code
stringlengths 2.5k
6.36M
| kind
stringclasses 2
values | parsed_code
stringlengths 0
404k
| quality_prob
float64 0
0.98
| learning_prob
float64 0.03
1
|
|---|---|---|---|---|
```
##importing libraries
import nltk
nltk.download('punkt')
nltk.download('wordnet')
from nltk.stem import WordNetLemmatizer
lemmatizer = WordNetLemmatizer()
import json
import pickle
import numpy as np
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Dense, Activation, Dropout
from tensorflow.keras.optimizers import SGD
import random
##initializing chatbot training
words=[]
classes = []
documents = []
ignore_words = ['?', '!','#','&','(',')','a','are']
data_file = open('dataset2.json').read()
intents = json.loads(data_file)
for intent in intents['intents']:
for pattern in intent['patterns']:
# take each word and tokenize it
w = nltk.word_tokenize(pattern)
words.extend(w)
# adding documents
documents.append((w, intent['tag']))
# adding classes to our class list
if intent['tag'] not in classes:
classes.append(intent['tag'])
words = [lemmatizer.lemmatize(w.lower()) for w in words if w not in ignore_words]
# sort in alphabetical order
words = sorted(list(set(words)))
classes = sorted(list(set(classes)))
print (len(documents), "documents")
print (len(classes), "classes", classes)
print (len(words), "unique lemmatized words", words)
# save them into pickle file
pickle.dump(words,open('dictionary.pkl','wb'))
pickle.dump(classes,open('pattern_classes.pkl','wb'))
##Building Model
training = []
output_empty = [0] * len(classes)
for doc in documents:
# initializing bag of words
bag = []
# list of tokenized words for the pattern
pattern_words = doc[0]
# lemmatize each word - create base word, in attempt to represent related words
pattern_words = [lemmatizer.lemmatize(word.lower()) for word in pattern_words]
# create our bag of words array with 1, if word match found in current pattern
for w in words:
bag.append(1) if w in pattern_words else bag.append(0)
# output is a '0' for each tag and '1' for current tag (for each pattern)
output_row = list(output_empty)
output_row[classes.index(doc[1])] = 1
training.append([bag, output_row])
# shuffle our features and turn into np.array
random.shuffle(training)
training = np.array(training)
# create train and test lists.
# X -> patterns, Y -> intents
train_x = list(training[:,0])
train_y = list(training[:,1])
```
### May not need to run the below cell, if chatbot_model.h5 already created
```
# Create model - 3 layers. First layer 128 neurons, second layer 64 neurons and 3rd output layer contains number of neurons
# equal to number of intents to predict output intent with softmax
model = Sequential()
model.add(Dense(128, input_shape=(len(train_x[0]),), activation='relu'))
model.add(Dropout(0.5))
model.add(Dense(64, activation='relu'))
model.add(Dropout(0.5))
# softmax get highest probability -> only 1 output
model.add(Dense(len(train_y[0]), activation='softmax'))
# Model optimization
# Compile model. SGD with Nesterov accelerated gradient gives good results for this model
sgd = SGD(lr=0.01, decay=1e-6, momentum=0.9, nesterov=True)
model.compile(loss='categorical_crossentropy', optimizer=sgd, metrics=['accuracy'])
#fitting and saving the model
hist = model.fit(np.array(train_x), np.array(train_y), epochs=35, batch_size=5, verbose=1)
model.save('chatbot_model.h5', hist)
print("model created")
#print("predict ",model.predict(train_x, batch_size =5, verbose =1))
#print("predict class",model.predict_classes(train_x, batch_size =5, verbose =1))
#print("predict proba",model.predict_proba(train_x, batch_size =5, verbose =1))
##Building Chatbot GUI
from keras.models import load_model
model = load_model('chatbot_model.h5')
import json
import random
intents = json.loads(open('dataset2.json').read())
words = pickle.load(open('dictionary.pkl','rb'))
classes = pickle.load(open('pattern_classes.pkl','rb'))
import re
QuestionFlag = "Havent ask"
specialcase = " "
category = "null"
Storage_msg = " "
Remember_msg = " "
questionAns = []
interest = ["Accounting", "Broadcasting", "Food Science", "Psychology", "Public Relations", "Business Administration","Human Resource Management","Information Technology","Computer Science","Engineering","Architecture","Quantity Surveying","Hotel Management","Culinary Arts","Advertising","Journalism","Finance","English"]
questionArr = ["Q1","Q2","Q3","Q4","Q5","Q6","Q7","Q8","Q9","Q10","Q11","Q12","Q13","Q14","Q15","Q16","Q17","Q18"]
eduLevel = 0
# Convert word number to integer number
def word_to_number(word):
from word2number import w2n
return w2n.word_to_num(word)
def clean_up_sentence(sentence):
global Remember_msg
global category
global specialcase
global QuestionFlag
global eduLevel
global questionAns
# Age - Education level e.g. Pre-U, Undergraduate, Postgraduate
if (QuestionFlag == "Asked"):
specialcase = "age"
QuestionFlag = "null"
if (sentence.isnumeric() == False):
sentence = word_to_number(sentence)
sentence = str(sentence)
x = re.findall("[0-5][0-9]", sentence)
print("Age: ",x[0])
if(int(x[0]) <= 17):
category = "too young"
elif(18 <= int(x[0]) <= 19 ):
category = "pre-u"
eduLevel = 1
elif(20 <= int(x[0]) <= 23 ):
category = "undergraduate"
eduLevel = 2
elif(int(x[0]) >= 24):
category = "postgraduate"
eduLevel = 3
# Run Questions
for x in range(len(interest)):
if (QuestionFlag == questionArr[x]):
specialcase = questionArr[x]
QuestionFlag = "null"
if (sentence.isnumeric()):
questionAns.append(int(sentence))
print(questionAns)
# All Questions Done, run GaussianNB, get the suitable course name
if (QuestionFlag == "finish question"):
QuestionFlag = "null"
specialcase = "finish question"
import pandas as pd
df = pd.read_csv('nbDatasets.csv')
cols = list(df.columns)
features = df[cols[1:-1]]
result = df["course"]
# Import LabelEncoder
from sklearn import preprocessing
#creating labelEncoder
le = preprocessing.LabelEncoder()
# Converting string labels into numbers.
result_encoded=le.fit_transform(result)
# print(result_encoded)
#Import Gaussian Naive Bayes model
from sklearn.naive_bayes import GaussianNB
#Create a Gaussian Classifier
model = GaussianNB()
# Train the model using the training sets
model.fit(features,result_encoded)
# clean questionAns list in second run of questionnaire
if len(questionAns) > 18:
del questionAns [:18]
#Predict Output
predicted= model.predict([questionAns])
transform = le.inverse_transform(predicted)
transform = transform[0]
Remember_msg += " " + transform
sentence = sentence + " " + Remember_msg
sentence_words = nltk.word_tokenize(sentence)
sentence_words = [lemmatizer.lemmatize(word.lower()) for word in sentence_words]
print(sentence_words)
return sentence_words
sentence = sentence + " " + Remember_msg
print(sentence)
sentence_words = nltk.word_tokenize(sentence)
sentence_words = [lemmatizer.lemmatize(word.lower()) for word in sentence_words]
print(sentence_words)
return sentence_words
def words_bag(sentence, words, show_details=True):
# tokenize the pattern
sentence_words = clean_up_sentence(sentence)
# bag of words - matrix of N words, vocabulary matrix
bag = [0]*len(words)
for sentenceWord in sentence_words:
for i,word in enumerate(words):
if word == sentenceWord:
# assign 1 if current word is in the vocabulary position
bag[i] = 1
if show_details:
print ("found in bag: %s" % word)
return(np.array(bag))
def predict_class(sentence, model):
p = words_bag(sentence, words,show_details=False)
res = model.predict(np.array([p]))[0]
ERROR_THRESHOLD = 0.25
# filter out predictions below a threshold
results = [[i,r] for i,r in enumerate(res) if r>ERROR_THRESHOLD]
# sort by strength of probability
# get the highest one only -> Only one output
results.sort(key=lambda x: x[1], reverse=True)
return_list = []
for r in results:
return_list.append({"intent": classes[r[0]], "probability": str(r[1])})
return return_list
def getResponse(ints, intents):
global specialcase
global Remember_msg
print("-------ints: " ,ints)
print("-------tags: ", ints[0]['intent'])
tag = ints[0]['intent']
list_of_intents = intents['intents']
for i in list_of_intents:
if(i['tag']== tag):
if (eduLevel == 1 or eduLevel == 2 or eduLevel == 3):
try:
result = i['responses'][eduLevel-1]
except:
result = random.choice(i['responses'])
else:
result = random.choice(i['responses'])
break
return result
def chatbot_response(msg):
global specialcase
global QuestionFlag
global questionAns
global Storage_msg
global Remember_msg
global eduLevel
ints = predict_class(msg, model)
#print("edu leveL: ", eduLevel)
res = getResponse(ints, intents)
# Get Age
if (res[-1] == '{'):
if (QuestionFlag == "Havent ask"):
QuestionFlag = "Asked"
Storage_msg = msg
res = "Oh before that, may i know your age please?"
else:
res = res[:-1]
# Run the likert scale questions
if (res[-1] == 'x'):
Remember_msg = " " #clear the Remember_msg
QuestionFlag = "Q1"
res = "Answer the question with scale 1 to 5, 1 - Most disagree, 2 - Disagree, 3 - Average, 4 - Agree, 5 - Most agree \nAre you a detail oriented, number sensitive, trustworthy, good in calculation person?"
if (specialcase != "null"):
if (specialcase == "age"):
# Age and education level responses
if (category == "too young"):
res += " but you are too young for now, come consult me when your are > 17 years old"
elif (category == "pre-u"):
res += " and based on your age, i bet you are a Pre University student, hence I only display Pre University Courses for you"
Remember_msg += " pre-u"
eduLevel = 1
elif (category == "undergraduate"):
res += " and based on your age, i bet you are an Undergraduate student, hence I only display Undergraduate Courses for you "
Remember_msg += " undergraduate"
eduLevel = 2
elif (category == "postgraduate"):
res += " and based on your age, i bet you are a Postgraduate student, hence I only display Postgraduate Courses for you "
Remember_msg += " postgraduate"
eduLevel = 3
# Ask Questions
if (specialcase == "Q1"):
QuestionFlag = "Q2"
res = "Are you interested in broadcasting e.g. filming, cinematography, production and multimedia?"
if (specialcase == "Q2"):
QuestionFlag = "Q3"
res = "Are you interested in chemistry, biology and composition of foods?"
if (specialcase == "Q3"):
QuestionFlag = "Q4"
res = "Are you interested in helping people to solve their problems patiently?"
if (specialcase == "Q4"):
QuestionFlag = "Q5"
res = "Are you interested in public relations which buidls mutually beneficial relationships between organization and their publics?"
if (specialcase == "Q5"):
QuestionFlag = "Q6"
res = "Are you interested in operating a business?"
if (specialcase == "Q6"):
QuestionFlag = "Q7"
res = "Are you interested in managing people where we called it as human resource management?"
if (specialcase == "Q7"):
QuestionFlag = "Q8"
res = "Are you interested in coding, programming, computer security or develop software?"
if (specialcase == "Q8"):
QuestionFlag = "Q9"
res = "Are you interested in analyze data, computer science or develop computer system application?"
if (specialcase == "Q9"):
QuestionFlag = "Q10"
res = "Are you interested in engineering e.g. electronic, mechanical, mechatronic and telecommunication?"
if (specialcase == "Q10"):
QuestionFlag = "Q11"
res = "Are you interested in interior design or architercture?"
if (specialcase == "Q11"):
QuestionFlag = "Q12"
res = "Are you interested in managing the costs and budgets of a project?"
if (specialcase == "Q12"):
QuestionFlag = "Q13"
res = "Are you interested in learning the technical skills in food and beverage, room management, managerial and personal skills?"
if (specialcase == "Q13"):
QuestionFlag = "Q14"
res = "Are you interested in cooking?"
if (specialcase == "Q14"):
QuestionFlag = "Q15"
res = "Are you interested in learning the skills of advertising, media and communication?"
if (specialcase == "Q15"):
QuestionFlag = "Q16"
res = "Are you interested in become a journalists, writers or reporters?"
if (specialcase == "Q16"):
QuestionFlag = "Q17"
res = "Are you interested in managing the financial assets?"
if (specialcase == "Q17"):
QuestionFlag = "Q18"
res = "Are you interested in english education, english drama or english studies?"
if (specialcase == "Q18"):
QuestionFlag = "finish question"
res = "Based on the questions you answered just now, here are the recommended course(s). Type ok to display.\n"
print("remember msg: ",Remember_msg)
print(specialcase)
return res
#Creating GUI with tkinter
import tkinter
from tkinter import *
import cv2
countRes = 0
def send():
global specialcase
global Storage_msg
global countRes
msg = EntryBox.get("1.0",'end-1c').strip()
EntryBox.delete("0.0",END)
if msg != '':
ChatLog.config(state=NORMAL)
# User input
ChatLog.insert(END, "You: " + msg + '\n\n')
# Bot responses
res = chatbot_response(msg)
ChatLog.insert(END, "Bot: " + res + '\n\n')
# Display hot courses
countRes += 1
if countRes == 1:
ChatLog.insert(END, "Bot: Here are some hot courses: Accounting\n"+
"\t\t\t Interior design\n"+
"\t\t\t Data Science\n"+
"\t\t\t Broadcasting\n"+
"\t\t\t Business administration\n"+
"\t\t\t Food Science\n"+
"\t\t\t Finance\n"+
"\t\t\t Hotel Management\n"+
"\t\t\t Psychology" + '\n\n')
ChatLog.insert(END, "Bot: May I know what course you interested in?" +'\n\n')
# print(specialcase)
# print(Storage_msg)
if(specialcase == 'age'):
specialcase = "null"
res = chatbot_response(Storage_msg)
ChatLog.insert(END, "Bot: " + res + '\n\n')
ChatLog.config(state=DISABLED)
ChatLog.yview(END)
# Tkinter Initialization
base = Tk()
base.title("TARUC Course Recommender Bot")
base.geometry("545x500")
base.resizable(width=FALSE, height=FALSE)
# Background
img = cv2.imread("chat_bg.png")
cv2.imwrite("chat_bg.png",img)
background = PhotoImage(file="chat_bg.png")
background_Label = Label(base, image = background)
background_Label.place(x=0,y=0)
#background_Label.pack()
#Create Chat window
ChatLog = Text(base, bd=0, height="10", width="150", font="Arial")
ChatLog.config(foreground="#4361ee", font=("Verdana", 12 ))
ChatLog.insert(END, "Bot: Welcome to TARUC Course Recommender System." + '\n\n')
ChatLog.config(state=DISABLED)
#Create scrollbar to Chat window
scrollbar = Scrollbar(base, command=ChatLog.yview, cursor="heart")
ChatLog['yscrollcommand'] = scrollbar.set
#Create Button to send message
SendButton = Button(base, font=("Verdana",12,'bold'), text="Send", width="12", height=5,
bd=0, bg="#4361ee", activebackground="#023e8a",fg='#ffffff',
command= send)
#Create the input box to enter message
EntryBox = Text(base, bd=0, bg="white",width="29", height="5", font="Arial")
#EntryBox.bind("<Return>", send)
#Place all components on the screen
scrollbar.place(x=525,y=6, height=376)
ChatLog.place(x=6,y=6, height=386, width=520)
EntryBox.place(x=6, y=401, height=90, width=400)
SendButton.place(x=405, y=401, height=90)
base.mainloop()
```
|
github_jupyter
|
##importing libraries
import nltk
nltk.download('punkt')
nltk.download('wordnet')
from nltk.stem import WordNetLemmatizer
lemmatizer = WordNetLemmatizer()
import json
import pickle
import numpy as np
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Dense, Activation, Dropout
from tensorflow.keras.optimizers import SGD
import random
##initializing chatbot training
words=[]
classes = []
documents = []
ignore_words = ['?', '!','#','&','(',')','a','are']
data_file = open('dataset2.json').read()
intents = json.loads(data_file)
for intent in intents['intents']:
for pattern in intent['patterns']:
# take each word and tokenize it
w = nltk.word_tokenize(pattern)
words.extend(w)
# adding documents
documents.append((w, intent['tag']))
# adding classes to our class list
if intent['tag'] not in classes:
classes.append(intent['tag'])
words = [lemmatizer.lemmatize(w.lower()) for w in words if w not in ignore_words]
# sort in alphabetical order
words = sorted(list(set(words)))
classes = sorted(list(set(classes)))
print (len(documents), "documents")
print (len(classes), "classes", classes)
print (len(words), "unique lemmatized words", words)
# save them into pickle file
pickle.dump(words,open('dictionary.pkl','wb'))
pickle.dump(classes,open('pattern_classes.pkl','wb'))
##Building Model
training = []
output_empty = [0] * len(classes)
for doc in documents:
# initializing bag of words
bag = []
# list of tokenized words for the pattern
pattern_words = doc[0]
# lemmatize each word - create base word, in attempt to represent related words
pattern_words = [lemmatizer.lemmatize(word.lower()) for word in pattern_words]
# create our bag of words array with 1, if word match found in current pattern
for w in words:
bag.append(1) if w in pattern_words else bag.append(0)
# output is a '0' for each tag and '1' for current tag (for each pattern)
output_row = list(output_empty)
output_row[classes.index(doc[1])] = 1
training.append([bag, output_row])
# shuffle our features and turn into np.array
random.shuffle(training)
training = np.array(training)
# create train and test lists.
# X -> patterns, Y -> intents
train_x = list(training[:,0])
train_y = list(training[:,1])
# Create model - 3 layers. First layer 128 neurons, second layer 64 neurons and 3rd output layer contains number of neurons
# equal to number of intents to predict output intent with softmax
model = Sequential()
model.add(Dense(128, input_shape=(len(train_x[0]),), activation='relu'))
model.add(Dropout(0.5))
model.add(Dense(64, activation='relu'))
model.add(Dropout(0.5))
# softmax get highest probability -> only 1 output
model.add(Dense(len(train_y[0]), activation='softmax'))
# Model optimization
# Compile model. SGD with Nesterov accelerated gradient gives good results for this model
sgd = SGD(lr=0.01, decay=1e-6, momentum=0.9, nesterov=True)
model.compile(loss='categorical_crossentropy', optimizer=sgd, metrics=['accuracy'])
#fitting and saving the model
hist = model.fit(np.array(train_x), np.array(train_y), epochs=35, batch_size=5, verbose=1)
model.save('chatbot_model.h5', hist)
print("model created")
#print("predict ",model.predict(train_x, batch_size =5, verbose =1))
#print("predict class",model.predict_classes(train_x, batch_size =5, verbose =1))
#print("predict proba",model.predict_proba(train_x, batch_size =5, verbose =1))
##Building Chatbot GUI
from keras.models import load_model
model = load_model('chatbot_model.h5')
import json
import random
intents = json.loads(open('dataset2.json').read())
words = pickle.load(open('dictionary.pkl','rb'))
classes = pickle.load(open('pattern_classes.pkl','rb'))
import re
QuestionFlag = "Havent ask"
specialcase = " "
category = "null"
Storage_msg = " "
Remember_msg = " "
questionAns = []
interest = ["Accounting", "Broadcasting", "Food Science", "Psychology", "Public Relations", "Business Administration","Human Resource Management","Information Technology","Computer Science","Engineering","Architecture","Quantity Surveying","Hotel Management","Culinary Arts","Advertising","Journalism","Finance","English"]
questionArr = ["Q1","Q2","Q3","Q4","Q5","Q6","Q7","Q8","Q9","Q10","Q11","Q12","Q13","Q14","Q15","Q16","Q17","Q18"]
eduLevel = 0
# Convert word number to integer number
def word_to_number(word):
from word2number import w2n
return w2n.word_to_num(word)
def clean_up_sentence(sentence):
global Remember_msg
global category
global specialcase
global QuestionFlag
global eduLevel
global questionAns
# Age - Education level e.g. Pre-U, Undergraduate, Postgraduate
if (QuestionFlag == "Asked"):
specialcase = "age"
QuestionFlag = "null"
if (sentence.isnumeric() == False):
sentence = word_to_number(sentence)
sentence = str(sentence)
x = re.findall("[0-5][0-9]", sentence)
print("Age: ",x[0])
if(int(x[0]) <= 17):
category = "too young"
elif(18 <= int(x[0]) <= 19 ):
category = "pre-u"
eduLevel = 1
elif(20 <= int(x[0]) <= 23 ):
category = "undergraduate"
eduLevel = 2
elif(int(x[0]) >= 24):
category = "postgraduate"
eduLevel = 3
# Run Questions
for x in range(len(interest)):
if (QuestionFlag == questionArr[x]):
specialcase = questionArr[x]
QuestionFlag = "null"
if (sentence.isnumeric()):
questionAns.append(int(sentence))
print(questionAns)
# All Questions Done, run GaussianNB, get the suitable course name
if (QuestionFlag == "finish question"):
QuestionFlag = "null"
specialcase = "finish question"
import pandas as pd
df = pd.read_csv('nbDatasets.csv')
cols = list(df.columns)
features = df[cols[1:-1]]
result = df["course"]
# Import LabelEncoder
from sklearn import preprocessing
#creating labelEncoder
le = preprocessing.LabelEncoder()
# Converting string labels into numbers.
result_encoded=le.fit_transform(result)
# print(result_encoded)
#Import Gaussian Naive Bayes model
from sklearn.naive_bayes import GaussianNB
#Create a Gaussian Classifier
model = GaussianNB()
# Train the model using the training sets
model.fit(features,result_encoded)
# clean questionAns list in second run of questionnaire
if len(questionAns) > 18:
del questionAns [:18]
#Predict Output
predicted= model.predict([questionAns])
transform = le.inverse_transform(predicted)
transform = transform[0]
Remember_msg += " " + transform
sentence = sentence + " " + Remember_msg
sentence_words = nltk.word_tokenize(sentence)
sentence_words = [lemmatizer.lemmatize(word.lower()) for word in sentence_words]
print(sentence_words)
return sentence_words
sentence = sentence + " " + Remember_msg
print(sentence)
sentence_words = nltk.word_tokenize(sentence)
sentence_words = [lemmatizer.lemmatize(word.lower()) for word in sentence_words]
print(sentence_words)
return sentence_words
def words_bag(sentence, words, show_details=True):
# tokenize the pattern
sentence_words = clean_up_sentence(sentence)
# bag of words - matrix of N words, vocabulary matrix
bag = [0]*len(words)
for sentenceWord in sentence_words:
for i,word in enumerate(words):
if word == sentenceWord:
# assign 1 if current word is in the vocabulary position
bag[i] = 1
if show_details:
print ("found in bag: %s" % word)
return(np.array(bag))
def predict_class(sentence, model):
p = words_bag(sentence, words,show_details=False)
res = model.predict(np.array([p]))[0]
ERROR_THRESHOLD = 0.25
# filter out predictions below a threshold
results = [[i,r] for i,r in enumerate(res) if r>ERROR_THRESHOLD]
# sort by strength of probability
# get the highest one only -> Only one output
results.sort(key=lambda x: x[1], reverse=True)
return_list = []
for r in results:
return_list.append({"intent": classes[r[0]], "probability": str(r[1])})
return return_list
def getResponse(ints, intents):
global specialcase
global Remember_msg
print("-------ints: " ,ints)
print("-------tags: ", ints[0]['intent'])
tag = ints[0]['intent']
list_of_intents = intents['intents']
for i in list_of_intents:
if(i['tag']== tag):
if (eduLevel == 1 or eduLevel == 2 or eduLevel == 3):
try:
result = i['responses'][eduLevel-1]
except:
result = random.choice(i['responses'])
else:
result = random.choice(i['responses'])
break
return result
def chatbot_response(msg):
global specialcase
global QuestionFlag
global questionAns
global Storage_msg
global Remember_msg
global eduLevel
ints = predict_class(msg, model)
#print("edu leveL: ", eduLevel)
res = getResponse(ints, intents)
# Get Age
if (res[-1] == '{'):
if (QuestionFlag == "Havent ask"):
QuestionFlag = "Asked"
Storage_msg = msg
res = "Oh before that, may i know your age please?"
else:
res = res[:-1]
# Run the likert scale questions
if (res[-1] == 'x'):
Remember_msg = " " #clear the Remember_msg
QuestionFlag = "Q1"
res = "Answer the question with scale 1 to 5, 1 - Most disagree, 2 - Disagree, 3 - Average, 4 - Agree, 5 - Most agree \nAre you a detail oriented, number sensitive, trustworthy, good in calculation person?"
if (specialcase != "null"):
if (specialcase == "age"):
# Age and education level responses
if (category == "too young"):
res += " but you are too young for now, come consult me when your are > 17 years old"
elif (category == "pre-u"):
res += " and based on your age, i bet you are a Pre University student, hence I only display Pre University Courses for you"
Remember_msg += " pre-u"
eduLevel = 1
elif (category == "undergraduate"):
res += " and based on your age, i bet you are an Undergraduate student, hence I only display Undergraduate Courses for you "
Remember_msg += " undergraduate"
eduLevel = 2
elif (category == "postgraduate"):
res += " and based on your age, i bet you are a Postgraduate student, hence I only display Postgraduate Courses for you "
Remember_msg += " postgraduate"
eduLevel = 3
# Ask Questions
if (specialcase == "Q1"):
QuestionFlag = "Q2"
res = "Are you interested in broadcasting e.g. filming, cinematography, production and multimedia?"
if (specialcase == "Q2"):
QuestionFlag = "Q3"
res = "Are you interested in chemistry, biology and composition of foods?"
if (specialcase == "Q3"):
QuestionFlag = "Q4"
res = "Are you interested in helping people to solve their problems patiently?"
if (specialcase == "Q4"):
QuestionFlag = "Q5"
res = "Are you interested in public relations which buidls mutually beneficial relationships between organization and their publics?"
if (specialcase == "Q5"):
QuestionFlag = "Q6"
res = "Are you interested in operating a business?"
if (specialcase == "Q6"):
QuestionFlag = "Q7"
res = "Are you interested in managing people where we called it as human resource management?"
if (specialcase == "Q7"):
QuestionFlag = "Q8"
res = "Are you interested in coding, programming, computer security or develop software?"
if (specialcase == "Q8"):
QuestionFlag = "Q9"
res = "Are you interested in analyze data, computer science or develop computer system application?"
if (specialcase == "Q9"):
QuestionFlag = "Q10"
res = "Are you interested in engineering e.g. electronic, mechanical, mechatronic and telecommunication?"
if (specialcase == "Q10"):
QuestionFlag = "Q11"
res = "Are you interested in interior design or architercture?"
if (specialcase == "Q11"):
QuestionFlag = "Q12"
res = "Are you interested in managing the costs and budgets of a project?"
if (specialcase == "Q12"):
QuestionFlag = "Q13"
res = "Are you interested in learning the technical skills in food and beverage, room management, managerial and personal skills?"
if (specialcase == "Q13"):
QuestionFlag = "Q14"
res = "Are you interested in cooking?"
if (specialcase == "Q14"):
QuestionFlag = "Q15"
res = "Are you interested in learning the skills of advertising, media and communication?"
if (specialcase == "Q15"):
QuestionFlag = "Q16"
res = "Are you interested in become a journalists, writers or reporters?"
if (specialcase == "Q16"):
QuestionFlag = "Q17"
res = "Are you interested in managing the financial assets?"
if (specialcase == "Q17"):
QuestionFlag = "Q18"
res = "Are you interested in english education, english drama or english studies?"
if (specialcase == "Q18"):
QuestionFlag = "finish question"
res = "Based on the questions you answered just now, here are the recommended course(s). Type ok to display.\n"
print("remember msg: ",Remember_msg)
print(specialcase)
return res
#Creating GUI with tkinter
import tkinter
from tkinter import *
import cv2
countRes = 0
def send():
global specialcase
global Storage_msg
global countRes
msg = EntryBox.get("1.0",'end-1c').strip()
EntryBox.delete("0.0",END)
if msg != '':
ChatLog.config(state=NORMAL)
# User input
ChatLog.insert(END, "You: " + msg + '\n\n')
# Bot responses
res = chatbot_response(msg)
ChatLog.insert(END, "Bot: " + res + '\n\n')
# Display hot courses
countRes += 1
if countRes == 1:
ChatLog.insert(END, "Bot: Here are some hot courses: Accounting\n"+
"\t\t\t Interior design\n"+
"\t\t\t Data Science\n"+
"\t\t\t Broadcasting\n"+
"\t\t\t Business administration\n"+
"\t\t\t Food Science\n"+
"\t\t\t Finance\n"+
"\t\t\t Hotel Management\n"+
"\t\t\t Psychology" + '\n\n')
ChatLog.insert(END, "Bot: May I know what course you interested in?" +'\n\n')
# print(specialcase)
# print(Storage_msg)
if(specialcase == 'age'):
specialcase = "null"
res = chatbot_response(Storage_msg)
ChatLog.insert(END, "Bot: " + res + '\n\n')
ChatLog.config(state=DISABLED)
ChatLog.yview(END)
# Tkinter Initialization
base = Tk()
base.title("TARUC Course Recommender Bot")
base.geometry("545x500")
base.resizable(width=FALSE, height=FALSE)
# Background
img = cv2.imread("chat_bg.png")
cv2.imwrite("chat_bg.png",img)
background = PhotoImage(file="chat_bg.png")
background_Label = Label(base, image = background)
background_Label.place(x=0,y=0)
#background_Label.pack()
#Create Chat window
ChatLog = Text(base, bd=0, height="10", width="150", font="Arial")
ChatLog.config(foreground="#4361ee", font=("Verdana", 12 ))
ChatLog.insert(END, "Bot: Welcome to TARUC Course Recommender System." + '\n\n')
ChatLog.config(state=DISABLED)
#Create scrollbar to Chat window
scrollbar = Scrollbar(base, command=ChatLog.yview, cursor="heart")
ChatLog['yscrollcommand'] = scrollbar.set
#Create Button to send message
SendButton = Button(base, font=("Verdana",12,'bold'), text="Send", width="12", height=5,
bd=0, bg="#4361ee", activebackground="#023e8a",fg='#ffffff',
command= send)
#Create the input box to enter message
EntryBox = Text(base, bd=0, bg="white",width="29", height="5", font="Arial")
#EntryBox.bind("<Return>", send)
#Place all components on the screen
scrollbar.place(x=525,y=6, height=376)
ChatLog.place(x=6,y=6, height=386, width=520)
EntryBox.place(x=6, y=401, height=90, width=400)
SendButton.place(x=405, y=401, height=90)
base.mainloop()
| 0.350533 | 0.488161 |
# Assingment "Assignment" System for DCT Academy's Code Platform
```
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import functools
from sqlalchemy import create_engine
import sklearn
from sklearn.decomposition import TruncatedSVD
engine = create_engine('postgresql+psycopg2://postgres:sudhanvasud@localhost/postgres')
print(engine.table_names())
```
## Creating list of dataframe of all tables, a dictionary mapping to corresponding dataframe
```
# Dictionary of all the tables and their columns
table_columns = {}
# Dictionary of all dataframes mapped with table names
df_all = {}
# List of all dataframes of all tables
df_list = []
for table in engine.table_names():
df = pd.read_sql(table, engine)
df_all[table] = df
df_list.append(df)
table_columns[table] = list(df.columns)
```
## Get all student/user assignments
### Merge submissions, assignments, taggings, tags
```
user_submissions = df_all['submissions'] \
.merge(df_all['assignments'], left_on='assignment_id', right_on='id') \
.merge(df_all['taggings'], left_on='assignment_id', right_on='taggable_id') \
.merge(df_all['tags'], left_on='tag_id', right_on='id')
user_submissions.drop(['statement', 'output', 'language', 'created_at_x', 'updated_at_x', 'is_checked',
'body', 'url', 'created_at_y', 'updated_at_y', 'user_id_y', 'source', 'tagger_type', 'created_at'], axis=1, inplace=True)
user_submissions.sort_index(axis=1, inplace=True)
user_submissions.to_html('all-users.html')
user_submissions['time_in_minutes'] = user_submissions['time_in_seconds'] / 60
quant = user_submissions['time_in_minutes'].quantile(0.99)
shreekar_sub = user_submissions[user_submissions['time_in_minutes'] < quant]
shreekar_sub['time_in_minutes'].plot(kind='hist')
plt.show()
```
## Create a difficulty ration column based on the time taken to solve
```
user_submissions['difficult_ratio'] = user_submissions['time_in_minutes'] / user_submissions['minutes']
user_submissions.columns
final_user_submissions = user_submissions[['user_id_x', 'difficult_ratio', 'title', 'code']]
final_user_submissions.head()
final_user_submissions.groupby('user_id_x')['difficult_ratio'].count().sort_values(ascending=False).head()
filt = final_user_submissions['code'] == 'DCT8eb1'
final_user_submissions[filt]['title'].unique()
difficulty_crosstab = final_user_submissions.pivot_table(values='difficult_ratio', index='user_id_x', columns='title', fill_value=0)
difficulty_crosstab[difficulty_crosstab <= 0] = 0
difficulty_crosstab[difficulty_crosstab > 1000] = (difficulty_crosstab.mean()).mean()
difficulty_crosstab.fillna(difficulty_crosstab.mean(), inplace=True)
difficulty_crosstab.head()
```
## Trasposing the matrix
```
difficulty_crosstab.shape
X = difficulty_crosstab.T
X.shape
SVD = TruncatedSVD(random_state=17)
resultant_matrix = SVD.fit_transform(X)
resultant_matrix.shape
```
## Generating a correlation matrix
```
corr_mat = np.corrcoef(resultant_matrix)
corr_mat.shape
corr_mat_df = pd.DataFrame(corr_mat)
corr_mat_df.to_html('corr.html')
```
## Finding Difficulty From the Correlation Matrix
```
assignment_titles = difficulty_crosstab.columns
assignment_list = list(assignment_titles)
merge_strings = assignment_list.index('Wheel of Fortune')
merge_strings
corr_merge_strings = corr_mat[merge_strings]
corr_merge_strings.shape
```
## Recommending a Correlated Assignment
```
print(len(list(assignment_titles[(corr_merge_strings <= 1.0) & (corr_merge_strings > 0.9)])))
list(assignment_titles[(corr_merge_strings <= 1.0) & (corr_merge_strings > 0.9)])
```
|
github_jupyter
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import functools
from sqlalchemy import create_engine
import sklearn
from sklearn.decomposition import TruncatedSVD
engine = create_engine('postgresql+psycopg2://postgres:sudhanvasud@localhost/postgres')
print(engine.table_names())
# Dictionary of all the tables and their columns
table_columns = {}
# Dictionary of all dataframes mapped with table names
df_all = {}
# List of all dataframes of all tables
df_list = []
for table in engine.table_names():
df = pd.read_sql(table, engine)
df_all[table] = df
df_list.append(df)
table_columns[table] = list(df.columns)
user_submissions = df_all['submissions'] \
.merge(df_all['assignments'], left_on='assignment_id', right_on='id') \
.merge(df_all['taggings'], left_on='assignment_id', right_on='taggable_id') \
.merge(df_all['tags'], left_on='tag_id', right_on='id')
user_submissions.drop(['statement', 'output', 'language', 'created_at_x', 'updated_at_x', 'is_checked',
'body', 'url', 'created_at_y', 'updated_at_y', 'user_id_y', 'source', 'tagger_type', 'created_at'], axis=1, inplace=True)
user_submissions.sort_index(axis=1, inplace=True)
user_submissions.to_html('all-users.html')
user_submissions['time_in_minutes'] = user_submissions['time_in_seconds'] / 60
quant = user_submissions['time_in_minutes'].quantile(0.99)
shreekar_sub = user_submissions[user_submissions['time_in_minutes'] < quant]
shreekar_sub['time_in_minutes'].plot(kind='hist')
plt.show()
user_submissions['difficult_ratio'] = user_submissions['time_in_minutes'] / user_submissions['minutes']
user_submissions.columns
final_user_submissions = user_submissions[['user_id_x', 'difficult_ratio', 'title', 'code']]
final_user_submissions.head()
final_user_submissions.groupby('user_id_x')['difficult_ratio'].count().sort_values(ascending=False).head()
filt = final_user_submissions['code'] == 'DCT8eb1'
final_user_submissions[filt]['title'].unique()
difficulty_crosstab = final_user_submissions.pivot_table(values='difficult_ratio', index='user_id_x', columns='title', fill_value=0)
difficulty_crosstab[difficulty_crosstab <= 0] = 0
difficulty_crosstab[difficulty_crosstab > 1000] = (difficulty_crosstab.mean()).mean()
difficulty_crosstab.fillna(difficulty_crosstab.mean(), inplace=True)
difficulty_crosstab.head()
difficulty_crosstab.shape
X = difficulty_crosstab.T
X.shape
SVD = TruncatedSVD(random_state=17)
resultant_matrix = SVD.fit_transform(X)
resultant_matrix.shape
corr_mat = np.corrcoef(resultant_matrix)
corr_mat.shape
corr_mat_df = pd.DataFrame(corr_mat)
corr_mat_df.to_html('corr.html')
assignment_titles = difficulty_crosstab.columns
assignment_list = list(assignment_titles)
merge_strings = assignment_list.index('Wheel of Fortune')
merge_strings
corr_merge_strings = corr_mat[merge_strings]
corr_merge_strings.shape
print(len(list(assignment_titles[(corr_merge_strings <= 1.0) & (corr_merge_strings > 0.9)])))
list(assignment_titles[(corr_merge_strings <= 1.0) & (corr_merge_strings > 0.9)])
| 0.317003 | 0.83901 |
## Face Recognition – Unlock Your Computer With Your Face!
### Step 1 - Create Training Data
```
import cv2
import numpy as np
# Load HAAR face classifier
face_classifier = cv2.CascadeClassifier('Haarcascades/haarcascade_frontalface_default.xml')
# Load functions
def face_extractor(img):
# Function detects faces and returns the cropped face
# If no face detected, it returns the input image
gray = cv2.cvtColor(img,cv2.COLOR_BGR2GRAY)
faces = face_classifier.detectMultiScale(gray, 1.3, 5)
if faces is ():
return None
# Crop all faces found
for (x,y,w,h) in faces:
cropped_face = img[y:y+h, x:x+w]
return cropped_face
# Initialize Webcam
cap = cv2.VideoCapture(0)
count = 0
# Collect 100 samples of your face from webcam input
while True:
ret, frame = cap.read()
if face_extractor(frame) is not None:
count += 1
face = cv2.resize(face_extractor(frame), (200, 200))
face = cv2.cvtColor(face, cv2.COLOR_BGR2GRAY)
# Save file in specified directory with unique name
file_name_path = './faces/user/' + str(count) + '.jpg'
cv2.imwrite(file_name_path, face)
# Put count on images and display live count
cv2.putText(face, str(count), (50, 50), cv2.FONT_HERSHEY_COMPLEX, 1, (0,255,0), 2)
cv2.imshow('Face Cropper', face)
else:
print("Face not found")
pass
if cv2.waitKey(1) == 13 or count == 100: #13 is the Enter Key
break
cap.release()
cv2.destroyAllWindows()
print("Collecting Samples Complete")
```
### Step 2 - Train Model
```
import cv2
import numpy as np
from os import listdir
from os.path import isfile, join
# Get the training data we previously made
data_path = './faces/user/'
onlyfiles = [f for f in listdir(data_path) if isfile(join(data_path, f))]
# Create arrays for training data and labels
Training_Data, Labels = [], []
# Open training images in our datapath
# Create a numpy array for training data
for i, files in enumerate(onlyfiles):
image_path = data_path + onlyfiles[i]
images = cv2.imread(image_path, cv2.IMREAD_GRAYSCALE)
Training_Data.append(np.asarray(images, dtype=np.uint8))
Labels.append(i)
# Create a numpy array for both training data and labels
Labels = np.asarray(Labels, dtype=np.int32)
# Initialize facial recognizer
model = cv2.createLBPHFaceRecognizer()
# NOTE: For OpenCV 3.0 use cv2.face.createLBPHFaceRecognizer()
# Let's train our model
model.train(np.asarray(Training_Data), np.asarray(Labels))
print("Model trained sucessefully")
```
### Step 3 - Run Our Facial Recognition
```
import cv2
import numpy as np
face_classifier = cv2.CascadeClassifier('Haarcascades/haarcascade_frontalface_default.xml')
def face_detector(img, size=0.5):
# Convert image to grayscale
gray = cv2.cvtColor(img,cv2.COLOR_BGR2GRAY)
faces = face_classifier.detectMultiScale(gray, 1.3, 5)
if faces is ():
return img, []
for (x,y,w,h) in faces:
cv2.rectangle(img,(x,y),(x+w,y+h),(0,255,255),2)
roi = img[y:y+h, x:x+w]
roi = cv2.resize(roi, (200, 200))
return img, roi
# Open Webcam
cap = cv2.VideoCapture(0)
while True:
ret, frame = cap.read()
image, face = face_detector(frame)
try:
face = cv2.cvtColor(face, cv2.COLOR_BGR2GRAY)
# Pass face to prediction model
# "results" comprises of a tuple containing the label and the confidence value
results = model.predict(face)
if results[1] < 500:
confidence = int( 100 * (1 - (results[1])/400) )
display_string = str(confidence) + '% Confident it is User'
cv2.putText(image, display_string, (100, 120), cv2.FONT_HERSHEY_COMPLEX, 1, (255,120,150), 2)
if confidence > 75:
cv2.putText(image, "Unlocked", (250, 450), cv2.FONT_HERSHEY_COMPLEX, 1, (0,255,0), 2)
cv2.imshow('Face Recognition', image )
else:
cv2.putText(image, "Locked", (250, 450), cv2.FONT_HERSHEY_COMPLEX, 1, (0,0,255), 2)
cv2.imshow('Face Recognition', image )
except:
cv2.putText(image, "No Face Found", (220, 120) , cv2.FONT_HERSHEY_COMPLEX, 1, (0,0,255), 2)
cv2.putText(image, "Locked", (250, 450), cv2.FONT_HERSHEY_COMPLEX, 1, (0,0,255), 2)
cv2.imshow('Face Recognition', image )
pass
if cv2.waitKey(1) == 13: #13 is the Enter Key
break
cap.release()
cv2.destroyAllWindows()
```
|
github_jupyter
|
import cv2
import numpy as np
# Load HAAR face classifier
face_classifier = cv2.CascadeClassifier('Haarcascades/haarcascade_frontalface_default.xml')
# Load functions
def face_extractor(img):
# Function detects faces and returns the cropped face
# If no face detected, it returns the input image
gray = cv2.cvtColor(img,cv2.COLOR_BGR2GRAY)
faces = face_classifier.detectMultiScale(gray, 1.3, 5)
if faces is ():
return None
# Crop all faces found
for (x,y,w,h) in faces:
cropped_face = img[y:y+h, x:x+w]
return cropped_face
# Initialize Webcam
cap = cv2.VideoCapture(0)
count = 0
# Collect 100 samples of your face from webcam input
while True:
ret, frame = cap.read()
if face_extractor(frame) is not None:
count += 1
face = cv2.resize(face_extractor(frame), (200, 200))
face = cv2.cvtColor(face, cv2.COLOR_BGR2GRAY)
# Save file in specified directory with unique name
file_name_path = './faces/user/' + str(count) + '.jpg'
cv2.imwrite(file_name_path, face)
# Put count on images and display live count
cv2.putText(face, str(count), (50, 50), cv2.FONT_HERSHEY_COMPLEX, 1, (0,255,0), 2)
cv2.imshow('Face Cropper', face)
else:
print("Face not found")
pass
if cv2.waitKey(1) == 13 or count == 100: #13 is the Enter Key
break
cap.release()
cv2.destroyAllWindows()
print("Collecting Samples Complete")
import cv2
import numpy as np
from os import listdir
from os.path import isfile, join
# Get the training data we previously made
data_path = './faces/user/'
onlyfiles = [f for f in listdir(data_path) if isfile(join(data_path, f))]
# Create arrays for training data and labels
Training_Data, Labels = [], []
# Open training images in our datapath
# Create a numpy array for training data
for i, files in enumerate(onlyfiles):
image_path = data_path + onlyfiles[i]
images = cv2.imread(image_path, cv2.IMREAD_GRAYSCALE)
Training_Data.append(np.asarray(images, dtype=np.uint8))
Labels.append(i)
# Create a numpy array for both training data and labels
Labels = np.asarray(Labels, dtype=np.int32)
# Initialize facial recognizer
model = cv2.createLBPHFaceRecognizer()
# NOTE: For OpenCV 3.0 use cv2.face.createLBPHFaceRecognizer()
# Let's train our model
model.train(np.asarray(Training_Data), np.asarray(Labels))
print("Model trained sucessefully")
import cv2
import numpy as np
face_classifier = cv2.CascadeClassifier('Haarcascades/haarcascade_frontalface_default.xml')
def face_detector(img, size=0.5):
# Convert image to grayscale
gray = cv2.cvtColor(img,cv2.COLOR_BGR2GRAY)
faces = face_classifier.detectMultiScale(gray, 1.3, 5)
if faces is ():
return img, []
for (x,y,w,h) in faces:
cv2.rectangle(img,(x,y),(x+w,y+h),(0,255,255),2)
roi = img[y:y+h, x:x+w]
roi = cv2.resize(roi, (200, 200))
return img, roi
# Open Webcam
cap = cv2.VideoCapture(0)
while True:
ret, frame = cap.read()
image, face = face_detector(frame)
try:
face = cv2.cvtColor(face, cv2.COLOR_BGR2GRAY)
# Pass face to prediction model
# "results" comprises of a tuple containing the label and the confidence value
results = model.predict(face)
if results[1] < 500:
confidence = int( 100 * (1 - (results[1])/400) )
display_string = str(confidence) + '% Confident it is User'
cv2.putText(image, display_string, (100, 120), cv2.FONT_HERSHEY_COMPLEX, 1, (255,120,150), 2)
if confidence > 75:
cv2.putText(image, "Unlocked", (250, 450), cv2.FONT_HERSHEY_COMPLEX, 1, (0,255,0), 2)
cv2.imshow('Face Recognition', image )
else:
cv2.putText(image, "Locked", (250, 450), cv2.FONT_HERSHEY_COMPLEX, 1, (0,0,255), 2)
cv2.imshow('Face Recognition', image )
except:
cv2.putText(image, "No Face Found", (220, 120) , cv2.FONT_HERSHEY_COMPLEX, 1, (0,0,255), 2)
cv2.putText(image, "Locked", (250, 450), cv2.FONT_HERSHEY_COMPLEX, 1, (0,0,255), 2)
cv2.imshow('Face Recognition', image )
pass
if cv2.waitKey(1) == 13: #13 is the Enter Key
break
cap.release()
cv2.destroyAllWindows()
| 0.641422 | 0.805594 |
# VacationPy
----
#### Note
* Keep an eye on your API usage. Use https://developers.google.com/maps/reporting/gmp-reporting as reference for how to monitor your usage and billing.
* Instructions have been included for each segment. You do not have to follow them exactly, but they are included to help you think through the steps.
```
# Dependencies and Setup
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
import requests
import gmaps
import os
# Import API key
from api_keys import g_key
```
### Store Part I results into DataFrame
* Load the csv exported in Part I to a DataFrame
```
weather = pd.read_csv("../output_Data/Weather Data 2")
weather
weather.describe()
```
### Humidity Heatmap
* Configure gmaps.
* Use the Lat and Lng as locations and Humidity as the weight.
* Add Heatmap layer to map.
```
# Configure maps
gmaps.configure(api_key=g_key)
#latitude and longitude
locations = weather[["Lat", "Lng"]]
# Humidity
humidity = weather["Humidity"]
#Heatmap
fig = gmaps.figure(center=(46.0, -5.0), zoom_level=2)
max_intensity = np.max(humidity)
#Heat layer
heat_layer = gmaps.heatmap_layer(locations, weights = humidity, dissipating=False, max_intensity=100, point_radius=3)
#Add layer
fig.add_layer(heat_layer)
fig
```
### Create new DataFrame fitting weather criteria
* Narrow down the cities to fit weather conditions.
* Drop any rows with null values.
```
city_weather = weather.loc[(weather["Wind Speed"]<=10) & (weather["Cloudiness"]==0) & (weather["Max Temp"]>=65) & \
(weather["Max Temp"]>=80)].dropna()
city_weather
```
### Hotel Map
* Store into variable named `hotel_df`.
* Add a "Hotel Name" column to the DataFrame.
* Set parameters to search for hotels with 5000 meters.
* Hit the Google Places API for each city's coordinates.
* Store the first Hotel result into the DataFrame.
* Plot markers on top of the heatmap.
```
# Variable hotel_df
hotel_df = city_weather.loc[:,["City", "County", "Lat", "Lng"]]
# Adding column "Hotel Name" to Dataframe
hotel_df["Hotel Name"] = ""
hotel_df
hotel_df.rename(columns={"County": "Country"})
base_url = "https://maps.googleapis.com/maps/api/place/nearbysearch/json"
params ={"type" : "hotel",
"Keyword" : "hotel",
"radius" : 5000,
"key" : g_key}
for index, row in hotel_df.iterrows():
#retrieve city name, lat, lng from df
lat = row["Lat"]
lng = row["Lng"]
city_name = row["City"]
# adding keyword to pramas dict
params["location"] = f"{lat}, {lng}"
# Url and API request
print(f"Retrieving Results for Index {index}: {city_name}.")
response = requests.get(base_url, params=params).json()
# result
results = response["results"]
# Dataframe to hold hotel names
try:
print(f"Closest hotel in {city_name} is {results[0]['name']}.")
hotel_df.loc[index, "Hotel Name"]= results[0]['name']
#show missing result, if no hotel found
except (KeyError, IndexError):
print("Missing result...... skipping.")
print("--------------------------")
# Wait before making another api request to avoid error
#time.sleep(1)
print("-------- End------------------")
# NOTE: Do not change any of the code in this cell
# Using the template add the hotel marks to the heatmap
info_box_template = """
<dl>
<dt>Name</dt><dd>{Hotel Name}</dd>
<dt>City</dt><dd>{City}</dd>
<dt>County</dt><dd>{County}</dd>
</dl>
"""
# Store the DataFrame Row
# NOTE: be sure to update with your DataFrame name
hotel_info = [info_box_template.format(**row) for index, row in hotel_df.iterrows()]
locations = hotel_df[["Lat", "Lng"]]
# Add marker layer ontop of heat map
markers = gmaps.marker_layer(locations, info_box_content = hotel_info)
fig.add_layer(markers)
# Display figure
fig
```
|
github_jupyter
|
# Dependencies and Setup
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
import requests
import gmaps
import os
# Import API key
from api_keys import g_key
weather = pd.read_csv("../output_Data/Weather Data 2")
weather
weather.describe()
# Configure maps
gmaps.configure(api_key=g_key)
#latitude and longitude
locations = weather[["Lat", "Lng"]]
# Humidity
humidity = weather["Humidity"]
#Heatmap
fig = gmaps.figure(center=(46.0, -5.0), zoom_level=2)
max_intensity = np.max(humidity)
#Heat layer
heat_layer = gmaps.heatmap_layer(locations, weights = humidity, dissipating=False, max_intensity=100, point_radius=3)
#Add layer
fig.add_layer(heat_layer)
fig
city_weather = weather.loc[(weather["Wind Speed"]<=10) & (weather["Cloudiness"]==0) & (weather["Max Temp"]>=65) & \
(weather["Max Temp"]>=80)].dropna()
city_weather
# Variable hotel_df
hotel_df = city_weather.loc[:,["City", "County", "Lat", "Lng"]]
# Adding column "Hotel Name" to Dataframe
hotel_df["Hotel Name"] = ""
hotel_df
hotel_df.rename(columns={"County": "Country"})
base_url = "https://maps.googleapis.com/maps/api/place/nearbysearch/json"
params ={"type" : "hotel",
"Keyword" : "hotel",
"radius" : 5000,
"key" : g_key}
for index, row in hotel_df.iterrows():
#retrieve city name, lat, lng from df
lat = row["Lat"]
lng = row["Lng"]
city_name = row["City"]
# adding keyword to pramas dict
params["location"] = f"{lat}, {lng}"
# Url and API request
print(f"Retrieving Results for Index {index}: {city_name}.")
response = requests.get(base_url, params=params).json()
# result
results = response["results"]
# Dataframe to hold hotel names
try:
print(f"Closest hotel in {city_name} is {results[0]['name']}.")
hotel_df.loc[index, "Hotel Name"]= results[0]['name']
#show missing result, if no hotel found
except (KeyError, IndexError):
print("Missing result...... skipping.")
print("--------------------------")
# Wait before making another api request to avoid error
#time.sleep(1)
print("-------- End------------------")
# NOTE: Do not change any of the code in this cell
# Using the template add the hotel marks to the heatmap
info_box_template = """
<dl>
<dt>Name</dt><dd>{Hotel Name}</dd>
<dt>City</dt><dd>{City}</dd>
<dt>County</dt><dd>{County}</dd>
</dl>
"""
# Store the DataFrame Row
# NOTE: be sure to update with your DataFrame name
hotel_info = [info_box_template.format(**row) for index, row in hotel_df.iterrows()]
locations = hotel_df[["Lat", "Lng"]]
# Add marker layer ontop of heat map
markers = gmaps.marker_layer(locations, info_box_content = hotel_info)
fig.add_layer(markers)
# Display figure
fig
| 0.460532 | 0.880695 |
<!--BOOK_INFORMATION-->
<img align="left" style="padding-right:10px;" src="figures/PDSH-cover-small.png">
*This notebook contains an excerpt from the [Python Data Science Handbook](http://shop.oreilly.com/product/0636920034919.do) by Jake VanderPlas; the content is available [on GitHub](https://github.com/jakevdp/PythonDataScienceHandbook).*
*The text is released under the [CC-BY-NC-ND license](https://creativecommons.org/licenses/by-nc-nd/3.0/us/legalcode), and code is released under the [MIT license](https://opensource.org/licenses/MIT). If you find this content useful, please consider supporting the work by [buying the book](http://shop.oreilly.com/product/0636920034919.do)!*
<!--NAVIGATION-->
< [Feature Engineering](05.04-Feature-Engineering.ipynb) | [Contents](Index.ipynb) | [In Depth: Linear Regression](05.06-Linear-Regression.ipynb) >
# In Depth: Naive Bayes Classification
The previous four sections have given a general overview of the concepts of machine learning.
In this section and the ones that follow, we will be taking a closer look at several specific algorithms for supervised and unsupervised learning, starting here with naive Bayes classification.
Naive Bayes models are a group of extremely fast and simple classification algorithms that are often suitable for very high-dimensional datasets.
Because they are so fast and have so few tunable parameters, they end up being very useful as a quick-and-dirty baseline for a classification problem.
This section will focus on an intuitive explanation of how naive Bayes classifiers work, followed by a couple examples of them in action on some datasets.
## Bayesian Classification
Naive Bayes classifiers are built on Bayesian classification methods.
These rely on Bayes's theorem, which is an equation describing the relationship of conditional probabilities of statistical quantities.
In Bayesian classification, we're interested in finding the probability of a label given some observed features, which we can write as $P(L~|~{\rm features})$.
Bayes's theorem tells us how to express this in terms of quantities we can compute more directly:
$$
P(L~|~{\rm features}) = \frac{P({\rm features}~|~L)P(L)}{P({\rm features})}
$$
If we are trying to decide between two labels—let's call them $L_1$ and $L_2$—then one way to make this decision is to compute the ratio of the posterior probabilities for each label:
$$
\frac{P(L_1~|~{\rm features})}{P(L_2~|~{\rm features})} = \frac{P({\rm features}~|~L_1)}{P({\rm features}~|~L_2)}\frac{P(L_1)}{P(L_2)}
$$
All we need now is some model by which we can compute $P({\rm features}~|~L_i)$ for each label.
Such a model is called a *generative model* because it specifies the hypothetical random process that generates the data.
Specifying this generative model for each label is the main piece of the training of such a Bayesian classifier.
The general version of such a training step is a very difficult task, but we can make it simpler through the use of some simplifying assumptions about the form of this model.
This is where the "naive" in "naive Bayes" comes in: if we make very naive assumptions about the generative model for each label, we can find a rough approximation of the generative model for each class, and then proceed with the Bayesian classification.
Different types of naive Bayes classifiers rest on different naive assumptions about the data, and we will examine a few of these in the following sections.
We begin with the standard imports:
```
%matplotlib inline
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
```
## Gaussian Naive Bayes
Perhaps the easiest naive Bayes classifier to understand is Gaussian naive Bayes.
In this classifier, the assumption is that *data from each label is drawn from a simple Gaussian distribution*.
Imagine that you have the following data:
```
from sklearn.datasets import make_blobs
X, y = make_blobs(100, 2, centers=2, random_state=2, cluster_std=1.5)
plt.scatter(X[:, 0], X[:, 1], c=y, s=50, cmap='RdBu');
```
One extremely fast way to create a simple model is to assume that the data is described by a Gaussian distribution with no covariance between dimensions.
This model can be fit by simply finding the mean and standard deviation of the points within each label, which is all you need to define such a distribution.
The result of this naive Gaussian assumption is shown in the following figure:

[figure source in Appendix](06.00-Figure-Code.ipynb#Gaussian-Naive-Bayes)
The ellipses here represent the Gaussian generative model for each label, with larger probability toward the center of the ellipses.
With this generative model in place for each class, we have a simple recipe to compute the likelihood $P({\rm features}~|~L_1)$ for any data point, and thus we can quickly compute the posterior ratio and determine which label is the most probable for a given point.
This procedure is implemented in Scikit-Learn's ``sklearn.naive_bayes.GaussianNB`` estimator:
```
from sklearn.naive_bayes import GaussianNB
model = GaussianNB()
model.fit(X, y);
```
Now let's generate some new data and predict the label:
```
rng = np.random.RandomState(0)
Xnew = [-6, -14] + [14, 18] * rng.rand(2000, 2)
ynew = model.predict(Xnew)
```
Now we can plot this new data to get an idea of where the decision boundary is:
```
plt.scatter(X[:, 0], X[:, 1], c=y, s=50, cmap='RdBu')
lim = plt.axis()
plt.scatter(Xnew[:, 0], Xnew[:, 1], c=ynew, s=20, cmap='RdBu', alpha=0.1)
plt.axis(lim);
```
We see a slightly curved boundary in the classifications—in general, the boundary in Gaussian naive Bayes is quadratic.
A nice piece of this Bayesian formalism is that it naturally allows for probabilistic classification, which we can compute using the ``predict_proba`` method:
```
yprob = model.predict_proba(Xnew)
yprob[-8:].round(2)
```
The columns give the posterior probabilities of the first and second label, respectively.
If you are looking for estimates of uncertainty in your classification, Bayesian approaches like this can be a useful approach.
Of course, the final classification will only be as good as the model assumptions that lead to it, which is why Gaussian naive Bayes often does not produce very good results.
Still, in many cases—especially as the number of features becomes large—this assumption is not detrimental enough to prevent Gaussian naive Bayes from being a useful method.
## Multinomial Naive Bayes
The Gaussian assumption just described is by no means the only simple assumption that could be used to specify the generative distribution for each label.
Another useful example is multinomial naive Bayes, where the features are assumed to be generated from a simple multinomial distribution.
The multinomial distribution describes the probability of observing counts among a number of categories, and thus multinomial naive Bayes is most appropriate for features that represent counts or count rates.
The idea is precisely the same as before, except that instead of modeling the data distribution with the best-fit Gaussian, we model the data distribuiton with a best-fit multinomial distribution.
### Example: Classifying Text
One place where multinomial naive Bayes is often used is in text classification, where the features are related to word counts or frequencies within the documents to be classified.
We discussed the extraction of such features from text in [Feature Engineering](05.04-Feature-Engineering.ipynb); here we will use the sparse word count features from the 20 Newsgroups corpus to show how we might classify these short documents into categories.
Let's download the data and take a look at the target names:
```
from sklearn.datasets import fetch_20newsgroups
data = fetch_20newsgroups()
data.target_names
```
For simplicity here, we will select just a few of these categories, and download the training and testing set:
```
categories = ['talk.religion.misc', 'soc.religion.christian',
'sci.space', 'comp.graphics']
train = fetch_20newsgroups(subset='train', categories=categories)
test = fetch_20newsgroups(subset='test', categories=categories)
```
Here is a representative entry from the data:
```
print(train.data[5])
```
In order to use this data for machine learning, we need to be able to convert the content of each string into a vector of numbers.
For this we will use the TF-IDF vectorizer (discussed in [Feature Engineering](05.04-Feature-Engineering.ipynb)), and create a pipeline that attaches it to a multinomial naive Bayes classifier:
```
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.naive_bayes import MultinomialNB
from sklearn.pipeline import make_pipeline
model = make_pipeline(TfidfVectorizer(), MultinomialNB())
```
With this pipeline, we can apply the model to the training data, and predict labels for the test data:
```
model.fit(train.data, train.target)
labels = model.predict(test.data)
```
Now that we have predicted the labels for the test data, we can evaluate them to learn about the performance of the estimator.
For example, here is the confusion matrix between the true and predicted labels for the test data:
```
from sklearn.metrics import confusion_matrix
mat = confusion_matrix(test.target, labels)
sns.heatmap(mat.T, square=True, annot=True, fmt='d', cbar=False,
xticklabels=train.target_names, yticklabels=train.target_names)
plt.xlabel('true label')
plt.ylabel('predicted label');
```
Evidently, even this very simple classifier can successfully separate space talk from computer talk, but it gets confused between talk about religion and talk about Christianity.
This is perhaps an expected area of confusion!
The very cool thing here is that we now have the tools to determine the category for *any* string, using the ``predict()`` method of this pipeline.
Here's a quick utility function that will return the prediction for a single string:
```
def predict_category(s, train=train, model=model):
pred = model.predict([s])
return train.target_names[pred[0]]
```
Let's try it out:
```
predict_category('sending a payload to the ISS')
predict_category('discussing islam vs atheism')
predict_category('determining the screen resolution')
```
Remember that this is nothing more sophisticated than a simple probability model for the (weighted) frequency of each word in the string; nevertheless, the result is striking.
Even a very naive algorithm, when used carefully and trained on a large set of high-dimensional data, can be surprisingly effective.
## When to Use Naive Bayes
Because naive Bayesian classifiers make such stringent assumptions about data, they will generally not perform as well as a more complicated model.
That said, they have several advantages:
- They are extremely fast for both training and prediction
- They provide straightforward probabilistic prediction
- They are often very easily interpretable
- They have very few (if any) tunable parameters
These advantages mean a naive Bayesian classifier is often a good choice as an initial baseline classification.
If it performs suitably, then congratulations: you have a very fast, very interpretable classifier for your problem.
If it does not perform well, then you can begin exploring more sophisticated models, with some baseline knowledge of how well they should perform.
Naive Bayes classifiers tend to perform especially well in one of the following situations:
- When the naive assumptions actually match the data (very rare in practice)
- For very well-separated categories, when model complexity is less important
- For very high-dimensional data, when model complexity is less important
The last two points seem distinct, but they actually are related: as the dimension of a dataset grows, it is much less likely for any two points to be found close together (after all, they must be close in *every single dimension* to be close overall).
This means that clusters in high dimensions tend to be more separated, on average, than clusters in low dimensions, assuming the new dimensions actually add information.
For this reason, simplistic classifiers like naive Bayes tend to work as well or better than more complicated classifiers as the dimensionality grows: once you have enough data, even a simple model can be very powerful.
<!--NAVIGATION-->
< [Feature Engineering](05.04-Feature-Engineering.ipynb) | [Contents](Index.ipynb) | [In Depth: Linear Regression](05.06-Linear-Regression.ipynb) >
|
github_jupyter
|
%matplotlib inline
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.datasets import make_blobs
X, y = make_blobs(100, 2, centers=2, random_state=2, cluster_std=1.5)
plt.scatter(X[:, 0], X[:, 1], c=y, s=50, cmap='RdBu');
from sklearn.naive_bayes import GaussianNB
model = GaussianNB()
model.fit(X, y);
rng = np.random.RandomState(0)
Xnew = [-6, -14] + [14, 18] * rng.rand(2000, 2)
ynew = model.predict(Xnew)
plt.scatter(X[:, 0], X[:, 1], c=y, s=50, cmap='RdBu')
lim = plt.axis()
plt.scatter(Xnew[:, 0], Xnew[:, 1], c=ynew, s=20, cmap='RdBu', alpha=0.1)
plt.axis(lim);
yprob = model.predict_proba(Xnew)
yprob[-8:].round(2)
from sklearn.datasets import fetch_20newsgroups
data = fetch_20newsgroups()
data.target_names
categories = ['talk.religion.misc', 'soc.religion.christian',
'sci.space', 'comp.graphics']
train = fetch_20newsgroups(subset='train', categories=categories)
test = fetch_20newsgroups(subset='test', categories=categories)
print(train.data[5])
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.naive_bayes import MultinomialNB
from sklearn.pipeline import make_pipeline
model = make_pipeline(TfidfVectorizer(), MultinomialNB())
model.fit(train.data, train.target)
labels = model.predict(test.data)
from sklearn.metrics import confusion_matrix
mat = confusion_matrix(test.target, labels)
sns.heatmap(mat.T, square=True, annot=True, fmt='d', cbar=False,
xticklabels=train.target_names, yticklabels=train.target_names)
plt.xlabel('true label')
plt.ylabel('predicted label');
def predict_category(s, train=train, model=model):
pred = model.predict([s])
return train.target_names[pred[0]]
predict_category('sending a payload to the ISS')
predict_category('discussing islam vs atheism')
predict_category('determining the screen resolution')
| 0.678859 | 0.990624 |
# Support Vector Machine For 2-class Linear Separable Question
## Decision Boundary
for the positive class: $$w^TX + b \geq 0$$
for the negative class: $$w^TX + b < 0$$
Problem? There many solutions, and which one should be chosen?
## Idea of SVM
Core Idea: Find the decision boundary, while maximize the <b>margin</b>
Then it will have two boundary, upper boundary and lower boundary
for the upper boundary: $$w^TX+b \geq 1$$
for the lower boundary: $$w^TX+b < 1$$
They can be merged into one: $$(w^TX+b)^TY \geq 1$$
Then, we have three areas. The negative class is below or on the lower boundary, whereas the positive class is higher or on the upper boundary. And there is an area without any samples (this area should be maximize). The samples on the boundary are called <b>support vector</b>
## The Distance Between Two Planes/Lines
The distance between the marginal boundary and the decision boundary: $$\frac{|b-(b-1)|}{\sqrt{||w||^2}} = \frac{1}{||w||}$$
The distance between the margins: $$\frac{2}{||w||}$$
maximize the above <==> minimize the following: $$\frac{1}{2}||w||^2$$
## Task
What we want to do now is finding a decision boundary which owns the largest distance of margins:
$$min_{w} \frac{1}{2}||w||^2 \ subject \ to \ (w^TX+b)^TY \geq 1$$
## Derivative Task by "Lagrange Multiplier"
Minimize:
$$\ell(w,b,a)=\frac{1}{2}||w||^2 - \sum_i a_i(y_i(w^Tx_i+b)-1)$$
Solve:
$$\frac{\partial \ell(w,b,a)}{\partial w}=0 \to w = \sum_i^l a_iy_ix_i, \ x_i \in {SV}$$
$l$ is the number of support vectors (note: sometimes, you may see $n$ instead of $l$, because $a_i=0$ for the samples not on the marginal boundaries)
$a_i$ is the lagrange multipliers
## VC Dimension and Bounding True Error
VC dimension: https://www.zhihu.com/question/23418822
VC for SVC: $$h\leq min(\frac{R^2}{\rho^2},p)+1$$
Bounding True Error: silde 31,week4_a, TU Delft CS4220
## Error Estimation
By leave-one-out cross validation, the true error can be bounded by: $\varepsilon_{LOO} \le \frac{\# support \ vectors}{N}$
# for Linear Nonseparable Question
## Adding Slack
<img src="./week4_figure/outlier1.png" width="75%">
<br>For linear separable one: $$(w^TX+b)^TY \geq 1$$
<br>We can make the marginal boundaries more flexible for <b>some of the points</b>: $$(w^Tx_i+b)^Ty_i \geq 1-\xi_i,\ i={1,2,...n}$$
or
$$(w^TX+b)^TY \geq 1-\xi$$
<br>slack variable $\xi$:</br>
* large slack variable like (a), $\xi_i \ge 1$
* small slack variable like (b), $1 > \xi_i > 0$
* no slack variable for the correct points, $\xi_i = 0$
* 软误差(sum of the slack variable. they can be ragarded as a kind of error, as we ingore/misclassify them in order to build our margianl boundaries), $\sum_i^n \xi$
* panalty, C to constarin the slack
<br> new model:
$$
min \frac{1}{2}||w||^2+C\sum_i^n \xi_i
\\
s.t. (w^TX+b)^TY \geq 1-\xi
\\
\xi_i \ge 0, \ for i={1,2,...,n}
$$
<br> new optimization problem:
$$
max_{\alpha} \sum_{i=1}^n \alpha_i - \frac{1}{2} \sum_{i,j=1}^n \alpha_i \alpha_j y_i y_j x_i^T x_j
\\
s.t. 0 \le \alpha_i \le C
\\
\sum_{j=1}^m \alpha_i y_i =0
$$
$$\to f(X) = w^TX+b = (\sum_{i=1}^n a_iy_ix_i)^TX + b$$
<br>reference: https://blog.csdn.net/guoziqing506/article/details/81120354
## Mapping To A Higher Dimenssion

