code
stringlengths 2.5k
6.36M
| kind
stringclasses 2
values | parsed_code
stringlengths 0
404k
| quality_prob
float64 0
0.98
| learning_prob
float64 0.03
1
|
|---|---|---|---|---|
# Anomaly Detection Using Gaussian Distribution
_Source: 🤖[Homemade Machine Learning](https://github.com/trekhleb/homemade-machine-learning) repository_
> ☝Before moving on with this demo you might want to take a look at:
> - 📗[Math behind the Anomaly Detection](https://github.com/trekhleb/homemade-machine-learning/tree/master/homemade/anomaly_detection)
> - ⚙️[Gaussian Anomaly Detection Source Code](https://github.com/trekhleb/homemade-machine-learning/blob/master/homemade/anomaly_detection/gaussian_anomaly_detection.py)
**Anomaly detection** (also **outlier detection**) is the identification of rare items, events or observations which raise suspicions by differing significantly from the majority of the data.
The **normal** (or **Gaussian**) distribution is a very common continuous probability distribution. Normal distributions are important in statistics and are often used in the natural and social sciences to represent real-valued random variables whose distributions are not known. A random variable with a Gaussian distribution is said to be normally distributed and is called a normal deviate.
> **Demo Project:** In this demo we will build a model that will find anomalies in server operational parameters such as `Latency` and `Throughput`.
```
# To make debugging of logistic_regression module easier we enable imported modules autoreloading feature.
# By doing this you may change the code of logistic_regression library and all these changes will be available here.
%load_ext autoreload
%autoreload 2
# Add project root folder to module loading paths.
import sys
sys.path.append('../..')
```
### Import Dependencies
- [pandas](https://pandas.pydata.org/) - library that we will use for loading and displaying the data in a table
- [numpy](http://www.numpy.org/) - library that we will use for linear algebra operations
- [matplotlib](https://matplotlib.org/) - library that we will use for plotting the data
- [anomaly_detection](https://github.com/trekhleb/homemade-machine-learning/blob/master/homemade/anomaly_detection/gaussian_anomaly_detection.py) - custom implementation of anomaly detection using Gaussian distribution.
```
# Import 3rd party dependencies.
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
# Import custom Gaussian anomaly detection implementation.
from homemade.anomaly_detection import GaussianAnomalyDetection
```
### Load the Data
In this demo we will use the dataset with server operational parameters such as `Latency` and `Throughput` and will try to find anomalies in them.
```
# Load the data.
pd_data = pd.read_csv('../../data/server-operational-params.csv')
# Print the data table.
pd_data.head(10)
# Print histograms for each feature to see how they vary.
histohrams = pd_data[['Latency (ms)', 'Throughput (mb/s)']].hist(grid=False, figsize=(10,4))
```
### Plot the Data
Let's plot `Throughput(Latency)` dependency and see if the distribution is similar to Gaussian one.
```
# Extract first two column from the dataset.
data = pd_data[['Latency (ms)', 'Throughput (mb/s)']].values
# Plot the data.
plt.scatter(data[:, 0], data[:, 1], alpha=0.6)
plt.xlabel('Latency (ms)')
plt.ylabel('Throughput (mb/s)')
plt.title('Server Operational Params')
plt.show()
```
### Find Anomalies Using Gaussian Distribution
Let's try to use our custom anomaly detection implementation using Gaussian distribution.
```
# Init Gaussian anomaly instance.
gaussian_anomaly = GaussianAnomalyDetection(data)
# Let's see Gaussian estimation parameters.
print('mu')
print(gaussian_anomaly.mu_param)
print('\n')
print('sigma^2')
print(gaussian_anomaly.sigma_squared)
```
### Visualize the Fit
Let's draw a contour plots that will represent our Gaussian distribution for the dataset.
```
# Create a 3D grid to build a contour plots.
# Create ranges along X and Y axes.
latency_from = 0
latency_to = 35
throughput_from = 0
throughput_to = 35
step = 0.5
latency_range = np.arange(latency_from, latency_to, step)
throughput_range = np.arange(throughput_from, throughput_to, step)
# Create X and Y grids.
(latency_grid, throughput_grid) = np.meshgrid(latency_range, throughput_range)
# Flatten latency and throughput grids.
flat_latency_grid = latency_grid.flatten().reshape((latency_grid.size, 1))
flat_throughput_grid = throughput_grid.flatten().reshape((throughput_grid.size, 1))
# Joing latency and throughput flatten grids together to form all combinations of latency and throughput.
combinations = np.hstack((flat_latency_grid, flat_throughput_grid))
# Now let's calculate the probabilities for every combination of latency and throughput.
flat_probabilities = gaussian_anomaly.multivariate_gaussian(combinations)
# Resghape probabilities back to matrix in order to build contours.
probabilities = flat_probabilities.reshape(latency_grid.shape)
# Let's build plot our original dataset.
plt.scatter(data[:, 0], data[:, 1], alpha=0.6)
plt.xlabel('Latency (ms)')
plt.ylabel('Throughput (mb/s)')
plt.title('Server Operational Params')
# On top of our original dataset let's plot probability contours.
plt.contour(latency_grid, throughput_grid, probabilities, levels=10)
# Display the plot.
plt.show()
```
### Select best threshold
Now, in order to decide which examples should be counted as an anomaly we need to decide which probability threshold to choose. We could do it intuitively but since we have all data examples labeled in our dataset let's use that data to calculate the best threshold.
```
# Extract the information about which example is anomaly and which is not.
num_examples = data.shape[0]
labels = pd_data['Anomaly'].values.reshape((num_examples, 1))
# Returns the density of the multivariate normal at each data point (row) of X dataset.
probabilities = gaussian_anomaly.multivariate_gaussian(data)
# Let's go through many possible thresholds and pick the one with the highest F1 score.
(epsilon, f1, precision_history, recall_history, f1_history) = gaussian_anomaly.select_threshold(
labels, probabilities
)
print('Best epsilon:')
print(epsilon)
print('\n')
print('Best F1 score:')
print(f1)
```
### Plot Precision/Recall Progress
Let's now plot precision, reacall and F1 score changes for every iteration.
```
# Make the plot a little bit bigger than default one.
plt.figure(figsize=(15, 5))
# Plot precission history.
plt.subplot(1, 3, 1)
plt.xlabel('Iteration')
plt.ylabel('Value')
plt.title('Precission Progress')
plt.plot(precision_history)
# Plot recall history.
plt.subplot(1, 3, 2)
plt.xlabel('Iteration')
plt.ylabel('Value')
plt.title('Recall Progress')
plt.plot(recall_history)
# Plot F1 history.
plt.subplot(1, 3, 3)
plt.xlabel('Iteration')
plt.ylabel('Value')
plt.title('F1 Progress')
plt.plot(f1_history)
# Display all plots.
plt.show()
```
### Fing Outliers
Since now we have calculated best `epsilon` we may find outliers.
```
# Find indices of data examples with probabilities less than the best epsilon.
outliers_indices = np.where(probabilities < epsilon)[0]
# Plot original data.
plt.scatter(data[:, 0], data[:, 1], alpha=0.6, label='Dataset')
plt.xlabel('Latency (ms)')
plt.ylabel('Throughput (mb/s)')
plt.title('Server Operational Params')
# Plot the outliers.
plt.scatter(data[outliers_indices, 0], data[outliers_indices, 1], alpha=0.6, c='red', label='Outliers')
# Display plots.
plt.legend()
plt.plot()
```
|
github_jupyter
|
# To make debugging of logistic_regression module easier we enable imported modules autoreloading feature.
# By doing this you may change the code of logistic_regression library and all these changes will be available here.
%load_ext autoreload
%autoreload 2
# Add project root folder to module loading paths.
import sys
sys.path.append('../..')
# Import 3rd party dependencies.
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
# Import custom Gaussian anomaly detection implementation.
from homemade.anomaly_detection import GaussianAnomalyDetection
# Load the data.
pd_data = pd.read_csv('../../data/server-operational-params.csv')
# Print the data table.
pd_data.head(10)
# Print histograms for each feature to see how they vary.
histohrams = pd_data[['Latency (ms)', 'Throughput (mb/s)']].hist(grid=False, figsize=(10,4))
# Extract first two column from the dataset.
data = pd_data[['Latency (ms)', 'Throughput (mb/s)']].values
# Plot the data.
plt.scatter(data[:, 0], data[:, 1], alpha=0.6)
plt.xlabel('Latency (ms)')
plt.ylabel('Throughput (mb/s)')
plt.title('Server Operational Params')
plt.show()
# Init Gaussian anomaly instance.
gaussian_anomaly = GaussianAnomalyDetection(data)
# Let's see Gaussian estimation parameters.
print('mu')
print(gaussian_anomaly.mu_param)
print('\n')
print('sigma^2')
print(gaussian_anomaly.sigma_squared)
# Create a 3D grid to build a contour plots.
# Create ranges along X and Y axes.
latency_from = 0
latency_to = 35
throughput_from = 0
throughput_to = 35
step = 0.5
latency_range = np.arange(latency_from, latency_to, step)
throughput_range = np.arange(throughput_from, throughput_to, step)
# Create X and Y grids.
(latency_grid, throughput_grid) = np.meshgrid(latency_range, throughput_range)
# Flatten latency and throughput grids.
flat_latency_grid = latency_grid.flatten().reshape((latency_grid.size, 1))
flat_throughput_grid = throughput_grid.flatten().reshape((throughput_grid.size, 1))
# Joing latency and throughput flatten grids together to form all combinations of latency and throughput.
combinations = np.hstack((flat_latency_grid, flat_throughput_grid))
# Now let's calculate the probabilities for every combination of latency and throughput.
flat_probabilities = gaussian_anomaly.multivariate_gaussian(combinations)
# Resghape probabilities back to matrix in order to build contours.
probabilities = flat_probabilities.reshape(latency_grid.shape)
# Let's build plot our original dataset.
plt.scatter(data[:, 0], data[:, 1], alpha=0.6)
plt.xlabel('Latency (ms)')
plt.ylabel('Throughput (mb/s)')
plt.title('Server Operational Params')
# On top of our original dataset let's plot probability contours.
plt.contour(latency_grid, throughput_grid, probabilities, levels=10)
# Display the plot.
plt.show()
# Extract the information about which example is anomaly and which is not.
num_examples = data.shape[0]
labels = pd_data['Anomaly'].values.reshape((num_examples, 1))
# Returns the density of the multivariate normal at each data point (row) of X dataset.
probabilities = gaussian_anomaly.multivariate_gaussian(data)
# Let's go through many possible thresholds and pick the one with the highest F1 score.
(epsilon, f1, precision_history, recall_history, f1_history) = gaussian_anomaly.select_threshold(
labels, probabilities
)
print('Best epsilon:')
print(epsilon)
print('\n')
print('Best F1 score:')
print(f1)
# Make the plot a little bit bigger than default one.
plt.figure(figsize=(15, 5))
# Plot precission history.
plt.subplot(1, 3, 1)
plt.xlabel('Iteration')
plt.ylabel('Value')
plt.title('Precission Progress')
plt.plot(precision_history)
# Plot recall history.
plt.subplot(1, 3, 2)
plt.xlabel('Iteration')
plt.ylabel('Value')
plt.title('Recall Progress')
plt.plot(recall_history)
# Plot F1 history.
plt.subplot(1, 3, 3)
plt.xlabel('Iteration')
plt.ylabel('Value')
plt.title('F1 Progress')
plt.plot(f1_history)
# Display all plots.
plt.show()
# Find indices of data examples with probabilities less than the best epsilon.
outliers_indices = np.where(probabilities < epsilon)[0]
# Plot original data.
plt.scatter(data[:, 0], data[:, 1], alpha=0.6, label='Dataset')
plt.xlabel('Latency (ms)')
plt.ylabel('Throughput (mb/s)')
plt.title('Server Operational Params')
# Plot the outliers.
plt.scatter(data[outliers_indices, 0], data[outliers_indices, 1], alpha=0.6, c='red', label='Outliers')
# Display plots.
plt.legend()
plt.plot()
| 0.797083 | 0.991187 |
# Part 2 - Making materials from elements
As we saw in Part 1, materials can be defined in OpenMC using isotopes. However, materials can also be made from elements - this is more concise and still supports isotopic enrichment.
This python notebook allows users to create different materials from elements using OpenMC.
The following code block is a simple example of creating a material (water H2O) from elements. (Note how Hydrogen and Oxygen elements have been specified rather than each specific isotope).
```
import openmc
# Making water from elements
water_mat = openmc.Material(name='water')
water_mat.add_element('H', 2.0, percent_type='ao')
water_mat.add_element('O', 1.0, percent_type='ao')
water_mat.set_density('g/cm3', 0.99821)
water_mat
```
The next code block is an example of making a ceramic breeder material.
```
# Making Li4SiO4 from elements
Li4SiO4_mat = openmc.Material(name='lithium_orthosilicate')
Li4SiO4_mat.add_element('Li', 4.0, percent_type='ao')
Li4SiO4_mat.add_element('Si', 1.0, percent_type='ao')
Li4SiO4_mat.add_element('O', 4.0, percent_type='ao')
Li4SiO4_mat.set_density('g/cm3', 2.32)
Li4SiO4_mat
```
It is also possible to enrich specific isotopes while still benefitting from the concise code of making materials from elements.
Here is an example of making the same ceramic breeder material but this time with Li6 enrichment.
```
# Making enriched Li4SiO4 from elements with enrichment of Li6 enrichment
Li4SiO4_mat = openmc.Material(name='lithium_orthosilicate')
Li4SiO4_mat.add_element('Li', 4.0, percent_type='ao',
enrichment=60,
enrichment_target='Li6',
enrichment_type='ao'
)
Li4SiO4_mat.add_element('Si', 1.0, percent_type='ao')
Li4SiO4_mat.add_element('O', 4.0, percent_type='ao')
Li4SiO4_mat.set_density('g/cm3', 2.32) # this would actually be lower than 2.32 g/cm3 but this would need calculating
Li4SiO4_mat
```
In the case of materials that can be represented as a chemical formula (e.g. 'H2O', 'Li4SiO4') there is an even more concise way of making these materials by using their chemical formula.
```
# making Li4SiO4 from a formula
Li4SiO4_mat = openmc.Material(name='lithium_orthosilicate')
Li4SiO4_mat.add_elements_from_formula('Li4SiO4')
Li4SiO4_mat
```
This add_elements_from_formula (which was added to OpenMC source code by myself) can also support enrichment.
```
# making Li4SiO4 from a formula with enrichment
Li4SiO4_mat = openmc.Material(name='lithium_orthosilicate')
Li4SiO4_mat.add_elements_from_formula('Li4SiO4',
enrichment=60,
enrichment_target='Li6',
enrichment_type='ao'
)
Li4SiO4_mat
```
Making more detailed materials such as a low activation steel Eurofer would require about 20 elements. While this is fewer user inputs than making the material from isotopes it is still quite a lot of coding for the user. Unfortunately, they cannot be input as a chemical formula either.
**Learning Outcomes for Part 2:**
- Materials can be made in OpenMC using element fractions and densities.
- Making materials from elements is more concise than making materials from isotopes.
- If materials can be represented as a chemical formula, OpenMC also offers a way to construct those materials from that.
- Making materials from elements also supports isotopic enrichment.
|
github_jupyter
|
import openmc
# Making water from elements
water_mat = openmc.Material(name='water')
water_mat.add_element('H', 2.0, percent_type='ao')
water_mat.add_element('O', 1.0, percent_type='ao')
water_mat.set_density('g/cm3', 0.99821)
water_mat
# Making Li4SiO4 from elements
Li4SiO4_mat = openmc.Material(name='lithium_orthosilicate')
Li4SiO4_mat.add_element('Li', 4.0, percent_type='ao')
Li4SiO4_mat.add_element('Si', 1.0, percent_type='ao')
Li4SiO4_mat.add_element('O', 4.0, percent_type='ao')
Li4SiO4_mat.set_density('g/cm3', 2.32)
Li4SiO4_mat
# Making enriched Li4SiO4 from elements with enrichment of Li6 enrichment
Li4SiO4_mat = openmc.Material(name='lithium_orthosilicate')
Li4SiO4_mat.add_element('Li', 4.0, percent_type='ao',
enrichment=60,
enrichment_target='Li6',
enrichment_type='ao'
)
Li4SiO4_mat.add_element('Si', 1.0, percent_type='ao')
Li4SiO4_mat.add_element('O', 4.0, percent_type='ao')
Li4SiO4_mat.set_density('g/cm3', 2.32) # this would actually be lower than 2.32 g/cm3 but this would need calculating
Li4SiO4_mat
# making Li4SiO4 from a formula
Li4SiO4_mat = openmc.Material(name='lithium_orthosilicate')
Li4SiO4_mat.add_elements_from_formula('Li4SiO4')
Li4SiO4_mat
# making Li4SiO4 from a formula with enrichment
Li4SiO4_mat = openmc.Material(name='lithium_orthosilicate')
Li4SiO4_mat.add_elements_from_formula('Li4SiO4',
enrichment=60,
enrichment_target='Li6',
enrichment_type='ao'
)
Li4SiO4_mat
| 0.410402 | 0.97824 |
```
import numpy as np
import pandas as pd
from timeit import timeit
```
# Enhancing performance
We first look at native code compilation. Here we show 3 common methods for doing this using `numba` JIT compilation, `cython` AOT compilation, and direct wrapping of C++ code using `pybind11`. In general, `numba` is the simplest to use, while you have the most flexibility with `pybind11`. Which approach gives the best performance generally requires some experimentation.
Then we review common methods for concurrent execution of embarrassingly parallel code using `multiprocessing`, `concurrent.futures` and `joblib`. Comparison of performance using processes and threads is made, with a brief explanation of the Global Interpreter Lock (GIL).
More details for each of the libraries used to improve performance is provided in the course notebooks.
## Python
```
def cdist(xs, ys):
"""Returns pairwise distance between row vectors in xs and ys.
xs has shape (m, p)
ys has shape (n, p)
Return value has shape (m, n)
"""
m, p = xs.shape
n, p = ys.shape
res = np.empty((m, n))
for i in range(m):
for j in range(n):
res[i, j] = np.sqrt(np.sum((ys[j] - xs[i])**2))
return res
```
### Sanity check
```
xs = np.arange(6).reshape(-1,2).astype('float')
ys = np.arange(4).reshape(-1, 2).astype('float')
zs = cdist(xs, ys)
zs
%timeit -r 3 -n 10 cdist(xs, ys)
m = 1000
n = 1000
p = 100
X = np.random.random((m, p))
Y = np.random.random((n, p))
%%time
Z = cdist(X, Y)
t0 = timeit(lambda : cdist(X, Y), number=1)
```
## Using `numba`
```
from numba import jit, njit
@njit
def cdist_numba(xs, ys):
"""Returns pairwise distance between row vectors in xs and ys.
xs has shape (m, p)
ys has shape (n, p)
Return value has shape (m, n)
"""
m, p = xs.shape
n, p = ys.shape
res = np.empty((m, n))
for i in range(m):
for j in range(n):
res[i, j] = np.sqrt(np.sum((ys[j] - xs[i])**2))
return res
```
Check
```
assert(np.allclose(cdist(xs, ys), cdist_numba(xs, ys)))
%%time
Z = cdist_numba(X, Y)
t_numba = timeit(lambda : cdist_numba(X, Y), number=1)
```
### Unrolling
We can help `numba` by unrolling the code.
```
@njit
def cdist_numba1(xs, ys):
"""Returns pairwise distance between row vectors in xs and ys.
xs has shape (m, p)
ys has shape (n, p)
Return value has shape (m, n)
"""
m, p = xs.shape
n, p = ys.shape
res = np.empty((m, n))
for i in range(m):
for j in range(n):
s = 0
for k in range(p):
s += (ys[j,k] - xs[i,k])**2
res[i, j] = np.sqrt(s)
return res
```
Check
```
assert(np.allclose(cdist(xs, ys), cdist_numba1(xs, ys)))
%%time
Z = cdist_numba1(X, Y)
t_numba1 = timeit(lambda : cdist_numba1(X, Y), number=1)
```
## Using `cython`
```
%load_ext cython
%%cython -a
import numpy as np
def cdist_cython(xs, ys):
"""Returns pairwise distance between row vectors in xs and ys.
xs has shape (m, p)
ys has shape (n, p)
Return value has shape (m, n)
"""
m, p = xs.shape
n, p = ys.shape
res = np.empty((m, n))
for i in range(m):
for j in range(n):
res[i, j] = np.sqrt(np.sum((ys[j] - xs[i])**2))
return res
```
Check
```
assert(np.allclose(cdist(xs, ys), cdist_cython(xs, ys)))
%%time
Z = cdist_cython(X, Y)
t_cython = timeit(lambda : cdist_cython(X, Y), number=1)
%%cython -a
import cython
import numpy as np
from libc.math cimport sqrt, pow
@cython.boundscheck(False)
@cython.wraparound(False)
def cdist_cython1(double[:, :] xs, double[:, :] ys):
"""Returns pairwise distance between row vectors in xs and ys.
xs has shape (m, p)
ys has shape (n, p)
Return value has shape (m, n)
"""
cdef int m, n, p
m = xs.shape[0]
n = ys.shape[0]
p = xs.shape[1]
cdef double[:, :] res = np.empty((m, n))
cdef int i, j
cdef double s
for i in range(m):
for j in range(n):
s = 0.0
for k in range(p):
s += pow(ys[j,k] - xs[i,k], 2)
res[i, j] = sqrt(s)
return res
```
Check
```
assert(np.allclose(cdist(xs, ys), cdist_cython(xs, ys)))
%%time
Z = cdist_cython1(X, Y)
t_cython1 = timeit(lambda : cdist_cython1(X, Y), number=1)
perf = pd.DataFrame(dict(
methods = ['python', 'numba', 'numba1', 'cython', 'cython1'],
times = [t0, t_numba, t_numba1, t_cython, t_cython1],
))
perf['speed-up'] = np.around(perf['times'][0]/perf['times'], 1)
perf
```
## Using multiple cores
The standard implementation of Python uses a Global Interpreter Lock (GIL). This means that only one thread can be run at any one time, and multiple threads work by time-slicing. Hence multi-threaded code with lots of latency can result in speed-ups, but multi-threaded code which is computationally intensive will not see any speed-up. For numerically intensive code, parallel code needs to be run in separate processes to see speed-ups.
First we see how to split the computation into pieces using a loop.
```
xs
ys
cdist(xs, ys)
res = np.concatenate([cdist(x, ys) for x in np.split(xs, 3, 0)])
res
%%time
Z = cdist(X, Y)
```
### Using `multiprocessing`
```
from multiprocessing import Pool
%%time
with Pool(processes=4) as p:
Z1 = p.starmap(cdist, [(X_, Y) for X_ in np.split(X, 100, 0)])
Z1 = np.concatenate(Z1)
```
Check
```
np.testing.assert_allclose(Z, Z1)
```
### Using `concurrent.futures
```
from concurrent.futures import ProcessPoolExecutor, ThreadPoolExecutor
def cdist_(args):
return cdist(*args)
%%time
with ProcessPoolExecutor(max_workers=4) as pool:
Z2 = list(pool.map(cdist_, [(X_, Y) for X_ in np.split(X, 100, 0)]))
Z2 = np.concatenate(Z2)
```
Check
```
np.testing.assert_allclose(Z, Z2)
```
### Using `joblib`
`joblib` provides parallel processing using a comprehension syntax.
```
from joblib import Parallel, delayed
%%time
Z3 = Parallel(n_jobs=4)(delayed(cdist)(X_, Y) for X_ in np.split(X, 100, 0))
Z3 = np.concatenate(Z3)
```
Check
```
np.testing.assert_allclose(Z, Z3)
```
### Using threads
Note that there is no gain with using multiple threads for computationally intensive tasks because of the GIL.
```
%%time
with ThreadPoolExecutor(max_workers=4) as pool:
Z4 = list(pool.map(cdist_, [(X_, Y) for X_ in np.split(X, 100, 0)]))
Z4 = np.concatenate(Z4)
```
Check
```
np.testing.assert_allclose(Z, Z4)
```
|
github_jupyter
|
import numpy as np
import pandas as pd
from timeit import timeit
def cdist(xs, ys):
"""Returns pairwise distance between row vectors in xs and ys.
xs has shape (m, p)
ys has shape (n, p)
Return value has shape (m, n)
"""
m, p = xs.shape
n, p = ys.shape
res = np.empty((m, n))
for i in range(m):
for j in range(n):
res[i, j] = np.sqrt(np.sum((ys[j] - xs[i])**2))
return res
xs = np.arange(6).reshape(-1,2).astype('float')
ys = np.arange(4).reshape(-1, 2).astype('float')
zs = cdist(xs, ys)
zs
%timeit -r 3 -n 10 cdist(xs, ys)
m = 1000
n = 1000
p = 100
X = np.random.random((m, p))
Y = np.random.random((n, p))
%%time
Z = cdist(X, Y)
t0 = timeit(lambda : cdist(X, Y), number=1)
from numba import jit, njit
@njit
def cdist_numba(xs, ys):
"""Returns pairwise distance between row vectors in xs and ys.
xs has shape (m, p)
ys has shape (n, p)
Return value has shape (m, n)
"""
m, p = xs.shape
n, p = ys.shape
res = np.empty((m, n))
for i in range(m):
for j in range(n):
res[i, j] = np.sqrt(np.sum((ys[j] - xs[i])**2))
return res
assert(np.allclose(cdist(xs, ys), cdist_numba(xs, ys)))
%%time
Z = cdist_numba(X, Y)
t_numba = timeit(lambda : cdist_numba(X, Y), number=1)
@njit
def cdist_numba1(xs, ys):
"""Returns pairwise distance between row vectors in xs and ys.
xs has shape (m, p)
ys has shape (n, p)
Return value has shape (m, n)
"""
m, p = xs.shape
n, p = ys.shape
res = np.empty((m, n))
for i in range(m):
for j in range(n):
s = 0
for k in range(p):
s += (ys[j,k] - xs[i,k])**2
res[i, j] = np.sqrt(s)
return res
assert(np.allclose(cdist(xs, ys), cdist_numba1(xs, ys)))
%%time
Z = cdist_numba1(X, Y)
t_numba1 = timeit(lambda : cdist_numba1(X, Y), number=1)
%load_ext cython
%%cython -a
import numpy as np
def cdist_cython(xs, ys):
"""Returns pairwise distance between row vectors in xs and ys.
xs has shape (m, p)
ys has shape (n, p)
Return value has shape (m, n)
"""
m, p = xs.shape
n, p = ys.shape
res = np.empty((m, n))
for i in range(m):
for j in range(n):
res[i, j] = np.sqrt(np.sum((ys[j] - xs[i])**2))
return res
assert(np.allclose(cdist(xs, ys), cdist_cython(xs, ys)))
%%time
Z = cdist_cython(X, Y)
t_cython = timeit(lambda : cdist_cython(X, Y), number=1)
%%cython -a
import cython
import numpy as np
from libc.math cimport sqrt, pow
@cython.boundscheck(False)
@cython.wraparound(False)
def cdist_cython1(double[:, :] xs, double[:, :] ys):
"""Returns pairwise distance between row vectors in xs and ys.
xs has shape (m, p)
ys has shape (n, p)
Return value has shape (m, n)
"""
cdef int m, n, p
m = xs.shape[0]
n = ys.shape[0]
p = xs.shape[1]
cdef double[:, :] res = np.empty((m, n))
cdef int i, j
cdef double s
for i in range(m):
for j in range(n):
s = 0.0
for k in range(p):
s += pow(ys[j,k] - xs[i,k], 2)
res[i, j] = sqrt(s)
return res
assert(np.allclose(cdist(xs, ys), cdist_cython(xs, ys)))
%%time
Z = cdist_cython1(X, Y)
t_cython1 = timeit(lambda : cdist_cython1(X, Y), number=1)
perf = pd.DataFrame(dict(
methods = ['python', 'numba', 'numba1', 'cython', 'cython1'],
times = [t0, t_numba, t_numba1, t_cython, t_cython1],
))
perf['speed-up'] = np.around(perf['times'][0]/perf['times'], 1)
perf
xs
ys
cdist(xs, ys)
res = np.concatenate([cdist(x, ys) for x in np.split(xs, 3, 0)])
res
%%time
Z = cdist(X, Y)
from multiprocessing import Pool
%%time
with Pool(processes=4) as p:
Z1 = p.starmap(cdist, [(X_, Y) for X_ in np.split(X, 100, 0)])
Z1 = np.concatenate(Z1)
np.testing.assert_allclose(Z, Z1)
from concurrent.futures import ProcessPoolExecutor, ThreadPoolExecutor
def cdist_(args):
return cdist(*args)
%%time
with ProcessPoolExecutor(max_workers=4) as pool:
Z2 = list(pool.map(cdist_, [(X_, Y) for X_ in np.split(X, 100, 0)]))
Z2 = np.concatenate(Z2)
np.testing.assert_allclose(Z, Z2)
from joblib import Parallel, delayed
%%time
Z3 = Parallel(n_jobs=4)(delayed(cdist)(X_, Y) for X_ in np.split(X, 100, 0))
Z3 = np.concatenate(Z3)
np.testing.assert_allclose(Z, Z3)
%%time
with ThreadPoolExecutor(max_workers=4) as pool:
Z4 = list(pool.map(cdist_, [(X_, Y) for X_ in np.split(X, 100, 0)]))
Z4 = np.concatenate(Z4)
np.testing.assert_allclose(Z, Z4)
| 0.643777 | 0.946523 |
```
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from matplotlib.ticker import MaxNLocator
from enum import Enum
%matplotlib inline
class Labels(Enum):
ALL = 0,
TOXIC = 1,
SEVERE_TOXIC = 2,
OBSCENE = 3,
THREAT = 4,
INSULT = 5,
IDENTITY_HATE = 6
class YoutoxicLabels(Enum):
ALL = 0,
TOXIC = 1,
ABUSIVE = 2,
HATE_SPEECH = 3
class DataSets(Enum):
TRAIN = 0,
VALIDATION = 1
class Metrics(Enum):
ROC_AUC = 0,
F1 = 1
def calculate_mean_std(eval_scores, data_set, label, metric):
current_scores = eval_scores[:, data_set.value, :, label.value, metric.value]
if current_scores.ndim == 3:
best_epochs = current_scores.max(axis=2)
elif current_scores.ndim == 2:
best_epochs = current_scores.max(axis=1)
#print(best_epochs.shape)
return best_epochs.mean(), best_epochs.std()
def calculated_all_metrics(scores):
roc_all_mean, roc_all_std = calculate_mean_std(scores, DataSets.VALIDATION, Labels.ALL, Metrics.ROC_AUC)
roc_c1_mean, roc_c1_std = calculate_mean_std(scores, DataSets.VALIDATION, Labels.TOXIC, Metrics.ROC_AUC)
roc_c2_mean, roc_c2_std = calculate_mean_std(scores, DataSets.VALIDATION, Labels.SEVERE_TOXIC, Metrics.ROC_AUC)
roc_c3_mean, roc_c3_std = calculate_mean_std(scores, DataSets.VALIDATION, Labels.OBSCENE, Metrics.ROC_AUC)
roc_c4_mean, roc_c4_std = calculate_mean_std(scores, DataSets.VALIDATION, Labels.THREAT, Metrics.ROC_AUC)
roc_c5_mean, roc_c5_std = calculate_mean_std(scores, DataSets.VALIDATION, Labels.INSULT, Metrics.ROC_AUC)
roc_c6_mean, roc_c6_std = calculate_mean_std(scores, DataSets.VALIDATION, Labels.IDENTITY_HATE, Metrics.ROC_AUC)
f1_all_mean, f1_all_std = calculate_mean_std(scores, DataSets.VALIDATION, Labels.ALL, Metrics.F1)
f1_c1_mean, f1_c1_std = calculate_mean_std(scores, DataSets.VALIDATION, Labels.TOXIC, Metrics.F1)
f1_c2_mean, f1_c2_std = calculate_mean_std(scores, DataSets.VALIDATION, Labels.SEVERE_TOXIC, Metrics.F1)
f1_c3_mean, f1_c3_std = calculate_mean_std(scores, DataSets.VALIDATION, Labels.OBSCENE, Metrics.F1)
f1_c4_mean, f1_c4_std = calculate_mean_std(scores, DataSets.VALIDATION, Labels.THREAT, Metrics.F1)
f1_c5_mean, f1_c5_std = calculate_mean_std(scores, DataSets.VALIDATION, Labels.INSULT, Metrics.F1)
f1_c6_mean, f1_c6_std = calculate_mean_std(scores, DataSets.VALIDATION, Labels.IDENTITY_HATE, Metrics.F1)
print("ROC AUC over 5 runs:")
print("{:20s} {:.4f} ±{:.4f}".format('All labels:', roc_all_mean, roc_all_std))
print("{:20s} {:.4f} ±{:.4f}".format('Toxic:', roc_c1_mean, roc_c1_std))
print("{:20s} {:.4f} ±{:.4f}".format('Severe toxic:', roc_c2_mean, roc_c2_std))
print("{:20s} {:.4f} ±{:.4f}".format('Obscene:', roc_c3_mean, roc_c3_std))
print("{:20s} {:.4f} ±{:.4f}".format('Threat:', roc_c4_mean, roc_c4_std))
print("{:20s} {:.4f} ±{:.4f}".format('Insult:', roc_c5_mean, roc_c5_std))
print("{:20s} {:.4f} ±{:.4f}".format('Identity hate:', roc_c6_mean, roc_c6_std))
print("F1 over 5 runs:")
print("{:20s} {:.4f} ±{:.4f}".format('All labels:', f1_all_mean, f1_all_std))
print("{:20s} {:.4f} ±{:.4f}".format('Toxic:', f1_c1_mean, f1_c1_std))
print("{:20s} {:.4f} ±{:.4f}".format('Severe toxic:', f1_c2_mean, f1_c2_std))
print("{:20s} {:.4f} ±{:.4f}".format('Obscene:', f1_c3_mean, f1_c3_std))
print("{:20s} {:.4f} ±{:.4f}".format('Threat:', f1_c4_mean, f1_c4_std))
print("{:20s} {:.4f} ±{:.4f}".format('Insult:', f1_c5_mean, f1_c5_std))
print("{:20s} {:.4f} ±{:.4f}".format('Identity hate:', f1_c6_mean, f1_c6_std))
def calculate_all_youtoxic_metrics(scores):
roc_all_mean, roc_all_std = calculate_mean_std(scores, DataSets.VALIDATION, YoutoxicLabels.ALL, Metrics.ROC_AUC)
roc_c1_mean, roc_c1_std = calculate_mean_std(scores, DataSets.VALIDATION, YoutoxicLabels.TOXIC, Metrics.ROC_AUC)
roc_c2_mean, roc_c2_std = calculate_mean_std(scores, DataSets.VALIDATION, YoutoxicLabels.ABUSIVE, Metrics.ROC_AUC)
roc_c3_mean, roc_c3_std = calculate_mean_std(scores, DataSets.VALIDATION, YoutoxicLabels.HATE_SPEECH, Metrics.ROC_AUC)
f1_all_mean, f1_all_std = calculate_mean_std(scores, DataSets.VALIDATION, YoutoxicLabels.ALL, Metrics.F1)
f1_c1_mean, f1_c1_std = calculate_mean_std(scores, DataSets.VALIDATION, YoutoxicLabels.TOXIC, Metrics.F1)
f1_c2_mean, f1_c2_std = calculate_mean_std(scores, DataSets.VALIDATION, YoutoxicLabels.ABUSIVE, Metrics.F1)
f1_c3_mean, f1_c3_std = calculate_mean_std(scores, DataSets.VALIDATION, YoutoxicLabels.HATE_SPEECH, Metrics.F1)
print("ROC AUC over 5 runs:")
print("{:20s} {:.4f} ±{:.4f}".format('All labels:', roc_all_mean, roc_all_std))
print("{:20s} {:.4f} ±{:.4f}".format('Toxic:', roc_c1_mean, roc_c1_std))
print("{:20s} {:.4f} ±{:.4f}".format('Severe toxic:', roc_c2_mean, roc_c2_std))
print("{:20s} {:.4f} ±{:.4f}".format('Obscene:', roc_c3_mean, roc_c3_std))
print("F1 over 5 runs:")
print("{:20s} {:.4f} ±{:.4f}".format('All labels:', f1_all_mean, f1_all_std))
print("{:20s} {:.4f} ±{:.4f}".format('Toxic:', f1_c1_mean, f1_c1_std))
print("{:20s} {:.4f} ±{:.4f}".format('Severe toxic:', f1_c2_mean, f1_c2_std))
print("{:20s} {:.4f} ±{:.4f}".format('Obscene:', f1_c3_mean, f1_c3_std))
def plot_learningcurve(scores, label, metric):
train_scores = scores[:, DataSets.TRAIN.value, :, label.value, metric.value].mean(axis=1)
val_scores = scores[:, DataSets.VALIDATION.value, :, label.value, metric.value].mean(axis=1)
fig = plt.figure()
ax = fig.add_subplot(111)
ax.xaxis.set_major_locator(MaxNLocator(integer=True))
ax.plot(range(1,len(val_scores[0])+1),val_scores[0])
ax.plot(range(1,len(val_scores[0])+1),train_scores[0])
fig.show()
```
# CNN Architecture Experiments
### Singlelayer CNN
```
ex1_scores = np.load('data/scores/cnn_simple/scores_1543539380.5547197.npy')
ex1_scores.shape
calculated_all_metrics(ex1_scores)
plot_learningcurve(ex1_scores, Labels.ALL, Metrics.ROC_AUC)
```
### Singlelayer CNN with multiple window sizes
```
ex2_scores = np.load('data/scores/cnn_multiwindowsizes/scores_1543641482.1065862.npy')
ex2_scores.shape
calculated_all_metrics(ex2_scores)
plot_learningcurve(ex2_scores, Labels.ALL, Metrics.ROC_AUC)
```
### Multilayer CNN
```
ex3_scores = np.load('data/scores/cnn_multilayer/scores_1543686279.5532448.npy')
ex3_scores.shape
calculated_all_metrics(ex3_scores)
plot_learningcurve(ex3_scores, Labels.ALL, Metrics.ROC_AUC)
```
### Dilated CNN
```
ex4_scores = np.load('data/scores/cnn_dilated/scores_1545399367.0580235.npy')
ex4_scores.shape
calculated_all_metrics(ex4_scores)
plot_learningcurve(ex4_scores, Labels.ALL, Metrics.ROC_AUC)
```
# Preprocessing comparison
### Baseline
```
ep1_scores = np.load('data/scores/preprocessing/e1_scores_1544101761.8759387.npy')
ep1_scores.shape
calculated_all_metrics(ep1_scores)
plot_learningcurve(ep1_scores, Labels.ALL, Metrics.ROC_AUC)
```
### Strip more than 3 of the same characters in a row
```
ep2_scores = np.load('data/scores/preprocessing/e2_scores_1544182966.0022054.npy')
ep2_scores.shape
calculated_all_metrics(ep2_scores)
plot_learningcurve(ep2_scores, Labels.ALL, Metrics.ROC_AUC)
```
### Remove all punctuation
```
ep3_scores = np.load('data/scores/preprocessing/e3_scores_1545309175.410995.npy')
ep3_scores.shape
calculated_all_metrics(ep3_scores)
plot_learningcurve(ep3_scores, Labels.ALL, Metrics.ROC_AUC)
```
### Remove all punctuation except for .,!?
```
ep4_scores = np.load('data/scores/preprocessing/e4_scores_1545326105.8022852.npy')
ep4_scores.shape
calculated_all_metrics(ep4_scores)
plot_learningcurve(ep4_scores, Labels.ALL, Metrics.ROC_AUC)
```
### Stemming
```
ep5_scores = np.load('data/scores/preprocessing/e5_scores_1544714914.6546211.npy')
ep5_scores.shape
calculated_all_metrics(ep5_scores)
plot_learningcurve(ep5_scores, Labels.ALL, Metrics.ROC_AUC)
```
### Lemmatization
```
ep6_scores = np.load('data/scores/preprocessing/e6_scores_1544744071.8302839.npy')
ep6_scores.shape
calculated_all_metrics(ep6_scores)
plot_learningcurve(ep6_scores, Labels.ALL, Metrics.ROC_AUC)
```
### Replace unknown tokens
```
ep7_scores = np.load('data/scores/preprocessing/e7_scores_1544790681.2920856.npy')
ep7_scores.shape
calculated_all_metrics(ep7_scores)
plot_learningcurve(ep7_scores, Labels.ALL, Metrics.ROC_AUC)
```
### Replace punctuation except for .,!? + Lemmatization
```
ep8_scores = np.load('data/scores/preprocessing/e8_scores_1547583857.9355667.npy')
ep8_scores.shape
calculated_all_metrics(ep8_scores)
plot_learningcurve(ep8_scores, Labels.ALL, Metrics.ROC_AUC)
```
### Replace punctuation except for .,!? + Stemming
```
ep9_scores = np.load('data/scores/preprocessing/e9_scores_1547548853.502363.npy')
ep9_scores.shape
calculated_all_metrics(ep9_scores)
plot_learningcurve(ep9_scores, Labels.ALL, Metrics.ROC_AUC)
```
### Replace punctuation + Stemming
```
ep10_scores = np.load('data/scores/preprocessing/e10_scores_1547600264.255076.npy')
ep10_scores.shape
calculated_all_metrics(ep10_scores)
plot_learningcurve(ep10_scores, Labels.ALL, Metrics.ROC_AUC)
```
### Replace punctuation + lemmatization
```
ep11_scores = np.load('data/scores/preprocessing/e11_scores_1547637074.9931095.npy')
ep11_scores.shape
calculated_all_metrics(ep11_scores)
plot_learningcurve(ep11_scores, Labels.ALL, Metrics.ROC_AUC)
```
### Replace unknown tokens + lemmatization
```
ep12_scores = np.load('data/scores/preprocessing/e12_scores_1547834804.8858764.npy')
ep12_scores.shape
calculated_all_metrics(ep12_scores)
plot_learningcurve(ep12_scores, Labels.ALL, Metrics.ROC_AUC)
```
# YouToxic experiments
### Youtoxic with transfer learning
```
eyt1_scores = np.load('data/scores/youtoxic/youtoxic_1546250048.7388098.npy')
eyt1_scores.shape
calculate_all_youtoxic_metrics(eyt1_scores)
plot_learningcurve(eyt1_scores, YoutoxicLabels.ALL, Metrics.ROC_AUC)
```
### YouToxic from scratch
```
eyt2_scores = np.load('data/scores/youtoxic/youtoxic_fromscratch_1547712976.138965.npy')
eyt2_scores.shape
calculate_all_youtoxic_metrics(eyt2_scores)
plot_learningcurve(eyt2_scores, YoutoxicLabels.ALL, Metrics.ROC_AUC)
```
### YouToxic transfer learning with smaller learning rate
```
eyt3_scores = np.load('data/scores/youtoxic/youtoxic_smalllr_1548095533.3741865.npy')
eyt3_scores.shape
calculate_all_youtoxic_metrics(eyt3_scores)
plot_learningcurve(eyt3_scores, YoutoxicLabels.ALL, Metrics.ROC_AUC)
```
|
github_jupyter
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from matplotlib.ticker import MaxNLocator
from enum import Enum
%matplotlib inline
class Labels(Enum):
ALL = 0,
TOXIC = 1,
SEVERE_TOXIC = 2,
OBSCENE = 3,
THREAT = 4,
INSULT = 5,
IDENTITY_HATE = 6
class YoutoxicLabels(Enum):
ALL = 0,
TOXIC = 1,
ABUSIVE = 2,
HATE_SPEECH = 3
class DataSets(Enum):
TRAIN = 0,
VALIDATION = 1
class Metrics(Enum):
ROC_AUC = 0,
F1 = 1
def calculate_mean_std(eval_scores, data_set, label, metric):
current_scores = eval_scores[:, data_set.value, :, label.value, metric.value]
if current_scores.ndim == 3:
best_epochs = current_scores.max(axis=2)
elif current_scores.ndim == 2:
best_epochs = current_scores.max(axis=1)
#print(best_epochs.shape)
return best_epochs.mean(), best_epochs.std()
def calculated_all_metrics(scores):
roc_all_mean, roc_all_std = calculate_mean_std(scores, DataSets.VALIDATION, Labels.ALL, Metrics.ROC_AUC)
roc_c1_mean, roc_c1_std = calculate_mean_std(scores, DataSets.VALIDATION, Labels.TOXIC, Metrics.ROC_AUC)
roc_c2_mean, roc_c2_std = calculate_mean_std(scores, DataSets.VALIDATION, Labels.SEVERE_TOXIC, Metrics.ROC_AUC)
roc_c3_mean, roc_c3_std = calculate_mean_std(scores, DataSets.VALIDATION, Labels.OBSCENE, Metrics.ROC_AUC)
roc_c4_mean, roc_c4_std = calculate_mean_std(scores, DataSets.VALIDATION, Labels.THREAT, Metrics.ROC_AUC)
roc_c5_mean, roc_c5_std = calculate_mean_std(scores, DataSets.VALIDATION, Labels.INSULT, Metrics.ROC_AUC)
roc_c6_mean, roc_c6_std = calculate_mean_std(scores, DataSets.VALIDATION, Labels.IDENTITY_HATE, Metrics.ROC_AUC)
f1_all_mean, f1_all_std = calculate_mean_std(scores, DataSets.VALIDATION, Labels.ALL, Metrics.F1)
f1_c1_mean, f1_c1_std = calculate_mean_std(scores, DataSets.VALIDATION, Labels.TOXIC, Metrics.F1)
f1_c2_mean, f1_c2_std = calculate_mean_std(scores, DataSets.VALIDATION, Labels.SEVERE_TOXIC, Metrics.F1)
f1_c3_mean, f1_c3_std = calculate_mean_std(scores, DataSets.VALIDATION, Labels.OBSCENE, Metrics.F1)
f1_c4_mean, f1_c4_std = calculate_mean_std(scores, DataSets.VALIDATION, Labels.THREAT, Metrics.F1)
f1_c5_mean, f1_c5_std = calculate_mean_std(scores, DataSets.VALIDATION, Labels.INSULT, Metrics.F1)
f1_c6_mean, f1_c6_std = calculate_mean_std(scores, DataSets.VALIDATION, Labels.IDENTITY_HATE, Metrics.F1)
print("ROC AUC over 5 runs:")
print("{:20s} {:.4f} ±{:.4f}".format('All labels:', roc_all_mean, roc_all_std))
print("{:20s} {:.4f} ±{:.4f}".format('Toxic:', roc_c1_mean, roc_c1_std))
print("{:20s} {:.4f} ±{:.4f}".format('Severe toxic:', roc_c2_mean, roc_c2_std))
print("{:20s} {:.4f} ±{:.4f}".format('Obscene:', roc_c3_mean, roc_c3_std))
print("{:20s} {:.4f} ±{:.4f}".format('Threat:', roc_c4_mean, roc_c4_std))
print("{:20s} {:.4f} ±{:.4f}".format('Insult:', roc_c5_mean, roc_c5_std))
print("{:20s} {:.4f} ±{:.4f}".format('Identity hate:', roc_c6_mean, roc_c6_std))
print("F1 over 5 runs:")
print("{:20s} {:.4f} ±{:.4f}".format('All labels:', f1_all_mean, f1_all_std))
print("{:20s} {:.4f} ±{:.4f}".format('Toxic:', f1_c1_mean, f1_c1_std))
print("{:20s} {:.4f} ±{:.4f}".format('Severe toxic:', f1_c2_mean, f1_c2_std))
print("{:20s} {:.4f} ±{:.4f}".format('Obscene:', f1_c3_mean, f1_c3_std))
print("{:20s} {:.4f} ±{:.4f}".format('Threat:', f1_c4_mean, f1_c4_std))
print("{:20s} {:.4f} ±{:.4f}".format('Insult:', f1_c5_mean, f1_c5_std))
print("{:20s} {:.4f} ±{:.4f}".format('Identity hate:', f1_c6_mean, f1_c6_std))
def calculate_all_youtoxic_metrics(scores):
roc_all_mean, roc_all_std = calculate_mean_std(scores, DataSets.VALIDATION, YoutoxicLabels.ALL, Metrics.ROC_AUC)
roc_c1_mean, roc_c1_std = calculate_mean_std(scores, DataSets.VALIDATION, YoutoxicLabels.TOXIC, Metrics.ROC_AUC)
roc_c2_mean, roc_c2_std = calculate_mean_std(scores, DataSets.VALIDATION, YoutoxicLabels.ABUSIVE, Metrics.ROC_AUC)
roc_c3_mean, roc_c3_std = calculate_mean_std(scores, DataSets.VALIDATION, YoutoxicLabels.HATE_SPEECH, Metrics.ROC_AUC)
f1_all_mean, f1_all_std = calculate_mean_std(scores, DataSets.VALIDATION, YoutoxicLabels.ALL, Metrics.F1)
f1_c1_mean, f1_c1_std = calculate_mean_std(scores, DataSets.VALIDATION, YoutoxicLabels.TOXIC, Metrics.F1)
f1_c2_mean, f1_c2_std = calculate_mean_std(scores, DataSets.VALIDATION, YoutoxicLabels.ABUSIVE, Metrics.F1)
f1_c3_mean, f1_c3_std = calculate_mean_std(scores, DataSets.VALIDATION, YoutoxicLabels.HATE_SPEECH, Metrics.F1)
print("ROC AUC over 5 runs:")
print("{:20s} {:.4f} ±{:.4f}".format('All labels:', roc_all_mean, roc_all_std))
print("{:20s} {:.4f} ±{:.4f}".format('Toxic:', roc_c1_mean, roc_c1_std))
print("{:20s} {:.4f} ±{:.4f}".format('Severe toxic:', roc_c2_mean, roc_c2_std))
print("{:20s} {:.4f} ±{:.4f}".format('Obscene:', roc_c3_mean, roc_c3_std))
print("F1 over 5 runs:")
print("{:20s} {:.4f} ±{:.4f}".format('All labels:', f1_all_mean, f1_all_std))
print("{:20s} {:.4f} ±{:.4f}".format('Toxic:', f1_c1_mean, f1_c1_std))
print("{:20s} {:.4f} ±{:.4f}".format('Severe toxic:', f1_c2_mean, f1_c2_std))
print("{:20s} {:.4f} ±{:.4f}".format('Obscene:', f1_c3_mean, f1_c3_std))
def plot_learningcurve(scores, label, metric):
train_scores = scores[:, DataSets.TRAIN.value, :, label.value, metric.value].mean(axis=1)
val_scores = scores[:, DataSets.VALIDATION.value, :, label.value, metric.value].mean(axis=1)
fig = plt.figure()
ax = fig.add_subplot(111)
ax.xaxis.set_major_locator(MaxNLocator(integer=True))
ax.plot(range(1,len(val_scores[0])+1),val_scores[0])
ax.plot(range(1,len(val_scores[0])+1),train_scores[0])
fig.show()
ex1_scores = np.load('data/scores/cnn_simple/scores_1543539380.5547197.npy')
ex1_scores.shape
calculated_all_metrics(ex1_scores)
plot_learningcurve(ex1_scores, Labels.ALL, Metrics.ROC_AUC)
ex2_scores = np.load('data/scores/cnn_multiwindowsizes/scores_1543641482.1065862.npy')
ex2_scores.shape
calculated_all_metrics(ex2_scores)
plot_learningcurve(ex2_scores, Labels.ALL, Metrics.ROC_AUC)
ex3_scores = np.load('data/scores/cnn_multilayer/scores_1543686279.5532448.npy')
ex3_scores.shape
calculated_all_metrics(ex3_scores)
plot_learningcurve(ex3_scores, Labels.ALL, Metrics.ROC_AUC)
ex4_scores = np.load('data/scores/cnn_dilated/scores_1545399367.0580235.npy')
ex4_scores.shape
calculated_all_metrics(ex4_scores)
plot_learningcurve(ex4_scores, Labels.ALL, Metrics.ROC_AUC)
ep1_scores = np.load('data/scores/preprocessing/e1_scores_1544101761.8759387.npy')
ep1_scores.shape
calculated_all_metrics(ep1_scores)
plot_learningcurve(ep1_scores, Labels.ALL, Metrics.ROC_AUC)
ep2_scores = np.load('data/scores/preprocessing/e2_scores_1544182966.0022054.npy')
ep2_scores.shape
calculated_all_metrics(ep2_scores)
plot_learningcurve(ep2_scores, Labels.ALL, Metrics.ROC_AUC)
ep3_scores = np.load('data/scores/preprocessing/e3_scores_1545309175.410995.npy')
ep3_scores.shape
calculated_all_metrics(ep3_scores)
plot_learningcurve(ep3_scores, Labels.ALL, Metrics.ROC_AUC)
ep4_scores = np.load('data/scores/preprocessing/e4_scores_1545326105.8022852.npy')
ep4_scores.shape
calculated_all_metrics(ep4_scores)
plot_learningcurve(ep4_scores, Labels.ALL, Metrics.ROC_AUC)
ep5_scores = np.load('data/scores/preprocessing/e5_scores_1544714914.6546211.npy')
ep5_scores.shape
calculated_all_metrics(ep5_scores)
plot_learningcurve(ep5_scores, Labels.ALL, Metrics.ROC_AUC)
ep6_scores = np.load('data/scores/preprocessing/e6_scores_1544744071.8302839.npy')
ep6_scores.shape
calculated_all_metrics(ep6_scores)
plot_learningcurve(ep6_scores, Labels.ALL, Metrics.ROC_AUC)
ep7_scores = np.load('data/scores/preprocessing/e7_scores_1544790681.2920856.npy')
ep7_scores.shape
calculated_all_metrics(ep7_scores)
plot_learningcurve(ep7_scores, Labels.ALL, Metrics.ROC_AUC)
ep8_scores = np.load('data/scores/preprocessing/e8_scores_1547583857.9355667.npy')
ep8_scores.shape
calculated_all_metrics(ep8_scores)
plot_learningcurve(ep8_scores, Labels.ALL, Metrics.ROC_AUC)
ep9_scores = np.load('data/scores/preprocessing/e9_scores_1547548853.502363.npy')
ep9_scores.shape
calculated_all_metrics(ep9_scores)
plot_learningcurve(ep9_scores, Labels.ALL, Metrics.ROC_AUC)
ep10_scores = np.load('data/scores/preprocessing/e10_scores_1547600264.255076.npy')
ep10_scores.shape
calculated_all_metrics(ep10_scores)
plot_learningcurve(ep10_scores, Labels.ALL, Metrics.ROC_AUC)
ep11_scores = np.load('data/scores/preprocessing/e11_scores_1547637074.9931095.npy')
ep11_scores.shape
calculated_all_metrics(ep11_scores)
plot_learningcurve(ep11_scores, Labels.ALL, Metrics.ROC_AUC)
ep12_scores = np.load('data/scores/preprocessing/e12_scores_1547834804.8858764.npy')
ep12_scores.shape
calculated_all_metrics(ep12_scores)
plot_learningcurve(ep12_scores, Labels.ALL, Metrics.ROC_AUC)
eyt1_scores = np.load('data/scores/youtoxic/youtoxic_1546250048.7388098.npy')
eyt1_scores.shape
calculate_all_youtoxic_metrics(eyt1_scores)
plot_learningcurve(eyt1_scores, YoutoxicLabels.ALL, Metrics.ROC_AUC)
eyt2_scores = np.load('data/scores/youtoxic/youtoxic_fromscratch_1547712976.138965.npy')
eyt2_scores.shape
calculate_all_youtoxic_metrics(eyt2_scores)
plot_learningcurve(eyt2_scores, YoutoxicLabels.ALL, Metrics.ROC_AUC)
eyt3_scores = np.load('data/scores/youtoxic/youtoxic_smalllr_1548095533.3741865.npy')
eyt3_scores.shape
calculate_all_youtoxic_metrics(eyt3_scores)
plot_learningcurve(eyt3_scores, YoutoxicLabels.ALL, Metrics.ROC_AUC)
| 0.629319 | 0.486758 |
# Backpropagation
## Instructions
In this assignment, you will train a neural network to draw a curve.
The curve takes one input variable, the amount travelled along the curve from 0 to 1, and returns 2 outputs, the 2D coordinates of the position of points on the curve.
To help capture the complexity of the curve, we shall use two hidden layers in our network with 6 and 7 neurons respectively.

You will be asked to complete functions that calculate the Jacobian of the cost function, with respect to the weights and biases of the network. Your code will form part of a stochastic steepest descent algorithm that will train your network.
### Matrices in Python
Recall from assignments in the previous course in this specialisation that matrices can be multiplied together in two ways.
Element wise: when two matrices have the same dimensions, matrix elements in the same position in each matrix are multiplied together
In python this uses the '$*$' operator.
```python
A = B * C
```
Matrix multiplication: when the number of columns in the first matrix is the same as the number of rows in the second.
In python this uses the '$@$' operator
```python
A = B @ C
```
This assignment will not test which ones to use where, but it will use both in the starter code presented to you.
There is no need to change these or worry about their specifics.
### How to submit
To complete the assignment, edit the code in the cells below where you are told to do so.
Once you are finished and happy with it, press the **Submit Assignment** button at the top of this worksheet.
Test your code using the cells at the bottom of the notebook before you submit.
Please don't change any of the function names, as these will be checked by the grading script.
## Feed forward
In the following cell, we will define functions to set up our neural network.
Namely an activation function, $\sigma(z)$, it's derivative, $\sigma'(z)$, a function to initialise weights and biases, and a function that calculates each activation of the network using feed-forward.
Recall the feed-forward equations,
$$ \mathbf{a}^{(n)} = \sigma(\mathbf{z}^{(n)}) $$
$$ \mathbf{z}^{(n)} = \mathbf{W}^{(n)}\mathbf{a}^{(n-1)} + \mathbf{b}^{(n)} $$
In this worksheet we will use the *logistic function* as our activation function, rather than the more familiar $\tanh$.
$$ \sigma(\mathbf{z}) = \frac{1}{1 + \exp(-\mathbf{z})} $$
There is no need to edit the following cells.
They do not form part of the assessment.
You may wish to study how it works though.
**Run the following cells before continuing.**
```
%run "readonly/BackpropModule.ipynb"
# PACKAGE
import numpy as np
import matplotlib.pyplot as plt
# PACKAGE
# First load the worksheet dependencies.
# Here is the activation function and its derivative.
sigma = lambda z : 1 / (1 + np.exp(-z))
d_sigma = lambda z : np.cosh(z/2)**(-2) / 4
# This function initialises the network with it's structure, it also resets any training already done.
def reset_network (n1 = 6, n2 = 7, random=np.random) :
global W1, W2, W3, b1, b2, b3
W1 = random.randn(n1, 1) / 2
W2 = random.randn(n2, n1) / 2
W3 = random.randn(2, n2) / 2
b1 = random.randn(n1, 1) / 2
b2 = random.randn(n2, 1) / 2
b3 = random.randn(2, 1) / 2
# This function feeds forward each activation to the next layer. It returns all weighted sums and activations.
def network_function(a0) :
z1 = W1 @ a0 + b1
a1 = sigma(z1)
z2 = W2 @ a1 + b2
a2 = sigma(z2)
z3 = W3 @ a2 + b3
a3 = sigma(z3)
return a0, z1, a1, z2, a2, z3, a3
# This is the cost function of a neural network with respect to a training set.
def cost(x, y) :
return np.linalg.norm(network_function(x)[-1] - y)**2 / x.size
```
## Backpropagation
In the next cells, you will be asked to complete functions for the Jacobian of the cost function with respect to the weights and biases.
We will start with layer 3, which is the easiest, and work backwards through the layers.
We'll define our Jacobians as,
$$ \mathbf{J}_{\mathbf{W}^{(3)}} = \frac{\partial C}{\partial \mathbf{W}^{(3)}} $$
$$ \mathbf{J}_{\mathbf{b}^{(3)}} = \frac{\partial C}{\partial \mathbf{b}^{(3)}} $$
etc., where $C$ is the average cost function over the training set. i.e.,
$$ C = \frac{1}{N}\sum_k C_k $$
You calculated the following in the practice quizzes,
$$ \frac{\partial C}{\partial \mathbf{W}^{(3)}} =
\frac{\partial C}{\partial \mathbf{a}^{(3)}}
\frac{\partial \mathbf{a}^{(3)}}{\partial \mathbf{z}^{(3)}}
\frac{\partial \mathbf{z}^{(3)}}{\partial \mathbf{W}^{(3)}}
,$$
for the weight, and similarly for the bias,
$$ \frac{\partial C}{\partial \mathbf{b}^{(3)}} =
\frac{\partial C}{\partial \mathbf{a}^{(3)}}
\frac{\partial \mathbf{a}^{(3)}}{\partial \mathbf{z}^{(3)}}
\frac{\partial \mathbf{z}^{(3)}}{\partial \mathbf{b}^{(3)}}
.$$
With the partial derivatives taking the form,
$$ \frac{\partial C}{\partial \mathbf{a}^{(3)}} = 2(\mathbf{a}^{(3)} - \mathbf{y}) $$
$$ \frac{\partial \mathbf{a}^{(3)}}{\partial \mathbf{z}^{(3)}} = \sigma'({z}^{(3)})$$
$$ \frac{\partial \mathbf{z}^{(3)}}{\partial \mathbf{W}^{(3)}} = \mathbf{a}^{(2)}$$
$$ \frac{\partial \mathbf{z}^{(3)}}{\partial \mathbf{b}^{(3)}} = 1$$
We'll do the J_W3 ($\mathbf{J}_{\mathbf{W}^{(3)}}$) function for you, so you can see how it works.
You should then be able to adapt the J_b3 function, with help, yourself.
```
# GRADED FUNCTION
# Jacobian for the third layer weights. There is no need to edit this function.
def J_W3 (x, y) :
# First get all the activations and weighted sums at each layer of the network.
a0, z1, a1, z2, a2, z3, a3 = network_function(x)
# We'll use the variable J to store parts of our result as we go along, updating it in each line.
# Firstly, we calculate dC/da3, using the expressions above.
J = 2 * (a3 - y)
# Next multiply the result we've calculated by the derivative of sigma, evaluated at z3.
J = J * d_sigma(z3)
# Then we take the dot product (along the axis that holds the training examples) with the final partial derivative,
# i.e. dz3/dW3 = a2
# and divide by the number of training examples, for the average over all training examples.
J = J @ a2.T / x.size
# Finally return the result out of the function.
return J
# In this function, you will implement the jacobian for the bias.
# As you will see from the partial derivatives, only the last partial derivative is different.
# The first two partial derivatives are the same as previously.
# ===YOU SHOULD EDIT THIS FUNCTION===
def J_b3 (x, y) :
# As last time, we'll first set up the activations.
a0, z1, a1, z2, a2, z3, a3 = network_function(x)
# Next you should implement the first two partial derivatives of the Jacobian.
# ===COPY TWO LINES FROM THE PREVIOUS FUNCTION TO SET UP THE FIRST TWO JACOBIAN TERMS===
J = 2 * (a3 - y)
J = J * d_sigma(z3)
# For the final line, we don't need to multiply by dz3/db3, because that is multiplying by 1.
# We still need to sum over all training examples however.
# There is no need to edit this line.
J = np.sum(J, axis=1, keepdims=True) / x.size
return J
```
We'll next do the Jacobian for the Layer 2. The partial derivatives for this are,
$$ \frac{\partial C}{\partial \mathbf{W}^{(2)}} =
\frac{\partial C}{\partial \mathbf{a}^{(3)}}
\left(
\frac{\partial \mathbf{a}^{(3)}}{\partial \mathbf{a}^{(2)}}
\right)
\frac{\partial \mathbf{a}^{(2)}}{\partial \mathbf{z}^{(2)}}
\frac{\partial \mathbf{z}^{(2)}}{\partial \mathbf{W}^{(2)}}
,$$
$$ \frac{\partial C}{\partial \mathbf{b}^{(2)}} =
\frac{\partial C}{\partial \mathbf{a}^{(3)}}
\left(
\frac{\partial \mathbf{a}^{(3)}}{\partial \mathbf{a}^{(2)}}
\right)
\frac{\partial \mathbf{a}^{(2)}}{\partial \mathbf{z}^{(2)}}
\frac{\partial \mathbf{z}^{(2)}}{\partial \mathbf{b}^{(2)}}
.$$
This is very similar to the previous layer, with two exceptions:
* There is a new partial derivative, in parentheses, $\frac{\partial \mathbf{a}^{(3)}}{\partial \mathbf{a}^{(2)}}$
* The terms after the parentheses are now one layer lower.
Recall the new partial derivative takes the following form,
$$ \frac{\partial \mathbf{a}^{(3)}}{\partial \mathbf{a}^{(2)}} =
\frac{\partial \mathbf{a}^{(3)}}{\partial \mathbf{z}^{(3)}}
\frac{\partial \mathbf{z}^{(3)}}{\partial \mathbf{a}^{(2)}} =
\sigma'(\mathbf{z}^{(3)})
\mathbf{W}^{(3)}
$$
To show how this changes things, we will implement the Jacobian for the weight again and ask you to implement it for the bias.
```
# GRADED FUNCTION
# Compare this function to J_W3 to see how it changes.
# There is no need to edit this function.
def J_W2 (x, y) :
#The first two lines are identical to in J_W3.
a0, z1, a1, z2, a2, z3, a3 = network_function(x)
J = 2 * (a3 - y)
# the next two lines implement da3/da2, first σ' and then W3.
J = J * d_sigma(z3)
J = (J.T @ W3).T
# then the final lines are the same as in J_W3 but with the layer number bumped down.
J = J * d_sigma(z2)
J = J @ a1.T / x.size
return J
# As previously, fill in all the incomplete lines.
# ===YOU SHOULD EDIT THIS FUNCTION===
def J_b2 (x, y) :
a0, z1, a1, z2, a2, z3, a3 = network_function(x)
J = 2 * (a3 - y)
J = J * d_sigma(z3)
J = (J.T @ W3).T
J = J * d_sigma(z2)
J = np.sum(J, axis=1, keepdims=True) / x.size
return J
```
Layer 1 is very similar to Layer 2, but with an addition partial derivative term.
$$ \frac{\partial C}{\partial \mathbf{W}^{(1)}} =
\frac{\partial C}{\partial \mathbf{a}^{(3)}}
\left(
\frac{\partial \mathbf{a}^{(3)}}{\partial \mathbf{a}^{(2)}}
\frac{\partial \mathbf{a}^{(2)}}{\partial \mathbf{a}^{(1)}}
\right)
\frac{\partial \mathbf{a}^{(1)}}{\partial \mathbf{z}^{(1)}}
\frac{\partial \mathbf{z}^{(1)}}{\partial \mathbf{W}^{(1)}}
,$$
$$ \frac{\partial C}{\partial \mathbf{b}^{(1)}} =
\frac{\partial C}{\partial \mathbf{a}^{(3)}}
\left(
\frac{\partial \mathbf{a}^{(3)}}{\partial \mathbf{a}^{(2)}}
\frac{\partial \mathbf{a}^{(2)}}{\partial \mathbf{a}^{(1)}}
\right)
\frac{\partial \mathbf{a}^{(1)}}{\partial \mathbf{z}^{(1)}}
\frac{\partial \mathbf{z}^{(1)}}{\partial \mathbf{b}^{(1)}}
.$$
You should be able to adapt lines from the previous cells to complete **both** the weight and bias Jacobian.
```
# GRADED FUNCTION
# Fill in all incomplete lines.
# ===YOU SHOULD EDIT THIS FUNCTION===
def J_W1 (x, y) :
a0, z1, a1, z2, a2, z3, a3 = network_function(x)
J = 2 * (a3 - y)
J = J * d_sigma(z3)
J = (J.T @ W3).T
J = J * d_sigma(z2)
J = (J.T @ W2).T
J = J * d_sigma(z1)
J = J @ a0.T / x.size
return J
# Fill in all incomplete lines.
# ===YOU SHOULD EDIT THIS FUNCTION===
def J_b1 (x, y) :
a0, z1, a1, z2, a2, z3, a3 = network_function(x)
J = 2 * (a3 - y)
J = J * d_sigma(z3)
J = (J.T @ W3).T
J = J * d_sigma(z2)
J = (J.T @ W2).T
J = J * d_sigma(z1)
J = J*1
J = np.sum(J, axis=1, keepdims=True) / x.size
return J
```
## Test your code before submission
To test the code you've written above, run all previous cells (select each cell, then press the play button [ ▶| ] or press shift-enter).
You can then use the code below to test out your function.
You don't need to submit these cells; you can edit and run them as much as you like.
First, we generate training data, and generate a network with randomly assigned weights and biases.
```
x, y = training_data()
reset_network()
```
Next, if you've implemented the assignment correctly, the following code will iterate through a steepest descent algorithm using the Jacobians you have calculated.
The function will plot the training data (in green), and your neural network solutions in pink for each iteration, and orange for the last output.
It takes about 50,000 iterations to train this network.
We can split this up though - **10,000 iterations should take about a minute to run**.
Run the line below as many times as you like.
```
plot_training(x, y, iterations=20000, aggression=7, noise=1)
```
If you wish, you can change parameters of the steepest descent algorithm (We'll go into more details in future exercises), but you can change how many iterations are plotted, how agressive the step down the Jacobian is, and how much noise to add.
You can also edit the parameters of the neural network, i.e. to give it different amounts of neurons in the hidden layers by calling,
```python
reset_network(n1, n2)
```
Play around with the parameters, and save your favourite result for the discussion prompt - *I ❤️ backpropagation*.
|
github_jupyter
|
A = B * C
A = B @ C
%run "readonly/BackpropModule.ipynb"
# PACKAGE
import numpy as np
import matplotlib.pyplot as plt
# PACKAGE
# First load the worksheet dependencies.
# Here is the activation function and its derivative.
sigma = lambda z : 1 / (1 + np.exp(-z))
d_sigma = lambda z : np.cosh(z/2)**(-2) / 4
# This function initialises the network with it's structure, it also resets any training already done.
def reset_network (n1 = 6, n2 = 7, random=np.random) :
global W1, W2, W3, b1, b2, b3
W1 = random.randn(n1, 1) / 2
W2 = random.randn(n2, n1) / 2
W3 = random.randn(2, n2) / 2
b1 = random.randn(n1, 1) / 2
b2 = random.randn(n2, 1) / 2
b3 = random.randn(2, 1) / 2
# This function feeds forward each activation to the next layer. It returns all weighted sums and activations.
def network_function(a0) :
z1 = W1 @ a0 + b1
a1 = sigma(z1)
z2 = W2 @ a1 + b2
a2 = sigma(z2)
z3 = W3 @ a2 + b3
a3 = sigma(z3)
return a0, z1, a1, z2, a2, z3, a3
# This is the cost function of a neural network with respect to a training set.
def cost(x, y) :
return np.linalg.norm(network_function(x)[-1] - y)**2 / x.size
# GRADED FUNCTION
# Jacobian for the third layer weights. There is no need to edit this function.
def J_W3 (x, y) :
# First get all the activations and weighted sums at each layer of the network.
a0, z1, a1, z2, a2, z3, a3 = network_function(x)
# We'll use the variable J to store parts of our result as we go along, updating it in each line.
# Firstly, we calculate dC/da3, using the expressions above.
J = 2 * (a3 - y)
# Next multiply the result we've calculated by the derivative of sigma, evaluated at z3.
J = J * d_sigma(z3)
# Then we take the dot product (along the axis that holds the training examples) with the final partial derivative,
# i.e. dz3/dW3 = a2
# and divide by the number of training examples, for the average over all training examples.
J = J @ a2.T / x.size
# Finally return the result out of the function.
return J
# In this function, you will implement the jacobian for the bias.
# As you will see from the partial derivatives, only the last partial derivative is different.
# The first two partial derivatives are the same as previously.
# ===YOU SHOULD EDIT THIS FUNCTION===
def J_b3 (x, y) :
# As last time, we'll first set up the activations.
a0, z1, a1, z2, a2, z3, a3 = network_function(x)
# Next you should implement the first two partial derivatives of the Jacobian.
# ===COPY TWO LINES FROM THE PREVIOUS FUNCTION TO SET UP THE FIRST TWO JACOBIAN TERMS===
J = 2 * (a3 - y)
J = J * d_sigma(z3)
# For the final line, we don't need to multiply by dz3/db3, because that is multiplying by 1.
# We still need to sum over all training examples however.
# There is no need to edit this line.
J = np.sum(J, axis=1, keepdims=True) / x.size
return J
# GRADED FUNCTION
# Compare this function to J_W3 to see how it changes.
# There is no need to edit this function.
def J_W2 (x, y) :
#The first two lines are identical to in J_W3.
a0, z1, a1, z2, a2, z3, a3 = network_function(x)
J = 2 * (a3 - y)
# the next two lines implement da3/da2, first σ' and then W3.
J = J * d_sigma(z3)
J = (J.T @ W3).T
# then the final lines are the same as in J_W3 but with the layer number bumped down.
J = J * d_sigma(z2)
J = J @ a1.T / x.size
return J
# As previously, fill in all the incomplete lines.
# ===YOU SHOULD EDIT THIS FUNCTION===
def J_b2 (x, y) :
a0, z1, a1, z2, a2, z3, a3 = network_function(x)
J = 2 * (a3 - y)
J = J * d_sigma(z3)
J = (J.T @ W3).T
J = J * d_sigma(z2)
J = np.sum(J, axis=1, keepdims=True) / x.size
return J
# GRADED FUNCTION
# Fill in all incomplete lines.
# ===YOU SHOULD EDIT THIS FUNCTION===
def J_W1 (x, y) :
a0, z1, a1, z2, a2, z3, a3 = network_function(x)
J = 2 * (a3 - y)
J = J * d_sigma(z3)
J = (J.T @ W3).T
J = J * d_sigma(z2)
J = (J.T @ W2).T
J = J * d_sigma(z1)
J = J @ a0.T / x.size
return J
# Fill in all incomplete lines.
# ===YOU SHOULD EDIT THIS FUNCTION===
def J_b1 (x, y) :
a0, z1, a1, z2, a2, z3, a3 = network_function(x)
J = 2 * (a3 - y)
J = J * d_sigma(z3)
J = (J.T @ W3).T
J = J * d_sigma(z2)
J = (J.T @ W2).T
J = J * d_sigma(z1)
J = J*1
J = np.sum(J, axis=1, keepdims=True) / x.size
return J
x, y = training_data()
reset_network()
plot_training(x, y, iterations=20000, aggression=7, noise=1)
reset_network(n1, n2)
| 0.708213 | 0.993667 |
# Recursion
<p>
A recursive method is a method that calls itself. An iterative method is
a method that uses a loop to repeat an action. Anything that can be done
iteratively can be done recursively, and vice versa. Iterative algorithms
and methods are generally more efficient than recursive algorithms.
</p>
<p>
Recursion is based on two key problem solving concepts: divide and conquer
and self-similarity. A recursive solution solves a problem by solving a
smaller instance of the same problem. It solves this new problem by solving
an even smaller instance of the same problem. Eventually, the new problem will
be so small that its solution will either be obvious or known. This solution
will lead to the solution of the original problem.
</p>
<p>
A recursive definition consists of two parts: a recursive part in which the
nth value is defined in terms of the (n-1)th value, and a non recursive
boundary case or base case which defines a limiting condition. An infinite
repetition will result if a recursive definition is not properly bounded.
In a recursive algorithm, each recursive call must make progress toward the
bound, or base case. A recursion parameter is a parameter whose value is
used to control the progress of the recursion towards its bound.
</p>
<p>
Function call and return in Python uses a last-in-first-out protocol. As
each method call is made, a representation of the method call is place on
the method call stack. When a method returns, its block is removed from the
top of the stack.
</p>
<p>
Use an iterative algorithm instead of a recursive algorithm
whenever efficiency and memory usage are important design factors. When all
other factors are equal, choose the algorithm (recursive or iterative) that
is easiest to understand, develop, and maintain.
</p>
## Calculatating Factorial
A simple way to calculate it is using a for loop
```
def fact(n):
fact = 1
for i in range(1,n+1):
fact = fact * i
return fact
# Calculate 10!
fact(10)
```
Here is an example of a **recursive method** that calculates the factorial
of **n**.
- The base case occurs when n is equal to 0.
- We know that 0! is equal to 1. Otherwise we use the relationship n! = n * ( n - 1 )!
```
def fact(n):
if(n == 0):
return 1
else:
return n * fact(n - 1)
# Calculate 10!
fact(10)
```
## Fibonacci series
In mathematics there are recurrence relations that are defined recursively.
A recurrence relation defines a term in a sequence as a function of one
or more previous terms. One of the most famous of such recurrence
sequences is the Fibonacci series.
https://en.wikipedia.org/wiki/Fibonacci_number
Other than the first two terms in
this series, every term is defined as the sum of the previous two terms:
<pre>
F(1) = 1
F(2) = 1
F(n) = F(n-1) + F(n-2) for n > 2
1, 1, 2, 3, 5, 8, 13, 21, 34, 55, ...
</pre>
Here is the Python code that generates this series:
```
def fibonacci(n):
"""Iterative Implementation of Fibonacci number"""
a,b = 0,1
for i in range(n):
a,b = b,a+b
return a
print(fibonacci(10))
def fib(n):
if ((n == 1) or (n == 2)):
return 1
else:
return fib(n - 1) + fib (n - 2)
# fib(10)
fib(10)
```
Even though the series is defined recursively, the above code is extremely
inefficient in determining the terms in a Fibonacci series **(why?)**.
An iterative solution works best in this case.
However, there are sorting algorithms that use recursion that are extremely
efficient in what they do. One example of such a sorting algorithm is
MergeSort. Let us say you have a list of numbers to sort.
Then this algorithm can be stated as follows: Divide the list in half. Sort one
half, sort the other half and then merge the two sorted halves. You keep
dividing each half until you are down to one item. That item is sorted!
You then merge that item with another single item and work backwards merging
sorted sub-lists until you have the complete list.
This concept is used in Divide and Conquer algorithms.
https://en.wikipedia.org/wiki/Divide-and-conquer_algorithm
|
github_jupyter
|
def fact(n):
fact = 1
for i in range(1,n+1):
fact = fact * i
return fact
# Calculate 10!
fact(10)
def fact(n):
if(n == 0):
return 1
else:
return n * fact(n - 1)
# Calculate 10!
fact(10)
def fibonacci(n):
"""Iterative Implementation of Fibonacci number"""
a,b = 0,1
for i in range(n):
a,b = b,a+b
return a
print(fibonacci(10))
def fib(n):
if ((n == 1) or (n == 2)):
return 1
else:
return fib(n - 1) + fib (n - 2)
# fib(10)
fib(10)
| 0.313525 | 0.947866 |
# 注意,需要预先创建以下目录和文件
`../glove/glove.6B.100d.txt`:下载地址:<http://nlp.stanford.edu/data/glove.6B.zip>
# import
0. 读取数据、矩阵计算、路径计算等基础库
--------
1. 正则去标点等+大小写
2. 空格分词+去停用词
3. 词形统一
3. tf-idf编码方式
--------
4. 模型:使用keras自己构建CNN
5. 分析:混淆矩阵、准确率预测、训练耗时
6. 保存模型
## 改进点
- GPU加速
- 其他模型
- 参数
- 结果显示
- 函数封装
```
import csv
import numpy as np
import nltk
import re
from nltk import word_tokenize, pos_tag
from nltk.corpus import stopwords, wordnet
from nltk.stem import WordNetLemmatizer
from sklearn.feature_extraction.text import TfidfVectorizer
from keras.preprocessing.text import Tokenizer
from keras.preprocessing.sequence import pad_sequences
from keras.utils import to_categorical
from keras.layers import Dense, Input, GlobalMaxPooling1D
from keras.layers import Conv1D, MaxPooling1D, Embedding
from keras.models import Model
import time
```
# 读取数据集
使用csv读取文本文件,得到二维列表
```
"""获取原始数据"""
file_path = '../smsspamcollection/SMSSpamCollection'
smsFile = open(file_path, 'r', encoding='utf-8') ## 返回文件对象
sms = csv.reader(smsFile, delimiter='\t') ## 第一层:行列表;第二层:列列表
sms = list(sms)
smsFile.close()
"""显示原始数据"""
for line in sms[0:3]:
print(line)
```
# 预处理
## 定义预处理
```
def regUse(text):
text = re.sub(r"[,.?!\":]", '', text) # 去标点
text = re.sub(r"'\w*\s", ' ', text) # 去缩写
text = re.sub(r"#?&.{1,3};", '', text) # 去html符号
return text.lower()
def sampleSeg(text):
tokens = [word for word in word_tokenize(text) if word not in stopwords.words('english') and len(word)>=3]
return tokens
def get_wordnet_pos(treebank_tag):
if treebank_tag.startswith('J'):
return wordnet.ADJ
elif treebank_tag.startswith('V'):
return wordnet.VERB
elif treebank_tag.startswith('N'):
return wordnet.NOUN
elif treebank_tag.startswith('R'):
return wordnet.ADV
else:
return None
def lemSeg(tokens):
res = []
lemmatizer = WordNetLemmatizer()
for word, pos in pos_tag(tokens):
wordnet_pos = get_wordnet_pos(pos) or wordnet.NOUN
res.append(lemmatizer.lemmatize(word, pos=wordnet_pos))
return res
def preprocess(text):
text = regUse(text)
tokens = sampleSeg(text)
tokens = lemSeg(tokens)
return tokens ## 返回的是单词列表
```
## 实施预处理
```
sms_data = [] ## 每个元素是一个句子
sms_label = [] ## 每个元素是一个字符串0/1
label_num = {"spam":1, "ham":0} ## 垃圾邮件为1,正常邮件为0
start = time.perf_counter()
for line in sms:
sms_data.append(" ".join(preprocess(line[1])))
sms_label.append(label_num[line[0]])
elapsed = (time.perf_counter() - start)
print("预处理耗时:{:.2f}s".format(elapsed))
"""显示预处理结果"""
print("预处理后的结果示例:")
print(sms_data[0:3])
print(sms_label[0:3])
```
# 准备训练集、验证集
```
MAX_NUM_WORDS = 2000
tokenizer = Tokenizer(num_words=MAX_NUM_WORDS) ## 最终选取频率前MAX_NUM_WORDS个单词
tokenizer.fit_on_texts(sms_data)
MAX_SEQUENCE_LENGTH = 50 ## 长度超过MAX_SEQUENCE_LENGTH则截断,不足则补0
sequences = tokenizer.texts_to_sequences(sms_data) ## 是一个二维数值数组,每一个数值都是对应句子对应单词的**索引**
dataset = pad_sequences(sequences, maxlen=MAX_SEQUENCE_LENGTH)
labels = to_categorical(np.asarray(sms_label)) ## 将label转为独热编码形式
"""打乱"""
indices = np.arange(dataset.shape[0])
np.random.shuffle(indices)
dataset = dataset[indices]
labels = labels[indices]
"""划分"""
size_dataset = len(dataset)
size_trainset = int(round(size_dataset*0.7))
print("训练集大小:{}".format(size_trainset))
print("验证集大小:{}".format(size_dataset - size_trainset))
x_train = dataset[0:size_trainset]
y_train = labels[0:size_trainset]
x_val = dataset[size_trainset+1: size_dataset]
y_val = labels[size_trainset+1: size_dataset]
```
# 构建模型
## 构建embedding层
### 1.准备预训练好的词典
```
embedding_dic = {}
file_path = '../glove/glove.6B.100d.txt'
start = time.perf_counter()
with open(file_path, encoding="utf-8") as f:
for line in f:
values = line.split()
word = values[0]
coefs = np.asarray(values[1:], dtype='float32')
embedding_dic[word] = coefs
elapsed = (time.perf_counter() - start)
print("准备词典耗时:{:.2f}s".format(elapsed))
```
### 2.准备embedding_matrix
embedding_matrix是本文中所有(或者选取高频部分)单词对应的词向量
```
"""选取部分单词"""
word_index = tokenizer.word_index ## 得到一个字典,key是选择的单词,value是它的索引
print("共有{}个单词,示例:".format(len(word_index)))
print(list(word_index.keys())[0:5], list(word_index.values())[0:5])
"""准备这些单词的embedding_matrix"""
EMBEDDING_DIM = 100 ## 令词向量的维度是100
num_words = min(MAX_NUM_WORDS, len(word_index) + 1) ## 为什么要加一?
embedding_matrix = np.zeros((num_words, EMBEDDING_DIM))
start = time.perf_counter()
for word, i in word_index.items():
if i >= MAX_NUM_WORDS:
continue
embedding_vector = embedding_dic.get(word)
if embedding_vector is not None: ## 单词在emmbeding_dic中存在时
embedding_matrix[i] = embedding_vector
elapsed = (time.perf_counter() - start)
print("准备embedding_matrix耗时:{:.2f}s".format(elapsed))
```
### 3.构建embedding layer
```
embedding_layer = Embedding(input_dim=num_words, # 词汇表单词数量
output_dim=EMBEDDING_DIM, # 词向量维度
weights=[embedding_matrix],
input_length=MAX_SEQUENCE_LENGTH,
trainable=False) # 词向量矩阵不进行训练
```
## 构建、连接其他层
```
sequence_input = Input(shape=(MAX_SEQUENCE_LENGTH,), dtype='int32') # 占位。
embedded_sequences = embedding_layer(sequence_input) # 返回 句子个数*50*100
x = Conv1D(128, 5, activation='relu')(embedded_sequences)
x = MaxPooling1D(2)(x)
x = Conv1D(128, 5, activation='relu')(x)
x = MaxPooling1D(2)(x)
x = Conv1D(128, 5, activation='relu')(x)
x = MaxPooling1D(2)(x)
x = GlobalMaxPooling1D()(x)
x = Dense(128, activation='relu')(x)
preds = Dense(len(label_num), activation='softmax')(x)
model = Model(sequence_input, preds)
model.compile(loss='categorical_crossentropy', optimizer='rmsprop', metrics=['acc'])
model.summary()
```
# 应用模型
```
model.fit(x_train, y_train, batch_size=16, epochs=5, validation_data=(x_val, y_val))
```
# 显示性能指标
## 1.格式转换函数
```
"""将y_val转为y_val_label"""
# y_val[0:3]示例:[[0. 1.]
# [1. 0.]
# [1. 0.]]
# y_val_label[0:3]示例:['spam' 'ham' 'ham']
def TOy_val_label(y_val):
y_val_label = []
spam = np.array([0., 1.]) ## [0 1]表示垃圾邮件?
ham = np.array([1., 0.]) ## [1 0]表示正常邮件
for line in y_val:
if all(line == spam):
y_val_label.append("spam")
else:
y_val_label.append("ham")
y_val_label = np.array(y_val_label)
return y_val_label
"""将y_pred转为y_pred_label"""
# y_pred[0:3]示例:[[9.9905199e-01 9.4802002e-04]
# [9.8692465e-01 1.3075325e-02]
# [1.0000000e+00 0.0000000e+00]]
# y_pred_label[0:3]示例:['ham' 'ham' 'ham']
def TOy_pred_label(y_pred):
y_pred_label = []
y_pred_index = np.argmax(y_pred, axis=1)
for line in y_pred_index:
if line == 0:
y_pred_label.append("ham")
else:
y_pred_label.append("spam")
y_pred_label = np.array(y_pred_label)
return y_pred_label
```
## 2.性能显示函数
```
from sklearn.metrics import confusion_matrix
from sklearn.metrics import classification_report
import matplotlib.pyplot as plt
from sklearn.model_selection import cross_val_predict
from sklearn.metrics import roc_curve, auc
def show_model(y_val_label, y_pred_label):
cm = confusion_matrix(y_val_label, y_pred_label)
cr = classification_report(y_val_label, y_pred_label)
print('混淆矩阵:')
print(cm)
print('分类结果:')
print(cr)
def print_AUC(y_val_label, y_pred):
y_scores = y_pred[:, 1]
fpr,tpr,threshold = roc_curve(y_val_label, y_scores, pos_label='spam')
roc_auc = auc(fpr,tpr)
plt.figure()
lw = 2
plt.figure(figsize=(5,5))
plt.plot(fpr, tpr, color='darkorange',
lw=lw, label='ROC curve (area = %0.2f)' % roc_auc)
plt.plot([0, 1], [0, 1], color='navy', lw=lw, linestyle='--')
plt.xlim([0.0, 1.0])
plt.ylim([0.0, 1.05])
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
plt.title('Receiver operating characteristic example')
plt.legend(loc="lower right")
plt.show()
```
## 3.开始评估
```
y_pred = model.predict(x_val,batch_size = 16)
y_val_label = TOy_val_label(y_val)
y_pred_label = TOy_pred_label(y_pred)
show_model(y_val_label, y_pred_label)
print_AUC(y_val_label, y_pred)
```
|
github_jupyter
|
import csv
import numpy as np
import nltk
import re
from nltk import word_tokenize, pos_tag
from nltk.corpus import stopwords, wordnet
from nltk.stem import WordNetLemmatizer
from sklearn.feature_extraction.text import TfidfVectorizer
from keras.preprocessing.text import Tokenizer
from keras.preprocessing.sequence import pad_sequences
from keras.utils import to_categorical
from keras.layers import Dense, Input, GlobalMaxPooling1D
from keras.layers import Conv1D, MaxPooling1D, Embedding
from keras.models import Model
import time
"""获取原始数据"""
file_path = '../smsspamcollection/SMSSpamCollection'
smsFile = open(file_path, 'r', encoding='utf-8') ## 返回文件对象
sms = csv.reader(smsFile, delimiter='\t') ## 第一层:行列表;第二层:列列表
sms = list(sms)
smsFile.close()
"""显示原始数据"""
for line in sms[0:3]:
print(line)
def regUse(text):
text = re.sub(r"[,.?!\":]", '', text) # 去标点
text = re.sub(r"'\w*\s", ' ', text) # 去缩写
text = re.sub(r"#?&.{1,3};", '', text) # 去html符号
return text.lower()
def sampleSeg(text):
tokens = [word for word in word_tokenize(text) if word not in stopwords.words('english') and len(word)>=3]
return tokens
def get_wordnet_pos(treebank_tag):
if treebank_tag.startswith('J'):
return wordnet.ADJ
elif treebank_tag.startswith('V'):
return wordnet.VERB
elif treebank_tag.startswith('N'):
return wordnet.NOUN
elif treebank_tag.startswith('R'):
return wordnet.ADV
else:
return None
def lemSeg(tokens):
res = []
lemmatizer = WordNetLemmatizer()
for word, pos in pos_tag(tokens):
wordnet_pos = get_wordnet_pos(pos) or wordnet.NOUN
res.append(lemmatizer.lemmatize(word, pos=wordnet_pos))
return res
def preprocess(text):
text = regUse(text)
tokens = sampleSeg(text)
tokens = lemSeg(tokens)
return tokens ## 返回的是单词列表
sms_data = [] ## 每个元素是一个句子
sms_label = [] ## 每个元素是一个字符串0/1
label_num = {"spam":1, "ham":0} ## 垃圾邮件为1,正常邮件为0
start = time.perf_counter()
for line in sms:
sms_data.append(" ".join(preprocess(line[1])))
sms_label.append(label_num[line[0]])
elapsed = (time.perf_counter() - start)
print("预处理耗时:{:.2f}s".format(elapsed))
"""显示预处理结果"""
print("预处理后的结果示例:")
print(sms_data[0:3])
print(sms_label[0:3])
MAX_NUM_WORDS = 2000
tokenizer = Tokenizer(num_words=MAX_NUM_WORDS) ## 最终选取频率前MAX_NUM_WORDS个单词
tokenizer.fit_on_texts(sms_data)
MAX_SEQUENCE_LENGTH = 50 ## 长度超过MAX_SEQUENCE_LENGTH则截断,不足则补0
sequences = tokenizer.texts_to_sequences(sms_data) ## 是一个二维数值数组,每一个数值都是对应句子对应单词的**索引**
dataset = pad_sequences(sequences, maxlen=MAX_SEQUENCE_LENGTH)
labels = to_categorical(np.asarray(sms_label)) ## 将label转为独热编码形式
"""打乱"""
indices = np.arange(dataset.shape[0])
np.random.shuffle(indices)
dataset = dataset[indices]
labels = labels[indices]
"""划分"""
size_dataset = len(dataset)
size_trainset = int(round(size_dataset*0.7))
print("训练集大小:{}".format(size_trainset))
print("验证集大小:{}".format(size_dataset - size_trainset))
x_train = dataset[0:size_trainset]
y_train = labels[0:size_trainset]
x_val = dataset[size_trainset+1: size_dataset]
y_val = labels[size_trainset+1: size_dataset]
embedding_dic = {}
file_path = '../glove/glove.6B.100d.txt'
start = time.perf_counter()
with open(file_path, encoding="utf-8") as f:
for line in f:
values = line.split()
word = values[0]
coefs = np.asarray(values[1:], dtype='float32')
embedding_dic[word] = coefs
elapsed = (time.perf_counter() - start)
print("准备词典耗时:{:.2f}s".format(elapsed))
"""选取部分单词"""
word_index = tokenizer.word_index ## 得到一个字典,key是选择的单词,value是它的索引
print("共有{}个单词,示例:".format(len(word_index)))
print(list(word_index.keys())[0:5], list(word_index.values())[0:5])
"""准备这些单词的embedding_matrix"""
EMBEDDING_DIM = 100 ## 令词向量的维度是100
num_words = min(MAX_NUM_WORDS, len(word_index) + 1) ## 为什么要加一?
embedding_matrix = np.zeros((num_words, EMBEDDING_DIM))
start = time.perf_counter()
for word, i in word_index.items():
if i >= MAX_NUM_WORDS:
continue
embedding_vector = embedding_dic.get(word)
if embedding_vector is not None: ## 单词在emmbeding_dic中存在时
embedding_matrix[i] = embedding_vector
elapsed = (time.perf_counter() - start)
print("准备embedding_matrix耗时:{:.2f}s".format(elapsed))
embedding_layer = Embedding(input_dim=num_words, # 词汇表单词数量
output_dim=EMBEDDING_DIM, # 词向量维度
weights=[embedding_matrix],
input_length=MAX_SEQUENCE_LENGTH,
trainable=False) # 词向量矩阵不进行训练
sequence_input = Input(shape=(MAX_SEQUENCE_LENGTH,), dtype='int32') # 占位。
embedded_sequences = embedding_layer(sequence_input) # 返回 句子个数*50*100
x = Conv1D(128, 5, activation='relu')(embedded_sequences)
x = MaxPooling1D(2)(x)
x = Conv1D(128, 5, activation='relu')(x)
x = MaxPooling1D(2)(x)
x = Conv1D(128, 5, activation='relu')(x)
x = MaxPooling1D(2)(x)
x = GlobalMaxPooling1D()(x)
x = Dense(128, activation='relu')(x)
preds = Dense(len(label_num), activation='softmax')(x)
model = Model(sequence_input, preds)
model.compile(loss='categorical_crossentropy', optimizer='rmsprop', metrics=['acc'])
model.summary()
model.fit(x_train, y_train, batch_size=16, epochs=5, validation_data=(x_val, y_val))
"""将y_val转为y_val_label"""
# y_val[0:3]示例:[[0. 1.]
# [1. 0.]
# [1. 0.]]
# y_val_label[0:3]示例:['spam' 'ham' 'ham']
def TOy_val_label(y_val):
y_val_label = []
spam = np.array([0., 1.]) ## [0 1]表示垃圾邮件?
ham = np.array([1., 0.]) ## [1 0]表示正常邮件
for line in y_val:
if all(line == spam):
y_val_label.append("spam")
else:
y_val_label.append("ham")
y_val_label = np.array(y_val_label)
return y_val_label
"""将y_pred转为y_pred_label"""
# y_pred[0:3]示例:[[9.9905199e-01 9.4802002e-04]
# [9.8692465e-01 1.3075325e-02]
# [1.0000000e+00 0.0000000e+00]]
# y_pred_label[0:3]示例:['ham' 'ham' 'ham']
def TOy_pred_label(y_pred):
y_pred_label = []
y_pred_index = np.argmax(y_pred, axis=1)
for line in y_pred_index:
if line == 0:
y_pred_label.append("ham")
else:
y_pred_label.append("spam")
y_pred_label = np.array(y_pred_label)
return y_pred_label
from sklearn.metrics import confusion_matrix
from sklearn.metrics import classification_report
import matplotlib.pyplot as plt
from sklearn.model_selection import cross_val_predict
from sklearn.metrics import roc_curve, auc
def show_model(y_val_label, y_pred_label):
cm = confusion_matrix(y_val_label, y_pred_label)
cr = classification_report(y_val_label, y_pred_label)
print('混淆矩阵:')
print(cm)
print('分类结果:')
print(cr)
def print_AUC(y_val_label, y_pred):
y_scores = y_pred[:, 1]
fpr,tpr,threshold = roc_curve(y_val_label, y_scores, pos_label='spam')
roc_auc = auc(fpr,tpr)
plt.figure()
lw = 2
plt.figure(figsize=(5,5))
plt.plot(fpr, tpr, color='darkorange',
lw=lw, label='ROC curve (area = %0.2f)' % roc_auc)
plt.plot([0, 1], [0, 1], color='navy', lw=lw, linestyle='--')
plt.xlim([0.0, 1.0])
plt.ylim([0.0, 1.05])
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
plt.title('Receiver operating characteristic example')
plt.legend(loc="lower right")
plt.show()
y_pred = model.predict(x_val,batch_size = 16)
y_val_label = TOy_val_label(y_val)
y_pred_label = TOy_pred_label(y_pred)
show_model(y_val_label, y_pred_label)
print_AUC(y_val_label, y_pred)
| 0.50415 | 0.764254 |
# UT2000 Data Processing
For the MADS framework paper, we need to present the data. This notebooks helps reorganize the data so that it should be clear for anyone that picks it up.
```
import warnings
warnings.filterwarnings('ignore')
import os
import os.path
from os import path
from datetime import datetime, timedelta
import pytz
import matplotlib.pyplot as plt
import seaborn as sns
import matplotlib.dates as mdates
import pandas as pd
import numpy as np
```
# ID Crossover
We need the IDs from the multiply modalities to cross-reference the participants.
```
ids_1000 = pd.read_csv('../data/raw/ut1000/admin/id_crossover.csv')
# limiting so we don't have repeats with ut2000
ids_1000 = ids_1000[ids_1000['record'] < 2000]
ids_1000.head()
ids_2000 = pd.read_csv('../data/raw/ut2000/admin/id_crossover.csv')
ids_2000.head()
# combining ut1000 and 2000 records
ids = ids_1000.append(ids_2000)
```
# Fitbit Data
The Fitbit data is combined into a single CSV that contains all participants for a certain datatype. I need to import each file, separate by beiwe ID, and then export again.
```
data_dir = '../data/raw/ut2000/fitbit/'
export_dir = '../data/processed/MADS_framework_data/'
local = pytz.timezone ("America/Chicago")
# looping through all the files
for file in os.listdir(data_dir):
if file[-1] == 'v': # checking if csv file
fitbit_label = file.split('merged')[0][:-1]
if fitbit_label == 'activitylogs':
df = pd.read_csv(data_dir+file,parse_dates=[[1,2]])
df.set_index('Id',inplace=True)
else:
df = pd.read_csv(data_dir+file,index_col=0)
for col in df.columns:
if col in ['Time','ActivityHour','ActivityMinute','Date_StartTime']:
df['UTC time'] = pd.to_datetime(df[col],errors='coerce') - timedelta(hours=6)
fitbit_ids = df.index.unique().values
for fid in fitbit_ids:
df_byid = df[df.index.values == fid]
try:
bid = ids[ids['record'] == fid]['beiwe'].values[0]
df_byid['beiwe'] = bid
df_byid['fitbit'] = fid
if path.isdir(export_dir + bid + '/Fitbit/'):
df_byid.to_csv(export_dir+bid+'/Fitbit/'+fitbit_label+'.csv')
else:
os.mkdir(f'../data/processed/MADS_framework_data/{bid}')
os.mkdir(f'../data/processed/MADS_framework_data/{bid}/Fitbit')
df_byid.to_csv(export_dir+bid+'/Fitbit/'+fitbit_label+'.csv')
except Exception as inst:
print(f'File {fitbit_label} - Beiwe ID {bid} - Exception {inst}')
```
# Beacon Data
The beacon data might not as easy to coax into a format that works. The beacon data in the UT1000 file does not present any useful data so we can skip straight to the UT2000 data.
```
# headers for datafiles - pms, sgp, sht
headers = [['UTC time','pm1','pm2p5','pm10',
'std1','std2p5','std10',
'pc0p3','pc0p5','pc1','pc2p5','pc5','pc10'],
['UTC time','eco2','tvoc'],
['UTC time','rh','tc']
]
dirs = ['PM','TVOC','TRH']
data_dir = '../data/raw/ut2000/beacon'
export_dir = '../data/processed/MADS_framework_data'
for beacon in os.listdir(data_dir):
if beacon in ['beacon-d3-00','beacon-d3-01','beacon-d3-02']:
print(f'No processing data for beacon {beacon}')
else:
print(f'Reading for beacon {beacon}')
for sensor, header, sensor_dir in zip(['pms5003','sgp30','sht31d'], headers, dirs):
for file in os.listdir(f'{data_dir}/{beacon}/bevo/{sensor}/'):
df = pd.read_csv(f'{data_dir}/{beacon}/bevo/{sensor}/{file}',names=header)
beacon_no = int(beacon.split('-')[2])
if sensor == 'pms5003':
df = df[['UTC time','pm1','pm2p5','pm10']]
df.columns = ['UTC time','pm10','pm2.5','pm1']
try:
bid = ids_2000[ids_2000['beacon'] == beacon_no]
bid = bid['beiwe'].values[0]
df['beiwe'] = bid
df['beacon'] = beacon_no
df.set_index('UTC time',inplace=True)
if path.isdir(f'{export_dir}/{bid}/BEVO/{sensor_dir}/'):
df.to_csv(f'{export_dir}/{bid}/BEVO/{sensor_dir}/{file}')
elif not path.isdir(f'{export_dir}/{bid}'):
os.mkdir(f'{export_dir}/{bid}/')
os.mkdir(f'{export_dir}/{bid}/BEVO/')
os.mkdir(f'{export_dir}/{bid}/BEVO/{sensor_dir}/')
df.to_csv(f'{export_dir}/{bid}/BEVO/{sensor_dir}/{file}')
elif not path.isdir(f'{export_dir}/{bid}/BEVO/'):
os.mkdir(f'{export_dir}/{bid}/BEVO/')
os.mkdir(f'{export_dir}/{bid}/BEVO/{sensor_dir}/')
df.to_csv(f'{export_dir}/{bid}/BEVO/{sensor_dir}/{file}')
else:
os.mkdir(f'{export_dir}/{bid}/BEVO/{sensor_dir}/')
df.to_csv(f'{export_dir}/{bid}/BEVO/{sensor_dir}/{file}')
except Exception as inst:
print(f'{inst} - Beacon {beacon_no} not deployed')
```
|
github_jupyter
|
import warnings
warnings.filterwarnings('ignore')
import os
import os.path
from os import path
from datetime import datetime, timedelta
import pytz
import matplotlib.pyplot as plt
import seaborn as sns
import matplotlib.dates as mdates
import pandas as pd
import numpy as np
ids_1000 = pd.read_csv('../data/raw/ut1000/admin/id_crossover.csv')
# limiting so we don't have repeats with ut2000
ids_1000 = ids_1000[ids_1000['record'] < 2000]
ids_1000.head()
ids_2000 = pd.read_csv('../data/raw/ut2000/admin/id_crossover.csv')
ids_2000.head()
# combining ut1000 and 2000 records
ids = ids_1000.append(ids_2000)
data_dir = '../data/raw/ut2000/fitbit/'
export_dir = '../data/processed/MADS_framework_data/'
local = pytz.timezone ("America/Chicago")
# looping through all the files
for file in os.listdir(data_dir):
if file[-1] == 'v': # checking if csv file
fitbit_label = file.split('merged')[0][:-1]
if fitbit_label == 'activitylogs':
df = pd.read_csv(data_dir+file,parse_dates=[[1,2]])
df.set_index('Id',inplace=True)
else:
df = pd.read_csv(data_dir+file,index_col=0)
for col in df.columns:
if col in ['Time','ActivityHour','ActivityMinute','Date_StartTime']:
df['UTC time'] = pd.to_datetime(df[col],errors='coerce') - timedelta(hours=6)
fitbit_ids = df.index.unique().values
for fid in fitbit_ids:
df_byid = df[df.index.values == fid]
try:
bid = ids[ids['record'] == fid]['beiwe'].values[0]
df_byid['beiwe'] = bid
df_byid['fitbit'] = fid
if path.isdir(export_dir + bid + '/Fitbit/'):
df_byid.to_csv(export_dir+bid+'/Fitbit/'+fitbit_label+'.csv')
else:
os.mkdir(f'../data/processed/MADS_framework_data/{bid}')
os.mkdir(f'../data/processed/MADS_framework_data/{bid}/Fitbit')
df_byid.to_csv(export_dir+bid+'/Fitbit/'+fitbit_label+'.csv')
except Exception as inst:
print(f'File {fitbit_label} - Beiwe ID {bid} - Exception {inst}')
# headers for datafiles - pms, sgp, sht
headers = [['UTC time','pm1','pm2p5','pm10',
'std1','std2p5','std10',
'pc0p3','pc0p5','pc1','pc2p5','pc5','pc10'],
['UTC time','eco2','tvoc'],
['UTC time','rh','tc']
]
dirs = ['PM','TVOC','TRH']
data_dir = '../data/raw/ut2000/beacon'
export_dir = '../data/processed/MADS_framework_data'
for beacon in os.listdir(data_dir):
if beacon in ['beacon-d3-00','beacon-d3-01','beacon-d3-02']:
print(f'No processing data for beacon {beacon}')
else:
print(f'Reading for beacon {beacon}')
for sensor, header, sensor_dir in zip(['pms5003','sgp30','sht31d'], headers, dirs):
for file in os.listdir(f'{data_dir}/{beacon}/bevo/{sensor}/'):
df = pd.read_csv(f'{data_dir}/{beacon}/bevo/{sensor}/{file}',names=header)
beacon_no = int(beacon.split('-')[2])
if sensor == 'pms5003':
df = df[['UTC time','pm1','pm2p5','pm10']]
df.columns = ['UTC time','pm10','pm2.5','pm1']
try:
bid = ids_2000[ids_2000['beacon'] == beacon_no]
bid = bid['beiwe'].values[0]
df['beiwe'] = bid
df['beacon'] = beacon_no
df.set_index('UTC time',inplace=True)
if path.isdir(f'{export_dir}/{bid}/BEVO/{sensor_dir}/'):
df.to_csv(f'{export_dir}/{bid}/BEVO/{sensor_dir}/{file}')
elif not path.isdir(f'{export_dir}/{bid}'):
os.mkdir(f'{export_dir}/{bid}/')
os.mkdir(f'{export_dir}/{bid}/BEVO/')
os.mkdir(f'{export_dir}/{bid}/BEVO/{sensor_dir}/')
df.to_csv(f'{export_dir}/{bid}/BEVO/{sensor_dir}/{file}')
elif not path.isdir(f'{export_dir}/{bid}/BEVO/'):
os.mkdir(f'{export_dir}/{bid}/BEVO/')
os.mkdir(f'{export_dir}/{bid}/BEVO/{sensor_dir}/')
df.to_csv(f'{export_dir}/{bid}/BEVO/{sensor_dir}/{file}')
else:
os.mkdir(f'{export_dir}/{bid}/BEVO/{sensor_dir}/')
df.to_csv(f'{export_dir}/{bid}/BEVO/{sensor_dir}/{file}')
except Exception as inst:
print(f'{inst} - Beacon {beacon_no} not deployed')
| 0.111434 | 0.670713 |
```
import pandas as pd
import os
import pickle
import numpy as np
import scipy.sparse as sp
import scipy.io as spio
import matplotlib.pyplot as plt
import isolearn.io as isoio
import isolearn.keras as isol
#Load APA plasmid data (random mpra)
file_path = '../../../../aparent/data/apa_plasmid_data/apa_plasmid_data'
plasmid_dict = isoio.load(file_path)
plasmid_df = plasmid_dict['plasmid_df']
plasmid_cuts = plasmid_dict['plasmid_cuts']
print("len(plasmid_df) = " + str(len(plasmid_df)))
prox_c = np.ravel(plasmid_cuts[:, 180+70+6:180+70+6+35].sum(axis=-1))
total_c = np.ravel(plasmid_cuts[:, 180:180+205].sum(axis=-1)) + np.ravel(plasmid_cuts[:, -1].todense())
plasmid_df['proximal_count_from_cuts'] = prox_c
plasmid_df['total_count_from_cuts'] = total_c
#Filter data (positive set)
kept_libraries = [20]
min_count = 10
min_usage = 0.80
plasmid_df_pos = plasmid_df.copy()
keep_index = np.nonzero(plasmid_df_pos.sublibrary.isin(["doubledope_5prime_0"]))[0]
plasmid_df_pos = plasmid_df_pos.iloc[keep_index].copy()
if kept_libraries is not None :
keep_index = np.nonzero(plasmid_df_pos.library_index.isin(kept_libraries))[0]
plasmid_df_pos = plasmid_df_pos.iloc[keep_index].copy()
keep_index = np.nonzero(plasmid_df_pos.seq.str.slice(70, 76).isin(['ATTAAA', 'AATAAA']))[0]
plasmid_df_pos = plasmid_df_pos.iloc[keep_index].copy()
keep_index = np.nonzero(~plasmid_df_pos.seq.str.slice(155, 161).isin(['ATTAAA', 'AATAAA', 'AGTAAA', 'ACTAAA']))[0]
plasmid_df_pos = plasmid_df_pos.iloc[keep_index].copy()
if min_count is not None :
keep_index = np.nonzero(plasmid_df_pos.total_count_from_cuts >= min_count)[0]
plasmid_df_pos = plasmid_df_pos.iloc[keep_index].copy()
if min_usage is not None :
keep_index = np.nonzero(plasmid_df_pos.proximal_count_from_cuts / plasmid_df_pos.total_count_from_cuts >= min_usage)[0]
plasmid_df_pos = plasmid_df_pos.iloc[keep_index].copy()
print("len(plasmid_df_pos) = " + str(len(plasmid_df_pos)) + " (filtered)")
#Filter data (negative set)
kept_libraries = [20]
min_count = 4
max_usage = 0.20
plasmid_df_neg = plasmid_df.copy()
keep_index = np.nonzero(plasmid_df_neg.sublibrary.isin(["doubledope_5prime_0"]))[0]
plasmid_df_neg = plasmid_df_neg.iloc[keep_index].copy()
if kept_libraries is not None :
keep_index = np.nonzero(plasmid_df_neg.library_index.isin(kept_libraries))[0]
plasmid_df_neg = plasmid_df_neg.iloc[keep_index].copy()
keep_index = np.nonzero(plasmid_df_neg.seq.str.slice(70, 76).isin(['ATTAAA', 'AATAAA']))[0]
plasmid_df_neg = plasmid_df_neg.iloc[keep_index].copy()
keep_index = np.nonzero(~plasmid_df_neg.seq.str.slice(155, 161).isin(['ATTAAA', 'AATAAA', 'AGTAAA', 'ACTAAA']))[0]
plasmid_df_neg = plasmid_df_neg.iloc[keep_index].copy()
if min_count is not None :
keep_index = np.nonzero(plasmid_df_neg.total_count_from_cuts >= min_count)[0]
plasmid_df_neg = plasmid_df_neg.iloc[keep_index].copy()
if max_usage is not None :
keep_index = np.nonzero(plasmid_df_neg.proximal_count_from_cuts / plasmid_df_neg.total_count_from_cuts <= max_usage)[0]
plasmid_df_neg = plasmid_df_neg.iloc[keep_index].copy()
print("len(plasmid_df_neg) = " + str(len(plasmid_df_neg)) + " (filtered)")
data_df = pd.concat([plasmid_df_pos, plasmid_df_neg]).copy().reset_index(drop=True)
shuffle_index = np.arange(len(data_df))
np.random.shuffle(shuffle_index)
data_df = data_df.iloc[shuffle_index].copy().reset_index(drop=True)
data_df['proximal_usage'] = data_df.proximal_count_from_cuts / data_df.total_count_from_cuts
data_df['proximal_usage'].hist(bins=50)
#Store cached filtered dataframe
pickle.dump({'data_df' : data_df}, open('apa_doubledope_cached_set.pickle', 'wb'))
data_df[['padded_seq', 'proximal_usage']].to_csv('apa_doubledope_cached_set.csv', sep='\t', index=False)
```
|
github_jupyter
|
import pandas as pd
import os
import pickle
import numpy as np
import scipy.sparse as sp
import scipy.io as spio
import matplotlib.pyplot as plt
import isolearn.io as isoio
import isolearn.keras as isol
#Load APA plasmid data (random mpra)
file_path = '../../../../aparent/data/apa_plasmid_data/apa_plasmid_data'
plasmid_dict = isoio.load(file_path)
plasmid_df = plasmid_dict['plasmid_df']
plasmid_cuts = plasmid_dict['plasmid_cuts']
print("len(plasmid_df) = " + str(len(plasmid_df)))
prox_c = np.ravel(plasmid_cuts[:, 180+70+6:180+70+6+35].sum(axis=-1))
total_c = np.ravel(plasmid_cuts[:, 180:180+205].sum(axis=-1)) + np.ravel(plasmid_cuts[:, -1].todense())
plasmid_df['proximal_count_from_cuts'] = prox_c
plasmid_df['total_count_from_cuts'] = total_c
#Filter data (positive set)
kept_libraries = [20]
min_count = 10
min_usage = 0.80
plasmid_df_pos = plasmid_df.copy()
keep_index = np.nonzero(plasmid_df_pos.sublibrary.isin(["doubledope_5prime_0"]))[0]
plasmid_df_pos = plasmid_df_pos.iloc[keep_index].copy()
if kept_libraries is not None :
keep_index = np.nonzero(plasmid_df_pos.library_index.isin(kept_libraries))[0]
plasmid_df_pos = plasmid_df_pos.iloc[keep_index].copy()
keep_index = np.nonzero(plasmid_df_pos.seq.str.slice(70, 76).isin(['ATTAAA', 'AATAAA']))[0]
plasmid_df_pos = plasmid_df_pos.iloc[keep_index].copy()
keep_index = np.nonzero(~plasmid_df_pos.seq.str.slice(155, 161).isin(['ATTAAA', 'AATAAA', 'AGTAAA', 'ACTAAA']))[0]
plasmid_df_pos = plasmid_df_pos.iloc[keep_index].copy()
if min_count is not None :
keep_index = np.nonzero(plasmid_df_pos.total_count_from_cuts >= min_count)[0]
plasmid_df_pos = plasmid_df_pos.iloc[keep_index].copy()
if min_usage is not None :
keep_index = np.nonzero(plasmid_df_pos.proximal_count_from_cuts / plasmid_df_pos.total_count_from_cuts >= min_usage)[0]
plasmid_df_pos = plasmid_df_pos.iloc[keep_index].copy()
print("len(plasmid_df_pos) = " + str(len(plasmid_df_pos)) + " (filtered)")
#Filter data (negative set)
kept_libraries = [20]
min_count = 4
max_usage = 0.20
plasmid_df_neg = plasmid_df.copy()
keep_index = np.nonzero(plasmid_df_neg.sublibrary.isin(["doubledope_5prime_0"]))[0]
plasmid_df_neg = plasmid_df_neg.iloc[keep_index].copy()
if kept_libraries is not None :
keep_index = np.nonzero(plasmid_df_neg.library_index.isin(kept_libraries))[0]
plasmid_df_neg = plasmid_df_neg.iloc[keep_index].copy()
keep_index = np.nonzero(plasmid_df_neg.seq.str.slice(70, 76).isin(['ATTAAA', 'AATAAA']))[0]
plasmid_df_neg = plasmid_df_neg.iloc[keep_index].copy()
keep_index = np.nonzero(~plasmid_df_neg.seq.str.slice(155, 161).isin(['ATTAAA', 'AATAAA', 'AGTAAA', 'ACTAAA']))[0]
plasmid_df_neg = plasmid_df_neg.iloc[keep_index].copy()
if min_count is not None :
keep_index = np.nonzero(plasmid_df_neg.total_count_from_cuts >= min_count)[0]
plasmid_df_neg = plasmid_df_neg.iloc[keep_index].copy()
if max_usage is not None :
keep_index = np.nonzero(plasmid_df_neg.proximal_count_from_cuts / plasmid_df_neg.total_count_from_cuts <= max_usage)[0]
plasmid_df_neg = plasmid_df_neg.iloc[keep_index].copy()
print("len(plasmid_df_neg) = " + str(len(plasmid_df_neg)) + " (filtered)")
data_df = pd.concat([plasmid_df_pos, plasmid_df_neg]).copy().reset_index(drop=True)
shuffle_index = np.arange(len(data_df))
np.random.shuffle(shuffle_index)
data_df = data_df.iloc[shuffle_index].copy().reset_index(drop=True)
data_df['proximal_usage'] = data_df.proximal_count_from_cuts / data_df.total_count_from_cuts
data_df['proximal_usage'].hist(bins=50)
#Store cached filtered dataframe
pickle.dump({'data_df' : data_df}, open('apa_doubledope_cached_set.pickle', 'wb'))
data_df[['padded_seq', 'proximal_usage']].to_csv('apa_doubledope_cached_set.csv', sep='\t', index=False)
| 0.16238 | 0.249322 |
# Ch `09`: Concept `03`
## Convolution Neural Network
Load data from CIFAR-10.
```
import numpy as np
import matplotlib.pyplot as plt
import cifar_tools
import tensorflow as tf
learning_rate = 0.001
names, data, labels = \
cifar_tools.read_data('./cifar-10-batches-py')
```
Define the placeholders and variables for the CNN model:
```
x = tf.placeholder(tf.float32, [None, 24 * 24])
y = tf.placeholder(tf.float32, [None, len(names)])
W1 = tf.Variable(tf.random_normal([5, 5, 1, 64]))
b1 = tf.Variable(tf.random_normal([64]))
W2 = tf.Variable(tf.random_normal([5, 5, 64, 64]))
b2 = tf.Variable(tf.random_normal([64]))
W3 = tf.Variable(tf.random_normal([6*6*64, 1024]))
b3 = tf.Variable(tf.random_normal([1024]))
W_out = tf.Variable(tf.random_normal([1024, len(names)]))
b_out = tf.Variable(tf.random_normal([len(names)]))
```
Define helper functions for the convolution and maxpool layers:
```
def conv_layer(x, W, b):
conv = tf.nn.conv2d(x, W, strides=[1, 1, 1, 1], padding='SAME')
conv_with_b = tf.nn.bias_add(conv, b)
conv_out = tf.nn.relu(conv_with_b)
return conv_out
def maxpool_layer(conv, k=2):
return tf.nn.max_pool(conv, ksize=[1, k, k, 1], strides=[1, k, k, 1], padding='SAME')
```
The CNN model is defined all within the following method:
```
def model():
x_reshaped = tf.reshape(x, shape=[-1, 24, 24, 1])
conv_out1 = conv_layer(x_reshaped, W1, b1)
maxpool_out1 = maxpool_layer(conv_out1)
norm1 = tf.nn.lrn(maxpool_out1, 4, bias=1.0, alpha=0.001 / 9.0, beta=0.75)
conv_out2 = conv_layer(norm1, W2, b2)
norm2 = tf.nn.lrn(conv_out2, 4, bias=1.0, alpha=0.001 / 9.0, beta=0.75)
maxpool_out2 = maxpool_layer(norm2)
maxpool_reshaped = tf.reshape(maxpool_out2, [-1, W3.get_shape().as_list()[0]])
local = tf.add(tf.matmul(maxpool_reshaped, W3), b3)
local_out = tf.nn.relu(local)
out = tf.add(tf.matmul(local_out, W_out), b_out)
return out
```
Here's the cost function to train the classifier.
```
model_op = model()
cost = tf.reduce_mean(tf.nn.softmax_cross_entropy_with_logits(model_op, y))
train_op = tf.train.AdamOptimizer(learning_rate=learning_rate).minimize(cost)
correct_pred = tf.equal(tf.argmax(model_op, 1), tf.argmax(y, 1))
accuracy = tf.reduce_mean(tf.cast(correct_pred, tf.float32))
```
Let's train the classifier on our data:
```
with tf.Session() as sess:
sess.run(tf.initialize_all_variables())
onehot_labels = tf.one_hot(labels, len(names), on_value=1., off_value=0., axis=-1)
onehot_vals = sess.run(onehot_labels)
batch_size = len(data) / 200
print('batch size', batch_size)
for j in range(0, 1000):
print('EPOCH', j)
for i in range(0, len(data), batch_size):
batch_data = data[i:i+batch_size, :]
batch_onehot_vals = onehot_vals[i:i+batch_size, :]
_, accuracy_val = sess.run([train_op, accuracy], feed_dict={x: batch_data, y: batch_onehot_vals})
if i % 1000 == 0:
print(i, accuracy_val)
print('DONE WITH EPOCH')
```
|
github_jupyter
|
import numpy as np
import matplotlib.pyplot as plt
import cifar_tools
import tensorflow as tf
learning_rate = 0.001
names, data, labels = \
cifar_tools.read_data('./cifar-10-batches-py')
x = tf.placeholder(tf.float32, [None, 24 * 24])
y = tf.placeholder(tf.float32, [None, len(names)])
W1 = tf.Variable(tf.random_normal([5, 5, 1, 64]))
b1 = tf.Variable(tf.random_normal([64]))
W2 = tf.Variable(tf.random_normal([5, 5, 64, 64]))
b2 = tf.Variable(tf.random_normal([64]))
W3 = tf.Variable(tf.random_normal([6*6*64, 1024]))
b3 = tf.Variable(tf.random_normal([1024]))
W_out = tf.Variable(tf.random_normal([1024, len(names)]))
b_out = tf.Variable(tf.random_normal([len(names)]))
def conv_layer(x, W, b):
conv = tf.nn.conv2d(x, W, strides=[1, 1, 1, 1], padding='SAME')
conv_with_b = tf.nn.bias_add(conv, b)
conv_out = tf.nn.relu(conv_with_b)
return conv_out
def maxpool_layer(conv, k=2):
return tf.nn.max_pool(conv, ksize=[1, k, k, 1], strides=[1, k, k, 1], padding='SAME')
def model():
x_reshaped = tf.reshape(x, shape=[-1, 24, 24, 1])
conv_out1 = conv_layer(x_reshaped, W1, b1)
maxpool_out1 = maxpool_layer(conv_out1)
norm1 = tf.nn.lrn(maxpool_out1, 4, bias=1.0, alpha=0.001 / 9.0, beta=0.75)
conv_out2 = conv_layer(norm1, W2, b2)
norm2 = tf.nn.lrn(conv_out2, 4, bias=1.0, alpha=0.001 / 9.0, beta=0.75)
maxpool_out2 = maxpool_layer(norm2)
maxpool_reshaped = tf.reshape(maxpool_out2, [-1, W3.get_shape().as_list()[0]])
local = tf.add(tf.matmul(maxpool_reshaped, W3), b3)
local_out = tf.nn.relu(local)
out = tf.add(tf.matmul(local_out, W_out), b_out)
return out
model_op = model()
cost = tf.reduce_mean(tf.nn.softmax_cross_entropy_with_logits(model_op, y))
train_op = tf.train.AdamOptimizer(learning_rate=learning_rate).minimize(cost)
correct_pred = tf.equal(tf.argmax(model_op, 1), tf.argmax(y, 1))
accuracy = tf.reduce_mean(tf.cast(correct_pred, tf.float32))
with tf.Session() as sess:
sess.run(tf.initialize_all_variables())
onehot_labels = tf.one_hot(labels, len(names), on_value=1., off_value=0., axis=-1)
onehot_vals = sess.run(onehot_labels)
batch_size = len(data) / 200
print('batch size', batch_size)
for j in range(0, 1000):
print('EPOCH', j)
for i in range(0, len(data), batch_size):
batch_data = data[i:i+batch_size, :]
batch_onehot_vals = onehot_vals[i:i+batch_size, :]
_, accuracy_val = sess.run([train_op, accuracy], feed_dict={x: batch_data, y: batch_onehot_vals})
if i % 1000 == 0:
print(i, accuracy_val)
print('DONE WITH EPOCH')
| 0.612773 | 0.962848 |
```
import numpy as np
import pandas as pd
```
The dataset I got was a txt file, so I will tranform it to a csv file for the future use.
```
data_txt = np.loadtxt('./dataset/X_train.txt')
data_txtDF = pd.DataFrame(data_txt)
data_txtDF.to_csv('./dataset/X_tran.csv',index=False)
train_X = pd.read_csv('./dataset/X_tran.csv')
train_X
```
Now we start to do the pre-process and the feature learning.
```
train_X.describe()
from sklearn import preprocessing as sklpp
from sklearn import decomposition as skldecomp
```
We do the preprocess to the dataset to have a centered dataset for future use.
Here we first use preprocessing package to center this sample matrix.
```
mean_datascaler = sklpp.StandardScaler(with_mean=True, with_std=False)
ctd_train_X = mean_datascaler.fit_transform(train_X)
```
After we have the centered matrix, we can do the dimensionality reduction to this matrix. Here we want the samples to have two demensionality so that we can show the result eaier.
```
data_pca = skldecomp.PCA(n_components=0.90, svd_solver='full')
skl_features = data_pca.fit_transform(ctd_train_X)
```
Now we can see that the dimensionality of the sample matrix has been reduced from 561 to 2. And we have a variance ratio about 67%.
```
print(np.sum(data_pca.explained_variance_ratio_))
print(skl_features.shape)
```
With the pre-processed data samples we can start our work in clustering.
```
from sklearn import cluster as cluster
```
This dataset records 30 volunteers perform 5 activities (WALKING, WALKING_UPSTAIRS, WALKING_DOWNSTAIRS, SITTING, STANDING, LAYING). So it is supposed to be divided into 6 clusters.
```
y_pred = cluster.KMeans(n_clusters=6).fit_predict(skl_features)
import matplotlib.pyplot as plt
plt.scatter(skl_features[:, 0], skl_features[:, 33], c=y_pred)
plt.show()
```
The following graph shows the original distribution of
```
l_Y = np.array(label_Y).T.astype(int)
plt.scatter(skl_features[:, 0], skl_features[:, 33], c=l_Y[0])
plt.show()
data_txt = np.loadtxt('./dataset/y_train.txt')
data_txtDF = pd.DataFrame(data_txt)
data_txtDF.to_csv('./dataset/y_tran.csv',index=False)
label_Y = pd.read_csv('./dataset/y_tran.csv')
label_Y
```
0:78;
1:0;
2:66;
3:3;
4:101;
5:14;
```
label_Y.columns=['Y']
np.argwhere(y_pred == 4).T[0]
series=[]
for i in range(0,6):
print('pre label------',i,'------')
position = np.argwhere(y_pred == i).T[0]
one_se = []
for pos in position:
one_se.append(label_Y.loc[pos].iloc[0])
series.append(one_se)
print('most label')
print(max(one_se,key=one_se.count))
tmp0 = series[1]
print('rate 2 = ', tmp0.count(4)/len(tmp0))
tmp5 = series[2]
print('rate 5 4 = ', tmp5.count(4)/len(tmp5))
print('rate 5 5 = ', tmp5.count(5)/len(tmp5))
print('rate 5 6 = ', tmp5.count(6)/len(tmp5))
print('pre 2 is 4')
s3=series[3]
s5=series[5]
print('rate 3 3 =', s3.count(1)/len(s3))
print('rate 5 3 =', s5.count(3)/len(s5))
print('rate 5 2 =', s5.count(2)/len(s5))
print('rate 5 1 =', s5.count(1)/len(s5))
pre_Y = [2,5,4,3,6,1]
accr = []
for i in range(0,6):
accr.append(series[i].count(pre_Y[i])/len(series[i]))
print(accr)
print(sum(accr)/6)
```
|
github_jupyter
|
import numpy as np
import pandas as pd
data_txt = np.loadtxt('./dataset/X_train.txt')
data_txtDF = pd.DataFrame(data_txt)
data_txtDF.to_csv('./dataset/X_tran.csv',index=False)
train_X = pd.read_csv('./dataset/X_tran.csv')
train_X
train_X.describe()
from sklearn import preprocessing as sklpp
from sklearn import decomposition as skldecomp
mean_datascaler = sklpp.StandardScaler(with_mean=True, with_std=False)
ctd_train_X = mean_datascaler.fit_transform(train_X)
data_pca = skldecomp.PCA(n_components=0.90, svd_solver='full')
skl_features = data_pca.fit_transform(ctd_train_X)
print(np.sum(data_pca.explained_variance_ratio_))
print(skl_features.shape)
from sklearn import cluster as cluster
y_pred = cluster.KMeans(n_clusters=6).fit_predict(skl_features)
import matplotlib.pyplot as plt
plt.scatter(skl_features[:, 0], skl_features[:, 33], c=y_pred)
plt.show()
l_Y = np.array(label_Y).T.astype(int)
plt.scatter(skl_features[:, 0], skl_features[:, 33], c=l_Y[0])
plt.show()
data_txt = np.loadtxt('./dataset/y_train.txt')
data_txtDF = pd.DataFrame(data_txt)
data_txtDF.to_csv('./dataset/y_tran.csv',index=False)
label_Y = pd.read_csv('./dataset/y_tran.csv')
label_Y
label_Y.columns=['Y']
np.argwhere(y_pred == 4).T[0]
series=[]
for i in range(0,6):
print('pre label------',i,'------')
position = np.argwhere(y_pred == i).T[0]
one_se = []
for pos in position:
one_se.append(label_Y.loc[pos].iloc[0])
series.append(one_se)
print('most label')
print(max(one_se,key=one_se.count))
tmp0 = series[1]
print('rate 2 = ', tmp0.count(4)/len(tmp0))
tmp5 = series[2]
print('rate 5 4 = ', tmp5.count(4)/len(tmp5))
print('rate 5 5 = ', tmp5.count(5)/len(tmp5))
print('rate 5 6 = ', tmp5.count(6)/len(tmp5))
print('pre 2 is 4')
s3=series[3]
s5=series[5]
print('rate 3 3 =', s3.count(1)/len(s3))
print('rate 5 3 =', s5.count(3)/len(s5))
print('rate 5 2 =', s5.count(2)/len(s5))
print('rate 5 1 =', s5.count(1)/len(s5))
pre_Y = [2,5,4,3,6,1]
accr = []
for i in range(0,6):
accr.append(series[i].count(pre_Y[i])/len(series[i]))
print(accr)
print(sum(accr)/6)
| 0.194942 | 0.875787 |
[Index](Index.ipynb) - [Back](Output Widget.ipynb) - [Next](Widget Styling.ipynb)
# Widget Events
## Special events
The `Button` is not used to represent a data type. Instead the button widget is used to handle mouse clicks. The `on_click` method of the `Button` can be used to register function to be called when the button is clicked. The doc string of the `on_click` can be seen below.
```
import ipywidgets as widgets
print(widgets.Button.on_click.__doc__)
```
### Example
Since button clicks are stateless, they are transmitted from the front-end to the back-end using custom messages. By using the `on_click` method, a button that prints a message when it has been clicked is shown below. To capture `print`s (or any other kind of output) and ensure it is displayed, be sure to send it to an `Output` widget (or put the information you want to display into an `HTML` widget).
```
from IPython.display import display
button = widgets.Button(description="Click Me!")
output = widgets.Output()
display(button, output)
def on_button_clicked(b):
with output:
print("Button clicked.")
button.on_click(on_button_clicked)
```
## Traitlet events
Widget properties are IPython traitlets and traitlets are eventful. To handle changes, the `observe` method of the widget can be used to register a callback. The doc string for `observe` can be seen below.
```
print(widgets.Widget.observe.__doc__)
```
### Signatures
Mentioned in the doc string, the callback registered must have the signature `handler(change)` where `change` is a dictionary holding the information about the change.
Using this method, an example of how to output an `IntSlider`'s value as it is changed can be seen below.
```
int_range = widgets.IntSlider()
output2 = widgets.Output()
display(int_range, output2)
def on_value_change(change):
with output2:
print(change['new'])
int_range.observe(on_value_change, names='value')
```
## Linking Widgets
Often, you may want to simply link widget attributes together. Synchronization of attributes can be done in a simpler way than by using bare traitlets events.
### Linking traitlets attributes in the kernel
The first method is to use the `link` and `dlink` functions from the `traitlets` module (these two functions are re-exported by the `ipywidgets` module for convenience). This only works if we are interacting with a live kernel.
```
caption = widgets.Label(value='The values of slider1 and slider2 are synchronized')
sliders1, slider2 = widgets.IntSlider(description='Slider 1'),\
widgets.IntSlider(description='Slider 2')
l = widgets.link((sliders1, 'value'), (slider2, 'value'))
display(caption, sliders1, slider2)
caption = widgets.Label(value='Changes in source values are reflected in target1')
source, target1 = widgets.IntSlider(description='Source'),\
widgets.IntSlider(description='Target 1')
dl = widgets.dlink((source, 'value'), (target1, 'value'))
display(caption, source, target1)
```
Function `traitlets.link` and `traitlets.dlink` return a `Link` or `DLink` object. The link can be broken by calling the `unlink` method.
```
l.unlink()
dl.unlink()
```
### Registering callbacks to trait changes in the kernel
Since attributes of widgets on the Python side are traitlets, you can register handlers to the change events whenever the model gets updates from the front-end.
The handler passed to observe will be called with one change argument. The change object holds at least a `type` key and a `name` key, corresponding respectively to the type of notification and the name of the attribute that triggered the notification.
Other keys may be passed depending on the value of `type`. In the case where type is `change`, we also have the following keys:
- `owner` : the HasTraits instance
- `old` : the old value of the modified trait attribute
- `new` : the new value of the modified trait attribute
- `name` : the name of the modified trait attribute.
```
caption = widgets.Label(value='The values of range1 and range2 are synchronized')
slider = widgets.IntSlider(min=-5, max=5, value=1, description='Slider')
def handle_slider_change(change):
caption.value = 'The slider value is ' + (
'negative' if change.new < 0 else 'nonnegative'
)
slider.observe(handle_slider_change, names='value')
display(caption, slider)
```
### Linking widgets attributes from the client side
When synchronizing traitlets attributes, you may experience a lag because of the latency due to the roundtrip to the server side. You can also directly link widget attributes in the browser using the link widgets, in either a unidirectional or a bidirectional fashion.
Javascript links persist when embedding widgets in html web pages without a kernel.
```
caption = widgets.Label(value='The values of range1 and range2 are synchronized')
range1, range2 = widgets.IntSlider(description='Range 1'),\
widgets.IntSlider(description='Range 2')
l = widgets.jslink((range1, 'value'), (range2, 'value'))
display(caption, range1, range2)
caption = widgets.Label(value='Changes in source_range values are reflected in target_range1')
source_range, target_range1 = widgets.IntSlider(description='Source range'),\
widgets.IntSlider(description='Target range 1')
dl = widgets.jsdlink((source_range, 'value'), (target_range1, 'value'))
display(caption, source_range, target_range1)
```
Function `widgets.jslink` returns a `Link` widget. The link can be broken by calling the `unlink` method.
```
# l.unlink()
# dl.unlink()
```
### The difference between linking in the kernel and linking in the client
Linking in the kernel means linking via python. If two sliders are linked in the kernel, when one slider is changed the browser sends a message to the kernel (python in this case) updating the changed slider, the link widget in the kernel then propagates the change to the other slider object in the kernel, and then the other slider's kernel object sends a message to the browser to update the other slider's views in the browser. If the kernel is not running (as in a static web page), then the controls will not be linked.
Linking using jslink (i.e., on the browser side) means constructing the link in Javascript. When one slider is changed, Javascript running in the browser changes the value of the other slider in the browser, without needing to communicate with the kernel at all. If the sliders are attached to kernel objects, each slider will update their kernel-side objects independently.
To see the difference between the two, go to the [static version of this page in the ipywidgets documentation](http://ipywidgets.readthedocs.io/en/latest/examples/Widget%20Events.html) and try out the sliders near the bottom. The ones linked in the kernel with `link` and `dlink` are no longer linked, but the ones linked in the browser with `jslink` and `jsdlink` are still linked.
## Continuous updates
Some widgets offer a choice with their `continuous_update` attribute between continually updating values or only updating values when a user submits the value (for example, by pressing Enter or navigating away from the control). In the next example, we see the "Delayed" controls only transmit their value after the user finishes dragging the slider or submitting the textbox. The "Continuous" controls continually transmit their values as they are changed. Try typing a two-digit number into each of the text boxes, or dragging each of the sliders, to see the difference.
```
a = widgets.IntSlider(description="Delayed", continuous_update=False)
b = widgets.IntText(description="Delayed", continuous_update=False)
c = widgets.IntSlider(description="Continuous", continuous_update=True)
d = widgets.IntText(description="Continuous", continuous_update=True)
widgets.link((a, 'value'), (b, 'value'))
widgets.link((a, 'value'), (c, 'value'))
widgets.link((a, 'value'), (d, 'value'))
widgets.VBox([a,b,c,d])
```
Sliders, `Text`, and `Textarea` controls default to `continuous_update=True`. `IntText` and other text boxes for entering integer or float numbers default to `continuous_update=False` (since often you'll want to type an entire number before submitting the value by pressing enter or navigating out of the box).
## Debouncing
When trait changes trigger a callback that performs a heavy computation, you may want to not do the computation as often as the value is updated. For instance, if the trait is driven by a slider which has its `continuous_update` set to `True`, the user will trigger a bunch of computations in rapid succession.
Debouncing solves this problem by delaying callback execution until the value has not changed for a certain time, after which the callback is called with the latest value. The effect is that the callback is only called when the trait pauses changing for a certain amount of time.
Debouncing can be implemented using an asynchronous loop or threads. We show an asynchronous solution below, which is more suited for ipywidgets. If you would like to instead use threads to do the debouncing, replace the `Timer` class with `from threading import Timer`.
```
import asyncio
class Timer:
def __init__(self, timeout, callback):
self._timeout = timeout
self._callback = callback
async def _job(self):
await asyncio.sleep(self._timeout)
self._callback()
def start(self):
self._task = asyncio.ensure_future(self._job())
def cancel(self):
self._task.cancel()
def debounce(wait):
""" Decorator that will postpone a function's
execution until after `wait` seconds
have elapsed since the last time it was invoked. """
def decorator(fn):
timer = None
def debounced(*args, **kwargs):
nonlocal timer
def call_it():
fn(*args, **kwargs)
if timer is not None:
timer.cancel()
timer = Timer(wait, call_it)
timer.start()
return debounced
return decorator
```
Here is how we use the `debounce` function as a decorator. Try changing the value of the slider. The text box will only update after the slider has paused for about 0.2 seconds.
```
slider = widgets.IntSlider()
text = widgets.IntText()
@debounce(0.2)
def value_changed(change):
text.value = change.new
slider.observe(value_changed, 'value')
widgets.VBox([slider, text])
```
## Throttling
Throttling is another technique that can be used to limit callbacks. Whereas debouncing ignores calls to a function if a certain amount of time has not passed since the last (attempt of) call to the function, throttling will just limit the rate of calls. This ensures that the function is regularly called.
We show an synchronous solution below. Likewise, you can replace the `Timer` class with `from threading import Timer` if you want to use threads instead of asynchronous programming.
```
import asyncio
from time import time
def throttle(wait):
""" Decorator that prevents a function from being called
more than once every wait period. """
def decorator(fn):
time_of_last_call = 0
scheduled, timer = False, None
new_args, new_kwargs = None, None
def throttled(*args, **kwargs):
nonlocal new_args, new_kwargs, time_of_last_call, scheduled, timer
def call_it():
nonlocal new_args, new_kwargs, time_of_last_call, scheduled, timer
time_of_last_call = time()
fn(*new_args, **new_kwargs)
scheduled = False
time_since_last_call = time() - time_of_last_call
new_args, new_kwargs = args, kwargs
if not scheduled:
scheduled = True
new_wait = max(0, wait - time_since_last_call)
timer = Timer(new_wait, call_it)
timer.start()
return throttled
return decorator
```
To see how different it behaves compared to the debouncer, here is the same slider example with its throttled value displayed in the text box. Notice how much more interactive it is, while still limiting the callback rate.
```
slider = widgets.IntSlider()
text = widgets.IntText()
@throttle(0.2)
def value_changed(change):
text.value = change.new
slider.observe(value_changed, 'value')
widgets.VBox([slider, text])
```
[Index](Index.ipynb) - [Back](Output Widget.ipynb) - [Next](Widget Styling.ipynb)
|
github_jupyter
|
import ipywidgets as widgets
print(widgets.Button.on_click.__doc__)
from IPython.display import display
button = widgets.Button(description="Click Me!")
output = widgets.Output()
display(button, output)
def on_button_clicked(b):
with output:
print("Button clicked.")
button.on_click(on_button_clicked)
print(widgets.Widget.observe.__doc__)
int_range = widgets.IntSlider()
output2 = widgets.Output()
display(int_range, output2)
def on_value_change(change):
with output2:
print(change['new'])
int_range.observe(on_value_change, names='value')
caption = widgets.Label(value='The values of slider1 and slider2 are synchronized')
sliders1, slider2 = widgets.IntSlider(description='Slider 1'),\
widgets.IntSlider(description='Slider 2')
l = widgets.link((sliders1, 'value'), (slider2, 'value'))
display(caption, sliders1, slider2)
caption = widgets.Label(value='Changes in source values are reflected in target1')
source, target1 = widgets.IntSlider(description='Source'),\
widgets.IntSlider(description='Target 1')
dl = widgets.dlink((source, 'value'), (target1, 'value'))
display(caption, source, target1)
l.unlink()
dl.unlink()
caption = widgets.Label(value='The values of range1 and range2 are synchronized')
slider = widgets.IntSlider(min=-5, max=5, value=1, description='Slider')
def handle_slider_change(change):
caption.value = 'The slider value is ' + (
'negative' if change.new < 0 else 'nonnegative'
)
slider.observe(handle_slider_change, names='value')
display(caption, slider)
caption = widgets.Label(value='The values of range1 and range2 are synchronized')
range1, range2 = widgets.IntSlider(description='Range 1'),\
widgets.IntSlider(description='Range 2')
l = widgets.jslink((range1, 'value'), (range2, 'value'))
display(caption, range1, range2)
caption = widgets.Label(value='Changes in source_range values are reflected in target_range1')
source_range, target_range1 = widgets.IntSlider(description='Source range'),\
widgets.IntSlider(description='Target range 1')
dl = widgets.jsdlink((source_range, 'value'), (target_range1, 'value'))
display(caption, source_range, target_range1)
# l.unlink()
# dl.unlink()
a = widgets.IntSlider(description="Delayed", continuous_update=False)
b = widgets.IntText(description="Delayed", continuous_update=False)
c = widgets.IntSlider(description="Continuous", continuous_update=True)
d = widgets.IntText(description="Continuous", continuous_update=True)
widgets.link((a, 'value'), (b, 'value'))
widgets.link((a, 'value'), (c, 'value'))
widgets.link((a, 'value'), (d, 'value'))
widgets.VBox([a,b,c,d])
import asyncio
class Timer:
def __init__(self, timeout, callback):
self._timeout = timeout
self._callback = callback
async def _job(self):
await asyncio.sleep(self._timeout)
self._callback()
def start(self):
self._task = asyncio.ensure_future(self._job())
def cancel(self):
self._task.cancel()
def debounce(wait):
""" Decorator that will postpone a function's
execution until after `wait` seconds
have elapsed since the last time it was invoked. """
def decorator(fn):
timer = None
def debounced(*args, **kwargs):
nonlocal timer
def call_it():
fn(*args, **kwargs)
if timer is not None:
timer.cancel()
timer = Timer(wait, call_it)
timer.start()
return debounced
return decorator
slider = widgets.IntSlider()
text = widgets.IntText()
@debounce(0.2)
def value_changed(change):
text.value = change.new
slider.observe(value_changed, 'value')
widgets.VBox([slider, text])
import asyncio
from time import time
def throttle(wait):
""" Decorator that prevents a function from being called
more than once every wait period. """
def decorator(fn):
time_of_last_call = 0
scheduled, timer = False, None
new_args, new_kwargs = None, None
def throttled(*args, **kwargs):
nonlocal new_args, new_kwargs, time_of_last_call, scheduled, timer
def call_it():
nonlocal new_args, new_kwargs, time_of_last_call, scheduled, timer
time_of_last_call = time()
fn(*new_args, **new_kwargs)
scheduled = False
time_since_last_call = time() - time_of_last_call
new_args, new_kwargs = args, kwargs
if not scheduled:
scheduled = True
new_wait = max(0, wait - time_since_last_call)
timer = Timer(new_wait, call_it)
timer.start()
return throttled
return decorator
slider = widgets.IntSlider()
text = widgets.IntText()
@throttle(0.2)
def value_changed(change):
text.value = change.new
slider.observe(value_changed, 'value')
widgets.VBox([slider, text])
| 0.431345 | 0.958148 |
# Explore network traffic dump
We will explore a network traffic trace file and perform some initial analysis.
You will need following tools:
* **Pandas**, a Python library for analysing data <http://pandas.pydata.org/>
* **tshark**, the command line version of the Wireshark sniffer <https://www.wireshark.org/>
* **Matplotlib**, a Python plotting library <http://matplotlib.org/>
## Download the traffic trace file
First, download the traffic trace file "traffic_dump.pcap", and this IPython notebook "Network Traffic Exploration.ipynb" from Blackboard, and save them into the same directory.
```
!ls -l
```
## Convert PCAP to a CSV using tshark
We can use the `tshark` command from the Wireshark tool suite to read the PCAP file and convert it into a tab-separated file. This might not be very fast, but it is very flexible, because all of Wireshark's diplay filters can be used to select the packets that we are interested in.
```
!tshark --help
```
Let's read the traffic_dump.pcap into tshark, and display frame number and frame length of each captured packet.
* -n: no name resolution
* -r: read from file, not a live capture
* -T fields: use table fields as the output format
* -E header=y: display header
* -e: field to display
```
!tshark -n -r traffic_dump.pcap -T fields -Eheader=y -e frame.number -e frame.len > frame.len
```
Let's have a look at the file:
```
!head -10 frame.len
```
Use Pandas to read the table into a DataFrame object:
```
import pandas as pd
df=pd.read_table("frame.len")
```
Take a look of the data frame.
```
df
```
Some statistics about the frame length:
```
df["frame.len"].describe()
```
The min and max are compliant with Ethernet specifications.
## Plotting
For a better overview, we plot the frame length over time.
We initialise IPython to show inline graphics:
```
import matplotlib.pyplot as plt
%matplotlib inline
%pylab inline
```
Set a figure size in inches:
```
figsize(10,6)
```
Pandas automatically uses Matplotlib for plotting. We plot with small dots and an alpha channel of 0.2:
```
df["frame.len"].plot(style=".", alpha=0.2)
title("Frame length")
ylabel("bytes")
xlabel("frame number")
```
So there are always lots of small packets (< 100 bytes) and lots of large packets (> 1400 bytes). Some bursts of packets with other sizes (around 400 bytes, 1000 bytes, etc.) can be clearly seen.
### A nice Python function to help read data
Here is a convenient helper function that reads the given fields into a Pandas DataFrame:
```
import subprocess
import datetime
import pandas as pd
def mydateparser(time_in_secs):
return datetime.datetime.fromtimestamp(float(time_in_secs))
def read_traffic(filename, fields=[], display_filter="",
timeseries=False, strict=False):
""" Read PCAP file into Pandas DataFrame object.
Uses tshark command-line tool from Wireshark.
filename: Name or full path of the PCAP file to read
fields: List of fields to include as columns
display_filter: Additional filter to restrict frames
strict: Only include frames that contain all given fields
(Default: false)
timeseries: Create DatetimeIndex from frame.time_epoch
(Default: false)
Syntax for fields and display_filter is specified in
Wireshark's Display Filter Reference:
http://www.wireshark.org/docs/dfref/
"""
if timeseries:
fields = ["frame.time_epoch"] + fields
fieldspec = " ".join("-e %s" % f for f in fields)
display_filters = fields if strict else []
if display_filter:
display_filters.append(display_filters)
filterspec = "-Y '%s'" % " and ".join(f for f in display_filters)
options = "-r %s -n -T fields -Eheader=y" % filename
cmd = "tshark %s %s %s" % (options, filterspec, fieldspec)
print(cmd)
proc = subprocess.Popen(cmd, shell = True,
stdout=subprocess.PIPE)
if timeseries:
df = pd.read_table(proc.stdout,
index_col = "frame.time_epoch",
parse_dates=True,
date_parser=mydateparser)
else:
df = pd.read_table(proc.stdout)
return df
```
## Bandwidth Usage
We want to know how much bandwidth is used every second. To do that, we need to sum up the frame lengths for every second.
First use our helper function to read the traffic trace into a DataFrame:
```
framelen=read_traffic("traffic_dump.pcap", ["frame.len"], timeseries=True)
#framelen
```
Then we re-sample the timeseries into buckets of 1 second, summing over the lengths of all frames that were captured in that second:
```
#bytes_per_second=framelen.resample("S", how="sum")
bytes_per_second=framelen.resample("S").sum()
```
Here are the first 5 rows. We get NaN for those timestamps where no frames were captured:
```
bytes_per_second.head()
bytes_per_second.plot()
```
## Analyze the trace at TCP level
Let's try to replicate the TCP Time-Sequence Graph
```
fields=["tcp.stream", "ip.src", "ip.dst", "tcp.seq", "tcp.ack", "tcp.window_size", "tcp.len"]
ts=read_traffic("traffic_dump.pcap", fields, timeseries=True, strict=True)
ts
```
Now we have to select a TCP stream to analyse. As an example, we just pick stream number 10:
```
stream=ts[ts["tcp.stream"] == 10]
stream
```
Add a column that shows who sent the packet (client or server).
The fancy lambda expression is a function that distinguishes between the client and the server side of the stream by comparing the source IP address with the source IP address of the first packet in the stream (for TCP steams that should have been sent by the client).
```
stream = stream.copy()
stream["type"] = stream.apply(lambda x: "client" if x["ip.src"] == stream.iloc[0]["ip.src"] else "server", axis=1)
#print stream.to_string()
stream
client_stream=stream[stream.type == "client"]
client_stream
client_stream["tcp.seq"].plot(style="r-o")
```
Notice that the x-axis shows the real timestamps.
For comparison, change the x-axis to be the packet number in the stream:
```
client_stream.index = arange(len(client_stream))
client_stream["tcp.seq"].plot(style="r-o")
```
Looks different of course.
## Bytes per stream
```
per_stream=ts.groupby("tcp.stream")
per_stream.head()
bytes_per_stream = per_stream["tcp.len"].sum()
bytes_per_stream.head()
bytes_per_stream.plot()
bytes_per_stream.max()
biggest_stream=bytes_per_stream.idxmax()
biggest_stream
bytes_per_stream.loc[biggest_stream]
```
|
github_jupyter
|
!ls -l
!tshark --help
!tshark -n -r traffic_dump.pcap -T fields -Eheader=y -e frame.number -e frame.len > frame.len
!head -10 frame.len
import pandas as pd
df=pd.read_table("frame.len")
df
df["frame.len"].describe()
import matplotlib.pyplot as plt
%matplotlib inline
%pylab inline
figsize(10,6)
df["frame.len"].plot(style=".", alpha=0.2)
title("Frame length")
ylabel("bytes")
xlabel("frame number")
import subprocess
import datetime
import pandas as pd
def mydateparser(time_in_secs):
return datetime.datetime.fromtimestamp(float(time_in_secs))
def read_traffic(filename, fields=[], display_filter="",
timeseries=False, strict=False):
""" Read PCAP file into Pandas DataFrame object.
Uses tshark command-line tool from Wireshark.
filename: Name or full path of the PCAP file to read
fields: List of fields to include as columns
display_filter: Additional filter to restrict frames
strict: Only include frames that contain all given fields
(Default: false)
timeseries: Create DatetimeIndex from frame.time_epoch
(Default: false)
Syntax for fields and display_filter is specified in
Wireshark's Display Filter Reference:
http://www.wireshark.org/docs/dfref/
"""
if timeseries:
fields = ["frame.time_epoch"] + fields
fieldspec = " ".join("-e %s" % f for f in fields)
display_filters = fields if strict else []
if display_filter:
display_filters.append(display_filters)
filterspec = "-Y '%s'" % " and ".join(f for f in display_filters)
options = "-r %s -n -T fields -Eheader=y" % filename
cmd = "tshark %s %s %s" % (options, filterspec, fieldspec)
print(cmd)
proc = subprocess.Popen(cmd, shell = True,
stdout=subprocess.PIPE)
if timeseries:
df = pd.read_table(proc.stdout,
index_col = "frame.time_epoch",
parse_dates=True,
date_parser=mydateparser)
else:
df = pd.read_table(proc.stdout)
return df
framelen=read_traffic("traffic_dump.pcap", ["frame.len"], timeseries=True)
#framelen
#bytes_per_second=framelen.resample("S", how="sum")
bytes_per_second=framelen.resample("S").sum()
bytes_per_second.head()
bytes_per_second.plot()
fields=["tcp.stream", "ip.src", "ip.dst", "tcp.seq", "tcp.ack", "tcp.window_size", "tcp.len"]
ts=read_traffic("traffic_dump.pcap", fields, timeseries=True, strict=True)
ts
stream=ts[ts["tcp.stream"] == 10]
stream
stream = stream.copy()
stream["type"] = stream.apply(lambda x: "client" if x["ip.src"] == stream.iloc[0]["ip.src"] else "server", axis=1)
#print stream.to_string()
stream
client_stream=stream[stream.type == "client"]
client_stream
client_stream["tcp.seq"].plot(style="r-o")
client_stream.index = arange(len(client_stream))
client_stream["tcp.seq"].plot(style="r-o")
per_stream=ts.groupby("tcp.stream")
per_stream.head()
bytes_per_stream = per_stream["tcp.len"].sum()
bytes_per_stream.head()
bytes_per_stream.plot()
bytes_per_stream.max()
biggest_stream=bytes_per_stream.idxmax()
biggest_stream
bytes_per_stream.loc[biggest_stream]
| 0.35209 | 0.970799 |
```
from scipy.io import loadmat
from scipy.optimize import curve_fit
import matplotlib as mpl
import matplotlib.pyplot as plt
import numpy as np
from mpl_toolkits.mplot3d import Axes3D
from numpy import trapz
def cm2inch(value):
return value/2.54
#axes.xaxis.set_tick_params(direction='in', which='both')
#axes.yaxis.set_tick_params(direction='in', which='both')
mpl.rcParams["xtick.direction"] = "in"
mpl.rcParams["ytick.direction"] = "in"
mpl.rcParams["lines.markeredgecolor"] = "k"
mpl.rcParams["lines.markeredgewidth"] = 1
mpl.rcParams["figure.dpi"] = 130
from matplotlib import rc
#rc('font', family='serif')
rc('text', usetex=True)
rc('xtick', labelsize='medium')
rc('ytick', labelsize='medium')
def cm2inch(value):
return value/2.54
def gauss_function(x, a, x0, sigma):
return a*np.exp(-(x-x0)**2/(2*sigma**2))
def pdf(data, bins = 10, density = True):
pdf, bins_edge = np.histogram(data, bins = bins, density = density)
bins_center = (bins_edge[0:-1] + bins_edge[1:]) / 2
return pdf, bins_center
def format_dataset(dataset):
for i in dataset.keys():
try:
dataset[i] = np.squeeze(dataset[i])
except:
continue
return dataset
dataset_15 = loadmat("15kpa/data_graphs.mat")
dataset_15 = format_dataset(dataset_15)
dataset_28_1 = loadmat("28kPa/12-2910/data_graphs.mat")
dataset_28_1 = format_dataset(dataset_28_1)
dataset_28_2 = loadmat("28kPa/14-2910/data_graphs.mat")
dataset_28_2 = format_dataset(dataset_28_2)
dataset_28_3 = loadmat("28kPa/13-2910/data_graphs.mat")
dataset_28_3 = format_dataset(dataset_28_3)
a = 1.5e-6
def F_corr(alpha,z):
return -alpha * D(z) * 4e-21 * D_1_prime(z)
def D_1_prime(z):
return (2 * r * (2 * r**2 + 12 * r * z + 21 * z**2))/(2 * r** + 9 * r * z + 6 * z**2)**2
def D(z):
return (6*z**2 + 9*r*z + 2*r**2) / (6*z**2 + 2 * r * z)
def F_corr(alpha,z):
return -alpha * D(z) * 4e-21 * D_1_prime(z)
def F_corr(alpha,z):
return - 4e-21 * alpha *(42*a*z*z + 24*a*a*z + 4*a*a*a)/(36*(z**4) + 66*a*(z**3) + 30*a*a*z*z + 4*(a**3)*z)
fig = plt.figure(figsize=((cm2inch(16),cm2inch(8))))
plt.plot(dataset_15["z_F_EHD_exp"][dataset_15["z_F_EHD_exp"]>4e-8],dataset_15["F_EHD_exp"][dataset_15["z_F_EHD_exp"]>4e-8]*1e15 + 1*1e15*F_corr(1,dataset_15["z_F_EHD_exp"][dataset_15["z_F_EHD_exp"]>4e-8]), "o", label="$G = 15$ kPa")
plt.plot(dataset_15["z_F_EHD_th"][dataset_15["z_F_EHD_th"] > 4e-8],dataset_15["F_EHD_th"][dataset_15["z_F_EHD_th"] > 4e-8]*1e15 , color="tab:blue")
plt.plot(dataset_28_1["z_F_EHD_exp"][dataset_28_1["z_F_EHD_exp"] > 4e-8],dataset_28_1["F_EHD_exp"][dataset_28_1["z_F_EHD_exp"] > 4e-8]*1e15 + 1*1e15*F_corr(1,dataset_28_1["z_F_EHD_exp"][dataset_28_1["z_F_EHD_exp"] > 4e-8]) , "o", label="$G= 28$ kPa")
ax=plt.gca()
ax.loglog(dataset_28_1["z_F_EHD_th"][dataset_28_1["z_F_EHD_th"] > 4e-8],dataset_28_1["F_EHD_th"][dataset_28_1["z_F_EHD_th"] > 4e-8]*1e15, color="tab:orange")
plt.plot(dataset_28_2["z_F_EHD_exp"][dataset_28_2["z_F_EHD_exp"] > 4e-8],dataset_28_2["F_EHD_exp"][dataset_28_2["z_F_EHD_exp"] > 4e-8]*1e15 +1* 1e15*F_corr(1,dataset_28_2["z_F_EHD_exp"][dataset_28_2["z_F_EHD_exp"] > 4e-8]), "o",color="tab:orange")
from mpltools import annotation
annotation.slope_marker((4e-7, 0.5), (-5, 2), ax=ax, invert=True, size_frac=0.12, text_kwargs={"usetex":True})
plt.legend()
plt.ylabel("$F_\mathrm{NC} $ (fN)")
plt.xlabel("$z$ ($\mathrm{\mu}$m)")
plt.ylim([1e-1,None])
plt.xlim([0.4e-7,1e-6])
plt.tight_layout(pad=0.2)
plt.savefig("EHD_force.pdf")
fig = plt.figure(figsize=((cm2inch(6*1.68),cm2inch(6))))
plt.plot(dataset_15["z_F_EHD_exp"][dataset_15["z_F_EHD_exp"]>4e-8],(dataset_15["F_EHD_exp"][dataset_15["z_F_EHD_exp"]>4e-8] + F_corr(1,dataset_15["z_F_EHD_exp"][dataset_15["z_F_EHD_exp"]>4e-8]))*15e3*np.sqrt(1.5e-6)/(0.001**2), "o", label="$G=15$ kPa")
plt.gca()
plt.plot(dataset_28_1["z_F_EHD_exp"][dataset_28_1["z_F_EHD_exp"] > 4e-8],(dataset_28_1["F_EHD_exp"][dataset_28_1["z_F_EHD_exp"] > 4e-8] + F_corr(1,dataset_28_1["z_F_EHD_exp"][dataset_28_1["z_F_EHD_exp"] > 4e-8])) *28e3*np.sqrt(1.5e-6)/(0.001**2), "o", label="$G=28$ kPa")
plt.loglog(dataset_28_1["z_F_EHD_th"][dataset_28_1["z_F_EHD_th"] > 4e-8],(dataset_28_1["F_EHD_th"][dataset_28_1["z_F_EHD_th"] > 4e-8] )*28e3*np.sqrt(1.5e-6)/(0.001**2), color="black")
plt.plot(dataset_28_2["z_F_EHD_exp"][dataset_28_2["z_F_EHD_exp"] > 4e-8],(dataset_28_2["F_EHD_exp"][dataset_28_2["z_F_EHD_exp"] > 4e-8] +F_corr(1,dataset_28_2["z_F_EHD_exp"][dataset_28_2["z_F_EHD_exp"] > 4e-8]))*28e3*np.sqrt(1.5e-6)/(0.001**2), "d",color="tab:orange")
annotation.slope_marker((3.5e-7, 4e-8), (-5, 2), ax=plt.gca(), invert=True, size_frac=0.12)
plt.legend()
plt.ylabel("$F_\mathrm{NC} Ga^{\\frac{1}{2}} \eta ^{-2}$ ($\mathrm{ m^{5/2} s ^{-2}}$)")
plt.xlabel("$z$ ($\mathrm{\mu}$m)")
plt.ylim([1e-8,None])
plt.xlim([4e-8,1e-6])
plt.tight_layout(pad=0.2)
plt.savefig("EHD_force_rescale.svg")
fig = plt.figure(figsize=((cm2inch(16),cm2inch(8))))
plt.plot(dataset_15["z_F_EHD_exp"][dataset_15["z_F_EHD_exp"]>4e-8],dataset_15["F_EHD_exp"][dataset_15["z_F_EHD_exp"]>4e-8]*1e15*15e3*np.sqrt(1.5e-6), "o", label="15 kPa")
#plt.plot(dataset_15["z_F_EHD_th"][dataset_15["z_F_EHD_th"] > 4e-8],dataset_15["F_EHD_th"][dataset_15["z_F_EHD_th"] > 4e-8]*1e15 + 1.5, color="tab:blue")
plt.plot(dataset_28_1["z_F_EHD_exp"][dataset_28_1["z_F_EHD_exp"] > 4e-8],dataset_28_1["F_EHD_exp"][dataset_28_1["z_F_EHD_exp"] > 4e-8]*1e15*28e3*np.sqrt(1.5e-6), "o", label="28 kPa")
plt.loglog(dataset_28_1["z_F_EHD_th"][dataset_28_1["z_F_EHD_th"] > 4e-8],dataset_28_1["F_EHD_th"][dataset_28_1["z_F_EHD_th"] > 4e-8]*1e15*28e3*np.sqrt(1.5e-6) + 1.5*28e3*np.sqrt(1.5e-6), color="black")
plt.plot(dataset_28_2["z_F_EHD_exp"][dataset_28_2["z_F_EHD_exp"] > 4e-8],dataset_28_2["F_EHD_exp"][dataset_28_2["z_F_EHD_exp"] > 4e-8]*1e15*28e3*np.sqrt(1.5e-6), "d", label="28 kPa",color="tab:orange")
plt.plot(dataset_28_3["z_F_EHD_exp"][dataset_28_3["z_F_EHD_exp"] > 4e-8],dataset_28_3["F_EHD_exp"][dataset_28_3["z_F_EHD_exp"] > 4e-8]*1e15*28e3*np.sqrt(1.5e-6), "s", label="28 kPa",color="tab:orange")
plt.legend()
plt.ylabel("$F_{NC} E\sqrt{a} $ ($\mathrm{fN^2 m^{3/2}}$)")
plt.xlabel("$z$ ($\mathrm{\mu}$m)")
#plt.ylim([-20,100])
#lt.xlim([1e-2,10])v
plt.savefig("EHD_force_rescale.pdf")
```
|
github_jupyter
|
from scipy.io import loadmat
from scipy.optimize import curve_fit
import matplotlib as mpl
import matplotlib.pyplot as plt
import numpy as np
from mpl_toolkits.mplot3d import Axes3D
from numpy import trapz
def cm2inch(value):
return value/2.54
#axes.xaxis.set_tick_params(direction='in', which='both')
#axes.yaxis.set_tick_params(direction='in', which='both')
mpl.rcParams["xtick.direction"] = "in"
mpl.rcParams["ytick.direction"] = "in"
mpl.rcParams["lines.markeredgecolor"] = "k"
mpl.rcParams["lines.markeredgewidth"] = 1
mpl.rcParams["figure.dpi"] = 130
from matplotlib import rc
#rc('font', family='serif')
rc('text', usetex=True)
rc('xtick', labelsize='medium')
rc('ytick', labelsize='medium')
def cm2inch(value):
return value/2.54
def gauss_function(x, a, x0, sigma):
return a*np.exp(-(x-x0)**2/(2*sigma**2))
def pdf(data, bins = 10, density = True):
pdf, bins_edge = np.histogram(data, bins = bins, density = density)
bins_center = (bins_edge[0:-1] + bins_edge[1:]) / 2
return pdf, bins_center
def format_dataset(dataset):
for i in dataset.keys():
try:
dataset[i] = np.squeeze(dataset[i])
except:
continue
return dataset
dataset_15 = loadmat("15kpa/data_graphs.mat")
dataset_15 = format_dataset(dataset_15)
dataset_28_1 = loadmat("28kPa/12-2910/data_graphs.mat")
dataset_28_1 = format_dataset(dataset_28_1)
dataset_28_2 = loadmat("28kPa/14-2910/data_graphs.mat")
dataset_28_2 = format_dataset(dataset_28_2)
dataset_28_3 = loadmat("28kPa/13-2910/data_graphs.mat")
dataset_28_3 = format_dataset(dataset_28_3)
a = 1.5e-6
def F_corr(alpha,z):
return -alpha * D(z) * 4e-21 * D_1_prime(z)
def D_1_prime(z):
return (2 * r * (2 * r**2 + 12 * r * z + 21 * z**2))/(2 * r** + 9 * r * z + 6 * z**2)**2
def D(z):
return (6*z**2 + 9*r*z + 2*r**2) / (6*z**2 + 2 * r * z)
def F_corr(alpha,z):
return -alpha * D(z) * 4e-21 * D_1_prime(z)
def F_corr(alpha,z):
return - 4e-21 * alpha *(42*a*z*z + 24*a*a*z + 4*a*a*a)/(36*(z**4) + 66*a*(z**3) + 30*a*a*z*z + 4*(a**3)*z)
fig = plt.figure(figsize=((cm2inch(16),cm2inch(8))))
plt.plot(dataset_15["z_F_EHD_exp"][dataset_15["z_F_EHD_exp"]>4e-8],dataset_15["F_EHD_exp"][dataset_15["z_F_EHD_exp"]>4e-8]*1e15 + 1*1e15*F_corr(1,dataset_15["z_F_EHD_exp"][dataset_15["z_F_EHD_exp"]>4e-8]), "o", label="$G = 15$ kPa")
plt.plot(dataset_15["z_F_EHD_th"][dataset_15["z_F_EHD_th"] > 4e-8],dataset_15["F_EHD_th"][dataset_15["z_F_EHD_th"] > 4e-8]*1e15 , color="tab:blue")
plt.plot(dataset_28_1["z_F_EHD_exp"][dataset_28_1["z_F_EHD_exp"] > 4e-8],dataset_28_1["F_EHD_exp"][dataset_28_1["z_F_EHD_exp"] > 4e-8]*1e15 + 1*1e15*F_corr(1,dataset_28_1["z_F_EHD_exp"][dataset_28_1["z_F_EHD_exp"] > 4e-8]) , "o", label="$G= 28$ kPa")
ax=plt.gca()
ax.loglog(dataset_28_1["z_F_EHD_th"][dataset_28_1["z_F_EHD_th"] > 4e-8],dataset_28_1["F_EHD_th"][dataset_28_1["z_F_EHD_th"] > 4e-8]*1e15, color="tab:orange")
plt.plot(dataset_28_2["z_F_EHD_exp"][dataset_28_2["z_F_EHD_exp"] > 4e-8],dataset_28_2["F_EHD_exp"][dataset_28_2["z_F_EHD_exp"] > 4e-8]*1e15 +1* 1e15*F_corr(1,dataset_28_2["z_F_EHD_exp"][dataset_28_2["z_F_EHD_exp"] > 4e-8]), "o",color="tab:orange")
from mpltools import annotation
annotation.slope_marker((4e-7, 0.5), (-5, 2), ax=ax, invert=True, size_frac=0.12, text_kwargs={"usetex":True})
plt.legend()
plt.ylabel("$F_\mathrm{NC} $ (fN)")
plt.xlabel("$z$ ($\mathrm{\mu}$m)")
plt.ylim([1e-1,None])
plt.xlim([0.4e-7,1e-6])
plt.tight_layout(pad=0.2)
plt.savefig("EHD_force.pdf")
fig = plt.figure(figsize=((cm2inch(6*1.68),cm2inch(6))))
plt.plot(dataset_15["z_F_EHD_exp"][dataset_15["z_F_EHD_exp"]>4e-8],(dataset_15["F_EHD_exp"][dataset_15["z_F_EHD_exp"]>4e-8] + F_corr(1,dataset_15["z_F_EHD_exp"][dataset_15["z_F_EHD_exp"]>4e-8]))*15e3*np.sqrt(1.5e-6)/(0.001**2), "o", label="$G=15$ kPa")
plt.gca()
plt.plot(dataset_28_1["z_F_EHD_exp"][dataset_28_1["z_F_EHD_exp"] > 4e-8],(dataset_28_1["F_EHD_exp"][dataset_28_1["z_F_EHD_exp"] > 4e-8] + F_corr(1,dataset_28_1["z_F_EHD_exp"][dataset_28_1["z_F_EHD_exp"] > 4e-8])) *28e3*np.sqrt(1.5e-6)/(0.001**2), "o", label="$G=28$ kPa")
plt.loglog(dataset_28_1["z_F_EHD_th"][dataset_28_1["z_F_EHD_th"] > 4e-8],(dataset_28_1["F_EHD_th"][dataset_28_1["z_F_EHD_th"] > 4e-8] )*28e3*np.sqrt(1.5e-6)/(0.001**2), color="black")
plt.plot(dataset_28_2["z_F_EHD_exp"][dataset_28_2["z_F_EHD_exp"] > 4e-8],(dataset_28_2["F_EHD_exp"][dataset_28_2["z_F_EHD_exp"] > 4e-8] +F_corr(1,dataset_28_2["z_F_EHD_exp"][dataset_28_2["z_F_EHD_exp"] > 4e-8]))*28e3*np.sqrt(1.5e-6)/(0.001**2), "d",color="tab:orange")
annotation.slope_marker((3.5e-7, 4e-8), (-5, 2), ax=plt.gca(), invert=True, size_frac=0.12)
plt.legend()
plt.ylabel("$F_\mathrm{NC} Ga^{\\frac{1}{2}} \eta ^{-2}$ ($\mathrm{ m^{5/2} s ^{-2}}$)")
plt.xlabel("$z$ ($\mathrm{\mu}$m)")
plt.ylim([1e-8,None])
plt.xlim([4e-8,1e-6])
plt.tight_layout(pad=0.2)
plt.savefig("EHD_force_rescale.svg")
fig = plt.figure(figsize=((cm2inch(16),cm2inch(8))))
plt.plot(dataset_15["z_F_EHD_exp"][dataset_15["z_F_EHD_exp"]>4e-8],dataset_15["F_EHD_exp"][dataset_15["z_F_EHD_exp"]>4e-8]*1e15*15e3*np.sqrt(1.5e-6), "o", label="15 kPa")
#plt.plot(dataset_15["z_F_EHD_th"][dataset_15["z_F_EHD_th"] > 4e-8],dataset_15["F_EHD_th"][dataset_15["z_F_EHD_th"] > 4e-8]*1e15 + 1.5, color="tab:blue")
plt.plot(dataset_28_1["z_F_EHD_exp"][dataset_28_1["z_F_EHD_exp"] > 4e-8],dataset_28_1["F_EHD_exp"][dataset_28_1["z_F_EHD_exp"] > 4e-8]*1e15*28e3*np.sqrt(1.5e-6), "o", label="28 kPa")
plt.loglog(dataset_28_1["z_F_EHD_th"][dataset_28_1["z_F_EHD_th"] > 4e-8],dataset_28_1["F_EHD_th"][dataset_28_1["z_F_EHD_th"] > 4e-8]*1e15*28e3*np.sqrt(1.5e-6) + 1.5*28e3*np.sqrt(1.5e-6), color="black")
plt.plot(dataset_28_2["z_F_EHD_exp"][dataset_28_2["z_F_EHD_exp"] > 4e-8],dataset_28_2["F_EHD_exp"][dataset_28_2["z_F_EHD_exp"] > 4e-8]*1e15*28e3*np.sqrt(1.5e-6), "d", label="28 kPa",color="tab:orange")
plt.plot(dataset_28_3["z_F_EHD_exp"][dataset_28_3["z_F_EHD_exp"] > 4e-8],dataset_28_3["F_EHD_exp"][dataset_28_3["z_F_EHD_exp"] > 4e-8]*1e15*28e3*np.sqrt(1.5e-6), "s", label="28 kPa",color="tab:orange")
plt.legend()
plt.ylabel("$F_{NC} E\sqrt{a} $ ($\mathrm{fN^2 m^{3/2}}$)")
plt.xlabel("$z$ ($\mathrm{\mu}$m)")
#plt.ylim([-20,100])
#lt.xlim([1e-2,10])v
plt.savefig("EHD_force_rescale.pdf")
| 0.310904 | 0.585931 |
# Exploring Notes & Queries
This chapter, and several following ones, will describe how to create various search context for 19th century issues of *Notes & Queries*. These include:
- a monolithic PDF of index issues up to 1900;
- a searchable database of index issues up to 1900;
- a full text searchable database of non-index issues up to 1900.
Original scans of the original publication, as well as automatically extracted search text, are available, for free, from the Internet Archive.
## Working With Documents From the Internet Archive
The Internet Archive – [`archive.org`](https://archive.org/) – is an incredible resource. Amongst other things, it is home to a large number of out-of-copyright digitised books scanned by the Google Book project as well as other book scanning initiatives.
In this unbook, I will explore various ways in which can build tools around the Internet Archive and documents retrieved from it.
## Searching the Internet Archive
Many people will be familiar with the web interface to the [Internet Archive](https://archive.org) (and I suspect many more are not aware of the existence of the Internet Archive at all). This provides tools for discovering documents available in the archive, previewing the scanned versions of them, and even searching within them.
