tag('nodes', f'{mpi.node_count}')
run.tag('exp', 'lr 3e-5 ')
print("Submitted run")
```
|
github_jupyter
|
# For automatic reloading of modified libraries
%reload_ext autoreload
%autoreload 2
# Regular python libraries
import os
import requests
import sys
import json
import statistics
import torch
# AzureML libraries
import azureml
import azureml.core
from azureml.core import Experiment, Workspace, Datastore, ScriptRunConfig
from azureml.core.compute import ComputeTarget, AmlCompute
from azureml.core.compute_target import ComputeTargetException
from azureml.core.conda_dependencies import CondaDependencies
from azureml.core.container_registry import ContainerRegistry
from azureml.core.runconfig import MpiConfiguration, RunConfiguration, DEFAULT_GPU_IMAGE
from azureml.train.estimator import Estimator
from azureml.widgets import RunDetails
# Check core SDK version number
print("SDK version:", azureml.core.VERSION)
subscription_id = '<subscription_id>'
resource_group = '<resource_group>'
workspace_name = '<workspace_name>'
ws = Workspace(subscription_id, resource_group, workspace_name)
ws_details = ws.get_details()
print('Name:\t\t{}\nLocation:\t{}'
.format(ws_details['name'],
ws_details['location']))
from azureml.core import Datastore
ds = ws.get_default_datastore()
#ds = Datastore.get(ws,'default')
#ds = Datastore.get(ws,'workspaceblobstore')
print('Datastore name: ' + ds.name,
'Container name: ' + ds.container_name,
'Datastore type: ' + ds.datastore_type,
'Workspace name: ' + ds.workspace.name, sep = '\n')
# ws.get_default_datastore().container_name
# ws.datastores
# Create the compute cluster
gpu_cluster_name = "<cluster name>"
# Verify that the cluster doesn't exist already
try:
gpu_compute_target = ComputeTarget(workspace=ws, name=gpu_cluster_name)
if gpu_compute_target.provisioning_state == 'Failed':
gpu_compute_target.delete()
gpu_compute_target.wait_for_completion(show_output=True)
raise ComputeTargetException('failed cluster')
print('Found existing compute target.')
except ComputeTargetException:
print('Creating a new compute target...')
compute_config = AmlCompute.provisioning_configuration(vm_priority='lowpriority' ,
vm_size='Standard_NC24rs_v3',
min_nodes=0, max_nodes=16)
# ^^^ Change to min_nodes=8 and max_nodes=64 when testing is completed^^^
# create the cluster
gpu_compute_target = ComputeTarget.create(ws, gpu_cluster_name, compute_config)
gpu_compute_target.wait_for_completion(show_output=True)
# Use the 'status' property to get a detailed status for the current cluster.
print(gpu_compute_target.status.serialize())
from azureml.train.dnn import PyTorch
from azureml.core.runconfig import RunConfiguration
from azureml.core.container_registry import ContainerRegistry
run_user_managed = RunConfiguration()
run_user_managed.environment.python.user_managed_dependencies = True
script_name = 'test.py'
codepath = '.'
from azureml.core import Dataset
from azureml.data import OutputFileDatasetConfig
#create input/output datasets
def ds_input_path(path):
return Dataset.File.from_files(ds.path(path))
def ds_output_path(path):
return OutputFileDatasetConfig(destination=(ds, path))
def get_args():
all_params_default = [
'--data.train_filepath', './train_germ/train.tsv',
'--data.val_filepath', './val_germ/dev.tsv',
'--config_path', 'config_germ.yaml',
'--ckpt.model_state_save_dir', './mod_ckpts',
'--ckpt.save_dir', './ckpts'
]
return all_params_default
from azureml.core import Environment
myenv = Environment(name="myenv")
myenv.docker.base_image = 'jonrsleep/elr2:latest'
myenv.python.interpreter_path = '/opt/miniconda/envs/elr2/bin/python'
myenv.python.user_managed_dependencies = True
mpi = MpiConfiguration()
mpi.process_count_per_node = 1 #NC SKU has 4 GPU's per node
mpi.node_count = 1 #scale to the amount of nodes you'd like
config = ScriptRunConfig(source_directory=codepath,
script=script_name,
arguments = get_args(),
compute_target=gpu_compute_target,
environment=myenv,
distributed_job_config=mpi)
experiment_name = 'marlin_ner_train_plugin_germ'
experiment = Experiment(ws, name=experiment_name)
run = experiment.submit(config)
run.tag('nodes', f'{mpi.node_count}')
run.tag('exp', 'lr 3e-5 ')
print("Submitted run")
# distrib eval test
RunDetails(run).show()
import torch
from collections import OrderedDict
state_dict = torch.load('marlin_0.bin', map_location='cpu')
##Modify to point to model
new_dict = OrderedDict((key.replace('model.',''), value) for key, value in state_dict['module_interface_state'].items() if key.startswith('model.') )
#print(new_dict.keys())
torch.save(new_dict, 'marlin_model.bin')
script_name = 'test.py'
codepath = '.'
from azureml.core import Dataset
from azureml.data import OutputFileDatasetConfig
#create input/output datasets
def ds_input_path(path):
return Dataset.File.from_files(ds.path(path))
def ds_output_path(path):
return OutputFileDatasetConfig(destination=(ds, path))
def get_args():
all_params_default = [
'--data.train_filepath', './train_germ/train.tsv',
'--data.val_filepath', './val_germ/dev.tsv',
'--config_path', 'config_germ.yaml',
'--model.model_path', '< Modify to point to model directory>',
'--model.model_file', 'marlin_model.bin'
]
return all_params_default
from azureml.core import Environment
myenv = Environment(name="myenv")
myenv.docker.base_image = 'jonrsleep/elr2:latest'
myenv.python.interpreter_path = '/opt/miniconda/envs/elr2/bin/python'
myenv.python.user_managed_dependencies = True
mpi = MpiConfiguration()
mpi.process_count_per_node = 1 #NC SKU has 4 GPU's per node
mpi.node_count = 1 #scale to the amount of nodes you'd like
config = ScriptRunConfig(source_directory=codepath,
script=script_name,
arguments = get_args(),
compute_target=gpu_compute_target,
environment=myenv,
distributed_job_config=mpi)
experiment_name = 'marlin_ner_train_plugin_germ_inf'
experiment = Experiment(ws, name=experiment_name)
run = experiment.submit(config)
run.tag('nodes', f'{mpi.node_count}')
run.tag('exp', 'lr 3e-5 ')
print("Submitted run")
| 0.367838 | 0.588416 |
# Qiskit Pulseで高エネルギー状態へアクセスする
ほとんどの量子アルゴリズム/アプリケーションでは、$|0\rangle$と$|1\rangle$によって張られた2次元空間で計算が実行されます。ただし、IBMのハードウェアでは、通常は使用されない、より高いエネルギー状態も存在します。このセクションでは、Qiskit Pulseを使ってこれらの状態を探索することにフォーカスを当てます。特に、$|2\rangle$ 状態を励起し、$|0\rangle$、$|1\rangle$、$|2\rangle$の状態を分類するための識別器を作成する方法を示します。
このノートブックを読む前に、[前の章](./calibrating-qubits-openpulse.html)を読むことをお勧めします。また、Qiskit Pulseのスペック(Ref [1](#refs))も読むことをお勧めします。
### 物理学的背景
ここで、IBMの量子ハードウェアの多くの基礎となっている、トランズモンキュービットの物理学的な背景を説明します。このシステムには、ジョセフソン接合とコンデンサーで構成される超伝導回路が含まれています。超伝導回路に不慣れな方は、[こちらのレビュー](https://arxiv.org/pdf/1904.06560.pdf) (Ref. [2](#refs))を参照してください。このシステムのハミルトニアンは以下で与えられます。
$$
H = 4 E_C n^2 - E_J \cos(\phi),
$$
ここで、$E_C, E_J$はコンデンサーのエネルギーとジョセフソンエネルギーを示し、$n$は減衰した電荷数演算子で、$\phi$はジャンクションのために減衰した磁束です。$\hbar=1$として扱います。
トランズモンキュービットは$\phi$が小さい領域で定義されるため、$E_J \cos(\phi)$をテイラー級数で展開できます(定数項を無視します)。
$$
E_J \cos(\phi) \approx \frac{1}{2} E_J \phi^2 - \frac{1}{24} E_J \phi^4 + \mathcal{O}(\phi^6).
$$
$\phi$の二次の項$\phi^2$は、標準の調和振動子を定義します。その他の追加の項はそれぞれ非調和性をもたらします。
$n \sim (a-a^\dagger), \phi \sim (a+a^\dagger)$の関係を使うと($a^\dagger,a$は生成消滅演算子)、システムは以下のハミルトニアンを持つダフィング(Duffing)振動子に似ていることを示せます。
$$
H = \omega a^\dagger a + \frac{\alpha}{2} a^\dagger a^\dagger a a,
$$
$\omega$は、$0\rightarrow1$の励起周波数($\omega \equiv \omega^{0\rightarrow1}$)を与え、$\alpha$は$0\rightarrow1$の周波数と$1\rightarrow2$の周波数の間の非調和です。必要に応じて駆動の条件を追加できます。
標準の2次元部分空間へ特化したい場合は、$|\alpha|$ を十分に大きくとるか、高エネルギー状態を抑制する特別な制御テクニックを使います。
## 目次
0. [はじめに](#importing)
1. [0と1の状態の識別](#discrim01)
1. [0->1の周波数スイープ](#freqsweep01)
2. [0->1のラビ実験](#rabi01)
3. [0,1の識別器を構築する](#builddiscrim01)
2. [0,1,2の状態の識別](#discrim012)
1. [1->2の周波数の計算](#freq12)
1. [サイドバンド法を使った1->2の周波数スイープ](#sideband12)
2. [1->2のラビ実験](#rabi12)
3. [0,1,2の識別器を構築する](#builddiscrim012)
4. [参考文献](#refs)
## 0. はじめに <a id="importing"></a>
まず、依存関係をインポートし、いくつかのデフォルトの変数を定義します。量子ビット0を実験に使います。公開されている単一量子ビットデバイスである`ibmq_armonk`で実験を行います。
```
import numpy as np
import matplotlib.pyplot as plt
from scipy.optimize import curve_fit
from scipy.signal import find_peaks
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis
from sklearn.model_selection import train_test_split
import qiskit.pulse as pulse
import qiskit.pulse.library as pulse_lib
from qiskit.compiler import assemble
from qiskit.pulse.library import SamplePulse
from qiskit.tools.monitor import job_monitor
import warnings
warnings.filterwarnings('ignore')
from qiskit.tools.jupyter import *
%matplotlib inline
from qiskit import IBMQ
IBMQ.load_account()
provider = IBMQ.get_provider(hub='ibm-q', group='open', project='main')
backend = provider.get_backend('ibmq_armonk')
backend_config = backend.configuration()
assert backend_config.open_pulse, "Backend doesn't support Pulse"
dt = backend_config.dt
backend_defaults = backend.defaults()
# 単位変換係数 -> すべてのバックエンドのプロパティーがSI単位系(Hz, sec, etc)で返される
GHz = 1.0e9 # Gigahertz
MHz = 1.0e6 # Megahertz
us = 1.0e-6 # Microseconds
ns = 1.0e-9 # Nanoseconds
qubit = 0 # 分析に使う量子ビット
default_qubit_freq = backend_defaults.qubit_freq_est[qubit] # デフォルトの量子ビット周波数単位はHz
print(f"Qubit {qubit} has an estimated frequency of {default_qubit_freq/ GHz} GHz.")
#(各デバイスに固有の)データをスケーリング
scale_factor = 1e-14
# 実験のショット回数
NUM_SHOTS = 1024
### 必要なチャネルを収集する
drive_chan = pulse.DriveChannel(qubit)
meas_chan = pulse.MeasureChannel(qubit)
acq_chan = pulse.AcquireChannel(qubit)
```
いくつか便利な関数を追加で定義します。
```
def get_job_data(job, average):
"""すでに実行されているジョブからデータを取得します。
引数:
job (Job): データが必要なジョブ
average (bool): Trueの場合、データが平均であると想定してデータを取得。
Falseの場合、シングルショット用と想定してデータを取得。
返し値:
list: ジョブの結果データを含むリスト
"""
job_results = job.result(timeout=120) # タイムアウトパラメーターは120秒にセット
result_data = []
for i in range(len(job_results.results)):
if average: # 平均データを得る
result_data.append(job_results.get_memory(i)[qubit]*scale_factor)
else: # シングルデータを得る
result_data.append(job_results.get_memory(i)[:, qubit]*scale_factor)
return result_data
def get_closest_multiple_of_16(num):
"""16の倍数に最も近いものを計算します。
パルスが使えるデバイスが16サンプルの倍数の期間が必要なためです。
"""
return (int(num) - (int(num)%16))
```
次に、駆動パルスと測定のためのいくつかのデフォルトパラメーターを含めます。命令スケジュールマップから(バックエンドデフォルトから)`measure`コマンドをプルして、新しいキャリブレーションでアップデートされるようにします。
```
# 駆動パルスのパラメーター (us = マイクロ秒)
drive_sigma_us = 0.075 # ガウシアンの実際の幅を決めます
drive_samples_us = drive_sigma_us*8 # 切り捨てパラメーター
# ガウシアンには自然な有限長がないためです。
drive_sigma = get_closest_multiple_of_16(drive_sigma_us * us /dt) # ガウシアンの幅の単位はdt
drive_samples = get_closest_multiple_of_16(drive_samples_us * us /dt) # 切り捨てパラメーターの単位はdt
# この量子ビットに必要な測定マップインデックスを見つける
meas_map_idx = None
for i, measure_group in enumerate(backend_config.meas_map):
if qubit in measure_group:
meas_map_idx = i
break
assert meas_map_idx is not None, f"Couldn't find qubit {qubit} in the meas_map!"
# 命令スケジュールマップからデフォルトの測定パルスを取得
inst_sched_map = backend_defaults.instruction_schedule_map
measure = inst_sched_map.get('measure', qubits=backend_config.meas_map[meas_map_idx])
```
## 1. $|0\rangle$ と $|1\rangle$の状態の識別 <a id="discrim01"></a>
このセクションでは、標準の$|0\rangle$と$|1\rangle$の状態の識別器を構築します。識別器のジョブは、`meas_level=1`の複素数データを取得し、標準の$|0\rangle$の$|1\rangle$の状態(`meas_level=2`)に分類することです。これは、前の[章](./calibrating-qubits-openpulse.html)の多くと同じ作業です。この結果は、このNotebookがフォーカスしている高エネルギー状態に励起するために必要です。
### 1A. 0->1 周波数のスイープ <a id="freqsweep01"></a>
識別器の構築の最初のステップは、前の章でやったのと同じように、我々の量子ビット周波数をキャリブレーションすることです。
```
def create_ground_freq_sweep_program(freqs, drive_power):
"""基底状態を励起して周波数掃引を行うプログラムを作成します。
ドライブパワーに応じて、これは0->1の周波数なのか、または0->2の周波数なのかを明らかにすることができます。
引数:
freqs (np.ndarray(dtype=float)):スイープする周波数のNumpy配列。
drive_power (float):ドライブ振幅の値。
レイズ:
ValueError:75を超える頻度を使用すると発生します。
現在、これを実行しようとすると、バックエンドでエラーが投げられます。
戻り値:
Qobj:基底状態の周波数掃引実験のプログラム。
"""
if len(freqs) > 75:
raise ValueError("You can only run 75 schedules at a time.")
# スイープ情報を表示
print(f"The frequency sweep will go from {freqs[0] / GHz} GHz to {freqs[-1]/ GHz} GHz \
using {len(freqs)} frequencies. The drive power is {drive_power}.")
# 駆動パルスを定義
ground_sweep_drive_pulse = pulse_lib.gaussian(duration=drive_samples,
sigma=drive_sigma,
amp=drive_power,
name='ground_sweep_drive_pulse')
# スイープのための周波数を定義
schedule = pulse.Schedule(name='Frequency sweep starting from ground state.')
schedule |= pulse.Play(ground_sweep_drive_pulse, drive_chan)
schedule |= measure << schedule.duration
# define frequencies for the sweep
schedule_freqs = [{drive_chan: freq} for freq in freqs]
# プログラムを組み立てる
# 注:それぞれが同じことを行うため、必要なスケジュールは1つだけです;
# スケジュールごとに、ドライブをミックスダウンするLO周波数が変化します
# これにより周波数掃引が可能になります
ground_freq_sweep_program = assemble(schedule,
backend=backend,
meas_level=1,
meas_return='avg',
shots=NUM_SHOTS,
schedule_los=schedule_freqs)
return ground_freq_sweep_program
# 75個の周波数で推定周波数の周りに40MHzを掃引します
num_freqs = 75
ground_sweep_freqs = default_qubit_freq + np.linspace(-20*MHz, 20*MHz, num_freqs)
ground_freq_sweep_program = create_ground_freq_sweep_program(ground_sweep_freqs, drive_power=0.3)
ground_freq_sweep_job = backend.run(ground_freq_sweep_program)
print(ground_freq_sweep_job.job_id())
job_monitor(ground_freq_sweep_job)
# ジョブのデータ(平均)を取得する
ground_freq_sweep_data = get_job_data(ground_freq_sweep_job, average=True)
```
データをローレンツ曲線に適合させ、キャリブレーションされた周波数を抽出します。
```
def fit_function(x_values, y_values, function, init_params):
"""Fit a function using scipy curve_fit."""
fitparams, conv = curve_fit(function, x_values, y_values, init_params)
y_fit = function(x_values, *fitparams)
return fitparams, y_fit
# Hz単位でのフィッティングをします
(ground_sweep_fit_params,
ground_sweep_y_fit) = fit_function(ground_sweep_freqs,
ground_freq_sweep_data,
lambda x, A, q_freq, B, C: (A / np.pi) * (B / ((x - q_freq)**2 + B**2)) + C,
[7, 4.975*GHz, 1*GHz, 3*GHz] # フィッティングのための初期パラメーター
)
# 注:シグナルの実数部のみをプロットしています
plt.scatter(ground_sweep_freqs/GHz, ground_freq_sweep_data, color='black')
plt.plot(ground_sweep_freqs/GHz, ground_sweep_y_fit, color='red')
plt.xlim([min(ground_sweep_freqs/GHz), max(ground_sweep_freqs/GHz)])
plt.xlabel("Frequency [GHz]", fontsize=15)
plt.ylabel("Measured Signal [a.u.]", fontsize=15)
plt.title("0->1 Frequency Sweep", fontsize=15)
plt.show()
_, cal_qubit_freq, _, _ = ground_sweep_fit_params
print(f"We've updated our qubit frequency estimate from "
f"{round(default_qubit_freq/GHz, 7)} GHz to {round(cal_qubit_freq/GHz, 7)} GHz.")