## Kernal Function
$$
max_{\alpha} \sum_{i=1}^n \alpha_i - \frac{1}{2} \sum_{i,j=1}^n \alpha_i \alpha_j y_i y_j x_i^T x_j
\\
s.t. 0 \le \alpha_i \le C
\\
\sum_{j=1}^m \alpha_i y_i =0
$$
$$
f(z) = \sum_{i=1}^n a_iy_iK(x_i,z) + b
$$
<br> brilliant video provided by MIT: </br>
<br>https://www.youtube.com/watch?v=_PwhiWxHK8o&ab_channel=MITOpenCourseWare
# Exercise
```
import numpy as np
import prtools as pr
import matplotlib.pyplot as plt
from jupyterthemes import jtplot
jtplot.style(theme="grade3")
```
## exercise 4.1
```
plt.figure(figsize=(8,8))
x = np.array([[0,1], [0,3], [2,0]])
y = np.array([0, 0, 1])
plt.scatter(x[:,0], x[:,1], c=y, marker="x")
plt.plot([2,0], [0,1])
plt.show()
```
(a)
totally 2 support vectors on the margins
```
plt.figure(figsize=(8,8))
x = np.array([[0,-1], [0,3], [2,0]])
y = np.array([0, 0, 1])
plt.scatter(x[:,0], x[:,1], c=y, marker="x")
plt.plot([2,0], [0,0])
plt.show()
```
(b)
totally 3 support vectors on the margins
## Exercise 4.2
```
from sklearn.preprocessing import minmax_scale
from sklearn.preprocessing import maxabs_scale
from sklearn.preprocessing import normalize
plt.figure(figsize=(16,8))
plt.subplot(1,2,1)
x = np.array([[1,1], [2,0], [0,0], [1,0]])
y = np.array([0, 0, 1, 2])
plt.scatter(x[:,0], x[:,1], c=["black", "black", "green", "#ff1493"], marker="x", s=100)
train = pr.prdataset(x[:3], y[:3])
c = pr.svc(train, ("linear", 0, 20))
pr.plotc(c)
plt.subplot(1,2,2)
x_rescale = normalize(x)
plt.scatter(x_rescale[:,0], x_rescale[:,1], c=["black", "black", "green", "#ff1493"], marker="x", s=100)
train_re = pr.prdataset(x_rescale[:3], y[:3])
c = pr.svc(train_re, ("linear", 0, 20))
pr.plotc(c)
plt.show()
```
(a)
??????
Is it noly sensitive to the non-linear scaling?
## Exercise 4.3
```
plt.figure(figsize=(8,8))
x = np.array([[0,1], [2,4], [1,0]])
y = np.array([0, 0, 1])
data = pr.prdataset(x, y)
w = pr.ldc(data, 1)
w2 = pr.svc(data,("linear", 0, 10))
plt.scatter(x[:,0], x[:,1], c=y, marker="x")
pr.plotc(w)
pr.plotc(w2)
plt.legend()
plt.show()
```
(a)
?????
The two solutions will be the same when <b>the number of support vectors is three in the 2D case.</b> LDA will always have three “support vectors” in this 2D setting
## Exercise 4.4
## Exercise 4.5
(a)
$$exp(-(x-y)^2)=exp(-x^2)exp(-y^2)exp(2xy)=exp(-x^2)exp(-y^2)\sum \frac{(2xy)^n}{n!}$$
$$\phi(x)=exp(-x)\sum \frac{\sqrt{2}x^n}{n!}$$
$$\phi(y)=exp(-y)\sum \frac{\sqrt{2}y^n}{n!}$$
(b)
$\infty$
## Exercise 4.6
(a)
$$||x - \frac{1}{N_C} \sum_{x_i^C \in N_C} x_i^C||^2
=
<x- \frac{1}{N_C} \sum_{x_i^C \in N_C} x_i^C,
x- \frac{1}{N_C} \sum_{x_i^C \in N_C} x_i^C>$$
(b)
$$
<x- \frac{1}{N_C} \sum_{x_i^C \in N_C} x_i^C,
x- \frac{1}{N_C} \sum_{x_i^C \in N_C} x_i^C>
\\
= x^2 - \frac{2}{N_C} \sum_{x_i^C \in N_C} x_i^C x + \frac{1}{N_C^2} \sum_{x_i^C \in N_C} \sum_{x_j^C \in N_C} x_i^C x_j^C
$$
$$
\frac
{x^2 - \frac{2}{N_C} \sum_{x_i^C \in N_C} x_i^C x + \frac{1}{N_C^2} \sum_{x_i^C \in N_C} \sum_{x_j^C \in N_C} x_i^C x_j^C}
{x^2 - \frac{2}{N_K} \sum_{x_i^K \in N_K} x_i^K x + \frac{1}{N_K^2} \sum_{x_i^K \in N_K} \sum_{x_j^K \in N_K} x_i^K x_j^K}=1
$$
$$
\frac{1}{N_C} \sum_{x_i^C \in N_C} x_i^C x -
\frac{1}{N_K} \sum_{x_i^K \in N_K} x_i^K x = c
$$
$$
\frac{1}{N_C} \sum_{x_i^C \in N_C} K(x, x_i^C) -
\frac{1}{N_K} \sum_{x_i^K \in N_K} K(x, x_i^K) = c
$$
## Exercise 4.7
(a)
```
a = pr.gendatb(n=[20,20], s=1)
plt.figure(figsize=(40,25))
widths = [0.1, 0.5, 0.75, 1, 2.5, 5, 7.5, 10]
for i in range(len(widths)):
svc = pr.svc(a, ("rbf", widths[i], 10))
plt.subplot(2,4,i+1)
pr.scatterd(a)
pr.plotc(svc)
plt.title("widths="+str(widths[i]))
plt.show()
```
(b)
<img src="./week4_figure/4-72.png" width="75%">
<img src="./week4_figure/2-110.png" width="75%">
(c)
For SVC, only the support vectors are taken into consideration.
## Exercise 4.8
(a)
```
a = pr.gendatb(n=[200,200], s=1)
s = np.array([0.2,0.5,1.0,2.0,5.0,7.0,10.0,25.0])
e = np.zeros(len(s))
for i in range(len(s)):
e[i] = pr.prcrossval(a, pr.svc([], ("rbf", s[i], 5)), k=10).mean()
plt.figure(figsize=(15,10))
plt.plot(s, e, "-D")
plt.title("Validation Curve")
plt.xlabel("s")
plt.ylabel("error")
plt.show()
```
## Exercise 4.9
## Exercise 4.10
## Exercise 4.11
## Exercise 4.12
|
github_jupyter
|
import numpy as np
import prtools as pr
import matplotlib.pyplot as plt
from jupyterthemes import jtplot
jtplot.style(theme="grade3")
plt.figure(figsize=(8,8))
x = np.array([[0,1], [0,3], [2,0]])
y = np.array([0, 0, 1])
plt.scatter(x[:,0], x[:,1], c=y, marker="x")
plt.plot([2,0], [0,1])
plt.show()
plt.figure(figsize=(8,8))
x = np.array([[0,-1], [0,3], [2,0]])
y = np.array([0, 0, 1])
plt.scatter(x[:,0], x[:,1], c=y, marker="x")
plt.plot([2,0], [0,0])
plt.show()
from sklearn.preprocessing import minmax_scale
from sklearn.preprocessing import maxabs_scale
from sklearn.preprocessing import normalize
plt.figure(figsize=(16,8))
plt.subplot(1,2,1)
x = np.array([[1,1], [2,0], [0,0], [1,0]])
y = np.array([0, 0, 1, 2])
plt.scatter(x[:,0], x[:,1], c=["black", "black", "green", "#ff1493"], marker="x", s=100)
train = pr.prdataset(x[:3], y[:3])
c = pr.svc(train, ("linear", 0, 20))
pr.plotc(c)
plt.subplot(1,2,2)
x_rescale = normalize(x)
plt.scatter(x_rescale[:,0], x_rescale[:,1], c=["black", "black", "green", "#ff1493"], marker="x", s=100)
train_re = pr.prdataset(x_rescale[:3], y[:3])
c = pr.svc(train_re, ("linear", 0, 20))
pr.plotc(c)
plt.show()
plt.figure(figsize=(8,8))
x = np.array([[0,1], [2,4], [1,0]])
y = np.array([0, 0, 1])
data = pr.prdataset(x, y)
w = pr.ldc(data, 1)
w2 = pr.svc(data,("linear", 0, 10))
plt.scatter(x[:,0], x[:,1], c=y, marker="x")
pr.plotc(w)
pr.plotc(w2)
plt.legend()
plt.show()
a = pr.gendatb(n=[20,20], s=1)
plt.figure(figsize=(40,25))
widths = [0.1, 0.5, 0.75, 1, 2.5, 5, 7.5, 10]
for i in range(len(widths)):
svc = pr.svc(a, ("rbf", widths[i], 10))
plt.subplot(2,4,i+1)
pr.scatterd(a)
pr.plotc(svc)
plt.title("widths="+str(widths[i]))
plt.show()
a = pr.gendatb(n=[200,200], s=1)
s = np.array([0.2,0.5,1.0,2.0,5.0,7.0,10.0,25.0])
e = np.zeros(len(s))
for i in range(len(s)):
e[i] = pr.prcrossval(a, pr.svc([], ("rbf", s[i], 5)), k=10).mean()
plt.figure(figsize=(15,10))
plt.plot(s, e, "-D")
plt.title("Validation Curve")
plt.xlabel("s")
plt.ylabel("error")
plt.show()
| 0.413122 | 0.991058 |
99% Accuracy in 30 Minutes
==============
Using the following code, I was able to achieve a score 99.071% on the Kaggle digit recognizer challenge, with a total training time of roughly 30 minutes on my Core i5 MacBook Pro. As of writing this, I'm sitting in 47th place overall.
```
import pyneural
import numpy as np
import pandas as pd
import scipy as sp
import scipy.ndimage as nd
# load the training data and transform it into usable numpy arrays
# Note: you may need to change the path to the data
training_set = pd.read_csv('~/kaggle/digits/train.csv')
labels = np.array(training_set)[:, 0]
features = np.array(training_set)[:, 1:].astype(float) / 255.0
n_rows = features.shape[0]
n_features = features.shape[1]
n_labels = 10
n_steps = 5
labels_expanded = np.zeros((n_rows, n_labels))
for i in xrange(n_rows):
labels_expanded[i][labels[i]] = 1
```
Expanding the training set is the key to success
---------------------------------------------
I had managed to get over 98% accuracy on the test set with PyNeural in the past, but my ability to crack 99% was thanks to insight gained from [this excellent blog post](http://nicklocascio.com/neural-net-mnist-kaggle/).
The MNIST training set supplied in the Kaggle competition is, by computer vision standards, relatively small, so overfitting is a big issue. Nick Locascio points out in his blog post above that you can greatly, and almost trivially, expand the training set using simple transformations of the given training examples.
Nick uses vertical and horizontal shifts, or "nudges", and rotations to multiply the number of training examples. I used the `scipy.ndimage` to perform these transformations, and further expanded on these by adding horizontal and vertical scalings as well.
You could increase the training set further by repeating these transformations with different degrees of shifting, rotation, and scaling; I would imagine there is some point of marginal returns, but I don't actually know where that is.
```
# transform the training set into 42000 28x28 pixel "images"
ims = features.reshape((42000 , 28, 28))
# shift each image down, up, right, and left, respectively, by 2 pixels
dshift = nd.shift(ims, (0, 2, 0), order=0).reshape((42000, 784))
ushift = nd.shift(ims, (0, -2, 0), order=0).reshape((42000, 784))
rshift = nd.shift(ims, (0, 0, 2), order=0).reshape((42000, 784))
lshift = nd.shift(ims, (0, 0, -2), order=0).reshape((42000, 784))
# rotate each image by 15 degrees both counter-clockwise and clockwise
lrotate = nd.rotate(ims, 15, axes=(1,2), reshape=False, prefilter=False).reshape((42000, 784))
rrotate = nd.rotate(ims, -15, axes=(1,2), reshape=False, prefilter=False).reshape((42000, 784))
# scale each image by 1.5 in both the vertical and horizontal directions
vscale = nd.zoom(ims, (1, 1.5, 1), order=0, prefilter=False)[:, 7:-7, :].reshape((42000, 784))
hscale = nd.zoom(ims, (1, 1, 1.5), order=0, prefilter=False)[:, :, 7:-7].reshape((42000, 784))
# combine each of the transformations along with the original training set into a super set
new_features = np.vstack((features, dshift, ushift, rshift, lshift, lrotate, rrotate, vscale, hscale))
new_labels = np.vstack(9 * [labels_expanded])
```
Training the Neural Net
----------------------
To achieve my results, I used a neural network with two hidden layers of 400 nodes each and trained it over 65 epochs using mini-batches of size 100, a learning rate of 0.01, no L2 regularization, and no decay of the learning rate.
```
nn = pyneural.NeuralNet([784, 400, 400, 10])
nn.train(new_features, new_labels, 65, 100, 0.01, 0.0, 1.0)
# check the accuracy on the training set
preds = nn.predict_label(new_features)
correct = np.sum(preds == np.hstack(9 * [labels]))
print "%f%% percent correct " % (100.0 * correct / new_features.shape[0])
# load the test set and make our predictions
test_set = pd.read_csv('~/kaggle/digits/test.csv')
test_features = np.array(test_set).astype(float) / 255.0
test_preds = nn.predict_label(test_features)
# save our predictions to a csv file
df = pd.DataFrame({'ImageId': np.arange(1, len(test_preds) + 1), 'Label': test_preds})
df.to_csv('/Users/taylor/kaggle/digits/exp_set4.csv', index=False)
```
|
github_jupyter
|
import pyneural
import numpy as np
import pandas as pd
import scipy as sp
import scipy.ndimage as nd
# load the training data and transform it into usable numpy arrays
# Note: you may need to change the path to the data
training_set = pd.read_csv('~/kaggle/digits/train.csv')
labels = np.array(training_set)[:, 0]
features = np.array(training_set)[:, 1:].astype(float) / 255.0
n_rows = features.shape[0]
n_features = features.shape[1]
n_labels = 10
n_steps = 5
labels_expanded = np.zeros((n_rows, n_labels))
for i in xrange(n_rows):
labels_expanded[i][labels[i]] = 1
# transform the training set into 42000 28x28 pixel "images"
ims = features.reshape((42000 , 28, 28))
# shift each image down, up, right, and left, respectively, by 2 pixels
dshift = nd.shift(ims, (0, 2, 0), order=0).reshape((42000, 784))
ushift = nd.shift(ims, (0, -2, 0), order=0).reshape((42000, 784))
rshift = nd.shift(ims, (0, 0, 2), order=0).reshape((42000, 784))
lshift = nd.shift(ims, (0, 0, -2), order=0).reshape((42000, 784))
# rotate each image by 15 degrees both counter-clockwise and clockwise
lrotate = nd.rotate(ims, 15, axes=(1,2), reshape=False, prefilter=False).reshape((42000, 784))
rrotate = nd.rotate(ims, -15, axes=(1,2), reshape=False, prefilter=False).reshape((42000, 784))
# scale each image by 1.5 in both the vertical and horizontal directions
vscale = nd.zoom(ims, (1, 1.5, 1), order=0, prefilter=False)[:, 7:-7, :].reshape((42000, 784))
hscale = nd.zoom(ims, (1, 1, 1.5), order=0, prefilter=False)[:, :, 7:-7].reshape((42000, 784))
# combine each of the transformations along with the original training set into a super set
new_features = np.vstack((features, dshift, ushift, rshift, lshift, lrotate, rrotate, vscale, hscale))
new_labels = np.vstack(9 * [labels_expanded])
nn = pyneural.NeuralNet([784, 400, 400, 10])
nn.train(new_features, new_labels, 65, 100, 0.01, 0.0, 1.0)
# check the accuracy on the training set
preds = nn.predict_label(new_features)
correct = np.sum(preds == np.hstack(9 * [labels]))
print "%f%% percent correct " % (100.0 * correct / new_features.shape[0])
# load the test set and make our predictions
test_set = pd.read_csv('~/kaggle/digits/test.csv')
test_features = np.array(test_set).astype(float) / 255.0
test_preds = nn.predict_label(test_features)
# save our predictions to a csv file
df = pd.DataFrame({'ImageId': np.arange(1, len(test_preds) + 1), 'Label': test_preds})
df.to_csv('/Users/taylor/kaggle/digits/exp_set4.csv', index=False)
| 0.496338 | 0.901704 |
```
import os
import json
import pandas as pd
import seaborn as sns
import numpy as np
import math
import pandas as pd
import matplotlib.pyplot as plt
from utils.analysys_utils import list_experiments_in_dir
from utils.derivatives import first_derivative
import torch
sns.set()
experiments_path = os.path.join(os.getcwd(), "runs", "derivative_reg", "sidarthe_extended", "Italy")
figs_path = os.path.join(experiments_path, "figs")
if not os.path.exists(figs_path):
os.makedirs(figs_path)
experiments = list_experiments_in_dir(experiments_path)
def first_derivative_loss(params):
der_loss_total = torch.zeros(1)
for key, param in params.items():
param = torch.tensor(param)
der = first_derivative(param, 1)
der_loss_total = der_loss_total + 0.5 * torch.pow(der, 2)
return torch.mean(der_loss_total).numpy()
x = {
'alpha': [1., 3., 5., 7., 9.]
}
print(first_derivative_loss(x))
# generate data for graphs
"""# take as baseline the exp with 0. regularization
base_val_loss = experiments[0]['final']['best_epoch']['val_loss']
base_der_loss = 0."""
data = []
indexes = []
for exp in experiments:
try: #avoid NaNs
val_loss = exp['final']['best_epoch']['val_loss']
except:
val_loss = np.nan
der_1st_reg = exp['settings']['der_1st_reg']
val_loss = exp['final']['best_epoch']['val_loss']
if der_1st_reg != 0.:
der_1st_loss = exp['final']['best_epoch']['losses']['der_1st']
der_1st_norm = der_1st_loss / der_1st_reg
initial_der_loss = float(first_derivative_loss(exp['settings']['initial_values']))
initial_der_norm = initial_der_loss / der_1st_reg
else:
params = exp['final']['params']
der_1st_loss = der_1st_norm = float(first_derivative_loss(params))
initial_der_loss = float(first_derivative_loss(exp['settings']['initial_values']))
initial_der_norm = initial_der_loss
data.append({
'val_loss': val_loss,
'der_1st_reg': der_1st_reg,
'der_1st': der_1st_loss,
'initial_der_1st': initial_der_loss,
'der_1st_norm': der_1st_norm,
'initial_der_1st_norm': initial_der_norm
})
indexes.append(der_1st_reg)
index = pd.Index(indexes)
df = pd.DataFrame(data, index=index)
d_index = index.sort_values()
df['der_1st_norm']
# plot reg weight vs validation loss
pl, ax = plt.subplots()
#ax.set_yscale('log')
ax.set_xscale('log')
sns.lineplot(data=df, x='der_1st_reg', y='val_loss', ax=ax, legend='brief', label="Regularized")
# add band for baseline (no momentum)
band_data = []
df_0d = df.loc[0.]
for i in range(0,len(df_0d)):
val_loss = df_0d.iloc[i]['val_loss']
band_data.append({
'val_loss': val_loss,
'd': d_index[0]
})
band_data.append({
'val_loss': val_loss,
'd': d_index[-1]
})
band_df = pd.DataFrame(band_data)
plot = sns.lineplot(data=band_df, x='d', ax=ax, y='val_loss', label='None', legend='brief', color='r')
plot.set(xlabel="Derivative weight", ylabel="Validation loss")
plot.get_figure().savefig(os.path.join(figs_path, "val_loss.pdf"),bbox_inches='tight')
# plot reg weight vs der_loss
pl, ax = plt.subplots()
#ax.set_yscale('log')
ax.set_xscale('log')
plot = sns.lineplot(data=df, x='der_1st_reg', y='der_1st_norm', ax=ax, legend='brief', label="Regularized")
#plot = sns.lineplot(data=df, x='der_1st_reg', y='initial_der_1st_norm', ax=ax, legend='brief', label="Regularized (initial)")
plot.set(xlabel="Derivative Regularization", ylabel="Derivative loss")
# add band for baseline (no momentum)
band_data = []
df_0d = df.loc[0.]
for i in range(0,len(df_0d)):
der_1st_loss = df_0d.iloc[i]['der_1st']
der_1st_norm = df_0d.iloc[i]['der_1st_norm']
initial_der_1st_norm = df_0d.iloc[i]['initial_der_1st_norm']
band_data.append({
'der_1st': der_1st_loss,
'der_1st_norm': der_1st_norm,
'initial_der_1st_norm': initial_der_1st_norm,
'd': d_index[0]
})
band_data.append({
'der_1st': der_1st_loss,
'der_1st_norm': der_1st_norm,
'initial_der_1st_norm': initial_der_1st_norm,
'd': d_index[-1]
})
band_df = pd.DataFrame(band_data)
plot = sns.lineplot(data=band_df, x='d', ax=ax, y='der_1st_norm', label='None', legend='brief', color='r')
plot.set(xlabel="Derivative weight", ylabel="Derivative loss")
plot.get_figure().savefig(os.path.join(figs_path, "der_loss.pdf"), bbox_inches='tight')
sns.color_palette()
der_values = [0., 1e5, 1e8, 1e11]
pl, ax = plt.subplots()
#ax.set_yscale('log')
for der in der_values:
df_d = df.loc[der]
plot = sns.scatterplot(data=df_d, x='der_1st_norm', y='val_loss', label=f"{der:.0e}", ax=ax)
plot.set(xlabel="Derivative loss", ylabel="Validation loss")
plot.get_legend().set_title("Weight")
pl, ax = plt.subplots()
#ax.set_yscale('log')
plot = sns.boxplot(data=df, x='der_1st_reg', y='der_1st_norm')
```
|
github_jupyter
|
import os
import json
import pandas as pd
import seaborn as sns
import numpy as np
import math
import pandas as pd
import matplotlib.pyplot as plt
from utils.analysys_utils import list_experiments_in_dir
from utils.derivatives import first_derivative
import torch
sns.set()
experiments_path = os.path.join(os.getcwd(), "runs", "derivative_reg", "sidarthe_extended", "Italy")
figs_path = os.path.join(experiments_path, "figs")
if not os.path.exists(figs_path):
os.makedirs(figs_path)
experiments = list_experiments_in_dir(experiments_path)
def first_derivative_loss(params):
der_loss_total = torch.zeros(1)
for key, param in params.items():
param = torch.tensor(param)
der = first_derivative(param, 1)
der_loss_total = der_loss_total + 0.5 * torch.pow(der, 2)
return torch.mean(der_loss_total).numpy()
x = {
'alpha': [1., 3., 5., 7., 9.]
}
print(first_derivative_loss(x))
# generate data for graphs
"""# take as baseline the exp with 0. regularization
base_val_loss = experiments[0]['final']['best_epoch']['val_loss']
base_der_loss = 0."""
data = []
indexes = []
for exp in experiments:
try: #avoid NaNs
val_loss = exp['final']['best_epoch']['val_loss']
except:
val_loss = np.nan
der_1st_reg = exp['settings']['der_1st_reg']
val_loss = exp['final']['best_epoch']['val_loss']
if der_1st_reg != 0.:
der_1st_loss = exp['final']['best_epoch']['losses']['der_1st']
der_1st_norm = der_1st_loss / der_1st_reg
initial_der_loss = float(first_derivative_loss(exp['settings']['initial_values']))
initial_der_norm = initial_der_loss / der_1st_reg
else:
params = exp['final']['params']
der_1st_loss = der_1st_norm = float(first_derivative_loss(params))
initial_der_loss = float(first_derivative_loss(exp['settings']['initial_values']))
initial_der_norm = initial_der_loss
data.append({
'val_loss': val_loss,
'der_1st_reg': der_1st_reg,
'der_1st': der_1st_loss,
'initial_der_1st': initial_der_loss,
'der_1st_norm': der_1st_norm,
'initial_der_1st_norm': initial_der_norm
})
indexes.append(der_1st_reg)
index = pd.Index(indexes)
df = pd.DataFrame(data, index=index)
d_index = index.sort_values()
df['der_1st_norm']
# plot reg weight vs validation loss
pl, ax = plt.subplots()
#ax.set_yscale('log')
ax.set_xscale('log')
sns.lineplot(data=df, x='der_1st_reg', y='val_loss', ax=ax, legend='brief', label="Regularized")
# add band for baseline (no momentum)
band_data = []
df_0d = df.loc[0.]
for i in range(0,len(df_0d)):
val_loss = df_0d.iloc[i]['val_loss']
band_data.append({
'val_loss': val_loss,
'd': d_index[0]
})
band_data.append({
'val_loss': val_loss,
'd': d_index[-1]
})
band_df = pd.DataFrame(band_data)
plot = sns.lineplot(data=band_df, x='d', ax=ax, y='val_loss', label='None', legend='brief', color='r')
plot.set(xlabel="Derivative weight", ylabel="Validation loss")
plot.get_figure().savefig(os.path.join(figs_path, "val_loss.pdf"),bbox_inches='tight')
# plot reg weight vs der_loss
pl, ax = plt.subplots()
#ax.set_yscale('log')
ax.set_xscale('log')
plot = sns.lineplot(data=df, x='der_1st_reg', y='der_1st_norm', ax=ax, legend='brief', label="Regularized")
#plot = sns.lineplot(data=df, x='der_1st_reg', y='initial_der_1st_norm', ax=ax, legend='brief', label="Regularized (initial)")
plot.set(xlabel="Derivative Regularization", ylabel="Derivative loss")
# add band for baseline (no momentum)
band_data = []
df_0d = df.loc[0.]
for i in range(0,len(df_0d)):
der_1st_loss = df_0d.iloc[i]['der_1st']
der_1st_norm = df_0d.iloc[i]['der_1st_norm']
initial_der_1st_norm = df_0d.iloc[i]['initial_der_1st_norm']
band_data.append({
'der_1st': der_1st_loss,
'der_1st_norm': der_1st_norm,
'initial_der_1st_norm': initial_der_1st_norm,
'd': d_index[0]
})
band_data.append({
'der_1st': der_1st_loss,
'der_1st_norm': der_1st_norm,
'initial_der_1st_norm': initial_der_1st_norm,
'd': d_index[-1]
})
band_df = pd.DataFrame(band_data)
plot = sns.lineplot(data=band_df, x='d', ax=ax, y='der_1st_norm', label='None', legend='brief', color='r')
plot.set(xlabel="Derivative weight", ylabel="Derivative loss")
plot.get_figure().savefig(os.path.join(figs_path, "der_loss.pdf"), bbox_inches='tight')
sns.color_palette()
der_values = [0., 1e5, 1e8, 1e11]
pl, ax = plt.subplots()
#ax.set_yscale('log')
for der in der_values:
df_d = df.loc[der]
plot = sns.scatterplot(data=df_d, x='der_1st_norm', y='val_loss', label=f"{der:.0e}", ax=ax)
plot.set(xlabel="Derivative loss", ylabel="Validation loss")
plot.get_legend().set_title("Weight")
pl, ax = plt.subplots()
#ax.set_yscale('log')
plot = sns.boxplot(data=df, x='der_1st_reg', y='der_1st_norm')
| 0.500977 | 0.273526 |
Based entirely on Abishek's, Bluefool's and Bojan's kernels.
For no ther reason than to reproduce the results.
No original contribution whatsoever.
Didn't do any new stuff other than merging two cells and got LB score 0.399 :P
You are welcome.
Links to orginal kernel https://www.kaggle.com/tunguz/annoying-ab-shreck-and-bluetooth
https://www.kaggle.com/domcastro/let-s-annoy-abhishek
https://www.kaggle.com/abhishek/maybe-something-interesting-here
EAS - All I have done is change the parameters for the LGB. I am also abandoning the OptimizedRounder and have hand-tuned the coefficients to match the distribution of the test predictions to the distribution of the train targets.
** Change Log **
* v19 - added regularization lambda (0.2)
* v21 - lowered lambda (0.05)
* v22 - lowered lambda again (0.02)
* v23 - lambda up to (0.075)
* v24 - lambda back to 0.05, commenting out tfidf code that causes error
* v25 - uncommenting commented out code
* v26 - fixed bug that was throwing error for tf-idf code
* v27 - examining code that had been throwing error, fixing the error lowers the score
* v28 - passing list of categorical features
* v29 - tuning params
* v30 - using more tfidf features
* v31, 32 - playing with tfidf params
* v35 - back to 120 tfidf svd features
* v36 - reverting to baseline and not using OptimizedRounder, hand-tuning coefs instead
```
import json
import scipy as sp
import pandas as pd
import numpy as np
from functools import partial
from math import sqrt
from sklearn.metrics import cohen_kappa_score, mean_squared_error
from sklearn.metrics import confusion_matrix as sk_cmatrix
from sklearn.model_selection import StratifiedKFold
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.decomposition import TruncatedSVD
from collections import Counter
import lightgbm as lgb
np.random.seed(369)
# The following 3 functions have been taken from Ben Hamner's github repository
# https://github.com/benhamner/Metrics
def confusion_matrix(rater_a, rater_b, min_rating=None, max_rating=None):
"""
Returns the confusion matrix between rater's ratings
"""
assert(len(rater_a) == len(rater_b))
if min_rating is None:
min_rating = min(rater_a + rater_b)
if max_rating is None:
max_rating = max(rater_a + rater_b)
num_ratings = int(max_rating - min_rating + 1)
conf_mat = [[0 for i in range(num_ratings)]
for j in range(num_ratings)]
for a, b in zip(rater_a, rater_b):
conf_mat[a - min_rating][b - min_rating] += 1
return conf_mat
def histogram(ratings, min_rating=None, max_rating=None):
"""
Returns the counts of each type of rating that a rater made
"""
if min_rating is None:
min_rating = min(ratings)
if max_rating is None:
max_rating = max(ratings)
num_ratings = int(max_rating - min_rating + 1)
hist_ratings = [0 for x in range(num_ratings)]
for r in ratings:
hist_ratings[r - min_rating] += 1
return hist_ratings
def quadratic_weighted_kappa(y, y_pred):
"""
Calculates the quadratic weighted kappa
axquadratic_weighted_kappa calculates the quadratic weighted kappa
value, which is a measure of inter-rater agreement between two raters
that provide discrete numeric ratings. Potential values range from -1
(representing complete disagreement) to 1 (representing complete
agreement). A kappa value of 0 is expected if all agreement is due to
chance.
quadratic_weighted_kappa(rater_a, rater_b), where rater_a and rater_b
each correspond to a list of integer ratings. These lists must have the
same length.
The ratings should be integers, and it is assumed that they contain
the complete range of possible ratings.
quadratic_weighted_kappa(X, min_rating, max_rating), where min_rating
is the minimum possible rating, and max_rating is the maximum possible
rating
"""
rater_a = y
rater_b = y_pred
min_rating=None
max_rating=None
rater_a = np.array(rater_a, dtype=int)
rater_b = np.array(rater_b, dtype=int)
assert(len(rater_a) == len(rater_b))
if min_rating is None:
min_rating = min(min(rater_a), min(rater_b))
if max_rating is None:
max_rating = max(max(rater_a), max(rater_b))
conf_mat = confusion_matrix(rater_a, rater_b,
min_rating, max_rating)
num_ratings = len(conf_mat)
num_scored_items = float(len(rater_a))
hist_rater_a = histogram(rater_a, min_rating, max_rating)
hist_rater_b = histogram(rater_b, min_rating, max_rating)
numerator = 0.0
denominator = 0.0
for i in range(num_ratings):
for j in range(num_ratings):
expected_count = (hist_rater_a[i] * hist_rater_b[j]
/ num_scored_items)
d = pow(i - j, 2.0) / pow(num_ratings - 1, 2.0)
numerator += d * conf_mat[i][j] / num_scored_items
denominator += d * expected_count / num_scored_items
return (1.0 - numerator / denominator)
class OptimizedRounder(object):
def __init__(self):
self.coef_ = 0
def _kappa_loss(self, coef, X, y):
X_p = np.copy(X)
for i, pred in enumerate(X_p):
if pred < coef[0]:
X_p[i] = 0
elif pred >= coef[0] and pred < coef[1]:
X_p[i] = 1
elif pred >= coef[1] and pred < coef[2]:
X_p[i] = 2
elif pred >= coef[2] and pred < coef[3]:
X_p[i] = 3
else:
X_p[i] = 4
ll = quadratic_weighted_kappa(y, X_p)
return -ll
def fit(self, X, y):
loss_partial = partial(self._kappa_loss, X=X, y=y)
initial_coef = [0.5, 1.5, 2.5, 3.5]
self.coef_ = sp.optimize.minimize(loss_partial, initial_coef, method='nelder-mead')
def predict(self, X, coef):
X_p = np.copy(X)
for i, pred in enumerate(X_p):
if pred < coef[0]:
X_p[i] = 0
elif pred >= coef[0] and pred < coef[1]:
X_p[i] = 1
elif pred >= coef[1] and pred < coef[2]:
X_p[i] = 2
elif pred >= coef[2] and pred < coef[3]:
X_p[i] = 3
else:
X_p[i] = 4
return X_p
def coefficients(self):
return self.coef_['x']
def rmse(actual, predicted):
return sqrt(mean_squared_error(actual, predicted))
```
## Load Data
```
print('Train')
train = pd.read_csv("../input/train/train.csv")
print(train.shape)
print('Test')
test = pd.read_csv("../input/test/test.csv")
print(test.shape)
print('Breeds')
breeds = pd.read_csv("../input/breed_labels.csv")
print(breeds.shape)
print('Colors')
colors = pd.read_csv("../input/color_labels.csv")
print(colors.shape)
print('States')
states = pd.read_csv("../input/state_labels.csv")
print(states.shape)
target = train['AdoptionSpeed']
train_id = train['PetID']
test_id = test['PetID']
train.drop(['AdoptionSpeed', 'PetID'], axis=1, inplace=True)
test.drop(['PetID'], axis=1, inplace=True)
doc_sent_mag = []
doc_sent_score = []
nf_count = 0
for pet in train_id:
try:
with open('../input/train_sentiment/' + pet + '.json', 'r') as f:
sentiment = json.load(f)
doc_sent_mag.append(sentiment['documentSentiment']['magnitude'])
doc_sent_score.append(sentiment['documentSentiment']['score'])
except FileNotFoundError:
nf_count += 1
doc_sent_mag.append(-1)
doc_sent_score.append(-1)
train.loc[:, 'doc_sent_mag'] = doc_sent_mag
train.loc[:, 'doc_sent_score'] = doc_sent_score
doc_sent_mag = []
doc_sent_score = []
nf_count = 0
for pet in test_id:
try:
with open('../input/test_sentiment/' + pet + '.json', 'r') as f:
sentiment = json.load(f)
doc_sent_mag.append(sentiment['documentSentiment']['magnitude'])
doc_sent_score.append(sentiment['documentSentiment']['score'])
except FileNotFoundError:
nf_count += 1
doc_sent_mag.append(-1)
doc_sent_score.append(-1)
test.loc[:, 'doc_sent_mag'] = doc_sent_mag
test.loc[:, 'doc_sent_score'] = doc_sent_score
train_desc = train.Description.fillna("none").values
test_desc = test.Description.fillna("none").values
tfv = TfidfVectorizer(min_df=2, max_features=None,
strip_accents='unicode', analyzer='word', token_pattern=r'(?u)\b\w+\b',
ngram_range=(1, 3), use_idf=1, smooth_idf=1, sublinear_tf=1,
)
# Fit TFIDF
tfv.fit(list(train_desc))
X = tfv.transform(train_desc)
X_test = tfv.transform(test_desc)
print("X (tfidf):", X.shape)
svd = TruncatedSVD(n_components=200)
svd.fit(X)
# print(svd.explained_variance_ratio_.sum())
# print(svd.explained_variance_ratio_)
X = svd.transform(X)
print("X (svd):", X.shape)
# X = pd.DataFrame(X, columns=['svd_{}'.format(i) for i in range(120)])
# train = pd.concat((train, X), axis=1)
# X_test = svd.transform(X_test)
# X_test = pd.DataFrame(X_test, columns=['svd_{}'.format(i) for i in range(120)])
# test = pd.concat((test, X_test), axis=1)
print("train:", train.shape)
## WITHOUT ERROR FIXED
train_desc = train.Description.fillna("none").values
test_desc = test.Description.fillna("none").values
tfv = TfidfVectorizer(min_df=3, max_features=10000,
strip_accents='unicode', analyzer='word', token_pattern=r'\w{1,}',
ngram_range=(1, 3), use_idf=1, smooth_idf=1, sublinear_tf=1,
stop_words = 'english')
# Fit TFIDF
tfv.fit(list(train_desc))
X = tfv.transform(train_desc)
X_test = tfv.transform(test_desc)
print("X (tfidf):", X.shape)
svd = TruncatedSVD(n_components=120)
svd.fit(X)
# print(svd.explained_variance_ratio_.sum())
# print(svd.explained_variance_ratio_)
X = svd.transform(X)
print("X (svd):", X.shape)
X = pd.DataFrame(X, columns=['svd_{}'.format(i) for i in range(120)])
train = pd.concat((train, X), axis=1)
X_test = svd.transform(X_test)
X_test = pd.DataFrame(X_test, columns=['svd_{}'.format(i) for i in range(120)])
test = pd.concat((test, X_test), axis=1)
print("train:", train.shape)
vertex_xs = []
vertex_ys = []
bounding_confidences = []
bounding_importance_fracs = []
dominant_blues = []
dominant_greens = []
dominant_reds = []
dominant_pixel_fracs = []
dominant_scores = []
label_descriptions = []
label_scores = []
nf_count = 0
nl_count = 0
for pet in train_id:
try:
with open('../input/train_metadata/' + pet + '-1.json', 'r') as f:
data = json.load(f)
vertex_x = data['cropHintsAnnotation']['cropHints'][0]['boundingPoly']['vertices'][2]['x']
vertex_xs.append(vertex_x)
vertex_y = data['cropHintsAnnotation']['cropHints'][0]['boundingPoly']['vertices'][2]['y']
vertex_ys.append(vertex_y)
bounding_confidence = data['cropHintsAnnotation']['cropHints'][0]['confidence']
bounding_confidences.append(bounding_confidence)
bounding_importance_frac = data['cropHintsAnnotation']['cropHints'][0].get('importanceFraction', -1)
bounding_importance_fracs.append(bounding_importance_frac)
dominant_blue = data['imagePropertiesAnnotation']['dominantColors']['colors'][0]['color']['blue']
dominant_blues.append(dominant_blue)
dominant_green = data['imagePropertiesAnnotation']['dominantColors']['colors'][0]['color']['green']
dominant_greens.append(dominant_green)
dominant_red = data['imagePropertiesAnnotation']['dominantColors']['colors'][0]['color']['red']
dominant_reds.append(dominant_red)
dominant_pixel_frac = data['imagePropertiesAnnotation']['dominantColors']['colors'][0]['pixelFraction']
dominant_pixel_fracs.append(dominant_pixel_frac)
dominant_score = data['imagePropertiesAnnotation']['dominantColors']['colors'][0]['score']
dominant_scores.append(dominant_score)
if data.get('labelAnnotations'):
label_description = data['labelAnnotations'][0]['description']
label_descriptions.append(label_description)
label_score = data['labelAnnotations'][0]['score']
label_scores.append(label_score)
else:
nl_count += 1
label_descriptions.append('nothing')
label_scores.append(-1)
except FileNotFoundError:
nf_count += 1
vertex_xs.append(-1)
vertex_ys.append(-1)
bounding_confidences.append(-1)
bounding_importance_fracs.append(-1)
dominant_blues.append(-1)
dominant_greens.append(-1)
dominant_reds.append(-1)
dominant_pixel_fracs.append(-1)
dominant_scores.append(-1)
label_descriptions.append('nothing')
label_scores.append(-1)
print(nf_count)
print(nl_count)
train.loc[:, 'vertex_x'] = vertex_xs
train.loc[:, 'vertex_y'] = vertex_ys
train.loc[:, 'bounding_confidence'] = bounding_confidences
train.loc[:, 'bounding_importance'] = bounding_importance_fracs
train.loc[:, 'dominant_blue'] = dominant_blues
train.loc[:, 'dominant_green'] = dominant_greens
train.loc[:, 'dominant_red'] = dominant_reds
train.loc[:, 'dominant_pixel_frac'] = dominant_pixel_fracs
train.loc[:, 'dominant_score'] = dominant_scores
train.loc[:, 'label_description'] = label_descriptions
train.loc[:, 'label_score'] = label_scores
vertex_xs = []
vertex_ys = []
bounding_confidences = []
bounding_importance_fracs = []
dominant_blues = []
dominant_greens = []
dominant_reds = []
dominant_pixel_fracs = []
dominant_scores = []
label_descriptions = []
label_scores = []
nf_count = 0
nl_count = 0
for pet in test_id:
try:
with open('../input/test_metadata/' + pet + '-1.json', 'r') as f:
data = json.load(f)
vertex_x = data['cropHintsAnnotation']['cropHints'][0]['boundingPoly']['vertices'][2]['x']
vertex_xs.append(vertex_x)
vertex_y = data['cropHintsAnnotation']['cropHints'][0]['boundingPoly']['vertices'][2]['y']
vertex_ys.append(vertex_y)
bounding_confidence = data['cropHintsAnnotation']['cropHints'][0]['confidence']
bounding_confidences.append(bounding_confidence)
bounding_importance_frac = data['cropHintsAnnotation']['cropHints'][0].get('importanceFraction', -1)
bounding_importance_fracs.append(bounding_importance_frac)
dominant_blue = data['imagePropertiesAnnotation']['dominantColors']['colors'][0]['color']['blue']
dominant_blues.append(dominant_blue)
dominant_green = data['imagePropertiesAnnotation']['dominantColors']['colors'][0]['color']['green']
dominant_greens.append(dominant_green)
dominant_red = data['imagePropertiesAnnotation']['dominantColors']['colors'][0]['color']['red']
dominant_reds.append(dominant_red)
dominant_pixel_frac = data['imagePropertiesAnnotation']['dominantColors']['colors'][0]['pixelFraction']
dominant_pixel_fracs.append(dominant_pixel_frac)
dominant_score = data['imagePropertiesAnnotation']['dominantColors']['colors'][0]['score']
dominant_scores.append(dominant_score)
if data.get('labelAnnotations'):
label_description = data['labelAnnotations'][0]['description']
label_descriptions.append(label_description)
label_score = data['labelAnnotations'][0]['score']
label_scores.append(label_score)
else:
nl_count += 1
label_descriptions.append('nothing')
label_scores.append(-1)
except FileNotFoundError:
nf_count += 1
vertex_xs.append(-1)
vertex_ys.append(-1)
bounding_confidences.append(-1)
bounding_importance_fracs.append(-1)
dominant_blues.append(-1)
dominant_greens.append(-1)
dominant_reds.append(-1)
dominant_pixel_fracs.append(-1)
dominant_scores.append(-1)
label_descriptions.append('nothing')
label_scores.append(-1)
print(nf_count)
test.loc[:, 'vertex_x'] = vertex_xs
test.loc[:, 'vertex_y'] = vertex_ys
test.loc[:, 'bounding_confidence'] = bounding_confidences
test.loc[:, 'bounding_importance'] = bounding_importance_fracs
test.loc[:, 'dominant_blue'] = dominant_blues
test.loc[:, 'dominant_green'] = dominant_greens
test.loc[:, 'dominant_red'] = dominant_reds
test.loc[:, 'dominant_pixel_frac'] = dominant_pixel_fracs
test.loc[:, 'dominant_score'] = dominant_scores
test.loc[:, 'label_description'] = label_descriptions
test.loc[:, 'label_score'] = label_scores
train.drop(['Name', 'RescuerID', 'Description'], axis=1, inplace=True)
test.drop(['Name', 'RescuerID', 'Description'], axis=1, inplace=True)
numeric_cols = ['Age', 'Quantity', 'Fee', 'VideoAmt', 'PhotoAmt', 'AdoptionSpeed', 'doc_sent_mag', 'doc_sent_score', 'dominant_score', 'dominant_pixel_frac', 'dominant_red', 'dominant_green', 'dominant_blue', 'bounding_importance', 'bounding_confidence', 'vertex_x', 'vertex_y', 'label_score'] + ['svd_{}'.format(i) for i in range(120)]
cat_cols = list(set(train.columns) - set(numeric_cols))
train.loc[:, cat_cols] = train[cat_cols].astype('category')
test.loc[:, cat_cols] = test[cat_cols].astype('category')
print(train.shape)
print(test.shape)
# get the categorical features
foo = train.dtypes
cat_feature_names = foo[foo == "category"]
cat_features = [train.columns.get_loc(c) for c in train.columns if c in cat_feature_names]
N_SPLITS = 3
def run_cv_model(train, test, target, model_fn, params={}, eval_fn=None, label='model'):
kf = StratifiedKFold(n_splits=N_SPLITS, random_state=42, shuffle=True)
fold_splits = kf.split(train, target)
cv_scores = []
qwk_scores = []
pred_full_test = 0
pred_train = np.zeros((train.shape[0], N_SPLITS))
all_coefficients = np.zeros((N_SPLITS, 4))
feature_importance_df = pd.DataFrame()
i = 1
for dev_index, val_index in fold_splits:
print('Started ' + label + ' fold ' + str(i) + '/' + str(N_SPLITS))
if isinstance(train, pd.DataFrame):
dev_X, val_X = train.iloc[dev_index], train.iloc[val_index]
dev_y, val_y = target[dev_index], target[val_index]
else:
dev_X, val_X = train[dev_index], train[val_index]
dev_y, val_y = target[dev_index], target[val_index]
params2 = params.copy()
pred_val_y, pred_test_y, importances, coefficients, qwk = model_fn(dev_X, dev_y, val_X, val_y, test, params2)
pred_full_test = pred_full_test + pred_test_y
pred_train[val_index] = pred_val_y
all_coefficients[i-1, :] = coefficients
if eval_fn is not None:
cv_score = eval_fn(val_y, pred_val_y)
cv_scores.append(cv_score)
qwk_scores.append(qwk)
print(label + ' cv score {}: RMSE {} QWK {}'.format(i, cv_score, qwk))
fold_importance_df = pd.DataFrame()
fold_importance_df['feature'] = train.columns.values
fold_importance_df['importance'] = importances
fold_importance_df['fold'] = i
feature_importance_df = pd.concat([feature_importance_df, fold_importance_df], axis=0)
i += 1
print('{} cv RMSE scores : {}'.format(label, cv_scores))
print('{} cv mean RMSE score : {}'.format(label, np.mean(cv_scores)))
print('{} cv std RMSE score : {}'.format(label, np.mean(cv_scores)))
print('{} cv QWK scores : {}'.format(label, qwk_scores))
print('{} cv mean QWK score : {}'.format(label, np.mean(qwk_scores)))
print('{} cv std QWK score : {}'.format(label, np.std(qwk_scores)))
pred_full_test = pred_full_test / float(N_SPLITS)
results = {'label': label,
'train': pred_train, 'test': pred_full_test,
'cv': cv_scores, 'qwk': qwk_scores,
'importance': feature_importance_df,
'coefficients': all_coefficients}
return results
params = {'application': 'regression',
'boosting': 'gbdt',
'metric': 'rmse',
'num_leaves': 70,
'max_depth': 8,
'learning_rate': 0.02,
'bagging_fraction': 0.85,
'feature_fraction': 0.8,
'min_split_gain': 0.02,
'min_child_samples': 150,
'min_child_weight': 0.02,
'lambda_l2': 0.05,
'verbosity': -1,
'data_random_seed': 17,
'early_stop': 100,
'verbose_eval': 100,
'num_rounds': 10000}
def runLGB(train_X, train_y, test_X, test_y, test_X2, params):
print('Prep LGB')
d_train = lgb.Dataset(train_X, label=train_y)
d_valid = lgb.Dataset(test_X, label=test_y)
watchlist = [d_train, d_valid]
print('Train LGB')
num_rounds = params.pop('num_rounds')
verbose_eval = params.pop('verbose_eval')
early_stop = None
if params.get('early_stop'):
early_stop = params.pop('early_stop')
model = lgb.train(params,
train_set=d_train,
num_boost_round=num_rounds,
valid_sets=watchlist,
verbose_eval=verbose_eval,
categorical_feature=list(cat_features),
early_stopping_rounds=early_stop)
print('Predict 1/2')
pred_test_y = model.predict(test_X, num_iteration=model.best_iteration)
optR = OptimizedRounder()
optR.fit(pred_test_y, test_y)
coefficients = optR.coefficients()
pred_test_y_k = optR.predict(pred_test_y, coefficients)
print("Valid Counts = ", Counter(test_y))
print("Predicted Counts = ", Counter(pred_test_y_k))
print("Coefficients = ", coefficients)
qwk = quadratic_weighted_kappa(test_y, pred_test_y_k)
print("QWK = ", qwk)
print('Predict 2/2')
pred_test_y2 = model.predict(test_X2, num_iteration=model.best_iteration)
return pred_test_y.reshape(-1, 1), pred_test_y2.reshape(-1, 1), model.feature_importance(), coefficients, qwk
results = run_cv_model(train, test, target, runLGB, params, rmse, 'lgb')
imports = results['importance'].groupby('feature')['feature', 'importance'].mean().reset_index()
imports.sort_values('importance', ascending=False)
optR = OptimizedRounder()
coefficients_ = np.mean(results['coefficients'], axis=0)
print(coefficients_)
# manually adjust coefs
coefficients_[0] = 1.645
coefficients_[1] = 2.115
coefficients_[3] = 2.84
train_predictions = [r[0] for r in results['train']]
train_predictions = optR.predict(train_predictions, coefficients_).astype(int)
Counter(train_predictions)
optR = OptimizedRounder()
coefficients_ = np.mean(results['coefficients'], axis=0)
print(coefficients_)
# manually adjust coefs
coefficients_[0] = 1.645
coefficients_[1] = 2.115
coefficients_[3] = 2.84
test_predictions = [r[0] for r in results['test']]
test_predictions = optR.predict(test_predictions, coefficients_).astype(int)
Counter(test_predictions)
print("True Distribution:")
print(pd.value_counts(target, normalize=True).sort_index())
print("Test Predicted Distribution:")
print(pd.value_counts(test_predictions, normalize=True).sort_index())
print("Train Predicted Distribution:")
print(pd.value_counts(train_predictions, normalize=True).sort_index())
pd.DataFrame(sk_cmatrix(target, train_predictions), index=list(range(5)), columns=list(range(5)))
quadratic_weighted_kappa(target, train_predictions)
rmse(target, [r[0] for r in results['train']])
submission = pd.DataFrame({'PetID': test_id, 'AdoptionSpeed': test_predictions})
submission.head()
submission.to_csv('submission.csv', index=False)
```
|
github_jupyter
|
import json
import scipy as sp
import pandas as pd
import numpy as np
from functools import partial
from math import sqrt
from sklearn.metrics import cohen_kappa_score, mean_squared_error
from sklearn.metrics import confusion_matrix as sk_cmatrix
from sklearn.model_selection import StratifiedKFold
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.decomposition import TruncatedSVD
from collections import Counter
import lightgbm as lgb
np.random.seed(369)
# The following 3 functions have been taken from Ben Hamner's github repository
# https://github.com/benhamner/Metrics
def confusion_matrix(rater_a, rater_b, min_rating=None, max_rating=None):
"""
Returns the confusion matrix between rater's ratings
"""
assert(len(rater_a) == len(rater_b))
if min_rating is None:
min_rating = min(rater_a + rater_b)
if max_rating is None:
max_rating = max(rater_a + rater_b)
num_ratings = int(max_rating - min_rating + 1)
conf_mat = [[0 for i in range(num_ratings)]
for j in range(num_ratings)]
for a, b in zip(rater_a, rater_b):
conf_mat[a - min_rating][b - min_rating] += 1
return conf_mat
def histogram(ratings, min_rating=None, max_rating=None):
"""
Returns the counts of each type of rating that a rater made
"""
if min_rating is None:
min_rating = min(ratings)
if max_rating is None:
max_rating = max(ratings)
num_ratings = int(max_rating - min_rating + 1)
hist_ratings = [0 for x in range(num_ratings)]
for r in ratings:
hist_ratings[r - min_rating] += 1
return hist_ratings
def quadratic_weighted_kappa(y, y_pred):
"""
Calculates the quadratic weighted kappa
axquadratic_weighted_kappa calculates the quadratic weighted kappa
value, which is a measure of inter-rater agreement between two raters
that provide discrete numeric ratings. Potential values range from -1
(representing complete disagreement) to 1 (representing complete
agreement). A kappa value of 0 is expected if all agreement is due to
chance.
quadratic_weighted_kappa(rater_a, rater_b), where rater_a and rater_b
each correspond to a list of integer ratings. These lists must have the
same length.
The ratings should be integers, and it is assumed that they contain
the complete range of possible ratings.
quadratic_weighted_kappa(X, min_rating, max_rating), where min_rating
is the minimum possible rating, and max_rating is the maximum possible
rating
"""
rater_a = y
rater_b = y_pred
min_rating=None
max_rating=None
rater_a = np.array(rater_a, dtype=int)
rater_b = np.array(rater_b, dtype=int)
assert(len(rater_a) == len(rater_b))
if min_rating is None:
min_rating = min(min(rater_a), min(rater_b))
if max_rating is None:
max_rating = max(max(rater_a), max(rater_b))
conf_mat = confusion_matrix(rater_a, rater_b,
min_rating, max_rating)
num_ratings = len(conf_mat)
num_scored_items = float(len(rater_a))
hist_rater_a = histogram(rater_a, min_rating, max_rating)
hist_rater_b = histogram(rater_b, min_rating, max_rating)
numerator = 0.0
denominator = 0.0
for i in range(num_ratings):
for j in range(num_ratings):
expected_count = (hist_rater_a[i] * hist_rater_b[j]
/ num_scored_items)
d = pow(i - j, 2.0) / pow(num_ratings - 1, 2.0)
numerator += d * conf_mat[i][j] / num_scored_items
denominator += d * expected_count / num_scored_items
return (1.0 - numerator / denominator)
class OptimizedRounder(object):
def __init__(self):
self.coef_ = 0
def _kappa_loss(self, coef, X, y):
X_p = np.copy(X)
for i, pred in enumerate(X_p):
if pred < coef[0]:
X_p[i] = 0
elif pred >= coef[0] and pred < coef[1]:
X_p[i] = 1
elif pred >= coef[1] and pred < coef[2]:
X_p[i] = 2
elif pred >= coef[2] and pred < coef[3]:
X_p[i] = 3
else:
X_p[i] = 4
ll = quadratic_weighted_kappa(y, X_p)
return -ll
def fit(self, X, y):
loss_partial = partial(self._kappa_loss, X=X, y=y)
initial_coef = [0.5, 1.5, 2.5, 3.5]
self.coef_ = sp.optimize.minimize(loss_partial, initial_coef, method='nelder-mead')
def predict(self, X, coef):
X_p = np.copy(X)
for i, pred in enumerate(X_p):
if pred < coef[0]:
X_p[i] = 0
elif pred >= coef[0] and pred < coef[1]:
X_p[i] = 1
elif pred >= coef[1] and pred < coef[2]:
X_p[i] = 2
elif pred >= coef[2] and pred < coef[3]:
X_p[i] = 3
else:
X_p[i] = 4
return X_p
def coefficients(self):
return self.coef_['x']
def rmse(actual, predicted):
return sqrt(mean_squared_error(actual, predicted))
print('Train')
train = pd.read_csv("../input/train/train.csv")
print(train.shape)
print('Test')
test = pd.read_csv("../input/test/test.csv")
print(test.shape)
print('Breeds')
breeds = pd.read_csv("../input/breed_labels.csv")
print(breeds.shape)
print('Colors')
colors = pd.read_csv("../input/color_labels.csv")
print(colors.shape)
print('States')
states = pd.read_csv("../input/state_labels.csv")
print(states.shape)
target = train['AdoptionSpeed']
train_id = train['PetID']
test_id = test['PetID']
train.drop(['AdoptionSpeed', 'PetID'], axis=1, inplace=True)
test.drop(['PetID'], axis=1, inplace=True)
doc_sent_mag = []
doc_sent_score = []
nf_count = 0
for pet in train_id:
try:
with open('../input/train_sentiment/' + pet + '.json', 'r') as f:
sentiment = json.load(f)
doc_sent_mag.append(sentiment['documentSentiment']['magnitude'])
doc_sent_score.append(sentiment['documentSentiment']['score'])
except FileNotFoundError:
nf_count += 1
doc_sent_mag.append(-1)
doc_sent_score.append(-1)
train.loc[:, 'doc_sent_mag'] = doc_sent_mag
train.loc[:, 'doc_sent_score'] = doc_sent_score
doc_sent_mag = []
doc_sent_score = []
nf_count = 0
for pet in test_id:
try:
with open('../input/test_sentiment/' + pet + '.json', 'r') as f:
sentiment = json.load(f)
doc_sent_mag.append(sentiment['documentSentiment']['magnitude'])
doc_sent_score.append(sentiment['documentSentiment']['score'])
except FileNotFoundError:
nf_count += 1
doc_sent_mag.append(-1)
doc_sent_score.append(-1)
test.loc[:, 'doc_sent_mag'] = doc_sent_mag
test.loc[:, 'doc_sent_score'] = doc_sent_score
train_desc = train.Description.fillna("none").values
test_desc = test.Description.fillna("none").values
tfv = TfidfVectorizer(min_df=2, max_features=None,
strip_accents='unicode', analyzer='word', token_pattern=r'(?u)\b\w+\b',
ngram_range=(1, 3), use_idf=1, smooth_idf=1, sublinear_tf=1,
)
# Fit TFIDF
tfv.fit(list(train_desc))
X = tfv.transform(train_desc)
X_test = tfv.transform(test_desc)
print("X (tfidf):", X.shape)
svd = TruncatedSVD(n_components=200)
svd.fit(X)
# print(svd.explained_variance_ratio_.sum())
# print(svd.explained_variance_ratio_)
X = svd.transform(X)
print("X (svd):", X.shape)
# X = pd.DataFrame(X, columns=['svd_{}'.format(i) for i in range(120)])
# train = pd.concat((train, X), axis=1)
# X_test = svd.transform(X_test)
# X_test = pd.DataFrame(X_test, columns=['svd_{}'.format(i) for i in range(120)])
# test = pd.concat((test, X_test), axis=1)
print("train:", train.shape)
## WITHOUT ERROR FIXED
train_desc = train.Description.fillna("none").values
test_desc = test.Description.fillna("none").values
tfv = TfidfVectorizer(min_df=3, max_features=10000,
strip_accents='unicode', analyzer='word', token_pattern=r'\w{1,}',
ngram_range=(1, 3), use_idf=1, smooth_idf=1, sublinear_tf=1,
stop_words = 'english')
# Fit TFIDF
tfv.fit(list(train_desc))
X = tfv.transform(train_desc)
X_test = tfv.transform(test_desc)
print("X (tfidf):", X.shape)
svd = TruncatedSVD(n_components=120)
svd.fit(X)
# print(svd.explained_variance_ratio_.sum())
# print(svd.explained_variance_ratio_)
X = svd.transform(X)
print("X (svd):", X.shape)
X = pd.DataFrame(X, columns=['svd_{}'.format(i) for i in range(120)])
train = pd.concat((train, X), axis=1)
X_test = svd.transform(X_test)
X_test = pd.DataFrame(X_test, columns=['svd_{}'.format(i) for i in range(120)])
test = pd.concat((test, X_test), axis=1)
print("train:", train.shape)
vertex_xs = []
vertex_ys = []
bounding_confidences = []
bounding_importance_fracs = []
dominant_blues = []
dominant_greens = []
dominant_reds = []
dominant_pixel_fracs = []
dominant_scores = []
label_descriptions = []
label_scores = []
nf_count = 0
nl_count = 0
for pet in train_id:
try:
with open('../input/train_metadata/' + pet + '-1.json', 'r') as f:
data = json.load(f)
vertex_x = data['cropHintsAnnotation']['cropHints'][0]['boundingPoly']['vertices'][2]['x']
vertex_xs.append(vertex_x)
vertex_y = data['cropHintsAnnotation']['cropHints'][0]['boundingPoly']['vertices'][2]['y']
vertex_ys.append(vertex_y)
bounding_confidence = data['cropHintsAnnotation']['cropHints'][0]['confidence']
bounding_confidences.append(bounding_confidence)
bounding_importance_frac = data['cropHintsAnnotation']['cropHints'][0].get('importanceFraction', -1)
bounding_importance_fracs.append(bounding_importance_frac)
dominant_blue = data['imagePropertiesAnnotation']['dominantColors']['colors'][0]['color']['blue']
dominant_blues.append(dominant_blue)
dominant_green = data['imagePropertiesAnnotation']['dominantColors']['colors'][0]['color']['green']
dominant_greens.append(dominant_green)
dominant_red = data['imagePropertiesAnnotation']['dominantColors']['colors'][0]['color']['red']
dominant_reds.append(dominant_red)
dominant_pixel_frac = data['imagePropertiesAnnotation']['dominantColors']['colors'][0]['pixelFraction']
dominant_pixel_fracs.append(dominant_pixel_frac)
dominant_score = data['imagePropertiesAnnotation']['dominantColors']['colors'][0]['score']
dominant_scores.append(dominant_score)
if data.get('labelAnnotations'):
label_description = data['labelAnnotations'][0]['description']
label_descriptions.append(label_description)
label_score = data['labelAnnotations'][0]['score']
label_scores.append(label_score)
else:
nl_count += 1
label_descriptions.append('nothing')
label_scores.append(-1)
except FileNotFoundError:
nf_count += 1
vertex_xs.append(-1)
vertex_ys.append(-1)
bounding_confidences.append(-1)
bounding_importance_fracs.append(-1)
dominant_blues.append(-1)
dominant_greens.append(-1)
dominant_reds.append(-1)
dominant_pixel_fracs.append(-1)
dominant_scores.append(-1)
label_descriptions.append('nothing')
label_scores.append(-1)
print(nf_count)
print(nl_count)
train.loc[:, 'vertex_x'] = vertex_xs
train.loc[:, 'vertex_y'] = vertex_ys
train.loc[:, 'bounding_confidence'] = bounding_confidences
train.loc[:, 'bounding_importance'] = bounding_importance_fracs
train.loc[:, 'dominant_blue'] = dominant_blues
train.loc[:, 'dominant_green'] = dominant_greens
train.loc[:, 'dominant_red'] = dominant_reds
train.loc[:, 'dominant_pixel_frac'] = dominant_pixel_fracs
train.loc[:, 'dominant_score'] = dominant_scores
train.loc[:, 'label_description'] = label_descriptions
train.loc[:, 'label_score'] = label_scores
vertex_xs = []
vertex_ys = []
bounding_confidences = []
bounding_importance_fracs = []
dominant_blues = []
dominant_greens = []
dominant_reds = []
dominant_pixel_fracs = []
dominant_scores = []
label_descriptions = []
label_scores = []
nf_count = 0
nl_count = 0
for pet in test_id:
try:
with open('../input/test_metadata/' + pet + '-1.json', 'r') as f:
data = json.load(f)
vertex_x = data['cropHintsAnnotation']['cropHints'][0]['boundingPoly']['vertices'][2]['x']
vertex_xs.append(vertex_x)
vertex_y = data['cropHintsAnnotation']['cropHints'][0]['boundingPoly']['vertices'][2]['y']
vertex_ys.append(vertex_y)
bounding_confidence = data['cropHintsAnnotation']['cropHints'][0]['confidence']
bounding_confidences.append(bounding_confidence)
bounding_importance_frac = data['cropHintsAnnotation']['cropHints'][0].get('importanceFraction', -1)
bounding_importance_fracs.append(bounding_importance_frac)
dominant_blue = data['imagePropertiesAnnotation']['dominantColors']['colors'][0]['color']['blue']
dominant_blues.append(dominant_blue)
dominant_green = data['imagePropertiesAnnotation']['dominantColors']['colors'][0]['color']['green']
dominant_greens.append(dominant_green)
dominant_red = data['imagePropertiesAnnotation']['dominantColors']['colors'][0]['color']['red']
dominant_reds.append(dominant_red)
dominant_pixel_frac = data['imagePropertiesAnnotation']['dominantColors']['colors'][0]['pixelFraction']
dominant_pixel_fracs.append(dominant_pixel_frac)
dominant_score = data['imagePropertiesAnnotation']['dominantColors']['colors'][0]['score']
dominant_scores.append(dominant_score)
if data.get('labelAnnotations'):
label_description = data['labelAnnotations'][0]['description']
label_descriptions.append(label_description)
label_score = data['labelAnnotations'][0]['score']
label_scores.append(label_score)
else:
nl_count += 1
label_descriptions.append('nothing')
label_scores.append(-1)
except FileNotFoundError:
nf_count += 1
vertex_xs.append(-1)
vertex_ys.append(-1)
bounding_confidences.append(-1)
bounding_importance_fracs.append(-1)
dominant_blues.append(-1)
dominant_greens.append(-1)
dominant_reds.append(-1)
dominant_pixel_fracs.append(-1)
dominant_scores.append(-1)
label_descriptions.append('nothing')
label_scores.append(-1)
print(nf_count)
test.loc[:, 'vertex_x'] = vertex_xs
test.loc[:, 'vertex_y'] = vertex_ys
test.loc[:, 'bounding_confidence'] = bounding_confidences
test.loc[:, 'bounding_importance'] = bounding_importance_fracs
test.loc[:, 'dominant_blue'] = dominant_blues
test.loc[:, 'dominant_green'] = dominant_greens
test.loc[:, 'dominant_red'] = dominant_reds
test.loc[:, 'dominant_pixel_frac'] = dominant_pixel_fracs
test.loc[:, 'dominant_score'] = dominant_scores
test.loc[:, 'label_description'] = label_descriptions
test.loc[:, 'label_score'] = label_scores
train.drop(['Name', 'RescuerID', 'Description'], axis=1, inplace=True)
test.drop(['Name', 'RescuerID', 'Description'], axis=1, inplace=True)
numeric_cols = ['Age', 'Quantity', 'Fee', 'VideoAmt', 'PhotoAmt', 'AdoptionSpeed', 'doc_sent_mag', 'doc_sent_score', 'dominant_score', 'dominant_pixel_frac', 'dominant_red', 'dominant_green', 'dominant_blue', 'bounding_importance', 'bounding_confidence', 'vertex_x', 'vertex_y', 'label_score'] + ['svd_{}'.format(i) for i in range(120)]
cat_cols = list(set(train.columns) - set(numeric_cols))
train.loc[:, cat_cols] = train[cat_cols].astype('category')
test.loc[:, cat_cols] = test[cat_cols].astype('category')
print(train.shape)
print(test.shape)
# get the categorical features
foo = train.dtypes
cat_feature_names = foo[foo == "category"]
cat_features = [train.columns.get_loc(c) for c in train.columns if c in cat_feature_names]
N_SPLITS = 3
def run_cv_model(train, test, target, model_fn, params={}, eval_fn=None, label='model'):
kf = StratifiedKFold(n_splits=N_SPLITS, random_state=42, shuffle=True)
fold_splits = kf.split(train, target)
cv_scores = []
qwk_scores = []
pred_full_test = 0
pred_train = np.zeros((train.shape[0], N_SPLITS))
all_coefficients = np.zeros((N_SPLITS, 4))
feature_importance_df = pd.DataFrame()
i = 1
for dev_index, val_index in fold_splits:
print('Started ' + label + ' fold ' + str(i) + '/' + str(N_SPLITS))
if isinstance(train, pd.DataFrame):
dev_X, val_X = train.iloc[dev_index], train.iloc[val_index]
dev_y, val_y = target[dev_index], target[val_index]
else:
dev_X, val_X = train[dev_index], train[val_index]
dev_y, val_y = target[dev_index], target[val_index]
params2 = params.copy()
pred_val_y, pred_test_y, importances, coefficients, qwk = model_fn(dev_X, dev_y, val_X, val_y, test, params2)
pred_full_test = pred_full_test + pred_test_y
pred_train[val_index] = pred_val_y
all_coefficients[i-1, :] = coefficients
if eval_fn is not None:
cv_score = eval_fn(val_y, pred_val_y)
cv_scores.append(cv_score)
qwk_scores.append(qwk)
print(label + ' cv score {}: RMSE {} QWK {}'.format(i, cv_score, qwk))
fold_importance_df = pd.DataFrame()
fold_importance_df['feature'] = train.columns.values
fold_importance_df['importance'] = importances
fold_importance_df['fold'] = i
feature_importance_df = pd.concat([feature_importance_df, fold_importance_df], axis=0)
i += 1
print('{} cv RMSE scores : {}'.format(label, cv_scores))
print('{} cv mean RMSE score : {}'.format(label, np.mean(cv_scores)))
print('{} cv std RMSE score : {}'.format(label, np.mean(cv_scores)))
print('{} cv QWK scores : {}'.format(label, qwk_scores))
print('{} cv mean QWK score : {}'.format(label, np.mean(qwk_scores)))
print('{} cv std QWK score : {}'.format(label, np.std(qwk_scores)))
pred_full_test = pred_full_test / float(N_SPLITS)
results = {'label': label,
'train': pred_train, 'test': pred_full_test,
'cv': cv_scores, 'qwk': qwk_scores,
'importance': feature_importance_df,
'coefficients': all_coefficients}
return results
params = {'application': 'regression',
'boosting': 'gbdt',
'metric': 'rmse',
'num_leaves': 70,
'max_depth': 8,
'learning_rate': 0.02,
'bagging_fraction': 0.85,
'feature_fraction': 0.8,
'min_split_gain': 0.02,
'min_child_samples': 150,
'min_child_weight': 0.02,
'lambda_l2': 0.05,
'verbosity': -1,
'data_random_seed': 17,
'early_stop': 100,
'verbose_eval': 100,
'num_rounds': 10000}
def runLGB(train_X, train_y, test_X, test_y, test_X2, params):
print('Prep LGB')
d_train = lgb.Dataset(train_X, label=train_y)
d_valid = lgb.Dataset(test_X, label=test_y)
watchlist = [d_train, d_valid]
print('Train LGB')
num_rounds = params.pop('num_rounds')
verbose_eval = params.pop('verbose_eval')
early_stop = None
if params.get('early_stop'):
early_stop = params.pop('early_stop')
model = lgb.train(params,
train_set=d_train,
num_boost_round=num_rounds,
valid_sets=watchlist,
verbose_eval=verbose_eval,
categorical_feature=list(cat_features),
early_stopping_rounds=early_stop)
print('Predict 1/2')
pred_test_y = model.predict(test_X, num_iteration=model.best_iteration)
optR = OptimizedRounder()
optR.fit(pred_test_y, test_y)
coefficients = optR.coefficients()
pred_test_y_k = optR.predict(pred_test_y, coefficients)
print("Valid Counts = ", Counter(test_y))
print("Predicted Counts = ", Counter(pred_test_y_k))
print("Coefficients = ", coefficients)
qwk = quadratic_weighted_kappa(test_y, pred_test_y_k)
print("QWK = ", qwk)
print('Predict 2/2')
pred_test_y2 = model.predict(test_X2, num_iteration=model.best_iteration)
return pred_test_y.reshape(-1, 1), pred_test_y2.reshape(-1, 1), model.feature_importance(), coefficients, qwk
results = run_cv_model(train, test, target, runLGB, params, rmse, 'lgb')
imports = results['importance'].groupby('feature')['feature', 'importance'].mean().reset_index()
imports.sort_values('importance', ascending=False)
optR = OptimizedRounder()
coefficients_ = np.mean(results['coefficients'], axis=0)
print(coefficients_)
# manually adjust coefs
coefficients_[0] = 1.645
coefficients_[1] = 2.115
coefficients_[3] = 2.84
train_predictions = [r[0] for r in results['train']]
train_predictions = optR.predict(train_predictions, coefficients_).astype(int)
Counter(train_predictions)
optR = OptimizedRounder()
coefficients_ = np.mean(results['coefficients'], axis=0)
print(coefficients_)
# manually adjust coefs
coefficients_[0] = 1.645
coefficients_[1] = 2.115
coefficients_[3] = 2.84
test_predictions = [r[0] for r in results['test']]
test_predictions = optR.predict(test_predictions, coefficients_).astype(int)
Counter(test_predictions)
print("True Distribution:")
print(pd.value_counts(target, normalize=True).sort_index())
print("Test Predicted Distribution:")
print(pd.value_counts(test_predictions, normalize=True).sort_index())
print("Train Predicted Distribution:")
print(pd.value_counts(train_predictions, normalize=True).sort_index())
pd.DataFrame(sk_cmatrix(target, train_predictions), index=list(range(5)), columns=list(range(5)))
quadratic_weighted_kappa(target, train_predictions)
rmse(target, [r[0] for r in results['train']])
submission = pd.DataFrame({'PetID': test_id, 'AdoptionSpeed': test_predictions})
submission.head()
submission.to_csv('submission.csv', index=False)
| 0.662469 | 0.783057 |
# Probabilistic Graphical Models
## Introduction
* <a href="https://www.coursera.org/learn/probabilistic-graphical-models"> Daphne Koller: Probabilistic Graphical Models: Representation </a>
### Generative Models v/s Discriminative models:
* Generative: Model the joint distribution of all the random variables (X) i.e P(X).
* Discriminative: Model the conditional probability of some of the variables (say Y), given other ones (say X) i.e P(Y|X)
* **Q**. Suppose $ | Y | = n $ and $ | X | = m $. Assume all the random variables are binary. How many parameters would you have to learn if you want to build a:
* Discriminative model?
* Generative model?
* You can obtain a discriminative model from a generative one.
* Discriminative models involve lesser complexity than the Generative ones.
### Why Probabilistic Models?
* Probabilistic models capture the uncertainty in the result as well.
* Some people believe that there's an inherent stochasticity in nature. So, everything is probabilistic.
* Probability theory has been well studied. Can utilize the Statistical learning toolbox.
### Why Graph Representation?
* Sparse Parametrization
* Efficient Algorithms to reason from the data.
### Two types of models:
* Bayesian Networks: Directed Models
* Markov Networks: Undirected Models
### Where have these been used?
* Bayesian Networks: Medical Diagonsis
* Markov Networks: Image Segmentation
## Quiz
**Q1**
<img src="./files/Q1.png" width=70%> </img>
---------------
**Q2.**
<img src="./files/bayesian_net.png" width=80%> </img>
* Calculate:
$ P(d^0, i^1, g^3, s^1, l^1) $
$ P(d^0, i^1,l^1) $
$ P(G | i^0, d^0) $
$ P(G | i^0) $
---------------
<img src="./files/Q2.png" width=50%> </img>
**Q3** Calculate P(Accident = 1|President = 1) and P(Accident = 1| Traffic=1, President = 1)
----------------
## Learning Bayesian Networks
* Chapter 6 - Paramaeter learning: Binary Variables from <a href="https://www.amazon.com/Learning-Bayesian-Networks-Richard-Neapolitan/dp/0130125342"> Learning Bayesian Networks</a>.
### Structure learning v/s parameter learning
* Two types of learning: Structure and Parameter learning. Both of them can be learnt from data.
* Linear Regression. The use of regularization as a means to achieve structure learning and parameter learning simultaneously.
* Here we'll use that we have structure given to us. All we need to do is learn the Parameters.
### Beta distribution
* $$ \Gamma(x+1)=\int_{0}^{\infty}t^{x}e^{-t} $$
* $$ \Gamma(x+1)=x \; \Gamma(x) $$
* $$ B(x,y)=\int_{0}^{1}t^{x-1}(1-t)^{y-1} dt = \frac{\Gamma(x) \; \Gamma(y)}{\Gamma(x+y)} $$
* $$ Beta(x \; | \; \alpha, \beta) = \frac{x^{\alpha-1} \; \; (1-x)^{ \; \beta-1} }{ B(\alpha,\beta) } $$
### Coin: Single node Bayesian Net
* You have a coin. Assume that it is a biased coin and $ Side \sim Bernoulli(f). $
<img src="./files/single_node.png" width=30%/>
* Your task is to learn $ f $. Let $ F $ be a random variable for the parameter $ f $. You have certain beliefs about the value of parameter $ f $ and you encode it in the prior $ F \sim Beta(f; \alpha, \beta) $.
* **Q.** Suppose your Prior is $ F \sim Beta(f; a, b) $. What is $ P(Side=heads) $ ?
#### Theorem 6.1
Suppose $ X $ is a random variable with two values 0 and 1, $ F $ is another random variable such that
$$ P(X = 1 | F = f) = f $$
Then,
$$ P(X = 1) = E(F) $$
-------------------------------------------------
Now in order to learn the parameter, you let's model $ F $ along with the our variable $ X $.
This is called as the **Augmented Bayesian Network** representation of our earlier Bayesian Net.
Let's $ F $ follow the density function $ \rho $ which is called the **prior density function of the parameters**.
Now by the Bayes' rule we have $$ P(F=f\;|\;D=d) = \frac{P(D=d \;|\; F=f) \rho (f)} {P(D=d)} $$
In this way, we'll learn the distribution of $ F $ from the data.
<img src="./files/augmented_net.png" width=20%/>
--------------------------------------------------
#### Definition 6.2
Suppose
1. We have a set of random variables (or random vectors) $ D = { X^{(1)} , X^{(2)} , . . . X^{(M)} } $ such that each $ X^{(h)} $ has the same space.
2. There is a random variable $ F $ with density function $ \rho $ such that the $X^{(h)}$ s
are I.I.D. for all values $ f $ of $ F $.
Then $ D $ is called a sample of size $ M $ with parameter $ F $.
Given a sample, the density function $ \rho $ is called the **prior density function of the parameters** relative to the sample. It represents our prior belief concerning the unknown parameters.
Given a sample, the marginal distribution of each $ X^{(h)} $ is the same. This distribution is called the **prior distribution** relative to the sample. It represents our prior belief concerning each trial.
--------------------------------------------------
#### Definition 6.3
Suppose we have a sample of size $ M $ such that
1. each $ X^{(h)} $ has space $ {0, 1} $;
2. $ F $ has space $ [0, 1] $, and for $ 1 ≤ h ≤ M \quad P (X^{(h)} = 1| f) = f $.
Then $D$ is called a binomial sample of size $M$ with parameter $F$.
<img src="./files/sample.png" width=50%/>
#### Theorem 6.2
Suppose
1. $D$ is a binomial sample of size $M$ with parameter $F$.
2. We have a set of values $d = {x^{(1)} , x^{(2)} , . . . x^{(M)}}$ of the variables in $D$ (The set $d$ is called our data set (or simply data)).
3. $s$ is the number of variables in $d$ equal to $1$.
4. $t$ is the number of variables in $d$ equal to $0$.
Then
$ P(d) = E(F^s (1 − F)^t) $.
** Proof **: Marginalization: $$ P(D = d) = \int_0^1 P(D=d \;|\; F = f) \rho(F = f) df $$
#### Corollary 6.2
If the conditions in Theorem 6.2 hold, and $F$ has a beta distribution with parameters $ a, b, N = a + b$, then,
$$ P(d) = \frac{\Gamma (N)}{\Gamma(N + M)} \frac{\Gamma(a + s) \Gamma (b + t)}{\Gamma(a) \Gamma(b)} $$
**Proof**: Application of Lemma 6.4
#### Lemma 6.4
Suppose $F$ has a beta distribution with parameters $a, b$ and $ N = a + b$. $s$ and $t$ are two integers ≥ 0, and $M = s + t$. Then
$$ E[F^s \; [1 − F]^t] = \frac{\Gamma (N)}{\Gamma(N + M)} \frac{\Gamma(a + s) \Gamma (b + t)}{\Gamma(a) \Gamma(b)} $$
#### Lemma 6.5
Suppose $F$ has a beta distribution with parameters $a, b$ and $ N = a + b$. $s$ and $t$ are two integers ≥ 0, and $M = s + t$. Then
$$\frac{f^s (1 − f)^t \rho(f)}{E(F^s [1 − F]^t )} = Beta(f ; a + s, b + t) $$
#### Theorem 6.3
If the conditions in Theorem 6.2 hold, then
$$ ρ(f|d) = \frac{f^s (1 − f )^t ρ(f)}{E(F^s[1 − F ]^t)} $$
where $\rho(f|d) $ denotes the density function of $F$ conditional on $D = d$.
### Conclusion:
#### Corollary 6.3
Suppose the conditions in Theorem 6.2 hold, and F has a $ Beta $ distribution with parameters $a, b$ and $ N = a + b$. That is, $$ ρ(f) = Beta(f ; a, b) $$
Then, $$ ρ(f |d) = beta(f ; a + s, b + t) $$
#### Theorem 6.4
Suppose the conditions in Theorem 6.2 hold, and we create a binomial sample of size $M + 1$ by adding another variable $X^{(M+1)} $ . Then if $ D $ is the binomial sample of size $ M $, the updated distribution relative to the sample and data $ d $ is given by
$$P(X^{(M +1)} = 1| d) = E(F |d) $$
#### Corollary 6.4
If the conditions in Theorem 6.4 hold and F has a beta distribution with parameters $a, b$ and N = a + b, then
$$ P(X^{(M +1)} = 1| d) = \frac{a + s}{N + M} = \frac{a + s} {a + s + b + t} $$
### Next class
* Marginalization (Some Problems)
* Connections and building blocks of Bayesian Networks
* Conditional Independences: d - Separation
* Multinomial distribution
|
github_jupyter
|
# Probabilistic Graphical Models
## Introduction
* <a href="https://www.coursera.org/learn/probabilistic-graphical-models"> Daphne Koller: Probabilistic Graphical Models: Representation </a>
### Generative Models v/s Discriminative models:
* Generative: Model the joint distribution of all the random variables (X) i.e P(X).
* Discriminative: Model the conditional probability of some of the variables (say Y), given other ones (say X) i.e P(Y|X)
* **Q**. Suppose $ | Y | = n $ and $ | X | = m $. Assume all the random variables are binary. How many parameters would you have to learn if you want to build a:
* Discriminative model?
* Generative model?
* You can obtain a discriminative model from a generative one.
* Discriminative models involve lesser complexity than the Generative ones.
### Why Probabilistic Models?
* Probabilistic models capture the uncertainty in the result as well.
* Some people believe that there's an inherent stochasticity in nature. So, everything is probabilistic.
* Probability theory has been well studied. Can utilize the Statistical learning toolbox.
### Why Graph Representation?
* Sparse Parametrization
* Efficient Algorithms to reason from the data.
### Two types of models:
* Bayesian Networks: Directed Models
* Markov Networks: Undirected Models
### Where have these been used?
* Bayesian Networks: Medical Diagonsis
* Markov Networks: Image Segmentation
## Quiz
**Q1**
<img src="./files/Q1.png" width=70%> </img>
---------------
**Q2.**
<img src="./files/bayesian_net.png" width=80%> </img>
* Calculate:
$ P(d^0, i^1, g^3, s^1, l^1) $
$ P(d^0, i^1,l^1) $
$ P(G | i^0, d^0) $
$ P(G | i^0) $
---------------
<img src="./files/Q2.png" width=50%> </img>
**Q3** Calculate P(Accident = 1|President = 1) and P(Accident = 1| Traffic=1, President = 1)
----------------
## Learning Bayesian Networks
* Chapter 6 - Paramaeter learning: Binary Variables from <a href="https://www.amazon.com/Learning-Bayesian-Networks-Richard-Neapolitan/dp/0130125342"> Learning Bayesian Networks</a>.
### Structure learning v/s parameter learning
* Two types of learning: Structure and Parameter learning. Both of them can be learnt from data.
* Linear Regression. The use of regularization as a means to achieve structure learning and parameter learning simultaneously.
* Here we'll use that we have structure given to us. All we need to do is learn the Parameters.
### Beta distribution
* $$ \Gamma(x+1)=\int_{0}^{\infty}t^{x}e^{-t} $$
* $$ \Gamma(x+1)=x \; \Gamma(x) $$
* $$ B(x,y)=\int_{0}^{1}t^{x-1}(1-t)^{y-1} dt = \frac{\Gamma(x) \; \Gamma(y)}{\Gamma(x+y)} $$
* $$ Beta(x \; | \; \alpha, \beta) = \frac{x^{\alpha-1} \; \; (1-x)^{ \; \beta-1} }{ B(\alpha,\beta) } $$
### Coin: Single node Bayesian Net
* You have a coin. Assume that it is a biased coin and $ Side \sim Bernoulli(f). $
<img src="./files/single_node.png" width=30%/>
* Your task is to learn $ f $. Let $ F $ be a random variable for the parameter $ f $. You have certain beliefs about the value of parameter $ f $ and you encode it in the prior $ F \sim Beta(f; \alpha, \beta) $.
* **Q.** Suppose your Prior is $ F \sim Beta(f; a, b) $. What is $ P(Side=heads) $ ?
#### Theorem 6.1
Suppose $ X $ is a random variable with two values 0 and 1, $ F $ is another random variable such that
$$ P(X = 1 | F = f) = f $$
Then,
$$ P(X = 1) = E(F) $$
-------------------------------------------------
Now in order to learn the parameter, you let's model $ F $ along with the our variable $ X $.
This is called as the **Augmented Bayesian Network** representation of our earlier Bayesian Net.
Let's $ F $ follow the density function $ \rho $ which is called the **prior density function of the parameters**.
Now by the Bayes' rule we have $$ P(F=f\;|\;D=d) = \frac{P(D=d \;|\; F=f) \rho (f)} {P(D=d)} $$
In this way, we'll learn the distribution of $ F $ from the data.
<img src="./files/augmented_net.png" width=20%/>
--------------------------------------------------
#### Definition 6.2
Suppose
1. We have a set of random variables (or random vectors) $ D = { X^{(1)} , X^{(2)} , . . . X^{(M)} } $ such that each $ X^{(h)} $ has the same space.
2. There is a random variable $ F $ with density function $ \rho $ such that the $X^{(h)}$ s
are I.I.D. for all values $ f $ of $ F $.
Then $ D $ is called a sample of size $ M $ with parameter $ F $.
Given a sample, the density function $ \rho $ is called the **prior density function of the parameters** relative to the sample. It represents our prior belief concerning the unknown parameters.
Given a sample, the marginal distribution of each $ X^{(h)} $ is the same. This distribution is called the **prior distribution** relative to the sample. It represents our prior belief concerning each trial.
--------------------------------------------------
#### Definition 6.3
Suppose we have a sample of size $ M $ such that
1. each $ X^{(h)} $ has space $ {0, 1} $;
2. $ F $ has space $ [0, 1] $, and for $ 1 ≤ h ≤ M \quad P (X^{(h)} = 1| f) = f $.
Then $D$ is called a binomial sample of size $M$ with parameter $F$.
<img src="./files/sample.png" width=50%/>
#### Theorem 6.2
Suppose
1. $D$ is a binomial sample of size $M$ with parameter $F$.
2. We have a set of values $d = {x^{(1)} , x^{(2)} , . . . x^{(M)}}$ of the variables in $D$ (The set $d$ is called our data set (or simply data)).
3. $s$ is the number of variables in $d$ equal to $1$.
4. $t$ is the number of variables in $d$ equal to $0$.
Then
$ P(d) = E(F^s (1 − F)^t) $.
** Proof **: Marginalization: $$ P(D = d) = \int_0^1 P(D=d \;|\; F = f) \rho(F = f) df $$
#### Corollary 6.2
If the conditions in Theorem 6.2 hold, and $F$ has a beta distribution with parameters $ a, b, N = a + b$, then,
$$ P(d) = \frac{\Gamma (N)}{\Gamma(N + M)} \frac{\Gamma(a + s) \Gamma (b + t)}{\Gamma(a) \Gamma(b)} $$
**Proof**: Application of Lemma 6.4
#### Lemma 6.4
Suppose $F$ has a beta distribution with parameters $a, b$ and $ N = a + b$. $s$ and $t$ are two integers ≥ 0, and $M = s + t$. Then
$$ E[F^s \; [1 − F]^t] = \frac{\Gamma (N)}{\Gamma(N + M)} \frac{\Gamma(a + s) \Gamma (b + t)}{\Gamma(a) \Gamma(b)} $$
#### Lemma 6.5
Suppose $F$ has a beta distribution with parameters $a, b$ and $ N = a + b$. $s$ and $t$ are two integers ≥ 0, and $M = s + t$. Then
$$\frac{f^s (1 − f)^t \rho(f)}{E(F^s [1 − F]^t )} = Beta(f ; a + s, b + t) $$
#### Theorem 6.3
If the conditions in Theorem 6.2 hold, then
$$ ρ(f|d) = \frac{f^s (1 − f )^t ρ(f)}{E(F^s[1 − F ]^t)} $$
where $\rho(f|d) $ denotes the density function of $F$ conditional on $D = d$.
### Conclusion:
#### Corollary 6.3
Suppose the conditions in Theorem 6.2 hold, and F has a $ Beta $ distribution with parameters $a, b$ and $ N = a + b$. That is, $$ ρ(f) = Beta(f ; a, b) $$
Then, $$ ρ(f |d) = beta(f ; a + s, b + t) $$
#### Theorem 6.4
Suppose the conditions in Theorem 6.2 hold, and we create a binomial sample of size $M + 1$ by adding another variable $X^{(M+1)} $ . Then if $ D $ is the binomial sample of size $ M $, the updated distribution relative to the sample and data $ d $ is given by
$$P(X^{(M +1)} = 1| d) = E(F |d) $$
#### Corollary 6.4
If the conditions in Theorem 6.4 hold and F has a beta distribution with parameters $a, b$ and N = a + b, then
$$ P(X^{(M +1)} = 1| d) = \frac{a + s}{N + M} = \frac{a + s} {a + s + b + t} $$
### Next class
* Marginalization (Some Problems)
* Connections and building blocks of Bayesian Networks
* Conditional Independences: d - Separation
* Multinomial distribution
| 0.888976 | 0.989161 |
In this exercise, we'll build out a mock version of the system described above. We won't use a real database and some of the components will be simple mocks (we won't trade real money on real trading platforms), but it will help you get a feel for designing a pipeline-based system. We'll build out the Scraper, Cleaner, Deduplicator, Analyzer, and DecisionMaker components of the system we described previously.
1. Define some example URLs of news articles that might be interesting as shown in the code below.
```
uber_url = "https://www.reuters.com/article/us-uber-lawsuit-california/uber-is-sued-over-resistance-to-california-gig-employment-law-idUSKCN1VX1VE"
apple_url = "https://www.reuters.com/article/us-apple-macbook/apple-refreshes-macbook-pro-laptop-with-16-inch-screen-idUSKBN1XN1V8"
apple_url2 = "https://www.reuters.com/article/us-apple-macbook/apple-refreshes-macbook-pro-laptop-with-16-inch-screen-idUSKBN1XN1V8"
article_urls = [uber_url, apple_url, apple_url2]
```
We define only three URLs, all from a single source. Two of them are the same. In the last line, we add all three URLs to an array.
2. Import the `requests` `string` libraries and the `Counter` module from `collections`, as shown in the code below.
```
import requests
import string
from collections import Counter
```
We imported three libraries that we'll use in various components.
3. Define a Scraper class that can fetch news articles and extract the full contents, including HTML, as shown in the code below.
```
class Scraper:
def fetch_news(self, urls):
article_contents = []
for url in urls:
try:
contents = requests.get(url).text
article_contents.append(contents)
except Exception as e:
print(e)
return article_contents
```
Here we defined a Scraper class which can fetch news articles. We give it a list of urls, and it loops through these. For each url, it attempts to fetch it with the `requests` library and adds the contents from that page to an array. Finally, it returns the array. Note that the Scraper class takes in an array of URLs (which we have) and outputs an array containing the contents of the page.
4. Define an `is_clean` function which we'll use in our `Cleaner` module, as shown in the code below.
```
def is_clean(word):
blacklist = {"var", "img", "e", "void"}
if not word:
return False
if word in blacklist:
return False
for i, letter in enumerate(word):
if i > 0 and letter in string.ascii_uppercase:
return False
if letter not in string.ascii_letters:
return False
return True
```
This function is outside of the main `Cleaner` module. It looks at a word and decides if it is part of an article or not. We use a very naive method for this. If the word is in our blacklist, we discard it, as it is probably part of the JavaScript content of the article. If the word is blank, we also discard it.
If both of the above tests pass, we check if the word has any uppercase letters that are not the first letter. If it does, it is probably a function name. Finally, we check if all of the letters in the word are part of the English alphabet. If any other characters are present, we discard the word.
5. Define the full Cleaner module which uses the `is_clean` function, as shown in the code below.
```
class Cleaner:
def clean_articles(self, articles):
clean_articles = []
for article in articles:
clean_words = []
try:
for word in article.split(" "):
if is_clean(word):
clean_words.append(word)
except Exception as e:
print(e)
clean_articles.append(' '.join(clean_words))
return clean_articles
```
In this code, we define a `Cleaner` module with a `clean_articles` function. This function takes the list of artiles that the Scraper produced and loops through it. For each article, it breaks it into words, and keeps only the clean words. It then joins these together again, adds them to a different array, and finally returns the array of cleaned articles.
6. Create the Deduplicator module as shown in the code below.
```
class Deduplicator:
def deduplicate_articles(self, articles):
seen_articles = set()
deduplicated_articles = []
for article in articles:
if hash(article) in seen_articles:
continue
else:
seen_articles.add(hash(article))
deduplicated_articles.append(article)
return deduplicated_articles
```
This module takes in a list of clean articles and checks if any of them are duplicated. It keeps only a single copy of each one and returns a new list, without any duplicates.
7. Create the Analyzer module, as shown in the code below.
```
class Analyzer:
good_words = {"unveiled", "available", "faster", "stable"}
bad_words = {"sued", "defiance", "violation"}
def extract_entities_and_sentiment(self, articles):
entity_score_pairs = []
for article in articles:
score = 0
entities = []
for word in article.split(" "):
if word[0] == word[0].upper():
entities.append(word)
if word.lower() in self.good_words:
score += 1
elif word.lower() in self.bad_words:
score -= 1
main_entities = [i[0] for i in Counter(entities).most_common(2)]
entity_score_pair = (main_entities, score)
entity_score_pairs.append(entity_score_pair)
return entity_score_pairs
```
The Analyzer module is a bit more complicated than the previous modules becuase it has two jobs: two extract the entities from an aritcle, and to extract a sentiment score. We first define two very limited lists of 'good words' and 'bad words'. If the article is talking about a new product being unveiled, that is a good sign. If a company is being sued, that is probably bad. We then define a function that loops through each clean article (with duplicates already removed) and looks at every word.
For each word, it checks if the word is an entity (it guesses that it is if it starts with capital letter). It then checks if the word is regarded as a 'good' or 'bad' word. If it is good, it adds 1 to the score. If it is bad, it removes one. If the word does not appear in either list, it leaves the score as is.
Finally, it finds the two most common entities mentioned in the article, and creates a data structure with both of these entities and the overall score. It returns this as output.
8. Now create the Decision Maker module using the following code.
```
class DecisionMaker:
target_companies = set(['Apple', 'Uber', 'Google'])
def make_decisions(self, entity_score_pairs):
decisions = []
for entities, score in entity_score_pairs:
for entity in entities:
if entity in self.target_companies:
quantity = abs(score)
order = "Buy" if score > 0 else "Sell"
decision = (order, quantity, entity)
decisions.append(decision)
return decisions
```
This module has a set of target companies. These are the companies whose stock we want to trade. It takes as input the entity and score pairs that we created in the Analyzer module and turns these into structured trading decisions. If the score is positive for a given entity, it buys that stock. If it is negative, it sells the stock. The more positive or negative the score is, the more stock it buys or sells. It returns a list of decisions as output.
9. Initialise all components by running the following code.
```
scraper = Scraper()
cleaner = Cleaner()
deduplicator = Deduplicator()
analyzer = Analyzer()
decision_maker = DecisionMaker()
```
We created all 5 components, and they are now ready to be tested.
10. Fetch the news articles with the scraper and print out an excerpt, by running the following code.
```
contents = scraper.fetch_news(article_urls)
contents[0][:500]
```
We ran our Scraper and output the first 500 characters of the first article. We can see it fetched content, but that this is messy and full of HTML tags and other information that is not part of the article.
11. Pass these article to the cleaner for cleaning, as in the following code.
```
clean_articles = cleaner.clean_articles(contents)
clean_articles[0][:500]
```
We ran our cleaner and output the first 500 characters of the first article. We can see that a lot of the junk is removed. It isn't perfect as there is still some content in the beginning that is not from the article, but cleaning is a tricky task and we at least we can see that the real content appears near the beginning.
12. Check how many articles we have, run the deduplicator and then check the count of the articles again, as in the code below.
```
print(len(clean_articles))
deduplicated = deduplicator.deduplicate_articles(clean_articles)
print(len(deduplicated))
```
We printed out the length of our `clean_articles` and noted that we have 3, one for each of our original URLs. We then ran our deduplicator, which removed the duplicate article, leaving us with the text of 2 articles.
13. Run our analyzer on our clean deduplicated articles, as shown in the code below.
```
entity_score_pairs = analyzer.extract_entities_and_sentiment(deduplicated)
print(entity_score_pairs)
```
We ran the analyzer on our articles. We can see that it figured out that the first article was mainly about Uber and California and that it had a negative sentiment. The second article was mainly about Apple and "Pro" (the article talks a lot about the new Macbook Pro), and has a positive sentiment.
Pass this information to our Decision Maker to create trade instructions, as shown in the code below.
```
decisions = decision_maker.make_decisions(entity_score_pairs)
print(decisions)
```
We created two decisions from our entity and sentiment pairs. The decision maker wants to sell 18 shares of Uber and buy 16 shares of Apple.
All of our components are very naive and would not work well in a real-world case. The Scraper downloads articles that we give it, but can't find them for itself. The cleaner doesn't even attempt to parse HTML, and keeps a lot of content that is not relevant. It also discards a lot of "real" words: those with punctuation, brand names with capitals in the middle of a word, and more.
Our deduplicator only deals with exact duplicates, but in real cases, often there are small differences between articles that are almost the same. Therefore hashing is not a good strategy here. Our Analyzer uses some hand picked wordlists that are relevant mainly to the articles that we chose, and it has a very naive entity extractor, relying only on capital letters.
Finally, our decision maker does not take information from all articles into account. If there is one very positive article on Apple and 10 slightly negative ones, it might still decide to buy more stock than it sells.
With all of the above in mind, we still can see that the system is both very modular and pipeline based. Each component is responsible only for a single aspect of the entire system, and any of them can be improved without affecting the others, as long as the input and output formats remain the same. The output of each component is fed as input into another component, leaving us with a very neat pipeline.
This is great for maintaining and understanding the system, but also good for reproducing results. Often in machine learning systems, reproducibility is important, and by having a structured pipeline you can always feed the same data in to get the same results out.
|
github_jupyter
|
uber_url = "https://www.reuters.com/article/us-uber-lawsuit-california/uber-is-sued-over-resistance-to-california-gig-employment-law-idUSKCN1VX1VE"
apple_url = "https://www.reuters.com/article/us-apple-macbook/apple-refreshes-macbook-pro-laptop-with-16-inch-screen-idUSKBN1XN1V8"
apple_url2 = "https://www.reuters.com/article/us-apple-macbook/apple-refreshes-macbook-pro-laptop-with-16-inch-screen-idUSKBN1XN1V8"
article_urls = [uber_url, apple_url, apple_url2]
import requests
import string
from collections import Counter
class Scraper:
def fetch_news(self, urls):
article_contents = []
for url in urls:
try:
contents = requests.get(url).text
article_contents.append(contents)
except Exception as e:
print(e)
return article_contents
def is_clean(word):
blacklist = {"var", "img", "e", "void"}
if not word:
return False
if word in blacklist:
return False
for i, letter in enumerate(word):
if i > 0 and letter in string.ascii_uppercase:
return False
if letter not in string.ascii_letters:
return False
return True
class Cleaner:
def clean_articles(self, articles):
clean_articles = []
for article in articles:
clean_words = []
try:
for word in article.split(" "):
if is_clean(word):
clean_words.append(word)
except Exception as e:
print(e)
clean_articles.append(' '.join(clean_words))
return clean_articles
class Deduplicator:
def deduplicate_articles(self, articles):
seen_articles = set()
deduplicated_articles = []
for article in articles:
if hash(article) in seen_articles:
continue
else:
seen_articles.add(hash(article))
deduplicated_articles.append(article)
return deduplicated_articles
class Analyzer:
good_words = {"unveiled", "available", "faster", "stable"}
bad_words = {"sued", "defiance", "violation"}
def extract_entities_and_sentiment(self, articles):
entity_score_pairs = []
for article in articles:
score = 0
entities = []
for word in article.split(" "):
if word[0] == word[0].upper():
entities.append(word)
if word.lower() in self.good_words:
score += 1
elif word.lower() in self.bad_words:
score -= 1
main_entities = [i[0] for i in Counter(entities).most_common(2)]
entity_score_pair = (main_entities, score)
entity_score_pairs.append(entity_score_pair)
return entity_score_pairs
class DecisionMaker:
target_companies = set(['Apple', 'Uber', 'Google'])
def make_decisions(self, entity_score_pairs):
decisions = []
for entities, score in entity_score_pairs:
for entity in entities:
if entity in self.target_companies:
quantity = abs(score)
order = "Buy" if score > 0 else "Sell"
decision = (order, quantity, entity)
decisions.append(decision)
return decisions
scraper = Scraper()
cleaner = Cleaner()
deduplicator = Deduplicator()
analyzer = Analyzer()
decision_maker = DecisionMaker()
contents = scraper.fetch_news(article_urls)
contents[0][:500]
clean_articles = cleaner.clean_articles(contents)
clean_articles[0][:500]
print(len(clean_articles))
deduplicated = deduplicator.deduplicate_articles(clean_articles)
print(len(deduplicated))
entity_score_pairs = analyzer.extract_entities_and_sentiment(deduplicated)
print(entity_score_pairs)
decisions = decision_maker.make_decisions(entity_score_pairs)
print(decisions)
| 0.301979 | 0.940517 |
```
#재귀호출을 이용한 이분 탐색
#리스트에서 특정 숫자 위치 찾기(이분 탐색과 재귀 호출)
#입력: 리스트a, 찾는값 x
#출력: 특정 숫자를 찾이면 그 값의 위치, 찾지 못하면 -1
#리스트 a의 어디부터(start) 어디까지(end)가 탐색 범위인지 지정하여
#그 범위 안에서 x를 찾는 재귀 함수
def binary_search_sub(a, x, start, end):
#종료 조건: 남은 탐색 범위가 비었으면 종료
if start > end:
return -1
mid = (start + end)//2 #탐색 범위의 중간 위치
if x == a[mid]: #발견!
return mid
elif x > a[mid]: #찾는 값이 더 크면 중간을 기준으로 오른쪽 값을 대상으로 재귀 호출
return binary_search_sub(a, x, mid+1, end)
else: #찾는 값이 더 작으면 중간을 기준으로 왼쪽 값을 대상으로 재귀 호출
return binary_search_sub(a, x, start, mid-1)
return -1 #찾지 못했을때
#리스트 전체(0~len(a)-1)대상으로 재귀 호출 함수 호출
def binary_search(a, x):
return binary_search_sub(a, x, 0, len(a)-1)
d= [1,4,9,16,25,36,49,64,81]
print(binary_search(d,36))
print(binary_search(d,50))
```
이진 탐색트리 : 탐색트리의 기본,
이진 트리 이면서 같은 값을 갖는 노드가 없어야함.
왼쪽 서브 트리에 있는 모든 데이터는 현재 노드의 값보다 작고, 오른쪽 서브 트리에있는 모든 노드의 데이터는 현재 노드의 값보다 큼
각 노드는 최대 두 개의 자식을 가짐.
서브 트리의 예
서브트리도 역시 이진 탐색 트리의 성질을 만족해야 하며
루트의 왼쪽 자식은 루트보다 작고 오른쪽 자식은 루트보다 큼
삽입정렬
```
def insertion_sort(data):
for index in range(len(data)-1):
for index2 in range(index +1, 0, -1):
if data[index2] < data[index2-1]:
data[index2], data[index2-1] = data[index2-1], data[index2]
else:
break
return data
import random
data_list = random.sample(range(100),50)
data_list
print(insertion_sort(data_list))
len(insertion_sort(data_list))
```
쉽게 설명한 삽입 정렬 알고리즘
```
#쉽게 설명한 삽입 정렬
#입력: 리스트a
#출력: 정렬된 새 리스트
#리스트 r 에서 v가 들어가야 할 위치를 돌려주는 함수
def find_ins_idx(r,v):
#이미 정렬된 리스트 r의 자료를 앞에서부터 차례로 확인하야
for i in range(0,len(r)):
#v값보다 i번 위치에 있는 자료값이 크면
#v가 그 값 바로 앞에 놓여야 정렬 순서가 유지됨
if v < r[i]:
return i
#적절한 위치를 못 찾았을 때는
#v가 r의 모든 자료보다 크다는 뜻이므로 맨 뒤에 삽입
return len(r)
def ins_sort(a):
result = [] #새 리스트를 만들어 정렬된 값을 저장
while a: #기존 리스트에 값이 남아 있는동안 반복
value = a.pop(0) #기존 리스트에서 한개를 꺼냄
ins_idx = find_ins_idx(result, value) #꺼낸 값이 들어갈 적당한 위치 찾기
result.insert(ins_idx, value) # 찾은 위치에 값 삽입(이후 값은 한 칸씩 밀려남)
return result
d = [2,4,5,1,3]
print(ins_sort(d))
```
일반적인 삽입 정렬 알고리즘
```
#삽입 정렬
#입력: 리스트a
#출력: 없음(입력으로 주어진 a가 정렬됨)
def ins_sort(a):
n = len(a)
for i in range(1, n): #1부터 n-1까지
#i번 위치의 값을 key로 저장
key = a[i]
#j를 i 바로 왼쪽 위치로 저장
j = i -1
#리스트의 j번 위치에 있는 값과 key를 비교해 key가 삽입될 적절한 위치를 찾음
while j>=0 and a[j]> key:
a[j+1]= a[j] #삽입할 공간이 생기도록 값을 오른쪽으로 한칸 이동
j-= 1
a[j+1] = key #찾은 삽입 위치에 key를 저장.
d=[2,4,5,1,3]
ins_sort(d)
print(d)
#d.sort()
#print(d)
#d.reverse()
#print(d)
#삽입 정렬
#입력: 리스트a
#출력: 없음(입력으로 주어진 a가 정렬됨)
def ins_sort(a):
n = len(a)
for i in range(1, n): #1부터 n-1까지
#i번 위치의 값을 key로 저장
key = a[i]
#j를 i 바로 왼쪽 위치로 저장
j = i -1
#리스트의 j번 위치에 있는 값과 key를 비교해 key가 삽입될 적절한 위치를 찾음
while j>=0 and a[j]< key:
a[j+1]= a[j] #삽입할 공간이 생기도록 값을 오른쪽으로 한칸 이동
j-= 1
a[j+1] = key #찾은 삽입 위치에 key를 저장.
d=[2,4,5,1,3]
ins_sort(d)
print(d)
```
|
github_jupyter
|
#재귀호출을 이용한 이분 탐색
#리스트에서 특정 숫자 위치 찾기(이분 탐색과 재귀 호출)
#입력: 리스트a, 찾는값 x
#출력: 특정 숫자를 찾이면 그 값의 위치, 찾지 못하면 -1
#리스트 a의 어디부터(start) 어디까지(end)가 탐색 범위인지 지정하여
#그 범위 안에서 x를 찾는 재귀 함수
def binary_search_sub(a, x, start, end):
#종료 조건: 남은 탐색 범위가 비었으면 종료
if start > end:
return -1
mid = (start + end)//2 #탐색 범위의 중간 위치
if x == a[mid]: #발견!
return mid
elif x > a[mid]: #찾는 값이 더 크면 중간을 기준으로 오른쪽 값을 대상으로 재귀 호출
return binary_search_sub(a, x, mid+1, end)
else: #찾는 값이 더 작으면 중간을 기준으로 왼쪽 값을 대상으로 재귀 호출
return binary_search_sub(a, x, start, mid-1)
return -1 #찾지 못했을때
#리스트 전체(0~len(a)-1)대상으로 재귀 호출 함수 호출
def binary_search(a, x):
return binary_search_sub(a, x, 0, len(a)-1)
d= [1,4,9,16,25,36,49,64,81]
print(binary_search(d,36))
print(binary_search(d,50))
def insertion_sort(data):
for index in range(len(data)-1):
for index2 in range(index +1, 0, -1):
if data[index2] < data[index2-1]:
data[index2], data[index2-1] = data[index2-1], data[index2]
else:
break
return data
import random
data_list = random.sample(range(100),50)
data_list
print(insertion_sort(data_list))
len(insertion_sort(data_list))
#쉽게 설명한 삽입 정렬
#입력: 리스트a
#출력: 정렬된 새 리스트
#리스트 r 에서 v가 들어가야 할 위치를 돌려주는 함수
def find_ins_idx(r,v):
#이미 정렬된 리스트 r의 자료를 앞에서부터 차례로 확인하야
for i in range(0,len(r)):
#v값보다 i번 위치에 있는 자료값이 크면
#v가 그 값 바로 앞에 놓여야 정렬 순서가 유지됨
if v < r[i]:
return i
#적절한 위치를 못 찾았을 때는
#v가 r의 모든 자료보다 크다는 뜻이므로 맨 뒤에 삽입
return len(r)
def ins_sort(a):
result = [] #새 리스트를 만들어 정렬된 값을 저장
while a: #기존 리스트에 값이 남아 있는동안 반복
value = a.pop(0) #기존 리스트에서 한개를 꺼냄
ins_idx = find_ins_idx(result, value) #꺼낸 값이 들어갈 적당한 위치 찾기
result.insert(ins_idx, value) # 찾은 위치에 값 삽입(이후 값은 한 칸씩 밀려남)
return result
d = [2,4,5,1,3]
print(ins_sort(d))
#삽입 정렬
#입력: 리스트a
#출력: 없음(입력으로 주어진 a가 정렬됨)
def ins_sort(a):
n = len(a)
for i in range(1, n): #1부터 n-1까지
#i번 위치의 값을 key로 저장
key = a[i]
#j를 i 바로 왼쪽 위치로 저장
j = i -1
#리스트의 j번 위치에 있는 값과 key를 비교해 key가 삽입될 적절한 위치를 찾음
while j>=0 and a[j]> key:
a[j+1]= a[j] #삽입할 공간이 생기도록 값을 오른쪽으로 한칸 이동
j-= 1
a[j+1] = key #찾은 삽입 위치에 key를 저장.
d=[2,4,5,1,3]
ins_sort(d)
print(d)
#d.sort()
#print(d)
#d.reverse()
#print(d)
#삽입 정렬
#입력: 리스트a
#출력: 없음(입력으로 주어진 a가 정렬됨)
def ins_sort(a):
n = len(a)
for i in range(1, n): #1부터 n-1까지
#i번 위치의 값을 key로 저장
key = a[i]
#j를 i 바로 왼쪽 위치로 저장
j = i -1
#리스트의 j번 위치에 있는 값과 key를 비교해 key가 삽입될 적절한 위치를 찾음
while j>=0 and a[j]< key:
a[j+1]= a[j] #삽입할 공간이 생기도록 값을 오른쪽으로 한칸 이동
j-= 1
a[j+1] = key #찾은 삽입 위치에 key를 저장.
d=[2,4,5,1,3]
ins_sort(d)
print(d)
| 0.112515 | 0.918407 |
# EDA Case Study: House Price
### Task Description
House Prices is a classical Kaggle competition. The task is to predicts final price of each house. For more detail, refer to https://www.kaggle.com/c/house-prices-advanced-regression-techniques/.
### Goal of this notebook
As it is a famous competition, there exists lots of excelent analysis on how to do eda and how to build model for this task. See https://www.kaggle.com/khandelwallaksya/house-prices-eda for a reference. In this notebook, we will show how dataprep.eda can simply the eda process using a few lines of code.
In conclusion:
* **Understand the problem**. We'll look at each variable and do a philosophical analysis about their meaning and importance for this problem.
* **Univariable study**. We'll just focus on the dependent variable ('SalePrice') and try to know a little bit more about it.
* **Multivariate study**. We'll try to understand how the dependent variable and independent variables relate.
* **Basic cleaning**. We'll clean the dataset and handle the missing data, outliers and categorical variables.
### Import libraries
```
from dataprep.eda import plot
from dataprep.eda import plot_correlation
from dataprep.eda import plot_missing
from dataprep.datasets import load_dataset
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
sns.set(style="whitegrid", color_codes=True)
sns.set(font_scale=1)
```
### Load data
```
houses = load_dataset("house_prices_train")
houses.head()
houses_test = load_dataset("house_prices_test")
houses_test.head()
houses.shape
```
There are total 1460 tuples, each tuple contains 80 features and 1 target value.
```
houses_test.shape
```
### Variable identification
```
plot(houses)
```
### Overview of the data
We could get the following information:
* **Variable**-Variable name
* **Type**-There are 43 categorical columns and 38 numerical columns.
* **Missing value**-How many missing values each column contains. For instance, Fence contains 80.8% * 1460 = 1180 missing tuples. Usually, some model does not allow the input data contains missing value such as SVM, we have to clean the data before we utilize it.
* **Target Value**-The distribution of target value (SalePrice). According to the distribution of the target value, we could get the information that the target value is numerical and the distribution of the target value conforms to the norm distribution. Thus, we are not confronted with imbalanced classes problem. It is really great.
* **Guess**-According to the columns' name, we reckon GrLivArea, YearBuilt and OverallQual are likely to be correlated to the target value (SalePrice).
### Correlation in data
```
plot_correlation(houses, "SalePrice")
plot_correlation(houses, "SalePrice", value_range=[0.5, 1])
```
OverallQual, GrLivArea, GarageCars, GarageArea, TotalBsmtSF, 1stFlrSF, FullBath, TotRmsAbvGrd, YearBuilt, YearRemodAdd have more than 0.5 Pearson correlation with SalePrice.
OverallQual, GrLivArea, GarageCars, YearBuilt, GarageArea, FullBath, TotalBsmtSF, GarageYrBlt, 1stFlrSF, YearRemodAdd, TotRmsAbvGrd and Fireplaces have more than 0.5 Spearman correlation with SalePrice.
OverallQual, GarageCars, GrLivArea and FullBath have more than 0.5 KendallTau correlation with SalePrice.
EnclosedPorch and KitchenAbvGr have little negative correlation with target variable.
These can prove to be important features to predict SalePrice.
### Heatmap
```
plot_correlation(houses)
```
### In summary
In my opinion, this heatmap is the best way to get a quick overview of features' relationships.
At first sight, there are two red colored squares that get my attention. The first one refers to the 'TotalBsmtSF' and '1stFlrSF' variables, and the second one refers to the 'GarageX' variables. Both cases show how significant the correlation is between these variables. Actually, this correlation is so strong that it can indicate a situation of multicollinearity. If we think about these variables, we can conclude that they give almost the same information so multicollinearity really occurs. Heatmaps are great to detect this kind of situations and in problems dominated by feature selection, like ours, they are an essential tool.
Another thing that got my attention was the 'SalePrice' correlations. We can see our well-known 'GrLivArea', 'TotalBsmtSF', and 'OverallQual', but we can also see many other variables that should be taken into account. That's what we will do next.
```
plot_correlation(houses[["SalePrice","OverallQual","GrLivArea","GarageCars",
"GarageArea","GarageYrBlt","TotalBsmtSF","1stFlrSF","FullBath",
"TotRmsAbvGrd","YearBuilt","YearRemodAdd"]])
```
As we saw above there are few feature which shows high multicollinearity from heatmap. Lets focus on red squares on diagonal line and few on the sides.
SalePrice and OverallQual
GarageArea and GarageCars
TotalBsmtSF and 1stFlrSF
GrLiveArea and TotRmsAbvGrd
YearBulit and GarageYrBlt
We have to create a single feature from them before we use them as predictors.
```
plot_correlation(houses, value_range=[0.5, 1])
plot_correlation(houses, k=30)
```
**Attribute Pair Correlation**
7 (GarageArea, GarageCars) 0.882475
11 (GarageYrBlt, YearBuilt) 0.825667
15 (GrLivArea, TotRmsAbvGrd) 0.825489
18 (1stFlrSF, TotalBsmtSF) 0.819530
19 (2ndFlrSF, GrLivArea) 0.687501
9 (BedroomAbvGr, TotRmsAbvGrd) 0.676620
0 (BsmtFinSF1, BsmtFullBath) 0.649212
2 (GarageYrBlt, YearRemodAdd) 0.642277
24 (FullBath, GrLivArea) 0.630012
8 (2ndFlrSF, TotRmsAbvGrd) 0.616423
1 (2ndFlrSF, HalfBath) 0.609707
4 (GarageCars, OverallQual) 0.600671
16 (GrLivArea, OverallQual) 0.593007
23 (YearBuilt, YearRemodAdd) 0.592855
22 (GarageCars, GarageYrBlt) 0.588920
12 (OverallQual, YearBuilt) 0.572323
5 (1stFlrSF, GrLivArea) 0.566024
25 (GarageArea, GarageYrBlt) 0.564567
6 (GarageArea, OverallQual) 0.562022
17 (FullBath, TotRmsAbvGrd) 0.554784
13 (OverallQual, YearRemodAdd) 0.550684
14 (FullBath, OverallQual) 0.550600
3 (GarageYrBlt, OverallQual) 0.547766
10 (GarageCars, YearBuilt) 0.537850
27 (OverallQual, TotalBsmtSF) 0.537808
20 (BsmtFinSF1, TotalBsmtSF) 0.522396
21 (BedroomAbvGr, GrLivArea) 0.521270
26 (2ndFlrSF, BedroomAbvGr) 0.502901
This shows multicollinearity. In regression, "multicollinearity" refers to features that are correlated with other features. Multicollinearity occurs when your model includes multiple factors that are correlated not just to your target variable, but also to each other.
Problem:
Multicollinearity increases the standard errors of the coefficients. That means, multicollinearity makes some variables statistically insignificant when they should be significant.
To avoid this we can do 3 things:
Completely remove those variables
Make new feature by adding them or by some other operation.
Use PCA, which will reduce feature set to small number of non-collinear features.
Reference:http://blog.minitab.com/blog/understanding-statistics/handling-multicollinearity-in-regression-analysis
### Univariate Analysis
How 1 single variable is distributed in numeric range. What is statistical summary of it. Is it positively skewed or negatively.
```
plot(houses, "SalePrice")
```
### Pivotal Features
```
plot_correlation(houses, "OverallQual", "SalePrice")
plot(houses, "OverallQual", "SalePrice")
plot(houses, "GarageCars", "SalePrice")
plot(houses, "Fireplaces", "SalePrice")
plot(houses, "GrLivArea", "SalePrice")
plot(houses, "TotalBsmtSF", "SalePrice")
plot(houses, "YearBuilt", "SalePrice")
```
### In summary
Based on the above analysis, we can conclude that:
'GrLivArea' and 'TotalBsmtSF' seem to be linearly related with 'SalePrice'. Both relationships are positive, which means that as one variable increases, the other also increases. In the case of 'TotalBsmtSF', we can see that the slope of the linear relationship is particularly high.
'OverallQual' and 'YearBuilt' also seem to be related with 'SalePrice'. The relationship seems to be stronger in the case of 'OverallQual', where the box plot shows how sales prices increase with the overall quality.
We just analysed four variables, but there are many other that we should analyse. The trick here seems to be the choice of the right features (feature selection) and not the definition of complex relationships between them (feature engineering).
That said, let's separate the wheat from the chaff.
### Missing Value Imputation
Missing values in the training data set can affect prediction or classification of a model negatively.
Also some machine learning algorithms can't accept missing data eg. SVM, Neural Network.
But filling missing values with mean/median/mode or using another predictive model to predict missing values is also a prediction which may not be 100% accurate, instead you can use models like Decision Trees and Random Forest which handle missing values very well.
Some of this part is based on this kernel: https://www.kaggle.com/bisaria/house-prices-advanced-regression-techniques/handling-missing-data
```
plot_missing(houses)
# plot_missing(houses, "BsmtQual")
basement_cols=['BsmtQual','BsmtCond','BsmtExposure','BsmtFinType1','BsmtFinType2','BsmtFinSF1','BsmtFinSF2']
houses[basement_cols][houses['BsmtQual'].isnull()==True]
```
All categorical variables contains NAN whereas continuous ones have 0. So that means there is no basement for those houses. we can replace it with 'None'.
```
for col in basement_cols:
if 'FinSF'not in col:
houses[col] = houses[col].fillna('None')
# plot_missing(houses, "FireplaceQu")
houses["FireplaceQu"] = houses["FireplaceQu"].fillna('None')
pd.crosstab(houses.Fireplaces, houses.FireplaceQu)
garage_cols=['GarageType','GarageQual','GarageCond','GarageYrBlt','GarageFinish','GarageCars','GarageArea']
houses[garage_cols][houses['GarageType'].isnull()==True]
```
All garage related features are missing values in same rows. that means we can replace categorical variables with None and continuous ones with 0.
```
for col in garage_cols:
if houses[col].dtype==np.object:
houses[col] = houses[col].fillna('None')
else:
houses[col] = houses[col].fillna(0)
```
|
github_jupyter
|
from dataprep.eda import plot
from dataprep.eda import plot_correlation
from dataprep.eda import plot_missing
from dataprep.datasets import load_dataset
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
sns.set(style="whitegrid", color_codes=True)
sns.set(font_scale=1)
houses = load_dataset("house_prices_train")
houses.head()
houses_test = load_dataset("house_prices_test")
houses_test.head()
houses.shape
houses_test.shape
plot(houses)
plot_correlation(houses, "SalePrice")
plot_correlation(houses, "SalePrice", value_range=[0.5, 1])
plot_correlation(houses)
plot_correlation(houses[["SalePrice","OverallQual","GrLivArea","GarageCars",
"GarageArea","GarageYrBlt","TotalBsmtSF","1stFlrSF","FullBath",
"TotRmsAbvGrd","YearBuilt","YearRemodAdd"]])
plot_correlation(houses, value_range=[0.5, 1])
plot_correlation(houses, k=30)
plot(houses, "SalePrice")
plot_correlation(houses, "OverallQual", "SalePrice")
plot(houses, "OverallQual", "SalePrice")
plot(houses, "GarageCars", "SalePrice")
plot(houses, "Fireplaces", "SalePrice")
plot(houses, "GrLivArea", "SalePrice")
plot(houses, "TotalBsmtSF", "SalePrice")
plot(houses, "YearBuilt", "SalePrice")
plot_missing(houses)
# plot_missing(houses, "BsmtQual")
basement_cols=['BsmtQual','BsmtCond','BsmtExposure','BsmtFinType1','BsmtFinType2','BsmtFinSF1','BsmtFinSF2']
houses[basement_cols][houses['BsmtQual'].isnull()==True]
for col in basement_cols:
if 'FinSF'not in col:
houses[col] = houses[col].fillna('None')
# plot_missing(houses, "FireplaceQu")
houses["FireplaceQu"] = houses["FireplaceQu"].fillna('None')
pd.crosstab(houses.Fireplaces, houses.FireplaceQu)
garage_cols=['GarageType','GarageQual','GarageCond','GarageYrBlt','GarageFinish','GarageCars','GarageArea']
houses[garage_cols][houses['GarageType'].isnull()==True]
for col in garage_cols:
if houses[col].dtype==np.object:
houses[col] = houses[col].fillna('None')
else:
houses[col] = houses[col].fillna(0)
| 0.548915 | 0.957358 |
# Deep Q-Network (DQN)
---
In this notebook, you will implement a DQN agent with OpenAI Gym's LunarLander-v2 environment.
### 1. Import the Necessary Packages
```
import gym
import random
import torch
import numpy as np
from collections import deque
import matplotlib.pyplot as plt
%matplotlib inline
```
### 2. Instantiate the Environment and Agent
Initialize the environment in the code cell below.
```
env = gym.make('LunarLander-v2')
env.seed(0)
print('State shape: ', env.observation_space.shape)
print('Number of actions: ', env.action_space.n)
```
Please refer to the instructions in `Deep_Q_Network.ipynb` if you would like to write your own DQN agent. Otherwise, run the code cell below to load the solution files.
```
from dqn_agent import Agent
agent = Agent(state_size=8, action_size=4, seed=0)
# watch an untrained agent
state = env.reset()
for j in range(200):
action = agent.act(state)
env.render()
state, reward, done, _ = env.step(action)
if done:
break
env.close()
```
### 3. Train the Agent with DQN
Run the code cell below to train the agent from scratch. You are welcome to amend the supplied values of the parameters in the function, to try to see if you can get better performance!
Alternatively, you can skip to the next step below (**4. Watch a Smart Agent!**), to load the saved model weights from a pre-trained agent.
```
def dqn(n_episodes=2000, max_t=1000, eps_start=1.0, eps_end=0.01, eps_decay=0.995):
"""Deep Q-Learning.
Params
======
n_episodes (int): maximum number of training episodes
max_t (int): maximum number of timesteps per episode
eps_start (float): starting value of epsilon, for epsilon-greedy action selection
eps_end (float): minimum value of epsilon
eps_decay (float): multiplicative factor (per episode) for decreasing epsilon
"""
scores = [] # list containing scores from each episode
scores_window = deque(maxlen=100) # last 100 scores
eps = eps_start # initialize epsilon
for i_episode in range(1, n_episodes+1):
state = env.reset()
score = 0
for t in range(max_t):
action = agent.act(state, eps)
next_state, reward, done, _ = env.step(action)
agent.step(state, action, reward, next_state, done)
state = next_state
score += reward
if done:
break
scores_window.append(score) # save most recent score
scores.append(score) # save most recent score
eps = max(eps_end, eps_decay*eps) # decrease epsilon
print('\rEpisode {}\tAverage Score: {:.2f}'.format(i_episode, np.mean(scores_window)), end="")
if i_episode % 100 == 0:
print('\rEpisode {}\tAverage Score: {:.2f}'.format(i_episode, np.mean(scores_window)))
if np.mean(scores_window)>=200.0:
print('\nEnvironment solved in {:d} episodes!\tAverage Score: {:.2f}'.format(i_episode-100, np.mean(scores_window)))
torch.save(agent.qnetwork_local.state_dict(), 'checkpoint.pth')
break
return scores
scores = dqn()
# plot the scores
fig = plt.figure()
ax = fig.add_subplot(111)
plt.plot(np.arange(len(scores)), scores)
plt.ylabel('Score')
plt.xlabel('Episode #')
plt.show()
```
### 4. Watch a Smart Agent!
In the next code cell, you will load the trained weights from file to watch a smart agent!
```
# load the weights from file
agent.qnetwork_local.load_state_dict(torch.load('checkpoint.pth'))
for i in range(3):
state = env.reset()
for j in range(200):
action = agent.act(state)
env.render()
state, reward, done, _ = env.step(action)
if done:
break
env.close()
```
### 5. Explore
In this exercise, you have implemented a DQN agent and demonstrated how to use it to solve an OpenAI Gym environment. To continue your learning, you are encouraged to complete any (or all!) of the following tasks:
- Amend the various hyperparameters and network architecture to see if you can get your agent to solve the environment faster. Once you build intuition for the hyperparameters that work well with this environment, try solving a different OpenAI Gym task with discrete actions!
- You may like to implement some improvements such as prioritized experience replay, Double DQN, or Dueling DQN!
- Write a blog post explaining the intuition behind the DQN algorithm and demonstrating how to use it to solve an RL environment of your choosing.
|
github_jupyter
|
import gym
import random
import torch
import numpy as np
from collections import deque
import matplotlib.pyplot as plt
%matplotlib inline
env = gym.make('LunarLander-v2')
env.seed(0)
print('State shape: ', env.observation_space.shape)
print('Number of actions: ', env.action_space.n)
from dqn_agent import Agent
agent = Agent(state_size=8, action_size=4, seed=0)
# watch an untrained agent
state = env.reset()
for j in range(200):
action = agent.act(state)
env.render()
state, reward, done, _ = env.step(action)
if done:
break
env.close()
def dqn(n_episodes=2000, max_t=1000, eps_start=1.0, eps_end=0.01, eps_decay=0.995):
"""Deep Q-Learning.
Params
======
n_episodes (int): maximum number of training episodes
max_t (int): maximum number of timesteps per episode
eps_start (float): starting value of epsilon, for epsilon-greedy action selection
eps_end (float): minimum value of epsilon
eps_decay (float): multiplicative factor (per episode) for decreasing epsilon
"""
scores = [] # list containing scores from each episode
scores_window = deque(maxlen=100) # last 100 scores
eps = eps_start # initialize epsilon
for i_episode in range(1, n_episodes+1):
state = env.reset()
score = 0
for t in range(max_t):
action = agent.act(state, eps)
next_state, reward, done, _ = env.step(action)
agent.step(state, action, reward, next_state, done)
state = next_state
score += reward
if done:
break
scores_window.append(score) # save most recent score
scores.append(score) # save most recent score
eps = max(eps_end, eps_decay*eps) # decrease epsilon
print('\rEpisode {}\tAverage Score: {:.2f}'.format(i_episode, np.mean(scores_window)), end="")
if i_episode % 100 == 0:
print('\rEpisode {}\tAverage Score: {:.2f}'.format(i_episode, np.mean(scores_window)))
if np.mean(scores_window)>=200.0:
print('\nEnvironment solved in {:d} episodes!\tAverage Score: {:.2f}'.format(i_episode-100, np.mean(scores_window)))
torch.save(agent.qnetwork_local.state_dict(), 'checkpoint.pth')
break
return scores
scores = dqn()
# plot the scores
fig = plt.figure()
ax = fig.add_subplot(111)
plt.plot(np.arange(len(scores)), scores)
plt.ylabel('Score')
plt.xlabel('Episode #')
plt.show()
# load the weights from file
agent.qnetwork_local.load_state_dict(torch.load('checkpoint.pth'))
for i in range(3):
state = env.reset()
for j in range(200):
action = agent.act(state)
env.render()
state, reward, done, _ = env.step(action)
if done:
break
env.close()
| 0.608478 | 0.953923 |
```
from IPython.display import clear_output
def display_board(board):
clear_output()
print(board[7] + '|' + board[8] + '|' + board[9])
print('-----')
print(board[4] + '|' + board[5] + '|' + board[6])
print('-----')
print(board[1] + '|' + board[2] + '|' + board[3])
#test_board = ['#', 'X', '0', 'X', '0', 'X', '0', 'X', '0', 'X']
test_board = [' ']*10
display_board(test_board)
# display_board(test_board)
def player_input():
marker = ''
# Asking Player 1 to Choose X or 0
while marker != 'X' and marker != '0':
marker = input('Player 1, Choose X or 0: ').upper()
if marker == 'X':
return ('X', '0')
else:
return ('0', 'X')
player1_marker, player2_marker = player_input()
player2_marker
player1_marker
def place_marker(board, marker, position):
board[position] = marker
def win_check(board, mark):
# Winning Tic Tac Toe
# All Rows, and Check to see if they all share the same marker?
# All Columns and Check to see if they all share the same marker?
# 2 Diagonals and Check to see if they all share the same marker?
return ((board[7] == mark and board[8] == mark and board[9] == mark) or
(board[4] == mark and board[5] == mark and board[6] == mark) or
(board[1] == mark and board[2] == mark and board[3] == mark) or
(board[7] == mark and board[4] == mark and board[1] == mark) or
(board[8] == mark and board[5] == mark and board[2] == mark) or
(board[9] == mark and board[6] == mark and board[3] == mark) or
(board[7] == mark and board[5] == mark and board[3] == mark) or
(board[9] == mark and board[5] == mark and board[1] == mark))
test_board = ['#', 'X', '0', 'X', '0', 'X', '0', 'X', '0', 'X']
display_board(test_board)
win_check(test_board, 'X')
import random
def choose_first():
flip = random.randint(0, 1)
if flip == 0:
return 'Player 1'
else:
return 'Player 2'
def space_check(board, position):
return board[position] == ' '
def full_board_check(board):
for i in range(1, 10):
if space_check(board, i):
return False
# Board is Full if we return True
return True
def player_choice(board):
position = 0
while position not in [1, 2, 3, 4, 5, 6, 7, 8, 9] or not space_check(board, position):
position = int(input('Chose a Postion: (1 - 9) '))
return position
def replay():
choice = input("Play Again? Enter Yes or No")
return choice == 'Yes'
print('Welcome to Tic Tac Toe')
while True:
the_board = [' '] * 10
player1_marker, player2_marker = player_input()
turn = choose_first()
print(turn + ' Will go first')
play_game = input('Read to Play? y or n?')
if play_game == 'y':
game_on = True
else:
game_on = False
while game_on:
if turn == 'Player 1':
display_board(the_board)
position = player_choice(the_board)
place_marker(the_board, player1_marker, position)
if win_check(the_board, player1_marker):
display_board(the_board)
print('Yo!! Player 1 has won')
game_on = False
else:
if full_board_check(the_board):
display_board(the_board)
print('Tie Game!')
game_on = False
else:
turn = 'Player 2'
else:
display_board(the_board)
position = player_choice(the_board)
place_marker(the_board, player2_marker, position)
if win_check(the_board, player2_marker):
display_board(the_board)
print('Yo!! Player 2 has won')
game_on = False
else:
if full_board_check(the_board):
display_board(the_board)
print('Tie Game!')
game_on = False
else:
turn = 'Player 1'
if not replay():
break
clear()
```
|
github_jupyter
|
from IPython.display import clear_output
def display_board(board):
clear_output()
print(board[7] + '|' + board[8] + '|' + board[9])
print('-----')
print(board[4] + '|' + board[5] + '|' + board[6])
print('-----')
print(board[1] + '|' + board[2] + '|' + board[3])
#test_board = ['#', 'X', '0', 'X', '0', 'X', '0', 'X', '0', 'X']
test_board = [' ']*10
display_board(test_board)
# display_board(test_board)
def player_input():
marker = ''
# Asking Player 1 to Choose X or 0
while marker != 'X' and marker != '0':
marker = input('Player 1, Choose X or 0: ').upper()
if marker == 'X':
return ('X', '0')
else:
return ('0', 'X')
player1_marker, player2_marker = player_input()
player2_marker
player1_marker
def place_marker(board, marker, position):
board[position] = marker
def win_check(board, mark):
# Winning Tic Tac Toe
# All Rows, and Check to see if they all share the same marker?
# All Columns and Check to see if they all share the same marker?
# 2 Diagonals and Check to see if they all share the same marker?
return ((board[7] == mark and board[8] == mark and board[9] == mark) or
(board[4] == mark and board[5] == mark and board[6] == mark) or
(board[1] == mark and board[2] == mark and board[3] == mark) or
(board[7] == mark and board[4] == mark and board[1] == mark) or
(board[8] == mark and board[5] == mark and board[2] == mark) or
(board[9] == mark and board[6] == mark and board[3] == mark) or
(board[7] == mark and board[5] == mark and board[3] == mark) or
(board[9] == mark and board[5] == mark and board[1] == mark))
test_board = ['#', 'X', '0', 'X', '0', 'X', '0', 'X', '0', 'X']
display_board(test_board)
win_check(test_board, 'X')
import random
def choose_first():
flip = random.randint(0, 1)
if flip == 0:
return 'Player 1'
else:
return 'Player 2'
def space_check(board, position):
return board[position] == ' '
def full_board_check(board):
for i in range(1, 10):
if space_check(board, i):
return False
# Board is Full if we return True
return True
def player_choice(board):
position = 0
while position not in [1, 2, 3, 4, 5, 6, 7, 8, 9] or not space_check(board, position):
position = int(input('Chose a Postion: (1 - 9) '))
return position
def replay():
choice = input("Play Again? Enter Yes or No")
return choice == 'Yes'
print('Welcome to Tic Tac Toe')
while True:
the_board = [' '] * 10
player1_marker, player2_marker = player_input()
turn = choose_first()
print(turn + ' Will go first')
play_game = input('Read to Play? y or n?')
if play_game == 'y':
game_on = True
else:
game_on = False
while game_on:
if turn == 'Player 1':
display_board(the_board)
position = player_choice(the_board)
place_marker(the_board, player1_marker, position)
if win_check(the_board, player1_marker):
display_board(the_board)
print('Yo!! Player 1 has won')
game_on = False
else:
if full_board_check(the_board):
display_board(the_board)
print('Tie Game!')
game_on = False
else:
turn = 'Player 2'
else:
display_board(the_board)
position = player_choice(the_board)
place_marker(the_board, player2_marker, position)
if win_check(the_board, player2_marker):
display_board(the_board)
print('Yo!! Player 2 has won')
game_on = False
else:
if full_board_check(the_board):
display_board(the_board)
print('Tie Game!')
game_on = False
else:
turn = 'Player 1'
if not replay():
break
clear()
| 0.241221 | 0.488344 |
# Linked Birth / Infant Death Records, 2007-2018
Exploring relationships between prenatal care, and gestational age, and infant death rate
Data source: [Linked Birth / Infant Death Records, 2007-2018](https://wonder.cdc.gov/lbd-current.html) from [CDC Wonder](https://wonder.cdc.gov/)
```
import warnings
import numpy as np
import matplotlib.cm as cm
import matplotlib.colors as colors
import matplotlib.pyplot as plt
import pandas as pd
import seaborn as sns
from scipy.stats import ttest_ind
import statsmodels.api as sm
warnings.filterwarnings('ignore')
viridis_week = cm.ScalarMappable(norm=colors.Normalize(vmin=28, vmax=40), cmap='viridis')
viridis_month = cm.ScalarMappable(norm=colors.Normalize(vmin=1, vmax=9), cmap='viridis_r')
```
## Data Cleaning
```
def filter_values(row):
if row.isna().any():
return True
if 'Unreliable' in row['Death Rate']:
return True
if 'Not Applicable' in row['Death Rate']:
return True
if float(row['Births']) == 0.0:
return True
return False
def get_prenatal(row):
return row['Month Prenatal Care Began Code'] != 0
prenatal = pd.read_csv('../../data/CDC/death_rate.txt', sep='\t')
prenatal.drop(columns=['Notes', 'Year of Death Code'], inplace=True)
prenatal['Drop'] = prenatal.apply(filter_values, axis=1)
prenatal = prenatal[prenatal['Drop'] == False].drop(columns='Drop')
prenatal['Prenatal Care'] = prenatal.apply(get_prenatal, axis=1)
prenatal = prenatal.astype({
'Month Prenatal Care Began Code': int,
'Year of Death': int,
'OE Gestational Age Weekly Code': int,
'Deaths': int,
'Births': int,
'Death Rate': float
})
subset = prenatal[(prenatal['Year of Death'] >= 2016) &
(prenatal['OE Gestational Age Weekly Code'] >= 28) &
(prenatal['OE Gestational Age Weekly Code'] <= 40)]
```
## Is there a difference between infant death rates with or without prenatal care?
```
df1 = subset.groupby(['Year of Death', 'OE Gestational Age Weekly Code', 'Prenatal Care'])[['Deaths', 'Births']].sum()
df1['Death Rate'] = df1.apply(lambda row: 1000*row['Deaths']/row['Births'], axis=1)
year_list = []
week_list = []
care_list = []
rate_list = []
log_list = []
for idx, row in df1.iterrows():
year_list.append(row.name[0])
week_list.append(row.name[1])
care_list.append(row.name[2])
rate_list.append(row['Death Rate'])
log_list.append(np.log(row['Death Rate']))
df2 = pd.DataFrame(data={
'Year of Death': year_list,
'Gestational Week': week_list,
'Prenatal Care': care_list,
'Death Rate': rate_list,
'Log Death Rate': log_list
})
fig, ax = plt.subplots(1, 2, figsize=(12, 4))
sns.boxplot(x='Prenatal Care', y='Death Rate', data=df2, ax=ax[0])
ax[0].set_ylabel('Death Rate (per 1,000)')
ax[0].set_title('2016-2018, Weeks 28-40')
sns.boxplot(x='Gestational Week', y='Death Rate', hue='Prenatal Care', data=df2, ax=ax[1])
ax[1].set_xlabel('OE Gestational Age (Week)')
ax[1].set_ylabel('Death Rate (per 1,000)')
ax[1].set_title('2016-2018')
plt.savefig('../../results/death_rate_1.png', bbox_inches='tight', dpi=300)
care = df2[df2['Prenatal Care'] == True]
nocare = df2[df2['Prenatal Care'] == False]
print('00', ttest_ind(care['Death Rate'], nocare['Death Rate'], equal_var=False))
for week in df2['Gestational Week'].unique():
care_sub = care[care['Gestational Week'] == week]
nocare_sub = nocare[nocare['Gestational Week'] == week]
print(week, ttest_ind(care_sub['Death Rate'], nocare_sub['Death Rate'], equal_var=False))
```
## Can we fit a model to the data above?
```
df2['Prenatal Binary'] = df2.apply(lambda row: int(row['Prenatal Care']), axis=1)
df2['Week Centered'] = df2['Gestational Week'] - df2['Gestational Week'].mean()
df2['Cross Term'] = df2.apply(lambda row: row['Week Centered']*row['Prenatal Binary'], axis=1)
model = sm.OLS(df2['Log Death Rate'], sm.add_constant(df2[['Week Centered', 'Prenatal Binary', 'Cross Term']]))
res = model.fit()
res.summary()
fig, ax = plt.subplots(1, 2, figsize=(12, 4))
sns.scatterplot(x='Gestational Week', y='Death Rate', hue='Prenatal Care', data=df2, ax=ax[0])
sns.scatterplot(x='Gestational Week', y='Log Death Rate', hue='Prenatal Care', data=df2, ax=ax[1])
x = np.linspace(28, 40)
x_centered = x - df2['Gestational Week'].mean()
y_f = np.polyval([res.params[1], res.params[0]], x_centered)
ax[0].plot(x, np.exp(y_f))
ax[1].plot(x, y_f)
y_t = np.polyval([res.params[3] + res.params[1], res.params[0]], x_centered) + res.params[2]
ax[0].plot(x, np.exp(y_t))
ax[1].plot(x, y_t)
ax[0].set_xlabel('OE Gestational Age (Week)')
ax[1].set_xlabel('OE Gestational Age (Week)')
ax[0].set_ylabel('Death Rate (per 1,000)')
ax[1].set_ylabel('Log Death Rate (per 1,000)')
plt.savefig('../../results/death_rate_2.png', bbox_inches='tight', dpi=300)
```
## Does the month you start prenatal care matter?
```
df = subset[subset['Prenatal Care'] == True]
df['Week Centered'] = df['OE Gestational Age Weekly Code'] - df['OE Gestational Age Weekly Code'].mean()
df['Month Centered'] = df['Month Prenatal Care Began Code'] - df['Month Prenatal Care Began Code'].mean()
df['Cross Term'] = df.apply(lambda row: row['Week Centered']*row['Month Centered'], axis=1)
df['Log Death Rate'] = df.apply(lambda row: np.log(row['Death Rate']), axis=1)
model = sm.OLS(df['Log Death Rate'], sm.add_constant(df[['Week Centered', 'Month Centered', 'Cross Term']]))
res = model.fit()
res.summary()
fig, ax = plt.subplots(1, 2, figsize=(12, 4))
sns.scatterplot(x='Month Prenatal Care Began Code', y='Death Rate', hue='OE Gestational Age Weekly',
palette='viridis', data=df, ax=ax[0], alpha=0.5, legend=False)
sns.scatterplot(x='Month Prenatal Care Began Code', y='Log Death Rate', hue='OE Gestational Age Weekly',
palette='viridis', data=df, ax=ax[1], alpha=0.5)
for week in df['OE Gestational Age Weekly Code'].unique():
x = np.linspace(1, week/4)
x_centered = x - df['Month Prenatal Care Began Code'].mean()
week_centered = week - df['OE Gestational Age Weekly Code'].mean()
y = res.params[0] + res.params[1]*week_centered + res.params[2]*x_centered + \
res.params[3]*week_centered*x_centered
ax[0].plot(x, np.exp(y), c=viridis_week.to_rgba(week))
ax[1].plot(x, y, c=viridis_week.to_rgba(week))
ax[0].set_xlabel('Month Prenatal Care Began')
ax[1].set_xlabel('Month Prenatal Care Began')
ax[0].set_ylabel('Death Rate (per 1,000)')
ax[1].set_ylabel('Log Death Rate (per 1,000)')
ax[1].legend(title='OE Gestational Age (Week)', bbox_to_anchor=(1.6, 1));
fig, ax = plt.subplots(1, 2, figsize=(12, 4))
sns.scatterplot(x='OE Gestational Age Weekly Code', y='Death Rate', hue='Month Prenatal Care Began',
palette='viridis_r', data=df, ax=ax[0], alpha=0.5, legend=False)
sns.scatterplot(x='OE Gestational Age Weekly Code', y='Log Death Rate', hue='Month Prenatal Care Began',
palette='viridis_r', data=df, ax=ax[1], alpha=0.5)
for month in df['Month Prenatal Care Began Code'].unique():
x = np.linspace(max(28, 4*month), 41)
x_centered = x - df['OE Gestational Age Weekly Code'].mean()
month_centered = month - df['Month Prenatal Care Began Code'].mean()
y = res.params[0] + res.params[1]*x_centered + res.params[2]*month_centered + \
res.params[3]*month_centered*x_centered
ax[0].plot(x, np.exp(y), c=viridis_month.to_rgba(month))
ax[1].plot(x, y, c=viridis_month.to_rgba(month))
ax[0].set_xlabel('OE Gestational Age (Week)')
ax[1].set_xlabel('OE Gestational Age (Week)')
ax[0].set_ylabel('Death Rate (per 1,000)')
ax[1].set_ylabel('Log Death Rate (per 1,000)')
ax[1].legend(title='Month Prenatal Care Began', bbox_to_anchor=(1.6, 1));
model_me = sm.MixedLM(df['Log Death Rate'].values, # dependent variable
df['Month Centered'].values, # fixed effects covariates
df['Week Centered'].values) # groups
res_me = model_me.fit()
res_me.summary(xname_fe=['Month Centered'], xname_re=['Week Centered'])
fig, ax = plt.subplots(1, 2, figsize=(11, 4))
sns.scatterplot(x='Month Prenatal Care Began Code', y='Death Rate', hue='OE Gestational Age Weekly',
palette='viridis', data=df, ax=ax[0], alpha=0.5, legend=False)
sns.scatterplot(x='Month Prenatal Care Began Code', y='Log Death Rate', hue='OE Gestational Age Weekly',
palette='viridis', data=df, ax=ax[1], alpha=0.5)
for week in df['OE Gestational Age Weekly Code'].unique():
x = np.linspace(1, week/4)
x_centered = x - df['Month Prenatal Care Began Code'].mean()
week_centered = week - df['OE Gestational Age Weekly Code'].mean()
y = res_me.random_effects[week_centered][0] + res_me.params[0]*x_centered
ax[0].plot(x, np.exp(y), c=viridis_week.to_rgba(week))
ax[1].plot(x, y, c=viridis_week.to_rgba(week))
ax[0].set_xlabel('Month Prenatal Care Began')
ax[1].set_xlabel('Month Prenatal Care Began')
ax[0].set_ylabel('Death Rate (per 1,000)')
ax[1].set_ylabel('Log Death Rate (per 1,000)')
ax[1].legend(title='OE Gestational Age (Week)', bbox_to_anchor=(1.6, 1))
plt.savefig('../../results/death_rate_3.png', bbox_inches='tight', dpi=300)
fig, ax = plt.subplots(1, 3, figsize=(18, 4))
sns.scatterplot(x='OE Gestational Age Weekly Code', y='Death Rate', hue='Month Prenatal Care Began',
palette='viridis_r', data=df, ax=ax[0], alpha=0.5, legend=False)
sns.scatterplot(x='OE Gestational Age Weekly Code', y='Log Death Rate', hue='Month Prenatal Care Began',
palette='viridis_r', data=df, ax=ax[1], alpha=0.5)
for month in df['Month Prenatal Care Began Code'].unique():
x = np.arange(max(28, 4*month), 41)
x_centered = x - df['OE Gestational Age Weekly Code'].mean()
month_centered = month - df['Month Prenatal Care Began Code'].mean()
y = [res_me.random_effects[xi][0] for xi in x_centered] + res_me.params[0]*month_centered
ax[0].plot(x, np.exp(y), 'x', c=viridis_month.to_rgba(month))
ax[1].plot(x, y, 'x', c=viridis_month.to_rgba(month))
ax[0].set_xlabel('OE Gestational Age (Week)')
ax[1].set_xlabel('OE Gestational Age (Week)')
ax[0].set_ylabel('Death Rate (per 1,000)')
ax[1].set_ylabel('Log Death Rate (per 1,000)')
ax[1].legend(title='Month Prenatal Care Began', bbox_to_anchor=(1.6, 1))
ax[2].set_visible(False)
plt.savefig('../../results/death_rate_4.png', bbox_inches='tight', dpi=300)
```
|
github_jupyter
|
import warnings
import numpy as np
import matplotlib.cm as cm
import matplotlib.colors as colors
import matplotlib.pyplot as plt
import pandas as pd
import seaborn as sns
from scipy.stats import ttest_ind
import statsmodels.api as sm
warnings.filterwarnings('ignore')
viridis_week = cm.ScalarMappable(norm=colors.Normalize(vmin=28, vmax=40), cmap='viridis')
viridis_month = cm.ScalarMappable(norm=colors.Normalize(vmin=1, vmax=9), cmap='viridis_r')
def filter_values(row):
if row.isna().any():
return True
if 'Unreliable' in row['Death Rate']:
return True
if 'Not Applicable' in row['Death Rate']:
return True
if float(row['Births']) == 0.0:
return True
return False
def get_prenatal(row):
return row['Month Prenatal Care Began Code'] != 0
prenatal = pd.read_csv('../../data/CDC/death_rate.txt', sep='\t')
prenatal.drop(columns=['Notes', 'Year of Death Code'], inplace=True)
prenatal['Drop'] = prenatal.apply(filter_values, axis=1)
prenatal = prenatal[prenatal['Drop'] == False].drop(columns='Drop')
prenatal['Prenatal Care'] = prenatal.apply(get_prenatal, axis=1)
prenatal = prenatal.astype({
'Month Prenatal Care Began Code': int,
'Year of Death': int,
'OE Gestational Age Weekly Code': int,
'Deaths': int,
'Births': int,
'Death Rate': float
})
subset = prenatal[(prenatal['Year of Death'] >= 2016) &
(prenatal['OE Gestational Age Weekly Code'] >= 28) &
(prenatal['OE Gestational Age Weekly Code'] <= 40)]
df1 = subset.groupby(['Year of Death', 'OE Gestational Age Weekly Code', 'Prenatal Care'])[['Deaths', 'Births']].sum()
df1['Death Rate'] = df1.apply(lambda row: 1000*row['Deaths']/row['Births'], axis=1)
year_list = []
week_list = []
care_list = []
rate_list = []
log_list = []
for idx, row in df1.iterrows():
year_list.append(row.name[0])
week_list.append(row.name[1])
care_list.append(row.name[2])
rate_list.append(row['Death Rate'])
log_list.append(np.log(row['Death Rate']))
df2 = pd.DataFrame(data={
'Year of Death': year_list,
'Gestational Week': week_list,
'Prenatal Care': care_list,
'Death Rate': rate_list,
'Log Death Rate': log_list
})
fig, ax = plt.subplots(1, 2, figsize=(12, 4))
sns.boxplot(x='Prenatal Care', y='Death Rate', data=df2, ax=ax[0])
ax[0].set_ylabel('Death Rate (per 1,000)')
ax[0].set_title('2016-2018, Weeks 28-40')
sns.boxplot(x='Gestational Week', y='Death Rate', hue='Prenatal Care', data=df2, ax=ax[1])
ax[1].set_xlabel('OE Gestational Age (Week)')
ax[1].set_ylabel('Death Rate (per 1,000)')
ax[1].set_title('2016-2018')
plt.savefig('../../results/death_rate_1.png', bbox_inches='tight', dpi=300)
care = df2[df2['Prenatal Care'] == True]
nocare = df2[df2['Prenatal Care'] == False]
print('00', ttest_ind(care['Death Rate'], nocare['Death Rate'], equal_var=False))
for week in df2['Gestational Week'].unique():
care_sub = care[care['Gestational Week'] == week]
nocare_sub = nocare[nocare['Gestational Week'] == week]
print(week, ttest_ind(care_sub['Death Rate'], nocare_sub['Death Rate'], equal_var=False))
df2['Prenatal Binary'] = df2.apply(lambda row: int(row['Prenatal Care']), axis=1)
df2['Week Centered'] = df2['Gestational Week'] - df2['Gestational Week'].mean()
df2['Cross Term'] = df2.apply(lambda row: row['Week Centered']*row['Prenatal Binary'], axis=1)
model = sm.OLS(df2['Log Death Rate'], sm.add_constant(df2[['Week Centered', 'Prenatal Binary', 'Cross Term']]))
res = model.fit()
res.summary()
fig, ax = plt.subplots(1, 2, figsize=(12, 4))
sns.scatterplot(x='Gestational Week', y='Death Rate', hue='Prenatal Care', data=df2, ax=ax[0])
sns.scatterplot(x='Gestational Week', y='Log Death Rate', hue='Prenatal Care', data=df2, ax=ax[1])
x = np.linspace(28, 40)
x_centered = x - df2['Gestational Week'].mean()
y_f = np.polyval([res.params[1], res.params[0]], x_centered)
ax[0].plot(x, np.exp(y_f))
ax[1].plot(x, y_f)
y_t = np.polyval([res.params[3] + res.params[1], res.params[0]], x_centered) + res.params[2]
ax[0].plot(x, np.exp(y_t))
ax[1].plot(x, y_t)
ax[0].set_xlabel('OE Gestational Age (Week)')
ax[1].set_xlabel('OE Gestational Age (Week)')
ax[0].set_ylabel('Death Rate (per 1,000)')
ax[1].set_ylabel('Log Death Rate (per 1,000)')
plt.savefig('../../results/death_rate_2.png', bbox_inches='tight', dpi=300)
df = subset[subset['Prenatal Care'] == True]
df['Week Centered'] = df['OE Gestational Age Weekly Code'] - df['OE Gestational Age Weekly Code'].mean()
df['Month Centered'] = df['Month Prenatal Care Began Code'] - df['Month Prenatal Care Began Code'].mean()
df['Cross Term'] = df.apply(lambda row: row['Week Centered']*row['Month Centered'], axis=1)
df['Log Death Rate'] = df.apply(lambda row: np.log(row['Death Rate']), axis=1)
model = sm.OLS(df['Log Death Rate'], sm.add_constant(df[['Week Centered', 'Month Centered', 'Cross Term']]))
res = model.fit()
res.summary()
fig, ax = plt.subplots(1, 2, figsize=(12, 4))
sns.scatterplot(x='Month Prenatal Care Began Code', y='Death Rate', hue='OE Gestational Age Weekly',
palette='viridis', data=df, ax=ax[0], alpha=0.5, legend=False)
sns.scatterplot(x='Month Prenatal Care Began Code', y='Log Death Rate', hue='OE Gestational Age Weekly',
palette='viridis', data=df, ax=ax[1], alpha=0.5)
for week in df['OE Gestational Age Weekly Code'].unique():
x = np.linspace(1, week/4)
x_centered = x - df['Month Prenatal Care Began Code'].mean()
week_centered = week - df['OE Gestational Age Weekly Code'].mean()
y = res.params[0] + res.params[1]*week_centered + res.params[2]*x_centered + \
res.params[3]*week_centered*x_centered
ax[0].plot(x, np.exp(y), c=viridis_week.to_rgba(week))
ax[1].plot(x, y, c=viridis_week.to_rgba(week))
ax[0].set_xlabel('Month Prenatal Care Began')
ax[1].set_xlabel('Month Prenatal Care Began')
ax[0].set_ylabel('Death Rate (per 1,000)')
ax[1].set_ylabel('Log Death Rate (per 1,000)')
ax[1].legend(title='OE Gestational Age (Week)', bbox_to_anchor=(1.6, 1));
fig, ax = plt.subplots(1, 2, figsize=(12, 4))
sns.scatterplot(x='OE Gestational Age Weekly Code', y='Death Rate', hue='Month Prenatal Care Began',
palette='viridis_r', data=df, ax=ax[0], alpha=0.5, legend=False)
sns.scatterplot(x='OE Gestational Age Weekly Code', y='Log Death Rate', hue='Month Prenatal Care Began',
palette='viridis_r', data=df, ax=ax[1], alpha=0.5)
for month in df['Month Prenatal Care Began Code'].unique():
x = np.linspace(max(28, 4*month), 41)
x_centered = x - df['OE Gestational Age Weekly Code'].mean()
month_centered = month - df['Month Prenatal Care Began Code'].mean()
y = res.params[0] + res.params[1]*x_centered + res.params[2]*month_centered + \
res.params[3]*month_centered*x_centered
ax[0].plot(x, np.exp(y), c=viridis_month.to_rgba(month))
ax[1].plot(x, y, c=viridis_month.to_rgba(month))
ax[0].set_xlabel('OE Gestational Age (Week)')
ax[1].set_xlabel('OE Gestational Age (Week)')
ax[0].set_ylabel('Death Rate (per 1,000)')
ax[1].set_ylabel('Log Death Rate (per 1,000)')
ax[1].legend(title='Month Prenatal Care Began', bbox_to_anchor=(1.6, 1));
model_me = sm.MixedLM(df['Log Death Rate'].values, # dependent variable
df['Month Centered'].values, # fixed effects covariates
df['Week Centered'].values) # groups
res_me = model_me.fit()
res_me.summary(xname_fe=['Month Centered'], xname_re=['Week Centered'])
fig, ax = plt.subplots(1, 2, figsize=(11, 4))
sns.scatterplot(x='Month Prenatal Care Began Code', y='Death Rate', hue='OE Gestational Age Weekly',
palette='viridis', data=df, ax=ax[0], alpha=0.5, legend=False)
sns.scatterplot(x='Month Prenatal Care Began Code', y='Log Death Rate', hue='OE Gestational Age Weekly',
palette='viridis', data=df, ax=ax[1], alpha=0.5)
for week in df['OE Gestational Age Weekly Code'].unique():
x = np.linspace(1, week/4)
x_centered = x - df['Month Prenatal Care Began Code'].mean()
week_centered = week - df['OE Gestational Age Weekly Code'].mean()
y = res_me.random_effects[week_centered][0] + res_me.params[0]*x_centered
ax[0].plot(x, np.exp(y), c=viridis_week.to_rgba(week))
ax[1].plot(x, y, c=viridis_week.to_rgba(week))
ax[0].set_xlabel('Month Prenatal Care Began')
ax[1].set_xlabel('Month Prenatal Care Began')
ax[0].set_ylabel('Death Rate (per 1,000)')
ax[1].set_ylabel('Log Death Rate (per 1,000)')
ax[1].legend(title='OE Gestational Age (Week)', bbox_to_anchor=(1.6, 1))
plt.savefig('../../results/death_rate_3.png', bbox_inches='tight', dpi=300)
fig, ax = plt.subplots(1, 3, figsize=(18, 4))
sns.scatterplot(x='OE Gestational Age Weekly Code', y='Death Rate', hue='Month Prenatal Care Began',
palette='viridis_r', data=df, ax=ax[0], alpha=0.5, legend=False)
sns.scatterplot(x='OE Gestational Age Weekly Code', y='Log Death Rate', hue='Month Prenatal Care Began',
palette='viridis_r', data=df, ax=ax[1], alpha=0.5)
for month in df['Month Prenatal Care Began Code'].unique():
x = np.arange(max(28, 4*month), 41)
x_centered = x - df['OE Gestational Age Weekly Code'].mean()
month_centered = month - df['Month Prenatal Care Began Code'].mean()
y = [res_me.random_effects[xi][0] for xi in x_centered] + res_me.params[0]*month_centered
ax[0].plot(x, np.exp(y), 'x', c=viridis_month.to_rgba(month))
ax[1].plot(x, y, 'x', c=viridis_month.to_rgba(month))
ax[0].set_xlabel('OE Gestational Age (Week)')
ax[1].set_xlabel('OE Gestational Age (Week)')
ax[0].set_ylabel('Death Rate (per 1,000)')
ax[1].set_ylabel('Log Death Rate (per 1,000)')
ax[1].legend(title='Month Prenatal Care Began', bbox_to_anchor=(1.6, 1))
ax[2].set_visible(False)
plt.savefig('../../results/death_rate_4.png', bbox_inches='tight', dpi=300)
| 0.42179 | 0.900792 |
# Laboratorio 9
```
import pandas as pd
import altair as alt
import matplotlib.pyplot as plt
import numpy as np
from vega_datasets import data
alt.themes.enable('opaque')
%matplotlib inline
```
En este laboratorio utilizaremos un conjunto de datos _famoso_, el GapMinder. Esta es una versión reducida que solo considera países, ingresos, salud y población.
¿Hay alguna forma natural de agrupar a estos países?
```
gapminder = data.gapminder_health_income()
gapminder.head()
```
## Ejercicio 1
(1 pto.)
Realiza un Análisis exploratorio, como mínimo un `describe` del dataframe y una visualización adecuada, por ejemplo un _scatter matrix_ con los valores numéricos.
```
gapminder.describe()
alt.Chart(gapminder).mark_circle( color='red', opacity=0.7).encode(
alt.X(alt.repeat("column"), type='quantitative'),
alt.Y(alt.repeat("row"), type='quantitative'),
).properties(
width=200,
height=200
).repeat(
row=['income', 'health', 'population'],
column=['population', 'health', 'income']
)
```
__Pregunta:__ ¿Hay alguna variable que te entregue indicios a simple vista donde se puedan separar países en grupos?
__Respuesta:__ A simple vista population podria decir que hay 2 clusters (pues en cada grafico que contiene a population hay una masa de paises en un sector y 2 paises en otro sector), sin embargo en los otros graficos no se ven clusters a simple vista.
## Ejercicio 2
(1 pto.)
Aplicar un escalamiento a los datos antes de aplicar nuestro algoritmo de clustering. Para ello, definir la variable `X_raw` que corresponde a un `numpy.array` con los valores del dataframe `gapminder` en las columnas _income_, _health_ y _population_. Luego, definir la variable `X` que deben ser los datos escalados de `X_raw`.
```
from sklearn.preprocessing import StandardScaler
X_raw = gapminder.drop("country",axis=1).to_numpy()
X = StandardScaler().fit_transform(X_raw)
```
## Ejercicio 3
(1 pto.)
Definir un _estimator_ `KMeans` con `k=3` y `random_state=42`, luego ajustar con `X` y finalmente, agregar los _labels_ obtenidos a una nueva columna del dataframe `gapminder` llamada `cluster`. Finalmente, realizar el mismo gráfico del principio pero coloreado por los clusters obtenidos.
```
from sklearn.cluster import KMeans
k = 3
kmeans = KMeans(n_clusters=k)
kmeans.fit(X)
clusters = kmeans.labels_
gapminder["cluster"] = clusters
alt.Chart(gapminder).mark_circle().encode(
alt.X(alt.repeat("column"), type='quantitative'),
alt.Y(alt.repeat("row"), type='quantitative'),
color=alt.Color('cluster', scale = alt.Scale(scheme = 'plasma'))
).properties(
width=200,
height=200
).repeat(
row=['income', 'health', 'population'],
column=['population', 'health', 'income']
)
```
## Ejercicio 4
(1 pto.)
__Regla del codo__
__¿Cómo escoger la mejor cantidad de _clusters_?__
En este ejercicio hemos utilizado que el número de clusters es igual a 3. El ajuste del modelo siempre será mejor al aumentar el número de clusters, pero ello no significa que el número de clusters sea el apropiado. De hecho, si tenemos que ajustar $n$ puntos, claramente tomar $n$ clusters generaría un ajuste perfecto, pero no permitiría representar si existen realmente agrupaciones de datos.
Cuando no se conoce el número de clusters a priori, se utiliza la [regla del codo](https://jarroba.com/seleccion-del-numero-optimo-clusters/), que indica que el número más apropiado es aquel donde "cambia la pendiente" de decrecimiento de la la suma de las distancias a los clusters para cada punto, en función del número de clusters.
A continuación se provee el código para el caso de clustering sobre los datos estandarizados, leídos directamente de un archivo preparado especialmente.En la línea que se declara `kmeans` dentro del ciclo _for_ debes definir un estimador K-Means, con `k` clusters y `random_state` 42. Recuerda aprovechar de ajustar el modelo en una sola línea.
```
elbow = pd.Series(name="inertia", dtype="float64").rename_axis(index="k")
for k in range(1, 10):
kmeans = KMeans(k, random_state=42).fit(X)
elbow.loc[k] = kmeans.inertia_ # Inertia: Sum of distances of samples to their closest cluster center
elbow = elbow.reset_index()
alt.Chart(elbow).mark_line(point=True).encode(
x="k:O",
y="inertia:Q"
).properties(
height=600,
width=800
)
```
__Pregunta:__ Considerando los datos (países) y el gráfico anterior, ¿Cuántos clusters escogerías?
__Respuesta:__ Escogeria 4 clusters, pues es el primer cambio de pendiente. Si bien en k=5 hay otro cambio notorio de pendiente este parece estabilizarse para k>5.
|
github_jupyter
|
import pandas as pd
import altair as alt
import matplotlib.pyplot as plt
import numpy as np
from vega_datasets import data
alt.themes.enable('opaque')
%matplotlib inline
gapminder = data.gapminder_health_income()
gapminder.head()
gapminder.describe()
alt.Chart(gapminder).mark_circle( color='red', opacity=0.7).encode(
alt.X(alt.repeat("column"), type='quantitative'),
alt.Y(alt.repeat("row"), type='quantitative'),
).properties(
width=200,
height=200
).repeat(
row=['income', 'health', 'population'],
column=['population', 'health', 'income']
)
from sklearn.preprocessing import StandardScaler
X_raw = gapminder.drop("country",axis=1).to_numpy()
X = StandardScaler().fit_transform(X_raw)
from sklearn.cluster import KMeans
k = 3
kmeans = KMeans(n_clusters=k)
kmeans.fit(X)
clusters = kmeans.labels_
gapminder["cluster"] = clusters
alt.Chart(gapminder).mark_circle().encode(
alt.X(alt.repeat("column"), type='quantitative'),
alt.Y(alt.repeat("row"), type='quantitative'),
color=alt.Color('cluster', scale = alt.Scale(scheme = 'plasma'))
).properties(
width=200,
height=200
).repeat(
row=['income', 'health', 'population'],
column=['population', 'health', 'income']
)
elbow = pd.Series(name="inertia", dtype="float64").rename_axis(index="k")
for k in range(1, 10):
kmeans = KMeans(k, random_state=42).fit(X)
elbow.loc[k] = kmeans.inertia_ # Inertia: Sum of distances of samples to their closest cluster center
elbow = elbow.reset_index()
alt.Chart(elbow).mark_line(point=True).encode(
x="k:O",
y="inertia:Q"
).properties(
height=600,
width=800
)
| 0.604749 | 0.971645 |
# Nearest Neighbour Analysis
One commonly used GIS task is to be able to find the nearest neighbour. For instance, you might have a single Point object
representing your home location, and then another set of locations representing e.g. public transport stops. Then, quite typical question is *"which of the stops is closest one to my home?"*
This is a typical nearest neighbour analysis, where the aim is to find the closest geometry to another geometry.
In Python this kind of analysis can be done with shapely function called ``nearest_points()`` that [returns a tuple of the nearest points in the input geometries](https://shapely.readthedocs.io/en/latest/manual.html#shapely.ops.nearest_points).
## Nearest point using Shapely
Let's start by testing how we can find the nearest Point using the ``nearest_points()`` function of Shapely.
Let's create an origin Point and a few destination Points and find out the closest destination.
```
from shapely.geometry import Point, MultiPoint
from shapely.ops import nearest_points
orig = Point(1, 1.67)
dest1, dest2, dest3 = Point(0, 1.45), Point(2, 2), Point(0, 2.5)
```
To be able to find out the closest destination point from the origin, we need to create a MultiPoint object from the destination points.
```
destinations = MultiPoint([dest1, dest2, dest3])
print(destinations)
```
Okey, now we can see that all the destination points are represented as a single MultiPoint object.
- Now we can find out the nearest destination point by using ``nearest_points()`` function.
```
nearest_geoms = nearest_points(orig, destinations)
near_idx0 = nearest_geoms[0]
near_idx1 = nearest_geoms[1]
print(nearest_geoms)
print(near_idx0)
print(near_idx1)
```
As you can see the ``nearest_points()`` function returns a tuple of geometries where the first item is the geometry
of our origin point and the second item (at index 1) is the actual nearest geometry from the destination points.
Hence, the closest destination point seems to be the one located at coordinates (0, 1.45).
This is the basic logic how we can find the nearest point from a set of points.
## Nearest points using Geopandas
Of course, the previous example is not really useful yet. Hence, next I show, how it is possible to find nearest points
from a set of origin points to a set of destination points using GeoDataFrames. Here, we will use the ``PKS_suuralueet.kml`` district data, and the ``addresses.shp`` address points from previous sections.
- First we need to create a function that takes advantage of the previous function but is tailored to work with two GeoDataFrames.
```
def nearest(row, geom_union, df1, df2, geom1_col='geometry', geom2_col='geometry', src_column=None):
"""Find the nearest point and return the corresponding value from specified column."""
# Find the geometry that is closest
nearest = df2[geom2_col] == nearest_points(row[geom1_col], geom_union)[1]
# Get the corresponding value from df2 (matching is based on the geometry)
value = df2[nearest][src_column].get_values()[0]
return value
```
Next we read the address data and the Helsinki districts data and find out the closest address to the centroid of each district.
```
# Import geopandas
import geopandas as gpd
# Define filepaths
fp1 = "data/PKS_suuralue.kml"
fp2 = "data/addresses.shp"
# Enable KML driver
gpd.io.file.fiona.drvsupport.supported_drivers['KML'] = 'rw'
# Read in data with geopandas
df1 = gpd.read_file(fp1, driver='KML')
df2 = gpd.read_file(fp2)
```
Create unary union from the address points, which basically creates a MultiPoint object from the Point geometries.
```
unary_union = df2.unary_union
print(unary_union)
```
Calculate the centroids for each district area.
```
df1['centroid'] = df1.centroid
df1.head()
```
Okey now we are ready to use our function and find closest Points (taking the value from id column) from df2 to df1 centroids.
Let's store the id of the nearest address into a new column `"nearest_id"` in df1:
```
df1['nearest_id'] = df1.apply(nearest, geom_union=unary_union, df1=df1, df2=df2, geom1_col='centroid', src_column='id', axis=1)
df1.head(20)
```
That's it! Now we found the closest point for each centroid and got the ``id`` value from our addresses into the ``df1`` GeoDataFrame.
|
github_jupyter
|
from shapely.geometry import Point, MultiPoint
from shapely.ops import nearest_points
orig = Point(1, 1.67)
dest1, dest2, dest3 = Point(0, 1.45), Point(2, 2), Point(0, 2.5)
destinations = MultiPoint([dest1, dest2, dest3])
print(destinations)
nearest_geoms = nearest_points(orig, destinations)
near_idx0 = nearest_geoms[0]
near_idx1 = nearest_geoms[1]
print(nearest_geoms)
print(near_idx0)
print(near_idx1)
def nearest(row, geom_union, df1, df2, geom1_col='geometry', geom2_col='geometry', src_column=None):
"""Find the nearest point and return the corresponding value from specified column."""
# Find the geometry that is closest
nearest = df2[geom2_col] == nearest_points(row[geom1_col], geom_union)[1]
# Get the corresponding value from df2 (matching is based on the geometry)
value = df2[nearest][src_column].get_values()[0]
return value
# Import geopandas
import geopandas as gpd
# Define filepaths
fp1 = "data/PKS_suuralue.kml"
fp2 = "data/addresses.shp"
# Enable KML driver
gpd.io.file.fiona.drvsupport.supported_drivers['KML'] = 'rw'
# Read in data with geopandas
df1 = gpd.read_file(fp1, driver='KML')
df2 = gpd.read_file(fp2)
unary_union = df2.unary_union
print(unary_union)
df1['centroid'] = df1.centroid
df1.head()
df1['nearest_id'] = df1.apply(nearest, geom_union=unary_union, df1=df1, df2=df2, geom1_col='centroid', src_column='id', axis=1)
df1.head(20)
| 0.7773 | 0.992237 |
```
# Importacao de pacotes
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
from ising2d import *
# Configuracoes de plot para o relatorio
plt.rcParams['figure.dpi'] = 72*2
plt.rcParams['figure.figsize'] = (0.7*5.95114, 0.7*5.95114)
plt.rcParams['font.size'] = 10
plt.rcParams['axes.labelsize'] = 9
plt.rcParams['xtick.labelsize'] = 9
plt.rcParams['ytick.labelsize'] = 9
plt.rcParams['legend.fontsize'] = 9
plt.rcParams['lines.linewidth'] = 1
np.seterr(divide = 'ignore', invalid = 'ignore'); # ignora warnings sobre divisao por zero (no calculo da probabilidade de flipar os spins para T = 0)
```
## Versao interativa da simulacao
```
# Inicializacao das variaveis da simulacao
L = 100 # largura do grid de spins
J = 1 # constante de interacao entre os spins
h = 0 # constante de interacao do spin com o campo magnetico externo
Nsteps = 2000 # numero de passos de Monte Carlo da simulacao
T = 1 # temperatura da simulacao (kB = 1)
start = choice([-1, +1], (L, L)) # configuracao inicial dos spins (aleatoria)
steps = [start] # vetor de arrays de spins para cada passo de Monte Carlo
for k in range(Nsteps):
steps.append(ising_step(steps[-1].copy(), T, J, h))
display_ising_sequence(steps);
```
### Figuras dos estados inicial e final para a simulacao anterior
```
fig, axes = plt.subplots(ncols = 2)
axes[0].imshow(steps[0], cmap = 'binary')
axes[0].axis('off')
axes[0].set_title('(a) n = {:d}'.format(0), fontsize = 10)
axes[1].imshow(steps[-1], cmap = 'binary')
axes[1].axis('off')
axes[1].set_title('(b) n = {:d}'.format(Nsteps), fontsize = 10)
plt.savefig('../relatorio/img/evolucao-grid-100x100.eps', format = 'eps', dpi = 300)
plt.show()
```
## Graficos da evolucao da magnetizacao por sitio e da energia por sitio a cada passo de Monte Carlo
```
L = 100 # largura do grid de spins
J = 1 # constante de interacao entre os spins
h = 0 # constante de interacao do spin com o campo magnetico externo
TC = 2*J/np.log(1 + np.sqrt(2)) # temperatura critica (solucao analitica)
vecT = [0.001, 2, TC, 4] # temperaturas das curvas
Nsteps = 1000 # numero de passos de Monte Carlo da simulacao
fig, axes = plt.subplots(nrows = 2, sharex = True)
axes[0].set_ylabel('Magnetizacao por sitio')
axes[0].set_ylim(-1.1, 1.1)
axes[1].set_xlabel('Passo')
axes[1].set_ylabel('Energia por sitio')
for T in vecT:
m = np.zeros(Nsteps) # vetor de magnetizacoes de cada passo de Monte Carlo
e = np.zeros(Nsteps) # vetor de energias de cada passo de Monte Carlo
steps = [choice([-1, +1], (L, L))] # vetor de arrays de spins para cada passo de Monte Carlo
for k in range(Nsteps):
steps.append(ising_step(steps[-1].copy(), T, J, h))
m[k] = magnetization_per_site(steps[k])
e[k] = energy_per_site(steps[k], J, h)
axes[0].plot(m, label = 'T/kB = {:.3f}'.format(T))
axes[1].plot(e, label = 'T/kB = {:.3f}'.format(T))
axes[0].legend(loc = 'best')
axes[1].legend(loc = 'best')
plt.tight_layout()
plt.savefig('../relatorio/img/evolucao-mc-temperaturas.eps', format = 'eps', dpi = 300)
plt.show()
```
## Graficos da evolucao da magnetizacao por sitio e da energia por sitio em funcao da temperatura para varias larguras
```
J = 1 # constante de interacao entre os spins
h = 0 # constante de interacao do spin com o campo magnetico externo
TC = 2*J/np.log(1 + np.sqrt(2)) # temperatura critica (solucao analitica)
Tmin = 1e-17 # temperatura proxima de zero
Tmax = 5 # temperatura maxima
DeltaT = 0.5 # passo em temperaturas
T = np.arange(Tmin, Tmax + DeltaT, DeltaT)
NT = len(T)
vecL = [10, 20, 30] # tamanho do array de spins
vecNsteps = [5000, 10000, 15000] # numero de passos de Monte Carlo
vectrans = [1000, 2000, 3000] # tamanho do transiente a ser removido em numero de passos
# Plot da funcao exata (solucao de Onsager)
linT = np.linspace(Tmin, Tmax, 1000, endpoint = True)
mexact = np.zeros(len(linT), dtype = np.float128) # solucao exata
for k in range(len(linT)):
if linT[k] < TC:
mexact[k] = (1 - np.sinh(2*J/linT[k])**(-4))**(1/8)
fig, axes = plt.subplots(nrows = 2, sharex = True)
axes[0].set_ylabel('Magnetizacao por sitio')
axes[0].set_ylim(-0.1, 1.1)
axes[1].set_xlabel('Temperatura / kB')
axes[1].set_ylabel('Energia por sitio')
axes[0].plot(linT, mexact, 'k--', label = 'Onsager')
# Obtem curvas de magnetizacao e energia por temperatura para alguns tamanhos de grid
for L, Nsteps, ntrans in zip(vecL, vecNsteps, vectrans):
m = np.zeros(NT) # vetor de magnetizacoes medias de cada temperatura
e = np.zeros(NT) # vetor de energias medias de cada temperatura
for k in range(NT):
thism = np.zeros(Nsteps) # vetor de magnetizacoes de cada passo de Monte Carlo
thise = np.zeros(Nsteps) # vetor de energias de cada passo de Monte Carlo
steps = [choice([-1, +1], (L, L))] # vetor de arrays de spins para cada passo de Monte Carlo
for n in range(Nsteps):
steps.append(ising_step(steps[-1].copy(), T[k], J, h))
thism[n] = magnetization_per_site(steps[n])
thise[n] = energy_per_site(steps[n], J, h)
m[k] = np.mean(np.abs(thism[ntrans:]))
e[k] = np.mean(thise[ntrans:])
axes[0].plot(T, m, '-o', markersize = 3, label = 'L = {:d}'.format(L))
axes[1].plot(T, e, '-o', markersize = 3, label = 'L = {:d}'.format(L))
axes[0].legend(loc = 'best')
axes[1].legend(loc = 'best')
plt.savefig('../relatorio/img/simulacao-3-larguras.eps', format = 'eps', dpi = 300)
plt.show()
```
## Graficos da evolucao da magnetizacao por sitio e da energia por sitio para cada passo de Monte Carlo com h = 1
```
L = 100 # largura do grid de spins
J = 1 # constante de interacao entre os spins
h = 1 # constante de interacao do spin com o campo magnetico externo
TC = 2*J/np.log(1 + np.sqrt(2)) # temperatura critica (solucao analitica)
vecT = [0.001, 2, TC, 4] # temperaturas das curvas
Nsteps = 1000 # numero de passos de Monte Carlo da simulacao
fig, axes = plt.subplots(nrows = 2, sharex = True)
axes[0].set_ylabel('Magnetizacao por sitio')
axes[0].set_ylim(-1.1, 1.1)
axes[1].set_xlabel('Passo')
axes[1].set_ylabel('Energia por sitio')
for T in vecT:
m = np.zeros(Nsteps) # vetor de magnetizacoes de cada passo de Monte Carlo
e = np.zeros(Nsteps) # vetor de energias de cada passo de Monte Carlo
steps = [choice([-1, +1], (L, L))] # vetor de arrays de spins para cada passo de Monte Carlo
for k in range(Nsteps):
steps.append(ising_step(steps[-1].copy(), T, J, h))
m[k] = magnetization_per_site(steps[k])
e[k] = energy_per_site(steps[k], J, h)
axes[0].plot(m, label = 'T/kB = {:.3f}'.format(T))
axes[1].plot(e, label = 'T/kB = {:.3f}'.format(T))
axes[0].legend(loc = 'best')
axes[1].legend(loc = 'best')
plt.tight_layout()
plt.savefig('../relatorio/img/evolucao-h=1-mc-temperaturas.eps', format = 'eps', dpi = 300)
plt.show()
J = 1 # constante de interacao entre os spins
vech = [0, 0.2, 1] # constante de interacao do spin com o campo magnetico externo
TC = 2*J/np.log(1 + np.sqrt(2)) # temperatura critica (solucao analitica)
Tmin = 1e-17 # temperatura proxima de zero
Tmax = 5 # temperatura maxima
DeltaT = 0.5 # passo em temperaturas
T = np.arange(Tmin, Tmax + DeltaT, DeltaT)
NT = len(T)
L = 10 # tamanho do array de spins
Nsteps = 20000 # numero de passos de Monte Carlo
trans = 1000 # tamanho do transiente a ser removido em numero de passos
# Plot da funcao exata (solucao de Onsager)
linT = np.linspace(Tmin, Tmax, 1000, endpoint = True)
mexact = np.zeros(len(linT), dtype = np.float128) # solucao exata
for k in range(len(linT)):
if linT[k] < TC:
mexact[k] = (1 - np.sinh(2*J/linT[k])**(-4))**(1/8)
fig, axes = plt.subplots(nrows = 2, sharex = True)
axes[0].set_ylabel('Magnetizacao por sitio')
axes[0].set_ylim(-0.1, 1.1)
axes[1].set_xlabel('Temperatura / kB')
axes[1].set_ylabel('Energia por sitio')
axes[0].plot(linT, mexact, 'k--', label = 'Onsager')
# Obtem curvas de magnetizacao e energia por temperatura para alguns tamanhos de grid
for h in vech:
m = np.zeros(NT) # vetor de magnetizacoes medias de cada temperatura
e = np.zeros(NT) # vetor de energias medias de cada temperatura
for k in range(NT):
thism = np.zeros(Nsteps) # vetor de magnetizacoes de cada passo de Monte Carlo
thise = np.zeros(Nsteps) # vetor de energias de cada passo de Monte Carlo
steps = [choice([-1, +1], (L, L))] # vetor de arrays de spins para cada passo de Monte Carlo
for n in range(Nsteps):
steps.append(ising_step(steps[-1].copy(), T[k], J, h))
thism[n] = magnetization_per_site(steps[n])
thise[n] = energy_per_site(steps[n], J, h)
m[k] = np.mean(np.abs(thism[trans:]))
e[k] = np.mean(thise[trans:])
axes[0].plot(T, m, '-o', markersize = 3, label = 'h = {:.3f}'.format(h))
axes[1].plot(T, e, '-o', markersize = 3, label = 'h = {:.3f}'.format(h))
axes[0].legend(loc = 3)
axes[1].legend(loc = 'best')
plt.savefig('../relatorio/img/simulacao-h.eps', format = 'eps', dpi = 300)
plt.show()
```
|
github_jupyter
|
# Importacao de pacotes
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
from ising2d import *
# Configuracoes de plot para o relatorio
plt.rcParams['figure.dpi'] = 72*2
plt.rcParams['figure.figsize'] = (0.7*5.95114, 0.7*5.95114)
plt.rcParams['font.size'] = 10
plt.rcParams['axes.labelsize'] = 9
plt.rcParams['xtick.labelsize'] = 9
plt.rcParams['ytick.labelsize'] = 9
plt.rcParams['legend.fontsize'] = 9
plt.rcParams['lines.linewidth'] = 1
np.seterr(divide = 'ignore', invalid = 'ignore'); # ignora warnings sobre divisao por zero (no calculo da probabilidade de flipar os spins para T = 0)
# Inicializacao das variaveis da simulacao
L = 100 # largura do grid de spins
J = 1 # constante de interacao entre os spins
h = 0 # constante de interacao do spin com o campo magnetico externo
Nsteps = 2000 # numero de passos de Monte Carlo da simulacao
T = 1 # temperatura da simulacao (kB = 1)
start = choice([-1, +1], (L, L)) # configuracao inicial dos spins (aleatoria)
steps = [start] # vetor de arrays de spins para cada passo de Monte Carlo
for k in range(Nsteps):
steps.append(ising_step(steps[-1].copy(), T, J, h))
display_ising_sequence(steps);
fig, axes = plt.subplots(ncols = 2)
axes[0].imshow(steps[0], cmap = 'binary')
axes[0].axis('off')
axes[0].set_title('(a) n = {:d}'.format(0), fontsize = 10)
axes[1].imshow(steps[-1], cmap = 'binary')
axes[1].axis('off')
axes[1].set_title('(b) n = {:d}'.format(Nsteps), fontsize = 10)
plt.savefig('../relatorio/img/evolucao-grid-100x100.eps', format = 'eps', dpi = 300)
plt.show()
L = 100 # largura do grid de spins
J = 1 # constante de interacao entre os spins
h = 0 # constante de interacao do spin com o campo magnetico externo
TC = 2*J/np.log(1 + np.sqrt(2)) # temperatura critica (solucao analitica)
vecT = [0.001, 2, TC, 4] # temperaturas das curvas
Nsteps = 1000 # numero de passos de Monte Carlo da simulacao
fig, axes = plt.subplots(nrows = 2, sharex = True)
axes[0].set_ylabel('Magnetizacao por sitio')
axes[0].set_ylim(-1.1, 1.1)
axes[1].set_xlabel('Passo')
axes[1].set_ylabel('Energia por sitio')
for T in vecT:
m = np.zeros(Nsteps) # vetor de magnetizacoes de cada passo de Monte Carlo
e = np.zeros(Nsteps) # vetor de energias de cada passo de Monte Carlo
steps = [choice([-1, +1], (L, L))] # vetor de arrays de spins para cada passo de Monte Carlo
for k in range(Nsteps):
steps.append(ising_step(steps[-1].copy(), T, J, h))
m[k] = magnetization_per_site(steps[k])
e[k] = energy_per_site(steps[k], J, h)
axes[0].plot(m, label = 'T/kB = {:.3f}'.format(T))
axes[1].plot(e, label = 'T/kB = {:.3f}'.format(T))
axes[0].legend(loc = 'best')
axes[1].legend(loc = 'best')
plt.tight_layout()
plt.savefig('../relatorio/img/evolucao-mc-temperaturas.eps', format = 'eps', dpi = 300)
plt.show()
J = 1 # constante de interacao entre os spins
h = 0 # constante de interacao do spin com o campo magnetico externo
TC = 2*J/np.log(1 + np.sqrt(2)) # temperatura critica (solucao analitica)
Tmin = 1e-17 # temperatura proxima de zero
Tmax = 5 # temperatura maxima
DeltaT = 0.5 # passo em temperaturas
T = np.arange(Tmin, Tmax + DeltaT, DeltaT)
NT = len(T)
vecL = [10, 20, 30] # tamanho do array de spins
vecNsteps = [5000, 10000, 15000] # numero de passos de Monte Carlo
vectrans = [1000, 2000, 3000] # tamanho do transiente a ser removido em numero de passos
# Plot da funcao exata (solucao de Onsager)
linT = np.linspace(Tmin, Tmax, 1000, endpoint = True)
mexact = np.zeros(len(linT), dtype = np.float128) # solucao exata
for k in range(len(linT)):
if linT[k] < TC:
mexact[k] = (1 - np.sinh(2*J/linT[k])**(-4))**(1/8)
fig, axes = plt.subplots(nrows = 2, sharex = True)
axes[0].set_ylabel('Magnetizacao por sitio')
axes[0].set_ylim(-0.1, 1.1)
axes[1].set_xlabel('Temperatura / kB')
axes[1].set_ylabel('Energia por sitio')
axes[0].plot(linT, mexact, 'k--', label = 'Onsager')
# Obtem curvas de magnetizacao e energia por temperatura para alguns tamanhos de grid
for L, Nsteps, ntrans in zip(vecL, vecNsteps, vectrans):
m = np.zeros(NT) # vetor de magnetizacoes medias de cada temperatura
e = np.zeros(NT) # vetor de energias medias de cada temperatura
for k in range(NT):
thism = np.zeros(Nsteps) # vetor de magnetizacoes de cada passo de Monte Carlo
thise = np.zeros(Nsteps) # vetor de energias de cada passo de Monte Carlo
steps = [choice([-1, +1], (L, L))] # vetor de arrays de spins para cada passo de Monte Carlo
for n in range(Nsteps):
steps.append(ising_step(steps[-1].copy(), T[k], J, h))
thism[n] = magnetization_per_site(steps[n])
thise[n] = energy_per_site(steps[n], J, h)
m[k] = np.mean(np.abs(thism[ntrans:]))
e[k] = np.mean(thise[ntrans:])
axes[0].plot(T, m, '-o', markersize = 3, label = 'L = {:d}'.format(L))
axes[1].plot(T, e, '-o', markersize = 3, label = 'L = {:d}'.format(L))
axes[0].legend(loc = 'best')
axes[1].legend(loc = 'best')
plt.savefig('../relatorio/img/simulacao-3-larguras.eps', format = 'eps', dpi = 300)
plt.show()
L = 100 # largura do grid de spins
J = 1 # constante de interacao entre os spins
h = 1 # constante de interacao do spin com o campo magnetico externo
TC = 2*J/np.log(1 + np.sqrt(2)) # temperatura critica (solucao analitica)
vecT = [0.001, 2, TC, 4] # temperaturas das curvas
Nsteps = 1000 # numero de passos de Monte Carlo da simulacao
fig, axes = plt.subplots(nrows = 2, sharex = True)
axes[0].set_ylabel('Magnetizacao por sitio')
axes[0].set_ylim(-1.1, 1.1)
axes[1].set_xlabel('Passo')
axes[1].set_ylabel('Energia por sitio')
for T in vecT:
m = np.zeros(Nsteps) # vetor de magnetizacoes de cada passo de Monte Carlo
e = np.zeros(Nsteps) # vetor de energias de cada passo de Monte Carlo
steps = [choice([-1, +1], (L, L))] # vetor de arrays de spins para cada passo de Monte Carlo
for k in range(Nsteps):
steps.append(ising_step(steps[-1].copy(), T, J, h))
m[k] = magnetization_per_site(steps[k])
e[k] = energy_per_site(steps[k], J, h)
axes[0].plot(m, label = 'T/kB = {:.3f}'.format(T))
axes[1].plot(e, label = 'T/kB = {:.3f}'.format(T))
axes[0].legend(loc = 'best')
axes[1].legend(loc = 'best')
plt.tight_layout()
plt.savefig('../relatorio/img/evolucao-h=1-mc-temperaturas.eps', format = 'eps', dpi = 300)
plt.show()
J = 1 # constante de interacao entre os spins
vech = [0, 0.2, 1] # constante de interacao do spin com o campo magnetico externo
TC = 2*J/np.log(1 + np.sqrt(2)) # temperatura critica (solucao analitica)
Tmin = 1e-17 # temperatura proxima de zero
Tmax = 5 # temperatura maxima
DeltaT = 0.5 # passo em temperaturas
T = np.arange(Tmin, Tmax + DeltaT, DeltaT)
NT = len(T)
L = 10 # tamanho do array de spins
Nsteps = 20000 # numero de passos de Monte Carlo
trans = 1000 # tamanho do transiente a ser removido em numero de passos
# Plot da funcao exata (solucao de Onsager)
linT = np.linspace(Tmin, Tmax, 1000, endpoint = True)
mexact = np.zeros(len(linT), dtype = np.float128) # solucao exata
for k in range(len(linT)):
if linT[k] < TC:
mexact[k] = (1 - np.sinh(2*J/linT[k])**(-4))**(1/8)
fig, axes = plt.subplots(nrows = 2, sharex = True)
axes[0].set_ylabel('Magnetizacao por sitio')
axes[0].set_ylim(-0.1, 1.1)
axes[1].set_xlabel('Temperatura / kB')
axes[1].set_ylabel('Energia por sitio')
axes[0].plot(linT, mexact, 'k--', label = 'Onsager')
# Obtem curvas de magnetizacao e energia por temperatura para alguns tamanhos de grid
for h in vech:
m = np.zeros(NT) # vetor de magnetizacoes medias de cada temperatura
e = np.zeros(NT) # vetor de energias medias de cada temperatura
for k in range(NT):
thism = np.zeros(Nsteps) # vetor de magnetizacoes de cada passo de Monte Carlo
thise = np.zeros(Nsteps) # vetor de energias de cada passo de Monte Carlo
steps = [choice([-1, +1], (L, L))] # vetor de arrays de spins para cada passo de Monte Carlo
for n in range(Nsteps):
steps.append(ising_step(steps[-1].copy(), T[k], J, h))
thism[n] = magnetization_per_site(steps[n])
thise[n] = energy_per_site(steps[n], J, h)
m[k] = np.mean(np.abs(thism[trans:]))
e[k] = np.mean(thise[trans:])
axes[0].plot(T, m, '-o', markersize = 3, label = 'h = {:.3f}'.format(h))
axes[1].plot(T, e, '-o', markersize = 3, label = 'h = {:.3f}'.format(h))
axes[0].legend(loc = 3)
axes[1].legend(loc = 'best')
plt.savefig('../relatorio/img/simulacao-h.eps', format = 'eps', dpi = 300)
plt.show()
| 0.486332 | 0.904524 |
```
import heapq
import numpy as np
import pandas as pd
from matplotlib import pyplot as plt
from sklearn.datasets import fetch_20newsgroups
from sklearn.model_selection import StratifiedKFold
%matplotlib inline
```
# Задача классификации текстов
Задача классификации текстов заключается в том, чтобы определить по документу его класс.
В данном случае предлагается рассмотреть в качестве документов - письма, заранее отклассифицированных по 20 темам.
```
all_categories = fetch_20newsgroups().target_names
all_categories
```
Возьмём всего 3 темы, но из одного раздела (документы из близких тем сложнее отличать друг от друга)
```
categories = [
'sci.electronics',
'sci.space',
'sci.med'
]
train_data = fetch_20newsgroups(subset='train',
categories=categories,
remove=('headers', 'footers', 'quotes'))
test_data = fetch_20newsgroups(subset='test',
categories=categories,
remove=('headers', 'footers', 'quotes'))
```
## Векторизация текстов
**Вопрос: как описать текстовые документы пространством признаков?**
**Идея №1**: мешок слов (bag-of-words) - каждый документ или текст выглядит как неупорядоченный набор слов без сведений о связях между ними.
<img src='https://st2.depositphotos.com/2454953/9959/i/450/depositphotos_99593622-stock-photo-holidays-travel-bag-word-cloud.jpg'>
**Идея №2**: создаём вектор "слов", каждая компонента отвечает отдельному слову.
Для векторизации текстов воспользуемся [CountVectorizer](https://scikit-learn.org/stable/modules/generated/sklearn.feature_extraction.text.CountVectorizer.html). Можно всячески варировать извлечение признаков (убирать редкие слова, убирать частые слова, убирать слова общей лексики, брать биграмы и т.д.)
```
from sklearn.feature_extraction.text import CountVectorizer, TfidfVectorizer
CountVectorizer()
count_vectorizer = CountVectorizer(min_df=5, ngram_range=(1, 2))
sparse_feature_matrix = count_vectorizer.fit_transform(train_data.data)
sparse_feature_matrix
num_2_words = {
v: k
for k, v in count_vectorizer.vocabulary_.items()
}
```
Слова с наибольшим положительным весом, являются характерными словами темы
```
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import accuracy_score, f1_score
from sklearn.metrics.scorer import make_scorer
from sklearn.model_selection import cross_val_score, GridSearchCV
```
Воспользуемся `macro`-average для оценки качества решения в задаче многоклассовой классификации.
```
f_scorer = make_scorer(f1_score, average='macro')
```
Обучим логистическую регрессию для предсказания темы документа
```
algo = LogisticRegression(C=0.00001)
algo.fit(sparse_feature_matrix, train_data.target)
W = algo.coef_.shape[1]
for c in algo.classes_:
topic_words = [
num_2_words[w_num]
for w_num in heapq.nlargest(10, range(W), key=lambda w: algo.coef_[c, w])
]
print(', '.join(topic_words))
```
Сравним качество на обучающей и отложенной выборках.
```
algo.fit(sparse_feature_matrix, train_data.target)
f_scorer(algo, sparse_feature_matrix, train_data.target)
f_scorer(algo, count_vectorizer.transform(test_data.data), test_data.target)
```
Значения f-меры получились очень низкие.
**Вопрос:** в чём причина?
```
plt.hist(algo.coef_[0], bins=500)
plt.xlim([-0.0006, 0.0006])
plt.show()
```
** Какую выбрать метрику для регуляризации? **
```
algo = LogisticRegression(penalty='l1', C=0.1)
arr = cross_val_score(algo, sparse_feature_matrix, train_data.target, cv=5, scoring=f_scorer)
print(arr)
print(np.mean(arr))
algo.fit(sparse_feature_matrix, train_data.target)
f_scorer(algo, sparse_feature_matrix, train_data.target)
f_scorer(algo, count_vectorizer.transform(test_data.data), test_data.target)
```
Подберём оптимальное значение параметра регуляризации
```
def grid_plot(x, y, x_label, title, y_label='f_measure'):
plt.figure(figsize=(12, 6))
plt.grid(True),
plt.plot(x, y, 'go-')
plt.xlabel(x_label)
plt.ylabel(y_label)
plt.title(title)
print(*map(float, np.logspace(-2, 2, 10)))
lr_grid = {
'C': np.logspace(-2, 2, 10),
}
gs = GridSearchCV(LogisticRegression(penalty='l1'), lr_grid, scoring=f_scorer, cv=5, n_jobs=5)
%time gs.fit(sparse_feature_matrix, train_data.target)
print("best_params: {}, best_score: {}".format(gs.best_params_, gs.best_score_))
```
Рассмотрим график:
```
grid_plot(
lr_grid['C'], gs.cv_results_['mean_test_score'], 'C - coefficient of regularization', 'LogReg(penalty=l1)'
)
lr_grid = {
'C': np.linspace(1, 20, 40),
}
gs = GridSearchCV(LogisticRegression(penalty='l1'), lr_grid, scoring=f_scorer, cv=5, n_jobs=5)
%time gs.fit(sparse_feature_matrix, train_data.target)
print("best_params: {}, best_score: {}".format(gs.best_params_, gs.best_score_))
grid_plot(
lr_grid['C'], gs.cv_results_['mean_test_score'], 'C - coefficient of regularization', 'LogReg(penalty=l1)'
)
lr_final = LogisticRegression(penalty='l1', C=10)
%time lr_final.fit(sparse_feature_matrix, train_data.target)
accuracy_score(lr_final.predict(sparse_feature_matrix), train_data.target)
f_scorer(lr_final, sparse_feature_matrix, train_data.target)
accuracy_score(lr_final.predict(count_vectorizer.transform(test_data.data)), test_data.target)
f_scorer(lr_final, count_vectorizer.transform(test_data.data), test_data.target)
```
## Регуляризация вместе с векторизацией признаков
Чтобы не делать векторизацию и обучение раздельно, есть удобный класс Pipeline. Он позволяет объединить в цепочку последовательность действий
```
from sklearn.pipeline import Pipeline
pipeline = Pipeline([
("vectorizer", CountVectorizer(min_df=5, ngram_range=(1, 2))),
("algo", LogisticRegression())
])
pipeline.fit(train_data.data, train_data.target)
f_scorer(pipeline, train_data.data, train_data.target)
f_scorer(pipeline, test_data.data, test_data.target)
```
Значения такие же как мы получали ранее, делая шаги раздельно.
```
from sklearn.pipeline import make_pipeline
```
При кроссвалидации нужно, чтобы CountVectorizer не обучался на тесте (иначе объекты становятся зависимыми). Pipeline позволяет это просто сделать.
```
pipeline = make_pipeline(CountVectorizer(min_df=5, ngram_range=(1, 2)), LogisticRegression())
arr = cross_val_score(pipeline, train_data.data, train_data.target, cv=5, scoring=f_scorer)
print(arr)
print(np.mean(arr))
```
В Pipeline можно добавлять новые шаги препроцессинга данных
```
from sklearn.feature_extraction.text import TfidfTransformer
pipeline = make_pipeline(CountVectorizer(min_df=5, ngram_range=(1, 2)), TfidfTransformer(), LogisticRegression())
arr = cross_val_score(pipeline, train_data.data, train_data.target, cv=5, scoring=f_scorer)
print(arr)
print(np.mean(arr))
pipeline.fit(train_data.data, train_data.target)
accuracy_score(pipeline.predict(train_data.data), train_data.target)
f_scorer(pipeline, train_data.data, train_data.target)
accuracy_score(pipeline.predict(test_data.data), test_data.target)
f_scorer(pipeline, test_data.data, test_data.target)
```
Качество стало немного лучше
# Классификация сообщений чатов
В качестве задания предлагается построить модель классификации текстов, соответствующих сообщениям из чатов по ML, Python и знакомствам.
**Данные** можно взять с <a src="https://www.kaggle.com/c/tfstextclassification">соревнования на Kaggle</a>, проведенное в рамках курса "Диалоговые системы" в Тинькофф. Прямая [ссылка](https://www.dropbox.com/s/8wckwzfy63ajxpm/tfstextclassification.zip?dl=0) на скачивание.
```
data_path = 'data/{}'
df = pd.read_csv('data/train.csv')
```
### Первичный анализ данных
```
print(df.shape)
df.head()
label = 0
print('Label: ', label, '\n'+'='*100+'\n')
print(*df[df['label'] == label].sample(10).text, sep='\n'+'-'*100+'\n\n')
label = 1
print('Label: ', label, '\n'+'='*100+'\n')
print(*df[df['label'] == label].sample(10).text, sep='\n'+'-'*100+'\n\n')
label = 2
print('Label: ', label, '\n'+'='*100+'\n')
print(*df[df['label'] == label].sample(10).text, sep='\n'+'-'*100+'\n\n')
```
### Разделим данные на train/test
```
skf = StratifiedKFold(3, random_state=37)
train_index, test_index = next(skf.split(df.text, df.label))
train_df, test_df = df.iloc[train_index], df.iloc[test_index]
print(train_df.shape, test_df.shape)
train_df.head()
test_df.head()
```
## Baseline
```
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.decomposition import TruncatedSVD
from xgboost.sklearn import XGBClassifier
```
Преобразуем данные
```
X_train = train_df.text
y_train = train_df.label
print(X_train.shape)
X_test = test_df.text
y_test = test_df.label
print(X_test.shape)
```
Подготовим pipeline
```
pipeline = Pipeline([
("vectorizer", CountVectorizer()),
("clf", DecisionTreeClassifier()),
])
```
Обучим классификатор
```
%%time
clf = pipeline
clf.fit(X_train, y_train)
```
Оценим качество
```
print("Train_acc: {:.4f}, train_f-measure: {:.4f}".format(
accuracy_score(clf.predict(X_train), y_train),
f_scorer(clf, X_train, y_train)
))
print("Test_acc: {:.4f}, test_f-measure: {:.4f}".format(
accuracy_score(clf.predict(X_test), y_test),
f_scorer(clf, X_test, y_test)
))
```
### Your turn
Как видим, наша модель переобучилась. Для получения лучших результатов попробуйте воспользоваться более хитрыми и походящими инструментами.
1. Попробуйте поработать с параметрами `CountVectorizer`.
2. Попробуйте воспользоваться TF-IDF для кодирования текстовой информации ([ссылка](https://scikit-learn.org/stable/modules/generated/sklearn.feature_extraction.text.TfidfVectorizer.html)).
3. Попробуйте воспользоваться другими моделями и средствами снижения размерности.
Формальный критерий успешности выполнения данного задания:
* Проведен честный эксперимент с апробацией различных методов (>=3)
* Полученный алгоритм не выказывает явных следов переобучения (качество на train и test не различаются более, чем на 0.03 условных попугая)
* Test accuracy >= 0.835, f1-score >= 0.815
**0.5 extra points**
При частичном выполнении данного задания можно получить неполный балл. Качественный эксперимент, не побивший пороговые значения оценивается неполным баллом на усмотрение преподавателя. При получении пороговых score'ов случайным образом (без анализа) бонус не засчитывается.
|
github_jupyter
|
import heapq
import numpy as np
import pandas as pd
from matplotlib import pyplot as plt
from sklearn.datasets import fetch_20newsgroups
from sklearn.model_selection import StratifiedKFold
%matplotlib inline
all_categories = fetch_20newsgroups().target_names
all_categories
categories = [
'sci.electronics',
'sci.space',
'sci.med'
]
train_data = fetch_20newsgroups(subset='train',
categories=categories,
remove=('headers', 'footers', 'quotes'))
test_data = fetch_20newsgroups(subset='test',
categories=categories,
remove=('headers', 'footers', 'quotes'))
from sklearn.feature_extraction.text import CountVectorizer, TfidfVectorizer
CountVectorizer()
count_vectorizer = CountVectorizer(min_df=5, ngram_range=(1, 2))
sparse_feature_matrix = count_vectorizer.fit_transform(train_data.data)
sparse_feature_matrix
num_2_words = {
v: k
for k, v in count_vectorizer.vocabulary_.items()
}
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import accuracy_score, f1_score
from sklearn.metrics.scorer import make_scorer
from sklearn.model_selection import cross_val_score, GridSearchCV
f_scorer = make_scorer(f1_score, average='macro')
algo = LogisticRegression(C=0.00001)
algo.fit(sparse_feature_matrix, train_data.target)
W = algo.coef_.shape[1]
for c in algo.classes_:
topic_words = [
num_2_words[w_num]
for w_num in heapq.nlargest(10, range(W), key=lambda w: algo.coef_[c, w])
]
print(', '.join(topic_words))
algo.fit(sparse_feature_matrix, train_data.target)
f_scorer(algo, sparse_feature_matrix, train_data.target)
f_scorer(algo, count_vectorizer.transform(test_data.data), test_data.target)
plt.hist(algo.coef_[0], bins=500)
plt.xlim([-0.0006, 0.0006])
plt.show()
algo = LogisticRegression(penalty='l1', C=0.1)
arr = cross_val_score(algo, sparse_feature_matrix, train_data.target, cv=5, scoring=f_scorer)
print(arr)
print(np.mean(arr))
algo.fit(sparse_feature_matrix, train_data.target)
f_scorer(algo, sparse_feature_matrix, train_data.target)
f_scorer(algo, count_vectorizer.transform(test_data.data), test_data.target)
def grid_plot(x, y, x_label, title, y_label='f_measure'):
plt.figure(figsize=(12, 6))
plt.grid(True),
plt.plot(x, y, 'go-')
plt.xlabel(x_label)
plt.ylabel(y_label)
plt.title(title)
print(*map(float, np.logspace(-2, 2, 10)))
lr_grid = {
'C': np.logspace(-2, 2, 10),
}
gs = GridSearchCV(LogisticRegression(penalty='l1'), lr_grid, scoring=f_scorer, cv=5, n_jobs=5)
%time gs.fit(sparse_feature_matrix, train_data.target)
print("best_params: {}, best_score: {}".format(gs.best_params_, gs.best_score_))
grid_plot(
lr_grid['C'], gs.cv_results_['mean_test_score'], 'C - coefficient of regularization', 'LogReg(penalty=l1)'
)
lr_grid = {
'C': np.linspace(1, 20, 40),
}
gs = GridSearchCV(LogisticRegression(penalty='l1'), lr_grid, scoring=f_scorer, cv=5, n_jobs=5)
%time gs.fit(sparse_feature_matrix, train_data.target)
print("best_params: {}, best_score: {}".format(gs.best_params_, gs.best_score_))
grid_plot(
lr_grid['C'], gs.cv_results_['mean_test_score'], 'C - coefficient of regularization', 'LogReg(penalty=l1)'
)
lr_final = LogisticRegression(penalty='l1', C=10)
%time lr_final.fit(sparse_feature_matrix, train_data.target)
accuracy_score(lr_final.predict(sparse_feature_matrix), train_data.target)
f_scorer(lr_final, sparse_feature_matrix, train_data.target)
accuracy_score(lr_final.predict(count_vectorizer.transform(test_data.data)), test_data.target)
f_scorer(lr_final, count_vectorizer.transform(test_data.data), test_data.target)
from sklearn.pipeline import Pipeline
pipeline = Pipeline([
("vectorizer", CountVectorizer(min_df=5, ngram_range=(1, 2))),
("algo", LogisticRegression())
])
pipeline.fit(train_data.data, train_data.target)
f_scorer(pipeline, train_data.data, train_data.target)
f_scorer(pipeline, test_data.data, test_data.target)
from sklearn.pipeline import make_pipeline
pipeline = make_pipeline(CountVectorizer(min_df=5, ngram_range=(1, 2)), LogisticRegression())
arr = cross_val_score(pipeline, train_data.data, train_data.target, cv=5, scoring=f_scorer)
print(arr)
print(np.mean(arr))
from sklearn.feature_extraction.text import TfidfTransformer
pipeline = make_pipeline(CountVectorizer(min_df=5, ngram_range=(1, 2)), TfidfTransformer(), LogisticRegression())
arr = cross_val_score(pipeline, train_data.data, train_data.target, cv=5, scoring=f_scorer)
print(arr)
print(np.mean(arr))
pipeline.fit(train_data.data, train_data.target)
accuracy_score(pipeline.predict(train_data.data), train_data.target)
f_scorer(pipeline, train_data.data, train_data.target)
accuracy_score(pipeline.predict(test_data.data), test_data.target)
f_scorer(pipeline, test_data.data, test_data.target)
data_path = 'data/{}'
df = pd.read_csv('data/train.csv')
print(df.shape)
df.head()
label = 0
print('Label: ', label, '\n'+'='*100+'\n')
print(*df[df['label'] == label].sample(10).text, sep='\n'+'-'*100+'\n\n')
label = 1
print('Label: ', label, '\n'+'='*100+'\n')
print(*df[df['label'] == label].sample(10).text, sep='\n'+'-'*100+'\n\n')
label = 2
print('Label: ', label, '\n'+'='*100+'\n')
print(*df[df['label'] == label].sample(10).text, sep='\n'+'-'*100+'\n\n')
skf = StratifiedKFold(3, random_state=37)
train_index, test_index = next(skf.split(df.text, df.label))
train_df, test_df = df.iloc[train_index], df.iloc[test_index]
print(train_df.shape, test_df.shape)
train_df.head()
test_df.head()
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.decomposition import TruncatedSVD
from xgboost.sklearn import XGBClassifier
X_train = train_df.text
y_train = train_df.label
print(X_train.shape)
X_test = test_df.text
y_test = test_df.label
print(X_test.shape)
pipeline = Pipeline([
("vectorizer", CountVectorizer()),
("clf", DecisionTreeClassifier()),
])
%%time
clf = pipeline
clf.fit(X_train, y_train)
print("Train_acc: {:.4f}, train_f-measure: {:.4f}".format(
accuracy_score(clf.predict(X_train), y_train),
f_scorer(clf, X_train, y_train)
))
print("Test_acc: {:.4f}, test_f-measure: {:.4f}".format(
accuracy_score(clf.predict(X_test), y_test),
f_scorer(clf, X_test, y_test)
))
| 0.635222 | 0.903038 |
<h1>Table of Contents<span class="tocSkip"></span></h1>
<div class="toc"><ul class="toc-item"><li><span><a href="#Segunda-práctica:-Aspectos-prácticos-de-las-redes-neuronales" data-toc-modified-id="Segunda-práctica:-Aspectos-prácticos-de-las-redes-neuronales-1"><span class="toc-item-num">1 </span>Segunda práctica: Aspectos prácticos de las redes neuronales</a></span><ul class="toc-item"><li><ul class="toc-item"><li><ul class="toc-item"><li><span><a href="#Obtención-de-los-datos-y-pre-processing" data-toc-modified-id="Obtención-de-los-datos-y-pre-processing-1.0.0.1"><span class="toc-item-num">1.0.0.1 </span>Obtención de los datos y pre-processing</a></span></li></ul></li></ul></li><li><span><a href="#Consideraciones-iniciales" data-toc-modified-id="Consideraciones-iniciales-1.1"><span class="toc-item-num">1.1 </span>Consideraciones iniciales</a></span><ul class="toc-item"><li><span><a href="#Train-validation-test-split" data-toc-modified-id="Train-validation-test-split-1.1.1"><span class="toc-item-num">1.1.1 </span>Train-validation-test split</a></span></li><li><span><a href="#Un-error-común-con-modelos-de-Keras" data-toc-modified-id="Un-error-común-con-modelos-de-Keras-1.1.2"><span class="toc-item-num">1.1.2 </span>Un error común con modelos de Keras</a></span></li><li><span><a href="#Análisis-de-resultados" data-toc-modified-id="Análisis-de-resultados-1.1.3"><span class="toc-item-num">1.1.3 </span>Análisis de resultados</a></span></li></ul></li><li><span><a href="#1.-Unidades-de-activación" data-toc-modified-id="1.-Unidades-de-activación-1.2"><span class="toc-item-num">1.2 </span>1. Unidades de activación</a></span></li><li><span><a href="#2.-Inicialización-de-parámetros" data-toc-modified-id="2.-Inicialización-de-parámetros-1.3"><span class="toc-item-num">1.3 </span>2. Inicialización de parámetros</a></span></li><li><span><a href="#3.-Optimizadores" data-toc-modified-id="3.-Optimizadores-1.4"><span class="toc-item-num">1.4 </span>3. Optimizadores</a></span></li><li><span><a href="#4.-Regularización-y-red-final-(2.5-puntos)" data-toc-modified-id="4.-Regularización-y-red-final-(2.5-puntos)-1.5"><span class="toc-item-num">1.5 </span>4. Regularización y red final <em>(2.5 puntos)</em></a></span><ul class="toc-item"><li><span><a href="#Evaluación-del-modelo-en-datos-de-test" data-toc-modified-id="Evaluación-del-modelo-en-datos-de-test-1.5.1"><span class="toc-item-num">1.5.1 </span>Evaluación del modelo en datos de test</a></span></li></ul></li></ul></li></ul></div>
# Segunda práctica: Aspectos prácticos de las redes neuronales
En esta segunda parte, vamos a continuar desarrollando el problema de Fashion MNIST, con el objetivo de entender los aspectos prácticos del entrenamiento de redes neuronales.
El código utilizado para contestar tiene que quedar claramente reflejado en el Notebook. Puedes crear nuevas cells si así lo deseas para estructurar tu código y sus salidas. A la hora de entregar el notebook, **asegúrate de que los resultados de ejecutar tu código han quedado guardados**.
```
# Puedes añadir todos los imports adicionales que necesites aquí
import keras
from keras.datasets import fashion_mnist
from keras.models import Sequential
from keras.layers import Dense, Dropout, Flatten
import tensorflow as tf
import matplotlib
import matplotlib.pyplot as plt
```
#### Obtención de los datos y pre-processing
```
(x_train, y_train), (x_test, y_test) = fashion_mnist.load_data()
x_train = x_train / 255.0
x_test = x_test / 255.0
```
## Consideraciones iniciales
### Train-validation-test split
En todos los modelos que entrenemos, vamos a partir los datos de training (x_train) en dos sets: training y validación. De este modo, al final tendremos tres datasets distintos: training, validation, y test. Esta es una estrategia común en el aprendizaje automático, en la que los datos de test (o held-out data) se
"esconden" hasta el final. Los datos de validación se utilizan para estimar cómo de bien están funcionando nuestros modelos y para observar si estamos cayendo en overfitting. Esto nos permite cambiar hiperparámetros y probar distintas arquitecturas **sabiendo que no estamos utilizando información del test set para "optimizar" los resultados en éste** (si eligiéramos nuestro mejor modelo en base a los resultados de test, estaríamos "haciendo trampas", ya que se ha utilizado la información contenida en éste para elegir el modelo y las métricas reportadas serían optimistas).
Para utilizar un split training-validation data durante el entrenamiento, podemos partir nosotros mismos los datos o dejar que Keras lo haga. Podéis ver cómo hacer estas particiones en la documentación de *fit*.
**Requisito: En todos los entrenamientos de esta práctica, se requiere utilizar el 20% de los datos en x_train como conjunto de datos de validación**
### Un error común con modelos de Keras
En esta práctica entrenaremos varios modelos para comparar resultados. Un error común en Keras es no instanciar un nuevo modelo cada vez que hacemos un nuevo entrenamiento. Al hacer
*model = Sequential()*
*model.add(lo que sea) # Definición del modelo*
*model.fit()*
si queremos entrenar un nuevo modelo o el mismo modelo otra vez, es necesario volver a inicializar el modelo con model = Sequential(). Si olvidamos este paso y volvemos a hacer fit(), el modelo seguirá entrenando por donde se quedó en el último fit().
### Análisis de resultados
A la hora de escribir las respuestas y los análisis pedidos, es importante presentar las conclusiones de manera adecuada a partir de lo visto en nuestros experimentos. Los Jupyter Notebook son una herramienta imprescindible para *data scientists* e ingenieros de Machine Learning para presentar los resultados, incluyendo soporte para incluir gráficas y elementos visuales. Podéis explicar vuestras observaciones del modo que consideréis adecuado, si bien recomendamos la utilización de gráficas para evaluar los entrenamientos y comparar resultados.
Como ayuda, las siguientes funciones pueden resultar interesantes a la hora de evaluar resultados. Todas ellas utilizan el objeto *history* que podéis obtener como salida del método *fit()* de Keras:
history = model.fit(x_train, y_train, ...)
Por supuesto, podéis modificarlas y utilizarlas como prefiráis para crear vuestros propios informes.
```
def plot_acc(history, title="Model Accuracy"):
"""Imprime una gráfica mostrando la accuracy por epoch obtenida en un entrenamiento"""
plt.plot(history.history['accuracy'])
plt.plot(history.history['val_accuracy'])
plt.title(title)
plt.ylabel('Accuracy')
plt.xlabel('Epoch')
plt.legend(['Train', 'Val'], loc='upper left')
plt.show()
def plot_loss(history, title="Model Loss"):
"""Imprime una gráfica mostrando la pérdida por epoch obtenida en un entrenamiento"""
plt.plot(history.history['loss'])
plt.plot(history.history['val_loss'])
plt.title(title)
plt.ylabel('Loss')
plt.xlabel('Epoch')
plt.legend(['Train', 'Val'], loc='upper right')
plt.show()
def plot_compare_losses(history1, history2, name1="Red 1",
name2="Red 2", title="Graph title"):
"""Compara losses de dos entrenamientos con nombres name1 y name2"""
plt.plot(history1.history['loss'], color="green")
plt.plot(history1.history['val_loss'], 'r--', color="green")
plt.plot(history2.history['loss'], color="blue")
plt.plot(history2.history['val_loss'], 'r--', color="blue")
plt.title(title)
plt.ylabel('Loss')
plt.xlabel('Epoch')
plt.legend(['Train ' + name1, 'Val ' + name1,
'Train ' + name2, 'Val ' + name2],
loc='upper right')
plt.show()
def plot_compare_accs(history1, history2, name1="Red 1",
name2="Red 2", title="Graph title"):
"""Compara accuracies de dos entrenamientos con nombres name1 y name2"""
plt.plot(history1.history['accuracy'], color="green")
plt.plot(history1.history['val_accuracy'], 'r--', color="green")
plt.plot(history2.history['accuracy'], color="blue")
plt.plot(history2.history['val_accuracy'], 'r--', color="blue")
plt.title(title)
plt.ylabel('Accuracy')
plt.xlabel('Epoch')
plt.legend(['Train ' + name1, 'Val ' + name1,
'Train ' + name2, 'Val ' + name2],
loc='lower right')
plt.show()
# Nota: podéis cambiar los números aquí presentes y ejecutar esta línea si queréis cambiar el tamaño
# de las gráficas
# matplotlib.rcParams['figure.figsize'] = [8, 8]
```
## 1. Unidades de activación
En este ejercicio, vamos a evaluar la importancia de utilizar las unidades de activación adecuadas. Las funciones de activación como sigmoid han dejado de utilizarse en favor de otras unidades como ReLU.
**Ejercicio 1 *(2.5 puntos)***: Partiendo de una red sencilla como la desarrollada en el Trabajo 1, escribir un breve análisis comparando la utilización de unidades sigmoid y ReLU (por ejemplo, se pueden comentar aspectos como velocidad de convergencia, métricas obtenidas...). Explicar por qué pueden darse estas diferencias. Opcionalmente, comparar con otras activaciones disponibles en Keras.
*Pista: Usando redes más grandes se hace más sencillo apreciar las diferencias. Es mejor utilizar al menos 3 o 4 capas densas.*
```
## Definición de parámetros generales
hidden_units_1 = 128
hidden_units_2 = 64
hidden_units_3 = 64
dropout_rate = 0.25
batch_size = 64
epochs = 20
validation_split = 0.20
metrics = ['accuracy']
## Obteniendo las etiquetas y convertiendo los valores de las categorías en un vector one-hot
import numpy as np
from keras.utils import to_categorical
labels, counts = np.unique(y_train, return_counts=True)
y_train = to_categorical(y_train)
y_test = to_categorical(y_test)
## Método que construye un modelo con:
## - 1 capa Flatten de entrada para aplanar las imágenes de entrenamiento
## - 3 capas densas ocultas con función de activación "activation_function"
## - 1 capa Dropout para borrar pesos de forma aleatoria durante el entrenamiento, para evitar el sobreajuste
## - 1 capa densa de salida con función de activación softmax
def build_model(x_train, labels, activation_function='relu', loss_function='categorical_crossentropy',
optimizer = 'adam', initializer = 'glorot_uniform'):
model = Sequential()
model.add(Flatten(input_shape=(x_train.shape[1] , x_train.shape[2])))
model.add(Dense(hidden_units_1, activation=activation_function, kernel_initializer=initializer))
model.add(Dense(hidden_units_2, activation=activation_function))
model.add(Dense(hidden_units_3, activation=activation_function))
model.add(Dropout(dropout_rate))
model.add(Dense(len(labels), activation='softmax'))
model.compile(loss=loss_function,
optimizer=optimizer,
metrics=metrics)
return model
## Obteniendo el modelo con función de activación Sigmoid, función de perdidad Binary Crossentropy y optimizador SGD.
from keras.optimizers import SGD
from keras.utils import plot_model
model_sigmoid = build_model(x_train, labels, 'sigmoid', 'binary_crossentropy', SGD(learning_rate = 0.1, momentum=0.9))
model_sigmoid.summary()
plot_model(model_sigmoid, show_shapes=True)
```
**Comentario:** Se muestra una sola vez la información del modelo ya que la estructura de la red, el número de capas y el número nodos por capa, y el número de parámetros por capa y total es el mismo que en todos los modelos que se construyen en esta pregunta.
```
## Entrenando modelo
history_sigmoid = model_sigmoid.fit(x_train, y_train,
validation_split=validation_split,
batch_size=batch_size,
epochs=epochs)
## Dibujar gráfica de los valores de Accuracy y Loss del modelo entrenado con Sigmoid
plot_acc(history_sigmoid, "Accuracy en el entrenamiento con Sigmoid")
plot_loss(history_sigmoid, "Loss en el entrenamiento con Sigmoid")
## Obteniendo el modelo con función de activación ReLU, función de perdidad Categorical Crossentropy y optimizador Adam.
model_relu = build_model(x_train, labels)
## Entrenando modelo
history_relu = model_relu.fit(x_train, y_train,
validation_split=validation_split,
batch_size=batch_size,
epochs=epochs)
## Dibujar gráfica de los valores de Accuracy y Loss del modelo entrenado con ReLU
plot_acc(history_relu, "Accuracy en el entrenamiento con ReLU")
plot_loss(history_relu, "Loss en el entrenamiento con ReLU")
```
**Respuesta:** En este ejercicio he entrenado con los mismos parámetros globales (números de nodos por cada capa, tasa de dropout para el entrenamiento, entre otros) a dos modelos, donde en uno de ellos la función de activación es *Sigmoid*, y el otro modelo su función de activación es *ReLU*. Además, en cada modelo uso sus **respectivos optimizadores y funciones de perdidas** más adecuados ajustando un poco sus parámetros.
Como resultado del entrenamiento, se observa que el modelo con la función de activación **ReLU tiene un resultado más aceptable si nos referimos al accuracy**, sin embargo, el valor loss respecto de los datos de validación es alto. Una característica negativa vista de este modelo es que parece que sufre de **overfitting** porque el accuracy y loss de los datos de validación se alejan respecto a sus análogos de los datos de entrenamiento, es decir, *esta memorizando muy bien los registros de entrenamiento, pero no es capaz de predecir bien nuevos registros no vistos antes*. Otra observación que se destaca es que para la cantidad de epocas definidas (20) se puede apreciar la **evolución del entrenamiento** llegando a valores casi estables en las últimas epocas.
Respecto al modelo que fue entrenado con la función de activación *Sigmoid*, he tenido que ajustar los parámetros de la función de perdida, porque **no estaba encontrando los mínimos locales**. Con los nuevos parámetros de la tasa de aprendizaje y el momentum, se obtiene un resultado bueno de accuracy pero peor al de ReLU, si embargo, la ventaja se resalta en **los valores de loss que mejoran sustancialmente a los de ReLU**, 0.069 (Sigmoid) vs 0.1903 (ReLU). En este modelo se puede ver también que *no se sufre de overfitting*, es decir, el modelo predice con mucha similitud los datos de entrenamiento y los de validación.
```
## Obteniendo el modelo con función de activación Tangente Hiperbólica, función de perdidad MSE y optimizador Adam.
model_tanh = build_model(x_train, labels, 'tanh', 'mean_squared_error')
## Entrenando modelo
history_tanh = model_tanh.fit(x_train, y_train,
validation_split=validation_split,
batch_size=batch_size,
epochs=epochs)
## Dibujar gráfica de los valores de Accuracy y Loss del modelo entrenado con Tanh
plot_compare_losses(history_relu, history_tanh, "ReLU", "Tanh", "Comparación del valor Loss entre ReLU y Tanh")
plot_compare_accs(history_relu, history_tanh, "ReLU", "Tanh", "Comparación del valor Accuracy entre ReLU y Tanh")
```
**Respuesta:** En este nuevo modelo entrenado con la función de activación Tanh vemos que se produce *una tendencia del valor accuracy similar al de ReLU*, sin embargo **mejora considerablemente el valor loss** tanto en datos de entrenamiento, como en datos de validación. Este modelo también tiene tendencia a sufrir de **overfitting** al igual que el modelo de ReLU.
```
## Obteniendo el modelo con función de activación Softplus, función de perdidad Categorical Crossentropy y optimizador Adam.
model_softplus = build_model(x_train, labels, 'softplus', 'categorical_crossentropy')
## Entrenando modelo
history_softplus = model_softplus.fit(x_train, y_train,
validation_split=validation_split,
batch_size=batch_size,
epochs=epochs)
## Dibujar gráfica de los valores de Accuracy y Loss del modelo entrenado con Softplus
plot_compare_losses(history_relu, history_softplus, "ReLU", "Softplus", "Comparación del valor Loss entre ReLU y Softplus")
plot_compare_accs(history_relu, history_softplus, "ReLU", "Softplus", "Comparación del valor Accuracy entre ReLU y Softplus")
```
**Respuesta:** Usar la función **softplus** (una acercamiento a la funcioón ReLU con una curva suave) **no mejora el resultado de ReLU**, e incluso reproduce casi exactamente sus resultados, pero si se observa más suavidad en la gráfica, es decir, tiene menor varianza respecto a ReLU.
## 2. Inicialización de parámetros
En este ejercicio, vamos a evaluar la importancia de una correcta inicialización de parámetros en una red neuronal.
**Ejercicio 2 *(2.5 puntos)***: Partiendo de una red similar a la del ejercicio anterior (usando ya ReLUs), comentar las diferencias que se aprecian en el entrenamiento al utilizar distintas estrategias de inicialización de parámetros. Para ello, inicializar todas las capas con las siguientes estrategias, disponibles en Keras, y analizar sus diferencias:
* Inicialización con ceros.
* Inicialización con una variable aleatoria normal.
* Inicialización con los valores por defecto de Keras para una capa Dense (estrategia *glorot uniform*)
```
from tensorflow.keras import initializers
## Obteniendo el modelo con función de activación ReLU, función de perdidad Categorical Crossentropy, optimizador Adam e inicializador de ceros.
model_relu_zeros = build_model(x_train, labels, initializer=initializers.Zeros())
## Entrenando modelo
history_relu_zeros = model_relu_zeros.fit(x_train, y_train,
validation_split=validation_split,
batch_size=batch_size,
epochs=epochs)
## Obteniendo el modelo con función de activación ReLU, función de perdidad Categorical Crossentropy, optimizador Adam e inicializador de ceros.
model_relu_random = build_model(x_train, labels, initializer=initializers.RandomNormal(mean=0.0, stddev=1.0))
## Entrenando modelo
history_relu_random = model_relu_random.fit(x_train, y_train,
validation_split=validation_split,
batch_size=batch_size,
epochs=epochs)
## Dibujar gráfica de los valores de Accuracy y Loss para los modelos entrenado con ReLU, el verde con inicializador "glorot_uniform" y el azul con inicializador de pesos en ceros.
plot_compare_losses(history_relu, history_relu_zeros, "ReLU GorotUniform", "ReLU RandomNormal", "Comparación del valor Loss entre inicialización glorot_uniform e inicialización con ceros")
plot_compare_accs(history_relu, history_relu_zeros, "ReLU GorotUniform", "ReLU RandomNormal", "Comparación del valor Accuracy entre inicialización glorot_uniform e inicialización con ceros")
## Dibujar gráfica de los valores de Accuracy y Loss para los modelos entrenado con ReLU, el verde con inicializador "glorot_uniform" y el azul con inicializador de la distribución normal.
plot_compare_losses(history_relu, history_relu_random, "ReLU GorotUniform", "ReLU RandomNormal", "Comparación del valor Loss entre inicialización glorot_uniform e inicialización con pesos aleatorios de la distribución normal")
plot_compare_accs(history_relu, history_relu_random, "ReLU GorotUniform", "ReLU RandomNormal", "Comparación del valor Accuracy entre inicialización glorot_uniform e inicialización con pesos aleatorios de la distribución normal")
```
**Respuesta**:
## 3. Optimizadores
**Ejercicio 3 *(2.5 puntos)***: Partiendo de una red similar a la del ejercicio anterior (utilizando la mejor estrategia de inicialización observada), comparar y analizar las diferencias que se observan al entrenar con varios de los optimizadores vistos en clase, incluyendo SGD como optimizador básico (se puede explorar el espacio de hiperparámetros de cada optimizador, aunque para optimizadores más avanzados del estilo de adam y RMSprop es buena idea dejar los valores por defecto provistos por Keras).
```
## Obteniendo el modelo con función de activación ReLU, función de perdidad Categorical Crossentropy y optimizador SGD.
model_relu_sgd = build_model(x_train, labels, optimizer=SGD(learning_rate = 0.01, momentum=0.9))
## Entrenando modelo
history_relu_sgd = model_relu_sgd.fit(x_train, y_train,
validation_split=validation_split,
batch_size=batch_size,
epochs=epochs)
## Dibujar gráfica de los valores de Accuracy y Loss para los modelos entrenado con ReLU, el verde con inicializador "glorot_uniform" y el azul con inicializador de la distribución normal.
plot_compare_losses(history_relu, history_relu_sgd, "ReLU Adam", "ReLU SGD", "Comparación del valor Loss entre optimizador Adam y SGD")
plot_compare_accs(history_relu, history_relu_sgd, "ReLU Adam", "ReLU SGD", "Comparación del valor Accuracy entre optimizador Adam y SGD")
from keras.optimizers import RMSprop
## Obteniendo el modelo con función de activación ReLU, función de perdidad Categorical Crossentropy y optimizador RMSprop.
model_relu_rmsprop = build_model(x_train, labels, optimizer=RMSprop())
## Entrenando modelo
history_relu_rmsprop = model_relu_rmsprop.fit(x_train, y_train,
validation_split=validation_split,
batch_size=batch_size,
epochs=epochs)
## Dibujar gráfica de los valores de Accuracy y Loss para los modelos entrenado con ReLU, el verde con optimizador "Adam" y el azul con RMSprop.
plot_compare_losses(history_relu, history_relu_rmsprop, "ReLU Adam", "ReLU RMSpro", "Comparación del valor Loss entre optimizador Adam y RMSpro")
plot_compare_accs(history_relu, history_relu_rmsprop, "ReLU Adam", "ReLU RMSpro", "Comparación del valor Accuracy entre optimizador Adam y RMSpro")
from keras.optimizers import Nadam
## Obteniendo el modelo con función de activación ReLU, función de perdidad Categorical Crossentropy y optimizador Nadam.
model_relu_nadam = build_model(x_train, labels, optimizer=Nadam())
## Entrenando modelo
history_relu_nadam = model_relu_nadam.fit(x_train, y_train,
validation_split=validation_split,
batch_size=batch_size,
epochs=epochs)
## Dibujar gráfica de los valores de Accuracy y Loss para los modelos entrenado con ReLU, el verde con optimizador Adam y el azul con Nadam.
plot_compare_losses(history_relu, history_relu_nadam, "ReLU Adam", "ReLU Nadam", "Comparación del valor Loss entre optimizador Adam y Nadam")
plot_compare_accs(history_relu, history_relu_nadam, "ReLU Adam", "ReLU Nadam", "Comparación del valor Accuracy entre optimizador Adam y Nadam")
```
**Respuesta**: He probado tres tipos de optimizadores distintos al usado en el modelo construido con la función de activación ReLU de la pregunta anterior. Comparando los resultados para cada optimizador, se observa que el mejor resultado ha sido con el optimizador **NADAM**, que mejora ligeramente al optimizador ADAM.
Se podría decir que el *peor otimizador* para este modelo y grupo de datos de entrenamiento fue el RMSprop. Para el caso del optimizador SGD, se ha tenido que **buscar nuevos valores para sus parámetros** que mejora el resultado que genera los parámetros por defecto.
## 4. Regularización y red final *(2.5 puntos)*
**Ejercicio 4.1**: Entrenar una red final que sea capaz de obtener una accuracy en el validation set cercana al 90%. Para ello, combinar todo lo aprendido anteriormente y utilizar técnicas de regularización para evitar overfitting. Algunos de los elementos que pueden tenerse en cuenta son los siguientes.
* Número de capas y neuronas por capa
* Optimizadores y sus parámetros
* Batch size
* Unidades de activación
* Uso de capas dropout, regularización L2, regularización L1...
* Early stopping (se puede aplicar como un callback de Keras, o se puede ver un poco "a ojo" cuándo el modelo empieza a caer en overfitting y seleccionar el número de epochs necesarias)
* Batch normalization
Si los modelos entrenados anteriormente ya se acercaban al valor requerido de accuracy, probar distintas estrategias igualmente y comentar los resultados.
Explicar brevemente la estrategia seguida y los modelos probados para obtener el modelo final, que debe verse entrenado en este Notebook. No es necesario guardar el entrenamiento de todos los modelos que se han probado, es suficiente con explicar cómo se ha llegado al modelo final.
```
## Método que construye un modelo final de la práctica con:
## - 1 capa Flatten de entrada para aplanar las imágenes de entrenamiento
## - 3 capas densas ocultas con función de activación "relu"
## - 1 capa Dropout para borrar pesos de forma aleatoria durante el entrenamiento, para evitar el sobreajuste
## - 1 capa densa de salida con función de activación softmax
def build_final_model(x_train, labels):
model = Sequential()
model.add(Flatten(input_shape=(x_train.shape[1] , x_train.shape[2])))
model.add(Dense(128, activation='softplus'))
model.add(Dropout(0.3))
model.add(Dense(256, activation='softplus'))
model.add(Dropout(0.3))
model.add(Dense(256, activation='softplus'))
model.add(Dropout(0.3))
model.add(Dense(128, activation='softplus'))
model.add(Dropout(0.3))
model.add(Dense(len(labels), activation='softmax'))
model.summary()
plot_model(model, show_shapes=True)
model.compile(loss='categorical_crossentropy',
optimizer='nadam',
metrics=['accuracy'])
return model
from keras.callbacks import EarlyStopping
## Obteniendo el modelo final.
final_model = build_final_model(x_train, labels)
## Definiendo la parada del entrenamiento
callback = [EarlyStopping(monitor='val_accuracy', patience=10)]
## Entrenando modelo
history_final = final_model.fit(x_train, y_train,
validation_split=validation_split,
batch_size=64,
epochs=80,
callbacks=callback)
## Dibujar gráfica de los valores de Accuracy y Loss del modelo entrenado con Sigmoid
plot_acc(history_final, "Accuracy en el entrenamiento del modelo final")
plot_loss(history_final, "Loss en el entrenamiento del modelo final")
```
**Respuesta**: Para mejorar los resultados del entrenamiento, concretamente la predicción sobre los valores de validación, se han aplicado algunas técnicas que mejoran la predicción y la reducen algunos problemas como el overfitting.
Entre las principales técnicas o herramientas usadas tenemos, selección de número de capas ocultas y nodos por capa, función de activación, función de perdida, optimizador, incorporación de capas de Dropout, etc.
* Se ha ajustado el número de capas y el número de nodos para mejorar el valor de **val_accuracy**.
* Se inicio con una función de activación ReLU, pero despúes se obtuvo mejores resultados con **Softplus**.
* Se mantuvo la función de perdida **categorical_crossentropy** porque el resto no mejoraba el resultado.
* A partir del resultado de la pregunta anterior se ha decidido usar el optimizador **NADAM**.
* Dado que se producía overffing con pocas epicas, se ha añadido capas **Dropout** con una tasa de 0.3.
* También se ha creado un callback de tipo **EarlyStopping** con un valor de paciencia de *10* para detener el entramiento cuando ya no mejore el valor de *val_accuracy*. En el último entrenamiento no se ha detenido antes de las 80 epocas.
* Finalmente se han ajustado los valores de **batch_size** y **epochs** para conseguir llegar al *90% de val_accuracy*.
Después de aplicar todos los pasos descritos previamente, se ha logrado conseguir pocas iteraciones que superen el *90% de val_accuracy* pero la mayoría estuvieron en **89%**.
### Evaluación del modelo en datos de test
Una vez elegido el que creemos que es nuestro mejor modelo a partir de la estimación que hemos visto en los datos de validación, es hora de utilizar los datos de test para ver cómo se comporta nuestro modelo ante nuevos datos. Si hemos hecho bien las cosas, este número debería ser parecido al valor de nuestra estimación vista en los datos de validación.
**Pregunta 4.2**. Utilizando nuestro mejor modelo, obtener la accuracy resultante en el dataset de test. Comentar este resultado.
```
loss, acc = final_model.evaluate(x_test, y_test, batch_size=64)
print("\nTest loss: %.1f%%" % (100.0 * loss))
print("Test accuracy: %.1f%%" % (100.0 * acc))
result = final_model.predict(x_test)
from sklearn.metrics import confusion_matrix
confusion_matrix(np.argmax(y_test, axis=1), np.argmax(result, axis=1))
from sklearn.metrics import classification_report
print(classification_report(np.argmax(y_test, axis=1), np.argmax(result, axis=1)))
```
**Respuesta**: El valor del accuracy conseguido con los datos de pruebas es del **89.7%**, muy similar a los valores de val_accuracy conseguidos en el entrenamiento.
* Las prendas que **mejor predijo** el modelo a partir de los datos de prueba son el **Trouser, Ankle boot y Sandal**, ya que tienen una forma diferentes al resto de prendas.
* En el tipo de prenda en que **más problemas tuvo al predecir fue Shirt**, porque es el que más tiene parecido con el resto de prendas, por ejemplo *T-Shirt, Pullover, Dress y Coat*.
|
github_jupyter
|
# Puedes añadir todos los imports adicionales que necesites aquí
import keras
from keras.datasets import fashion_mnist
from keras.models import Sequential
from keras.layers import Dense, Dropout, Flatten
import tensorflow as tf
import matplotlib
import matplotlib.pyplot as plt
(x_train, y_train), (x_test, y_test) = fashion_mnist.load_data()
x_train = x_train / 255.0
x_test = x_test / 255.0
def plot_acc(history, title="Model Accuracy"):
"""Imprime una gráfica mostrando la accuracy por epoch obtenida en un entrenamiento"""
plt.plot(history.history['accuracy'])
plt.plot(history.history['val_accuracy'])
plt.title(title)
plt.ylabel('Accuracy')
plt.xlabel('Epoch')
plt.legend(['Train', 'Val'], loc='upper left')
plt.show()
def plot_loss(history, title="Model Loss"):
"""Imprime una gráfica mostrando la pérdida por epoch obtenida en un entrenamiento"""
plt.plot(history.history['loss'])
plt.plot(history.history['val_loss'])
plt.title(title)
plt.ylabel('Loss')
plt.xlabel('Epoch')
plt.legend(['Train', 'Val'], loc='upper right')
plt.show()
def plot_compare_losses(history1, history2, name1="Red 1",
name2="Red 2", title="Graph title"):
"""Compara losses de dos entrenamientos con nombres name1 y name2"""
plt.plot(history1.history['loss'], color="green")
plt.plot(history1.history['val_loss'], 'r--', color="green")
plt.plot(history2.history['loss'], color="blue")
plt.plot(history2.history['val_loss'], 'r--', color="blue")
plt.title(title)
plt.ylabel('Loss')
plt.xlabel('Epoch')
plt.legend(['Train ' + name1, 'Val ' + name1,
'Train ' + name2, 'Val ' + name2],
loc='upper right')
plt.show()
def plot_compare_accs(history1, history2, name1="Red 1",
name2="Red 2", title="Graph title"):
"""Compara accuracies de dos entrenamientos con nombres name1 y name2"""
plt.plot(history1.history['accuracy'], color="green")
plt.plot(history1.history['val_accuracy'], 'r--', color="green")
plt.plot(history2.history['accuracy'], color="blue")
plt.plot(history2.history['val_accuracy'], 'r--', color="blue")
plt.title(title)
plt.ylabel('Accuracy')
plt.xlabel('Epoch')
plt.legend(['Train ' + name1, 'Val ' + name1,
'Train ' + name2, 'Val ' + name2],
loc='lower right')
plt.show()
# Nota: podéis cambiar los números aquí presentes y ejecutar esta línea si queréis cambiar el tamaño
# de las gráficas
# matplotlib.rcParams['figure.figsize'] = [8, 8]
## Definición de parámetros generales
hidden_units_1 = 128
hidden_units_2 = 64
hidden_units_3 = 64
dropout_rate = 0.25
batch_size = 64
epochs = 20
validation_split = 0.20
metrics = ['accuracy']
## Obteniendo las etiquetas y convertiendo los valores de las categorías en un vector one-hot
import numpy as np
from keras.utils import to_categorical
labels, counts = np.unique(y_train, return_counts=True)
y_train = to_categorical(y_train)
y_test = to_categorical(y_test)
## Método que construye un modelo con:
## - 1 capa Flatten de entrada para aplanar las imágenes de entrenamiento
## - 3 capas densas ocultas con función de activación "activation_function"
## - 1 capa Dropout para borrar pesos de forma aleatoria durante el entrenamiento, para evitar el sobreajuste
## - 1 capa densa de salida con función de activación softmax
def build_model(x_train, labels, activation_function='relu', loss_function='categorical_crossentropy',
optimizer = 'adam', initializer = 'glorot_uniform'):
model = Sequential()
model.add(Flatten(input_shape=(x_train.shape[1] , x_train.shape[2])))
model.add(Dense(hidden_units_1, activation=activation_function, kernel_initializer=initializer))
model.add(Dense(hidden_units_2, activation=activation_function))
model.add(Dense(hidden_units_3, activation=activation_function))
model.add(Dropout(dropout_rate))
model.add(Dense(len(labels), activation='softmax'))
model.compile(loss=loss_function,
optimizer=optimizer,
metrics=metrics)
return model
## Obteniendo el modelo con función de activación Sigmoid, función de perdidad Binary Crossentropy y optimizador SGD.
from keras.optimizers import SGD
from keras.utils import plot_model
model_sigmoid = build_model(x_train, labels, 'sigmoid', 'binary_crossentropy', SGD(learning_rate = 0.1, momentum=0.9))
model_sigmoid.summary()
plot_model(model_sigmoid, show_shapes=True)
## Entrenando modelo
history_sigmoid = model_sigmoid.fit(x_train, y_train,
validation_split=validation_split,
batch_size=batch_size,
epochs=epochs)
## Dibujar gráfica de los valores de Accuracy y Loss del modelo entrenado con Sigmoid
plot_acc(history_sigmoid, "Accuracy en el entrenamiento con Sigmoid")
plot_loss(history_sigmoid, "Loss en el entrenamiento con Sigmoid")
## Obteniendo el modelo con función de activación ReLU, función de perdidad Categorical Crossentropy y optimizador Adam.
model_relu = build_model(x_train, labels)
## Entrenando modelo
history_relu = model_relu.fit(x_train, y_train,
validation_split=validation_split,
batch_size=batch_size,
epochs=epochs)
## Dibujar gráfica de los valores de Accuracy y Loss del modelo entrenado con ReLU
plot_acc(history_relu, "Accuracy en el entrenamiento con ReLU")
plot_loss(history_relu, "Loss en el entrenamiento con ReLU")
## Obteniendo el modelo con función de activación Tangente Hiperbólica, función de perdidad MSE y optimizador Adam.
model_tanh = build_model(x_train, labels, 'tanh', 'mean_squared_error')
## Entrenando modelo
history_tanh = model_tanh.fit(x_train, y_train,
validation_split=validation_split,
batch_size=batch_size,
epochs=epochs)
## Dibujar gráfica de los valores de Accuracy y Loss del modelo entrenado con Tanh
plot_compare_losses(history_relu, history_tanh, "ReLU", "Tanh", "Comparación del valor Loss entre ReLU y Tanh")
plot_compare_accs(history_relu, history_tanh, "ReLU", "Tanh", "Comparación del valor Accuracy entre ReLU y Tanh")
## Obteniendo el modelo con función de activación Softplus, función de perdidad Categorical Crossentropy y optimizador Adam.
model_softplus = build_model(x_train, labels, 'softplus', 'categorical_crossentropy')
## Entrenando modelo
history_softplus = model_softplus.fit(x_train, y_train,
validation_split=validation_split,
batch_size=batch_size,
epochs=epochs)
## Dibujar gráfica de los valores de Accuracy y Loss del modelo entrenado con Softplus
plot_compare_losses(history_relu, history_softplus, "ReLU", "Softplus", "Comparación del valor Loss entre ReLU y Softplus")
plot_compare_accs(history_relu, history_softplus, "ReLU", "Softplus", "Comparación del valor Accuracy entre ReLU y Softplus")
from tensorflow.keras import initializers
## Obteniendo el modelo con función de activación ReLU, función de perdidad Categorical Crossentropy, optimizador Adam e inicializador de ceros.
model_relu_zeros = build_model(x_train, labels, initializer=initializers.Zeros())
## Entrenando modelo
history_relu_zeros = model_relu_zeros.fit(x_train, y_train,
validation_split=validation_split,
batch_size=batch_size,
epochs=epochs)
## Obteniendo el modelo con función de activación ReLU, función de perdidad Categorical Crossentropy, optimizador Adam e inicializador de ceros.
model_relu_random = build_model(x_train, labels, initializer=initializers.RandomNormal(mean=0.0, stddev=1.0))
## Entrenando modelo
history_relu_random = model_relu_random.fit(x_train, y_train,
validation_split=validation_split,
batch_size=batch_size,
epochs=epochs)
## Dibujar gráfica de los valores de Accuracy y Loss para los modelos entrenado con ReLU, el verde con inicializador "glorot_uniform" y el azul con inicializador de pesos en ceros.
plot_compare_losses(history_relu, history_relu_zeros, "ReLU GorotUniform", "ReLU RandomNormal", "Comparación del valor Loss entre inicialización glorot_uniform e inicialización con ceros")
plot_compare_accs(history_relu, history_relu_zeros, "ReLU GorotUniform", "ReLU RandomNormal", "Comparación del valor Accuracy entre inicialización glorot_uniform e inicialización con ceros")
## Dibujar gráfica de los valores de Accuracy y Loss para los modelos entrenado con ReLU, el verde con inicializador "glorot_uniform" y el azul con inicializador de la distribución normal.
plot_compare_losses(history_relu, history_relu_random, "ReLU GorotUniform", "ReLU RandomNormal", "Comparación del valor Loss entre inicialización glorot_uniform e inicialización con pesos aleatorios de la distribución normal")
plot_compare_accs(history_relu, history_relu_random, "ReLU GorotUniform", "ReLU RandomNormal", "Comparación del valor Accuracy entre inicialización glorot_uniform e inicialización con pesos aleatorios de la distribución normal")
## Obteniendo el modelo con función de activación ReLU, función de perdidad Categorical Crossentropy y optimizador SGD.
model_relu_sgd = build_model(x_train, labels, optimizer=SGD(learning_rate = 0.01, momentum=0.9))
## Entrenando modelo
history_relu_sgd = model_relu_sgd.fit(x_train, y_train,
validation_split=validation_split,
batch_size=batch_size,
epochs=epochs)
## Dibujar gráfica de los valores de Accuracy y Loss para los modelos entrenado con ReLU, el verde con inicializador "glorot_uniform" y el azul con inicializador de la distribución normal.
plot_compare_losses(history_relu, history_relu_sgd, "ReLU Adam", "ReLU SGD", "Comparación del valor Loss entre optimizador Adam y SGD")
plot_compare_accs(history_relu, history_relu_sgd, "ReLU Adam", "ReLU SGD", "Comparación del valor Accuracy entre optimizador Adam y SGD")
from keras.optimizers import RMSprop
## Obteniendo el modelo con función de activación ReLU, función de perdidad Categorical Crossentropy y optimizador RMSprop.
model_relu_rmsprop = build_model(x_train, labels, optimizer=RMSprop())
## Entrenando modelo
history_relu_rmsprop = model_relu_rmsprop.fit(x_train, y_train,
validation_split=validation_split,
batch_size=batch_size,
epochs=epochs)
## Dibujar gráfica de los valores de Accuracy y Loss para los modelos entrenado con ReLU, el verde con optimizador "Adam" y el azul con RMSprop.
plot_compare_losses(history_relu, history_relu_rmsprop, "ReLU Adam", "ReLU RMSpro", "Comparación del valor Loss entre optimizador Adam y RMSpro")
plot_compare_accs(history_relu, history_relu_rmsprop, "ReLU Adam", "ReLU RMSpro", "Comparación del valor Accuracy entre optimizador Adam y RMSpro")
from keras.optimizers import Nadam
## Obteniendo el modelo con función de activación ReLU, función de perdidad Categorical Crossentropy y optimizador Nadam.
model_relu_nadam = build_model(x_train, labels, optimizer=Nadam())
## Entrenando modelo
history_relu_nadam = model_relu_nadam.fit(x_train, y_train,
validation_split=validation_split,
batch_size=batch_size,
epochs=epochs)
## Dibujar gráfica de los valores de Accuracy y Loss para los modelos entrenado con ReLU, el verde con optimizador Adam y el azul con Nadam.
plot_compare_losses(history_relu, history_relu_nadam, "ReLU Adam", "ReLU Nadam", "Comparación del valor Loss entre optimizador Adam y Nadam")
plot_compare_accs(history_relu, history_relu_nadam, "ReLU Adam", "ReLU Nadam", "Comparación del valor Accuracy entre optimizador Adam y Nadam")
## Método que construye un modelo final de la práctica con:
## - 1 capa Flatten de entrada para aplanar las imágenes de entrenamiento
## - 3 capas densas ocultas con función de activación "relu"
## - 1 capa Dropout para borrar pesos de forma aleatoria durante el entrenamiento, para evitar el sobreajuste
## - 1 capa densa de salida con función de activación softmax
def build_final_model(x_train, labels):
model = Sequential()
model.add(Flatten(input_shape=(x_train.shape[1] , x_train.shape[2])))
model.add(Dense(128, activation='softplus'))
model.add(Dropout(0.3))
model.add(Dense(256, activation='softplus'))
model.add(Dropout(0.3))
model.add(Dense(256, activation='softplus'))
model.add(Dropout(0.3))
model.add(Dense(128, activation='softplus'))
model.add(Dropout(0.3))
model.add(Dense(len(labels), activation='softmax'))
model.summary()
plot_model(model, show_shapes=True)
model.compile(loss='categorical_crossentropy',
optimizer='nadam',
metrics=['accuracy'])
return model
from keras.callbacks import EarlyStopping
## Obteniendo el modelo final.
final_model = build_final_model(x_train, labels)
## Definiendo la parada del entrenamiento
callback = [EarlyStopping(monitor='val_accuracy', patience=10)]
## Entrenando modelo
history_final = final_model.fit(x_train, y_train,
validation_split=validation_split,
batch_size=64,
epochs=80,
callbacks=callback)
## Dibujar gráfica de los valores de Accuracy y Loss del modelo entrenado con Sigmoid
plot_acc(history_final, "Accuracy en el entrenamiento del modelo final")
plot_loss(history_final, "Loss en el entrenamiento del modelo final")
loss, acc = final_model.evaluate(x_test, y_test, batch_size=64)
print("\nTest loss: %.1f%%" % (100.0 * loss))
print("Test accuracy: %.1f%%" % (100.0 * acc))
result = final_model.predict(x_test)
from sklearn.metrics import confusion_matrix
confusion_matrix(np.argmax(y_test, axis=1), np.argmax(result, axis=1))
from sklearn.metrics import classification_report
print(classification_report(np.argmax(y_test, axis=1), np.argmax(result, axis=1)))
| 0.865579 | 0.922203 |
## This notebook shows how to use Orchestrator APIs for user experiments
```
import os
from fabrictestbed.slice_manager import SliceManager, Status, SliceState
import json
ssh_key_file_priv=os.environ['HOME']+"/.ssh/id_rsa"
ssh_key_file_pub=os.environ['HOME']+"/.ssh/id_rsa.pub"
ssh_key_pub = None
with open (ssh_key_file_pub, "r") as myfile:
ssh_key_pub=myfile.read()
ssh_key_pub=ssh_key_pub.strip()
slice_name='Slice-l2bridge-ded-tag'
```
## Create Slice Manager Object
Users can request tokens with different Project and Scopes by altering `project_name` and `scope` parameters in the refresh call below.
```
slice_manager = SliceManager()
```
### Orchestrator API example to query for available resources
```
status, advertised_topology = slice_manager.resources()
print(f"Status: {status}")
if status == Status.OK:
print(f"Toplogy: {advertised_topology}")
else:
print(f"Error: {advertised_topology}")
if status == Status.OK:
advertised_topology.draw()
```
## Create Slice
In Release 1.0, user is expected to create tagged interface and assign the IP addresses manually. Please use the example comands indicated below:
### Run on Node N1
```
ip link add link eth1 name eth1.200 type vlan id 200
ip link set dev eth1.200 up
ip addr add 192.168.10.51/24 dev eth1.200
```
### Run on Node N2
```
ip link add link eth1 name eth1.200 type vlan id 200
ip link set dev eth1.200 up
ip addr add 192.168.10.52/24 dev eth1.200
```
```
from fabrictestbed.slice_editor import ExperimentTopology, Capacities, ComponentType, ComponentModelType, ServiceType
# Create topology
t = ExperimentTopology()
# Add node
n1 = t.add_node(name='n1', site='LBNL')
# Set capacities
cap = Capacities()
cap.set_fields(core=2, ram=6, disk=10)
# Set Properties
n1.set_properties(capacities=cap, image_type='qcow2', image_ref='default_centos_8')
# Add PCI devices
n1.add_component(ctype=ComponentType.NVME, model='P4510', name='c1')
# Add node
n2 = t.add_node(name='n2', site='LBNL')
# Set properties
n2.set_properties(capacities=cap, image_type='qcow2', image_ref='default_centos_8')
# Dedicated Cards
n1.add_component(model_type=ComponentModelType.SmartNIC_ConnectX_6, name='n1-nic1')
n2.add_component(model_type=ComponentModelType.SmartNIC_ConnectX_5, name='n2-nic1')
# For Tagged Bridge, specify VLAN
for i in t.interface_list:
if_labels = i.get_property(pname="labels")
if_labels.vlan = "200"
i.set_properties(labels=if_labels)
# L2Bridge Service
t.add_network_service(name='bridge1', nstype=ServiceType.L2Bridge, interfaces=t.interface_list)
# Generate Slice Graph
slice_graph = t.serialize()
# Request slice from Orchestrator
status, reservations = slice_manager.create(slice_name=slice_name, slice_graph=slice_graph, ssh_key=ssh_key_pub)
print("Response Status {}".format(status))
if status == Status.OK:
print("Reservations created {}".format(reservations))
else:
print(f"Failure: {reservations}")
```
## Query Slices
```
status, slices = slice_manager.slices(excludes=[SliceState.Closing, SliceState.Dead])
print("Response Status {}".format(status))
if status == Status.OK:
print("Slices {}".format(slices))
slice_object=list(filter(lambda s: s.slice_name == slice_name, slices))[0]
else:
print(f"Failure: {slices}")
```
## Query Slivers
```
status, slivers = slice_manager.slivers(slice_object=slice_object)
print("Response Status {}".format(status))
if status == Status.OK:
print("Slivers {}".format(slivers))
else:
print(f"Failure: {slivers}")
```
## Sliver Status
```
for s in slivers:
status, sliver_status = slice_manager.sliver_status(sliver=s)
print("Response Status {}".format(status))
if status == Status.OK:
print()
print("Sliver Status {}".format(sliver_status))
print()
```
## Delete Slice
```
status, result = slice_manager.delete(slice_object=slice_object)
print("Response Status {}".format(status))
print("Response received {}".format(result))
```
|
github_jupyter
|
import os
from fabrictestbed.slice_manager import SliceManager, Status, SliceState
import json
ssh_key_file_priv=os.environ['HOME']+"/.ssh/id_rsa"
ssh_key_file_pub=os.environ['HOME']+"/.ssh/id_rsa.pub"
ssh_key_pub = None
with open (ssh_key_file_pub, "r") as myfile:
ssh_key_pub=myfile.read()
ssh_key_pub=ssh_key_pub.strip()
slice_name='Slice-l2bridge-ded-tag'
slice_manager = SliceManager()
status, advertised_topology = slice_manager.resources()
print(f"Status: {status}")
if status == Status.OK:
print(f"Toplogy: {advertised_topology}")
else:
print(f"Error: {advertised_topology}")
if status == Status.OK:
advertised_topology.draw()
ip link add link eth1 name eth1.200 type vlan id 200
ip link set dev eth1.200 up
ip addr add 192.168.10.51/24 dev eth1.200
ip link add link eth1 name eth1.200 type vlan id 200
ip link set dev eth1.200 up
ip addr add 192.168.10.52/24 dev eth1.200
from fabrictestbed.slice_editor import ExperimentTopology, Capacities, ComponentType, ComponentModelType, ServiceType
# Create topology
t = ExperimentTopology()
# Add node
n1 = t.add_node(name='n1', site='LBNL')
# Set capacities
cap = Capacities()
cap.set_fields(core=2, ram=6, disk=10)
# Set Properties
n1.set_properties(capacities=cap, image_type='qcow2', image_ref='default_centos_8')
# Add PCI devices
n1.add_component(ctype=ComponentType.NVME, model='P4510', name='c1')
# Add node
n2 = t.add_node(name='n2', site='LBNL')
# Set properties
n2.set_properties(capacities=cap, image_type='qcow2', image_ref='default_centos_8')
# Dedicated Cards
n1.add_component(model_type=ComponentModelType.SmartNIC_ConnectX_6, name='n1-nic1')
n2.add_component(model_type=ComponentModelType.SmartNIC_ConnectX_5, name='n2-nic1')
# For Tagged Bridge, specify VLAN
for i in t.interface_list:
if_labels = i.get_property(pname="labels")
if_labels.vlan = "200"
i.set_properties(labels=if_labels)
# L2Bridge Service
t.add_network_service(name='bridge1', nstype=ServiceType.L2Bridge, interfaces=t.interface_list)
# Generate Slice Graph
slice_graph = t.serialize()
# Request slice from Orchestrator
status, reservations = slice_manager.create(slice_name=slice_name, slice_graph=slice_graph, ssh_key=ssh_key_pub)
print("Response Status {}".format(status))
if status == Status.OK:
print("Reservations created {}".format(reservations))
else:
print(f"Failure: {reservations}")
status, slices = slice_manager.slices(excludes=[SliceState.Closing, SliceState.Dead])
print("Response Status {}".format(status))
if status == Status.OK:
print("Slices {}".format(slices))
slice_object=list(filter(lambda s: s.slice_name == slice_name, slices))[0]
else:
print(f"Failure: {slices}")
status, slivers = slice_manager.slivers(slice_object=slice_object)
print("Response Status {}".format(status))
if status == Status.OK:
print("Slivers {}".format(slivers))
else:
print(f"Failure: {slivers}")
for s in slivers:
status, sliver_status = slice_manager.sliver_status(sliver=s)
print("Response Status {}".format(status))
if status == Status.OK:
print()
print("Sliver Status {}".format(sliver_status))
print()
status, result = slice_manager.delete(slice_object=slice_object)
print("Response Status {}".format(status))
print("Response received {}".format(result))
| 0.230833 | 0.605274 |
## Validating Configuration Settings with Batfish
Network engineers routinely need to validate configuration settings of various devices in their network. In a multi-vendor network, this validation can be hard and few tools exist today to enable this basic task. However, the vendor-independent models of Batfish and its querying mechanisms make such validation almost trivial.
In this notebook, we show how to validate configuration settings with Batfish. More specifically, we examine how the configuration of NTP servers can be validated. The same validation scenarios can be performed for other configuration settings of nodes (such as dns servers, tacacs servers, snmp communities, VRFs, etc.) interfaces (such as MTU, bandwidth, input and output access lists, state, etc.), VRFs, BGP and OSPF sessions, and more.
Check out a video demo of this notebook [here](https://youtu.be/qOXRaVs1Uz4).