At times, the search inside a book can be a bit hit and miss, in part depending on the quality of the scanned images and the ability of the OCR tools - where "OCR" stands for "optical character recognition" - to convert the pictures of text into actual text. Which is to say, *searchable* text.
One of the advantages of creating our own database is that as well as having the corpus available locally, we can use various fuzzy search tools to find partial matches to text to supplement our full text search activities.
To work with the archive, we'll use the Python programming language. This lets us write instructions for our machine helpers to follow. One of the machine helpers comes in the form of the [`internetarchive` Python package](https://archive.org/services/docs/api/internetarchive/index.html), a collection of routines that can access the Internet Archive at the programming, rather than human user interface, level.
*The human level interface simply provides graphical tools that we can understand, such as menu items and toolbar buttons. Selecting or clicking these simply invokes machine level commands in a useable-for-us way. Writing program code lets us call those commands directly, in a textual way, rather than visually, by clicking menu items and buttons. Copying and pasting simple text instructions that can be used to perform a particular function is often quite straightforward. Modifying such commands may also be relatively straightforward. (For example, given a block of code that downloads a file from a web location using code of the form `download_file("https://example.com/this_file.pdf")`, you could probably work out how to download a file from `http://another.example.com/myfile.pdf`.) Creating graphical user interfaces is hard. Graphical user interfaces also constrains users to using just the functions and features that the designers and developers of the user interface chose to support in the user interface, in just the way that the user interface allows. Being able to instruct a machine using code, even copied and pasted code, gives the end-use far more power over the machine.*
Within any particular programming language, *packages* are often used to bundle together various tools and functions that can be used to support particular activities or tasks, or work with particular resources or resource types.
One of the most useful tools within the Internet Archive package is the `search_items()` function, which lets us search the Internet Archive.
```
# If we haven't already installed the package into our computing environment,
# we need to download it and install it.
#%pip install internetarchive
# Load in a function to search the archive
from internetarchive import search_items
# We are going to build up a list of search results
items = []
```
### Item Metadata
At the data level, the Internet Archive has *metadata*, or "data about data" that provides key information or summary information about each data record. For example, works can be organised as part of different collections via `collection` elements such as `collection:"pub_notes-and-queries"`.
For periodicals, there may also be a publication identifier associated with the periodical (for example, `sim_pubid:1250`) or metadata identifying which *volume* or *issue* a particular edition of a periodical may be.
In the following bit of code, we search over the *Notes & Queries* collection, retrieving data about each item in the collection.
This is quite a large collection, so to run a query that retrieves all the items in it may take a considerable amount of time. Instead, we can limit the search to issues published in a particular year, and further limit the query to only retrieve a certain number of records.
```
# We can use a programming loop to search for items, iterate through the items
# and retrieve a record for each one
# The enumerate() command will loop trhough all the items, returnin a running count of items
# returned, as well as each separate item
# The count starts at 0...
for count, item in enumerate(search_items('collection:"pub_notes-and-queries" AND year:1867').iter_as_items()):
# Display thecount, the item identifier and title
print(count, item.identifier, item.metadata['title'])
# If we see item with count value of at least 3, which is to say, the fourth item,
# (we start counting at zero, remember...)
if count >= 3:
# Then break out of this loop
break
```
As well as the "offical" collection, some copies of *Notes and Queries* from other providers are also available in the Internet Archive. For example, there are some submissions from *Project Gutenberg*.
The following retrieves an item obtained from the `gutenberg` collection, which is to say, *Project Gutenberg*, and previews its metadata:
```
from internetarchive import get_item
# Retrieve an item from its unique identifier
item = get_item('notesandqueriesi13536gut')
# And display its metadata
item.metadata
```
The items in the `pub_notes-and-queries` collection have much more metadata available, including `volume` and `issue` data, and the identifiers for the `previous` and `next` issue.
In some cases, the identifier values may be human readable, if you look closely enough. For example, *Notes and Queries* was published weekly, typically with two volumes per year, and an index for each. In the `pub_notes-and-queries` collections, the identifier for Volume 11, issue 262, published on January 5th, 1867, is `sim_notes-and-queries_1867-01-05_11_262`; and the identifier for the index of volume 12, published in throughout the second half of 1867, is `sim_notes-and-queries_1867_12_index`.
### Available Files
As well as the data record, certain other files may be associated with that item such as PDF scans, or files containing the raw scanned text of the document.
We have already seen how we can retrieve an item given it's identifier, but let's see it in action again:
```
item = get_item("sim_notes-and-queries_1867_12_index")
item.metadata['title'], item.identifier
```
We can make a call from this data item to return a list of the files associated with that item, and display their file formats:
```
for file_item in item.get_files():
print(file_item.format)
```
For this item, then, we can get a PDF document, a file containing the search text, a record with information about page numbers, an XML version of the original scanned version, some image scans, and various other things containing who knows what!
### A Complete List of *Notes & Queries* Issues
To help us work with the *pub_notes-and-queries* collection, let's construct a local copy of the most important metadata associated with each item in the collection, specifically the item identifier, date and title, as well as the volume and issue. (*Notes and Queries* also has a higher level of organisation, a *Series*, which means that volume and issue numbers can actually recycle, so by itself, a particular `(volume, issue)` pair does not identify a unique item, but a `(series, volume, issue)` or `(year, volume, issue)` triple does.)
For convenience, we might also collect the *previous* and *next* item identifiers, as well as a flag that tells us whether access is restricted or not. (For 19th century editions, there are no restrictions; but for more recent 20th century editions, access may be limited to library shelf access).
As we construct various tools for working with the Internet Archive and various files downloaded from it, it will be useful to also save those tools in a way that we can can make use of them.
The Python programming language supports a simple mechanism for bundling files into "packages" simply by including files in directory that is marked as a package directory. The simplest way to mark a directory as a Python package is simple to create an empty file called `__init__.py` inside it.
So let's create a package called `ia_utils` by creating a directory of that name containing an empty `__init__.py` file:
```
from pathlib import Path
# Create the directory if it doesnlt already exist
ia_utils = Path("ia_utils")
ia_utils.mkdir(exist_ok=True)
# Create the blank file
Path( ia_utils / "__init__.py" ).touch()
```
```{note}
The `pathlib` package contains powerful tools for working with directories, files, and file paths.
```
The following cell contains a set of instructions bundled together to define a *function* under a unique function name. Functions provide us with a shorthand way of writing a set of instructions once, then calling on them repeatedly via their function name.
In particular, the function takes in an item metadata record, tidies it up a little and returns just the fields we are interested in.
In the following cell, we use some magic to write the contents of the cell to a package file; in the next cell after that, we import the function from the file. This provides us with a convenient way of saving code to a file that we can also reuse elsewhere.
```
%%writefile ia_utils/out_ia_metadata.py
import csv
def out_ia_metadata(item):
"""Retrieve a subset of item metadata and return it as a list."""
# This is a nested function that looks up piece of metadata if it exists
# If it doesn't exist, we set it to ''
def _get(_item, field):
return _item[field] if field in _item else ''
#item = get_item(i.identifier)
identifier = item.metadata['identifier']
date = _get(item.metadata, 'date')
title = _get(item.metadata, 'title')
volume =_get(item.metadata, 'volume')
issue = _get(item.metadata, 'issue')
prev_ = _get(item.metadata, 'previous_item')
next_ = _get(item.metadata, 'next_item')
restricted = _get(item.metadata,'access-restricted-item')
return [identifier, date, title, volume, issue, prev_, next_, restricted]
```
Now we can import the function from the package. And so can other notebooks.
```
from ia_utils.out_ia_metadata import out_ia_metadata
```
```{admonition} Tracking Updates to the Function
:class: dropdown
If we update the function and rewrite the file, the `from...import..` line will not normally reload the (updated) function if the function has already been imported.
There are two ways round this:
- load the file in and run it, rather than importing the package, using a magic command of the form `%run -i ia_utils/out_ia_metadata.py`
- configure the notebook at the start by running `%load_ext autoreload ; %autoreload 2` (see the [documentation](https://ipython.readthedocs.io/en/stable/config/extensions/autoreload.html)).
```
Here's what the data retrieved from an item record by the `out_ia_metadata` function looks like:
```
# Get an item record form its identifier
item = get_item("sim_notes-and-queries_1867_12_index")
# Display the key metadata
out_ia_metadata(item)
```
We can now build up a list of lists containing the key metadata for all editions of *Notes of Queries* in the `pub_notes-and-queries` collection.
Our recipe will proceed in the following three steps:
- search for all the items in the collection;
- build up a list of records where item contains the key metadata, extracted from the full record using the `out_ia_metadata()` function;
- open a file (*nandq_internet_archive.txt*), give it a column header line, and write the key metadata records to it, one record per line.
The file will be written in "CSV" format (comma separarated variable), a simple text format for describing tabular data. CSV files can be read by spreadsheet applications, as well as other tools, and use comma separators to identify "columns" of information in each row.
```
# The name of the file we'll write our csv data to
csv_fn = "nandq_internet_archive.txt"
```
The file takes quite a long time to assemble (we need to download several thousand metadata records), so we only want to do it once.
So let's check to see if the file exists (if it does, we won't try to recreate it:
```
from pathlib import Path
csv_file_exists = Path(csv_fn).is_file()
```
Conveniently, identifiers for all the issues of *Notes and Queries* held by the Internet Archive can be retrieved via the `pub_notes-and-queries` collection.
The return object is an iterator with individual results that take the form `{'identifier': 'sim_notes-and-queries_1849-11-03_1_1'}` and from which we can obtain unique identifiers:
```
# Find records for all items in the collection
items = search_items('collection:"pub_notes-and-queries"')
```
The following incantation constructs one list from the members of another. In particular, we iterate through each item in the `pub_notes-and-queries`, extract the identifier, retrieve the corresponding metadata record (`get_item()`), create our own corresponding metadata record (`out_ia_metadata()`) and add it to a new list.
In all, there are several thousand records to download, and each takes a noticeable time, so rather than just sitting watching a progress bar for an hour, go and grab a meal rather than a coffee...
```
# The tqdm package provides a convenient progress bar
# for tracking progress through looped actions
from tqdm.notebook import tqdm
# If a local file containing the data doesn't already exist,
# then grab the data...
if not csv_file_exists:
# Our list of custom metadata records
csv_items = []
for i in tqdm(items):
id_val = i["identifier"]
metadata_record = get_item(id_val)
custom_metadata_record = out_ia_metadata( metadata_record )
csv_items.append( custom_metadata_record )
# We should perhaps incrementally write the CSV file as we go along
# or incrementally save the data to a simple local database
# If something goes wrong during the downloads, then at least
# we won;t have lost everything..
```
We can now open the CSV file and write the data to it:
```
# If a local file containing the data doesn't already exist,
# then grab the data...
if not csv_file_exists:
with open(csv_fn, 'w') as outfile:
print(f"Writing data to file {csv_fn}")
# Create a "CSV writer" object that can write to the file
csv_write = csv.writer(outfile)
# Write a header row at the top of the file
csv_write.writerow(['id','date','title','vol','iss','prev_id', 'next_id','restricted'])
# Then write out list of essential metadata items out, one record per row
csv_write.writerows(csv_items)
# Update the file exists flag
csv_file_exists = Path(csv_fn).is_file()
```
We can use a simple Linux command line tool (`head`) to show the top five lines of the file:
```
!head -n 5 nandq_internet_archive.txt
```
So, with some idea of what's available to us, data wise, and file wise, what can we start to do with it?
## Generating a Monolithic PDF Index for *Notes & Queries* Up To 1900
If we want to search for items in *Notes and Queries* "manually", one of the most effective ways is to look up items in the volume indexes. With two volumes a year, this means checking almost 100 separate documents if we want to look up 19th century references. (That's not quite true: from the 1890s, indexes were produced that started to to aggregate indices over several years.)
So how might we go about producing a single index PDF for 19th c. editions of *Notes & Queries*? As a conjoined set of original index PDFs, this wouldn't provide us with unified index terms - a search on an index item would return separate entries for each volume index in which the term appeared – but it would mean we only needed to search one PDF document.
We'll use the Python `csv` package to simplify saving and load the data:
```
import csv
```
To begin with, we can load in our list of *Notes and Queries* record data downloaded from the Internet Archive.
```
%%writefile ia_utils/open_metadata_records.py
import csv
# Specify the file name we want to read data in from
def open_metadata_records(fn='nandq_internet_archive.txt'):
"""Open and read metadata records file."""
with open(fn, 'r') as f:
# We are going to load the data into a data structure known as a dictionary, or dict
# Each item in the dictionary contains several elements as `key:value` pairs
# The key matches the column name in the CSV data file,
# along with the corresponding value in a given item row
# Read the data in
csv_data = csv.DictReader(f)
# And convert it to a list of data records
data_records = list(csv_data)
return data_records
# Import that function from the package we just wrote it to
from ia_utils.open_metadata_records import open_metadata_records
```
Let's grab the metadata records from our saved file:
```
data_records = open_metadata_records()
# Preview the first record (index count starts at 0)
# The object returned is a dictionary / dict
data_records[0]
```
## Populating a Database With Record Metadata
Let's start by creating a table in the database that can store our metadata data records, as loaded in from the data file.
```
from sqlite_utils import Database
db_name = "nq_demo.db"
# While developing the script, recreate database each time...
db = Database(db_name, recreate=True)
%%writefile ia_utils/create_db_table_metadata.py
import datetime
def create_db_table_metadata(db, drop=True):
# If we want to remove the table completely, we can drop it
if drop:
db["metadata"].drop(ignore=True)
db["metadata"].create({
"id": str,
"date": str,
"datetime": datetime.datetime, # Use an actual time representation
"series": str,
"vol": str,
"iss": str,
"title": str,
"next_id": str,
"prev_id": str,
"is_index": bool, # Is the record an index record
"restricted": str, # should really be boolean
}, pk=("id"))
```
Now we can load the function back in from out package and call it:
```
from ia_utils.create_db_table_metadata import create_db_table_metadata
create_db_table_metadata(db)
```
We need to do a little tidying of the records, but then we can add them directly to the database:
```
%%writefile ia_utils/add_patched_metadata_records_to_db.py
from tqdm.notebook import tqdm
import dateparser
def add_patched_metadata_records_to_db(db, data_records):
"""Add metadata records to database."""
# Patch records to include a parsed datetime element
for record in tqdm(data_records):
# Parse the raw date into a date object
# Need to handle a YYYY - YYYY exception
# If we detect this form, use the last year for the record
if len(record['date'].split()[0]) > 1:
record['datetime'] = dateparser.parse(record['date'].split()[-1])
else:
record['datetime'] = dateparser.parse(record['date'])
record['is_index'] = 'index' in record['title'].lower() # We assign the result of a logical test
# Add records to the database
db["metadata"].insert_all(data_records)
```
Let's call that function and add our metadata data records:
```
from ia_utils.add_patched_metadata_records_to_db import add_patched_metadata_records_to_db
add_patched_metadata_records_to_db(db, data_records)
```
We can then query the data, for example return the first rows:
```
from pandas import read_sql
q = "SELECT * FROM metadata LIMIT 5"
read_sql(q, db.conn)
```
Or we could return the identifiers for index issues between 1875 and 1877:
```
q = """
SELECT id, title
FROM metadata
WHERE is_index = 1
-- Extract the year
AND strftime('%Y', datetime) >= '1875'
AND strftime('%Y', datetime) <= '1877'
"""
read_sql(q, db.conn)
```
By inspection of the list of index entries, we note that at some point cumulative indexes over a set of years, as well as volume level indexes, were made available. Cumulative indexes include:
- Notes and Queries 1892 - 1897: Vol 1-12 Index
- Notes and Queries 1898 - 1903: Vol 1-12 Index
- Notes and Queries 1904 - 1909: Vol 1-12 Index
- Notes and Queries 1910 - 1915: Vol 1-12 Index
In this first pass, we shall just ignore the cumulative indexes.
At this point, it is not clear where we might reliably obtain the series information from.
To make the data easier to work with, we can parse the date as a date thing (technical term!;-) using tools in the Python `dateparser` package:
```
import dateparser
```
The parsed data provides ways of comparing dates, extracting month and year, and so on.
```
indexes = []
# Get index records up to 1900
max_year = 1900
for record in data_records:
# Only look at index records
# exclude cumulative indexes
if 'index' in record['id'] and "cumulative" not in record['id']:
# Need to handle a YYYY - YYYY exception
# If we detect it, ignore it
if len(record['date'].split()) > 1:
continue
# Parse the year into a date object
# Then filter by year
if dateparser.parse(record['date'].split()[0]).year >= max_year:
break
indexes.append(record)
# Preview the first three index records
indexes[:3]
```
To generate the complete PDF index, we need to do several things:
- iterate through the list of index records;
- for each one, download the associated PDF to a directory;
- merge all the downloaded files into a single PDF;
- optionally, delete the original PDF files.
### Working With PDF Files Downloaded from the Internet Archive
We can download files from the Internet Archive using the `internetarchive.download()` function. This takes a list of items via a `formats` parameter for the files we want to download. For example, we might want to download the "Text PDF" (a PDF file with full text search), or a simple text file containing just the OCR captured text (`OCR Search Text`), or both.
We can also specify the directory into which the files are downloaded.
Let's import the packages that help simplify this task, and create a path to our desired download directory:
```
# Import the necessary packages
from internetarchive import download
```
To keep our files organised, we'll create a directory into which we can download the files:
```
# Create download dir file path
dirname = 'ia-downloads'
p = Path(dirname)
```
One of the ways we can work with the data is to process it using Python programming code.
For example, we can iterate through the index records and download the required files:
```
# Use tqdm for provide a progress bar
for record in tqdm(indexes):
_id = record['id']
# Download PDF - this may take time to retrieve / download
# This downloads to a directory with the same name as the record id
# The file name is akin to ${id}.pdf
download(_id, destdir=p, silent = True,
formats=["Text PDF", "OCR Search Text"])
```
To create single monolithic PDF, we can use another fragment of code to iterate through the downloaded PDF files, adding each one to a single merged PDF file object. We can also create and insert a reference page between each of the original documents to provide provenance if the is no date on the index pages.
Let's start by seeing how to create a simple PDF page. The `reportlab` Python package provides various tools for creating simple PDF documents:
```
#%pip install --upgrade reportlab
from reportlab.pdfgen.canvas import Canvas
```
For example, we can create a simple single page document that we can add index metadata to and then insert in between the pages of each index issue:
```
# Create a page canvas
test_pdf = "test-page.pdf"
canvas = Canvas(test_pdf)
# Write something on the page at a particular location
# In this case, let's use the title from the first index record
txt = indexes[0]['title']
# Co-ordinate origin is bottom left of the page
# Scale is points, where 72 points = 1 inch
canvas.drawString(72, 10*72, txt)
# Save the page
canvas.save()
```
Now we can preview the test page:
```
from IPython.display import IFrame
IFrame(test_pdf, width=600, height=500)
```
A simple function lets us generate a simple page rendering a short text string:
```
def make_pdf_page(txt, fn="test_pdf.pdf"):
""""""
canvas = Canvas(fn)
# Write something on the page at a partcular location
# Co-ordinate origin is bottom left of the page
# Scale is points, where 72 points = 1 inch
canvas.drawString(72, 10*72, txt)
# Save the page
canvas.save()
return fn
```
Let's now create our monolithic index with metadata page inserts.
The `PyPDF2` package contains various tools for splitting and combining PDF documents:
```
from PyPDF2 import PdfFileReader, PdfFileMerger
```
We can use it merge our separate index cover page and index issue documents, for example:
```
# Create a merged PDF file creating object
output = PdfFileMerger()
# Generate a monolithic PDF index file by concatenating the pages
# from each individual PDF index file
# Use tqdm for provide a progress bar
for record in tqdm(indexes):
# Generate some metadata:
txt = record['title']
metadata_pdf = make_pdf_page(txt)
# Add this to the output document
output.append(metadata_pdf)
# Delete the metadata file
Path(metadata_pdf).unlink()
# Get the record ID
_id = record['id']
# Locate the file and merge it into the monolithic PDF
output.append((p / _id / f'{_id}.pdf').as_posix())
# Write merged PDF file
with open("notes_and_queries_big_index.pdf", "wb") as output_stream:
output.write(output_stream)
output = None
```
The resulting PDF document is a large document (about 100MB) that collects all the separate indexes in one place, although not as a single, *reconciled* index: if the same index terms exist in multiple index documents, there will be multiple occurrences of that term in the longer document.
However, if we do need a PDF reference to the index, it is useful to have to hand.
|
github_jupyter
|
# If we haven't already installed the package into our computing environment,
# we need to download it and install it.
#%pip install internetarchive
# Load in a function to search the archive
from internetarchive import search_items
# We are going to build up a list of search results
items = []
# We can use a programming loop to search for items, iterate through the items
# and retrieve a record for each one
# The enumerate() command will loop trhough all the items, returnin a running count of items
# returned, as well as each separate item
# The count starts at 0...
for count, item in enumerate(search_items('collection:"pub_notes-and-queries" AND year:1867').iter_as_items()):
# Display thecount, the item identifier and title
print(count, item.identifier, item.metadata['title'])
# If we see item with count value of at least 3, which is to say, the fourth item,
# (we start counting at zero, remember...)
if count >= 3:
# Then break out of this loop
break
from internetarchive import get_item
# Retrieve an item from its unique identifier
item = get_item('notesandqueriesi13536gut')
# And display its metadata
item.metadata
item = get_item("sim_notes-and-queries_1867_12_index")
item.metadata['title'], item.identifier
for file_item in item.get_files():
print(file_item.format)
from pathlib import Path
# Create the directory if it doesnlt already exist
ia_utils = Path("ia_utils")
ia_utils.mkdir(exist_ok=True)
# Create the blank file
Path( ia_utils / "__init__.py" ).touch()
The following cell contains a set of instructions bundled together to define a *function* under a unique function name. Functions provide us with a shorthand way of writing a set of instructions once, then calling on them repeatedly via their function name.
In particular, the function takes in an item metadata record, tidies it up a little and returns just the fields we are interested in.
In the following cell, we use some magic to write the contents of the cell to a package file; in the next cell after that, we import the function from the file. This provides us with a convenient way of saving code to a file that we can also reuse elsewhere.
Now we can import the function from the package. And so can other notebooks.
Here's what the data retrieved from an item record by the `out_ia_metadata` function looks like:
We can now build up a list of lists containing the key metadata for all editions of *Notes of Queries* in the `pub_notes-and-queries` collection.
Our recipe will proceed in the following three steps:
- search for all the items in the collection;
- build up a list of records where item contains the key metadata, extracted from the full record using the `out_ia_metadata()` function;
- open a file (*nandq_internet_archive.txt*), give it a column header line, and write the key metadata records to it, one record per line.
The file will be written in "CSV" format (comma separarated variable), a simple text format for describing tabular data. CSV files can be read by spreadsheet applications, as well as other tools, and use comma separators to identify "columns" of information in each row.
The file takes quite a long time to assemble (we need to download several thousand metadata records), so we only want to do it once.
So let's check to see if the file exists (if it does, we won't try to recreate it:
Conveniently, identifiers for all the issues of *Notes and Queries* held by the Internet Archive can be retrieved via the `pub_notes-and-queries` collection.
The return object is an iterator with individual results that take the form `{'identifier': 'sim_notes-and-queries_1849-11-03_1_1'}` and from which we can obtain unique identifiers:
The following incantation constructs one list from the members of another. In particular, we iterate through each item in the `pub_notes-and-queries`, extract the identifier, retrieve the corresponding metadata record (`get_item()`), create our own corresponding metadata record (`out_ia_metadata()`) and add it to a new list.
In all, there are several thousand records to download, and each takes a noticeable time, so rather than just sitting watching a progress bar for an hour, go and grab a meal rather than a coffee...
We can now open the CSV file and write the data to it:
We can use a simple Linux command line tool (`head`) to show the top five lines of the file:
So, with some idea of what's available to us, data wise, and file wise, what can we start to do with it?
## Generating a Monolithic PDF Index for *Notes & Queries* Up To 1900
If we want to search for items in *Notes and Queries* "manually", one of the most effective ways is to look up items in the volume indexes. With two volumes a year, this means checking almost 100 separate documents if we want to look up 19th century references. (That's not quite true: from the 1890s, indexes were produced that started to to aggregate indices over several years.)
So how might we go about producing a single index PDF for 19th c. editions of *Notes & Queries*? As a conjoined set of original index PDFs, this wouldn't provide us with unified index terms - a search on an index item would return separate entries for each volume index in which the term appeared – but it would mean we only needed to search one PDF document.
We'll use the Python `csv` package to simplify saving and load the data:
To begin with, we can load in our list of *Notes and Queries* record data downloaded from the Internet Archive.
Let's grab the metadata records from our saved file:
## Populating a Database With Record Metadata
Let's start by creating a table in the database that can store our metadata data records, as loaded in from the data file.
Now we can load the function back in from out package and call it:
We need to do a little tidying of the records, but then we can add them directly to the database:
Let's call that function and add our metadata data records:
We can then query the data, for example return the first rows:
Or we could return the identifiers for index issues between 1875 and 1877:
By inspection of the list of index entries, we note that at some point cumulative indexes over a set of years, as well as volume level indexes, were made available. Cumulative indexes include:
- Notes and Queries 1892 - 1897: Vol 1-12 Index
- Notes and Queries 1898 - 1903: Vol 1-12 Index
- Notes and Queries 1904 - 1909: Vol 1-12 Index
- Notes and Queries 1910 - 1915: Vol 1-12 Index
In this first pass, we shall just ignore the cumulative indexes.
At this point, it is not clear where we might reliably obtain the series information from.
To make the data easier to work with, we can parse the date as a date thing (technical term!;-) using tools in the Python `dateparser` package:
The parsed data provides ways of comparing dates, extracting month and year, and so on.
To generate the complete PDF index, we need to do several things:
- iterate through the list of index records;
- for each one, download the associated PDF to a directory;
- merge all the downloaded files into a single PDF;
- optionally, delete the original PDF files.
### Working With PDF Files Downloaded from the Internet Archive
We can download files from the Internet Archive using the `internetarchive.download()` function. This takes a list of items via a `formats` parameter for the files we want to download. For example, we might want to download the "Text PDF" (a PDF file with full text search), or a simple text file containing just the OCR captured text (`OCR Search Text`), or both.
We can also specify the directory into which the files are downloaded.
Let's import the packages that help simplify this task, and create a path to our desired download directory:
To keep our files organised, we'll create a directory into which we can download the files:
One of the ways we can work with the data is to process it using Python programming code.
For example, we can iterate through the index records and download the required files:
To create single monolithic PDF, we can use another fragment of code to iterate through the downloaded PDF files, adding each one to a single merged PDF file object. We can also create and insert a reference page between each of the original documents to provide provenance if the is no date on the index pages.
Let's start by seeing how to create a simple PDF page. The `reportlab` Python package provides various tools for creating simple PDF documents:
For example, we can create a simple single page document that we can add index metadata to and then insert in between the pages of each index issue:
Now we can preview the test page:
A simple function lets us generate a simple page rendering a short text string:
Let's now create our monolithic index with metadata page inserts.
The `PyPDF2` package contains various tools for splitting and combining PDF documents:
We can use it merge our separate index cover page and index issue documents, for example:
| 0.826922 | 0.991067 |
# Working with Multi-Output models
In this notebook, we'll be performing multi-output tasks with the help of DFFML. DFFML makes it so that users can use a range of different models to perform multi-output tasks with the usual worklow, but instantiating models with a list of targets/labels!
**Import Packages**
Let us import dffml and other packages that we might need.
```
import asyncio
import nest_asyncio
import numpy as np
from sklearn.datasets import load_linnerud
from sklearn.datasets import make_multilabel_classification
from dffml import *
```
To use asyncio in a notebook, we need to use `nest_asycio.apply()`
```
nest_asyncio.apply()
```
## Multi-Output Regression
To perform multi-output regression tasks, we can select any scikit regression model and pass our multi-output dataset onto the model.
### Build our Dataset
For our dataset, we will be generating a random multi-output dataset
```
X, y = load_linnerud(return_X_y=True)
data = np.concatenate((X, y), axis=1)
print(X.shape, y.shape)
print(data)
data = [
{"x1": row[0], "x2": row[1], "x3": row[2], "y1": row[3], "y2": row[4], "y3": row[5]}
for row in data
]
data
```
### Instantiate our Model with parameters
Dffml makes it quite easy to load multiple models dynamically using the `Model.load()` function. All the entrypoints for models available in DFFML can be found at the [Model Plugins Page](../../plugins/dffml_model.rst).
**Note:** All(and only) [Scikit Regressors and Classifiers](../../plugins/dffml_model.rst#dffml-model-scikit) support multioutput.
After that, you just have to parameterize the loaded model and it is ready to train!
Note that we will set `predict` in the model config to `Features`, a list of features ie. the targets, instead of a single feature. This sets the output targets in the model causing it to work as a Multi-Output model.
```
ScikitRidgeModel = Model.load("scikitridge")
features = Features(
Feature("x1", int, 1), Feature("x2", int, 1), Feature("x3", int, 1),
)
predict_features = Features(
Feature("y1", int, 1), Feature("y2", int, 1), Feature("y3", int, 1),
)
multi_reg_model = ScikitRidgeModel(
features=features, predict=predict_features, location="scikitridgemulti",
)
print(type(predict_features))
```
### Train our Model
Finally, our model is ready to be trained using the `high-level` API. Let's make sure to pass each record as a parameter by simply using the unpacking operator(*).
```
await train(multi_reg_model, *data)
```
### Test our Models
To test our model, we'll use the `accuracy()` function in the `high-level` API.
Just like models, the scorers can be dynamically loaded in a similar fashion.
We will evaluate our model using the Mean Squared Error Scorer by passing `"meansqrerr"` to `AccuracyScorer.load()`, which is an entrypoint for a scikit scorer.
All the available entrypoints for scorers can be found at the [Scorers Plugin Page](../../plugins/dffml_accuracy.rst).
```
MeanSquaredError = AccuracyScorer.load("meansqrerr")
scorer = MeanSquaredError()
print("Score:", await accuracy(multi_reg_model, scorer, predict_features, *data))
```
### Make predictions using our Model.
Let's make predictions and see what they look like for each model using the `predict` function in the `high-level` API.
Note that the `predict` function returns an asynciterator of a `Record` object that contains a tuple of record.key, features and predictions.
```
async for i, features, prediction in predict(multi_reg_model, *data):
print(prediction)
```
## Multi-Output Classifier
To perform multi-output classification tasks, we can select any scikit classification model and pass our multi-output dataset onto the model.
### Build our Dataset
We again utilize sklearn to build our multi-output dataset for our classification task.
```
X, y = make_multilabel_classification(n_classes=3, n_features=3, random_state=1)
data = np.concatenate((X, y), axis=1)
data = [
{"x1": row[0], "x2": row[1], "x3": row[2], "y1": row[3], "y2": row[4], "y3": row[5]}
for row in data
]
data[0:5]
```
### Instantiate our Model with parameters
Dffml makes it quite easy to load multiple models dynamically using the `Model.load()` function. All the entrypoints for models available in DFFML can be found at the [Model Plugins Page](../../plugins/dffml_model.rst). After that, you just have to parameterize the loaded model and it is ready to train!
Note that we will set `predict` in the model config to `Features`, a list of features ie. the targets, instead of a single feature. This sets the output targets in the model causing it to work as a Multi-Output model.
```
ScikitETCModel = Model.load("scikitetc")
features = Features(
Feature("x1", int, 1), Feature("x2", int, 1), Feature("x3", int, 1),
)
predict_features = Features(
Feature("y1", int, 1), Feature("y2", int, 1), Feature("y3", int, 1),
)
multi_classif_model = ScikitETCModel(
features=features, predict=predict_features, location="scikitetcmulti",
)
```
### Train our Model
Finally, our model is ready to be trained using the `high-level` API. Let's make sure to pass each record as a parameter by simply using the unpacking operator(*).
```
await train(multi_classif_model, *data)
```
### Test our Models
To test our model, we'll again use the `accuracy()` function in the `high-level` API.
This time, we evaluate our model using the Accuracy Scorer by passing `"acscore"` to `AccuracyScorer.load()`, which is also an entrypoint for a scikit scorer.
All the available entrypoints for scorers can be found at the [Scorers Plugin Page](../../plugins/dffml_accuracy.rst).
```
Accuracy = AccuracyScorer.load("acscore")
scorer = Accuracy()
print("Accuracy:", await accuracy(multi_classif_model, scorer, predict_features, *data))
```
### Make predictions using our Model.
Let's make predictions and see what they look like using the `predict` function in the `high-level` API.
Note that the `predict` function returns an asynciterator of a `Record` object that contains a tuple of record.key, features and predictions.
```
async for i, features, prediction in predict(multi_classif_model, *data):
print(prediction)
```
|
github_jupyter
|
import asyncio
import nest_asyncio
import numpy as np
from sklearn.datasets import load_linnerud
from sklearn.datasets import make_multilabel_classification
from dffml import *
nest_asyncio.apply()
X, y = load_linnerud(return_X_y=True)
data = np.concatenate((X, y), axis=1)
print(X.shape, y.shape)
print(data)
data = [
{"x1": row[0], "x2": row[1], "x3": row[2], "y1": row[3], "y2": row[4], "y3": row[5]}
for row in data
]
data
ScikitRidgeModel = Model.load("scikitridge")
features = Features(
Feature("x1", int, 1), Feature("x2", int, 1), Feature("x3", int, 1),
)
predict_features = Features(
Feature("y1", int, 1), Feature("y2", int, 1), Feature("y3", int, 1),
)
multi_reg_model = ScikitRidgeModel(
features=features, predict=predict_features, location="scikitridgemulti",
)
print(type(predict_features))
await train(multi_reg_model, *data)
MeanSquaredError = AccuracyScorer.load("meansqrerr")
scorer = MeanSquaredError()
print("Score:", await accuracy(multi_reg_model, scorer, predict_features, *data))
async for i, features, prediction in predict(multi_reg_model, *data):
print(prediction)
X, y = make_multilabel_classification(n_classes=3, n_features=3, random_state=1)
data = np.concatenate((X, y), axis=1)
data = [
{"x1": row[0], "x2": row[1], "x3": row[2], "y1": row[3], "y2": row[4], "y3": row[5]}
for row in data
]
data[0:5]
ScikitETCModel = Model.load("scikitetc")
features = Features(
Feature("x1", int, 1), Feature("x2", int, 1), Feature("x3", int, 1),
)
predict_features = Features(
Feature("y1", int, 1), Feature("y2", int, 1), Feature("y3", int, 1),
)
multi_classif_model = ScikitETCModel(
features=features, predict=predict_features, location="scikitetcmulti",
)
await train(multi_classif_model, *data)
Accuracy = AccuracyScorer.load("acscore")
scorer = Accuracy()
print("Accuracy:", await accuracy(multi_classif_model, scorer, predict_features, *data))
async for i, features, prediction in predict(multi_classif_model, *data):
print(prediction)
| 0.45423 | 0.986533 |
# Nuclear Energetics
## Learning Objectives
- Define: exothermic and endothermic reactions
- Differentiate exothermic and endothermic reactions
- Calculate binding energies
- Recognize the noation for various binary reactions
- Recognize the physics in common binary reactions
- Define the relationship between the Q value, mass, and energy in a reaction
- Explain the role of energy conservation in binary reactions
- Explain the role of charge conservation in binary reactions
- Calculate Q values for various reactions
## Endothermic and Exothermic Reactions
In any reaction (nuclear, mechanical, chemical) energy can be either emitted or absorbed.
- When energy is emitted by the reaction, this is called **_exothermic_**.
- When energy is absorbed into the reaction, this is called **_endothermic_**.
If we consider the energy in relation to mass via Einstein's equivalence, then it's very clear that a change in the mass of reactants will result in a change in energy ($\Delta E = \Delta M c^2$).
\begin{align}
\mbox{reactants} &\rightarrow \mbox{products}\\
A + B + \cdots &\rightarrow C + D + \cdots\\
\Delta M = (M_A + M_B + \cdots) &- (M_C + M_D + \cdots)\\
\implies \Delta E &= \left[(M_A + M_B + \cdots) - (M_C + M_D + \cdots)\right]c^2\\
\end{align}
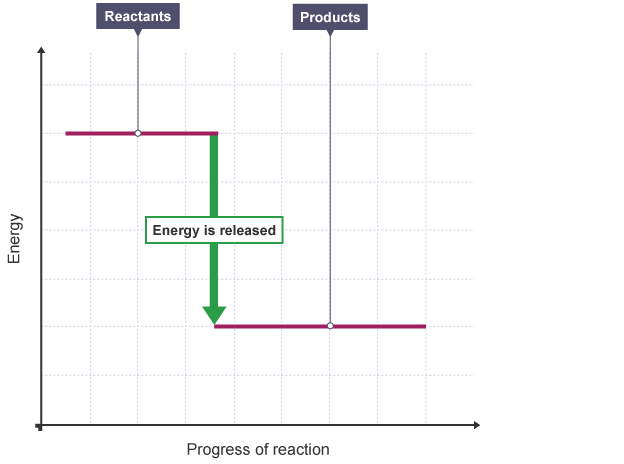
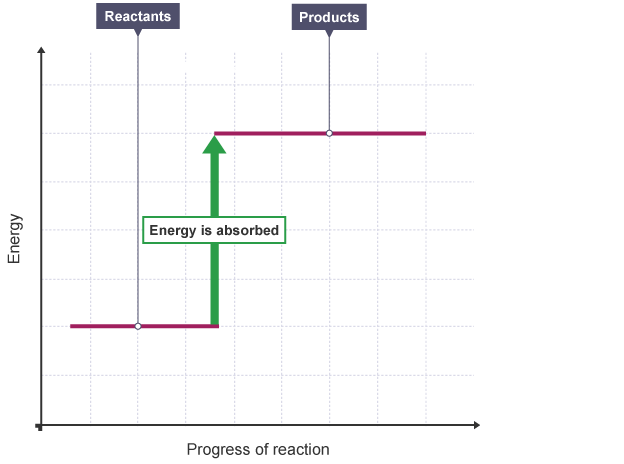
<center>(credit: BBC)</center>
## Exercise: Think-pair-share
Which of the following are exothermic and which are endothermic?
Easy ones:
- Boiling Water
- Lighting a match
- Freezing an ice cube
- Melting an ice cube
- Snow formation in the clouds
- Conversion of frost to water vapor on the grass
Harder ones:
- Formation of ion pairs (exothermic)
- Separation of ion pairs (endothermic)
The most important one:
- nuclear fission
# Binding Energy
For ease, let's imagine just two reactants and one product.
\begin{align}
\mbox{reactants} &\rightarrow \mbox{products}\\
A + B &\rightarrow C \\
\Delta M = (mass(A) + mass(B)) &- (mass(C))\\
\implies \Delta E &= \left[(mass(A) + mass(B) - (mass(C))\right]c^2\\
&= BE
\end{align}
## Nuclear and Atomic Masses
\begin{align}
M\left(^A_ZX\right) &= \mbox{rest mass of an atom}\\
m\left(^A_ZX\right) &= \mbox{rest mass of its nucleus}
\end{align}
All Z electrons must be bound to the atom, so the atomic and nuclear masses have the following relationship:
\begin{align}
M\left(^A_ZX\right) &=m\left(^A_ZX\right) + Zm_e - \frac{BE_{Ze}}{c^2}
\end{align}
Your book points out that it requires 13.6 eV to ionize the hydrogen atom. This electron binding energy represents a mass change of $BE_{1e/c^2} = 13.6 (eV)/9.315 \times 10^8 (eV/u) = 1.4 \times 10^{−8} u$.
## Binding Energy of the nucleus
A nucleus, with Z protons and N=A-Z neutrons will have a formation reaction that looks like:
\begin{align}
Z \mbox{ protons } + (A − Z) \mbox{ neutrons }\rightarrow \mbox{ nucleus}\left(^A_ZX\right) + BE.
\end{align}
It can be helpful to define the term $BE/c^2$:
\begin{align}
\frac{BE}{c^2} &=Zm_p +(A−Z)m_n − m\left(^A_ZX\right)\\
\end{align}
We can arrive at the nuclear binding energy from this:
\begin{align}
BE\left(^A_ZX\right) = \left[ZM(\left(^1_1H\right) + (A-Z)m_n - M\left(^A_ZX\right)\right]c^2
\end{align}
The above equation neglects the electron binding energies because:
- the terms tend to cancel
- electron binding energies are millions of times smaller than the nuclear binding energy.
### Exercise: Nuclear binding energy
From your book, example 4.1: What is the binding energy of an alpha particle?
\begin{align}
\frac{BE}{c^2} &=Zm_p +(A−Z)m_n − m\left(^A_ZX\right)\\
\implies\frac{BE}{c^2} &=\left[ZM(\left(^1_1H\right) + (A-Z)m_n - M\left(^A_ZX\right)\right]\\
&=\left[2M(\left(^1_1H\right) + 2m_n - M\left(^4_2He\right)\right]\\
&=\left[2\left(1.0078250\right) + 2\left(1.0086649\right) - 4.0026032\right]\\
\end{align}
Thus:
\begin{align}
BE\left(^4_2 He\right) &= \frac{BE}{c^2}\times 931.5 (MeV/u) \\
&= 28.30 MeV.
\end{align}
## Binding Energy per nucleon
The BE/A versus A curve immediately suggests two ways to extract energy from the nucleus.

Binding energy curve **(average binding energy per nucleon in MeV against number of nucleons in nucleus)** for a number of relatively common (abundant) isotopes (not chosen systematically; almost anything with an occurence of over .2 was chosen though a few exceptions are in there, such as U235). A few important ones for the purposes of nuclear fusion and nuclear fission are marked, as well as iron-56, which sits at the highest point on this graph and cannot yield energy from fusion or fission.
From your book (Shultis and Faw)
> Most isotopes of an element are radioactive, i.e., their nuclei are unstable. To better understand what makes a nucleus stable, it is instructive to plot the BE per nucleon, BE/A, of all the isotopes of an element. For example, if one calculates the BE/A for all the isotopes of calcium (Z = 20), as is done in Example 4.2 for one isotope, and plots BE/A versus the neutron number N, a plot like that shown in Fig. 4.2 is obtained. The BE/A rapidly rises as N increase from A = 14 and reaches a broad maximum before rapidly decreasing as N increases above 30. It is in the broad maximum where the BE/A is highest that one finds the stable isotopes. The reason for the zig-zag shape in the maximum region is due to the fact that an isotope with an even number of neutrons tends to have a higher BE/A compared
### Fusion
**Two light nuclei $\longrightarrow$ one heavy nucleus**
\begin{align}
^{2}_1H + ^2_1H \longrightarrow ^{4}_2He
\end{align}
### Fission
**One heavy nucleus $\longrightarrow$ two lighter nuclei**
\begin{align}
^{235}_{92}U + ^1_0n \longrightarrow \left(^{236}_{92}U\right)^* \longrightarrow ^{139}_{56}Ba + ^{94}_{36}Kr + 3^1_0n
\end{align}
<a title="JWB at en.wikipedia [CC BY 3.0
(https://creativecommons.org/licenses/by/3.0
) or GFDL (http://www.gnu.org/copyleft/fdl.html)], via Wikimedia Commons" href="https://commons.wikimedia.org/wiki/File:ThermalFissionYield.svg"><img width="512" alt="ThermalFissionYield" src="https://upload.wikimedia.org/wikipedia/commons/thumb/6/68/ThermalFissionYield.svg/512px-ThermalFissionYield.svg.png"></a>
## Nucleon Separation Energy
The energy required to remove a single nucleon from a nucleus is called "separation energy". Consider the addition of a single neutron to form the nucleus of $^A_ZX$, i.e.,
\begin{align}
^{A-1}_ZX + ^1_0n \rightarrow ^{A}_ZX.
\end{align}
$S_n\left(^A_ZX\right)$ is the energy released in this reaction and is equivalent to the energy required to remove (or separate) a single neutron from the nucleus $^A_ZX$.
\begin{align}
S_n\left(^A_ZX\right) &= [m\left(^{A-1}_ZX\right) + m_n − m\left(^{A}_ZX\right)]c^2 \\
&\simeq [M\left(^{A-1}_ZX\right) + m_n − M\left(^{A}_ZX\right)]c^2.
\end{align}
If we make the substition for nuclear binding energy:
\begin{align}
BE\left(^A_ZX\right) = \left[ZM(\left(^1_1H\right) + (A-Z)m_n - M\left(^A_ZX\right)\right]c^2
\end{align}
We can arrive at the separation energy:
\begin{align}
S_n\left(^A_ZX\right) &= BE\left(^A_ZX\right) - BE\left(^{A-1}_ZX\right)
\end{align}
## Exercise: Nucleon Separation Energy
(Example 4.3 in Shultis and Faw)
What is the binding energy of the last neutron in $^{16}_8O$? This is the energy released in the reaction:
\begin{align}
^{15}_8O + ^1_0n \rightarrow ^{16}_8O
\end{align}
So, we can use the equation above and plug in the mass values we can find in the book appendix.
\begin{align}
S_n\left(^{A}_ZX\right) &= [m\left(^{A-1}_ZX\right) + m_n − m\left(^{A}_ZX\right)]c^2 \\
&\simeq [M\left(^{A-1}_ZX\right) + m_n − M\left(^{A}_ZX\right)]c^2\\
\implies S_n &= [m\left(^{15}_8O\right) + m_n − m\left(^{16}_8O\right)]c^2 \\
&\simeq [M\left(^{15}_8X\right) + m_n − M\left(^{16}_8X\right)]c^2\\
&=[15.0030654 + 1.00866492 − 15.9949146] u \times 931.5 MeV/u\\
&= 15.66 MeV.
\end{align}
This is a very high value for a single nucleon.
### Think-Pair-Share
What does this high value mean about the comparative stability of $^{16}_8O$ and $^{15}_8O$?
### $S_p$
The equation for the separation energy of the proton is very similar to the separation energy of the neutron.
\begin{align}
S_p\left(^{A}_ZX\right) &= [m\left(^{A-1}_{Z-1}X\right) + m_p − m\left(^{A}_ZX\right)]c^2 \\
&\simeq [M\left(^{A-1}_{Z-1}X\right) + M\left(^1_1H\right) − M\left(^{A}_ZX\right)]c^2\\
\end{align}
## Exercise: $BE_e$ in $^1_1H$
A problem example from Shultis and Faw follows below.
**Energies of chemical reactions can typically not be calculated by finding the difference between the masses of the reactants and the products because the mass must be known to 10 or more significant figures.**
However, the mass of the proton and hydrogen atom are known to 10 significant figures. Let's estimate the binding energy of the electron $BE_e$ in the $^1_1H$ atom and compare this result to what the Bohr model predicts.
Discuss this comparison.
The binding energy reaction can be written as
\begin{align}
^{1}_1p + ^0_{-1}e \rightarrow ^{1}_1H + BE_e
\end{align}
We use mass values found in Table A.1 and Appendix B to find the binding energy:
\begin{align}
BE_e &= \left[m_p + m_e - M\left(^1_1H\right)\right]\left(u\right) \times 931.5(MeV/u)\\
&= \left[1.0072764669 + 5.48579909\times 10^{-4} - 1.0078250321\right]\left(u\right) \times 931.5(MeV/u)\\
&= 1.3887 \times 10^{−5} MeV\\
&= \boxed{13.887 eV}.
\end{align}
Recall that $BE_e = 13.606 eV$. Although the estimate of 13.887 is correct to 2 significant figures. To get a better estimate, the proton and hydrogen atomic masses must be known to at least 13 significant figures, an accuracy beyond present day technology.
## Binary Nuclear Reactions
We will refer to the reaction $x+X\longrightarrow Y+ y$ as :
\begin{align}
(x, y)
\end{align}
which is shorthand for $X(x,y)Y$. The very long form of this reaction is:
\begin{align}
^{A}_{Z}X + ^{A}_{Z}x \longrightarrow ^{A}_{Z}Y + ^{A}_{Z}y
\end{align}
### $(\alpha, p)$
This was first seen in air by Rutherford. Protons were produced when alpha particles were absorbed by nitrogen in the air to make oxygen. That example follows.
\begin{align}
^{4}_{2}He + ^{14}_{7}N \longrightarrow ^{17}_{8}O + ^{1}_{1}H
\end{align}
### $(\alpha, n)$
This was first seen with beryllium by Chadwick. Neutrons were produced when alpha particles were absorbed by beryllium in the air to make carbon. That example follows.
\begin{align}
^{4}_{2}He + ^{9}_{4}Be \longrightarrow ^{12}_{6}C + ^{1}_{0}n
\end{align}
### $(\gamma, n)$
Neutrons can also be produced when very energetic photons strike a nucleus. An example follows.
\begin{align}
^\gamma + ^{2}_{1}H \longrightarrow ^{1}_{1}H + ^{1}_{0}n
\end{align}
### $(p, \gamma)$
This is typically called "radiative capture" of a proton. An example follows.
\begin{align}
^{1}_{1}H + ^{7}_{3}Li \longrightarrow ^{8}_{4}Be + \gamma
\end{align}
Interestingly, in this example, the resulting beryllium nucleus is poorly bound and becomes two alphas.
### $(\gamma, \alpha n)$
Sometimes, more than two products can be produced. An example follows.
\begin{align}
\gamma + ^{17}_{8}O \longrightarrow ^{12}_{6}C + ^{4}_{2}He + ^{1}_{0}n
\end{align}
### $(n, p)$
Fast neutrons are very common in nculear reactor cores and can produce liberate protons. An example follows.
\begin{align}
^{1}_{0}n + ^{16}_{8}O \longrightarrow ^{16}_{7}N + ^{1}_{1}p
\end{align}
### $(n, \gamma)$
Most free neutrons in a reactor are fated to be radiatively captured.
\begin{align}
^{1}_{0}n + ^{238}_{92}U \longrightarrow ^{239}_{92}U + \gamma
\end{align}
## Q-Value For a Reaction
Energy is conserved in nuclear reactions. Specifically, the _total energy_ is conserved.
The total energy, recall, includes rest mass energy as well as kinetic energy.
\begin{align}
\sum_i \left[E_i + m_ic^2\right] &= \sum_i\left[E'_i + m'_ic^2\right]\\
\mbox{where }&\\
E_i &= \mbox{the kinetic energy of the ith initial particle }\\
E'_i &=\mbox{the kinetic energy of the ith final particle }\\
m_i &= \mbox{rest mass of the ith initial particle}\\
m'_i &= \mbox{rest mass of the ith final particle}\\
\end{align}
Changes in kinetic energy between the initial and final states must be balanced by equivalent changes in total rest mass. **The Q-value is the kinetic energy gained in the reaction.**
\begin{align}
Q &= (\mbox{KE of final particles}) − (\mbox{KE of initial particles})\\
&= \left(\sum_iE'_i\right) − \left(\sum_iE_i\right) \\
\end{align}
**The Q-value is equivalently the rest mass energy lost in the reaction.**
\begin{align}
Q &= (\mbox{rest mass of initial particles})c^2 − (\mbox{rest mass of final particles})c^2\\
&= \left(\sum_im_i\right)c^2 − \left(\sum_im'_i\right)c^2 \\
\end{align}
### Think-pair-share: Exothermic or Endothermic
If $Q<0$, is the reaction exothermic or endothermic?
### Q-value in binary reactions
For an $X(x, y)Y$ reaction:
\begin{align}
Q &= \left(E_y + E_Y \right) − \left(E_x + E_X\right) \\
&= \left( m_x + m_X\right)c^2 − \left( m_y + m_Y\right)c^2 \\
\end{align}
**If proton numbers (and therefore electron masses) are conserved,** we can use the neutral atom masses and simplify this to:
\begin{align}
Q &= \left(E_y + E_Y \right) − \left(E_x + E_X\right) \\
&= \left( M_x + M_X\right)c^2 − \left( M_y + M_Y\right)c^2 \\
\end{align}
### Q-value in decay reactions
For a radioactive decay reaction:
\begin{align}
X\longrightarrow Y+ y
\end{align}
The corresponding Q-value is:
\begin{align}
Q&=(E_y +E_Y)\\
&=[m_X −(m_y +m_Y )]c^2 \\
\end{align}
Note that radioactive decay, being spontaneous, is **always exothermic**. So, $Q>0$ for all decay reactions.
### Think-pair-share
- Can we make the same simplification as above for all decay reactions?
- If yes, why?
- If not, why not?
### Total Charge Conservation
Charge is always conserved in nuclear reactions. Thus, we might get into trouble with reactions that drive a change in electron number. The mass of the electron should be managed carefully in these cases. In an $n, p$ reaction, a proton is ejected from the nucleus by the incident neutron. During this process, an orbital electron is usually also lost from the resulting product atom. Thus, we must include an extra electron in the equation.
**Does the electron $_{-1}^0e$ belong on the right hand side (products) or the left hand side (reactants) of the following reaction?**
\begin{align}
^1_0n + ^{16}_8O \longrightarrow ^{16}_7N + ^1_1p
\end{align}
### Special case for changing proton number
The above procedure is further complicated in cases when the number of protons and the number of neutrons are not conserved. These reactions involve the nuclear weak force.
**The nuclear _weak force_ alters neutrons into protons and vice versa, usually inside the nucleus.**
The details are provided in an example in your book. Effectively, the neutrinos which moderate this reaction must be considered among the products and reactants.
## Special case for exited products
Many of the reactions we're most interested in as nuclear engineers involve an intermediate excited compound nucleus.
Let's consider Example 4.6 in your book regarding the reaction $^{10}B + ^1_0n \longrightarrow \left(^7Li\right)^*$. In short hand, this is an $(n,\alpha)$ reaction. Specifically, $^{10}B(n,\alpha)^7Li^∗$. The product, $\left(^7Li\right)^*$ is left in an excited state. Specifically it is left 0.48 MeV above its ground state.
The ordinary neutral atomic masses approach would look like this:
\begin{align}
Q &= \left[m_n + M\left(^{10}_5B\right) − M\left(^{4}_2He\right) - M\left(^{7}_3Li\right)^*\right]c^2 \\
\end{align}
But, the mass tabulated for $M\left(^{7}_3Li\right)$ is the ground state mass. So if we want to use that ground state mass to arrive at the solution, the extra energy contributing to the excited state needs to be accounted for.
\begin{align}
Q &= \left[m_n + M\left(^{10}_5B\right) − M\left(^{4}_2He\right) - M\left(^{7}_3Li\right)\right]c^2 -0.48MeV\\
\end{align}
## An aside on Florence and Mangkhut


[Duke shuts down Brunswick ahead of Florence](https://www.reuters.com/article/us-storm-florence-duke-nuclear/duke-starts-to-shut-north-carolina-brunswick-nuclear-plant-ahead-of-florence-idUSKCN1LT1ZT)
[Yangjiang and Taishan in path of Typhoon Mangkhut](https://www.scmp.com/news/china/society/article/2164363/chinese-nuclear-power-plant-path-super-typhoon-mangkhut)
|
github_jupyter
|
# Nuclear Energetics
## Learning Objectives
- Define: exothermic and endothermic reactions
- Differentiate exothermic and endothermic reactions
- Calculate binding energies
- Recognize the noation for various binary reactions
- Recognize the physics in common binary reactions
- Define the relationship between the Q value, mass, and energy in a reaction
- Explain the role of energy conservation in binary reactions
- Explain the role of charge conservation in binary reactions
- Calculate Q values for various reactions
## Endothermic and Exothermic Reactions
In any reaction (nuclear, mechanical, chemical) energy can be either emitted or absorbed.
- When energy is emitted by the reaction, this is called **_exothermic_**.
- When energy is absorbed into the reaction, this is called **_endothermic_**.
If we consider the energy in relation to mass via Einstein's equivalence, then it's very clear that a change in the mass of reactants will result in a change in energy ($\Delta E = \Delta M c^2$).
\begin{align}
\mbox{reactants} &\rightarrow \mbox{products}\\
A + B + \cdots &\rightarrow C + D + \cdots\\
\Delta M = (M_A + M_B + \cdots) &- (M_C + M_D + \cdots)\\
\implies \Delta E &= \left[(M_A + M_B + \cdots) - (M_C + M_D + \cdots)\right]c^2\\
\end{align}


<center>(credit: BBC)</center>
## Exercise: Think-pair-share
Which of the following are exothermic and which are endothermic?
Easy ones:
- Boiling Water
- Lighting a match
- Freezing an ice cube
- Melting an ice cube
- Snow formation in the clouds
- Conversion of frost to water vapor on the grass
Harder ones:
- Formation of ion pairs (exothermic)
- Separation of ion pairs (endothermic)
The most important one:
- nuclear fission
# Binding Energy
For ease, let's imagine just two reactants and one product.
\begin{align}
\mbox{reactants} &\rightarrow \mbox{products}\\
A + B &\rightarrow C \\
\Delta M = (mass(A) + mass(B)) &- (mass(C))\\
\implies \Delta E &= \left[(mass(A) + mass(B) - (mass(C))\right]c^2\\
&= BE
\end{align}
## Nuclear and Atomic Masses
\begin{align}
M\left(^A_ZX\right) &= \mbox{rest mass of an atom}\\
m\left(^A_ZX\right) &= \mbox{rest mass of its nucleus}
\end{align}
All Z electrons must be bound to the atom, so the atomic and nuclear masses have the following relationship:
\begin{align}
M\left(^A_ZX\right) &=m\left(^A_ZX\right) + Zm_e - \frac{BE_{Ze}}{c^2}
\end{align}
Your book points out that it requires 13.6 eV to ionize the hydrogen atom. This electron binding energy represents a mass change of $BE_{1e/c^2} = 13.6 (eV)/9.315 \times 10^8 (eV/u) = 1.4 \times 10^{−8} u$.
## Binding Energy of the nucleus
A nucleus, with Z protons and N=A-Z neutrons will have a formation reaction that looks like:
\begin{align}
Z \mbox{ protons } + (A − Z) \mbox{ neutrons }\rightarrow \mbox{ nucleus}\left(^A_ZX\right) + BE.
\end{align}
It can be helpful to define the term $BE/c^2$:
\begin{align}
\frac{BE}{c^2} &=Zm_p +(A−Z)m_n − m\left(^A_ZX\right)\\
\end{align}
We can arrive at the nuclear binding energy from this:
\begin{align}
BE\left(^A_ZX\right) = \left[ZM(\left(^1_1H\right) + (A-Z)m_n - M\left(^A_ZX\right)\right]c^2
\end{align}
The above equation neglects the electron binding energies because:
- the terms tend to cancel
- electron binding energies are millions of times smaller than the nuclear binding energy.
### Exercise: Nuclear binding energy
From your book, example 4.1: What is the binding energy of an alpha particle?
\begin{align}
\frac{BE}{c^2} &=Zm_p +(A−Z)m_n − m\left(^A_ZX\right)\\
\implies\frac{BE}{c^2} &=\left[ZM(\left(^1_1H\right) + (A-Z)m_n - M\left(^A_ZX\right)\right]\\
&=\left[2M(\left(^1_1H\right) + 2m_n - M\left(^4_2He\right)\right]\\
&=\left[2\left(1.0078250\right) + 2\left(1.0086649\right) - 4.0026032\right]\\
\end{align}
Thus:
\begin{align}
BE\left(^4_2 He\right) &= \frac{BE}{c^2}\times 931.5 (MeV/u) \\
&= 28.30 MeV.
\end{align}
## Binding Energy per nucleon
The BE/A versus A curve immediately suggests two ways to extract energy from the nucleus.

Binding energy curve **(average binding energy per nucleon in MeV against number of nucleons in nucleus)** for a number of relatively common (abundant) isotopes (not chosen systematically; almost anything with an occurence of over .2 was chosen though a few exceptions are in there, such as U235). A few important ones for the purposes of nuclear fusion and nuclear fission are marked, as well as iron-56, which sits at the highest point on this graph and cannot yield energy from fusion or fission.
From your book (Shultis and Faw)
> Most isotopes of an element are radioactive, i.e., their nuclei are unstable. To better understand what makes a nucleus stable, it is instructive to plot the BE per nucleon, BE/A, of all the isotopes of an element. For example, if one calculates the BE/A for all the isotopes of calcium (Z = 20), as is done in Example 4.2 for one isotope, and plots BE/A versus the neutron number N, a plot like that shown in Fig. 4.2 is obtained. The BE/A rapidly rises as N increase from A = 14 and reaches a broad maximum before rapidly decreasing as N increases above 30. It is in the broad maximum where the BE/A is highest that one finds the stable isotopes. The reason for the zig-zag shape in the maximum region is due to the fact that an isotope with an even number of neutrons tends to have a higher BE/A compared
### Fusion
**Two light nuclei $\longrightarrow$ one heavy nucleus**
\begin{align}
^{2}_1H + ^2_1H \longrightarrow ^{4}_2He
\end{align}
### Fission
**One heavy nucleus $\longrightarrow$ two lighter nuclei**
\begin{align}
^{235}_{92}U + ^1_0n \longrightarrow \left(^{236}_{92}U\right)^* \longrightarrow ^{139}_{56}Ba + ^{94}_{36}Kr + 3^1_0n
\end{align}
<a title="JWB at en.wikipedia [CC BY 3.0
(https://creativecommons.org/licenses/by/3.0
) or GFDL (http://www.gnu.org/copyleft/fdl.html)], via Wikimedia Commons" href="https://commons.wikimedia.org/wiki/File:ThermalFissionYield.svg"><img width="512" alt="ThermalFissionYield" src="https://upload.wikimedia.org/wikipedia/commons/thumb/6/68/ThermalFissionYield.svg/512px-ThermalFissionYield.svg.png"></a>
## Nucleon Separation Energy
The energy required to remove a single nucleon from a nucleus is called "separation energy". Consider the addition of a single neutron to form the nucleus of $^A_ZX$, i.e.,
\begin{align}
^{A-1}_ZX + ^1_0n \rightarrow ^{A}_ZX.
\end{align}
$S_n\left(^A_ZX\right)$ is the energy released in this reaction and is equivalent to the energy required to remove (or separate) a single neutron from the nucleus $^A_ZX$.
\begin{align}
S_n\left(^A_ZX\right) &= [m\left(^{A-1}_ZX\right) + m_n − m\left(^{A}_ZX\right)]c^2 \\
&\simeq [M\left(^{A-1}_ZX\right) + m_n − M\left(^{A}_ZX\right)]c^2.
\end{align}
If we make the substition for nuclear binding energy:
\begin{align}
BE\left(^A_ZX\right) = \left[ZM(\left(^1_1H\right) + (A-Z)m_n - M\left(^A_ZX\right)\right]c^2
\end{align}
We can arrive at the separation energy:
\begin{align}
S_n\left(^A_ZX\right) &= BE\left(^A_ZX\right) - BE\left(^{A-1}_ZX\right)
\end{align}
## Exercise: Nucleon Separation Energy
(Example 4.3 in Shultis and Faw)
What is the binding energy of the last neutron in $^{16}_8O$? This is the energy released in the reaction:
\begin{align}
^{15}_8O + ^1_0n \rightarrow ^{16}_8O
\end{align}
So, we can use the equation above and plug in the mass values we can find in the book appendix.
\begin{align}
S_n\left(^{A}_ZX\right) &= [m\left(^{A-1}_ZX\right) + m_n − m\left(^{A}_ZX\right)]c^2 \\
&\simeq [M\left(^{A-1}_ZX\right) + m_n − M\left(^{A}_ZX\right)]c^2\\
\implies S_n &= [m\left(^{15}_8O\right) + m_n − m\left(^{16}_8O\right)]c^2 \\
&\simeq [M\left(^{15}_8X\right) + m_n − M\left(^{16}_8X\right)]c^2\\
&=[15.0030654 + 1.00866492 − 15.9949146] u \times 931.5 MeV/u\\
&= 15.66 MeV.
\end{align}
This is a very high value for a single nucleon.
### Think-Pair-Share
What does this high value mean about the comparative stability of $^{16}_8O$ and $^{15}_8O$?
### $S_p$
The equation for the separation energy of the proton is very similar to the separation energy of the neutron.
\begin{align}
S_p\left(^{A}_ZX\right) &= [m\left(^{A-1}_{Z-1}X\right) + m_p − m\left(^{A}_ZX\right)]c^2 \\
&\simeq [M\left(^{A-1}_{Z-1}X\right) + M\left(^1_1H\right) − M\left(^{A}_ZX\right)]c^2\\
\end{align}
## Exercise: $BE_e$ in $^1_1H$
A problem example from Shultis and Faw follows below.
**Energies of chemical reactions can typically not be calculated by finding the difference between the masses of the reactants and the products because the mass must be known to 10 or more significant figures.**
However, the mass of the proton and hydrogen atom are known to 10 significant figures. Let's estimate the binding energy of the electron $BE_e$ in the $^1_1H$ atom and compare this result to what the Bohr model predicts.
Discuss this comparison.
The binding energy reaction can be written as
\begin{align}
^{1}_1p + ^0_{-1}e \rightarrow ^{1}_1H + BE_e
\end{align}
We use mass values found in Table A.1 and Appendix B to find the binding energy:
\begin{align}
BE_e &= \left[m_p + m_e - M\left(^1_1H\right)\right]\left(u\right) \times 931.5(MeV/u)\\
&= \left[1.0072764669 + 5.48579909\times 10^{-4} - 1.0078250321\right]\left(u\right) \times 931.5(MeV/u)\\
&= 1.3887 \times 10^{−5} MeV\\
&= \boxed{13.887 eV}.
\end{align}
Recall that $BE_e = 13.606 eV$. Although the estimate of 13.887 is correct to 2 significant figures. To get a better estimate, the proton and hydrogen atomic masses must be known to at least 13 significant figures, an accuracy beyond present day technology.
## Binary Nuclear Reactions
We will refer to the reaction $x+X\longrightarrow Y+ y$ as :
\begin{align}
(x, y)
\end{align}
which is shorthand for $X(x,y)Y$. The very long form of this reaction is:
\begin{align}
^{A}_{Z}X + ^{A}_{Z}x \longrightarrow ^{A}_{Z}Y + ^{A}_{Z}y
\end{align}
### $(\alpha, p)$
This was first seen in air by Rutherford. Protons were produced when alpha particles were absorbed by nitrogen in the air to make oxygen. That example follows.
\begin{align}
^{4}_{2}He + ^{14}_{7}N \longrightarrow ^{17}_{8}O + ^{1}_{1}H
\end{align}
### $(\alpha, n)$
This was first seen with beryllium by Chadwick. Neutrons were produced when alpha particles were absorbed by beryllium in the air to make carbon. That example follows.
\begin{align}
^{4}_{2}He + ^{9}_{4}Be \longrightarrow ^{12}_{6}C + ^{1}_{0}n
\end{align}
### $(\gamma, n)$
Neutrons can also be produced when very energetic photons strike a nucleus. An example follows.
\begin{align}
^\gamma + ^{2}_{1}H \longrightarrow ^{1}_{1}H + ^{1}_{0}n
\end{align}
### $(p, \gamma)$
This is typically called "radiative capture" of a proton. An example follows.
\begin{align}
^{1}_{1}H + ^{7}_{3}Li \longrightarrow ^{8}_{4}Be + \gamma
\end{align}
Interestingly, in this example, the resulting beryllium nucleus is poorly bound and becomes two alphas.
### $(\gamma, \alpha n)$
Sometimes, more than two products can be produced. An example follows.
\begin{align}
\gamma + ^{17}_{8}O \longrightarrow ^{12}_{6}C + ^{4}_{2}He + ^{1}_{0}n
\end{align}
### $(n, p)$
Fast neutrons are very common in nculear reactor cores and can produce liberate protons. An example follows.
\begin{align}
^{1}_{0}n + ^{16}_{8}O \longrightarrow ^{16}_{7}N + ^{1}_{1}p
\end{align}
### $(n, \gamma)$
Most free neutrons in a reactor are fated to be radiatively captured.
\begin{align}
^{1}_{0}n + ^{238}_{92}U \longrightarrow ^{239}_{92}U + \gamma
\end{align}
## Q-Value For a Reaction
Energy is conserved in nuclear reactions. Specifically, the _total energy_ is conserved.
The total energy, recall, includes rest mass energy as well as kinetic energy.
\begin{align}
\sum_i \left[E_i + m_ic^2\right] &= \sum_i\left[E'_i + m'_ic^2\right]\\
\mbox{where }&\\
E_i &= \mbox{the kinetic energy of the ith initial particle }\\
E'_i &=\mbox{the kinetic energy of the ith final particle }\\
m_i &= \mbox{rest mass of the ith initial particle}\\
m'_i &= \mbox{rest mass of the ith final particle}\\
\end{align}
Changes in kinetic energy between the initial and final states must be balanced by equivalent changes in total rest mass. **The Q-value is the kinetic energy gained in the reaction.**
\begin{align}
Q &= (\mbox{KE of final particles}) − (\mbox{KE of initial particles})\\
&= \left(\sum_iE'_i\right) − \left(\sum_iE_i\right) \\
\end{align}
**The Q-value is equivalently the rest mass energy lost in the reaction.**
\begin{align}
Q &= (\mbox{rest mass of initial particles})c^2 − (\mbox{rest mass of final particles})c^2\\
&= \left(\sum_im_i\right)c^2 − \left(\sum_im'_i\right)c^2 \\
\end{align}
### Think-pair-share: Exothermic or Endothermic
If $Q<0$, is the reaction exothermic or endothermic?
### Q-value in binary reactions
For an $X(x, y)Y$ reaction:
\begin{align}
Q &= \left(E_y + E_Y \right) − \left(E_x + E_X\right) \\
&= \left( m_x + m_X\right)c^2 − \left( m_y + m_Y\right)c^2 \\
\end{align}
**If proton numbers (and therefore electron masses) are conserved,** we can use the neutral atom masses and simplify this to:
\begin{align}
Q &= \left(E_y + E_Y \right) − \left(E_x + E_X\right) \\
&= \left( M_x + M_X\right)c^2 − \left( M_y + M_Y\right)c^2 \\
\end{align}
### Q-value in decay reactions
For a radioactive decay reaction:
\begin{align}
X\longrightarrow Y+ y
\end{align}
The corresponding Q-value is:
\begin{align}
Q&=(E_y +E_Y)\\
&=[m_X −(m_y +m_Y )]c^2 \\
\end{align}
Note that radioactive decay, being spontaneous, is **always exothermic**. So, $Q>0$ for all decay reactions.
### Think-pair-share
- Can we make the same simplification as above for all decay reactions?
- If yes, why?
- If not, why not?
### Total Charge Conservation
Charge is always conserved in nuclear reactions. Thus, we might get into trouble with reactions that drive a change in electron number. The mass of the electron should be managed carefully in these cases. In an $n, p$ reaction, a proton is ejected from the nucleus by the incident neutron. During this process, an orbital electron is usually also lost from the resulting product atom. Thus, we must include an extra electron in the equation.
**Does the electron $_{-1}^0e$ belong on the right hand side (products) or the left hand side (reactants) of the following reaction?**
\begin{align}
^1_0n + ^{16}_8O \longrightarrow ^{16}_7N + ^1_1p
\end{align}
### Special case for changing proton number
The above procedure is further complicated in cases when the number of protons and the number of neutrons are not conserved. These reactions involve the nuclear weak force.
**The nuclear _weak force_ alters neutrons into protons and vice versa, usually inside the nucleus.**
The details are provided in an example in your book. Effectively, the neutrinos which moderate this reaction must be considered among the products and reactants.
## Special case for exited products
Many of the reactions we're most interested in as nuclear engineers involve an intermediate excited compound nucleus.
Let's consider Example 4.6 in your book regarding the reaction $^{10}B + ^1_0n \longrightarrow \left(^7Li\right)^*$. In short hand, this is an $(n,\alpha)$ reaction. Specifically, $^{10}B(n,\alpha)^7Li^∗$. The product, $\left(^7Li\right)^*$ is left in an excited state. Specifically it is left 0.48 MeV above its ground state.
The ordinary neutral atomic masses approach would look like this:
\begin{align}
Q &= \left[m_n + M\left(^{10}_5B\right) − M\left(^{4}_2He\right) - M\left(^{7}_3Li\right)^*\right]c^2 \\
\end{align}
But, the mass tabulated for $M\left(^{7}_3Li\right)$ is the ground state mass. So if we want to use that ground state mass to arrive at the solution, the extra energy contributing to the excited state needs to be accounted for.
\begin{align}
Q &= \left[m_n + M\left(^{10}_5B\right) − M\left(^{4}_2He\right) - M\left(^{7}_3Li\right)\right]c^2 -0.48MeV\\
\end{align}
## An aside on Florence and Mangkhut



[Duke shuts down Brunswick ahead of Florence](https://www.reuters.com/article/us-storm-florence-duke-nuclear/duke-starts-to-shut-north-carolina-brunswick-nuclear-plant-ahead-of-florence-idUSKCN1LT1ZT)
[Yangjiang and Taishan in path of Typhoon Mangkhut](https://www.scmp.com/news/china/society/article/2164363/chinese-nuclear-power-plant-path-super-typhoon-mangkhut)
| 0.910259 | 0.96799 |
```
%%sh
apt-get update
apt-get -y install graphviz
pip install -q graphviz
import pandas as pd
dataset = pd.read_csv('housing.csv')
# Move 'medv' column to front
dataset = pd.concat([dataset['medv'], dataset.drop(['medv'], axis=1)], axis=1)
from sklearn.model_selection import train_test_split
training_dataset, validation_dataset = train_test_split(dataset, test_size=0.1)
print(training_dataset.shape)
print(validation_dataset.shape)
training_dataset.to_csv('training_dataset.csv', index=False, header=False)
validation_dataset.to_csv('validation_dataset.csv', index=False, header=False)
import sagemaker
print(sagemaker.__version__)
sess = sagemaker.Session()
bucket = sess.default_bucket()
prefix = 'boston-housing'
training_data_path = sess.upload_data(path='training_dataset.csv', key_prefix=prefix + '/input/training')
validation_data_path = sess.upload_data(path='validation_dataset.csv', key_prefix=prefix + '/input/validation')
print(training_data_path)
print(validation_data_path)
from sagemaker import get_execution_role
from sagemaker.image_uris import retrieve
from sagemaker.estimator import Estimator
region = sess.boto_session.region_name
container = retrieve('linear-learner', region)
print(container)
ll_estimator = Estimator(container,
role=get_execution_role() ,
instance_count=1,
instance_type='ml.m5.large',
output_path='s3://{}/{}/output'.format(bucket, prefix)
)
ll_estimator.set_hyperparameters(
predictor_type='regressor',
mini_batch_size=32)
from sagemaker import TrainingInput
training_data_channel = TrainingInput(s3_data=training_data_path, content_type='text/csv')
validation_data_channel = TrainingInput(s3_data=validation_data_path, content_type='text/csv')
ll_data = {'train': training_data_channel, 'validation': validation_data_channel}
ll_estimator.fit(ll_data)
%%bash -s $ll_estimator.model_data
aws s3 cp $1 .
tar xvfz model.tar.gz
unzip -o model_algo-1
import json
sym_json = json.load(open('mx-mod-symbol.json'))
sym_json_string = json.dumps(sym_json)
import mxnet as mx
from mxnet import gluon
net = gluon.nn.SymbolBlock(
outputs=mx.sym.load_json(sym_json_string),
inputs=mx.sym.var('data'))
mx.viz.plot_network(
net(mx.sym.var('data'))[0],
node_attrs={'shape':'oval','fixedsize':'false'})
net.load_parameters('mx-mod-0000.params', allow_missing=True)
net.collect_params().initialize()
test_sample = mx.nd.array([0.00632,18.00,2.310,0,0.5380,6.5750,65.20,4.0900,1,296.0,15.30,4.98])
test_sample
response = net(test_sample)
print(response)
```
|
github_jupyter
|
%%sh
apt-get update
apt-get -y install graphviz
pip install -q graphviz
import pandas as pd
dataset = pd.read_csv('housing.csv')
# Move 'medv' column to front
dataset = pd.concat([dataset['medv'], dataset.drop(['medv'], axis=1)], axis=1)
from sklearn.model_selection import train_test_split
training_dataset, validation_dataset = train_test_split(dataset, test_size=0.1)
print(training_dataset.shape)
print(validation_dataset.shape)
training_dataset.to_csv('training_dataset.csv', index=False, header=False)
validation_dataset.to_csv('validation_dataset.csv', index=False, header=False)
import sagemaker
print(sagemaker.__version__)
sess = sagemaker.Session()
bucket = sess.default_bucket()
prefix = 'boston-housing'
training_data_path = sess.upload_data(path='training_dataset.csv', key_prefix=prefix + '/input/training')
validation_data_path = sess.upload_data(path='validation_dataset.csv', key_prefix=prefix + '/input/validation')
print(training_data_path)
print(validation_data_path)
from sagemaker import get_execution_role
from sagemaker.image_uris import retrieve
from sagemaker.estimator import Estimator
region = sess.boto_session.region_name
container = retrieve('linear-learner', region)
print(container)
ll_estimator = Estimator(container,
role=get_execution_role() ,
instance_count=1,
instance_type='ml.m5.large',
output_path='s3://{}/{}/output'.format(bucket, prefix)
)
ll_estimator.set_hyperparameters(
predictor_type='regressor',
mini_batch_size=32)
from sagemaker import TrainingInput
training_data_channel = TrainingInput(s3_data=training_data_path, content_type='text/csv')
validation_data_channel = TrainingInput(s3_data=validation_data_path, content_type='text/csv')
ll_data = {'train': training_data_channel, 'validation': validation_data_channel}
ll_estimator.fit(ll_data)
%%bash -s $ll_estimator.model_data
aws s3 cp $1 .
tar xvfz model.tar.gz
unzip -o model_algo-1
import json
sym_json = json.load(open('mx-mod-symbol.json'))
sym_json_string = json.dumps(sym_json)
import mxnet as mx
from mxnet import gluon
net = gluon.nn.SymbolBlock(
outputs=mx.sym.load_json(sym_json_string),
inputs=mx.sym.var('data'))
mx.viz.plot_network(
net(mx.sym.var('data'))[0],
node_attrs={'shape':'oval','fixedsize':'false'})
net.load_parameters('mx-mod-0000.params', allow_missing=True)
net.collect_params().initialize()
test_sample = mx.nd.array([0.00632,18.00,2.310,0,0.5380,6.5750,65.20,4.0900,1,296.0,15.30,4.98])
test_sample
response = net(test_sample)
print(response)
| 0.376738 | 0.22448 |
```
from PIL import Image
from IPython.display import display
import random
import json
# Each image is made up a series of traits
# The weightings for each trait drive the rarity and add up to 100%
background = ["Blue", "Orange", "Purple", "Red", "Yellow"]
background_weights = [30, 40, 15, 5, 10]
circle = ["Blue", "Green", "Orange", "Red", "Yellow"]
circle_weights = [30, 40, 15, 5, 10]
square = ["Blue", "Green", "Orange", "Red", "Yellow"]
square_weights = [30, 40, 15, 5, 10]
# Dictionary variable for each trait.
# Eech trait corresponds to its file name
background_files = {
"Blue": "blue",
"Orange": "orange",
"Purple": "purple",
"Red": "red",
"Yellow": "yellow",
}
circle_files = {
"Blue": "blue-circle",
"Green": "green-circle",
"Orange": "orange-circle",
"Red": "red-circle",
"Yellow": "yellow-circle"
}
square_files = {
"Blue": "blue-square",
"Green": "green-square",
"Orange": "orange-square",
"Red": "red-square",
"Yellow": "yellow-square"
}
## Generate Traits
TOTAL_IMAGES = 30 # Number of random unique images we want to generate
all_images = []
# A recursive function to generate unique image combinations
def create_new_image():
new_image = {} #
# For each trait category, select a random trait based on the weightings
new_image ["Background"] = random.choices(background, background_weights)[0]
new_image ["Circle"] = random.choices(circle, circle_weights)[0]
new_image ["Square"] = random.choices(square, square_weights)[0]
if new_image in all_images:
return create_new_image()
else:
return new_image
# Generate the unique combinations based on trait weightings
for i in range(TOTAL_IMAGES):
new_trait_image = create_new_image()
all_images.append(new_trait_image)
# Returns true if all images are unique
def all_images_unique(all_images):
seen = list()
return not any(i in seen or seen.append(i) for i in all_images)
print("Are all images unique?", all_images_unique(all_images))
# Add token Id to each image
i = 0
for item in all_images:
item["tokenId"] = i
i = i + 1
print(all_images)
# Get Trait Counts
background_count = {}
for item in background:
background_count[item] = 0
circle_count = {}
for item in circle:
circle_count[item] = 0
square_count = {}
for item in square:
square_count[item] = 0
for image in all_images:
background_count[image["Background"]] += 1
circle_count[image["Circle"]] += 1
square_count[image["Square"]] += 1
print(background_count)
print(circle_count)
print(square_count)
#### Generate Metadata for all Traits
METADATA_FILE_NAME = './metadata/all-traits.json';
with open(METADATA_FILE_NAME, 'w') as outfile:
json.dump(all_images, outfile, indent=4)
#### Generate Images
for item in all_images:
im1 = Image.open(f'./trait-layers/backgrounds/{background_files[item["Background"]]}.jpg').convert('RGBA')
im2 = Image.open(f'./trait-layers/circles/{circle_files[item["Circle"]]}.png').convert('RGBA')
im3 = Image.open(f'./trait-layers/squares/{square_files[item["Square"]]}.png').convert('RGBA')
#Create each composite
com1 = Image.alpha_composite(im1, im2)
com2 = Image.alpha_composite(com1, im3)
#Convert to RGB
rgb_im = com2.convert('RGB')
file_name = str(item["tokenId"]) + ".png"
rgb_im.save("./images/" + file_name)
#### Generate Metadata for each Image
f = open('./metadata/all-traits.json',)
data = json.load(f)
IMAGES_BASE_URI = "ADD_IMAGES_BASE_URI_HERE"
PROJECT_NAME = "ADD_PROJECT_NAME_HERE"
def getAttribute(key, value):
return {
"trait_type": key,
"value": value
}
for i in data:
token_id = i['tokenId']
token = {
"image": IMAGES_BASE_URI + str(token_id) + '.png',
"tokenId": token_id,
"name": PROJECT_NAME + ' ' + str(token_id),
"attributes": []
}
token["attributes"].append(getAttribute("Background", i["Background"]))
token["attributes"].append(getAttribute("Circle", i["Circle"]))
token["attributes"].append(getAttribute("Square", i["Square"]))
with open('./metadata/' + str(token_id), 'w') as outfile:
json.dump(token, outfile, indent=4)
f.close()
```
|
github_jupyter
|
from PIL import Image
from IPython.display import display
import random
import json
# Each image is made up a series of traits
# The weightings for each trait drive the rarity and add up to 100%
background = ["Blue", "Orange", "Purple", "Red", "Yellow"]
background_weights = [30, 40, 15, 5, 10]
circle = ["Blue", "Green", "Orange", "Red", "Yellow"]
circle_weights = [30, 40, 15, 5, 10]
square = ["Blue", "Green", "Orange", "Red", "Yellow"]
square_weights = [30, 40, 15, 5, 10]
# Dictionary variable for each trait.
# Eech trait corresponds to its file name
background_files = {
"Blue": "blue",
"Orange": "orange",
"Purple": "purple",
"Red": "red",
"Yellow": "yellow",
}
circle_files = {
"Blue": "blue-circle",
"Green": "green-circle",
"Orange": "orange-circle",
"Red": "red-circle",
"Yellow": "yellow-circle"
}
square_files = {
"Blue": "blue-square",
"Green": "green-square",
"Orange": "orange-square",
"Red": "red-square",
"Yellow": "yellow-square"
}
## Generate Traits
TOTAL_IMAGES = 30 # Number of random unique images we want to generate
all_images = []
# A recursive function to generate unique image combinations
def create_new_image():
new_image = {} #
# For each trait category, select a random trait based on the weightings
new_image ["Background"] = random.choices(background, background_weights)[0]
new_image ["Circle"] = random.choices(circle, circle_weights)[0]
new_image ["Square"] = random.choices(square, square_weights)[0]
if new_image in all_images:
return create_new_image()
else:
return new_image
# Generate the unique combinations based on trait weightings
for i in range(TOTAL_IMAGES):
new_trait_image = create_new_image()
all_images.append(new_trait_image)
# Returns true if all images are unique
def all_images_unique(all_images):
seen = list()
return not any(i in seen or seen.append(i) for i in all_images)
print("Are all images unique?", all_images_unique(all_images))
# Add token Id to each image
i = 0
for item in all_images:
item["tokenId"] = i
i = i + 1
print(all_images)
# Get Trait Counts
background_count = {}
for item in background:
background_count[item] = 0
circle_count = {}
for item in circle:
circle_count[item] = 0
square_count = {}
for item in square:
square_count[item] = 0
for image in all_images:
background_count[image["Background"]] += 1
circle_count[image["Circle"]] += 1
square_count[image["Square"]] += 1
print(background_count)
print(circle_count)
print(square_count)
#### Generate Metadata for all Traits
METADATA_FILE_NAME = './metadata/all-traits.json';
with open(METADATA_FILE_NAME, 'w') as outfile:
json.dump(all_images, outfile, indent=4)
#### Generate Images
for item in all_images:
im1 = Image.open(f'./trait-layers/backgrounds/{background_files[item["Background"]]}.jpg').convert('RGBA')
im2 = Image.open(f'./trait-layers/circles/{circle_files[item["Circle"]]}.png').convert('RGBA')
im3 = Image.open(f'./trait-layers/squares/{square_files[item["Square"]]}.png').convert('RGBA')
#Create each composite
com1 = Image.alpha_composite(im1, im2)
com2 = Image.alpha_composite(com1, im3)
#Convert to RGB
rgb_im = com2.convert('RGB')
file_name = str(item["tokenId"]) + ".png"
rgb_im.save("./images/" + file_name)
#### Generate Metadata for each Image
f = open('./metadata/all-traits.json',)
data = json.load(f)
IMAGES_BASE_URI = "ADD_IMAGES_BASE_URI_HERE"
PROJECT_NAME = "ADD_PROJECT_NAME_HERE"
def getAttribute(key, value):
return {
"trait_type": key,
"value": value
}
for i in data:
token_id = i['tokenId']
token = {
"image": IMAGES_BASE_URI + str(token_id) + '.png',
"tokenId": token_id,
"name": PROJECT_NAME + ' ' + str(token_id),
"attributes": []
}
token["attributes"].append(getAttribute("Background", i["Background"]))
token["attributes"].append(getAttribute("Circle", i["Circle"]))
token["attributes"].append(getAttribute("Square", i["Square"]))
with open('./metadata/' + str(token_id), 'w') as outfile:
json.dump(token, outfile, indent=4)
f.close()
| 0.473901 | 0.231766 |
```
import pandas as pd
import matplotlib.pyplot as plt
import numpy as np
import scipy as sp
import glob
import re, os
!pwd
#collate data
df_s_full = pd.DataFrame()
data_s = []
first_row_s = []
data_c = []
first_row_c = []
to_be_deleted = []
index_s = set()
index_c = set()
for infile in glob.glob("data_apollo/*_sim_s.csv"):
df = pd.read_csv(infile)
data_s.append(df)
first_row_s.append(list(df.iloc[0]))
numbers = re.findall('\d+',infile)
index_s.add(int(numbers[0]))
for infile in glob.glob("data_apollo/*_sim_c.csv"):
df = pd.read_csv(infile)
data_c.append(df)
first_row_c.append(list(df.iloc[0]))
numbers = re.findall('\d+',infile)
index_c.add(int(numbers[0]))
df_s_full = pd.concat(data_s)
df_s_first = pd.DataFrame(first_row_s)
df_c_full = pd.concat(data_c)
df_c_first = pd.DataFrame(first_row_c)
if len(to_be_deleted)>0:
import re, os
numbers = re.findall('\d+',string)
indexs = [int(n) for n in numbers]
for ind in indexs:
os.remove(f'data_apollo/{ind}_sim_s.csv')
os.remove(f'data_apollo/{ind}_sim_c.csv')
index_s - index_c
index_c - index_s
if 0:
df_s_full.to_csv('data_apollo/df_s_full.csv', index=False)
df_c_full.to_csv('data_apollo/df_c_full.csv', index=False)
df_s_full.describe()
df_c_full.describe()
df_c_first.describe()
df_s_first.describe()
df_s_first.hist(bins=30, figsize=(10,15), layout=(5,1))
plt.matshow(df_s_first.corr())
plt.colorbar()
plt.matshow(df_s_full.corr())
plt.colorbar()
plt.set_cmap('coolwarm')
df_s_full.hist(bins=100, figsize=(10,15), layout=(5,1))
plt.scatter(df_s_full['0'],df_s_full['2'],marker='.',alpha=0.01)
plt.scatter(df_s_full['1'],df_s_full['3'],marker='.',alpha=0.005)
plt.scatter(df_s_full['2'],df_s_full['3'],marker='.',alpha=0.005)
plt.scatter(df_s_full['0'],df_s_full['1'],marker='.',alpha=0.005)
ax = plt.gca()
ax.set_aspect('equal', 'datalim')
plt.grid()
plt.scatter(df_s_full['0'],df_s_full['1'],marker='.',alpha=0.005)
ax = plt.gca()
plt.ylim([0, 500])
plt.xlim([-100, 100])
ax.set_aspect('equal', 'datalim')
plt.grid()
plt.scatter((df_s_full['0']**2+df_s_full['1']**2)**0.5,(df_s_full['2']**2+df_s_full['3']**2)**0.5,marker='.',alpha=0.01)
df_s_full.shape
df_c_full.shape
plt.scatter((df_s_full['0']**2+df_s_full['1']**2)**0.5,df_c_full['0'],marker='.',alpha=0.01)
plt.scatter((df_s_full['2']**2+df_s_full['3']**2)**0.5,df_c_full['0'],marker='.',alpha=0.01)
plt.figure()
for ind in range(2000):
try:
df = pd.read_csv(f'data_apollo/{ind}_sim_c.csv')
plt.plot(df['1'])
except:
pass
ax = plt.gca()
plt.grid()
df_c_full.hist(bins=100, figsize=(10,6), layout=(2,1))
df_c_full['1'].describe()
plt.scatter(df_c_full['1'],df_c_full['0'],marker='.',alpha=0.01)
plt.grid()
import re
import os
index_s = []
index_c = []
for infile in glob.glob("data_apollo/*_sim_s.csv"):
numbers = re.findall('\d+',infile)
index_s.append(int(numbers[0]))
for infile in glob.glob("data_apollo/*_sim_c.csv"):
numbers = re.findall('\d+',infile)
index_c.append(int(numbers[0]))
index_s
index_c
len(index_s)
len(index_c)
```
|
github_jupyter
|
import pandas as pd
import matplotlib.pyplot as plt
import numpy as np
import scipy as sp
import glob
import re, os
!pwd
#collate data
df_s_full = pd.DataFrame()
data_s = []
first_row_s = []
data_c = []
first_row_c = []
to_be_deleted = []
index_s = set()
index_c = set()
for infile in glob.glob("data_apollo/*_sim_s.csv"):
df = pd.read_csv(infile)
data_s.append(df)
first_row_s.append(list(df.iloc[0]))
numbers = re.findall('\d+',infile)
index_s.add(int(numbers[0]))
for infile in glob.glob("data_apollo/*_sim_c.csv"):
df = pd.read_csv(infile)
data_c.append(df)
first_row_c.append(list(df.iloc[0]))
numbers = re.findall('\d+',infile)
index_c.add(int(numbers[0]))
df_s_full = pd.concat(data_s)
df_s_first = pd.DataFrame(first_row_s)
df_c_full = pd.concat(data_c)
df_c_first = pd.DataFrame(first_row_c)
if len(to_be_deleted)>0:
import re, os
numbers = re.findall('\d+',string)
indexs = [int(n) for n in numbers]
for ind in indexs:
os.remove(f'data_apollo/{ind}_sim_s.csv')
os.remove(f'data_apollo/{ind}_sim_c.csv')
index_s - index_c
index_c - index_s
if 0:
df_s_full.to_csv('data_apollo/df_s_full.csv', index=False)
df_c_full.to_csv('data_apollo/df_c_full.csv', index=False)
df_s_full.describe()
df_c_full.describe()
df_c_first.describe()
df_s_first.describe()
df_s_first.hist(bins=30, figsize=(10,15), layout=(5,1))
plt.matshow(df_s_first.corr())
plt.colorbar()
plt.matshow(df_s_full.corr())
plt.colorbar()
plt.set_cmap('coolwarm')
df_s_full.hist(bins=100, figsize=(10,15), layout=(5,1))
plt.scatter(df_s_full['0'],df_s_full['2'],marker='.',alpha=0.01)
plt.scatter(df_s_full['1'],df_s_full['3'],marker='.',alpha=0.005)
plt.scatter(df_s_full['2'],df_s_full['3'],marker='.',alpha=0.005)
plt.scatter(df_s_full['0'],df_s_full['1'],marker='.',alpha=0.005)
ax = plt.gca()
ax.set_aspect('equal', 'datalim')
plt.grid()
plt.scatter(df_s_full['0'],df_s_full['1'],marker='.',alpha=0.005)
ax = plt.gca()
plt.ylim([0, 500])
plt.xlim([-100, 100])
ax.set_aspect('equal', 'datalim')
plt.grid()
plt.scatter((df_s_full['0']**2+df_s_full['1']**2)**0.5,(df_s_full['2']**2+df_s_full['3']**2)**0.5,marker='.',alpha=0.01)
df_s_full.shape
df_c_full.shape
plt.scatter((df_s_full['0']**2+df_s_full['1']**2)**0.5,df_c_full['0'],marker='.',alpha=0.01)
plt.scatter((df_s_full['2']**2+df_s_full['3']**2)**0.5,df_c_full['0'],marker='.',alpha=0.01)
plt.figure()
for ind in range(2000):
try:
df = pd.read_csv(f'data_apollo/{ind}_sim_c.csv')
plt.plot(df['1'])
except:
pass
ax = plt.gca()
plt.grid()
df_c_full.hist(bins=100, figsize=(10,6), layout=(2,1))
df_c_full['1'].describe()
plt.scatter(df_c_full['1'],df_c_full['0'],marker='.',alpha=0.01)
plt.grid()
import re
import os
index_s = []
index_c = []
for infile in glob.glob("data_apollo/*_sim_s.csv"):
numbers = re.findall('\d+',infile)
index_s.append(int(numbers[0]))
for infile in glob.glob("data_apollo/*_sim_c.csv"):
numbers = re.findall('\d+',infile)
index_c.append(int(numbers[0]))
index_s
index_c
len(index_s)
len(index_c)
| 0.13658 | 0.246148 |
# Arrays in numpy: indexing, ufuncs, broadcasting
Last week you learned why numpy was created. This week we are going to dig a little deeper in this fundamental piece of the scientific python ecosystem.
This chapter contains a lot of new concepts and tools, and I'm aware that you won't be able to remember all of them at once. My objective here is to help you to better understand the numpy documentation when you'll need it, by being prepared for new semantics like "advanced indexing" or "ufunc" (universal function). There are entire books about numpy (I've listed some in the introduction), and my personal recommendation is to learn it on the go (i.e. step by step, task after task). I still hope that this chapter (even if too short for such an important material) will help a little bit.
<h1>Table of Contents<span class="tocSkip"></span></h1>
<div class="toc"><ul class="toc-item"><li><span><a href="#Arrays-in-numpy:-indexing,-ufuncs,-broadcasting" data-toc-modified-id="Arrays-in-numpy:-indexing,-ufuncs,-broadcasting-14"><span class="toc-item-num">14 </span>Arrays in numpy: indexing, ufuncs, broadcasting</a></span><ul class="toc-item"><li><span><a href="#Anatomy-of-a-ndarray" data-toc-modified-id="Anatomy-of-a-ndarray-14.1"><span class="toc-item-num">14.1 </span>Anatomy of a ndarray</a></span></li><li><span><a href="#Creating-ndarrays" data-toc-modified-id="Creating-ndarrays-14.2"><span class="toc-item-num">14.2 </span>Creating ndarrays</a></span><ul class="toc-item"><li><span><a href="#np.emtpy,-np.zeros,-np.ones,-np.full" data-toc-modified-id="np.emtpy,-np.zeros,-np.ones,-np.full-14.2.1"><span class="toc-item-num">14.2.1 </span><code>np.emtpy</code>, <code>np.zeros</code>, <code>np.ones</code>, <code>np.full</code></a></span></li><li><span><a href="#np.array" data-toc-modified-id="np.array-14.2.2"><span class="toc-item-num">14.2.2 </span><code>np.array</code></a></span></li><li><span><a href="#np.copy" data-toc-modified-id="np.copy-14.2.3"><span class="toc-item-num">14.2.3 </span><code>np.copy</code></a></span></li><li><span><a href="#np.arange" data-toc-modified-id="np.arange-14.2.4"><span class="toc-item-num">14.2.4 </span><code>np.arange</code></a></span></li><li><span><a href="#np.linspace" data-toc-modified-id="np.linspace-14.2.5"><span class="toc-item-num">14.2.5 </span><code>np.linspace</code></a></span></li></ul></li><li><span><a href="#The-shape-of-ndarrays" data-toc-modified-id="The-shape-of-ndarrays-14.3"><span class="toc-item-num">14.3 </span>The shape of ndarrays</a></span></li><li><span><a href="#Indexing" data-toc-modified-id="Indexing-14.4"><span class="toc-item-num">14.4 </span>Indexing</a></span><ul class="toc-item"><li><span><a href="#Slicing" data-toc-modified-id="Slicing-14.4.1"><span class="toc-item-num">14.4.1 </span>Slicing</a></span></li><li><span><a href="#Basic-vs-Advanced-indexing" data-toc-modified-id="Basic-vs-Advanced-indexing-14.4.2"><span class="toc-item-num">14.4.2 </span>Basic vs Advanced indexing</a></span></li><li><span><a href="#Integer-indexing" data-toc-modified-id="Integer-indexing-14.4.3"><span class="toc-item-num">14.4.3 </span>Integer indexing</a></span></li><li><span><a href="#Boolean-indexing:-indexing-based-on-a-condition" data-toc-modified-id="Boolean-indexing:-indexing-based-on-a-condition-14.4.4"><span class="toc-item-num">14.4.4 </span>Boolean indexing: indexing based on a condition</a></span></li></ul></li><li><span><a href="#Universal-functions" data-toc-modified-id="Universal-functions-14.5"><span class="toc-item-num">14.5 </span>Universal functions</a></span></li><li><span><a href="#Broadcasting" data-toc-modified-id="Broadcasting-14.6"><span class="toc-item-num">14.6 </span>Broadcasting</a></span></li><li><span><a href="#Take-home-points" data-toc-modified-id="Take-home-points-14.7"><span class="toc-item-num">14.7 </span>Take home points</a></span></li><li><span><a href="#Addendum:-numpy-versus-other-scientific-languages" data-toc-modified-id="Addendum:-numpy-versus-other-scientific-languages-14.8"><span class="toc-item-num">14.8 </span>Addendum: numpy versus other scientific languages</a></span></li><li><span><a href="#What's-next?" data-toc-modified-id="What's-next?-14.9"><span class="toc-item-num">14.9 </span>What's next?</a></span></li><li><span><a href="#License" data-toc-modified-id="License-14.10"><span class="toc-item-num">14.10 </span>License</a></span></li></ul></li></ul></div>
## Anatomy of a ndarray
*From the [numpy reference](https://docs.scipy.org/doc/numpy/reference/arrays.html):*
N-dimensional [ndarray](https://docs.scipy.org/doc/numpy/reference/arrays.ndarray.html#arrays-ndarray)'s are the core structure of the numpy library. It is a multidimensional container of items of the same type and size. The number of dimensions and items in an array is defined by its shape, which is a tuple of N positive integers that specify the number of items in each dimension. The type of items in the array is specified by a separate data-type object (dtype), one of which is associated with each ndarray.
All ndarrays are homogenous: every item takes up the same size block of memory, and all blocks are interpreted in exactly the same way.
An item extracted from an array, e.g., by indexing, is represented by a Python object whose type is one of the array scalar types built in NumPy.
**Figure:** Conceptual diagram showing the relationship between the three fundamental objects used to describe the data in an array: 1) the ndarray itself, 2) the data-type object that describes the layout of a single fixed-size element of the array, 3) the array-scalar Python object that is returned when a single element of the array is accessed.
```
import numpy as np
x = np.array([[1, 2, 3], [4, 5, 6]], np.int32)
type(x)
x.dtype # x is of type ndarray, but the data it contains is not
```
Numpy was created to work with numbers, and large arrays of numbers with the same type. Some of ``ndarray``'s attributes give us information about the memory they need:
```
print(x.itemsize) # size of one element (np.int32) in bytes
print(x.nbytes) # size of 2 * 3 = 6 elements in bytes
```
The "shape" of an array formalizes how the data values are accessed or printed:
```
x.shape
print(x)
```
However, the data is one-dimensional and contiguous *in memory*. I.e. the 1D segment of computer memory stored by the array is combined with an indexing scheme that maps N-dimensional indexes into the location of an item in the block. Concretely, this means that there is no difference in the memory layout of these two arrays:
```
a = np.arange(9)
b = a.reshape((3, 3)) # what does "reshape" do?
```
Both are internally stored using a one dimensional memory block. The difference lies in the way numpy gives access to the internal data:
```
a[4], b[1, 1]
```
This means that elementwise operations have the same execution speed for N dimensional arrays as for 1-D arrays. Now, let's replace an element from array b:
```
b[1, 1] = 99
b
```
Since the indexing ``[1, 1]`` is on the left-hand side of the assignment operator, we modified b **in-place**. Numpy located the block of memory that needed to be changed and replaced it with another number. Since all these numbers are integers with a fixed memory size, this is not a problem. But what happens if we try to assign another data type?
```
b[1, 1] = 999.99
b
```
The float is converted to an integer! This is a dangerous "feature" of numpy and should be used with care.
Another extremely important mechanism of ndarrays is their internal handling of data. Indeed:
```
a
```
What happened here? *Modifying the data in b also modified the data in a*! More precisely, both arrays share the same internal data: b is a **view** of the data owned by a.
```
b.base is a
a.base is None
np.shares_memory(a, b)
```
This allows for memory efficient reshaping and vector operations on numpy arrays, but is a source of confusion for many. in this lecture, we will look into more detail which operations return a view and which return a copy.
## Creating ndarrays
There are [many ways](https://docs.scipy.org/doc/numpy/reference/routines.array-creation.html#array-creation-routines) to create numpy arrays. The functions you will use most often are:
### ``np.emtpy``, ``np.zeros``, ``np.ones``, ``np.full``
These three work the same way, The first argument defines the shape of the array:
```
a = np.ones((2, 3, 4))
a
```
The ``dtype`` kwarg specifies the type:
```
np.zeros(2, dtype=np.bool)
a = np.empty((2, 3), dtype=np.float32)
a.dtype
```
What is an "empty" array by the way?
```
a
```
What are all these numbers? As it turns out, they are completely unpredictable: with ``np.empty``, numpy just takes a free slot of memory somewhere, and doesn't change the bits in it. Computers are smart enough: when deleting a variable, they are just removing the pointer (the address) to this series of bits, not deleting them (this would cost too much time). The same is true for the "delete" function in your operating system by the way: even after "deleting" a file, be aware that a motivated hacker can find and recover your data.
Why using ``np.empty`` instead of ``np.zeros``? Mostly for performance reasons. With ``np.empty``, numpy spares the step of setting all the underlying bits to zero:
```
%timeit np.empty(20000)
%timeit np.ones(20000)
```
So at least a factor 10 faster on my laptop. If you know that your are going to use your array as data storage and fill it later on, it *might* be a good idea to use ``np.empty``. In practice, however, performance doesn't matter that much and avoiding bugs is more important: initialize your arrays with NaNs in order to easily find out if all values were actually set by your program after completion:
```
np.full((2, 3), np.NaN)
```
### ``np.array``
``np.array`` converts existing data to a ndarray:
```
np.array([[1, 2, 3], [4, 5, 6]], np.float64)
```
But be aware that it doesn't behave like the python equivalent ``list``!
```
list('abcd'), np.array('abcd')
```
### ``np.copy``
When a variable is assigned to another variable in python, it creates **a new reference to the object it contains, NOT a copy**:
```
a = np.zeros(3)
b = a
b[1] = 1
a # ups, didn't want to do that!
```
<img src="../img/logo_ex.png" align="left" style="width:1em; height:1em;"> **Question**: if you learned programming with another language (Matlab, R), compare this behavior to the language you used before.
This is why ``np.copy`` is useful:
```
a = np.zeros(3)
b = a.copy() # same as np.copy(a)
b[1] = 1
a # ah!
```
### ``np.arange``
```
np.arange(10)
```
Be careful! the start and stop arguments define the half open interval ``[start, stop[``:
```
np.arange(3, 15, 3)
np.arange(3, 15.00000001, 3)
```
### ``np.linspace``
Regularly spaced intervals between two values (both limits are included this time):
```
np.linspace(0, 1, 11)
```
## The shape of ndarrays
Is numpy [row-major or column-major order](https://en.wikipedia.org/wiki/Row-_and_column-major_order)?
The default order in numpy is that of the C-language: **row-major**. This means that the array:
```
a = np.array([[1, 2, 3],
[4, 5, 6]])
a
```
has two rows and three columns, the rows being listed first:
```
a.shape
```
and that the internal representation in memory is sorted by rows:
```
a.flatten()
```
A consequence is that reading data out of rows (first dimension in the array) is usually a bit faster than reading out of columns:
```
t = np.zeros((1000, 1000))
%timeit t[[0], :]
%timeit t[:, [0]]
```
This is different from some other languages like FORTRAN, Matlab, R, or IDL. In my opinion though, most of the time it doesn't matter much to remember whether it is row- or column-major, as long as you remember which dimension is what. Still, this can be a source of confusion at first, especially if you come from one of these column-major languages.
Most datasets are then going to store the data in a way that allows faster retrieval along the dimension which is read most often: for geophysical datasets, this is often going to be the "time" dimension, i.e. the size of the time dimension is going to give the size of `array.shape[0]`.
My personal way to deal with this when I started using numpy is that I always thought of numpy being in the "wrong" order. If you have data defined in four dimensions (x, y, z, t in the "intuitive", mathematical order), then it is often stored in numpy with the shape (t, z, y, x).
Remember the 4D arrays that we use a lot in the climate lecture: they were stored on disk as netCDF files, and their description read:
```
double t(month, level, latitude, longitude) ;
t:least_significant_digit = 2LL ;
t:units = "K" ;
t:long_name = "Temperature" ;
```
which is also the order used by numpy:
```
# you can't do this at home!
import netCDF4
with netCDF4.Dataset('ERA-Int-MonthlyAvg-4D-T.nc') as nc:
temp = nc.variables['t'][:]
temp.shape
```
This order might be different in other languages. It's just a convention!
Looping over time and z (which happens more often than looping over x or y) is as easy as:
```
for time_slice in temp:
for z_slice in time_slice:
# Do something useful with your 2d spatial slice
assert z_slice.shape == (241, 480)
```
There is one notable exception to this rule though. RGB images:
```
from scipy import misc
import matplotlib.pyplot as plt
%matplotlib inline
img = misc.face()
plt.imshow(img);
img.shape
```
``img`` is in RGB space, and (in my humble opinion) the third dimension (the channel) *should* be at the first place in order to be consistent with the z, y, x order of the temperature variable above. Because of this strange order, if you want to unpack (or loop over) each color channel you'll have to do the counter intuitive:
```
numpy_ordered_img = np.rollaxis(img, 2)
R, G, B = numpy_ordered_img
f, (ax1, ax2, ax3) = plt.subplots(1, 3, figsize=(12, 4))
ax1.imshow(R, cmap='Reds'); ax2.imshow(G, cmap='Greens'); ax3.imshow(B, cmap='Blues');
```
I guess there must be historical reasons for this choice
Anyways, with some experience you'll see that there is *always* a way to get numpy arrays in the shape you want them. It is sometimes going to be confusing and you are going to need some googling skills (like the ``np.rollaxis`` trick from above), but you'll manage. All the tools at your disposition for theses purposes are listed in the [array manipulation](https://docs.scipy.org/doc/numpy-1.14.0/reference/routines.array-manipulation.html) documentation page.
*Parenthesis: note that numpy also allows to use the column-major order you may be familiar with:*
```
a = np.array([[1, 2, 3],
[4, 5, 6]])
a.flatten(order='F')
```
*I **don't** recommend to take this path unless really necessary: sooner or later you are going to regret it. If you really need to flatten an array this way, I recommend the more numpy-like:*
```
a.T.flatten()
```
## Indexing
**Indexing** refers to the act of accessing values in an array by their **index**, i.e. their position in the array.
There are many ways to index arrays, and the [numpy documentation](https://docs.scipy.org/doc/numpy-1.14.0/reference/arrays.indexing.html) about the subject is excellent. Here we will just revise some of the most important aspects of it.
### Slicing
The common way to do **slicing** is by using the following syntax:
```
x = np.array([0, 1, 2, 3, 4, 5, 6, 7, 8, 9])
x[1:7:2]
```
The ``start:stop:step`` syntax is actually creating a python ``slice`` object. The statement above is therefore the **literal version** of the less concise:
```
x[slice(1, 7, 2)]
```
The ``step`` can be used to reverse (flip) the order of elements in an array:
```
x[::-1]
```
Inverting the elements that way is *very* fast. It is not significantly slower than reading the data in order:
```
%timeit x[::-1]
%timeit x[::1]
```
How can that be? Again, it has something to do with the internal memory layout of an array. **Slicing always returns a view** of an array. That is:
```
x = np.array([0, 1, 2, 3, 4, 5, 6, 7, 8, 9], dtype=np.float)
y = x[::-1]
y[0] = np.NaN
x # ups, I also changed x, but at position 9!
```
Or:
```
x = np.array([0, 1, 2, 3, 4, 5, 6, 7, 8, 9], dtype=np.float)
y = x[2:4]
y[0] = np.NaN
x # ups, I also changed x, but at position 2!
```
It is *very* important to keep the view mechanism in mind when writing numpy code. It is a great advantage for performance considerations, but it might lead to unexpected results if you are not careful!
### Basic vs Advanced indexing
Throughout the numpy documentation there is a clear distinction between the terms **basic slicing/indexing** and **advanced indexing**. The numpy developers are insisting on this one because there is a crucial difference between the two:
- **basic slicing/indexing always returns a view**
- **advanced indexing always returns a copy**
Slicing with **a slice object** (constructed by ``start:stop:step`` notation inside of brackets, or ``slice(start, stop, step)``) **is always basic** and returns a view:
```
x = np.array([[1, 2, 3],
[4, 5, 6]])
x[::-1, ::2].base is x
```
**Indexing with an integer is basic** and returns a view:
```
x[:, 2].base is x
x[(slice(0, 1, 1), 2)].base is x
```
In Python, ``x[(exp1, exp2, ..., expN)]`` is equivalent to ``x[exp1, exp2, ..., expN]``; the latter is just "syntactic sugar" for the former.
The obvious exception of the "integer indexing returns a view" rule is when the returned object is a scalar (scalars aren't arrays and cannot be a view of an array):
```
x[1, 2].base is x
```
**Advanced indexing** is triggered when the selection occurs over an ``ndarray`` (as opposed to basic indexing where selection happens with slices and/or integers). There are two types of advanced indexing: integer and boolean.
### Integer indexing
**Integer indexing** happens when selecting data points based on their coordinates:
```
x[[0, 1, 1], [0, 2, 0]]
```
<img src="../img/logo_ex.png" align="left" style="width:1em; height:1em;"> **Question**: Integer indexing is also called "positional indexing". Why?
Let's try to get the corner elements of a 4x3 array:
```
x = np.array([[ 0, 1, 2],
[ 3, 4, 5],
[ 6, 7, 8],
[ 9, 10, 11]])
ycoords = [0, 0, 3, 3]
xcoords = [0, 2, 0, 2]
x[ycoords, xcoords]
```
It may be easier for you to see the indexing command as such: we are indexing the array at 4 locations: ``(0, 0)``, ``(0, 2)``, ``(3, 0)`` and ``(3, 2)``. Therefore, the output array is of length 4 and keeps the order of the coordinate points of course.
A useful feature of advanced indexing is that the shape of the indexers is conserved by the output:
```
ycoords = [[0 , 0],
[-1, -1]]
xcoords = [[ 0, -1],
[ 0, -1]]
x[ycoords, xcoords]
```
Unlike basic indexing, integer indexing doesn't return a view. We have two ways to test if this is the case:
```
x = np.array([1, 2, 3, 4])
y = x[[1, 3]]
y.base is x # y doesn't share memory with x
y[0] = 999 # changing y doesn't alter x
x
```
### Boolean indexing: indexing based on a condition
Instead of integers, booleans can be used for indexing:
```
a = np.array([1, 2, 3, 4])
a[[True, False, True, False]]
```
Unlike integer-based indexing, **the shape of the indexer and the array must match** (except when broadcasting occurs, see below).
The most frequent application of boolean indexing is to select values based on a condition:
```
x = np.array([[ 0, 1, 2],
[ 3, 4, 5],
[ 6, 7, 8],
[ 9, 10, 11]])
x[x >= 8]
```
<img src="../img/logo_ex.png" align="left" style="width:1em; height:1em;"> **Question**: What is the shape of ``x >= 8``? Try it! And try another command, too: ``~ (x >= 8)``.
As you can see, boolean indexing in this case returns a 1D array of the same length as the number of ``True`` in the indexer.
Another way to do indexing based on a condition is to use the [np.nonzero](https://docs.scipy.org/doc/numpy-1.14.0/reference/generated/numpy.nonzero.html) function:
```
nz = np.nonzero(x >= 8)
nz
```
This creates a tuple object of integer arrays specifying the location of where the condition is True, thus it is directly applicable as an indexer:
```
x[nz]
```
In practice there is no difference between ``x[x >= 8]`` and ``x[np.nonzero(x >= 8)]``, but the former is faster in most cases. ``np.nonzero`` is still very useful if you want to get access to the *location* of where certain conditions are met in an array. Both are using advanced indexing, thus returning a copy of the array.
## Universal functions
A universal function (or ``ufunc`` for short) is a function that operates on ``ndarrays`` in an element-by-element fashion. ``ufuncs`` are a core element of the numpy library, and you already used them without noticing: arithmetic operations like multiplication or addition are ``ufuncs``, and trigonometric operations like ``np.sin`` or ``np.cos`` are ``ufuncs`` as well.
Numpy ``ufuncs`` are coded in C, which means that they can apply repeated operations on array elements much faster than their python equivalent. Numpy users use ``ufuncs`` to **vectorize** their code. Exercise [#04-02, Monte-Carlo estimation of $\pi$](13-Assignment-04.ipynb#vector) was an example of such a vectorization process: there were two possible solutions to the problem of estimating $\pi$: one of them contains a for-loop, while the vectorized solution didn't require any loop.
Note that some ``ufuncs`` are hidden from you: calling ``a + b`` on ``ndarrays`` is actually calling ``np.add`` internally. How it is possible for numpy to mess around with the Python syntax in such a way is going to be the topic of another lecture.
The numpy documentation lists [all available ufuncs](https://docs.scipy.org/doc/numpy-1.14.0/reference/ufuncs.html#available-ufuncs) to date. Have a quick look at them, just to see how many there are!
## Broadcasting
*Copyright note: much of the content of this section (including images) is copied from the [EricsBroadcastingDoc](http://scipy.github.io/old-wiki/pages/EricsBroadcastingDoc) page on SciPy.*
When two arrays have the same shape, multiplying them using the multiply ufunc is easy:
```
a = np.array([1.0, 2.0, 3.0])
b = np.array([2.0, 2.0, 2.0])
```
If the shape of the two arrays do not match, however, numpy will raise a ``ValueError``:
```
a = np.array([0.0, 10.0, 20.0, 30.0])
b = np.array([1.0, 2.0, 3.0])
a + b
```
But what does *"could not be broadcast together"* actually mean? **Broadcasting** is a term which is quite specific to numpy. From the [documentation]( https://docs.scipy.org/doc/numpy-1.14.0/user/basics.broadcasting.html): "broadcasting describes how numpy treats arrays with different shapes during arithmetic operations". In which cases does numpy allow arrays of different shape to be associated together via universal functions?
The simplest example is surely the multiplication with a scalar:
```
a = np.array([1, 2, 3])
b = 2.
a * b
```
The action of broadcasting can schematically represented as a "stretching" of the scalar so that the target array b is as large as the array a:
<img src="../img/numpy/image0013830.gif" align='left'>
The rule governing whether two arrays have compatible shapes for broadcasting can be expressed in a single sentence:
**The Broadcasting Rule: in order to broadcast, the size of the trailing axes for both arrays in an operation must either be the same size or one of them must be one.**
For example, let's multiply an array of shape(4, 3) with an array of shape (3) (both trailing axes are of length 3):
```
a = np.array([[ 0, 0, 0],
[10, 10, 10],
[20, 20, 20],
[30, 30, 30]])
b = np.array([0, 1, 2])
a + b
```
Schematically, the array is stretched in the dimension which is missing to fill the gap between the two shapes:
<img src="../img/numpy/image0020619.gif" align='left'>
Broadcasting provides a convenient way of taking the outer product (or any other outer operation) of two arrays. The following example shows an outer addition operation of two 1D arrays that produces the same result as above:
```
a = np.array([0, 10, 20, 30])
b = np.array([0, 1, 2])
a.reshape((4, 1)) + b
```
<img src="../img/numpy/image004de9e.gif" align='left'>
In this case, broadcasting stretches both arrays to form an output array larger than either of the initial arrays.
Note: a convenient syntax for the reshaping operation above is following:
```
a[..., np.newaxis]
a[..., np.newaxis].shape
```
where [np.newaxis](https://docs.scipy.org/doc/numpy-1.13.0/reference/arrays.indexing.html#numpy.newaxis) is used to increase the dimension of the existing array by one more dimension where needed.
Broadcasting is quite useful when writing vectorized functions that apply on vectors and matrices. You will often use broadcasting when working with statistical or physical models dealing with high-dimensional arrays.
## Take home points
- numpy is the core library of the scientific python stack. It is used by many (many) companies and researchers worldwide, and its documentation is good. Use it! There is a [user guide](https://docs.scipy.org/doc/numpy/user/) to get you started, but the [reference](https://docs.scipy.org/doc/numpy/reference/) is more complete.
- "views" allow numpy to spare memory by giving various variables access to the same data. This is good for memory optimization, but error prone: always keep track of the variables pointing to the same data!
- basic indexing operations (slices or single integer indexers) return a view, advanced indexing operations (boolean or integer arrays) return a copy of the data
- universal functions ("ufuncs") is a fancy name for vectorized operations in numpy. You will see the term ufunc quite often in the documentation
- using broadcasting you can operate on arrays with different shapes in a very elegant manner. The rule of broadcasting is simple: in order to broadcast, the size of the trailing axes for both arrays in an operation must either be the same size or one of them must be one.
## Addendum: numpy versus other scientific languages
If you come from a vectorized array language like Matlab or R, most of the information above sounds like "giving fancy names" to things you already used all the time. On top of that, numpy is quite verbose: a ``1:10`` in Matlab becomes a ``np.arange(1., 11)`` in numpy, and ``[ 1 2 3; 4 5 6 ]`` becomes ``np.array([[1.,2.,3.], [4.,5.,6.]])``.
All of this is true and I won't argue about it. It all boils down to the fact that python was not written as a scientific language, and that the scientific tools have been glued together around it. I didn't like it at first either, and I'm still not a big fan of all this verbosity.
What I like, however, is that this syntax is very explicit and clear. Numpy uses the strength and flexibility of python to offer a great number of simple and complex tools to the scientific community: the flourishing ecosystem of packages developing around numpy is a good sign that its upsides are more important than its downsides.
## What's next?
Back to the [table of contents](00-Introduction.ipynb#ctoc), or [jump to the next chapter](15-Scientific-Python.ipynb).
## License
<a href="https://creativecommons.org/licenses/by/4.0/" target="_blank">
<img align="left" src="https://mirrors.creativecommons.org/presskit/buttons/88x31/svg/by.svg"/>
</a>
|
github_jupyter
|
import numpy as np
x = np.array([[1, 2, 3], [4, 5, 6]], np.int32)
type(x)
x.dtype # x is of type ndarray, but the data it contains is not
print(x.itemsize) # size of one element (np.int32) in bytes
print(x.nbytes) # size of 2 * 3 = 6 elements in bytes
x.shape
print(x)
a = np.arange(9)
b = a.reshape((3, 3)) # what does "reshape" do?
a[4], b[1, 1]
b[1, 1] = 99
b
b[1, 1] = 999.99
b
a
b.base is a
a.base is None
np.shares_memory(a, b)
a = np.ones((2, 3, 4))
a
np.zeros(2, dtype=np.bool)
a = np.empty((2, 3), dtype=np.float32)
a.dtype
a
%timeit np.empty(20000)
%timeit np.ones(20000)
np.full((2, 3), np.NaN)
np.array([[1, 2, 3], [4, 5, 6]], np.float64)
list('abcd'), np.array('abcd')
a = np.zeros(3)
b = a
b[1] = 1
a # ups, didn't want to do that!
a = np.zeros(3)
b = a.copy() # same as np.copy(a)
b[1] = 1
a # ah!
np.arange(10)
np.arange(3, 15, 3)
np.arange(3, 15.00000001, 3)
np.linspace(0, 1, 11)
a = np.array([[1, 2, 3],
[4, 5, 6]])
a
a.shape
a.flatten()
t = np.zeros((1000, 1000))
%timeit t[[0], :]
%timeit t[:, [0]]
double t(month, level, latitude, longitude) ;
t:least_significant_digit = 2LL ;
t:units = "K" ;
t:long_name = "Temperature" ;
# you can't do this at home!
import netCDF4
with netCDF4.Dataset('ERA-Int-MonthlyAvg-4D-T.nc') as nc:
temp = nc.variables['t'][:]
temp.shape
for time_slice in temp:
for z_slice in time_slice:
# Do something useful with your 2d spatial slice
assert z_slice.shape == (241, 480)
from scipy import misc
import matplotlib.pyplot as plt
%matplotlib inline
img = misc.face()
plt.imshow(img);
img.shape
numpy_ordered_img = np.rollaxis(img, 2)
R, G, B = numpy_ordered_img
f, (ax1, ax2, ax3) = plt.subplots(1, 3, figsize=(12, 4))
ax1.imshow(R, cmap='Reds'); ax2.imshow(G, cmap='Greens'); ax3.imshow(B, cmap='Blues');
a = np.array([[1, 2, 3],
[4, 5, 6]])
a.flatten(order='F')
a.T.flatten()
x = np.array([0, 1, 2, 3, 4, 5, 6, 7, 8, 9])
x[1:7:2]
x[slice(1, 7, 2)]
x[::-1]
%timeit x[::-1]
%timeit x[::1]
x = np.array([0, 1, 2, 3, 4, 5, 6, 7, 8, 9], dtype=np.float)
y = x[::-1]
y[0] = np.NaN
x # ups, I also changed x, but at position 9!
x = np.array([0, 1, 2, 3, 4, 5, 6, 7, 8, 9], dtype=np.float)
y = x[2:4]
y[0] = np.NaN
x # ups, I also changed x, but at position 2!
x = np.array([[1, 2, 3],
[4, 5, 6]])
x[::-1, ::2].base is x
x[:, 2].base is x
x[(slice(0, 1, 1), 2)].base is x
x[1, 2].base is x
x[[0, 1, 1], [0, 2, 0]]
x = np.array([[ 0, 1, 2],
[ 3, 4, 5],
[ 6, 7, 8],
[ 9, 10, 11]])
ycoords = [0, 0, 3, 3]
xcoords = [0, 2, 0, 2]
x[ycoords, xcoords]
ycoords = [[0 , 0],
[-1, -1]]
xcoords = [[ 0, -1],
[ 0, -1]]
x[ycoords, xcoords]
x = np.array([1, 2, 3, 4])
y = x[[1, 3]]
y.base is x # y doesn't share memory with x
y[0] = 999 # changing y doesn't alter x
x
a = np.array([1, 2, 3, 4])
a[[True, False, True, False]]
x = np.array([[ 0, 1, 2],
[ 3, 4, 5],
[ 6, 7, 8],
[ 9, 10, 11]])
x[x >= 8]
nz = np.nonzero(x >= 8)
nz
x[nz]
a = np.array([1.0, 2.0, 3.0])
b = np.array([2.0, 2.0, 2.0])
a = np.array([0.0, 10.0, 20.0, 30.0])
b = np.array([1.0, 2.0, 3.0])
a + b
a = np.array([1, 2, 3])
b = 2.
a * b
a = np.array([[ 0, 0, 0],
[10, 10, 10],
[20, 20, 20],
[30, 30, 30]])
b = np.array([0, 1, 2])
a + b
a = np.array([0, 10, 20, 30])
b = np.array([0, 1, 2])
a.reshape((4, 1)) + b
a[..., np.newaxis]
a[..., np.newaxis].shape
| 0.302391 | 0.937211 |
```
import numpy as np
import matplotlib.pyplot as plt
from scipy import special
##load data
path = r"C:/Users/14131/Desktop";
data = [];
p = ["1p0", "1p5", "2p0"];
for i in range(3):
data.append(np.loadtxt(path + r"/g_" + p[i] + ".txt"));
#data form
#real_part
#imag_part
#real_part
#...
#delta t = 0.5
#handle data
for i in range(3):
data[i] = data[i][0:800:2] + 1j * data[i][1:800:2];
color_cycle = [u'#1f77b4', u'#ff7f0e', u'#2ca02c', u'#d62728', u'#9467bd', u'#8c564b', u'#e377c2', u'#7f7f7f', u'#bcbd22', u'#17becf'];
#Semi-classical results by Sachdev
#Only useful for g = 1.5 and g = 2.0
t= np.arange(0, 200, 0.5);
delta = [-1.0, -2.0];
K=[];
R = [];
for i in range(2):
K = (np.power(abs(delta[i]), 0.25) * np.exp(1j * delta[i] * t) * np.sqrt(1/ (2 * np.pi *1j * abs(delta[i]) *t[:] )));
tau = 1/(2.0 * 1.0/20.0 / np.pi * np.exp(-abs(delta[i]) * 20));
R.append(K[:] * np.exp(t[:] / tau));
#Get the <sigma(-t)sigma(0)> by complex conjugate.
full_corre = [];
for i in range(3):
temp = np.zeros(1 + 399 * 2, dtype = complex);
for j in range(400):
temp[399 - j] = data[i][j].conjugate();
temp[399 + j] = data[i][j];
full_corre.append(temp);
#fourier transformation
omega = [];
for i in range(3):
temp = np.fft.fft(full_corre[i]);
temp = np.fft.fftshift(temp);
omega.append(temp);
print(len(data[0]))
#plot figures
#g = 1.0
plt.figure(dpi = 150);
plt.plot(np.arange(0,200, 0.5), data[0].real, color = color_cycle[0], label = r"Re");
plt.scatter(np.arange(0,200, 0.5)[0::10], data[0].real[0::10], facecolor= "None", edgecolor = color_cycle[0]);
plt.plot(np.arange(0,200, 0.5), data[0].imag, color = color_cycle[1], label = r"Im");
plt.scatter(np.arange(0,200, 0.5)[0::10], data[0].imag[0::10], facecolor= "None", edgecolor = color_cycle[1]);
plt.xlabel(r"$t$", fontsize = 13);
plt.ylabel(r"$\langle \sigma_z(t)\sigma_z \rangle$", fontsize = 13);
plt.legend(fontsize = 13);
plt.show();
#g = 1.0; S(w);
plt.figure(dpi = 150);
plt.plot(np.arange(-2 * np.pi, 2 * np.pi, 4 * np.pi / (399 * 2 + 1)), abs(omega[0]));
plt.xlabel(r"$\omega$", fontsize = 13);
plt.ylabel(r"$Abs.S(\omega)$", fontsize = 13);
plt.show();
#g = 1.5
plt.figure(dpi = 150);
plt.plot(np.arange(0,200, 0.5), abs(data[1]), color = color_cycle[0], label = r"analytical");
plt.plot(np.arange(0,200, 0.5), abs(R[0]), color = color_cycle[1], label = r"semi-classical");
#plt.scatter(np.arange(0,200.5, 0.5)[0::10], abs(data[1])[0::10], facecolor= "None", edgecolor = color_cycle[0]);
plt.xlabel(r"$t$", fontsize = 13);
plt.ylabel(r"$Abs.\langle \sigma_z(t)\sigma_z \rangle$", fontsize = 13);
plt.legend();
plt.show();
#g = 1.5; S(w);
plt.figure(dpi = 150);
plt.plot(np.arange(-2 * np.pi, 2 * np.pi, 4 * np.pi / (399 * 2 + 1)), abs(omega[1]));
plt.xlabel(r"$\omega$", fontsize = 13);
plt.ylabel(r"$Abs.S(\omega)$", fontsize = 13);
plt.show();
#g = 2.0
plt.figure(dpi = 150);
plt.plot(np.arange(0,200, 0.5), abs(data[2]), color = color_cycle[0], label = r"analytical");
plt.plot(np.arange(0,200, 0.5), abs(R[1]), color = color_cycle[1], label = r"semi-classical");
#plt.scatter(np.arange(0,200.5, 0.5)[0::10], abs(data[1])[0::10], facecolor= "None", edgecolor = color_cycle[0]);
plt.xlabel(r"$t$", fontsize = 13);
plt.ylabel(r"$Abs.\langle \sigma_z(t)\sigma_z \rangle$", fontsize = 13);
plt.legend();
plt.show();
#g = 2.0; S(w);
plt.figure(dpi = 150);
plt.plot(np.arange(-2 * np.pi, 2 * np.pi, 4 * np.pi / (399 * 2 + 1)), abs(omega[2]));
plt.xlabel(r"$\omega$", fontsize = 13);
plt.ylabel(r"$Abs.S(\omega)$", fontsize = 13);
plt.show();
#load data
path = r"C:/Users/14131/Desktop";
data1 = [];
p = ["1p0", "1p5", "2p0"];
for i in range(3):
data1.append(np.loadtxt(path + r"/g_" + p[i]+"_tau_N_501.txt"));
#data form : correlation function in imaginary time are real
#real
#real
#...
g = [1.0, 1.5, 2.0];
plt.figure(dpi = 150);
for i in range(3):
plt.plot(np.arange(0, 20.05, 0.05), data1[i], color = color_cycle[i], label = r"$g = $" + str(g[i]));
plt.scatter(np.arange(0, 20.05, 0.05)[0::10], data1[i][0::10], facecolor = "None", edgecolor = color_cycle[i])
plt.legend();
plt.ylabel(r"$\langle \sigma_z(\tau)\sigma_z \rangle$", fontsize = 13);
plt.xlabel(r"$\tau$", fontsize = 13)
plt.show();
```
|
github_jupyter
|
import numpy as np
import matplotlib.pyplot as plt
from scipy import special
##load data
path = r"C:/Users/14131/Desktop";
data = [];
p = ["1p0", "1p5", "2p0"];
for i in range(3):
data.append(np.loadtxt(path + r"/g_" + p[i] + ".txt"));
#data form
#real_part
#imag_part
#real_part
#...
#delta t = 0.5
#handle data
for i in range(3):
data[i] = data[i][0:800:2] + 1j * data[i][1:800:2];
color_cycle = [u'#1f77b4', u'#ff7f0e', u'#2ca02c', u'#d62728', u'#9467bd', u'#8c564b', u'#e377c2', u'#7f7f7f', u'#bcbd22', u'#17becf'];
#Semi-classical results by Sachdev
#Only useful for g = 1.5 and g = 2.0
t= np.arange(0, 200, 0.5);
delta = [-1.0, -2.0];
K=[];
R = [];
for i in range(2):
K = (np.power(abs(delta[i]), 0.25) * np.exp(1j * delta[i] * t) * np.sqrt(1/ (2 * np.pi *1j * abs(delta[i]) *t[:] )));
tau = 1/(2.0 * 1.0/20.0 / np.pi * np.exp(-abs(delta[i]) * 20));
R.append(K[:] * np.exp(t[:] / tau));
#Get the <sigma(-t)sigma(0)> by complex conjugate.
full_corre = [];
for i in range(3):
temp = np.zeros(1 + 399 * 2, dtype = complex);
for j in range(400):
temp[399 - j] = data[i][j].conjugate();
temp[399 + j] = data[i][j];
full_corre.append(temp);
#fourier transformation
omega = [];
for i in range(3):
temp = np.fft.fft(full_corre[i]);
temp = np.fft.fftshift(temp);
omega.append(temp);
print(len(data[0]))
#plot figures
#g = 1.0
plt.figure(dpi = 150);
plt.plot(np.arange(0,200, 0.5), data[0].real, color = color_cycle[0], label = r"Re");
plt.scatter(np.arange(0,200, 0.5)[0::10], data[0].real[0::10], facecolor= "None", edgecolor = color_cycle[0]);
plt.plot(np.arange(0,200, 0.5), data[0].imag, color = color_cycle[1], label = r"Im");
plt.scatter(np.arange(0,200, 0.5)[0::10], data[0].imag[0::10], facecolor= "None", edgecolor = color_cycle[1]);
plt.xlabel(r"$t$", fontsize = 13);
plt.ylabel(r"$\langle \sigma_z(t)\sigma_z \rangle$", fontsize = 13);
plt.legend(fontsize = 13);
plt.show();
#g = 1.0; S(w);
plt.figure(dpi = 150);
plt.plot(np.arange(-2 * np.pi, 2 * np.pi, 4 * np.pi / (399 * 2 + 1)), abs(omega[0]));
plt.xlabel(r"$\omega$", fontsize = 13);
plt.ylabel(r"$Abs.S(\omega)$", fontsize = 13);
plt.show();
#g = 1.5
plt.figure(dpi = 150);
plt.plot(np.arange(0,200, 0.5), abs(data[1]), color = color_cycle[0], label = r"analytical");
plt.plot(np.arange(0,200, 0.5), abs(R[0]), color = color_cycle[1], label = r"semi-classical");
#plt.scatter(np.arange(0,200.5, 0.5)[0::10], abs(data[1])[0::10], facecolor= "None", edgecolor = color_cycle[0]);
plt.xlabel(r"$t$", fontsize = 13);
plt.ylabel(r"$Abs.\langle \sigma_z(t)\sigma_z \rangle$", fontsize = 13);
plt.legend();
plt.show();
#g = 1.5; S(w);
plt.figure(dpi = 150);
plt.plot(np.arange(-2 * np.pi, 2 * np.pi, 4 * np.pi / (399 * 2 + 1)), abs(omega[1]));
plt.xlabel(r"$\omega$", fontsize = 13);
plt.ylabel(r"$Abs.S(\omega)$", fontsize = 13);
plt.show();
#g = 2.0
plt.figure(dpi = 150);
plt.plot(np.arange(0,200, 0.5), abs(data[2]), color = color_cycle[0], label = r"analytical");
plt.plot(np.arange(0,200, 0.5), abs(R[1]), color = color_cycle[1], label = r"semi-classical");
#plt.scatter(np.arange(0,200.5, 0.5)[0::10], abs(data[1])[0::10], facecolor= "None", edgecolor = color_cycle[0]);
plt.xlabel(r"$t$", fontsize = 13);
plt.ylabel(r"$Abs.\langle \sigma_z(t)\sigma_z \rangle$", fontsize = 13);
plt.legend();
plt.show();
#g = 2.0; S(w);
plt.figure(dpi = 150);
plt.plot(np.arange(-2 * np.pi, 2 * np.pi, 4 * np.pi / (399 * 2 + 1)), abs(omega[2]));
plt.xlabel(r"$\omega$", fontsize = 13);
plt.ylabel(r"$Abs.S(\omega)$", fontsize = 13);
plt.show();
#load data
path = r"C:/Users/14131/Desktop";
data1 = [];
p = ["1p0", "1p5", "2p0"];
for i in range(3):
data1.append(np.loadtxt(path + r"/g_" + p[i]+"_tau_N_501.txt"));
#data form : correlation function in imaginary time are real
#real
#real
#...
g = [1.0, 1.5, 2.0];
plt.figure(dpi = 150);
for i in range(3):
plt.plot(np.arange(0, 20.05, 0.05), data1[i], color = color_cycle[i], label = r"$g = $" + str(g[i]));
plt.scatter(np.arange(0, 20.05, 0.05)[0::10], data1[i][0::10], facecolor = "None", edgecolor = color_cycle[i])
plt.legend();
plt.ylabel(r"$\langle \sigma_z(\tau)\sigma_z \rangle$", fontsize = 13);
plt.xlabel(r"$\tau$", fontsize = 13)
plt.show();
| 0.250271 | 0.53607 |
```
import os
import numpy as np
import scipy.stats as stats
from statsmodels.robust.scale import mad
from statsmodels.graphics.gofplots import qqplot
import pandas as pd
from matplotlib import pyplot
import flowio
%matplotlib inline
time_channel = "Time"
data_channel = "SSC-A"
data_dir = "/home/swhite/Projects/flowClean_testing/data"
diff_roll = 0.01
final_roll = 0.02
k = 2.0
k2 = 3.0
figure_out_dir = "flow_rate_qc_square"
fig_size = (16, 4)
def find_channel_index(channel_dict, pnn_text):
for k, v in channel_dict.iteritems():
if v['PnN'] == pnn_text:
index = int(k) - 1
return index
def calculate_flow_rate(events, time_index, roll):
time_diff = np.diff(events[:, time_index])
time_diff = np.insert(time_diff, 0, 0)
time_diff_mean = pd.rolling_mean(time_diff, roll, min_periods=1)
min_diff = time_diff_mean[time_diff_mean > 0].min()
time_diff_mean[time_diff_mean == 0] = min_diff
flow_rate = 1 / time_diff_mean
return flow_rate
def plot_channel(file_name, x, y, x_label, y_label):
pre_scale = 0.003
my_cmap = pyplot.cm.get_cmap('jet')
my_cmap.set_under('w', alpha=0)
bins = int(np.sqrt(x.shape[0]))
fig = pyplot.figure(figsize=(16, 8))
ax = fig.add_subplot(1, 1, 1)
ax.set_title(file_name, fontsize=16)
ax.set_xlabel(x_label, fontsize=14)
ax.set_ylabel(y_label, fontsize=14)
ax.hist2d(
x,
np.arcsinh(y * pre_scale),
bins=[bins, bins],
cmap=my_cmap,
vmin=0.9
)
fig.tight_layout()
pyplot.show()
def plot_flow_rate(
file_name,
flow_rate,
event_idx,
hline=None,
trendline=None,
x_lim=None,
y_lim=None,
save_figure=False
):
fig = pyplot.figure(figsize=fig_size)
ax = fig.add_subplot(1, 1, 1)
ax.set_title(file_name, fontsize=16)
ax.set_xlabel("Event", fontsize=14)
ax.set_ylabel("Flow rate (events/ms)", fontsize=14)
if x_lim is None:
ax.set_xlim([0, max(event_idx)])
else:
ax.set_xlim([x_lim[0], x_lim[1]])
if y_lim is not None:
ax.set_ylim([y_lim[0], y_lim[1]])
ax.plot(
event_idx,
flow_rate,
c='darkslateblue'
)
if hline is not None:
ax.axhline(hline, linestyle='-', linewidth=1, c='coral')
if trendline is not None:
ax.plot(event_idx, trendline, c='cornflowerblue')
fig.tight_layout()
if save_figure:
fig_name = "".join([file_name, '_a_', 'flow_rate.png'])
fig_path = "/".join([figure_out_dir, fig_name])
pyplot.savefig(fig_path)
pyplot.show()
def plot_deviation(
file_name,
flow_rate,
event_indices,
diff,
stable_diff,
smooth_stable_diff,
threshold,
save_figure=False
):
fig = pyplot.figure(figsize=fig_size)
ax = fig.add_subplot(1, 1, 1)
ax.set_title(file_name, fontsize=16)
ax.set_xlim([0, len(flow_rate)])
#pyplot.ylim([0, 5])
ax.set_xlabel("Event", fontsize=14)
ax.set_ylabel("Deviation (log)", fontsize=14)
ax.plot(
event_indices,
np.log10(1 + diff),
c='coral',
alpha=0.6,
linewidth=1
)
ax.plot(
event_indices,
np.log10(1 + stable_diff),
c='cornflowerblue',
alpha=0.6,
linewidth=1
)
ax.plot(
event_indices,
np.log10(1 + smooth_stable_diff),
c='darkslateblue',
alpha=1.0,
linewidth=1
)
ax.axhline(np.log10(1 + threshold), linestyle='dashed', linewidth=2, c='crimson')
fig.tight_layout()
if save_figure:
fig_name = "".join([file_name, '_b_', 'deviation.png'])
fig_path = "/".join([figure_out_dir, fig_name])
pyplot.savefig(fig_path)
pyplot.show()
def plot_channel_good_vs_bad(
file_name,
channel_data,
time_data,
channel_name,
good_event_map,
bi_ex=True,
save_figure=False,
drop_negative=False
):
pre_scale = 0.003
good_cmap = pyplot.cm.get_cmap('jet')
good_cmap.set_under('w', alpha=0)
bad_cmap = pyplot.cm.get_cmap('jet')
bad_cmap.set_under('w', alpha=0)
x_good = time_data[good_event_map]
x_bad = time_data[~good_event_map]
if bi_ex:
y_good = np.arcsinh(channel_data[good_event_map] * pre_scale)
y_bad = np.arcsinh(channel_data[~good_event_map] * pre_scale)
else:
y_good = channel_data[good_event_map]
y_bad = channel_data[~good_event_map]
# if drop_negative:
# pos_good_indices = y_good > 0
# y_good = y_good[pos_good_indices]
# x_good = x_good[pos_good_indices]
# pos_bad_indices = y_bad > 0
# y_bad = y_bad[pos_bad_indices]
# x_bad = x_bad[pos_bad_indices]
bins_good = int(np.sqrt(good_event_map.shape[0]))
bins_bad = bins_good
if bins_good >= y_good.shape[0]:
print("Good values less than bins: %s" % file_name)
return
if bins_bad >= y_bad.shape[0]:
print("Bad values less than bins: %s" % file_name)
return
fig = pyplot.figure(figsize=fig_size)
ax = fig.add_subplot(1, 1, 1)
ax.set_title(file_name, fontsize=16)
ax.set_xlabel('Time', fontsize=14)
ax.set_ylabel(channel_name, fontsize=14)
ax.hist2d(
x_good,
y_good,
bins=[bins_good, bins_good],
cmap=good_cmap,
vmin=0.9
)
ax.hist2d(
x_bad,
y_bad,
bins=[bins_bad, bins_bad],
cmap=bad_cmap,
vmin=0.9,
alpha=0.3
)
if drop_negative:
ax.set_ylim(ymin=0)
fig.tight_layout()
if save_figure:
fig_name = "".join([file_name, '_c_', 'filtered_events.png'])
fig_path = "/".join([figure_out_dir, fig_name])
pyplot.savefig(fig_path)
pyplot.show()
def clean(fcs_file, save_figures=False):
fd = flowio.FlowData("/".join([data_dir, fcs_file]))
events = np.reshape(fd.events, (-1, fd.channel_count))
time_index = find_channel_index(fd.channels, time_channel)
diff_roll_count = int(diff_roll * events.shape[0])
flow_rate = calculate_flow_rate(events, time_index, diff_roll_count)
median = np.median(flow_rate)
median_diff = np.abs(flow_rate - median)
threshold = k * mad(median_diff)
initial_good_events = median_diff < threshold
event_indices = np.arange(0, len(flow_rate))
good_event_indices = event_indices[initial_good_events]
line_regress = stats.linregress(good_event_indices, flow_rate[initial_good_events])
linear_fit = (line_regress.slope * event_indices) + line_regress.intercept
stable_diff = np.abs(flow_rate - linear_fit)
final_threshold = k2 * mad(stable_diff)
final_w = int(final_roll * stable_diff.shape[0])
smoothed_diff = pd.rolling_mean(stable_diff, window=final_w, min_periods=1, center=True)
final_good_events = smoothed_diff < final_threshold
plot_flow_rate(
fd.name,
flow_rate,
event_indices,
hline=median,
trendline=linear_fit,
save_figure=save_figures
)
plot_deviation(
fd.name,
flow_rate,
event_indices,
median_diff,
stable_diff,
smoothed_diff,
final_threshold,
save_figure=save_figures
)
data_channel_index = find_channel_index(fd.channels, data_channel)
plot_channel_good_vs_bad(
fd.name,
events[:, data_channel_index],
events[:, time_index],
data_channel,
final_good_events,
save_figure=save_figures,
drop_negative=True
)
return final_good_events
if not os.path.isdir(figure_out_dir):
os.mkdir(figure_out_dir)
save_fig = False
fcs_files = os.listdir(data_dir)
fcs_files = sorted(fcs_files)
fd = flowio.FlowData("/".join([data_dir, fcs_files[7]]))
events = np.reshape(fd.events, (-1, fd.channel_count))
plot_channel(fd.name, events[:, 14], events[:, 3], 'Time', 'SSC-A')
f = fcs_files[7]
fd = flowio.FlowData("/".join([data_dir, f]))
events = np.reshape(fd.events, (-1, fd.channel_count))
good = clean(f, save_figures=False)
n_bins = 50
for channel in fd.channels:
channel_name = fd.channels[channel]['PnN']
fig = pyplot.figure(figsize=(16, 4))
ax = fig.add_subplot(1, 1, 1)
good_events = np.arcsinh(events[good, int(channel) - 1] * 0.003)
bad_events = np.arcsinh(events[~good, int(channel) - 1] * 0.003)
#ax.hist([good_events, bad_events], n_bins, normed=1, histtype='bar', stacked=True, color=['cornflowerblue', 'coral'])
ax.hist([bad_events, good_events], n_bins, normed=1, histtype='step', color=['coral', 'cornflowerblue'])
ax.set_title(channel_name)
pyplot.tight_layout()
pyplot.show()
x = np.random.randn(1000, 3)
x.shape
save_fig=True
for fcs_file in fcs_files:
clean(fcs_file, save_figures=save_fig)
```
|
github_jupyter
|
import os
import numpy as np
import scipy.stats as stats
from statsmodels.robust.scale import mad
from statsmodels.graphics.gofplots import qqplot
import pandas as pd
from matplotlib import pyplot
import flowio
%matplotlib inline
time_channel = "Time"
data_channel = "SSC-A"
data_dir = "/home/swhite/Projects/flowClean_testing/data"
diff_roll = 0.01
final_roll = 0.02
k = 2.0
k2 = 3.0
figure_out_dir = "flow_rate_qc_square"
fig_size = (16, 4)
def find_channel_index(channel_dict, pnn_text):
for k, v in channel_dict.iteritems():
if v['PnN'] == pnn_text:
index = int(k) - 1
return index
def calculate_flow_rate(events, time_index, roll):
time_diff = np.diff(events[:, time_index])
time_diff = np.insert(time_diff, 0, 0)
time_diff_mean = pd.rolling_mean(time_diff, roll, min_periods=1)
min_diff = time_diff_mean[time_diff_mean > 0].min()
time_diff_mean[time_diff_mean == 0] = min_diff
flow_rate = 1 / time_diff_mean
return flow_rate
def plot_channel(file_name, x, y, x_label, y_label):
pre_scale = 0.003
my_cmap = pyplot.cm.get_cmap('jet')
my_cmap.set_under('w', alpha=0)
bins = int(np.sqrt(x.shape[0]))
fig = pyplot.figure(figsize=(16, 8))
ax = fig.add_subplot(1, 1, 1)
ax.set_title(file_name, fontsize=16)
ax.set_xlabel(x_label, fontsize=14)
ax.set_ylabel(y_label, fontsize=14)
ax.hist2d(
x,
np.arcsinh(y * pre_scale),
bins=[bins, bins],
cmap=my_cmap,
vmin=0.9
)
fig.tight_layout()
pyplot.show()
def plot_flow_rate(
file_name,
flow_rate,
event_idx,
hline=None,
trendline=None,
x_lim=None,
y_lim=None,
save_figure=False
):
fig = pyplot.figure(figsize=fig_size)
ax = fig.add_subplot(1, 1, 1)
ax.set_title(file_name, fontsize=16)
ax.set_xlabel("Event", fontsize=14)
ax.set_ylabel("Flow rate (events/ms)", fontsize=14)
if x_lim is None:
ax.set_xlim([0, max(event_idx)])
else:
ax.set_xlim([x_lim[0], x_lim[1]])
if y_lim is not None:
ax.set_ylim([y_lim[0], y_lim[1]])
ax.plot(
event_idx,
flow_rate,
c='darkslateblue'
)
if hline is not None:
ax.axhline(hline, linestyle='-', linewidth=1, c='coral')
if trendline is not None:
ax.plot(event_idx, trendline, c='cornflowerblue')
fig.tight_layout()
if save_figure:
fig_name = "".join([file_name, '_a_', 'flow_rate.png'])
fig_path = "/".join([figure_out_dir, fig_name])
pyplot.savefig(fig_path)
pyplot.show()
def plot_deviation(
file_name,
flow_rate,
event_indices,
diff,
stable_diff,
smooth_stable_diff,
threshold,
save_figure=False
):
fig = pyplot.figure(figsize=fig_size)
ax = fig.add_subplot(1, 1, 1)
ax.set_title(file_name, fontsize=16)
ax.set_xlim([0, len(flow_rate)])
#pyplot.ylim([0, 5])
ax.set_xlabel("Event", fontsize=14)
ax.set_ylabel("Deviation (log)", fontsize=14)
ax.plot(
event_indices,
np.log10(1 + diff),
c='coral',
alpha=0.6,
linewidth=1
)
ax.plot(
event_indices,
np.log10(1 + stable_diff),
c='cornflowerblue',
alpha=0.6,
linewidth=1
)
ax.plot(
event_indices,
np.log10(1 + smooth_stable_diff),
c='darkslateblue',
alpha=1.0,
linewidth=1
)
ax.axhline(np.log10(1 + threshold), linestyle='dashed', linewidth=2, c='crimson')
fig.tight_layout()
if save_figure:
fig_name = "".join([file_name, '_b_', 'deviation.png'])
fig_path = "/".join([figure_out_dir, fig_name])
pyplot.savefig(fig_path)
pyplot.show()
def plot_channel_good_vs_bad(
file_name,
channel_data,
time_data,
channel_name,
good_event_map,
bi_ex=True,
save_figure=False,
drop_negative=False
):
pre_scale = 0.003
good_cmap = pyplot.cm.get_cmap('jet')
good_cmap.set_under('w', alpha=0)
bad_cmap = pyplot.cm.get_cmap('jet')
bad_cmap.set_under('w', alpha=0)
x_good = time_data[good_event_map]
x_bad = time_data[~good_event_map]
if bi_ex:
y_good = np.arcsinh(channel_data[good_event_map] * pre_scale)
y_bad = np.arcsinh(channel_data[~good_event_map] * pre_scale)
else:
y_good = channel_data[good_event_map]
y_bad = channel_data[~good_event_map]
# if drop_negative:
# pos_good_indices = y_good > 0
# y_good = y_good[pos_good_indices]
# x_good = x_good[pos_good_indices]
# pos_bad_indices = y_bad > 0
# y_bad = y_bad[pos_bad_indices]
# x_bad = x_bad[pos_bad_indices]
bins_good = int(np.sqrt(good_event_map.shape[0]))
bins_bad = bins_good
if bins_good >= y_good.shape[0]:
print("Good values less than bins: %s" % file_name)
return
if bins_bad >= y_bad.shape[0]:
print("Bad values less than bins: %s" % file_name)
return
fig = pyplot.figure(figsize=fig_size)
ax = fig.add_subplot(1, 1, 1)
ax.set_title(file_name, fontsize=16)
ax.set_xlabel('Time', fontsize=14)
ax.set_ylabel(channel_name, fontsize=14)
ax.hist2d(
x_good,
y_good,
bins=[bins_good, bins_good],
cmap=good_cmap,
vmin=0.9
)
ax.hist2d(
x_bad,
y_bad,
bins=[bins_bad, bins_bad],
cmap=bad_cmap,
vmin=0.9,
alpha=0.3
)
if drop_negative:
ax.set_ylim(ymin=0)
fig.tight_layout()
if save_figure:
fig_name = "".join([file_name, '_c_', 'filtered_events.png'])
fig_path = "/".join([figure_out_dir, fig_name])
pyplot.savefig(fig_path)
pyplot.show()
def clean(fcs_file, save_figures=False):
fd = flowio.FlowData("/".join([data_dir, fcs_file]))
events = np.reshape(fd.events, (-1, fd.channel_count))
time_index = find_channel_index(fd.channels, time_channel)
diff_roll_count = int(diff_roll * events.shape[0])
flow_rate = calculate_flow_rate(events, time_index, diff_roll_count)
median = np.median(flow_rate)
median_diff = np.abs(flow_rate - median)
threshold = k * mad(median_diff)
initial_good_events = median_diff < threshold
event_indices = np.arange(0, len(flow_rate))
good_event_indices = event_indices[initial_good_events]
line_regress = stats.linregress(good_event_indices, flow_rate[initial_good_events])
linear_fit = (line_regress.slope * event_indices) + line_regress.intercept
stable_diff = np.abs(flow_rate - linear_fit)
final_threshold = k2 * mad(stable_diff)
final_w = int(final_roll * stable_diff.shape[0])
smoothed_diff = pd.rolling_mean(stable_diff, window=final_w, min_periods=1, center=True)
final_good_events = smoothed_diff < final_threshold
plot_flow_rate(
fd.name,
flow_rate,
event_indices,
hline=median,
trendline=linear_fit,
save_figure=save_figures
)
plot_deviation(
fd.name,
flow_rate,
event_indices,
median_diff,
stable_diff,
smoothed_diff,
final_threshold,
save_figure=save_figures
)
data_channel_index = find_channel_index(fd.channels, data_channel)
plot_channel_good_vs_bad(
fd.name,
events[:, data_channel_index],
events[:, time_index],
data_channel,
final_good_events,
save_figure=save_figures,
drop_negative=True
)
return final_good_events
if not os.path.isdir(figure_out_dir):
os.mkdir(figure_out_dir)
save_fig = False
fcs_files = os.listdir(data_dir)
fcs_files = sorted(fcs_files)
fd = flowio.FlowData("/".join([data_dir, fcs_files[7]]))
events = np.reshape(fd.events, (-1, fd.channel_count))
plot_channel(fd.name, events[:, 14], events[:, 3], 'Time', 'SSC-A')
f = fcs_files[7]
fd = flowio.FlowData("/".join([data_dir, f]))
events = np.reshape(fd.events, (-1, fd.channel_count))
good = clean(f, save_figures=False)
n_bins = 50
for channel in fd.channels:
channel_name = fd.channels[channel]['PnN']
fig = pyplot.figure(figsize=(16, 4))
ax = fig.add_subplot(1, 1, 1)
good_events = np.arcsinh(events[good, int(channel) - 1] * 0.003)
bad_events = np.arcsinh(events[~good, int(channel) - 1] * 0.003)
#ax.hist([good_events, bad_events], n_bins, normed=1, histtype='bar', stacked=True, color=['cornflowerblue', 'coral'])
ax.hist([bad_events, good_events], n_bins, normed=1, histtype='step', color=['coral', 'cornflowerblue'])
ax.set_title(channel_name)
pyplot.tight_layout()
pyplot.show()
x = np.random.randn(1000, 3)
x.shape
save_fig=True
for fcs_file in fcs_files:
clean(fcs_file, save_figures=save_fig)
| 0.343122 | 0.418935 |
```
import time
import numpy as np
import pandas as pd
import random
# Install Simon
!pip install git+https://github.com/algorine/simon
from Simon import Simon
from Simon.Encoder import Encoder
from Simon.LengthStandardizer import DataLengthStandardizerRaw
### Read-in the emails and print some basic statistics
# Enron
EnronEmails = pd.read_csv('../input/enron-email-bodies/enron_emails_body.csv',dtype='str', header=None)
print("The size of the Enron emails dataframe is:")
print(EnronEmails.shape)
print("Ten Enron emails are:")
print(EnronEmails.loc[:10])
# Spam
SpamEmails = pd.read_csv('../input/fraudulent-email-bodies/fraudulent_emails_body.csv',encoding="ISO-8859-1",dtype='str', header=None)
print("The size of the Spam emails dataframe is:")
print(SpamEmails.shape)
print("Ten Spam emails are:")
print(SpamEmails.loc[:10])
# Some hyper-parameters for the CNN we will use
maxlen = 20 # max length of each tabular cell <==> max number of characters in a line
max_cells = 50 # max number of cells in a column <==> max number of email lines
p_threshold = 0.5 # prediction threshold probability
Nsamp = 5000
nb_epoch = 20
#batch_size = 8
#checkpoint_dir = "trained_models/"
# Convert everything to lower-case, put one sentence per column in a tabular
# structure
ProcessedEnronEmails=[row.lower().split('\n') for row in EnronEmails.iloc[:,1]]
#print("3 Enron emails after Processing (in list form) are:")
#print((ProcessedEnronEmails[:3]))
EnronEmails = pd.DataFrame(random.sample(ProcessedEnronEmails,Nsamp)).transpose()
EnronEmails = DataLengthStandardizerRaw(EnronEmails,max_cells)
#print("Ten Enron emails after Processing (in DataFrame form) are:")
#print((EnronEmails[:10]))
print("Enron email dataframe after Processing shape:")
print(EnronEmails.shape)
ProcessedSpamEmails=[row.lower().split('/n') for row in SpamEmails.iloc[:,1]]
#print("3 Spam emails after Processing (in list form) are:")
#print((ProcessedSpamEmails[:3]))
SpamEmails = pd.DataFrame(random.sample(ProcessedSpamEmails,Nsamp)).transpose()
SpamEmails = DataLengthStandardizerRaw(SpamEmails,max_cells)
#print("Ten Spam emails after Processing (in DataFrame form) are:")
#print((SpamEmails[:10]))
print("Spam email dataframe after Processing shape:")
print(SpamEmails.shape)
# Encode labels and data
Categories = ['spam','notspam']
encoder = Encoder(categories=Categories)
header = ([['spam',]]*Nsamp)
header.extend(([['notspam',]]*Nsamp))
#print(header)
import time
batch_size = 32
checkpoint_dir = ""
raw_data = np.column_stack((SpamEmails,EnronEmails)).T
print("DEBUG::raw_data:")
print(raw_data)
encoder.process(raw_data, max_cells)
X, y = encoder.encode_data(raw_data, header, maxlen)
print("DEBUG::X")
print(X)
print("DEBUG::y")
print(y)
Classifier = Simon(encoder=encoder)
data = Classifier.setup_test_sets(X, y)
category_count = y.shape[1]
model = Classifier.generate_model(maxlen, max_cells, category_count)
model.compile(loss='binary_crossentropy',optimizer='adam', metrics=['binary_accuracy'])
start = time.time()
history = Classifier.train_model(batch_size, checkpoint_dir, model, nb_epoch, data)
end = time.time()
print("Time for training is %f sec"%(end-start))
config = { 'encoder' : encoder,
'checkpoint' : Classifier.get_best_checkpoint(checkpoint_dir) }
Classifier.save_config(config, checkpoint_dir)
Classifier.plot_loss(history) #comment out on docker images...
pred_headers = Classifier.evaluate_model(max_cells, model, data, encoder, p_threshold)
#print("DEBUG::The predicted headers are:")
#print(pred_headers)
#print("DEBUG::The actual headers are:")
#print(header)
import os
os.listdir(".")
from IPython.display import HTML
def create_download_link(title = "Download file", filename = "data.csv"):
html = '<a href={filename}>{title}</a>'
html = html.format(title=title,filename=filename)
return HTML(html)
# create a link to download the dataframe which was saved with .to_csv method
# create_download_link(filename='text-class.10-0.15.hdf5')
```
|
github_jupyter
|
import time
import numpy as np
import pandas as pd
import random
# Install Simon
!pip install git+https://github.com/algorine/simon
from Simon import Simon
from Simon.Encoder import Encoder
from Simon.LengthStandardizer import DataLengthStandardizerRaw
### Read-in the emails and print some basic statistics
# Enron
EnronEmails = pd.read_csv('../input/enron-email-bodies/enron_emails_body.csv',dtype='str', header=None)
print("The size of the Enron emails dataframe is:")
print(EnronEmails.shape)
print("Ten Enron emails are:")
print(EnronEmails.loc[:10])
# Spam
SpamEmails = pd.read_csv('../input/fraudulent-email-bodies/fraudulent_emails_body.csv',encoding="ISO-8859-1",dtype='str', header=None)
print("The size of the Spam emails dataframe is:")
print(SpamEmails.shape)
print("Ten Spam emails are:")
print(SpamEmails.loc[:10])
# Some hyper-parameters for the CNN we will use
maxlen = 20 # max length of each tabular cell <==> max number of characters in a line
max_cells = 50 # max number of cells in a column <==> max number of email lines
p_threshold = 0.5 # prediction threshold probability
Nsamp = 5000
nb_epoch = 20
#batch_size = 8
#checkpoint_dir = "trained_models/"
# Convert everything to lower-case, put one sentence per column in a tabular
# structure
ProcessedEnronEmails=[row.lower().split('\n') for row in EnronEmails.iloc[:,1]]
#print("3 Enron emails after Processing (in list form) are:")
#print((ProcessedEnronEmails[:3]))
EnronEmails = pd.DataFrame(random.sample(ProcessedEnronEmails,Nsamp)).transpose()
EnronEmails = DataLengthStandardizerRaw(EnronEmails,max_cells)
#print("Ten Enron emails after Processing (in DataFrame form) are:")
#print((EnronEmails[:10]))
print("Enron email dataframe after Processing shape:")
print(EnronEmails.shape)
ProcessedSpamEmails=[row.lower().split('/n') for row in SpamEmails.iloc[:,1]]
#print("3 Spam emails after Processing (in list form) are:")
#print((ProcessedSpamEmails[:3]))
SpamEmails = pd.DataFrame(random.sample(ProcessedSpamEmails,Nsamp)).transpose()
SpamEmails = DataLengthStandardizerRaw(SpamEmails,max_cells)
#print("Ten Spam emails after Processing (in DataFrame form) are:")
#print((SpamEmails[:10]))
print("Spam email dataframe after Processing shape:")
print(SpamEmails.shape)
# Encode labels and data
Categories = ['spam','notspam']
encoder = Encoder(categories=Categories)
header = ([['spam',]]*Nsamp)
header.extend(([['notspam',]]*Nsamp))
#print(header)
import time
batch_size = 32
checkpoint_dir = ""
raw_data = np.column_stack((SpamEmails,EnronEmails)).T
print("DEBUG::raw_data:")
print(raw_data)
encoder.process(raw_data, max_cells)
X, y = encoder.encode_data(raw_data, header, maxlen)
print("DEBUG::X")
print(X)
print("DEBUG::y")
print(y)
Classifier = Simon(encoder=encoder)
data = Classifier.setup_test_sets(X, y)
category_count = y.shape[1]
model = Classifier.generate_model(maxlen, max_cells, category_count)
model.compile(loss='binary_crossentropy',optimizer='adam', metrics=['binary_accuracy'])
start = time.time()
history = Classifier.train_model(batch_size, checkpoint_dir, model, nb_epoch, data)
end = time.time()
print("Time for training is %f sec"%(end-start))
config = { 'encoder' : encoder,
'checkpoint' : Classifier.get_best_checkpoint(checkpoint_dir) }
Classifier.save_config(config, checkpoint_dir)
Classifier.plot_loss(history) #comment out on docker images...
pred_headers = Classifier.evaluate_model(max_cells, model, data, encoder, p_threshold)
#print("DEBUG::The predicted headers are:")
#print(pred_headers)
#print("DEBUG::The actual headers are:")
#print(header)
import os
os.listdir(".")
from IPython.display import HTML
def create_download_link(title = "Download file", filename = "data.csv"):
html = '<a href={filename}>{title}</a>'
html = html.format(title=title,filename=filename)
return HTML(html)
# create a link to download the dataframe which was saved with .to_csv method
# create_download_link(filename='text-class.10-0.15.hdf5')
| 0.268558 | 0.288698 |
# Introduction to XGBoost Spark with GPU
Taxi is an example of xgboost regressor. In this notebook, we will show you how to load data, train the xgboost model and use this model to predict "fare_amount" of your taxi trip. Comparing to original XGBoost Spark codes, there're only two API differences.
## Load libraries
First we load some common libraries that both GPU version and CPU version xgboost will use:
```
import ml.dmlc.xgboost4j.scala.spark.{XGBoostRegressor, XGBoostRegressionModel}
import org.apache.spark.sql.SparkSession
import org.apache.spark.ml.evaluation.RegressionEvaluator
import org.apache.spark.sql.types.{DoubleType, IntegerType, StructField, StructType}
```
what is new to xgboost-spark users is only `rapids.GpuDataReader`
```
import ml.dmlc.xgboost4j.scala.spark.rapids.{GpuDataReader, GpuDataset}
```
Some libraries needed for CPU version are not needed in GPU version any more. The extra libraries needed for CPU are like below:
```scala
import org.apache.spark.ml.feature.VectorAssembler
import org.apache.spark.sql.DataFrame
import org.apache.spark.sql.functions._
import org.apache.spark.sql.types.FloatType
```
## Set your dataset path
```
// Set the paths of datasets for training and prediction
// You need to update them to your real paths!
val trainPath = "/data/taxi/csv/train/"
val evalPath = "/data/taxi/csv/eval/"
```
## Set the schema of the dataset
For Taxi example, the data has 16 columns: 15 features and 1 label. "fare_amount" is set to the label column. The schema will be used to help load data in the future. We also defined some key parameters used in xgboost training process. We also set some basic xgboost parameters here.
```
lazy val schema =
StructType(Array(
StructField("vendor_id", DoubleType),
StructField("passenger_count", DoubleType),
StructField("trip_distance", DoubleType),
StructField("pickup_longitude", DoubleType),
StructField("pickup_latitude", DoubleType),
StructField("rate_code", DoubleType),
StructField("store_and_fwd", DoubleType),
StructField("dropoff_longitude", DoubleType),
StructField("dropoff_latitude", DoubleType),
StructField(labelName, DoubleType),
StructField("hour", DoubleType),
StructField("year", IntegerType),
StructField("month", IntegerType),
StructField("day", DoubleType),
StructField("day_of_week", DoubleType),
StructField("is_weekend", DoubleType)
))
val labelName = "fare_amount"
lazy val paramMap = Map(
"learning_rate" -> 0.05,
"max_depth" -> 8,
"subsample" -> 0.8,
"gamma" -> 1,
"num_round" -> 500
)
```
## Create a new spark session and load data
we must create a new spark session to continue all spark operations. It will also be used to initilize the `GpuDataReader` which is a data reader powered by GPU.
NOTE: in this notebook, we have uploaded dependency jars when installing toree kernel. If we don't upload them at installation time, we can also upload in notebook by [%AddJar magic](https://toree.incubator.apache.org/docs/current/user/faq/). However, there's one restriction for `%AddJar`: the jar uploaded can only be available when `AddJar` is called after a new spark session is created. We must use it as below:
```scala
import org.apache.spark.sql.SparkSession
val spark = SparkSession.builder().appName("Taxi-GPU").getOrCreate
%AddJar file:/data/libs/cudf-0.8-Beta-cuda10.jar
%AddJar file:/data/libs/xgboost4j-0.9_on_Rapids-cuda10.jar
%AddJar file:/data/libs/xgboost4j-spark-0.9_on_Rapids-cuda10.jar
// ...
```
```
val spark = SparkSession.builder().appName("Taxi-GPU").getOrCreate
```
Here's the first API difference, we now use GpuDataReader to load dataset. Similar to original Spark data loading API, GpuDataReader also uses chaining call of "option", "schema","csv". For CPU verions data reader, the code is like below:
```scala
val dataReader = spark.read
```
`featureNames` is used to tell xgboost which columns are `features`, and which column is `label`.
```
val reader = new GpuDataReader(spark).option("header", true).schema(schema)
val featureNames = schema.filter(_.name != labelName).map(_.name)
```
## Initialize XGBoostClassifier
The second API difference is `setFeaturesCol` in CPU version vs `setFeaturesCols` in GPU version. setFeaturesCol accepts a String that indicates which vectorized column is the feature column. It requires `VectorAssembler` to help vectorize all feature columns into one. setFeaturesCols accepts a list of strings so that we don't need VectorAssembler any more. So GPU verion help reduce the preparation codes before you train your xgboost model.
CPU version:
```scala
object Vectorize {
def apply(df: DataFrame, featureNames: Seq[String], labelName: String): DataFrame = {
val toFloat = df.schema.map(f => col(f.name).cast(FloatType))
new VectorAssembler()
.setInputCols(featureNames.toArray)
.setOutputCol("features")
.transform(df.select(toFloat:_*))
.select(col("features"), col(labelName))
}
}
val reader = spark.read.option("header", true).schema(schema)
var trainSet = reader.csv(trainPath)
var evalSet = reader.csv(evalPath)
trainSet = Vectorize(trainSet, featureNames, labelColName)
evalSet = Vectorize(evalSet, featureNames, labelColName)
```
While with GpuDataReader, `VectorAssembler` is not needed any more. We can simply read data by:
```
val trainSet = reader.csv(trainPath)
val evalSet = reader.csv(evalPath)
```
## Add XGBoost parameters for GPU version
Another modification is `num_workers` should be set to the number of machines with GPU in Spark cluster, while it can be set to the number of your CPU cores in CPU version
```scala
// difference in parameters
"tree_method" -> "hist",
"num_workers" -> 12
```
```
val xgbParamFinal = paramMap ++ Map("tree_method" -> "gpu_hist", "num_workers" -> 1)
```
## Initialize XGBoostClassifier
The second API difference is `setFeaturesCol` in CPU version vs `setFeaturesCols` in GPU version. `setFeaturesCol` accepts a String that indicates which vectorized column is the feature column. It requires `VectorAssembler` to help vectorize all feature columns into one. setFeaturesCols accepts a list of strings so that we don't need VectorAssembler any more. So GPU verion help reduce the preparation codes before you train your xgboost model.
CPU version:
```scala
val xgbClassifier = new XGBoostClassifier(xgbParamFinal)
.setLabelCol(labelColName)
.setFeaturesCol("features")
```
```
val xgbRegressor = new XGBoostRegressor(xgbParamFinal)
.setLabelCol(labelName)
.setFeaturesCols(featureNames)
```
## Benchmark and train
The benchmark object is for calculating training time. We will use it to compare with xgboost in CPU version.
```
object Benchmark {
def time[R](phase: String)(block: => R): (R, Float) = {
val t0 = System.currentTimeMillis
val result = block // call-by-name
val t1 = System.currentTimeMillis
println("Elapsed time [" + phase + "]: " + ((t1 - t0).toFloat / 1000) + "s")
(result, (t1 - t0).toFloat / 1000)
}
}
// start training
val (model, _) = Benchmark.time("train") {
xgbRegressor.fit(trainSet)
}
```
## Transformation and evaluation
We use `evalSet` to evaluate our model and use some key columns to show our predictions. Finally we use `MulticlassClassificationEvaluator` to calculate an overall accuracy of our predictions.
```
// start transform
val (prediction, _) = Benchmark.time("transform") {
val ret = model.transform(evalSet).cache()
ret.foreachPartition(_ => ())
ret
}
prediction.select("vendor_id", "passenger_count", "trip_distance", labelName, "prediction").show(10)
val evaluator = new RegressionEvaluator().setLabelCol(labelName)
val (rmse, _) = Benchmark.time("evaluation") {
evaluator.evaluate(prediction)
}
println(s"RMSE == $rmse")
```
## Save the model to disk and load model
We save the model to disk and then load it to memory. We can use the loaded model to do a new prediction.
```
model.write.overwrite.save("/data/model/taxi")
val modelFromDisk = XGBoostRegressionModel.load("/data/model/taxi")
val (results2, _) = Benchmark.time("transform2") {
modelFromDisk.transform(evalSet)
}
results2.select("vendor_id", "passenger_count", "trip_distance", labelName, "prediction").show(5)
spark.close()
```
|
github_jupyter
|
import ml.dmlc.xgboost4j.scala.spark.{XGBoostRegressor, XGBoostRegressionModel}
import org.apache.spark.sql.SparkSession
import org.apache.spark.ml.evaluation.RegressionEvaluator
import org.apache.spark.sql.types.{DoubleType, IntegerType, StructField, StructType}
import ml.dmlc.xgboost4j.scala.spark.rapids.{GpuDataReader, GpuDataset}
import org.apache.spark.ml.feature.VectorAssembler
import org.apache.spark.sql.DataFrame
import org.apache.spark.sql.functions._
import org.apache.spark.sql.types.FloatType
// Set the paths of datasets for training and prediction
// You need to update them to your real paths!
val trainPath = "/data/taxi/csv/train/"
val evalPath = "/data/taxi/csv/eval/"
lazy val schema =
StructType(Array(
StructField("vendor_id", DoubleType),
StructField("passenger_count", DoubleType),
StructField("trip_distance", DoubleType),
StructField("pickup_longitude", DoubleType),
StructField("pickup_latitude", DoubleType),
StructField("rate_code", DoubleType),
StructField("store_and_fwd", DoubleType),
StructField("dropoff_longitude", DoubleType),
StructField("dropoff_latitude", DoubleType),
StructField(labelName, DoubleType),
StructField("hour", DoubleType),
StructField("year", IntegerType),
StructField("month", IntegerType),
StructField("day", DoubleType),
StructField("day_of_week", DoubleType),
StructField("is_weekend", DoubleType)
))
val labelName = "fare_amount"
lazy val paramMap = Map(
"learning_rate" -> 0.05,
"max_depth" -> 8,
"subsample" -> 0.8,
"gamma" -> 1,
"num_round" -> 500
)
import org.apache.spark.sql.SparkSession
val spark = SparkSession.builder().appName("Taxi-GPU").getOrCreate
%AddJar file:/data/libs/cudf-0.8-Beta-cuda10.jar
%AddJar file:/data/libs/xgboost4j-0.9_on_Rapids-cuda10.jar
%AddJar file:/data/libs/xgboost4j-spark-0.9_on_Rapids-cuda10.jar
// ...
val spark = SparkSession.builder().appName("Taxi-GPU").getOrCreate
val dataReader = spark.read
val reader = new GpuDataReader(spark).option("header", true).schema(schema)
val featureNames = schema.filter(_.name != labelName).map(_.name)
object Vectorize {
def apply(df: DataFrame, featureNames: Seq[String], labelName: String): DataFrame = {
val toFloat = df.schema.map(f => col(f.name).cast(FloatType))
new VectorAssembler()
.setInputCols(featureNames.toArray)
.setOutputCol("features")
.transform(df.select(toFloat:_*))
.select(col("features"), col(labelName))
}
}
val reader = spark.read.option("header", true).schema(schema)
var trainSet = reader.csv(trainPath)
var evalSet = reader.csv(evalPath)
trainSet = Vectorize(trainSet, featureNames, labelColName)
evalSet = Vectorize(evalSet, featureNames, labelColName)
val trainSet = reader.csv(trainPath)
val evalSet = reader.csv(evalPath)
// difference in parameters
"tree_method" -> "hist",
"num_workers" -> 12
val xgbParamFinal = paramMap ++ Map("tree_method" -> "gpu_hist", "num_workers" -> 1)
val xgbClassifier = new XGBoostClassifier(xgbParamFinal)
.setLabelCol(labelColName)
.setFeaturesCol("features")
val xgbRegressor = new XGBoostRegressor(xgbParamFinal)
.setLabelCol(labelName)
.setFeaturesCols(featureNames)
object Benchmark {
def time[R](phase: String)(block: => R): (R, Float) = {
val t0 = System.currentTimeMillis
val result = block // call-by-name
val t1 = System.currentTimeMillis
println("Elapsed time [" + phase + "]: " + ((t1 - t0).toFloat / 1000) + "s")
(result, (t1 - t0).toFloat / 1000)
}
}
// start training
val (model, _) = Benchmark.time("train") {
xgbRegressor.fit(trainSet)
}
// start transform
val (prediction, _) = Benchmark.time("transform") {
val ret = model.transform(evalSet).cache()
ret.foreachPartition(_ => ())
ret
}
prediction.select("vendor_id", "passenger_count", "trip_distance", labelName, "prediction").show(10)
val evaluator = new RegressionEvaluator().setLabelCol(labelName)
val (rmse, _) = Benchmark.time("evaluation") {
evaluator.evaluate(prediction)
}
println(s"RMSE == $rmse")
model.write.overwrite.save("/data/model/taxi")
val modelFromDisk = XGBoostRegressionModel.load("/data/model/taxi")
val (results2, _) = Benchmark.time("transform2") {
modelFromDisk.transform(evalSet)
}
results2.select("vendor_id", "passenger_count", "trip_distance", labelName, "prediction").show(5)
spark.close()
| 0.390243 | 0.949856 |
# Character Sequence to Sequence
In this notebook, we'll build a model that takes in a sequence of letters, and outputs a sorted version of that sequence. We'll do that using what we've learned so far about Sequence to Sequence models. This notebook was updated to work with TensorFlow 1.1 and builds on the work of Dave Currie. Check out Dave's post [Text Summarization with Amazon Reviews](https://medium.com/towards-data-science/text-summarization-with-amazon-reviews-41801c2210b).