```
### 1B. 0->1 のラビ実験 <a id="rabi01"></a>
次に、$0\rightarrow1 ~ \pi$パルスの振幅を計算するラビ実験を実行します。$\pi$パルスは、$|0\rangle$から$|1\rangle$の状態へ移動させるパルス(ブロッホ球上での$\pi$回転)だということを思い出してください。
```
# 実験の構成
num_rabi_points = 50 # 実験の数(つまり、掃引の振幅)
# 反復する駆動パルスの振幅値:0から0.75まで等間隔に配置された50の振幅
drive_amp_min = 0
drive_amp_max = 0.75
drive_amps = np.linspace(drive_amp_min, drive_amp_max, num_rabi_points)
# スケジュールを作成
rabi_01_schedules = []
# 駆動振幅すべてにわたってループ
for ii, drive_amp in enumerate(drive_amps):
# 駆動パルス
rabi_01_pulse = pulse_lib.gaussian(duration=drive_samples,
amp=drive_amp,
sigma=drive_sigma,
name='rabi_01_pulse_%d' % ii)
# スケジュールにコマンドを追加
schedule = pulse.Schedule(name='Rabi Experiment at drive amp = %s' % drive_amp)
schedule |= pulse.Play(rabi_01_pulse, drive_chan)
schedule |= measure << schedule.duration # 測定をドライブパルスの後にシフト
rabi_01_schedules.append(schedule)
# プログラムにスケジュールを組み込む
# 注:較正された周波数で駆動します。
rabi_01_expt_program = assemble(rabi_01_schedules,
backend=backend,
meas_level=1,
meas_return='avg',
shots=NUM_SHOTS,
schedule_los=[{drive_chan: cal_qubit_freq}]
* num_rabi_points)
rabi_01_job = backend.run(rabi_01_expt_program)
print(rabi_01_job.job_id())
job_monitor(rabi_01_job)
# ジョブのデータ(平均)を取得する
rabi_01_data = get_job_data(rabi_01_job, average=True)
def baseline_remove(values):
"""Center data around 0."""
return np.array(values) - np.mean(values)
# 注:データの実数部のみがプロットされます
rabi_01_data = np.real(baseline_remove(rabi_01_data))
(rabi_01_fit_params,
rabi_01_y_fit) = fit_function(drive_amps,
rabi_01_data,
lambda x, A, B, drive_01_period, phi: (A*np.cos(2*np.pi*x/drive_01_period - phi) + B),
[4, -4, 0.5, 0])
plt.scatter(drive_amps, rabi_01_data, color='black')
plt.plot(drive_amps, rabi_01_y_fit, color='red')
drive_01_period = rabi_01_fit_params[2]
# piの振幅計算でphiを計算
pi_amp_01 = (drive_01_period/2/np.pi) *(np.pi+rabi_01_fit_params[3])
plt.axvline(pi_amp_01, color='red', linestyle='--')
plt.axvline(pi_amp_01+drive_01_period/2, color='red', linestyle='--')
plt.annotate("", xy=(pi_amp_01+drive_01_period/2, 0), xytext=(pi_amp_01,0), arrowprops=dict(arrowstyle="<->", color='red'))
plt.annotate("$\pi$", xy=(pi_amp_01-0.03, 0.1), color='red')
plt.xlabel("Drive amp [a.u.]", fontsize=15)
plt.ylabel("Measured signal [a.u.]", fontsize=15)
plt.title('0->1 Rabi Experiment', fontsize=15)
plt.show()
print(f"Pi Amplitude (0->1) = {pi_amp_01}")
```
この結果を使って、$0\rightarrow1$ $\pi$パルスを定義します。
```
pi_pulse_01 = pulse_lib.gaussian(duration=drive_samples,
amp=pi_amp_01,
sigma=drive_sigma,
name='pi_pulse_01')
```
### 1C. 0,1 の識別器を構築する <a id="builddiscrim01"></a>
これで、キャリブレーションされた周波数と$\pi$パルスを得たので、$|0\rangle$と$1\rangle$の状態の識別器を構築できます。識別器は、IQ平面において`meas_level=1`のデータを取って、それを$|0\rangle$または$1\rangle$を判別することで機能します。
$|0\rangle$と$|1\rangle$の状態は、IQ平面上で重心として知られているコヒーレントな円形の"ブロブ"を形成します。重心の中心は、各状態の正確なノイズのないIQポイントを定義します。周囲の雲は、様々なノイズ源から生成されたデータの分散を示します。
$|0\rangle$と$|1\rangle$間を識別(判別)するために、機械学習のテクニック、線形判別分析を適用します。この方法は量子ビットの状態を判別する一般的なテクニックです。
最初のステップは、重心データを得ることです。そのために、2つのスケジュールを定義します(システムが$|0\rangle$の状態から始まることを思い出しましょう。):
1. $|0\rangle$の状態を直接測定します($|0\rangle$の重心を得ます)。
2. $\pi$パルスを適用して、測定します($|1\rangle$の重心を得ます)。
```
# 2つのスケジュールを作る
# 基底状態のスケジュール
zero_schedule = pulse.Schedule(name="zero schedule")
zero_schedule |= measure
# 励起状態のスケジュール
one_schedule = pulse.Schedule(name="one schedule")
one_schedule |= pulse.Play(pi_pulse_01, drive_chan)
one_schedule |= measure << one_schedule.duration
# スケジュールをプログラムにアセンブルする
IQ_01_program = assemble([zero_schedule, one_schedule],
backend=backend,
meas_level=1,
meas_return='single',
shots=NUM_SHOTS,
schedule_los=[{drive_chan: cal_qubit_freq}] * 2)
IQ_01_job = backend.run(IQ_01_program)
print(IQ_01_job.job_id())
job_monitor(IQ_01_job)
# (単一の)ジョブデータを取得します;0と1に分割します
IQ_01_data = get_job_data(IQ_01_job, average=False)
zero_data = IQ_01_data[0]
one_data = IQ_01_data[1]
def IQ_01_plot(x_min, x_max, y_min, y_max):
"""Helper function for plotting IQ plane for |0>, |1>. Limits of plot given
as arguments."""
# 0のデータは青でプロット
plt.scatter(np.real(zero_data), np.imag(zero_data),
s=5, cmap='viridis', c='blue', alpha=0.5, label=r'$|0\rangle$')
# 1のデータは赤でプロット
plt.scatter(np.real(one_data), np.imag(one_data),
s=5, cmap='viridis', c='red', alpha=0.5, label=r'$|1\rangle$')
# 0状態と1状態の平均に大きなドットをプロットします。
mean_zero = np.mean(zero_data) # 実部と虚部両方の平均を取ります。
mean_one = np.mean(one_data)
plt.scatter(np.real(mean_zero), np.imag(mean_zero),
s=200, cmap='viridis', c='black',alpha=1.0)
plt.scatter(np.real(mean_one), np.imag(mean_one),
s=200, cmap='viridis', c='black',alpha=1.0)
plt.xlim(x_min, x_max)
plt.ylim(y_min,y_max)
plt.legend()
plt.ylabel('I [a.u.]', fontsize=15)
plt.xlabel('Q [a.u.]', fontsize=15)
plt.title("0-1 discrimination", fontsize=15)
```
以下のように、IQプロットを表示します。青の重心は$|0\rangle$状態で、赤の重心は$|1\rangle$状態です。(注:プロットが見えないときは、Notebookを再実行してください。)
```
x_min = -5
x_max = 15
y_min = -5
y_max = 10
IQ_01_plot(x_min, x_max, y_min, y_max)
```
さて、実際に識別器を構築する時が来ました。先に述べたように、線形判別分析(Linear Discriminant Analysis, LDA)と呼ばれる機械学習のテクニックを使います。LDAは、任意のデータセットをカテゴリーのセット(ここでは$|0\rangle$と$|1\rangle$)に分類するために、各カテゴリーの平均の間の距離を最大化し、各カテゴリーの分散を最小化します。より詳しくは、[こちら](https://scikit-learn.org/stable/modules/lda_qda.html#id4) (Ref. [3](#refs))をご覧ください。
LDAは、セパラトリックス(separatrix)と呼ばれるラインを生成します。与えられたデータポイントがどちら側のセパラトリックスにあるかに応じて、それがどのカテゴリーに属しているかを判別できます。我々の場合、セパラトリックスの片側が$|0\rangle$状態で、もう一方の側が$|1\rangle$の状態です。
我々は、最初の半分のデータを学習用に使い、残りの半分をテスト用に使います。LDAの実装のために`scikit.learn`を使います:将来のリリースでは、この機能は、Qiskit-Ignisに直接実装されてリリースされる予定です([ここ](https://github.com/Qiskit/qiskit-ignis/tree/master/qiskit/ignis/measurement/discriminator)を参照)。
結果データを判別に適したフォーマットになるように再形成します。
```
def reshape_complex_vec(vec):
"""
複素数ベクトルvecを取り込んで、実際のimagエントリーを含む2d配列を返します。
これは学習に必要なデータです。
Args:
vec (list):データの複素数ベクトル
戻り値:
list:(real(vec], imag(vec))で指定されたエントリー付きのベクトル
"""
length = len(vec)
vec_reshaped = np.zeros((length, 2))
for i in range(len(vec)):
vec_reshaped[i]=[np.real(vec[i]), np.imag(vec[i])]
return vec_reshaped
# IQベクトルを作成します(実部と虚部で構成されています)
zero_data_reshaped = reshape_complex_vec(zero_data)
one_data_reshaped = reshape_complex_vec(one_data)
IQ_01_data = np.concatenate((zero_data_reshaped, one_data_reshaped))
print(IQ_01_data.shape) # IQデータの形を確認します
```
次に、学習用データとテスト用データを分割します。期待される結果(基底状態のスケジュールは`0`の配列、励起状態のスケジュールは`1`の配列)を含む状態ベクトルを使ってテストします。
```
#(テスト用に)0と1でベクトルを構築する
state_01 = np.zeros(NUM_SHOTS) # shotsは実験の回数
state_01 = np.concatenate((state_01, np.ones(NUM_SHOTS)))
print(len(state_01))
# データをシャッフルしてトレーニングセットとテストセットに分割します
IQ_01_train, IQ_01_test, state_01_train, state_01_test = train_test_split(IQ_01_data, state_01, test_size=0.5)
```
最後に、モデルを設定して、学習します。学習精度が表示されます。
```
# LDAをセットアップします
LDA_01 = LinearDiscriminantAnalysis()
LDA_01.fit(IQ_01_train, state_01_train)
# シンプルなデータでテストします
print(LDA_01.predict([[0,0], [10, 0]]))
# 精度を計算します
score_01 = LDA_01.score(IQ_01_test, state_01_test)
print(score_01)
```
最後のステップは、セパラトリックスをプロットすることです。
```
# セパラトリックスを表示データの上にプロットします
def separatrixPlot(lda, x_min, x_max, y_min, y_max, shots):
nx, ny = shots, shots
xx, yy = np.meshgrid(np.linspace(x_min, x_max, nx),
np.linspace(y_min, y_max, ny))
Z = lda.predict_proba(np.c_[xx.ravel(), yy.ravel()])
Z = Z[:, 1].reshape(xx.shape)
plt.contour(xx, yy, Z, [0.5], linewidths=2., colors='black')
IQ_01_plot(x_min, x_max, y_min, y_max)
separatrixPlot(LDA_01, x_min, x_max, y_min, y_max, NUM_SHOTS)
```
セラパトリックスのどちらのサイドがどの重心(つまり状態)に対応しているか確認します。IQ平面上の点が与えられると、このモデルはセラパトリックスのどちらの側にそれが置かれているかチェックし、対応する状態を返します。
## 2. $|0\rangle$, $|1\rangle$, $|2\rangle$ の状態の識別 <a id="discrim012"></a>
$0, 1$の識別器をキャリブレーションしたので、高エネルギー状態の励起に移ります。特に、$|2\rangle$の状態の励起にフォーカスし、$|0\rangle$と$|1\rangle$と$|2\rangle$の状態をそれぞれのIQデータポイントから判別する識別器を構築することに焦点を当てます。さらに高い状態($|3\rangle$、$|4\rangle$など)の手順も同様ですが、明示的にテストはしていません。
高い状態の識別器を構築する手順は以下の通りです:
1. $1\rightarrow2$周波数を計算します。
2. $1\rightarrow2$のための$\pi$パルスの振幅を得るためにラビ実験を行います。そのためには、まず、$0\rightarrow1$ $\pi$パルスを適用して、$|0\rangle$から$|1\rangle$の状態にします。次に、上記で得た$1\rightarrow2$周波数において、駆動振幅のスイープを行います。
3. 3つのスケジュールを構成します:\
a. 0スケジュール:基底状態を測定するだけです。\
b. 1スケジュール:$0\rightarrow1$ $\pi$パルスを適用し、測定します。\
c. 2スケジュール:$0\rightarrow1$ $\pi$パルスを適用し、次に$1\rightarrow2$ $\pi$パルスを適用し測定します。
4. 各スケジュールのデータを学習用データとテスト用データのセットに分け、判別用のLDAモデルを構築します。
### 2A. 1->2 周波数の計算 <a id="freq12"></a>
キャリブレーションの最初のステップは、$1\rightarrow2$ の状態に移行するために必要な周波数を計算することです。これを行うには2つの方法があります:
1. 基底状態から周波数をスイープし、非常に高い電力をかけます。印加電力が十分に高い場合には、2つのピークが観測されます。1つはセクション [1](#discrim01)で見つかった $0\rightarrow1$周波数で、もう一つは、$0\rightarrow2$周波数です。$1\rightarrow2$周波数は2つの差を取ることで得られます。残念ながら、`ibmq_armonk`では、最大駆動電力$1.0$はこの遷移を起こすのに十分ではありません。代わりに、2番目の方法を使います。
2. $0\rightarrow1$ $\pi$パルスを適用して、$|1\rangle$状態を励起します。その後、$|1\rangle$状態のさらに上の励起に対して、周波数スイープを実行します。$0\rightarrow1$周波数より低いところで、$1\rightarrow2$周波数に対応した単一ピークが観測されるはずです。
#### サイドバンド法を使用した1->2 の周波数スイープ <a id="sideband12"></a>
上記の2番目の方法に従いましょう。$0\rightarrow 1$ $\pi$パルスを駆動するために、ローカル共振(local oscilattor, LO)周波数が必要です。これは、キャリブレーションされた$0\rightarrow1$周波数`cal_qubit_freq`(セクション[1](#discrim01)のラビ$\pi$パルスの構築を参照)によって与えられます。ただし、$1\rightarrow2$周波数の範囲をスイープするために、LO周波数を変化させる必要があります。残念ながら、Pulseのスペックでは、各スケジュールごとに、一つのLO周波数が必要です。
これを解決するには、LO周波数を`cal_qubit_freq`にセットし、を`freq-cal_qubit_freq`で$1\rightarrow2$パルスの上にサイン関数を乗算します。ここで`freq`は目的のスキャン周波数です。知られているように、正弦波サイドバンドを適用すると、プログラムのアセンブル時に手動で設定せずにLO周波数を変更可能です。
```
def apply_sideband(pulse, freq):
"""freq周波数でこのパルスに正弦波サイドバンドを適用します。
引数:
pulse (SamplePulse):対象のパルス。
freq (float):スイープを適用するLO周波数。
戻り値:
SamplePulse:サイドバンドが適用されたパルス(freqとcal_qubit_freqの差で振動します)。
"""
# 時間は0からdt*drive_samplesで、2*pi*f*tの形の正弦波引数になります
t_samples = np.linspace(0, dt*drive_samples, drive_samples)
sine_pulse = np.sin(2*np.pi*(freq-cal_qubit_freq)*t_samples) # no amp for the sine
# サイドバンドが適用されたサンプルパルスを作成
# 注:sq_pulse.samplesを実数にし、要素ごとに乗算する必要があります
sideband_pulse = SamplePulse(np.multiply(np.real(pulse.samples), sine_pulse), name='sideband_pulse')
return sideband_pulse
```
プログラムをアセンブルするためのロジックをメソッドにラップして、プログラムを実行します。
```
def create_excited_freq_sweep_program(freqs, drive_power):
"""|1>状態を励起することにより、周波数掃引を行うプログラムを作成します。
これにより、1-> 2の周波数を取得できます。
較正された量子ビット周波数を使用して、piパルスを介して|0>から|1>の状態になります。
|1>から|2>への周波数掃引を行うには、正弦係数を掃引駆動パルスに追加することにより、サイドバンド法を使用します。
引数:
freqs (np.ndarray(dtype=float)):掃引周波数のNumpy配列。
drive_power (float):駆動振幅の値。
レイズ:
ValueError:75を超える頻度を使用するとスローされます; 現在、75個を超える周波数を試行すると、
バックエンドでエラーがスローされます。
戻り値:
Qobj:周波数掃引実験用のプログラム。
"""
if len(freqs) > 75:
raise ValueError("You can only run 75 schedules at a time.")
print(f"The frequency sweep will go from {freqs[0] / GHz} GHz to {freqs[-1]/ GHz} GHz \
using {len(freqs)} frequencies. The drive power is {drive_power}.")
base_12_pulse = pulse_lib.gaussian(duration=drive_samples,
sigma=drive_sigma,
amp=drive_power,
name='base_12_pulse')
schedules = []
for jj, freq in enumerate(freqs):
# ガウシアンパルスにサイドバンドを追加
freq_sweep_12_pulse = apply_sideband(base_12_pulse, freq)
# スケジュールのコマンドを追加
schedule = pulse.Schedule(name="Frequency = {}".format(freq))
# 0->1のパルス、掃引パルスの周波数、測定を追加
schedule |= pulse.Play(pi_pulse_01, drive_chan)
schedule |= pulse.Play(freq_sweep_12_pulse, drive_chan) << schedule.duration
schedule |= measure << schedule.duration # 駆動パルスの後に測定をシフト
schedules.append(schedule)
num_freqs = len(freqs)
# スケジュールを表示します
display(schedules[-1].draw(channels=[drive_chan, meas_chan], label=True, scale=1.0))
# 周波数掃引プログラムを組み込みます
# 注:LOは各スケジュールでのcal_qubit_freqです;サイドバンドによって組み込みます
excited_freq_sweep_program = assemble(schedules,
backend=backend,
meas_level=1,
meas_return='avg',
shots=NUM_SHOTS,
schedule_los=[{drive_chan: cal_qubit_freq}]
* num_freqs)
return excited_freq_sweep_program
# 0->1周波数より下で1->2の周波数を見つけるために400 MHzを掃引します
num_freqs = 75
excited_sweep_freqs = cal_qubit_freq + np.linspace(-400*MHz, 30*MHz, num_freqs)
excited_freq_sweep_program = create_excited_freq_sweep_program(excited_sweep_freqs, drive_power=0.3)
# 確認のためにスケジュールの一例をプロットします
excited_freq_sweep_job = backend.run(excited_freq_sweep_program)
print(excited_freq_sweep_job.job_id())
job_monitor(excited_freq_sweep_job)
# (平均の)ジョブデータを取得します
excited_freq_sweep_data = get_job_data(excited_freq_sweep_job, average=True)
# 注:シグナルの実部だけをプロットします
plt.scatter(excited_sweep_freqs/GHz, excited_freq_sweep_data, color='black')
plt.xlim([min(excited_sweep_freqs/GHz)+0.01, max(excited_sweep_freqs/GHz)]) # ignore min point (is off)
plt.xlabel("Frequency [GHz]", fontsize=15)
plt.ylabel("Measured Signal [a.u.]", fontsize=15)
plt.title("1->2 Frequency Sweep (first pass)", fontsize=15)
plt.show()
```
最小値が$4.64$ GHz近辺に見られます。いくつかの偽の最大値がありますが、それらは、$1\rightarrow2$周波数には大きすぎます。最小値が$1\rightarrow2$周波数に対応します。
相対最小関数を使って、この点の値を正確に計算します。これで、$1\rightarrow2$周波数の推定値が得られます。
```
# output_dataに相対的最小周波数を表示します;高さは下限(絶対値)を示します
def rel_maxima(freqs, output_data, height):
"""output_dataに相対的な最小周波数を出力します(ピークを確認できます);
高さは上限(絶対値)を示します。
高さを正しく設定しないと、ピークが無視されます。
引数:
freqs (list):周波数リスト
output_data (list):結果のシグナルのリスト
height (float):ピークの上限(絶対値)
戻り値:
list:相対的な最小周波数を含むリスト
"""
peaks, _ = find_peaks(output_data, height)
print("Freq. dips: ", freqs[peaks])
return freqs[peaks]
maxima = rel_maxima(excited_sweep_freqs, np.real(excited_freq_sweep_data), 10)
approx_12_freq = maxima
```
上記で得られた推定値を使って、より正確な掃引を行います(つまり、大幅に狭い範囲で掃引を行います)。これによって、$1\rightarrow2$周波数のより正確な値を得ることができます。上下$20$ MHzをスイープします。
```
# 狭い範囲での掃引
num_freqs = 75
refined_excited_sweep_freqs = approx_12_freq + np.linspace(-20*MHz, 20*MHz, num_freqs)
refined_excited_freq_sweep_program = create_excited_freq_sweep_program(refined_excited_sweep_freqs, drive_power=0.3)
refined_excited_freq_sweep_job = backend.run(refined_excited_freq_sweep_program)
print(refined_excited_freq_sweep_job.job_id())
job_monitor(refined_excited_freq_sweep_job)
# より正確な(平均)データを取得する
refined_excited_freq_sweep_data = get_job_data(refined_excited_freq_sweep_job, average=True)
```
標準ローレンツ曲線を用いて、このより正確な信号をプロットしてフィットします。
```
# Hzの単位でフィッティングする
(refined_excited_sweep_fit_params,
refined_excited_sweep_y_fit) = fit_function(refined_excited_sweep_freqs,
refined_excited_freq_sweep_data,
lambda x, A, q_freq, B, C: (A / np.pi) * (B / ((x - q_freq)**2 + B**2)) + C,
[-12, 4.625*GHz, 0.05*GHz, 3*GHz] # フィッティングのための初期パラメーター
)
# 注:シグナルの実数部のみをプロットしています
plt.scatter(refined_excited_sweep_freqs/GHz, refined_excited_freq_sweep_data, color='black')
plt.plot(refined_excited_sweep_freqs/GHz, refined_excited_sweep_y_fit, color='red')
plt.xlim([min(refined_excited_sweep_freqs/GHz), max(refined_excited_sweep_freqs/GHz)])
plt.xlabel("Frequency [GHz]", fontsize=15)
plt.ylabel("Measured Signal [a.u.]", fontsize=15)
plt.title("1->2 Frequency Sweep (refined pass)", fontsize=15)
plt.show()
_, qubit_12_freq, _, _ = refined_excited_sweep_fit_params
print(f"Our updated estimate for the 1->2 transition frequency is "
f"{round(qubit_12_freq/GHz, 7)} GHz.")