### Initializing our Network and Snapshot
`SNAPSHOT_PATH` below can be updated to point to a custom snapshot directory, see the [Batfish instructions](https://github.com/batfish/batfish/wiki/Packaging-snapshots-for-analysis) for how to package data for analysis.<br>
More example networks are available in the [networks](https://github.com/batfish/batfish/tree/master/networks) folder of the Batfish repository.
```
# Import packages and load questions
%run startup.py
# Initialize a network and snapshot
NETWORK_NAME = "example_network"
SNAPSHOT_NAME = "example_snapshot"
SNAPSHOT_PATH = "networks/example"
bf_set_network(NETWORK_NAME)
bf_init_snapshot(SNAPSHOT_PATH, name=SNAPSHOT_NAME, overwrite=True)
```
The network snapshot that we initialized above is illustrated below. You can download/view devices' configuration files [here](https://github.com/batfish/pybatfish/tree/master/jupyter_notebooks/networks/example). We will focus on the validation for the six **border** routers.
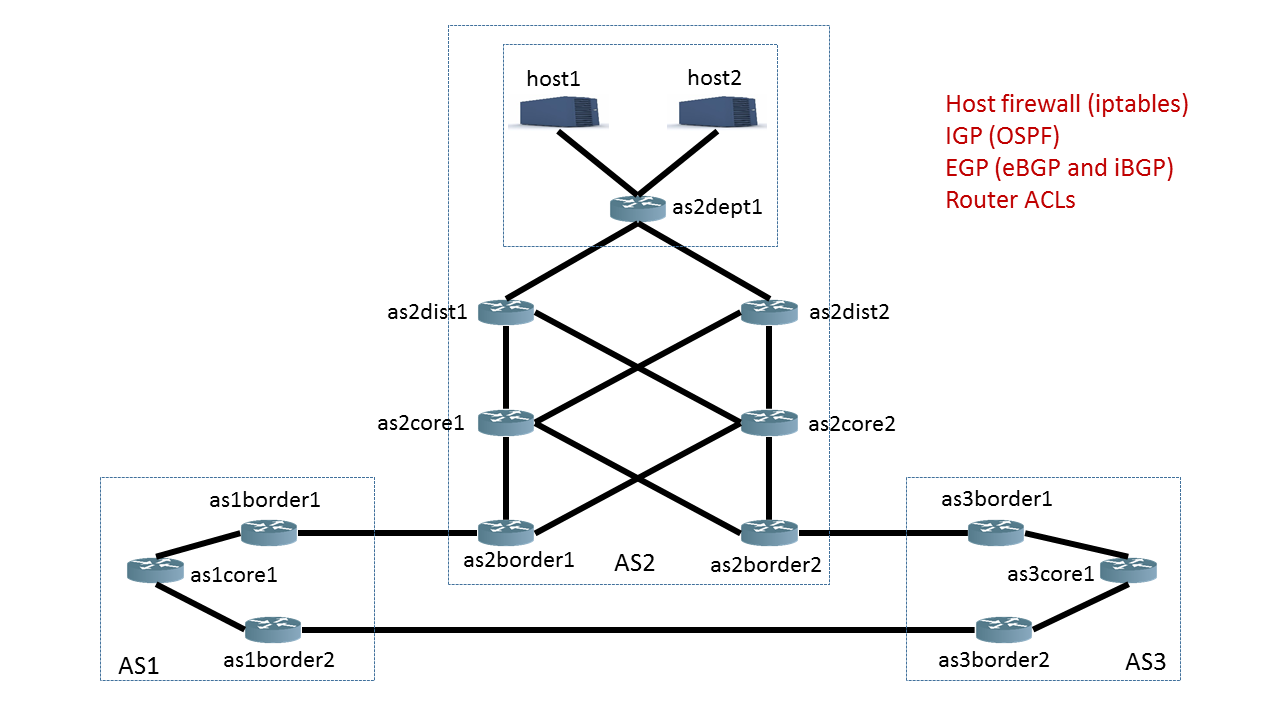
### Extracting configured NTP servers
This can be done using the `nodeProperties()` question.
```
# Set the property that we want to extract
COL_NAME = "NTP_Servers"
# Extract NTP servers for border routers
node_props = bfq.nodeProperties(
nodes=".*border.*",
properties=COL_NAME).answer().frame()
node_props
```
The `.frame()` function call above returns a [Pandas](https://pandas.pydata.org/pandas-docs/stable/) data frame that contains the answer.
### Validating NTP Servers Configuration
Depending on the network's policy, there are several possible validation scenarios for NTP-servers configuration:
1. Every node has at least one NTP server configured.
2. Every node has at least one NTP server configured from the reference set.
3. Every node has the reference set of NTP servers configured.
4. Every node has NTP servers that match those in a per-node database.
We demonstrate each scenario below.
#### Validation scenario 1: Every node has at least one NTP server configured
Now that we have the list of NTP servers, let's check if at least one server is configured on the border routers. We accomplish that by using ([lambda expressions](https://docs.python.org/3/reference/expressions.html#lambda)) to identify nodes where the list is empty.
```
# Find nodes that have no NTP servers configured
ns_violators = node_props[node_props[COL_NAME].apply(
lambda x: len(x) == 0)]
ns_violators
```
#### Validation scenario 2: Every node has at least one NTP server configured from the reference set.
Now if we want to validate that configured _NTP servers_ should contain at least one _NTP server_ from a reference set, we can use the command below. It identifies any node whose configured set of _NTP servers_ does not overlap with the reference set at all.
```
# Define the reference set of NTP servers
ref_ntp_servers = set(["23.23.23.23"])
# Find nodes that have no NTP server in common with the reference set
ns_violators = node_props[node_props[COL_NAME].apply(
lambda x: len(ref_ntp_servers.intersection(set(x))) == 0)]
ns_violators
```
Because `as1border1` has no configured NTP servers, it clearly violates our assertion, and so does `as2border2` which has a configured server but not one that is present in the reference set.
#### Validation scenario 3: Every node has the reference set of NTP servers configured
A common use case for validating _NTP servers_ involves checking that the set of _NTP servers_ exactly matches a desired reference set. Such validation is quite straightforward as well.
```
# Find violating nodes whose configured NTP servers do not match the reference set
ns_violators = node_props[node_props[COL_NAME].apply(
lambda x: ref_ntp_servers != set(x))]
ns_violators
```
As we can see, all border nodes violate this condition.
A slightly advanced version of pandas filtering can also show us which configured _NTP servers_ are missing or extra (compared to the reference set) at each node.
```
# Find extra and missing servers at each node
ns_extra = node_props[COL_NAME].map(lambda x: set(x) - ref_ntp_servers)
ns_missing = node_props[COL_NAME].map(lambda x: ref_ntp_servers - set(x))
# Join these columns up with the node columns for a complete view
diff_df = pd.concat([node_props["Node"],
ns_extra.rename('extra-{}'.format(COL_NAME)),
ns_missing.rename('missing-{}'.format(COL_NAME))],
axis=1)
diff_df
```
#### Validation scenario 4: Every node has _NTP servers_ that match those in a per-node database.
Every node should match its reference set of _NTP Servers_ which may be stored in an external database. This check enables easy validation of configuration settings that differ acorss nodes.
We assume data from the database is fetched in the following format, where node names are dictionary keys and specific properties are defined in a property-keyed dictionary per node.
```
# Mock reference-node-data, presumably taken from an external database
database = {'as1border1': {'NTP_Servers': ['23.23.23.23'],
'DNS_Servers': ['1.1.1.1']},
'as1border2': {'NTP_Servers': ['23.23.23.23'],
'DNS_Servers': ['1.1.1.1']},
'as2border1': {'NTP_Servers': ['18.18.18.18', '23.23.23.23'],
'DNS_Servers': ['2.2.2.2']},
'as2border2': {'NTP_Servers': ['18.18.18.18'],
'DNS_Servers': ['1.1.1.1']},
'as3border1': {'NTP_Servers': ['18.18.18.18', '23.23.23.23'],
'DNS_Servers': ['2.2.2.2']},
'as3border2': {'NTP_Servers': ['18.18.18.18', '23.23.23.23'],
'DNS_Servers': ['2.2.2.2']},
}
```
Note that there is an extra property in this dictionary that we don't care about comparing right now: `dns-server`. We will filter out this property below, before comparing the data from `Batfish` to that in the database.
After a little massaging, the database and `Batfish` data can be compared to generate two sets of servers: missing (i.e., present in the database but not in the configurations) and extra (i.e., present in the configurations but not in the database).
```
# Transpose database data so each node has its own row
database_df = pd.DataFrame(data=database).transpose()
# Index on node for easier comparison
df_node_props = node_props.set_index('Node')
# Select only columns present in node_props (get rid of the extra dns-servers column)
df_db_node_props = database_df[df_node_props.columns].copy()
# Convert server lists into sets to support arithmetic below
df_node_props[COL_NAME] = df_node_props[COL_NAME].apply(set)
df_db_node_props[COL_NAME] = df_db_node_props[COL_NAME].apply(set)
# Figure out what servers are in the configs but not the database and vice versa
missing_servers = (df_db_node_props - df_node_props).rename(
columns={COL_NAME: 'missing-{}'.format(COL_NAME)})
extra_servers = (df_node_props - df_db_node_props).rename(
columns={COL_NAME: 'extra-{}'.format(COL_NAME)})
result = pd.concat([missing_servers, extra_servers], axis=1, sort=False)
# removing the index name for clearer output
del result.index.name
result
```
### Continue exploring
We showed you how to extract the database of configured _NTP servers_ for every node and how to test that the settings are correct for a variety of desired test configurations. The underlying principles can be applied to other network configurations, such as [interfaceProperties](https://pybatfish.readthedocs.io/en/latest/questions.html#pybatfish.question.bfq.interfaceProperties), [bgpProcessConfiguration](https://pybatfish.readthedocs.io/en/latest/questions.html#pybatfish.question.bfq.bgpProcessConfiguration), [ospfProperties](https://pybatfish.readthedocs.io/en/latest/questions.html#pybatfish.question.bfq.ospfProperties) etc.
For example `interfaceProperties()` question can be used to fetch properties like interface MTU using a simple command.
```
# Extract interface MTU for Ethernet0/0 interfaces on border routers
interface_mtu = bfq.interfaceProperties(
nodes=".*border.*",
interfaces="Ethernet0/0",
properties="MTU").answer().frame()
interface_mtu
```
***
### Get involved with the Batfish community!
Start interacting through [Slack](https://join.slack.com/t/batfish-org/shared_invite/enQtMzA0Nzg2OTAzNzQ1LTUxOTJlY2YyNTVlNGQ3MTJkOTIwZTU2YjY3YzRjZWFiYzE4ODE5ODZiNjA4NGI5NTJhZmU2ZTllOTMwZDhjMzA) or [GitHub](https://github.com/batfish/batfish) to know more. We would love to talk with you about Batfish or your Network!
|
github_jupyter
|
# Import packages and load questions
%run startup.py
# Initialize a network and snapshot
NETWORK_NAME = "example_network"
SNAPSHOT_NAME = "example_snapshot"
SNAPSHOT_PATH = "networks/example"
bf_set_network(NETWORK_NAME)
bf_init_snapshot(SNAPSHOT_PATH, name=SNAPSHOT_NAME, overwrite=True)
# Set the property that we want to extract
COL_NAME = "NTP_Servers"
# Extract NTP servers for border routers
node_props = bfq.nodeProperties(
nodes=".*border.*",
properties=COL_NAME).answer().frame()
node_props
# Find nodes that have no NTP servers configured
ns_violators = node_props[node_props[COL_NAME].apply(
lambda x: len(x) == 0)]
ns_violators
# Define the reference set of NTP servers
ref_ntp_servers = set(["23.23.23.23"])
# Find nodes that have no NTP server in common with the reference set
ns_violators = node_props[node_props[COL_NAME].apply(
lambda x: len(ref_ntp_servers.intersection(set(x))) == 0)]
ns_violators
# Find violating nodes whose configured NTP servers do not match the reference set
ns_violators = node_props[node_props[COL_NAME].apply(
lambda x: ref_ntp_servers != set(x))]
ns_violators
# Find extra and missing servers at each node
ns_extra = node_props[COL_NAME].map(lambda x: set(x) - ref_ntp_servers)
ns_missing = node_props[COL_NAME].map(lambda x: ref_ntp_servers - set(x))
# Join these columns up with the node columns for a complete view
diff_df = pd.concat([node_props["Node"],
ns_extra.rename('extra-{}'.format(COL_NAME)),
ns_missing.rename('missing-{}'.format(COL_NAME))],
axis=1)
diff_df
# Mock reference-node-data, presumably taken from an external database
database = {'as1border1': {'NTP_Servers': ['23.23.23.23'],
'DNS_Servers': ['1.1.1.1']},
'as1border2': {'NTP_Servers': ['23.23.23.23'],
'DNS_Servers': ['1.1.1.1']},
'as2border1': {'NTP_Servers': ['18.18.18.18', '23.23.23.23'],
'DNS_Servers': ['2.2.2.2']},
'as2border2': {'NTP_Servers': ['18.18.18.18'],
'DNS_Servers': ['1.1.1.1']},
'as3border1': {'NTP_Servers': ['18.18.18.18', '23.23.23.23'],
'DNS_Servers': ['2.2.2.2']},
'as3border2': {'NTP_Servers': ['18.18.18.18', '23.23.23.23'],
'DNS_Servers': ['2.2.2.2']},
}
# Transpose database data so each node has its own row
database_df = pd.DataFrame(data=database).transpose()
# Index on node for easier comparison
df_node_props = node_props.set_index('Node')
# Select only columns present in node_props (get rid of the extra dns-servers column)
df_db_node_props = database_df[df_node_props.columns].copy()
# Convert server lists into sets to support arithmetic below
df_node_props[COL_NAME] = df_node_props[COL_NAME].apply(set)
df_db_node_props[COL_NAME] = df_db_node_props[COL_NAME].apply(set)
# Figure out what servers are in the configs but not the database and vice versa
missing_servers = (df_db_node_props - df_node_props).rename(
columns={COL_NAME: 'missing-{}'.format(COL_NAME)})
extra_servers = (df_node_props - df_db_node_props).rename(
columns={COL_NAME: 'extra-{}'.format(COL_NAME)})
result = pd.concat([missing_servers, extra_servers], axis=1, sort=False)
# removing the index name for clearer output
del result.index.name
result
# Extract interface MTU for Ethernet0/0 interfaces on border routers
interface_mtu = bfq.interfaceProperties(
nodes=".*border.*",
interfaces="Ethernet0/0",
properties="MTU").answer().frame()
interface_mtu
| 0.594904 | 0.946745 |
**[Deep Learning Home Page](https://www.kaggle.com/learn/deep-learning)**
---
# Intro
You don't directly choose the numbers to go into your convolutions for deep learning... instead the deep learning technique determines what convolutions will be useful from the data (as part of model-training). We'll come back to how the model does that soon.

But looking closely at convolutions and how they are applied to your image will improve your intuition for these models, how they work, and how to debug them when they don't work.
**Let's get started.**
# Exercises
We'll use some small utilty functions to visualize raw images and some results of your code. Execute the next cell to load the utility functions.
```
# set up code checking
from learntools.core import binder
binder.bind(globals())
from learntools.deep_learning.exercise_1 import *
print("Setup Complete")
```
<hr/>
## Exercise 1
In the video, you saw a convolution that detected horizontal lines. That convolution shows up again in the code cell below.
Run the cell to see a raw image as well as the output from applying this convolution to the image. `load_my_image` and `visualize_conv` are utility functions provided for this exercise.
```
horizontal_line_conv = [[1, 1],
[-1, -1]]
original_image = load_my_image()
visualize_conv(original_image, horizontal_line_conv)
# inspect the source of hidden functions
from inspect import getsource
print(getsource(load_my_image))
```
Now it's your turn. Instead of a horizontal line detector, you will create a vertical line detector.
**Replace the underscores with numbers to make a vertical line detector and uncomment both lines of code in the cell below. Then run **
```
vertical_line_conv = [[1, -1],
[1, -1]]
# check your answer
q_1.check()
visualize_conv(original_image, vertical_line_conv)
```
If you'd like a hint or the solution, uncomment the appropriate line below.
```
# q_1.hint()
# q_1.solution()
```
<hr/>
## Exercise 2
The convolutions you've seen are 2x2. But you could have larger convolutions. They could be 3x3, 4x4, etc. They don't even have to be square. Nothing prevents using a 4x7 convolution.
Compare the number of visual patterns that can be captured by small convolutions. Which of the following is true?
- There are more visual patterns that can be captured by large convolutions
- There are fewer visual patterns that can be captured by large convolutions
- The number of visual patterns that can be captured by large convolutions is the same as the number of visual patterns that can be captured by small convolutions?
Once you think you know the answer, check it by uncommenting and running the line below.
```
# check your answer (Run this code cell to receive credit!)
q_2.solution()
```
# Keep Going
Now you are ready to **[combine convolutions into powerful models](https://www.kaggle.com/dansbecker/building-models-from-convolutions).** These models are fun to work with, so keep going.
---
**[Deep Learning Home Page](https://www.kaggle.com/learn/deep-learning)**
*Have questions or comments? Visit the [Learn Discussion forum](https://www.kaggle.com/learn-forum) to chat with other Learners.*
|
github_jupyter
|
# set up code checking
from learntools.core import binder
binder.bind(globals())
from learntools.deep_learning.exercise_1 import *
print("Setup Complete")
horizontal_line_conv = [[1, 1],
[-1, -1]]
original_image = load_my_image()
visualize_conv(original_image, horizontal_line_conv)
# inspect the source of hidden functions
from inspect import getsource
print(getsource(load_my_image))
vertical_line_conv = [[1, -1],
[1, -1]]
# check your answer
q_1.check()
visualize_conv(original_image, vertical_line_conv)
# q_1.hint()
# q_1.solution()
# check your answer (Run this code cell to receive credit!)
q_2.solution()
| 0.436622 | 0.988268 |
<div class="alert block alert-info alert">
# <center> Scientific Programming in Python
## <center>Karl N. Kirschner<br>Bonn-Rhein-Sieg University of Applied Sciences<br>Sankt Augustin, Germany
# <center> Course Introduction
<hr style="border:2px solid gray"></hr>
## A sneak-peak into our future
```
import matplotlib.pyplot as plt
x_values = [1, 2, 3, 4, 7]
y_values = [1, 4, 9, 16, 49]
plt.figure()
plt.plot(x_values, y_values,
marker='.', markersize=24,
linewidth=5, linestyle='-',
color='red')
plt.xlabel("X Label (unit)")
plt.ylabel("Y Label (unit)")
plt.show()
```
# Keys to success in this course
### <font color='dodgerblue'>$\textrm{C}^3$</font>: code is witten <font color='dodgerblue'>concisely</font>, with a <font color='dodgerblue'>clear</font> thought process, and placed into <font color='dodgerblue'>context</font>
1. **Concise**, cleanly written code and output
- Easy to read and understand
- Reduced chances of introduction programmer errror
- Easy to debug
2. **Clear** thought process with logical grouping of code
- User-defined functions that contain a single concept
- Promotes usability and reusability in future code (i.e. user-defined functions)
3. **Context** for the code's a) purpose and b) usage are provided
- Block comments (e.g user-defined functions) - purpose and usage
- Jupyter-notebook markdown language (citations, data interpretation)
### <font color='dodgerblue'>K.I.S.S.</font>: <font color='dodgerblue'>K</font>eep <font color='dodgerblue'>I</font>t (i.e. coding) <font color='dodgerblue'>S</font>imple & <font color='dodgerblue'>S</font>mart
- $\textrm{C}^3$ - concise, clear and context
- Use of built-in functions over libraries with large overhead
- User-defined functions for reproducibility, reuse, error reduction and isolating ideas
<font color='dodgerblue'>$\textrm{C}^3$ and K.I.S.S. are the same keys to success that are found in **all** scientific working.</font>
<hr style="border:2px solid gray"> </hr>
## Scientific Programming
**Definition**
1. Programming whose goal is for scientific usage (e.g. workflows, data analysis) and visualization.
2. Learning to program is an academic, scholarly manner (i.e. wissenschaftliches Arbeit).
**3 Ways to Think About It**
1. **Usage**: to perform mathematics (simple numerical computations to some complex math models)
2. **Practice**: to create while maintaining good scholarship
- knowing what is state-of-the-art
- careful, clear and supportive
- "A machine requires precise instructions, which users often fail to supply"[1]
3. **Target**: to support science (doing research, data support and analysis)
- "Scientists commonly use languages such as Python and R to conduct and **automate analyses**, because in this way they can **speed data crunching**, **increase reproducibility**, protect data from accidental deletion or alteration and handle data sets that would overwhelm commercial applications."[1]
- Create workflows to help do the research
- Create simulations (increasingly becoming more important in research)
- exploratory: for understanding raw data
- supportive: for strengthening interpretations of the data
- predictive: creating new ideas
[1] Baker, Monya. "Scientific computing: code alert." Nature 541, no. 7638 (2017): 563-565.
<hr style="border:2px solid gray"> </hr>
## Why is this Important?
- "Societally important **science relies on models and the software implementing them**. The scientific community must ensure that the findings and recommendations put forth based on **those models conform to the highest scientific expectation**" <br> L. N. Joppa et al. , “Troubling Trends in Scientific Software Use,” Science, 340(6134), 814–815, 2013.
<br><br>
**Positive Example:**
ChemRxiv (a pre-print science paper archive) sent out an email on March 20th, 2021 highlighting the recent research submissions on the Coronavirus. 9 out of the 10 highlights had a significant amount of computer modeling (computational chemistry). In other words, computer models were the first to get some research results that target the pandemic.

**Negative Example:**
- 2001 & 2005 Science peer-reviewed article <br> G. Changet al., “Retraction,” Science, 314(5807), 1875, 2006

## Why Python?
- Accessible and readable (especially to people outside of computer science)
- Natural scientists
- Engineers
- Powerful due to the number of libraries available that are created by domain experts and programmers
- Good for creating larger programs - create functions that do specific tasks
- Call bash commands from inside of python (via "import os")
- Popular

Source: https://redmonk.com - ca. 90000 respondants, 2020
***
## What Disciplines use Python - Its "Main" Catagories (as of 2018)
For a Pyhton job, one should known a) python's core and b) one of the following catagories:
- Web - flask, django, html, javascript
- Data engineering (collecting data) - sql, airflow, luigi
- Software engineering - git, unit testing, large codebases
- Cyber security - requests, volatility, pyew, peepdf, penetration tests
- **Data science / scientific python** (seeks new knowledge) - Numpy, Pandas, Matplotlib, SciPy, PyTorch, TensorFlow
<hr style="border:2px solid gray"> </hr>
## Misc. Background
- General-purpose programming language
- Python interpreter: python3 (python2 is out-of-date)
- Contains programming common concepts like
- statements
- expressions
- operators
- modules
- methods
- classes
- Contains Lists (e.g. movies = [ 'Star Wars', 'X-men', 'Forest Gump'])
- Has IDE (Integrated Devlopment Environment): IDLE, PyCharm, atom, sublime
- OS independent
- Good for researchers dealing with lots of data (i.e. scientists)
- Bad for developing fast parallel programs
<hr style="border:2px solid gray"> </hr>
## Getting Python3
- Have Python (and desired libraries) installed onto your computer
- It may already be installed
- If not: https://www.python.org/downloads
<hr style="border:2px solid gray"> </hr>
## Create and execute a code (and demos)
1. Start a Python3 in a Terminal
- Exit: 'exit()' or 'Cntl D' ('Strg D')
Example:
- Linux: 'Menu' -> 'Terminal' -> 'python3'
- Macintosh: 'Application' -> 'Utilities' -> 'Terminal' -> 'python3'
- Windows: ? -> 'python3'
- Warning: Done in real-time, and thus is not saved
2. Create a 'program_name.py' script using a text program and command-line execute
- Simple (e.g. texteditor, gedit) - warning, can be problematic
- Sophisticated
- [Sublime](https://www.sublimetext.com/)
- [PyCharm](https://www.jetbrains.com/pycharm/download)
- other integrated development environment [IDE]
- Execuate: In a terminal, type 'python program_name.py'
3. <font color='dodgerblue'>**Google's Colaboratory**</font>: https://colab.research.google.com
4. <font color='dodgerblue'>**Jupyter Notebooks**</font>: https://jupyter.org - Recommended for novice and experienced programmers
<hr style="border:2px solid gray"> </hr>
## Important Information
1. Lectures will be ca. 90 minutes long
2. We will make use of LEA, including handing in projects
3. I am available through university email - I tend to respond to these fairly quickly
4. Individual/group online meetings can be made upon request
5. We will use the WebEx software, and adjust if needed
## Instructions for this class
### Coding homework/projects:
1. Will be handed in as a **Jupyter Notebook** file
2. Include you name at the top of the notebook
<hr style="border:2px solid gray"> </hr>
### Grades
**See the course syllabus on LEA**
**Homework/problems will be given points from 0-100**<br>
**Mark given for point range**<br>
1.0: 100 -- 95<br>
1.3: 94 -- 90<br>
1.7: 89 -- 85<br>
2.0: 84 -- 80<br>
2.3: 79 -- 75<br>
2.7: 74 -- 70<br>
3.0: 69 -- 65<br>
3.3: 64 -- 60<br>
3.7: 59 -- 55<br>
4.0: 54 -- 50<br>
5.0: 50 -- 0<br>
**Marks Definition**<br>
1.0: extremely well done work (i.e. "perfect" to "excellent")<br>
1.3 -- 1.7: well done work, with minor blemishes (i.e. "very good" to "good")<br>
2.0 -- 2.3: decently done work (i.e. "good")<br>
2.7 -- 3.3: the work has some flaws, but overall is okay (i.e. "average")<br>
3.7 -- 4.0: notably flawed work, but still acceptable (i.e. "sufficient").<br>
5.0: very poorly done work (i.e. "insufficient")<br>
**5 Grading Catagories** that are weighted (i.e. relative importance)
1. Code quality & design (weighted by 4)
- 6 pts: Solution/goal is well thought out
- 3 pts: Solution/goal is partially planned
- 0 pts: Solution/goal is unrefined
2. Code execution & results (weighted by 4)
- 6 pts: Code runs correctly in its entirety
- 3 pts: Code runs, but only partially correct
- 0 pts: Code doesn’t run, or doesn’t give proper output
3. Assignment requirements (weighted by 4)
- 6 pts: All of the requirements were fulfilled
- 3 pts: Half of the requirements were met
- 0 pts: Very little or none of the requirements were met
4. Scientific programming (weighted by 4)
- 6 pts: Code completely follows scientific programming concepts
- 3 pts: Code somewhat follows scientific programming concepts
- 0 pts: Code does not conform to scientific programming concepts
5. Creativity (weighted by 1)
- 4 pts: Code shows significant problem solving skills, creativity, or robustness
- 2 pts: Code shows an average approach to problem solving, creativity, or robustness
- 0 pts: Code does not show notable problem solving skills, creativity, or robustness

|
github_jupyter
|
import matplotlib.pyplot as plt
x_values = [1, 2, 3, 4, 7]
y_values = [1, 4, 9, 16, 49]
plt.figure()
plt.plot(x_values, y_values,
marker='.', markersize=24,
linewidth=5, linestyle='-',
color='red')
plt.xlabel("X Label (unit)")
plt.ylabel("Y Label (unit)")
plt.show()
| 0.464659 | 0.947088 |
## Subject Selection Experiments disorder data - Srinivas (handle: thewickedaxe)
### Initial Data Cleaning
```
# Standard
import pandas as pd
import numpy as np
%matplotlib inline
import matplotlib.pyplot as plt
# Dimensionality reduction and Clustering
from sklearn.decomposition import PCA
from sklearn.cluster import KMeans
from sklearn.cluster import MeanShift, estimate_bandwidth
from sklearn import manifold, datasets
from itertools import cycle
# Plotting tools and classifiers
from matplotlib.colors import ListedColormap
from sklearn.linear_model import LogisticRegression
from sklearn.cross_validation import train_test_split
from sklearn import preprocessing
from sklearn.datasets import make_moons, make_circles, make_classification
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.naive_bayes import GaussianNB
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis as LDA
from sklearn.discriminant_analysis import QuadraticDiscriminantAnalysis as QDA
from sklearn import cross_validation
from sklearn.cross_validation import LeaveOneOut
# Let's read the data in and clean it
def get_NaNs(df):
columns = list(df.columns.get_values())
row_metrics = df.isnull().sum(axis=1)
rows_with_na = []
for i, x in enumerate(row_metrics):
if x > 0: rows_with_na.append(i)
return rows_with_na
def remove_NaNs(df):
rows_with_na = get_NaNs(df)
cleansed_df = df.drop(df.index[rows_with_na], inplace=False)
return cleansed_df
initial_data = pd.DataFrame.from_csv('Data_Adults_1_reduced_inv3.csv')
cleansed_df = remove_NaNs(initial_data)
# Let's also get rid of nominal data
numerics = ['int16', 'int32', 'int64', 'float16', 'float32', 'float64']
X = cleansed_df.select_dtypes(include=numerics)
print X.shape
# Let's now clean columns getting rid of certain columns that might not be important to our analysis
cols2drop = ['GROUP_ID', 'doa', 'Baseline_header_id', 'Concentration_header_id', 'Baseline_Reading_id',
'Concentration_Reading_id']
X = X.drop(cols2drop, axis=1, inplace=False)
print X.shape
# For our studies children skew the data, it would be cleaner to just analyse adults
X = X.loc[X['Age'] >= 18]
Y = X.loc[X['race_id'] == 1]
X = X.loc[X['Gender_id'] == 1]
print X.shape
print Y.shape
```
### Extracting the samples we are interested in
```
# Let's extract ADHd and Bipolar patients (mutually exclusive)
ADHD_men = X.loc[X['ADHD'] == 1]
ADHD_men = ADHD_men.loc[ADHD_men['Bipolar'] == 0]
BP_men = X.loc[X['Bipolar'] == 1]
BP_men = BP_men.loc[BP_men['ADHD'] == 0]
ADHD_cauc = Y.loc[Y['ADHD'] == 1]
ADHD_cauc = ADHD_cauc.loc[ADHD_cauc['Bipolar'] == 0]
BP_cauc = Y.loc[Y['Bipolar'] == 1]
BP_cauc = BP_cauc.loc[BP_cauc['ADHD'] == 0]
print ADHD_men.shape
print BP_men.shape
print ADHD_cauc.shape
print BP_cauc.shape
# Keeping a backup of the data frame object because numpy arrays don't play well with certain scikit functions
ADHD_men = pd.DataFrame(ADHD_men.drop(['Patient_ID', 'Gender_id', 'ADHD', 'Bipolar'], axis = 1, inplace = False))
BP_men = pd.DataFrame(BP_men.drop(['Patient_ID', 'Gender_id', 'ADHD', 'Bipolar'], axis = 1, inplace = False))
ADHD_cauc = pd.DataFrame(ADHD_cauc.drop(['Patient_ID', 'race_id', 'ADHD', 'Bipolar'], axis = 1, inplace = False))
BP_cauc = pd.DataFrame(BP_cauc.drop(['Patient_ID', 'race_id', 'ADHD', 'Bipolar'], axis = 1, inplace = False))
```
### Dimensionality reduction
#### Manifold Techniques
##### ISOMAP
```
combined1 = pd.concat([ADHD_men, BP_men])
combined2 = pd.concat([ADHD_cauc, BP_cauc])
print combined1.shape
print combined2.shape
combined1 = preprocessing.scale(combined1)
combined2 = preprocessing.scale(combined2)
combined1 = manifold.Isomap(20, 20).fit_transform(combined1)
ADHD_men_iso = combined1[:946]
BP_men_iso = combined1[946:]
combined2 = manifold.Isomap(20, 20).fit_transform(combined2)
ADHD_cauc_iso = combined2[:992]
BP_cauc_iso = combined2[992:]
```
### Clustering and other grouping experiments
#### K-Means clustering - iso
```
data1 = pd.concat([pd.DataFrame(ADHD_men_iso), pd.DataFrame(BP_men_iso)])
data2 = pd.concat([pd.DataFrame(ADHD_cauc_iso), pd.DataFrame(BP_cauc_iso)])
print data1.shape
print data2.shape
kmeans = KMeans(n_clusters=2)
kmeans.fit(data1.get_values())
labels1 = kmeans.labels_
centroids1 = kmeans.cluster_centers_
print('Estimated number of clusters: %d' % len(centroids1))
for label in [0, 1]:
ds = data1.get_values()[np.where(labels1 == label)]
plt.plot(ds[:,0], ds[:,1], '.')
lines = plt.plot(centroids1[label,0], centroids1[label,1], 'o')
kmeans = KMeans(n_clusters=2)
kmeans.fit(data2.get_values())
labels2 = kmeans.labels_
centroids2 = kmeans.cluster_centers_
print('Estimated number of clusters: %d' % len(centroids2))
for label in [0, 1]:
ds2 = data2.get_values()[np.where(labels2 == label)]
plt.plot(ds2[:,0], ds2[:,1], '.')
lines = plt.plot(centroids2[label,0], centroids2[label,1], 'o')
```
### Classification Experiments
Let's experiment with a bunch of classifiers
```
ADHD_men_iso = pd.DataFrame(ADHD_men_iso)
BP_men_iso = pd.DataFrame(BP_men_iso)
ADHD_cauc_iso = pd.DataFrame(ADHD_cauc_iso)
BP_cauc_iso = pd.DataFrame(BP_cauc_iso)
BP_men_iso['ADHD-Bipolar'] = 0
ADHD_men_iso['ADHD-Bipolar'] = 1
BP_cauc_iso['ADHD-Bipolar'] = 0
ADHD_cauc_iso['ADHD-Bipolar'] = 1
data1 = pd.concat([ADHD_men_iso, BP_men_iso])
data2 = pd.concat([ADHD_cauc_iso, BP_cauc_iso])
class_labels1 = data1['ADHD-Bipolar']
class_labels2 = data2['ADHD-Bipolar']
data1 = data1.drop(['ADHD-Bipolar'], axis = 1, inplace = False)
data2 = data2.drop(['ADHD-Bipolar'], axis = 1, inplace = False)
data1 = data1.get_values()
data2 = data2.get_values()
# Leave one Out cross validation
def leave_one_out(classifier, values, labels):
leave_one_out_validator = LeaveOneOut(len(values))
classifier_metrics = cross_validation.cross_val_score(classifier, values, labels, cv=leave_one_out_validator)
accuracy = classifier_metrics.mean()
deviation = classifier_metrics.std()
return accuracy, deviation
rf = RandomForestClassifier(n_estimators = 22)
qda = QDA()
lda = LDA()
gnb = GaussianNB()
classifier_accuracy_list = []
classifiers = [(rf, "Random Forest"), (lda, "LDA"), (qda, "QDA"), (gnb, "Gaussian NB")]
for classifier, name in classifiers:
accuracy, deviation = leave_one_out(classifier, data1, class_labels1)
print '%s accuracy is %0.4f (+/- %0.3f)' % (name, accuracy, deviation)
classifier_accuracy_list.append((name, accuracy))
for classifier, name in classifiers:
accuracy, deviation = leave_one_out(classifier, data2, class_labels2)
print '%s accuracy is %0.4f (+/- %0.3f)' % (name, accuracy, deviation)
classifier_accuracy_list.append((name, accuracy))
```
|
github_jupyter
|
# Standard
import pandas as pd
import numpy as np
%matplotlib inline
import matplotlib.pyplot as plt
# Dimensionality reduction and Clustering
from sklearn.decomposition import PCA
from sklearn.cluster import KMeans
from sklearn.cluster import MeanShift, estimate_bandwidth
from sklearn import manifold, datasets
from itertools import cycle
# Plotting tools and classifiers
from matplotlib.colors import ListedColormap
from sklearn.linear_model import LogisticRegression
from sklearn.cross_validation import train_test_split
from sklearn import preprocessing
from sklearn.datasets import make_moons, make_circles, make_classification
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.naive_bayes import GaussianNB
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis as LDA
from sklearn.discriminant_analysis import QuadraticDiscriminantAnalysis as QDA
from sklearn import cross_validation
from sklearn.cross_validation import LeaveOneOut
# Let's read the data in and clean it
def get_NaNs(df):
columns = list(df.columns.get_values())
row_metrics = df.isnull().sum(axis=1)
rows_with_na = []
for i, x in enumerate(row_metrics):
if x > 0: rows_with_na.append(i)
return rows_with_na
def remove_NaNs(df):
rows_with_na = get_NaNs(df)
cleansed_df = df.drop(df.index[rows_with_na], inplace=False)
return cleansed_df
initial_data = pd.DataFrame.from_csv('Data_Adults_1_reduced_inv3.csv')
cleansed_df = remove_NaNs(initial_data)
# Let's also get rid of nominal data
numerics = ['int16', 'int32', 'int64', 'float16', 'float32', 'float64']
X = cleansed_df.select_dtypes(include=numerics)
print X.shape
# Let's now clean columns getting rid of certain columns that might not be important to our analysis
cols2drop = ['GROUP_ID', 'doa', 'Baseline_header_id', 'Concentration_header_id', 'Baseline_Reading_id',
'Concentration_Reading_id']
X = X.drop(cols2drop, axis=1, inplace=False)
print X.shape
# For our studies children skew the data, it would be cleaner to just analyse adults
X = X.loc[X['Age'] >= 18]
Y = X.loc[X['race_id'] == 1]
X = X.loc[X['Gender_id'] == 1]
print X.shape
print Y.shape
# Let's extract ADHd and Bipolar patients (mutually exclusive)
ADHD_men = X.loc[X['ADHD'] == 1]
ADHD_men = ADHD_men.loc[ADHD_men['Bipolar'] == 0]
BP_men = X.loc[X['Bipolar'] == 1]
BP_men = BP_men.loc[BP_men['ADHD'] == 0]
ADHD_cauc = Y.loc[Y['ADHD'] == 1]
ADHD_cauc = ADHD_cauc.loc[ADHD_cauc['Bipolar'] == 0]
BP_cauc = Y.loc[Y['Bipolar'] == 1]
BP_cauc = BP_cauc.loc[BP_cauc['ADHD'] == 0]
print ADHD_men.shape
print BP_men.shape
print ADHD_cauc.shape
print BP_cauc.shape
# Keeping a backup of the data frame object because numpy arrays don't play well with certain scikit functions
ADHD_men = pd.DataFrame(ADHD_men.drop(['Patient_ID', 'Gender_id', 'ADHD', 'Bipolar'], axis = 1, inplace = False))
BP_men = pd.DataFrame(BP_men.drop(['Patient_ID', 'Gender_id', 'ADHD', 'Bipolar'], axis = 1, inplace = False))
ADHD_cauc = pd.DataFrame(ADHD_cauc.drop(['Patient_ID', 'race_id', 'ADHD', 'Bipolar'], axis = 1, inplace = False))
BP_cauc = pd.DataFrame(BP_cauc.drop(['Patient_ID', 'race_id', 'ADHD', 'Bipolar'], axis = 1, inplace = False))
combined1 = pd.concat([ADHD_men, BP_men])
combined2 = pd.concat([ADHD_cauc, BP_cauc])
print combined1.shape
print combined2.shape
combined1 = preprocessing.scale(combined1)
combined2 = preprocessing.scale(combined2)
combined1 = manifold.Isomap(20, 20).fit_transform(combined1)
ADHD_men_iso = combined1[:946]
BP_men_iso = combined1[946:]
combined2 = manifold.Isomap(20, 20).fit_transform(combined2)
ADHD_cauc_iso = combined2[:992]
BP_cauc_iso = combined2[992:]
data1 = pd.concat([pd.DataFrame(ADHD_men_iso), pd.DataFrame(BP_men_iso)])
data2 = pd.concat([pd.DataFrame(ADHD_cauc_iso), pd.DataFrame(BP_cauc_iso)])
print data1.shape
print data2.shape
kmeans = KMeans(n_clusters=2)
kmeans.fit(data1.get_values())
labels1 = kmeans.labels_
centroids1 = kmeans.cluster_centers_
print('Estimated number of clusters: %d' % len(centroids1))
for label in [0, 1]:
ds = data1.get_values()[np.where(labels1 == label)]
plt.plot(ds[:,0], ds[:,1], '.')
lines = plt.plot(centroids1[label,0], centroids1[label,1], 'o')
kmeans = KMeans(n_clusters=2)
kmeans.fit(data2.get_values())
labels2 = kmeans.labels_
centroids2 = kmeans.cluster_centers_
print('Estimated number of clusters: %d' % len(centroids2))
for label in [0, 1]:
ds2 = data2.get_values()[np.where(labels2 == label)]
plt.plot(ds2[:,0], ds2[:,1], '.')
lines = plt.plot(centroids2[label,0], centroids2[label,1], 'o')
ADHD_men_iso = pd.DataFrame(ADHD_men_iso)
BP_men_iso = pd.DataFrame(BP_men_iso)
ADHD_cauc_iso = pd.DataFrame(ADHD_cauc_iso)
BP_cauc_iso = pd.DataFrame(BP_cauc_iso)
BP_men_iso['ADHD-Bipolar'] = 0
ADHD_men_iso['ADHD-Bipolar'] = 1
BP_cauc_iso['ADHD-Bipolar'] = 0
ADHD_cauc_iso['ADHD-Bipolar'] = 1
data1 = pd.concat([ADHD_men_iso, BP_men_iso])
data2 = pd.concat([ADHD_cauc_iso, BP_cauc_iso])
class_labels1 = data1['ADHD-Bipolar']
class_labels2 = data2['ADHD-Bipolar']
data1 = data1.drop(['ADHD-Bipolar'], axis = 1, inplace = False)
data2 = data2.drop(['ADHD-Bipolar'], axis = 1, inplace = False)
data1 = data1.get_values()
data2 = data2.get_values()
# Leave one Out cross validation
def leave_one_out(classifier, values, labels):
leave_one_out_validator = LeaveOneOut(len(values))
classifier_metrics = cross_validation.cross_val_score(classifier, values, labels, cv=leave_one_out_validator)
accuracy = classifier_metrics.mean()
deviation = classifier_metrics.std()
return accuracy, deviation
rf = RandomForestClassifier(n_estimators = 22)
qda = QDA()
lda = LDA()
gnb = GaussianNB()
classifier_accuracy_list = []
classifiers = [(rf, "Random Forest"), (lda, "LDA"), (qda, "QDA"), (gnb, "Gaussian NB")]
for classifier, name in classifiers:
accuracy, deviation = leave_one_out(classifier, data1, class_labels1)
print '%s accuracy is %0.4f (+/- %0.3f)' % (name, accuracy, deviation)
classifier_accuracy_list.append((name, accuracy))
for classifier, name in classifiers:
accuracy, deviation = leave_one_out(classifier, data2, class_labels2)
print '%s accuracy is %0.4f (+/- %0.3f)' % (name, accuracy, deviation)
classifier_accuracy_list.append((name, accuracy))
| 0.677047 | 0.883085 |
<p align="center" style="text-align:center">
<a style="display:inline-block" href="https://github.com/MKarimi21/University-of-Bojnurd/tree/master/Data-Mining" target="_blank">
<img src="http://rozup.ir/view/3132924/python-logo.png" alt="Python Examples" width="90" height="90">
</a>
<a style="display:inline-block" href="https://github.com/MKarimi21/University-of-Bojnurd" target="_blank">
<img src="http://rozup.ir/view/3132926/University_of_Bojnord_logo.png" alt="Python Examples" width="100" height="120">
</a>
<a style="display:inline-block" href="https://github.com/MKarimi21/University-of-Bojnurd/tree/master/Data-Mining" target="_blank">
<img src="http://rozup.ir/view/3132925/tlg_group.jpg" alt="Python Examples" width="80" height="80">
</a>
</p>
</br>
</br>
<p align="center" style="text-align:center">
<a href="https://github.com/MKarimi21/University-of-Bojnurd/blob/master/LICENSE" target="_blank" style="display:inline-flex"><img src="https://img.shields.io/badge/Licence-MIT-blue?style=flat-square" target="_blank"></a>
<a href="https://www.mr-karimi.ir" target="_blank" style="display:inline-flex"><img src="https://img.shields.io/badge/Create--by-MKarimi-red?style=flat-square" target="_blank"></a>
</p>
<h2 align="center"> Data Mining </h2>
<h3 align="center" style="font-family:tahoma">
درس داده کاوی
</h3>
<h4 align="center" style="font-family:tahoma; font-weight:300">
<b>
تمرین های درس داده کاوی، الگوریتم ها و کاربرد ها - دکتر مربی
</b>
<br>
<br>
دانشجو: مصطفی کریمی
</h4>
<h4 align="center" style="font-family:tahoma; font-weight:300">
<br>
<br>
<b>
#============== پروژه سوم و چهارم ==============#
</b>
<br>
<br>
</h4>
<h4 align="right" style="direction:rtl; font-family:tahoma; font-weight:300">
یک فایل شامل متغیر کمی و متغیر اسمی با حجم بیش از 200 داده که لااقل یکی از متغیر های کمی شامل گمشده نیز باشد انتخاب کرده.
</h4>
### Description
About data set
This database contains 76 attributes, but all published experiments refer to using a subset of 14 of them. In particular, the Cleveland database is the only one that has been used by ML researchers to
this date.The "target" field refers to the presence of heart disease in the patient. It is integer valued 0 = no/less chance of heart attack and 1 = more chance of heart attack
Attribute Information
> - 1) age
> - 2) sex
> - 3) chest pain type (4 values)
> - 4) resting blood pressure
> - 5) serum cholestoral in mg/dl
> - 6) fasting blood sugar > 120 mg/dl
> - 7) resting electrocardiographic results (values 0,1,2)
> - 8) maximum heart rate achieved
> - 9) exercise induced angina
> - 10) oldpeak = ST depression induced by exercise relative to rest
> - 11) the slope of the peak exercise ST segment
> - 12) number of major vessels (0-3) colored by flourosopy
> - 13) thal: 0 = normal; 1 = fixed defect; 2 = reversable defect
> - 14) target: 0= less chance of heart attack 1= more chance of heart attack
Reference: [Kaggle](https://www.kaggle.com/nareshbhat/health-care-data-set-on-heart-attack-possibility)
```
import pandas as pd
heart = pd.read_csv("heart.csv")
print(heart[1:5])
heart.info()
```
<h4 align="right" style="direction:rtl; font-family:tahoma; font-weight:300">
فراخوانی داده ها و انتقال آن ها در ماتریس های مشخص
</h4>
```
heart_age = [heart['age'][i] for i in range(303)]
heart_sex = [heart['sex_n'][i] for i in range(303)]
heart_chol = [heart['chol'][i] for i in range(303)]
print("\t Age of Person: \n{}\n\n\t Sex of Person:\n{}\n\n\t Serum Cholestoral:\n{}".format(heart_age, heart_sex, heart_chol))
import random as rd
r = [rd.randint(0, 302) for i in range(3)]
print(r,"\n")
[print("Age Person {} is {}, gender is {} and cholestoral is {} \n".format(r[i], heart_age[r[i]], heart_sex[r[i]], heart_chol[r[i]])) for i in range(3)]
```
<h4 align="right" style="direction:rtl; font-family:tahoma; font-weight:300">
داده های گمشده در متغیر های کمی را جایگذاری کنید و در متغیر های جدید ذخیره کنید.
</h4>
<h4 align="right" style="direction:rtl; font-family:tahoma; font-weight:300">
بررسی برای سن افراد
</h4>
```
import matplotlib.pyplot as plt
import numpy as np
bins = np.arange(20, max(heart_age)+20, 2)
a = plt.hist(heart_age, bins = bins, facecolor='g')
plt.xlabel("Age of Person")
plt.ylabel("Count")
plt.title("Histogram of Person Heart Age")
plt.text(20, 25, r'$M-Karimi$')
plt.ylim(0, 38)
plt.grid(True)
plt.show()
import seaborn as sb
sb.distplot(heart_age);
```
<h4 align="right" style="direction:rtl; font-family:tahoma; font-weight:300">
بررسی برای میزان کلسترول
</h4>
```
import matplotlib.pyplot as plt
import numpy as np
import seaborn as sb
bins = np.arange(0, max(heart_chol)+1, 10)
a = plt.hist(heart_chol, bins = bins, facecolor='g')
plt.xlabel("Cholestoral of Person")
plt.ylabel("Count")
plt.title("Histogram of Person Cholestoral")
plt.text(20, 25, r'$M-Karimi$')
plt.ylim(0, 33)
plt.grid(True)
plt.show()
sb.distplot(heart_chol);
```
<h4 align="right" style="direction:rtl; font-family:tahoma; font-weight:300">
پراکندی داده ها یا Scatterplots
</h4>
```
plt.scatter(data = heart, x = 'age', y = 'chol');
plt.xlabel("Age")
plt.ylabel("Cholestoral");
sb.regplot(data = heart, x = 'age', y = 'chol');
```
<h4 align="right" style="direction:rtl; font-family:tahoma; font-weight:300">
ایجاد دامنه پراکندگی و جایگزینی آن با میانه کلسترول
</h4>
```
import statistics as stc
h_median = stc.median(heart_chol)
print("Median is: {}".format(h_median))
h_Qua = stc.quantiles(heart_chol)
h_IQR = h_Qua[2] - h_Qua[0]
print("Quantile: {}".format(h_Qua))
h_DisIQR = h_IQR * 1.5
print("Distance of IQR: {}".format(h_DisIQR))
h_IQLow = h_Qua[0] - h_DisIQR
h_IQHig = h_Qua[2] + h_DisIQR
print("Low is [* {} *] and High is [* {} *]".format(h_IQLow, h_IQHig))
```
<h4 align="right" style="direction:rtl; font-family:tahoma; font-weight:300">
حذف نوییز ها از داده های کلسترول و جایگزینی آن ها با میانه
</h4>
```
import statistics as stc
h_median = stc.median(heart_chol)
heart_newChol = [0]*303
print(heart_newChol,"\n")
for i in range(len(heart_chol)):
if heart_chol[i] > h_IQLow and heart_chol[i] < h_IQHig:
heart_newChol[i] = heart_chol[i]
else:
heart_newChol[i] = h_median
print(heart_newChol)
import seaborn as sb
print(heart_newChol)
sb.distplot(heart_newChol);
sb.regplot(data = heart, x = 'age', y = 'chol_n');
```
<h4 align="right" style="direction:rtl; font-family:tahoma; font-weight:300">
محاسبه رگرسیون بعد از تمیز کاری
</h4>
$$ y = w_{1}x_{1} + w_{2}x_{2} + w_{3}x_{3} + b$$
$$ \left\{\begin{matrix}
x_{1} = age\\
x_{2} = cholestoral\\
x_{3} = fasting - blood - sugar
\end{matrix}\right.
$$
$$
y = target
$$
```
import numpy as np
from sklearn.linear_model import LinearRegression
x = []
for i in range(len(heart_newChol)):
x_m = []
x_m.append(heart['age'][i])
x_m.append(heart['chol_n'][i])
x_m.append(heart['fbs'][i])
x.append(x_m)
y = []
for j in range(len(heart_newChol)):
y.append(heart['target'][j])
x, y = np.array(x), np.array(y)
print("\n\t Way 1: X = \n\n{}\n\n\t Way 2: Y = \n\n{}".format(x, y))
print(type(x), np.shape(x))
```
<h4 align="right" style="direction:rtl; font-family:tahoma; font-weight:300">
حل مدل با رگرسیون
</h4>
```
model = LinearRegression().fit(x, y)
```
<h4 align="right" style="direction:rtl; font-family:tahoma; font-weight:300">
نمایش خروجی
</h4>
```
r_sq = model.score(x, y)
intercept = model.intercept_
slop = model.coef_
print(" Coefficient of Determination: {} \n Intercept:\t\t\t{} \n Slop:\t\t\t\t{}".format(r_sq, intercept, slop))
```
<h4 align="right" style="direction:rtl; font-family:tahoma; font-weight:300">
پیش بینی
</h4>
```
a = ['Age', 'Cholesteral', 'Fasting Blood Sugar']
x_new = [int(input("Input {}:\n".format(a[i]))) for i in range(3)]
# x_n = np.array(x_new)
# y_new = model.predict(x_new)
# print(y_new)
y_n = [x_new[j]*slop[j] for j in range(3)]
y_pred = (sum(y_n)*r_sq) + intercept
print("Prediction: {}".format(y_pred))
```
<h4 align="right" style="direction:rtl; font-family:tahoma; font-weight:300">
محاسبه آزمون کای دو
</h4>
<h4 align="right" style="direction:rtl; font-family:tahoma; font-weight:300">
آزمون برای میزان کلسترول
</h4>
```
print(heart['chol_n'])
Mn_h = min(heart['chol_n'])
Mx_h = max(heart['chol_n'])
print("Min: ",Mn_h, "| Max: ", Mx_h)
Domain = (Mx_h - Mn_h) / 5
print("Range: ", Domain)
D_ch = [0] * 6
D_ch[0] = Mn_h
for i in range(1, 6):
D_ch[i] += D_ch[i-1] + Domain
print(D_ch)
```
|
github_jupyter
|
import pandas as pd
heart = pd.read_csv("heart.csv")
print(heart[1:5])
heart.info()
heart_age = [heart['age'][i] for i in range(303)]
heart_sex = [heart['sex_n'][i] for i in range(303)]
heart_chol = [heart['chol'][i] for i in range(303)]
print("\t Age of Person: \n{}\n\n\t Sex of Person:\n{}\n\n\t Serum Cholestoral:\n{}".format(heart_age, heart_sex, heart_chol))
import random as rd
r = [rd.randint(0, 302) for i in range(3)]
print(r,"\n")
[print("Age Person {} is {}, gender is {} and cholestoral is {} \n".format(r[i], heart_age[r[i]], heart_sex[r[i]], heart_chol[r[i]])) for i in range(3)]
import matplotlib.pyplot as plt
import numpy as np
bins = np.arange(20, max(heart_age)+20, 2)
a = plt.hist(heart_age, bins = bins, facecolor='g')
plt.xlabel("Age of Person")
plt.ylabel("Count")
plt.title("Histogram of Person Heart Age")
plt.text(20, 25, r'$M-Karimi$')
plt.ylim(0, 38)
plt.grid(True)
plt.show()
import seaborn as sb
sb.distplot(heart_age);
import matplotlib.pyplot as plt
import numpy as np
import seaborn as sb
bins = np.arange(0, max(heart_chol)+1, 10)
a = plt.hist(heart_chol, bins = bins, facecolor='g')
plt.xlabel("Cholestoral of Person")
plt.ylabel("Count")
plt.title("Histogram of Person Cholestoral")
plt.text(20, 25, r'$M-Karimi$')
plt.ylim(0, 33)
plt.grid(True)
plt.show()
sb.distplot(heart_chol);
plt.scatter(data = heart, x = 'age', y = 'chol');
plt.xlabel("Age")
plt.ylabel("Cholestoral");
sb.regplot(data = heart, x = 'age', y = 'chol');
import statistics as stc
h_median = stc.median(heart_chol)
print("Median is: {}".format(h_median))
h_Qua = stc.quantiles(heart_chol)
h_IQR = h_Qua[2] - h_Qua[0]
print("Quantile: {}".format(h_Qua))
h_DisIQR = h_IQR * 1.5
print("Distance of IQR: {}".format(h_DisIQR))
h_IQLow = h_Qua[0] - h_DisIQR
h_IQHig = h_Qua[2] + h_DisIQR
print("Low is [* {} *] and High is [* {} *]".format(h_IQLow, h_IQHig))
import statistics as stc
h_median = stc.median(heart_chol)
heart_newChol = [0]*303
print(heart_newChol,"\n")
for i in range(len(heart_chol)):
if heart_chol[i] > h_IQLow and heart_chol[i] < h_IQHig:
heart_newChol[i] = heart_chol[i]
else:
heart_newChol[i] = h_median
print(heart_newChol)
import seaborn as sb
print(heart_newChol)
sb.distplot(heart_newChol);
sb.regplot(data = heart, x = 'age', y = 'chol_n');
import numpy as np
from sklearn.linear_model import LinearRegression
x = []
for i in range(len(heart_newChol)):
x_m = []
x_m.append(heart['age'][i])
x_m.append(heart['chol_n'][i])
x_m.append(heart['fbs'][i])
x.append(x_m)
y = []
for j in range(len(heart_newChol)):
y.append(heart['target'][j])
x, y = np.array(x), np.array(y)
print("\n\t Way 1: X = \n\n{}\n\n\t Way 2: Y = \n\n{}".format(x, y))
print(type(x), np.shape(x))
model = LinearRegression().fit(x, y)
r_sq = model.score(x, y)
intercept = model.intercept_
slop = model.coef_
print(" Coefficient of Determination: {} \n Intercept:\t\t\t{} \n Slop:\t\t\t\t{}".format(r_sq, intercept, slop))
a = ['Age', 'Cholesteral', 'Fasting Blood Sugar']
x_new = [int(input("Input {}:\n".format(a[i]))) for i in range(3)]
# x_n = np.array(x_new)
# y_new = model.predict(x_new)
# print(y_new)
y_n = [x_new[j]*slop[j] for j in range(3)]
y_pred = (sum(y_n)*r_sq) + intercept
print("Prediction: {}".format(y_pred))
print(heart['chol_n'])
Mn_h = min(heart['chol_n'])
Mx_h = max(heart['chol_n'])
print("Min: ",Mn_h, "| Max: ", Mx_h)
Domain = (Mx_h - Mn_h) / 5
print("Range: ", Domain)
D_ch = [0] * 6
D_ch[0] = Mn_h
for i in range(1, 6):
D_ch[i] += D_ch[i-1] + Domain
print(D_ch)
| 0.096909 | 0.938688 |
```
import pickle
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
import cv2
cv2.__version__
import imageio
import PIL
imageio.__version__
PIL.__version__
import scipy.ndimage
imageio.plugins.freeimage.download()
```
### Investigate loading of tower mask data
```
#load dictionary of masks (pixel locations of correctly identified towers)
with open("masks_dict.pkl", "rb") as md:
final_mask_dict = pickle.load(md)
len(final_mask_dict)
final_mask_dict.keys()
final_mask_dict['frame18135_splash.jpg'].shape
final_mask_dict['frame12048_splash.jpg'].shape
final_mask_dict['frame12048_splash.jpg'][1,1,:]
for img_file in final_mask_dict.keys():
print(final_mask_dict[img_file].shape)
unique, counts = np.unique(final_mask_dict['frame12048_splash.jpg'], return_counts=True)
print(np.asarray((unique,counts)).T)
unique, counts = np.unique(final_mask_dict['frame12048_splash.jpg'][:,:,0], return_counts=True)
print(np.asarray((unique,counts)).T)
unique, counts = np.unique(final_mask_dict['frame12048_splash.jpg'][:,:,1], return_counts=True)
print(np.asarray((unique,counts)).T)
plt.imshow(final_mask_dict['frame12048_splash.jpg'][:,:,0])
plt.imshow(final_mask_dict['frame12048_splash.jpg'][:,:,1])
#remove second (incorrect) splash from frame12048
temp_dict = {}
temp_dict['frame12048_splash.jpg'] = final_mask_dict['frame12048_splash.jpg']
temp_dict['frame12048_splash.jpg'] = np.delete(temp_dict['frame12048_splash.jpg'],1,2)
temp_dict['frame12048_splash.jpg'].shape
temp_dict.keys()
plt.imshow(temp_dict['frame12048_splash.jpg'][:,:,0])
#identify locations (indices) of pixels where splash is located
np.argwhere(temp_dict['frame12048_splash.jpg'][:,:,0]==True)
splash_arr = np.argwhere(temp_dict['frame12048_splash.jpg'][:,:,0]==True)
```
### Investigate how to work with depth prediction arrays
```
#load depth prediction array for one image
with open("../../../../FCRN-DepthPrediction/tensorflow/pred_array.pkl", "rb") as pa:
pred_array = pickle.load(pa)
pred_array.shape
pred_array[:,1,1,:]
pred_array[:,100,200,:]
pred_array_sqz = pred_array[0,:,:,:]
pred_array_sqz.shape
pred_array_sqz[100,500]
pred_arr_sqz = pred_array[0,:,:,0]
pred_arr_sqz.shape
```
#### Save array as EXR file and load it back from EXR to understand effect on array values
```
imageio.imwrite('./pred_arr_sqz_float.exr',pred_arr_sqz)
image_in = imageio.imread('./pred_arr_sqz_float.exr')
type(image_in)
assert image_in.dtype == np.float32
image_in.shape
image_in[100,500]
pred_arr_sqz[100,500]
np.argwhere(pred_arr_sqz==1.0653863)
np.argwhere(pred_arr_sqz==1.0654297)
plt.imshow(pred_arr_sqz)
plt.imshow(image_in)
```
**CONCLUSION:** Saving and loading array from EXR modifies original array values
#### Use scipy.ndimage.interpolation.zoom to enlarge array without first saving it and loading it from EXR file
```
im2 = scipy.ndimage.interpolation.zoom(pred_arr_sqz, 2.)
im2.shape
im2[100,100]
plt.imshow(im2)
im3 = scipy.ndimage.interpolation.zoom(pred_array_sqz, 2.)
im3.shape
im3[100,100,:]
plt.imshow(im3[:,:,0])
```
#### Resize and plot array created from reading in of EXR file (using OpenCV)
```
r_in,c_in = image_in.shape
res_in = cv2.resize(image_in, (2*c_in, 2*r_in), interpolation = cv2.INTER_CUBIC)
res_in.shape
plt.imshow(res_in)
res_in[100,100]
```
**CONCLUSION:** OpenCV can resize this kind of array (created from loading EXR file) while still keeping float values, but the process of saving array to EXR file and loading it from EXR modifies values.
#### Save prediction array to EXR and load it using OpenCV, then resize it using OpenCV
```
cv2.imwrite('pred_array_sqz.exr',pred_array_sqz)
img = cv2.imread("pred_array_sqz.exr")
r,c = img.shape[:2]
r, c
img[100,400,:]
res = cv2.resize(img, (2*c, 2*r), interpolation = cv2.INTER_CUBIC)
type(res)
res.shape
plt.imshow(res)
res[100,400,:]
unique, counts = np.unique(res[:,:,0], return_counts=True)
print(np.asarray((unique,counts)).T)
```
**CONCLUSION**: Using OpenCV directly on a float array does not work as OpenCV converts values from EXR file to integers
#### Enlarge array using OpenCV, but without first saving it to EXR file
```
r, c = pred_arr_sqz.shape
r, c
im4 = cv2.resize(pred_arr_sqz, (c*2, r*2))
im4.shape
im4[100,100]
im5 = cv2.resize(pred_arr_sqz, (1920,1080), interpolation = cv2.INTER_CUBIC)
im5.shape
im5[100,100]
plt.imshow(im5)
```
**CONCLUSION:** Using OpenCV directly to resize a float array works and allows you to specify the exact dimensions of resized image
#### Test identifying pixels of mask on a depth prediction array and taking a trimmed mean of those pixel values
```
splash_arr.shape
tuple(map(tuple,splash_arr))
im5[tuple(splash_arr.T)]
from scipy.stats import trim_mean
trim_mean(im5[tuple(splash_arr.T)],0.1)
arr_for_trim_mean = im5[tuple(splash_arr.T)]
trim_mean(arr_for_trim_mean,0.1)
```
|
github_jupyter
|
import pickle
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
import cv2
cv2.__version__
import imageio
import PIL
imageio.__version__
PIL.__version__
import scipy.ndimage
imageio.plugins.freeimage.download()
#load dictionary of masks (pixel locations of correctly identified towers)
with open("masks_dict.pkl", "rb") as md:
final_mask_dict = pickle.load(md)
len(final_mask_dict)
final_mask_dict.keys()
final_mask_dict['frame18135_splash.jpg'].shape
final_mask_dict['frame12048_splash.jpg'].shape
final_mask_dict['frame12048_splash.jpg'][1,1,:]
for img_file in final_mask_dict.keys():
print(final_mask_dict[img_file].shape)
unique, counts = np.unique(final_mask_dict['frame12048_splash.jpg'], return_counts=True)
print(np.asarray((unique,counts)).T)
unique, counts = np.unique(final_mask_dict['frame12048_splash.jpg'][:,:,0], return_counts=True)
print(np.asarray((unique,counts)).T)
unique, counts = np.unique(final_mask_dict['frame12048_splash.jpg'][:,:,1], return_counts=True)
print(np.asarray((unique,counts)).T)
plt.imshow(final_mask_dict['frame12048_splash.jpg'][:,:,0])
plt.imshow(final_mask_dict['frame12048_splash.jpg'][:,:,1])
#remove second (incorrect) splash from frame12048
temp_dict = {}
temp_dict['frame12048_splash.jpg'] = final_mask_dict['frame12048_splash.jpg']
temp_dict['frame12048_splash.jpg'] = np.delete(temp_dict['frame12048_splash.jpg'],1,2)
temp_dict['frame12048_splash.jpg'].shape
temp_dict.keys()
plt.imshow(temp_dict['frame12048_splash.jpg'][:,:,0])
#identify locations (indices) of pixels where splash is located
np.argwhere(temp_dict['frame12048_splash.jpg'][:,:,0]==True)
splash_arr = np.argwhere(temp_dict['frame12048_splash.jpg'][:,:,0]==True)
#load depth prediction array for one image
with open("../../../../FCRN-DepthPrediction/tensorflow/pred_array.pkl", "rb") as pa:
pred_array = pickle.load(pa)
pred_array.shape
pred_array[:,1,1,:]
pred_array[:,100,200,:]
pred_array_sqz = pred_array[0,:,:,:]
pred_array_sqz.shape
pred_array_sqz[100,500]
pred_arr_sqz = pred_array[0,:,:,0]
pred_arr_sqz.shape
imageio.imwrite('./pred_arr_sqz_float.exr',pred_arr_sqz)
image_in = imageio.imread('./pred_arr_sqz_float.exr')
type(image_in)
assert image_in.dtype == np.float32
image_in.shape
image_in[100,500]
pred_arr_sqz[100,500]
np.argwhere(pred_arr_sqz==1.0653863)
np.argwhere(pred_arr_sqz==1.0654297)
plt.imshow(pred_arr_sqz)
plt.imshow(image_in)
im2 = scipy.ndimage.interpolation.zoom(pred_arr_sqz, 2.)
im2.shape
im2[100,100]
plt.imshow(im2)
im3 = scipy.ndimage.interpolation.zoom(pred_array_sqz, 2.)
im3.shape
im3[100,100,:]
plt.imshow(im3[:,:,0])
r_in,c_in = image_in.shape
res_in = cv2.resize(image_in, (2*c_in, 2*r_in), interpolation = cv2.INTER_CUBIC)
res_in.shape
plt.imshow(res_in)
res_in[100,100]
cv2.imwrite('pred_array_sqz.exr',pred_array_sqz)
img = cv2.imread("pred_array_sqz.exr")
r,c = img.shape[:2]
r, c
img[100,400,:]
res = cv2.resize(img, (2*c, 2*r), interpolation = cv2.INTER_CUBIC)
type(res)
res.shape
plt.imshow(res)
res[100,400,:]
unique, counts = np.unique(res[:,:,0], return_counts=True)
print(np.asarray((unique,counts)).T)
r, c = pred_arr_sqz.shape
r, c
im4 = cv2.resize(pred_arr_sqz, (c*2, r*2))
im4.shape
im4[100,100]
im5 = cv2.resize(pred_arr_sqz, (1920,1080), interpolation = cv2.INTER_CUBIC)
im5.shape
im5[100,100]
plt.imshow(im5)
splash_arr.shape
tuple(map(tuple,splash_arr))
im5[tuple(splash_arr.T)]
from scipy.stats import trim_mean
trim_mean(im5[tuple(splash_arr.T)],0.1)
arr_for_trim_mean = im5[tuple(splash_arr.T)]
trim_mean(arr_for_trim_mean,0.1)
| 0.263315 | 0.742445 |
## 1. Problem definition
Start with explaining the problem and how you plan the address it using machine learning.
## 2. Data description
Explain the source of data, what each record represents, number of records, and detailed descriptions of all variables including at least a one-sentence description and data type. Specify which variables will be used in analysis.
Source of Data: We downloaded this data set from Kaggle, which is a subset of a Cleaveland hospital database.
<br>
Record Represenation: Each record represents individual characteristics of an single patient and if they have heart disease or not.
<br>
Number of Records: 303
<br>
Variable Descriptions:
<br>
$\;\;\;\;\;\;$age: The patient's age (years); dtype=int64
<br>
$\;\;\;\;\;\;$sex: The patient's gender (1=male, 0=female); dtype=int64
<br>
$\;\;\;\;\;\;$cp: The patient's experienced chest pain (Value 0: typical angina, Value 1: atypical angina, Value 2: non-anginal pain, Value 3: asymptomatic); $\;\;\;\;\;\;$dtype=int64
<br>
$\;\;\;\;\;\;$trestbps: The patient's resting blood pressure (mm HG on admission to hospital); dtype=int64
<br>
$\;\;\;\;\;\;$chol: The patient's cholesterol measurement in mg/dl; dtype=int64
<br>
$\;\;\;\;\;\;$fbs: The patient's fasting blood sugar (> 120 mg/dl, 1 = true; 0 = false); dtype=int64
<br>
$\;\;\;\;\;\;$restecg: Resting electrocardiographic measurement (0 = normal, 1 = having ST-T wave abnormality, 2 = showing probable or definite left ventricular $\;\;\;\;\;\;$hypertrophy by Estes' criteria); dtype=int64
<br>
$\;\;\;\;\;\;$thalach: The patient's maximum heart rate achieved; dtype=int64
<br>
$\;\;\;\;\;\;$exang: The patient's exercise induced angina (1=yes, 0=no); dtype=int64
<br>
$\;\;\;\;\;\;$oldpeak: ST depression induced by exercise relative to rest ('ST' relates to positions on the ECG plot); dtype=int64
<br>
$\;\;\;\;\;\;$slope: the slope of the peak exercise ST segment (Value 1: upsloping, Value 2: flat, Value 3: downsloping)); dtype=int64
<br>
$\;\;\;\;\;\;$ca: The number of major vessels (0-3); dtype=int64
<br>
$\;\;\;\;\;\;$thal: The patient's presence of a blood disorder called thalssemia (3 = normal; 6 = fixed defect; 7 = reversable defect); dtype=int64
<br>
$\;\;\;\;\;\;$target: The patient's presence of heart disease (0 = no, 1 = yes); dtype=int64
<br>
We will exclude variables "oldpeak" and "slope".
## 3. Method of analysis
Explain the selected method (classification, regression, or clustering).
<ul><li>Classification: identify the label (i.e., dependent variable) and all predictors.
<li>Regression: identify the label (i.e., dependent variable) and all predictors.
<li>Clustering: explain what kind of clusters you expect to find and how those clusters would help you solve the stated problem.
## 4. Loading data
Load your dataset using a relative path.
```
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
import pandas as pd
from scipy import stats
import seaborn as sns
from sklearn import preprocessing
from sklearn.linear_model import LogisticRegression, LogisticRegressionCV
from sklearn.model_selection import train_test_split
import statsmodels.api as sm
import matplotlib.pylab as plt
from dmba import classificationSummary, gainsChart, liftChart
from dmba.metric import AIC_score
from sklearn.metrics import classification_report
df = pd.read_csv('clean_data.csv')
df = df.drop(df.columns[0],axis=1)
from sklearn.metrics import confusion_matrix, accuracy_score
from sklearn.neighbors import NearestNeighbors, KNeighborsClassifier
from sklearn.tree import DecisionTreeClassifier, DecisionTreeRegressor
from sklearn.ensemble import RandomForestClassifier, GradientBoostingClassifier
from sklearn.model_selection import train_test_split, cross_val_score, GridSearchCV
from sklearn.metrics import roc_curve, roc_auc_score
```
Import libraries and dataset. We dropped first column because it represented IDs.
```
df.head()
```
## 5. Descriptive statistics
Run descriptive statistics. Explain how the output will guide your analysis.
Using this descriptive chart below, we emphasize on taking note of which columns are numeric and which are categorical by looking at the max. We also take note of the basic characteristics of our patients, sex and gender, to base our analysis on.
```
df.describe()
```
We aggregate our data around our target variable to take note of the averages of the medical characteristics between those who have heart disease and those that don't. This gives us a good idea of what medical characterisitics are higher, lower, or most prevalent for those who have heart disease and those without.
```
df.groupby('target').mean()
```
## 6. Missing values and outliers
Explain the steps that you plan to take to handle missing values and any potential outliers. Run code that handles missing values and outliers.
## Missing Values
There aren't any missing values so we do not to perform any method to handle them such as dropping or imputation.
```
df.isna().sum()
```
## Outliers
We will use a formula to find the z-scores of all the columns in the dataset that is less than the threshold of 3 standard deviations from the mean. The values that are greater than 3 will be considered outliers and will be removed.
```
df = df[(np.abs(stats.zscore(df)) < 3).all(axis=1)]
```
## 7. Data visualization
Please see the project description for requirements.
## Numerical Data
```
%matplotlib inline
sns.distplot(df['age'])
```
The above cell creates a histogram of ages.
The peak age is about 60 years which represents the most common age. The data spread is from about 29 years to 76 years. Age is slightly skewed to the left because the mean of all ages is less than the mode.
```
sns.distplot(df['trestbps'])
```
The above cell creates a histogram of resting blood pressures.
The peak resting blood pressure is about 120mm Hg 137mm Hg. The data spread is from about 94mm Hg to 180mm Hg. Resting blood pressure is slightly to the right because the mean of all resting blood pressures is higher than the mode.
```
sns.distplot(df['chol'])
```
The above cell creates a histogram of cholesterol levels.
The peak cholesterol is about 250mg/dl. The data spread is from about 126mg/dl to 394mg/dl.
```
sns.distplot(df['thalach'])
```
The above cell creates a histogram of maximum heart rates.
The peak maximum heart rate achieved is about 160 beats per minute. The data spread is from about 96 beats per minute to 202 beats per minute. Maximum heart rate achieved is skewed to the left because the mean of maximum heart rates is less than the mode.
```
sns.distplot(df['ca'])
```
The above cell creates a histogram of number of blood vessels.
The peak number of major vessels is 0. The data is spread from 0 vessels to 4 vessels.
## Categorical Variables
```
sns.countplot(df['sex'])
```
The above cell creates a bar chart of sex.
There are 199 males and 90 females.
```
sns.countplot(df['cp'])
```
The above cell creates a bar chart of type of chest pain.
137 participants had typical angina, 48 had atypical angina, 81 had non-anginal pain, 23 were asymptomatic. (cp = type of pain)
```
sns.countplot(df['fbs'])
```
The above cell creates a bar chart visualizing how many people had under and over 120mg/dl fasting blood sugar.
248 participants had under 120mg/dl fasting blood sugar and 41 participants had over 120mg/dl fasting blood sugar.
```
sns.countplot(df['exang'])
```
The above cell creates a bar chart of who had exercise induced angina and who did not.
194 participants did not have exercise induced angina and 95 participants did have exercise induced angina.
```
sns.countplot(df['thal'])
```
The above cell creates a bar chart that shows who had normal thalassemia, fixed defect thalassemia, or reversable defect thalassemia.
18 participants had normal thalassemia, 161 had fixed defect thalassemia, and 110 had reversable defect thalassemia.
```
sns.countplot(df['target'])
```
The above cell creates a bar chart that shows who had heart disease and who did not.
131 participants did not have heart disease and 158 participants did have heart disease.
## Aggregation Charts
Here we do create a cross tab visualization showing the frequency of heart disease by sex (male and female). Here we see that even though our data is mostly men, women have a higher proportion of heart disease.
```
fig, ax = plt.subplots(figsize=(10, 6))
pd.crosstab(df.sex, df.target).plot(kind="bar",
color=["pink", 'navy'],
figsize=(10, 6),
ax=ax);
ax.set(xlabel="Sex (Female-0 , Male-1)",
ylabel="Heart Disease Frequeny",
title="Heart disease frequency for sex");
plt.xticks(rotation=0);
ax.legend(['Negative','Positive'],title ="Target");
```
Here we do a scatterplot to see the spread of patients with heart disease and don't have heart disease with the max heart rate that they acheived. We see that as patients get older they seem to be less prone to heart disease and have a lower max heart rate than younger patients.
```
fig, ax = plt.subplots(figsize=(10, 6))
scatter = ax.scatter(x=df.age,
y= df.thalach,
c=df.target,
cmap='bwr');
ax.set(xlabel="Age",ylabel="Max Heaer Rate Achieved",title="Heart Disease in function of Age and Max_Heart_Rate ")
ax.legend(*scatter.legend_elements(),title="Target");
plt.xticks(rotation=0);
```
Here we do another cross tab showing the heart disease frequency for each type of chest pain.
```
fig, ax = plt.subplots(figsize=(10, 6))
pd.crosstab(df.cp,df.target,).plot.bar(color=["salmon","lightblue"],ax=ax)
ax.set(xlabel="Chest Pain type",
ylabel="Heart Disease Frequeny",
title="Heart Disease frequency per chest pain type");
plt.xticks(rotation=0);
ax.legend(['Negative','Positive'],title ="Heart Disease");
```
## 8. Correlation analysis
Generate a correlation matrix and interpret it. Is multicollinearity an issue?
```
corrMatrix = df.corr()
plt.figure(figsize=(15,20))
sns.heatmap(corrMatrix, annot=True, vmin=-1.0, vmax=1.0)
plt.show()
```
Collinearity is a linear association between two predictors. Multicollinearity is a situation where two or more predictors are highly linearly related. In general, an absolute correlation coefficient of >0.7 among two or more predictors indicates the presence of multicollinearity.
For our data, we don't see any significantly high instances of collinearity or mutlicollinearity that would hurt out model by increasing the variance of our coefficient estiates.
## 9. Data preprocessing
In this step you conduct preprocessing. Different algorithms require different types of preprocessing so you may need to replicate this step multiple times for different models.
### 9.1. Dummies
Explain why or why not you need to create dummies. Create dummies below if needed.
We will create dummies for our variables that are categorical. We will rename the values for these variables to make the dummy variables easier to understand and interpret. We will also drop the first dummy varible to reduce the amount of variables at the end.
```
df['sex'][df['sex'] == 0] = 'female'
df['sex'][df['sex'] == 1] = 'male'
df['cp'][df['cp'] == 0] = 'typical angina'
df['cp'][df['cp'] == 1] = 'atypical angina'
df['cp'][df['cp'] == 2] = 'non-anginal pain'
df['cp'][df['cp'] == 3] = 'asymptomatic'
df['fbs'][df['fbs'] == 0] = 'lower than 120mg/ml'
df['fbs'][df['fbs'] == 1] = 'greater than 120mg/ml'
df['exang'][df['exang'] == 0] = 'no'
df['exang'][df['exang'] == 1] = 'yes'
df['thal'][df['thal'] == 1] = 'normal'
df['thal'][df['thal'] == 2] = 'fixed defect'
df['thal'][df['thal'] == 3] = 'reversable defect'
df['sex'] = df['sex'].astype('category')
df['cp'] =df['cp'].astype('category')
df['fbs'] = df['fbs'].astype('category')
df['exang'] = df['exang'].astype('category')
df['thal'] = df['thal'].astype('category')
df = pd.get_dummies(df, drop_first=False)
```
Here we will use all the columns in our dataset as our predictors except our target variable and our target variable as our outcome variable.
```
predictors = list(df.columns)
predictors.remove('target')
outcome = 'target'
```
Let's take a look at our data types.
```
df.info()
```
### 9.2. Normalization
Explain why or why not you need to normalize the data. Normalize it below if needed.
Here we will lay out how we met different preprocessing requirements for each method. We will use two dataframes for further analysis. One dataframe that is normalized with our dummy variables and one that is not normalized with our dummy variables. We need to normalize for logistic regression and KNN. We need to normalize for logistic regression because our numerical columns are on different scales. For example, age is typically from 0-100 years and cholesterol is typically from 100-400 mg/dl. We normalized for KNN because KNN does not make assumptions about the distribution of our data.
```
norm_df = df.copy()
```
Creating a copy of our dataset.
```
cols_to_norm = ['age','trestbps', 'chol', 'thalach', 'ca']
norm_df[cols_to_norm] = norm_df[cols_to_norm].apply(lambda x: (x - x.min()) / (x.max() - x.min()))
```
Normalize numerical columns by applying a lamda function.
```
norm_df.head()
```
## 10. Modeling
Please refer to project description for the requirements.
## Logistic Regression
```
X = norm_df[predictors]
y = norm_df[outcome]
train_X, valid_X, train_y, valid_y = train_test_split(X, y, test_size=0.4, random_state=1)
logit_reg = LogisticRegression(penalty="l2", C=1e42, solver='liblinear')
logit_reg.fit(train_X, train_y)
LR_score = logit_reg.score(train_X, train_y)
ylr = logit_reg.predict(train_X)
pd.set_option('display.width', 95)
pd.set_option('precision',3)
pd.set_option('max_columns', 33)
print('intercept ', logit_reg.intercept_[0])
print(pd.DataFrame({'coeff': logit_reg.coef_[0]}, index=X.columns).transpose())
print()
print('AIC', AIC_score(valid_y, logit_reg.predict(valid_X), df=len(train_X.columns) + 1))
pd.reset_option('display.width')
pd.reset_option('precision')
pd.reset_option('max_columns')
```
Assigned predictors to X and outcome to y.
Separate into train and test partitions.
Run logistic regression.
Format how the output will look and calculate intercept.
Format how output will look and calculate coefficients and AIC score.
```
classificationSummary(train_y, logit_reg.predict(train_X))
classificationSummary(valid_y, logit_reg.predict(valid_X))
```
Run confusion matrix on training and testing sets.
```
print (classification_report(train_y, ylr))
```
Run classification report to find precision and F-1 score.
## Random Forest
```
X = df.drop(columns=['target'])
y = df['target']
```
Assign X to all columns but target. Assign y to target.
```
df.head()
train_X, valid_X, train_y, valid_y = train_test_split(X, y, test_size=0.4, random_state=1)
rf = RandomForestClassifier(n_estimators=500, random_state=1)
rf.fit(train_X, train_y)
LR_score = rf.score(train_X, train_y)
ylr = rf.predict(train_X)
```
Split data into train and test partitions.
Run Random Forest.
```
importances = rf.feature_importances_
std = np.std([tree.feature_importances_ for tree in rf.estimators_], axis=0)
df1 = pd.DataFrame({'feature': train_X.columns, 'importance': importances, 'std': std})
df1 = df1.sort_values('importance')
print(df1)
ax = df1.plot(kind='barh', xerr='std', x='feature', legend=False)
ax.set_ylabel('')
plt.tight_layout()
plt.show()
```
Define importances from results from Random Forest.
Define standard deviation from results from Random Forest.
Create new data frame from results and order by importance.
Show dataframe.
Set Axes, legend, and how importances and std are formatted on the chart.
Show chart.
```
classificationSummary(valid_y, rf.predict(valid_X))
```
Run confusion matrix.
```
print (classification_report(train_y, ylr))
```
Run classification report to see precision and F1-score.
## KNN
Split our data into training and validation sets.
```
trainData, validData = train_test_split(norm_df, test_size=0.4, random_state=26)
print(trainData.shape, validData.shape)
```
Further split our data
```
train_X = trainData[predictors]
train_y = trainData[outcome]
valid_X = validData[predictors]
valid_y = validData[outcome]
```
Train a classifier for different values of k (1-15), test performance on validation set, then convert results to a pandas data frame in order to see accuracy for each value of k.
```
results = []
for k in range(1, 16):
knn = KNeighborsClassifier(n_neighbors=k).fit(train_X, train_y)
results.append({
'k': k,
'accuracy': accuracy_score(valid_y, knn.predict(valid_X))
})
results = pd.DataFrame(results)
print(results)
```
We see that the first k with the highest accuracy is k=10, so we will fit a KNN model with k=10. We further visualize what our error rate is with other k values to best determine what is the most ideal value for k.
```
error = []
# Calculating error for K values between 1 and 40
for i in range(1, 20):
knn = KNeighborsClassifier(n_neighbors=i)
knn.fit(train_X, train_y)
pred_i = knn.predict(valid_X)
error.append(np.mean(pred_i != valid_y))
%matplotlib inline
plt.figure(figsize=(8, 6))
plt.plot(range(1, 20), error, color='red', linestyle='dashed', marker='o',
markerfacecolor='blue', markersize=10)
plt.title('Error Rate K Value')
plt.xlabel('K Value')
plt.ylabel('Mean Error')
```
Fit KNN model to k=10
```
knn = KNeighborsClassifier(n_neighbors=10)
knn.fit(train_X,train_y)
knn_prediction = knn.predict(valid_X)
```
Run classification report to find precision and F-1 score.
```
print(classification_report(knn_prediction,valid_y))
```
## ROC Curve
Here we assign our predictors to X and our target variable to y for splitting.
```
X = norm_df[predictors]
y = norm_df[outcome]
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=.4, random_state=1)
```
We define a list of our methods that we will use in a for loop to fit to our data. We will then append the results of our methods to a dataframe to plot. We will plot all our methods on an ROC curve to determine what the best model is.
```
classifiers = [LogisticRegression(),
KNeighborsClassifier(),
RandomForestClassifier()]
# Define a result table as a DataFrame
result_table = pd.DataFrame(columns=['classifiers', 'fpr','tpr','auc'])
# Train the models and record the results
for cls in classifiers:
model = cls.fit(X_train, y_train)
yproba = model.predict_proba(X_test)[::,1]
fpr, tpr, _ = roc_curve(y_test, yproba)
auc = roc_auc_score(y_test, yproba)
result_table = result_table.append({'classifiers':cls.__class__.__name__,
'fpr':fpr,
'tpr':tpr,
'auc':auc}, ignore_index=True)
# Set name of the classifiers as index labels
result_table.set_index('classifiers', inplace=True)
fig = plt.figure(figsize=(8,6))
for i in result_table.index:
plt.plot(result_table.loc[i]['fpr'],
result_table.loc[i]['tpr'],
label="{}, AUC={:.3f}".format(i, result_table.loc[i]['auc']))
plt.plot([0,1], [0,1], color='orange', linestyle='--')
plt.xticks(np.arange(0.0, 1.1, step=0.1))
plt.xlabel("False Positive Rate", fontsize=15)
plt.yticks(np.arange(0.0, 1.1, step=0.1))
plt.ylabel("True Positive Rate", fontsize=15)
plt.title('ROC Curve Analysis', fontweight='bold', fontsize=15)
plt.legend(prop={'size':13}, loc='lower right')
plt.show()
```
By acknowledging the top left most curve, we see that our best model is Logstic Regression. Logistic Regression also has the highest AUC curve.
|
github_jupyter
|
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
import pandas as pd
from scipy import stats
import seaborn as sns
from sklearn import preprocessing
from sklearn.linear_model import LogisticRegression, LogisticRegressionCV
from sklearn.model_selection import train_test_split
import statsmodels.api as sm
import matplotlib.pylab as plt
from dmba import classificationSummary, gainsChart, liftChart
from dmba.metric import AIC_score
from sklearn.metrics import classification_report
df = pd.read_csv('clean_data.csv')
df = df.drop(df.columns[0],axis=1)
from sklearn.metrics import confusion_matrix, accuracy_score
from sklearn.neighbors import NearestNeighbors, KNeighborsClassifier
from sklearn.tree import DecisionTreeClassifier, DecisionTreeRegressor
from sklearn.ensemble import RandomForestClassifier, GradientBoostingClassifier
from sklearn.model_selection import train_test_split, cross_val_score, GridSearchCV
from sklearn.metrics import roc_curve, roc_auc_score
df.head()
df.describe()
df.groupby('target').mean()
df.isna().sum()
df = df[(np.abs(stats.zscore(df)) < 3).all(axis=1)]
%matplotlib inline
sns.distplot(df['age'])
sns.distplot(df['trestbps'])
sns.distplot(df['chol'])
sns.distplot(df['thalach'])
sns.distplot(df['ca'])
sns.countplot(df['sex'])
sns.countplot(df['cp'])
sns.countplot(df['fbs'])
sns.countplot(df['exang'])
sns.countplot(df['thal'])
sns.countplot(df['target'])
fig, ax = plt.subplots(figsize=(10, 6))
pd.crosstab(df.sex, df.target).plot(kind="bar",
color=["pink", 'navy'],
figsize=(10, 6),
ax=ax);
ax.set(xlabel="Sex (Female-0 , Male-1)",
ylabel="Heart Disease Frequeny",
title="Heart disease frequency for sex");
plt.xticks(rotation=0);
ax.legend(['Negative','Positive'],title ="Target");
fig, ax = plt.subplots(figsize=(10, 6))
scatter = ax.scatter(x=df.age,
y= df.thalach,
c=df.target,
cmap='bwr');
ax.set(xlabel="Age",ylabel="Max Heaer Rate Achieved",title="Heart Disease in function of Age and Max_Heart_Rate ")
ax.legend(*scatter.legend_elements(),title="Target");
plt.xticks(rotation=0);
fig, ax = plt.subplots(figsize=(10, 6))
pd.crosstab(df.cp,df.target,).plot.bar(color=["salmon","lightblue"],ax=ax)
ax.set(xlabel="Chest Pain type",
ylabel="Heart Disease Frequeny",
title="Heart Disease frequency per chest pain type");
plt.xticks(rotation=0);
ax.legend(['Negative','Positive'],title ="Heart Disease");
corrMatrix = df.corr()
plt.figure(figsize=(15,20))
sns.heatmap(corrMatrix, annot=True, vmin=-1.0, vmax=1.0)
plt.show()
df['sex'][df['sex'] == 0] = 'female'
df['sex'][df['sex'] == 1] = 'male'
df['cp'][df['cp'] == 0] = 'typical angina'
df['cp'][df['cp'] == 1] = 'atypical angina'
df['cp'][df['cp'] == 2] = 'non-anginal pain'
df['cp'][df['cp'] == 3] = 'asymptomatic'
df['fbs'][df['fbs'] == 0] = 'lower than 120mg/ml'
df['fbs'][df['fbs'] == 1] = 'greater than 120mg/ml'
df['exang'][df['exang'] == 0] = 'no'
df['exang'][df['exang'] == 1] = 'yes'
df['thal'][df['thal'] == 1] = 'normal'
df['thal'][df['thal'] == 2] = 'fixed defect'
df['thal'][df['thal'] == 3] = 'reversable defect'
df['sex'] = df['sex'].astype('category')
df['cp'] =df['cp'].astype('category')
df['fbs'] = df['fbs'].astype('category')
df['exang'] = df['exang'].astype('category')
df['thal'] = df['thal'].astype('category')
df = pd.get_dummies(df, drop_first=False)
predictors = list(df.columns)
predictors.remove('target')
outcome = 'target'
df.info()
norm_df = df.copy()
cols_to_norm = ['age','trestbps', 'chol', 'thalach', 'ca']
norm_df[cols_to_norm] = norm_df[cols_to_norm].apply(lambda x: (x - x.min()) / (x.max() - x.min()))
norm_df.head()
X = norm_df[predictors]
y = norm_df[outcome]
train_X, valid_X, train_y, valid_y = train_test_split(X, y, test_size=0.4, random_state=1)
logit_reg = LogisticRegression(penalty="l2", C=1e42, solver='liblinear')
logit_reg.fit(train_X, train_y)
LR_score = logit_reg.score(train_X, train_y)
ylr = logit_reg.predict(train_X)
pd.set_option('display.width', 95)
pd.set_option('precision',3)
pd.set_option('max_columns', 33)
print('intercept ', logit_reg.intercept_[0])
print(pd.DataFrame({'coeff': logit_reg.coef_[0]}, index=X.columns).transpose())
print()
print('AIC', AIC_score(valid_y, logit_reg.predict(valid_X), df=len(train_X.columns) + 1))
pd.reset_option('display.width')
pd.reset_option('precision')
pd.reset_option('max_columns')
classificationSummary(train_y, logit_reg.predict(train_X))
classificationSummary(valid_y, logit_reg.predict(valid_X))
print (classification_report(train_y, ylr))
X = df.drop(columns=['target'])
y = df['target']
df.head()
train_X, valid_X, train_y, valid_y = train_test_split(X, y, test_size=0.4, random_state=1)
rf = RandomForestClassifier(n_estimators=500, random_state=1)
rf.fit(train_X, train_y)
LR_score = rf.score(train_X, train_y)
ylr = rf.predict(train_X)
importances = rf.feature_importances_
std = np.std([tree.feature_importances_ for tree in rf.estimators_], axis=0)
df1 = pd.DataFrame({'feature': train_X.columns, 'importance': importances, 'std': std})
df1 = df1.sort_values('importance')
print(df1)
ax = df1.plot(kind='barh', xerr='std', x='feature', legend=False)
ax.set_ylabel('')
plt.tight_layout()
plt.show()
classificationSummary(valid_y, rf.predict(valid_X))
print (classification_report(train_y, ylr))
trainData, validData = train_test_split(norm_df, test_size=0.4, random_state=26)
print(trainData.shape, validData.shape)
train_X = trainData[predictors]
train_y = trainData[outcome]
valid_X = validData[predictors]
valid_y = validData[outcome]
results = []
for k in range(1, 16):
knn = KNeighborsClassifier(n_neighbors=k).fit(train_X, train_y)
results.append({
'k': k,
'accuracy': accuracy_score(valid_y, knn.predict(valid_X))
})
results = pd.DataFrame(results)
print(results)
error = []
# Calculating error for K values between 1 and 40
for i in range(1, 20):
knn = KNeighborsClassifier(n_neighbors=i)
knn.fit(train_X, train_y)
pred_i = knn.predict(valid_X)
error.append(np.mean(pred_i != valid_y))
%matplotlib inline
plt.figure(figsize=(8, 6))
plt.plot(range(1, 20), error, color='red', linestyle='dashed', marker='o',
markerfacecolor='blue', markersize=10)
plt.title('Error Rate K Value')
plt.xlabel('K Value')
plt.ylabel('Mean Error')
knn = KNeighborsClassifier(n_neighbors=10)
knn.fit(train_X,train_y)
knn_prediction = knn.predict(valid_X)
print(classification_report(knn_prediction,valid_y))
X = norm_df[predictors]
y = norm_df[outcome]
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=.4, random_state=1)
classifiers = [LogisticRegression(),
KNeighborsClassifier(),
RandomForestClassifier()]
# Define a result table as a DataFrame
result_table = pd.DataFrame(columns=['classifiers', 'fpr','tpr','auc'])
# Train the models and record the results
for cls in classifiers:
model = cls.fit(X_train, y_train)
yproba = model.predict_proba(X_test)[::,1]
fpr, tpr, _ = roc_curve(y_test, yproba)
auc = roc_auc_score(y_test, yproba)
result_table = result_table.append({'classifiers':cls.__class__.__name__,
'fpr':fpr,
'tpr':tpr,
'auc':auc}, ignore_index=True)
# Set name of the classifiers as index labels
result_table.set_index('classifiers', inplace=True)
fig = plt.figure(figsize=(8,6))
for i in result_table.index:
plt.plot(result_table.loc[i]['fpr'],
result_table.loc[i]['tpr'],
label="{}, AUC={:.3f}".format(i, result_table.loc[i]['auc']))
plt.plot([0,1], [0,1], color='orange', linestyle='--')
plt.xticks(np.arange(0.0, 1.1, step=0.1))
plt.xlabel("False Positive Rate", fontsize=15)
plt.yticks(np.arange(0.0, 1.1, step=0.1))
plt.ylabel("True Positive Rate", fontsize=15)
plt.title('ROC Curve Analysis', fontweight='bold', fontsize=15)
plt.legend(prop={'size':13}, loc='lower right')
plt.show()
| 0.472197 | 0.989399 |
## Calculate wet bulb temperature
Approaches to calculate wet bulb temperature are discussed in [Knox et al. (2017)](https://journals.ametsoc.org/bams/article/98/9/1897/70218/Two-Simple-and-Accurate-Approximations-for-Wet).
```
import xarray as xr
import numpy as np
from matplotlib import pyplot as plt
from metpy import calc as mpcalc
from metpy.units import units as mpunits
from numba import jit
### Iterative formula originally from NWS website : https://www.weather.gov/epz/wxcalc_dewpoint
### and written in python here: https://github.com/Unidata/MetPy/issues/1006#issuecomment-464409522
def esubx(temp):
e_x = 6.112 * np.exp((17.67 * temp) / (temp + 243.5))
return e_x
@jit(nopython=True)
def wet_bulb_temperature_point(tw_guess,
temp_c,
pressure_mb):
# e = esubx(tw_guess)
e = 6.112 * np.exp((17.67 * tw_guess) / (tw_guess + 243.5))
increment = 10
e_diff = 1
prev_sign = 1
while abs(e_diff) > 0.05:
e_w_guess = 6.112 * np.exp((17.67 * tw_guess) / (tw_guess + 243.5))
e_guess = e_w_guess - pressure_mb * (temp_c - tw_guess) * 0.00066 * (1 + (0.00115 * tw_guess))
e_diff = e - e_guess
if e_diff == 0:
break
else:
if e_diff < 0:
cursign = -1
if cursign != prev_sign:
prev_sign = cursign
increment = increment/10
else:
increment = increment
else:
cursign = 1
if cursign != prev_sign:
prev_sign = cursign
increment = increment/10
else:
increment = increment
if abs(e_diff) <= 0.05:
break
else:
tw_guess = tw_guess + increment * prev_sign
wetbulb = tw_guess
return wetbulb
## Some other functions from here: https://anaconda.org/ahuang11/wet_bulb_temperature/notebook
def stull_wtb(tmp, rlh):
# https://journals.ametsoc.org/doi/full/10.1175/JAMC-D-11-0143.1
term1_stull = tmp * np.arctan(0.151977 * np.sqrt(rlh + 8.313659))
term2_stull = np.arctan(tmp + rlh)
term3_stull = np.arctan(rlh - 1.676331)
term4_stull = 0.00391838 * np.power(rlh, 1.5) * np.arctan(0.023101 * rlh)
term5_stull = 4.686035
return term1_stull + term2_stull - term3_stull + term4_stull - term5_stull
def half_wtb(tmp, dpt):
# https://journals.ametsoc.org/doi/10.1175/BAMS-D-16-0246.1
return 0.5 * (tmp + dpt)
def third_wtb(tmp, dpt):
# https://journals.ametsoc.org/doi/10.1175/BAMS-D-16-0246.1
return 2 / 3 * tmp + 1 / 3 * dpt
@jit(nopython=True)
def wet_bulb_temperature(tw_guess,temp_c,pressure_mb):
ni,nj,nt = tw_guess.shape
twb = np.empty(shape=(ni,nj,nt))
for i in range(ni):
for j in range(nj):
for t in range(nt):
twb[i,j,t] = wet_bulb_temperature_point(tw_guess[i,j,t],temp_c[i,j,t],pressure_mb[i,j,t])
return twb
def mpcalc_wet_bulb_temperature_from_specific_humidity(specific_humidity, temperature, pressure):
qvr = specific_humidity/(1-specific_humidity)
e = mpcalc.vapor_pressure(pressure,qvr*mpunits('kg/kg'))
td = mpcalc.dewpoint(e)
twb = mpcalc.wet_bulb_temperature(pressure,temperature,td)
return twb
# Point to model data
rootdir = '/local/projects/GFDL_LEs/ATM/RCP85/'
suffix = '.rcp85.ens101.1950_1959.nc'
variables = ['sphum_k24','t_ref']
ds = xr.Dataset()
for var in variables:
print(var)
ds[var] = xr.open_dataarray(rootdir+var+suffix).squeeze()
# Load pressure (time variable is slightly off in raw data)
var = 'ps'
print(var)
ps = xr.open_dataarray(rootdir+var+suffix).squeeze()
tmin = ds['sphum_k24']['time'].min('time').values
tmax = ds['sphum_k24']['time'].max('time').values
ds['ps'] = ps.sel(time=slice(tmin,tmax))
# drop the pfull variable
ds = ds.drop('pfull')
# Specify temperature, pressure and specific humidity
ni = ds.sizes['lon']
nj = ds.sizes['lat']
nt = ds.sizes['time']
lons = np.array([55,56])#np.arange(0,ni)
lats = np.array([35,36])#np.arange(0,nj)
times = np.arange(0,100)
temp_c = ds['t_ref'].transpose('lon','lat','time')[lons,lats,times].values-273.15
pressure_mb = ds['ps'].transpose('lon','lat','time')[lons,lats,times].values/100
sphum = ds['sphum_k24'].transpose('lon','lat','time')[lons,lats,times].values
# Take a first guess at a temperature, starting from the dewpoint
mixing_ratio = sphum/(1-sphum)
e = mpcalc.vapor_pressure(pressure_mb*mpunits('mbar'),mixing_ratio*mpunits('kg/kg'))
dewpoint = mpcalc.dewpoint(e).magnitude
relative_humidity = mpcalc.relative_humidity_from_specific_humidity(sphum, temp_c*mpunits('degC'), pressure_mb*mpunits('mbar')).magnitude
# Take a first guess at the wet bulb temperature
# tw_guess = stull_wtb(temp_c,relative_humidity)
tw_guess = third_wtb(temp_c, dewpoint)
twb_metpy = mpcalc.wet_bulb_temperature(pressure_mb.squeeze()*mpunits('mbar'),temp_c.squeeze()*mpunits('degC'), dewpoint.squeeze()*mpunits('degC'))
twb_metpy.shape
twb = wet_bulb_temperature(tw_guess,temp_c,pressure_mb).squeeze()
twb.shape
plt.plot(twb_metpy[0,0,:].magnitude)
plt.plot(twb[0,0,:])
plt.plot(tw_guess[0,0,:])
im = plt.pcolormesh(twb)
plt.colorbar(im)
im = plt.pcolormesh((twb_metpy.magnitude-twb))
plt.colorbar(im)
```
|
github_jupyter
|
import xarray as xr
import numpy as np
from matplotlib import pyplot as plt
from metpy import calc as mpcalc
from metpy.units import units as mpunits
from numba import jit
### Iterative formula originally from NWS website : https://www.weather.gov/epz/wxcalc_dewpoint
### and written in python here: https://github.com/Unidata/MetPy/issues/1006#issuecomment-464409522
def esubx(temp):
e_x = 6.112 * np.exp((17.67 * temp) / (temp + 243.5))
return e_x
@jit(nopython=True)
def wet_bulb_temperature_point(tw_guess,
temp_c,
pressure_mb):
# e = esubx(tw_guess)
e = 6.112 * np.exp((17.67 * tw_guess) / (tw_guess + 243.5))
increment = 10
e_diff = 1
prev_sign = 1
while abs(e_diff) > 0.05:
e_w_guess = 6.112 * np.exp((17.67 * tw_guess) / (tw_guess + 243.5))
e_guess = e_w_guess - pressure_mb * (temp_c - tw_guess) * 0.00066 * (1 + (0.00115 * tw_guess))
e_diff = e - e_guess
if e_diff == 0:
break
else:
if e_diff < 0:
cursign = -1
if cursign != prev_sign:
prev_sign = cursign
increment = increment/10
else:
increment = increment
else:
cursign = 1
if cursign != prev_sign:
prev_sign = cursign
increment = increment/10
else:
increment = increment
if abs(e_diff) <= 0.05:
break
else:
tw_guess = tw_guess + increment * prev_sign
wetbulb = tw_guess
return wetbulb
## Some other functions from here: https://anaconda.org/ahuang11/wet_bulb_temperature/notebook
def stull_wtb(tmp, rlh):
# https://journals.ametsoc.org/doi/full/10.1175/JAMC-D-11-0143.1
term1_stull = tmp * np.arctan(0.151977 * np.sqrt(rlh + 8.313659))
term2_stull = np.arctan(tmp + rlh)
term3_stull = np.arctan(rlh - 1.676331)
term4_stull = 0.00391838 * np.power(rlh, 1.5) * np.arctan(0.023101 * rlh)
term5_stull = 4.686035
return term1_stull + term2_stull - term3_stull + term4_stull - term5_stull
def half_wtb(tmp, dpt):
# https://journals.ametsoc.org/doi/10.1175/BAMS-D-16-0246.1
return 0.5 * (tmp + dpt)
def third_wtb(tmp, dpt):
# https://journals.ametsoc.org/doi/10.1175/BAMS-D-16-0246.1
return 2 / 3 * tmp + 1 / 3 * dpt
@jit(nopython=True)
def wet_bulb_temperature(tw_guess,temp_c,pressure_mb):
ni,nj,nt = tw_guess.shape
twb = np.empty(shape=(ni,nj,nt))
for i in range(ni):
for j in range(nj):
for t in range(nt):
twb[i,j,t] = wet_bulb_temperature_point(tw_guess[i,j,t],temp_c[i,j,t],pressure_mb[i,j,t])
return twb
def mpcalc_wet_bulb_temperature_from_specific_humidity(specific_humidity, temperature, pressure):
qvr = specific_humidity/(1-specific_humidity)
e = mpcalc.vapor_pressure(pressure,qvr*mpunits('kg/kg'))
td = mpcalc.dewpoint(e)
twb = mpcalc.wet_bulb_temperature(pressure,temperature,td)
return twb
# Point to model data
rootdir = '/local/projects/GFDL_LEs/ATM/RCP85/'
suffix = '.rcp85.ens101.1950_1959.nc'
variables = ['sphum_k24','t_ref']
ds = xr.Dataset()
for var in variables:
print(var)
ds[var] = xr.open_dataarray(rootdir+var+suffix).squeeze()
# Load pressure (time variable is slightly off in raw data)
var = 'ps'
print(var)
ps = xr.open_dataarray(rootdir+var+suffix).squeeze()
tmin = ds['sphum_k24']['time'].min('time').values
tmax = ds['sphum_k24']['time'].max('time').values
ds['ps'] = ps.sel(time=slice(tmin,tmax))
# drop the pfull variable
ds = ds.drop('pfull')
# Specify temperature, pressure and specific humidity
ni = ds.sizes['lon']
nj = ds.sizes['lat']
nt = ds.sizes['time']
lons = np.array([55,56])#np.arange(0,ni)
lats = np.array([35,36])#np.arange(0,nj)
times = np.arange(0,100)
temp_c = ds['t_ref'].transpose('lon','lat','time')[lons,lats,times].values-273.15
pressure_mb = ds['ps'].transpose('lon','lat','time')[lons,lats,times].values/100
sphum = ds['sphum_k24'].transpose('lon','lat','time')[lons,lats,times].values
# Take a first guess at a temperature, starting from the dewpoint
mixing_ratio = sphum/(1-sphum)
e = mpcalc.vapor_pressure(pressure_mb*mpunits('mbar'),mixing_ratio*mpunits('kg/kg'))
dewpoint = mpcalc.dewpoint(e).magnitude
relative_humidity = mpcalc.relative_humidity_from_specific_humidity(sphum, temp_c*mpunits('degC'), pressure_mb*mpunits('mbar')).magnitude
# Take a first guess at the wet bulb temperature
# tw_guess = stull_wtb(temp_c,relative_humidity)
tw_guess = third_wtb(temp_c, dewpoint)
twb_metpy = mpcalc.wet_bulb_temperature(pressure_mb.squeeze()*mpunits('mbar'),temp_c.squeeze()*mpunits('degC'), dewpoint.squeeze()*mpunits('degC'))
twb_metpy.shape
twb = wet_bulb_temperature(tw_guess,temp_c,pressure_mb).squeeze()
twb.shape
plt.plot(twb_metpy[0,0,:].magnitude)
plt.plot(twb[0,0,:])
plt.plot(tw_guess[0,0,:])
im = plt.pcolormesh(twb)
plt.colorbar(im)
im = plt.pcolormesh((twb_metpy.magnitude-twb))
plt.colorbar(im)
| 0.497559 | 0.81615 |
# Sequence to Sequence Learning
:label:`sec_seq2seq`
As we have seen in :numref:`sec_machine_translation`,
in machine translation
both the input and output are a variable-length sequence.
To address this type of problem,
we have designed a general encoder-decoder architecture
in :numref:`sec_encoder-decoder`.
In this section,
we will
use two RNNs to design
the encoder and the decoder of
this architecture
and apply it to *sequence to sequence* learning
for machine translation
:cite:`Sutskever.Vinyals.Le.2014,Cho.Van-Merrienboer.Gulcehre.ea.2014`.
Following the design principle
of the encoder-decoder architecture,
the RNN encoder can
take a variable-length sequence as the input and transforms it into a fixed-shape hidden state.
In other words,
information of the input (source) sequence
is *encoded* in the hidden state of the RNN encoder.
To generate the output sequence token by token,
a separate RNN decoder
can predict the next token based on
what tokens have been seen (such as in language modeling) or generated,
together with the encoded information of the input sequence.
:numref:`fig_seq2seq` illustrates
how to use two RNNs
for sequence to sequence learning
in machine translation.

:label:`fig_seq2seq`
In :numref:`fig_seq2seq`,
the special "<eos>" token
marks the end of the sequence.
The model can stop making predictions
once this token is generated.
At the initial time step of the RNN decoder,
there are two special design decisions.
First, the special beginning-of-sequence "<bos>" token is an input.
Second,
the final hidden state of the RNN encoder is used
to initiate the hidden state of the decoder.
In designs such as :cite:`Sutskever.Vinyals.Le.2014`,
this is exactly
how the encoded input sequence information
is fed into the decoder for generating the output (target) sequence.
In some other designs such as :cite:`Cho.Van-Merrienboer.Gulcehre.ea.2014`,
the final hidden state of the encoder
is also fed into the decoder as
part of the inputs
at every time step as shown in :numref:`fig_seq2seq`.
Similar to the training of language models in
:numref:`sec_language_model`,
we can allow the labels to be the original output sequence,
shifted by one token:
"<bos>", "Ils", "regardent", "." $\rightarrow$
"Ils", "regardent", ".", "<eos>".
In the following,
we will explain the design of :numref:`fig_seq2seq`
in greater detail.
We will train this model for machine translation
on the English-French dataset as introduced in
:numref:`sec_machine_translation`.
```
import collections
from d2l import mxnet as d2l
import math
from mxnet import np, npx, init, gluon, autograd
from mxnet.gluon import nn, rnn
npx.set_np()
```
## Encoder
Technically speaking,
the encoder transforms an input sequence of variable length into a fixed-shape *context variable* $\mathbf{c}$, and encodes the input sequence information in this context variable.
As depicted in :numref:`fig_seq2seq`,
we can use an RNN to design the encoder.
Let us consider a sequence example (batch size: 1).
Suppose that
the input sequence is $x_1, \ldots, x_T$, such that $x_t$ is the $t^{\mathrm{th}}$ token in the input text sequence.
At time step $t$, the RNN transforms
the input feature vector $\mathbf{x}_t$ for $x_t$
and the hidden state $\mathbf{h} _{t-1}$ from the previous time step
into the current hidden state $\mathbf{h}_t$.
We can use a function $f$ to express the transformation of the RNN's recurrent layer:
$$\mathbf{h}_t = f(\mathbf{x}_t, \mathbf{h}_{t-1}). $$
In general,
the encoder transforms the hidden states at
all the time steps
into the context variable through a customized function $q$:
$$\mathbf{c} = q(\mathbf{h}_1, \ldots, \mathbf{h}_T).$$
For example, when choosing $q(\mathbf{h}_1, \ldots, \mathbf{h}_T) = \mathbf{h}_T$ such as in :numref:`fig_seq2seq`,
the context variable is just the hidden state $\mathbf{h}_T$
of the input sequence at the final time step.
So far we have used a unidirectional RNN
to design the encoder,
where
a hidden state only depends on
the input subsequence at and before the time step of the hidden state.
We can also construct encoders using bidirectional RNNs. In this case, a hidden state depends on
the subsequence before and after the time step (including the input at the current time step), which encodes the information of the entire sequence.
Now let us implement the RNN encoder.
Note that we use an *embedding layer*
to obtain the feature vector for each token in the input sequence.
The weight
of an embedding layer
is a matrix
whose number of rows equals to the size of the input vocabulary (`vocab_size`)
and number of columns equals to the feature vector's dimension (`embed_size`).
For any input token index $i$,
the embedding layer
fetches the $i^{\mathrm{th}}$ row (starting from 0) of the weight matrix
to return its feature vector.
Besides,
here we choose a multilayer GRU to
implement the encoder.
```
#@save
class Seq2SeqEncoder(d2l.Encoder):
"""The RNN encoder for sequence to sequence learning."""
def __init__(self, vocab_size, embed_size, num_hiddens, num_layers,
dropout=0, **kwargs):
super(Seq2SeqEncoder, self).__init__(**kwargs)
# Embedding layer
self.embedding = nn.Embedding(vocab_size, embed_size)
self.rnn = rnn.GRU(num_hiddens, num_layers, dropout=dropout)
def forward(self, X, *args):
# The output `X` shape: (`batch_size`, `num_steps`, `embed_size`)
X = self.embedding(X)
# In RNN models, the first axis corresponds to time steps
X = X.swapaxes(0, 1)
state = self.rnn.begin_state(batch_size=X.shape[1], ctx=X.ctx)
output, state = self.rnn(X, state)
# `output` shape: (`num_steps`, `batch_size`, `num_hiddens`)
# `state[0]` shape: (`num_layers`, `batch_size`, `num_hiddens`)
return output, state
```
The returned variables of recurrent layers
have been explained in :numref:`sec_rnn-concise`.
Let us still use a concrete example
to illustrate the above encoder implementation.
Below
we instantiate a two-layer GRU encoder
whose number of hidden units is 16.
Given
a minibatch of sequence inputs `X`
(batch size: 4, number of time steps: 7),
the hidden states of the last layer
at all the time steps
(`output` return by the encoder's recurrent layers)
are a tensor
of shape
(number of time steps, batch size, number of hidden units).
```
encoder = Seq2SeqEncoder(vocab_size=10, embed_size=8, num_hiddens=16,
num_layers=2)
encoder.initialize()
X = np.zeros((4, 7))
output, state = encoder(X)
output.shape
```
Since a GRU is employed here,
the shape of the multilayer hidden states
at the final time step
is
(number of hidden layers, batch size, number of hidden units).
If an LSTM is used,
memory cell information will also be contained in `state`.
```
len(state), state[0].shape
```
## Decoder
:label:`sec_seq2seq_decoder`
As we just mentioned,
the context variable $\mathbf{c}$ of the encoder's output encodes the entire input sequence $x_1, \ldots, x_T$. Given the output sequence $y_1, y_2, \ldots, y_{T'}$ from the training dataset,
for each time step $t'$
(the symbol differs from the time step $t$ of input sequences or encoders),
the probability of the decoder output $y_{t'}$
is conditional
on the previous output subsequence
$y_1, \ldots, y_{t'-1}$ and
the context variable $\mathbf{c}$, i.e., $P(y_{t'} \mid y_1, \ldots, y_{t'-1}, \mathbf{c})$.
To model this conditional probability on sequences,
we can use another RNN as the decoder.
At any time step $t^\prime$ on the output sequence,
the RNN takes the output $y_{t^\prime-1}$ from the previous time step
and the context variable $\mathbf{c}$ as its input,
then transforms
them and
the previous hidden state $\mathbf{s}_{t^\prime-1}$
into the
hidden state $\mathbf{s}_{t^\prime}$ at the current time step.
As a result, we can use a function $g$ to express the transformation of the decoder's hidden layer:
$$\mathbf{s}_{t^\prime} = g(y_{t^\prime-1}, \mathbf{c}, \mathbf{s}_{t^\prime-1}).$$
:eqlabel:`eq_seq2seq_s_t`
After obtaining the hidden state of the decoder,
we can use an output layer and the softmax operation to compute the conditional probability distribution
$P(y_{t^\prime} \mid y_1, \ldots, y_{t^\prime-1}, \mathbf{c})$ for the output at time step $t^\prime$.
Following :numref:`fig_seq2seq`,
when implementing the decoder as follows,
we directly use the hidden state at the final time step
of the encoder
to initialize the hidden state of the decoder.
This requires that the RNN encoder and the RNN decoder have the same number of layers and hidden units.
To further incorporate the encoded input sequence information,
the context variable is concatenated
with the decoder input at all the time steps.
To predict the probability distribution of the output token,
a fully-connected layer is used to transform
the hidden state at the final layer of the RNN decoder.
```
class Seq2SeqDecoder(d2l.Decoder):
"""The RNN decoder for sequence to sequence learning."""
def __init__(self, vocab_size, embed_size, num_hiddens, num_layers,
dropout=0, **kwargs):
super(Seq2SeqDecoder, self).__init__(**kwargs)
self.embedding = nn.Embedding(vocab_size, embed_size)
self.rnn = rnn.GRU(num_hiddens, num_layers, dropout=dropout)
self.dense = nn.Dense(vocab_size, flatten=False)
def init_state(self, enc_outputs, *args):
return enc_outputs[1]
def forward(self, X, state):
# The output `X` shape: (`num_steps`, `batch_size`, `embed_size`)
X = self.embedding(X).swapaxes(0, 1)
# `context` shape: (`batch_size`, `num_hiddens`)
context = state[0][-1]
# Broadcast `context` so it has the same `num_steps` as `X`
context = np.broadcast_to(context, (
X.shape[0], context.shape[0], context.shape[1]))
X_and_context = np.concatenate((X, context), 2)
output, state = self.rnn(X_and_context, state)
output = self.dense(output).swapaxes(0, 1)
# `output` shape: (`batch_size`, `num_steps`, `vocab_size`)
# `state[0]` shape: (`num_layers`, `batch_size`, `num_hiddens`)
return output, state
```
To illustrate the implemented decoder,
below we instantiate it with the same hyperparameters from the aforementioned encoder.
As we can see, the output shape of the decoder becomes (batch size, number of time steps, vocabulary size),
where the last dimension of the tensor stores the predicted token distribution.
```
decoder = Seq2SeqDecoder(vocab_size=10, embed_size=8, num_hiddens=16,
num_layers=2)
decoder.initialize()
state = decoder.init_state(encoder(X))
output, state = decoder(X, state)
output.shape, len(state), state[0].shape
```
To summarize,
the layers in the above RNN encoder-decoder model are illustrated in :numref:`fig_seq2seq_details`.

:label:`fig_seq2seq_details`
## Loss Function
At each time step, the decoder
predicts a probability distribution for the output tokens.
Similar to language modeling,
we can apply softmax to obtain the distribution
and calculate the cross-entropy loss for optimization.
Recall :numref:`sec_machine_translation`
that the special padding tokens
are appended to the end of sequences
so sequences of varying lengths
can be efficiently loaded
in minibatches of the same shape.
However,
prediction of padding tokens
should be excluded from loss calculations.
To this end,
we can use the following
`sequence_mask` function
to mask irrelevant entries with zero values
so later
multiplication of any irrelevant prediction
with zero equals to zero.
For example,
if the valid length of two sequences
excluding padding tokens
are one and two, respectively,
the remaining entries after
the first one
and the first two entries are cleared to zeros.
```
X = np.array([[1, 2, 3], [4, 5, 6]])
npx.sequence_mask(X, np.array([1, 2]), True, axis=1)
```
We can also mask all the entries across the last
few axes.
If you like, you may even specify
to replace such entries with a non-zero value.
```
X = np.ones((2, 3, 4))
npx.sequence_mask(X, np.array([1, 2]), True, value=-1, axis=1)
```
Now we can extend the softmax cross-entropy loss
to allow the masking of irrelevant predictions.
Initially,
masks for all the predicted tokens are set to one.
Once the valid length is given,
the mask corresponding to any padding token
will be cleared to zero.
In the end,
the loss for all the tokens
will be multipled by the mask to filter out
irrelevant predictions of padding tokens in the loss.
```
#@save
class MaskedSoftmaxCELoss(gluon.loss.SoftmaxCELoss):
"""The softmax cross-entropy loss with masks."""
# `pred` shape: (`batch_size`, `num_steps`, `vocab_size`)
# `label` shape: (`batch_size`, `num_steps`)
# `valid_len` shape: (`batch_size`,)
def forward(self, pred, label, valid_len):
# `weights` shape: (`batch_size`, `num_steps`, 1)
weights = np.expand_dims(np.ones_like(label), axis=-1)
weights = npx.sequence_mask(weights, valid_len, True, axis=1)
return super(MaskedSoftmaxCELoss, self).forward(pred, label, weights)
```
For a sanity check, we can create three identical sequences.
Then we can
specify that the valid lengths of these sequences
are 4, 2, and 0, respectively.
As a result,
the loss of the first sequence
should be twice as large as that of the second sequence,
while the third sequence should have a zero loss.
```
loss = MaskedSoftmaxCELoss()
loss(np.ones((3, 4, 10)), np.ones((3, 4)), np.array([4, 2, 0]))
```
## Training
:label:`sec_seq2seq_training`
In the following training loop,
we concatenate the special beginning-of-sequence token
and the original output sequence excluding the final token as
the input to the decoder, as shown in :numref:`fig_seq2seq`.
This is called *teacher forcing* because
the original output sequence (token labels) is fed into the decoder.
Alternatively,
we could also feed the *predicted* token
from the previous time step
as the current input to the decoder.
```
#@save
def train_seq2seq(net, data_iter, lr, num_epochs, tgt_vocab, device):
"""Train a model for sequence to sequence."""
net.initialize(init.Xavier(), force_reinit=True, ctx=device)
trainer = gluon.Trainer(net.collect_params(), 'adam',
{'learning_rate': lr})
loss = MaskedSoftmaxCELoss()
animator = d2l.Animator(xlabel='epoch', ylabel='loss',
xlim=[10, num_epochs])
for epoch in range(num_epochs):
timer = d2l.Timer()
metric = d2l.Accumulator(2) # Sum of training loss, no. of tokens
for batch in data_iter:
X, X_valid_len, Y, Y_valid_len = [
x.as_in_ctx(device) for x in batch]
bos = np.array(
[tgt_vocab['<bos>']] * Y.shape[0], ctx=device).reshape(-1, 1)
dec_input = np.concatenate([bos, Y[:, :-1]], 1) # Teacher forcing
with autograd.record():
Y_hat, _ = net(X, dec_input, X_valid_len)
l = loss(Y_hat, Y, Y_valid_len)
l.backward()
d2l.grad_clipping(net, 1)
num_tokens = Y_valid_len.sum()
trainer.step(num_tokens)
metric.add(l.sum(), num_tokens)
if (epoch + 1) % 10 == 0:
animator.add(epoch + 1, (metric[0] / metric[1],))
print(f'loss {metric[0] / metric[1]:.3f}, {metric[1] / timer.stop():.1f} '
f'tokens/sec on {str(device)}')
```
Now we can create and train an RNN encoder-decoder model
for sequence to sequence learning on the machine translation dataset.
```
embed_size, num_hiddens, num_layers, dropout = 32, 32, 2, 0.1
batch_size, num_steps = 64, 10
lr, num_epochs, device = 0.005, 300, d2l.try_gpu()
train_iter, src_vocab, tgt_vocab = d2l.load_data_nmt(batch_size, num_steps)
encoder = Seq2SeqEncoder(
len(src_vocab), embed_size, num_hiddens, num_layers, dropout)
decoder = Seq2SeqDecoder(
len(tgt_vocab), embed_size, num_hiddens, num_layers, dropout)
net = d2l.EncoderDecoder(encoder, decoder)
train_seq2seq(net, train_iter, lr, num_epochs, tgt_vocab, device)
```
## Prediction
To predict the output sequence
token by token,
at each decoder time step
the predicted token from the previous
time step is fed into the decoder as an input.
Similar to training,
at the initial time step
the beginning-of-sequence ("<bos>") token
is fed into the decoder.
This prediction process
is illustrated in :numref:`fig_seq2seq_predict`.
When the end-of-sequence ("<eos>") token is predicted,
the prediction of the output sequence is complete.