<img src="images/sequence-to-sequence.jpg"/>
## Dataset
The dataset lives in the /data/ folder. At the moment, it is made up of the following files:
* **letters_source.txt**: The list of input letter sequences. Each sequence is its own line.
* **letters_target.txt**: The list of target sequences we'll use in the training process. Each sequence here is a response to the input sequence in letters_source.txt with the same line number.
```
import numpy as np
import time
import helper
source_path = 'data/letters_source.txt'
target_path = 'data/letters_target.txt'
source_sentences = helper.load_data(source_path)
target_sentences = helper.load_data(target_path)
```
Let's start by examining the current state of the dataset. `source_sentences` contains the entire input sequence file as text delimited by newline symbols.
```
source_sentences[:50].split('\n')
```
`source_sentences` contains the entire output sequence file as text delimited by newline symbols. Each line corresponds to the line from `source_sentences`. `source_sentences` contains a sorted characters of the line.
```
target_sentences[:50].split('\n')
```
## Preprocess
To do anything useful with it, we'll need to turn the each string into a list of characters:
<img src="images/source_and_target_arrays.png"/>
Then convert the characters to their int values as declared in our vocabulary:
```
def extract_character_vocab(data):
special_words = ['<PAD>', '<UNK>', '<GO>', '<EOS>']
set_words = set([character for line in data.split('\n') for character in line])
int_to_vocab = {word_i: word for word_i, word in enumerate(special_words + list(set_words))}
vocab_to_int = {word: word_i for word_i, word in int_to_vocab.items()}
return int_to_vocab, vocab_to_int
# Build int2letter and letter2int dicts
source_int_to_letter, source_letter_to_int = extract_character_vocab(source_sentences)
target_int_to_letter, target_letter_to_int = extract_character_vocab(target_sentences)
# Convert characters to ids
source_letter_ids = [[source_letter_to_int.get(letter, source_letter_to_int['<UNK>']) for letter in line] for line in source_sentences.split('\n')]
target_letter_ids = [[target_letter_to_int.get(letter, target_letter_to_int['<UNK>']) for letter in line] + [target_letter_to_int['<EOS>']] for line in target_sentences.split('\n')]
print("Example source sequence")
print(source_letter_ids[:3])
print("\n")
print("Example target sequence")
print(target_letter_ids[:3])
```
This is the final shape we need them to be in. We can now proceed to building the model.
## Model
#### Check the Version of TensorFlow
This will check to make sure you have the correct version of TensorFlow
```
from distutils.version import LooseVersion
import tensorflow as tf
from tensorflow.python.layers.core import Dense
# Check TensorFlow Version
assert LooseVersion(tf.__version__) >= LooseVersion('1.1'), 'Please use TensorFlow version 1.1 or newer'
print('TensorFlow Version: {}'.format(tf.__version__))
```
### Hyperparameters
```
# Number of Epochs
epochs = 60
# Batch Size
batch_size = 128
# RNN Size
rnn_size = 50
# Number of Layers
num_layers = 2
# Embedding Size
encoding_embedding_size = 15
decoding_embedding_size = 15
# Learning Rate
learning_rate = 0.001
```
### Input
```
def get_model_inputs():
input_data = tf.placeholder(tf.int32, [None, None], name='input')
targets = tf.placeholder(tf.int32, [None, None], name='targets')
lr = tf.placeholder(tf.float32, name='learning_rate')
target_sequence_length = tf.placeholder(tf.int32, (None,), name='target_sequence_length')
max_target_sequence_length = tf.reduce_max(target_sequence_length, name='max_target_len')
source_sequence_length = tf.placeholder(tf.int32, (None,), name='source_sequence_length')
return input_data, targets, lr, target_sequence_length, max_target_sequence_length, source_sequence_length
```
### Sequence to Sequence Model
We can now stat defining the functions that will build the seq2seq model. We are building it from the bottom up with the following components:
2.1 Encoder
- Embedding
- Encoder cell
2.2 Decoder
1- Process decoder inputs
2- Set up the decoder
- Embedding
- Decoder cell
- Dense output layer
- Training decoder
- Inference decoder
2.3 Seq2seq model connecting the encoder and decoder
2.4 Build the training graph hooking up the model with the
optimizer
### 2.1 Encoder
The first bit of the model we'll build is the encoder. Here, we'll embed the input data, construct our encoder, then pass the embedded data to the encoder.
- Embed the input data using [`tf.contrib.layers.embed_sequence`](https://www.tensorflow.org/api_docs/python/tf/contrib/layers/embed_sequence)
<img src="images/embed_sequence.png" />
- Pass the embedded input into a stack of RNNs. Save the RNN state and ignore the output.
<img src="images/encoder.png" />
```
def encoding_layer(input_data, rnn_size, num_layers,
source_sequence_length, source_vocab_size,
encoding_embedding_size):
# Encoder embedding
enc_embed_input = tf.contrib.layers.embed_sequence(input_data, source_vocab_size, encoding_embedding_size)
# RNN cell
def make_cell(rnn_size):
enc_cell = tf.contrib.rnn.LSTMCell(rnn_size,
initializer=tf.random_uniform_initializer(-0.1, 0.1, seed=2))
return enc_cell
enc_cell = tf.contrib.rnn.MultiRNNCell([make_cell(rnn_size) for _ in range(num_layers)])
enc_output, enc_state = tf.nn.dynamic_rnn(enc_cell, enc_embed_input, sequence_length=source_sequence_length, dtype=tf.float32)
return enc_output, enc_state
```
## 2.2 Decoder
The decoder is probably the most involved part of this model. The following steps are needed to create it:
1- Process decoder inputs
2- Set up the decoder components
- Embedding
- Decoder cell
- Dense output layer
- Training decoder
- Inference decoder
### Process Decoder Input
In the training process, the target sequences will be used in two different places:
1. Using them to calculate the loss
2. Feeding them to the decoder during training to make the model more robust.
Now we need to address the second point. Let's assume our targets look like this in their letter/word form (we're doing this for readibility. At this point in the code, these sequences would be in int form):
<img src="images/targets_1.png"/>
We need to do a simple transformation on the tensor before feeding it to the decoder:
1- We will feed an item of the sequence to the decoder at each time step. Think about the last timestep -- where the decoder outputs the final word in its output. The input to that step is the item before last from the target sequence. The decoder has no use for the last item in the target sequence in this scenario. So we'll need to remove the last item.
We do that using tensorflow's tf.strided_slice() method. We hand it the tensor, and the index of where to start and where to end the cutting.
<img src="images/strided_slice_1.png"/>
2- The first item in each sequence we feed to the decoder has to be GO symbol. So We'll add that to the beginning.
<img src="images/targets_add_go.png"/>
Now the tensor is ready to be fed to the decoder. It looks like this (if we convert from ints to letters/symbols):
<img src="images/targets_after_processing_1.png"/>
```
# Process the input we'll feed to the decoder
def process_decoder_input(target_data, vocab_to_int, batch_size):
'''Remove the last word id from each batch and concat the <GO> to the begining of each batch'''
ending = tf.strided_slice(target_data, [0, 0], [batch_size, -1], [1, 1])
dec_input = tf.concat([tf.fill([batch_size, 1], vocab_to_int['<GO>']), ending], 1)
return dec_input
```
### Set up the decoder components
- Embedding
- Decoder cell
- Dense output layer
- Training decoder
- Inference decoder
#### 1- Embedding
Now that we have prepared the inputs to the training decoder, we need to embed them so they can be ready to be passed to the decoder.
We'll create an embedding matrix like the following then have tf.nn.embedding_lookup convert our input to its embedded equivalent:
<img src="images/embeddings.png" />
#### 2- Decoder Cell
Then we declare our decoder cell. Just like the encoder, we'll use an tf.contrib.rnn.LSTMCell here as well.
We need to declare a decoder for the training process, and a decoder for the inference/prediction process. These two decoders will share their parameters (so that all the weights and biases that are set during the training phase can be used when we deploy the model).
First, we'll need to define the type of cell we'll be using for our decoder RNNs. We opted for LSTM.
#### 3- Dense output layer
Before we move to declaring our decoders, we'll need to create the output layer, which will be a tensorflow.python.layers.core.Dense layer that translates the outputs of the decoder to logits that tell us which element of the decoder vocabulary the decoder is choosing to output at each time step.
#### 4- Training decoder
Essentially, we'll be creating two decoders which share their parameters. One for training and one for inference. The two are similar in that both created using tf.contrib.seq2seq.**BasicDecoder** and tf.contrib.seq2seq.**dynamic_decode**. They differ, however, in that we feed the the target sequences as inputs to the training decoder at each time step to make it more robust.
We can think of the training decoder as looking like this (except that it works with sequences in batches):
<img src="images/sequence-to-sequence-training-decoder.png"/>
The training decoder **does not** feed the output of each time step to the next. Rather, the inputs to the decoder time steps are the target sequence from the training dataset (the orange letters).
#### 5- Inference decoder
The inference decoder is the one we'll use when we deploy our model to the wild.
<img src="images/sequence-to-sequence-inference-decoder.png"/>
We'll hand our encoder hidden state to both the training and inference decoders and have it process its output. TensorFlow handles most of the logic for us. We just have to use the appropriate methods from tf.contrib.seq2seq and supply them with the appropriate inputs.
```
def decoding_layer(target_letter_to_int, decoding_embedding_size, num_layers, rnn_size,
target_sequence_length, max_target_sequence_length, enc_state, dec_input):
# 1. Decoder Embedding
target_vocab_size = len(target_letter_to_int)
dec_embeddings = tf.Variable(tf.random_uniform([target_vocab_size, decoding_embedding_size]))
dec_embed_input = tf.nn.embedding_lookup(dec_embeddings, dec_input)
# 2. Construct the decoder cell
def make_cell(rnn_size):
dec_cell = tf.contrib.rnn.LSTMCell(rnn_size,
initializer=tf.random_uniform_initializer(-0.1, 0.1, seed=2))
return dec_cell
dec_cell = tf.contrib.rnn.MultiRNNCell([make_cell(rnn_size) for _ in range(num_layers)])
# 3. Dense layer to translate the decoder's output at each time
# step into a choice from the target vocabulary
output_layer = Dense(target_vocab_size,
kernel_initializer = tf.truncated_normal_initializer(mean = 0.0, stddev=0.1))
# 4. Set up a training decoder and an inference decoder
# Training Decoder
with tf.variable_scope("decode"):
# Helper for the training process. Used by BasicDecoder to read inputs.
training_helper = tf.contrib.seq2seq.TrainingHelper(inputs=dec_embed_input,
sequence_length=target_sequence_length,
time_major=False)
# Basic decoder
training_decoder = tf.contrib.seq2seq.BasicDecoder(dec_cell,
training_helper,
enc_state,
output_layer)
# Perform dynamic decoding using the decoder
training_decoder_output, _ = tf.contrib.seq2seq.dynamic_decode(training_decoder,
impute_finished=True,
maximum_iterations=max_target_sequence_length)
# 5. Inference Decoder
# Reuses the same parameters trained by the training process
with tf.variable_scope("decode", reuse=True):
start_tokens = tf.tile(tf.constant([target_letter_to_int['<GO>']], dtype=tf.int32), [batch_size], name='start_tokens')
# Helper for the inference process.
inference_helper = tf.contrib.seq2seq.GreedyEmbeddingHelper(dec_embeddings,
start_tokens,
target_letter_to_int['<EOS>'])
# Basic decoder
inference_decoder = tf.contrib.seq2seq.BasicDecoder(dec_cell,
inference_helper,
enc_state,
output_layer)
# Perform dynamic decoding using the decoder
inference_decoder_output, _ = tf.contrib.seq2seq.dynamic_decode(inference_decoder,
impute_finished=True,
maximum_iterations=max_target_sequence_length)
return training_decoder_output, inference_decoder_output
```
## 2.3 Seq2seq model
Let's now go a step above, and hook up the encoder and decoder using the methods we just declared
```
def seq2seq_model(input_data, targets, lr, target_sequence_length,
max_target_sequence_length, source_sequence_length,
source_vocab_size, target_vocab_size,
enc_embedding_size, dec_embedding_size,
rnn_size, num_layers):
# Pass the input data through the encoder. We'll ignore the encoder output, but use the state
_, enc_state = encoding_layer(input_data,
rnn_size,
num_layers,
source_sequence_length,
source_vocab_size,
encoding_embedding_size)
# Prepare the target sequences we'll feed to the decoder in training mode
dec_input = process_decoder_input(targets, target_letter_to_int, batch_size)
# Pass encoder state and decoder inputs to the decoders
training_decoder_output, inference_decoder_output = decoding_layer(target_letter_to_int,
decoding_embedding_size,
num_layers,
rnn_size,
target_sequence_length,
max_target_sequence_length,
enc_state,
dec_input)
return training_decoder_output, inference_decoder_output
```
Model outputs *training_decoder_output* and *inference_decoder_output* both contain a 'rnn_output' logits tensor that looks like this:
<img src="images/logits.png"/>
The logits we get from the training tensor we'll pass to tf.contrib.seq2seq.**sequence_loss()** to calculate the loss and ultimately the gradient.
```
# Build the graph
train_graph = tf.Graph()
# Set the graph to default to ensure that it is ready for training
with train_graph.as_default():
# Load the model inputs
input_data, targets, lr, target_sequence_length, max_target_sequence_length, source_sequence_length = get_model_inputs()
# Create the training and inference logits
training_decoder_output, inference_decoder_output = seq2seq_model(input_data,
targets,
lr,
target_sequence_length,
max_target_sequence_length,
source_sequence_length,
len(source_letter_to_int),
len(target_letter_to_int),
encoding_embedding_size,
decoding_embedding_size,
rnn_size,
num_layers)
# Create tensors for the training logits and inference logits
training_logits = tf.identity(training_decoder_output.rnn_output, 'logits')
inference_logits = tf.identity(inference_decoder_output.sample_id, name='predictions')
# Create the weights for sequence_loss
masks = tf.sequence_mask(target_sequence_length, max_target_sequence_length, dtype=tf.float32, name='masks')
with tf.name_scope("optimization"):
# Loss function
cost = tf.contrib.seq2seq.sequence_loss(
training_logits,
targets,
masks)
# Optimizer
optimizer = tf.train.AdamOptimizer(lr)
# Gradient Clipping
gradients = optimizer.compute_gradients(cost)
capped_gradients = [(tf.clip_by_value(grad, -5., 5.), var) for grad, var in gradients if grad is not None]
train_op = optimizer.apply_gradients(capped_gradients)
```
## Get Batches
There's little processing involved when we retreive the batches. This is a simple example assuming batch_size = 2
Source sequences (it's actually in int form, we're showing the characters for clarity):
<img src="images/source_batch.png" />
Target sequences (also in int, but showing letters for clarity):
<img src="images/target_batch.png" />
```
def pad_sentence_batch(sentence_batch, pad_int):
"""Pad sentences with <PAD> so that each sentence of a batch has the same length"""
max_sentence = max([len(sentence) for sentence in sentence_batch])
return [sentence + [pad_int] * (max_sentence - len(sentence)) for sentence in sentence_batch]
def get_batches(targets, sources, batch_size, source_pad_int, target_pad_int):
"""Batch targets, sources, and the lengths of their sentences together"""
for batch_i in range(0, len(sources)//batch_size):
start_i = batch_i * batch_size
sources_batch = sources[start_i:start_i + batch_size]
targets_batch = targets[start_i:start_i + batch_size]
pad_sources_batch = np.array(pad_sentence_batch(sources_batch, source_pad_int))
pad_targets_batch = np.array(pad_sentence_batch(targets_batch, target_pad_int))
# Need the lengths for the _lengths parameters
pad_targets_lengths = []
for target in pad_targets_batch:
pad_targets_lengths.append(len(target))
pad_source_lengths = []
for source in pad_sources_batch:
pad_source_lengths.append(len(source))
yield pad_targets_batch, pad_sources_batch, pad_targets_lengths, pad_source_lengths
```
## Train
We're now ready to train our model. If you run into OOM (out of memory) issues during training, try to decrease the batch_size.
```
# Split data to training and validation sets
train_source = source_letter_ids[batch_size:]
train_target = target_letter_ids[batch_size:]
valid_source = source_letter_ids[:batch_size]
valid_target = target_letter_ids[:batch_size]
(valid_targets_batch, valid_sources_batch, valid_targets_lengths, valid_sources_lengths) = next(get_batches(valid_target, valid_source, batch_size,
source_letter_to_int['<PAD>'],
target_letter_to_int['<PAD>']))
display_step = 20 # Check training loss after every 20 batches
checkpoint = "best_model.ckpt"
with tf.Session(graph=train_graph) as sess:
sess.run(tf.global_variables_initializer())
for epoch_i in range(1, epochs+1):
for batch_i, (targets_batch, sources_batch, targets_lengths, sources_lengths) in enumerate(
get_batches(train_target, train_source, batch_size,
source_letter_to_int['<PAD>'],
target_letter_to_int['<PAD>'])):
# Training step
_, loss = sess.run(
[train_op, cost],
{input_data: sources_batch,
targets: targets_batch,
lr: learning_rate,
target_sequence_length: targets_lengths,
source_sequence_length: sources_lengths})
# Debug message updating us on the status of the training
if batch_i % display_step == 0 and batch_i > 0:
# Calculate validation cost
validation_loss = sess.run(
[cost],
{input_data: valid_sources_batch,
targets: valid_targets_batch,
lr: learning_rate,
target_sequence_length: valid_targets_lengths,
source_sequence_length: valid_sources_lengths})
print('Epoch {:>3}/{} Batch {:>4}/{} - Loss: {:>6.3f} - Validation loss: {:>6.3f}'
.format(epoch_i,
epochs,
batch_i,
len(train_source) // batch_size,
loss,
validation_loss[0]))
# Save Model
saver = tf.train.Saver()
saver.save(sess, checkpoint)
print('Model Trained and Saved')
```
## Prediction
```
def source_to_seq(text):
'''Prepare the text for the model'''
sequence_length = 7
return [source_letter_to_int.get(word, source_letter_to_int['<UNK>']) for word in text]+ [source_letter_to_int['<PAD>']]*(sequence_length-len(text))
input_sentence = 'hello'
text = source_to_seq(input_sentence)
checkpoint = "./best_model.ckpt"
loaded_graph = tf.Graph()
with tf.Session(graph=loaded_graph) as sess:
# Load saved model
loader = tf.train.import_meta_graph(checkpoint + '.meta')
loader.restore(sess, checkpoint)
input_data = loaded_graph.get_tensor_by_name('input:0')
logits = loaded_graph.get_tensor_by_name('predictions:0')
source_sequence_length = loaded_graph.get_tensor_by_name('source_sequence_length:0')
target_sequence_length = loaded_graph.get_tensor_by_name('target_sequence_length:0')
#Multiply by batch_size to match the model's input parameters
answer_logits = sess.run(logits, {input_data: [text]*batch_size,
target_sequence_length: [len(text)]*batch_size,
source_sequence_length: [len(text)]*batch_size})[0]
pad = source_letter_to_int["<PAD>"]
print('Original Text:', input_sentence)
print('\nSource')
print(' Word Ids: {}'.format([i for i in text]))
print(' Input Words: {}'.format(" ".join([source_int_to_letter[i] for i in text])))
print('\nTarget')
print(' Word Ids: {}'.format([i for i in answer_logits if i != pad]))
print(' Response Words: {}'.format(" ".join([target_int_to_letter[i] for i in answer_logits if i != pad])))
```
|
github_jupyter
|
import numpy as np
import time
import helper
source_path = 'data/letters_source.txt'
target_path = 'data/letters_target.txt'
source_sentences = helper.load_data(source_path)
target_sentences = helper.load_data(target_path)
source_sentences[:50].split('\n')
target_sentences[:50].split('\n')
def extract_character_vocab(data):
special_words = ['<PAD>', '<UNK>', '<GO>', '<EOS>']
set_words = set([character for line in data.split('\n') for character in line])
int_to_vocab = {word_i: word for word_i, word in enumerate(special_words + list(set_words))}
vocab_to_int = {word: word_i for word_i, word in int_to_vocab.items()}
return int_to_vocab, vocab_to_int
# Build int2letter and letter2int dicts
source_int_to_letter, source_letter_to_int = extract_character_vocab(source_sentences)
target_int_to_letter, target_letter_to_int = extract_character_vocab(target_sentences)
# Convert characters to ids
source_letter_ids = [[source_letter_to_int.get(letter, source_letter_to_int['<UNK>']) for letter in line] for line in source_sentences.split('\n')]
target_letter_ids = [[target_letter_to_int.get(letter, target_letter_to_int['<UNK>']) for letter in line] + [target_letter_to_int['<EOS>']] for line in target_sentences.split('\n')]
print("Example source sequence")
print(source_letter_ids[:3])
print("\n")
print("Example target sequence")
print(target_letter_ids[:3])
from distutils.version import LooseVersion
import tensorflow as tf
from tensorflow.python.layers.core import Dense
# Check TensorFlow Version
assert LooseVersion(tf.__version__) >= LooseVersion('1.1'), 'Please use TensorFlow version 1.1 or newer'
print('TensorFlow Version: {}'.format(tf.__version__))
# Number of Epochs
epochs = 60
# Batch Size
batch_size = 128
# RNN Size
rnn_size = 50
# Number of Layers
num_layers = 2
# Embedding Size
encoding_embedding_size = 15
decoding_embedding_size = 15
# Learning Rate
learning_rate = 0.001
def get_model_inputs():
input_data = tf.placeholder(tf.int32, [None, None], name='input')
targets = tf.placeholder(tf.int32, [None, None], name='targets')
lr = tf.placeholder(tf.float32, name='learning_rate')
target_sequence_length = tf.placeholder(tf.int32, (None,), name='target_sequence_length')
max_target_sequence_length = tf.reduce_max(target_sequence_length, name='max_target_len')
source_sequence_length = tf.placeholder(tf.int32, (None,), name='source_sequence_length')
return input_data, targets, lr, target_sequence_length, max_target_sequence_length, source_sequence_length
def encoding_layer(input_data, rnn_size, num_layers,
source_sequence_length, source_vocab_size,
encoding_embedding_size):
# Encoder embedding
enc_embed_input = tf.contrib.layers.embed_sequence(input_data, source_vocab_size, encoding_embedding_size)
# RNN cell
def make_cell(rnn_size):
enc_cell = tf.contrib.rnn.LSTMCell(rnn_size,
initializer=tf.random_uniform_initializer(-0.1, 0.1, seed=2))
return enc_cell
enc_cell = tf.contrib.rnn.MultiRNNCell([make_cell(rnn_size) for _ in range(num_layers)])
enc_output, enc_state = tf.nn.dynamic_rnn(enc_cell, enc_embed_input, sequence_length=source_sequence_length, dtype=tf.float32)
return enc_output, enc_state
# Process the input we'll feed to the decoder
def process_decoder_input(target_data, vocab_to_int, batch_size):
'''Remove the last word id from each batch and concat the <GO> to the begining of each batch'''
ending = tf.strided_slice(target_data, [0, 0], [batch_size, -1], [1, 1])
dec_input = tf.concat([tf.fill([batch_size, 1], vocab_to_int['<GO>']), ending], 1)
return dec_input
def decoding_layer(target_letter_to_int, decoding_embedding_size, num_layers, rnn_size,
target_sequence_length, max_target_sequence_length, enc_state, dec_input):
# 1. Decoder Embedding
target_vocab_size = len(target_letter_to_int)
dec_embeddings = tf.Variable(tf.random_uniform([target_vocab_size, decoding_embedding_size]))
dec_embed_input = tf.nn.embedding_lookup(dec_embeddings, dec_input)
# 2. Construct the decoder cell
def make_cell(rnn_size):
dec_cell = tf.contrib.rnn.LSTMCell(rnn_size,
initializer=tf.random_uniform_initializer(-0.1, 0.1, seed=2))
return dec_cell
dec_cell = tf.contrib.rnn.MultiRNNCell([make_cell(rnn_size) for _ in range(num_layers)])
# 3. Dense layer to translate the decoder's output at each time
# step into a choice from the target vocabulary
output_layer = Dense(target_vocab_size,
kernel_initializer = tf.truncated_normal_initializer(mean = 0.0, stddev=0.1))
# 4. Set up a training decoder and an inference decoder
# Training Decoder
with tf.variable_scope("decode"):
# Helper for the training process. Used by BasicDecoder to read inputs.
training_helper = tf.contrib.seq2seq.TrainingHelper(inputs=dec_embed_input,
sequence_length=target_sequence_length,
time_major=False)
# Basic decoder
training_decoder = tf.contrib.seq2seq.BasicDecoder(dec_cell,
training_helper,
enc_state,
output_layer)
# Perform dynamic decoding using the decoder
training_decoder_output, _ = tf.contrib.seq2seq.dynamic_decode(training_decoder,
impute_finished=True,
maximum_iterations=max_target_sequence_length)
# 5. Inference Decoder
# Reuses the same parameters trained by the training process
with tf.variable_scope("decode", reuse=True):
start_tokens = tf.tile(tf.constant([target_letter_to_int['<GO>']], dtype=tf.int32), [batch_size], name='start_tokens')
# Helper for the inference process.
inference_helper = tf.contrib.seq2seq.GreedyEmbeddingHelper(dec_embeddings,
start_tokens,
target_letter_to_int['<EOS>'])
# Basic decoder
inference_decoder = tf.contrib.seq2seq.BasicDecoder(dec_cell,
inference_helper,
enc_state,
output_layer)
# Perform dynamic decoding using the decoder
inference_decoder_output, _ = tf.contrib.seq2seq.dynamic_decode(inference_decoder,
impute_finished=True,
maximum_iterations=max_target_sequence_length)
return training_decoder_output, inference_decoder_output
def seq2seq_model(input_data, targets, lr, target_sequence_length,
max_target_sequence_length, source_sequence_length,
source_vocab_size, target_vocab_size,
enc_embedding_size, dec_embedding_size,
rnn_size, num_layers):
# Pass the input data through the encoder. We'll ignore the encoder output, but use the state
_, enc_state = encoding_layer(input_data,
rnn_size,
num_layers,
source_sequence_length,
source_vocab_size,
encoding_embedding_size)
# Prepare the target sequences we'll feed to the decoder in training mode
dec_input = process_decoder_input(targets, target_letter_to_int, batch_size)
# Pass encoder state and decoder inputs to the decoders
training_decoder_output, inference_decoder_output = decoding_layer(target_letter_to_int,
decoding_embedding_size,
num_layers,
rnn_size,
target_sequence_length,
max_target_sequence_length,
enc_state,
dec_input)
return training_decoder_output, inference_decoder_output
# Build the graph
train_graph = tf.Graph()
# Set the graph to default to ensure that it is ready for training
with train_graph.as_default():
# Load the model inputs
input_data, targets, lr, target_sequence_length, max_target_sequence_length, source_sequence_length = get_model_inputs()
# Create the training and inference logits
training_decoder_output, inference_decoder_output = seq2seq_model(input_data,
targets,
lr,
target_sequence_length,
max_target_sequence_length,
source_sequence_length,
len(source_letter_to_int),
len(target_letter_to_int),
encoding_embedding_size,
decoding_embedding_size,
rnn_size,
num_layers)
# Create tensors for the training logits and inference logits
training_logits = tf.identity(training_decoder_output.rnn_output, 'logits')
inference_logits = tf.identity(inference_decoder_output.sample_id, name='predictions')
# Create the weights for sequence_loss
masks = tf.sequence_mask(target_sequence_length, max_target_sequence_length, dtype=tf.float32, name='masks')
with tf.name_scope("optimization"):
# Loss function
cost = tf.contrib.seq2seq.sequence_loss(
training_logits,
targets,
masks)
# Optimizer
optimizer = tf.train.AdamOptimizer(lr)
# Gradient Clipping
gradients = optimizer.compute_gradients(cost)
capped_gradients = [(tf.clip_by_value(grad, -5., 5.), var) for grad, var in gradients if grad is not None]
train_op = optimizer.apply_gradients(capped_gradients)
def pad_sentence_batch(sentence_batch, pad_int):
"""Pad sentences with <PAD> so that each sentence of a batch has the same length"""
max_sentence = max([len(sentence) for sentence in sentence_batch])
return [sentence + [pad_int] * (max_sentence - len(sentence)) for sentence in sentence_batch]
def get_batches(targets, sources, batch_size, source_pad_int, target_pad_int):
"""Batch targets, sources, and the lengths of their sentences together"""
for batch_i in range(0, len(sources)//batch_size):
start_i = batch_i * batch_size
sources_batch = sources[start_i:start_i + batch_size]
targets_batch = targets[start_i:start_i + batch_size]
pad_sources_batch = np.array(pad_sentence_batch(sources_batch, source_pad_int))
pad_targets_batch = np.array(pad_sentence_batch(targets_batch, target_pad_int))
# Need the lengths for the _lengths parameters
pad_targets_lengths = []
for target in pad_targets_batch:
pad_targets_lengths.append(len(target))
pad_source_lengths = []
for source in pad_sources_batch:
pad_source_lengths.append(len(source))
yield pad_targets_batch, pad_sources_batch, pad_targets_lengths, pad_source_lengths
# Split data to training and validation sets
train_source = source_letter_ids[batch_size:]
train_target = target_letter_ids[batch_size:]
valid_source = source_letter_ids[:batch_size]
valid_target = target_letter_ids[:batch_size]
(valid_targets_batch, valid_sources_batch, valid_targets_lengths, valid_sources_lengths) = next(get_batches(valid_target, valid_source, batch_size,
source_letter_to_int['<PAD>'],
target_letter_to_int['<PAD>']))
display_step = 20 # Check training loss after every 20 batches
checkpoint = "best_model.ckpt"
with tf.Session(graph=train_graph) as sess:
sess.run(tf.global_variables_initializer())
for epoch_i in range(1, epochs+1):
for batch_i, (targets_batch, sources_batch, targets_lengths, sources_lengths) in enumerate(
get_batches(train_target, train_source, batch_size,
source_letter_to_int['<PAD>'],
target_letter_to_int['<PAD>'])):
# Training step
_, loss = sess.run(
[train_op, cost],
{input_data: sources_batch,
targets: targets_batch,
lr: learning_rate,
target_sequence_length: targets_lengths,
source_sequence_length: sources_lengths})
# Debug message updating us on the status of the training
if batch_i % display_step == 0 and batch_i > 0:
# Calculate validation cost
validation_loss = sess.run(
[cost],
{input_data: valid_sources_batch,
targets: valid_targets_batch,
lr: learning_rate,
target_sequence_length: valid_targets_lengths,
source_sequence_length: valid_sources_lengths})
print('Epoch {:>3}/{} Batch {:>4}/{} - Loss: {:>6.3f} - Validation loss: {:>6.3f}'
.format(epoch_i,
epochs,
batch_i,
len(train_source) // batch_size,
loss,
validation_loss[0]))
# Save Model
saver = tf.train.Saver()
saver.save(sess, checkpoint)
print('Model Trained and Saved')
def source_to_seq(text):
'''Prepare the text for the model'''
sequence_length = 7
return [source_letter_to_int.get(word, source_letter_to_int['<UNK>']) for word in text]+ [source_letter_to_int['<PAD>']]*(sequence_length-len(text))
input_sentence = 'hello'
text = source_to_seq(input_sentence)
checkpoint = "./best_model.ckpt"
loaded_graph = tf.Graph()
with tf.Session(graph=loaded_graph) as sess:
# Load saved model
loader = tf.train.import_meta_graph(checkpoint + '.meta')
loader.restore(sess, checkpoint)
input_data = loaded_graph.get_tensor_by_name('input:0')
logits = loaded_graph.get_tensor_by_name('predictions:0')
source_sequence_length = loaded_graph.get_tensor_by_name('source_sequence_length:0')
target_sequence_length = loaded_graph.get_tensor_by_name('target_sequence_length:0')
#Multiply by batch_size to match the model's input parameters
answer_logits = sess.run(logits, {input_data: [text]*batch_size,
target_sequence_length: [len(text)]*batch_size,
source_sequence_length: [len(text)]*batch_size})[0]
pad = source_letter_to_int["<PAD>"]
print('Original Text:', input_sentence)
print('\nSource')
print(' Word Ids: {}'.format([i for i in text]))
print(' Input Words: {}'.format(" ".join([source_int_to_letter[i] for i in text])))
print('\nTarget')
print(' Word Ids: {}'.format([i for i in answer_logits if i != pad]))
print(' Response Words: {}'.format(" ".join([target_int_to_letter[i] for i in answer_logits if i != pad])))
| 0.583441 | 0.98163 |
# Runtime Metrics / Tags Example
## Prerequisites
* Kind cluster with Seldon Installed
* curl
* s2i
* seldon-core-analytics
## Setup Seldon Core
Use the setup notebook to [Setup Cluster](seldon_core_setup.ipynb) to setup Seldon Core with an ingress.
```
!kubectl create namespace seldon
!kubectl config set-context $(kubectl config current-context) --namespace=seldon
```
## Install Seldon Core Analytics
```
!helm install seldon-core-analytics ../../../helm-charts/seldon-core-analytics -n seldon-system --wait
```
## Define Model
```
%%writefile Model.py
import logging
from seldon_core.user_model import SeldonResponse
def reshape(x):
if len(x.shape) < 2:
return x.reshape(1, -1)
else:
return x
class Model:
def predict(self, features, names=[], meta={}):
X = reshape(features)
logging.info(f"model features: {features}")
logging.info(f"model names: {names}")
logging.info(f"model meta: {meta}")
logging.info(f"model X: {X}")
runtime_metrics = [{"type": "COUNTER", "key": "instance_counter", "value": len(X)}]
runtime_tags = {"runtime": "tag", "shared": "right one"}
return SeldonResponse(data=X, metrics=runtime_metrics, tags=runtime_tags)
def metrics(self):
return [{"type": "COUNTER", "key": "requests_counter", "value": 1}]
def tags(self):
return {"static": "tag", "shared": "not right one"}
```
## Build Image and load into kind cluster
```
%%bash
s2i build -E ENVIRONMENT_REST . seldonio/seldon-core-s2i-python37:1.3.0-dev runtime-metrics-tags:0.1
kind load docker-image runtime-metrics-tags:0.1
```
## Deploy Model
```
%%writefile deployment.yaml
apiVersion: machinelearning.seldon.io/v1
kind: SeldonDeployment
metadata:
name: seldon-model-runtime-data
spec:
name: test-deployment
predictors:
- componentSpecs:
- spec:
containers:
- image: runtime-metrics-tags:0.1
name: my-model
graph:
name: my-model
type: MODEL
name: example
replicas: 1
!kubectl apply -f deployment.yaml
!kubectl rollout status deploy/$(kubectl get deploy -l seldon-deployment-id=seldon-model-runtime-data -o jsonpath='{.items[0].metadata.name}')
```
## Send few inference requests
```
%%bash
curl -s -H 'Content-Type: application/json' -d '{"data": {"ndarray": [[1, 2, 3]]}}' \
http://localhost:8003/seldon/seldon/seldon-model-runtime-data/api/v1.0/predictions
%%bash
curl -s -H 'Content-Type: application/json' -d '{"data": {"ndarray": [[1, 2, 3], [4, 5, 6]]}}' \
http://localhost:8003/seldon/seldon/seldon-model-runtime-data/api/v1.0/predictions
```
## Check metrics
```
import json
metrics =! kubectl run --quiet=true -it --rm curlmetrics --image=tutum/curl --restart=Never -- \
curl -s seldon-core-analytics-prometheus-seldon.seldon-system/api/v1/query?query=instance_counter_total
json.loads(metrics[0])["data"]["result"][0]["value"][1]
metrics =! kubectl run --quiet=true -it --rm curlmetrics --image=tutum/curl --restart=Never -- \
curl -s seldon-core-analytics-prometheus-seldon.seldon-system/api/v1/query?query=requests_counter_total
json.loads(metrics[0])["data"]["result"][0]["value"][1]
```
## Cleanup
```
!kubectl delete -f deployment.yaml
!helm delete seldon-core-analytics --namespace seldon-system
```
|
github_jupyter
|
!kubectl create namespace seldon
!kubectl config set-context $(kubectl config current-context) --namespace=seldon
!helm install seldon-core-analytics ../../../helm-charts/seldon-core-analytics -n seldon-system --wait
%%writefile Model.py
import logging
from seldon_core.user_model import SeldonResponse
def reshape(x):
if len(x.shape) < 2:
return x.reshape(1, -1)
else:
return x
class Model:
def predict(self, features, names=[], meta={}):
X = reshape(features)
logging.info(f"model features: {features}")
logging.info(f"model names: {names}")
logging.info(f"model meta: {meta}")
logging.info(f"model X: {X}")
runtime_metrics = [{"type": "COUNTER", "key": "instance_counter", "value": len(X)}]
runtime_tags = {"runtime": "tag", "shared": "right one"}
return SeldonResponse(data=X, metrics=runtime_metrics, tags=runtime_tags)
def metrics(self):
return [{"type": "COUNTER", "key": "requests_counter", "value": 1}]
def tags(self):
return {"static": "tag", "shared": "not right one"}
%%bash
s2i build -E ENVIRONMENT_REST . seldonio/seldon-core-s2i-python37:1.3.0-dev runtime-metrics-tags:0.1
kind load docker-image runtime-metrics-tags:0.1
%%writefile deployment.yaml
apiVersion: machinelearning.seldon.io/v1
kind: SeldonDeployment
metadata:
name: seldon-model-runtime-data
spec:
name: test-deployment
predictors:
- componentSpecs:
- spec:
containers:
- image: runtime-metrics-tags:0.1
name: my-model
graph:
name: my-model
type: MODEL
name: example
replicas: 1
!kubectl apply -f deployment.yaml
!kubectl rollout status deploy/$(kubectl get deploy -l seldon-deployment-id=seldon-model-runtime-data -o jsonpath='{.items[0].metadata.name}')
%%bash
curl -s -H 'Content-Type: application/json' -d '{"data": {"ndarray": [[1, 2, 3]]}}' \
http://localhost:8003/seldon/seldon/seldon-model-runtime-data/api/v1.0/predictions
%%bash
curl -s -H 'Content-Type: application/json' -d '{"data": {"ndarray": [[1, 2, 3], [4, 5, 6]]}}' \
http://localhost:8003/seldon/seldon/seldon-model-runtime-data/api/v1.0/predictions
import json
metrics =! kubectl run --quiet=true -it --rm curlmetrics --image=tutum/curl --restart=Never -- \
curl -s seldon-core-analytics-prometheus-seldon.seldon-system/api/v1/query?query=instance_counter_total
json.loads(metrics[0])["data"]["result"][0]["value"][1]
metrics =! kubectl run --quiet=true -it --rm curlmetrics --image=tutum/curl --restart=Never -- \
curl -s seldon-core-analytics-prometheus-seldon.seldon-system/api/v1/query?query=requests_counter_total
json.loads(metrics[0])["data"]["result"][0]["value"][1]
!kubectl delete -f deployment.yaml
!helm delete seldon-core-analytics --namespace seldon-system
| 0.323915 | 0.870597 |
<a href="https://colab.research.google.com/github/CreatorClement/b575f20/blob/master/project_group_7.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
Group Homework - Project2 - Drug annotation of 23andme report
Group 7 - Tasmine Clement, Charles Zhang, Shengchao Zhao, Julia Rosander
Due Date: Dec 1, 2020
<b>Tasmine Clement Answers</b>
*What was your biggest challenge in this project? (regarding writing code and not only)*
In the beginning it took time to navigate the websites to make sure I had the correct files and all of the docs for the project. The biggest challenge was realizing that even if my code 'works' there may be unintended consequences of trying to fix an error with my corrections (like deleting more lines than necessary in this case).
*What did you learn while working on this project? (regarding writing code and not only)*
I learned that when working with unknown data, it is very valuable to take some time to explore the data to become familiar with how things are organized because small details can cause errors in code.
*If you had more time on the project what other question(s) would you like to answer? (at least one question is required)*
What disorders/diseases are most common for genetic variants with drug effects? I also wonder at what levels the author determined associations to be significant.
<b>Julia Rosander Answers </b>
*What was your biggest challenge in this project? (regarding writing code and not only)*
The biggest challenge was ensuring that the code worked correctly and was also the most efficient way to accomplish the analysis of drug associations.
*What did you learn while working on this project? (regarding writing code and not only)*
Utilizing dataframes correctly can be tricky prior to doing an exploratory analysis of the data. It is important to ensure that you understand your methods before applying them to new datasets.
*If you had more time on the project what other question(s) would you like to answer? (at least one question is required)*
Are there other confounding variables that could affect the interpretation of drug associations that is not inlcuded within this dataset? This could explored through researching drug associations and running more analysis.
<b>Charles Zhang Answers</b>
*What was your biggest challenge in this project? (regarding writing code and not only)*
Trying to get all the data accurately incorporated into a pandas dataframe when one line contains an error. It was already challenging when I knew that there was an error and had an idea of where it was, but I can see it being very difficult if it's an unknown dataset.
*What did you learn while working on this project? (regarding writing code and not only)*
Pandas dataframes are super useful and I would use it over other forms of data management like Excel or JMP when proficient.
*If you had more time on the project what other question(s) would you like to answer? (at least one question is required)*
In what ethnicities are the variants most commonly found in? How closely are the significant drug associations tied to ethnicity?
<b>Shengchao Zhao Answers</b>
*What was your biggest challenge in this project? (regarding writing code and not only)*
The biggest challenge was to catch up Zhang's progress as he was so fast in this assignment. The code part isn't hard at all for it's somehow done once in the homework.
*What did you learn while working on this project? (regarding writing code and not only)*
The errors that are detected by the editor, no matter how much there is, are the best errors; the errors that aren't detected by the editor, and the code doesn't work, are the fine errors; the errors that aren't detected by the editor, and the code works, are the worst errors.
*If you had more time on the project what other question(s) would you like to answer? (at least one question is required)*
If we can use deep learning to look for paterns.
```
import pandas as pd
```
First, to connect the variants to the information available in the PharmGKB data we needed the var_drug_ann.tsv available in the annotations.zip archive under 'Variant and Clinical Annotations Data':
https://www.pharmgkb.org/downloads
```
var_drug_ann = pd.read_csv("var_drug_ann.tsv", sep='\t' )
var_drug_ann.columns = ['Annotation ID','Variant','GENE_SYMBOL','DRUG_NAME','PMID','PHENOTYPE_CATEGORY','SIGNIFICANCE','NOTES','SENTENCE','StudyParameters','ALLELE_PharmGKB','Chromosome']
#loads pharmgkb data into a pandas dataframe and renames the columns to that of the instructions
#The error on line 8156 was corrected by balancing the quotation marks beginning and ending at "The 15-year cumulative....allele (39% vs 19%)" for the Notes.
```
Retreive the 23andme 23andme_v5_hg19_ref.txt.gz raw data file regarding variants from the following github account: https://github.com/arrogantrobot/23andme2vcf
```
database = pd.read_csv("23andme_v5_hg19_ref.txt", sep='\t', names=["CHR", "POS", "dbSNP_ID", "ALLELE_23andme"])
#loads 23andme data into a pandas dataframe from the tab separated file and gives them column headers (not found in file)
joined = database.merge(var_drug_ann, left_on='dbSNP_ID', right_on='Variant')
#merges the 23andme and pharmgkb data using the column dbSNP_ID in the former and Variant in the latter
joined_filtered = joined.loc[(joined['SIGNIFICANCE'].str.lower() == 'yes') & (joined['PHENOTYPE_CATEGORY'].str.lower() == 'efficacy')].filter(items=['dbSNP_ID', 'GENE_SYMBOL', 'DRUG_NAME', 'NOTES', 'SENTENCE', 'ALLELE_PharmGKB', 'ALLELE_23andme'])
#simultaneously filters rows based on if significance is yes (lowered, in case there is yes, Yes, YES, etc) and if phenotype_category is 'efficacy'. Also only includes the columns we were asked to keep.
joined_filtered.to_csv("23andme_PharmGKB_map.tsv", sep="\t",index=False)
#writes to file
drug_data = pd.DataFrame()
for index,row in joined_filtered.itterows():
drug_data.append(row['GENE_SYMBOL'], row['DRUG_NAME']]])
#parses two columns from previously filtered dataset and appends to an object
list_dbSNPs = []
for index,row in joined_filtered.itterows():
list_dbSNPs.append(row['dbSNP_ID'])
drug_data = drug_data.append(list_dbSNPs)
#Creates a list of dbSNP IDs and appends to the previously created dataset
drug_data.to_csv("drug_data_23andme_PharmGKB_map.tsv", sep=";",index=False)
#creates tab separated file of the previous three columns in the dataset
```
|
github_jupyter
|
import pandas as pd
var_drug_ann = pd.read_csv("var_drug_ann.tsv", sep='\t' )
var_drug_ann.columns = ['Annotation ID','Variant','GENE_SYMBOL','DRUG_NAME','PMID','PHENOTYPE_CATEGORY','SIGNIFICANCE','NOTES','SENTENCE','StudyParameters','ALLELE_PharmGKB','Chromosome']
#loads pharmgkb data into a pandas dataframe and renames the columns to that of the instructions
#The error on line 8156 was corrected by balancing the quotation marks beginning and ending at "The 15-year cumulative....allele (39% vs 19%)" for the Notes.
database = pd.read_csv("23andme_v5_hg19_ref.txt", sep='\t', names=["CHR", "POS", "dbSNP_ID", "ALLELE_23andme"])
#loads 23andme data into a pandas dataframe from the tab separated file and gives them column headers (not found in file)
joined = database.merge(var_drug_ann, left_on='dbSNP_ID', right_on='Variant')
#merges the 23andme and pharmgkb data using the column dbSNP_ID in the former and Variant in the latter
joined_filtered = joined.loc[(joined['SIGNIFICANCE'].str.lower() == 'yes') & (joined['PHENOTYPE_CATEGORY'].str.lower() == 'efficacy')].filter(items=['dbSNP_ID', 'GENE_SYMBOL', 'DRUG_NAME', 'NOTES', 'SENTENCE', 'ALLELE_PharmGKB', 'ALLELE_23andme'])
#simultaneously filters rows based on if significance is yes (lowered, in case there is yes, Yes, YES, etc) and if phenotype_category is 'efficacy'. Also only includes the columns we were asked to keep.
joined_filtered.to_csv("23andme_PharmGKB_map.tsv", sep="\t",index=False)
#writes to file
drug_data = pd.DataFrame()
for index,row in joined_filtered.itterows():
drug_data.append(row['GENE_SYMBOL'], row['DRUG_NAME']]])
#parses two columns from previously filtered dataset and appends to an object
list_dbSNPs = []
for index,row in joined_filtered.itterows():
list_dbSNPs.append(row['dbSNP_ID'])
drug_data = drug_data.append(list_dbSNPs)
#Creates a list of dbSNP IDs and appends to the previously created dataset
drug_data.to_csv("drug_data_23andme_PharmGKB_map.tsv", sep=";",index=False)
#creates tab separated file of the previous three columns in the dataset
| 0.243822 | 0.938576 |
# Environmental Sound Classification using Deep Learning - <ins>Keras</ins>
## >> Urban sound classification with CNNs
* [0. Load the preprocessed data](#zero-bullet)
* [1. Data augmentation](#first-bullet)
* [2. CNN model](#second-bullet)
* [3. Helper functions](#third-bullet)
* [4. 10-Fold Cross Validation](#fourth-bullet)
* [5. Results](#fifth-bullet)
---
```
import numpy as np
import pandas as pd
from keras import regularizers, activations
from keras.models import Sequential
from keras.layers import Dense, Dropout, Flatten, Activation, Conv2D, MaxPooling2D, GlobalAveragePooling2D
from keras.utils import np_utils, to_categorical
from tensorflow.keras.preprocessing.image import ImageDataGenerator
from datetime import datetime
from matplotlib import pyplot as plt
%matplotlib inline
USE_GOOGLE_COLAB = True
if USE_GOOGLE_COLAB:
from google.colab import drive
drive.mount('/content/gdrive')
# change the current working directory
%cd gdrive/'My Drive'/US8K
else:
%cd US8K
```
---
## 0. Load the preprocessed data <a name="zero-bullet"></a>
```
us8k_df = pd.read_pickle("us8k_df.pkl")
us8k_df.head()
```
---
## 1. Data augmentation <a name="first-bullet"></a>
```
def init_data_aug():
train_datagen = ImageDataGenerator(
featurewise_center=True,
featurewise_std_normalization=True,
fill_mode = 'constant',
cval=-80.0,
width_shift_range=0.1,
height_shift_range=0.0)
val_datagen = ImageDataGenerator(
featurewise_center=True,
featurewise_std_normalization=True,
fill_mode = 'constant',
cval=-80.0)
return train_datagen, val_datagen
```
---
## 2. CNN model <a name="second-bullet"></a>
```
def init_model():
model1 = Sequential()
#layer-1
model1.add(Conv2D(filters=24, kernel_size=5, input_shape=(128, 128, 1),
kernel_regularizer=regularizers.l2(1e-3)))
model1.add(MaxPooling2D(pool_size=(3,3), strides=3))
model1.add(Activation(activations.relu))
#layer-2
model1.add(Conv2D(filters=36, kernel_size=4, padding='valid', kernel_regularizer=regularizers.l2(1e-3)))
model1.add(MaxPooling2D(pool_size=(2,2), strides=2))
model1.add(Activation(activations.relu))
#layer-3
model1.add(Conv2D(filters=48, kernel_size=3, padding='valid'))
model1.add(Activation(activations.relu))
model1.add(GlobalAveragePooling2D())
#layer-4 (1st dense layer)
model1.add(Dense(60, activation='relu'))
model1.add(Dropout(0.5))
#layer-5 (2nd dense layer)
model1.add(Dense(10, activation='softmax'))
# compile
model1.compile(loss='categorical_crossentropy', metrics=['accuracy'], optimizer='adam')
return model1
model = init_model()
model.summary()
```
---
## 3. Helper functions <a name="third-bullet"></a>
```
def train_test_split(fold_k, data, X_dim=(128, 128, 1)):
X_train = np.stack(data[data.fold != fold_k].melspectrogram.to_numpy())
X_test = np.stack(data[data.fold == fold_k].melspectrogram.to_numpy())
y_train = data[data.fold != fold_k].label.to_numpy()
y_test = data[data.fold == fold_k].label.to_numpy()
XX_train = X_train.reshape(X_train.shape[0], *X_dim)
XX_test = X_test.reshape(X_test.shape[0], *X_dim)
yy_train = to_categorical(y_train)
yy_test = to_categorical(y_test)
return XX_train, XX_test, yy_train, yy_test
def process_fold(fold_k, data, epochs=100, num_batch_size=32):
# split the data
X_train, X_test, y_train, y_test = train_test_split(fold_k, data)
# init data augmention
train_datagen, val_datagen = init_data_aug()
# fit augmentation
train_datagen.fit(X_train)
val_datagen.fit(X_train)
# init model
model = init_model()
# pre-training accuracy
score = model.evaluate(val_datagen.flow(X_test, y_test, batch_size=num_batch_size), verbose=0)
print("Pre-training accuracy: %.4f%%\n" % (100 * score[1]))
# train the model
start = datetime.now()
history = model.fit(train_datagen.flow(X_train, y_train, batch_size=num_batch_size),
steps_per_epoch=len(X_train) / num_batch_size,
epochs=epochs,
validation_data=val_datagen.flow(X_test, y_test, batch_size=num_batch_size))
end = datetime.now()
print("Training completed in time: ", end - start, '\n')
return history
def show_results(tot_history):
"""Show accuracy and loss graphs for train and test sets."""
for i, history in enumerate(tot_history):
print('\n({})'.format(i+1))
plt.figure(figsize=(15,5))
plt.subplot(121)
plt.plot(history.history['accuracy'])
plt.plot(history.history['val_accuracy'])
plt.grid(linestyle='--')
plt.ylabel('Accuracy')
plt.xlabel('Epoch')
plt.legend(['train', 'validation'], loc='upper left')
plt.subplot(122)
plt.plot(history.history['loss'])
plt.plot(history.history['val_loss'])
plt.grid(linestyle='--')
plt.ylabel('Loss')
plt.xlabel('Epoch')
plt.legend(['train', 'validation'], loc='upper left')
plt.show()
print('\tMax validation accuracy: %.4f %%' % (np.max(history.history['val_accuracy']) * 100))
print('\tMin validation loss: %.5f' % np.min(history.history['val_loss']))
```
---
## 4. 10-Fold Cross Validation <a name="fourth-bullet"></a>
* [fold-1](#fold-1)
* [fold-2](#fold-2)
* [fold-3](#fold-3)
* [fold-4](#fold-4)
* [fold-5](#fold-5)
* [fold-6](#fold-6)
* [fold-7](#fold-7)
* [fold-8](#fold-8)
* [fold-9](#fold-9)
* [fold-10](#fold-10)
### fold-1 <a name="fold-1"></a>
```
FOLD_K = 1
REPEAT = 1
history1 = []
for i in range(REPEAT):
print('-'*80)
print("\n({})\n".format(i+1))
history = process_fold(FOLD_K, us8k_df, epochs=100)
history1.append(history)
show_results(history1)
```
### fold-2 <a name="fold-2"></a>
```
FOLD_K = 2
REPEAT = 1
history2 = []
for i in range(REPEAT):
print('-'*80)
print("\n({})\n".format(i+1))
history = process_fold(FOLD_K, us8k_df, epochs=100)
history2.append(history)
show_results(history2)
```
### fold-3 <a name="fold-3"></a>
```
FOLD_K = 3
REPEAT = 1
history3 = []
for i in range(REPEAT):
print('-'*80)
print("\n({})\n".format(i+1))
history = process_fold(FOLD_K, us8k_df, epochs=100)
history3.append(history)
show_results(history3)
```
### fold-4 <a name="fold-4"></a>
```
FOLD_K = 4
REPEAT = 1
history4 = []
for i in range(REPEAT):
print('-'*80)
print("\n({})\n".format(i+1))
history = process_fold(FOLD_K, us8k_df, epochs=100)
history4.append(history)
show_results(history4)
```
### fold-5 <a name="fold-5"></a>
```
FOLD_K = 5
REPEAT = 1
history5 = []
for i in range(REPEAT):
print('-'*80)
print("\n({})\n".format(i+1))
history = process_fold(FOLD_K, us8k_df, epochs=100)
history5.append(history)
show_results(history5)
```
### fold-6 <a name="fold-6"></a>
```
FOLD_K = 6
REPEAT = 1
history6 = []
for i in range(REPEAT):
print('-'*80)
print("\n({})\n".format(i+1))
history = process_fold(FOLD_K, us8k_df, epochs=100)
history6.append(history)
show_results(history6)
```
### fold-7 <a name="fold-7"></a>
```
FOLD_K = 7
REPEAT = 1
history7 = []
for i in range(REPEAT):
print('-'*80)
print("\n({})\n".format(i+1))
history = process_fold(FOLD_K, us8k_df, epochs=100)
history7.append(history)
show_results(history7)
```
### fold-8 <a name="fold-8"></a>
```
FOLD_K = 8
REPEAT = 1
history8 = []
for i in range(REPEAT):
print('-'*80)
print("\n({})\n".format(i+1))
history = process_fold(FOLD_K, us8k_df, epochs=100)
history8.append(history)
show_results(history8)
```
### fold-9 <a name="fold-9"></a>
```
FOLD_K = 9
REPEAT = 1
history9 = []
for i in range(REPEAT):
print('-'*80)
print("\n({})\n".format(i+1))
history = process_fold(FOLD_K, us8k_df, epochs=100)
history9.append(history)
show_results(history9)
```
### fold-10 <a name="fold-10"></a>
```
FOLD_K = 10
REPEAT = 1
history10 = []
for i in range(REPEAT):
print('-'*80)
print("\n({})\n".format(i+1))
history = process_fold(FOLD_K, us8k_df, epochs=100)
history10.append(history)
show_results(history10)
```
---
## 5. Results <a name="fifth-bullet"></a>
### 10-fold cross validation:
|fold|accuracy| loss
|---|:-:|:-:|
|fold-1| 0.82|0.739|
|fold-2|0.72|0.998|
|fold-3|0.70|1.055|
|fold-4|0.70|1.134|
|fold-5|0.80|0.741|
|fold-6|0.72|1.027|
|fold-7|0.70|0.962|
|fold-8|0.74|1.040|
|fold-9|0.77|0.833|
|fold-10|0.80|0.713|
|**Total**|**0.75**|**0.924**|
|
github_jupyter
|
import numpy as np
import pandas as pd
from keras import regularizers, activations
from keras.models import Sequential
from keras.layers import Dense, Dropout, Flatten, Activation, Conv2D, MaxPooling2D, GlobalAveragePooling2D
from keras.utils import np_utils, to_categorical
from tensorflow.keras.preprocessing.image import ImageDataGenerator
from datetime import datetime
from matplotlib import pyplot as plt
%matplotlib inline
USE_GOOGLE_COLAB = True
if USE_GOOGLE_COLAB:
from google.colab import drive
drive.mount('/content/gdrive')
# change the current working directory
%cd gdrive/'My Drive'/US8K
else:
%cd US8K
us8k_df = pd.read_pickle("us8k_df.pkl")
us8k_df.head()
def init_data_aug():
train_datagen = ImageDataGenerator(
featurewise_center=True,
featurewise_std_normalization=True,
fill_mode = 'constant',
cval=-80.0,
width_shift_range=0.1,
height_shift_range=0.0)
val_datagen = ImageDataGenerator(
featurewise_center=True,
featurewise_std_normalization=True,
fill_mode = 'constant',
cval=-80.0)
return train_datagen, val_datagen
def init_model():
model1 = Sequential()
#layer-1
model1.add(Conv2D(filters=24, kernel_size=5, input_shape=(128, 128, 1),
kernel_regularizer=regularizers.l2(1e-3)))
model1.add(MaxPooling2D(pool_size=(3,3), strides=3))
model1.add(Activation(activations.relu))
#layer-2
model1.add(Conv2D(filters=36, kernel_size=4, padding='valid', kernel_regularizer=regularizers.l2(1e-3)))
model1.add(MaxPooling2D(pool_size=(2,2), strides=2))
model1.add(Activation(activations.relu))
#layer-3
model1.add(Conv2D(filters=48, kernel_size=3, padding='valid'))
model1.add(Activation(activations.relu))
model1.add(GlobalAveragePooling2D())
#layer-4 (1st dense layer)
model1.add(Dense(60, activation='relu'))
model1.add(Dropout(0.5))
#layer-5 (2nd dense layer)
model1.add(Dense(10, activation='softmax'))
# compile
model1.compile(loss='categorical_crossentropy', metrics=['accuracy'], optimizer='adam')
return model1
model = init_model()
model.summary()
def train_test_split(fold_k, data, X_dim=(128, 128, 1)):
X_train = np.stack(data[data.fold != fold_k].melspectrogram.to_numpy())
X_test = np.stack(data[data.fold == fold_k].melspectrogram.to_numpy())
y_train = data[data.fold != fold_k].label.to_numpy()
y_test = data[data.fold == fold_k].label.to_numpy()
XX_train = X_train.reshape(X_train.shape[0], *X_dim)
XX_test = X_test.reshape(X_test.shape[0], *X_dim)
yy_train = to_categorical(y_train)
yy_test = to_categorical(y_test)
return XX_train, XX_test, yy_train, yy_test
def process_fold(fold_k, data, epochs=100, num_batch_size=32):
# split the data
X_train, X_test, y_train, y_test = train_test_split(fold_k, data)
# init data augmention
train_datagen, val_datagen = init_data_aug()
# fit augmentation
train_datagen.fit(X_train)
val_datagen.fit(X_train)
# init model
model = init_model()
# pre-training accuracy
score = model.evaluate(val_datagen.flow(X_test, y_test, batch_size=num_batch_size), verbose=0)
print("Pre-training accuracy: %.4f%%\n" % (100 * score[1]))
# train the model
start = datetime.now()
history = model.fit(train_datagen.flow(X_train, y_train, batch_size=num_batch_size),
steps_per_epoch=len(X_train) / num_batch_size,
epochs=epochs,
validation_data=val_datagen.flow(X_test, y_test, batch_size=num_batch_size))
end = datetime.now()
print("Training completed in time: ", end - start, '\n')
return history
def show_results(tot_history):
"""Show accuracy and loss graphs for train and test sets."""
for i, history in enumerate(tot_history):
print('\n({})'.format(i+1))
plt.figure(figsize=(15,5))
plt.subplot(121)
plt.plot(history.history['accuracy'])
plt.plot(history.history['val_accuracy'])
plt.grid(linestyle='--')
plt.ylabel('Accuracy')
plt.xlabel('Epoch')
plt.legend(['train', 'validation'], loc='upper left')
plt.subplot(122)
plt.plot(history.history['loss'])
plt.plot(history.history['val_loss'])
plt.grid(linestyle='--')
plt.ylabel('Loss')
plt.xlabel('Epoch')
plt.legend(['train', 'validation'], loc='upper left')
plt.show()
print('\tMax validation accuracy: %.4f %%' % (np.max(history.history['val_accuracy']) * 100))
print('\tMin validation loss: %.5f' % np.min(history.history['val_loss']))
FOLD_K = 1
REPEAT = 1
history1 = []
for i in range(REPEAT):
print('-'*80)
print("\n({})\n".format(i+1))
history = process_fold(FOLD_K, us8k_df, epochs=100)
history1.append(history)
show_results(history1)
FOLD_K = 2
REPEAT = 1
history2 = []
for i in range(REPEAT):
print('-'*80)
print("\n({})\n".format(i+1))
history = process_fold(FOLD_K, us8k_df, epochs=100)
history2.append(history)
show_results(history2)
FOLD_K = 3
REPEAT = 1
history3 = []
for i in range(REPEAT):
print('-'*80)
print("\n({})\n".format(i+1))
history = process_fold(FOLD_K, us8k_df, epochs=100)
history3.append(history)
show_results(history3)
FOLD_K = 4
REPEAT = 1
history4 = []
for i in range(REPEAT):
print('-'*80)
print("\n({})\n".format(i+1))
history = process_fold(FOLD_K, us8k_df, epochs=100)
history4.append(history)
show_results(history4)
FOLD_K = 5
REPEAT = 1
history5 = []
for i in range(REPEAT):
print('-'*80)
print("\n({})\n".format(i+1))
history = process_fold(FOLD_K, us8k_df, epochs=100)
history5.append(history)
show_results(history5)
FOLD_K = 6
REPEAT = 1
history6 = []
for i in range(REPEAT):
print('-'*80)
print("\n({})\n".format(i+1))
history = process_fold(FOLD_K, us8k_df, epochs=100)
history6.append(history)
show_results(history6)
FOLD_K = 7
REPEAT = 1
history7 = []
for i in range(REPEAT):
print('-'*80)
print("\n({})\n".format(i+1))
history = process_fold(FOLD_K, us8k_df, epochs=100)
history7.append(history)
show_results(history7)
FOLD_K = 8
REPEAT = 1
history8 = []
for i in range(REPEAT):
print('-'*80)
print("\n({})\n".format(i+1))
history = process_fold(FOLD_K, us8k_df, epochs=100)
history8.append(history)
show_results(history8)
FOLD_K = 9
REPEAT = 1
history9 = []
for i in range(REPEAT):
print('-'*80)
print("\n({})\n".format(i+1))
history = process_fold(FOLD_K, us8k_df, epochs=100)
history9.append(history)
show_results(history9)
FOLD_K = 10
REPEAT = 1
history10 = []
for i in range(REPEAT):
print('-'*80)
print("\n({})\n".format(i+1))
history = process_fold(FOLD_K, us8k_df, epochs=100)
history10.append(history)
show_results(history10)
| 0.702122 | 0.897336 |
## Transfer Learning
Let's pick a well-trained model and see how it did on the test data:
```
import os
import logging
import numpy as np
import tensorflow as tf
from tensorflow.keras import layers
from tensorflow.keras import regularizers
import tools.train as train
import tools.plot as plot
# Suppress tensorflow warnings about internal deprecations
logger = tf.get_logger()
logger.setLevel(logging.ERROR)
# Count data
files = ("../data/mitbih_train.csv", "../data/mitbih_test.csv")
inputs_mit, labels_mit, sparse_labels_mit, df_mit = train.preprocess(*files, fft=False)
# Add a dimension for "channels"
for key in inputs_mit:
inputs_mit[key] = tf.expand_dims(inputs_mit[key], axis=2)
train.class_count(df_mit)
# Load in a model
modelpath = os.path.join("..", "models", "20190812-181641", "nofft", "wavenet.h5") # A specific run stored locally
model = tf.keras.models.load_model(modelpath)
# See model performance
test_pred = np.argmax(model.predict(inputs_mit["test"]), axis=1)
plot.plot_cm(sparse_labels_mit["test"], test_pred, classes=np.array(["N", "S", "V", "F", "Q"]), normalize=True)
```
Now we'll freeze all of the layers in the model except for the last classifying (dense) layer.
```
for layer in model.layers[:-1]:
layer.trainable = False
# Put in a dummy compile to see the frozen layers
model.compile(optimizer="Nadam", loss="categorical_crossentropy")
```
Let's split the PTB dataset into train and test sets (if we haven't already) and load them in with our custom API:
```
train.split_test_train(
["../data/ptbdb_abnormal.csv", "../data/ptbdb_normal.csv"],
os.path.join("..", "data", "ptbdb"),
0.2,
)
# Count data
files = ("../data/ptbdb_train.csv", "../data/ptbdb_test.csv")
inputs_ptb, labels_ptb, sparse_labels_ptb, df_ptb = train.preprocess(*files, fft=False)
# Add a dimension for "channels"
for key in inputs_ptb:
inputs_ptb[key] = tf.expand_dims(inputs_ptb[key], axis=2)
train.class_count(df_ptb)
```
Now let's use the flattened convolutional output as input to a few fully-connected layers, and train the mostly-frozen model on the new data (we'll apply weights to the loss function due to the imbalanced dataset):
```
largest_class_count = df_ptb["train"].groupby("Classes").size().max()
class_weights = np.divide(largest_class_count, df_ptb["train"].groupby("Classes").size().to_numpy())
print("Weighting the classes:", class_weights)
config = {
"optimizer": "Nadam",
"loss": "binary_crossentropy",
"class_weights": class_weights,
"batch_size": 50,
"val_split": 0.1,
"epochs": 300,
"verbose": 0,
"patience": 300,
"metrics": ["accuracy"],
"regularizer": regularizers.l1_l2(l1=0.001, l2=0.01),
}
conv_output = model.layers[-2].output
new_output = layers.Dense(94, activation="relu", kernel_regularizer=config.get("regularizer"))(conv_output)
new_output = layers.Dense(94, activation="relu", kernel_regularizer=config.get("regularizer"))(new_output)
new_output = layers.Dropout(0.8, name="transfer_dropout")(new_output)
new_output = layers.Dense(1, activation="sigmoid")(new_output)
transferred_model = tf.keras.Model(inputs=model.layers[0].input, outputs=new_output, name="transferred_conv1d")
# Train it
history = train.train(transferred_model, inputs_ptb, sparse_labels_ptb, config)
plot.plot_fit_history(history)
test_pred = transferred_model.predict(inputs_ptb["test"])
plot.plot_pr_curve(
np.expand_dims(test_pred, axis=1),
np.expand_dims(sparse_labels_ptb["test"], axis=1),
["Normal", "Abnormal"],
savefile="../presentation/wavenet_transfer_pr.png",
)
test_pred[test_pred > 0.5] = 1
test_pred[test_pred <= 0.5] = 0
plot.plot_cm(
sparse_labels_ptb["test"],
test_pred,
classes=np.array(["Normal", "Abnormal"]),
normalize=True,
savefile="../presentation/wavenet_transfer_cm.png",
)
```
Now let's look at the more standard residual CNN.
```
# Load in a model
modelpath = os.path.join("..", "models", "20190812-202306", "nofft", "CNN.h5") # A specific run on Jeffmin's computer
model = tf.keras.models.load_model(modelpath)
# See model performance
test_pred = np.argmax(model.predict(inputs_mit["test"]), axis=1)
plot.plot_cm(sparse_labels_mit["test"], test_pred, classes=np.array(["N", "S", "V", "F", "Q"]), normalize=True)
for layer in model.layers[:-1]:
layer.trainable = False
largest_class_count = df_ptb["train"].groupby("Classes").size().max()
class_weights = np.divide(largest_class_count, df_ptb["train"].groupby("Classes").size().to_numpy())
print("Weighting the classes:", class_weights)
config = {
"optimizer": "Nadam",
"loss": "binary_crossentropy",
"class_weights": class_weights,
"batch_size": 50,
"val_split": 0.1,
"epochs": 300,
"verbose": 0,
"patience": 300,
"metrics": ["accuracy"],
"regularizer": regularizers.l1_l2(l1=0.001, l2=0.01),
}
conv_output = model.layers[-4].output
new_output = layers.Flatten()(conv_output)
new_output = layers.Dense(94, activation="relu", kernel_regularizer=config.get("regularizer"))(new_output)
new_output = layers.Dense(94, activation="relu", kernel_regularizer=config.get("regularizer"))(new_output)
new_output = layers.Dropout(0.8, name="transfer_dropout")(new_output)
new_output = layers.Dense(1, activation="sigmoid")(new_output)
transferred_model = tf.keras.Model(inputs=model.layers[0].input, outputs=new_output, name="transferred_conv1d")
# Train it
history = train.train(transferred_model, inputs_ptb, sparse_labels_ptb, config)
plot.plot_fit_history(history)
test_pred = transferred_model.predict(inputs_ptb["test"])
plot.plot_pr_curve(
np.expand_dims(test_pred, axis=1),
np.expand_dims(sparse_labels_ptb["test"], axis=1),
["Normal", "Abnormal"],
savefile="../presentation/CNN_transfer_pr.png",
)
test_pred[test_pred > 0.5] = 1
test_pred[test_pred <= 0.5] = 0
plot.plot_cm(
sparse_labels_ptb["test"],
test_pred,
classes=np.array(["Normal", "Abnormal"]),
normalize=True,
savefile="../presentation/CNN_transfer_cm.png",
)
```
|
github_jupyter
|
import os
import logging
import numpy as np
import tensorflow as tf
from tensorflow.keras import layers
from tensorflow.keras import regularizers
import tools.train as train
import tools.plot as plot
# Suppress tensorflow warnings about internal deprecations
logger = tf.get_logger()
logger.setLevel(logging.ERROR)
# Count data
files = ("../data/mitbih_train.csv", "../data/mitbih_test.csv")
inputs_mit, labels_mit, sparse_labels_mit, df_mit = train.preprocess(*files, fft=False)
# Add a dimension for "channels"
for key in inputs_mit:
inputs_mit[key] = tf.expand_dims(inputs_mit[key], axis=2)
train.class_count(df_mit)
# Load in a model
modelpath = os.path.join("..", "models", "20190812-181641", "nofft", "wavenet.h5") # A specific run stored locally
model = tf.keras.models.load_model(modelpath)
# See model performance
test_pred = np.argmax(model.predict(inputs_mit["test"]), axis=1)
plot.plot_cm(sparse_labels_mit["test"], test_pred, classes=np.array(["N", "S", "V", "F", "Q"]), normalize=True)
for layer in model.layers[:-1]:
layer.trainable = False
# Put in a dummy compile to see the frozen layers
model.compile(optimizer="Nadam", loss="categorical_crossentropy")
train.split_test_train(
["../data/ptbdb_abnormal.csv", "../data/ptbdb_normal.csv"],
os.path.join("..", "data", "ptbdb"),
0.2,
)
# Count data
files = ("../data/ptbdb_train.csv", "../data/ptbdb_test.csv")
inputs_ptb, labels_ptb, sparse_labels_ptb, df_ptb = train.preprocess(*files, fft=False)
# Add a dimension for "channels"
for key in inputs_ptb:
inputs_ptb[key] = tf.expand_dims(inputs_ptb[key], axis=2)
train.class_count(df_ptb)
largest_class_count = df_ptb["train"].groupby("Classes").size().max()
class_weights = np.divide(largest_class_count, df_ptb["train"].groupby("Classes").size().to_numpy())
print("Weighting the classes:", class_weights)
config = {
"optimizer": "Nadam",
"loss": "binary_crossentropy",
"class_weights": class_weights,
"batch_size": 50,
"val_split": 0.1,
"epochs": 300,
"verbose": 0,
"patience": 300,
"metrics": ["accuracy"],
"regularizer": regularizers.l1_l2(l1=0.001, l2=0.01),
}
conv_output = model.layers[-2].output
new_output = layers.Dense(94, activation="relu", kernel_regularizer=config.get("regularizer"))(conv_output)
new_output = layers.Dense(94, activation="relu", kernel_regularizer=config.get("regularizer"))(new_output)
new_output = layers.Dropout(0.8, name="transfer_dropout")(new_output)
new_output = layers.Dense(1, activation="sigmoid")(new_output)
transferred_model = tf.keras.Model(inputs=model.layers[0].input, outputs=new_output, name="transferred_conv1d")
# Train it
history = train.train(transferred_model, inputs_ptb, sparse_labels_ptb, config)
plot.plot_fit_history(history)
test_pred = transferred_model.predict(inputs_ptb["test"])
plot.plot_pr_curve(
np.expand_dims(test_pred, axis=1),
np.expand_dims(sparse_labels_ptb["test"], axis=1),
["Normal", "Abnormal"],
savefile="../presentation/wavenet_transfer_pr.png",
)
test_pred[test_pred > 0.5] = 1
test_pred[test_pred <= 0.5] = 0
plot.plot_cm(
sparse_labels_ptb["test"],
test_pred,
classes=np.array(["Normal", "Abnormal"]),
normalize=True,
savefile="../presentation/wavenet_transfer_cm.png",
)
# Load in a model
modelpath = os.path.join("..", "models", "20190812-202306", "nofft", "CNN.h5") # A specific run on Jeffmin's computer
model = tf.keras.models.load_model(modelpath)
# See model performance
test_pred = np.argmax(model.predict(inputs_mit["test"]), axis=1)
plot.plot_cm(sparse_labels_mit["test"], test_pred, classes=np.array(["N", "S", "V", "F", "Q"]), normalize=True)
for layer in model.layers[:-1]:
layer.trainable = False
largest_class_count = df_ptb["train"].groupby("Classes").size().max()
class_weights = np.divide(largest_class_count, df_ptb["train"].groupby("Classes").size().to_numpy())
print("Weighting the classes:", class_weights)
config = {
"optimizer": "Nadam",
"loss": "binary_crossentropy",
"class_weights": class_weights,
"batch_size": 50,
"val_split": 0.1,
"epochs": 300,
"verbose": 0,
"patience": 300,
"metrics": ["accuracy"],
"regularizer": regularizers.l1_l2(l1=0.001, l2=0.01),
}
conv_output = model.layers[-4].output
new_output = layers.Flatten()(conv_output)
new_output = layers.Dense(94, activation="relu", kernel_regularizer=config.get("regularizer"))(new_output)
new_output = layers.Dense(94, activation="relu", kernel_regularizer=config.get("regularizer"))(new_output)
new_output = layers.Dropout(0.8, name="transfer_dropout")(new_output)
new_output = layers.Dense(1, activation="sigmoid")(new_output)
transferred_model = tf.keras.Model(inputs=model.layers[0].input, outputs=new_output, name="transferred_conv1d")
# Train it
history = train.train(transferred_model, inputs_ptb, sparse_labels_ptb, config)
plot.plot_fit_history(history)
test_pred = transferred_model.predict(inputs_ptb["test"])
plot.plot_pr_curve(
np.expand_dims(test_pred, axis=1),
np.expand_dims(sparse_labels_ptb["test"], axis=1),
["Normal", "Abnormal"],
savefile="../presentation/CNN_transfer_pr.png",
)
test_pred[test_pred > 0.5] = 1
test_pred[test_pred <= 0.5] = 0
plot.plot_cm(
sparse_labels_ptb["test"],
test_pred,
classes=np.array(["Normal", "Abnormal"]),
normalize=True,
savefile="../presentation/CNN_transfer_cm.png",
)
| 0.809201 | 0.855429 |
```
%matplotlib inline
import numpy as np
import tensorflow as tf
import matplotlib.pyplot as plt
sess = tf.InteractiveSession()
image = np.array([[[[1],[2]],
[[3],[4]]]], dtype=np.float32)
print(image.shape)
plt.imshow(image.reshape(2,2), cmap='Greys')
```
## Conv Layer
weight.shape = 2 filters (1 , 1 , 1)


(TensorFlow For Machine Intelligence: A hands-on introduction to learning algorithms
by Sam Abrahams et al.)
```
print("imag:\n", image)
weight = tf.constant([[[[2., 0.5]]]])
print("weight.shape", weight.shape)
conv2d = tf.nn.conv2d(image, weight, strides=[1, 1, 1, 1], padding='SAME')
conv2d_img = conv2d.eval()
conv2d_img = np.swapaxes(conv2d_img, 0, 3)
for i, one_img in enumerate(conv2d_img):
print(one_img.reshape(2,2))
plt.subplot(1,2,i+1), plt.imshow(one_img.reshape(2,2), cmap='gray')
```
## MAX POOLING


```
image = np.array([[[[4],[3]],
[[2],[1]]]], dtype=np.float32)
pool = tf.nn.max_pool(image, ksize=[1, 2, 2, 1],
strides=[1, 1, 1, 1], padding='VALID')
print(pool.shape)
print(pool.eval())
```
## SAME: Zero paddings

```
image = np.array([[[[4],[3]],
[[2],[1]]]], dtype=np.float32)
pool = tf.nn.max_pool(image, ksize=[1, 2, 2, 1],
strides=[1, 1, 1, 1], padding='SAME')
print(pool.shape)
print(pool.eval())
from tensorflow.examples.tutorials.mnist import input_data
mnist = input_data.read_data_sets("MNIST_data/", one_hot=True)
# Check out https://www.tensorflow.org/get_started/mnist/beginners for
# more information about the mnist dataset
img = mnist.train.images[0].reshape(28,28)
plt.imshow(img, cmap='gray')
sess = tf.InteractiveSession()
img = img.reshape(-1,28,28,1)
W1 = tf.Variable(tf.random_normal([3, 3, 1, 5], stddev=0.01))
conv2d = tf.nn.conv2d(img, W1, strides=[1, 2, 2, 1], padding='SAME')
print(conv2d)
sess.run(tf.global_variables_initializer())
conv2d_img = conv2d.eval()
conv2d_img = np.swapaxes(conv2d_img, 0, 3)
for i, one_img in enumerate(conv2d_img):
plt.subplot(1,5,i+1), plt.imshow(one_img.reshape(14,14), cmap='gray')
pool = tf.nn.max_pool(conv2d, ksize=[1, 2, 2, 1], strides=[
1, 2, 2, 1], padding='SAME')
print(pool)
sess.run(tf.global_variables_initializer())
pool_img = pool.eval()
pool_img = np.swapaxes(pool_img, 0, 3)
for i, one_img in enumerate(pool_img):
plt.subplot(1,5,i+1), plt.imshow(one_img.reshape(7, 7), cmap='gray')
```
|
github_jupyter
|
%matplotlib inline
import numpy as np
import tensorflow as tf
import matplotlib.pyplot as plt
sess = tf.InteractiveSession()
image = np.array([[[[1],[2]],
[[3],[4]]]], dtype=np.float32)
print(image.shape)
plt.imshow(image.reshape(2,2), cmap='Greys')
print("imag:\n", image)
weight = tf.constant([[[[2., 0.5]]]])
print("weight.shape", weight.shape)
conv2d = tf.nn.conv2d(image, weight, strides=[1, 1, 1, 1], padding='SAME')
conv2d_img = conv2d.eval()
conv2d_img = np.swapaxes(conv2d_img, 0, 3)
for i, one_img in enumerate(conv2d_img):
print(one_img.reshape(2,2))
plt.subplot(1,2,i+1), plt.imshow(one_img.reshape(2,2), cmap='gray')
image = np.array([[[[4],[3]],
[[2],[1]]]], dtype=np.float32)
pool = tf.nn.max_pool(image, ksize=[1, 2, 2, 1],
strides=[1, 1, 1, 1], padding='VALID')
print(pool.shape)
print(pool.eval())
image = np.array([[[[4],[3]],
[[2],[1]]]], dtype=np.float32)
pool = tf.nn.max_pool(image, ksize=[1, 2, 2, 1],
strides=[1, 1, 1, 1], padding='SAME')
print(pool.shape)
print(pool.eval())
from tensorflow.examples.tutorials.mnist import input_data
mnist = input_data.read_data_sets("MNIST_data/", one_hot=True)
# Check out https://www.tensorflow.org/get_started/mnist/beginners for
# more information about the mnist dataset
img = mnist.train.images[0].reshape(28,28)
plt.imshow(img, cmap='gray')
sess = tf.InteractiveSession()
img = img.reshape(-1,28,28,1)
W1 = tf.Variable(tf.random_normal([3, 3, 1, 5], stddev=0.01))
conv2d = tf.nn.conv2d(img, W1, strides=[1, 2, 2, 1], padding='SAME')
print(conv2d)
sess.run(tf.global_variables_initializer())
conv2d_img = conv2d.eval()
conv2d_img = np.swapaxes(conv2d_img, 0, 3)
for i, one_img in enumerate(conv2d_img):
plt.subplot(1,5,i+1), plt.imshow(one_img.reshape(14,14), cmap='gray')
pool = tf.nn.max_pool(conv2d, ksize=[1, 2, 2, 1], strides=[
1, 2, 2, 1], padding='SAME')
print(pool)
sess.run(tf.global_variables_initializer())
pool_img = pool.eval()
pool_img = np.swapaxes(pool_img, 0, 3)
for i, one_img in enumerate(pool_img):
plt.subplot(1,5,i+1), plt.imshow(one_img.reshape(7, 7), cmap='gray')
| 0.760384 | 0.934335 |
# Implementing Different Layers
We will illustrate how to use different types of layers in TensorFlow
The layers of interest are:
1. Convolutional Layer
2. Activation Layer
3. Max-Pool Layer
4. Fully Connected Layer
We will generate two different data sets for this script, a 1-D data set (row of data) and a 2-D data set (similar to picture)
```
import tensorflow as tf
import matplotlib.pyplot as plt
import csv
import os
import random
import numpy as np
import random
from tensorflow.python.framework import ops
```
```
#---------------------------------------------------|
#-------------------1D-data-------------------------|
#---------------------------------------------------|
```
```
# Create graph session
ops.reset_default_graph()
sess = tf.Session()
# parameters for the run
data_size = 25
conv_size = 5
maxpool_size = 5
stride_size = 1
# ensure reproducibility
seed=13
np.random.seed(seed)
tf.set_random_seed(seed)
# Generate 1D data
data_1d = np.random.normal(size=data_size)
# Placeholder
x_input_1d = tf.placeholder(dtype=tf.float32, shape=[data_size])
#--------Convolution--------
def conv_layer_1d(input_1d, my_filter,stride):
# TensorFlow's 'conv2d()' function only works with 4D arrays:
# [batch#, width, height, channels], we have 1 batch, and
# width = 1, but height = the length of the input, and 1 channel.
# So next we create the 4D array by inserting dimension 1's.
input_2d = tf.expand_dims(input_1d, 0)
input_3d = tf.expand_dims(input_2d, 0)
input_4d = tf.expand_dims(input_3d, 3)
# Perform convolution with stride = 1, if we wanted to increase the stride,
# to say '2', then strides=[1,1,2,1]
convolution_output = tf.nn.conv2d(input_4d, filter=my_filter, strides=[1,1,stride,1], padding="VALID")
# Get rid of extra dimensions
conv_output_1d = tf.squeeze(convolution_output)
return(conv_output_1d)
# Create filter for convolution.
my_filter = tf.Variable(tf.random_normal(shape=[1,conv_size,1,1]))
# Create convolution layer
my_convolution_output = conv_layer_1d(x_input_1d, my_filter,stride=stride_size)
#--------Activation--------
def activation(input_1d):
return(tf.nn.relu(input_1d))
# Create activation layer
my_activation_output = activation(my_convolution_output)
#--------Max Pool--------
def max_pool(input_1d, width,stride):
# Just like 'conv2d()' above, max_pool() works with 4D arrays.
# [batch_size=1, width=1, height=num_input, channels=1]
input_2d = tf.expand_dims(input_1d, 0)
input_3d = tf.expand_dims(input_2d, 0)
input_4d = tf.expand_dims(input_3d, 3)
# Perform the max pooling with strides = [1,1,1,1]
# If we wanted to increase the stride on our data dimension, say by
# a factor of '2', we put strides = [1, 1, 2, 1]
# We will also need to specify the width of the max-window ('width')
pool_output = tf.nn.max_pool(input_4d, ksize=[1, 1, width, 1],
strides=[1, 1, stride, 1],
padding='VALID')
# Get rid of extra dimensions
pool_output_1d = tf.squeeze(pool_output)
return(pool_output_1d)
my_maxpool_output = max_pool(my_activation_output, width=maxpool_size,stride=stride_size)
#--------Fully Connected--------
def fully_connected(input_layer, num_outputs):
# First we find the needed shape of the multiplication weight matrix:
# The dimension will be (length of input) by (num_outputs)
weight_shape = tf.squeeze(tf.stack([tf.shape(input_layer),[num_outputs]]))
# Initialize such weight
weight = tf.random_normal(weight_shape, stddev=0.1)
# Initialize the bias
bias = tf.random_normal(shape=[num_outputs])
# Make the 1D input array into a 2D array for matrix multiplication
input_layer_2d = tf.expand_dims(input_layer, 0)
# Perform the matrix multiplication and add the bias
full_output = tf.add(tf.matmul(input_layer_2d, weight), bias)
# Get rid of extra dimensions
full_output_1d = tf.squeeze(full_output)
return(full_output_1d)
my_full_output = fully_connected(my_maxpool_output, 5)
# Run graph
# Initialize Variables
init = tf.global_variables_initializer()
sess.run(init)
feed_dict = {x_input_1d: data_1d}
print('>>>> 1D Data <<<<')
# Convolution Output
print('Input = array of length %d' % (x_input_1d.shape.as_list()[0]))
print('Convolution w/ filter, length = %d, stride size = %d, results in an array of length %d:' %
(conv_size,stride_size,my_convolution_output.shape.as_list()[0]))
print(sess.run(my_convolution_output, feed_dict=feed_dict))
# Activation Output
print('\nInput = above array of length %d' % (my_convolution_output.shape.as_list()[0]))
print('ReLU element wise returns an array of length %d:' % (my_activation_output.shape.as_list()[0]))
print(sess.run(my_activation_output, feed_dict=feed_dict))
# Max Pool Output
print('\nInput = above array of length %d' % (my_activation_output.shape.as_list()[0]))
print('MaxPool, window length = %d, stride size = %d, results in the array of length %d' %
(maxpool_size,stride_size,my_maxpool_output.shape.as_list()[0]))
print(sess.run(my_maxpool_output, feed_dict=feed_dict))
# Fully Connected Output
print('\nInput = above array of length %d' % (my_maxpool_output.shape.as_list()[0]))
print('Fully connected layer on all 4 rows with %d outputs:' %
(my_full_output.shape.as_list()[0]))
print(sess.run(my_full_output, feed_dict=feed_dict))
```
```
#---------------------------------------------------|
#-------------------2D-data-------------------------|
#---------------------------------------------------|
```
```
# Reset Graph
ops.reset_default_graph()
sess = tf.Session()
# parameters for the run
row_size = 10
col_size = 10
conv_size = 2
conv_stride_size = 2
maxpool_size = 2
maxpool_stride_size = 1
# ensure reproducibility
seed=13
np.random.seed(seed)
tf.set_random_seed(seed)
#Generate 2D data
data_size = [row_size,col_size]
data_2d = np.random.normal(size=data_size)
#--------Placeholder--------
x_input_2d = tf.placeholder(dtype=tf.float32, shape=data_size)
# Convolution
def conv_layer_2d(input_2d, my_filter,stride_size):
# TensorFlow's 'conv2d()' function only works with 4D arrays:
# [batch#, width, height, channels], we have 1 batch, and
# 1 channel, but we do have width AND height this time.
# So next we create the 4D array by inserting dimension 1's.
input_3d = tf.expand_dims(input_2d, 0)
input_4d = tf.expand_dims(input_3d, 3)
# Note the stride difference below!
convolution_output = tf.nn.conv2d(input_4d, filter=my_filter,
strides=[1,stride_size,stride_size,1], padding="VALID")
# Get rid of unnecessary dimensions
conv_output_2d = tf.squeeze(convolution_output)
return(conv_output_2d)
# Create Convolutional Filter
my_filter = tf.Variable(tf.random_normal(shape=[conv_size,conv_size,1,1]))
# Create Convolutional Layer
my_convolution_output = conv_layer_2d(x_input_2d, my_filter,stride_size=conv_stride_size)
#--------Activation--------
def activation(input_1d):
return(tf.nn.relu(input_1d))
# Create Activation Layer
my_activation_output = activation(my_convolution_output)
#--------Max Pool--------
def max_pool(input_2d, width, height,stride):
# Just like 'conv2d()' above, max_pool() works with 4D arrays.
# [batch_size=1, width=given, height=given, channels=1]
input_3d = tf.expand_dims(input_2d, 0)
input_4d = tf.expand_dims(input_3d, 3)
# Perform the max pooling with strides = [1,1,1,1]
# If we wanted to increase the stride on our data dimension, say by
# a factor of '2', we put strides = [1, 2, 2, 1]
pool_output = tf.nn.max_pool(input_4d, ksize=[1, height, width, 1],
strides=[1, stride, stride, 1],
padding='VALID')
# Get rid of unnecessary dimensions
pool_output_2d = tf.squeeze(pool_output)
return(pool_output_2d)
# Create Max-Pool Layer
my_maxpool_output = max_pool(my_activation_output,
width=maxpool_size, height=maxpool_size,stride=maxpool_stride_size)
#--------Fully Connected--------
def fully_connected(input_layer, num_outputs):
# In order to connect our whole W byH 2d array, we first flatten it out to
# a W times H 1D array.
flat_input = tf.reshape(input_layer, [-1])
# We then find out how long it is, and create an array for the shape of
# the multiplication weight = (WxH) by (num_outputs)
weight_shape = tf.squeeze(tf.stack([tf.shape(flat_input),[num_outputs]]))
# Initialize the weight
weight = tf.random_normal(weight_shape, stddev=0.1)
# Initialize the bias
bias = tf.random_normal(shape=[num_outputs])
# Now make the flat 1D array into a 2D array for multiplication
input_2d = tf.expand_dims(flat_input, 0)
# Multiply and add the bias
full_output = tf.add(tf.matmul(input_2d, weight), bias)
# Get rid of extra dimension
full_output_2d = tf.squeeze(full_output)
return(full_output_2d)
# Create Fully Connected Layer
my_full_output = fully_connected(my_maxpool_output, 5)
# Run graph
# Initialize Variables
init = tf.global_variables_initializer()
sess.run(init)
feed_dict = {x_input_2d: data_2d}
print('>>>> 2D Data <<<<')
# Convolution Output
print('Input = %s array' % (x_input_2d.shape.as_list()))
print('%s Convolution, stride size = [%d, %d] , results in the %s array' %
(my_filter.get_shape().as_list()[:2],conv_stride_size,conv_stride_size,my_convolution_output.shape.as_list()))
print(sess.run(my_convolution_output, feed_dict=feed_dict))
# Activation Output
print('\nInput = the above %s array' % (my_convolution_output.shape.as_list()))
print('ReLU element wise returns the %s array' % (my_activation_output.shape.as_list()))
print(sess.run(my_activation_output, feed_dict=feed_dict))
# Max Pool Output
print('\nInput = the above %s array' % (my_activation_output.shape.as_list()))
print('MaxPool, stride size = [%d, %d], results in %s array' %
(maxpool_stride_size,maxpool_stride_size,my_maxpool_output.shape.as_list()))
print(sess.run(my_maxpool_output, feed_dict=feed_dict))
# Fully Connected Output
print('\nInput = the above %s array' % (my_maxpool_output.shape.as_list()))
print('Fully connected layer on all %d rows results in %s outputs:' %
(my_maxpool_output.shape.as_list()[0],my_full_output.shape.as_list()[0]))
print(sess.run(my_full_output, feed_dict=feed_dict))
tested; G
```
|
github_jupyter
|
import tensorflow as tf
import matplotlib.pyplot as plt
import csv
import os
import random
import numpy as np
import random
from tensorflow.python.framework import ops
#---------------------------------------------------|
#-------------------1D-data-------------------------|
#---------------------------------------------------|
# Create graph session
ops.reset_default_graph()
sess = tf.Session()
# parameters for the run
data_size = 25
conv_size = 5
maxpool_size = 5
stride_size = 1
# ensure reproducibility
seed=13
np.random.seed(seed)
tf.set_random_seed(seed)
# Generate 1D data
data_1d = np.random.normal(size=data_size)
# Placeholder
x_input_1d = tf.placeholder(dtype=tf.float32, shape=[data_size])
#--------Convolution--------
def conv_layer_1d(input_1d, my_filter,stride):
# TensorFlow's 'conv2d()' function only works with 4D arrays:
# [batch#, width, height, channels], we have 1 batch, and
# width = 1, but height = the length of the input, and 1 channel.
# So next we create the 4D array by inserting dimension 1's.
input_2d = tf.expand_dims(input_1d, 0)
input_3d = tf.expand_dims(input_2d, 0)
input_4d = tf.expand_dims(input_3d, 3)
# Perform convolution with stride = 1, if we wanted to increase the stride,
# to say '2', then strides=[1,1,2,1]
convolution_output = tf.nn.conv2d(input_4d, filter=my_filter, strides=[1,1,stride,1], padding="VALID")
# Get rid of extra dimensions
conv_output_1d = tf.squeeze(convolution_output)
return(conv_output_1d)
# Create filter for convolution.
my_filter = tf.Variable(tf.random_normal(shape=[1,conv_size,1,1]))
# Create convolution layer
my_convolution_output = conv_layer_1d(x_input_1d, my_filter,stride=stride_size)
#--------Activation--------
def activation(input_1d):
return(tf.nn.relu(input_1d))
# Create activation layer
my_activation_output = activation(my_convolution_output)
#--------Max Pool--------
def max_pool(input_1d, width,stride):
# Just like 'conv2d()' above, max_pool() works with 4D arrays.
# [batch_size=1, width=1, height=num_input, channels=1]
input_2d = tf.expand_dims(input_1d, 0)
input_3d = tf.expand_dims(input_2d, 0)
input_4d = tf.expand_dims(input_3d, 3)
# Perform the max pooling with strides = [1,1,1,1]
# If we wanted to increase the stride on our data dimension, say by
# a factor of '2', we put strides = [1, 1, 2, 1]
# We will also need to specify the width of the max-window ('width')
pool_output = tf.nn.max_pool(input_4d, ksize=[1, 1, width, 1],
strides=[1, 1, stride, 1],
padding='VALID')
# Get rid of extra dimensions
pool_output_1d = tf.squeeze(pool_output)
return(pool_output_1d)
my_maxpool_output = max_pool(my_activation_output, width=maxpool_size,stride=stride_size)
#--------Fully Connected--------
def fully_connected(input_layer, num_outputs):
# First we find the needed shape of the multiplication weight matrix:
# The dimension will be (length of input) by (num_outputs)
weight_shape = tf.squeeze(tf.stack([tf.shape(input_layer),[num_outputs]]))
# Initialize such weight
weight = tf.random_normal(weight_shape, stddev=0.1)
# Initialize the bias
bias = tf.random_normal(shape=[num_outputs])
# Make the 1D input array into a 2D array for matrix multiplication
input_layer_2d = tf.expand_dims(input_layer, 0)
# Perform the matrix multiplication and add the bias
full_output = tf.add(tf.matmul(input_layer_2d, weight), bias)
# Get rid of extra dimensions
full_output_1d = tf.squeeze(full_output)
return(full_output_1d)
my_full_output = fully_connected(my_maxpool_output, 5)
# Run graph
# Initialize Variables
init = tf.global_variables_initializer()
sess.run(init)
feed_dict = {x_input_1d: data_1d}
print('>>>> 1D Data <<<<')
# Convolution Output
print('Input = array of length %d' % (x_input_1d.shape.as_list()[0]))
print('Convolution w/ filter, length = %d, stride size = %d, results in an array of length %d:' %
(conv_size,stride_size,my_convolution_output.shape.as_list()[0]))
print(sess.run(my_convolution_output, feed_dict=feed_dict))
# Activation Output
print('\nInput = above array of length %d' % (my_convolution_output.shape.as_list()[0]))
print('ReLU element wise returns an array of length %d:' % (my_activation_output.shape.as_list()[0]))
print(sess.run(my_activation_output, feed_dict=feed_dict))
# Max Pool Output
print('\nInput = above array of length %d' % (my_activation_output.shape.as_list()[0]))
print('MaxPool, window length = %d, stride size = %d, results in the array of length %d' %
(maxpool_size,stride_size,my_maxpool_output.shape.as_list()[0]))
print(sess.run(my_maxpool_output, feed_dict=feed_dict))
# Fully Connected Output
print('\nInput = above array of length %d' % (my_maxpool_output.shape.as_list()[0]))
print('Fully connected layer on all 4 rows with %d outputs:' %
(my_full_output.shape.as_list()[0]))
print(sess.run(my_full_output, feed_dict=feed_dict))
#---------------------------------------------------|
#-------------------2D-data-------------------------|
#---------------------------------------------------|
# Reset Graph
ops.reset_default_graph()
sess = tf.Session()
# parameters for the run
row_size = 10
col_size = 10
conv_size = 2
conv_stride_size = 2
maxpool_size = 2
maxpool_stride_size = 1
# ensure reproducibility
seed=13
np.random.seed(seed)
tf.set_random_seed(seed)
#Generate 2D data
data_size = [row_size,col_size]
data_2d = np.random.normal(size=data_size)
#--------Placeholder--------
x_input_2d = tf.placeholder(dtype=tf.float32, shape=data_size)
# Convolution
def conv_layer_2d(input_2d, my_filter,stride_size):
# TensorFlow's 'conv2d()' function only works with 4D arrays:
# [batch#, width, height, channels], we have 1 batch, and
# 1 channel, but we do have width AND height this time.
# So next we create the 4D array by inserting dimension 1's.
input_3d = tf.expand_dims(input_2d, 0)
input_4d = tf.expand_dims(input_3d, 3)
# Note the stride difference below!
convolution_output = tf.nn.conv2d(input_4d, filter=my_filter,
strides=[1,stride_size,stride_size,1], padding="VALID")
# Get rid of unnecessary dimensions
conv_output_2d = tf.squeeze(convolution_output)
return(conv_output_2d)
# Create Convolutional Filter
my_filter = tf.Variable(tf.random_normal(shape=[conv_size,conv_size,1,1]))
# Create Convolutional Layer
my_convolution_output = conv_layer_2d(x_input_2d, my_filter,stride_size=conv_stride_size)
#--------Activation--------
def activation(input_1d):
return(tf.nn.relu(input_1d))
# Create Activation Layer
my_activation_output = activation(my_convolution_output)
#--------Max Pool--------
def max_pool(input_2d, width, height,stride):
# Just like 'conv2d()' above, max_pool() works with 4D arrays.
# [batch_size=1, width=given, height=given, channels=1]
input_3d = tf.expand_dims(input_2d, 0)
input_4d = tf.expand_dims(input_3d, 3)
# Perform the max pooling with strides = [1,1,1,1]
# If we wanted to increase the stride on our data dimension, say by
# a factor of '2', we put strides = [1, 2, 2, 1]
pool_output = tf.nn.max_pool(input_4d, ksize=[1, height, width, 1],
strides=[1, stride, stride, 1],
padding='VALID')
# Get rid of unnecessary dimensions
pool_output_2d = tf.squeeze(pool_output)
return(pool_output_2d)
# Create Max-Pool Layer
my_maxpool_output = max_pool(my_activation_output,
width=maxpool_size, height=maxpool_size,stride=maxpool_stride_size)
#--------Fully Connected--------
def fully_connected(input_layer, num_outputs):
# In order to connect our whole W byH 2d array, we first flatten it out to
# a W times H 1D array.
flat_input = tf.reshape(input_layer, [-1])
# We then find out how long it is, and create an array for the shape of
# the multiplication weight = (WxH) by (num_outputs)
weight_shape = tf.squeeze(tf.stack([tf.shape(flat_input),[num_outputs]]))
# Initialize the weight
weight = tf.random_normal(weight_shape, stddev=0.1)
# Initialize the bias
bias = tf.random_normal(shape=[num_outputs])
# Now make the flat 1D array into a 2D array for multiplication
input_2d = tf.expand_dims(flat_input, 0)
# Multiply and add the bias
full_output = tf.add(tf.matmul(input_2d, weight), bias)
# Get rid of extra dimension
full_output_2d = tf.squeeze(full_output)
return(full_output_2d)
# Create Fully Connected Layer
my_full_output = fully_connected(my_maxpool_output, 5)
# Run graph
# Initialize Variables
init = tf.global_variables_initializer()
sess.run(init)
feed_dict = {x_input_2d: data_2d}
print('>>>> 2D Data <<<<')
# Convolution Output
print('Input = %s array' % (x_input_2d.shape.as_list()))
print('%s Convolution, stride size = [%d, %d] , results in the %s array' %
(my_filter.get_shape().as_list()[:2],conv_stride_size,conv_stride_size,my_convolution_output.shape.as_list()))
print(sess.run(my_convolution_output, feed_dict=feed_dict))
# Activation Output
print('\nInput = the above %s array' % (my_convolution_output.shape.as_list()))
print('ReLU element wise returns the %s array' % (my_activation_output.shape.as_list()))
print(sess.run(my_activation_output, feed_dict=feed_dict))
# Max Pool Output
print('\nInput = the above %s array' % (my_activation_output.shape.as_list()))
print('MaxPool, stride size = [%d, %d], results in %s array' %
(maxpool_stride_size,maxpool_stride_size,my_maxpool_output.shape.as_list()))
print(sess.run(my_maxpool_output, feed_dict=feed_dict))
# Fully Connected Output
print('\nInput = the above %s array' % (my_maxpool_output.shape.as_list()))
print('Fully connected layer on all %d rows results in %s outputs:' %
(my_maxpool_output.shape.as_list()[0],my_full_output.shape.as_list()[0]))
print(sess.run(my_full_output, feed_dict=feed_dict))
tested; G
| 0.721743 | 0.963265 |
```
import os
import sys
import warnings
warnings.filterwarnings('ignore')
# fichier utils.py
sys.path.append(os.path.dirname(os.path.dirname(os.path.abspath('utils.py'))))
import utils
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.model_selection import train_test_split
from collections import Counter
import matplotlib.pyplot as plt
from bs4 import BeautifulSoup
import numpy as np
import pandas as pd
import pickle
import random
import string
import json
%config InlineBackend.figure_format = 'retina'
%matplotlib inline
df = utils.load_data() # nettoyage du dataset
```
## Un peu d'exploration de l'anamyse des données
```
# distribution des cyber trolls vs non-cyber trolls
counter = Counter(df.label)
plt.title('Distribution des CyberTrolls - set d\'entrainement')
plt.bar(list(counter.keys())[0], list(counter.values())[0], align='center', color='g', label='Non Cyber-Agressive')
plt.bar(list(counter.keys())[1], list(counter.values())[1], align='center', color='r', label='Cyber-Agressive')
plt.xticks(list(set(df.label)))
plt.legend()
plt.show()
# Quelques commentaires
utils.sample_data(df, n=10)
# Mots plus courant par étiquette
trolls = Counter(' '.join(list(df[df.label == 1].text)).split())
non_trolls = Counter(' '.join(list(df[df.label == 0].text)).split())
print('Cyber Trolls')
print(*trolls.most_common()[:5], sep='\n')
print('\nNon Cyber Trolls')
print(*non_trolls.most_common()[:5], sep='\n')
## Sauvegarde du vectorizer dans ./model_assets
vectorizer=utils.build_encoder(df.text, True, True)
utils.persist_vectorizer(vectorizer, 'test_v.0.0')
```
## extracteurs de fonctions de texte
```
# encodage bag of words
enc = utils.build_encoder(df.text, count_vectorizer=True)
count_vectorized = enc.fit_transform(df.text).toarray()
# encodage tf-idf
enc = utils.build_encoder(df.text, tf_idf=True)
tf_idf = enc.fit_transform(df.text).toarray()
print(tf_idf.shape == count_vectorized.shape)
```
# Model dev
```
X_train, X_test, y_train, y_test = train_test_split(count_vectorized, df.label)
# Etiquettes des sets d'entrainements et de test
fig, axs = plt.subplots(1, 2, figsize=(15,5))
train_count = Counter(y_train)
axs[0].set_title('Distribution des CyberTrolls - set d\'entrainement')
axs[0].bar(list(train_count.keys())[0], list(train_count.values())[0], align='center', color='g', label='Non Cyber-Agressive')
axs[0].bar(list(train_count.keys())[1], list(train_count.values())[1], align='center', color='r', label='Cyber-Agressive')
axs[0].set_xticks(list(set(y_train)))
axs[0].legend()
test_count = Counter(y_test)
axs[1].set_title('Distribution de CyberTrolls - set de test')
axs[1].bar(list(test_count.keys())[0], list(test_count.values())[0], align='center', color='g', label='Non Cyber-Agressive')
axs[1].bar(list(test_count.keys())[1], list(test_count.values())[1], align='center', color='r', label='Cyber-Agressive')
axs[1].set_xticks(list(set(y_test)))
axs[1].legend()
plt.show()
from BaseModel import SVM
# svc params
params = {'C': np.logspace(-5, 5, 5)}
data = {'X_train': X_train, 'X_test': X_test, 'y_train': y_train, 'y_test': y_test}
clf = SVM(description='dev')
clf.train(data=data, **params)
clf.display_results(data)
```
## Sauvegarde du model et du vectorizer
```
utils.persist_model(clf, 'test_v.0')
utils.persist_model(clf, 'test_v.0')
data['X_train'].shape
```
|
github_jupyter
|
import os
import sys
import warnings
warnings.filterwarnings('ignore')
# fichier utils.py
sys.path.append(os.path.dirname(os.path.dirname(os.path.abspath('utils.py'))))
import utils
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.model_selection import train_test_split
from collections import Counter
import matplotlib.pyplot as plt
from bs4 import BeautifulSoup
import numpy as np
import pandas as pd
import pickle
import random
import string
import json
%config InlineBackend.figure_format = 'retina'
%matplotlib inline
df = utils.load_data() # nettoyage du dataset
# distribution des cyber trolls vs non-cyber trolls
counter = Counter(df.label)
plt.title('Distribution des CyberTrolls - set d\'entrainement')
plt.bar(list(counter.keys())[0], list(counter.values())[0], align='center', color='g', label='Non Cyber-Agressive')
plt.bar(list(counter.keys())[1], list(counter.values())[1], align='center', color='r', label='Cyber-Agressive')
plt.xticks(list(set(df.label)))
plt.legend()
plt.show()
# Quelques commentaires
utils.sample_data(df, n=10)
# Mots plus courant par étiquette
trolls = Counter(' '.join(list(df[df.label == 1].text)).split())
non_trolls = Counter(' '.join(list(df[df.label == 0].text)).split())
print('Cyber Trolls')
print(*trolls.most_common()[:5], sep='\n')
print('\nNon Cyber Trolls')
print(*non_trolls.most_common()[:5], sep='\n')
## Sauvegarde du vectorizer dans ./model_assets
vectorizer=utils.build_encoder(df.text, True, True)
utils.persist_vectorizer(vectorizer, 'test_v.0.0')
# encodage bag of words
enc = utils.build_encoder(df.text, count_vectorizer=True)
count_vectorized = enc.fit_transform(df.text).toarray()
# encodage tf-idf
enc = utils.build_encoder(df.text, tf_idf=True)
tf_idf = enc.fit_transform(df.text).toarray()
print(tf_idf.shape == count_vectorized.shape)
X_train, X_test, y_train, y_test = train_test_split(count_vectorized, df.label)
# Etiquettes des sets d'entrainements et de test
fig, axs = plt.subplots(1, 2, figsize=(15,5))
train_count = Counter(y_train)
axs[0].set_title('Distribution des CyberTrolls - set d\'entrainement')
axs[0].bar(list(train_count.keys())[0], list(train_count.values())[0], align='center', color='g', label='Non Cyber-Agressive')
axs[0].bar(list(train_count.keys())[1], list(train_count.values())[1], align='center', color='r', label='Cyber-Agressive')
axs[0].set_xticks(list(set(y_train)))
axs[0].legend()
test_count = Counter(y_test)
axs[1].set_title('Distribution de CyberTrolls - set de test')
axs[1].bar(list(test_count.keys())[0], list(test_count.values())[0], align='center', color='g', label='Non Cyber-Agressive')
axs[1].bar(list(test_count.keys())[1], list(test_count.values())[1], align='center', color='r', label='Cyber-Agressive')
axs[1].set_xticks(list(set(y_test)))
axs[1].legend()
plt.show()
from BaseModel import SVM
# svc params
params = {'C': np.logspace(-5, 5, 5)}
data = {'X_train': X_train, 'X_test': X_test, 'y_train': y_train, 'y_test': y_test}
clf = SVM(description='dev')
clf.train(data=data, **params)
clf.display_results(data)
utils.persist_model(clf, 'test_v.0')
utils.persist_model(clf, 'test_v.0')
data['X_train'].shape
| 0.226099 | 0.593727 |
```
!pip install keras
!pip install tensorflow
import pandas as pd
import re
import pickle
from tqdm import tqdm_notebook
from bs4 import BeautifulSoup
import matplotlib.pyplot as plt
from sklearn.feature_extraction.text import CountVectorizer,TfidfTransformer
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.naive_bayes import MultinomialNB
from sklearn.linear_model import SGDClassifier
from sklearn import svm
from nltk.stem.porter import PorterStemmer
from nltk.corpus import stopwords
from nltk.tokenize import word_tokenize
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
from sklearn.metrics import precision_score
from sklearn.metrics import recall_score
import os
import tensorflow
os.environ['KERAS_BACKEND'] = 'tensorflow'
import keras
from keras.models import Sequential
from keras.layers import Dense, Embedding, LSTM, SpatialDropout1D
from keras.preprocessing.text import Tokenizer
from keras.preprocessing.sequence import pad_sequences
import pandas as pd
from termcolor import colored
```
## Read data from sentiment 140
```
sentiment = pd.read_csv("sentiment140.csv", encoding = "ISO-8859-1")
sentiment.columns=['polarity','tweetid','date','nq','author','tweet']
sentiment = sentiment.drop(["tweetid","date","nq","author"],axis=1)
```
## Functions for cleaning the tweets
```
stop_words = set(stopwords.words('english'))
stop_words.remove("not")
def cleanText(s):
bad_chars = [';', ':', '!', '*', '(' , ')', '&','[',']','.','?','{','}',',']
non_ascii = "".join(i for i in s if ord(i)< 128)
html_decoded_string = BeautifulSoup(non_ascii, "lxml")
string = html_decoded_string.string
non_name = " ".join((filter(lambda x:x[0]!='@', string.split())))
non_badchars = ''.join(filter(lambda i: i not in bad_chars, non_name))
non_links = re.sub(r"http\S+", "", non_badchars)
non_websites = re.sub(r"www.[^ ]+","",non_links)
non_numbers = re.sub(r"[0-9]+","",non_websites)
clean = stopwords_stem(non_numbers)
clean = decontracted(clean)
return clean
def decontracted(phrase):
# specific
phrase = re.sub(r"won\'t", "will not", phrase)
phrase = re.sub(r"can\'t", "can not", phrase)
# general
phrase = re.sub(r"n\'t", " not", phrase)
phrase = re.sub(r"\'re", " are", phrase)
phrase = re.sub(r"\'s", " is", phrase)
phrase = re.sub(r"\'d", " would", phrase)
phrase = re.sub(r"\'ll", " will", phrase)
phrase = re.sub(r"\'t", " not", phrase)
phrase = re.sub(r"\'ve", " have", phrase)
phrase = re.sub(r"\'m", " am", phrase)
return phrase
def stopwords_stem(document):
words = word_tokenize(document)
words = removeStopwords(words)
words = stemWords(words)
result = ""
for word in words:
result += word + " "
return result
def removeStopwords(document):
result = []
for word in document:
if word not in stop_words:
result.append(word)
return result
def stemWords(document):
"""Stem words in list of tokenized words"""
stemmer = PorterStemmer()
result = []
for word in document:
stem = stemmer.stem(word)
result.append(stem)
return result
```
## Cleaning tweets from dataframe and transform to list
#### Can skip if load from disk (below)
```
tweets = sentiment.tweet
print("Cleaning Tweets: ")
tweets = [cleanText(t) for t in tqdm_notebook(tweets)]
polarity = sentiment.polarity
```
### Save the processed tweets for later use
```
with open('tweets.pickle', 'wb') as f:
pickle.dump(tweets, f)
with open('polarity.pickle', 'wb') as f:
pickle.dump(polarity, f)
```
### Load from saved files
```
with open('tweets.pickle', 'rb') as f:
tweets = pickle.load(f)
with open('polarity.pickle', 'rb') as f:
polarity = pickle.load(f)
for i in range(10):
print(sentiment.tweet[i])
print(tweets[i]+"\n")
sentiment.polarity.value_counts().plot(kind="bar",subplots="True")
plt.show()
```
# TrainTestSplitToCSV
```
# Train test split
print(colored("Splitting train and test dataset into 80:20", "yellow"))
X_train, X_test, y_train, y_test = train_test_split(tweets, polarity, test_size = 0.33, random_state = 123)
train_dataset = pd.DataFrame({
'Tweet': X_train,
'Sentiment': y_train
})
print(colored("Train data distribution:", "yellow"))
print(train_dataset['Sentiment'].value_counts())
test_dataset = pd.DataFrame({
'Tweet': X_test,
'Sentiment': y_test
})
print(colored("Test data distribution:", "yellow"))
print(test_dataset['Sentiment'].value_counts())
print(colored("Split complete", "yellow"))
# Save train data
print(colored("Saving train data", "yellow"))
train_dataset.to_csv('C:/Users/Phu Wai Paing/Desktop/Course Books/Text and Web Mining/TrainTestSplitCSV/train.csv', index = False)
print(colored("Train data saved to TrainTestSplitCSV/train.csv", "green"))
# Save test data
print(colored("Saving test data", "yellow"))
test_dataset.to_csv('C:/Users/Phu Wai Paing/Desktop/Course Books/Text and Web Mining/TrainTestSplitCSV/test.csv', index = False)
print(colored("Test data saved to TrainTestSplitCSV/test.csv", "green"))
```
# LSTM
```
# Load data
print(colored("Loading train and test data", "yellow"))
train_data = pd.read_csv('C:/Users/Phu Wai Paing/Desktop/Course Books/Text and Web Mining/TrainTestSplitCSV/train.csv')
test_data = pd.read_csv('C:/Users/Phu Wai Paing/Desktop/Course Books/Text and Web Mining/TrainTestSplitCSV/test.csv')
print(colored("Data loaded", "yellow"))
# Tokenization
print(colored("Tokenizing and padding data", "yellow"))
tokenizer = Tokenizer(num_words = 2000, split = ' ')
tokenizer.fit_on_texts(train_data['Tweet'].astype(str).values)
train_tweets = tokenizer.texts_to_sequences(train_data['Tweet'].astype(str).values)
max_len = max([len(i) for i in train_tweets])
train_tweets = pad_sequences(train_tweets, maxlen = max_len)
test_tweets = tokenizer.texts_to_sequences(test_data['Tweet'].astype(str).values)
test_tweets = pad_sequences(test_tweets, maxlen = max_len)
print(colored("Tokenizing and padding complete", "yellow"))
# Building the model
print(colored("Creating the LSTM model", "yellow"))
model = Sequential()
model.add(Embedding(2000, 128, input_length = train_tweets.shape[1]))
model.add(SpatialDropout1D(0.4))
model.add(LSTM(256, dropout = 0.2))
model.add(Dense(2, activation = 'softmax'))
model.compile(loss = 'categorical_crossentropy', optimizer = 'adam', metrics = ['accuracy'])
model.summary()
# Training the model
print(colored("Training the LSTM model", "green"))
history = model.fit(train_tweets, pd.get_dummies(train_data['Sentiment']).values, epochs = 1, batch_size = 128, validation_split = 0.2)
print(colored(history, "green"))
# Testing the model
print(colored("Testing the LSTM model", "green"))
score, accuracy = model.evaluate(test_tweets, pd.get_dummies(test_data['Sentiment']).values, batch_size = 128)
print("Test accuracy: {}".format(accuracy))
```
### Count Vectorizer
```
print("Running Count Vectorizer: ")
count_vectorizer = CountVectorizer(binary=True)
train_vector = count_vectorizer.fit_transform(tweets)
print("Train Test Split: ")
X_train, X_test, y_train, y_test = train_test_split(train_vector, polarity, test_size=0.33, random_state=123)
print("\nTraining Naive Bayes:")
clf = MultinomialNB().fit(X_train, y_train)
print("Testing: ")
predicted = clf.predict(X_test)
print("Accuracy" , accuracy_score(y_test, predicted))
print("Precision" , precision_score(y_test, predicted, pos_label=4))
print("\nTraining SVM:")
svmclf = svm.LinearSVC().fit(X_train, y_train)
print("Testing: ")
predicted = svmclf.predict(X_test)
print("\nAccuracy" , accuracy_score(y_test, predicted))
print("Precision" , precision_score(y_test, predicted, pos_label=4))
```
### TF-IDF
```
print("Running TF-IDF Vectorizer: ")
tfidf_vectorizer = TfidfVectorizer()
train_vector = tfidf_vectorizer.fit_transform(tweets)
print("Train Test Split: ")
X_train, X_test, y_train, y_test = train_test_split(train_vector, polarity, test_size=0.33, random_state=123)
print("\nTraining Naive Bayes:")
nbclf = MultinomialNB().fit(X_train, y_train)
print("Testing: ")
predicted = nbclf.predict(X_test)
print("Accuracy" , accuracy_score(y_test, predicted))
print("Precision" , precision_score(y_test, predicted, pos_label=4))
print("\nTraining SVM:")
svmclf = svm.LinearSVC().fit(X_train, y_train)
print("Testing: ")
predicted = svmclf.predict(X_test)
print("\nAccuracy" , accuracy_score(y_test, predicted))
print("Precision" , precision_score(y_test, predicted, pos_label=4))
```
### TF-IDF N-gram
```
print("Running TF-IDF with N-gram Vectorizer")
ngram = (1,2)
tfidf_ngram_vectorizer = TfidfVectorizer(ngram_range = ngram)
train_vector = tfidf_ngram_vectorizer.fit_transform(tweets)
print("Train Test Split: ")
X_train, X_test, y_train, y_test = train_test_split(train_vector, polarity, test_size=0.33, random_state=123)
print("\nTraining Naive Bayes:")
ngclf = MultinomialNB().fit(X_train, y_train)
print("Testing: ")
predicted = ngclf.predict(X_test)
print("Accuracy" , accuracy_score(y_test, predicted))
print("Precision" , precision_score(y_test, predicted, pos_label=4))
print("\nTraining SVM:")
svmclf = svm.LinearSVC().fit(X_train, y_train)
print("Testing: ")
predicted = svmclf.predict(X_test)
print("\nAccuracy" , accuracy_score(y_test, predicted))
print("Precision" , precision_score(y_test, predicted, pos_label=4))
```
|
github_jupyter
|
!pip install keras
!pip install tensorflow
import pandas as pd
import re
import pickle
from tqdm import tqdm_notebook
from bs4 import BeautifulSoup
import matplotlib.pyplot as plt
from sklearn.feature_extraction.text import CountVectorizer,TfidfTransformer
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.naive_bayes import MultinomialNB
from sklearn.linear_model import SGDClassifier
from sklearn import svm
from nltk.stem.porter import PorterStemmer
from nltk.corpus import stopwords
from nltk.tokenize import word_tokenize
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
from sklearn.metrics import precision_score
from sklearn.metrics import recall_score
import os
import tensorflow
os.environ['KERAS_BACKEND'] = 'tensorflow'
import keras
from keras.models import Sequential
from keras.layers import Dense, Embedding, LSTM, SpatialDropout1D
from keras.preprocessing.text import Tokenizer
from keras.preprocessing.sequence import pad_sequences
import pandas as pd
from termcolor import colored
sentiment = pd.read_csv("sentiment140.csv", encoding = "ISO-8859-1")
sentiment.columns=['polarity','tweetid','date','nq','author','tweet']
sentiment = sentiment.drop(["tweetid","date","nq","author"],axis=1)
stop_words = set(stopwords.words('english'))
stop_words.remove("not")
def cleanText(s):
bad_chars = [';', ':', '!', '*', '(' , ')', '&','[',']','.','?','{','}',',']
non_ascii = "".join(i for i in s if ord(i)< 128)
html_decoded_string = BeautifulSoup(non_ascii, "lxml")
string = html_decoded_string.string
non_name = " ".join((filter(lambda x:x[0]!='@', string.split())))
non_badchars = ''.join(filter(lambda i: i not in bad_chars, non_name))
non_links = re.sub(r"http\S+", "", non_badchars)
non_websites = re.sub(r"www.[^ ]+","",non_links)
non_numbers = re.sub(r"[0-9]+","",non_websites)
clean = stopwords_stem(non_numbers)
clean = decontracted(clean)
return clean
def decontracted(phrase):
# specific
phrase = re.sub(r"won\'t", "will not", phrase)
phrase = re.sub(r"can\'t", "can not", phrase)
# general
phrase = re.sub(r"n\'t", " not", phrase)
phrase = re.sub(r"\'re", " are", phrase)
phrase = re.sub(r"\'s", " is", phrase)
phrase = re.sub(r"\'d", " would", phrase)
phrase = re.sub(r"\'ll", " will", phrase)
phrase = re.sub(r"\'t", " not", phrase)
phrase = re.sub(r"\'ve", " have", phrase)
phrase = re.sub(r"\'m", " am", phrase)
return phrase
def stopwords_stem(document):
words = word_tokenize(document)
words = removeStopwords(words)
words = stemWords(words)
result = ""
for word in words:
result += word + " "
return result
def removeStopwords(document):
result = []
for word in document:
if word not in stop_words:
result.append(word)
return result
def stemWords(document):
"""Stem words in list of tokenized words"""
stemmer = PorterStemmer()
result = []
for word in document:
stem = stemmer.stem(word)
result.append(stem)
return result
tweets = sentiment.tweet
print("Cleaning Tweets: ")
tweets = [cleanText(t) for t in tqdm_notebook(tweets)]
polarity = sentiment.polarity
with open('tweets.pickle', 'wb') as f:
pickle.dump(tweets, f)
with open('polarity.pickle', 'wb') as f:
pickle.dump(polarity, f)
with open('tweets.pickle', 'rb') as f:
tweets = pickle.load(f)
with open('polarity.pickle', 'rb') as f:
polarity = pickle.load(f)
for i in range(10):
print(sentiment.tweet[i])
print(tweets[i]+"\n")
sentiment.polarity.value_counts().plot(kind="bar",subplots="True")
plt.show()
# Train test split
print(colored("Splitting train and test dataset into 80:20", "yellow"))
X_train, X_test, y_train, y_test = train_test_split(tweets, polarity, test_size = 0.33, random_state = 123)
train_dataset = pd.DataFrame({
'Tweet': X_train,
'Sentiment': y_train
})
print(colored("Train data distribution:", "yellow"))
print(train_dataset['Sentiment'].value_counts())
test_dataset = pd.DataFrame({
'Tweet': X_test,
'Sentiment': y_test
})
print(colored("Test data distribution:", "yellow"))
print(test_dataset['Sentiment'].value_counts())
print(colored("Split complete", "yellow"))
# Save train data
print(colored("Saving train data", "yellow"))
train_dataset.to_csv('C:/Users/Phu Wai Paing/Desktop/Course Books/Text and Web Mining/TrainTestSplitCSV/train.csv', index = False)
print(colored("Train data saved to TrainTestSplitCSV/train.csv", "green"))
# Save test data
print(colored("Saving test data", "yellow"))
test_dataset.to_csv('C:/Users/Phu Wai Paing/Desktop/Course Books/Text and Web Mining/TrainTestSplitCSV/test.csv', index = False)
print(colored("Test data saved to TrainTestSplitCSV/test.csv", "green"))
# Load data
print(colored("Loading train and test data", "yellow"))
train_data = pd.read_csv('C:/Users/Phu Wai Paing/Desktop/Course Books/Text and Web Mining/TrainTestSplitCSV/train.csv')
test_data = pd.read_csv('C:/Users/Phu Wai Paing/Desktop/Course Books/Text and Web Mining/TrainTestSplitCSV/test.csv')
print(colored("Data loaded", "yellow"))
# Tokenization
print(colored("Tokenizing and padding data", "yellow"))
tokenizer = Tokenizer(num_words = 2000, split = ' ')
tokenizer.fit_on_texts(train_data['Tweet'].astype(str).values)
train_tweets = tokenizer.texts_to_sequences(train_data['Tweet'].astype(str).values)
max_len = max([len(i) for i in train_tweets])
train_tweets = pad_sequences(train_tweets, maxlen = max_len)
test_tweets = tokenizer.texts_to_sequences(test_data['Tweet'].astype(str).values)
test_tweets = pad_sequences(test_tweets, maxlen = max_len)
print(colored("Tokenizing and padding complete", "yellow"))
# Building the model
print(colored("Creating the LSTM model", "yellow"))
model = Sequential()
model.add(Embedding(2000, 128, input_length = train_tweets.shape[1]))
model.add(SpatialDropout1D(0.4))
model.add(LSTM(256, dropout = 0.2))
model.add(Dense(2, activation = 'softmax'))
model.compile(loss = 'categorical_crossentropy', optimizer = 'adam', metrics = ['accuracy'])
model.summary()
# Training the model
print(colored("Training the LSTM model", "green"))
history = model.fit(train_tweets, pd.get_dummies(train_data['Sentiment']).values, epochs = 1, batch_size = 128, validation_split = 0.2)
print(colored(history, "green"))
# Testing the model
print(colored("Testing the LSTM model", "green"))
score, accuracy = model.evaluate(test_tweets, pd.get_dummies(test_data['Sentiment']).values, batch_size = 128)
print("Test accuracy: {}".format(accuracy))
print("Running Count Vectorizer: ")
count_vectorizer = CountVectorizer(binary=True)
train_vector = count_vectorizer.fit_transform(tweets)
print("Train Test Split: ")
X_train, X_test, y_train, y_test = train_test_split(train_vector, polarity, test_size=0.33, random_state=123)
print("\nTraining Naive Bayes:")
clf = MultinomialNB().fit(X_train, y_train)
print("Testing: ")
predicted = clf.predict(X_test)
print("Accuracy" , accuracy_score(y_test, predicted))
print("Precision" , precision_score(y_test, predicted, pos_label=4))
print("\nTraining SVM:")
svmclf = svm.LinearSVC().fit(X_train, y_train)
print("Testing: ")
predicted = svmclf.predict(X_test)
print("\nAccuracy" , accuracy_score(y_test, predicted))
print("Precision" , precision_score(y_test, predicted, pos_label=4))
print("Running TF-IDF Vectorizer: ")
tfidf_vectorizer = TfidfVectorizer()
train_vector = tfidf_vectorizer.fit_transform(tweets)
print("Train Test Split: ")
X_train, X_test, y_train, y_test = train_test_split(train_vector, polarity, test_size=0.33, random_state=123)
print("\nTraining Naive Bayes:")
nbclf = MultinomialNB().fit(X_train, y_train)
print("Testing: ")
predicted = nbclf.predict(X_test)
print("Accuracy" , accuracy_score(y_test, predicted))
print("Precision" , precision_score(y_test, predicted, pos_label=4))
print("\nTraining SVM:")
svmclf = svm.LinearSVC().fit(X_train, y_train)
print("Testing: ")
predicted = svmclf.predict(X_test)
print("\nAccuracy" , accuracy_score(y_test, predicted))
print("Precision" , precision_score(y_test, predicted, pos_label=4))
print("Running TF-IDF with N-gram Vectorizer")
ngram = (1,2)
tfidf_ngram_vectorizer = TfidfVectorizer(ngram_range = ngram)
train_vector = tfidf_ngram_vectorizer.fit_transform(tweets)
print("Train Test Split: ")
X_train, X_test, y_train, y_test = train_test_split(train_vector, polarity, test_size=0.33, random_state=123)
print("\nTraining Naive Bayes:")
ngclf = MultinomialNB().fit(X_train, y_train)
print("Testing: ")
predicted = ngclf.predict(X_test)
print("Accuracy" , accuracy_score(y_test, predicted))
print("Precision" , precision_score(y_test, predicted, pos_label=4))
print("\nTraining SVM:")
svmclf = svm.LinearSVC().fit(X_train, y_train)
print("Testing: ")
predicted = svmclf.predict(X_test)
print("\nAccuracy" , accuracy_score(y_test, predicted))
print("Precision" , precision_score(y_test, predicted, pos_label=4))
| 0.457379 | 0.431165 |
# Name
Data preparation by deleting a cluster in Cloud Dataproc
# Label
Cloud Dataproc, cluster, GCP, Cloud Storage, Kubeflow, Pipeline
# Summary
A Kubeflow Pipeline component to delete a cluster in Cloud Dataproc.
## Intended use
Use this component at the start of a Kubeflow Pipeline to delete a temporary Cloud Dataproc cluster to run Cloud Dataproc jobs as steps in the pipeline. This component is usually used with an [exit handler](https://github.com/kubeflow/pipelines/blob/master/samples/basic/exit_handler.py) to run at the end of a pipeline.
## Runtime arguments
| Argument | Description | Optional | Data type | Accepted values | Default |
|----------|-------------|----------|-----------|-----------------|---------|
| project_id | The Google Cloud Platform (GCP) project ID that the cluster belongs to. | No | GCPProjectID | | |
| region | The Cloud Dataproc region in which to handle the request. | No | GCPRegion | | |
| name | The name of the cluster to delete. | No | String | | |
| wait_interval | The number of seconds to pause between polling the operation. | Yes | Integer | | 30 |
## Cautions & requirements
To use the component, you must:
* Set up a GCP project by following this [guide](https://cloud.google.com/dataproc/docs/guides/setup-project).
* Run the component under a secret [Kubeflow user service account](https://www.kubeflow.org/docs/started/getting-started-gke/#gcp-service-accounts) in a Kubeflow cluster. For example:
```
component_op(...).apply(gcp.use_gcp_secret('user-gcp-sa'))
```
* Grant the Kubeflow user service account the role `roles/dataproc.editor` on the project.
## Detailed description
This component deletes a Dataproc cluster by using [Dataproc delete cluster REST API](https://cloud.google.com/dataproc/docs/reference/rest/v1/projects.regions.clusters/delete).
Follow these steps to use the component in a pipeline:
1. Install the Kubeflow Pipeline SDK:
```
%%capture --no-stderr
KFP_PACKAGE = 'https://storage.googleapis.com/ml-pipeline/release/0.1.14/kfp.tar.gz'
!pip3 install $KFP_PACKAGE --upgrade
```
2. Load the component using KFP SDK
```
import kfp.components as comp
dataproc_delete_cluster_op = comp.load_component_from_url(
'https://raw.githubusercontent.com/kubeflow/pipelines/d0aa15dfb3ff618e8cd1b03f86804ec4307fd9c2/components/gcp/dataproc/delete_cluster/component.yaml')
help(dataproc_delete_cluster_op)
```
### Sample
Note: The following sample code works in an IPython notebook or directly in Python code. See the sample code below to learn how to execute the template.
#### Prerequisites
[Create a Dataproc cluster](https://cloud.google.com/dataproc/docs/guides/create-cluster) before running the sample code.
#### Set sample parameters
```
PROJECT_ID = '<Please put your project ID here>'
CLUSTER_NAME = '<Please put your existing cluster name here>'
REGION = 'us-central1'
EXPERIMENT_NAME = 'Dataproc - Delete Cluster'
```
#### Example pipeline that uses the component
```
import kfp.dsl as dsl
import kfp.gcp as gcp
import json
@dsl.pipeline(
name='Dataproc delete cluster pipeline',
description='Dataproc delete cluster pipeline'
)
def dataproc_delete_cluster_pipeline(
project_id = PROJECT_ID,
region = REGION,
name = CLUSTER_NAME
):
dataproc_delete_cluster_op(
project_id=project_id,
region=region,
name=name).apply(gcp.use_gcp_secret('user-gcp-sa'))
```
#### Compile the pipeline
```
pipeline_func = dataproc_delete_cluster_pipeline
pipeline_filename = pipeline_func.__name__ + '.zip'
import kfp.compiler as compiler
compiler.Compiler().compile(pipeline_func, pipeline_filename)
```
#### Submit the pipeline for execution
```
#Specify pipeline argument values
arguments = {}
#Get or create an experiment and submit a pipeline run
import kfp
client = kfp.Client()
experiment = client.create_experiment(EXPERIMENT_NAME)
#Submit a pipeline run
run_name = pipeline_func.__name__ + ' run'
run_result = client.run_pipeline(experiment.id, run_name, pipeline_filename, arguments)
```
## References
* [Component Python code](https://github.com/kubeflow/pipelines/blob/master/component_sdk/python/kfp_component/google/dataproc/_delete_cluster.py)
* [Component Docker file](https://github.com/kubeflow/pipelines/blob/master/components/gcp/container/Dockerfile)
* [Sample notebook](https://github.com/kubeflow/pipelines/blob/master/components/gcp/dataproc/delete_cluster/sample.ipynb)
* [Dataproc delete cluster REST API](https://cloud.google.com/dataproc/docs/reference/rest/v1/projects.regions.clusters/delete)
## License
By deploying or using this software you agree to comply with the [AI Hub Terms of Service](https://aihub.cloud.google.com/u/0/aihub-tos) and the [Google APIs Terms of Service](https://developers.google.com/terms/). To the extent of a direct conflict of terms, the AI Hub Terms of Service will control.
|
github_jupyter
|
component_op(...).apply(gcp.use_gcp_secret('user-gcp-sa'))
```
* Grant the Kubeflow user service account the role `roles/dataproc.editor` on the project.
## Detailed description
This component deletes a Dataproc cluster by using [Dataproc delete cluster REST API](https://cloud.google.com/dataproc/docs/reference/rest/v1/projects.regions.clusters/delete).
Follow these steps to use the component in a pipeline:
1. Install the Kubeflow Pipeline SDK:
2. Load the component using KFP SDK
### Sample
Note: The following sample code works in an IPython notebook or directly in Python code. See the sample code below to learn how to execute the template.
#### Prerequisites
[Create a Dataproc cluster](https://cloud.google.com/dataproc/docs/guides/create-cluster) before running the sample code.
#### Set sample parameters
#### Example pipeline that uses the component
#### Compile the pipeline
#### Submit the pipeline for execution
| 0.826362 | 0.945951 |
# Iris Data Set Analysis with Tensorflow Estimators
Estimators from tensorflow is much easier to use, but we sacrifice some level of customization of the model.
```
import pandas as pd
```
## Get the Data
```
df = pd.read_csv('mk028-project_iris_data_analysis_with_tensorflow_and_estimators/iris.csv')
df.head()
```
In tensorflow column names cannot have spaces or special characters between them.
```
df.columns = ['sepal_length',
'sepal_width',
'petal_length',
'petal_width',
'target']
df.head()
```
`target` column must be integer for tensorflow, because it is binary type.
```
X = df.drop('target',
axis=1)
y = df['target'].apply(int)
y.head()
```
## Train Test Split
```
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X,
y,
test_size=0.3)
```
## Estimators
```
import tensorflow as tf
```
### Feature Columns
Create `feat_cols` for the tensorflow estimator.
```
feat_cols = []
[feat_cols.append(tf.feature_column.numeric_column(col)) for col in X.columns]
feat_cols
```
### Create the DNN(Deep Neural Network) Estimator
`hidden_units` define the number of neurons for each hidden neural layer.
```
classifier = tf.estimator.DNNClassifier(hidden_units=[10, 20, 10],
n_classes=3,
feature_columns=feat_cols)
```
### Input Function
Create input functions for training data. `num_epochs` means maximum number of repeats.
```
tr_input_func = tf.estimator.inputs.pandas_input_fn(x=X_train,
y=y_train,
batch_size=10,
num_epochs=5,
shuffle=True)
```
Now we train the `classifier` with the input function.
```
classifier.train(input_fn=tr_input_func,
steps=50)
```
## Model Evaluation
Now we create input functions for test(prediction) data.
```
pred_input_func = tf.estimator.inputs.pandas_input_fn(x=X_test,
batch_size=len(X_test),
shuffle=False)
```
Now we predict with the trained classifier by using prediction input function.
```
predictions = list(classifier.predict(input_fn=pred_input_func))
predictions[:4]
final_predictions = []
[final_predictions.append(p["class_ids"][0]) for p in predictions]
final_predictions[:5]
```
Now create a classification report and a Confusion Matrix.
```
from sklearn.metrics import classification_report, confusion_matrix, accuracy_score
print(classification_report(y_test, final_predictions))
print(confusion_matrix(y_test, final_predictions))
print(accuracy_score(y_test, final_predictions))
```
|
github_jupyter
|
import pandas as pd
df = pd.read_csv('mk028-project_iris_data_analysis_with_tensorflow_and_estimators/iris.csv')
df.head()
df.columns = ['sepal_length',
'sepal_width',
'petal_length',
'petal_width',
'target']
df.head()
X = df.drop('target',
axis=1)
y = df['target'].apply(int)
y.head()
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X,
y,
test_size=0.3)
import tensorflow as tf
feat_cols = []
[feat_cols.append(tf.feature_column.numeric_column(col)) for col in X.columns]
feat_cols
classifier = tf.estimator.DNNClassifier(hidden_units=[10, 20, 10],
n_classes=3,
feature_columns=feat_cols)
tr_input_func = tf.estimator.inputs.pandas_input_fn(x=X_train,
y=y_train,
batch_size=10,
num_epochs=5,
shuffle=True)
classifier.train(input_fn=tr_input_func,
steps=50)
pred_input_func = tf.estimator.inputs.pandas_input_fn(x=X_test,
batch_size=len(X_test),
shuffle=False)
predictions = list(classifier.predict(input_fn=pred_input_func))
predictions[:4]
final_predictions = []
[final_predictions.append(p["class_ids"][0]) for p in predictions]
final_predictions[:5]
from sklearn.metrics import classification_report, confusion_matrix, accuracy_score
print(classification_report(y_test, final_predictions))
print(confusion_matrix(y_test, final_predictions))
print(accuracy_score(y_test, final_predictions))
| 0.559531 | 0.961389 |
```
import numpy as np
import scipy.stats as spstats
from scipy import signal
import pickle
from multiprocessing import Pool
import multiprocessing
import scipy.sparse as sparse
import seaborn as sns
from potentials import IndependentPotential
from baselines import GenerateSigma,construct_ESVM_kernel,set_function
from optimize import Run_eval_test,optimize_parallel_new
from samplers import MCMC_sampler,Generate_train
from utils import *
N_train = 1*10**4 # Number of samples on which we optimize
N_test = 1*10**4 # Number of samples
n_traj_train = 1
n_traj_test = 100 # Number of independent MCMC trajectories for test
f_type = "cos_sum"
sampler = {"sampler":"3rd_poly","burn_type":"full","main_type":"full"}# Sampling method
d = 10
typ = sampler["sampler"]
#mu = np.array([0.0],dtype = float)
#Sigma = 1.0
mu = np.zeros(d, dtype = float)
Sigma = np.eye(d)
#Sigma = GenerateSigma(d,rand_seed = 777,eps = 0.1) #covariation matrix
"""
params_distr = {
"mu":mu,
"Sigma":Sigma,
"d":d
}
params_distr = {
"mu":0.0,
"lambda":1.0,
"d":d
}
"""
params_distr = {
"b":2.0,
"d":d
}
Cur_pot = IndependentPotential(typ,params_distr)
#Cur_pot = GausMixtureSame(Sigma,mu,p)
#Cur_pot = GausMixtureIdent(mu,p)
rand_seed = 777
traj,traj_grad = Cur_pot.sample(rand_seed,N_train)
traj = np.expand_dims(traj,axis=0)
traj_grad = np.expand_dims(traj_grad,axis=0)
print(traj.shape)
print(traj_grad.shape)
inds_arr = np.array([0]) # Taking the second index (not intercept)
params = None
f_vals = set_function(f_type,traj,inds_arr,params)
print(f_vals.shape)
W_train_spec = None
W_test_spec = None
degree = 5 #degree of the polynomial
opt_structure_train = {
"W":W_train_spec,
"n_restarts": 5, # Number of restarts during optimization,
"sigma": 3.0, # Deviation of starting points
"tol": 1e-6, # Tolerance (for the norm of gradient)
"alpha": 0.0, # Ridge penalty for 2nd order control functionals
"beta": 1e2 # smoothing parameter in the softmax
}
methods = ["EVM","LS","MAX"]
coef_dict_k_deg = optimize_parallel_new(degree,inds_arr,f_vals,traj,traj_grad,opt_structure_train,methods)
```
### Coefficients for given training methods
```
print(coef_dict_k_deg["EVM"])
print(coef_dict_k_deg["LS"])
print(coef_dict_k_deg["MAX"])
```
### Test
```
#Create a dictionary and put respective matrices into it
test_params = {
"W":W_test_spec,
"step":None,
"burn_in":None,
"n_test":N_test,
"dim":d
}
nbcores = multiprocessing.cpu_count()
trav = Pool(nbcores)
res = trav.starmap(Run_eval_test, [(i,degree,sampler,methods,inds_arr,Cur_pot,test_params,coef_dict_k_deg,params,f_type) for i in range (n_traj_test)])
trav.close()
methods_enh = ['Vanilla'] + methods
print(methods_enh)
ints_result = {key: [] for key in methods_enh}
vars_result = {key: [] for key in methods_enh}
for i in range(len(res)):
for j in range(len(methods_enh)):
ints_result[methods_enh[j]].append(res[i][0][methods_enh[j]][0])
vars_result[methods_enh[j]].append(res[i][1][methods_enh[j]][0])
for key in methods_enh:
ints_result[key] = np.asarray(ints_result[key])
vars_result[key] = np.asarray(vars_result[key])
```
### Results
```
print("Estimators")
for i in range(len(methods_enh)):
print(methods_enh[i])
print("mean: ",np.mean(ints_result[methods_enh[i]],axis=0))
print("std: ",np.std(ints_result[methods_enh[i]],axis=0))
print("max deviation: ",np.max(np.abs(ints_result[methods_enh[i]] - np.mean(ints_result[methods_enh[i]]))))
print("Variances")
for i in range(len(methods_enh)):
print(methods_enh[i])
print(np.mean(vars_result[methods_enh[i]],axis=0))
#save results
res_dict = {"int":ints_result,"var":vars_result}
np.save("Results/15_09/MC_Pareto_sum_cos_traj_1_d_10_beta_1e-1_train_5e2_test_1e4_deg_3.npy",res_dict)
```
### Plot results
```
var_ind = 0
title = ""
labels = ['Vanilla', 'EVM', 'LS','MAX']
# Box plot
data = [res_dict['int'][method][:,var_ind] for method in labels]
boxplot_ind(data, title, labels)
var_ind = 0
title = ""
labels = ['EVM','MAX']
data = [res_dict['int'][method][:,var_ind] for method in labels]
boxplot_ind(data, title, labels)
var_ind = 0 #Index to plot
title = ""
labels = ['ULA \nwith EVM','ULA\nwith ESVM']
data = [results[:,0,4,var_ind]-1.25,results[:,0,2,var_ind]-1.25]
boxplot_ind(data, title, labels)
vars_vanilla = results[:,1,0,:]
vars_esvm_1st = results[:,1,1,:]
vars_esvm_2nd = results[:,1,2,:]
vars_evm_1st = results[:,1,3,:]
vars_evm_2nd = results[:,1,4,:]
print("average VRF for 1st order EVM:",np.mean(vars_vanilla)/np.mean(vars_evm_1st))
print("average VRF for 2nd order EVM:",np.mean(vars_vanilla)/np.mean(vars_evm_2nd))
print("average VRF for 1st order ESVM:",np.mean(vars_vanilla)/np.mean(vars_esvm_1st))
print("average VRF for 2nd order ESVM:",np.mean(vars_vanilla)/np.mean(vars_esvm_2nd))
```
|
github_jupyter
|
import numpy as np
import scipy.stats as spstats
from scipy import signal
import pickle
from multiprocessing import Pool
import multiprocessing
import scipy.sparse as sparse
import seaborn as sns
from potentials import IndependentPotential
from baselines import GenerateSigma,construct_ESVM_kernel,set_function
from optimize import Run_eval_test,optimize_parallel_new
from samplers import MCMC_sampler,Generate_train
from utils import *
N_train = 1*10**4 # Number of samples on which we optimize
N_test = 1*10**4 # Number of samples
n_traj_train = 1
n_traj_test = 100 # Number of independent MCMC trajectories for test
f_type = "cos_sum"
sampler = {"sampler":"3rd_poly","burn_type":"full","main_type":"full"}# Sampling method
d = 10
typ = sampler["sampler"]
#mu = np.array([0.0],dtype = float)
#Sigma = 1.0
mu = np.zeros(d, dtype = float)
Sigma = np.eye(d)
#Sigma = GenerateSigma(d,rand_seed = 777,eps = 0.1) #covariation matrix
"""
params_distr = {
"mu":mu,
"Sigma":Sigma,
"d":d
}
params_distr = {
"mu":0.0,
"lambda":1.0,
"d":d
}
"""
params_distr = {
"b":2.0,
"d":d
}
Cur_pot = IndependentPotential(typ,params_distr)
#Cur_pot = GausMixtureSame(Sigma,mu,p)
#Cur_pot = GausMixtureIdent(mu,p)
rand_seed = 777
traj,traj_grad = Cur_pot.sample(rand_seed,N_train)
traj = np.expand_dims(traj,axis=0)
traj_grad = np.expand_dims(traj_grad,axis=0)
print(traj.shape)
print(traj_grad.shape)
inds_arr = np.array([0]) # Taking the second index (not intercept)
params = None
f_vals = set_function(f_type,traj,inds_arr,params)
print(f_vals.shape)
W_train_spec = None
W_test_spec = None
degree = 5 #degree of the polynomial
opt_structure_train = {
"W":W_train_spec,
"n_restarts": 5, # Number of restarts during optimization,
"sigma": 3.0, # Deviation of starting points
"tol": 1e-6, # Tolerance (for the norm of gradient)
"alpha": 0.0, # Ridge penalty for 2nd order control functionals
"beta": 1e2 # smoothing parameter in the softmax
}
methods = ["EVM","LS","MAX"]
coef_dict_k_deg = optimize_parallel_new(degree,inds_arr,f_vals,traj,traj_grad,opt_structure_train,methods)
print(coef_dict_k_deg["EVM"])
print(coef_dict_k_deg["LS"])
print(coef_dict_k_deg["MAX"])
#Create a dictionary and put respective matrices into it
test_params = {
"W":W_test_spec,
"step":None,
"burn_in":None,
"n_test":N_test,
"dim":d
}
nbcores = multiprocessing.cpu_count()
trav = Pool(nbcores)
res = trav.starmap(Run_eval_test, [(i,degree,sampler,methods,inds_arr,Cur_pot,test_params,coef_dict_k_deg,params,f_type) for i in range (n_traj_test)])
trav.close()
methods_enh = ['Vanilla'] + methods
print(methods_enh)
ints_result = {key: [] for key in methods_enh}
vars_result = {key: [] for key in methods_enh}
for i in range(len(res)):
for j in range(len(methods_enh)):
ints_result[methods_enh[j]].append(res[i][0][methods_enh[j]][0])
vars_result[methods_enh[j]].append(res[i][1][methods_enh[j]][0])
for key in methods_enh:
ints_result[key] = np.asarray(ints_result[key])
vars_result[key] = np.asarray(vars_result[key])
print("Estimators")
for i in range(len(methods_enh)):
print(methods_enh[i])
print("mean: ",np.mean(ints_result[methods_enh[i]],axis=0))
print("std: ",np.std(ints_result[methods_enh[i]],axis=0))
print("max deviation: ",np.max(np.abs(ints_result[methods_enh[i]] - np.mean(ints_result[methods_enh[i]]))))
print("Variances")
for i in range(len(methods_enh)):
print(methods_enh[i])
print(np.mean(vars_result[methods_enh[i]],axis=0))
#save results
res_dict = {"int":ints_result,"var":vars_result}
np.save("Results/15_09/MC_Pareto_sum_cos_traj_1_d_10_beta_1e-1_train_5e2_test_1e4_deg_3.npy",res_dict)
var_ind = 0
title = ""
labels = ['Vanilla', 'EVM', 'LS','MAX']
# Box plot
data = [res_dict['int'][method][:,var_ind] for method in labels]
boxplot_ind(data, title, labels)
var_ind = 0
title = ""
labels = ['EVM','MAX']
data = [res_dict['int'][method][:,var_ind] for method in labels]
boxplot_ind(data, title, labels)
var_ind = 0 #Index to plot
title = ""
labels = ['ULA \nwith EVM','ULA\nwith ESVM']
data = [results[:,0,4,var_ind]-1.25,results[:,0,2,var_ind]-1.25]
boxplot_ind(data, title, labels)
vars_vanilla = results[:,1,0,:]
vars_esvm_1st = results[:,1,1,:]
vars_esvm_2nd = results[:,1,2,:]
vars_evm_1st = results[:,1,3,:]
vars_evm_2nd = results[:,1,4,:]
print("average VRF for 1st order EVM:",np.mean(vars_vanilla)/np.mean(vars_evm_1st))
print("average VRF for 2nd order EVM:",np.mean(vars_vanilla)/np.mean(vars_evm_2nd))
print("average VRF for 1st order ESVM:",np.mean(vars_vanilla)/np.mean(vars_esvm_1st))
print("average VRF for 2nd order ESVM:",np.mean(vars_vanilla)/np.mean(vars_esvm_2nd))
| 0.382372 | 0.587233 |
### Bi-directional LSTM with max pooling
Author: Jeanne Elizabeth Daniel
November 2019
We make use of a bi-directional LSTM networks that extends the modelling capabilities of the vanilla LSTM. This approach is similar to that of InferSent (Conneau et al. 2017) where the authors combine bi-directional LSTM models with pooling layers to produce high-quality sentence embeddings. In addition to InferSent, we attach a dense classification layer after the pooling layers.
```
import sys
import os
#sys.path.append(os.path.join(\"..\")) # path to source relative to current directory"
import numpy as np
import gensim
import preprocess_data
import pandas as pd
import tensorflow as tf
physical_devices = tf.config.experimental.list_physical_devices("GPU")
tf.config.experimental.set_memory_growth(physical_devices[0], True)
from tensorflow.keras.models import Model
from tensorflow.keras.preprocessing import sequence
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import GlobalAveragePooling1D, GlobalMaxPooling1D
from tensorflow.keras.layers import Dense, Dropout, Embedding, LSTM, Bidirectional, TimeDistributed, Input, Flatten, AdditiveAttention
from tensorflow.keras.callbacks import EarlyStopping, ModelCheckpoint
data = pd.read_csv('dataset_7B', delimiter = ';', engine = 'python')
data_text = data.loc[data['set'] == 'Train'][['helpdesk_question']]
number_of_classes = data.loc[data['set'] == 'Train']['helpdesk_reply'].value_counts().shape[0]
data = data[['helpdesk_question', 'helpdesk_reply', 'set', 'low_resource']]
responses = pd.DataFrame(data.loc[data['set'] == 'Train']['helpdesk_reply'].value_counts()).reset_index()
responses['reply'] = responses['index']
responses['index'] = responses.index
responses = dict(responses.set_index('reply')['index'])
len(responses)
data_text['index'] = data_text.index
documents = data_text
dictionary = preprocess_data.create_dictionary(data_text, 1, 0.25, 95000) #our entire vocabulary
df_train = data.loc[data['set'] == 'Train']
df_train = df_train.reset_index()[['helpdesk_question', 'helpdesk_reply']]
df_valid = data.loc[data['set'] == 'Valid']
df_valid = df_valid.reset_index()[['helpdesk_question', 'helpdesk_reply']]
df_test = data.loc[data['set'] == 'Test']
df_test = df_test.reset_index()[['helpdesk_question', 'helpdesk_reply']]
df_LR = data.loc[(data['set'] == 'Test') & (data['low_resource'] == 'True') ]
df_LR = df_LR.reset_index()[['helpdesk_question', 'helpdesk_reply']]
df_train.shape
unique_words = dictionary
len(unique_words) + 1
max_length = 30
min_token_length = 0
word_to_id, id_to_word = preprocess_data.create_lookup_tables(unique_words)
```
#### Transforming the input sentence into a sequence of word IDs
```
train_x_word_ids = []
for question in df_train['helpdesk_question'].apply(preprocess_data.preprocess_question,
args = [unique_words, min_token_length]):
word_ids = preprocess_data.transform_sequence_to_word_ids(question, word_to_id)
train_x_word_ids.append(np.array(word_ids, dtype = float))
train_x_word_ids = np.stack(train_x_word_ids)
print(train_x_word_ids.shape)
val_x_word_ids = []
for question in data['helpdesk_question'].loc[data['set'] == 'Valid'].apply(preprocess_data.preprocess_question,
args = [unique_words, min_token_length]):
word_ids = preprocess_data.transform_sequence_to_word_ids(question, word_to_id)
val_x_word_ids.append(np.array(word_ids, dtype = float))
val_x_word_ids = np.stack(val_x_word_ids)
test_x_word_ids = []
for question in data['helpdesk_question'].loc[data['set'] == 'Test'].apply(preprocess_data.preprocess_question,
args = [unique_words, min_token_length]):
word_ids = preprocess_data.transform_sequence_to_word_ids(question, word_to_id)
test_x_word_ids.append(np.array(word_ids, dtype = float))
test_x_word_ids = np.stack(test_x_word_ids)
LR_x_word_ids = []
for question in data['helpdesk_question'].loc[(data['set'] == 'Test') &
(data['low_resource'] == 'True')].apply(preprocess_data.preprocess_question,
args = [unique_words, min_token_length]):
word_ids = preprocess_data.transform_sequence_to_word_ids(question, word_to_id)
LR_x_word_ids.append(np.array(word_ids, dtype = float))
LR_x_word_ids = np.stack(LR_x_word_ids)
def get_dummies(reply, all_responses):
""" Constructs a one-hot vector for replies
Args:
reply: query item
all_responses: dict containing all the template responses with their corresponding IDs
Return:
a one-hot vector where the corresponding ID of the reply is the one-hot index
"""
Y = np.zeros(len(all_responses), dtype = int)
Y[all_responses[reply]] += 1
return Y
train_y = np.array(list(df_train['helpdesk_reply'].apply(get_dummies, args = [responses])))
valid_y = np.array(list(df_valid['helpdesk_reply'].apply(get_dummies, args = [responses])))
test_y = np.array(list(df_test['helpdesk_reply'].apply(get_dummies, args = [responses])))
LR_y = np.array(list(df_LR['helpdesk_reply'].apply(get_dummies, args = [responses])))
train_x_word_ids = train_x_word_ids.reshape(train_x_word_ids.shape[:-1])
val_x_word_ids = val_x_word_ids.reshape(val_x_word_ids.shape[:-1])
test_x_word_ids = test_x_word_ids.reshape(test_x_word_ids.shape[:-1])
LR_x_word_ids = LR_x_word_ids.reshape(LR_x_word_ids.shape[:-1])
```
#### Transform vectors where the input sentence yields a sequence of length 0
```
train_zero_vectors = np.where(train_x_word_ids.sum(axis = 1) == 0.0)[0]
for t in range(train_zero_vectors.shape[0]):
train_x_word_ids[train_zero_vectors[t]][0] += 1
val_zero_vectors = np.where(val_x_word_ids.sum(axis = 1) == 0.0)[0]
for t in range(val_zero_vectors.shape[0]):
val_x_word_ids[val_zero_vectors[t]][0] += 1
```
#### Bi-directional LSTM with max pooling
The network consists of an embedding layer, followed by a dropout layer. This is followed by an bi-directional LSTM layer that outputs a variable-length sequence of embedding vectors. To construct a single sentence embedding from the sequence we use max pooling. The sentence embedding is then fed to a classification layer. We train with a dropout rate of 0.5 and batch size of 32. During training we use early stopping and Adadelta as our optimization algorithm. This network has an embedding of size 300 and 128 hidden units in the biLSTM network.
```
def bilstm_max_pooling_network(max_features, input_length=30, embed_dim=100, lstm_units=512):
""" Constructs a bi-directional LSTM network with max pooling
Args:
max_features: size of vocabulary
input_length: length of input sequence
embed_dim: dimension of the embedding vector
lstm_units: number of hidden units in biLSTM
Returns:
An biLSTM model
"""
inputs = Input(shape=(input_length, ))
x = Embedding(max_features, output_dim=embed_dim, input_length=input_length, mask_zero=True)(inputs)
x = (Dropout(rate = 0.5))(x)
x = Bidirectional(LSTM(lstm_units, activation = 'tanh', return_sequences=True,
dropout=0.25, recurrent_dropout=0.5))(x)
x = GlobalMaxPooling1D()(x)
outputs = Dense(89, activation='softmax')(x)
return Model(inputs=inputs, outputs=outputs)
max_features = len(unique_words) + 1
model = bilstm_max_pooling_network(max_features, embed_dim=300, input_length=30, lstm_units = 128)
model.summary()
```
### Training
```
es = EarlyStopping(monitor='val_accuracy', verbose=1, restore_best_weights=True, patience=10)
model.compile(loss='categorical_crossentropy',
optimizer=tf.keras.optimizers.Adadelta(learning_rate=0.5, rho=0.95),
metrics=['accuracy'])
model.fit(train_x_word_ids, train_y,
batch_size=32,
epochs=500,
callbacks=[es],
validation_data=[val_x_word_ids, valid_y])
```
### Test score
```
def classifier_score_top_1(word_ids, y_true, model):
""" Computes top-1 classification accuracy for model.
Args:
word_ids: matrix where each row is
y_true: true labels
model: trained model
Returns:
None
"""
score = 0
probs = model.predict(word_ids)
for i in range(word_ids.shape[0]):
if y_true[i].argmax() == np.argsort(probs[i])[-1]:
score += 1
print("Overall Accuracy:", score/word_ids.shape[0])
classifier_score_top_1(test_x_word_ids, test_y, model)
```
### LR test score
```
classifier_score_top_1(LR_x_word_ids, LR_y, model)
test_y[0].argmax()
```
### Top-5 accuracy
```
def classifier_score_top_5(word_ids, y_true, model):
""" Computes top-5 classification accuracy for model.
Args:
word_ids: matrix where each row is
y_true: true labels
model: trained model
Returns:
None
"""
score = 0
probs = model.predict(word_ids)
for i in range(word_ids.shape[0]):
if y_true[i].argmax() in np.argsort(probs[i])[-5:]:
score += 1
print("Overall Accuracy:", score/word_ids.shape[0])
classifier_score_top_5(test_x_word_ids, test_y, model)
classifier_score_top_5(LR_x_word_ids, LR_y, model)
```
|
github_jupyter
|
import sys
import os
#sys.path.append(os.path.join(\"..\")) # path to source relative to current directory"
import numpy as np
import gensim
import preprocess_data
import pandas as pd
import tensorflow as tf
physical_devices = tf.config.experimental.list_physical_devices("GPU")
tf.config.experimental.set_memory_growth(physical_devices[0], True)
from tensorflow.keras.models import Model
from tensorflow.keras.preprocessing import sequence
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import GlobalAveragePooling1D, GlobalMaxPooling1D
from tensorflow.keras.layers import Dense, Dropout, Embedding, LSTM, Bidirectional, TimeDistributed, Input, Flatten, AdditiveAttention
from tensorflow.keras.callbacks import EarlyStopping, ModelCheckpoint
data = pd.read_csv('dataset_7B', delimiter = ';', engine = 'python')
data_text = data.loc[data['set'] == 'Train'][['helpdesk_question']]
number_of_classes = data.loc[data['set'] == 'Train']['helpdesk_reply'].value_counts().shape[0]
data = data[['helpdesk_question', 'helpdesk_reply', 'set', 'low_resource']]
responses = pd.DataFrame(data.loc[data['set'] == 'Train']['helpdesk_reply'].value_counts()).reset_index()
responses['reply'] = responses['index']
responses['index'] = responses.index
responses = dict(responses.set_index('reply')['index'])
len(responses)
data_text['index'] = data_text.index
documents = data_text
dictionary = preprocess_data.create_dictionary(data_text, 1, 0.25, 95000) #our entire vocabulary
df_train = data.loc[data['set'] == 'Train']
df_train = df_train.reset_index()[['helpdesk_question', 'helpdesk_reply']]
df_valid = data.loc[data['set'] == 'Valid']
df_valid = df_valid.reset_index()[['helpdesk_question', 'helpdesk_reply']]
df_test = data.loc[data['set'] == 'Test']
df_test = df_test.reset_index()[['helpdesk_question', 'helpdesk_reply']]
df_LR = data.loc[(data['set'] == 'Test') & (data['low_resource'] == 'True') ]
df_LR = df_LR.reset_index()[['helpdesk_question', 'helpdesk_reply']]
df_train.shape
unique_words = dictionary
len(unique_words) + 1
max_length = 30
min_token_length = 0
word_to_id, id_to_word = preprocess_data.create_lookup_tables(unique_words)
train_x_word_ids = []
for question in df_train['helpdesk_question'].apply(preprocess_data.preprocess_question,
args = [unique_words, min_token_length]):
word_ids = preprocess_data.transform_sequence_to_word_ids(question, word_to_id)
train_x_word_ids.append(np.array(word_ids, dtype = float))
train_x_word_ids = np.stack(train_x_word_ids)
print(train_x_word_ids.shape)
val_x_word_ids = []
for question in data['helpdesk_question'].loc[data['set'] == 'Valid'].apply(preprocess_data.preprocess_question,
args = [unique_words, min_token_length]):
word_ids = preprocess_data.transform_sequence_to_word_ids(question, word_to_id)
val_x_word_ids.append(np.array(word_ids, dtype = float))
val_x_word_ids = np.stack(val_x_word_ids)
test_x_word_ids = []
for question in data['helpdesk_question'].loc[data['set'] == 'Test'].apply(preprocess_data.preprocess_question,
args = [unique_words, min_token_length]):
word_ids = preprocess_data.transform_sequence_to_word_ids(question, word_to_id)
test_x_word_ids.append(np.array(word_ids, dtype = float))
test_x_word_ids = np.stack(test_x_word_ids)
LR_x_word_ids = []
for question in data['helpdesk_question'].loc[(data['set'] == 'Test') &
(data['low_resource'] == 'True')].apply(preprocess_data.preprocess_question,
args = [unique_words, min_token_length]):
word_ids = preprocess_data.transform_sequence_to_word_ids(question, word_to_id)
LR_x_word_ids.append(np.array(word_ids, dtype = float))
LR_x_word_ids = np.stack(LR_x_word_ids)
def get_dummies(reply, all_responses):
""" Constructs a one-hot vector for replies
Args:
reply: query item
all_responses: dict containing all the template responses with their corresponding IDs
Return:
a one-hot vector where the corresponding ID of the reply is the one-hot index
"""
Y = np.zeros(len(all_responses), dtype = int)
Y[all_responses[reply]] += 1
return Y
train_y = np.array(list(df_train['helpdesk_reply'].apply(get_dummies, args = [responses])))
valid_y = np.array(list(df_valid['helpdesk_reply'].apply(get_dummies, args = [responses])))
test_y = np.array(list(df_test['helpdesk_reply'].apply(get_dummies, args = [responses])))
LR_y = np.array(list(df_LR['helpdesk_reply'].apply(get_dummies, args = [responses])))
train_x_word_ids = train_x_word_ids.reshape(train_x_word_ids.shape[:-1])
val_x_word_ids = val_x_word_ids.reshape(val_x_word_ids.shape[:-1])
test_x_word_ids = test_x_word_ids.reshape(test_x_word_ids.shape[:-1])
LR_x_word_ids = LR_x_word_ids.reshape(LR_x_word_ids.shape[:-1])
train_zero_vectors = np.where(train_x_word_ids.sum(axis = 1) == 0.0)[0]
for t in range(train_zero_vectors.shape[0]):
train_x_word_ids[train_zero_vectors[t]][0] += 1
val_zero_vectors = np.where(val_x_word_ids.sum(axis = 1) == 0.0)[0]
for t in range(val_zero_vectors.shape[0]):
val_x_word_ids[val_zero_vectors[t]][0] += 1
def bilstm_max_pooling_network(max_features, input_length=30, embed_dim=100, lstm_units=512):
""" Constructs a bi-directional LSTM network with max pooling
Args:
max_features: size of vocabulary
input_length: length of input sequence
embed_dim: dimension of the embedding vector
lstm_units: number of hidden units in biLSTM
Returns:
An biLSTM model
"""
inputs = Input(shape=(input_length, ))
x = Embedding(max_features, output_dim=embed_dim, input_length=input_length, mask_zero=True)(inputs)
x = (Dropout(rate = 0.5))(x)
x = Bidirectional(LSTM(lstm_units, activation = 'tanh', return_sequences=True,
dropout=0.25, recurrent_dropout=0.5))(x)
x = GlobalMaxPooling1D()(x)
outputs = Dense(89, activation='softmax')(x)
return Model(inputs=inputs, outputs=outputs)
max_features = len(unique_words) + 1
model = bilstm_max_pooling_network(max_features, embed_dim=300, input_length=30, lstm_units = 128)
model.summary()
es = EarlyStopping(monitor='val_accuracy', verbose=1, restore_best_weights=True, patience=10)
model.compile(loss='categorical_crossentropy',
optimizer=tf.keras.optimizers.Adadelta(learning_rate=0.5, rho=0.95),
metrics=['accuracy'])
model.fit(train_x_word_ids, train_y,
batch_size=32,
epochs=500,
callbacks=[es],
validation_data=[val_x_word_ids, valid_y])
def classifier_score_top_1(word_ids, y_true, model):
""" Computes top-1 classification accuracy for model.
Args:
word_ids: matrix where each row is
y_true: true labels
model: trained model
Returns:
None
"""
score = 0
probs = model.predict(word_ids)
for i in range(word_ids.shape[0]):
if y_true[i].argmax() == np.argsort(probs[i])[-1]:
score += 1
print("Overall Accuracy:", score/word_ids.shape[0])
classifier_score_top_1(test_x_word_ids, test_y, model)
classifier_score_top_1(LR_x_word_ids, LR_y, model)
test_y[0].argmax()
def classifier_score_top_5(word_ids, y_true, model):
""" Computes top-5 classification accuracy for model.
Args:
word_ids: matrix where each row is
y_true: true labels
model: trained model
Returns:
None
"""
score = 0
probs = model.predict(word_ids)
for i in range(word_ids.shape[0]):
if y_true[i].argmax() in np.argsort(probs[i])[-5:]:
score += 1
print("Overall Accuracy:", score/word_ids.shape[0])
classifier_score_top_5(test_x_word_ids, test_y, model)
classifier_score_top_5(LR_x_word_ids, LR_y, model)
| 0.36693 | 0.806243 |
# Arm Reacher with Continuous Control
---
move a double-jointed arm towards target locations in the [Reacher](https://github.com/Unity-Technologies/ml-agents/blob/master/docs/Learning-Environment-Examples.md#reacher) environment.
---
You can download the environment matching your operation system from one of the following links:
* Linux: [click here](https://s3-us-west-1.amazonaws.com/udacity-drlnd/P2/Reacher/one_agent/Reacher_Linux.zip)
* Mac OSX: [click here](https://s3-us-west-1.amazonaws.com/udacity-drlnd/P2/Reacher/one_agent/Reacher.app.zip)
* Windows (32-bit): [click here](https://s3-us-west-1.amazonaws.com/udacity-drlnd/P2/Reacher/one_agent/Reacher_Windows_x86.zip)
* Windows (64-bit): [click here](https://s3-us-west-1.amazonaws.com/udacity-drlnd/P2/Reacher/one_agent/Reacher_Windows_x86_64.zip)
## 1. Start the Environment
If necessary install following packages: **matplotlib**, **numpy**, **torch**, **unityagents**.
If needed uncoment the following cell and install **matplotlib**
```
import sys
# !{sys.executable} -m pip install matplotlib
```
If needed uncoment the following cell and install numpy
```
# !{sys.executable} -m pip install numpy
```
If needed uncoment the following cell and install torch. We will build the deep Q-Networks using torch.
```
#!{sys.executable} -m pip install torch
```
If needed uncoment the following cell and install unityagents. This package is needed to run the downloaded Unity Environment.
```
#!{sys.executable} -m pip install unityagents
```
Import the packages.
```
import random
import torch
import numpy as np
from collections import deque
import matplotlib.pyplot as plt
from unityagents import UnityEnvironment
from agent import Agent, Brain
seed = 10
```
Next, we will start the environment!
Before running the code cell below, change the `unityEnvPath` parameter to match the location of the Unity environment that you downloaded.
- **Mac**: `"path/to/Reacher.app"`
- **Windows** (x86): `"path/to/Reacher_Windows_x86/Reacher.exe"`
- **Windows** (x86_64): `"path/to/Reacher_Windows_x86_64/Reacher.exe"`
- **Linux** (x86): `"path/to/Reacher_Linux/Reacher.x86"`
- **Linux** (x86_64): `"path/to/Reacher_Linux/Reacher.x86_64"`
- **Linux** (x86, headless): `"path/to/Reacher_Linux_NoVis/Reacher.x86"`
- **Linux** (x86_64, headless): `"path/to/Reacher_Linux_NoVis/Reacher.x86_64"`
```
unityEnvPath = 'path/to/Reacher/Environment...'
env = UnityEnvironment(file_name=unityEnvPath,no_graphics=True,seed=seed)
# get the default brain
brain_name = env.brain_names[0]
brain = env.brains[brain_name]
```
## 2. Examine the State and Action Spaces
Before starting with the training let's examine the environment.
The **state space** is `33` dimensional corresponding to `position, rotation, velocity, angular velocities` of the arm.
The **actions** `4` dimensional corresponding to `torque applicable to two joints`. Every entry in the action vector must be a number between `-1` and `1`.
A **reward** of `+0.1` is provided for each step that the agent's hand is in the goal location.
```
# reset the environment
env_info = env.reset(train_mode=True)[brain_name]
# number of agents in the environment
num_agents = len(env_info.agents)
print('Number of agents:', num_agents)
# number of actions
action_size = brain.vector_action_space_size
print('Number of actions:', action_size)
# examine the state space
state = env_info.vector_observations[0]
state_size = len(state)
print('States have length:', state_size)
print('States look like:\n', state)
```
## 3. Train your agent
The goal of your agent is to maintain its position at the target location for as many time steps as possible. The environment is solved if an average score of 30 is maintained over 100 episodes.
To solve the environment we use the method of deep deterministic policy gradients (DDPG). The algorithm is implemented in the `Brain` class.