```
### 2B. 1->2 ラビ実験 <a id="rabi12"></a>
これで、$1\rightarrow2$周波数の良い推定が得られたので、$1\rightarrow2$遷移のための$\pi$パルス振幅を得るためのラビ実験を行います。そのために、$0\rightarrow1$ $\pi$ パルスを適用してから、$1\rightarrow2$周波数において駆動振幅をスイープします(サイドバンド法を使います)。
```
# 実験の構成
num_rabi_points = 75 # 実験数(つまり掃引する振幅)
# 駆動振幅の繰り返し値:0から1.0の間で均等に配置された75個の振幅
drive_amp_min = 0
drive_amp_max = 1.0
drive_amps = np.linspace(drive_amp_min, drive_amp_max, num_rabi_points)
# スケジュールの作成
rabi_12_schedules = []
# すべての駆動振幅をループします
for ii, drive_amp in enumerate(drive_amps):
base_12_pulse = pulse_lib.gaussian(duration=drive_samples,
sigma=drive_sigma,
amp=drive_amp,
name='base_12_pulse')
# 1->2の周波数においてサイドバンドを適用
rabi_12_pulse = apply_sideband(base_12_pulse, qubit_12_freq)
# スケジュールにコマンドを追加
schedule = pulse.Schedule(name='Rabi Experiment at drive amp = %s' % drive_amp)
schedule |= pulse.Play(pi_pulse_01, drive_chan) # 0->1
schedule |= pulse.Play(rabi_12_pulse, drive_chan) << schedule.duration # 1->2のラビパルス
schedule |= measure << schedule.duration # 駆動パルスの後に測定をシフト
rabi_12_schedules.append(schedule)
# プログラムにスケジュールを組み込みます
# 注:LO周波数はcal_qubit_freqであり、0->1のpiパルスを作ります;
# サイドバンドを使って、1->2のパルス用に変更されます
rabi_12_expt_program = assemble(rabi_12_schedules,
backend=backend,
meas_level=1,
meas_return='avg',
shots=NUM_SHOTS,
schedule_los=[{drive_chan: cal_qubit_freq}]
* num_rabi_points)
rabi_12_job = backend.run(rabi_12_expt_program)
print(rabi_12_job.job_id())
job_monitor(rabi_12_job)
# ジョブデータ(平均)を取得します
rabi_12_data = get_job_data(rabi_12_job, average=True)
```
We plot and fit our data as before.
```
# 注:信号の実部のみプロットします。
rabi_12_data = np.real(baseline_remove(rabi_12_data))
(rabi_12_fit_params,
rabi_12_y_fit) = fit_function(drive_amps,
rabi_12_data,
lambda x, A, B, drive_12_period, phi: (A*np.cos(2*np.pi*x/drive_12_period - phi) + B),
[3, 0.5, 0.9, 0])
plt.scatter(drive_amps, rabi_12_data, color='black')
plt.plot(drive_amps, rabi_12_y_fit, color='red')
drive_12_period = rabi_12_fit_params[2]
# piパルス用の振幅のためにphiを考慮します
pi_amp_12 = (drive_12_period/2/np.pi) *(np.pi+rabi_12_fit_params[3])
plt.axvline(pi_amp_12, color='red', linestyle='--')
plt.axvline(pi_amp_12+drive_12_period/2, color='red', linestyle='--')
plt.annotate("", xy=(pi_amp_12+drive_12_period/2, 0), xytext=(pi_amp_12,0), arrowprops=dict(arrowstyle="<->", color='red'))
plt.annotate("$\pi$", xy=(pi_amp_12-0.03, 0.1), color='red')
plt.xlabel("Drive amp [a.u.]", fontsize=15)
plt.ylabel("Measured signal [a.u.]", fontsize=15)
plt.title('Rabi Experiment (1->2)', fontsize=20)
plt.show()
print(f"Our updated estimate for the 1->2 transition frequency is "
f"{round(qubit_12_freq/GHz, 7)} GHz.")
print(f"Pi Amplitude (1->2) = {pi_amp_12}")
```
この情報を使って、$1\rightarrow2$ $\pi$パルスを定義できます。(必ず、$1\rightarrow2$周波数でサイドバンドを追加してください。)
```
pi_pulse_12 = pulse_lib.gaussian(duration=drive_samples,
amp=pi_amp_12,
sigma=drive_sigma,
name='pi_pulse_12')
# このパルスがサイドバンドであることを再確認してください
pi_pulse_12 = apply_sideband(pi_pulse_12, qubit_12_freq)
```
### 2C. 0,1,2の識別器を構築する <a id="builddiscrim012"></a>
とうとう、 $|0\rangle$と$|1\rangle$と$|2\rangle$状態の識別器を構築できます。手順はセクション[1](#discrim01)と同様ですが、$|2\rangle$状態のためにスケジュールを追加します。
3つのスケジュールがあります。(再度、私たちのシステムが$|0\rangle$から開始することを思い出してください):
1. $|0\rangle$状態を直接測定します。($|0\rangle$の重心を得ます。)
2. $0\rightarrow1$ $\pi$パルスを適用し、測定します。($|1\rangle$の重心を得ます。)
3. $0\rightarrow1$ $\pi$パルスを適用した後、$1\rightarrow2$ $\pi$パルスを適用しそして測定します。($|2\rangle$の重心を得ます。)
```
# 3つのスケジュールを作ります
# 基底状態のスケジュール
zero_schedule = pulse.Schedule(name="zero schedule")
zero_schedule |= measure
# 励起状態のスケジュール
one_schedule = pulse.Schedule(name="one schedule")
one_schedule |= pulse.Play(pi_pulse_01, drive_chan)
one_schedule |= measure << one_schedule.duration
# 励起状態のスケジュール
two_schedule = pulse.Schedule(name="two schedule")
two_schedule |= pulse.Play(pi_pulse_01, drive_chan)
two_schedule |= pulse.Play(pi_pulse_12, drive_chan) << two_schedule.duration
two_schedule |= measure << two_schedule.duration
```
プログラムを構築し、IQ平面上に重心をプロットします。
```
# プログラムにスケジュールを組み込みます
IQ_012_program = assemble([zero_schedule, one_schedule, two_schedule],
backend=backend,
meas_level=1,
meas_return='single',
shots=NUM_SHOTS,
schedule_los=[{drive_chan: cal_qubit_freq}] * 3)
IQ_012_job = backend.run(IQ_012_program)
print(IQ_012_job.job_id())
job_monitor(IQ_012_job)
# (単一の)ジョブデータを取得します;0,1,2に分割します
IQ_012_data = get_job_data(IQ_012_job, average=False)
zero_data = IQ_012_data[0]
one_data = IQ_012_data[1]
two_data = IQ_012_data[2]
def IQ_012_plot(x_min, x_max, y_min, y_max):
"""0、1、2のIQ平面をプロットするための補助関数。引数としてプロットの制限を与えます。
"""
# 0のデータは青でプロット
plt.scatter(np.real(zero_data), np.imag(zero_data),
s=5, cmap='viridis', c='blue', alpha=0.5, label=r'$|0\rangle$')
# 1のデータは赤でプロット
plt.scatter(np.real(one_data), np.imag(one_data),
s=5, cmap='viridis', c='red', alpha=0.5, label=r'$|1\rangle$')
# 2のデータは緑でプロット
plt.scatter(np.real(two_data), np.imag(two_data),
s=5, cmap='viridis', c='green', alpha=0.5, label=r'$|2\rangle$')
# 0、1、2の状態の結果の平均を大きなドットでプロット
mean_zero = np.mean(zero_data) # 実部と虚部それぞれの平均をとる
mean_one = np.mean(one_data)
mean_two = np.mean(two_data)
plt.scatter(np.real(mean_zero), np.imag(mean_zero),
s=200, cmap='viridis', c='black',alpha=1.0)
plt.scatter(np.real(mean_one), np.imag(mean_one),
s=200, cmap='viridis', c='black',alpha=1.0)
plt.scatter(np.real(mean_two), np.imag(mean_two),
s=200, cmap='viridis', c='black',alpha=1.0)
plt.xlim(x_min, x_max)
plt.ylim(y_min,y_max)
plt.legend()
plt.ylabel('I [a.u.]', fontsize=15)
plt.xlabel('Q [a.u.]', fontsize=15)
plt.title("0-1-2 discrimination", fontsize=15)
x_min = -10
x_max = 20
y_min = -25
y_max = 10
IQ_012_plot(x_min, x_max, y_min, y_max)
```
今回は、$|2\rangle$状態に対応した3個目の重心が観測されます。(注:プロットが見えない場合は、Notebookを再実行してください。)
このデータで、識別器を構築します。再び`scikit.learn` を使って線形判別分析(LDA)を使います。
LDAのためにデータを形成することから始めます。
```
# IQベクトルを作成します(実部と虚部で構成されています)
zero_data_reshaped = reshape_complex_vec(zero_data)
one_data_reshaped = reshape_complex_vec(one_data)
two_data_reshaped = reshape_complex_vec(two_data)
IQ_012_data = np.concatenate((zero_data_reshaped, one_data_reshaped, two_data_reshaped))
print(IQ_012_data.shape) # IQデータの形を確認します
```
次に、学習用データとテスト用データを分割します(前回と同じように半分ずつです)。テスト用データは、0スケジュールの場合、`0`の配列が含まれたベクトルで、1スケジュールの場合、`1`の配列が含まれたベクトルで、2スケジュールの場合`2`の配列が含まれたベクトルです。
```
# (テスト用に)0と1と2の値が含まれたベクトルを構築します
state_012 = np.zeros(NUM_SHOTS) # 実験のショット数
state_012 = np.concatenate((state_012, np.ones(NUM_SHOTS)))
state_012 = np.concatenate((state_012, 2*np.ones(NUM_SHOTS)))
print(len(state_012))
# データをシャッフルして学習用セットとテスト用セットに分割します
IQ_012_train, IQ_012_test, state_012_train, state_012_test = train_test_split(IQ_012_data, state_012, test_size=0.5)
```
最後に、モデルを設定して学習します。学習の精度が出力されます。
```
# LDAを設定します
LDA_012 = LinearDiscriminantAnalysis()
LDA_012.fit(IQ_012_train, state_012_train)
# シンプルなデータでテストします
print(LDA_012.predict([[0, 0], [-10, 0], [-15, -5]]))
# 精度を計算します
score_012 = LDA_012.score(IQ_012_test, state_012_test)
print(score_012)
```
最後のステップは、セパラトリックスのプロットです。
```
IQ_012_plot(x_min, x_max, y_min, y_max)
separatrixPlot(LDA_012, x_min, x_max, y_min, y_max, NUM_SHOTS)
```
3つの重心を得たので、セラパトリックスは線ではなく、2つの線の組み合わせを含む曲線になります。$|0\rangle$、$|1\rangle$と$|2\rangle$の状態を区別するために、私たちのモデルは、IQ上の点がセラパトリックスのどの側にあるかどこにあるかチェックし、それに応じて分類します。
## 3. 参考文献 <a id="refs"></a>
1. D. C. McKay, T. Alexander, L. Bello, M. J. Biercuk, L. Bishop, J. Chen, J. M. Chow, A. D. C ́orcoles, D. Egger, S. Filipp, J. Gomez, M. Hush, A. Javadi-Abhari, D. Moreda, P. Nation, B. Paulovicks, E. Winston, C. J. Wood, J. Wootton, and J. M. Gambetta, “Qiskit backend specifications for OpenQASM and OpenPulse experiments,” 2018, https://arxiv.org/abs/1809.03452.
2. Krantz, P. et al. “A Quantum Engineer’s Guide to Superconducting Qubits.” Applied Physics Reviews 6.2 (2019): 021318, https://arxiv.org/abs/1904.06560.
3. Scikit-learn: Machine Learning in Python, Pedregosa et al., JMLR 12, pp. 2825-2830, 2011, https://scikit-learn.org/stable/modules/lda_qda.html#id4.
```
import qiskit.tools.jupyter
%qiskit_version_table
```
|
github_jupyter
|
import numpy as np
import matplotlib.pyplot as plt
from scipy.optimize import curve_fit
from scipy.signal import find_peaks
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis
from sklearn.model_selection import train_test_split
import qiskit.pulse as pulse
import qiskit.pulse.library as pulse_lib
from qiskit.compiler import assemble
from qiskit.pulse.library import SamplePulse
from qiskit.tools.monitor import job_monitor
import warnings
warnings.filterwarnings('ignore')
from qiskit.tools.jupyter import *
%matplotlib inline
from qiskit import IBMQ
IBMQ.load_account()
provider = IBMQ.get_provider(hub='ibm-q', group='open', project='main')
backend = provider.get_backend('ibmq_armonk')
backend_config = backend.configuration()
assert backend_config.open_pulse, "Backend doesn't support Pulse"
dt = backend_config.dt
backend_defaults = backend.defaults()
# 単位変換係数 -> すべてのバックエンドのプロパティーがSI単位系(Hz, sec, etc)で返される
GHz = 1.0e9 # Gigahertz
MHz = 1.0e6 # Megahertz
us = 1.0e-6 # Microseconds
ns = 1.0e-9 # Nanoseconds
qubit = 0 # 分析に使う量子ビット
default_qubit_freq = backend_defaults.qubit_freq_est[qubit] # デフォルトの量子ビット周波数単位はHz
print(f"Qubit {qubit} has an estimated frequency of {default_qubit_freq/ GHz} GHz.")
#(各デバイスに固有の)データをスケーリング
scale_factor = 1e-14
# 実験のショット回数
NUM_SHOTS = 1024
### 必要なチャネルを収集する
drive_chan = pulse.DriveChannel(qubit)
meas_chan = pulse.MeasureChannel(qubit)
acq_chan = pulse.AcquireChannel(qubit)
def get_job_data(job, average):
"""すでに実行されているジョブからデータを取得します。
引数:
job (Job): データが必要なジョブ
average (bool): Trueの場合、データが平均であると想定してデータを取得。
Falseの場合、シングルショット用と想定してデータを取得。
返し値:
list: ジョブの結果データを含むリスト
"""
job_results = job.result(timeout=120) # タイムアウトパラメーターは120秒にセット
result_data = []
for i in range(len(job_results.results)):
if average: # 平均データを得る
result_data.append(job_results.get_memory(i)[qubit]*scale_factor)
else: # シングルデータを得る
result_data.append(job_results.get_memory(i)[:, qubit]*scale_factor)
return result_data
def get_closest_multiple_of_16(num):
"""16の倍数に最も近いものを計算します。
パルスが使えるデバイスが16サンプルの倍数の期間が必要なためです。
"""
return (int(num) - (int(num)%16))
# 駆動パルスのパラメーター (us = マイクロ秒)
drive_sigma_us = 0.075 # ガウシアンの実際の幅を決めます
drive_samples_us = drive_sigma_us*8 # 切り捨てパラメーター
# ガウシアンには自然な有限長がないためです。
drive_sigma = get_closest_multiple_of_16(drive_sigma_us * us /dt) # ガウシアンの幅の単位はdt
drive_samples = get_closest_multiple_of_16(drive_samples_us * us /dt) # 切り捨てパラメーターの単位はdt
# この量子ビットに必要な測定マップインデックスを見つける
meas_map_idx = None
for i, measure_group in enumerate(backend_config.meas_map):
if qubit in measure_group:
meas_map_idx = i
break
assert meas_map_idx is not None, f"Couldn't find qubit {qubit} in the meas_map!"
# 命令スケジュールマップからデフォルトの測定パルスを取得
inst_sched_map = backend_defaults.instruction_schedule_map
measure = inst_sched_map.get('measure', qubits=backend_config.meas_map[meas_map_idx])
def create_ground_freq_sweep_program(freqs, drive_power):
"""基底状態を励起して周波数掃引を行うプログラムを作成します。
ドライブパワーに応じて、これは0->1の周波数なのか、または0->2の周波数なのかを明らかにすることができます。
引数:
freqs (np.ndarray(dtype=float)):スイープする周波数のNumpy配列。
drive_power (float):ドライブ振幅の値。
レイズ:
ValueError:75を超える頻度を使用すると発生します。
現在、これを実行しようとすると、バックエンドでエラーが投げられます。
戻り値:
Qobj:基底状態の周波数掃引実験のプログラム。
"""
if len(freqs) > 75:
raise ValueError("You can only run 75 schedules at a time.")
# スイープ情報を表示
print(f"The frequency sweep will go from {freqs[0] / GHz} GHz to {freqs[-1]/ GHz} GHz \
using {len(freqs)} frequencies. The drive power is {drive_power}.")
# 駆動パルスを定義
ground_sweep_drive_pulse = pulse_lib.gaussian(duration=drive_samples,
sigma=drive_sigma,
amp=drive_power,
name='ground_sweep_drive_pulse')
# スイープのための周波数を定義
schedule = pulse.Schedule(name='Frequency sweep starting from ground state.')
schedule |= pulse.Play(ground_sweep_drive_pulse, drive_chan)
schedule |= measure << schedule.duration
# define frequencies for the sweep
schedule_freqs = [{drive_chan: freq} for freq in freqs]
# プログラムを組み立てる
# 注:それぞれが同じことを行うため、必要なスケジュールは1つだけです;
# スケジュールごとに、ドライブをミックスダウンするLO周波数が変化します
# これにより周波数掃引が可能になります
ground_freq_sweep_program = assemble(schedule,
backend=backend,
meas_level=1,
meas_return='avg',
shots=NUM_SHOTS,
schedule_los=schedule_freqs)
return ground_freq_sweep_program
# 75個の周波数で推定周波数の周りに40MHzを掃引します
num_freqs = 75
ground_sweep_freqs = default_qubit_freq + np.linspace(-20*MHz, 20*MHz, num_freqs)
ground_freq_sweep_program = create_ground_freq_sweep_program(ground_sweep_freqs, drive_power=0.3)
ground_freq_sweep_job = backend.run(ground_freq_sweep_program)
print(ground_freq_sweep_job.job_id())
job_monitor(ground_freq_sweep_job)
# ジョブのデータ(平均)を取得する
ground_freq_sweep_data = get_job_data(ground_freq_sweep_job, average=True)
def fit_function(x_values, y_values, function, init_params):
"""Fit a function using scipy curve_fit."""
fitparams, conv = curve_fit(function, x_values, y_values, init_params)
y_fit = function(x_values, *fitparams)
return fitparams, y_fit
# Hz単位でのフィッティングをします
(ground_sweep_fit_params,
ground_sweep_y_fit) = fit_function(ground_sweep_freqs,
ground_freq_sweep_data,
lambda x, A, q_freq, B, C: (A / np.pi) * (B / ((x - q_freq)**2 + B**2)) + C,
[7, 4.975*GHz, 1*GHz, 3*GHz] # フィッティングのための初期パラメーター
)
# 注:シグナルの実数部のみをプロットしています
plt.scatter(ground_sweep_freqs/GHz, ground_freq_sweep_data, color='black')
plt.plot(ground_sweep_freqs/GHz, ground_sweep_y_fit, color='red')
plt.xlim([min(ground_sweep_freqs/GHz), max(ground_sweep_freqs/GHz)])
plt.xlabel("Frequency [GHz]", fontsize=15)
plt.ylabel("Measured Signal [a.u.]", fontsize=15)
plt.title("0->1 Frequency Sweep", fontsize=15)
plt.show()
_, cal_qubit_freq, _, _ = ground_sweep_fit_params
print(f"We've updated our qubit frequency estimate from "
f"{round(default_qubit_freq/GHz, 7)} GHz to {round(cal_qubit_freq/GHz, 7)} GHz.")