:label:`fig_seq2seq_predict`
We will introduce different
strategies for sequence generation in
:numref:`sec_beam-search`.
```
#@save
def predict_seq2seq(net, src_sentence, src_vocab, tgt_vocab, num_steps,
device, save_attention_weights=False):
"""Predict for sequence to sequence."""
src_tokens = src_vocab[src_sentence.lower().split(' ')] + [
src_vocab['<eos>']]
enc_valid_len = np.array([len(src_tokens)], ctx=device)
src_tokens = d2l.truncate_pad(src_tokens, num_steps, src_vocab['<pad>'])
# Add the batch axis
enc_X = np.expand_dims(np.array(src_tokens, ctx=device), axis=0)
enc_outputs = net.encoder(enc_X, enc_valid_len)
dec_state = net.decoder.init_state(enc_outputs, enc_valid_len)
# Add the batch axis
dec_X = np.expand_dims(np.array([tgt_vocab['<bos>']], ctx=device), axis=0)
output_seq, attention_weight_seq = [], []
for _ in range(num_steps):
Y, dec_state = net.decoder(dec_X, dec_state)
# We use the token with the highest prediction likelihood as the input
# of the decoder at the next time step
dec_X = Y.argmax(axis=2)
pred = dec_X.squeeze(axis=0).astype('int32').item()
# Save attention weights (to be covered later)
if save_attention_weights:
attention_weight_seq.append(net.decoder.attention_weights)
# Once the end-of-sequence token is predicted, the generation of the
# output sequence is complete
if pred == tgt_vocab['<eos>']:
break
output_seq.append(pred)
return ' '.join(tgt_vocab.to_tokens(output_seq)), attention_weight_seq
```
## Evaluation of Predicted Sequences
We can evaluate a predicted sequence
by comparing it with the
label sequence (the ground-truth).
BLEU (Bilingual Evaluation Understudy),
though originally proposed for evaluating
machine translation results :cite:`Papineni.Roukos.Ward.ea.2002`,
has been extensively used in measuring
the quality of output sequences for different applications.
In principle, for any $n$-grams in the predicted sequence,
BLEU evaluates whether this $n$-grams appears
in the label sequence.
Denote by $p_n$
the precision of $n$-grams,
which is
the ratio of
the number of matched $n$-grams in
the predicted and label sequences
to
the number of $n$-grams in the predicted sequence.
To explain,
given a label sequence $A$, $B$, $C$, $D$, $E$, $F$,
and a predicted sequence $A$, $B$, $B$, $C$, $D$,
we have $p_1 = 4/5$, $p_2 = 3/4$, $p_3 = 1/3$, and $p_4 = 0$.
Besides,
let $\mathrm{len}_{\text{label}}$ and $\mathrm{len}_{\text{pred}}$
be
the numbers of tokens in the label sequence and the predicted sequence, respectively.
Then, BLEU is defined as
$$ \exp\left(\min\left(0, 1 - \frac{\mathrm{len}_{\text{label}}}{\mathrm{len}_{\text{pred}}}\right)\right) \prod_{n=1}^k p_n^{1/2^n},$$
:eqlabel:`eq_bleu`
where $k$ is the longest $n$-grams for matching.
Based on the definition of BLEU in :eqref:`eq_bleu`,
whenever the predicted sequence is the same as the label sequence, BLEU is 1.
Moreover,
since matching longer $n$-grams is more difficult,
BLEU assigns a greater weight
to a longer $n$-gram precision.
Specifically, when $p_n$ is fixed,
$p_n^{1/2^n}$ increases as $n$ grows (the original paper uses $p_n^{1/n}$).
Furthermore,
since
predicting shorter sequences
tends to obtain a higher $p_n$ value,
the coefficient before the multiplication term in :eqref:`eq_bleu`
penalizes shorter predicted sequences.
For example, when $k=2$,
given the label sequence $A$, $B$, $C$, $D$, $E$, $F$ and the predicted sequence $A$, $B$,
although $p_1 = p_2 = 1$, the penalty factor $\exp(1-6/2) \approx 0.14$ lowers the BLEU.
We implement the BLEU measure as follows.
```
def bleu(pred_seq, label_seq, k): #@save
"""Compute the BLEU."""
pred_tokens, label_tokens = pred_seq.split(' '), label_seq.split(' ')
len_pred, len_label = len(pred_tokens), len(label_tokens)
score = math.exp(min(0, 1 - len_label / len_pred))
for n in range(1, k + 1):
num_matches, label_subs = 0, collections.defaultdict(int)
for i in range(len_label - n + 1):
label_subs[''.join(label_tokens[i: i + n])] += 1
for i in range(len_pred - n + 1):
if label_subs[''.join(pred_tokens[i: i + n])] > 0:
num_matches += 1
label_subs[''.join(pred_tokens[i: i + n])] -= 1
score *= math.pow(num_matches / (len_pred - n + 1), math.pow(0.5, n))
return score
```
In the end,
we use the trained RNN encoder-decoder
to translate a few English sentences into French
and compute the BLEU of the results.
```
engs = ['go .', "i lost .", 'he\'s calm .', 'i\'m home .']
fras = ['va !', 'j\'ai perdu .', 'il est calme .', 'je suis chez moi .']
for eng, fra in zip(engs, fras):
translation, attention_weight_seq = predict_seq2seq(
net, eng, src_vocab, tgt_vocab, num_steps, device)
print(f'{eng} => {translation}, bleu {bleu(translation, fra, k=2):.3f}')
```
## Summary
* Following the design of the encoder-decoder architecture, we can use two RNNs to design a model for sequence to sequence learning.
* When implementing the encoder and the decoder, we can use multilayer RNNs.
* We can use masks to filter out irrelevant computations, such as when calculating the loss.
* In encoder-decoder training, the teacher forcing approach feeds original output sequences (in contrast to predictions) into the decoder.
* BLEU is a popular measure for evaluating output sequences by matching $n$-grams between the predicted sequence and the label sequence.
## Exercises
1. Can you adjust the hyperparameters to improve the translation results?
1. Rerun the experiment without using masks in the loss calculation. What results do you observe? Why?
1. If the encoder and the decoder differ in the number of layers or the number of hidden units, how can we initialize the hidden state of the decoder?
1. In training, replace teacher forcing with feeding the prediction at the previous time step into the decoder. How does this influence the performance?
1. Rerun the experiment by replacing GRU with LSTM.
1. Are there any other ways to design the output layer of the decoder?
[Discussions](https://discuss.d2l.ai/t/345)
|
github_jupyter
|
import collections
from d2l import mxnet as d2l
import math
from mxnet import np, npx, init, gluon, autograd
from mxnet.gluon import nn, rnn
npx.set_np()
#@save
class Seq2SeqEncoder(d2l.Encoder):
"""The RNN encoder for sequence to sequence learning."""
def __init__(self, vocab_size, embed_size, num_hiddens, num_layers,
dropout=0, **kwargs):
super(Seq2SeqEncoder, self).__init__(**kwargs)
# Embedding layer
self.embedding = nn.Embedding(vocab_size, embed_size)
self.rnn = rnn.GRU(num_hiddens, num_layers, dropout=dropout)
def forward(self, X, *args):
# The output `X` shape: (`batch_size`, `num_steps`, `embed_size`)
X = self.embedding(X)
# In RNN models, the first axis corresponds to time steps
X = X.swapaxes(0, 1)
state = self.rnn.begin_state(batch_size=X.shape[1], ctx=X.ctx)
output, state = self.rnn(X, state)
# `output` shape: (`num_steps`, `batch_size`, `num_hiddens`)
# `state[0]` shape: (`num_layers`, `batch_size`, `num_hiddens`)
return output, state
encoder = Seq2SeqEncoder(vocab_size=10, embed_size=8, num_hiddens=16,
num_layers=2)
encoder.initialize()
X = np.zeros((4, 7))
output, state = encoder(X)
output.shape
len(state), state[0].shape
class Seq2SeqDecoder(d2l.Decoder):
"""The RNN decoder for sequence to sequence learning."""
def __init__(self, vocab_size, embed_size, num_hiddens, num_layers,
dropout=0, **kwargs):
super(Seq2SeqDecoder, self).__init__(**kwargs)
self.embedding = nn.Embedding(vocab_size, embed_size)
self.rnn = rnn.GRU(num_hiddens, num_layers, dropout=dropout)
self.dense = nn.Dense(vocab_size, flatten=False)
def init_state(self, enc_outputs, *args):
return enc_outputs[1]
def forward(self, X, state):
# The output `X` shape: (`num_steps`, `batch_size`, `embed_size`)
X = self.embedding(X).swapaxes(0, 1)
# `context` shape: (`batch_size`, `num_hiddens`)
context = state[0][-1]
# Broadcast `context` so it has the same `num_steps` as `X`
context = np.broadcast_to(context, (
X.shape[0], context.shape[0], context.shape[1]))
X_and_context = np.concatenate((X, context), 2)
output, state = self.rnn(X_and_context, state)
output = self.dense(output).swapaxes(0, 1)
# `output` shape: (`batch_size`, `num_steps`, `vocab_size`)
# `state[0]` shape: (`num_layers`, `batch_size`, `num_hiddens`)
return output, state
decoder = Seq2SeqDecoder(vocab_size=10, embed_size=8, num_hiddens=16,
num_layers=2)
decoder.initialize()
state = decoder.init_state(encoder(X))
output, state = decoder(X, state)
output.shape, len(state), state[0].shape
X = np.array([[1, 2, 3], [4, 5, 6]])
npx.sequence_mask(X, np.array([1, 2]), True, axis=1)
X = np.ones((2, 3, 4))
npx.sequence_mask(X, np.array([1, 2]), True, value=-1, axis=1)
#@save
class MaskedSoftmaxCELoss(gluon.loss.SoftmaxCELoss):
"""The softmax cross-entropy loss with masks."""
# `pred` shape: (`batch_size`, `num_steps`, `vocab_size`)
# `label` shape: (`batch_size`, `num_steps`)
# `valid_len` shape: (`batch_size`,)
def forward(self, pred, label, valid_len):
# `weights` shape: (`batch_size`, `num_steps`, 1)
weights = np.expand_dims(np.ones_like(label), axis=-1)
weights = npx.sequence_mask(weights, valid_len, True, axis=1)
return super(MaskedSoftmaxCELoss, self).forward(pred, label, weights)
loss = MaskedSoftmaxCELoss()
loss(np.ones((3, 4, 10)), np.ones((3, 4)), np.array([4, 2, 0]))
#@save
def train_seq2seq(net, data_iter, lr, num_epochs, tgt_vocab, device):
"""Train a model for sequence to sequence."""
net.initialize(init.Xavier(), force_reinit=True, ctx=device)
trainer = gluon.Trainer(net.collect_params(), 'adam',
{'learning_rate': lr})
loss = MaskedSoftmaxCELoss()
animator = d2l.Animator(xlabel='epoch', ylabel='loss',
xlim=[10, num_epochs])
for epoch in range(num_epochs):
timer = d2l.Timer()
metric = d2l.Accumulator(2) # Sum of training loss, no. of tokens
for batch in data_iter:
X, X_valid_len, Y, Y_valid_len = [
x.as_in_ctx(device) for x in batch]
bos = np.array(
[tgt_vocab['<bos>']] * Y.shape[0], ctx=device).reshape(-1, 1)
dec_input = np.concatenate([bos, Y[:, :-1]], 1) # Teacher forcing
with autograd.record():
Y_hat, _ = net(X, dec_input, X_valid_len)
l = loss(Y_hat, Y, Y_valid_len)
l.backward()
d2l.grad_clipping(net, 1)
num_tokens = Y_valid_len.sum()
trainer.step(num_tokens)
metric.add(l.sum(), num_tokens)
if (epoch + 1) % 10 == 0:
animator.add(epoch + 1, (metric[0] / metric[1],))
print(f'loss {metric[0] / metric[1]:.3f}, {metric[1] / timer.stop():.1f} '
f'tokens/sec on {str(device)}')
embed_size, num_hiddens, num_layers, dropout = 32, 32, 2, 0.1
batch_size, num_steps = 64, 10
lr, num_epochs, device = 0.005, 300, d2l.try_gpu()
train_iter, src_vocab, tgt_vocab = d2l.load_data_nmt(batch_size, num_steps)
encoder = Seq2SeqEncoder(
len(src_vocab), embed_size, num_hiddens, num_layers, dropout)
decoder = Seq2SeqDecoder(
len(tgt_vocab), embed_size, num_hiddens, num_layers, dropout)
net = d2l.EncoderDecoder(encoder, decoder)
train_seq2seq(net, train_iter, lr, num_epochs, tgt_vocab, device)
#@save
def predict_seq2seq(net, src_sentence, src_vocab, tgt_vocab, num_steps,
device, save_attention_weights=False):
"""Predict for sequence to sequence."""
src_tokens = src_vocab[src_sentence.lower().split(' ')] + [
src_vocab['<eos>']]
enc_valid_len = np.array([len(src_tokens)], ctx=device)
src_tokens = d2l.truncate_pad(src_tokens, num_steps, src_vocab['<pad>'])
# Add the batch axis
enc_X = np.expand_dims(np.array(src_tokens, ctx=device), axis=0)
enc_outputs = net.encoder(enc_X, enc_valid_len)
dec_state = net.decoder.init_state(enc_outputs, enc_valid_len)
# Add the batch axis
dec_X = np.expand_dims(np.array([tgt_vocab['<bos>']], ctx=device), axis=0)
output_seq, attention_weight_seq = [], []
for _ in range(num_steps):
Y, dec_state = net.decoder(dec_X, dec_state)
# We use the token with the highest prediction likelihood as the input
# of the decoder at the next time step
dec_X = Y.argmax(axis=2)
pred = dec_X.squeeze(axis=0).astype('int32').item()
# Save attention weights (to be covered later)
if save_attention_weights:
attention_weight_seq.append(net.decoder.attention_weights)
# Once the end-of-sequence token is predicted, the generation of the
# output sequence is complete
if pred == tgt_vocab['<eos>']:
break
output_seq.append(pred)
return ' '.join(tgt_vocab.to_tokens(output_seq)), attention_weight_seq
def bleu(pred_seq, label_seq, k): #@save
"""Compute the BLEU."""
pred_tokens, label_tokens = pred_seq.split(' '), label_seq.split(' ')
len_pred, len_label = len(pred_tokens), len(label_tokens)
score = math.exp(min(0, 1 - len_label / len_pred))
for n in range(1, k + 1):
num_matches, label_subs = 0, collections.defaultdict(int)
for i in range(len_label - n + 1):
label_subs[''.join(label_tokens[i: i + n])] += 1
for i in range(len_pred - n + 1):
if label_subs[''.join(pred_tokens[i: i + n])] > 0:
num_matches += 1
label_subs[''.join(pred_tokens[i: i + n])] -= 1
score *= math.pow(num_matches / (len_pred - n + 1), math.pow(0.5, n))
return score
engs = ['go .', "i lost .", 'he\'s calm .', 'i\'m home .']
fras = ['va !', 'j\'ai perdu .', 'il est calme .', 'je suis chez moi .']
for eng, fra in zip(engs, fras):
translation, attention_weight_seq = predict_seq2seq(
net, eng, src_vocab, tgt_vocab, num_steps, device)
print(f'{eng} => {translation}, bleu {bleu(translation, fra, k=2):.3f}')
| 0.828384 | 0.993307 |
# <img style="float: left; padding-right: 10px; width: 45px" src="https://raw.githubusercontent.com/Harvard-IACS/2018-CS109A/master/content/styles/iacs.png"> APCOMP 295 Advanced Practical Data Science
## Exercise 3: Dask
**Harvard University**<br/>
**Fall 2020**<br/>
**Instructors**: Pavlos Protopapas
<hr style="height:2pt">
**Each assignment is graded out of 5 points. The topic for this assignment is Dask.**
**Due:** 09/29/2020 10:15 AM EDT
**Submit:** We won't be re running your notebooks, please ensure output is visible in the notebook.
## Question 1: Reflection on Exercise 2 (1 points)
In exercise 2, we developed our app, tested it locally, afterward we deploy it with docker and finally with minikube and/or gcloud. Please answer following questions in 1-3 sentences.
<br/>
(A) For the local code files:<br/>
1. Which port is `maindb.py` listening at? <br/> <b>8082</b>
2. Which port is `task1.py` listening at? <br/> <b>8081</b>
<br/>
(B) For the Docker implementation we built two images: `webapp:db` and `task1:frontend`
<br>
1. For `webapp:db` which port is exposed by the container? <b>8082</b> Are you binding any port from host (i.e your laptop) to container port ? <b>no</b> <br/>
<br/>
2. For `task1:frontend` which port is exposed by the container? <b>8081</b> Are you binding any port from host (i.e your laptop) to container port ? <b>30000</b> <br/>
<br/>
3. There's an environment variable in `Docker_task1frontend` named `DB_HOST`. How is that getting utilized? <br/>
Reminder: We used these 2 commands to run our images (or used docker-compose) - <br/> `docker run --name mywebdb -d --network appNetwork webapp:db` <br/>
`docker run --name fe -d -p 5000:8081 -e DB_HOST=mywebdb --network appNetwork task1:frontend` <br/>
Hint: Check `task1.py`
<b>the environment var `DB_HOST` is passed in as a command line argument to `task1.py`. `task1.py` uses it as part of `db_url` for sending requests to it, and it is defaulted to `0.0.0.0`.</b>
<br/>
(C) For the minikube/gcloud implementation, <br/>
1. How is the environment variable `DB_HOST` getting populated ?
<b>it is read from a `database_host` setting from k8s config map</b>
2. How are we able to access our front end via browser ? <br/>
by loading `http://localhost:30000`
3. What is the purpose of `webapp-db-service` in `webapp_db_deployment_k8s.yaml` ?<br/>
<b>it is specifying the logically name of the db service to be matched with config map. it is then passed into web frontend.</b>
## Question 2: Compute Pi with a Slowly Converging Series (1 points)
Leibniz published one of the oldest known series in 1676. While this is easy to understand and derive, it converges very slowly.
https://en.wikipedia.org/wiki/Leibniz_formula_for_%CF%80 <br/>
$$\frac{\pi}{4} = 1 - \frac{1}{3} + \frac{1}{5} - \frac{1}{7} ...$$
While this is a genuinely cruel way to compute the value of $\pi$, it’s a fun opportunity to use brute force on a problem instead of thinking.
Compute $\pi$ using at least four billion terms in this sequence. Compare your time taken with numpy and dask. On my mac, with numpy this took 44 seconds and with dask it took 5.7 seconds.
*Hint:* Use dask array
```
import numpy as np
num_terms = 100000000
def pi_arr(arr):
return (-2*(arr%2 ==0)+1)/(2*arr-1)*4
%%time
import dask.array as da
pi_arr(da.arange(1, num_terms, chunks=num_terms/4)).sum().compute()
%%time
pi_arr(np.arange(1, num_terms)).sum()
```
## Question 3: Filter Parking Tickets Dataset (1 points)
Please download the data set from https://www.kaggle.com/new-york-city/nyc-parking-tickets. According to the documentation for the parking tickets data set, the column called ‘Plate Type’ consists mainly of two different types: ‘PAS’ and ‘COM’, presumably for passenger and commercial vehicles, respectively. Maybe the rest are the famous parking tickets from the UN diplomats, who take advantage of diplomatic immunity not to pay their fines.
Create a filtered Dask DataFrame with only the commercial plates.
Persist it, so it is available in memory for future computations. Count the number of summonses in 2017 issued to commercial plate types. Compute them as a percentage of the total data set.
*Hint*: This is easy; it is only about 5-7 lines of code.
```
import dask
import dask.dataframe as dd
df_2017 = dd.read_csv('data/Parking_Violations_Issued_-_Fiscal_Year_2017.csv')
df_2017_COM = df_2017[df_2017['Plate Type'] == 'COM']
df_2017_COM.persist()
print(f'commerical vehicle parking ticket percentage {len(df_2017_COM)*100/len(df_2017)}%')
```
## Question 4 : Build a Cluster with Dask and Jupyter Lab using Helm (2 points)
Your challenge is to build a Dask Cluster with 5 nodes (a scheduler, a server to deploy a jupyter
notebook, and 3 workers) using Kubernetes on Google Cloud and Helm. Helm is a popular
Kubernetes package manager currently maintained by the Cloud Native Computing Foundation
(CNCF). Similar to Docker, Helm has hundreds of images (called charts) ready for deployment. We
used Helm version 3.2.4 to test this part of the question. Original instructions to install dask on kubernetes via helm can be found [here](https://docs.dask.org/en/latest/setup/kubernetes-helm.html#kubernetes-helm-single).
**Step 0:** Install [helm](https://helm.sh/docs/intro/install/) and add helm chart. Helm uses a packaging format called charts. A chart is a collection of files that describe a related set of Kubernetes resources. `brew install helm` worked on mac for installing mac. To add helm chart use `helm repo add dask https://helm.dask.org/` <br/>
**Step 1:** Create a Google Cloud cluster with 5 nodes using the CLI to manage Kubernetes service. <br/>
`export PROJECT_ID=<your project id>` <br/>
`gcloud config set project $PROJECT_ID` <br/>
`gcloud config list` - This is to ensure you have right account, zone and project set.<br/>
`gcloud container clusters create mydask-cluster --num-nodes 5`
**Submit a screenshot** <br/>
Do not change the number of nodes or default machine type when you create the cluster.

**Step 2:** Use the `values.yaml` file provided to you. We have customized this file to add a few packages and loadbalancers, you can find the original [here](https://github.com/dask/helm-chart/tree/master/dask).
Use Helm to copy charts in each pod
`helm install my-release -f values.yaml dask/dask` <br/> <br/>
**Question :** Please compare the original and modified YAML files. What changes did we make to the load balancers? Why did we make them?
<b>the scheduler and jupyter notebook service type is changed from ClusterIP to LoadBalancer, so that these services can be exposed outside the cluster.</b>
<br/>
**Step 3:** Check if all the pods are running and services are up (this may take few minutes)
Submit a screenshot of `kubectl get pods` and `kubectl get services`

**Step 4:**Copy external-ip of dask-jupyter, access jupyter from your browser (password: dask)
Also copy the external-ip of dask-scheduler and access the dask dashboard from your browser.
```
➜ exercise3 git:(master) ✗
echo tcp://$DASK_SCHEDULER:$DASK_SCHEDULER_PORT -- Dask Client connection
echo http://$DASK_SCHEDULER_UI_IP:$DASK_SCHEDULER_UI_PORT -- Dask dashboard
echo http://$JUPYTER_NOTEBOOK_IP:$JUPYTER_NOTEBOOK_PORT -- Jupyter notebook
tcp://34.73.111.217:8786 -- Dask Client connection
http://34.73.111.217:80 -- Dask dashboard
http://35.185.65.225:80 -- Jupyter notebook
```
**Step 5:** We have provided `helm_gcsfs.ipynb` for this part - this is the same exercise you have seen in lecture 4 but reads from the google cloud storage bucket. `gcsfs` is python based file-system interface to Google Cloud Storage. We have already installed `gcsfs` package on our cluster using `values.yaml`. First we will upload the dataset on Google Cloud Storage bucket and then run `helm_gcsfs.ipynb`. <br/>
Now we are going to create a Google Cloud Storage Bucket (and then upload the dataset `Parking_Violations_Issued_-_Fiscal_Year_2017.csv`) - On cloud console search for `storage`. Mine looks like this -

(i) Create a bucket, give it a name and leave the defaults as is. Upload `Parking_Violations_Issued_-_Fiscal_Year_2017.csv` within this bucket. <br/>
(ii) Once uploaded, click on the file and then click `Edit Permissions`. `Add Entry` - `Public` - `All Users` - `Reader` and save. <br/> Ideally we do not want to upload large datsets, we should mount the bucket and download the dataset from kaggle. For the purpose of this homework we will directly upload the file to google cloud storage bucket. **(Submit a screenshot)**
Example: <br/>
<br/>

**Question :** Why is this step necessary, why don't we just directly upload the .csv file on Jupyter lab ?
<b>because it is making the file publicly accessible from the google cloud. if we updated it to Jupyter lab, it will stay inside a container of a pod. when the container is removed, the data will be lost; so that next time we need ot reference it, we will have to upload it again.</b>
<br/>
(iii) Upload the attached `helm_gcsfs.ipynb` to jupyter lab. In `helm_gcsfs.ipynb` change the name of your project and the name of the bucket. Run the code, download your `helm_gcsfs.ipynb` and submit along with this notebook. Also **submit 2-3 screenshot** of dask dashboard - we would like to see some computation happening, so take the screenshot while your program is running. <br/>
**Question :** How much time did helm_gcfs.ipynb took (time is reported in the notebook) ? <br/>
Example screenshots:


### <font color=red> Step 6: Delete your cluster </font>
`gcloud container clusters delete mydask-cluster`
Optional: Repeat step 1-6 with 6 workers - did it take same amount of time or less time ? Hint: change values.yaml to have 6 workers
Fun fact about user [jovyan](https://github.com/jupyter/docker-stacks/issues/358)
|
github_jupyter
|
import numpy as np
num_terms = 100000000
def pi_arr(arr):
return (-2*(arr%2 ==0)+1)/(2*arr-1)*4
%%time
import dask.array as da
pi_arr(da.arange(1, num_terms, chunks=num_terms/4)).sum().compute()
%%time
pi_arr(np.arange(1, num_terms)).sum()
import dask
import dask.dataframe as dd
df_2017 = dd.read_csv('data/Parking_Violations_Issued_-_Fiscal_Year_2017.csv')
df_2017_COM = df_2017[df_2017['Plate Type'] == 'COM']
df_2017_COM.persist()
print(f'commerical vehicle parking ticket percentage {len(df_2017_COM)*100/len(df_2017)}%')
➜ exercise3 git:(master) ✗
echo tcp://$DASK_SCHEDULER:$DASK_SCHEDULER_PORT -- Dask Client connection
echo http://$DASK_SCHEDULER_UI_IP:$DASK_SCHEDULER_UI_PORT -- Dask dashboard
echo http://$JUPYTER_NOTEBOOK_IP:$JUPYTER_NOTEBOOK_PORT -- Jupyter notebook
tcp://34.73.111.217:8786 -- Dask Client connection
http://34.73.111.217:80 -- Dask dashboard
http://35.185.65.225:80 -- Jupyter notebook
| 0.162446 | 0.944485 |
<a href="https://colab.research.google.com/github/hrai/M4-Data-Analysis/blob/master/Assignment_2.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
#Analysis of M4 dataset - Assignment 2
###Hangjit Rai (12749723)
##Introduction
M4 has been extended and new tolls have been added between Parramatta and Homebush to reduce travel times in the West with increased lanes.
It is a massive project undertaken by the New South Wales government to make the commute easier and faster for the residents of Sydney. Various entry and exit points have been established within this section of M4 known as toll gantry. Since it was opened, traffic data have been collected which is openly available to the public.
The data was available in CSV format on the Transport NSW website. There were 12 files for each month from July 2018 to June 2019 (financial year 2019). The data has been collected in 15 minutes bursts. A data dictionary was also provided for easier understanding of information in the CSV files. This data has been used to create a forecasting model for the number of vehicles using the motorway.
Using this model, the number of vehicles using the motorway can be forecast so that the government can take necessary measures to expand the highway. Additionally, higher toll charges can be applied at peak times to discourage drivers from using the motorway and adding to traffic jam. It also lets an insight into the quiet times of the day so that repair work can be conducted during those hours minimising the disruption to motorway users.
##Exploration
The data was converted into python notebook and analysed using pandas and numpy and visualised with matplotlib and seaborn. The dataset comprised of the following information - 'DateTime', 'AssetID', 'FinancialQtrID', 'IntervalEnd', 'Version', 'VehicleClass', 'TollPointID', 'GantryDirection', 'GantryLocation', 'GantryGPSLatitude', 'GantryGPSLongitude', 'GantryType', 'TotalVolume'.
The data set was split into four quarters for the financial year 2019. There were only two categories of vehicles namely *car* and *truck*. There were three types of gantries namely 'Mainline', 'Ramp' and 'Mainline + G'. Also 11 different gantry locations were mentioned in the data set.
Upon analysis, it was seen that there are more cars than trucks using the motorway. The data set didn't comprise of any missing fields. Some of the columns were renamed so it was easier to reference and use them. It was observed that a number of attributes were correlated so they were removed.
After running some aggregations, it was visible that most of the vehicles used Ramp gantry type followed by Mainline and finally Mainline+G. A trend was observed when the data was spotted on a chart. The number of vehicles using the M4 dropped on the weekends. Compared to Saturday, less vehicles were reported to be using M4 on Sunday. Also a huge drop was observed in the traffic between Christmas and New Years Day. This meant that a number of people were outside of Sydney at the time resulting in an overall drop in the usage of the highway. On the contrary, a scatter plot showed that over the 12-month period February and March saw the highest number of users of M4.
Data from a single day plotted on to a bar chart gave some interesting insights. It was clear to see the rise and fall of traffic in and out of the motorway. The traffic slowly started to pick up at 5 in the morning, peaking at 12:30 and slowly going down until 2:30 p.m. and then started picking up again until 5 p.m. to a new higher peak after which it started plummeting until 8 p.m. Then there was an irregular flow of traffic until midnight.
##Methodology
M4 data set was recorded over regular time intervals with a frequency of 15 mins, therefore the data set was a time-based data. It was, in other words a time series. As we are only using one value or attribute for forecasting it falls under univariate time series forecasting. ARIMA model which stands for 'autoregressive integrated moving average' is a class of combination of models that is used for forecasting time series. The models are flexible enough to be used with non-seasonal time series.
Rolling mean and standard deviation were created to check the stationarity of the time series. Using numpy’s log method, a graph was generated that showed the ruling mean and standard deviation.
DateTime column was created by merging *date* and *time interval* fields. For the analysis purposes, DateTime and TrafficVolume columns were selected from the original data set. The test and train datasets were split with 33:27 ratio. DateTime column was set as the index of the new data frame. The data was then fed into the ARIMA model and fitted to create the forecasting model. Upon printing the summary, low error of 0.070 or less was observed. Also, the coefficient of the constant was 0.0004
##Evaluation
ARIMA comes with a number of methods to test the model and its accuracy. A convenience method called summary generates the ARIMA model results with the coefficient and standard errors. Also, residuals can be created from the fitted model and plotted on the kde chart.
The residuals were plotted on kde plot. Invoking the *describe()* method on *residuals* data frame yielded the following summary:
Aggregation | Value
--- | ---
Count | 736824
Mean | -0.000184
Std | 159.973717
Min | -1276.74957
25% | -45.53026
50% | -1.045925
75% | 34.526041
Max | 1155.036415
Finally, the model was used for forecasting the test data. The plot that was created showed that the prediction was linear. Over the whole period of 1 year, there was no significant rise in the number of people using M4. That's why the forecast produced by the model is more or less linear.
##Conclusion
Time series is a unique type of data that has its own complexities and requires a different analysis process. Depending on the type of the data set whether it's univariate or multivariate, different algorithms need to be selected for the analysis. ARIMA model which is a combination of multiple techniques produces satisfying results. M4 data that was used in this report was analysed and a prediction model was created from ARIMA model. Because the overall trend in the data was more or less horizontal, the prediction model’s accuracy couldn’t be ascertained effectively. If the data set provided becomes more varied then the model could be able to find patterns in the time series and make proper productions.
##Ethical Issues
Artificial intelligence is built on the ability of computer systems to find patterns in huge amounts of data. This is unbeknownst to the knowledge of people, a number of companies have been collecting information and selling them to make profits. Ethical issues arise when there is no strict guidelines and laws around the collection and usage of personal information. The M4 data that has been collected can be used to personally identify the drivers or the owners of the vehicles. If let's say Joe uses M4 every morning to go to work then it will be easy enough to find this pattern in the data. This information is a valuable information. Care should be taken to not disclose personally-identifying information when the data is being collected and stored. Data breaches may cause the information to land in the wrong hands and unintended damages.
There have been incidents with ethical issues related to AI. Amazon used an AI system to create to manage recruitment process but because the tech industry is primarily male-driven, the system was favouring male candidates over female candidates. There are ethical concerns around targeted marketing which have been used by big multinational companies like Target and Kmart. Targeted online advertising is another facet bearing the ethical dilemma. As long as these issues are addressed, we will be able to reap the benefits of AI.
##Data Analysis
###Loading all M4 data for 2018-19
```
import pandas as pd
import numpy as np
import seaborn as sns
from pandas import DataFrame
from statsmodels.tsa.arima_model import ARIMA
import matplotlib.pyplot as plt
urls=[
'https://raw.githubusercontent.com/hrai/M4-Data-Analysis/master/data/M4_passages_2018-07.csv',
'https://raw.githubusercontent.com/hrai/M4-Data-Analysis/master/data/M4_passages_2018-08.csv',
'https://raw.githubusercontent.com/hrai/M4-Data-Analysis/master/data/M4_passages_2018-09.csv',
'https://raw.githubusercontent.com/hrai/M4-Data-Analysis/master/data/M4_passages_2018-10.csv',
'https://raw.githubusercontent.com/hrai/M4-Data-Analysis/master/data/M4_passages_2018-11.csv',
'https://raw.githubusercontent.com/hrai/M4-Data-Analysis/master/data/M4_passages_2018-12.csv',
'https://raw.githubusercontent.com/hrai/M4-Data-Analysis/master/data/M4_passages_2019-01.csv',
'https://raw.githubusercontent.com/hrai/M4-Data-Analysis/master/data/M4_passages_2019-02.csv',
'https://raw.githubusercontent.com/hrai/M4-Data-Analysis/master/data/M4_passages_2019-03.csv',
'https://raw.githubusercontent.com/hrai/M4-Data-Analysis/master/data/M4_passages_2019-04.csv',
'https://raw.githubusercontent.com/hrai/M4-Data-Analysis/master/data/M4_passages_2019-05.csv',
'https://raw.githubusercontent.com/hrai/M4-Data-Analysis/master/data/M4_passages_2019-06.csv'
]
df = pd.concat(pd.read_csv(url, parse_dates={'DateTime': ['Date', 'IntervalStart']}) for url in urls)
count_row,count_col=df.shape
print('Total rows: ' + str(count_row))
df[:10]
total_vehicles=df['TotalVolume'].sum()
total_vehicles
df.columns
total_cars = df.loc[df['VehicleClass']=='Car', 'VehicleClass'].value_counts()
print(f"total cars rows {total_cars}")
total_trucks = df.loc[df['VehicleClass']=='Truck', 'VehicleClass'].value_counts()
print(f"total trucks rows {total_trucks}")
```
### Check for null values
```
df.isnull().values.any()
df.values
```
###Rename column
```
df.rename(columns={'TotalVolume': 'TrafficVolume', 'VehicleClass':'VehicleType'}, inplace=True)
df.columns
```
###Scatter plot of latitude and longitude
```
plt.scatter(x=df['GantryGPSLatitude'], y=df['GantryGPSLongitude'])
plt.show()
```
###Correlation heatmap
```
corr = df.corr()
sns.heatmap(corr, xticklabels=corr.columns, yticklabels=corr.columns, cmap=sns.diverging_palette(220, 10, as_cmap=True))
```
###Checking latitute and longitude against GantryLocation
```
temp_df=df[['GantryLocation','GantryGPSLatitude','GantryGPSLongitude','TollPointID']].groupby(by=['GantryLocation'])
print(temp_df.first())
```
###Checking unique values
```
print(df['AssetID'].unique())
print(df['Version'].unique())
print(df['FinancialQtrID'].unique())
locs=df['GantryLocation'].unique()
print(locs)
print(locs.size)
print(df['VehicleType'].unique())
print(df['GantryType'].unique())
```
###Drop unwanted columns like latitude and longitude
```
df.drop(['GantryGPSLatitude', 'GantryGPSLongitude','AssetID', 'Version','TollPointID','FinancialQtrID'], axis = 1, inplace=True)
df.columns
```
###Grouping data
```
group_by_gantry_type = df[['GantryType', 'VehicleType','TrafficVolume']].groupby(by=['GantryType', 'VehicleType'])
group_by_gantry_type.head(2)
```
###Getting average and total counts from the grouping
```
car_data_avg = group_by_gantry_type.mean()
car_data_count = group_by_gantry_type.count()
print(car_data_avg.head(2))
car_data_count
```
###Plotting aggregates onto bar chart - Most of the vehicles are recorded via Ramp
```
car_data_count.sort_values(by='TrafficVolume').plot(kind='barh')
# Draw Plot
def plot_df(df, x, y, title="", xlabel='Date', ylabel='Traffic volume', dpi=100):
plt.figure(figsize=(16,5), dpi=dpi)
plt.plot(x, y, color='tab:red')
plt.gca().set(title=title, xlabel=xlabel, ylabel=ylabel)
plt.show()
plot_df(df, x=df['DateTime'], y=df['TrafficVolume'], title='Flow of traffic in M4.')
```
###Scatter plot - Traffic in days of week
```
df['Day'] = df['DateTime'].dt.day_name()
sns.pairplot(x_vars=["Day"], y_vars=["TrafficVolume"],
data=df, height=7)
```
###Scatter plot - Traffic in 12 months
```
df['Month'] = df['DateTime'].map(lambda x: x.strftime('%Y-%m'))
sns.pairplot(x_vars=["Month"], y_vars=["TrafficVolume"],
data=df, height=11)
```
###Date filters for a day's data
```
start_date=pd.to_datetime('1 Jun 2019 00:00AM')
end_date=pd.to_datetime('1 Jun 2019 11:59PM')
#apply date filter
mask = (df['DateTime'] > start_date) & (df['DateTime'] <= end_date)
df_day=df.loc[mask]
#check unique value in month column
df_day.Month.unique()
df_day['Time'] = df_day['DateTime'].map(lambda x: x.strftime('%H:%M'))
df_day.drop(columns=['DateTime'], inplace=True)
df_day.head()
plt.figure(figsize=(18,25))
sns.barplot(x='TrafficVolume', y='Time', data=df_day)
```
###Method to create rolling mean/std
```
def test_stationarity_of_timeseries(timeseries):
#Determing rolling statistics
rolmean = timeseries.rolling(12).mean()
rolstd = timeseries.rolling(12).std()
#Plot rolling statistics:
orig = plt.plot(timeseries, color='blue',label='Original')
mean = plt.plot(rolmean, color='red', label='Rolling Mean')
std = plt.plot(rolstd, color='black', label = 'Rolling Std')
plt.legend(loc='best')
plt.title('Rolling Mean & Standard Deviation')
plt.show(block=False)
```
###Creating rolling mean/std
```
df_stationarity=df[['DateTime','TrafficVolume']].set_index('DateTime')
test_stationarity_of_timeseries(df_stationarity)
df_stationarity_log = np.log(df_stationarity)
test_stationarity_of_timeseries(df_stationarity_log)
```
###Creating Test and Train datasets
```
df_prediction=df[['DateTime','TrafficVolume']]
df_prediction.set_index('DateTime', inplace=True)
train_size = int(len(df_prediction) * 0.66)
train, test = df_prediction[0:train_size], df_prediction[train_size:]
train.head(5)
# fit model
model = ARIMA(train.values.astype(float), order=(2,2,0))
fitted_model = model.fit(disp=0)
print(fitted_model.summary())
# plot residual errors
residuals = DataFrame(fitted_model.resid)
residuals.plot(kind='kde')
plt.show()
print(residuals.describe())
# Plot residual errors
residuals.plot(title="Residuals")
plt.show()
# Actual vs Fitted
fitted_model.plot_predict()
plt.show()
# Forecast
test_rows_count=test.shape[0]
fc, se, conf = fitted_model.forecast(test_rows_count, alpha=0.05) # 95% conf
# Make as pandas series
fc_series = pd.Series(fc, index=test.index)
lower_series = pd.Series(conf[:, 0], index=test.index)
upper_series = pd.Series(conf[:, 1], index=test.index)
# Plot
plt.figure(figsize=(12,5), dpi=100)
plt.plot(train, label='training')
plt.plot(test, label='actual')
plt.plot(fc_series, label='forecast')
plt.fill_between(lower_series.index, lower_series, upper_series,
color='k', alpha=.15)
plt.title('Forecast vs Actuals')
plt.legend(loc='upper left', fontsize=8)
plt.show()
```
##GitHub URL Link
https://github.com/hrai/M4-Data-Analysis/blob/master/Assignment_2.ipynb
|
github_jupyter
|
import pandas as pd
import numpy as np
import seaborn as sns
from pandas import DataFrame
from statsmodels.tsa.arima_model import ARIMA
import matplotlib.pyplot as plt
urls=[
'https://raw.githubusercontent.com/hrai/M4-Data-Analysis/master/data/M4_passages_2018-07.csv',
'https://raw.githubusercontent.com/hrai/M4-Data-Analysis/master/data/M4_passages_2018-08.csv',
'https://raw.githubusercontent.com/hrai/M4-Data-Analysis/master/data/M4_passages_2018-09.csv',
'https://raw.githubusercontent.com/hrai/M4-Data-Analysis/master/data/M4_passages_2018-10.csv',
'https://raw.githubusercontent.com/hrai/M4-Data-Analysis/master/data/M4_passages_2018-11.csv',
'https://raw.githubusercontent.com/hrai/M4-Data-Analysis/master/data/M4_passages_2018-12.csv',
'https://raw.githubusercontent.com/hrai/M4-Data-Analysis/master/data/M4_passages_2019-01.csv',
'https://raw.githubusercontent.com/hrai/M4-Data-Analysis/master/data/M4_passages_2019-02.csv',
'https://raw.githubusercontent.com/hrai/M4-Data-Analysis/master/data/M4_passages_2019-03.csv',
'https://raw.githubusercontent.com/hrai/M4-Data-Analysis/master/data/M4_passages_2019-04.csv',
'https://raw.githubusercontent.com/hrai/M4-Data-Analysis/master/data/M4_passages_2019-05.csv',
'https://raw.githubusercontent.com/hrai/M4-Data-Analysis/master/data/M4_passages_2019-06.csv'
]
df = pd.concat(pd.read_csv(url, parse_dates={'DateTime': ['Date', 'IntervalStart']}) for url in urls)
count_row,count_col=df.shape
print('Total rows: ' + str(count_row))
df[:10]
total_vehicles=df['TotalVolume'].sum()
total_vehicles
df.columns
total_cars = df.loc[df['VehicleClass']=='Car', 'VehicleClass'].value_counts()
print(f"total cars rows {total_cars}")
total_trucks = df.loc[df['VehicleClass']=='Truck', 'VehicleClass'].value_counts()
print(f"total trucks rows {total_trucks}")
df.isnull().values.any()
df.values
df.rename(columns={'TotalVolume': 'TrafficVolume', 'VehicleClass':'VehicleType'}, inplace=True)
df.columns
plt.scatter(x=df['GantryGPSLatitude'], y=df['GantryGPSLongitude'])
plt.show()
corr = df.corr()
sns.heatmap(corr, xticklabels=corr.columns, yticklabels=corr.columns, cmap=sns.diverging_palette(220, 10, as_cmap=True))
temp_df=df[['GantryLocation','GantryGPSLatitude','GantryGPSLongitude','TollPointID']].groupby(by=['GantryLocation'])
print(temp_df.first())
print(df['AssetID'].unique())
print(df['Version'].unique())
print(df['FinancialQtrID'].unique())
locs=df['GantryLocation'].unique()
print(locs)
print(locs.size)
print(df['VehicleType'].unique())
print(df['GantryType'].unique())
df.drop(['GantryGPSLatitude', 'GantryGPSLongitude','AssetID', 'Version','TollPointID','FinancialQtrID'], axis = 1, inplace=True)
df.columns
group_by_gantry_type = df[['GantryType', 'VehicleType','TrafficVolume']].groupby(by=['GantryType', 'VehicleType'])
group_by_gantry_type.head(2)
car_data_avg = group_by_gantry_type.mean()
car_data_count = group_by_gantry_type.count()
print(car_data_avg.head(2))
car_data_count
car_data_count.sort_values(by='TrafficVolume').plot(kind='barh')
# Draw Plot
def plot_df(df, x, y, title="", xlabel='Date', ylabel='Traffic volume', dpi=100):
plt.figure(figsize=(16,5), dpi=dpi)
plt.plot(x, y, color='tab:red')
plt.gca().set(title=title, xlabel=xlabel, ylabel=ylabel)
plt.show()
plot_df(df, x=df['DateTime'], y=df['TrafficVolume'], title='Flow of traffic in M4.')
df['Day'] = df['DateTime'].dt.day_name()
sns.pairplot(x_vars=["Day"], y_vars=["TrafficVolume"],
data=df, height=7)
df['Month'] = df['DateTime'].map(lambda x: x.strftime('%Y-%m'))
sns.pairplot(x_vars=["Month"], y_vars=["TrafficVolume"],
data=df, height=11)
start_date=pd.to_datetime('1 Jun 2019 00:00AM')
end_date=pd.to_datetime('1 Jun 2019 11:59PM')
#apply date filter
mask = (df['DateTime'] > start_date) & (df['DateTime'] <= end_date)
df_day=df.loc[mask]
#check unique value in month column
df_day.Month.unique()
df_day['Time'] = df_day['DateTime'].map(lambda x: x.strftime('%H:%M'))
df_day.drop(columns=['DateTime'], inplace=True)
df_day.head()
plt.figure(figsize=(18,25))
sns.barplot(x='TrafficVolume', y='Time', data=df_day)
def test_stationarity_of_timeseries(timeseries):
#Determing rolling statistics
rolmean = timeseries.rolling(12).mean()
rolstd = timeseries.rolling(12).std()
#Plot rolling statistics:
orig = plt.plot(timeseries, color='blue',label='Original')
mean = plt.plot(rolmean, color='red', label='Rolling Mean')
std = plt.plot(rolstd, color='black', label = 'Rolling Std')
plt.legend(loc='best')
plt.title('Rolling Mean & Standard Deviation')
plt.show(block=False)
df_stationarity=df[['DateTime','TrafficVolume']].set_index('DateTime')
test_stationarity_of_timeseries(df_stationarity)
df_stationarity_log = np.log(df_stationarity)
test_stationarity_of_timeseries(df_stationarity_log)
df_prediction=df[['DateTime','TrafficVolume']]
df_prediction.set_index('DateTime', inplace=True)
train_size = int(len(df_prediction) * 0.66)
train, test = df_prediction[0:train_size], df_prediction[train_size:]
train.head(5)
# fit model
model = ARIMA(train.values.astype(float), order=(2,2,0))
fitted_model = model.fit(disp=0)
print(fitted_model.summary())
# plot residual errors
residuals = DataFrame(fitted_model.resid)
residuals.plot(kind='kde')
plt.show()
print(residuals.describe())
# Plot residual errors
residuals.plot(title="Residuals")
plt.show()
# Actual vs Fitted
fitted_model.plot_predict()
plt.show()
# Forecast
test_rows_count=test.shape[0]
fc, se, conf = fitted_model.forecast(test_rows_count, alpha=0.05) # 95% conf
# Make as pandas series
fc_series = pd.Series(fc, index=test.index)
lower_series = pd.Series(conf[:, 0], index=test.index)
upper_series = pd.Series(conf[:, 1], index=test.index)
# Plot
plt.figure(figsize=(12,5), dpi=100)
plt.plot(train, label='training')
plt.plot(test, label='actual')
plt.plot(fc_series, label='forecast')
plt.fill_between(lower_series.index, lower_series, upper_series,
color='k', alpha=.15)
plt.title('Forecast vs Actuals')
plt.legend(loc='upper left', fontsize=8)
plt.show()
| 0.455441 | 0.986138 |
# LSTM Stock Predictor Using Fear and Greed Index
In this notebook, you will build and train a custom LSTM RNN that uses a 10 day window of Bitcoin fear and greed index values to predict the 11th day closing price.
You will need to:
1. Prepare the data for training and testing
2. Build and train a custom LSTM RNN
3. Evaluate the performance of the model
## Data Preparation
In this section, you will need to prepare the training and testing data for the model. The model will use a rolling 10 day window to predict the 11th day closing price.
You will need to:
1. Use the `window_data` function to generate the X and y values for the model.
2. Split the data into 70% training and 30% testing
3. Apply the MinMaxScaler to the X and y values
4. Reshape the X_train and X_test data for the model. Note: The required input format for the LSTM is:
```python
reshape((X_train.shape[0], X_train.shape[1], 1))
```
```
import numpy as np
import pandas as pd
import hvplot.pandas
# Set the random seed for reproducibility
# Note: This is for the homework solution, but it is good practice to comment this out and run multiple experiments to evaluate your model
from numpy.random import seed
seed(1)
from tensorflow import random
random.set_seed(2)
# Load the fear and greed sentiment data for Bitcoin
df = pd.read_csv("Resources/btc_sentiment.csv", index_col="date", infer_datetime_format=True, parse_dates=True)
df = df.drop(columns="fng_classification")
df.head()
# Load the historical closing prices for Bitcoin
df2 = pd.read_csv("Resources/btc_historic.csv", index_col="Date", infer_datetime_format=True, parse_dates=True)['Close']
df2 = df2.sort_index()
df2.tail()
# Join the data into a single DataFrame
df = df.join(df2, how="inner")
df.tail()
df.head()
# This function accepts the column number for the features (X) and the target (y)
# It chunks the data up with a rolling window of Xt-n to predict Xt
# It returns a numpy array of X any y
def window_data(df, window, feature_col_number, target_col_number):
X = []
y = []
for i in range(len(df) - window - 1):
features = df.iloc[i:(i + window), feature_col_number]
target = df.iloc[(i + window), target_col_number]
X.append(features)
y.append(target)
return np.array(X), np.array(y).reshape(-1, 1)
# Predict Closing Prices using a 10 day window of previous fng values
# Then, experiment with window sizes anywhere from 1 to 10 and see how the model performance changes
window_size = 10
# Column index 0 is the 'fng_value' column
# Column index 1 is the `Close` column
feature_column = 0
target_column = 1
X, y = window_data(df, window_size, feature_column, target_column)
# Use 70% of the data for training and the remainder for testing
split = int(0.7 * len(X))
X_train = X[: split]
X_test = X[split:]
y_train = y[: split]
y_test = y[split:]
from sklearn.preprocessing import MinMaxScaler
# Use the MinMaxScaler to scale data between 0 and 1.
scaler = MinMaxScaler()
scaler.fit(X)
X_train_scaler = scaler.transform(X_train)
X_test_scaler = scaler.transform(X_test)
scaler.fit(y)
y_train = scaler.transform(y_train)
y_test = scaler.transform(y_test)
# Reshape the features for the model
X_train = X_train_scaler.reshape((X_train_scaler.shape[0], X_train_scaler.shape[1], 1))
X_test = X_test_scaler.reshape((X_test_scaler.shape[0], X_test_scaler.shape[1], 1))
print (f"X_train sample values:\n{X_train[:5]} \n")
print (f"X_test sample values:\n{X_test[:5]}")
```
---
## Build and Train the LSTM RNN
In this section, you will design a custom LSTM RNN and fit (train) it using the training data.
You will need to:
1. Define the model architecture
2. Compile the model
3. Fit the model to the training data
### Hints:
You will want to use the same model architecture and random seed for both notebooks. This is necessary to accurately compare the performance of the FNG model vs the closing price model.
```
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import LSTM, Dense, Dropout
# Build the LSTM model.
# The return sequences need to be set to True if you are adding additional LSTM layers, but
# You don't have to do this for the final layer.
# Note: The dropouts help prevent overfitting
# Note: The input shape is the number of time steps and the number of indicators
# Note: Batching inputs has a different input shape of Samples/TimeSteps/Features
model = Sequential()
number_units = 30
dropout_fraction = 0.2
# Layer 1
model.add(LSTM(
units=number_units,
return_sequences=True,
input_shape=(X_train.shape[1], 1))
)
model.add(Dropout(dropout_fraction))
# Layer 2
model.add(LSTM(units=number_units, return_sequences=True))
model.add(Dropout(dropout_fraction))
# Layer 3
model.add(LSTM(units=number_units))
model.add(Dropout(dropout_fraction))
# Output layer
model.add(Dense(1))
# Compile the model
model.compile(optimizer="adam", loss="mean_squared_error")
# Summarize the model
model.summary()
# Train the model
# Use at least 10 epochs
# Do not shuffle the data
# Experiement with the batch size, but a smaller batch size is recommended
model.fit(X_train, y_train, epochs=10, shuffle=False, batch_size=4, verbose=1)
```
---
## Model Performance
In this section, you will evaluate the model using the test data.
You will need to:
1. Evaluate the model using the `X_test` and `y_test` data.
2. Use the X_test data to make predictions
3. Create a DataFrame of Real (y_test) vs predicted values.
4. Plot the Real vs predicted values as a line chart
### Hints
Remember to apply the `inverse_transform` function to the predicted and y_test values to recover the actual closing prices.
```
# Evaluate the model
model.evaluate(X_test, y_test)
# Batch size of 4 minimizes the loss function value.
# Make some predictions
predicted = model.predict(X_test)
# Recover the original prices instead of the scaled version
predicted_prices = scaler.inverse_transform(predicted)
real_prices = scaler.inverse_transform(y_test.reshape(-1, 1))
# Create a DataFrame of Real and Predicted values
stocks = pd.DataFrame({
"Real": real_prices.ravel(),
"Predicted": predicted_prices.ravel()
}, index = df.index[-len(real_prices): ])
stocks.head()
# Plot the real vs predicted values as a line chart
stocks.plot()
# The model struggles to predict future prices based on fng values.
# Between the two, LSTM predictions based on the closing prices show better trend following, just not at the same scale.
```
|
github_jupyter
|
reshape((X_train.shape[0], X_train.shape[1], 1))
import numpy as np
import pandas as pd
import hvplot.pandas
# Set the random seed for reproducibility
# Note: This is for the homework solution, but it is good practice to comment this out and run multiple experiments to evaluate your model
from numpy.random import seed
seed(1)
from tensorflow import random
random.set_seed(2)
# Load the fear and greed sentiment data for Bitcoin
df = pd.read_csv("Resources/btc_sentiment.csv", index_col="date", infer_datetime_format=True, parse_dates=True)
df = df.drop(columns="fng_classification")
df.head()
# Load the historical closing prices for Bitcoin
df2 = pd.read_csv("Resources/btc_historic.csv", index_col="Date", infer_datetime_format=True, parse_dates=True)['Close']
df2 = df2.sort_index()
df2.tail()
# Join the data into a single DataFrame
df = df.join(df2, how="inner")
df.tail()
df.head()
# This function accepts the column number for the features (X) and the target (y)
# It chunks the data up with a rolling window of Xt-n to predict Xt
# It returns a numpy array of X any y
def window_data(df, window, feature_col_number, target_col_number):
X = []
y = []
for i in range(len(df) - window - 1):
features = df.iloc[i:(i + window), feature_col_number]
target = df.iloc[(i + window), target_col_number]
X.append(features)
y.append(target)
return np.array(X), np.array(y).reshape(-1, 1)
# Predict Closing Prices using a 10 day window of previous fng values
# Then, experiment with window sizes anywhere from 1 to 10 and see how the model performance changes
window_size = 10
# Column index 0 is the 'fng_value' column
# Column index 1 is the `Close` column
feature_column = 0
target_column = 1
X, y = window_data(df, window_size, feature_column, target_column)
# Use 70% of the data for training and the remainder for testing
split = int(0.7 * len(X))
X_train = X[: split]
X_test = X[split:]
y_train = y[: split]
y_test = y[split:]
from sklearn.preprocessing import MinMaxScaler
# Use the MinMaxScaler to scale data between 0 and 1.
scaler = MinMaxScaler()
scaler.fit(X)
X_train_scaler = scaler.transform(X_train)
X_test_scaler = scaler.transform(X_test)
scaler.fit(y)
y_train = scaler.transform(y_train)
y_test = scaler.transform(y_test)
# Reshape the features for the model
X_train = X_train_scaler.reshape((X_train_scaler.shape[0], X_train_scaler.shape[1], 1))
X_test = X_test_scaler.reshape((X_test_scaler.shape[0], X_test_scaler.shape[1], 1))
print (f"X_train sample values:\n{X_train[:5]} \n")
print (f"X_test sample values:\n{X_test[:5]}")
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import LSTM, Dense, Dropout
# Build the LSTM model.
# The return sequences need to be set to True if you are adding additional LSTM layers, but
# You don't have to do this for the final layer.
# Note: The dropouts help prevent overfitting
# Note: The input shape is the number of time steps and the number of indicators
# Note: Batching inputs has a different input shape of Samples/TimeSteps/Features
model = Sequential()
number_units = 30
dropout_fraction = 0.2
# Layer 1
model.add(LSTM(
units=number_units,
return_sequences=True,
input_shape=(X_train.shape[1], 1))
)
model.add(Dropout(dropout_fraction))
# Layer 2
model.add(LSTM(units=number_units, return_sequences=True))
model.add(Dropout(dropout_fraction))
# Layer 3
model.add(LSTM(units=number_units))
model.add(Dropout(dropout_fraction))
# Output layer
model.add(Dense(1))
# Compile the model
model.compile(optimizer="adam", loss="mean_squared_error")
# Summarize the model
model.summary()
# Train the model
# Use at least 10 epochs
# Do not shuffle the data
# Experiement with the batch size, but a smaller batch size is recommended
model.fit(X_train, y_train, epochs=10, shuffle=False, batch_size=4, verbose=1)
# Evaluate the model
model.evaluate(X_test, y_test)
# Batch size of 4 minimizes the loss function value.
# Make some predictions
predicted = model.predict(X_test)
# Recover the original prices instead of the scaled version
predicted_prices = scaler.inverse_transform(predicted)
real_prices = scaler.inverse_transform(y_test.reshape(-1, 1))
# Create a DataFrame of Real and Predicted values
stocks = pd.DataFrame({
"Real": real_prices.ravel(),
"Predicted": predicted_prices.ravel()
}, index = df.index[-len(real_prices): ])
stocks.head()
# Plot the real vs predicted values as a line chart
stocks.plot()
# The model struggles to predict future prices based on fng values.
# Between the two, LSTM predictions based on the closing prices show better trend following, just not at the same scale.
| 0.879522 | 0.979393 |
### Módulo 2 - continuação no arquivo .ipynb
### 2.3 Plotando dados no Jupyter Notebook
Continuamos o Módulo 2 em um Jupyter Notebook. Essas páginas possibilitam a disposição de texto MarkDown (como este) e código Python (algumas caixas abaixo). Vamos anotar nossa análise com porções de MarkDown, mas o que cada comando específico faz será anotado com comentários no próprio código. Os comentários são precedidos do caracter '#' que indica que aquela porção é um comentário e portanto não será executado como código. Diferente de uma IDE, os Notebooks **não são** ambientes ideais para testes, experimentos ou instalação de programas, mas são excelentes para descrição e organização de análises. Os Notebooks suportam uma variedade de linguagens, inclusive bash, então é possível comandar o Terminal por aqui. Basta botar um `!` na frente do comando.
```
cd ~/Bio/tutorial_v2
!ls
```
Há muito o que se fazer com os Jupyter Notebooks, e é muito bom ficar confortável com eles! Bom, mas o que queremos agora é usa-los para realizar uma análise de dados que poderia caber a qualquer estudante de biologia. Vamos usar o famoso conjunto de dados Iris, que está em um arquivo .csv em nossa pasta local. Arquivos .csv ou .tsv (*comma* ou *tab-separated values*) são arquivos de texto usados para representar tabelas. Uma tabela do Excel, por exemplo, pode ser exportada como .csv.
O uso do **Pandas** possibilita importar a tabela com os dados diretamente em um objeto do tipo *Dataframe*. Vamos importar a biblioteca Pandas com o comando `import` e abreviar ela como `pd`. Também vamos importar a biblioteca `matplotlib.pyplot` e abreviar como `plt`. Ambas essas abreviações são universais para esses pacotes. A razão que importamos `matplotlib.pyplot` ao invés de `matplotlib` é porque `pyplot` é uma **API**, um programa que serve como uma interface para 'conectar' os pacotes Pandas e Matplotlib.
```
# Importando as bibliotecas e suas respectivas abreviações
import pandas as pd
import matplotlib.pyplot as plt
```
Agora já temos todos os métodos dessas bibliotecas à nossa disposição. Só falta importar os dados. Para isso, o Pandas oferece o método read_csv, que converte um arquivo .csv em dataframe. Vamos começar com um arquivo bem simples, o [dest.txt](../data/dest.txt).
```
# Vamos dar uma olhadinha nos dados.
!cat data/dest.txt
# O comando pd.read_csv lê os dados e armazena em um dataframe "df". Precisamos especificar a separação por tab ("\t")
df = pd.read_csv("data/dest.txt", sep="\t")
plt.plot(df["Volume_mL"], df["Temperatura_C"])
plt.show()
plt.plot(df["Volume_mL"], df["Temperatura_C"])
plt.ylim((0, 150))
plt.xlabel("Volume (mL)")
plt.ylabel("Temperatura (ºC)")
plt.show()
```
Agora, vamos importar um conjunto de dados mais complexo, o "Iris". Vamos armazenar esse dataframe em uma variável `iris`.
```
# Vamos armazenar esse objeto Pandas em uma variável 'iris'
iris = pd.read_csv('../data/iris.csv')
print(iris)
```
#### Nota sobre importar arquivos
Aqui é importante notar o que fizemos ao importar o conjunto Iris. Utilizamos o **método** ```pd.read_csv()```. Esse método serve apenas para arquivos do tipo CSV (comma-separated values). Esse formato de arquivo é bem comum, basicamente é um arquivo de texto contendo um tabela com valores separados por vírgulas (*commas*), ou qualquer outro *delimiter*. Uma planilha do Excel, por exemplo, pode ser salva em .csv. Se nosso arquivo tivesse os valores separados pelo caracter ";", precisamos "explicar" isso. Nosso comando seria ```pd.read_csv('data/iris.csv', delimiter = ';')```. O método ```pd.read_csv()``` tem um **argumento** obrigatório, que é o arquivo que será importado. O argumento "delimiter" é "," a não ser que especificado.
A biblioteca Pandas é útil porque ela transforma arquivos em *Dataframes*, que são **objetos** que permitem uma fácil visualização e manipulação dos dados. Logo, depois do comando acima, temos um **objeto** (um Pandas dataframe) armazenado em uma variável (```iris```).
Como o formato CSV é bem comum, sua importação é simples. Quando trabalhamos com outros formatos de arquivo, devemos nos atentar a como importa-los. Aqui existem muitas possibilidades, podemos coletar dados da web, importar dados de outro ambiente (como R, por exemplo), se estamos com um formato incomum, talvez precisaremos de algum pacote que converta ele em algo que o Pandas possa entender. Que tipo de dado você tem?
```
# Quando verificamos que tipo de dado é 'iris', vemos que o Pandas já converte o arquivo .csv em um Dataframe.
type(iris)
# Outra possibilidade de visualização é com os comandos head() e tail() ou describe().
iris.head()
iris.tail()
iris.describe() # species não foi incluído pois contém strings, e não floats.
# Podemos selecionar colunas específicas com colchetes e a string que dá nome à coluna.
iris['species']
```
Repare a diferença entre um **método**, que é invocado com parênteses, como read_csv( ), e um **atributo** de um objeto, que é invocado com colchetes, como ['species']. 'species' é uma coluna, portanto, um atributo do objeto "iris".
```
len(iris['species'])
# ou com iloc[], que permite escolher as linhas:
iris['species'].iloc[10:51]
```
Utilizamos o Pandas para importar nosso arquivo .csv em um *dataframe* armazenado na variável `iris`. Além de importar, o Pandas também possui o método `plot()`. Escolhemos nossos eixos bem como o tipo de Plot.
```
# Vamos tentar um scatter plot simples com o método plot(). Repare nos parâmetros definindo x, y e 'kind'
iris.plot(x='PetalLengthCm', y='SepalLengthCm', kind = 'scatter')
# Agora um pouco mais sofisticado, com cores.
# ax é um objeto da biblioteca Matplotlib, que corresponde a um eixo ou porção de uma figura
# Vamos projetar três plots, um para cada espécie, porém todos no mesmo eixo (ax)
# Também é possível adicionar labels e um título para o eixo.
ax = iris[iris.species=='Iris-setosa'].plot.scatter(x='PetalLengthCm', y='PetalWidthCm',
color='red', label='setosa')
iris[iris.species=='Iris-versicolor'].plot.scatter(x='PetalLengthCm', y='PetalWidthCm',
color='green', label='versicolor', ax=ax)
iris[iris.species=='Iris-virginica'].plot.scatter(x='PetalLengthCm', y='PetalWidthCm',
color='blue', label='virginica', ax=ax)
ax.set_title("scatter")
```
Esse último snippet foi retirado [deste notebook Kaggle.](https://www.kaggle.com/xuhewen/iris-dataset-visualization-and-machine-learning)
Na verdade, o Pandas possui toda uma biblioteca de plotagem (pandas.plotting) que funciona por meio da API `matplotlib.pyplot`. Temos, por exemplo, os métodos `radviz()` e `parallel_coordinates()`, usados para visualizar dados multivariados. Exemplos retirados da [galeria do Pandas.](http://pandas.pydata.org/pandas-docs/version/0.13/visualization.html)
```
# Precisamos importar os métodos da biblioteca plotting do Pandas
from pandas.plotting import radviz, parallel_coordinates
# O método requere somente o dataframe e a seleção da coluna com nomes das classes (strings)
radviz(iris,'species')
# Vamos experimentar com o outro método, parallel_coordinates()
parallel_coordinates(iris, 'species')
# Aparentemente, o radviz() foi melhor que este pois ilustrou a diferença em 'SepalWidth' observada no scatterplot.
```
Até agora, vimos como:
* Abrir o Jupyter Notebook
* Importar as bibliotecas Pandas e Matplotlib.pyplot
* Usar o método do Pandas `pd.read_csv()`para importar o .csv como um Dataframe.
* Selecionar porções do Dataframe com métodos como `head()` ou `iloc[]`.
* Usar o Matplotlib para visualizar plots do Pandas.
Conhecendo e usando as bibliotecas que se aprende a linguagem; existem galerias no site do [Pandas](http://pandas.pydata.org/pandas-docs/version/0.13/visualization.html) e [Matplotlib](https://matplotlib.org/users/pyplot_tutorial.html) com diversos exemplos, entretanto é melhor concentrar em um curso de fundamentos de Python antes. Abaixo exemplos da segunda galeria. Repare que com bibliotecas como Pandas e [NumPy](http://www.numpy.org/) (uma dependência do Pandas), é possível criar 'séries' numéricas para exemplos de plots.
```
plt.plot([1,2,3,4])
plt.ylabel('some numbers')
plt.show()
# Vamos importar a biblioteca NumPy como np
import numpy as np
# Como mencionado anteriormente, podemos usa-la para criar uma série numérica.
t = np.arange(0., 5., 0.2)
# E plotar essa série de diferentes formas
plt.plot(t, t, 'r--', t, t**2, 'bs', t, t**3, 'g^')
plt.show()
# Aqui temos um exemplo mais avançado de como fazer um plot com múltiplas figuras
# Essa linha estabelece uma função que gera os valores que serão plotados.
def f(t):
return np.exp(-t) * np.cos(2*np.pi*t)
# t1 e t2 são séries numéricas que serviram de argumento para a função f(t).
t1 = np.arange(0.0, 5.0, 0.1)
t2 = np.arange(0.0, 5.0, 0.02)
# Queremos UMA figura
plt.figure(1)
# Com DOIS subplots. Na segunda linha vamos plotar o primeiro subplot
plt.subplot(211)
plt.plot(t1, f(t1), 'bo', t2, f(t2), 'k')
# Plotando o segundo subplot
plt.subplot(212)
plt.plot(t2, np.cos(2*np.pi*t2), 'r--')
# Mostrar a figura
plt.show()
```
### Conclusão do Módulo 2
Enquanto no Módulo 1 vimos as ferramentas essenciais para registrar (editor de texto) e executar (Terminal) linhas de código (em Python), no Módulo 2 aprendemos como gerenciar pacotes e ambientes para podermos tirar o melhor proveito da nossa linguagem. Fizemos uso de três bibliotecas (Jupyter, Pandas e Matplotlib) para importar o conjunto de dados "Iris" e visualiza-lo com diferentes métodos.
|
github_jupyter
|
cd ~/Bio/tutorial_v2
!ls
# Importando as bibliotecas e suas respectivas abreviações
import pandas as pd
import matplotlib.pyplot as plt
# Vamos dar uma olhadinha nos dados.
!cat data/dest.txt
# O comando pd.read_csv lê os dados e armazena em um dataframe "df". Precisamos especificar a separação por tab ("\t")
df = pd.read_csv("data/dest.txt", sep="\t")
plt.plot(df["Volume_mL"], df["Temperatura_C"])
plt.show()
plt.plot(df["Volume_mL"], df["Temperatura_C"])
plt.ylim((0, 150))
plt.xlabel("Volume (mL)")
plt.ylabel("Temperatura (ºC)")
plt.show()
# Vamos armazenar esse objeto Pandas em uma variável 'iris'
iris = pd.read_csv('../data/iris.csv')
print(iris)
# Quando verificamos que tipo de dado é 'iris', vemos que o Pandas já converte o arquivo .csv em um Dataframe.
type(iris)
# Outra possibilidade de visualização é com os comandos head() e tail() ou describe().
iris.head()
iris.tail()
iris.describe() # species não foi incluído pois contém strings, e não floats.
# Podemos selecionar colunas específicas com colchetes e a string que dá nome à coluna.
iris['species']
len(iris['species'])
# ou com iloc[], que permite escolher as linhas:
iris['species'].iloc[10:51]
# Vamos tentar um scatter plot simples com o método plot(). Repare nos parâmetros definindo x, y e 'kind'
iris.plot(x='PetalLengthCm', y='SepalLengthCm', kind = 'scatter')
# Agora um pouco mais sofisticado, com cores.
# ax é um objeto da biblioteca Matplotlib, que corresponde a um eixo ou porção de uma figura
# Vamos projetar três plots, um para cada espécie, porém todos no mesmo eixo (ax)
# Também é possível adicionar labels e um título para o eixo.
ax = iris[iris.species=='Iris-setosa'].plot.scatter(x='PetalLengthCm', y='PetalWidthCm',
color='red', label='setosa')
iris[iris.species=='Iris-versicolor'].plot.scatter(x='PetalLengthCm', y='PetalWidthCm',
color='green', label='versicolor', ax=ax)
iris[iris.species=='Iris-virginica'].plot.scatter(x='PetalLengthCm', y='PetalWidthCm',
color='blue', label='virginica', ax=ax)
ax.set_title("scatter")
# Precisamos importar os métodos da biblioteca plotting do Pandas
from pandas.plotting import radviz, parallel_coordinates
# O método requere somente o dataframe e a seleção da coluna com nomes das classes (strings)
radviz(iris,'species')
# Vamos experimentar com o outro método, parallel_coordinates()
parallel_coordinates(iris, 'species')
# Aparentemente, o radviz() foi melhor que este pois ilustrou a diferença em 'SepalWidth' observada no scatterplot.
plt.plot([1,2,3,4])
plt.ylabel('some numbers')
plt.show()
# Vamos importar a biblioteca NumPy como np
import numpy as np
# Como mencionado anteriormente, podemos usa-la para criar uma série numérica.
t = np.arange(0., 5., 0.2)
# E plotar essa série de diferentes formas
plt.plot(t, t, 'r--', t, t**2, 'bs', t, t**3, 'g^')
plt.show()
# Aqui temos um exemplo mais avançado de como fazer um plot com múltiplas figuras
# Essa linha estabelece uma função que gera os valores que serão plotados.
def f(t):
return np.exp(-t) * np.cos(2*np.pi*t)
# t1 e t2 são séries numéricas que serviram de argumento para a função f(t).
t1 = np.arange(0.0, 5.0, 0.1)
t2 = np.arange(0.0, 5.0, 0.02)
# Queremos UMA figura
plt.figure(1)
# Com DOIS subplots. Na segunda linha vamos plotar o primeiro subplot
plt.subplot(211)
plt.plot(t1, f(t1), 'bo', t2, f(t2), 'k')
# Plotando o segundo subplot
plt.subplot(212)
plt.plot(t2, np.cos(2*np.pi*t2), 'r--')
# Mostrar a figura
plt.show()
| 0.504394 | 0.95452 |
# Lambda School Data Science Module 144
## Real-world Experiment Design
[Induction experiment, Wikipedia](https://commons.wikimedia.org/wiki/File:Induction_experiment.png)
## Prepare - Learn about JavaScript and Google Analytics
Python is great - but with web applications, it's impossible to avoid JavaScript. The lingua franca of the web, JavaScript runs in all browsers, and thus all front-end code must either be JS or transpiled to it. As a data scientist you don't have to learn JavaScript - but you do have to be aware of it, and being able to figure out snippets of it is an invaluable skill to connect your skills with real-world applications.
So, we leave the warm comfort of Python, and venture to a bigger world - check out the [LambdaSchool/AB-Demo repo](https://github.com/LambdaSchool/AB-Demo) and [live experiment](https://lambdaschool.github.io/AB-Demo/) before class.
Additionally, sign up for [Google Analytics](https://www.google.com/analytics) - if you're not sure on the steps or what "property" to give it, you can put a placeholder or wait until the live lecture. Google also has [Analytics documentation](https://support.google.com/analytics/) that is worth a look.
Note - if you use any of the various tracker blocking techniques, it's quite likely you won't show up in Google Analytics. You'll have to disable them to be able to fully test your experiment.
## Live Lecture - Using Google Analytics with a live A/B test
Again we won't do much Python here, but we'll put a few notes and results in the notebook as we go.
## Assignment - Set up your own A/B test!
For a baseline, a straight fork of the Lambda School repo is OK. Getting that working with your own Analytics profile is already a task. But if you get through that, stretch goals:
1. Explore Google Analytics - it's big and changes frequently, but powerful (can track conversions and events, flows, etc.)
2. Customize the experiment to be more interesting/different (try colors!)
3. Check out the various tools for setting up A/B experiments (e.g. [Optimizely](https://www.optimizely.com/) and [alternatives](https://alternativeto.net/software/optimizely/))
4. Try to get enough traffic to actually have more real data (don't spam people, but do share with friends)
5. If you do get more traffic, don't just apply a t-test - dig into the results and use both math and writing to describe your findings
Additionally, today it is a good idea to go back and review the frequentist hypothesis testing material from the first two modules. And if you feel on top of things - you can use your newfound GitHub Pages and Google Analytics skills to build/iterate a portfolio page, and maybe even instrument it with Analytics!
## Resources
- [Demo Google Analytics Data](https://support.google.com/analytics/answer/6367342?hl=en) - an Analytics profile you can add to your account with real data from the Google swag store
- [Design of Experiment](https://explorable.com/design-of-experiment) - an essay summarizing some of the things to be aware of when designing and running an experiment
|
github_jupyter
|
# Lambda School Data Science Module 144
## Real-world Experiment Design

[Induction experiment, Wikipedia](https://commons.wikimedia.org/wiki/File:Induction_experiment.png)
## Prepare - Learn about JavaScript and Google Analytics
Python is great - but with web applications, it's impossible to avoid JavaScript. The lingua franca of the web, JavaScript runs in all browsers, and thus all front-end code must either be JS or transpiled to it. As a data scientist you don't have to learn JavaScript - but you do have to be aware of it, and being able to figure out snippets of it is an invaluable skill to connect your skills with real-world applications.
So, we leave the warm comfort of Python, and venture to a bigger world - check out the [LambdaSchool/AB-Demo repo](https://github.com/LambdaSchool/AB-Demo) and [live experiment](https://lambdaschool.github.io/AB-Demo/) before class.
Additionally, sign up for [Google Analytics](https://www.google.com/analytics) - if you're not sure on the steps or what "property" to give it, you can put a placeholder or wait until the live lecture. Google also has [Analytics documentation](https://support.google.com/analytics/) that is worth a look.
Note - if you use any of the various tracker blocking techniques, it's quite likely you won't show up in Google Analytics. You'll have to disable them to be able to fully test your experiment.
## Live Lecture - Using Google Analytics with a live A/B test
Again we won't do much Python here, but we'll put a few notes and results in the notebook as we go.
## Assignment - Set up your own A/B test!
For a baseline, a straight fork of the Lambda School repo is OK. Getting that working with your own Analytics profile is already a task. But if you get through that, stretch goals:
1. Explore Google Analytics - it's big and changes frequently, but powerful (can track conversions and events, flows, etc.)
2. Customize the experiment to be more interesting/different (try colors!)
3. Check out the various tools for setting up A/B experiments (e.g. [Optimizely](https://www.optimizely.com/) and [alternatives](https://alternativeto.net/software/optimizely/))
4. Try to get enough traffic to actually have more real data (don't spam people, but do share with friends)
5. If you do get more traffic, don't just apply a t-test - dig into the results and use both math and writing to describe your findings
Additionally, today it is a good idea to go back and review the frequentist hypothesis testing material from the first two modules. And if you feel on top of things - you can use your newfound GitHub Pages and Google Analytics skills to build/iterate a portfolio page, and maybe even instrument it with Analytics!
## Resources
- [Demo Google Analytics Data](https://support.google.com/analytics/answer/6367342?hl=en) - an Analytics profile you can add to your account with real data from the Google swag store
- [Design of Experiment](https://explorable.com/design-of-experiment) - an essay summarizing some of the things to be aware of when designing and running an experiment
| 0.717309 | 0.940408 |
```
#install addons for
!pip install tensorflow-addons
!pip install transformers[tf-cpu]
from transformers import BertTokenizer, TFBertModel
from tensorflow import keras
from tensorflow.keras import layers
import tensorflow as tf
import numpy as np
from sklearn.model_selection import train_test_split
import matplotlib.pyplot as plt
from tensorflow.keras.layers import Embedding, Dense, Input, Dropout, LayerNormalization, GlobalAveragePooling1D, Flatten
from tensorflow_addons.layers import MultiHeadAttention
from tensorflow.keras.preprocessing.text import Tokenizer
from tensorflow.keras.preprocessing.sequence import pad_sequences
import csv
import os
import shutil
# Download the data from Kaggle https://www.kaggle.com/lakshmi25npathi/imdb-dataset-of-50k-movie-reviews/tasks?taskId=588
# Load the 1mdb datasets from Kaggle
kaggle_imdb_file = 'datasets/IMDB_Dataset.csv'
data_x = []
data_y = []
with open(kaggle_imdb_file, 'r') as csvfile:
filereader = csv.reader(csvfile, delimiter=',', dialect='excel')
next(filereader)
for row in filereader:
data_x.append(row[0])
label = 1 if row[1] == 'positive' else 0
data_y.append(label)
# Prepare the data into trainable format
data_x = np.array(data_x)
data_y = np.array(data_y)
X_train, X_test, y_train, y_test = train_test_split(data_x, data_y, test_size=0.2, random_state=29)
# Generate Pre-trained outputs
tokenizer = BertTokenizer.from_pretrained('bert-base-uncased')
bert_model = TFBertModel.from_pretrained("bert-base-uncased")
n_dim = 768
def compute_bert_pt(text_data, batch_size):
i = 0;
output = tf.reshape(tf.constant([]), (0, n_dim))
while i*batch_size < text_data.shape[0]:
start = i*batch_size
end = (i+1)*batch_size
if end > text_data.shape[0]:
end = text_data.shape[0]
batch_pt = bert_model(tokenizer(text_data[start:end].tolist(), max_length = 512, pad_to_max_length=True, truncation=True, return_tensors='tf'))['pooler_output']
output = tf.concat([output, batch_pt], 0)
print(f'Processing sample {start} to {end} ')
i+=1
return output
X_train_pt = compute_bert_pt(X_train, 100)
X_test_pt = compute_bert_pt(X_test, 100)
print(X_train_pt.shape)
print(y_train.shape)
inputs = tf.keras.layers.Input(shape=(n_dim), dtype=tf.float32)
outputs = tf.keras.layers.Dropout(0.1)(inputs)
outputs = tf.keras.layers.Dense(128, activation='relu')(inputs)
outputs = tf.keras.layers.Dropout(0.1)(outputs)
outputs = tf.keras.layers.Dense(1, activation='sigmoid')(outputs)
model = tf.keras.Model(inputs, outputs)
print(model.summary())
model.compile("adam", "binary_crossentropy", metrics=["accuracy"])
history = model.fit(X_train_pt, y_train, batch_size=32, epochs=8, validation_data=(X_test_pt, y_test))
```
|
github_jupyter
|
#install addons for
!pip install tensorflow-addons
!pip install transformers[tf-cpu]
from transformers import BertTokenizer, TFBertModel
from tensorflow import keras
from tensorflow.keras import layers
import tensorflow as tf
import numpy as np
from sklearn.model_selection import train_test_split
import matplotlib.pyplot as plt
from tensorflow.keras.layers import Embedding, Dense, Input, Dropout, LayerNormalization, GlobalAveragePooling1D, Flatten
from tensorflow_addons.layers import MultiHeadAttention
from tensorflow.keras.preprocessing.text import Tokenizer
from tensorflow.keras.preprocessing.sequence import pad_sequences
import csv
import os
import shutil
# Download the data from Kaggle https://www.kaggle.com/lakshmi25npathi/imdb-dataset-of-50k-movie-reviews/tasks?taskId=588
# Load the 1mdb datasets from Kaggle
kaggle_imdb_file = 'datasets/IMDB_Dataset.csv'
data_x = []
data_y = []
with open(kaggle_imdb_file, 'r') as csvfile:
filereader = csv.reader(csvfile, delimiter=',', dialect='excel')
next(filereader)
for row in filereader:
data_x.append(row[0])
label = 1 if row[1] == 'positive' else 0
data_y.append(label)
# Prepare the data into trainable format
data_x = np.array(data_x)
data_y = np.array(data_y)
X_train, X_test, y_train, y_test = train_test_split(data_x, data_y, test_size=0.2, random_state=29)
# Generate Pre-trained outputs
tokenizer = BertTokenizer.from_pretrained('bert-base-uncased')
bert_model = TFBertModel.from_pretrained("bert-base-uncased")
n_dim = 768
def compute_bert_pt(text_data, batch_size):
i = 0;
output = tf.reshape(tf.constant([]), (0, n_dim))
while i*batch_size < text_data.shape[0]:
start = i*batch_size
end = (i+1)*batch_size
if end > text_data.shape[0]:
end = text_data.shape[0]
batch_pt = bert_model(tokenizer(text_data[start:end].tolist(), max_length = 512, pad_to_max_length=True, truncation=True, return_tensors='tf'))['pooler_output']
output = tf.concat([output, batch_pt], 0)
print(f'Processing sample {start} to {end} ')
i+=1
return output
X_train_pt = compute_bert_pt(X_train, 100)
X_test_pt = compute_bert_pt(X_test, 100)
print(X_train_pt.shape)
print(y_train.shape)
inputs = tf.keras.layers.Input(shape=(n_dim), dtype=tf.float32)
outputs = tf.keras.layers.Dropout(0.1)(inputs)
outputs = tf.keras.layers.Dense(128, activation='relu')(inputs)
outputs = tf.keras.layers.Dropout(0.1)(outputs)
outputs = tf.keras.layers.Dense(1, activation='sigmoid')(outputs)
model = tf.keras.Model(inputs, outputs)
print(model.summary())
model.compile("adam", "binary_crossentropy", metrics=["accuracy"])
history = model.fit(X_train_pt, y_train, batch_size=32, epochs=8, validation_data=(X_test_pt, y_test))
| 0.773644 | 0.293825 |
<a href="https://colab.research.google.com/github/kaindoh/Sendy-zindi-project/blob/master/sendy.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
```
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn.preprocessing import StandardScaler, RobustScaler, MinMaxScaler
from sklearn.model_selection import train_test_split, KFold, StratifiedKFold, GridSearchCV, RandomizedSearchCV,cross_val_score
from sklearn.ensemble import RandomForestRegressor, AdaBoostRegressor, GradientBoostingRegressor,ExtraTreesRegressor
from xgboost import XGBRFRegressor
import lightgbm as lgb
import xgboost as xgb
import requests
from io import StringIO
import hyperopt
import datetime
import datetime as dt
import warnings
warnings.filterwarnings('ignore')
train = 'https://drive.google.com/file/d/1ZP9pFBATu38l97Tut5hKLvpzKRLFXX_P/view?usp=sharing'
test = 'https://drive.google.com/file/d/1_aElMoEIRs55avOafA7U1_YXEuaDBXLh/view?usp=sharing'
submission = 'https://drive.google.com/file/d/1mqXS8euMqF9_bhTEU6O9cLHoX2FI_5HD/view?usp=sharing'
dictionary = 'https://drive.google.com/file/d/1juqltwSs6OXQgJJEhTxD7Gm443fnLpCp/view?usp=sharing'
riders = 'https://drive.google.com/file/d/19-aVgAcKRxX_Tk9StUQMNeAUVi0ZTo9K/view?usp=sharing'
def read_csv(url):
url = 'https://drive.google.com/uc?export=download&id=' + url.split('/')[-2]
csv_raw = requests.get(url).text
csv = StringIO(csv_raw)
return csv
df = pd.read_csv(read_csv(train))
df1 = pd.read_csv(read_csv(test))
sub = pd.read_csv(read_csv(submission))
dictionary = pd.read_csv(read_csv(dictionary))
riders = pd.read_csv(read_csv(riders))
```
### Cleaning column names
```
df.columns
df.columns = df.columns.str.lower().str.replace(' ', '_').str.replace('-', '_').str.replace('=', '_')
df.columns = df.columns.str.replace('__', '_')
df.columns = df.columns.str.replace('(', '').str.replace(')', '')
df.columns = df.columns.str.replace('__', '_')
df.columns
# Arrival at Destination - Day of Month
# Arrival at Destination - Weekday (Mo = 1)
# Arrival at Destination - Time
# Time from Pickup to Arrival
df.drop(['arrival_at_destination_day_of_month', 'arrival_at_destination_weekday_mo_1','arrival_at_destination_time'],axis=1,inplace=True)
df1.columns = df1.columns.str.lower().str.replace(' ', '_').str.replace('-', '_').str.replace('=', '_')
df1.columns = df1.columns.str.replace('__', '_')
df1.columns = df1.columns.str.replace('(', '').str.replace(')', '')
df1.columns = df1.columns.str.replace('__', '_')
df1.columns
```
### Ordering the dataset
```
df.columns
# #Stripping the order no to have whole numbers
# df['order_no']= df['order_no'].str.replace('Order_No_', '')
# #Changing the data types
# df['order_no']=df['order_no'].astype(int)
# df.sort_values(by=['order_no'], ascending= True)
# df1['order_no']= df1['order_no'].str.replace('Order_No_', '')
# #Changing the data types
# df1['order_no']=df1['order_no'].astype(int)
# df1.sort_values(by=['order_no'], ascending= True)
df['personal_or_business']=df['personal_or_business'].astype("category")
df1['personal_or_business']=df1['personal_or_business'].astype("category")
```
### Filling Temperature missing values
```
df['temperature'].fillna(df['temperature'].mean(), inplace= True)
df1['temperature'].fillna(df['temperature'].mean(), inplace= True)
# #Forward Fill Temprature Column
# df1['temperature'].fillna(method='ffill', inplace= True)
# #Backward Fill Temprature column
# df1['temperature'].fillna(method='bfill', inplace= True)
#Filling in the Precipitation column with 0
df['precipitation_in_millimeters'].fillna(0, inplace= True)
#Filling in the Precipitation column with 0
df1['precipitation_in_millimeters'].fillna(0, inplace= True)
df.head()
df1.head()
df.isnull().sum()
df1.isnull().sum()
```
### Creating hour and minute variables
```
time_cols = ['placement_time', 'confirmation_time', 'pickup_time']
for col in time_cols:
df[col] = pd.to_datetime(df[col])
df[col.split('_')[0] + '_hour'] = df[col].dt.hour
df[col.split('_')[0] + '_minute'] = df[col].dt.minute
df[col] = [time.time() for time in df[col]]
time_cols = ['placement_time', 'confirmation_time', 'pickup_time']
for col in time_cols:
df1[col] = pd.to_datetime(df1[col])
df1[col.split('_')[0] + '_hour'] = df1[col].dt.hour
df1[col.split('_')[0] + '_minute'] = df1[col].dt.minute
df1[col] = [time.time() for time in df1[col]]
time_col = ["arrival_at_pickup_time"]
for col in time_col:
df[col] = pd.to_datetime(df[col])
df[col + '_hour'] = df[col].dt.hour
df[col + '_minute'] = df[col].dt.minute
df[col] = [time.time() for time in df[col]]
time_col = ["arrival_at_pickup_time",]
for col in time_col:
df1[col] = pd.to_datetime(df1[col])
df1[col + '_hour'] = df1[col].dt.hour
df1[col + '_minute'] = df1[col].dt.minute
df1[col] = [time.time() for time in df1[col]]
df.head()
```
### Creating peak and offpeak variables
```
df.columns
#7-5
hour_cols = ['placement_hour', 'confirmation_hour', 'pickup_hour',"arrival_at_pickup_time_hour"]
for col in hour_cols:
df[col.split('_')[0] + '_ap'] = df[col].apply(lambda x: 1 if( x >= 7 & x <= 17) else 0)
#7-5
hour_cols = ['placement_hour', 'confirmation_hour', 'pickup_hour',"arrival_at_pickup_time_hour"]
for col in hour_cols:
df1[col.split('_')[0] + '_ap'] = df1[col].apply(lambda x: 1 if( x >= 7 & x <= 17) else 0)
```
### Creating new cluster variables
```
from sklearn.cluster import KMeans
kmeans = KMeans(n_clusters = 15, init ='k-means++')
kmeans.fit(df[['pickup_lat', 'pickup_long']]) # Compute k-means clustering.
df['pickup_cluster_label'] = kmeans.fit_predict(df[['pickup_lat', 'pickup_long']])
centers1 = kmeans.cluster_centers_ # Coordinates of cluster centers.
labels1 = kmeans.predict(df[['pickup_lat', 'pickup_long']]) # Labels of each point
df['pickup_cluster_label'] = df['pickup_cluster_label'].astype('category')
kmeans = KMeans(n_clusters = 15, init ='k-means++')
kmeans.fit(df[['destination_lat', 'destination_long']]) # Compute k-means clustering.
df['destination_cluster_label'] = kmeans.fit_predict(df[['destination_lat', 'destination_long']])
centers = kmeans.cluster_centers_ # Coordinates of cluster centers.
labels = kmeans.predict(df[['destination_lat', 'destination_long']]) # Labels of each point
df['destination_cluster_label'] =df['destination_cluster_label'].astype('category')
from sklearn.cluster import KMeans
kmeans = KMeans(n_clusters = 15, init ='k-means++')
kmeans.fit(df1[['pickup_lat', 'pickup_long']]) # Compute k-means clustering.
df1['pickup_cluster_label'] = kmeans.fit_predict(df1[['pickup_lat', 'pickup_long']])
centers1 = kmeans.cluster_centers_ # Coordinates of cluster centers.
labels1 = kmeans.predict(df1[['pickup_lat', 'pickup_long']]) # Labels of each point
df1['pickup_cluster_label'] = df1['pickup_cluster_label'].astype('category')
kmeans = KMeans(n_clusters = 15, init ='k-means++')
kmeans.fit(df1[['destination_lat', 'destination_long']]) # Compute k-means clustering.
df1['destination_cluster_label'] = kmeans.fit_predict(df1[['destination_lat', 'destination_long']])
centers = kmeans.cluster_centers_ # Coordinates of cluster centers.
labels = kmeans.predict(df1[['destination_lat', 'destination_long']]) # Labels of each point
df1['destination_cluster_label'] =df1['destination_cluster_label'].astype('category')
df.head()
```
# Cyclic of time
```
# x = df.drop(['user_id','vehicle_type', 'confirmation_time', 'arrival_at_pickup_time','pickup_time','placement_time','arrival_at_destination_day_of_month', 'arrival_at_destination_weekday_mo_1', 'arrival_at_destination_time','rider_id'],axis=1)
# X = pd.get_dummies(x)
# y = df['time_from_pickup_to_arrival']
# train,test,y_train,y_test = train_test_split(X,y,random_state =0 ,test_size = 0.2)
train = df.copy()
test = df1.copy()
test.head()
test.dtypes
train['placement_hourfloat']=train.placement_hour+train.placement_minute/60.0
train['confirmation_hourfloat']=train.confirmation_hour+train.confirmation_minute/60.0
train['pickup_hourfloat']=train.pickup_hour+train.pickup_minute/60.0
train['arrival_hourfloat']=train.arrival_at_pickup_time_hour+train.arrival_at_pickup_time_minute/60.0
train['placement_x']=np.sin(2.*np.pi*train.placement_hourfloat/24.)
train['confirmation_x']=np.sin(2.*np.pi*train.confirmation_hourfloat/24.)
train['arrival_x']=np.sin(2.*np.pi*train.arrival_hourfloat/24.)
train['pickup_x']=np.sin(2.*np.pi*train.pickup_hourfloat/24.)
train['placement_y']=np.cos(2.*np.pi*train.placement_hourfloat/24.)
train['confirmation_y']=np.cos(2.*np.pi*train.confirmation_hourfloat/24.)
train['arrival_y']=np.cos(2.*np.pi*train.arrival_hourfloat/24.)
train['pickup_y']=np.cos(2.*np.pi*train.pickup_hourfloat/24.)
# TEST
test['placement_hourfloat']=test.placement_hour+test.placement_minute/60.0
test['confirmation_hourfloat']=test.confirmation_hour+test.confirmation_minute/60.0
test['pickup_hourfloat']=test.pickup_hour+test.pickup_minute/60.0
test['arrival_hourfloat']=test.arrival_at_pickup_time_hour+test.arrival_at_pickup_time_minute/60.0
test['placement_x']=np.sin(2.*np.pi*test.placement_hourfloat/24.)
test['confirmation_x']=np.sin(2.*np.pi*test.confirmation_hourfloat/24.)
test['arrival_x']=np.sin(2.*np.pi*test.arrival_hourfloat/24.)
test['pickup_x']=np.sin(2.*np.pi*test.pickup_hourfloat/24.)
test['placement_y']=np.cos(2.*np.pi*test.placement_hourfloat/24.)
test['confirmation_y']=np.cos(2.*np.pi*test.confirmation_hourfloat/24.)
test['arrival_y']=np.cos(2.*np.pi*test.arrival_hourfloat/24.)
test['pickup_y']=np.cos(2.*np.pi*test.pickup_hourfloat/24.)
train.columns
x = train.drop(['time_from_pickup_to_arrival','order_no','user_id','vehicle_type', 'confirmation_time', 'arrival_at_pickup_time','pickup_time','placement_time','rider_id'],axis=1)
X = pd.get_dummies(x)
y = train["time_from_pickup_to_arrival"]
test1 = test.drop(['user_id','vehicle_type','order_no', 'confirmation_time', 'arrival_at_pickup_time','pickup_time','placement_time','rider_id'],axis=1)
test2 = pd.get_dummies(test1)
from sklearn.decomposition import PCA
from sklearn.preprocessing import StandardScaler,MinMaxScaler
X.shape
test2.shape
sc =StandardScaler()
X = sc.fit_transform(X)
test = sc.transform(test2)
pca =PCA()
X = pca.fit_transform(X)
test = pca.transform(test2)
pca =PCA()
X = pca.fit_transform(X)
test = pca.transform(test2)
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis as LDA
# lda =LDA()
# X = lda.fit_transform(X,y)
# test = lda.transform(test2)
clf = lgb.LGBMRegressor(learning_rate= 0.1, min_data_in_leaf= 300, n_estimators= 100, num_leaves= 15, objective='regression', reg_alpha= 0.02)
clf.fit(X,y)
pred = clf.predict(test)
clf_output = pd.DataFrame({"Order_No":sub['Order_No'],
"Time from Pickup to Arrival": pred })
clf_output.to_csv("submission6.csv", index=False)
```
|
github_jupyter
|
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn.preprocessing import StandardScaler, RobustScaler, MinMaxScaler
from sklearn.model_selection import train_test_split, KFold, StratifiedKFold, GridSearchCV, RandomizedSearchCV,cross_val_score
from sklearn.ensemble import RandomForestRegressor, AdaBoostRegressor, GradientBoostingRegressor,ExtraTreesRegressor
from xgboost import XGBRFRegressor
import lightgbm as lgb
import xgboost as xgb
import requests
from io import StringIO
import hyperopt
import datetime
import datetime as dt
import warnings
warnings.filterwarnings('ignore')
train = 'https://drive.google.com/file/d/1ZP9pFBATu38l97Tut5hKLvpzKRLFXX_P/view?usp=sharing'
test = 'https://drive.google.com/file/d/1_aElMoEIRs55avOafA7U1_YXEuaDBXLh/view?usp=sharing'
submission = 'https://drive.google.com/file/d/1mqXS8euMqF9_bhTEU6O9cLHoX2FI_5HD/view?usp=sharing'
dictionary = 'https://drive.google.com/file/d/1juqltwSs6OXQgJJEhTxD7Gm443fnLpCp/view?usp=sharing'
riders = 'https://drive.google.com/file/d/19-aVgAcKRxX_Tk9StUQMNeAUVi0ZTo9K/view?usp=sharing'
def read_csv(url):
url = 'https://drive.google.com/uc?export=download&id=' + url.split('/')[-2]
csv_raw = requests.get(url).text
csv = StringIO(csv_raw)
return csv
df = pd.read_csv(read_csv(train))
df1 = pd.read_csv(read_csv(test))
sub = pd.read_csv(read_csv(submission))
dictionary = pd.read_csv(read_csv(dictionary))
riders = pd.read_csv(read_csv(riders))
df.columns
df.columns = df.columns.str.lower().str.replace(' ', '_').str.replace('-', '_').str.replace('=', '_')
df.columns = df.columns.str.replace('__', '_')
df.columns = df.columns.str.replace('(', '').str.replace(')', '')
df.columns = df.columns.str.replace('__', '_')
df.columns
# Arrival at Destination - Day of Month
# Arrival at Destination - Weekday (Mo = 1)
# Arrival at Destination - Time
# Time from Pickup to Arrival
df.drop(['arrival_at_destination_day_of_month', 'arrival_at_destination_weekday_mo_1','arrival_at_destination_time'],axis=1,inplace=True)
df1.columns = df1.columns.str.lower().str.replace(' ', '_').str.replace('-', '_').str.replace('=', '_')
df1.columns = df1.columns.str.replace('__', '_')
df1.columns = df1.columns.str.replace('(', '').str.replace(')', '')
df1.columns = df1.columns.str.replace('__', '_')
df1.columns
df.columns
# #Stripping the order no to have whole numbers
# df['order_no']= df['order_no'].str.replace('Order_No_', '')
# #Changing the data types
# df['order_no']=df['order_no'].astype(int)
# df.sort_values(by=['order_no'], ascending= True)
# df1['order_no']= df1['order_no'].str.replace('Order_No_', '')
# #Changing the data types
# df1['order_no']=df1['order_no'].astype(int)
# df1.sort_values(by=['order_no'], ascending= True)
df['personal_or_business']=df['personal_or_business'].astype("category")
df1['personal_or_business']=df1['personal_or_business'].astype("category")
df['temperature'].fillna(df['temperature'].mean(), inplace= True)
df1['temperature'].fillna(df['temperature'].mean(), inplace= True)
# #Forward Fill Temprature Column
# df1['temperature'].fillna(method='ffill', inplace= True)
# #Backward Fill Temprature column
# df1['temperature'].fillna(method='bfill', inplace= True)
#Filling in the Precipitation column with 0
df['precipitation_in_millimeters'].fillna(0, inplace= True)
#Filling in the Precipitation column with 0
df1['precipitation_in_millimeters'].fillna(0, inplace= True)
df.head()
df1.head()
df.isnull().sum()
df1.isnull().sum()
time_cols = ['placement_time', 'confirmation_time', 'pickup_time']
for col in time_cols:
df[col] = pd.to_datetime(df[col])
df[col.split('_')[0] + '_hour'] = df[col].dt.hour
df[col.split('_')[0] + '_minute'] = df[col].dt.minute
df[col] = [time.time() for time in df[col]]
time_cols = ['placement_time', 'confirmation_time', 'pickup_time']
for col in time_cols:
df1[col] = pd.to_datetime(df1[col])
df1[col.split('_')[0] + '_hour'] = df1[col].dt.hour
df1[col.split('_')[0] + '_minute'] = df1[col].dt.minute
df1[col] = [time.time() for time in df1[col]]
time_col = ["arrival_at_pickup_time"]
for col in time_col:
df[col] = pd.to_datetime(df[col])
df[col + '_hour'] = df[col].dt.hour
df[col + '_minute'] = df[col].dt.minute
df[col] = [time.time() for time in df[col]]
time_col = ["arrival_at_pickup_time",]
for col in time_col:
df1[col] = pd.to_datetime(df1[col])
df1[col + '_hour'] = df1[col].dt.hour
df1[col + '_minute'] = df1[col].dt.minute
df1[col] = [time.time() for time in df1[col]]
df.head()
df.columns
#7-5
hour_cols = ['placement_hour', 'confirmation_hour', 'pickup_hour',"arrival_at_pickup_time_hour"]
for col in hour_cols:
df[col.split('_')[0] + '_ap'] = df[col].apply(lambda x: 1 if( x >= 7 & x <= 17) else 0)
#7-5
hour_cols = ['placement_hour', 'confirmation_hour', 'pickup_hour',"arrival_at_pickup_time_hour"]
for col in hour_cols:
df1[col.split('_')[0] + '_ap'] = df1[col].apply(lambda x: 1 if( x >= 7 & x <= 17) else 0)
from sklearn.cluster import KMeans
kmeans = KMeans(n_clusters = 15, init ='k-means++')
kmeans.fit(df[['pickup_lat', 'pickup_long']]) # Compute k-means clustering.
df['pickup_cluster_label'] = kmeans.fit_predict(df[['pickup_lat', 'pickup_long']])
centers1 = kmeans.cluster_centers_ # Coordinates of cluster centers.
labels1 = kmeans.predict(df[['pickup_lat', 'pickup_long']]) # Labels of each point
df['pickup_cluster_label'] = df['pickup_cluster_label'].astype('category')
kmeans = KMeans(n_clusters = 15, init ='k-means++')
kmeans.fit(df[['destination_lat', 'destination_long']]) # Compute k-means clustering.
df['destination_cluster_label'] = kmeans.fit_predict(df[['destination_lat', 'destination_long']])
centers = kmeans.cluster_centers_ # Coordinates of cluster centers.
labels = kmeans.predict(df[['destination_lat', 'destination_long']]) # Labels of each point
df['destination_cluster_label'] =df['destination_cluster_label'].astype('category')
from sklearn.cluster import KMeans
kmeans = KMeans(n_clusters = 15, init ='k-means++')
kmeans.fit(df1[['pickup_lat', 'pickup_long']]) # Compute k-means clustering.
df1['pickup_cluster_label'] = kmeans.fit_predict(df1[['pickup_lat', 'pickup_long']])
centers1 = kmeans.cluster_centers_ # Coordinates of cluster centers.
labels1 = kmeans.predict(df1[['pickup_lat', 'pickup_long']]) # Labels of each point
df1['pickup_cluster_label'] = df1['pickup_cluster_label'].astype('category')
kmeans = KMeans(n_clusters = 15, init ='k-means++')
kmeans.fit(df1[['destination_lat', 'destination_long']]) # Compute k-means clustering.
df1['destination_cluster_label'] = kmeans.fit_predict(df1[['destination_lat', 'destination_long']])
centers = kmeans.cluster_centers_ # Coordinates of cluster centers.
labels = kmeans.predict(df1[['destination_lat', 'destination_long']]) # Labels of each point
df1['destination_cluster_label'] =df1['destination_cluster_label'].astype('category')
df.head()
# x = df.drop(['user_id','vehicle_type', 'confirmation_time', 'arrival_at_pickup_time','pickup_time','placement_time','arrival_at_destination_day_of_month', 'arrival_at_destination_weekday_mo_1', 'arrival_at_destination_time','rider_id'],axis=1)
# X = pd.get_dummies(x)
# y = df['time_from_pickup_to_arrival']
# train,test,y_train,y_test = train_test_split(X,y,random_state =0 ,test_size = 0.2)
train = df.copy()
test = df1.copy()
test.head()
test.dtypes
train['placement_hourfloat']=train.placement_hour+train.placement_minute/60.0
train['confirmation_hourfloat']=train.confirmation_hour+train.confirmation_minute/60.0
train['pickup_hourfloat']=train.pickup_hour+train.pickup_minute/60.0
train['arrival_hourfloat']=train.arrival_at_pickup_time_hour+train.arrival_at_pickup_time_minute/60.0
train['placement_x']=np.sin(2.*np.pi*train.placement_hourfloat/24.)
train['confirmation_x']=np.sin(2.*np.pi*train.confirmation_hourfloat/24.)
train['arrival_x']=np.sin(2.*np.pi*train.arrival_hourfloat/24.)
train['pickup_x']=np.sin(2.*np.pi*train.pickup_hourfloat/24.)
train['placement_y']=np.cos(2.*np.pi*train.placement_hourfloat/24.)
train['confirmation_y']=np.cos(2.*np.pi*train.confirmation_hourfloat/24.)
train['arrival_y']=np.cos(2.*np.pi*train.arrival_hourfloat/24.)
train['pickup_y']=np.cos(2.*np.pi*train.pickup_hourfloat/24.)
# TEST
test['placement_hourfloat']=test.placement_hour+test.placement_minute/60.0
test['confirmation_hourfloat']=test.confirmation_hour+test.confirmation_minute/60.0
test['pickup_hourfloat']=test.pickup_hour+test.pickup_minute/60.0
test['arrival_hourfloat']=test.arrival_at_pickup_time_hour+test.arrival_at_pickup_time_minute/60.0
test['placement_x']=np.sin(2.*np.pi*test.placement_hourfloat/24.)
test['confirmation_x']=np.sin(2.*np.pi*test.confirmation_hourfloat/24.)
test['arrival_x']=np.sin(2.*np.pi*test.arrival_hourfloat/24.)
test['pickup_x']=np.sin(2.*np.pi*test.pickup_hourfloat/24.)
test['placement_y']=np.cos(2.*np.pi*test.placement_hourfloat/24.)
test['confirmation_y']=np.cos(2.*np.pi*test.confirmation_hourfloat/24.)
test['arrival_y']=np.cos(2.*np.pi*test.arrival_hourfloat/24.)
test['pickup_y']=np.cos(2.*np.pi*test.pickup_hourfloat/24.)
train.columns
x = train.drop(['time_from_pickup_to_arrival','order_no','user_id','vehicle_type', 'confirmation_time', 'arrival_at_pickup_time','pickup_time','placement_time','rider_id'],axis=1)
X = pd.get_dummies(x)
y = train["time_from_pickup_to_arrival"]
test1 = test.drop(['user_id','vehicle_type','order_no', 'confirmation_time', 'arrival_at_pickup_time','pickup_time','placement_time','rider_id'],axis=1)
test2 = pd.get_dummies(test1)
from sklearn.decomposition import PCA
from sklearn.preprocessing import StandardScaler,MinMaxScaler
X.shape
test2.shape
sc =StandardScaler()
X = sc.fit_transform(X)
test = sc.transform(test2)
pca =PCA()
X = pca.fit_transform(X)
test = pca.transform(test2)
pca =PCA()
X = pca.fit_transform(X)
test = pca.transform(test2)
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis as LDA
# lda =LDA()
# X = lda.fit_transform(X,y)
# test = lda.transform(test2)
clf = lgb.LGBMRegressor(learning_rate= 0.1, min_data_in_leaf= 300, n_estimators= 100, num_leaves= 15, objective='regression', reg_alpha= 0.02)
clf.fit(X,y)
pred = clf.predict(test)
clf_output = pd.DataFrame({"Order_No":sub['Order_No'],
"Time from Pickup to Arrival": pred })
clf_output.to_csv("submission6.csv", index=False)
| 0.52074 | 0.826222 |
```
import os
os.chdir('..')
os.chdir('..')
print(os.getcwd())
import rsnapsim as rss
import numpy as np
os.chdir('rsnapsim')
os.chdir('interactive_notebooks')
import numpy as np
import matplotlib.pyplot as plt
import time
poi_strs, poi_objs, tagged_pois,raw_seq = rss.seqmanip.open_seq_file('../gene_files/H2B_withTags.txt')
poi = tagged_pois['1'][0] #protein object
poi.tag_epitopes['T_Flag'] = [10,20,30,40,50,60,70]
poi.tag_epitopes['T_Hemagglutinin'] = [300,330,340,350]
plt.style.use('dark_background')
plt.rcParams['figure.dpi'] = 120
plt.rcParams['lines.linewidth'] = 1
plt.rcParams['axes.linewidth'] = 1.5
plt.rcParams['font.size'] = 15
plt.rcParams['axes.grid'] = False
colors = ['#00ff51', '#00f7ff']
rss.solver.protein=poi
t = np.linspace(0,500,501)
poi.visualize_probe(colors=['#00ff51', '#00f7ff'])
sttime = time.time()
ssa_soln = rss.solver.solve_ssa(poi.kelong,t,ki=.033,n_traj=20)
solvetime = time.time()-sttime
print(ssa_soln.intensity_vec.shape)
plt.plot(np.mean(ssa_soln.intensity_vec[0],axis=1),color='#00ff51',alpha=.8)
plt.plot(np.mean(ssa_soln.intensity_vec[1],axis=1),color='#00f7ff',alpha=.8)
plt.xlabel('time')
plt.ylabel('intensity')
print("Low memory, no recording: solved in %f seconds" % solvetime)
```
## Autocovariances with individual means
```
acov,err_acov = rss.inta.get_autocov(ssa_soln.intensity_vec,norm='ind')
plt.plot(np.mean(acov[0],axis=1),color=colors[0]);plt.plot(np.mean(acov[1],axis=1),color=colors[1])
plt.plot(np.mean(acov[0],axis=1) - err_acov[0],'--',color=colors[0]);plt.plot(np.mean(acov[1],axis=1)- err_acov[1],'--',color=colors[1])
plt.plot(np.mean(acov[0],axis=1)+ err_acov[0],'--',color=colors[0]);plt.plot(np.mean(acov[1],axis=1)+ err_acov[1],'--',color=colors[1])
plt.plot([0,500],[0,0],'r--')
plt.xlim([0,100])
plt.xlabel('tau')
plt.ylabel('G(tau)')
#normalized by G0
acc,acc_err = rss.inta.get_autocorr(acov)
n_traj = acc.shape[-1]
err_acov = 1.0/np.sqrt(n_traj)*np.std(acc,ddof=1,axis=2)
plt.plot(np.mean(acc[0],axis=1),color=colors[0]);plt.plot(np.mean(acc[1],axis=1),color=colors[1])
plt.plot(np.mean(acc[0],axis=1) - err_acov[0],'--',color=colors[0]);plt.plot(np.mean(acc[1],axis=1)- err_acov[1],'--',color=colors[1])
plt.plot(np.mean(acc[0],axis=1)+ err_acov[0],'--',color=colors[0]);plt.plot(np.mean(acc[1],axis=1)+ err_acov[1],'--',color=colors[1])
plt.plot([0,500],[0,0],'r--')
plt.xlim([0,100])
plt.xlabel('tau')
plt.ylabel('G(tau)')
```
## Global means
```
acov,err_acov = rss.inta.get_autocov(ssa_soln.intensity_vec,norm='global')
plt.plot(np.mean(acov[0],axis=1),color='seagreen');plt.plot(np.mean(acov[1],axis=1),color='violet')
plt.plot(np.mean(acov[0],axis=1) - err_acov[0],'--',color='seagreen');plt.plot(np.mean(acov[1],axis=1)- err_acov[1],'--',color='violet')
plt.plot(np.mean(acov[0],axis=1)+ err_acov[0],'--',color='seagreen');plt.plot(np.mean(acov[1],axis=1)+ err_acov[1],'--',color='violet')
plt.plot([0,500],[0,0],'r--')
plt.xlim([0,100])
#normalized by G0
acc,acc_error = rss.inta.get_autocorr(acov,g0='G1')
mean_acc = np.mean(acc,axis=2)
plt.plot(mean_acc[0],color='seagreen');plt.plot(mean_acc[1],color='violet')
plt.plot(np.mean(acc[0],axis=1) - acc_error[0],'--',color='seagreen');plt.plot(np.mean(acc[1],axis=1)- acc_error[1],'--',color='violet')
plt.plot(np.mean(acc[0],axis=1)+ acc_error[0],'--',color='seagreen');plt.plot(np.mean(acc[1],axis=1)+ acc_error[1],'--',color='violet')
plt.plot([0,500],[0,0],'r--')
plt.xlim([0,100])
```
## Cross correlations
```
cross_corr,err_cc,inds = rss.inta.get_crosscorr(ssa_soln.intensity_vec,norm='indiv')
plt.figure()
s11_cc = np.mean(cross_corr[0],axis=1)
s12_cc = np.mean(cross_corr[1],axis=1)
s21_cc = np.mean(cross_corr[2],axis=1)
s22_cc = np.mean(cross_corr[3],axis=1)
plt.plot(s11_cc/s11_cc[500],color=colors[0] );
plt.plot(s21_cc/s21_cc[500],color='#ff00ee');
plt.plot(s22_cc/s22_cc[500],color=colors[1]);
plt.plot(s11_cc/s11_cc[500] - err_cc[0]/s11_cc[500],'--',color=colors[0] );
plt.plot(s11_cc/s11_cc[500] + err_cc[0]/s11_cc[500],'--',color=colors[0] );
plt.plot(s21_cc/s21_cc[500] - err_cc[2]/s21_cc[500] ,'--',color='#ff00ee' );
plt.plot(s21_cc/s21_cc[500] + err_cc[2]/s21_cc[500] ,'--',color='#ff00ee');
plt.plot(s22_cc/s22_cc[500] - s22_cc[3]/s22_cc[500],'--',color=colors[1] );
plt.plot(s22_cc/s22_cc[500] + s22_cc[3]/s22_cc[500],'--',color=colors[1] );
plt.plot([500,500],[0,1.1],'r--')
plt.plot([400,600],[0,0],'r--')
plt.legend(['00','10','11' ])
plt.xlim([400,600])
plt.xlabel('tau')
plt.ylabel('G(tau)')
```
## normalization modes
| norm | effect |
| :- | :-: |
| global | subtract all intensities by the global mean intensity before correlation |
| individual | subtract all intensities by the trajectory mean intensity before correlation |
| raw | do nothing, correlate the intensities as they are |
## G0
| norm | effect |
| :- | :-: |
| global_max | divide correlations by the global maximum point |
| individual_max | divide correlations by the individual trajectory maximum point |
| global_center | divide correlations by the global average of the center point of the correlation |
| individual_center | divide all correlations by the trajectory center point value |
| None | do nothing, do not normalize the correlations by anything|
```
cross_corr,err_cc,inds = rss.inta.get_crosscorr(ssa_soln.intensity_vec,norm='indiv',g0=None)
plt.figure()
plt.plot(cross_corr[0], color = colors[0],alpha=.5)
plt.plot(cross_corr[2],color = '#ff00ee',alpha=.5)
plt.plot(cross_corr[3], color = colors[1],alpha=.5)
s11_cc = np.mean(cross_corr[0],axis=1)
s12_cc = np.mean(cross_corr[1],axis=1)
s21_cc = np.mean(cross_corr[2],axis=1)
s22_cc = np.mean(cross_corr[3],axis=1)
plt.plot([500,500],[0,1.1],'r--')
plt.plot([400,600],[0,0],'r--')
plt.legend(['00','10','11' ])
plt.xlim([400,600])
plt.xlabel('tau')
plt.ylabel('G(tau)')
cross_corr,err_cc,inds = rss.inta.get_crosscorr(ssa_soln.intensity_vec,norm='global',g0='indiv_max')
plt.figure()
plt.plot(cross_corr[0], color = colors[0],alpha=.5)
plt.plot(cross_corr[2],color = '#ff00ee',alpha=.5)
plt.plot(cross_corr[3], color = colors[1],alpha=.5)
s11_cc = np.mean(cross_corr[0],axis=1)
s12_cc = np.mean(cross_corr[1],axis=1)
s21_cc = np.mean(cross_corr[2],axis=1)
s22_cc = np.mean(cross_corr[3],axis=1)
plt.plot([500,500],[0,1.1],'r--')
plt.plot([400,600],[0,0],'r--')
plt.legend(['00','10','11' ])
plt.xlim([400,600])
plt.xlabel('tau')
plt.ylabel('G(tau)')
```
|
github_jupyter
|
import os
os.chdir('..')
os.chdir('..')
print(os.getcwd())
import rsnapsim as rss
import numpy as np
os.chdir('rsnapsim')
os.chdir('interactive_notebooks')
import numpy as np
import matplotlib.pyplot as plt
import time
poi_strs, poi_objs, tagged_pois,raw_seq = rss.seqmanip.open_seq_file('../gene_files/H2B_withTags.txt')
poi = tagged_pois['1'][0] #protein object
poi.tag_epitopes['T_Flag'] = [10,20,30,40,50,60,70]
poi.tag_epitopes['T_Hemagglutinin'] = [300,330,340,350]
plt.style.use('dark_background')
plt.rcParams['figure.dpi'] = 120
plt.rcParams['lines.linewidth'] = 1
plt.rcParams['axes.linewidth'] = 1.5
plt.rcParams['font.size'] = 15
plt.rcParams['axes.grid'] = False
colors = ['#00ff51', '#00f7ff']
rss.solver.protein=poi
t = np.linspace(0,500,501)
poi.visualize_probe(colors=['#00ff51', '#00f7ff'])
sttime = time.time()
ssa_soln = rss.solver.solve_ssa(poi.kelong,t,ki=.033,n_traj=20)
solvetime = time.time()-sttime
print(ssa_soln.intensity_vec.shape)
plt.plot(np.mean(ssa_soln.intensity_vec[0],axis=1),color='#00ff51',alpha=.8)
plt.plot(np.mean(ssa_soln.intensity_vec[1],axis=1),color='#00f7ff',alpha=.8)
plt.xlabel('time')
plt.ylabel('intensity')
print("Low memory, no recording: solved in %f seconds" % solvetime)
acov,err_acov = rss.inta.get_autocov(ssa_soln.intensity_vec,norm='ind')
plt.plot(np.mean(acov[0],axis=1),color=colors[0]);plt.plot(np.mean(acov[1],axis=1),color=colors[1])
plt.plot(np.mean(acov[0],axis=1) - err_acov[0],'--',color=colors[0]);plt.plot(np.mean(acov[1],axis=1)- err_acov[1],'--',color=colors[1])
plt.plot(np.mean(acov[0],axis=1)+ err_acov[0],'--',color=colors[0]);plt.plot(np.mean(acov[1],axis=1)+ err_acov[1],'--',color=colors[1])
plt.plot([0,500],[0,0],'r--')
plt.xlim([0,100])
plt.xlabel('tau')
plt.ylabel('G(tau)')
#normalized by G0
acc,acc_err = rss.inta.get_autocorr(acov)
n_traj = acc.shape[-1]
err_acov = 1.0/np.sqrt(n_traj)*np.std(acc,ddof=1,axis=2)
plt.plot(np.mean(acc[0],axis=1),color=colors[0]);plt.plot(np.mean(acc[1],axis=1),color=colors[1])
plt.plot(np.mean(acc[0],axis=1) - err_acov[0],'--',color=colors[0]);plt.plot(np.mean(acc[1],axis=1)- err_acov[1],'--',color=colors[1])
plt.plot(np.mean(acc[0],axis=1)+ err_acov[0],'--',color=colors[0]);plt.plot(np.mean(acc[1],axis=1)+ err_acov[1],'--',color=colors[1])
plt.plot([0,500],[0,0],'r--')
plt.xlim([0,100])
plt.xlabel('tau')
plt.ylabel('G(tau)')
acov,err_acov = rss.inta.get_autocov(ssa_soln.intensity_vec,norm='global')
plt.plot(np.mean(acov[0],axis=1),color='seagreen');plt.plot(np.mean(acov[1],axis=1),color='violet')
plt.plot(np.mean(acov[0],axis=1) - err_acov[0],'--',color='seagreen');plt.plot(np.mean(acov[1],axis=1)- err_acov[1],'--',color='violet')
plt.plot(np.mean(acov[0],axis=1)+ err_acov[0],'--',color='seagreen');plt.plot(np.mean(acov[1],axis=1)+ err_acov[1],'--',color='violet')
plt.plot([0,500],[0,0],'r--')
plt.xlim([0,100])
#normalized by G0
acc,acc_error = rss.inta.get_autocorr(acov,g0='G1')
mean_acc = np.mean(acc,axis=2)
plt.plot(mean_acc[0],color='seagreen');plt.plot(mean_acc[1],color='violet')
plt.plot(np.mean(acc[0],axis=1) - acc_error[0],'--',color='seagreen');plt.plot(np.mean(acc[1],axis=1)- acc_error[1],'--',color='violet')
plt.plot(np.mean(acc[0],axis=1)+ acc_error[0],'--',color='seagreen');plt.plot(np.mean(acc[1],axis=1)+ acc_error[1],'--',color='violet')
plt.plot([0,500],[0,0],'r--')
plt.xlim([0,100])
cross_corr,err_cc,inds = rss.inta.get_crosscorr(ssa_soln.intensity_vec,norm='indiv')
plt.figure()
s11_cc = np.mean(cross_corr[0],axis=1)
s12_cc = np.mean(cross_corr[1],axis=1)
s21_cc = np.mean(cross_corr[2],axis=1)
s22_cc = np.mean(cross_corr[3],axis=1)
plt.plot(s11_cc/s11_cc[500],color=colors[0] );
plt.plot(s21_cc/s21_cc[500],color='#ff00ee');
plt.plot(s22_cc/s22_cc[500],color=colors[1]);
plt.plot(s11_cc/s11_cc[500] - err_cc[0]/s11_cc[500],'--',color=colors[0] );
plt.plot(s11_cc/s11_cc[500] + err_cc[0]/s11_cc[500],'--',color=colors[0] );
plt.plot(s21_cc/s21_cc[500] - err_cc[2]/s21_cc[500] ,'--',color='#ff00ee' );
plt.plot(s21_cc/s21_cc[500] + err_cc[2]/s21_cc[500] ,'--',color='#ff00ee');
plt.plot(s22_cc/s22_cc[500] - s22_cc[3]/s22_cc[500],'--',color=colors[1] );
plt.plot(s22_cc/s22_cc[500] + s22_cc[3]/s22_cc[500],'--',color=colors[1] );
plt.plot([500,500],[0,1.1],'r--')
plt.plot([400,600],[0,0],'r--')
plt.legend(['00','10','11' ])
plt.xlim([400,600])
plt.xlabel('tau')
plt.ylabel('G(tau)')
cross_corr,err_cc,inds = rss.inta.get_crosscorr(ssa_soln.intensity_vec,norm='indiv',g0=None)
plt.figure()
plt.plot(cross_corr[0], color = colors[0],alpha=.5)
plt.plot(cross_corr[2],color = '#ff00ee',alpha=.5)
plt.plot(cross_corr[3], color = colors[1],alpha=.5)
s11_cc = np.mean(cross_corr[0],axis=1)
s12_cc = np.mean(cross_corr[1],axis=1)
s21_cc = np.mean(cross_corr[2],axis=1)
s22_cc = np.mean(cross_corr[3],axis=1)
plt.plot([500,500],[0,1.1],'r--')
plt.plot([400,600],[0,0],'r--')
plt.legend(['00','10','11' ])
plt.xlim([400,600])
plt.xlabel('tau')
plt.ylabel('G(tau)')
cross_corr,err_cc,inds = rss.inta.get_crosscorr(ssa_soln.intensity_vec,norm='global',g0='indiv_max')
plt.figure()
plt.plot(cross_corr[0], color = colors[0],alpha=.5)
plt.plot(cross_corr[2],color = '#ff00ee',alpha=.5)
plt.plot(cross_corr[3], color = colors[1],alpha=.5)
s11_cc = np.mean(cross_corr[0],axis=1)
s12_cc = np.mean(cross_corr[1],axis=1)
s21_cc = np.mean(cross_corr[2],axis=1)
s22_cc = np.mean(cross_corr[3],axis=1)
plt.plot([500,500],[0,1.1],'r--')
plt.plot([400,600],[0,0],'r--')
plt.legend(['00','10','11' ])
plt.xlim([400,600])
plt.xlabel('tau')
plt.ylabel('G(tau)')
| 0.168651 | 0.675659 |
# word cloud generation
# word cloud- word cloud is a cloud of words in different sizes which represents the frequency or the importance of each word.
# Here, we will see the step by step process to generate word cloud
# points to be covered
1.How to create a basic word cloud from a text document.
2.Adjusting color,size and number of text inside word cloud
3.you can create your word cloud in any shape
4.you can create your word cloud in any color pattern
# Prerequisites
1. numpy
https://docs.scipy.org/doc/numpy-1.15.0/user/install.html
2. pandas
https://pandas.pydata.org/pandas-docs/stable/install.html
3. matplotlib
https://matplotlib.org/users/installing.html
4. pillow
https://pillow.readthedocs.io/en/3.0.x/installation.html
5. word cloud
https://anaconda.org/conda-forge/wordcloud
# How to create a basic word cloud from a text document
```
import nltk
import matplotlib.pyplot as plt
from wordcloud import WordCloud, STOPWORDS
from nltk.corpus import stopwords
from nltk.tokenize import word_tokenize
from nltk.corpus import words
#nltk.download('words')
f=open('tom','r')
stop_words = set(stopwords.words('english'))
#print(stop_words)
word_tokens = word_tokenize(f.read())
#filtered_sentence = [w for w in word_tokens if not w in stop_words]
filtered_sentence ="";
special_chr=[',','{','}','[',']','(',')','#','*']
for w in word_tokens:
if w not in stop_words and w not in special_chr:
filtered_sentence+=w+" "
#print(filtered_sentence)
# Create and generate a word cloud image:
wordcloud = WordCloud().generate(filtered_sentence)
# Display the generated image:
plt.imshow(wordcloud, interpolation='bilinear')
plt.axis("off")
plt.show()
```
# Adjusting color,size and number of text inside word cloud
```
wordcloud = WordCloud(width=600,height=400,min_font_size=7,background_color='black').generate(filtered_sentence)
# Display the generated image:
plt.imshow(wordcloud, interpolation='bilinear')
plt.axis("off")
plt.show()
```
# you can create your word cloud in any shape
```
import numpy as np
from PIL import Image
wave_mask = np.array(Image.open( "twitter.jpg"))
wordcloud = WordCloud(mask=wave_mask,width=900,height=400,min_font_size=7,background_color='black').generate(filtered_sentence)
# Display the generated image:
plt.imshow(wordcloud, interpolation='bilinear')
plt.axis("off")
plt.show()
```
you can create your word cloud in any color pattern
# you can create your word cloud in any color pattern
```
import numpy as np
from PIL import Image
wave_mask = np.array(Image.open( "twitter.jpg"))
wordcloud = WordCloud(mask=wave_mask,width=900,background_color='white',height=400,min_font_size=7).generate(filtered_sentence)
# Display the generated image:
plt.imshow(wordcloud, interpolation='bilinear')
plt.axis("off")
plt.show()
```
|
github_jupyter
|
import nltk
import matplotlib.pyplot as plt
from wordcloud import WordCloud, STOPWORDS
from nltk.corpus import stopwords
from nltk.tokenize import word_tokenize
from nltk.corpus import words
#nltk.download('words')
f=open('tom','r')
stop_words = set(stopwords.words('english'))
#print(stop_words)
word_tokens = word_tokenize(f.read())
#filtered_sentence = [w for w in word_tokens if not w in stop_words]
filtered_sentence ="";
special_chr=[',','{','}','[',']','(',')','#','*']
for w in word_tokens:
if w not in stop_words and w not in special_chr:
filtered_sentence+=w+" "
#print(filtered_sentence)
# Create and generate a word cloud image:
wordcloud = WordCloud().generate(filtered_sentence)
# Display the generated image:
plt.imshow(wordcloud, interpolation='bilinear')
plt.axis("off")
plt.show()
wordcloud = WordCloud(width=600,height=400,min_font_size=7,background_color='black').generate(filtered_sentence)
# Display the generated image:
plt.imshow(wordcloud, interpolation='bilinear')
plt.axis("off")
plt.show()
import numpy as np
from PIL import Image
wave_mask = np.array(Image.open( "twitter.jpg"))
wordcloud = WordCloud(mask=wave_mask,width=900,height=400,min_font_size=7,background_color='black').generate(filtered_sentence)
# Display the generated image:
plt.imshow(wordcloud, interpolation='bilinear')
plt.axis("off")
plt.show()
import numpy as np
from PIL import Image
wave_mask = np.array(Image.open( "twitter.jpg"))
wordcloud = WordCloud(mask=wave_mask,width=900,background_color='white',height=400,min_font_size=7).generate(filtered_sentence)
# Display the generated image:
plt.imshow(wordcloud, interpolation='bilinear')
plt.axis("off")
plt.show()
| 0.217254 | 0.799521 |
# Lecture 48: Global Activation Pooling
### Dataset used:- [ALL-IDB:Acute Lymphoblastic Leukemia Image Database for Image Processing](https://homes.di.unimi.it/scotti/all/)
Follow the instructions provided in the linked website to download the dataset. After downloading, extract the files to the current directory (same folder as your codes).
```
%matplotlib inline
import os
import time
import copy
import tqdm
import torch
import random
import numpy as np
import torch.nn as nn
from PIL import Image
import torch.optim as optim
from torchvision import models
import torch.nn.functional as F
import matplotlib.pyplot as plt
from torch.utils.data import TensorDataset,DataLoader
print(torch.__version__) # This code has been updated for PyTorch 1.0.0
```
### Load Data
```
Datapath = 'ALL_IDB2/img/'
listing = os.listdir(Datapath)
random.shuffle(listing)
# ALL_IDB2 dataset has 260 images in total
TrainImages = torch.FloatTensor(200,3,224,224)
TrainLabels = torch.LongTensor(200)
TestImages = torch.FloatTensor(60,3,224,224)
TestLabels = torch.LongTensor(60)
# First 200 images are used for training and the remaining 60 for testing
img_no = 0
for file in listing:
im=Image.open(Datapath + file)
im = im.resize((224,224))
im = np.array(im)
if img_no < 200:
TrainImages[img_no] = torch.from_numpy(im).transpose(0,2).unsqueeze(0)
TrainLabels[img_no] = int(listing[img_no][6:7])
else:
TestImages[img_no - 200] = torch.from_numpy(im).transpose(0,2).unsqueeze(0)
TestLabels[img_no - 200] = int(listing[img_no][6:7])
img_no = img_no + 1
print(TrainImages.size())
print(TrainLabels.size())
print(TestImages.size())
print(TestLabels.size())
# Check availability of GPU
use_gpu = torch.cuda.is_available()
# use_gpu = False # Uncomment in case of GPU memory error
if use_gpu:
print('GPU is available!')
device = "cuda"
pinMem = True
else:
print('GPU is not available!')
device = "cpu"
pinMem = False
# Creating pytorch dataset
trainDataset = TensorDataset(TrainImages, TrainLabels)
testDataset = TensorDataset(TestImages, TestLabels)
# Creating dataloader
BatchSize = 32
trainLoader = DataLoader(trainDataset, batch_size=BatchSize, shuffle=True,num_workers=4, pin_memory=pinMem)
testLoader = DataLoader(testDataset, batch_size=BatchSize, shuffle=False,num_workers=4, pin_memory=pinMem)
```
### Initialize the network
```
# ResNet18
net = models.resnet18(pretrained=True)
num_ftrs = net.fc.in_features
net.fc = nn.Linear(num_ftrs, 2)
print(net)
net = net.to(device)
```
### Define loss function and optimizer
```
criterion = nn.NLLLoss() # Negative Log-Likelihood
optimizer = optim.SGD(net.parameters(), lr=1e-3 , momentum=0.9) # Stochastic gradient descent
```
### Train the network
```
iterations = 15
trainLoss = []
trainAcc = []
testLoss = []
testAcc = []
start = time.time()
for epoch in range(iterations):
epochStart = time.time()
runningLoss = 0
runningCorr = 0
net.train() # For training
for data in tqdm.tqdm_notebook(trainLoader):
inputs,labels = data
inputs, labels = inputs.to(device), labels.long().to(device)
inputs = inputs/255.0
# Feed-forward input data through the network
outputs = net(inputs)
# Compute loss/error
loss = criterion(F.log_softmax(outputs,dim=1), labels)
_, predicted = torch.max(outputs.data, 1)
# Initialize gradients to zero
optimizer.zero_grad()
# Backpropagate loss and compute gradients
loss.backward()
# Update the network parameters
optimizer.step()
# Accumulate loss per batch
runningLoss += loss.item()
# Accumuate correct predictions per batch
runningCorr += (predicted == labels.data).sum()
avgTrainLoss = runningLoss/(200.0/BatchSize)
avgTrainAcc = 100*float(runningCorr)/200.0
trainLoss.append(avgTrainLoss)
trainAcc.append(avgTrainAcc)
# Evaluating performance on test set for each epoch
net.eval() # For testing
test_runningCorr = 0
test_runningLoss = 0
with torch.no_grad():
for data in testLoader:
inputs,labels = data
inputs, labels = inputs.to(device), labels.long().to(device)
inputs = inputs/255
outputs = net(inputs)
_, predicted = torch.max(outputs.data, 1)
# Compute loss/error
loss = criterion(F.log_softmax(outputs,dim=1), labels)
# Accumulate loss per batch
test_runningLoss += loss.item()
# Accumuate correct predictions per batch
test_runningCorr += (predicted == labels.data).sum()
avgTestLoss = test_runningLoss/(60.0/BatchSize)
avgTestAcc = 100*float(test_runningCorr)/60.0
testAcc.append(avgTestAcc)
testLoss.append(avgTestLoss)
# Plotting Loss vs Epochs
fig1 = plt.figure(1)
plt.plot(range(epoch+1),trainLoss,'r--',label='train')
plt.plot(range(epoch+1),testLoss,'g--',label='test')
if epoch==0:
plt.legend(loc='upper left')
plt.xlabel('Epochs')
plt.ylabel('Loss')
# Plotting testing accuracy vs Epochs
fig2 = plt.figure(2)
plt.plot(range(epoch+1),trainAcc,'r-',label='train')
plt.plot(range(epoch+1),testAcc,'g-',label='test')
if epoch==0:
plt.legend(loc='upper left')
plt.xlabel('Epochs')
plt.ylabel('Accuracy')
epochEnd = time.time()-epochStart
print('At Iteration: {:.0f} /{:.0f} ; Training Loss: {:.6f} ; Training Acc: {:.3f} ; Time consumed: {:.0f}m {:.0f}s '\
.format(epoch + 1,iterations,avgTrainLoss,avgTrainAcc,epochEnd//60,epochEnd%60))
print('At Iteration: {:.0f} /{:.0f} ; Testing Loss: {:.6f} ; Testing Acc: {:.3f} ; Time consumed: {:.0f}m {:.0f}s '\
.format(epoch + 1,iterations,avgTestLoss,avgTestAcc,epochEnd//60,epochEnd%60))
end = time.time()-start
print('Training completed in {:.0f}m {:.0f}s'.format(end//60,end%60))
# Extracting the convolutional layers of the network
conv_net = nn.Sequential(*list(net.children())[:-2])
print(conv_net)
# Copying weights of the final layer for obtaining the segmented output
weights = copy.deepcopy(net.fc.weight.data)
if use_gpu:
weights = weights.cpu()
weights = weights.numpy()
print(weights.shape)
# Loading one sample image for testing
testPath = 'ALL_IDB1/img/'
testImages = os.listdir(testPath)
img1 = plt.imread(testPath+testImages[0])
testInput = torch.from_numpy(img1).transpose(0,2).transpose(1,2).unsqueeze(0).float().to(device)
# Feed-forward
out = conv_net(testInput)
# Visualization
if use_gpu:
out_np = out.squeeze(0).data.cpu().numpy()
else:
out_np = out.squeeze(0).data.numpy()
mask1 = np.ones(out_np.shape)
for n1 in range(512):
mask1[n1] = weights[0,n1]*mask1[n1]
outImg1 = np.sum(np.multiply(mask1,out_np),axis=0)
# Averaged activation map
plt.figure()
plt.subplot(121)
plt.imshow(np.sum(out_np,axis=0)/512,cmap='gray')
# Weighted-sum activation map
plt.subplot(122)
plt.imshow(outImg1,cmap='gray')
# Activation maps chosen at random
plt.figure()
randIdx = np.random.randint(0,511,4)
plt.subplot(141)
plt.imshow(out_np[randIdx[0]],cmap='gray')
plt.subplot(142)
plt.imshow(out_np[randIdx[1]],cmap='gray')
plt.subplot(143)
plt.imshow(out_np[randIdx[2]],cmap='gray')
plt.subplot(144)
plt.imshow(out_np[randIdx[3]],cmap='gray')
plt.imshow(img1)
```
|
github_jupyter
|
%matplotlib inline
import os
import time
import copy
import tqdm
import torch
import random
import numpy as np
import torch.nn as nn
from PIL import Image
import torch.optim as optim
from torchvision import models
import torch.nn.functional as F
import matplotlib.pyplot as plt
from torch.utils.data import TensorDataset,DataLoader
print(torch.__version__) # This code has been updated for PyTorch 1.0.0
Datapath = 'ALL_IDB2/img/'
listing = os.listdir(Datapath)
random.shuffle(listing)
# ALL_IDB2 dataset has 260 images in total
TrainImages = torch.FloatTensor(200,3,224,224)
TrainLabels = torch.LongTensor(200)
TestImages = torch.FloatTensor(60,3,224,224)
TestLabels = torch.LongTensor(60)
# First 200 images are used for training and the remaining 60 for testing
img_no = 0
for file in listing:
im=Image.open(Datapath + file)
im = im.resize((224,224))
im = np.array(im)
if img_no < 200:
TrainImages[img_no] = torch.from_numpy(im).transpose(0,2).unsqueeze(0)
TrainLabels[img_no] = int(listing[img_no][6:7])
else:
TestImages[img_no - 200] = torch.from_numpy(im).transpose(0,2).unsqueeze(0)
TestLabels[img_no - 200] = int(listing[img_no][6:7])
img_no = img_no + 1
print(TrainImages.size())
print(TrainLabels.size())
print(TestImages.size())
print(TestLabels.size())
# Check availability of GPU
use_gpu = torch.cuda.is_available()
# use_gpu = False # Uncomment in case of GPU memory error
if use_gpu:
print('GPU is available!')
device = "cuda"
pinMem = True
else:
print('GPU is not available!')
device = "cpu"
pinMem = False
# Creating pytorch dataset
trainDataset = TensorDataset(TrainImages, TrainLabels)
testDataset = TensorDataset(TestImages, TestLabels)
# Creating dataloader
BatchSize = 32
trainLoader = DataLoader(trainDataset, batch_size=BatchSize, shuffle=True,num_workers=4, pin_memory=pinMem)
testLoader = DataLoader(testDataset, batch_size=BatchSize, shuffle=False,num_workers=4, pin_memory=pinMem)
# ResNet18
net = models.resnet18(pretrained=True)
num_ftrs = net.fc.in_features
net.fc = nn.Linear(num_ftrs, 2)
print(net)
net = net.to(device)
criterion = nn.NLLLoss() # Negative Log-Likelihood
optimizer = optim.SGD(net.parameters(), lr=1e-3 , momentum=0.9) # Stochastic gradient descent
iterations = 15
trainLoss = []
trainAcc = []
testLoss = []
testAcc = []
start = time.time()
for epoch in range(iterations):
epochStart = time.time()
runningLoss = 0
runningCorr = 0
net.train() # For training
for data in tqdm.tqdm_notebook(trainLoader):
inputs,labels = data
inputs, labels = inputs.to(device), labels.long().to(device)
inputs = inputs/255.0
# Feed-forward input data through the network
outputs = net(inputs)
# Compute loss/error
loss = criterion(F.log_softmax(outputs,dim=1), labels)
_, predicted = torch.max(outputs.data, 1)
# Initialize gradients to zero
optimizer.zero_grad()
# Backpropagate loss and compute gradients
loss.backward()
# Update the network parameters
optimizer.step()
# Accumulate loss per batch
runningLoss += loss.item()
# Accumuate correct predictions per batch
runningCorr += (predicted == labels.data).sum()
avgTrainLoss = runningLoss/(200.0/BatchSize)
avgTrainAcc = 100*float(runningCorr)/200.0
trainLoss.append(avgTrainLoss)
trainAcc.append(avgTrainAcc)
# Evaluating performance on test set for each epoch
net.eval() # For testing
test_runningCorr = 0
test_runningLoss = 0
with torch.no_grad():
for data in testLoader:
inputs,labels = data
inputs, labels = inputs.to(device), labels.long().to(device)
inputs = inputs/255
outputs = net(inputs)
_, predicted = torch.max(outputs.data, 1)
# Compute loss/error
loss = criterion(F.log_softmax(outputs,dim=1), labels)
# Accumulate loss per batch
test_runningLoss += loss.item()
# Accumuate correct predictions per batch
test_runningCorr += (predicted == labels.data).sum()
avgTestLoss = test_runningLoss/(60.0/BatchSize)
avgTestAcc = 100*float(test_runningCorr)/60.0
testAcc.append(avgTestAcc)
testLoss.append(avgTestLoss)
# Plotting Loss vs Epochs
fig1 = plt.figure(1)
plt.plot(range(epoch+1),trainLoss,'r--',label='train')
plt.plot(range(epoch+1),testLoss,'g--',label='test')
if epoch==0:
plt.legend(loc='upper left')
plt.xlabel('Epochs')
plt.ylabel('Loss')
# Plotting testing accuracy vs Epochs
fig2 = plt.figure(2)
plt.plot(range(epoch+1),trainAcc,'r-',label='train')
plt.plot(range(epoch+1),testAcc,'g-',label='test')
if epoch==0:
plt.legend(loc='upper left')
plt.xlabel('Epochs')
plt.ylabel('Accuracy')
epochEnd = time.time()-epochStart
print('At Iteration: {:.0f} /{:.0f} ; Training Loss: {:.6f} ; Training Acc: {:.3f} ; Time consumed: {:.0f}m {:.0f}s '\
.format(epoch + 1,iterations,avgTrainLoss,avgTrainAcc,epochEnd//60,epochEnd%60))
print('At Iteration: {:.0f} /{:.0f} ; Testing Loss: {:.6f} ; Testing Acc: {:.3f} ; Time consumed: {:.0f}m {:.0f}s '\
.format(epoch + 1,iterations,avgTestLoss,avgTestAcc,epochEnd//60,epochEnd%60))
end = time.time()-start
print('Training completed in {:.0f}m {:.0f}s'.format(end//60,end%60))
# Extracting the convolutional layers of the network
conv_net = nn.Sequential(*list(net.children())[:-2])
print(conv_net)
# Copying weights of the final layer for obtaining the segmented output
weights = copy.deepcopy(net.fc.weight.data)
if use_gpu:
weights = weights.cpu()
weights = weights.numpy()
print(weights.shape)
# Loading one sample image for testing
testPath = 'ALL_IDB1/img/'
testImages = os.listdir(testPath)
img1 = plt.imread(testPath+testImages[0])
testInput = torch.from_numpy(img1).transpose(0,2).transpose(1,2).unsqueeze(0).float().to(device)
# Feed-forward
out = conv_net(testInput)
# Visualization
if use_gpu:
out_np = out.squeeze(0).data.cpu().numpy()
else:
out_np = out.squeeze(0).data.numpy()
mask1 = np.ones(out_np.shape)
for n1 in range(512):
mask1[n1] = weights[0,n1]*mask1[n1]
outImg1 = np.sum(np.multiply(mask1,out_np),axis=0)
# Averaged activation map
plt.figure()
plt.subplot(121)
plt.imshow(np.sum(out_np,axis=0)/512,cmap='gray')
# Weighted-sum activation map
plt.subplot(122)
plt.imshow(outImg1,cmap='gray')
# Activation maps chosen at random
plt.figure()
randIdx = np.random.randint(0,511,4)
plt.subplot(141)
plt.imshow(out_np[randIdx[0]],cmap='gray')
plt.subplot(142)
plt.imshow(out_np[randIdx[1]],cmap='gray')
plt.subplot(143)
plt.imshow(out_np[randIdx[2]],cmap='gray')
plt.subplot(144)
plt.imshow(out_np[randIdx[3]],cmap='gray')
plt.imshow(img1)
| 0.736116 | 0.918444 |
```
# https://griddb.net/en/blog/data-visualization-with-python-matplotlib-and-griddb/
# https://www.statsmodels.org/stable/generated/statsmodels.tsa.seasonal.seasonal_decompose.html
# https://dziganto.github.io/python/time%20series/Introduction-to-Time-Series/#:~:text=Trend%2C%20as%20its%20name%20suggests,them%20as%20a%20noise%20component.
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import os
import csv
from statsmodels.tsa.seasonal import seasonal_decompose
import statsmodels.api as sm
input_path = r'C:\Users\baharudin.ahmad\Documents\analytics\bitcoin_analytics\dataset'
output_path = r'C:\Users\baharudin.ahmad\Documents\analytics\t'
file_1 = 'BTC_USD_investing_com.csv'
file_2 = 'BTC_USD_yahoo_finance.csv'
df1 = pd.read_csv(os.path.join(input_path, file_1), encoding='utf-8')
df2 = pd.read_csv(os.path.join(input_path, file_2), encoding='utf-8')
df1 # dataset from investing.com
df2 # dataset from yahoo finance
#df1 = df1.sort_values(by=['Date'])
df2_top = df2.tail(300)
df2['Date'] = pd.to_datetime(df2['Date'], format='%Y-%m-%d')
df2.info()
df2 = df2.set_index("Date")
df2.plot(subplots=True, figsize=(20,24))
#plt.savefig('stocks.png')
plt.show()
df2 = df2[df2['Open'].notnull()]
df2.isnull().sum()
```
### Dickey Fuller Test
```
from statsmodels.tsa.stattools import adfuller
# Perform Dickey-Fuller test:
# low p-value: means that we reject the null hypothesis and classify it as stationary.
# high p-value: cannot reject the null hypothesis and classify non-stationary series.
print ('Results of Dickey-Fuller Test:')
dftest = adfuller(df2['Volume'])
# Extract and display test results
dfoutput = pd.Series(dftest[0:4], index=['Test Statistic','p-value','#Lags Used','Number of Observations Used'])
for key,value in dftest[4].items():
dfoutput['Critical Value (%s)'%key] = value
print (dfoutput)
```
### Decomposition (Original, Trend, Seasonality, Residual)
```
# Plot Decomposition (Original, Trend, Seasonality, Residual)
# Note: You have to have at least twice as many observations in your data as the period that you want to test; i.e. if you want to set the period to 20 then you need at least 40 observations).
decomp = seasonal_decompose(df2[['Close']], period= 360)
est_trend = decomp.trend # Trend is overall direction of the data
est_seasonal = decomp.seasonal # Seasonality is the periodic component
est_residual = decomp.resid # Residuals are random fluctuations. You can think of them as a noise component.
fig, axes = plt.subplots(4, 1)
fig.set_figheight(10)
fig.set_figwidth(15)
axes[0].plot(df2['Close'], label='Original', color='b')
axes[0].legend(loc='upper left')
axes[1].plot(est_trend, label='Trend', color='g')
axes[1].legend(loc='upper left')
axes[2].plot(est_seasonal, label='Seasonality',color='g')
axes[2].legend(loc='upper left')
axes[3].plot(est_residual, label='Residual',color='g')
axes[3].legend(loc='upper left')
plt.suptitle('Decomposition: Close', fontsize=20)
# Plot Decomposition (Original, Trend, Seasonality, Residual)
decomp = seasonal_decompose(df2[['Volume']], period= 360)
est_trend = decomp.trend # Trend is overall direction of the data
est_seasonal = decomp.seasonal # Seasonality is the periodic component
est_residual = decomp.resid # Residuals are random fluctuations. You can think of them as a noise component.
fig, axes = plt.subplots(4, 1)
fig.set_figheight(10)
fig.set_figwidth(15)
axes[0].plot(df2['Close'], label='Original', color='b')
axes[0].legend(loc='upper left')
axes[1].plot(est_trend, label='Trend', color='g')
axes[1].legend(loc='upper left')
axes[2].plot(est_seasonal, label='Seasonality',color='g')
axes[2].legend(loc='upper left')
axes[3].plot(est_residual, label='Residual',color='g')
axes[3].legend(loc='upper left')
plt.suptitle('Decomposition: Volume', fontsize=20)
```
### Auto-Correlation (ACF)
```
# Plot Auto-Correlation (ACF)
# Autocorrelation is the process of comparing a time series to its past; the more autocorrelated a time series is the easier it will be to predict.
sm.graphics.tsa.plot_acf(df2['Close'], lags=35)
plt.title('Autocorrelation: Close', fontsize=20)
# Plot Auto-Correlation (ACF)
sm.graphics.tsa.plot_acf(df2['Volume'], lags=35)
plt.title('Autocorrelation: Volume', fontsize=20)
```
### Plot residual stats
```
# line plot
est_residual.plot()
plt.show()
# histogram plot
est_residual.hist(bins=20)
plt.show()
```
|
github_jupyter
|
# https://griddb.net/en/blog/data-visualization-with-python-matplotlib-and-griddb/
# https://www.statsmodels.org/stable/generated/statsmodels.tsa.seasonal.seasonal_decompose.html
# https://dziganto.github.io/python/time%20series/Introduction-to-Time-Series/#:~:text=Trend%2C%20as%20its%20name%20suggests,them%20as%20a%20noise%20component.
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import os
import csv
from statsmodels.tsa.seasonal import seasonal_decompose
import statsmodels.api as sm
input_path = r'C:\Users\baharudin.ahmad\Documents\analytics\bitcoin_analytics\dataset'
output_path = r'C:\Users\baharudin.ahmad\Documents\analytics\t'
file_1 = 'BTC_USD_investing_com.csv'
file_2 = 'BTC_USD_yahoo_finance.csv'
df1 = pd.read_csv(os.path.join(input_path, file_1), encoding='utf-8')
df2 = pd.read_csv(os.path.join(input_path, file_2), encoding='utf-8')
df1 # dataset from investing.com
df2 # dataset from yahoo finance
#df1 = df1.sort_values(by=['Date'])
df2_top = df2.tail(300)
df2['Date'] = pd.to_datetime(df2['Date'], format='%Y-%m-%d')
df2.info()
df2 = df2.set_index("Date")
df2.plot(subplots=True, figsize=(20,24))
#plt.savefig('stocks.png')
plt.show()
df2 = df2[df2['Open'].notnull()]
df2.isnull().sum()
from statsmodels.tsa.stattools import adfuller
# Perform Dickey-Fuller test:
# low p-value: means that we reject the null hypothesis and classify it as stationary.
# high p-value: cannot reject the null hypothesis and classify non-stationary series.
print ('Results of Dickey-Fuller Test:')
dftest = adfuller(df2['Volume'])
# Extract and display test results
dfoutput = pd.Series(dftest[0:4], index=['Test Statistic','p-value','#Lags Used','Number of Observations Used'])
for key,value in dftest[4].items():
dfoutput['Critical Value (%s)'%key] = value
print (dfoutput)
# Plot Decomposition (Original, Trend, Seasonality, Residual)
# Note: You have to have at least twice as many observations in your data as the period that you want to test; i.e. if you want to set the period to 20 then you need at least 40 observations).
decomp = seasonal_decompose(df2[['Close']], period= 360)
est_trend = decomp.trend # Trend is overall direction of the data
est_seasonal = decomp.seasonal # Seasonality is the periodic component
est_residual = decomp.resid # Residuals are random fluctuations. You can think of them as a noise component.
fig, axes = plt.subplots(4, 1)
fig.set_figheight(10)
fig.set_figwidth(15)
axes[0].plot(df2['Close'], label='Original', color='b')
axes[0].legend(loc='upper left')
axes[1].plot(est_trend, label='Trend', color='g')
axes[1].legend(loc='upper left')
axes[2].plot(est_seasonal, label='Seasonality',color='g')
axes[2].legend(loc='upper left')
axes[3].plot(est_residual, label='Residual',color='g')
axes[3].legend(loc='upper left')
plt.suptitle('Decomposition: Close', fontsize=20)
# Plot Decomposition (Original, Trend, Seasonality, Residual)
decomp = seasonal_decompose(df2[['Volume']], period= 360)
est_trend = decomp.trend # Trend is overall direction of the data
est_seasonal = decomp.seasonal # Seasonality is the periodic component
est_residual = decomp.resid # Residuals are random fluctuations. You can think of them as a noise component.
fig, axes = plt.subplots(4, 1)
fig.set_figheight(10)
fig.set_figwidth(15)
axes[0].plot(df2['Close'], label='Original', color='b')
axes[0].legend(loc='upper left')
axes[1].plot(est_trend, label='Trend', color='g')
axes[1].legend(loc='upper left')
axes[2].plot(est_seasonal, label='Seasonality',color='g')
axes[2].legend(loc='upper left')
axes[3].plot(est_residual, label='Residual',color='g')
axes[3].legend(loc='upper left')
plt.suptitle('Decomposition: Volume', fontsize=20)
# Plot Auto-Correlation (ACF)
# Autocorrelation is the process of comparing a time series to its past; the more autocorrelated a time series is the easier it will be to predict.
sm.graphics.tsa.plot_acf(df2['Close'], lags=35)
plt.title('Autocorrelation: Close', fontsize=20)
# Plot Auto-Correlation (ACF)
sm.graphics.tsa.plot_acf(df2['Volume'], lags=35)
plt.title('Autocorrelation: Volume', fontsize=20)
# line plot
est_residual.plot()
plt.show()
# histogram plot
est_residual.hist(bins=20)
plt.show()
| 0.838018 | 0.82828 |
## Getting Dataset
```
from google.colab import drive
drive.mount('/content/drive')
```
The dataset is present as a zip file. So we need to unzip the file. At this step we need to give the **source path** and the **destination path**. The file will be unzipped and stored at the destination path.
```
!unzip "/content/drive/My Drive/food_classifer_dataset.zip" -d "/content/"
```
# Importing Important Libraries
```
# this is going to help in data processing
from keras.preprocessing.image import ImageDataGenerator
#these are going to help in the model building.
from keras.models import Sequential
from keras.layers import Conv2D, MaxPooling2D, BatchNormalization
from keras.layers import Activation, Dropout, Flatten, Dense
```
# Setting up Variables
```
# dimensions of our images.
img_width, img_height = 150, 150
#setting up the directories
train_data_dir = '/content/classifer_dataset/train'
validation_data_dir = '/content/classifer_dataset/validate'
#setting up the batchsizes.
nb_train_samples = 8400
nb_validation_samples = 1600
epochs = 50
batch_size = 16
```
Here we set the values in different variables which will be used later. We also need to save the input dimension because we need to give that in the model.
```
input_shape = (img_width, img_height, 3)
```
# Building the CNN model
Here we are going to do build sequential model in keras. We will simply keep on adding the layers which we want to.
```
model = Sequential()
model.add(Conv2D(32, (3, 3), activation='relu', input_shape=input_shape))
model.add(BatchNormalization())
model.add(MaxPooling2D(pool_size=(2, 2)))
model.add(Dropout(0.25))
model.add(Conv2D(64, (3, 3), activation='relu'))
model.add(BatchNormalization())
model.add(MaxPooling2D(pool_size=(2, 2)))
model.add(Dropout(0.25))
model.add(Conv2D(128, (3, 3), activation='relu'))
model.add(BatchNormalization())
model.add(MaxPooling2D(pool_size=(2, 2)))
model.add(Dropout(0.25))
model.add(Flatten())
model.add(Dense(512, activation='relu'))
model.add(BatchNormalization())
model.add(Dropout(0.5))
model.add(Dense(1, activation='sigmoid'))
#configuring the model
model.compile(loss='binary_crossentropy',
optimizer='rmsprop',
metrics=['accuracy'])
#to print a summary representation of your model
model.summary()
```
# Data Augmentation
Here different operations will be used for data augmentation and also data processing.
```
# this is the augmentation configuration we will use for training
train_datagen = ImageDataGenerator(
rescale=1./ 255,
shear_range=0.2,
zoom_range=0.2,
horizontal_flip=True)
#this generates batches of augment data for training
train_generator = train_datagen.flow_from_directory(
train_data_dir,
target_size=(img_width, img_height),
batch_size=batch_size,
class_mode='binary')
# this is the augmentation configuration we will use for validating
val_datagen = ImageDataGenerator(rescale=1./255)
#this generates batches of augment data for validating
validation_generator = val_datagen.flow_from_directory(
validation_data_dir,
target_size=(img_width, img_height),
batch_size=batch_size,
class_mode='binary')
```
# Training the model (Forward + Back Propagation)
Now our data is ready to be sent in the model. So here we will train the model. This training includes both forward and backward propagation. The model will be trained for 50 epochs. You are free to change the number.
```
model.fit_generator(
train_generator,
steps_per_epoch=nb_train_samples // batch_size,
epochs=epochs,
validation_data=validation_generator,
validation_steps=nb_validation_samples // batch_size)
```
This training took around 95 minutes. Once the model ims ready, just save the architecture and trained parameters of the model.
# Saving Model
```
#to save the weights in the model as a HDFS file
model.save_weights('model_weight.h5')
#to save the architecture of the model as a json file
with open('model_architecture.json','w') as f:
f.write(model.to_json())
```
# Restoring the Model
Here we are going to see how to restore any model whose parameters and architecture files are with us.
```
from keras.models import load_model
from keras.models import model_from_json
# Model reconstruction from JSON file
with open('/content/food_model_architecture.json', 'r') as f:
model = model_from_json(f.read())
# Load weights into the new model
model.load_weights('/content/first_try.h5')
```
# Testing the model
Hurray!!!!!!
Congratulations!! Your 1st Computer Vision Model is ready. Its time to taste the food.
```
import numpy as np
from keras.preprocessing import image
test_image=image.load_img('/content/images (1).jfif',target_size=(img_width,img_height))
test_image=image.img_to_array(test_image)
test_image=np.expand_dims(test_image,axis=0)
result=model.predict(test_image)
print(result)
if result[0][0]==1.0:
prediction='SoftDrink'
else:
prediction='Pizza'
print("You got a "+ prediction + " Yupeeeeeeeeeeeeeeeeeeeeeeeeeeeeee!!!")
```
|
github_jupyter
|
from google.colab import drive
drive.mount('/content/drive')
!unzip "/content/drive/My Drive/food_classifer_dataset.zip" -d "/content/"
# this is going to help in data processing
from keras.preprocessing.image import ImageDataGenerator
#these are going to help in the model building.
from keras.models import Sequential
from keras.layers import Conv2D, MaxPooling2D, BatchNormalization
from keras.layers import Activation, Dropout, Flatten, Dense
# dimensions of our images.
img_width, img_height = 150, 150
#setting up the directories
train_data_dir = '/content/classifer_dataset/train'
validation_data_dir = '/content/classifer_dataset/validate'
#setting up the batchsizes.
nb_train_samples = 8400
nb_validation_samples = 1600
epochs = 50
batch_size = 16
input_shape = (img_width, img_height, 3)
model = Sequential()
model.add(Conv2D(32, (3, 3), activation='relu', input_shape=input_shape))
model.add(BatchNormalization())
model.add(MaxPooling2D(pool_size=(2, 2)))
model.add(Dropout(0.25))
model.add(Conv2D(64, (3, 3), activation='relu'))
model.add(BatchNormalization())
model.add(MaxPooling2D(pool_size=(2, 2)))
model.add(Dropout(0.25))
model.add(Conv2D(128, (3, 3), activation='relu'))
model.add(BatchNormalization())
model.add(MaxPooling2D(pool_size=(2, 2)))
model.add(Dropout(0.25))
model.add(Flatten())
model.add(Dense(512, activation='relu'))
model.add(BatchNormalization())
model.add(Dropout(0.5))
model.add(Dense(1, activation='sigmoid'))
#configuring the model
model.compile(loss='binary_crossentropy',
optimizer='rmsprop',
metrics=['accuracy'])
#to print a summary representation of your model
model.summary()
# this is the augmentation configuration we will use for training
train_datagen = ImageDataGenerator(
rescale=1./ 255,
shear_range=0.2,
zoom_range=0.2,
horizontal_flip=True)
#this generates batches of augment data for training
train_generator = train_datagen.flow_from_directory(
train_data_dir,
target_size=(img_width, img_height),
batch_size=batch_size,
class_mode='binary')
# this is the augmentation configuration we will use for validating
val_datagen = ImageDataGenerator(rescale=1./255)
#this generates batches of augment data for validating
validation_generator = val_datagen.flow_from_directory(
validation_data_dir,
target_size=(img_width, img_height),
batch_size=batch_size,
class_mode='binary')
model.fit_generator(
train_generator,
steps_per_epoch=nb_train_samples // batch_size,
epochs=epochs,
validation_data=validation_generator,
validation_steps=nb_validation_samples // batch_size)
#to save the weights in the model as a HDFS file
model.save_weights('model_weight.h5')
#to save the architecture of the model as a json file
with open('model_architecture.json','w') as f:
f.write(model.to_json())
from keras.models import load_model
from keras.models import model_from_json
# Model reconstruction from JSON file
with open('/content/food_model_architecture.json', 'r') as f:
model = model_from_json(f.read())
# Load weights into the new model
model.load_weights('/content/first_try.h5')
import numpy as np
from keras.preprocessing import image
test_image=image.load_img('/content/images (1).jfif',target_size=(img_width,img_height))
test_image=image.img_to_array(test_image)
test_image=np.expand_dims(test_image,axis=0)
result=model.predict(test_image)
print(result)
if result[0][0]==1.0:
prediction='SoftDrink'
else:
prediction='Pizza'
print("You got a "+ prediction + " Yupeeeeeeeeeeeeeeeeeeeeeeeeeeeeee!!!")
| 0.646683 | 0.908618 |
<center><strong>A Journey Into Math For Machine Learning</strong></center>
<center><strong>机器学习之数学之旅</strong></center>
<center><strong>之 自适应增强$adaboost$$(adaptive \ boosting)$</strong></center>