Now we can define the training function:
```
def trainAgent(n_episodes=1000, max_t=1000, startNoise = 0.1, endNoise = 0.01, noiseDecay = 1):
scores_deque = deque(maxlen=100)
scores_list = []
max_score = -np.Inf
noise = startNoise/noiseDecay
for i_episode in range(1, n_episodes+1):
# Per episode we have to reset the environment.
env_info = env.reset(train_mode=True)[brain_name]
# Per episode we have to get the current state.
states = env_info.vector_observations
noise = max(endNoise,noise*noiseDecay)
agent.reset()
scores = np.zeros(num_agents)
for t in range(max_t):
actions = agent.act(states,noise)
env_info = env.step(actions)[brain_name]
# We obtain the next state.
next_states = env_info.vector_observations
# We get the reward.
rewards = env_info.rewards
# We get the done status.
dones = env_info.local_done
agent.step(states, actions, rewards, next_states, dones)
states = next_states
scores += rewards
if np.any(dones):
break
score = np.mean(scores)
scores_deque.append(score)
scores_list.append(score)
print('\rEpisode {}\tAverage Score: {:.2f}\tScore: {:.2f}'.format(i_episode, np.mean(scores_deque), score))
if i_episode % 100 == 0:
torch.save(agentBrain.actorLocal.state_dict(), 'checkpoint_actor.pth')
torch.save(agentBrain.criticLocal.state_dict(), 'checkpoint_critic.pth')
print('\rEpisode {}\tAverage Score: {:.2f}'.format(i_episode, np.mean(scores_deque)))
# if np.mean(scores_deque) > 30:
# torch.save(agentBrain.actorLocal.state_dict(), 'checkpoint_actor.pth')
# torch.save(agentBrain.criticLocal.state_dict(), 'checkpoint_critic.pth')
# print('\rEpisode {}\tAverage Score: {:.2f}'.format(i_episode, np.mean(scores_deque)))
# print('\n\nEnvironment solved in ',i_episode,' episodes')
# break
return scores_list
```
We first create an instance of the `Brain` class and adjust the parameters. Then we create an instance of the agent itself.
```
agentBrain = Brain(stateSize=state_size, actionSize=action_size,
gamma = 0.99,
actorLearningRate = 2e-4,
criticLearningRate = 2e-4,
actorSoftHardUpdatePace = 5e-3,
criticSoftHardUpdatePace = 5e-3,
dnnUpdatePace = 4,
bufferSize = int(1e6),
batchSize = 128,
batchEpochs = 1,
weightDecay = 0,
seed = seed)
agent = Agent(agentBrain)
```
Start the trainig
```
scores = trainAgent()
def plotScores(scores,meanOver = 100):
"""
Plot the scores
"""
yLimMin = -0.5
scores = np.array(scores)
runMean = np.convolve(scores, np.ones((meanOver,))/meanOver,mode='valid')[1:]
mean13 = np.argwhere(runMean>30) + meanOver
score13 = np.argwhere(scores>30)
fig = plt.figure()
ax = fig.add_subplot(111)
plt.plot(np.arange(len(scores)), scores)
plt.plot(np.arange(meanOver,len(scores)), runMean)
plt.plot([0, len(scores)],[30,30],'r')
plt.plot([mean13[0],mean13[0]],[-5,30],'r')
plt.text(mean13[0]-100,yLimMin+1,str(mean13[0]),color = 'r')
#plt.scatter(score13,scores[score13],color='r')
plt.ylabel('Score')
plt.xlabel('Episode #')
plt.xlim([0,len(scores)])
plt.ylim([yLimMin,42])
plt.show()
plotScores(scores)
```
### 5. Watch a Smart Agent!
Now we can load the trained weights from the files and watch the performance of the trained agent.
```
agentBrain.actorLocal.load_state_dict(torch.load('checkpoint_actor.pth'))
agentBrain.criticLocal.load_state_dict(torch.load('checkpoint_critic.pth'))
env_info = env.reset(train_mode=False)[brain_name] # reset the environment
states = env_info.vector_observations # get the current state(s)
scores = np.zeros(num_agents) # initialize the score(s)
while True:
actions = agent.act(states) # select the action(s)
env_info = env.step(actions)[brain_name] # send the action(s) to the environment
next_states = env_info.vector_observations # get the next state(s)
rewards = env_info.rewards # get the reward(s)
dones = env_info.local_done # see if episode has finished
scores += rewards # update the score(s)
states = next_states # roll over the state to next time step
if np.any(dones): # exit loop if episode is finished
break
print("Score: {}".format(scores)) # print the score(s) of the episode
```
Close the environment
```
env.close()
```
|
github_jupyter
|
import sys
# !{sys.executable} -m pip install matplotlib
# !{sys.executable} -m pip install numpy
#!{sys.executable} -m pip install torch
#!{sys.executable} -m pip install unityagents
import random
import torch
import numpy as np
from collections import deque
import matplotlib.pyplot as plt
from unityagents import UnityEnvironment
from agent import Agent, Brain
seed = 10
unityEnvPath = 'path/to/Reacher/Environment...'
env = UnityEnvironment(file_name=unityEnvPath,no_graphics=True,seed=seed)
# get the default brain
brain_name = env.brain_names[0]
brain = env.brains[brain_name]
# reset the environment
env_info = env.reset(train_mode=True)[brain_name]
# number of agents in the environment
num_agents = len(env_info.agents)
print('Number of agents:', num_agents)
# number of actions
action_size = brain.vector_action_space_size
print('Number of actions:', action_size)
# examine the state space
state = env_info.vector_observations[0]
state_size = len(state)
print('States have length:', state_size)
print('States look like:\n', state)
def trainAgent(n_episodes=1000, max_t=1000, startNoise = 0.1, endNoise = 0.01, noiseDecay = 1):
scores_deque = deque(maxlen=100)
scores_list = []
max_score = -np.Inf
noise = startNoise/noiseDecay
for i_episode in range(1, n_episodes+1):
# Per episode we have to reset the environment.
env_info = env.reset(train_mode=True)[brain_name]
# Per episode we have to get the current state.
states = env_info.vector_observations
noise = max(endNoise,noise*noiseDecay)
agent.reset()
scores = np.zeros(num_agents)
for t in range(max_t):
actions = agent.act(states,noise)
env_info = env.step(actions)[brain_name]
# We obtain the next state.
next_states = env_info.vector_observations
# We get the reward.
rewards = env_info.rewards
# We get the done status.
dones = env_info.local_done
agent.step(states, actions, rewards, next_states, dones)
states = next_states
scores += rewards
if np.any(dones):
break
score = np.mean(scores)
scores_deque.append(score)
scores_list.append(score)
print('\rEpisode {}\tAverage Score: {:.2f}\tScore: {:.2f}'.format(i_episode, np.mean(scores_deque), score))
if i_episode % 100 == 0:
torch.save(agentBrain.actorLocal.state_dict(), 'checkpoint_actor.pth')
torch.save(agentBrain.criticLocal.state_dict(), 'checkpoint_critic.pth')
print('\rEpisode {}\tAverage Score: {:.2f}'.format(i_episode, np.mean(scores_deque)))
# if np.mean(scores_deque) > 30:
# torch.save(agentBrain.actorLocal.state_dict(), 'checkpoint_actor.pth')
# torch.save(agentBrain.criticLocal.state_dict(), 'checkpoint_critic.pth')
# print('\rEpisode {}\tAverage Score: {:.2f}'.format(i_episode, np.mean(scores_deque)))
# print('\n\nEnvironment solved in ',i_episode,' episodes')
# break
return scores_list
agentBrain = Brain(stateSize=state_size, actionSize=action_size,
gamma = 0.99,
actorLearningRate = 2e-4,
criticLearningRate = 2e-4,
actorSoftHardUpdatePace = 5e-3,
criticSoftHardUpdatePace = 5e-3,
dnnUpdatePace = 4,
bufferSize = int(1e6),
batchSize = 128,
batchEpochs = 1,
weightDecay = 0,
seed = seed)
agent = Agent(agentBrain)
scores = trainAgent()
def plotScores(scores,meanOver = 100):
"""
Plot the scores
"""
yLimMin = -0.5
scores = np.array(scores)
runMean = np.convolve(scores, np.ones((meanOver,))/meanOver,mode='valid')[1:]
mean13 = np.argwhere(runMean>30) + meanOver
score13 = np.argwhere(scores>30)
fig = plt.figure()
ax = fig.add_subplot(111)
plt.plot(np.arange(len(scores)), scores)
plt.plot(np.arange(meanOver,len(scores)), runMean)
plt.plot([0, len(scores)],[30,30],'r')
plt.plot([mean13[0],mean13[0]],[-5,30],'r')
plt.text(mean13[0]-100,yLimMin+1,str(mean13[0]),color = 'r')
#plt.scatter(score13,scores[score13],color='r')
plt.ylabel('Score')
plt.xlabel('Episode #')
plt.xlim([0,len(scores)])
plt.ylim([yLimMin,42])
plt.show()
plotScores(scores)
agentBrain.actorLocal.load_state_dict(torch.load('checkpoint_actor.pth'))
agentBrain.criticLocal.load_state_dict(torch.load('checkpoint_critic.pth'))
env_info = env.reset(train_mode=False)[brain_name] # reset the environment
states = env_info.vector_observations # get the current state(s)
scores = np.zeros(num_agents) # initialize the score(s)
while True:
actions = agent.act(states) # select the action(s)
env_info = env.step(actions)[brain_name] # send the action(s) to the environment
next_states = env_info.vector_observations # get the next state(s)
rewards = env_info.rewards # get the reward(s)
dones = env_info.local_done # see if episode has finished
scores += rewards # update the score(s)
states = next_states # roll over the state to next time step
if np.any(dones): # exit loop if episode is finished
break
print("Score: {}".format(scores)) # print the score(s) of the episode
env.close()
| 0.30715 | 0.966474 |
# Introduction to NLP tasks
The main goal of Natural Language Processing is to convert free text to / from a formal representation that allows us to process the linguistic data algorithmically.
There are two directions:
* **Analysis**: text $\rightarrow$ formal representation
* **Generation**: formal representation $\rightarrow$ text
### The linguistic pipeline
The tasks we are going to cover:
* **Tokenization**
* Morphological analysis / **POS tagging**
* Syntactic parsing
* Semantic parsing
These tasks are usually incorporated in a **linguistic pipeline**:

### Sequentiality
Natural language is inherently sequential:
* Words are sequences of characters
* Sentences are sequences of words
* Paragraphs are sequences of sentences
* Documents are sequences of paragraphs.
As such, all the tasks above are implemented with algorithms that work on *sequences*:
* Dynamic programming (algorithmic)
* Sequence classification (machine learning)
### NLP Software
#### [Stanford CoreNLP](http://nlp.stanford.edu:8080/corenlp/)
* Written in Java; can be used via the API, GUI, command line or a web service.
* Supports several major languages (other than English): Chinese, Spanish, Arabic, French and German.
#### [Spacy](https://spacy.io/)
* Written in Python; very simple API
* Supports 27+ languages (to some extent)
#### [e-magyar](http://e-magyar.hu/hu/)
* A pipeline that includes most state-of-the-art NLP tools written for Hungarian
* Java + XML; most components are very similar to CoreNLP's
## [Tokenization](https://nlp.stanford.edu/IR-book/html/htmledition/tokenization-1.html)
The computer does not know anything about words or any human language. We have to tell it what the (basic) units are.
<!-- From the data point of view, written language is a sequence of characters. To introduce one of the earliest and most fundamental task in NLP, we consider the text as the list of words. Breaking the text into words is not always that easy as in the example, that is called __tokenization__. We deal with tokenized texts for now. -->
#### Tokenization
Split the string into **tokens**:
```Python
In[1]: tokenize("Mr. and Mrs. Dursley of Number Four, Privet Drive, were proud to say that they were perfectly normal, thank you very much.")
Out[1]: ['Mr.', 'and', 'Mrs.', 'Dursley', 'of', 'Number', 'Four', ',', 'Privet', 'Drive', ',', 'were', 'proud', 'to', 'say','that', 'they', 'were', 'perfectly', 'normal', ',', 'thank', 'you', 'very', 'much', '.']
```
A **token** can be
* a word
* a punctuation mark
`str.split()` is not enough:
* `"much."` $\rightarrow$ `"much" + "."`
* `"Mr."` $\rightarrow$ `"Mr."`
#### Sentence splitting
Similarly: split the tokenized text into sentences.
```Python
In[1]: ssplit(['Me', 'Tarzan', '.', 'You', 'Jane', '.'])
Out[2]: [['Me', 'Tarzan', '.'],
['You', 'Jane', '.']]
```
#### Conversion to IDs
To the computer, words are just abstract labels. Usually we put the words into a vocabulary and assign an integer ID to all of them.
```
vocabulary = {w:i for i, w in enumerate(set(x.split()))}
print(vocabulary)
print("------------------")
print(words)
print([vocabulary[w] for w in words])
```
For now, this is what we have to work with, but later you will learn more about representing words (other than meaningless IDs).
## (POS) Tagging
### Part-of-speech (POS)
Words can be put into categories according to their grammatical properties, or the role they can play in a sentence.
The category into a particular word falls is its **part-of-speech**.
<!--A __Part-of-speech__ tag is a property of words. More precisely, we put words into categories according what role they play in a given sentence.-->
#### Example
| The|dog|saw| a|cat|.|
|----|---|---|--|---|---|
| determiner|noun|verb| determiner|noun|punctuation|
<!-- The categorization is aimed to label interchangeable words with the same label. For example __cat__ could be replaced with __dog__, __boat__ or any other nouns, but not with a verb.
-->
Interchangeable words should end up in the same category.
* One can change "_saw_" to "_smelled_" or "_cat_" to "_mouse_" without loss of grammaticality, but not e.g. "_cat_" to "_smelled_".
* On the other hand, "_saw_" could also be changed to "_walked_": POS categories do not take semantics into consideration.
The correct POS depends on the context, not only the word itself.
<!--|You|talk|the|talk|but|do|you|walk|the|walk|?|
|---|----|---|----|---|--|---|----|---|----|-|
|pronoun|verb|determiner|noun|conjunction|verb|pronoun|verb|determiner|noun|punctuation|-->
|You|talk|the|talk|but|do|
|---|----|---|----|---|--|
|pronoun|verb|determiner|noun|conjunction|verb|
|**you**|**walk**|**the**|**walk**|**?**|
|pronoun|verb|determiner|noun|punctuation|
### POS tagging
... is a task to label every word in a given text with the appropriate POS **tags**. An algorithm for that will be our very first NLP task!
### Tagsets
* The tags themselves are the result of linguistic/NLP consensus
* there are several conventions for them.
* From computational point of view, there is no definition for POS tags
* Benchmark datasets are the definition (gold standard) of what the correct tags are.
From [Universal Dependencies](https://github.com/UniversalDependencies/UD_English) using [Universal POS tags](http://universaldependencies.org/u/pos/all.html):
|This|killing|of|a|respected|cleric|will|be|
|---|----|---|---|---|----|---|---|
|DET|NOUN|ADP|DET|ADJ|NOUN|AUX|AUX|
|**causing**|**us**|**trouble**|**for**|**years**|**to**|**come**|**.**|
|VERB|PRON|NOUN|ADP|NOUN|PART|VERB|PUNCT|
The Universal tagset is language independent, except some language specific features. For example the words _"cleric"_ and _"gazdaság"_ are both NOUNs. In English _"a"_ and _"the"_ are determiners, in Hungarian _"a"_ and _"az"_ have similar grammatical functions.
Or a [Hungarian one](https://github.com/UniversalDependencies/UD_Hungarian)
|A|gazdaság|ilyen|mértékű|fejlődését|több|folyamat|gerjeszti|.|
|-|--------|-----|-------|----------|----|--------|---------|-|
|DET|NOUN|DET|ADJ|NOUN|DET|NOUN|VERB|PUNCT|
From [UMBC webbase](http://ebiquity.umbc.edu/resource/html/id/351) corpus using [Penn Treebank tagset](https://ling.upenn.edu/courses/Fall_2003/ling001/penn_treebank_pos.html):
|Well| ,| let| me| just| say| there| is| n't| too|
|---|--|---|----|---|----|---|----|---|---|
|RB |, |VB |PRP |RB |VBP |EX |VBZ |RB |RB |
|**much**|**I**| **can**| **tell**| **you**| **right**| **at**| **the**| **moment**| **.**|
|JJ |PRP |MD |VB |PRP |RB |IN |DT |NN |.|
The latter is just for English, note the EX (existential _there_) tag and the token _"n't"_ after _"is"_ with tag RB.
These are also tokenized!
## Tagging in general
<!-- In general, a tagging task requires a labeled dataset: a list of symbols with a list of corresponding target symbols.
The "sentence" is a list of source symbols (tokens or words), and the target symbols are from a (more-or-less) well defined set.
-->
|$word_1$| $word_2$| $word_3$| $\ldots$|
|---|--|---|----|
|$tag_1$ |$tag_2$ |$tag_3$ |$\ldots$ |
> __Definition__ (Token)
> _The atoms of interest, in our case the words of a text._
> __Definition__ (Corpus)
> _A list of tokens_
> __Definition__ (Tagset)
> _A finite (usually small) set of symbols, which are linguistically defined properties of tokens._
> __Definition__ (Labeled corpus)
> _A List of (token, tag) pairs, where the tags are from a given tagset._
> __Definition__ (Tagging)
> _Take a given (unlabeled) corpus and tagset. The_ tagging _is the task of assigning tags to tokens._
Sometimes a corpus is split at sentence boundaries, which means that sentences can be processed separately.
Otherwise, a sentence boundary is just a special punctuation or end-of-sentence symbol.
There are cases when the tag of a word cannot be deduced within a sentence, only in context of other sentences.
### Approaches
If the tags were algorithmically well-defined, then implementing that definition would result a 100% correct tagger without any further ado. Needless to mention, this is not the case.
There are two main approaches to tagging.
#### Rule based
Based on manually created rules.
* written by linguists; requires great effort; expensive
* high precision, low recall
* example: English Constraint Grammar (from [Apertium](https://github.com/apertium/apertium-eng/blob/master/apertium-eng.eng.rlx)).
```
#Lemmas that are N or V: Inf if preposition precedes it
SELECT Inf IF (0 Inf) (0 N) (-1 To) ;
SELECT Inf IF (0 Inf) (0 N) (-1 Adv) (-2 To) ;
```
#### __Statistical__
Based on machine learning.
* the models are trained on annotated gold standard corpora
* Penn Treebank (PTB, for English)
* Szeged Corpus (for Hungarian)
* "_statistical_" because the model is conditioned on the training corpus
#### Terminology:
<!-- , we split the data into two parts. One for training, this is at the disposal of our algorithm.
The other part of the split is for testing. The correct labels are stripped off of the test set and are compared to the output of the algorithm.-->
> __Annotating__
> _The human labor of making the gold dataset. Manually processing sentences and label them correctly._
> __Gold data__
> _Annotated corpus, usually annotated with serious efforts and for a specific task._
> __Silver data__
> _Not that good quality or automatically labeled data._
<!-- Sometimes we call the correctly labeled dataset __gold data__, usually annotated with serious efforts. If a given data is not perfectly labeled, or come from unknown origin, or the labels themselves are questionable, them we can talk about __silver data__. Silver is not always correct, but without any better at hand, they are used as training data. Sometimes silver data is acquired with automated or semi-automated techniques rather than human annotation.
It is worth mentioning that one can evaluate a model on the training data, but it does not tell much about the correctness of the algorithm. If you train an algorithm and use the training set in the prediction, thats called __test on train__. Every sound algorithm is expected to perform well (if not perfectly) on training data, since that data was available during the design/making of the algorithm. -->
### Evaluation
Lets suppose that one has a tagger and performed the tagging on the test set.
|token|$w_1$|$w_2$|$\ldots$|
|:--|-----|-----|--------|
|gold labels|$l_1$|$l_2$|$\ldots$|
|predicted labels|$p_1$|$p_2$|$\ldots$|
* The predicted and gold labels are compared to each other.
* The performance of a tagger can be measured several ways:
* per-token accuracy:
$$\frac{\# \text{ correct labels}}{\# \text{ words}}$$
* per-sentence accuracy:
$$\frac{\# \text{ sentences with all correct labels}}{\# \text{ sentences}}$$
* unknown word accuracy:
$$\frac{\# \text{ correct labels of OOV words}}{\# \text{ OOV words}}$$
OOV is out-of-vocabulary, words that were seen in test time, but not in training time.
### Other tagging tasks
Beside POS tagging, there are several other tasks.
#### NER
Named entity recognition.
> <u>Uber</u> isn't pulling out of <u>Quebec</u> just yet.<br>
> <u>Bill Gates</u> buys <u>Apple</u>. <u>DJIA</u> crashing!
* Mark names of entities in text
* people
* places
* organizations
* A very important task in _information extraction_. Helps identify what real word entities play role in a given sentence.
In the above example the target labels are just $\{0,1\}$, i.e. marking whether it is a named entity or not. There are more detailed NER tagsets which tells you what that entity is (plus one tag for _"not a named entity"_).
|tagset|# of tags| language independent | |
|:---|:--|:--:|:--|
|CoNLL| 4 |yes|Person, Location, Organization, Misc|
|MUC-7| 7 | yes|also Date, Time, Numbers, Percent|
|Penn Treebank|22|no|Animal, Cardinal, Date, Disease,...|
It is a different task to match various forms of the same entity, like: _USA_, _United States of America_, _The States_, _'merica_.
#### NP-chunking
| He | reckons |the current account deficit| will narrow| to |only £1.8 billion |in|September|.|
|:--:|:-------:|:-------------------------:|:----------:|:--:|:----------------:|:-:|:------:|:-:|
|NP | |NP | | | NP | | NP| |
The task is to find __noun phrases__:
* refer to (not necesserily named) entities or things
* correspond to grammatical roles (subject, object...) in the sentence
It is often called __shallow parsing__ because it finds some syntactic components of the sentence, but not the whole structure.
### Naive methods
Here we discuss some naive approaches and common mistakes.
##### The tag (label) of a word is not a function of the word itself.
* It depends on the context, the surrounding words.
* Most of the words can have several part-of-speech tags.
* In English noun-verbs are common: _work_, _talk_, _walk_.
##### Names entities are not always proper nouns
Neither start with capital letters.
Counter examples:
* the States
* von Neumann
Sentences start with capital letters, irrespective of whether the first word is a named entity.
##### There is no comprehensive list of all named entities
* Let's suppose that one wants to collect every famous person, geographic place, company, trademark and title (real and fictional ever) in a list.
* That list will never be comprehensive, since a well known thing can be mentioned in an unusual or abbreviated form.
* And the list would became obsolete very soon, since new movies, books, famous person and firms are born.
* Still, lists provide a good starting point for statistical models and the creation of silver standard corpora.
### Challenges
In this section we discuss some of the main challenges / difficulties of tagging.
#### Training data
* Bad quality or insufficient training data.
* Luckily, the aforementioned tasks have well established gold standards. Still,
* every dataset has mistakes in it.
* gold standard corpora are usually very small
* English: PTB (1M) $\ll$ UMBC Webbase (3B)
* Hungarian: Szeged (1.5M) $\ll$ Webcorpus (500M) $<$ MNSZ (1B)
There is a trade-off between the quality and quantity. Human annotated data are of higher quality but lower in quantity.
#### Evaluation
* Without proper evaluation, no way of knowing how good the model is.
* Given a certain amount of gold data, you have to decide how to cut it into train and testing.
* If you have no test data, you can examine the output of your algorithm yourself, but
* lacks statistical perspective
* getting 10 of your favorite sentences right doesn't mean that your algorithm is any good!
* this is called __testing by glance__ and you are advised to avoid it.
<!-- Without any test data, you can compete your algorithm against humans, or see it for yourself. This kind of manual test is expensive in human time and lacks statistical perspective if you don't use enough annotators. Getting 10 of your favorite sentences right doesn't mean that your algorithm rocks!
-->
This is a serious problem in machine translation, where you don't even have a correct automated testing.
<!-- A sentence can have several equally good translations, so if your algorithm does not give you the desired result, that doesn't mean that the translation was wrong. It's difficult to even compare sentences and tell that they mean a similar thing, if they have a different wording. -->
#### Linguistic changes
* Healthy languages changes over time:
* new words and expressions are introduced every day
* the grammar also changes, but much slower
* New linguistic theories are also proposed
* No (or very small) gold standard corpora for the less popular ones
* Conversion of gold standards
* Not always possible
* Usually the quality is lower
## Hidden Markov Model (HMM)
### A simple POS tagger
* Let's write a very simple tagger.
* Given the corpus, establish
* a vocabulary $V$ with every word in it
* and the set of labels $L$
* The labeled corpus is a list of pairs in $V\times L$.
* Usually $|V|\approx 10^5, 10^6$ and $|L|$ is never more than $100$
The model: a lookup table which assigns a POS tag to a word based on the list of $n$ previous words.
```
from itertools import chain
from collections import defaultdict, OrderedDict
# Two inputs: the corpus and n
corpus=list(zip(["The", "quick", "brown", "fox", "jumps", "over", "the", "lazy", "dog", "."],
["DET", "ADJ", "ADJ", "NOUN", "VERB", "ADP", "DET", "ADJ", "NOUN", "PUNCT"]))
n = 3
print('The corpus:', corpus, sep='\n')
pos_lookup = OrderedDict([(tuple(corpus[j][0] for j in range(max(i - 2, 0), i + 1)),
corpus[i][1])
for i in range(len(corpus))])
print('\nThe lookup table:')
pos_lookup
```
#### Markov model
The keys in the table are called **n-grams**.
The lookup table is a **Markov model**:
* _Model_: given a word and $n-1$ previous words, the POS tag can be looked (if the n-gram is in the corpus)
* _Markov assumption_: the POS tag depends only on the last $n$ words
Context can help:
> work $\rightarrow$ it can be noun or verb<br>
> my <u>work</u> $\rightarrow$ noun<br>
> I <u>work</u> $\rightarrow$ verb<br>
The longer the context, the better the prediction $-$ and larger the memory requirements.
#### Other models
Clearly, the Markov assumption does not hold for natural language, which has _long distance dependencies_. More powerful models exist:
* You can use the words after the target word
* but not the gold tags, not even before the target word.
* Temporarily, you can use previously predicted labels as if they were correct
* but in the end, test prediction cannot use any gold labels.
The algorithm is as follows:
* look up a given word with the surrounding words.
* If the sequence is in the table, then use the corresponding tag, or any of the tags.
* If not, then make a guess (usually: NOUN).
```
pos_lookup2 = OrderedDict()
for i in range(len(corpus)):
words_before = tuple(corpus[j][0] for j in range(max(i - 2, 0), i))
word_of_interest = corpus[i][0]
words_after = tuple(corpus[j][0] for j in range(i + 1, min(i + 3, len(corpus))))
if (words_before, word_of_interest, words_after) in pos_lookup2:
pos_lookup2[(words_before, word_of_interest, words_after)] |= {corpus[i][1]}
else:
pos_lookup2[(words_before, word_of_interest, words_after)] = {corpus[i][1]}
pos_lookup2
```
#### Problems
* **Out-of-vocabulary** (**OOV**) words:
* the word in the test set, but not in the train set
* an important metric in evaluation
* **Data sparsity**:
* a word can occur in many contexts, and not all can be in the train set
* inevitable: a language can generate infinitely many sentences
* countermeasures: _back off_ to smaller contexts (or just the target word)
For example if one takes the word _"the"_, then it is most certainly a determiner. However take the following segment:
> ... Steve Jobs met the Dalai Lama ...
This context might not have occured in the whole history of English (until now!), so it's not in our model. We need to back off:
> "Steve Jobs met", "the", "Dalai Lama" $\rightarrow$<br/>
> "Steve Jobs met", "the", "" $\rightarrow$<br/>
> "Jobs met", "the", "" $\rightarrow$<br/>
> "met", "the", "" $\rightarrow$<br/>
> "", "the", "" $\rightarrow$
Luckily in this case, $P(DET|the)\approx1$.
### Hidden Markov Model
* Like the Markov model, we take only the $n$ preceding tokens into consideration
* The idea behind the model is very different:
* We imagine an automaton that is always in a **(hidden) state**
* In each state, it emits something we can observe
* The task is to find out which is _the most probable_ state sequence that generates the observations
* In the POS tagging context,
* The words in the text are the **observed events**
* The POS tags are the hidden states
That is, we will think of a sentence as the sequence of POS tags, for which the actual choice of words is rather arbitrary.
|PRON|VERB |PRON|PREP|NOUN|PUNCT|
|---|-----|-----|----|--------|--|
| I | saw | you | in | school | .|
|You| saw | me | in | school | .|
|You| met | me | in | school | .|
|You| met | me | in | work | .|
|We| met |him | at | work | .|
We are presented with one of the sentences and the task is to reconstruct the POS sequence at the top.
#### Probabilistic model
The HMM is a probabilistic model. Translating "looking for the tag sequence that best explains the sentence" to mathematical notation gives us
$$
{\arg\max}_{l_i\in L}\mathbb{P}(w_1, w_2, \ldots w_N \ | \ l_1, l_2 \ldots l_N)
$$
, i.e.
* $\mathbb{P}(w_1, w_2, \ldots w_N \ | \ l_1, l_2 \ldots l_N)$ is the probability that a particular $l_1, \ldots, l_N$ sequence generated the sentence $w_1, \ldots, w_N$
* ${\arg\max}_{l_i}$ looks for the $l$ sequence that produces the highest probability.
<!-- THIS SHOULD GO TO ML BASICS NEXT YEAR.
* We are given a list of probabilities ($N$ is the sentence length)
$$
\mathbb{P}(w_1, w_2, \ldots w_N, l_1, l_2 \ldots l_N)
$$
For example,
$$
\mathbb{P}(\text{the}, \text{dog}, \text{saw}, \text{the}, \text{cat}, \text{DET}, \text{NOUN}, \text{VERB}, \text{DET}, \text{NOUN})
$$ -->
Without any restriction on the probability,
* the only way to find the best state sequence is to compute all probabilities
* this incurs $\mathcal{O}(|L|^N)$ complexity
* for a sentence of 15 words, this is already around 14M numbers to compute!
In HMM, we make three assumptions to make the computation tractable:
1. The current POS tag only depends on a fix window of $n$ tokens to the past; $n$ is a parameter of the model (the _Markov assumption_)
1. The current word only depends on the current POS tag
1. The words are generated independently of one another
| I | saw | you | in | school | .|
|---|:-----|-----|----:|--------|--|
|DET|VERB |PRON|PREP|NOUN|PUNCT|
| | | | $w_i$ |||
| |$l_{i-2}$|$l_{i-1}$|$l_i$|||
| | [ |window| ] ||
With this, the probability can be decomposed as:
$$
\mathbb{P}(w_1, w_2, \ldots w_N \ | \ l_1, l_2 \ldots l_N) =
\prod_{i=1}^N\mathbb{P}(l_i \ |\ l_{i-n+1}, l_{i-n+2}\ldots l_{i-1})\cdot\mathbb{P}(w_i \ | \ l_i)
$$
* The term $\mathbb{P}(l_i \ |\ l_{i-n+1},l_{i-n+2}\ldots l_{i-1})$ is the **transition probability**: the probability of tag $l_i$, given the previous $n-1$ tags.
* The terms $\mathbb{P}(w_i|l_i)$ are the **emission probabilities**: the probability of a word given its POS tag.
#### Training
<a id="hmm_training"></a>
The transition probabilities are estimated from the n-gram frequencies in the corpus.
$$\mathbb{P}(l_n\ |\ l_1,l_2\ldots l_{n-1})\approx \frac{\#\{l_1,l_2\ldots l_{n-1},l_n\}}{\#\{l_1,l_2\ldots l_{n-1}\}}$$
If the index not positive (at the beginning of sentences), then we simply omit it and fall back to a lower n-gram order. If normally we are estimating trigrams ($n=3$), we fall back to bi- and unigrams:
$$
\begin{split}
\mathbb{P}(l_2\ |\ l_0, l_1) & \approx \mathbb{P}(l_2\ |\ l_1) & = \frac{\#\{l_1,l_2\}}{\#\{l_1\}} \quad \text{and } \\
\mathbb{P}(l_1\ |\ l_{-1},l_0) & \approx \mathbb{P}(l_1) & = \frac{\#\{l_1\}}{\text{# of words}}
\end{split}
$$
Emission probabilities are computed similarly:
$$ \mathbb{P}(w_i|l_i) \approx \frac{\#\{\text{word }w_i \text{ with the tag }l_i\}}{\#\{l_i\}}$$
The effect of data sparsity on HMM is much smaller than on an n-gram model:
* The size of the lookup table depends on the size of the tagset ($\mathcal{O}(|L|^n)$, not $\mathcal{O}(|V|^n)$)
* The size of the tagset is way smaller than the vocabulary ($|L| \ll |V|$)
* This results in a much denser table.
<!-- For example if you see the word _"dog"_ many times in the corpus, and never with the POS tag _DET_ then you can be pretty sure that it will never have that tag. -->
#### Inference
* At prediction (inference) time, given a sequence of words, we need to find the ${\arg\max}$.
* The naive algorithm would just compute the probabilities for all $l_1, \ldots, l_N$, incurring $\mathcal{O}(|L|^N)$ complexity.
* Because of how we decomposed the probability, we can use a more efficient algorithm.
**Dynamic Programming (DP)** algorithms solve complex problems by breaking it up into smaller sub-problems.
#### Viterbi algorithm
The **Viterbi algorithm** has two phases:
* **Forward phase**: goes through the sentence from beginning to end and fills the probability and backpointer tables;
* **Backward phase**: follows the backpointers to find the most probably state sequence.
We detail the case of bigrams, $n=2$. The input of the algorithm:
* the sentence $W = \{w_1, \ldots,w_N\}$
* the hidden state space $S=\{s_1, \ldots , s_{|L|}\}$ (the POS tags)
* the transition probabilities $T_{i,j} = \mathbb{P}(l_t=s_j|l_{t-1}=s_i)$
* the emission probabilities $E_{i,j} = \mathbb{P}(w_j|l_t=s_i)$
The algorithm outputs two ($|L| \times N$) tables:
* $\pi[i,j]$: the probability of the most likely hidden state sequence that ends with $l_j=s_i$
* $B[i,j]$: the backpointers along the most probable transitions
Since the algorithm only fills these two tables, the complexity is only $\mathcal{O}(N \cdot |L|^n)$.
#### Example
We demonstrate the algorithm on the sentence
|DET|NOUN|VERB|DET|NOUN|
|---|---|---|-|---|
|the|dog|saw|a|cat|
The tables $\pi$ and $B$ then look like (the stars show the state sequence we are looking for):
| |the|dog|saw|a|cat|
|-|---|---|---|-|---|
|**DET**| *| | | *| |
|**NOUN**|| *| | | *|
|**VERB**|| | *| | |
The tables are filled left-to-right, column-by-column.
$\pi$ implements the computation:
$$
\mathbb{P}(w_1, w_2, \ldots w_N \ | \ l_1, l_2 \ldots l_N) =
\prod_{t=1}^N\mathbb{P}(l_t \ |\ l_{t-1})\cdot\mathbb{P}(w_t \ | \ l_t)
$$
Note this is the same as before, but
* $n = 2$, so the transition probability is simpler
* the running index to $t$
In particular, the cell $\pi[i,j]$ records the probability of
* the most likely state sequence $l_1, \ldots, l_j$
* that produced the words $w_1, \ldots, w_j$.
To compute that probability, we need
* the probability of the path thus far. Since we don't know the previous state, we need to consider all cells in the previous column $\pi[*,j-1]$
* the emission probability for the current cell $E_{i,j}$
* the transition probabilities between the current cell and a cell in the previous column $T_{*,i}$
* finally, we take the maximum of the above.
Putting that to equations,
$$
\begin{split}
\pi[i,j] & = \max_{l_1, \ldots, l_j}\mathbb{P}(w_1, \ldots, w_j \ |\ l_1, \ldots, l_j) \\
& = \max_{l_t, \forall t}\prod_{t=1}^j\mathbb{P}(l_t \ |\ l_{t-1})\cdot\mathbb{P}(w_t \ | \ l_t) \\
& = \max_{l_t, \forall t} \prod_{t=1}^{j-1}\mathbb{P}(l_t \ |\ l_{t-1})\cdot\mathbb{P}(w_t \ | \ l_t)
& \max_k \mathbb{P}(l_j=s_i | l_{j-1}=s_k) & \cdot \mathbb{P}(w_j | l_j=s_i) \\
& = \max_k \pi[k,j-1] & \cdot \mathbb{P}(l_j=s_i | l_{j-1}=s_k) & \cdot \mathbb{P}(w_j | l_j=s_i) \\
& = \max_k \pi[k,j-1] & \cdot T_{k,j} & \cdot E_{i,j}
\end{split}
$$
In the end, the maximum value in the last column $\max_{i \in |L|}\pi[i,N]$ is the probability of the most likely tag sequence.
* This tells us the most probable state (POS tag) to end the sequence.
* But what about the rest?
<a id="aha">This</a> is where table $B$ enters the picture:
* when computing $\pi[i,j]$, we note which state transition $k \rightarrow i$ resulted in the highest probability
* we store that information in $B[i,j]$ as **backpointer** to $B[k, j-1]$.
In the backward phase, all we have to do is follow the backpointers from $B[i,N]$ ($i$ is where $\pi[i,N]$ takes its maximum) to find all most probable state sequence.
The algorithm in pseudocode:
<!-- set $\pi[i,0] = 1, \forall i \in 1 \ldots |L|$<br/>-->
> begin for $j = 1 \ldots N$<br/>
> $\quad$ begin for $i = 1 \ldots |L|$<br/>
> $\quad \quad$ $\pi[i,j] = \max_{k \in 1:|L|} \pi[k,j-1]\cdot T_{k,i} \cdot E_{i,j}$<br/>
> $\quad \quad$ $B[i,j]={\arg\max}_k \pi[k,j-1] \cdot T_{k,i}$<br/>
> $\quad$ end for<br/>
> end for
#### Notes about the algorithm
1. When computing $T_{i,1}$, there is no state to transition from. Use the unigram probabilities in this case.
1. Note that the uni- and bigrams collected in the [HMM Training slide](#hmm_training) are not the general uni- or bigram distributions, as we only collect $n-1$, $n-2$, etc.-grams at the beginning of sentences. This is the reason why you can use the unigram distribution for $T_{i,1}$ $-$ and also why you should use them **only** there.
1. The pages above described the Viterbi algorithm for bigram ($n=2$) transition probabilities. When going to $n=3$ (and beyond):
* the transition probabilities have the form $\mathbb{P}(l_t\ |\ l_{t-1},l_{t-2})$
* at the beginning of the sentence, use the unigram distribution for the first word and the bigram for the second
* consequently, the tables must have $n$ dimensions: $|L|\times|L|\times N$ for $n=3$, etc.
* let's say that the second dimension corresponds to the current tag, the first to the previous one
* the update rule then changes as: $\pi[i,j,k] = \max_l \pi[l,j,k-1] \cdot T_{l,i,j} \cdot E_{i,j}$
1. See more: https://courses.engr.illinois.edu/cs447/fa2017/Slides/Lecture07.pdf
|
github_jupyter
|
In[1]: tokenize("Mr. and Mrs. Dursley of Number Four, Privet Drive, were proud to say that they were perfectly normal, thank you very much.")
Out[1]: ['Mr.', 'and', 'Mrs.', 'Dursley', 'of', 'Number', 'Four', ',', 'Privet', 'Drive', ',', 'were', 'proud', 'to', 'say','that', 'they', 'were', 'perfectly', 'normal', ',', 'thank', 'you', 'very', 'much', '.']
In[1]: ssplit(['Me', 'Tarzan', '.', 'You', 'Jane', '.'])
Out[2]: [['Me', 'Tarzan', '.'],
['You', 'Jane', '.']]
vocabulary = {w:i for i, w in enumerate(set(x.split()))}
print(vocabulary)
print("------------------")
print(words)
print([vocabulary[w] for w in words])
#Lemmas that are N or V: Inf if preposition precedes it
SELECT Inf IF (0 Inf) (0 N) (-1 To) ;
SELECT Inf IF (0 Inf) (0 N) (-1 Adv) (-2 To) ;
from itertools import chain
from collections import defaultdict, OrderedDict
# Two inputs: the corpus and n
corpus=list(zip(["The", "quick", "brown", "fox", "jumps", "over", "the", "lazy", "dog", "."],
["DET", "ADJ", "ADJ", "NOUN", "VERB", "ADP", "DET", "ADJ", "NOUN", "PUNCT"]))
n = 3
print('The corpus:', corpus, sep='\n')
pos_lookup = OrderedDict([(tuple(corpus[j][0] for j in range(max(i - 2, 0), i + 1)),
corpus[i][1])
for i in range(len(corpus))])
print('\nThe lookup table:')
pos_lookup
pos_lookup2 = OrderedDict()
for i in range(len(corpus)):
words_before = tuple(corpus[j][0] for j in range(max(i - 2, 0), i))
word_of_interest = corpus[i][0]
words_after = tuple(corpus[j][0] for j in range(i + 1, min(i + 3, len(corpus))))
if (words_before, word_of_interest, words_after) in pos_lookup2:
pos_lookup2[(words_before, word_of_interest, words_after)] |= {corpus[i][1]}
else:
pos_lookup2[(words_before, word_of_interest, words_after)] = {corpus[i][1]}
pos_lookup2
| 0.320502 | 0.975785 |
# Self-Driving Car Engineer Nanodegree
## Deep Learning
## Project: Build a Traffic Sign Recognition Classifier
In this project, I utilized TensorFlow and Python to implement a deep learning pipeline to classify traffic signs for self-driving cars. I used convolutional neural networks to dramatically improve performance in image classification. I achieved a 94% testing accuracy with the help of various machine learning techniques such as regularization, dropout, pooling.
---
## Step 0: Load The Data
```
# Load pickled data
import pickle
training_file = "../data/train.p"
validation_file= "../data/valid.p"
testing_file = "../data/test.p"
with open(training_file, mode='rb') as f:
train = pickle.load(f)
with open(validation_file, mode='rb') as f:
valid = pickle.load(f)
with open(testing_file, mode='rb') as f:
test = pickle.load(f)
x_train, y_train = train['features'], train['labels']
x_valid, y_valid = valid['features'], valid['labels']
x_test, y_test = test['features'], test['labels']
print("Done")
# Convert to numpy arrays
import numpy as np
np_x_train = np.array(x_train)
np_y_train = np.array(y_train)
np_x_valid = np.array(x_valid)
np_y_valid = np.array(y_valid)
np_x_test = np.array(x_test)
np_y_test = np.array(y_test)
print("Done")
```
---
## Step 1: Dataset Summary & Exploration
The pickled data is a dictionary with 4 key/value pairs:
- `'features'` is a 4D array containing raw pixel data of the traffic sign images, (num examples, width, height, channels).
- `'labels'` is a 1D array containing the label/class id of the traffic sign. The file `signnames.csv` contains id -> name mappings for each id.
- `'sizes'` is a list containing tuples, (width, height) representing the original width and height the image.
- `'coords'` is a list containing tuples, (x1, y1, x2, y2) representing coordinates of a bounding box around the sign in the image. **THESE COORDINATES ASSUME THE ORIGINAL IMAGE. THE PICKLED DATA CONTAINS RESIZED VERSIONS (32 by 32) OF THESE IMAGES**
```
import numpy as np
# Number of training examples
n_train = len(np_x_train)
# Number of validation examples
n_validation = len(np_x_valid)
# Number of testing examples.
n_test = len(np_x_test)
# What's the shape of an traffic sign image?
image_shape = np_x_train.shape
# How many unique classes/labels there are in the dataset.
n_classes = len(set(np.hstack((np_y_train, np_y_valid, np_y_test))))
print("Number of training examples =", n_train)
print("Number of validation examples =", n_validation)
print("Number of testing examples =", n_test)
print("Image data shape =", image_shape)
print("Number of classes =", n_classes)
```
### Exploratory visualization of the dataset
```
# Load class names
import pandas as pd
df = pd.read_csv("signnames.csv")
class_names = df["SignName"]
print(class_names[:5])
### Data exploration visualization code goes here.
### Feel free to use as many code cells as needed.
import matplotlib.pyplot as plt
from PIL import Image
# Visualizations will be shown in the notebook.
%matplotlib inline
from random import sample
# Show some train images
sample_inds = sample(range(0, len(np_x_train)), 25)
np_x_train_samples = np_x_train[sample_inds]
np_y_train_samples = np_y_train[sample_inds]
plt.figure(figsize=(15, 15))
for i in range(25):
plt.subplot(5, 5, i + 1)
plt.xticks([])
plt.yticks([])
plt.grid(False)
plt.imshow(np_x_train_samples[i], cmap = plt.cm.binary)
plt.xlabel(class_names[np_y_train_samples[i]])
plt.show()
np_y_train = np.array(y_train)
plt.hist(np_y_train, bins=n_classes - 1)
np_y_test = np.array(y_test)
plt.hist(np_y_test,bins=n_classes - 1)
```
----
## Step 2: Design and Test a Model Architecture
Design and implement a deep learning model that learns to recognize traffic signs. Train and test your model on the [German Traffic Sign Dataset](http://benchmark.ini.rub.de/?section=gtsrb&subsection=dataset).
### Pre-process the Data Set
```
def normalize(image_set):
""" normalize so that image data has mean zero and equal variance """
return (image_set - 128.0) / 128.0
train_X_norm = normalize(np_x_train)
valid_X_norm = normalize(np_x_valid)
test_X_norm = normalize(np_x_test)
```
### Model Architecture
```
# Tunable parameters
dropout_rate = 0.2
from tensorflow.keras import datasets, layers, models
import matplotlib.pyplot as plt
import tensorflow as tf
model = models.Sequential()
model.add(layers.Conv2D(32, (3, 3), activation='relu', input_shape=(32, 32, 3)))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Dropout(dropout_rate, input_shape=(15, 15, 32)))
model.add(layers.Conv2D(64, (3, 3), activation='relu'))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Dropout(dropout_rate, input_shape=(6, 6, 64)))
model.add(layers.Conv2D(64, (3, 3), activation='relu'))
model.add(layers.Dropout(dropout_rate, input_shape=(4, 4, 64)))
model.add(layers.Flatten())
model.add(layers.Dense(128, activation='relu'))
model.add(layers.Dense(43))
# print model summary
model.summary()
```
### Train, Validate and Test the Model
A validation set can be used to assess how well the model is performing. A low accuracy on the training and validation
sets imply underfitting. A high accuracy on the training set but low accuracy on the validation set implies overfitting.
```
### Train the model
model.compile(optimizer='adam',
loss=tf.keras.losses.SparseCategoricalCrossentropy(from_logits=True),
metrics=['accuracy'])
history = model.fit(train_X_norm, np_y_train, epochs=10,
validation_data=(valid_X_norm, np_y_valid))
plt.plot(history.history['accuracy'], label='accuracy')
plt.plot(history.history['val_accuracy'], label = 'val_accuracy')
plt.xlabel('Epoch')
plt.ylabel('Accuracy')
plt.ylim([0.5, 1])
plt.legend(loc='lower right')
# evaluate the model
test_loss, test_acc = model.evaluate(test_X_norm, np_y_test, verbose=2)
print("Test Accuracy = {:.3f}".format(test_acc))
```
---
## Step 3: Test a Model on New Images
Test the model on five pictures of German traffic signs from the web.
### Load and Output the Images
```
test_image_file_names = os.listdir("./images/web")
print(test_image_file_names)
### Load 25 test images
image_file_names = os.listdir("./images/web")
plt.figure(figsize=(15, 15))
images = []
for image_file_name in image_file_names:
image = plt.imread("./images/web/" + image_file_name)
images.append(image)
for i in range(0, 5):
plt.subplot(1, 5, i + 1)
plt.xticks([])
plt.yticks([])
plt.grid(False)
plt.imshow(images[i], cmap = plt.cm.binary)
i += 1
plt.show()
```
### Predict the Sign Type for Each Image
```
print(np.array(images).shape)
### Run the predictions here and use the model to output the prediction for each image.
pred = model.predict(np.array(images))
print(pred[0])
```
### Analyze Performance
```
### Calculate the accuracy for these 5 new images.
def get_sample_test_accuracy(prediction, actual):
res = 0
size = len(prediction)
for i in range(0, size):
if prediction[i] == actual[i]:
res += 1
return res / size
prediction = np.argmax(pred, axis=1)
actual = [1, 34, 3, 38, 18]
print("Test accuracy on the 5 samples is: ", get_sample_test_accuracy(prediction, actual))
```
### Output Top 5 Softmax Probabilities For Each Image Found on the Web
```
### Print out the top five softmax probabilities for the predictions on the German traffic sign images found on the web.
print(tf.nn.top_k(tf.nn.softmax(pred), 5))
```
|
github_jupyter
|
# Load pickled data
import pickle
training_file = "../data/train.p"
validation_file= "../data/valid.p"
testing_file = "../data/test.p"
with open(training_file, mode='rb') as f:
train = pickle.load(f)
with open(validation_file, mode='rb') as f:
valid = pickle.load(f)
with open(testing_file, mode='rb') as f:
test = pickle.load(f)
x_train, y_train = train['features'], train['labels']
x_valid, y_valid = valid['features'], valid['labels']
x_test, y_test = test['features'], test['labels']
print("Done")
# Convert to numpy arrays
import numpy as np
np_x_train = np.array(x_train)
np_y_train = np.array(y_train)
np_x_valid = np.array(x_valid)
np_y_valid = np.array(y_valid)
np_x_test = np.array(x_test)
np_y_test = np.array(y_test)
print("Done")
import numpy as np
# Number of training examples
n_train = len(np_x_train)
# Number of validation examples
n_validation = len(np_x_valid)
# Number of testing examples.
n_test = len(np_x_test)
# What's the shape of an traffic sign image?
image_shape = np_x_train.shape
# How many unique classes/labels there are in the dataset.
n_classes = len(set(np.hstack((np_y_train, np_y_valid, np_y_test))))
print("Number of training examples =", n_train)
print("Number of validation examples =", n_validation)
print("Number of testing examples =", n_test)
print("Image data shape =", image_shape)
print("Number of classes =", n_classes)
# Load class names
import pandas as pd
df = pd.read_csv("signnames.csv")
class_names = df["SignName"]
print(class_names[:5])
### Data exploration visualization code goes here.
### Feel free to use as many code cells as needed.
import matplotlib.pyplot as plt
from PIL import Image
# Visualizations will be shown in the notebook.
%matplotlib inline
from random import sample
# Show some train images
sample_inds = sample(range(0, len(np_x_train)), 25)
np_x_train_samples = np_x_train[sample_inds]
np_y_train_samples = np_y_train[sample_inds]
plt.figure(figsize=(15, 15))
for i in range(25):
plt.subplot(5, 5, i + 1)
plt.xticks([])
plt.yticks([])
plt.grid(False)
plt.imshow(np_x_train_samples[i], cmap = plt.cm.binary)
plt.xlabel(class_names[np_y_train_samples[i]])
plt.show()
np_y_train = np.array(y_train)
plt.hist(np_y_train, bins=n_classes - 1)
np_y_test = np.array(y_test)
plt.hist(np_y_test,bins=n_classes - 1)
def normalize(image_set):
""" normalize so that image data has mean zero and equal variance """
return (image_set - 128.0) / 128.0
train_X_norm = normalize(np_x_train)
valid_X_norm = normalize(np_x_valid)
test_X_norm = normalize(np_x_test)
# Tunable parameters
dropout_rate = 0.2
from tensorflow.keras import datasets, layers, models
import matplotlib.pyplot as plt
import tensorflow as tf
model = models.Sequential()
model.add(layers.Conv2D(32, (3, 3), activation='relu', input_shape=(32, 32, 3)))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Dropout(dropout_rate, input_shape=(15, 15, 32)))
model.add(layers.Conv2D(64, (3, 3), activation='relu'))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Dropout(dropout_rate, input_shape=(6, 6, 64)))
model.add(layers.Conv2D(64, (3, 3), activation='relu'))
model.add(layers.Dropout(dropout_rate, input_shape=(4, 4, 64)))
model.add(layers.Flatten())
model.add(layers.Dense(128, activation='relu'))
model.add(layers.Dense(43))
# print model summary
model.summary()
### Train the model
model.compile(optimizer='adam',
loss=tf.keras.losses.SparseCategoricalCrossentropy(from_logits=True),
metrics=['accuracy'])
history = model.fit(train_X_norm, np_y_train, epochs=10,
validation_data=(valid_X_norm, np_y_valid))
plt.plot(history.history['accuracy'], label='accuracy')
plt.plot(history.history['val_accuracy'], label = 'val_accuracy')
plt.xlabel('Epoch')
plt.ylabel('Accuracy')
plt.ylim([0.5, 1])
plt.legend(loc='lower right')
# evaluate the model
test_loss, test_acc = model.evaluate(test_X_norm, np_y_test, verbose=2)
print("Test Accuracy = {:.3f}".format(test_acc))
test_image_file_names = os.listdir("./images/web")
print(test_image_file_names)
### Load 25 test images
image_file_names = os.listdir("./images/web")
plt.figure(figsize=(15, 15))
images = []
for image_file_name in image_file_names:
image = plt.imread("./images/web/" + image_file_name)
images.append(image)
for i in range(0, 5):
plt.subplot(1, 5, i + 1)
plt.xticks([])
plt.yticks([])
plt.grid(False)
plt.imshow(images[i], cmap = plt.cm.binary)
i += 1
plt.show()
print(np.array(images).shape)
### Run the predictions here and use the model to output the prediction for each image.
pred = model.predict(np.array(images))
print(pred[0])
### Calculate the accuracy for these 5 new images.
def get_sample_test_accuracy(prediction, actual):
res = 0
size = len(prediction)
for i in range(0, size):
if prediction[i] == actual[i]:
res += 1
return res / size
prediction = np.argmax(pred, axis=1)
actual = [1, 34, 3, 38, 18]
print("Test accuracy on the 5 samples is: ", get_sample_test_accuracy(prediction, actual))
### Print out the top five softmax probabilities for the predictions on the German traffic sign images found on the web.
print(tf.nn.top_k(tf.nn.softmax(pred), 5))
| 0.765593 | 0.983518 |
```
import pandas as pd
from pathlib import Path
from sklearn.ensemble import RandomForestRegressor
from sklearn.pipeline import make_pipeline
from imblearn import over_sampling as ovs
from sklearn.decomposition import PCA
import matplotlib.pyplot as plt
import seaborn as sns
import numpy as np
from sklearn.metrics import mean_squared_error,mean_absolute_error,r2_score
from sklearn.preprocessing import StandardScaler
from sklearn import metrics
from sklearn.externals import joblib
from sklearn.feature_selection import VarianceThreshold
from sklearn.model_selection import GridSearchCV
from sklearn.model_selection import train_test_split
%matplotlib inline
sns.set_context(context="paper")
def plot_test_reg(y_true,y_pred):
from matplotlib.offsetbox import AnchoredText
rmse_test = np.round(np.sqrt(mean_squared_error(y_true, y_pred)), 3)
df_pred = pd.DataFrame({"Predicted ddG(kcal/mol)": y_pred, "Actual ddG(kcal/mol)": y_true})
pearsonr_test = round(df_pred.corr().iloc[0,1],3)
g = sns.regplot(x="Actual ddG(kcal/mol)", y="Predicted ddG(kcal/mol)",data=df_pred)
plt.title("Predicted vs Experimental ddG (Test set: %s complexes)"%y_true.shape[0])
anchored_text1 = AnchoredText("pearsonr = %s" %pearsonr_test, loc=1)
anchored_text2 = AnchoredText("RMSE = %s" %rmse_test, loc=4)
g.add_artist(anchored_text1)
g.add_artist(anchored_text2)
from sklearn.model_selection import learning_curve
def learning_curves(estimator, features, target, cv):
train_sizes, train_scores, validation_scores = learning_curve(
estimator, features, target,train_sizes = np.linspace(.1, 1.0, 10),
cv = cv, scoring = 'neg_mean_squared_error',n_jobs=-1)
train_scores_mean = -train_scores.mean(axis = 1)
validation_scores_mean = -validation_scores.mean(axis = 1)
plt.plot(train_sizes, np.sqrt(train_scores_mean), label = 'Training error')
plt.plot(train_sizes, np.sqrt(validation_scores_mean), label = 'Validation error')
plt.ylabel('RMSE', fontsize = 14)
plt.xlabel('Training set size', fontsize = 14)
title = 'Learning curve'
plt.title(title, fontsize = 18, y = 1.03)
plt.legend()
plt.ylim(0,3)
ABPRED_DIR = Path().cwd().parent
DATA = ABPRED_DIR / "data"
#dataframe final
df_final = pd.read_csv(DATA/"../data/DF_features_400_2019.csv",index_col=0)
# Quitar modelos por homologia deltraining set
#df_final_onlyHM = df_final.loc[df_final.index.str.startswith("HM")]
#df_final= df_final.loc[~df_final.index.str.startswith("HM")]
index_ddg8 = (df_final['ddG(kcal/mol)']==8)
df_final = df_final.loc[-index_ddg8]
#testiar eliminando estructuras con ddg menor o igual a -4 kcal/mol , outliers
index_ddg_4 = (df_final['ddG(kcal/mol)'] <= -4)
df_final = df_final.loc[-index_ddg_4]
pdb_names = df_final.index
features_names = df_final.drop('ddG(kcal/mol)',axis=1).columns
import math
def deltaG_to_Kd(delg):
Kd_value = math.exp((delg*1000)/(1.98*298.15))
return Kd_value
#X = df_final_f1 # con filtrado de varianza
X = df_final.drop('ddG(kcal/mol)',axis=1)
y = df_final['ddG(kcal/mol)']
#y_pKd = -y.apply(deltaG_to_Kd).apply(math.log10)
#y = y_pKd
X_train, X_test, y_train, y_test = train_test_split(X, y,train_size=0.8,random_state=12)
```
max_depth=7 , min_samples_leaf=1, min_samples_split=2,
```
#1)
scaler = StandardScaler()
#2)
rfr = RandomForestRegressor()
#3) Crear pipeline
pipeline1 = make_pipeline(scaler ,rfr)
# Create the parameter grid based on the results of random search
param_grid = {
'randomforestregressor__bootstrap': [True],
'randomforestregressor__max_depth': [2,5,15,25,30,None],
'randomforestregressor__max_features': ["log2","sqrt","auto"],
'randomforestregressor__max_leaf_nodes':[50,60,70,80,90],
'randomforestregressor__min_samples_leaf': [1, 2, 5],
'randomforestregressor__min_samples_split': [2,5,10,15],
'randomforestregressor__n_estimators': [100,300,500,1000],
'randomforestregressor__random_state':[1],
}
grid1 = GridSearchCV(pipeline1,param_grid, verbose=1,scoring="neg_mean_squared_error",cv=10,n_jobs=-1)
# fit
grid1.fit(X_train, y_train)
print(grid1.best_params_)
print('Training score (r2): {}'.format(grid1.best_estimator_.score(X_train, y_train)))
print('Test score (r2): {}'.format(grid1.best_estimator_.score(X_test, y_test)))
y_pred = grid1.best_estimator_.predict(X_test)
print("\nRoot mean square error for test dataset: {}".format(np.round(np.sqrt(mean_squared_error(y_test, y_pred)), 2)))
print("pearson corr:\n",np.corrcoef(y_pred,y_test)[0][1])
plot_test_reg(y_pred=y_pred,y_true=y_test)
learning_curves(grid1.best_estimator_,X,y,10)
```
# Apply SMOTE, only on train data, and test only one with best parameters
```
# forma 1
df_final['class'] = np.where((df_final['ddG(kcal/mol)'] < -1) | (df_final['ddG(kcal/mol)'] > 3.0),1,0)
# forma 2
#df_final['class'] = np.where((df_final['ddG(kcal/mol)'] < 0),1,0)
print("Before OverSampling, counts of label '1': {}".format(sum(df_final['class'] == 1)))
print("Before OverSampling, counts of label '0': {}".format(sum(df_final['class'] == 0)))
X = df_final.drop('class',axis=1)
y = df_final['class']
X_train, X_test, y_train, y_test = train_test_split(X, y,stratify = y,train_size=0.7,random_state=12)
#smote = ovs.SMOTE(random_state=12)
#smote = ovs.BorderlineSMOTE(random_state=12)
adasyn = ovs.ADASYN(random_state=12,n_neighbors=5)
X_train_re , y_train_re = adasyn.fit_sample(X_train,y_train)
#X_train_re , y_train_re = smote.fit_sample(X_train,y_train)
# back to originalk shape and target
X_train_normal = X_train_re[:,:-1]
y_train_normal = X_train_re[:,-1]
X_test_normal = X_test.iloc[:,:-1]
y_test_normal = X_test.iloc[:,-1]
#1)
scaler = StandardScaler()
#2)
rfr = RandomForestRegressor(bootstrap=True, max_depth=None,max_features='sqrt',max_leaf_nodes=90,
min_samples_leaf=1,min_samples_split=2, n_estimators=500,random_state=1)
#3) Crear pipeline
pipe = make_pipeline(scaler ,rfr)
pipe.fit(X_train_normal,y_train_normal)
print('Training score (r2): {}'.format(pipe.score(X_train_normal, y_train_normal)))
print('Test score (r2): {}'.format(pipe.score(X_test_normal, y_test_normal)))
y_test_pred = pipe.predict(X_test_normal)
y_train_pred = pipe.predict(X_train_normal)
print("\nRoot mean square error for train dataset: {}".format(np.round(np.sqrt(mean_squared_error(y_train_normal, y_train_pred)), 2)))
print("\nRoot mean square error for test dataset: {}".format(np.round(np.sqrt(mean_squared_error(y_test_normal, y_test_pred)), 2)))
print("pearson corr:\n",np.corrcoef(y_test_pred,y_test_normal)[0][1])
plot_test_reg(y_pred=y_test_pred,y_true=y_test_normal)
learning_curves(pipe,X_train_normal,y_train_normal,10)
grid_search.fit(X_train, y_train)
model = grid_search.best_estimator_
model.fit(X_train, y_train)
joblib.dump(model, '../../../../GDrive/RFmodel_400.10cv.v4.pkl')
f = open("rf_models400F_Fullsearch.txt","a")
print(grid_search.best_estimator_)
f.write('10 fold\n'+str(grid_search.best_estimator_))
print('\nBest GridSearchCV Score 5fold : ' + str(-grid_search.best_score_))
model = grid_search.best_estimator_
model.fit(X_train, y_train)
y_pred_train = model.predict(X_train)
y_pred_test = model.predict(X_test)
#print("OOB Score : " + str(model.oob_score_)) # yg diitung r2 score
print('Train MSE : ' + str(mean_squared_error(y_train, y_pred_train)))
print('Train MAE : ' + str(mean_absolute_error(y_train, y_pred_train)))
print('Train R^2 : ' + str(r2_score(y_train, y_pred_train)))
print('Test R^2 : ' + str(r2_score(y_test, y_pred_test)))
print('Test MSE : ' + str(mean_squared_error(y_test, y_pred_test)))
print('Test MAE : ' + str(mean_absolute_error(y_test, y_pred_test)))
print("\nRoot mean square error for test dataset: {}".format(np.round(np.sqrt(mean_squared_error(y_test, y_pred_test)), 2)))
print("pearson corr:\n",np.corrcoef(y_pred_test,y_test)[0][1])
f.write('\nTrain MSE : ' + str(mean_squared_error(y_train, y_pred_train)))
f.write('\nTrain MAE : ' + str(mean_absolute_error(y_train, y_pred_train)))
f.write('\nTrain R^2 : ' + str(r2_score(y_train, y_pred_train)))
f.write('\nTest R^2 : ' + str(r2_score(y_test, y_pred_test)))
f.write('\nTest MSE : ' + str(mean_squared_error(y_test, y_pred_test)))
f.write('\nTest MAE : ' + str(mean_absolute_error(y_test, y_pred_test)))
f.write("\nRoot mean square error for test dataset: {}".format(np.round(np.sqrt(mean_squared_error(y_test, y_pred_test)), 2)))
f.write("\npearson corr:\n"+str(np.corrcoef(y_pred_test,y_test)))
f.close()
```
## Salvar modelo final, entrenado con el total de lso datos
```
# save final model
final_model = grid_search.best_estimator_
final_model.fit(X,y)
joblib.dump(final_model, 'RFmodel_400.v2.pkl')
rmse_test = np.round(np.sqrt(mean_squared_error(y_test, y_pred_test)), 3)
df_pred = pd.DataFrame({"Predicted ddG(kcal/mol)": y_pred_test, "Actual ddG(kcal/mol)": y_test.values})
pearsonr_test = round(df_pred.corr().iloc[0,1],3)
g = sns.regplot(x="Actual ddG(kcal/mol)", y="Predicted ddG(kcal/mol)",data=df_pred)
plt.title("Predicted vs Experimental ddG (Independent set: 123 complexes)")
plt.text(-2,3,"pearsonr = %s" %pearsonr_test)
plt.text(4.5,-0.5,"RMSE = %s" %rmse_test)
#plt.savefig("RFmodel_300_testfit.png",dpi=600)
df_train_pred = pd.DataFrame({"Predicted ddG(kcal/mol)": y_train, "Actual ddG(kcal/mol)": y_pred_train})
pearsonr_train = round(df_train_pred.corr().iloc[0,1],3)
rmse_train = np.round(np.sqrt(mean_squared_error(y_train, y_pred_train)), 3)
g = sns.regplot(x="Actual ddG(kcal/mol)", y="Predicted ddG(kcal/mol)",data=df_train_pred)
plt.text(-0.4,6.5,"pearsonr = %s" %pearsonr_train)
plt.text(3.5,-2.5,"RMSE = %s" %rmse_train)
plt.title("Predicted vs Experimental ddG (Train set: 492 complexes)")
#plt.savefig("RFmodel_300_trainfit.png",dpi=600)
importances = list(model.feature_importances_)
feature_list = df_final.columns
# List of tuples with variable and importance
feature_importances = [(feature, round(importance, 4)) for feature, importance in zip(feature_list, importances)]
# Sort the feature importances by most important first
feature_importances = sorted(feature_importances, key = lambda x: x[1], reverse = True)
# Print out the feature and importances
[print('Variable: {:20} Importance: {}'.format(*pair)) for pair in feature_importances]
idx_features = model.feature_importances_.argsort()[::-1]
plt.figure(figsize=(15,4))
plt.bar(np.arange(X_train.shape[1]), model.feature_importances_[idx_features])
plt.xticks(range(len(feature_list)),feature_list[idx_features])
plt.autoscale(enable=True, axis='x', tight=True)
plt.xlabel(u"Feature importance")
#plt.savefig("RFmodel_300_50features",dpi=600,bbox_inches="tight")
residual = y_test.values - y_pred_test
plt.scatter(x=y_pred_test.T, y=residual.T)
from sklearn.model_selection import learning_curve
def learning_curves(estimator, features, target, cv):
train_sizes, train_scores, validation_scores = learning_curve(
estimator, features, target,train_sizes = np.linspace(.1, 1.0, 10),
cv = cv, scoring = 'neg_mean_squared_error',n_jobs=-1)
train_scores_mean = -train_scores.mean(axis = 1)
validation_scores_mean = -validation_scores.mean(axis = 1)
plt.plot(train_sizes, np.sqrt(train_scores_mean), label = 'Training error')
plt.plot(train_sizes, np.sqrt(validation_scores_mean), label = 'Validation error')
plt.ylabel('RMSE', fontsize = 14)
plt.xlabel('Training set size', fontsize = 14)
title = 'Learning curve'
plt.title(title, fontsize = 18, y = 1.03)
plt.legend()
plt.ylim(0,3)
learning_curves(model,X,y,5)
#plt.savefig("RFmodel_300_learnincurve.png",dpi=600,bbox_inches="tight")
blind_test_mCSM = pd.read_csv("../benchmark/mCSM-AB-blind_test.predictions.csv",sep="\t")
blind_test_toCompare =blind_test_mCSM.loc[blind_test_mCSM["Type"]=="Forward"]
blind_test_toCompare
df_final.describe()
X_onlyHM, y_onlyHM = df_final_onlyHM.drop("ddG(kcal/mol)",axis=1),df_final_onlyHM["ddG(kcal/mol)"]
X_onlyHM_f1 = sel.transform(X_onlyHM)
y_onlyHM_pred = model.predict(X_onlyHM_f1)
blind_test_toCompare.loc[:,["DDG","PRED"]].corr()
y_onlyHM_predicted = model.predict(X_onlyHM)
np.corrcoef([y_onlyHM_pred,y_onlyHM])
```
|
github_jupyter
|
import pandas as pd
from pathlib import Path
from sklearn.ensemble import RandomForestRegressor
from sklearn.pipeline import make_pipeline
from imblearn import over_sampling as ovs
from sklearn.decomposition import PCA
import matplotlib.pyplot as plt
import seaborn as sns
import numpy as np
from sklearn.metrics import mean_squared_error,mean_absolute_error,r2_score
from sklearn.preprocessing import StandardScaler
from sklearn import metrics
from sklearn.externals import joblib
from sklearn.feature_selection import VarianceThreshold
from sklearn.model_selection import GridSearchCV
from sklearn.model_selection import train_test_split
%matplotlib inline
sns.set_context(context="paper")
def plot_test_reg(y_true,y_pred):
from matplotlib.offsetbox import AnchoredText
rmse_test = np.round(np.sqrt(mean_squared_error(y_true, y_pred)), 3)
df_pred = pd.DataFrame({"Predicted ddG(kcal/mol)": y_pred, "Actual ddG(kcal/mol)": y_true})
pearsonr_test = round(df_pred.corr().iloc[0,1],3)
g = sns.regplot(x="Actual ddG(kcal/mol)", y="Predicted ddG(kcal/mol)",data=df_pred)
plt.title("Predicted vs Experimental ddG (Test set: %s complexes)"%y_true.shape[0])
anchored_text1 = AnchoredText("pearsonr = %s" %pearsonr_test, loc=1)
anchored_text2 = AnchoredText("RMSE = %s" %rmse_test, loc=4)
g.add_artist(anchored_text1)
g.add_artist(anchored_text2)
from sklearn.model_selection import learning_curve
def learning_curves(estimator, features, target, cv):
train_sizes, train_scores, validation_scores = learning_curve(
estimator, features, target,train_sizes = np.linspace(.1, 1.0, 10),
cv = cv, scoring = 'neg_mean_squared_error',n_jobs=-1)
train_scores_mean = -train_scores.mean(axis = 1)
validation_scores_mean = -validation_scores.mean(axis = 1)
plt.plot(train_sizes, np.sqrt(train_scores_mean), label = 'Training error')
plt.plot(train_sizes, np.sqrt(validation_scores_mean), label = 'Validation error')
plt.ylabel('RMSE', fontsize = 14)
plt.xlabel('Training set size', fontsize = 14)
title = 'Learning curve'
plt.title(title, fontsize = 18, y = 1.03)
plt.legend()
plt.ylim(0,3)
ABPRED_DIR = Path().cwd().parent
DATA = ABPRED_DIR / "data"
#dataframe final
df_final = pd.read_csv(DATA/"../data/DF_features_400_2019.csv",index_col=0)
# Quitar modelos por homologia deltraining set
#df_final_onlyHM = df_final.loc[df_final.index.str.startswith("HM")]
#df_final= df_final.loc[~df_final.index.str.startswith("HM")]
index_ddg8 = (df_final['ddG(kcal/mol)']==8)
df_final = df_final.loc[-index_ddg8]
#testiar eliminando estructuras con ddg menor o igual a -4 kcal/mol , outliers
index_ddg_4 = (df_final['ddG(kcal/mol)'] <= -4)
df_final = df_final.loc[-index_ddg_4]
pdb_names = df_final.index
features_names = df_final.drop('ddG(kcal/mol)',axis=1).columns
import math
def deltaG_to_Kd(delg):
Kd_value = math.exp((delg*1000)/(1.98*298.15))
return Kd_value
#X = df_final_f1 # con filtrado de varianza
X = df_final.drop('ddG(kcal/mol)',axis=1)
y = df_final['ddG(kcal/mol)']
#y_pKd = -y.apply(deltaG_to_Kd).apply(math.log10)
#y = y_pKd
X_train, X_test, y_train, y_test = train_test_split(X, y,train_size=0.8,random_state=12)
#1)
scaler = StandardScaler()
#2)
rfr = RandomForestRegressor()
#3) Crear pipeline
pipeline1 = make_pipeline(scaler ,rfr)
# Create the parameter grid based on the results of random search
param_grid = {
'randomforestregressor__bootstrap': [True],
'randomforestregressor__max_depth': [2,5,15,25,30,None],
'randomforestregressor__max_features': ["log2","sqrt","auto"],
'randomforestregressor__max_leaf_nodes':[50,60,70,80,90],
'randomforestregressor__min_samples_leaf': [1, 2, 5],
'randomforestregressor__min_samples_split': [2,5,10,15],
'randomforestregressor__n_estimators': [100,300,500,1000],
'randomforestregressor__random_state':[1],
}
grid1 = GridSearchCV(pipeline1,param_grid, verbose=1,scoring="neg_mean_squared_error",cv=10,n_jobs=-1)
# fit
grid1.fit(X_train, y_train)
print(grid1.best_params_)
print('Training score (r2): {}'.format(grid1.best_estimator_.score(X_train, y_train)))
print('Test score (r2): {}'.format(grid1.best_estimator_.score(X_test, y_test)))
y_pred = grid1.best_estimator_.predict(X_test)
print("\nRoot mean square error for test dataset: {}".format(np.round(np.sqrt(mean_squared_error(y_test, y_pred)), 2)))
print("pearson corr:\n",np.corrcoef(y_pred,y_test)[0][1])
plot_test_reg(y_pred=y_pred,y_true=y_test)
learning_curves(grid1.best_estimator_,X,y,10)
# forma 1
df_final['class'] = np.where((df_final['ddG(kcal/mol)'] < -1) | (df_final['ddG(kcal/mol)'] > 3.0),1,0)
# forma 2
#df_final['class'] = np.where((df_final['ddG(kcal/mol)'] < 0),1,0)
print("Before OverSampling, counts of label '1': {}".format(sum(df_final['class'] == 1)))
print("Before OverSampling, counts of label '0': {}".format(sum(df_final['class'] == 0)))
X = df_final.drop('class',axis=1)
y = df_final['class']
X_train, X_test, y_train, y_test = train_test_split(X, y,stratify = y,train_size=0.7,random_state=12)
#smote = ovs.SMOTE(random_state=12)
#smote = ovs.BorderlineSMOTE(random_state=12)
adasyn = ovs.ADASYN(random_state=12,n_neighbors=5)
X_train_re , y_train_re = adasyn.fit_sample(X_train,y_train)
#X_train_re , y_train_re = smote.fit_sample(X_train,y_train)
# back to originalk shape and target
X_train_normal = X_train_re[:,:-1]
y_train_normal = X_train_re[:,-1]
X_test_normal = X_test.iloc[:,:-1]
y_test_normal = X_test.iloc[:,-1]
#1)
scaler = StandardScaler()
#2)
rfr = RandomForestRegressor(bootstrap=True, max_depth=None,max_features='sqrt',max_leaf_nodes=90,
min_samples_leaf=1,min_samples_split=2, n_estimators=500,random_state=1)
#3) Crear pipeline
pipe = make_pipeline(scaler ,rfr)
pipe.fit(X_train_normal,y_train_normal)
print('Training score (r2): {}'.format(pipe.score(X_train_normal, y_train_normal)))
print('Test score (r2): {}'.format(pipe.score(X_test_normal, y_test_normal)))
y_test_pred = pipe.predict(X_test_normal)
y_train_pred = pipe.predict(X_train_normal)
print("\nRoot mean square error for train dataset: {}".format(np.round(np.sqrt(mean_squared_error(y_train_normal, y_train_pred)), 2)))
print("\nRoot mean square error for test dataset: {}".format(np.round(np.sqrt(mean_squared_error(y_test_normal, y_test_pred)), 2)))
print("pearson corr:\n",np.corrcoef(y_test_pred,y_test_normal)[0][1])
plot_test_reg(y_pred=y_test_pred,y_true=y_test_normal)
learning_curves(pipe,X_train_normal,y_train_normal,10)
grid_search.fit(X_train, y_train)
model = grid_search.best_estimator_
model.fit(X_train, y_train)
joblib.dump(model, '../../../../GDrive/RFmodel_400.10cv.v4.pkl')
f = open("rf_models400F_Fullsearch.txt","a")
print(grid_search.best_estimator_)
f.write('10 fold\n'+str(grid_search.best_estimator_))
print('\nBest GridSearchCV Score 5fold : ' + str(-grid_search.best_score_))
model = grid_search.best_estimator_
model.fit(X_train, y_train)
y_pred_train = model.predict(X_train)
y_pred_test = model.predict(X_test)
#print("OOB Score : " + str(model.oob_score_)) # yg diitung r2 score
print('Train MSE : ' + str(mean_squared_error(y_train, y_pred_train)))
print('Train MAE : ' + str(mean_absolute_error(y_train, y_pred_train)))
print('Train R^2 : ' + str(r2_score(y_train, y_pred_train)))
print('Test R^2 : ' + str(r2_score(y_test, y_pred_test)))
print('Test MSE : ' + str(mean_squared_error(y_test, y_pred_test)))
print('Test MAE : ' + str(mean_absolute_error(y_test, y_pred_test)))
print("\nRoot mean square error for test dataset: {}".format(np.round(np.sqrt(mean_squared_error(y_test, y_pred_test)), 2)))
print("pearson corr:\n",np.corrcoef(y_pred_test,y_test)[0][1])
f.write('\nTrain MSE : ' + str(mean_squared_error(y_train, y_pred_train)))
f.write('\nTrain MAE : ' + str(mean_absolute_error(y_train, y_pred_train)))
f.write('\nTrain R^2 : ' + str(r2_score(y_train, y_pred_train)))
f.write('\nTest R^2 : ' + str(r2_score(y_test, y_pred_test)))
f.write('\nTest MSE : ' + str(mean_squared_error(y_test, y_pred_test)))
f.write('\nTest MAE : ' + str(mean_absolute_error(y_test, y_pred_test)))
f.write("\nRoot mean square error for test dataset: {}".format(np.round(np.sqrt(mean_squared_error(y_test, y_pred_test)), 2)))
f.write("\npearson corr:\n"+str(np.corrcoef(y_pred_test,y_test)))
f.close()
# save final model
final_model = grid_search.best_estimator_
final_model.fit(X,y)
joblib.dump(final_model, 'RFmodel_400.v2.pkl')
rmse_test = np.round(np.sqrt(mean_squared_error(y_test, y_pred_test)), 3)
df_pred = pd.DataFrame({"Predicted ddG(kcal/mol)": y_pred_test, "Actual ddG(kcal/mol)": y_test.values})
pearsonr_test = round(df_pred.corr().iloc[0,1],3)
g = sns.regplot(x="Actual ddG(kcal/mol)", y="Predicted ddG(kcal/mol)",data=df_pred)
plt.title("Predicted vs Experimental ddG (Independent set: 123 complexes)")
plt.text(-2,3,"pearsonr = %s" %pearsonr_test)
plt.text(4.5,-0.5,"RMSE = %s" %rmse_test)
#plt.savefig("RFmodel_300_testfit.png",dpi=600)
df_train_pred = pd.DataFrame({"Predicted ddG(kcal/mol)": y_train, "Actual ddG(kcal/mol)": y_pred_train})
pearsonr_train = round(df_train_pred.corr().iloc[0,1],3)
rmse_train = np.round(np.sqrt(mean_squared_error(y_train, y_pred_train)), 3)
g = sns.regplot(x="Actual ddG(kcal/mol)", y="Predicted ddG(kcal/mol)",data=df_train_pred)
plt.text(-0.4,6.5,"pearsonr = %s" %pearsonr_train)
plt.text(3.5,-2.5,"RMSE = %s" %rmse_train)
plt.title("Predicted vs Experimental ddG (Train set: 492 complexes)")
#plt.savefig("RFmodel_300_trainfit.png",dpi=600)
importances = list(model.feature_importances_)
feature_list = df_final.columns
# List of tuples with variable and importance
feature_importances = [(feature, round(importance, 4)) for feature, importance in zip(feature_list, importances)]
# Sort the feature importances by most important first
feature_importances = sorted(feature_importances, key = lambda x: x[1], reverse = True)
# Print out the feature and importances
[print('Variable: {:20} Importance: {}'.format(*pair)) for pair in feature_importances]
idx_features = model.feature_importances_.argsort()[::-1]
plt.figure(figsize=(15,4))
plt.bar(np.arange(X_train.shape[1]), model.feature_importances_[idx_features])
plt.xticks(range(len(feature_list)),feature_list[idx_features])
plt.autoscale(enable=True, axis='x', tight=True)
plt.xlabel(u"Feature importance")
#plt.savefig("RFmodel_300_50features",dpi=600,bbox_inches="tight")
residual = y_test.values - y_pred_test
plt.scatter(x=y_pred_test.T, y=residual.T)
from sklearn.model_selection import learning_curve
def learning_curves(estimator, features, target, cv):
train_sizes, train_scores, validation_scores = learning_curve(
estimator, features, target,train_sizes = np.linspace(.1, 1.0, 10),
cv = cv, scoring = 'neg_mean_squared_error',n_jobs=-1)
train_scores_mean = -train_scores.mean(axis = 1)
validation_scores_mean = -validation_scores.mean(axis = 1)
plt.plot(train_sizes, np.sqrt(train_scores_mean), label = 'Training error')
plt.plot(train_sizes, np.sqrt(validation_scores_mean), label = 'Validation error')
plt.ylabel('RMSE', fontsize = 14)
plt.xlabel('Training set size', fontsize = 14)
title = 'Learning curve'
plt.title(title, fontsize = 18, y = 1.03)
plt.legend()
plt.ylim(0,3)
learning_curves(model,X,y,5)
#plt.savefig("RFmodel_300_learnincurve.png",dpi=600,bbox_inches="tight")
blind_test_mCSM = pd.read_csv("../benchmark/mCSM-AB-blind_test.predictions.csv",sep="\t")
blind_test_toCompare =blind_test_mCSM.loc[blind_test_mCSM["Type"]=="Forward"]
blind_test_toCompare
df_final.describe()
X_onlyHM, y_onlyHM = df_final_onlyHM.drop("ddG(kcal/mol)",axis=1),df_final_onlyHM["ddG(kcal/mol)"]
X_onlyHM_f1 = sel.transform(X_onlyHM)
y_onlyHM_pred = model.predict(X_onlyHM_f1)
blind_test_toCompare.loc[:,["DDG","PRED"]].corr()
y_onlyHM_predicted = model.predict(X_onlyHM)
np.corrcoef([y_onlyHM_pred,y_onlyHM])
| 0.63861 | 0.700383 |
# [Ateliers: Technologies de l'intelligence Artificielle](https://github.com/wikistat/AI-Frameworks)
<center>
<a href="http://www.insa-toulouse.fr/" ><img src="http://www.math.univ-toulouse.fr/~besse/Wikistat/Images/logo-insa.jpg" style="float:left; max-width: 120px; display: inline" alt="INSA"/></a>
<a href="http://wikistat.fr/" ><img src="http://www.math.univ-toulouse.fr/~besse/Wikistat/Images/wikistat.jpg" width=400, style="max-width: 150px; display: inline" alt="Wikistat"/></a>
<a href="http://www.math.univ-toulouse.fr/" ><img src="http://www.math.univ-toulouse.fr/~besse/Wikistat/Images/logo_imt.jpg" width=400, style="float:right; display: inline" alt="IMT"/> </a>
</center>
# Traitement Naturel du Langage (NLP) : Catégorisation de Produits Cdiscount
Il s'agit d'une version simplifiée du concours proposé par Cdiscount et paru sur le site [datascience.net](https://www.datascience.net/fr/challenge). Les données d'apprentissage sont accessibles sur demande auprès de Cdiscount mais les solutions de l'échantillon test du concours ne sont pas et ne seront pas rendues publiques. Un échantillon test est donc construit pour l'usage de ce tutoriel. L'objectif est de prévoir la catégorie d'un produit à partir de son descriptif (*text mining*). Seule la catégorie principale (1er niveau, 47 classes) est prédite au lieu des trois niveaux demandés dans le concours. L'objectif est plutôt de comparer les performances des méthodes et technologies en fonction de la taille de la base d'apprentissage ainsi que d'illustrer sur un exemple complexe le prétraitement de données textuelles.
Le jeux de données complet (15M produits) permet un test en vrai grandeur du **passage à l'échelle volume** des phases de préparation (*munging*), vectorisation (hashage, TF-IDF) et d'apprentissage en fonction de la technologie utilisée.
La synthèse des résultats obtenus est développée par [Besse et al. 2016](https://hal.archives-ouvertes.fr/hal-01350099) (section 5).
## Partie 2-2 Catégorisation des Produits Cdiscount avec [SparkML](https://spark.apache.org/docs/latest/ml-guide.html) de <a href="http://spark.apache.org/"><img src="http://spark.apache.org/images/spark-logo-trademark.png" style="max-width: 100px; display: inline" alt="Spark"/></a>
Le principal objectif est de comparer les performances: temps de calcul, qualité des résultats, des principales technologies; ici PySpark avec la librairie SparkML. Il s'agit d'un problème de fouille de texte qui enchaîne nécessairement plusieurs étapes et le choix de la meilleure stratégie est fonction de l'étape:
- Spark pour la préparation des données: nettoyage, racinisaiton
- Python Scikit-learn pour la transformaiton suivante (TF-IDF) et l'apprentissage notamment avec la régresison logistique qui conduit aux meilleurs résultats.
L'objectif est ici de comparer les performances des méthodes et technologies en fonction de la taille de la base d'apprentissage. La stratégie de sous ou sur échantillonnage des catégories qui permet d'améliorer la prévision n'a pas été mise en oeuvre.
* L'exemple est présenté avec la possibilité de sous-échantillonner afin de réduire les temps de calcul.
* L'échantillon réduit peut encore l'être puis, après "nettoyage", séparé en 2 parties: apprentissage et test.
* Les données textuelles de l'échantillon d'apprentissage sont, "racinisées", "hashées", "vectorisées" avant modélisation.
* Les mêmes transformations, notamment (hashage et TF-IDF) évaluées sur l'échantillon d'apprentissage sont appliquées à l'échantillon test.
* Un seul modèle est estimé par régression logistique "multimodal", plus précisément et implicitement, un modèle par classe.
* Différents paramètres: de vectorisation (hashage, TF-IDF), paramètres de la régression logistique (pénalisation L1) pourraient encore être optimisés.
## Librairies
```
sc.version
# Importation des packages génériques et ceux
# des librairie ML et MLlib
##Nettoyage
import nltk
import re
##Liste
from numpy import array
import pandas as pd
##Temps
import time
##Row and Vector
from pyspark.sql import Row
from pyspark.ml.linalg import Vectors
##Hashage et vectorisation
from pyspark.ml.feature import HashingTF
from pyspark.ml.feature import IDF
##Regression logistique
from pyspark.ml.classification import LogisticRegression
##Decision Tree
from pyspark.ml.classification import DecisionTreeClassifier
##Random Forest
from pyspark.ml.classification import RandomForestClassifier
##Pour la création des DataFrames
from pyspark.sql import SQLContext
from pyspark.sql.types import *
```
## Nettoyage des données
### Lecture des données
"dezippé" le fichier *data/cdiscount_train.csv.zip* avant d'éxecuter la cellule suivante.
```
sqlContext = SQLContext(sc)
RowDF = sqlContext.read.format('com.databricks.spark.csv').options(header='true', inferschema='true').load('data/cdiscount_train.csv')
RowDF.take(2)
```
### Extraction sous-échantillon
```
# Taux de sous-échantillonnage des données pour tester le programme de préparation
# sur un petit jeu de données
taux_donnees=0.99
dataEchDF,data_drop = RowDF.randomSplit([taux_donnees,1-taux_donnees])
```
Les instructions précédentes ne sont pas exécutées (exécution paresseuse ou *lazy*). Elles ne le sont que lors d'un traitement explicite.
```
ts=time.time()
size = dataEchDF.count()
te=time.time()
rt_count = te-ts
print("Comptage prend %d s, pour une taille de %d" %(rt_count, size)) #63s
```
taux | Taille M | Temps
-----|--------|------
0.01 | 0.157 | 51
0.1 | 1.57 | 49
0.4 | 6.29 | 53
0.8 | 12.6 | 60
1 | 15.7 | 64
### Nettoyage
Afin de limiter la dimension de l'espace des variables ou *features* tout en conservant les informations essentielles, il est nécessaire de nettoyer les données en appliquant plusieurs étapes:
* **Tokenizer**: Les textes de descrpitions sont transformés en liste de mots. Chaque mot est écrit en minuscule, les termes numériques, de ponctuation et autres symboles sont supprimés.
* **StopWord**: 155 mots-courants, et donc non informatif, de la langue française sont supprimés (STOPWORDS). Ex: le, la, du, alors, etc...
* **Stemmer**: Chaque mot est "racinisé". La racinisation transforme un mot en son radical ou sa racine. Par exemple, les mots: cheval, chevaux, chevalier, chevalerie, chevaucher sont tous remplacés par "cheva".
La librairie pyspark.ml ne supporte actuellement pas de fonction de stemming pour pyspark. Pour cette étape nous faisons donc appel à la librairie nltk de python et la fonction *nltk.stem.SnowballStemmer*.
```
import nltk
from pyspark.sql.types import ArrayType
from pyspark.sql.functions import udf,col
from pyspark.ml.feature import RegexTokenizer, StopWordsRemover
# liste des mots à supprimer
STOPWORDS = set(nltk.corpus.stopwords.words('french'))
# Fonction tokenizer qui permet de remplacer un long texte par une liste de mot
regexTokenizer = RegexTokenizer(inputCol="Description", outputCol="tokenizedDescr", pattern="[^a-z_]",
minTokenLength=3, gaps=True)
dataTokenized = regexTokenizer.transform(dataEchDF)
# Fonction StopWordsRemover qui permet de supprimer des mots
remover = StopWordsRemover(inputCol="tokenizedDescr", outputCol="tokenizedRemovedDescr", stopWords = list(STOPWORDS))
dataTokenizedRemoved = remover.transform(dataTokenized)
# La fonction de stemming de spark existe aujourd'hui qu'en scala et ne possède pas de wrapper en python.
# Cette étape doit donc être effectué en utilisant des librairies python et notamment nltk
STEMMER = nltk.stem.SnowballStemmer('french')
def clean_text(tokens):
tokens_stem = [ STEMMER.stem(token) for token in tokens]
return tokens_stem
udfCleanText = udf(lambda lt : clean_text(lt), ArrayType(StringType()))
dataClean = dataTokenizedRemoved.withColumn("cleanDescr", udfCleanText(col('tokenizedRemovedDescr')))
dataClean.take(2)
# Create a new rdd with the resulte of the clean_text function on the description
ts=time.time()
size = dataClean.count()
te=time.time()
rt = te-ts
print("Nettoyage prend %d s, pour la taille %d" %(rt, size)) #64s
```
## Préparation des données
### Construction des Labels
Convertir les 47 labels d'origine (Catégorie 1), qui ne sont pas dans un format acceptable (string) par les fonctions de sparkML, en des entiers numérotés de 0 a 46 nécessaires lors de l'application de la régression logistique.
Cette étape peut être facilement réalisé a l'aide de la fonction *StringIndexer*.
```
ts=time.time()
from pyspark.ml.feature import StringIndexer
indexer = StringIndexer(inputCol="Categorie1", outputCol="categoryIndex")
dataCleanindexed = indexer.fit(dataClean).transform(dataClean)
te=time.time()
rt = te-ts
print("Index car prend %d s, taille %d" %(rt, size)) #67
```
### Création des DataFrame train and Test
Une fois extrait l'échantillon test, l'échantillon d'apprentissage peut être sous échantillonné.
```
tauxEch=0.8
(trainTotDF, testDF) = dataCleanindexed.randomSplit([tauxEch, 1-tauxEch])
n_test = trainTotDF.count()
print(n_test) # 314441
```
### Sous-échantillonnage
Si toutes les données sont préparées, il est possible de sous-échantillonner l'échantillon d'apprentissage pour étudier l'impact de sa taille sur l'erreur de prévision.
```
tauxApp=0.9
(trainDF, testDF)=trainTotDF.randomSplit([tauxApp, 1-tauxApp])
n_train=trainDF.count()
print(n_train) # 1131577
```
## 4 Construction des *features* (TF-IDF)
### 4.1 Introduction
La vectorisation, c'est à dire la construction des *features* à partir de la liste des mots se fait en 2 étapes:
* Le hashage permet de réduire l'espace des variables (taille du dictionnaire) en un nombre limité et fixé a priori *n_hash* de *features*. Il repose sur la définition d'une *hash function*, $h$ qui à un indice $j$ défini dans l'espace des entiers naturels, renvoie un indice $i=h(j)$ dans dans l'espace réduit (1 à *n_hash*) des *features*. Ainsi le poids de l'indice $i$, du nouvel espace, est l'association de tous les poids d'indice $j$ tels que $i=h(j)$ de l'espace originale. Ici, les poids sont associés d'après la méthode décrit par [Weinberger et al. (2009)](https://arxiv.org/pdf/0902.2206.pdf).
*N.B.* *$h$ n'est pas généré aléatoirement. Ainsi pour un même fichier d'apprentissage (ou de test) et pour un même entier n_hash, le résultat de la fonction de hashage est identique*
* Le *TF-IDF*. Le TF-IDF permet de faire ressortir l'importance relative de chaque mot $m$ (ou couples de mots consécutifs) dans un texte-produit ou un descriptif $d$, par rapport à la liste entière des produits. La fonction $TF(m,d)$ compte le nombre d'occurences du mot $m$ dans le descriptif $d$. La fonction $IDF(m)$ mesure l'importance du terme dans l'ensemble des documents ou descriptifs en donnant plus de poids aux termes les moins fréquents car considérés comme les plus discriminants (motivation analogue à celle de la métrique du chi2 en anamlyse des correspondance). $IDF(m,l)=\log\frac{D}{f(m)}$ où $D$ est le nombre de documents, la taille de l'échantillon d'apprentissage, et $f(m)$ le nombre de documents ou descriptifs contenant le mot $m$. La nouvelle variable ou *features* est $V_m(l)=TF(m,l)\times IDF(m,l)$.
* Comme pour les transformations des variables quantitatives (centrage, réduction), la même transformation c'est-à-dire les mêmes pondérations, est calculée sur l'achantillon d'apprentissage et appliquée à celui de test.
### 4.2 Traitement
Dans le traitement qui suit, étape de hashage et calcul de $TF$ sont associés dans la même fonction.
Les transformations *hashage* et *tf-idf* calculées sur l'apprentissage sont appliquées à l'échantillon test.
```
trainDF.take(2)
ts=time.time()
# Term Frequency
hashing_tf = HashingTF(inputCol="cleanDescr", outputCol='tf', numFeatures=10000)
trainTfDF = hashing_tf.transform(trainDF)
# Inverse Document Frequency
idf = IDF(inputCol=hashing_tf.getOutputCol(), outputCol="tfidf")
idf_model = idf.fit(trainTfDF)
trainTfIdfDF = idf_model.transform(trainTfDF)
# application à l'échantillon tesy
testTfDF = hashing_tf.transform(testDF)
testTfIdfDF = idf_model.transform(testTfDF)
te=time.time()
rt = te-ts
print("Hashage et vectorisation prennent %d s, taille %d" %(rt, n_train)) #456 s, taille 1131577
```
## 4 Modélisation et test
Le [calepin](http://www.math.univ-toulouse.fr/~besse/Wikistat/Notebooks/Tuto_Notebook_Cdiscount.html) python a clairement montré la meilleure performance de la régression logistique dans ce problème. C'est d'ailleurs ce modèle ou une *pyramide de logistiques* qui est industriellement implémentée.
### 4.1 Estimation du modèle logistique
Le paramètre de pénalisation (lasso) est pris par défaut sans optimisation.
```
### Configuraiton des paramètres de la méthode
time_start=time.time()
lr = LogisticRegression(maxIter=100, regParam=0.01, fitIntercept=False, tol=0.0001,
family = "multinomial", elasticNetParam=0.0, featuresCol="tfidf", labelCol="categoryIndex") #0 for L2 penalty, 1 for L1 penalty
### Génération du modèle
model_lr = lr.fit(trainTfIdfDF)
time_end=time.time()
time_lrm=(time_end - time_start)
print("LR prend %d s" %(time_lrm)) # (104s avec taux=1)
```
### 4.2 Estimation de l'erreur sur l'échantillon test
```
predictionsDF = model_lr.transform(testTfIdfDF)
labelsAndPredictions = predictionsDF.select("categoryIndex","prediction").collect()
nb_good_prediction = sum([r[0]==r[1] for r in labelsAndPredictions])
testErr = 1-nb_good_prediction/n_test
print('Test Error = ' + str(testErr)) # (0.08 avec taux =1)
```
Taille M| Temps | Erreur
-------|-------|--------
1.131 | 786 | 0.94
|
github_jupyter
|
sc.version
# Importation des packages génériques et ceux
# des librairie ML et MLlib
##Nettoyage
import nltk
import re
##Liste
from numpy import array
import pandas as pd
##Temps
import time
##Row and Vector
from pyspark.sql import Row
from pyspark.ml.linalg import Vectors
##Hashage et vectorisation
from pyspark.ml.feature import HashingTF
from pyspark.ml.feature import IDF
##Regression logistique
from pyspark.ml.classification import LogisticRegression
##Decision Tree
from pyspark.ml.classification import DecisionTreeClassifier
##Random Forest
from pyspark.ml.classification import RandomForestClassifier
##Pour la création des DataFrames
from pyspark.sql import SQLContext
from pyspark.sql.types import *
sqlContext = SQLContext(sc)
RowDF = sqlContext.read.format('com.databricks.spark.csv').options(header='true', inferschema='true').load('data/cdiscount_train.csv')
RowDF.take(2)
# Taux de sous-échantillonnage des données pour tester le programme de préparation
# sur un petit jeu de données
taux_donnees=0.99
dataEchDF,data_drop = RowDF.randomSplit([taux_donnees,1-taux_donnees])
ts=time.time()
size = dataEchDF.count()
te=time.time()
rt_count = te-ts
print("Comptage prend %d s, pour une taille de %d" %(rt_count, size)) #63s
import nltk
from pyspark.sql.types import ArrayType
from pyspark.sql.functions import udf,col
from pyspark.ml.feature import RegexTokenizer, StopWordsRemover
# liste des mots à supprimer
STOPWORDS = set(nltk.corpus.stopwords.words('french'))
# Fonction tokenizer qui permet de remplacer un long texte par une liste de mot
regexTokenizer = RegexTokenizer(inputCol="Description", outputCol="tokenizedDescr", pattern="[^a-z_]",
minTokenLength=3, gaps=True)
dataTokenized = regexTokenizer.transform(dataEchDF)
# Fonction StopWordsRemover qui permet de supprimer des mots
remover = StopWordsRemover(inputCol="tokenizedDescr", outputCol="tokenizedRemovedDescr", stopWords = list(STOPWORDS))
dataTokenizedRemoved = remover.transform(dataTokenized)
# La fonction de stemming de spark existe aujourd'hui qu'en scala et ne possède pas de wrapper en python.
# Cette étape doit donc être effectué en utilisant des librairies python et notamment nltk
STEMMER = nltk.stem.SnowballStemmer('french')
def clean_text(tokens):
tokens_stem = [ STEMMER.stem(token) for token in tokens]
return tokens_stem
udfCleanText = udf(lambda lt : clean_text(lt), ArrayType(StringType()))
dataClean = dataTokenizedRemoved.withColumn("cleanDescr", udfCleanText(col('tokenizedRemovedDescr')))
dataClean.take(2)
# Create a new rdd with the resulte of the clean_text function on the description
ts=time.time()
size = dataClean.count()
te=time.time()
rt = te-ts
print("Nettoyage prend %d s, pour la taille %d" %(rt, size)) #64s
ts=time.time()
from pyspark.ml.feature import StringIndexer
indexer = StringIndexer(inputCol="Categorie1", outputCol="categoryIndex")
dataCleanindexed = indexer.fit(dataClean).transform(dataClean)
te=time.time()
rt = te-ts
print("Index car prend %d s, taille %d" %(rt, size)) #67
tauxEch=0.8
(trainTotDF, testDF) = dataCleanindexed.randomSplit([tauxEch, 1-tauxEch])
n_test = trainTotDF.count()
print(n_test) # 314441
tauxApp=0.9
(trainDF, testDF)=trainTotDF.randomSplit([tauxApp, 1-tauxApp])
n_train=trainDF.count()
print(n_train) # 1131577
trainDF.take(2)
ts=time.time()
# Term Frequency
hashing_tf = HashingTF(inputCol="cleanDescr", outputCol='tf', numFeatures=10000)
trainTfDF = hashing_tf.transform(trainDF)
# Inverse Document Frequency
idf = IDF(inputCol=hashing_tf.getOutputCol(), outputCol="tfidf")
idf_model = idf.fit(trainTfDF)
trainTfIdfDF = idf_model.transform(trainTfDF)
# application à l'échantillon tesy
testTfDF = hashing_tf.transform(testDF)
testTfIdfDF = idf_model.transform(testTfDF)
te=time.time()
rt = te-ts
print("Hashage et vectorisation prennent %d s, taille %d" %(rt, n_train)) #456 s, taille 1131577
### Configuraiton des paramètres de la méthode
time_start=time.time()
lr = LogisticRegression(maxIter=100, regParam=0.01, fitIntercept=False, tol=0.0001,
family = "multinomial", elasticNetParam=0.0, featuresCol="tfidf", labelCol="categoryIndex") #0 for L2 penalty, 1 for L1 penalty
### Génération du modèle
model_lr = lr.fit(trainTfIdfDF)
time_end=time.time()
time_lrm=(time_end - time_start)
print("LR prend %d s" %(time_lrm)) # (104s avec taux=1)
predictionsDF = model_lr.transform(testTfIdfDF)
labelsAndPredictions = predictionsDF.select("categoryIndex","prediction").collect()
nb_good_prediction = sum([r[0]==r[1] for r in labelsAndPredictions])
testErr = 1-nb_good_prediction/n_test
print('Test Error = ' + str(testErr)) # (0.08 avec taux =1)
| 0.398172 | 0.949201 |
Demo script to build a tower in Coppeliasim, using PyRep
```
from pyrep import PyRep
from pyrep.robots.arms.panda import Panda
from pyrep.robots.end_effectors.panda_gripper import PandaGripper
import numpy as np
from matplotlib import pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
from IPython import display
from pyrep.objects.shape import Shape
from pyrep.const import PrimitiveShape
from scipy.spatial.transform import Rotation as R
import math
from copy import copy
pr = PyRep()
pr.launch('../../assets/scene_panda.ttt', headless=False)
agent = Panda()
gripper = PandaGripper()
home_pos = agent.get_tip().get_position()
home_orient = agent.get_tip().get_orientation()
def grasp(grip=False):
if grip:
pos = 0.1
else:
pos = 0.9
actuated = False
ims = []
states = []
while not actuated:
actuated = gripper.actuate(pos,0.1)
im0,im1 = get_image()
ims.append((im0,im1))
states.append(agent.get_tip().get_pose())
return ims,states
def move_above_object(object_name='',offset=0.05):
pos = agent.get_object(object_name).get_position()
pos[2] = pos[2] + offset
orient = [-np.pi,0,np.pi/2]
path = agent.get_path(position=pos,euler=orient)
done = False
ims = []
states = []
while not done:
done = path.step()
im0,im1 = get_image()
ims.append((im0,im1))
states.append(agent.get_tip().get_pose())
return ims,states
def clear_view():
path = agent.get_path(position=home_pos,euler=home_orient)
done = False
ims = []
states = []
while not done:
done = path.step()
im0,im1 = get_image()
ims.append((im0,im1))
states.append(agent.get_tip().get_pose())
return ims,states
def get_image():
cam = agent.get_object('Vision_sensor_front')
im0 = cam.capture_rgb()
cam1 = agent.get_object('Vision_sensor')
im1 = cam1.capture_rgb()
pr.step()
return im0, im1
pr.start()
obj_ids = np.arange(6)
np.random.shuffle(obj_ids)
grasp(grip=False)
gripper.release()
ims = []
states = []
for j in range(1,6):
object_name = 'Cuboid%d'%obj_ids[j]
i,s = move_above_object(object_name,offset=0.08)
ims = ims + i
states = states + s
i,s = move_above_object(object_name,offset=0)
ims = ims + i
states = states + s
i,s = grasp(grip=True)
ims = ims + i
states = states + s
gripper.grasp(agent.get_object(object_name))
i,s = move_above_object(object_name,offset=0.08)
ims = ims + i
states = states + s
object_name = 'Cuboid%d'%obj_ids[j-1]
i,s = move_above_object(object_name,offset=0.15)
ims = ims + i
states = states + s
i,s = move_above_object(object_name,offset=0.05)
ims = ims + i
states = states + s
i,s = grasp(grip=False)
ims = ims + i
states = states + s
gripper.release()
i,s = move_above_object(object_name,offset=0.2)
ims = ims + i
states = states + s
#clear_view()
plt.cla()
plt.clf()
plt.subplot(1,2,1)
plt.imshow(ims[-1][0])
plt.subplot(1,2,2)
plt.imshow(ims[-1][1])
display.clear_output(wait=True)
display.display(plt.gcf())
pr.stop()
for i,im in enumerate(ims):
if i %10==0:
plt.cla()
plt.clf()
plt.subplot(2,2,1)
plt.imshow(im[0])
plt.subplot(2,2,2)
plt.imshow(im[1])
plt.subplot(2,1,2)
plt.plot(np.vstack(states)[0:i,:])
display.clear_output(wait=True)
display.display(plt.gcf())
```
|
github_jupyter
|
from pyrep import PyRep
from pyrep.robots.arms.panda import Panda
from pyrep.robots.end_effectors.panda_gripper import PandaGripper
import numpy as np
from matplotlib import pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
from IPython import display
from pyrep.objects.shape import Shape
from pyrep.const import PrimitiveShape
from scipy.spatial.transform import Rotation as R
import math
from copy import copy
pr = PyRep()
pr.launch('../../assets/scene_panda.ttt', headless=False)
agent = Panda()
gripper = PandaGripper()
home_pos = agent.get_tip().get_position()
home_orient = agent.get_tip().get_orientation()
def grasp(grip=False):
if grip:
pos = 0.1
else:
pos = 0.9
actuated = False
ims = []
states = []
while not actuated:
actuated = gripper.actuate(pos,0.1)
im0,im1 = get_image()
ims.append((im0,im1))
states.append(agent.get_tip().get_pose())
return ims,states
def move_above_object(object_name='',offset=0.05):
pos = agent.get_object(object_name).get_position()
pos[2] = pos[2] + offset
orient = [-np.pi,0,np.pi/2]
path = agent.get_path(position=pos,euler=orient)
done = False
ims = []
states = []
while not done:
done = path.step()
im0,im1 = get_image()
ims.append((im0,im1))
states.append(agent.get_tip().get_pose())
return ims,states
def clear_view():
path = agent.get_path(position=home_pos,euler=home_orient)
done = False
ims = []
states = []
while not done:
done = path.step()
im0,im1 = get_image()
ims.append((im0,im1))
states.append(agent.get_tip().get_pose())
return ims,states
def get_image():
cam = agent.get_object('Vision_sensor_front')
im0 = cam.capture_rgb()
cam1 = agent.get_object('Vision_sensor')
im1 = cam1.capture_rgb()
pr.step()
return im0, im1
pr.start()
obj_ids = np.arange(6)
np.random.shuffle(obj_ids)
grasp(grip=False)
gripper.release()
ims = []
states = []
for j in range(1,6):
object_name = 'Cuboid%d'%obj_ids[j]
i,s = move_above_object(object_name,offset=0.08)
ims = ims + i
states = states + s
i,s = move_above_object(object_name,offset=0)
ims = ims + i
states = states + s
i,s = grasp(grip=True)
ims = ims + i
states = states + s
gripper.grasp(agent.get_object(object_name))
i,s = move_above_object(object_name,offset=0.08)
ims = ims + i
states = states + s
object_name = 'Cuboid%d'%obj_ids[j-1]
i,s = move_above_object(object_name,offset=0.15)
ims = ims + i
states = states + s
i,s = move_above_object(object_name,offset=0.05)
ims = ims + i
states = states + s
i,s = grasp(grip=False)
ims = ims + i
states = states + s
gripper.release()
i,s = move_above_object(object_name,offset=0.2)
ims = ims + i
states = states + s
#clear_view()
plt.cla()
plt.clf()
plt.subplot(1,2,1)
plt.imshow(ims[-1][0])
plt.subplot(1,2,2)
plt.imshow(ims[-1][1])
display.clear_output(wait=True)
display.display(plt.gcf())
pr.stop()
for i,im in enumerate(ims):
if i %10==0:
plt.cla()
plt.clf()
plt.subplot(2,2,1)
plt.imshow(im[0])
plt.subplot(2,2,2)
plt.imshow(im[1])
plt.subplot(2,1,2)
plt.plot(np.vstack(states)[0:i,:])
display.clear_output(wait=True)
display.display(plt.gcf())
| 0.275227 | 0.708276 |
# Bi-LSTM on AGnews
## Librairies
```
# !pip install transformers==4.8.2
# !pip install datasets==1.7.0
import os
import sys
import time
import pickle
import torch
import torch.nn as nn
import torch.optim as optim
from datasets import load_dataset, Dataset, concatenate_datasets
from transformers import AutoTokenizer
from transformers.data.data_collator import DataCollatorWithPadding
from esntorch.utils.embedding import EmbeddingModel
device = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
device
```
## Global variables
```
MODEL_FILE = '~/Results/Bi-LSTM/Bi-LSTM/AGnews-4_bilstm-128_10ep_model.pt'
RESULTS_FILE = '~/Results/Bi-LSTM/Bi-LSTM/AGnews-4_bilstm-128_10ep_results.pkl'
CACHE_DIR = 'cache_dir/' # put your path here
```
## Dataset
```
# rename correct column as 'labels': depends on the dataset you load
def load_and_enrich_dataset(dataset_name, split, cache_dir):
dataset = load_dataset(dataset_name, split=split, cache_dir=CACHE_DIR)
dataset = dataset.rename_column('label', 'labels') # 'label-fine'
dataset = dataset.map(lambda e: tokenizer(e['text'], truncation=True, padding=False), batched=True)
dataset.set_format(type='torch', columns=['input_ids', 'attention_mask', 'labels'])
def add_lengths(sample):
sample["lengths"] = sum(sample["input_ids"] != 0)
return sample
dataset = dataset.map(add_lengths, batched=False)
return dataset
tokenizer = AutoTokenizer.from_pretrained('bert-base-uncased')
full_train_dataset = load_and_enrich_dataset('ag_news', split='train', cache_dir=CACHE_DIR).sort("lengths") # toriving/sst5
train_val_datasets = full_train_dataset.train_test_split(train_size=0.8, shuffle=True)
train_dataset = train_val_datasets['train'].sort("lengths")
val_dataset = train_val_datasets['test'].sort("lengths")
test_dataset = load_and_enrich_dataset('ag_news', split='test', cache_dir=CACHE_DIR).sort("lengths")
dataset_d = {
'full_train': full_train_dataset,
'train': train_dataset,
'val': val_dataset,
'test': test_dataset
}
dataloader_d = {}
for k, v in dataset_d.items():
dataloader_d[k] = torch.utils.data.DataLoader(v, batch_size=256, collate_fn=DataCollatorWithPadding(tokenizer))
dataset_d
```
## Model
```
class RNN(nn.Module):
def __init__(self, embedding_dim,
hidden_dim, output_dim, n_layers,
bidirectional, dropout):
super().__init__()
self.embedding = EmbeddingModel('bert-base-uncased', device)
# recurrent hidden layers (LSTM)
self.rnn = nn.LSTM(embedding_dim,
hidden_dim,
num_layers=n_layers,
bidirectional=bidirectional,
dropout=dropout)
# fully connected output layer (fc)
self.fc = nn.Linear(hidden_dim * 2, output_dim)
self.dropout = nn.Dropout(dropout)
def forward(self, batch):
packed_embedded = self.dropout(self.embedding.get_embedding(batch))
packed_output, (hidden, cell) = self.rnn(packed_embedded)
hidden = self.dropout(torch.cat((hidden[-2,:,:], hidden[-1,:,:]), dim = 1))
#hidden = [batch size, hid dim * num directions]
return self.fc(hidden)
output_dim = len(set(full_train_dataset['labels'].tolist()))
output_dim
# Parameters
# INPUT_DIM = len(TEXT.vocab)
EMBEDDING_DIM = 768
HIDDEN_DIM = 128 # 256 was default
OUTPUT_DIM = output_dim # 1 (binary) or output_dim (*** multiclass ***)
N_LAYERS = 1 # 2 was default
BIDIRECTIONAL = True
DROPOUT = 0.5
# PAD_IDX = TEXT.vocab.stoi[TEXT.pad_token]
# Model
model = RNN(EMBEDDING_DIM,
HIDDEN_DIM,
OUTPUT_DIM,
N_LAYERS,
BIDIRECTIONAL,
DROPOUT)
def count_parameters(model):
return sum(p.numel() for p in model.parameters() if p.requires_grad)
print(f'The model has {count_parameters(model):,} trainable parameters')
```
## Training
```
# Adam optimizer
optimizer = optim.Adam(model.parameters())
# Criterion
# criterion = nn.BCEWithLogitsLoss() # binary
criterion = nn.CrossEntropyLoss() # *** multiclass ***
# Put model and criterion to device
model = model.to(device)
criterion = criterion.to(device)
# binary accuracy
def binary_accuracy(preds, y):
"""
Returns accuracy per batch, i.e. if you get 8/10 right, this returns 0.8, NOT 8
"""
# round predictions to the closest integer
rounded_preds = torch.round(torch.sigmoid(preds)) # thresholding output neuron
correct = (rounded_preds == y).float() # convert into float for division
acc = correct.sum() / len(correct)
return acc
# *** multiclass ***
def categorical_accuracy(preds, y):
"""
Returns accuracy per batch, i.e. if you get 8/10 right, this returns 0.8, NOT 8
"""
max_preds = preds.argmax(dim = 1, keepdim = True) # get the index of the max probability
correct = max_preds.squeeze(1).eq(y)
return correct.sum().item() / torch.FloatTensor([y.shape[0]])
def train(model, iterator, optimizer, criterion):
epoch_loss = 0
epoch_acc = 0
model.train()
for batch in iterator:
optimizer.zero_grad()
predictions_raw = model(batch)
predictions = predictions_raw.squeeze(1)
loss = criterion(predictions, batch['labels'])
# acc = binary_accuracy(predictions, batch.label) # *** binary ***
acc = categorical_accuracy(predictions, batch['labels']) # *** multiclass ***
loss.backward()
optimizer.step()
epoch_loss += loss.item()
epoch_acc += acc.item()
return epoch_loss / len(iterator), epoch_acc / len(iterator)
def evaluate(model, iterator, criterion):
epoch_loss = 0
epoch_acc = 0
model.eval()
with torch.no_grad():
for batch in iterator:
predictions = model(batch).squeeze(1)
loss = criterion(predictions, batch['labels'])
# acc = binary_accuracy(predictions, batch.label) # *** binary ***
acc = categorical_accuracy(predictions, batch['labels']) # *** multiclass ***
epoch_loss += loss.item()
epoch_acc += acc.item()
return epoch_loss / len(iterator), epoch_acc / len(iterator)
import time
def epoch_time(start_time, end_time):
elapsed_time = end_time - start_time
elapsed_mins = int(elapsed_time / 60)
elapsed_secs = int(elapsed_time - (elapsed_mins * 60))
return elapsed_mins, elapsed_secs
N_EPOCHS = 10 # 30
best_valid_loss = float('inf')
t0 = time.time()
for epoch in range(N_EPOCHS):
start_time = time.time()
train_loss, train_acc = train(model, dataloader_d['train'], optimizer, criterion)
valid_loss, valid_acc = evaluate(model, dataloader_d['val'], criterion)
end_time = time.time()
epoch_mins, epoch_secs = epoch_time(start_time, end_time)
if valid_loss < best_valid_loss:
best_valid_loss = valid_loss
torch.save(model.state_dict(), MODEL_FILE)
print(f'Epoch: {epoch+1:02} | Epoch Time: {epoch_mins}m {epoch_secs}s')
print(f'\tTrain Loss: {train_loss:.3f} | Train Acc: {train_acc*100:.2f}%')
print(f'\t Val. Loss: {valid_loss:.3f} | Val. Acc: {valid_acc*100:.2f}%')
t1 = time.time()
train_time = t1 - t0
print('training time: {}'.format(train_time))
model = RNN(EMBEDDING_DIM,
HIDDEN_DIM,
OUTPUT_DIM,
N_LAYERS,
BIDIRECTIONAL,
DROPOUT)
model.load_state_dict(torch.load(MODEL_FILE))
model.eval().to(device)
test_loss, test_acc = evaluate(model, dataloader_d['test'], criterion)
print(f'Test Loss: {test_loss:.3f} | Test Acc: {test_acc*100:.2f}%')
results = test_acc, train_time, count_parameters(model)
results
with open(RESULTS_FILE, 'wb') as fh:
pickle.dump(results, fh)
```
|
github_jupyter
|
# !pip install transformers==4.8.2
# !pip install datasets==1.7.0
import os
import sys
import time
import pickle
import torch
import torch.nn as nn
import torch.optim as optim
from datasets import load_dataset, Dataset, concatenate_datasets
from transformers import AutoTokenizer
from transformers.data.data_collator import DataCollatorWithPadding
from esntorch.utils.embedding import EmbeddingModel
device = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
device
MODEL_FILE = '~/Results/Bi-LSTM/Bi-LSTM/AGnews-4_bilstm-128_10ep_model.pt'
RESULTS_FILE = '~/Results/Bi-LSTM/Bi-LSTM/AGnews-4_bilstm-128_10ep_results.pkl'
CACHE_DIR = 'cache_dir/' # put your path here
# rename correct column as 'labels': depends on the dataset you load
def load_and_enrich_dataset(dataset_name, split, cache_dir):
dataset = load_dataset(dataset_name, split=split, cache_dir=CACHE_DIR)
dataset = dataset.rename_column('label', 'labels') # 'label-fine'
dataset = dataset.map(lambda e: tokenizer(e['text'], truncation=True, padding=False), batched=True)
dataset.set_format(type='torch', columns=['input_ids', 'attention_mask', 'labels'])
def add_lengths(sample):
sample["lengths"] = sum(sample["input_ids"] != 0)
return sample
dataset = dataset.map(add_lengths, batched=False)
return dataset
tokenizer = AutoTokenizer.from_pretrained('bert-base-uncased')
full_train_dataset = load_and_enrich_dataset('ag_news', split='train', cache_dir=CACHE_DIR).sort("lengths") # toriving/sst5
train_val_datasets = full_train_dataset.train_test_split(train_size=0.8, shuffle=True)
train_dataset = train_val_datasets['train'].sort("lengths")
val_dataset = train_val_datasets['test'].sort("lengths")
test_dataset = load_and_enrich_dataset('ag_news', split='test', cache_dir=CACHE_DIR).sort("lengths")
dataset_d = {
'full_train': full_train_dataset,
'train': train_dataset,
'val': val_dataset,
'test': test_dataset
}
dataloader_d = {}
for k, v in dataset_d.items():
dataloader_d[k] = torch.utils.data.DataLoader(v, batch_size=256, collate_fn=DataCollatorWithPadding(tokenizer))
dataset_d
class RNN(nn.Module):
def __init__(self, embedding_dim,
hidden_dim, output_dim, n_layers,
bidirectional, dropout):
super().__init__()
self.embedding = EmbeddingModel('bert-base-uncased', device)
# recurrent hidden layers (LSTM)
self.rnn = nn.LSTM(embedding_dim,
hidden_dim,
num_layers=n_layers,
bidirectional=bidirectional,
dropout=dropout)
# fully connected output layer (fc)
self.fc = nn.Linear(hidden_dim * 2, output_dim)
self.dropout = nn.Dropout(dropout)
def forward(self, batch):
packed_embedded = self.dropout(self.embedding.get_embedding(batch))
packed_output, (hidden, cell) = self.rnn(packed_embedded)
hidden = self.dropout(torch.cat((hidden[-2,:,:], hidden[-1,:,:]), dim = 1))
#hidden = [batch size, hid dim * num directions]
return self.fc(hidden)
output_dim = len(set(full_train_dataset['labels'].tolist()))
output_dim
# Parameters
# INPUT_DIM = len(TEXT.vocab)
EMBEDDING_DIM = 768
HIDDEN_DIM = 128 # 256 was default
OUTPUT_DIM = output_dim # 1 (binary) or output_dim (*** multiclass ***)
N_LAYERS = 1 # 2 was default
BIDIRECTIONAL = True
DROPOUT = 0.5
# PAD_IDX = TEXT.vocab.stoi[TEXT.pad_token]
# Model
model = RNN(EMBEDDING_DIM,
HIDDEN_DIM,
OUTPUT_DIM,
N_LAYERS,
BIDIRECTIONAL,
DROPOUT)
def count_parameters(model):
return sum(p.numel() for p in model.parameters() if p.requires_grad)
print(f'The model has {count_parameters(model):,} trainable parameters')
# Adam optimizer
optimizer = optim.Adam(model.parameters())
# Criterion
# criterion = nn.BCEWithLogitsLoss() # binary
criterion = nn.CrossEntropyLoss() # *** multiclass ***
# Put model and criterion to device
model = model.to(device)
criterion = criterion.to(device)
# binary accuracy
def binary_accuracy(preds, y):
"""
Returns accuracy per batch, i.e. if you get 8/10 right, this returns 0.8, NOT 8
"""
# round predictions to the closest integer
rounded_preds = torch.round(torch.sigmoid(preds)) # thresholding output neuron
correct = (rounded_preds == y).float() # convert into float for division
acc = correct.sum() / len(correct)
return acc
# *** multiclass ***
def categorical_accuracy(preds, y):
"""
Returns accuracy per batch, i.e. if you get 8/10 right, this returns 0.8, NOT 8
"""
max_preds = preds.argmax(dim = 1, keepdim = True) # get the index of the max probability
correct = max_preds.squeeze(1).eq(y)
return correct.sum().item() / torch.FloatTensor([y.shape[0]])
def train(model, iterator, optimizer, criterion):
epoch_loss = 0
epoch_acc = 0
model.train()
for batch in iterator:
optimizer.zero_grad()
predictions_raw = model(batch)
predictions = predictions_raw.squeeze(1)
loss = criterion(predictions, batch['labels'])
# acc = binary_accuracy(predictions, batch.label) # *** binary ***
acc = categorical_accuracy(predictions, batch['labels']) # *** multiclass ***
loss.backward()
optimizer.step()
epoch_loss += loss.item()
epoch_acc += acc.item()
return epoch_loss / len(iterator), epoch_acc / len(iterator)
def evaluate(model, iterator, criterion):
epoch_loss = 0
epoch_acc = 0
model.eval()
with torch.no_grad():
for batch in iterator:
predictions = model(batch).squeeze(1)
loss = criterion(predictions, batch['labels'])
# acc = binary_accuracy(predictions, batch.label) # *** binary ***
acc = categorical_accuracy(predictions, batch['labels']) # *** multiclass ***
epoch_loss += loss.item()
epoch_acc += acc.item()
return epoch_loss / len(iterator), epoch_acc / len(iterator)
import time
def epoch_time(start_time, end_time):
elapsed_time = end_time - start_time
elapsed_mins = int(elapsed_time / 60)
elapsed_secs = int(elapsed_time - (elapsed_mins * 60))
return elapsed_mins, elapsed_secs
N_EPOCHS = 10 # 30
best_valid_loss = float('inf')
t0 = time.time()
for epoch in range(N_EPOCHS):
start_time = time.time()
train_loss, train_acc = train(model, dataloader_d['train'], optimizer, criterion)
valid_loss, valid_acc = evaluate(model, dataloader_d['val'], criterion)
end_time = time.time()
epoch_mins, epoch_secs = epoch_time(start_time, end_time)
if valid_loss < best_valid_loss:
best_valid_loss = valid_loss
torch.save(model.state_dict(), MODEL_FILE)
print(f'Epoch: {epoch+1:02} | Epoch Time: {epoch_mins}m {epoch_secs}s')
print(f'\tTrain Loss: {train_loss:.3f} | Train Acc: {train_acc*100:.2f}%')
print(f'\t Val. Loss: {valid_loss:.3f} | Val. Acc: {valid_acc*100:.2f}%')
t1 = time.time()
train_time = t1 - t0
print('training time: {}'.format(train_time))
model = RNN(EMBEDDING_DIM,
HIDDEN_DIM,
OUTPUT_DIM,
N_LAYERS,
BIDIRECTIONAL,
DROPOUT)
model.load_state_dict(torch.load(MODEL_FILE))
model.eval().to(device)
test_loss, test_acc = evaluate(model, dataloader_d['test'], criterion)
print(f'Test Loss: {test_loss:.3f} | Test Acc: {test_acc*100:.2f}%')
results = test_acc, train_time, count_parameters(model)
results
with open(RESULTS_FILE, 'wb') as fh:
pickle.dump(results, fh)
| 0.770206 | 0.819207 |
```
import json
import time
from base64 import b64decode
from io import BytesIO, StringIO
from IPython import get_ipython
from IPython.core.magic import register_cell_magic
from IPython.utils.capture import capture_output
from IPython.core.interactiveshell import InteractiveShell
from manim import *
params = f" -v WARNING -r {80*4},{40*4} -s --disable_caching Example"
import PIL
cell_counter = 0
@register_cell_magic
def capture_png(line, cell):
global cell_counter
cell_counter += 1
# init capturing cell output
get_ipython().run_cell_magic(
'capture',
' --no-stderr --no-stdout result',
cell
)
argument_array = line.split('--style')
post_path = argument_array[0].strip()
chapter_name_underscore = chapter_name.replace(" ", "_")
path = f"gallery_assets/{chapter_name_underscore}_{cell_counter:03}_{post_path}"
# path = path.split(".png")[0] + str(time.time_ns()) + ".png" time stemps not needed
if not path:
raise ValueError('No path found!')
style = str(*argument_array[1:])
style = style.strip()
style = style.strip('"') # remove quotes
default_style = ""
style = default_style + style
raw_code_block = cell
code_block = ""
for codeline in StringIO(raw_code_block):
if "#NOT" in codeline:
pass
else:
code_block += codeline
new_codeblock = ""
for codeline in StringIO(code_block):
if "#ONLY" in codeline:
codeline= codeline.replace("#ONLY", "")
if codeline.startswith(" "): # delete the indention for manim -> TODO this can be made prettier!
codeline = codeline[8:]
new_codeblock += codeline
else:
pass
if new_codeblock: # checks if there are lines that include "#ONLY"
code_block = new_codeblock
code_block = code_block.replace("'", "'") # make sure that javascript can read the single quote character
code_block = code_block.strip("\n")
with open(joson_file_path, "r") as jsonFile:
data = json.load(jsonFile)
if not chapter_name in data:
data[chapter_name] = []
chapter_content = data[chapter_name]
chapter_content.append(
{"image_path": path,
"celltype": "Normal",
"css": style,
"code": code_block})
data[chapter_name] = chapter_content
with open(joson_file_path, "w") as jsonFile:
json.dump(data, jsonFile, indent=2, sort_keys=False)
shell = InteractiveShell.instance()
# save the output
with capture_output(stdout=False, stderr=False, display=True) as result:
shell.run_cell(cell) # idea by @krassowski
# save image
for output in result.outputs:
display(output)
data = output.data
if 'image/png' in data:
png_bytes = data['image/png']
if isinstance(png_bytes, str):
png_bytes = b64decode(png_bytes)
assert isinstance(png_bytes, bytes)
bytes_io = BytesIO(png_bytes)
image = PIL.Image.open(bytes_io)
image.save(path, 'png')
joson_file_path = "gallery_assets/gallery_parameters.json"
chapter_name = "Text"
%%capture_png Text1.png
%%manim $params
class Example(ZoomedScene):
def construct(self):
mob = Text("Hello", color=WHITE) #ONLY
self.add(mob)
self.camera.frame.scale(1/8)
%%capture_png Text2.png
%%manim $params
class Example(ZoomedScene):
def construct(self):
mob = Text("World", color=YELLOW,slant=ITALIC) #ONLY
self.add(mob)
self.camera.frame.scale(1/8)
%%capture_png Underline.png
%%manim $params
class Example(ZoomedScene):
def construct(self):
t = Tex("YES") #ONLY
ul = Underline(t, color=YELLOW) #ONLY
self.add(t, ul)
self.camera.frame.scale(1/8)
%%capture_png Bold.png
%%manim $params
class Example(ZoomedScene):
def construct(self):
mob = Text("NO", color=YELLOW,weight=BOLD) #ONLY
self.add(mob)
self.camera.frame.scale(1/8)
%%capture_png MathTex.png
%%manim $params
class Example(ZoomedScene):
def construct(self):
mob = MathTex(r"\Psi(x)", color=WHITE) #ONLY
self.add(mob)
self.camera.frame.scale(1/8)
%%capture_png MathTex_stix2.png
%%manim $params
class Example(ZoomedScene):
def construct(self):
stix2 = TexTemplate() #ONLY
stix2.add_to_preamble(r"\usepackage{stix2}", prepend= True) #ONLY
MathTex.set_default(tex_template=stix2) #ONLY
mob = MathTex(r"\Psi(x)", color=WHITE) #ONLY
self.add(mob)
self.camera.frame.scale(1/8)
%%capture_png MathTex2.png
%%manim $params
MathTex.set_default()
class Example(ZoomedScene):
def construct(self):
mob = MathTex(r"e^x",substrings_to_isolate="x") #ONLY
mob.set_color_by_tex("x", YELLOW) #ONLY
self.add(mob)
self.camera.frame.scale(1/10)
%%capture_png Check.png
%%manim $params
class Example(ZoomedScene):
def construct(self):
check= Tex(r"\checkmark", color= GREEN, stroke_width=8).scale(3) #ONLY
check.stretch(0.8, dim = 1) #ONLY
check.stretch(1.1, dim = 0) #ONLY
self.camera.frame.scale(1/7)
self.add(check)
image_full_name = "Check.png"
```
|
github_jupyter
|
import json
import time
from base64 import b64decode
from io import BytesIO, StringIO
from IPython import get_ipython
from IPython.core.magic import register_cell_magic
from IPython.utils.capture import capture_output
from IPython.core.interactiveshell import InteractiveShell
from manim import *
params = f" -v WARNING -r {80*4},{40*4} -s --disable_caching Example"
import PIL
cell_counter = 0
@register_cell_magic
def capture_png(line, cell):
global cell_counter
cell_counter += 1
# init capturing cell output
get_ipython().run_cell_magic(
'capture',
' --no-stderr --no-stdout result',
cell
)
argument_array = line.split('--style')
post_path = argument_array[0].strip()
chapter_name_underscore = chapter_name.replace(" ", "_")
path = f"gallery_assets/{chapter_name_underscore}_{cell_counter:03}_{post_path}"
# path = path.split(".png")[0] + str(time.time_ns()) + ".png" time stemps not needed
if not path:
raise ValueError('No path found!')
style = str(*argument_array[1:])
style = style.strip()
style = style.strip('"') # remove quotes
default_style = ""
style = default_style + style
raw_code_block = cell
code_block = ""
for codeline in StringIO(raw_code_block):
if "#NOT" in codeline:
pass
else:
code_block += codeline
new_codeblock = ""
for codeline in StringIO(code_block):
if "#ONLY" in codeline:
codeline= codeline.replace("#ONLY", "")
if codeline.startswith(" "): # delete the indention for manim -> TODO this can be made prettier!
codeline = codeline[8:]
new_codeblock += codeline
else:
pass
if new_codeblock: # checks if there are lines that include "#ONLY"
code_block = new_codeblock
code_block = code_block.replace("'", "'") # make sure that javascript can read the single quote character
code_block = code_block.strip("\n")
with open(joson_file_path, "r") as jsonFile:
data = json.load(jsonFile)
if not chapter_name in data:
data[chapter_name] = []
chapter_content = data[chapter_name]
chapter_content.append(
{"image_path": path,
"celltype": "Normal",
"css": style,
"code": code_block})
data[chapter_name] = chapter_content
with open(joson_file_path, "w") as jsonFile:
json.dump(data, jsonFile, indent=2, sort_keys=False)
shell = InteractiveShell.instance()
# save the output
with capture_output(stdout=False, stderr=False, display=True) as result:
shell.run_cell(cell) # idea by @krassowski
# save image
for output in result.outputs:
display(output)
data = output.data
if 'image/png' in data:
png_bytes = data['image/png']
if isinstance(png_bytes, str):
png_bytes = b64decode(png_bytes)
assert isinstance(png_bytes, bytes)
bytes_io = BytesIO(png_bytes)
image = PIL.Image.open(bytes_io)
image.save(path, 'png')
joson_file_path = "gallery_assets/gallery_parameters.json"
chapter_name = "Text"
%%capture_png Text1.png
%%manim $params
class Example(ZoomedScene):
def construct(self):
mob = Text("Hello", color=WHITE) #ONLY
self.add(mob)
self.camera.frame.scale(1/8)
%%capture_png Text2.png
%%manim $params
class Example(ZoomedScene):
def construct(self):
mob = Text("World", color=YELLOW,slant=ITALIC) #ONLY
self.add(mob)
self.camera.frame.scale(1/8)
%%capture_png Underline.png
%%manim $params
class Example(ZoomedScene):
def construct(self):
t = Tex("YES") #ONLY
ul = Underline(t, color=YELLOW) #ONLY
self.add(t, ul)
self.camera.frame.scale(1/8)
%%capture_png Bold.png
%%manim $params
class Example(ZoomedScene):
def construct(self):
mob = Text("NO", color=YELLOW,weight=BOLD) #ONLY
self.add(mob)
self.camera.frame.scale(1/8)
%%capture_png MathTex.png
%%manim $params
class Example(ZoomedScene):
def construct(self):
mob = MathTex(r"\Psi(x)", color=WHITE) #ONLY
self.add(mob)
self.camera.frame.scale(1/8)
%%capture_png MathTex_stix2.png
%%manim $params
class Example(ZoomedScene):
def construct(self):
stix2 = TexTemplate() #ONLY
stix2.add_to_preamble(r"\usepackage{stix2}", prepend= True) #ONLY
MathTex.set_default(tex_template=stix2) #ONLY
mob = MathTex(r"\Psi(x)", color=WHITE) #ONLY
self.add(mob)
self.camera.frame.scale(1/8)
%%capture_png MathTex2.png
%%manim $params
MathTex.set_default()
class Example(ZoomedScene):
def construct(self):
mob = MathTex(r"e^x",substrings_to_isolate="x") #ONLY
mob.set_color_by_tex("x", YELLOW) #ONLY
self.add(mob)
self.camera.frame.scale(1/10)
%%capture_png Check.png
%%manim $params
class Example(ZoomedScene):
def construct(self):
check= Tex(r"\checkmark", color= GREEN, stroke_width=8).scale(3) #ONLY
check.stretch(0.8, dim = 1) #ONLY
check.stretch(1.1, dim = 0) #ONLY
self.camera.frame.scale(1/7)
self.add(check)
image_full_name = "Check.png"
| 0.314261 | 0.226463 |
```
import warnings
warnings.filterwarnings('ignore')
import numpy as np
import pandas as pd
from pathlib import Path
from collections import Counter
from sklearn.metrics import balanced_accuracy_score
from sklearn.metrics import confusion_matrix
from imblearn.metrics import classification_report_imbalanced
```
# Read the CSV and Perform Basic Data Cleaning
```
# https://help.lendingclub.com/hc/en-us/articles/215488038-What-do-the-different-Note-statuses-mean-
columns = [
"loan_amnt", "int_rate", "installment", "home_ownership",
"annual_inc", "verification_status", "issue_d", "loan_status",
"pymnt_plan", "dti", "delinq_2yrs", "inq_last_6mths",
"open_acc", "pub_rec", "revol_bal", "total_acc",
"initial_list_status", "out_prncp", "out_prncp_inv", "total_pymnt",
"total_pymnt_inv", "total_rec_prncp", "total_rec_int", "total_rec_late_fee",
"recoveries", "collection_recovery_fee", "last_pymnt_amnt", "next_pymnt_d",
"collections_12_mths_ex_med", "policy_code", "application_type", "acc_now_delinq",
"tot_coll_amt", "tot_cur_bal", "open_acc_6m", "open_act_il",
"open_il_12m", "open_il_24m", "mths_since_rcnt_il", "total_bal_il",
"il_util", "open_rv_12m", "open_rv_24m", "max_bal_bc",
"all_util", "total_rev_hi_lim", "inq_fi", "total_cu_tl",
"inq_last_12m", "acc_open_past_24mths", "avg_cur_bal", "bc_open_to_buy",
"bc_util", "chargeoff_within_12_mths", "delinq_amnt", "mo_sin_old_il_acct",
"mo_sin_old_rev_tl_op", "mo_sin_rcnt_rev_tl_op", "mo_sin_rcnt_tl", "mort_acc",
"mths_since_recent_bc", "mths_since_recent_inq", "num_accts_ever_120_pd", "num_actv_bc_tl",
"num_actv_rev_tl", "num_bc_sats", "num_bc_tl", "num_il_tl",
"num_op_rev_tl", "num_rev_accts", "num_rev_tl_bal_gt_0",
"num_sats", "num_tl_120dpd_2m", "num_tl_30dpd", "num_tl_90g_dpd_24m",
"num_tl_op_past_12m", "pct_tl_nvr_dlq", "percent_bc_gt_75", "pub_rec_bankruptcies",
"tax_liens", "tot_hi_cred_lim", "total_bal_ex_mort", "total_bc_limit",
"total_il_high_credit_limit", "hardship_flag", "debt_settlement_flag"
]
target = ["loan_status"]
# Load the data
file_path = Path('./resources/LoanStats_2019Q1.csv.zip')
df = pd.read_csv(file_path, skiprows=1)[:-2]
df = df.loc[:, columns].copy()
# Drop the null columns where all values are null
df = df.dropna(axis='columns', how='all')
# Drop the null rows
df = df.dropna()
# Remove the `Issued` loan status
issued_mask = df['loan_status'] != 'Issued'
df = df.loc[issued_mask]
# convert interest rate to numerical
df['int_rate'] = df['int_rate'].str.replace('%', '')
df['int_rate'] = df['int_rate'].astype('float') / 100
# Convert the target column values to low_risk and high_risk based on their values
x = {'Current': 'low_risk'}
df = df.replace(x)
x = dict.fromkeys(['Late (31-120 days)', 'Late (16-30 days)', 'Default', 'In Grace Period'], 'high_risk')
df = df.replace(x)
df.reset_index(inplace=True, drop=True)
df.head()
```
# Split the Data into Training and Testing
```
# Create our features
X = df.copy()
X.drop(target, axis=1, inplace = True)
# Create our target
y = df[target]
X.describe()
X = pd.get_dummies(X)
# Check the balance of our target values
y['loan_status'].value_counts()
# Split the X and y into X_train, X_test, y_train, y_test
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test= train_test_split(X,y,random_state=1)
```
## Data Pre-Processing
Scale the training and testing data using the `StandardScaler` from `sklearn`. Remember that when scaling the data, you only scale the features data (`X_train` and `X_testing`).
```
# Create the StandardScaler instance
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
# Fit the Standard Scaler with the training data
# When fitting scaling functions, only train on the training dataset
X_scaler = scaler.fit(X_train)
# Scale the training and testing data
X_train_scaled = X_scaler.transform(X_train)
X_test_scaled = X_scaler.transform(X_test)
```
# Ensemble Learners
In this section, you will compare two ensemble algorithms to determine which algorithm results in the best performance. You will train a Balanced Random Forest Classifier and an Easy Ensemble classifier . For each algorithm, be sure to complete the folliowing steps:
1. Train the model using the training data.
2. Calculate the balanced accuracy score from sklearn.metrics.
3. Print the confusion matrix from sklearn.metrics.
4. Generate a classication report using the `imbalanced_classification_report` from imbalanced-learn.
5. For the Balanced Random Forest Classifier onely, print the feature importance sorted in descending order (most important feature to least important) along with the feature score
Note: Use a random state of 1 for each algorithm to ensure consistency between tests
### Balanced Random Forest Classifier
```
# Resample the training data with the BalancedRandomForestClassifier
from imblearn.ensemble import BalancedRandomForestClassifier
brf = BalancedRandomForestClassifier(n_estimators = 100, random_state = 1)
brf.fit(X_train, y_train)
y_pred_brf = brf.predict(X_test)
# Calculated the balanced accuracy score
balanced_accuracy_score(y_test, y_pred_brf)
# Display the confusion matrix
confusion_matrix(y_test, y_pred_brf)
# Print the imbalanced classification report
print(classification_report_imbalanced(y_test, y_pred_brf))
# List the features sorted in descending order by feature importance
importance = brf.feature_importances_
indice = np.argsort(importance)[::-1]
for i in range(X.shape[1]):
print(f"{X.columns.values[i]}: ({importance[indice[i]]})")
```
### Easy Ensemble Classifier
```
# Train the EasyEnsembleClassifier
from imblearn.ensemble import EasyEnsembleClassifier
eec_model = EasyEnsembleClassifier(n_estimators = 100, random_state =1)
eec_model.fit(X_train, y_train)
ypred_eec = eec_model.predict(X_test)
# Calculated the balanced accuracy score
balanced_accuracy_score(y_test, ypred_eec)
# Display the confusion matrix
print(confusion_matrix(y_test, ypred_eec))
# Print the imbalanced classification report
print(classification_report_imbalanced(y_test, ypred_eec))
```
# Question:
## Which model had the best balanced accuracy score?
Easy Ensemble Model had a more balance accuracy. Score: 0.9316600714093861
## Which model had the best recall score?
Random forest: .87 average; Easy Ensemble: high risk - .92, low risk - .94
## Which model had the best geometric mean score?
Easy Ensemble
## What are the top three features?
Top 3 features are: total_pymnt_inv, total_pymnt, total_rec_prncp
|
github_jupyter
|
import warnings
warnings.filterwarnings('ignore')
import numpy as np
import pandas as pd
from pathlib import Path
from collections import Counter
from sklearn.metrics import balanced_accuracy_score
from sklearn.metrics import confusion_matrix
from imblearn.metrics import classification_report_imbalanced
# https://help.lendingclub.com/hc/en-us/articles/215488038-What-do-the-different-Note-statuses-mean-
columns = [
"loan_amnt", "int_rate", "installment", "home_ownership",
"annual_inc", "verification_status", "issue_d", "loan_status",
"pymnt_plan", "dti", "delinq_2yrs", "inq_last_6mths",
"open_acc", "pub_rec", "revol_bal", "total_acc",
"initial_list_status", "out_prncp", "out_prncp_inv", "total_pymnt",
"total_pymnt_inv", "total_rec_prncp", "total_rec_int", "total_rec_late_fee",
"recoveries", "collection_recovery_fee", "last_pymnt_amnt", "next_pymnt_d",
"collections_12_mths_ex_med", "policy_code", "application_type", "acc_now_delinq",
"tot_coll_amt", "tot_cur_bal", "open_acc_6m", "open_act_il",
"open_il_12m", "open_il_24m", "mths_since_rcnt_il", "total_bal_il",
"il_util", "open_rv_12m", "open_rv_24m", "max_bal_bc",
"all_util", "total_rev_hi_lim", "inq_fi", "total_cu_tl",
"inq_last_12m", "acc_open_past_24mths", "avg_cur_bal", "bc_open_to_buy",
"bc_util", "chargeoff_within_12_mths", "delinq_amnt", "mo_sin_old_il_acct",
"mo_sin_old_rev_tl_op", "mo_sin_rcnt_rev_tl_op", "mo_sin_rcnt_tl", "mort_acc",
"mths_since_recent_bc", "mths_since_recent_inq", "num_accts_ever_120_pd", "num_actv_bc_tl",
"num_actv_rev_tl", "num_bc_sats", "num_bc_tl", "num_il_tl",
"num_op_rev_tl", "num_rev_accts", "num_rev_tl_bal_gt_0",
"num_sats", "num_tl_120dpd_2m", "num_tl_30dpd", "num_tl_90g_dpd_24m",
"num_tl_op_past_12m", "pct_tl_nvr_dlq", "percent_bc_gt_75", "pub_rec_bankruptcies",
"tax_liens", "tot_hi_cred_lim", "total_bal_ex_mort", "total_bc_limit",
"total_il_high_credit_limit", "hardship_flag", "debt_settlement_flag"
]
target = ["loan_status"]
# Load the data
file_path = Path('./resources/LoanStats_2019Q1.csv.zip')
df = pd.read_csv(file_path, skiprows=1)[:-2]
df = df.loc[:, columns].copy()
# Drop the null columns where all values are null
df = df.dropna(axis='columns', how='all')
# Drop the null rows
df = df.dropna()
# Remove the `Issued` loan status
issued_mask = df['loan_status'] != 'Issued'
df = df.loc[issued_mask]
# convert interest rate to numerical
df['int_rate'] = df['int_rate'].str.replace('%', '')
df['int_rate'] = df['int_rate'].astype('float') / 100
# Convert the target column values to low_risk and high_risk based on their values
x = {'Current': 'low_risk'}
df = df.replace(x)
x = dict.fromkeys(['Late (31-120 days)', 'Late (16-30 days)', 'Default', 'In Grace Period'], 'high_risk')
df = df.replace(x)
df.reset_index(inplace=True, drop=True)
df.head()
# Create our features
X = df.copy()
X.drop(target, axis=1, inplace = True)
# Create our target
y = df[target]
X.describe()
X = pd.get_dummies(X)
# Check the balance of our target values
y['loan_status'].value_counts()
# Split the X and y into X_train, X_test, y_train, y_test
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test= train_test_split(X,y,random_state=1)
# Create the StandardScaler instance
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
# Fit the Standard Scaler with the training data
# When fitting scaling functions, only train on the training dataset
X_scaler = scaler.fit(X_train)
# Scale the training and testing data
X_train_scaled = X_scaler.transform(X_train)
X_test_scaled = X_scaler.transform(X_test)
# Resample the training data with the BalancedRandomForestClassifier
from imblearn.ensemble import BalancedRandomForestClassifier
brf = BalancedRandomForestClassifier(n_estimators = 100, random_state = 1)
brf.fit(X_train, y_train)
y_pred_brf = brf.predict(X_test)
# Calculated the balanced accuracy score
balanced_accuracy_score(y_test, y_pred_brf)
# Display the confusion matrix
confusion_matrix(y_test, y_pred_brf)
# Print the imbalanced classification report
print(classification_report_imbalanced(y_test, y_pred_brf))
# List the features sorted in descending order by feature importance
importance = brf.feature_importances_
indice = np.argsort(importance)[::-1]
for i in range(X.shape[1]):
print(f"{X.columns.values[i]}: ({importance[indice[i]]})")
# Train the EasyEnsembleClassifier
from imblearn.ensemble import EasyEnsembleClassifier
eec_model = EasyEnsembleClassifier(n_estimators = 100, random_state =1)
eec_model.fit(X_train, y_train)
ypred_eec = eec_model.predict(X_test)
# Calculated the balanced accuracy score
balanced_accuracy_score(y_test, ypred_eec)
# Display the confusion matrix
print(confusion_matrix(y_test, ypred_eec))
# Print the imbalanced classification report
print(classification_report_imbalanced(y_test, ypred_eec))
| 0.597138 | 0.758622 |
<a href="https://colab.research.google.com/github/jameschapman19/cca_zoo/blob/master/tutorial_notebooks/cca_zoo_tutorial.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
```
!pip install cca-zoo[deep,probabilistic]
!pip install scipy --upgrade
# Imports
import numpy as np
from cca_zoo.data import Noisy_MNIST_Dataset
import itertools
import os
import matplotlib.pyplot as plt
from torch.utils.data import Subset
from torch import optim
# Load MNIST Data
os.chdir('..')
N = 500
dataset = Noisy_MNIST_Dataset(mnist_type='FashionMNIST', train=True)
ids = np.arange(min(2 * N, len(dataset)))
np.random.shuffle(ids)
train_ids, val_ids = np.array_split(ids, 2)
val_dataset = Subset(dataset, val_ids)
train_dataset = Subset(dataset, train_ids)
test_dataset = Noisy_MNIST_Dataset(mnist_type='FashionMNIST', train=False)
test_ids = np.arange(min(N, len(test_dataset)))
np.random.shuffle(test_ids)
test_dataset = Subset(test_dataset, test_ids)
train_view_1, train_view_2, train_rotations, train_OH_labels, train_labels = train_dataset.dataset.to_numpy(
train_dataset.indices)
val_view_1, val_view_2, val_rotations, val_OH_labels, val_labels = val_dataset.dataset.to_numpy(val_dataset.indices)
test_view_1, test_view_2, test_rotations, test_OH_labels, test_labels = test_dataset.dataset.to_numpy(
test_dataset.indices)
# Settings
# The number of latent dimensions across models
latent_dims = 2
# The number of folds used for cross-validation/hyperparameter tuning
cv_folds = 5
# For running hyperparameter tuning in parallel (0 if not)
jobs = 2
# Number of iterations for iterative algorithms
max_iter = 2
# number of epochs for deep models
epochs = 50
```
# Canonical Correlation Analysis
```
from cca_zoo.models import CCA, CCA_ALS
"""
### Linear CCA by eigendecomposition
"""
linear_cca = CCA(latent_dims=latent_dims)
linear_cca.fit(train_view_1, train_view_2)
linear_cca_results = np.stack(
(linear_cca.train_correlations[0, 1], linear_cca.predict_corr(test_view_1, test_view_2)[0, 1]))
"""
### Linear CCA by alternating least squares (can pass more than 2 views)
"""
linear_cca_als = CCA_ALS(latent_dims=latent_dims)
linear_cca_als.fit(train_view_1, train_view_2)
linear_cca_als_results = np.stack(
(linear_cca_als.train_correlations[0, 1], linear_cca_als.predict_corr(test_view_1, test_view_2)[0, 1]))
```
# Partial Least Squares
```
from cca_zoo.models import PLS
"""
### PLS with scikit-learn (only permits 2 views)
"""
pls = PLS(latent_dims=latent_dims)
pls.fit(train_view_1, train_view_2)
pls_results = np.stack(
(pls.train_correlations[0, 1], pls.predict_corr(test_view_1, test_view_2)[0, 1]))
```
# Extension to multiple views
```
from cca_zoo.models import GCCA, MCCA
"""
### (Regularized) Generalized CCA(can pass more than 2 views)
"""
# small ammount of regularisation added since data is not full rank
c=[0.5,0.5,0.5]
gcca = GCCA(latent_dims=latent_dims,c=c)
gcca.fit(train_view_1, train_view_2,train_view_1)
gcca_results = np.stack((gcca.train_correlations[0, 1], gcca.predict_corr(test_view_1, test_view_2)[0, 1]))
"""
### (Regularized) Multiset CCA(can pass more than 2 views)
"""
mcca = MCCA(latent_dims=latent_dims, c=c)
mcca.fit(train_view_1, train_view_2,train_view_1)
mcca_results = np.stack((mcca.train_correlations[0, 1], mcca.predict_corr(test_view_1, test_view_2)[0, 1]))
"""
### Multiset CCA by alternating least squares
"""
mcca_als = CCA_ALS(latent_dims=latent_dims, max_iter=max_iter)
mcca_als.fit(train_view_1, train_view_2,train_view_1)
mcca_als_results = np.stack(
(mcca_als.train_correlations[0, 1], mcca_als.predict_corr(test_view_1, test_view_2)[0, 1]))
"""
### Multiset PLS by alternating least squares
"""
mcca_pls = PLS(latent_dims=latent_dims, max_iter=max_iter)
mcca_pls.fit(train_view_1, train_view_2,train_view_1)
mcca_pls_results = np.stack(
(mcca_als.train_correlations[0, 1], mcca_pls.predict_corr(test_view_1, test_view_2)[0, 1]))
```
# Tensor CCA
```
from cca_zoo.models import TCCA
"""
### (Regularized) Tensor CCA(can pass more than 2 views)
"""
tcca = TCCA(latent_dims=latent_dims, c=c)
tcca.fit(train_view_1[:,:100], train_view_2[:,:100],train_view_1[:,:100])
tcca_results = np.stack((tcca.train_correlations[0, 1], tcca.predict_corr(test_view_1[:,:100], test_view_2[:,:100])[0, 1]))
```
# Weighted GCCA/Missing Observation GCCA
```
#observation_matrix
K = np.ones((3, N))
K[0, 200:] = 0
K[1, :100] = 0
#view weights
view_weights=[1,2,1.2]
c=[0.5,0.5,0.5]
gcca = GCCA(latent_dims=latent_dims,c=c,view_weights=view_weights)
gcca.fit(train_view_1, train_view_2,train_view_1,K=K)
gcca_results = np.stack((gcca.train_correlations[0, 1], gcca.predict_corr(test_view_1, test_view_2)[0, 1]))
```
# Rgularised CCA solutions based on alternating minimisation/alternating least squares
We implement Witten's penalized matrix decomposition form of sparse CCA using 'pmd'
We implement Waaijenborg's penalized CCA using elastic net using 'elastic'
We implement Mai's sparse CCA using 'scca'
Furthermore, any of these methods can be extended to multiple views. Witten describes this method explicitly.
```
from cca_zoo.models import rCCA, PMD,SCCA,ElasticCCA
"""
### Ridge CCA (can pass more than 2 views)
"""
c1 = [0.1, 0.3, 0.7, 0.9]
c2 = [0.1, 0.3, 0.7, 0.9]
param_candidates = {'c': list(itertools.product(c1, c2))}
ridge = rCCA(latent_dims=latent_dims).gridsearch_fit(
train_view_1,
train_view_2,
param_candidates=param_candidates,
folds=cv_folds,
verbose=True, jobs=jobs,
plot=True)
ridge_results = np.stack((ridge.train_correlations[0, 1, :], ridge.predict_corr(test_view_1, test_view_2)[0, 1, :]))
"""
### Sparse CCA (Penalized Matrix Decomposition) (can pass more than 2 views)
"""
# PMD
c1 = [1, 3, 7, 9]
c2 = [1, 3, 7, 9]
param_candidates = {'c': list(itertools.product(c1, c2))}
pmd = PMD(latent_dims=latent_dims, max_iter=max_iter).gridsearch_fit(
train_view_1,
train_view_2,
param_candidates=param_candidates,
folds=cv_folds,
verbose=True, jobs=jobs,
plot=True)
pmd_results = np.stack((pmd.train_correlations[0, 1, :], pmd.predict_corr(test_view_1, test_view_2)[0, 1, :]))
"""
### Sparse CCA (can pass more than 2 views)
"""
# Sparse CCA
c1 = [0.00001, 0.0001]
c2 = [0.00001, 0.0001]
param_candidates = {'c': list(itertools.product(c1, c2))}
scca = SCCA(latent_dims=latent_dims, max_iter=max_iter).gridsearch_fit(
train_view_1,
train_view_2,
param_candidates=param_candidates,
folds=cv_folds,
verbose=True,
jobs=jobs, plot=True)
scca_results = np.stack(
(scca.train_correlations[0, 1, :], scca.predict_corr(test_view_1, test_view_2)[0, 1, :]))
"""
### Elastic CCA (can pass more than 2 views)
"""
# Elastic CCA
c1 = [0.001, 0.0001]
c2 = [0.001, 0.0001]
l1_1 = [0.01, 0.1]
l1_2 = [0.01, 0.1]
param_candidates = {'c': list(itertools.product(c1, c2)), 'l1_ratio': list(itertools.product(l1_1, l1_2))}
elastic = ElasticCCA(latent_dims=latent_dims,
max_iter=max_iter).gridsearch_fit(train_view_1,
train_view_2,
param_candidates=param_candidates,
folds=cv_folds,
verbose=True,
jobs=jobs,
plot=True)
elastic_results = np.stack(
(elastic.train_correlations[0, 1, :], elastic.predict_corr(test_view_1, test_view_2)[0, 1, :]))
```
# Kernel CCA
```
from cca_zoo.models import KCCA
"""
### Kernel CCA
Similarly, we can use kernel CCA methods with [method='kernel']
We can use different kernels and their associated parameters in a similar manner to before
- regularized linear kernel CCA: parameters : 'kernel'='linear', 0<'c'<1
- polynomial kernel CCA: parameters : 'kernel'='poly', 'degree', 0<'c'<1
- gaussian rbf kernel CCA: parameters : 'kernel'='gaussian', 'sigma', 0<'c'<1
"""
# %%
# r-kernel cca
c1 = [0.9, 0.99]
c2 = [0.9, 0.99]
param_candidates = {'kernel': [['linear', 'linear']], 'c': list(itertools.product(c1, c2))}
kernel_reg = KCCA(latent_dims=latent_dims).gridsearch_fit(train_view_1, train_view_2,
folds=cv_folds,
param_candidates=param_candidates,
verbose=True, jobs=jobs,
plot=True)
kernel_reg_results = np.stack((
kernel_reg.train_correlations[0, 1, :],
kernel_reg.predict_corr(test_view_1, test_view_2)[0, 1, :]))
# kernel cca (poly)
degree1 = [2, 3]
degree2 = [2, 3]
param_candidates = {'kernel': [['poly', 'poly']], 'degree': list(itertools.product(degree1, degree2)),
'c': list(itertools.product(c1, c2))}
kernel_poly = KCCA(latent_dims=latent_dims).gridsearch_fit(train_view_1, train_view_2,
folds=cv_folds,
param_candidates=param_candidates,
verbose=True, jobs=jobs,
plot=True)
kernel_poly_results = np.stack((
kernel_poly.train_correlations[0, 1, :],
kernel_poly.predict_corr(test_view_1, test_view_2)[0, 1, :]))
# kernel cca (gaussian)
gamma1 = [1e+1, 1e+2, 1e+3]
gamma2 = [1e+1, 1e+2, 1e+3]
param_candidates = {'kernel': [['rbf', 'rbf']], 'gamma': list(itertools.product(gamma1, gamma2)),
'c': list(itertools.product(c1, c2))}
kernel_gaussian = KCCA(latent_dims=latent_dims).gridsearch_fit(train_view_1, train_view_2,
folds=cv_folds,
param_candidates=param_candidates,
verbose=True, jobs=jobs,
plot=True)
kernel_gaussian_results = np.stack((
kernel_gaussian.train_correlations[0, 1, :],
kernel_gaussian.predict_corr(test_view_1, test_view_2)[0, 1, :]))
```
# Deep CCA
DCCA can be optimized using Andrew's original tracenorm objective or Wang's DCCA by nonlinear orthogonal iterations using the argument als=True.
```
"""
### Deep Learning
We also have deep CCA methods (and autoencoder variants)
- Deep CCA (DCCA)
- Deep Canonically Correlated Autoencoders (DCCAE)
We introduce a Config class from configuration.py. This contains a number of default settings for running DCCA.
"""
from cca_zoo.deepmodels import Dee
# %%
# DCCA
print('DCCA')
encoder_1 = deep_models.Encoder(latent_dims=latent_dims, feature_size=784)
encoder_2 = deep_models.Encoder(latent_dims=latent_dims, feature_size=784)
dcca_model = DCCA(latent_dims=latent_dims, encoders=[encoder_1, encoder_2])
dcca_model = DeepWrapper(dcca_model)
dcca_model.fit(train_dataset, val_dataset=val_dataset, epochs=epochs)
dcca_results = np.stack((dcca_model.train_correlations[0, 1], dcca_model.predict_corr(test_dataset)[0, 1]))
from cca_zoo import dcca_noi
# DCCA_NOI
# Note that als=True
print('DCCA by non-linear orthogonal iterations')
encoder_1 = deep_models.Encoder(latent_dims=latent_dims, feature_size=784)
encoder_2 = deep_models.Encoder(latent_dims=latent_dims, feature_size=784)
dcca_noi_model = dcca_noi.DCCA_NOI(latent_dims=latent_dims, encoders=[encoder_1, encoder_2])
dcca_noi_model = deepwrapper.DeepWrapper(dcca_noi_model)
dcca_noi_model.fit(train_dataset, val_dataset=val_dataset, epochs=epochs)
dcca_noi_results = np.stack(
(dcca_noi_model.train_correlations[0, 1], dcca_noi_model.predict_corr(test_dataset)[0, 1]))
```
# DCCA with custom optimizers and schedulers
```
# DCCA
optimizers = [optim.Adam(encoder_1.parameters(), lr=1e-4), optim.Adam(encoder_2.parameters(), lr=1e-4)]
schedulers = [optim.lr_scheduler.CosineAnnealingLR(optimizers[0], 1),
optim.lr_scheduler.ReduceLROnPlateau(optimizers[1])]
dcca_model = dcca.DCCA(latent_dims=latent_dims, encoders=[encoder_1, encoder_2],
objective=objectives.CCA, optimizers=optimizers, schedulers=schedulers)
# hidden_layer_sizes are shown explicitly but these are also the defaults
dcca_model = deepwrapper.DeepWrapper(dcca_model)
dcca_model.fit(train_dataset, val_dataset=val_dataset,epochs=20)
```
# DGCCA and DMCCA for more than 2 views
The only change we need to make is to the objective argument to perform DGCCA and DMCCA.
```
# DGCCA
print('DGCCA')
encoder_1 = deep_models.Encoder(latent_dims=latent_dims, feature_size=784)
encoder_2 = deep_models.Encoder(latent_dims=latent_dims, feature_size=784)
dgcca_model = dcca.DCCA(latent_dims=latent_dims, encoders=[encoder_1, encoder_2], objective=objectives.GCCA)
dgcca_model = deepwrapper.DeepWrapper(dgcca_model)
dgcca_model.fit(train_dataset, val_dataset=val_dataset, epochs=epochs)
dgcca_results = np.stack(
(dgcca_model.train_correlations[0, 1], dgcca_model.predict_corr(test_dataset)[0, 1]))
# DMCCA
print('DMCCA')
encoder_1 = deep_models.Encoder(latent_dims=latent_dims, feature_size=784)
encoder_2 = deep_models.Encoder(latent_dims=latent_dims, feature_size=784)
dmcca_model = dcca.DCCA(latent_dims=latent_dims, encoders=[encoder_1, encoder_2], objective=objectives.MCCA)
dmcca_model = deepwrapper.DeepWrapper(dmcca_model)
dmcca_model.fit(train_dataset, val_dataset=val_dataset, epochs=epochs)
dmcca_results = np.stack(
(dmcca_model.train_correlations[0, 1], dmcca_model.predict_corr(test_dataset)[0, 1]))
```
# Deep Canonically Correlated Autoencoders
We need to add decoders in order to model deep canonically correlated autoencoders and we also use the DCCAE class which inherits from DCCA
```
from cca_zoo import dccae
# DCCAE
print('DCCAE')
encoder_1 = deep_models.Encoder(latent_dims=latent_dims, feature_size=784)
encoder_2 = deep_models.Encoder(latent_dims=latent_dims, feature_size=784)
decoder_1 = deep_models.Decoder(latent_dims=latent_dims, feature_size=784)
decoder_2 = deep_models.Decoder(latent_dims=latent_dims, feature_size=784)
dccae_model = dccae.DCCAE(latent_dims=latent_dims, encoders=[encoder_1, encoder_2], decoders=[decoder_1, decoder_2])
dccae_model = deepwrapper.DeepWrapper(dccae_model)
#can also pass a tuple of numpy arrays
dccae_model.fit((train_view_1, train_view_2), epochs=epochs)
dccae_results = np.stack(
(dccae_model.train_correlations[0, 1], dccae_model.predict_corr(test_dataset)[0, 1]))
```
# Deep Variational CCA
```
"""
### Deep Variational Learning
Finally we have Deep Variational CCA methods.
- Deep Variational CCA (DVCCA)
- Deep Variational CCA - private (DVVCA_p)
These are both implemented by the DVCCA class with private=True/False and both_encoders=True/False. If both_encoders,
the encoder to the shared information Q(z_shared|x) is modelled for both x_1 and x_2 whereas if both_encoders is false
it is modelled for x_1 as in the paper
"""
from cca_zoo import dvcca
# %%
# DVCCA (technically bi-DVCCA)
print('DVCCA')
encoder_1 = deep_models.Encoder(latent_dims=latent_dims, feature_size=784, variational=True)
encoder_2 = deep_models.Encoder(latent_dims=latent_dims, feature_size=784, variational=True)
decoder_1 = deep_models.Decoder(latent_dims=latent_dims, feature_size=784, norm_output=True)
decoder_2 = deep_models.Decoder(latent_dims=latent_dims, feature_size=784, norm_output=True)
dvcca_model = dvcca.DVCCA(latent_dims=latent_dims, encoders=[encoder_1, encoder_2], decoders=[decoder_1, decoder_2],
private=False)
dvcca_model = deepwrapper.DeepWrapper(dvcca_model)
dvcca_model.fit(train_dataset, val_dataset=val_dataset, epochs=epochs)
dvcca_model_results = np.stack(
(dvcca_model.train_correlations[0, 1], dvcca_model.predict_corr(test_dataset)[0, 1]))
# DVCCA_private (technically bi-DVCCA_private)
print('DVCCA_private')
encoder_1 = deep_models.Encoder(latent_dims=latent_dims, feature_size=784, variational=True)
encoder_2 = deep_models.Encoder(latent_dims=latent_dims, feature_size=784, variational=True)
private_encoder_1 = deep_models.Encoder(latent_dims=latent_dims, feature_size=784, variational=True)
private_encoder_2 = deep_models.Encoder(latent_dims=latent_dims, feature_size=784, variational=True)
decoder_1 = deep_models.Decoder(latent_dims=latent_dims * 2, feature_size=784, norm_output=True)
decoder_2 = deep_models.Decoder(latent_dims=latent_dims * 2, feature_size=784, norm_output=True)
dvccap_model = dvcca.DVCCA(latent_dims=latent_dims, encoders=[encoder_1, encoder_2], decoders=[decoder_1, decoder_2],
private_encoders=[private_encoder_1, private_encoder_2], private=True)
dvccap_model = deepwrapper.DeepWrapper(dvccap_model)
dvccap_model.fit(train_dataset, val_dataset=val_dataset, epochs=epochs)
dvccap_model_results = np.stack(
(dvccap_model.train_correlations[0, 1], dvccap_model.predict_corr(test_dataset)[0, 1]))
```
# Convolutional Deep CCA (and using other architectures)
We provide a standard CNN encoder and decoder but users can build their own encoders and decoders by inheriting BaseEncoder and BaseDecoder for seamless integration with the pipeline
```
print('Convolutional DCCA')
encoder_1 = deep_models.CNNEncoder(latent_dims=latent_dims, channels=[3, 3])
encoder_2 = deep_models.CNNEncoder(latent_dims=latent_dims, channels=[3, 3])
dcca_conv_model = dcca.DCCA(latent_dims=latent_dims, encoders=[encoder_1, encoder_2])
dcca_conv_model = deepwrapper.DeepWrapper(dcca_conv_model)
# to change the models used change the cfg.encoder_models. We implement a CNN_Encoder and CNN_decoder as well
# as some based on brainnet architecture in cca_zoo.deep_models. Equally you could pass your own encoder/decoder models
dcca_conv_model.fit((train_view_1.reshape((-1, 1, 28, 28)), train_view_2.reshape((-1, 1, 28, 28))), epochs=epochs)
dcca_conv_results = np.stack(
(dcca_conv_model.train_correlations[0, 1], dcca_conv_model.predict_corr((test_view_1.reshape((-1, 1, 28, 28)),
test_view_2.reshape(
(-1, 1, 28, 28))))[0, 1]))
```
# DTCCA
```
from cca_zoo import dtcca
# %%
# DTCCA
print('DTCCA')
encoder_1 = deep_models.Encoder(latent_dims=latent_dims, feature_size=784)
encoder_2 = deep_models.Encoder(latent_dims=latent_dims, feature_size=784)
dtcca_model = dtcca.DTCCA(latent_dims=latent_dims, encoders=[encoder_1, encoder_2])
dtcca_model = deepwrapper.DeepWrapper(dtcca_model)
dtcca_model.fit(train_dataset, val_dataset=val_dataset, epochs=epochs)
dtcca_results = np.stack((dtcca_model.train_correlations[0, 1], dtcca_model.predict_corr(test_dataset)[0, 1]))
```
# Generate Some Plots
```
"""
### Make results plot to compare methods
"""
# %%
all_results = np.stack(
[linear_cca_results, gcca_results, mcca_results, pls_results, pmd_results, elastic_results,
scca_results, kernel_reg_results, kernel_poly_results,
kernel_gaussian_results, dcca_results, dgcca_results, dmcca_results, dvcca_model_results,
dcca_conv_results, dtcca_results],
axis=0)
all_labels = ['linear', 'gcca', 'mcca', 'pls', 'pmd', 'elastic', 'scca', 'linear kernel', 'polynomial kernel',
'gaussian kernel', 'deep CCA', 'deep generalized CCA', 'deep multiset CCA', 'deep VCCA',
'deep convolutional cca', 'DTCCA']
from cca_zoo import plot_utils
plot_utils.plot_results(all_results, all_labels)
plt.show()
```
|
github_jupyter
|
!pip install cca-zoo[deep,probabilistic]
!pip install scipy --upgrade
# Imports
import numpy as np
from cca_zoo.data import Noisy_MNIST_Dataset
import itertools
import os
import matplotlib.pyplot as plt
from torch.utils.data import Subset
from torch import optim
# Load MNIST Data
os.chdir('..')
N = 500
dataset = Noisy_MNIST_Dataset(mnist_type='FashionMNIST', train=True)
ids = np.arange(min(2 * N, len(dataset)))
np.random.shuffle(ids)
train_ids, val_ids = np.array_split(ids, 2)
val_dataset = Subset(dataset, val_ids)
train_dataset = Subset(dataset, train_ids)
test_dataset = Noisy_MNIST_Dataset(mnist_type='FashionMNIST', train=False)
test_ids = np.arange(min(N, len(test_dataset)))
np.random.shuffle(test_ids)
test_dataset = Subset(test_dataset, test_ids)
train_view_1, train_view_2, train_rotations, train_OH_labels, train_labels = train_dataset.dataset.to_numpy(
train_dataset.indices)
val_view_1, val_view_2, val_rotations, val_OH_labels, val_labels = val_dataset.dataset.to_numpy(val_dataset.indices)
test_view_1, test_view_2, test_rotations, test_OH_labels, test_labels = test_dataset.dataset.to_numpy(
test_dataset.indices)
# Settings
# The number of latent dimensions across models
latent_dims = 2
# The number of folds used for cross-validation/hyperparameter tuning
cv_folds = 5
# For running hyperparameter tuning in parallel (0 if not)
jobs = 2
# Number of iterations for iterative algorithms
max_iter = 2
# number of epochs for deep models
epochs = 50
from cca_zoo.models import CCA, CCA_ALS
"""
### Linear CCA by eigendecomposition
"""
linear_cca = CCA(latent_dims=latent_dims)
linear_cca.fit(train_view_1, train_view_2)
linear_cca_results = np.stack(
(linear_cca.train_correlations[0, 1], linear_cca.predict_corr(test_view_1, test_view_2)[0, 1]))
"""
### Linear CCA by alternating least squares (can pass more than 2 views)
"""
linear_cca_als = CCA_ALS(latent_dims=latent_dims)
linear_cca_als.fit(train_view_1, train_view_2)
linear_cca_als_results = np.stack(
(linear_cca_als.train_correlations[0, 1], linear_cca_als.predict_corr(test_view_1, test_view_2)[0, 1]))
from cca_zoo.models import PLS
"""
### PLS with scikit-learn (only permits 2 views)
"""
pls = PLS(latent_dims=latent_dims)
pls.fit(train_view_1, train_view_2)
pls_results = np.stack(
(pls.train_correlations[0, 1], pls.predict_corr(test_view_1, test_view_2)[0, 1]))
from cca_zoo.models import GCCA, MCCA
"""
### (Regularized) Generalized CCA(can pass more than 2 views)
"""
# small ammount of regularisation added since data is not full rank
c=[0.5,0.5,0.5]
gcca = GCCA(latent_dims=latent_dims,c=c)
gcca.fit(train_view_1, train_view_2,train_view_1)
gcca_results = np.stack((gcca.train_correlations[0, 1], gcca.predict_corr(test_view_1, test_view_2)[0, 1]))
"""
### (Regularized) Multiset CCA(can pass more than 2 views)
"""
mcca = MCCA(latent_dims=latent_dims, c=c)
mcca.fit(train_view_1, train_view_2,train_view_1)
mcca_results = np.stack((mcca.train_correlations[0, 1], mcca.predict_corr(test_view_1, test_view_2)[0, 1]))
"""
### Multiset CCA by alternating least squares
"""
mcca_als = CCA_ALS(latent_dims=latent_dims, max_iter=max_iter)
mcca_als.fit(train_view_1, train_view_2,train_view_1)
mcca_als_results = np.stack(
(mcca_als.train_correlations[0, 1], mcca_als.predict_corr(test_view_1, test_view_2)[0, 1]))
"""
### Multiset PLS by alternating least squares
"""
mcca_pls = PLS(latent_dims=latent_dims, max_iter=max_iter)
mcca_pls.fit(train_view_1, train_view_2,train_view_1)
mcca_pls_results = np.stack(
(mcca_als.train_correlations[0, 1], mcca_pls.predict_corr(test_view_1, test_view_2)[0, 1]))
from cca_zoo.models import TCCA
"""
### (Regularized) Tensor CCA(can pass more than 2 views)
"""
tcca = TCCA(latent_dims=latent_dims, c=c)
tcca.fit(train_view_1[:,:100], train_view_2[:,:100],train_view_1[:,:100])
tcca_results = np.stack((tcca.train_correlations[0, 1], tcca.predict_corr(test_view_1[:,:100], test_view_2[:,:100])[0, 1]))
#observation_matrix
K = np.ones((3, N))
K[0, 200:] = 0
K[1, :100] = 0
#view weights
view_weights=[1,2,1.2]
c=[0.5,0.5,0.5]
gcca = GCCA(latent_dims=latent_dims,c=c,view_weights=view_weights)
gcca.fit(train_view_1, train_view_2,train_view_1,K=K)
gcca_results = np.stack((gcca.train_correlations[0, 1], gcca.predict_corr(test_view_1, test_view_2)[0, 1]))
from cca_zoo.models import rCCA, PMD,SCCA,ElasticCCA
"""
### Ridge CCA (can pass more than 2 views)
"""
c1 = [0.1, 0.3, 0.7, 0.9]
c2 = [0.1, 0.3, 0.7, 0.9]
param_candidates = {'c': list(itertools.product(c1, c2))}
ridge = rCCA(latent_dims=latent_dims).gridsearch_fit(
train_view_1,
train_view_2,
param_candidates=param_candidates,
folds=cv_folds,
verbose=True, jobs=jobs,
plot=True)
ridge_results = np.stack((ridge.train_correlations[0, 1, :], ridge.predict_corr(test_view_1, test_view_2)[0, 1, :]))
"""
### Sparse CCA (Penalized Matrix Decomposition) (can pass more than 2 views)
"""
# PMD
c1 = [1, 3, 7, 9]
c2 = [1, 3, 7, 9]
param_candidates = {'c': list(itertools.product(c1, c2))}
pmd = PMD(latent_dims=latent_dims, max_iter=max_iter).gridsearch_fit(
train_view_1,
train_view_2,
param_candidates=param_candidates,
folds=cv_folds,
verbose=True, jobs=jobs,
plot=True)
pmd_results = np.stack((pmd.train_correlations[0, 1, :], pmd.predict_corr(test_view_1, test_view_2)[0, 1, :]))
"""
### Sparse CCA (can pass more than 2 views)
"""
# Sparse CCA
c1 = [0.00001, 0.0001]
c2 = [0.00001, 0.0001]
param_candidates = {'c': list(itertools.product(c1, c2))}
scca = SCCA(latent_dims=latent_dims, max_iter=max_iter).gridsearch_fit(
train_view_1,
train_view_2,
param_candidates=param_candidates,
folds=cv_folds,
verbose=True,
jobs=jobs, plot=True)
scca_results = np.stack(
(scca.train_correlations[0, 1, :], scca.predict_corr(test_view_1, test_view_2)[0, 1, :]))
"""
### Elastic CCA (can pass more than 2 views)
"""
# Elastic CCA
c1 = [0.001, 0.0001]
c2 = [0.001, 0.0001]
l1_1 = [0.01, 0.1]
l1_2 = [0.01, 0.1]
param_candidates = {'c': list(itertools.product(c1, c2)), 'l1_ratio': list(itertools.product(l1_1, l1_2))}
elastic = ElasticCCA(latent_dims=latent_dims,
max_iter=max_iter).gridsearch_fit(train_view_1,
train_view_2,
param_candidates=param_candidates,
folds=cv_folds,
verbose=True,
jobs=jobs,
plot=True)
elastic_results = np.stack(
(elastic.train_correlations[0, 1, :], elastic.predict_corr(test_view_1, test_view_2)[0, 1, :]))
from cca_zoo.models import KCCA
"""
### Kernel CCA
Similarly, we can use kernel CCA methods with [method='kernel']
We can use different kernels and their associated parameters in a similar manner to before
- regularized linear kernel CCA: parameters : 'kernel'='linear', 0<'c'<1
- polynomial kernel CCA: parameters : 'kernel'='poly', 'degree', 0<'c'<1
- gaussian rbf kernel CCA: parameters : 'kernel'='gaussian', 'sigma', 0<'c'<1
"""
# %%
# r-kernel cca
c1 = [0.9, 0.99]
c2 = [0.9, 0.99]
param_candidates = {'kernel': [['linear', 'linear']], 'c': list(itertools.product(c1, c2))}
kernel_reg = KCCA(latent_dims=latent_dims).gridsearch_fit(train_view_1, train_view_2,
folds=cv_folds,
param_candidates=param_candidates,
verbose=True, jobs=jobs,
plot=True)
kernel_reg_results = np.stack((
kernel_reg.train_correlations[0, 1, :],
kernel_reg.predict_corr(test_view_1, test_view_2)[0, 1, :]))
# kernel cca (poly)
degree1 = [2, 3]
degree2 = [2, 3]
param_candidates = {'kernel': [['poly', 'poly']], 'degree': list(itertools.product(degree1, degree2)),
'c': list(itertools.product(c1, c2))}
kernel_poly = KCCA(latent_dims=latent_dims).gridsearch_fit(train_view_1, train_view_2,
folds=cv_folds,
param_candidates=param_candidates,
verbose=True, jobs=jobs,
plot=True)
kernel_poly_results = np.stack((
kernel_poly.train_correlations[0, 1, :],
kernel_poly.predict_corr(test_view_1, test_view_2)[0, 1, :]))
# kernel cca (gaussian)
gamma1 = [1e+1, 1e+2, 1e+3]
gamma2 = [1e+1, 1e+2, 1e+3]
param_candidates = {'kernel': [['rbf', 'rbf']], 'gamma': list(itertools.product(gamma1, gamma2)),
'c': list(itertools.product(c1, c2))}
kernel_gaussian = KCCA(latent_dims=latent_dims).gridsearch_fit(train_view_1, train_view_2,
folds=cv_folds,
param_candidates=param_candidates,
verbose=True, jobs=jobs,
plot=True)
kernel_gaussian_results = np.stack((
kernel_gaussian.train_correlations[0, 1, :],
kernel_gaussian.predict_corr(test_view_1, test_view_2)[0, 1, :]))
"""
### Deep Learning
We also have deep CCA methods (and autoencoder variants)
- Deep CCA (DCCA)
- Deep Canonically Correlated Autoencoders (DCCAE)
We introduce a Config class from configuration.py. This contains a number of default settings for running DCCA.