# 実験の構成
num_rabi_points = 50 # 実験の数(つまり、掃引の振幅)
# 反復する駆動パルスの振幅値:0から0.75まで等間隔に配置された50の振幅
drive_amp_min = 0
drive_amp_max = 0.75
drive_amps = np.linspace(drive_amp_min, drive_amp_max, num_rabi_points)
# スケジュールを作成
rabi_01_schedules = []
# 駆動振幅すべてにわたってループ
for ii, drive_amp in enumerate(drive_amps):
# 駆動パルス
rabi_01_pulse = pulse_lib.gaussian(duration=drive_samples,
amp=drive_amp,
sigma=drive_sigma,
name='rabi_01_pulse_%d' % ii)
# スケジュールにコマンドを追加
schedule = pulse.Schedule(name='Rabi Experiment at drive amp = %s' % drive_amp)
schedule |= pulse.Play(rabi_01_pulse, drive_chan)
schedule |= measure << schedule.duration # 測定をドライブパルスの後にシフト
rabi_01_schedules.append(schedule)
# プログラムにスケジュールを組み込む
# 注:較正された周波数で駆動します。
rabi_01_expt_program = assemble(rabi_01_schedules,
backend=backend,
meas_level=1,
meas_return='avg',
shots=NUM_SHOTS,
schedule_los=[{drive_chan: cal_qubit_freq}]
* num_rabi_points)
rabi_01_job = backend.run(rabi_01_expt_program)
print(rabi_01_job.job_id())
job_monitor(rabi_01_job)
# ジョブのデータ(平均)を取得する
rabi_01_data = get_job_data(rabi_01_job, average=True)
def baseline_remove(values):
"""Center data around 0."""
return np.array(values) - np.mean(values)
# 注:データの実数部のみがプロットされます
rabi_01_data = np.real(baseline_remove(rabi_01_data))
(rabi_01_fit_params,
rabi_01_y_fit) = fit_function(drive_amps,
rabi_01_data,
lambda x, A, B, drive_01_period, phi: (A*np.cos(2*np.pi*x/drive_01_period - phi) + B),
[4, -4, 0.5, 0])
plt.scatter(drive_amps, rabi_01_data, color='black')
plt.plot(drive_amps, rabi_01_y_fit, color='red')
drive_01_period = rabi_01_fit_params[2]
# piの振幅計算でphiを計算
pi_amp_01 = (drive_01_period/2/np.pi) *(np.pi+rabi_01_fit_params[3])
plt.axvline(pi_amp_01, color='red', linestyle='--')
plt.axvline(pi_amp_01+drive_01_period/2, color='red', linestyle='--')
plt.annotate("", xy=(pi_amp_01+drive_01_period/2, 0), xytext=(pi_amp_01,0), arrowprops=dict(arrowstyle="<->", color='red'))
plt.annotate("$\pi$", xy=(pi_amp_01-0.03, 0.1), color='red')
plt.xlabel("Drive amp [a.u.]", fontsize=15)
plt.ylabel("Measured signal [a.u.]", fontsize=15)
plt.title('0->1 Rabi Experiment', fontsize=15)
plt.show()
print(f"Pi Amplitude (0->1) = {pi_amp_01}")
pi_pulse_01 = pulse_lib.gaussian(duration=drive_samples,
amp=pi_amp_01,
sigma=drive_sigma,
name='pi_pulse_01')
# 2つのスケジュールを作る
# 基底状態のスケジュール
zero_schedule = pulse.Schedule(name="zero schedule")
zero_schedule |= measure
# 励起状態のスケジュール
one_schedule = pulse.Schedule(name="one schedule")
one_schedule |= pulse.Play(pi_pulse_01, drive_chan)
one_schedule |= measure << one_schedule.duration
# スケジュールをプログラムにアセンブルする
IQ_01_program = assemble([zero_schedule, one_schedule],
backend=backend,
meas_level=1,
meas_return='single',
shots=NUM_SHOTS,
schedule_los=[{drive_chan: cal_qubit_freq}] * 2)
IQ_01_job = backend.run(IQ_01_program)
print(IQ_01_job.job_id())
job_monitor(IQ_01_job)
# (単一の)ジョブデータを取得します;0と1に分割します
IQ_01_data = get_job_data(IQ_01_job, average=False)
zero_data = IQ_01_data[0]
one_data = IQ_01_data[1]
def IQ_01_plot(x_min, x_max, y_min, y_max):
"""Helper function for plotting IQ plane for |0>, |1>. Limits of plot given
as arguments."""
# 0のデータは青でプロット
plt.scatter(np.real(zero_data), np.imag(zero_data),
s=5, cmap='viridis', c='blue', alpha=0.5, label=r'$|0\rangle$')
# 1のデータは赤でプロット
plt.scatter(np.real(one_data), np.imag(one_data),
s=5, cmap='viridis', c='red', alpha=0.5, label=r'$|1\rangle$')
# 0状態と1状態の平均に大きなドットをプロットします。
mean_zero = np.mean(zero_data) # 実部と虚部両方の平均を取ります。
mean_one = np.mean(one_data)
plt.scatter(np.real(mean_zero), np.imag(mean_zero),
s=200, cmap='viridis', c='black',alpha=1.0)
plt.scatter(np.real(mean_one), np.imag(mean_one),
s=200, cmap='viridis', c='black',alpha=1.0)
plt.xlim(x_min, x_max)
plt.ylim(y_min,y_max)
plt.legend()
plt.ylabel('I [a.u.]', fontsize=15)
plt.xlabel('Q [a.u.]', fontsize=15)
plt.title("0-1 discrimination", fontsize=15)
x_min = -5
x_max = 15
y_min = -5
y_max = 10
IQ_01_plot(x_min, x_max, y_min, y_max)
def reshape_complex_vec(vec):
"""
複素数ベクトルvecを取り込んで、実際のimagエントリーを含む2d配列を返します。
これは学習に必要なデータです。
Args:
vec (list):データの複素数ベクトル
戻り値:
list:(real(vec], imag(vec))で指定されたエントリー付きのベクトル
"""
length = len(vec)
vec_reshaped = np.zeros((length, 2))
for i in range(len(vec)):
vec_reshaped[i]=[np.real(vec[i]), np.imag(vec[i])]
return vec_reshaped
# IQベクトルを作成します(実部と虚部で構成されています)
zero_data_reshaped = reshape_complex_vec(zero_data)
one_data_reshaped = reshape_complex_vec(one_data)
IQ_01_data = np.concatenate((zero_data_reshaped, one_data_reshaped))
print(IQ_01_data.shape) # IQデータの形を確認します
#(テスト用に)0と1でベクトルを構築する
state_01 = np.zeros(NUM_SHOTS) # shotsは実験の回数
state_01 = np.concatenate((state_01, np.ones(NUM_SHOTS)))
print(len(state_01))
# データをシャッフルしてトレーニングセットとテストセットに分割します
IQ_01_train, IQ_01_test, state_01_train, state_01_test = train_test_split(IQ_01_data, state_01, test_size=0.5)
# LDAをセットアップします
LDA_01 = LinearDiscriminantAnalysis()
LDA_01.fit(IQ_01_train, state_01_train)
# シンプルなデータでテストします
print(LDA_01.predict([[0,0], [10, 0]]))
# 精度を計算します
score_01 = LDA_01.score(IQ_01_test, state_01_test)
print(score_01)
# セパラトリックスを表示データの上にプロットします
def separatrixPlot(lda, x_min, x_max, y_min, y_max, shots):
nx, ny = shots, shots
xx, yy = np.meshgrid(np.linspace(x_min, x_max, nx),
np.linspace(y_min, y_max, ny))
Z = lda.predict_proba(np.c_[xx.ravel(), yy.ravel()])
Z = Z[:, 1].reshape(xx.shape)
plt.contour(xx, yy, Z, [0.5], linewidths=2., colors='black')
IQ_01_plot(x_min, x_max, y_min, y_max)
separatrixPlot(LDA_01, x_min, x_max, y_min, y_max, NUM_SHOTS)
def apply_sideband(pulse, freq):
"""freq周波数でこのパルスに正弦波サイドバンドを適用します。
引数:
pulse (SamplePulse):対象のパルス。
freq (float):スイープを適用するLO周波数。
戻り値:
SamplePulse:サイドバンドが適用されたパルス(freqとcal_qubit_freqの差で振動します)。
"""
# 時間は0からdt*drive_samplesで、2*pi*f*tの形の正弦波引数になります
t_samples = np.linspace(0, dt*drive_samples, drive_samples)
sine_pulse = np.sin(2*np.pi*(freq-cal_qubit_freq)*t_samples) # no amp for the sine
# サイドバンドが適用されたサンプルパルスを作成
# 注:sq_pulse.samplesを実数にし、要素ごとに乗算する必要があります
sideband_pulse = SamplePulse(np.multiply(np.real(pulse.samples), sine_pulse), name='sideband_pulse')
return sideband_pulse
def create_excited_freq_sweep_program(freqs, drive_power):
"""|1>状態を励起することにより、周波数掃引を行うプログラムを作成します。
これにより、1-> 2の周波数を取得できます。
較正された量子ビット周波数を使用して、piパルスを介して|0>から|1>の状態になります。
|1>から|2>への周波数掃引を行うには、正弦係数を掃引駆動パルスに追加することにより、サイドバンド法を使用します。
引数:
freqs (np.ndarray(dtype=float)):掃引周波数のNumpy配列。
drive_power (float):駆動振幅の値。
レイズ:
ValueError:75を超える頻度を使用するとスローされます; 現在、75個を超える周波数を試行すると、
バックエンドでエラーがスローされます。
戻り値:
Qobj:周波数掃引実験用のプログラム。
"""
if len(freqs) > 75:
raise ValueError("You can only run 75 schedules at a time.")
print(f"The frequency sweep will go from {freqs[0] / GHz} GHz to {freqs[-1]/ GHz} GHz \
using {len(freqs)} frequencies. The drive power is {drive_power}.")
base_12_pulse = pulse_lib.gaussian(duration=drive_samples,
sigma=drive_sigma,
amp=drive_power,
name='base_12_pulse')
schedules = []
for jj, freq in enumerate(freqs):
# ガウシアンパルスにサイドバンドを追加
freq_sweep_12_pulse = apply_sideband(base_12_pulse, freq)
# スケジュールのコマンドを追加
schedule = pulse.Schedule(name="Frequency = {}".format(freq))
# 0->1のパルス、掃引パルスの周波数、測定を追加
schedule |= pulse.Play(pi_pulse_01, drive_chan)
schedule |= pulse.Play(freq_sweep_12_pulse, drive_chan) << schedule.duration
schedule |= measure << schedule.duration # 駆動パルスの後に測定をシフト
schedules.append(schedule)
num_freqs = len(freqs)
# スケジュールを表示します
display(schedules[-1].draw(channels=[drive_chan, meas_chan], label=True, scale=1.0))
# 周波数掃引プログラムを組み込みます
# 注:LOは各スケジュールでのcal_qubit_freqです;サイドバンドによって組み込みます
excited_freq_sweep_program = assemble(schedules,
backend=backend,
meas_level=1,
meas_return='avg',
shots=NUM_SHOTS,
schedule_los=[{drive_chan: cal_qubit_freq}]
* num_freqs)
return excited_freq_sweep_program
# 0->1周波数より下で1->2の周波数を見つけるために400 MHzを掃引します
num_freqs = 75
excited_sweep_freqs = cal_qubit_freq + np.linspace(-400*MHz, 30*MHz, num_freqs)
excited_freq_sweep_program = create_excited_freq_sweep_program(excited_sweep_freqs, drive_power=0.3)
# 確認のためにスケジュールの一例をプロットします
excited_freq_sweep_job = backend.run(excited_freq_sweep_program)
print(excited_freq_sweep_job.job_id())
job_monitor(excited_freq_sweep_job)
# (平均の)ジョブデータを取得します
excited_freq_sweep_data = get_job_data(excited_freq_sweep_job, average=True)
# 注:シグナルの実部だけをプロットします
plt.scatter(excited_sweep_freqs/GHz, excited_freq_sweep_data, color='black')
plt.xlim([min(excited_sweep_freqs/GHz)+0.01, max(excited_sweep_freqs/GHz)]) # ignore min point (is off)
plt.xlabel("Frequency [GHz]", fontsize=15)
plt.ylabel("Measured Signal [a.u.]", fontsize=15)
plt.title("1->2 Frequency Sweep (first pass)", fontsize=15)
plt.show()
# output_dataに相対的最小周波数を表示します;高さは下限(絶対値)を示します
def rel_maxima(freqs, output_data, height):
"""output_dataに相対的な最小周波数を出力します(ピークを確認できます);
高さは上限(絶対値)を示します。
高さを正しく設定しないと、ピークが無視されます。
引数:
freqs (list):周波数リスト
output_data (list):結果のシグナルのリスト
height (float):ピークの上限(絶対値)
戻り値:
list:相対的な最小周波数を含むリスト
"""
peaks, _ = find_peaks(output_data, height)
print("Freq. dips: ", freqs[peaks])
return freqs[peaks]
maxima = rel_maxima(excited_sweep_freqs, np.real(excited_freq_sweep_data), 10)
approx_12_freq = maxima
# 狭い範囲での掃引
num_freqs = 75
refined_excited_sweep_freqs = approx_12_freq + np.linspace(-20*MHz, 20*MHz, num_freqs)
refined_excited_freq_sweep_program = create_excited_freq_sweep_program(refined_excited_sweep_freqs, drive_power=0.3)
refined_excited_freq_sweep_job = backend.run(refined_excited_freq_sweep_program)
print(refined_excited_freq_sweep_job.job_id())
job_monitor(refined_excited_freq_sweep_job)
# より正確な(平均)データを取得する
refined_excited_freq_sweep_data = get_job_data(refined_excited_freq_sweep_job, average=True)
# Hzの単位でフィッティングする
(refined_excited_sweep_fit_params,
refined_excited_sweep_y_fit) = fit_function(refined_excited_sweep_freqs,
refined_excited_freq_sweep_data,
lambda x, A, q_freq, B, C: (A / np.pi) * (B / ((x - q_freq)**2 + B**2)) + C,
[-12, 4.625*GHz, 0.05*GHz, 3*GHz] # フィッティングのための初期パラメーター
)
# 注:シグナルの実数部のみをプロットしています
plt.scatter(refined_excited_sweep_freqs/GHz, refined_excited_freq_sweep_data, color='black')
plt.plot(refined_excited_sweep_freqs/GHz, refined_excited_sweep_y_fit, color='red')
plt.xlim([min(refined_excited_sweep_freqs/GHz), max(refined_excited_sweep_freqs/GHz)])
plt.xlabel("Frequency [GHz]", fontsize=15)
plt.ylabel("Measured Signal [a.u.]", fontsize=15)
plt.title("1->2 Frequency Sweep (refined pass)", fontsize=15)
plt.show()
_, qubit_12_freq, _, _ = refined_excited_sweep_fit_params
print(f"Our updated estimate for the 1->2 transition frequency is "
f"{round(qubit_12_freq/GHz, 7)} GHz.")
# 実験の構成
num_rabi_points = 75 # 実験数(つまり掃引する振幅)
# 駆動振幅の繰り返し値:0から1.0の間で均等に配置された75個の振幅
drive_amp_min = 0
drive_amp_max = 1.0
drive_amps = np.linspace(drive_amp_min, drive_amp_max, num_rabi_points)
# スケジュールの作成
rabi_12_schedules = []
# すべての駆動振幅をループします
for ii, drive_amp in enumerate(drive_amps):
base_12_pulse = pulse_lib.gaussian(duration=drive_samples,
sigma=drive_sigma,
amp=drive_amp,
name='base_12_pulse')
# 1->2の周波数においてサイドバンドを適用
rabi_12_pulse = apply_sideband(base_12_pulse, qubit_12_freq)
# スケジュールにコマンドを追加
schedule = pulse.Schedule(name='Rabi Experiment at drive amp = %s' % drive_amp)
schedule |= pulse.Play(pi_pulse_01, drive_chan) # 0->1
schedule |= pulse.Play(rabi_12_pulse, drive_chan) << schedule.duration # 1->2のラビパルス
schedule |= measure << schedule.duration # 駆動パルスの後に測定をシフト
rabi_12_schedules.append(schedule)
# プログラムにスケジュールを組み込みます
# 注:LO周波数はcal_qubit_freqであり、0->1のpiパルスを作ります;
# サイドバンドを使って、1->2のパルス用に変更されます
rabi_12_expt_program = assemble(rabi_12_schedules,
backend=backend,
meas_level=1,
meas_return='avg',
shots=NUM_SHOTS,
schedule_los=[{drive_chan: cal_qubit_freq}]
* num_rabi_points)
rabi_12_job = backend.run(rabi_12_expt_program)
print(rabi_12_job.job_id())
job_monitor(rabi_12_job)
# ジョブデータ(平均)を取得します
rabi_12_data = get_job_data(rabi_12_job, average=True)
# 注:信号の実部のみプロットします。
rabi_12_data = np.real(baseline_remove(rabi_12_data))
(rabi_12_fit_params,
rabi_12_y_fit) = fit_function(drive_amps,
rabi_12_data,
lambda x, A, B, drive_12_period, phi: (A*np.cos(2*np.pi*x/drive_12_period - phi) + B),
[3, 0.5, 0.9, 0])
plt.scatter(drive_amps, rabi_12_data, color='black')
plt.plot(drive_amps, rabi_12_y_fit, color='red')
drive_12_period = rabi_12_fit_params[2]
# piパルス用の振幅のためにphiを考慮します
pi_amp_12 = (drive_12_period/2/np.pi) *(np.pi+rabi_12_fit_params[3])
plt.axvline(pi_amp_12, color='red', linestyle='--')
plt.axvline(pi_amp_12+drive_12_period/2, color='red', linestyle='--')
plt.annotate("", xy=(pi_amp_12+drive_12_period/2, 0), xytext=(pi_amp_12,0), arrowprops=dict(arrowstyle="<->", color='red'))
plt.annotate("$\pi$", xy=(pi_amp_12-0.03, 0.1), color='red')
plt.xlabel("Drive amp [a.u.]", fontsize=15)
plt.ylabel("Measured signal [a.u.]", fontsize=15)
plt.title('Rabi Experiment (1->2)', fontsize=20)
plt.show()
print(f"Our updated estimate for the 1->2 transition frequency is "
f"{round(qubit_12_freq/GHz, 7)} GHz.")