今天的主题是**集成学习**中的$adaboost$, 或称作$adaptive \ boosting$, 首先我们来建立一种概念, 什么是$adaptive \ boosting$:
$adaboost$是**集成学习**的一种, 意思是建立多个**弱分类器**, 然后用这些弱分类器的**线性加权组合**来形成一个**强分类器**. 什么是弱分类器呢, 就是只比胡猜稍微好一点的分类器, 训练这些弱分类器是一个迭代的过程, 在这个过程里, 下一个弱分类器总是更加关注上一个弱分类器没有分好的数据样本, 以弥补之前弱分类器的不足, $adaboost$就是类似"三个臭皮匠顶个诸葛亮"的算法.
课程预览:
(一). 建立一种直觉——$adaboost$的可视化;
(二). 从零推导$adaboost$;
(三). $adaboost$的特性;
(四). 代码解读.
首先我们先来通过可视化的方法来建立一种直觉.
```
from demonstration import *
import plotly.graph_objs as go
from plotly.offline import download_plotlyjs, init_notebook_mode, plot, iplot
# 在jupyter notebook之内画图
init_notebook_mode(connected=True)
```
我们首先造一些数据, 并把它们画出来:
```
from sklearn.datasets import make_gaussian_quantiles
# 画两个正态分布的群
X1, y1 = make_gaussian_quantiles(cov=2.,
n_samples=200, n_features=2,
n_classes=2, random_state=1)
X2, y2 = make_gaussian_quantiles(mean=(3, 3), cov=1.5,
n_samples=300, n_features=2,
n_classes=2, random_state=1)
X=np.concatenate([X1,X2])
y=np.concatenate([y1,1-y2])
plt.figure(figsize=(5, 5))
plt.scatter(X[:, 0], X[:, 1], c=y, cmap=plt.cm.RdBu)
y[np.where(y == 0)] =-1
A = Adaboost_Demonstration(X, y)
```
每次运行\_\_next\_\_函数, 都会新增加一个弱分类器, 并对当前的强分类器的分类边界进行可视化
```
A.__next__(plot=True, plot_mode="3d")
```
(二). 从零推导$adaboost$:
我们首先有一个弱分类器$h(x)$, 它输出的值非$1$即$-1$, $h(x) \in \{1, -1\}$, $adaboost$的思路是把这些弱分类器的预测结果都加起来, 然后取他们的均值, 在我们的推导过程中, 假设我们要解决的问题是一个二项分类问题, 则我们要把这些弱分类器都加起来取$sign$, 则有:
$$H(x)=sign(\sum_{t=1}^T h^t(x))$$
上式中$H(x)$是一个强分类器, $x$是数据或特征值, $t$指的是弱分类器的个数, 上式的意思是对弱分类器的输出值求和并取$sign$, $H(x) \in \{1, -1\}$
然后我们发现只对弱分类器求和好像少了点什么, 没错, 我们要赋予每个弱分类器不同的权重:
$$H(x)=sign(\sum_{t=1}^T \alpha^t h^t(x))$$
上式中$\alpha$指的是第$t$个弱分类器的权重.
在$adaboost$中, 这么多弱分类器, 是凭什么依据划分的呢, 其实每个弱分类器关注的是前面弱分类器没有分对的数据样本, 比如说第$t$个弱分类器犯了一些错误, 导致有些数据样本没有分对, 那么第$t+1$个弱分类器就会更加关注前面没有分对的数据点, 尝试弥补第$t$个弱分类器的不足.
假设我们有一些数据样本$\{(x_{1},y_{1}),\ldots ,(x_{N},y_{N})\}$, 其中$y_{i}\in \{-1,1\}$, $N$是数据样本的个数, 我们要解决的是二项分类问题.
我们需要给每一个数据样本赋一个权重, 以控制每个弱分类器应该更加关注哪些数据样本, 我们需要让下一个弱分类器更加关注它前面的弱分类器没有分对的数据点, 设$x_i$在第$t$个弱分类器上的权重为$w^t_i$, 我们首先初始化, 在第$1$个弱分类器上所有数据点的权重都是一样的, 即:
$$w^{t=1}_i=\frac{1}{N}$$
初始化错误率$\epsilon$为, 比如说$100$个数据点, 分错了40个, 则$\epsilon=0.4$, $\epsilon$取值在$0$到$1$之间:
$$\epsilon=\sum_{y_i \neq h_i(x)}\frac{1}{N} \quad \epsilon \in [0, 1]$$
将$w_i$代入$\epsilon$, 得出:
$$\epsilon=\sum_{y_i \neq h_i(x)}w_i ; \quad \sum w_i =1\tag{eq. 1}$$
那我们怎么确定下一个弱分类器需要关注哪些数据样本呢? 当我们定义好当前的弱分类器$h^t(x)$之后, 做以下操作:
$$w^{t+1}_{i}=\frac{w^{t}_{i}}{Z}e^{-\alpha^t h^t(x_i) y_i}$$
上式中$Z$是归一化因子, 因为我们想要$\sum w_i =1$, 怎么理解这个式子呢, 首先$\alpha$是正数, 当$h^t(x_i)=y_i$的时候, $h^t(x_i)y_i$的是$1$, 否则是$-1$, 不难看出当当前弱分类器$h^t(x_i)$预测正确的时候, $w^{t+1}=$会变小, 而错误的时候, $w^{t+1}$会变大:
$${\displaystyle w^{t+1}_{i}={\begin{cases} \frac{1}{Z} w_{i}^{t}e^{-\alpha ^{t}}&{\text{if }}y_{i}=h^{t}(x_{i}),
\\ \frac{1}{Z} w_{i}^{t}e^{\alpha ^{t}}&{\text{if }}{y_{i} \neq h^{t}(x_{i})}.\end{cases}}}\tag{eq. 2}$$
注意我们暂且不管$Z$.
我们现在要求所有未归一的权重的和, 并求出可以使它最小的$\alpha$, 我们把未归一的权重的和定义为$E$, 因为分错的样本输入到下一个弱分类器的权重(未归一)为$w_{i}^{t}e^{\alpha ^{t}}$(见$eq.2$), 则分错的样本越多$E$会越大, 相反分对的样本越多$E$越小.
$$E=
\sum _{y_{i}=h^{t}(x_{i})}w_{i}^{t}e^{-\alpha ^{t}}+\sum _{y_{i}\neq h^{t}(x_{i})}w_{i}^{t}e^{\alpha ^{t}}$$
上式左边加上$\sum _{y_{i}\neq h^{t}(x_{i})} w_{i}^{t} e^{-\alpha ^{t}}$, 右边再减去.
$$E=
\sum _{i=1}^{N}w_{i}^{t} e^{-\alpha ^{t}}+\sum _{y_{i}\neq h^{t}(x_{i})}w_{i}^{t}(e^{\alpha ^{t}} - e^{-\alpha ^{t}})$$
上式中可看出, 上式左边跟我们当前的弱分类器$h^{t}(x)$无关, 当前的弱分类器只对右式起作用, 也就是右式控制着$E$的大小, 为了得到我们想要的$\alpha$, 我们要求$E$对于$\alpha^{t}$的导数, 并把它设为$0$, 求出$\alpha^{t}$的值, 注意下边我们会把等式左边省略掉, 因为它可看作一个常数.
$${\frac {d E}{d\alpha ^{t}}}=-\sum _{y_{i}= h^{t}(x_{i})}w_{i}^{t}e^{-\alpha ^{t}}+\sum _{y_{i}\neq h^{t}(x_{i})}w_{i}^{t}e^{\alpha ^{t}}=0$$
解上面的式子, 这里省略过程, 因为比较简单, 大概解法是将$e^{\alpha ^{t}}$和$e^{-\alpha ^{t}}$提到求和符号外面来, 同时取$\log$, 化简, 得到$\alpha^{t}$:
$$\alpha ^{t}={\dfrac {1}{2}}\log \left({\dfrac {\sum _{y_{i}=h^{t}(x_{i})}w_{i}^{t}}{\sum _{y_{i}\neq h^{t}(x_{i})}w_{i}^t}}\right)={\dfrac {1}{2}}\log \left(\dfrac {1-\epsilon^{t}}{\epsilon^{t}}\right) $$
将$\alpha ^{t}$带入到$(eq. 2)$里, 得到下式, 注意这里用到了$e^{log_e(x)}=x$:
$${\displaystyle w^{t+1}_{i}=\frac{1}{Z} w_{i}^{t}{\begin{cases} \sqrt{\frac{\epsilon^{t}}{1-\epsilon^{t}}} &{\text{if }}y_{i}=h^{t}(x_{i}),
\\ \sqrt{\frac{1-\epsilon^{t}}{\epsilon^{t}}} &{\text{if }}{y_{i} \neq h^{t}(x_{i})}.\end{cases}}}\tag{eq. 3}$$
我们求$Z$, $Z$是所有数据样本权重的和也就是所有$w^{t+1}_{i}$取值的和:
$$Z=\sqrt{\frac{\epsilon^{t}}{1-\epsilon^{t}}} \sum _{y_{i}= h^{t}(x_{i})}w^{t}_i +
\sqrt{\frac{1-\epsilon^{t}}{\epsilon^{t}}} \sum _{y_{i}\neq h^{t}(x_{i})}w^{t}_i$$
将$\epsilon=\sum_{y_i \neq h_i(x)}w_i$代入上式, 得出:
$$Z=\sqrt{\frac{\epsilon^{t}}{1-\epsilon^{t}}} (1-\epsilon^t) +
\sqrt{\frac{1-\epsilon^{t}}{\epsilon^{t}}} \epsilon^t=2 \sqrt{\epsilon^t(1-\epsilon^t)}$$
将$Z$代入$(eq.3)$里:
$${\displaystyle w^{t+1}_{i}={\begin{cases} \frac{w_{i}^{t}}{2(1-\epsilon^{t})} &{\text{if }}y_{i}=h^{t}(x_{i}),
\\ \frac{w_{i}^{t}}{2\epsilon^{t}} &{\text{if }}{y_{i} \neq h^{t}(x_{i})}.\end{cases}}}\tag{eq. 4}$$
我们通过上式还是很难看出$w^{t+1}_i$的意义到底是什么, 没关系, 接下来就恍然大悟了, 下面我们求数据点中分类正确的样本和分类错误样本各自$w^{t+1}_i$的合:
$$\sum_{y_{i}=h^{t}(x_{i})}w^{t+1}_i=\frac{\sum_{y_{i}=h^{t}(x_{i})} w_{i}^{t}}{2(1-\epsilon^{t})}$$
我们又知道$\sum_{y_{i}\neq h^{t}(x_{i})} w_{i}^{t}=\epsilon^t$, 得出$\sum_{y_{i}=h^{t}(x_{i})} w_{i}^{t}=1-\epsilon^t$, 代入上式:
$$\sum_{y_{i}=h^{t}(x_{i})}w^{t+1}_i=\frac{1-\epsilon^t}{2(1-\epsilon^{t})}=\frac{1}{2}$$
又因为$\sum w_i =1$, 得出:
$$\sum_{y_{i} \neq h^{t}(x_{i})}w^{t+1}_i=1-\sum_{y_{i}=h^{t}(x_{i})}w^{t+1}_i=1-\frac{1}{2}=\frac{1}{2}$$
上式的意义是, 在我们每次增加一个新弱分类器的时候, 前面的弱分类器分错的样本的权重占总样本权重的$\frac{1}{2}$, 前面的弱分类器分对的样本的权重也占总样本权重的$\frac{1}{2}$
我们把$adaboost$的计算步骤梳理一下:
1. 初始化$w^1_i=\frac{1}{N}$;
2. 在第$t$步, 选择可以最小化当前错误率$\epsilon$的弱分类器, 并添加这个弱分类器, 以降低错误率(如果还没有弱分类器, 那就创建一个), 并计算$\alpha^t$, $\alpha ^{t}={\dfrac {1}{2}}\log \left(\dfrac {1-\epsilon^{t}}{\epsilon^{t}}\right)$, 定义新的弱分类器为$f^t(x)=\alpha^t h^t(x))$
3. 计算新的数据样本权重$w^{t+1}_i$, 见$(eq. 4)$
4. 回到第2, 直到收敛.
5. 得到的模型为: $H(x)=sign(\sum_{t=1}^T \alpha^t h^t(x))$
(三). $adaboost$的特性:
1. 易用, 需要调节的参数少;
2. 弱分类器可以是任何模型, 比如逻辑回归, 线性回归等等, 最常用的是决策树桩;
3. 对数据敏感, 如果数据有很多噪音, 则$adaboost$会倾向于拟合噪音而最终导致模型的过拟合, 为了解决这样的问题, 可以给每一个弱分类器一个学习率权重, 让每个弱分类器最强分类器造成的影响小一些, 或做交叉验证, 让迭代的过程在合适的时机停止;
4. 可以用来做特征选择, 举个例子吧, 比如说要评估人群的信用风险, 然后你会有很多这些人的资料, 比如说性别年龄, 居住地址, 收入, 有无不动产等等, 如果你用$adaboost$的集成方法来做分类, 在迭代的每一步你需要用一个弱分类器划分数据样本, 假设我们用决策树桩, 决策树桩每次只能选择一个特征值进行划分, 在建模完成之后, 看一下模型里的弱分类器都是靠那些特征进行分类的, 就可以得出每种特征的重要性, 方便我们做特征选择, 剔除多余的特征, 增加计算效率, 减少噪音;
5. 注意, 在推导的过程中, 我们对强分类器输出的结果取$sign$, 实际中我们可以不单纯取$sign$, 而是找一个分类最好的阈值, 或者说边界, 这个阈值不一定为$0$.
(四). 代码解读.
请参照B站或youtube视频讲解, 或参考demonstration.py文件内注释.
|
github_jupyter
|
from demonstration import *
import plotly.graph_objs as go
from plotly.offline import download_plotlyjs, init_notebook_mode, plot, iplot
# 在jupyter notebook之内画图
init_notebook_mode(connected=True)
from sklearn.datasets import make_gaussian_quantiles
# 画两个正态分布的群
X1, y1 = make_gaussian_quantiles(cov=2.,
n_samples=200, n_features=2,
n_classes=2, random_state=1)
X2, y2 = make_gaussian_quantiles(mean=(3, 3), cov=1.5,
n_samples=300, n_features=2,
n_classes=2, random_state=1)
X=np.concatenate([X1,X2])
y=np.concatenate([y1,1-y2])
plt.figure(figsize=(5, 5))
plt.scatter(X[:, 0], X[:, 1], c=y, cmap=plt.cm.RdBu)
y[np.where(y == 0)] =-1
A = Adaboost_Demonstration(X, y)
A.__next__(plot=True, plot_mode="3d")
| 0.73307 | 0.975554 |
<table class="ee-notebook-buttons" align="left">
<td><a target="_blank" href="https://github.com/giswqs/earthengine-py-notebooks/tree/master/Visualization/nlcd_land_cover.ipynb"><img width=32px src="https://www.tensorflow.org/images/GitHub-Mark-32px.png" /> View source on GitHub</a></td>
<td><a target="_blank" href="https://nbviewer.jupyter.org/github/giswqs/earthengine-py-notebooks/blob/master/Visualization/nlcd_land_cover.ipynb"><img width=26px src="https://upload.wikimedia.org/wikipedia/commons/thumb/3/38/Jupyter_logo.svg/883px-Jupyter_logo.svg.png" />Notebook Viewer</a></td>
<td><a target="_blank" href="https://mybinder.org/v2/gh/giswqs/earthengine-py-notebooks/master?filepath=Visualization/nlcd_land_cover.ipynb"><img width=58px src="https://mybinder.org/static/images/logo_social.png" />Run in binder</a></td>
<td><a target="_blank" href="https://colab.research.google.com/github/giswqs/earthengine-py-notebooks/blob/master/Visualization/nlcd_land_cover.ipynb"><img src="https://www.tensorflow.org/images/colab_logo_32px.png" /> Run in Google Colab</a></td>
</table>
## Install Earth Engine API
Install the [Earth Engine Python API](https://developers.google.com/earth-engine/python_install) and [geehydro](https://github.com/giswqs/geehydro). The **geehydro** Python package builds on the [folium](https://github.com/python-visualization/folium) package and implements several methods for displaying Earth Engine data layers, such as `Map.addLayer()`, `Map.setCenter()`, `Map.centerObject()`, and `Map.setOptions()`.
The magic command `%%capture` can be used to hide output from a specific cell. Uncomment these lines if you are running this notebook for the first time.
```
# %%capture
# !pip install earthengine-api
# !pip install geehydro
```
Import libraries
```
import ee
import folium
import geehydro
```
Authenticate and initialize Earth Engine API. You only need to authenticate the Earth Engine API once. Uncomment the line `ee.Authenticate()`
if you are running this notebook for the first time or if you are getting an authentication error.
```
# ee.Authenticate()
ee.Initialize()
```
## Create an interactive map
This step creates an interactive map using [folium](https://github.com/python-visualization/folium). The default basemap is the OpenStreetMap. Additional basemaps can be added using the `Map.setOptions()` function.
The optional basemaps can be `ROADMAP`, `SATELLITE`, `HYBRID`, `TERRAIN`, or `ESRI`.
```
Map = folium.Map(location=[40, -100], zoom_start=4)
Map.setOptions('HYBRID')
```
## Add Earth Engine Python script
```
dataset = ee.Image('USGS/NLCD/NLCD2016')
landcover = ee.Image(dataset.select('landcover'))
landcoverVis = {
'min': 0.0,
'max': 95.0,
'palette': [
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'466b9f',
'd1def8',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'dec5c5',
'd99282',
'eb0000',
'ab0000',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'b3ac9f',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'68ab5f',
'1c5f2c',
'b5c58f',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'af963c',
'ccb879',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'dfdfc2',
'd1d182',
'a3cc51',
'82ba9e',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'dcd939',
'ab6c28',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'b8d9eb',
'000000',
'000000',
'000000',
'000000',
'6c9fb8'
],
}
Map.setCenter(-95, 38, 5)
Map.addLayer(landcover, landcoverVis, 'Landcover')
```
## Display Earth Engine data layers
```
Map.setControlVisibility(layerControl=True, fullscreenControl=True, latLngPopup=True)
Map
```
|
github_jupyter
|
# %%capture
# !pip install earthengine-api
# !pip install geehydro
import ee
import folium
import geehydro
# ee.Authenticate()
ee.Initialize()
Map = folium.Map(location=[40, -100], zoom_start=4)
Map.setOptions('HYBRID')
dataset = ee.Image('USGS/NLCD/NLCD2016')
landcover = ee.Image(dataset.select('landcover'))
landcoverVis = {
'min': 0.0,
'max': 95.0,
'palette': [
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'466b9f',
'd1def8',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'dec5c5',
'd99282',
'eb0000',
'ab0000',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'b3ac9f',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'68ab5f',
'1c5f2c',
'b5c58f',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'af963c',
'ccb879',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'dfdfc2',
'd1d182',
'a3cc51',
'82ba9e',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'dcd939',
'ab6c28',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'000000',
'b8d9eb',
'000000',
'000000',
'000000',
'000000',
'6c9fb8'
],
}
Map.setCenter(-95, 38, 5)
Map.addLayer(landcover, landcoverVis, 'Landcover')
Map.setControlVisibility(layerControl=True, fullscreenControl=True, latLngPopup=True)
Map
| 0.370567 | 0.955981 |
# Machine Learning and Statistics for Physicists
Material for a [UC Irvine](https://uci.edu/) course offered by the [Department of Physics and Astronomy](https://www.physics.uci.edu/).
Content is maintained on [github](github.com/dkirkby/MachineLearningStatistics) and distributed under a [BSD3 license](https://opensource.org/licenses/BSD-3-Clause).
##### ► [View table of contents](Contents.ipynb)
```
%matplotlib inline
import matplotlib.pyplot as plt
import seaborn as sns; sns.set()
import numpy as np
import pandas as pd
```
## Tensor Computing
Most practical algorithms of ML can be decomposed into small steps where the calculations are expressed with linear algebra, i.e., linear combinations of scalars, vectors and matrices.
For example, a neural network can be built from layers that each calculate
$$
\mathbf{x}_\text{out} = \max(0, W \mathbf{x}_\text{in} + \mathbf{b}) \; ,
$$
where $W$ is a matrix, and boldface symbols represent vectors. In typical applications, $\mathbf{x}_\text{out}$ and $\mathbf{x}_\text{in}$ are derived from **data** while $W$ and $\mathbf{b}$ are considered **model parameters**. (This expression is not strictly linear: why?)
The python numeric and list types can represent arbitrary scalars, vectors, and matrices, but are designed for flexibility instead of efficiency.
Numpy is instead optimized for the special case where all list elements are numeric values of the same type, which can be organized and accessed very efficiently in memory, with a specialized array type with lots of nice features. One downside of this approach is that most of builtin math functions are duplicated (e.g., `math.sin` and `np.sin`) to work with numpy arrays.
**EXERCISE:** Complete the function below using numpy to evaluate the neural-network layer defined above:
```
def xout(W, xin, b):
return np.maximum(0, W.dot(xin) + b)
def xout(W, xin, b):
# Add your solution here
return 0
```
### Terminology
We frequently use $\mathbf{r} = (x, y, z)$ in physics to represent an *arbitrary* position in three (continuous) dimensions.
In numpy, we cannot represent an *arbitrary* position but can easily represent a *specific* position, for example:
```
rvec = np.array([0.1, -0.2, 0.3])
```
However, `rvec` has only one (discrete) dimension, which we use to access its three array elements with indices 0,1,2:
```
rvec[0], rvec[1], rvec[2]
```
Note how we use the term **dimension** differently in these two cases!
All numpy arrays have a `shape` property that specifies the range of indices allowed for each of their (discrete) dimensions:
```
rvec.shape
rvec.ndim
```
Compare with a matrix represented in numpy:
```
matrix = np.identity(3)
print(matrix)
matrix[1, 0], matrix[1, 1]
matrix.shape
matrix.ndim
```
Numpy supports arrays with any (finite) number of (discrete) dimensions. The general name for these arrays is a **tensor** (so, scalars, vectors and matrices are tensors). For example:
```
tensor = np.ones((2, 3, 4))
print(tensor)
tensor[0, 0, 0], tensor[1, 2, 3]
tensor.shape
tensor.ndim
```
Tensors are used in physics also: for example, the tensor expression $g^{il} \Gamma^m_{ki} x^k$ arises in [contravariant derivatives in General Relativity](https://en.wikipedia.org/wiki/Christoffel_symbols#Covariant_derivatives_of_tensors). What are the **dimensions** of $g$, $\Gamma$ and $x$ in this expression? Note that numpy tensors do not make any distinction between upper or lower indices.
The numpy dimension is sometimes also referred to as the **rank**, but note that [array rank](https://en.wikipedia.org/wiki/Rank_(computer_programming)) is similar to but subtly different from [linear algebra rank](https://en.wikipedia.org/wiki/Rank_(linear_algebra)).
### Fundamental Operations
#### Tensor Creation
The most common ways you will create new arrays are:
- Filled with a simple sequence of constant values
- Filled with (reproducible) random values
- Calculated as a mathematical function of existing arrays.
```
# Regular sequence of values
shape = (3, 4)
c1 = np.zeros(shape)
c2 = np.ones(shape)
c3 = np.full(shape, -1)
c4 = np.arange(12)
# Reproducible "random" numbers
gen = np.random.RandomState(seed=123)
r1 = gen.uniform(size=shape)
r2 = gen.normal(loc=-1, scale=2, size=shape)
# Calculated as function of existing array.
f1 = r1 * np.sin(r2) ** c3
```
All the values contained within a tensors have the same [data type](https://docs.scipy.org/doc/numpy-1.15.0/user/basics.types.html), which you can inspect:
```
c1.dtype, c4.dtype
```
**EXERCISE:** Try to guess the `dtype` of `c3`, `r2` and `f1`, then check your answer. Deep learning often uses smaller (32 or 16 bit) float data types: what advantages and disadvantages might this have?
**SOLUTION:** The `zeros` and `ones` functions default to `float64`, but `full` uses the type of the provided constant value. Integers are automatically promoted to floats in mixed expressions.
```
c3.dtype, r2.dtype, f1.dtype
```
Smaller floats allow more efficient use of limited (GPU) memory and faster calculations, at the cost of some accuracy. Since the training of a deep neural network is inherently noisy, this is generally a good tradeoff.
#### Tensor Reshaping
It is often useful to [reshape](https://docs.scipy.org/doc/numpy-1.15.0/reference/generated/numpy.reshape.html) a tensor without changing its total size, which can be done very efficiently since the layout of the tensor values in memory does not need to be changed:
```
c4.reshape(c1.shape)
```
**EXERCISE:** Predict the result of `c4.reshape(2, 3, 2)` then check your answer.
```
c4.reshape(2, 3, 2)
```
#### Tensor Broadcasting
The real power of tensor computing comes from expressions like this:
```
# Add one to each element.
c4 + 1
# Scale each column of the 3x4 ones matrix by a different value.
np.ones(shape=(3, 4)) * np.arange(4)
```
The results are not surprising in these examples, but something non-trivial is going on behind the scenes to make this work since we are combining tensors with different shapes. This is called [broadcasting](https://docs.scipy.org/doc/numpy-1.15.0/user/basics.broadcasting.html) and has specific rules for how to handle less obvious cases.
Broadcasting serves two purposes:
- It allows more compact and easier to understand "vectorized" expressions, where loops over elements in each dimension are implicit.
- It enables automatic optimizations to take advantage of the available hardware, since explicit python loops are generally a bottleneck.
Not all expressions can be automatically broadcast, even if they seem to make sense. For example:
```
# Scale each row of the 3x4 ones matrix by a different value.
try:
np.ones(shape=(3, 4)) * np.arange(3)
except ValueError as e:
print(e)
```
However, you can usually reshape the inputs to get the desired result:
```
np.ones(shape=(3, 4)) * np.arange(3).reshape(3, 1)
```
To experiment with broadcasting rules, define a function to try broadcasting two arbitrary tensor shapes:
```
def broadcast(shape1, shape2):
array1 = np.ones(shape1)
array2 = np.ones(shape2)
try:
array12 = array1 + array2
print('shapes {} {} broadcast to {}'.format(shape1, shape2, array12.shape))
except ValueError as e:
print(e)
broadcast((1, 3), (3,))
broadcast((1, 2), (3,))
```
**EXERCISE:** Predict the results of the following then check your answers:
```
broadcast((3, 1, 2), (3, 2))
broadcast((2, 1, 3), (3, 2))
broadcast((3,), (2, 1))
broadcast((3,), (1, 2))
broadcast((3,), (1, 3))
```
```
broadcast((3, 1, 2), (3, 2))
broadcast((2, 1, 3), (3, 2))
broadcast((3,), (2, 1))
broadcast((3,), (1, 2))
broadcast((3,), (1, 3))
```
### Tensor Frameworks
#### Numpy
Numpy is an example of a framework for tensor computing that is widely supported and requires no special hardware. However, it still offers significant performance improvements by eliminating explicit python loops and using memory efficiently.
For example, let's calculate the opening angle separation between two unit vectors, each specified with (lat, lon) angles in radians (or RA,DEC for astronomers, as implemented [here](https://desisurvey.readthedocs.io/en/latest/api.html#desisurvey.utils.separation_matrix)). The [Haversine formula](https://en.wikipedia.org/wiki/Haversine_formula) is a good way to calculate this quantity.
Generate a large number of random unit vectors for benchmarking (are these uniformly distributed on the sphere?)
```
def generate(N, seed=123):
gen = np.random.RandomState(seed=123)
lats = gen.uniform(low=-np.pi / 2, high=+np.pi / 2, size=N)
lons = gen.uniform(low=0, high=2 * np.pi, size=N)
plt.plot(lons, lats, '.')
return lats, lons
lats, lons = generate(N=1000)
```
Use explicit python loops to calculate the (square) matrix of separation angles between all pairs of unit vectors:
```
def separation_matrix_loops():
# Allocate memory for the matrix.
N = len(lats)
matrix = np.empty((N, N))
for i, (lat1, lon1) in enumerate(zip(lats, lons)):
for j, (lat2, lon2) in enumerate(zip(lats, lons)):
# Evaluate the Haversine formula for matrix element [i, j].
matrix[i, j] = 2 * np.arcsin(np.sqrt(
np.sin(0.5 * (lat2 - lat1)) ** 2 +
np.cos(lat1) * np.cos(lat2) * np.sin(0.5 * (lon2 - lon1)) ** 2))
return matrix
%time S1 = separation_matrix_loops()
```
Now calculate the same separations using numpy implicit loops:
```
def separation_matrix_numpy():
lat1, lat2 = lats, lats.reshape(-1, 1)
lon1, lon2 = lons, lons.reshape(-1, 1)
return 2 * np.arcsin(np.sqrt(
np.sin(0.5 * (lat2 - lat1)) ** 2 +
np.cos(lat1) * np.cos(lat2) * np.sin(0.5 * (lon2 - lon1)) ** 2))
%time S2 = separation_matrix_numpy()
```
Check that both calculations give the same results:
```
np.allclose(S1, S2)
```
Since this is so much faster, increase the amount of computation (and memory) 100x for a better benchmark:
```
lats, lons = generate(N=10000)
%time S2 = separation_matrix_numpy()
```
Therefore using implicit numpy loops speeds up the calculation by a factor of about 6.8 / 0.02 = 340. Since we are using the efficient numpy arrays in both cases, the speed up is entirely due to the loops!
#### Other Frameworks: PyTorch and TensorFlow
Machine learning relies heavily on frameworks that copy the successful numpy design for tensor computing, while adding some important new features:
- Automatic hardware acceleration.
- Automatic calculation of derivatives.
- Efficient deployment to other platforms (mobile, cloud).
Unlike numpy, the default type in these frameworks is usually a 32-bit float, rather than a 64-bit float.
The two most popular tensor computing frameworks for machine learning today are [PyTorch](https://pytorch.org/) and [TensorFlow](https://www.tensorflow.org/). Both are large open-source projects, primarily developed by facebook (pytorch) and google (tensorflow). These frameworks were originally quite different, with pytorch preferred for research and tensorflow preferred for large-scale deployment, but they are gradually converging towards similar a feature set.
Below, we repeat our calculation of the separation matrix with both of these frameworks. You will notice that the new features come with some additional complexity.
#### PyTorch Example
```
import torch
device = torch.device("cuda") if torch.cuda.is_available() else "cpu"
print(f'Using device: {device}.')
lons_pt = torch.tensor(lons, device=device)
lats_pt = torch.tensor(lats, device=device)
def separation_matrix_torch():
lat1, lat2 = lats_pt, lats_pt.reshape(-1, 1)
lon1, lon2 = lons_pt, lons_pt.reshape(-1, 1)
return 2 * torch.asin(torch.sqrt(
torch.sin(0.5 * (lat2 - lat1)) ** 2 +
torch.cos(lat1) * torch.cos(lat2) * torch.sin(0.5 * (lon2 - lon1)) ** 2))
%time S3 = separation_matrix_torch()
np.allclose(S2, S3.numpy())
```
#### TensorFlow Example
```
import tensorflow as tf
tf.enable_eager_execution()
device = 'GPU:0' if tf.test.is_gpu_available() else 'CPU:0'
print(f'Using device: {device}.')
with tf.device(device):
lons_tf = tf.constant(lons)
lats_tf = tf.constant(lats)
def separation_matrix_tensorflow():
lat1, lat2 = lats_tf, tf.reshape(lats_tf, [-1, 1])
lon1, lon2 = lons_tf, tf.reshape(lons_tf, [-1, 1])
return 2 * tf.asin(tf.sqrt(
tf.sin(0.5 * (lat2 - lat1)) ** 2 +
tf.cos(lat1) * tf.cos(lat2) * tf.sin(0.5 * (lon2 - lon1)) ** 2))
%time S4 = separation_matrix_tensorflow()
np.allclose(S2, S4.numpy())
```
#### Hardware Acceleration
Tensor computing can be sped up significantly (10-100x) using hardware that is optimized to perform tensor computing by distributing simple calculations ("kernels") across many independent processors ("cores") running in parallel.
The original driver for such hardware was to accelerate the 3D geometry calculations required to render real time 3D graphics, leading to the first [Graphics Processing Units (GPUs)[https://en.wikipedia.org/wiki/Graphics_processing_unit] in the 1990s. More recently, GPUs have been adopted for purely numerical calculations, with no display attached, leading to the development of specialized programming languages such as [CUDA](https://en.wikipedia.org/wiki/CUDA) and [OpenCL](https://en.wikipedia.org/wiki/OpenCL).
Currently, one vendor (Nvidia) dominates the use of GPUs for ML with its proprietary CUDA language. Google has also introduced an even more specialized [TPU](https://en.wikipedia.org/wiki/Tensor_processing_unit) architecture.
The table below shows some benchmarks for the separation matrix problem, running on different hardware with different frameworks. The speed ups obtained using PyTorch and TensorFlow with a GPU are typical. The two frameworks provide comparable GPU performance overall, but can differ on specific problems.
| Test | Laptop |Server(GPU) | Collab(CPU) | Collab(GPU) |
|------------|--------|------------|-------------|-------------|
| numpy | 2.08s | 1.17s | 10.5s | 10.3s |
| torch | 7.32s | 48.7ms | --- | --- |
| tensorflow | --- | --- | 9.11s | 246ms |
| ratio | 3.5 | 1 / 24 | 0.87 | 1 / 41 |
To benefit from this hardware, you can either add a GPU to a linux server, or use a cloud computing platform.
Cloud computing is the easiest way to get started. There are some free options, but generally you have to "pay as you go" to do a useful amount of work. Some good starting points are:
- [Google Collaboratory](https://colab.research.google.com/): free research tool with a jupyter notebook front end.
- [PaperSpace](https://www.paperspace.com/): reasonably priced and simple to get started.
- [Amazon Web Services](https://aws.amazon.com/ec2/): free to try, very flexible and relatively complex.
**Note: this is not a complete list, and pricing and capabilities are rapidly changing.**
If you are considering building your own GPU server, start [here](http://timdettmers.com/2018/11/05/which-gpu-for-deep-learning/). As of Nov 2018, the recommended GPU for most users is the [RTX 2070](https://www.nvidia.com/en-us/geforce/graphics-cards/rtx-2070/). A single server can host 4 GPUs. Here is a single water-cooled [RTX 2080 Ti](https://www.nvidia.com/en-us/geforce/graphics-cards/rtx-2080-ti/) GPU installed in my office:

### Automatic Derivatives
In addition to hardware acceleration, a key feature of tensor computing frameworks for ML is their ability to automate the calculation of derivatives, which then enable efficient and accurate gradient-based optimization algorithms.
In general, a derivate can be implemented in software three ways:
- Analytically (using paper or mathematica) then copied into code: this is the most efficient and accurate but least generalizable.
- Numerically, with [finite difference equations](https://en.wikipedia.org/wiki/Finite_difference): this is the least efficient and accurate, but most generalizable.
- [Automatically](https://en.wikipedia.org/wiki/Automatic_differentiation): a hybrid approach where a small set of primitive functions (sin, cos, log, ...) are handled analytically, then the derivatives of expressions using these primitives are computed on the fly using the chain rule, product rule, etc. This is efficient and accurate, but requires that expressions are built entirely from primitives that support AD.
As a concrete example calculate the Gaussian expression
$$
y(x) = e^{-x^2}
$$
in PyTorch:
```
x = torch.linspace(-5, 5, 20, requires_grad=True)
y = torch.exp(-x ** 2)
y
```
We specify `requires_grad=True` to enable AD for all tensors that depend on `x` (so just `y` in this case). To calculate partial derivatives ("gradients") of `y` wrt `x`, use:
```
y.backward(torch.ones_like(y))
```
The tensor `x.grad` now contains $y'(x)$ at each value of `x`:
```
x.grad
x_n = x.detach().numpy()
yp_n = x.grad.detach().numpy()
y_n = y.detach().numpy()
plt.plot(x_n, y_n, 'o--', label='$y(x)$')
plt.plot(x_n, yp_n, 'o:', label='$y^\prime(x)$')
plt.legend();
```
Note that the derivatives are calculate to full machine precision and not affected by the coarse spacing in $x$.
### Higher-Level APIs for Tensor Computing
Although TensorFlow and PyTorch are both similar to numpy, they have different APIs so you are forced to choose one to take advantage of their unique features. However, for many calculations they are interchangeable, and a new ecosystem of higher-level APIs is growing to support this. For example, check out:
- [Tensorly](http://tensorly.org/stable/index.html): *"Tensor learning in python"*. Includes powerful [decomposition](https://arxiv.org/abs/1711.10781) (generalized PCA) and regression algorithms.
- [einops](https://github.com/arogozhnikov/einops): *"Deep learning operations reinvented"*. Supports compact expressions for complex indexing operations ([np.einsum](https://docs.scipy.org/doc/numpy/reference/generated/numpy.einsum.html) on steroids).
Neither of these packages are included in the MLS conda environment, but I encourage you to experiment with them if you want to write framework-independent tensor code.
|
github_jupyter
|
%matplotlib inline
import matplotlib.pyplot as plt
import seaborn as sns; sns.set()
import numpy as np
import pandas as pd
def xout(W, xin, b):
return np.maximum(0, W.dot(xin) + b)
def xout(W, xin, b):
# Add your solution here
return 0
rvec = np.array([0.1, -0.2, 0.3])
rvec[0], rvec[1], rvec[2]
rvec.shape
rvec.ndim
matrix = np.identity(3)
print(matrix)
matrix[1, 0], matrix[1, 1]
matrix.shape
matrix.ndim
tensor = np.ones((2, 3, 4))
print(tensor)
tensor[0, 0, 0], tensor[1, 2, 3]
tensor.shape
tensor.ndim
# Regular sequence of values
shape = (3, 4)
c1 = np.zeros(shape)
c2 = np.ones(shape)
c3 = np.full(shape, -1)
c4 = np.arange(12)
# Reproducible "random" numbers
gen = np.random.RandomState(seed=123)
r1 = gen.uniform(size=shape)
r2 = gen.normal(loc=-1, scale=2, size=shape)
# Calculated as function of existing array.
f1 = r1 * np.sin(r2) ** c3
c1.dtype, c4.dtype
c3.dtype, r2.dtype, f1.dtype
c4.reshape(c1.shape)
c4.reshape(2, 3, 2)
# Add one to each element.
c4 + 1
# Scale each column of the 3x4 ones matrix by a different value.
np.ones(shape=(3, 4)) * np.arange(4)
# Scale each row of the 3x4 ones matrix by a different value.
try:
np.ones(shape=(3, 4)) * np.arange(3)
except ValueError as e:
print(e)
np.ones(shape=(3, 4)) * np.arange(3).reshape(3, 1)
def broadcast(shape1, shape2):
array1 = np.ones(shape1)
array2 = np.ones(shape2)
try:
array12 = array1 + array2
print('shapes {} {} broadcast to {}'.format(shape1, shape2, array12.shape))
except ValueError as e:
print(e)
broadcast((1, 3), (3,))
broadcast((1, 2), (3,))
broadcast((3, 1, 2), (3, 2))
broadcast((2, 1, 3), (3, 2))
broadcast((3,), (2, 1))
broadcast((3,), (1, 2))
broadcast((3,), (1, 3))
broadcast((3, 1, 2), (3, 2))
broadcast((2, 1, 3), (3, 2))
broadcast((3,), (2, 1))
broadcast((3,), (1, 2))
broadcast((3,), (1, 3))
def generate(N, seed=123):
gen = np.random.RandomState(seed=123)
lats = gen.uniform(low=-np.pi / 2, high=+np.pi / 2, size=N)
lons = gen.uniform(low=0, high=2 * np.pi, size=N)
plt.plot(lons, lats, '.')
return lats, lons
lats, lons = generate(N=1000)
def separation_matrix_loops():
# Allocate memory for the matrix.
N = len(lats)
matrix = np.empty((N, N))
for i, (lat1, lon1) in enumerate(zip(lats, lons)):
for j, (lat2, lon2) in enumerate(zip(lats, lons)):
# Evaluate the Haversine formula for matrix element [i, j].
matrix[i, j] = 2 * np.arcsin(np.sqrt(
np.sin(0.5 * (lat2 - lat1)) ** 2 +
np.cos(lat1) * np.cos(lat2) * np.sin(0.5 * (lon2 - lon1)) ** 2))
return matrix
%time S1 = separation_matrix_loops()
def separation_matrix_numpy():
lat1, lat2 = lats, lats.reshape(-1, 1)
lon1, lon2 = lons, lons.reshape(-1, 1)
return 2 * np.arcsin(np.sqrt(
np.sin(0.5 * (lat2 - lat1)) ** 2 +
np.cos(lat1) * np.cos(lat2) * np.sin(0.5 * (lon2 - lon1)) ** 2))
%time S2 = separation_matrix_numpy()
np.allclose(S1, S2)
lats, lons = generate(N=10000)
%time S2 = separation_matrix_numpy()
import torch
device = torch.device("cuda") if torch.cuda.is_available() else "cpu"
print(f'Using device: {device}.')
lons_pt = torch.tensor(lons, device=device)
lats_pt = torch.tensor(lats, device=device)
def separation_matrix_torch():
lat1, lat2 = lats_pt, lats_pt.reshape(-1, 1)
lon1, lon2 = lons_pt, lons_pt.reshape(-1, 1)
return 2 * torch.asin(torch.sqrt(
torch.sin(0.5 * (lat2 - lat1)) ** 2 +
torch.cos(lat1) * torch.cos(lat2) * torch.sin(0.5 * (lon2 - lon1)) ** 2))
%time S3 = separation_matrix_torch()
np.allclose(S2, S3.numpy())
import tensorflow as tf
tf.enable_eager_execution()
device = 'GPU:0' if tf.test.is_gpu_available() else 'CPU:0'
print(f'Using device: {device}.')
with tf.device(device):
lons_tf = tf.constant(lons)
lats_tf = tf.constant(lats)
def separation_matrix_tensorflow():
lat1, lat2 = lats_tf, tf.reshape(lats_tf, [-1, 1])
lon1, lon2 = lons_tf, tf.reshape(lons_tf, [-1, 1])
return 2 * tf.asin(tf.sqrt(
tf.sin(0.5 * (lat2 - lat1)) ** 2 +
tf.cos(lat1) * tf.cos(lat2) * tf.sin(0.5 * (lon2 - lon1)) ** 2))
%time S4 = separation_matrix_tensorflow()
np.allclose(S2, S4.numpy())
x = torch.linspace(-5, 5, 20, requires_grad=True)
y = torch.exp(-x ** 2)
y
y.backward(torch.ones_like(y))
x.grad
x_n = x.detach().numpy()
yp_n = x.grad.detach().numpy()
y_n = y.detach().numpy()
plt.plot(x_n, y_n, 'o--', label='$y(x)$')
plt.plot(x_n, yp_n, 'o:', label='$y^\prime(x)$')
plt.legend();
| 0.631594 | 0.995085 |
```
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
import os
from IPython.display import display
# Packages for custom plot styles
from matplotlib import rc
from matplotlib.gridspec import GridSpec
rc("font", family="serif", size=11)
folder = "../../data/models/"
file = "m10y024/M1.0_Y0.24_0212.data"
df = pd.read_csv(folder+file, skiprows=5, delim_whitespace=True)
#display(df)
plt.figure(1, figsize=(9,4))
plt.subplot(121)
plt.plot(df['log_Teff'], df['log_L'])
plt.plot(df.loc[(df.log_R > 0.3)].log_Teff, df.loc[(df.log_R > 0.3)].log_L)
plt.gca().invert_xaxis()
plt.xlabel('Effective temperature (log($T_{eff}$))')
plt.ylabel('Luminosity ($\log(L/L_{\odot})$)')
plt.subplot(122)
plt.plot(df['star_age'], df['log_R'])
plt.plot(df.loc[(df.log_R > 0.3)].star_age, df.loc[(df.log_R > 0.3)].log_R)
plt.plot(df.loc[(df.log_R > 1.2)].star_age, df.loc[(df.log_R > 1.2)].log_R)
plt.scatter(df.loc[(df.log_center_T > 7.65)].star_age, df.loc[(df.log_center_T > 7.65)].log_R)
plt.xlim(1.4e10,)
plt.xlabel('Star Age')
plt.ylabel('Radius (log(R))')
plt.show()
folder = "../../data/models/"
file = "gyre/M1.0_Y0.24_0209_7.summary.txt"
dg = pd.read_csv(folder+file, skiprows=5, delim_whitespace=True)#, names=['l', 'n', 'f0', 'f0im', 'e_norm'])
#dg['Re(freq)'] = dg['Re(freq)']*2*np.pi
dnuavg = np.mean(np.diff(dg['Re(freq)']))
print(dnuavg)
display(dg.head(10))
plt.figure(3, figsize=(9,4))
plt.subplot(121)
plt.scatter(dg['n_pg'], dg['Re(freq)'])
plt.subplot(122)
plt.scatter(dg['Re(freq)'] % dnuavg, dg['Re(freq)'])
plt.show()
file = "m10y024/M1.0_Y0.24_0212.index"
dh = pd.read_csv(folder+file, skiprows=1, delim_whitespace=True, names=['model','priority','profile'])
prof1 = dh.loc[(dh.priority == 1)]['model'].min()
prof2 = dh.loc[(dh.priority == 1)]['model'].max()
prof = [prof1, prof2]
df = pd.read_csv(folder + 'm10y024/M1.0_Y0.24_0212.data', skiprows=5, delim_whitespace=True)
pts = pd.read_csv(folder + 'm10y024/M1.0_Y0.24_0212.index', skiprows=1,
names=['model', 'priority', 'profile'], delim_whitespace=True)
pts = pts.loc[(pts.priority == 1)]['model']
# This section of code is a test of a modification to the run_star_extras.f file
# that outputs a profile file when the star increases in radius in 0.1 log_R units.
# The conditions applied also allow multiple profiles to be produced for the region
# around the RGB bump.
# NOTE: This test appears to have been successful, and will now be applied to the
# grid of models.
plt.figure(1, figsize=(6,7))
df = df.loc[(df.he_core_mass != 0.0)]
plt.plot(df['log_Teff'], df['log_L'], label='')
#plt.scatter(df.loc[(df.model_number == 1500)]['log_Teff'],
# df.loc[(df.model_number == 1500)]['log_L'], marker='x')
#plt.scatter(df.loc[(np.abs(df['log_R']-0.85) < 0.01)]['log_Teff'],
# df.loc[(np.abs(df['log_R']-0.85) < 0.01)]['log_L'], alpha=0.5)
for i in pts:
label = df.loc[(df.model_number == i)]['log_R'].values
plt.scatter(df.loc[(df.model_number == i)]['log_Teff'],
df.loc[(df.model_number == i)]['log_L'],
label = f'{label[0]:.1f}')
plt.gca().invert_xaxis()
plt.xlabel(r'Effective Temperature ($\log(T_{eff}$)')
plt.ylabel(r'Luminosity ($\log(L/L_{\odot}$)')
plt.legend(title=r'Radius ($\log$R)')
plt.show()
import glob
#folder = "~/Documents/mesa/models/"
#files = sorted(os.listdir('/home/daniel/Documents/mesa/models/'))
#dfs = [pd.read_csv(folder+str(i), skiprows=5, delim_whitespace=True) for i in files]
directory = '../../data/models/'
folders = sorted(os.listdir(directory))
folders.pop(0)
files = [glob.glob(directory + i + '/*.data') for i in folders]
profs = [glob.glob(directory + i + '/*.index') for i in folders]
import itertools
colours = itertools.cycle(('r', 'orange', 'olive', 'teal', 'sienna', 'indigo'))#, 'black'))
## Need to develop tool for taking Y values and masses and allocating colours/linestyles (to-do)
Y = 0.24
M = 1.0
plt.figure(1, figsize=(6,7))
for i,j in zip(files, profs):
df = pd.read_csv(i[0], skiprows=5, delim_whitespace=True)
df = df.loc[(df.he_core_mass != 0.0)]
dg = pd.read_csv(j[0], skiprows=1,
names=['model', 'priority', 'profile'], delim_whitespace=True)
dg = dg.loc[(dg.priority == 1) & (dg.profile == 4)]['model']
pt = dg.values
if not pt:
continue
else:
if str(i[0][42:45]) == str(M):
colour = next(colours)
label = 'model '+str(int(pt))
plt.scatter(df.loc[(df.model_number == int(pt))]['log_Teff'],
df.loc[(df.model_number == int(pt))]['log_L'], c=colour, label=label)
label = str(i[0][47:51])
#label = str(i[0][42:45])
plt.plot(df['log_Teff'], df['log_L'], c=colour, zorder=0,
alpha=0.5, label=label)
plt.gca().invert_xaxis()
plt.xlabel(r'Effective Temperature ($\log(T_{eff})$)')
plt.ylabel(r'Luminosity ($\log(L/L_{\odot})$)')
#plt.legend(['0.24', '0.26', '0.28', '0.32', '0.36', '0.40'], title=r'$Y_{init}$')
plt.legend()
plt.show()
```
|
github_jupyter
|
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
import os
from IPython.display import display
# Packages for custom plot styles
from matplotlib import rc
from matplotlib.gridspec import GridSpec
rc("font", family="serif", size=11)
folder = "../../data/models/"
file = "m10y024/M1.0_Y0.24_0212.data"
df = pd.read_csv(folder+file, skiprows=5, delim_whitespace=True)
#display(df)
plt.figure(1, figsize=(9,4))
plt.subplot(121)
plt.plot(df['log_Teff'], df['log_L'])
plt.plot(df.loc[(df.log_R > 0.3)].log_Teff, df.loc[(df.log_R > 0.3)].log_L)
plt.gca().invert_xaxis()
plt.xlabel('Effective temperature (log($T_{eff}$))')
plt.ylabel('Luminosity ($\log(L/L_{\odot})$)')
plt.subplot(122)
plt.plot(df['star_age'], df['log_R'])
plt.plot(df.loc[(df.log_R > 0.3)].star_age, df.loc[(df.log_R > 0.3)].log_R)
plt.plot(df.loc[(df.log_R > 1.2)].star_age, df.loc[(df.log_R > 1.2)].log_R)
plt.scatter(df.loc[(df.log_center_T > 7.65)].star_age, df.loc[(df.log_center_T > 7.65)].log_R)
plt.xlim(1.4e10,)
plt.xlabel('Star Age')
plt.ylabel('Radius (log(R))')
plt.show()
folder = "../../data/models/"
file = "gyre/M1.0_Y0.24_0209_7.summary.txt"
dg = pd.read_csv(folder+file, skiprows=5, delim_whitespace=True)#, names=['l', 'n', 'f0', 'f0im', 'e_norm'])
#dg['Re(freq)'] = dg['Re(freq)']*2*np.pi
dnuavg = np.mean(np.diff(dg['Re(freq)']))
print(dnuavg)
display(dg.head(10))
plt.figure(3, figsize=(9,4))
plt.subplot(121)
plt.scatter(dg['n_pg'], dg['Re(freq)'])
plt.subplot(122)
plt.scatter(dg['Re(freq)'] % dnuavg, dg['Re(freq)'])
plt.show()
file = "m10y024/M1.0_Y0.24_0212.index"
dh = pd.read_csv(folder+file, skiprows=1, delim_whitespace=True, names=['model','priority','profile'])
prof1 = dh.loc[(dh.priority == 1)]['model'].min()
prof2 = dh.loc[(dh.priority == 1)]['model'].max()
prof = [prof1, prof2]
df = pd.read_csv(folder + 'm10y024/M1.0_Y0.24_0212.data', skiprows=5, delim_whitespace=True)
pts = pd.read_csv(folder + 'm10y024/M1.0_Y0.24_0212.index', skiprows=1,
names=['model', 'priority', 'profile'], delim_whitespace=True)
pts = pts.loc[(pts.priority == 1)]['model']
# This section of code is a test of a modification to the run_star_extras.f file
# that outputs a profile file when the star increases in radius in 0.1 log_R units.
# The conditions applied also allow multiple profiles to be produced for the region
# around the RGB bump.
# NOTE: This test appears to have been successful, and will now be applied to the
# grid of models.
plt.figure(1, figsize=(6,7))
df = df.loc[(df.he_core_mass != 0.0)]
plt.plot(df['log_Teff'], df['log_L'], label='')
#plt.scatter(df.loc[(df.model_number == 1500)]['log_Teff'],
# df.loc[(df.model_number == 1500)]['log_L'], marker='x')
#plt.scatter(df.loc[(np.abs(df['log_R']-0.85) < 0.01)]['log_Teff'],
# df.loc[(np.abs(df['log_R']-0.85) < 0.01)]['log_L'], alpha=0.5)
for i in pts:
label = df.loc[(df.model_number == i)]['log_R'].values
plt.scatter(df.loc[(df.model_number == i)]['log_Teff'],
df.loc[(df.model_number == i)]['log_L'],
label = f'{label[0]:.1f}')
plt.gca().invert_xaxis()
plt.xlabel(r'Effective Temperature ($\log(T_{eff}$)')
plt.ylabel(r'Luminosity ($\log(L/L_{\odot}$)')
plt.legend(title=r'Radius ($\log$R)')
plt.show()
import glob
#folder = "~/Documents/mesa/models/"
#files = sorted(os.listdir('/home/daniel/Documents/mesa/models/'))
#dfs = [pd.read_csv(folder+str(i), skiprows=5, delim_whitespace=True) for i in files]
directory = '../../data/models/'
folders = sorted(os.listdir(directory))
folders.pop(0)
files = [glob.glob(directory + i + '/*.data') for i in folders]
profs = [glob.glob(directory + i + '/*.index') for i in folders]
import itertools
colours = itertools.cycle(('r', 'orange', 'olive', 'teal', 'sienna', 'indigo'))#, 'black'))
## Need to develop tool for taking Y values and masses and allocating colours/linestyles (to-do)
Y = 0.24
M = 1.0
plt.figure(1, figsize=(6,7))
for i,j in zip(files, profs):
df = pd.read_csv(i[0], skiprows=5, delim_whitespace=True)
df = df.loc[(df.he_core_mass != 0.0)]
dg = pd.read_csv(j[0], skiprows=1,
names=['model', 'priority', 'profile'], delim_whitespace=True)
dg = dg.loc[(dg.priority == 1) & (dg.profile == 4)]['model']
pt = dg.values
if not pt:
continue
else:
if str(i[0][42:45]) == str(M):
colour = next(colours)
label = 'model '+str(int(pt))
plt.scatter(df.loc[(df.model_number == int(pt))]['log_Teff'],
df.loc[(df.model_number == int(pt))]['log_L'], c=colour, label=label)
label = str(i[0][47:51])
#label = str(i[0][42:45])
plt.plot(df['log_Teff'], df['log_L'], c=colour, zorder=0,
alpha=0.5, label=label)
plt.gca().invert_xaxis()
plt.xlabel(r'Effective Temperature ($\log(T_{eff})$)')
plt.ylabel(r'Luminosity ($\log(L/L_{\odot})$)')
#plt.legend(['0.24', '0.26', '0.28', '0.32', '0.36', '0.40'], title=r'$Y_{init}$')
plt.legend()
plt.show()
| 0.416441 | 0.432003 |
```
import numpy as np
from sympy.solvers import solve
from sympy import Symbol
import sympy
import matplotlib.pyplot as plt
```
#### Реализовать явный метод Эйлера
```
def euler_method(f, t0, tn, tau, y0):
eps = tau / 10000
while t0 < tn and abs(t0 - tn) > eps:
y0 += tau * f(t0, y0)
t0 += tau
return y0
```
##### Euler method testing
Function: $y'(t) = t\sqrt{y(t)}$ \
Initials: $t_0=0, \ y_0=y(t_0)=y(0)=1$ \
Step: $\tau=0.1$
```
t = np.arange(0, 11, 1, int)
y1 = np.array([euler_method(lambda t, y: t * y ** 0.5, 0, i, 0.1, 1) for i in t])
y2 = (t ** 2 + 4) ** 2 / 16
plt.plot(t, y1, label='estimated', marker='.')
plt.plot(t, y2, label='calculated', marker='.')
plt.grid(linestyle='--')
plt.title("Euler method")
plt.xlabel('t')
plt.ylabel('y')
plt.legend()
plt.show()
for _t, _y1, _y2 in zip(t, y1, y2):
print(f"t = {_t}:\n\tEstimated: {_y1}\n\tCalculated: {_y2}\n")
t = np.arange(0, 11, 1, int)
y1 = [euler_method(lambda t, y: t * y ** 0.5, 0, t[0], 0.1, 1)]
for i in range(1, len(t)):
y1 += [euler_method(lambda t, y: t * y ** 0.5, t[i - 1], t[i], 0.1, y1[-1])]
y2 = (t ** 2 + 4) ** 2 / 16
plt.plot(t, y1, label='estimated', marker='.')
plt.plot(t, y2, label='calculated', marker='.')
plt.grid(linestyle='--')
plt.title("Euler method")
plt.xlabel('t')
plt.ylabel('y')
plt.legend()
plt.show()
for _t, _y1, _y2 in zip(t, y1, y2):
print(f"t = {_t}:\n\tEstimated: {_y1}\n\tCalculated: {_y2}\n")
```
##### Implicit Euler method testing
Function: $y'(t) = t\sqrt{y(t)}$ \
Initials: $t_0=0, \ y_0=y(t_0)=y(0)=1$ \
Step: $\tau=0.1$
#### Реализовать неявный метод Эйлера
```
def implicit_euler_method(f, t0, tn, tau, y0):
eps = tau / 10000
while t0 + tau < tn and abs(tn - t0) > eps:
t0 += tau
y = Symbol('y')
y0 = solve(y - tau * f(t0, y) - y0, y)[0]
return y0
t = np.arange(0, 11, 1, int)
y1 = [implicit_euler_method(lambda t, y: t * y ** 0.5, 0, t[0], 0.1, 1)]
for i in range(1, len(t)):
y1 += [implicit_euler_method(lambda t, y: t * y ** 0.5, t[i - 1], t[i], 0.1, y1[-1])]
y2 = (t ** 2 + 4) ** 2 / 16
plt.plot(t, y1, label='estimated', marker='.')
plt.plot(t, y2, label='calculated', marker='.')
plt.grid(linestyle='--')
plt.title("Implicit Euler method")
plt.xlabel('t')
plt.ylabel('y')
plt.legend()
plt.show()
for _t, _y1, _y2 in zip(t, y1, y2):
print(f"t = {_t}:\n\tEstimated: {_y1}\n\tCalculated: {_y2}\n")
```
#### Реализовать метод Рунге-Кутты 4 порядка
```
def runge_kutta(f, t0, tn, tau, y0):
eps = tau / 10000
while t0 < tn and (tn - t0) > eps:
k1 = f(t0, y0)
k2 = f(t0 + tau / 2, y0 + tau * k1 / 2)
k3 = f(t0 + tau / 2, y0 + tau * k2 / 2)
k4 = f(t0 + tau, y0 + tau * k3)
y0 += tau / 6 * (k1 + 2 * k2 + 2 * k3 + k4)
t0 += tau
return y0
t = np.arange(0, 11, 1, int)
y1 = [runge_kutta(lambda t, y: t * y ** 0.5, 0, t[0], 0.1, 1)]
for i in range(1, len(t)):
y1 += [runge_kutta(lambda t, y: t * y ** 0.5, t[i - 1], t[i], 0.1, y1[-1])]
y2 = (t ** 2 + 4) ** 2 / 16
plt.plot(t, y1, label='estimated', marker='.')
plt.plot(t, y2, label='calculated', marker='.')
plt.grid(linestyle='--')
plt.title("Runge-Kutta method")
plt.xlabel('t')
plt.ylabel('y')
plt.legend()
plt.show()
for _t, _y1, _y2 in zip(t, y1, y2):
print(f"t = {_t}:\n\tEstimated: {_y1}\n\tCalculated: {_y2}\n")
```
|
github_jupyter
|
import numpy as np
from sympy.solvers import solve
from sympy import Symbol
import sympy
import matplotlib.pyplot as plt
def euler_method(f, t0, tn, tau, y0):
eps = tau / 10000
while t0 < tn and abs(t0 - tn) > eps:
y0 += tau * f(t0, y0)
t0 += tau
return y0
t = np.arange(0, 11, 1, int)
y1 = np.array([euler_method(lambda t, y: t * y ** 0.5, 0, i, 0.1, 1) for i in t])
y2 = (t ** 2 + 4) ** 2 / 16
plt.plot(t, y1, label='estimated', marker='.')
plt.plot(t, y2, label='calculated', marker='.')
plt.grid(linestyle='--')
plt.title("Euler method")
plt.xlabel('t')
plt.ylabel('y')
plt.legend()
plt.show()
for _t, _y1, _y2 in zip(t, y1, y2):
print(f"t = {_t}:\n\tEstimated: {_y1}\n\tCalculated: {_y2}\n")
t = np.arange(0, 11, 1, int)
y1 = [euler_method(lambda t, y: t * y ** 0.5, 0, t[0], 0.1, 1)]
for i in range(1, len(t)):
y1 += [euler_method(lambda t, y: t * y ** 0.5, t[i - 1], t[i], 0.1, y1[-1])]
y2 = (t ** 2 + 4) ** 2 / 16
plt.plot(t, y1, label='estimated', marker='.')
plt.plot(t, y2, label='calculated', marker='.')
plt.grid(linestyle='--')
plt.title("Euler method")
plt.xlabel('t')
plt.ylabel('y')
plt.legend()
plt.show()
for _t, _y1, _y2 in zip(t, y1, y2):
print(f"t = {_t}:\n\tEstimated: {_y1}\n\tCalculated: {_y2}\n")
def implicit_euler_method(f, t0, tn, tau, y0):
eps = tau / 10000
while t0 + tau < tn and abs(tn - t0) > eps:
t0 += tau
y = Symbol('y')
y0 = solve(y - tau * f(t0, y) - y0, y)[0]
return y0
t = np.arange(0, 11, 1, int)
y1 = [implicit_euler_method(lambda t, y: t * y ** 0.5, 0, t[0], 0.1, 1)]
for i in range(1, len(t)):
y1 += [implicit_euler_method(lambda t, y: t * y ** 0.5, t[i - 1], t[i], 0.1, y1[-1])]
y2 = (t ** 2 + 4) ** 2 / 16
plt.plot(t, y1, label='estimated', marker='.')
plt.plot(t, y2, label='calculated', marker='.')
plt.grid(linestyle='--')
plt.title("Implicit Euler method")
plt.xlabel('t')
plt.ylabel('y')
plt.legend()
plt.show()
for _t, _y1, _y2 in zip(t, y1, y2):
print(f"t = {_t}:\n\tEstimated: {_y1}\n\tCalculated: {_y2}\n")
def runge_kutta(f, t0, tn, tau, y0):
eps = tau / 10000
while t0 < tn and (tn - t0) > eps:
k1 = f(t0, y0)
k2 = f(t0 + tau / 2, y0 + tau * k1 / 2)
k3 = f(t0 + tau / 2, y0 + tau * k2 / 2)
k4 = f(t0 + tau, y0 + tau * k3)
y0 += tau / 6 * (k1 + 2 * k2 + 2 * k3 + k4)
t0 += tau
return y0
t = np.arange(0, 11, 1, int)
y1 = [runge_kutta(lambda t, y: t * y ** 0.5, 0, t[0], 0.1, 1)]
for i in range(1, len(t)):
y1 += [runge_kutta(lambda t, y: t * y ** 0.5, t[i - 1], t[i], 0.1, y1[-1])]
y2 = (t ** 2 + 4) ** 2 / 16
plt.plot(t, y1, label='estimated', marker='.')
plt.plot(t, y2, label='calculated', marker='.')
plt.grid(linestyle='--')
plt.title("Runge-Kutta method")
plt.xlabel('t')
plt.ylabel('y')
plt.legend()
plt.show()
for _t, _y1, _y2 in zip(t, y1, y2):
print(f"t = {_t}:\n\tEstimated: {_y1}\n\tCalculated: {_y2}\n")
| 0.4856 | 0.920254 |
# Signans - ASL Detector
```
import tensorflow as tf
import tensorflow_hub as hub
import threading
import cv2
import numpy as np
import sys
import os
import time
import six
import argparse
from object_detection.utils import config_util
from object_detection.protos import pipeline_pb2
from google.protobuf import text_format
from gtts import gTTS
from IPython.display import Audio
from object_detection.utils import label_map_util
from object_detection.utils import visualization_utils as viz_utils
from object_detection.builders import model_builder
from autocorrect import Speller
from pydub import AudioSegment
from pydub.playback import play
from google.cloud import translate_v2 as translate
from random import *
google = os.path.join(os.getcwd(), "stately-vector.json")
%env GOOGLE_APPLICATION_CREDENTIALS= google
config_file = os.path.join('TensorFlow-Model', 'pipeline.config')
config = config_util.get_configs_from_pipeline_file(config_file)
# Loading the model
detection_model = model_builder.build(model_config=config['model'], is_training=False)
ckpt = tf.compat.v2.train.Checkpoint(model=detection_model)
ckpt.restore(os.path.join('TensorFlow-Model', 'checkpoint', 'ckpt-0')).expect_partial()
@tf.function
def detect_fn(image):
image, shapes = detection_model.preprocess(image)
prediction_dict = detection_model.predict(image, shapes)
detections = detection_model.postprocess(prediction_dict, shapes)
return detections
category_index = label_map_util.create_category_index_from_labelmap(os.path.join('label_map.pbtxt'))
# OPEN CV capture
cap = cv2.VideoCapture(0)
width = int(cap.get(cv2.CAP_PROP_FRAME_WIDTH))
height = int(cap.get(cv2.CAP_PROP_FRAME_HEIGHT))
```
### Set Variables Below
```
output_language = "en-uk"
detection_speed = 2
lastest_reading = ""
phrase = ""
top_domain = {'pt-br':'com.br',
'pt':'pt',
'fr':'fr',
'fr-ca':'ca',
'en-uk':'co.uk',
'en':'com',
'es':'es',
'es-mx':'com.mx',
'zh-CN' : 'com'
}
lang = output_language.split('-')[0]
def play_audio(voice_file):
sound = AudioSegment.from_mp3(voice_file)
play(sound)
os.remove(voice_file)
def get_lastest_reading():
global phrase
while get_lastest_reading_thread.is_alive():
global lastest_reading
label = lastest_reading
if label != "":
if label == "dot":
if phrase != "":
spell = Speller()
phrase = spell(phrase)
if lang != "en":
translate_client = translate.Client()
translated_test = translate_client.translate(phrase, target_language=lang)["translatedText"]
voice = gTTS( text=translated_test, lang=lang, tld=top_domain[output_language], slow=False)
else:
voice = gTTS( text=phrase, lang=lang, tld=top_domain[output_language], slow=False)
voice_file = "voice-{}.mp3".format(str(randint(1, 9999999)))
voice.save(voice_file)
play_audio_thread = threading.Thread(target=play_audio, args=[voice_file])
play_audio_thread.daemon = True
play_audio_thread.start()
print ("audio:" + phrase)
phrase = ""
elif label == "space":
phrase += " "
spell = Speller()
phrase = spell(phrase)
print (phrase)
else:
phrase += label
print (label)
time.sleep(detection_speed)
lastest_reading = ""
get_lastest_reading_thread = threading.Thread(target=get_lastest_reading)
get_lastest_reading_thread.daemon = True
while True:
ret, frame = cap.read()
image_np = np.array(frame)
input_tensor = tf.convert_to_tensor(np.expand_dims(image_np, 0), dtype=tf.float32)
detections = detect_fn(input_tensor)
num_detections = int(detections.pop('num_detections'))
detections = {key: value[0, :num_detections].numpy()
for key, value in detections.items()}
detections['num_detections'] = num_detections
detections['detection_classes'] = detections['detection_classes'].astype(np.int64)
image_np_with_detections = image_np.copy()
# Accuracy of 85%
if detections['detection_scores'][0] > 0.85:
index = detections['detection_classes'][0]+1
label = category_index[index]['name']
cv2.putText(image_np_with_detections, label.upper() ,(50,150), cv2.FONT_HERSHEY_SIMPLEX, 3,(0, 0, 0),6,cv2.LINE_AA)
cv2.putText(image_np_with_detections, str(detections['detection_scores'][0]*100)[0:4]+"%" ,(50,250), cv2.FONT_HERSHEY_SIMPLEX, 2,(0, 0, 0),6,cv2.LINE_AA)
if lastest_reading == "":
if not get_lastest_reading_thread.is_alive():
get_lastest_reading_thread.start()
#Update the latest reading
lastest_reading = label
cv2.putText(image_np_with_detections,phrase.replace(" ", "-"),(10,50), cv2.FONT_HERSHEY_SIMPLEX, 1,(0, 0, 0),3,cv2.LINE_AA)
cv2.imshow('ASL - Signans', cv2.resize(image_np_with_detections, (640, 480)))
# Press Q to exit
if cv2.waitKey(1) & 0xFF == ord('q'):
cap.release()
break
# Press B to delete last letter
if cv2.waitKey(1) & 0xFF == ord('b'):
if len(phrase) >= 1:
phrase = phrase[0: -1]
else:
phrase = ""
cap.release()
```
|
github_jupyter
|
import tensorflow as tf
import tensorflow_hub as hub
import threading
import cv2
import numpy as np
import sys
import os
import time
import six
import argparse
from object_detection.utils import config_util
from object_detection.protos import pipeline_pb2
from google.protobuf import text_format
from gtts import gTTS
from IPython.display import Audio
from object_detection.utils import label_map_util
from object_detection.utils import visualization_utils as viz_utils
from object_detection.builders import model_builder
from autocorrect import Speller
from pydub import AudioSegment
from pydub.playback import play
from google.cloud import translate_v2 as translate
from random import *
google = os.path.join(os.getcwd(), "stately-vector.json")
%env GOOGLE_APPLICATION_CREDENTIALS= google
config_file = os.path.join('TensorFlow-Model', 'pipeline.config')
config = config_util.get_configs_from_pipeline_file(config_file)
# Loading the model
detection_model = model_builder.build(model_config=config['model'], is_training=False)
ckpt = tf.compat.v2.train.Checkpoint(model=detection_model)
ckpt.restore(os.path.join('TensorFlow-Model', 'checkpoint', 'ckpt-0')).expect_partial()
@tf.function
def detect_fn(image):
image, shapes = detection_model.preprocess(image)
prediction_dict = detection_model.predict(image, shapes)
detections = detection_model.postprocess(prediction_dict, shapes)
return detections
category_index = label_map_util.create_category_index_from_labelmap(os.path.join('label_map.pbtxt'))
# OPEN CV capture
cap = cv2.VideoCapture(0)
width = int(cap.get(cv2.CAP_PROP_FRAME_WIDTH))
height = int(cap.get(cv2.CAP_PROP_FRAME_HEIGHT))
output_language = "en-uk"
detection_speed = 2
lastest_reading = ""
phrase = ""
top_domain = {'pt-br':'com.br',
'pt':'pt',
'fr':'fr',
'fr-ca':'ca',
'en-uk':'co.uk',
'en':'com',
'es':'es',
'es-mx':'com.mx',
'zh-CN' : 'com'
}
lang = output_language.split('-')[0]
def play_audio(voice_file):
sound = AudioSegment.from_mp3(voice_file)
play(sound)
os.remove(voice_file)
def get_lastest_reading():
global phrase
while get_lastest_reading_thread.is_alive():
global lastest_reading
label = lastest_reading
if label != "":
if label == "dot":
if phrase != "":
spell = Speller()
phrase = spell(phrase)
if lang != "en":
translate_client = translate.Client()
translated_test = translate_client.translate(phrase, target_language=lang)["translatedText"]
voice = gTTS( text=translated_test, lang=lang, tld=top_domain[output_language], slow=False)
else:
voice = gTTS( text=phrase, lang=lang, tld=top_domain[output_language], slow=False)
voice_file = "voice-{}.mp3".format(str(randint(1, 9999999)))
voice.save(voice_file)
play_audio_thread = threading.Thread(target=play_audio, args=[voice_file])
play_audio_thread.daemon = True
play_audio_thread.start()
print ("audio:" + phrase)
phrase = ""
elif label == "space":
phrase += " "
spell = Speller()
phrase = spell(phrase)
print (phrase)
else:
phrase += label
print (label)
time.sleep(detection_speed)
lastest_reading = ""
get_lastest_reading_thread = threading.Thread(target=get_lastest_reading)
get_lastest_reading_thread.daemon = True
while True:
ret, frame = cap.read()
image_np = np.array(frame)
input_tensor = tf.convert_to_tensor(np.expand_dims(image_np, 0), dtype=tf.float32)
detections = detect_fn(input_tensor)
num_detections = int(detections.pop('num_detections'))
detections = {key: value[0, :num_detections].numpy()
for key, value in detections.items()}
detections['num_detections'] = num_detections
detections['detection_classes'] = detections['detection_classes'].astype(np.int64)
image_np_with_detections = image_np.copy()
# Accuracy of 85%
if detections['detection_scores'][0] > 0.85:
index = detections['detection_classes'][0]+1
label = category_index[index]['name']
cv2.putText(image_np_with_detections, label.upper() ,(50,150), cv2.FONT_HERSHEY_SIMPLEX, 3,(0, 0, 0),6,cv2.LINE_AA)
cv2.putText(image_np_with_detections, str(detections['detection_scores'][0]*100)[0:4]+"%" ,(50,250), cv2.FONT_HERSHEY_SIMPLEX, 2,(0, 0, 0),6,cv2.LINE_AA)
if lastest_reading == "":
if not get_lastest_reading_thread.is_alive():
get_lastest_reading_thread.start()
#Update the latest reading
lastest_reading = label
cv2.putText(image_np_with_detections,phrase.replace(" ", "-"),(10,50), cv2.FONT_HERSHEY_SIMPLEX, 1,(0, 0, 0),3,cv2.LINE_AA)
cv2.imshow('ASL - Signans', cv2.resize(image_np_with_detections, (640, 480)))
# Press Q to exit
if cv2.waitKey(1) & 0xFF == ord('q'):
cap.release()
break
# Press B to delete last letter
if cv2.waitKey(1) & 0xFF == ord('b'):
if len(phrase) >= 1:
phrase = phrase[0: -1]
else:
phrase = ""
cap.release()
| 0.373304 | 0.50061 |
# Training metrics
*Metrics* for training fastai models are simply functions that take `input` and `target` tensors, and return some metric of interest for training. You can write your own metrics by defining a function of that type, and passing it to [`Learner`](/basic_train.html#Learner) in the [code]metrics[/code] parameter, or use one of the following pre-defined functions.
```
from fastai.gen_doc.nbdoc import *
from fastai import *
```
## Predefined metrics:
```
show_doc(accuracy)
show_doc(accuracy_thresh, doc_string=False)
```
Compute accuracy when `y_pred` and `y_true` for multi-label models, based on comparing predictions to `thresh`, `sigmoid` will be applied to `y_pred` if the corresponding flag is True.
```
show_doc(dice)
show_doc(fbeta)
```
See the [F1 score wikipedia page](https://en.wikipedia.org/wiki/F1_score) for details.
```
show_doc(exp_rmspe)
```
## Creating your own metric
Creating a new metric can be as simple as creating a new function. If you metric is an average over the total number of elements in your dataset, just write the function that will compute it on a batch (taking `pred` and `targ` as arguments). It will then be automatically averaged over the batches (taking their different sizes into acount).
Sometimes metrics aren't simple averages however. If we take the example of precision for instance, we have to divide the number of true positives by the number of predictions we made for that class. This isn't an average over the number of elements we have in the dataset, we only consider those where we made a positive prediction for a specific thing. Computing the precision for each batch, then averaging them will yield to a result that may be close to the real value, but won't be it exactly (and it really depends on how you deal with special case of 0 positive predicitions).
This why in fastai, every metric is implemented as a callback. If you pass a regular function, the library transforms it to a proper callback called `AverageCallback`. The callback metrics are only called during the validation phase, and only for the following events:
- <code>on_epoch_begin</code> (for initialization)
- <code>on_batch_begin</code> (if we need to have a look at the input/target and maybe modify them)
- <code>on_batch_end</code> (to analyze the last results and update our computation)
- <code>on_epoch_end</code>(to wrap up the final result that should be stored in `.metric`)
As an example, is here the exact implementation of the [`AverageMetric`](/callback.html#AverageMetric) callback that transforms a function like [`accuracy`](/metrics.html#accuracy) into a metric callback.
```
class AverageMetric(Callback):
def __init__(self, func):
self.func, self.name = func, func.__name__
def on_epoch_begin(self, **kwargs):
self.val, self.count = 0.,0
def on_batch_end(self, last_output, last_target, train, **kwargs):
self.count += last_target.size(0)
self.val += last_target.size(0) * self.func(last_output, last_target).detach().item()
def on_epoch_end(self, **kwargs):
self.metric = self.val/self.count
```
And here is another example that properly computes the precision for a given class.
```
class Precision(Callback):
def on_epoch_begin(self, **kwargs):
self.correct, self.total = 0, 0
def on_batch_end(self, last_output, last_target, **kwargs):
preds = last_output.argmax(1)
self.correct += ((preds==0) * (last_target==0)).float().sum()
self.total += (preds==0).float().sum()
def on_epoch_end(self, **kwargs):
self.metric = self.correct/self.total
```
## Undocumented Methods - Methods moved below this line will intentionally be hidden
## New Methods - Please document or move to the undocumented section
|
github_jupyter
|
from fastai.gen_doc.nbdoc import *
from fastai import *
show_doc(accuracy)
show_doc(accuracy_thresh, doc_string=False)
show_doc(dice)
show_doc(fbeta)
show_doc(exp_rmspe)
class AverageMetric(Callback):
def __init__(self, func):
self.func, self.name = func, func.__name__
def on_epoch_begin(self, **kwargs):
self.val, self.count = 0.,0
def on_batch_end(self, last_output, last_target, train, **kwargs):
self.count += last_target.size(0)
self.val += last_target.size(0) * self.func(last_output, last_target).detach().item()
def on_epoch_end(self, **kwargs):
self.metric = self.val/self.count
class Precision(Callback):
def on_epoch_begin(self, **kwargs):
self.correct, self.total = 0, 0
def on_batch_end(self, last_output, last_target, **kwargs):
preds = last_output.argmax(1)
self.correct += ((preds==0) * (last_target==0)).float().sum()
self.total += (preds==0).float().sum()
def on_epoch_end(self, **kwargs):
self.metric = self.correct/self.total
| 0.601008 | 0.984471 |
# Table of Contents
<p><div class="lev1"><a href="#Login"><span class="toc-item-num">1 </span>Login</a></div><div class="lev1"><a href="#Get-Machines-and-Users"><span class="toc-item-num">2 </span>Get Machines and Users</a></div><div class="lev1"><a href="#Plotting-code"><span class="toc-item-num">3 </span>Plotting code</a></div>
```
import sys
import os
import json
import time
import getpass
import pymongo
import datetime
import getpass
import numpy as np
#mpl for plotting
import matplotlib.pyplot as plt
%matplotlib inline
import matplotlib.cm as cm
import matplotlib.gridspec as gridspec
import matplotlib as mpl
```
# Login
```
mongo_client = pymongo.MongoClient(host='localhost', port=27018)
mongo_client['admin'].authenticate('admin',getpass.getpass('Please enter a password: '))
x = mongo_client['data']['machine_list'].find()
machines = []
for x_i in x:
machines.append(x_i['machine'])
```
# Get Machines and Users
```
x = mongo_client['data']['machine_list'].find()
machines = []
for x_i in x:
machines.append(x_i['machine'])
x = mongo_client['data']['user_list'].find()
users = []
for x_i in x:
users.append(x_i['user'])
start_date = datetime.datetime(2016, 11, 9)
#end_date = datetime.datetime(2016, 10, 27)
end_date = datetime.datetime.now()
machine = 'einhorn'
data = list(mongo_client['data']['load_info'].find({'machine' : machine, 'date' : {'$gt' : start_date, '$lt' : end_date}}).sort('date', pymongo.DESCENDING))
plot_gpu_load(data)
plot_gpu_memory_load(data)
plot_cpu_load(data)
plot_cpu_memory_load(data)
```
# Plotting code
* Add the max memory and swap into legend / titel. Think about changes max etc could change!
* Multi machine plots.
* Change report across time!
* HDD info.
* Change color for non supported. http://stackoverflow.com/questions/31345489/pyplot-change-color-of-line-if-data-is-less-than-zero
```
def plot_gpu_load(data):
machine = data[0]['machine']
gpus = set()
times = []
for d in data:
gpus.update(d.get('gpu', {}).keys())
times.append(d['date'])
xy = {}
for g in gpus:
xy[g] = []
for d in data:
t = d['date']
if d.get('error', False) == False:
for g,v in d['gpu'].items():
xy[g].append((t,v.get('load',-4)))
else:
for g in gpus:
xy[g].append((t, np.nan))
k = list(xy.keys())
fig, axes = plt.subplots(figsize=(15,5))
axes.set_title('GPU load on {} [{:s} - {:s} ]'.format(machine, np.min(times).strftime("%d.%m.%y %H:%M"), np.max(times).strftime("%d.%m.%y %H:%M")))
axes.set_ylim((-5, 105))
for p in k:
x, y = zip(*xy[p])
l = axes.plot(x,y, label='{} Load [%]'.format(p))
axes.fill_between(x,y, facecolor=l[0].get_color(), alpha=0.5)
fig.autofmt_xdate()
axes.legend(loc="upper left", bbox_to_anchor=(1,1))
def plot_cpu_load(data):
machine = data[0]['machine']
times = []
for d in data:
times.append(d['date'])
xy = []
for d in data:
t = d['date']
if d.get('error', False) == False:
xy.append((t,d['cpu'].get('load',-4)))
else:
xy.append((t, np.nan))
fig, axes = plt.subplots(figsize=(15,5))
axes.set_title('CPU load on {} [{:s} - {:s} ]'.format(machine, np.min(times).strftime("%d.%m.%y %H:%M"), np.max(times).strftime("%d.%m.%y %H:%M")))
axes.set_ylim((-5, 105))
x, y = zip(*xy)
l = axes.plot(x,y, label='CPU Load [%]')
axes.fill_between(x,y, facecolor=l[0].get_color(), alpha=0.5)
fig.autofmt_xdate()
axes.legend(loc="upper left", bbox_to_anchor=(1,1))
def plot_cpu_memory_load(data):
machine = data[0]['machine']
times = []
for d in data:
times.append(d['date'])
xy_mem = []
xy_ram = []
for d in data:
t = d['date']
if d.get('error', False) == False:
xy_mem.append((t,d['cpu'].get('used_ram',-4)*100/d['cpu'].get('max_ram',1)))
xy_ram.append((t,d['cpu'].get('used_swap',-4)*100/d['cpu'].get('max_swap',1)))
else:
xy_mem.append((t, np.nan))
xy_ram.append((t, np.nan))
fig, axes = plt.subplots(figsize=(15,5))
axes.set_title('Memory load on {} [{:s} - {:s} ]'.format(machine, np.min(times).strftime("%d.%m.%y %H:%M"), np.max(times).strftime("%d.%m.%y %H:%M")))
axes.set_ylim((-5, 105))
x, y = zip(*xy_mem)
l = axes.plot(x,y, color='green', label='Memory[%]')
axes.fill_between(x,y, facecolor=l[0].get_color(), alpha=0.5)
x, y = zip(*xy_ram)
l = axes.plot(x,y, color='red', label='Swap[%]')
axes.fill_between(x,y, facecolor=l[0].get_color(), alpha=0.5)
fig.autofmt_xdate()
axes.legend(loc="upper left", bbox_to_anchor=(1,1))
def plot_gpu_memory_load(data):
machine = data[0]['machine']
gpus = set()
times = []
for d in data:
gpus.update(d.get('gpu', {}).keys())
times.append(d['date'])
gpus_users = {g : set() for g in list(gpus)}
for d in data:
gs = d.get('gpu', {}).keys()
for g in gs:
if d['gpu'][g]['proc_info_support']:
gpus_users[g].update(set(d['gpu'][g]['proc_info'].keys()))
xy = {}
for k, v in gpus_users.items():
xy[k] = {'total' : []}
for u in list(v):
xy[k][u] = []
for d in data:
t = d['date']
if d.get('error', False) == False:
for g,v in d['gpu'].items():
max_mem = v['max_mem']
xy[g]['total'].append((t,100.0*v['cur_mem']/max_mem))
for u in gpus_users[g]:
xy[g][u].append((t,100.0*v.get('proc_info',{}).get(u,0)/max_mem))
else:
for g in gpus:
xy[g]['total'].append((t,np.nan))
for u in gpus_users[g]:
xy[g][u].append((t, np.nan))
k = list(xy.keys())
fig, axes = plt.subplots(len(k),1,figsize=(15,5*len(k)))
if type(axes) is not np.ndarray:
axes = np.asarray([axes])
for i, g in enumerate(sorted(k)):
axes[i].set_title('{} memory load on {} [{:s} - {:s} ]'.format(g, machine, np.min(times).strftime("%d.%m.%y %H:%M"), np.max(times).strftime("%d.%m.%y %H:%M")))
axes[i].set_ylim((-5, 105))
x, y = zip(*xy[g]['total'])
l = axes[i].plot(x,y, label='Total mem [%]', lw=0.2)
axes[i].fill_between(x,y, facecolor=l[0].get_color(), alpha=0.15)
prev = 0
mem_sum = 0*np.asarray(y)
for u in gpus_users[g]:
x, y = zip(*xy[g][u])
mem_sum += np.asarray(y)
l = axes[i].plot(x,mem_sum, label='{} mem [%]'.format(u), lw=0.5)
axes[i].fill_between(x,mem_sum, prev, facecolor=l[0].get_color(), alpha=0.7)
prev = mem_sum.copy()
fig.autofmt_xdate()
axes[i].legend(loc="upper left", bbox_to_anchor=(1,1))
return None
for p in k:
x, y = zip(*xy[p])
l = axes.plot(x,y, label='{} Load [%]'.format(p))
x, y = zip(*xy_mem)
l = axes.plot(x,y, color='green', label='Memory[%]')
axes.fill_between(x,y, facecolor=l[0].get_color(), alpha=0.5)
x, y = zip(*xy_ram)
l = axes.plot(x,y, color='red', label='Swap[%]')
axes.fill_between(x,y, facecolor=l[0].get_color(), alpha=0.5)
fig.autofmt_xdate()
axes.legend(loc="upper left", bbox_to_anchor=(1,1))
```
|
github_jupyter
|
import sys
import os
import json
import time
import getpass
import pymongo
import datetime
import getpass
import numpy as np
#mpl for plotting
import matplotlib.pyplot as plt
%matplotlib inline
import matplotlib.cm as cm
import matplotlib.gridspec as gridspec
import matplotlib as mpl
mongo_client = pymongo.MongoClient(host='localhost', port=27018)
mongo_client['admin'].authenticate('admin',getpass.getpass('Please enter a password: '))
x = mongo_client['data']['machine_list'].find()
machines = []
for x_i in x:
machines.append(x_i['machine'])
x = mongo_client['data']['machine_list'].find()
machines = []
for x_i in x:
machines.append(x_i['machine'])
x = mongo_client['data']['user_list'].find()
users = []
for x_i in x:
users.append(x_i['user'])
start_date = datetime.datetime(2016, 11, 9)
#end_date = datetime.datetime(2016, 10, 27)
end_date = datetime.datetime.now()
machine = 'einhorn'
data = list(mongo_client['data']['load_info'].find({'machine' : machine, 'date' : {'$gt' : start_date, '$lt' : end_date}}).sort('date', pymongo.DESCENDING))
plot_gpu_load(data)
plot_gpu_memory_load(data)
plot_cpu_load(data)
plot_cpu_memory_load(data)
def plot_gpu_load(data):
machine = data[0]['machine']
gpus = set()
times = []
for d in data:
gpus.update(d.get('gpu', {}).keys())
times.append(d['date'])
xy = {}
for g in gpus:
xy[g] = []
for d in data:
t = d['date']
if d.get('error', False) == False:
for g,v in d['gpu'].items():
xy[g].append((t,v.get('load',-4)))
else:
for g in gpus:
xy[g].append((t, np.nan))
k = list(xy.keys())
fig, axes = plt.subplots(figsize=(15,5))
axes.set_title('GPU load on {} [{:s} - {:s} ]'.format(machine, np.min(times).strftime("%d.%m.%y %H:%M"), np.max(times).strftime("%d.%m.%y %H:%M")))
axes.set_ylim((-5, 105))
for p in k:
x, y = zip(*xy[p])
l = axes.plot(x,y, label='{} Load [%]'.format(p))
axes.fill_between(x,y, facecolor=l[0].get_color(), alpha=0.5)
fig.autofmt_xdate()
axes.legend(loc="upper left", bbox_to_anchor=(1,1))
def plot_cpu_load(data):
machine = data[0]['machine']
times = []
for d in data:
times.append(d['date'])
xy = []
for d in data:
t = d['date']
if d.get('error', False) == False:
xy.append((t,d['cpu'].get('load',-4)))
else:
xy.append((t, np.nan))
fig, axes = plt.subplots(figsize=(15,5))
axes.set_title('CPU load on {} [{:s} - {:s} ]'.format(machine, np.min(times).strftime("%d.%m.%y %H:%M"), np.max(times).strftime("%d.%m.%y %H:%M")))
axes.set_ylim((-5, 105))
x, y = zip(*xy)
l = axes.plot(x,y, label='CPU Load [%]')
axes.fill_between(x,y, facecolor=l[0].get_color(), alpha=0.5)
fig.autofmt_xdate()
axes.legend(loc="upper left", bbox_to_anchor=(1,1))
def plot_cpu_memory_load(data):
machine = data[0]['machine']
times = []
for d in data:
times.append(d['date'])
xy_mem = []
xy_ram = []
for d in data:
t = d['date']
if d.get('error', False) == False:
xy_mem.append((t,d['cpu'].get('used_ram',-4)*100/d['cpu'].get('max_ram',1)))
xy_ram.append((t,d['cpu'].get('used_swap',-4)*100/d['cpu'].get('max_swap',1)))
else:
xy_mem.append((t, np.nan))
xy_ram.append((t, np.nan))
fig, axes = plt.subplots(figsize=(15,5))
axes.set_title('Memory load on {} [{:s} - {:s} ]'.format(machine, np.min(times).strftime("%d.%m.%y %H:%M"), np.max(times).strftime("%d.%m.%y %H:%M")))
axes.set_ylim((-5, 105))
x, y = zip(*xy_mem)
l = axes.plot(x,y, color='green', label='Memory[%]')
axes.fill_between(x,y, facecolor=l[0].get_color(), alpha=0.5)
x, y = zip(*xy_ram)
l = axes.plot(x,y, color='red', label='Swap[%]')
axes.fill_between(x,y, facecolor=l[0].get_color(), alpha=0.5)
fig.autofmt_xdate()
axes.legend(loc="upper left", bbox_to_anchor=(1,1))
def plot_gpu_memory_load(data):
machine = data[0]['machine']
gpus = set()
times = []
for d in data:
gpus.update(d.get('gpu', {}).keys())
times.append(d['date'])
gpus_users = {g : set() for g in list(gpus)}
for d in data:
gs = d.get('gpu', {}).keys()
for g in gs:
if d['gpu'][g]['proc_info_support']:
gpus_users[g].update(set(d['gpu'][g]['proc_info'].keys()))
xy = {}
for k, v in gpus_users.items():
xy[k] = {'total' : []}
for u in list(v):
xy[k][u] = []
for d in data:
t = d['date']
if d.get('error', False) == False:
for g,v in d['gpu'].items():
max_mem = v['max_mem']
xy[g]['total'].append((t,100.0*v['cur_mem']/max_mem))
for u in gpus_users[g]:
xy[g][u].append((t,100.0*v.get('proc_info',{}).get(u,0)/max_mem))
else:
for g in gpus:
xy[g]['total'].append((t,np.nan))
for u in gpus_users[g]:
xy[g][u].append((t, np.nan))
k = list(xy.keys())
fig, axes = plt.subplots(len(k),1,figsize=(15,5*len(k)))
if type(axes) is not np.ndarray:
axes = np.asarray([axes])
for i, g in enumerate(sorted(k)):
axes[i].set_title('{} memory load on {} [{:s} - {:s} ]'.format(g, machine, np.min(times).strftime("%d.%m.%y %H:%M"), np.max(times).strftime("%d.%m.%y %H:%M")))
axes[i].set_ylim((-5, 105))
x, y = zip(*xy[g]['total'])
l = axes[i].plot(x,y, label='Total mem [%]', lw=0.2)
axes[i].fill_between(x,y, facecolor=l[0].get_color(), alpha=0.15)
prev = 0
mem_sum = 0*np.asarray(y)
for u in gpus_users[g]:
x, y = zip(*xy[g][u])
mem_sum += np.asarray(y)
l = axes[i].plot(x,mem_sum, label='{} mem [%]'.format(u), lw=0.5)
axes[i].fill_between(x,mem_sum, prev, facecolor=l[0].get_color(), alpha=0.7)
prev = mem_sum.copy()
fig.autofmt_xdate()
axes[i].legend(loc="upper left", bbox_to_anchor=(1,1))
return None
for p in k:
x, y = zip(*xy[p])
l = axes.plot(x,y, label='{} Load [%]'.format(p))
x, y = zip(*xy_mem)
l = axes.plot(x,y, color='green', label='Memory[%]')
axes.fill_between(x,y, facecolor=l[0].get_color(), alpha=0.5)
x, y = zip(*xy_ram)
l = axes.plot(x,y, color='red', label='Swap[%]')
axes.fill_between(x,y, facecolor=l[0].get_color(), alpha=0.5)
fig.autofmt_xdate()
axes.legend(loc="upper left", bbox_to_anchor=(1,1))
| 0.150153 | 0.81231 |
<a href="https://colab.research.google.com/github/adowaconan/LevelUpPythonTutorial/blob/master/1_3_Numpy_numerical_computation.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# Numpy makes most of the work in Python much easier than dealing with lists, tuples, and sets.
```
import numpy as np
from matplotlib import pyplot as plt # just to visualize the arrays, will be introduced later
complex_list = [[ 0, 1, 2, 3, 4],
[ 5, 6, 7, 8, 9],
[10, 11, 12, 13, 14]]
print(complex_list)
complex_array = np.arange(15).reshape(3, 5)
print(complex_array)
complex_array.shape # 3 rows and 5 columns
complex_array.ndim
type(complex_array)
```
# convert lists to arrays
```
a = [1,2,3]
print(a)
print(type(a)) # type of class
b = np.array(a)
print(b)
print(type(b)) # type of class
print(b.dtype) # tyep of the data in the class
a = [1.0,2,3]
print(a)
print(type(a))
b = np.array(a)
print(b)
print(type(b))
print(b.dtype)
a = [1.0,2,3]
print(a)
print(type(a))
b = np.array(a).astype(int)
print(b)
print(type(b))
print(b.dtype)
```
# Something we do in Matlab: create zeros, ones, eyes, but only in numpy can we create empty containers
```
np.zeros((2,5))
np.ones((2,5))
np.eye(5)
```
## Although you see that these are ones, they have no values. The ones merely fill the container/spaces
```
np.empty((2,5))
```
Did you notice most of the functions take tuples as the argument for array size?
# Create a list from 0 to 10
## Wrong count because Python counts from 0, thus from 0 to 10, so we have 11 values
```
np.arange(10)
np.arange(11)
```
# Create a list from 0 to 10 with specified step sizes
```
np.arange(start = 0, stop = 10.1, step = 0.5)
```
# Create a list from 0 to 10 with unknown step sizes (but know how many elements we want in between)
```
np.linspace(start = 0, stop = 10, num = 26,)
```
# 1D vector
```
np.arange(6)
```
# 2D matrix
```
np.arange(12).reshape(4,3) # 4 rows and 3 columns
```
## Transpose
```
a = np.arange(12).reshape(4,3)
a.T
```
# 3D array
```
np.arange(24).reshape(2,3,4) # 2 slice, 3 rows, and 4 columns
```
# One main reason we like arrays better than lists: calculation
```
a = [1,2,3]
b = [2,3,4]
print(b - a)
a = np.array([1,2,3])
b = np.array([2,3,4])
print(b - a)
```
## Elementwise multiplication
```
a * b
```
## Matrix product
```
a @ b
a.dot(b)
```
# Random number generator
```
a = np.random.normal(loc = 0.,# mean
scale = 1., # standard deviation
size = 100,)
print(a)
plt.hist(a)
```
# Pseudo random generator
```
for i in range(10):
print(np.random.randint(1,10))
for i in range(10):
np.random.seed(12345)
print(np.random.randint(1,10))
```
# Sample from a given array
```
# this is equivalent to flip a fair coin 100 times
a = np.random.choice([0,1],size = 100, replace = True, p = [0.5,0.5])
print(a)
# this is equivalent to roll a fair die 100 times
a = np.random.choice(np.arange(start = 1, stop = 7),size = 100, replace = True, p = [1./6] * 6)
print(a)
```
# Reorder a vector
```
a = np.arange(20)
print(a)
np.random.shuffle(a)
print(a)
```
# Iterate through a N-dimensional array: you will iterate through the first dimension (which usually are the rows)
```
a = np.arange(100).reshape(20,5)
print(a)
for ii,row in enumerate(a):
print('row ',ii,row)
```
# Concatenation, stack: putting multiple arrays together
## Simple concatenation
```
a = np.arange(6)
b = np.arange(7)
print(a,b)
print(np.concatenate([a,b]))
```
## Stacking
### Row stacking
```
a = np.arange(6).reshape(2,3)
b = np.arange(8).reshape(2,4)
print(a)
print(b)
print(np.hstack([a,b]))
```
### Column stacking
```
a = np.arange(6).reshape(2,3)
b = np.arange(8).reshape(2,4)
print(a)
print(b)
print(np.vstack([a,b]))
a = np.arange(6).reshape(3,2)
b = np.arange(8).reshape(4,2)
print(a)
print(b)
print(np.vstack([a,b]))
a = np.arange(7)
b = np.arange(7)
print(a)
print(b)
print(np.vstack([a,b]))
```
# Another reason we like arrays better than lists
```
a = [[False, False, False, False],
[False, True, True, True],
[ True, True, True, True]]
sum(a)
print('how many True in each row: ',np.sum(a,0))
print('how many True in each column: ',np.sum(a,1))
print('how many True in total: ', np.sum(a))
```
# Implement resampling-based t-test only using numpy
```
mean = 0.015,
std = 0.026
a = np.random.normal(mean,std,size = 26)
print(a)
plt.hist(a)
```
## Is variable `a` significantly different from 0?
### [resampling method](http://www.stat.ucla.edu/~rgould/110as02/bshypothesis.pdf)
```
baseline = 0
n_permutations = 5000
n_simulations = 100
experiment_mean = np.mean(a)
rescale_experiment_mean_to_zero = a - np.mean(a) + baseline
# to generate a distribution of null hypothesis that centers at 0 but with the
# same shape of distribution as variable a
# 1st dimension: number of samples same as variable a
# 2nd dimension: within one permutation resampling, we perform resampling number
# of samples same as variable a, but also repeat this n_permutations times
# 3rd dimension: repeat the permutation n_simulation times to estimate if the
# permutation is effective
resamples = np.random.choice(rescale_experiment_mean_to_zero,
size = (a.shape[0],n_permutations,n_simulations),
replace = True)
# take the average over the 1st dimension because we only care about if the mean
# of the null distribution along each permutation and each simulation
resamples = resamples.mean(0)
pvalues = (np.sum(np.abs(resamples) >= np.abs(experiment_mean),axis=0)+1.) / (n_permutations + 1.) # to avoid divided by zero
print(pvalues)
plt.hist(pvalues)
```
## The same idea can be used to implement pair-sample t-test with resampling method
```
```
|
github_jupyter
|
import numpy as np
from matplotlib import pyplot as plt # just to visualize the arrays, will be introduced later
complex_list = [[ 0, 1, 2, 3, 4],
[ 5, 6, 7, 8, 9],
[10, 11, 12, 13, 14]]
print(complex_list)
complex_array = np.arange(15).reshape(3, 5)
print(complex_array)
complex_array.shape # 3 rows and 5 columns
complex_array.ndim
type(complex_array)
a = [1,2,3]
print(a)
print(type(a)) # type of class
b = np.array(a)
print(b)
print(type(b)) # type of class
print(b.dtype) # tyep of the data in the class
a = [1.0,2,3]
print(a)
print(type(a))
b = np.array(a)
print(b)
print(type(b))
print(b.dtype)
a = [1.0,2,3]
print(a)
print(type(a))
b = np.array(a).astype(int)
print(b)
print(type(b))
print(b.dtype)
np.zeros((2,5))
np.ones((2,5))
np.eye(5)
np.empty((2,5))
np.arange(10)
np.arange(11)
np.arange(start = 0, stop = 10.1, step = 0.5)
np.linspace(start = 0, stop = 10, num = 26,)
np.arange(6)
np.arange(12).reshape(4,3) # 4 rows and 3 columns
a = np.arange(12).reshape(4,3)
a.T
np.arange(24).reshape(2,3,4) # 2 slice, 3 rows, and 4 columns
a = [1,2,3]
b = [2,3,4]
print(b - a)
a = np.array([1,2,3])
b = np.array([2,3,4])
print(b - a)
a * b
a @ b
a.dot(b)
a = np.random.normal(loc = 0.,# mean
scale = 1., # standard deviation
size = 100,)
print(a)
plt.hist(a)
for i in range(10):
print(np.random.randint(1,10))
for i in range(10):
np.random.seed(12345)
print(np.random.randint(1,10))
# this is equivalent to flip a fair coin 100 times
a = np.random.choice([0,1],size = 100, replace = True, p = [0.5,0.5])
print(a)
# this is equivalent to roll a fair die 100 times
a = np.random.choice(np.arange(start = 1, stop = 7),size = 100, replace = True, p = [1./6] * 6)
print(a)
a = np.arange(20)
print(a)
np.random.shuffle(a)
print(a)
a = np.arange(100).reshape(20,5)
print(a)
for ii,row in enumerate(a):
print('row ',ii,row)
a = np.arange(6)
b = np.arange(7)
print(a,b)
print(np.concatenate([a,b]))
a = np.arange(6).reshape(2,3)
b = np.arange(8).reshape(2,4)
print(a)
print(b)
print(np.hstack([a,b]))
a = np.arange(6).reshape(2,3)
b = np.arange(8).reshape(2,4)
print(a)
print(b)
print(np.vstack([a,b]))
a = np.arange(6).reshape(3,2)
b = np.arange(8).reshape(4,2)
print(a)
print(b)
print(np.vstack([a,b]))
a = np.arange(7)
b = np.arange(7)
print(a)
print(b)
print(np.vstack([a,b]))
a = [[False, False, False, False],
[False, True, True, True],
[ True, True, True, True]]
sum(a)
print('how many True in each row: ',np.sum(a,0))
print('how many True in each column: ',np.sum(a,1))
print('how many True in total: ', np.sum(a))
mean = 0.015,
std = 0.026
a = np.random.normal(mean,std,size = 26)
print(a)
plt.hist(a)
baseline = 0
n_permutations = 5000
n_simulations = 100
experiment_mean = np.mean(a)
rescale_experiment_mean_to_zero = a - np.mean(a) + baseline
# to generate a distribution of null hypothesis that centers at 0 but with the
# same shape of distribution as variable a
# 1st dimension: number of samples same as variable a
# 2nd dimension: within one permutation resampling, we perform resampling number
# of samples same as variable a, but also repeat this n_permutations times
# 3rd dimension: repeat the permutation n_simulation times to estimate if the
# permutation is effective
resamples = np.random.choice(rescale_experiment_mean_to_zero,
size = (a.shape[0],n_permutations,n_simulations),
replace = True)
# take the average over the 1st dimension because we only care about if the mean
# of the null distribution along each permutation and each simulation
resamples = resamples.mean(0)
pvalues = (np.sum(np.abs(resamples) >= np.abs(experiment_mean),axis=0)+1.) / (n_permutations + 1.) # to avoid divided by zero
print(pvalues)
plt.hist(pvalues)
| 0.318167 | 0.97859 |
```
import pandas as pd
import pytz, datetime
import pytz
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
from scipy import stats
from sklearn.linear_model import LinearRegression
from sklearn import metrics
gdas = pd.read_csv('teffDatavAbr2021.csv',delimiter=r",+", skiprows=1, names=['id','date','teff', 'ts', 'ps'],lineterminator='\n')
gdas .drop(columns=['id'])
gdas.info()
gdas['date']=pd.to_datetime(gdas.date, format='%Y-%m-%d %H:%M:%S',utc=True)
gdas.set_index(['date'],drop=True, inplace=True)
path='datos2021/'
filenamePy=path+'flux.json'
py = pd.read_json (filenamePy, lines=True)
py['datetime']=pd.to_datetime(py.hora, unit='s',utc=True)
py.set_index(['datetime'],drop=True, inplace=True)
py= py.iloc[1:]
py.head()
py=py.resample('H').sum()
py['conteo']=py['conteo'].values/(0.0512*0.028)/3600
py['canal_a']=py['canal_a'].values/(1.364*0.028)/3600
py['canal_b']=py['canal_b'].values/(0.0512*0.028)/3600
py['ratio']=py['canal_a'].values - py['conteo'].values
flagPy1=np.logical_and(py.index>="2021-04-6",py.index<="2021-04-20")
py=py[flagPy1]
sup=np.percentile(py['conteo'].values, 95)
inf=np.percentile(py['conteo'].values, 6)
flag=np.logical_and(py['conteo'].values<sup,py['conteo'].values>inf)
py=py[flag]
py.tail()
fig, axes = plt.subplots(1,1, figsize=(10,2), sharex=True)
py['conteo'].plot(subplots=True,marker='.', markersize=4, linestyle='None', ax=axes)
axes.set_ylabel('Contaje')
axes.set_xlabel('Date')
plt.tight_layout()
#plt.savefig('pngs/SerieCrudaContaje.png')
fig, axes = plt.subplots(1,1, figsize=(10,2), sharex=True)
gdas['ps'].plot(subplots=True,marker='.', markersize=4, linestyle='None', ax=axes)
axes.set_ylabel('Pres')
axes.set_xlabel('Date')
plt.tight_layout()
#plt.savefig('pngs/GDASTefSerieCrudaPress.png')
fig, axes = plt.subplots(1,1, figsize=(10,2), sharex=True)
gdas['teff'].plot(subplots=True,marker='.', markersize=4, linestyle='None', ax=axes)
axes.set_ylabel('Temp')
axes.set_xlabel('Date')
plt.tight_layout()
#plt.savefig('pngs/GDASTefSerieCrudaTemp.png')
#flagB=counts.eficiencia>=0.7
countsClean=py
combined = gdas.join(countsClean['conteo'], how='outer')
combined=combined.drop(combined.index[0])
#combined[18000:].head()
#remove data
#flag=np.logical_and(combined.index>="2021-3-24",combined.index<="2021-3-25 ")
flag=np.logical_and(combined.index>="2021-04-6",combined.index<="2021-04-19")
#normalize delta
Im=combined['conteo'].dropna().mean()
combined['dIoIm']=(combined['conteo'][flag]-Im)/(Im)*100
P0=combined['ps'][flag].dropna().mean()
combined['dP']=(combined['ps'][flag]-P0)
P0
Tg=combined['teff'][flag].dropna().mean()
combined['dTg']=(combined['teff'][flag]-Tg)
var = ['dIoIm', 'dP']
axes = combined[var][flag].plot(marker='.', markersize=5 , linestyle='None', figsize=(11, 9), subplots=True)
#axes = combined[var].plot(marker='.', alpha=1, linestyle='None', figsize=(11, 9), subplots=True)
for ax,v in zip(axes,var):
ax.set_ylabel(v)
#plt.savefig('pngs/GDASTefSerieDeltaIComOutrliers.png')
combined=combined[flag]
#remove outliers
var='dIoIm'
distance = 1.5 * (np.percentile(combined[var][~np.isnan(combined[var])], 75) - np.percentile(combined[var][~np.isnan(combined[var])], 25))
fA=combined[var]< distance + np.percentile(combined[var][~np.isnan(combined[var])], 75)
fB=combined[var] > np.percentile(combined[var][~np.isnan(combined[var])], 25) - distance
fC=np.isnan(combined[var])
combinedNew=combined[np.logical_or(np.logical_and(fA,fB),fC)]
flagNa=~np.isnan(combinedNew['dIoIm'].values)
combinedNew = combinedNew.apply(
pd.Series.interpolate,
args=('index',)
)
combinedNew=combinedNew[flagNa]
var = ['dIoIm', 'dP','dTg']
axes = combinedNew[var].plot(marker='.',markersize=4.5, linestyle='None', alpha=0.6, figsize=(11, 9), subplots=True)
#axes = combined[var].plot(marker='.', alpha=1, linestyle='None', figsize=(11, 9), subplots=True)
for ax,v in zip(axes,var):
ax.set_ylabel(v)
#plt.savefig('pngs/GDASTefSeries.png')
combinedNewHour=combinedNew.resample('H').mean()
combinedNewHour['dIoIm']
combinedNewHour=combinedNewHour.dropna()
var = ['dIoIm', 'dP','dTg']
axes = combinedNewHour[var].plot(marker='.',markersize=5, linestyle='None', alpha=1.6, figsize=(11, 9), subplots=True)
#axes = combined[var].plot(marker='.', alpha=1, linestyle='None', figsize=(11, 9), subplots=True)
for ax,v in zip(axes,var):
ax.set_ylabel(v)
#plt.savefig('pngs/GDASTefSeriesDeltaIDeltaPDeltaTgPorHOra.png')
lm = LinearRegression()
X=combinedNewHour['dP'].values.reshape(-1,1)
Y=combinedNewHour['dIoIm']
lm.fit(X,Y)
print(lm.intercept_)
print(lm.coef_)
pred=lm.predict(X)
print(np.sum(np.square(np.square(pred - Y))))
print("R-squared value of this fit:",round(metrics.r2_score(Y,pred),3))
combinedNewHour['dP'].corr(combinedNewHour['dIoIm'])
combinedNew['dP'].corr(combinedNew['dIoIm'])
sns.regplot(combinedNewHour['dP'].values.reshape(-1,1),combinedNewHour['dIoIm'],scatter_kws={'alpha':0.1})
#plt.savefig('pngs/GDASTefscatterdPvsDIoIm.png')
lm.coef_
#combinedNewHour['dIoImPC']=combinedNewHour['dIoIm']-pred
combinedNewHour['dIoImPC']=combinedNewHour['dIoIm']-lm.coef_*combinedNewHour['dP']
sns.regplot(combinedNewHour['dP'].values.reshape(-1,1),combinedNewHour['dIoImPC'],scatter_kws={'alpha':0.1})
#plt.savefig('pngs/GDASTefscatterdPvsDIoImPC.png')
combinedNewHour['dP'].corr(combinedNewHour['dIoImPC'])
lmT = LinearRegression()
XT=combinedNewHour['dTg'].values.reshape(-1,1)
YT=combinedNewHour['dIoImPC']
lmT.fit(XT,YT)
print(lmT.intercept_)
print(lmT.coef_)
predT=lmT.predict(X)
print(np.sum(np.square(np.square(predT - YT))))
print(lmT.coef_)
#combinedNewHour['dIoImPC']=combinedNewHour['dIoIm']-pred
combinedNewHour['dIoImPTC']=combinedNewHour['dIoImPC']-lmT.coef_*combinedNewHour['dTg']
sns.regplot(combinedNewHour['dTg'].values.reshape(-1,1),combinedNewHour['dIoImPC'],scatter_kws={'alpha':0.1})
#plt.savefig('pngs/GDASTefscatterdTgvsDIoImPC.png')
sns.regplot(combinedNewHour['dTg'].values.reshape(-1,1),combinedNewHour['dIoImPTC'],scatter_kws={'alpha':0.1})
#plt.savefig('pngs/GDASTefscatterdTgvsDIoImPC.png')
var = ['dIoIm', 'dP','dTg','dIoImPC','dIoImPTC']
axes = combinedNewHour[var].plot(marker='.',markersize=2.5, linestyle='None', alpha=0.9, figsize=(11, 9), subplots=True)
#axes = combined[var].plot(marker='.', alpha=1, linestyle='None', figsize=(11, 9), subplots=True)
for ax,v in zip(axes,var):
ax.set_ylabel(v)
#plt.savefig('pngs/GDASTefSeriesDIoImvsdPvsDIoImPTC.png')
combinedNewHour.head()
combinedNewHour.tail()
combinedNewHour['Year'] =combinedNewHour.index.year
combinedNewHour['Month'] = combinedNewHour.index.month
combinedNewHour['day'] = combinedNewHour.index.day
combinedNewHour['hour'] = combinedNewHour.index.hour
combinedNewHour['hour3'] = combinedNewHour.index.hour-3
#fig, ax = plt.subplots(figsize=(15,7))
#combinedNew.groupby(['hour']).mean()['count'].plot(ax=ax,marker='.',markersize=5, linestyle='none',legend=True,label='Muons',alpha=1, figsize=(11, 5),ax=ax)
ax = plt.gca()
combinedNewHour.groupby(['hour3']).mean()['dIoImPTC'].plot(marker='.',markersize=10,ylim=(-1.5,1.5), linestyle='dashed',legend=True,label='Muons',alpha=1, figsize=(11, 5),ax=ax)
ax_secondary =combinedNewHour.groupby(['hour']).mean()['dP'].plot(ax=ax,marker='.',markersize=10, label='Pressure', linestyle='dotted',
legend=True, secondary_y=True, color='g')
ax.set_ylabel('dIoImPTC')
#plt.savefig('GDASTefDayHourdIoImTPChourlyPres.png')
```
|
github_jupyter
|
import pandas as pd
import pytz, datetime
import pytz
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
from scipy import stats
from sklearn.linear_model import LinearRegression
from sklearn import metrics
gdas = pd.read_csv('teffDatavAbr2021.csv',delimiter=r",+", skiprows=1, names=['id','date','teff', 'ts', 'ps'],lineterminator='\n')
gdas .drop(columns=['id'])
gdas.info()
gdas['date']=pd.to_datetime(gdas.date, format='%Y-%m-%d %H:%M:%S',utc=True)
gdas.set_index(['date'],drop=True, inplace=True)
path='datos2021/'
filenamePy=path+'flux.json'
py = pd.read_json (filenamePy, lines=True)
py['datetime']=pd.to_datetime(py.hora, unit='s',utc=True)
py.set_index(['datetime'],drop=True, inplace=True)
py= py.iloc[1:]
py.head()
py=py.resample('H').sum()
py['conteo']=py['conteo'].values/(0.0512*0.028)/3600
py['canal_a']=py['canal_a'].values/(1.364*0.028)/3600
py['canal_b']=py['canal_b'].values/(0.0512*0.028)/3600
py['ratio']=py['canal_a'].values - py['conteo'].values
flagPy1=np.logical_and(py.index>="2021-04-6",py.index<="2021-04-20")
py=py[flagPy1]
sup=np.percentile(py['conteo'].values, 95)
inf=np.percentile(py['conteo'].values, 6)
flag=np.logical_and(py['conteo'].values<sup,py['conteo'].values>inf)
py=py[flag]
py.tail()
fig, axes = plt.subplots(1,1, figsize=(10,2), sharex=True)
py['conteo'].plot(subplots=True,marker='.', markersize=4, linestyle='None', ax=axes)
axes.set_ylabel('Contaje')
axes.set_xlabel('Date')
plt.tight_layout()
#plt.savefig('pngs/SerieCrudaContaje.png')
fig, axes = plt.subplots(1,1, figsize=(10,2), sharex=True)
gdas['ps'].plot(subplots=True,marker='.', markersize=4, linestyle='None', ax=axes)
axes.set_ylabel('Pres')
axes.set_xlabel('Date')
plt.tight_layout()
#plt.savefig('pngs/GDASTefSerieCrudaPress.png')
fig, axes = plt.subplots(1,1, figsize=(10,2), sharex=True)
gdas['teff'].plot(subplots=True,marker='.', markersize=4, linestyle='None', ax=axes)
axes.set_ylabel('Temp')
axes.set_xlabel('Date')
plt.tight_layout()
#plt.savefig('pngs/GDASTefSerieCrudaTemp.png')
#flagB=counts.eficiencia>=0.7
countsClean=py
combined = gdas.join(countsClean['conteo'], how='outer')
combined=combined.drop(combined.index[0])
#combined[18000:].head()
#remove data
#flag=np.logical_and(combined.index>="2021-3-24",combined.index<="2021-3-25 ")
flag=np.logical_and(combined.index>="2021-04-6",combined.index<="2021-04-19")
#normalize delta
Im=combined['conteo'].dropna().mean()
combined['dIoIm']=(combined['conteo'][flag]-Im)/(Im)*100
P0=combined['ps'][flag].dropna().mean()
combined['dP']=(combined['ps'][flag]-P0)
P0
Tg=combined['teff'][flag].dropna().mean()
combined['dTg']=(combined['teff'][flag]-Tg)
var = ['dIoIm', 'dP']
axes = combined[var][flag].plot(marker='.', markersize=5 , linestyle='None', figsize=(11, 9), subplots=True)
#axes = combined[var].plot(marker='.', alpha=1, linestyle='None', figsize=(11, 9), subplots=True)
for ax,v in zip(axes,var):
ax.set_ylabel(v)
#plt.savefig('pngs/GDASTefSerieDeltaIComOutrliers.png')
combined=combined[flag]
#remove outliers
var='dIoIm'
distance = 1.5 * (np.percentile(combined[var][~np.isnan(combined[var])], 75) - np.percentile(combined[var][~np.isnan(combined[var])], 25))
fA=combined[var]< distance + np.percentile(combined[var][~np.isnan(combined[var])], 75)
fB=combined[var] > np.percentile(combined[var][~np.isnan(combined[var])], 25) - distance
fC=np.isnan(combined[var])
combinedNew=combined[np.logical_or(np.logical_and(fA,fB),fC)]
flagNa=~np.isnan(combinedNew['dIoIm'].values)
combinedNew = combinedNew.apply(
pd.Series.interpolate,
args=('index',)
)
combinedNew=combinedNew[flagNa]
var = ['dIoIm', 'dP','dTg']
axes = combinedNew[var].plot(marker='.',markersize=4.5, linestyle='None', alpha=0.6, figsize=(11, 9), subplots=True)
#axes = combined[var].plot(marker='.', alpha=1, linestyle='None', figsize=(11, 9), subplots=True)
for ax,v in zip(axes,var):
ax.set_ylabel(v)
#plt.savefig('pngs/GDASTefSeries.png')
combinedNewHour=combinedNew.resample('H').mean()
combinedNewHour['dIoIm']
combinedNewHour=combinedNewHour.dropna()
var = ['dIoIm', 'dP','dTg']
axes = combinedNewHour[var].plot(marker='.',markersize=5, linestyle='None', alpha=1.6, figsize=(11, 9), subplots=True)
#axes = combined[var].plot(marker='.', alpha=1, linestyle='None', figsize=(11, 9), subplots=True)
for ax,v in zip(axes,var):
ax.set_ylabel(v)
#plt.savefig('pngs/GDASTefSeriesDeltaIDeltaPDeltaTgPorHOra.png')
lm = LinearRegression()
X=combinedNewHour['dP'].values.reshape(-1,1)
Y=combinedNewHour['dIoIm']
lm.fit(X,Y)
print(lm.intercept_)
print(lm.coef_)
pred=lm.predict(X)
print(np.sum(np.square(np.square(pred - Y))))
print("R-squared value of this fit:",round(metrics.r2_score(Y,pred),3))
combinedNewHour['dP'].corr(combinedNewHour['dIoIm'])
combinedNew['dP'].corr(combinedNew['dIoIm'])
sns.regplot(combinedNewHour['dP'].values.reshape(-1,1),combinedNewHour['dIoIm'],scatter_kws={'alpha':0.1})
#plt.savefig('pngs/GDASTefscatterdPvsDIoIm.png')
lm.coef_
#combinedNewHour['dIoImPC']=combinedNewHour['dIoIm']-pred
combinedNewHour['dIoImPC']=combinedNewHour['dIoIm']-lm.coef_*combinedNewHour['dP']
sns.regplot(combinedNewHour['dP'].values.reshape(-1,1),combinedNewHour['dIoImPC'],scatter_kws={'alpha':0.1})
#plt.savefig('pngs/GDASTefscatterdPvsDIoImPC.png')
combinedNewHour['dP'].corr(combinedNewHour['dIoImPC'])
lmT = LinearRegression()
XT=combinedNewHour['dTg'].values.reshape(-1,1)
YT=combinedNewHour['dIoImPC']
lmT.fit(XT,YT)
print(lmT.intercept_)
print(lmT.coef_)
predT=lmT.predict(X)
print(np.sum(np.square(np.square(predT - YT))))
print(lmT.coef_)
#combinedNewHour['dIoImPC']=combinedNewHour['dIoIm']-pred
combinedNewHour['dIoImPTC']=combinedNewHour['dIoImPC']-lmT.coef_*combinedNewHour['dTg']
sns.regplot(combinedNewHour['dTg'].values.reshape(-1,1),combinedNewHour['dIoImPC'],scatter_kws={'alpha':0.1})
#plt.savefig('pngs/GDASTefscatterdTgvsDIoImPC.png')
sns.regplot(combinedNewHour['dTg'].values.reshape(-1,1),combinedNewHour['dIoImPTC'],scatter_kws={'alpha':0.1})
#plt.savefig('pngs/GDASTefscatterdTgvsDIoImPC.png')
var = ['dIoIm', 'dP','dTg','dIoImPC','dIoImPTC']
axes = combinedNewHour[var].plot(marker='.',markersize=2.5, linestyle='None', alpha=0.9, figsize=(11, 9), subplots=True)
#axes = combined[var].plot(marker='.', alpha=1, linestyle='None', figsize=(11, 9), subplots=True)
for ax,v in zip(axes,var):
ax.set_ylabel(v)
#plt.savefig('pngs/GDASTefSeriesDIoImvsdPvsDIoImPTC.png')
combinedNewHour.head()
combinedNewHour.tail()
combinedNewHour['Year'] =combinedNewHour.index.year
combinedNewHour['Month'] = combinedNewHour.index.month
combinedNewHour['day'] = combinedNewHour.index.day
combinedNewHour['hour'] = combinedNewHour.index.hour
combinedNewHour['hour3'] = combinedNewHour.index.hour-3
#fig, ax = plt.subplots(figsize=(15,7))
#combinedNew.groupby(['hour']).mean()['count'].plot(ax=ax,marker='.',markersize=5, linestyle='none',legend=True,label='Muons',alpha=1, figsize=(11, 5),ax=ax)
ax = plt.gca()
combinedNewHour.groupby(['hour3']).mean()['dIoImPTC'].plot(marker='.',markersize=10,ylim=(-1.5,1.5), linestyle='dashed',legend=True,label='Muons',alpha=1, figsize=(11, 5),ax=ax)
ax_secondary =combinedNewHour.groupby(['hour']).mean()['dP'].plot(ax=ax,marker='.',markersize=10, label='Pressure', linestyle='dotted',
legend=True, secondary_y=True, color='g')
ax.set_ylabel('dIoImPTC')
#plt.savefig('GDASTefDayHourdIoImTPChourlyPres.png')
| 0.319546 | 0.469095 |
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from keras.preprocessing.text import Tokenizer
from keras.preprocessing.sequence import pad_sequences
from sklearn.model_selection import train_test_split
from tensorflow.keras import utils as np_utils
from keras.models import Sequential
from keras.layers import Embedding, LSTM, Dense, Bidirectional
from sklearn.metrics import confusion_matrix, classification_report
from tqdm import tqdm
import re
os.environ['TF_CPP_MIN_LOG_LEVEL'] = '1'
# tf.get_logger().setLevel('INFO')
train_df = pd.read_json('../../data/raw/raw_nyt/raw_train.json', lines = True)
print(train_df.columns)
train_df.head()
#rel_sets -> relation_sets
def extract_relations(df):
relations = []
for rel_sets in df['relationMentions']:
relations.append(rel_sets[-1]['label'])
return relations
train_df['relations'] = extract_relations(train_df)
set(train_df['relations'])
plt.style.use('dark_background')
train_df['relations'].value_counts().plot.barh(title='Relation Counts', logx = True, figsize=(15,10))
plt.xlabel('Frequencies', fontsize=18)
plt.ylabel('Relations', fontsize=18)
plt.savefig('../../reports/figures/raw_nyt_train_data_relationFrequencies.png')
train_df['sentText'].head()
def get_x(df):
X = []
for data in df['sentText']:
X.append(data.lower())
return X
X = get_x(train_df)
for i in range(5):
print(X[i], end="\n\n")
def find_max_len(x, max_len):
if(max_len < len(x.split())):
max_len = len(x.split())
return max_len
max_len = 0
for x in train_df['sentText']:
max_len = find_max_len(x, max_len)
print("Max Length = ", max_len)
n_most_common_words = 65000 # vocabulary size
max_len = 100 # Max Length of Sequence
tokenizer = Tokenizer(num_words=n_most_common_words, filters='!"#$%&()*+,-./:;=?@[]^_`{|}~', lower=True)
tokenizer.fit_on_texts(train_df['sentText'].values)
sequences = tokenizer.texts_to_sequences(train_df['sentText'].values)
word_index = tokenizer.word_index
print('Found %s unique tokens.' % len(word_index))
X = pad_sequences(sequences, maxlen=max_len)
print([i for i in sequences[0]])
print("Found", len(train_df.relations.unique()), "relations")
dictionary = {}
rev_dictionary = {}
for i, rel in enumerate(train_df.relations.unique()):
#map relation to index
dictionary[rel] = i
#map index to relation
rev_dictionary[i] = rel
print("Length of Relations = ", len(dictionary))
for key, values in dictionary.items():
print(values, " -----> ", key)
# encoding relations -- (relations to numbers)
encoded_relations = []
for rel in train_df['relations']:
encoded_relations.append(dictionary[rel])
train_df['encoded_relations'] = encoded_relations
train_df.head()
y = np_utils.to_categorical(train_df['encoded_relations'], num_classes=len(train_df.encoded_relations.unique()))
X_train, X_test, y_train, y_test = train_test_split(X , y, test_size=0.20, random_state=42)
embedding_size = 300
model = Sequential()
model.add(Embedding(n_most_common_words, embedding_size, input_length=X.shape[1]))
model.add(Bidirectional(LSTM(128, dropout=0.7, recurrent_dropout=0.7)))
model.add(Dense(y.shape[1], activation='softmax'))
model.compile(optimizer='adam', loss='categorical_crossentropy', metrics=['acc'])
print(model.summary())
model.fit(X_train, y_train, epochs=10, batch_size=128, validation_split=0.2)
prediction_probas = model.predict(X_test)
predictions = [np.argmax(pred) for pred in prediction_probas]
print(predictions[:10])
y_test_dup=np.argmax(y_test, axis=1)
print(confusion_matrix(y_test_dup, predictions))
print(classification_report(y_test_dup, predictions, digits=3))
# Testing ------
seq = tokenizer.texts_to_sequences(['Steve Jobs Founded Apple'])
test_sent = pad_sequences(seq, max_len)
test_sent_pred = model.predict(test_sent)
# Output -----
print(rev_dictionary[np.argmax(test_sent_pred)])
model.save('../../models/tensorflow_bidirectional_model.h5')
# Read Write File
def write_modelsfile(dictionary):
'''
write dictionary to models_list file in models directory
'''
dict_file = open("models_list", "wb")
pickle.dump(dictionary, dict_file)
f.close()
def read_modelsfile():
'''
returns dictionary containing the links of models
'''
L = None
f = open("models_list", 'rb')
while True:
try:
L = pickle.load(f)
break
except EOFError:
print("Completed reading details")
f.close()
return L
```
|
github_jupyter
|
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from keras.preprocessing.text import Tokenizer
from keras.preprocessing.sequence import pad_sequences
from sklearn.model_selection import train_test_split
from tensorflow.keras import utils as np_utils
from keras.models import Sequential
from keras.layers import Embedding, LSTM, Dense, Bidirectional
from sklearn.metrics import confusion_matrix, classification_report
from tqdm import tqdm
import re
os.environ['TF_CPP_MIN_LOG_LEVEL'] = '1'
# tf.get_logger().setLevel('INFO')
train_df = pd.read_json('../../data/raw/raw_nyt/raw_train.json', lines = True)
print(train_df.columns)
train_df.head()
#rel_sets -> relation_sets
def extract_relations(df):
relations = []
for rel_sets in df['relationMentions']:
relations.append(rel_sets[-1]['label'])
return relations
train_df['relations'] = extract_relations(train_df)
set(train_df['relations'])
plt.style.use('dark_background')
train_df['relations'].value_counts().plot.barh(title='Relation Counts', logx = True, figsize=(15,10))
plt.xlabel('Frequencies', fontsize=18)
plt.ylabel('Relations', fontsize=18)
plt.savefig('../../reports/figures/raw_nyt_train_data_relationFrequencies.png')
train_df['sentText'].head()
def get_x(df):
X = []
for data in df['sentText']:
X.append(data.lower())
return X
X = get_x(train_df)
for i in range(5):
print(X[i], end="\n\n")
def find_max_len(x, max_len):
if(max_len < len(x.split())):
max_len = len(x.split())
return max_len
max_len = 0
for x in train_df['sentText']:
max_len = find_max_len(x, max_len)
print("Max Length = ", max_len)
n_most_common_words = 65000 # vocabulary size
max_len = 100 # Max Length of Sequence
tokenizer = Tokenizer(num_words=n_most_common_words, filters='!"#$%&()*+,-./:;=?@[]^_`{|}~', lower=True)
tokenizer.fit_on_texts(train_df['sentText'].values)
sequences = tokenizer.texts_to_sequences(train_df['sentText'].values)
word_index = tokenizer.word_index
print('Found %s unique tokens.' % len(word_index))
X = pad_sequences(sequences, maxlen=max_len)
print([i for i in sequences[0]])
print("Found", len(train_df.relations.unique()), "relations")
dictionary = {}
rev_dictionary = {}
for i, rel in enumerate(train_df.relations.unique()):
#map relation to index
dictionary[rel] = i
#map index to relation
rev_dictionary[i] = rel
print("Length of Relations = ", len(dictionary))
for key, values in dictionary.items():
print(values, " -----> ", key)
# encoding relations -- (relations to numbers)
encoded_relations = []
for rel in train_df['relations']:
encoded_relations.append(dictionary[rel])
train_df['encoded_relations'] = encoded_relations
train_df.head()
y = np_utils.to_categorical(train_df['encoded_relations'], num_classes=len(train_df.encoded_relations.unique()))
X_train, X_test, y_train, y_test = train_test_split(X , y, test_size=0.20, random_state=42)
embedding_size = 300
model = Sequential()
model.add(Embedding(n_most_common_words, embedding_size, input_length=X.shape[1]))
model.add(Bidirectional(LSTM(128, dropout=0.7, recurrent_dropout=0.7)))
model.add(Dense(y.shape[1], activation='softmax'))
model.compile(optimizer='adam', loss='categorical_crossentropy', metrics=['acc'])
print(model.summary())
model.fit(X_train, y_train, epochs=10, batch_size=128, validation_split=0.2)
prediction_probas = model.predict(X_test)
predictions = [np.argmax(pred) for pred in prediction_probas]
print(predictions[:10])
y_test_dup=np.argmax(y_test, axis=1)
print(confusion_matrix(y_test_dup, predictions))
print(classification_report(y_test_dup, predictions, digits=3))
# Testing ------
seq = tokenizer.texts_to_sequences(['Steve Jobs Founded Apple'])
test_sent = pad_sequences(seq, max_len)
test_sent_pred = model.predict(test_sent)
# Output -----
print(rev_dictionary[np.argmax(test_sent_pred)])
model.save('../../models/tensorflow_bidirectional_model.h5')
# Read Write File
def write_modelsfile(dictionary):
'''
write dictionary to models_list file in models directory
'''
dict_file = open("models_list", "wb")
pickle.dump(dictionary, dict_file)
f.close()
def read_modelsfile():
'''
returns dictionary containing the links of models
'''
L = None
f = open("models_list", 'rb')
while True:
try:
L = pickle.load(f)
break
except EOFError:
print("Completed reading details")
f.close()
return L
| 0.505371 | 0.320276 |
```
from sklearn.datasets import load_boston
boston = load_boston() # データセットの読み込み
import pandas as pd
boston_df = pd.DataFrame(boston.data, columns = boston.feature_names) # 説明変数(boston.data)をDataFrameに保存
boston_df['MEDV'] = boston.target # 目的変数(boston.target)もDataFrameに追加
boston.data
boston.target
boston_df
import matplotlib.pyplot as plt
%matplotlib inline
plt.scatter(boston_df['RM'], boston_df['MEDV']) # 平均部屋数と住宅価格の散布図をプロット
plt.title('Scatter Plot of RM vs MEDV') # 図のタイトル
plt.xlabel('Average number of rooms [RM]') # x軸のラベル
plt.ylabel('Prices in $1000\'s [MEDV]') # y軸のラベル
plt.grid() # グリッド線を表示
plt.show() # 図の表示
boston_df[['RM','MEDV']].corr()
from sklearn.linear_model import LinearRegression
lr = LinearRegression()
X = boston_df[['RM']].values # 説明変数(Numpyの配列)
Y = boston_df['MEDV'].values # 目的変数(Numpyの配列)
lr.fit(X, Y) # 線形モデルの重みを学習
boston_df[['RM']]
print('coefficient = ', lr.coef_[0]) # 説明変数の係数を出力
print('intercept = ', lr.intercept_) # 切片を出力
plt.scatter(X, Y, color = 'blue') # 説明変数と目的変数のデータ点の散布図をプロット
plt.plot(X, lr.predict(X), color = 'red') # 回帰直線をプロット
plt.title('Regression Line') # 図のタイトル
plt.xlabel('Average number of rooms [RM]') # x軸のラベル
plt.ylabel('Prices in $1000\'s [MEDV]') # y軸のラベル
plt.grid() # グリッド線を表示
plt.show() # 図の表示
from sklearn.model_selection import train_test_split
X_train, X_test, Y_train, Y_test = train_test_split(X, Y, train_size = 0.7, test_size = 0.3, random_state = 0) # データを学習用と検証用に分割
lr = LinearRegression()
lr.fit(X_train, Y_train) # 線形モデルの重みを学習
Y_pred = lr.predict(X_test) # 検証データを用いて目的変数を予測
plt.scatter(Y_pred, Y_pred - Y_test, color = 'blue') # 残差をプロット
plt.hlines(y = 0, xmin = -10, xmax = 50, color = 'black') # x軸に沿った直線をプロット
plt.title('Residual Plot') # 図のタイトル
plt.xlabel('Predicted Values') # x軸のラベル
plt.ylabel('Residuals') # y軸のラベル
plt.grid() # グリッド線を表示
plt.show() # 図の表示
from sklearn.metrics import mean_squared_error
Y_train_pred = lr.predict(X_train) # 学習データに対する目的変数を予測
print('MSE train data: ', mean_squared_error(Y_train, Y_train_pred)) # 学習データを用いたときの平均二乗誤差を出力
print('MSE test data: ', mean_squared_error(Y_test, Y_pred))
from sklearn.metrics import r2_score
print('r^2 train data: ', r2_score(Y_train, Y_train_pred))
print('r^2 test data: ', r2_score(Y_test, Y_pred))
lr = LinearRegression()
X = boston_df[['RM', 'LSTAT']].values # 説明変数(Numpyの配列)
Y = boston_df['MEDV'].values # 目的変数(Numpyの配列)
lr.fit(X, Y) # 線形モデルの重みを学習
boston_df[['RM', 'LSTAT']]
x = boston_df['RM']
y = boston_df['LSTAT']
z = boston_df['MEDV']
plt.scatter(x, y, z, color='blue')
print(x)
from mpl_toolkits.mplot3d import Axes3D
fig = plt.figure(figsize=(8,8))
ax = fig.add_subplot(111, projection='3d')
ax.scatter(x, y, z, color='blue')
```
|
github_jupyter
|
from sklearn.datasets import load_boston
boston = load_boston() # データセットの読み込み
import pandas as pd
boston_df = pd.DataFrame(boston.data, columns = boston.feature_names) # 説明変数(boston.data)をDataFrameに保存
boston_df['MEDV'] = boston.target # 目的変数(boston.target)もDataFrameに追加
boston.data
boston.target
boston_df
import matplotlib.pyplot as plt
%matplotlib inline
plt.scatter(boston_df['RM'], boston_df['MEDV']) # 平均部屋数と住宅価格の散布図をプロット
plt.title('Scatter Plot of RM vs MEDV') # 図のタイトル
plt.xlabel('Average number of rooms [RM]') # x軸のラベル
plt.ylabel('Prices in $1000\'s [MEDV]') # y軸のラベル
plt.grid() # グリッド線を表示
plt.show() # 図の表示
boston_df[['RM','MEDV']].corr()
from sklearn.linear_model import LinearRegression
lr = LinearRegression()
X = boston_df[['RM']].values # 説明変数(Numpyの配列)
Y = boston_df['MEDV'].values # 目的変数(Numpyの配列)
lr.fit(X, Y) # 線形モデルの重みを学習
boston_df[['RM']]
print('coefficient = ', lr.coef_[0]) # 説明変数の係数を出力
print('intercept = ', lr.intercept_) # 切片を出力
plt.scatter(X, Y, color = 'blue') # 説明変数と目的変数のデータ点の散布図をプロット
plt.plot(X, lr.predict(X), color = 'red') # 回帰直線をプロット
plt.title('Regression Line') # 図のタイトル
plt.xlabel('Average number of rooms [RM]') # x軸のラベル
plt.ylabel('Prices in $1000\'s [MEDV]') # y軸のラベル
plt.grid() # グリッド線を表示
plt.show() # 図の表示
from sklearn.model_selection import train_test_split
X_train, X_test, Y_train, Y_test = train_test_split(X, Y, train_size = 0.7, test_size = 0.3, random_state = 0) # データを学習用と検証用に分割
lr = LinearRegression()
lr.fit(X_train, Y_train) # 線形モデルの重みを学習
Y_pred = lr.predict(X_test) # 検証データを用いて目的変数を予測
plt.scatter(Y_pred, Y_pred - Y_test, color = 'blue') # 残差をプロット
plt.hlines(y = 0, xmin = -10, xmax = 50, color = 'black') # x軸に沿った直線をプロット
plt.title('Residual Plot') # 図のタイトル
plt.xlabel('Predicted Values') # x軸のラベル
plt.ylabel('Residuals') # y軸のラベル
plt.grid() # グリッド線を表示
plt.show() # 図の表示
from sklearn.metrics import mean_squared_error
Y_train_pred = lr.predict(X_train) # 学習データに対する目的変数を予測
print('MSE train data: ', mean_squared_error(Y_train, Y_train_pred)) # 学習データを用いたときの平均二乗誤差を出力
print('MSE test data: ', mean_squared_error(Y_test, Y_pred))
from sklearn.metrics import r2_score
print('r^2 train data: ', r2_score(Y_train, Y_train_pred))
print('r^2 test data: ', r2_score(Y_test, Y_pred))
lr = LinearRegression()
X = boston_df[['RM', 'LSTAT']].values # 説明変数(Numpyの配列)
Y = boston_df['MEDV'].values # 目的変数(Numpyの配列)
lr.fit(X, Y) # 線形モデルの重みを学習
boston_df[['RM', 'LSTAT']]
x = boston_df['RM']
y = boston_df['LSTAT']
z = boston_df['MEDV']
plt.scatter(x, y, z, color='blue')
print(x)
from mpl_toolkits.mplot3d import Axes3D
fig = plt.figure(figsize=(8,8))
ax = fig.add_subplot(111, projection='3d')
ax.scatter(x, y, z, color='blue')
| 0.602529 | 0.90657 |
# Convolutional Neural Networks (LeNet)
We are now ready to put all of the tools together
to deploy your first fully-functional convolutional neural network.
In our first encounter with image data we applied a multilayer perceptron
to pictures of clothing in the Fashion-MNIST data set.
Each image in Fashion-MNIST consisted of
a two-dimensional $28 \times 28$ matrix.
To make this data amenable to multilayer perceptrons
which anticipate receiving inputs as one-dimensional fixed-length vectors,
we first flattened each image, yielding vectors of length 784,
before processing them with a series of fully-connected layers.
Now that we have introduced convolutional layers,
we can keep the image in its original spatially-organized grid,
processing it with a series of successive convolutional layers.
Moreover, because we are using convolutional layers,
we can enjoy a considerable savings in the number of parameters required.
In this section, we will introduce one of the first
published convolutional neural networks
whose benefit was first demonstrated by Yann Lecun,
then a researcher at AT&T Bell Labs,
for the purpose of recognizing handwritten digits in images—[LeNet5](http://yann.lecun.com/exdb/lenet/).
In the 90s, their experiments with LeNet gave the first compelling evidence
that it was possible to train convolutional neural networks
by backpropagation.
Their model achieved outstanding results at the time
(only matched by Support Vector Machines at the time)
and was adopted to recognize digits for processing deposits in ATM machines.
Some ATMs still run the code
that Yann and his colleague Leon Bottou wrote in the 1990s!
## LeNet
In a rough sense, we can think LeNet as consisting of two parts:
(i) a block of convolutional layers; and
(ii) a block of fully-connected layers.
Before getting into the weeds, let's briefly review the model in
```
from IPython.display import Image
Image(filename='../img/lenet.png')
```
Data flow in LeNet 5. The input is a handwritten digit, the output a probabilitiy over 10 possible outcomes.
The basic units in the convolutional block are a convolutional layer
and a subsequent average pooling layer
(note that max-pooling works better,
but it had not been invented in the 90s yet).
The convolutional layer is used to recognize
the spatial patterns in the image,
such as lines and the parts of objects,
and the subsequent average pooling layer
is used to reduce the dimensionality.
The convolutional layer block is composed of
repeated stacks of these two basic units.
Each convolutional layer uses a $5\times 5$ kernel
and processes each output with a sigmoid activation function
(again, note that ReLUs are now known to work more reliably,
but had not been invented yet).
The first convolutional layer has 6 output channels,
and second convolutional layer increases channel depth further to 16.
However, coinciding with this increase in the number of channels,
the height and width are shrunk considerably.
Therefore, increasing the number of output channels
makes the parameter sizes of the two convolutional layers similar.
The two average pooling layers are of size $2\times 2$ and take stride 2
(note that this means they are non-overlapping).
In other words, the pooling layer downsamples the representation
to be precisely *one quarter* the pre-pooling size.
The convolutional block emits an output with size given by
(batch size, channel, height, width).
Before we can pass the convolutional block's output
to the fully-connected block, we must flatten
each example in the mini-batch.
In other words, we take this 4D input and tansform it into the 2D
input expected by fully-connected layers:
as a reminder, the first dimension indexes the examples in the mini-batch
and the second gives the flat vector representation of each example.
LeNet's fully-connected layer block has three fully-connected layers,
with 120, 84, and 10 outputs, respectively.
Because we are still performing classification,
the 10 dimensional output layer corresponds
to the number of possible output classes.
While getting to the point
where you truly understand
what's going on inside LeNet
may have taken a bit of work,
you can see below that implementing it
in a modern deep learning library
is remarkably simple.
Again, we'll rely on the Sequential class.
```
import sys
sys.path.insert(0, '..')
import d2l
import torch
import torch.nn as nn
import torch.optim as optim
import time
class Flatten(torch.nn.Module):
def forward(self, x):
return x.view(x.shape[0], -1)
class Reshape(torch.nn.Module):
def forward(self, x):
return x.view(-1,1,28,28)
net = torch.nn.Sequential(
Reshape(),
nn.Conv2d(in_channels=1, out_channels=6, kernel_size=5, padding=2),
nn.Sigmoid(),
nn.AvgPool2d(kernel_size=2, stride=2),
nn.Conv2d(in_channels=6, out_channels=16, kernel_size=5),
nn.Sigmoid(),
nn.AvgPool2d(kernel_size=2, stride=2),
Flatten(),
nn.Linear(in_features=16*5*5, out_features=120),
nn.Sigmoid(),
nn.Linear(120, 84),
nn.Sigmoid(),
nn.Linear(84, 10)
)
```
As compared to the original network, we took the liberty of replacing the Gaussian activation in the last layer by a regular linear layer, which tends to be significantly more convenient to train. Other than that, this network matches the historical definition of LeNet5. Next, we feed a single-channel example of size $28 \times 28$ into the network and perform a forward computation layer by layer printing the output shape at each layer
to make sure we understand what's happening here.
```
X = torch.randn(size=(1,1,28,28), dtype = torch.float32)
for layer in net:
X = layer(X)
print(layer.__class__.__name__,'output shape: \t',X.shape)
```
Note that the height and width of the representation
at each layer throughout the convolutional block is reduced
(compared to the previous layer).
The convolutional layer uses a kernel
with a height and width of 5,
which with only $2$ pixels of padding in the first convolutional layer
and none in the second convolutional layer
leads to reductions in both height and width by 2 and 4 pixels, respectively.
Moreover each pooling layer halves the height and width.
However, as we go up the stack of layers,
the number of channels increases layer-over-layer
from 1 in the input to 6 after the first convolutional layer
and 16 after the second layer.
Then, the fully-connected layer reduces dimensionality layer by layer,
until emitting an output that matches the number of image classes.
```
Image(filename="../img/lenet-vert.png")
```
Compressed notation for LeNet5
## Data Acquisition and Training
Now that we've implemented the model,
we might as well run some experiments
to see what we can accomplish with the LeNet model.
While it might serve nostalgia
to train LeNet on the original MNIST OCR dataset,
that dataset has become too easy,
with MLPs getting over 98% accuracy,
so it would be hard to see the benefits of convolutional networks.
Thus we will stick with Fashion-MNIST as our dataset
because while it has the same shape ($28\times28$ images),
this dataset is notably more challenging.
```
batch_size = 256
train_iter, test_iter = d2l.load_data_fashion_mnist(batch_size=batch_size)
```
While convolutional networks may have few parameters, they can still be significantly more expensive to compute than a similarly deep multilayer perceptron so if you have access to a GPU, this might be a good time to put it into action to speed up training.
Here’s a simple function that we can use to detect whether we have a GPU. In it, we try to allocate gpu0 if available using `torch.cuda.is_available()` method. Otherwise, we stick with the CPU.
```
# This function has been saved in the d2l package for future use
def try_gpu():
"""If GPU is available, return torch.device as cuda:0; else return torch.device as cpu."""
if torch.cuda.is_available():
device = torch.device('cuda:0')
else:
device = torch.device('cpu')
return device
device = try_gpu()
device
```
For evaluation, we need to make a slight modification
to the `evaluate_accuracy` function that we described
when implementing the softmax from scratch (`chapter_softmax_scratch`).
Since the full dataset lives on the CPU,
we need to copy it to the GPU before we can compute our models.
This is accomplished via the `.to(device)` described in `chapter_use_gpu`.
Note that we accumulate the errors on the device
where the data eventually lives (in `acc`).
This avoids intermediate copy operations that might harm performance.
```
# This function has been saved in the d2l package for future use. The function
# will be gradually improved. Its complete implementation will be discussed in
# the "Image Augmentation" section
def evaluate_accuracy(data_iter, net,device=torch.device('cpu')):
"""Evaluate accuracy of a model on the given data set."""
acc_sum,n = torch.tensor([0],dtype=torch.float32,device=device),0
for X,y in data_iter:
# If device is the GPU, copy the data to the GPU.
X,y = X.to(device),y.to(device)
net.eval()
with torch.no_grad():
y = y.long()
acc_sum += torch.sum((torch.argmax(net(X), dim=1) == y))
n += y.shape[0]
return acc_sum.item()/n
```
We also need to update our training function to deal with GPUs.
Unlike `train_ch3` defined in `chapter_softmax_scratch`, we now need to move each batch of data
to our designated device (hopefully, the GPU)
prior to making the forward and backward passes.
```
# This function has been saved in the d2l package for future use
def train_ch5(net, train_iter, test_iter,criterion, num_epochs, batch_size, device,lr=None):
"""Train and evaluate a model with CPU or GPU."""
print('training on', device)
net.to(device)
optimizer = optim.SGD(net.parameters(), lr=lr)
for epoch in range(num_epochs):
train_l_sum = torch.tensor([0.0],dtype=torch.float32,device=device)
train_acc_sum = torch.tensor([0.0],dtype=torch.float32,device=device)
n, start = 0, time.time()
for X, y in train_iter:
net.train()
optimizer.zero_grad()
X,y = X.to(device),y.to(device)
y_hat = net(X)
loss = criterion(y_hat, y)
loss.backward()
optimizer.step()
with torch.no_grad():
y = y.long()
train_l_sum += loss.float()
train_acc_sum += (torch.sum((torch.argmax(y_hat, dim=1) == y))).float()
n += y.shape[0]
test_acc = evaluate_accuracy(test_iter, net,device)
print('epoch %d, loss %.4f, train acc %.3f, test acc %.3f, '
'time %.1f sec'
% (epoch + 1, train_l_sum/n, train_acc_sum/n, test_acc,
time.time() - start))
```
We initialize the model parameters on the device indicated by `device`,
this time using the Xavier initializer.
The loss function and the training algorithm
still use the cross-entropy loss function
and mini-batch stochastic gradient descent.
```
lr, num_epochs = 0.9, 5
def init_weights(m):
if type(m) == nn.Linear or type(m) == nn.Conv2d:
torch.nn.init.xavier_uniform_(m.weight)
net.apply(init_weights)
net = net.to(device)
criterion = nn.CrossEntropyLoss()
train_ch5(net, train_iter, test_iter, criterion,num_epochs, batch_size,device, lr)
```
## Summary
* A convolutional neural network (in short, ConvNet) is a network using convolutional layers.
* In a ConvNet we alternate between convolutions, nonlinearities and often also pooling operations.
* Ultimately the resolution is reduced prior to emitting an output via one (or more) dense layers.
* LeNet was the first successful deployment of such a network.
## Exercises
1. Replace the average pooling with max pooling. What happens?
1. Try to construct a more complex network based on LeNet to improve its accuracy.
* Adjust the convolution window size.
* Adjust the number of output channels.
* Adjust the activation function (ReLU?).
* Adjust the number of convolution layers.
* Adjust the number of fully connected layers.
* Adjust the learning rates and other training details (initialization, epochs, etc.)
1. Try out the improved network on the original MNIST dataset.
1. Display the activations of the first and second layer of LeNet for different inputs (e.g. sweaters, coats).
## References
[1] LeCun, Y., Bottou, L., Bengio, Y., & Haffner, P. (1998). Gradient-based learning applied to document recognition. Proceedings of the IEEE, 86(11), 2278-2324.
|
github_jupyter
|
from IPython.display import Image
Image(filename='../img/lenet.png')
import sys
sys.path.insert(0, '..')
import d2l
import torch
import torch.nn as nn
import torch.optim as optim
import time
class Flatten(torch.nn.Module):
def forward(self, x):
return x.view(x.shape[0], -1)
class Reshape(torch.nn.Module):
def forward(self, x):
return x.view(-1,1,28,28)
net = torch.nn.Sequential(
Reshape(),
nn.Conv2d(in_channels=1, out_channels=6, kernel_size=5, padding=2),
nn.Sigmoid(),
nn.AvgPool2d(kernel_size=2, stride=2),
nn.Conv2d(in_channels=6, out_channels=16, kernel_size=5),
nn.Sigmoid(),
nn.AvgPool2d(kernel_size=2, stride=2),
Flatten(),
nn.Linear(in_features=16*5*5, out_features=120),
nn.Sigmoid(),
nn.Linear(120, 84),
nn.Sigmoid(),
nn.Linear(84, 10)
)
X = torch.randn(size=(1,1,28,28), dtype = torch.float32)
for layer in net:
X = layer(X)
print(layer.__class__.__name__,'output shape: \t',X.shape)
Image(filename="../img/lenet-vert.png")
batch_size = 256
train_iter, test_iter = d2l.load_data_fashion_mnist(batch_size=batch_size)
# This function has been saved in the d2l package for future use
def try_gpu():
"""If GPU is available, return torch.device as cuda:0; else return torch.device as cpu."""
if torch.cuda.is_available():
device = torch.device('cuda:0')
else:
device = torch.device('cpu')
return device
device = try_gpu()
device
# This function has been saved in the d2l package for future use. The function
# will be gradually improved. Its complete implementation will be discussed in
# the "Image Augmentation" section
def evaluate_accuracy(data_iter, net,device=torch.device('cpu')):
"""Evaluate accuracy of a model on the given data set."""
acc_sum,n = torch.tensor([0],dtype=torch.float32,device=device),0
for X,y in data_iter:
# If device is the GPU, copy the data to the GPU.
X,y = X.to(device),y.to(device)
net.eval()
with torch.no_grad():
y = y.long()
acc_sum += torch.sum((torch.argmax(net(X), dim=1) == y))
n += y.shape[0]
return acc_sum.item()/n
# This function has been saved in the d2l package for future use
def train_ch5(net, train_iter, test_iter,criterion, num_epochs, batch_size, device,lr=None):
"""Train and evaluate a model with CPU or GPU."""
print('training on', device)
net.to(device)
optimizer = optim.SGD(net.parameters(), lr=lr)
for epoch in range(num_epochs):
train_l_sum = torch.tensor([0.0],dtype=torch.float32,device=device)
train_acc_sum = torch.tensor([0.0],dtype=torch.float32,device=device)
n, start = 0, time.time()
for X, y in train_iter:
net.train()
optimizer.zero_grad()
X,y = X.to(device),y.to(device)
y_hat = net(X)
loss = criterion(y_hat, y)
loss.backward()
optimizer.step()
with torch.no_grad():
y = y.long()
train_l_sum += loss.float()
train_acc_sum += (torch.sum((torch.argmax(y_hat, dim=1) == y))).float()
n += y.shape[0]
test_acc = evaluate_accuracy(test_iter, net,device)
print('epoch %d, loss %.4f, train acc %.3f, test acc %.3f, '
'time %.1f sec'
% (epoch + 1, train_l_sum/n, train_acc_sum/n, test_acc,
time.time() - start))
lr, num_epochs = 0.9, 5
def init_weights(m):
if type(m) == nn.Linear or type(m) == nn.Conv2d:
torch.nn.init.xavier_uniform_(m.weight)
net.apply(init_weights)
net = net.to(device)
criterion = nn.CrossEntropyLoss()
train_ch5(net, train_iter, test_iter, criterion,num_epochs, batch_size,device, lr)
| 0.778481 | 0.994631 |
# Boosting-based estimator
In this notebook, we will present a second family of ensemble method known as boosting. We will first give an intuitive example of how boosting works. It will follow by an introduction to gradient boosting decision tree models.
```
# temporary fix to avoid spurious warning raised in scikit-learn 1.0.0
# it will be solved in scikit-learn 1.0.1
import warnings
warnings.filterwarnings("ignore", message="X has feature names.*")
warnings.filterwarnings("ignore", message="X does not have valid feature names.*")
```
## Introduction to boosting
We will first give an intuive explanation on the principle of boosting. In the previous notebook, we saw that bagging was creating several datasets with a little variation using bootstrapping. Then an estimator was trained on each of these different datasets and the different results were aggregated. In boosting, the paradigm is different: the estimators will be trained on the same dataset. Thus, to combine them, we will train an estimator to correct the error of all previous estimators. So, we have a sequence of estimators instead of independent estimators.
Let's give an example on a classification dataset.
```
import pandas as pd
data = pd.read_csv("../datasets/penguins_classification.csv")
data["Species"] = data["Species"].astype("category")
X, y = data[["Culmen Length (mm)", "Culmen Depth (mm)"]], data["Species"]
import seaborn as sns
sns.set_context("poster")
import matplotlib.pyplot as plt
_, ax = plt.subplots(figsize=(8, 6))
_ = data.plot.scatter(
x="Culmen Length (mm)",
y="Culmen Depth (mm)",
c="Species",
s=80,
cmap=plt.cm.viridis,
ax=ax,
)
```
In this dataset, we have three species of penguin and we want to distinguish them based on the culmen depth and length.
Let's start to train a shallow decision tree classifier.
```
from sklearn.tree import DecisionTreeClassifier
tree = DecisionTreeClassifier(max_depth=2, random_state=0)
tree.fit(X, y)
```
We will check qualitatively the statistical performance of our model by looking at the decision boundary and point-out the misclassified samples.
```
import numpy as np
target_predicted = tree.predict(X)
mask_misclassified = y != target_predicted
from helper.plotting import DecisionBoundaryDisplay
_, ax = plt.subplots(figsize=(8, 6))
# plot the decision boundaries
display = DecisionBoundaryDisplay.from_estimator(
tree, X, response_method="predict", cmap=plt.cm.viridis,
alpha=0.4, ax=ax,
)
# plot the original dataset
data.plot.scatter(
x="Culmen Length (mm)",
y="Culmen Depth (mm)",
c="Species",
s=80,
cmap=plt.cm.viridis,
alpha=0.5,
edgecolor="black",
ax=ax,
)
# plot the misclassified samples
data[mask_misclassified].plot.scatter(
x="Culmen Length (mm)",
y="Culmen Depth (mm)",
c="black",
s=200,
marker="+",
ax=ax,
)
_ = plt.title("Decision tree predictions \nwith misclassified samples "
"highlighted")
```
We observe that our decision tree is making a couple of error for some Gentoo and Adelie samples. What we would be interested in now is to train a new decision tree but that should only focus on the misclassified samples this time. Scikit-learn exposes a `sample_weight` parameter in the `fit` method that allows to give more weight to some specific sample. We will use this parameter to only focus our new decision tree on the misclassified samples.
```
sample_weight = mask_misclassified.astype(np.float64)
tree = DecisionTreeClassifier(max_depth=2, random_state=0)
tree.fit(X, y, sample_weight=sample_weight)
```
Let's check the decision boundary of this newly trained decision tree classifier.
```
_, ax = plt.subplots(figsize=(8, 6))
# plot the decision boundaries
display = DecisionBoundaryDisplay.from_estimator(
tree, X, response_method="predict", cmap=plt.cm.viridis,
alpha=0.4,
ax=ax,
)
# plot the original dataset
data.plot.scatter(
x="Culmen Length (mm)",
y="Culmen Depth (mm)",
c="Species",
s=80,
cmap=plt.cm.viridis,
alpha=0.5,
edgecolor="black",
ax=ax,
)
# plot the misclassified samples
data[mask_misclassified].plot.scatter(
x="Culmen Length (mm)",
y="Culmen Depth (mm)",
c="black",
s=200,
marker="+",
ax=ax,
)
_ = plt.title("Decision tree predictions \nwith misclassified samples "
"highlighted")
target_predicted = tree.predict(X)
mask_new_misclassifier = y != target_predicted
remaining_misclassified_samples_idx = (
mask_misclassified & mask_new_misclassifier
)
print(f"Number of samples previously misclassified and "
f"still misclassified: {remaining_misclassified_samples_idx.sum()}")
```
We can observe that the previously misclassified samples are well classified now. However, it comes at the cost of misclassifying some other samples. We could continue by training a serie of decision tree classifiers. However, at some point, we need to find a way to combine them. One way could be that we could trust more or less a classifier depending on the ratio of good classification on the full training set.
```
ensemble_weight = [
(y.size - mask_misclassified.sum()) / y.size,
(y.size - mask_new_misclassifier.sum()) / y.size,
]
ensemble_weight
```
In our example, the first classification has a good accuracy and we will trust it more than the second classifier. Thus, we could make a linear combination of the different decision tree classifiers.
The algorithm that we just did is a simplification of an algorithm known as `AdaBoostClassifier`.
<div class="alert alert-success">
<b>EXERCISE:</b>:
<br>
<ul>
<li>Train a <tt>sklearn.ensemble.AdaBoostClassifier</tt> with 3 estimators and where the base estimator is a <tt>DecisionTreeClassifier</tt> with a <tt>max_depth=3</tt>.</li>
<li>Once this classifier trained, access the fitted attribute <tt>estimators_</tt> that contains the different decision tree classifiers and plot their decision boundary.</li>
<li>What are the weights associated with each decision tree classifiers.</li>
</ul>
</div>
```
# %load solutions/solution_20.py
from sklearn.ensemble import AdaBoostClassifier
base_estimator = DecisionTreeClassifier(max_depth=3, random_state=0)
adaboost = AdaBoostClassifier(
base_estimator=base_estimator,
n_estimators=3, algorithm="SAMME",
random_state=0
)
adaboost.fit(X, y)
# %load solutions/solution_21.py
for boosting_round, tree in enumerate(adaboost.estimators_):
_, ax = plt.subplots(figsize=(8, 6))
display = DecisionBoundaryDisplay.from_estimator(
tree, X, response_method="predict", cmap=plt.cm.viridis,
alpha=0.4,
ax=ax,
)
data.plot.scatter(
x="Culmen Length (mm)",
y="Culmen Depth (mm)",
c="Species",
s=80,
cmap=plt.cm.viridis,
alpha=0.5,
edgecolor="black",
ax=ax,
)
_ = ax.set_title(f"Decision tree trained at round {boosting_round}")
# %load solutions/solution_22.py
print(f"Weight of each classifier: {adaboost.estimator_weights_}")
# %load solutions/solution_23.py
print(f"Error of each classifier: {adaboost.estimator_errors_}")
```
## Gradient Boosting Decision Trees
AdaBoost predictors are less use nowadays in practice. Instead, gradient boosting decision trees are used and have been demonstrated to be better models.
In gradient boosting, each estimator will be a decision tree regressor even in classification. Using a regression tree allows to get a continuous residuals. Each estimator to be added in the sequence of estimator will be trained on the residuals of the previous estimators. In addition, there are a couple of parameters allowing to correct more or less fast the residuals from previous estimators.
Let's illustrate such model on a classification task.
```
from sklearn.model_selection import train_test_split
data = pd.read_csv("../datasets/adult-census-numeric-all.csv")
X, y = data.drop(columns="class"), data["class"]
X_train, X_test, y_train, y_test = train_test_split(
X, y, random_state=0
)
from sklearn.ensemble import GradientBoostingClassifier
classifier = GradientBoostingClassifier(n_estimators=5)
classifier.fit(X_train, y_train)
classifier.score(X_test, y_test)
```
We can inspect the different underlying estimators to show that we used indeed decisiont tree regressor even in a classification setting.
```
classifier.estimators_
from sklearn.tree import plot_tree
_, ax = plt.subplots(figsize=(20, 8))
_ = plot_tree(
classifier.estimators_[0][0],
feature_names=X_train.columns,
ax=ax,
)
```
### Histogram gradient boosting decision trees
<div class="alert alert-success">
<b>ACCELERATE GRADIENT BOOSTING</b>:
<ul>
<li>Which solution would you use to accelerate the training speed of gradient boosting algorithm.</li>
</ul>
</div>
## Short introduction of `KBinsDiscretizer`
We will show a trick to accelerate gradient boosting and more generally decision tree. When presenting decision trees, we mentioned that a split is chosen among all possible available splits that are defined by the unique values available in a given feature. One can reduce the amount of splits by binning the values of a feature beforehand and only consider the bin edges as potential edge. Since gradient boosting is ensembling several model, the lack of available splits will be attenuated by the size of the ensemble.
Here, we show that you can bin a dataset in scikit-learn using the `KBinsDiscritizer`.
```
from sklearn.preprocessing import KBinsDiscretizer
discretizer = KBinsDiscretizer(
n_bins=10, encode="ordinal", strategy="uniform"
)
X_trans = discretizer.fit_transform(X)
X_trans
[len(np.unique(col)) for col in X_trans.T]
```
Here, we decided to use 10 bins for each features.
<div class="alert alert-success">
<b>EXERCISE</b>:
<ul>
<li>Create a pipeline composed of a <tt>KBinsDiscretizer</tt> followed by a <tt>GradientBoostingClassfier</tt>.</li>
<li>Compare the training time with the vanilla <tt>GradientBoostingClassifier</tt>.</li>
</ul>
</div>
```
from sklearn.pipeline import make_pipeline
n_estimators = 100
%%time
make_pipeline(discretizer, GradientBoostingClassifier(n_estimators=n_estimators)).fit(X, y)
%%time
GradientBoostingClassifier(n_estimators=n_estimators).fit(X, y)
# %load solutions/solution_24.py
# %load solutions/solution_25.py
# %load solutions/solution_26.py
# %load solutions/solution_27.py
```
Scikit-learn provides `HistGradientBoostingClassifier` which is an approximate gradient boosting algorithm similar to `lightgbm` and `xgboost`.
```
%%time
from sklearn.ensemble import HistGradientBoostingClassifier
clf = HistGradientBoostingClassifier(max_iter=200, max_bins=10)
clf.fit(X_train, y_train)
clf.score(X_test, y_test)
```
### Hyperparameters
For gradient-boosting, parameters are coupled, so we cannot set the parameters one after the other anymore. The important parameters are `n_estimators`, `max_depth`, and `learning_rate`.
Let’s first discuss the `max_depth` parameter. We saw in the section on gradient-boosting that the algorithm fits the error of the previous tree in the ensemble. Thus, fitting fully grown trees will be detrimental. Indeed, the first tree of the ensemble would perfectly fit (overfit) the data and thus no subsequent tree would be required, since there would be no residuals. Therefore, the tree used in gradient-boosting should have a low depth, typically between 3 to 8 levels. Having very weak learners at each step will help reducing overfitting.
With this consideration in mind, the deeper the trees, the faster the residuals will be corrected and less learners are required. Therefore, `n_estimators` should be increased if `max_depth` is lower.
Finally, we have overlooked the impact of the `learning_rate` parameter until now. When fitting the residuals, we would like the tree to try to correct all possible errors or only a fraction of them. The learning-rate allows you to control this behaviour. A small learning-rate value would only correct the residuals of very few samples. If a large learning-rate is set (e.g., 1), we would fit the residuals of all samples. So, with a very low learning-rate, we will need more estimators to correct the overall error. However, a too large learning-rate tends to obtain an overfitted ensemble, similar to having a too large tree depth.
We will come back in the next chapter how to find the best set of hyperparameters in practice.
An option that is useful in histogram gradient boosting is the `early_stopping` parameter. This will split the data internally during `fit` and use a validation set to assess the improvement of adding a new decision tree regressor. If the model detect that adding new estimators will not improve the statistical performance of the model, it will stop the `fit` process. Let's check in practice how it works:
```
model = HistGradientBoostingClassifier(early_stopping=True, max_iter=1_000)
model.fit(X_train, y_train)
```
We requested 1,000 decision trees that is more than we actually need to fit the data at hand. We can now check the number of trees that has been added in the model:
```
model.n_iter_
```
We see that the gradient boosting stopped the learning process after 127 decision trees.
|
github_jupyter
|
# temporary fix to avoid spurious warning raised in scikit-learn 1.0.0
# it will be solved in scikit-learn 1.0.1
import warnings
warnings.filterwarnings("ignore", message="X has feature names.*")
warnings.filterwarnings("ignore", message="X does not have valid feature names.*")
import pandas as pd
data = pd.read_csv("../datasets/penguins_classification.csv")
data["Species"] = data["Species"].astype("category")
X, y = data[["Culmen Length (mm)", "Culmen Depth (mm)"]], data["Species"]
import seaborn as sns
sns.set_context("poster")
import matplotlib.pyplot as plt
_, ax = plt.subplots(figsize=(8, 6))
_ = data.plot.scatter(
x="Culmen Length (mm)",
y="Culmen Depth (mm)",
c="Species",
s=80,
cmap=plt.cm.viridis,
ax=ax,
)
from sklearn.tree import DecisionTreeClassifier
tree = DecisionTreeClassifier(max_depth=2, random_state=0)
tree.fit(X, y)
import numpy as np
target_predicted = tree.predict(X)
mask_misclassified = y != target_predicted
from helper.plotting import DecisionBoundaryDisplay
_, ax = plt.subplots(figsize=(8, 6))
# plot the decision boundaries
display = DecisionBoundaryDisplay.from_estimator(
tree, X, response_method="predict", cmap=plt.cm.viridis,
alpha=0.4, ax=ax,
)
# plot the original dataset
data.plot.scatter(
x="Culmen Length (mm)",
y="Culmen Depth (mm)",
c="Species",
s=80,
cmap=plt.cm.viridis,
alpha=0.5,
edgecolor="black",
ax=ax,
)
# plot the misclassified samples
data[mask_misclassified].plot.scatter(
x="Culmen Length (mm)",
y="Culmen Depth (mm)",
c="black",
s=200,
marker="+",
ax=ax,
)
_ = plt.title("Decision tree predictions \nwith misclassified samples "
"highlighted")
sample_weight = mask_misclassified.astype(np.float64)
tree = DecisionTreeClassifier(max_depth=2, random_state=0)
tree.fit(X, y, sample_weight=sample_weight)
_, ax = plt.subplots(figsize=(8, 6))
# plot the decision boundaries
display = DecisionBoundaryDisplay.from_estimator(
tree, X, response_method="predict", cmap=plt.cm.viridis,
alpha=0.4,
ax=ax,
)
# plot the original dataset
data.plot.scatter(
x="Culmen Length (mm)",
y="Culmen Depth (mm)",
c="Species",
s=80,
cmap=plt.cm.viridis,
alpha=0.5,
edgecolor="black",
ax=ax,
)
# plot the misclassified samples
data[mask_misclassified].plot.scatter(
x="Culmen Length (mm)",
y="Culmen Depth (mm)",
c="black",
s=200,
marker="+",
ax=ax,
)
_ = plt.title("Decision tree predictions \nwith misclassified samples "
"highlighted")
target_predicted = tree.predict(X)
mask_new_misclassifier = y != target_predicted
remaining_misclassified_samples_idx = (
mask_misclassified & mask_new_misclassifier
)
print(f"Number of samples previously misclassified and "
f"still misclassified: {remaining_misclassified_samples_idx.sum()}")
ensemble_weight = [
(y.size - mask_misclassified.sum()) / y.size,
(y.size - mask_new_misclassifier.sum()) / y.size,
]
ensemble_weight
# %load solutions/solution_20.py
from sklearn.ensemble import AdaBoostClassifier
base_estimator = DecisionTreeClassifier(max_depth=3, random_state=0)
adaboost = AdaBoostClassifier(
base_estimator=base_estimator,
n_estimators=3, algorithm="SAMME",
random_state=0
)
adaboost.fit(X, y)
# %load solutions/solution_21.py
for boosting_round, tree in enumerate(adaboost.estimators_):
_, ax = plt.subplots(figsize=(8, 6))
display = DecisionBoundaryDisplay.from_estimator(
tree, X, response_method="predict", cmap=plt.cm.viridis,
alpha=0.4,
ax=ax,
)
data.plot.scatter(
x="Culmen Length (mm)",
y="Culmen Depth (mm)",
c="Species",
s=80,
cmap=plt.cm.viridis,
alpha=0.5,
edgecolor="black",
ax=ax,
)
_ = ax.set_title(f"Decision tree trained at round {boosting_round}")
# %load solutions/solution_22.py
print(f"Weight of each classifier: {adaboost.estimator_weights_}")
# %load solutions/solution_23.py
print(f"Error of each classifier: {adaboost.estimator_errors_}")
from sklearn.model_selection import train_test_split
data = pd.read_csv("../datasets/adult-census-numeric-all.csv")
X, y = data.drop(columns="class"), data["class"]
X_train, X_test, y_train, y_test = train_test_split(
X, y, random_state=0
)
from sklearn.ensemble import GradientBoostingClassifier
classifier = GradientBoostingClassifier(n_estimators=5)
classifier.fit(X_train, y_train)
classifier.score(X_test, y_test)
classifier.estimators_
from sklearn.tree import plot_tree
_, ax = plt.subplots(figsize=(20, 8))
_ = plot_tree(
classifier.estimators_[0][0],
feature_names=X_train.columns,
ax=ax,
)
from sklearn.preprocessing import KBinsDiscretizer
discretizer = KBinsDiscretizer(
n_bins=10, encode="ordinal", strategy="uniform"
)
X_trans = discretizer.fit_transform(X)
X_trans
[len(np.unique(col)) for col in X_trans.T]
from sklearn.pipeline import make_pipeline
n_estimators = 100
%%time
make_pipeline(discretizer, GradientBoostingClassifier(n_estimators=n_estimators)).fit(X, y)
%%time
GradientBoostingClassifier(n_estimators=n_estimators).fit(X, y)
# %load solutions/solution_24.py
# %load solutions/solution_25.py
# %load solutions/solution_26.py
# %load solutions/solution_27.py
%%time
from sklearn.ensemble import HistGradientBoostingClassifier
clf = HistGradientBoostingClassifier(max_iter=200, max_bins=10)
clf.fit(X_train, y_train)
clf.score(X_test, y_test)
model = HistGradientBoostingClassifier(early_stopping=True, max_iter=1_000)
model.fit(X_train, y_train)
model.n_iter_
| 0.645455 | 0.982541 |
# London Start-up Map
This notebook contains the code performed for the analysis in <a href="https://medium.com/@quantscoop/london-animated-start-up-map-2011-2020-3ae5a709edf9">this blog post</a>.
The end result is a map of London that displays relative start-up formation vs cessation:
<img src="animation.gif"/>
### Packages
```
%%capture
!pip install pgeocode
!pip install plotly-express
!pip install numpy
!pip install matplotlib
!pip install pandas
!pip install imageio
!pip install visvis
%%capture
import pandas as pd
import datetime
from datetime import date
from dateutil.relativedelta import relativedelta
import pgeocode
import json
import numpy as np
import plotly.express as px
import copy
from matplotlib import cm
import matplotlib.pyplot as plt
import urllib.request
import requests
import string
import time
import pickle
import random
import io
import os
import sys
import collections
import imageio
import glob
import visvis as vv
```
## Downloading London Postcode GEO-code JSON
We will download the JSON file for the geographical boundaries of London postcodes from <a href="https://github.com/sjwhitworth/london_geojson/blob/master/london_postcodes.json">here</a>.
```
london_geojson_url = "https://raw.githubusercontent.com/sjwhitworth/london_geojson/master/london_postcodes.json"
urllib.request.urlretrieve(london_geojson_url,"london_postcodes.json")
with open("london_postcodes.json","r") as file:
data = json.load(file)
```
## Scraping Companies House Data
We will now scrap the details of London-based companies from the Companies House website.
You will need an API key, which you can set-up <a href="https://developer.companieshouse.gov.uk/api/docs/">here</a>.
```
%%capture
api_key = "api_key"
url = "https://api.companieshouse.gov.uk/company/"
request_counter = 0
attempted_company_codes = []
scrape_start = 7500000
scrape_stop = 12480000
scraping_attempts = int(0) # Change to a larger number e.g 10000
current_company_id = 0
if (os.path.exists("companies_house_data.txt")):
dataframe = pd.read_table("companies_house_data.txt",delimiter="\t")
else:
dataframe = pd.DataFrame(columns=['number','formed','active_until','postcode'])
dataframe.set_index('number')
for counter in range(scraping_attempts):
while (current_company_id in attempted_company_codes):
current_company_id = random.randint(scrape_start,scrape_stop)
attempted_company_codes.append(current_company_id)
str_company_id = str(current_company_id).zfill(8)
request_result = requests.get(url = url + str_company_id, auth = (api_key,""))
request_counter +=1
json_fail = False
json_result = request_result.json()
try:
sys.stdout = io.StringIO()
print(json_result)
sys.stdout = sys.__stdout__
except:
json_fail = True
# Sleep when requests close to rate limit.
if (request_counter > 550):
request_counter = 0
time.sleep(300)
print("sleeping for 5 min")
print("Counter: {}".format(counter))
contains_registered_office = 'registered_office_address' in json_result
is_situated_in_london = contains_registered_office and \
'locality' in json_result['registered_office_address'] \
and 'London' in json_result['registered_office_address']['locality']
contains_company_status = 'company_status' in json_result
contains_errors = 'errors' in json_result
contains_postal_code = contains_registered_office and 'postal_code' in json_result['registered_office_address']
is_error_free = json_fail == False and contains_errors == False
if (is_error_free and contains_postal_code and is_situated_in_london and contains_company_status):
cur_dict = {}
cur_dict['number'] = json_result['company_number']
cur_dict['formed'] = json_result['date_of_creation']
cur_dict['active_until'] = json_result['date_of_cessation'] if 'date_of_cessation' in json_result else "2020-12-01"
cur_dict['postcode'] = cessation,json_result['registered_office_address']['postal_code']
dataframe.append(cur_dict,ignore_index=True)
```
Aggregate the set of existing postcodes in the JSON and apply reformatting for further down the line:
```
postcodes_extant = set()
for counter in range(len(data['features'])):
data['features'][counter]['id'] = data['features'][counter]['properties']['Name']
postcodes_extant.add(data['features'][counter]['properties']['Name'])
```
Next, to compare the Companies House company postcode data vs. the London postcodes, we will use this utility function:
```
def get_closest_postcode(postcode, postcodes_list):
if (" " in postcode and postcode.split()[0] in postcodes_list):
return postcode.split()[0]
postcode = ''.join(postcode.split())
longest = ""
for pc in postcodes_list:
result = postcode.find(pc)
if (result == 0 and len(pc) > len(longest)):
longest = pc
return longest
postcodes_list = list(postcodes_extant)
dataframe['postcode'] = [get_closest_postcode(val.upper(), postcodes_list) for val in dataframe['postcode'].values.tolist()]
```
Now, lets transform the company creation vs. cessation dates to python datetime format:
```
dataframe['dt1'] = [datetime.date(int(val.split("-")[0]), int(val.split("-")[1]),1) for val in dataframe['formed'].values.tolist()]
dataframe['dt2'] = [datetime.date(int(val.split("-")[0]), int(val.split("-")[1]),1) for val in dataframe['active_until'].values.tolist()]
earliest = min(dataframe['dt1'].tolist())
latest = max(dataframe['dt1'].tolist())
```
And another utility function for getting the time-difference between months:
```
def get_months_dif(d1,d2):
return d1.month - d2.month + 12*(d1.year-d2.year)
```
Now, lets create a dictionary that maps postcodes to how many companies were
active in a given period (represented as a numpy array):
```
total_months = get_months_dif(latest,earliest)
postcode_dict = collections.defaultdict(lambda:np.zeros((total_months+1)))
total = np.zeros((total_months+1))
total2 = np.zeros((total_months+1))
for index,row in dataframe.iterrows():
matrix = np.zeros((total_months+1))
start_months = get_months_dif(row['dt1'],earliest)
end_months = get_months_dif(min(row['dt2'],latest),earliest)
matrix[start_months:end_months] = 1.0
postcode_dict[row['postcode']] += matrix
test = np.zeros((total_months+1))
test2 = np.zeros((total_months+1))
test[end_months] = 1
test2[start_months] = 1
total += matrix
total2 += test2
postcode_vals = postcode_dict.values()
max_val = 0
#We will take the square root of the number of active companies for visual purposes
for i in postcode_vals:
max_val = np.power(max(max_val,np.max(i,axis=0)),0.5)
```
Now lets create a mapping of postcodes to transformed net-active company values:
```
plotting_values = [pd.DataFrame.from_dict({'id': list(postcode_dict.keys()), 'val': [float(np.power(val[index],0.5)) for val in list(postcode_dict.values())]}) for index in range(total_months)]
```
And finally, we will plot each month of the start-up map using the plotly express choropleth map,
and save them as PNGs, which are then combined with imageio into an animated .gif image.
```
viridis = cm.get_cmap('viridis', int(max_val)+1)
viridis.colors
def get_col(viridis_val):
return 'rgb({},{},{})'.format(int(viridis_val[0]*255),int(viridis_val[1]*255),int(viridis_val[2]*255))
colorscale = {}
colorscale2 = ((0.0,get_col(viridis.colors[0])),)
for index in range(1,int(max_val)+1):
colorscale[float(index)] = get_col(viridis.colors[index])
colorscale2 += ((float(index),get_col(viridis.colors[index])),)
for index in range(total_months):
day = earliest + relativedelta(months=index)
fig = px.choropleth_mapbox(plotting_values[index], geojson=data, locations='id', color='val',
color_continuous_scale ="RdYlGn",
range_color=(0, float(max_val)),
mapbox_style='carto-positron',
zoom=10, center = {"lat": 51.5073, "lon": -0.1277},
opacity=0.1,
labels={'val':'(sqrt) Net Active'}
)
fig.update_layout(title=" London: (sqrt of) Active Companies per Month, {}-{}".format(day.year,day.month), title_font_size=24)
fig.layout.coloraxis.autocolorscale= False
fig.layout.coloraxis.cauto= False
fig.layout.coloraxis.cmin= 0.0
fig.layout.coloraxis.cmax= float(max_val)
fig.write_image("./images_output/image_{}_{}_{}.png".format(day.year,day.month//10,day.month%10))
images = []
imageio.plugins.freeimage.download()
for filename in sorted(glob.glob("./images_output/*.png")):
images.append(imageio.imread(filename))
images.append(imageio.imread(filename))
imageio.mimsave('animation.gif', images, 'GIF-FI')
```
|
github_jupyter
|
%%capture
!pip install pgeocode
!pip install plotly-express
!pip install numpy
!pip install matplotlib
!pip install pandas
!pip install imageio
!pip install visvis
%%capture
import pandas as pd
import datetime
from datetime import date
from dateutil.relativedelta import relativedelta
import pgeocode
import json
import numpy as np
import plotly.express as px
import copy
from matplotlib import cm
import matplotlib.pyplot as plt
import urllib.request
import requests
import string
import time
import pickle
import random
import io
import os
import sys
import collections
import imageio
import glob
import visvis as vv
london_geojson_url = "https://raw.githubusercontent.com/sjwhitworth/london_geojson/master/london_postcodes.json"
urllib.request.urlretrieve(london_geojson_url,"london_postcodes.json")
with open("london_postcodes.json","r") as file:
data = json.load(file)
%%capture
api_key = "api_key"
url = "https://api.companieshouse.gov.uk/company/"
request_counter = 0
attempted_company_codes = []
scrape_start = 7500000
scrape_stop = 12480000
scraping_attempts = int(0) # Change to a larger number e.g 10000
current_company_id = 0
if (os.path.exists("companies_house_data.txt")):
dataframe = pd.read_table("companies_house_data.txt",delimiter="\t")
else:
dataframe = pd.DataFrame(columns=['number','formed','active_until','postcode'])
dataframe.set_index('number')
for counter in range(scraping_attempts):
while (current_company_id in attempted_company_codes):
current_company_id = random.randint(scrape_start,scrape_stop)
attempted_company_codes.append(current_company_id)
str_company_id = str(current_company_id).zfill(8)
request_result = requests.get(url = url + str_company_id, auth = (api_key,""))
request_counter +=1
json_fail = False
json_result = request_result.json()
try:
sys.stdout = io.StringIO()
print(json_result)
sys.stdout = sys.__stdout__
except:
json_fail = True
# Sleep when requests close to rate limit.
if (request_counter > 550):
request_counter = 0
time.sleep(300)
print("sleeping for 5 min")
print("Counter: {}".format(counter))
contains_registered_office = 'registered_office_address' in json_result
is_situated_in_london = contains_registered_office and \
'locality' in json_result['registered_office_address'] \
and 'London' in json_result['registered_office_address']['locality']
contains_company_status = 'company_status' in json_result
contains_errors = 'errors' in json_result
contains_postal_code = contains_registered_office and 'postal_code' in json_result['registered_office_address']
is_error_free = json_fail == False and contains_errors == False
if (is_error_free and contains_postal_code and is_situated_in_london and contains_company_status):
cur_dict = {}
cur_dict['number'] = json_result['company_number']
cur_dict['formed'] = json_result['date_of_creation']
cur_dict['active_until'] = json_result['date_of_cessation'] if 'date_of_cessation' in json_result else "2020-12-01"
cur_dict['postcode'] = cessation,json_result['registered_office_address']['postal_code']
dataframe.append(cur_dict,ignore_index=True)
postcodes_extant = set()
for counter in range(len(data['features'])):
data['features'][counter]['id'] = data['features'][counter]['properties']['Name']
postcodes_extant.add(data['features'][counter]['properties']['Name'])
def get_closest_postcode(postcode, postcodes_list):
if (" " in postcode and postcode.split()[0] in postcodes_list):
return postcode.split()[0]
postcode = ''.join(postcode.split())
longest = ""
for pc in postcodes_list:
result = postcode.find(pc)
if (result == 0 and len(pc) > len(longest)):
longest = pc
return longest
postcodes_list = list(postcodes_extant)
dataframe['postcode'] = [get_closest_postcode(val.upper(), postcodes_list) for val in dataframe['postcode'].values.tolist()]
dataframe['dt1'] = [datetime.date(int(val.split("-")[0]), int(val.split("-")[1]),1) for val in dataframe['formed'].values.tolist()]
dataframe['dt2'] = [datetime.date(int(val.split("-")[0]), int(val.split("-")[1]),1) for val in dataframe['active_until'].values.tolist()]
earliest = min(dataframe['dt1'].tolist())
latest = max(dataframe['dt1'].tolist())
def get_months_dif(d1,d2):
return d1.month - d2.month + 12*(d1.year-d2.year)
total_months = get_months_dif(latest,earliest)
postcode_dict = collections.defaultdict(lambda:np.zeros((total_months+1)))
total = np.zeros((total_months+1))
total2 = np.zeros((total_months+1))
for index,row in dataframe.iterrows():
matrix = np.zeros((total_months+1))
start_months = get_months_dif(row['dt1'],earliest)
end_months = get_months_dif(min(row['dt2'],latest),earliest)
matrix[start_months:end_months] = 1.0
postcode_dict[row['postcode']] += matrix
test = np.zeros((total_months+1))
test2 = np.zeros((total_months+1))
test[end_months] = 1
test2[start_months] = 1
total += matrix
total2 += test2
postcode_vals = postcode_dict.values()
max_val = 0
#We will take the square root of the number of active companies for visual purposes
for i in postcode_vals:
max_val = np.power(max(max_val,np.max(i,axis=0)),0.5)
plotting_values = [pd.DataFrame.from_dict({'id': list(postcode_dict.keys()), 'val': [float(np.power(val[index],0.5)) for val in list(postcode_dict.values())]}) for index in range(total_months)]
viridis = cm.get_cmap('viridis', int(max_val)+1)
viridis.colors
def get_col(viridis_val):
return 'rgb({},{},{})'.format(int(viridis_val[0]*255),int(viridis_val[1]*255),int(viridis_val[2]*255))
colorscale = {}
colorscale2 = ((0.0,get_col(viridis.colors[0])),)
for index in range(1,int(max_val)+1):
colorscale[float(index)] = get_col(viridis.colors[index])
colorscale2 += ((float(index),get_col(viridis.colors[index])),)
for index in range(total_months):
day = earliest + relativedelta(months=index)
fig = px.choropleth_mapbox(plotting_values[index], geojson=data, locations='id', color='val',
color_continuous_scale ="RdYlGn",
range_color=(0, float(max_val)),
mapbox_style='carto-positron',
zoom=10, center = {"lat": 51.5073, "lon": -0.1277},
opacity=0.1,
labels={'val':'(sqrt) Net Active'}
)
fig.update_layout(title=" London: (sqrt of) Active Companies per Month, {}-{}".format(day.year,day.month), title_font_size=24)
fig.layout.coloraxis.autocolorscale= False
fig.layout.coloraxis.cauto= False
fig.layout.coloraxis.cmin= 0.0
fig.layout.coloraxis.cmax= float(max_val)
fig.write_image("./images_output/image_{}_{}_{}.png".format(day.year,day.month//10,day.month%10))
images = []
imageio.plugins.freeimage.download()
for filename in sorted(glob.glob("./images_output/*.png")):
images.append(imageio.imread(filename))
images.append(imageio.imread(filename))
imageio.mimsave('animation.gif', images, 'GIF-FI')
| 0.201185 | 0.777131 |
```
import pygraphviz as pgv
from IPython.core.display import Image
```
## The cell below comes from the Orthology notebook
```
import requests
ensembl_server = 'http://rest.ensembl.org'
def do_request(server, service, *args, **kwargs):
params = ''
for a in args:
if a is not None:
params += '/' + a
req = requests.get('%s/%s%s' % (server, service, params),
params=kwargs,
headers={'Content-Type': 'application/json'})
if not req.ok:
req.raise_for_status()
return req.json()
lct_id = 'ENSG00000115850'
refs = do_request(ensembl_server, 'xrefs/id', lct_id, external_db='GO', all_levels='1')
print(len(refs))
print(refs[0].keys())
for ref in refs:
go_id = ref['primary_id']
details = do_request(ensembl_server, 'ontology/id', go_id)
print('%s %s %s' % (go_id, details['namespace'], ref['description']))
print('%s\n' % details['definition'])
go_id = 'GO:0000016'
my_data = do_request(ensembl_server, 'ontology/id', go_id)
for k, v in my_data.items():
if k == 'parents':
for parent in v:
print(parent)
parent_id = parent['accession']
else:
print('%s: %s' % (k, str(v)))
parent_data = do_request(ensembl_server, 'ontology/id', parent_id)
print(parent_id, len(parent_data['children']))
refs = do_request(ensembl_server, 'ontology/ancestors/chart', go_id)
for go, entry in refs.items():
print(go)
term = entry['term']
print('%s %s' % (term['name'], term['definition']))
is_a = entry.get('is_a', [])
print('\t is a: %s\n' % ', '.join([x['accession'] for x in is_a]))
def get_upper(go_id):
parents = {}
node_data = {}
refs = do_request(ensembl_server, 'ontology/ancestors/chart', go_id)
for ref, entry in refs.items():
my_data = do_request(ensembl_server, 'ontology/id', ref)
node_data[ref] = {'name': entry['term']['name'], 'children': my_data['children']}
try:
parents[ref] = [x['accession'] for x in entry['is_a']]
except KeyError:
pass # Top of hierarchy
return parents, node_data
parents, node_data = get_upper(go_id)
g = pgv.AGraph(directed=True)
for ofs, ofs_parents in parents.items():
ofs_text = '%s\n(%s)' % (node_data[ofs]['name'].replace(', ', '\n'), ofs)
for parent in ofs_parents:
parent_text = '%s\n(%s)' % (node_data[parent]['name'].replace(', ', '\n'), parent)
children = node_data[parent]['children']
if len(children) < 3:
for child in children:
if child['accession'] in node_data:
continue
g.add_edge(parent_text, child['accession'])
else:
g.add_edge(parent_text, '...%d...' % (len(children) - 1))
g.add_edge(parent_text, ofs_text)
print(g)
g.graph_attr['label']='Ontology tree for Lactase activity'
g.node_attr['shape']='rectangle'
g.layout(prog='dot')
g.draw('graph.png')
Image("graph.png")
print(go_id)
refs = do_request(ensembl_server, 'ontology/descendants', go_id)
for go in refs:
print(go['accession'], go['name'], go['definition'])
```
|
github_jupyter
|
import pygraphviz as pgv
from IPython.core.display import Image
import requests
ensembl_server = 'http://rest.ensembl.org'
def do_request(server, service, *args, **kwargs):
params = ''
for a in args:
if a is not None:
params += '/' + a
req = requests.get('%s/%s%s' % (server, service, params),
params=kwargs,
headers={'Content-Type': 'application/json'})
if not req.ok:
req.raise_for_status()
return req.json()
lct_id = 'ENSG00000115850'
refs = do_request(ensembl_server, 'xrefs/id', lct_id, external_db='GO', all_levels='1')
print(len(refs))
print(refs[0].keys())
for ref in refs:
go_id = ref['primary_id']
details = do_request(ensembl_server, 'ontology/id', go_id)
print('%s %s %s' % (go_id, details['namespace'], ref['description']))
print('%s\n' % details['definition'])
go_id = 'GO:0000016'
my_data = do_request(ensembl_server, 'ontology/id', go_id)
for k, v in my_data.items():
if k == 'parents':
for parent in v:
print(parent)
parent_id = parent['accession']
else:
print('%s: %s' % (k, str(v)))
parent_data = do_request(ensembl_server, 'ontology/id', parent_id)
print(parent_id, len(parent_data['children']))
refs = do_request(ensembl_server, 'ontology/ancestors/chart', go_id)
for go, entry in refs.items():
print(go)
term = entry['term']
print('%s %s' % (term['name'], term['definition']))
is_a = entry.get('is_a', [])
print('\t is a: %s\n' % ', '.join([x['accession'] for x in is_a]))
def get_upper(go_id):
parents = {}
node_data = {}
refs = do_request(ensembl_server, 'ontology/ancestors/chart', go_id)
for ref, entry in refs.items():
my_data = do_request(ensembl_server, 'ontology/id', ref)
node_data[ref] = {'name': entry['term']['name'], 'children': my_data['children']}
try:
parents[ref] = [x['accession'] for x in entry['is_a']]
except KeyError:
pass # Top of hierarchy
return parents, node_data
parents, node_data = get_upper(go_id)
g = pgv.AGraph(directed=True)
for ofs, ofs_parents in parents.items():
ofs_text = '%s\n(%s)' % (node_data[ofs]['name'].replace(', ', '\n'), ofs)
for parent in ofs_parents:
parent_text = '%s\n(%s)' % (node_data[parent]['name'].replace(', ', '\n'), parent)
children = node_data[parent]['children']
if len(children) < 3:
for child in children:
if child['accession'] in node_data:
continue
g.add_edge(parent_text, child['accession'])
else:
g.add_edge(parent_text, '...%d...' % (len(children) - 1))
g.add_edge(parent_text, ofs_text)
print(g)
g.graph_attr['label']='Ontology tree for Lactase activity'
g.node_attr['shape']='rectangle'
g.layout(prog='dot')
g.draw('graph.png')
Image("graph.png")
print(go_id)
refs = do_request(ensembl_server, 'ontology/descendants', go_id)
for go in refs:
print(go['accession'], go['name'], go['definition'])
| 0.148911 | 0.364297 |
# Stanza: A Tutorial on the Python CoreNLP Interface


While the Stanza library implements accurate neural network modules for basic functionalities such as part-of-speech tagging and dependency parsing, the [Stanford CoreNLP Java library](https://stanfordnlp.github.io/CoreNLP/) has been developed for years and offers more complementary features such as coreference resolution and relation extraction. To unlock these features, the Stanza library also offers an officially maintained Python interface to the CoreNLP Java library. This interface allows you to get NLP anntotations from CoreNLP by writing native Python code.
This tutorial walks you through the installation, setup and basic usage of this Python CoreNLP interface. If you want to learn how to use the neural network components in Stanza, please refer to other tutorials.
## 1. Installation
Before the installation starts, please make sure that you have Python 3 and Java installed on your computer. Since Colab already has them installed, we'll skip this procedure in this notebook.
### Installing Stanza
Installing and importing Stanza are as simple as running the following commands:
```
# Install stanza; note that the prefix "!" is not needed if you are running in a terminal
!pip install stanza==1.0.0
# Import stanza
import stanza
```
### Setting up Stanford CoreNLP
In order for the interface to work, the Stanford CoreNLP library has to be installed and a `CORENLP_HOME` environment variable has to be pointed to the installation location.
**Note**: if you are want to use the interface in a terminal (instead of a Colab notebook), you can properly set the `CORENLP_HOME` environment variable with:
```bash
export CORENLP_HOME=path_to_corenlp
```
Here we instead set this variable with the Python `os` library, simply because `export` command is not well-supported in Colab notebook.
```
# Download the Stanford CoreNLP Java library and unzip it to a ./corenlp folder
!echo "Downloading CoreNLP..."
!wget "http://nlp.stanford.edu/software/stanford-corenlp-full-2018-10-05.zip" -O corenlp.zip
!unzip corenlp.zip
!mv ./stanford-corenlp-full-2018-10-05 ./corenlp
# Set the CORENLP_HOME environment variable to point to the installation location
import os
os.environ["CORENLP_HOME"] = "./corenlp"
```
## 2. Annotating Text with CoreNLP Interface
### Constructing CoreNLPClient
At a high level, the CoreNLP Python interface works by first starting a background Java CoreNLP server process, and then initializing a client instance in Python which can pass the text to the background server process, and accept the returned annotation results.
We wrap these functionalities in a `CoreNLPClient` class. Therefore, we need to start by importing this class from Stanza.
```
# Import client module
from stanza.server import CoreNLPClient
```
After the import is done, we can construct a `CoreNLPClient` instance. The constructor method takes a Python list of annotator names as argument. Here let's explore some basic annotators including tokenization, sentence split, part-of-speech tagging, lemmatization and named entity recognition (NER).
Additionally, the client constructor accepts a `memory` argument, which specify how much memory will be allocated to the background Java process. An `endpoint` option can be used to specify a port number used by the communication between the server and the client. The default port is 9000, however, since this port is token in Colab, we'll manually set it to 9001 in the following example.
For more options in constructing the clients, please refer to the [CoreNLP Client Options List](https://stanfordnlp.github.io/stanza/corenlp_client.html#corenlp-client-options).
```
# Construct a CoreNLPClient with some basic annotators, a memory allocation of 4GB, and port number 9001
client = CoreNLPClient(annotators=['tokenize','ssplit', 'pos', 'lemma', 'ner'], memory='4G', endpoint='http://localhost:9001')
print(client)
# Start the background server and wait for some time
# Note that in practice this is totally optional, as by default the server will be started when the first annotation is performed
client.start()
import time; time.sleep(10)
```
Now if you print the background processes, you should be able to find the Java CoreNLP server running.
```
# Print background processes and look for java
!ps -o pid,cmd | grep java
```
### Annotating Text
Annotating a piece of text is as simple as passing the text into an `annotate` function of the client object. After the annotation is complete, a `Document` object will be returned with all annotations.
Note that although in general annotations are very fast, the first annotation might take a while to complete in the notebook. Please stay patient.
```
# Annotate some text
text = "Albert Einstein was a German-born theoretical physicist. He developed the theory of relativity."
document = client.annotate(text)
print(type(document))
```
## 3. Accessing Annotations
Annotations can be accessed from the returned `Document` object.
A `Document` contains a list of `Sentence`s, which contain a list of `Token`s. Here let's first explore the annotations stored in all tokens.
```
# Iterate over all tokens in all sentences, and print out the word, lemma, pos and ner tags
print("{:12s}\t{:12s}\t{:6s}\t{}".format("Word", "Lemma", "POS", "NER"))
for i, sent in enumerate(document.sentence):
print("[Sentence {}]".format(i+1))
for t in sent.token:
print("{:12s}\t{:12s}\t{:6s}\t{}".format(t.word, t.lemma, t.pos, t.ner))
print("")
```
Alternatively, you can also browse the NER results by iterating over entity mentions over the sentences. For example:
```
# Iterate over all detected entity mentions
print("{:30s}\t{}".format("Mention", "Type"))
for sent in document.sentence:
for m in sent.mentions:
print("{:30s}\t{}".format(m.entityMentionText, m.entityType))
```
To print all annotations a sentence, token or mention has, you can simply print the corresponding obejct.
```
# Print annotations of a token
print(document.sentence[0].token[0])
# Print annotations of a mention
print(document.sentence[0].mentions[0])
```
**Note**: Since the Stanza CoreNLP client interface simply ports the CoreNLP annotation results to native Python objects, for a comprehensive lists of available annotators and how their annotation results can be accessed, you will need to visit the [Stanford CoreNLP website](https://stanfordnlp.github.io/CoreNLP/).
## 4. Shutting Down the CoreNLP Server
To shut down the background CoreNLP server process, simply call the `stop` function of the client. Note that once a server is shutdown, you'll have to restart the server with the `start()` function before any annotation is requested.
```
# Shut down the background CoreNLP server
client.stop()
time.sleep(10)
!ps -o pid,cmd | grep java
```
### More Information
For more information on how to use the `CoreNLPClient`, please go to the [CoreNLPClient documentation page](https://stanfordnlp.github.io/stanza/corenlp_client.html).
## 5. Other Resources
- [Stanza Homepage](https://stanfordnlp.github.io/stanza/)
- [FAQs](https://stanfordnlp.github.io/stanza/faq.html)
- [GitHub Repo](https://github.com/stanfordnlp/stanza)
- [Reporting Issues](https://github.com/stanfordnlp/stanza/issues)
|
github_jupyter
|
# Install stanza; note that the prefix "!" is not needed if you are running in a terminal
!pip install stanza==1.0.0
# Import stanza
import stanza
export CORENLP_HOME=path_to_corenlp
# Download the Stanford CoreNLP Java library and unzip it to a ./corenlp folder
!echo "Downloading CoreNLP..."
!wget "http://nlp.stanford.edu/software/stanford-corenlp-full-2018-10-05.zip" -O corenlp.zip
!unzip corenlp.zip
!mv ./stanford-corenlp-full-2018-10-05 ./corenlp
# Set the CORENLP_HOME environment variable to point to the installation location
import os
os.environ["CORENLP_HOME"] = "./corenlp"
# Import client module
from stanza.server import CoreNLPClient
# Construct a CoreNLPClient with some basic annotators, a memory allocation of 4GB, and port number 9001
client = CoreNLPClient(annotators=['tokenize','ssplit', 'pos', 'lemma', 'ner'], memory='4G', endpoint='http://localhost:9001')
print(client)
# Start the background server and wait for some time
# Note that in practice this is totally optional, as by default the server will be started when the first annotation is performed
client.start()
import time; time.sleep(10)
# Print background processes and look for java
!ps -o pid,cmd | grep java
# Annotate some text
text = "Albert Einstein was a German-born theoretical physicist. He developed the theory of relativity."
document = client.annotate(text)
print(type(document))
# Iterate over all tokens in all sentences, and print out the word, lemma, pos and ner tags
print("{:12s}\t{:12s}\t{:6s}\t{}".format("Word", "Lemma", "POS", "NER"))
for i, sent in enumerate(document.sentence):
print("[Sentence {}]".format(i+1))
for t in sent.token:
print("{:12s}\t{:12s}\t{:6s}\t{}".format(t.word, t.lemma, t.pos, t.ner))
print("")
# Iterate over all detected entity mentions
print("{:30s}\t{}".format("Mention", "Type"))
for sent in document.sentence:
for m in sent.mentions:
print("{:30s}\t{}".format(m.entityMentionText, m.entityType))
# Print annotations of a token
print(document.sentence[0].token[0])
# Print annotations of a mention
print(document.sentence[0].mentions[0])
# Shut down the background CoreNLP server
client.stop()
time.sleep(10)
!ps -o pid,cmd | grep java
| 0.430866 | 0.986017 |
# Tune hyperparameters in your custom training loop
**Authors:** Tom O'Malley, Haifeng Jin<br>
**Date created:** 2019/10/28<br>
**Last modified:** 2022/01/12<br>
**Description:** Use `HyperModel.fit()` to tune training hyperparameters (such as batch size).
```
!pip install keras-tuner -q
```
## Introduction
The `HyperModel` class in KerasTuner provides a convenient way to define your
search space in a reusable object. You can override `HyperModel.build()` to
define and hypertune the model itself. To hypertune the training process (e.g.
by selecting the proper batch size, number of training epochs, or data
augmentation setup), you can override `HyperModel.fit()`, where you can access:
- The `hp` object, which is an instance of `keras_tuner.HyperParameters`
- The model built by `HyperModel.build()`
A basic example is shown in the "tune model training" section of
[Getting Started with KerasTuner](https://keras.io/guides/keras_tuner/getting_started/#tune-model-training).
## Tuning the custom training loop
In this guide, we will subclass the `HyperModel` class and write a custom
training loop by overriding `HyperModel.fit()`. For how to write a custom
training loop with Keras, you can refer to the guide
[Writing a training loop from scratch](https://keras.io/guides/writing_a_training_loop_from_scratch/).
First, we import the libraries we need, and we create datasets for training and
validation. Here, we just use some random data for demonstration purposes.
```
import keras_tuner
import tensorflow as tf
from tensorflow import keras
import numpy as np
x_train = np.random.rand(1000, 28, 28, 1)
y_train = np.random.randint(0, 10, (1000, 1))
x_val = np.random.rand(1000, 28, 28, 1)
y_val = np.random.randint(0, 10, (1000, 1))
```
Then, we subclass the `HyperModel` class as `MyHyperModel`. In
`MyHyperModel.build()`, we build a simple Keras model to do image
classification for 10 different classes. `MyHyperModel.fit()` accepts several
arguments. Its signature is shown below:
```python
def fit(self, hp, model, x, y, validation_data, callbacks=None, **kwargs):
```
* The `hp` argument is for defining the hyperparameters.
* The `model` argument is the model returned by `MyHyperModel.build()`.
* `x`, `y`, and `validation_data` are all custom-defined arguments. We will
pass our data to them by calling `tuner.search(x=x, y=y,
validation_data=(x_val, y_val))` later. You can define any number of them and
give custom names.
* The `callbacks` argument was intended to be used with `model.fit()`.
KerasTuner put some helpful Keras callbacks in it, for example, the callback
for checkpointing the model at its best epoch.
We will manually call the callbacks in the custom training loop. Before we
can call them, we need to assign our model to them with the following code so
that they have access to the model for checkpointing.
```py
for callback in callbacks:
callback.model = model
```
In this example, we only called the `on_epoch_end()` method of the callbacks
to help us checkpoint the model. You may also call other callback methods
if needed. If you don't need to save the model, you don't need to use the
callbacks.
In the custom training loop, we tune the batch size of the dataset as we wrap
the NumPy data into a `tf.data.Dataset`. Note that you can tune any
preprocessing steps here as well. We also tune the learning rate of the
optimizer.
We will use the validation loss as the evaluation metric for the model. To
compute the mean validation loss, we will use `keras.metrics.Mean()`, which
averages the validation loss across the batches. We need to return the
validation loss for the tuner to make a record.
```
class MyHyperModel(keras_tuner.HyperModel):
def build(self, hp):
"""Builds a convolutional model."""
inputs = keras.Input(shape=(28, 28, 1))
x = keras.layers.Flatten()(inputs)
x = keras.layers.Dense(
units=hp.Choice("units", [32, 64, 128]), activation="relu"
)(x)
outputs = keras.layers.Dense(10)(x)
return keras.Model(inputs=inputs, outputs=outputs)
def fit(self, hp, model, x, y, validation_data, callbacks=None, **kwargs):
# Convert the datasets to tf.data.Dataset.
batch_size = hp.Int("batch_size", 32, 128, step=32, default=64)
train_ds = tf.data.Dataset.from_tensor_slices((x_train, y_train)).batch(
batch_size
)
validation_data = tf.data.Dataset.from_tensor_slices(validation_data).batch(
batch_size
)
# Define the optimizer.
optimizer = keras.optimizers.Adam(
hp.Float("learning_rate", 1e-4, 1e-2, sampling="log", default=1e-3)
)
loss_fn = keras.losses.SparseCategoricalCrossentropy(from_logits=True)
# The metric to track validation loss.
epoch_loss_metric = keras.metrics.Mean()
# Function to run the train step.
@tf.function
def run_train_step(images, labels):
with tf.GradientTape() as tape:
logits = model(images)
loss = loss_fn(labels, logits)
# Add any regularization losses.
if model.losses:
loss += tf.math.add_n(model.losses)
gradients = tape.gradient(loss, model.trainable_variables)
optimizer.apply_gradients(zip(gradients, model.trainable_variables))
# Function to run the validation step.
@tf.function
def run_val_step(images, labels):
logits = model(images)
loss = loss_fn(labels, logits)
# Update the metric.
epoch_loss_metric.update_state(loss)
# Assign the model to the callbacks.
for callback in callbacks:
callback.model = model
# Record the best validation loss value
best_epoch_loss = float("inf")
# The custom training loop.
for epoch in range(2):
print(f"Epoch: {epoch}")
# Iterate the training data to run the training step.
for images, labels in train_ds:
run_train_step(images, labels)
# Iterate the validation data to run the validation step.
for images, labels in validation_data:
run_val_step(images, labels)
# Calling the callbacks after epoch.
epoch_loss = float(epoch_loss_metric.result().numpy())
for callback in callbacks:
# The "my_metric" is the objective passed to the tuner.
callback.on_epoch_end(epoch, logs={"my_metric": epoch_loss})
epoch_loss_metric.reset_states()
print(f"Epoch loss: {epoch_loss}")
best_epoch_loss = min(best_epoch_loss, epoch_loss)
# Return the evaluation metric value.
return best_epoch_loss
```
Now, we can initialize the tuner. Here, we use `Objective("my_metric", "min")`
as our metric to be minimized. The objective name should be consistent with the
one you use as the key in the `logs` passed to the 'on_epoch_end()' method of
the callbacks. The callbacks need to use this value in the `logs` to find the
best epoch to checkpoint the model.
```
tuner = keras_tuner.RandomSearch(
objective=keras_tuner.Objective("my_metric", "min"),
max_trials=2,
hypermodel=MyHyperModel(),
directory="results",
project_name="custom_training",
overwrite=True,
)
```
We start the search by passing the arguments we defined in the signature of
`MyHyperModel.fit()` to `tuner.search()`.
```
tuner.search(x=x_train, y=y_train, validation_data=(x_val, y_val))
```
Finally, we can retrieve the results.
```
best_hps = tuner.get_best_hyperparameters()[0]
print(best_hps.values)
best_model = tuner.get_best_models()[0]
best_model.summary()
```
In summary, to tune the hyperparameters in your custom training loop, you just
override `HyperModel.fit()` to train the model and return the evaluation
results. With the provided callbacks, you can easily save the trained models at
their best epochs and load the best models later.
To find out more about the basics of KerasTuner, please see
[Getting Started with KerasTuner](https://keras.io/guides/keras_tuner/getting_started/).
|
github_jupyter
|
!pip install keras-tuner -q
import keras_tuner
import tensorflow as tf
from tensorflow import keras
import numpy as np
x_train = np.random.rand(1000, 28, 28, 1)
y_train = np.random.randint(0, 10, (1000, 1))
x_val = np.random.rand(1000, 28, 28, 1)
y_val = np.random.randint(0, 10, (1000, 1))
def fit(self, hp, model, x, y, validation_data, callbacks=None, **kwargs):
for callback in callbacks:
callback.model = model
class MyHyperModel(keras_tuner.HyperModel):
def build(self, hp):
"""Builds a convolutional model."""
inputs = keras.Input(shape=(28, 28, 1))
x = keras.layers.Flatten()(inputs)
x = keras.layers.Dense(
units=hp.Choice("units", [32, 64, 128]), activation="relu"
)(x)
outputs = keras.layers.Dense(10)(x)
return keras.Model(inputs=inputs, outputs=outputs)
def fit(self, hp, model, x, y, validation_data, callbacks=None, **kwargs):
# Convert the datasets to tf.data.Dataset.
batch_size = hp.Int("batch_size", 32, 128, step=32, default=64)
train_ds = tf.data.Dataset.from_tensor_slices((x_train, y_train)).batch(
batch_size
)
validation_data = tf.data.Dataset.from_tensor_slices(validation_data).batch(
batch_size
)
# Define the optimizer.
optimizer = keras.optimizers.Adam(
hp.Float("learning_rate", 1e-4, 1e-2, sampling="log", default=1e-3)
)
loss_fn = keras.losses.SparseCategoricalCrossentropy(from_logits=True)
# The metric to track validation loss.
epoch_loss_metric = keras.metrics.Mean()
# Function to run the train step.
@tf.function
def run_train_step(images, labels):
with tf.GradientTape() as tape:
logits = model(images)
loss = loss_fn(labels, logits)
# Add any regularization losses.
if model.losses:
loss += tf.math.add_n(model.losses)
gradients = tape.gradient(loss, model.trainable_variables)
optimizer.apply_gradients(zip(gradients, model.trainable_variables))
# Function to run the validation step.
@tf.function
def run_val_step(images, labels):
logits = model(images)
loss = loss_fn(labels, logits)
# Update the metric.
epoch_loss_metric.update_state(loss)
# Assign the model to the callbacks.
for callback in callbacks:
callback.model = model
# Record the best validation loss value
best_epoch_loss = float("inf")
# The custom training loop.
for epoch in range(2):
print(f"Epoch: {epoch}")
# Iterate the training data to run the training step.
for images, labels in train_ds:
run_train_step(images, labels)
# Iterate the validation data to run the validation step.
for images, labels in validation_data:
run_val_step(images, labels)
# Calling the callbacks after epoch.
epoch_loss = float(epoch_loss_metric.result().numpy())
for callback in callbacks:
# The "my_metric" is the objective passed to the tuner.
callback.on_epoch_end(epoch, logs={"my_metric": epoch_loss})
epoch_loss_metric.reset_states()
print(f"Epoch loss: {epoch_loss}")
best_epoch_loss = min(best_epoch_loss, epoch_loss)
# Return the evaluation metric value.
return best_epoch_loss
tuner = keras_tuner.RandomSearch(
objective=keras_tuner.Objective("my_metric", "min"),
max_trials=2,
hypermodel=MyHyperModel(),
directory="results",
project_name="custom_training",
overwrite=True,
)
tuner.search(x=x_train, y=y_train, validation_data=(x_val, y_val))
best_hps = tuner.get_best_hyperparameters()[0]
print(best_hps.values)
best_model = tuner.get_best_models()[0]
best_model.summary()
| 0.928384 | 0.961929 |
This notebook shows the how tallies can be combined (added, subtracted, multiplied, etc.) using the Python API in order to create derived tallies. Since no covariance information is obtained, it is assumed that tallies are completely independent of one another when propagating uncertainties. The target problem is a simple pin cell.
```
import glob
from IPython.display import Image
import numpy as np
import openmc
```
## Generate Input Files
First we need to define materials that will be used in the problem. We'll create three materials for the fuel, water, and cladding of the fuel pin.
```
# 1.6 enriched fuel
fuel = openmc.Material(name='1.6% Fuel')
fuel.set_density('g/cm3', 10.31341)
fuel.add_nuclide('U235', 3.7503e-4)
fuel.add_nuclide('U238', 2.2625e-2)
fuel.add_nuclide('O16', 4.6007e-2)
# borated water
water = openmc.Material(name='Borated Water')
water.set_density('g/cm3', 0.740582)
water.add_nuclide('H1', 4.9457e-2)
water.add_nuclide('O16', 2.4732e-2)
water.add_nuclide('B10', 8.0042e-6)
# zircaloy
zircaloy = openmc.Material(name='Zircaloy')
zircaloy.set_density('g/cm3', 6.55)
zircaloy.add_nuclide('Zr90', 7.2758e-3)
```
With our three materials, we can now create a materials file object that can be exported to an actual XML file.
```
# Instantiate a Materials collection
materials_file = openmc.Materials([fuel, water, zircaloy])
# Export to "materials.xml"
materials_file.export_to_xml()
```
Now let's move on to the geometry. Our problem will have three regions for the fuel, the clad, and the surrounding coolant. The first step is to create the bounding surfaces -- in this case two cylinders and six planes.
```
# Create cylinders for the fuel and clad
fuel_outer_radius = openmc.ZCylinder(x0=0.0, y0=0.0, R=0.39218)
clad_outer_radius = openmc.ZCylinder(x0=0.0, y0=0.0, R=0.45720)
# Create boundary planes to surround the geometry
# Use both reflective and vacuum boundaries to make life interesting
min_x = openmc.XPlane(x0=-0.63, boundary_type='reflective')
max_x = openmc.XPlane(x0=+0.63, boundary_type='reflective')
min_y = openmc.YPlane(y0=-0.63, boundary_type='reflective')
max_y = openmc.YPlane(y0=+0.63, boundary_type='reflective')
min_z = openmc.ZPlane(z0=-100., boundary_type='vacuum')
max_z = openmc.ZPlane(z0=+100., boundary_type='vacuum')
```
With the surfaces defined, we can now create cells that are defined by intersections of half-spaces created by the surfaces.
```
# Create a Universe to encapsulate a fuel pin
pin_cell_universe = openmc.Universe(name='1.6% Fuel Pin')
# Create fuel Cell
fuel_cell = openmc.Cell(name='1.6% Fuel')
fuel_cell.fill = fuel
fuel_cell.region = -fuel_outer_radius
pin_cell_universe.add_cell(fuel_cell)
# Create a clad Cell
clad_cell = openmc.Cell(name='1.6% Clad')
clad_cell.fill = zircaloy
clad_cell.region = +fuel_outer_radius & -clad_outer_radius
pin_cell_universe.add_cell(clad_cell)
# Create a moderator Cell
moderator_cell = openmc.Cell(name='1.6% Moderator')
moderator_cell.fill = water
moderator_cell.region = +clad_outer_radius
pin_cell_universe.add_cell(moderator_cell)
```
OpenMC requires that there is a "root" universe. Let us create a root cell that is filled by the pin cell universe and then assign it to the root universe.
```
# Create root Cell
root_cell = openmc.Cell(name='root cell')
root_cell.fill = pin_cell_universe
# Add boundary planes
root_cell.region = +min_x & -max_x & +min_y & -max_y & +min_z & -max_z
# Create root Universe
root_universe = openmc.Universe(universe_id=0, name='root universe')
root_universe.add_cell(root_cell)
```
We now must create a geometry that is assigned a root universe, put the geometry into a geometry file, and export it to XML.
```
# Create Geometry and set root Universe
geometry = openmc.Geometry(root_universe)
# Export to "geometry.xml"
geometry.export_to_xml()
```
With the geometry and materials finished, we now just need to define simulation parameters. In this case, we will use 5 inactive batches and 15 active batches each with 2500 particles.
```
# OpenMC simulation parameters
batches = 20
inactive = 5
particles = 2500
# Instantiate a Settings object
settings_file = openmc.Settings()
settings_file.batches = batches
settings_file.inactive = inactive
settings_file.particles = particles
settings_file.output = {'tallies': True}
# Create an initial uniform spatial source distribution over fissionable zones
bounds = [-0.63, -0.63, -100., 0.63, 0.63, 100.]
uniform_dist = openmc.stats.Box(bounds[:3], bounds[3:], only_fissionable=True)
settings_file.source = openmc.source.Source(space=uniform_dist)
# Export to "settings.xml"
settings_file.export_to_xml()
```
Let us also create a plot file that we can use to verify that our pin cell geometry was created successfully.
```
# Instantiate a Plot
plot = openmc.Plot(plot_id=1)
plot.filename = 'materials-xy'
plot.origin = [0, 0, 0]
plot.width = [1.26, 1.26]
plot.pixels = [250, 250]
plot.color_by = 'material'
# Instantiate a Plots collection and export to "plots.xml"
plot_file = openmc.Plots([plot])
plot_file.export_to_xml()
```
With the plots.xml file, we can now generate and view the plot. OpenMC outputs plots in .ppm format, which can be converted into a compressed format like .png with the convert utility.
```
# Run openmc in plotting mode
openmc.plot_geometry(output=False)
# Convert OpenMC's funky ppm to png
!convert materials-xy.ppm materials-xy.png
# Display the materials plot inline
Image(filename='materials-xy.png')
```
As we can see from the plot, we have a nice pin cell with fuel, cladding, and water! Before we run our simulation, we need to tell the code what we want to tally. The following code shows how to create a variety of tallies.
```
# Instantiate an empty Tallies object
tallies_file = openmc.Tallies()
# Create Tallies to compute microscopic multi-group cross-sections
# Instantiate energy filter for multi-group cross-section Tallies
energy_filter = openmc.EnergyFilter([0., 0.625, 20.0e6])
# Instantiate flux Tally in moderator and fuel
tally = openmc.Tally(name='flux')
tally.filters = [openmc.CellFilter([fuel_cell, moderator_cell])]
tally.filters.append(energy_filter)
tally.scores = ['flux']
tallies_file.append(tally)
# Instantiate reaction rate Tally in fuel
tally = openmc.Tally(name='fuel rxn rates')
tally.filters = [openmc.CellFilter(fuel_cell)]
tally.filters.append(energy_filter)
tally.scores = ['nu-fission', 'scatter']
tally.nuclides = ['U238', 'U235']
tallies_file.append(tally)
# Instantiate reaction rate Tally in moderator
tally = openmc.Tally(name='moderator rxn rates')
tally.filters = [openmc.CellFilter(moderator_cell)]
tally.filters.append(energy_filter)
tally.scores = ['absorption', 'total']
tally.nuclides = ['O16', 'H1']
tallies_file.append(tally)
# Instantiate a tally mesh
mesh = openmc.Mesh(mesh_id=1)
mesh.type = 'regular'
mesh.dimension = [1, 1, 1]
mesh.lower_left = [-0.63, -0.63, -100.]
mesh.width = [1.26, 1.26, 200.]
meshsurface_filter = openmc.MeshSurfaceFilter(mesh)
# Instantiate thermal, fast, and total leakage tallies
leak = openmc.Tally(name='leakage')
leak.filters = [meshsurface_filter]
leak.scores = ['current']
tallies_file.append(leak)
thermal_leak = openmc.Tally(name='thermal leakage')
thermal_leak.filters = [meshsurface_filter, openmc.EnergyFilter([0., 0.625])]
thermal_leak.scores = ['current']
tallies_file.append(thermal_leak)
fast_leak = openmc.Tally(name='fast leakage')
fast_leak.filters = [meshsurface_filter, openmc.EnergyFilter([0.625, 20.0e6])]
fast_leak.scores = ['current']
tallies_file.append(fast_leak)
# K-Eigenvalue (infinity) tallies
fiss_rate = openmc.Tally(name='fiss. rate')
abs_rate = openmc.Tally(name='abs. rate')
fiss_rate.scores = ['nu-fission']
abs_rate.scores = ['absorption']
tallies_file += (fiss_rate, abs_rate)
# Resonance Escape Probability tallies
therm_abs_rate = openmc.Tally(name='therm. abs. rate')
therm_abs_rate.scores = ['absorption']
therm_abs_rate.filters = [openmc.EnergyFilter([0., 0.625])]
tallies_file.append(therm_abs_rate)
# Thermal Flux Utilization tallies
fuel_therm_abs_rate = openmc.Tally(name='fuel therm. abs. rate')
fuel_therm_abs_rate.scores = ['absorption']
fuel_therm_abs_rate.filters = [openmc.EnergyFilter([0., 0.625]),
openmc.CellFilter([fuel_cell])]
tallies_file.append(fuel_therm_abs_rate)
# Fast Fission Factor tallies
therm_fiss_rate = openmc.Tally(name='therm. fiss. rate')
therm_fiss_rate.scores = ['nu-fission']
therm_fiss_rate.filters = [openmc.EnergyFilter([0., 0.625])]
tallies_file.append(therm_fiss_rate)
# Instantiate energy filter to illustrate Tally slicing
fine_energy_filter = openmc.EnergyFilter(np.logspace(np.log10(1e-2), np.log10(20.0e6), 10))
# Instantiate flux Tally in moderator and fuel
tally = openmc.Tally(name='need-to-slice')
tally.filters = [openmc.CellFilter([fuel_cell, moderator_cell])]
tally.filters.append(fine_energy_filter)
tally.scores = ['nu-fission', 'scatter']
tally.nuclides = ['H1', 'U238']
tallies_file.append(tally)
# Export to "tallies.xml"
tallies_file.export_to_xml()
```
Now we a have a complete set of inputs, so we can go ahead and run our simulation.
```
# Run OpenMC!
openmc.run()
```
## Tally Data Processing
Our simulation ran successfully and created a statepoint file with all the tally data in it. We begin our analysis here loading the statepoint file and 'reading' the results. By default, the tally results are not read into memory because they might be large, even large enough to exceed the available memory on a computer.
```
# Load the statepoint file
sp = openmc.StatePoint('statepoint.20.h5')
```
We have a tally of the total fission rate and the total absorption rate, so we can calculate k-eff as:
$$k_{eff} = \frac{\langle \nu \Sigma_f \phi \rangle}{\langle \Sigma_a \phi \rangle + \langle L \rangle}$$
In this notation, $\langle \cdot \rangle^a_b$ represents an OpenMC that is integrated over region $a$ and energy range $b$. If $a$ or $b$ is not reported, it means the value represents an integral over all space or all energy, respectively.
```
# Get the fission and absorption rate tallies
fiss_rate = sp.get_tally(name='fiss. rate')
abs_rate = sp.get_tally(name='abs. rate')
# Get the leakage tally
leak = sp.get_tally(name='leakage')
leak = leak.summation(filter_type=openmc.MeshSurfaceFilter, remove_filter=True)
# Compute k-infinity using tally arithmetic
keff = fiss_rate / (abs_rate + leak)
keff.get_pandas_dataframe()
```
Notice that even though the neutron production rate, absorption rate, and current are separate tallies, we still get a first-order estimate of the uncertainty on the quotient of them automatically!
Often in textbooks you'll see k-eff represented using the six-factor formula $$k_{eff} = p \epsilon f \eta P_{FNL} P_{TNL}.$$ Let's analyze each of these factors, starting with the resonance escape probability which is defined as $$p=\frac{\langle\Sigma_a\phi\rangle_T + \langle L \rangle_T}{\langle\Sigma_a\phi\rangle + \langle L \rangle_T}$$ where the subscript $T$ means thermal energies.
```
# Compute resonance escape probability using tally arithmetic
therm_abs_rate = sp.get_tally(name='therm. abs. rate')
thermal_leak = sp.get_tally(name='thermal leakage')
thermal_leak = thermal_leak.summation(filter_type=openmc.MeshSurfaceFilter, remove_filter=True)
res_esc = (therm_abs_rate + thermal_leak) / (abs_rate + thermal_leak)
res_esc.get_pandas_dataframe()
```
The fast fission factor can be calculated as
$$\epsilon=\frac{\langle\nu\Sigma_f\phi\rangle}{\langle\nu\Sigma_f\phi\rangle_T}$$
```
# Compute fast fission factor factor using tally arithmetic
therm_fiss_rate = sp.get_tally(name='therm. fiss. rate')
fast_fiss = fiss_rate / therm_fiss_rate
fast_fiss.get_pandas_dataframe()
```
The thermal flux utilization is calculated as
$$f=\frac{\langle\Sigma_a\phi\rangle^F_T}{\langle\Sigma_a\phi\rangle_T}$$
where the superscript $F$ denotes fuel.
```
# Compute thermal flux utilization factor using tally arithmetic
fuel_therm_abs_rate = sp.get_tally(name='fuel therm. abs. rate')
therm_util = fuel_therm_abs_rate / therm_abs_rate
therm_util.get_pandas_dataframe()
```
The next factor is the number of fission neutrons produced per absorption in fuel, calculated as $$\eta = \frac{\langle \nu\Sigma_f\phi \rangle_T}{\langle \Sigma_a \phi \rangle^F_T}$$
```
# Compute neutrons produced per absorption (eta) using tally arithmetic
eta = therm_fiss_rate / fuel_therm_abs_rate
eta.get_pandas_dataframe()
```
There are two leakage factors to account for fast and thermal leakage. The fast non-leakage probability is computed as $$P_{FNL} = \frac{\langle \Sigma_a\phi \rangle + \langle L \rangle_T}{\langle \Sigma_a \phi \rangle + \langle L \rangle}$$
```
p_fnl = (abs_rate + thermal_leak) / (abs_rate + leak)
p_fnl.get_pandas_dataframe()
```
The final factor is the thermal non-leakage probability and is computed as $$P_{TNL} = \frac{\langle \Sigma_a\phi \rangle_T}{\langle \Sigma_a \phi \rangle_T + \langle L \rangle_T}$$
```
p_tnl = therm_abs_rate / (therm_abs_rate + thermal_leak)
p_tnl.get_pandas_dataframe()
```
Now we can calculate $k_{eff}$ using the product of the factors form the four-factor formula.
```
keff = res_esc * fast_fiss * therm_util * eta * p_fnl * p_tnl
keff.get_pandas_dataframe()
```
We see that the value we've obtained here has exactly the same mean as before. However, because of the way it was calculated, the standard deviation appears to be larger.
Let's move on to a more complicated example now. Before we set up tallies to get reaction rates in the fuel and moderator in two energy groups for two different nuclides. We can use tally arithmetic to divide each of these reaction rates by the flux to get microscopic multi-group cross sections.
```
# Compute microscopic multi-group cross-sections
flux = sp.get_tally(name='flux')
flux = flux.get_slice(filters=[openmc.CellFilter], filter_bins=[(fuel_cell.id,)])
fuel_rxn_rates = sp.get_tally(name='fuel rxn rates')
mod_rxn_rates = sp.get_tally(name='moderator rxn rates')
fuel_xs = fuel_rxn_rates / flux
fuel_xs.get_pandas_dataframe()
```
We see that when the two tallies with multiple bins were divided, the derived tally contains the outer product of the combinations. If the filters/scores are the same, no outer product is needed. The `get_values(...)` method allows us to obtain a subset of tally scores. In the following example, we obtain just the neutron production microscopic cross sections.
```
# Show how to use Tally.get_values(...) with a CrossScore
nu_fiss_xs = fuel_xs.get_values(scores=['(nu-fission / flux)'])
print(nu_fiss_xs)
```
The same idea can be used not only for scores but also for filters and nuclides.
```
# Show how to use Tally.get_values(...) with a CrossScore and CrossNuclide
u235_scatter_xs = fuel_xs.get_values(nuclides=['(U235 / total)'],
scores=['(scatter / flux)'])
print(u235_scatter_xs)
# Show how to use Tally.get_values(...) with a CrossFilter and CrossScore
fast_scatter_xs = fuel_xs.get_values(filters=[openmc.EnergyFilter],
filter_bins=[((0.625, 20.0e6),)],
scores=['(scatter / flux)'])
print(fast_scatter_xs)
```
A more advanced method is to use `get_slice(...)` to create a new derived tally that is a subset of an existing tally. This has the benefit that we can use `get_pandas_dataframe()` to see the tallies in a more human-readable format.
```
# "Slice" the nu-fission data into a new derived Tally
nu_fission_rates = fuel_rxn_rates.get_slice(scores=['nu-fission'])
nu_fission_rates.get_pandas_dataframe()
# "Slice" the H-1 scatter data in the moderator Cell into a new derived Tally
need_to_slice = sp.get_tally(name='need-to-slice')
slice_test = need_to_slice.get_slice(scores=['scatter'], nuclides=['H1'],
filters=[openmc.CellFilter], filter_bins=[(moderator_cell.id,)])
slice_test.get_pandas_dataframe()
```
|
github_jupyter
|
import glob
from IPython.display import Image
import numpy as np
import openmc
# 1.6 enriched fuel
fuel = openmc.Material(name='1.6% Fuel')
fuel.set_density('g/cm3', 10.31341)
fuel.add_nuclide('U235', 3.7503e-4)
fuel.add_nuclide('U238', 2.2625e-2)
fuel.add_nuclide('O16', 4.6007e-2)
# borated water
water = openmc.Material(name='Borated Water')
water.set_density('g/cm3', 0.740582)
water.add_nuclide('H1', 4.9457e-2)
water.add_nuclide('O16', 2.4732e-2)
water.add_nuclide('B10', 8.0042e-6)
# zircaloy
zircaloy = openmc.Material(name='Zircaloy')
zircaloy.set_density('g/cm3', 6.55)
zircaloy.add_nuclide('Zr90', 7.2758e-3)
# Instantiate a Materials collection
materials_file = openmc.Materials([fuel, water, zircaloy])
# Export to "materials.xml"
materials_file.export_to_xml()
# Create cylinders for the fuel and clad
fuel_outer_radius = openmc.ZCylinder(x0=0.0, y0=0.0, R=0.39218)
clad_outer_radius = openmc.ZCylinder(x0=0.0, y0=0.0, R=0.45720)
# Create boundary planes to surround the geometry
# Use both reflective and vacuum boundaries to make life interesting
min_x = openmc.XPlane(x0=-0.63, boundary_type='reflective')
max_x = openmc.XPlane(x0=+0.63, boundary_type='reflective')
min_y = openmc.YPlane(y0=-0.63, boundary_type='reflective')
max_y = openmc.YPlane(y0=+0.63, boundary_type='reflective')
min_z = openmc.ZPlane(z0=-100., boundary_type='vacuum')
max_z = openmc.ZPlane(z0=+100., boundary_type='vacuum')
# Create a Universe to encapsulate a fuel pin
pin_cell_universe = openmc.Universe(name='1.6% Fuel Pin')
# Create fuel Cell
fuel_cell = openmc.Cell(name='1.6% Fuel')
fuel_cell.fill = fuel
fuel_cell.region = -fuel_outer_radius
pin_cell_universe.add_cell(fuel_cell)
# Create a clad Cell
clad_cell = openmc.Cell(name='1.6% Clad')
clad_cell.fill = zircaloy
clad_cell.region = +fuel_outer_radius & -clad_outer_radius
pin_cell_universe.add_cell(clad_cell)
# Create a moderator Cell
moderator_cell = openmc.Cell(name='1.6% Moderator')
moderator_cell.fill = water
moderator_cell.region = +clad_outer_radius
pin_cell_universe.add_cell(moderator_cell)
# Create root Cell
root_cell = openmc.Cell(name='root cell')
root_cell.fill = pin_cell_universe
# Add boundary planes
root_cell.region = +min_x & -max_x & +min_y & -max_y & +min_z & -max_z
# Create root Universe
root_universe = openmc.Universe(universe_id=0, name='root universe')
root_universe.add_cell(root_cell)
# Create Geometry and set root Universe
geometry = openmc.Geometry(root_universe)
# Export to "geometry.xml"
geometry.export_to_xml()
# OpenMC simulation parameters
batches = 20
inactive = 5
particles = 2500
# Instantiate a Settings object
settings_file = openmc.Settings()
settings_file.batches = batches
settings_file.inactive = inactive
settings_file.particles = particles
settings_file.output = {'tallies': True}
# Create an initial uniform spatial source distribution over fissionable zones
bounds = [-0.63, -0.63, -100., 0.63, 0.63, 100.]
uniform_dist = openmc.stats.Box(bounds[:3], bounds[3:], only_fissionable=True)
settings_file.source = openmc.source.Source(space=uniform_dist)
# Export to "settings.xml"
settings_file.export_to_xml()
# Instantiate a Plot
plot = openmc.Plot(plot_id=1)
plot.filename = 'materials-xy'
plot.origin = [0, 0, 0]
plot.width = [1.26, 1.26]
plot.pixels = [250, 250]
plot.color_by = 'material'
# Instantiate a Plots collection and export to "plots.xml"
plot_file = openmc.Plots([plot])
plot_file.export_to_xml()
# Run openmc in plotting mode
openmc.plot_geometry(output=False)
# Convert OpenMC's funky ppm to png
!convert materials-xy.ppm materials-xy.png
# Display the materials plot inline
Image(filename='materials-xy.png')
# Instantiate an empty Tallies object
tallies_file = openmc.Tallies()
# Create Tallies to compute microscopic multi-group cross-sections
# Instantiate energy filter for multi-group cross-section Tallies
energy_filter = openmc.EnergyFilter([0., 0.625, 20.0e6])
# Instantiate flux Tally in moderator and fuel
tally = openmc.Tally(name='flux')
tally.filters = [openmc.CellFilter([fuel_cell, moderator_cell])]
tally.filters.append(energy_filter)
tally.scores = ['flux']
tallies_file.append(tally)
# Instantiate reaction rate Tally in fuel
tally = openmc.Tally(name='fuel rxn rates')
tally.filters = [openmc.CellFilter(fuel_cell)]
tally.filters.append(energy_filter)
tally.scores = ['nu-fission', 'scatter']
tally.nuclides = ['U238', 'U235']
tallies_file.append(tally)
# Instantiate reaction rate Tally in moderator
tally = openmc.Tally(name='moderator rxn rates')
tally.filters = [openmc.CellFilter(moderator_cell)]
tally.filters.append(energy_filter)
tally.scores = ['absorption', 'total']
tally.nuclides = ['O16', 'H1']
tallies_file.append(tally)
# Instantiate a tally mesh
mesh = openmc.Mesh(mesh_id=1)
mesh.type = 'regular'
mesh.dimension = [1, 1, 1]
mesh.lower_left = [-0.63, -0.63, -100.]
mesh.width = [1.26, 1.26, 200.]
meshsurface_filter = openmc.MeshSurfaceFilter(mesh)
# Instantiate thermal, fast, and total leakage tallies
leak = openmc.Tally(name='leakage')
leak.filters = [meshsurface_filter]
leak.scores = ['current']
tallies_file.append(leak)
thermal_leak = openmc.Tally(name='thermal leakage')
thermal_leak.filters = [meshsurface_filter, openmc.EnergyFilter([0., 0.625])]
thermal_leak.scores = ['current']
tallies_file.append(thermal_leak)
fast_leak = openmc.Tally(name='fast leakage')
fast_leak.filters = [meshsurface_filter, openmc.EnergyFilter([0.625, 20.0e6])]
fast_leak.scores = ['current']
tallies_file.append(fast_leak)
# K-Eigenvalue (infinity) tallies
fiss_rate = openmc.Tally(name='fiss. rate')
abs_rate = openmc.Tally(name='abs. rate')
fiss_rate.scores = ['nu-fission']
abs_rate.scores = ['absorption']
tallies_file += (fiss_rate, abs_rate)
# Resonance Escape Probability tallies
therm_abs_rate = openmc.Tally(name='therm. abs. rate')
therm_abs_rate.scores = ['absorption']
therm_abs_rate.filters = [openmc.EnergyFilter([0., 0.625])]
tallies_file.append(therm_abs_rate)
# Thermal Flux Utilization tallies
fuel_therm_abs_rate = openmc.Tally(name='fuel therm. abs. rate')
fuel_therm_abs_rate.scores = ['absorption']
fuel_therm_abs_rate.filters = [openmc.EnergyFilter([0., 0.625]),
openmc.CellFilter([fuel_cell])]
tallies_file.append(fuel_therm_abs_rate)
# Fast Fission Factor tallies
therm_fiss_rate = openmc.Tally(name='therm. fiss. rate')
therm_fiss_rate.scores = ['nu-fission']
therm_fiss_rate.filters = [openmc.EnergyFilter([0., 0.625])]
tallies_file.append(therm_fiss_rate)
# Instantiate energy filter to illustrate Tally slicing
fine_energy_filter = openmc.EnergyFilter(np.logspace(np.log10(1e-2), np.log10(20.0e6), 10))
# Instantiate flux Tally in moderator and fuel
tally = openmc.Tally(name='need-to-slice')
tally.filters = [openmc.CellFilter([fuel_cell, moderator_cell])]
tally.filters.append(fine_energy_filter)
tally.scores = ['nu-fission', 'scatter']
tally.nuclides = ['H1', 'U238']
tallies_file.append(tally)
# Export to "tallies.xml"
tallies_file.export_to_xml()
# Run OpenMC!
openmc.run()
# Load the statepoint file
sp = openmc.StatePoint('statepoint.20.h5')
# Get the fission and absorption rate tallies
fiss_rate = sp.get_tally(name='fiss. rate')
abs_rate = sp.get_tally(name='abs. rate')
# Get the leakage tally
leak = sp.get_tally(name='leakage')
leak = leak.summation(filter_type=openmc.MeshSurfaceFilter, remove_filter=True)
# Compute k-infinity using tally arithmetic
keff = fiss_rate / (abs_rate + leak)
keff.get_pandas_dataframe()
# Compute resonance escape probability using tally arithmetic
therm_abs_rate = sp.get_tally(name='therm. abs. rate')
thermal_leak = sp.get_tally(name='thermal leakage')
thermal_leak = thermal_leak.summation(filter_type=openmc.MeshSurfaceFilter, remove_filter=True)
res_esc = (therm_abs_rate + thermal_leak) / (abs_rate + thermal_leak)
res_esc.get_pandas_dataframe()
# Compute fast fission factor factor using tally arithmetic
therm_fiss_rate = sp.get_tally(name='therm. fiss. rate')
fast_fiss = fiss_rate / therm_fiss_rate
fast_fiss.get_pandas_dataframe()
# Compute thermal flux utilization factor using tally arithmetic
fuel_therm_abs_rate = sp.get_tally(name='fuel therm. abs. rate')
therm_util = fuel_therm_abs_rate / therm_abs_rate
therm_util.get_pandas_dataframe()
# Compute neutrons produced per absorption (eta) using tally arithmetic
eta = therm_fiss_rate / fuel_therm_abs_rate
eta.get_pandas_dataframe()
p_fnl = (abs_rate + thermal_leak) / (abs_rate + leak)
p_fnl.get_pandas_dataframe()
p_tnl = therm_abs_rate / (therm_abs_rate + thermal_leak)
p_tnl.get_pandas_dataframe()
keff = res_esc * fast_fiss * therm_util * eta * p_fnl * p_tnl
keff.get_pandas_dataframe()
# Compute microscopic multi-group cross-sections
flux = sp.get_tally(name='flux')
flux = flux.get_slice(filters=[openmc.CellFilter], filter_bins=[(fuel_cell.id,)])
fuel_rxn_rates = sp.get_tally(name='fuel rxn rates')
mod_rxn_rates = sp.get_tally(name='moderator rxn rates')
fuel_xs = fuel_rxn_rates / flux
fuel_xs.get_pandas_dataframe()
# Show how to use Tally.get_values(...) with a CrossScore
nu_fiss_xs = fuel_xs.get_values(scores=['(nu-fission / flux)'])
print(nu_fiss_xs)
# Show how to use Tally.get_values(...) with a CrossScore and CrossNuclide
u235_scatter_xs = fuel_xs.get_values(nuclides=['(U235 / total)'],
scores=['(scatter / flux)'])
print(u235_scatter_xs)
# Show how to use Tally.get_values(...) with a CrossFilter and CrossScore
fast_scatter_xs = fuel_xs.get_values(filters=[openmc.EnergyFilter],
filter_bins=[((0.625, 20.0e6),)],
scores=['(scatter / flux)'])
print(fast_scatter_xs)
# "Slice" the nu-fission data into a new derived Tally
nu_fission_rates = fuel_rxn_rates.get_slice(scores=['nu-fission'])
nu_fission_rates.get_pandas_dataframe()
# "Slice" the H-1 scatter data in the moderator Cell into a new derived Tally
need_to_slice = sp.get_tally(name='need-to-slice')
slice_test = need_to_slice.get_slice(scores=['scatter'], nuclides=['H1'],
filters=[openmc.CellFilter], filter_bins=[(moderator_cell.id,)])
slice_test.get_pandas_dataframe()
| 0.55447 | 0.95594 |
# "COVID-19 vaccine tweet sentiment analysis with fastai - part 1"
> "This is part one of a two-part NLP series where we carry out sentiment analysis on COVID-19 vaccine tweets. In this part we follow the ULMFiT approach with fastai to create a Twitter language model, then use this to fine-tune a tweet sentiment classification model."
- toc: true
- branch: master
- badges: true
- comments: true
- author: Tom Whelan
- categories: [fastai, NLP, sentiment analysis, PyTorch]
- image: images/twitter-sentiment-analysis.jpg
# Introduction
In this post we will create a model to perform sentiment analysis on tweets about COVID-19 vaccines using the [`fastai`](https://docs.fast.ai/) library. I will provide a brief overview of the process here, but a much more in-depth explanation of NLP with [`fastai`](https://docs.fast.ai/) can be found in [lesson 8](https://course.fast.ai/videos/?lesson=8) of the [`fastai`](https://docs.fast.ai/) course. In [part 2](https://thomaswhelan.com/fastai/nlp/sentiment%20analysis/pytorch/visualisation/2021/03/17/covid-19-vaccine-tweet-sentiment-analysis-with-fastai-part-2.html) we will use the model for analysis, looking at changes in tweet sentiment over time and how that relates to the progress of vaccination in different countries.
# Transfer learning in NLP - the ULMFiT approach
We will be making use of *transfer learning* to help us create a model to analyse tweet sentiment. The idea behind transfer learning is that neural networks learn information that generalises to new problems, [particularly the early layers of the network](https://arxiv.org/pdf/1311.2901.pdf). In computer vision, for example, we can take a model that was trained on the ImageNet dataset to recognise different features of images such as circles, then apply that to a smaller dataset and *fine-tune* the model to be more suited to a specific task (e.g. classifying images as cats or dogs). This technique allows us to train neural networks much faster and with far less data than we would otherwise need.
In 2018 [a paper](https://arxiv.org/abs/1801.06146) introduced a transfer learning technique for NLP called 'Universal Language Model Fine-Tuning' (ULMFiT). The approach is as follows:
1. Train a *language model* to predict the next word in a sentence. This step is already done for us; with [`fastai`](https://docs.fast.ai/) we can download a model that has been pre-trained for this task on millions of Wikipedia articles. A good language model already knows a lot about how language works in general - for instance, given the sentence 'Tokyo is the capital of', the model might predict 'Japan' as the next word. In this case the model understands that Tokyo is closely related to Japan and that 'capital' refers to 'city' here instead of 'upper-case' or 'money'.
2. Fine-tune the language model to a more specific task. The pre-trained language model is good at understanding Wikipedia English, but Twitter English is a bit different. We can take the information the Wikipedia model has learned and apply that to a Twitter dataset to get a Twitter language model that is good at predicting the next word in a tweet.
3. Fine-tune a *classification model* to identify sentiment using the pre-trained language model. The idea here is that since our language model already knows a lot about Twitter English, it's not a huge leap from there to train a classifier that understands that 'love' refers to positive sentiment and 'hate' refers to negative sentiment. If we tried to train a classifier without using a pre-trained model it would have to learn the whole language from scratch first, which would be very difficult and time consuming.

This notebook will walk through steps 2 and 3 with [`fastai`](https://docs.fast.ai/). We will then apply the model to unlabelled COVID-19 vaccine tweets and save the results for analysis in [part 2](https://thomaswhelan.com/fastai/nlp/sentiment%20analysis/pytorch/visualisation/2021/03/17/covid-19-vaccine-tweet-sentiment-analysis-with-fastai-part-2.html).
> Important: You will need a GPU to train models with [`fastai`](https://course.fast.ai/start_colab), but fortunately for us Google Colab provides us with access to one for free! To use it, select 'Runtime' from the menu at the top of the notebook, then 'Change runtime type', and ensure your hardware accelerator is set to 'GPU' before continuing!
# Data preparation
This is a write-up of a submission I made for several [Kaggle tasks](https://www.kaggle.com/gpreda/all-covid19-vaccines-tweets/tasks). The tasks are still open and accepting new entries at the time of writing if you want to enter as well! On Kaggle the data is already readily available when using their notebook servers; however, we are using Google Colab today, so we will need to access the [Kaggle API](https://www.kaggle.com/docs/api) to download the data.
> Note: Kaggle also have free GPU credits if you prefer to work on their notebook servers instead.
## Getting the data from Kaggle
The first step is to create an API token. To do this, the steps are as follows:
1. Go to 'Account' on Kaggle and scroll down to the 'API' section.
2. Expire all current API tokens by clicking 'Expire API Token'.
3. Click 'Create New API Token', which will automatically download a file called `kaggle.json`.
4. Upload the `kaggle.json` file using the file uploader widget below.
```
#collapse-output
# See https://neptune.ai/blog/google-colab-dealing-with-files for more tips on working with files in Colab
from google.colab import files
uploaded = files.upload()
```
Next, we need to install the Kaggle API.
> Note: The API is already preinstalled in Google Colab, but sometimes it's an outdated version, so it's best to upgrade it in case.
```
#collapse-output
!pip uninstall -q -y kaggle
!pip install -q --upgrade pip
!pip install -q --upgrade kaggle
```
The [API docs](https://github.com/Kaggle/kaggle-api) tell us that we need to ensure `kaggle.json` is in the location `~/.kaggle/kaggle.json`, so let's make the directory and move the file.
```
#collapse-output
# https://www.machinelearningmindset.com/kaggle-dataset-in-google-colab/
!mkdir -p ~/.kaggle
!cp kaggle.json ~/.kaggle/
# Check the file in its new directory
!ls /root/.kaggle/
# Check the file permission
!ls -l ~/.kaggle/kaggle.json
#Change the file permission
# chmod 600 file – owner can read and write
# chmod 700 file – owner can read, write and execute
!chmod 600 ~/.kaggle/kaggle.json
```
Now we can download the data using `!kaggle dataset download -d username-of-dataset-creator/name-of-dataset`.
> Note: There is also an API download command on the dataset page that you can copy/paste instead.
```
#collapse-output
# We will be using two datasets for this part, as well as a third dataset for part 2
# To save time in part 2 I'm going to download them all now and save locally
!kaggle datasets download -d gpreda/all-covid19-vaccines-tweets
!kaggle datasets download -d maxjon/complete-tweet-sentiment-extraction-data
!kaggle datasets download -d gpreda/covid-world-vaccination-progress
```
The files will be downloaded in `.zip` format, so let's unzip them.
```
#collapse-output
# To unzip you can use the following:
#!mkdir folder_name
#!unzip anyfile.zip -d folder_name
# Or unzip all
!unzip -q \*.zip
```
## Loading and cleaning the data
As with `kaggle`, an older version of [`fastai`](https://docs.fast.ai/) is preinstalled in Colab, so we will need to upgrade it first.
> Important: Make a note of the [`fastai`](https://docs.fast.ai/) version you are using, since any models you create and save will need to be run using the same version later.
```
#collapse-output
! [ -e /content ] && pip install -Uqq fastai # upgrade fastai on colab
import fastai; fastai.__version__
```
Let's import [`fastai`](https://docs.fast.ai/)'s [`text`](https://docs.fast.ai/tutorial.text.html) module and take a look at our data.
> Tip: If you use `import *`, useful libraries like `pandas` and `numpy` will also be imported at the same time!
```
from fastai.text.all import *
vax_tweets = pd.read_csv('vaccination_all_tweets.csv')
vax_tweets[['date', 'text', 'hashtags', 'user_followers']].head()
```
We could use the `text` column of this dataset to train a Twitter language model, but since our end goal is sentiment analysis we will need to find another dataset that also contains sentiment labels to train our classifier. Let's use ['Complete Tweet Sentiment Extraction Data'](https://www.kaggle.com/maxjon/complete-tweet-sentiment-extraction-data), which contains 40,000 tweets labelled as either negative, neutral or positive sentiment. For more accurate results you could use the ['sentiment140'](https://www.kaggle.com/kazanova/sentiment140) dataset instead, which contains 1.6m tweets labelled as either positive or negative.
```
tweets = pd.read_csv('tweet_dataset.csv')
tweets[['old_text', 'new_sentiment']].head()
```
For our language model, the only input we need is the tweet text. As we will see in a moment [`fastai`](https://docs.fast.ai/) can handle text preprocessing and tokenization for us, but it might be a good idea to remove things like twitter handles, urls, hashtags and emojis first. You could experiment with leaving these in for your own models and see how it affects the results. There are also some rows with blank tweets which need to be removed.
We ideally want the language model to learn not just about tweet language, but more specifically about vaccine tweet language. We can therefore use text from both datasets as input for the language model. For the classification model we need to remove all rows with missing sentiment, however.
```
# Code via https://www.kaggle.com/garyongguanjie/comments-analysis
def de_emojify(inputString):
return inputString.encode('ascii', 'ignore').decode('ascii')
# Code via https://www.kaggle.com/pawanbhandarkar/generate-smarter-word-clouds-with-log-likelihood
def tweet_proc(df, text_col='text'):
df['orig_text'] = df[text_col]
# Remove twitter handles
df[text_col] = df[text_col].apply(lambda x:re.sub('@[^\s]+','',x))
# Remove URLs
df[text_col] = df[text_col].apply(lambda x:re.sub(r"http\S+", "", x))
# Remove emojis
df[text_col] = df[text_col].apply(de_emojify)
# Remove hashtags
df[text_col] = df[text_col].apply(lambda x:re.sub(r'\B#\S+','',x))
return df[df[text_col]!='']
# Clean the text data and combine the dfs
tweets = tweets[['old_text', 'new_sentiment']].rename(columns={'old_text':'text', 'new_sentiment':'sentiment'})
vax_tweets['sentiment'] = np.nan
tweets = tweet_proc(tweets)
vax_tweets = tweet_proc(vax_tweets)
df_lm = tweets[['text', 'sentiment']].append(vax_tweets[['text', 'sentiment']])
df_clas = df_lm.dropna(subset=['sentiment'])
print(len(df_lm), len(df_clas))
df_clas.head()
```
# Training a language model
To train our language model we can use self-supervised learning; we just need to give the model some text as an independent variable and [`fastai`](https://docs.fast.ai/) will automatically preprocess it and create a dependent variable for us. We can do this in one line of code using the [`DataLoaders`](https://docs.fast.ai/data.core.html#DataLoaders) class, which converts our input data into a [`DataLoader`](https://docs.fast.ai/data.load.html#DataLoader) object that can be used as an input to a [`fastai`](https://docs.fast.ai/) [`Learner`](https://docs.fast.ai/learner.html#Learner).
```
#collapse-output
dls_lm = TextDataLoaders.from_df(df_lm, text_col='text', is_lm=True, valid_pct=0.1)
```
Here we told [`fastai`](https://docs.fast.ai/) that we are working with text data, which is contained in the `text` column of a [`pandas`](https://pandas.pydata.org/docs/) [`DataFrame`](https://pandas.pydata.org/docs/reference/api/pandas.DataFrame.html) called `df_lm`. We set [`is_lm=True`](https://docs.fast.ai/text.data.html#TextDataLoaders) since we want to train a language model, so [`fastai`](https://docs.fast.ai/) needs to label the input data for us. Finally, we told [`fastai`](https://docs.fast.ai/) to hold out a random 10% of our data for a validation set using [`valid_pct=0.1`](https://docs.fast.ai/text.data.html#TextDataLoaders).
Let's take a look at the first two rows of the [`DataLoader`](https://docs.fast.ai/data.load.html#DataLoader) using [`show_batch`](https://docs.fast.ai/data.core.html#TfmdDL.show_batch).
```
dls_lm.show_batch(max_n=2)
```
We have a new column, `text_`, which is `text` offset by one. This is the dependent variable [`fastai`](https://docs.fast.ai/) created for us. By default [`fastai`](https://docs.fast.ai/) uses *word tokenization*, which splits the text on spaces and punctuation marks and breaks up words like *can't* into two separate tokens. [`fastai`](https://docs.fast.ai/) also has some special tokens starting with 'xx' that are designed to make things easier for the model; for example [`xxmaj`](https://docs.fast.ai/text.data.html) indicates that the next word begins with a capital letter and [`xxunk`](https://docs.fast.ai/text.data.html) represents an unknown word that doesn't appear in the vocabulary very often. You could experiment with *subword tokenization* instead, which will split the text on commonly occuring groups of letters instead of spaces. This might help if you wanted to leave hashtags in since they often contain multiple words joined together with no spaces, e.g. #CovidVaccine. The [`fastai`](https://docs.fast.ai/) tokenization process is explained in much more detail [here](https://youtu.be/WjnwWeGjZcM?t=626) for those interested.
## Fine-tuning the language model
The next step is to create a language model using [`language_model_learner`](https://docs.fast.ai/text.learner.html#language_model_learner).
```
learn = language_model_learner(dls_lm, AWD_LSTM, drop_mult=0.3,
metrics=[accuracy, Perplexity()]).to_fp16()
```
Here we passed [`language_model_learner`](https://docs.fast.ai/text.learner.html#language_model_learner) our [`DataLoaders`](https://docs.fast.ai/data.core.html#DataLoaders), `dls_lm`, and the pre-trained [RNN](https://www.simplilearn.com/tutorials/deep-learning-tutorial/rnn) model, [*AWD_LSTM*](https://docs.fast.ai/text.models.awdlstm.html), which is built into [`fastai`](https://docs.fast.ai/). [`drop_mult`](https://docs.fast.ai/text.learner.html#text_classifier_learner) is a multiplier applied to all [dropouts](https://machinelearningmastery.com/dropout-for-regularizing-deep-neural-networks/) in the AWD_LSTM model to reduce overfitting. For example, by default [`fastai`](https://docs.fast.ai/)'s AWD_LSTM applies [`EmbeddingDropout`](https://docs.fast.ai/text.models.awdlstm.html#EmbeddingDropout) with 10% probability (at the time of writing), but we told [`fastai`](https://docs.fast.ai/) that we want to reduce that to 3%. The [`metrics`](https://docs.fast.ai/metrics.html) we want to track are *perplexity*, which is the exponential of the loss (in this case cross entropy loss), and *accuracy*, which tells us how often our model predicts the next word correctly. We can also train with fp16 to use less memory and speed up the training process.
We can find a good learning rate for training using [`lr_find`](https://docs.fast.ai/callback.schedule.html#Learner.lr_find) and use that to fit our model.
```
learn.lr_find()
```
When we created our [`Learner`](https://docs.fast.ai/learner.html#Learner) the embeddings from the pre-trained AWD_LSTM model were merged with random embeddings added for words that weren't in the vocabulary. The pre-trained layers were also automatically frozen for us. Using [`fit_one_cycle`](https://docs.fast.ai/callback.schedule.html#Learner.fit_one_cycle) with our [`Learner`](https://docs.fast.ai/learner.html#Learner) will train only the *new random embeddings* (i.e. words that are in our Twitter vocab but not the Wikipedia vocab) in the last layer of the neural network.
```
learn.fit_one_cycle(1, 3e-2)
```
After one epoch our language model is predicting the next word in a tweet around 25% of the time - not too bad! We can [`unfreeze`](https://docs.fast.ai/learner.html#Learner.unfreeze) the entire model, find a more suitable learning rate and train for a few more epochs to improve the accuracy further.
```
learn.unfreeze()
learn.lr_find()
learn.fit_one_cycle(4, 1e-3)
```
After a bit more training we can predict the next word in a tweet around 29% of the time. Let's test the model out by using it to write some random tweets (in this case it will generate some text following 'I love').
```
# Text generation using the language model
TEXT = "I love"
N_WORDS = 30
N_SENTENCES = 2
print("\n".join(learn.predict(TEXT, N_WORDS, temperature=0.75) for _ in range(N_SENTENCES)))
```
Some interesting results there! Let's save the model *encoder* so we can use it to fine-tune our classifier. The encoder is all of the model except for the final layer, which converts activations to probabilities of picking each token in the vocabulary. We want to keep the knowledge the model has learned about tweet language but we won't be using our classifier to predict the next word in a sentence, so we won't need the final layer any more.
```
learn.save_encoder('finetuned_lm')
```
# Training a sentiment classifier
To get the [`DataLoaders`](https://docs.fast.ai/data.core.html#DataLoaders) for our classifier let's use the [`DataBlock`](https://docs.fast.ai/tutorial.datablock.html#Text) API this time, which is more customisable.
```
#collapse-output
dls_clas = DataBlock(
blocks = (TextBlock.from_df('text', seq_len=dls_lm.seq_len, vocab=dls_lm.vocab),
CategoryBlock),
get_x=ColReader('text'),
get_y=ColReader('sentiment'),
splitter=RandomSplitter()
).dataloaders(df_clas, bs=64)
```
To use the API, [`fastai`](https://docs.fast.ai/) needs the following:
* [`blocks`](https://docs.fast.ai/data.block.html#TransformBlock):
* [`TextBlock`](https://docs.fast.ai/text.data.html#TextBlock): Our x variable will be text contained in a [`pandas`](https://pandas.pydata.org/docs/) [`DataFrame`](https://pandas.pydata.org/docs/reference/api/pandas.DataFrame.html). We want to use the same sequence length and vocab as the language model [`DataLoaders`](https://docs.fast.ai/data.core.html#DataLoaders) so we can make use of our pre-trained model.
* [`CategoryBlock`](https://docs.fast.ai/data.block.html#CategoryBlock): Our y variable will be a single-label category (negative, neutral or positive sentiment).
* [`get_x`](https://docs.fast.ai/data.transforms.html#ColReader), [`get_y`](https://docs.fast.ai/data.transforms.html#ColReader): Get data for the model by reading the `text` and `sentiment` columns from the [`DataFrame`](https://pandas.pydata.org/docs/reference/api/pandas.DataFrame.html).
* [`splitter`](https://docs.fast.ai/data.transforms.html#RandomSplitter): We will use [`RandomSplitter()`](https://docs.fast.ai/data.transforms.html#RandomSplitter) to randomly split the data into a training set (80% by default) and a validation set (20%).
* [`dataloaders`](https://docs.fast.ai/data.block#DataBlock.dataloaders): Builds the [`DataLoaders`](https://docs.fast.ai/data.core.html#DataLoaders) using the [`DataBlock`](https://docs.fast.ai/tutorial.datablock.html#Text) template we just defined, the *df_clas* [`DataFrame`](https://pandas.pydata.org/docs/reference/api/pandas.DataFrame.html) and a batch size of 64.
We can call show batch as before; this time the dependent variable is sentiment.
```
dls_clas.show_batch(max_n=2)
```
Initialising the [`Learner`](https://docs.fast.ai/learner.html#Learner) is similar to before, but in this case we want a [`text_classifier_learner`](https://docs.fast.ai/text.learner.html#text_classifier_learner).
```
learn = text_classifier_learner(dls_clas, AWD_LSTM, drop_mult=0.5, metrics=accuracy).to_fp16()
```
Finally, we want to load the encoder from the language model we trained earlier, so our classifier uses pre-trained weights.
```
learn = learn.load_encoder('finetuned_lm')
```
## Fine-tuning the classifier
Now we can train the classifier using *discriminative learning rates* and *gradual unfreezing*, which has been found to give better results for this type of model. First let's freeze all but the last layer:
```
learn.fit_one_cycle(1, 3e-2)
```
Now freeze all but the last two layers:
```
learn.freeze_to(-2)
learn.fit_one_cycle(1, slice(1e-2/(2.6**4),1e-2))
```
Now all but the last three:
```
learn.freeze_to(-3)
learn.fit_one_cycle(1, slice(5e-3/(2.6**4),5e-3))
```
Finally, let's unfreeze the entire model and train a bit more:
```
learn.unfreeze()
learn.fit_one_cycle(3, slice(1e-3/(2.6**4),1e-3))
learn.save('classifier')
```
Our model correctly predicts sentiment around 77% of the time. We could perhaps do better with a larger dataset as mentioned earlier, or different model hyperparameters. It might be worth experimenting with this yourself to see if you can improve the accuracy.
We can quickly sense check the model by calling [`predict`](https://docs.fast.ai/learner.html#Learner.predict), which returns the predicted sentiment, the index of the prediction and predicted probabilities for negative, neutral and positive sentiment.
```
learn.predict("I love")
learn.predict("I hate")
```
# Classifying unlabelled tweets
To carry out sentiment analysis on the vaccine tweets, we can add them to the [`DataLoaders`](https://docs.fast.ai/data.core.html#DataLoaders) as a test set:
```
pred_dl = dls_clas.test_dl(vax_tweets['text'])
```
We can then make predictions using [`get_preds`](https://docs.fast.ai/learner.html#Learner.get_preds):
```
preds = learn.get_preds(dl=pred_dl)
```
Finally, we can save the results for analysis later.
```
# Get predicted sentiment
vax_tweets['sentiment'] = preds[0].argmax(dim=-1)
vax_tweets['sentiment'] = vax_tweets['sentiment'].map({0:'negative', 1:'neutral', 2:'positive'})
# Convert dates
vax_tweets['date'] = pd.to_datetime(vax_tweets['date'], errors='coerce').dt.date
# Save to csv
vax_tweets.to_csv('vax_tweets_inc_sentiment.csv')
```
# Conclusion
[`fastai`](https://docs.fast.ai/) make NLP really easy, and we were able to get quite good results with a limited dataset and not a lot of training time by using the ULMFiT approach. To summarise, the steps are:
1. Fine-tune a language model to predict the next word in a tweet, using a model pre-trained on Wikipedia.
2. Fine-tune a classification model to predict tweet sentiment using the pre-trained language model.
3. Apply the classifier to unlabelled tweets to analyse sentiment.
In [part 2](https://thomaswhelan.com/fastai/nlp/sentiment%20analysis/pytorch/visualisation/2021/03/17/covid-19-vaccine-tweet-sentiment-analysis-with-fastai-part-2.html) we will use our new model for analysis, investigating the overall sentiment of each vaccine, how sentiment changes over time and the relationship between sentiment and vaccination progress in different countries.
I hope you found this useful, and thanks very much to [Gabriel Preda](https://www.kaggle.com/gpreda) for providing the data!
{{ 'Cover image via https://www.analyticsvidhya.com/blog/2018/07/hands-on-sentiment-analysis-dataset-python/' | fndetail: 1 }}
```
```
|
github_jupyter
|
#collapse-output
# See https://neptune.ai/blog/google-colab-dealing-with-files for more tips on working with files in Colab
from google.colab import files
uploaded = files.upload()
#collapse-output
!pip uninstall -q -y kaggle
!pip install -q --upgrade pip
!pip install -q --upgrade kaggle
#collapse-output
# https://www.machinelearningmindset.com/kaggle-dataset-in-google-colab/
!mkdir -p ~/.kaggle
!cp kaggle.json ~/.kaggle/
# Check the file in its new directory
!ls /root/.kaggle/
# Check the file permission
!ls -l ~/.kaggle/kaggle.json
#Change the file permission
# chmod 600 file – owner can read and write
# chmod 700 file – owner can read, write and execute
!chmod 600 ~/.kaggle/kaggle.json
#collapse-output
# We will be using two datasets for this part, as well as a third dataset for part 2
# To save time in part 2 I'm going to download them all now and save locally
!kaggle datasets download -d gpreda/all-covid19-vaccines-tweets
!kaggle datasets download -d maxjon/complete-tweet-sentiment-extraction-data
!kaggle datasets download -d gpreda/covid-world-vaccination-progress
#collapse-output
# To unzip you can use the following:
#!mkdir folder_name
#!unzip anyfile.zip -d folder_name
# Or unzip all
!unzip -q \*.zip
#collapse-output
! [ -e /content ] && pip install -Uqq fastai # upgrade fastai on colab
import fastai; fastai.__version__
from fastai.text.all import *
vax_tweets = pd.read_csv('vaccination_all_tweets.csv')
vax_tweets[['date', 'text', 'hashtags', 'user_followers']].head()
tweets = pd.read_csv('tweet_dataset.csv')
tweets[['old_text', 'new_sentiment']].head()
# Code via https://www.kaggle.com/garyongguanjie/comments-analysis
def de_emojify(inputString):
return inputString.encode('ascii', 'ignore').decode('ascii')
# Code via https://www.kaggle.com/pawanbhandarkar/generate-smarter-word-clouds-with-log-likelihood
def tweet_proc(df, text_col='text'):
df['orig_text'] = df[text_col]
# Remove twitter handles
df[text_col] = df[text_col].apply(lambda x:re.sub('@[^\s]+','',x))
# Remove URLs
df[text_col] = df[text_col].apply(lambda x:re.sub(r"http\S+", "", x))
# Remove emojis
df[text_col] = df[text_col].apply(de_emojify)
# Remove hashtags
df[text_col] = df[text_col].apply(lambda x:re.sub(r'\B#\S+','',x))
return df[df[text_col]!='']
# Clean the text data and combine the dfs
tweets = tweets[['old_text', 'new_sentiment']].rename(columns={'old_text':'text', 'new_sentiment':'sentiment'})
vax_tweets['sentiment'] = np.nan
tweets = tweet_proc(tweets)
vax_tweets = tweet_proc(vax_tweets)
df_lm = tweets[['text', 'sentiment']].append(vax_tweets[['text', 'sentiment']])
df_clas = df_lm.dropna(subset=['sentiment'])
print(len(df_lm), len(df_clas))
df_clas.head()
#collapse-output
dls_lm = TextDataLoaders.from_df(df_lm, text_col='text', is_lm=True, valid_pct=0.1)
dls_lm.show_batch(max_n=2)
learn = language_model_learner(dls_lm, AWD_LSTM, drop_mult=0.3,
metrics=[accuracy, Perplexity()]).to_fp16()
learn.lr_find()
learn.fit_one_cycle(1, 3e-2)
learn.unfreeze()
learn.lr_find()
learn.fit_one_cycle(4, 1e-3)
# Text generation using the language model
TEXT = "I love"
N_WORDS = 30
N_SENTENCES = 2
print("\n".join(learn.predict(TEXT, N_WORDS, temperature=0.75) for _ in range(N_SENTENCES)))
learn.save_encoder('finetuned_lm')
#collapse-output
dls_clas = DataBlock(
blocks = (TextBlock.from_df('text', seq_len=dls_lm.seq_len, vocab=dls_lm.vocab),
CategoryBlock),
get_x=ColReader('text'),
get_y=ColReader('sentiment'),
splitter=RandomSplitter()
).dataloaders(df_clas, bs=64)
dls_clas.show_batch(max_n=2)
learn = text_classifier_learner(dls_clas, AWD_LSTM, drop_mult=0.5, metrics=accuracy).to_fp16()
learn = learn.load_encoder('finetuned_lm')
learn.fit_one_cycle(1, 3e-2)
learn.freeze_to(-2)
learn.fit_one_cycle(1, slice(1e-2/(2.6**4),1e-2))
learn.freeze_to(-3)
learn.fit_one_cycle(1, slice(5e-3/(2.6**4),5e-3))
learn.unfreeze()
learn.fit_one_cycle(3, slice(1e-3/(2.6**4),1e-3))
learn.save('classifier')
learn.predict("I love")
learn.predict("I hate")
pred_dl = dls_clas.test_dl(vax_tweets['text'])
preds = learn.get_preds(dl=pred_dl)
# Get predicted sentiment
vax_tweets['sentiment'] = preds[0].argmax(dim=-1)
vax_tweets['sentiment'] = vax_tweets['sentiment'].map({0:'negative', 1:'neutral', 2:'positive'})
# Convert dates
vax_tweets['date'] = pd.to_datetime(vax_tweets['date'], errors='coerce').dt.date
# Save to csv
vax_tweets.to_csv('vax_tweets_inc_sentiment.csv')
| 0.563258 | 0.951549 |
# Diseño de software para cómputo científico
----
Property Based Testing

## Property base testing

Paper seminal:
> Claessen, K., & Hughes, J. (2011). QuickCheck: a lightweight tool for random testing of Haskell programs. Acm sigplan notices, 46(4), 53-64.
<small>Fuente: <a href="https://www.freecodecamp.org/news/intro-to-property-based-testing-in-python-6321e0c2f8b/">https://www.freecodecamp.org/news/intro-to-property-based-testing-in-python-6321e0c2f8b/</a></small>
## Part 1: Example-based testing
- Given a test input `I`
- When passed to function under test
- should return an output `O`

Probemos esta máquina con testing basado en ejemplos
- take a blue-coloured raw plastic (fixed test data)
- feed the plastic to machine
- expect a blue-coloured plastic ball as output (fixed test output)
## Lo mismo pero con un programa
```
# test_example.py
def sum(num1, num2):
"""It returns sum of two numbers"""
return num1 + num2
def test_sum():
assert sum(1, 2) == 3
!pytest 10_PBT/test_example.py -v
```
## Este test es suficiente para **validar** la funcionalidad de `sum()`?
- La respuesta simple es no.
- Podemos usar parametrización de pytest para mejorar esto
```
# test_param_example.py
def sum(num1, num2):
"""It returns sum of two numbers"""
return num1 + num2
import pytest
@pytest.mark.parametrize('num1, num2, expected',
[(3, 5, 8), (-2, -2, -4), (-1, 5, 4), (3, -5, -2), (0, 5, 5) ]
)
def test_sum(num1, num2, expected):
assert sum(num1, num2) == expected
!pytest 10_PBT/test_param_example.py -v
```
Pero es esencialmente más de lo mismo. Lo cual nos lleva a al primer problema de los testings basados en ejemplos
## Issue 1: La exhaustividad depende de la persona que escribe los tests
- La persona puede querer escribir 5, 50 o 500 tests.
- Pero nunca puede estar seguro si logro probar todo. (sobre todos los edge cases)
Lo cual nos lleva al segundo problema
## Issue 2: Los tests no son robustos por el entendimiento ambiguo de los requerimientos
Nos dieron todos los detalles de la función sum?
- Que tipo de input espera la función?
- Como debe comportarse la funcion frente a inputs inesperados?
- Que tipo de output debe retornar nuestra función?
## Issue 2: Los tests no son robustos por el entendimiento ambiguo de los requerimientos
O si lo ponemos en contexto de nuestro código:
- `num1` y `num2` tienen que ser `int` o `float`? pueden ser `str`?
- Hay cotas minimas y maximas para `num1` y `num2`?
- Como manejo `None`?
- El retorno tiene que ser `int` o `float`?
- Cuales ecenarios justifican mensajes de errores?
- Hay algun ecenario donde pueda engañar al test?
```
# test_buggy_example.py
def sum(num1, num2):
"""Buggy logic"""
results = {
(3, 5): 8, (-2, -2): -4,
(-1, 5): 4, (3, -5): -2, (0, 5): 5}
return results.get((num1, num2))
import pytest
@pytest.mark.parametrize(
'num1, num2, expected',
[(3, 5, 8), (-2, -2, -4), (-1, 5, 4), (3, -5, -2), (0, 5, 5)])
def test_sum(num1, num2, expected):
assert sum(num1, num2) == expected
!pytest 10_PBT/test_buggy_example.py
```
## Part 2: Property-based testing
- Los frameworks de Property based testing verifican la veracidad de propiedades
- Una propiedad es un statement del tipo:
```python
for all (x, y, …)
such as precondition(x, y, …) holds
property(x, y, …) is true.
```
Asi el *aproach* en propiedades para la maquina de hacer pelotas seria:
- Dada una gran coleccion de plasticos.
- Aseguraque todos este coloreados.
- Y la salida tiene que tener las siguientes propiedades:
- Es esferica.
- Es coloreada con el mismo color del input.
## Hypotesis testing
```
!pip install hypothesis
# test_property_example.py
def sum(num1, num2):
"""It returns sum of two numbers"""
return num1 + num2
from hypothesis import given
from hypothesis import strategies as st
@given(st.integers(), st.integers())
def test_sum(num1, num2):
assert sum(num1, num2) == num1 + num2
!pytest 10_PBT/test_example.py -s
```
## Hypotesis testing - Verbose
```
# test_property_v_example.py
from hypothesis import given, settings, Verbosity, example
from hypothesis import strategies as st
@settings(verbosity=Verbosity.verbose)
@given(st.integers(), st.integers())
@example(1, 2)
@example(0, 0)
@example(10000, 2000000)
def test_sum(num1, num2):
assert sum(num1, num2) == num1 + num2
!pytest 10_PBT/test_property_v_example.py -v -s
```
## Las estrategias son desgraciadas
```
st.lists(st.text(), min_size=5, unique=True).example()
st.dictionaries(st.characters(), st.floats(), min_size=3).example()
```
Sobre las estrategias: https://hypothesis.readthedocs.io/en/latest/data.html
## Hay estrategias para el stack científico
```
from hypothesis.extra import numpy as npst
npst.arrays(float, (3, 4), elements=None, fill=None, unique=True).example()
from hypothesis.extra import pandas as pdst
pdst.data_frames([
pdst.column('A', dtype=int, unique=True),
pdst.column('B', dtype=float, unique=True)]).example()
```
## Pensando en propiedades reales
- Al parecer la suma es conmutativa.
- Sumar cero a un valor devuelve el mismo valor
- Si sumamos dos numeros y restamos uno de los dos, nos da el otro.
```
from hypothesis import given, settings, Verbosity
from hypothesis import strategies as st
@settings(verbosity=Verbosity.verbose)
@given(st.integers(), st.integers())
def test_sum(num1, num2):
assert sum(num1, 0) == num1
assert sum(0, num2) == num2
assert sum(num1, num2) == sum(num2, num1)
assert sum(num1, num2) - num1 == num2
```
## Shrinking
- Hasta ahora todo es razonable de programar.
- Pero la inferencia es solo parte del truco.
```
@settings(verbosity=Verbosity.verbose)
@given(st.integers(), st.integers())
def test_sum(num1, num2):
assert sum(num1, 0) == num1
assert sum(num1, num2) == sum(num2, num1)
assert num1 <= 30
!pytest 10_PBT/test_property_srink_example.py
```
Hypotesis trata de encontrar el ejemplo minimo entendible para un ser humano
### Estrategias custom
```
import attr
@attr.s(hash=True)
class Persona:
name: str = attr.ib()
edad: int = attr.ib()
class Persona:
def __init__(self, name, edad):
self.name = name
self.edad = edad
l = [1]
hash("hola")
PersonaStrategy = st.lists(st.builds(
Persona,
st.text(min_size=5, max_size=10),
st.integers(min_value=0, max_value=80)), min_size=3, unique=True)
@PersonaStrategy
def test_persona(persona):
assert persona.edad <= 120
assert persona.edad >= 0
PersonaMalaEdadStrategy = st.lists(st.builds(
Persona,
st.text(min_size=5, max_size=10),
st.integers(), min_size=3, unique=True))
@PersonaMalaEdadStrategy()
def test_persona_mala(p):
with pytest.raises(ValueError)
PersonaStrategy.example()
```
### Estrategias custom (con composición)
```
import attr
@attr.s(hash=True)
class Persona:
name: str = attr.ib()
edad: int = attr.ib()
mayor_de_edad: bool = attr.ib()
@st.composite
def persona_stategy(draw):
name = draw(st.text(min_size=5, max_size=80))
age = draw(st.integers(min_value=0, max_value=17))
mayor_de_edad = draw(st.booleans())
return Persona(name, age, mayor_de_edad)
persona_stategy().example()
```
|
github_jupyter
|
# test_example.py
def sum(num1, num2):
"""It returns sum of two numbers"""
return num1 + num2
def test_sum():
assert sum(1, 2) == 3
!pytest 10_PBT/test_example.py -v
# test_param_example.py
def sum(num1, num2):
"""It returns sum of two numbers"""
return num1 + num2
import pytest
@pytest.mark.parametrize('num1, num2, expected',
[(3, 5, 8), (-2, -2, -4), (-1, 5, 4), (3, -5, -2), (0, 5, 5) ]
)
def test_sum(num1, num2, expected):
assert sum(num1, num2) == expected
!pytest 10_PBT/test_param_example.py -v
# test_buggy_example.py
def sum(num1, num2):
"""Buggy logic"""
results = {
(3, 5): 8, (-2, -2): -4,
(-1, 5): 4, (3, -5): -2, (0, 5): 5}
return results.get((num1, num2))
import pytest
@pytest.mark.parametrize(
'num1, num2, expected',
[(3, 5, 8), (-2, -2, -4), (-1, 5, 4), (3, -5, -2), (0, 5, 5)])
def test_sum(num1, num2, expected):
assert sum(num1, num2) == expected
!pytest 10_PBT/test_buggy_example.py
for all (x, y, …)
such as precondition(x, y, …) holds
property(x, y, …) is true.
!pip install hypothesis
# test_property_example.py
def sum(num1, num2):
"""It returns sum of two numbers"""
return num1 + num2
from hypothesis import given
from hypothesis import strategies as st
@given(st.integers(), st.integers())
def test_sum(num1, num2):
assert sum(num1, num2) == num1 + num2
!pytest 10_PBT/test_example.py -s
# test_property_v_example.py
from hypothesis import given, settings, Verbosity, example
from hypothesis import strategies as st
@settings(verbosity=Verbosity.verbose)
@given(st.integers(), st.integers())
@example(1, 2)
@example(0, 0)
@example(10000, 2000000)
def test_sum(num1, num2):
assert sum(num1, num2) == num1 + num2
!pytest 10_PBT/test_property_v_example.py -v -s
st.lists(st.text(), min_size=5, unique=True).example()
st.dictionaries(st.characters(), st.floats(), min_size=3).example()
from hypothesis.extra import numpy as npst
npst.arrays(float, (3, 4), elements=None, fill=None, unique=True).example()
from hypothesis.extra import pandas as pdst
pdst.data_frames([
pdst.column('A', dtype=int, unique=True),
pdst.column('B', dtype=float, unique=True)]).example()
from hypothesis import given, settings, Verbosity
from hypothesis import strategies as st
@settings(verbosity=Verbosity.verbose)
@given(st.integers(), st.integers())
def test_sum(num1, num2):
assert sum(num1, 0) == num1
assert sum(0, num2) == num2
assert sum(num1, num2) == sum(num2, num1)
assert sum(num1, num2) - num1 == num2
@settings(verbosity=Verbosity.verbose)
@given(st.integers(), st.integers())
def test_sum(num1, num2):
assert sum(num1, 0) == num1
assert sum(num1, num2) == sum(num2, num1)
assert num1 <= 30
!pytest 10_PBT/test_property_srink_example.py
import attr
@attr.s(hash=True)
class Persona:
name: str = attr.ib()
edad: int = attr.ib()
class Persona:
def __init__(self, name, edad):
self.name = name
self.edad = edad
l = [1]
hash("hola")
PersonaStrategy = st.lists(st.builds(
Persona,
st.text(min_size=5, max_size=10),
st.integers(min_value=0, max_value=80)), min_size=3, unique=True)
@PersonaStrategy
def test_persona(persona):
assert persona.edad <= 120
assert persona.edad >= 0
PersonaMalaEdadStrategy = st.lists(st.builds(
Persona,
st.text(min_size=5, max_size=10),
st.integers(), min_size=3, unique=True))
@PersonaMalaEdadStrategy()
def test_persona_mala(p):
with pytest.raises(ValueError)
PersonaStrategy.example()
import attr
@attr.s(hash=True)
class Persona:
name: str = attr.ib()
edad: int = attr.ib()
mayor_de_edad: bool = attr.ib()
@st.composite
def persona_stategy(draw):
name = draw(st.text(min_size=5, max_size=80))
age = draw(st.integers(min_value=0, max_value=17))
mayor_de_edad = draw(st.booleans())
return Persona(name, age, mayor_de_edad)
persona_stategy().example()
| 0.58747 | 0.972441 |
```
import scipy.io
import scipy.misc
import tensorflow as tf
import numpy as np
from matplotlib import pyplot as plt
import random
import os
try:
from os import scandir, walk
except ImportError:
from scandir import scandir, walk
IMAGE_HEIGHT = 256
IMAGE_WIDTH = 256
TO_TRAIN_PATH = '2500_TRAIN/'
GROUND_TRUTH_PATH = 'long_pants2500_TRUTH/'
VALIDATION_PATH = '997_Train/'
def conv2d_batch_relu(input, kernel_size, stride, num_filter, scope):
with tf.variable_scope(scope):
stride_shape = [1, stride, stride, 1]
filter_shape = [kernel_size, kernel_size, input.get_shape()[3], num_filter]
W = tf.get_variable('w', filter_shape, tf.float32, tf.random_normal_initializer(0.0, 0.02))
b = tf.get_variable('b', [1, 1, 1, num_filter], initializer=tf.constant_initializer(0.0))
conv_2d = tf.nn.conv2d(input, W, stride_shape, padding='SAME') + b
batch = tf.layers.batch_normalization(conv_2d)
relu = tf.nn.relu(batch)
print(scope, ' output dim: ', relu.shape)
return relu
def conv2d_transpose_batch_relu(input, kernel_size, stride, num_filter, output_dim, scope):
with tf.variable_scope(scope):
stride_shape = [1, stride, stride, 1]
shape = input.get_shape().as_list()
filter_shape = [kernel_size, kernel_size, num_filter, shape[3]]
output_shape = [shape[0], output_dim, output_dim, num_filter]
W = tf.get_variable('w', filter_shape, tf.float32, tf.random_normal_initializer(0.0, 0.02))
b = tf.get_variable('b', [1, 1, 1, num_filter], initializer=tf.constant_initializer(0.0))
conv_2d = tf.nn.conv2d_transpose(input, W, output_shape, stride_shape)
batch = tf.layers.batch_normalization(conv_2d)
relu = tf.nn.relu(batch)
print(scope, ' output dim: ', relu.shape)
return relu
def max_pool(input, kernel_size, stride):
ksize = [1, kernel_size, kernel_size, 1]
strides = [1, stride, stride, 1]
pool = tf.nn.max_pool(input, ksize=ksize, strides=strides, padding='SAME')
print('Max Pool Layer: ', pool.shape)
return pool
def unsample(input, outputdim):
unsample = tf.image.resize_nearest_neighbor(input, outputdim)
print('Unsampling Layer: ', unsample.shape)
return unsample
class SegmentationNN:
def __init__(self):
self.num_epoch = 10
self.batch_size = 10
self.input = tf.placeholder(tf.float32, [self.batch_size, IMAGE_HEIGHT, IMAGE_WIDTH, 3])
self.label = tf.placeholder(tf.float32, [self.batch_size, IMAGE_HEIGHT, IMAGE_WIDTH, 1])
self.output = self.build_model(self.input)
print(self.output.shape)
self.log_step = 50
self.lr = 1e-4
self.loss = self._loss(self.output, self.label)
self.accuracy = self._accuracy(self.output, self.label)
self.optimizer = self._optimizer()
def load_data(self, TO_TRAIN_PATH, GROUD_TRUTH_PATH):
to_train = []
count=0
label = []
for file in scandir(TO_TRAIN_PATH):
if file.name.endswith('jpg') or file.name.endswith('png') and file.is_file():
image = scipy.misc.imread(file.path)
image = scipy.misc.imresize(image, (IMAGE_HEIGHT, IMAGE_WIDTH))
to_train.append(image)
image = scipy.misc.imread((GROUND_TRUTH_PATH + file.name).replace('jpg', 'png'))
image = scipy.misc.imresize(image, (IMAGE_HEIGHT, IMAGE_WIDTH))
image = np.reshape(image, (IMAGE_HEIGHT, IMAGE_WIDTH, 1))
label.append(image)
count = count + 1
self.training_set = to_train
self.label_set = label
self.num_training = count
return to_train, label
def load_validation(self, VALIDATION_PATH):
validation = []
for file in scandir(VALIDATION_PATH):
if file.name.endswith('jpg') or file.name.endswith('png') and file.is_file():
image = scipy.misc.imread(file.path)
image = scipy.misc.imresize(image, (IMAGE_HEIGHT, IMAGE_WIDTH))
validation.append(image)
self.validation_set = validation
return validation
def build_model(self, input):
conv1 = conv2d_batch_relu(input, 7, 2, 64, 'conv_1_1')
conv2 = conv2d_batch_relu(conv1, 7, 1, 64, 'conv_1_2')
max_pool_1 = max_pool(conv2, 3, 2)
conv3 = conv2d_batch_relu(max_pool_1, 7, 2, 64, 'conv_1_3')
conv4 = conv2d_batch_relu(conv3, 7, 1, 64, 'conv_1_4')
max_pool_2 = max_pool(conv4, 3, 2)
conv5 = conv2d_batch_relu(max_pool_2, 7, 2, 64, 'conv_1_5')
conv6 = conv2d_batch_relu(conv5, 7, 1, 64, 'conv_1_6')
max_pool_3 = max_pool(conv6, 3, 2)
unsampled_1 = unsample(max_pool_3, [8,8])
conv1 = conv2d_transpose_batch_relu(unsampled_1, 7, 1, 64, 8, 'conv_2_1')
conv2 = conv2d_transpose_batch_relu(conv1, 7, 2, 64, 16, 'conv_2_2')
unsampled_2 = unsample(conv2, [32,32])
conv3 = conv2d_transpose_batch_relu(unsampled_2, 7, 1, 64, 32, 'conv_2_3')
conv4 = conv2d_transpose_batch_relu(conv3, 7, 2, 64, 64, 'conv_2_4')
unsampled_3 = unsample(conv5, [128,128])
conv5 = conv2d_transpose_batch_relu(unsampled_3, 7, 1, 64, 128, 'conv_2_5')
conv6 = conv2d_transpose_batch_relu(conv5, 7, 2, 1, 256, 'conv_2_6')
return conv6
def _loss(self, logits, labels):
return tf.reduce_mean(tf.squared_difference(logits, labels))
def _accuracy(self, logits, labels):
return tf.reduce_mean(tf.divide(tf.abs(logits - labels), labels))
def _optimizer(self):
return tf.train.AdamOptimizer(learning_rate = self.lr).minimize(self.loss)
def train(self, sess):
iteration = 0
losses = []
accuracies = []
for epoch in range(self.num_epoch):
for it in range(self.num_training // self.batch_size):
fetches = [self.optimizer, self.loss]
_input = self.training_set[it*self.batch_size: (it+1)*self.batch_size]
_label = self.label_set[it*self.batch_size: (it+1)*self.batch_size]
feed_dict = {
self.input: _input,
self.label: _label
}
_, loss, accuracy = sess.run([self.optimizer, self.loss, self.accuracy], feed_dict = feed_dict)
losses.append(loss)
accuracies.append(accuracy)
if iteration%self.log_step is 0:
print('iteration: {} loss: {}, accuracy: {}'.format(iteration, loss, accuracy))
iteration = iteration + 1
feed_dict = {
self.input: self.validation_set[0: self.batch_size]
}
generated_image = sess.run([self.output], feed_dict = feed_dict)
images = np.concatenate(generated_image)
images = images[:,:,:,0]
images = np.reshape(images, (self.batch_size*IMAGE_HEIGHT, IMAGE_WIDTH))
save_path = 'output/epoch_{}.jpg'.format(epoch + 1)
scipy.misc.imsave(save_path, images)
tf.reset_default_graph()
model1 = SegmentationNN()
trainning_set, label_set = model1.load_data(TO_TRAIN_PATH, GROUND_TRUTH_PATH)
validation_set = model1.load_validation(VALIDATION_PATH)
print(model1.num_training)
plt.imshow(model1.validation_set[10])
plt.show()
tf.reset_default_graph()
with tf.Session() as sess:
model = SegmentationNN()
sess.run(tf.global_variables_initializer())
model.train(sess)
```
|
github_jupyter
|
import scipy.io
import scipy.misc
import tensorflow as tf
import numpy as np
from matplotlib import pyplot as plt
import random
import os
try:
from os import scandir, walk
except ImportError:
from scandir import scandir, walk
IMAGE_HEIGHT = 256
IMAGE_WIDTH = 256
TO_TRAIN_PATH = '2500_TRAIN/'
GROUND_TRUTH_PATH = 'long_pants2500_TRUTH/'
VALIDATION_PATH = '997_Train/'
def conv2d_batch_relu(input, kernel_size, stride, num_filter, scope):
with tf.variable_scope(scope):
stride_shape = [1, stride, stride, 1]
filter_shape = [kernel_size, kernel_size, input.get_shape()[3], num_filter]
W = tf.get_variable('w', filter_shape, tf.float32, tf.random_normal_initializer(0.0, 0.02))
b = tf.get_variable('b', [1, 1, 1, num_filter], initializer=tf.constant_initializer(0.0))
conv_2d = tf.nn.conv2d(input, W, stride_shape, padding='SAME') + b
batch = tf.layers.batch_normalization(conv_2d)
relu = tf.nn.relu(batch)
print(scope, ' output dim: ', relu.shape)
return relu
def conv2d_transpose_batch_relu(input, kernel_size, stride, num_filter, output_dim, scope):
with tf.variable_scope(scope):
stride_shape = [1, stride, stride, 1]
shape = input.get_shape().as_list()
filter_shape = [kernel_size, kernel_size, num_filter, shape[3]]
output_shape = [shape[0], output_dim, output_dim, num_filter]
W = tf.get_variable('w', filter_shape, tf.float32, tf.random_normal_initializer(0.0, 0.02))
b = tf.get_variable('b', [1, 1, 1, num_filter], initializer=tf.constant_initializer(0.0))
conv_2d = tf.nn.conv2d_transpose(input, W, output_shape, stride_shape)
batch = tf.layers.batch_normalization(conv_2d)
relu = tf.nn.relu(batch)
print(scope, ' output dim: ', relu.shape)
return relu
def max_pool(input, kernel_size, stride):
ksize = [1, kernel_size, kernel_size, 1]
strides = [1, stride, stride, 1]
pool = tf.nn.max_pool(input, ksize=ksize, strides=strides, padding='SAME')
print('Max Pool Layer: ', pool.shape)
return pool
def unsample(input, outputdim):
unsample = tf.image.resize_nearest_neighbor(input, outputdim)
print('Unsampling Layer: ', unsample.shape)
return unsample
class SegmentationNN:
def __init__(self):
self.num_epoch = 10
self.batch_size = 10
self.input = tf.placeholder(tf.float32, [self.batch_size, IMAGE_HEIGHT, IMAGE_WIDTH, 3])
self.label = tf.placeholder(tf.float32, [self.batch_size, IMAGE_HEIGHT, IMAGE_WIDTH, 1])
self.output = self.build_model(self.input)
print(self.output.shape)
self.log_step = 50
self.lr = 1e-4
self.loss = self._loss(self.output, self.label)
self.accuracy = self._accuracy(self.output, self.label)
self.optimizer = self._optimizer()
def load_data(self, TO_TRAIN_PATH, GROUD_TRUTH_PATH):
to_train = []
count=0
label = []
for file in scandir(TO_TRAIN_PATH):
if file.name.endswith('jpg') or file.name.endswith('png') and file.is_file():
image = scipy.misc.imread(file.path)
image = scipy.misc.imresize(image, (IMAGE_HEIGHT, IMAGE_WIDTH))
to_train.append(image)
image = scipy.misc.imread((GROUND_TRUTH_PATH + file.name).replace('jpg', 'png'))
image = scipy.misc.imresize(image, (IMAGE_HEIGHT, IMAGE_WIDTH))
image = np.reshape(image, (IMAGE_HEIGHT, IMAGE_WIDTH, 1))
label.append(image)
count = count + 1
self.training_set = to_train
self.label_set = label
self.num_training = count
return to_train, label
def load_validation(self, VALIDATION_PATH):
validation = []
for file in scandir(VALIDATION_PATH):
if file.name.endswith('jpg') or file.name.endswith('png') and file.is_file():
image = scipy.misc.imread(file.path)
image = scipy.misc.imresize(image, (IMAGE_HEIGHT, IMAGE_WIDTH))
validation.append(image)
self.validation_set = validation
return validation
def build_model(self, input):
conv1 = conv2d_batch_relu(input, 7, 2, 64, 'conv_1_1')
conv2 = conv2d_batch_relu(conv1, 7, 1, 64, 'conv_1_2')
max_pool_1 = max_pool(conv2, 3, 2)
conv3 = conv2d_batch_relu(max_pool_1, 7, 2, 64, 'conv_1_3')
conv4 = conv2d_batch_relu(conv3, 7, 1, 64, 'conv_1_4')
max_pool_2 = max_pool(conv4, 3, 2)
conv5 = conv2d_batch_relu(max_pool_2, 7, 2, 64, 'conv_1_5')
conv6 = conv2d_batch_relu(conv5, 7, 1, 64, 'conv_1_6')
max_pool_3 = max_pool(conv6, 3, 2)
unsampled_1 = unsample(max_pool_3, [8,8])
conv1 = conv2d_transpose_batch_relu(unsampled_1, 7, 1, 64, 8, 'conv_2_1')
conv2 = conv2d_transpose_batch_relu(conv1, 7, 2, 64, 16, 'conv_2_2')
unsampled_2 = unsample(conv2, [32,32])
conv3 = conv2d_transpose_batch_relu(unsampled_2, 7, 1, 64, 32, 'conv_2_3')
conv4 = conv2d_transpose_batch_relu(conv3, 7, 2, 64, 64, 'conv_2_4')
unsampled_3 = unsample(conv5, [128,128])
conv5 = conv2d_transpose_batch_relu(unsampled_3, 7, 1, 64, 128, 'conv_2_5')
conv6 = conv2d_transpose_batch_relu(conv5, 7, 2, 1, 256, 'conv_2_6')
return conv6
def _loss(self, logits, labels):
return tf.reduce_mean(tf.squared_difference(logits, labels))
def _accuracy(self, logits, labels):
return tf.reduce_mean(tf.divide(tf.abs(logits - labels), labels))
def _optimizer(self):
return tf.train.AdamOptimizer(learning_rate = self.lr).minimize(self.loss)
def train(self, sess):
iteration = 0
losses = []
accuracies = []
for epoch in range(self.num_epoch):
for it in range(self.num_training // self.batch_size):
fetches = [self.optimizer, self.loss]
_input = self.training_set[it*self.batch_size: (it+1)*self.batch_size]
_label = self.label_set[it*self.batch_size: (it+1)*self.batch_size]
feed_dict = {
self.input: _input,
self.label: _label
}
_, loss, accuracy = sess.run([self.optimizer, self.loss, self.accuracy], feed_dict = feed_dict)
losses.append(loss)
accuracies.append(accuracy)
if iteration%self.log_step is 0:
print('iteration: {} loss: {}, accuracy: {}'.format(iteration, loss, accuracy))
iteration = iteration + 1
feed_dict = {
self.input: self.validation_set[0: self.batch_size]
}
generated_image = sess.run([self.output], feed_dict = feed_dict)
images = np.concatenate(generated_image)
images = images[:,:,:,0]
images = np.reshape(images, (self.batch_size*IMAGE_HEIGHT, IMAGE_WIDTH))
save_path = 'output/epoch_{}.jpg'.format(epoch + 1)
scipy.misc.imsave(save_path, images)
tf.reset_default_graph()
model1 = SegmentationNN()
trainning_set, label_set = model1.load_data(TO_TRAIN_PATH, GROUND_TRUTH_PATH)
validation_set = model1.load_validation(VALIDATION_PATH)
print(model1.num_training)
plt.imshow(model1.validation_set[10])
plt.show()
tf.reset_default_graph()
with tf.Session() as sess:
model = SegmentationNN()
sess.run(tf.global_variables_initializer())
model.train(sess)
| 0.641871 | 0.414425 |
# Circular hole probllem
In this notebook an analytic solution for a disk-shaped hole is constructed.
The domain defined as
$$
\Omega = \{ x \in \mathbb{R^2} : 0 < a < |x| < A\}
$$
Let
$$
\Gamma^r = \{ x \in \mathbb{R^2} : |x| = r\},
$$
then $\partial\Omega = \Gamma^a \cup \Gamma^A$.
## Poisson
Sought is the solution to the problem
$$
\begin{aligned}
\nabla^2 u = 0, & \quad x \in \Omega, \\
\frac{\partial u}{\partial r} = 0, & \quad x \in \Gamma^a, \\
u = g, & \quad x \in \Gamma^A, \\
\end{aligned}
$$
where the Laplacian in polar coordinates is
$$
\nabla^2 = \frac{\partial^2}{\partial r^2}
+ \frac{1}{r}\frac{\partial}{\partial r}
+ \frac{1}{r^2}\frac{\partial^2}{\partial \theta^2}
$$
and $g$ is chosen such that the solution is easy to construct :-)
```
from sympy import *
from util import lua_code
# Laplacian in polar coordinates
def laplacian(f):
return f.diff(r, 2) + f.diff(r)/r + f.diff(t, 2)/r**2
```
Solution ansatz:
```
r, t = symbols('r theta')
a, B = symbols('a B')
u = r**2 * sin(2*t) + B * sin(2*t) / r**2
u
```
Check Laplacian
```
simplify(laplacian(u))
```
Check inner circle boundary
```
u.diff(r).subs(r, a)
```
Hence,
```
u = u.subs(B, a**4)
u.diff(r).subs(r, a)
```
We set $g=u$ on the outer circle boundary.
Gradient:
```
u_x = simplify(u.diff(r) * cos(t) - u.diff(t) * sin(t) / r)
u_x
u_y = simplify(u.diff(r) * sin(t) + u.diff(t) * cos(t) / r)
u_y
```
## Elasticity
For a plane-strain problem, we assume there exists a biharmonic function $\phi$ such that [1]
\begin{align}
2\mu u_r &= -\frac{\partial \phi}{\partial r} + (1-\nu)r\frac{\partial\psi}{\partial \theta} \\
2\mu u_\theta &= -\frac{1}{r}\frac{\partial \phi}{\partial \theta} + (1-\nu)r^2\frac{\partial\psi}{\partial r} \\
\sigma_{rr} &= \frac{1}{r}\frac{\partial \phi}{\partial r} +
\frac{1}{r^2}\frac{\partial^2 \phi}{\partial \theta^2} \\
\sigma_{\theta\theta} &= \frac{\partial^2 \phi}{\partial r^2} \\
\sigma_{r\theta} &= -\frac{\partial }{\partial r} \left(
\frac{1}{r}\frac{\partial \phi}{\partial\theta}
\right) \\
\end{align}
where $\mu$ is the shear modulus, $\nu$ is Poisson's ratio, and $\phi$ and $\psi$ are related by
$$
\nabla^2\phi = \frac{\partial}{\partial r}\left(r\frac{\partial\psi}{\partial\theta}\right)
$$
Remark: Plane-stress is modelled in [1], therefore the formula are adjusted here for plane-strain.
[1] "Stress Singularites Resulting From Various Boundary Conditions in Angular Corners of Plates in Extension", M. L. Williams, 1952, Journal of Applied Mechanics, 19 (4), pp. 526-528.
Sought is the solution to the problem
$$
\begin{aligned}
\sigma_{rr} = 0, & \quad x \in \Gamma^a, \\
\sigma_{r\theta} = 0, & \quad x \in \Gamma^a, \\
u_r = g_r, & \quad x \in \Gamma^A, \\
u_\theta = g_\theta, & \quad x \in \Gamma^A, \\
\end{aligned}
$$
for $\phi,\psi$ such that the equations of elasticity are satisfied.
Solution ansatz (from J.R. Barber, Elasticity, 2002):
```
r, t = symbols('r theta')
a, S, mu, nu = symbols('a S mu nu')
A = S*a**2
B = -S*a**4/2
phi = -S * r**2 * sin(2*t) / 2 + A * sin(2*t) + B * sin(2*t) / r**2
phi
```
Check that phi is biharmonic.
```
laplacian(simplify(laplacian(phi)))
```
Compute stresses
```
srr = simplify(phi.diff(r) / r + phi.diff(t, 2) / r**2)
stt = simplify(phi.diff(t, 2))
srt = simplify(-(phi.diff(t) / r).diff(r))
```
Check boundary condition on inner circle.
```
(srr.subs(r, a), srt.subs(r, a))
```
Find $\psi$ from compatbility condition, disregarding integration constants.
```
psi = integrate(integrate(simplify(laplacian(phi)), r) / r, t)
psi
```
Check compatbility condition.
```
simplify((r*psi.diff(t)).diff(r) - laplacian(phi))
```
Compute displacement
```
ur = simplify(-phi.diff(r) + (1 - nu) * r * psi.diff(t)) / (2*mu)
ut = simplify(-phi.diff(t) / r + (1 - nu) * r**2 * psi.diff(r)) / (2*mu)
ur
lua_code(ur)
ut
lua_code(ut)
```
We set $g_r=u_r$ and $g_\theta = u_\theta$ on the outer circle boundary.
Compute Jacobian
```
J = {}
J['ur_r'] = simplify(ur.diff(r))
J['ur_t'] = simplify(ur.diff(t))
J['ut_r'] = simplify(ut.diff(r))
J['ut_t'] = simplify(ut.diff(t))
for key, value in J.items():
print(f'local {key} = {lua_code(value)}')
```
|
github_jupyter
|
from sympy import *
from util import lua_code
# Laplacian in polar coordinates
def laplacian(f):
return f.diff(r, 2) + f.diff(r)/r + f.diff(t, 2)/r**2
r, t = symbols('r theta')
a, B = symbols('a B')
u = r**2 * sin(2*t) + B * sin(2*t) / r**2
u
simplify(laplacian(u))
u.diff(r).subs(r, a)
u = u.subs(B, a**4)
u.diff(r).subs(r, a)
u_x = simplify(u.diff(r) * cos(t) - u.diff(t) * sin(t) / r)
u_x
u_y = simplify(u.diff(r) * sin(t) + u.diff(t) * cos(t) / r)
u_y
r, t = symbols('r theta')
a, S, mu, nu = symbols('a S mu nu')
A = S*a**2
B = -S*a**4/2
phi = -S * r**2 * sin(2*t) / 2 + A * sin(2*t) + B * sin(2*t) / r**2
phi
laplacian(simplify(laplacian(phi)))
srr = simplify(phi.diff(r) / r + phi.diff(t, 2) / r**2)
stt = simplify(phi.diff(t, 2))
srt = simplify(-(phi.diff(t) / r).diff(r))
(srr.subs(r, a), srt.subs(r, a))
psi = integrate(integrate(simplify(laplacian(phi)), r) / r, t)
psi
simplify((r*psi.diff(t)).diff(r) - laplacian(phi))
ur = simplify(-phi.diff(r) + (1 - nu) * r * psi.diff(t)) / (2*mu)
ut = simplify(-phi.diff(t) / r + (1 - nu) * r**2 * psi.diff(r)) / (2*mu)
ur
lua_code(ur)
ut
lua_code(ut)
J = {}
J['ur_r'] = simplify(ur.diff(r))
J['ur_t'] = simplify(ur.diff(t))
J['ut_r'] = simplify(ut.diff(r))
J['ut_t'] = simplify(ut.diff(t))
for key, value in J.items():
print(f'local {key} = {lua_code(value)}')
| 0.543106 | 0.994309 |
```
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
sns.set()
%pylab inline
pylab.rcParams['figure.figsize'] = (4, 4)
# Avoid inaccurate floating values (for inverse matrices in dot product for instance)
# See https://stackoverflow.com/questions/24537791/numpy-matrix-inversion-rounding-errors
np.set_printoptions(suppress=True)
def plotVectors(vecs, cols, alpha=1):
"""
Plot set of vectors.
Parameters
----------
vecs : array-like
Coordinates of the vectors to plot. Each vectors is in an array. For
instance: [[1, 3], [2, 2]] can be used to plot 2 vectors.
cols : array-like
Colors of the vectors. For instance: ['red', 'blue'] will display the
first vector in red and the second in blue.
alpha : float
Opacity of vectors
Returns:
fig : instance of matplotlib.figure.Figure
The figure of the vectors
"""
plt.figure()
plt.axvline(x=0, color='#A9A9A9', zorder=0)
plt.axhline(y=0, color='#A9A9A9', zorder=0)
for i in range(len(vecs)):
x = np.concatenate([[0,0],vecs[i]])
plt.quiver([x[0]],
[x[1]],
[x[2]],
[x[3]],
angles='xy', scale_units='xy', scale=1, color=cols[i],
alpha=alpha)
```
$$
\newcommand\bs[1]{\boldsymbol{#1}}
$$
# Introduction
We will see how to represent systems of equations graphically, how to interpret the number of solutions of a system, what is linear combination and more. As usual, we will use Numpy/Matplotlib as a tool to experiment these concepts and hopefully gain a more concrete understanding.
# Linear Dependence, Independence and Span
Since it is all about systems of linear equations, let's start again with the set of equations:
$$\bs{Ax}=\bs{b}$$
This system corresponds to:
$$
A_{1,1}x_1 + A_{1,2}x_2 + \cdots + A_{1,n}x_n = b_1 \\\\
A_{2,1}x_1 + A_{2,2}x_2 + \cdots + A_{2,n}x_n = b_2 \\\\
\cdots \\\\
A_{m,1}x_1 + A_{m,2}x_2 + \cdots + A_{m,n}x_n = b_n
$$
So we have multiple equations with multiple unknowns. We know $A_{1,1}...A_{m,n}$ and $b_1...b_n$. To solve the system we need to find the values of the variables $x_1...x_n$ that satisfies all equations.
# Number of solutions
The first thing to ask when we face such a system of equations is: what is the number of solutions ?
Three cases can represent the number of solutions of the system of equations $\bs{Ax}=\bs{b}$.
1. No solution
2. 1 solution
3. An infinite number of solutions
## Why there can't be more than 1 solution and less than an infinite number of solutions ?
### Intuition
Simply because we deal with **linear** systems! Two lines can't cross more than once.
To be able to visualize it, let's take two dimensions and two equations. The solutions of the system correspond to the intersection of the lines. One option is that the two lines never cross (parallel). Another option is that they cross once. And finally, the last option is that they cross everywhere (superimposed):
<img src="images/number-solutions-system-equations.png" width="700" alt="Examples of systems of equations with 0, 1 and an infinite number of solutions" title="System of equations with 0, 1 and an infinite number of solutions">
<em>A system of equations has no solution, 1 solution or an infinite number of solutions</em>
<span class='pquote'>
Two lines can't cross more than once but can be either parallel or superimposed
</span>
### Proof
Let's imagine that $\bs{x}$ and $\bs{y}$ are two solutions of our system. This means that
$$
\begin{cases}
\bs{Ax}=\bs{b}\\\\
\bs{Ay}=\bs{b}
\end{cases}
$$
In that case, we will see that $\bs{z}=\alpha \bs{x} + (1-\alpha \bs{y})$ is also a solution for any value of $\alpha$. If $\bs{z}$ is a solution, we can say that $\bs{Az}=\bs{b}$. Indeed, if we plug $\bs{z}$ into the left hand side of the equation we obtain:
$$
\begin{align*}
\bs{Az}&=\bs{A}(\alpha x + (1-\alpha y))\\\\
&=\bs{Ax}\alpha + \bs{A}(1-\alpha y)\\\\
&=\bs{Ax}\alpha + \bs{Ay}(1-\alpha)
\end{align*}
$$
And since $\bs{Ax}=\bs{Ay}=\bs{b}$. This leads to:
$$
\begin{align*}
\bs{Az}&=\bs{b}\alpha + \bs{b}(1-\alpha)\\\\
&=\bs{b}\alpha + \bs{b}-\bs{b}\alpha\\\\
&=\bs{b}
\end{align*}
$$
So $\bs{z}$ is also a solution.
# Matrix representation of the system
As we saw it, the equation $\bs{Ax}=\bs{b}$ can be represented by a matrix $\bs{A}$ containing the weigths of each variable and a vector $\bs{x}$ containing each variable. The product of $\bs{A}$ and $\bs{x}$ gives $\bs{b}$ that is another vector of size $m$:
$$
\begin{bmatrix}
A_{1,1} & A_{1,2} & \cdots & A_{1,n} \\\\
A_{2,1} & A_{2,2} & \cdots & A_{2,n} \\\\
\cdots & \cdots & \cdots & \cdots \\\\
A_{m,1} & A_{m,2} & \cdots & A_{m,n}
\end{bmatrix}
\times
\begin{bmatrix}
x_1 \\\\
x_2 \\\\
\cdots \\\\
x_n
\end{bmatrix}
=
\begin{bmatrix}
b_1 \\\\
b_2 \\\\
\cdots \\\\
b_m
\end{bmatrix}
$$
Which corresponds to the set of linear equations
$$
A_{1,1}x_1 + A_{1,2}x_2 + \cdots + A_{1,n}x_n = b_1 \\\\
A_{2,1}x_1 + A_{2,2}x_2 + \cdots + A_{2,n}x_n = b_2 \\\\
\cdots \\\\
A_{m,1}x_1 + A_{m,2}x_2 + \cdots + A_{m,n}x_n = b_n
$$
Here are some intuitions about what is represented by these matrices. The number of columns of $\bs{A}$ is the number of dimensions of our vector space. It is the number $n$ of directions we can travel by. The number of solutions of our linear system corresponds to the number of ways we can reach $\bs{b}$ by travelling through our $n$ dimensions.
But to understand this, we need to underline that two possibilities exist to represent the system of equations: ***the row figure*** and ***the column figure***.
# Graphical views: Row and column figures
I recommend to look at [this video lesson of Gilbert Strang](http://ia802205.us.archive.org/18/items/MIT18.06S05_MP4/01.mp4). It provides a very nice intuition about these two ways of looking at a system of linear equations.
When you are looking to the matrix $\bs{A}$:
$$
\bs{A}=\begin{bmatrix}
A_{1,1} & A_{1,2} & \cdots & A_{1,n} \\\\
A_{2,1} & A_{2,2} & \cdots & A_{2,n} \\\\
\cdots & \cdots & \cdots & \cdots \\\\
A_{m,1} & A_{m,2} & \cdots & A_{m,n}
\end{bmatrix}
$$
You can consider its rows or its columns separately. Recall that the values are the weights corresponding to each variable. Each row synthetizes one equation. Each column is the set of weights given to 1 variable.
It is possible to draw a different graphical represention of the set of equations looking at the rows or at the columns.
## Graphical view 1: the row figure
The row figure is maybe more usual because it is the representation used when we have only one equation. It can now be extended to an infinite number of equations and unknowns (even if it would be hard to represent a 9-dimensional hyperplane in a 10-dimensional space...).
We said that the solutions of the linear system of equations are the sets of values of $x_1...x_n$ that satisfies all equations, that is to say, the values taken by the unknowns. For instance, in the case of $\bs{A}$ being a ($2 \times 2$) matrix ($n=m=2$) the equations correspond to lines in a 2-dimensional space and the solution of the system is the intersection of these lines.
Note that associating one direction in space to one parameter is only one way to represent the equations. There are number of ways to represent more than 3 parameters systems. For instance, you can add colors to have the representation of a fourth dimension. It is all about **representation**.
<img src="images/representing-features.png" width="900" alt="Different ways of representing features" title="Feature representation">
<em>Graphical representations of features</em>
### Overdetermined and underdetermined systems
A linear system of equations can be viewed as a set of $(n-1)$-dimensional hyperplanes in a *n*-dimensional space. So the linear system can be characterized with its number of equations ($m$) and the number of unknown variables ($n$).
- If there are more equations than unknows the system is called **overdetermined**. In the following example we can see a system of 3 equations (represented by 3 lines) and 2 unknowns (corresponding to 2 dimensions). In this example there is no solution since there is no point belonging to the three lines:
<img src="images/overdetermined-system-linear-equations.png" width="300" alt="Example of an overdetermined system of linear equations with no solution" title="Example of an overdetermined system of linear equations with no solution">
<em>Example of an overdetermined system of linear equations with no solution</em>
- If there is more unknowns than equations the system is called **underdetermined**. In the following picture, there is only 1 equation (1 line) and 2 dimensions. Each point that is on the line is a solution of the system. In this case there is an infinite number of solutions:
<img src="images/underdetermined-system-linear-equations.png" width="300" alt="Example of an underdetermined system of linear equations with an infinite number of solutions" title="Example of an underdetermined system of linear equations with an infinite number of solutions">
<em>Example of an underdetermined system of linear equations with an infinite number of solutions</em>
Let's see few examples of these different cases to clarify that.
### Example 1.
$m=1$, $n=2$: **1 equation and 2 variables**
$$
A_{1,1}x_1 + A_{1,2}x_2 = b_1
$$
The graphical interpretation of $n=2$ is that we have a 2-D space. So we can represent it with 2 axes. Since our hyperplane is of $n-1$-dimensional, we have a 1-D hyperplane. This is simply a line. As $m=1$, we have only one equation. This means that we have only one line characterizing our linear system.
Note that the last equation can also be written in a way that may be more usual:
$$
y = ax + b
$$
with $y$ corresponding to $x_2$, $x$ corresponding to $x_1$, $a$ corresponding to $A_{1,1}$ and $A_{1,2}=1$.
For this first example we will take the following equation:
$$
y = 2x + 1
$$
Let's draw the line of this equation with Numpy and Matplotlib
```
x = np.arange(-10, 10)
y = 2*x + 1
plt.figure()
plt.plot(x, y)
plt.xlim(-2, 10)
plt.ylim(-2, 10)
# draw axes
plt.axvline(x=0, color='#A9A9A9')
plt.axhline(y=0, color='#A9A9A9')
plt.show()
plt.close()
```
#### Solutions
The solutions of this linear system correspond to the value of $x$ and $y$ such as $y=2x+1$. Graphically, it corresponds to each point on the line so there is an infinite number of solutions. For instance, one solution is $x=0$ and $y=1$, or $x=1$ and $y=3$ and so on.
### Example 2.
*m*=2, *n*=2: **2 equations and 2 unknowns**
$$
A_{1,1}x_1 + A_{1,2}x_2 = b_1\\\\
A_{2,1}x_1 + A_{2,2}x_2 = b_2
$$
The graphical interpretation of this system is that we still have lines in a 2-D space. However this time there are 2 lines since there are 2 equations.
Let's take these equations as example:
$$
\begin{cases}
y = 2x + 1\\\\
y = 6x - 2
\end{cases}
$$
```
x = np.arange(-10, 10)
y = 2*x + 1
y1 = 6*x - 2
plt.figure()
plt.plot(x, y)
plt.plot(x, y1)
plt.xlim(-2, 10)
plt.ylim(-2, 10)
# draw axes
plt.axvline(x=0, color='#A9A9A9')
plt.axhline(y=0, color='#A9A9A9')
plt.show()
plt.close()
```
As we have seen, with 2 lines in a 2-D space, there are multiple possible cases. On the above figure, the two lines are crossing so there is one unique solution. If they are superimposed (same equation or equivalent, *cf*. linear dependance bellow) there are a infinite number of solutions since each points of the lines corresponds to an intersection. If they are parallel, there is no solution.
The same thing can be observed with other values of $m$ (number of equations) and $n$ (number of dimensions). For instance, two 2-D planes in a 3-D space can be superposed (infinitely many solutions), or crossed (infinitely many solutions since their crossing is a line), or parallel (no solution).
### Example 3.
*m*=3, *n*=2: **3 equations and 2 unknowns**
$$
A_{1,1}x_1 + A_{1,2}x_2 = b_1\\\\
A_{2,1}x_1 + A_{2,2}x_2 = b_2\\\\
A_{3,1}x_1 + A_{3,2}x_2 = b_3
$$
The same idea stands with more than 2 equations in a 2-D space. In that example we have the following 3 equations:
$$
\begin{cases}
y = 2x + 1\\\\
y = 6x - 2\\\\
y = \frac{1}{10}x+6
\end{cases}
$$
```
x = np.arange(-10, 10)
y = 2*x + 1
y1 = 6*x - 2
y2 = 0.1*x+6
plt.figure()
plt.plot(x, y)
plt.plot(x, y1)
plt.plot(x, y2)
plt.xlim(-2, 10)
plt.ylim(-2, 10)
# draw axes
plt.axvline(x=0, color='#A9A9A9')
plt.axhline(y=0, color='#A9A9A9')
plt.show()
plt.close()
```
In the above case, there is 3 equations and no solution because there is no point in space that is on each of these lines.
## Linear combination
Before going to the column figure, we need to talk about linear combination. The linear combination of 2 vectors corresponds to their weighted sum.
### Example 4.
Let's take two vectors
$$
\vec{u}=
\begin{bmatrix}
1 \\\\
3
\end{bmatrix}
$$
and
$$
\vec{v}=
\begin{bmatrix}
2 \\\\
1
\end{bmatrix}
$$
These two vectors have 2 dimensions and thus contain coordinates in 2-D.
The linear combination of $\vec{u}$ and $\vec{v}$ is
$$
a\vec{u}+b\vec{v}= a
\begin{bmatrix}
1 \\\\
3
\end{bmatrix} + b\begin{bmatrix}
2 \\\\
1
\end{bmatrix}
$$
with $a$ and $b$ the weights of the vectors.
Graphically, the vectors are added to reach a specific point in space. For example if $a=2$ and $b=1$:
$$
2\vec{u}+\vec{v}= 2
\begin{bmatrix}
1 \\\\
3
\end{bmatrix} +
\begin{bmatrix}
2 \\\\
1
\end{bmatrix} =
\begin{bmatrix}
2 \cdot 1 + 2 \\\\
2 \cdot 3 + 1
\end{bmatrix} =
\begin{bmatrix}
4 \\\\
7
\end{bmatrix}
$$
The sum of $\vec{u}$ and $\vec{v}$ is a vector that will reach the point of corrdinates $(4, 7)$. To show that on a plot, I will use the custom function `plotVectors()`. It takes a set of coordinates and an array of colors as input and plot the corresponding vectors. So let's plot $\vec{u}$ and $\vec{v}$:
```
orange = '#FF9A13'
blue = '#1190FF'
plotVectors([[1, 3], [2, 1]], [orange, blue])
plt.xlim(0, 5)
plt.ylim(0, 5)
```
We will now add these vectors and their weights. This gives:
```
# Weigths of the vectors
a = 2
b = 1
# Start and end coordinates of the vectors
u = [0,0,1,3]
v = [2,6,2,1]
plt.quiver([u[0], a*u[0], b*v[0]],
[u[1], a*u[1], b*v[1]],
[u[2], a*u[2], b*v[2]],
[u[3], a*u[3], b*v[3]],
angles='xy', scale_units='xy', scale=1, color=[orange, orange, blue])
plt.xlim(-1, 8)
plt.ylim(-1, 8)
# Draw axes
plt.axvline(x=0, color='#A9A9A9')
plt.axhline(y=0, color='#A9A9A9')
plt.scatter(4,7,marker='x',s=50)
# Draw the name of the vectors
plt.text(-0.5, 2, r'$\vec{u}$', color=orange, size=18)
plt.text(0.5, 4.5, r'$\vec{u}$', color=orange, size=18)
plt.text(2.5, 7, r'$\vec{v}$', color=blue, size=18)
plt.show()
plt.close()
```
We can see that we end up with the coordinates ($4$, $7$).
## Span
Take the vectors $\vec{u}$ and $\vec{v}$ from the previous example and think about all the points you can reach by their combination changing $a$ and $b$. This set of points is the span of the set of vectors $\{\vec{u}, \vec{v}\}$.
```
# Defining u and v
u = [1, 3]
v = [2, 1]
# Ploting a sample of the set of points in the span of u and v
for a in range(-10, 10):
for b in range(-10, 10):
plt.scatter(u[0] * a + v[0] * b, u[1] * a + v[1] * b, marker='.', color=blue)
# Defining x and y sizes
plt.xlim(-50, 50)
plt.ylim(-50, 50)
# Draw axes
plt.axvline(x=0, color='#A9A9A9')
plt.axhline(y=0, color='#A9A9A9')
plt.show()
plt.close()
```
## Note on spaces and subspaces
(For more details see Strang (2006), p.70)
The space of a vector determines all the values that can be taken by this vector. The vector spaces are denoted $\mathbb{R}$ because the values are real numbers. If there are multiple dimensions the space is denoted $\mathbb{R}^n$ with $n$ corresponding to the number of dimensions. For instance $\mathbb{R}^2$ is the space of the usual $x$-$y$ plane where $x$ and $y$ values are real numbers.
If you take a 2-dimensional plane in $\mathbb{R}^3$ (3-dimensional space), this plane is a **subspace** of your original $\mathbb{R}^3$ space. On the same manner, if you start with a $\mathbb{R}^2$ space and take a line in this space, this line is a subspace of the original space.
The linear combination of vectors gives vectors in the original space. Every linear combination of vectors inside a space will stay in this space. For instance, if you take 2 lines in a $\mathbb{R}^2$ space, any linear combinations will give you a vector in the same $\mathbb{R}^2$ space.
<span class='pquote'>
The linear combination of vectors gives vectors in the original space
</span>
## Graphical view 2: the column figure
It is also possible to represent the set of equations by considering that the solution vector $\bs{b}$ corresponds to a linear combination of each columns multiplied by their weights.
From the set of equations:
$$
A_{1,1}x_1 + A_{1,2}x_2 + A_{1,n}x_n = b_1 \\\\
A_{2,1}x_1 + A_{2,2}x_2 + A_{2,n}x_n = b_2 \\\\
\cdots \\\\
A_{m,1}x_1 + A_{m,2}x_2 + A_{m,n}x_n = b_m
$$
The column form is then:
$$
x_1
\begin{bmatrix}
A_{1,1}\\\\
A_{2,1}\\\\
A_{m,1}
\end{bmatrix}
+
x_2
\begin{bmatrix}
A_{1,2}\\\\
A_{2,2}\\\\
A_{m,2}
\end{bmatrix}
+
x_n
\begin{bmatrix}
A_{1,n}\\\\
A_{2,n}\\\\
A_{m,n}
\end{bmatrix}
=
\begin{bmatrix}
b_1\\\\
b_2\\\\
b_m
\end{bmatrix}
$$
On a graphical point of view, we have to travel from the origin (zero on every dimensions) to the point of coordinate $\bs{b}$. The columns of $\bs{A}$ give us the directions we can travel by and their weights are the length of the way in that direction.
<span class='pquote'>
The columns of $\bs{A}$ give us the directions we can travel by and their weights are the length of the way in each direction.
</span>
### Example 5.
$m=2$, $n=2$: 2 equations and 2 variables
$$
A_{1,1}x_1 + A_{1,2}x_2 = b_1\\\\
A_{2,1}x_1 + A_{2,2}x_2 = b_2
$$
$$
\begin{cases}
y = \frac{1}{2}x+1\\\\
y = -x + 4
\end{cases}
\Leftrightarrow
\begin{cases}
\frac{1}{2}x-y = -1\\\\
x+y=4
\end{cases}
$$
So here is the matrix $\bs{A}$:
$$
\bs{A}=
\begin{bmatrix}
\frac{1}{2} & -1 \\\\
1 & 1
\end{bmatrix}
$$
The column figure gives us:
$$
x
\begin{bmatrix}
\frac{1}{2} \\\\
1
\end{bmatrix}
+
y
\begin{bmatrix}
-1 \\\\
1
\end{bmatrix}
=
\begin{bmatrix}
-1 \\\\
4
\end{bmatrix}
$$
The goal is to find the value of the weights ($x$ and $y$) for which the linear combination of the vector
$$
\begin{bmatrix}
\frac{1}{2} \\\\
1
\end{bmatrix}
$$
and
$$
\begin{bmatrix}
-1 \\\\
1
\end{bmatrix}
$$
gives the vector
$$
\begin{bmatrix}
-1 \\\\
4
\end{bmatrix}
$$
We will solve the system graphically by plotting the equations and looking for their intersection:
```
x = np.arange(-10, 10)
y = 0.5*x + 1
y1 = -x + 4
plt.figure()
plt.plot(x, y)
plt.plot(x, y1)
plt.xlim(-2, 10)
plt.ylim(-2, 10)
# draw axes
plt.axvline(x=0, color='#A9A9A9')
plt.axhline(y=0, color='#A9A9A9')
plt.show()
plt.close()
```
We can see that the solution (the intersection of the lines representing our two equations) is $x=2$ and $y=2$. This means that the linear combination is the following:
$$
2
\begin{bmatrix}
\frac{1}{2} \\\\
1
\end{bmatrix}
+
2
\begin{bmatrix}
-1 \\\\
1
\end{bmatrix}
=
\begin{bmatrix}
-1 \\\\
4
\end{bmatrix}
$$
Let's say that
$$
\vec{u}=
\begin{bmatrix}
\frac{1}{2} \\\\
1
\end{bmatrix}
$$
and
$$
\vec{v}=
\begin{bmatrix}
-1 \\\\
1
\end{bmatrix}
$$
To talk in term of the column figure we can reach the point of coordinates $(-1, 4)$ if we add two times the vector $\vec{u}$ and two times the vector $\vec{v}$. Let's check that:
```
u = [0,0,0.5,1]
u_bis = [u[2],u[3],u[2],u[3]]
v = [2*u[2],2*u[3],-1,1]
v_bis = [2*u[2]-1,2*u[3]+1,v[2],v[3]]
plt.quiver([u[0], u_bis[0], v[0], v_bis[0]],
[u[1], u_bis[1], v[1], v_bis[1]],
[u[2], u_bis[2], v[2], v_bis[2]],
[u[3], u_bis[3], v[3], v_bis[3]],
angles='xy', scale_units='xy', scale=1, color=[blue, blue, orange, orange])
# plt.rc('text', usetex=True)
plt.xlim(-1.5, 2)
plt.ylim(-0.5, 4.5)
# draw axes
plt.axvline(x=0, color='#A9A9A9')
plt.axhline(y=0, color='#A9A9A9')
plt.scatter(-1,4,marker='x',s=50)
plt.text(0, 0.5, r'$\vec{u}$', color=blue, size=18)
plt.text(0.5, 1.5, r'$\vec{u}$', color=blue, size=18)
plt.text(0.5, 2.7, r'$\vec{v}$', color=orange, size=18)
plt.text(-0.8, 3, r'$\vec{v}$', color=orange, size=18)
plt.show()
plt.close()
```
We can see that it is working! We arrive to the point ($-1$, $4$).
## Determine if the system has one and only one solution for every value of $\bs{b}$
We will now see how to determine if a system of equations has one and only one solution. Note that this is only the general cases. This can be split into two requirements:
1. The system must have at least one solution
2. Then, the system must have **only** one solution
### Requirement 1. Underdetermined system: the system must have at least one solution for each value of $\bs{b}$: $n\geq m$
<span class='pquote'>
An underdetermined system of equations is a system with less equations than unknowns
</span>
If we want our system to have one and only one solution a first requirement is that $n$ must not be bigger than $m$.
Let's take the example of a ($2\times 3$) matrix that corresponds to a set of 2 equations with 3 unknowns variables:
<div>
$$
\begin{cases}
8x+y+z=1\\\\
x+y+z=1
\end{cases}
$$
</div>
<div>
$$
x
\begin{bmatrix}
8 \\\\
1
\end{bmatrix}
+
y
\begin{bmatrix}
1 \\\\
1
\end{bmatrix}
+
z
\begin{bmatrix}
1 \\\\
1
\end{bmatrix}
=
\begin{bmatrix}
1 \\\\
1
\end{bmatrix}
$$
</div>
Here is the representation of the planes plotted with the help of this [website](https://technology.cpm.org/general/3dgraph/):
<img src="images/intersection-2-planes-line.png" alt="Plot showing two planes. The intersection of the two planes is a line" title="The intersection of the two planes is a line" width="500">
<em>The intersection of the two planes is a line</em>
We can see that in the best case the two planes are not parallel and there are solutions to the set of equations. It means that it exists some points that rely on both planes. But we can also see that there is inevitably an infinite number of points on the intersection (a line that we can see on the figure). We need a third plane to have a unique solution.
### Requirement 2. Overdetermined system: the system must have **only** one solution for each value of $\bs{b}$: $m\geq n$
<span class='pquote'>
An overdetermined system of equations is a system with more equations than unknowns
</span>
The column figure is helpful to understand why the linear system has usually no solution if $n$ (the number of unknowns) is smaller than $m$ (the number of equations). Let's add 1 equation to the above system in order to end up with a ($3\times2$) matrix (3 equations and 2 unknowns):
<div>
$$
\begin{cases}
y = \frac{1}{2}x+1\\\\
y = -x + 4\\\\
y = 7x + 2
\end{cases}
\Leftrightarrow
\begin{cases}
\frac{1}{2}x-y = -1\\\\
x+y=4\\\\
7x-y=2
\end{cases}
$$
</div>
This corresponds to:
<div>
$$
x
\begin{bmatrix}
\frac{1}{2} \\\\
1 \\\\
7
\end{bmatrix}
+
y
\begin{bmatrix}
-1 \\\\
1 \\\\
-1
\end{bmatrix}
=
\begin{bmatrix}
-1 \\\\
4 \\\\
2
\end{bmatrix}
$$
</div>
So we are still traveling in our 2-dimensional space (see the plot of the column space above) but the point that we are looking for is defined by 3 dimensions. There are cases where the third coordinate does not rely on our 2-dimensional $x$-$y$ plane. In that case no solution exists.
<span class='pquote'>
We are traveling in a 2D space but the solution is defined by 3 dimensions. If the third coordinate does not rely on our 2D $x$-$y$ plane then there is no solution.
</span>
### Linear dependence
The number of columns can thus provide information on the number of solutions. But the number that we have to take into account is the number of **linearly independent** columns. Columns are linearly dependent if one of them is a linear combination of the others. Thinking in the column picture, the direction of two linearly dependent vectors is the same. This doesn't add a dimension that we can use to travel and reach $\bs{b}$.
Here is an example of linear system containing linear dependency:
$$
\begin{cases}
y = 2x+6\\\\
y = 2x
\end{cases}
\Leftrightarrow
\begin{cases}
2x-y = -6\\\\
2x-y=0
\end{cases}
$$
The row figure shows that the system has no solution:
```
x = np.arange(-10, 10)
y = 2*x + 6
y1 = 2*x
plt.figure()
plt.plot(x, y)
plt.plot(x, y1)
plt.xlim(-2, 10)
plt.ylim(-2, 10)
# draw axes
plt.axvline(x=0, color='#A9A9A9')
plt.axhline(y=0, color='#A9A9A9')
plt.show()
plt.close()
```
Since the lines are parallel, there is no point at their intersection.
The column figure illustrates the point as well:
$$
x
\begin{bmatrix}
2 \\\\
2
\end{bmatrix}
+
y
\begin{bmatrix}
-1 \\\\
-1
\end{bmatrix}
=
\begin{bmatrix}
-6 \\\\
0
\end{bmatrix}
$$
```
u = [0,0,2,2]
v = [0,0,-1,-1]
plt.quiver([u[0], v[0]],
[u[1], v[1]],
[u[2], v[2]],
[u[3], v[3]],
angles='xy', scale_units='xy', scale=1, color=[blue, orange])
plt.xlim(-7, 3)
plt.ylim(-2, 3)
# draw axes
plt.axvline(x=0, color='#A9A9A9')
plt.axhline(y=0, color='#A9A9A9')
plt.scatter(-6,0,marker='x',s=150)
plt.text(-6, 0.5, r'$b$', color='b', size=18)
plt.show()
plt.close()
```
We would like to go to $b$ but the only path we can take is the blue/orange line. The second equation doesn't provide us with a new direction to take since it is just a linear combination of the first one.
Thus, an overdetermined system of independant equations has at most 1 solution.
### Square matrix
How could we satisfy both requirements ($m\geq n$ and $n\geq m$): we must have $m=n$!
The resulting of all of this is that the system needs a **square matrix** $\bs{A}$ ($m=n$) with linearly independant columns to have a unique solution for every values of $\bs{b}$.
<span class='pquote'>
The system needs a **square matrix** $\bs{A}$ ($m=n$) with linearly independant columns to have a unique solution for every values of $\bs{b}$
</span>
The inverse of a matrix exists only if the set of equations has one and only one solution for each value of $\bs{b}$ because:
- The matrix $\bs{A}$ cannot have more than 1 inverse. Imagine that $\bs{A}$ has 2 inverses $\bs{B}$ and $\bs{C}$ such as $\bs{AB}=\bs{I}$ and $\bs{AC}=\bs{I}$. This would mean that $\bs{B}=\bs{C}$.
- The solution of the system $\bs{Ax}=\bs{b}$ is $\bs{x}=\bs{A} ^{-1} \bs{b}$. So if there are multiple solutions, there are multiple inverses and the first point is not met.
|
github_jupyter
|
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
sns.set()
%pylab inline
pylab.rcParams['figure.figsize'] = (4, 4)
# Avoid inaccurate floating values (for inverse matrices in dot product for instance)
# See https://stackoverflow.com/questions/24537791/numpy-matrix-inversion-rounding-errors
np.set_printoptions(suppress=True)
def plotVectors(vecs, cols, alpha=1):
"""
Plot set of vectors.
Parameters
----------
vecs : array-like
Coordinates of the vectors to plot. Each vectors is in an array. For
instance: [[1, 3], [2, 2]] can be used to plot 2 vectors.
cols : array-like
Colors of the vectors. For instance: ['red', 'blue'] will display the
first vector in red and the second in blue.
alpha : float
Opacity of vectors
Returns:
fig : instance of matplotlib.figure.Figure
The figure of the vectors
"""
plt.figure()
plt.axvline(x=0, color='#A9A9A9', zorder=0)
plt.axhline(y=0, color='#A9A9A9', zorder=0)
for i in range(len(vecs)):
x = np.concatenate([[0,0],vecs[i]])
plt.quiver([x[0]],
[x[1]],
[x[2]],
[x[3]],
angles='xy', scale_units='xy', scale=1, color=cols[i],
alpha=alpha)
x = np.arange(-10, 10)
y = 2*x + 1
plt.figure()
plt.plot(x, y)
plt.xlim(-2, 10)
plt.ylim(-2, 10)
# draw axes
plt.axvline(x=0, color='#A9A9A9')
plt.axhline(y=0, color='#A9A9A9')
plt.show()
plt.close()
x = np.arange(-10, 10)
y = 2*x + 1
y1 = 6*x - 2
plt.figure()
plt.plot(x, y)
plt.plot(x, y1)
plt.xlim(-2, 10)
plt.ylim(-2, 10)
# draw axes
plt.axvline(x=0, color='#A9A9A9')
plt.axhline(y=0, color='#A9A9A9')
plt.show()
plt.close()
x = np.arange(-10, 10)
y = 2*x + 1
y1 = 6*x - 2
y2 = 0.1*x+6
plt.figure()
plt.plot(x, y)
plt.plot(x, y1)
plt.plot(x, y2)
plt.xlim(-2, 10)
plt.ylim(-2, 10)
# draw axes
plt.axvline(x=0, color='#A9A9A9')
plt.axhline(y=0, color='#A9A9A9')
plt.show()
plt.close()
orange = '#FF9A13'
blue = '#1190FF'
plotVectors([[1, 3], [2, 1]], [orange, blue])
plt.xlim(0, 5)
plt.ylim(0, 5)
# Weigths of the vectors
a = 2
b = 1
# Start and end coordinates of the vectors
u = [0,0,1,3]
v = [2,6,2,1]
plt.quiver([u[0], a*u[0], b*v[0]],
[u[1], a*u[1], b*v[1]],
[u[2], a*u[2], b*v[2]],
[u[3], a*u[3], b*v[3]],
angles='xy', scale_units='xy', scale=1, color=[orange, orange, blue])
plt.xlim(-1, 8)
plt.ylim(-1, 8)
# Draw axes
plt.axvline(x=0, color='#A9A9A9')
plt.axhline(y=0, color='#A9A9A9')
plt.scatter(4,7,marker='x',s=50)
# Draw the name of the vectors
plt.text(-0.5, 2, r'$\vec{u}$', color=orange, size=18)
plt.text(0.5, 4.5, r'$\vec{u}$', color=orange, size=18)
plt.text(2.5, 7, r'$\vec{v}$', color=blue, size=18)
plt.show()
plt.close()
# Defining u and v
u = [1, 3]
v = [2, 1]
# Ploting a sample of the set of points in the span of u and v
for a in range(-10, 10):
for b in range(-10, 10):
plt.scatter(u[0] * a + v[0] * b, u[1] * a + v[1] * b, marker='.', color=blue)
# Defining x and y sizes
plt.xlim(-50, 50)
plt.ylim(-50, 50)
# Draw axes
plt.axvline(x=0, color='#A9A9A9')
plt.axhline(y=0, color='#A9A9A9')
plt.show()
plt.close()
x = np.arange(-10, 10)
y = 0.5*x + 1
y1 = -x + 4
plt.figure()
plt.plot(x, y)
plt.plot(x, y1)
plt.xlim(-2, 10)
plt.ylim(-2, 10)
# draw axes
plt.axvline(x=0, color='#A9A9A9')
plt.axhline(y=0, color='#A9A9A9')
plt.show()
plt.close()
u = [0,0,0.5,1]
u_bis = [u[2],u[3],u[2],u[3]]
v = [2*u[2],2*u[3],-1,1]
v_bis = [2*u[2]-1,2*u[3]+1,v[2],v[3]]
plt.quiver([u[0], u_bis[0], v[0], v_bis[0]],
[u[1], u_bis[1], v[1], v_bis[1]],
[u[2], u_bis[2], v[2], v_bis[2]],
[u[3], u_bis[3], v[3], v_bis[3]],
angles='xy', scale_units='xy', scale=1, color=[blue, blue, orange, orange])
# plt.rc('text', usetex=True)
plt.xlim(-1.5, 2)
plt.ylim(-0.5, 4.5)
# draw axes
plt.axvline(x=0, color='#A9A9A9')
plt.axhline(y=0, color='#A9A9A9')
plt.scatter(-1,4,marker='x',s=50)
plt.text(0, 0.5, r'$\vec{u}$', color=blue, size=18)
plt.text(0.5, 1.5, r'$\vec{u}$', color=blue, size=18)
plt.text(0.5, 2.7, r'$\vec{v}$', color=orange, size=18)
plt.text(-0.8, 3, r'$\vec{v}$', color=orange, size=18)
plt.show()
plt.close()
x = np.arange(-10, 10)
y = 2*x + 6
y1 = 2*x
plt.figure()
plt.plot(x, y)
plt.plot(x, y1)
plt.xlim(-2, 10)
plt.ylim(-2, 10)
# draw axes
plt.axvline(x=0, color='#A9A9A9')
plt.axhline(y=0, color='#A9A9A9')
plt.show()
plt.close()
u = [0,0,2,2]
v = [0,0,-1,-1]
plt.quiver([u[0], v[0]],
[u[1], v[1]],
[u[2], v[2]],
[u[3], v[3]],
angles='xy', scale_units='xy', scale=1, color=[blue, orange])
plt.xlim(-7, 3)
plt.ylim(-2, 3)
# draw axes
plt.axvline(x=0, color='#A9A9A9')
plt.axhline(y=0, color='#A9A9A9')
plt.scatter(-6,0,marker='x',s=150)
plt.text(-6, 0.5, r'$b$', color='b', size=18)
plt.show()
plt.close()
| 0.88526 | 0.953708 |
<h1>Table of Contents<span class="tocSkip"></span></h1>
<div class="toc"><ul class="toc-item"><li><span><a href="#Import-pandas,-numpy,-and-matplotlib,-and-load-the-COVID-case-totals-data" data-toc-modified-id="Import-pandas,-numpy,-and-matplotlib,-and-load-the-COVID-case-totals-data-1"><span class="toc-item-num">1 </span>Import pandas, numpy, and matplotlib, and load the COVID case totals data</a></span></li><li><span><a href="#Let's-remind-ourselves-of-the-structure-of-the-data" data-toc-modified-id="Let's-remind-ourselves-of-the-structure-of-the-data-2"><span class="toc-item-num">2 </span>Let's remind ourselves of the structure of the data</a></span></li><li><span><a href="#Get-the-descriptive-statistics-on-the-COVID-totals-and-demographic-columns" data-toc-modified-id="Get-the-descriptive-statistics-on-the-COVID-totals-and-demographic-columns-3"><span class="toc-item-num">3 </span>Get the descriptive statistics on the COVID totals and demographic columns</a></span></li><li><span><a href="#Take-a-closer-look-at-the-distribution-of-values-for-the-cases-and-deaths-columns" data-toc-modified-id="Take-a-closer-look-at-the-distribution-of-values-for-the-cases-and-deaths-columns-4"><span class="toc-item-num">4 </span>Take a closer look at the distribution of values for the cases and deaths columns</a></span></li><li><span><a href="#View-the-distribution-of-total-cases" data-toc-modified-id="View-the-distribution-of-total-cases-5"><span class="toc-item-num">5 </span>View the distribution of total cases</a></span></li></ul></div>
# Import pandas, numpy, and matplotlib, and load the COVID case totals data
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import watermark
%load_ext watermark
%watermark -n -v -iv
pd.set_option('display.width', 75)
pd.set_option('display.max_columns', 7)
pd.set_option('display.max_rows', 20)
pd.options.display.float_format = '{:,.2f}'.format
covidtotals = pd.read_csv('data/covidtotals.csv', parse_dates=['lastdate'])
covidtotals.set_index('iso_code', inplace=True)
```
# Let's remind ourselves of the structure of the data
```
covidtotals.shape
covidtotals.sample(2, random_state=1).T
```
# Get the descriptive statistics on the COVID totals and demographic columns
```
covidtotals.describe()
```
# Take a closer look at the distribution of values for the cases and deaths columns
```
totvars = [
'location', 'total_cases', 'total_deaths', 'total_cases_pm',
'total_deaths_pm'
]
covidtotals[totvars].quantile(np.arange(0.0, 1.1, 0.1))
```
# View the distribution of total cases
```
plt.hist(covidtotals['total_cases'] / 1000, bins=12)
plt.title('Total Covid Cases')
plt.xlabel('Cases')
plt.ylabel('Number of Countries')
plt.show()
```
|
github_jupyter
|
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import watermark
%load_ext watermark
%watermark -n -v -iv
pd.set_option('display.width', 75)
pd.set_option('display.max_columns', 7)
pd.set_option('display.max_rows', 20)
pd.options.display.float_format = '{:,.2f}'.format
covidtotals = pd.read_csv('data/covidtotals.csv', parse_dates=['lastdate'])
covidtotals.set_index('iso_code', inplace=True)
covidtotals.shape
covidtotals.sample(2, random_state=1).T
covidtotals.describe()
totvars = [
'location', 'total_cases', 'total_deaths', 'total_cases_pm',
'total_deaths_pm'
]
covidtotals[totvars].quantile(np.arange(0.0, 1.1, 0.1))
plt.hist(covidtotals['total_cases'] / 1000, bins=12)
plt.title('Total Covid Cases')
plt.xlabel('Cases')
plt.ylabel('Number of Countries')
plt.show()
| 0.260389 | 0.890485 |
# Spark High-Level API