"""
from cca_zoo.deepmodels import Dee
# %%
# DCCA
print('DCCA')
encoder_1 = deep_models.Encoder(latent_dims=latent_dims, feature_size=784)
encoder_2 = deep_models.Encoder(latent_dims=latent_dims, feature_size=784)
dcca_model = DCCA(latent_dims=latent_dims, encoders=[encoder_1, encoder_2])
dcca_model = DeepWrapper(dcca_model)
dcca_model.fit(train_dataset, val_dataset=val_dataset, epochs=epochs)
dcca_results = np.stack((dcca_model.train_correlations[0, 1], dcca_model.predict_corr(test_dataset)[0, 1]))
from cca_zoo import dcca_noi
# DCCA_NOI
# Note that als=True
print('DCCA by non-linear orthogonal iterations')
encoder_1 = deep_models.Encoder(latent_dims=latent_dims, feature_size=784)
encoder_2 = deep_models.Encoder(latent_dims=latent_dims, feature_size=784)
dcca_noi_model = dcca_noi.DCCA_NOI(latent_dims=latent_dims, encoders=[encoder_1, encoder_2])
dcca_noi_model = deepwrapper.DeepWrapper(dcca_noi_model)
dcca_noi_model.fit(train_dataset, val_dataset=val_dataset, epochs=epochs)
dcca_noi_results = np.stack(
(dcca_noi_model.train_correlations[0, 1], dcca_noi_model.predict_corr(test_dataset)[0, 1]))
# DCCA
optimizers = [optim.Adam(encoder_1.parameters(), lr=1e-4), optim.Adam(encoder_2.parameters(), lr=1e-4)]
schedulers = [optim.lr_scheduler.CosineAnnealingLR(optimizers[0], 1),
optim.lr_scheduler.ReduceLROnPlateau(optimizers[1])]
dcca_model = dcca.DCCA(latent_dims=latent_dims, encoders=[encoder_1, encoder_2],
objective=objectives.CCA, optimizers=optimizers, schedulers=schedulers)
# hidden_layer_sizes are shown explicitly but these are also the defaults
dcca_model = deepwrapper.DeepWrapper(dcca_model)
dcca_model.fit(train_dataset, val_dataset=val_dataset,epochs=20)
# DGCCA
print('DGCCA')
encoder_1 = deep_models.Encoder(latent_dims=latent_dims, feature_size=784)
encoder_2 = deep_models.Encoder(latent_dims=latent_dims, feature_size=784)
dgcca_model = dcca.DCCA(latent_dims=latent_dims, encoders=[encoder_1, encoder_2], objective=objectives.GCCA)
dgcca_model = deepwrapper.DeepWrapper(dgcca_model)
dgcca_model.fit(train_dataset, val_dataset=val_dataset, epochs=epochs)
dgcca_results = np.stack(
(dgcca_model.train_correlations[0, 1], dgcca_model.predict_corr(test_dataset)[0, 1]))
# DMCCA
print('DMCCA')
encoder_1 = deep_models.Encoder(latent_dims=latent_dims, feature_size=784)
encoder_2 = deep_models.Encoder(latent_dims=latent_dims, feature_size=784)
dmcca_model = dcca.DCCA(latent_dims=latent_dims, encoders=[encoder_1, encoder_2], objective=objectives.MCCA)
dmcca_model = deepwrapper.DeepWrapper(dmcca_model)
dmcca_model.fit(train_dataset, val_dataset=val_dataset, epochs=epochs)
dmcca_results = np.stack(
(dmcca_model.train_correlations[0, 1], dmcca_model.predict_corr(test_dataset)[0, 1]))
from cca_zoo import dccae
# DCCAE
print('DCCAE')
encoder_1 = deep_models.Encoder(latent_dims=latent_dims, feature_size=784)
encoder_2 = deep_models.Encoder(latent_dims=latent_dims, feature_size=784)
decoder_1 = deep_models.Decoder(latent_dims=latent_dims, feature_size=784)
decoder_2 = deep_models.Decoder(latent_dims=latent_dims, feature_size=784)
dccae_model = dccae.DCCAE(latent_dims=latent_dims, encoders=[encoder_1, encoder_2], decoders=[decoder_1, decoder_2])
dccae_model = deepwrapper.DeepWrapper(dccae_model)
#can also pass a tuple of numpy arrays
dccae_model.fit((train_view_1, train_view_2), epochs=epochs)
dccae_results = np.stack(
(dccae_model.train_correlations[0, 1], dccae_model.predict_corr(test_dataset)[0, 1]))
"""
### Deep Variational Learning
Finally we have Deep Variational CCA methods.
- Deep Variational CCA (DVCCA)
- Deep Variational CCA - private (DVVCA_p)
These are both implemented by the DVCCA class with private=True/False and both_encoders=True/False. If both_encoders,
the encoder to the shared information Q(z_shared|x) is modelled for both x_1 and x_2 whereas if both_encoders is false
it is modelled for x_1 as in the paper
"""
from cca_zoo import dvcca
# %%
# DVCCA (technically bi-DVCCA)
print('DVCCA')
encoder_1 = deep_models.Encoder(latent_dims=latent_dims, feature_size=784, variational=True)
encoder_2 = deep_models.Encoder(latent_dims=latent_dims, feature_size=784, variational=True)
decoder_1 = deep_models.Decoder(latent_dims=latent_dims, feature_size=784, norm_output=True)
decoder_2 = deep_models.Decoder(latent_dims=latent_dims, feature_size=784, norm_output=True)
dvcca_model = dvcca.DVCCA(latent_dims=latent_dims, encoders=[encoder_1, encoder_2], decoders=[decoder_1, decoder_2],
private=False)
dvcca_model = deepwrapper.DeepWrapper(dvcca_model)
dvcca_model.fit(train_dataset, val_dataset=val_dataset, epochs=epochs)
dvcca_model_results = np.stack(
(dvcca_model.train_correlations[0, 1], dvcca_model.predict_corr(test_dataset)[0, 1]))
# DVCCA_private (technically bi-DVCCA_private)
print('DVCCA_private')
encoder_1 = deep_models.Encoder(latent_dims=latent_dims, feature_size=784, variational=True)
encoder_2 = deep_models.Encoder(latent_dims=latent_dims, feature_size=784, variational=True)
private_encoder_1 = deep_models.Encoder(latent_dims=latent_dims, feature_size=784, variational=True)
private_encoder_2 = deep_models.Encoder(latent_dims=latent_dims, feature_size=784, variational=True)
decoder_1 = deep_models.Decoder(latent_dims=latent_dims * 2, feature_size=784, norm_output=True)
decoder_2 = deep_models.Decoder(latent_dims=latent_dims * 2, feature_size=784, norm_output=True)
dvccap_model = dvcca.DVCCA(latent_dims=latent_dims, encoders=[encoder_1, encoder_2], decoders=[decoder_1, decoder_2],
private_encoders=[private_encoder_1, private_encoder_2], private=True)
dvccap_model = deepwrapper.DeepWrapper(dvccap_model)
dvccap_model.fit(train_dataset, val_dataset=val_dataset, epochs=epochs)
dvccap_model_results = np.stack(
(dvccap_model.train_correlations[0, 1], dvccap_model.predict_corr(test_dataset)[0, 1]))
print('Convolutional DCCA')
encoder_1 = deep_models.CNNEncoder(latent_dims=latent_dims, channels=[3, 3])
encoder_2 = deep_models.CNNEncoder(latent_dims=latent_dims, channels=[3, 3])
dcca_conv_model = dcca.DCCA(latent_dims=latent_dims, encoders=[encoder_1, encoder_2])
dcca_conv_model = deepwrapper.DeepWrapper(dcca_conv_model)
# to change the models used change the cfg.encoder_models. We implement a CNN_Encoder and CNN_decoder as well
# as some based on brainnet architecture in cca_zoo.deep_models. Equally you could pass your own encoder/decoder models
dcca_conv_model.fit((train_view_1.reshape((-1, 1, 28, 28)), train_view_2.reshape((-1, 1, 28, 28))), epochs=epochs)
dcca_conv_results = np.stack(
(dcca_conv_model.train_correlations[0, 1], dcca_conv_model.predict_corr((test_view_1.reshape((-1, 1, 28, 28)),
test_view_2.reshape(
(-1, 1, 28, 28))))[0, 1]))
from cca_zoo import dtcca
# %%
# DTCCA
print('DTCCA')
encoder_1 = deep_models.Encoder(latent_dims=latent_dims, feature_size=784)
encoder_2 = deep_models.Encoder(latent_dims=latent_dims, feature_size=784)
dtcca_model = dtcca.DTCCA(latent_dims=latent_dims, encoders=[encoder_1, encoder_2])
dtcca_model = deepwrapper.DeepWrapper(dtcca_model)
dtcca_model.fit(train_dataset, val_dataset=val_dataset, epochs=epochs)
dtcca_results = np.stack((dtcca_model.train_correlations[0, 1], dtcca_model.predict_corr(test_dataset)[0, 1]))
"""
### Make results plot to compare methods
"""
# %%
all_results = np.stack(
[linear_cca_results, gcca_results, mcca_results, pls_results, pmd_results, elastic_results,
scca_results, kernel_reg_results, kernel_poly_results,
kernel_gaussian_results, dcca_results, dgcca_results, dmcca_results, dvcca_model_results,
dcca_conv_results, dtcca_results],
axis=0)
all_labels = ['linear', 'gcca', 'mcca', 'pls', 'pmd', 'elastic', 'scca', 'linear kernel', 'polynomial kernel',
'gaussian kernel', 'deep CCA', 'deep generalized CCA', 'deep multiset CCA', 'deep VCCA',
'deep convolutional cca', 'DTCCA']
from cca_zoo import plot_utils
plot_utils.plot_results(all_results, all_labels)
plt.show()
| 0.79732 | 0.968471 |
# Lista 00 - Revisão Python e Numpy
[NumPy](http://numpy.org) é um pacote incrivelmente poderoso em Python, onipresente em qualquer projeto de ciência de dados. Possui forte integração com o [Pandas](http://pandas.pydata.org), outra ferramenta que iremos abordar na matéria. NumPy adiciona suporte para matrizes multidimensionais e funções matemáticas que permitem que você execute facilmente cálculos de álgebra linear. Este notebook será uma coleção de exemplos de álgebra linear computados usando NumPy.
## Numpy
Para fazer uso de Numpy precisamos importar a biblioteca
```
# -*- coding: utf8
import numpy as np
```
Quando pensamos no lado prático de ciência de dados, um aspecto chave que ajuda na implementação de novos algoritmos é a vetorização. De forma simples, vetorização consiste do uso de tipos como **escalar**, **vetor** e **matriz** para realizar uma computação mais eficaz (em tempo de execução).
Uma matriz é uma coleção de valores, normalmente representada por uma grade 𝑚 × 𝑛, onde 𝑚 é o número de linhas e 𝑛 é o número de colunas. Os comprimentos das arestas 𝑚 e 𝑛 não precisam ser necessariamente diferentes. Se tivermos 𝑚 = 𝑛, chamamos isso de matriz quadrada. Um caso particularmente interessante de uma matriz é quando 𝑚 = 1 ou 𝑛 = 1. Nesse caso, temos um caso especial de uma matriz que chamamos de vetor. Embora haja um objeto de matriz em NumPy, faremos tudo usando matrizes NumPy porque elas podem ter dimensões maiores que 2.
1. **Escalar:** Um vetor de zero dimensões
```
1
```
2. **Vetor:** Representa uma dimensão
Abaixo vamos criar um vetor simples. Inicialmente, vamos criar uma lista.
```
data_list = [3.5, 5, 2, 8, 4.2]
```
Observe o tipo da mesma.
```
type(data_list)
```
Embora vetores e listas sejam parecidos, vetores Numpy são otimizados para operações de Álgebra Linear. Ciência de Dados faz bastante uso de tais operações, sendo este um dos motivos da dependência em Numpy.
Abaixo criamos um vetor.
```
data = np.array(data_list)
print(data)
print(type(data))
```
Observe como podemos somar o mesmo com um número. Não é possível fazer tal operação com listas.
```
data + 7
```
3. **Matrizes:** Representam duas dimensões.
```
X = np.array([[2, 4],
[1, 3]])
X
```
Podemos indexar as matrizes e os vetores.
```
data[0]
X[0, 1] # aqui é primeira linha, segunda coluna
```
Podemos também criar vetores/matrizes de números aleatórios
```
X = np.random.randn(4, 3) # Gera números aleatórios de uma normal
print(X)
```
### Indexando
Pegando a primeira linha
```
X[0] # observe que 0 é a linha 1, compare com o X[0, 1] de antes.
X[1] # segunda
X[2] # terceira
```
Observe como todos os tipos retornados são `array`. Array é o nome genérico de Numpy para vetores e matrizes.
`X[:, c]` pega uma coluna
```
X[:, 0]
X[:, 1]
```
`X[um_vetor]` pega as linhas da matriz. `X[:, um_vetor]` pega as colunas
```
X[[0, 0, 1]] # observe que pego a primeira linha, indexada por 0, duas vezes
```
Abaixo pego a segunda a primeira coluna
```
X[:, [1, 0]]
```
### Indexação Booleana
`X[vetor_booleano]` retorna as linhas (ou colunas quando X[:, vetor_booleano]) onde o vetor é true
```
X[[True, False, True, False]]
X[:, [False, True, True]]
```
### Reshape, Flatten e Ravel
Todo vetor ou matriz pode ser redimensionado. Observe como uma matriz abaixo de 9x8=72 elementos. Podemos redimensionar os mesmos para outros arrays de tamanho 72.
```
X = np.random.randn(9, 8)
```
Criando uma matriz de 18x4.
```
X.reshape((18, 4))
```
Ou um vetor de 72
```
X.reshape(72)
```
A chamada flatten e ravel faz a mesma coisa, criam uma visão de uma dimensão da matriz.
```
X.flatten()
X.ravel()
```
As funções incorporadas ao NumPy podem ser facilmente chamadas em matrizes. A maioria das funções são aplicadas a um elemento de array (como a multiplicação escalar). Por exemplo, se chamarmos `log()` em um array, o logaritmo será obtido de cada elemento.
```
np.log(data)
```
Mean tira a média
```
np.mean(data)
```
Algumas funções podem ser chamadas direto no vetor, nem todas serão assim. O importante é ler a [documentação](http://numpy.org) e aprender. Com um pouco de prática você vai se acostumando.
```
data.mean()
```
Abaixo temos a mediana,
```
np.median(data) # por exemplo, não existe data.median(). Faz sentido? Não. Mas é assim.
```
Em matrizes as funções operam em todos os elemntos.
```
np.median(X)
X.mean()
np.log(X + 10)
```
Porém, caso você queira a media de linhas ou colunas use `axis`. Antes, vamos ver o tamanho do vetor.
```
X.shape
np.mean(X, axis=0) # média das colunas. como temos 8 colunas, temos 8 elementos.
np.mean(X, axis=0).shape
np.mean(X, axis=1) # média das linhas
np.mean(X, axis=1).shape
```
Lembre-se que eixo 0 é coluna. Eixo 1 é linas.
### Multiplicação de Matrizes
Para transpor uma matriz fazemos uso de .T
```
X.shape
X.T.shape
X.T
```
Para multiplicar matrizes, do ponto de visto de multiplicação matricial como definido na álgebra linear, fazemos uso de `@`.
```
X @ X.T
```
O uso de `*` realiza uma operação ponto a ponto
```
X * X
```
Observe a diferença de tamanhos
```
(X * X).shape
(X @ X.T).shape
```
**Pense:** Para o nosso `X` de tamanho `(9, 8)`, qual o motivo de `X * X.T` não funcionar? Qual o motivo de `X @ X` não funcionar?
## Correção Automática
Nossa correção automática depende das funções abaixo. Tais funções comparam valores que serão computados pelo seu código com uma saída esperada. Normalmente, vocês não fazer uso de tais funções em notebooks como este. Porém, elas são chave em ambientes de testes automáticos (fora do nosso escopo).
Observe como algumas funções comparam valores e outras comparam vetores. Além do mais, temos funções para comparar dentro de algumas casas decimais.
```
from numpy.testing import assert_almost_equal
from numpy.testing import assert_equal
from numpy.testing import assert_array_almost_equal
from numpy.testing import assert_array_equal
# caso você mude um dos valores vamos receber um erro!
assert_array_equal(2, 2)
# caso você mude um dos valores vamos receber um erro!
assert_array_equal([1, 2], [1, 2])
# caso você mude um dos valores vamos receber um erro!
assert_almost_equal(3.1415, 3.14, 1)
```
Caso você mude um dos valores abaixo vamos receber um erro! Como o abaixo.
```
-----------------------------------------------------------------------
AssertionError Traceback (most recent call last)
<ipython-input-10-396672d880f2> in <module>
----> 1 assert_equal(2, 3) # caso você mude um dos valores vamos receber um erro!
~/miniconda3/lib/python3.7/site-packages/numpy/testing/_private/utils.py in assert_equal(actual, desired, err_msg, verbose)
413 # Explicitly use __eq__ for comparison, gh-2552
414 if not (desired == actual):
--> 415 raise AssertionError(msg)
416
417 except (DeprecationWarning, FutureWarning) as e:
AssertionError:
Items are not equal:
ACTUAL: 2
DESIRED: 3
```
É essencial que todo seu código execute sem erros! Portanto, antes de submeter clique em `Kernel` no menu acima. Depois clique em `Restart & Execute All.`
**Garanta que o notebook executa até o fim!** Isto é, sem erros como o acima.
## Funções em Python
Para criar uma função em Python fazemos uso da palavra-chave:
```python
def
```
Todos nossos exercícios farão uso de funções. **Mantenha a assinatura das funções exatamente como requisitado, a correção automática depende disso.** Abaixo, temos um exempo de uma função que imprime algo na tela!
```
def print_something(txt):
print(f'Você passou o argumento: {txt}')
print_something('DCC 212')
```
Podemos também dizer o tipo do argumento, porém faremos pouco uso disto em ICD.
```
def print_something(txt: str):
print(f'Você passou o argumento: {txt}')
print_something('DCC 212')
```
Abaixo temos uma função que soma, a soma, dois vetores
```
def sum_of_sum_vectors(array_1, array_2):
return (array_1 + array_2).sum()
x = np.array([1, 2])
y = np.array([1, 2])
sum_of_sum_vectors(x, y)
```
Abaixo temos um teste, tais testes vão avaliar o seu código. Nem todos estão aqui no notebook!
```
assert_equal(6, sum_of_sum_vectors(x, y))
```
## Exercício 01
Inicialmente, crie uma função que recebe duas listas de numéros, converte as duas para um vetor numpy usando `np.array` e retorna o produto interno das duas listas.
__Dicas:__
1. Tente fazer um código sem nenhum **for**! Ou seja, numpy permite operações em vetores e matrizes, onde: `np.array([1, 2]) + np.array([2, 2]) = np.array([3, 4])`.
__Funções:__
1. `np.sum(array)` soma os elementos do array. `array.sum()` tem o mesmo efeito!
```
def inner(array_1, array_2):
# Seu código aqui!
# Apague o return None abaixo e mude para seu retorno
return None
x1 = np.array([2, 4, 8])
x2 = np.array([10, 100, 1000])
assert_equal(20 + 400 + 8000, inner(x1, x2))
```
## Exercício 02
Implemente uma função utilizando numpy que recebe duas matrizes, multiplica as duas e retorne o valor médio das células da multiplicação. Por exemplo, ao multiplicar:
```
[1 2]
[3 4]
com
[2 1]
[1 2]
temos
[4 5 ]
[10 11]
onde a média de [4, 5, 10, 11] é
7.5, sua resposta final!
```
__Dicas:__
1. Use o operador @ para multiplicar matrizes!
```
def medmult(X_1, X_2):
# Seu código aqui!
# Apague o return None abaixo e mude para seu retorno
return None
X = np.array([1, 2, 3, 4]).reshape(2, 2)
Y = np.array([2, 1, 1, 2]).reshape(2, 2)
assert_equal(7.5, medmult(X, Y))
```
|
github_jupyter
|
# -*- coding: utf8
import numpy as np
1
data_list = [3.5, 5, 2, 8, 4.2]
type(data_list)
data = np.array(data_list)
print(data)
print(type(data))
data + 7
X = np.array([[2, 4],
[1, 3]])
X
data[0]
X[0, 1] # aqui é primeira linha, segunda coluna
X = np.random.randn(4, 3) # Gera números aleatórios de uma normal
print(X)
X[0] # observe que 0 é a linha 1, compare com o X[0, 1] de antes.
X[1] # segunda
X[2] # terceira
X[:, 0]
X[:, 1]
X[[0, 0, 1]] # observe que pego a primeira linha, indexada por 0, duas vezes
X[:, [1, 0]]
X[[True, False, True, False]]
X[:, [False, True, True]]
X = np.random.randn(9, 8)
X.reshape((18, 4))
X.reshape(72)
X.flatten()
X.ravel()
np.log(data)
np.mean(data)
data.mean()
np.median(data) # por exemplo, não existe data.median(). Faz sentido? Não. Mas é assim.
np.median(X)
X.mean()
np.log(X + 10)
X.shape
np.mean(X, axis=0) # média das colunas. como temos 8 colunas, temos 8 elementos.
np.mean(X, axis=0).shape
np.mean(X, axis=1) # média das linhas
np.mean(X, axis=1).shape
X.shape
X.T.shape
X.T
X @ X.T
X * X
(X * X).shape
(X @ X.T).shape
from numpy.testing import assert_almost_equal
from numpy.testing import assert_equal
from numpy.testing import assert_array_almost_equal
from numpy.testing import assert_array_equal
# caso você mude um dos valores vamos receber um erro!
assert_array_equal(2, 2)
# caso você mude um dos valores vamos receber um erro!
assert_array_equal([1, 2], [1, 2])
# caso você mude um dos valores vamos receber um erro!
assert_almost_equal(3.1415, 3.14, 1)
-----------------------------------------------------------------------
AssertionError Traceback (most recent call last)
<ipython-input-10-396672d880f2> in <module>
----> 1 assert_equal(2, 3) # caso você mude um dos valores vamos receber um erro!
~/miniconda3/lib/python3.7/site-packages/numpy/testing/_private/utils.py in assert_equal(actual, desired, err_msg, verbose)
413 # Explicitly use __eq__ for comparison, gh-2552
414 if not (desired == actual):
--> 415 raise AssertionError(msg)
416
417 except (DeprecationWarning, FutureWarning) as e:
AssertionError:
Items are not equal:
ACTUAL: 2
DESIRED: 3
```
É essencial que todo seu código execute sem erros! Portanto, antes de submeter clique em `Kernel` no menu acima. Depois clique em `Restart & Execute All.`
**Garanta que o notebook executa até o fim!** Isto é, sem erros como o acima.
## Funções em Python
Para criar uma função em Python fazemos uso da palavra-chave:
Todos nossos exercícios farão uso de funções. **Mantenha a assinatura das funções exatamente como requisitado, a correção automática depende disso.** Abaixo, temos um exempo de uma função que imprime algo na tela!
Podemos também dizer o tipo do argumento, porém faremos pouco uso disto em ICD.
Abaixo temos uma função que soma, a soma, dois vetores
Abaixo temos um teste, tais testes vão avaliar o seu código. Nem todos estão aqui no notebook!
## Exercício 01
Inicialmente, crie uma função que recebe duas listas de numéros, converte as duas para um vetor numpy usando `np.array` e retorna o produto interno das duas listas.
__Dicas:__
1. Tente fazer um código sem nenhum **for**! Ou seja, numpy permite operações em vetores e matrizes, onde: `np.array([1, 2]) + np.array([2, 2]) = np.array([3, 4])`.
__Funções:__
1. `np.sum(array)` soma os elementos do array. `array.sum()` tem o mesmo efeito!
## Exercício 02
Implemente uma função utilizando numpy que recebe duas matrizes, multiplica as duas e retorne o valor médio das células da multiplicação. Por exemplo, ao multiplicar:
__Dicas:__
1. Use o operador @ para multiplicar matrizes!
| 0.404272 | 0.987412 |
##### Import the GIS module and other needed Python modules
The IPython.display module has some helper functions that the Python API takes advantage of for displaying objects like item details and maps in the notebook.
```
from arcgis.gis import GIS
from getpass import getpass
from IPython.display import display
```
##### Create the GIS object and point it to AGOL
```
# Get username and password
username = input('Username: ')
password = getpass(prompt='Password: ')
# Connect to portal
gis = GIS("https://arcgis.com/", username, password)
```
##### Test the connection
The output here is an example of the Python API taking advantage of IPython.display. The *me* property gives you a direct shortcut to the *User* object for the logged in user.
```
me = gis.users.me
me
```
You can also search for other users.
```
username = input('Username: ')
user = gis.users.get(username)
user
```
##### User properties
There are a number of properties that can be accessed the *user* resource. The full list can be found here: [User](http://resources.arcgis.com/en/help/arcgis-rest-api/#/User/02r3000000m2000000/).
```
print("Description: {}\nEmail: {}\nLast Name: {}\nFirst Name: {}\nName: {}\nLevel: {}\nMFA Enabled: {}".format(me.description, me.email, me.firstName, me.lastName, me.fullName, me.level, me.mfaEnabled))
user_groups = me.groups
print("Member of " + str(len(user_groups)) + " groups")
```
The Python *time* module can be used to display time values in human readable form.
ArcGIS Online stores time in **milliseconds** since the epoch, but the Python time library is expecting **seconds** since the epoch (aka Unix time, epoch time), so the AGOL times need to be divided by 1000 when using the *time* module.
```
import time
created_time = time.localtime(me.created/1000)
print("Created: {}/{}/{}".format(created_time[1], created_time[2], created_time[0]))
last_accessed = time.localtime(me.lastLogin/1000)
print("Last active: {}/{}/{}".format(last_accessed[1], last_accessed[2], last_accessed[0]))
```
##### Searching for user accounts
Parameters here: http://resources.arcgis.com/en/help/arcgis-rest-api/index.html#//02r30000009v000000
and here: http://resources.arcgis.com/en/help/arcgis-rest-api/#/Search_reference/02r3000000mn000000/
The *gis.users.search* method allows you to perform search operations using wildcards.
```
tpt_users = gis.users.search('username:tpt_*',max_users=10000)
len(tpt_users)
```
The power of Python can also be used to search for specific patterns.
```
# Get a list of all users
all_users = gis.users.search(None, max_users=500)
len(all_users)
# Use list comprehension to create a subset list of disabled users
disabled_users = [user for user in all_users if user.disabled == True]
len(disabled_users)
```
##### Finding the items owned by users
Items are either in a folder or not. The latter are called root items. There can only be one folder level, so only one level needs to be traversed.
```
from operator import attrgetter
for user in sorted(disabled_users, key=attrgetter('lastName', 'firstName')):
print(user.fullName + " (" + user.username + ")")
# user.items() returns a list of root items.
total_items = len(user.items())
print("Root items:" + str(total_items))
# user.folders returns a list of folders
folders = user.folders
print("Folders: " + str(len(folders)))
for folder in folders:
# The folder parameter on user.items() returns a list of items in the given folder
folder_items = len(user.items(folder = folder))
print("Folder " + folder["title"] + " items:" + str(folder_items))
total_items += folder_items
print("Total items: " + str(total_items))
print("=" * 25)
```
|
github_jupyter
|
from arcgis.gis import GIS
from getpass import getpass
from IPython.display import display
# Get username and password
username = input('Username: ')
password = getpass(prompt='Password: ')
# Connect to portal
gis = GIS("https://arcgis.com/", username, password)
me = gis.users.me
me
username = input('Username: ')
user = gis.users.get(username)
user
print("Description: {}\nEmail: {}\nLast Name: {}\nFirst Name: {}\nName: {}\nLevel: {}\nMFA Enabled: {}".format(me.description, me.email, me.firstName, me.lastName, me.fullName, me.level, me.mfaEnabled))
user_groups = me.groups
print("Member of " + str(len(user_groups)) + " groups")
import time
created_time = time.localtime(me.created/1000)
print("Created: {}/{}/{}".format(created_time[1], created_time[2], created_time[0]))
last_accessed = time.localtime(me.lastLogin/1000)
print("Last active: {}/{}/{}".format(last_accessed[1], last_accessed[2], last_accessed[0]))
tpt_users = gis.users.search('username:tpt_*',max_users=10000)
len(tpt_users)
# Get a list of all users
all_users = gis.users.search(None, max_users=500)
len(all_users)
# Use list comprehension to create a subset list of disabled users
disabled_users = [user for user in all_users if user.disabled == True]
len(disabled_users)
from operator import attrgetter
for user in sorted(disabled_users, key=attrgetter('lastName', 'firstName')):
print(user.fullName + " (" + user.username + ")")
# user.items() returns a list of root items.
total_items = len(user.items())
print("Root items:" + str(total_items))
# user.folders returns a list of folders
folders = user.folders
print("Folders: " + str(len(folders)))
for folder in folders:
# The folder parameter on user.items() returns a list of items in the given folder
folder_items = len(user.items(folder = folder))
print("Folder " + folder["title"] + " items:" + str(folder_items))
total_items += folder_items
print("Total items: " + str(total_items))
print("=" * 25)
| 0.238462 | 0.906239 |
```
import numpy as np
import matplotlib.pyplot as plt
from keras.models import Sequential
from keras.layers import Dense, Dropout, Flatten, Conv2D, MaxPool2D
from keras.datasets import mnist
%matplotlib inline
```
'%matplotlib inline' is a 'Magic' command to get the plots inline with the notebook.
### Loading the Dataset from Keras
```
(x_train, y_train), (x_test, y_test) = mnist.load_data()
```
### Visualising the Data
```
np.random.seed(3)
n_rows = 2
n_cols = 3
fig, ax = plt.subplots(nrows=n_rows,ncols=n_cols)
fig.suptitle("Dataset", size=18)
for i in range(n_rows):
for j in range(n_cols):
ax[i][j].imshow(x_train[np.random.randint(0,6000)])
```
### Preprocessing the Data:
Preprocessing includes scaling the data to [-1,1] and getting zero mean. Also, as the Conv2D requires number of channels as a dimension, we reshaped the dataset to include the one and only grayscal channel.
```
def preprocess(dataset):
dataset = (dataset - np.mean(dataset))/255
dataset = dataset.reshape(-1,28,28,1)
return dataset
x_train = preprocess(x_train)
x_test = preprocess(x_test)
```
### 'One-hot'-ing the Labels
One-hot vectors are used for multiclass classification. In one-hot vectors, only one bit would be set which corresponds to the correct category it belongs to. Rest are zero. Although here, an inbuilt function to_categorical from keras.utils can be used.
```
def onehot(labels):
return np.array([[float(i==data) for i in range(10)] for data in labels])
y_train = onehot(y_train)
y_test = onehot(y_test)
```
### Defining the Model and its Architecture:
The Model defined is a very simple one with two Convolutional Layers and two Dense layers. Dropout layers were used for regularization. Since there are 10 labels (0 to 9), the last layer has 10 output nodes. Softmax activation gives us the probabilities directly.
```
model = Sequential()
model.add(Conv2D(filters=32, kernel_size=5, padding='SAME', activation='relu', input_shape=(28,28,1)))
model.add(MaxPool2D(padding = 'SAME'))
model.add(Conv2D(filters=32, kernel_size=5, padding='SAME', activation='relu'))
model.add(MaxPool2D())
model.add(Dropout(0.25))
model.add(Flatten())
model.add(Dense(256, activation='relu'))
model.add(Dense(10, activation='softmax'))
```
Let us now see what our model looks like. Keras has a very simple method called summary() that outputs the layers, their shapes and the number of parameters in it.
```
model.summary()
```
### Compiling the Model and Training:
```
model.compile(optimizer='adam', loss='categorical_crossentropy', metrics=['accuracy'])
history = model.fit(x_train, y_train, batch_size=128, epochs=10, verbose=1, validation_data=(x_test, y_test))
```
### Using the Model on a test dataset and Calculating the Accuracy:
```
model.evaluate(x_test, y_test, verbose=1)
```
### Visualising Predictions
```
np.random.seed(3)
n_rows = 2
n_cols = 3
fig, ax = plt.subplots(nrows=n_rows,ncols=n_cols)
fig.suptitle("Predictions", size=18)
ec = (0, 0, 0)
fc = (1, 1, 1)
for i in range(n_rows):
for j in range(n_cols):
k = np.random.randint(0,6000)
ax[i][j].imshow(x_test[k].reshape((28,28)))
temp = np.argmax(model.predict(np.expand_dims(x_test[k], axis=0)))
ax[i][j].text(s="Predicted Value: {}".format(temp), x=0.75, y=0,
bbox=dict(boxstyle="round", ec=ec, fc=fc))
plt.setp(ax, xticks=[], yticks=[])
```
|
github_jupyter
|
import numpy as np
import matplotlib.pyplot as plt
from keras.models import Sequential
from keras.layers import Dense, Dropout, Flatten, Conv2D, MaxPool2D
from keras.datasets import mnist
%matplotlib inline
(x_train, y_train), (x_test, y_test) = mnist.load_data()
np.random.seed(3)
n_rows = 2
n_cols = 3
fig, ax = plt.subplots(nrows=n_rows,ncols=n_cols)
fig.suptitle("Dataset", size=18)
for i in range(n_rows):
for j in range(n_cols):
ax[i][j].imshow(x_train[np.random.randint(0,6000)])
def preprocess(dataset):
dataset = (dataset - np.mean(dataset))/255
dataset = dataset.reshape(-1,28,28,1)
return dataset
x_train = preprocess(x_train)
x_test = preprocess(x_test)
def onehot(labels):
return np.array([[float(i==data) for i in range(10)] for data in labels])
y_train = onehot(y_train)
y_test = onehot(y_test)
model = Sequential()
model.add(Conv2D(filters=32, kernel_size=5, padding='SAME', activation='relu', input_shape=(28,28,1)))
model.add(MaxPool2D(padding = 'SAME'))
model.add(Conv2D(filters=32, kernel_size=5, padding='SAME', activation='relu'))
model.add(MaxPool2D())
model.add(Dropout(0.25))
model.add(Flatten())
model.add(Dense(256, activation='relu'))
model.add(Dense(10, activation='softmax'))
model.summary()
model.compile(optimizer='adam', loss='categorical_crossentropy', metrics=['accuracy'])
history = model.fit(x_train, y_train, batch_size=128, epochs=10, verbose=1, validation_data=(x_test, y_test))
model.evaluate(x_test, y_test, verbose=1)
np.random.seed(3)
n_rows = 2
n_cols = 3
fig, ax = plt.subplots(nrows=n_rows,ncols=n_cols)
fig.suptitle("Predictions", size=18)
ec = (0, 0, 0)
fc = (1, 1, 1)
for i in range(n_rows):
for j in range(n_cols):
k = np.random.randint(0,6000)
ax[i][j].imshow(x_test[k].reshape((28,28)))
temp = np.argmax(model.predict(np.expand_dims(x_test[k], axis=0)))
ax[i][j].text(s="Predicted Value: {}".format(temp), x=0.75, y=0,
bbox=dict(boxstyle="round", ec=ec, fc=fc))
plt.setp(ax, xticks=[], yticks=[])
| 0.833426 | 0.963057 |
# Overview videos
## Video: Kay/Gallant dataset
```
# @title Video: Steinmetz dataset
from ipywidgets import widgets
out2 = widgets.Output()
with out2:
from IPython.display import IFrame
class BiliVideo(IFrame):
def __init__(self, id, page=1, width=400, height=300, **kwargs):
self.id=id
src = 'https://player.bilibili.com/player.html?bvid={0}&page={1}'.format(id, page)
super(BiliVideo, self).__init__(src, width, height, **kwargs)
video = BiliVideo(id="", width=730, height=410, fs=1)
print('Video available at https://www.bilibili.com/video/{0}'.format(video.id))
display(video)
out1 = widgets.Output()
with out1:
from IPython.display import YouTubeVideo
video = YouTubeVideo(id="LdJkLyw4yzg", width=730, height=410, fs=1, rel=0)
print('Video available at https://youtube.com/watch?v=' + video.id)
display(video)
out = widgets.Tab([out1, out2])
out.set_title(0, 'Youtube')
out.set_title(1, 'Bilibili')
display(out)
```
## Video: HCP dataset
```
# @title Video: Steinmetz dataset
from ipywidgets import widgets
out2 = widgets.Output()
with out2:
from IPython.display import IFrame
class BiliVideo(IFrame):
def __init__(self, id, page=1, width=400, height=300, **kwargs):
self.id=id
src = 'https://player.bilibili.com/player.html?bvid={0}&page={1}'.format(id, page)
super(BiliVideo, self).__init__(src, width, height, **kwargs)
video = BiliVideo(id="", width=730, height=410, fs=1)
print('Video available at https://www.bilibili.com/video/{0}'.format(video.id))
display(video)
out1 = widgets.Output()
with out1:
from IPython.display import YouTubeVideo
video = YouTubeVideo(id="iOCcY0QFMS4", width=730, height=410, fs=1, rel=0)
print('Video available at https://youtube.com/watch?v=' + video.id)
display(video)
out = widgets.Tab([out1, out2])
out.set_title(0, 'Youtube')
out.set_title(1, 'Bilibili')
display(out)
```
## Video: HCP dataset (2021 video)
```
# @title Video: Steinmetz dataset
from ipywidgets import widgets
out2 = widgets.Output()
with out2:
from IPython.display import IFrame
class BiliVideo(IFrame):
def __init__(self, id, page=1, width=400, height=300, **kwargs):
self.id=id
src = 'https://player.bilibili.com/player.html?bvid={0}&page={1}'.format(id, page)
super(BiliVideo, self).__init__(src, width, height, **kwargs)
video = BiliVideo(id="", width=730, height=410, fs=1)
print('Video available at https://www.bilibili.com/video/{0}'.format(video.id))
display(video)
out1 = widgets.Output()
with out1:
from IPython.display import YouTubeVideo
video = YouTubeVideo(id="nssSiCmbjxw", width=730, height=410, fs=1, rel=0)
print('Video available at https://youtube.com/watch?v=' + video.id)
display(video)
out = widgets.Tab([out1, out2])
out.set_title(0, 'Youtube')
out.set_title(1, 'Bilibili')
display(out)
```
## Video: FSLcourse task fMRI
```
# @title Video: Steinmetz dataset
from ipywidgets import widgets
out2 = widgets.Output()
with out2:
from IPython.display import IFrame
class BiliVideo(IFrame):
def __init__(self, id, page=1, width=400, height=300, **kwargs):
self.id=id
src = 'https://player.bilibili.com/player.html?bvid={0}&page={1}'.format(id, page)
super(BiliVideo, self).__init__(src, width, height, **kwargs)
video = BiliVideo(id="", width=730, height=410, fs=1)
print('Video available at https://www.bilibili.com/video/{0}'.format(video.id))
display(video)
out1 = widgets.Output()
with out1:
from IPython.display import YouTubeVideo
video = YouTubeVideo(id="ZI-xFYubENw", width=730, height=410, fs=1, rel=0)
print('Video available at https://youtube.com/watch?v=' + video.id)
display(video)
out = widgets.Tab([out1, out2])
out.set_title(0, 'Youtube')
out.set_title(1, 'Bilibili')
display(out)
```
## Video: HCP retinotopy
```
# @title Video: Steinmetz dataset
from ipywidgets import widgets
out2 = widgets.Output()
with out2:
from IPython.display import IFrame
class BiliVideo(IFrame):
def __init__(self, id, page=1, width=400, height=300, **kwargs):
self.id=id
src = 'https://player.bilibili.com/player.html?bvid={0}&page={1}'.format(id, page)
super(BiliVideo, self).__init__(src, width, height, **kwargs)
video = BiliVideo(id="", width=730, height=410, fs=1)
print('Video available at https://www.bilibili.com/video/{0}'.format(video.id))
display(video)
out1 = widgets.Output()
with out1:
from IPython.display import YouTubeVideo
video = YouTubeVideo(id="nssSiCmbjxw", width=730, height=410, fs=1, rel=0)
print('Video available at https://youtube.com/watch?v=' + video.id)
display(video)
out = widgets.Tab([out1, out2])
out.set_title(0, 'Youtube')
out.set_title(1, 'Bilibili')
display(out)
```
## Video: Algonauts dataset
```
# @title Video: Steinmetz dataset
from ipywidgets import widgets
out2 = widgets.Output()
with out2:
from IPython.display import IFrame
class BiliVideo(IFrame):
def __init__(self, id, page=1, width=400, height=300, **kwargs):
self.id=id
src = 'https://player.bilibili.com/player.html?bvid={0}&page={1}'.format(id, page)
super(BiliVideo, self).__init__(src, width, height, **kwargs)
video = BiliVideo(id="", width=730, height=410, fs=1)
print('Video available at https://www.bilibili.com/video/{0}'.format(video.id))
display(video)
out1 = widgets.Output()
with out1:
from IPython.display import YouTubeVideo
video = YouTubeVideo(id="TID48cMcneo", width=730, height=410, fs=1, rel=0)
print('Video available at https://youtube.com/watch?v=' + video.id)
display(video)
out = widgets.Tab([out1, out2])
out.set_title(0, 'Youtube')
out.set_title(1, 'Bilibili')
display(out)
```
## Video: Bonner dataset
```
# @title Video: Steinmetz dataset
from ipywidgets import widgets
out2 = widgets.Output()
with out2:
from IPython.display import IFrame
class BiliVideo(IFrame):
def __init__(self, id, page=1, width=400, height=300, **kwargs):
self.id=id
src = 'https://player.bilibili.com/player.html?bvid={0}&page={1}'.format(id, page)
super(BiliVideo, self).__init__(src, width, height, **kwargs)
video = BiliVideo(id="", width=730, height=410, fs=1)
print('Video available at https://www.bilibili.com/video/{0}'.format(video.id))
display(video)
out1 = widgets.Output()
with out1:
from IPython.display import YouTubeVideo
video = YouTubeVideo(id="7NggvUlobQQ", width=730, height=410, fs=1, rel=0)
print('Video available at https://youtube.com/watch?v=' + video.id)
display(video)
out = widgets.Tab([out1, out2])
out.set_title(0, 'Youtube')
out.set_title(1, 'Bilibili')
display(out)
```
## Video: Cichy dataset
```
# @title Video: Steinmetz dataset
from ipywidgets import widgets
out2 = widgets.Output()
with out2:
from IPython.display import IFrame
class BiliVideo(IFrame):
def __init__(self, id, page=1, width=400, height=300, **kwargs):
self.id=id
src = 'https://player.bilibili.com/player.html?bvid={0}&page={1}'.format(id, page)
super(BiliVideo, self).__init__(src, width, height, **kwargs)
video = BiliVideo(id="", width=730, height=410, fs=1)
print('Video available at https://www.bilibili.com/video/{0}'.format(video.id))
display(video)
out1 = widgets.Output()
with out1:
from IPython.display import YouTubeVideo
video = YouTubeVideo(id="I3_nA_6mq1g", width=730, height=410, fs=1, rel=0)
print('Video available at https://youtube.com/watch?v=' + video.id)
display(video)
out = widgets.Tab([out1, out2])
out.set_title(0, 'Youtube')
out.set_title(1, 'Bilibili')
display(out)
```
|
github_jupyter
|
# @title Video: Steinmetz dataset
from ipywidgets import widgets
out2 = widgets.Output()
with out2:
from IPython.display import IFrame
class BiliVideo(IFrame):
def __init__(self, id, page=1, width=400, height=300, **kwargs):
self.id=id
src = 'https://player.bilibili.com/player.html?bvid={0}&page={1}'.format(id, page)
super(BiliVideo, self).__init__(src, width, height, **kwargs)
video = BiliVideo(id="", width=730, height=410, fs=1)
print('Video available at https://www.bilibili.com/video/{0}'.format(video.id))
display(video)
out1 = widgets.Output()
with out1:
from IPython.display import YouTubeVideo
video = YouTubeVideo(id="LdJkLyw4yzg", width=730, height=410, fs=1, rel=0)
print('Video available at https://youtube.com/watch?v=' + video.id)
display(video)
out = widgets.Tab([out1, out2])
out.set_title(0, 'Youtube')
out.set_title(1, 'Bilibili')
display(out)
# @title Video: Steinmetz dataset
from ipywidgets import widgets
out2 = widgets.Output()
with out2:
from IPython.display import IFrame
class BiliVideo(IFrame):
def __init__(self, id, page=1, width=400, height=300, **kwargs):
self.id=id
src = 'https://player.bilibili.com/player.html?bvid={0}&page={1}'.format(id, page)
super(BiliVideo, self).__init__(src, width, height, **kwargs)
video = BiliVideo(id="", width=730, height=410, fs=1)
print('Video available at https://www.bilibili.com/video/{0}'.format(video.id))
display(video)
out1 = widgets.Output()
with out1:
from IPython.display import YouTubeVideo
video = YouTubeVideo(id="iOCcY0QFMS4", width=730, height=410, fs=1, rel=0)
print('Video available at https://youtube.com/watch?v=' + video.id)
display(video)
out = widgets.Tab([out1, out2])
out.set_title(0, 'Youtube')
out.set_title(1, 'Bilibili')
display(out)
# @title Video: Steinmetz dataset
from ipywidgets import widgets
out2 = widgets.Output()
with out2:
from IPython.display import IFrame
class BiliVideo(IFrame):
def __init__(self, id, page=1, width=400, height=300, **kwargs):
self.id=id
src = 'https://player.bilibili.com/player.html?bvid={0}&page={1}'.format(id, page)
super(BiliVideo, self).__init__(src, width, height, **kwargs)
video = BiliVideo(id="", width=730, height=410, fs=1)
print('Video available at https://www.bilibili.com/video/{0}'.format(video.id))
display(video)
out1 = widgets.Output()
with out1:
from IPython.display import YouTubeVideo
video = YouTubeVideo(id="nssSiCmbjxw", width=730, height=410, fs=1, rel=0)
print('Video available at https://youtube.com/watch?v=' + video.id)
display(video)
out = widgets.Tab([out1, out2])
out.set_title(0, 'Youtube')
out.set_title(1, 'Bilibili')
display(out)
# @title Video: Steinmetz dataset
from ipywidgets import widgets
out2 = widgets.Output()
with out2:
from IPython.display import IFrame
class BiliVideo(IFrame):
def __init__(self, id, page=1, width=400, height=300, **kwargs):
self.id=id
src = 'https://player.bilibili.com/player.html?bvid={0}&page={1}'.format(id, page)
super(BiliVideo, self).__init__(src, width, height, **kwargs)
video = BiliVideo(id="", width=730, height=410, fs=1)
print('Video available at https://www.bilibili.com/video/{0}'.format(video.id))
display(video)
out1 = widgets.Output()
with out1:
from IPython.display import YouTubeVideo
video = YouTubeVideo(id="ZI-xFYubENw", width=730, height=410, fs=1, rel=0)
print('Video available at https://youtube.com/watch?v=' + video.id)
display(video)
out = widgets.Tab([out1, out2])
out.set_title(0, 'Youtube')
out.set_title(1, 'Bilibili')
display(out)
# @title Video: Steinmetz dataset
from ipywidgets import widgets
out2 = widgets.Output()
with out2:
from IPython.display import IFrame
class BiliVideo(IFrame):
def __init__(self, id, page=1, width=400, height=300, **kwargs):
self.id=id
src = 'https://player.bilibili.com/player.html?bvid={0}&page={1}'.format(id, page)
super(BiliVideo, self).__init__(src, width, height, **kwargs)
video = BiliVideo(id="", width=730, height=410, fs=1)
print('Video available at https://www.bilibili.com/video/{0}'.format(video.id))
display(video)
out1 = widgets.Output()
with out1:
from IPython.display import YouTubeVideo
video = YouTubeVideo(id="nssSiCmbjxw", width=730, height=410, fs=1, rel=0)
print('Video available at https://youtube.com/watch?v=' + video.id)
display(video)
out = widgets.Tab([out1, out2])
out.set_title(0, 'Youtube')
out.set_title(1, 'Bilibili')
display(out)
# @title Video: Steinmetz dataset
from ipywidgets import widgets
out2 = widgets.Output()
with out2:
from IPython.display import IFrame
class BiliVideo(IFrame):
def __init__(self, id, page=1, width=400, height=300, **kwargs):
self.id=id
src = 'https://player.bilibili.com/player.html?bvid={0}&page={1}'.format(id, page)
super(BiliVideo, self).__init__(src, width, height, **kwargs)
video = BiliVideo(id="", width=730, height=410, fs=1)
print('Video available at https://www.bilibili.com/video/{0}'.format(video.id))
display(video)
out1 = widgets.Output()
with out1:
from IPython.display import YouTubeVideo
video = YouTubeVideo(id="TID48cMcneo", width=730, height=410, fs=1, rel=0)
print('Video available at https://youtube.com/watch?v=' + video.id)
display(video)
out = widgets.Tab([out1, out2])
out.set_title(0, 'Youtube')
out.set_title(1, 'Bilibili')
display(out)
# @title Video: Steinmetz dataset
from ipywidgets import widgets
out2 = widgets.Output()
with out2:
from IPython.display import IFrame
class BiliVideo(IFrame):
def __init__(self, id, page=1, width=400, height=300, **kwargs):
self.id=id
src = 'https://player.bilibili.com/player.html?bvid={0}&page={1}'.format(id, page)
super(BiliVideo, self).__init__(src, width, height, **kwargs)
video = BiliVideo(id="", width=730, height=410, fs=1)
print('Video available at https://www.bilibili.com/video/{0}'.format(video.id))
display(video)
out1 = widgets.Output()
with out1:
from IPython.display import YouTubeVideo
video = YouTubeVideo(id="7NggvUlobQQ", width=730, height=410, fs=1, rel=0)
print('Video available at https://youtube.com/watch?v=' + video.id)
display(video)
out = widgets.Tab([out1, out2])
out.set_title(0, 'Youtube')
out.set_title(1, 'Bilibili')
display(out)
# @title Video: Steinmetz dataset
from ipywidgets import widgets
out2 = widgets.Output()
with out2:
from IPython.display import IFrame
class BiliVideo(IFrame):
def __init__(self, id, page=1, width=400, height=300, **kwargs):
self.id=id
src = 'https://player.bilibili.com/player.html?bvid={0}&page={1}'.format(id, page)
super(BiliVideo, self).__init__(src, width, height, **kwargs)
video = BiliVideo(id="", width=730, height=410, fs=1)
print('Video available at https://www.bilibili.com/video/{0}'.format(video.id))
display(video)
out1 = widgets.Output()
with out1:
from IPython.display import YouTubeVideo
video = YouTubeVideo(id="I3_nA_6mq1g", width=730, height=410, fs=1, rel=0)
print('Video available at https://youtube.com/watch?v=' + video.id)
display(video)
out = widgets.Tab([out1, out2])
out.set_title(0, 'Youtube')
out.set_title(1, 'Bilibili')
display(out)
| 0.472683 | 0.690129 |
# FTE/BTE Experiment for Aircraft & Birdsnap
---
This experiment investigates the ability of progressive learning on tranferring knowledge across different datasets. Two datasets, the [FGVC-Aircraft-2013b](https://www.robots.ox.ac.uk/~vgg/data/fgvc-aircraft/), an image dataset of aircrafts, and [Birdsnap](http://thomasberg.org/), an image dataset of birds, have been used. The information of the datasets has been listed below.
| Dataset |# images | # labels | # images per label (minimum) |
| --------------- |:-----------------:| :-------:|---------------------------:|
| fgvc-aircraft-2013b | 10000 | 100 | 100 |
| birdSnap | 49829 | 500 | 69 (100 for most species) |
Before the experiment, we uniform the images by padding them into squares and then by resizing them to $32\times32$ (pixels). The code for dataset preprocessing and the resized images can be found [here](https://github.com/chenyugoal/Dataset_Preprocessing).
Considering the difference between the two datasets in the number of images, we choose a proportion of each dataset to use so that the number of classes and the number of images per class are equal.
```
import numpy as np
from joblib import Parallel, delayed
```
### Load tasks
The processed dataset can be downloaded [here](https://github.com/chenyugoal/Dataset_Preprocessing).
After running the blocks of code below, we will get the splitted tasks. We have 10 tasks in total, with first 5 set up from Aircraft and the last 5 set up from Birdsnap. Each task has 20 labels to classify. For each label, there are 90 samples.
Therefore, the total number of samples is:
$10\times20\times90=18000$
```
path_aircraft_x_all = 'F:/Programming/Python/NDD/Dataset_Preprocessing/data/aircraft_x_all.npy'
path_aircraft_y_all = 'F:/Programming/Python/NDD/Dataset_Preprocessing/data/aircraft_y_all.npy'
path_birdsnap_x_all = 'F:/Programming/Python/NDD/Dataset_Preprocessing/data/birdsnap_x_all.npy'
path_birdsnap_y_all = 'F:/Programming/Python/NDD/Dataset_Preprocessing/data/birdsnap_y_all.npy'
from functions.fte_bte_aircraft_bird_functions import load_tasks
train_x_task, test_x_task, train_y_task, test_y_task = load_tasks(path_aircraft_x_all,
path_aircraft_y_all,
path_birdsnap_x_all,
path_birdsnap_y_all)
```
### Sample images
Let's take a look at images from Aircraft and Birdsnap by running the block below.
```
from functions.fte_bte_aircraft_bird_functions import show_image
show_image(train_x_task)
```
### Run progressive learning
Here we provide two options of implementations of progressive learning:
- omnidirectional forest (Odif), which uses uncertainty forests as the base representer
- omnidirectional networks (Odin), which uses a deep network as the base representer.
Use `odif` for omnidirectional forest and `odin` for omnidirectional networks.
```
from functions.fte_bte_aircraft_bird_functions import single_experiment
model = 'odif' # Choose 'odif' or 'odin'
ntrees = 10 # Number of trees
num_repetition = 30
accuracy_list = Parallel(n_jobs=6)(delayed(single_experiment)(train_x_task,test_x_task,train_y_task,test_y_task,ntrees,model) for _ in range(num_repetition))
accuracy_all_avg = np.average(accuracy_list, axis = 0)
```
### Calculate and plot transfer efficiency
```
from functions.fte_bte_aircraft_bird_functions import calculate_results
err, bte, fte, te = calculate_results(accuracy_all_avg)
from functions.fte_bte_aircraft_bird_functions import plot_all
plot_all(err, bte, fte, te)
```
|
github_jupyter
|
import numpy as np
from joblib import Parallel, delayed
path_aircraft_x_all = 'F:/Programming/Python/NDD/Dataset_Preprocessing/data/aircraft_x_all.npy'
path_aircraft_y_all = 'F:/Programming/Python/NDD/Dataset_Preprocessing/data/aircraft_y_all.npy'
path_birdsnap_x_all = 'F:/Programming/Python/NDD/Dataset_Preprocessing/data/birdsnap_x_all.npy'
path_birdsnap_y_all = 'F:/Programming/Python/NDD/Dataset_Preprocessing/data/birdsnap_y_all.npy'
from functions.fte_bte_aircraft_bird_functions import load_tasks
train_x_task, test_x_task, train_y_task, test_y_task = load_tasks(path_aircraft_x_all,
path_aircraft_y_all,
path_birdsnap_x_all,
path_birdsnap_y_all)
from functions.fte_bte_aircraft_bird_functions import show_image
show_image(train_x_task)
from functions.fte_bte_aircraft_bird_functions import single_experiment
model = 'odif' # Choose 'odif' or 'odin'
ntrees = 10 # Number of trees
num_repetition = 30
accuracy_list = Parallel(n_jobs=6)(delayed(single_experiment)(train_x_task,test_x_task,train_y_task,test_y_task,ntrees,model) for _ in range(num_repetition))
accuracy_all_avg = np.average(accuracy_list, axis = 0)
from functions.fte_bte_aircraft_bird_functions import calculate_results
err, bte, fte, te = calculate_results(accuracy_all_avg)
from functions.fte_bte_aircraft_bird_functions import plot_all
plot_all(err, bte, fte, te)
| 0.290477 | 0.973139 |
```
import torch
import numpy as np
import pandas as pd
import torch.nn as nn
import torch.optim as optim
import torch.nn.functional as F
import matplotlib.pyplot as plt
from pathlib import Path
from scipy import signal
from scipy.io import wavfile
from scipy.fftpack import fft
from torch.autograd import Variable
from sklearn.metrics import precision_score
from torchvision import transforms, datasets
from torch.utils.data import Dataset, DataLoader
from sklearn.model_selection import train_test_split
import warnings
warnings.filterwarnings("ignore")
%matplotlib inline
```
# Data processing
```
legal_labels = 'yes no up down left right on off stop go silence unknown'.split()
train_data_path = Path('./train/audio')
valid_subset_path = Path('./train/validation_list.txt')
def unzip2(values):
a, b = zip(*values)
return list(a), list(b)
class SoundDataset(Dataset):
def __init__(self, directory, subset_path=None, transform=None):
self.directory = directory
self.transform = transform
self._subset = self._load_subset(subset_path) if subset_path else None
self._labels, self._sounds = self._list_of_wavs(directory)
self._transformed_labels = self._transform_labels(self._labels)
def __len__(self):
return len(self._labels)
def __getitem__(self, idx):
label, sound = self._labels[idx], self._sounds[idx]
sample_rate, samples = wavfile.read(self.directory/label/sound)
label = self._transformed_labels[idx]
sample = {'sample_rate': sample_rate, 'samples': samples, 'label': label}
if self.transform is not None:
sample = self.transform(sample)
return sample
def _is_needed(self, name):
return (name in self._subset) if self._subset is not None else True
def _list_of_wavs(self, directory, ext='wav'):
return unzip2(path.parts[-2:] for path in directory.glob(f'*/*.{ext}') if self._is_needed(path.parts[-1]))
@staticmethod
def _load_subset(file):
return set(path.split('/')[1] for path in file.read_text().split('\n'))
@staticmethod
def _transform_labels(labels):
nlabels = []
for label in labels:
if label == '_background_noise_':
nlabels.append('silence')
elif label not in legal_labels:
nlabels.append('unknown')
else:
nlabels.append(label)
return np.array(pd.get_dummies(pd.Series(nlabels)), dtype=np.float32)
class RandomChop:
def __init__(self, length=16_000):
self.length = length
def __call__(self, sample):
sample_rate, samples, label = sample['sample_rate'], sample['samples'], sample['label']
samples = self._pad_audio(samples)
if len(samples) > self.length:
samples = self._chop_audio(samples)
return {'sample_rate': sample_rate, 'samples': samples, 'label': label}
def _pad_audio(self, samples):
if len(samples) >= self.length:
return samples
return np.pad(samples, pad_width=(self.length - len(samples), 0), mode='constant', constant_values=(0, 0))
def _chop_audio(self, samples):
start = np.random.randint(0, len(samples) - self.length)
return samples[start : start + self.length]
class Specgram:
def __init__(self, sample_rate=8_000):
self.sample_rate = sample_rate
def __call__(self, sample):
sample_rate, samples, label = sample['sample_rate'], sample['samples'], sample['label']
resampled = signal.resample(samples, int(self.sample_rate / sample_rate * samples.shape[0]))
_, _, specgram = self._log_specgram(resampled, sample_rate=self.sample_rate)
specgram = specgram.reshape(1, specgram.shape[0], specgram.shape[1])
return {'samples': specgram, 'label': label}
@staticmethod
def _log_specgram(audio, sample_rate, window_size=20, step_size=10, eps=1e-10):
nperseg = int(round(window_size * sample_rate / 1e3))
noverlap = int(round(step_size * sample_rate / 1e3))
freqs, times, spec = signal.spectrogram(audio,
fs=sample_rate,
window='hann',
nperseg=nperseg,
noverlap=noverlap,
detrend=False)
return freqs, times, np.log(spec.T.astype(np.float32) + eps)
class ToTensor:
def __call__(self, sample):
samples, label = sample['samples'], sample['label']
return {'samples': torch.from_numpy(samples), 'label': torch.from_numpy(label)}
data_transform = transforms.Compose([RandomChop(), Specgram(), ToTensor()])
train_dataset = SoundDataset(train_data_path, transform=data_transform)
valid_dataset = SoundDataset(train_data_path, valid_subset_path, transform=data_transform)
train_dataloader = torch.utils.data.DataLoader(train_dataset, batch_size=32, shuffle=True, num_workers=4)
valid_dataloader = torch.utils.data.DataLoader(valid_dataset, batch_size=32, shuffle=True, num_workers=4)
```
# Model
```
dropout = nn.Dropout(p=0.30)
class ConvRes(nn.Module):
def __init__(self, insize, outsize):
super(ConvRes, self).__init__()
self.math = nn.Sequential(
nn.BatchNorm2d(insize),
torch.nn.Conv2d(insize, outsize, kernel_size=2, padding=2),
nn.PReLU(),
)
def forward(self, x):
return self.math(x)
class ConvCNN(nn.Module):
def __init__(self, insize, outsize, kernel_size=7, padding=2, pool=2, avg=True):
super(ConvCNN, self).__init__()
self.avg = avg
self.math = torch.nn.Sequential(
torch.nn.Conv2d(insize, outsize, kernel_size=kernel_size, padding=padding),
torch.nn.BatchNorm2d(outsize),
torch.nn.LeakyReLU(),
torch.nn.MaxPool2d(pool, pool),
)
self.avgpool = torch.nn.AvgPool2d(pool, pool)
def forward(self, x):
x = self.math(x)
if self.avg is True:
x = self.avgpool(x)
return x
class Net(nn.Module):
def __init__(self):
super(Net, self).__init__()
self.avgpool = nn.AdaptiveAvgPool2d(1)
self.cnn1 = ConvCNN(1, 32, kernel_size=7, pool=4, avg=False)
self.cnn2 = ConvCNN(32, 32, kernel_size=5, pool=2, avg=True)
self.cnn3 = ConvCNN(32, 32, kernel_size=5, pool=2, avg=True)
self.res1 = ConvRes(32, 64)
self.features = nn.Sequential(
self.cnn1,
dropout,
self.cnn2,
self.cnn3,
self.res1,
)
self.classifier = torch.nn.Sequential(
nn.Linear(1024, 12),
)
self.sig = nn.Sigmoid()
def forward(self, x):
x = self.features(x)
x = x.view(x.size(0), -1)
x = self.classifier(x)
return x
```
# Testing
```
LR = 0.005
MOMENTUM = 0.9
model = Net()
loss_func = torch.nn.BCEWithLogitsLoss()
optimizer = torch.optim.Adam(model.parameters(), lr=LR, weight_decay=5e-5) # L2 regularization
def train(epoch, dataloader, log_steps=250):
n_total = dataloader.batch_size * len(dataloader)
last_batch = len(dataloader)
loss_history = []
loss_batch_history = []
model.train()
for batch_idx, batch in enumerate(dataloader, 1):
data, target = Variable(batch['samples']), Variable(batch['label'])
optimizer.zero_grad()
preds = model(data)
loss = loss_func(preds, target)
loss.backward()
optimizer.step()
loss_batch_history.append(loss.data[0])
if batch_idx % log_steps == 0 or batch_idx == last_batch:
loss_history.append(np.mean(loss_batch_history))
loss_batch_history = []
n_samples = min(batch_idx * dataloader.batch_size, n_total)
progress = 100. * n_samples / n_total
print(f'Train Epoch: {epoch} [{n_samples}/{n_total} ({progress:.0f}%)]\tLoss: {loss.data[0]:.6f}')
return loss_history
%%time
loss_history = []
for epoch in range(1, 5):
print("Epoch %d" % epoch)
loss_history += train(epoch, train_dataloader)
plt.plot(loss_history);
def evaluate(dataloader):
y_true = np.zeros(0)
y_pred = np.zeros(0)
for batch_idx, batch in enumerate(dataloader, 1):
data, target = Variable(batch['samples']), batch['label'].numpy()
y_true = np.concatenate((y_true, np.argmax(target, axis=1)))
y_pred = np.concatenate((y_pred, np.argmax(model(data).data.numpy(), axis=1)))
return y_true, y_pred
y_true, y_pred = evaluate(valid_dataloader)
precision = precision_score(y_true, y_pred, average='macro')
print(f'Precision: {precision: 0.3f}')
```
|
github_jupyter
|
import torch
import numpy as np
import pandas as pd
import torch.nn as nn
import torch.optim as optim
import torch.nn.functional as F
import matplotlib.pyplot as plt
from pathlib import Path
from scipy import signal
from scipy.io import wavfile
from scipy.fftpack import fft
from torch.autograd import Variable
from sklearn.metrics import precision_score
from torchvision import transforms, datasets
from torch.utils.data import Dataset, DataLoader
from sklearn.model_selection import train_test_split
import warnings
warnings.filterwarnings("ignore")
%matplotlib inline
legal_labels = 'yes no up down left right on off stop go silence unknown'.split()
train_data_path = Path('./train/audio')
valid_subset_path = Path('./train/validation_list.txt')
def unzip2(values):
a, b = zip(*values)
return list(a), list(b)
class SoundDataset(Dataset):
def __init__(self, directory, subset_path=None, transform=None):
self.directory = directory
self.transform = transform
self._subset = self._load_subset(subset_path) if subset_path else None
self._labels, self._sounds = self._list_of_wavs(directory)
self._transformed_labels = self._transform_labels(self._labels)
def __len__(self):
return len(self._labels)
def __getitem__(self, idx):
label, sound = self._labels[idx], self._sounds[idx]
sample_rate, samples = wavfile.read(self.directory/label/sound)
label = self._transformed_labels[idx]
sample = {'sample_rate': sample_rate, 'samples': samples, 'label': label}
if self.transform is not None:
sample = self.transform(sample)
return sample
def _is_needed(self, name):
return (name in self._subset) if self._subset is not None else True
def _list_of_wavs(self, directory, ext='wav'):
return unzip2(path.parts[-2:] for path in directory.glob(f'*/*.{ext}') if self._is_needed(path.parts[-1]))
@staticmethod
def _load_subset(file):
return set(path.split('/')[1] for path in file.read_text().split('\n'))
@staticmethod
def _transform_labels(labels):
nlabels = []
for label in labels:
if label == '_background_noise_':
nlabels.append('silence')
elif label not in legal_labels:
nlabels.append('unknown')
else:
nlabels.append(label)
return np.array(pd.get_dummies(pd.Series(nlabels)), dtype=np.float32)
class RandomChop:
def __init__(self, length=16_000):
self.length = length
def __call__(self, sample):
sample_rate, samples, label = sample['sample_rate'], sample['samples'], sample['label']
samples = self._pad_audio(samples)
if len(samples) > self.length:
samples = self._chop_audio(samples)
return {'sample_rate': sample_rate, 'samples': samples, 'label': label}
def _pad_audio(self, samples):
if len(samples) >= self.length:
return samples
return np.pad(samples, pad_width=(self.length - len(samples), 0), mode='constant', constant_values=(0, 0))
def _chop_audio(self, samples):
start = np.random.randint(0, len(samples) - self.length)
return samples[start : start + self.length]
class Specgram:
def __init__(self, sample_rate=8_000):
self.sample_rate = sample_rate
def __call__(self, sample):
sample_rate, samples, label = sample['sample_rate'], sample['samples'], sample['label']
resampled = signal.resample(samples, int(self.sample_rate / sample_rate * samples.shape[0]))
_, _, specgram = self._log_specgram(resampled, sample_rate=self.sample_rate)
specgram = specgram.reshape(1, specgram.shape[0], specgram.shape[1])
return {'samples': specgram, 'label': label}
@staticmethod
def _log_specgram(audio, sample_rate, window_size=20, step_size=10, eps=1e-10):
nperseg = int(round(window_size * sample_rate / 1e3))
noverlap = int(round(step_size * sample_rate / 1e3))
freqs, times, spec = signal.spectrogram(audio,
fs=sample_rate,
window='hann',
nperseg=nperseg,
noverlap=noverlap,
detrend=False)
return freqs, times, np.log(spec.T.astype(np.float32) + eps)
class ToTensor:
def __call__(self, sample):
samples, label = sample['samples'], sample['label']
return {'samples': torch.from_numpy(samples), 'label': torch.from_numpy(label)}
data_transform = transforms.Compose([RandomChop(), Specgram(), ToTensor()])
train_dataset = SoundDataset(train_data_path, transform=data_transform)
valid_dataset = SoundDataset(train_data_path, valid_subset_path, transform=data_transform)
train_dataloader = torch.utils.data.DataLoader(train_dataset, batch_size=32, shuffle=True, num_workers=4)
valid_dataloader = torch.utils.data.DataLoader(valid_dataset, batch_size=32, shuffle=True, num_workers=4)
dropout = nn.Dropout(p=0.30)
class ConvRes(nn.Module):
def __init__(self, insize, outsize):
super(ConvRes, self).__init__()
self.math = nn.Sequential(
nn.BatchNorm2d(insize),
torch.nn.Conv2d(insize, outsize, kernel_size=2, padding=2),
nn.PReLU(),
)
def forward(self, x):
return self.math(x)
class ConvCNN(nn.Module):
def __init__(self, insize, outsize, kernel_size=7, padding=2, pool=2, avg=True):
super(ConvCNN, self).__init__()
self.avg = avg
self.math = torch.nn.Sequential(
torch.nn.Conv2d(insize, outsize, kernel_size=kernel_size, padding=padding),
torch.nn.BatchNorm2d(outsize),
torch.nn.LeakyReLU(),
torch.nn.MaxPool2d(pool, pool),
)
self.avgpool = torch.nn.AvgPool2d(pool, pool)
def forward(self, x):
x = self.math(x)
if self.avg is True:
x = self.avgpool(x)
return x
class Net(nn.Module):
def __init__(self):
super(Net, self).__init__()
self.avgpool = nn.AdaptiveAvgPool2d(1)
self.cnn1 = ConvCNN(1, 32, kernel_size=7, pool=4, avg=False)
self.cnn2 = ConvCNN(32, 32, kernel_size=5, pool=2, avg=True)
self.cnn3 = ConvCNN(32, 32, kernel_size=5, pool=2, avg=True)
self.res1 = ConvRes(32, 64)
self.features = nn.Sequential(
self.cnn1,
dropout,
self.cnn2,
self.cnn3,
self.res1,
)
self.classifier = torch.nn.Sequential(
nn.Linear(1024, 12),
)
self.sig = nn.Sigmoid()
def forward(self, x):
x = self.features(x)
x = x.view(x.size(0), -1)
x = self.classifier(x)
return x
LR = 0.005
MOMENTUM = 0.9
model = Net()
loss_func = torch.nn.BCEWithLogitsLoss()
optimizer = torch.optim.Adam(model.parameters(), lr=LR, weight_decay=5e-5) # L2 regularization
def train(epoch, dataloader, log_steps=250):
n_total = dataloader.batch_size * len(dataloader)
last_batch = len(dataloader)
loss_history = []
loss_batch_history = []
model.train()
for batch_idx, batch in enumerate(dataloader, 1):
data, target = Variable(batch['samples']), Variable(batch['label'])
optimizer.zero_grad()
preds = model(data)
loss = loss_func(preds, target)
loss.backward()
optimizer.step()
loss_batch_history.append(loss.data[0])
if batch_idx % log_steps == 0 or batch_idx == last_batch:
loss_history.append(np.mean(loss_batch_history))
loss_batch_history = []
n_samples = min(batch_idx * dataloader.batch_size, n_total)
progress = 100. * n_samples / n_total
print(f'Train Epoch: {epoch} [{n_samples}/{n_total} ({progress:.0f}%)]\tLoss: {loss.data[0]:.6f}')
return loss_history
%%time
loss_history = []
for epoch in range(1, 5):
print("Epoch %d" % epoch)
loss_history += train(epoch, train_dataloader)
plt.plot(loss_history);
def evaluate(dataloader):
y_true = np.zeros(0)
y_pred = np.zeros(0)
for batch_idx, batch in enumerate(dataloader, 1):
data, target = Variable(batch['samples']), batch['label'].numpy()
y_true = np.concatenate((y_true, np.argmax(target, axis=1)))
y_pred = np.concatenate((y_pred, np.argmax(model(data).data.numpy(), axis=1)))
return y_true, y_pred
y_true, y_pred = evaluate(valid_dataloader)
precision = precision_score(y_true, y_pred, average='macro')
print(f'Precision: {precision: 0.3f}')
| 0.850251 | 0.674242 |
# Decision Trees for Classification Explained
#### Build Decision Tree in classification and regression algorithm to build a model.
```
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
import warnings
warnings.filterwarnings("ignore")
# yahoo finance is used to fetch data
import yfinance as yf
yf.pdr_override()
```
#### Getting the histocial data from yahoo finance
```
# input
symbol = 'AMD'
start = '2012-01-01'
end = '2021-09-05'
# Read data
dataset = yf.download(symbol,start,end)
# View Columns
dataset.head()
```
#### Create more data
```
# Create more data
dataset['Increase_Decrease'] = np.where(dataset['Volume'].shift(-1) > dataset['Volume'],1,-1)
dataset['Buy_Sell_on_Open'] = np.where(dataset['Open'].shift(-1) > dataset['Open'],1,-1)
dataset['Buy_Sell'] = np.where(dataset['Adj Close'].shift(-1) > dataset['Adj Close'],1,-1)
dataset['Return'] = dataset['Adj Close'].pct_change()
dataset = dataset.dropna()
dataset['Up_Down'] = np.where(dataset['Return'].shift(-1) > dataset['Return'],'Up','Down')
dataset.head()
dataset.shape
predictors_list = ['Open', 'High','Low', 'Adj Close', 'Return', 'Volume']
X = dataset[predictors_list]
X.tail()
y = dataset['Buy_Sell']
y.name
X.columns
```
#### test_size
the size of the test data set, in this case, 30% of the data for the tests and, therefore, 70% for the training.
#### random_state
Since the sampling is random, this parameter allows us to reproduce the same randomness in each execution.
#### stratify
To ensure that the training and test sample data are proportional, we set the parameter to yes. This means that, for example, if there are more days with positive than negative return, the training and test samples will keep the same proportion.
```
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=200, stratify=y)
train_length = int(len(dataset)*0.80)
X_train = X[:train_length]
X_test = X[train_length:]
y_train = y[:train_length]
y_test = y[train_length:]
print (X_train.shape, y_train.shape)
print (X_test.shape, y_test.shape)
from sklearn.tree import DecisionTreeClassifier
clf = DecisionTreeClassifier(criterion='gini', max_depth=3, min_samples_leaf=3)
clf.fit(X_train, y_train)
from mlxtend.evaluate import bias_variance_decomp
avg_expected_loss, avg_bias, avg_var = bias_variance_decomp(clf, X_train.values, y_train.values, X_test.values, y_test.values, loss='mse', random_seed=123)
print('MSE: %.3f' % avg_expected_loss)
print('Bias: %.3f' % avg_bias)
print('Variance: %.3f' % avg_var)
# Prediction
prediction = clf.predict(X_test)
# Evaluation
from sklearn import metrics
print("DecisionTrees's Accuracy: ", metrics.accuracy_score(y_test, prediction))
# Accuracy Score without Sklearn
boolian = (y_test==prediction)
accuracy = sum(boolian)/y_test.size
accuracy
from sklearn import tree
import graphviz
dot_data = tree.export_graphviz(clf, out_file=None,
feature_names=X.columns,
class_names=X.columns)
dot_data = tree.export_graphviz(clf, out_file=None,filled=True,feature_names=predictors_list)
graphviz.Source(dot_data)
def plot_curve(ticks, train_scores, test_scores):
train_scores_mean = -1 * np.mean(train_scores, axis=1)
train_scores_std = -1 * np.std(train_scores, axis=1)
test_scores_mean = -1 * np.mean(test_scores, axis=1)
test_scores_std = -1 * np.std(test_scores, axis=1)
plt.figure()
plt.fill_between(ticks,
train_scores_mean - train_scores_std,
train_scores_mean + train_scores_std, alpha=0.1, color="b")
plt.fill_between(ticks,
test_scores_mean - test_scores_std,
test_scores_mean + test_scores_std, alpha=0.1, color="r")
plt.plot(ticks, train_scores_mean, 'b-', label='Training score')
plt.plot(ticks, test_scores_mean, 'r-', label='Test score')
plt.legend(fancybox=True, facecolor='w')
return plt.gca()
def plot_validation_curve(clf, X, y, param_name, param_range, scoring='roc_auc'):
plt.xkcd()
ax = plot_curve(param_range, *validation_curve(clf, X, y, cv=num_folds,
scoring=scoring,
param_name=param_name,
param_range=param_range, n_jobs=-1))
ax.set_title('')
ax.set_xticklabels([])
ax.set_yticklabels([])
ax.set_xlim(2,12)
ax.set_ylim(-0.97, -0.83)
ax.set_ylabel('Error')
ax.set_xlabel('Model complexity')
ax.text(9, -0.94, 'Overfitting', fontsize=22)
ax.text(3, -0.94, 'Underfitting', fontsize=22)
ax.axvline(7, ls='--')
plt.tight_layout()
from sklearn.model_selection import validation_curve
num_folds = 7
plot_validation_curve(clf, X_train, y_train, param_name='max_depth', param_range=range(10,16))
```
#### Compare the scores of testing on training and testing data. If the scores are close to equal, is likely underfitting; however, if they are far apart, is likely overfitting.
```
from sklearn import cross_validation
for i in range(5):
X_train, X_test, y_train, y_test = cross_validation.train_test_split(X, y, test_size=0.2)
clf = DecisionTreeClassifier(criterion='gini', max_depth=3, min_samples_leaf=3).fit(X_train, y_train)
print("Test score", clf.score(X_test, y_test))
print("Train score", clf.score(X_train, y_train))
from sklearn.metrics import classification_report
print(classification_report(y_test, prediction, target_names=y.name))
from sklearn.metrics import confusion_matrix
# Calculate the confusion matrix
conf_matrix = confusion_matrix(y_true=y_test, y_pred=prediction)
fig, ax = plt.subplots(figsize=(5, 5))
ax.matshow(conf_matrix, cmap=plt.cm.Oranges, alpha=0.3)
for i in range(conf_matrix.shape[0]):
for j in range(conf_matrix.shape[1]):
ax.text(x=j, y=i,s=conf_matrix[i, j], va='center', ha='center', size='xx-large')
plt.xlabel('Predictions', fontsize=18)
plt.ylabel('Actuals', fontsize=18)
plt.title('Confusion Matrix', fontsize=18)
plt.show()
from sklearn.metrics import precision_score, recall_score, f1_score, accuracy_score
print('Precision: %.3f' % precision_score(y_test, prediction))
print('Recall: %.3f' % recall_score(y_test, prediction))
print('Accuracy: %.3f' % accuracy_score(y_test, prediction))
print('F1 Score: %.3f' % f1_score(y_test, prediction))
```
|
github_jupyter
|
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
import warnings
warnings.filterwarnings("ignore")
# yahoo finance is used to fetch data
import yfinance as yf
yf.pdr_override()
# input
symbol = 'AMD'
start = '2012-01-01'
end = '2021-09-05'
# Read data
dataset = yf.download(symbol,start,end)
# View Columns
dataset.head()
# Create more data
dataset['Increase_Decrease'] = np.where(dataset['Volume'].shift(-1) > dataset['Volume'],1,-1)
dataset['Buy_Sell_on_Open'] = np.where(dataset['Open'].shift(-1) > dataset['Open'],1,-1)
dataset['Buy_Sell'] = np.where(dataset['Adj Close'].shift(-1) > dataset['Adj Close'],1,-1)
dataset['Return'] = dataset['Adj Close'].pct_change()
dataset = dataset.dropna()
dataset['Up_Down'] = np.where(dataset['Return'].shift(-1) > dataset['Return'],'Up','Down')
dataset.head()
dataset.shape
predictors_list = ['Open', 'High','Low', 'Adj Close', 'Return', 'Volume']
X = dataset[predictors_list]
X.tail()
y = dataset['Buy_Sell']
y.name
X.columns
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=200, stratify=y)
train_length = int(len(dataset)*0.80)
X_train = X[:train_length]
X_test = X[train_length:]
y_train = y[:train_length]
y_test = y[train_length:]
print (X_train.shape, y_train.shape)
print (X_test.shape, y_test.shape)
from sklearn.tree import DecisionTreeClassifier
clf = DecisionTreeClassifier(criterion='gini', max_depth=3, min_samples_leaf=3)
clf.fit(X_train, y_train)
from mlxtend.evaluate import bias_variance_decomp
avg_expected_loss, avg_bias, avg_var = bias_variance_decomp(clf, X_train.values, y_train.values, X_test.values, y_test.values, loss='mse', random_seed=123)
print('MSE: %.3f' % avg_expected_loss)
print('Bias: %.3f' % avg_bias)
print('Variance: %.3f' % avg_var)
# Prediction
prediction = clf.predict(X_test)
# Evaluation
from sklearn import metrics
print("DecisionTrees's Accuracy: ", metrics.accuracy_score(y_test, prediction))
# Accuracy Score without Sklearn
boolian = (y_test==prediction)
accuracy = sum(boolian)/y_test.size
accuracy
from sklearn import tree
import graphviz
dot_data = tree.export_graphviz(clf, out_file=None,
feature_names=X.columns,
class_names=X.columns)
dot_data = tree.export_graphviz(clf, out_file=None,filled=True,feature_names=predictors_list)
graphviz.Source(dot_data)
def plot_curve(ticks, train_scores, test_scores):
train_scores_mean = -1 * np.mean(train_scores, axis=1)
train_scores_std = -1 * np.std(train_scores, axis=1)
test_scores_mean = -1 * np.mean(test_scores, axis=1)
test_scores_std = -1 * np.std(test_scores, axis=1)
plt.figure()
plt.fill_between(ticks,
train_scores_mean - train_scores_std,
train_scores_mean + train_scores_std, alpha=0.1, color="b")
plt.fill_between(ticks,
test_scores_mean - test_scores_std,
test_scores_mean + test_scores_std, alpha=0.1, color="r")
plt.plot(ticks, train_scores_mean, 'b-', label='Training score')
plt.plot(ticks, test_scores_mean, 'r-', label='Test score')
plt.legend(fancybox=True, facecolor='w')
return plt.gca()
def plot_validation_curve(clf, X, y, param_name, param_range, scoring='roc_auc'):
plt.xkcd()
ax = plot_curve(param_range, *validation_curve(clf, X, y, cv=num_folds,
scoring=scoring,
param_name=param_name,
param_range=param_range, n_jobs=-1))
ax.set_title('')
ax.set_xticklabels([])
ax.set_yticklabels([])
ax.set_xlim(2,12)
ax.set_ylim(-0.97, -0.83)
ax.set_ylabel('Error')
ax.set_xlabel('Model complexity')
ax.text(9, -0.94, 'Overfitting', fontsize=22)
ax.text(3, -0.94, 'Underfitting', fontsize=22)
ax.axvline(7, ls='--')
plt.tight_layout()
from sklearn.model_selection import validation_curve
num_folds = 7
plot_validation_curve(clf, X_train, y_train, param_name='max_depth', param_range=range(10,16))
from sklearn import cross_validation
for i in range(5):
X_train, X_test, y_train, y_test = cross_validation.train_test_split(X, y, test_size=0.2)
clf = DecisionTreeClassifier(criterion='gini', max_depth=3, min_samples_leaf=3).fit(X_train, y_train)
print("Test score", clf.score(X_test, y_test))
print("Train score", clf.score(X_train, y_train))
from sklearn.metrics import classification_report
print(classification_report(y_test, prediction, target_names=y.name))
from sklearn.metrics import confusion_matrix
# Calculate the confusion matrix
conf_matrix = confusion_matrix(y_true=y_test, y_pred=prediction)
fig, ax = plt.subplots(figsize=(5, 5))
ax.matshow(conf_matrix, cmap=plt.cm.Oranges, alpha=0.3)
for i in range(conf_matrix.shape[0]):
for j in range(conf_matrix.shape[1]):
ax.text(x=j, y=i,s=conf_matrix[i, j], va='center', ha='center', size='xx-large')
plt.xlabel('Predictions', fontsize=18)
plt.ylabel('Actuals', fontsize=18)
plt.title('Confusion Matrix', fontsize=18)
plt.show()
from sklearn.metrics import precision_score, recall_score, f1_score, accuracy_score
print('Precision: %.3f' % precision_score(y_test, prediction))
print('Recall: %.3f' % recall_score(y_test, prediction))
print('Accuracy: %.3f' % accuracy_score(y_test, prediction))
print('F1 Score: %.3f' % f1_score(y_test, prediction))
| 0.636579 | 0.930868 |
# Functional Linear Regression
> "Linear regression augmented through interpolation and regularization techniques."
- toc: false
- branch: master
- badges: true
- comments: true
- image: images/FDA.png
- hide: false
- search_exclude: false
Covariance estimation is a problem of great interest in many different disciplines, including machine learning, signal processing, economics and bioinformatics. In many applications the number of variables is very large, e.g., in the tens or hundreds of thousands, leading to a number of covariance parameters that greatly exceeds the number of observations. To address this problem constraints are frequently imposed on the covariance to reduce the number of parameters in the model. For example, the Glasso model of Yuan and Lin and Banerjee et al [<sup>1</sup>](#fn1) imposes sparsity constraints on the covariance. The Kronecker product model of Dutilleul and Werner et al [<sup>2</sup>](#fn2) assumes that the covariance can be represented as the Kronecker product of two lower dimensional covariance matrices. Here we will implement a combination of these two aproaches.
Here is our problem setting:
A combustion engine produces gas with polluting substances such as nitrogen oxides (NOx).Gas emission control regulations have been set up to protect the environment. The NOx Storage Catalyst (NSC) is an emission control system by which the exhaust gas is treated after the combustion process in two phases: adsorption and regeneration. During the regeneration phase, the engine control unit is programmed to maintain the combustion process in a rich air-to-fuel status. The average relative air/fuel ratio is the indicator of a correct regeneration phase. Our goal is to predict this value, using the information from eleven sensors. To do so, we are going to use group lasso regression.
List of on-board sensorsair aspirated per cylinder
- engine rotational speed
- total quantity of fuel injected
- low presure EGR valve
- inner torque
- accelerator pedal position
- aperture ratio of inlet valve
- downstreem intercooler preasure
- fuel in the 2nd pre-injection
- vehicle velocity
First we will write the problem that we want to solve in mathematical notation.
$$ \underset{\beta_g \in \mathbb{R}}{armin} \ \left \| \sum_{g \in G}\left [ X_g\beta_g \right ]-y\right \|_2^2 + \lambda_1\left | \beta \right |_1 + \lambda_2\sum_{g \in G}\sqrt[]{d_g}\left \| \beta_g \right \|_2 $$
Where
$$ $$
$ X_g \in \mathbb{R}^{n x d_g}$ is the data matrix for each sensor's covariates which compose group $g$,
$ \beta_g $ is the B spline coefficients for group $g$,
$ y \in \mathbb{R}^{n}$ is the air/fuel ratio target,
$ n$ is the number of measurements,
$d_g$ is the dimensionality of group $g$,
$\lambda_1 $ is the parameter-wise regularization penalty,
$\lambda_2$ is the group-wise regularization penalty,
$ G $ is the set of all groups for all sensors
Now on to the code. We will use group lasso to learn the B-spline coefficients. We will use B-splines with 8 knots to reduce the dimensionality of the problem. Ultimately, we want to determine which sensors are correlated with the air/fuel ratio? Also, we want to predict the air/fuel ratio for the observations in the test dataset.
```
from scipy import interpolate
import group_lasso
import sklearn.linear_model as lm
from sklearn.model_selection import GridSearchCV, RandomizedSearchCV
x_train = loadmat('NSC.mat')['x']
y_train = loadmat('NSC.mat')['y']
x_test = loadmat('NSC.test.mat')['x_test']
y_test = loadmat('NSC.test.mat')['y_test']
for i in range(len(x_train[0])):
plt.figure(figsize=(15,8))
pd.DataFrame(x_train[0][i]).plot(legend=False, title=f"Sensor {i}")
def transformation(data):
coefficients = []
x = np.linspace(0, 203, 203)
knots = np.linspace(0, 203, 10) [1:-1]
for i,d in enumerate(data):
t, c, k = interpolate.splrep(x, d, task=-1, t=knots, k=2)
coefficients.append(np.trim_zeros(c, trim='b')[:-1])
return np.array(coefficients)
def standardize(data):
results = []
for i in data:
temp = scaler.fit_transform(i)
results.append(temp)
return results
scaler = StandardScaler()
Y_train = transformation(scaler.fit_transform(y_train)).ravel()
Y_test = transformation(scaler.fit_transform(y_test)).ravel()
X_train = np.hstack(np.array([transformation(i) for i in standardize(x_train[0])]))
X_test = np.hstack(np.array([transformation(i) for i in standardize(x_test[0])]))
identity = np.identity(10)
```
Kronecker Products
```
final_train = np.kron(X_train, identity)
final_test = np.kron(X_test, identity)
g = [[i]*100 for i in range(1,11)]
groups = np.array([item for sublist in g for item in sublist])
gl = group_lasso.GroupLasso(
groups=groups,
group_reg=0,
l1_reg=0,
fit_intercept=True,
scale_reg="none",
supress_warning=True,
tol=1e-5
)
lambdas, _, _ = lm.lasso_path(final_train, Y_train)
CV = RandomizedSearchCV(estimator=gl, param_distributions={'group_reg': lambdas[::5]}, scoring='neg_mean_squared_error', n_iter=100, verbose=2)
CV.fit(final_train, Y_train)
coef = gl.coef_.ravel().reshape(100, 10)
coef_base = X_train@coef
coef_df = pd.DataFrame(coef_base)
print("Best lambda:", CV.best_params_['group_reg'])
print("Coefficients Correlated to Target")
coef_df.corrwith(pd.DataFrame(Y_train.reshape(150,10)))
```
It appears sensors 2 and 7 have the greatest correlation to the air fuel ration
```
_y = pd.DataFrame(Y_train.reshape(150,10))
for sensor in [2, 7]:
plt.figure(figsize=(15,8))
plt.scatter(coef_df[sensor], _y[sensor])
plt.title(f"Correlation of sensor {sensor} and air/fuel ratio")
plt.xlabel(f"Sensor {sensor}")
plt.ylabel("Air/fuel ratio")
coef_df[2].plot(title='Coefficients for sensor 2')
coef_df[7].plot(title='Coefficients for sensor 7')
predicted = CV.predict(final_test)
print("Mean Square Prediction Error:", sum((Y_test - predicted)**2))
```
________________
<span id="fn1"> Yuan et al. "Model Selection and Estimation in Regression With Grouped Variables," Journal of the Royal Statistical Society Series B. (2006): 49-67.</span>
<span id="fn2"> Tsiligkaridis et al. "Convergence Properties of Kronecker Graphical Lasso Algorithms," IEEE (2013).</span>
|
github_jupyter
|
from scipy import interpolate
import group_lasso
import sklearn.linear_model as lm
from sklearn.model_selection import GridSearchCV, RandomizedSearchCV
x_train = loadmat('NSC.mat')['x']
y_train = loadmat('NSC.mat')['y']
x_test = loadmat('NSC.test.mat')['x_test']
y_test = loadmat('NSC.test.mat')['y_test']
for i in range(len(x_train[0])):
plt.figure(figsize=(15,8))
pd.DataFrame(x_train[0][i]).plot(legend=False, title=f"Sensor {i}")
def transformation(data):
coefficients = []
x = np.linspace(0, 203, 203)
knots = np.linspace(0, 203, 10) [1:-1]
for i,d in enumerate(data):
t, c, k = interpolate.splrep(x, d, task=-1, t=knots, k=2)
coefficients.append(np.trim_zeros(c, trim='b')[:-1])
return np.array(coefficients)
def standardize(data):
results = []
for i in data:
temp = scaler.fit_transform(i)
results.append(temp)
return results
scaler = StandardScaler()
Y_train = transformation(scaler.fit_transform(y_train)).ravel()
Y_test = transformation(scaler.fit_transform(y_test)).ravel()
X_train = np.hstack(np.array([transformation(i) for i in standardize(x_train[0])]))
X_test = np.hstack(np.array([transformation(i) for i in standardize(x_test[0])]))
identity = np.identity(10)
final_train = np.kron(X_train, identity)
final_test = np.kron(X_test, identity)
g = [[i]*100 for i in range(1,11)]
groups = np.array([item for sublist in g for item in sublist])
gl = group_lasso.GroupLasso(
groups=groups,
group_reg=0,
l1_reg=0,
fit_intercept=True,
scale_reg="none",
supress_warning=True,
tol=1e-5
)
lambdas, _, _ = lm.lasso_path(final_train, Y_train)
CV = RandomizedSearchCV(estimator=gl, param_distributions={'group_reg': lambdas[::5]}, scoring='neg_mean_squared_error', n_iter=100, verbose=2)
CV.fit(final_train, Y_train)
coef = gl.coef_.ravel().reshape(100, 10)
coef_base = X_train@coef
coef_df = pd.DataFrame(coef_base)
print("Best lambda:", CV.best_params_['group_reg'])
print("Coefficients Correlated to Target")
coef_df.corrwith(pd.DataFrame(Y_train.reshape(150,10)))
_y = pd.DataFrame(Y_train.reshape(150,10))
for sensor in [2, 7]:
plt.figure(figsize=(15,8))
plt.scatter(coef_df[sensor], _y[sensor])
plt.title(f"Correlation of sensor {sensor} and air/fuel ratio")
plt.xlabel(f"Sensor {sensor}")
plt.ylabel("Air/fuel ratio")
coef_df[2].plot(title='Coefficients for sensor 2')
coef_df[7].plot(title='Coefficients for sensor 7')
predicted = CV.predict(final_test)
print("Mean Square Prediction Error:", sum((Y_test - predicted)**2))
| 0.493653 | 0.986649 |
___
<a href='http://www.pieriandata.com'> <img src='../Pierian_Data_Logo.png' /></a>
___
# Python Crash Course Exercises - Solutions
This is an optional exercise to test your understanding of Python Basics. If you find this extremely challenging, then you probably are not ready for the rest of this course yet and don't have enough programming experience to continue. I would suggest you take another course more geared towards complete beginners, such as [Complete Python Bootcamp](https://www.udemy.com/complete-python-bootcamp/?couponCode=PY20)
## Exercises
Answer the questions or complete the tasks outlined in bold below, use the specific method described if applicable.
** What is 7 to the power of 4?**
```
7**4
```
** Split this string:**
s = "Hi there Sam!"
**into a list. **
```
s = 'Hi there Sam!'
s.split()
```
** Given the variables:**
planet = "Earth"
diameter = 12742
** Use .format() to print the following string: **
The diameter of Earth is 12742 kilometers.
```
planet = "Earth"
diameter = 12742
print("The diameter of {} is {} kilometers.".format(planet,diameter))
```
** Given this nested list, use indexing to grab the word "hello" **
```
lst = [1,2,[3,4],[5,[100,200,['hello']],23,11],1,7]
lst[3][1][2][0]
```
** Given this nest dictionary grab the word "hello". Be prepared, this will be annoying/tricky **
```
d = {'k1':[1,2,3,{'tricky':['oh','man','inception',{'target':[1,2,3,'hello']}]}]}
d['k1'][3]['tricky'][3]['target'][3]
```
** What is the main difference between a tuple and a list? **
```
# Tuple is immutable
```
** Create a function that grabs the email website domain from a string in the form: **
user@domain.com
**So for example, passing "user@domain.com" would return: domain.com**
```
def domainGet(email):
return email.split('@')[-1]
domainGet('user@domain.com')
```
** Create a basic function that returns True if the word 'dog' is contained in the input string. Don't worry about edge cases like a punctuation being attached to the word dog, but do account for capitalization. **
```
def findDog(st):
return 'dog' in st.lower().split()
findDog('Is there a dog here?')
```
** Create a function that counts the number of times the word "dog" occurs in a string. Again ignore edge cases. **
```
def countDog(st):
count = 0
for word in st.lower().split():
if word == 'dog':
count += 1
return count
countDog('This dog runs faster than the other dog dude!')
```
** Use lambda expressions and the filter() function to filter out words from a list that don't start with the letter 's'. For example:**
seq = ['soup','dog','salad','cat','great']
**should be filtered down to:**
['soup','salad']
```
seq = ['soup','dog','salad','cat','great']
list(filter(lambda word: word[0]=='s',seq))
```
### Final Problem
**You are driving a little too fast, and a police officer stops you. Write a function
to return one of 3 possible results: "No ticket", "Small ticket", or "Big Ticket".
If your speed is 60 or less, the result is "No Ticket". If speed is between 61
and 80 inclusive, the result is "Small Ticket". If speed is 81 or more, the result is "Big Ticket". Unless it is your birthday (encoded as a boolean value in the parameters of the function) -- on your birthday, your speed can be 5 higher in all
cases. **
```
def caught_speeding(speed, is_birthday):
if is_birthday:
speeding = speed - 5
else:
speeding = speed
if speeding > 80:
return 'Big Ticket'
elif speeding > 60:
return 'Small Ticket'
else:
return 'No Ticket'
caught_speeding(81,True)
caught_speeding(81,False)
```
# Great job!
|
github_jupyter
|
7**4
s = 'Hi there Sam!'
s.split()
planet = "Earth"
diameter = 12742
print("The diameter of {} is {} kilometers.".format(planet,diameter))
lst = [1,2,[3,4],[5,[100,200,['hello']],23,11],1,7]
lst[3][1][2][0]
d = {'k1':[1,2,3,{'tricky':['oh','man','inception',{'target':[1,2,3,'hello']}]}]}
d['k1'][3]['tricky'][3]['target'][3]
# Tuple is immutable
def domainGet(email):
return email.split('@')[-1]
domainGet('user@domain.com')
def findDog(st):
return 'dog' in st.lower().split()
findDog('Is there a dog here?')
def countDog(st):
count = 0
for word in st.lower().split():
if word == 'dog':
count += 1
return count
countDog('This dog runs faster than the other dog dude!')
seq = ['soup','dog','salad','cat','great']
list(filter(lambda word: word[0]=='s',seq))
def caught_speeding(speed, is_birthday):
if is_birthday:
speeding = speed - 5
else:
speeding = speed
if speeding > 80:
return 'Big Ticket'
elif speeding > 60:
return 'Small Ticket'
else:
return 'No Ticket'
caught_speeding(81,True)
caught_speeding(81,False)
| 0.206414 | 0.976288 |
## The SIR model with two age groups
The partitioning of the population can be refined to include other attributes relevant to the disease. One of the most important of these is the age. Let us assume we partition the population into two age groups, children and adults, and label them by the index $i=1,2$. Children can catch the infection from other children or from adults; likewise, adults can catch the infection from other adults or from children. Calling their respective rates of infection $\lambda_1(t)$ and $\lambda_2(t)$ we get
\begin{align}
\lambda_1(t) = \beta(C_{11}\frac{I_1}{N_1} + C_{12}\frac{I_2}{N_2})S_1\\
\lambda_2(t) = \beta(C_{21}\frac{I_1}{N_1} + C_{22}\frac{I_2}{N_2})S_2
\end{align}
where $C_{ij}$ are contact matrices, quantifying how much each age group interacts with the other. The ordinary differential equations of this age-structured SIR model are
\begin{align}
\dot S_i &= -\lambda_i(t)S_i \\
\dot I_i &= \lambda(t)_iI_i - \gamma I_i \\
\dot R_i &= \gamma I_i
\end{align}
Again, for each $i$ the sum $N_i = S_i + I_i + R_i$ remains constant. What do we expect qualitatively ? The group that has a greater rate will catch the disease faster and catch more of it. This depends on how the entries of the contact matrix are distributed.
This example integrates the above equations to **epidemic curve** for both the children and the adults. We see that they have unequal rates of infection.
```
%%capture
## compile PyRoss for this notebook
import os
owd = os.getcwd()
os.chdir('../')
%run setup.py install
os.chdir(owd)
%matplotlib inline
import numpy as np
import pyross
import matplotlib.pyplot as plt
M = 2 # the population has two age groups
N = 1000000 # and this is the total population
beta = 0.0131 # infection rate
gIa = 1./7 # recovery rate of asymptomatic infectives
gIs = 1./7 # recovery rate of asymptomatic infectives
alpha = 0 # fraction of asymptomatic infectives
fsa = 1 # the self-isolation parameter
Ni = np.zeros((M)) # population in each group
fi = np.zeros((M)) # fraction of population in age age group
# set the age structure
fi = np.array((0.25, 0.75))
for i in range(M):
Ni[i] = fi[i]*N
# set the contact structure
C = np.array(([18., 9.], [3., 12.]))
Ia_0 = np.array((1,1)) # each age group has asymptomatic infectives
Is_0 = np.array((1,1)) # and also symptomatic infectives
R_0 = np.array((0,0)) # there are no recovered individuals initially
S_0 = Ni - (Ia_0 + Is_0 + R_0)
# matrix for linearised dynamics
L = np.zeros((M, M))
for i in range(M):
for j in range(M):
L[i,j]=C[i,j]*Ni[i]/Ni[j]
L = (alpha*beta/gIs)*L
# the basic reproductive ratio
r0 = np.max(np.linalg.eigvals(L))
print("The basic reproductive ratio for these parameters is", r0)
# duration of simulation and data file
Tf=200; Nf=2000; filename='this.mat'
# the contact structure is independent of time
def contactMatrix(t):
return C
# instantiate model
parameters = {'alpha':alpha, 'beta':beta, 'gIa':gIa, 'gIs':gIs,'fsa':fsa}
model = pyross.models.SIR(parameters, M, Ni)
# simulate model
data=model.simulate(S_0, Ia_0, Is_0, contactMatrix, Tf, Nf, filename)
IK = data.get('X')[:,2*M].flatten()
IA = data.get('X')[:,2*M+1].flatten()
t = data.get('t')
fig = plt.figure(num=None, figsize=(10, 8), dpi=80, facecolor='w', edgecolor='k')
plt.rcParams.update({'font.size': 22})
plt.fill_between(t, 0, IK/Ni[0], color="#348ABD", alpha=0.3)
plt.plot(t, IK/Ni[0], '-', color="#348ABD", label='$Children$', lw=4)
plt.fill_between(t, 0, IA/Ni[1], color='#A60628', alpha=0.3)
plt.plot(t, IA/Ni[1], '-', color='#A60628', label='$Adults$', lw=4)
plt.legend(fontsize=26); plt.grid()
plt.autoscale(enable=True, axis='x', tight=True)
```
|
github_jupyter
|
%%capture
## compile PyRoss for this notebook
import os
owd = os.getcwd()
os.chdir('../')
%run setup.py install
os.chdir(owd)
%matplotlib inline
import numpy as np
import pyross
import matplotlib.pyplot as plt
M = 2 # the population has two age groups
N = 1000000 # and this is the total population
beta = 0.0131 # infection rate
gIa = 1./7 # recovery rate of asymptomatic infectives
gIs = 1./7 # recovery rate of asymptomatic infectives
alpha = 0 # fraction of asymptomatic infectives
fsa = 1 # the self-isolation parameter
Ni = np.zeros((M)) # population in each group
fi = np.zeros((M)) # fraction of population in age age group
# set the age structure
fi = np.array((0.25, 0.75))
for i in range(M):
Ni[i] = fi[i]*N
# set the contact structure
C = np.array(([18., 9.], [3., 12.]))
Ia_0 = np.array((1,1)) # each age group has asymptomatic infectives
Is_0 = np.array((1,1)) # and also symptomatic infectives
R_0 = np.array((0,0)) # there are no recovered individuals initially
S_0 = Ni - (Ia_0 + Is_0 + R_0)
# matrix for linearised dynamics
L = np.zeros((M, M))
for i in range(M):
for j in range(M):
L[i,j]=C[i,j]*Ni[i]/Ni[j]
L = (alpha*beta/gIs)*L
# the basic reproductive ratio
r0 = np.max(np.linalg.eigvals(L))
print("The basic reproductive ratio for these parameters is", r0)
# duration of simulation and data file
Tf=200; Nf=2000; filename='this.mat'
# the contact structure is independent of time
def contactMatrix(t):
return C
# instantiate model
parameters = {'alpha':alpha, 'beta':beta, 'gIa':gIa, 'gIs':gIs,'fsa':fsa}
model = pyross.models.SIR(parameters, M, Ni)
# simulate model
data=model.simulate(S_0, Ia_0, Is_0, contactMatrix, Tf, Nf, filename)
IK = data.get('X')[:,2*M].flatten()
IA = data.get('X')[:,2*M+1].flatten()
t = data.get('t')
fig = plt.figure(num=None, figsize=(10, 8), dpi=80, facecolor='w', edgecolor='k')
plt.rcParams.update({'font.size': 22})
plt.fill_between(t, 0, IK/Ni[0], color="#348ABD", alpha=0.3)
plt.plot(t, IK/Ni[0], '-', color="#348ABD", label='$Children$', lw=4)
plt.fill_between(t, 0, IA/Ni[1], color='#A60628', alpha=0.3)
plt.plot(t, IA/Ni[1], '-', color='#A60628', label='$Adults$', lw=4)
plt.legend(fontsize=26); plt.grid()
plt.autoscale(enable=True, axis='x', tight=True)
| 0.510496 | 0.977001 |
```
%matplotlib notebook
import numpy as np
import matplotlib.pyplot as plt
import matplotlib
matplotlib.rc("image", cmap="gray", interpolation="nearest")
```
First load the patterns, and display them. There are a few numbers, plus "HI", plus a smiley face:
```
f = np.load("patterns.npz")
# extract the labels and data
labels = sorted(f.keys())
data = np.array([f[key].reshape((10, 10)).T.ravel() for key in labels])
# reverse numbers
data[labels.index('num1')] = 1 - data[labels.index('num1')]
data[labels.index('num2')] = 1 - data[labels.index('num2')]
data[labels.index('num3')] = 1 - data[labels.index('num3')]
data[labels.index('num4')] = 1 - data[labels.index('num4')]
# display them
fig, axes = plt.subplots(1, 7)
for j in range(7):
axes[j].imshow(data[j].reshape((10, 10)))
axes[j].set_xticks([])
axes[j].set_yticks([])
fig.set_size_inches(10, 2, forward=True)
plt.tight_layout()
```
Define some helper functions:
```
def sigmoid(x):
"""Sigmoid function"""
return 1.0 / (1.0 + np.exp(-x))
def dsigmoid(x):
"""Derivative of the sigmoid function"""
return sigmoid(x) * (1 - sigmoid(x))
def add_noise(X0, p=0.5):
"""Adds random zero-masking noise"""
X1 = X0.copy()
idx = np.random.rand(*X1.shape) < 0.5
X1[idx] = 0
return X1
def init_plotting(data, M, N):
"""Initialize figure and subplots"""
fig, axes = plt.subplots(4, 7)
axes = axes.T
ims = np.empty(axes.shape, dtype=object)
for j in range(k):
ims[j, 0] = axes[j, 0].imshow(data[j].reshape((10, 10)))
ims[j, 1] = axes[j, 1].imshow(M[j].reshape((10, 10)))
ims[j, 2] = axes[j, 2].imshow(N[:, j].reshape((10, 10)))
ims[j, 3] = axes[j, 3].imshow(data[j].reshape((10, 10)))
for ax in axes.flat:
ax.set_xticks([])
ax.set_yticks([])
axes[0, 0].set_ylabel("Corrupted input")
axes[0, 1].set_ylabel("Encoder weights")
axes[0, 2].set_ylabel("Decoder weights")
axes[0, 3].set_ylabel("Decoded image")
fig.set_size_inches(12, 7, forward=True)
plt.tight_layout()
return fig, axes, ims
def draw(fig, data, M, bM, N, bN):
"""Draw the current weights, as well as an example
corrupted input and reconstruction from that input.
"""
k = data.shape[0]
for j in range(k):
X1 = add_noise(data[[j]])
# compute hidden representation
Y = sigmoid(np.dot(M, X1.T) + bM)
# reconstruct the input
X2 = sigmoid(np.dot(N, Y) + bN).T
ims[j, 0].set_data(X1.reshape((10, 10)))
ims[j, 1].set_data(M[j].reshape((10, 10)))
ims[j, 2].set_data(N[:, j].reshape((10, 10)))
ims[j, 3].set_data(X2.reshape((10, 10)))
fig.canvas.draw()
alpha = 0.01
M = np.random.randn(7, 100)
bM = np.random.randn(7, 1)
N = np.random.randn(100, 7)
bN = np.random.randn(100, 1)
k = data.shape[0]
fig, axes, ims = init_plotting(data, M, N)
for i in range(10000):
for j in range(k):
X0 = data[[j]]
X1 = add_noise(data[[j]])
# compute hidden representation
Y = sigmoid(np.dot(M, X1.T) + bM)
# reconstruct the input
X2 = sigmoid(np.dot(N, Y) + bN).T
# do backpropagation to compute weight
# updates
d1 = 2 * (X0 - X2) * dsigmoid(X2)
dN = alpha * np.dot(d1.T, Y.T)
dbN = alpha * d1.sum(axis=1)[:, None]
d0 = np.dot(d1, N).T * dsigmoid(Y)
dM = alpha * np.dot(d0, X0)
dbM = alpha * d0.sum(axis=1)[:, None]
# update weights
N += dN
bN += dbN
M += dM
bM += dbM
# update plots every 100 steps
if (i % 100) == 0:
draw(fig, data, M, bM, N, bN)
```
|
github_jupyter
|
%matplotlib notebook
import numpy as np
import matplotlib.pyplot as plt
import matplotlib
matplotlib.rc("image", cmap="gray", interpolation="nearest")
f = np.load("patterns.npz")
# extract the labels and data
labels = sorted(f.keys())
data = np.array([f[key].reshape((10, 10)).T.ravel() for key in labels])
# reverse numbers
data[labels.index('num1')] = 1 - data[labels.index('num1')]
data[labels.index('num2')] = 1 - data[labels.index('num2')]
data[labels.index('num3')] = 1 - data[labels.index('num3')]
data[labels.index('num4')] = 1 - data[labels.index('num4')]
# display them
fig, axes = plt.subplots(1, 7)
for j in range(7):
axes[j].imshow(data[j].reshape((10, 10)))
axes[j].set_xticks([])
axes[j].set_yticks([])
fig.set_size_inches(10, 2, forward=True)
plt.tight_layout()
def sigmoid(x):
"""Sigmoid function"""
return 1.0 / (1.0 + np.exp(-x))
def dsigmoid(x):
"""Derivative of the sigmoid function"""
return sigmoid(x) * (1 - sigmoid(x))
def add_noise(X0, p=0.5):
"""Adds random zero-masking noise"""
X1 = X0.copy()
idx = np.random.rand(*X1.shape) < 0.5
X1[idx] = 0
return X1
def init_plotting(data, M, N):
"""Initialize figure and subplots"""
fig, axes = plt.subplots(4, 7)
axes = axes.T
ims = np.empty(axes.shape, dtype=object)
for j in range(k):
ims[j, 0] = axes[j, 0].imshow(data[j].reshape((10, 10)))
ims[j, 1] = axes[j, 1].imshow(M[j].reshape((10, 10)))
ims[j, 2] = axes[j, 2].imshow(N[:, j].reshape((10, 10)))
ims[j, 3] = axes[j, 3].imshow(data[j].reshape((10, 10)))
for ax in axes.flat:
ax.set_xticks([])
ax.set_yticks([])
axes[0, 0].set_ylabel("Corrupted input")
axes[0, 1].set_ylabel("Encoder weights")
axes[0, 2].set_ylabel("Decoder weights")
axes[0, 3].set_ylabel("Decoded image")
fig.set_size_inches(12, 7, forward=True)
plt.tight_layout()
return fig, axes, ims
def draw(fig, data, M, bM, N, bN):
"""Draw the current weights, as well as an example
corrupted input and reconstruction from that input.
"""
k = data.shape[0]
for j in range(k):
X1 = add_noise(data[[j]])
# compute hidden representation
Y = sigmoid(np.dot(M, X1.T) + bM)
# reconstruct the input
X2 = sigmoid(np.dot(N, Y) + bN).T
ims[j, 0].set_data(X1.reshape((10, 10)))
ims[j, 1].set_data(M[j].reshape((10, 10)))
ims[j, 2].set_data(N[:, j].reshape((10, 10)))
ims[j, 3].set_data(X2.reshape((10, 10)))
fig.canvas.draw()
alpha = 0.01
M = np.random.randn(7, 100)
bM = np.random.randn(7, 1)
N = np.random.randn(100, 7)
bN = np.random.randn(100, 1)
k = data.shape[0]
fig, axes, ims = init_plotting(data, M, N)
for i in range(10000):
for j in range(k):
X0 = data[[j]]
X1 = add_noise(data[[j]])
# compute hidden representation
Y = sigmoid(np.dot(M, X1.T) + bM)
# reconstruct the input
X2 = sigmoid(np.dot(N, Y) + bN).T
# do backpropagation to compute weight
# updates
d1 = 2 * (X0 - X2) * dsigmoid(X2)
dN = alpha * np.dot(d1.T, Y.T)
dbN = alpha * d1.sum(axis=1)[:, None]
d0 = np.dot(d1, N).T * dsigmoid(Y)
dM = alpha * np.dot(d0, X0)
dbM = alpha * d0.sum(axis=1)[:, None]
# update weights
N += dN
bN += dbN
M += dM
bM += dbM
# update plots every 100 steps
if (i % 100) == 0:
draw(fig, data, M, bM, N, bN)
| 0.646906 | 0.950134 |
<script async src="https://www.googletagmanager.com/gtag/js?id=UA-59152712-8"></script>
<script>
window.dataLayer = window.dataLayer || [];
function gtag(){dataLayer.push(arguments);}
gtag('js', new Date());
gtag('config', 'UA-59152712-8');
</script>
# Start-to-Finish Example: Unit Testing `GiRaFFE_NRPy`: Interpolating Metric Face-Values
## Author: Patrick Nelson
## This module Validates the `FCVAL` routine for `GiRaFFE`.
**Notebook Status:** <font color='green'><b>Validated</b></font>
**Validation Notes:** This module will validate the routines in [Tutorial-GiRaFFE_NRPy-Metric_Face_Values](Tutorial-GiRaFFE_NRPy-Metric_Face_Values.ipynb).
### NRPy+ Source Code for this module:
* [GiRaFFE_NRPy/GiRaFFE_NRPy_Metric_Face_Values.py](../../edit/in_progress/GiRaFFE_NRPy/GiRaFFE_NRPy_Metric_Face_Values.py) [\[**tutorial**\]](Tutorial-GiRaFFE_NRPy-FCVAL.ipynb) Generates the driver to compute the magnetic field from the vector potential in arbitrary spactimes.
## Introduction:
This notebook validates the code that will interpolate the metric gridfunctions on cell faces. These values, along with the reconstruction of primitive variables on the faces, are necessary for the Riemann solvers to compute the fluxes through the cell faces.
It is, in general, good coding practice to unit test functions individually to verify that they produce the expected and intended output. We will generate test data with arbitrarily-chosen analytic functions and calculate gridfunctions at the cell centers on a small numeric grid. We will then compute the values on the cell faces in two ways: first, with our interpolator, then second, we will shift the grid and compute them analytically. Then, we will rerun the function at a finer resolution. Finally, we will compare the results of the two runs to show third-order convergence.
<a id='toc'></a>
# Table of Contents
$$\label{toc}$$
This notebook is organized as follows
1. [Step 1](#setup): Set up core functions and parameters for unit testing the FCVAL algorithm
1. [Step 1.a](#expressions) Write expressions for the metric gridfunctions
1. [Step 1.b](#ccodekernels) Generate C functions to calculate the gridfunctions
1. [Step 1.c](#free_parameters) Set free parameters in the code
1. [Step 2](#mainc): `FCVAL_unit_test.c`: The Main C Code
1. [Step 2.a](#compile_run): Compile and run the code
1. [Step 3](#convergence): Code validation: Verify that relative error in numerical solution converges to zero at the expected order
1. [Step 4](#latex_pdf_output): Output this notebook to $\LaTeX$-formatted PDF file
<a id='setup'></a>
# Step 1: Set up core functions and parameters for unit testing the FCVAL algorithm \[Back to [top](#toc)\]
$$\label{setup}$$
We'll start by appending the relevant paths to `sys.path` so that we can access sympy modules in other places. Then, we'll import NRPy+ core functionality and set up a directory in which to carry out our test.
```
import shutil, os, sys # Standard Python modules for multiplatform OS-level functions
# First, we'll add the parent directory to the list of directories Python will check for modules.
nrpy_dir_path = os.path.join("..")
if nrpy_dir_path not in sys.path:
sys.path.append(nrpy_dir_path)
nrpy_dir_path = os.path.join("..","..")
if nrpy_dir_path not in sys.path:
sys.path.append(nrpy_dir_path)
from outputC import * # NRPy+: Core C code output module
import finite_difference as fin # NRPy+: Finite difference C code generation module
import NRPy_param_funcs as par # NRPy+: Parameter interface
import grid as gri # NRPy+: Functions having to do with numerical grids
import loop as lp # NRPy+: Generate C code loops
import indexedexp as ixp # NRPy+: Symbolic indexed expression (e.g., tensors, vectors, etc.) support
import reference_metric as rfm # NRPy+: Reference metric support
import cmdline_helper as cmd # NRPy+: Multi-platform Python command-line interface
out_dir = "Validation/"
cmd.mkdir(out_dir)
thismodule = "Start_to_Finish_UnitTest-GiRaFFE_NRPy-Metric_Face_Values"
# Set the finite-differencing order to 2
par.set_parval_from_str("finite_difference::FD_CENTDERIVS_ORDER", 2)
```
<a id='expressions'></a>
## Step 1.a: Write expressions for the metric gridfunctions \[Back to [top](#toc)\]
$$\label{expressions}$$
Now, we'll choose some functions with arbitrary forms to generate test data. We'll need to set ten gridfunctions, so expressions are being pulled from several previously written unit tests.
\begin{align}
\gamma_{xx} &= ax^3 + by^3 + cz^3 + dy^2 + ez^2 + f \\
\gamma_{yy} &= gx^3 + hy^3 + lz^3 + mx^2 + nz^2 + p \\
\gamma_{zz} &= px^3 + qy^3 + rz^3 + sx^2 + ty^2 + u. \\
\gamma_{xy} &= a \exp\left(-\left((x-b)^2+(y-c)^2+(z-d)^2\right)\right) \\
\gamma_{xz} &= f \exp\left(-\left((x-g)^2+(y-h)^2+(z-l)^2\right)\right) \\
\gamma_{yz} &= m \exp\left(-\left((x-n)^2+(y-o)^2+(z-p)^2\right)\right), \\
\beta^x &= \frac{2}{\pi} \arctan(ax + by + cz) \\
\beta^y &= \frac{2}{\pi} \arctan(bx + cy + az) \\
\beta^z &= \frac{2}{\pi} \arctan(cx + ay + bz) \\
\alpha &= 1 - \frac{1}{2+x^2+y^2+z^2} \\
\end{align}
```
a,b,c,d,e,f,g,h,l,m,n,o,p,q,r,s,t,u = par.Cparameters("REAL",thismodule,["a","b","c","d","e","f","g","h","l","m","n","o","p","q","r","s","t","u"],1e300)
M_PI = par.Cparameters("#define",thismodule,["M_PI"], "")
gammaDD = ixp.register_gridfunctions_for_single_rank2("AUXEVOL","gammaDD","sym01",DIM=3)
betaU = ixp.register_gridfunctions_for_single_rank1("AUXEVOL","betaU",DIM=3)
alpha = gri.register_gridfunctions("AUXEVOL","alpha")
par.set_parval_from_str("reference_metric::CoordSystem","Cartesian")
rfm.reference_metric()
x = rfm.xxCart[0]
y = rfm.xxCart[1]
z = rfm.xxCart[2]
gammaDD[0][0] = a*x**3 + b*y**3 + c*z**3 + d*y**2 + e*z**2 + f
gammaDD[1][1] = g*x**3 + h*y**3 + l*z**3 + m*x**2 + n*z**2 + o
gammaDD[2][2] = p*x**3 + q*y**3 + r*z**3 + s*x**2 + t*y**2 + u
gammaDD[0][1] = a * sp.exp(-((x-b)**2 + (y-c)**2 + (z-d)**2))
gammaDD[0][2] = f * sp.exp(-((x-g)**2 + (y-h)**2 + (z-l)**2))
gammaDD[1][2] = m * sp.exp(-((x-n)**2 + (y-o)**2 + (z-p)**2))
betaU[0] = (sp.sympify(2.0)/M_PI) * sp.atan(a*x + b*y + c*z)
betaU[1] = (sp.sympify(2.0)/M_PI) * sp.atan(b*x + c*y + a*z)
betaU[2] = (sp.sympify(2.0)/M_PI) * sp.atan(c*x + a*y + b*z)
alpha = sp.sympify(1.0) - sp.sympify(1.0) / (sp.sympify(2.0) + x**2 + y**2 + z**2)
```
<a id='ccodekernels'></a>
## Step 1.b: Generate C functions to calculate the gridfunctions \[Back to [top](#toc)\]
$$\label{ccodekernels}$$
Here, we will use the NRPy+ function `outCfunction()` to generate C code that will calculate our metric gridfunctions over an entire grid. We will also call the function to generate the function we are testing.
```
metric_gfs_to_print = [\
lhrh(lhs=gri.gfaccess("aux_gfs","gammaDD00"),rhs=gammaDD[0][0]),\
lhrh(lhs=gri.gfaccess("aux_gfs","gammaDD01"),rhs=gammaDD[0][1]),\
lhrh(lhs=gri.gfaccess("aux_gfs","gammaDD02"),rhs=gammaDD[0][2]),\
lhrh(lhs=gri.gfaccess("aux_gfs","gammaDD11"),rhs=gammaDD[1][1]),\
lhrh(lhs=gri.gfaccess("aux_gfs","gammaDD12"),rhs=gammaDD[1][2]),\
lhrh(lhs=gri.gfaccess("aux_gfs","gammaDD22"),rhs=gammaDD[2][2]),\
lhrh(lhs=gri.gfaccess("aux_gfs","betaU0"),rhs=betaU[0]),\
lhrh(lhs=gri.gfaccess("aux_gfs","betaU1"),rhs=betaU[1]),\
lhrh(lhs=gri.gfaccess("aux_gfs","betaU2"),rhs=betaU[2]),\
lhrh(lhs=gri.gfaccess("aux_gfs","alpha"),rhs=alpha),\
]
desc = "Calculate the metric gridfunctions"
name = "calculate_metric_gfs"
outCfunction(
outfile = os.path.join(out_dir,name+".h"), desc=desc, name=name,
params ="const paramstruct *restrict params,REAL *restrict xx[3],REAL *restrict auxevol_gfs",
body = fin.FD_outputC("returnstring",metric_gfs_to_print,params="outCverbose=False").replace("IDX4","IDX4S"),
loopopts="AllPoints,Read_xxs")
import GiRaFFE_NRPy.GiRaFFE_NRPy_Metric_Face_Values as FCVAL
FCVAL.GiRaFFE_NRPy_FCVAL(out_dir)
```
<a id='free_parameters'></a>
## Step 1.c: Set free parameters in the code \[Back to [top](#toc)\]
$$\label{free_parameters}$$
We also need to create the files that interact with NRPy's C parameter interface.
```
# Step 3.d.i: Generate declare_Cparameters_struct.h, set_Cparameters_default.h, and set_Cparameters[-SIMD].h
# par.generate_Cparameters_Ccodes(os.path.join(out_dir))
# Step 3.d.ii: Set free_parameters.h
with open(os.path.join(out_dir,"free_parameters.h"),"w") as file:
file.write("""
// Override parameter defaults with values based on command line arguments and NGHOSTS.
params.Nxx0 = atoi(argv[1]);
params.Nxx1 = atoi(argv[2]);
params.Nxx2 = atoi(argv[3]);
params.Nxx_plus_2NGHOSTS0 = params.Nxx0 + 2*NGHOSTS;
params.Nxx_plus_2NGHOSTS1 = params.Nxx1 + 2*NGHOSTS;
params.Nxx_plus_2NGHOSTS2 = params.Nxx2 + 2*NGHOSTS;
// Step 0d: Set up space and time coordinates
// Step 0d.i: Declare \Delta x^i=dxx{0,1,2} and invdxx{0,1,2}, as well as xxmin[3] and xxmax[3]:
const REAL xxmin[3] = {-1.0,-1.0,-1.0};
const REAL xxmax[3] = { 1.0, 1.0, 1.0};
params.dxx0 = (xxmax[0] - xxmin[0]) / ((REAL)params.Nxx0);
params.dxx1 = (xxmax[1] - xxmin[1]) / ((REAL)params.Nxx1);
params.dxx2 = (xxmax[2] - xxmin[2]) / ((REAL)params.Nxx2);
printf("dxx0,dxx1,dxx2 = %.5e,%.5e,%.5e\\n",params.dxx0,params.dxx1,params.dxx2);
params.invdx0 = 1.0 / params.dxx0;
params.invdx1 = 1.0 / params.dxx1;
params.invdx2 = 1.0 / params.dxx2;
\n""")
# Generates declare_Cparameters_struct.h, set_Cparameters_default.h, and set_Cparameters[-SIMD].h
par.generate_Cparameters_Ccodes(os.path.join(out_dir))
```
<a id='mainc'></a>
# Step 2: `FCVAL_unit_test.c`: The Main C Code \[Back to [top](#toc)\]
$$\label{mainc}$$
```
%%writefile $out_dir/FCVAL_unit_test.c
// These are common packages that we are likely to need.
#include "stdio.h"
#include "stdlib.h"
#include "math.h"
#include "string.h" // Needed for strncmp, etc.
#include "stdint.h" // Needed for Windows GCC 6.x compatibility
#include <time.h> // Needed to set a random seed.
#define REAL double
#include "declare_Cparameters_struct.h"
const int NGHOSTS = 3;
REAL a,b,c,d,e,f,g,h,l,m,n,o,p,q,r,s,t,u;
// Standard NRPy+ memory access:
#define IDX4S(g,i,j,k) \
( (i) + Nxx_plus_2NGHOSTS0 * ( (j) + Nxx_plus_2NGHOSTS1 * ( (k) + Nxx_plus_2NGHOSTS2 * (g) ) ) )
// Give gridfunctions their names:
#define GAMMADD00GF 0
#define GAMMADD01GF 1
#define GAMMADD02GF 2
#define GAMMADD11GF 3
#define GAMMADD12GF 4
#define GAMMADD22GF 5
#define BETAU0GF 6
#define BETAU1GF 7
#define BETAU2GF 8
#define ALPHAGF 9
#define GAMMA_FACEDD00GF 10
#define GAMMA_FACEDD01GF 11
#define GAMMA_FACEDD02GF 12
#define GAMMA_FACEDD11GF 13
#define GAMMA_FACEDD12GF 14
#define GAMMA_FACEDD22GF 15
#define BETA_FACEU0GF 16
#define BETA_FACEU1GF 17
#define BETA_FACEU2GF 18
#define ALPHA_FACEGF 19
#define NUM_AUXEVOL_GFS 20
#include "calculate_metric_gfs.h"
#include "interpolate_metric_gfs_to_cell_faces.h"
int main(int argc, const char *argv[]) {
paramstruct params;
#include "set_Cparameters_default.h"
// Step 0c: Set free parameters, overwriting Cparameters defaults
// by hand or with command-line input, as desired.
#include "free_parameters.h"
#include "set_Cparameters-nopointer.h"
// Step 0e: Set up cell-centered Cartesian coordinate grids
REAL *xx[3];
xx[0] = (REAL *)malloc(sizeof(REAL)*Nxx_plus_2NGHOSTS0);
xx[1] = (REAL *)malloc(sizeof(REAL)*Nxx_plus_2NGHOSTS1);
xx[2] = (REAL *)malloc(sizeof(REAL)*Nxx_plus_2NGHOSTS2);
for(int j=0;j<Nxx_plus_2NGHOSTS0;j++) xx[0][j] = xxmin[0] + (j-NGHOSTS)*dxx0;
for(int j=0;j<Nxx_plus_2NGHOSTS1;j++) xx[1][j] = xxmin[1] + (j-NGHOSTS)*dxx1;
for(int j=0;j<Nxx_plus_2NGHOSTS2;j++) xx[2][j] = xxmin[2] + (j-NGHOSTS)*dxx2;
//for(int i=0;i<Nxx_plus_2NGHOSTS0;i++) printf("xx[0][%d] = %.15e\n",i,xx[0][i]);
// This is the array to which we'll write the NRPy+ variables.
REAL *auxevol_gfs = (REAL *)malloc(sizeof(REAL) * NUM_AUXEVOL_GFS * Nxx_plus_2NGHOSTS2 * Nxx_plus_2NGHOSTS1 * Nxx_plus_2NGHOSTS0);
// And another for exact data:
REAL *auxevol_exact_gfs = (REAL *)malloc(sizeof(REAL) * NUM_AUXEVOL_GFS * Nxx_plus_2NGHOSTS2 * Nxx_plus_2NGHOSTS1 * Nxx_plus_2NGHOSTS0);
// Generate some random coefficients. Leave the random seed on its default for consistency between trials.
a = (double)(rand()%20)/5.0;
f = (double)(rand()%20)/5.0;
m = (double)(rand()%20)/5.0;
b = (double)(rand()%10-5)/100.0;
c = (double)(rand()%10-5)/100.0;
d = (double)(rand()%10-5)/100.0;
g = (double)(rand()%10-5)/100.0;
h = (double)(rand()%10-5)/100.0;
l = (double)(rand()%10-5)/100.0;
n = (double)(rand()%10-5)/100.0;
o = (double)(rand()%10-5)/100.0;
p = (double)(rand()%10-5)/100.0;
// First, calculate the test data on our grid:
calculate_metric_gfs(¶ms,xx,auxevol_gfs);
// Now, run our function:
int flux_dirn = 0;
interpolate_metric_gfs_to_cell_faces(¶ms,auxevol_gfs,flux_dirn);
// For comparison, shift the grid half a gridpoint, then calculate the test data again.
REAL dxx[3] = {dxx0,dxx1,dxx2};
REAL Nxxp2NG[3] = {Nxx_plus_2NGHOSTS0,Nxx_plus_2NGHOSTS1,Nxx_plus_2NGHOSTS2};
for(int i=0;i<Nxxp2NG[flux_dirn];i++) {
xx[flux_dirn][i] -= dxx[flux_dirn]/2.0;
}
calculate_metric_gfs(¶ms,xx,auxevol_exact_gfs);
char filename[100];
sprintf(filename,"out%d-numer.txt",Nxx0);
FILE *out2D = fopen(filename, "w");
for(int i2=0;i2<Nxx_plus_2NGHOSTS2;i2++) for(int i1=0;i1<Nxx_plus_2NGHOSTS1;i1++) for(int i0=0;i0<Nxx_plus_2NGHOSTS0;i0++) {
// We print the difference between approximate and exact numbers.
fprintf(out2D,"%.16e\t %e %e %e\n",
auxevol_gfs[IDX4S(BETA_FACEU0GF,i0,i1,i2)]-auxevol_exact_gfs[IDX4S(BETAU0GF,i0,i1,i2)],
xx[0][i0],xx[1][i1],xx[2][i2]
);
}
fclose(out2D);
}
```
<a id='compile_run'></a>
## Step 2.a: Compile and run the code \[Back to [top](#toc)\]
$$\label{compile_run}$$
Now that we have our file, we can compile it and run the executable.
```
import time
print("Now compiling, should take ~2 seconds...\n")
start = time.time()
cmd.C_compile(os.path.join(out_dir,"FCVAL_unit_test.c"), os.path.join(out_dir,"FCVAL_unit_test"))
end = time.time()
print("Finished in "+str(end-start)+" seconds.\n\n")
print("Now running...\n")
start = time.time()
!./Validation/FCVAL_unit_test 10 10 10
# To do a convergence test, we'll also need a second grid with twice the resolution.
!./Validation/FCVAL_unit_test 20 20 20
end = time.time()
print("Finished in "+str(end-start)+" seconds.\n\n")
```
<a id='convergence'></a>
# Step 3: Code validation: Verify that relative error in numerical solution converges to zero at the expected order \[Back to [top](#toc)\]
$$\label{convergence}$$
Here, we import the data at two resolutions and wrote to text files. This data consists of the absolute error of a metric gridfunction at each in the grid. We'll plot a portion of this data along the axis at the lower resolution along with that same data at the higher resolution scaled to demonstrate that this error converges to 0 at the expected rate. Since our algorithm uses a third-order polynomial, we expect fourth-order convergence here.
```
import numpy as np
import matplotlib.pyplot as plt
plt.rcParams.update({'font.size': 12})
Data1 = np.loadtxt("out10-numer.txt")
Data2 = np.loadtxt("out20-numer.txt")
def IDX4(i,j,k,Nxx_plus_2NGHOSTS0,Nxx_plus_2NGHOSTS1,Nxx_plus_2NGHOSTS2):
return (i) + Nxx_plus_2NGHOSTS0 * ( (j) + Nxx_plus_2NGHOSTS1 * ( (k) + Nxx_plus_2NGHOSTS2 * (0) ) )
x1 = np.zeros(10)
a1 = np.zeros(10)
for i in range(10):
x1[i] = Data1[IDX4(i+3,8,8,16,16,16),1]
a1[i] = Data1[IDX4(i+3,8,8,16,16,16),0]
x2 = np.zeros(20)
a2 = np.zeros(20)
for i in range(20):
x2[i] = Data2[IDX4(i+3,13,13,26,26,26),1]
a2[i] = Data2[IDX4(i+3,13,13,26,26,26),0]
plt.figure()
a = plt.plot(x1,a1,'.',label="dx")
b = plt.plot(x2,a2*(2**4),label="dx/2, times (20/10)^4")
plt.legend()
plt.xlabel("x")
plt.ylabel("alpha")
plt.show()
```
<a id='latex_pdf_output'></a>
# Step 4: Output this notebook to $\LaTeX$-formatted PDF file \[Back to [top](#toc)\]
$$\label{latex_pdf_output}$$
The following code cell converts this Jupyter notebook into a proper, clickable $\LaTeX$-formatted PDF file. After the cell is successfully run, the generated PDF may be found in the root NRPy+ tutorial directory, with filename
[Tutorial-Start_to_Finish-GiRaFFE_NRPy-Metric_Face_Values.pdf](Tutorial-Start_to_Finish-GiRaFFE_NRPy-Metric_Face_Values.pdf) (Note that clicking on this link may not work; you may need to open the PDF file through another means.)
```
!jupyter nbconvert --to latex --template latex_nrpy_style.tplx --log-level='WARN' Tutorial-Start_to_Finish-GiRaFFE_NRPy-Metric_Face_Values.ipynb
!pdflatex -interaction=batchmode Tutorial-Start_to_Finish-GiRaFFE_NRPy-Metric_Face_Values.tex
!pdflatex -interaction=batchmode Tutorial-Start_to_Finish-GiRaFFE_NRPy-Metric_Face_Values.tex
!pdflatex -interaction=batchmode Tutorial-Start_to_Finish-GiRaFFE_NRPy-Metric_Face_Values.tex
!rm -f Tut*.out Tut*.aux Tut*.log
```
|
github_jupyter
|
import shutil, os, sys # Standard Python modules for multiplatform OS-level functions
# First, we'll add the parent directory to the list of directories Python will check for modules.
nrpy_dir_path = os.path.join("..")
if nrpy_dir_path not in sys.path:
sys.path.append(nrpy_dir_path)
nrpy_dir_path = os.path.join("..","..")
if nrpy_dir_path not in sys.path:
sys.path.append(nrpy_dir_path)
from outputC import * # NRPy+: Core C code output module
import finite_difference as fin # NRPy+: Finite difference C code generation module
import NRPy_param_funcs as par # NRPy+: Parameter interface
import grid as gri # NRPy+: Functions having to do with numerical grids
import loop as lp # NRPy+: Generate C code loops
import indexedexp as ixp # NRPy+: Symbolic indexed expression (e.g., tensors, vectors, etc.) support
import reference_metric as rfm # NRPy+: Reference metric support
import cmdline_helper as cmd # NRPy+: Multi-platform Python command-line interface
out_dir = "Validation/"
cmd.mkdir(out_dir)
thismodule = "Start_to_Finish_UnitTest-GiRaFFE_NRPy-Metric_Face_Values"
# Set the finite-differencing order to 2
par.set_parval_from_str("finite_difference::FD_CENTDERIVS_ORDER", 2)
a,b,c,d,e,f,g,h,l,m,n,o,p,q,r,s,t,u = par.Cparameters("REAL",thismodule,["a","b","c","d","e","f","g","h","l","m","n","o","p","q","r","s","t","u"],1e300)
M_PI = par.Cparameters("#define",thismodule,["M_PI"], "")
gammaDD = ixp.register_gridfunctions_for_single_rank2("AUXEVOL","gammaDD","sym01",DIM=3)
betaU = ixp.register_gridfunctions_for_single_rank1("AUXEVOL","betaU",DIM=3)
alpha = gri.register_gridfunctions("AUXEVOL","alpha")
par.set_parval_from_str("reference_metric::CoordSystem","Cartesian")
rfm.reference_metric()
x = rfm.xxCart[0]
y = rfm.xxCart[1]
z = rfm.xxCart[2]
gammaDD[0][0] = a*x**3 + b*y**3 + c*z**3 + d*y**2 + e*z**2 + f
gammaDD[1][1] = g*x**3 + h*y**3 + l*z**3 + m*x**2 + n*z**2 + o
gammaDD[2][2] = p*x**3 + q*y**3 + r*z**3 + s*x**2 + t*y**2 + u
gammaDD[0][1] = a * sp.exp(-((x-b)**2 + (y-c)**2 + (z-d)**2))
gammaDD[0][2] = f * sp.exp(-((x-g)**2 + (y-h)**2 + (z-l)**2))
gammaDD[1][2] = m * sp.exp(-((x-n)**2 + (y-o)**2 + (z-p)**2))
betaU[0] = (sp.sympify(2.0)/M_PI) * sp.atan(a*x + b*y + c*z)
betaU[1] = (sp.sympify(2.0)/M_PI) * sp.atan(b*x + c*y + a*z)
betaU[2] = (sp.sympify(2.0)/M_PI) * sp.atan(c*x + a*y + b*z)
alpha = sp.sympify(1.0) - sp.sympify(1.0) / (sp.sympify(2.0) + x**2 + y**2 + z**2)
metric_gfs_to_print = [\
lhrh(lhs=gri.gfaccess("aux_gfs","gammaDD00"),rhs=gammaDD[0][0]),\
lhrh(lhs=gri.gfaccess("aux_gfs","gammaDD01"),rhs=gammaDD[0][1]),\
lhrh(lhs=gri.gfaccess("aux_gfs","gammaDD02"),rhs=gammaDD[0][2]),\
lhrh(lhs=gri.gfaccess("aux_gfs","gammaDD11"),rhs=gammaDD[1][1]),\
lhrh(lhs=gri.gfaccess("aux_gfs","gammaDD12"),rhs=gammaDD[1][2]),\
lhrh(lhs=gri.gfaccess("aux_gfs","gammaDD22"),rhs=gammaDD[2][2]),\
lhrh(lhs=gri.gfaccess("aux_gfs","betaU0"),rhs=betaU[0]),\
lhrh(lhs=gri.gfaccess("aux_gfs","betaU1"),rhs=betaU[1]),\
lhrh(lhs=gri.gfaccess("aux_gfs","betaU2"),rhs=betaU[2]),\
lhrh(lhs=gri.gfaccess("aux_gfs","alpha"),rhs=alpha),\
]
desc = "Calculate the metric gridfunctions"
name = "calculate_metric_gfs"
outCfunction(
outfile = os.path.join(out_dir,name+".h"), desc=desc, name=name,
params ="const paramstruct *restrict params,REAL *restrict xx[3],REAL *restrict auxevol_gfs",
body = fin.FD_outputC("returnstring",metric_gfs_to_print,params="outCverbose=False").replace("IDX4","IDX4S"),
loopopts="AllPoints,Read_xxs")
import GiRaFFE_NRPy.GiRaFFE_NRPy_Metric_Face_Values as FCVAL
FCVAL.GiRaFFE_NRPy_FCVAL(out_dir)
# Step 3.d.i: Generate declare_Cparameters_struct.h, set_Cparameters_default.h, and set_Cparameters[-SIMD].h
# par.generate_Cparameters_Ccodes(os.path.join(out_dir))
# Step 3.d.ii: Set free_parameters.h
with open(os.path.join(out_dir,"free_parameters.h"),"w") as file:
file.write("""
// Override parameter defaults with values based on command line arguments and NGHOSTS.
params.Nxx0 = atoi(argv[1]);
params.Nxx1 = atoi(argv[2]);
params.Nxx2 = atoi(argv[3]);
params.Nxx_plus_2NGHOSTS0 = params.Nxx0 + 2*NGHOSTS;
params.Nxx_plus_2NGHOSTS1 = params.Nxx1 + 2*NGHOSTS;
params.Nxx_plus_2NGHOSTS2 = params.Nxx2 + 2*NGHOSTS;
// Step 0d: Set up space and time coordinates
// Step 0d.i: Declare \Delta x^i=dxx{0,1,2} and invdxx{0,1,2}, as well as xxmin[3] and xxmax[3]:
const REAL xxmin[3] = {-1.0,-1.0,-1.0};
const REAL xxmax[3] = { 1.0, 1.0, 1.0};
params.dxx0 = (xxmax[0] - xxmin[0]) / ((REAL)params.Nxx0);
params.dxx1 = (xxmax[1] - xxmin[1]) / ((REAL)params.Nxx1);
params.dxx2 = (xxmax[2] - xxmin[2]) / ((REAL)params.Nxx2);
printf("dxx0,dxx1,dxx2 = %.5e,%.5e,%.5e\\n",params.dxx0,params.dxx1,params.dxx2);
params.invdx0 = 1.0 / params.dxx0;
params.invdx1 = 1.0 / params.dxx1;
params.invdx2 = 1.0 / params.dxx2;
\n""")
# Generates declare_Cparameters_struct.h, set_Cparameters_default.h, and set_Cparameters[-SIMD].h
par.generate_Cparameters_Ccodes(os.path.join(out_dir))
%%writefile $out_dir/FCVAL_unit_test.c
// These are common packages that we are likely to need.
#include "stdio.h"
#include "stdlib.h"
#include "math.h"
#include "string.h" // Needed for strncmp, etc.
#include "stdint.h" // Needed for Windows GCC 6.x compatibility
#include <time.h> // Needed to set a random seed.
#define REAL double
#include "declare_Cparameters_struct.h"
const int NGHOSTS = 3;
REAL a,b,c,d,e,f,g,h,l,m,n,o,p,q,r,s,t,u;
// Standard NRPy+ memory access:
#define IDX4S(g,i,j,k) \
( (i) + Nxx_plus_2NGHOSTS0 * ( (j) + Nxx_plus_2NGHOSTS1 * ( (k) + Nxx_plus_2NGHOSTS2 * (g) ) ) )
// Give gridfunctions their names:
#define GAMMADD00GF 0
#define GAMMADD01GF 1
#define GAMMADD02GF 2
#define GAMMADD11GF 3
#define GAMMADD12GF 4
#define GAMMADD22GF 5
#define BETAU0GF 6
#define BETAU1GF 7
#define BETAU2GF 8
#define ALPHAGF 9
#define GAMMA_FACEDD00GF 10
#define GAMMA_FACEDD01GF 11
#define GAMMA_FACEDD02GF 12
#define GAMMA_FACEDD11GF 13
#define GAMMA_FACEDD12GF 14
#define GAMMA_FACEDD22GF 15
#define BETA_FACEU0GF 16
#define BETA_FACEU1GF 17
#define BETA_FACEU2GF 18
#define ALPHA_FACEGF 19
#define NUM_AUXEVOL_GFS 20
#include "calculate_metric_gfs.h"
#include "interpolate_metric_gfs_to_cell_faces.h"
int main(int argc, const char *argv[]) {
paramstruct params;
#include "set_Cparameters_default.h"
// Step 0c: Set free parameters, overwriting Cparameters defaults
// by hand or with command-line input, as desired.
#include "free_parameters.h"
#include "set_Cparameters-nopointer.h"
// Step 0e: Set up cell-centered Cartesian coordinate grids
REAL *xx[3];
xx[0] = (REAL *)malloc(sizeof(REAL)*Nxx_plus_2NGHOSTS0);
xx[1] = (REAL *)malloc(sizeof(REAL)*Nxx_plus_2NGHOSTS1);
xx[2] = (REAL *)malloc(sizeof(REAL)*Nxx_plus_2NGHOSTS2);
for(int j=0;j<Nxx_plus_2NGHOSTS0;j++) xx[0][j] = xxmin[0] + (j-NGHOSTS)*dxx0;
for(int j=0;j<Nxx_plus_2NGHOSTS1;j++) xx[1][j] = xxmin[1] + (j-NGHOSTS)*dxx1;
for(int j=0;j<Nxx_plus_2NGHOSTS2;j++) xx[2][j] = xxmin[2] + (j-NGHOSTS)*dxx2;
//for(int i=0;i<Nxx_plus_2NGHOSTS0;i++) printf("xx[0][%d] = %.15e\n",i,xx[0][i]);
// This is the array to which we'll write the NRPy+ variables.
REAL *auxevol_gfs = (REAL *)malloc(sizeof(REAL) * NUM_AUXEVOL_GFS * Nxx_plus_2NGHOSTS2 * Nxx_plus_2NGHOSTS1 * Nxx_plus_2NGHOSTS0);
// And another for exact data:
REAL *auxevol_exact_gfs = (REAL *)malloc(sizeof(REAL) * NUM_AUXEVOL_GFS * Nxx_plus_2NGHOSTS2 * Nxx_plus_2NGHOSTS1 * Nxx_plus_2NGHOSTS0);
// Generate some random coefficients. Leave the random seed on its default for consistency between trials.
a = (double)(rand()%20)/5.0;
f = (double)(rand()%20)/5.0;
m = (double)(rand()%20)/5.0;
b = (double)(rand()%10-5)/100.0;
c = (double)(rand()%10-5)/100.0;
d = (double)(rand()%10-5)/100.0;
g = (double)(rand()%10-5)/100.0;
h = (double)(rand()%10-5)/100.0;
l = (double)(rand()%10-5)/100.0;
n = (double)(rand()%10-5)/100.0;
o = (double)(rand()%10-5)/100.0;
p = (double)(rand()%10-5)/100.0;
// First, calculate the test data on our grid:
calculate_metric_gfs(¶ms,xx,auxevol_gfs);
// Now, run our function:
int flux_dirn = 0;
interpolate_metric_gfs_to_cell_faces(¶ms,auxevol_gfs,flux_dirn);
// For comparison, shift the grid half a gridpoint, then calculate the test data again.
REAL dxx[3] = {dxx0,dxx1,dxx2};
REAL Nxxp2NG[3] = {Nxx_plus_2NGHOSTS0,Nxx_plus_2NGHOSTS1,Nxx_plus_2NGHOSTS2};
for(int i=0;i<Nxxp2NG[flux_dirn];i++) {
xx[flux_dirn][i] -= dxx[flux_dirn]/2.0;
}
calculate_metric_gfs(¶ms,xx,auxevol_exact_gfs);
char filename[100];
sprintf(filename,"out%d-numer.txt",Nxx0);
FILE *out2D = fopen(filename, "w");
for(int i2=0;i2<Nxx_plus_2NGHOSTS2;i2++) for(int i1=0;i1<Nxx_plus_2NGHOSTS1;i1++) for(int i0=0;i0<Nxx_plus_2NGHOSTS0;i0++) {
// We print the difference between approximate and exact numbers.
fprintf(out2D,"%.16e\t %e %e %e\n",
auxevol_gfs[IDX4S(BETA_FACEU0GF,i0,i1,i2)]-auxevol_exact_gfs[IDX4S(BETAU0GF,i0,i1,i2)],
xx[0][i0],xx[1][i1],xx[2][i2]
);
}
fclose(out2D);
}
import time
print("Now compiling, should take ~2 seconds...\n")
start = time.time()
cmd.C_compile(os.path.join(out_dir,"FCVAL_unit_test.c"), os.path.join(out_dir,"FCVAL_unit_test"))
end = time.time()
print("Finished in "+str(end-start)+" seconds.\n\n")
print("Now running...\n")
start = time.time()
!./Validation/FCVAL_unit_test 10 10 10
# To do a convergence test, we'll also need a second grid with twice the resolution.
!./Validation/FCVAL_unit_test 20 20 20
end = time.time()
print("Finished in "+str(end-start)+" seconds.\n\n")
import numpy as np
import matplotlib.pyplot as plt
plt.rcParams.update({'font.size': 12})
Data1 = np.loadtxt("out10-numer.txt")
Data2 = np.loadtxt("out20-numer.txt")
def IDX4(i,j,k,Nxx_plus_2NGHOSTS0,Nxx_plus_2NGHOSTS1,Nxx_plus_2NGHOSTS2):
return (i) + Nxx_plus_2NGHOSTS0 * ( (j) + Nxx_plus_2NGHOSTS1 * ( (k) + Nxx_plus_2NGHOSTS2 * (0) ) )
x1 = np.zeros(10)
a1 = np.zeros(10)
for i in range(10):
x1[i] = Data1[IDX4(i+3,8,8,16,16,16),1]
a1[i] = Data1[IDX4(i+3,8,8,16,16,16),0]
x2 = np.zeros(20)
a2 = np.zeros(20)
for i in range(20):
x2[i] = Data2[IDX4(i+3,13,13,26,26,26),1]
a2[i] = Data2[IDX4(i+3,13,13,26,26,26),0]
plt.figure()
a = plt.plot(x1,a1,'.',label="dx")
b = plt.plot(x2,a2*(2**4),label="dx/2, times (20/10)^4")
plt.legend()
plt.xlabel("x")
plt.ylabel("alpha")
plt.show()
!jupyter nbconvert --to latex --template latex_nrpy_style.tplx --log-level='WARN' Tutorial-Start_to_Finish-GiRaFFE_NRPy-Metric_Face_Values.ipynb
!pdflatex -interaction=batchmode Tutorial-Start_to_Finish-GiRaFFE_NRPy-Metric_Face_Values.tex
!pdflatex -interaction=batchmode Tutorial-Start_to_Finish-GiRaFFE_NRPy-Metric_Face_Values.tex
!pdflatex -interaction=batchmode Tutorial-Start_to_Finish-GiRaFFE_NRPy-Metric_Face_Values.tex
!rm -f Tut*.out Tut*.aux Tut*.log
| 0.312895 | 0.941007 |
```
import pandas as pd # dataset handling
import numpy as np
import geopandas as gpd # geodataset handling
from keplergl import KeplerGl # geospatial visualization
import datetime
import json
import itertools
from prophet import Prophet
from prophet.serialize import model_from_json, model_to_json
from sklearn.metrics import mean_squared_error
import warnings
warnings.filterwarnings("ignore")
def create_regressor_column(ds, train_col, test_col, location_id):
"""Get a regressor of train or test data for corresponding timestamp
Args:
ds (datetime): timestamp
train_col (string): column name of regressor in train data
test_col (string): column name of regressor in test data
Returns:
float: regressor value for given timestamp
"""
if ds in df_prophet_reg['ds'].values:
return df_prophet_reg[df_prophet_reg['ds'] == ds][train_col].values[0]
elif ds in weather['Frankfurt']['timestamp'].values:
if location_id <= 124:
return weather['Frankfurt'][(weather['Frankfurt']['timestamp'] == ds)][test_col].values[0]
else:
return weather['Bremen'][(weather['Bremen']['timestamp'] == ds)][test_col].values[0]
else:
return np.nan
def forecast_dict_to_df(forecast: dict) -> pd.DataFrame:
"""Convert forecast dictionary to dataframe
Args:
forecast (dict): forecast dictionary from all locations' prophet models
Returns:
pd.DataFrame: DataFrame with all forecasts
"""
forecasts = pd.DataFrame(columns=['ds', 'yhat', 'location_id'])
for location in list(forecast.keys()):
forecast[location]['location_id'] = location
forecasts = pd.concat([forecasts, forecast[location]], axis=0)
return forecasts
def rmse_per_location(df_rmse: pd.DataFrame, forecast_horizon=168):
"""Calculate RMSE per location
Args:
df_rmse (pd.DataFrame): dataframe with y_true and y_hat values, timestamp and location_id
forecast_horizon (int): forecast horizon (in hours)
Returns:
pd.DataFrame: dataframe with RMSE train and test values per location
"""
locs = df_rmse.location_id.unique().tolist()
rmse = pd.DataFrame(columns=['RMSE_train', 'RMSE_test', 'PM2p5_mean'], index=locs)
for loc in locs:
df_loc = df_rmse[df_rmse.location_id == loc]
df_loc.dropna(inplace=True)
rmse.loc[loc, 'RMSE_train'] = mean_squared_error(df_loc['PM2p5'][:-forecast_horizon], df_loc['yhat'][:-forecast_horizon], squared=False)
rmse.loc[loc, 'RMSE_test'] = mean_squared_error(df_loc['PM2p5'][-forecast_horizon:], df_loc['yhat'][-forecast_horizon:], squared= False)
rmse.loc[loc, 'PM2p5_mean'] = df_loc['PM2p5'][:-forecast_horizon].mean()
rmse.reset_index(inplace=True)
rmse.rename(columns={'index': 'location_id'}, inplace=True)
return rmse
```
## Importing latest Sensor and Meteomatics data
```
# load meteomatics data for the cities of interest
cities = ['Bremen', 'Frankfurt']
# set the day of the downloaded data
day = datetime.datetime(2022, 3, 20).date()
weather = {}
for city in cities:
weather[city] = pd.read_csv(f'../data/Meteomatics/auto_processed_weather_forecast_{city}_{day}.csv')
weather[city].timestamp = pd.to_datetime(weather[city].timestamp)
df = pd.read_csv(f'../data/cleaned_sensors_dwd_2022-03-28.csv', index_col=0)
df.timestamp = pd.to_datetime(df.timestamp)
df_test_eval = df[(df.timestamp >= weather['Bremen'].timestamp.min()) & (df.timestamp <= weather['Bremen'].timestamp.min() + datetime.timedelta(days=7))][['location_id', 'timestamp', 'PM2p5']]
```
## Train Prophet on all available data
```
# set locations
locations = list(df.location_id.unique())
# initialize model
seasonality_mode='additive'
yearly_seasonality=True
weekly_seasonality=True
daily_seasonality=True
growth='logistic' # 'logistic'
# n_changepoints=25
# changepoint_prior_scale = 0.6 #default 0.05
params = {'changepoint_range': [0.8],
'changepoint_prior_scale': [0.3], #default 0.05
#'prior_scale': [0.25, 0.3, 0.35]
'seasonality_prior_scale': [10.0]
}
params_grid = [dict(zip(params.keys(), v)) for v in itertools.product(*params.values())]
temp_flag = True
press_flag = True
windsp_flag = True
winddir_flag = True
precip_flag = True
temp_prior_scale = 0.3
press_prior_scale = 0.3
windsp_prior_scale = 0.3
winddir_prior_scale = 0.3
precip_prior_scale = 0.3
if not temp_flag:
temp_prior_scale = None
if not press_flag:
press_prior_scale = None
if not windsp_flag:
windsp_prior_scale = None
if not winddir_flag:
winddir_prior_scale = None
if not precip_flag:
precip_prior_scale = None
params_grid
rmse_per_run = {}
runnumber = 0
runs = {}
for para in params_grid:
runnumber += 1
models = {}
limits = pd.DataFrame(index=locations, columns=['cap', 'floor'])
forecasts = {}
for location in locations:
# reduce data to one location
df_prophet_reg = df.query(f'location_id == {location}')[['location_id', 'timestamp','PM2p5', 'temperature', 'pressure', 'wind_speed', 'wind_direction', 'precip']]
df_prophet_reg = df_prophet_reg.sort_values(['timestamp'], axis=0)
# rename columns to expected format for prophet
df_prophet_reg.rename(columns={'timestamp': 'ds', 'PM2p5': 'y', 'temperature': 'temp', 'pressure': 'press', 'wind_speed': 'windsp', 'wind_direction': 'winddir', 'precip': 'precip'}, inplace=True)
# prophet can not handle nans in dataframe
# df_prophet_reg_all = df_prophet_reg.copy()
# Comment in if using Meteomatics data, for DWD comment out
df_prophet_reg = df_prophet_reg[df_prophet_reg.ds < weather['Bremen'].timestamp.min()]
df_train = df_prophet_reg[df_prophet_reg.ds < weather['Bremen'].timestamp.min()]
df_train.dropna(inplace=True, subset=['temp', 'press', 'windsp', 'precip', 'winddir'])
df_average = df_train.query(f'location_id == {location}')[['ds','y']]
df_average['PM2p5_average'] = (df_average.y.shift(2) + df_average.y.shift(1) + df_average.y + df_average.y.shift(-1) + df_average.y.shift(-2)) / 5
# add cap column for to set growth = logistic
cap = df_average.PM2p5_average.quantile(0.99)
floor = df_average.PM2p5_average.min()
df_train['cap'] = cap
df_train['floor'] = floor
df_train.drop('location_id', inplace=True, axis=1)
model = Prophet(seasonality_mode=seasonality_mode, yearly_seasonality=yearly_seasonality, weekly_seasonality=weekly_seasonality, daily_seasonality=daily_seasonality,
growth=growth, **para)
# add regressors
model.add_regressor('temp', standardize=True, prior_scale=temp_prior_scale)
model.add_regressor('press', standardize=True, prior_scale=press_prior_scale)
model.add_regressor('windsp', standardize=True, prior_scale=windsp_prior_scale)
model.add_regressor('winddir', standardize=True, prior_scale=winddir_prior_scale)
model.add_regressor('precip', standardize=True, prior_scale=precip_prior_scale)
# fit model
model.fit(df_train)
models[location] = model
future = model.make_future_dataframe(periods=168, freq='H')
# add regressors
future['temp'] = future['ds'].apply(create_regressor_column, args=('temp', 'temperature', location))
future['press'] = future['ds'].apply(create_regressor_column, args=('press', 'pressure', location))
future['windsp'] = future['ds'].apply(create_regressor_column, args=('windsp', 'wind_speed', location))
future['winddir'] = future['ds'].apply(create_regressor_column, args=('winddir', 'wind_direction', location))
future['precip'] = future['ds'].apply(create_regressor_column, args=('precip', 'precip', location))
future['cap'] = cap
future['floor'] = floor
# drop nans
future.dropna(inplace=True)
# predict
forecasts[location] = model.predict(future)
# combine forecasts into one dataframe
df_forecasts = forecast_dict_to_df(forecasts)
df_eval = df[(df.timestamp >= df_forecasts.ds.min()) & (df.timestamp <= df_forecasts.ds.max())][['location_id', 'timestamp', 'PM2p5']]
df_eval.rename(columns={'timestamp': 'ds'}, inplace=True)
df_rmse = df_forecasts[['location_id','ds', 'yhat']].merge(df_eval, on=['location_id', 'ds'], how='left')
runs[runnumber] = list(para.values())
rmse_per_run[runnumber] = rmse_per_location(df_rmse)
# save model and forecasts
for location_id in locations:
model_json = model_to_json(models[location_id])
with open(f'../models/final_model_meteo_{location_id}.json', 'w') as f:
json.dump(model_json, f)
forecasts[location_id].to_csv(f'../models/final_forecast_meteo_{location_id}.csv')
```
## RMSE plots
```
import seaborn as sns
import matplotlib.pyplot as plt
i=0
mean_rmses = []
plt.figure(figsize=(20,15))
for frame in list(rmse_per_run.values()):
#display(frame.sort_values(by='RMSE_train', ascending=True).head(10))
p = sns.scatterplot(data = frame, x='PM2p5_mean', y='RMSE_train')
mean_rmses.append(frame[['RMSE_train']].mean())
display(frame.mean())
df_dwd_rmses = frame.copy()
df_meteomatics_rmses
```
# Fill future dataframe with meteomatics data
## Visualization with Kepler.gl
```
# get GPS data and merge with forecast dataframe since kepler.gl needs GPS data
df_gps = df[['location_id', 'lat', 'lon']].groupby(['location_id']).first().reset_index()
df_kepler = df_forecasts.merge(df_gps, how='left', on='location_id')[['ds', 'yhat', 'location_id', 'lat', 'lon']]
df_kepler.dropna(subset=['yhat'], inplace=True)
# make dummies
timestamps = pd.Series(df_kepler['ds'].unique(), name='ds')
dummies = pd.DataFrame(data={
'location_id': -1,
'lat': [0, 90],
'lon': [0, 90],
'yhat': [0, 50]
})
dummies = dummies.merge(timestamps, how='cross')
df_kepler = pd.concat([df_kepler, dummies])
# Adjust plotting time horizon
df_kepler = df_kepler.sort_values('ds').query('ds > "2022-03"')
# Create bins for PM2p5 for Hexbin plotting
pm2p5_bins = np.append(0,np.arange(0, 50, 5))
pm2p5_labels = pm2p5_bins
pm2p5_labels[0] = -1
pm2p5_bins = np.append(pm2p5_bins, 1000)
pm2p5_bins[0] = -20
print(pm2p5_bins)
df_kepler['PM2p5_bins'] = pd.cut(df_kepler['yhat'], bins=pm2p5_bins, labels=pm2p5_labels).astype(int)
# Make the geo DataFrame
gdf_sensors = gpd.GeoDataFrame(
df_kepler,
geometry=gpd.points_from_xy(
x=df_kepler['lon'],
y=df_kepler['lat']
)
)
# Creating a Datetime column (Kepler is funny about datetimes)
gdf_sensors['timestamp'] = pd.to_datetime(gdf_sensors['ds'])
gdf_sensors['timestamp'] = gdf_sensors['timestamp'].dt.strftime('%Y-%m-%d %H:%M:%S')
# Selecting only columns we need
gdf_sensors = gdf_sensors[[
'yhat', 'lon', 'lat', 'geometry', 'timestamp', 'PM2p5_bins', 'location_id'
]]
gdf_sensors
gdf_sensors[gdf_sensors.yhat < 0].location_id.unique()
%run config.py
map_config = config
kepler_map = KeplerGl(
height=800,
data={
'Sensors': gdf_sensors,
}, config=map_config
)
kepler_map
```
### Export for Kepler.gl or Unfolded online use
```
df_kepler.to_csv('../data/kepler.csv', index=False)
```
|
github_jupyter
|
import pandas as pd # dataset handling
import numpy as np
import geopandas as gpd # geodataset handling
from keplergl import KeplerGl # geospatial visualization
import datetime
import json
import itertools
from prophet import Prophet
from prophet.serialize import model_from_json, model_to_json
from sklearn.metrics import mean_squared_error
import warnings
warnings.filterwarnings("ignore")
def create_regressor_column(ds, train_col, test_col, location_id):
"""Get a regressor of train or test data for corresponding timestamp
Args:
ds (datetime): timestamp
train_col (string): column name of regressor in train data
test_col (string): column name of regressor in test data
Returns:
float: regressor value for given timestamp
"""
if ds in df_prophet_reg['ds'].values:
return df_prophet_reg[df_prophet_reg['ds'] == ds][train_col].values[0]
elif ds in weather['Frankfurt']['timestamp'].values:
if location_id <= 124:
return weather['Frankfurt'][(weather['Frankfurt']['timestamp'] == ds)][test_col].values[0]
else:
return weather['Bremen'][(weather['Bremen']['timestamp'] == ds)][test_col].values[0]
else:
return np.nan
def forecast_dict_to_df(forecast: dict) -> pd.DataFrame:
"""Convert forecast dictionary to dataframe
Args:
forecast (dict): forecast dictionary from all locations' prophet models
Returns:
pd.DataFrame: DataFrame with all forecasts
"""
forecasts = pd.DataFrame(columns=['ds', 'yhat', 'location_id'])
for location in list(forecast.keys()):
forecast[location]['location_id'] = location
forecasts = pd.concat([forecasts, forecast[location]], axis=0)
return forecasts
def rmse_per_location(df_rmse: pd.DataFrame, forecast_horizon=168):
"""Calculate RMSE per location
Args:
df_rmse (pd.DataFrame): dataframe with y_true and y_hat values, timestamp and location_id
forecast_horizon (int): forecast horizon (in hours)
Returns:
pd.DataFrame: dataframe with RMSE train and test values per location
"""
locs = df_rmse.location_id.unique().tolist()
rmse = pd.DataFrame(columns=['RMSE_train', 'RMSE_test', 'PM2p5_mean'], index=locs)
for loc in locs:
df_loc = df_rmse[df_rmse.location_id == loc]
df_loc.dropna(inplace=True)
rmse.loc[loc, 'RMSE_train'] = mean_squared_error(df_loc['PM2p5'][:-forecast_horizon], df_loc['yhat'][:-forecast_horizon], squared=False)
rmse.loc[loc, 'RMSE_test'] = mean_squared_error(df_loc['PM2p5'][-forecast_horizon:], df_loc['yhat'][-forecast_horizon:], squared= False)
rmse.loc[loc, 'PM2p5_mean'] = df_loc['PM2p5'][:-forecast_horizon].mean()
rmse.reset_index(inplace=True)
rmse.rename(columns={'index': 'location_id'}, inplace=True)
return rmse
# load meteomatics data for the cities of interest
cities = ['Bremen', 'Frankfurt']
# set the day of the downloaded data
day = datetime.datetime(2022, 3, 20).date()
weather = {}
for city in cities:
weather[city] = pd.read_csv(f'../data/Meteomatics/auto_processed_weather_forecast_{city}_{day}.csv')
weather[city].timestamp = pd.to_datetime(weather[city].timestamp)
df = pd.read_csv(f'../data/cleaned_sensors_dwd_2022-03-28.csv', index_col=0)
df.timestamp = pd.to_datetime(df.timestamp)
df_test_eval = df[(df.timestamp >= weather['Bremen'].timestamp.min()) & (df.timestamp <= weather['Bremen'].timestamp.min() + datetime.timedelta(days=7))][['location_id', 'timestamp', 'PM2p5']]
# set locations
locations = list(df.location_id.unique())
# initialize model
seasonality_mode='additive'
yearly_seasonality=True
weekly_seasonality=True
daily_seasonality=True
growth='logistic' # 'logistic'
# n_changepoints=25
# changepoint_prior_scale = 0.6 #default 0.05
params = {'changepoint_range': [0.8],
'changepoint_prior_scale': [0.3], #default 0.05
#'prior_scale': [0.25, 0.3, 0.35]
'seasonality_prior_scale': [10.0]
}
params_grid = [dict(zip(params.keys(), v)) for v in itertools.product(*params.values())]
temp_flag = True
press_flag = True
windsp_flag = True
winddir_flag = True
precip_flag = True
temp_prior_scale = 0.3
press_prior_scale = 0.3
windsp_prior_scale = 0.3
winddir_prior_scale = 0.3
precip_prior_scale = 0.3
if not temp_flag:
temp_prior_scale = None
if not press_flag:
press_prior_scale = None
if not windsp_flag:
windsp_prior_scale = None
if not winddir_flag:
winddir_prior_scale = None
if not precip_flag:
precip_prior_scale = None
params_grid
rmse_per_run = {}
runnumber = 0
runs = {}
for para in params_grid:
runnumber += 1
models = {}
limits = pd.DataFrame(index=locations, columns=['cap', 'floor'])
forecasts = {}
for location in locations:
# reduce data to one location
df_prophet_reg = df.query(f'location_id == {location}')[['location_id', 'timestamp','PM2p5', 'temperature', 'pressure', 'wind_speed', 'wind_direction', 'precip']]
df_prophet_reg = df_prophet_reg.sort_values(['timestamp'], axis=0)
# rename columns to expected format for prophet
df_prophet_reg.rename(columns={'timestamp': 'ds', 'PM2p5': 'y', 'temperature': 'temp', 'pressure': 'press', 'wind_speed': 'windsp', 'wind_direction': 'winddir', 'precip': 'precip'}, inplace=True)
# prophet can not handle nans in dataframe
# df_prophet_reg_all = df_prophet_reg.copy()
# Comment in if using Meteomatics data, for DWD comment out
df_prophet_reg = df_prophet_reg[df_prophet_reg.ds < weather['Bremen'].timestamp.min()]
df_train = df_prophet_reg[df_prophet_reg.ds < weather['Bremen'].timestamp.min()]
df_train.dropna(inplace=True, subset=['temp', 'press', 'windsp', 'precip', 'winddir'])
df_average = df_train.query(f'location_id == {location}')[['ds','y']]
df_average['PM2p5_average'] = (df_average.y.shift(2) + df_average.y.shift(1) + df_average.y + df_average.y.shift(-1) + df_average.y.shift(-2)) / 5
# add cap column for to set growth = logistic
cap = df_average.PM2p5_average.quantile(0.99)
floor = df_average.PM2p5_average.min()
df_train['cap'] = cap
df_train['floor'] = floor
df_train.drop('location_id', inplace=True, axis=1)
model = Prophet(seasonality_mode=seasonality_mode, yearly_seasonality=yearly_seasonality, weekly_seasonality=weekly_seasonality, daily_seasonality=daily_seasonality,
growth=growth, **para)
# add regressors
model.add_regressor('temp', standardize=True, prior_scale=temp_prior_scale)
model.add_regressor('press', standardize=True, prior_scale=press_prior_scale)
model.add_regressor('windsp', standardize=True, prior_scale=windsp_prior_scale)
model.add_regressor('winddir', standardize=True, prior_scale=winddir_prior_scale)
model.add_regressor('precip', standardize=True, prior_scale=precip_prior_scale)
# fit model
model.fit(df_train)
models[location] = model
future = model.make_future_dataframe(periods=168, freq='H')
# add regressors
future['temp'] = future['ds'].apply(create_regressor_column, args=('temp', 'temperature', location))
future['press'] = future['ds'].apply(create_regressor_column, args=('press', 'pressure', location))
future['windsp'] = future['ds'].apply(create_regressor_column, args=('windsp', 'wind_speed', location))
future['winddir'] = future['ds'].apply(create_regressor_column, args=('winddir', 'wind_direction', location))
future['precip'] = future['ds'].apply(create_regressor_column, args=('precip', 'precip', location))
future['cap'] = cap
future['floor'] = floor
# drop nans
future.dropna(inplace=True)
# predict
forecasts[location] = model.predict(future)
# combine forecasts into one dataframe
df_forecasts = forecast_dict_to_df(forecasts)
df_eval = df[(df.timestamp >= df_forecasts.ds.min()) & (df.timestamp <= df_forecasts.ds.max())][['location_id', 'timestamp', 'PM2p5']]
df_eval.rename(columns={'timestamp': 'ds'}, inplace=True)
df_rmse = df_forecasts[['location_id','ds', 'yhat']].merge(df_eval, on=['location_id', 'ds'], how='left')
runs[runnumber] = list(para.values())
rmse_per_run[runnumber] = rmse_per_location(df_rmse)
# save model and forecasts
for location_id in locations:
model_json = model_to_json(models[location_id])
with open(f'../models/final_model_meteo_{location_id}.json', 'w') as f:
json.dump(model_json, f)
forecasts[location_id].to_csv(f'../models/final_forecast_meteo_{location_id}.csv')
import seaborn as sns
import matplotlib.pyplot as plt
i=0
mean_rmses = []
plt.figure(figsize=(20,15))
for frame in list(rmse_per_run.values()):
#display(frame.sort_values(by='RMSE_train', ascending=True).head(10))
p = sns.scatterplot(data = frame, x='PM2p5_mean', y='RMSE_train')
mean_rmses.append(frame[['RMSE_train']].mean())
display(frame.mean())
df_dwd_rmses = frame.copy()
df_meteomatics_rmses
# get GPS data and merge with forecast dataframe since kepler.gl needs GPS data
df_gps = df[['location_id', 'lat', 'lon']].groupby(['location_id']).first().reset_index()
df_kepler = df_forecasts.merge(df_gps, how='left', on='location_id')[['ds', 'yhat', 'location_id', 'lat', 'lon']]
df_kepler.dropna(subset=['yhat'], inplace=True)
# make dummies
timestamps = pd.Series(df_kepler['ds'].unique(), name='ds')
dummies = pd.DataFrame(data={
'location_id': -1,
'lat': [0, 90],
'lon': [0, 90],
'yhat': [0, 50]
})
dummies = dummies.merge(timestamps, how='cross')
df_kepler = pd.concat([df_kepler, dummies])
# Adjust plotting time horizon
df_kepler = df_kepler.sort_values('ds').query('ds > "2022-03"')
# Create bins for PM2p5 for Hexbin plotting
pm2p5_bins = np.append(0,np.arange(0, 50, 5))
pm2p5_labels = pm2p5_bins
pm2p5_labels[0] = -1
pm2p5_bins = np.append(pm2p5_bins, 1000)
pm2p5_bins[0] = -20
print(pm2p5_bins)
df_kepler['PM2p5_bins'] = pd.cut(df_kepler['yhat'], bins=pm2p5_bins, labels=pm2p5_labels).astype(int)
# Make the geo DataFrame
gdf_sensors = gpd.GeoDataFrame(
df_kepler,
geometry=gpd.points_from_xy(
x=df_kepler['lon'],
y=df_kepler['lat']
)
)
# Creating a Datetime column (Kepler is funny about datetimes)
gdf_sensors['timestamp'] = pd.to_datetime(gdf_sensors['ds'])
gdf_sensors['timestamp'] = gdf_sensors['timestamp'].dt.strftime('%Y-%m-%d %H:%M:%S')
# Selecting only columns we need
gdf_sensors = gdf_sensors[[
'yhat', 'lon', 'lat', 'geometry', 'timestamp', 'PM2p5_bins', 'location_id'
]]
gdf_sensors
gdf_sensors[gdf_sensors.yhat < 0].location_id.unique()
%run config.py
map_config = config
kepler_map = KeplerGl(
height=800,
data={
'Sensors': gdf_sensors,
}, config=map_config
)
kepler_map
df_kepler.to_csv('../data/kepler.csv', index=False)
| 0.715623 | 0.75435 |
```
import matplotlib.pyplot as plt
from matplotlib import style
style.use('ggplot')
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.animation as animation
import time
X = np.array([[1, 2],
[1.5, 1.8],
[5, 8 ],
[8, 8],
[1, 0.6],
[9,11]])
plt.scatter(X[:,0], X[:,1], s=90)
plt.show()
colors = 10*["g","r","c","b","k"]
f= [1,2,3,0]
v=f.index(min(f))
print(v)
class K_Means:
fig = plt.figure()
ax1 = fig.add_subplot(1,1,1)
def __init__(self, k=2, tol=0.001, max_iter=300):
self.k = k
self.tol = tol
self.max_iter = max_iter
def fit(self,data):
#centroid dict
self.centroids = {}
# since k=2 we will select first two points from the data and we will declare that as a centroid
for i in range(self.k):
self.centroids[i] = data[i]
# we will run this loop for 300 times (300 iteration)
for i in range(self.max_iter):
self.classifications = {} #{0: [], 1: []}
for i in range(self.k):
self.classifications[i] = []# 0 and 1 ke elements ki list
for featureset in data: #finding distance from centroid , finding mini value , putting them in classification
distances = [np.linalg.norm(featureset - self.centroids[centroid]) for centroid in
self.centroids]
classification = distances.index(min(distances)) #find the index of the min distance
self.classifications[classification].append(featureset)
prev_centroids = dict(self.centroids)
for classification in self.classifications:
self.centroids[classification] = np.average(self.classifications[classification],axis=0)
#print(self.centroids)
optimized = True
for c in self.centroids:
original_centroid = prev_centroids[c]
current_centroid = self.centroids[c]
if np.sum((current_centroid-original_centroid)/original_centroid*100.0) < self.tol:
break
def predict(self,data):
distances = [np.linalg.norm(data-self.centroids[centroid]) for centroid in self.centroids]
classification = distances.index(min(distances))
return classification
clf = K_Means()
clf.fit(X)
for centroid in clf.centroids:
plt.scatter(clf.centroids[centroid][0], clf.centroids[centroid][1],
marker="o", color="k", s=50, linewidths=1)
for classification in clf.classifications:
color = colors[classification]
for featureset in clf.classifications[classification]:
plt.scatter(featureset[0], featureset[1], marker="x", color=color, s=50, linewidths=1)
plt.show()
import numpy as np
import matplotlib.pyplot as plt
# evenly sampled time at 200ms intervals
t = np.arange(0., 5., 1)
# red dashes, blue squares and green triangles
plt.plot(t, t, 'r--', t, t**2, 'bs', t, t**3, 'g^')
plt.show()
import matplotlib.pyplot as plt
import matplotlib.animation as animation
import time
fig = plt.figure()
ax1 = fig.add_subplot(1,1,1)
def animate(i):
pullData = open("sampleText.txt","r").read()
dataArray = pullData.split('\n')
xar = []
yar = []
for eachLine in dataArray:
if len(eachLine)>1:
x,y = eachLine.split(',')
xar.append(int(x))
yar.append(int(y))
ax1.clear()
ax1.plot(xar,yar)
ani = animation.FuncAnimation(fig, animate, interval=1000)
plt.show()
```
|
github_jupyter
|
import matplotlib.pyplot as plt
from matplotlib import style
style.use('ggplot')
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.animation as animation
import time
X = np.array([[1, 2],
[1.5, 1.8],
[5, 8 ],
[8, 8],
[1, 0.6],
[9,11]])
plt.scatter(X[:,0], X[:,1], s=90)
plt.show()
colors = 10*["g","r","c","b","k"]
f= [1,2,3,0]
v=f.index(min(f))
print(v)
class K_Means:
fig = plt.figure()
ax1 = fig.add_subplot(1,1,1)
def __init__(self, k=2, tol=0.001, max_iter=300):
self.k = k
self.tol = tol
self.max_iter = max_iter
def fit(self,data):
#centroid dict
self.centroids = {}
# since k=2 we will select first two points from the data and we will declare that as a centroid
for i in range(self.k):
self.centroids[i] = data[i]
# we will run this loop for 300 times (300 iteration)
for i in range(self.max_iter):
self.classifications = {} #{0: [], 1: []}
for i in range(self.k):
self.classifications[i] = []# 0 and 1 ke elements ki list
for featureset in data: #finding distance from centroid , finding mini value , putting them in classification
distances = [np.linalg.norm(featureset - self.centroids[centroid]) for centroid in
self.centroids]
classification = distances.index(min(distances)) #find the index of the min distance
self.classifications[classification].append(featureset)
prev_centroids = dict(self.centroids)
for classification in self.classifications:
self.centroids[classification] = np.average(self.classifications[classification],axis=0)
#print(self.centroids)
optimized = True
for c in self.centroids:
original_centroid = prev_centroids[c]
current_centroid = self.centroids[c]
if np.sum((current_centroid-original_centroid)/original_centroid*100.0) < self.tol:
break
def predict(self,data):
distances = [np.linalg.norm(data-self.centroids[centroid]) for centroid in self.centroids]
classification = distances.index(min(distances))
return classification
clf = K_Means()
clf.fit(X)
for centroid in clf.centroids:
plt.scatter(clf.centroids[centroid][0], clf.centroids[centroid][1],
marker="o", color="k", s=50, linewidths=1)
for classification in clf.classifications:
color = colors[classification]
for featureset in clf.classifications[classification]:
plt.scatter(featureset[0], featureset[1], marker="x", color=color, s=50, linewidths=1)
plt.show()
import numpy as np
import matplotlib.pyplot as plt
# evenly sampled time at 200ms intervals
t = np.arange(0., 5., 1)
# red dashes, blue squares and green triangles
plt.plot(t, t, 'r--', t, t**2, 'bs', t, t**3, 'g^')
plt.show()
import matplotlib.pyplot as plt
import matplotlib.animation as animation
import time
fig = plt.figure()
ax1 = fig.add_subplot(1,1,1)
def animate(i):
pullData = open("sampleText.txt","r").read()
dataArray = pullData.split('\n')
xar = []
yar = []
for eachLine in dataArray:
if len(eachLine)>1:
x,y = eachLine.split(',')
xar.append(int(x))
yar.append(int(y))
ax1.clear()
ax1.plot(xar,yar)
ani = animation.FuncAnimation(fig, animate, interval=1000)
plt.show()
| 0.325735 | 0.67981 |
# Swish-based classifier using cosine-annealed LR with restarts and data augmentation
- Swish activation, 4 layers, 100 neurons per layer
- LR using cosine-annealing with restarts and cycle multiplicity of 2
- Data is augmentaed via phi rotations, and transvers and longitudinal flips
- Validation score use ensemble of 10 models weighted by loss
### Import modules
```
%matplotlib inline
%reload_ext autoreload
%autoreload 2
from __future__ import division
import sys
import os
sys.path.append('../')
from modules.basics import *
from modules.class_basics import *
DATA_PATH = Path('../data/')
```
## Options
```
with open(DATA_PATH/'feats.pkl', 'rb') as fin:
train_feats = pickle.load(fin)
n_models = 10
patience = 2
max_epochs = 200
ensemble_size = 10
ensemble_mode = 'loss'
compile_args = {'loss':'binary_crossentropy', 'optimizer':'adam'}
train_params = {'epochs' : 1, 'batch_size' : 256, 'verbose' : 0}
model_params = {'version':'modelSwish', 'n_in':len(train_feats), 'compile_args':compile_args, 'mode':'classifier'}
print ("\nTraining on", len(train_feats), "features:", [var for var in train_feats])
```
## Import data
```
with open(DATA_PATH/'input_pipe.pkl', 'rb') as fin:
input_pipe = pickle.load(fin)
train_fy = FoldYielder(h5py.File(DATA_PATH/'train.hdf5', "r+"))
```
## Determine LR
```
lr_finder = fold_lr_find(train_fy, get_model, model_params, train_params,
lr_bounds=[1e-5,1e-1], train_on_weights=True, verbose=0)
```
## Train classifier
```
model_gen_params = {**model_params, 'compile_args':{**compile_args, 'lr':2e-3}}
use_callbacks = {'CosAnnealLR':{'cycle_mult':2, 'reverse':False, 'redux_decay':True}}
ams_args = {'n_total':250000, 'br':10, 'delta_b':0}
plots = ['history', 'lr']
results, histories = fold_train_model(train_fy, n_models,
model_gen_params=model_gen_params,
train_params=train_params, train_on_weights=True,
use_callbacks=use_callbacks, ams_args=ams_args,
max_epochs=max_epochs, patience=patience,
verbose=1, plots=plots)
```
The impact of data augmentation is pretty clear. Comparing the training here to that of the the CRL Swish model without augmentation we can see that we effectively gain another LR cycles worth of training epochs before we start overfitting, which allows the networks to reach much lower looses (3.18e-5 c.f. 3.23e-5) and a higher AMSs (3.98 c.f. 3.71)
## Construct ensemble
```
with open('train_weights/resultsFile.pkl', 'rb') as fin:
results = pickle.load(fin)
ensemble, weights = assemble_ensemble(results, ensemble_size, ensemble_mode, compile_args)
```
## Response on validation data
```
val_fy = FoldYielder(h5py.File(DATA_PATH/'val.hdf5', "r+"))
fold_ensemble_predict(ensemble, weights, val_fy, ensemble_size=ensemble_size, verbose=1)
print('Testing ROC AUC: unweighted {}, weighted {}'.format(roc_auc_score(get_feature('targets', val_fy.source), get_feature('pred', val_fy.source)),
roc_auc_score(get_feature('targets', val_fy.source), get_feature('pred', val_fy.source), sample_weight=get_feature('weights', val_fy.source))))
ams_scan_slow(convert_to_df(val_fy.source), br=10, w_factor=250000/50000)
%%time
bootstrap_mean_calc_ams(convert_to_df(val_fy.source), br=10, w_factor=250000/50000, N=512)
```
# Test scoring
```
test_fy = FoldYielder(h5py.File(DATA_PATH/'testing.hdf5', "r+"))
%%time
fold_ensemble_predict(ensemble, weights, test_fy, ensemble_size=ensemble_size, verbose=1)
score_test_data(test_fy.source, 0.9654454513220116)
```
# Save/Load
```
name = "weights/Swish_CLR_TTA"
saveEnsemble(name, ensemble, weights, compile_args, overwrite=1)
ensemble, weights, compile_args, _, _ = loadEnsemble(name)
```
|
github_jupyter
|
%matplotlib inline
%reload_ext autoreload
%autoreload 2
from __future__ import division
import sys
import os
sys.path.append('../')
from modules.basics import *
from modules.class_basics import *
DATA_PATH = Path('../data/')
with open(DATA_PATH/'feats.pkl', 'rb') as fin:
train_feats = pickle.load(fin)
n_models = 10
patience = 2
max_epochs = 200
ensemble_size = 10
ensemble_mode = 'loss'
compile_args = {'loss':'binary_crossentropy', 'optimizer':'adam'}
train_params = {'epochs' : 1, 'batch_size' : 256, 'verbose' : 0}
model_params = {'version':'modelSwish', 'n_in':len(train_feats), 'compile_args':compile_args, 'mode':'classifier'}
print ("\nTraining on", len(train_feats), "features:", [var for var in train_feats])
with open(DATA_PATH/'input_pipe.pkl', 'rb') as fin:
input_pipe = pickle.load(fin)
train_fy = FoldYielder(h5py.File(DATA_PATH/'train.hdf5', "r+"))
lr_finder = fold_lr_find(train_fy, get_model, model_params, train_params,
lr_bounds=[1e-5,1e-1], train_on_weights=True, verbose=0)
model_gen_params = {**model_params, 'compile_args':{**compile_args, 'lr':2e-3}}
use_callbacks = {'CosAnnealLR':{'cycle_mult':2, 'reverse':False, 'redux_decay':True}}
ams_args = {'n_total':250000, 'br':10, 'delta_b':0}
plots = ['history', 'lr']
results, histories = fold_train_model(train_fy, n_models,
model_gen_params=model_gen_params,
train_params=train_params, train_on_weights=True,
use_callbacks=use_callbacks, ams_args=ams_args,
max_epochs=max_epochs, patience=patience,
verbose=1, plots=plots)
with open('train_weights/resultsFile.pkl', 'rb') as fin:
results = pickle.load(fin)
ensemble, weights = assemble_ensemble(results, ensemble_size, ensemble_mode, compile_args)
val_fy = FoldYielder(h5py.File(DATA_PATH/'val.hdf5', "r+"))
fold_ensemble_predict(ensemble, weights, val_fy, ensemble_size=ensemble_size, verbose=1)
print('Testing ROC AUC: unweighted {}, weighted {}'.format(roc_auc_score(get_feature('targets', val_fy.source), get_feature('pred', val_fy.source)),
roc_auc_score(get_feature('targets', val_fy.source), get_feature('pred', val_fy.source), sample_weight=get_feature('weights', val_fy.source))))
ams_scan_slow(convert_to_df(val_fy.source), br=10, w_factor=250000/50000)
%%time
bootstrap_mean_calc_ams(convert_to_df(val_fy.source), br=10, w_factor=250000/50000, N=512)
test_fy = FoldYielder(h5py.File(DATA_PATH/'testing.hdf5', "r+"))
%%time
fold_ensemble_predict(ensemble, weights, test_fy, ensemble_size=ensemble_size, verbose=1)
score_test_data(test_fy.source, 0.9654454513220116)
name = "weights/Swish_CLR_TTA"
saveEnsemble(name, ensemble, weights, compile_args, overwrite=1)
ensemble, weights, compile_args, _, _ = loadEnsemble(name)
| 0.391057 | 0.838415 |
# WeatherPy
----
#### Note
* Instructions have been included for each segment. You do not have to follow them exactly, but they are included to help you think through the steps.