print(f"Pi Amplitude (1->2) = {pi_amp_12}")
pi_pulse_12 = pulse_lib.gaussian(duration=drive_samples,
amp=pi_amp_12,
sigma=drive_sigma,
name='pi_pulse_12')
# このパルスがサイドバンドであることを再確認してください
pi_pulse_12 = apply_sideband(pi_pulse_12, qubit_12_freq)
# 3つのスケジュールを作ります
# 基底状態のスケジュール
zero_schedule = pulse.Schedule(name="zero schedule")
zero_schedule |= measure
# 励起状態のスケジュール
one_schedule = pulse.Schedule(name="one schedule")
one_schedule |= pulse.Play(pi_pulse_01, drive_chan)
one_schedule |= measure << one_schedule.duration
# 励起状態のスケジュール
two_schedule = pulse.Schedule(name="two schedule")
two_schedule |= pulse.Play(pi_pulse_01, drive_chan)
two_schedule |= pulse.Play(pi_pulse_12, drive_chan) << two_schedule.duration
two_schedule |= measure << two_schedule.duration
# プログラムにスケジュールを組み込みます
IQ_012_program = assemble([zero_schedule, one_schedule, two_schedule],
backend=backend,
meas_level=1,
meas_return='single',
shots=NUM_SHOTS,
schedule_los=[{drive_chan: cal_qubit_freq}] * 3)
IQ_012_job = backend.run(IQ_012_program)
print(IQ_012_job.job_id())
job_monitor(IQ_012_job)
# (単一の)ジョブデータを取得します;0,1,2に分割します
IQ_012_data = get_job_data(IQ_012_job, average=False)
zero_data = IQ_012_data[0]
one_data = IQ_012_data[1]
two_data = IQ_012_data[2]
def IQ_012_plot(x_min, x_max, y_min, y_max):
"""0、1、2のIQ平面をプロットするための補助関数。引数としてプロットの制限を与えます。
"""
# 0のデータは青でプロット
plt.scatter(np.real(zero_data), np.imag(zero_data),
s=5, cmap='viridis', c='blue', alpha=0.5, label=r'$|0\rangle$')
# 1のデータは赤でプロット
plt.scatter(np.real(one_data), np.imag(one_data),
s=5, cmap='viridis', c='red', alpha=0.5, label=r'$|1\rangle$')
# 2のデータは緑でプロット
plt.scatter(np.real(two_data), np.imag(two_data),
s=5, cmap='viridis', c='green', alpha=0.5, label=r'$|2\rangle$')
# 0、1、2の状態の結果の平均を大きなドットでプロット
mean_zero = np.mean(zero_data) # 実部と虚部それぞれの平均をとる
mean_one = np.mean(one_data)
mean_two = np.mean(two_data)
plt.scatter(np.real(mean_zero), np.imag(mean_zero),
s=200, cmap='viridis', c='black',alpha=1.0)
plt.scatter(np.real(mean_one), np.imag(mean_one),
s=200, cmap='viridis', c='black',alpha=1.0)
plt.scatter(np.real(mean_two), np.imag(mean_two),
s=200, cmap='viridis', c='black',alpha=1.0)
plt.xlim(x_min, x_max)
plt.ylim(y_min,y_max)
plt.legend()
plt.ylabel('I [a.u.]', fontsize=15)
plt.xlabel('Q [a.u.]', fontsize=15)
plt.title("0-1-2 discrimination", fontsize=15)
x_min = -10
x_max = 20
y_min = -25
y_max = 10
IQ_012_plot(x_min, x_max, y_min, y_max)
# IQベクトルを作成します(実部と虚部で構成されています)
zero_data_reshaped = reshape_complex_vec(zero_data)
one_data_reshaped = reshape_complex_vec(one_data)
two_data_reshaped = reshape_complex_vec(two_data)
IQ_012_data = np.concatenate((zero_data_reshaped, one_data_reshaped, two_data_reshaped))
print(IQ_012_data.shape) # IQデータの形を確認します
# (テスト用に)0と1と2の値が含まれたベクトルを構築します
state_012 = np.zeros(NUM_SHOTS) # 実験のショット数
state_012 = np.concatenate((state_012, np.ones(NUM_SHOTS)))
state_012 = np.concatenate((state_012, 2*np.ones(NUM_SHOTS)))
print(len(state_012))
# データをシャッフルして学習用セットとテスト用セットに分割します
IQ_012_train, IQ_012_test, state_012_train, state_012_test = train_test_split(IQ_012_data, state_012, test_size=0.5)
# LDAを設定します
LDA_012 = LinearDiscriminantAnalysis()
LDA_012.fit(IQ_012_train, state_012_train)
# シンプルなデータでテストします
print(LDA_012.predict([[0, 0], [-10, 0], [-15, -5]]))
# 精度を計算します
score_012 = LDA_012.score(IQ_012_test, state_012_test)
print(score_012)
IQ_012_plot(x_min, x_max, y_min, y_max)
separatrixPlot(LDA_012, x_min, x_max, y_min, y_max, NUM_SHOTS)
import qiskit.tools.jupyter
%qiskit_version_table
| 0.473414 | 0.958265 |
In this notebook, we read the 'lif' files that resulted from manual annotation in labelIMG. They provide a bounding box that marks the 'OS' in the image. We want to use these as input for a classifier that segments the OS. We could generate a train data set by making patches that are centered around the center of the bbox, and patches that are completely outside of the bbox. The resulting classifier can classify a pixel by looking at the patch around it. This gives 'heatmap' of os-iness of the pixels. Hopefully we can take the maximum of the heatmap as the center of OS.
```
import matplotlib.pyplot as plt
%matplotlib inline
from bs4 import BeautifulSoup
import os
import cv2
lif_path = '/media/sf_VBox_Shared/kaggle/cervical-cancer/labels/'
image_path = '/media/sf_VBox_Shared/kaggle/cervical-cancer/processed/'
types = ['Type_1', 'Type_2', 'Type_3']
def get_bbox(path):
with open(path, 'r') as f:
soup = BeautifulSoup(f, 'lxml')
box = soup.find('bndbox')
keys = ['xmin', 'xmax', 'ymin', 'ymax']
return {key:int(box.find(key).contents[0]) for key in keys}
bboxes = []
for typ in types:
for fn in os.listdir(os.path.join(lif_path, typ)):
bbox = get_bbox(os.path.join(lif_path, typ, fn))
bbox['width'] = bbox['xmax'] - bbox['xmin']
bbox['height'] = bbox['ymax'] - bbox['ymin']
bbox['area'] = bbox['width'] * bbox['height']
fn_image = fn.replace('.lif', '.jpg')
img = cv2.imread(os.path.join(image_path, typ, fn_image))
w, h, c = img.shape
bbox['img_dim'] = (w, h, c)
bbox['rel_xmin'] = bbox['xmin'] / float(w)
bbox['rel_xmax'] = bbox['xmax'] / float(w)
bbox['rel_ymin'] = bbox['ymin'] / float(h)
bbox['rel_ymax'] = bbox['ymax'] / float(h)
bbox['rel_width'] = bbox['rel_xmax'] - bbox['rel_xmin']
bbox['rel_height'] = bbox['rel_ymax'] - bbox['rel_ymin']
bboxes.append(bbox)
import pandas as pd
bboxes_df = pd.DataFrame(bboxes)
bboxes_df.head()
print('min:', bboxes_df[['width', 'height']].min())
print('max:', bboxes_df[['width', 'height']].max())
bboxes_df[['width', 'height']].boxplot();
plt.show()
bboxes_df[['area']].boxplot();
plt.show()
print('min:', bboxes_df[['rel_width', 'rel_height']].min())
print('max:', bboxes_df[['rel_width', 'rel_height']].max())
bboxes_df[['rel_width', 'rel_height']].boxplot();
plt.show()
plt.scatter(bboxes_df['width'], bboxes_df['height'])
plt.scatter(bboxes_df['rel_width'], bboxes_df['rel_height'])
plt.scatter(bboxes_df['rel_width'], [d[0] for d in bboxes_df['img_dim']])
img = cv2.imread(image_path)
img = cv2.cvtColor(img, cv2.COLOR_BGR2RGB)
img.shape
plt.imshow(img)
rect_img = cv2.rectangle(img, (bbox['xmin'], bbox['ymin']), (bbox['xmax'], bbox['ymax']), (0, 255, 0))
plt.imshow(rect_img)
```
|
github_jupyter
|
import matplotlib.pyplot as plt
%matplotlib inline
from bs4 import BeautifulSoup
import os
import cv2
lif_path = '/media/sf_VBox_Shared/kaggle/cervical-cancer/labels/'
image_path = '/media/sf_VBox_Shared/kaggle/cervical-cancer/processed/'
types = ['Type_1', 'Type_2', 'Type_3']
def get_bbox(path):
with open(path, 'r') as f:
soup = BeautifulSoup(f, 'lxml')
box = soup.find('bndbox')
keys = ['xmin', 'xmax', 'ymin', 'ymax']
return {key:int(box.find(key).contents[0]) for key in keys}
bboxes = []
for typ in types:
for fn in os.listdir(os.path.join(lif_path, typ)):
bbox = get_bbox(os.path.join(lif_path, typ, fn))
bbox['width'] = bbox['xmax'] - bbox['xmin']
bbox['height'] = bbox['ymax'] - bbox['ymin']
bbox['area'] = bbox['width'] * bbox['height']
fn_image = fn.replace('.lif', '.jpg')
img = cv2.imread(os.path.join(image_path, typ, fn_image))
w, h, c = img.shape
bbox['img_dim'] = (w, h, c)
bbox['rel_xmin'] = bbox['xmin'] / float(w)
bbox['rel_xmax'] = bbox['xmax'] / float(w)
bbox['rel_ymin'] = bbox['ymin'] / float(h)
bbox['rel_ymax'] = bbox['ymax'] / float(h)
bbox['rel_width'] = bbox['rel_xmax'] - bbox['rel_xmin']
bbox['rel_height'] = bbox['rel_ymax'] - bbox['rel_ymin']
bboxes.append(bbox)
import pandas as pd
bboxes_df = pd.DataFrame(bboxes)
bboxes_df.head()
print('min:', bboxes_df[['width', 'height']].min())
print('max:', bboxes_df[['width', 'height']].max())
bboxes_df[['width', 'height']].boxplot();
plt.show()
bboxes_df[['area']].boxplot();
plt.show()
print('min:', bboxes_df[['rel_width', 'rel_height']].min())
print('max:', bboxes_df[['rel_width', 'rel_height']].max())
bboxes_df[['rel_width', 'rel_height']].boxplot();
plt.show()
plt.scatter(bboxes_df['width'], bboxes_df['height'])
plt.scatter(bboxes_df['rel_width'], bboxes_df['rel_height'])
plt.scatter(bboxes_df['rel_width'], [d[0] for d in bboxes_df['img_dim']])
img = cv2.imread(image_path)
img = cv2.cvtColor(img, cv2.COLOR_BGR2RGB)
img.shape
plt.imshow(img)
rect_img = cv2.rectangle(img, (bbox['xmin'], bbox['ymin']), (bbox['xmax'], bbox['ymax']), (0, 255, 0))
plt.imshow(rect_img)
| 0.213131 | 0.811639 |
# Summary
1. [Test PyTorch GPU](#1.-Test-PyTorch-GPU)
2. [Test fastai](#2.-Test-fastai)
3. [List package versions](#Test)
## 1. Test PyTorch GPU
```
import torch
torch.cuda.is_available()
!nvidia-smi
a = torch.zeros(100,100, device=torch.device('cuda'))
a.shape, a.device
```
## 2. Test fastai
```
from fastai.vision.all import *
path = untar_data(URLs.CAMVID_TINY)
codes = np.loadtxt(path/'codes.txt', dtype=str)
fnames = get_image_files(path/"images")
def label_func(fn): return path/"labels"/f"{fn.stem}_P{fn.suffix}"
dls = SegmentationDataLoaders.from_label_func(
path, bs=8, fnames=fnames, label_func=label_func, codes=codes
)
dls.show_batch(max_n=2)
learn = unet_learner(dls, resnet18, pretrained=False)
learn.fine_tune(1)
!rm -r /home/default/.fastai/data/camvid_tiny
```
## 3. List package versions
```
from platform import python_version
from matplotlib import __version__ as matplotlib_v
from notebook import __version__ as note_v
from numpy import __version__ as numpy_v
from pandas import __version__ as pandas_v
from PIL import __version__ as pillow_v
from pip import __version__ as pip_v
from sklearn import __version__ as scikit_learn_v
from scipy import __version__ as scipy_v
from spacy import __version__ as spacy_v
from torch import __version__ as torch_v
from torchvision import __version__ as torchvision_v
from subprocess import check_output
fastai_info = check_output('git -C ~/fastai log -n 1 --pretty=format:"%H %ci"', shell=True).decode('utf-8').split(' ', 1)
fastcore_info = check_output('git -C ~/fastcore log -n 1 --pretty=format:"%H %ci"', shell=True).decode('utf-8').split(' ', 1)
nbdev_info = check_output('git -C ~/nbdev log -n 1 --pretty=format:"%H %ci"', shell=True).decode('utf-8').split(' ', 1)
package_versions = {
'python': {
'matplotlib' : matplotlib_v,
'notebook' : note_v,
'numpy' : numpy_v,
'pandas' : pandas_v,
'pillow' : pillow_v,
'pip' : pip_v,
'python' : python_version(),
'scikit-learn': scikit_learn_v,
'scipy' : scipy_v,
'spacy' : spacy_v,
},
'pytorch': {
'pytorch' : torch_v,
'torchvision': torchvision_v,
},
'fastai': {
'fastai' : fastai_info,
'fastcore': fastcore_info,
'nbdev' : nbdev_info,
},
}
for type, packages in package_versions.items():
if type in ('python', 'pytorch'):
print(f'{type}:')
for i,k in packages.items():
print(f' {i}: {k}')
if type in ('fastai'):
print(f'{type}:')
for i,k in packages.items():
print(f' {i}:')
print(f'\tHash = {k[0]}')
print(f'\tTime = {k[1]}')
```
|
github_jupyter
|
import torch
torch.cuda.is_available()
!nvidia-smi
a = torch.zeros(100,100, device=torch.device('cuda'))
a.shape, a.device
from fastai.vision.all import *
path = untar_data(URLs.CAMVID_TINY)
codes = np.loadtxt(path/'codes.txt', dtype=str)
fnames = get_image_files(path/"images")
def label_func(fn): return path/"labels"/f"{fn.stem}_P{fn.suffix}"
dls = SegmentationDataLoaders.from_label_func(
path, bs=8, fnames=fnames, label_func=label_func, codes=codes
)
dls.show_batch(max_n=2)
learn = unet_learner(dls, resnet18, pretrained=False)
learn.fine_tune(1)
!rm -r /home/default/.fastai/data/camvid_tiny
from platform import python_version
from matplotlib import __version__ as matplotlib_v
from notebook import __version__ as note_v
from numpy import __version__ as numpy_v
from pandas import __version__ as pandas_v
from PIL import __version__ as pillow_v
from pip import __version__ as pip_v
from sklearn import __version__ as scikit_learn_v
from scipy import __version__ as scipy_v
from spacy import __version__ as spacy_v
from torch import __version__ as torch_v
from torchvision import __version__ as torchvision_v
from subprocess import check_output
fastai_info = check_output('git -C ~/fastai log -n 1 --pretty=format:"%H %ci"', shell=True).decode('utf-8').split(' ', 1)
fastcore_info = check_output('git -C ~/fastcore log -n 1 --pretty=format:"%H %ci"', shell=True).decode('utf-8').split(' ', 1)
nbdev_info = check_output('git -C ~/nbdev log -n 1 --pretty=format:"%H %ci"', shell=True).decode('utf-8').split(' ', 1)
package_versions = {
'python': {
'matplotlib' : matplotlib_v,
'notebook' : note_v,
'numpy' : numpy_v,
'pandas' : pandas_v,
'pillow' : pillow_v,
'pip' : pip_v,
'python' : python_version(),
'scikit-learn': scikit_learn_v,
'scipy' : scipy_v,
'spacy' : spacy_v,
},
'pytorch': {
'pytorch' : torch_v,
'torchvision': torchvision_v,
},
'fastai': {
'fastai' : fastai_info,
'fastcore': fastcore_info,
'nbdev' : nbdev_info,
},
}
for type, packages in package_versions.items():
if type in ('python', 'pytorch'):
print(f'{type}:')
for i,k in packages.items():
print(f' {i}: {k}')
if type in ('fastai'):
print(f'{type}:')
for i,k in packages.items():
print(f' {i}:')
print(f'\tHash = {k[0]}')
print(f'\tTime = {k[1]}')
| 0.468791 | 0.808181 |
# Introduction
We'll validate our extraction of mental functions from article texts by comparing the term occurrences to manual annotations of articles in the BrainMap database.