Source: https://static.packt-cdn.com/products/9781785888359/graphics/8bffbd94-04f7-46e3-a1e9-0d6046d2dcab.png
## Overview of Spark SQL
Creating DataFrames
- RDD
- createDataFrame()
- Text file
- read.text()
- JSON file
- read.json()
- read,json(RDD)
- Parquet file
- read.parquet()
- Table in a relational database
- Temporary table in Spark
DataFrame to RDD
- rdd()
Schemas
- Inferring schemas
- Why it is not optimal practice
- Specifying schemas
- Using StructType and StructField
- Using DDL string (schema = “author STRING, title STRING, pages INT”)
- Metadata
- printSchema()
- columns()
- dtypes()
- Actions
- show()
- Transforms
- select() and alias()
- drop()
- filter() / where()
- distinct()
- dropDuplicates()
- sample
- sampleBy()
- limit()
- orderBy() / sort()
- groupBy()
Operations that return an RDD
- rdd.map()
- rdd.flatMap()
pyspark.sql.functions module
- String functions
- Math functions
- Statistics functions
- Date functions
- Hashing functions
- Algorithms (sounded, levenstein)
- Windowing functions
User defined functions
- udf()
- pandas_udf()
Multiple DataFrames
- join(other, on, how)
- union(), unionAll()
- intersect()
- subtract()
Persistence
- cache()
- persist(:
- unpersist()
- cacheTable()
- clearCache()
- repartition()
- coalesce()
Output
- write.csv()
- write.parquet()
- write.json()
Spark SQL
- df.createOrReplaceTempView
- sql()
- table()
```
from pyspark.sql import SparkSession
import pyspark.sql.functions as F
import pyspark.sql.types as T
spark = (
SparkSession.builder
.master("local")
.appName("BIOS-823")
.config("spark.executor.cores", 4)
.getOrCreate()
)
spark.version
spark.conf.get('spark.executor.cores')
```
## Create a Spark DataFrame
```
df = spark.range(3)
df.show(3)
%%file data/test.csv
number,letter
0,a
1,c
2,b
3,a
4,b
5,c
6,a
7,a
8,a
9,b
10,b
11,c
12,c
13,b
14,b
```
#### Implicit schema
```
df = (
spark.read.
format('csv').
option('header', 'true').
option('inferSchema', 'true').
load('csv/test.csv')
)
df.show(3)
df.printSchema()
```
#### Explicit schema
For production use, you should provide an explicit schema to reduce risk of error.
```
schema = T.StructType([
T.StructField("number", T.DoubleType()),
T.StructField("letter", T.StringType()),
])
df = (
spark.read.
format('csv').
option('header', 'true').
schema(schema).
load('csv/test.csv')
)
df.show(3)
df.printSchema()
```
#### Alternative way to specify schema
You can use SQL DDL syntax to specify a schema as well.
```
schema = 'number DOUBLE, letter STRING'
df_altschema = (
spark.read.
format('csv').
option('header', 'true').
schema(schema=schema).
load('csv/test.csv')
)
df_altschema.take(3)
df_altschema.printSchema()
```
### Persist
```
df.cache()
```
## Data manipulation
### Select
```
df.select('number').show(3)
from pyspark.sql.functions import col, expr
df.select(col('number').alias('index')).show(3)
df.select(expr('number as x')).show(3)
df.withColumnRenamed('number', 'x').show(3)
```
## Filter
```
df.filter('number % 2 == 0').show(3)
df.filter("number % 2 == 0 AND letter == 'a'").show(3)
```
## Sort
```
df.sort(df.number.desc()).show(3)
df.sort('number', ascending=False).show(3)
df.orderBy(df.letter.desc()).show(3)
```
## Transform
```
df.selectExpr('number*2 as x').show(3)
df.selectExpr('number', 'letter', 'number*2 as x').show(3)
df.withColumn('x', expr('number*2')).show(3)
```
## Sumarize
```
import pyspark.sql.functions as F
df.agg(F.min('number'),
F.max('number'),
F.min('letter'),
F.max('letter')).show()
```
## Group by
```
(
df.groupby('letter').
agg(F.mean('number'), F.stddev_samp('number')).show()
)
```
## Window functions
```
from pyspark.sql.window import Window
ws = (
Window.partitionBy('letter').
orderBy(F.desc('number')).
rowsBetween(Window.unboundedPreceding, Window.currentRow)
)
df.groupby('letter').agg(F.sum('number')).show()
df.show()
(
df.select('letter', F.sum('number').
over(ws).
alias('rank')).show()
)
```
## SQL
```
df.createOrReplaceTempView('df_table')
spark.sql('''SELECT * FROM df_table''').show(3)
spark.sql('''
SELECT letter, mean(number) AS mean,
stddev_samp(number) AS sd from df_table
WHERE number % 2 = 0
GROUP BY letter
ORDER BY letter DESC
''').show()
```
## String operatons
```
from pyspark.sql.functions import split, lower, explode
import pandas as pd
s = spark.createDataFrame(
pd.DataFrame(
dict(sents=('Thing 1 and Thing 2',
'The Quick Brown Fox'))))
s.show()
from pyspark.sql.functions import regexp_replace
s1 = (
s.select(explode(split(lower(expr('sents')), ' '))).
sort('col')
)
s1.show()
s1.groupBy('col').count().show()
s.createOrReplaceTempView('s_table')
spark.sql('''
SELECT regexp_replace(sents, 'T.*?g', 'FOO')
FROM s_table
''').show()
```
### Numeric operations
```
from pyspark.sql.functions import log1p, randn
df.selectExpr('number', 'log1p(number)', 'letter').show(3)
(
df.selectExpr('number', 'randn() as random').
stat.corr('number', 'random')
)
```
### Date and time
```
dt = (
spark.range(3).
withColumn('today', F.current_date()).
withColumn('tomorrow', F.date_add('today', 1)).
withColumn('time', F.current_timestamp())
)
dt.show()
dt.printSchema()
```
### Nulls
```
%%file data/test_null.csv
number,letter
0,a
1,
2,b
3,a
4,b
5,
6,a
7,a
8,
9,b
10,b
11,c
12,
13,b
14,b
dn = (
spark.read.
format('csv').
option('header', 'true').
option('inferSchema', 'true').
load('csv/test_null.csv')
)
dn.printSchema()
dn.show()
dn.na.drop().show()
dn.na.fill('Missing').show()
```
## UDF
To avoid degrading performance, avoid using UDF if you can use the functions in `pyspark.sql.functions`. If you must use UDFs, prefer `pandas_udf` to `udf` where possible.
```
from pyspark.sql.functions import udf, pandas_udf
```
### Standard Python UDF
```
@udf('double')
def square(x):
return x**2
df.select('number', square('number')).show(3)
```
### Pandas UDF
This can be tricky to set up. I use Oracle Java SDK v11 and set the following environment variables.
```bash
export JAVA_HOME=$(/usr/libexec/java_home -v 11)
export JAVA_TOOL_OPTIONS="-Dio.netty.tryReflectionSetAccessible=true"
```
```
@pandas_udf('double')
def scale(x):
return (x - x.mean())/x.std()
df.select(scale('number')).show(3)
```
#### Grouped agg
```
import warnings
warnings.simplefilter('ignore', UserWarning)
@pandas_udf('double', F.PandasUDFType.GROUPED_AGG)
def gmean(x):
return x.mean()
df.groupby('letter').agg(gmean('number')).show()
```
### Spark 3
Use type hints rather than specify pandas UDF type
See [blog](https://databricks.com/blog/2020/05/20/new-pandas-udfs-and-python-type-hints-in-the-upcoming-release-of-apache-spark-3-0.html)
```
@pandas_udf('double')
def gmean1(x: pd.Series) -> float:
return x.mean()
df.groupby('letter').agg(gmean1('number')).show()
```
#### Grouped map
```
@pandas_udf(df.schema, F.PandasUDFType.GROUPED_MAP)
def gscale(pdf):
return pdf.assign(
number = (pdf.number - pdf.number.mean()) /
pdf.number.std())
df.groupby('letter').apply(gscale).show()
```
### Spark 3
Use the new `pandas` function API. Currently, type annotations are not used in the function API.
See [blog](https://databricks.com/blog/2020/05/20/new-pandas-udfs-and-python-type-hints-in-the-upcoming-release-of-apache-spark-3-0.html)
#### `applyinPandas`
This implements the `split-apply-combine` pattern. Method of grouped DataFrame.
- Variant 1: Function takes a single DataFrame input
- Variant 2: Function takes a tuple of keys, and a DataFrame input
```
def gscale1(pdf: pd.DataFrame) -> pd.DataFrame:
num = pdf.number
return pdf.assign(
number = (num - num.mean()) / num.std())
df.groupby('letter').applyInPandas(gscale1, schema=df.schema).show()
def gsum(key, pdf):
return pd.DataFrame([key + (pdf.number.sum(),)])
df.groupby('letter').applyInPandas(gsum, 'letter string, number long').show()
```
Of course, you do not need a UDF in this example!
```
df.groupBy('letter').sum().show()
```
So shoudl only be used for truly custom functions.
```
def func(pdf: pd.DataFrame) -> int:
return (pdf.number.astype('str').str.len()).sum()
def gcustom(key, pdf):
return pd.DataFrame([key + (func(pdf),)])
df.groupby('letter').applyInPandas(gcustom, 'letter string, number long').show()
```
#### `mapinPandas`
This works on iterators. Method of DataFrame. Can be used to implement a filter.
```
def even(it):
for pdf in it:
yield pdf[pdf.number % 2 == 0]
df.mapInPandas(even, 'letter string, number long').show()
```
## Joins
```
names = 'ann ann bob bob chcuk'.split()
courses = '821 823 821 824 823'.split()
pdf1 = pd.DataFrame(dict(name=names, course=courses))
pdf1
course_id = '821 822 823 824 825'.split()
course_names = 'Unix Python R Spark GLM'.split()
pdf2 = pd.DataFrame(dict(course_id=course_id, name=course_names))
pdf2
df1 = spark.createDataFrame(pdf1)
df2 = spark.createDataFrame(pdf2)
df1.join(df2, df1.course == df2.course_id, how='inner').show()
df1.join(df2, df1.course == df2.course_id, how='right').show()
```
## DataFrame conversions
```
sc = spark.sparkContext
rdd = sc.parallelize([('ann', 23), ('bob', 34)])
df = spark.createDataFrame(rdd, schema='name STRING, age INT')
df.show()
df.rdd.map(lambda x: (x[0], x[1]**2)).collect()
df.rdd.mapValues(lambda x: x**2).collect()
df.toPandas()
```
|
github_jupyter
|
from pyspark.sql import SparkSession
import pyspark.sql.functions as F
import pyspark.sql.types as T
spark = (
SparkSession.builder
.master("local")
.appName("BIOS-823")
.config("spark.executor.cores", 4)
.getOrCreate()
)
spark.version
spark.conf.get('spark.executor.cores')
df = spark.range(3)
df.show(3)
%%file data/test.csv
number,letter
0,a
1,c
2,b
3,a
4,b
5,c
6,a
7,a
8,a
9,b
10,b
11,c
12,c
13,b
14,b
df = (
spark.read.
format('csv').
option('header', 'true').
option('inferSchema', 'true').
load('csv/test.csv')
)
df.show(3)
df.printSchema()
schema = T.StructType([
T.StructField("number", T.DoubleType()),
T.StructField("letter", T.StringType()),
])
df = (
spark.read.
format('csv').
option('header', 'true').
schema(schema).
load('csv/test.csv')
)
df.show(3)
df.printSchema()
schema = 'number DOUBLE, letter STRING'
df_altschema = (
spark.read.
format('csv').
option('header', 'true').
schema(schema=schema).
load('csv/test.csv')
)
df_altschema.take(3)
df_altschema.printSchema()
df.cache()
df.select('number').show(3)
from pyspark.sql.functions import col, expr
df.select(col('number').alias('index')).show(3)
df.select(expr('number as x')).show(3)
df.withColumnRenamed('number', 'x').show(3)
df.filter('number % 2 == 0').show(3)
df.filter("number % 2 == 0 AND letter == 'a'").show(3)
df.sort(df.number.desc()).show(3)
df.sort('number', ascending=False).show(3)
df.orderBy(df.letter.desc()).show(3)
df.selectExpr('number*2 as x').show(3)
df.selectExpr('number', 'letter', 'number*2 as x').show(3)
df.withColumn('x', expr('number*2')).show(3)
import pyspark.sql.functions as F
df.agg(F.min('number'),
F.max('number'),
F.min('letter'),
F.max('letter')).show()
(
df.groupby('letter').
agg(F.mean('number'), F.stddev_samp('number')).show()
)
from pyspark.sql.window import Window
ws = (
Window.partitionBy('letter').
orderBy(F.desc('number')).
rowsBetween(Window.unboundedPreceding, Window.currentRow)
)
df.groupby('letter').agg(F.sum('number')).show()
df.show()
(
df.select('letter', F.sum('number').
over(ws).
alias('rank')).show()
)
df.createOrReplaceTempView('df_table')
spark.sql('''SELECT * FROM df_table''').show(3)
spark.sql('''
SELECT letter, mean(number) AS mean,
stddev_samp(number) AS sd from df_table
WHERE number % 2 = 0
GROUP BY letter
ORDER BY letter DESC
''').show()
from pyspark.sql.functions import split, lower, explode
import pandas as pd
s = spark.createDataFrame(
pd.DataFrame(
dict(sents=('Thing 1 and Thing 2',
'The Quick Brown Fox'))))
s.show()
from pyspark.sql.functions import regexp_replace
s1 = (
s.select(explode(split(lower(expr('sents')), ' '))).
sort('col')
)
s1.show()
s1.groupBy('col').count().show()
s.createOrReplaceTempView('s_table')
spark.sql('''
SELECT regexp_replace(sents, 'T.*?g', 'FOO')
FROM s_table
''').show()
from pyspark.sql.functions import log1p, randn
df.selectExpr('number', 'log1p(number)', 'letter').show(3)
(
df.selectExpr('number', 'randn() as random').
stat.corr('number', 'random')
)
dt = (
spark.range(3).
withColumn('today', F.current_date()).
withColumn('tomorrow', F.date_add('today', 1)).
withColumn('time', F.current_timestamp())
)
dt.show()
dt.printSchema()
%%file data/test_null.csv
number,letter
0,a
1,
2,b
3,a
4,b
5,
6,a
7,a
8,
9,b
10,b
11,c
12,
13,b
14,b
dn = (
spark.read.
format('csv').
option('header', 'true').
option('inferSchema', 'true').
load('csv/test_null.csv')
)
dn.printSchema()
dn.show()
dn.na.drop().show()
dn.na.fill('Missing').show()
from pyspark.sql.functions import udf, pandas_udf
@udf('double')
def square(x):
return x**2
df.select('number', square('number')).show(3)
export JAVA_HOME=$(/usr/libexec/java_home -v 11)
export JAVA_TOOL_OPTIONS="-Dio.netty.tryReflectionSetAccessible=true"
@pandas_udf('double')
def scale(x):
return (x - x.mean())/x.std()
df.select(scale('number')).show(3)
import warnings
warnings.simplefilter('ignore', UserWarning)
@pandas_udf('double', F.PandasUDFType.GROUPED_AGG)
def gmean(x):
return x.mean()
df.groupby('letter').agg(gmean('number')).show()
@pandas_udf('double')
def gmean1(x: pd.Series) -> float:
return x.mean()
df.groupby('letter').agg(gmean1('number')).show()
@pandas_udf(df.schema, F.PandasUDFType.GROUPED_MAP)
def gscale(pdf):
return pdf.assign(
number = (pdf.number - pdf.number.mean()) /
pdf.number.std())
df.groupby('letter').apply(gscale).show()
def gscale1(pdf: pd.DataFrame) -> pd.DataFrame:
num = pdf.number
return pdf.assign(
number = (num - num.mean()) / num.std())
df.groupby('letter').applyInPandas(gscale1, schema=df.schema).show()
def gsum(key, pdf):
return pd.DataFrame([key + (pdf.number.sum(),)])
df.groupby('letter').applyInPandas(gsum, 'letter string, number long').show()
df.groupBy('letter').sum().show()
def func(pdf: pd.DataFrame) -> int:
return (pdf.number.astype('str').str.len()).sum()
def gcustom(key, pdf):
return pd.DataFrame([key + (func(pdf),)])
df.groupby('letter').applyInPandas(gcustom, 'letter string, number long').show()
def even(it):
for pdf in it:
yield pdf[pdf.number % 2 == 0]
df.mapInPandas(even, 'letter string, number long').show()
names = 'ann ann bob bob chcuk'.split()
courses = '821 823 821 824 823'.split()
pdf1 = pd.DataFrame(dict(name=names, course=courses))
pdf1
course_id = '821 822 823 824 825'.split()
course_names = 'Unix Python R Spark GLM'.split()
pdf2 = pd.DataFrame(dict(course_id=course_id, name=course_names))
pdf2
df1 = spark.createDataFrame(pdf1)
df2 = spark.createDataFrame(pdf2)
df1.join(df2, df1.course == df2.course_id, how='inner').show()
df1.join(df2, df1.course == df2.course_id, how='right').show()
sc = spark.sparkContext
rdd = sc.parallelize([('ann', 23), ('bob', 34)])
df = spark.createDataFrame(rdd, schema='name STRING, age INT')
df.show()
df.rdd.map(lambda x: (x[0], x[1]**2)).collect()
df.rdd.mapValues(lambda x: x**2).collect()
df.toPandas()
| 0.508544 | 0.869105 |
```
import pandas as pd
df=pd.read_csv('car data.csv')
df.shape
print(df['Seller_Type'].unique())
print(df['Fuel_Type'].unique())
print(df['Transmission'].unique())
print(df['Owner'].unique())
##check missing values
df.isnull().sum()
df.describe()
final_dataset=df[['Year','Selling_Price','Present_Price','Kms_Driven','Fuel_Type','Seller_Type','Transmission','Owner']]
final_dataset.head()
final_dataset['Current Year']=2020
final_dataset.head()
final_dataset['no_year']=final_dataset['Current Year']- final_dataset['Year']
final_dataset.head()
final_dataset.drop(['Year'],axis=1,inplace=True)
final_dataset.head()
final_dataset=pd.get_dummies(final_dataset,drop_first=True)
final_dataset.head()
final_dataset.head()
final_dataset=final_dataset.drop(['Current Year'],axis=1)
final_dataset.head()
final_dataset.corr()
import seaborn as sns
sns.pairplot(final_dataset)
import seaborn as sns
#get correlations of each features in dataset
corrmat = df.corr()
top_corr_features = corrmat.index
plt.figure(figsize=(20,20))
#plot heat map
g=sns.heatmap(df[top_corr_features].corr(),annot=True,cmap="RdYlGn")
X=final_dataset.iloc[:,1:]
y=final_dataset.iloc[:,0]
X['Owner'].unique()
X.head()
y.head()
### Feature Importance
from sklearn.ensemble import ExtraTreesRegressor
import matplotlib.pyplot as plt
model = ExtraTreesRegressor()
model.fit(X,y)
print(model.feature_importances_)
#plot graph of feature importances for better visualization
feat_importances = pd.Series(model.feature_importances_, index=X.columns)
feat_importances.nlargest(5).plot(kind='barh')
plt.show()
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state=0)
from sklearn.ensemble import RandomForestRegressor
regressor=RandomForestRegressor()
n_estimators = [int(x) for x in np.linspace(start = 100, stop = 1200, num = 12)]
print(n_estimators)
from sklearn.model_selection import RandomizedSearchCV
#Randomized Search CV
# Number of trees in random forest
n_estimators = [int(x) for x in np.linspace(start = 100, stop = 1200, num = 12)]
# Number of features to consider at every split
max_features = ['auto', 'sqrt']
# Maximum number of levels in tree
max_depth = [int(x) for x in np.linspace(5, 30, num = 6)]
# max_depth.append(None)
# Minimum number of samples required to split a node
min_samples_split = [2, 5, 10, 15, 100]
# Minimum number of samples required at each leaf node
min_samples_leaf = [1, 2, 5, 10]
# Create the random grid
random_grid = {'n_estimators': n_estimators,
'max_features': max_features,
'max_depth': max_depth,
'min_samples_split': min_samples_split,
'min_samples_leaf': min_samples_leaf}
print(random_grid)
# Use the random grid to search for best hyperparameters
# First create the base model to tune
rf = RandomForestRegressor()
# Random search of parameters, using 3 fold cross validation,
# search across 100 different combinations
rf_random = RandomizedSearchCV(estimator = rf, param_distributions = random_grid,scoring='neg_mean_squared_error', n_iter = 10, cv = 5, verbose=2, random_state=42, n_jobs = 1)
rf_random.fit(X_train,y_train)
rf_random.best_params_
rf_random.best_score_
predictions=rf_random.predict(X_test)
sns.distplot(y_test-predictions)
plt.scatter(y_test,predictions)
from sklearn import metrics
print('MAE:', metrics.mean_absolute_error(y_test, predictions))
print('MSE:', metrics.mean_squared_error(y_test, predictions))
print('RMSE:', np.sqrt(metrics.mean_squared_error(y_test, predictions)))
import pickle
# open a file, where you ant to store the data
file = open('random_forest_regression_model.pkl', 'wb')
# dump information to that file
pickle.dump(rf_random, file)
```
|
github_jupyter
|
import pandas as pd
df=pd.read_csv('car data.csv')
df.shape
print(df['Seller_Type'].unique())
print(df['Fuel_Type'].unique())
print(df['Transmission'].unique())
print(df['Owner'].unique())
##check missing values
df.isnull().sum()
df.describe()
final_dataset=df[['Year','Selling_Price','Present_Price','Kms_Driven','Fuel_Type','Seller_Type','Transmission','Owner']]
final_dataset.head()
final_dataset['Current Year']=2020
final_dataset.head()
final_dataset['no_year']=final_dataset['Current Year']- final_dataset['Year']
final_dataset.head()
final_dataset.drop(['Year'],axis=1,inplace=True)
final_dataset.head()
final_dataset=pd.get_dummies(final_dataset,drop_first=True)
final_dataset.head()
final_dataset.head()
final_dataset=final_dataset.drop(['Current Year'],axis=1)
final_dataset.head()
final_dataset.corr()
import seaborn as sns
sns.pairplot(final_dataset)
import seaborn as sns
#get correlations of each features in dataset
corrmat = df.corr()
top_corr_features = corrmat.index
plt.figure(figsize=(20,20))
#plot heat map
g=sns.heatmap(df[top_corr_features].corr(),annot=True,cmap="RdYlGn")
X=final_dataset.iloc[:,1:]
y=final_dataset.iloc[:,0]
X['Owner'].unique()
X.head()
y.head()
### Feature Importance
from sklearn.ensemble import ExtraTreesRegressor
import matplotlib.pyplot as plt
model = ExtraTreesRegressor()
model.fit(X,y)
print(model.feature_importances_)
#plot graph of feature importances for better visualization
feat_importances = pd.Series(model.feature_importances_, index=X.columns)
feat_importances.nlargest(5).plot(kind='barh')
plt.show()
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state=0)
from sklearn.ensemble import RandomForestRegressor
regressor=RandomForestRegressor()
n_estimators = [int(x) for x in np.linspace(start = 100, stop = 1200, num = 12)]
print(n_estimators)
from sklearn.model_selection import RandomizedSearchCV
#Randomized Search CV
# Number of trees in random forest
n_estimators = [int(x) for x in np.linspace(start = 100, stop = 1200, num = 12)]
# Number of features to consider at every split
max_features = ['auto', 'sqrt']
# Maximum number of levels in tree
max_depth = [int(x) for x in np.linspace(5, 30, num = 6)]
# max_depth.append(None)
# Minimum number of samples required to split a node
min_samples_split = [2, 5, 10, 15, 100]
# Minimum number of samples required at each leaf node
min_samples_leaf = [1, 2, 5, 10]
# Create the random grid
random_grid = {'n_estimators': n_estimators,
'max_features': max_features,
'max_depth': max_depth,
'min_samples_split': min_samples_split,
'min_samples_leaf': min_samples_leaf}
print(random_grid)
# Use the random grid to search for best hyperparameters
# First create the base model to tune
rf = RandomForestRegressor()
# Random search of parameters, using 3 fold cross validation,
# search across 100 different combinations
rf_random = RandomizedSearchCV(estimator = rf, param_distributions = random_grid,scoring='neg_mean_squared_error', n_iter = 10, cv = 5, verbose=2, random_state=42, n_jobs = 1)
rf_random.fit(X_train,y_train)
rf_random.best_params_
rf_random.best_score_
predictions=rf_random.predict(X_test)
sns.distplot(y_test-predictions)
plt.scatter(y_test,predictions)
from sklearn import metrics
print('MAE:', metrics.mean_absolute_error(y_test, predictions))
print('MSE:', metrics.mean_squared_error(y_test, predictions))
print('RMSE:', np.sqrt(metrics.mean_squared_error(y_test, predictions)))
import pickle
# open a file, where you ant to store the data
file = open('random_forest_regression_model.pkl', 'wb')
# dump information to that file
pickle.dump(rf_random, file)
| 0.451327 | 0.391697 |
## Generating facies realizations with GeostatsPy
We generate here a set of facies realizations (sand/ shale) using the fantastic geostatistical library _geostatspy_ (https://github.com/GeostatsGuy/GeostatsPy) by Michael Pyrcz (University of Texas, Austin).
The example is modified from his notebook
`PythonNumericalDemos/GeostatsPy_sisim.ipynb`
```
import os # to set current working directory
import numpy as np # arrays and matrix math
import pandas as pd # DataFrames
import matplotlib.pyplot as plt # plotting
import geostatspy.GSLIB as GSLIB
import geostatspy.geostats as geostats
import pickle
```
### Load Data
We are using a data set that is also provided by Michael Pyrcz on https://github.com/GeostatsGuy/GeoDataSets
Two options here:
1. Generate data from scratch (from original data file
2. Load processed and adjusted data (with three facices)
```
# selection option
data_type = 'processed' # 'processed' or 'original'
if data_type == 'original':
df = pd.read_csv("sample_data_MV_biased.csv") # read a .csv file in as a DataFrame
df.describe() # summary statistics
df = df.sample(50) # extract 50 samples
df = df.reset_index() # reset the record index
# adjust
vals = df[["X", "Y", "Facies"]].values
idx = np.where(vals[:,2] == 0)[0]
select = np.random.choice(idx, size=10, replace=False)
# set new facies
for s in select:
vals[s,2] = 2
df['Facies'] = vals[:,2]
# save generated data
# add random number to avoid acciental overwriting:
num = np.random.randint(100000)
f = open("data_frame_three_facies_%d.pkl" % num, mode='wb')
pickle.dump(df, f)
f.close()
elif data_type == 'processed':
df = pickle.load(open("data_frame_three_facies_2.pkl", 'rb'))
df.head()
df.plot.scatter(x="X", y="Y", c="Facies", colormap='viridis')
```
### Generating Realizations
As a first step, we define the global model domain, the number of facies (with proportions) and the facies variograms:
```
# Sequential Indicator Simulation with Simple Kriging Multiple Realizations
nx = 64; ny = 64; xsiz = 1000./nx; ysiz = 1000./ny; xmn = 10.0; ymn = 10.0; nxdis = 1; nydis = 1
ndmin = 0; ndmax = 10; nodmax = 10; radius = 400; skmean = 0.5
tmin = -999; tmax = 999
dummy_trend = np.zeros((11,11)) # the current version requires trend input - if wrong size it is ignored
ncut = 3 # number of facies
if ncut == 2: # standard version
thresh = [0,1] # the facies categories (use consistent order)
gcdf = [0.4,0.6] # the global proportions of the categories
varios = [] # the variogram list
varios.append(GSLIB.make_variogram(nug=0.0,nst=1,it1=1,cc1=1.0,azi1=90,hmaj1=400,hmin1=100)) # shale indicator variogram
# varios.append(GSLIB.make_variogram(nug=0.0,nst=1,it1=1,cc1=1.0,azi1=0,hmaj1=100,hmin1=100)) # sand indicator variogram
varios.append(GSLIB.make_variogram(nug=0.0,nst=1,it1=1,cc1=1.0,azi1=90,hmaj1=400,hmin1=100)) # sand indicator variogram
if ncut == 3: # adjusted for three facies (FW)
thresh = [0,1,2] # the facies categories (use consistent order)
# gcdf = [0.4,0.6] # the global proportions of the categories
gcdf = [0.1,0.1,0.8] # the global proportions of the categories
varios = [] # the variogram list
varios.append(GSLIB.make_variogram(nug=0.0,nst=1,it1=1,cc1=0.8,cc2=0.2,azi1=90,hmaj1=400,hmin1=100)) # shale indicator variogram
# varios.append(GSLIB.make_variogram(nug=0.0,nst=1,it1=1,cc1=1.0,azi1=0,hmaj1=100,hmin1=100)) # sand indicator variogram
varios.append(GSLIB.make_variogram(nug=0.0,nst=1,it1=1,cc1=0.8,cc2=0.2,azi1=90,hmaj1=400,hmin1=100)) # sand indicator variogram
varios.append(GSLIB.make_variogram(nug=0.0,nst=1,it1=1,cc1=0.8,cc2=0.2,azi1=90,hmaj1=100,hmin1=100)) # facies 2 indicator variogram
```
Before we generate realizations, we set the random seed for later reproducibility (Technical side note: seeds need to be generated for subsequent implementation of GSLib functions):
```
n_realizations = 10
i_save = 10 # save steps
np.random.seed(seed = 12345)
seeds = np.random.choice(10000, n_realizations, replace=False)
seeds
```
We now can generate realizations (approx. 10s per realization for settings above!):
```
realizations = np.empty(shape=(n_realizations, nx, ny))
for i in range(n_realizations):
# Added just for better checks during offline run:
print("\n")
print(80*"*")
print("\n\n\t Realization %d \n\n"% i)
print(80*"*")
print("\n")
realizations[i,:,:] = geostats.sisim(df,'X','Y','Facies',ivtype=0,koption=0,ncut=ncut,thresh=thresh,gcdf=gcdf,trend=dummy_trend,
tmin=tmin,tmax=tmax,zmin=0.0,zmax=1.0,ltail=1,ltpar=1,middle=1,mpar=0,utail=1,utpar=2,
nx=nx,xmn=xmn,xsiz=xsiz,ny=ny,ymn=ymn,ysiz=ysiz,seed=seeds[i],
ndmin=ndmin,ndmax=ndmax,nodmax=nodmax,mults=1,nmult=3,noct=-1,radius=radius,ktype=0,vario=varios)
# save every i_save iterations:
if np.mod(i,i_save) == 0:
# add random int to filename to avoid overwriting
num = np.random.randint(100000)
f = open("facies_realizations_ncut_f3_aniso_64x64_%05d.pkl" % num, mode='wb')
print("Save to file facies_realizations_ncut_f3_aniso_64x64_%05d.pkl" % num)
pickle.dump(realizations, f)
f.close()
plt.imshow(realizations[0])
plt.colorbar()
```
### Plot realizations
Just to get an impression, here a plot of some realizations:
```
plt.figure(figsize=(12,6))
for i in range(1, 9):
plt.subplot(2, 4, i)
plt.imshow(realizations[i])
```
...and here simply the sum:
```
plt.imshow(np.sum(realizations, axis=0))
plt.colorbar()
```
...and the (estimated) probability for facies=1:
### Save realizations
Save realizations for further use later
```
import pickle
# add random int to filename to avoid overwriting
num = np.random.randint(100000)
f = open("facies_realizations_ncut_f3_aniso_64x64_%05d.pkl" % num, mode='wb')
pickle.dump(realizations, f)
f.close()
```
### Show conditioning points
It is also possible to show the conditioning points using proper GSLib functions - see github page of GeostatsPy (commented out here for now).
```
xmin = 0.0; xmax = 1000.0; ymin = 0.0; ymax = 1000.0; cmap = plt.cm.inferno # plotting parameters
plt.subplot(131) # plot the results
GSLIB.locpix_st(np.sum(realizations, axis=0)/n_realizations,xmin,xmax,ymin,ymax,xsiz,-.4,1.0,df,'X','Y','Facies','Sequential Indicator Simulation - Realization 1','X(m)','Y(m)','Facies',cmap)
plt.subplot(132) # plot the results
GSLIB.locpix_st(realizations[1],xmin,xmax,ymin,ymax,xsiz,-.4,1.0,df,'X','Y','Facies','Sequential Indicator Simulation - Realization 2','X(m)','Y(m)','Facies',cmap)
plt.subplot(133) # plot the results
GSLIB.locpix_st(realizations[2],xmin,xmax,ymin,ymax,xsiz,-.4,1.0,df,'X','Y','Facies','Sequential Indicator Simulation - Realization 3','X(m)','Y(m)','Facies',cmap)
plt.subplots_adjust(left=0.0, bottom=0.0, right=4.0, top=1.5, wspace=0.2, hspace=0.2)
plt.show()
```
|
github_jupyter
|
import os # to set current working directory
import numpy as np # arrays and matrix math
import pandas as pd # DataFrames
import matplotlib.pyplot as plt # plotting
import geostatspy.GSLIB as GSLIB
import geostatspy.geostats as geostats
import pickle
# selection option
data_type = 'processed' # 'processed' or 'original'
if data_type == 'original':
df = pd.read_csv("sample_data_MV_biased.csv") # read a .csv file in as a DataFrame
df.describe() # summary statistics
df = df.sample(50) # extract 50 samples
df = df.reset_index() # reset the record index
# adjust
vals = df[["X", "Y", "Facies"]].values
idx = np.where(vals[:,2] == 0)[0]
select = np.random.choice(idx, size=10, replace=False)
# set new facies
for s in select:
vals[s,2] = 2
df['Facies'] = vals[:,2]
# save generated data
# add random number to avoid acciental overwriting:
num = np.random.randint(100000)
f = open("data_frame_three_facies_%d.pkl" % num, mode='wb')
pickle.dump(df, f)
f.close()
elif data_type == 'processed':
df = pickle.load(open("data_frame_three_facies_2.pkl", 'rb'))
df.head()
df.plot.scatter(x="X", y="Y", c="Facies", colormap='viridis')
# Sequential Indicator Simulation with Simple Kriging Multiple Realizations
nx = 64; ny = 64; xsiz = 1000./nx; ysiz = 1000./ny; xmn = 10.0; ymn = 10.0; nxdis = 1; nydis = 1
ndmin = 0; ndmax = 10; nodmax = 10; radius = 400; skmean = 0.5
tmin = -999; tmax = 999
dummy_trend = np.zeros((11,11)) # the current version requires trend input - if wrong size it is ignored
ncut = 3 # number of facies
if ncut == 2: # standard version
thresh = [0,1] # the facies categories (use consistent order)
gcdf = [0.4,0.6] # the global proportions of the categories
varios = [] # the variogram list
varios.append(GSLIB.make_variogram(nug=0.0,nst=1,it1=1,cc1=1.0,azi1=90,hmaj1=400,hmin1=100)) # shale indicator variogram
# varios.append(GSLIB.make_variogram(nug=0.0,nst=1,it1=1,cc1=1.0,azi1=0,hmaj1=100,hmin1=100)) # sand indicator variogram
varios.append(GSLIB.make_variogram(nug=0.0,nst=1,it1=1,cc1=1.0,azi1=90,hmaj1=400,hmin1=100)) # sand indicator variogram
if ncut == 3: # adjusted for three facies (FW)
thresh = [0,1,2] # the facies categories (use consistent order)
# gcdf = [0.4,0.6] # the global proportions of the categories
gcdf = [0.1,0.1,0.8] # the global proportions of the categories
varios = [] # the variogram list
varios.append(GSLIB.make_variogram(nug=0.0,nst=1,it1=1,cc1=0.8,cc2=0.2,azi1=90,hmaj1=400,hmin1=100)) # shale indicator variogram
# varios.append(GSLIB.make_variogram(nug=0.0,nst=1,it1=1,cc1=1.0,azi1=0,hmaj1=100,hmin1=100)) # sand indicator variogram
varios.append(GSLIB.make_variogram(nug=0.0,nst=1,it1=1,cc1=0.8,cc2=0.2,azi1=90,hmaj1=400,hmin1=100)) # sand indicator variogram
varios.append(GSLIB.make_variogram(nug=0.0,nst=1,it1=1,cc1=0.8,cc2=0.2,azi1=90,hmaj1=100,hmin1=100)) # facies 2 indicator variogram
n_realizations = 10
i_save = 10 # save steps
np.random.seed(seed = 12345)
seeds = np.random.choice(10000, n_realizations, replace=False)
seeds
realizations = np.empty(shape=(n_realizations, nx, ny))
for i in range(n_realizations):
# Added just for better checks during offline run:
print("\n")
print(80*"*")
print("\n\n\t Realization %d \n\n"% i)
print(80*"*")
print("\n")
realizations[i,:,:] = geostats.sisim(df,'X','Y','Facies',ivtype=0,koption=0,ncut=ncut,thresh=thresh,gcdf=gcdf,trend=dummy_trend,
tmin=tmin,tmax=tmax,zmin=0.0,zmax=1.0,ltail=1,ltpar=1,middle=1,mpar=0,utail=1,utpar=2,
nx=nx,xmn=xmn,xsiz=xsiz,ny=ny,ymn=ymn,ysiz=ysiz,seed=seeds[i],
ndmin=ndmin,ndmax=ndmax,nodmax=nodmax,mults=1,nmult=3,noct=-1,radius=radius,ktype=0,vario=varios)
# save every i_save iterations:
if np.mod(i,i_save) == 0:
# add random int to filename to avoid overwriting
num = np.random.randint(100000)
f = open("facies_realizations_ncut_f3_aniso_64x64_%05d.pkl" % num, mode='wb')
print("Save to file facies_realizations_ncut_f3_aniso_64x64_%05d.pkl" % num)
pickle.dump(realizations, f)
f.close()
plt.imshow(realizations[0])
plt.colorbar()
plt.figure(figsize=(12,6))
for i in range(1, 9):
plt.subplot(2, 4, i)
plt.imshow(realizations[i])
plt.imshow(np.sum(realizations, axis=0))
plt.colorbar()
import pickle
# add random int to filename to avoid overwriting
num = np.random.randint(100000)
f = open("facies_realizations_ncut_f3_aniso_64x64_%05d.pkl" % num, mode='wb')
pickle.dump(realizations, f)
f.close()
xmin = 0.0; xmax = 1000.0; ymin = 0.0; ymax = 1000.0; cmap = plt.cm.inferno # plotting parameters
plt.subplot(131) # plot the results
GSLIB.locpix_st(np.sum(realizations, axis=0)/n_realizations,xmin,xmax,ymin,ymax,xsiz,-.4,1.0,df,'X','Y','Facies','Sequential Indicator Simulation - Realization 1','X(m)','Y(m)','Facies',cmap)
plt.subplot(132) # plot the results
GSLIB.locpix_st(realizations[1],xmin,xmax,ymin,ymax,xsiz,-.4,1.0,df,'X','Y','Facies','Sequential Indicator Simulation - Realization 2','X(m)','Y(m)','Facies',cmap)
plt.subplot(133) # plot the results
GSLIB.locpix_st(realizations[2],xmin,xmax,ymin,ymax,xsiz,-.4,1.0,df,'X','Y','Facies','Sequential Indicator Simulation - Realization 3','X(m)','Y(m)','Facies',cmap)
plt.subplots_adjust(left=0.0, bottom=0.0, right=4.0, top=1.5, wspace=0.2, hspace=0.2)
plt.show()
| 0.425009 | 0.900966 |
Resources Used
- wget.download('https://tensorflow-object-detection-api-tutorial.readthedocs.io/en/latest/_downloads/da4babe668a8afb093cc7776d7e630f3/generate_tfrecord.py')
- Setup https://tensorflow-object-detection-api-tutorial.readthedocs.io/en/latest/install.html
# 0. Setup Paths
```
WORKSPACE_PATH = 'Tensorflow/workspace'
SCRIPTS_PATH = 'Tensorflow/scripts'
APIMODEL_PATH = 'Tensorflow/models'
ANNOTATION_PATH = WORKSPACE_PATH+'/annotations/'
IMAGE_PATH = WORKSPACE_PATH+'/images'
MODEL_PATH = WORKSPACE_PATH+'/models'
PRETRAINED_MODEL_PATH = WORKSPACE_PATH+'/pre-trained-models'
CONFIG_PATH = MODEL_PATH+'/my_ssd_mobnet/pipeline.config'
CHECKPOINT_PATH = MODEL_PATH+'/my_ssd_mobnet/'
```
# 1. Create Label Map
```
labels = [{'name':'Hello', 'id':1},
{'name':'Yes', 'id':2},
{'name':'No', 'id':3},
{'name':'Thanks', 'id':4},
{'name':'I Love You', 'id':5},
]
with open(ANNOTATION_PATH+'label_map.pbtxt', 'w') as f:
for label in labels:
f.write('item { \n')
f.write('\tname:\'{}\'\n'.format(label['name']))
f.write('\tid:{}\n'.format(label['id']))
f.write('}\n')
```
# 2. Create TF records
```
!python {SCRIPTS_PATH + '/generate_tfrecord.py'} -x {IMAGE_PATH + '/train'} -l {ANNOTATION_PATH + '/label_map.pbtxt'} -o {ANNOTATION_PATH + '/train.record'}
!python {SCRIPTS_PATH + '/generate_tfrecord.py'} -x{IMAGE_PATH + '/test'} -l {ANNOTATION_PATH + '/label_map.pbtxt'} -o {ANNOTATION_PATH + '/test.record'}
```
# 3. Download TF Models Pretrained Models from Tensorflow Model Zoo
```
!cd Tensorflow && git clone https://github.com/tensorflow/models
#wget.download('http://download.tensorflow.org/models/object_detection/tf2/20200711/ssd_mobilenet_v2_fpnlite_320x320_coco17_tpu-8.tar.gz')
#!mv ssd_mobilenet_v2_fpnlite_320x320_coco17_tpu-8.tar.gz {PRETRAINED_MODEL_PATH}
#!cd {PRETRAINED_MODEL_PATH} && tar -zxvf ssd_mobilenet_v2_fpnlite_320x320_coco17_tpu-8.tar.gz
```
# 4. Copy Model Config to Training Folder
```
CUSTOM_MODEL_NAME = 'my_ssd_mobnet'
!mkdir {'Tensorflow/workspace/models/my_ssd_mobnet'}
!cp {PRETRAINED_MODEL_PATH+'/ssd_mobilenet_v2_fpnlite_320x320_coco17_tpu-8/pipeline.config'} {MODEL_PATH+'/my_ssd_mobnet'}
```
# 5. Update Config For Transfer Learning
```
import tensorflow as tf
from object_detection.utils import config_util
from object_detection.protos import pipeline_pb2
from google.protobuf import text_format
CONFIG_PATH = MODEL_PATH+'/'+CUSTOM_MODEL_NAME+'/pipeline.config'
print(CONFIG_PATH)
config = config_util.get_configs_from_pipeline_file(CONFIG_PATH)
config
pipeline_config = pipeline_pb2.TrainEvalPipelineConfig()
with tf.io.gfile.GFile(CONFIG_PATH, "r") as f:
proto_str = f.read()
text_format.Merge(proto_str, pipeline_config)
pipeline_config.model.ssd.num_classes = 5
pipeline_config.train_config.batch_size = 4
pipeline_config.train_config.fine_tune_checkpoint = PRETRAINED_MODEL_PATH+'/ssd_mobilenet_v2_fpnlite_320x320_coco17_tpu-8/checkpoint/ckpt-0'
pipeline_config.train_config.fine_tune_checkpoint_type = "detection"
pipeline_config.train_input_reader.label_map_path= ANNOTATION_PATH + '/label_map.pbtxt'
pipeline_config.train_input_reader.tf_record_input_reader.input_path[:] = [ANNOTATION_PATH + '/train.record']
pipeline_config.eval_input_reader[0].label_map_path = ANNOTATION_PATH + '/label_map.pbtxt'
pipeline_config.eval_input_reader[0].tf_record_input_reader.input_path[:] = [ANNOTATION_PATH + '/test.record']
config_text = text_format.MessageToString(pipeline_config)
with tf.io.gfile.GFile(CONFIG_PATH, "wb") as f:
f.write(config_text)
```
# 6. Train the model
```
print("""python {}/research/object_detection/model_main_tf2.py --model_dir={}/{} --pipeline_config_path={}/{}/pipeline.config --num_train_steps=20000""".format(APIMODEL_PATH, MODEL_PATH,CUSTOM_MODEL_NAME,MODEL_PATH,CUSTOM_MODEL_NAME))
```
# 7. Load Train Model From Checkpoint
```
import os
from object_detection.utils import label_map_util
from object_detection.utils import visualization_utils as viz_utils
from object_detection.builders import model_builder
# Load pipeline config and build a detection model
configs = config_util.get_configs_from_pipeline_file(CONFIG_PATH)
detection_model = model_builder.build(model_config=configs['model'], is_training=False)
# Restore checkpoint
ckpt = tf.compat.v2.train.Checkpoint(model=detection_model)
ckpt.restore(os.path.join(CHECKPOINT_PATH, 'ckpt-6')).expect_partial()
@tf.function
def detect_fn(image):
image, shapes = detection_model.preprocess(image)
prediction_dict = detection_model.predict(image, shapes)
detections = detection_model.postprocess(prediction_dict, shapes)
return detections
```
# 8. Detect in Real-Time
```
import cv2
import numpy as np
category_index = label_map_util.create_category_index_from_labelmap(ANNOTATION_PATH+'/label_map.pbtxt')
cap.release()
# Setup capture
cap = cv2.VideoCapture(0)
width = int(cap.get(cv2.CAP_PROP_FRAME_WIDTH))
height = int(cap.get(cv2.CAP_PROP_FRAME_HEIGHT))
while True:
ret, frame = cap.read()
image_np = np.array(frame)
input_tensor = tf.convert_to_tensor(np.expand_dims(image_np, 0), dtype=tf.float32)
detections = detect_fn(input_tensor)
num_detections = int(detections.pop('num_detections'))
detections = {key: value[0, :num_detections].numpy()
for key, value in detections.items()}
detections['num_detections'] = num_detections
# detection_classes should be ints.
detections['detection_classes'] = detections['detection_classes'].astype(np.int64)
label_id_offset = 1
image_np_with_detections = image_np.copy()
viz_utils.visualize_boxes_and_labels_on_image_array(
image_np_with_detections,
detections['detection_boxes'],
detections['detection_classes']+label_id_offset,
detections['detection_scores'],
category_index,
use_normalized_coordinates=True,
max_boxes_to_draw=5,
min_score_thresh=.5,
agnostic_mode=False)
cv2.imshow('object detection', cv2.resize(image_np_with_detections, (800, 600)))
if cv2.waitKey(1) & 0xFF == ord('q'):
cap.release()
break
detections = detect_fn(input_tensor)
from matplotlib import pyplot as plt
```
|
github_jupyter
|
WORKSPACE_PATH = 'Tensorflow/workspace'
SCRIPTS_PATH = 'Tensorflow/scripts'
APIMODEL_PATH = 'Tensorflow/models'
ANNOTATION_PATH = WORKSPACE_PATH+'/annotations/'
IMAGE_PATH = WORKSPACE_PATH+'/images'
MODEL_PATH = WORKSPACE_PATH+'/models'
PRETRAINED_MODEL_PATH = WORKSPACE_PATH+'/pre-trained-models'
CONFIG_PATH = MODEL_PATH+'/my_ssd_mobnet/pipeline.config'
CHECKPOINT_PATH = MODEL_PATH+'/my_ssd_mobnet/'
labels = [{'name':'Hello', 'id':1},
{'name':'Yes', 'id':2},
{'name':'No', 'id':3},
{'name':'Thanks', 'id':4},
{'name':'I Love You', 'id':5},
]
with open(ANNOTATION_PATH+'label_map.pbtxt', 'w') as f:
for label in labels:
f.write('item { \n')
f.write('\tname:\'{}\'\n'.format(label['name']))
f.write('\tid:{}\n'.format(label['id']))
f.write('}\n')
!python {SCRIPTS_PATH + '/generate_tfrecord.py'} -x {IMAGE_PATH + '/train'} -l {ANNOTATION_PATH + '/label_map.pbtxt'} -o {ANNOTATION_PATH + '/train.record'}
!python {SCRIPTS_PATH + '/generate_tfrecord.py'} -x{IMAGE_PATH + '/test'} -l {ANNOTATION_PATH + '/label_map.pbtxt'} -o {ANNOTATION_PATH + '/test.record'}
!cd Tensorflow && git clone https://github.com/tensorflow/models
#wget.download('http://download.tensorflow.org/models/object_detection/tf2/20200711/ssd_mobilenet_v2_fpnlite_320x320_coco17_tpu-8.tar.gz')
#!mv ssd_mobilenet_v2_fpnlite_320x320_coco17_tpu-8.tar.gz {PRETRAINED_MODEL_PATH}
#!cd {PRETRAINED_MODEL_PATH} && tar -zxvf ssd_mobilenet_v2_fpnlite_320x320_coco17_tpu-8.tar.gz
CUSTOM_MODEL_NAME = 'my_ssd_mobnet'
!mkdir {'Tensorflow/workspace/models/my_ssd_mobnet'}
!cp {PRETRAINED_MODEL_PATH+'/ssd_mobilenet_v2_fpnlite_320x320_coco17_tpu-8/pipeline.config'} {MODEL_PATH+'/my_ssd_mobnet'}
import tensorflow as tf
from object_detection.utils import config_util
from object_detection.protos import pipeline_pb2
from google.protobuf import text_format
CONFIG_PATH = MODEL_PATH+'/'+CUSTOM_MODEL_NAME+'/pipeline.config'
print(CONFIG_PATH)
config = config_util.get_configs_from_pipeline_file(CONFIG_PATH)
config
pipeline_config = pipeline_pb2.TrainEvalPipelineConfig()
with tf.io.gfile.GFile(CONFIG_PATH, "r") as f:
proto_str = f.read()
text_format.Merge(proto_str, pipeline_config)
pipeline_config.model.ssd.num_classes = 5
pipeline_config.train_config.batch_size = 4
pipeline_config.train_config.fine_tune_checkpoint = PRETRAINED_MODEL_PATH+'/ssd_mobilenet_v2_fpnlite_320x320_coco17_tpu-8/checkpoint/ckpt-0'
pipeline_config.train_config.fine_tune_checkpoint_type = "detection"
pipeline_config.train_input_reader.label_map_path= ANNOTATION_PATH + '/label_map.pbtxt'
pipeline_config.train_input_reader.tf_record_input_reader.input_path[:] = [ANNOTATION_PATH + '/train.record']
pipeline_config.eval_input_reader[0].label_map_path = ANNOTATION_PATH + '/label_map.pbtxt'
pipeline_config.eval_input_reader[0].tf_record_input_reader.input_path[:] = [ANNOTATION_PATH + '/test.record']
config_text = text_format.MessageToString(pipeline_config)
with tf.io.gfile.GFile(CONFIG_PATH, "wb") as f:
f.write(config_text)
print("""python {}/research/object_detection/model_main_tf2.py --model_dir={}/{} --pipeline_config_path={}/{}/pipeline.config --num_train_steps=20000""".format(APIMODEL_PATH, MODEL_PATH,CUSTOM_MODEL_NAME,MODEL_PATH,CUSTOM_MODEL_NAME))
import os
from object_detection.utils import label_map_util
from object_detection.utils import visualization_utils as viz_utils
from object_detection.builders import model_builder
# Load pipeline config and build a detection model
configs = config_util.get_configs_from_pipeline_file(CONFIG_PATH)
detection_model = model_builder.build(model_config=configs['model'], is_training=False)
# Restore checkpoint
ckpt = tf.compat.v2.train.Checkpoint(model=detection_model)
ckpt.restore(os.path.join(CHECKPOINT_PATH, 'ckpt-6')).expect_partial()
@tf.function
def detect_fn(image):
image, shapes = detection_model.preprocess(image)
prediction_dict = detection_model.predict(image, shapes)
detections = detection_model.postprocess(prediction_dict, shapes)
return detections
import cv2
import numpy as np
category_index = label_map_util.create_category_index_from_labelmap(ANNOTATION_PATH+'/label_map.pbtxt')
cap.release()
# Setup capture
cap = cv2.VideoCapture(0)
width = int(cap.get(cv2.CAP_PROP_FRAME_WIDTH))
height = int(cap.get(cv2.CAP_PROP_FRAME_HEIGHT))
while True:
ret, frame = cap.read()
image_np = np.array(frame)
input_tensor = tf.convert_to_tensor(np.expand_dims(image_np, 0), dtype=tf.float32)
detections = detect_fn(input_tensor)
num_detections = int(detections.pop('num_detections'))
detections = {key: value[0, :num_detections].numpy()
for key, value in detections.items()}
detections['num_detections'] = num_detections
# detection_classes should be ints.
detections['detection_classes'] = detections['detection_classes'].astype(np.int64)
label_id_offset = 1
image_np_with_detections = image_np.copy()
viz_utils.visualize_boxes_and_labels_on_image_array(
image_np_with_detections,
detections['detection_boxes'],
detections['detection_classes']+label_id_offset,
detections['detection_scores'],
category_index,
use_normalized_coordinates=True,
max_boxes_to_draw=5,
min_score_thresh=.5,
agnostic_mode=False)
cv2.imshow('object detection', cv2.resize(image_np_with_detections, (800, 600)))
if cv2.waitKey(1) & 0xFF == ord('q'):
cap.release()
break
detections = detect_fn(input_tensor)
from matplotlib import pyplot as plt
| 0.553023 | 0.655005 |
```
# Import libraries and modules
import datetime
import matplotlib.pyplot as plt
import numpy as np
import os
import tensorflow as tf
print(np.__version__)
print(tf.__version__)
np.set_printoptions(threshold=np.inf)
```
# Local Development
## Arguments
```
arguments = {}
# File arguments.
arguments["train_file_pattern"] = "gs://machine-learning-1234-bucket/gan/data/mnist/train*.tfrecord"
arguments["eval_file_pattern"] = "gs://machine-learning-1234-bucket/gan/data/mnist/test*.tfrecord"
arguments["output_dir"] = "gs://machine-learning-1234-bucket/gan/dcgan/tf2/trained_model"
# Training parameters.
arguments["tf_version"] = 2.2
arguments["use_graph_mode"] = True
arguments["distribution_strategy"] = ""
arguments["write_summaries"] = False
arguments["num_epochs"] = 30
arguments["train_dataset_length"] = 60000
arguments["train_batch_size"] = 32
arguments["log_step_count_steps"] = 100
arguments["save_summary_steps"] = 100
arguments["save_checkpoints_steps"] = 10000
arguments["keep_checkpoint_max"] = 10
arguments["input_fn_autotune"] = False
# Eval parameters.
arguments["eval_batch_size"] = 32
arguments["eval_steps"] = 100
# Image parameters.
arguments["height"] = 28
arguments["width"] = 28
arguments["depth"] = 1
# Generator parameters.
arguments["latent_size"] = 512
arguments["generator_projection_dims"] = [7, 7, 256]
arguments["generator_num_filters"] = [128, 128, arguments["depth"]]
arguments["generator_kernel_sizes"] = [4, 4, 3]
arguments["generator_strides"] = [2, 2, 1]
arguments["generator_leaky_relu_alpha"] = 0.2
arguments["generator_final_activation"] = "tanh"
arguments["generator_l1_regularization_scale"] = 0.
arguments["generator_l2_regularization_scale"] = 0.
arguments["generator_optimizer"] = "Adam"
arguments["generator_learning_rate"] = 0.0002
arguments["generator_adam_beta1"] = 0.5
arguments["generator_adam_beta2"] = 0.999
arguments["generator_adam_epsilon"] = 1e-8
arguments["generator_clip_gradients"] = None
arguments["generator_train_steps"] = 1
# Discriminator hyperparameters.
arguments["discriminator_num_filters"] = [64, 128, 128, 256]
arguments["discriminator_kernel_sizes"] = [3, 3, 3, 3]
arguments["discriminator_strides"] = [1, 2, 2, 2]
arguments["discriminator_dropout_rates"] = [0.3, 0.3, 0.3, 0.3]
arguments["discriminator_leaky_relu_alpha"] = 0.2
arguments["discriminator_l1_regularization_scale"] = 0.
arguments["discriminator_l2_regularization_scale"] = 0.
arguments["discriminator_optimizer"] = "Adam"
arguments["discriminator_learning_rate"] = 0.0002
arguments["discriminator_adam_beta1"] = 0.5
arguments["discriminator_adam_beta2"] = 0.999
arguments["discriminator_adam_epsilon"] = 1e-8
arguments["discriminator_clip_gradients"] = None
arguments["discriminator_train_steps"] = 1
arguments["label_smoothing"] = 0.9
```
## input.py
```
def preprocess_image(image):
"""Preprocess image tensor.
Args:
image: tensor, input image with shape
[batch_size, height, width, depth].
Returns:
Preprocessed image tensor with shape
[batch_size, height, width, depth].
"""
# Convert from [0, 255] -> [-1.0, 1.0] floats.
image = tf.cast(x=image, dtype=tf.float32) * (2. / 255) - 1.0
return image
def decode_example(protos, params):
"""Decodes TFRecord file into tensors.
Given protobufs, decode into image and label tensors.
Args:
protos: protobufs from TFRecord file.
params: dict, user passed parameters.
Returns:
Image and label tensors.
"""
# Create feature schema map for protos.
features = {
"image_raw": tf.io.FixedLenFeature(shape=[], dtype=tf.string),
"label": tf.io.FixedLenFeature(shape=[], dtype=tf.int64)
}
# Parse features from tf.Example.
parsed_features = tf.io.parse_single_example(
serialized=protos, features=features
)
# Convert from a scalar string tensor (whose single string has
# length height * width * depth) to a uint8 tensor with shape
# [height * width * depth].
image = tf.io.decode_raw(
input_bytes=parsed_features["image_raw"], out_type=tf.uint8
)
# Reshape flattened image back into normal dimensions.
image = tf.reshape(
tensor=image,
shape=[params["height"], params["width"], params["depth"]]
)
# Preprocess image.
image = preprocess_image(image=image)
# Convert label from a scalar uint8 tensor to an int32 scalar.
label = tf.cast(x=parsed_features["label"], dtype=tf.int32)
return {"image": image}, label
def read_dataset(filename, batch_size, params, training):
"""Reads TF Record data using tf.data, doing necessary preprocessing.
Given filename, mode, batch size, and other parameters, read TF Record
dataset using Dataset API, apply necessary preprocessing, and return an
input function to the Estimator API.
Args:
filename: str, file pattern that to read into our tf.data dataset.
batch_size: int, number of examples per batch.
params: dict, dictionary of user passed parameters.
training: bool, if training or not.
Returns:
An input function.
"""
def _input_fn():
"""Wrapper input function used by Estimator API to get data tensors.
Returns:
Batched dataset object of dictionary of feature tensors and label
tensor.
"""
# Create list of files that match pattern.
file_list = tf.data.Dataset.list_files(file_pattern=filename)
# Create dataset from file list.
dataset = tf.data.TFRecordDataset(
filenames=file_list,
num_parallel_reads=(
tf.contrib.data.AUTOTUNE
if params["input_fn_autotune"]
else None
)
)
# Shuffle and repeat if training with fused op.
if training:
dataset = dataset.apply(
tf.data.experimental.shuffle_and_repeat(
buffer_size=50 * batch_size,
count=None # indefinitely
)
)
# Decode CSV file into a features dictionary of tensors, then batch.
dataset = dataset.apply(
tf.data.experimental.map_and_batch(
map_func=lambda x: decode_example(
protos=x,
params=params
),
batch_size=batch_size,
num_parallel_calls=(
tf.contrib.data.AUTOTUNE
if params["input_fn_autotune"]
else None
)
)
)
# Prefetch data to improve latency.
dataset = dataset.prefetch(
buffer_size=(
tf.data.experimental.AUTOTUNE
if params["input_fn_autotune"]
else 1
)
)
return dataset
return _input_fn
train_ds = read_dataset(
filename=arguments["train_file_pattern"],
batch_size=arguments["train_batch_size"],
params=arguments,
training=True
)()
train_ds
eval_ds = read_dataset(
filename=arguments["eval_file_pattern"],
batch_size=arguments["eval_batch_size"],
params=arguments,
training=False
)()
eval_ds
```
## generators.py
```
class Generator(object):
"""Generator that takes latent vector input and outputs image.
Fields:
name: str, name of `Generator`.
kernel_regularizer: `l1_l2_regularizer` object, regularizar for
kernel variables.
bias_regularizer: `l1_l2_regularizer` object, regularizar for bias
variables.
params: dict, user passed parameters.
model: instance of generator `Model`.
"""
def __init__(
self,
input_shape,
kernel_regularizer,
bias_regularizer,
name,
params):
"""Instantiates and builds generator network.
Args:
input_shape: tuple, shape of latent vector input of shape
[batch_size, latent_size].
kernel_regularizer: `l1_l2_regularizer` object, regularizar for
kernel variables.
bias_regularizer: `l1_l2_regularizer` object, regularizar for bias
variables.
name: str, name of generator.
params: dict, user passed parameters.
"""
# Set name of generator.
self.name = name
# Store regularizers.
self.kernel_regularizer = kernel_regularizer
self.bias_regularizer = bias_regularizer
# Store parameters.
self.params = params
# Instantiate generator `Model`.
self.model = self._define_generator(input_shape)
def _project_latent_vectors(self, latent_vectors):
"""Defines generator network.
Args:
latent_vectors: tensor, latent vector inputs of shape
[batch_size, latent_size].
Returns:
Projected image of latent vector inputs.
"""
projection_height = self.params["generator_projection_dims"][0]
projection_width = self.params["generator_projection_dims"][1]
projection_depth = self.params["generator_projection_dims"][2]
# shape = (
# batch_size,
# projection_height * projection_width * projection_depth
# )
projection = tf.keras.layers.Dense(
units=projection_height * projection_width * projection_depth,
activation=None,
kernel_regularizer=self.kernel_regularizer,
bias_regularizer=self.bias_regularizer,
name="projection_dense_layer"
)(inputs=latent_vectors)
projection_leaky_relu = tf.keras.layers.LeakyReLU(
alpha=self.params["generator_leaky_relu_alpha"],
name="projection_leaky_relu"
)(inputs=projection)
# Add batch normalization to keep the inputs from blowing up.
# shape = (
# batch_size,
# projection_height * projection_width * projection_depth
# )
projection_batch_norm = tf.keras.layers.BatchNormalization(
name="projection_batch_norm"
)(inputs=projection_leaky_relu)
# Reshape projection into "image".
# shape = (
# batch_size,
# projection_height,
# projection_width,
# projection_depth
# )
projected_image = tf.reshape(
tensor=projection_batch_norm,
shape=[
-1, projection_height, projection_width, projection_depth
],
name="projected_image"
)
return projected_image
def _define_generator(
self, input_shape):
"""Defines generator network.
Args:
input_shape: tuple, shape of latent vector input of shape
[batch_size, latent_size].
Returns:
Instance of `Model` object.
"""
# Create the input layer to generator.
# shape = (batch_size, latent_size)
inputs = tf.keras.Input(
shape=input_shape, name="{}_inputs".format(self.name)
)
# Dictionary containing possible final activations.
final_activation_set = {"sigmoid", "relu", "tanh"}
# Project latent vectors.
network = self._project_latent_vectors(latent_vectors=inputs)
# Iteratively build upsampling layers.
for i in range(len(self.params["generator_num_filters"]) - 1):
# Add conv transpose layers with given params per layer.
# shape = (
# batch_size,
# generator_kernel_sizes[i - 1] * generator_strides[i],
# generator_kernel_sizes[i - 1] * generator_strides[i],
# generator_num_filters[i]
# )
network = tf.keras.layers.Conv2DTranspose(
filters=self.params["generator_num_filters"][i],
kernel_size=self.params["generator_kernel_sizes"][i],
strides=self.params["generator_strides"][i],
padding="same",
activation=None,
kernel_regularizer=self.kernel_regularizer,
bias_regularizer=self.bias_regularizer,
name="{}_layers_conv2d_tranpose_{}".format(self.name, i)
)(inputs=network)
network = tf.keras.layers.LeakyReLU(
alpha=self.params["generator_leaky_relu_alpha"],
name="{}_leaky_relu_{}".format(self.name, i)
)(inputs=network)
# Add batch normalization to keep the inputs from blowing up.
network = tf.keras.layers.BatchNormalization(
name="{}_layers_batch_norm_{}".format(self.name, i)
)(inputs=network)
# Final conv2d transpose layer for image output.
# shape = (batch_size, height, width, depth)
fake_images = tf.keras.layers.Conv2DTranspose(
filters=self.params["generator_num_filters"][-1],
kernel_size=self.params["generator_kernel_sizes"][-1],
strides=self.params["generator_strides"][-1],
padding="same",
activation=(
self.params["generator_final_activation"].lower()
if self.params["generator_final_activation"].lower()
in final_activation_set
else None
),
kernel_regularizer=self.kernel_regularizer,
bias_regularizer=self.bias_regularizer,
name="{}_layers_conv2d_tranpose_fake_images".format(self.name)
)(inputs=network)
# Resize fake images to match real images in case of mismatch.
height = self.params["height"]
width = self.params["width"]
fake_images = tf.keras.layers.Lambda(
function=lambda x: tf.image.resize(
images=x, size=[height, width], method="bilinear"
),
name="{}_resize_fake_images".format(self.name)
)(inputs=fake_images)
# Define model.
model = tf.keras.Model(
inputs=inputs, outputs=fake_images, name=self.name
)
return model
def get_model(self):
"""Returns generator's `Model` object.
Returns:
Generator's `Model` object.
"""
return self.model
def get_generator_loss(
self,
global_batch_size,
fake_logits,
global_step,
summary_file_writer
):
"""Gets generator loss.
Args:
global_batch_size: int, global batch size for distribution.
fake_logits: tensor, shape of
[batch_size, 1].
global_step: int, current global step for training.
summary_file_writer: summary file writer.
Returns:
Tensor of generator's total loss of shape [].
"""
if self.params["distribution_strategy"]:
# Calculate base generator loss.
generator_loss = tf.nn.compute_average_loss(
per_example_loss=tf.keras.losses.BinaryCrossentropy(
from_logits=True,
reduction=tf.keras.losses.Reduction.NONE
)(
y_true=tf.ones_like(input=fake_logits), y_pred=fake_logits
),
global_batch_size=global_batch_size
)
# Get regularization losses.
generator_reg_loss = tf.nn.scale_regularization_loss(
regularization_loss=sum(self.model.losses)
)
else:
# Calculate base generator loss.
generator_loss = tf.keras.losses.BinaryCrossentropy(
from_logits=True
)(
y_true=tf.ones_like(input=fake_logits), y_pred=fake_logits
)
# Get regularization losses.
generator_reg_loss = sum(self.model.losses)
# Combine losses for total losses.
generator_total_loss = tf.math.add(
x=generator_loss,
y=generator_reg_loss,
name="generator_total_loss"
)
if self.params["write_summaries"]:
# Add summaries for TensorBoard.
with summary_file_writer.as_default():
with tf.summary.record_if(
condition=tf.equal(
x=tf.math.floormod(
x=global_step,
y=self.params["save_summary_steps"]
), y=0
)
):
tf.summary.scalar(
name="losses/generator_loss",
data=generator_loss,
step=global_step
)
tf.summary.scalar(
name="losses/generator_reg_loss",
data=generator_reg_loss,
step=global_step
)
tf.summary.scalar(
name="optimized_losses/generator_total_loss",
data=generator_total_loss,
step=global_step
)
summary_file_writer.flush()
return generator_total_loss
# Instantiate generator.
dcgan_generator = Generator(
input_shape=(arguments["latent_size"]),
kernel_regularizer=tf.keras.regularizers.l1_l2(
l1=arguments["generator_l1_regularization_scale"],
l2=arguments["generator_l2_regularization_scale"]
),
bias_regularizer=None,
name="generator",
params=arguments
)
# Define generator model.
generator_model = dcgan_generator.get_model()
# Summarize generator model.
generator_model.summary()
generator_model.__dict__
# Plot generator model.
tf.keras.utils.plot_model(
model=generator_model,
to_file="generator_model.png",
show_shapes=True,
show_layer_names=True
)
noise = tf.random.normal(shape=[10, arguments["latent_size"]])
fake_images = generator_model(inputs=noise, training=False)
plt.imshow(fake_images[0, :, :, 0], cmap='gray')
```
## discriminators.py
```
class Discriminator(object):
"""Discriminator that takes image input and outputs logits.
Fields:
name: str, name of `Discriminator`.
kernel_regularizer: `l1_l2_regularizer` object, regularizar for
kernel variables.
bias_regularizer: `l1_l2_regularizer` object, regularizar for bias
variables.
params: dict, user passed parameters.
model: instance of discriminator `Model`.
"""
def __init__(
self,
input_shape,
kernel_regularizer,
bias_regularizer,
name,
params):
"""Instantiates and builds discriminator network.
Args:
input_shape: tuple, shape of image vector input of shape
[batch_size, height * width * depth].
kernel_regularizer: `l1_l2_regularizer` object, regularizar for
kernel variables.
bias_regularizer: `l1_l2_regularizer` object, regularizar for bias
variables.
name: str, name of discriminator.
params: dict, user passed parameters.
"""
# Set name of discriminator.
self.name = name
# Store regularizers.
self.kernel_regularizer = kernel_regularizer
self.bias_regularizer = bias_regularizer
# Store parameters.
self.params = params
# Instantiate discriminator `Model`.
self.model = self._define_discriminator(input_shape)
def _define_discriminator(self, input_shape):
"""Defines discriminator network.
Args:
input_shape: tuple, shape of image vector input of shape
[batch_size, height * width * depth].
Returns:
Instance of `Model` object.
"""
# Create the input layer to discriminator.
# shape = (batch_size, height * width * depth)
inputs = tf.keras.Input(
shape=input_shape,
name="{}_inputs".format(self.name)
)
network = inputs
# Iteratively build downsampling layers.
for i in range(len(self.params["discriminator_num_filters"])):
# Add convolutional layers with given params per layer.
# shape = (
# batch_size,
# discriminator_kernel_sizes[i - 1] / discriminator_strides[i],
# discriminator_kernel_sizes[i - 1] / discriminator_strides[i],
# discriminator_num_filters[i]
# )
network = tf.keras.layers.Conv2D(
filters=self.params["discriminator_num_filters"][i],
kernel_size=self.params["discriminator_kernel_sizes"][i],
strides=self.params["discriminator_strides"][i],
padding="same",
activation=None,
kernel_regularizer=self.kernel_regularizer,
bias_regularizer=self.bias_regularizer,
name="{}_layers_conv2d_{}".format(self.name, i)
)(inputs=network)
network = tf.keras.layers.LeakyReLU(
alpha=self.params["discriminator_leaky_relu_alpha"],
name="{}_leaky_relu_{}".format(self.name, i)
)(inputs=network)
# Add some dropout for better regularization and stability.
network = tf.keras.layers.Dropout(
rate=self.params["discriminator_dropout_rates"][i],
name="{}_layers_dropout_{}".format(self.name, i)
)(inputs=network)
# Flatten network output.
# shape = (
# batch_size,
# (discriminator_kernel_sizes[-2] / discriminator_strides[-1]) ** 2 * discriminator_num_filters[-1]
# )
network = tf.keras.layers.Flatten()(inputs=network)
# Final linear layer for logits.
# shape = (batch_size, 1)
logits = tf.keras.layers.Dense(
units=1,
activation=None,
kernel_regularizer=self.kernel_regularizer,
bias_regularizer=self.bias_regularizer,
name="{}_layers_dense_logits".format(self.name)
)(inputs=network)
# Define model.
model = tf.keras.Model(
inputs=inputs, outputs=logits, name=self.name
)
return model
def get_model(self):
"""Returns discriminator's `Model` object.
Returns:
Discriminator's `Model` object.
"""
return self.model
def get_discriminator_loss(
self,
global_batch_size,
fake_logits,
real_logits,
global_step,
summary_file_writer
):
"""Gets discriminator loss.
Args:
global_batch_size: int, global batch size for distribution.
fake_logits: tensor, shape of
[batch_size, 1].
real_logits: tensor, shape of
[batch_size, 1].
global_step: int, current global step for training.
summary_file_writer: summary file writer.
Returns:
Tensor of discriminator's total loss of shape [].
"""
if self.params["distribution_strategy"]:
# Calculate base discriminator loss.
discriminator_real_loss = tf.nn.compute_average_loss(
per_example_loss=tf.keras.losses.BinaryCrossentropy(
from_logits=True,
label_smoothing=self.params["label_smoothing"],
reduction=tf.keras.losses.Reduction.NONE
)(
y_true=tf.ones_like(input=real_logits), y_pred=real_logits
),
global_batch_size=global_batch_size
)
discriminator_fake_loss = tf.nn.compute_average_loss(
per_example_loss=tf.keras.losses.BinaryCrossentropy(
from_logits=True,
reduction=tf.keras.losses.Reduction.NONE
)(
y_true=tf.zeros_like(input=fake_logits), y_pred=fake_logits
),
global_batch_size=global_batch_size
)
else:
# Calculate base discriminator loss.
discriminator_real_loss = tf.keras.losses.BinaryCrossentropy(
from_logits=True,
label_smoothing=self.params["label_smoothing"]
)(
y_true=tf.ones_like(input=real_logits), y_pred=real_logits
)
discriminator_fake_loss = tf.keras.losses.BinaryCrossentropy(
from_logits=True
)(
y_true=tf.zeros_like(input=fake_logits), y_pred=fake_logits
)
discriminator_loss = tf.math.add(
x=discriminator_real_loss,
y=discriminator_fake_loss,
name="discriminator_loss"
)
if self.params["distribution_strategy"]:
# Get regularization losses.
discriminator_reg_loss = tf.nn.scale_regularization_loss(
regularization_loss=sum(self.model.losses)
)
else:
# Get regularization losses.
discriminator_reg_loss = sum(self.model.losses)
# Combine losses for total losses.
discriminator_total_loss = tf.math.add(
x=discriminator_loss,
y=discriminator_reg_loss,
name="discriminator_total_loss"
)
if self.params["write_summaries"]:
# Add summaries for TensorBoard.
with summary_file_writer.as_default():
with tf.summary.record_if(
condition=tf.equal(
x=tf.math.floormod(
x=global_step,
y=self.params["save_summary_steps"]
), y=0
)
):
tf.summary.scalar(
name="losses/discriminator_real_loss",
data=discriminator_real_loss,
step=global_step
)
tf.summary.scalar(
name="losses/discriminator_fake_loss",
data=discriminator_fake_loss,
step=global_step
)
tf.summary.scalar(
name="losses/discriminator_loss",
data=discriminator_loss,
step=global_step
)
tf.summary.scalar(
name="losses/discriminator_reg_loss",
data=discriminator_reg_loss,
step=global_step
)
tf.summary.scalar(
name="optimized_losses/discriminator_total_loss",
data=discriminator_total_loss,
step=global_step
)
summary_file_writer.flush()
return discriminator_total_loss
# Instantiate discriminator.
dcgan_discriminator = Discriminator(
input_shape=(arguments["height"], arguments["width"], arguments["depth"]),
kernel_regularizer=tf.keras.regularizers.l1_l2(
l1=arguments["discriminator_l1_regularization_scale"],
l2=arguments["discriminator_l2_regularization_scale"]
),
bias_regularizer=None,
name="discriminator",
params=arguments
)
# Define discriminator model.
discriminator_model = dcgan_discriminator.get_model()
# Summarize discriminator model.
discriminator_model.summary()
discriminator_model.__dict__
# Plot discriminator model.
tf.keras.utils.plot_model(
model=discriminator_model,
to_file="discriminator_model.png",
show_shapes=True,
show_layer_names=True
)
images = tf.random.normal(
shape=[10, arguments["height"], arguments["width"], arguments["depth"]]
)
logits = discriminator_model(inputs=images, training=False)
logits.shape
```
## train_and_evaluate_loop.py
```
class TrainAndEvaluateLoop(object):
"""Train and evaluate loop trainer.
Fields:
params: dict, user passed parameters.
network_objects: dict, instances of `Generator` and `Discriminator`
network objects.
network_models: dict, instances of Keras `Model`s for each network.
optimizers: dict, instances of Keras `Optimizer`s for each network.
strategy: instance of tf.distribute.strategy.
global_batch_size: int, the global batch size after summing batch
sizes across replicas.
global_step: tf.Variable, the global step counter across epochs and
steps within epoch.
checkpoint_manager: instance of `tf.train.CheckpointManager`.
summary_file_writer: instance of tf.summary.create_file_writer for
summaries for TensorBoard.
"""
def __init__(self, params):
"""Instantiate trainer.
Args:
params: dict, user passed parameters.
"""
self.params = params
self.network_objects = {}
self.network_models = {}
self.optimizers = {}
self.strategy = None
self.global_batch_size = None
self.global_step = tf.Variable(
initial_value=tf.zeros(shape=[], dtype=tf.int64),
trainable=False,
name="global_step"
)
self.checkpoint_manager = None
self.summary_file_writer = None
@tf.function
def increment_global_step(self):
self.global_step.assign_add(
delta=tf.ones(shape=[], dtype=tf.int64)
)
# train_and_eval.py
def generator_loss_phase(self, mode, training):
"""Gets fake logits and loss for generator.
Args:
mode: str, what mode currently in: TRAIN or EVAL.
training: bool, if model should be training.
Returns:
Fake logits of shape [batch_size, 1] and generator loss of shape
[].
"""
batch_size = (
self.params["train_batch_size"]
if mode == "TRAIN"
else self.params["eval_batch_size"]
)
# Create random noise latent vector for each batch example.
Z = tf.random.normal(
shape=[batch_size, self.params["latent_size"]],
mean=0.0,
stddev=1.0,
dtype=tf.float32
)
# Get generated image from generator network from gaussian noise.
fake_images = self.network_models["generator"](
inputs=Z, training=training
)
if self.params["write_summaries"] and mode == "TRAIN":
# Add summaries for TensorBoard.
with self.summary_file_writer.as_default():
with tf.summary.record_if(
condition=tf.equal(
x=tf.math.floormod(
x=self.global_step,
y=self.params["save_summary_steps"]
), y=0
)
):
tf.summary.image(
name="fake_images",
data=fake_images,
step=self.global_step,
max_outputs=5
)
self.summary_file_writer.flush()
# Get fake logits from discriminator using generator's output image.
fake_logits = self.network_models["discriminator"](
inputs=fake_images, training=training
)
# Get generator total loss.
generator_total_loss = (
self.network_objects["generator"].get_generator_loss(
global_batch_size=self.global_batch_size,
fake_logits=fake_logits,
global_step=self.global_step,
summary_file_writer=self.summary_file_writer
)
)
return fake_logits, generator_total_loss
def discriminator_loss_phase(self, real_images, fake_logits, training):
"""Gets real logits and loss for discriminator.
Args:
real_images: tensor, real images of shape
[batch_size, height * width * depth].
fake_logits: tensor, discriminator logits of fake images of shape
[batch_size, 1].
training: bool, if in training mode.
Returns:
Real logits of shape [batch_size, 1] and discriminator loss of
shape [].
"""
# Get real logits from discriminator using real image.
real_logits = self.network_models["discriminator"](
inputs=real_images, training=training
)
# Get discriminator total loss.
discriminator_total_loss = (
self.network_objects["discriminator"].get_discriminator_loss(
global_batch_size=self.global_batch_size,
fake_logits=fake_logits,
real_logits=real_logits,
global_step=self.global_step,
summary_file_writer=self.summary_file_writer
)
)
return real_logits, discriminator_total_loss
# train.py
def get_variables_and_gradients(self, loss, gradient_tape, scope):
"""Gets variables and gradients from model wrt. loss.
Args:
loss: tensor, shape of [].
gradient_tape: instance of `GradientTape`.
scope: str, the name of the network of interest.
Returns:
Lists of network's variables and gradients.
"""
# Get trainable variables.
variables = self.network_models[scope].trainable_variables
# Get gradients from gradient tape.
gradients = gradient_tape.gradient(
target=loss, sources=variables
)
# Clip gradients.
if self.params["{}_clip_gradients".format(scope)]:
gradients, _ = tf.clip_by_global_norm(
t_list=gradients,
clip_norm=params["{}_clip_gradients".format(scope)],
name="{}_clip_by_global_norm_gradients".format(scope)
)
# Add variable names back in for identification.
gradients = [
tf.identity(
input=g,
name="{}_{}_gradients".format(scope, v.name[:-2])
)
if tf.is_tensor(x=g) else g
for g, v in zip(gradients, variables)
]
return variables, gradients
def create_variable_and_gradient_histogram_summaries(
self, variables, gradients, scope
):
"""Creates variable and gradient histogram summaries.
Args:
variables: list, network's trainable variables.
gradients: list, gradients of network's trainable variables wrt.
loss.
scope: str, the name of the network of interest.
"""
if self.params["write_summaries"]:
# Add summaries for TensorBoard.
with self.summary_file_writer.as_default():
with tf.summary.record_if(
condition=tf.equal(
x=tf.math.floormod(
x=self.global_step,
y=self.params["save_summary_steps"]
), y=0
)
):
for v, g in zip(variables, gradients):
tf.summary.histogram(
name="{}_variables/{}".format(
scope, v.name[:-2]
),
data=v,
step=self.global_step
)
if tf.is_tensor(x=g):
tf.summary.histogram(
name="{}_gradients/{}".format(
scope, v.name[:-2]
),
data=g,
step=self.global_step
)
self.summary_file_writer.flush()
def get_select_loss_variables_and_gradients(self, real_images, scope):
"""Gets selected network's loss, variables, and gradients.
Args:
real_images: tensor, real images of shape
[batch_size, height * width * depth].
scope: str, the name of the network of interest.
Returns:
Selected network's loss, variables, and gradients.
"""
with tf.GradientTape() as gen_tape, tf.GradientTape() as dis_tape:
# Get fake logits from generator.
fake_logits, generator_loss = self.generator_loss_phase(
mode="TRAIN", training=True
)
# Get discriminator loss.
_, discriminator_loss = self.discriminator_loss_phase(
real_images, fake_logits, training=True
)
# Create empty dicts to hold loss, variables, gradients.
loss_dict = {}
vars_dict = {}
grads_dict = {}
# Loop over generator and discriminator.
for (loss, gradient_tape, scope_name) in zip(
[generator_loss, discriminator_loss],
[gen_tape, dis_tape],
["generator", "discriminator"]
):
# Get variables and gradients from generator wrt. loss.
variables, gradients = self.get_variables_and_gradients(
loss, gradient_tape, scope_name
)
# Add loss, variables, and gradients to dictionaries.
loss_dict[scope_name] = loss
vars_dict[scope_name] = variables
grads_dict[scope_name] = gradients
# Create variable and gradient histogram summaries.
self.create_variable_and_gradient_histogram_summaries(
variables, gradients, scope_name
)
return loss_dict[scope], vars_dict[scope], grads_dict[scope]
def train_network(self, variables, gradients, scope):
"""Trains network variables using gradients with optimizer.
Args:
variables: list, network's trainable variables.
gradients: list, gradients of network's trainable variables wrt.
loss.
scope: str, the name of the network of interest.
"""
# Zip together gradients and variables.
grads_and_vars = zip(gradients, variables)
# Applying gradients to variables using optimizer.
self.optimizers[scope].apply_gradients(grads_and_vars=grads_and_vars)
def train_discriminator(self, features):
"""Trains discriminator network.
Args:
features: dict, feature tensors from input function.
Returns:
Discriminator loss tensor.
"""
# Extract real images from features dictionary.
real_images = features["image"]
# Get gradients for training by running inputs through networks.
loss, variables, gradients = (
self.get_select_loss_variables_and_gradients(
real_images, scope="discriminator"
)
)
# Train discriminator network.
self.train_network(variables, gradients, scope="discriminator")
return loss
def train_generator(self, features):
"""Trains generator network.
Args:
features: dict, feature tensors from input function.
Returns:
Generator loss tensor.
"""
# Extract real images from features dictionary.
real_images = features["image"]
# Get gradients for training by running inputs through networks.
loss, variables, gradients = (
self.get_select_loss_variables_and_gradients(
real_images, scope="generator"
)
)
# Train generator network.
self.train_network(variables, gradients, scope="generator")
return loss
# instantiate_model.py
def instantiate_network_objects(self):
"""Instantiates generator and discriminator with parameters.
"""
# Instantiate generator.
self.network_objects["generator"] = Generator(
input_shape=(self.params["latent_size"]),
kernel_regularizer=tf.keras.regularizers.l1_l2(
l1=self.params["generator_l1_regularization_scale"],
l2=self.params["generator_l2_regularization_scale"]
),
bias_regularizer=None,
name="generator",
params=self.params
)
# Instantiate discriminator.
self.network_objects["discriminator"] = Discriminator(
input_shape=(
self.params["height"],
self.params["width"],
self.params["depth"]
),
kernel_regularizer=tf.keras.regularizers.l1_l2(
l1=self.params["discriminator_l1_regularization_scale"],
l2=self.params["discriminator_l2_regularization_scale"]
),
bias_regularizer=None,
name="discriminator",
params=self.params
)
def instantiate_optimizer(self, scope):
"""Instantiates optimizer with parameters.
Args:
scope: str, the name of the network of interest.
"""
# Create optimizer map.
optimizers = {
"Adadelta": tf.keras.optimizers.Adadelta,
"Adagrad": tf.keras.optimizers.Adagrad,
"Adam": tf.keras.optimizers.Adam,
"Adamax": tf.keras.optimizers.Adamax,
"Ftrl": tf.keras.optimizers.Ftrl,
"Nadam": tf.keras.optimizers.Nadam,
"RMSprop": tf.keras.optimizers.RMSprop,
"SGD": tf.keras.optimizers.SGD
}
# Get optimizer and instantiate it.
if self.params["{}_optimizer".format(scope)] == "Adam":
optimizer = optimizers[self.params["{}_optimizer".format(scope)]](
learning_rate=self.params["{}_learning_rate".format(scope)],
beta_1=self.params["{}_adam_beta1".format(scope)],
beta_2=self.params["{}_adam_beta2".format(scope)],
epsilon=self.params["{}_adam_epsilon".format(scope)],
name="{}_{}_optimizer".format(
scope, self.params["{}_optimizer".format(scope)].lower()
)
)
else:
optimizer = optimizers[self.params["{}_optimizer".format(scope)]](
learning_rate=self.params["{}_learning_rate".format(scope)],
name="{}_{}_optimizer".format(
scope, self.params["{}_optimizer".format(scope)].lower()
)
)
self.optimizers[scope] = optimizer
def instantiate_model_objects(self):
"""Instantiate model network objects, network models, and optimizers.
"""
# Instantiate generator and discriminator objects.
self.instantiate_network_objects()
# Get generator and discriminator `Model`s.
self.network_models["generator"] = (
self.network_objects["generator"].get_model()
)
self.network_models["discriminator"] = (
self.network_objects["discriminator"].get_model()
)
# Instantiate generator optimizer.
self.instantiate_optimizer(scope="generator")
# Instantiate discriminator optimizer.
self.instantiate_optimizer(scope="discriminator")
# model.py
def get_train_eval_datasets(self, num_replicas):
"""Gets train and eval datasets.
Args:
num_replicas: int, number of device replicas.
Returns:
Train and eval datasets.
"""
train_dataset = read_dataset(
filename=self.params["train_file_pattern"],
batch_size=self.params["train_batch_size"] * num_replicas,
params=self.params,
training=True
)()
eval_dataset = read_dataset(
filename=self.params["eval_file_pattern"],
batch_size=self.params["eval_batch_size"] * num_replicas,
params=self.params,
training=False
)()
if self.params["eval_steps"]:
eval_dataset = eval_dataset.take(count=self.params["eval_steps"])
return train_dataset, eval_dataset
def create_checkpoint_machinery(self):
"""Creates checkpoint machinery needed to save & restore checkpoints.
"""
# Create checkpoint instance.
checkpoint_dir = os.path.join(
self.params["output_dir"], "checkpoints"
)
checkpoint_prefix = os.path.join(checkpoint_dir, "ckpt")
checkpoint = tf.train.Checkpoint(
generator_model=self.network_models["generator"],
discriminator_model=self.network_models["discriminator"],
generator_optimizer=self.optimizers["generator"],
discriminator_optimizer=self.optimizers["discriminator"]
)
# Create checkpoint manager.
self.checkpoint_manager = tf.train.CheckpointManager(
checkpoint=checkpoint,
directory=checkpoint_dir,
max_to_keep=self.params["keep_checkpoint_max"],
step_counter=self.global_step,
checkpoint_interval=self.params["save_checkpoints_steps"]
)
# Restore any prior checkpoints.
status = checkpoint.restore(
save_path=self.checkpoint_manager.latest_checkpoint
)
def distributed_eager_discriminator_train_step(self, features):
"""Perform one distributed, eager discriminator train step.
Args:
features: dict, feature tensors from input function.
Returns:
Reduced loss tensor for chosen network across replicas.
"""
if self.params["tf_version"] > 2.1:
run_function = self.strategy.run
else:
run_function = self.strategy.experimental_run_v2
per_replica_losses = run_function(
fn=self.train_discriminator, kwargs={"features": features}
)
return self.strategy.reduce(
reduce_op=tf.distribute.ReduceOp.SUM,
value=per_replica_losses,
axis=None
)
def non_distributed_eager_discriminator_train_step(self, features):
"""Perform one non-distributed, eager discriminator train step.
Args:
features: dict, feature tensors from input function.
Returns:
Reduced loss tensor for chosen network across replicas.
"""
return self.train_discriminator(features=features)
@tf.function
def distributed_graph_discriminator_train_step(self, features):
"""Perform one distributed, graph discriminator train step.
Args:
features: dict, feature tensors from input function.
Returns:
Reduced loss tensor for chosen network across replicas.
"""
if self.params["tf_version"] > 2.1:
run_function = self.strategy.run
else:
run_function = self.strategy.experimental_run_v2
per_replica_losses = run_function(
fn=self.train_discriminator, kwargs={"features": features}
)
return self.strategy.reduce(
reduce_op=tf.distribute.ReduceOp.SUM,
value=per_replica_losses,
axis=None
)
@tf.function
def non_distributed_graph_discriminator_train_step(self, features):
"""Perform one non-distributed, graph discriminator train step.
Args:
features: dict, feature tensors from input function.
Returns:
Reduced loss tensor for chosen network across replicas.
"""
return self.train_discriminator(features=features)
def distributed_eager_generator_train_step(self, features):
"""Perform one distributed, eager generator train step.
Args:
features: dict, feature tensors from input function.
Returns:
Reduced loss tensor for chosen network across replicas.
"""
if self.params["tf_version"] > 2.1:
run_function = self.strategy.run
else:
run_function = self.strategy.experimental_run_v2
per_replica_losses = run_function(
fn=self.train_generator, kwargs={"features": features}
)
return self.strategy.reduce(
reduce_op=tf.distribute.ReduceOp.SUM,
value=per_replica_losses,
axis=None
)
def non_distributed_eager_generator_train_step(self, features):
"""Perform one non-distributed, eager generator train step.
Args:
features: dict, feature tensors from input function.
Returns:
Reduced loss tensor for chosen network across replicas.
"""
return self.train_generator(features=features)
@tf.function
def distributed_graph_generator_train_step(self, features):
"""Perform one distributed, graph generator train step.
Args:
features: dict, feature tensors from input function.
Returns:
Reduced loss tensor for chosen network across replicas.
"""
if self.params["tf_version"] > 2.1:
run_function = self.strategy.run
else:
run_function = self.strategy.experimental_run_v2
per_replica_losses = run_function(
fn=self.train_generator, kwargs={"features": features}
)
return self.strategy.reduce(
reduce_op=tf.distribute.ReduceOp.SUM,
value=per_replica_losses,
axis=None
)
@tf.function
def non_distributed_graph_generator_train_step(self, features):
"""Perform one non-distributed, graph generator train step.
Args:
features: dict, feature tensors from input function.
Returns:
Reduced loss tensor for chosen network across replicas.
"""
return self.train_generator(features=features)
def log_step_loss(self, epoch, epoch_step, loss):
"""Logs step information and loss.
Args:
epoch: int, current iteration fully through the dataset.
epoch_step: int, number of batches through epoch.
loss: float, the loss of the model at the current step.
"""
if self.global_step % self.params["log_step_count_steps"] == 0:
print(
"epoch = {}, global_step = {}, epoch_step = {}, loss = {}".format(
epoch, self.global_step, epoch_step, loss
)
)
def training_loop(self, steps_per_epoch, train_dataset_iter):
"""Logs step information and loss.
Args:
steps_per_epoch: int, number of steps/batches to take each epoch.
train_dataset_iter: iterator, training dataset iterator.
"""
# Get correct train function based on parameters.
if self.strategy:
if self.params["use_graph_mode"]:
discriminator_train_step_fn = (
self.distributed_graph_discriminator_train_step
)
generator_train_step_fn = (
self.distributed_graph_generator_train_step
)
else:
discriminator_train_step_fn = (
self.distributed_eager_discriminator_train_step
)
generator_train_step_fn = (
self.distributed_eager_generator_train_step
)
else:
if self.params["use_graph_mode"]:
discriminator_train_step_fn = (
self.non_distributed_graph_discriminator_train_step
)
generator_train_step_fn = (
self.non_distributed_graph_generator_train_step
)
else:
discriminator_train_step_fn = (
self.non_distributed_eager_discriminator_train_step
)
generator_train_step_fn = (
self.non_distributed_eager_generator_train_step
)
for epoch in range(self.params["num_epochs"]):
for epoch_step in range(steps_per_epoch):
# Train model on batch of features and get loss.
features, labels = next(train_dataset_iter)
# Determine if it is time to train generator or discriminator.
cycle_step = self.global_step % (
self.params["discriminator_train_steps"] + self.params["generator_train_steps"]
)
# Conditionally choose to train generator or discriminator subgraph.
if cycle_step < self.params["discriminator_train_steps"]:
loss = discriminator_train_step_fn(features=features)
else:
loss = generator_train_step_fn(features=features)
# Log step information and loss.
self.log_step_loss(epoch, epoch_step, loss)
# Checkpoint model every save_checkpoints_steps steps.
self.checkpoint_manager.save(
checkpoint_number=self.global_step, check_interval=True
)
# Increment global step.
self.increment_global_step()
def training_loop_end_save_model(self):
"""Saving model when training loop ends.
"""
# Write final checkpoint.
self.checkpoint_manager.save(
checkpoint_number=self.global_step, check_interval=False
)
# Export SavedModel for serving.
export_path = os.path.join(
self.params["output_dir"],
"export",
datetime.datetime.now().strftime("%Y%m%d%H%M%S")
)
# Signature will be serving_default.
tf.saved_model.save(
obj=self.network_models["generator"], export_dir=export_path
)
def train_block(self, train_dataset, eval_dataset):
"""Training block setups training, then loops through datasets.
Args:
train_dataset: instance of `Dataset` for training data.
eval_dataset: instance of `Dataset` for evaluation data.
"""
# Create iterators of datasets.
train_dataset_iter = iter(train_dataset)
eval_dataset_iter = iter(eval_dataset)
steps_per_epoch = (
self.params["train_dataset_length"] // self.global_batch_size
)
# Instantiate model objects.
self.instantiate_model_objects()
# Create checkpoint machinery to save/restore checkpoints.
self.create_checkpoint_machinery()
# Create summary file writer.
self.summary_file_writer = tf.summary.create_file_writer(
logdir=os.path.join(self.params["output_dir"], "summaries"),
name="summary_file_writer"
)
# Run training loop.
self.training_loop(steps_per_epoch, train_dataset_iter)
# Save model at end of training loop.
self.training_loop_end_save_model()
def train_and_evaluate(self):
"""Trains and evaluates Keras model.
Args:
args: dict, user passed parameters.
Returns:
Generator's `Model` object for in-memory predictions.
"""
if self.params["distribution_strategy"]:
# If the list of devices is not specified in the
# Strategy constructor, it will be auto-detected.
if self.params["distribution_strategy"] == "Mirrored":
self.strategy = tf.distribute.MirroredStrategy()
print(
"Number of devices = {}".format(
self.strategy.num_replicas_in_sync
)
)
# Set global batch size for training.
self.global_batch_size = (
self.params["train_batch_size"] * self.strategy.num_replicas_in_sync
)
# Get input datasets. Batch size is split evenly between replicas.
train_dataset, eval_dataset = self.get_train_eval_datasets(
num_replicas=self.strategy.num_replicas_in_sync
)
with self.strategy.scope():
# Create distributed datasets.
train_dist_dataset = (
self.strategy.experimental_distribute_dataset(
dataset=train_dataset
)
)
eval_dist_dataset = (
self.strategy.experimental_distribute_dataset(
dataset=eval_dataset
)
)
# Training block setups training, then loops through datasets.
self.train_block(
train_dataset=train_dist_dataset,
eval_dataset=eval_dist_dataset
)
else:
# Set global batch size for training.
self.global_batch_size = self.params["train_batch_size"]
# Get input datasets.
train_dataset, eval_dataset = self.get_train_eval_datasets(
num_replicas=1
)
# Training block setups training, then loops through datasets.
self.train_block(
train_dataset=train_dataset, eval_dataset=eval_dataset
)
```
## Run model
```
os.environ["OUTPUT_DIR"] = arguments["output_dir"]
%%bash
gsutil -m rm -rf ${OUTPUT_DIR}
train_and_evaluate_loop = TrainAndEvaluateLoop(params=arguments)
train_and_evaluate_loop.train_and_evaluate()
!gsutil ls ${OUTPUT_DIR}
!gsutil ls ${OUTPUT_DIR}/checkpoints
!gsutil ls ${OUTPUT_DIR}/summaries
```
## Prediction
```
!gsutil ls ${OUTPUT_DIR}/export
loaded = tf.saved_model.load(
export_dir=os.path.join(
arguments["output_dir"], "export", "20200731075031"
)
)
print(list(loaded.signatures.keys()))
infer = loaded.signatures["serving_default"]
print(infer.structured_outputs)
Z = tf.random.normal(shape=(10, 512))
predictions = infer(Z)
predictions.keys()
```
Convert image back to the original scale.
```
generated_images = np.clip(
a=tf.cast(
x=((predictions["generator_resize_fake_images"] + 1.0) * (255. / 2)),
dtype=tf.int32
),
a_min=0,
a_max=255
)
print(generated_images.shape)
def plot_images(images):
"""Plots images.
Args:
images: np.array, array of images of
[num_images, image_size, image_size, num_channels].
"""
num_images = len(images)
plt.figure(figsize=(20, 20))
for i in range(num_images):
image = images[i]
plt.subplot(1, num_images, i + 1)
plt.xticks([])
plt.yticks([])
plt.grid(False)
plt.imshow(
tf.reshape(image, image.shape[:-1]),
cmap="gray_r"
)
plt.show()
plot_images(generated_images)
```
|
github_jupyter
|
# Import libraries and modules
import datetime
import matplotlib.pyplot as plt
import numpy as np
import os
import tensorflow as tf
print(np.__version__)
print(tf.__version__)
np.set_printoptions(threshold=np.inf)
arguments = {}
# File arguments.
arguments["train_file_pattern"] = "gs://machine-learning-1234-bucket/gan/data/mnist/train*.tfrecord"
arguments["eval_file_pattern"] = "gs://machine-learning-1234-bucket/gan/data/mnist/test*.tfrecord"
arguments["output_dir"] = "gs://machine-learning-1234-bucket/gan/dcgan/tf2/trained_model"
# Training parameters.
arguments["tf_version"] = 2.2
arguments["use_graph_mode"] = True
arguments["distribution_strategy"] = ""
arguments["write_summaries"] = False
arguments["num_epochs"] = 30
arguments["train_dataset_length"] = 60000
arguments["train_batch_size"] = 32
arguments["log_step_count_steps"] = 100
arguments["save_summary_steps"] = 100
arguments["save_checkpoints_steps"] = 10000
arguments["keep_checkpoint_max"] = 10
arguments["input_fn_autotune"] = False
# Eval parameters.
arguments["eval_batch_size"] = 32
arguments["eval_steps"] = 100
# Image parameters.
arguments["height"] = 28
arguments["width"] = 28
arguments["depth"] = 1
# Generator parameters.
arguments["latent_size"] = 512
arguments["generator_projection_dims"] = [7, 7, 256]
arguments["generator_num_filters"] = [128, 128, arguments["depth"]]
arguments["generator_kernel_sizes"] = [4, 4, 3]
arguments["generator_strides"] = [2, 2, 1]
arguments["generator_leaky_relu_alpha"] = 0.2
arguments["generator_final_activation"] = "tanh"
arguments["generator_l1_regularization_scale"] = 0.
arguments["generator_l2_regularization_scale"] = 0.
arguments["generator_optimizer"] = "Adam"
arguments["generator_learning_rate"] = 0.0002
arguments["generator_adam_beta1"] = 0.5
arguments["generator_adam_beta2"] = 0.999
arguments["generator_adam_epsilon"] = 1e-8
arguments["generator_clip_gradients"] = None
arguments["generator_train_steps"] = 1
# Discriminator hyperparameters.
arguments["discriminator_num_filters"] = [64, 128, 128, 256]
arguments["discriminator_kernel_sizes"] = [3, 3, 3, 3]
arguments["discriminator_strides"] = [1, 2, 2, 2]
arguments["discriminator_dropout_rates"] = [0.3, 0.3, 0.3, 0.3]
arguments["discriminator_leaky_relu_alpha"] = 0.2
arguments["discriminator_l1_regularization_scale"] = 0.
arguments["discriminator_l2_regularization_scale"] = 0.
arguments["discriminator_optimizer"] = "Adam"
arguments["discriminator_learning_rate"] = 0.0002
arguments["discriminator_adam_beta1"] = 0.5
arguments["discriminator_adam_beta2"] = 0.999
arguments["discriminator_adam_epsilon"] = 1e-8
arguments["discriminator_clip_gradients"] = None
arguments["discriminator_train_steps"] = 1
arguments["label_smoothing"] = 0.9
def preprocess_image(image):
"""Preprocess image tensor.
Args:
image: tensor, input image with shape
[batch_size, height, width, depth].
Returns:
Preprocessed image tensor with shape
[batch_size, height, width, depth].
"""
# Convert from [0, 255] -> [-1.0, 1.0] floats.
image = tf.cast(x=image, dtype=tf.float32) * (2. / 255) - 1.0
return image
def decode_example(protos, params):
"""Decodes TFRecord file into tensors.
Given protobufs, decode into image and label tensors.
Args:
protos: protobufs from TFRecord file.
params: dict, user passed parameters.
Returns:
Image and label tensors.
"""
# Create feature schema map for protos.
features = {
"image_raw": tf.io.FixedLenFeature(shape=[], dtype=tf.string),
"label": tf.io.FixedLenFeature(shape=[], dtype=tf.int64)
}
# Parse features from tf.Example.
parsed_features = tf.io.parse_single_example(
serialized=protos, features=features
)
# Convert from a scalar string tensor (whose single string has
# length height * width * depth) to a uint8 tensor with shape
# [height * width * depth].
image = tf.io.decode_raw(
input_bytes=parsed_features["image_raw"], out_type=tf.uint8
)
# Reshape flattened image back into normal dimensions.
image = tf.reshape(
tensor=image,
shape=[params["height"], params["width"], params["depth"]]
)
# Preprocess image.
image = preprocess_image(image=image)
# Convert label from a scalar uint8 tensor to an int32 scalar.
label = tf.cast(x=parsed_features["label"], dtype=tf.int32)
return {"image": image}, label
def read_dataset(filename, batch_size, params, training):
"""Reads TF Record data using tf.data, doing necessary preprocessing.
Given filename, mode, batch size, and other parameters, read TF Record
dataset using Dataset API, apply necessary preprocessing, and return an
input function to the Estimator API.
Args:
filename: str, file pattern that to read into our tf.data dataset.
batch_size: int, number of examples per batch.
params: dict, dictionary of user passed parameters.
training: bool, if training or not.
Returns:
An input function.
"""
def _input_fn():
"""Wrapper input function used by Estimator API to get data tensors.
Returns:
Batched dataset object of dictionary of feature tensors and label
tensor.
"""
# Create list of files that match pattern.
file_list = tf.data.Dataset.list_files(file_pattern=filename)
# Create dataset from file list.
dataset = tf.data.TFRecordDataset(
filenames=file_list,
num_parallel_reads=(
tf.contrib.data.AUTOTUNE
if params["input_fn_autotune"]
else None
)
)
# Shuffle and repeat if training with fused op.
if training:
dataset = dataset.apply(
tf.data.experimental.shuffle_and_repeat(
buffer_size=50 * batch_size,
count=None # indefinitely
)
)
# Decode CSV file into a features dictionary of tensors, then batch.
dataset = dataset.apply(
tf.data.experimental.map_and_batch(
map_func=lambda x: decode_example(
protos=x,
params=params
),
batch_size=batch_size,
num_parallel_calls=(
tf.contrib.data.AUTOTUNE
if params["input_fn_autotune"]
else None
)
)
)
# Prefetch data to improve latency.
dataset = dataset.prefetch(
buffer_size=(
tf.data.experimental.AUTOTUNE
if params["input_fn_autotune"]
else 1
)
)
return dataset
return _input_fn
train_ds = read_dataset(
filename=arguments["train_file_pattern"],
batch_size=arguments["train_batch_size"],
params=arguments,
training=True
)()
train_ds
eval_ds = read_dataset(
filename=arguments["eval_file_pattern"],
batch_size=arguments["eval_batch_size"],
params=arguments,
training=False
)()
eval_ds
class Generator(object):
"""Generator that takes latent vector input and outputs image.
Fields:
name: str, name of `Generator`.
kernel_regularizer: `l1_l2_regularizer` object, regularizar for
kernel variables.
bias_regularizer: `l1_l2_regularizer` object, regularizar for bias
variables.
params: dict, user passed parameters.
model: instance of generator `Model`.
"""
def __init__(
self,
input_shape,
kernel_regularizer,
bias_regularizer,
name,
params):
"""Instantiates and builds generator network.
Args:
input_shape: tuple, shape of latent vector input of shape
[batch_size, latent_size].
kernel_regularizer: `l1_l2_regularizer` object, regularizar for
kernel variables.
bias_regularizer: `l1_l2_regularizer` object, regularizar for bias
variables.
name: str, name of generator.
params: dict, user passed parameters.
"""
# Set name of generator.
self.name = name
# Store regularizers.
self.kernel_regularizer = kernel_regularizer
self.bias_regularizer = bias_regularizer
# Store parameters.
self.params = params
# Instantiate generator `Model`.
self.model = self._define_generator(input_shape)
def _project_latent_vectors(self, latent_vectors):
"""Defines generator network.
Args:
latent_vectors: tensor, latent vector inputs of shape
[batch_size, latent_size].
Returns:
Projected image of latent vector inputs.
"""
projection_height = self.params["generator_projection_dims"][0]
projection_width = self.params["generator_projection_dims"][1]
projection_depth = self.params["generator_projection_dims"][2]
# shape = (
# batch_size,
# projection_height * projection_width * projection_depth
# )
projection = tf.keras.layers.Dense(
units=projection_height * projection_width * projection_depth,
activation=None,
kernel_regularizer=self.kernel_regularizer,
bias_regularizer=self.bias_regularizer,
name="projection_dense_layer"
)(inputs=latent_vectors)
projection_leaky_relu = tf.keras.layers.LeakyReLU(
alpha=self.params["generator_leaky_relu_alpha"],
name="projection_leaky_relu"
)(inputs=projection)
# Add batch normalization to keep the inputs from blowing up.
# shape = (
# batch_size,
# projection_height * projection_width * projection_depth
# )
projection_batch_norm = tf.keras.layers.BatchNormalization(
name="projection_batch_norm"
)(inputs=projection_leaky_relu)
# Reshape projection into "image".
# shape = (
# batch_size,
# projection_height,
# projection_width,
# projection_depth
# )
projected_image = tf.reshape(
tensor=projection_batch_norm,
shape=[
-1, projection_height, projection_width, projection_depth
],
name="projected_image"
)
return projected_image
def _define_generator(
self, input_shape):
"""Defines generator network.
Args:
input_shape: tuple, shape of latent vector input of shape
[batch_size, latent_size].
Returns:
Instance of `Model` object.
"""
# Create the input layer to generator.
# shape = (batch_size, latent_size)
inputs = tf.keras.Input(
shape=input_shape, name="{}_inputs".format(self.name)
)
# Dictionary containing possible final activations.
final_activation_set = {"sigmoid", "relu", "tanh"}
# Project latent vectors.
network = self._project_latent_vectors(latent_vectors=inputs)
# Iteratively build upsampling layers.
for i in range(len(self.params["generator_num_filters"]) - 1):
# Add conv transpose layers with given params per layer.
# shape = (
# batch_size,
# generator_kernel_sizes[i - 1] * generator_strides[i],
# generator_kernel_sizes[i - 1] * generator_strides[i],
# generator_num_filters[i]
# )
network = tf.keras.layers.Conv2DTranspose(
filters=self.params["generator_num_filters"][i],
kernel_size=self.params["generator_kernel_sizes"][i],
strides=self.params["generator_strides"][i],
padding="same",
activation=None,
kernel_regularizer=self.kernel_regularizer,
bias_regularizer=self.bias_regularizer,
name="{}_layers_conv2d_tranpose_{}".format(self.name, i)
)(inputs=network)
network = tf.keras.layers.LeakyReLU(
alpha=self.params["generator_leaky_relu_alpha"],
name="{}_leaky_relu_{}".format(self.name, i)
)(inputs=network)
# Add batch normalization to keep the inputs from blowing up.
network = tf.keras.layers.BatchNormalization(
name="{}_layers_batch_norm_{}".format(self.name, i)
)(inputs=network)
# Final conv2d transpose layer for image output.
# shape = (batch_size, height, width, depth)
fake_images = tf.keras.layers.Conv2DTranspose(
filters=self.params["generator_num_filters"][-1],
kernel_size=self.params["generator_kernel_sizes"][-1],
strides=self.params["generator_strides"][-1],
padding="same",
activation=(
self.params["generator_final_activation"].lower()
if self.params["generator_final_activation"].lower()
in final_activation_set
else None
),
kernel_regularizer=self.kernel_regularizer,
bias_regularizer=self.bias_regularizer,
name="{}_layers_conv2d_tranpose_fake_images".format(self.name)
)(inputs=network)
# Resize fake images to match real images in case of mismatch.
height = self.params["height"]
width = self.params["width"]
fake_images = tf.keras.layers.Lambda(
function=lambda x: tf.image.resize(
images=x, size=[height, width], method="bilinear"
),
name="{}_resize_fake_images".format(self.name)
)(inputs=fake_images)
# Define model.
model = tf.keras.Model(
inputs=inputs, outputs=fake_images, name=self.name
)
return model
def get_model(self):
"""Returns generator's `Model` object.
Returns:
Generator's `Model` object.
"""
return self.model
def get_generator_loss(
self,
global_batch_size,
fake_logits,
global_step,
summary_file_writer
):
"""Gets generator loss.
Args:
global_batch_size: int, global batch size for distribution.
fake_logits: tensor, shape of
[batch_size, 1].
global_step: int, current global step for training.
summary_file_writer: summary file writer.
Returns:
Tensor of generator's total loss of shape [].
"""
if self.params["distribution_strategy"]:
# Calculate base generator loss.
generator_loss = tf.nn.compute_average_loss(
per_example_loss=tf.keras.losses.BinaryCrossentropy(
from_logits=True,
reduction=tf.keras.losses.Reduction.NONE
)(
y_true=tf.ones_like(input=fake_logits), y_pred=fake_logits
),
global_batch_size=global_batch_size
)
# Get regularization losses.
generator_reg_loss = tf.nn.scale_regularization_loss(
regularization_loss=sum(self.model.losses)
)
else:
# Calculate base generator loss.
generator_loss = tf.keras.losses.BinaryCrossentropy(
from_logits=True
)(
y_true=tf.ones_like(input=fake_logits), y_pred=fake_logits
)
# Get regularization losses.
generator_reg_loss = sum(self.model.losses)
# Combine losses for total losses.
generator_total_loss = tf.math.add(
x=generator_loss,
y=generator_reg_loss,
name="generator_total_loss"
)
if self.params["write_summaries"]:
# Add summaries for TensorBoard.
with summary_file_writer.as_default():
with tf.summary.record_if(
condition=tf.equal(
x=tf.math.floormod(
x=global_step,
y=self.params["save_summary_steps"]
), y=0
)
):
tf.summary.scalar(
name="losses/generator_loss",
data=generator_loss,
step=global_step
)
tf.summary.scalar(
name="losses/generator_reg_loss",
data=generator_reg_loss,
step=global_step
)
tf.summary.scalar(
name="optimized_losses/generator_total_loss",
data=generator_total_loss,
step=global_step
)
summary_file_writer.flush()
return generator_total_loss
# Instantiate generator.
dcgan_generator = Generator(
input_shape=(arguments["latent_size"]),
kernel_regularizer=tf.keras.regularizers.l1_l2(
l1=arguments["generator_l1_regularization_scale"],
l2=arguments["generator_l2_regularization_scale"]
),
bias_regularizer=None,
name="generator",
params=arguments
)
# Define generator model.
generator_model = dcgan_generator.get_model()
# Summarize generator model.
generator_model.summary()
generator_model.__dict__
# Plot generator model.
tf.keras.utils.plot_model(
model=generator_model,
to_file="generator_model.png",
show_shapes=True,
show_layer_names=True
)
noise = tf.random.normal(shape=[10, arguments["latent_size"]])
fake_images = generator_model(inputs=noise, training=False)
plt.imshow(fake_images[0, :, :, 0], cmap='gray')
class Discriminator(object):
"""Discriminator that takes image input and outputs logits.
Fields:
name: str, name of `Discriminator`.
kernel_regularizer: `l1_l2_regularizer` object, regularizar for
kernel variables.
bias_regularizer: `l1_l2_regularizer` object, regularizar for bias
variables.
params: dict, user passed parameters.
model: instance of discriminator `Model`.
"""
def __init__(
self,
input_shape,
kernel_regularizer,
bias_regularizer,
name,
params):
"""Instantiates and builds discriminator network.
Args:
input_shape: tuple, shape of image vector input of shape
[batch_size, height * width * depth].
kernel_regularizer: `l1_l2_regularizer` object, regularizar for
kernel variables.
bias_regularizer: `l1_l2_regularizer` object, regularizar for bias
variables.
name: str, name of discriminator.
params: dict, user passed parameters.
"""
# Set name of discriminator.
self.name = name
# Store regularizers.
self.kernel_regularizer = kernel_regularizer
self.bias_regularizer = bias_regularizer
# Store parameters.
self.params = params
# Instantiate discriminator `Model`.
self.model = self._define_discriminator(input_shape)
def _define_discriminator(self, input_shape):
"""Defines discriminator network.
Args:
input_shape: tuple, shape of image vector input of shape
[batch_size, height * width * depth].
Returns:
Instance of `Model` object.
"""
# Create the input layer to discriminator.
# shape = (batch_size, height * width * depth)
inputs = tf.keras.Input(
shape=input_shape,
name="{}_inputs".format(self.name)
)
network = inputs
# Iteratively build downsampling layers.
for i in range(len(self.params["discriminator_num_filters"])):
# Add convolutional layers with given params per layer.
# shape = (
# batch_size,
# discriminator_kernel_sizes[i - 1] / discriminator_strides[i],
# discriminator_kernel_sizes[i - 1] / discriminator_strides[i],
# discriminator_num_filters[i]
# )
network = tf.keras.layers.Conv2D(
filters=self.params["discriminator_num_filters"][i],
kernel_size=self.params["discriminator_kernel_sizes"][i],
strides=self.params["discriminator_strides"][i],
padding="same",
activation=None,
kernel_regularizer=self.kernel_regularizer,
bias_regularizer=self.bias_regularizer,
name="{}_layers_conv2d_{}".format(self.name, i)
)(inputs=network)
network = tf.keras.layers.LeakyReLU(
alpha=self.params["discriminator_leaky_relu_alpha"],
name="{}_leaky_relu_{}".format(self.name, i)
)(inputs=network)
# Add some dropout for better regularization and stability.
network = tf.keras.layers.Dropout(
rate=self.params["discriminator_dropout_rates"][i],
name="{}_layers_dropout_{}".format(self.name, i)
)(inputs=network)
# Flatten network output.
# shape = (
# batch_size,
# (discriminator_kernel_sizes[-2] / discriminator_strides[-1]) ** 2 * discriminator_num_filters[-1]
# )
network = tf.keras.layers.Flatten()(inputs=network)
# Final linear layer for logits.
# shape = (batch_size, 1)
logits = tf.keras.layers.Dense(
units=1,
activation=None,
kernel_regularizer=self.kernel_regularizer,
bias_regularizer=self.bias_regularizer,
name="{}_layers_dense_logits".format(self.name)
)(inputs=network)
# Define model.
model = tf.keras.Model(
inputs=inputs, outputs=logits, name=self.name
)
return model
def get_model(self):
"""Returns discriminator's `Model` object.
Returns:
Discriminator's `Model` object.
"""
return self.model
def get_discriminator_loss(
self,
global_batch_size,
fake_logits,
real_logits,
global_step,
summary_file_writer
):
"""Gets discriminator loss.
Args:
global_batch_size: int, global batch size for distribution.
fake_logits: tensor, shape of
[batch_size, 1].
real_logits: tensor, shape of
[batch_size, 1].
global_step: int, current global step for training.
summary_file_writer: summary file writer.
Returns:
Tensor of discriminator's total loss of shape [].
"""
if self.params["distribution_strategy"]:
# Calculate base discriminator loss.
discriminator_real_loss = tf.nn.compute_average_loss(
per_example_loss=tf.keras.losses.BinaryCrossentropy(
from_logits=True,
label_smoothing=self.params["label_smoothing"],
reduction=tf.keras.losses.Reduction.NONE
)(
y_true=tf.ones_like(input=real_logits), y_pred=real_logits
),
global_batch_size=global_batch_size
)
discriminator_fake_loss = tf.nn.compute_average_loss(
per_example_loss=tf.keras.losses.BinaryCrossentropy(
from_logits=True,
reduction=tf.keras.losses.Reduction.NONE
)(
y_true=tf.zeros_like(input=fake_logits), y_pred=fake_logits
),
global_batch_size=global_batch_size
)
else:
# Calculate base discriminator loss.
discriminator_real_loss = tf.keras.losses.BinaryCrossentropy(
from_logits=True,
label_smoothing=self.params["label_smoothing"]
)(
y_true=tf.ones_like(input=real_logits), y_pred=real_logits
)
discriminator_fake_loss = tf.keras.losses.BinaryCrossentropy(
from_logits=True
)(
y_true=tf.zeros_like(input=fake_logits), y_pred=fake_logits
)
discriminator_loss = tf.math.add(
x=discriminator_real_loss,
y=discriminator_fake_loss,
name="discriminator_loss"
)
if self.params["distribution_strategy"]:
# Get regularization losses.
discriminator_reg_loss = tf.nn.scale_regularization_loss(
regularization_loss=sum(self.model.losses)
)
else:
# Get regularization losses.
discriminator_reg_loss = sum(self.model.losses)
# Combine losses for total losses.
discriminator_total_loss = tf.math.add(
x=discriminator_loss,
y=discriminator_reg_loss,
name="discriminator_total_loss"
)
if self.params["write_summaries"]:
# Add summaries for TensorBoard.
with summary_file_writer.as_default():
with tf.summary.record_if(
condition=tf.equal(
x=tf.math.floormod(
x=global_step,
y=self.params["save_summary_steps"]
), y=0
)
):
tf.summary.scalar(
name="losses/discriminator_real_loss",
data=discriminator_real_loss,
step=global_step
)
tf.summary.scalar(
name="losses/discriminator_fake_loss",
data=discriminator_fake_loss,
step=global_step
)
tf.summary.scalar(
name="losses/discriminator_loss",
data=discriminator_loss,
step=global_step
)
tf.summary.scalar(
name="losses/discriminator_reg_loss",
data=discriminator_reg_loss,
step=global_step
)
tf.summary.scalar(
name="optimized_losses/discriminator_total_loss",
data=discriminator_total_loss,
step=global_step
)
summary_file_writer.flush()
return discriminator_total_loss
# Instantiate discriminator.
dcgan_discriminator = Discriminator(
input_shape=(arguments["height"], arguments["width"], arguments["depth"]),
kernel_regularizer=tf.keras.regularizers.l1_l2(
l1=arguments["discriminator_l1_regularization_scale"],
l2=arguments["discriminator_l2_regularization_scale"]
),
bias_regularizer=None,
name="discriminator",
params=arguments
)
# Define discriminator model.
discriminator_model = dcgan_discriminator.get_model()
# Summarize discriminator model.
discriminator_model.summary()
discriminator_model.__dict__
# Plot discriminator model.
tf.keras.utils.plot_model(
model=discriminator_model,
to_file="discriminator_model.png",
show_shapes=True,
show_layer_names=True
)
images = tf.random.normal(
shape=[10, arguments["height"], arguments["width"], arguments["depth"]]
)
logits = discriminator_model(inputs=images, training=False)
logits.shape
class TrainAndEvaluateLoop(object):
"""Train and evaluate loop trainer.
Fields:
params: dict, user passed parameters.
network_objects: dict, instances of `Generator` and `Discriminator`
network objects.
network_models: dict, instances of Keras `Model`s for each network.
optimizers: dict, instances of Keras `Optimizer`s for each network.
strategy: instance of tf.distribute.strategy.
global_batch_size: int, the global batch size after summing batch
sizes across replicas.
global_step: tf.Variable, the global step counter across epochs and
steps within epoch.
checkpoint_manager: instance of `tf.train.CheckpointManager`.
summary_file_writer: instance of tf.summary.create_file_writer for
summaries for TensorBoard.
"""
def __init__(self, params):
"""Instantiate trainer.
Args:
params: dict, user passed parameters.
"""
self.params = params
self.network_objects = {}
self.network_models = {}
self.optimizers = {}
self.strategy = None
self.global_batch_size = None
self.global_step = tf.Variable(
initial_value=tf.zeros(shape=[], dtype=tf.int64),
trainable=False,
name="global_step"
)
self.checkpoint_manager = None
self.summary_file_writer = None
@tf.function
def increment_global_step(self):
self.global_step.assign_add(
delta=tf.ones(shape=[], dtype=tf.int64)
)
# train_and_eval.py
def generator_loss_phase(self, mode, training):
"""Gets fake logits and loss for generator.
Args:
mode: str, what mode currently in: TRAIN or EVAL.
training: bool, if model should be training.
Returns:
Fake logits of shape [batch_size, 1] and generator loss of shape
[].
"""
batch_size = (
self.params["train_batch_size"]
if mode == "TRAIN"
else self.params["eval_batch_size"]
)
# Create random noise latent vector for each batch example.
Z = tf.random.normal(
shape=[batch_size, self.params["latent_size"]],
mean=0.0,
stddev=1.0,
dtype=tf.float32
)
# Get generated image from generator network from gaussian noise.
fake_images = self.network_models["generator"](
inputs=Z, training=training
)
if self.params["write_summaries"] and mode == "TRAIN":
# Add summaries for TensorBoard.
with self.summary_file_writer.as_default():
with tf.summary.record_if(
condition=tf.equal(
x=tf.math.floormod(
x=self.global_step,
y=self.params["save_summary_steps"]
), y=0
)
):
tf.summary.image(
name="fake_images",
data=fake_images,
step=self.global_step,
max_outputs=5
)
self.summary_file_writer.flush()
# Get fake logits from discriminator using generator's output image.
fake_logits = self.network_models["discriminator"](
inputs=fake_images, training=training
)
# Get generator total loss.
generator_total_loss = (
self.network_objects["generator"].get_generator_loss(
global_batch_size=self.global_batch_size,
fake_logits=fake_logits,
global_step=self.global_step,
summary_file_writer=self.summary_file_writer
)
)
return fake_logits, generator_total_loss
def discriminator_loss_phase(self, real_images, fake_logits, training):
"""Gets real logits and loss for discriminator.
Args:
real_images: tensor, real images of shape
[batch_size, height * width * depth].
fake_logits: tensor, discriminator logits of fake images of shape
[batch_size, 1].
training: bool, if in training mode.
Returns:
Real logits of shape [batch_size, 1] and discriminator loss of
shape [].
"""
# Get real logits from discriminator using real image.
real_logits = self.network_models["discriminator"](
inputs=real_images, training=training
)
# Get discriminator total loss.
discriminator_total_loss = (
self.network_objects["discriminator"].get_discriminator_loss(
global_batch_size=self.global_batch_size,
fake_logits=fake_logits,
real_logits=real_logits,
global_step=self.global_step,
summary_file_writer=self.summary_file_writer
)
)
return real_logits, discriminator_total_loss
# train.py
def get_variables_and_gradients(self, loss, gradient_tape, scope):
"""Gets variables and gradients from model wrt. loss.
Args:
loss: tensor, shape of [].
gradient_tape: instance of `GradientTape`.
scope: str, the name of the network of interest.
Returns:
Lists of network's variables and gradients.
"""
# Get trainable variables.
variables = self.network_models[scope].trainable_variables
# Get gradients from gradient tape.
gradients = gradient_tape.gradient(
target=loss, sources=variables
)
# Clip gradients.
if self.params["{}_clip_gradients".format(scope)]:
gradients, _ = tf.clip_by_global_norm(
t_list=gradients,
clip_norm=params["{}_clip_gradients".format(scope)],
name="{}_clip_by_global_norm_gradients".format(scope)
)
# Add variable names back in for identification.
gradients = [
tf.identity(
input=g,
name="{}_{}_gradients".format(scope, v.name[:-2])
)
if tf.is_tensor(x=g) else g
for g, v in zip(gradients, variables)
]
return variables, gradients
def create_variable_and_gradient_histogram_summaries(
self, variables, gradients, scope
):
"""Creates variable and gradient histogram summaries.
Args:
variables: list, network's trainable variables.
gradients: list, gradients of network's trainable variables wrt.
loss.
scope: str, the name of the network of interest.
"""
if self.params["write_summaries"]:
# Add summaries for TensorBoard.
with self.summary_file_writer.as_default():
with tf.summary.record_if(
condition=tf.equal(
x=tf.math.floormod(
x=self.global_step,
y=self.params["save_summary_steps"]
), y=0
)
):
for v, g in zip(variables, gradients):
tf.summary.histogram(
name="{}_variables/{}".format(
scope, v.name[:-2]
),
data=v,
step=self.global_step
)
if tf.is_tensor(x=g):
tf.summary.histogram(
name="{}_gradients/{}".format(
scope, v.name[:-2]
),
data=g,
step=self.global_step
)
self.summary_file_writer.flush()
def get_select_loss_variables_and_gradients(self, real_images, scope):
"""Gets selected network's loss, variables, and gradients.
Args:
real_images: tensor, real images of shape
[batch_size, height * width * depth].
scope: str, the name of the network of interest.
Returns:
Selected network's loss, variables, and gradients.
"""
with tf.GradientTape() as gen_tape, tf.GradientTape() as dis_tape:
# Get fake logits from generator.
fake_logits, generator_loss = self.generator_loss_phase(
mode="TRAIN", training=True
)
# Get discriminator loss.
_, discriminator_loss = self.discriminator_loss_phase(
real_images, fake_logits, training=True
)
# Create empty dicts to hold loss, variables, gradients.
loss_dict = {}
vars_dict = {}
grads_dict = {}
# Loop over generator and discriminator.
for (loss, gradient_tape, scope_name) in zip(
[generator_loss, discriminator_loss],
[gen_tape, dis_tape],
["generator", "discriminator"]
):
# Get variables and gradients from generator wrt. loss.
variables, gradients = self.get_variables_and_gradients(
loss, gradient_tape, scope_name
)
# Add loss, variables, and gradients to dictionaries.
loss_dict[scope_name] = loss
vars_dict[scope_name] = variables
grads_dict[scope_name] = gradients
# Create variable and gradient histogram summaries.
self.create_variable_and_gradient_histogram_summaries(
variables, gradients, scope_name
)
return loss_dict[scope], vars_dict[scope], grads_dict[scope]
def train_network(self, variables, gradients, scope):
"""Trains network variables using gradients with optimizer.
Args:
variables: list, network's trainable variables.
gradients: list, gradients of network's trainable variables wrt.
loss.
scope: str, the name of the network of interest.
"""
# Zip together gradients and variables.
grads_and_vars = zip(gradients, variables)
# Applying gradients to variables using optimizer.
self.optimizers[scope].apply_gradients(grads_and_vars=grads_and_vars)
def train_discriminator(self, features):
"""Trains discriminator network.
Args:
features: dict, feature tensors from input function.
Returns:
Discriminator loss tensor.
"""
# Extract real images from features dictionary.
real_images = features["image"]
# Get gradients for training by running inputs through networks.
loss, variables, gradients = (
self.get_select_loss_variables_and_gradients(
real_images, scope="discriminator"
)
)
# Train discriminator network.
self.train_network(variables, gradients, scope="discriminator")
return loss
def train_generator(self, features):
"""Trains generator network.
Args:
features: dict, feature tensors from input function.
Returns:
Generator loss tensor.
"""
# Extract real images from features dictionary.
real_images = features["image"]
# Get gradients for training by running inputs through networks.
loss, variables, gradients = (
self.get_select_loss_variables_and_gradients(
real_images, scope="generator"
)
)
# Train generator network.
self.train_network(variables, gradients, scope="generator")
return loss
# instantiate_model.py
def instantiate_network_objects(self):
"""Instantiates generator and discriminator with parameters.
"""
# Instantiate generator.
self.network_objects["generator"] = Generator(
input_shape=(self.params["latent_size"]),
kernel_regularizer=tf.keras.regularizers.l1_l2(
l1=self.params["generator_l1_regularization_scale"],
l2=self.params["generator_l2_regularization_scale"]
),
bias_regularizer=None,
name="generator",
params=self.params
)
# Instantiate discriminator.
self.network_objects["discriminator"] = Discriminator(
input_shape=(
self.params["height"],
self.params["width"],
self.params["depth"]
),
kernel_regularizer=tf.keras.regularizers.l1_l2(
l1=self.params["discriminator_l1_regularization_scale"],
l2=self.params["discriminator_l2_regularization_scale"]
),
bias_regularizer=None,
name="discriminator",
params=self.params
)
def instantiate_optimizer(self, scope):
"""Instantiates optimizer with parameters.
Args:
scope: str, the name of the network of interest.
"""
# Create optimizer map.
optimizers = {
"Adadelta": tf.keras.optimizers.Adadelta,
"Adagrad": tf.keras.optimizers.Adagrad,
"Adam": tf.keras.optimizers.Adam,
"Adamax": tf.keras.optimizers.Adamax,
"Ftrl": tf.keras.optimizers.Ftrl,
"Nadam": tf.keras.optimizers.Nadam,
"RMSprop": tf.keras.optimizers.RMSprop,
"SGD": tf.keras.optimizers.SGD
}
# Get optimizer and instantiate it.
if self.params["{}_optimizer".format(scope)] == "Adam":
optimizer = optimizers[self.params["{}_optimizer".format(scope)]](
learning_rate=self.params["{}_learning_rate".format(scope)],
beta_1=self.params["{}_adam_beta1".format(scope)],
beta_2=self.params["{}_adam_beta2".format(scope)],
epsilon=self.params["{}_adam_epsilon".format(scope)],
name="{}_{}_optimizer".format(
scope, self.params["{}_optimizer".format(scope)].lower()
)
)
else:
optimizer = optimizers[self.params["{}_optimizer".format(scope)]](
learning_rate=self.params["{}_learning_rate".format(scope)],
name="{}_{}_optimizer".format(
scope, self.params["{}_optimizer".format(scope)].lower()
)
)
self.optimizers[scope] = optimizer
def instantiate_model_objects(self):
"""Instantiate model network objects, network models, and optimizers.
"""
# Instantiate generator and discriminator objects.
self.instantiate_network_objects()
# Get generator and discriminator `Model`s.
self.network_models["generator"] = (
self.network_objects["generator"].get_model()
)
self.network_models["discriminator"] = (
self.network_objects["discriminator"].get_model()
)
# Instantiate generator optimizer.
self.instantiate_optimizer(scope="generator")
# Instantiate discriminator optimizer.
self.instantiate_optimizer(scope="discriminator")
# model.py
def get_train_eval_datasets(self, num_replicas):
"""Gets train and eval datasets.
Args:
num_replicas: int, number of device replicas.
Returns:
Train and eval datasets.
"""
train_dataset = read_dataset(
filename=self.params["train_file_pattern"],
batch_size=self.params["train_batch_size"] * num_replicas,
params=self.params,
training=True
)()
eval_dataset = read_dataset(
filename=self.params["eval_file_pattern"],
batch_size=self.params["eval_batch_size"] * num_replicas,
params=self.params,
training=False
)()
if self.params["eval_steps"]:
eval_dataset = eval_dataset.take(count=self.params["eval_steps"])
return train_dataset, eval_dataset
def create_checkpoint_machinery(self):
"""Creates checkpoint machinery needed to save & restore checkpoints.
"""
# Create checkpoint instance.
checkpoint_dir = os.path.join(
self.params["output_dir"], "checkpoints"
)
checkpoint_prefix = os.path.join(checkpoint_dir, "ckpt")
checkpoint = tf.train.Checkpoint(
generator_model=self.network_models["generator"],
discriminator_model=self.network_models["discriminator"],
generator_optimizer=self.optimizers["generator"],
discriminator_optimizer=self.optimizers["discriminator"]
)
# Create checkpoint manager.
self.checkpoint_manager = tf.train.CheckpointManager(
checkpoint=checkpoint,
directory=checkpoint_dir,
max_to_keep=self.params["keep_checkpoint_max"],
step_counter=self.global_step,
checkpoint_interval=self.params["save_checkpoints_steps"]
)
# Restore any prior checkpoints.
status = checkpoint.restore(
save_path=self.checkpoint_manager.latest_checkpoint
)
def distributed_eager_discriminator_train_step(self, features):
"""Perform one distributed, eager discriminator train step.
Args:
features: dict, feature tensors from input function.
Returns:
Reduced loss tensor for chosen network across replicas.
"""
if self.params["tf_version"] > 2.1:
run_function = self.strategy.run
else:
run_function = self.strategy.experimental_run_v2
per_replica_losses = run_function(
fn=self.train_discriminator, kwargs={"features": features}
)
return self.strategy.reduce(
reduce_op=tf.distribute.ReduceOp.SUM,
value=per_replica_losses,
axis=None
)
def non_distributed_eager_discriminator_train_step(self, features):
"""Perform one non-distributed, eager discriminator train step.
Args:
features: dict, feature tensors from input function.
Returns:
Reduced loss tensor for chosen network across replicas.
"""
return self.train_discriminator(features=features)
@tf.function
def distributed_graph_discriminator_train_step(self, features):
"""Perform one distributed, graph discriminator train step.
Args:
features: dict, feature tensors from input function.
Returns:
Reduced loss tensor for chosen network across replicas.
"""
if self.params["tf_version"] > 2.1:
run_function = self.strategy.run
else:
run_function = self.strategy.experimental_run_v2
per_replica_losses = run_function(
fn=self.train_discriminator, kwargs={"features": features}
)
return self.strategy.reduce(
reduce_op=tf.distribute.ReduceOp.SUM,
value=per_replica_losses,
axis=None
)
@tf.function
def non_distributed_graph_discriminator_train_step(self, features):
"""Perform one non-distributed, graph discriminator train step.
Args:
features: dict, feature tensors from input function.
Returns:
Reduced loss tensor for chosen network across replicas.
"""
return self.train_discriminator(features=features)
def distributed_eager_generator_train_step(self, features):
"""Perform one distributed, eager generator train step.
Args:
features: dict, feature tensors from input function.
Returns:
Reduced loss tensor for chosen network across replicas.
"""
if self.params["tf_version"] > 2.1:
run_function = self.strategy.run
else:
run_function = self.strategy.experimental_run_v2
per_replica_losses = run_function(
fn=self.train_generator, kwargs={"features": features}
)
return self.strategy.reduce(
reduce_op=tf.distribute.ReduceOp.SUM,
value=per_replica_losses,
axis=None
)
def non_distributed_eager_generator_train_step(self, features):
"""Perform one non-distributed, eager generator train step.
Args:
features: dict, feature tensors from input function.
Returns:
Reduced loss tensor for chosen network across replicas.
"""
return self.train_generator(features=features)
@tf.function
def distributed_graph_generator_train_step(self, features):
"""Perform one distributed, graph generator train step.
Args:
features: dict, feature tensors from input function.
Returns:
Reduced loss tensor for chosen network across replicas.
"""
if self.params["tf_version"] > 2.1:
run_function = self.strategy.run
else:
run_function = self.strategy.experimental_run_v2
per_replica_losses = run_function(
fn=self.train_generator, kwargs={"features": features}
)
return self.strategy.reduce(
reduce_op=tf.distribute.ReduceOp.SUM,
value=per_replica_losses,
axis=None
)
@tf.function
def non_distributed_graph_generator_train_step(self, features):
"""Perform one non-distributed, graph generator train step.
Args:
features: dict, feature tensors from input function.
Returns:
Reduced loss tensor for chosen network across replicas.
"""
return self.train_generator(features=features)
def log_step_loss(self, epoch, epoch_step, loss):
"""Logs step information and loss.
Args:
epoch: int, current iteration fully through the dataset.
epoch_step: int, number of batches through epoch.
loss: float, the loss of the model at the current step.
"""
if self.global_step % self.params["log_step_count_steps"] == 0:
print(
"epoch = {}, global_step = {}, epoch_step = {}, loss = {}".format(
epoch, self.global_step, epoch_step, loss
)
)
def training_loop(self, steps_per_epoch, train_dataset_iter):
"""Logs step information and loss.
Args:
steps_per_epoch: int, number of steps/batches to take each epoch.
train_dataset_iter: iterator, training dataset iterator.
"""
# Get correct train function based on parameters.
if self.strategy:
if self.params["use_graph_mode"]:
discriminator_train_step_fn = (
self.distributed_graph_discriminator_train_step
)
generator_train_step_fn = (
self.distributed_graph_generator_train_step
)
else:
discriminator_train_step_fn = (
self.distributed_eager_discriminator_train_step
)
generator_train_step_fn = (
self.distributed_eager_generator_train_step
)
else:
if self.params["use_graph_mode"]:
discriminator_train_step_fn = (
self.non_distributed_graph_discriminator_train_step
)
generator_train_step_fn = (
self.non_distributed_graph_generator_train_step
)
else:
discriminator_train_step_fn = (
self.non_distributed_eager_discriminator_train_step
)
generator_train_step_fn = (
self.non_distributed_eager_generator_train_step
)
for epoch in range(self.params["num_epochs"]):
for epoch_step in range(steps_per_epoch):
# Train model on batch of features and get loss.
features, labels = next(train_dataset_iter)
# Determine if it is time to train generator or discriminator.
cycle_step = self.global_step % (
self.params["discriminator_train_steps"] + self.params["generator_train_steps"]
)
# Conditionally choose to train generator or discriminator subgraph.
if cycle_step < self.params["discriminator_train_steps"]:
loss = discriminator_train_step_fn(features=features)
else:
loss = generator_train_step_fn(features=features)
# Log step information and loss.
self.log_step_loss(epoch, epoch_step, loss)
# Checkpoint model every save_checkpoints_steps steps.
self.checkpoint_manager.save(
checkpoint_number=self.global_step, check_interval=True
)
# Increment global step.
self.increment_global_step()
def training_loop_end_save_model(self):
"""Saving model when training loop ends.
"""
# Write final checkpoint.
self.checkpoint_manager.save(
checkpoint_number=self.global_step, check_interval=False
)
# Export SavedModel for serving.
export_path = os.path.join(
self.params["output_dir"],
"export",
datetime.datetime.now().strftime("%Y%m%d%H%M%S")
)
# Signature will be serving_default.
tf.saved_model.save(
obj=self.network_models["generator"], export_dir=export_path
)
def train_block(self, train_dataset, eval_dataset):
"""Training block setups training, then loops through datasets.
Args:
train_dataset: instance of `Dataset` for training data.
eval_dataset: instance of `Dataset` for evaluation data.
"""
# Create iterators of datasets.
train_dataset_iter = iter(train_dataset)
eval_dataset_iter = iter(eval_dataset)
steps_per_epoch = (
self.params["train_dataset_length"] // self.global_batch_size
)
# Instantiate model objects.
self.instantiate_model_objects()
# Create checkpoint machinery to save/restore checkpoints.
self.create_checkpoint_machinery()
# Create summary file writer.
self.summary_file_writer = tf.summary.create_file_writer(
logdir=os.path.join(self.params["output_dir"], "summaries"),
name="summary_file_writer"
)
# Run training loop.
self.training_loop(steps_per_epoch, train_dataset_iter)
# Save model at end of training loop.
self.training_loop_end_save_model()
def train_and_evaluate(self):
"""Trains and evaluates Keras model.
Args:
args: dict, user passed parameters.
Returns:
Generator's `Model` object for in-memory predictions.
"""
if self.params["distribution_strategy"]:
# If the list of devices is not specified in the
# Strategy constructor, it will be auto-detected.
if self.params["distribution_strategy"] == "Mirrored":
self.strategy = tf.distribute.MirroredStrategy()
print(
"Number of devices = {}".format(
self.strategy.num_replicas_in_sync
)
)
# Set global batch size for training.
self.global_batch_size = (
self.params["train_batch_size"] * self.strategy.num_replicas_in_sync
)
# Get input datasets. Batch size is split evenly between replicas.
train_dataset, eval_dataset = self.get_train_eval_datasets(
num_replicas=self.strategy.num_replicas_in_sync
)
with self.strategy.scope():
# Create distributed datasets.
train_dist_dataset = (
self.strategy.experimental_distribute_dataset(
dataset=train_dataset
)
)
eval_dist_dataset = (
self.strategy.experimental_distribute_dataset(
dataset=eval_dataset
)
)
# Training block setups training, then loops through datasets.
self.train_block(
train_dataset=train_dist_dataset,
eval_dataset=eval_dist_dataset
)
else:
# Set global batch size for training.
self.global_batch_size = self.params["train_batch_size"]
# Get input datasets.
train_dataset, eval_dataset = self.get_train_eval_datasets(
num_replicas=1
)
# Training block setups training, then loops through datasets.
self.train_block(
train_dataset=train_dataset, eval_dataset=eval_dataset
)
os.environ["OUTPUT_DIR"] = arguments["output_dir"]
%%bash
gsutil -m rm -rf ${OUTPUT_DIR}
train_and_evaluate_loop = TrainAndEvaluateLoop(params=arguments)
train_and_evaluate_loop.train_and_evaluate()
!gsutil ls ${OUTPUT_DIR}
!gsutil ls ${OUTPUT_DIR}/checkpoints
!gsutil ls ${OUTPUT_DIR}/summaries
!gsutil ls ${OUTPUT_DIR}/export
loaded = tf.saved_model.load(
export_dir=os.path.join(
arguments["output_dir"], "export", "20200731075031"
)
)
print(list(loaded.signatures.keys()))
infer = loaded.signatures["serving_default"]
print(infer.structured_outputs)
Z = tf.random.normal(shape=(10, 512))
predictions = infer(Z)
predictions.keys()
generated_images = np.clip(
a=tf.cast(
x=((predictions["generator_resize_fake_images"] + 1.0) * (255. / 2)),
dtype=tf.int32
),
a_min=0,
a_max=255
)
print(generated_images.shape)
def plot_images(images):
"""Plots images.
Args:
images: np.array, array of images of
[num_images, image_size, image_size, num_channels].
"""
num_images = len(images)
plt.figure(figsize=(20, 20))
for i in range(num_images):
image = images[i]
plt.subplot(1, num_images, i + 1)
plt.xticks([])
plt.yticks([])
plt.grid(False)
plt.imshow(
tf.reshape(image, image.shape[:-1]),
cmap="gray_r"
)
plt.show()
plot_images(generated_images)
| 0.76145 | 0.734239 |
```
import sys
import os
import random
# Counter
from collections import Counter
# Package
import somhos.resources.dataset as rd
import somhos.resources.queries as rq
import somhos.methods.useful as mu
from somhos.methods.useful import save_pickle, load_pickle
from somhos.config.paths import *
```
Default path
```
prefix_path = "../../"
data_path = get_relative_path(prefix_path, V9GAMMA_PATH)
os.path.exists(data_path)
samples_content = load_pickle(get_relative_path(data_path, DOCS_SAMPLES_CONTENT))
samples_content['index980808']
from gensim import corpora
import logging
logging.basicConfig(format='%(asctime)s : %(levelname)s : %(message)s', level=logging.INFO)
doc_directory = {}
doc_inverse_directory = []
texts_tokens = []
texts_keyphrases = []
for i, (k, doc_content) in enumerate(samples_content.items()):
doc_directory[k] = i
doc_inverse_directory.append(k)
texts_tokens.append(doc_content['tokens'])
texts_keyphrases.append(doc_content['kps-normalized'])
dictionary_tokens = corpora.Dictionary(texts_tokens)
dictionary_keyphrases = corpora.Dictionary(texts_keyphrases)
corpus_tokens = [dictionary_tokens.doc2idx(text) for text in texts_tokens]
corpus_keyphrases = [dictionary_keyphrases.doc2idx(text) for text in texts_keyphrases]
corpus_bag_of_words = [dictionary_tokens.doc2bow(text) for text in texts_tokens]
corpus_bag_of_keyphrases = [dictionary_keyphrases.doc2bow(text) for text in texts_keyphrases]
example = 1723
print(len(doc_inverse_directory), doc_inverse_directory[example])
print(len(doc_directory), doc_directory[doc_inverse_directory[example]])
print("\nCorpus tokens\n")
print(len(corpus_tokens), corpus_tokens[example])
print(len(corpus_bag_of_words), corpus_bag_of_words[example])
print("\nCorpus keyphrases\n")
print(len(corpus_keyphrases), corpus_keyphrases[example])
print(len(corpus_bag_of_keyphrases), corpus_bag_of_keyphrases[example])
save_pickle(doc_directory, get_relative_path(data_path, DOC_DIRECTORY))
save_pickle(doc_inverse_directory, get_relative_path(data_path, DOC_INVERSE_DIRECTORY))
save_pickle(dictionary_tokens, get_relative_path(data_path, DICTIONARY_TOKENS))
save_pickle(dictionary_keyphrases, get_relative_path(data_path, DICTIONARY_KEYPHRASES))
save_pickle(corpus_tokens, get_relative_path(data_path, CORPUS_TOKENS))
save_pickle(corpus_bag_of_words, get_relative_path(data_path, CORPUS_BAG_OF_WORDS))
save_pickle(corpus_keyphrases, get_relative_path(data_path, CORPUS_KEYPHRASES))
save_pickle(corpus_bag_of_keyphrases, get_relative_path(data_path, CORPUS_BAG_OF_KEYPHRASES))
```
|
github_jupyter
|
import sys
import os
import random
# Counter
from collections import Counter
# Package
import somhos.resources.dataset as rd
import somhos.resources.queries as rq
import somhos.methods.useful as mu
from somhos.methods.useful import save_pickle, load_pickle
from somhos.config.paths import *
prefix_path = "../../"
data_path = get_relative_path(prefix_path, V9GAMMA_PATH)
os.path.exists(data_path)
samples_content = load_pickle(get_relative_path(data_path, DOCS_SAMPLES_CONTENT))
samples_content['index980808']
from gensim import corpora
import logging
logging.basicConfig(format='%(asctime)s : %(levelname)s : %(message)s', level=logging.INFO)
doc_directory = {}
doc_inverse_directory = []
texts_tokens = []
texts_keyphrases = []
for i, (k, doc_content) in enumerate(samples_content.items()):
doc_directory[k] = i
doc_inverse_directory.append(k)
texts_tokens.append(doc_content['tokens'])
texts_keyphrases.append(doc_content['kps-normalized'])
dictionary_tokens = corpora.Dictionary(texts_tokens)
dictionary_keyphrases = corpora.Dictionary(texts_keyphrases)
corpus_tokens = [dictionary_tokens.doc2idx(text) for text in texts_tokens]
corpus_keyphrases = [dictionary_keyphrases.doc2idx(text) for text in texts_keyphrases]
corpus_bag_of_words = [dictionary_tokens.doc2bow(text) for text in texts_tokens]
corpus_bag_of_keyphrases = [dictionary_keyphrases.doc2bow(text) for text in texts_keyphrases]
example = 1723
print(len(doc_inverse_directory), doc_inverse_directory[example])
print(len(doc_directory), doc_directory[doc_inverse_directory[example]])
print("\nCorpus tokens\n")
print(len(corpus_tokens), corpus_tokens[example])
print(len(corpus_bag_of_words), corpus_bag_of_words[example])
print("\nCorpus keyphrases\n")
print(len(corpus_keyphrases), corpus_keyphrases[example])
print(len(corpus_bag_of_keyphrases), corpus_bag_of_keyphrases[example])
save_pickle(doc_directory, get_relative_path(data_path, DOC_DIRECTORY))
save_pickle(doc_inverse_directory, get_relative_path(data_path, DOC_INVERSE_DIRECTORY))
save_pickle(dictionary_tokens, get_relative_path(data_path, DICTIONARY_TOKENS))
save_pickle(dictionary_keyphrases, get_relative_path(data_path, DICTIONARY_KEYPHRASES))
save_pickle(corpus_tokens, get_relative_path(data_path, CORPUS_TOKENS))
save_pickle(corpus_bag_of_words, get_relative_path(data_path, CORPUS_BAG_OF_WORDS))
save_pickle(corpus_keyphrases, get_relative_path(data_path, CORPUS_KEYPHRASES))
save_pickle(corpus_bag_of_keyphrases, get_relative_path(data_path, CORPUS_BAG_OF_KEYPHRASES))
| 0.15925 | 0.26016 |
#### New to Plotly?
"Plotly's Python library is free and open source! [Get started](https://plot.ly/python/getting-started/) by downloading the client and [reading the primer](https://plot.ly/python/getting-started/).\n",
"<br>You can set up Plotly to work in [online](https://plot.ly/python/getting-started/#initialization-for-online-plotting) or [offline](https://plot.ly/python/getting-started/#initialization-for-offline-plotting) mode, or in [jupyter notebooks](https://plot.ly/python/getting-started/#start-plotting-online).\n",
"<br>We also have a quick-reference [cheatsheet](https://images.plot.ly/plotly-documentation/images/python_cheat_sheet.pdf) (new!) to help you get started!"
```
import plotly
plotly.__version__
```
### Introduction
Funnel charts are often used to represent data in different stages of a business process. It’s an important mechanism in Business Intelligence to identify potential problem areas of a process. For example, it’s used to observe the revenue or loss in a sales process for each stage, and displays values that are decreasing progressively. Each stage is illustrated as a percentage of the total of all values.
### Basic Funnel Plot
```
import plotly.plotly as py
from plotly import graph_objs as go
trace1 = go.Funnel(
y = ["Website visit", "Downloads", "Potential customers", "Requested price", "invoice sent"],
x = [39, 27.4, 20.6, 11, 2])
layout = go.Layout(
title = "Annual Sales",
margin = {"l": 200, "r": 200})
py.iplot(go.Figure([trace1],layout), filename = "basic_funnel")
```
### Setting Marker Size and Color
This example uses [textposition](https://plot.ly/python/reference/#scatter-textposition) and [textinfo](https://plot.ly/python/reference/#funnel-textinfo) to determine information apears on the graph, and shows how to customize the bars.
```
import plotly.plotly as py
from plotly import graph_objs as go
trace1 = go.Funnel(
y = ["Website visit", "Downloads", "Potential customers", "Requested price", "Finalized"],
x = [39, 27.4, 20.6, 11, 2],
textposition = "inside",
textinfo = "value+percent initial",
opacity = 0.65, marker = {"color": ["deepskyblue", "lightsalmon", "tan", "teal", "silver"],
"line": {"width": [4, 2, 2, 3, 1, 1], "color": ["wheat", "wheat", "blue", "wheat", "wheat"]}},
connector = {"line": {"color": "royalblue", "dash": "dot", "width": 3}})
layout = go.Layout(margin = {"l": 200, "r": 200 })
py.iplot(go.Figure([trace1], layout), filename = "Style_funnel")
```
### Stack Funnel Plot
```
import plotly.plotly as py
from plotly import graph_objs as go
trace1 = go.Funnel(
name = 'Montreal',
y = ["Website visit", "Downloads", "Potential customers", "Requested price"],
x = [120, 60, 30, 20],
textinfo = "value+percent initial")
trace2 = go.Funnel(
name = 'Toronto',
orientation = "h",
y = ["Website visit", "Downloads", "Potential customers", "Requested price", "invoice sent"],
x = [100, 60, 40, 30, 20],
textposition = "inside",
textinfo = "value+percent previous")
trace3 = go.Funnel(
name = 'Vancouver',
orientation = "h",
y = ["Website visit", "Downloads", "Potential customers", "Requested price", "invoice sent", "Finalized"],
x = [90, 70, 50, 30, 10, 5],
textposition = "outside",
textinfo = "value+percent total")
layout = go.Layout(margin = {"l": 200 , "r": 200}, funnelmode = "stack", showlegend = True)
py.iplot(go.Figure([trace1, trace2, trace3], layout), filename = "funnel_stack")
```
#### Basic Funnelarea Plot
```
import plotly.plotly as py
from plotly import graph_objs as go
trace = go.Funnelarea(values = [5, 4, 3, 2, 1])
layout = go.Layout(margin = go.layout.Margin(l= 200, r = 200))
py.iplot(go.Figure([trace], layout), filename = "basic_funnelarea")
```
#### Set Marker Size and Color in Funnelarea Plot
```
import plotly.plotly as py
from plotly import graph_objs as go
trace = go.Funnelarea(
values = [5, 4, 3, 2, 1], text = ["The 1st","The 2nd", "The 3rd", "The 4th", "The 5th"],
marker = {"colors": ["deepskyblue", "lightsalmon", "tan", "teal", "silver"],
"line": {"color": ["wheat", "wheat", "blue", "wheat", "wheat"], "width": [0, 1, 5, 0, 4]}},
textfont = {"family": "Old Standard TT, serif", "size": 13, "color": "black"}, opacity = 0.65)
layout = go.Layout(margin = {"l": 200, "r": 200})
py.iplot(go.Figure([trace], layout), filename = "stylefunnelarea")
```
#### Multi Funnel
```
import plotly.plotly as py
from plotly import graph_objs as go
trace1 = go.Funnelarea(
scalegroup = "first", values = [500, 450, 340, 230, 220, 110], textinfo = "value",
title = {"position": "top center", "text": "Sales for Sale Person A in U.S."},
domain = {"x": [0, 0.5], "y": [0, 0.5]})
trace2 = go.Funnelarea(
scalegroup = "first", values = [600, 500, 400, 300, 200, 100], textinfo = "value",
title = {"position": "top center", "text": "Sales of Sale Person B in Canada"},
domain = {"x": [0, 0.5], "y": [0.55, 1]})
trace3 = go.Funnelarea(
scalegroup = "second", values = [510, 480, 440, 330, 220, 100], textinfo = "value",
title = {"position": "top left", "text": "Sales of Sale Person A in Canada"},
domain = {"x": [0.55, 1], "y": [0, 0.5]})
trace4 = go.Funnelarea(
scalegroup = "second", values = [360, 250, 240, 130, 120, 60],
textinfo = "value", title = {"position": "top left", "text": "Sales of Sale Person B in U.S."},
domain = {"x": [0.55, 1], "y": [0.55, 1]})
layout = go.Layout(
margin = {"l": 200, "r": 200}, shapes = [
{"x0": 0, "x1": 0.5, "y0": 0, "y1": 0.5},
{"x0": 0, "x1": 0.5, "y0": 0.55, "y1": 1},
{"x0": 0.55, "x1": 1, "y0": 0, "y1": 0.5},
{"x0": 0.55, "x1": 1, "y0": 0.55, "y1": 1}])
py.iplot(go.Figure([trace1, trace2, trace3, trace4], layout), filename = "scale_group")
```
#### Reference
See https://plot.ly/python/reference/#funnel for more information and chart attribute options!
```
from IPython.display import display, HTML
display(HTML('<link href="//fonts.googleapis.com/css?family=Open+Sans:600,400,300,200|Inconsolata|Ubuntu+Mono:400,700" rel="stylesheet" type="text/css" />'))
display(HTML('<link rel="stylesheet" type="text/css" href="http://help.plot.ly/documentation/all_static/css/ipython-notebook-custom.css">'))
! pip3 install git+https://github.com/plotly/publisher.git --upgrade
import publisher
publisher.publish(
'funnel-chart.ipynb', 'python/funnel-charts/', 'Funnel Chart',
'How to make funnel-chart plots in Python with Plotly.',
title = 'Python Funnel Chart | Plotly',
has_thumbnail='true', thumbnail='thumbnail/funnel.jpg',
language='python',
# page_type='example_index', // note this is only if you want the tutorial to appear on the main page: plot.ly/python
display_as='basic', order=6.3, ipynb='~notebook_demo/293',
uses_plotly_offline=False)
```
|
github_jupyter
|
import plotly
plotly.__version__
import plotly.plotly as py
from plotly import graph_objs as go
trace1 = go.Funnel(
y = ["Website visit", "Downloads", "Potential customers", "Requested price", "invoice sent"],
x = [39, 27.4, 20.6, 11, 2])
layout = go.Layout(
title = "Annual Sales",
margin = {"l": 200, "r": 200})
py.iplot(go.Figure([trace1],layout), filename = "basic_funnel")
import plotly.plotly as py
from plotly import graph_objs as go
trace1 = go.Funnel(
y = ["Website visit", "Downloads", "Potential customers", "Requested price", "Finalized"],
x = [39, 27.4, 20.6, 11, 2],
textposition = "inside",
textinfo = "value+percent initial",
opacity = 0.65, marker = {"color": ["deepskyblue", "lightsalmon", "tan", "teal", "silver"],
"line": {"width": [4, 2, 2, 3, 1, 1], "color": ["wheat", "wheat", "blue", "wheat", "wheat"]}},
connector = {"line": {"color": "royalblue", "dash": "dot", "width": 3}})
layout = go.Layout(margin = {"l": 200, "r": 200 })
py.iplot(go.Figure([trace1], layout), filename = "Style_funnel")
import plotly.plotly as py
from plotly import graph_objs as go
trace1 = go.Funnel(
name = 'Montreal',
y = ["Website visit", "Downloads", "Potential customers", "Requested price"],
x = [120, 60, 30, 20],
textinfo = "value+percent initial")
trace2 = go.Funnel(
name = 'Toronto',
orientation = "h",
y = ["Website visit", "Downloads", "Potential customers", "Requested price", "invoice sent"],
x = [100, 60, 40, 30, 20],
textposition = "inside",
textinfo = "value+percent previous")
trace3 = go.Funnel(
name = 'Vancouver',
orientation = "h",
y = ["Website visit", "Downloads", "Potential customers", "Requested price", "invoice sent", "Finalized"],
x = [90, 70, 50, 30, 10, 5],
textposition = "outside",
textinfo = "value+percent total")
layout = go.Layout(margin = {"l": 200 , "r": 200}, funnelmode = "stack", showlegend = True)
py.iplot(go.Figure([trace1, trace2, trace3], layout), filename = "funnel_stack")
import plotly.plotly as py
from plotly import graph_objs as go
trace = go.Funnelarea(values = [5, 4, 3, 2, 1])
layout = go.Layout(margin = go.layout.Margin(l= 200, r = 200))
py.iplot(go.Figure([trace], layout), filename = "basic_funnelarea")
import plotly.plotly as py
from plotly import graph_objs as go
trace = go.Funnelarea(
values = [5, 4, 3, 2, 1], text = ["The 1st","The 2nd", "The 3rd", "The 4th", "The 5th"],
marker = {"colors": ["deepskyblue", "lightsalmon", "tan", "teal", "silver"],
"line": {"color": ["wheat", "wheat", "blue", "wheat", "wheat"], "width": [0, 1, 5, 0, 4]}},
textfont = {"family": "Old Standard TT, serif", "size": 13, "color": "black"}, opacity = 0.65)
layout = go.Layout(margin = {"l": 200, "r": 200})
py.iplot(go.Figure([trace], layout), filename = "stylefunnelarea")
import plotly.plotly as py
from plotly import graph_objs as go
trace1 = go.Funnelarea(
scalegroup = "first", values = [500, 450, 340, 230, 220, 110], textinfo = "value",
title = {"position": "top center", "text": "Sales for Sale Person A in U.S."},
domain = {"x": [0, 0.5], "y": [0, 0.5]})
trace2 = go.Funnelarea(
scalegroup = "first", values = [600, 500, 400, 300, 200, 100], textinfo = "value",
title = {"position": "top center", "text": "Sales of Sale Person B in Canada"},
domain = {"x": [0, 0.5], "y": [0.55, 1]})
trace3 = go.Funnelarea(
scalegroup = "second", values = [510, 480, 440, 330, 220, 100], textinfo = "value",
title = {"position": "top left", "text": "Sales of Sale Person A in Canada"},
domain = {"x": [0.55, 1], "y": [0, 0.5]})
trace4 = go.Funnelarea(
scalegroup = "second", values = [360, 250, 240, 130, 120, 60],
textinfo = "value", title = {"position": "top left", "text": "Sales of Sale Person B in U.S."},
domain = {"x": [0.55, 1], "y": [0.55, 1]})
layout = go.Layout(
margin = {"l": 200, "r": 200}, shapes = [
{"x0": 0, "x1": 0.5, "y0": 0, "y1": 0.5},
{"x0": 0, "x1": 0.5, "y0": 0.55, "y1": 1},
{"x0": 0.55, "x1": 1, "y0": 0, "y1": 0.5},
{"x0": 0.55, "x1": 1, "y0": 0.55, "y1": 1}])
py.iplot(go.Figure([trace1, trace2, trace3, trace4], layout), filename = "scale_group")
from IPython.display import display, HTML
display(HTML('<link href="//fonts.googleapis.com/css?family=Open+Sans:600,400,300,200|Inconsolata|Ubuntu+Mono:400,700" rel="stylesheet" type="text/css" />'))
display(HTML('<link rel="stylesheet" type="text/css" href="http://help.plot.ly/documentation/all_static/css/ipython-notebook-custom.css">'))
! pip3 install git+https://github.com/plotly/publisher.git --upgrade
import publisher
publisher.publish(
'funnel-chart.ipynb', 'python/funnel-charts/', 'Funnel Chart',
'How to make funnel-chart plots in Python with Plotly.',
title = 'Python Funnel Chart | Plotly',
has_thumbnail='true', thumbnail='thumbnail/funnel.jpg',
language='python',
# page_type='example_index', // note this is only if you want the tutorial to appear on the main page: plot.ly/python
display_as='basic', order=6.3, ipynb='~notebook_demo/293',
uses_plotly_offline=False)
| 0.541166 | 0.97419 |
# Harmonizome ETL: Pathway Commons Pathways
Created by: Charles Dai <br>
Credit to: Moshe Silverstein
Data Source Home: https://www.pathwaycommons.org/ <br>
Data Source Download: http://www.pathwaycommons.org/archives/PC2/v12/
```
# appyter init
from appyter import magic
magic.init(lambda _=globals: _())
import sys
import os
from datetime import date
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
%matplotlib inline
import harmonizome.utility_functions as uf
import harmonizome.lookup as lookup
%load_ext autoreload
%autoreload 2
```
### Notebook Information
```
print('This notebook was run on:', date.today(), '\nPython version:', sys.version)
```
# Initialization
```
%%appyter hide_code
{% do SectionField(
name='data',
title='Upload Data',
img='load_icon.png'
) %}
{% do SectionField(
name='settings',
title='Settings',
img='setting_icon.png'
) %}
%%appyter code_eval
{% do DescriptionField(
name='description',
text='The example below was sourced from <a href="http://www.pathwaycommons.org/archives/PC2/v12/" target="_blank">www.pathwaycommons.org</a>. If clicking on the example does not work, it should be downloaded directly from the source website.',
section='data'
) %}
{% set df_file = FileField(
constraint='.*\.txt.gz$',
name='phenotype_gene_list',
label='Phenotypes to Genes (txt.gz)',
default='PathwayCommons12.All.hgnc.txt.gz',
examples={
'PathwayCommons12.All.hgnc.txt.gz': 'http://www.pathwaycommons.org/archives/PC2/v12/PathwayCommons12.All.hgnc.txt.gz'
},
section='data'
) %}
%%appyter code_eval
{% set attribute = ChoiceField(
name='attribute',
label='Attribute',
choices={
'Pathways': "'Pathways'",
'Protein-Protein Interactions': "'PPI'"
},
default='Protein-Protein Interactions',
section='settings'
) %}
```
### Load Mapping Dictionaries
```
symbol_lookup, geneid_lookup = lookup.get_lookups()
```
### Output Path
```
%%appyter code_exec
output_name = 'pathway_commons_' + {{attribute}}.lower()
path = 'Output/Pathway-Commons-' + {{attribute}}
if not os.path.exists(path):
os.makedirs(path)
```
# Load Data
```
%%appyter code_exec
df = pd.read_csv(
{{df_file}},
sep='\t', usecols=['PARTICIPANT_A', 'PARTICIPANT_B', 'PATHWAY_NAMES']
)
df.head()
df.shape
```
# Pre-process Data
```
# Some participant B have multiple names, take first for gene symbol
df['PARTICIPANT_B'] = df['PARTICIPANT_B'].map(lambda x: x.split(';')[0])
```
## Get Relevant Data
```
%%appyter code_exec
if {{attribute}} == 'Pathways':
df = pd.concat([
df[['PARTICIPANT_B', 'PATHWAY_NAMES']].rename(columns={'PARTICIPANT_B':'PARTICIPANT_A'}),
df[['PARTICIPANT_A', 'PATHWAY_NAMES']]
])
df.columns = ['Gene Symbol', 'Pathway']
elif {{attribute}} == 'PPI':
df = df[['PARTICIPANT_A', 'PARTICIPANT_B']]
df.columns = ['Protein A', 'Protein B']
df = df.dropna().set_index(df.columns[0])
df.head()
```
# Filter Data
## Map Gene Symbols to Up-to-date Approved Gene Symbols
```
%%appyter code_exec
if {{attribute}} == 'PPI':
df = uf.map_symbols(df.reset_index().set_index('Protein B'),
symbol_lookup)
df = df.reset_index().set_index('Protein A')
df = uf.map_symbols(df, symbol_lookup, remove_duplicates=True)
df.shape
```
# Analyze Data
## Create Binary Matrix
```
binary_matrix = uf.binary_matrix(df)
binary_matrix.head()
binary_matrix.shape
uf.save_data(binary_matrix, path, output_name + '_binary_matrix',
compression='npz', dtype=np.uint8)
```
## Create Gene List
```
gene_list = uf.gene_list(binary_matrix, geneid_lookup)
gene_list.head()
gene_list.shape
uf.save_data(gene_list, path, output_name + '_gene_list',
ext='tsv', compression='gzip', index=False)
```
## Create Attribute List
```
attribute_list = uf.attribute_list(binary_matrix)
attribute_list.head()
attribute_list.shape
uf.save_data(attribute_list, path, output_name + '_attribute_list',
ext='tsv', compression='gzip')
```
## Create Gene and Attribute Set Libraries
```
uf.save_setlib(binary_matrix, 'gene', 'up', path, output_name + '_gene_up_set')
uf.save_setlib(binary_matrix, 'attribute', 'up', path,
output_name + '_attribute_up_set')
```
## Create Attribute Similarity Matrix
```
attribute_similarity_matrix = uf.similarity_matrix(binary_matrix.T, 'jaccard', sparse=True)
attribute_similarity_matrix.head()
uf.save_data(attribute_similarity_matrix, path,
output_name + '_attribute_similarity_matrix',
compression='npz', symmetric=True, dtype=np.float32)
```
## Create Gene Similarity Matrix
```
gene_similarity_matrix = uf.similarity_matrix(binary_matrix, 'jaccard', sparse=True)
gene_similarity_matrix.head()
uf.save_data(gene_similarity_matrix, path,
output_name + '_gene_similarity_matrix',
compression='npz', symmetric=True, dtype=np.float32)
```
## Create Gene-Attribute Edge List
```
edge_list = uf.edge_list(binary_matrix)
uf.save_data(edge_list, path, output_name + '_edge_list',
ext='tsv', compression='gzip')
```
# Create Downloadable Save File
```
uf.archive(path)
```
### Link to download output files: [click here](./output_archive.zip)
|
github_jupyter
|
# appyter init
from appyter import magic
magic.init(lambda _=globals: _())
import sys
import os
from datetime import date
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
%matplotlib inline
import harmonizome.utility_functions as uf
import harmonizome.lookup as lookup
%load_ext autoreload
%autoreload 2
print('This notebook was run on:', date.today(), '\nPython version:', sys.version)
%%appyter hide_code
{% do SectionField(
name='data',
title='Upload Data',
img='load_icon.png'
) %}
{% do SectionField(
name='settings',
title='Settings',
img='setting_icon.png'
) %}
%%appyter code_eval
{% do DescriptionField(
name='description',
text='The example below was sourced from <a href="http://www.pathwaycommons.org/archives/PC2/v12/" target="_blank">www.pathwaycommons.org</a>. If clicking on the example does not work, it should be downloaded directly from the source website.',
section='data'
) %}
{% set df_file = FileField(
constraint='.*\.txt.gz$',
name='phenotype_gene_list',
label='Phenotypes to Genes (txt.gz)',
default='PathwayCommons12.All.hgnc.txt.gz',
examples={
'PathwayCommons12.All.hgnc.txt.gz': 'http://www.pathwaycommons.org/archives/PC2/v12/PathwayCommons12.All.hgnc.txt.gz'
},
section='data'
) %}
%%appyter code_eval
{% set attribute = ChoiceField(
name='attribute',
label='Attribute',
choices={
'Pathways': "'Pathways'",
'Protein-Protein Interactions': "'PPI'"
},
default='Protein-Protein Interactions',
section='settings'
) %}
symbol_lookup, geneid_lookup = lookup.get_lookups()
%%appyter code_exec
output_name = 'pathway_commons_' + {{attribute}}.lower()
path = 'Output/Pathway-Commons-' + {{attribute}}
if not os.path.exists(path):
os.makedirs(path)
%%appyter code_exec
df = pd.read_csv(
{{df_file}},
sep='\t', usecols=['PARTICIPANT_A', 'PARTICIPANT_B', 'PATHWAY_NAMES']
)
df.head()
df.shape
# Some participant B have multiple names, take first for gene symbol
df['PARTICIPANT_B'] = df['PARTICIPANT_B'].map(lambda x: x.split(';')[0])
%%appyter code_exec
if {{attribute}} == 'Pathways':
df = pd.concat([
df[['PARTICIPANT_B', 'PATHWAY_NAMES']].rename(columns={'PARTICIPANT_B':'PARTICIPANT_A'}),
df[['PARTICIPANT_A', 'PATHWAY_NAMES']]
])
df.columns = ['Gene Symbol', 'Pathway']
elif {{attribute}} == 'PPI':
df = df[['PARTICIPANT_A', 'PARTICIPANT_B']]
df.columns = ['Protein A', 'Protein B']
df = df.dropna().set_index(df.columns[0])
df.head()
%%appyter code_exec
if {{attribute}} == 'PPI':
df = uf.map_symbols(df.reset_index().set_index('Protein B'),
symbol_lookup)
df = df.reset_index().set_index('Protein A')
df = uf.map_symbols(df, symbol_lookup, remove_duplicates=True)
df.shape
binary_matrix = uf.binary_matrix(df)
binary_matrix.head()
binary_matrix.shape
uf.save_data(binary_matrix, path, output_name + '_binary_matrix',
compression='npz', dtype=np.uint8)
gene_list = uf.gene_list(binary_matrix, geneid_lookup)
gene_list.head()
gene_list.shape
uf.save_data(gene_list, path, output_name + '_gene_list',
ext='tsv', compression='gzip', index=False)
attribute_list = uf.attribute_list(binary_matrix)
attribute_list.head()
attribute_list.shape
uf.save_data(attribute_list, path, output_name + '_attribute_list',
ext='tsv', compression='gzip')
uf.save_setlib(binary_matrix, 'gene', 'up', path, output_name + '_gene_up_set')
uf.save_setlib(binary_matrix, 'attribute', 'up', path,
output_name + '_attribute_up_set')
attribute_similarity_matrix = uf.similarity_matrix(binary_matrix.T, 'jaccard', sparse=True)
attribute_similarity_matrix.head()
uf.save_data(attribute_similarity_matrix, path,
output_name + '_attribute_similarity_matrix',
compression='npz', symmetric=True, dtype=np.float32)
gene_similarity_matrix = uf.similarity_matrix(binary_matrix, 'jaccard', sparse=True)
gene_similarity_matrix.head()
uf.save_data(gene_similarity_matrix, path,
output_name + '_gene_similarity_matrix',
compression='npz', symmetric=True, dtype=np.float32)
edge_list = uf.edge_list(binary_matrix)
uf.save_data(edge_list, path, output_name + '_edge_list',
ext='tsv', compression='gzip')
uf.archive(path)
| 0.350977 | 0.803829 |
# Reproducing plot from Body Orientation post-processed data set
The goal is to visualize the results from the Body Orientation assesment experiment. For doing so, the function to create the plot and the dataset are provided.
First, import the libraries to create the plot. The plot contains location points with the body orientation represented by an arrow. The **Wedge** represents the field of view of the depth camera.
```
import matplotlib
import matplotlib.cm as cm
import matplotlib.colors as mcolors
import matplotlib.colorbar as mcolorbar
import matplotlib.pyplot as plt
from matplotlib.patches import Wedge
%matplotlib widget
```
Additionally, to process the data set it is necessary to import the following libraries:
```
import seaborn as sns
import sklearn
import pandas as pd
import numpy as np
```
Next, we define the plotting configuration function. The function handles the titles and the colors per body orientation **type**.
```
def plot_conf(type):
choices = {'back': 'dodgerblue', 'backd': 'navy', 'frontal': 'seagreen', 'frontald': 'lightseagreen'}
titles = {'sider': 'Side Right',
'backdr': 'Back Diagonal Right',
'back': 'Back',
'backdl': 'Back Diagonal Left',
'sidel': 'Side Left',
'frontaldl': 'Frontal Diagonal Left',
'frontal': 'Frontal',
'frontaldr': 'Frontal Diagonal Right',
'pairinter':'Pair view intersection',
'formations': 'Group movement stops'
}
ranges = {'sider': '[-22.5°,0°),[0°,22.5°)',
'backdr': '[22.5°,67.5°)',
'back': '[67.5°,112.5°)',
'backdl': '[112.5°,157.5°)',
'sidel': '[157.5°,180°),(-180°,157.5°]',
'frontaldl': '[-157.5°, -112.5°)',
'frontal': '[-112.5°, -67.5°)',
'frontaldr': '[-67.5°, -22.5°)'
}
shade_color = choices.get(type, 'purple')
title = titles.get(type, 'Body')
range_title=ranges.get(type,' ')
return shade_color, title, range_title
```
For drawing the calculated body orientation from the skeleton joints, we calculate the vectors for the quiver function. Later, we setup the depth sensor's field of view.
```
def angle_cloud(data_kinect, shade_color, title,range_title, type):
U = np.cos((data_kinect['re_body_angle']) )
V = np.sin((data_kinect['re_body_angle']) )
sns.set()
norm = matplotlib.colors.Normalize()
norm.autoscale(data_kinect['re_body_angle'])
cm = matplotlib.cm.viridis
sm = matplotlib.cm.ScalarMappable(cmap=cm, norm=norm)
sm.set_array([])
fig, ax = plt.subplots()
ax.quiver(data_kinect['shl_x'], data_kinect['shl_y'], U, V, angles=data_kinect['re_body_angle'], color=cm(norm(data_kinect['re_body_angle'])), units='xy',pivot='middle')
ax.axis(xmin=-3, xmax=3)
ax.axis(ymin=-0.5, ymax=4.5)
fov = Wedge(center=(0, 0), r=4.895, theta1=55, theta2=125, color=shade_color, alpha=0.05)
ax.add_artist(fov)
plt.ylabel('(Y) Distance from Origin')
plt.xlabel('(X) Distance from Origin')
plt.title('Body Orientation' +type+': '+title+'\n Accepted angle range '+range_title)
cax, _ = mcolorbar.make_axes(plt.gca())
cb = mcolorbar.ColorbarBase(cax, cmap=matplotlib.cm.viridis, norm=norm)
cb.set_label('Body Orientation angle')
return plt
```
The data is available in the folder *data*. It's a postprocessed file containing information about the body detected:
- ID_exp: identifier experiment
- date_exp: date and time of the sample
- Description: evaluated body orientation
- ID_subject: unique body identifier
- height: body height
- time: hh:mm:ss.ms formated time for the sample
- joints: object with coordinates x,y,z for each body joint.
- posture: detected body posture
- re_body_angle: calculated body angle
- origin_x and y: transformed location coordinates in the global reference system
- Coordinates for shoulder joints (sh_x,sh_y) where *r* refers to right and *l* to left.
- Coordinates for spine (sp_x, y)
```
data_kinect = pd.read_csv("data/frontaldl_processed_data_frontaldl.csv",sep=';')
data_kinect.reset_index(drop=True)
print(data_kinect)
```
To create the plot, specify the type of Body Orientation that would be processed. In this case, we have the *Frontal Diagonal Left* dataset. With this, the colors and titles are defined:
```
type='frontaldl'
shade_color, title, range_title= plot_conf(type)
angle_cloud(data_kinect, shade_color, title, range_title, type).show()
```
|
github_jupyter
|
import matplotlib
import matplotlib.cm as cm
import matplotlib.colors as mcolors
import matplotlib.colorbar as mcolorbar
import matplotlib.pyplot as plt
from matplotlib.patches import Wedge
%matplotlib widget
import seaborn as sns
import sklearn
import pandas as pd
import numpy as np
def plot_conf(type):
choices = {'back': 'dodgerblue', 'backd': 'navy', 'frontal': 'seagreen', 'frontald': 'lightseagreen'}
titles = {'sider': 'Side Right',
'backdr': 'Back Diagonal Right',
'back': 'Back',
'backdl': 'Back Diagonal Left',
'sidel': 'Side Left',
'frontaldl': 'Frontal Diagonal Left',
'frontal': 'Frontal',
'frontaldr': 'Frontal Diagonal Right',
'pairinter':'Pair view intersection',
'formations': 'Group movement stops'
}
ranges = {'sider': '[-22.5°,0°),[0°,22.5°)',
'backdr': '[22.5°,67.5°)',
'back': '[67.5°,112.5°)',
'backdl': '[112.5°,157.5°)',
'sidel': '[157.5°,180°),(-180°,157.5°]',
'frontaldl': '[-157.5°, -112.5°)',
'frontal': '[-112.5°, -67.5°)',
'frontaldr': '[-67.5°, -22.5°)'
}
shade_color = choices.get(type, 'purple')
title = titles.get(type, 'Body')
range_title=ranges.get(type,' ')
return shade_color, title, range_title
def angle_cloud(data_kinect, shade_color, title,range_title, type):
U = np.cos((data_kinect['re_body_angle']) )
V = np.sin((data_kinect['re_body_angle']) )
sns.set()
norm = matplotlib.colors.Normalize()
norm.autoscale(data_kinect['re_body_angle'])
cm = matplotlib.cm.viridis
sm = matplotlib.cm.ScalarMappable(cmap=cm, norm=norm)
sm.set_array([])
fig, ax = plt.subplots()
ax.quiver(data_kinect['shl_x'], data_kinect['shl_y'], U, V, angles=data_kinect['re_body_angle'], color=cm(norm(data_kinect['re_body_angle'])), units='xy',pivot='middle')
ax.axis(xmin=-3, xmax=3)
ax.axis(ymin=-0.5, ymax=4.5)
fov = Wedge(center=(0, 0), r=4.895, theta1=55, theta2=125, color=shade_color, alpha=0.05)
ax.add_artist(fov)
plt.ylabel('(Y) Distance from Origin')
plt.xlabel('(X) Distance from Origin')
plt.title('Body Orientation' +type+': '+title+'\n Accepted angle range '+range_title)
cax, _ = mcolorbar.make_axes(plt.gca())
cb = mcolorbar.ColorbarBase(cax, cmap=matplotlib.cm.viridis, norm=norm)
cb.set_label('Body Orientation angle')
return plt
data_kinect = pd.read_csv("data/frontaldl_processed_data_frontaldl.csv",sep=';')
data_kinect.reset_index(drop=True)
print(data_kinect)
type='frontaldl'
shade_color, title, range_title= plot_conf(type)
angle_cloud(data_kinect, shade_color, title, range_title, type).show()
| 0.705379 | 0.944944 |
# Convolutional Autoencoder
Sticking with the MNIST dataset, let's improve our autoencoder's performance using convolutional layers. Again, loading modules and the data.
```
%matplotlib inline
import numpy as np
import tensorflow as tf
import matplotlib.pyplot as plt
from tensorflow.examples.tutorials.mnist import input_data
mnist = input_data.read_data_sets('MNIST_data', validation_size=0)
img = mnist.train.images[2]
plt.imshow(img.reshape((28, 28)), cmap='Greys_r')
```
## Network Architecture
The encoder part of the network will be a typical convolutional pyramid. Each convolutional layer will be followed by a max-pooling layer to reduce the dimensions of the layers. The decoder though might be something new to you. The decoder needs to convert from a narrow representation to a wide reconstructed image. For example, the representation could be a 4x4x8 max-pool layer. This is the output of the encoder, but also the input to the decoder. We want to get a 28x28x1 image out from the decoder so we need to work our way back up from the narrow decoder input layer. A schematic of the network is shown below.
<img src='assets/convolutional_autoencoder.png' width=500px>
Here our final encoder layer has size 4x4x8 = 128. The original images have size 28x28 = 784, so the encoded vector is roughly 16% the size of the original image. These are just suggested sizes for each of the layers. Feel free to change the depths and sizes, but remember our goal here is to find a small representation of the input data.
### What's going on with the decoder
Okay, so the decoder has these "Upsample" layers that you might not have seen before. First off, I'll discuss a bit what these layers *aren't*. Usually, you'll see **transposed convolution** layers used to increase the width and height of the layers. They work almost exactly the same as convolutional layers, but in reverse. A stride in the input layer results in a larger stride in the transposed convolution layer. For example, if you have a 3x3 kernel, a 3x3 patch in the input layer will be reduced to one unit in a convolutional layer. Comparatively, one unit in the input layer will be expanded to a 3x3 path in a transposed convolution layer. The TensorFlow API provides us with an easy way to create the layers, [`tf.nn.conv2d_transpose`](https://www.tensorflow.org/api_docs/python/tf/nn/conv2d_transpose).
However, transposed convolution layers can lead to artifacts in the final images, such as checkerboard patterns. This is due to overlap in the kernels which can be avoided by setting the stride and kernel size equal. In [this Distill article](http://distill.pub/2016/deconv-checkerboard/) from Augustus Odena, *et al*, the authors show that these checkerboard artifacts can be avoided by resizing the layers using nearest neighbor or bilinear interpolation (upsampling) followed by a convolutional layer. In TensorFlow, this is easily done with [`tf.image.resize_images`](https://www.tensorflow.org/versions/r1.1/api_docs/python/tf/image/resize_images), followed by a convolution. Be sure to read the Distill article to get a better understanding of deconvolutional layers and why we're using upsampling.
> **Exercise:** Build the network shown above. Remember that a convolutional layer with strides of 1 and 'same' padding won't reduce the height and width. That is, if the input is 28x28 and the convolution layer has stride = 1 and 'same' padding, the convolutional layer will also be 28x28. The max-pool layers are used the reduce the width and height. A stride of 2 will reduce the size by a factor of 2. Odena *et al* claim that nearest neighbor interpolation works best for the upsampling, so make sure to include that as a parameter in `tf.image.resize_images` or use [`tf.image.resize_nearest_neighbor`]( `https://www.tensorflow.org/api_docs/python/tf/image/resize_nearest_neighbor). For convolutional layers, use [`tf.layers.conv2d`](https://www.tensorflow.org/api_docs/python/tf/layers/conv2d). For example, you would write `conv1 = tf.layers.conv2d(inputs, 32, (5,5), padding='same', activation=tf.nn.relu)` for a layer with a depth of 32, a 5x5 kernel, stride of (1,1), padding is 'same', and a ReLU activation. Similarly, for the max-pool layers, use [`tf.layers.max_pooling2d`](https://www.tensorflow.org/api_docs/python/tf/layers/max_pooling2d).
```
learning_rate = 0.001
# Input and target placeholders
inputs_ = tf.placeholder(tf.float32, shape=(None, 28, 28, 1))
targets_ = tf.placeholder(tf.float32, shape=(None, 28, 28, 1))
### Encoder
conv1 = tf.layers.conv2d(inputs=inputs_, filters=16, kernel_size=(5,5), strides=(1,1), padding='same',activation=tf.nn.relu)
# Now 28x28x16
maxpool1 = tf.layers.max_pooling2d(inputs=conv1, pool_size=(2,2), strides=(2,2), padding='same')
# Now 14x14x16
conv2 = tf.layers.conv2d(inputs=maxpool1, filters=8, strides=(1,1), kernel_size=(3,3), padding='same',activation=tf.nn.relu)
# Now 14x14x8
maxpool2 = tf.layers.max_pooling2d(inputs=conv2, pool_size=(2,2), strides=(2,2), padding='same')
# Now 7x7x8
conv3 = tf.layers.conv2d(inputs=maxpool2, strides=(1,1), kernel_size=(3,3), filters=8, padding='same',activation=tf.nn.relu)
# Now 7x7x8
encoded = tf.layers.max_pooling2d(inputs=conv3, pool_size=(2,2), strides=(2,2), padding='same')
# Now 4x4x8
### Decoder
upsample1 = tf.image.resize_nearest_neighbor(images=encoded, size=(7,7))
# Now 7x7x8
conv4 = tf.layers.conv2d(inputs=upsample1, kernel_size=(3,3), filters=8, strides=(1,1), padding='same',activation=tf.nn.relu)
# Now 7x7x8
upsample2 = tf.image.resize_nearest_neighbor(images=conv4, size=(14, 14))
# Now 14x14x8
conv5 = tf.layers.conv2d(inputs=upsample2, kernel_size=(3,3), filters=8, strides=(1,1), padding='same',activation=tf.nn.relu)
# Now 14x14x8
upsample3 = tf.image.resize_nearest_neighbor(images=conv5, size=(28, 28))
# Now 28x28x8
conv6 = tf.layers.conv2d(inputs=upsample3, kernel_size=(3,3), filters=16, strides=(1,1), padding='same',activation=tf.nn.relu)
# Now 28x28x16
logits = tf.layers.conv2d(inputs=conv6, kernel_size=(3,3), filters=1, strides=(1,1), padding='same')
#Now 28x28x1
# Pass logits through sigmoid to get reconstructed image
decoded = tf.nn.sigmoid(logits)
# Pass logits through sigmoid and calculate the cross-entropy loss
loss = tf.nn.sigmoid_cross_entropy_with_logits(labels=targets_, logits=logits)
# Get cost and define the optimizer
cost = tf.reduce_mean(loss)
opt = tf.train.AdamOptimizer(learning_rate).minimize(cost)
```
## Training
As before, here we'll train the network. Instead of flattening the images though, we can pass them in as 28x28x1 arrays.
```
sess = tf.Session(config=tf.ConfigProto(log_device_placement=True))
epochs = 20
batch_size = 200
sess.run(tf.global_variables_initializer())
for e in range(epochs):
for ii in range(mnist.train.num_examples//batch_size):
batch = mnist.train.next_batch(batch_size)
imgs = batch[0].reshape((-1, 28, 28, 1))
batch_cost, _ = sess.run([cost, opt], feed_dict={inputs_: imgs,
targets_: imgs})
print("Epoch: {}/{}...".format(e+1, epochs),
"Training loss: {:.4f}".format(batch_cost))
fig, axes = plt.subplots(nrows=2, ncols=10, sharex=True, sharey=True, figsize=(20,4))
in_imgs = mnist.test.images[:10]
reconstructed = sess.run(decoded, feed_dict={inputs_: in_imgs.reshape((10, 28, 28, 1))})
for images, row in zip([in_imgs, reconstructed], axes):
for img, ax in zip(images, row):
ax.imshow(img.reshape((28, 28)), cmap='Greys_r')
ax.get_xaxis().set_visible(False)
ax.get_yaxis().set_visible(False)
fig.tight_layout(pad=0.1)
sess.close()
```
## Denoising
As I've mentioned before, autoencoders like the ones you've built so far aren't too useful in practive. However, they can be used to denoise images quite successfully just by training the network on noisy images. We can create the noisy images ourselves by adding Gaussian noise to the training images, then clipping the values to be between 0 and 1. We'll use noisy images as input and the original, clean images as targets. Here's an example of the noisy images I generated and the denoised images.