```
# Dependencies and Setup
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
import requests
import time
from scipy.stats import linregress
# Import API key
from api_keys import weather_api_key
# Incorporated citipy to determine city based on latitude and longitude
from citipy import citipy
# Output File (CSV)
output_data_file = "output_data/cities.csv"
# Range of latitudes and longitudes
lat_range = (-90, 90)
lng_range = (-180, 180)
```
## Generate Cities List
```
# List for holding lat_lngs and cities
lat_lngs = []
cities = []
# Create a set of random lat and lng combinations
lats = np.random.uniform(lat_range[0], lat_range[1], size=1500)
lngs = np.random.uniform(lng_range[0], lng_range[1], size=1500)
lat_lngs = zip(lats, lngs)
# Identify nearest city for each lat, lng combination
for lat_lng in lat_lngs:
city = citipy.nearest_city(lat_lng[0], lat_lng[1]).city_name
# If the city is unique, then add it to a our cities list
if city not in cities:
cities.append(city)
# Print the city count to confirm sufficient count
len(cities)
```
### Perform API Calls
* Perform a weather check on each city using a series of successive API calls.
* Include a print log of each city as it'sbeing processed (with the city number and city name).
```
# Save config information
url = "http://api.openweathermap.org/data/2.5/weather?"
units = "imperial"
number = 1
city_name = []
lat = []
lng = []
temp = []
humid = []
clouds = []
wind = []
for city in cities:
try:
city_data = (requests.get(url + '&q=' + city + '&appid=' + weather_api_key)).json()
city_name.append(city_data['name'])
lat.append(city_data['coord']['lat'])
lng.append(city_data['coord']['lon'])
temp.append(city_data['main']['temp'])
humid.append(city_data['main']['humidity'])
clouds.append(city_data['clouds']['all'])
wind.append(city_data['wind']['speed'])
print(f'City number {number} of {len(cities)} complete. | Added {city}')
number = number + 1
except KeyError:
print(f'Missing data in city number {number} of {len(cities)}. | Skipping {city}')
number = number + 1
```
### Convert Raw Data to DataFrame
* Export the city data into a .csv.
* Display the DataFrame
```
city_data_df = pd.DataFrame({'City': city_name,
'Latitude': lat,
'Longitude': lng,
'Temperature': temp,
'Humidity': humid,
'Cloudiness': clouds,
'Wind Speed': wind})
pd.DataFrame.to_csv(city_data_df, 'city_data.csv')
city_data_df.head()
print(city_data_df)
```
## Inspect the data and remove the cities where the humidity > 100%.
----
Skip this step if there are no cities that have humidity > 100%.
```
# Get the indices of cities that have humidity over 100%.
humid_cities = city_data_df[city_data_df['Humidity'] > 100].index
print(humid_cities)
# Make a new DataFrame equal to the city data to drop all humidity outliers by index.
# Passing "inplace=False" will make a copy of the city_data DataFrame, which we call "clean_city_data".
clean_city_df = city_data_df.drop(humid_cities, inplace=False)
clean_city_df.head()
```
## Plotting the Data
* Use proper labeling of the plots using plot titles (including date of analysis) and axes labels.
* Save the plotted figures as .pngs.
```
from datetime import date
```
## Latitude vs. Temperature Plot
```
plt.scatter(clean_city_df['Latitude'], clean_city_df['Temperature'])
plt.title(f'City Latitude vs. Temperature {date.today()}')
plt.xlabel('Latitude')
plt.ylabel('Temperature (F)')
plt.grid(True)
plt.savefig('lat_temp.png', bbox_inches='tight')
```
## Latitude vs. Humidity Plot
```
plt.scatter(clean_city_df['Latitude'], clean_city_df['Humidity'])
plt.title(f'City Latitude vs. Humidity {date.today()}')
plt.xlabel('Latitude')
plt.ylabel('Humidity (%)')
plt.grid(True)
plt.savefig('lat_humid.png', bbox_inches='tight')
```
## Latitude vs. Cloudiness Plot
```
plt.scatter(clean_city_df['Latitude'], clean_city_df['Cloudiness'])
plt.title(f'City Latitude vs. Cloudiness {date.today()}')
plt.xlabel('Latitude')
plt.ylabel('Cloudiness (%)')
plt.grid(True)
plt.savefig('lat_cloud.png', bbox_inches='tight')
```
## Latitude vs. Wind Speed Plot
```
plt.scatter(clean_city_df['Latitude'], clean_city_df['Wind Speed'])
plt.title(f'City Latitude vs. Wind Speed {date.today()}')
plt.xlabel('Latitude')
plt.ylabel('Wind Speed (mph)')
plt.grid(True)
plt.savefig('lat_wind.png', bbox_inches='tight')
```
## Linear Regression
```
nothern = clean_city_df.loc[clean_city_df["Latitude"] >= 0.0]
nothern.reset_index(inplace=False)
southern = clean_city_df.loc[clean_city_df["Latitude"] < 0.0]
southern.reset_index(inplace=False)
def plotLinearRegression(xdata,ydata,xlbl,ylbl,lblpos,ifig):
(slope, intercept, rvalue, pvalue, stderr) = linregress(xdata, ydata)
print(f"The r-squared is: {rvalue}")
regress_values = xdata * slope + intercept
line_eq = "y = " + str(round(slope,2)) + "x + " + str(round(intercept,2))
plt.scatter(xdata,ydata)
plt.plot(xdata,regress_values,"r-")
plt.annotate(line_eq,lblpos,fontsize=15,color="red")
plt.xlabel(xlbl)
plt.ylabel(ylbl)
plt.show()
```
#### Northern Hemisphere - Max Temp vs. Latitude Linear Regression
```
xlbl = "Latitude"
ylbl = "Temperature"
lblpos = (0,25)
plotLinearRegression(nothern[xlbl],nothern[ylbl],xlbl,ylbl,lblpos,5)
```
#### Southern Hemisphere - Max Temp vs. Latitude Linear Regression
```
xlbl = "Latitude"
ylbl = "Temperature"
lblpos = (-55,90)
plotLinearRegression(southern[xlbl],southern[ylbl],xlbl,ylbl,lblpos,6)
```
#### Northern Hemisphere - Humidity (%) vs. Latitude Linear Regression
```
xlbl = "Latitude"
ylbl = "Humidity"
lblpos = (45,10)
plotLinearRegression(nothern[xlbl],nothern[ylbl],xlbl,ylbl,lblpos,7)
```
#### Southern Hemisphere - Humidity (%) vs. Latitude Linear Regression
```
xlbl = "Latitude"
ylbl = "Humidity"
lblpos = (-55,15)
plotLinearRegression(southern[xlbl],southern[ylbl],xlbl,ylbl,lblpos,8)
```
#### Northern Hemisphere - Cloudiness (%) vs. Latitude Linear Regression
```
xlbl = "Latitude"
ylbl = "Cloudiness"
lblpos = (20,40)
plotLinearRegression(nothern[xlbl],nothern[ylbl],xlbl,ylbl,lblpos,9)
```
#### Southern Hemisphere - Cloudiness (%) vs. Latitude Linear Regression
```
xlbl = "Latitude"
ylbl = "Cloudiness"
lblpos = (-55,50)
plotLinearRegression(southern[xlbl],southern[ylbl],xlbl,ylbl,lblpos,10)
```
#### Northern Hemisphere - Wind Speed (mph) vs. Latitude Linear Regression
```
xlbl = "Latitude"
ylbl = "Wind Speed"
lblpos = (0,30)
plotLinearRegression(nothern[xlbl],nothern[ylbl],xlbl,ylbl,lblpos,11)
```
#### Southern Hemisphere - Wind Speed (mph) vs. Latitude Linear Regression
```
xlbl = "Latitude"
ylbl = "Wind Speed"
lblpos = (-25,33)
plotLinearRegression(southern[xlbl],southern[ylbl],xlbl,ylbl,lblpos,12)
print('The tempurature with any given area does seem to have a coorelation with the latitude. There does not seem to be a coorelation between the latitude and the cloud coverage in a given region. Windspeed is also negligable when compared to latitude.')
```
|
github_jupyter
|
# Dependencies and Setup
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
import requests
import time
from scipy.stats import linregress
# Import API key
from api_keys import weather_api_key
# Incorporated citipy to determine city based on latitude and longitude
from citipy import citipy
# Output File (CSV)
output_data_file = "output_data/cities.csv"
# Range of latitudes and longitudes
lat_range = (-90, 90)
lng_range = (-180, 180)
# List for holding lat_lngs and cities
lat_lngs = []
cities = []
# Create a set of random lat and lng combinations
lats = np.random.uniform(lat_range[0], lat_range[1], size=1500)
lngs = np.random.uniform(lng_range[0], lng_range[1], size=1500)
lat_lngs = zip(lats, lngs)
# Identify nearest city for each lat, lng combination
for lat_lng in lat_lngs:
city = citipy.nearest_city(lat_lng[0], lat_lng[1]).city_name
# If the city is unique, then add it to a our cities list
if city not in cities:
cities.append(city)
# Print the city count to confirm sufficient count
len(cities)
# Save config information
url = "http://api.openweathermap.org/data/2.5/weather?"
units = "imperial"
number = 1
city_name = []
lat = []
lng = []
temp = []
humid = []
clouds = []
wind = []
for city in cities:
try:
city_data = (requests.get(url + '&q=' + city + '&appid=' + weather_api_key)).json()
city_name.append(city_data['name'])
lat.append(city_data['coord']['lat'])
lng.append(city_data['coord']['lon'])
temp.append(city_data['main']['temp'])
humid.append(city_data['main']['humidity'])
clouds.append(city_data['clouds']['all'])
wind.append(city_data['wind']['speed'])
print(f'City number {number} of {len(cities)} complete. | Added {city}')
number = number + 1
except KeyError:
print(f'Missing data in city number {number} of {len(cities)}. | Skipping {city}')
number = number + 1
city_data_df = pd.DataFrame({'City': city_name,
'Latitude': lat,
'Longitude': lng,
'Temperature': temp,
'Humidity': humid,
'Cloudiness': clouds,
'Wind Speed': wind})
pd.DataFrame.to_csv(city_data_df, 'city_data.csv')
city_data_df.head()
print(city_data_df)
# Get the indices of cities that have humidity over 100%.
humid_cities = city_data_df[city_data_df['Humidity'] > 100].index
print(humid_cities)
# Make a new DataFrame equal to the city data to drop all humidity outliers by index.
# Passing "inplace=False" will make a copy of the city_data DataFrame, which we call "clean_city_data".
clean_city_df = city_data_df.drop(humid_cities, inplace=False)
clean_city_df.head()
from datetime import date
plt.scatter(clean_city_df['Latitude'], clean_city_df['Temperature'])
plt.title(f'City Latitude vs. Temperature {date.today()}')
plt.xlabel('Latitude')
plt.ylabel('Temperature (F)')
plt.grid(True)
plt.savefig('lat_temp.png', bbox_inches='tight')
plt.scatter(clean_city_df['Latitude'], clean_city_df['Humidity'])
plt.title(f'City Latitude vs. Humidity {date.today()}')
plt.xlabel('Latitude')
plt.ylabel('Humidity (%)')
plt.grid(True)
plt.savefig('lat_humid.png', bbox_inches='tight')
plt.scatter(clean_city_df['Latitude'], clean_city_df['Cloudiness'])
plt.title(f'City Latitude vs. Cloudiness {date.today()}')
plt.xlabel('Latitude')
plt.ylabel('Cloudiness (%)')
plt.grid(True)
plt.savefig('lat_cloud.png', bbox_inches='tight')
plt.scatter(clean_city_df['Latitude'], clean_city_df['Wind Speed'])
plt.title(f'City Latitude vs. Wind Speed {date.today()}')
plt.xlabel('Latitude')
plt.ylabel('Wind Speed (mph)')
plt.grid(True)
plt.savefig('lat_wind.png', bbox_inches='tight')
nothern = clean_city_df.loc[clean_city_df["Latitude"] >= 0.0]
nothern.reset_index(inplace=False)
southern = clean_city_df.loc[clean_city_df["Latitude"] < 0.0]
southern.reset_index(inplace=False)
def plotLinearRegression(xdata,ydata,xlbl,ylbl,lblpos,ifig):
(slope, intercept, rvalue, pvalue, stderr) = linregress(xdata, ydata)
print(f"The r-squared is: {rvalue}")
regress_values = xdata * slope + intercept
line_eq = "y = " + str(round(slope,2)) + "x + " + str(round(intercept,2))
plt.scatter(xdata,ydata)
plt.plot(xdata,regress_values,"r-")
plt.annotate(line_eq,lblpos,fontsize=15,color="red")
plt.xlabel(xlbl)
plt.ylabel(ylbl)
plt.show()
xlbl = "Latitude"
ylbl = "Temperature"
lblpos = (0,25)
plotLinearRegression(nothern[xlbl],nothern[ylbl],xlbl,ylbl,lblpos,5)
xlbl = "Latitude"
ylbl = "Temperature"
lblpos = (-55,90)
plotLinearRegression(southern[xlbl],southern[ylbl],xlbl,ylbl,lblpos,6)
xlbl = "Latitude"
ylbl = "Humidity"
lblpos = (45,10)
plotLinearRegression(nothern[xlbl],nothern[ylbl],xlbl,ylbl,lblpos,7)
xlbl = "Latitude"
ylbl = "Humidity"
lblpos = (-55,15)
plotLinearRegression(southern[xlbl],southern[ylbl],xlbl,ylbl,lblpos,8)
xlbl = "Latitude"
ylbl = "Cloudiness"
lblpos = (20,40)
plotLinearRegression(nothern[xlbl],nothern[ylbl],xlbl,ylbl,lblpos,9)
xlbl = "Latitude"
ylbl = "Cloudiness"
lblpos = (-55,50)
plotLinearRegression(southern[xlbl],southern[ylbl],xlbl,ylbl,lblpos,10)
xlbl = "Latitude"
ylbl = "Wind Speed"
lblpos = (0,30)
plotLinearRegression(nothern[xlbl],nothern[ylbl],xlbl,ylbl,lblpos,11)
xlbl = "Latitude"
ylbl = "Wind Speed"
lblpos = (-25,33)
plotLinearRegression(southern[xlbl],southern[ylbl],xlbl,ylbl,lblpos,12)
print('The tempurature with any given area does seem to have a coorelation with the latitude. There does not seem to be a coorelation between the latitude and the cloud coverage in a given region. Windspeed is also negligable when compared to latitude.')
| 0.425605 | 0.836154 |
# Data Checks
- Schema checks: Making sure that only the columns that are expected are provided.
- Datum checks:
- Looking for missing values
- Ensuring that expected value ranges are correct
- Statistical checks:
- Visual check of data distributions.
- Correlations between columns.
- Statistical distribution checks.
# Roles in Data Analysis
- **Data Provider:** Someone who's collected and/or curated the data.
- **Data Analyst:** The person who is analyzing the data.
Sometimes they're the same person; at other times they're not. Tasks related to testing can often be assigned to either role, but there are some tasks more naturally suited to each.
# Schema Checks
Schema checks are all about making sure that the data columns that you want to have are all present, and that they have the expected data types.
The way data are provided to you should be in two files. The first file is the actual data matrix. The second file should be a metadata specification file, minimally containing the name of the CSV file it describes, and the list of columns present. Why the duplication? The list of columns is basically an implicit contract between your data provider and you, and provides a verifiable way of describing the data matrix's columns.
We're going to use a few datasets from Boston's open data repository. Let's first take a look at Boston's annual budget data, while pretending we're the person who curated the data, the "data provider".
```
%load_ext autoreload
%autoreload 2
%matplotlib inline
%config InlineBackend.figure_format = 'retina'
```
## A bit of basic `pandas`
Let's first start by reading in the CSV file as a `pandas.DataFrame()`.
```
import pandas as pd
df = pd.read_csv('data/boston_budget.csv')
df.head()
```
To get the columns of a DataFrame object `df`, call `df.columns`. This is a list-like object that can be iterated over.
```
df.columns
```
## YAML Files
Describe data in a human-friendly & computer-readable format. The `environment.yml` file in your downloaded repository is also a YAML file, by the way!
Structure:
```yaml
key1: value
key2:
- value1
- value2
- subkey1:
- value3
```
Example YAML-formatted schema:
```yaml
filename: boston_budget.csv
column_names:
- "Fiscal Year"
- "Service (cabinet)"
- "Department"
- "Program #"
...
- "Fund"
- "Amount"
```
YAML-formatted text can be read as dictionaries.
```
spec = """
filename: boston_budget.csv
columns:
- "Fiscal Year"
- "Service (Cabinet)"
- "Department"
- "Program #"
- "Program"
- "Expense Type"
- "ACCT #"
- "Expense Category (Account)"
- "Fund"
- "Amount"
"""
import yaml
metadata = yaml.load(spec)
metadata
```
You can also take dictionaries, and return YAML-formatted text.
```
print(yaml.dump(metadata))
```
By having things YAML formatted, you preserve human-readability and computer-readability simultaneously.
Providing metadata should be something already done when doing analytics; YAML-format is a strong suggestion, but YAML schema will depend on use case.
Let's now switch roles, and pretend that we're on side of the "analyst" and are no longer the "data provider".
How would you check that the columns match the spec? Basically, check that every element in `df.columns` is present inside the `metadata['columns']` list.
## Exercise
Inside `test_datafuncs.py`, write a utility function, `check_schema(df, meta_columns)` that tests whether every column in a DataFrame is present in some metadata spec file. It should accept two arguments:
- `df`: a `pandas.DataFrame`
- `meta_columns`: A list of columns from the metadata spec.
```python
def check_schema(df, meta_columns):
for col in df.columns:
assert col in meta_columns, f'"{col}" not in metadata column spec'
```
In your test file, outside the function definition, write another test function, `test_budget_schemas()`, explicitly runs a test for just the budget data.
```python
def test_budget_schemas():
columns = read_metadata('data/metadata_budget.yml')['columns']
df = pd.read_csv('data/boston_budget.csv')
check_schema(df, columns)
```
Now, run the test. Do you get the following error? Can you spot the error?
```bash
def check_schema(df, meta_columns):
for col in df.columns:
> assert col in meta_columns, f'"{col}" not in metadata column spec'
E AssertionError: " Amount" not in metadata column spec
E assert ' Amount' in ['Fiscal Year', 'Service (Cabinet)', 'Department', 'Program #', 'Program', 'Expense Type', ...]
test_datafuncs_soln.py:63: AssertionError
=================================== 1 failed, 7 passed in 0.91 seconds ===================================
```
If there is even a slight mis-spelling, this kind of check will help you pinpoint where that is. Note how the "Amount" column is spelled with an extra space.
At this point, I would contact the data provider to correct errors like this.
It is a logical practice to keep one schema spec file per table provided to you. However, it is also possible to take advantage of YAML "documents" to keep multiple schema specs inside a single YAML file.
The choice is yours - in cases where there are a lot of data files, it may make sense (for the sake of file-system sanity) to keep all of the specs in multiple files that represent logical groupings of data.
## Exercise: Write `YAML` metadata spec.
Put yourself in the shoes of a data provider. Take the `boston_ei.csv` file in the `data/` directory, and make a schema spec file for that file.
```
spec_ei = """
filename: boston_ei.csv
columns:
- Year
- Month
- logan_passengers
- logan_intl_flights
- hotel_occup_rate
- hotel_avg_daily_rate
- total_jobs
- unemp_rate
- labor_force_part_rate
- pipeline_unit
- pipeline_total_dev_cost
- pipeline_sqft
- pipeline_const_jobs
- foreclosure_pet
- foreclosure_deeds
- med_housing_price
- housing_sales_vol
- new_housing_const_permits
- new-affordable_housing_permits
"""
import yaml
metadata_ei = yaml.load(spec_ex)
metadata_ei
print(yaml.dump(metadata_ei))
```
## Exercise: Write test for metadata spec.
Next, put yourself in the shoes of a data analyst. Take the schema spec file and write a test for it.
```
def check_schema(df, schema):
for col in df.columns:
assert col in metadata_ei, f'"{col}" not in metadata_ei column spec'
def test_budget_ei_schemas():
columns = yaml.load(open('data/metadata_ei.yml'), Loader=yaml.FullLoader)['columns']
df = pd.read_csv('data/boston_ei.csv')
check_schema(df, columns)
test_budget_ei_schemas()
```
## Exercise: Auto YAML Spec.
Inside `datafuncs.py`, write a function with the signature `autospec(handle)` that takes in a file path, and does the following:
- Create a dictionary, with two keys:
- a "filename" key, whose value only records the filename (and not the full file path),
- a "columns" key, whose value records the list of columns in the dataframe.
- Converts the dictionary to a YAML string
- Writes the YAML string to disk.
## Optional Exercise: Write meta-test
Now, let's go "meta". Write a "meta-test" that ensures that every CSV file in the `data/` directory has a schema file associated with it. (The function need not check each schema.) Until we finish filling out the rest of the exercises, this test can be allowed to fail, and we can mark it as a test to skip by marking it with an `@skip` decorator:
```python
@pytest.mark.skip(reason="no way of currently testing this")
def test_my_func():
...
```
## Notes
- The point here is to have a trusted copy of schema apart from data file. YAML not necessarily only way!
- If no schema provided, manually create one; this is exploratory data analysis anyways - no effort wasted!
# Datum Checks
Now that we're done with the schema checks, let's do some sanity checks on the data as well. This is my personal favourite too, as some of the activities here overlap with the early stages of exploratory data analysis.
We're going to switch datasets here, and move to a 'corrupted' version of the Boston Economic Indicators dataset. Its file path is: `./data/boston_ei-corrupt.csv`.
```
import pandas as pd
import seaborn as sns
sns.set_style('white')
%matplotlib inline
df = pd.read_csv('data/boston_ei-corrupt.csv')
df.head()
```
### Demo: Visual Diagnostics
We can use a package called `missingno`, which gives us a quick visual view of the completeness of the data. This is a good starting point for deciding whether you need to manually comb through the data or not.
```
# First, we check for missing data.
import missingno as msno
msno.matrix(df)
```
Immediately it's clear that there's a number of rows with empty values! Nothing beats a quick visual check like this one.
We can get a table version of this using another package called `pandas_summary`.
```
# We can do the same using pandas-summary.
from pandas_summary import DataFrameSummary
dfs = DataFrameSummary(df)
dfs.summary()
```
`dfs.summary()` returns a Pandas DataFrame; this means we can write tests for data completeness!
## Exercise: Test for data completeness.
Write a test named `check_data_completeness(df)` that takes in a DataFrame and confirms that there's no missing data from the `pandas-summary` output. Then, write a corresponding `test_boston_ei()` that tests the schema for the Boston Economic Indicators dataframe.
```python
# In test_datafuncs.py
from pandas_summary import DataFrameSummary
def check_data_completeness(df):
df_summary = DataFrameSummary(df).summary()
for col in df_summary.columns:
assert df_summary.loc['missing', col] == 0, f'{col} has missing values'
def test_boston_ei():
df = pd.read_csv('data/boston_ei.csv')
check_data_completeness(df)
```
## Exercise: Test for value correctness.
In the Economic Indicators dataset, there are four "rate" columns: `['labor_force_part_rate', 'hotel_occup_rate', 'hotel_avg_daily_rate', 'unemp_rate']`, which must have values between 0 and 1.
Add a utility function to `test_datafuncs.py`, `check_data_range(data, lower=0, upper=1)`, which checks the range of the data such that:
- `data` is a list-like object.
- `data <= upper`
- `data >= lower`
- `upper` and `lower` have default values of 1 and 0 respectively.
Then, add to the `test_boston_ei()` function tests for each of these four columns, using the `check_data_range()` function.
```python
# In test_datafuncs.py
def check_data_range(data, lower=0, upper=1):
assert min(data) >= lower, f"minimum value less than {lower}"
assert max(data) <= upper, f"maximum value greater than {upper}"
def test_boston_ei():
df = pd.read_csv('data/boston_ei.csv')
check_data_completeness(df)
zero_one_cols = ['labor_force_part_rate', 'hotel_occup_rate',
'hotel_avg_daily_rate', 'unemp_rate']
for col in zero_one_cols:
check_data_range(df['labor_force_part_rate'])
```
## Distributions
Most of what is coming is going to be a demonstration of the kinds of tools that are potentially useful for you. Feel free to relax from coding, as these aren't necessarily obviously automatable.
### Numerical Data
We can take the EDA portion further, by doing an empirical cumulative distribution plot for each data column.
```
import numpy as np
def compute_dimensions(length):
"""
Given an integer, compute the "square-est" pair of dimensions for plotting.
Examples:
- length: 17 => rows: 4, cols: 5
- length: 14 => rows: 4, cols: 4
This is a utility function; can be tested separately.
"""
sqrt = np.sqrt(length)
floor = int(np.floor(sqrt))
ceil = int(np.ceil(sqrt))
if floor ** 2 >= length:
return (floor, floor)
elif floor * ceil >= length:
return (floor, ceil)
else:
return (ceil, ceil)
compute_dimensions(length=17)
assert compute_dimensions(17) == (4, 5)
assert compute_dimensions(16) == (4, 4)
assert compute_dimensions(15) == (4, 4)
assert compute_dimensions(11) == (3, 4)
# Next, let's visualize the empirical CDF for each column of data.
import matplotlib.pyplot as plt
def empirical_cumdist(data, ax, title=None):
"""
Plots the empirical cumulative distribution of values.
"""
x, y = np.sort(data), np.arange(1, len(data)+1) / len(data)
ax.scatter(x, y)
ax.set_title(title)
data_cols = [i for i in df.columns if i not in ['Year', 'Month']]
n_rows, n_cols = compute_dimensions(len(data_cols))
fig = plt.figure(figsize=(n_cols*3, n_rows*3))
from matplotlib.gridspec import GridSpec
gs = GridSpec(n_rows, n_cols)
for i, col in enumerate(data_cols):
ax = plt.subplot(gs[i])
empirical_cumdist(df[col], ax, title=col)
plt.tight_layout()
plt.show()
```
It's often a good idea to **standardize** numerical data (that aren't count data). The term **standardize** often refers to the statistical procedure of subtracting the mean and dividing by the standard deviation, yielding an empirical distribution of data centered on 0 and having standard deviation of 1.
### Exercise
Write a test for a function that standardizes a column of data. Then, write the function.
**Note:** This function is also implemented in the `scikit-learn` library as part of their `preprocessing` module. However, in case an engineering decision that you make is that you don't want to import an entire library just to use one function, you can re-implement it on your own.
```python
def standard_scaler(x):
return (x - x.mean()) / x.std()
def test_standard_scaler(x):
std = standard_scaler(x)
assert np.allclose(std.mean(), 0)
assert np.allclose(std.std(), 1)
```
### Exercise
Now, plot the grid of standardized values.
```
data_cols = [i for i in df.columns if i not in ['Year', 'Month']]
n_rows, n_cols = compute_dimensions(len(data_cols))
fig = plt.figure(figsize=(n_cols*3, n_rows*3))
from matplotlib.gridspec import GridSpec
gs = GridSpec(n_rows, n_cols)
for i, col in enumerate(data_cols):
ax = plt.subplot(gs[i])
empirical_cumdist(standard_scaler(df[col]), ax, title=col)
plt.tight_layout()
plt.show()
```
### Exercise
Did we just copy/paste the function?! It's time to stop doing this. Let's refactor the code into a function that can be called.
### Categorical Data
For categorical-type data, we can plot the empirical distribution as well. (This example uses the `smartphone_sanitization.csv` dataset.)
```
from collections import Counter
def empirical_catdist(data, ax, title=None):
d = Counter(data)
print(d)
x = range(len(d.keys()))
labels = list(d.keys())
y = list(d.values())
ax.bar(x, y)
ax.set_xticks(x)
ax.set_xticklabels(labels)
smartphone_df = pd.read_csv('data/smartphone_sanitization.csv')
fig = plt.figure()
ax = fig.add_subplot(1,1,1)
empirical_catdist(smartphone_df['site'], ax=ax)
```
# Statistical Checks
- Report on deviations from normality.
## Normality?!
- The Gaussian (Normal) distribution is commonly assumed in downstream statistical procedures, e.g. outlier detection.
- We can test for normality by using a K-S test.
## K-S test
From Wikipedia:
> In statistics, the Kolmogorov–Smirnov test (K–S test or KS test) is a nonparametric test of the equality of continuous, one-dimensional probability distributions that can be used to compare a sample with a reference probability distribution (one-sample K–S test), or to compare two samples (two-sample K–S test). It is named after Andrey Kolmogorov and Nikolai Smirnov.

```
from scipy.stats import ks_2samp
import numpy.random as npr
# Simulate a normal distribution with 10000 draws.
normal_rvs = npr.normal(size=10000)
result = ks_2samp(normal_rvs, df['labor_force_part_rate'].dropna())
result.pvalue < 0.05
fig = plt.figure()
ax = fig.add_subplot(111)
empirical_cumdist(normal_rvs, ax=ax)
empirical_cumdist(df['hotel_occup_rate'], ax=ax)
```
## Exercise
Re-create the panel of cumulative distribution plots, this time adding on the Normal distribution, and annotating the p-value of the K-S test in the title.
```
data_cols = [i for i in df.columns if i not in ['Year', 'Month']]
n_rows, n_cols = compute_dimensions(len(data_cols))
fig = plt.figure(figsize=(n_cols*3, n_rows*3))
from matplotlib.gridspec import GridSpec
gs = GridSpec(n_rows, n_cols)
for i, col in enumerate(data_cols):
ax = plt.subplot(gs[i])
test = ks_2samp(normal_rvs, standard_scaler(df[col]))
empirical_cumdist(normal_rvs, ax)
empirical_cumdist(standard_scaler(df[col]), ax, title=f"{col}, p={round(test.pvalue, 2)}")
plt.tight_layout()
plt.show()
```
|
github_jupyter
|
%load_ext autoreload
%autoreload 2
%matplotlib inline
%config InlineBackend.figure_format = 'retina'
import pandas as pd
df = pd.read_csv('data/boston_budget.csv')
df.head()
df.columns
key1: value
key2:
- value1
- value2
- subkey1:
- value3
filename: boston_budget.csv
column_names:
- "Fiscal Year"
- "Service (cabinet)"
- "Department"
- "Program #"
...
- "Fund"
- "Amount"
spec = """
filename: boston_budget.csv
columns:
- "Fiscal Year"
- "Service (Cabinet)"
- "Department"
- "Program #"
- "Program"
- "Expense Type"
- "ACCT #"
- "Expense Category (Account)"
- "Fund"
- "Amount"
"""
import yaml
metadata = yaml.load(spec)
metadata
print(yaml.dump(metadata))
def check_schema(df, meta_columns):
for col in df.columns:
assert col in meta_columns, f'"{col}" not in metadata column spec'
def test_budget_schemas():
columns = read_metadata('data/metadata_budget.yml')['columns']
df = pd.read_csv('data/boston_budget.csv')
check_schema(df, columns)
def check_schema(df, meta_columns):
for col in df.columns:
> assert col in meta_columns, f'"{col}" not in metadata column spec'
E AssertionError: " Amount" not in metadata column spec
E assert ' Amount' in ['Fiscal Year', 'Service (Cabinet)', 'Department', 'Program #', 'Program', 'Expense Type', ...]
test_datafuncs_soln.py:63: AssertionError
=================================== 1 failed, 7 passed in 0.91 seconds ===================================
spec_ei = """
filename: boston_ei.csv
columns:
- Year
- Month
- logan_passengers
- logan_intl_flights
- hotel_occup_rate
- hotel_avg_daily_rate
- total_jobs
- unemp_rate
- labor_force_part_rate
- pipeline_unit
- pipeline_total_dev_cost
- pipeline_sqft
- pipeline_const_jobs
- foreclosure_pet
- foreclosure_deeds
- med_housing_price
- housing_sales_vol
- new_housing_const_permits
- new-affordable_housing_permits
"""
import yaml
metadata_ei = yaml.load(spec_ex)
metadata_ei
print(yaml.dump(metadata_ei))
def check_schema(df, schema):
for col in df.columns:
assert col in metadata_ei, f'"{col}" not in metadata_ei column spec'
def test_budget_ei_schemas():
columns = yaml.load(open('data/metadata_ei.yml'), Loader=yaml.FullLoader)['columns']
df = pd.read_csv('data/boston_ei.csv')
check_schema(df, columns)
test_budget_ei_schemas()
@pytest.mark.skip(reason="no way of currently testing this")
def test_my_func():
...
import pandas as pd
import seaborn as sns
sns.set_style('white')
%matplotlib inline
df = pd.read_csv('data/boston_ei-corrupt.csv')
df.head()
# First, we check for missing data.
import missingno as msno
msno.matrix(df)
# We can do the same using pandas-summary.
from pandas_summary import DataFrameSummary
dfs = DataFrameSummary(df)
dfs.summary()
# In test_datafuncs.py
from pandas_summary import DataFrameSummary
def check_data_completeness(df):
df_summary = DataFrameSummary(df).summary()
for col in df_summary.columns:
assert df_summary.loc['missing', col] == 0, f'{col} has missing values'
def test_boston_ei():
df = pd.read_csv('data/boston_ei.csv')
check_data_completeness(df)
# In test_datafuncs.py
def check_data_range(data, lower=0, upper=1):
assert min(data) >= lower, f"minimum value less than {lower}"
assert max(data) <= upper, f"maximum value greater than {upper}"
def test_boston_ei():
df = pd.read_csv('data/boston_ei.csv')
check_data_completeness(df)
zero_one_cols = ['labor_force_part_rate', 'hotel_occup_rate',
'hotel_avg_daily_rate', 'unemp_rate']
for col in zero_one_cols:
check_data_range(df['labor_force_part_rate'])
import numpy as np
def compute_dimensions(length):
"""
Given an integer, compute the "square-est" pair of dimensions for plotting.
Examples:
- length: 17 => rows: 4, cols: 5
- length: 14 => rows: 4, cols: 4
This is a utility function; can be tested separately.
"""
sqrt = np.sqrt(length)
floor = int(np.floor(sqrt))
ceil = int(np.ceil(sqrt))
if floor ** 2 >= length:
return (floor, floor)
elif floor * ceil >= length:
return (floor, ceil)
else:
return (ceil, ceil)
compute_dimensions(length=17)
assert compute_dimensions(17) == (4, 5)
assert compute_dimensions(16) == (4, 4)
assert compute_dimensions(15) == (4, 4)
assert compute_dimensions(11) == (3, 4)
# Next, let's visualize the empirical CDF for each column of data.
import matplotlib.pyplot as plt
def empirical_cumdist(data, ax, title=None):
"""
Plots the empirical cumulative distribution of values.
"""
x, y = np.sort(data), np.arange(1, len(data)+1) / len(data)
ax.scatter(x, y)
ax.set_title(title)
data_cols = [i for i in df.columns if i not in ['Year', 'Month']]
n_rows, n_cols = compute_dimensions(len(data_cols))
fig = plt.figure(figsize=(n_cols*3, n_rows*3))
from matplotlib.gridspec import GridSpec
gs = GridSpec(n_rows, n_cols)
for i, col in enumerate(data_cols):
ax = plt.subplot(gs[i])
empirical_cumdist(df[col], ax, title=col)
plt.tight_layout()
plt.show()
def standard_scaler(x):
return (x - x.mean()) / x.std()
def test_standard_scaler(x):
std = standard_scaler(x)
assert np.allclose(std.mean(), 0)
assert np.allclose(std.std(), 1)
data_cols = [i for i in df.columns if i not in ['Year', 'Month']]
n_rows, n_cols = compute_dimensions(len(data_cols))
fig = plt.figure(figsize=(n_cols*3, n_rows*3))
from matplotlib.gridspec import GridSpec
gs = GridSpec(n_rows, n_cols)
for i, col in enumerate(data_cols):
ax = plt.subplot(gs[i])
empirical_cumdist(standard_scaler(df[col]), ax, title=col)
plt.tight_layout()
plt.show()
from collections import Counter
def empirical_catdist(data, ax, title=None):
d = Counter(data)
print(d)
x = range(len(d.keys()))
labels = list(d.keys())
y = list(d.values())
ax.bar(x, y)
ax.set_xticks(x)
ax.set_xticklabels(labels)
smartphone_df = pd.read_csv('data/smartphone_sanitization.csv')
fig = plt.figure()
ax = fig.add_subplot(1,1,1)
empirical_catdist(smartphone_df['site'], ax=ax)
from scipy.stats import ks_2samp
import numpy.random as npr
# Simulate a normal distribution with 10000 draws.
normal_rvs = npr.normal(size=10000)
result = ks_2samp(normal_rvs, df['labor_force_part_rate'].dropna())
result.pvalue < 0.05
fig = plt.figure()
ax = fig.add_subplot(111)
empirical_cumdist(normal_rvs, ax=ax)
empirical_cumdist(df['hotel_occup_rate'], ax=ax)
data_cols = [i for i in df.columns if i not in ['Year', 'Month']]
n_rows, n_cols = compute_dimensions(len(data_cols))
fig = plt.figure(figsize=(n_cols*3, n_rows*3))
from matplotlib.gridspec import GridSpec
gs = GridSpec(n_rows, n_cols)
for i, col in enumerate(data_cols):
ax = plt.subplot(gs[i])
test = ks_2samp(normal_rvs, standard_scaler(df[col]))
empirical_cumdist(normal_rvs, ax)
empirical_cumdist(standard_scaler(df[col]), ax, title=f"{col}, p={round(test.pvalue, 2)}")
plt.tight_layout()
plt.show()
| 0.673084 | 0.965185 |
# Validation
We will talk about different validation techniques used to do *model selection*.
The goald of model selection is to use the data available to choose the model (from a class/set of models) which will work best *in the future*. One important cornerstone of model selection is that **historical performance does not imply future performance**. As we saw before, optimizing for historical performance can often cause overfitting where the model's future performance is far far worse than the historical performance. The term **generalization error** is used to describe the error of a model on previously unseen data (or future data).
## Information Based Model Selection
We start with the discussion with one way to select models: we would like the model which has the minimum *information loss* to the true reality. We will denote truth as $f$ and have a set of models $g_i( x, \theta )$ where $\theta$ denotes the model parameters.
We define the difference in information between two distributions as:
$$ KL( f || g ) = \int_x f(x) \cdot log\left(\frac{f(x)}{g(x)}\right) $$
This is the Kullback–Leibler divergence and denotes the amount of information which must be added to g to get to f (otehrwise the information which is lost by using g when f is reality).
### models have data associated with them as training/learning
In general machine learning algorithms are trained with data (henve the learning part). So we will explicitly denote this data *seen* as $y$. This data is actual data seen from reality, so we have $y \sim f$ since y is a sample dataset from reality $f$. Now we would like to select the model using the data $y$ that minimizes the informatio loss from relaity, so:
$$ \min \int_x f(x) \cdot log\left( \frac{f(x)}{g(x, \theta(y)} \right) = \min \int_x f(x) \cdot log( f(x) ) - \int_x f(x) \cdot log( g(x, \theta(y)) ) $$
We see that the first term involves only realityf $f$ and so we can ignore it since it is a constant with respects to the different possible $g_i( x, \theta(y) )$ models we are selecting from. This means we end up minimizing:
$$ \min - \int_x f(x) \cdot log[g(x,\theta(y))] $$
Let us define $\theta_0$ as the parameters for $g$ which minimize hte information loss according to the above equation. This is in essence the _best_ $\theta$ for hte model in terms of information loss.
### The unknown reality
We don't really know $f$ (otherwise we would not be trying to model it ). So, we will try to minimize the *expected* information loss in terms of the data we have seen, to get:
$$ \min E_y\left[ - \int_x f(x) \cdot log[ g(x,\theta(y)) ] \right] = \min - E_y \left[ E_x \left[ g(x,\theta(y) ) \right] \right] $$
one idea is to try to jsut use the maximum likelihood estimatro (MLE) for $\theta_0$. We will call the MLE $\hat{\theta}(y)$, and plug it in as the minimizer to get
$$ E_y \left[ E_x \left[ g(x,\hat{\theta}(y) ) \right] \right] = log[ L(\hat{\theta}(y) | data) ]$$
Where $L(\theta|data)$ is the likelihood of MLE estimate. However, it turns out the this estimate is _BIASED_.
### Unbiasing MLE : The AIC
It can be shown that, under certain conditions, the bias of MLE $\hat{\theta}$ is approsimaltely $K$, where $K$ = #of parameters in the model. This leads us to a unbiased estimator of the information loss of a model $g_i$ as:
$$ loss = -log( L(\hat{\theta}) | data ) + K $$
This is the Akaike Information Criteria (AIC) for a set of models. We choose the model with the minimum AIC as our estimate of hte model which has the smallest information loss.
## Estimation of Future Performance
Another way to choose a model is to actually try to estimate the future performance of the model. In the ideal world, we would have accessto $f$ directly and could in fact as for a sample dataset $y \sim f$ at any time. Then we could simply as for enough sample datasets to truly characterize the performance of a model $g(x,\theta(y))$ in the reality $f$.
This basic ideas leads us to _cross validation_ as an option. Here, we take our entire single dataset $y \sim f$ and split it into a *train* and a *test* group. Denote the test group as $y_{test}$ and the train group as $y_{train}$. Now we judge the performance of any particular model $g(x,\theta(y_{train})$ using the *test* group, so:
$$performance = g( y_{test}, \theta(y_{train}) )$$
### K-Folds, Leave-one-out, and deterministic splitting
Now, since this is an estimate of performance, we might want to take more than one measurement of teh performance. Rather than splitting into a single test/train grouping, we can split the dataset into a set of _folds_ (sections) and use each section as a test dataset while using hte other folds as training data. In this way we get k observations of the performace. In hte extreme case we get the leave-one-out cross validation where each datapoint is it's own fold and we test the performance over each single datapoint being a test point. For such k-fold cross-validation we generally choose the model with the bet _expected performance_ given the observed performances from the folds.
A second approach is to randomly sample test and training datasets from $y$ a number of times and estimate the performance as the average performance over the samples test/train. This allows us to not have to itertave over all of the k-folds. It also allows us to generate test/train datasets that are repesentative of the original dataset but are no where in it (if we use sampling with replamcenet, we can make some arguments around the distribution over the test/train if the original dataset was sampled from reality and was large enough).
## Direct Measure of Generalizability
We will now consider teh setting of _emprirical risk minimization_. Suppose that we have a *supervised* learning algorithm. There exists a distribution over data point $x$ and label $y$ defined as $P(x,y)$. Now our algorithm has a training dataset $Z=\{(x_i,y_i)\} \sim P$ of $|Z| = n$ samples. We further have a *loss function* $L(\hat{y},y)$ which measures the difference of a predicted $\hat{y}$ from a true $y$. We define the _risk_ as the expectation of the loss function:
$$R(g) = E_{(x,y) \sim P}\left[ L( g(x), y ) \right]$$
However, we do not acutally known $P$ and so we will minimize the _empirical risk_:
$$ R_{emp}(g) = \frac{1}{n}\sum_Z L( g(x_i), y_i ) $$
We will choose the model $g$ which minimizes this empirical risk.
In this setting, there is a direct connection between a version of _stability_ and a notion of _generalizability_ in a strict sense: the more _stable_ an model the mode _general_ it is (so higher stability implies lower generalization error).
|
github_jupyter
|
# Validation
We will talk about different validation techniques used to do *model selection*.
The goald of model selection is to use the data available to choose the model (from a class/set of models) which will work best *in the future*. One important cornerstone of model selection is that **historical performance does not imply future performance**. As we saw before, optimizing for historical performance can often cause overfitting where the model's future performance is far far worse than the historical performance. The term **generalization error** is used to describe the error of a model on previously unseen data (or future data).
## Information Based Model Selection
We start with the discussion with one way to select models: we would like the model which has the minimum *information loss* to the true reality. We will denote truth as $f$ and have a set of models $g_i( x, \theta )$ where $\theta$ denotes the model parameters.
We define the difference in information between two distributions as:
$$ KL( f || g ) = \int_x f(x) \cdot log\left(\frac{f(x)}{g(x)}\right) $$
This is the Kullback–Leibler divergence and denotes the amount of information which must be added to g to get to f (otehrwise the information which is lost by using g when f is reality).
### models have data associated with them as training/learning
In general machine learning algorithms are trained with data (henve the learning part). So we will explicitly denote this data *seen* as $y$. This data is actual data seen from reality, so we have $y \sim f$ since y is a sample dataset from reality $f$. Now we would like to select the model using the data $y$ that minimizes the informatio loss from relaity, so:
$$ \min \int_x f(x) \cdot log\left( \frac{f(x)}{g(x, \theta(y)} \right) = \min \int_x f(x) \cdot log( f(x) ) - \int_x f(x) \cdot log( g(x, \theta(y)) ) $$
We see that the first term involves only realityf $f$ and so we can ignore it since it is a constant with respects to the different possible $g_i( x, \theta(y) )$ models we are selecting from. This means we end up minimizing:
$$ \min - \int_x f(x) \cdot log[g(x,\theta(y))] $$
Let us define $\theta_0$ as the parameters for $g$ which minimize hte information loss according to the above equation. This is in essence the _best_ $\theta$ for hte model in terms of information loss.
### The unknown reality
We don't really know $f$ (otherwise we would not be trying to model it ). So, we will try to minimize the *expected* information loss in terms of the data we have seen, to get:
$$ \min E_y\left[ - \int_x f(x) \cdot log[ g(x,\theta(y)) ] \right] = \min - E_y \left[ E_x \left[ g(x,\theta(y) ) \right] \right] $$
one idea is to try to jsut use the maximum likelihood estimatro (MLE) for $\theta_0$. We will call the MLE $\hat{\theta}(y)$, and plug it in as the minimizer to get
$$ E_y \left[ E_x \left[ g(x,\hat{\theta}(y) ) \right] \right] = log[ L(\hat{\theta}(y) | data) ]$$
Where $L(\theta|data)$ is the likelihood of MLE estimate. However, it turns out the this estimate is _BIASED_.
### Unbiasing MLE : The AIC
It can be shown that, under certain conditions, the bias of MLE $\hat{\theta}$ is approsimaltely $K$, where $K$ = #of parameters in the model. This leads us to a unbiased estimator of the information loss of a model $g_i$ as:
$$ loss = -log( L(\hat{\theta}) | data ) + K $$
This is the Akaike Information Criteria (AIC) for a set of models. We choose the model with the minimum AIC as our estimate of hte model which has the smallest information loss.
## Estimation of Future Performance
Another way to choose a model is to actually try to estimate the future performance of the model. In the ideal world, we would have accessto $f$ directly and could in fact as for a sample dataset $y \sim f$ at any time. Then we could simply as for enough sample datasets to truly characterize the performance of a model $g(x,\theta(y))$ in the reality $f$.
This basic ideas leads us to _cross validation_ as an option. Here, we take our entire single dataset $y \sim f$ and split it into a *train* and a *test* group. Denote the test group as $y_{test}$ and the train group as $y_{train}$. Now we judge the performance of any particular model $g(x,\theta(y_{train})$ using the *test* group, so:
$$performance = g( y_{test}, \theta(y_{train}) )$$
### K-Folds, Leave-one-out, and deterministic splitting
Now, since this is an estimate of performance, we might want to take more than one measurement of teh performance. Rather than splitting into a single test/train grouping, we can split the dataset into a set of _folds_ (sections) and use each section as a test dataset while using hte other folds as training data. In this way we get k observations of the performace. In hte extreme case we get the leave-one-out cross validation where each datapoint is it's own fold and we test the performance over each single datapoint being a test point. For such k-fold cross-validation we generally choose the model with the bet _expected performance_ given the observed performances from the folds.
A second approach is to randomly sample test and training datasets from $y$ a number of times and estimate the performance as the average performance over the samples test/train. This allows us to not have to itertave over all of the k-folds. It also allows us to generate test/train datasets that are repesentative of the original dataset but are no where in it (if we use sampling with replamcenet, we can make some arguments around the distribution over the test/train if the original dataset was sampled from reality and was large enough).
## Direct Measure of Generalizability
We will now consider teh setting of _emprirical risk minimization_. Suppose that we have a *supervised* learning algorithm. There exists a distribution over data point $x$ and label $y$ defined as $P(x,y)$. Now our algorithm has a training dataset $Z=\{(x_i,y_i)\} \sim P$ of $|Z| = n$ samples. We further have a *loss function* $L(\hat{y},y)$ which measures the difference of a predicted $\hat{y}$ from a true $y$. We define the _risk_ as the expectation of the loss function:
$$R(g) = E_{(x,y) \sim P}\left[ L( g(x), y ) \right]$$
However, we do not acutally known $P$ and so we will minimize the _empirical risk_:
$$ R_{emp}(g) = \frac{1}{n}\sum_Z L( g(x_i), y_i ) $$
We will choose the model $g$ which minimizes this empirical risk.
In this setting, there is a direct connection between a version of _stability_ and a notion of _generalizability_ in a strict sense: the more _stable_ an model the mode _general_ it is (so higher stability implies lower generalization error).
| 0.834677 | 0.990533 |
```
import socket
from datetime import datetime
from random import getrandbits
from ipaddress import IPv4Address
from netaddr import IPNetwork, IPAddress
import threading
import requests
from urllib.request import urlopen
import json
print('''Don't Mess With Network List from Mirai:
127.0.0.0/8 # Loopback
0.0.0.0/8 # Invalid address space
3.0.0.0/8 # General Electric (GE)
15.0.0.0/7 # Hewlett-Packard (HP)
56.0.0.0/8 # US Postal Service
10.0.0.0/8 # Internal network
192.168.0.0/16 # Internal network
172.16.0.0/14 # Internal network
100.64.0.0/10 # IANA NAT reserved
169.254.0.0/16 # IANA NAT reserved
198.18.0.0/15 # IANA Special use
224.0.0.0/4 # Multicast
6.0.0.0/7 # Department of Defense
11.0.0.0/8 # Department of Defense
21.0.0.0/8 # Department of Defense
22.0.0.0/8 # Department of Defense
26.0.0.0/8 # Department of Defense
28.0.0.0/7 # Department of Defense
30.0.0.0/8 # Department of Defense
33.0.0.0/8 # Department of Defense
55.0.0.0/8 # Department of Defense
214.0.0.0/7 # Department of Defense''')
dontMessWithList = ['127.0.0.0/8', '0.0.0.0/8', '3.0.0.0/8', '15.0.0.0/7', '56.0.0.0/8', '10.0.0.0/8', '192.168.0.0/16', '172.16.0.0/14', '100.64.0.0/10', '169.254.0.0/16', '198.18.0.0/15', '224.0.0.0/4', '6.0.0.0/7', '11.0.0.0/8', '21.0.0.0/8', '22.0.0.0/8', '26.0.0.0/8', '28.0.0.0/7', '30.0.0.0/8', '33.0.0.0/8', '55.0.0.0/8', '214.0.0.0/7']
def generate_rand_ip():
ip_in_bits = getrandbits(32)
ip_addr = IPv4Address(ip_in_bits)
rand_ip_addr = str(ip_addr)
return rand_ip_addr
def check_ip(ip_addr):
for network in dontMessWithList:
if IPAddress(ip_addr) in IPNetwork(network):
return False
return True
print('Some standard ports: \nTR-069:\t7547\nUPnP:\t1900\nXMPP:\t5222\nCoAP:\t5683\nMQTT:\t1883/8883')
def TCP_connect(ip, port_number, delay, output):
TCPsock = socket.socket(socket.AF_INET, socket.SOCK_STREAM)
TCPsock.setsockopt(socket.SOL_SOCKET, socket.SO_REUSEADDR, 1)
TCPsock.settimeout(delay)
try:
TCPsock.connect((ip, port_number))
output[port_number] = 'Listening'
except:
output[port_number] = ''
def scan_ports(host_ip, delay):
# print("-" * 60)
# print("Please wait, scanning remote host", host_ip)
# print("-" * 60)
t1 = datetime.now()
threads = []
output = {}
common_iot_ports = [21, 22, 23, 25, 80, 81, 82, 83, 84, 88, 137, 143, 443, 445, 554, 631, 1080, 1883, 1900, 2000, 2323, 4433, 4443, 4567, 5222, 5683, 7474, 7547, 8000, 8023, 8080, 8081, 8443, 8088, 8883, 8888, 9000, 9090, 9999, 10000, 37777, 49152]
for i in common_iot_ports:
t = threading.Thread(target=TCP_connect, args=(host_ip, i, delay, output))
threads.append(t)
for i in range(len(common_iot_ports)):
threads[i].start()
for i in range(len(common_iot_ports)):
threads[i].join()
total_listening = 0
ports_listening = []
for i in range(len(common_iot_ports)):
if output[common_iot_ports[i]] == 'Listening':
total_listening += 1
ports_listening.append(common_iot_ports[i])
# print(str(common_iot_ports[i]) + ': ' + output[common_iot_ports[i]])
# t2 = datetime.now()
# total = t2 - t1
# print('Scanning completed in:', total)
if total_listening > 0:
print("IP:", host_ip)
print(total_listening, ports_listening)
return total_listening, ports_listening
def check_if_phue_bulb(ip_addr, port):
url = 'http://' + ip_addr + ':' + str(port)
try:
r = requests.get(url, verify=False, timeout=2)
return r.headers
r = urlopen(url, timeout=3, verify=False)
string = r.read().decode('utf-8')
json_obj = json.loads(string)
return json_obj
except Exception as e:
return "Empty json object"
import pickle
def storeData(obj, filename):
pickleFile = open(filename, 'wb')
pickle.dump(obj, pickleFile)
pickleFile.close()
def loadData(filename):
pickleFile = open(filename, 'rb')
obj = pickle.load(pickleFile)
pickleFile.close()
return obj
# Every one of them is a set
ips_checked = loadData("ips_checked.dat")
hikvision_camera_addr = loadData("hikvision_camera_addr.dat")
sonicWall_firewall_addr = loadData("sonicWall_firewall_addr.dat")
netgear_router_addr = loadData("netgear_router_addr.dat")
TR069_protocolDevice_addr = loadData("TR069_protocolDevice_addr.dat")
lighttpd_protocolDevice_addr = loadData("lighttpd_protocolDevice_addr.dat")
Huawei_router_addr = loadData("Huawei_router_addr.dat")
kangle_addr = loadData("kangle_addr.dat")
tplink_router_addr = loadData("tplink_router_addr.dat")
app_web_server_addr = loadData("app_web_server_addr.dat")
try:
while True:
ip_addr = generate_rand_ip()
if ip_addr not in ips_checked:
ips_checked.add(ip_addr)
else:
continue
# ip_addr = '73.162.12.235'
if check_ip(ip_addr):
a, b = scan_ports(ip_addr, 2)
if a > 0:
for port in b:
# print("Reading port:", port)
json_obj = check_if_phue_bulb(ip_addr, port)
# print(json_obj)
if json_obj != "Empty json object":
print(json_obj)
rh = json.dumps(json_obj.__dict__['_store'])
print(rh)
if 'Hikvision'.lower() in rh.lower() or 'DVRDVS'.lower() in rh.lower():
hikvision_camera_addr.add(ip_addr + ":" + str(port))
elif 'SonicWALL'.lower() in rh.lower():
sonicWall_firewall_addr.add(ip_addr + ":" + str(port))
elif 'NETGEAR'.lower() in rh.lower():
netgear_router_addr.add(ip_addr + ":" + str(port))
elif 'TR069'.lower() in rh.lower() or 'gSOAP'.lower() in rh.lower() or 'TR-069'.lower() in rh.lower():
TR069_protocolDevice_addr.add(ip_addr + ":" + str(port))
elif 'lighttpd'.lower() in rh.lower():
lighttpd_protocolDevice_addr.add(ip_addr + ":" + str(port))
elif 'HuaweiHomeGateway'.lower() in rh.lower():
Huawei_router_addr.add(ip_addr + ":" + str(port))
elif 'kangle'.lower() in rh.lower():
kangle_addr.add(ip_addr + ":" + str(port))
elif 'TP-LINK'.lower() in rh.lower():
tplink_router_addr.add(ip_addr + ":" + str(port))
elif 'App-webs'.lower() in rh.lower():
app_web_server_addr.add(ip_addr + ":" + str(port))
if 'name' in json_obj:
if json_obj['name'] == 'Philips hue':
print(("*" * 10) + 'Philips hue bulb is found.' + ("*" * 10))
# print()
# else:
# print(ip_addr, "is in excluded ip list.")
# break
except KeyboardInterrupt:
pass
print("Total unique IPs checked:", len(ips_checked))
print("Total unique hikvision cameras found:", len(hikvision_camera_addr))
print("Total unique sonicWall firewall found:", len(sonicWall_firewall_addr))
print("Total unique netgear router found:", len(netgear_router_addr))
print("Total unique TR069_protocolDevice found:", len(TR069_protocolDevice_addr))
print("Total unique lighttpd_protocolDevice found:", len(lighttpd_protocolDevice_addr))
print("Total unique Huawei_router found:", len(Huawei_router_addr))
print("Total unique kangle found:", len(kangle_addr))
print("Total unique tplink_router found:", len(tplink_router_addr))
print("Total unique app_web_server found:", len(app_web_server_addr))
print(hikvision_camera_addr)
print(sonicWall_firewall_addr)
print(netgear_router_addr)
print(TR069_protocolDevice_addr)
print(lighttpd_protocolDevice_addr)
print(Huawei_router_addr)
print(kangle_addr)
print(tplink_router_addr)
print(app_web_server_addr)
storeData(ips_checked, "ips_checked.dat")
storeData(hikvision_camera_addr, "hikvision_camera_addr.dat")
storeData(sonicWall_firewall_addr, "sonicWall_firewall_addr.dat")
storeData(netgear_router_addr, "netgear_router_addr.dat")
storeData(TR069_protocolDevice_addr, "TR069_protocolDevice_addr.dat")
storeData(lighttpd_protocolDevice_addr, "lighttpd_protocolDevice_addr.dat")
storeData(Huawei_router_addr, "Huawei_router_addr.dat")
storeData(kangle_addr, "kangle_addr.dat")
storeData(tplink_router_addr, "tplink_router_addr.dat")
storeData(app_web_server_addr, "app_web_server_addr.dat")
from collections import defaultdict
port_dict = defaultdict(lambda : 0)
for i in hikvision_camera_addr:
port_dict[i.split(':')[1]] += 1
for i in sonicWall_firewall_addr:
port_dict[i.split(':')[1]] += 1
for i in netgear_router_addr:
port_dict[i.split(':')[1]] += 1
for i in TR069_protocolDevice_addr:
port_dict[i.split(':')[1]] += 1
for i in lighttpd_protocolDevice_addr:
port_dict[i.split(':')[1]] += 1
for i in Huawei_router_addr:
port_dict[i.split(':')[1]] += 1
for i in kangle_addr:
port_dict[i.split(':')[1]] += 1
for i in tplink_router_addr:
port_dict[i.split(':')[1]] += 1
for i in app_web_server_addr:
port_dict[i.split(':')[1]] += 1
print(port_dict)
```
|
github_jupyter
|
import socket
from datetime import datetime
from random import getrandbits
from ipaddress import IPv4Address
from netaddr import IPNetwork, IPAddress
import threading
import requests
from urllib.request import urlopen
import json
print('''Don't Mess With Network List from Mirai:
127.0.0.0/8 # Loopback
0.0.0.0/8 # Invalid address space
3.0.0.0/8 # General Electric (GE)
15.0.0.0/7 # Hewlett-Packard (HP)
56.0.0.0/8 # US Postal Service
10.0.0.0/8 # Internal network
192.168.0.0/16 # Internal network
172.16.0.0/14 # Internal network
100.64.0.0/10 # IANA NAT reserved
169.254.0.0/16 # IANA NAT reserved
198.18.0.0/15 # IANA Special use
224.0.0.0/4 # Multicast
6.0.0.0/7 # Department of Defense
11.0.0.0/8 # Department of Defense
21.0.0.0/8 # Department of Defense
22.0.0.0/8 # Department of Defense
26.0.0.0/8 # Department of Defense
28.0.0.0/7 # Department of Defense
30.0.0.0/8 # Department of Defense
33.0.0.0/8 # Department of Defense
55.0.0.0/8 # Department of Defense
214.0.0.0/7 # Department of Defense''')
dontMessWithList = ['127.0.0.0/8', '0.0.0.0/8', '3.0.0.0/8', '15.0.0.0/7', '56.0.0.0/8', '10.0.0.0/8', '192.168.0.0/16', '172.16.0.0/14', '100.64.0.0/10', '169.254.0.0/16', '198.18.0.0/15', '224.0.0.0/4', '6.0.0.0/7', '11.0.0.0/8', '21.0.0.0/8', '22.0.0.0/8', '26.0.0.0/8', '28.0.0.0/7', '30.0.0.0/8', '33.0.0.0/8', '55.0.0.0/8', '214.0.0.0/7']
def generate_rand_ip():
ip_in_bits = getrandbits(32)
ip_addr = IPv4Address(ip_in_bits)
rand_ip_addr = str(ip_addr)
return rand_ip_addr
def check_ip(ip_addr):
for network in dontMessWithList:
if IPAddress(ip_addr) in IPNetwork(network):
return False
return True
print('Some standard ports: \nTR-069:\t7547\nUPnP:\t1900\nXMPP:\t5222\nCoAP:\t5683\nMQTT:\t1883/8883')
def TCP_connect(ip, port_number, delay, output):
TCPsock = socket.socket(socket.AF_INET, socket.SOCK_STREAM)
TCPsock.setsockopt(socket.SOL_SOCKET, socket.SO_REUSEADDR, 1)
TCPsock.settimeout(delay)
try:
TCPsock.connect((ip, port_number))
output[port_number] = 'Listening'
except:
output[port_number] = ''
def scan_ports(host_ip, delay):
# print("-" * 60)
# print("Please wait, scanning remote host", host_ip)
# print("-" * 60)
t1 = datetime.now()
threads = []
output = {}
common_iot_ports = [21, 22, 23, 25, 80, 81, 82, 83, 84, 88, 137, 143, 443, 445, 554, 631, 1080, 1883, 1900, 2000, 2323, 4433, 4443, 4567, 5222, 5683, 7474, 7547, 8000, 8023, 8080, 8081, 8443, 8088, 8883, 8888, 9000, 9090, 9999, 10000, 37777, 49152]
for i in common_iot_ports:
t = threading.Thread(target=TCP_connect, args=(host_ip, i, delay, output))
threads.append(t)
for i in range(len(common_iot_ports)):
threads[i].start()
for i in range(len(common_iot_ports)):
threads[i].join()
total_listening = 0
ports_listening = []
for i in range(len(common_iot_ports)):
if output[common_iot_ports[i]] == 'Listening':
total_listening += 1
ports_listening.append(common_iot_ports[i])
# print(str(common_iot_ports[i]) + ': ' + output[common_iot_ports[i]])
# t2 = datetime.now()
# total = t2 - t1
# print('Scanning completed in:', total)
if total_listening > 0:
print("IP:", host_ip)
print(total_listening, ports_listening)
return total_listening, ports_listening
def check_if_phue_bulb(ip_addr, port):
url = 'http://' + ip_addr + ':' + str(port)
try:
r = requests.get(url, verify=False, timeout=2)
return r.headers
r = urlopen(url, timeout=3, verify=False)
string = r.read().decode('utf-8')
json_obj = json.loads(string)
return json_obj
except Exception as e:
return "Empty json object"
import pickle
def storeData(obj, filename):
pickleFile = open(filename, 'wb')
pickle.dump(obj, pickleFile)
pickleFile.close()
def loadData(filename):
pickleFile = open(filename, 'rb')
obj = pickle.load(pickleFile)
pickleFile.close()
return obj
# Every one of them is a set
ips_checked = loadData("ips_checked.dat")
hikvision_camera_addr = loadData("hikvision_camera_addr.dat")
sonicWall_firewall_addr = loadData("sonicWall_firewall_addr.dat")
netgear_router_addr = loadData("netgear_router_addr.dat")
TR069_protocolDevice_addr = loadData("TR069_protocolDevice_addr.dat")
lighttpd_protocolDevice_addr = loadData("lighttpd_protocolDevice_addr.dat")
Huawei_router_addr = loadData("Huawei_router_addr.dat")
kangle_addr = loadData("kangle_addr.dat")
tplink_router_addr = loadData("tplink_router_addr.dat")
app_web_server_addr = loadData("app_web_server_addr.dat")
try:
while True:
ip_addr = generate_rand_ip()
if ip_addr not in ips_checked:
ips_checked.add(ip_addr)
else:
continue
# ip_addr = '73.162.12.235'
if check_ip(ip_addr):
a, b = scan_ports(ip_addr, 2)
if a > 0:
for port in b:
# print("Reading port:", port)
json_obj = check_if_phue_bulb(ip_addr, port)
# print(json_obj)
if json_obj != "Empty json object":
print(json_obj)
rh = json.dumps(json_obj.__dict__['_store'])
print(rh)
if 'Hikvision'.lower() in rh.lower() or 'DVRDVS'.lower() in rh.lower():
hikvision_camera_addr.add(ip_addr + ":" + str(port))
elif 'SonicWALL'.lower() in rh.lower():
sonicWall_firewall_addr.add(ip_addr + ":" + str(port))
elif 'NETGEAR'.lower() in rh.lower():
netgear_router_addr.add(ip_addr + ":" + str(port))
elif 'TR069'.lower() in rh.lower() or 'gSOAP'.lower() in rh.lower() or 'TR-069'.lower() in rh.lower():
TR069_protocolDevice_addr.add(ip_addr + ":" + str(port))
elif 'lighttpd'.lower() in rh.lower():
lighttpd_protocolDevice_addr.add(ip_addr + ":" + str(port))
elif 'HuaweiHomeGateway'.lower() in rh.lower():
Huawei_router_addr.add(ip_addr + ":" + str(port))
elif 'kangle'.lower() in rh.lower():
kangle_addr.add(ip_addr + ":" + str(port))
elif 'TP-LINK'.lower() in rh.lower():
tplink_router_addr.add(ip_addr + ":" + str(port))
elif 'App-webs'.lower() in rh.lower():
app_web_server_addr.add(ip_addr + ":" + str(port))
if 'name' in json_obj:
if json_obj['name'] == 'Philips hue':
print(("*" * 10) + 'Philips hue bulb is found.' + ("*" * 10))
# print()
# else:
# print(ip_addr, "is in excluded ip list.")
# break
except KeyboardInterrupt:
pass
print("Total unique IPs checked:", len(ips_checked))
print("Total unique hikvision cameras found:", len(hikvision_camera_addr))
print("Total unique sonicWall firewall found:", len(sonicWall_firewall_addr))
print("Total unique netgear router found:", len(netgear_router_addr))
print("Total unique TR069_protocolDevice found:", len(TR069_protocolDevice_addr))
print("Total unique lighttpd_protocolDevice found:", len(lighttpd_protocolDevice_addr))
print("Total unique Huawei_router found:", len(Huawei_router_addr))
print("Total unique kangle found:", len(kangle_addr))
print("Total unique tplink_router found:", len(tplink_router_addr))
print("Total unique app_web_server found:", len(app_web_server_addr))
print(hikvision_camera_addr)
print(sonicWall_firewall_addr)
print(netgear_router_addr)
print(TR069_protocolDevice_addr)
print(lighttpd_protocolDevice_addr)
print(Huawei_router_addr)
print(kangle_addr)
print(tplink_router_addr)
print(app_web_server_addr)
storeData(ips_checked, "ips_checked.dat")
storeData(hikvision_camera_addr, "hikvision_camera_addr.dat")
storeData(sonicWall_firewall_addr, "sonicWall_firewall_addr.dat")
storeData(netgear_router_addr, "netgear_router_addr.dat")
storeData(TR069_protocolDevice_addr, "TR069_protocolDevice_addr.dat")
storeData(lighttpd_protocolDevice_addr, "lighttpd_protocolDevice_addr.dat")
storeData(Huawei_router_addr, "Huawei_router_addr.dat")
storeData(kangle_addr, "kangle_addr.dat")
storeData(tplink_router_addr, "tplink_router_addr.dat")
storeData(app_web_server_addr, "app_web_server_addr.dat")
from collections import defaultdict
port_dict = defaultdict(lambda : 0)
for i in hikvision_camera_addr:
port_dict[i.split(':')[1]] += 1
for i in sonicWall_firewall_addr:
port_dict[i.split(':')[1]] += 1
for i in netgear_router_addr:
port_dict[i.split(':')[1]] += 1
for i in TR069_protocolDevice_addr:
port_dict[i.split(':')[1]] += 1
for i in lighttpd_protocolDevice_addr:
port_dict[i.split(':')[1]] += 1
for i in Huawei_router_addr:
port_dict[i.split(':')[1]] += 1
for i in kangle_addr:
port_dict[i.split(':')[1]] += 1
for i in tplink_router_addr:
port_dict[i.split(':')[1]] += 1
for i in app_web_server_addr:
port_dict[i.split(':')[1]] += 1
print(port_dict)
| 0.067439 | 0.089494 |
# Exceptions
Exceptions which are events that can modify the *flow* of control through a program.
In Python, exceptions are triggered automatically on errors, and they can be triggered and intercepted by your code.
They are processed by **four** statements we’ll study in this notebook, the first of which has two variations (listed separately here) and the last of which was an optional extension until Python 2.6 and 3.0:
* `try/except`:
* Catch and recover from exceptions raised by Python, or by you
* `try/finally`:
* Perform cleanup actions, whether exceptions occur or not.
* `raise`:
* Trigger an exception manually in your code.
* `assert`:
* Conditionally trigger an exception in your code.
* `with/as`:
* Implement context managers in Python 2.6, 3.0, and later (optional in 2.5).
# `try/except` Statement
```
try:
statements # Run this main action first
except name1:
# Run if name1 is raised during try block
statements
except (name2, name3):
# Run if any of these exceptions occur
statements
except name4 as var:
# Run if name4 is raised, assign instance raised to var
statements
except: # Run for all other exceptions raised
statements
else:
statements # Run if no exception was raised during try block
```
```
list_of_numbers = [number for number in range(1, 100)]
print(list_of_numbers)
dictionary_of_numbers = {}
for number in list_of_numbers:
dictionary_of_numbers[number**2] = number
try:
index = list_of_numbers.index(2)
value = dictionary_of_numbers[index]
except (ValueError, KeyError):
print('Error Raised, but Controlled! ')
else:
# This executes ONLY if no exception is raised
print('Getting number at position %d : %d' % (index, value))
finally:
# Do cleanup operations
print('Cleaning UP')
```
# `try/finally` Statement
The other flavor of the try statement is a specialization that has to do with finalization (a.k.a. termination) actions. If a finally clause is included in a try, Python will always run its block of statements “on the way out” of the try statement, whether an exception occurred while the try block was running or not.
In it's general form, it is:
```
try:
statements # Run this action first
finally:
statements # Always run this code on the way out
```
<a name="ctx"></a>
# `with/as` Context Managers
Python 2.6 and 3.0 introduced a new exception-related statement—the with, and its optional as clause. This statement is designed to work with context manager objects, which support a new method-based protocol, similar in spirit to the way that iteration tools work with methods of the iteration protocol.
## Context Manager Intro
### Basic Usage:
```
with expression [as variable]:
with-block
```
### Classical Usage
```python
with open(r'C:\misc\data') as myfile:
for line in myfile:
print(line)
# ...more code here...
```
... even using multiple context managers:
```python
with open('script1.py') as f1, open('script2.py') as f2:
for (linenum, (line1, line2)) in enumerate(zip(f1, f2)):
if line1 != line2:
print('%s\n%r\n%r' % (linenum, line1, line2))
```
### How it works
1. The expression is evaluated,resulting in an object known as a **context manager** that must have `__enter__` and `__exit__` methods
2. The context manager’s `__enter__` method is called. The value it returns is assigned to the variable in the as clause if present, or simply discarded otherwise
3. The code in the nested with block is executed.
4. If the with block raises an exception, the `__exit__(type,value,traceback)` method is called with the exception details. These are the same three values returned by `sys.exc_info` (Python function). If this method returns a `false` value, the exception is **re-raised**; otherwise, the exception is terminated. The exception should normally be reraised so that it is propagated outside the with statement.
5. If the with block does not raise an exception, the `__exit__` method is still called, but its type, value, and traceback arguments are all passed in as `None`.
## Usage with Exceptions
```
class TraceBlock:
def message(self, arg):
print('running ' + arg)
def __enter__(self):
print('starting with block')
return self
def __exit__(self, exc_type, exc_value, exc_tb):
if exc_type is None:
print('exited normally\n')
else:
print('raise an exception! ' + str(exc_type))
return False # Propagate
with TraceBlock() as action:
action.message('test 1')
print('reached')
with TraceBlock() as action:
action.message('test 2')
raise TypeError()
print('not reached')
```
## User Defined Exceptions
```
class AlreadyGotOne(Exception):
pass
def gail():
raise AlreadyGotOne()
try:
gail()
except AlreadyGotOne:
print('got exception')
class Career(Exception):
def __init__(self, job, *args, **kwargs):
super(Career, self).__init__(*args, **kwargs)
self._job = job
def __str__(self):
return 'So I became a waiter of {}'.format(self._job)
raise Career('Engineer')
```
|
github_jupyter
|
try:
statements # Run this main action first
except name1:
# Run if name1 is raised during try block
statements
except (name2, name3):
# Run if any of these exceptions occur
statements
except name4 as var:
# Run if name4 is raised, assign instance raised to var
statements
except: # Run for all other exceptions raised
statements
else:
statements # Run if no exception was raised during try block
list_of_numbers = [number for number in range(1, 100)]
print(list_of_numbers)
dictionary_of_numbers = {}
for number in list_of_numbers:
dictionary_of_numbers[number**2] = number
try:
index = list_of_numbers.index(2)
value = dictionary_of_numbers[index]
except (ValueError, KeyError):
print('Error Raised, but Controlled! ')
else:
# This executes ONLY if no exception is raised
print('Getting number at position %d : %d' % (index, value))
finally:
# Do cleanup operations
print('Cleaning UP')
try:
statements # Run this action first
finally:
statements # Always run this code on the way out
with expression [as variable]:
with-block
with open(r'C:\misc\data') as myfile:
for line in myfile:
print(line)
# ...more code here...
with open('script1.py') as f1, open('script2.py') as f2:
for (linenum, (line1, line2)) in enumerate(zip(f1, f2)):
if line1 != line2:
print('%s\n%r\n%r' % (linenum, line1, line2))
class TraceBlock:
def message(self, arg):
print('running ' + arg)
def __enter__(self):
print('starting with block')
return self
def __exit__(self, exc_type, exc_value, exc_tb):
if exc_type is None:
print('exited normally\n')
else:
print('raise an exception! ' + str(exc_type))
return False # Propagate
with TraceBlock() as action:
action.message('test 1')
print('reached')
with TraceBlock() as action:
action.message('test 2')
raise TypeError()
print('not reached')
class AlreadyGotOne(Exception):
pass
def gail():
raise AlreadyGotOne()
try:
gail()
except AlreadyGotOne:
print('got exception')
class Career(Exception):
def __init__(self, job, *args, **kwargs):
super(Career, self).__init__(*args, **kwargs)
self._job = job
def __str__(self):
return 'So I became a waiter of {}'.format(self._job)
raise Career('Engineer')
| 0.254324 | 0.860193 |
# Table of Contents
* [Loading Data](#loading-data)
* [Statistical Summary and Class Breakdown](#statistical-summary)
* [Dropping Rows with Missing Values](#dropping-rows)
* [Data Imputation](#data-imputation)
* [Min-max Scaling](#min-max-scaling)
* [standardization](#standardization)
* [Robust Scaling](#robust-scaling)
* [Categorical Data](#categorical-data)
* [Feature Engineering](#feature-engineering)
* [Univariate Selection](#univariate-selection)
* [Recursive Feature Elimination](#recursive-feature-elimination)
* [Dimensionality Reduction](#dimensionality-reduction)
* [Hold-out Validation](#hold-out-validation)
* [Cross Validation](#cross-validation)
* [Confusion Matrix](#confusion-matrix)
* [Classification Report](#classification-report)
* [Grid Search](#grid-search)
```
# Initialization
%matplotlib inline
from warnings import filterwarnings
filterwarnings('ignore')
```
## Loading Data <a class="anchor" id="loading-data"></a>
```
# Load data from CSV file
from pandas import read_csv
from numpy import set_printoptions
from sklearn.preprocessing import MinMaxScaler
names = ['preg', 'plas', 'pres', 'skin', 'test', 'mass', 'pedi', 'age', 'class']
df = read_csv('pima-indians-diabetes.data.csv', names=names)
df.head(5)
array = df.values
# separate data into features and targets
X = array[:,0:8]
y = array[:,8]
print(X.shape, y.shape)
```
## Statistical Summary and Class Breakdown <a class="anchor" id="statistical-summary"></a>
```
# Print statistical summary and class breakdown
from pandas import read_csv
names = ['preg', 'plas', 'pres', 'skin', 'test', 'mass', 'pedi', 'age', 'class']
df = read_csv('pima-indians-diabetes.data.csv', names=names)
print(df.shape) # print the dimension of the data
print(df.describe()) # print the statistical summary of the data
class_counts = df.groupby('class').size()
print(class_counts) # print the class breakdown of the data
```
## Dropping Rows with Missing Values <a class="anchor" id="dropping-rows"></a>
```
# Handling missing values by dropping data samples with missing values
import pandas as pd
df = pd.DataFrame({'Age': [17, 23, 'x', 38, 54, 67, 32],
'Height': [160, 172, 150, 165, 163, 158, 175],
'Weight':[50, 68, 43, 52, 47, 49, 'x']})
df.replace({'x': None}, inplace=True) # replace missing values (x) with NaN
print(df)
df.dropna(inplace=True) # drop rows with NaN
print(df)
```
## Data Imputation <a class="anchor" id="data-imputation"></a>
```
# Handling missing values by imputing missing values with statistic
import pandas as pd
df = pd.DataFrame({'Age': [17, 23, 'x', 38, 54, 67, 32],
'Height': [160, 172, 150, 165, 163, 158, 175],
'Weight':[50, 68, 43, 52, 47, 49, 'x']})
df.replace({'x': None}, inplace=True)
df['Age'].fillna(df['Age'].median(), inplace=True) # replace NaN with median
df['Weight'].fillna(df['Weight'].mean(), inplace=True) # replace NaN with mean
print(df)
```
## Min-max Scaling <a class="anchor" id="min-max-scaling"></a>
```
# Scale data (between 0 and 1)
from pandas import read_csv
from sklearn.preprocessing import MinMaxScaler
names = ['preg', 'plas', 'pres', 'skin', 'test', 'mass', 'pedi', 'age', 'class']
df = read_csv('pima-indians-diabetes.data.csv', names=names)
array = df.values
# separate array into input and output components
X = array[:,:-1]
y = array[:,-1]
scaler = MinMaxScaler(feature_range=(0, 1))
scaler.fit(X)
scaledX = scaler.transform(X)
# Preview the scaled data
print(scaledX[:5,:])
```
## Standardization <a class="anchor" id="standardization"></a>
```
# Standardize data (0 mean, 1 stdev)
from sklearn.preprocessing import StandardScaler
from pandas import read_csv
names = ['preg', 'plas', 'pres', 'skin', 'test', 'mass', 'pedi', 'age', 'class']
df = read_csv('pima-indians-diabetes.data.csv', names=names)
array = df.values
# separate array into input and output components
X = array[:, :-1]
y = array[:, -1]
scaler = StandardScaler()
scaledX = scaler.fit_transform(X)
# Preview transformed data
print(scaledX[:5, :])
```
## Robust Scaling <a class="anchor" id="robust-scaling"></a>
```
# Robust scaling (0 median, 1 IQR)
from sklearn.preprocessing import RobustScaler
from pandas import read_csv
names = ['preg', 'plas', 'pres', 'skin', 'test', 'mass', 'pedi', 'age', 'class']
df = read_csv('pima-indians-diabetes.data.csv', names=names)
array = df.values
# separate array into input and output components
X = array[:,:-1]
y = array[:,-1]
scaler = RobustScaler()
scaledX = scaler.fit_transform(X)
# Preview transformed data
print(scaledX[0:5,:])
```
## Categorical Data <a class="anchor" id="categorical-data"></a>
```
# Handling categorical data
import pandas as pd
df0 = pd.DataFrame({'year':[2015, 2017, 2013, 2018, 2020],
'make':['Toyota', 'Honda', 'Perodua', 'Hyundai', 'Toyota'],
'engine':[1.5, 1.8, 1.3, 1.6, 1.8],
'review':['moderate', 'good', 'poor', 'moderate', 'good']})
mapping = {'poor':1, 'moderate':2, 'good':3}
df0['review'] = df0['review'].map(mapping) # encode ordinal data
df0 = pd.get_dummies(df0) # encode nominal data
print(df0)
```
## Feature Engineering <a class="anchor" id="feature-engineering"></a>
```
# Create 2 new features
from pandas import read_csv
from sklearn.preprocessing import MinMaxScaler
names = ['preg', 'plas', 'pres', 'skin', 'test', 'mass', 'pedi', 'age', 'class']
df = read_csv('pima-indians-diabetes.data.csv', names=names)
win_size = 3
df['plas_pres'] = df['plas'] + df['pres'] # new feature 1
df['mass_ave'] = df['mass'].rolling(win_size).mean() # new feature 2
df.head()
```
## Univariate Selection <a class="anchor" id="univariate-selection"></a>
```
# Feature Selection with Univariate Selection
from pandas import read_csv
from sklearn.feature_selection import SelectKBest
# load data
names = ['preg', 'plas', 'pres', 'skin', 'test', 'mass', 'pedi', 'age', 'class']
df = read_csv('pima-indians-diabetes.data.csv', names=names)
array = df.values
X = array[:, :-1]
y = array[:, -1]
selector = SelectKBest(k=4)
features = selector.fit_transform(X, y)
selected = selector.get_support()
# Show selected features
print([names[i] for i in range(len(names)-1) if selected[i]])
```
## Recursive Feature Elimination <a class="anchor" id="recursive-feature-elimination"></a>
```
# Feature Selection with RFE
from pandas import read_csv
from sklearn.feature_selection import RFE
from sklearn.tree import DecisionTreeClassifier
filename = 'pima-indians-diabetes.data.csv'
names = ['preg', 'plas', 'pres', 'skin', 'test', 'mass', 'pedi', 'age', 'class']
df = read_csv(filename, names=names)
array = df.values
X = array[:, :-1]
y = array[:, -1]
model = DecisionTreeClassifier()
rfe = RFE(model, 3)
features = rfe.fit_transform(X, y)
selected = rfe.get_support()
# Show selected features
print([names[i] for i in range(len(names)-1) if selected[i]])
```
## Dimensionality Reduction <a class="anchor" id="dimensionality-reduction"></a>
```
# Dimensionality Reduction with PCA
from pandas import read_csv
from sklearn.decomposition import PCA
names = ['preg', 'plas', 'pres', 'skin', 'test', 'mass', 'pedi', 'age', 'class']
df = read_csv('pima-indians-diabetes.data.csv', names=names)
array = df.values
X = array[:, :-1]
y = array[:, -1]
pca = PCA(n_components=3)
features = pca.fit_transform(X)
# summarize components
print("Explained Variance: %s" % pca.explained_variance_ratio_)
```
## Hold-out Validation <a class="anchor" id="hold-out-validation"></a>
```
# Evaluate using a train and a test set
from pandas import read_csv
from sklearn.model_selection import train_test_split as split
from sklearn.neighbors import KNeighborsClassifier
names = ['preg', 'plas', 'pres', 'skin', 'test', 'mass', 'pedi', 'age', 'class']
df = read_csv('pima-indians-diabetes.data.csv', names=names)
array = df.values
X = array[:, :-1]
y = array[:, -1]
X_train, X_test, y_train, y_test = split(X, y, test_size=0.33, random_state=42)
model = KNeighborsClassifier()
model.fit(X_train, y_train)
result = model.score(X_test, y_test)
print(f"Accuracy: {100 * result:.2f} %")
```
## Cross Validation <a class="anchor" id="cross-validation"></a>
```
# Evaluate using Cross Validation
from pandas import read_csv
from sklearn.model_selection import KFold, cross_val_score
from sklearn.neighbors import KNeighborsClassifier
names = ['preg', 'plas', 'pres', 'skin', 'test', 'mass', 'pedi', 'age', 'class']
df = read_csv('pima-indians-diabetes.data.csv', names=names)
array = df.values
X = array[:, :-1]
y = array[:, -1]
model = KNeighborsClassifier()
kfold = KFold(n_splits=5, shuffle=True, random_state=42)
results = cross_val_score(model, X, y, cv=kfold)
print(f"Accuracy: {100*results.mean():.2f} % ({100*results.std():.2f})")
```
## Confusion Matrix <a class="anchor" id="confusion-matrix"></a>
```
# Plot Confusion Matrix
from pandas import read_csv
from sklearn.model_selection import train_test_split as split
from sklearn.neighbors import KNeighborsClassifier
from sklearn.metrics import plot_confusion_matrix
names = ['preg', 'plas', 'pres', 'skin', 'test', 'mass', 'pedi', 'age', 'class']
df = read_csv('pima-indians-diabetes.data.csv', names=names)
array = df.values
X = array[:, :-1]
y = array[:, -1]
X_train, X_test, y_train, y_test = split(X, y, test_size=0.33, random_state=42)
model = KNeighborsClassifier()
model.fit(X_train, y_train)
plot_confusion_matrix(model, X_test, y_test)
```
## Classification Report <a class="anchor" id="classification-report"></a>
```
# Classification report
from pandas import read_csv
from sklearn.model_selection import train_test_split as split
from sklearn.neighbors import KNeighborsClassifier
from yellowbrick.classifier import ClassificationReport
names = ['preg', 'plas', 'pres', 'skin', 'test', 'mass', 'pedi', 'age', 'class']
df = read_csv('pima-indians-diabetes.data.csv', names=names)
array = df.values
X = array[:, :-1]
y = array[:, -1]
X_train, X_test, y_train, y_test = split(X, y, test_size=0.33, random_state=42)
model = KNeighborsClassifier()
model.fit(X_train, y_train)
report = ClassificationReport(model)
report.score(X_test, y_test)
report.show()
```
## Grid Search <a class="anchor" id="grid-search"></a>
```
# Grid Search for Hyperparameter Tuning
from pandas import read_csv
from sklearn.svm import SVC
from sklearn.model_selection import GridSearchCV
names = ['preg', 'plas', 'pres', 'skin', 'test', 'mass', 'pedi', 'age', 'class']
df = read_csv('pima-indians-diabetes.data.csv', names=names)
array = df.values
X = array[:, :-1]
y = array[:, -1]
params = dict(C=[0.001, 0.01, 0.1, 1, 10], gamma=[0.001, 0.01, 0.1, 1, 10])
model = SVC()
grid = GridSearchCV(model, params, n_jobs=-1, verbose=2)
grid.fit(X, y)
print(grid.best_score_)
print(grid.best_params_)
```
|
github_jupyter
|
# Initialization
%matplotlib inline
from warnings import filterwarnings
filterwarnings('ignore')
# Load data from CSV file
from pandas import read_csv
from numpy import set_printoptions
from sklearn.preprocessing import MinMaxScaler
names = ['preg', 'plas', 'pres', 'skin', 'test', 'mass', 'pedi', 'age', 'class']
df = read_csv('pima-indians-diabetes.data.csv', names=names)
df.head(5)
array = df.values
# separate data into features and targets
X = array[:,0:8]
y = array[:,8]
print(X.shape, y.shape)
# Print statistical summary and class breakdown
from pandas import read_csv
names = ['preg', 'plas', 'pres', 'skin', 'test', 'mass', 'pedi', 'age', 'class']
df = read_csv('pima-indians-diabetes.data.csv', names=names)
print(df.shape) # print the dimension of the data
print(df.describe()) # print the statistical summary of the data
class_counts = df.groupby('class').size()
print(class_counts) # print the class breakdown of the data
# Handling missing values by dropping data samples with missing values
import pandas as pd
df = pd.DataFrame({'Age': [17, 23, 'x', 38, 54, 67, 32],
'Height': [160, 172, 150, 165, 163, 158, 175],
'Weight':[50, 68, 43, 52, 47, 49, 'x']})
df.replace({'x': None}, inplace=True) # replace missing values (x) with NaN
print(df)
df.dropna(inplace=True) # drop rows with NaN
print(df)
# Handling missing values by imputing missing values with statistic
import pandas as pd
df = pd.DataFrame({'Age': [17, 23, 'x', 38, 54, 67, 32],
'Height': [160, 172, 150, 165, 163, 158, 175],
'Weight':[50, 68, 43, 52, 47, 49, 'x']})
df.replace({'x': None}, inplace=True)
df['Age'].fillna(df['Age'].median(), inplace=True) # replace NaN with median
df['Weight'].fillna(df['Weight'].mean(), inplace=True) # replace NaN with mean
print(df)
# Scale data (between 0 and 1)
from pandas import read_csv
from sklearn.preprocessing import MinMaxScaler
names = ['preg', 'plas', 'pres', 'skin', 'test', 'mass', 'pedi', 'age', 'class']
df = read_csv('pima-indians-diabetes.data.csv', names=names)
array = df.values
# separate array into input and output components
X = array[:,:-1]
y = array[:,-1]
scaler = MinMaxScaler(feature_range=(0, 1))
scaler.fit(X)
scaledX = scaler.transform(X)
# Preview the scaled data
print(scaledX[:5,:])
# Standardize data (0 mean, 1 stdev)
from sklearn.preprocessing import StandardScaler
from pandas import read_csv
names = ['preg', 'plas', 'pres', 'skin', 'test', 'mass', 'pedi', 'age', 'class']
df = read_csv('pima-indians-diabetes.data.csv', names=names)
array = df.values
# separate array into input and output components
X = array[:, :-1]
y = array[:, -1]
scaler = StandardScaler()
scaledX = scaler.fit_transform(X)
# Preview transformed data
print(scaledX[:5, :])
# Robust scaling (0 median, 1 IQR)
from sklearn.preprocessing import RobustScaler
from pandas import read_csv
names = ['preg', 'plas', 'pres', 'skin', 'test', 'mass', 'pedi', 'age', 'class']
df = read_csv('pima-indians-diabetes.data.csv', names=names)
array = df.values
# separate array into input and output components
X = array[:,:-1]
y = array[:,-1]
scaler = RobustScaler()
scaledX = scaler.fit_transform(X)
# Preview transformed data
print(scaledX[0:5,:])
# Handling categorical data
import pandas as pd
df0 = pd.DataFrame({'year':[2015, 2017, 2013, 2018, 2020],
'make':['Toyota', 'Honda', 'Perodua', 'Hyundai', 'Toyota'],
'engine':[1.5, 1.8, 1.3, 1.6, 1.8],
'review':['moderate', 'good', 'poor', 'moderate', 'good']})
mapping = {'poor':1, 'moderate':2, 'good':3}
df0['review'] = df0['review'].map(mapping) # encode ordinal data
df0 = pd.get_dummies(df0) # encode nominal data
print(df0)
# Create 2 new features
from pandas import read_csv
from sklearn.preprocessing import MinMaxScaler
names = ['preg', 'plas', 'pres', 'skin', 'test', 'mass', 'pedi', 'age', 'class']
df = read_csv('pima-indians-diabetes.data.csv', names=names)
win_size = 3
df['plas_pres'] = df['plas'] + df['pres'] # new feature 1
df['mass_ave'] = df['mass'].rolling(win_size).mean() # new feature 2
df.head()
# Feature Selection with Univariate Selection
from pandas import read_csv
from sklearn.feature_selection import SelectKBest
# load data
names = ['preg', 'plas', 'pres', 'skin', 'test', 'mass', 'pedi', 'age', 'class']
df = read_csv('pima-indians-diabetes.data.csv', names=names)
array = df.values
X = array[:, :-1]
y = array[:, -1]
selector = SelectKBest(k=4)
features = selector.fit_transform(X, y)
selected = selector.get_support()
# Show selected features
print([names[i] for i in range(len(names)-1) if selected[i]])
# Feature Selection with RFE
from pandas import read_csv
from sklearn.feature_selection import RFE
from sklearn.tree import DecisionTreeClassifier
filename = 'pima-indians-diabetes.data.csv'
names = ['preg', 'plas', 'pres', 'skin', 'test', 'mass', 'pedi', 'age', 'class']
df = read_csv(filename, names=names)
array = df.values
X = array[:, :-1]
y = array[:, -1]
model = DecisionTreeClassifier()
rfe = RFE(model, 3)
features = rfe.fit_transform(X, y)
selected = rfe.get_support()
# Show selected features
print([names[i] for i in range(len(names)-1) if selected[i]])
# Dimensionality Reduction with PCA
from pandas import read_csv
from sklearn.decomposition import PCA
names = ['preg', 'plas', 'pres', 'skin', 'test', 'mass', 'pedi', 'age', 'class']
df = read_csv('pima-indians-diabetes.data.csv', names=names)
array = df.values
X = array[:, :-1]
y = array[:, -1]
pca = PCA(n_components=3)
features = pca.fit_transform(X)
# summarize components
print("Explained Variance: %s" % pca.explained_variance_ratio_)
# Evaluate using a train and a test set
from pandas import read_csv
from sklearn.model_selection import train_test_split as split
from sklearn.neighbors import KNeighborsClassifier
names = ['preg', 'plas', 'pres', 'skin', 'test', 'mass', 'pedi', 'age', 'class']
df = read_csv('pima-indians-diabetes.data.csv', names=names)
array = df.values
X = array[:, :-1]
y = array[:, -1]
X_train, X_test, y_train, y_test = split(X, y, test_size=0.33, random_state=42)
model = KNeighborsClassifier()
model.fit(X_train, y_train)
result = model.score(X_test, y_test)
print(f"Accuracy: {100 * result:.2f} %")
# Evaluate using Cross Validation
from pandas import read_csv
from sklearn.model_selection import KFold, cross_val_score
from sklearn.neighbors import KNeighborsClassifier
names = ['preg', 'plas', 'pres', 'skin', 'test', 'mass', 'pedi', 'age', 'class']
df = read_csv('pima-indians-diabetes.data.csv', names=names)
array = df.values
X = array[:, :-1]
y = array[:, -1]
model = KNeighborsClassifier()
kfold = KFold(n_splits=5, shuffle=True, random_state=42)
results = cross_val_score(model, X, y, cv=kfold)
print(f"Accuracy: {100*results.mean():.2f} % ({100*results.std():.2f})")
# Plot Confusion Matrix
from pandas import read_csv
from sklearn.model_selection import train_test_split as split
from sklearn.neighbors import KNeighborsClassifier
from sklearn.metrics import plot_confusion_matrix
names = ['preg', 'plas', 'pres', 'skin', 'test', 'mass', 'pedi', 'age', 'class']
df = read_csv('pima-indians-diabetes.data.csv', names=names)
array = df.values
X = array[:, :-1]
y = array[:, -1]
X_train, X_test, y_train, y_test = split(X, y, test_size=0.33, random_state=42)
model = KNeighborsClassifier()
model.fit(X_train, y_train)
plot_confusion_matrix(model, X_test, y_test)
# Classification report
from pandas import read_csv
from sklearn.model_selection import train_test_split as split
from sklearn.neighbors import KNeighborsClassifier
from yellowbrick.classifier import ClassificationReport
names = ['preg', 'plas', 'pres', 'skin', 'test', 'mass', 'pedi', 'age', 'class']
df = read_csv('pima-indians-diabetes.data.csv', names=names)
array = df.values
X = array[:, :-1]
y = array[:, -1]
X_train, X_test, y_train, y_test = split(X, y, test_size=0.33, random_state=42)
model = KNeighborsClassifier()
model.fit(X_train, y_train)
report = ClassificationReport(model)
report.score(X_test, y_test)
report.show()
# Grid Search for Hyperparameter Tuning
from pandas import read_csv
from sklearn.svm import SVC
from sklearn.model_selection import GridSearchCV
names = ['preg', 'plas', 'pres', 'skin', 'test', 'mass', 'pedi', 'age', 'class']
df = read_csv('pima-indians-diabetes.data.csv', names=names)
array = df.values
X = array[:, :-1]
y = array[:, -1]
params = dict(C=[0.001, 0.01, 0.1, 1, 10], gamma=[0.001, 0.01, 0.1, 1, 10])
model = SVC()
grid = GridSearchCV(model, params, n_jobs=-1, verbose=2)
grid.fit(X, y)
print(grid.best_score_)
print(grid.best_params_)
| 0.716814 | 0.955899 |
```
import numpy as np
class Wangzhe(object):
def __init__(self):
pass
def duizhanfangshi(self):
print('请选择人机或者多人对战!')
a = input('输入')
print(a)
self.a=a
def renwu(self):
print('请从典韦、赵云、鲁班中挑选一个人物!')
b = input('输入')
print(b)
self.b=b
def shuxing(self):
if self.b == '典韦':
print("典韦 战力6666 防御力5555")
elif self.b == '赵云':
print("赵云 战力9999 防御力8888")
else:
print("鲁班 战力11111 防御力33333")
def shengcheng(self):
print("随机生成对战玩家")
c = np.random.choice(['典韦','赵云','鲁班'])
print(c)
def kaishi(self):
print("请输入开始")
d = input('输入')
if d == "开始":
print("输入正确")
else:
print("输入有误!")
def jiazai(self):
print('正在加载,请耐心等待!')
jomei = Wangzhe()
jomei.duizhanfangshi()
jomei.renwu()
jomei.shuxing()
jomei.shengcheng()
jomei.kaishi()
jomei.jiazai()
def getpentagonalNumber(n):
a=0
c=0
for i in range(1,n,1):
a=int(i*(3*i-1)/2)
if c%10==0:
print()
print(a,end=" ")
c+=1
n=int(input("输入数字"))
getpentagonalNumber(n)
def sumdigits(n):
y=n%10
a=n//10%10
n=n//100
sum=y+a+n
return sum
n=int(input("输入"))
sumdigits(n)
def displaysortednumbers(a,b,c):
if a > b:
a,b = b,a
if a > c:
a,c = c,a
if b > c:
b,c = c,b
return a,b,c
a,b,c = map(int,input('输入三个整数').split(" "))
displaysortednumbers(a,b,c)
def numberofdayinyear():
c=31+28+31+30+31+30+31+31+30+31+30+31
for i in range(2010,2021):
if i%4 == 0 and i%100 != 0 or i%400 == 0:
print('%d年有%d天'%(i,c+1))
else:
print('%d年有%d天'%(i,c))
numberofdayinyear()
def printchars(ch1,ch2,n):
count = 0
for i in range(ord(ch1),ord(ch2)+1):
print(chr(i),end=" ")
count += 1
if count % n == 0:
print()
ch1 = input('请输入第一个字符:')
ch2 = input('请输入第二个字符:')
n = int(input('请输入每行打印的个数:'))
printchars(ch1,ch2,n)
import math
def distance(x1,y1,x2,y2):
a=x2-x1
b=y2-y1
c=math.sqrt((a**2)+(b**2))
return c
x1,y1,x2,y2=map(int,input('请输入坐标:').split())
distance(x1,y1,x2,y2)
import time
localtime = time.asctime(time.localtime(time.time()))
print("本地时间为:",localtime)
import random
def shaizi():
a=random.choice([1,2,3,4,5,6])
b=random.choice([1,2,3,4,5,6])
if a+b==2 or a+b==3 or a+b==12:
print('%d + %d = %d'%(a,b,a+b))
print('你输了')
elif a+b==7 or a+b==11:
print('%d + %d = %d' %(a,b,a+b))
print('你赢了')
else:
print('%d + %d = %d' %(a,b,a+b))
c=random.choice([1,2,3,4,5,6])
d=random.choice([1,2,3,4,5,6])
if c+d==7:
print('%d + %d = %d' %(c,d,c+d))
print('你输了')
elif c+d==a+b:
print('%d + %d = %d' %(c,d,c+d))
print('你赢了')
shaizi()
import math
def prime():
num=[]
i=2
for i in range(2,32):
j-2
for j in range(2,i):
if(i%j==0):
break
else:
num.append(i)
return num
def misen():
print('p 2^p-1')
a=prime()
for p in range(1,32):
for i in range(11):
if (2**p) 1--a[i]:
print("%d %d" %(p,a[i]))
misen()
```
|
github_jupyter
|
import numpy as np
class Wangzhe(object):
def __init__(self):
pass
def duizhanfangshi(self):
print('请选择人机或者多人对战!')
a = input('输入')
print(a)
self.a=a
def renwu(self):
print('请从典韦、赵云、鲁班中挑选一个人物!')
b = input('输入')
print(b)
self.b=b
def shuxing(self):
if self.b == '典韦':
print("典韦 战力6666 防御力5555")
elif self.b == '赵云':
print("赵云 战力9999 防御力8888")
else:
print("鲁班 战力11111 防御力33333")
def shengcheng(self):
print("随机生成对战玩家")
c = np.random.choice(['典韦','赵云','鲁班'])
print(c)
def kaishi(self):
print("请输入开始")
d = input('输入')
if d == "开始":
print("输入正确")
else:
print("输入有误!")
def jiazai(self):
print('正在加载,请耐心等待!')
jomei = Wangzhe()
jomei.duizhanfangshi()
jomei.renwu()
jomei.shuxing()
jomei.shengcheng()
jomei.kaishi()
jomei.jiazai()
def getpentagonalNumber(n):
a=0
c=0
for i in range(1,n,1):
a=int(i*(3*i-1)/2)
if c%10==0:
print()
print(a,end=" ")
c+=1
n=int(input("输入数字"))
getpentagonalNumber(n)
def sumdigits(n):
y=n%10
a=n//10%10
n=n//100
sum=y+a+n
return sum
n=int(input("输入"))
sumdigits(n)
def displaysortednumbers(a,b,c):
if a > b:
a,b = b,a
if a > c:
a,c = c,a
if b > c:
b,c = c,b
return a,b,c
a,b,c = map(int,input('输入三个整数').split(" "))
displaysortednumbers(a,b,c)
def numberofdayinyear():
c=31+28+31+30+31+30+31+31+30+31+30+31
for i in range(2010,2021):
if i%4 == 0 and i%100 != 0 or i%400 == 0:
print('%d年有%d天'%(i,c+1))
else:
print('%d年有%d天'%(i,c))
numberofdayinyear()
def printchars(ch1,ch2,n):
count = 0
for i in range(ord(ch1),ord(ch2)+1):
print(chr(i),end=" ")
count += 1
if count % n == 0:
print()
ch1 = input('请输入第一个字符:')
ch2 = input('请输入第二个字符:')
n = int(input('请输入每行打印的个数:'))
printchars(ch1,ch2,n)
import math
def distance(x1,y1,x2,y2):
a=x2-x1
b=y2-y1
c=math.sqrt((a**2)+(b**2))
return c
x1,y1,x2,y2=map(int,input('请输入坐标:').split())
distance(x1,y1,x2,y2)
import time
localtime = time.asctime(time.localtime(time.time()))
print("本地时间为:",localtime)
import random
def shaizi():
a=random.choice([1,2,3,4,5,6])
b=random.choice([1,2,3,4,5,6])
if a+b==2 or a+b==3 or a+b==12:
print('%d + %d = %d'%(a,b,a+b))
print('你输了')
elif a+b==7 or a+b==11:
print('%d + %d = %d' %(a,b,a+b))
print('你赢了')
else:
print('%d + %d = %d' %(a,b,a+b))
c=random.choice([1,2,3,4,5,6])
d=random.choice([1,2,3,4,5,6])
if c+d==7:
print('%d + %d = %d' %(c,d,c+d))
print('你输了')
elif c+d==a+b:
print('%d + %d = %d' %(c,d,c+d))
print('你赢了')
shaizi()
import math
def prime():
num=[]
i=2
for i in range(2,32):
j-2
for j in range(2,i):
if(i%j==0):
break
else:
num.append(i)
return num
def misen():
print('p 2^p-1')
a=prime()
for p in range(1,32):
for i in range(11):
if (2**p) 1--a[i]:
print("%d %d" %(p,a[i]))
misen()
| 0.061473 | 0.192881 |
# Linear Regression Implementation from Scratch
:label:`sec_linear_scratch`
Now that you understand the key ideas behind linear regression,
we can begin to work through a hands-on implementation in code.
In this section, (**we will implement the entire method from scratch,
including the data pipeline, the model,
the loss function, and the minibatch stochastic gradient descent optimizer.**)
While modern deep learning frameworks can automate nearly all of this work,
implementing things from scratch is the only way
to make sure that you really know what you are doing.
Moreover, when it comes time to customize models,
defining our own layers or loss functions,
understanding how things work under the hood will prove handy.
In this section, we will rely only on tensors and auto differentiation.
Afterwards, we will introduce a more concise implementation,
taking advantage of bells and whistles of deep learning frameworks.
```
%matplotlib inline
import random
import tensorflow as tf
from d2l import tensorflow as d2l
```
## Generating the Dataset
To keep things simple, we will [**construct an artificial dataset
according to a linear model with additive noise.**]
Our task will be to recover this model's parameters
using the finite set of examples contained in our dataset.
We will keep the data low-dimensional so we can visualize it easily.
In the following code snippet, we generate a dataset
containing 1000 examples, each consisting of 2 features
sampled from a standard normal distribution.
Thus our synthetic dataset will be a matrix
$\mathbf{X}\in \mathbb{R}^{1000 \times 2}$.
(**The true parameters generating our dataset will be
$\mathbf{w} = [2, -3.4]^\top$ and $b = 4.2$,
and**) our synthetic labels will be assigned according
to the following linear model with the noise term $\epsilon$:
(**$$\mathbf{y}= \mathbf{X} \mathbf{w} + b + \mathbf\epsilon.$$**)
You could think of $\epsilon$ as capturing potential
measurement errors on the features and labels.
We will assume that the standard assumptions hold and thus
that $\epsilon$ obeys a normal distribution with mean of 0.
To make our problem easy, we will set its standard deviation to 0.01.
The following code generates our synthetic dataset.
```
def synthetic_data(w, b, num_examples): #@save
"""Generate y = Xw + b + noise."""
X = tf.zeros((num_examples, w.shape[0]))
X += tf.random.normal(shape=X.shape)
y = tf.matmul(X, tf.reshape(w, (-1, 1))) + b
y += tf.random.normal(shape=y.shape, stddev=0.01)
y = tf.reshape(y, (-1, 1))
return X, y
true_w = tf.constant([2, -3.4])
true_b = 4.2
features, labels = synthetic_data(true_w, true_b, 1000)
```
Note that [**each row in `features` consists of a 2-dimensional data example
and that each row in `labels` consists of a 1-dimensional label value (a scalar).**]
```
print('features:', features[0], '\nlabel:', labels[0])
```
By generating a scatter plot using the second feature `features[:, 1]` and `labels`,
we can clearly observe the linear correlation between the two.
```
d2l.set_figsize()
# The semicolon is for displaying the plot only
d2l.plt.scatter(features[:, (1)].numpy(), labels.numpy(), 1);
```
## Reading the Dataset
Recall that training models consists of
making multiple passes over the dataset,
grabbing one minibatch of examples at a time,
and using them to update our model.
Since this process is so fundamental
to training machine learning algorithms,
it is worth defining a utility function
to shuffle the dataset and access it in minibatches.
In the following code, we [**define the `data_iter` function**] (~~that~~)
to demonstrate one possible implementation of this functionality.
The function (**takes a batch size, a matrix of features,
and a vector of labels, yielding minibatches of the size `batch_size`.**)
Each minibatch consists of a tuple of features and labels.
```
def data_iter(batch_size, features, labels):
num_examples = len(features)
indices = list(range(num_examples))
# The examples are read at random, in no particular order
random.shuffle(indices)
for i in range(0, num_examples, batch_size):
j = tf.constant(indices[i:min(i + batch_size, num_examples)])
yield tf.gather(features, j), tf.gather(labels, j)
```
In general, note that we want to use reasonably sized minibatches
to take advantage of the GPU hardware,
which excels at parallelizing operations.
Because each example can be fed through our models in parallel
and the gradient of the loss function for each example can also be taken in parallel,
GPUs allow us to process hundreds of examples in scarcely more time
than it might take to process just a single example.
To build some intuition, let us read and print
the first small batch of data examples.
The shape of the features in each minibatch tells us
both the minibatch size and the number of input features.
Likewise, our minibatch of labels will have a shape given by `batch_size`.
```
batch_size = 10
for X, y in data_iter(batch_size, features, labels):
print(X, '\n', y)
break
```
As we run the iteration, we obtain distinct minibatches
successively until the entire dataset has been exhausted (try this).
While the iteration implemented above is good for didactic purposes,
it is inefficient in ways that might get us in trouble on real problems.
For example, it requires that we load all the data in memory
and that we perform lots of random memory access.
The built-in iterators implemented in a deep learning framework
are considerably more efficient and they can deal
with both data stored in files and data fed via data streams.
## Initializing Model Parameters
[**Before we can begin optimizing our model's parameters**] by minibatch stochastic gradient descent,
(**we need to have some parameters in the first place.**)
In the following code, we initialize weights by sampling
random numbers from a normal distribution with mean 0
and a standard deviation of 0.01, and setting the bias to 0.
```
w = tf.Variable(tf.random.normal(shape=(2, 1), mean=0, stddev=0.01),
trainable=True)
b = tf.Variable(tf.zeros(1), trainable=True)
```
After initializing our parameters,
our next task is to update them until
they fit our data sufficiently well.
Each update requires taking the gradient
of our loss function with respect to the parameters.
Given this gradient, we can update each parameter
in the direction that may reduce the loss.
Since nobody wants to compute gradients explicitly
(this is tedious and error prone),
we use automatic differentiation,
as introduced in :numref:`sec_autograd`, to compute the gradient.
## Defining the Model
Next, we must [**define our model,
relating its inputs and parameters to its outputs.**]
Recall that to calculate the output of the linear model,
we simply take the matrix-vector dot product
of the input features $\mathbf{X}$ and the model weights $\mathbf{w}$,
and add the offset $b$ to each example.
Note that below $\mathbf{Xw}$ is a vector and $b$ is a scalar.
Recall the broadcasting mechanism as described in :numref:`subsec_broadcasting`.
When we add a vector and a scalar,
the scalar is added to each component of the vector.
```
def linreg(X, w, b): #@save
"""The linear regression model."""
return tf.matmul(X, w) + b
```
## Defining the Loss Function
Since [**updating our model requires taking
the gradient of our loss function,**]
we ought to (**define the loss function first.**)
Here we will use the squared loss function
as described in :numref:`sec_linear_regression`.
In the implementation, we need to transform the true value `y`
into the predicted value's shape `y_hat`.
The result returned by the following function
will also have the same shape as `y_hat`.
```
def squared_loss(y_hat, y): #@save
"""Squared loss."""
return (y_hat - tf.reshape(y, y_hat.shape))**2 / 2
```
## Defining the Optimization Algorithm
As we discussed in :numref:`sec_linear_regression`,
linear regression has a closed-form solution.
However, this is not a book about linear regression:
it is a book about deep learning.
Since none of the other models that this book introduces
can be solved analytically, we will take this opportunity to introduce your first working example of
minibatch stochastic gradient descent.
[~~Despite linear regression has a closed-form solution, other models in this book don't. Here we introduce minibatch stochastic gradient descent.~~]
At each step, using one minibatch randomly drawn from our dataset,
we will estimate the gradient of the loss with respect to our parameters.
Next, we will update our parameters
in the direction that may reduce the loss.
The following code applies the minibatch stochastic gradient descent update,
given a set of parameters, a learning rate, and a batch size.
The size of the update step is determined by the learning rate `lr`.
Because our loss is calculated as a sum over the minibatch of examples,
we normalize our step size by the batch size (`batch_size`),
so that the magnitude of a typical step size
does not depend heavily on our choice of the batch size.
```
def sgd(params, grads, lr, batch_size): #@save
"""Minibatch stochastic gradient descent."""
for param, grad in zip(params, grads):
param.assign_sub(lr * grad / batch_size)
```
## Training
Now that we have all of the parts in place,
we are ready to [**implement the main training loop.**]
It is crucial that you understand this code
because you will see nearly identical training loops
over and over again throughout your career in deep learning.
In each iteration, we will grab a minibatch of training examples,
and pass them through our model to obtain a set of predictions.
After calculating the loss, we initiate the backwards pass through the network,
storing the gradients with respect to each parameter.
Finally, we will call the optimization algorithm `sgd`
to update the model parameters.
In summary, we will execute the following loop:
* Initialize parameters $(\mathbf{w}, b)$
* Repeat until done
* Compute gradient $\mathbf{g} \leftarrow \partial_{(\mathbf{w},b)} \frac{1}{|\mathcal{B}|} \sum_{i \in \mathcal{B}} l(\mathbf{x}^{(i)}, y^{(i)}, \mathbf{w}, b)$
* Update parameters $(\mathbf{w}, b) \leftarrow (\mathbf{w}, b) - \eta \mathbf{g}$
In each *epoch*,
we will iterate through the entire dataset
(using the `data_iter` function) once
passing through every example in the training dataset
(assuming that the number of examples is divisible by the batch size).
The number of epochs `num_epochs` and the learning rate `lr` are both hyperparameters,
which we set here to 3 and 0.03, respectively.
Unfortunately, setting hyperparameters is tricky
and requires some adjustment by trial and error.
We elide these details for now but revise them
later in
:numref:`chap_optimization`.
```
lr = 0.03
num_epochs = 3
net = linreg
loss = squared_loss
for epoch in range(num_epochs):
for X, y in data_iter(batch_size, features, labels):
with tf.GradientTape() as g:
l = loss(net(X, w, b), y) # Minibatch loss in `X` and `y`
# Compute gradient on l with respect to [`w`, `b`]
dw, db = g.gradient(l, [w, b])
# Update parameters using their gradient
sgd([w, b], [dw, db], lr, batch_size)
train_l = loss(net(features, w, b), labels)
print(f'epoch {epoch + 1}, loss {float(tf.reduce_mean(train_l)):f}')
```
In this case, because we synthesized the dataset ourselves,
we know precisely what the true parameters are.
Thus, we can [**evaluate our success in training
by comparing the true parameters
with those that we learned**] through our training loop.
Indeed they turn out to be very close to each other.
```
print(f'error in estimating w: {true_w - tf.reshape(w, true_w.shape)}')
print(f'error in estimating b: {true_b - b}')
```
Note that we should not take it for granted
that we are able to recover the parameters perfectly.
However, in machine learning, we are typically less concerned
with recovering true underlying parameters,
and more concerned with parameters that lead to highly accurate prediction.
Fortunately, even on difficult optimization problems,
stochastic gradient descent can often find remarkably good solutions,
owing partly to the fact that, for deep networks,
there exist many configurations of the parameters
that lead to highly accurate prediction.
## Summary
* We saw how a deep network can be implemented and optimized from scratch, using just tensors and auto differentiation, without any need for defining layers or fancy optimizers.
* This section only scratches the surface of what is possible. In the following sections, we will describe additional models based on the concepts that we have just introduced and learn how to implement them more concisely.
## Exercises
1. What would happen if we were to initialize the weights to zero. Would the algorithm still work?
1. Assume that you are
[Georg Simon Ohm](https://en.wikipedia.org/wiki/Georg_Ohm) trying to come up
with a model between voltage and current. Can you use auto differentiation to learn the parameters of your model?
1. Can you use [Planck's Law](https://en.wikipedia.org/wiki/Planck%27s_law) to determine the temperature of an object using spectral energy density?
1. What are the problems you might encounter if you wanted to compute the second derivatives? How would you fix them?
1. Why is the `reshape` function needed in the `squared_loss` function?
1. Experiment using different learning rates to find out how fast the loss function value drops.
1. If the number of examples cannot be divided by the batch size, what happens to the `data_iter` function's behavior?
[Discussions](https://discuss.d2l.ai/t/201)
|
github_jupyter
|
%matplotlib inline
import random
import tensorflow as tf
from d2l import tensorflow as d2l
def synthetic_data(w, b, num_examples): #@save
"""Generate y = Xw + b + noise."""
X = tf.zeros((num_examples, w.shape[0]))
X += tf.random.normal(shape=X.shape)
y = tf.matmul(X, tf.reshape(w, (-1, 1))) + b
y += tf.random.normal(shape=y.shape, stddev=0.01)
y = tf.reshape(y, (-1, 1))
return X, y
true_w = tf.constant([2, -3.4])
true_b = 4.2
features, labels = synthetic_data(true_w, true_b, 1000)
print('features:', features[0], '\nlabel:', labels[0])
d2l.set_figsize()
# The semicolon is for displaying the plot only
d2l.plt.scatter(features[:, (1)].numpy(), labels.numpy(), 1);
def data_iter(batch_size, features, labels):
num_examples = len(features)
indices = list(range(num_examples))
# The examples are read at random, in no particular order
random.shuffle(indices)
for i in range(0, num_examples, batch_size):
j = tf.constant(indices[i:min(i + batch_size, num_examples)])
yield tf.gather(features, j), tf.gather(labels, j)
batch_size = 10
for X, y in data_iter(batch_size, features, labels):
print(X, '\n', y)
break
w = tf.Variable(tf.random.normal(shape=(2, 1), mean=0, stddev=0.01),
trainable=True)
b = tf.Variable(tf.zeros(1), trainable=True)
def linreg(X, w, b): #@save
"""The linear regression model."""
return tf.matmul(X, w) + b
def squared_loss(y_hat, y): #@save
"""Squared loss."""
return (y_hat - tf.reshape(y, y_hat.shape))**2 / 2
def sgd(params, grads, lr, batch_size): #@save
"""Minibatch stochastic gradient descent."""
for param, grad in zip(params, grads):
param.assign_sub(lr * grad / batch_size)
lr = 0.03
num_epochs = 3
net = linreg
loss = squared_loss
for epoch in range(num_epochs):
for X, y in data_iter(batch_size, features, labels):
with tf.GradientTape() as g:
l = loss(net(X, w, b), y) # Minibatch loss in `X` and `y`
# Compute gradient on l with respect to [`w`, `b`]
dw, db = g.gradient(l, [w, b])
# Update parameters using their gradient
sgd([w, b], [dw, db], lr, batch_size)
train_l = loss(net(features, w, b), labels)
print(f'epoch {epoch + 1}, loss {float(tf.reduce_mean(train_l)):f}')
print(f'error in estimating w: {true_w - tf.reshape(w, true_w.shape)}')
print(f'error in estimating b: {true_b - b}')
| 0.874198 | 0.994281 |
# ipywidgets
```
from __future__ import print_function
import sys
import pandas as pd
from io import StringIO
import numpy as np
import ipywidgets as widgets
from IPython.display import display
out = widgets.Output(layout={'border': '1px solid black'})
up = widgets.FileUpload(accept="", multiple=False)
eraser = widgets.SelectMultiple(
options=['tab','"'],
value=['tab'],
#rows=10,
description='Eraser: ',
disabled=False)
rows = widgets.IntSlider(
value=0,
step=1,
description='# of lines:',
disabled=False,
continuous_update=False,
orientation='horizontal',
readout=True,
readout_format='d')
button_preview = widgets.Button(
description='Preview',
disabled=False,
button_style='info',
tooltip='Click to Preview',
icon='search')
accordion = widgets.Accordion(children=[up])
accordion.set_title(0, 'File Selection')
accordion_box = widgets.VBox([
accordion,
widgets.HBox([button_preview]),
out
])
def content_parser():
if up.value == {}:
with out:
print('No CSV loaded')
else:
typ, content = "", ""
up_value = up.value
for i in up_value.keys():
typ = up_value[i]["metadata"]["type"]
if typ == "application/vnd.ms-excel":
content = up_value[i]["content"]
content_str = str(content, 'utf-8')
if eraser.value != {}:
for val in eraser.value:
if val == "tab":
content_str = content_str.replace("\t","")
else:
content_str = content_str.replace(val,"")
if content_str != "":
str_io = StringIO(content_str)
return str_io
def df_converter():
content = content_parser()
if content is not None:
df = pd.read_csv(content, index_col=False, skiprows=rows.value)
return df
else:
return None
def preview():
df = df_converter()
with out:
out.clear_output()
print('\n -----Now this is how your DF looks like:----- \n')
if df is not None:
print(df.head(10))
else:
print('Configuration is wrong/missing...')
def preview_clicked(b):
preview()
button_preview.on_click(preview_clicked)
accordion_box
# up.value
from IPython.display import HTML
HTML('''
<!-- Button -->
<form action="javascript:code_toggle()">
<input type="submit" value="Click here to toggle on/off the raw code.">
</form>
<!-- Script -->
<script>
// Replace global?
var code_show = true;
/**
* Simple function for toggle button.
*
* @param {void} - Nothing.
* @return {void} - Nothing.
*/
function code_toggle() {
if (code_show){
$('div.input').hide();
} else {
$('div.input').show();
}
// Set boolean to toggle back
code_show = !code_show;
}
$( document ).ready(code_toggle);
</script>
''')
```
|
github_jupyter
|
from __future__ import print_function
import sys
import pandas as pd
from io import StringIO
import numpy as np
import ipywidgets as widgets
from IPython.display import display
out = widgets.Output(layout={'border': '1px solid black'})
up = widgets.FileUpload(accept="", multiple=False)
eraser = widgets.SelectMultiple(
options=['tab','"'],
value=['tab'],
#rows=10,
description='Eraser: ',
disabled=False)
rows = widgets.IntSlider(
value=0,
step=1,
description='# of lines:',
disabled=False,
continuous_update=False,
orientation='horizontal',
readout=True,
readout_format='d')
button_preview = widgets.Button(
description='Preview',
disabled=False,
button_style='info',
tooltip='Click to Preview',
icon='search')
accordion = widgets.Accordion(children=[up])
accordion.set_title(0, 'File Selection')
accordion_box = widgets.VBox([
accordion,
widgets.HBox([button_preview]),
out
])
def content_parser():
if up.value == {}:
with out:
print('No CSV loaded')
else:
typ, content = "", ""
up_value = up.value
for i in up_value.keys():
typ = up_value[i]["metadata"]["type"]
if typ == "application/vnd.ms-excel":
content = up_value[i]["content"]
content_str = str(content, 'utf-8')
if eraser.value != {}:
for val in eraser.value:
if val == "tab":
content_str = content_str.replace("\t","")
else:
content_str = content_str.replace(val,"")
if content_str != "":
str_io = StringIO(content_str)
return str_io
def df_converter():
content = content_parser()
if content is not None:
df = pd.read_csv(content, index_col=False, skiprows=rows.value)
return df
else:
return None
def preview():
df = df_converter()
with out:
out.clear_output()
print('\n -----Now this is how your DF looks like:----- \n')
if df is not None:
print(df.head(10))
else:
print('Configuration is wrong/missing...')
def preview_clicked(b):
preview()
button_preview.on_click(preview_clicked)
accordion_box
# up.value
from IPython.display import HTML
HTML('''
<!-- Button -->
<form action="javascript:code_toggle()">
<input type="submit" value="Click here to toggle on/off the raw code.">
</form>
<!-- Script -->
<script>
// Replace global?
var code_show = true;
/**
* Simple function for toggle button.
*
* @param {void} - Nothing.
* @return {void} - Nothing.
*/
function code_toggle() {
if (code_show){
$('div.input').hide();
} else {
$('div.input').show();
}
// Set boolean to toggle back
code_show = !code_show;
}
$( document ).ready(code_toggle);
</script>
''')
| 0.399226 | 0.226891 |
## Sentiment Classification on Large Movie Reviews
[Sentiment Analysis](https://en.wikipedia.org/wiki/Sentiment_analysis) is understood as a classic natural language processing problem. In this example, a large moview review dataset was chosen from IMDB to do a sentiment classification task with some deep learning approaches. The labeled data set consists of 50,000 [IMDB](http://www.imdb.com/) movie reviews (good or bad), in which 25000 highly polar movie reviews for training, and 25,000 for testing. The dataset is originally collected by Stanford researchers and was used in a [2011 paper](http://ai.stanford.edu/~amaas/papers/wvSent_acl2011.pdf), and the highest accuray of 88.33% was achieved without using the unbalanced data. This example illustrates some deep learning approaches to do the sentiment classification with [BigDL](https://github.com/intel-analytics/BigDL) python API.
### Load the IMDB Dataset
The IMDB dataset need to be loaded into BigDL, note that the dataset has been pre-processed, and each review was encoded as a sequence of integers. Each integer represents the index of the overall frequency of dataset, for instance, '5' means the 5-th most frequent words occured in the data. It is very convinient to filter the words by some conditions, for example, to filter only the top 5,000 most common word and/or eliminate the top 30 most common words. Let's define functions to load the pre-processed data.
```
from bigdl.dataset import base
import numpy as np
def download_imdb(dest_dir):
"""Download pre-processed IMDB movie review data
:argument
dest_dir: destination directory to store the data
:return
The absolute path of the stored data
"""
file_name = "imdb.npz"
file_abs_path = base.maybe_download(file_name,
dest_dir,
'https://s3.amazonaws.com/text-datasets/imdb.npz')
return file_abs_path
def load_imdb(dest_dir='/tmp/.bigdl/dataset'):
"""Load IMDB dataset.
:argument
dest_dir: where to cache the data (relative to `~/.bigdl/dataset`).
:return
the train, test separated IMDB dataset.
"""
path = download_imdb(dest_dir)
f = np.load(path)
x_train = f['x_train']
y_train = f['y_train']
x_test = f['x_test']
y_test = f['y_test']
f.close()
return (x_train, y_train), (x_test, y_test)
print('Processing text dataset')
(x_train, y_train), (x_test, y_test) = load_imdb()
print('finished processing text')
```
In order to set a proper max sequence length, we need to go througth the property of the data and see the length distribution of each sentence in the dataset. A box and whisker plot is shown below for reviewing the length distribution in words.
```
import matplotlib
matplotlib.use('Agg')
%pylab inline
# Summarize review length
from matplotlib import pyplot
print("Review length: ")
X = np.concatenate((x_train, x_test), axis=0)
result = [len(x) for x in X]
print("Mean %.2f words (%f)" % (np.mean(result), np.std(result)))
# plot review length
# Create a figure instance
fig = pyplot.figure(1, figsize=(6, 6))
pyplot.boxplot(result)
pyplot.show()
```
Looking the box and whisker plot, the max length of a sample in words is 500, and the mean and median are below 250. According to the plot, we can probably cover the mass of the distribution with a clipped length of 400 to 500. Here we set the max sequence length of each sample as 500.
The corresponding vocabulary sorted by frequency is also required, for further embedding the words with pre-trained vectors. The downloaded vocabulary is in {word: index}, where each word as a key and the index as a value. It needs to be transformed into {index: word} format.
Let's define a function to obtain the vocabulary.
```
import json
def get_word_index(dest_dir='/tmp/.bigdl/dataset', ):
"""Retrieves the dictionary mapping word indices back to words.
:argument
path: where to cache the data (relative to `~/.bigdl/dataset`).
:return
The word index dictionary.
"""
file_name = "imdb_word_index.json"
path = base.maybe_download(file_name,
dest_dir,
source_url='https://s3.amazonaws.com/text-datasets/imdb_word_index.json')
f = open(path)
data = json.load(f)
f.close()
return data
print('Processing vocabulary')
word_idx = get_word_index()
idx_word = {v:k for k,v in word_idx.items()}
print('finished processing vocabulary')
```
### Text pre-processing
Before we train the network, some pre-processing steps need to be applied to the dataset.
Next let's go through the mechanisms that used to be applied to the data.
* We insert a `start_char` at the beginning of each sentence to mark the start point. We set it as `2` here, and each other word index will plus a constant `index_from` to differentiate some 'helper index' (eg. `start_char`, `oov_char`, etc.).
* A `max_words` variable is defined as the maximum index number (the least frequent word) included in the sequence. If the word index number is larger than `max_words`, it will be replaced by a out-of-vocabulary number `oov_char`, which is `3` here.
* Each word index sequence is restricted to the same length. We used left-padding here, which means the right (end) of the sequence will be keep as many as possible and drop the left (head) of the sequence if its length is more than pre-defined `sequence_len`, or padding the left (head) of the sequence with `padding_value`.
```
def replace_oov(x, oov_char, max_words):
"""
Replace the words out of vocabulary with `oov_char`
:param x: a sequence
:param max_words: the max number of words to include
:param oov_char: words out of vocabulary because of exceeding the `max_words`
limit will be replaced by this character
:return: The replaced sequence
"""
return [oov_char if w >= max_words else w for w in x]
def pad_sequence(x, fill_value, length):
"""
Pads each sequence to the same length
:param x: a sequence
:param fill_value: pad the sequence with this value
:param length: pad sequence to the length
:return: the padded sequence
"""
if len(x) >= length:
return x[(len(x) - length):]
else:
return [fill_value] * (length - len(x)) + x
def to_sample(features, label):
"""
Wrap the `features` and `label` to a training sample object
:param features: features of a sample
:param label: label of a sample
:return: a sample object including features and label
"""
return Sample.from_ndarray(np.array(features, dtype='float'), np.array(label))
padding_value = 1
start_char = 2
oov_char = 3
index_from = 3
max_words = 5000
sequence_len = 500
print('start transformation')
from bigdl.util.common import get_spark_context
sc = get_spark_context()
train_rdd = sc.parallelize(zip(x_train, y_train), 2) \
.map(lambda record: ([start_char] + [w + index_from for w in record[0]], record[1])) \
.map(lambda record: (replace_oov(record[0], oov_char, max_words), record[1])) \
.map(lambda record: (pad_sequence(record[0], padding_value, sequence_len), record[1])) \
.map(lambda record: to_sample(record[0], record[1]))
test_rdd = sc.parallelize(zip(x_test, y_test), 2) \
.map(lambda record: ([start_char] + [w + index_from for w in record[0]], record[1])) \
.map(lambda record: (replace_oov(record[0], oov_char, max_words), record[1])) \
.map(lambda record: (pad_sequence(record[0], padding_value, sequence_len), record[1])) \
.map(lambda record: to_sample(record[0], record[1]))
print('finish transformation')
```
### Word Embedding
[Word embedding](https://en.wikipedia.org/wiki/Word_embedding) is a recent breakthrough in natural language field. The key idea is to encode words and phrases into distributed representations in the format of word vectors, which means each word is represented as a vector. There are two widely used word vector training alogirhms, one is published by Google called [word to vector](https://arxiv.org/abs/1310.4546), the other is published by Standford called [Glove](https://nlp.stanford.edu/projects/glove/). In this example, pre-trained glove is loaded into a lookup table and will be fine-tuned during the training process. BigDL provides a method to download and load glove in `news20` package.
```
from bigdl.dataset import news20
import itertools
embedding_dim = 100
print('loading glove')
glove = news20.get_glove_w2v(source_dir='/tmp/.bigdl/dataset', dim=embedding_dim)
print('finish loading glove')
```
For each word whose index less than the `max_word` should try to match its embedding and store in an array.
With regard to those words which can not be found in glove, we randomly sample it from a [-0.05, 0.05] uniform distribution.
BigDL usually use a `LookupTable` layer to do word embedding, so the matrix will be loaded to the LookupTable by seting the weight.
```
print('processing glove')
w2v = [glove.get(idx_word.get(i - index_from), np.random.uniform(-0.05, 0.05, embedding_dim))
for i in range(1, max_words + 1)]
w2v = np.array(list(itertools.chain(*np.array(w2v, dtype='float'))), dtype='float') \
.reshape([max_words, embedding_dim])
print('finish processing glove')
```
### Build models
Next, let's build some deep learning models for the sentiment classification.
As an example, several deep learning models are illustrated for tutorial, comparison and demonstration.
**LSTM**, **GRU**, **Bi-LSTM**, **CNN** and **CNN + LSTM** models are implemented as options. To decide which model to use, just assign model_type the corresponding string.
```
from bigdl.nn.layer import *
from bigdl.util.common import *
p = 0.2
def build_model(w2v):
model = Sequential()
embedding = LookupTable(max_words, embedding_dim)
embedding.set_weights([w2v])
model.add(embedding)
if model_type.lower() == "gru":
model.add(Recurrent()
.add(GRU(embedding_dim, 128, p))) \
.add(Select(2, -1))
elif model_type.lower() == "lstm":
model.add(Recurrent()
.add(LSTM(embedding_dim, 128, p)))\
.add(Select(2, -1))
elif model_type.lower() == "bi_lstm":
model.add(BiRecurrent(CAddTable())
.add(LSTM(embedding_dim, 128, p)))\
.add(Select(2, -1))
elif model_type.lower() == "cnn":
model.add(Transpose([(2, 3)]))\
.add(Dropout(p))\
.add(Reshape([embedding_dim, 1, sequence_len]))\
.add(SpatialConvolution(embedding_dim, 128, 5, 1))\
.add(ReLU())\
.add(SpatialMaxPooling(sequence_len - 5 + 1, 1, 1, 1))\
.add(Reshape([128]))
elif model_type.lower() == "cnn_lstm":
model.add(Transpose([(2, 3)]))\
.add(Dropout(p))\
.add(Reshape([embedding_dim, 1, sequence_len])) \
.add(SpatialConvolution(embedding_dim, 64, 5, 1)) \
.add(ReLU()) \
.add(SpatialMaxPooling(4, 1, 1, 1)) \
.add(Squeeze(3)) \
.add(Transpose([(2, 3)])) \
.add(Recurrent()
.add(LSTM(64, 128, p))) \
.add(Select(2, -1))
model.add(Linear(128, 100))\
.add(Dropout(0.2))\
.add(ReLU())\
.add(Linear(100, 1))\
.add(Sigmoid())
return model
```
### Optimization
`Optimizer` need to be created to optimise the model.
Here we use the `CNN` model.
```
from bigdl.optim.optimizer import *
from bigdl.nn.criterion import *
# max_epoch = 4
max_epoch = 1
batch_size = 64
model_type = 'gru'
init_engine()
optimizer = Optimizer(
model=build_model(w2v),
training_rdd=train_rdd,
criterion=BCECriterion(),
end_trigger=MaxEpoch(max_epoch),
batch_size=batch_size,
optim_method=Adam())
optimizer.set_validation(
batch_size=batch_size,
val_rdd=test_rdd,
trigger=EveryEpoch(),
val_method=Top1Accuracy())
```
To make the training process be visualized by TensorBoard, training summaries should be saved as a format of logs.
```
import datetime as dt
logdir = '/tmp/.bigdl/'
app_name = 'adam-' + dt.datetime.now().strftime("%Y%m%d-%H%M%S")
train_summary = TrainSummary(log_dir=logdir, app_name=app_name)
train_summary.set_summary_trigger("Parameters", SeveralIteration(50))
val_summary = ValidationSummary(log_dir=logdir, app_name=app_name)
optimizer.set_train_summary(train_summary)
optimizer.set_val_summary(val_summary)
```
Now, let's start training!
```
%%time
train_model = optimizer.optimize()
print ("Optimization Done.")
```
### Test
Validation accuracy is shown in the training log, here let's get the accuracy on validation set by hand.
Predict the `test_rdd` (validation set data), and obtain the predicted label and ground truth label in the list.
```
predictions = train_model.predict(test_rdd)
def map_predict_label(l):
if l > 0.5:
return 1
else:
return 0
def map_groundtruth_label(l):
return l.to_ndarray()[0]
y_pred = np.array([ map_predict_label(s) for s in predictions.collect()])
y_true = np.array([map_groundtruth_label(s.label) for s in test_rdd.collect()])
```
Then let's see the prediction accuracy on validation set.
```
correct = 0
for i in range(0, y_pred.size):
if (y_pred[i] == y_true[i]):
correct += 1
accuracy = float(correct) / y_pred.size
print ('Prediction accuracy on validation set is: ', accuracy)
```
Show the confusion matrix
```
matplotlib.use('Agg')
%pylab inline
import matplotlib.pyplot as plt
import seaborn as sn
import pandas as pd
from sklearn.metrics import confusion_matrix
cm = confusion_matrix(y_true, y_pred)
cm.shape
df_cm = pd.DataFrame(cm)
plt.figure(figsize = (5,4))
sn.heatmap(df_cm, annot=True,fmt='d')
```
Because of the limitation of ariticle length, not all the results of optional models can be shown respectively. Please try other provided optional models to see the results. If you are interested in optimizing the results, try different training parameters which may make inpacts on the result, such as the max sequence length, batch size, training epochs, preprocessing schemes, optimization methods and so on. Among the models, CNN training would be much quicker. Note that the LSTM and it variants (eg. GRU) are difficult to train, even a unsuitable batch size may cause the model not converge. In addition it is prone to overfitting, please try different dropout threshold and/or add regularizers.
|
github_jupyter
|
from bigdl.dataset import base
import numpy as np
def download_imdb(dest_dir):
"""Download pre-processed IMDB movie review data
:argument
dest_dir: destination directory to store the data
:return
The absolute path of the stored data
"""
file_name = "imdb.npz"
file_abs_path = base.maybe_download(file_name,
dest_dir,
'https://s3.amazonaws.com/text-datasets/imdb.npz')
return file_abs_path
def load_imdb(dest_dir='/tmp/.bigdl/dataset'):
"""Load IMDB dataset.
:argument
dest_dir: where to cache the data (relative to `~/.bigdl/dataset`).
:return
the train, test separated IMDB dataset.
"""
path = download_imdb(dest_dir)
f = np.load(path)
x_train = f['x_train']
y_train = f['y_train']
x_test = f['x_test']
y_test = f['y_test']
f.close()
return (x_train, y_train), (x_test, y_test)
print('Processing text dataset')
(x_train, y_train), (x_test, y_test) = load_imdb()
print('finished processing text')
import matplotlib
matplotlib.use('Agg')
%pylab inline
# Summarize review length
from matplotlib import pyplot
print("Review length: ")
X = np.concatenate((x_train, x_test), axis=0)
result = [len(x) for x in X]
print("Mean %.2f words (%f)" % (np.mean(result), np.std(result)))
# plot review length
# Create a figure instance
fig = pyplot.figure(1, figsize=(6, 6))
pyplot.boxplot(result)
pyplot.show()
import json
def get_word_index(dest_dir='/tmp/.bigdl/dataset', ):
"""Retrieves the dictionary mapping word indices back to words.
:argument
path: where to cache the data (relative to `~/.bigdl/dataset`).
:return
The word index dictionary.
"""
file_name = "imdb_word_index.json"
path = base.maybe_download(file_name,
dest_dir,
source_url='https://s3.amazonaws.com/text-datasets/imdb_word_index.json')
f = open(path)
data = json.load(f)
f.close()
return data
print('Processing vocabulary')
word_idx = get_word_index()
idx_word = {v:k for k,v in word_idx.items()}
print('finished processing vocabulary')
def replace_oov(x, oov_char, max_words):
"""
Replace the words out of vocabulary with `oov_char`
:param x: a sequence
:param max_words: the max number of words to include
:param oov_char: words out of vocabulary because of exceeding the `max_words`
limit will be replaced by this character
:return: The replaced sequence
"""
return [oov_char if w >= max_words else w for w in x]
def pad_sequence(x, fill_value, length):
"""
Pads each sequence to the same length
:param x: a sequence
:param fill_value: pad the sequence with this value
:param length: pad sequence to the length
:return: the padded sequence
"""
if len(x) >= length:
return x[(len(x) - length):]
else:
return [fill_value] * (length - len(x)) + x
def to_sample(features, label):
"""
Wrap the `features` and `label` to a training sample object
:param features: features of a sample
:param label: label of a sample
:return: a sample object including features and label
"""
return Sample.from_ndarray(np.array(features, dtype='float'), np.array(label))
padding_value = 1
start_char = 2
oov_char = 3
index_from = 3
max_words = 5000
sequence_len = 500
print('start transformation')
from bigdl.util.common import get_spark_context
sc = get_spark_context()
train_rdd = sc.parallelize(zip(x_train, y_train), 2) \
.map(lambda record: ([start_char] + [w + index_from for w in record[0]], record[1])) \
.map(lambda record: (replace_oov(record[0], oov_char, max_words), record[1])) \
.map(lambda record: (pad_sequence(record[0], padding_value, sequence_len), record[1])) \
.map(lambda record: to_sample(record[0], record[1]))
test_rdd = sc.parallelize(zip(x_test, y_test), 2) \
.map(lambda record: ([start_char] + [w + index_from for w in record[0]], record[1])) \
.map(lambda record: (replace_oov(record[0], oov_char, max_words), record[1])) \
.map(lambda record: (pad_sequence(record[0], padding_value, sequence_len), record[1])) \
.map(lambda record: to_sample(record[0], record[1]))
print('finish transformation')
from bigdl.dataset import news20
import itertools
embedding_dim = 100
print('loading glove')
glove = news20.get_glove_w2v(source_dir='/tmp/.bigdl/dataset', dim=embedding_dim)
print('finish loading glove')
print('processing glove')
w2v = [glove.get(idx_word.get(i - index_from), np.random.uniform(-0.05, 0.05, embedding_dim))
for i in range(1, max_words + 1)]
w2v = np.array(list(itertools.chain(*np.array(w2v, dtype='float'))), dtype='float') \
.reshape([max_words, embedding_dim])
print('finish processing glove')
from bigdl.nn.layer import *
from bigdl.util.common import *
p = 0.2
def build_model(w2v):
model = Sequential()
embedding = LookupTable(max_words, embedding_dim)
embedding.set_weights([w2v])
model.add(embedding)
if model_type.lower() == "gru":
model.add(Recurrent()
.add(GRU(embedding_dim, 128, p))) \
.add(Select(2, -1))
elif model_type.lower() == "lstm":
model.add(Recurrent()
.add(LSTM(embedding_dim, 128, p)))\
.add(Select(2, -1))
elif model_type.lower() == "bi_lstm":
model.add(BiRecurrent(CAddTable())
.add(LSTM(embedding_dim, 128, p)))\
.add(Select(2, -1))
elif model_type.lower() == "cnn":
model.add(Transpose([(2, 3)]))\
.add(Dropout(p))\
.add(Reshape([embedding_dim, 1, sequence_len]))\
.add(SpatialConvolution(embedding_dim, 128, 5, 1))\
.add(ReLU())\
.add(SpatialMaxPooling(sequence_len - 5 + 1, 1, 1, 1))\
.add(Reshape([128]))
elif model_type.lower() == "cnn_lstm":
model.add(Transpose([(2, 3)]))\
.add(Dropout(p))\
.add(Reshape([embedding_dim, 1, sequence_len])) \
.add(SpatialConvolution(embedding_dim, 64, 5, 1)) \
.add(ReLU()) \
.add(SpatialMaxPooling(4, 1, 1, 1)) \
.add(Squeeze(3)) \
.add(Transpose([(2, 3)])) \
.add(Recurrent()
.add(LSTM(64, 128, p))) \
.add(Select(2, -1))
model.add(Linear(128, 100))\
.add(Dropout(0.2))\
.add(ReLU())\
.add(Linear(100, 1))\
.add(Sigmoid())
return model
from bigdl.optim.optimizer import *
from bigdl.nn.criterion import *
# max_epoch = 4
max_epoch = 1
batch_size = 64
model_type = 'gru'
init_engine()
optimizer = Optimizer(
model=build_model(w2v),
training_rdd=train_rdd,
criterion=BCECriterion(),
end_trigger=MaxEpoch(max_epoch),
batch_size=batch_size,
optim_method=Adam())
optimizer.set_validation(
batch_size=batch_size,
val_rdd=test_rdd,
trigger=EveryEpoch(),
val_method=Top1Accuracy())
import datetime as dt
logdir = '/tmp/.bigdl/'
app_name = 'adam-' + dt.datetime.now().strftime("%Y%m%d-%H%M%S")
train_summary = TrainSummary(log_dir=logdir, app_name=app_name)
train_summary.set_summary_trigger("Parameters", SeveralIteration(50))
val_summary = ValidationSummary(log_dir=logdir, app_name=app_name)
optimizer.set_train_summary(train_summary)
optimizer.set_val_summary(val_summary)
%%time
train_model = optimizer.optimize()
print ("Optimization Done.")
predictions = train_model.predict(test_rdd)
def map_predict_label(l):
if l > 0.5:
return 1
else:
return 0
def map_groundtruth_label(l):
return l.to_ndarray()[0]
y_pred = np.array([ map_predict_label(s) for s in predictions.collect()])
y_true = np.array([map_groundtruth_label(s.label) for s in test_rdd.collect()])
correct = 0
for i in range(0, y_pred.size):
if (y_pred[i] == y_true[i]):
correct += 1
accuracy = float(correct) / y_pred.size
print ('Prediction accuracy on validation set is: ', accuracy)
matplotlib.use('Agg')
%pylab inline
import matplotlib.pyplot as plt
import seaborn as sn
import pandas as pd
from sklearn.metrics import confusion_matrix
cm = confusion_matrix(y_true, y_pred)
cm.shape
df_cm = pd.DataFrame(cm)
plt.figure(figsize = (5,4))
sn.heatmap(df_cm, annot=True,fmt='d')
| 0.744378 | 0.984956 |
# Predicting Boston Housing Prices
## Updating a model using SageMaker
_Deep Learning Nanodegree Program | Deployment_
---
In this notebook, we will continue working with the [Boston Housing Dataset](https://www.cs.toronto.edu/~delve/data/boston/bostonDetail.html). Our goal in this notebook will be to train two different models and to use SageMaker to switch a deployed endpoint from using one model to the other. One of the benefits of using SageMaker to do this is that we can make the change without interrupting service. What this means is that we can continue sending data to the endpoint and at no point will that endpoint disappear.
## General Outline
Typically, when using a notebook instance with SageMaker, you will proceed through the following steps. Of course, not every step will need to be done with each project. Also, there is quite a lot of room for variation in many of the steps, as you will see throughout these lessons.
1. Download or otherwise retrieve the data.
2. Process / Prepare the data.
3. Upload the processed data to S3.
4. Train a chosen model.
5. Test the trained model (typically using a batch transform job).
6. Deploy the trained model.
7. Use the deployed model.
In this notebook we will be skipping step 5, testing the model. In addition, we will perform steps 4, 6 and 7 multiple times with different models.
```
# Make sure that we use SageMaker 1.x
!pip install sagemaker==1.72.0
```
## Step 0: Setting up the notebook
We begin by setting up all of the necessary bits required to run our notebook. To start that means loading all of the Python modules we will need.
```
%matplotlib inline
import os
import numpy as np
import pandas as pd
from pprint import pprint
import matplotlib.pyplot as plt
from time import gmtime, strftime
from sklearn.datasets import load_boston
import sklearn.model_selection
```
In addition to the modules above, we need to import the various bits of SageMaker that we will be using.
```
import sagemaker
from sagemaker import get_execution_role
from sagemaker.amazon.amazon_estimator import get_image_uri
from sagemaker.predictor import csv_serializer
# This is an object that represents the SageMaker session that we are currently operating in. This
# object contains some useful information that we will need to access later such as our region.
session = sagemaker.Session()
# This is an object that represents the IAM role that we are currently assigned. When we construct
# and launch the training job later we will need to tell it what IAM role it should have. Since our
# use case is relatively simple we will simply assign the training job the role we currently have.
role = get_execution_role()
```
## Step 1: Downloading the data
Fortunately, this dataset can be retrieved using sklearn and so this step is relatively straightforward.
```
boston = load_boston()
```
## Step 2: Preparing and splitting the data
Given that this is clean tabular data, we don't need to do any processing. However, we do need to split the rows in the dataset up into train, test and validation sets.
```
# First we package up the input data and the target variable (the median value) as pandas dataframes. This
# will make saving the data to a file a little easier later on.
X_bos_pd = pd.DataFrame(boston.data, columns=boston.feature_names)
Y_bos_pd = pd.DataFrame(boston.target)
# We split the dataset into 2/3 training and 1/3 testing sets.
X_train, X_test, Y_train, Y_test = sklearn.model_selection.train_test_split(X_bos_pd, Y_bos_pd, test_size=0.33)
# Then we split the training set further into 2/3 training and 1/3 validation sets.
X_train, X_val, Y_train, Y_val = sklearn.model_selection.train_test_split(X_train, Y_train, test_size=0.33)
```
## Step 3: Uploading the training and validation files to S3
When a training job is constructed using SageMaker, a container is executed which performs the training operation. This container is given access to data that is stored in S3. This means that we need to upload the data we want to use for training to S3. We can use the SageMaker API to do this and hide some of the details.
### Save the data locally
First we need to create the train and validation csv files which we will then upload to S3.
```
# This is our local data directory. We need to make sure that it exists.
data_dir = '../data/boston'
if not os.path.exists(data_dir):
os.makedirs(data_dir)
# We use pandas to save our train and validation data to csv files. Note that we make sure not to include header
# information or an index as this is required by the built in algorithms provided by Amazon. Also, it is assumed
# that the first entry in each row is the target variable.
pd.concat([Y_val, X_val], axis=1).to_csv(os.path.join(data_dir, 'validation.csv'), header=False, index=False)
pd.concat([Y_train, X_train], axis=1).to_csv(os.path.join(data_dir, 'train.csv'), header=False, index=False)
```
### Upload to S3
Since we are currently running inside of a SageMaker session, we can use the object which represents this session to upload our data to the 'default' S3 bucket. Note that it is good practice to provide a custom prefix (essentially an S3 folder) to make sure that you don't accidentally interfere with data uploaded from some other notebook or project.
```
prefix = 'boston-update-endpoints'
val_location = session.upload_data(os.path.join(data_dir, 'validation.csv'), key_prefix=prefix)
train_location = session.upload_data(os.path.join(data_dir, 'train.csv'), key_prefix=prefix)
```
## Step 4 (A): Train the XGBoost model
Now that we have the training and validation data uploaded to S3, we can construct our XGBoost model and train it. We will be making use of the high level SageMaker API to do this which will make the resulting code a little easier to read at the cost of some flexibility.
To construct an estimator, the object which we wish to train, we need to provide the location of a container which contains the training code. Since we are using a built in algorithm this container is provided by Amazon. However, the full name of the container is a bit lengthy and depends on the region that we are operating in. Fortunately, SageMaker provides a useful utility method called `get_image_uri` that constructs the image name for us.
To use the `get_image_uri` method we need to provide it with our current region, which can be obtained from the session object, and the name of the algorithm we wish to use. In this notebook we will be using XGBoost however you could try another algorithm if you wish. The list of built in algorithms can be found in the list of [Common Parameters](https://docs.aws.amazon.com/sagemaker/latest/dg/sagemaker-algo-docker-registry-paths.html).
```
# As stated above, we use this utility method to construct the image name for the training container.
xgb_container = get_image_uri(session.boto_region_name, 'xgboost')
# Now that we know which container to use, we can construct the estimator object.
xgb = sagemaker.estimator.Estimator(xgb_container, # The name of the training container
role, # The IAM role to use (our current role in this case)
train_instance_count=1, # The number of instances to use for training
train_instance_type='ml.m4.xlarge', # The type of instance ot use for training
output_path='s3://{}/{}/output'.format(session.default_bucket(), prefix),
# Where to save the output (the model artifacts)
sagemaker_session=session) # The current SageMaker session
```
Before asking SageMaker to begin the training job, we should probably set any model specific hyperparameters. There are quite a few that can be set when using the XGBoost algorithm, below are just a few of them. If you would like to change the hyperparameters below or modify additional ones you can find additional information on the [XGBoost hyperparameter page](https://docs.aws.amazon.com/sagemaker/latest/dg/xgboost_hyperparameters.html)
```
xgb.set_hyperparameters(max_depth=5,
eta=0.2,
gamma=4,
min_child_weight=6,
subsample=0.8,
objective='reg:linear',
early_stopping_rounds=10,
num_round=200)
```
Now that we have our estimator object completely set up, it is time to train it. To do this we make sure that SageMaker knows our input data is in csv format and then execute the `fit` method.
```
# This is a wrapper around the location of our train and validation data, to make sure that SageMaker
# knows our data is in csv format.
s3_input_train = sagemaker.s3_input(s3_data=train_location, content_type='text/csv')
s3_input_validation = sagemaker.s3_input(s3_data=val_location, content_type='text/csv')
xgb.fit({'train': s3_input_train, 'validation': s3_input_validation})
```
## Step 5: Test the trained model
We will be skipping this step for now.
## Step 6 (A): Deploy the trained model
Even though we used the high level approach to construct and train the XGBoost model, we will be using the lower level approach to deploy it. One of the reasons for this is so that we have additional control over how the endpoint is constructed. This will be a little more clear later on when construct more advanced endpoints.
### Build the model
Of course, before we can deploy the model, we need to first create it. The `fit` method that we used earlier created some model artifacts and we can use these to construct a model object.
```
# Remember that a model needs to have a unique name
xgb_model_name = "boston-update-xgboost-model" + strftime("%Y-%m-%d-%H-%M-%S", gmtime())
# We also need to tell SageMaker which container should be used for inference and where it should
# retrieve the model artifacts from. In our case, the xgboost container that we used for training
# can also be used for inference and the model artifacts come from the previous call to fit.
xgb_primary_container = {
"Image": xgb_container,
"ModelDataUrl": xgb.model_data
}
# And lastly we construct the SageMaker model
xgb_model_info = session.sagemaker_client.create_model(
ModelName = xgb_model_name,
ExecutionRoleArn = role,
PrimaryContainer = xgb_primary_container)
```
### Create the endpoint configuration
Once we have a model we can start putting together the endpoint. Recall that to do this we need to first create an endpoint configuration, essentially the blueprint that SageMaker will use to build the endpoint itself.
```
# As before, we need to give our endpoint configuration a name which should be unique
xgb_endpoint_config_name = "boston-update-xgboost-endpoint-config-" + strftime("%Y-%m-%d-%H-%M-%S", gmtime())
# And then we ask SageMaker to construct the endpoint configuration
xgb_endpoint_config_info = session.sagemaker_client.create_endpoint_config(
EndpointConfigName = xgb_endpoint_config_name,
ProductionVariants = [{
"InstanceType": "ml.m4.xlarge",
"InitialVariantWeight": 1,
"InitialInstanceCount": 1,
"ModelName": xgb_model_name,
"VariantName": "XGB-Model"
}])
```
### Deploy the endpoint
Now that the endpoint configuration has been created, we can ask SageMaker to build our endpoint.
**Note:** This is a friendly (repeated) reminder that you are about to deploy an endpoint. Make sure that you shut it down once you've finished with it!
```
# Again, we need a unique name for our endpoint
endpoint_name = "boston-update-endpoint-" + strftime("%Y-%m-%d-%H-%M-%S", gmtime())
# And then we can deploy our endpoint
endpoint_info = session.sagemaker_client.create_endpoint(
EndpointName = endpoint_name,
EndpointConfigName = xgb_endpoint_config_name)
endpoint_dec = session.wait_for_endpoint(endpoint_name)
```
## Step 7 (A): Use the model
Now that our model is trained and deployed we can send some test data to it and evaluate the results.
```
response = session.sagemaker_runtime_client.invoke_endpoint(
EndpointName = endpoint_name,
ContentType = 'text/csv',
Body = ','.join(map(str, X_test.values[0])))
pprint(response)
result = response['Body'].read().decode("utf-8")
pprint(result)
Y_test.values[0]
```
## Shut down the endpoint
Now that we know that the XGBoost endpoint works, we can shut it down. We will make use of it again later.
```
session.sagemaker_client.delete_endpoint(EndpointName = endpoint_name)
```
## Step 4 (B): Train the Linear model
Suppose we are working in an environment where the XGBoost model that we trained earlier is becoming too costly. Perhaps the number of calls to our endpoint has increased and the length of time it takes to perform inference with the XGBoost model is becoming problematic.
A possible solution might be to train a simpler model to see if it performs nearly as well. In our case, we will construct a linear model. The process of doing this is the same as for creating the XGBoost model that we created earlier, although there are different hyperparameters that we need to set.
```
# Similar to the XGBoost model, we will use the utility method to construct the image name for the training container.
linear_container = get_image_uri(session.boto_region_name, 'linear-learner')
# Now that we know which container to use, we can construct the estimator object.
linear = sagemaker.estimator.Estimator(linear_container, # The name of the training container
role, # The IAM role to use (our current role in this case)
train_instance_count=1, # The number of instances to use for training
train_instance_type='ml.m4.xlarge', # The type of instance ot use for training
output_path='s3://{}/{}/output'.format(session.default_bucket(), prefix),
# Where to save the output (the model artifacts)
sagemaker_session=session) # The current SageMaker session
```
Before asking SageMaker to train our model, we need to set some hyperparameters. In this case we will be using a linear model so the number of hyperparameters we need to set is much fewer. For more details see the [Linear model hyperparameter page](https://docs.aws.amazon.com/sagemaker/latest/dg/ll_hyperparameters.html)
```
linear.set_hyperparameters(feature_dim=13, # Our data has 13 feature columns
predictor_type='regressor', # We wish to create a regression model
mini_batch_size=200) # Here we set how many samples to look at in each iteration
```
Now that the hyperparameters have been set, we can ask SageMaker to fit the linear model to our data.
```
linear.fit({'train': s3_input_train, 'validation': s3_input_validation})
```
## Step 6 (B): Deploy the trained model
Similar to the XGBoost model, now that we've fit the model we need to deploy it. Also like the XGBoost model, we will use the lower level approach so that we have more control over the endpoint that gets created.
### Build the model
Of course, before we can deploy the model, we need to first create it. The `fit` method that we used earlier created some model artifacts and we can use these to construct a model object.