# Load the data
```
import os
import pandas as pd
import numpy as np
```
## BrainMap metadata
```
df = pd.read_csv("../data/metadata.csv", encoding="latin-1")
df = df.loc[df["SOURCE"] == "BrainMap"]
len(df)
pmids = df["PMID"].astype(int)
df["PMID"] = pmids
df.head(3)
```
## Document-term matrix
```
dtm = pd.read_csv("../data/text/dtm_190325.csv.gz",
compression="gzip", index_col=0)
dtm = dtm.loc[pmids]
len(dtm)
dtm.head(3)
dtm.max()[:5]
```
# Load the BrainMap taxonomy
```
def process_annotations(annotations):
annotations = annotations.replace("[", "").replace("]", "")
annotations = annotations.replace("'", "").replace(" ", "").split(",")
annotations = [a for a in annotations if "." in a]
return annotations
```
## Behavioral subdomains
```
subdomains = []
for i, row in df.iterrows():
subdomains += process_annotations(row["BEHAVIORAL_DOMAIN"])
subdomains = sorted(list(set(subdomains)))
subdomains[:5]
```
## Behavioral domains
```
domains = sorted(list(set([s.split(".")[0] for s in subdomains])))
domains
```
## Behavioral taxonomy
```
taxonomy = {domain: [s for s in subdomains if s.startswith(domain)] for domain in domains}
taxonomy
```
# Load the BrainMap annotations
```
dam = pd.DataFrame(0, index=pmids, columns=subdomains)
for pmid in pmids:
annotations = process_annotations(df.loc[df["PMID"] == pmid, "BEHAVIORAL_DOMAIN"].values[0])
for subdomain in annotations:
dam.loc[pmid, subdomain] = dam.loc[pmid, subdomain] + 1
dam.head()
```
# Reweight by TF-IDF
```
def tfidf(df):
# Rows are documents, columns are terms
# Inverse document frequencies
doccount = float(df.shape[0])
freqs = df.astype(bool).sum(axis=0)
idfs = np.log(doccount / freqs)
idfs[np.isinf(idfs)] = 0.0 # log(0) = 0
# Term frequencies
terms_in_doc = df.sum(axis=1)
tfs = (df.T / terms_in_doc).T
# TF-IDF reweighting
X = tfs * idfs
# Fill NA with 0
X = X.fillna(0.0)
return X
dtm_tfidf = tfidf(dtm)
dam_tfidf = tfidf(dam)
dtm_tfidf.head(3)
```
# Associate terms with domains
```
from scipy.stats import pearsonr
from statsmodels.stats.multitest import multipletests
```
## Drop terms that never occurred
```
dtm_tfidf = dtm_tfidf.loc[:, (dtm_tfidf != 0).any(axis=0)]
dtm_tfidf.shape
```
## Drop subdomains annotating < 100 articles
```
subdomains = [subdomain for subdomain in subdomains if dam[subdomain].astype(bool).sum() > 0]
subdomains
domains = sorted(list(set([s.split(".")[0] for s in subdomains])))
domains
```
## Compute associations and FDRs
```
r_file = "data/domains_terms_r.csv"
p_file = "data/domains_terms_p.csv"
fdr_file = "data/domains_terms_fdr.csv"
if not os.path.exists(r_file) or not os.path.exists(p_file) or not os.path.exists(fdr_file):
terms = list(dtm_tfidf.columns)
r = pd.DataFrame(index=terms, columns=subdomains)
p = pd.DataFrame(index=terms, columns=subdomains)
fdr = pd.DataFrame(index=terms, columns=subdomains)
for subdomain in subdomains:
print("Processing {}".format(subdomain))
for term in terms:
rtest = pearsonr(dtm_tfidf[term], dam_tfidf[subdomain])
r.loc[term, subdomain] = rtest[0]
p.loc[term, subdomain] = rtest[1]
fdr[subdomain] = multipletests(p[subdomain], method="fdr_bh")[1]
r.to_csv(r_file)
p.to_csv(p_file)
fdr.to_csv(fdr_file)
else:
r = pd.read_csv(r_file, index_col=0)
p = pd.read_csv(p_file, index_col=0)
fdr = pd.read_csv(fdr_file, index_col=0)
r["Action.Execution"].sort_values(ascending=False)[:10]
```
## Threshold associations (r > 0, FDR < 0.001)
```
r_thres = r
r_thres[r_thres < 0] = 0
r_thres[fdr > 0.001] = 0
r_thres.head()
```
## Plot word clouds
```
%matplotlib inline
# Hex color mappings
c = {"red": "#CE7D69", "orange": "#BA7E39", "yellow": "#CEBE6D",
"chartreuse": "#AEC87C", "green": "#77B58A", "blue": "#7597D0",
"magenta": "#B07EB6", "purple": "#7D74A3", "brown": "#846B43", "pink": "#CF7593"}
# Palette for framework
palette = [c["red"], c["green"], c["blue"], c["yellow"], c["purple"], c["orange"]]
colors = []
for i, domain in enumerate(domains):
colors += [palette[i]] * len([s for s in taxonomy[domain] if s in subdomains])
len(colors) == len(subdomains)
def plot_wordclouds(df, path="", prefix="", font="../style/Arial Unicode.ttf",
print_fig=True, width=550):
from wordcloud import WordCloud
import matplotlib.pyplot as plt
df.index = [term.replace("_", " ") for term in df.index]
for i, title in enumerate(df.columns):
def color_func(word, font_size, position, orientation,
random_state=None, idx=0, **kwargs):
# Adapted from https://amueller.github.io/word_cloud/auto_examples/a_new_hope.html
return colors[i]
title_df = df.loc[df[title] > 0, title]
tkn2freq = {t: f for t, f in zip(title_df.index, title_df.values)}
cloud = WordCloud(background_color="rgba(255, 255, 255, 0)", mode="RGB",
max_font_size=100, prefer_horizontal=1, scale=20, margin=3,
width=width, height=width, font_path=font,
random_state=42).generate_from_frequencies(frequencies=tkn2freq)
fig = plt.figure()
plt.axis("off")
plt.imshow(cloud.recolor(color_func=color_func, random_state=42))
file_name = "{}wordcloud/{}{}.png".format(path, prefix, title.replace("/", ""))
plt.savefig(file_name, dpi=500, bbox_inches="tight")
if print_fig:
print(title)
plt.show()
plt.close()
plot_wordclouds(r_thres, path="figures/brainmap/")
```
|
github_jupyter
|
import os
import pandas as pd
import numpy as np
df = pd.read_csv("../data/metadata.csv", encoding="latin-1")
df = df.loc[df["SOURCE"] == "BrainMap"]
len(df)
pmids = df["PMID"].astype(int)
df["PMID"] = pmids
df.head(3)
dtm = pd.read_csv("../data/text/dtm_190325.csv.gz",
compression="gzip", index_col=0)
dtm = dtm.loc[pmids]
len(dtm)
dtm.head(3)
dtm.max()[:5]
def process_annotations(annotations):
annotations = annotations.replace("[", "").replace("]", "")
annotations = annotations.replace("'", "").replace(" ", "").split(",")
annotations = [a for a in annotations if "." in a]
return annotations
subdomains = []
for i, row in df.iterrows():
subdomains += process_annotations(row["BEHAVIORAL_DOMAIN"])
subdomains = sorted(list(set(subdomains)))
subdomains[:5]
domains = sorted(list(set([s.split(".")[0] for s in subdomains])))
domains
taxonomy = {domain: [s for s in subdomains if s.startswith(domain)] for domain in domains}
taxonomy
dam = pd.DataFrame(0, index=pmids, columns=subdomains)
for pmid in pmids:
annotations = process_annotations(df.loc[df["PMID"] == pmid, "BEHAVIORAL_DOMAIN"].values[0])
for subdomain in annotations:
dam.loc[pmid, subdomain] = dam.loc[pmid, subdomain] + 1
dam.head()
def tfidf(df):
# Rows are documents, columns are terms
# Inverse document frequencies
doccount = float(df.shape[0])
freqs = df.astype(bool).sum(axis=0)
idfs = np.log(doccount / freqs)
idfs[np.isinf(idfs)] = 0.0 # log(0) = 0
# Term frequencies
terms_in_doc = df.sum(axis=1)
tfs = (df.T / terms_in_doc).T
# TF-IDF reweighting
X = tfs * idfs
# Fill NA with 0
X = X.fillna(0.0)
return X
dtm_tfidf = tfidf(dtm)
dam_tfidf = tfidf(dam)
dtm_tfidf.head(3)
from scipy.stats import pearsonr
from statsmodels.stats.multitest import multipletests
dtm_tfidf = dtm_tfidf.loc[:, (dtm_tfidf != 0).any(axis=0)]
dtm_tfidf.shape
subdomains = [subdomain for subdomain in subdomains if dam[subdomain].astype(bool).sum() > 0]
subdomains
domains = sorted(list(set([s.split(".")[0] for s in subdomains])))
domains
r_file = "data/domains_terms_r.csv"
p_file = "data/domains_terms_p.csv"
fdr_file = "data/domains_terms_fdr.csv"
if not os.path.exists(r_file) or not os.path.exists(p_file) or not os.path.exists(fdr_file):
terms = list(dtm_tfidf.columns)
r = pd.DataFrame(index=terms, columns=subdomains)
p = pd.DataFrame(index=terms, columns=subdomains)
fdr = pd.DataFrame(index=terms, columns=subdomains)
for subdomain in subdomains:
print("Processing {}".format(subdomain))
for term in terms:
rtest = pearsonr(dtm_tfidf[term], dam_tfidf[subdomain])
r.loc[term, subdomain] = rtest[0]
p.loc[term, subdomain] = rtest[1]
fdr[subdomain] = multipletests(p[subdomain], method="fdr_bh")[1]
r.to_csv(r_file)
p.to_csv(p_file)
fdr.to_csv(fdr_file)
else:
r = pd.read_csv(r_file, index_col=0)
p = pd.read_csv(p_file, index_col=0)
fdr = pd.read_csv(fdr_file, index_col=0)
r["Action.Execution"].sort_values(ascending=False)[:10]
r_thres = r
r_thres[r_thres < 0] = 0
r_thres[fdr > 0.001] = 0
r_thres.head()
%matplotlib inline
# Hex color mappings
c = {"red": "#CE7D69", "orange": "#BA7E39", "yellow": "#CEBE6D",
"chartreuse": "#AEC87C", "green": "#77B58A", "blue": "#7597D0",
"magenta": "#B07EB6", "purple": "#7D74A3", "brown": "#846B43", "pink": "#CF7593"}
# Palette for framework
palette = [c["red"], c["green"], c["blue"], c["yellow"], c["purple"], c["orange"]]
colors = []
for i, domain in enumerate(domains):
colors += [palette[i]] * len([s for s in taxonomy[domain] if s in subdomains])
len(colors) == len(subdomains)
def plot_wordclouds(df, path="", prefix="", font="../style/Arial Unicode.ttf",
print_fig=True, width=550):
from wordcloud import WordCloud
import matplotlib.pyplot as plt
df.index = [term.replace("_", " ") for term in df.index]
for i, title in enumerate(df.columns):
def color_func(word, font_size, position, orientation,
random_state=None, idx=0, **kwargs):
# Adapted from https://amueller.github.io/word_cloud/auto_examples/a_new_hope.html
return colors[i]
title_df = df.loc[df[title] > 0, title]
tkn2freq = {t: f for t, f in zip(title_df.index, title_df.values)}
cloud = WordCloud(background_color="rgba(255, 255, 255, 0)", mode="RGB",
max_font_size=100, prefer_horizontal=1, scale=20, margin=3,
width=width, height=width, font_path=font,
random_state=42).generate_from_frequencies(frequencies=tkn2freq)
fig = plt.figure()
plt.axis("off")
plt.imshow(cloud.recolor(color_func=color_func, random_state=42))
file_name = "{}wordcloud/{}{}.png".format(path, prefix, title.replace("/", ""))
plt.savefig(file_name, dpi=500, bbox_inches="tight")
if print_fig:
print(title)
plt.show()
plt.close()
plot_wordclouds(r_thres, path="figures/brainmap/")
| 0.468547 | 0.857231 |
# 函数
- 函数可以用来定义可重复代码,组织和简化
- 一般来说一个函数在实际开发中为一个小功能
- 一个类为一个大功能
- 同样函数的长度不要超过一屏
Python中的所有函数实际上都是有返回值(return None),
如果你没有设置return,那么Python将不显示None.
如果你设置return,那么将返回出return这个值.
```
def HJN():
print('Hello')
return 1000
b=HJN()
print(b)
HJN
def panduan(number):
if number % 2 == 0:
print('O')
else:
print('J')
panduan(number=1)
panduan(2)
```
## 定义一个函数
def function_name(list of parameters):
do something

- 以前使用的random 或者range 或者print.. 其实都是函数或者类
函数的参数如果有默认值的情况,当你调用该函数的时候:
可以不给予参数值,那么就会走该参数的默认值
否则的话,就走你给予的参数值.
```
import random
def hahah():
n = random.randint(0,5)
while 1:
N = eval(input('>>'))
if n == N:
print('smart')
break
elif n < N:
print('太小了')
elif n > N:
print('太大了')
```
## 调用一个函数
- functionName()
- "()" 就代表调用
```
def H():
print('hahaha')
def B():
H()
B()
def A(f):
f()
A(B)
```

## 带返回值和不带返回值的函数
- return 返回的内容
- return 返回多个值
- 一般情况下,在多个函数协同完成一个功能的时候,那么将会有返回值

- 当然也可以自定义返回None
## EP:

```
def main():
print(min(min(5,6),(51,6)))
def min(n1,n2):
a = n1
if n2 < a:
a = n2
main()
```
## 类型和关键字参数
- 普通参数
- 多个参数
- 默认值参数
- 不定长参数
## 普通参数
## 多个参数
## 默认值参数
## 强制命名
```
def U(str_):
xiaoxie = 0
for i in str_:
ASCII = ord(i)
if 97<=ASCII<=122:
xiaoxie +=1
elif xxxx:
daxie += 1
elif xxxx:
shuzi += 1
return xiaoxie,daxie,shuzi
U('HJi12')
```
## 不定长参数
- \*args
> - 不定长,来多少装多少,不装也是可以的
- 返回的数据类型是元组
- args 名字是可以修改的,只是我们约定俗成的是args
- \**kwargs
> - 返回的字典
- 输入的一定要是表达式(键值对)
- name,\*args,name2,\**kwargs 使用参数名
```
def TT(a,b)
def TT(*args,**kwargs):
print(kwargs)
print(args)
TT(1,2,3,4,6,a=100,b=1000)
{'key':'value'}
TT(1,2,4,5,7,8,9,)
def B(name1,nam3):
pass
B(name1=100,2)
def sum_(*args,A='sum'):
res = 0
count = 0
for i in args:
res +=i
count += 1
if A == "sum":
return res
elif A == "mean":
mean = res / count
return res,mean
else:
print(A,'还未开放')
sum_(-1,0,1,4,A='var')
'aHbK134'.__iter__
b = 'asdkjfh'
for i in b :
print(i)
2,5
2 + 22 + 222 + 2222 + 22222
```
## 变量的作用域
- 局部变量 local
- 全局变量 global
- globals 函数返回一个全局变量的字典,包括所有导入的变量
- locals() 函数会以字典类型返回当前位置的全部局部变量。
```
a = 1000
b = 10
def Y():
global a,b
a += 100
print(a)
Y()
def YY(a1):
a1 += 100
print(a1)
YY(a)
print(a)
```
## 注意:
- global :在进行赋值操作的时候需要声明
- 官方解释:This is because when you make an assignment to a variable in a scope, that variable becomes local to that scope and shadows any similarly named variable in the outer scope.
- 
# Homework
- 1

```
def getPentagonalNumber(n):
return n*(3*n-1)/2
count =0
for n in range(1,101):
if count <9:
print( "%.0f "%getPentagonalNumber(n),end="")
count += 1
else:
print( "%.0f"%getPentagonalNumber(n))
count = 0
```
- 2

```
def sumDigits(n):
i = n // 100
j = n // 10 % 10
k = n % 10
sum = (i + j + k)
print(sum)
sumDigits(234)
```
- 3

```
def displaySortedNumbers(num1,num2,num3):
x = [num1,num2,num3]
x.sort()
print(x)
displaySortedNumbers(2,62,4)
```
- 4

- 5

```
def printChars(ch1,ch2,numberPerLine):
a = ord(ch1)
b = ord(ch2)
count = 0
for i in range(a,b+1):
count += 1
print(chr(i),end="")
if count % numberPerLine == 0:
print('')
printChars('A','Z',10)
```
- 6

```
import numpy as np
def is_leap_year(year):
return (year % 4 == 0 and year % 100 != 0) or year % 400 == 0
def numberOfDaysInAYear(year):
month_day = np.array([31,28,31,30,31,30,31,31,30,31,30,31])
if is_leap_year(year):
month_day[1] = 29
print(np.sum(month_day))
numberOfDaysInAYear(2001)
```
- 7

```
def distance(x1,y1,x2,y2):
import math
l = math.sqrt((x1-x2)*(x1-x2)+(y1-y2)*(y1-y2))
print('两点间距离为: %.2f'%l)
distance(7,7,9,9)
```
- 8

```
def js():
for a in range(2,32):
for i in range(2,a):
if(a%i==0):
break
else:
p=2 **a-1
print('%d %d '%(a,p))
js()
```
- 9


```
def js():
import time
localtime = time.asctime(time.localtime(time.time()))
print("Curretn date and time is ", localtime)
js()
```
- 10

```
def game():
import random
x = random.choice([1,2,3,4,5,6])
y = random.choice([1,2,3,4,5,6])
z = 0
sum = x + y
print('you rolled %d + %d = %d'%(x,y,sum))
if sum == 2 or sum == 3 or sum ==12 or sum == x :
print('you lose')
if sum == 7 or sum == 11:
print('you win')
else:
print('the point is %d' %sum)
z = sum
game()
```
- 11
### 去网上寻找如何用Python代码发送邮件
```
# -*- coding: UTF-8 -*-
import sys, os, re, urllib, urlparse
import smtplib
import traceback
from email.mime.text import MIMEText
from email.mime.multipart import MIMEMultipart
def sendmail(subject,msg,toaddrs,fromaddr,smtpaddr,password):
'''''
@subject:邮件主题
@msg:邮件内容
@toaddrs:收信人的邮箱地址
@fromaddr:发信人的邮箱地址
@smtpaddr:smtp服务地址,可以在邮箱看,比如163邮箱为smtp.163.com
@password:发信人的邮箱密码
'''
mail_msg = MIMEMultipart()
if not isinstance(subject,unicode):
subject = unicode(subject, 'utf-8')
mail_msg['Subject'] = subject
mail_msg['From'] =fromaddr
mail_msg['To'] = ','.join(toaddrs)
mail_msg.attach(MIMEText(msg, 'html', 'utf-8'))
try:
s = smtplib.SMTP()
s.connect(smtpaddr) #连接smtp服务器
s.login(fromaddr,password) #登录邮箱
s.sendmail(fromaddr, toaddrs, mail_msg.as_string()) #发送邮件
s.quit()
except Exception,e:
print "Error: unable to send email"
print traceback.format_exc()
if __name__ == '__main__':
fromaddr = "xxxxxxxx@163.com"
smtpaddr = "smtp.163.com"
toaddrs = ["xxxxxxxx@qq.com","xxxxxxxxx@163.com"]
subject = "测试邮件"
password = "xxxxxxxx"
msg = "测试一下"
sendmail(subject,msg,toaddrs,fromaddr,smtpaddr,password)
```
|
github_jupyter
|
def HJN():
print('Hello')
return 1000
b=HJN()
print(b)
HJN
def panduan(number):
if number % 2 == 0:
print('O')
else:
print('J')
panduan(number=1)
panduan(2)
import random
def hahah():
n = random.randint(0,5)
while 1:
N = eval(input('>>'))
if n == N:
print('smart')
break
elif n < N:
print('太小了')
elif n > N:
print('太大了')
def H():
print('hahaha')
def B():
H()
B()
def A(f):
f()
A(B)
def main():
print(min(min(5,6),(51,6)))
def min(n1,n2):
a = n1
if n2 < a:
a = n2
main()
def U(str_):
xiaoxie = 0
for i in str_:
ASCII = ord(i)
if 97<=ASCII<=122:
xiaoxie +=1
elif xxxx:
daxie += 1
elif xxxx:
shuzi += 1
return xiaoxie,daxie,shuzi
U('HJi12')
def TT(a,b)
def TT(*args,**kwargs):
print(kwargs)
print(args)
TT(1,2,3,4,6,a=100,b=1000)
{'key':'value'}
TT(1,2,4,5,7,8,9,)
def B(name1,nam3):
pass
B(name1=100,2)
def sum_(*args,A='sum'):
res = 0
count = 0
for i in args:
res +=i
count += 1
if A == "sum":
return res
elif A == "mean":
mean = res / count
return res,mean
else:
print(A,'还未开放')
sum_(-1,0,1,4,A='var')
'aHbK134'.__iter__
b = 'asdkjfh'
for i in b :
print(i)
2,5
2 + 22 + 222 + 2222 + 22222
a = 1000
b = 10
def Y():
global a,b
a += 100
print(a)
Y()
def YY(a1):
a1 += 100
print(a1)
YY(a)
print(a)
def getPentagonalNumber(n):
return n*(3*n-1)/2
count =0
for n in range(1,101):
if count <9:
print( "%.0f "%getPentagonalNumber(n),end="")
count += 1
else:
print( "%.0f"%getPentagonalNumber(n))
count = 0
def sumDigits(n):
i = n // 100
j = n // 10 % 10
k = n % 10
sum = (i + j + k)
print(sum)
sumDigits(234)
def displaySortedNumbers(num1,num2,num3):
x = [num1,num2,num3]
x.sort()
print(x)
displaySortedNumbers(2,62,4)
def printChars(ch1,ch2,numberPerLine):
a = ord(ch1)
b = ord(ch2)
count = 0
for i in range(a,b+1):
count += 1
print(chr(i),end="")
if count % numberPerLine == 0:
print('')
printChars('A','Z',10)
import numpy as np
def is_leap_year(year):
return (year % 4 == 0 and year % 100 != 0) or year % 400 == 0
def numberOfDaysInAYear(year):
month_day = np.array([31,28,31,30,31,30,31,31,30,31,30,31])
if is_leap_year(year):
month_day[1] = 29
print(np.sum(month_day))
numberOfDaysInAYear(2001)
def distance(x1,y1,x2,y2):
import math
l = math.sqrt((x1-x2)*(x1-x2)+(y1-y2)*(y1-y2))
print('两点间距离为: %.2f'%l)
distance(7,7,9,9)
def js():
for a in range(2,32):
for i in range(2,a):
if(a%i==0):
break
else:
p=2 **a-1
print('%d %d '%(a,p))
js()
def js():
import time
localtime = time.asctime(time.localtime(time.time()))
print("Curretn date and time is ", localtime)
js()
def game():
import random
x = random.choice([1,2,3,4,5,6])
y = random.choice([1,2,3,4,5,6])
z = 0
sum = x + y
print('you rolled %d + %d = %d'%(x,y,sum))
if sum == 2 or sum == 3 or sum ==12 or sum == x :
print('you lose')
if sum == 7 or sum == 11:
print('you win')
else:
print('the point is %d' %sum)
z = sum
game()
# -*- coding: UTF-8 -*-
import sys, os, re, urllib, urlparse
import smtplib
import traceback
from email.mime.text import MIMEText
from email.mime.multipart import MIMEMultipart
def sendmail(subject,msg,toaddrs,fromaddr,smtpaddr,password):
'''''
@subject:邮件主题
@msg:邮件内容
@toaddrs:收信人的邮箱地址
@fromaddr:发信人的邮箱地址
@smtpaddr:smtp服务地址,可以在邮箱看,比如163邮箱为smtp.163.com
@password:发信人的邮箱密码
'''
mail_msg = MIMEMultipart()
if not isinstance(subject,unicode):
subject = unicode(subject, 'utf-8')
mail_msg['Subject'] = subject
mail_msg['From'] =fromaddr
mail_msg['To'] = ','.join(toaddrs)
mail_msg.attach(MIMEText(msg, 'html', 'utf-8'))
try:
s = smtplib.SMTP()
s.connect(smtpaddr) #连接smtp服务器
s.login(fromaddr,password) #登录邮箱
s.sendmail(fromaddr, toaddrs, mail_msg.as_string()) #发送邮件
s.quit()
except Exception,e:
print "Error: unable to send email"
print traceback.format_exc()
if __name__ == '__main__':
fromaddr = "xxxxxxxx@163.com"
smtpaddr = "smtp.163.com"
toaddrs = ["xxxxxxxx@qq.com","xxxxxxxxx@163.com"]
subject = "测试邮件"
password = "xxxxxxxx"
msg = "测试一下"
sendmail(subject,msg,toaddrs,fromaddr,smtpaddr,password)
| 0.143668 | 0.758555 |
# A 10-minute Tutorial on How to Use `HFTA`
This notebook demonstrates the way to integrate HFTA into a simple MNIST training example.
## Setup
Install the HFTA library from GitHub.
```
!pip install git+https://github.com/UofT-EcoSystem/hfta
```
### Demo with a benchmark
Here is a demo run on one of the benchmarks provided in the `hfta` GitHub repo to make sure HFTA is properly installed.
Check [here](https://github.com/UofT-EcoSystem/hfta/tree/main/examples/mobilenet) for the code of this example (MobileNet V2).
```
# We need to sync down the GitHub repo to run the benchmarks
!git clone https://github.com/UofT-EcoSystem/hfta
# Run the MobileNet V2 benchmark
!python hfta/examples/mobilenet/main.py --version v2 --epochs 5 --amp --eval --dataset cifar10 --device cuda --lr 0.01 0.02 0.03 --hfta
```
Now, let's learn how to leverage HFTA on a normal PyTorch model in the following sections with a simple example of training a convolutional neural network on the MNIST dataset.
## Train a MNIST model without HFTA
We train a simple neural network with two convolutional layers and two fully connected layers, together with some max pooling and dropout layers. This model is trained with the MNIST dataset to recognize hand-written images.
### Define the model in the usual way
```
import time
import random
import torch
import numpy as np
import torch.nn as nn
import torch.nn.functional as F
import torch.optim as optim
from torchvision import datasets, transforms
class Net(nn.Module):
def __init__(self):
super(Net, self).__init__()
self.conv1 = nn.Conv2d(1, 32, 3, 1)
self.conv2 = nn.Conv2d(32, 64, 3, 1)
self.max_pool2d = nn.MaxPool2d(2)
self.fc1 = nn.Linear(9216, 128)
self.fc2 = nn.Linear(128, 10)
self.dropout1 = nn.Dropout2d(0.25)
self.dropout2 = nn.Dropout2d(0.5)
def forward(self, x):
x = self.conv1(x)
x = F.relu(x)
x = self.conv2(x)
x = F.relu(x)
x = self.max_pool2d(x)
x = self.dropout1(x)
x = torch.flatten(x, -3)
x = self.fc1(x)
x = F.relu(x)
x = self.dropout2(x)
x = self.fc2(x)
output = F.log_softmax(x, dim=-1)
return output
```
### Define the training and testing loop for a single epoch
```
def train(config, model, device, train_loader, optimizer, epoch):
"""
config: a dict defined by users to control the experiment
See section: "Train the model"
model: class Net defined in the code block above
device: torch.device
train_loader: torch.utils.data.dataloader.DataLoader
optimizer: torch.optim
epoch: int
"""
model.train()
for batch_idx, (data, target) in enumerate(train_loader):
data, target = data.to(device), target.to(device)
optimizer.zero_grad()
output = model(data)
loss = F.nll_loss(output, target)
loss.backward()
optimizer.step()
if batch_idx % config["log_interval"] == 0:
print('Train Epoch: {} [{}/{} ({:.0f}%)]\tLoss: {:.6f}'.format(
epoch,
batch_idx * len(data),
len(train_loader.dataset),
100. * batch_idx / len(train_loader),
loss.item(),
))
if config["dry_run"]:
break
def test(model, device, test_loader):
"""
model: class Net defined in the code block above
device: torch.device
test_loader: torch.utils.data.dataloader.DataLoader
"""
model.eval()
test_loss = 0
correct = 0
with torch.no_grad():
for data, target in test_loader:
data, target = data.to(device), target.to(device)
N = target.size(0)
output = model(data)
test_loss += F.nll_loss(output, target,
reduction='none').view(-1, N).sum(dim=1)
pred = output.argmax(dim=1, keepdim=True)
correct += pred.eq(target.view_as(pred)).view(-1, N).sum(dim=1)
length = len(test_loader.dataset)
test_loss /= length
loss_str = ["%.4f" % e for e in test_loss]
correct_str = [
"%d/%d(%.2lf%%)" % (e, length, 100. * e / length) for e in correct
]
print('Test set: \tAverage loss: {}, \n \t\t\tAccuracy: {}\n'.format(
loss_str, correct_str))
```
### Define the main loop
```
def main(config):
"""
config: a dict defined by users to control the experiment
"""
random.seed(1)
np.random.seed(1)
torch.manual_seed(1)
device = torch.device(config["device"])
kwargs = {'batch_size': config["batch_size"]}
kwargs.update({'num_workers': 1, 'pin_memory': True, 'shuffle': True},)
transform = transforms.Compose(
[transforms.ToTensor(),
transforms.Normalize((0.1307,), (0.3081,))])
dataset1 = datasets.MNIST('./data',
train=True,
download=True,
transform=transform)
dataset2 = datasets.MNIST('./data', train=False, transform=transform)
train_loader = torch.utils.data.DataLoader(dataset1, **kwargs)
test_loader = torch.utils.data.DataLoader(dataset2, **kwargs)
model = Net().to(device)
optimizer = optim.Adadelta(
model.parameters(),
lr=config["lr"][0],
)
start = time.perf_counter()
for epoch in range(1, config["epochs"] + 1):
now = time.perf_counter()
train(config, model, device, train_loader, optimizer, epoch)
print('Epoch {} took {} s!'.format(epoch, time.perf_counter() - now))
end = time.perf_counter()
test(model, device, test_loader)
print('All jobs Finished, Each epoch took {} s on average!'.format(
(end - start) / config["epochs"]))
```
### Train the model
```
config = {
"device": "cuda", # choose from cuda and cpu
"batch_size": 64,
"lr": [1.0],
"gamma": 0.7,
"epochs": 4,
"seed": 1,
"log_interval": 500,
"dry_run": False,
"save_model": False,
}
print(config)
main(config)
```
## Improve hardware utilization with HFTA
### How to modify a mnist model to use HFTA?
Our convolutional neural network was training fine with MNIST, and that's great! However, with such a small model and a small batch size, the underlying accelerator (NVIDIA GPU in this case) is likely going to be under-utilized. Thus, how can we possibly increase the hardware utilization for this training workload?
If this training workload is used under a repetitive setting (e.g., hyper-parameter tuning or convergence stability testing), hardware utilization can be directly increased by horizontally fusing multiple training workloads together, such that multiple models are trained on the same accelerator (e.g., GPU) at the same time.
However, fusing training workloads manually could be cumbersome and error-prone. Thus, the HFTA library provides convenient utilities to facilitate the effort of horizontally fusing models. Now, let us take a look into how we can easily perform the horizontal model fusion.
Please check the comments in the code to understand what needs to be done. In this example, we fuse multiple models (where the number of models is controlled by the parameter `B`) with different learning rates together via HFTA to improve the hardware utilization.
Be aware that this is just a very simple example of tuning the learning rate. However, in general, many other use cases might be applicable. For example: testing the convergence with different seeds; trying different weight initializers; or even ensemble learning.
#### Modify the model
```
from __future__ import print_function
import sys
import time
import random
import torch
import numpy as np
import torch.nn as nn
import torch.nn.functional as F
import torch.optim as optim
from torchvision import datasets, transforms
# Use utilities from the hfta package to convert your operators and optimizors.
from hfta.ops import convert_ops
from hfta.optim import get_hfta_optim_for
class Net(nn.Module):
# When initializing the model, save the number of models that need to be fused
# (B), and convert the default operators to their HFTA version with
# convert_ops(B, list of operators).
# When passing 0 to B, we train the model as it is without enabling HFTA.
def __init__(self, B=0):
super(Net, self).__init__()
# Convert default operators to their HFTA version.
(Conv2d, MaxPool2d, Linear, Dropout2d) = convert_ops(
B,
nn.Conv2d,
nn.MaxPool2d,
nn.Linear,
nn.Dropout2d,
)
# Define the model with converted operators as if they were unchanged.
self.B = B
self.conv1 = Conv2d(1, 32, 3, 1)
self.conv2 = Conv2d(32, 64, 3, 1)
self.max_pool2d = MaxPool2d(2)
self.fc1 = Linear(9216, 128)
self.fc2 = Linear(128, 10)
self.dropout1 = Dropout2d(0.25)
self.dropout2 = Dropout2d(0.5)
# Minor modifications to the forward pass on special operators.
# Check the documentation of each operator for details.
# Now the shape of x is [batch size, B, 3, 28, 28].
# This means that the input images to all B models are concatenated along the
# channel dimension.
def forward(self, x):
x = self.conv1(x)
x = F.relu(x)
x = self.conv2(x)
x = F.relu(x)
x = self.max_pool2d(x)
x = self.dropout1(x)
x = torch.flatten(x, -3)
if self.B > 0:
# The output shape from flatten is [batch size, B, features], where
# features == channels * height * width from dropout1.
# However, fc1 expects the input shape to be [B, batch size, features].
# Thus, we need to transpose the first and second dimensions.
x = x.transpose(0, 1)
x = self.fc1(x)
x = F.relu(x)
x = self.dropout2(x)
x = self.fc2(x)
output = F.log_softmax(x, dim=-1)
return output
```
#### Modify the training and testing loop
```
def train(config, model, device, train_loader, optimizer, epoch, B):
"""
config: a dict defined by users to control the experiment
See section: "Train the model"
model: class Net defined in the code block above
device: torch.device
train_loader: torch.utils.data.dataloader.DataLoader
optimizer: torch.optim
epoch: int
B: int, the number of models to be fused. When B == 0, we train the original
model as it is without enabling HFTA.
"""
model.train()
for batch_idx, (data, target) in enumerate(train_loader):
data, target = data.to(device), target.to(device)
# Need to duplicate a single batch of input images into multiple batches to
# feed into the fused model.
if B > 0:
N = target.size(0)
data = data.unsqueeze(1).expand(-1, B, -1, -1, -1)
target = target.repeat(B)
optimizer.zero_grad()
output = model(data)
# Also need to modify the loss function to take consideration on the fused
# model.
# In the case:
# 1) the loss function is reduced by averaging along the batch dimension.
# 2) multiple models are horizontally fused via HFTA.
# To make sure the mathematically equivalent gradients are derived by
# ".backward()", we need to scale the loss value by B.
# You might refer to our paper for why such scaling is needed.
if B > 0:
loss = B * F.nll_loss(output.view(B * N, -1), target)
else:
loss = F.nll_loss(output, target)
loss.backward()
optimizer.step()
if batch_idx % config["log_interval"] == 0:
print('Train Epoch: {} [{}/{} ({:.0f}%)]\tLoss: {:.6f}'.format(
epoch, batch_idx * len(data), len(train_loader.dataset),
100. * batch_idx / len(train_loader), loss.item()))
if config["dry_run"]:
break
def test(model, device, test_loader, B):
"""
model: class Net defined in the code block above
device: torch.device
test_loader: torch.utils.data.dataloader.DataLoader
B: int, the number of models to be fused. When B == 0, we test the original
model as it is without enabling HFTA.
"""
model.eval()
test_loss = 0
correct = 0
with torch.no_grad():
for data, target in test_loader:
data, target = data.to(device), target.to(device)
N = target.size(0)
# Need to duplicate a single batch of input images into multiple batches
# to feed into the fused model.
if B > 0:
data = data.unsqueeze(1).expand(-1, B, -1, -1, -1)
target = target.repeat(B)
output = model(data)
# Change the shape of the output to align with the loss function.
if B > 0:
output = output.view(B * N, -1)
test_loss += F.nll_loss(output, target,
reduction='none').view(-1, N).sum(dim=1)
pred = output.argmax(dim=1, keepdim=True)
correct += pred.eq(target.view_as(pred)).view(-1, N).sum(dim=1)
length = len(test_loader.dataset)
test_loss /= length
loss_str = ["%.4f" % e for e in test_loss]
correct_str = [
"%d/%d(%.2lf%%)" % (e, length, 100. * e / length) for e in correct
]
print('Test set: \tAverage loss: {}, \n \t\t\tAccuracy: {}\n'.format(
loss_str, correct_str))
```
#### Modify the main loop
```
def main(config):
"""
config: a dict defined by users to control the experiment
"""
random.seed(config["seed"])
np.random.seed(config["seed"])
torch.manual_seed(config["seed"])
device = torch.device(config["device"])
kwargs = {'batch_size': config["batch_size"]}
kwargs.update({'num_workers': 1, 'pin_memory': True, 'shuffle': True},)
transform = transforms.Compose(
[transforms.ToTensor(),
transforms.Normalize((0.1307,), (0.3081,))])
# Determine the number of models that are horizontally fused together from the
# number of provided learning rates that need to be tested.
B = len(config["lr"]) if config["use_hfta"] else 0
dataset1 = datasets.MNIST('./data',
train=True,
download=True,
transform=transform)
dataset2 = datasets.MNIST('./data', train=False, transform=transform)
train_loader = torch.utils.data.DataLoader(dataset1, **kwargs)
test_loader = torch.utils.data.DataLoader(dataset2, **kwargs)
# Specify the number of models that need to be fused horizontally together (B)
# and create the fused model.
model = Net(B).to(device)
print('B={} lr={}'.format(B, config["lr"]), file=sys.stderr)
# Convert the default optimizor (PyTorch Adadelta) to its HFTA version with
# get_hfta_optim_for(<default>, B).
optimizer = get_hfta_optim_for(optim.Adadelta, B=B)(
model.parameters(),
lr=config["lr"] if B > 0 else config["lr"][0],
)
start = time.perf_counter()
for epoch in range(1, config["epochs"] + 1):
now = time.perf_counter()
train(config, model, device, train_loader, optimizer, epoch, B)
print('Epoch {} took {} s!'.format(epoch, time.perf_counter() - now))
end = time.perf_counter()
test(model, device, test_loader, B)
print('All jobs Finished, Each epoch took {} s on average!'.format(
(end - start) / (max(B, 1) * config["epochs"])))
```
### Train a single HFTA-enabled model with MNIST
Note that this run may be slightly slower than the non-HFTA version because enabling HFTA might lead to a small amount of overhead.
```
# Enable HFTA to train only a single model.
# Only 1 model is trained
config = {
"use_hfta": True,
"device": "cuda", # choose from cuda and cpu
"batch_size": 64,
"lr": [0.1],
"gamma": 0.7,
"epochs": 4,
"seed": 1,
"log_interval": 500,
"dry_run": False,
"save_model": False,
}
print(config)
main(config)
```
### Enable HFTA to train multiple models in the fused form
```
# Enable HFTA and fuse 6 models
config = {
"use_hfta": True,
"device": "cuda", # choose from cuda and cpu
"batch_size": 64,
"lr": [0.1, 0.2, 0.3, 0.4, 0.5, 0.6],
"gamma": 0.7,
"epochs": 4,
"seed": 1,
"log_interval": 500,
"dry_run": False,
"save_model": False,
}
print(config)
main(config)
```
From the [Train the model](https://colab.research.google.com/drive/1gSW6PpWAKfHI3GCxOmSrbBS5PFzh7HEl#scrollTo=35jv_fzP-llU) section above, we would know that, if we want to test 6 different learning rates and train each model on a separate GPU, we would need `11.14 * 6 = 66.84` GPU seconds per epoch on average. As we can see, with HFTA, we can reduce the average training time for testing 6 different learning rates to `3.41 * 6 = 20.46` GPU seconds per epoch, thus, improving the overall utilization of the GPU and reducing the overall training time by `66.84 / 20.46 = 3.27x`.
## Conclusion
Based on the time each epoch takes when training the non-HFTA and HFTA version of the same model, we can see that HFTA helps to increase the throughput of the training, especially on a large hardware. Check our [paper](https://arxiv.org/pdf/2102.02344.pdf) for more details.
|
github_jupyter
|
!pip install git+https://github.com/UofT-EcoSystem/hfta
# We need to sync down the GitHub repo to run the benchmarks
!git clone https://github.com/UofT-EcoSystem/hfta
# Run the MobileNet V2 benchmark
!python hfta/examples/mobilenet/main.py --version v2 --epochs 5 --amp --eval --dataset cifar10 --device cuda --lr 0.01 0.02 0.03 --hfta
import time
import random
import torch
import numpy as np
import torch.nn as nn
import torch.nn.functional as F
import torch.optim as optim
from torchvision import datasets, transforms
class Net(nn.Module):
def __init__(self):
super(Net, self).__init__()
self.conv1 = nn.Conv2d(1, 32, 3, 1)
self.conv2 = nn.Conv2d(32, 64, 3, 1)
self.max_pool2d = nn.MaxPool2d(2)
self.fc1 = nn.Linear(9216, 128)
self.fc2 = nn.Linear(128, 10)
self.dropout1 = nn.Dropout2d(0.25)
self.dropout2 = nn.Dropout2d(0.5)
def forward(self, x):
x = self.conv1(x)
x = F.relu(x)
x = self.conv2(x)
x = F.relu(x)
x = self.max_pool2d(x)
x = self.dropout1(x)
x = torch.flatten(x, -3)
x = self.fc1(x)
x = F.relu(x)
x = self.dropout2(x)
x = self.fc2(x)
output = F.log_softmax(x, dim=-1)
return output
def train(config, model, device, train_loader, optimizer, epoch):
"""
config: a dict defined by users to control the experiment
See section: "Train the model"
model: class Net defined in the code block above
device: torch.device
train_loader: torch.utils.data.dataloader.DataLoader
optimizer: torch.optim
epoch: int
"""
model.train()
for batch_idx, (data, target) in enumerate(train_loader):
data, target = data.to(device), target.to(device)
optimizer.zero_grad()
output = model(data)
loss = F.nll_loss(output, target)
loss.backward()
optimizer.step()
if batch_idx % config["log_interval"] == 0:
print('Train Epoch: {} [{}/{} ({:.0f}%)]\tLoss: {:.6f}'.format(
epoch,
batch_idx * len(data),
len(train_loader.dataset),
100. * batch_idx / len(train_loader),
loss.item(),
))
if config["dry_run"]:
break
def test(model, device, test_loader):
"""
model: class Net defined in the code block above
device: torch.device
test_loader: torch.utils.data.dataloader.DataLoader
"""
model.eval()
test_loss = 0
correct = 0
with torch.no_grad():
for data, target in test_loader:
data, target = data.to(device), target.to(device)
N = target.size(0)
output = model(data)
test_loss += F.nll_loss(output, target,
reduction='none').view(-1, N).sum(dim=1)
pred = output.argmax(dim=1, keepdim=True)
correct += pred.eq(target.view_as(pred)).view(-1, N).sum(dim=1)
length = len(test_loader.dataset)
test_loss /= length
loss_str = ["%.4f" % e for e in test_loss]
correct_str = [
"%d/%d(%.2lf%%)" % (e, length, 100. * e / length) for e in correct
]
print('Test set: \tAverage loss: {}, \n \t\t\tAccuracy: {}\n'.format(
loss_str, correct_str))
def main(config):
"""
config: a dict defined by users to control the experiment
"""
random.seed(1)
np.random.seed(1)
torch.manual_seed(1)
device = torch.device(config["device"])
kwargs = {'batch_size': config["batch_size"]}
kwargs.update({'num_workers': 1, 'pin_memory': True, 'shuffle': True},)
transform = transforms.Compose(
[transforms.ToTensor(),
transforms.Normalize((0.1307,), (0.3081,))])
dataset1 = datasets.MNIST('./data',
train=True,
download=True,
transform=transform)
dataset2 = datasets.MNIST('./data', train=False, transform=transform)
train_loader = torch.utils.data.DataLoader(dataset1, **kwargs)
test_loader = torch.utils.data.DataLoader(dataset2, **kwargs)
model = Net().to(device)
optimizer = optim.Adadelta(
model.parameters(),
lr=config["lr"][0],
)
start = time.perf_counter()
for epoch in range(1, config["epochs"] + 1):
now = time.perf_counter()
train(config, model, device, train_loader, optimizer, epoch)
print('Epoch {} took {} s!'.format(epoch, time.perf_counter() - now))
end = time.perf_counter()
test(model, device, test_loader)
print('All jobs Finished, Each epoch took {} s on average!'.format(
(end - start) / config["epochs"]))
config = {
"device": "cuda", # choose from cuda and cpu
"batch_size": 64,
"lr": [1.0],
"gamma": 0.7,
"epochs": 4,
"seed": 1,
"log_interval": 500,
"dry_run": False,
"save_model": False,
}
print(config)
main(config)
from __future__ import print_function
import sys
import time
import random
import torch
import numpy as np
import torch.nn as nn
import torch.nn.functional as F
import torch.optim as optim
from torchvision import datasets, transforms
# Use utilities from the hfta package to convert your operators and optimizors.
from hfta.ops import convert_ops
from hfta.optim import get_hfta_optim_for
class Net(nn.Module):