```
# First, we create a unique model name
linear_model_name = "boston-update-linear-model" + strftime("%Y-%m-%d-%H-%M-%S", gmtime())
# We also need to tell SageMaker which container should be used for inference and where it should
# retrieve the model artifacts from. In our case, the linear-learner container that we used for training
# can also be used for inference.
linear_primary_container = {
"Image": linear_container,
"ModelDataUrl": linear.model_data
}
# And lastly we construct the SageMaker model
linear_model_info = session.sagemaker_client.create_model(
ModelName = linear_model_name,
ExecutionRoleArn = role,
PrimaryContainer = linear_primary_container)
```
### Create the endpoint configuration
Once we have the model we can start putting together the endpoint by creating an endpoint configuration.
```
# As before, we need to give our endpoint configuration a name which should be unique
linear_endpoint_config_name = "boston-linear-endpoint-config-" + strftime("%Y-%m-%d-%H-%M-%S", gmtime())
# And then we ask SageMaker to construct the endpoint configuration
linear_endpoint_config_info = session.sagemaker_client.create_endpoint_config(
EndpointConfigName = linear_endpoint_config_name,
ProductionVariants = [{
"InstanceType": "ml.m4.xlarge",
"InitialVariantWeight": 1,
"InitialInstanceCount": 1,
"ModelName": linear_model_name,
"VariantName": "Linear-Model"
}])
```
### Deploy the endpoint
Now that the endpoint configuration has been created, we can ask SageMaker to build our endpoint.
**Note:** This is a friendly (repeated) reminder that you are about to deploy an endpoint. Make sure that you shut it down once you've finished with it!
```
# Again, we need a unique name for our endpoint
endpoint_name = "boston-update-endpoint-" + strftime("%Y-%m-%d-%H-%M-%S", gmtime())
# And then we can deploy our endpoint
endpoint_info = session.sagemaker_client.create_endpoint(
EndpointName = endpoint_name,
EndpointConfigName = linear_endpoint_config_name)
endpoint_dec = session.wait_for_endpoint(endpoint_name)
```
## Step 7 (B): Use the model
Just like with the XGBoost model, we will send some data to our endpoint to make sure that it is working properly. An important note is that the output format for the linear model is different from the XGBoost model.
```
response = session.sagemaker_runtime_client.invoke_endpoint(
EndpointName = endpoint_name,
ContentType = 'text/csv',
Body = ','.join(map(str, X_test.values[0])))
pprint(response)
result = response['Body'].read().decode("utf-8")
pprint(result)
Y_test.values[0]
```
## Shut down the endpoint
Now that we know that the Linear model's endpoint works, we can shut it down.
```
session.sagemaker_client.delete_endpoint(EndpointName = endpoint_name)
```
## Step 6 (C): Deploy a combined model
So far we've constructed two separate models which we could deploy and use. Before we talk about how we can change a deployed endpoint from one configuration to another, let's consider a slightly different situation. Suppose that before we switch from using only the XGBoost model to only the Linear model, we first want to do something like an A-B test, where we send some of the incoming data to the XGBoost model and some of the data to the Linear model.
Fortunately, SageMaker provides this functionality. And to actually get SageMaker to do this for us is not too different from deploying a model in the way that we've already done. The only difference is that we need to list more than one model in the production variants parameter of the endpoint configuration.
A reasonable question to ask is, how much data is sent to each of the models that I list in the production variants parameter? The answer is that it depends on the weight set for each model.
Suppose that we have $k$ models listed in the production variants and that each model $i$ is assigned the weight $w_i$. Then each model $i$ will receive $w_i / W$ of the traffic where $W = \sum_{i} w_i$.
In our case, since we have two models, the linear model and the XGBoost model, and each model has weight 1, we see that each model will get 1 / (1 + 1) = 1/2 of the data sent to the endpoint.
```
# As before, we need to give our endpoint configuration a name which should be unique
combined_endpoint_config_name = "boston-combined-endpoint-config-" + strftime("%Y-%m-%d-%H-%M-%S", gmtime())
# And then we ask SageMaker to construct the endpoint configuration
combined_endpoint_config_info = session.sagemaker_client.create_endpoint_config(
EndpointConfigName = combined_endpoint_config_name,
ProductionVariants = [
{ # First we include the linear model
"InstanceType": "ml.m4.xlarge",
"InitialVariantWeight": 1,
"InitialInstanceCount": 1,
"ModelName": linear_model_name,
"VariantName": "Linear-Model"
}, { # And next we include the xgb model
"InstanceType": "ml.m4.xlarge",
"InitialVariantWeight": 1,
"InitialInstanceCount": 1,
"ModelName": xgb_model_name,
"VariantName": "XGB-Model"
}])
```
Now that we've created the endpoint configuration, we can ask SageMaker to construct the endpoint.
**Note:** This is a friendly (repeated) reminder that you are about to deploy an endpoint. Make sure that you shut it down once you've finished with it!
```
# Again, we need a unique name for our endpoint
endpoint_name = "boston-update-endpoint-" + strftime("%Y-%m-%d-%H-%M-%S", gmtime())
# And then we can deploy our endpoint
endpoint_info = session.sagemaker_client.create_endpoint(
EndpointName = endpoint_name,
EndpointConfigName = combined_endpoint_config_name)
endpoint_dec = session.wait_for_endpoint(endpoint_name)
```
## Step 7 (C): Use the model
Now that we've constructed an endpoint which sends data to both the XGBoost model and the linear model we can send some data to the endpoint and see what sort of results we get back.
```
response = session.sagemaker_runtime_client.invoke_endpoint(
EndpointName = endpoint_name,
ContentType = 'text/csv',
Body = ','.join(map(str, X_test.values[0])))
pprint(response)
```
Since looking at a single response doesn't give us a clear look at what is happening, we can instead take a look at a few different responses to our endpoint
```
for rec in range(10):
response = session.sagemaker_runtime_client.invoke_endpoint(
EndpointName = endpoint_name,
ContentType = 'text/csv',
Body = ','.join(map(str, X_test.values[rec])))
pprint(response)
result = response['Body'].read().decode("utf-8")
print(result)
print(Y_test.values[rec])
```
If at some point we aren't sure about the properties of a deployed endpoint, we can use the `describe_endpoint` function to get SageMaker to return a description of the deployed endpoint.
```
pprint(session.sagemaker_client.describe_endpoint(EndpointName=endpoint_name))
```
## Updating an Endpoint
Now suppose that we've done our A-B test and the new linear model is working well enough. What we'd like to do now is to switch our endpoint from sending data to both the XGBoost model and the linear model to sending data only to the linear model.
Of course, we don't really want to shut down the endpoint to do this as doing so would interrupt service to whoever depends on our endpoint. Instead, we can ask SageMaker to **update** an endpoint to a new endpoint configuration.
What is actually happening is that SageMaker will set up a new endpoint with the new characteristics. Once this new endpoint is running, SageMaker will switch the old endpoint so that it now points at the newly deployed model, making sure that this happens seamlessly in the background.
```
session.sagemaker_client.update_endpoint(EndpointName=endpoint_name, EndpointConfigName=linear_endpoint_config_name)
```
To get a glimpse at what is going on, we can ask SageMaker to describe our in-use endpoint now, before the update process has completed. When we do so, we can see that the in-use endpoint still has the same characteristics it had before.
```
pprint(session.sagemaker_client.describe_endpoint(EndpointName=endpoint_name))
```
If we now wait for the update process to complete, and then ask SageMaker to describe the endpoint, it will return the characteristics of the new endpoint configuration.
```
endpoint_dec = session.wait_for_endpoint(endpoint_name)
pprint(session.sagemaker_client.describe_endpoint(EndpointName=endpoint_name))
```
## Shut down the endpoint
Now that we've finished, we need to make sure to shut down the endpoint.
```
session.sagemaker_client.delete_endpoint(EndpointName = endpoint_name)
```
## Optional: Clean up
The default notebook instance on SageMaker doesn't have a lot of excess disk space available. As you continue to complete and execute notebooks you will eventually fill up this disk space, leading to errors which can be difficult to diagnose. Once you are completely finished using a notebook it is a good idea to remove the files that you created along the way. Of course, you can do this from the terminal or from the notebook hub if you would like. The cell below contains some commands to clean up the created files from within the notebook.
```
# First we will remove all of the files contained in the data_dir directory
!rm $data_dir/*
# And then we delete the directory itself
!rmdir $data_dir
```
|
github_jupyter
|
# Make sure that we use SageMaker 1.x
!pip install sagemaker==1.72.0
%matplotlib inline
import os
import numpy as np
import pandas as pd
from pprint import pprint
import matplotlib.pyplot as plt
from time import gmtime, strftime
from sklearn.datasets import load_boston
import sklearn.model_selection
import sagemaker
from sagemaker import get_execution_role
from sagemaker.amazon.amazon_estimator import get_image_uri
from sagemaker.predictor import csv_serializer
# This is an object that represents the SageMaker session that we are currently operating in. This
# object contains some useful information that we will need to access later such as our region.
session = sagemaker.Session()
# This is an object that represents the IAM role that we are currently assigned. When we construct
# and launch the training job later we will need to tell it what IAM role it should have. Since our
# use case is relatively simple we will simply assign the training job the role we currently have.
role = get_execution_role()
boston = load_boston()
# First we package up the input data and the target variable (the median value) as pandas dataframes. This
# will make saving the data to a file a little easier later on.
X_bos_pd = pd.DataFrame(boston.data, columns=boston.feature_names)
Y_bos_pd = pd.DataFrame(boston.target)
# We split the dataset into 2/3 training and 1/3 testing sets.
X_train, X_test, Y_train, Y_test = sklearn.model_selection.train_test_split(X_bos_pd, Y_bos_pd, test_size=0.33)
# Then we split the training set further into 2/3 training and 1/3 validation sets.
X_train, X_val, Y_train, Y_val = sklearn.model_selection.train_test_split(X_train, Y_train, test_size=0.33)
# This is our local data directory. We need to make sure that it exists.
data_dir = '../data/boston'
if not os.path.exists(data_dir):
os.makedirs(data_dir)
# We use pandas to save our train and validation data to csv files. Note that we make sure not to include header
# information or an index as this is required by the built in algorithms provided by Amazon. Also, it is assumed
# that the first entry in each row is the target variable.
pd.concat([Y_val, X_val], axis=1).to_csv(os.path.join(data_dir, 'validation.csv'), header=False, index=False)
pd.concat([Y_train, X_train], axis=1).to_csv(os.path.join(data_dir, 'train.csv'), header=False, index=False)
prefix = 'boston-update-endpoints'
val_location = session.upload_data(os.path.join(data_dir, 'validation.csv'), key_prefix=prefix)
train_location = session.upload_data(os.path.join(data_dir, 'train.csv'), key_prefix=prefix)
# As stated above, we use this utility method to construct the image name for the training container.
xgb_container = get_image_uri(session.boto_region_name, 'xgboost')
# Now that we know which container to use, we can construct the estimator object.
xgb = sagemaker.estimator.Estimator(xgb_container, # The name of the training container
role, # The IAM role to use (our current role in this case)
train_instance_count=1, # The number of instances to use for training
train_instance_type='ml.m4.xlarge', # The type of instance ot use for training
output_path='s3://{}/{}/output'.format(session.default_bucket(), prefix),
# Where to save the output (the model artifacts)
sagemaker_session=session) # The current SageMaker session
xgb.set_hyperparameters(max_depth=5,
eta=0.2,
gamma=4,
min_child_weight=6,
subsample=0.8,
objective='reg:linear',
early_stopping_rounds=10,
num_round=200)
# This is a wrapper around the location of our train and validation data, to make sure that SageMaker
# knows our data is in csv format.
s3_input_train = sagemaker.s3_input(s3_data=train_location, content_type='text/csv')
s3_input_validation = sagemaker.s3_input(s3_data=val_location, content_type='text/csv')
xgb.fit({'train': s3_input_train, 'validation': s3_input_validation})
# Remember that a model needs to have a unique name
xgb_model_name = "boston-update-xgboost-model" + strftime("%Y-%m-%d-%H-%M-%S", gmtime())
# We also need to tell SageMaker which container should be used for inference and where it should
# retrieve the model artifacts from. In our case, the xgboost container that we used for training
# can also be used for inference and the model artifacts come from the previous call to fit.
xgb_primary_container = {
"Image": xgb_container,
"ModelDataUrl": xgb.model_data
}
# And lastly we construct the SageMaker model
xgb_model_info = session.sagemaker_client.create_model(
ModelName = xgb_model_name,
ExecutionRoleArn = role,
PrimaryContainer = xgb_primary_container)
# As before, we need to give our endpoint configuration a name which should be unique
xgb_endpoint_config_name = "boston-update-xgboost-endpoint-config-" + strftime("%Y-%m-%d-%H-%M-%S", gmtime())
# And then we ask SageMaker to construct the endpoint configuration
xgb_endpoint_config_info = session.sagemaker_client.create_endpoint_config(
EndpointConfigName = xgb_endpoint_config_name,
ProductionVariants = [{
"InstanceType": "ml.m4.xlarge",
"InitialVariantWeight": 1,
"InitialInstanceCount": 1,
"ModelName": xgb_model_name,
"VariantName": "XGB-Model"
}])
# Again, we need a unique name for our endpoint
endpoint_name = "boston-update-endpoint-" + strftime("%Y-%m-%d-%H-%M-%S", gmtime())
# And then we can deploy our endpoint
endpoint_info = session.sagemaker_client.create_endpoint(
EndpointName = endpoint_name,
EndpointConfigName = xgb_endpoint_config_name)
endpoint_dec = session.wait_for_endpoint(endpoint_name)
response = session.sagemaker_runtime_client.invoke_endpoint(
EndpointName = endpoint_name,
ContentType = 'text/csv',
Body = ','.join(map(str, X_test.values[0])))
pprint(response)
result = response['Body'].read().decode("utf-8")
pprint(result)
Y_test.values[0]
session.sagemaker_client.delete_endpoint(EndpointName = endpoint_name)
# Similar to the XGBoost model, we will use the utility method to construct the image name for the training container.
linear_container = get_image_uri(session.boto_region_name, 'linear-learner')
# Now that we know which container to use, we can construct the estimator object.
linear = sagemaker.estimator.Estimator(linear_container, # The name of the training container
role, # The IAM role to use (our current role in this case)
train_instance_count=1, # The number of instances to use for training
train_instance_type='ml.m4.xlarge', # The type of instance ot use for training
output_path='s3://{}/{}/output'.format(session.default_bucket(), prefix),
# Where to save the output (the model artifacts)
sagemaker_session=session) # The current SageMaker session
linear.set_hyperparameters(feature_dim=13, # Our data has 13 feature columns
predictor_type='regressor', # We wish to create a regression model
mini_batch_size=200) # Here we set how many samples to look at in each iteration
linear.fit({'train': s3_input_train, 'validation': s3_input_validation})
# First, we create a unique model name
linear_model_name = "boston-update-linear-model" + strftime("%Y-%m-%d-%H-%M-%S", gmtime())
# We also need to tell SageMaker which container should be used for inference and where it should
# retrieve the model artifacts from. In our case, the linear-learner container that we used for training
# can also be used for inference.
linear_primary_container = {
"Image": linear_container,
"ModelDataUrl": linear.model_data
}
# And lastly we construct the SageMaker model
linear_model_info = session.sagemaker_client.create_model(
ModelName = linear_model_name,
ExecutionRoleArn = role,
PrimaryContainer = linear_primary_container)
# As before, we need to give our endpoint configuration a name which should be unique
linear_endpoint_config_name = "boston-linear-endpoint-config-" + strftime("%Y-%m-%d-%H-%M-%S", gmtime())
# And then we ask SageMaker to construct the endpoint configuration
linear_endpoint_config_info = session.sagemaker_client.create_endpoint_config(
EndpointConfigName = linear_endpoint_config_name,
ProductionVariants = [{
"InstanceType": "ml.m4.xlarge",
"InitialVariantWeight": 1,
"InitialInstanceCount": 1,
"ModelName": linear_model_name,
"VariantName": "Linear-Model"
}])
# Again, we need a unique name for our endpoint
endpoint_name = "boston-update-endpoint-" + strftime("%Y-%m-%d-%H-%M-%S", gmtime())
# And then we can deploy our endpoint
endpoint_info = session.sagemaker_client.create_endpoint(
EndpointName = endpoint_name,
EndpointConfigName = linear_endpoint_config_name)
endpoint_dec = session.wait_for_endpoint(endpoint_name)
response = session.sagemaker_runtime_client.invoke_endpoint(
EndpointName = endpoint_name,
ContentType = 'text/csv',
Body = ','.join(map(str, X_test.values[0])))
pprint(response)
result = response['Body'].read().decode("utf-8")
pprint(result)
Y_test.values[0]
session.sagemaker_client.delete_endpoint(EndpointName = endpoint_name)
# As before, we need to give our endpoint configuration a name which should be unique
combined_endpoint_config_name = "boston-combined-endpoint-config-" + strftime("%Y-%m-%d-%H-%M-%S", gmtime())
# And then we ask SageMaker to construct the endpoint configuration
combined_endpoint_config_info = session.sagemaker_client.create_endpoint_config(
EndpointConfigName = combined_endpoint_config_name,
ProductionVariants = [
{ # First we include the linear model
"InstanceType": "ml.m4.xlarge",
"InitialVariantWeight": 1,
"InitialInstanceCount": 1,
"ModelName": linear_model_name,
"VariantName": "Linear-Model"
}, { # And next we include the xgb model
"InstanceType": "ml.m4.xlarge",
"InitialVariantWeight": 1,
"InitialInstanceCount": 1,
"ModelName": xgb_model_name,
"VariantName": "XGB-Model"
}])
# Again, we need a unique name for our endpoint
endpoint_name = "boston-update-endpoint-" + strftime("%Y-%m-%d-%H-%M-%S", gmtime())
# And then we can deploy our endpoint
endpoint_info = session.sagemaker_client.create_endpoint(
EndpointName = endpoint_name,
EndpointConfigName = combined_endpoint_config_name)
endpoint_dec = session.wait_for_endpoint(endpoint_name)
response = session.sagemaker_runtime_client.invoke_endpoint(
EndpointName = endpoint_name,
ContentType = 'text/csv',
Body = ','.join(map(str, X_test.values[0])))
pprint(response)
for rec in range(10):
response = session.sagemaker_runtime_client.invoke_endpoint(
EndpointName = endpoint_name,
ContentType = 'text/csv',
Body = ','.join(map(str, X_test.values[rec])))
pprint(response)
result = response['Body'].read().decode("utf-8")
print(result)
print(Y_test.values[rec])
pprint(session.sagemaker_client.describe_endpoint(EndpointName=endpoint_name))
session.sagemaker_client.update_endpoint(EndpointName=endpoint_name, EndpointConfigName=linear_endpoint_config_name)
pprint(session.sagemaker_client.describe_endpoint(EndpointName=endpoint_name))
endpoint_dec = session.wait_for_endpoint(endpoint_name)
pprint(session.sagemaker_client.describe_endpoint(EndpointName=endpoint_name))
session.sagemaker_client.delete_endpoint(EndpointName = endpoint_name)
# First we will remove all of the files contained in the data_dir directory
!rm $data_dir/*
# And then we delete the directory itself
!rmdir $data_dir
| 0.516352 | 0.985814 |
# Binary Image creation
A method or combination of methods (i.e., color transforms, gradients) has been used to create a binary image containing likely lane pixels
load all the library
```
import cv2
import numpy as np
import glob
import matplotlib.pyplot as plt
```
Below code will convert threshold a color frame in HSV space
```
def thresholdHSV(frame, min_values, max_values):
"""
Threshold a color frame in HSV space
"""
HSV = cv2.cvtColor(frame, cv2.COLOR_RGB2HLS)
min_th_ok = np.all(HSV > min_values, axis=2)
max_th_ok = np.all(HSV < max_values, axis=2)
out = np.logical_and(min_th_ok, max_th_ok)
return out
```
Below function Apply Sobel edge detection to an input frame, then threshold the result
```
def thresholdSobel(frame, kernel_size):
"""
Apply Sobel edge detection to an input frame, then threshold the result
"""
gray = cv2.cvtColor(frame, cv2.COLOR_BGR2GRAY)
sobel_x = cv2.Sobel(gray, cv2.CV_64F, 1, 0, ksize=kernel_size)
sobel_y = cv2.Sobel(gray, cv2.CV_64F, 0, 1, ksize=kernel_size)
sobel_mag = np.sqrt(sobel_x ** 2 + sobel_y ** 2)
sobel_mag = np.uint8(sobel_mag / np.max(sobel_mag) * 255)
_, sobel_mag = cv2.threshold(sobel_mag, 50, 1, cv2.THRESH_BINARY)
return sobel_mag.astype(bool)
```
Below function apply histogram equalization to an input frame, threshold it and return the (binary) result.
```
def getBinaryFromGrayscale(frame):
"""
Apply histogram equalization to an input frame, threshold it and return the (binary) result.
"""
gray = cv2.cvtColor(frame, cv2.COLOR_BGR2GRAY)
eq_global = cv2.equalizeHist(gray)
_, th = cv2.threshold(eq_global, thresh=250, maxval=255, type=cv2.THRESH_BINARY)
return th
```
Below funciton will return the all binary image-Convert an input frame to a binary image which highlight as most as possible the lane-lines
```
def binaryImage(img):
"""
Convert an input frame to a binary image which highlight as most as possible the lane-lines.
"""
yellow_HSV_th_min = np.array([0, 70, 70])
yellow_HSV_th_max = np.array([50, 255, 255])
binary_Images = []
h, w = img.shape[:2]
binary_Images.append(cv2.cvtColor(img, cv2.COLOR_BGR2RGB))
binary = np.zeros(shape=(h, w), dtype=np.uint8)
# highlight yellow lines by threshold in HSV color space
HSV_yellow_mask = thresholdHSV(img, yellow_HSV_th_min, yellow_HSV_th_max)
binary = np.logical_or(binary, HSV_yellow_mask)
# highlight white lines by thresholding the equalized frame
eq_white_mask = getBinaryFromGrayscale(img)
binary_Images.append(eq_white_mask)
binary = np.logical_or(binary, eq_white_mask)
binary_Images.append(HSV_yellow_mask)
# get Sobel binary mask (thresholded gradients)
sobel_mask = thresholdSobel(img, kernel_size=9)
binary_Images.append(sobel_mask)
binary = np.logical_or(binary, sobel_mask)
binary_Images.append(binary)
# apply a light morphology to "fill the gaps" in the binary image
kernel = np.ones((5, 5), np.uint8)
closing = cv2.morphologyEx(binary.astype(np.uint8), cv2.MORPH_CLOSE, kernel)
binary_Images.append(closing)
return binary_Images
```
Below funciton will show all possibale image of avialbe into test images
```
def showImages(images, cmap=None):
for i in range(len(images)):
for j in range(len(images[i])):
f, ax = plt.subplots(2,3)
f.set_facecolor('white')
ax[0, 0].imshow(images[i][j])
ax[0, 0].set_title('input_frame')
ax[0, 0].set_axis_off()
j = j + 1
ax[0, 1].imshow(images[i][j], cmap='gray')
ax[0, 1].set_title('white mask')
ax[0, 1].set_axis_off()
j = j + 1
ax[0, 2].imshow(images[i][j], cmap='gray')
ax[0, 2].set_title('yellow mask')
ax[0, 2].set_axis_off()
j = j + 1
ax[1, 0].imshow(images[i][j], cmap='gray')
ax[1, 0].set_title('sobel mask')
ax[1, 0].set_axis_off()
j = j + 1
ax[1, 1].imshow(images[i][j], cmap='gray')
ax[1, 1].set_title('before closure')
ax[1, 1].set_axis_off()
j = j + 1
ax[1, 2].imshow(images[i][j], cmap='gray')
ax[1, 2].set_title('after closure')
ax[1, 2].set_axis_off()
break
plt.show()
final_images = []
test_images = glob.glob('test_images/*.jpg')
for test_image in test_images:
img = cv2.imread(test_image)
final_images.append(binaryImage(img=img))
showImages(final_images)
```
|
github_jupyter
|
import cv2
import numpy as np
import glob
import matplotlib.pyplot as plt
def thresholdHSV(frame, min_values, max_values):
"""
Threshold a color frame in HSV space
"""
HSV = cv2.cvtColor(frame, cv2.COLOR_RGB2HLS)
min_th_ok = np.all(HSV > min_values, axis=2)
max_th_ok = np.all(HSV < max_values, axis=2)
out = np.logical_and(min_th_ok, max_th_ok)
return out
def thresholdSobel(frame, kernel_size):
"""
Apply Sobel edge detection to an input frame, then threshold the result
"""
gray = cv2.cvtColor(frame, cv2.COLOR_BGR2GRAY)
sobel_x = cv2.Sobel(gray, cv2.CV_64F, 1, 0, ksize=kernel_size)
sobel_y = cv2.Sobel(gray, cv2.CV_64F, 0, 1, ksize=kernel_size)
sobel_mag = np.sqrt(sobel_x ** 2 + sobel_y ** 2)
sobel_mag = np.uint8(sobel_mag / np.max(sobel_mag) * 255)
_, sobel_mag = cv2.threshold(sobel_mag, 50, 1, cv2.THRESH_BINARY)
return sobel_mag.astype(bool)
def getBinaryFromGrayscale(frame):
"""
Apply histogram equalization to an input frame, threshold it and return the (binary) result.
"""
gray = cv2.cvtColor(frame, cv2.COLOR_BGR2GRAY)
eq_global = cv2.equalizeHist(gray)
_, th = cv2.threshold(eq_global, thresh=250, maxval=255, type=cv2.THRESH_BINARY)
return th
def binaryImage(img):
"""
Convert an input frame to a binary image which highlight as most as possible the lane-lines.
"""
yellow_HSV_th_min = np.array([0, 70, 70])
yellow_HSV_th_max = np.array([50, 255, 255])
binary_Images = []
h, w = img.shape[:2]
binary_Images.append(cv2.cvtColor(img, cv2.COLOR_BGR2RGB))
binary = np.zeros(shape=(h, w), dtype=np.uint8)
# highlight yellow lines by threshold in HSV color space
HSV_yellow_mask = thresholdHSV(img, yellow_HSV_th_min, yellow_HSV_th_max)
binary = np.logical_or(binary, HSV_yellow_mask)
# highlight white lines by thresholding the equalized frame
eq_white_mask = getBinaryFromGrayscale(img)
binary_Images.append(eq_white_mask)
binary = np.logical_or(binary, eq_white_mask)
binary_Images.append(HSV_yellow_mask)
# get Sobel binary mask (thresholded gradients)
sobel_mask = thresholdSobel(img, kernel_size=9)
binary_Images.append(sobel_mask)
binary = np.logical_or(binary, sobel_mask)
binary_Images.append(binary)
# apply a light morphology to "fill the gaps" in the binary image
kernel = np.ones((5, 5), np.uint8)
closing = cv2.morphologyEx(binary.astype(np.uint8), cv2.MORPH_CLOSE, kernel)
binary_Images.append(closing)
return binary_Images
def showImages(images, cmap=None):
for i in range(len(images)):
for j in range(len(images[i])):
f, ax = plt.subplots(2,3)
f.set_facecolor('white')
ax[0, 0].imshow(images[i][j])
ax[0, 0].set_title('input_frame')
ax[0, 0].set_axis_off()
j = j + 1
ax[0, 1].imshow(images[i][j], cmap='gray')
ax[0, 1].set_title('white mask')
ax[0, 1].set_axis_off()
j = j + 1
ax[0, 2].imshow(images[i][j], cmap='gray')
ax[0, 2].set_title('yellow mask')
ax[0, 2].set_axis_off()
j = j + 1
ax[1, 0].imshow(images[i][j], cmap='gray')
ax[1, 0].set_title('sobel mask')
ax[1, 0].set_axis_off()
j = j + 1
ax[1, 1].imshow(images[i][j], cmap='gray')
ax[1, 1].set_title('before closure')
ax[1, 1].set_axis_off()
j = j + 1
ax[1, 2].imshow(images[i][j], cmap='gray')
ax[1, 2].set_title('after closure')
ax[1, 2].set_axis_off()
break
plt.show()
final_images = []
test_images = glob.glob('test_images/*.jpg')
for test_image in test_images:
img = cv2.imread(test_image)
final_images.append(binaryImage(img=img))
showImages(final_images)
| 0.654122 | 0.951369 |
# Reply to the Supreme Court
[Our task](../code-basics/sampling_problem) has been to reply to the Supreme
Court on their judgment in the appeal of Robert Swain.
Remember, Robert Swain appealed his death sentence, on the basis that the jury
selection was biased against Black people.
His trial jury of 12 people had no Black members.
The local population of eligible jurors was 26% Black.
If the jury had been representative, we would expect about 26 of 100 people to
be Black. That's around 1 in 4 (25%), so we would expect about one in four
jurors to be Black - so around 3 of 12.
The Supreme Court was not convinced that there was evidence of systematic bias. But, to start with the jurors - is it surprising that we expected around 3 Black jurors, but we got 0. Is the value 0 surprising, if each juror has 26% chance of being Black?
To answer this, we are going to go through a couple of steps.
The first is to build an *ideal model* of the world, where it *is true* that
each juror has a 26% of being Black. We sometimes call this our *ideal world*.
If you are used to statistical terms, this ideal model is our *null
hypothesis*.
Then we can *simulate* making many juries in this ideal world.
Finally we ask whether our simulated juries, from the ideal world, often give
us a count of zero Black jurors. If they don't, then we can say that we are
*surprised* by the value of 0, if the jury did arise from that real world. If
the value 0 is sufficiently unusual, we become suspicious that the real world
is rather different from our ideal world. We consider *rejecting* the ideal
world as a good model of the real world.
## The ideal world
Our *ideal model* (or null hypothesis) is a world where each juror has been
truly randomly selected from the eligible population. That is, for any one
juror, there is a 0.26 probability that they are Black.
## Simulations with the ideal model
To do a *simulation* with this ideal model, we will start by making one jury,
of 12 people, where it is really true that each juror has a 26% of being Black.
Not to pun, but we will call one simulation of a jury of 12 - *one trial*.
Then we simulate 10 juries of 12 people (do 10 trials), to get warmed up.
Finally we make 10000 juries, each of 12 people, and see what we get.
```
# Import the array library
import numpy as np
```
Here is one jury, and the number of Black people we get in our simulation.
```
# Make 12 random integers from 0 through 99
randoms = np.random.randint(0, 100, size=12)
# Say values < 26 correspond to black jurors.
# 26 of the numbers 0 through 99 are less than 26 (so 26% or p=0.26).
black_yn = randoms < 26
# We now have True for Black jurors and False otherwise.
# Count the number of Trues
np.count_nonzero(black_yn)
```
That is one estimate, for the number of Black people we can expect, if our
model is correct. Call this one *trial*. We can run that a few times to get a
range of values. If we run it only a few times, we might be unlucky, and get
some results that are not representative. It is safer to run it a huge number
of times, to make sure we've got an idea of the variation.
To start with, we will run 10 trials.
We get ready to store the results of each estimate.
```
# Make an array of 10 zeros, to store the results.
counts = np.zeros(10)
```
We repeat the code from the cell above, but now, we store each trial result
(count) in the `counts` array:
```
randoms = np.random.randint(0, 100, size=100)
black_yn = randoms < 26
count = np.count_nonzero(black_yn)
counts[0] = count
counts
```
Run the cell above a few times, perhaps with Control-Enter, to see the first value in the `counts` array changing.
Now we collect the result of 10 trials, by using a for loop.
```
# Make a new counts array of zeros to store the results.
counts = np.zeros(10)
for i in np.arange(10):
# This code is the same as the cell above, but indented,
# so we run it all, for each time through the for loop.
randoms = np.random.randint(0, 100, size=12)
black_yn = randoms < 26
count = np.count_nonzero(black_yn)
# Store the result at position i
counts[i] = count
counts
```
Each of these values is one estimate for how many Black jurors we should
expect, if our model is right. Already we get the feeling that 0 is rather
unlikely, if our model is correct. But - how unlikely?
To get a better estimate, let us do the same thing, but with 10000 juries, and
therefore, 10000 estimates.
```
# Make a new counts array of zeros to store the results.
counts = np.zeros(10000)
for i in np.arange(10000):
# This code is the same as the cell above, but indented,
# so we run it all, for each time through the for loop.
randoms = np.random.randint(0, 100, size=12)
black_yn = randoms < 26
count = np.count_nonzero(black_yn)
# Store the result at position i
counts[i] = count
counts
```
If you ran this cell yourself, you will notice that it runs very fast, in much
less than a second, on any reasonable computer.
We now have 10000 estimates, one for each row in the original array, and
therefore, one for each simulated jury.
Remember, the function `len` shows us the length of the array, and therefore,
the number of values in this one-dimensional array.
```
len(counts)
```
Next we want to have a look at the spread of these values. To do this, we plot
a histogram. Here is how to do that, in Python. Don't worry about the details, we will go into this more soon.
```
# Please don't worry about this bit of code for now.
# It sets up plotting in the notebook.
import matplotlib.pyplot as plt
%matplotlib inline
# Fancy plots
plt.style.use('fivethirtyeight')
```
Now show the histogram. Don't worry about the details of this command.
```
# Do the histogram of our 10000 estimates.
plt.hist(counts, bins=np.arange(13))
```
The histogram above is called the *sampling distribution*. The sampling distribution is the distribution of thing we are interested in (the number of Black jurors) given the ideal model, of completely random selection of jurors from a 26% Black population.
It looks as if 0 is a relatively uncommon value among our simulations. How many
times did we get a value of 0, in all our 10000 estimates?
```
counts_of_0 = counts == 0
n_zeros = np.count_nonzero(counts_of_0)
n_zeros
```
What *proportion* of jury simulations give a value of 0? We just divide the
number of times we see 0 by the number trials we made:
```
p = n_zeros / 10000
p
```
We have run an analysis assuming that the jurors were selected at random. On
that assumption, a count of 0 jurors in 12 is fairly uncommon, in the sense
that the proportion of times we see that result is:
```
p
```
In other words, our *estimate* of the *probability* of getting 0 Black people
in a jury of 12, is
```
p
```
What can we conclude? Only this: that in our ideal model world, where each
juror has a 26% chance of being Black, 0 is uncommon. This surprising result,
of 0, gives us some cause to wonder if our ideal model of the world is wrong.
One way it could be wrong, is if there was bias in jury selection, so it was
not true that each juror had a 26% of being Black.
|
github_jupyter
|
# Import the array library
import numpy as np
# Make 12 random integers from 0 through 99
randoms = np.random.randint(0, 100, size=12)
# Say values < 26 correspond to black jurors.
# 26 of the numbers 0 through 99 are less than 26 (so 26% or p=0.26).
black_yn = randoms < 26
# We now have True for Black jurors and False otherwise.
# Count the number of Trues
np.count_nonzero(black_yn)
# Make an array of 10 zeros, to store the results.
counts = np.zeros(10)
randoms = np.random.randint(0, 100, size=100)
black_yn = randoms < 26
count = np.count_nonzero(black_yn)
counts[0] = count
counts
# Make a new counts array of zeros to store the results.
counts = np.zeros(10)
for i in np.arange(10):
# This code is the same as the cell above, but indented,
# so we run it all, for each time through the for loop.
randoms = np.random.randint(0, 100, size=12)
black_yn = randoms < 26
count = np.count_nonzero(black_yn)
# Store the result at position i
counts[i] = count
counts
# Make a new counts array of zeros to store the results.
counts = np.zeros(10000)
for i in np.arange(10000):
# This code is the same as the cell above, but indented,
# so we run it all, for each time through the for loop.
randoms = np.random.randint(0, 100, size=12)
black_yn = randoms < 26
count = np.count_nonzero(black_yn)
# Store the result at position i
counts[i] = count
counts
len(counts)
# Please don't worry about this bit of code for now.
# It sets up plotting in the notebook.
import matplotlib.pyplot as plt
%matplotlib inline
# Fancy plots
plt.style.use('fivethirtyeight')
# Do the histogram of our 10000 estimates.
plt.hist(counts, bins=np.arange(13))
counts_of_0 = counts == 0
n_zeros = np.count_nonzero(counts_of_0)
n_zeros
p = n_zeros / 10000
p
p
p
| 0.471467 | 0.943919 |
```
import pandas as pd
import re
from collections import Counter
import matplotlib.pyplot as plt
from statistics import mean, median, mode
df_train = pd.read_csv(f'../../../code-mixed-enma/hasoc_task_1/ml-Hasoc-offensive-train.csv', sep='\t', header=None)
df_train.head()
df_train.shape
df_dev = pd.read_csv(f'../../../code-mixed-enma/hasoc_task_1/ml-Hasoc-offensive-dev.csv', sep='\t', header=None)
df_dev.head()
df_dev.shape
df_test = pd.read_csv(f'../../../code-mixed-enma/hasoc_task_1/ml_mixedscript_Hascoc_offensive_test_without_label - ml_mixedscript_Hascoc_offensive_test_without_label.csv', header=None)
df_test.head()
df_test.shape
import unicodedata as ud
latin_letters= {}
def is_latin(uchr):
try: return latin_letters[uchr]
except KeyError:
return latin_letters.setdefault(uchr, 'LATIN' in ud.name(uchr))
def only_roman_chars(unistr):
return all(is_latin(uchr)
for uchr in unistr
if uchr.isalpha()) # isalpha suggested by John Machin
count = 0
for index, row in df_train.iterrows():
if not only_roman_chars(row[1]):
print(index, row[0], row[1])
print('\n\n')
count +=1
print(count)
1-1062/3200
count = 0
for index, row in df_dev.iterrows():
if not only_roman_chars(row[1]):
print(index, row[0], row[1])
print('\n\n')
count +=1
print(count)
1 - count/len(df_dev)
count = 0
for index, row in df_test.iterrows():
if not only_roman_chars(row[1]):
print(index, row[0], row[1])
print('\n\n')
count +=1
print(1 - count/len(df_test))
# Class distribution in training set
D = Counter(df_train[0])
plt.rcParams["figure.figsize"] = (3,3)
plt.bar(range(len(D)), list(D.values()), align='center')
plt.xticks(range(len(D)), list(D.keys()))
plt.show()
# Min, Max, Avg no of sentences per class in Training set
print('Min no. of sentences: ', min(D.values()))
print('Max no. of sentences: ', max(D.values()))
print('Avg no. of sentences: ', mean(D.values()))
print('Median of sentences: ', median(D.values()))
# Class distribution in dev set
D = Counter(df_dev[0])
plt.rcParams["figure.figsize"] = (3,3)
plt.bar(range(len(D)), list(D.values()), align='center')
plt.xticks(range(len(D)), list(D.keys()))
plt.show()
# Min, Max, Avg no of sentences per class in dev set
print('Min no. of sentences: ', min(D.values()))
print('Max no. of sentences: ', max(D.values()))
print('Avg no. of sentences: ', mean(D.values()))
print('Median of sentences: ', median(D.values()))
def tokenize(s: str):
return s.split()
# variation in length of sentences in train set
len_of_tokens = []
for index, row in df_train.iterrows():
tokens = tokenize((row[1]).lower())
len_of_tokens.append(len(tokens))
print('Min no. of tokens: ', min(len_of_tokens))
print('Max no. of tokens: ', max(len_of_tokens))
print('Avg no. of tokens: ', mean(len_of_tokens))
print('Median of no. of tokens: ', median(len_of_tokens))
# variation in length of sentences in dev set
len_of_tokens = []
for index, row in df_dev.iterrows():
tokens = tokenize((row[1]).lower())
len_of_tokens.append(len(tokens))
print('Min no. of tokens: ', min(len_of_tokens))
print('Max no. of tokens: ', max(len_of_tokens))
print('Avg no. of tokens: ', mean(len_of_tokens))
print('Median of no. of tokens: ', median(len_of_tokens))
# variation in length of sentences in test set
len_of_tokens = []
for index, row in df_test.iterrows():
tokens = tokenize((row[1]).lower())
len_of_tokens.append(len(tokens))
print('Min no. of tokens: ', min(len_of_tokens))
print('Max no. of tokens: ', max(len_of_tokens))
print('Avg no. of tokens: ', mean(len_of_tokens))
print('Median of no. of tokens: ', median(len_of_tokens))
```
|
github_jupyter
|
import pandas as pd
import re
from collections import Counter
import matplotlib.pyplot as plt
from statistics import mean, median, mode
df_train = pd.read_csv(f'../../../code-mixed-enma/hasoc_task_1/ml-Hasoc-offensive-train.csv', sep='\t', header=None)
df_train.head()
df_train.shape
df_dev = pd.read_csv(f'../../../code-mixed-enma/hasoc_task_1/ml-Hasoc-offensive-dev.csv', sep='\t', header=None)
df_dev.head()
df_dev.shape
df_test = pd.read_csv(f'../../../code-mixed-enma/hasoc_task_1/ml_mixedscript_Hascoc_offensive_test_without_label - ml_mixedscript_Hascoc_offensive_test_without_label.csv', header=None)
df_test.head()
df_test.shape
import unicodedata as ud
latin_letters= {}
def is_latin(uchr):
try: return latin_letters[uchr]
except KeyError:
return latin_letters.setdefault(uchr, 'LATIN' in ud.name(uchr))
def only_roman_chars(unistr):
return all(is_latin(uchr)
for uchr in unistr
if uchr.isalpha()) # isalpha suggested by John Machin
count = 0
for index, row in df_train.iterrows():
if not only_roman_chars(row[1]):
print(index, row[0], row[1])
print('\n\n')
count +=1
print(count)
1-1062/3200
count = 0
for index, row in df_dev.iterrows():
if not only_roman_chars(row[1]):
print(index, row[0], row[1])
print('\n\n')
count +=1
print(count)
1 - count/len(df_dev)
count = 0
for index, row in df_test.iterrows():
if not only_roman_chars(row[1]):
print(index, row[0], row[1])
print('\n\n')
count +=1
print(1 - count/len(df_test))
# Class distribution in training set
D = Counter(df_train[0])
plt.rcParams["figure.figsize"] = (3,3)
plt.bar(range(len(D)), list(D.values()), align='center')
plt.xticks(range(len(D)), list(D.keys()))
plt.show()
# Min, Max, Avg no of sentences per class in Training set
print('Min no. of sentences: ', min(D.values()))
print('Max no. of sentences: ', max(D.values()))
print('Avg no. of sentences: ', mean(D.values()))
print('Median of sentences: ', median(D.values()))
# Class distribution in dev set
D = Counter(df_dev[0])
plt.rcParams["figure.figsize"] = (3,3)
plt.bar(range(len(D)), list(D.values()), align='center')
plt.xticks(range(len(D)), list(D.keys()))
plt.show()
# Min, Max, Avg no of sentences per class in dev set
print('Min no. of sentences: ', min(D.values()))
print('Max no. of sentences: ', max(D.values()))
print('Avg no. of sentences: ', mean(D.values()))
print('Median of sentences: ', median(D.values()))
def tokenize(s: str):
return s.split()
# variation in length of sentences in train set
len_of_tokens = []
for index, row in df_train.iterrows():
tokens = tokenize((row[1]).lower())
len_of_tokens.append(len(tokens))
print('Min no. of tokens: ', min(len_of_tokens))
print('Max no. of tokens: ', max(len_of_tokens))
print('Avg no. of tokens: ', mean(len_of_tokens))
print('Median of no. of tokens: ', median(len_of_tokens))
# variation in length of sentences in dev set
len_of_tokens = []
for index, row in df_dev.iterrows():
tokens = tokenize((row[1]).lower())
len_of_tokens.append(len(tokens))
print('Min no. of tokens: ', min(len_of_tokens))
print('Max no. of tokens: ', max(len_of_tokens))
print('Avg no. of tokens: ', mean(len_of_tokens))
print('Median of no. of tokens: ', median(len_of_tokens))
# variation in length of sentences in test set
len_of_tokens = []
for index, row in df_test.iterrows():
tokens = tokenize((row[1]).lower())
len_of_tokens.append(len(tokens))
print('Min no. of tokens: ', min(len_of_tokens))
print('Max no. of tokens: ', max(len_of_tokens))
print('Avg no. of tokens: ', mean(len_of_tokens))
print('Median of no. of tokens: ', median(len_of_tokens))
| 0.298594 | 0.369912 |

<a href="https://hub.callysto.ca/jupyter/hub/user-redirect/git-pull?repo=https%3A%2F%2Fgithub.com%2Fcallysto%2Fcurriculum-notebooks&branch=master&urlpath=notebooks/curriculum-notebooks/EnglishLanguageArts/ShakespeareStatistics/shakespeare-and-statistics.ipynb&depth=1" target="_parent"><img src="https://raw.githubusercontent.com/callysto/curriculum-notebooks/master/open-in-callysto-button.svg?sanitize=true" width="123" height="24" alt="Open in Callysto"></a>
# Shakespeare and Statistics

*Image from https://en.wikipedia.org/wiki/Droeshout_portrait*
Can art and science be combined? Natural language processing allows us to use the same statistical skills you might learn in a science class, such as counting up members of a population and looking at their distribution, to gain insight into the written word. Here's an example of what you can do. Let's consider the following question:
## What are the top 20 phrases in Shakespeare's Macbeth?
Normally when we study Shakespeare we critically read his plays and study the plot, characters, and themes. While this is definitely interesting and useful, we can gain very different insights by taking a multidisciplinary approach.
This is something we would probably never want to do if we had to do it by hand. Imagine getting out your clipboard, writing down every different word or phrase you come across and then counting how many times that same word or phrase reappears. Check out how quickly it can be done using Python code in this Jupyter notebook.
## Loading the text
There are many public domain works available at [Project Gutenberg](http://www.gutenberg.org). You can [search](http://www.gutenberg.org/ebooks) or [browse](http://www.gutenberg.org/catalog) to find works that you are interested in analysing.
We are going to search for the play *Macbeth* by William Shakespeare. On the **Download This eBook** page we'll copy the `Plain Text UTF-8` link, then use the `Requests` Python library to download it into a variable called `macbeth`. We can then refer to it by using `macbeth` at any point from here on
```
text_link = 'http://www.gutenberg.org/files/1533/1533-0.txt'
import requests
r = requests.get(text_link) # get the online book file
r.encoding = 'utf-8' # specify the type of text encoding in the file
macbeth = r.text.split('***')[2] # get the part after the header
macbeth = macbeth.replace("’","'").replace("“",'"').replace("”",'"') # replace any 'smart quotes'
```
For example, we can just print out the first 1000 letters to see that we've grabbed the correct document.
```
print(macbeth[0:1000])
```
Looks good! But that's a lot of reading to do. And a lot of phrases to count and keep track of. Here's where some Python libraries come into play.
## Crunching the text
`noun_phrases` will grab groups of words that have been identified as phrases containing nouns. This isn't always 100% correct. English can be a challenging language even for machines, and sometimes the files on [Project Gutenberg](http://www.gutenberg.org) contain errors that make it even harder, but it can usually do a pretty good job.
This code cell installs two Python libraries for natural language processing, [textblob](https://textblob.readthedocs.io/en/dev) and [nltk](https://www.nltk.org), then downloads a [corpora data file](http://www.nltk.org/nltk_data) that will allow us to process the text.
This may take a while to run. On the left you will see `In [*]:` while it is running. Once it finishes you should see the output printed on the screen.
```
!pip install textblob --user
!pip install nltk --user
import nltk
try:
nltk.data.find('tokenizers/punkt')
except LookupError:
nltk.download('punkt')
try:
nltk.data.find('tokenizers/brown')
except LookupError:
nltk.download('brown')
from textblob import TextBlob
macbeth_phrases = TextBlob(macbeth).noun_phrases
print(macbeth_phrases)
```
What you're seeing is no longer raw text. It's now a list of strings. How long is the list? Let's find out. `len` is short for "length", and it will tell you how many items are in any list.
```
len(macbeth_phrases)
```
Looks like we have over 3000 noun phrases. We don't yet know how many of them are repeated.
## Counting everything up
Here's where this starts to look like a real science project! Let's count the unique phrases and create a table of how many times they occur. They'll be sorted from most to least frequent.
```
import pandas as pd
unique_texts = list(set(macbeth_phrases))
text_counts = {text: macbeth_phrases.count(text) for text in unique_texts}
sorted_texts = sorted(text_counts.items(), key=lambda x: x[1], reverse=True)
macbeth_counts = pd.DataFrame(data=sorted_texts, columns=['text', 'count'])
macbeth_counts
```
There are a lot of them, so we'll use `.head(20)` which means show the top twenty. In these lists, the first item is always number 0.
```
macbeth_counts.head(20)
```
There we have it! The top 20 phrases in Macbeth! Let's put those in a plot.
## Plotting the results
You can do almost any kind of plot or other visual representation of observations like this you could think of in Callysto. We'll use the `cufflinks` library to produce a horizontal bar plot, ordered from most to least frequent word.
```
import cufflinks as cf
cf.go_offline()
macbeth_counts_sorted = macbeth_counts.head(20).sort_values(by='count', ascending=True)
macbeth_counts_sorted.iplot(kind='barh', x='text', y='count', title='Phrase Frequencies in MacBeth', xTitle='Count')
```
Surprise, surprise. *Macbeth* is the top phrase in Macbeth. Our main character is mentioned more than twice the number of times as the next most frequent phrase, *Macduff*, and more than three times the frequency that *Lady Macbeth* is mentioned.
## Thinking about the results
One of the first things we might realize from this simple investigation is the importance of proper nouns. Phrases containing the main characters occur far more frequently than other phrases, and the main character of the play is mentioned far more times than any other characters.
Are these observations particular to Macbeth? Or to Shakespeare's plays? Or are they more universal?
Now that we've gone through Macbeth, how hard could it be to look at other texts?
Let's define a function to to download an ebook from a text url, pull out all the noun phrases, count them up, and plot them. We can then use this for any ebook text that we would like to visualize.
```
def word_frequency_plot(text_url):
r = requests.get(text_url)
r.encoding = 'utf-8'
if 'gutenberg' in text_url:
text = r.text.split('***')[2]
else:
text = r.text
text = text.replace("’","'").replace("“",'"').replace("”",'"')
phrases = TextBlob(text).noun_phrases
unique_texts = list(set(phrases))
text_counts = {text: phrases.count(text) for text in unique_texts}
sorted_texts = sorted(text_counts.items(), key=lambda x: x[1], reverse=True)
counts = pd.DataFrame(data=sorted_texts, columns=['text', 'count'])
counts_sorted = counts.head(20).sort_values(by='count', ascending=True)
counts_sorted.iplot(kind='barh', x='text', y='count', title='Phrase Frequencies', xTitle='Count')
print('Word frequency plot function defined.')
```
## Looking at Hamlet
**Hamlet** can be found on [Project Gutenberg](http://www.gutenberg.org) under [EBook-No. 1524](http://www.gutenberg.org/ebooks/1524).
Run the following code to download **Hamlet**, pull out all the noun phrases, count them up, and plot them out.
```
word_frequency_plot('http://www.gutenberg.org/files/1524/1524-0.txt')
```
## Conclusion
Now that we have a function to plot word frequency, you can try this with other ebooks from [Project Gutenberg](http://www.gutenberg.org). You could also try running this on news articles or any other text, although there may be some tweaks required.
However this example is only scratching the surface of natural language processing. Libraries such as [spaCy](https://spacy.io) can be used to identify [parts of speech](https://spacy.io/api/annotation#pos-tagging), [vaderSentiment](https://github.com/cjhutto/vaderSentiment) can categorize text based on its tone, and [textstat](https://github.com/shivam5992/textstat) can check the readability and complexity of text. Examples can be found [here](https://github.com/callysto/interesting-problems/blob/main/notebooks/analysing-text-statistics.ipynb).
You could use these libraries to compare different writers or different times in history, see if you can trends in sentiment or [word usage](https://books.google.com/ngrams), or investigate changes in language and style.
[](https://github.com/callysto/curriculum-notebooks/blob/master/LICENSE.md)
|
github_jupyter
|
text_link = 'http://www.gutenberg.org/files/1533/1533-0.txt'
import requests
r = requests.get(text_link) # get the online book file
r.encoding = 'utf-8' # specify the type of text encoding in the file
macbeth = r.text.split('***')[2] # get the part after the header
macbeth = macbeth.replace("’","'").replace("“",'"').replace("”",'"') # replace any 'smart quotes'
print(macbeth[0:1000])
!pip install textblob --user
!pip install nltk --user
import nltk
try:
nltk.data.find('tokenizers/punkt')
except LookupError:
nltk.download('punkt')
try:
nltk.data.find('tokenizers/brown')
except LookupError:
nltk.download('brown')
from textblob import TextBlob
macbeth_phrases = TextBlob(macbeth).noun_phrases
print(macbeth_phrases)
len(macbeth_phrases)
import pandas as pd
unique_texts = list(set(macbeth_phrases))
text_counts = {text: macbeth_phrases.count(text) for text in unique_texts}
sorted_texts = sorted(text_counts.items(), key=lambda x: x[1], reverse=True)
macbeth_counts = pd.DataFrame(data=sorted_texts, columns=['text', 'count'])
macbeth_counts
macbeth_counts.head(20)
import cufflinks as cf
cf.go_offline()
macbeth_counts_sorted = macbeth_counts.head(20).sort_values(by='count', ascending=True)
macbeth_counts_sorted.iplot(kind='barh', x='text', y='count', title='Phrase Frequencies in MacBeth', xTitle='Count')
def word_frequency_plot(text_url):
r = requests.get(text_url)
r.encoding = 'utf-8'
if 'gutenberg' in text_url:
text = r.text.split('***')[2]
else:
text = r.text
text = text.replace("’","'").replace("“",'"').replace("”",'"')
phrases = TextBlob(text).noun_phrases
unique_texts = list(set(phrases))
text_counts = {text: phrases.count(text) for text in unique_texts}
sorted_texts = sorted(text_counts.items(), key=lambda x: x[1], reverse=True)
counts = pd.DataFrame(data=sorted_texts, columns=['text', 'count'])
counts_sorted = counts.head(20).sort_values(by='count', ascending=True)
counts_sorted.iplot(kind='barh', x='text', y='count', title='Phrase Frequencies', xTitle='Count')
print('Word frequency plot function defined.')
word_frequency_plot('http://www.gutenberg.org/files/1524/1524-0.txt')
| 0.430626 | 0.956715 |
# Train/fine-tune computer vision (CV) classifiers
In this notebook, we use the annotated images (see, e.g., notebooks `001` and `002`) to train/fine-tune CV classifiers.
```
# solve issue with autocomplete
%config Completer.use_jedi = False
%load_ext autoreload
%autoreload 2
%matplotlib inline
from mapreader import classifier
from mapreader import loadAnnotations
from mapreader import patchTorchDataset
import numpy as np
import torch
from torch import nn
import torchvision
from torchvision import transforms
from torchvision import models
```
## Read annotations
```
annotated_images = loadAnnotations()
annotated_images.load("./annotations_one_inch/rail_space_#kasra#.csv",
path2dir="./maps_tutorial/slice_50_50")
annotated_images.annotations.columns.tolist()
print(annotated_images)
# We need to shift these labels so that they start from 0:
annotated_images.adjust_labels(shiftby=-1)
# show sample images for target label (tar_label)
annotated_images.show_image_labels(tar_label=1, num_sample=6)
# show an image based on its index
annotated_images.show_image(indx=2)
```
### Split annotations into train/val or train/val/test
We use a stratified method for splitting the annotations, that is, each set contains approximately the same percentage of samples of each target label as the original set.
```
annotated_images.split_annotations(frac_train=0.7,
frac_val=0.15,
frac_test=0.15)
```
Dataframes for train, validation and test sets can be accessed via:
```python
annotated_images.train
annotated_images.val
annotated_images.test
```
```
annotated_images.train["label"].value_counts()
annotated_images.val["label"].value_counts()
annotated_images.test["label"].value_counts()
```
# Classifier
## Dataset
Define transformations to be applied to images before being used in training or validation/inference.
`patchTorchDataset` has some default transformations. However, it is possible to define your own transformations and pass them to `patchTorchDataset`:
```
# ------------------
# --- Transformation
# ------------------
# FOR INCEPTION
#resize2 = 299
# otherwise:
resize2 = 224
# mean and standard deviations of pixel intensities in
# all the patches in 6", second edition maps
normalize_mean = 1 - np.array([0.82860442, 0.82515008, 0.77019864])
normalize_std = 1 - np.array([0.1025585, 0.10527616, 0.10039222])
# other options:
# normalize_mean = [0.485, 0.456, 0.406]
# normalize_std = [0.229, 0.224, 0.225]
data_transforms = {
'train': transforms.Compose(
[transforms.Resize(resize2),
transforms.RandomApply([
transforms.RandomHorizontalFlip(p=0.5),
transforms.RandomVerticalFlip(p=0.5),
], p=0.5),
transforms.RandomApply([
transforms.GaussianBlur(21, sigma=(0.5, 5.0)),
], p=0.25),
transforms.RandomApply([
#transforms.RandomPerspective(distortion_scale=0.5, p=0.5),
transforms.Resize((50, 50)),
], p=0.25),
# transforms.RandomApply([
# transforms.RandomAffine(180, translate=None, scale=None, shear=20),
# ], p=0.25),
transforms.Resize(resize2),
transforms.ToTensor(),
transforms.Normalize(normalize_mean, normalize_std)
]),
'val': transforms.Compose(
[transforms.Resize(resize2),
transforms.ToTensor(),
transforms.Normalize(normalize_mean, normalize_std)
]),
}
```
Now, we can use these transformations to instantiate `patchTorchDataset`:
```
train_dataset = patchTorchDataset(annotated_images.train,
transform=data_transforms["train"])
valid_dataset = patchTorchDataset(annotated_images.val,
transform=data_transforms["val"])
test_dataset = patchTorchDataset(annotated_images.test,
transform=data_transforms["val"])
```
## Sampler
```
# -----------
# --- Sampler
# -----------
# We define a sampler as we have a highly imbalanced dataset
label_counts_dict = annotated_images.train["label"].value_counts().to_dict()
class_sample_count = []
for i in range(0, len(label_counts_dict)):
class_sample_count.append(label_counts_dict[i])
weights = 1. / (torch.Tensor(class_sample_count)/1.)
weights = weights.double()
print(f"Weights: {weights}")
train_sampler = torch.utils.data.sampler.WeightedRandomSampler(
weights[train_dataset.patchframe["label"].to_list()],
num_samples=len(train_dataset.patchframe))
valid_sampler = torch.utils.data.sampler.WeightedRandomSampler(
weights[valid_dataset.patchframe["label"].to_list()],
num_samples=len(valid_dataset.patchframe))
```
## Dataloader
```
myclassifier = classifier(device="default")
# myclassifier.load("./checkpoint_12.pkl")
batch_size = 8
# Add training dataset
myclassifier.add2dataloader(train_dataset,
set_name="train",
batch_size=batch_size,
# shuffle can be False as annotations have already been shuffled
shuffle=False,
num_workers=0,
sampler=train_sampler
)
# Add validation dataset
myclassifier.add2dataloader(valid_dataset,
set_name="val",
batch_size=batch_size,
shuffle=False,
num_workers=0,
#sampler=valid_sampler
)
myclassifier.print_classes_dl()
# set class names for plots
class_names = {0: "No",
1: "rail space"}
myclassifier.set_classnames(class_names)
myclassifier.print_classes_dl()
myclassifier.batch_info()
for bn in range(1, 3):
myclassifier.show_sample(set_name="train",
batch_number=bn,
print_batch_info=False)
```
## Load a (pretrained) PyTorch model and add it to `classifier`
Two methods to add a (pretrained) PyTorch model:
1. Define a model using `from torchvision import models`
2. Use `.initialize_model` method
### Method 1: Define a model using `from torchvision import models`
```
# # Choose a model from the supported PyTorch models
# model_ft = models.resnet18(pretrained=True)
# # Add FC based on the number of classes
# num_ftrs = model_ft.fc.in_features
# model_ft.fc = nn.Linear(num_ftrs, myclassifier.num_classes)
# # Add the model to myclassifier
# myclassifier.add_model(model_ft)
# myclassifier.model_summary()
```
### Method 2: use `.initialize_model`
```
myclassifier.del_model()
myclassifier.initialize_model("resnet18",
pretrained=True,
last_layer_num_classes="default",
add_model=True)
myclassifier.model_summary(only_trainable=False)
```
## (Un)freeze layers in the neural network architecture
```
# myclassifier.freeze_layers(["conv1.weight", "bn1.weight", "bn1.bias", "layer1*", "layer2*", "layer3*"])
# myclassifier.model_summary(only_trainable=False)
# myclassifier.unfreeze_layers(["layer3*"])
# myclassifier.model_summary(only_trainable=False)
# myclassifier.only_keep_layers(["fc.weight", "fc.bias"])
# myclassifier.model_summary(only_trainable=True)
```
## Define optimizer, scheduler and criterion
We can either use one learning rate for all the layers in the neural network or define layerwise learning rates, that is, the learning rate of each layer is different. This is normally used in fine-tuning pretrained models in which a smaller learning rate is assigned to the first layers.
`MapReader` has a `.layerwise_lr` method to define layerwise learning rates. By default, `MapReader` uses a linear function to distribute the learning rates (using `min_lr` for the first layer and `max_lr` for the last layer). The linear function can be changed using `ltype="geomspace"` argument.
```
list2optim = myclassifier.layerwise_lr(min_lr=1e-4, max_lr=1e-3)
# #list2optim = myclassifier.layerwise_lr(min_lr=1e-4, max_lr=1e-3, ltype="geomspace")
optim_param_dict = {
"lr": 1e-3,
"betas": (0.9, 0.999),
"eps": 1e-08,
"weight_decay": 0,
"amsgrad": False
}
# --- if list2optim is defined, e.g., by using `.layerwise_lr` method (see the previous cell):
myclassifier.initialize_optimizer(optim_type="adam",
params2optim=list2optim,
optim_param_dict=optim_param_dict,
add_optim=True)
# --- otherwise:
# myclassifier.initialize_optimizer(optim_type="adam",
# optim_param_dict=optim_param_dict,
# add_optim=True)
```
Other optimizers can also be used in the above cell, e.g.:
```python
optim_param_dict = {
"lr": 1e-3,
"momentum": 0,
"dampening": 0,
"weight_decay": 0,
"nesterov": False
}
myclassifier.initialize_optimizer(optim_type="sgd",
optim_param_dict=optim_param_dict,
add_optim=True)
```
```
scheduler_param_dict = {
"step_size": 10,
"gamma": 0.1,
"last_epoch": -1,
"verbose": False
}
myclassifier.initialize_scheduler(scheduler_type="steplr",
scheduler_param_dict=scheduler_param_dict,
add_scheduler=True)
```
Other schedulers can also be used in the above cell, e.g.:
```python
scheduler_param_dict = {
"max_lr": 1e-2,
"steps_per_epoch": len(myclassifier.dataloader["train"]),
"epochs": 5
}
myclassifier.initialize_scheduler(scheduler_type="OneCycleLR",
scheduler_param_dict=scheduler_param_dict,
add_scheduler=True)
```
```
# Add criterion
criterion = nn.CrossEntropyLoss()
myclassifier.add_criterion(criterion)
```
## Train/fine-tune a model
```
myclassifier.train_component_summary()
```
**Note:** it is possible to interrupt a training (using Kernel/Interrupt in Jupyter Notebook or ctrl+C).
```
myclassifier.train(num_epochs=5,
save_model_dir="./models_tutorial",
tensorboard_path="tboard_tutorial",
verbosity_level=0,
tmp_file_save_freq=2,
remove_after_load=False,
print_info_batch_freq=5)
```
### Plot results
```
list(myclassifier.metrics.keys())
myclassifier.plot_metric(y_axis=["epoch_loss_train", "epoch_loss_val"],
y_label="Loss",
legends=["Train", "Valid"],
colors=["k", "tab:red"])
myclassifier.plot_metric(y_axis=["epoch_rocauc_macro_train", "epoch_rocauc_macro_val"],
y_label="ROC AUC",
legends=["Train", "Valid"],
colors=["k", "tab:red"])
myclassifier.plot_metric(y_axis=["epoch_fscore_macro_train",
"epoch_fscore_macro_val",
"epoch_fscore_0_val",
"epoch_fscore_1_val"],
y_label="F-score",
legends=["Train",
"Valid",
"Valid (label: 0)",
"Valid (label: 1)",],
colors=["k", "tab:red", "tab:red", "tab:red"],
styles=["-", "-", "--", ":"],
markers=["o", "o", "", ""],
plt_yrange=[0, 100])
myclassifier.plot_metric(y_axis=["epoch_recall_macro_train",
"epoch_recall_macro_val",
"epoch_recall_0_val",
"epoch_recall_1_val"],
y_label="Recall",
legends=["Train",
"Valid",
"Valid (label: 0)",
"Valid (label: 1)",],
colors=["k", "tab:red", "tab:red", "tab:red"],
styles=["-", "-", "--", ":"],
markers=["o", "o", "", ""],
plt_yrange=[0, 100])
```
## Model inference on test set
Refer to the next notebook for details on model inference. Here, we use the test dataset (defined above) and run the trained model on that set.
```
# Add test dataset
myclassifier.add2dataloader(test_dataset,
set_name="test",
batch_size=batch_size,
shuffle=False,
num_workers=0)
# model inference
myclassifier.inference(set_name="test")
# Calculate metrics using inference outputs
myclassifier.calculate_add_metrics(myclassifier.orig_label,
myclassifier.pred_label,
myclassifier.pred_conf,
"test")
# list of calculated metrics (for test set):
[k for k in myclassifier.metrics if "test" in k]
print(myclassifier.metrics["epoch_fscore_micro_test"])
print(myclassifier.metrics["epoch_rocauc_weighted_test"])
myclassifier.inference_sample_results(num_samples=8,
class_index=1,
set_name="test",
min_conf=50,
max_conf=None)
```
|
github_jupyter
|
# solve issue with autocomplete
%config Completer.use_jedi = False
%load_ext autoreload
%autoreload 2
%matplotlib inline
from mapreader import classifier
from mapreader import loadAnnotations
from mapreader import patchTorchDataset
import numpy as np
import torch
from torch import nn
import torchvision
from torchvision import transforms
from torchvision import models
annotated_images = loadAnnotations()
annotated_images.load("./annotations_one_inch/rail_space_#kasra#.csv",
path2dir="./maps_tutorial/slice_50_50")
annotated_images.annotations.columns.tolist()
print(annotated_images)
# We need to shift these labels so that they start from 0:
annotated_images.adjust_labels(shiftby=-1)
# show sample images for target label (tar_label)
annotated_images.show_image_labels(tar_label=1, num_sample=6)
# show an image based on its index
annotated_images.show_image(indx=2)
annotated_images.split_annotations(frac_train=0.7,
frac_val=0.15,
frac_test=0.15)
annotated_images.train
annotated_images.val
annotated_images.test
annotated_images.train["label"].value_counts()
annotated_images.val["label"].value_counts()
annotated_images.test["label"].value_counts()
# ------------------
# --- Transformation
# ------------------
# FOR INCEPTION
#resize2 = 299
# otherwise:
resize2 = 224
# mean and standard deviations of pixel intensities in
# all the patches in 6", second edition maps
normalize_mean = 1 - np.array([0.82860442, 0.82515008, 0.77019864])
normalize_std = 1 - np.array([0.1025585, 0.10527616, 0.10039222])
# other options:
# normalize_mean = [0.485, 0.456, 0.406]
# normalize_std = [0.229, 0.224, 0.225]
data_transforms = {
'train': transforms.Compose(
[transforms.Resize(resize2),
transforms.RandomApply([
transforms.RandomHorizontalFlip(p=0.5),
transforms.RandomVerticalFlip(p=0.5),
], p=0.5),
transforms.RandomApply([
transforms.GaussianBlur(21, sigma=(0.5, 5.0)),
], p=0.25),
transforms.RandomApply([
#transforms.RandomPerspective(distortion_scale=0.5, p=0.5),
transforms.Resize((50, 50)),
], p=0.25),
# transforms.RandomApply([
# transforms.RandomAffine(180, translate=None, scale=None, shear=20),
# ], p=0.25),
transforms.Resize(resize2),
transforms.ToTensor(),
transforms.Normalize(normalize_mean, normalize_std)
]),
'val': transforms.Compose(
[transforms.Resize(resize2),
transforms.ToTensor(),
transforms.Normalize(normalize_mean, normalize_std)
]),
}
train_dataset = patchTorchDataset(annotated_images.train,
transform=data_transforms["train"])
valid_dataset = patchTorchDataset(annotated_images.val,
transform=data_transforms["val"])
test_dataset = patchTorchDataset(annotated_images.test,
transform=data_transforms["val"])
# -----------
# --- Sampler
# -----------
# We define a sampler as we have a highly imbalanced dataset
label_counts_dict = annotated_images.train["label"].value_counts().to_dict()
class_sample_count = []
for i in range(0, len(label_counts_dict)):
class_sample_count.append(label_counts_dict[i])
weights = 1. / (torch.Tensor(class_sample_count)/1.)
weights = weights.double()
print(f"Weights: {weights}")
train_sampler = torch.utils.data.sampler.WeightedRandomSampler(
weights[train_dataset.patchframe["label"].to_list()],
num_samples=len(train_dataset.patchframe))
valid_sampler = torch.utils.data.sampler.WeightedRandomSampler(
weights[valid_dataset.patchframe["label"].to_list()],
num_samples=len(valid_dataset.patchframe))
myclassifier = classifier(device="default")
# myclassifier.load("./checkpoint_12.pkl")
batch_size = 8
# Add training dataset
myclassifier.add2dataloader(train_dataset,
set_name="train",
batch_size=batch_size,
# shuffle can be False as annotations have already been shuffled
shuffle=False,
num_workers=0,
sampler=train_sampler
)
# Add validation dataset
myclassifier.add2dataloader(valid_dataset,
set_name="val",
batch_size=batch_size,
shuffle=False,
num_workers=0,
#sampler=valid_sampler
)
myclassifier.print_classes_dl()
# set class names for plots
class_names = {0: "No",
1: "rail space"}
myclassifier.set_classnames(class_names)
myclassifier.print_classes_dl()
myclassifier.batch_info()
for bn in range(1, 3):
myclassifier.show_sample(set_name="train",
batch_number=bn,
print_batch_info=False)
# # Choose a model from the supported PyTorch models
# model_ft = models.resnet18(pretrained=True)
# # Add FC based on the number of classes
# num_ftrs = model_ft.fc.in_features
# model_ft.fc = nn.Linear(num_ftrs, myclassifier.num_classes)
# # Add the model to myclassifier
# myclassifier.add_model(model_ft)
# myclassifier.model_summary()
myclassifier.del_model()
myclassifier.initialize_model("resnet18",
pretrained=True,
last_layer_num_classes="default",
add_model=True)
myclassifier.model_summary(only_trainable=False)
# myclassifier.freeze_layers(["conv1.weight", "bn1.weight", "bn1.bias", "layer1*", "layer2*", "layer3*"])
# myclassifier.model_summary(only_trainable=False)
# myclassifier.unfreeze_layers(["layer3*"])
# myclassifier.model_summary(only_trainable=False)
# myclassifier.only_keep_layers(["fc.weight", "fc.bias"])
# myclassifier.model_summary(only_trainable=True)
list2optim = myclassifier.layerwise_lr(min_lr=1e-4, max_lr=1e-3)
# #list2optim = myclassifier.layerwise_lr(min_lr=1e-4, max_lr=1e-3, ltype="geomspace")
optim_param_dict = {
"lr": 1e-3,
"betas": (0.9, 0.999),
"eps": 1e-08,
"weight_decay": 0,
"amsgrad": False
}
# --- if list2optim is defined, e.g., by using `.layerwise_lr` method (see the previous cell):
myclassifier.initialize_optimizer(optim_type="adam",
params2optim=list2optim,
optim_param_dict=optim_param_dict,
add_optim=True)
# --- otherwise:
# myclassifier.initialize_optimizer(optim_type="adam",
# optim_param_dict=optim_param_dict,
# add_optim=True)
optim_param_dict = {
"lr": 1e-3,
"momentum": 0,
"dampening": 0,
"weight_decay": 0,
"nesterov": False
}
myclassifier.initialize_optimizer(optim_type="sgd",
optim_param_dict=optim_param_dict,
add_optim=True)
scheduler_param_dict = {
"step_size": 10,
"gamma": 0.1,
"last_epoch": -1,
"verbose": False
}
myclassifier.initialize_scheduler(scheduler_type="steplr",
scheduler_param_dict=scheduler_param_dict,
add_scheduler=True)
scheduler_param_dict = {
"max_lr": 1e-2,
"steps_per_epoch": len(myclassifier.dataloader["train"]),
"epochs": 5
}
myclassifier.initialize_scheduler(scheduler_type="OneCycleLR",
scheduler_param_dict=scheduler_param_dict,
add_scheduler=True)
# Add criterion
criterion = nn.CrossEntropyLoss()
myclassifier.add_criterion(criterion)
myclassifier.train_component_summary()
myclassifier.train(num_epochs=5,
save_model_dir="./models_tutorial",
tensorboard_path="tboard_tutorial",
verbosity_level=0,
tmp_file_save_freq=2,
remove_after_load=False,
print_info_batch_freq=5)
list(myclassifier.metrics.keys())
myclassifier.plot_metric(y_axis=["epoch_loss_train", "epoch_loss_val"],
y_label="Loss",
legends=["Train", "Valid"],
colors=["k", "tab:red"])
myclassifier.plot_metric(y_axis=["epoch_rocauc_macro_train", "epoch_rocauc_macro_val"],
y_label="ROC AUC",
legends=["Train", "Valid"],
colors=["k", "tab:red"])
myclassifier.plot_metric(y_axis=["epoch_fscore_macro_train",
"epoch_fscore_macro_val",
"epoch_fscore_0_val",
"epoch_fscore_1_val"],
y_label="F-score",
legends=["Train",
"Valid",
"Valid (label: 0)",
"Valid (label: 1)",],
colors=["k", "tab:red", "tab:red", "tab:red"],
styles=["-", "-", "--", ":"],
markers=["o", "o", "", ""],
plt_yrange=[0, 100])
myclassifier.plot_metric(y_axis=["epoch_recall_macro_train",
"epoch_recall_macro_val",
"epoch_recall_0_val",
"epoch_recall_1_val"],
y_label="Recall",
legends=["Train",
"Valid",
"Valid (label: 0)",
"Valid (label: 1)",],
colors=["k", "tab:red", "tab:red", "tab:red"],
styles=["-", "-", "--", ":"],
markers=["o", "o", "", ""],
plt_yrange=[0, 100])
# Add test dataset
myclassifier.add2dataloader(test_dataset,
set_name="test",
batch_size=batch_size,
shuffle=False,
num_workers=0)
# model inference
myclassifier.inference(set_name="test")
# Calculate metrics using inference outputs
myclassifier.calculate_add_metrics(myclassifier.orig_label,
myclassifier.pred_label,
myclassifier.pred_conf,
"test")
# list of calculated metrics (for test set):
[k for k in myclassifier.metrics if "test" in k]
print(myclassifier.metrics["epoch_fscore_micro_test"])
print(myclassifier.metrics["epoch_rocauc_weighted_test"])
myclassifier.inference_sample_results(num_samples=8,
class_index=1,
set_name="test",
min_conf=50,
max_conf=None)
| 0.718594 | 0.938407 |
Plots: Campaign 8 control objects before/after PLD
Re-creating plots that I've lost the code for. These plots should show the campaign 8 control objects
```
from agn_everest.analysis import *
import seaborn as sns
import richardsplot
path = "object_lists/c8_control/"
df = pd.read_csv(path+"K2Campaign8_PSDslopes_GO8012-targets.csv.csv", skipinitialspace=False)
GOs = ['GO8012', 'GO8042', 'GO8077', 'GO8051']
colors = ['#3498db', '#e67e22', '#27ae60', '#e74c3c' ]
fig,ax = plt.subplots(1,2, figsize=(12,5), sharey=True, gridspec_kw = {'wspace':0, 'hspace':0})
for GO, c in zip(GOs, colors):
df = pd.read_csv(path+"K2Campaign8_PSDslopes_%s-targets.csv.csv"%GO, skipinitialspace=False)
# Before PLD
mask = (df['magnitude']!=' ')
mags = np.array(df['magnitude'][mask]).astype(float)
slope = np.array(df['PSD_m_raw_above_noise'][mask])
mask2 = ~np.isnan(slope)
mags = mags[mask2]
slope = slope[mask2]
ax[0].plot(mags, slope,'.', color=c, alpha = 0.15)#, label=GO)
sns.kdeplot(mags, slope, color=c, shade_lowest=False, shade=True, label=GO, ax=ax[0])
# After PLD
mask = (df['magnitude']!=' ')
mags = np.array(df['magnitude'][mask]).astype(float)
slope = np.array(df['PSD_m_cor_above_noise'][mask])
mask2 = ~np.isnan(slope)
mags = mags[mask2]
slope = slope[mask2]
ax[1].plot(mags, slope,'.', color=c, alpha = 0.15)#, label=GO)
sns.kdeplot(mags, slope, color=c, shade_lowest=False, shade=True, label=GO, ax=ax[1])
ax[0].plot(ax[0].get_xlim(), np.zeros(2), 'k--', alpha=0.5)
ax[0].plot(ax[0].get_xlim(), np.zeros(2)-2, 'k--', alpha=0.5)
ax[1].plot(ax[1].get_xlim(), np.zeros(2), 'k--', alpha=0.5)
ax[1].plot(ax[1].get_xlim(), np.zeros(2)-2, 'k--', alpha=0.5)
ax[0].set_ylabel("PSD Slope", fontsize=20)
#ax[0].legend()
#ax[1].set_ylabel("PSD Slope", fontsize=20)
#ax[1].set_xlabel("Kepler Magnitude", fontsize=20)
fig.text(0.5, 0.0, 'Kepler Magnitude', fontsize=20,ha='center')
ax[1].legend()
plt.tight_layout()
plt.savefig("submit/Plots/C8_control_PLD.pdf")
fig,ax = plt.subplots(2,1, figsize=(8,8), sharex=True, gridspec_kw = {'wspace':0, 'hspace':0})
m_list_raw = []
m_list_cor = []
# make histogram (of same thing I guess)
for GO, c in zip(GOs, colors):
df = pd.read_csv(path+"K2Campaign8_PSDslopes_%s-targets.csv.csv"%GO, skipinitialspace=False)
# Before PLD
mask = (df['magnitude']!=' ')
mags = np.array(df['magnitude'][mask]).astype(float)
slope = np.array(df['PSD_m_raw_above_noise'][mask])
mask2 = ~np.isnan(slope)
m_list_raw.append(slope[mask2])
# After PLD
mask = (df['magnitude']!=' ')
mags = np.array(df['magnitude'][mask]).astype(float)
slope = np.array(df['PSD_m_cor_above_noise'][mask])
mask2 = ~np.isnan(slope)
m_list_cor.append(slope[mask2])
ax[0].hist(m_list_raw , color=colors, histtype=u'step', linewidth=4, alpha=0.7)
ax[1].hist(m_list_cor , color=colors, histtype=u'step', linewidth=4, alpha=0.7)
ax[0].legend(GOs, loc='upper left')
ax[0].plot(np.zeros(2), ax[0].get_ylim(), 'k--', alpha=0.5)
ax[0].plot(np.zeros(2)-2, ax[0].get_ylim(), 'k--', alpha=0.5)
ax[1].plot(np.zeros(2), ax[1].get_ylim(), 'k--', alpha=0.5)
ax[1].plot(np.zeros(2)-2, ax[1].get_ylim(), 'k--', alpha=0.5)
ax[1].set_xlabel('PSD Slope', fontsize=20)
plt.tight_layout()
plt.savefig("submit/Plots/C8_control_PLD_hist.pdf")
```
---
Creating plot showing what can happen to a periodic object after PLD (doesn't happen to all of them)
```
epic=220182448
#epic = 220232812
campaign=8
#time = np.arange(3853)/48.0# time in days since start of campaign
%%time
# plotting
fig,ax = plt.subplots(2,2, figsize=(12,8), sharex='col', gridspec_kw = {'wspace':0.25,'hspace':0})
#
fake_omega = np.logspace(-6, -4, base=10)
fake_pow = fake_omega**(-2) / 1e9
ax[0,1].plot(fake_omega, fake_pow, ':', color='gray', label='$S \propto f^{-2}$', linewidth=1.5)
ax[1,1].plot(fake_omega, fake_pow, ':', color='gray', label='$S \propto f^{-2}$', linewidth=1.5)
# ----------------------------RAW------------------------------
lc_everest = everest.Everest(epic, season=campaign, mission='k2')
time = lc_everest.time.copy()
lc_raw = lc_everest.fraw.copy()
# interpolate bad cadences
cadno = np.arange(len(lc_everest.time))
mask = (np.isin(cadno, np.concatenate([lc_everest.nanmask, lc_everest.badmask, lc_everest.mask])))
interped_vals = np.interp(cadno[mask], cadno[~mask], lc_raw[~mask])
# replace spurious cadence values with the interpolated values
lc_raw[mask] = interped_vals
# calculate PSD slopes
freq_raw, power_raw = LS_PSD(time*86400, lc_raw, f = k2_freq)
noise_floor_days = 5
# noise floor are freqencies > X days, convert to Hz
noise_floor_mask = freq_raw>(2*np.pi/(noise_floor_days*86400))
m_raw, b_raw = np.polyfit(np.log10(freq_raw)[~noise_floor_mask], np.log10(power_raw)[~noise_floor_mask], 1)
m_rnoise, b_rnoise = np.polyfit(np.log10(freq_raw)[noise_floor_mask], np.log10(power_raw)[noise_floor_mask], 1)
f,power = LS_PSD(time*86400,lc_raw, f=k2_freq)
ax[0,0].plot(time,lc_raw, marker='.', ls='', label="EPIC %s Raw light curve"%epic, color='#34495e')
#ax[0,0].set_xlabel("Time - 2454833[BKJD days]")
ax[0,0].set_ylabel("Flux [$e^-s^{-1}$]")
ax[0,0].legend()
ax[0,1].plot(f,power, label="Raw PSD", marker='.', ls='', alpha=0.3, color='#34495e')
#ax[0,1].set_xlabel("frequency [Hz]")
ax[0,1].set_ylabel("power [$\mathrm{ppm}^2/\mathrm{Hz}$]")
ax[0,1].set_yscale("log")
ax[0,1].set_xscale("log")
ax[0,1].plot(freq_raw[~noise_floor_mask], 10**(np.log10(freq_raw[~noise_floor_mask])*m_raw+b_raw),
label="fit above noise floor m = %.2f"%m_raw, linewidth=3, alpha = 1, color='#3498db')
ax[0,1].plot(freq_raw[noise_floor_mask], 10**(np.log10(freq_raw[noise_floor_mask])*m_rnoise+b_rnoise),
label="fit below noise floor m = %.2f"%m_rnoise, linewidth=3, alpha = 1, color='#e74c3c')
ax[0,1].legend(loc='lower left')
# -----------------------EVEREST Corrected (2 CBVs)------------------------
tcorr, fcorr = lc_everest.time.copy(), lc_everest.flux.copy()
interped_vals = np.interp(cadno[mask], cadno[~mask], fcorr[~mask])
# replace spurious cadence values with the interpolated values
fcorr[mask] = interped_vals
f,power = LS_PSD(tcorr*86400,fcorr, f=k2_freq)
# calculate PSD slopes
freq_corr, power_corr = LS_PSD(tcorr*86400, fcorr, f = k2_freq)
noise_floor_days = 5
# noise floor are freqencies > X days, convert to Hz
noise_floor_mask = freq_corr>(2*np.pi/(noise_floor_days*86400))
m, b = np.polyfit(np.log10(freq_corr)[~noise_floor_mask], np.log10(power_corr)[~noise_floor_mask], 1)
m_noise, b_noise = np.polyfit(np.log10(freq_corr)[noise_floor_mask], np.log10(power_corr)[noise_floor_mask], 1)
ax[1,0].plot(tcorr, fcorr, marker='.', ls='', label="EPIC %s PLD light curve"%epic, color='#273c75')
ax[1,0].set_xlabel("Time - 2454833[BKJD days]")
ax[1,0].set_ylabel("Flux [$e^-s^{-1}$]")
ax[1,0].legend()
ax[1,1].plot(f,power, label="EVERST PLD PSD", marker='.', ls='', alpha=0.3, color='#273c75')
ax[1,1].set_xlabel("frequency [Hz]")
ax[1,1].set_ylabel("power [$\mathrm{ppm}^2/\mathrm{Hz}$]")
ax[1,1].set_yscale("log")
ax[1,1].set_xscale("log")
ax[1,1].plot(freq_raw[~noise_floor_mask], 10**(np.log10(freq_raw[~noise_floor_mask])*m+b),
label="fit above noise floor m = %.2f"%m, linewidth=3, alpha = 1, color='#3498db')
ax[1,1].plot(freq_raw[noise_floor_mask], 10**(np.log10(freq_raw[noise_floor_mask])*m_noise+b_noise),
label="fit below noise floor m = %.2f"%m_noise, linewidth=3, alpha = 1, color='#e74c3c')
ax[1,1].legend(loc='lower left')
plt.tight_layout()
plt.savefig("./submit/Plots/K2C8_PLD_Control_%s.pdf"%epic)
```
|
github_jupyter
|
from agn_everest.analysis import *
import seaborn as sns
import richardsplot
path = "object_lists/c8_control/"
df = pd.read_csv(path+"K2Campaign8_PSDslopes_GO8012-targets.csv.csv", skipinitialspace=False)
GOs = ['GO8012', 'GO8042', 'GO8077', 'GO8051']
colors = ['#3498db', '#e67e22', '#27ae60', '#e74c3c' ]
fig,ax = plt.subplots(1,2, figsize=(12,5), sharey=True, gridspec_kw = {'wspace':0, 'hspace':0})
for GO, c in zip(GOs, colors):
df = pd.read_csv(path+"K2Campaign8_PSDslopes_%s-targets.csv.csv"%GO, skipinitialspace=False)
# Before PLD
mask = (df['magnitude']!=' ')
mags = np.array(df['magnitude'][mask]).astype(float)
slope = np.array(df['PSD_m_raw_above_noise'][mask])
mask2 = ~np.isnan(slope)
mags = mags[mask2]
slope = slope[mask2]
ax[0].plot(mags, slope,'.', color=c, alpha = 0.15)#, label=GO)
sns.kdeplot(mags, slope, color=c, shade_lowest=False, shade=True, label=GO, ax=ax[0])
# After PLD
mask = (df['magnitude']!=' ')
mags = np.array(df['magnitude'][mask]).astype(float)
slope = np.array(df['PSD_m_cor_above_noise'][mask])
mask2 = ~np.isnan(slope)
mags = mags[mask2]
slope = slope[mask2]
ax[1].plot(mags, slope,'.', color=c, alpha = 0.15)#, label=GO)
sns.kdeplot(mags, slope, color=c, shade_lowest=False, shade=True, label=GO, ax=ax[1])
ax[0].plot(ax[0].get_xlim(), np.zeros(2), 'k--', alpha=0.5)
ax[0].plot(ax[0].get_xlim(), np.zeros(2)-2, 'k--', alpha=0.5)
ax[1].plot(ax[1].get_xlim(), np.zeros(2), 'k--', alpha=0.5)
ax[1].plot(ax[1].get_xlim(), np.zeros(2)-2, 'k--', alpha=0.5)
ax[0].set_ylabel("PSD Slope", fontsize=20)
#ax[0].legend()
#ax[1].set_ylabel("PSD Slope", fontsize=20)
#ax[1].set_xlabel("Kepler Magnitude", fontsize=20)
fig.text(0.5, 0.0, 'Kepler Magnitude', fontsize=20,ha='center')
ax[1].legend()
plt.tight_layout()
plt.savefig("submit/Plots/C8_control_PLD.pdf")
fig,ax = plt.subplots(2,1, figsize=(8,8), sharex=True, gridspec_kw = {'wspace':0, 'hspace':0})
m_list_raw = []
m_list_cor = []
# make histogram (of same thing I guess)
for GO, c in zip(GOs, colors):
df = pd.read_csv(path+"K2Campaign8_PSDslopes_%s-targets.csv.csv"%GO, skipinitialspace=False)
# Before PLD
mask = (df['magnitude']!=' ')
mags = np.array(df['magnitude'][mask]).astype(float)
slope = np.array(df['PSD_m_raw_above_noise'][mask])
mask2 = ~np.isnan(slope)
m_list_raw.append(slope[mask2])
# After PLD
mask = (df['magnitude']!=' ')
mags = np.array(df['magnitude'][mask]).astype(float)
slope = np.array(df['PSD_m_cor_above_noise'][mask])
mask2 = ~np.isnan(slope)
m_list_cor.append(slope[mask2])
ax[0].hist(m_list_raw , color=colors, histtype=u'step', linewidth=4, alpha=0.7)
ax[1].hist(m_list_cor , color=colors, histtype=u'step', linewidth=4, alpha=0.7)
ax[0].legend(GOs, loc='upper left')
ax[0].plot(np.zeros(2), ax[0].get_ylim(), 'k--', alpha=0.5)
ax[0].plot(np.zeros(2)-2, ax[0].get_ylim(), 'k--', alpha=0.5)
ax[1].plot(np.zeros(2), ax[1].get_ylim(), 'k--', alpha=0.5)
ax[1].plot(np.zeros(2)-2, ax[1].get_ylim(), 'k--', alpha=0.5)
ax[1].set_xlabel('PSD Slope', fontsize=20)
plt.tight_layout()
plt.savefig("submit/Plots/C8_control_PLD_hist.pdf")
epic=220182448
#epic = 220232812
campaign=8
#time = np.arange(3853)/48.0# time in days since start of campaign
%%time
# plotting
fig,ax = plt.subplots(2,2, figsize=(12,8), sharex='col', gridspec_kw = {'wspace':0.25,'hspace':0})
#
fake_omega = np.logspace(-6, -4, base=10)
fake_pow = fake_omega**(-2) / 1e9
ax[0,1].plot(fake_omega, fake_pow, ':', color='gray', label='$S \propto f^{-2}$', linewidth=1.5)
ax[1,1].plot(fake_omega, fake_pow, ':', color='gray', label='$S \propto f^{-2}$', linewidth=1.5)
# ----------------------------RAW------------------------------
lc_everest = everest.Everest(epic, season=campaign, mission='k2')
time = lc_everest.time.copy()
lc_raw = lc_everest.fraw.copy()
# interpolate bad cadences
cadno = np.arange(len(lc_everest.time))
mask = (np.isin(cadno, np.concatenate([lc_everest.nanmask, lc_everest.badmask, lc_everest.mask])))
interped_vals = np.interp(cadno[mask], cadno[~mask], lc_raw[~mask])
# replace spurious cadence values with the interpolated values
lc_raw[mask] = interped_vals
# calculate PSD slopes
freq_raw, power_raw = LS_PSD(time*86400, lc_raw, f = k2_freq)
noise_floor_days = 5
# noise floor are freqencies > X days, convert to Hz
noise_floor_mask = freq_raw>(2*np.pi/(noise_floor_days*86400))
m_raw, b_raw = np.polyfit(np.log10(freq_raw)[~noise_floor_mask], np.log10(power_raw)[~noise_floor_mask], 1)
m_rnoise, b_rnoise = np.polyfit(np.log10(freq_raw)[noise_floor_mask], np.log10(power_raw)[noise_floor_mask], 1)
f,power = LS_PSD(time*86400,lc_raw, f=k2_freq)
ax[0,0].plot(time,lc_raw, marker='.', ls='', label="EPIC %s Raw light curve"%epic, color='#34495e')
#ax[0,0].set_xlabel("Time - 2454833[BKJD days]")
ax[0,0].set_ylabel("Flux [$e^-s^{-1}$]")
ax[0,0].legend()
ax[0,1].plot(f,power, label="Raw PSD", marker='.', ls='', alpha=0.3, color='#34495e')
#ax[0,1].set_xlabel("frequency [Hz]")
ax[0,1].set_ylabel("power [$\mathrm{ppm}^2/\mathrm{Hz}$]")
ax[0,1].set_yscale("log")
ax[0,1].set_xscale("log")
ax[0,1].plot(freq_raw[~noise_floor_mask], 10**(np.log10(freq_raw[~noise_floor_mask])*m_raw+b_raw),
label="fit above noise floor m = %.2f"%m_raw, linewidth=3, alpha = 1, color='#3498db')
ax[0,1].plot(freq_raw[noise_floor_mask], 10**(np.log10(freq_raw[noise_floor_mask])*m_rnoise+b_rnoise),
label="fit below noise floor m = %.2f"%m_rnoise, linewidth=3, alpha = 1, color='#e74c3c')
ax[0,1].legend(loc='lower left')
# -----------------------EVEREST Corrected (2 CBVs)------------------------
tcorr, fcorr = lc_everest.time.copy(), lc_everest.flux.copy()
interped_vals = np.interp(cadno[mask], cadno[~mask], fcorr[~mask])
# replace spurious cadence values with the interpolated values
fcorr[mask] = interped_vals
f,power = LS_PSD(tcorr*86400,fcorr, f=k2_freq)
# calculate PSD slopes
freq_corr, power_corr = LS_PSD(tcorr*86400, fcorr, f = k2_freq)
noise_floor_days = 5
# noise floor are freqencies > X days, convert to Hz
noise_floor_mask = freq_corr>(2*np.pi/(noise_floor_days*86400))
m, b = np.polyfit(np.log10(freq_corr)[~noise_floor_mask], np.log10(power_corr)[~noise_floor_mask], 1)
m_noise, b_noise = np.polyfit(np.log10(freq_corr)[noise_floor_mask], np.log10(power_corr)[noise_floor_mask], 1)
ax[1,0].plot(tcorr, fcorr, marker='.', ls='', label="EPIC %s PLD light curve"%epic, color='#273c75')
ax[1,0].set_xlabel("Time - 2454833[BKJD days]")
ax[1,0].set_ylabel("Flux [$e^-s^{-1}$]")
ax[1,0].legend()
ax[1,1].plot(f,power, label="EVERST PLD PSD", marker='.', ls='', alpha=0.3, color='#273c75')
ax[1,1].set_xlabel("frequency [Hz]")
ax[1,1].set_ylabel("power [$\mathrm{ppm}^2/\mathrm{Hz}$]")
ax[1,1].set_yscale("log")
ax[1,1].set_xscale("log")
ax[1,1].plot(freq_raw[~noise_floor_mask], 10**(np.log10(freq_raw[~noise_floor_mask])*m+b),
label="fit above noise floor m = %.2f"%m, linewidth=3, alpha = 1, color='#3498db')
ax[1,1].plot(freq_raw[noise_floor_mask], 10**(np.log10(freq_raw[noise_floor_mask])*m_noise+b_noise),
label="fit below noise floor m = %.2f"%m_noise, linewidth=3, alpha = 1, color='#e74c3c')
ax[1,1].legend(loc='lower left')
plt.tight_layout()
plt.savefig("./submit/Plots/K2C8_PLD_Control_%s.pdf"%epic)
| 0.312895 | 0.706361 |
In the previous tutorials we introduced you to some basic finite fields
$$\mathbb{Z}_p$$
and the collection of polynomials (polynomial rings if we want to use jargon) over them
$$\mathbb{Z}_p[X]$$
While these finite fields are very interesting, they're often not the most convenient space to do cryptographic computations. Part of the reason is that they're kind of exotic. As humans, we don't expect numbers to wrap at 19 or 31 or whatever prime we choose. How about
$$\mathbb{Z}_{10}$$
then? Unfortunately, this isn't a finite field (since 10 factors into 2,5). It turns out that the interesting place to look is "binary" finite fields. What's a binary finite field? Well to give a simplistic explanation, it's any finite field where
$$1 + 1 = 0$$
The simplest example of a binary finite field is
$$\mathbb{Z}_2$$
Let's play with this finite field a bit.
```
import starks
from starks.modp import IntegersModP
mod2 = IntegersModP(2)
one = mod2(1)
one + one
```
Ok, this is neat. Let's play with this a bit more. Can we make an `AND` gate for example? Recall that an `AND` gate implements the logical operator `AND`
Luckily, it turns out that the `AND` gate is really simple to make! It's just a multiplication operation in $\mathbb{Z}_2$.
```
def AND(x, y):
return x * y
zero = mod2(0)
one = mod2(1)
print("AND(0, 0) = %s" % str(AND(zero, zero)))
print("AND(0, 1) = %s" % str(AND(zero, one)))
print("AND(1, 0) = %s" % str(AND(one, zero)))
print("AND(1, 1) = %s" % str(AND(one, one)))
```
This is pretty neat. Note that `AND` is equivalently represented as a simple polynomial in $\mathbb{Z}_2[X, y]$
$$\textrm{and}(x, y) = xy$$
Can we do this for other logical operators? Let's take a look at an `OR` gate
This one is a little trickier to write down as a function, but it's not too bad.
```
def OR(x, y):
return x + y - x * y
zero = mod2(0)
one = mod2(1)
print("OR(0, 0) = %s" % str(OR(zero, zero)))
print("OR(0, 1) = %s" % str(OR(zero, one)))
print("OR(1, 0) = %s" % str(OR(one, zero)))
print("OR(1, 1) = %s" % str(OR(one, one)))
```
Take a minute to study this function to understand why it makes sense. Note again that we can write `OR` as a polynomial
$$\textrm{OR}(x, y) = x + y - xy$$
How about `NOT`? Turns out this one is pretty simple to do too.
```
def NOT(x):
return (1 - x)
zero = mod2(0)
one = mod2(1)
print("NOT(0) = %s" % str(NOT(zero)))
print("NOT(1) = %s" % str(NOT(one)))
```
This is an interesting result since the three logical operators `OR`, `AND`, and `NOT` are "functionally complete". That means that any boolean circuit can be represented in terms of these operators. Since we've encoded these operators as polynomials over $\mathbb{Z}_2$, we now know we can represent any boolean circuit as a polynomial!
This is neat result, but it's not yet as useful as we'd like it. For one thing, we're operating on $\mathbb{Z}_2$, which is essentially a single bit. What if we want to represent a complex program, which accepts as input 64 bit words? We could represent 64-bit words as lists of 64 elements of $\mathbb{Z}_2$, but this is somewhat awkward. Isn't there a more elegant way to represent such words? How about the set of numbers $\mathbb{Z}/2^{64}$ modulo $2^{64}$? This seems to fit what we're looking for, but we know that it's not a finite field. Division doesn't work since $2^{64}$ has many prime factors.
It seems like we're stuck here. Luckily, there is a way to construct a finite field of size $2^{64}$. The technical construction is a little complex, so we're going to start by just putting some simple code examples in front of you to start.
```
from starks.polynomial import polynomials_over
from starks.finitefield import FiniteField
p = 2
m = 64
Zp = IntegersModP(p)
polysOver = polynomials_over(Zp)
#x^64 + x^4 + x^3 + x + 1 is irreducible (in fact primitive). Don't worry about what this means yet
coefficients = [Zp(0)] * 65
coefficients[0] = Zp(1)
coefficients[1] = Zp(1)
coefficients[3] = Zp(1)
coefficients[4] = Zp(1)
coefficients[64] = Zp(1)
poly = polysOver(coefficients)
field = FiniteField(p, m, polynomialModulus=poly)
```
This probably seems a little mysterious. What is this business with a primitive polynomial? What do polynomials have to do with anything here? You'll learn more about the details of these polynomials in the next part of this tutorial, but for now, let's treat these objects as givens and just play with some examples.
The first thing to note is that elements of a finite field are polynomials. Let's start with some really basic definitions.
```
A = field(polysOver([1,1,1]))
A
B = field(polysOver([1,0,1,1]))
B
```
For now, don't worry too much about the precise definitions of `A` and `B`. We'll come back and explain what these mean in the next tutorial. But let's just see that we can do all the usual things we'd expect to be able to do in a finite field with them.
```
A + B
A - B
A * B
A / B
```
We can add, subtract, multiply and divide these quantities. The precise mechanics of what these operations will be explained in more detail soon, but for now you can try running a few different operations.
How should you be thinking about binary finite fields? They seem to be these strange objects that are complicated polynomials. How does this tie into our earlier discussion about binary operations in $\mathbb{Z}_2$? Well, we now have a way to turn 64 bit words into these binary finite field objects. There's a lot of questions this linkage brings up. How do the addition, subtractions, multiplication, and division operations shown here compare with the "usual" notions that that processors do? It turns out this question is a little subtle. There are two different sets of arithmetic operations here, one belonging to the finite field, another which is the "usual" arithmetic. This is shading into research, so we won't say too much more on this topic yet.
## Exercises
1. Define a new finite field element `C` like we did `A` and `B` above. Try some basic arithmetic operations with `C`.
2. (Challenging) Suppose you are given a 64 bit word. For example, an 8 bit word would be `01110001`. A 64 bit word would be formed similarly. Can you see how to turn this word into a finite field element? Write a function that accepts a arbitrary 64 bit word and turns it into a finite field element. Hint: Turn the 64 bit word into a list of 0s and 1s of length 64.
|
github_jupyter
|
import starks
from starks.modp import IntegersModP
mod2 = IntegersModP(2)
one = mod2(1)
one + one
def AND(x, y):
return x * y
zero = mod2(0)
one = mod2(1)
print("AND(0, 0) = %s" % str(AND(zero, zero)))
print("AND(0, 1) = %s" % str(AND(zero, one)))
print("AND(1, 0) = %s" % str(AND(one, zero)))
print("AND(1, 1) = %s" % str(AND(one, one)))
def OR(x, y):
return x + y - x * y
zero = mod2(0)
one = mod2(1)
print("OR(0, 0) = %s" % str(OR(zero, zero)))
print("OR(0, 1) = %s" % str(OR(zero, one)))
print("OR(1, 0) = %s" % str(OR(one, zero)))
print("OR(1, 1) = %s" % str(OR(one, one)))
def NOT(x):
return (1 - x)
zero = mod2(0)
one = mod2(1)
print("NOT(0) = %s" % str(NOT(zero)))
print("NOT(1) = %s" % str(NOT(one)))
from starks.polynomial import polynomials_over
from starks.finitefield import FiniteField
p = 2
m = 64
Zp = IntegersModP(p)
polysOver = polynomials_over(Zp)
#x^64 + x^4 + x^3 + x + 1 is irreducible (in fact primitive). Don't worry about what this means yet
coefficients = [Zp(0)] * 65
coefficients[0] = Zp(1)
coefficients[1] = Zp(1)
coefficients[3] = Zp(1)
coefficients[4] = Zp(1)
coefficients[64] = Zp(1)
poly = polysOver(coefficients)
field = FiniteField(p, m, polynomialModulus=poly)
A = field(polysOver([1,1,1]))
A
B = field(polysOver([1,0,1,1]))
B
A + B
A - B
A * B
A / B
| 0.359252 | 0.974362 |
## Lambda Functions
Sometimes we want to use functions as arguments to other functions or define them in-line (within) other pieces of code. When this happens, it is sometimes desirable to define them **anonymously** as **lambda functions**. A lambda function is simply a function without a name. Let's first start by writing a simple function to add two numbers. We could write:
```
def add(num1, num2):
return num1 + num2
```
A lambda function is defined in a very similar way, but we use the `lambda` keyword instead. Lambda functions have the form `lambda arguments: expression`. Let's define the same function as a lambda function now.
```
lambda_add = lambda num1, num2: num1 + num2
```
How do we use it? Just like any normal function!
```
print(add(3, 17))
print(lambda_add(3, 17))
# We get the same answer using the lambda function.
# "3" gets assigned to "a", and "17" gets assigned to "b"
```
OK, so now we've seen how to define lambda functions, you might be wondering **Why use lambdas?**. The nice thing about lambda functions is that they can be defined **in-line.** That is, they can be defined within other pieces of code spontaneously, which makes them convenient and concise, particularly if they perform an operation that isn't reused elsewhere.
Let's look at a simple example to see why they might be useful. In Python, the `map(input function, input function arguments)` function takes as input
* a function
* that functions arguments
The reason it's called "map" is because it allows us to repeatedly apply, or "map", the input function if a *list* of arguments is passed in. Let's look at a simple example to get a better idea of what's going on:
```
# Let's start by defining a simple function times_two
# Notice that this function takes in a SINGLE argument
def times_two(number):
return number * 2
numbers = [1, 2, 3, 4, 5, 6]
# We pass our function, "times_two", as the first argument to "map".
# It then iterates like a for-loop over each number in "numbers",
# passing them as input to the "times_two" function and outputs
# the result.
numbers2 = list(map(times_two, numbers))
print(numbers2)
```
What exactly is happening? We have our function `times_two` that takes in a single argument, and yet it looks like we're passing it a list of numbers. What's actually happening is that `times_two` is getting called repeatedly, once for each number in the numbers list, e.g.
```
numbers = [1, 2, 3, 4, 5, 6]
list(map(times_two, numbers))
...
numbers2 = [times_two(1), times_two(2), times_two(3),
times_two(4), times_two(5), times_two(6)]
```
A simple implementation of `map` might look like:
```
def my_map(func, argument_list):
output = []
for argument in argument_list:
output.append(func(argument))
return output
print(my_map(times_two, numbers))
```
In reality, `map` is a bit more sophisticated than that, but that covers its basic functionality. What happens if the function you pass into `map` takes two arguments? We simply pass in another argument list. **Note:** The argument lists must be of the same length/size, though, otherwise there will be a mismatch and an error will occur.
```
# Let's start by defining a simple function times_multipler
def times_multipler(number, multiplier):
return number * multiplier
numbers = [1, 2, 3, 4, 5, 6]
multipliers = [2, 4, 6, 8, 10, 12]
numbers2 = list(map(times_multipler, numbers, multipliers))
print(numbers2)
```
It's often the case that we want to apply operations to lists of things, but we don't always want to define functions for these operations, especially if they're only going to be used once. **Lambda functions** are a more concise way to do exactly that. Let's look at the `times_two` example again and see how we can rewrite it. First, let's just establish that the following approach is *not* allowed:
```
numbers = [1, 2, 3, 4, 5, 6]
numbers2 = list(map(times_two, def times_two(number): return number * 2))
print(numbers2)
```
We can't define functions within other code in Python. We can, however, define lambda functions **in-line** just fine:
```
numbers = [1, 2, 3, 4, 5, 6]
numbers2 = list(map(lambda number: number * 2, numbers))
print(numbers2)
```
We can see that by writing a lambda function **in-line** inside the map function we've acheived the same result in a much more concise way! `lambda number: number * 2` serves as the function argument to the `map` function, meaning we don't have to define the `times_two` function to get the same result.
Now let's try some exercises!
### Exercises
You're given two lists, one of widths and one of heights. Write a function that iterates through both lists pairwise (1st element widths * 1st element heights, 2nd element widths * 2nd element heights, ...) and returns a list of areas.
```
widths = [2, 4, 5, 6, 7, 8]
heights = [4, 5, 3, 2, 5, 7]
```
Rewrite the function you wrote above using `map` and a lambda function.
|
github_jupyter
|
def add(num1, num2):
return num1 + num2
lambda_add = lambda num1, num2: num1 + num2
print(add(3, 17))
print(lambda_add(3, 17))
# We get the same answer using the lambda function.
# "3" gets assigned to "a", and "17" gets assigned to "b"
# Let's start by defining a simple function times_two
# Notice that this function takes in a SINGLE argument
def times_two(number):
return number * 2
numbers = [1, 2, 3, 4, 5, 6]
# We pass our function, "times_two", as the first argument to "map".
# It then iterates like a for-loop over each number in "numbers",
# passing them as input to the "times_two" function and outputs
# the result.
numbers2 = list(map(times_two, numbers))
print(numbers2)
numbers = [1, 2, 3, 4, 5, 6]
list(map(times_two, numbers))
...
numbers2 = [times_two(1), times_two(2), times_two(3),
times_two(4), times_two(5), times_two(6)]
def my_map(func, argument_list):
output = []
for argument in argument_list:
output.append(func(argument))
return output
print(my_map(times_two, numbers))
# Let's start by defining a simple function times_multipler
def times_multipler(number, multiplier):
return number * multiplier
numbers = [1, 2, 3, 4, 5, 6]
multipliers = [2, 4, 6, 8, 10, 12]
numbers2 = list(map(times_multipler, numbers, multipliers))
print(numbers2)
numbers = [1, 2, 3, 4, 5, 6]
numbers2 = list(map(times_two, def times_two(number): return number * 2))
print(numbers2)
numbers = [1, 2, 3, 4, 5, 6]
numbers2 = list(map(lambda number: number * 2, numbers))
print(numbers2)
widths = [2, 4, 5, 6, 7, 8]
heights = [4, 5, 3, 2, 5, 7]
| 0.404272 | 0.989572 |
# Programación Orientada a Objetos

```
# Crear un objeto del tipo lista, conjunto de varios elementos
variable = "Marco"
alumnos = ['Pepe', 'Fernando', 'Leticia', 'Maria', 'José']
# Creación de un objeto con diferentes atributos
mobile = {type: [None, {"k":"v"}]}
mobile = {
"marca" : "Samsung",
"SO": "Android",
"version" : "5.6",
"modelo" : "A22",
"screen_size" : 7,
"weight" : 200,
"color" : "silver",
"RAM" : 8,
"memoria" : 512,
"camara" : {
"frontal" : {"resolution":"15MP"},
"back" : [
{"type": "zoom",
"resolution": "25MP"},
{"type": "macro",
"resolution": "30MP"}
]
},
"bateria" : {"type" : "Lithium",
"MaxCharge" : 4500,
"charge":0.56},
"apps" : [
{
"name" : "Whatsapp",
"version" : "1.56.0",
"memoria_ocupada" : "15Mb",
"desarrollador" : "Meta"
},
{
"name" : "Gmail",
"version" : "123.56.0",
"memoria_ocupada" : "105Mb",
"desarrollador" : "Google"
}
]
}
# Definimos una función para cargar el móvil
def charge_phone(phone):
'''
La función toma un objeto y carga la bateria
Args:
----
phone obj dict positional
Return:
-----
mismo objeto dict con valor cambiado
'''
phone['bateria']['charge'] = 1
return phone
mobile
charge_phone()
charge_phone(mobile)
gmail_app = {'name': 'Gmail',
'version': '123.56.0',
'memoria_ocupada': '105Mb',
'desarrollador': 'Google'}
```
```
name : "TheBridgeDSapp"
vers: 1.0.1
desarrolador: TheBridge
memoria_ocupada: 200Mb
```
```
name : "YouTube"
vers: 10.50
desarrolador: Google
memoria_ocupada: 300Mb
```
```
type(mobile['apps'])
for i in mobile['apps']:
print(i)
[i for i in mobile['apps']]
gmail_app = {'name': 'Gmail',
'version': '13.56.0',
'memoria_ocupada': '105Mb',
'desarrollador': 'Google'}
```
```
# Crear dos funciones, una para update y otra para desinstalar una app
'name': 'Gmail',
'version': '12.56.0',
'memoria_ocupada': '105Mb',
'desarrollador': 'Google'
```
```
def install_app(phone, app):
IF lista...apps...> name == app['name'] Gmail, gmail gmaill g-mail
phone['apps'].append(app)
return phone['apps']
install_app(mobile, gmail_app)
def update_app(phone, app, version):
for i in phone['apps']:
if i['name'] == app:
i['version'] = version
update_app(mobile, 'Gmail', '13')
mobile
```
```
# Crear dos funciones, una para update y otra para desinstalar una app
'name': 'Gmail',
'version': '12.56.0',
'memoria_ocupada': '105Mb',
'desarrollador': 'Google'
```
```
mobile['apps'][1]['version'] = "13"
mobile
mobile.__dir__()
mobile['apps'][0]['name']
def remove_app(phone, appname):
for i in phone['apps']:
if ['name'] == appname:
print(i)
def update_app
for pos,i in enumerate(mobile['apps']):
if i['name'] == "Gmail":
mobile['apps'].pop(pos)
mobile['apps']
mobile['apps'].pop(i)
remove_app(mobile, 'Gmail')
def eliminar_app(phone,app):
cont = 0
for i in phone['apps']:
if i['name'] == app:
phone['apps'].pop(cont)
cont += 1
return phone
eliminar_app(mobile,'TheBridgeDSapp')
```
### Class
```
class MobilePhone():
'''
Docstring de una clase
'''
def __init__(self, phone):
self.phone = phone # convertimos el parámetro de entrada como variable global a nivel de clase
print("La clase MobilePhone ha inicializado")
def install_app(self, appname):
self.phone['apps'].append(appname)
return self.phone['apps']
def remove_app(self, appname):
for pos,i in enumerate(self.phone['apps']):
if i['name'] == appname:
self.phone['apps'].pop(pos)
return self.phone['apps']
# Instanciamos a un objeto la clase MobilePhone
telefono = MobilePhone(mobile)
youtube = {
"name" : "YouTube",
"vers": "10.50",
"desarrolador": "Google",
"memoria_ocupada": "300Mb"
}
telefono.install_app(youtube)
telefono.remove_app(youtube)
telefono
```
|
github_jupyter
|
# Crear un objeto del tipo lista, conjunto de varios elementos
variable = "Marco"
alumnos = ['Pepe', 'Fernando', 'Leticia', 'Maria', 'José']
# Creación de un objeto con diferentes atributos
mobile = {type: [None, {"k":"v"}]}
mobile = {
"marca" : "Samsung",
"SO": "Android",
"version" : "5.6",
"modelo" : "A22",
"screen_size" : 7,
"weight" : 200,
"color" : "silver",
"RAM" : 8,
"memoria" : 512,
"camara" : {
"frontal" : {"resolution":"15MP"},
"back" : [
{"type": "zoom",
"resolution": "25MP"},
{"type": "macro",
"resolution": "30MP"}
]
},
"bateria" : {"type" : "Lithium",
"MaxCharge" : 4500,
"charge":0.56},
"apps" : [
{
"name" : "Whatsapp",
"version" : "1.56.0",
"memoria_ocupada" : "15Mb",
"desarrollador" : "Meta"
},
{
"name" : "Gmail",
"version" : "123.56.0",
"memoria_ocupada" : "105Mb",
"desarrollador" : "Google"
}
]
}
# Definimos una función para cargar el móvil
def charge_phone(phone):
'''
La función toma un objeto y carga la bateria
Args:
----
phone obj dict positional
Return:
-----
mismo objeto dict con valor cambiado
'''
phone['bateria']['charge'] = 1
return phone
mobile
charge_phone()
charge_phone(mobile)
gmail_app = {'name': 'Gmail',
'version': '123.56.0',
'memoria_ocupada': '105Mb',
'desarrollador': 'Google'}
name : "TheBridgeDSapp"
vers: 1.0.1
desarrolador: TheBridge
memoria_ocupada: 200Mb
name : "YouTube"
vers: 10.50
desarrolador: Google
memoria_ocupada: 300Mb
type(mobile['apps'])
for i in mobile['apps']:
print(i)
[i for i in mobile['apps']]
gmail_app = {'name': 'Gmail',
'version': '13.56.0',
'memoria_ocupada': '105Mb',
'desarrollador': 'Google'}
# Crear dos funciones, una para update y otra para desinstalar una app
'name': 'Gmail',
'version': '12.56.0',
'memoria_ocupada': '105Mb',
'desarrollador': 'Google'
def install_app(phone, app):
IF lista...apps...> name == app['name'] Gmail, gmail gmaill g-mail
phone['apps'].append(app)
return phone['apps']
install_app(mobile, gmail_app)
def update_app(phone, app, version):
for i in phone['apps']:
if i['name'] == app:
i['version'] = version
update_app(mobile, 'Gmail', '13')
mobile
# Crear dos funciones, una para update y otra para desinstalar una app
'name': 'Gmail',
'version': '12.56.0',
'memoria_ocupada': '105Mb',
'desarrollador': 'Google'
mobile['apps'][1]['version'] = "13"
mobile
mobile.__dir__()
mobile['apps'][0]['name']
def remove_app(phone, appname):
for i in phone['apps']:
if ['name'] == appname:
print(i)
def update_app
for pos,i in enumerate(mobile['apps']):
if i['name'] == "Gmail":
mobile['apps'].pop(pos)
mobile['apps']
mobile['apps'].pop(i)
remove_app(mobile, 'Gmail')
def eliminar_app(phone,app):
cont = 0
for i in phone['apps']:
if i['name'] == app:
phone['apps'].pop(cont)
cont += 1
return phone
eliminar_app(mobile,'TheBridgeDSapp')
class MobilePhone():
'''
Docstring de una clase
'''
def __init__(self, phone):
self.phone = phone # convertimos el parámetro de entrada como variable global a nivel de clase
print("La clase MobilePhone ha inicializado")
def install_app(self, appname):
self.phone['apps'].append(appname)
return self.phone['apps']
def remove_app(self, appname):
for pos,i in enumerate(self.phone['apps']):
if i['name'] == appname:
self.phone['apps'].pop(pos)
return self.phone['apps']
# Instanciamos a un objeto la clase MobilePhone
telefono = MobilePhone(mobile)
youtube = {
"name" : "YouTube",
"vers": "10.50",
"desarrolador": "Google",
"memoria_ocupada": "300Mb"
}
telefono.install_app(youtube)
telefono.remove_app(youtube)
telefono
| 0.287568 | 0.764078 |
# Tensorflow Tutorial
## Tensors
### Constants
https://www.tensorflow.org/api_guides/python/constant_op
- `tf.constant()`
- `tf.zeros(), tf.zeros_initializer(), tf.zeros_like()`
- `tf.ones(), tf.ones_initializer(), tf.ones_like()`
- `tf.fill()`
- `tf.linspace(), tf.range()`
- `tf.random_uniform(), tf.random_normal(), tf.truncated_normal(), tf.random_gamma()`
- `tf.random_shuffle()`
### Variables
- `tf.Variable()`
### Placeholders
- `tf.placeholder()`
## Operations
## Graphs
```
import tensorflow as tf
import numpy as np
from datetime import datetime as dt
import os
def getNow():
now = dt.now().strftime('%Y-%m-%d--%H-%M-%S')
nowFolder = os.path.join('./logFolder', now)
return nowFolder
!tree logFolder
pwd
x = tf.constant(np.linspace(-10, 10, 5), dtype=tf.float32)
tf.reset_default_graph()
y = tf.Variable(np.arange(10), dtype=tf.float32)
folder = getNow()
with tf.Session() as sess:
writer = tf.summary.FileWriter(folder, sess.graph)
writer.close()
print('tensorboard --logdir={}'.format( folder ))
```
What happens when we do this [again](https://www.youtube.com/watch?v=r6inaBWSEdk)?
```
tf.reset_default_graph()
y = tf.Variable(np.arange(10), dtype=tf.float32)
m = tf.placeholder( dtype=tf.float32, shape=y.shape )
z = m + y
init = tf.global_variables_initializer()
```
Using the variables
```
folder = getNow()
with tf.Session() as sess:
writer = tf.summary.FileWriter(folder, sess.graph)
sess.run(init)
zVal = sess.run( z, feed_dict={ m: np.arange(10)*3 } )
writer.close()
print('tensorboard --logdir={}'.format( folder ))
zVal
```
In `feed_dict`, you can pass value for any tensor
```
folder = getNow()
with tf.Session() as sess:
writer = tf.summary.FileWriter(folder, sess.graph)
sess.run(init)
zVal = sess.run( z, feed_dict={
m: np.arange(10)*3,
y: np.ones( (10,) ) })
writer.close()
print('tensorboard --logdir={}'.format( folder ))
zVal
```
Getting the variable by name
```
z1 = tf.get_variable('Variable', (10,))
z2 = z1 * 3
folder = getNow()
init = tf.global_variables_initializer()
with tf.Session() as sess:
writer = tf.summary.FileWriter(folder, sess.graph)
sess.run(init)
zVal = sess.run( z, feed_dict={
m: np.arange(10)*3,
y: np.ones( (10,) ) })
writer.close()
print('tensorboard --logdir={}'.format( folder ))
z2 = tf.get_variable('bacd', (10,))
z3 = tf.Variable( np.array([5, 6, 7]), dtype=tf.float32, name='bobby' )
folder = getNow()
init = tf.global_variables_initializer()
with tf.Session() as sess:
writer = tf.summary.FileWriter(folder, sess.graph)
sess.run(init)
zVal = sess.run( z, feed_dict={
m: np.arange(10)*3,
y: np.ones( (10,) ) })
writer.close()
print('tensorboard --logdir={}'.format( folder ))
```
|
github_jupyter
|
import tensorflow as tf
import numpy as np
from datetime import datetime as dt
import os
def getNow():
now = dt.now().strftime('%Y-%m-%d--%H-%M-%S')
nowFolder = os.path.join('./logFolder', now)
return nowFolder
!tree logFolder
pwd
x = tf.constant(np.linspace(-10, 10, 5), dtype=tf.float32)
tf.reset_default_graph()
y = tf.Variable(np.arange(10), dtype=tf.float32)
folder = getNow()
with tf.Session() as sess:
writer = tf.summary.FileWriter(folder, sess.graph)
writer.close()
print('tensorboard --logdir={}'.format( folder ))
tf.reset_default_graph()
y = tf.Variable(np.arange(10), dtype=tf.float32)
m = tf.placeholder( dtype=tf.float32, shape=y.shape )
z = m + y
init = tf.global_variables_initializer()
folder = getNow()
with tf.Session() as sess:
writer = tf.summary.FileWriter(folder, sess.graph)
sess.run(init)
zVal = sess.run( z, feed_dict={ m: np.arange(10)*3 } )
writer.close()
print('tensorboard --logdir={}'.format( folder ))
zVal
folder = getNow()
with tf.Session() as sess:
writer = tf.summary.FileWriter(folder, sess.graph)
sess.run(init)
zVal = sess.run( z, feed_dict={
m: np.arange(10)*3,
y: np.ones( (10,) ) })
writer.close()
print('tensorboard --logdir={}'.format( folder ))
zVal
z1 = tf.get_variable('Variable', (10,))
z2 = z1 * 3
folder = getNow()
init = tf.global_variables_initializer()
with tf.Session() as sess:
writer = tf.summary.FileWriter(folder, sess.graph)
sess.run(init)
zVal = sess.run( z, feed_dict={
m: np.arange(10)*3,
y: np.ones( (10,) ) })
writer.close()
print('tensorboard --logdir={}'.format( folder ))
z2 = tf.get_variable('bacd', (10,))
z3 = tf.Variable( np.array([5, 6, 7]), dtype=tf.float32, name='bobby' )
folder = getNow()
init = tf.global_variables_initializer()
with tf.Session() as sess:
writer = tf.summary.FileWriter(folder, sess.graph)
sess.run(init)
zVal = sess.run( z, feed_dict={
m: np.arange(10)*3,
y: np.ones( (10,) ) })
writer.close()
print('tensorboard --logdir={}'.format( folder ))
| 0.360489 | 0.879613 |
# Navigation
---
In this notebook, you will learn how to use the Unity ML-Agents environment for the first project of the [Deep Reinforcement Learning Nanodegree](https://www.udacity.com/course/deep-reinforcement-learning-nanodegree--nd893).
### 1. Start the Environment
We begin by importing some necessary packages. If the code cell below returns an error, please revisit the project instructions to double-check that you have installed [Unity ML-Agents](https://github.com/Unity-Technologies/ml-agents/blob/master/docs/Installation.md) and [NumPy](http://www.numpy.org/).
```
from unityagents import UnityEnvironment
import numpy as np
import torch
import numpy as np
from collections import deque
import matplotlib.pyplot as plt
%matplotlib inline
```
Next, we will start the environment! **_Before running the code cell below_**, change the `file_name` parameter to match the location of the Unity environment that you downloaded.
- **Mac**: `"path/to/Banana.app"`
- **Windows** (x86): `"path/to/Banana_Windows_x86/Banana.exe"`
- **Windows** (x86_64): `"path/to/Banana_Windows_x86_64/Banana.exe"`
- **Linux** (x86): `"path/to/Banana_Linux/Banana.x86"`
- **Linux** (x86_64): `"path/to/Banana_Linux/Banana.x86_64"`
- **Linux** (x86, headless): `"path/to/Banana_Linux_NoVis/Banana.x86"`
- **Linux** (x86_64, headless): `"path/to/Banana_Linux_NoVis/Banana.x86_64"`
For instance, if you are using a Mac, then you downloaded `Banana.app`. If this file is in the same folder as the notebook, then the line below should appear as follows:
```
env = UnityEnvironment(file_name="Banana.app")
```
```
env = UnityEnvironment(file_name="Banana.app")
```
Environments contain **_brains_** which are responsible for deciding the actions of their associated agents. Here we check for the first brain available, and set it as the default brain we will be controlling from Python.
```
# get the default brain
brain_name = env.brain_names[0]
brain = env.brains[brain_name]
```
### 2. Examine the State and Action Spaces
The simulation contains a single agent that navigates a large environment. At each time step, it has four actions at its disposal:
- `0` - walk forward
- `1` - walk backward
- `2` - turn left
- `3` - turn right
The state space has `37` dimensions and contains the agent's velocity, along with ray-based perception of objects around agent's forward direction. A reward of `+1` is provided for collecting a yellow banana, and a reward of `-1` is provided for collecting a blue banana.
Run the code cell below to print some information about the environment.
```
# reset the environment
env_info = env.reset(train_mode=True)[brain_name]
# number of agents in the environment
print('Number of agents:', len(env_info.agents))
# number of actions
action_size = brain.vector_action_space_size
print('Number of actions:', action_size)
# examine the state space
state = env_info.vector_observations[0]
print('States look like:', state)
state_size = len(state)
print('States have length:', state_size)
```
### 3. Take Random Actions in the Environment
In the next code cell, you will learn how to use the Python API to control the agent and receive feedback from the environment.
Once this cell is executed, you will watch the agent's performance, if it selects an action (uniformly) at random with each time step. A window should pop up that allows you to observe the agent, as it moves through the environment.
Of course, as part of the project, you'll have to change the code so that the agent is able to use its experience to gradually choose better actions when interacting with the environment!
```
env_info = env.reset(train_mode=False)[brain_name] # reset the environment
state = env_info.vector_observations[0] # get the current state
score = 0 # initialize the score
while True:
action = np.random.randint(action_size) # select an action
env_info = env.step(action)[brain_name] # send the action to the environment
next_state = env_info.vector_observations[0] # get the next state
reward = env_info.rewards[0] # get the reward
done = env_info.local_done[0] # see if episode has finished
score += reward # update the score
state = next_state # roll over the state to next time step
if done: # exit loop if episode finished
break
print("Score: {}".format(score))
```
### 4. It's Your Turn!
Now it's your turn to train your own agent to solve the environment! When training the environment, set `train_mode=True`, so that the line for resetting the environment looks like the following:
```python
env_info = env.reset(train_mode=True)[brain_name]
```
### 4.0 Initialize Agent
```
from dqn_agent import Agent
agent = Agent(state_size=len(env_info.vector_observations[0]), action_size=brain.vector_action_space_size, seed=0)
```
### 4.1 Train
```
def dqn(n_episodes=2000, max_t=1000, eps_start=1.0, eps_end=0.01, eps_decay=0.995):
"""Deep Q-Learning.
Params
======
n_episodes (int): maximum number of training episodes
max_t (int): maximum number of timesteps per episode
eps_start (float): starting value of epsilon, for epsilon-greedy action selection
eps_end (float): minimum value of epsilon
eps_decay (float): multiplicative factor (per episode) for decreasing epsilon
"""
scores = [] # list containing scores from each episode
scores_window = deque(maxlen=100) # last 100 scores
eps = eps_start # initialize epsilon
for i_episode in range(1, n_episodes+1):
env_info = env.reset(train_mode=True)[brain_name]
state = env_info.vector_observations[0]
score = 0
for t in range(max_t):
action = agent.act(state, eps)
env_info = env.step(action)[brain_name]
next_state = env_info.vector_observations[0]
reward = env_info.rewards[0]
done = env_info.local_done[0]
agent.step(state, action, reward, next_state, done)
state = next_state
score += reward
if done:
break
scores_window.append(score) # save most recent score
scores.append(score) # save most recent score
eps = max(eps_end, eps_decay*eps) # decrease epsilon
print('\rEpisode {}\tAverage Score: {:.2f}'.format(i_episode, np.mean(scores_window)), end="")
if i_episode % 100 == 0:
print('\rEpisode {}\tAverage Score: {:.2f}'.format(i_episode, np.mean(scores_window)))
if np.mean(scores_window)>=13:
print('\nEnvironment solved in {:d} episodes!\tAverage Score: {:.2f}'.format(i_episode-100, np.mean(scores_window)))
torch.save(agent.qnetwork_local.state_dict(), 'checkpoint_double.pth')
break
return scores
def plot_scores(scores):
"""Plot list of scores
Params
======
scores (list): list of scores
"""
fig = plt.figure()
ax = fig.add_subplot(111)
plt.plot(np.arange(len(scores)), scores)
plt.ylabel('Score')
plt.xlabel('Episode #')
plt.savefig('dqn_scores.png')
scores = dqn()
plot_scores(scores)
```
### 4.2 Save checkpoint
```
torch.save(agent.qnetwork_local.state_dict(), 'checkpoint_double.pth')
```
### 4.3 Play with pretrained agent
```
# load the weights from file
from time import sleep
agent.qnetwork_local.load_state_dict(torch.load('checkpoint_double.pth'))
for i in range(5):
env_info = env.reset(train_mode=False)[brain_name]
state = env_info.vector_observations[0]
score = 0
for j in range(1000):
action = agent.act(state)
# env.render()
env_info = env.step(action)[brain_name]
next_state = env_info.vector_observations[0]
reward = env_info.rewards[0]
done = env_info.local_done[0]
score += reward
state = next_state
if done:
print('Game number: {}. Score: {}'.format(i, score))
# sleep(3)
break
env.close()
```
|
github_jupyter
|
from unityagents import UnityEnvironment
import numpy as np
import torch
import numpy as np
from collections import deque
import matplotlib.pyplot as plt
%matplotlib inline
env = UnityEnvironment(file_name="Banana.app")
env = UnityEnvironment(file_name="Banana.app")
# get the default brain
brain_name = env.brain_names[0]
brain = env.brains[brain_name]
# reset the environment
env_info = env.reset(train_mode=True)[brain_name]
# number of agents in the environment
print('Number of agents:', len(env_info.agents))
# number of actions
action_size = brain.vector_action_space_size
print('Number of actions:', action_size)
# examine the state space
state = env_info.vector_observations[0]
print('States look like:', state)
state_size = len(state)
print('States have length:', state_size)
env_info = env.reset(train_mode=False)[brain_name] # reset the environment
state = env_info.vector_observations[0] # get the current state
score = 0 # initialize the score
while True:
action = np.random.randint(action_size) # select an action
env_info = env.step(action)[brain_name] # send the action to the environment
next_state = env_info.vector_observations[0] # get the next state
reward = env_info.rewards[0] # get the reward
done = env_info.local_done[0] # see if episode has finished
score += reward # update the score
state = next_state # roll over the state to next time step
if done: # exit loop if episode finished
break
print("Score: {}".format(score))
env_info = env.reset(train_mode=True)[brain_name]
from dqn_agent import Agent
agent = Agent(state_size=len(env_info.vector_observations[0]), action_size=brain.vector_action_space_size, seed=0)
def dqn(n_episodes=2000, max_t=1000, eps_start=1.0, eps_end=0.01, eps_decay=0.995):
"""Deep Q-Learning.
Params
======
n_episodes (int): maximum number of training episodes
max_t (int): maximum number of timesteps per episode
eps_start (float): starting value of epsilon, for epsilon-greedy action selection
eps_end (float): minimum value of epsilon
eps_decay (float): multiplicative factor (per episode) for decreasing epsilon
"""
scores = [] # list containing scores from each episode
scores_window = deque(maxlen=100) # last 100 scores
eps = eps_start # initialize epsilon
for i_episode in range(1, n_episodes+1):
env_info = env.reset(train_mode=True)[brain_name]
state = env_info.vector_observations[0]
score = 0
for t in range(max_t):
action = agent.act(state, eps)
env_info = env.step(action)[brain_name]
next_state = env_info.vector_observations[0]
reward = env_info.rewards[0]
done = env_info.local_done[0]
agent.step(state, action, reward, next_state, done)
state = next_state
score += reward
if done:
break
scores_window.append(score) # save most recent score
scores.append(score) # save most recent score
eps = max(eps_end, eps_decay*eps) # decrease epsilon
print('\rEpisode {}\tAverage Score: {:.2f}'.format(i_episode, np.mean(scores_window)), end="")
if i_episode % 100 == 0:
print('\rEpisode {}\tAverage Score: {:.2f}'.format(i_episode, np.mean(scores_window)))
if np.mean(scores_window)>=13:
print('\nEnvironment solved in {:d} episodes!\tAverage Score: {:.2f}'.format(i_episode-100, np.mean(scores_window)))
torch.save(agent.qnetwork_local.state_dict(), 'checkpoint_double.pth')
break
return scores
def plot_scores(scores):
"""Plot list of scores
Params
======
scores (list): list of scores
"""
fig = plt.figure()
ax = fig.add_subplot(111)
plt.plot(np.arange(len(scores)), scores)
plt.ylabel('Score')
plt.xlabel('Episode #')
plt.savefig('dqn_scores.png')
scores = dqn()
plot_scores(scores)
torch.save(agent.qnetwork_local.state_dict(), 'checkpoint_double.pth')
# load the weights from file
from time import sleep
agent.qnetwork_local.load_state_dict(torch.load('checkpoint_double.pth'))
for i in range(5):
env_info = env.reset(train_mode=False)[brain_name]
state = env_info.vector_observations[0]
score = 0
for j in range(1000):
action = agent.act(state)
# env.render()
env_info = env.step(action)[brain_name]
next_state = env_info.vector_observations[0]
reward = env_info.rewards[0]
done = env_info.local_done[0]
score += reward
state = next_state
if done:
print('Game number: {}. Score: {}'.format(i, score))
# sleep(3)
break
env.close()
| 0.53777 | 0.981221 |
```
from qiskit import *
from qiskit.tools.visualization import plot_histogram
import numpy as np
```
# Exercise 4
* Hints: 08/04/2020
* Hand-in: 15/04/2020
* Solutions: 22/04/2020
* *There will be no penalty for late hand-in, but handing in on-time will allow theTAs to address your problems.*
## 1
Show that the Hadamard gate can be written in the following two forms
$$H = \frac{X+Z}{\sqrt{2}} \sim \exp\left(i \frac{\pi}{2} \, \frac{X+Z}{\sqrt{2}}\right).$$
Here $\sim$ is used to denote that the equality is valid up to a global phase, and hence that the resulting gates are physically equivalent.
Hint: it might even be easiest to prove that $e^{i\frac{\pi}{2} M} \sim M$ for any matrix whose eigenvalues are all $\pm 1$, and that such matrices uniquely satisfy $M^2=I$.
## 2
The Hadamard can be constructed from `rx` and `rz` operations as
$$ R_x(\theta) = e^{i\frac{\theta}{2} X}, ~~~ R_z(\theta) = e^{i\frac{\theta}{2} Z},\\ H \equiv \lim_{n\rightarrow\infty} \left( ~R_x\left(\frac{\theta}{n}\right) ~~R_z \left(\frac{\theta}{n}\right) ~\right)^n.$$
For some suitably chosen $\theta$. When implemented for finite $n$, the resulting gate will be an approximation to the Hadamard whose error decreases with $n$.
The following shows an example of this implemented with Qiskit with an incorrectly chosen value of $\theta$ (and with the global phase ignored).
* Determine the correct value of $\theta$.
* Show that the error (when using the correct value of $\theta$) decreases quadratically with $n$.
```
q = QuantumRegister(1)
c = ClassicalRegister(1)
error = {}
for n in range(1,11):
# Create a blank circuit
qc = QuantumCircuit(q,c)
# Implement an approximate Hadamard
theta = np.pi # here we incorrectly choose theta=pi
for j in range(n):
qc.rx(theta/n,q[0])
qc.rz(theta/n,q[0])
# We need to measure how good the above approximation is. Here's a simple way to do this.
# Step 1: Use a real hadamard to cancel the above approximation.
# For a good approximatuon, the qubit will return to state 0. For a bad one, it will end up as some superposition.
qc.h(q[0])
# Step 2: Run the circuit, and see how many times we get the outcome 1.
# Since it should return 0 with certainty, the fraction of 1s is a measure of the error.
qc.measure(q,c)
shots = 20000
job = execute(qc, Aer.get_backend('qasm_simulator'),shots=shots)
try:
error[n] = (job.result().get_counts()['1']/shots)
except:
pass
plot_histogram(error)
```
## 3
An improved version of the approximation can be found from,
$$H \equiv \lim_{n\rightarrow\infty} \left( ~ R_z \left(\frac{\theta}{2n}\right)~~ R_x\left(\frac{\theta}{n}\right) ~~ R_z \left(\frac{\theta}{2n}\right) ~\right)^n.$$
Implement this, and investigate the scaling of the error.
|
github_jupyter
|
from qiskit import *
from qiskit.tools.visualization import plot_histogram
import numpy as np
q = QuantumRegister(1)
c = ClassicalRegister(1)
error = {}
for n in range(1,11):
# Create a blank circuit
qc = QuantumCircuit(q,c)
# Implement an approximate Hadamard
theta = np.pi # here we incorrectly choose theta=pi
for j in range(n):
qc.rx(theta/n,q[0])
qc.rz(theta/n,q[0])
# We need to measure how good the above approximation is. Here's a simple way to do this.
# Step 1: Use a real hadamard to cancel the above approximation.
# For a good approximatuon, the qubit will return to state 0. For a bad one, it will end up as some superposition.
qc.h(q[0])
# Step 2: Run the circuit, and see how many times we get the outcome 1.
# Since it should return 0 with certainty, the fraction of 1s is a measure of the error.
qc.measure(q,c)
shots = 20000
job = execute(qc, Aer.get_backend('qasm_simulator'),shots=shots)
try:
error[n] = (job.result().get_counts()['1']/shots)
except:
pass
plot_histogram(error)
| 0.571049 | 0.98558 |
# Column Manipulations
Copyright (c) Microsoft Corporation. All rights reserved.<br>
Licensed under the MIT License.<br>
Azure ML Data Prep has many methods for manipulating columns, including basic CUD operations and several other more complex manipulations.
This notebook will focus primarily on data-agnostic operations. For all other column manipulation operations, we will link to their specific how-to guide.
## Table of Contents
[ColumnSelector](#ColumnSelector)<br>
[add_column](#add_column)<br>
[append_columns](#append_columns)<br>
[drop_columns](#drop_columns)<br>
[duplicate_column](#duplicate_column)<br>
[fuzzy_group_column](#fuzzy_group_column)<br>
[keep_columns](#keep_columns)<br>
[map_column](#map_column)<br>
[new_script_column](#new_script_column)<br>
[rename_columns](#rename_columns)<br>
<a id="ColumnSelector"></a>
## ColumnSelector
`ColumnSelector` is a Data Prep class that allows us to select columns by name. The idea is to be able to describe columns generally instead of explicitly, using a search term or regex expression, with various options.
Note that a `ColumnSelector` does not represent the columns they match themselves, but the selector of the described columns. Therefore if we use the same `ColumnSelector` on two different dataflows, we may get different results depending on the columns of each dataflow.
Column manipulations that can utilize `ColumnSelector` will be noted in their respective sections in this book.
```
from azureml.dataprep import auto_read_file
dflow = auto_read_file(path='../data/crime-dirty.csv')
dflow.head(5)
```
All parameters to a `ColumnSelector` are shown here for completeness. We will use `keep_columns` in our example, which will keep only the columns in the dataflow that we tell it to keep.
In the below example, we match all columns with the letter 'i'. Because we set `ignore_case` to false and `match_whole_word` to false, then any column that contains 'i' or 'I' will be selected.
```
from azureml.dataprep import ColumnSelector
column_selector = ColumnSelector(term="i",
use_regex=False,
ignore_case=True,
match_whole_word=False,
invert=False)
dflow_selected = dflow.keep_columns(column_selector)
dflow_selected.head(5)
```
If we set `invert` to true, we get the opposite of what we matched earlier.
```
column_selector = ColumnSelector(term="i",
use_regex=False,
ignore_case=True,
match_whole_word=False,
invert=True)
dflow_selected = dflow.keep_columns(column_selector)
dflow_selected.head(5)
```
If we change the search term to 'I' and set case sensitivity to true, we get only the handful of columns that contain an upper case 'I'.
```
column_selector = ColumnSelector(term="I",
use_regex=False,
ignore_case=False,
match_whole_word=False,
invert=False)
dflow_selected = dflow.keep_columns(column_selector)
dflow_selected.head(5)
```
And if we set `match_whole_word` to true, we get no results at all as there is no column called 'I'.
```
column_selector = ColumnSelector(term="I",
use_regex=False,
ignore_case=False,
match_whole_word=True,
invert=False)
dflow_selected = dflow.keep_columns(column_selector)
dflow_selected.head(5)
```
Finally, the `use_regex` flag dictates whether or not to treat the search term as a regex. It can be combined still with the other options.
Here we define all columns that begin with the capital letter 'I'.
```
column_selector = ColumnSelector(term="I.*",
use_regex=True,
ignore_case=True,
match_whole_word=True,
invert=False)
dflow_selected = dflow.keep_columns(column_selector)
dflow_selected.head(5)
```
<a id="add_column"></a>
## add_column
Please see [add-column-using-expression](add-column-using-expression.ipynb).
<a id="append_columns"></a>
## append_columns
Please see [append-columns-and-rows](append-columns-and-rows.ipynb).
<a id="drop_columns"></a>
## drop_columns
Data Prep supports dropping columns one or more columns in a single statement. Supports `ColumnSelector`.
```
from azureml.dataprep import auto_read_file
dflow = auto_read_file(path='../data/crime-dirty.csv')
dflow.head(5)
```
Note that there are 22 columns to begin with. We will now drop the 'ID' column and observe that the resulting dataflow contains 21 columns.
```
dflow_dropped = dflow.drop_columns('ID')
dflow_dropped.head(5)
```
We can also drop more than one column at once by passing a list of column names.
```
dflow_dropped = dflow_dropped.drop_columns(['IUCR', 'Description'])
dflow_dropped.head(5)
```
<a id="duplicate_column"></a>
## duplicate_column
Data Prep supports duplicating columns one or more columns in a single statement.
Duplicated columns are placed to the immediate right of their source column.
```
from azureml.dataprep import auto_read_file
dflow = auto_read_file(path='../data/crime-dirty.csv')
dflow.head(5)
```
We decide which column(s) to duplicate and what the new column name(s) should be with a key value pairing (dictionary).
```
dflow_dupe = dflow.duplicate_column({'ID': 'ID2', 'IUCR': 'IUCR_Clone'})
dflow_dupe.head(5)
```
<a id="fuzzy_group_column"></a>
## fuzzy_group_column
Please see [fuzzy-group](fuzzy-group.ipynb).
<a id="keep_columns"></a>
## keep_columns
Data Prep supports keeping one or more columns in a single statement. The resulting dataflow will contain only the column(s) specified; dropping all the other columns. Supports `ColumnSelector`.
```
from azureml.dataprep import auto_read_file
dflow = auto_read_file(path='../data/crime-dirty.csv')
dflow.head(5)
dflow_keep = dflow.keep_columns(['ID', 'Date', 'Description'])
dflow_keep.head(5)
```
Similar to `drop_columns`, we can pass a single column name or a list of them.
```
dflow_keep = dflow_keep.keep_columns('ID')
dflow_keep.head(5)
```
<a id="map_column"></a>
## map_column
Data Prep supports string mapping. For a column containing strings, we can provide specific mappings from an original value to a new value, and then produce a new column that contains the mapped values.
The mapped columns are placed to the immediate right of their source column.
```
from azureml.dataprep import auto_read_file
dflow = auto_read_file(path='../data/crime-dirty.csv')
dflow.head(5)
from azureml.dataprep import ReplacementsValue
replacements = [ReplacementsValue('THEFT', 'THEFT2'), ReplacementsValue('BATTERY', 'BATTERY!!!')]
dflow_mapped = dflow.map_column(column='Primary Type',
new_column_id='Primary Type V2',
replacements=replacements)
dflow_mapped.head(5)
```
<a id="new_script_column"></a>
## new_script_column
Please see [custom-python-transforms](custom-python-transforms.ipynb).
<a id="rename_columns"></a>
## rename_columns
Data Prep supports renaming one or more columns in a single statement.
```
from azureml.dataprep import auto_read_file
dflow = auto_read_file(path='../data/crime-dirty.csv')
dflow.head(5)
```
We decide which column(s) to rename and what the new column name(s) should be with a key value pairing (dictionary).
```
dflow_renamed = dflow.rename_columns({'ID': 'ID2', 'IUCR': 'IUCR_Clone'})
dflow_renamed.head(5)
```
|
github_jupyter
|
from azureml.dataprep import auto_read_file
dflow = auto_read_file(path='../data/crime-dirty.csv')
dflow.head(5)
from azureml.dataprep import ColumnSelector
column_selector = ColumnSelector(term="i",
use_regex=False,
ignore_case=True,
match_whole_word=False,
invert=False)
dflow_selected = dflow.keep_columns(column_selector)
dflow_selected.head(5)
column_selector = ColumnSelector(term="i",
use_regex=False,
ignore_case=True,
match_whole_word=False,
invert=True)
dflow_selected = dflow.keep_columns(column_selector)
dflow_selected.head(5)
column_selector = ColumnSelector(term="I",
use_regex=False,
ignore_case=False,
match_whole_word=False,
invert=False)
dflow_selected = dflow.keep_columns(column_selector)
dflow_selected.head(5)
column_selector = ColumnSelector(term="I",
use_regex=False,
ignore_case=False,
match_whole_word=True,
invert=False)
dflow_selected = dflow.keep_columns(column_selector)
dflow_selected.head(5)
column_selector = ColumnSelector(term="I.*",
use_regex=True,
ignore_case=True,
match_whole_word=True,
invert=False)
dflow_selected = dflow.keep_columns(column_selector)
dflow_selected.head(5)
from azureml.dataprep import auto_read_file
dflow = auto_read_file(path='../data/crime-dirty.csv')
dflow.head(5)
dflow_dropped = dflow.drop_columns('ID')
dflow_dropped.head(5)
dflow_dropped = dflow_dropped.drop_columns(['IUCR', 'Description'])
dflow_dropped.head(5)
from azureml.dataprep import auto_read_file
dflow = auto_read_file(path='../data/crime-dirty.csv')
dflow.head(5)
dflow_dupe = dflow.duplicate_column({'ID': 'ID2', 'IUCR': 'IUCR_Clone'})
dflow_dupe.head(5)
from azureml.dataprep import auto_read_file
dflow = auto_read_file(path='../data/crime-dirty.csv')
dflow.head(5)
dflow_keep = dflow.keep_columns(['ID', 'Date', 'Description'])
dflow_keep.head(5)
dflow_keep = dflow_keep.keep_columns('ID')
dflow_keep.head(5)
from azureml.dataprep import auto_read_file
dflow = auto_read_file(path='../data/crime-dirty.csv')
dflow.head(5)
from azureml.dataprep import ReplacementsValue
replacements = [ReplacementsValue('THEFT', 'THEFT2'), ReplacementsValue('BATTERY', 'BATTERY!!!')]
dflow_mapped = dflow.map_column(column='Primary Type',
new_column_id='Primary Type V2',
replacements=replacements)
dflow_mapped.head(5)
from azureml.dataprep import auto_read_file
dflow = auto_read_file(path='../data/crime-dirty.csv')
dflow.head(5)
dflow_renamed = dflow.rename_columns({'ID': 'ID2', 'IUCR': 'IUCR_Clone'})
dflow_renamed.head(5)
| 0.262558 | 0.96157 |
```
import numpy as np
import pandas as pd
from matplotlib import pyplot as plt
from tqdm import tqdm
%matplotlib inline
from torch.utils.data import Dataset, DataLoader
import torch
import torchvision
import torch.nn as nn
import torch.optim as optim
from torch.nn import functional as F
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
print(device)
```
# Generate dataset
```
y = np.random.randint(0,10,5000)
idx= []
for i in range(10):
print(i,sum(y==i))
idx.append(y==i)
x = np.zeros((5000,2))
x[idx[0],:] = np.random.multivariate_normal(mean = [5,5],cov=[[0.1,0],[0,0.1]],size=sum(idx[0]))
x[idx[1],:] = np.random.multivariate_normal(mean = [-6,7],cov=[[0.1,0],[0,0.1]],size=sum(idx[1]))
x[idx[2],:] = np.random.multivariate_normal(mean = [-5,-4],cov=[[0.1,0],[0,0.1]],size=sum(idx[2]))
# x[idx[0],:] = np.random.multivariate_normal(mean = [5,5],cov=[[0.1,0],[0,0.1]],size=sum(idx[0]))
# x[idx[1],:] = np.random.multivariate_normal(mean = [6,6],cov=[[0.1,0],[0,0.1]],size=sum(idx[1]))
# x[idx[2],:] = np.random.multivariate_normal(mean = [5.5,6.5],cov=[[0.1,0],[0,0.1]],size=sum(idx[2]))
x[idx[3],:] = np.random.multivariate_normal(mean = [-1,0],cov=[[0.1,0],[0,0.1]],size=sum(idx[3]))
x[idx[4],:] = np.random.multivariate_normal(mean = [0,2],cov=[[0.1,0],[0,0.1]],size=sum(idx[4]))
x[idx[5],:] = np.random.multivariate_normal(mean = [1,0],cov=[[0.1,0],[0,0.1]],size=sum(idx[5]))
x[idx[6],:] = np.random.multivariate_normal(mean = [0,-1],cov=[[0.1,0],[0,0.1]],size=sum(idx[6]))
x[idx[7],:] = np.random.multivariate_normal(mean = [0,0],cov=[[0.1,0],[0,0.1]],size=sum(idx[7]))
x[idx[8],:] = np.random.multivariate_normal(mean = [-0.5,-0.5],cov=[[0.1,0],[0,0.1]],size=sum(idx[8]))
x[idx[9],:] = np.random.multivariate_normal(mean = [0.4,0.2],cov=[[0.1,0],[0,0.1]],size=sum(idx[9]))
for i in range(10):
plt.scatter(x[idx[i],0],x[idx[i],1],label="class_"+str(i))
plt.legend(loc='center left', bbox_to_anchor=(1, 0.5))
foreground_classes = {'class_0','class_1', 'class_2'}
background_classes = {'class_3','class_4', 'class_5', 'class_6','class_7', 'class_8', 'class_9'}
fg_class = np.random.randint(0,3)
fg_idx = np.random.randint(0,9)
a = []
for i in range(9):
if i == fg_idx:
b = np.random.choice(np.where(idx[fg_class]==True)[0],size=1)
a.append(x[b])
print("foreground "+str(fg_class)+" present at " + str(fg_idx))
else:
bg_class = np.random.randint(3,10)
b = np.random.choice(np.where(idx[bg_class]==True)[0],size=1)
a.append(x[b])
print("background "+str(bg_class)+" present at " + str(i))
a = np.concatenate(a,axis=0)
print(a.shape)
print(fg_class , fg_idx)
a.shape
np.reshape(a,(18,1))
a=np.reshape(a,(3,6))
plt.imshow(a)
desired_num = 1000
mosaic_list_of_images =[]
mosaic_label = []
fore_idx=[]
for j in range(desired_num):
fg_class = np.random.randint(0,3)
fg_idx = 0
a = []
for i in range(9):
if i == fg_idx:
b = np.random.choice(np.where(idx[fg_class]==True)[0],size=1)
a.append(x[b])
# print("foreground "+str(fg_class)+" present at " + str(fg_idx))
else:
bg_class = np.random.randint(3,10)
b = np.random.choice(np.where(idx[bg_class]==True)[0],size=1)
a.append(x[b])
# print("background "+str(bg_class)+" present at " + str(i))
a = np.concatenate(a,axis=0)
mosaic_list_of_images.append(np.reshape(a,(18,1)))
mosaic_label.append(fg_class)
fore_idx.append(fg_idx)
mosaic_list_of_images = np.concatenate(mosaic_list_of_images,axis=1).T
# print(mosaic_list)
print(np.shape(mosaic_list_of_images))
print(np.shape(fore_idx))
print(np.shape(mosaic_label))
def create_avg_image_from_mosaic_dataset(mosaic_dataset,labels,foreground_index,dataset_number):
"""
mosaic_dataset : mosaic_dataset contains 9 images 32 x 32 each as 1 data point
labels : mosaic_dataset labels
foreground_index : contains list of indexes where foreground image is present so that using this we can take weighted average
dataset_number : will help us to tell what ratio of foreground image to be taken. for eg: if it is "j" then fg_image_ratio = j/9 , bg_image_ratio = (9-j)/8*9
"""
avg_image_dataset = []
cnt = 0
counter = np.array([0,0,0,0,0,0,0,0,0])
for i in range(len(mosaic_dataset)):
img = torch.zeros([18], dtype=torch.float64)
np.random.seed(dataset_number*10000 + i)
give_pref = foreground_index[i] #np.random.randint(0,9)
# print("outside", give_pref,foreground_index[i])
for j in range(9):
if j == give_pref:
img = img + mosaic_dataset[i][j]*dataset_number/9
else :
img = img + mosaic_dataset[i][j]*(9-dataset_number)/(8*9)
if give_pref == foreground_index[i] :
# print("equal are", give_pref,foreground_index[i])
cnt += 1
counter[give_pref] += 1
else :
counter[give_pref] += 1
avg_image_dataset.append(img)
print("number of correct averaging happened for dataset "+str(dataset_number)+" is "+str(cnt))
print("the averaging are done as ", counter)
return avg_image_dataset , labels , foreground_index
avg_image_dataset_1 , labels_1, fg_index_1 = create_avg_image_from_mosaic_dataset(mosaic_list_of_images, mosaic_label, fore_idx, 1)
avg_image_dataset_2 , labels_2, fg_index_2 = create_avg_image_from_mosaic_dataset(mosaic_list_of_images, mosaic_label, fore_idx, 2)
avg_image_dataset_3 , labels_3, fg_index_3 = create_avg_image_from_mosaic_dataset(mosaic_list_of_images, mosaic_label, fore_idx , 3)
avg_image_dataset_4 , labels_4, fg_index_4 = create_avg_image_from_mosaic_dataset(mosaic_list_of_images, mosaic_label, fore_idx , 4)
avg_image_dataset_5 , labels_5, fg_index_5 = create_avg_image_from_mosaic_dataset(mosaic_list_of_images, mosaic_label, fore_idx , 5)
avg_image_dataset_6 , labels_6, fg_index_6 = create_avg_image_from_mosaic_dataset(mosaic_list_of_images, mosaic_label, fore_idx , 6)
avg_image_dataset_7 , labels_7, fg_index_7 = create_avg_image_from_mosaic_dataset(mosaic_list_of_images, mosaic_label, fore_idx , 7)
avg_image_dataset_8 , labels_8, fg_index_8 = create_avg_image_from_mosaic_dataset(mosaic_list_of_images, mosaic_label, fore_idx , 8)
avg_image_dataset_9 , labels_9, fg_index_9 = create_avg_image_from_mosaic_dataset(mosaic_list_of_images, mosaic_label, fore_idx, 9)
class MosaicDataset(Dataset):
"""MosaicDataset dataset."""
def __init__(self, mosaic_list_of_images, mosaic_label):
"""
Args:
csv_file (string): Path to the csv file with annotations.
root_dir (string): Directory with all the images.
transform (callable, optional): Optional transform to be applied
on a sample.
"""
self.mosaic = mosaic_list_of_images
self.label = mosaic_label
#self.fore_idx = fore_idx
def __len__(self):
return len(self.label)
def __getitem__(self, idx):
return self.mosaic[idx] , self.label[idx] #, self.fore_idx[idx]
batch = 200
epochs = 300
# training_data = avg_image_dataset_5 #just change this and training_label to desired dataset for training
# training_label = labels_5
traindata_1 = MosaicDataset(avg_image_dataset_1, labels_1 )
trainloader_1 = DataLoader( traindata_1 , batch_size= batch ,shuffle=True)
traindata_2 = MosaicDataset(avg_image_dataset_2, labels_2 )
trainloader_2 = DataLoader( traindata_2 , batch_size= batch ,shuffle=True)
traindata_3 = MosaicDataset(avg_image_dataset_3, labels_3 )
trainloader_3 = DataLoader( traindata_3 , batch_size= batch ,shuffle=True)
traindata_4 = MosaicDataset(avg_image_dataset_4, labels_4 )
trainloader_4 = DataLoader( traindata_4 , batch_size= batch ,shuffle=True)
traindata_5 = MosaicDataset(avg_image_dataset_5, labels_5 )
trainloader_5 = DataLoader( traindata_5 , batch_size= batch ,shuffle=True)
traindata_6 = MosaicDataset(avg_image_dataset_6, labels_6 )
trainloader_6 = DataLoader( traindata_6 , batch_size= batch ,shuffle=True)
traindata_7 = MosaicDataset(avg_image_dataset_7, labels_7 )
trainloader_7 = DataLoader( traindata_7 , batch_size= batch ,shuffle=True)
traindata_8 = MosaicDataset(avg_image_dataset_8, labels_8 )
trainloader_8 = DataLoader( traindata_8 , batch_size= batch ,shuffle=True)
traindata_9 = MosaicDataset(avg_image_dataset_9, labels_9 )
trainloader_9 = DataLoader( traindata_9 , batch_size= batch ,shuffle=True)
testdata_1 = MosaicDataset(avg_image_dataset_1, labels_1 )
testloader_1 = DataLoader( testdata_1 , batch_size= batch ,shuffle=False)
testdata_2 = MosaicDataset(avg_image_dataset_2, labels_2 )
testloader_2 = DataLoader( testdata_2 , batch_size= batch ,shuffle=False)
testdata_3 = MosaicDataset(avg_image_dataset_3, labels_3 )
testloader_3 = DataLoader( testdata_3 , batch_size= batch ,shuffle=False)
testdata_4 = MosaicDataset(avg_image_dataset_4, labels_4 )
testloader_4 = DataLoader( testdata_4 , batch_size= batch ,shuffle=False)
testdata_5 = MosaicDataset(avg_image_dataset_5, labels_5 )
testloader_5 = DataLoader( testdata_5 , batch_size= batch ,shuffle=False)
testdata_6 = MosaicDataset(avg_image_dataset_6, labels_6 )
testloader_6 = DataLoader( testdata_6 , batch_size= batch ,shuffle=False)
testdata_7 = MosaicDataset(avg_image_dataset_7, labels_7 )
testloader_7 = DataLoader( testdata_7 , batch_size= batch ,shuffle=False)
testdata_8 = MosaicDataset(avg_image_dataset_8, labels_8 )
testloader_8 = DataLoader( testdata_8 , batch_size= batch ,shuffle=False)
testdata_9 = MosaicDataset(avg_image_dataset_9, labels_9 )
testloader_9 = DataLoader( testdata_9 , batch_size= batch ,shuffle=False)
class Wherenet(nn.Module):
def __init__(self):
super(Wherenet,self).__init__()
self.linear1 = nn.Linear(2,128)
self.linear2 = nn.Linear(128,256)
self.linear3 = nn.Linear(256,128)
self.linear4 = nn.Linear(128,64)
self.linear5 = nn.Linear(64,1)
def forward(self,z):
x = torch.zeros([batch,9],dtype=torch.float64)
y = torch.zeros([batch,2], dtype=torch.float64)
#x,y = x.to("cuda"),y.to("cuda")
for i in range(9):
x[:,i] = self.helper(z[:,2*i:2*i+2])[:,0]
#print(k[:,0].shape,x[:,i].shape)
x = F.softmax(x,dim=1) # alphas
x1 = x[:,0]
for i in range(9):
x1 = x[:,i]
#print()
y = y+torch.mul(x1[:,None],z[:,2*i:2*i+2])
return y , x
def helper(self,x):
x = F.relu(self.linear1(x))
x = F.relu(self.linear2(x))
x = F.relu(self.linear3(x))
x = F.relu(self.linear4(x))
x = self.linear5(x)
return x
trainiter = iter(trainloader_1)
input1,labels1 = trainiter.next()
input1.shape
where = Wherenet().double()
where = where
out_where,alphas = where(input1)
out_where.shape,alphas.shape
class Whatnet(nn.Module):
def __init__(self):
super(Whatnet,self).__init__()
self.linear1 = nn.Linear(2,128)
self.linear2 = nn.Linear(128,256)
self.linear3 = nn.Linear(256,128)
self.linear4 = nn.Linear(128,64)
self.linear5 = nn.Linear(64,3)
def forward(self,x):
x = F.relu(self.linear1(x))
x = F.relu(self.linear2(x))
x = F.relu(self.linear3(x))
x = F.relu(self.linear4(x))
x = self.linear5(x)
return x
# what = Whatnet().double()
# what(out_where)
def test_all(number, testloader,what, where):
correct = 0
total = 0
out = []
pred = []
with torch.no_grad():
for data in testloader:
inputs, labels = data
# images, labels = images.to("cuda"),labels.to("cuda")
out.append(labels.cpu().numpy())
avg_inp,alphas = where(inputs)
outputs = what(avg_inp)
_, predicted = torch.max(outputs.data, 1)
pred.append(predicted.cpu().numpy())
total += labels.size(0)
correct += (predicted == labels).sum().item()
print('Accuracy of the network on the 10000 test dataset %d: %d %%' % (number , 100 * correct / total))
def train_all(trainloader, ds_number, testloader_list):
print("--"*40)
print("training on data set ", ds_number)
where = Wherenet().double()
what = Whatnet().double()
criterion = nn.CrossEntropyLoss()
# optimizer_where = optim.SGD(where.parameters(), lr=0.001, momentum=0.9)
optimizer_where = optim.Adam(where.parameters(), lr=1e-2, betas=(0.9, 0.999), eps=1e-08, weight_decay=0, amsgrad=True)
# optimizer_what = optim.SGD(what.parameters(), lr=0.001, momentum=0.9)
optimizer_what = optim.Adam(what.parameters(), lr=1e-2, betas=(0.9, 0.999), eps=1e-08, weight_decay=0, amsgrad=True)
acti = []
loss_curi = []
epochs = 500
for epoch in range(epochs): # loop over the dataset multiple times
ep_lossi = []
running_loss = 0.0
for i, data in enumerate(trainloader, 0):
# get the inputs
inputs, labels = data
# inputs, labels = inputs.to("cuda"),labels.to("cuda")
# zero the parameter gradients
optimizer_what.zero_grad()
optimizer_where.zero_grad()
# forward + backward + optimize
avg_inp,alphas = where(inputs)
# print(avg_inp.shape)
outputs = what(avg_inp)
loss = criterion(outputs, labels)
loss.backward()
optimizer_what.step()
optimizer_where.step()
# print statistics
running_loss += loss.item()
mini = 4
if i % mini == mini-1: # print every 10 mini-batches
print('[%d, %5d] loss: %.3f' %
(epoch + 1, i + 1, running_loss / mini))
ep_lossi.append(running_loss/mini) # loss per minibatch
running_loss = 0.0
loss_curi.append(np.mean(ep_lossi)) #loss per epoch
if (np.mean(ep_lossi) <= 0.05):
break
print('Finished Training')
# torch.save(inc.state_dict(),"train_dataset_"+str(ds_number)+"_"+str(epochs)+".pt")
correct = 0
total = 0
with torch.no_grad():
for data in trainloader:
inputs, labels = data
# images, labels = images.to("cuda"), labels.to("cuda")
avg_inp,alphas = where(inputs)
outputs = what(avg_inp)
_, predicted = torch.max(outputs.data, 1)
total += labels.size(0)
correct += (predicted == labels).sum().item()
print('Accuracy of the network on the 10000 train images: %d %%' % ( 100 * correct / total))
for i, j in enumerate(testloader_list):
test_all(i+1, j,what, where)
print("--"*40)
return loss_curi
train_loss_all=[]
testloader_list= [ testloader_1, testloader_2, testloader_3, testloader_4, testloader_5, testloader_6,
testloader_7, testloader_8, testloader_9]
train_loss_all.append(train_all(trainloader_1, 1, testloader_list))
train_loss_all.append(train_all(trainloader_2, 2, testloader_list))
train_loss_all.append(train_all(trainloader_3, 3, testloader_list))
train_loss_all.append(train_all(trainloader_4, 4, testloader_list))
train_loss_all.append(train_all(trainloader_5, 5, testloader_list))
train_loss_all.append(train_all(trainloader_6, 6, testloader_list))
train_loss_all.append(train_all(trainloader_7, 7, testloader_list))
train_loss_all.append(train_all(trainloader_8, 8, testloader_list))
train_loss_all.append(train_all(trainloader_9, 9, testloader_list))
%matplotlib inline
for i,j in enumerate(train_loss_all):
plt.plot(j,label ="dataset "+str(i+1))
plt.xlabel("Epochs")
plt.ylabel("Training_loss")
plt.legend(loc='center left', bbox_to_anchor=(1, 0.5))
```
|
github_jupyter
|
import numpy as np
import pandas as pd
from matplotlib import pyplot as plt
from tqdm import tqdm
%matplotlib inline
from torch.utils.data import Dataset, DataLoader
import torch
import torchvision
import torch.nn as nn
import torch.optim as optim
from torch.nn import functional as F
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
print(device)
y = np.random.randint(0,10,5000)
idx= []
for i in range(10):
print(i,sum(y==i))
idx.append(y==i)
x = np.zeros((5000,2))
x[idx[0],:] = np.random.multivariate_normal(mean = [5,5],cov=[[0.1,0],[0,0.1]],size=sum(idx[0]))
x[idx[1],:] = np.random.multivariate_normal(mean = [-6,7],cov=[[0.1,0],[0,0.1]],size=sum(idx[1]))
x[idx[2],:] = np.random.multivariate_normal(mean = [-5,-4],cov=[[0.1,0],[0,0.1]],size=sum(idx[2]))
# x[idx[0],:] = np.random.multivariate_normal(mean = [5,5],cov=[[0.1,0],[0,0.1]],size=sum(idx[0]))
# x[idx[1],:] = np.random.multivariate_normal(mean = [6,6],cov=[[0.1,0],[0,0.1]],size=sum(idx[1]))
# x[idx[2],:] = np.random.multivariate_normal(mean = [5.5,6.5],cov=[[0.1,0],[0,0.1]],size=sum(idx[2]))
x[idx[3],:] = np.random.multivariate_normal(mean = [-1,0],cov=[[0.1,0],[0,0.1]],size=sum(idx[3]))
x[idx[4],:] = np.random.multivariate_normal(mean = [0,2],cov=[[0.1,0],[0,0.1]],size=sum(idx[4]))
x[idx[5],:] = np.random.multivariate_normal(mean = [1,0],cov=[[0.1,0],[0,0.1]],size=sum(idx[5]))
x[idx[6],:] = np.random.multivariate_normal(mean = [0,-1],cov=[[0.1,0],[0,0.1]],size=sum(idx[6]))
x[idx[7],:] = np.random.multivariate_normal(mean = [0,0],cov=[[0.1,0],[0,0.1]],size=sum(idx[7]))
x[idx[8],:] = np.random.multivariate_normal(mean = [-0.5,-0.5],cov=[[0.1,0],[0,0.1]],size=sum(idx[8]))
x[idx[9],:] = np.random.multivariate_normal(mean = [0.4,0.2],cov=[[0.1,0],[0,0.1]],size=sum(idx[9]))
for i in range(10):
plt.scatter(x[idx[i],0],x[idx[i],1],label="class_"+str(i))
plt.legend(loc='center left', bbox_to_anchor=(1, 0.5))
foreground_classes = {'class_0','class_1', 'class_2'}
background_classes = {'class_3','class_4', 'class_5', 'class_6','class_7', 'class_8', 'class_9'}
fg_class = np.random.randint(0,3)
fg_idx = np.random.randint(0,9)
a = []
for i in range(9):
if i == fg_idx:
b = np.random.choice(np.where(idx[fg_class]==True)[0],size=1)
a.append(x[b])
print("foreground "+str(fg_class)+" present at " + str(fg_idx))
else:
bg_class = np.random.randint(3,10)
b = np.random.choice(np.where(idx[bg_class]==True)[0],size=1)
a.append(x[b])
print("background "+str(bg_class)+" present at " + str(i))
a = np.concatenate(a,axis=0)
print(a.shape)
print(fg_class , fg_idx)
a.shape
np.reshape(a,(18,1))
a=np.reshape(a,(3,6))
plt.imshow(a)
desired_num = 1000
mosaic_list_of_images =[]
mosaic_label = []
fore_idx=[]
for j in range(desired_num):
fg_class = np.random.randint(0,3)
fg_idx = 0
a = []
for i in range(9):
if i == fg_idx:
b = np.random.choice(np.where(idx[fg_class]==True)[0],size=1)
a.append(x[b])
# print("foreground "+str(fg_class)+" present at " + str(fg_idx))
else:
bg_class = np.random.randint(3,10)
b = np.random.choice(np.where(idx[bg_class]==True)[0],size=1)
a.append(x[b])
# print("background "+str(bg_class)+" present at " + str(i))
a = np.concatenate(a,axis=0)
mosaic_list_of_images.append(np.reshape(a,(18,1)))
mosaic_label.append(fg_class)
fore_idx.append(fg_idx)
mosaic_list_of_images = np.concatenate(mosaic_list_of_images,axis=1).T
# print(mosaic_list)
print(np.shape(mosaic_list_of_images))
print(np.shape(fore_idx))
print(np.shape(mosaic_label))
def create_avg_image_from_mosaic_dataset(mosaic_dataset,labels,foreground_index,dataset_number):
"""
mosaic_dataset : mosaic_dataset contains 9 images 32 x 32 each as 1 data point
labels : mosaic_dataset labels
foreground_index : contains list of indexes where foreground image is present so that using this we can take weighted average
dataset_number : will help us to tell what ratio of foreground image to be taken. for eg: if it is "j" then fg_image_ratio = j/9 , bg_image_ratio = (9-j)/8*9
"""
avg_image_dataset = []
cnt = 0
counter = np.array([0,0,0,0,0,0,0,0,0])
for i in range(len(mosaic_dataset)):
img = torch.zeros([18], dtype=torch.float64)
np.random.seed(dataset_number*10000 + i)
give_pref = foreground_index[i] #np.random.randint(0,9)
# print("outside", give_pref,foreground_index[i])
for j in range(9):
if j == give_pref:
img = img + mosaic_dataset[i][j]*dataset_number/9
else :
img = img + mosaic_dataset[i][j]*(9-dataset_number)/(8*9)
if give_pref == foreground_index[i] :
# print("equal are", give_pref,foreground_index[i])
cnt += 1
counter[give_pref] += 1
else :
counter[give_pref] += 1
avg_image_dataset.append(img)
print("number of correct averaging happened for dataset "+str(dataset_number)+" is "+str(cnt))
print("the averaging are done as ", counter)
return avg_image_dataset , labels , foreground_index
avg_image_dataset_1 , labels_1, fg_index_1 = create_avg_image_from_mosaic_dataset(mosaic_list_of_images, mosaic_label, fore_idx, 1)
avg_image_dataset_2 , labels_2, fg_index_2 = create_avg_image_from_mosaic_dataset(mosaic_list_of_images, mosaic_label, fore_idx, 2)
avg_image_dataset_3 , labels_3, fg_index_3 = create_avg_image_from_mosaic_dataset(mosaic_list_of_images, mosaic_label, fore_idx , 3)
avg_image_dataset_4 , labels_4, fg_index_4 = create_avg_image_from_mosaic_dataset(mosaic_list_of_images, mosaic_label, fore_idx , 4)
avg_image_dataset_5 , labels_5, fg_index_5 = create_avg_image_from_mosaic_dataset(mosaic_list_of_images, mosaic_label, fore_idx , 5)
avg_image_dataset_6 , labels_6, fg_index_6 = create_avg_image_from_mosaic_dataset(mosaic_list_of_images, mosaic_label, fore_idx , 6)
avg_image_dataset_7 , labels_7, fg_index_7 = create_avg_image_from_mosaic_dataset(mosaic_list_of_images, mosaic_label, fore_idx , 7)
avg_image_dataset_8 , labels_8, fg_index_8 = create_avg_image_from_mosaic_dataset(mosaic_list_of_images, mosaic_label, fore_idx , 8)
avg_image_dataset_9 , labels_9, fg_index_9 = create_avg_image_from_mosaic_dataset(mosaic_list_of_images, mosaic_label, fore_idx, 9)
class MosaicDataset(Dataset):
"""MosaicDataset dataset."""
def __init__(self, mosaic_list_of_images, mosaic_label):
"""
Args:
csv_file (string): Path to the csv file with annotations.
root_dir (string): Directory with all the images.
transform (callable, optional): Optional transform to be applied
on a sample.