# When initializing the model, save the number of models that need to be fused
# (B), and convert the default operators to their HFTA version with
# convert_ops(B, list of operators).
# When passing 0 to B, we train the model as it is without enabling HFTA.
def __init__(self, B=0):
super(Net, self).__init__()
# Convert default operators to their HFTA version.
(Conv2d, MaxPool2d, Linear, Dropout2d) = convert_ops(
B,
nn.Conv2d,
nn.MaxPool2d,
nn.Linear,
nn.Dropout2d,
)
# Define the model with converted operators as if they were unchanged.
self.B = B
self.conv1 = Conv2d(1, 32, 3, 1)
self.conv2 = Conv2d(32, 64, 3, 1)
self.max_pool2d = MaxPool2d(2)
self.fc1 = Linear(9216, 128)
self.fc2 = Linear(128, 10)
self.dropout1 = Dropout2d(0.25)
self.dropout2 = Dropout2d(0.5)
# Minor modifications to the forward pass on special operators.
# Check the documentation of each operator for details.
# Now the shape of x is [batch size, B, 3, 28, 28].
# This means that the input images to all B models are concatenated along the
# channel dimension.
def forward(self, x):
x = self.conv1(x)
x = F.relu(x)
x = self.conv2(x)
x = F.relu(x)
x = self.max_pool2d(x)
x = self.dropout1(x)
x = torch.flatten(x, -3)
if self.B > 0:
# The output shape from flatten is [batch size, B, features], where
# features == channels * height * width from dropout1.
# However, fc1 expects the input shape to be [B, batch size, features].
# Thus, we need to transpose the first and second dimensions.
x = x.transpose(0, 1)
x = self.fc1(x)
x = F.relu(x)
x = self.dropout2(x)
x = self.fc2(x)
output = F.log_softmax(x, dim=-1)
return output
def train(config, model, device, train_loader, optimizer, epoch, B):
"""
config: a dict defined by users to control the experiment
See section: "Train the model"
model: class Net defined in the code block above
device: torch.device
train_loader: torch.utils.data.dataloader.DataLoader
optimizer: torch.optim
epoch: int
B: int, the number of models to be fused. When B == 0, we train the original
model as it is without enabling HFTA.
"""
model.train()
for batch_idx, (data, target) in enumerate(train_loader):
data, target = data.to(device), target.to(device)
# Need to duplicate a single batch of input images into multiple batches to
# feed into the fused model.
if B > 0:
N = target.size(0)
data = data.unsqueeze(1).expand(-1, B, -1, -1, -1)
target = target.repeat(B)
optimizer.zero_grad()
output = model(data)
# Also need to modify the loss function to take consideration on the fused
# model.
# In the case:
# 1) the loss function is reduced by averaging along the batch dimension.
# 2) multiple models are horizontally fused via HFTA.
# To make sure the mathematically equivalent gradients are derived by
# ".backward()", we need to scale the loss value by B.
# You might refer to our paper for why such scaling is needed.
if B > 0:
loss = B * F.nll_loss(output.view(B * N, -1), target)
else:
loss = F.nll_loss(output, target)
loss.backward()
optimizer.step()
if batch_idx % config["log_interval"] == 0:
print('Train Epoch: {} [{}/{} ({:.0f}%)]\tLoss: {:.6f}'.format(
epoch, batch_idx * len(data), len(train_loader.dataset),
100. * batch_idx / len(train_loader), loss.item()))
if config["dry_run"]:
break
def test(model, device, test_loader, B):
"""
model: class Net defined in the code block above
device: torch.device
test_loader: torch.utils.data.dataloader.DataLoader
B: int, the number of models to be fused. When B == 0, we test the original
model as it is without enabling HFTA.
"""
model.eval()
test_loss = 0
correct = 0
with torch.no_grad():
for data, target in test_loader:
data, target = data.to(device), target.to(device)
N = target.size(0)
# Need to duplicate a single batch of input images into multiple batches
# to feed into the fused model.
if B > 0:
data = data.unsqueeze(1).expand(-1, B, -1, -1, -1)
target = target.repeat(B)
output = model(data)
# Change the shape of the output to align with the loss function.
if B > 0:
output = output.view(B * N, -1)
test_loss += F.nll_loss(output, target,
reduction='none').view(-1, N).sum(dim=1)
pred = output.argmax(dim=1, keepdim=True)
correct += pred.eq(target.view_as(pred)).view(-1, N).sum(dim=1)
length = len(test_loader.dataset)
test_loss /= length
loss_str = ["%.4f" % e for e in test_loss]
correct_str = [
"%d/%d(%.2lf%%)" % (e, length, 100. * e / length) for e in correct
]
print('Test set: \tAverage loss: {}, \n \t\t\tAccuracy: {}\n'.format(
loss_str, correct_str))
def main(config):
"""
config: a dict defined by users to control the experiment
"""
random.seed(config["seed"])
np.random.seed(config["seed"])
torch.manual_seed(config["seed"])
device = torch.device(config["device"])
kwargs = {'batch_size': config["batch_size"]}
kwargs.update({'num_workers': 1, 'pin_memory': True, 'shuffle': True},)
transform = transforms.Compose(
[transforms.ToTensor(),
transforms.Normalize((0.1307,), (0.3081,))])
# Determine the number of models that are horizontally fused together from the
# number of provided learning rates that need to be tested.
B = len(config["lr"]) if config["use_hfta"] else 0
dataset1 = datasets.MNIST('./data',
train=True,
download=True,
transform=transform)
dataset2 = datasets.MNIST('./data', train=False, transform=transform)
train_loader = torch.utils.data.DataLoader(dataset1, **kwargs)
test_loader = torch.utils.data.DataLoader(dataset2, **kwargs)
# Specify the number of models that need to be fused horizontally together (B)
# and create the fused model.
model = Net(B).to(device)
print('B={} lr={}'.format(B, config["lr"]), file=sys.stderr)
# Convert the default optimizor (PyTorch Adadelta) to its HFTA version with
# get_hfta_optim_for(<default>, B).
optimizer = get_hfta_optim_for(optim.Adadelta, B=B)(
model.parameters(),
lr=config["lr"] if B > 0 else config["lr"][0],
)
start = time.perf_counter()
for epoch in range(1, config["epochs"] + 1):
now = time.perf_counter()
train(config, model, device, train_loader, optimizer, epoch, B)
print('Epoch {} took {} s!'.format(epoch, time.perf_counter() - now))
end = time.perf_counter()
test(model, device, test_loader, B)
print('All jobs Finished, Each epoch took {} s on average!'.format(
(end - start) / (max(B, 1) * config["epochs"])))
# Enable HFTA to train only a single model.
# Only 1 model is trained
config = {
"use_hfta": True,
"device": "cuda", # choose from cuda and cpu
"batch_size": 64,
"lr": [0.1],
"gamma": 0.7,
"epochs": 4,
"seed": 1,
"log_interval": 500,
"dry_run": False,
"save_model": False,
}
print(config)
main(config)
# Enable HFTA and fuse 6 models
config = {
"use_hfta": True,
"device": "cuda", # choose from cuda and cpu
"batch_size": 64,
"lr": [0.1, 0.2, 0.3, 0.4, 0.5, 0.6],
"gamma": 0.7,
"epochs": 4,
"seed": 1,
"log_interval": 500,
"dry_run": False,
"save_model": False,
}
print(config)
main(config)
| 0.918165 | 0.976152 |
# Heroes of Pymoli Data Analysis
```
import pandas as pd
import numpy as np
file = "./Resources/purchase_data.csv"
# Read Purchasing File and store into Pandas data frame
df_original = pd.read_csv(file)
df_original.head()
#df = df_original.set_index ("Purchase ID")
#df.head()
# number of players
#total_players = df["SN"].count()
#total_players.head()
total_players=len(df["SN"].unique())
pd.DataFrame({"Total Players":[total_players]})
# number of unique items
items=len(df["Item ID"].unique())
ave_price='S{:,.2f}'.format(df["Price"].mean())
purchase=len(df["Item ID"])
revenue='${:,.2f}'.format(df["Price"].sum())
pd.DataFrame({"Number of Unique Items": [items],
"Average Price": [ave_price],
"Number of Purchases": [purchase],
"Total Revenue" : [revenue]})
male_df=df[df["Gender"]=="Male"]
male_count=len(male_df["SN"].unique())
female_df=df[df["Gender"]=="Female"]
female_count=len(female_df["SN"].unique())
other_df=df[df["Gender"]=="Other / Non-Disclosed"]
other_count=len(other_df["SN"].unique())
male_percent = male_count/ total_players * 100
female_percent = female_count/ total_players * 100
other_percent = other_count/ total_players * 100
gender_df = pd.DataFrame({"Total Count":[male_count,female_count,other_count],
"Percentage of Players":[male_percent,female_percent,other_percent]},
index=["Male","Female","Other / Non-Disclosed"])
gender_df["Percentage of Players"]=gender_df["Percentage of Players"].map('{:,.2f}%'.format)
gender_df
grouped_df = df.groupby("Gender")
purchase_count = grouped_df["Item ID"].count()
avg_purchase_price = grouped_df["Price"].mean()
total_purchase_value = grouped_df["Price"].sum()
avg_person = [female_df ["Price"].sum()/female_count,
male_df ["Price"].sum()/male_count,
other_df ["Price"].sum()/other_count
]
purchasing_df=pd.DataFrame({"Purchase Count":purchase_count,
"Average Purchase Price":avg_purchase_price,
"Total Purchase Value":total_purchase_value,
"Avg Total Purchase per Person":avg_person},index=purchase_count.index)
purchasing_df["Average Purchase Price"]=purchasing_df["Average Purchase Price"].map("${:.2f}".format)
purchasing_df["Total Purchase Value"]=purchasing_df["Total Purchase Value"].map("${:.2f}".format)
purchasing_df["Avg Total Purchase per Person"]=purchasing_df["Avg Total Purchase per Person"].map("${:.2f}".format)
purchasing_df
sorted_df = df.sort_values(["SN"], ascending=True)
player_age = sorted_df.groupby("SN").first().reset_index()
bins=[0,9,14,19,24,29,34,39,100]
labels=["< 10","10-14","15-19","20-24","25-29","30-34","35-39","40 +"]
binned_data = pd.cut(player_age["Age"],bins,labels=labels)
age_df=pd.DataFrame({"Total Count" : binned_data.value_counts(),
"Percentage of Players" : binned_data.value_counts()/total_players * 100},
index=labels)
age_df["Percentage of Players"]=age_df["Percentage of Players"].map("{:.2f}%".format)
age_df
df["Age Group"]=pd.cut(df["Age"],bins,labels=labels,)
new_df=df.groupby("Age Group")
age_purchase=pd.DataFrame({"Purchase Count": new_df["Item ID"].count(),
"Average Purchase Price": new_df["Price"].mean(),
"Total Purchase Value": new_df["Price"].sum(),
"Avg Total Purchase per Person": new_df["Price"].sum()/binned_data.value_counts()})
age_purchase["Average Purchase Price"]=age_purchase["Average Purchase Price"].map("${:.2f}".format)
age_purchase["Total Purchase Value"]=age_purchase["Total Purchase Value"].map("${:.2f}".format)
age_purchase["Avg Total Purchase per Person"]=age_purchase["Avg Total Purchase per Person"].map("${:.2f}".format)
age_purchase
group_sn = df.groupby("SN")
top_df = pd.DataFrame({"Purchase Count":group_sn["Item ID"].count(),
"Average Purchase Price":group_sn["Price"].mean(),
"Total Purchase Value":group_sn["Price"].sum()})
top_df_sorted=top_df.sort_values("Total Purchase Value",ascending=False)
top_df_sorted["Average Purchase Price"]=top_df_sorted["Average Purchase Price"].map('${:.2f}'.format)
top_df_sorted["Total Purchase Value"]=top_df_sorted["Total Purchase Value"].map('${:.2f}'.format)
top_df_sorted.head()
popular_group = df[["Item ID","Item Name", "Price"]].groupby(["Item ID","Item Name"])
popular_df=pd.DataFrame({"Purchase Count":popular_group["Price"].count(),
"Item Price":popular_group["Price"].mean(),
"Total Purchase Value":popular_group["Price"].sum()})
popular_df_sorted=popular_df.sort_values("Purchase Count",ascending=False)
popular_df_sorted["Item Price"]=popular_df_sorted["Item Price"].map('${:.2f}'.format)
popular_df_sorted.head()
profitable_df_sorted=popular_df.sort_values("Total Purchase Value",ascending=False)
profitable_df_sorted["Item Price"]=profitable_df_sorted["Item Price"].map('${:.2f}'.format)
profitable_df_sorted["Total Purchase Value"]=profitable_df_sorted["Total Purchase Value"].map('${:.2f}'.format)
profitable_df_sorted.head()
```
|
github_jupyter
|
import pandas as pd
import numpy as np
file = "./Resources/purchase_data.csv"
# Read Purchasing File and store into Pandas data frame
df_original = pd.read_csv(file)
df_original.head()
#df = df_original.set_index ("Purchase ID")
#df.head()
# number of players
#total_players = df["SN"].count()
#total_players.head()
total_players=len(df["SN"].unique())
pd.DataFrame({"Total Players":[total_players]})
# number of unique items
items=len(df["Item ID"].unique())
ave_price='S{:,.2f}'.format(df["Price"].mean())
purchase=len(df["Item ID"])
revenue='${:,.2f}'.format(df["Price"].sum())
pd.DataFrame({"Number of Unique Items": [items],
"Average Price": [ave_price],
"Number of Purchases": [purchase],
"Total Revenue" : [revenue]})
male_df=df[df["Gender"]=="Male"]
male_count=len(male_df["SN"].unique())
female_df=df[df["Gender"]=="Female"]
female_count=len(female_df["SN"].unique())
other_df=df[df["Gender"]=="Other / Non-Disclosed"]
other_count=len(other_df["SN"].unique())
male_percent = male_count/ total_players * 100
female_percent = female_count/ total_players * 100
other_percent = other_count/ total_players * 100
gender_df = pd.DataFrame({"Total Count":[male_count,female_count,other_count],
"Percentage of Players":[male_percent,female_percent,other_percent]},
index=["Male","Female","Other / Non-Disclosed"])
gender_df["Percentage of Players"]=gender_df["Percentage of Players"].map('{:,.2f}%'.format)
gender_df
grouped_df = df.groupby("Gender")
purchase_count = grouped_df["Item ID"].count()
avg_purchase_price = grouped_df["Price"].mean()
total_purchase_value = grouped_df["Price"].sum()
avg_person = [female_df ["Price"].sum()/female_count,
male_df ["Price"].sum()/male_count,
other_df ["Price"].sum()/other_count
]
purchasing_df=pd.DataFrame({"Purchase Count":purchase_count,
"Average Purchase Price":avg_purchase_price,
"Total Purchase Value":total_purchase_value,
"Avg Total Purchase per Person":avg_person},index=purchase_count.index)
purchasing_df["Average Purchase Price"]=purchasing_df["Average Purchase Price"].map("${:.2f}".format)
purchasing_df["Total Purchase Value"]=purchasing_df["Total Purchase Value"].map("${:.2f}".format)
purchasing_df["Avg Total Purchase per Person"]=purchasing_df["Avg Total Purchase per Person"].map("${:.2f}".format)
purchasing_df
sorted_df = df.sort_values(["SN"], ascending=True)
player_age = sorted_df.groupby("SN").first().reset_index()
bins=[0,9,14,19,24,29,34,39,100]
labels=["< 10","10-14","15-19","20-24","25-29","30-34","35-39","40 +"]
binned_data = pd.cut(player_age["Age"],bins,labels=labels)
age_df=pd.DataFrame({"Total Count" : binned_data.value_counts(),
"Percentage of Players" : binned_data.value_counts()/total_players * 100},
index=labels)
age_df["Percentage of Players"]=age_df["Percentage of Players"].map("{:.2f}%".format)
age_df
df["Age Group"]=pd.cut(df["Age"],bins,labels=labels,)
new_df=df.groupby("Age Group")
age_purchase=pd.DataFrame({"Purchase Count": new_df["Item ID"].count(),
"Average Purchase Price": new_df["Price"].mean(),
"Total Purchase Value": new_df["Price"].sum(),
"Avg Total Purchase per Person": new_df["Price"].sum()/binned_data.value_counts()})
age_purchase["Average Purchase Price"]=age_purchase["Average Purchase Price"].map("${:.2f}".format)
age_purchase["Total Purchase Value"]=age_purchase["Total Purchase Value"].map("${:.2f}".format)
age_purchase["Avg Total Purchase per Person"]=age_purchase["Avg Total Purchase per Person"].map("${:.2f}".format)
age_purchase
group_sn = df.groupby("SN")
top_df = pd.DataFrame({"Purchase Count":group_sn["Item ID"].count(),
"Average Purchase Price":group_sn["Price"].mean(),
"Total Purchase Value":group_sn["Price"].sum()})
top_df_sorted=top_df.sort_values("Total Purchase Value",ascending=False)
top_df_sorted["Average Purchase Price"]=top_df_sorted["Average Purchase Price"].map('${:.2f}'.format)
top_df_sorted["Total Purchase Value"]=top_df_sorted["Total Purchase Value"].map('${:.2f}'.format)
top_df_sorted.head()
popular_group = df[["Item ID","Item Name", "Price"]].groupby(["Item ID","Item Name"])
popular_df=pd.DataFrame({"Purchase Count":popular_group["Price"].count(),
"Item Price":popular_group["Price"].mean(),
"Total Purchase Value":popular_group["Price"].sum()})
popular_df_sorted=popular_df.sort_values("Purchase Count",ascending=False)
popular_df_sorted["Item Price"]=popular_df_sorted["Item Price"].map('${:.2f}'.format)
popular_df_sorted.head()
profitable_df_sorted=popular_df.sort_values("Total Purchase Value",ascending=False)
profitable_df_sorted["Item Price"]=profitable_df_sorted["Item Price"].map('${:.2f}'.format)
profitable_df_sorted["Total Purchase Value"]=profitable_df_sorted["Total Purchase Value"].map('${:.2f}'.format)
profitable_df_sorted.head()
| 0.282988 | 0.502808 |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.