"""
self.mosaic = mosaic_list_of_images
self.label = mosaic_label
#self.fore_idx = fore_idx
def __len__(self):
return len(self.label)
def __getitem__(self, idx):
return self.mosaic[idx] , self.label[idx] #, self.fore_idx[idx]
batch = 200
epochs = 300
# training_data = avg_image_dataset_5 #just change this and training_label to desired dataset for training
# training_label = labels_5
traindata_1 = MosaicDataset(avg_image_dataset_1, labels_1 )
trainloader_1 = DataLoader( traindata_1 , batch_size= batch ,shuffle=True)
traindata_2 = MosaicDataset(avg_image_dataset_2, labels_2 )
trainloader_2 = DataLoader( traindata_2 , batch_size= batch ,shuffle=True)
traindata_3 = MosaicDataset(avg_image_dataset_3, labels_3 )
trainloader_3 = DataLoader( traindata_3 , batch_size= batch ,shuffle=True)
traindata_4 = MosaicDataset(avg_image_dataset_4, labels_4 )
trainloader_4 = DataLoader( traindata_4 , batch_size= batch ,shuffle=True)
traindata_5 = MosaicDataset(avg_image_dataset_5, labels_5 )
trainloader_5 = DataLoader( traindata_5 , batch_size= batch ,shuffle=True)
traindata_6 = MosaicDataset(avg_image_dataset_6, labels_6 )
trainloader_6 = DataLoader( traindata_6 , batch_size= batch ,shuffle=True)
traindata_7 = MosaicDataset(avg_image_dataset_7, labels_7 )
trainloader_7 = DataLoader( traindata_7 , batch_size= batch ,shuffle=True)
traindata_8 = MosaicDataset(avg_image_dataset_8, labels_8 )
trainloader_8 = DataLoader( traindata_8 , batch_size= batch ,shuffle=True)
traindata_9 = MosaicDataset(avg_image_dataset_9, labels_9 )
trainloader_9 = DataLoader( traindata_9 , batch_size= batch ,shuffle=True)
testdata_1 = MosaicDataset(avg_image_dataset_1, labels_1 )
testloader_1 = DataLoader( testdata_1 , batch_size= batch ,shuffle=False)
testdata_2 = MosaicDataset(avg_image_dataset_2, labels_2 )
testloader_2 = DataLoader( testdata_2 , batch_size= batch ,shuffle=False)
testdata_3 = MosaicDataset(avg_image_dataset_3, labels_3 )
testloader_3 = DataLoader( testdata_3 , batch_size= batch ,shuffle=False)
testdata_4 = MosaicDataset(avg_image_dataset_4, labels_4 )
testloader_4 = DataLoader( testdata_4 , batch_size= batch ,shuffle=False)
testdata_5 = MosaicDataset(avg_image_dataset_5, labels_5 )
testloader_5 = DataLoader( testdata_5 , batch_size= batch ,shuffle=False)
testdata_6 = MosaicDataset(avg_image_dataset_6, labels_6 )
testloader_6 = DataLoader( testdata_6 , batch_size= batch ,shuffle=False)
testdata_7 = MosaicDataset(avg_image_dataset_7, labels_7 )
testloader_7 = DataLoader( testdata_7 , batch_size= batch ,shuffle=False)
testdata_8 = MosaicDataset(avg_image_dataset_8, labels_8 )
testloader_8 = DataLoader( testdata_8 , batch_size= batch ,shuffle=False)
testdata_9 = MosaicDataset(avg_image_dataset_9, labels_9 )
testloader_9 = DataLoader( testdata_9 , batch_size= batch ,shuffle=False)
class Wherenet(nn.Module):
def __init__(self):
super(Wherenet,self).__init__()
self.linear1 = nn.Linear(2,128)
self.linear2 = nn.Linear(128,256)
self.linear3 = nn.Linear(256,128)
self.linear4 = nn.Linear(128,64)
self.linear5 = nn.Linear(64,1)
def forward(self,z):
x = torch.zeros([batch,9],dtype=torch.float64)
y = torch.zeros([batch,2], dtype=torch.float64)
#x,y = x.to("cuda"),y.to("cuda")
for i in range(9):
x[:,i] = self.helper(z[:,2*i:2*i+2])[:,0]
#print(k[:,0].shape,x[:,i].shape)
x = F.softmax(x,dim=1) # alphas
x1 = x[:,0]
for i in range(9):
x1 = x[:,i]
#print()
y = y+torch.mul(x1[:,None],z[:,2*i:2*i+2])
return y , x
def helper(self,x):
x = F.relu(self.linear1(x))
x = F.relu(self.linear2(x))
x = F.relu(self.linear3(x))
x = F.relu(self.linear4(x))
x = self.linear5(x)
return x
trainiter = iter(trainloader_1)
input1,labels1 = trainiter.next()
input1.shape
where = Wherenet().double()
where = where
out_where,alphas = where(input1)
out_where.shape,alphas.shape
class Whatnet(nn.Module):
def __init__(self):
super(Whatnet,self).__init__()
self.linear1 = nn.Linear(2,128)
self.linear2 = nn.Linear(128,256)
self.linear3 = nn.Linear(256,128)
self.linear4 = nn.Linear(128,64)
self.linear5 = nn.Linear(64,3)
def forward(self,x):
x = F.relu(self.linear1(x))
x = F.relu(self.linear2(x))
x = F.relu(self.linear3(x))
x = F.relu(self.linear4(x))
x = self.linear5(x)
return x
# what = Whatnet().double()
# what(out_where)
def test_all(number, testloader,what, where):
correct = 0
total = 0
out = []
pred = []
with torch.no_grad():
for data in testloader:
inputs, labels = data
# images, labels = images.to("cuda"),labels.to("cuda")
out.append(labels.cpu().numpy())
avg_inp,alphas = where(inputs)
outputs = what(avg_inp)
_, predicted = torch.max(outputs.data, 1)
pred.append(predicted.cpu().numpy())
total += labels.size(0)
correct += (predicted == labels).sum().item()
print('Accuracy of the network on the 10000 test dataset %d: %d %%' % (number , 100 * correct / total))
def train_all(trainloader, ds_number, testloader_list):
print("--"*40)
print("training on data set ", ds_number)
where = Wherenet().double()
what = Whatnet().double()
criterion = nn.CrossEntropyLoss()
# optimizer_where = optim.SGD(where.parameters(), lr=0.001, momentum=0.9)
optimizer_where = optim.Adam(where.parameters(), lr=1e-2, betas=(0.9, 0.999), eps=1e-08, weight_decay=0, amsgrad=True)
# optimizer_what = optim.SGD(what.parameters(), lr=0.001, momentum=0.9)
optimizer_what = optim.Adam(what.parameters(), lr=1e-2, betas=(0.9, 0.999), eps=1e-08, weight_decay=0, amsgrad=True)
acti = []
loss_curi = []
epochs = 500
for epoch in range(epochs): # loop over the dataset multiple times
ep_lossi = []
running_loss = 0.0
for i, data in enumerate(trainloader, 0):
# get the inputs
inputs, labels = data
# inputs, labels = inputs.to("cuda"),labels.to("cuda")
# zero the parameter gradients
optimizer_what.zero_grad()
optimizer_where.zero_grad()
# forward + backward + optimize
avg_inp,alphas = where(inputs)
# print(avg_inp.shape)
outputs = what(avg_inp)
loss = criterion(outputs, labels)
loss.backward()
optimizer_what.step()
optimizer_where.step()
# print statistics
running_loss += loss.item()
mini = 4
if i % mini == mini-1: # print every 10 mini-batches
print('[%d, %5d] loss: %.3f' %
(epoch + 1, i + 1, running_loss / mini))
ep_lossi.append(running_loss/mini) # loss per minibatch
running_loss = 0.0
loss_curi.append(np.mean(ep_lossi)) #loss per epoch
if (np.mean(ep_lossi) <= 0.05):
break
print('Finished Training')
# torch.save(inc.state_dict(),"train_dataset_"+str(ds_number)+"_"+str(epochs)+".pt")
correct = 0
total = 0
with torch.no_grad():
for data in trainloader:
inputs, labels = data
# images, labels = images.to("cuda"), labels.to("cuda")
avg_inp,alphas = where(inputs)
outputs = what(avg_inp)
_, predicted = torch.max(outputs.data, 1)
total += labels.size(0)
correct += (predicted == labels).sum().item()
print('Accuracy of the network on the 10000 train images: %d %%' % ( 100 * correct / total))
for i, j in enumerate(testloader_list):
test_all(i+1, j,what, where)
print("--"*40)
return loss_curi
train_loss_all=[]
testloader_list= [ testloader_1, testloader_2, testloader_3, testloader_4, testloader_5, testloader_6,
testloader_7, testloader_8, testloader_9]
train_loss_all.append(train_all(trainloader_1, 1, testloader_list))
train_loss_all.append(train_all(trainloader_2, 2, testloader_list))
train_loss_all.append(train_all(trainloader_3, 3, testloader_list))
train_loss_all.append(train_all(trainloader_4, 4, testloader_list))
train_loss_all.append(train_all(trainloader_5, 5, testloader_list))
train_loss_all.append(train_all(trainloader_6, 6, testloader_list))
train_loss_all.append(train_all(trainloader_7, 7, testloader_list))
train_loss_all.append(train_all(trainloader_8, 8, testloader_list))
train_loss_all.append(train_all(trainloader_9, 9, testloader_list))
%matplotlib inline
for i,j in enumerate(train_loss_all):
plt.plot(j,label ="dataset "+str(i+1))
plt.xlabel("Epochs")
plt.ylabel("Training_loss")
plt.legend(loc='center left', bbox_to_anchor=(1, 0.5))
| 0.194406 | 0.629006 |
# Plotting diphthongs
First, we import some standard numeric and plotting libraries, and set some basic defaults to apply to all following plots.
```
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
%matplotlib inline
plt.rc('figure', figsize=(16, 9)) # make plots wider
plt.rc('font', size=16) # make text bigger
```
Since vowel plots are pretty specialized, let's write a reusable function that will put our axes on the top and right sides, and invert the axis directions. We'll do this with a function called `garnish` that can be applied to any plot.
```
def garnish(ax):
ax.xaxis.tick_top()
ax.yaxis.tick_right()
ax.set_xlabel('F2')
ax.set_ylabel('F1')
ax.xaxis.set_label_position('top')
ax.yaxis.set_label_position('right')
ax.set_xlim(2800, 600)
ax.set_ylim(1100, 100)
return ax
```
## Lit review
Here's what some studies in the literature have done:
```
pd.read_csv('lit-review.csv')
```
## Load the data
```
formant_data = pd.read_csv('diphthongs-bVd.csv')
formant_data.head()
```
## Data cleaning
After loading the data, we notice that the `pct` column is strings. It would be easier if they were numbers (for sorting purposes), so let's convert them. Let's also reshape the data from "long format" (1 formant measurement per line) into "wide format" (1 record per line). The way we do this will have the side effect of dropping the `time` values (which we don't need anyway). The resulting table will have "heirarchical indexes" on both the columns and rows, which will make it easy to select whole spans of rows or columns at a time.
```
formant_data['pct'] = formant_data['pct'].map(lambda x: int(x.strip('%')))
wide_data = pd.pivot_table(formant_data, index=['ipa', 'filename'],
columns=['formant', 'pct'], values='formant_value')
wide_data.head(10)
```
it will be useful for later to have the IPA labels in both the table index *and* in their own column, so let's do that now. While we're at it, let's remove the heirarchical index names (`formant`, `pct`, etc), so our software doesn't get confused between the `ipa` column and the `ipa` sequence of row labels. Finally, for obscure reasons, indexing works best if the top level of heirarchical column names is in alphabetic order, so we'll do that now too.
```
idx = wide_data.index
wide_data.reset_index(0, inplace=True, col_level=0)
wide_data.index = idx
wide_data.columns.rename(['', ''], inplace=True)
wide_data.index.rename(['', ''], inplace=True)
wide_data = wide_data[['f1', 'f2', 'ipa']] # sort top-level
wide_data.head()
```
While we're munging data, let's add a column for color, and make the color the same for each instance of a given diphthong. Then we'll show an example of heirarchical indexing at work:
```
colors = plt.rcParams['axes.prop_cycle'].by_key()['color']
color_mapping = dict(zip(wide_data['ipa'].unique(), colors))
wide_data['color'] = wide_data['ipa'].replace(color_mapping)
# example of heirarchical indexing:
wide_data.loc[['aʊ', 'oʊ'], [('ipa', ''), ('color', ''), ('f1', 50), ('f2', 50)]]
```
## Plotting
Let's write a reusable function that will plot the vowels' IPA symbols instead of points.
```
def plot_vowel(f1, f2, vowel, ax=None, **kwargs):
if ax is None:
fig, ax = plt.subplots()
ax.text(f2, f1, vowel, ha='center', va='center', **kwargs)
ax = garnish(ax)
return ax
```
For starters, let's plot just the midpoint of each vowel, like we would do with monophthongs. We could do it all at once with a command like this:
```python
plot_vowels(wide_data[('f1', 50)], wide_data[('f2', 50)], wide_data['ipa'])
```
but it would be nicer if the color was the same for each token of the same diphthong. So we'll use a for-loop to do that:
```
fig, ax = plt.subplots()
for row in wide_data[[('f1', 50), ('f2', 50),
('ipa', ''), ('color', '')]].itertuples(index=False):
plot_vowel(row[0], row[1], row[2], color=row[3], ax=ax);
```
### Plotting 2 points (but which two?)
Now let's pick 2 timepoints in each diphthong. We'll want to connect the start- and end-points with a line, so we'll need a new plotting function.
```
def plot_diphthong(f1, f2, vowel, ax=None, **kwargs):
'''f1 and f2 should be matrices, where each column is one diphthong,
with later timepoints at higher row numbers'''
f1 = np.array(f1)
f2 = np.array(f2)
if ax is None:
fig, ax = plt.subplots()
ax.plot(f2, f1, **kwargs, linestyle='-', marker='o', markersize=3)
ax = plot_vowel(f1[0], f2[0], vowel, ax=ax)
ax = garnish(ax)
return ax
```
We could pick the `20%` and `80%` points, or `30%`-`70%`, or `10%`-`90%`... which is best?
```
pairs = [[5, 95], [10, 90], [15, 85], [20, 80], [25, 75], [30, 70]]
fig, axs = plt.subplots(2, 3, figsize=(16, 15))
for pair, ax in zip(pairs, axs.ravel()):
f1 = wide_data[[('f1', pair[0]), ('f1', pair[1])]]
f2 = wide_data[[('f2', pair[0]), ('f2', pair[1])]]
for this_f1, this_f2, ipa, color in zip(f1.values, f2.values,
wide_data['ipa'],
wide_data['color']):
ax = plot_diphthong(this_f1, this_f2, ipa, color=color, ax=ax)
ax.set_title('{} to {}'.format(*pair), y=1.1);
```
Looking at the red traces for oʊ, notice that the `30-70%` lines go up and to the right, whereas the `5-95%` lines go up and to the left. Let's plot several spans of just that vowel to get a closer look:
```
ou = wide_data.loc['oʊ', ['f1', 'f2', 'ipa', 'color']]
fig, ax = plt.subplots()
for pair in pairs:
for f1, f2, color in zip(ou[[('f1', pair[0]), ('f1', pair[1])]].values,
ou[[('f2', pair[0]), ('f2', pair[1])]].values,
colors):
ax = plot_diphthong(f1, f2, pair[0], ax=ax, color=color, linewidth=0.7)
ax = plot_vowel(f1[-1], f2[-1], pair[1], ax=ax);
```
This plot is a little crowded, but should make clear that you would have a very different impression of the direction of formant movement if you picked `30-70%` vs. `10-90%` or `5-95%`. We can plot the whole time course to get the full picture:
```
fig, ax = plt.subplots()
for f1, f2, ipa, color in zip(ou['f1'].values, ou['f2'].values,
ou['ipa'], colors):
ax = plot_diphthong(f1, f2, ipa, ax=ax, color=color);
```
Presumably all that fronting toward the end of the vowel is due to the following `/d/` consonant, but we can check the spectrogram just to be sure. The `scipy` and `matplotlib` modules both have spectrogram functions built in, but here we'll define a wrapper function that lets us use praat-style parameters like dynamic range in dB and window size in seconds (rather than in samples).
```
from scipy.io import wavfile
from matplotlib.colors import LogNorm
def draw_spectrogram(x, fs, window_dur=0.005, step_dur=None, dyn_range=70,
cmap=None, ax=None):
from scipy.signal import spectrogram, gaussian
# set default for step_dur, if unspecified
if step_dur is None:
step_dur = window_dur / np.sqrt(np.pi) / 8.
# convert window & step durations to numbers of samples
window_nsamp = int(window_dur * fs * 2)
step_nsamp = int(step_dur * fs)
# compute the window
window_sigma = (window_nsamp + 1) // 6
window = gaussian(window_nsamp, window_sigma)
# other arguments to compute spectrogram
noverlap = window_nsamp - step_nsamp
kwargs = dict(noverlap=noverlap, window=window, nperseg=window_nsamp, fs=fs)
freqs, times, power = spectrogram(x, detrend=False, mode='psd',
scaling='density', **kwargs)
# create the figure
if ax is None:
fig, ax = plt.subplots()
# default colormap
if cmap is None:
cmap = plt.get_cmap('Greys')
# other arguments to the figure
extent = (times.min(), times.max(), freqs.min(), freqs.max())
p_ref = 20e-3 # 20 micropascals
dB_max = 10 * np.log10(power.max() / p_ref)
vmin = p_ref * 10 ** ((dB_max - dyn_range) / 10)
ax.imshow(power, origin='lower', aspect='auto', cmap=cmap,
norm=LogNorm(), extent=extent, vmin=vmin, vmax=None)
return ax
```
We'll use the `wavio` module to import the audio file (`scipy` also has a function `scipy.io.wavfile.read()`, but it can't handle audio with 24 bits per sample).
```
import wavio
wav = wavio.read('audio/bVd/bowed1_11.wav')
wav_data = np.squeeze(wav.data)
sampling_frequency = wav.rate
```
Now to plot the spectrogram:
```
fig, ax = plt.subplots(figsize=(16, 6))
ax = draw_spectrogram(wav_data, sampling_frequency, ax=ax)
# draw an arrow
ax.annotate('F2 transition', xy=(0.45, 1650), xytext=(0.55, 2200),
color='r', fontsize=24, fontweight='bold',
arrowprops=dict(facecolor='r', edgecolor='r'));
```
Yep, looks like that raising of F2 is all right at the end, where we'd expect to see the effect of the following consonant.
Let's see what happens when we plot all the points for all the vowels. For good measure, let's average the tokens together, and add a mean track for each vowel type:
```
means = wide_data[['f1', 'f2', 'ipa']].groupby('ipa').aggregate('mean')
blank_labels = [''] * wide_data.shape[0]
fig, ax = plt.subplots()
for f1, f2, ipa, color in zip(wide_data['f1'].values, wide_data['f2'].values,
blank_labels, wide_data['color']):
ax = plot_diphthong(f1, f2, ipa, ax=ax, color=color, alpha=0.6, linewidth=0.7);
for f1, f2, ipa, color in zip(means['f1'].values, means['f2'].values,
means.index, colors):
ax = plot_diphthong(f1, f2, ipa, ax=ax, color=color, linewidth=2.5);
```
Nice, but a little messy... let's try smoothing with a moving average of length 3. When we do this, we lose the endpoints (5% and 95%) because there's nothing before/after them to average them with, so we'll remove those points.
```
rolled = wide_data[['f1', 'f2']].rolling(axis=1, window=3, min_periods=3,
center=True, win_type='boxcar').mean()
rolled['ipa'] = wide_data['ipa']
nan_columns = [(f, p) for p in (5, 95) for f in ('f1', 'f2')]
rolled.drop(nan_columns, axis=1, inplace=True)
fig, ax = plt.subplots()
for f1, f2, ipa, color in zip(rolled['f1'].values, rolled['f2'].values,
rolled['ipa'], wide_data['color']):
ax = plot_diphthong(f1, f2, ipa, ax=ax, color=color, alpha=0.6, linewidth=0.7);
```
## Issues to watch out for
Our data was fairly clean, but that won't always be the case. Let's load some data from /hVd/ tokens from the same talker.
```
# read in the data
formant_data_hvd = pd.read_csv('diphthongs-hVd.csv')
# convert percentage strings to numbers
formant_data_hvd['pct'] = formant_data_hvd['pct'].map(lambda x: int(x.strip('%')))
# convert long to wide format
wide_data_hvd = pd.pivot_table(formant_data_hvd, index=['ipa', 'filename'],
columns=['formant', 'pct'], values='formant_value')
idx = wide_data_hvd.index
wide_data_hvd.reset_index(0, inplace=True, col_level=0)
wide_data_hvd.index = idx
wide_data_hvd.columns.rename(['', ''], inplace=True)
wide_data_hvd.index.rename(['', ''], inplace=True)
wide_data_hvd = wide_data_hvd[['f1', 'f2', 'ipa']] # sort top-level
wide_data_hvd['color'] = wide_data_hvd['ipa'].replace(color_mapping)
# suppress labels for each token
blank_labels = [''] * wide_data_hvd.shape[0]
# plot tokens
fig, ax = plt.subplots()
for f1, f2, color, ipa in zip(wide_data_hvd['f1'].values,
wide_data_hvd['f2'].values,
wide_data_hvd['color'],
blank_labels):
ax = plot_diphthong(f1, f2, ipa, ax=ax, color=color, alpha=0.5, linewidth=0.6)
# plot means
means_hvd = wide_data_hvd[['f1', 'f2', 'ipa']].groupby('ipa').aggregate('mean')
for f1, f2, ipa, color in zip(means_hvd['f1'].values,
means_hvd['f2'].values,
means_hvd.index, colors):
ax = plot_diphthong(f1, f2, ipa, ax=ax, color=color, linewidth=2);
```
## Experimental
Another popular way of summarizing lots of tokens is to plot a ribbon showing the standard deviation around each data point. Since these plots are not true functions, mathematically speaking (i.e., they turn back on themselves in the horizontal direction), this is actually rather trickier than we might think. Here instead we show ellipses illustrating the variation at each timepoint.
```
def ellipse(x, y, ax, quant=0.6827, n_points=50, plot_eigvecs=False, **kwargs):
from scipy.stats import f
from matplotlib.patches import Polygon
mu = (x.mean(), y.mean())
# to reflect greater uncertainty with smaller numbers of observations,
# we use Hotelling's T-squared instead of chi-squared
ddf = len(x) - len(mu)
scale_factor = len(mu) * (len(x) - 1) / ddf
crit_rad = np.sqrt(scale_factor * f.ppf(quant, dfn=len(mu), dfd=ddf))
# compute eigens
cov = np.cov(x, y)
eigval, eigvec = np.linalg.eig(cov)
ixs = np.argsort(eigval)[::-1]
eigval = eigval[ixs]
eigvec = eigvec[:, ixs]
if plot_eigvecs:
# plot eigenvectors (major/minor axes of ellipses)
for val, vec in zip(eigval, eigvec.T):
ax.plot(*zip(mu, mu + vec * np.sqrt(val)), **kwargs)
ax.plot(*zip(mu, mu - vec * np.sqrt(val)), **kwargs)
else:
# plot ellipses
es = eigvec @ np.diag(np.sqrt(eigval))
theta = np.linspace(0, 2 * np.pi, n_points)
unit_circ = np.c_[np.cos(theta), np.sin(theta)].T
points = (np.array(mu)[:, np.newaxis] - es @ (crit_rad * unit_circ)).T
pgon = Polygon(xy=points, **kwargs)
ax.add_artist(pgon)
return ax
fig, ax = plt.subplots()
# plot ellipses
for _, df in wide_data[['f1', 'f2', 'color', 'ipa']].groupby('ipa'):
color = df[('color', '')].unique()[0]
for pct in np.arange(5, 100, 5):
f1 = df[('f1', pct)].values
f2 = df[('f2', pct)].values
ax = ellipse(f2, f1, ax, color=color, alpha=0.1, plot_eigvecs=False)
# plot mean lines
for f1, f2, ipa, color in zip(means['f1'].values, means['f2'].values,
means.index, colors):
ax = plot_diphthong(f1, f2, ipa, ax=ax, color=color);
```
|
github_jupyter
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
%matplotlib inline
plt.rc('figure', figsize=(16, 9)) # make plots wider
plt.rc('font', size=16) # make text bigger
def garnish(ax):
ax.xaxis.tick_top()
ax.yaxis.tick_right()
ax.set_xlabel('F2')
ax.set_ylabel('F1')
ax.xaxis.set_label_position('top')
ax.yaxis.set_label_position('right')
ax.set_xlim(2800, 600)
ax.set_ylim(1100, 100)
return ax
pd.read_csv('lit-review.csv')
formant_data = pd.read_csv('diphthongs-bVd.csv')
formant_data.head()
formant_data['pct'] = formant_data['pct'].map(lambda x: int(x.strip('%')))
wide_data = pd.pivot_table(formant_data, index=['ipa', 'filename'],
columns=['formant', 'pct'], values='formant_value')
wide_data.head(10)
idx = wide_data.index
wide_data.reset_index(0, inplace=True, col_level=0)
wide_data.index = idx
wide_data.columns.rename(['', ''], inplace=True)
wide_data.index.rename(['', ''], inplace=True)
wide_data = wide_data[['f1', 'f2', 'ipa']] # sort top-level
wide_data.head()
colors = plt.rcParams['axes.prop_cycle'].by_key()['color']
color_mapping = dict(zip(wide_data['ipa'].unique(), colors))
wide_data['color'] = wide_data['ipa'].replace(color_mapping)
# example of heirarchical indexing:
wide_data.loc[['aʊ', 'oʊ'], [('ipa', ''), ('color', ''), ('f1', 50), ('f2', 50)]]
def plot_vowel(f1, f2, vowel, ax=None, **kwargs):
if ax is None:
fig, ax = plt.subplots()
ax.text(f2, f1, vowel, ha='center', va='center', **kwargs)
ax = garnish(ax)
return ax
plot_vowels(wide_data[('f1', 50)], wide_data[('f2', 50)], wide_data['ipa'])
fig, ax = plt.subplots()
for row in wide_data[[('f1', 50), ('f2', 50),
('ipa', ''), ('color', '')]].itertuples(index=False):
plot_vowel(row[0], row[1], row[2], color=row[3], ax=ax);
def plot_diphthong(f1, f2, vowel, ax=None, **kwargs):
'''f1 and f2 should be matrices, where each column is one diphthong,
with later timepoints at higher row numbers'''
f1 = np.array(f1)
f2 = np.array(f2)
if ax is None:
fig, ax = plt.subplots()
ax.plot(f2, f1, **kwargs, linestyle='-', marker='o', markersize=3)
ax = plot_vowel(f1[0], f2[0], vowel, ax=ax)
ax = garnish(ax)
return ax
pairs = [[5, 95], [10, 90], [15, 85], [20, 80], [25, 75], [30, 70]]
fig, axs = plt.subplots(2, 3, figsize=(16, 15))
for pair, ax in zip(pairs, axs.ravel()):
f1 = wide_data[[('f1', pair[0]), ('f1', pair[1])]]
f2 = wide_data[[('f2', pair[0]), ('f2', pair[1])]]
for this_f1, this_f2, ipa, color in zip(f1.values, f2.values,
wide_data['ipa'],
wide_data['color']):
ax = plot_diphthong(this_f1, this_f2, ipa, color=color, ax=ax)
ax.set_title('{} to {}'.format(*pair), y=1.1);
ou = wide_data.loc['oʊ', ['f1', 'f2', 'ipa', 'color']]
fig, ax = plt.subplots()
for pair in pairs:
for f1, f2, color in zip(ou[[('f1', pair[0]), ('f1', pair[1])]].values,
ou[[('f2', pair[0]), ('f2', pair[1])]].values,
colors):
ax = plot_diphthong(f1, f2, pair[0], ax=ax, color=color, linewidth=0.7)
ax = plot_vowel(f1[-1], f2[-1], pair[1], ax=ax);
fig, ax = plt.subplots()
for f1, f2, ipa, color in zip(ou['f1'].values, ou['f2'].values,
ou['ipa'], colors):
ax = plot_diphthong(f1, f2, ipa, ax=ax, color=color);
from scipy.io import wavfile
from matplotlib.colors import LogNorm
def draw_spectrogram(x, fs, window_dur=0.005, step_dur=None, dyn_range=70,
cmap=None, ax=None):
from scipy.signal import spectrogram, gaussian
# set default for step_dur, if unspecified
if step_dur is None:
step_dur = window_dur / np.sqrt(np.pi) / 8.
# convert window & step durations to numbers of samples
window_nsamp = int(window_dur * fs * 2)
step_nsamp = int(step_dur * fs)
# compute the window
window_sigma = (window_nsamp + 1) // 6
window = gaussian(window_nsamp, window_sigma)
# other arguments to compute spectrogram
noverlap = window_nsamp - step_nsamp
kwargs = dict(noverlap=noverlap, window=window, nperseg=window_nsamp, fs=fs)
freqs, times, power = spectrogram(x, detrend=False, mode='psd',
scaling='density', **kwargs)
# create the figure
if ax is None:
fig, ax = plt.subplots()
# default colormap
if cmap is None:
cmap = plt.get_cmap('Greys')
# other arguments to the figure
extent = (times.min(), times.max(), freqs.min(), freqs.max())
p_ref = 20e-3 # 20 micropascals
dB_max = 10 * np.log10(power.max() / p_ref)
vmin = p_ref * 10 ** ((dB_max - dyn_range) / 10)
ax.imshow(power, origin='lower', aspect='auto', cmap=cmap,
norm=LogNorm(), extent=extent, vmin=vmin, vmax=None)
return ax
import wavio
wav = wavio.read('audio/bVd/bowed1_11.wav')
wav_data = np.squeeze(wav.data)
sampling_frequency = wav.rate
fig, ax = plt.subplots(figsize=(16, 6))
ax = draw_spectrogram(wav_data, sampling_frequency, ax=ax)
# draw an arrow
ax.annotate('F2 transition', xy=(0.45, 1650), xytext=(0.55, 2200),
color='r', fontsize=24, fontweight='bold',
arrowprops=dict(facecolor='r', edgecolor='r'));
means = wide_data[['f1', 'f2', 'ipa']].groupby('ipa').aggregate('mean')
blank_labels = [''] * wide_data.shape[0]
fig, ax = plt.subplots()
for f1, f2, ipa, color in zip(wide_data['f1'].values, wide_data['f2'].values,
blank_labels, wide_data['color']):
ax = plot_diphthong(f1, f2, ipa, ax=ax, color=color, alpha=0.6, linewidth=0.7);
for f1, f2, ipa, color in zip(means['f1'].values, means['f2'].values,
means.index, colors):
ax = plot_diphthong(f1, f2, ipa, ax=ax, color=color, linewidth=2.5);
rolled = wide_data[['f1', 'f2']].rolling(axis=1, window=3, min_periods=3,
center=True, win_type='boxcar').mean()
rolled['ipa'] = wide_data['ipa']
nan_columns = [(f, p) for p in (5, 95) for f in ('f1', 'f2')]
rolled.drop(nan_columns, axis=1, inplace=True)
fig, ax = plt.subplots()
for f1, f2, ipa, color in zip(rolled['f1'].values, rolled['f2'].values,
rolled['ipa'], wide_data['color']):
ax = plot_diphthong(f1, f2, ipa, ax=ax, color=color, alpha=0.6, linewidth=0.7);
# read in the data
formant_data_hvd = pd.read_csv('diphthongs-hVd.csv')
# convert percentage strings to numbers
formant_data_hvd['pct'] = formant_data_hvd['pct'].map(lambda x: int(x.strip('%')))
# convert long to wide format
wide_data_hvd = pd.pivot_table(formant_data_hvd, index=['ipa', 'filename'],
columns=['formant', 'pct'], values='formant_value')
idx = wide_data_hvd.index
wide_data_hvd.reset_index(0, inplace=True, col_level=0)
wide_data_hvd.index = idx
wide_data_hvd.columns.rename(['', ''], inplace=True)
wide_data_hvd.index.rename(['', ''], inplace=True)
wide_data_hvd = wide_data_hvd[['f1', 'f2', 'ipa']] # sort top-level
wide_data_hvd['color'] = wide_data_hvd['ipa'].replace(color_mapping)
# suppress labels for each token
blank_labels = [''] * wide_data_hvd.shape[0]
# plot tokens
fig, ax = plt.subplots()
for f1, f2, color, ipa in zip(wide_data_hvd['f1'].values,
wide_data_hvd['f2'].values,
wide_data_hvd['color'],
blank_labels):
ax = plot_diphthong(f1, f2, ipa, ax=ax, color=color, alpha=0.5, linewidth=0.6)
# plot means
means_hvd = wide_data_hvd[['f1', 'f2', 'ipa']].groupby('ipa').aggregate('mean')
for f1, f2, ipa, color in zip(means_hvd['f1'].values,
means_hvd['f2'].values,
means_hvd.index, colors):
ax = plot_diphthong(f1, f2, ipa, ax=ax, color=color, linewidth=2);
def ellipse(x, y, ax, quant=0.6827, n_points=50, plot_eigvecs=False, **kwargs):
from scipy.stats import f
from matplotlib.patches import Polygon
mu = (x.mean(), y.mean())
# to reflect greater uncertainty with smaller numbers of observations,
# we use Hotelling's T-squared instead of chi-squared
ddf = len(x) - len(mu)
scale_factor = len(mu) * (len(x) - 1) / ddf
crit_rad = np.sqrt(scale_factor * f.ppf(quant, dfn=len(mu), dfd=ddf))
# compute eigens
cov = np.cov(x, y)
eigval, eigvec = np.linalg.eig(cov)
ixs = np.argsort(eigval)[::-1]
eigval = eigval[ixs]
eigvec = eigvec[:, ixs]
if plot_eigvecs:
# plot eigenvectors (major/minor axes of ellipses)
for val, vec in zip(eigval, eigvec.T):
ax.plot(*zip(mu, mu + vec * np.sqrt(val)), **kwargs)
ax.plot(*zip(mu, mu - vec * np.sqrt(val)), **kwargs)
else:
# plot ellipses
es = eigvec @ np.diag(np.sqrt(eigval))
theta = np.linspace(0, 2 * np.pi, n_points)
unit_circ = np.c_[np.cos(theta), np.sin(theta)].T
points = (np.array(mu)[:, np.newaxis] - es @ (crit_rad * unit_circ)).T
pgon = Polygon(xy=points, **kwargs)
ax.add_artist(pgon)
return ax
fig, ax = plt.subplots()
# plot ellipses
for _, df in wide_data[['f1', 'f2', 'color', 'ipa']].groupby('ipa'):
color = df[('color', '')].unique()[0]
for pct in np.arange(5, 100, 5):
f1 = df[('f1', pct)].values
f2 = df[('f2', pct)].values
ax = ellipse(f2, f1, ax, color=color, alpha=0.1, plot_eigvecs=False)
# plot mean lines
for f1, f2, ipa, color in zip(means['f1'].values, means['f2'].values,
means.index, colors):
ax = plot_diphthong(f1, f2, ipa, ax=ax, color=color);
| 0.702938 | 0.966537 |
# Analysis of covariance (ANCOVA)
ANCOVA is used to compare one variable (**the dependent variable**) in two or more populations while considering other continuous variables.
These continuous varaibles, which are not part of the main experimental manipulation but have an influence on the dependent variable, are known as **covariates**.
## Loading Data
```
viagraData <- read.delim("data/ViagraCovariate.dat", stringsAsFactors = T, header = T)
viagraData$dose <- factor(viagraData$dose, levels = c(1:3), labels = c("Placebo","Low Dose", "High Dose"))
summary(viagraData)
viagraData
```
## Checking homogeneity of variance
```
library(car)
leveneTest(viagraData$libido, viagraData$dose, center=median)
```
Levene's test is **not significant**, which means that for these data the variances are very similar.
## Checking the predictor variable and covariates are independent
ANCOVA assumes that the predictor variable (groups) and covariates are independent.
We can test this by running an ANOVA with the covariate as the outcome.
```
summary( aov(partnerLibido ~ dose , data=viagraData) )
```
The F-ratio is **not significant**, so we can carry out the ANCOVA.
## Fitting an ANCOVA model
### Type I and Type III sum of squares
```
covariateFirst <- aov(libido ~ partnerLibido + dose, data=viagraData)
summary(covariateFirst)
doseFirst <- aov(libido ~ dose + partnerLibido, data=viagraData)
summary(doseFirst)
```
Note that the order in which we enter predicators into a model makes a difference to the effects in the overall ANOVA.
In model **libido ~ partnerLibido + dose**, **partnerLibido** is not significant and **dose** is significant.
In model **libido ~ dose + partnerLibido**, **dose** is not significant and **partnerLibido** is significant.
We can use Type III sums of squares (instead of Type I) by Anova() function to get consistent results.
```
Anova(covariateFirst, type="III")
Anova(doseFirst, type="III")
```
Note that although the results are consistent, but they are **not correct** because Type III sum of squares require **orthogonal contrasts** (The default dummy coding is nonorthogonal) .
### ANCOVA and Type I sum of squares
If we want Type I sum of squares, we should enter the covariates first, then the independent variables in ANCOVA.
So we should use **libido ~ partnerLibido + dose**.
```
viagraModel <- aov(libido ~ partnerLibido + dose, data=viagraData)
summary(doseFirst)
summary.lm(doseFirst)
```
### ANCOVA and Type III sum of squares
We must use orthogonal contrast to get correct results of Type III sum of squares.
```
contrasts(viagraData$dose)
contrasts(viagraData$dose) <- cbind(c(-2,1,1),c(0,-1,1))
contrasts(viagraData$dose)
viagraModel <- aov(libido ~ partnerLibido + dose, data=viagraData)
Anova(viagraModel, type ="III")
summary.lm(viagraModel)
```
## Testing for homogeneity of regression slopes
ANCOVA sassumes that the relationship bwtween the covariate and outcome variable (in this case **partnerLibido** and **dose**) should be similar at different levels of the predictor variable (in this case three **dose** groups).
```
hoRS <- aov(libido ~ partnerLibido * dose, data = viagraData)
Anova(hoRS, type="III")
```
The effect of interfaction of **partnerLibido** and **dose** (**partnerLibido:dose**) is significant, so the assumption can't hold.
```
```
|
github_jupyter
|
viagraData <- read.delim("data/ViagraCovariate.dat", stringsAsFactors = T, header = T)
viagraData$dose <- factor(viagraData$dose, levels = c(1:3), labels = c("Placebo","Low Dose", "High Dose"))
summary(viagraData)
viagraData
library(car)
leveneTest(viagraData$libido, viagraData$dose, center=median)
summary( aov(partnerLibido ~ dose , data=viagraData) )
covariateFirst <- aov(libido ~ partnerLibido + dose, data=viagraData)
summary(covariateFirst)
doseFirst <- aov(libido ~ dose + partnerLibido, data=viagraData)
summary(doseFirst)
Anova(covariateFirst, type="III")
Anova(doseFirst, type="III")
viagraModel <- aov(libido ~ partnerLibido + dose, data=viagraData)
summary(doseFirst)
summary.lm(doseFirst)
contrasts(viagraData$dose)
contrasts(viagraData$dose) <- cbind(c(-2,1,1),c(0,-1,1))
contrasts(viagraData$dose)
viagraModel <- aov(libido ~ partnerLibido + dose, data=viagraData)
Anova(viagraModel, type ="III")
summary.lm(viagraModel)
hoRS <- aov(libido ~ partnerLibido * dose, data = viagraData)
Anova(hoRS, type="III")
| 0.332419 | 0.984185 |
```
import importlib
import prepare_vocab
import train
import eval
import config
from data.loader import DataLoader
from utils import constant
importlib.reload(config)
vocab_params = config.VocabParameters()
training_params = config.TrainingParameters()
eval_params = config.EvalParameters()
opt = vars(vocab_params)
opt['num_class'] = len(constant.LABEL_TO_ID)
opt.update(vars(training_params))
vocab = prepare_vocab.prepare_voabulary (vocab_params)
from utils.vocab import Vocab
vocab_file = vocab_params.vocab_dir + '/vocab.pkl'
vocab = Vocab(vocab_file, load=True)
i = 0
for value in vocab.word2id:
i +=1
print(value)
if i >20:
break
# load data
print("Loading data from {} with batch size {}...".format(vocab_params.data_dir, training_params.batch_size))
train_batch = DataLoader(vocab_params.data_dir+ '/train.json', training_params.batch_size, opt, vocab, evaluation=False)
dev_batch = DataLoader(vocab_params.data_dir + '/dev.json', training_params.batch_size, opt, vocab, evaluation=True)
import json
with open(vocab_params.data_dir+ '/train.json') as infile:
json_data = json.load(infile)
len(json_data)
import pandas as pd
train_df = pd.DataFrame(json_data )
train_df_sorted = train_df.sort_values(by=['docid','id'], ascending = True)
train_df_sorted.head(10)
len(train_df)
import matplotlib.pyplot as plt
relation_df = train_df.groupby("relation")["id"].count().sort_values(ascending=False)
relation_df
%matplotlib inline
relation1_df = train_df[train_df["relation"] != "no_relation"].groupby("relation")["id"].count().sort_values(ascending=False)
plt.figure(figsize=(14,6))
plt.xticks( rotation=90)
plt.plot(relation1_df)
train_df.columns
```
## Expanding List columns to rows, so that we can analyze better
```
import numpy as np
def expand_list(df, list_columns, new_columns):
lens_of_lists = df[list_columns[0]].apply(len)
origin_rows = range(df.shape[0])
destination_rows = np.repeat(origin_rows, lens_of_lists)
non_list_cols = (
[idx for idx, col in enumerate(df.columns)
if col not in list_columns]
)
expanded_df = df.iloc[destination_rows, non_list_cols].copy()
for new_column, list_column in zip(new_columns,list_columns):
expanded_df[new_column] = (
[item for items in df[list_column] for item in items]
)
expanded_df.reset_index(inplace=True, drop=True)
return expanded_df
#usage
train_tokens_df = expand_list(train_df,['token','stanford_pos', 'stanford_ner', 'stanford_head', 'stanford_deprel'], ['token_splitted', 'pos','ner', 'head', 'deprel'] )
train_tokens_df.groupby('pos')['pos'].count().sort_values(ascending=False)
train_tokens_df.groupby('ner')['ner'].count().sort_values(ascending=False)
train_tokens_df.groupby('token_splitted')['token_splitted'].count().sort_values(ascending=False)
train_tokens_df.groupby('head')['head'].count().sort_values(ascending=False)
train_tokens_df.groupby('deprel')['deprel'].count().sort_values(ascending=False)
train_df[train_df["docid"]== "AFP_ENG_19960428.0415.LDC2007T07"]
```
## Creating a simple text document of all the sentences
```
def generate_docs(df, token_list_field, doc_id_field, file_path):
df_sorted = df.sort_values(by=[doc_id_field, "id"], inplace=False)
doc_id = df[doc_id_field][0]
doc_string = ""
f = open(file_path, "w")
for ind in df_sorted.index:
# still in the same document
if doc_id == df_sorted[doc_id_field][ind]:
doc_string += " ".join(df_sorted[token_list_field][ind])
else:
# print(doc_id, ": ", doc_string, "\n")
try:
f.write(doc_id + ": " + doc_string + "\n")
except Exception as inst:
print(doc_id, ind, ":", inst)
doc_string = ""
doc_id = df_sorted[doc_id_field][ind]
doc_string += " ".join(df_sorted[token_list_field][ind])
f.close()
file_path = vocab_params.data_dir + "/raw.txt"
generate_docs(train_df, "token", "docid", file_path)
# train_df["docid"][0]
# train_df.sort_values(by=["docid"], inplace=True)
# train_df
```
## Looking at the relationships that didn't work well in the baseline
```
i = 0
for token in train_df[train_df["relation"] == "per:employee_of"]["token"]:
print (" ".join(token), "\n \n")
i+=1
if i > 20:
break
i = 0
for token in train_df[train_df["relation"] == "org:alternate_names"]["token"]:
print (" ".join(token), "\n \n")
i+=1
if i > 20:
break
```
## Exploring with Spacy
```
import spacy
nlp = spacy.load("en_core_web_lg")
def generate_spacy_tokens(df, token_list_field, doc_id_field):
df_sorted = df.sort_values(by=[doc_id_field, "id"], inplace=False)
doc_id = df[doc_id_field][0]
doc_string = ""
spacy_tokens = []
for ind in df_sorted.index:
# still in the same document
if doc_id == df_sorted[doc_id_field][ind]:
doc_string += " ".join(df_sorted[token_list_field][ind])
else:
spacy_tokens.append(nlp(doc_string))
doc_string = ""
doc_id = df_sorted[doc_id_field][ind]
doc_string += " ".join(df_sorted[token_list_field][ind])
return(spacy_tokens)
len(train_df['docid'].unique())
spacy_tokens = generate_spacy_tokens(train_df, "token", "docid")
len(spacy_tokens)
spacy_tokens[0]
```
|
github_jupyter
|
import importlib
import prepare_vocab
import train
import eval
import config
from data.loader import DataLoader
from utils import constant
importlib.reload(config)
vocab_params = config.VocabParameters()
training_params = config.TrainingParameters()
eval_params = config.EvalParameters()
opt = vars(vocab_params)
opt['num_class'] = len(constant.LABEL_TO_ID)
opt.update(vars(training_params))
vocab = prepare_vocab.prepare_voabulary (vocab_params)
from utils.vocab import Vocab
vocab_file = vocab_params.vocab_dir + '/vocab.pkl'
vocab = Vocab(vocab_file, load=True)
i = 0
for value in vocab.word2id:
i +=1
print(value)
if i >20:
break
# load data
print("Loading data from {} with batch size {}...".format(vocab_params.data_dir, training_params.batch_size))
train_batch = DataLoader(vocab_params.data_dir+ '/train.json', training_params.batch_size, opt, vocab, evaluation=False)
dev_batch = DataLoader(vocab_params.data_dir + '/dev.json', training_params.batch_size, opt, vocab, evaluation=True)
import json
with open(vocab_params.data_dir+ '/train.json') as infile:
json_data = json.load(infile)
len(json_data)
import pandas as pd
train_df = pd.DataFrame(json_data )
train_df_sorted = train_df.sort_values(by=['docid','id'], ascending = True)
train_df_sorted.head(10)
len(train_df)
import matplotlib.pyplot as plt
relation_df = train_df.groupby("relation")["id"].count().sort_values(ascending=False)
relation_df
%matplotlib inline
relation1_df = train_df[train_df["relation"] != "no_relation"].groupby("relation")["id"].count().sort_values(ascending=False)
plt.figure(figsize=(14,6))
plt.xticks( rotation=90)
plt.plot(relation1_df)
train_df.columns
import numpy as np
def expand_list(df, list_columns, new_columns):
lens_of_lists = df[list_columns[0]].apply(len)
origin_rows = range(df.shape[0])
destination_rows = np.repeat(origin_rows, lens_of_lists)
non_list_cols = (
[idx for idx, col in enumerate(df.columns)
if col not in list_columns]
)
expanded_df = df.iloc[destination_rows, non_list_cols].copy()
for new_column, list_column in zip(new_columns,list_columns):
expanded_df[new_column] = (
[item for items in df[list_column] for item in items]
)
expanded_df.reset_index(inplace=True, drop=True)
return expanded_df
#usage
train_tokens_df = expand_list(train_df,['token','stanford_pos', 'stanford_ner', 'stanford_head', 'stanford_deprel'], ['token_splitted', 'pos','ner', 'head', 'deprel'] )
train_tokens_df.groupby('pos')['pos'].count().sort_values(ascending=False)
train_tokens_df.groupby('ner')['ner'].count().sort_values(ascending=False)
train_tokens_df.groupby('token_splitted')['token_splitted'].count().sort_values(ascending=False)
train_tokens_df.groupby('head')['head'].count().sort_values(ascending=False)
train_tokens_df.groupby('deprel')['deprel'].count().sort_values(ascending=False)
train_df[train_df["docid"]== "AFP_ENG_19960428.0415.LDC2007T07"]
def generate_docs(df, token_list_field, doc_id_field, file_path):
df_sorted = df.sort_values(by=[doc_id_field, "id"], inplace=False)
doc_id = df[doc_id_field][0]
doc_string = ""
f = open(file_path, "w")
for ind in df_sorted.index:
# still in the same document
if doc_id == df_sorted[doc_id_field][ind]:
doc_string += " ".join(df_sorted[token_list_field][ind])
else:
# print(doc_id, ": ", doc_string, "\n")
try:
f.write(doc_id + ": " + doc_string + "\n")
except Exception as inst:
print(doc_id, ind, ":", inst)
doc_string = ""
doc_id = df_sorted[doc_id_field][ind]
doc_string += " ".join(df_sorted[token_list_field][ind])
f.close()
file_path = vocab_params.data_dir + "/raw.txt"
generate_docs(train_df, "token", "docid", file_path)
# train_df["docid"][0]
# train_df.sort_values(by=["docid"], inplace=True)
# train_df
i = 0
for token in train_df[train_df["relation"] == "per:employee_of"]["token"]:
print (" ".join(token), "\n \n")
i+=1
if i > 20:
break
i = 0
for token in train_df[train_df["relation"] == "org:alternate_names"]["token"]:
print (" ".join(token), "\n \n")
i+=1
if i > 20:
break
import spacy
nlp = spacy.load("en_core_web_lg")
def generate_spacy_tokens(df, token_list_field, doc_id_field):
df_sorted = df.sort_values(by=[doc_id_field, "id"], inplace=False)
doc_id = df[doc_id_field][0]
doc_string = ""
spacy_tokens = []
for ind in df_sorted.index:
# still in the same document
if doc_id == df_sorted[doc_id_field][ind]:
doc_string += " ".join(df_sorted[token_list_field][ind])
else:
spacy_tokens.append(nlp(doc_string))
doc_string = ""
doc_id = df_sorted[doc_id_field][ind]
doc_string += " ".join(df_sorted[token_list_field][ind])
return(spacy_tokens)
len(train_df['docid'].unique())
spacy_tokens = generate_spacy_tokens(train_df, "token", "docid")
len(spacy_tokens)
spacy_tokens[0]
| 0.084995 | 0.407127 |
## Refactoring
Let's import first the context for this chapter.
```
from context import *
```
Let's put ourselves in an scenario - that you've probably been in before. Imagine you are changing a large piece of legacy code that's not well structured, introducing many changes at once, trying to keep in your head all the bits and pieces that need to be modified to make it all work again. And suddenly, your officemate comes and ask you to go for coffee... and you've lost all track of what you had in your head and need to start again.
Instead of doing so, we could use a more robust approach to go from nasty ugly code to clean code in a safer way.
### Refactoring
To refactor is to:
* Make a change to the design of some software
* Which improves the structure or readability
* But which leaves the actual behaviour of the program completely unchanged.
### A word from the Master
> Refactoring is a controlled technique for improving the design of an existing code base.
Its essence is applying a series of small behavior-preserving transformations, each of which "too small to be worth doing".
However the cumulative effect of each of these transformations is quite significant.
By doing them in small steps you reduce the risk of introducing errors.
You also avoid having the system broken while you are carrying out the restructuring -
which allows you to gradually refactor a system over an extended period of time.
-- Martin Fowler [Refactoring](https://martinfowler.com/books/refactoring.html) [[UCL library](https://ucl-new-primo.hosted.exlibrisgroup.com/primo-explore/fulldisplay?docid=UCL_LMS_DS21146093980004761)].
### List of known refactorings
The next few sections will present some known refactorings.
We'll show before and after code, present any new coding techniques needed to do the refactoring, and describe [*code smells*](https://en.wikipedia.org/wiki/Code_smell): how you know you need to refactor.
### Replace magic numbers with constants
Smell: Raw numbers appear in your code
Before:
```
data = [math.sin(x) for x in np.arange(0,3.141,3.141/100)]
result = [0]*100
for i in range(100):
for j in range(i+1, 100):
result[j] += data[i] * data[i-j] / 100
```
after:
```
resolution = 100
pi = 3.141
data = [math.sin(x) for x in np.arange(0, pi, pi/resolution)]
result = [0] * resolution
for i in range(resolution):
for j in range(i + 1, resolution):
result[j] += data[i] * data[i-j] / resolution
```
### Replace repeated code with a function
Smell: Fragments of repeated code appear.
Fragment of model where some birds are chasing each other: if the angle of view of one can see the prey, then start hunting, and if the other see the predator, then start running away.
Before:
```
if abs(hawk.facing - starling.facing) < hawk.viewport:
hawk.hunting()
if abs(starling.facing - hawk.facing) < starling.viewport:
starling.flee()
```
After:
```
def can_see(source, target):
return (source.facing - target.facing) < source.viewport
if can_see(hawk, starling):
hawk.hunting()
if can_see(starling, hawk):
starling.flee()
```
### Change of variable name
Smell: Code needs a comment to explain what it is for.
Before:
```
z = find(x,y)
if z:
ribe(x)
```
After:
```
gene = subsequence(chromosome, start_codon)
if gene:
transcribe(gene)
```
### Separate a complex expression into a local variable
Smell: An expression becomes long.
```
if ((my_name == your_name) and flag1 or flag2): do_something()
```
vs
```
same_names = (my_name == your_name)
flags_OK = flag1 or flag2
if same_names and flags_OK:
do_something()
```
### Replace loop with iterator
Smell: Loop variable is an integer from 1 to something.
Before:
```
sum = 0
for i in range(resolution):
sum += data[i]
```
After:
```
sum = 0
for value in data:
sum += value
```
### Replace hand-written code with library code
Smell: It feels like surely someone else must have done this at some point.
Before:
```
xcoords = [start + i * step for i in range(int((end - start) / step))]
```
After:
```
import numpy as np
xcoords = np.arange(start, end, step)
```
See [Numpy](http://docs.scipy.org/doc/numpy/reference/generated/numpy.arange.html),
[Pandas](http://pandas.pydata.org/).
### Replace set of arrays with array of structures
Smell: A function needs to work corresponding indices of several arrays:
Before:
```
def can_see(i, source_angles, target_angles, source_viewports):
return abs(source_angles[i] - target_angles[i]) < source_viewports[i]
```
After:
```
def can_see(source, target):
return (source["facing"] - target["facing"]) < source["viewport"]
```
Warning: this refactoring greatly improves readability but can make code slower,
depending on memory layout. Be careful.
### Replace constants with a configuration file
Smell: You need to change your code file to explore different research scenarios.
Before:
```
flight_speed = 2.0 # mph
bounds = [0, 0, 100, 100]
turning_circle = 3.0 # m
bird_counts = {"hawk": 5, "starling": 500}
```
After:
```
%%writefile config.yaml
bounds: [0, 0, 100, 100]
counts:
hawk: 5
starling: 500
speed: 2.0
turning_circle: 3.0
```
```
config = yaml.load(open("config.yaml"))
```
See [YAML](http://www.yaml.org/) and [PyYaml](http://pyyaml.org/),
and [Python's os module](https://docs.python.org/3/library/os.html).
### Replace global variables with function arguments
Smell: A global variable is assigned and then used inside a called function:
```
viewport = pi/4
if hawk.can_see(starling):
hawk.hunt(starling)
class Hawk(object):
def can_see(self, target):
return (self.facing - target.facing) < viewport
```
Becomes:
```
viewport = pi/4
if hawk.can_see(starling, viewport):
hawk.hunt(starling)
class Hawk(object):
def can_see(self, target, viewport):
return (self.facing - target.facing) < viewport
```
### Merge neighbouring loops
Smell: Two neighbouring loops have the same for statement
```
for bird in birds:
bird.build_nest()
for bird in birds:
bird.lay_eggs()
```
Becomes:
```
for bird in birds:
bird.build_nest()
bird.lay_eggs()
```
Though there may be a case where all the nests need to be built before the birds can start laying eggs.
### Break a large function into smaller units
* Smell: A function or subroutine no longer fits on a page in your editor.
* Smell: A line of code is indented more than three levels.
* Smell: A piece of code interacts with the surrounding code through just a few variables.
Before:
```
def do_calculation():
for predator in predators:
for prey in preys:
if predator.can_see(prey):
predator.hunt(prey)
if predator.can_reach(prey):
predator.eat(prey)
```
After:
```
def do_calculation():
for predator in predators:
for prey in preys:
predate(predator, prey)
def predate(predator, prey):
if predator.can_see(prey):
predator.hunt(prey)
if predator.can_reach(prey):
predator.eat(prey)
```
### Separate code concepts into files or modules
Smell: You find it hard to locate a piece of code.
Smell: You get a lot of version control conflicts.
Before:
```
class One(object):
pass
class Two(object):
def __init__():
self.child = One()
```
After:
```
%%writefile anotherfile.py
class One(object):
pass
from anotherfile import One
class Two(object):
def __init__():
self.child = One()
```
### Refactoring is a safe way to improve code
You may think you can see how to rewrite a whole codebase to be better.
However, you may well get lost halfway through the exercise.
By making the changes as small, reversible, incremental steps,
you can reach your target design more reliably.
### Tests and Refactoring
Badly structured code cannot be unit tested. There are no "units".
Before refactoring, ensure you have a robust regression test.
This will allow you to *Refactor with confidence*.
As you refactor, if you create any new units (functions, modules, classes),
add new tests for them.
### Refactoring Summary
* Replace magic numbers with constants
* Replace repeated code with a function
* Change of variable/function/class name
* Replace loop with iterator
* Replace hand-written code with library code
* Replace set of arrays with array of structures
* Replace constants with a configuration file
* Replace global variables with function arguments
* Break a large function into smaller units
* Separate code concepts into files or modules
And many more...
Read [The Refactoring Book](https://martinfowler.com/books/refactoring.html).
|
github_jupyter
|
from context import *
data = [math.sin(x) for x in np.arange(0,3.141,3.141/100)]
result = [0]*100
for i in range(100):
for j in range(i+1, 100):
result[j] += data[i] * data[i-j] / 100
resolution = 100
pi = 3.141
data = [math.sin(x) for x in np.arange(0, pi, pi/resolution)]
result = [0] * resolution
for i in range(resolution):
for j in range(i + 1, resolution):
result[j] += data[i] * data[i-j] / resolution
if abs(hawk.facing - starling.facing) < hawk.viewport:
hawk.hunting()
if abs(starling.facing - hawk.facing) < starling.viewport:
starling.flee()
def can_see(source, target):
return (source.facing - target.facing) < source.viewport
if can_see(hawk, starling):
hawk.hunting()
if can_see(starling, hawk):
starling.flee()
z = find(x,y)
if z:
ribe(x)
gene = subsequence(chromosome, start_codon)
if gene:
transcribe(gene)
if ((my_name == your_name) and flag1 or flag2): do_something()
same_names = (my_name == your_name)
flags_OK = flag1 or flag2
if same_names and flags_OK:
do_something()
sum = 0
for i in range(resolution):
sum += data[i]
sum = 0
for value in data:
sum += value
xcoords = [start + i * step for i in range(int((end - start) / step))]
import numpy as np
xcoords = np.arange(start, end, step)
def can_see(i, source_angles, target_angles, source_viewports):
return abs(source_angles[i] - target_angles[i]) < source_viewports[i]
def can_see(source, target):
return (source["facing"] - target["facing"]) < source["viewport"]
flight_speed = 2.0 # mph
bounds = [0, 0, 100, 100]
turning_circle = 3.0 # m
bird_counts = {"hawk": 5, "starling": 500}
%%writefile config.yaml
bounds: [0, 0, 100, 100]
counts:
hawk: 5
starling: 500
speed: 2.0
turning_circle: 3.0
config = yaml.load(open("config.yaml"))
viewport = pi/4
if hawk.can_see(starling):
hawk.hunt(starling)
class Hawk(object):
def can_see(self, target):
return (self.facing - target.facing) < viewport
viewport = pi/4
if hawk.can_see(starling, viewport):
hawk.hunt(starling)
class Hawk(object):
def can_see(self, target, viewport):
return (self.facing - target.facing) < viewport
for bird in birds:
bird.build_nest()
for bird in birds:
bird.lay_eggs()
for bird in birds:
bird.build_nest()
bird.lay_eggs()
def do_calculation():
for predator in predators:
for prey in preys:
if predator.can_see(prey):
predator.hunt(prey)
if predator.can_reach(prey):
predator.eat(prey)
def do_calculation():
for predator in predators:
for prey in preys:
predate(predator, prey)
def predate(predator, prey):
if predator.can_see(prey):
predator.hunt(prey)
if predator.can_reach(prey):
predator.eat(prey)
class One(object):
pass
class Two(object):
def __init__():
self.child = One()
%%writefile anotherfile.py
class One(object):
pass
from anotherfile import One
class Two(object):
def __init__():
self.child = One()
| 0.479747 | 0.975923 |
```
x = "my precious"
dummy = [x for x in "ABC"]
x
```
## 可迭代序列的拆包
```
a, b, *rest = range(5)
a, b, rest
x, *y, z = range(10)
x, y, z
*m, n, p, q = range(8)
m, n, p, q
metro_areas = [
('Tokyo','JP',36.933,(35.689722,139.691667)),
('Delhi NCR', 'IN', 21.935, (28.613889, 77.208889)),
('Mexico City', 'MX', 20.142, (19.433333, -99.133333)),
('New York-Newark', 'US', 20.104, (40.808611, -74.020386)),
('Sao Paulo', 'BR', 19.649, (-23.547778, -46.635833)),
]
print('{:15} | {:^9} | {:^9}'.format('', 'lat.', 'long.'))
print('{:15} | {:^9} | {:^9}'.format('', 'lat.', 'long.'))
fmt = '{:15} | {:9.4f} | {:9.4f}'
for name, cc, pop, (latitude, longitude) in metro_areas:
if longitude <= 0:
print(fmt.format(name, latitude, longitude))
```
## namedtuple
创建一个具名元组需要两个参数,一个是类名,另一个是类的各个 字段的名字。后者可以是由数个字符串组成的可迭代对象,或者是由空 格分隔开的字段名组成的字符串。
```
from collections import namedtuple
City = namedtuple('City', 'name country population coordinates')
tokyo = City('Tokyo', 'JP', 36.933, (35.689722, 139.691667))
tokyo
tokyo.population
tokyo.coordinates
tokyo[1]
```
_fields 属性是一个包含这个类所有字段名称的元组。
```
City._fields
```
### namedtuple的嵌套使用
```
LatLong = namedtuple('LatLong', 'lat long')
delhi_data = ('Delhi NCR', 'IN', 21.935, LatLong(28.613889, 77.208889))
```
_make() 通过接受一个可迭代对象来生成这个类的一个实例,它 的作用跟 City(*delhi_data) 是一样的。
```
delhi = City._make(delhi_data)
```
_asdict() 把具名元组以 collections.OrderedDict 的形式返 回,我们可以利用它来把元组里的信息友好地呈现出来。
```
delhi._asdict()
for key, value in delhi._asdict().items():
print(key + ":", value)
```
## 切片
```
invoice = """
... 0.....6................................40........52...55........
... 1909 Pimoroni PiBrella $17.50 3 $52.50
... 1489 6mm Tactile Switch x20 $4.95 2 $9.90
... 1510 Panavise Jr. - PV-201 $28.00 1 $28.00
... 1601 PiTFT Mini Kit 320x240 $34.95 1 $34.95
... """
invoice.split('\n')[2:]
```
使用slice()极大增强了代码的可读性
```
SKU = slice(0, 6)
DESCRIPTION = slice(6, 40)
UNIT_PRICE = slice(40, 52)
QUANTITY = slice(52, 55)
ITEM_TOTAL = slice(55, None)
line_items = invoice.split('\n')[2:]
for item in line_items:
print(item[UNIT_PRICE], item[DESCRIPTION])
l = list(range(10))
l
l[2:5] = [99]
l
l = list(range(10))
l[:4] = [11]
l
l = list(range(10))
print(l)
del l[2:5]
print(l)
```
## 对序列使用+和*
\+ 和 * 都遵循这个规律,不修改原有的操作对象,而是构建一个全新的 序列。
```
l = [1, 2, 3]
l * 5
l_1 = [[1, 2, 3]] * 3
l_1
l_1[0][1] = 99
l_1
```
另一种方式
```
l_3 = [[1] * 3] * 3
l_3
l_3[0][1] = 99
l_3
```
对比上面的创建列表和修改列表中的值
```
l_2 = [[1, 2, 3] for _ in range(3)]
l_2
l_2[0][1] = 99
```
## += / *=
可变序列的增量操作:
```
l = [1, 2, 3]
id(l)
l *= 2
l
id(l)
```
不可变序列的增量操作:
```
t = (1, 2, 3)
id(t)
t *= 2
t
id(t)
```
### 不要把可变对象放在元组里面
```
ls_ = (1, 2, [3, 4])
ls_[2] += [5, 6] # 虽然报错,但是仍然完成了操作
ls_
```
## 排序
### bisect为二分查找算法模块,其对有序序列的查找/插入十分有效率
```
import bisect
import sys
HAYSTACK = [1, 4, 5, 6, 8, 12, 15, 20, 21, 23, 23, 26, 29, 30]
NEEDLES = [0, 1, 2, 5, 8, 10, 22, 23, 29, 30, 31]
ROW_FMT = "{0:2d} @ {1:2d} {2}{0:<2d}"
def demo(bisect_fn):
for needle in reversed(NEEDLES):
position = bisect_fn(HAYSTACK, needle)
offset = position * " |"
print(ROW_FMT.format(needle, position, offset))
print("haystack ->", " ".join("%2d" % n for n in HAYSTACK))
demo(bisect.bisect)
print("haystack ->", " ".join("%2d" % n for n in HAYSTACK))
demo(bisect.bisect_left)
```
### bisect模块的查找函数可以用来代替index方法在长序列中快速查找一个元素的位置
```
def grade(score, breakpoints=[60, 70, 80, 90], grades="FDCBA"):
ix = bisect.bisect(breakpoints, score)
return grades[ix]
[grade(score) for score in [33, 99, 77, 70, 89, 60, 90, 100]]
```
### bisect.insort()对列表进行扩充,可以一直维持列表的有序状态
```
import random
my_list = []
for i in range(14):
random_val = random.randrange(14)
bisect.insort(my_list, random_val)
print("%2d ->" % random_val, my_list)
my_list
```
## 数组
```
from array import array
from random import random
# 'd' means 'double'
floats = array('d', (random() for i in range(10**7)))
floats[-1]
fp = open('floats.bin', 'wb')
floats.tofile(fp)
fp.close()
```
array.fromfile(f, n)
Read n items (as machine values) from the file object f and append them to the end of the array.
```
fp = open('floats.bin', 'rb')
floats2 = array('d')
floats2.fromfile(fp, 10**7)
fp.close()
floats[-1]
floats == floats2
```
## numpy
```
import numpy as np
a = np.arange(12)
a
a.shape
a.shape = 3, 4
a
```
## deque
```
from collections import deque
dq = deque(range(10), maxlen=10)
dq
dq = deque(range(10), maxlen=10)
dq.rotate(3)
dq
dq = deque(range(10), maxlen=10)
dq.rotate(-1)
dq
dq = deque(range(10), maxlen=10)
dq.appendleft(-1)
dq
dq = deque(range(10), maxlen=10)
dq.append(-1)
dq
dq = deque(range(10), maxlen=10)
dq.extend([11, 22, 33])
dq
dq = deque(range(10), maxlen=10)
dq.extendleft([11, 22, 33])
dq
dq.maxlen
```
|
github_jupyter
|
x = "my precious"
dummy = [x for x in "ABC"]
x
a, b, *rest = range(5)
a, b, rest
x, *y, z = range(10)
x, y, z
*m, n, p, q = range(8)
m, n, p, q
metro_areas = [
('Tokyo','JP',36.933,(35.689722,139.691667)),
('Delhi NCR', 'IN', 21.935, (28.613889, 77.208889)),
('Mexico City', 'MX', 20.142, (19.433333, -99.133333)),
('New York-Newark', 'US', 20.104, (40.808611, -74.020386)),
('Sao Paulo', 'BR', 19.649, (-23.547778, -46.635833)),
]
print('{:15} | {:^9} | {:^9}'.format('', 'lat.', 'long.'))
print('{:15} | {:^9} | {:^9}'.format('', 'lat.', 'long.'))
fmt = '{:15} | {:9.4f} | {:9.4f}'
for name, cc, pop, (latitude, longitude) in metro_areas:
if longitude <= 0:
print(fmt.format(name, latitude, longitude))
from collections import namedtuple
City = namedtuple('City', 'name country population coordinates')
tokyo = City('Tokyo', 'JP', 36.933, (35.689722, 139.691667))
tokyo
tokyo.population
tokyo.coordinates
tokyo[1]
City._fields
LatLong = namedtuple('LatLong', 'lat long')
delhi_data = ('Delhi NCR', 'IN', 21.935, LatLong(28.613889, 77.208889))
delhi = City._make(delhi_data)
delhi._asdict()
for key, value in delhi._asdict().items():
print(key + ":", value)
invoice = """
... 0.....6................................40........52...55........
... 1909 Pimoroni PiBrella $17.50 3 $52.50
... 1489 6mm Tactile Switch x20 $4.95 2 $9.90
... 1510 Panavise Jr. - PV-201 $28.00 1 $28.00
... 1601 PiTFT Mini Kit 320x240 $34.95 1 $34.95
... """
invoice.split('\n')[2:]
SKU = slice(0, 6)
DESCRIPTION = slice(6, 40)
UNIT_PRICE = slice(40, 52)
QUANTITY = slice(52, 55)
ITEM_TOTAL = slice(55, None)
line_items = invoice.split('\n')[2:]
for item in line_items:
print(item[UNIT_PRICE], item[DESCRIPTION])
l = list(range(10))
l
l[2:5] = [99]
l
l = list(range(10))
l[:4] = [11]
l
l = list(range(10))
print(l)
del l[2:5]
print(l)
l = [1, 2, 3]
l * 5
l_1 = [[1, 2, 3]] * 3
l_1
l_1[0][1] = 99
l_1
l_3 = [[1] * 3] * 3
l_3
l_3[0][1] = 99
l_3
l_2 = [[1, 2, 3] for _ in range(3)]
l_2
l_2[0][1] = 99
l = [1, 2, 3]
id(l)
l *= 2
l
id(l)
t = (1, 2, 3)
id(t)
t *= 2
t
id(t)
ls_ = (1, 2, [3, 4])
ls_[2] += [5, 6] # 虽然报错,但是仍然完成了操作
ls_
import bisect
import sys
HAYSTACK = [1, 4, 5, 6, 8, 12, 15, 20, 21, 23, 23, 26, 29, 30]
NEEDLES = [0, 1, 2, 5, 8, 10, 22, 23, 29, 30, 31]
ROW_FMT = "{0:2d} @ {1:2d} {2}{0:<2d}"
def demo(bisect_fn):
for needle in reversed(NEEDLES):
position = bisect_fn(HAYSTACK, needle)
offset = position * " |"
print(ROW_FMT.format(needle, position, offset))
print("haystack ->", " ".join("%2d" % n for n in HAYSTACK))
demo(bisect.bisect)
print("haystack ->", " ".join("%2d" % n for n in HAYSTACK))
demo(bisect.bisect_left)
def grade(score, breakpoints=[60, 70, 80, 90], grades="FDCBA"):
ix = bisect.bisect(breakpoints, score)
return grades[ix]
[grade(score) for score in [33, 99, 77, 70, 89, 60, 90, 100]]
import random
my_list = []
for i in range(14):
random_val = random.randrange(14)
bisect.insort(my_list, random_val)
print("%2d ->" % random_val, my_list)
my_list
from array import array
from random import random
# 'd' means 'double'
floats = array('d', (random() for i in range(10**7)))
floats[-1]
fp = open('floats.bin', 'wb')
floats.tofile(fp)
fp.close()
fp = open('floats.bin', 'rb')
floats2 = array('d')
floats2.fromfile(fp, 10**7)
fp.close()
floats[-1]
floats == floats2
import numpy as np
a = np.arange(12)
a
a.shape
a.shape = 3, 4
a
from collections import deque
dq = deque(range(10), maxlen=10)
dq
dq = deque(range(10), maxlen=10)
dq.rotate(3)
dq
dq = deque(range(10), maxlen=10)
dq.rotate(-1)
dq
dq = deque(range(10), maxlen=10)
dq.appendleft(-1)
dq
dq = deque(range(10), maxlen=10)
dq.append(-1)
dq
dq = deque(range(10), maxlen=10)
dq.extend([11, 22, 33])
dq
dq = deque(range(10), maxlen=10)
dq.extendleft([11, 22, 33])
dq
dq.maxlen
| 0.322206 | 0.797911 |
In this post we will work through performing the response time correction on oxygen observations following [Bittig et al. (2014)](https://doi.org/10.4319/lom.2014.12.617) on Argo data. The focus is more on accessing the proper variables within Argo than describing the actual correction. We will use [argopandas](https://github.com/ArgoCanada/argopandas) package to manage our data fetching from Argo, and use a function from [bgcArgoDMQC]((https://github.com/ArgoCanada/bgcArgoDMQC)) to do the response time correction. Other basic data manipulation and visualization will use the [pandas](https://pandas.pydata.org/), [numpy](https://numpy.org/), and [scipy](https://scipy.org/) packages, and [matplotlib](https://matplotlib.org/) and [seaborn](https://seaborn.pydata.org/) for plotting.
```
# conda install -c conda-forge argopandas bgcArgoDMQC
import numpy as np
import pandas as pd
from scipy.interpolate import interp1d
import matplotlib.pyplot as plt
import seaborn as sns
sns.set(style='ticks', palette='colorblind')
import argopandas as argo
from bgcArgoDMQC import correct_response_time
```
We will use float [7900589](https://fleetmonitoring.euro-argo.eu/float/7900589), an APEX float in the North Atlantic which has the intermediate parameter `MTIME`, defined as the relative time in fractional days since the date of the profile `JULD`.
```
flt = argo.float(7900589)
# grab core and bgc files for just the most recent cycle
core = flt.prof
bgc = flt.bio_prof
core = core[core.file == core.file.iloc[-2]]
bgc = bgc[bgc.file == bgc.file.iloc[-2]]
core
bgc
core_df = core.levels[['PRES', 'TEMP', 'PSAL']]
bgc_df = bgc.levels[['PRES', 'MTIME', 'DOXY']]
core_df
bgc_df
```
You will notice looking at the printout of `bgc_df` that there are a lot of NaN values. The valid `MTIME` and `DOXY` values are in the `N_PROF` dimension 2. There are a variety of reasons why there might be `N_PROF` > 1 dimensions in an Argo profile. Where that is not the subject I won't go into why, and frankly I only know the valid data is in `N_PROF` = 2 by inspecting the dataframe. The valid core data is in `N_PROF` = 0. If we simply tried to line these separate dataframes up into one, we would fail miserably since our time and oxygen data would not be aligned with our physical data. So instead, we will use the common pressure axis to interpolate onto a common axis.
```
# create a dataframe to store interpolated data in
df = pd.DataFrame()
# define a pressure axis to interpolate and a depth resolution
dP = 2.5
interp_pressure = np.arange(0, core_df['PRES'].max(), dP)
df['PRES'] = interp_pressure
# interpolate
for key, source in zip(['MTIME', 'TEMP', 'DOXY'], [bgc_df, core_df, bgc_df]):
ix = source[key].notna() # remove nan values that will mess with interp
f = interp1d(source['PRES'][ix], source[key][ix], bounds_error=False)
df[key] = f(interp_pressure)
df
```
Now we are almost ready to perform the time response correction, except that *we don't know what the time response of this optode is*. Without a reference data set like in [Bittig et al. (2014)](https://doi.org/10.4319/lom.2014.12.617) or consecutive up- and down-casts as in [Gordon et al. (2020)](https://doi.org/10.5194/bg-17-4119-2020), knowing the response time is not possible. For the purposes of demonstration we will choose a boundary layer thickness (an equivalent parameter, but independent of temperature unlike response time) of 120 micrometers (equivalent to a response time of 67.2 seconds at 20 degrees C).
```
Il = 100
df['DOXY_ADJUSTED'] = correct_response_time(df['MTIME'], df['DOXY'], df['TEMP'], Il)
df['DOXY_DELTA'] = df.DOXY - df.DOXY_ADJUSTED # change in oxygen
```
Finally, we'll plot the profiles to see the end result of the correction.
```
# melt the dataframe so that we can use hue keyword when plotting
df_melt = df.melt(id_vars=['PRES', 'MTIME', 'TEMP', 'DOXY_DELTA'], var_name='DOXY_STATUS', value_name='DOXY')
fig, axes = plt.subplots(1, 2, sharey=True)
sns.lineplot(x='DOXY', y='PRES', hue='DOXY_STATUS', data=df_melt, sort=False, ax=axes[0])
sns.lineplot(x='DOXY_DELTA', y='PRES', data=df, sort=False, ax=axes[1])
axes[0].legend(loc=3, fontsize=8)
axes[0].set_ylim((250, 0))
axes[0].set_ylabel('Pressure (dbar)')
axes[0].set_xlabel('Oxygen ($\\mathregular{\\mu}$mol kg$^{-1}$)')
axes[1].set_xlabel('$\\Delta$O$_2$ ($\\mathregular{\\mu}$mol kg$^{-1}$)')
fig.set_size_inches(fig.get_figwidth()*2/3, fig.get_figheight())
fig.savefig('../figures/time_response_example.png', dpi=350, bbox_inches='tight')
```
Some observations based on the above:
- It is important to recall that this is an *ascending* profile.
- The first thing your eye was likely drawn to is the large change 70m depth. I would wager that this single point is probably too dramatic, but also could be real as the gradient is strong there and oxygen would be favouring the higher side. This point makes me uncomfortable without reference data, but I can't say for sure that it is wrong.
- From 250-100m, oxygen is relatively linear. In this section of the profile, we see a slighly lower `DOXY_ADJUSTED` than the original `DOXY`. Since oxygen is *decreasing* as the float *ascends*, the float remembers the higher concentration from the deeper depth, and therefore slightly overestimates the true oxygen concentration.
- At points where there are "notches" in the original profile, those "notches" are amplified in the corrected on.
Thinking more generally about the wider Argo program, there are a few key questions:
- How would you include this adjusted data in the B-file? Would it go in the `DOXY_ADJUSTED` field, which currently is used for gain adjustment ([Johnson et al. (2015)](https://doi.org/10.1175/JTECH-D-15-0101.1)), or would it merit a different field?
- Assuming there is no reliable way to determine boundary layer thickness (time constant), should Argo correct using a generic one since the adjusted data will be "more correct" than the original, even if it is not perfect?
- Given a lack of reference data, how would you flag the above adjusted profile? Are there any points you don't believe that should be flagged as bad?
|
github_jupyter
|
# conda install -c conda-forge argopandas bgcArgoDMQC
import numpy as np
import pandas as pd
from scipy.interpolate import interp1d
import matplotlib.pyplot as plt
import seaborn as sns
sns.set(style='ticks', palette='colorblind')
import argopandas as argo
from bgcArgoDMQC import correct_response_time
flt = argo.float(7900589)
# grab core and bgc files for just the most recent cycle
core = flt.prof
bgc = flt.bio_prof
core = core[core.file == core.file.iloc[-2]]
bgc = bgc[bgc.file == bgc.file.iloc[-2]]
core
bgc
core_df = core.levels[['PRES', 'TEMP', 'PSAL']]
bgc_df = bgc.levels[['PRES', 'MTIME', 'DOXY']]
core_df
bgc_df
# create a dataframe to store interpolated data in
df = pd.DataFrame()
# define a pressure axis to interpolate and a depth resolution
dP = 2.5
interp_pressure = np.arange(0, core_df['PRES'].max(), dP)
df['PRES'] = interp_pressure
# interpolate
for key, source in zip(['MTIME', 'TEMP', 'DOXY'], [bgc_df, core_df, bgc_df]):
ix = source[key].notna() # remove nan values that will mess with interp
f = interp1d(source['PRES'][ix], source[key][ix], bounds_error=False)
df[key] = f(interp_pressure)
df
Il = 100
df['DOXY_ADJUSTED'] = correct_response_time(df['MTIME'], df['DOXY'], df['TEMP'], Il)
df['DOXY_DELTA'] = df.DOXY - df.DOXY_ADJUSTED # change in oxygen
# melt the dataframe so that we can use hue keyword when plotting
df_melt = df.melt(id_vars=['PRES', 'MTIME', 'TEMP', 'DOXY_DELTA'], var_name='DOXY_STATUS', value_name='DOXY')
fig, axes = plt.subplots(1, 2, sharey=True)
sns.lineplot(x='DOXY', y='PRES', hue='DOXY_STATUS', data=df_melt, sort=False, ax=axes[0])
sns.lineplot(x='DOXY_DELTA', y='PRES', data=df, sort=False, ax=axes[1])
axes[0].legend(loc=3, fontsize=8)
axes[0].set_ylim((250, 0))
axes[0].set_ylabel('Pressure (dbar)')
axes[0].set_xlabel('Oxygen ($\\mathregular{\\mu}$mol kg$^{-1}$)')
axes[1].set_xlabel('$\\Delta$O$_2$ ($\\mathregular{\\mu}$mol kg$^{-1}$)')
fig.set_size_inches(fig.get_figwidth()*2/3, fig.get_figheight())
fig.savefig('../figures/time_response_example.png', dpi=350, bbox_inches='tight')
| 0.393269 | 0.948489 |
# Publications markdown generator for academicpages
Takes a TSV of publications with metadata and converts them for use with [vgonzalezd.github.io](vgonzalezd.github.io). This is an interactive Jupyter notebook ([see more info here](http://jupyter-notebook-beginner-guide.readthedocs.io/en/latest/what_is_jupyter.html)). The core python code is also in `publications.py`. Run either from the `markdown_generator` folder after replacing `publications.tsv` with one containing your data.
TODO: Make this work with BibTex and other databases of citations, rather than Stuart's non-standard TSV format and citation style.
## Data format
The TSV needs to have the following columns: pub_date, title, venue, excerpt, citation, site_url, and paper_url, with a header at the top.
- `excerpt` and `paper_url` can be blank, but the others must have values.
- `pub_date` must be formatted as YYYY-MM-DD.
- `url_slug` will be the descriptive part of the .md file and the permalink URL for the page about the paper. The .md file will be `YYYY-MM-DD-[url_slug].md` and the permalink will be `https://[yourdomain]/publications/YYYY-MM-DD-[url_slug]`
This is how the raw file looks (it doesn't look pretty, use a spreadsheet or other program to edit and create).
```
!cat publications.tsv
```
## Import pandas
We are using the very handy pandas library for dataframes.
```
import pandas as pd
```
## Import TSV
Pandas makes this easy with the read_csv function. We are using a TSV, so we specify the separator as a tab, or `\t`.
I found it important to put this data in a tab-separated values format, because there are a lot of commas in this kind of data and comma-separated values can get messed up. However, you can modify the import statement, as pandas also has read_excel(), read_json(), and others.
```
publications = pd.read_csv("publications.tsv", sep="\t", header=0)
publications
```
## Escape special characters
YAML is very picky about how it takes a valid string, so we are replacing single and double quotes (and ampersands) with their HTML encoded equivilents. This makes them look not so readable in raw format, but they are parsed and rendered nicely.
```
html_escape_table = {
"&": "&",
'"': """,
"'": "'"
}
def html_escape(text):
"""Produce entities within text."""
return "".join(html_escape_table.get(c,c) for c in text)
```
## Creating the markdown files
This is where the heavy lifting is done. This loops through all the rows in the TSV dataframe, then starts to concatentate a big string (```md```) that contains the markdown for each type. It does the YAML metadata first, then does the description for the individual page.
```
import os
for row, item in publications.iterrows():
md_filename = str(item.pub_date) + "-" + item.url_slug + ".md"
html_filename = str(item.pub_date) + "-" + item.url_slug
year = item.pub_date[:4]
## YAML variables
md = "---\ntitle: \"" + item.title + '"\n'
md += """collection: publications"""
md += """\npermalink: /publication/""" + html_filename
if len(str(item.excerpt)) > 5:
md += "\nexcerpt: '" + html_escape(item.excerpt) + "'"
md += "\ndate: " + str(item.pub_date)
md += "\nvenue: '" + html_escape(item.venue) + "'"
if len(str(item.paper_url)) > 5:
md += "\npaperurl: '" + item.paper_url + "'"
md += "\ncitation: '" + html_escape(item.citation) + "'"
md += "\n---"
## Markdown description for individual page
if len(str(item.excerpt)) > 5:
md += "\n" + html_escape(item.excerpt) + "\n"
if len(str(item.paper_url)) > 5:
md += "\n[Download paper here](" + item.paper_url + ")\n"
md += "\nRecommended citation: " + item.citation
md_filename = os.path.basename(md_filename)
with open("../_publications/" + md_filename, 'w') as f:
f.write(md)
```
These files are in the publications directory, one directory below where we're working from.
```
!ls ../_publications/
!cat ../_publications/2009-10-01-paper-title-number-1.md
```
|
github_jupyter
|
!cat publications.tsv
import pandas as pd
publications = pd.read_csv("publications.tsv", sep="\t", header=0)
publications
html_escape_table = {
"&": "&",
'"': """,
"'": "'"
}
def html_escape(text):
"""Produce entities within text."""
return "".join(html_escape_table.get(c,c) for c in text)
import os
for row, item in publications.iterrows():
md_filename = str(item.pub_date) + "-" + item.url_slug + ".md"
html_filename = str(item.pub_date) + "-" + item.url_slug
year = item.pub_date[:4]
## YAML variables
md = "---\ntitle: \"" + item.title + '"\n'
md += """collection: publications"""
md += """\npermalink: /publication/""" + html_filename
if len(str(item.excerpt)) > 5:
md += "\nexcerpt: '" + html_escape(item.excerpt) + "'"
md += "\ndate: " + str(item.pub_date)
md += "\nvenue: '" + html_escape(item.venue) + "'"
if len(str(item.paper_url)) > 5:
md += "\npaperurl: '" + item.paper_url + "'"
md += "\ncitation: '" + html_escape(item.citation) + "'"
md += "\n---"
## Markdown description for individual page
if len(str(item.excerpt)) > 5:
md += "\n" + html_escape(item.excerpt) + "\n"
if len(str(item.paper_url)) > 5:
md += "\n[Download paper here](" + item.paper_url + ")\n"
md += "\nRecommended citation: " + item.citation
md_filename = os.path.basename(md_filename)
with open("../_publications/" + md_filename, 'w') as f:
f.write(md)
!ls ../_publications/
!cat ../_publications/2009-10-01-paper-title-number-1.md
| 0.38122 | 0.75674 |
<!--NAVIGATION-->
< [Jupyter Widgets Tutorial Introduction](00.00-introduction.ipynb) | [Contents](00.00-index.ipynb) | [Widgets without writing widgets: interact](02.00-Using-Interact.ipynb) >
# Overview
What you can accomplish with just Python has increased quite a bit in the last years as more sophisticated tools that plug in to the Jupyter widget ecosystem have been written.
One of those tools is [bqplot](https://github.com/bloomberg/bqplot/blob/master/examples/Index.ipynb), which provides a plotting tool in which the plot, and the lines, markers, labels and legend, all act as widgets. That required both Python *and* JavaScript. On the JavaScript side bqplot uses [d3](https://d3js.org/) to do the drawing in the browser.
The widely-used plotting library [matplotlib](https://matplotlib.org/3.2.2/contents.html) also has a widget interface. Use `%matplotlib widget` in the notebook to have interactive plots that are widgets. For more control, look at the documentation for [ipympl](https://github.com/matplotlib/ipympl) for more details on using it as a widget.
Another example is [ipyvolume](https://ipyvolume.readthedocs.io/en/latest/), which does three-dimensional renderings of point or volumetric data in the browser. It has both Python and JavaScript pieces but using requires only Python.
One last addition is in `ipywidgets` itself: the new `Output` widget can display any content which can be rendered in a Jupyter notebook. That means that anything you can show in a notebook you can include in a widget using only Python.
## Example 1: COVID dashboard (pure Python)
+ Dashboard: http://jupyter.mnstate.edu/COVID
+ Code: https://github.com/JuanCab/COVID_DataViz (see `Dashboard.ipynb`)
Orange boxes are [ipympl](); magenta box is [ipyleaflet](https://ipyleaflet.readthedocs.io/en/latest/); remaining widgets are from [ipywidgets](https://ipywidgets.readthedocs.io/en/stable/).
| | |
|----|----|
 | 
## Example 2: Binary star simulation (pure Python)
+ Green: [pythreejs](https://github.com/jupyter-widgets/pythreejs)
+ Blue: [bqplot](https://github.com/bloomberg/bqplot/blob/master/examples/Index.ipynb)
+ Everything else: [ipywidgets](https://github.com/jupyter-widgets/ipywidgets)
+ Serving it up to users during development on [mybinder.org](https://mybinder.org/)

### Source for this example (including links to binder): https://github.com/JuanCab/AstroInteractives
[Video](https://youtu.be/kbgST0uifvM)
## Remainder of this tutorial
+ Widget basics and core ipywidgets
+ Widget styling and layout
+ Widget events
+ Other widget libraries
<!--NAVIGATION-->
< [Jupyter Widgets Tutorial Introduction](00.00-introduction.ipynb) | [Contents](00.00-index.ipynb) | [Widgets without writing widgets: interact](02.00-Using-Interact.ipynb) >
|
github_jupyter
|
<!--NAVIGATION-->
< [Jupyter Widgets Tutorial Introduction](00.00-introduction.ipynb) | [Contents](00.00-index.ipynb) | [Widgets without writing widgets: interact](02.00-Using-Interact.ipynb) >
# Overview
What you can accomplish with just Python has increased quite a bit in the last years as more sophisticated tools that plug in to the Jupyter widget ecosystem have been written.
One of those tools is [bqplot](https://github.com/bloomberg/bqplot/blob/master/examples/Index.ipynb), which provides a plotting tool in which the plot, and the lines, markers, labels and legend, all act as widgets. That required both Python *and* JavaScript. On the JavaScript side bqplot uses [d3](https://d3js.org/) to do the drawing in the browser.
The widely-used plotting library [matplotlib](https://matplotlib.org/3.2.2/contents.html) also has a widget interface. Use `%matplotlib widget` in the notebook to have interactive plots that are widgets. For more control, look at the documentation for [ipympl](https://github.com/matplotlib/ipympl) for more details on using it as a widget.
Another example is [ipyvolume](https://ipyvolume.readthedocs.io/en/latest/), which does three-dimensional renderings of point or volumetric data in the browser. It has both Python and JavaScript pieces but using requires only Python.
One last addition is in `ipywidgets` itself: the new `Output` widget can display any content which can be rendered in a Jupyter notebook. That means that anything you can show in a notebook you can include in a widget using only Python.
## Example 1: COVID dashboard (pure Python)
+ Dashboard: http://jupyter.mnstate.edu/COVID
+ Code: https://github.com/JuanCab/COVID_DataViz (see `Dashboard.ipynb`)
Orange boxes are [ipympl](); magenta box is [ipyleaflet](https://ipyleaflet.readthedocs.io/en/latest/); remaining widgets are from [ipywidgets](https://ipywidgets.readthedocs.io/en/stable/).
| | |
|----|----|
 | 
## Example 2: Binary star simulation (pure Python)
+ Green: [pythreejs](https://github.com/jupyter-widgets/pythreejs)
+ Blue: [bqplot](https://github.com/bloomberg/bqplot/blob/master/examples/Index.ipynb)
+ Everything else: [ipywidgets](https://github.com/jupyter-widgets/ipywidgets)
+ Serving it up to users during development on [mybinder.org](https://mybinder.org/)

### Source for this example (including links to binder): https://github.com/JuanCab/AstroInteractives
[Video](https://youtu.be/kbgST0uifvM)
## Remainder of this tutorial
+ Widget basics and core ipywidgets
+ Widget styling and layout
+ Widget events
+ Other widget libraries
<!--NAVIGATION-->
< [Jupyter Widgets Tutorial Introduction](00.00-introduction.ipynb) | [Contents](00.00-index.ipynb) | [Widgets without writing widgets: interact](02.00-Using-Interact.ipynb) >
| 0.872279 | 0.959001 |
First, we need to import the main libraries that we will definitely use later.
```
import matplotlib.pyplot as plt
import tensorflow as tf
from matplotlib.image import imread
import sys, os, shutil
import numpy as np
from tqdm import tqdm
```
Train and test datasets must be seperated from each other, our is not right now. So let's seperate them. There is one issue that must be mentioned, there were not enough images in files of older ages etc 95,87 so I deleted the files that contain ages above 80. Also the model can be downloaded from kaggle, which is linked here:
https://www.kaggle.com/frabbisw/facial-age
```
ages = os.listdir("./face_age")
X = []
Y = []
for age in tqdm(ages):
ageint = int(age)
new_folder = "./face_age/{0}".format(age)
photos = os.listdir(new_folder)
for photo in photos:
img = imread("{0}/{1}".format(new_folder, photo))
X.append(img)
Y.append(ageint)
```
We need to create our augmented data which we can easily by the power of tensorflow
```
from sklearn.model_selection import train_test_split
print(type(X))
X = np.array(X, dtype=np.float16)
Y = np.array(Y)
X_train, X_test, y_train, y_test = train_test_split(X, Y, test_size=0.2, random_state=42)
```
Create our callback class and model
```
class myCallback(tf.keras.callbacks.Callback):
def on_epoch_end(self, epoch, logs={}):
if(logs.get('acc') > 0.95):
self.model.stop_training = True
callbacks = myCallback()
model = tf.keras.models.Sequential([
tf.keras.layers.Conv2D(32, (3,3), activation='relu', input_shape=(200,200,3)),
tf.keras.layers.MaxPooling2D(2,2),
tf.keras.layers.Conv2D(64, (3,3), activation='relu'),
tf.keras.layers.MaxPooling2D(2,2),
tf.keras.layers.Conv2D(64, (3,3), activation='relu'),
tf.keras.layers.MaxPooling2D(2,2),
tf.keras.layers.Conv2D(64, (3,3), activation='relu'),
tf.keras.layers.MaxPooling2D(2,2),
tf.keras.layers.Conv2D(64, (3,3), activation='relu'),
tf.keras.layers.MaxPooling2D(2,2),
tf.keras.layers.Conv2D(64, (3,3), activation='relu'),
tf.keras.layers.MaxPooling2D(2,2),
tf.keras.layers.Flatten(),
tf.keras.layers.Dense(256, activation='relu'),
tf.keras.layers.Dropout(0.2),
tf.keras.layers.Dense(1, activation='relu')
])
model.compile(optimizer='adam', loss='mse', metrics=['mae'])
r = model.fit(X_train, y_train, validation_data=(X_test,y_test), epochs=90)
plt.plot(r.history["loss"], label="loss")
plt.plot(r.history["val_loss"], label='val_loss')
plt.legend()
```
Let's save our model so we don't need to train it again and again
```
plt.plot(r.history["mae"], label="mae")
plt.plot(r.history["val_mae"], label="val_mae")
plt.legend()
model.save_weights('my_weights')
model.save('my_model')
```
Our model just works fine, let's test it. I tested the model with a photo of a pretty girl, who is 19.
```
test_img = imread("yas.png", 0) / 255
print(type(test_img))
test_img = tf.expand_dims(test_img, 0)
print(test_img.shape)
```
Model says she's only 16, seems she looks younger than she is lol
```
p = model.predict(test_img)
score = tf.nn.softmax(p[0])
print(str(int(np.round(p)[0][0])-3) + " - " + str(int(np.round(p)[0][0])+3))
```
|
github_jupyter
|
import matplotlib.pyplot as plt
import tensorflow as tf
from matplotlib.image import imread
import sys, os, shutil
import numpy as np
from tqdm import tqdm
ages = os.listdir("./face_age")
X = []
Y = []
for age in tqdm(ages):
ageint = int(age)
new_folder = "./face_age/{0}".format(age)
photos = os.listdir(new_folder)
for photo in photos:
img = imread("{0}/{1}".format(new_folder, photo))
X.append(img)
Y.append(ageint)
from sklearn.model_selection import train_test_split
print(type(X))
X = np.array(X, dtype=np.float16)
Y = np.array(Y)
X_train, X_test, y_train, y_test = train_test_split(X, Y, test_size=0.2, random_state=42)
class myCallback(tf.keras.callbacks.Callback):
def on_epoch_end(self, epoch, logs={}):
if(logs.get('acc') > 0.95):
self.model.stop_training = True
callbacks = myCallback()
model = tf.keras.models.Sequential([
tf.keras.layers.Conv2D(32, (3,3), activation='relu', input_shape=(200,200,3)),
tf.keras.layers.MaxPooling2D(2,2),
tf.keras.layers.Conv2D(64, (3,3), activation='relu'),
tf.keras.layers.MaxPooling2D(2,2),
tf.keras.layers.Conv2D(64, (3,3), activation='relu'),
tf.keras.layers.MaxPooling2D(2,2),
tf.keras.layers.Conv2D(64, (3,3), activation='relu'),
tf.keras.layers.MaxPooling2D(2,2),
tf.keras.layers.Conv2D(64, (3,3), activation='relu'),
tf.keras.layers.MaxPooling2D(2,2),
tf.keras.layers.Conv2D(64, (3,3), activation='relu'),
tf.keras.layers.MaxPooling2D(2,2),
tf.keras.layers.Flatten(),
tf.keras.layers.Dense(256, activation='relu'),
tf.keras.layers.Dropout(0.2),
tf.keras.layers.Dense(1, activation='relu')
])
model.compile(optimizer='adam', loss='mse', metrics=['mae'])
r = model.fit(X_train, y_train, validation_data=(X_test,y_test), epochs=90)
plt.plot(r.history["loss"], label="loss")
plt.plot(r.history["val_loss"], label='val_loss')
plt.legend()
plt.plot(r.history["mae"], label="mae")
plt.plot(r.history["val_mae"], label="val_mae")
plt.legend()
model.save_weights('my_weights')
model.save('my_model')
test_img = imread("yas.png", 0) / 255
print(type(test_img))
test_img = tf.expand_dims(test_img, 0)
print(test_img.shape)
p = model.predict(test_img)
score = tf.nn.softmax(p[0])
print(str(int(np.round(p)[0][0])-3) + " - " + str(int(np.round(p)[0][0])+3))
| 0.544559 | 0.909466 |
# Iterating over iterables
```
# Create a list of strings: flash
flash = ['jay garrick', 'barry allen', 'wally west', 'bart allen']
# Print each list item in flash using a for loop
for person in flash:
print(person)
# Create an iterator for flash: superspeed
superspeed=iter(flash)
# Print each item from the iterator
print(next(superspeed))
print(next(superspeed))
print(next(superspeed))
print(next(superspeed))
```
# Iterating over iterables
```
# Create an iterator for range(3): small_value
small_value = iter(range(3))
# Print the values in small_value
print(next(small_value))
print(next(small_value))
print(next(small_value))
# Loop over range(3) and print the values
for num in range(3):
print(num)
# Create an iterator for range(10 ** 100): googol
googol = iter(range(10**100))
# Print the first 5 values from googol
print(next(googol))
print(next(googol))
print(next(googol))
print(next(googol))
print(next(googol))
```
# Iterators as function arguments
```
# Create a range object: values
values = range(10,21)
# Print the range object
print(values)
# Create a list of integers: values_list
values_list = list(values)
# Print values_list
print(values_list)
# Get the sum of values: values_sum
values_sum = sum(values)
# Print values_sum
print(values_sum)
```
# Using enumerate
```
# Create a list of strings: mutants
mutants = ['charles xavier',
'bobby drake',
'kurt wagner',
'max eisenhardt',
'kitty pride']
# Create a list of tuples: mutant_list
mutant_list = list(enumerate(mutants))
# Print the list of tuples
print(mutant_list)
# Unpack and print the tuple pairs
for index1,value1 in enumerate(mutants):
print(index1, value1)
# Change the start index
for index2,value2 in enumerate(mutants,start=1) :
print(index2, value2)
```
# Using Zip
```
aliases= ['prof x', 'iceman', 'nightcrawler', 'magneto', 'shadowcat']
powers=['telepathy',
'thermokinesis',
'teleportation',
'magnetokinesis',
'intangibility']
# Create a list of tuples: mutant_data
mutant_data = list(zip(mutants,aliases,powers))
# Print the list of tuples
print(mutant_data)
# Create a zip object using the three lists: mutant_zip
mutant_zip = list(zip(mutants,aliases,powers))
# Print the zip object
print(mutant_zip)
# Unpack the zip object and print the tuple values
for value1,value2,value3 in mutant_zip:
print(value1, value2, value3)
```
# Using * and zip to 'unzip'
```
# Create a zip object from mutants and powers: z1
z1 = zip(mutants,powers)
# Print the tuples in z1 by unpacking with *
print(*z1)
# Re-create a zip object from mutants and powers: z1
z1 = zip(mutants,powers)
# 'Unzip' the tuples in z1 by unpacking with * and zip(): result1, result2
result1, result2 =zip(*z1)
# Check if unpacked tuples are equivalent to original tuples
print(result1 == mutants)
print(result2 == powers)
```
# Processing large amounts of Twitter data
```
import pandas as pd
# Initialize an empty dictionary: counts_dict
counts_dict={}
# Iterate over the file chunk by chunk
for chunk in pd.read_csv('https://assets.datacamp.com/production/course_1531/datasets/tweets.csv',chunksize=10):
# Iterate over the column in DataFrame
for entry in chunk['lang']:
if entry in counts_dict.keys():
counts_dict[entry] += 1
else:
counts_dict[entry] = 1
# Print the populated dictionary
print(counts_dict)
```
# Extracting information from large amount of twitter data
```
# Define count_entries()
def count_entries(csv_file,c_size,colname):
"""Return a dictionary with counts of
occurrences as value for each key."""
# Initialize an empty dictionary: counts_dict
counts_dict = {}
# Iterate over the file chunk by chunk
for chunk in pd.read_csv(csv_file,chunksize=c_size):
# Iterate over the column in DataFrame
for entry in chunk[colname]:
if entry in counts_dict.keys():
counts_dict[entry] += 1
else:
counts_dict[entry] = 1
# Return counts_dict
return counts_dict
# Call count_entries(): result_counts
result_counts =count_entries('https://assets.datacamp.com/production/course_1531/datasets/tweets.csv',10,'lang')
# Print result_counts
print(result_counts)
```
# Writing list comprehensions
```
# Create list comprehension: squares
squares = [i**2 for i in range(0,10)]
```
# Nested list comprehensions
```
# Create a 5 x 5 matrix using a list of lists: matrix
matrix = [[col for col in range(0,5)] for row in range(0,5)]
# Print the matrix
for row in matrix:
print(row)
```
# Using conditionals in comprehensions
```
# Create a list of strings: fellowship
fellowship = ['frodo', 'samwise', 'merry', 'aragorn', 'legolas', 'boromir', 'gimli']
# Create list comprehension: new_fellowship
new_fellowship = [member for member in fellowship if len(member)>=7]
# Print the new list
print(new_fellowship)
```
# Using conditionals in comprehensions
```
# Create a list of strings: fellowship
fellowship = ['frodo', 'samwise', 'merry', 'aragorn', 'legolas', 'boromir', 'gimli']
# Create list comprehension: new_fellowship
new_fellowship = [member if len(member)>=7 else '' for member in fellowship]
# Print the new list
print(new_fellowship)
```
# Dict comprehensions
```
# Create a list of strings: fellowship
fellowship = ['frodo', 'samwise', 'merry', 'aragorn', 'legolas', 'boromir', 'gimli']
# Create dict comprehension: new_fellowship
new_fellowship = {member:len(member) for member in fellowship}
# Print the new list
print(new_fellowship)
```
# Write your own generator expressions
```
# Create generator object: result
result = (num for num in range(0,31))
# Print the first 5 values
print(next(result))
print(next(result))
print(next(result))
print(next(result))
print(next(result))
# Print the rest of the values
for value in result:
print(value)
```
# Changing the output in generator expressions
```
# Create a list of strings: lannister
lannister = ['cersei', 'jaime', 'tywin', 'tyrion', 'joffrey']
# Create a generator object: lengths
lengths = (len(person) for person in lannister)
# Iterate over and print the values in lengths
for value in lengths:
print(value)
```
# Build a generator
```
# Create a list of strings
lannister = ['cersei', 'jaime', 'tywin', 'tyrion', 'joffrey']
# Define generator function get_lengths
def get_lengths(input_list):
"""Generator function that yields the
length of the strings in input_list."""
# Yield the length of a string
for person in input_list:
yield len(person)
# Print the values generated by get_lengths()
for value in get_lengths(lannister):
print(value)
```
# List comprehensions for time-stamped data
```
df=pd.read_csv('https://assets.datacamp.com/production/course_1531/datasets/tweets.csv')
# Extract the created_at column from df: tweet_time
tweet_time =df['created_at']
# Extract the clock time: tweet_clock_time
tweet_clock_time = [entry[11:19] for entry in tweet_time]
# Print the extracted times
print(tweet_clock_time)
```
# Conditional list comprehesions for time-stamped data
```
# Extract the created_at column from df: tweet_time
tweet_time = df['created_at']
# Extract the clock time: tweet_clock_time
tweet_clock_time = [entry[11:19] for entry in tweet_time if entry[17:19] == '19']
# Print the extracted times
print(tweet_clock_time)
```
# Dictionaries for data science
```
feature_names=['CountryName',
'CountryCode',
'IndicatorName',
'IndicatorCode',
'Year',
'Value']
row_vals=['Arab World',
'ARB',
'Adolescent fertility rate (births per 1,000 women ages 15-19)',
'SP.ADO.TFRT',
'1960',
'133.56090740552298']
# Zip lists: zipped_lists
zipped_lists = zip(feature_names,row_vals)
# Create a dictionary: rs_dict
rs_dict = dict(zipped_lists)
# Print the dictionary
print(rs_dict)
```
# Writing a function to help you
```
# Define lists2dict()
def lists2dict(list1,list2):
"""Return a dictionary where list1 provides
the keys and list2 provides the values."""
# Zip lists: zipped_lists
zipped_lists = zip(list1, list2)
# Create a dictionary: rs_dict
rs_dict = dict(zipped_lists)
# Return the dictionary
return rs_dict
# Call lists2dict: rs_fxn
rs_fxn = lists2dict(feature_names,row_vals)
# Print rs_fxn
print(rs_fxn)
```
# Using a list comprehension
```
row_lists=[['Arab World',
'ARB',
'Adolescent fertility rate (births per 1,000 women ages 15-19)',
'SP.ADO.TFRT',
'1960',
'133.56090740552298'],
['Arab World',
'ARB',
'Age dependency ratio (% of working-age population)',
'SP.POP.DPND',
'1960',
'87.7976011532547'],
['Arab World',
'ARB',
'Age dependency ratio, old (% of working-age population)',
'SP.POP.DPND.OL',
'1960',
'6.634579191565161'],
['Arab World',
'ARB',
'Age dependency ratio, young (% of working-age population)',
'SP.POP.DPND.YG',
'1960',
'81.02332950839141'],
['Arab World',
'ARB',
'Arms exports (SIPRI trend indicator values)',
'MS.MIL.XPRT.KD',
'1960',
'3000000.0'],
['Arab World',
'ARB',
'Arms imports (SIPRI trend indicator values)',
'MS.MIL.MPRT.KD',
'1960',
'538000000.0'],
['Arab World',
'ARB',
'Birth rate, crude (per 1,000 people)',
'SP.DYN.CBRT.IN',
'1960',
'47.697888095096395'],
['Arab World',
'ARB',
'CO2 emissions (kt)',
'EN.ATM.CO2E.KT',
'1960',
'59563.9892169935'],
['Arab World',
'ARB',
'CO2 emissions (metric tons per capita)',
'EN.ATM.CO2E.PC',
'1960',
'0.6439635478877049']]
# Print the first two lists in row_lists
print(row_lists[0])
print(row_lists[1])
# Turn list of lists into list of dicts: list_of_dicts
list_of_dicts = [lists2dict(feature_names, sublist) for sublist in row_lists]
# Print the first two dictionaries in list_of_dicts
print(list_of_dicts[0])
print(list_of_dicts[1])
```
# Turning this all into a DataFrame
```
# Import the pandas package
import pandas as pd
# Turn list of lists into list of dicts: list_of_dicts
list_of_dicts = [lists2dict(feature_names, sublist) for sublist in row_lists]
# Turn list of dicts into a DataFrame: df
df = pd.DataFrame(list_of_dicts)
# Print the head of the DataFrame
print(df.head())
```
|
github_jupyter
|
# Create a list of strings: flash
flash = ['jay garrick', 'barry allen', 'wally west', 'bart allen']
# Print each list item in flash using a for loop
for person in flash:
print(person)
# Create an iterator for flash: superspeed
superspeed=iter(flash)
# Print each item from the iterator
print(next(superspeed))
print(next(superspeed))
print(next(superspeed))
print(next(superspeed))
# Create an iterator for range(3): small_value
small_value = iter(range(3))
# Print the values in small_value
print(next(small_value))
print(next(small_value))
print(next(small_value))
# Loop over range(3) and print the values
for num in range(3):
print(num)
# Create an iterator for range(10 ** 100): googol
googol = iter(range(10**100))
# Print the first 5 values from googol
print(next(googol))
print(next(googol))
print(next(googol))
print(next(googol))
print(next(googol))
# Create a range object: values
values = range(10,21)
# Print the range object
print(values)
# Create a list of integers: values_list
values_list = list(values)
# Print values_list
print(values_list)
# Get the sum of values: values_sum
values_sum = sum(values)
# Print values_sum
print(values_sum)
# Create a list of strings: mutants
mutants = ['charles xavier',
'bobby drake',
'kurt wagner',
'max eisenhardt',
'kitty pride']
# Create a list of tuples: mutant_list
mutant_list = list(enumerate(mutants))
# Print the list of tuples
print(mutant_list)
# Unpack and print the tuple pairs
for index1,value1 in enumerate(mutants):
print(index1, value1)
# Change the start index
for index2,value2 in enumerate(mutants,start=1) :
print(index2, value2)
aliases= ['prof x', 'iceman', 'nightcrawler', 'magneto', 'shadowcat']
powers=['telepathy',
'thermokinesis',
'teleportation',
'magnetokinesis',
'intangibility']
# Create a list of tuples: mutant_data
mutant_data = list(zip(mutants,aliases,powers))
# Print the list of tuples
print(mutant_data)
# Create a zip object using the three lists: mutant_zip
mutant_zip = list(zip(mutants,aliases,powers))
# Print the zip object
print(mutant_zip)
# Unpack the zip object and print the tuple values
for value1,value2,value3 in mutant_zip:
print(value1, value2, value3)
# Create a zip object from mutants and powers: z1
z1 = zip(mutants,powers)
# Print the tuples in z1 by unpacking with *
print(*z1)
# Re-create a zip object from mutants and powers: z1
z1 = zip(mutants,powers)
# 'Unzip' the tuples in z1 by unpacking with * and zip(): result1, result2
result1, result2 =zip(*z1)
# Check if unpacked tuples are equivalent to original tuples
print(result1 == mutants)
print(result2 == powers)
import pandas as pd
# Initialize an empty dictionary: counts_dict
counts_dict={}
# Iterate over the file chunk by chunk
for chunk in pd.read_csv('https://assets.datacamp.com/production/course_1531/datasets/tweets.csv',chunksize=10):
# Iterate over the column in DataFrame
for entry in chunk['lang']:
if entry in counts_dict.keys():
counts_dict[entry] += 1
else:
counts_dict[entry] = 1
# Print the populated dictionary
print(counts_dict)
# Define count_entries()
def count_entries(csv_file,c_size,colname):
"""Return a dictionary with counts of
occurrences as value for each key."""
# Initialize an empty dictionary: counts_dict
counts_dict = {}
# Iterate over the file chunk by chunk
for chunk in pd.read_csv(csv_file,chunksize=c_size):
# Iterate over the column in DataFrame
for entry in chunk[colname]:
if entry in counts_dict.keys():
counts_dict[entry] += 1
else:
counts_dict[entry] = 1
# Return counts_dict
return counts_dict
# Call count_entries(): result_counts
result_counts =count_entries('https://assets.datacamp.com/production/course_1531/datasets/tweets.csv',10,'lang')
# Print result_counts
print(result_counts)
# Create list comprehension: squares
squares = [i**2 for i in range(0,10)]
# Create a 5 x 5 matrix using a list of lists: matrix
matrix = [[col for col in range(0,5)] for row in range(0,5)]
# Print the matrix
for row in matrix:
print(row)
# Create a list of strings: fellowship
fellowship = ['frodo', 'samwise', 'merry', 'aragorn', 'legolas', 'boromir', 'gimli']
# Create list comprehension: new_fellowship
new_fellowship = [member for member in fellowship if len(member)>=7]
# Print the new list
print(new_fellowship)
# Create a list of strings: fellowship
fellowship = ['frodo', 'samwise', 'merry', 'aragorn', 'legolas', 'boromir', 'gimli']
# Create list comprehension: new_fellowship
new_fellowship = [member if len(member)>=7 else '' for member in fellowship]
# Print the new list
print(new_fellowship)
# Create a list of strings: fellowship
fellowship = ['frodo', 'samwise', 'merry', 'aragorn', 'legolas', 'boromir', 'gimli']
# Create dict comprehension: new_fellowship
new_fellowship = {member:len(member) for member in fellowship}
# Print the new list
print(new_fellowship)
# Create generator object: result
result = (num for num in range(0,31))
# Print the first 5 values
print(next(result))
print(next(result))
print(next(result))
print(next(result))
print(next(result))
# Print the rest of the values
for value in result:
print(value)
# Create a list of strings: lannister
lannister = ['cersei', 'jaime', 'tywin', 'tyrion', 'joffrey']
# Create a generator object: lengths
lengths = (len(person) for person in lannister)
# Iterate over and print the values in lengths
for value in lengths:
print(value)
# Create a list of strings
lannister = ['cersei', 'jaime', 'tywin', 'tyrion', 'joffrey']
# Define generator function get_lengths
def get_lengths(input_list):
"""Generator function that yields the
length of the strings in input_list."""
# Yield the length of a string
for person in input_list:
yield len(person)
# Print the values generated by get_lengths()
for value in get_lengths(lannister):
print(value)
df=pd.read_csv('https://assets.datacamp.com/production/course_1531/datasets/tweets.csv')
# Extract the created_at column from df: tweet_time
tweet_time =df['created_at']
# Extract the clock time: tweet_clock_time
tweet_clock_time = [entry[11:19] for entry in tweet_time]
# Print the extracted times
print(tweet_clock_time)
# Extract the created_at column from df: tweet_time
tweet_time = df['created_at']
# Extract the clock time: tweet_clock_time
tweet_clock_time = [entry[11:19] for entry in tweet_time if entry[17:19] == '19']
# Print the extracted times
print(tweet_clock_time)
feature_names=['CountryName',
'CountryCode',
'IndicatorName',
'IndicatorCode',
'Year',
'Value']
row_vals=['Arab World',
'ARB',
'Adolescent fertility rate (births per 1,000 women ages 15-19)',
'SP.ADO.TFRT',
'1960',
'133.56090740552298']
# Zip lists: zipped_lists
zipped_lists = zip(feature_names,row_vals)
# Create a dictionary: rs_dict
rs_dict = dict(zipped_lists)
# Print the dictionary
print(rs_dict)
# Define lists2dict()
def lists2dict(list1,list2):
"""Return a dictionary where list1 provides
the keys and list2 provides the values."""
# Zip lists: zipped_lists
zipped_lists = zip(list1, list2)
# Create a dictionary: rs_dict
rs_dict = dict(zipped_lists)
# Return the dictionary
return rs_dict
# Call lists2dict: rs_fxn
rs_fxn = lists2dict(feature_names,row_vals)
# Print rs_fxn
print(rs_fxn)
row_lists=[['Arab World',
'ARB',
'Adolescent fertility rate (births per 1,000 women ages 15-19)',
'SP.ADO.TFRT',
'1960',
'133.56090740552298'],
['Arab World',
'ARB',
'Age dependency ratio (% of working-age population)',
'SP.POP.DPND',
'1960',
'87.7976011532547'],
['Arab World',
'ARB',
'Age dependency ratio, old (% of working-age population)',
'SP.POP.DPND.OL',
'1960',
'6.634579191565161'],
['Arab World',
'ARB',
'Age dependency ratio, young (% of working-age population)',
'SP.POP.DPND.YG',
'1960',
'81.02332950839141'],
['Arab World',
'ARB',
'Arms exports (SIPRI trend indicator values)',
'MS.MIL.XPRT.KD',
'1960',
'3000000.0'],
['Arab World',
'ARB',
'Arms imports (SIPRI trend indicator values)',
'MS.MIL.MPRT.KD',
'1960',
'538000000.0'],
['Arab World',
'ARB',
'Birth rate, crude (per 1,000 people)',
'SP.DYN.CBRT.IN',
'1960',
'47.697888095096395'],
['Arab World',
'ARB',
'CO2 emissions (kt)',
'EN.ATM.CO2E.KT',
'1960',
'59563.9892169935'],
['Arab World',
'ARB',
'CO2 emissions (metric tons per capita)',
'EN.ATM.CO2E.PC',
'1960',
'0.6439635478877049']]
# Print the first two lists in row_lists
print(row_lists[0])
print(row_lists[1])
# Turn list of lists into list of dicts: list_of_dicts
list_of_dicts = [lists2dict(feature_names, sublist) for sublist in row_lists]
# Print the first two dictionaries in list_of_dicts
print(list_of_dicts[0])
print(list_of_dicts[1])
# Import the pandas package
import pandas as pd
# Turn list of lists into list of dicts: list_of_dicts
list_of_dicts = [lists2dict(feature_names, sublist) for sublist in row_lists]
# Turn list of dicts into a DataFrame: df
df = pd.DataFrame(list_of_dicts)
# Print the head of the DataFrame
print(df.head())
| 0.617282 | 0.878991 |
# FLIR to PASCAL VOC Annotations
```
import os
import glob
from pathlib import Path
import json
from tqdm import tqdm
files_path = Path('./FLIR_ADAS/train/thermal_8_bit')
files = files_path.glob('../*.json')
files = list(files)
id_to_cat = {'3': 'car', '1': 'person', '2': 'bicycle', '17': 'dog'}
files_names = []
for each in range(len(files)):
with open(f'{files[each]}') as f:
ann = json.load(f)
files_names = [ sub['file_name'] for sub in ann["images"] ]
with open('flir_train.txt', 'w') as f:
for each in files_names:
f.write("%s\n" % each)
def getXMLAnn(ann, image_num):
st=""
for i,b in enumerate(ann):
if int(b['image_id']) < image_num:
continue
if int(b['image_id']) > image_num:
return st
if i == 0:
st += f'\
<object>\n \
<name>{id_to_cat[str(b["category_id"])]}</name>\n \
<pose>Frontal</pose>\n \
<truncated>0</truncated>\n \
<difficult>0</difficult>\n \
<occluded>0</occluded>\n \
<bndbox>\n \
<xmin>{b["bbox"][0]}</xmin>\n \
<xmax>{int(b["bbox"][0])+int(b["bbox"][2])}</xmax>\n \
<ymin>{b["bbox"][1]}</ymin>\n \
<ymax>{int(b["bbox"][1])+int(b["bbox"][3])}</ymax>\n \
</bndbox>\n \
</object> \n'
else:
st += f'\
<object>\n \
<name>{id_to_cat[str(b["category_id"])]}</name>\n \
<pose>Frontal</pose>\n \
<truncated>0</truncated>\n \
<difficult>0</difficult>\n \
<occluded>0</occluded>\n \
<bndbox>\n \
<xmin>{b["bbox"][0]}</xmin>\n \
<xmax>{int(b["bbox"][0])+int(b["bbox"][2])}</xmax>\n \
<ymin>{b["bbox"][1]}</ymin>\n \
<ymax>{int(b["bbox"][1])+int(b["bbox"][3])}</ymax>\n \
</bndbox>\n \
</object> \n'
return st
def toXML(ann, image_num):
x = f'\
<annotation>\n \
<folder>VOC2007</folder>\n \
<filename>{ann["images"][image_num]["file_name"]}</filename>\n \
<source>\n \
<database>The VOC2007 Database</database>\n \
<annotation>PASCAL VOC2007</annotation> \n \
</source>\n \
<size>\n \
<width>{ann["images"][image_num]["width"]}</width>\n \
<height>{ann["images"][image_num]["height"]}</height>\n \
<depth>3</depth>\n \
</size>\n \
<segmented>0</segmented>\n \
{getXMLAnn(ann["annotations"], image_num)} \
</annotation>\n'
return x
from tqdm import tqdm
for each in tqdm(range(len(files))):
with open(f'{files[each]}') as f:
ann = json.load(f)
for image_num in range(len(files_names)):
files_names[image_num] = files_names[image_num].strip('.jpeg')
with open(f'./Annotations_PVOC/{files_names[image_num]}.xml', 'w') as t:
t.write(toXML(ann,image_num))
```
## Validation Data Annotations
```
files_path = Path('./FLIR_ADAS/val/thermal_8_bit')
files = files_path.glob('../*.json')
files = list(files)
files_names = []
for each in range(len(files)):
with open(f'{files[each]}') as f:
ann = json.load(f)
files_names = [ sub['file_name'] for sub in ann["images"] ]
with open('flir_valid.txt', 'w') as f:
for each in files_names:
f.write("%s\n" % each)
```
#### In v1 of the dataset there is a problem in the validation annotations, annotation file name and the file tag inside the annotation do not match. We corrected this mistake before testing our models.
```
def getXMLAnn(ann, image_num):
st=""
for i,b in enumerate(ann):
if int(b['image_id']) < image_num:
continue
if int(b['image_id']) > image_num:
return st
if i == 0:
st += f'\
<object>\n \
<name>{id_to_cat[str(b["category_id"])]}</name>\n \
<pose>Frontal</pose>\n \
<truncated>0</truncated>\n \
<difficult>0</difficult>\n \
<occluded>0</occluded>\n \
<bndbox>\n \
<xmin>{b["bbox"][0]}</xmin>\n \
<xmax>{int(b["bbox"][0])+int(b["bbox"][2])}</xmax>\n \
<ymin>{b["bbox"][1]}</ymin>\n \
<ymax>{int(b["bbox"][1])+int(b["bbox"][3])}</ymax>\n \
</bndbox>\n \
</object> \n'
else:
st += f'\
<object>\n \
<name>{id_to_cat[str(b["category_id"])]}</name>\n \
<pose>Frontal</pose>\n \
<truncated>0</truncated>\n \
<difficult>0</difficult>\n \
<occluded>0</occluded>\n \
<bndbox>\n \
<xmin>{b["bbox"][0]}</xmin>\n \
<xmax>{int(b["bbox"][0])+int(b["bbox"][2])}</xmax>\n \
<ymin>{b["bbox"][1]}</ymin>\n \
<ymax>{int(b["bbox"][1])+int(b["bbox"][3])}</ymax>\n \
</bndbox>\n \
</object> \n'
return st
def toXML(ann, name, image_num):
x = f'\
<annotation>\n \
<folder>VOC2007</folder>\n \
<filename>{ann["images"][image_num]["file_name"]}</filename>\n \
<source>\n \
<database>The VOC2007 Database</database>\n \
<annotation>PASCAL VOC2007</annotation> \n \
</source>\n \
<size>\n \
<width>{ann["images"][image_num]["width"]}</width>\n \
<height>{ann["images"][image_num]["height"]}</height>\n \
<depth>3</depth>\n \
</size>\n \
<segmented>0</segmented>\n \
{getXMLAnn(ann["annotations"], image_num)} \
</annotation>\n'
return x
from tqdm import tqdm
for each in tqdm(range(len(files))):
with open(f'{files[each]}') as f:
ann = json.load(f)
for image_num in range(len(files_names)):
files_names[image_num] = files_names[image_num].strip('.jpeg')
with open(f'./Annotations_PVOC/{files_names[image_num]}.xml', 'w') as t:
t.write(toXML(ann, files[each], image_num))
```
|
github_jupyter
|
import os
import glob
from pathlib import Path
import json
from tqdm import tqdm
files_path = Path('./FLIR_ADAS/train/thermal_8_bit')
files = files_path.glob('../*.json')
files = list(files)
id_to_cat = {'3': 'car', '1': 'person', '2': 'bicycle', '17': 'dog'}
files_names = []
for each in range(len(files)):
with open(f'{files[each]}') as f:
ann = json.load(f)
files_names = [ sub['file_name'] for sub in ann["images"] ]
with open('flir_train.txt', 'w') as f:
for each in files_names:
f.write("%s\n" % each)
def getXMLAnn(ann, image_num):
st=""
for i,b in enumerate(ann):
if int(b['image_id']) < image_num:
continue
if int(b['image_id']) > image_num:
return st
if i == 0:
st += f'\
<object>\n \
<name>{id_to_cat[str(b["category_id"])]}</name>\n \
<pose>Frontal</pose>\n \
<truncated>0</truncated>\n \
<difficult>0</difficult>\n \
<occluded>0</occluded>\n \
<bndbox>\n \
<xmin>{b["bbox"][0]}</xmin>\n \
<xmax>{int(b["bbox"][0])+int(b["bbox"][2])}</xmax>\n \
<ymin>{b["bbox"][1]}</ymin>\n \
<ymax>{int(b["bbox"][1])+int(b["bbox"][3])}</ymax>\n \
</bndbox>\n \
</object> \n'
else:
st += f'\
<object>\n \
<name>{id_to_cat[str(b["category_id"])]}</name>\n \
<pose>Frontal</pose>\n \
<truncated>0</truncated>\n \
<difficult>0</difficult>\n \
<occluded>0</occluded>\n \
<bndbox>\n \
<xmin>{b["bbox"][0]}</xmin>\n \
<xmax>{int(b["bbox"][0])+int(b["bbox"][2])}</xmax>\n \
<ymin>{b["bbox"][1]}</ymin>\n \
<ymax>{int(b["bbox"][1])+int(b["bbox"][3])}</ymax>\n \
</bndbox>\n \
</object> \n'
return st
def toXML(ann, image_num):
x = f'\
<annotation>\n \
<folder>VOC2007</folder>\n \
<filename>{ann["images"][image_num]["file_name"]}</filename>\n \
<source>\n \
<database>The VOC2007 Database</database>\n \
<annotation>PASCAL VOC2007</annotation> \n \
</source>\n \
<size>\n \
<width>{ann["images"][image_num]["width"]}</width>\n \
<height>{ann["images"][image_num]["height"]}</height>\n \
<depth>3</depth>\n \
</size>\n \
<segmented>0</segmented>\n \
{getXMLAnn(ann["annotations"], image_num)} \
</annotation>\n'
return x
from tqdm import tqdm
for each in tqdm(range(len(files))):
with open(f'{files[each]}') as f:
ann = json.load(f)
for image_num in range(len(files_names)):
files_names[image_num] = files_names[image_num].strip('.jpeg')
with open(f'./Annotations_PVOC/{files_names[image_num]}.xml', 'w') as t:
t.write(toXML(ann,image_num))
files_path = Path('./FLIR_ADAS/val/thermal_8_bit')
files = files_path.glob('../*.json')
files = list(files)
files_names = []
for each in range(len(files)):
with open(f'{files[each]}') as f:
ann = json.load(f)
files_names = [ sub['file_name'] for sub in ann["images"] ]
with open('flir_valid.txt', 'w') as f:
for each in files_names:
f.write("%s\n" % each)
def getXMLAnn(ann, image_num):
st=""
for i,b in enumerate(ann):
if int(b['image_id']) < image_num:
continue
if int(b['image_id']) > image_num:
return st
if i == 0:
st += f'\
<object>\n \
<name>{id_to_cat[str(b["category_id"])]}</name>\n \
<pose>Frontal</pose>\n \
<truncated>0</truncated>\n \
<difficult>0</difficult>\n \
<occluded>0</occluded>\n \
<bndbox>\n \
<xmin>{b["bbox"][0]}</xmin>\n \
<xmax>{int(b["bbox"][0])+int(b["bbox"][2])}</xmax>\n \
<ymin>{b["bbox"][1]}</ymin>\n \
<ymax>{int(b["bbox"][1])+int(b["bbox"][3])}</ymax>\n \
</bndbox>\n \
</object> \n'
else:
st += f'\
<object>\n \
<name>{id_to_cat[str(b["category_id"])]}</name>\n \
<pose>Frontal</pose>\n \
<truncated>0</truncated>\n \
<difficult>0</difficult>\n \
<occluded>0</occluded>\n \
<bndbox>\n \
<xmin>{b["bbox"][0]}</xmin>\n \
<xmax>{int(b["bbox"][0])+int(b["bbox"][2])}</xmax>\n \
<ymin>{b["bbox"][1]}</ymin>\n \
<ymax>{int(b["bbox"][1])+int(b["bbox"][3])}</ymax>\n \
</bndbox>\n \
</object> \n'
return st
def toXML(ann, name, image_num):
x = f'\
<annotation>\n \
<folder>VOC2007</folder>\n \
<filename>{ann["images"][image_num]["file_name"]}</filename>\n \
<source>\n \
<database>The VOC2007 Database</database>\n \
<annotation>PASCAL VOC2007</annotation> \n \
</source>\n \
<size>\n \
<width>{ann["images"][image_num]["width"]}</width>\n \
<height>{ann["images"][image_num]["height"]}</height>\n \
<depth>3</depth>\n \
</size>\n \
<segmented>0</segmented>\n \
{getXMLAnn(ann["annotations"], image_num)} \
</annotation>\n'
return x
from tqdm import tqdm
for each in tqdm(range(len(files))):
with open(f'{files[each]}') as f:
ann = json.load(f)
for image_num in range(len(files_names)):
files_names[image_num] = files_names[image_num].strip('.jpeg')
with open(f'./Annotations_PVOC/{files_names[image_num]}.xml', 'w') as t:
t.write(toXML(ann, files[each], image_num))
| 0.16228 | 0.389779 |
# Simulation Analysis Graphics for Paper - Quartier Nord
by: Clayton Miller
Apr 13, 2016
```
%matplotlib inline
import esoreader
reload(esoreader)
import pandas as pd
import os
import seaborn as sns
import matplotlib.pyplot as plt
from pylab import *
import csv
from __future__ import division
sns.set_style("whitegrid")
```
# Get Co-Sim Data
```
# change this to point to where you checked out the GitHub project
PROJECT_PATH = r"/Users/Clayton/Dropbox/03-ETH/UMEM/Quartier_Nord_EPFL/"
# this is the output of the workflow
ESO_PATH = os.path.join(PROJECT_PATH,'cosimulation','quartier_nord.eso')
# yeah... we need an index for timeseries...
HOURS_IN_YEAR = pd.date_range('2013-01-01', '2013-12-31 T23:00', freq='H')
qn_cosim_eplus = esoreader.read_from_path(ESO_PATH)
Varlist = pd.DataFrame(qn_cosim_eplus.dd.variables)
Varlist = Varlist.T
uniquevarlist = Varlist#[2].unique()
qn_cosim_eplus.find_variable("Zone Ideal Loads")
#qn_cosim_eplus.to_frame("Zone Total Heating Energy")
heating = qn_cosim_eplus.to_frame('Zone Total Heating Energy', index=HOURS_IN_YEAR)#.sum(axis=1) , use_key_for_columns=False, frequency="HOURLY"
cooling = qn_cosim_eplus.to_frame('Zone Total Cooling Energy', index=HOURS_IN_YEAR)#.sum(axis=1) , use_key_for_columns=False
df = pd.DataFrame({'Co-Simulation EnergyPlus Heating': heating.sum(axis=1),
'Co-Simulation EnergyPlus Cooling': cooling.sum(axis=1)})
df_cosim_eplus = df*0.000000277777778
df_cosim_eplus.head()
```
# Get E+ Solo Data
```
# this is the output of the workflow
ESO_PATH = os.path.join(PROJECT_PATH, 'nord_only_EP', 'eplusout.eso')
#eplus_solo.to_frame("Zone Ideal Loads Supply Air Total Cooling Energy", frequency="HOURLY")
eplus_solo = esoreader.read_from_path(ESO_PATH)
heating = eplus_solo.to_frame('Zone Ideal Loads Supply Air Total Heating Energy', index=HOURS_IN_YEAR, frequency="HOURLY")#.sum(axis=1) use_key_for_columns=False,
cooling = eplus_solo.to_frame('Zone Ideal Loads Supply Air Total Cooling Energy', index=HOURS_IN_YEAR, frequency="HOURLY")#.sum(axis=1) use_key_for_columns=False,
df = pd.DataFrame({'Solo EnergyPlus Heating': heating.sum(axis=1),
'Solo EnergyPlus Cooling': cooling.sum(axis=1)})
df_solo_eplus = df*0.000000277777778
energyplus = pd.merge(df_solo_eplus, df_cosim_eplus, right_index=True, left_index=True, how='outer')
energyplus = energyplus[['Solo EnergyPlus Cooling','Co-Simulation EnergyPlus Cooling','Solo EnergyPlus Heating','Co-Simulation EnergyPlus Heating']]
energyplus.head()
```
# Get CitySim Data
```
def get_citysimdata_solo(path,key):
varlist = []
hours_in_year = pd.date_range('2013-01-01', '2013-12-31 T23:00', freq='H')
with open(path[1], 'r') as infile:
reader = csv.DictReader(infile, dialect=csv.Sniffer().sniff(infile.read(1000)))
infile.seek(0)
for row in reader:
varlist.append(float(row[key]))
#colname = key[0]+" "+path[0]
df = pd.DataFrame({key:pd.Series(varlist,index=hours_in_year)})
#outputdf = pd.DataFrame({path[0]+" Heating":df[df<0], path[0]+" Cooling":df[df>0]})
outputdf = pd.merge(np.abs(df[df<0]), df[df>0], right_index=True, left_index=True)
outputdf = outputdf.fillna(0)/1000
outputdf.columns = [path[0]+" Cooling",path[0]+" Heating"]
return outputdf
def get_citysimdata_cosim(path,key):
varlist = []
hours_in_year = pd.date_range('2013-01-01', '2013-12-31 T23:00', freq='H')
with open(path[1], 'r') as infile:
reader = csv.DictReader(infile, dialect=csv.Sniffer().sniff(infile.read(1000)))
infile.seek(0)
for row in reader:
varlist.append(float(row[key]))
#colname = key[0]+" "+path[0]
df = pd.DataFrame({key:pd.Series(varlist,index=hours_in_year)})
#outputdf = pd.DataFrame({path[0]+" Heating":df[df<0], path[0]+" Cooling":df[df>0]})
# outputdf = pd.merge(np.abs(df[df<0]), df[df>0], right_index=True, left_index=True)
# outputdf = outputdf.fillna(0)/1000
# outputdf.columns = [path[0]+" Cooling",path[0]+" Heating"]
return np.abs(df)/1000
citysim_solo = get_citysimdata_solo(['Solo CitySim',r'/Users/Clayton/Dropbox/03-ETH/UMEM/Quartier_Nord_EPFL/nord_only_CitySim/nord_only_CitySim_TH.out'], '1(1):1:Qs(Wh)')
# citysim_cosim3 = get_citysimdata(['Co-Simulation CitySim',r'/Users/Clayton/Dropbox/UMEM/Quartier_Nord_EPFL/cosimulation/quartier_nord_TH.out'], '3(3):3:Qs(Wh)')
# citysim_cosim1 = get_citysimdata(['Co-Simulation CitySim',r'/Users/Clayton/Dropbox/UMEM/Quartier_Nord_EPFL/cosimulation/quartier_nord_TH.out'], '1(1):1:Qs(Wh)')
# citysim_cosim4 = get_citysimdata(['Co-Simulation CitySim',r'/Users/Clayton/Dropbox/UMEM/Quartier_Nord_EPFL/cosimulation/quartier_nord_TH.out'], '4(4):4:Qs(Wh)')
# citysim_cosim5 = get_citysimdata(['Co-Simulation CitySim',r'/Users/Clayton/Dropbox/UMEM/Quartier_Nord_EPFL/cosimulation/quartier_nord_TH.out'], '5(5):5:Qs(Wh)')
# citysim_cosim1.plot(figsize=(15,2),title='CoSim 1(1):1:Qs(Wh)')
# citysim_cosim3.plot(figsize=(15,2),title='CoSim 3(3):3:Qs(Wh)')
# citysim_cosim4.plot(figsize=(15,2),title='CoSim 4(4):4:Qs(Wh)')
# citysim_cosim5.plot(figsize=(15,2),title='CoSim 5(5):5:Qs(Wh)')
# citysim_solo.plot(figsize=(15,3))
citysim_cosim_heating = get_citysimdata_cosim(['Co-Simulation CitySim',r'/Users/Clayton/Dropbox/03-ETH/UMEM/Quartier_Nord_EPFL/cosimulation/quartier_nord_TH.out'], '1(1):1:Heating(Wh)')
citysim_cosim_cooling = get_citysimdata_cosim(['Co-Simulation CitySim',r'/Users/Clayton/Dropbox/03-ETH/UMEM/Quartier_Nord_EPFL/cosimulation/quartier_nord_TH.out'], '1(1):1:Cooling(Wh)')
citysim_cosim_heating.columns = ["Co-Simulation CitySim Heating"]
citysim_cosim_cooling.columns = ["Co-Simulation CitySim Cooling"]
citysim_cosim = pd.merge(citysim_cosim_cooling, citysim_cosim_heating, right_index=True, left_index=True, how='outer')
#citysim_cosim
city_sim = pd.merge(citysim_cosim, citysim_solo, right_index=True, left_index=True)
city_sim.columns
city_sim = city_sim[['Solo CitySim Cooling','Co-Simulation CitySim Cooling','Solo CitySim Heating','Co-Simulation CitySim Heating']]
```
# Plot comparisons
```
def plot_comparison(df, colorpalette, filename, title, colabbrev, ylabel1, ylabel2):
current_palette = sns.color_palette(colorpalette, 4)
sns.set_palette(current_palette)
fig1 = plt.figure(figsize=[15,3])
fig1.suptitle(title, fontsize=14,y=1.02)
gs = GridSpec(100,100,bottom=0.18,left=0.18,right=0.88)
xfmt = ScalarFormatter()
xfmt.set_powerlimits((-3,3))
ax1 = fig1.add_subplot(gs[:,0:72])
styles1 = ['s-','^-','<-','-H']
df_toplot = df.resample("W",how='sum')
df_toplot.columns = colabbrev
df_toplot.plot(style=styles1, ax=ax1)
ax1.set_ylabel(ylabel1)
gca().yaxis.set_major_formatter(xfmt)
df_bar_combined = df.resample('A',how='sum')
df_bar_combined.columns = colabbrev
ax3 = fig1.add_subplot(gs[:,80:100])
gca().yaxis.set_major_formatter(xfmt)
df_bar_combined.T.plot(kind='bar',color=current_palette, legend=False, ax=ax3)
ax3.set_ylabel(ylabel2)
ax3.set_xticklabels(ax3.xaxis.get_majorticklabels(), rotation=30)
for tick in ax3.xaxis.get_majorticklabels():
tick.set_horizontalalignment("right")
plt.subplots_adjust(bottom=0.5)
plt.savefig("/Users/Clayton/umem-jbps-paper/latex_thirdsubmission_corrected/figures/"+filename,bbox_inches='tight')
plt.show()
```
# All CitySim -- this plot is redundant with the four plots below
```
plot_comparison(city_sim, "hls", "QN_CitySim.pdf", "EPFL Quartier Nord - CitySim CoSim and Solo Heating and Cooling",["Solo Cool","CoSim Cool","Solo Heat","CoSim Heat"], "Weekly Heating/Cooling [kWh]","Annual Heating/Cooling [kWh]")
plot_comparison(energyplus, "Set2", "QN_EnergyPlus.pdf", "EPFL Quartier Nord - EnergyPlus CoSim and Solo Heating and Cooling",["Solo Cool","CoSim Cool","Solo Heat","CoSim Heat"], "Weekly Heating/Cooling [kWh]","Annual Heating/Cooling [kWh]")
```
# EnergyPlus vs CitySim
```
def plot_comparison_2(df, colorpalette, filename, title, colabbrev, ylabel1, ylabel2):
current_palette = sns.color_palette(colorpalette, 4)
sns.set_palette(current_palette)
fig1 = plt.figure(figsize=[15,3])
fig1.suptitle(title, fontsize=14,y=1.02)
gs = GridSpec(100,100,bottom=0.18,left=0.18,right=0.88)
xfmt = ScalarFormatter()
xfmt.set_powerlimits((-3,3))
ax1 = fig1.add_subplot(gs[:,0:64])
styles1 = ['s-','^-','<-','-H']
df_toplot = df.resample("W",how='sum')
df_toplot.columns = colabbrev
df_toplot.plot(style=styles1, ax=ax1)
ax1.set_ylabel(ylabel1)
gca().yaxis.set_major_formatter(xfmt)
df_bar_combined = df.resample('A',how='sum')
df_bar_combined.columns = colabbrev
df_bar_solo = df_bar_combined[["CitySim Solo","Eplus Solo"]]
ax2 = fig1.add_subplot(gs[:,70:82])
gca().yaxis.set_major_formatter(xfmt)
df_bar_solo.T.plot(kind='bar',color=current_palette, legend=False, ax=ax2)
ax2.set_ylabel(ylabel2)
ax2.set_xticklabels(ax2.xaxis.get_majorticklabels(), rotation=30)
for tick in ax2.xaxis.get_majorticklabels():
tick.set_horizontalalignment("right")
diff = float((df_bar_solo.ix[:,1] - df_bar_solo.ix[:,0])/df_bar_solo.ix[:,0])
if diff >= 0:
annotation = '+{:.1%}'.format(diff)
else:
annotation = '{:.1%}'.format(diff)
# delta = int(df_bar_solo.ix[:,1] - df_bar_solo.ix[:,0])
# if delta >= 0:
# annotation_delta = '+{:,}'.format(delta)
# else:
# annotation_delta = '{:,}'.format(delta)
ax2.annotate(annotation, xy=(0, 10000), xytext=(0.7, df_bar_solo.ix[:,1][0]+(df_bar_solo.max().max() / 30)))
# ax2.annotate(annotation_delta, xy=(0, 10000), xytext=(0.5, df_bar_solo.ix[:,1][0]+(df_bar_solo.max().max() / 8)))
plt.ylim([0,df_bar_solo.max().max()+df_bar_solo.max().max()*.2])
df_bar_cosim = df_bar_combined[["CitySim CoSim","EPlus CoSim"]]
ax3 = fig1.add_subplot(gs[:,88:100])
gca().yaxis.set_major_formatter(xfmt)
df_bar_cosim.T.plot(kind='bar',color=current_palette[2:], legend=False, ax=ax3)
ax3.set_ylabel(ylabel2)
ax3.set_xticklabels(ax3.xaxis.get_majorticklabels(), rotation=30)
for tick in ax3.xaxis.get_majorticklabels():
tick.set_horizontalalignment("right")
diff = float((df_bar_cosim.ix[:,1] - df_bar_cosim.ix[:,0])/df_bar_cosim.ix[:,0])
if diff >= 0:
annotation = '+{:.1%}'.format(diff)
else:
annotation = '{:.1%}'.format(diff)
# delta = int(df_bar_cosim.ix[:,1] - df_bar_cosim.ix[:,0])
# if delta >= 0:
# annotation_delta = '+{:,}'.format(delta)
# else:
# annotation_delta = '{:,}'.format(delta)
ax3.annotate(annotation, xy=(0, 10000), xytext=(0.7, df_bar_cosim.ix[:,1][0]+(df_bar_cosim.max().max() / 30)))
# ax3.annotate(annotation_delta, xy=(0, 10000), xytext=(0.6, df_bar_cosim.ix[:,1][0]+(df_bar_cosim.max().max() / 8)))
plt.ylim([0,df_bar_cosim.max().max()+df_bar_cosim.max().max()*.2])
plt.subplots_adjust(bottom=0.5)
plt.savefig("/Users/Clayton/umem-jbps-paper/latex_thirdsubmission_corrected/figures/"+filename,bbox_inches='tight')
plt.show()
allheating = pd.merge(energyplus[["Co-Simulation EnergyPlus Heating","Solo EnergyPlus Heating"]], city_sim[["Co-Simulation CitySim Heating","Solo CitySim Heating"]],right_index=True, left_index=True)
allheating.head()
allheating = allheating[["Solo CitySim Heating","Solo EnergyPlus Heating","Co-Simulation CitySim Heating","Co-Simulation EnergyPlus Heating"]]
allheating.head()
plot_comparison_2(allheating, "spring", "QN_Heating.pdf", "EPFL Quartier Nord - EnergyPlus and CitySim CoSim and Solo Heating",["CitySim Solo","Eplus Solo","CitySim CoSim","EPlus CoSim"],"Weekly Heating [kWh]","Annual Heating [kWh]")
allcooling = pd.merge(energyplus[["Co-Simulation EnergyPlus Cooling","Solo EnergyPlus Cooling"]], city_sim[["Co-Simulation CitySim Cooling","Solo CitySim Cooling"]],right_index=True, left_index=True)
allcooling = allcooling[["Solo CitySim Cooling","Solo EnergyPlus Cooling","Co-Simulation CitySim Cooling","Co-Simulation EnergyPlus Cooling"]]
plot_comparison_2(allcooling, "winter", "QN_Cooling.pdf", "EPFL Quartier Nord - EnergyPlus and CitySim CoSim and Solo Cooling",["CitySim Solo","Eplus Solo","CitySim CoSim","EPlus CoSim"], "Weekly Cooling [kWh]","Annual Cooling [kWh]")
```
# Make the heating and cooling co-sim vs. solo charts
```
city_sim_annual = city_sim.resample('A',how='sum')
city_sim.columns
def pivot_day(df, cols, startdate, enddate):
df_forpiv = df.truncate(before=startdate, after=enddate).resample('2H', how='mean')
#df_cooling_forpiv.columns = ["1","2","3","4"]
df_forpiv['Date'] = df_forpiv.index.map(lambda t: t.date())
df_forpiv['Time'] = df_forpiv.index.map(lambda t: t.time())
df_pivot_1 = pd.pivot_table(df_forpiv, values=cols[0], index='Date', columns='Time')
df_pivot_2 = pd.pivot_table(df_forpiv, values=cols[1], index='Date', columns='Time')
# df_pivot_3 = pd.pivot_table(df_forpiv, values=cols[2], index='Date', columns='Time')
# df_pivot_4 = pd.pivot_table(df_forpiv, values=cols[3], index='Date', columns='Time')
df_pivot_sum = pd.DataFrame()
df_pivot_sum[cols[0]] = df_pivot_1.mean()#.plot(figsize=(20,8))
df_pivot_sum[cols[1]] = df_pivot_2.mean()#.plot()
# df_pivot_sum[cols[2]] = df_pivot_3.mean()#.plot()
# df_pivot_sum[cols[3]] = df_pivot_4.mean()#.plot()
return df_pivot_sum
def plot_cosim_vs_solo(df, df_pivot, colorpalette, ylabel1, ylabel2, ylabel3, filename, title, colnames):
current_palette = sns.color_palette(colorpalette, 2)
sns.set_palette(current_palette)
fig1 = plt.figure(figsize=[15,3])
fig1.suptitle(title, fontsize=14, y=1.03)
gs = GridSpec(100,100,bottom=0.18,left=0.18,right=0.88)
xfmt = ScalarFormatter()
xfmt.set_powerlimits((-3,3))
ax1 = fig1.add_subplot(gs[:,1:35])
styles1 = ['s-','^-','<-']
df.columns = colnames
df_pivot.columns = colnames
df_pivot.plot(style=styles1, ax=ax1, xticks=arange(0, 86400, 21600))
ax1.set_ylabel(ylabel1)
ax1.set_xlabel("Time of Day")
gca().yaxis.set_major_formatter(xfmt)
ax2 = fig1.add_subplot(gs[:,41:75])
styles1 = ['s-','^-','<-']
df.resample('M',how='sum').plot(style=styles1, ax=ax2)#.legend(loc='center left', bbox_to_anchor=(0, -0.5),), title="Monthly Total"
ax2.set_ylabel(ylabel2)
ax2.set_xlabel("Months of Year")
gca().yaxis.set_major_formatter(xfmt)
# df_netradiation.resample('M',how='sum').plot(style=styles1, ax=ax2, title="Monthly Total All Surfaces")#.legend(loc='center left', bbox_to_anchor=(0, -0.5),)
# ax2.set_ylabel("Monthly Net Thermal Radiation [J]")
# ax.yaxis.set_major_formatter(mtick.FormatStrFormatter('%.e'));
df_bar = df.resample('A',how='sum')
#df_bar.columns = ["CoSim","Solo"] #,"3","4"
ax3 = fig1.add_subplot(gs[:,81:100])
df_bar.T.plot(kind='bar',color=current_palette, legend=False, ax=ax3) #, title="Annual Total"
ax3.set_ylabel(ylabel3)
gca().yaxis.set_major_formatter(xfmt)
diff = float((df_bar.ix[:,1] - df_bar.ix[:,0])/df_bar.ix[:,0])
if diff >= 0:
annotation = '+{:.1%}'.format(diff)
else:
annotation = '{:.1%}'.format(diff)
# delta = int(df_bar.ix[:,1] - df_bar.ix[:,0])
# if delta >= 0:
# annotation_delta = '+{:,} kWh'.format(delta)
# else:
# annotation_delta = '{:,} kWh'.format(delta)
ax3.annotate(annotation, xy=(0, 10000), xytext=(0.8, df_bar["CoSim"][0]+(df_bar.max().max() / 30)))
# ax3.annotate(annotation_delta, xy=(0, 10000), xytext=(0.5, df_bar["CoSim"][0]+(df_bar.max().max() / 8)))
plt.ylim([0,df_bar.max().max()+df_bar.max().max()*.2])
# ax3.annotate(Exp2Annontate, xy=(0, 8000), xytext=(0.7, df_annual["2_C_NS"][0]+400))
# ax3.annotate(Exp3Annontate, xy=(0, 8000), xytext=(1.7, df_annual["3_S_WS"][0]+400))
# ax3.annotate(Exp4Annontate, xy=(0, 8000), xytext=(2.7, df_annual["4_C_WS"][0]+400))
# ax3.annotate(Exp2Annontate2, xy=(0, 8000), xytext=(0.8, df_annual["2_C_NS"][0]+3500))
# ax3.annotate(Exp3Annontate2, xy=(0, 8000), xytext=(1.8, df_annual["3_S_WS"][0]+3500))
# ax3.annotate(Exp4Annontate2, xy=(0, 8000), xytext=(2.8, df_annual["4_C_WS"][0]+3500))
# ax2.set_xticklabels(colnames, rotation=80)
plt.subplots_adjust(bottom=0.5)
# plt.tight_layout()
plt.savefig("/Users/Clayton/umem-jbps-paper/latex_thirdsubmission_corrected/figures/"+filename,bbox_inches='tight')
plt.show()
city_sim_cooling = city_sim[['Solo CitySim Cooling','Co-Simulation CitySim Cooling']]
city_sim_cooling_pivot = pivot_day(city_sim_cooling, list(city_sim_cooling.columns), '2013-07-01','2013-07-31')
plot_cosim_vs_solo(city_sim_cooling, city_sim_cooling_pivot, "winter", "July Avg. Daily Cooling [kWh]", "Monthly Cooling [kWh]", "Annual Cooling [kWh]", "QN_CitySim_Cooling.pdf","EPFL Quartier Nord CitySim CoSim vs Solo Cooling",["Solo","CoSim"])
city_sim_heating = city_sim[['Solo CitySim Heating','Co-Simulation CitySim Heating']]
city_sim_heating_pivot = pivot_day(city_sim_heating, list(city_sim_heating.columns), '2013-01-01','2013-01-31')
plot_cosim_vs_solo(city_sim_heating, city_sim_heating_pivot, "autumn", "Jan. Avg. Daily Heating [kWh]", "Monthly Heating [kWh]", "Annual Heating [kWh]", "QN_CitySim_Heating.pdf","EPFL Quartier Nord CitySim CoSim vs Solo Heating",["Solo","CoSim"])
energyplus_cooling = energyplus[['Solo EnergyPlus Cooling','Co-Simulation EnergyPlus Cooling']]
energyplus_cooling_pivot = pivot_day(energyplus_cooling, list(energyplus_cooling.columns), '2013-07-01','2013-07-31')
plot_cosim_vs_solo(energyplus_cooling, energyplus_cooling_pivot, "summer", "July Avg. Daily Cooling [kWh]", "Monthly Cooling [kWh]", "Annual Cooling [kWh]", "QN_EnergyPlus_Cooling.pdf","EPFL Quartier Nord EnergyPlus CoSim vs Solo Cooling",["Solo","CoSim"])
energyplus_heating = energyplus[['Solo EnergyPlus Heating','Co-Simulation EnergyPlus Heating']]
energyplus_heating_pivot = pivot_day(energyplus_heating, list(energyplus_heating.columns), '2013-01-01','2013-01-31')
plot_cosim_vs_solo(energyplus_heating, energyplus_heating_pivot, "spring", "Jan. Avg. Daily Heating [kWh]", "Monthly Heating [kWh]", "Annual Heating [kWh]", "QN_EnergyPlus_Heating.pdf","EPFL Quartier Nord EnergyPlus CoSim vs Solo Heating",["Solo","CoSim"])
```
|
github_jupyter
|
%matplotlib inline
import esoreader
reload(esoreader)
import pandas as pd
import os
import seaborn as sns
import matplotlib.pyplot as plt
from pylab import *
import csv
from __future__ import division
sns.set_style("whitegrid")
# change this to point to where you checked out the GitHub project
PROJECT_PATH = r"/Users/Clayton/Dropbox/03-ETH/UMEM/Quartier_Nord_EPFL/"
# this is the output of the workflow
ESO_PATH = os.path.join(PROJECT_PATH,'cosimulation','quartier_nord.eso')
# yeah... we need an index for timeseries...
HOURS_IN_YEAR = pd.date_range('2013-01-01', '2013-12-31 T23:00', freq='H')
qn_cosim_eplus = esoreader.read_from_path(ESO_PATH)
Varlist = pd.DataFrame(qn_cosim_eplus.dd.variables)
Varlist = Varlist.T
uniquevarlist = Varlist#[2].unique()
qn_cosim_eplus.find_variable("Zone Ideal Loads")
#qn_cosim_eplus.to_frame("Zone Total Heating Energy")
heating = qn_cosim_eplus.to_frame('Zone Total Heating Energy', index=HOURS_IN_YEAR)#.sum(axis=1) , use_key_for_columns=False, frequency="HOURLY"
cooling = qn_cosim_eplus.to_frame('Zone Total Cooling Energy', index=HOURS_IN_YEAR)#.sum(axis=1) , use_key_for_columns=False
df = pd.DataFrame({'Co-Simulation EnergyPlus Heating': heating.sum(axis=1),
'Co-Simulation EnergyPlus Cooling': cooling.sum(axis=1)})
df_cosim_eplus = df*0.000000277777778
df_cosim_eplus.head()
# this is the output of the workflow
ESO_PATH = os.path.join(PROJECT_PATH, 'nord_only_EP', 'eplusout.eso')
#eplus_solo.to_frame("Zone Ideal Loads Supply Air Total Cooling Energy", frequency="HOURLY")
eplus_solo = esoreader.read_from_path(ESO_PATH)
heating = eplus_solo.to_frame('Zone Ideal Loads Supply Air Total Heating Energy', index=HOURS_IN_YEAR, frequency="HOURLY")#.sum(axis=1) use_key_for_columns=False,
cooling = eplus_solo.to_frame('Zone Ideal Loads Supply Air Total Cooling Energy', index=HOURS_IN_YEAR, frequency="HOURLY")#.sum(axis=1) use_key_for_columns=False,
df = pd.DataFrame({'Solo EnergyPlus Heating': heating.sum(axis=1),
'Solo EnergyPlus Cooling': cooling.sum(axis=1)})
df_solo_eplus = df*0.000000277777778
energyplus = pd.merge(df_solo_eplus, df_cosim_eplus, right_index=True, left_index=True, how='outer')
energyplus = energyplus[['Solo EnergyPlus Cooling','Co-Simulation EnergyPlus Cooling','Solo EnergyPlus Heating','Co-Simulation EnergyPlus Heating']]
energyplus.head()
def get_citysimdata_solo(path,key):
varlist = []
hours_in_year = pd.date_range('2013-01-01', '2013-12-31 T23:00', freq='H')
with open(path[1], 'r') as infile:
reader = csv.DictReader(infile, dialect=csv.Sniffer().sniff(infile.read(1000)))
infile.seek(0)
for row in reader:
varlist.append(float(row[key]))
#colname = key[0]+" "+path[0]
df = pd.DataFrame({key:pd.Series(varlist,index=hours_in_year)})
#outputdf = pd.DataFrame({path[0]+" Heating":df[df<0], path[0]+" Cooling":df[df>0]})
outputdf = pd.merge(np.abs(df[df<0]), df[df>0], right_index=True, left_index=True)
outputdf = outputdf.fillna(0)/1000
outputdf.columns = [path[0]+" Cooling",path[0]+" Heating"]
return outputdf
def get_citysimdata_cosim(path,key):
varlist = []
hours_in_year = pd.date_range('2013-01-01', '2013-12-31 T23:00', freq='H')
with open(path[1], 'r') as infile:
reader = csv.DictReader(infile, dialect=csv.Sniffer().sniff(infile.read(1000)))
infile.seek(0)
for row in reader:
varlist.append(float(row[key]))
#colname = key[0]+" "+path[0]
df = pd.DataFrame({key:pd.Series(varlist,index=hours_in_year)})
#outputdf = pd.DataFrame({path[0]+" Heating":df[df<0], path[0]+" Cooling":df[df>0]})
# outputdf = pd.merge(np.abs(df[df<0]), df[df>0], right_index=True, left_index=True)
# outputdf = outputdf.fillna(0)/1000
# outputdf.columns = [path[0]+" Cooling",path[0]+" Heating"]
return np.abs(df)/1000
citysim_solo = get_citysimdata_solo(['Solo CitySim',r'/Users/Clayton/Dropbox/03-ETH/UMEM/Quartier_Nord_EPFL/nord_only_CitySim/nord_only_CitySim_TH.out'], '1(1):1:Qs(Wh)')
# citysim_cosim3 = get_citysimdata(['Co-Simulation CitySim',r'/Users/Clayton/Dropbox/UMEM/Quartier_Nord_EPFL/cosimulation/quartier_nord_TH.out'], '3(3):3:Qs(Wh)')
# citysim_cosim1 = get_citysimdata(['Co-Simulation CitySim',r'/Users/Clayton/Dropbox/UMEM/Quartier_Nord_EPFL/cosimulation/quartier_nord_TH.out'], '1(1):1:Qs(Wh)')
# citysim_cosim4 = get_citysimdata(['Co-Simulation CitySim',r'/Users/Clayton/Dropbox/UMEM/Quartier_Nord_EPFL/cosimulation/quartier_nord_TH.out'], '4(4):4:Qs(Wh)')
# citysim_cosim5 = get_citysimdata(['Co-Simulation CitySim',r'/Users/Clayton/Dropbox/UMEM/Quartier_Nord_EPFL/cosimulation/quartier_nord_TH.out'], '5(5):5:Qs(Wh)')
# citysim_cosim1.plot(figsize=(15,2),title='CoSim 1(1):1:Qs(Wh)')
# citysim_cosim3.plot(figsize=(15,2),title='CoSim 3(3):3:Qs(Wh)')
# citysim_cosim4.plot(figsize=(15,2),title='CoSim 4(4):4:Qs(Wh)')
# citysim_cosim5.plot(figsize=(15,2),title='CoSim 5(5):5:Qs(Wh)')
# citysim_solo.plot(figsize=(15,3))
citysim_cosim_heating = get_citysimdata_cosim(['Co-Simulation CitySim',r'/Users/Clayton/Dropbox/03-ETH/UMEM/Quartier_Nord_EPFL/cosimulation/quartier_nord_TH.out'], '1(1):1:Heating(Wh)')
citysim_cosim_cooling = get_citysimdata_cosim(['Co-Simulation CitySim',r'/Users/Clayton/Dropbox/03-ETH/UMEM/Quartier_Nord_EPFL/cosimulation/quartier_nord_TH.out'], '1(1):1:Cooling(Wh)')
citysim_cosim_heating.columns = ["Co-Simulation CitySim Heating"]
citysim_cosim_cooling.columns = ["Co-Simulation CitySim Cooling"]
citysim_cosim = pd.merge(citysim_cosim_cooling, citysim_cosim_heating, right_index=True, left_index=True, how='outer')
#citysim_cosim
city_sim = pd.merge(citysim_cosim, citysim_solo, right_index=True, left_index=True)
city_sim.columns
city_sim = city_sim[['Solo CitySim Cooling','Co-Simulation CitySim Cooling','Solo CitySim Heating','Co-Simulation CitySim Heating']]
def plot_comparison(df, colorpalette, filename, title, colabbrev, ylabel1, ylabel2):
current_palette = sns.color_palette(colorpalette, 4)
sns.set_palette(current_palette)
fig1 = plt.figure(figsize=[15,3])
fig1.suptitle(title, fontsize=14,y=1.02)
gs = GridSpec(100,100,bottom=0.18,left=0.18,right=0.88)
xfmt = ScalarFormatter()
xfmt.set_powerlimits((-3,3))
ax1 = fig1.add_subplot(gs[:,0:72])
styles1 = ['s-','^-','<-','-H']
df_toplot = df.resample("W",how='sum')
df_toplot.columns = colabbrev
df_toplot.plot(style=styles1, ax=ax1)
ax1.set_ylabel(ylabel1)
gca().yaxis.set_major_formatter(xfmt)
df_bar_combined = df.resample('A',how='sum')
df_bar_combined.columns = colabbrev
ax3 = fig1.add_subplot(gs[:,80:100])
gca().yaxis.set_major_formatter(xfmt)
df_bar_combined.T.plot(kind='bar',color=current_palette, legend=False, ax=ax3)
ax3.set_ylabel(ylabel2)
ax3.set_xticklabels(ax3.xaxis.get_majorticklabels(), rotation=30)
for tick in ax3.xaxis.get_majorticklabels():
tick.set_horizontalalignment("right")
plt.subplots_adjust(bottom=0.5)
plt.savefig("/Users/Clayton/umem-jbps-paper/latex_thirdsubmission_corrected/figures/"+filename,bbox_inches='tight')
plt.show()
plot_comparison(city_sim, "hls", "QN_CitySim.pdf", "EPFL Quartier Nord - CitySim CoSim and Solo Heating and Cooling",["Solo Cool","CoSim Cool","Solo Heat","CoSim Heat"], "Weekly Heating/Cooling [kWh]","Annual Heating/Cooling [kWh]")
plot_comparison(energyplus, "Set2", "QN_EnergyPlus.pdf", "EPFL Quartier Nord - EnergyPlus CoSim and Solo Heating and Cooling",["Solo Cool","CoSim Cool","Solo Heat","CoSim Heat"], "Weekly Heating/Cooling [kWh]","Annual Heating/Cooling [kWh]")
def plot_comparison_2(df, colorpalette, filename, title, colabbrev, ylabel1, ylabel2):
current_palette = sns.color_palette(colorpalette, 4)
sns.set_palette(current_palette)
fig1 = plt.figure(figsize=[15,3])
fig1.suptitle(title, fontsize=14,y=1.02)
gs = GridSpec(100,100,bottom=0.18,left=0.18,right=0.88)
xfmt = ScalarFormatter()
xfmt.set_powerlimits((-3,3))
ax1 = fig1.add_subplot(gs[:,0:64])
styles1 = ['s-','^-','<-','-H']
df_toplot = df.resample("W",how='sum')
df_toplot.columns = colabbrev
df_toplot.plot(style=styles1, ax=ax1)
ax1.set_ylabel(ylabel1)
gca().yaxis.set_major_formatter(xfmt)
df_bar_combined = df.resample('A',how='sum')
df_bar_combined.columns = colabbrev
df_bar_solo = df_bar_combined[["CitySim Solo","Eplus Solo"]]
ax2 = fig1.add_subplot(gs[:,70:82])
gca().yaxis.set_major_formatter(xfmt)
df_bar_solo.T.plot(kind='bar',color=current_palette, legend=False, ax=ax2)
ax2.set_ylabel(ylabel2)
ax2.set_xticklabels(ax2.xaxis.get_majorticklabels(), rotation=30)
for tick in ax2.xaxis.get_majorticklabels():
tick.set_horizontalalignment("right")
diff = float((df_bar_solo.ix[:,1] - df_bar_solo.ix[:,0])/df_bar_solo.ix[:,0])
if diff >= 0:
annotation = '+{:.1%}'.format(diff)
else:
annotation = '{:.1%}'.format(diff)
# delta = int(df_bar_solo.ix[:,1] - df_bar_solo.ix[:,0])
# if delta >= 0:
# annotation_delta = '+{:,}'.format(delta)
# else:
# annotation_delta = '{:,}'.format(delta)
ax2.annotate(annotation, xy=(0, 10000), xytext=(0.7, df_bar_solo.ix[:,1][0]+(df_bar_solo.max().max() / 30)))
# ax2.annotate(annotation_delta, xy=(0, 10000), xytext=(0.5, df_bar_solo.ix[:,1][0]+(df_bar_solo.max().max() / 8)))
plt.ylim([0,df_bar_solo.max().max()+df_bar_solo.max().max()*.2])
df_bar_cosim = df_bar_combined[["CitySim CoSim","EPlus CoSim"]]
ax3 = fig1.add_subplot(gs[:,88:100])
gca().yaxis.set_major_formatter(xfmt)
df_bar_cosim.T.plot(kind='bar',color=current_palette[2:], legend=False, ax=ax3)
ax3.set_ylabel(ylabel2)
ax3.set_xticklabels(ax3.xaxis.get_majorticklabels(), rotation=30)
for tick in ax3.xaxis.get_majorticklabels():
tick.set_horizontalalignment("right")
diff = float((df_bar_cosim.ix[:,1] - df_bar_cosim.ix[:,0])/df_bar_cosim.ix[:,0])
if diff >= 0:
annotation = '+{:.1%}'.format(diff)
else:
annotation = '{:.1%}'.format(diff)
# delta = int(df_bar_cosim.ix[:,1] - df_bar_cosim.ix[:,0])
# if delta >= 0:
# annotation_delta = '+{:,}'.format(delta)
# else:
# annotation_delta = '{:,}'.format(delta)
ax3.annotate(annotation, xy=(0, 10000), xytext=(0.7, df_bar_cosim.ix[:,1][0]+(df_bar_cosim.max().max() / 30)))
# ax3.annotate(annotation_delta, xy=(0, 10000), xytext=(0.6, df_bar_cosim.ix[:,1][0]+(df_bar_cosim.max().max() / 8)))
plt.ylim([0,df_bar_cosim.max().max()+df_bar_cosim.max().max()*.2])
plt.subplots_adjust(bottom=0.5)
plt.savefig("/Users/Clayton/umem-jbps-paper/latex_thirdsubmission_corrected/figures/"+filename,bbox_inches='tight')
plt.show()
allheating = pd.merge(energyplus[["Co-Simulation EnergyPlus Heating","Solo EnergyPlus Heating"]], city_sim[["Co-Simulation CitySim Heating","Solo CitySim Heating"]],right_index=True, left_index=True)
allheating.head()
allheating = allheating[["Solo CitySim Heating","Solo EnergyPlus Heating","Co-Simulation CitySim Heating","Co-Simulation EnergyPlus Heating"]]
allheating.head()
plot_comparison_2(allheating, "spring", "QN_Heating.pdf", "EPFL Quartier Nord - EnergyPlus and CitySim CoSim and Solo Heating",["CitySim Solo","Eplus Solo","CitySim CoSim","EPlus CoSim"],"Weekly Heating [kWh]","Annual Heating [kWh]")
allcooling = pd.merge(energyplus[["Co-Simulation EnergyPlus Cooling","Solo EnergyPlus Cooling"]], city_sim[["Co-Simulation CitySim Cooling","Solo CitySim Cooling"]],right_index=True, left_index=True)
allcooling = allcooling[["Solo CitySim Cooling","Solo EnergyPlus Cooling","Co-Simulation CitySim Cooling","Co-Simulation EnergyPlus Cooling"]]
plot_comparison_2(allcooling, "winter", "QN_Cooling.pdf", "EPFL Quartier Nord - EnergyPlus and CitySim CoSim and Solo Cooling",["CitySim Solo","Eplus Solo","CitySim CoSim","EPlus CoSim"], "Weekly Cooling [kWh]","Annual Cooling [kWh]")
city_sim_annual = city_sim.resample('A',how='sum')
city_sim.columns
def pivot_day(df, cols, startdate, enddate):
df_forpiv = df.truncate(before=startdate, after=enddate).resample('2H', how='mean')
#df_cooling_forpiv.columns = ["1","2","3","4"]
df_forpiv['Date'] = df_forpiv.index.map(lambda t: t.date())
df_forpiv['Time'] = df_forpiv.index.map(lambda t: t.time())
df_pivot_1 = pd.pivot_table(df_forpiv, values=cols[0], index='Date', columns='Time')
df_pivot_2 = pd.pivot_table(df_forpiv, values=cols[1], index='Date', columns='Time')
# df_pivot_3 = pd.pivot_table(df_forpiv, values=cols[2], index='Date', columns='Time')
# df_pivot_4 = pd.pivot_table(df_forpiv, values=cols[3], index='Date', columns='Time')
df_pivot_sum = pd.DataFrame()
df_pivot_sum[cols[0]] = df_pivot_1.mean()#.plot(figsize=(20,8))
df_pivot_sum[cols[1]] = df_pivot_2.mean()#.plot()
# df_pivot_sum[cols[2]] = df_pivot_3.mean()#.plot()
# df_pivot_sum[cols[3]] = df_pivot_4.mean()#.plot()
return df_pivot_sum
def plot_cosim_vs_solo(df, df_pivot, colorpalette, ylabel1, ylabel2, ylabel3, filename, title, colnames):
current_palette = sns.color_palette(colorpalette, 2)
sns.set_palette(current_palette)
fig1 = plt.figure(figsize=[15,3])
fig1.suptitle(title, fontsize=14, y=1.03)
gs = GridSpec(100,100,bottom=0.18,left=0.18,right=0.88)
xfmt = ScalarFormatter()
xfmt.set_powerlimits((-3,3))
ax1 = fig1.add_subplot(gs[:,1:35])
styles1 = ['s-','^-','<-']
df.columns = colnames
df_pivot.columns = colnames
df_pivot.plot(style=styles1, ax=ax1, xticks=arange(0, 86400, 21600))
ax1.set_ylabel(ylabel1)
ax1.set_xlabel("Time of Day")
gca().yaxis.set_major_formatter(xfmt)
ax2 = fig1.add_subplot(gs[:,41:75])
styles1 = ['s-','^-','<-']
df.resample('M',how='sum').plot(style=styles1, ax=ax2)#.legend(loc='center left', bbox_to_anchor=(0, -0.5),), title="Monthly Total"
ax2.set_ylabel(ylabel2)
ax2.set_xlabel("Months of Year")
gca().yaxis.set_major_formatter(xfmt)
# df_netradiation.resample('M',how='sum').plot(style=styles1, ax=ax2, title="Monthly Total All Surfaces")#.legend(loc='center left', bbox_to_anchor=(0, -0.5),)
# ax2.set_ylabel("Monthly Net Thermal Radiation [J]")
# ax.yaxis.set_major_formatter(mtick.FormatStrFormatter('%.e'));
df_bar = df.resample('A',how='sum')
#df_bar.columns = ["CoSim","Solo"] #,"3","4"
ax3 = fig1.add_subplot(gs[:,81:100])
df_bar.T.plot(kind='bar',color=current_palette, legend=False, ax=ax3) #, title="Annual Total"
ax3.set_ylabel(ylabel3)
gca().yaxis.set_major_formatter(xfmt)
diff = float((df_bar.ix[:,1] - df_bar.ix[:,0])/df_bar.ix[:,0])
if diff >= 0:
annotation = '+{:.1%}'.format(diff)
else:
annotation = '{:.1%}'.format(diff)
# delta = int(df_bar.ix[:,1] - df_bar.ix[:,0])
# if delta >= 0:
# annotation_delta = '+{:,} kWh'.format(delta)
# else:
# annotation_delta = '{:,} kWh'.format(delta)
ax3.annotate(annotation, xy=(0, 10000), xytext=(0.8, df_bar["CoSim"][0]+(df_bar.max().max() / 30)))
# ax3.annotate(annotation_delta, xy=(0, 10000), xytext=(0.5, df_bar["CoSim"][0]+(df_bar.max().max() / 8)))
plt.ylim([0,df_bar.max().max()+df_bar.max().max()*.2])
# ax3.annotate(Exp2Annontate, xy=(0, 8000), xytext=(0.7, df_annual["2_C_NS"][0]+400))
# ax3.annotate(Exp3Annontate, xy=(0, 8000), xytext=(1.7, df_annual["3_S_WS"][0]+400))
# ax3.annotate(Exp4Annontate, xy=(0, 8000), xytext=(2.7, df_annual["4_C_WS"][0]+400))
# ax3.annotate(Exp2Annontate2, xy=(0, 8000), xytext=(0.8, df_annual["2_C_NS"][0]+3500))
# ax3.annotate(Exp3Annontate2, xy=(0, 8000), xytext=(1.8, df_annual["3_S_WS"][0]+3500))
# ax3.annotate(Exp4Annontate2, xy=(0, 8000), xytext=(2.8, df_annual["4_C_WS"][0]+3500))
# ax2.set_xticklabels(colnames, rotation=80)
plt.subplots_adjust(bottom=0.5)
# plt.tight_layout()
plt.savefig("/Users/Clayton/umem-jbps-paper/latex_thirdsubmission_corrected/figures/"+filename,bbox_inches='tight')
plt.show()
city_sim_cooling = city_sim[['Solo CitySim Cooling','Co-Simulation CitySim Cooling']]
city_sim_cooling_pivot = pivot_day(city_sim_cooling, list(city_sim_cooling.columns), '2013-07-01','2013-07-31')
plot_cosim_vs_solo(city_sim_cooling, city_sim_cooling_pivot, "winter", "July Avg. Daily Cooling [kWh]", "Monthly Cooling [kWh]", "Annual Cooling [kWh]", "QN_CitySim_Cooling.pdf","EPFL Quartier Nord CitySim CoSim vs Solo Cooling",["Solo","CoSim"])
city_sim_heating = city_sim[['Solo CitySim Heating','Co-Simulation CitySim Heating']]
city_sim_heating_pivot = pivot_day(city_sim_heating, list(city_sim_heating.columns), '2013-01-01','2013-01-31')
plot_cosim_vs_solo(city_sim_heating, city_sim_heating_pivot, "autumn", "Jan. Avg. Daily Heating [kWh]", "Monthly Heating [kWh]", "Annual Heating [kWh]", "QN_CitySim_Heating.pdf","EPFL Quartier Nord CitySim CoSim vs Solo Heating",["Solo","CoSim"])
energyplus_cooling = energyplus[['Solo EnergyPlus Cooling','Co-Simulation EnergyPlus Cooling']]
energyplus_cooling_pivot = pivot_day(energyplus_cooling, list(energyplus_cooling.columns), '2013-07-01','2013-07-31')
plot_cosim_vs_solo(energyplus_cooling, energyplus_cooling_pivot, "summer", "July Avg. Daily Cooling [kWh]", "Monthly Cooling [kWh]", "Annual Cooling [kWh]", "QN_EnergyPlus_Cooling.pdf","EPFL Quartier Nord EnergyPlus CoSim vs Solo Cooling",["Solo","CoSim"])
energyplus_heating = energyplus[['Solo EnergyPlus Heating','Co-Simulation EnergyPlus Heating']]
energyplus_heating_pivot = pivot_day(energyplus_heating, list(energyplus_heating.columns), '2013-01-01','2013-01-31')
plot_cosim_vs_solo(energyplus_heating, energyplus_heating_pivot, "spring", "Jan. Avg. Daily Heating [kWh]", "Monthly Heating [kWh]", "Annual Heating [kWh]", "QN_EnergyPlus_Heating.pdf","EPFL Quartier Nord EnergyPlus CoSim vs Solo Heating",["Solo","CoSim"])
| 0.284874 | 0.733679 |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.