Since this is a harder problem for the network, we'll want to use deeper convolutional layers here, more feature maps. I suggest something like 32-32-16 for the depths of the convolutional layers in the encoder, and the same depths going backward through the decoder. Otherwise the architecture is the same as before.
> **Exercise:** Build the network for the denoising autoencoder. It's the same as before, but with deeper layers. I suggest 32-32-16 for the depths, but you can play with these numbers, or add more layers.
```
learning_rate = 0.001
inputs_ = tf.placeholder(tf.float32, (None, 28, 28, 1), name='inputs')
targets_ = tf.placeholder(tf.float32, (None, 28, 28, 1), name='targets')
### Encoder
conv1 = tf.layers.conv2d(inputs=inputs_, filters=32, kernel_size=(5,5), strides=(1,1), padding='same', activation=tf.nn.relu)
# Now 28x28x32
maxpool1 = tf.layers.max_pooling2d(inputs=conv1, pool_size=(2,2), padding='same',strides=(2,2))
# Now 14x14x32
conv2 = tf.layers.conv2d(inputs=maxpool1, filters=32, kernel_size=(3,3), strides=(1,1), padding='same', activation=tf.nn.relu)
# Now 14x14x32
maxpool2 = tf.layers.max_pooling2d(inputs=conv2, pool_size=(2,2), padding='same',strides=(2,2))
# Now 7x7x32
conv3 = tf.layers.conv2d(inputs=maxpool2, filters=16, kernel_size=(3,3), strides=(1,1), padding='same', activation=tf.nn.relu)
# Now 7x7x16
encoded = tf.layers.max_pooling2d(inputs=conv3, pool_size=(2,2), padding='same',strides=(2,2))
# Now 4x4x16
### Decoder
upsample1 = tf.image.resize_nearest_neighbor(images=encoded, size=(7,7))
# Now 7x7x16
conv4 = tf.layers.conv2d(inputs=upsample1, filters=16, kernel_size=(3,3), strides=(1,1), padding='same', activation=tf.nn.relu)
# Now 7x7x16
upsample2 = tf.image.resize_nearest_neighbor(images=conv4, size=(14,14))
# Now 14x14x16
conv5 = tf.layers.conv2d(inputs=upsample2, filters=32, kernel_size=(3,3), strides=(1,1), padding='same', activation=tf.nn.relu)
# Now 14x14x32
upsample3 = tf.image.resize_nearest_neighbor(images=conv5, size=(28,28))
# Now 28x28x32
conv6 = tf.layers.conv2d(inputs=upsample3, filters=32, strides=(1,1), kernel_size=(3,3), padding='same', activation=tf.nn.relu)
# Now 28x28x32
logits = tf.layers.conv2d(inputs=conv6, filters=1, strides=(1,1), kernel_size=(3,3), padding='same')
#Now 28x28x1
# Pass logits through sigmoid to get reconstructed image
decoded = tf.sigmoid(logits)
# Pass logits through sigmoid and calculate the cross-entropy loss
loss = tf.nn.sigmoid_cross_entropy_with_logits(labels=targets_, logits=logits)
# Get cost and define the optimizer
cost = tf.reduce_mean(loss)
opt = tf.train.AdamOptimizer(learning_rate).minimize(cost)
sess = tf.Session()
epochs = 10
batch_size = 200
# Set's how much noise we're adding to the MNIST images
noise_factor = 0.5
sess.run(tf.global_variables_initializer())
for e in range(epochs):
for ii in range(mnist.train.num_examples//batch_size):
batch = mnist.train.next_batch(batch_size)
# Get images from the batch
imgs = batch[0].reshape((-1, 28, 28, 1))
# Add random noise to the input images
noisy_imgs = imgs + noise_factor * np.random.randn(*imgs.shape)
# Clip the images to be between 0 and 1
noisy_imgs = np.clip(noisy_imgs, 0., 1.)
# Noisy images as inputs, original images as targets
batch_cost, _ = sess.run([cost, opt], feed_dict={inputs_: noisy_imgs,
targets_: imgs})
print("Epoch: {}/{}...".format(e+1, epochs),
"Training loss: {:.4f}".format(batch_cost))
```
## Checking out the performance
Here I'm adding noise to the test images and passing them through the autoencoder. It does a suprisingly great job of removing the noise, even though it's sometimes difficult to tell what the original number is.
```
fig, axes = plt.subplots(nrows=2, ncols=10, sharex=True, sharey=True, figsize=(20,4))
in_imgs = mnist.test.images[:10]
noisy_imgs = in_imgs + noise_factor * np.random.randn(*in_imgs.shape)
noisy_imgs = np.clip(noisy_imgs, 0., 1.)
reconstructed = sess.run(decoded, feed_dict={inputs_: noisy_imgs.reshape((10, 28, 28, 1))})
for images, row in zip([noisy_imgs, reconstructed], axes):
for img, ax in zip(images, row):
ax.imshow(img.reshape((28, 28)), cmap='Greys_r')
ax.get_xaxis().set_visible(False)
ax.get_yaxis().set_visible(False)
fig.tight_layout(pad=0.1)
```
|
github_jupyter
|
%matplotlib inline
import numpy as np
import tensorflow as tf
import matplotlib.pyplot as plt
from tensorflow.examples.tutorials.mnist import input_data
mnist = input_data.read_data_sets('MNIST_data', validation_size=0)
img = mnist.train.images[2]
plt.imshow(img.reshape((28, 28)), cmap='Greys_r')
learning_rate = 0.001
# Input and target placeholders
inputs_ = tf.placeholder(tf.float32, shape=(None, 28, 28, 1))
targets_ = tf.placeholder(tf.float32, shape=(None, 28, 28, 1))
### Encoder
conv1 = tf.layers.conv2d(inputs=inputs_, filters=16, kernel_size=(5,5), strides=(1,1), padding='same',activation=tf.nn.relu)
# Now 28x28x16
maxpool1 = tf.layers.max_pooling2d(inputs=conv1, pool_size=(2,2), strides=(2,2), padding='same')
# Now 14x14x16
conv2 = tf.layers.conv2d(inputs=maxpool1, filters=8, strides=(1,1), kernel_size=(3,3), padding='same',activation=tf.nn.relu)
# Now 14x14x8
maxpool2 = tf.layers.max_pooling2d(inputs=conv2, pool_size=(2,2), strides=(2,2), padding='same')
# Now 7x7x8
conv3 = tf.layers.conv2d(inputs=maxpool2, strides=(1,1), kernel_size=(3,3), filters=8, padding='same',activation=tf.nn.relu)
# Now 7x7x8
encoded = tf.layers.max_pooling2d(inputs=conv3, pool_size=(2,2), strides=(2,2), padding='same')
# Now 4x4x8
### Decoder
upsample1 = tf.image.resize_nearest_neighbor(images=encoded, size=(7,7))
# Now 7x7x8
conv4 = tf.layers.conv2d(inputs=upsample1, kernel_size=(3,3), filters=8, strides=(1,1), padding='same',activation=tf.nn.relu)
# Now 7x7x8
upsample2 = tf.image.resize_nearest_neighbor(images=conv4, size=(14, 14))
# Now 14x14x8
conv5 = tf.layers.conv2d(inputs=upsample2, kernel_size=(3,3), filters=8, strides=(1,1), padding='same',activation=tf.nn.relu)
# Now 14x14x8
upsample3 = tf.image.resize_nearest_neighbor(images=conv5, size=(28, 28))
# Now 28x28x8
conv6 = tf.layers.conv2d(inputs=upsample3, kernel_size=(3,3), filters=16, strides=(1,1), padding='same',activation=tf.nn.relu)
# Now 28x28x16
logits = tf.layers.conv2d(inputs=conv6, kernel_size=(3,3), filters=1, strides=(1,1), padding='same')
#Now 28x28x1
# Pass logits through sigmoid to get reconstructed image
decoded = tf.nn.sigmoid(logits)
# Pass logits through sigmoid and calculate the cross-entropy loss
loss = tf.nn.sigmoid_cross_entropy_with_logits(labels=targets_, logits=logits)
# Get cost and define the optimizer
cost = tf.reduce_mean(loss)
opt = tf.train.AdamOptimizer(learning_rate).minimize(cost)
sess = tf.Session(config=tf.ConfigProto(log_device_placement=True))
epochs = 20
batch_size = 200
sess.run(tf.global_variables_initializer())
for e in range(epochs):
for ii in range(mnist.train.num_examples//batch_size):
batch = mnist.train.next_batch(batch_size)
imgs = batch[0].reshape((-1, 28, 28, 1))
batch_cost, _ = sess.run([cost, opt], feed_dict={inputs_: imgs,
targets_: imgs})
print("Epoch: {}/{}...".format(e+1, epochs),
"Training loss: {:.4f}".format(batch_cost))
fig, axes = plt.subplots(nrows=2, ncols=10, sharex=True, sharey=True, figsize=(20,4))
in_imgs = mnist.test.images[:10]
reconstructed = sess.run(decoded, feed_dict={inputs_: in_imgs.reshape((10, 28, 28, 1))})
for images, row in zip([in_imgs, reconstructed], axes):
for img, ax in zip(images, row):
ax.imshow(img.reshape((28, 28)), cmap='Greys_r')
ax.get_xaxis().set_visible(False)
ax.get_yaxis().set_visible(False)
fig.tight_layout(pad=0.1)
sess.close()
learning_rate = 0.001
inputs_ = tf.placeholder(tf.float32, (None, 28, 28, 1), name='inputs')
targets_ = tf.placeholder(tf.float32, (None, 28, 28, 1), name='targets')
### Encoder
conv1 = tf.layers.conv2d(inputs=inputs_, filters=32, kernel_size=(5,5), strides=(1,1), padding='same', activation=tf.nn.relu)
# Now 28x28x32
maxpool1 = tf.layers.max_pooling2d(inputs=conv1, pool_size=(2,2), padding='same',strides=(2,2))
# Now 14x14x32
conv2 = tf.layers.conv2d(inputs=maxpool1, filters=32, kernel_size=(3,3), strides=(1,1), padding='same', activation=tf.nn.relu)
# Now 14x14x32
maxpool2 = tf.layers.max_pooling2d(inputs=conv2, pool_size=(2,2), padding='same',strides=(2,2))
# Now 7x7x32
conv3 = tf.layers.conv2d(inputs=maxpool2, filters=16, kernel_size=(3,3), strides=(1,1), padding='same', activation=tf.nn.relu)
# Now 7x7x16
encoded = tf.layers.max_pooling2d(inputs=conv3, pool_size=(2,2), padding='same',strides=(2,2))
# Now 4x4x16
### Decoder
upsample1 = tf.image.resize_nearest_neighbor(images=encoded, size=(7,7))
# Now 7x7x16
conv4 = tf.layers.conv2d(inputs=upsample1, filters=16, kernel_size=(3,3), strides=(1,1), padding='same', activation=tf.nn.relu)
# Now 7x7x16
upsample2 = tf.image.resize_nearest_neighbor(images=conv4, size=(14,14))
# Now 14x14x16
conv5 = tf.layers.conv2d(inputs=upsample2, filters=32, kernel_size=(3,3), strides=(1,1), padding='same', activation=tf.nn.relu)
# Now 14x14x32
upsample3 = tf.image.resize_nearest_neighbor(images=conv5, size=(28,28))
# Now 28x28x32
conv6 = tf.layers.conv2d(inputs=upsample3, filters=32, strides=(1,1), kernel_size=(3,3), padding='same', activation=tf.nn.relu)
# Now 28x28x32
logits = tf.layers.conv2d(inputs=conv6, filters=1, strides=(1,1), kernel_size=(3,3), padding='same')
#Now 28x28x1
# Pass logits through sigmoid to get reconstructed image
decoded = tf.sigmoid(logits)
# Pass logits through sigmoid and calculate the cross-entropy loss
loss = tf.nn.sigmoid_cross_entropy_with_logits(labels=targets_, logits=logits)
# Get cost and define the optimizer
cost = tf.reduce_mean(loss)
opt = tf.train.AdamOptimizer(learning_rate).minimize(cost)
sess = tf.Session()
epochs = 10
batch_size = 200
# Set's how much noise we're adding to the MNIST images
noise_factor = 0.5
sess.run(tf.global_variables_initializer())
for e in range(epochs):
for ii in range(mnist.train.num_examples//batch_size):
batch = mnist.train.next_batch(batch_size)
# Get images from the batch
imgs = batch[0].reshape((-1, 28, 28, 1))
# Add random noise to the input images
noisy_imgs = imgs + noise_factor * np.random.randn(*imgs.shape)
# Clip the images to be between 0 and 1
noisy_imgs = np.clip(noisy_imgs, 0., 1.)
# Noisy images as inputs, original images as targets
batch_cost, _ = sess.run([cost, opt], feed_dict={inputs_: noisy_imgs,
targets_: imgs})
print("Epoch: {}/{}...".format(e+1, epochs),
"Training loss: {:.4f}".format(batch_cost))
fig, axes = plt.subplots(nrows=2, ncols=10, sharex=True, sharey=True, figsize=(20,4))
in_imgs = mnist.test.images[:10]
noisy_imgs = in_imgs + noise_factor * np.random.randn(*in_imgs.shape)
noisy_imgs = np.clip(noisy_imgs, 0., 1.)
reconstructed = sess.run(decoded, feed_dict={inputs_: noisy_imgs.reshape((10, 28, 28, 1))})
for images, row in zip([noisy_imgs, reconstructed], axes):
for img, ax in zip(images, row):
ax.imshow(img.reshape((28, 28)), cmap='Greys_r')
ax.get_xaxis().set_visible(False)
ax.get_yaxis().set_visible(False)
fig.tight_layout(pad=0.1)
| 0.867654 | 0.994197 |
# PhaseNet Example
Applying GaMMA to associate PhaseNet picks
```
!pip install git+https://github.com/wayneweiqiang/GaMMA.git
import pandas as pd
from datetime import datetime, timedelta
from gamma import BayesianGaussianMixture, GaussianMixture
from gamma.utils import convert_picks_csv, association, from_seconds
import numpy as np
from sklearn.cluster import DBSCAN
from datetime import datetime, timedelta
import os
import json
import pickle
from tqdm import tqdm
```
## 1. Download and Read data
- Download test data: PhaseNet picks of the 2019 Ridgecrest earthquake sequence
1. picks file: picks.json
2. station information: stations.csv
3. events in SCSN catalog: events.csv
```bash
wget https://github.com/wayneweiqiang/GaMMA/releases/download/test_data/test_data.zip
unzip test_data.zip
```
```
!rm test_data.zip
!rm -rf test_data
!wget https://github.com/wayneweiqiang/GaMMA/releases/download/test_data/test_data.zip
!unzip test_data.zip
data_dir = lambda x: os.path.join("test_data", x)
station_csv = data_dir("stations.csv")
pick_json = data_dir("picks.json")
catalog_csv = data_dir("catalog_gamma.csv")
picks_csv = data_dir("picks_gamma.csv")
if not os.path.exists("figures"):
os.makedirs("figures")
figure_dir = lambda x: os.path.join("figures", x)
config = {'center': (-117.504, 35.705),
'xlim_degree': [-118.004, -117.004],
'ylim_degree': [35.205, 36.205],
'degree2km': 111.19492474777779,
'starttime': datetime(2019, 7, 4, 17, 0),
'endtime': datetime(2019, 7, 5, 0, 0)}
## read picks
picks = pd.read_json(pick_json)
picks["time_idx"] = picks["timestamp"].apply(lambda x: x.strftime("%Y-%m-%dT%H")) ## process by hours
## read stations
stations = pd.read_csv(station_csv, delimiter="\t")
stations = stations.rename(columns={"station":"id"})
stations["x(km)"] = stations["longitude"].apply(lambda x: (x - config["center"][0])*config["degree2km"])
stations["y(km)"] = stations["latitude"].apply(lambda x: (x - config["center"][1])*config["degree2km"])
stations["z(km)"] = stations["elevation(m)"].apply(lambda x: -x/1e3)
### setting GMMA configs
config["dims"] = ['x(km)', 'y(km)', 'z(km)']
config["use_dbscan"] = True
config["use_amplitude"] = True
config["x(km)"] = (np.array(config["xlim_degree"])-np.array(config["center"][0]))*config["degree2km"]
config["y(km)"] = (np.array(config["ylim_degree"])-np.array(config["center"][1]))*config["degree2km"]
config["z(km)"] = (0, 20)
config["vel"] = {"p":6.0, "s":6.0/1.75}
# DBSCAN
config["bfgs_bounds"] = ((config["x(km)"][0]-1, config["x(km)"][1]+1), #x
(config["y(km)"][0]-1, config["y(km)"][1]+1), #y
(0, config["z(km)"][1]+1), #x
(None, None)) #t
config["dbscan_eps"] = min(np.sqrt((stations["x(km)"].max()-stations["x(km)"].min())**2 +
(stations["y(km)"].max()-stations["y(km)"].min())**2)/(6.0/1.75), 6) #s
config["dbscan_min_samples"] = min(len(stations), 3)
# Filtering
config["min_picks_per_eq"] = min(len(stations)//2, 10)
config["oversample_factor"] = min(len(stations)//2, 10)
for k, v in config.items():
print(f"{k}: {v}")
```
## 2. Associaiton with GaMMA
```
pbar = tqdm(sorted(list(set(picks["time_idx"]))))
event_idx0 = 0 ## current earthquake index
assignments = []
if (len(picks) > 0) and (len(picks) < 5000):
data, locs, phase_type, phase_weight, phase_index = convert_picks_csv(picks, stations, config)
catalogs, assignments = association(data, locs, phase_type, phase_weight, len(stations), phase_index, event_idx0, config, pbar)
event_idx0 += len(catalogs)
else:
catalogs = []
for i, hour in enumerate(pbar):
picks_ = picks[picks["time_idx"] == hour]
if len(picks_) == 0:
continue
data, locs, phase_type, phase_weight, phase_index = convert_picks_csv(picks_, stations, config)
catalog, assign = association(data, locs, phase_type, phase_weight, len(stations), phase_index, event_idx0, config, pbar)
event_idx0 += len(catalog)
catalogs.extend(catalog)
assignments.extend(assign)
## create catalog
catalogs = pd.DataFrame(catalogs, columns=["time(s)"]+config["dims"]+["magnitude", "covariance"])
catalogs["time"] = catalogs["time(s)"].apply(lambda x: from_seconds(x))
catalogs["longitude"] = catalogs["x(km)"].apply(lambda x: x/config["degree2km"] + config["center"][0])
catalogs["latitude"] = catalogs["y(km)"].apply(lambda x: x/config["degree2km"] + config["center"][1])
catalogs["depth(m)"] = catalogs["z(km)"].apply(lambda x: x*1e3)
catalogs["event_idx"] = range(event_idx0)
if config["use_amplitude"]:
catalogs["covariance"] = catalogs["covariance"].apply(lambda x: f"{x[0][0]:.3f},{x[1][1]:.3f},{x[0][1]:.3f}")
else:
catalogs["covariance"] = catalogs["covariance"].apply(lambda x: f"{x[0][0]:.3f}")
with open(catalog_csv, 'w') as fp:
catalogs.to_csv(fp, sep="\t", index=False,
float_format="%.3f",
date_format='%Y-%m-%dT%H:%M:%S.%f',
columns=["time", "magnitude", "longitude", "latitude", "depth(m)", "covariance", "event_idx"])
catalogs = catalogs[['time', 'magnitude', 'longitude', 'latitude', 'depth(m)', 'covariance']]
## add assignment to picks
assignments = pd.DataFrame(assignments, columns=["pick_idx", "event_idx", "prob_gmma"])
picks = picks.join(assignments.set_index("pick_idx")).fillna(-1).astype({'event_idx': int})
with open(picks_csv, 'w') as fp:
picks.to_csv(fp, sep="\t", index=False,
date_format='%Y-%m-%dT%H:%M:%S.%f',
columns=["id", "timestamp", "type", "prob", "amp", "event_idx", "prob_gmma"])
```
## 3. Visualize results
Note that the location and magnitude are estimated during associaiton, which are not expected to have high accuracy.
```
import matplotlib.pyplot as plt
import matplotlib.dates as mdates
result_label="GaMMA"
catalog_label="SCSN"
stations = pd.read_csv(data_dir("stations.csv"), delimiter="\t")
events = pd.read_csv(data_dir("events.csv"), delimiter="\t")
events["time"] = events["time"].apply(lambda x: datetime.strptime(x, "%Y-%m-%dT%H:%M:%S.%f"))
catalog = pd.read_csv(data_dir("catalog_gamma.csv"), delimiter="\t")
catalog["time"] = catalog["time"].apply(lambda x: datetime.strptime(x, "%Y-%m-%dT%H:%M:%S.%f"))
catalog["covariance"] = catalog["covariance"].apply(lambda x: [float(i) for i in x.split(",")])
plt.figure()
plt.hist(catalog["time"], range=(config["starttime"], config["endtime"]), bins=24, edgecolor="k", alpha=1.0, linewidth=0.5, label=f"{result_label}: {len(catalog['time'])}")
plt.hist(events["time"], range=(config["starttime"], config["endtime"]), bins=24, edgecolor="k", alpha=1.0, linewidth=0.5, label=f"{catalog_label}: {len(events['time'])}")
plt.ylabel("Frequency")
plt.xlabel("Date")
plt.gca().autoscale(enable=True, axis='x', tight=True)
plt.gca().xaxis.set_major_formatter(mdates.DateFormatter('%m-%d:%H'))
plt.gcf().autofmt_xdate()
plt.legend()
plt.savefig(figure_dir("earthquake_number.png"), bbox_inches="tight", dpi=300)
plt.savefig(figure_dir("earthquake_number.pdf"), bbox_inches="tight")
plt.show();
fig = plt.figure(figsize=plt.rcParams["figure.figsize"]*np.array([1.5,1]))
box = dict(boxstyle='round', facecolor='white', alpha=1)
text_loc = [0.05, 0.92]
grd = fig.add_gridspec(ncols=2, nrows=2, width_ratios=[1.5, 1], height_ratios=[1,1])
fig.add_subplot(grd[:, 0])
plt.plot(catalog["longitude"], catalog["latitude"], '.',markersize=2, alpha=1.0)
plt.plot(events["longitude"], events["latitude"], '.', markersize=2, alpha=0.6)
plt.axis("scaled")
plt.xlim(np.array(config["xlim_degree"])+np.array([0.2,-0.27]))
plt.ylim(np.array(config["ylim_degree"])+np.array([0.2,-0.27]))
plt.xlabel("Latitude")
plt.ylabel("Longitude")
plt.gca().set_prop_cycle(None)
plt.plot(config["xlim_degree"][0]-10, config["ylim_degree"][0]-10, '.', markersize=10, label=f"{result_label}", rasterized=True)
plt.plot(config["xlim_degree"][0]-10, config["ylim_degree"][0]-10, '.', markersize=10, label=f"{catalog_label}", rasterized=True)
plt.plot(stations["longitude"], stations["latitude"], 'k^', markersize=5, alpha=0.7, label="Stations")
plt.legend(loc="lower right")
plt.text(text_loc[0], text_loc[1], '(i)', horizontalalignment='left', verticalalignment="top",
transform=plt.gca().transAxes, fontsize="large", fontweight="normal", bbox=box)
fig.add_subplot(grd[0, 1])
plt.plot(catalog["longitude"], catalog["depth(m)"]/1e3, '.', markersize=2, alpha=1.0, rasterized=True)
plt.plot(events["longitude"], events["depth(m)"]/1e3, '.', markersize=2, alpha=0.6, rasterized=True)
# plt.axis("scaled")
plt.xlim(np.array(config["xlim_degree"])+np.array([0.2,-0.27]))
plt.ylim([0,21])
plt.gca().invert_yaxis()
plt.xlabel("Longitude")
plt.ylabel("Depth (km)")
plt.gca().set_prop_cycle(None)
plt.plot(config["xlim_degree"][0]-10, 31, '.', markersize=10, label=f"{result_label}")
plt.plot(31, 31, '.', markersize=10, label=f"{catalog_label}")
plt.legend(loc="lower right")
plt.text(text_loc[0], text_loc[1], '(ii)', horizontalalignment='left', verticalalignment="top",
transform=plt.gca().transAxes, fontsize="large", fontweight="normal", bbox=box)
fig.add_subplot(grd[1, 1])
plt.plot(catalog["latitude"], catalog["depth(m)"]/1e3, '.', markersize=2, alpha=1.0, rasterized=True)
plt.plot(events["latitude"], events["depth(m)"]/1e3, '.', markersize=2, alpha=0.6, rasterized=True)
# plt.axis("scaled")
plt.xlim(np.array(config["ylim_degree"])+np.array([0.2,-0.27]))
plt.ylim([0,21])
plt.gca().invert_yaxis()
plt.xlabel("Latitude")
plt.ylabel("Depth (km)")
plt.gca().set_prop_cycle(None)
plt.plot(config["ylim_degree"][0]-10, 31, '.', markersize=10, label=f"{result_label}")
plt.plot(31, 31, '.', markersize=10, label=f"{catalog_label}")
plt.legend(loc="lower right")
plt.tight_layout()
plt.text(text_loc[0], text_loc[1], '(iii)', horizontalalignment='left', verticalalignment="top",
transform=plt.gca().transAxes, fontsize="large", fontweight="normal", bbox=box)
plt.savefig(figure_dir("earthquake_location.png"), bbox_inches="tight", dpi=300)
plt.savefig(figure_dir("earthquake_location.pdf"), bbox_inches="tight", dpi=300)
plt.show();
plt.figure()
plt.hist(catalog["magnitude"], range=(-1., events["magnitude"].max()), bins=25, alpha=1.0, edgecolor="k", linewidth=0.5, label=f"{result_label}: {len(catalog['magnitude'])}")
plt.hist(events["magnitude"], range=(-1., events["magnitude"].max()), bins=25, alpha=0.6, edgecolor="k", linewidth=0.5, label=f"{catalog_label}: {len(events['magnitude'])}")
plt.legend()
# plt.figure()
plt.xlim([-1,events["magnitude"].max()])
plt.xlabel("Magnitude")
plt.ylabel("Frequency")
plt.gca().set_yscale('log')
plt.savefig(figure_dir("earthquake_magnitude_frequency.png"), bbox_inches="tight", dpi=300)
plt.savefig(figure_dir("earthquake_magnitude_frequency.pdf"), bbox_inches="tight")
plt.show();
plt.figure()
plt.plot(catalog["time"], catalog["magnitude"], '.', markersize=5, alpha=1.0, rasterized=True)
plt.plot(events["time"], events["magnitude"], '.', markersize=5, alpha=0.8, rasterized=True)
plt.xlim(config["starttime"], config["endtime"])
ylim = plt.ylim()
plt.ylabel("Magnitude")
# plt.xlabel("Date")
plt.gca().xaxis.set_major_formatter(mdates.DateFormatter('%m-%d:%H'))
plt.gcf().autofmt_xdate()
plt.gca().set_prop_cycle(None)
plt.plot(config["starttime"], -10, '.', markersize=15, alpha=1.0, label=f"{result_label}: {len(catalog['magnitude'])}")
plt.plot(config["starttime"], -10, '.', markersize=15, alpha=1.0, label=f"{catalog_label}: {len(events['magnitude'])}")
plt.legend()
plt.ylim(ylim)
plt.grid()
plt.savefig(figure_dir("earthquake_magnitude_time.png"), bbox_inches="tight", dpi=300)
plt.savefig(figure_dir("earthquake_magnitude_time.pdf"), bbox_inches="tight", dpi=300)
plt.show();
covariance = np.array(catalog["covariance"].to_list())
fig = plt.figure(figsize=plt.rcParams["figure.figsize"]*np.array([0.8,1.1]))
box = dict(boxstyle='round', facecolor='white', alpha=1)
text_loc = [0.05, 0.90]
plt.subplot(311)
plt.plot(catalog["time"], covariance[:,0], '.', markersize=3.0, label="Travel-time")
plt.ylim([0, 3])
plt.ylabel(r"$\Sigma_{11}$ (s)$^2$")
plt.legend(loc="upper right")
plt.text(text_loc[0], text_loc[1], '(i)', horizontalalignment='left', verticalalignment="top",
transform=plt.gca().transAxes, fontsize="large", fontweight="normal", bbox=box)
plt.subplot(312)
plt.plot(catalog["time"], covariance[:,1], '.', markersize=3.0, label="Amplitude")
plt.ylim([0, 1])
plt.ylabel(r"$\Sigma_{22}$ ($\log10$ m/s)$^2$")
plt.legend(loc="upper right")
plt.text(text_loc[0], text_loc[1], '(ii)', horizontalalignment='left', verticalalignment="top",
transform=plt.gca().transAxes, fontsize="large", fontweight="normal", bbox=box)
plt.subplot(313)
plt.plot(catalog["time"], covariance[:,2], '.', markersize=3.0, label="Travel-time vs. Amplitude")
plt.ylabel(r"$\Sigma_{12}$")
plt.ylim([-0.5, 0.5])
plt.legend(loc="upper right")
plt.text(text_loc[0], text_loc[1], '(iii)', horizontalalignment='left', verticalalignment="top",
transform=plt.gca().transAxes, fontsize="large", fontweight="normal", bbox=box)
plt.gca().xaxis.set_major_formatter(mdates.DateFormatter('%m-%d:%H'))
plt.gcf().autofmt_xdate()
# plt.suptitle(r"Covariance Matrix ($\Sigma$) Coefficients")
plt.tight_layout()
plt.gcf().align_labels()
plt.savefig(figure_dir("covariance.png"), bbox_inches="tight", dpi=300)
plt.savefig(figure_dir("covariance.pdf"), bbox_inches="tight")
plt.show();
```
|
github_jupyter
|
!pip install git+https://github.com/wayneweiqiang/GaMMA.git
import pandas as pd
from datetime import datetime, timedelta
from gamma import BayesianGaussianMixture, GaussianMixture
from gamma.utils import convert_picks_csv, association, from_seconds
import numpy as np
from sklearn.cluster import DBSCAN
from datetime import datetime, timedelta
import os
import json
import pickle
from tqdm import tqdm
wget https://github.com/wayneweiqiang/GaMMA/releases/download/test_data/test_data.zip
unzip test_data.zip
!rm test_data.zip
!rm -rf test_data
!wget https://github.com/wayneweiqiang/GaMMA/releases/download/test_data/test_data.zip
!unzip test_data.zip
data_dir = lambda x: os.path.join("test_data", x)
station_csv = data_dir("stations.csv")
pick_json = data_dir("picks.json")
catalog_csv = data_dir("catalog_gamma.csv")
picks_csv = data_dir("picks_gamma.csv")
if not os.path.exists("figures"):
os.makedirs("figures")
figure_dir = lambda x: os.path.join("figures", x)
config = {'center': (-117.504, 35.705),
'xlim_degree': [-118.004, -117.004],
'ylim_degree': [35.205, 36.205],
'degree2km': 111.19492474777779,
'starttime': datetime(2019, 7, 4, 17, 0),
'endtime': datetime(2019, 7, 5, 0, 0)}
## read picks
picks = pd.read_json(pick_json)
picks["time_idx"] = picks["timestamp"].apply(lambda x: x.strftime("%Y-%m-%dT%H")) ## process by hours
## read stations
stations = pd.read_csv(station_csv, delimiter="\t")
stations = stations.rename(columns={"station":"id"})
stations["x(km)"] = stations["longitude"].apply(lambda x: (x - config["center"][0])*config["degree2km"])
stations["y(km)"] = stations["latitude"].apply(lambda x: (x - config["center"][1])*config["degree2km"])
stations["z(km)"] = stations["elevation(m)"].apply(lambda x: -x/1e3)
### setting GMMA configs
config["dims"] = ['x(km)', 'y(km)', 'z(km)']
config["use_dbscan"] = True
config["use_amplitude"] = True
config["x(km)"] = (np.array(config["xlim_degree"])-np.array(config["center"][0]))*config["degree2km"]
config["y(km)"] = (np.array(config["ylim_degree"])-np.array(config["center"][1]))*config["degree2km"]
config["z(km)"] = (0, 20)
config["vel"] = {"p":6.0, "s":6.0/1.75}
# DBSCAN
config["bfgs_bounds"] = ((config["x(km)"][0]-1, config["x(km)"][1]+1), #x
(config["y(km)"][0]-1, config["y(km)"][1]+1), #y
(0, config["z(km)"][1]+1), #x
(None, None)) #t
config["dbscan_eps"] = min(np.sqrt((stations["x(km)"].max()-stations["x(km)"].min())**2 +
(stations["y(km)"].max()-stations["y(km)"].min())**2)/(6.0/1.75), 6) #s
config["dbscan_min_samples"] = min(len(stations), 3)
# Filtering
config["min_picks_per_eq"] = min(len(stations)//2, 10)
config["oversample_factor"] = min(len(stations)//2, 10)
for k, v in config.items():
print(f"{k}: {v}")
pbar = tqdm(sorted(list(set(picks["time_idx"]))))
event_idx0 = 0 ## current earthquake index
assignments = []
if (len(picks) > 0) and (len(picks) < 5000):
data, locs, phase_type, phase_weight, phase_index = convert_picks_csv(picks, stations, config)
catalogs, assignments = association(data, locs, phase_type, phase_weight, len(stations), phase_index, event_idx0, config, pbar)
event_idx0 += len(catalogs)
else:
catalogs = []
for i, hour in enumerate(pbar):
picks_ = picks[picks["time_idx"] == hour]
if len(picks_) == 0:
continue
data, locs, phase_type, phase_weight, phase_index = convert_picks_csv(picks_, stations, config)
catalog, assign = association(data, locs, phase_type, phase_weight, len(stations), phase_index, event_idx0, config, pbar)
event_idx0 += len(catalog)
catalogs.extend(catalog)
assignments.extend(assign)
## create catalog
catalogs = pd.DataFrame(catalogs, columns=["time(s)"]+config["dims"]+["magnitude", "covariance"])
catalogs["time"] = catalogs["time(s)"].apply(lambda x: from_seconds(x))
catalogs["longitude"] = catalogs["x(km)"].apply(lambda x: x/config["degree2km"] + config["center"][0])
catalogs["latitude"] = catalogs["y(km)"].apply(lambda x: x/config["degree2km"] + config["center"][1])
catalogs["depth(m)"] = catalogs["z(km)"].apply(lambda x: x*1e3)
catalogs["event_idx"] = range(event_idx0)
if config["use_amplitude"]:
catalogs["covariance"] = catalogs["covariance"].apply(lambda x: f"{x[0][0]:.3f},{x[1][1]:.3f},{x[0][1]:.3f}")
else:
catalogs["covariance"] = catalogs["covariance"].apply(lambda x: f"{x[0][0]:.3f}")
with open(catalog_csv, 'w') as fp:
catalogs.to_csv(fp, sep="\t", index=False,
float_format="%.3f",
date_format='%Y-%m-%dT%H:%M:%S.%f',
columns=["time", "magnitude", "longitude", "latitude", "depth(m)", "covariance", "event_idx"])
catalogs = catalogs[['time', 'magnitude', 'longitude', 'latitude', 'depth(m)', 'covariance']]
## add assignment to picks
assignments = pd.DataFrame(assignments, columns=["pick_idx", "event_idx", "prob_gmma"])
picks = picks.join(assignments.set_index("pick_idx")).fillna(-1).astype({'event_idx': int})
with open(picks_csv, 'w') as fp:
picks.to_csv(fp, sep="\t", index=False,
date_format='%Y-%m-%dT%H:%M:%S.%f',
columns=["id", "timestamp", "type", "prob", "amp", "event_idx", "prob_gmma"])
import matplotlib.pyplot as plt
import matplotlib.dates as mdates
result_label="GaMMA"
catalog_label="SCSN"
stations = pd.read_csv(data_dir("stations.csv"), delimiter="\t")
events = pd.read_csv(data_dir("events.csv"), delimiter="\t")
events["time"] = events["time"].apply(lambda x: datetime.strptime(x, "%Y-%m-%dT%H:%M:%S.%f"))
catalog = pd.read_csv(data_dir("catalog_gamma.csv"), delimiter="\t")
catalog["time"] = catalog["time"].apply(lambda x: datetime.strptime(x, "%Y-%m-%dT%H:%M:%S.%f"))
catalog["covariance"] = catalog["covariance"].apply(lambda x: [float(i) for i in x.split(",")])
plt.figure()
plt.hist(catalog["time"], range=(config["starttime"], config["endtime"]), bins=24, edgecolor="k", alpha=1.0, linewidth=0.5, label=f"{result_label}: {len(catalog['time'])}")
plt.hist(events["time"], range=(config["starttime"], config["endtime"]), bins=24, edgecolor="k", alpha=1.0, linewidth=0.5, label=f"{catalog_label}: {len(events['time'])}")
plt.ylabel("Frequency")
plt.xlabel("Date")
plt.gca().autoscale(enable=True, axis='x', tight=True)
plt.gca().xaxis.set_major_formatter(mdates.DateFormatter('%m-%d:%H'))
plt.gcf().autofmt_xdate()
plt.legend()
plt.savefig(figure_dir("earthquake_number.png"), bbox_inches="tight", dpi=300)
plt.savefig(figure_dir("earthquake_number.pdf"), bbox_inches="tight")
plt.show();
fig = plt.figure(figsize=plt.rcParams["figure.figsize"]*np.array([1.5,1]))
box = dict(boxstyle='round', facecolor='white', alpha=1)
text_loc = [0.05, 0.92]
grd = fig.add_gridspec(ncols=2, nrows=2, width_ratios=[1.5, 1], height_ratios=[1,1])
fig.add_subplot(grd[:, 0])
plt.plot(catalog["longitude"], catalog["latitude"], '.',markersize=2, alpha=1.0)
plt.plot(events["longitude"], events["latitude"], '.', markersize=2, alpha=0.6)
plt.axis("scaled")
plt.xlim(np.array(config["xlim_degree"])+np.array([0.2,-0.27]))
plt.ylim(np.array(config["ylim_degree"])+np.array([0.2,-0.27]))
plt.xlabel("Latitude")
plt.ylabel("Longitude")
plt.gca().set_prop_cycle(None)
plt.plot(config["xlim_degree"][0]-10, config["ylim_degree"][0]-10, '.', markersize=10, label=f"{result_label}", rasterized=True)
plt.plot(config["xlim_degree"][0]-10, config["ylim_degree"][0]-10, '.', markersize=10, label=f"{catalog_label}", rasterized=True)
plt.plot(stations["longitude"], stations["latitude"], 'k^', markersize=5, alpha=0.7, label="Stations")
plt.legend(loc="lower right")
plt.text(text_loc[0], text_loc[1], '(i)', horizontalalignment='left', verticalalignment="top",
transform=plt.gca().transAxes, fontsize="large", fontweight="normal", bbox=box)
fig.add_subplot(grd[0, 1])
plt.plot(catalog["longitude"], catalog["depth(m)"]/1e3, '.', markersize=2, alpha=1.0, rasterized=True)
plt.plot(events["longitude"], events["depth(m)"]/1e3, '.', markersize=2, alpha=0.6, rasterized=True)
# plt.axis("scaled")
plt.xlim(np.array(config["xlim_degree"])+np.array([0.2,-0.27]))
plt.ylim([0,21])
plt.gca().invert_yaxis()
plt.xlabel("Longitude")
plt.ylabel("Depth (km)")
plt.gca().set_prop_cycle(None)
plt.plot(config["xlim_degree"][0]-10, 31, '.', markersize=10, label=f"{result_label}")
plt.plot(31, 31, '.', markersize=10, label=f"{catalog_label}")
plt.legend(loc="lower right")
plt.text(text_loc[0], text_loc[1], '(ii)', horizontalalignment='left', verticalalignment="top",
transform=plt.gca().transAxes, fontsize="large", fontweight="normal", bbox=box)
fig.add_subplot(grd[1, 1])
plt.plot(catalog["latitude"], catalog["depth(m)"]/1e3, '.', markersize=2, alpha=1.0, rasterized=True)
plt.plot(events["latitude"], events["depth(m)"]/1e3, '.', markersize=2, alpha=0.6, rasterized=True)
# plt.axis("scaled")
plt.xlim(np.array(config["ylim_degree"])+np.array([0.2,-0.27]))
plt.ylim([0,21])
plt.gca().invert_yaxis()
plt.xlabel("Latitude")
plt.ylabel("Depth (km)")
plt.gca().set_prop_cycle(None)
plt.plot(config["ylim_degree"][0]-10, 31, '.', markersize=10, label=f"{result_label}")
plt.plot(31, 31, '.', markersize=10, label=f"{catalog_label}")
plt.legend(loc="lower right")
plt.tight_layout()
plt.text(text_loc[0], text_loc[1], '(iii)', horizontalalignment='left', verticalalignment="top",
transform=plt.gca().transAxes, fontsize="large", fontweight="normal", bbox=box)
plt.savefig(figure_dir("earthquake_location.png"), bbox_inches="tight", dpi=300)
plt.savefig(figure_dir("earthquake_location.pdf"), bbox_inches="tight", dpi=300)
plt.show();
plt.figure()
plt.hist(catalog["magnitude"], range=(-1., events["magnitude"].max()), bins=25, alpha=1.0, edgecolor="k", linewidth=0.5, label=f"{result_label}: {len(catalog['magnitude'])}")
plt.hist(events["magnitude"], range=(-1., events["magnitude"].max()), bins=25, alpha=0.6, edgecolor="k", linewidth=0.5, label=f"{catalog_label}: {len(events['magnitude'])}")
plt.legend()
# plt.figure()
plt.xlim([-1,events["magnitude"].max()])
plt.xlabel("Magnitude")
plt.ylabel("Frequency")
plt.gca().set_yscale('log')
plt.savefig(figure_dir("earthquake_magnitude_frequency.png"), bbox_inches="tight", dpi=300)
plt.savefig(figure_dir("earthquake_magnitude_frequency.pdf"), bbox_inches="tight")
plt.show();
plt.figure()
plt.plot(catalog["time"], catalog["magnitude"], '.', markersize=5, alpha=1.0, rasterized=True)
plt.plot(events["time"], events["magnitude"], '.', markersize=5, alpha=0.8, rasterized=True)
plt.xlim(config["starttime"], config["endtime"])
ylim = plt.ylim()
plt.ylabel("Magnitude")
# plt.xlabel("Date")
plt.gca().xaxis.set_major_formatter(mdates.DateFormatter('%m-%d:%H'))
plt.gcf().autofmt_xdate()
plt.gca().set_prop_cycle(None)
plt.plot(config["starttime"], -10, '.', markersize=15, alpha=1.0, label=f"{result_label}: {len(catalog['magnitude'])}")
plt.plot(config["starttime"], -10, '.', markersize=15, alpha=1.0, label=f"{catalog_label}: {len(events['magnitude'])}")
plt.legend()
plt.ylim(ylim)
plt.grid()
plt.savefig(figure_dir("earthquake_magnitude_time.png"), bbox_inches="tight", dpi=300)
plt.savefig(figure_dir("earthquake_magnitude_time.pdf"), bbox_inches="tight", dpi=300)
plt.show();
covariance = np.array(catalog["covariance"].to_list())
fig = plt.figure(figsize=plt.rcParams["figure.figsize"]*np.array([0.8,1.1]))
box = dict(boxstyle='round', facecolor='white', alpha=1)
text_loc = [0.05, 0.90]
plt.subplot(311)
plt.plot(catalog["time"], covariance[:,0], '.', markersize=3.0, label="Travel-time")
plt.ylim([0, 3])
plt.ylabel(r"$\Sigma_{11}$ (s)$^2$")
plt.legend(loc="upper right")
plt.text(text_loc[0], text_loc[1], '(i)', horizontalalignment='left', verticalalignment="top",
transform=plt.gca().transAxes, fontsize="large", fontweight="normal", bbox=box)
plt.subplot(312)
plt.plot(catalog["time"], covariance[:,1], '.', markersize=3.0, label="Amplitude")
plt.ylim([0, 1])
plt.ylabel(r"$\Sigma_{22}$ ($\log10$ m/s)$^2$")
plt.legend(loc="upper right")
plt.text(text_loc[0], text_loc[1], '(ii)', horizontalalignment='left', verticalalignment="top",
transform=plt.gca().transAxes, fontsize="large", fontweight="normal", bbox=box)
plt.subplot(313)
plt.plot(catalog["time"], covariance[:,2], '.', markersize=3.0, label="Travel-time vs. Amplitude")
plt.ylabel(r"$\Sigma_{12}$")
plt.ylim([-0.5, 0.5])
plt.legend(loc="upper right")
plt.text(text_loc[0], text_loc[1], '(iii)', horizontalalignment='left', verticalalignment="top",
transform=plt.gca().transAxes, fontsize="large", fontweight="normal", bbox=box)
plt.gca().xaxis.set_major_formatter(mdates.DateFormatter('%m-%d:%H'))
plt.gcf().autofmt_xdate()
# plt.suptitle(r"Covariance Matrix ($\Sigma$) Coefficients")
plt.tight_layout()
plt.gcf().align_labels()
plt.savefig(figure_dir("covariance.png"), bbox_inches="tight", dpi=300)
plt.savefig(figure_dir("covariance.pdf"), bbox_inches="tight")
plt.show();
| 0.497315 | 0.759627 |
# SPARQL queries to RaDiPO ontology
```
%pip install rdflib
import rdflib
onto = rdflib.Graph()
onto.parse("C:/Users/Usuario/Documents/GitHub/RaDiPO/radipo.owl")
```
**1. Number of disorders (clinical entity)**
```
query1 = onto.query("""
PREFIX radipo:<https://w3id.org/def/radipo#>
SELECT (COUNT (DISTINCT ?disorder) AS ?num_disorders)
WHERE
{?disorder a radipo:Clinical_entity .
}""")
for row in query1:
print("Number of disorders: ", row.num_disorders)
```
**2. Number of proteins**
```
query = onto.query("""
PREFIX radipo:<https://w3id.org/def/radipo#>
SELECT (COUNT (DISTINCT ?protein) AS ?num_proteins)
WHERE
{?protein a radipo:Protein .
}""")
for row in query:
print("Number of proteins: ", row.num_proteins)
```
**3. Number of disorders with symptom "Intellectual disability"**
```
query = onto.query("""
PREFIX radipo:<https://w3id.org/def/radipo#>
PREFIX rdf:<http://www.w3.org/1999/02/22-rdf-syntax-ns#>
SELECT (COUNT (DISTINCT ?disorder) AS ?num_disorders)
WHERE
{?disorder a radipo:Clinical_entity ;
radipo:has_symptom ?symptom.
?symptom rdf:type radipo:Symptom ;
radipo:has_symptom_name ?symptom_name .
FILTER (CONTAINS(?symptom_name, "Intellectual disability"))
}""")
for row in query:
print("Number of disorders with symptom Intellectual disability: ", row.num_disorders)
```
**4. Ids of enzymes and the catalytic activity**
```
query = onto.query("""
PREFIX radipo:<https://w3id.org/def/radipo#>
PREFIX rdf:<http://www.w3.org/1999/02/22-rdf-syntax-ns#>
SELECT ?id ?activity
WHERE
{?protein rdf:type radipo:Enzyme ;
radipo:has_protein_id ?id ;
radipo:has_catalytic_activity ?activity .
} LIMIT 10""")
print("Disorder ID\tEnzyme activity")
for row in query:
print(row.id,"\t", row.activity)
```
**5. Drug name, id and phase related to a disorder **
```
query = onto.query("""
PREFIX radipo:<https://w3id.org/def/radipo#>
PREFIX rdf:<http://www.w3.org/1999/02/22-rdf-syntax-ns#>
SELECT ?disorder_name ?drug_name ?drug_id
WHERE
{
?disorder a radipo:Clinical_entity ;
radipo:is_associated_to_gene ?gene ;
radipo:has_disorder_name ?disorder_name .
?gene rdf:type radipo:Gene_with_protein ;
radipo:has_protein_product ?protein .
?protein radipo:is_target_of ?drug .
?drug radipo:has_drug_name ?drug_name ;
radipo:has_drug_id ?drug_id .
} LIMIT 10""")
print("Disorder name\tDrug name\tDrug ID")
for row in query:
print(row.disorder_name, row.drug_name, row.drug_id)
```
We can turn this into a table and use it for searching drugs related to a disease. The drugs are ralated to the protein related to the disorder, so their indications could not be the said disorder. However, there are research lines exploring how if these drugs can be applied for drug therapy in rare diseases too. This information could be a starting point of a research project for looking for drugs for a specific disorder.
**6.Number of consequences/disabilities**
```
query = onto.query("""
PREFIX radipo:<https://w3id.org/def/radipo#>
SELECT (COUNT (DISTINCT ?disability) AS ?num_dis)
WHERE
{?disability a radipo:Disability .
}""")
for row in query:
print("Number of consequences/disabilities: ", row.num_dis)
```
**7.Disorders whose patients have affected their writing skills**
```
query = onto.query("""
PREFIX radipo:<https://w3id.org/def/radipo#>
SELECT ?name
WHERE
{
?ability_class a radipo:Ability_affected ;
radipo:ability_specification ?ability .
FILTER (?ability = "Writing"^^xsd:string) .
?disability rdf:type radipo:Disability;
radipo:affects_ability ?ability_class .
?disorder radipo:is_associated_to_disability ?disability;
radipo:has_disorder_name ?name .
}""")
for row in query:
print(row.name)
```
**8. Number of disorders with patients that have the skill "Transferring oneself" affected.**
```
query = onto.query("""
PREFIX radipo:<https://w3id.org/def/radipo#>
SELECT (COUNT(DISTINCT ?disorder) AS ?num_dis)
WHERE
{
?ability_class a radipo:Ability_affected ;
radipo:ability_specification ?ability .
FILTER (?ability = "Transferring oneself"^^xsd:string) .
?disability rdf:type radipo:Disability;
radipo:affects_ability ?ability_class .
?disorder radipo:is_associated_to_disability ?disability.
}""")
for row in query:
print(row.num_dis)
```
Patients with these disability may need physical therapy, which is an information needed by health professionals
|
github_jupyter
|
%pip install rdflib
import rdflib
onto = rdflib.Graph()
onto.parse("C:/Users/Usuario/Documents/GitHub/RaDiPO/radipo.owl")
query1 = onto.query("""
PREFIX radipo:<https://w3id.org/def/radipo#>
SELECT (COUNT (DISTINCT ?disorder) AS ?num_disorders)
WHERE
{?disorder a radipo:Clinical_entity .
}""")
for row in query1:
print("Number of disorders: ", row.num_disorders)
query = onto.query("""
PREFIX radipo:<https://w3id.org/def/radipo#>
SELECT (COUNT (DISTINCT ?protein) AS ?num_proteins)
WHERE
{?protein a radipo:Protein .
}""")
for row in query:
print("Number of proteins: ", row.num_proteins)
query = onto.query("""
PREFIX radipo:<https://w3id.org/def/radipo#>
PREFIX rdf:<http://www.w3.org/1999/02/22-rdf-syntax-ns#>
SELECT (COUNT (DISTINCT ?disorder) AS ?num_disorders)
WHERE
{?disorder a radipo:Clinical_entity ;
radipo:has_symptom ?symptom.
?symptom rdf:type radipo:Symptom ;
radipo:has_symptom_name ?symptom_name .
FILTER (CONTAINS(?symptom_name, "Intellectual disability"))
}""")
for row in query:
print("Number of disorders with symptom Intellectual disability: ", row.num_disorders)
query = onto.query("""
PREFIX radipo:<https://w3id.org/def/radipo#>
PREFIX rdf:<http://www.w3.org/1999/02/22-rdf-syntax-ns#>
SELECT ?id ?activity
WHERE
{?protein rdf:type radipo:Enzyme ;
radipo:has_protein_id ?id ;
radipo:has_catalytic_activity ?activity .
} LIMIT 10""")
print("Disorder ID\tEnzyme activity")
for row in query:
print(row.id,"\t", row.activity)
query = onto.query("""
PREFIX radipo:<https://w3id.org/def/radipo#>
PREFIX rdf:<http://www.w3.org/1999/02/22-rdf-syntax-ns#>
SELECT ?disorder_name ?drug_name ?drug_id
WHERE
{
?disorder a radipo:Clinical_entity ;
radipo:is_associated_to_gene ?gene ;
radipo:has_disorder_name ?disorder_name .
?gene rdf:type radipo:Gene_with_protein ;
radipo:has_protein_product ?protein .
?protein radipo:is_target_of ?drug .
?drug radipo:has_drug_name ?drug_name ;
radipo:has_drug_id ?drug_id .
} LIMIT 10""")
print("Disorder name\tDrug name\tDrug ID")
for row in query:
print(row.disorder_name, row.drug_name, row.drug_id)
query = onto.query("""
PREFIX radipo:<https://w3id.org/def/radipo#>
SELECT (COUNT (DISTINCT ?disability) AS ?num_dis)
WHERE
{?disability a radipo:Disability .
}""")
for row in query:
print("Number of consequences/disabilities: ", row.num_dis)
query = onto.query("""
PREFIX radipo:<https://w3id.org/def/radipo#>
SELECT ?name
WHERE
{
?ability_class a radipo:Ability_affected ;
radipo:ability_specification ?ability .
FILTER (?ability = "Writing"^^xsd:string) .
?disability rdf:type radipo:Disability;
radipo:affects_ability ?ability_class .
?disorder radipo:is_associated_to_disability ?disability;
radipo:has_disorder_name ?name .
}""")
for row in query:
print(row.name)
query = onto.query("""
PREFIX radipo:<https://w3id.org/def/radipo#>
SELECT (COUNT(DISTINCT ?disorder) AS ?num_dis)
WHERE
{
?ability_class a radipo:Ability_affected ;
radipo:ability_specification ?ability .
FILTER (?ability = "Transferring oneself"^^xsd:string) .
?disability rdf:type radipo:Disability;
radipo:affects_ability ?ability_class .
?disorder radipo:is_associated_to_disability ?disability.
}""")
for row in query:
print(row.num_dis)
| 0.296756 | 0.894005 |
```
import os
import pandas as pd
import numpy as np
import xarray as xr
import matplotlib.pyplot as plt
from os.path import join
import yaml
%matplotlib inline
models_dir = "/glade/p/cisl/aiml/ggantos/200607/"
model_paths = sorted([x[0] for x in os.walk(models_dir)][1:])
models = list(range(0,54))
models.remove(48)
classifier_scores = pd.read_csv(join(models_dir, "cam_run5_models_0/dnn_classifier_scores.csv"))
regressor_scores = pd.read_csv(join(models_dir, "cam_run5_models_0/dnn_regressor_scores.csv"))
classifier_scores
l2_weight = [1.0e-3, 1.0e-4, 1.0e-5]
lrs = [0.001, 0.0001, 0.00001]
hidden_layers = [2, 3, 4]
activation = ['relu', 'tanh']
outputs_reg = ['qrtend_TAU_1', 'nctend_TAU_1', 'nrtend_TAU_-1', 'nrtend_TAU_1']
regs_dict = {}
for i in models:
y = yaml.load(open(f'../config/200607/cesm_tau_run5_full_train_nn_{i}.yml'), Loader=yaml.FullLoader)
regs_dict[i] = {}
regs_dict[i]['activation'] = y['classifier_networks']['activation']
regs_dict[i]['hidden_layers'] = y['classifier_networks']['hidden_layers']
regs_dict[i]['lr'] = y['classifier_networks']['lr']
regs_dict[i]['l2_weight'] = y['classifier_networks']['l2_weight']
regressions = {}
for out in outputs_reg:
regressions[out] = {}
ids = []
rmse = []
mae = []
r2 = []
hellinger = []
for i in models:
reg = pd.read_csv(join(models_dir, f"cam_run5_models_{i}/dnn_regressor_scores.csv"))
ids.append(i)
rmse.append(float(reg.loc[reg['Output'] == out]["rmse"]))
mae.append(float(reg.loc[reg['Output'] == out]["mae"]))
r2.append(float(reg.loc[reg['Output'] == out]["r2"]))
hellinger.append(float(reg.loc[reg['Output'] == out]["hellinger"]))
regressions[out]["ids"] = ids
regressions[out]["rmse"] = rmse
regressions[out]["mae"] = mae
regressions[out]["r2"] = r2
regressions[out]["hellinger"] = hellinger
tops = []
N = 10
f, (ax1, ax2, ax3, ax4) = plt.subplots(1, 4, sharey=True, figsize=(16,8))
for out, ax in zip(outputs_reg, (ax1, ax2, ax3, ax4)):
ax.plot(regressions[out]["rmse"], label="rmse")
top = sorted(range(len(regressions[out]["rmse"])), key = lambda sub: regressions[out]["rmse"][sub])[-N:]
tops.append(top)
for i in top:
ax.annotate(i, (i, regressions[out]["rmse"][i]))
ax.plot(regressions[out]["mae"], label="mae")
ax.plot(regressions[out]["r2"], label="r2")
top = sorted(range(len(regressions[out]["r2"])), key = lambda sub: regressions[out]["r2"][sub])[-N:]
tops.append(top)
for i in top:
ax.annotate(i, (i, regressions[out]["r2"][i]))
ax.plot(regressions[out]["hellinger"], label="hellinger")
plt.subplots_adjust(wspace=None, hspace=None)
plt.subplots_adjust(wspace = 0)
plt.legend(loc="best")
plt.show()
outputs_class = ['qrtend_TAU', 'nctend_TAU', 'nrtend_TAU']
classifications = {}
for out in outputs_class:
classifications[out] = {}
ids = []
accuracy = []
heidke = []
peirce = []
for i in models:
clss = pd.read_csv(join(models_dir, f"cam_run5_models_{i}/dnn_classifier_scores.csv"))
ids.append(i)
accuracy.append(float(clss.loc[clss['Output'] == out]["accuracy"]))
heidke.append(float(clss.loc[clss['Output'] == out]["heidke"]))
peirce.append(float(clss.loc[clss['Output'] == out]["peirce"]))
classifications[out]["ids"] = ids
classifications[out]["accuracy"] = accuracy
classifications[out]["heidke"] = heidke
classifications[out]["peirce"] = peirce
f, (ax1, ax2, ax3) = plt.subplots(1, 3, sharey=True, figsize=(15,8))
for out, ax in zip(outputs_class, (ax1, ax2, ax3)):
ax.plot(classifications[out]["accuracy"], label="accuracy")
top = sorted(range(len(classifications[out]["accuracy"])), key = lambda sub: classifications[out]["accuracy"][sub])[-N:]
tops.append(top)
for i in top:
ax.annotate(i, (i, classifications[out]["accuracy"][i]))
ax.plot(classifications[out]["heidke"], label="heidke")
ax.plot(classifications[out]["peirce"], label="peirce")
ax.set_title(out)
plt.subplots_adjust(wspace = 0)
plt.legend(loc="best")
plt.show()
tops = np.array(tops)
print(tops.shape)
tops
tops_intersection = set(tops[0]).intersection(*tops)
tops_intersection
unique_elements, counts_elements = np.unique(tops, return_counts=True)
for e, c in zip (unique_elements, counts_elements):
print (f"Element {e} has a frequency count of {c}")
tops_top = [11,14,17,20,23,26,49,52]
for i in tops_top:
print (regs_dict[i])
```
|
github_jupyter
|
import os
import pandas as pd
import numpy as np
import xarray as xr
import matplotlib.pyplot as plt
from os.path import join
import yaml
%matplotlib inline
models_dir = "/glade/p/cisl/aiml/ggantos/200607/"
model_paths = sorted([x[0] for x in os.walk(models_dir)][1:])
models = list(range(0,54))
models.remove(48)
classifier_scores = pd.read_csv(join(models_dir, "cam_run5_models_0/dnn_classifier_scores.csv"))
regressor_scores = pd.read_csv(join(models_dir, "cam_run5_models_0/dnn_regressor_scores.csv"))
classifier_scores
l2_weight = [1.0e-3, 1.0e-4, 1.0e-5]
lrs = [0.001, 0.0001, 0.00001]
hidden_layers = [2, 3, 4]
activation = ['relu', 'tanh']
outputs_reg = ['qrtend_TAU_1', 'nctend_TAU_1', 'nrtend_TAU_-1', 'nrtend_TAU_1']
regs_dict = {}
for i in models:
y = yaml.load(open(f'../config/200607/cesm_tau_run5_full_train_nn_{i}.yml'), Loader=yaml.FullLoader)
regs_dict[i] = {}
regs_dict[i]['activation'] = y['classifier_networks']['activation']
regs_dict[i]['hidden_layers'] = y['classifier_networks']['hidden_layers']
regs_dict[i]['lr'] = y['classifier_networks']['lr']
regs_dict[i]['l2_weight'] = y['classifier_networks']['l2_weight']
regressions = {}
for out in outputs_reg:
regressions[out] = {}
ids = []
rmse = []
mae = []
r2 = []
hellinger = []
for i in models:
reg = pd.read_csv(join(models_dir, f"cam_run5_models_{i}/dnn_regressor_scores.csv"))
ids.append(i)
rmse.append(float(reg.loc[reg['Output'] == out]["rmse"]))
mae.append(float(reg.loc[reg['Output'] == out]["mae"]))
r2.append(float(reg.loc[reg['Output'] == out]["r2"]))
hellinger.append(float(reg.loc[reg['Output'] == out]["hellinger"]))
regressions[out]["ids"] = ids
regressions[out]["rmse"] = rmse
regressions[out]["mae"] = mae
regressions[out]["r2"] = r2
regressions[out]["hellinger"] = hellinger
tops = []
N = 10
f, (ax1, ax2, ax3, ax4) = plt.subplots(1, 4, sharey=True, figsize=(16,8))
for out, ax in zip(outputs_reg, (ax1, ax2, ax3, ax4)):
ax.plot(regressions[out]["rmse"], label="rmse")
top = sorted(range(len(regressions[out]["rmse"])), key = lambda sub: regressions[out]["rmse"][sub])[-N:]
tops.append(top)
for i in top:
ax.annotate(i, (i, regressions[out]["rmse"][i]))
ax.plot(regressions[out]["mae"], label="mae")
ax.plot(regressions[out]["r2"], label="r2")
top = sorted(range(len(regressions[out]["r2"])), key = lambda sub: regressions[out]["r2"][sub])[-N:]
tops.append(top)
for i in top:
ax.annotate(i, (i, regressions[out]["r2"][i]))
ax.plot(regressions[out]["hellinger"], label="hellinger")
plt.subplots_adjust(wspace=None, hspace=None)
plt.subplots_adjust(wspace = 0)
plt.legend(loc="best")
plt.show()
outputs_class = ['qrtend_TAU', 'nctend_TAU', 'nrtend_TAU']
classifications = {}
for out in outputs_class:
classifications[out] = {}
ids = []
accuracy = []
heidke = []
peirce = []
for i in models:
clss = pd.read_csv(join(models_dir, f"cam_run5_models_{i}/dnn_classifier_scores.csv"))
ids.append(i)
accuracy.append(float(clss.loc[clss['Output'] == out]["accuracy"]))
heidke.append(float(clss.loc[clss['Output'] == out]["heidke"]))
peirce.append(float(clss.loc[clss['Output'] == out]["peirce"]))
classifications[out]["ids"] = ids
classifications[out]["accuracy"] = accuracy
classifications[out]["heidke"] = heidke
classifications[out]["peirce"] = peirce
f, (ax1, ax2, ax3) = plt.subplots(1, 3, sharey=True, figsize=(15,8))
for out, ax in zip(outputs_class, (ax1, ax2, ax3)):
ax.plot(classifications[out]["accuracy"], label="accuracy")
top = sorted(range(len(classifications[out]["accuracy"])), key = lambda sub: classifications[out]["accuracy"][sub])[-N:]
tops.append(top)
for i in top:
ax.annotate(i, (i, classifications[out]["accuracy"][i]))
ax.plot(classifications[out]["heidke"], label="heidke")
ax.plot(classifications[out]["peirce"], label="peirce")
ax.set_title(out)
plt.subplots_adjust(wspace = 0)
plt.legend(loc="best")
plt.show()
tops = np.array(tops)
print(tops.shape)
tops
tops_intersection = set(tops[0]).intersection(*tops)
tops_intersection
unique_elements, counts_elements = np.unique(tops, return_counts=True)
for e, c in zip (unique_elements, counts_elements):
print (f"Element {e} has a frequency count of {c}")
tops_top = [11,14,17,20,23,26,49,52]
for i in tops_top:
print (regs_dict[i])
| 0.307358 | 0.295138 |
# Distributions
Think Bayes, Second Edition
Copyright 2020 Allen B. Downey
License: [Attribution-NonCommercial-ShareAlike 4.0 International (CC BY-NC-SA 4.0)](https://creativecommons.org/licenses/by-nc-sa/4.0/)
```
# If we're running on Colab, install empiricaldist
# https://pypi.org/project/empiricaldist/
import sys
IN_COLAB = 'google.colab' in sys.modules
if IN_COLAB:
!pip install empiricaldist
# Get utils.py
import os
if not os.path.exists('utils.py'):
!wget https://github.com/AllenDowney/ThinkBayes2/raw/master/soln/utils.py
from utils import set_pyplot_params
set_pyplot_params()
```
In the previous chapter we used Bayes's Theorem to solve a cookie problem; then we solved it again using a Bayes table.
In this chapter, at the risk of testing your patience, we will solve it one more time using a `Pmf` object, which represents a "probability mass function".
I'll explain what that means, and why it is useful for Bayesian statistics.
We'll use `Pmf` objects to solve some more challenging problems and take one more step toward Bayesian statistics.
But we'll start with distributions.
## Distributions
In statistics a **distribution** is a set of possible outcomes and their corresponding probabilities.
For example, if you toss a coin, there are two possible outcomes with
approximately equal probability.
If you roll a six-sided die, the set of possible outcomes is the numbers 1 to 6, and the probability associated with each outcome is 1/6.
To represent distributions, we'll use a library called `empiricaldist`.
An "empirical" distribution is based on data, as opposed to a
theoretical distribution.
We'll use this library throughout the book. I'll introduce the basic features in this chapter and we'll see additional features later.
## Probability mass functions
If the outcomes in a distribution are discrete, we can describe the distribution with a **probability mass function**, or PMF, which is a function that maps from each possible outcome to its probability.
`empiricaldist` provides a class called `Pmf` that represents a
probability mass function.
To use `Pmf` you can import it like this:
```
from empiricaldist import Pmf
```
If that doesn't work, you might have to install `empiricaldist`; try running
```
!pip install empiricaldist
```
in a code cell or
```
pip install empiricaldist
```
in a terminal window.
The following example makes a `Pmf` that represents the outcome of a
coin toss.
```
coin = Pmf()
coin['heads'] = 1/2
coin['tails'] = 1/2
coin
```
`Pmf` creates an empty `Pmf` with no outcomes.
Then we can add new outcomes using the bracket operator.
In this example, the two outcomes are represented with strings, and they have the same probability, 0.5.
You can also make a `Pmf` from a sequence of possible outcomes.
The following example uses `Pmf.from_seq` to make a `Pmf` that represents a six-sided die.
```
die = Pmf.from_seq([1,2,3,4,5,6])
die
```
In this example, all outcomes in the sequence appear once, so they all have the same probability, $1/6$.
More generally, outcomes can appear more than once, as in the following example:
```
letters = Pmf.from_seq(list('Mississippi'))
letters
```
The letter `M` appears once out of 11 characters, so its probability is $1/11$.
The letter `i` appears 4 times, so its probability is $4/11$.
Since the letters in a string are not outcomes of a random process, I'll use the more general term "quantities" for the letters in the `Pmf`.
The `Pmf` class inherits from a Pandas `Series`, so anything you can do with a `Series`, you can also do with a `Pmf`.
For example, you can use the bracket operator to look up a quantity and get the corresponding probability.
```
letters['s']
```
In the word "Mississippi", about 36% of the letters are "s".
However, if you ask for the probability of a quantity that's not in the distribution, you get a `KeyError`.
```
try:
letters['t']
except KeyError as e:
print(type(e))
```
You can also call a `Pmf` as if it were a function, with a letter in parentheses.
```
letters('s')
```
If the quantity is in the distribution the results are the same.
But if it is not in the distribution, the result is `0`, not an error.
```
letters('t')
```
With parentheses, you can also provide a sequence of quantities and get a sequence of probabilities.
```
die([1,4,7])
```
The quantities in a `Pmf` can be strings, numbers, or any other type that can be stored in the index of a Pandas `Series`.
If you are familiar with Pandas, that will help you work with `Pmf` objects.
But I will explain what you need to know as we go along.
## The cookie problem
In this section I'll use a `Pmf` to solve the cookie problem from Section XX.
Here's the statement of the problem again:
> Suppose there are two bowls of cookies.
>
> * Bowl 1 contains 30 vanilla cookies and 10 chocolate cookies.
>
> * Bowl 2 contains 20 vanilla cookies and 20 chocolate cookies.
>
> Now suppose you choose one of the bowls at random and, without looking, choose a cookie at random. If the cookie is vanilla, what is the probability that it came from Bowl 1?
Here's a `Pmf` that represents the two hypotheses and their prior probabilities:
```
prior = Pmf.from_seq(['Bowl 1', 'Bowl 2'])
prior
```
This distribution, which contains the prior probability for each hypothesis, is called (wait for it) the **prior distribution**.
To update the distribution based on new data (the vanilla cookie),
we multiply the priors by the likelihoods. The likelihood
of drawing a vanilla cookie from Bowl 1 is `3/4`. The likelihood
for Bowl 2 is `1/2`.
```
likelihood_vanilla = [0.75, 0.5]
posterior = prior * likelihood_vanilla
posterior
```
The result is the unnormalized posteriors; that is, they don't add up to 1.
To make them add up to 1, we can use `normalize`, which is a method provided by `Pmf`.
```
posterior.normalize()
```
The return value from `normalize` is the total probability of the data, which is $5/8$.
`posterior`, which contains the posterior probability for each hypothesis, is called (wait now) the **posterior distribution**.
```
posterior
```
From the posterior distribution we can select the posterior probability for Bowl 1:
```
posterior('Bowl 1')
```
And the answer is 0.6.
One benefit of using `Pmf` objects is that it is easy to do successive updates with more data.
For example, suppose you put the first cookie back (so the contents of the bowls don't change) and draw again from the same bowl.
If the second cookie is also vanilla, we can do a second update like this:
```
posterior *= likelihood_vanilla
posterior.normalize()
posterior
```
Now the posterior probability for Bowl 1 is almost 70%.
But suppose we do the same thing again and get a chocolate cookie.
Here are the likelihoods for the new data:
```
likelihood_chocolate = [0.25, 0.5]
```
And here's the update.
```
posterior *= likelihood_chocolate
posterior.normalize()
posterior
```
Now the posterior probability for Bowl 1 is about 53%.
After two vanilla cookies and one chocolate, the posterior probabilities are close to 50/50.
## 101 Bowls
Next let's solve a cookie problem with 101 bowls:
* Bowl 0 contains 0% vanilla cookies,
* Bowl 1 contains 1% vanilla cookies,
* Bowl 2 contains 2% vanilla cookies,
and so on, up to
* Bowl 99 contains 99% vanilla cookies, and
* Bowl 100 contains all vanilla cookies.
As in the previous version, there are only two kinds of cookies, vanilla and chocolate. So Bowl 0 is all chocolate cookies, Bowl 1 is 99% chocolate, and so on.
Suppose we choose a bowl at random, choose a cookie at random, and it turns out to be vanilla. What is the probability that the cookie came from Bowl $x$, for each value of $x$?
To solve this problem, I'll use `np.arange` to make an array that represents 101 hypotheses, numbered from 0 to 100.
```
#id bowls101
import numpy as np
hypos = np.arange(101)
```
We can use this array to make the prior distribution:
```
prior = Pmf(1, hypos)
prior.normalize()
```
As this example shows, we can initialize a `Pmf` with two parameters.
The first parameter is the prior probability; the second parameter is a sequence of quantities.
In this example, the probabilities are all the same, so we only have to provide one of them; it gets "broadcast" across the hypotheses.
Since all hypotheses have the same prior probability, this distribution is **uniform**.
The likelihood of the data is the fraction of vanilla cookies in each bowl, which we can calculate using `hypos`:
```
likelihood_vanilla = hypos/100
```
Now we can compute the posterior distribution in the usual way:
```
posterior1 = prior * likelihood_vanilla
posterior1.normalize()
```
The following figure shows the prior distribution and the posterior distribution after one vanilla cookie.
```
from utils import decorate
def decorate_bowls(title):
decorate(xlabel='Bowl #',
ylabel='PMF',
title=title)
prior.plot(label='prior', color='C5')
posterior1.plot(label='posterior', color='C4')
decorate_bowls('Posterior after one vanilla cookie')
```
The posterior probability of Bowl 0 is 0 because it contains no vanilla cookies.
The posterior probability of Bowl 100 is the highest because it contain the most vanilla cookies.
In between, the shape of the posterior distribution is a line because the the likelihoods are proportional to the bowl numbers.
Now suppose we put the cookie back, draw again from the same bowl, and get another vanilla cookie.
Here's the update after the second cookie:
```
posterior2 = posterior1 * likelihood_vanilla
posterior2.normalize()
```
And here's what the posterior distribution looks like.
```
posterior2.plot(label='posterior', color='C4')
decorate_bowls('Posterior after two vanilla cookies')
```
After two vanilla cookies, the high-numbered bowls have the highest posterior probabilities because they contain the most vanilla cookies; the low-numbered bowls have the lowest probabilities.
But suppose we draw again and get a chocolate cookie.
Here's the update:
```
likelihood_chocolate = 1 - hypos/100
posterior3 = posterior2 * likelihood_chocolate
posterior3.normalize()
```
And here's the posterior distribution.
```
posterior3.plot(label='posterior', color='C4')
decorate_bowls('Posterior after 2 vanilla, 1 chocolate')
```
Now Bowl 100 has been eliminated because it contains no chocolate cookies.
But the high-numbered bowls are still more likely than the low-numbered bowls, because we have seen more vanilla cookies than chocolate.
In fact, the peak of the posterior distribution is at Bowl 67, which corresponds to the fraction of vanilla cookies in the data we've observed, $2/3$.
The quantity with the highest posterior probability is called the **MAP**, which stands for "maximum a posteori probability", where "a posteori" is unnecessary Latin for "posterior".
To compute the MAP, we can use the `Series` method `idxmax`:
```
posterior3.idxmax()
```
Or `Pmf` provides a more memorable name for the same thing:
```
posterior3.max_prob()
```
As you might suspect, this example isn't really about bowls; it's about estimating proportions.
Imagine that you have one bowl of cookies.
You don't know what fraction of cookies are vanilla, but you think it is equally likely to be any fraction from 0 to 1.
If you draw three cookies and two are vanilla, what proportion of cookies in the bowl do you think are vanilla?
The posterior distribution we just computed is the answer to that question.
We'll come back to estimating proportions in the next chapter.
But first let's use a `Pmf` to solve the dice problem.
## The dice problem
In the previous chapter we solved the dice problem using a Bayes table.
Here's the statement of the problem:
> Suppose I have a box with a 6-sided die, an 8-sided die, and a 12-sided die.
> I choose one of the dice at random, roll it, and report that the outcome is a 1.
> What is the probability that I chose the 6-sided die?
Let's solve it using a `Pmf`.
I'll use integers to represent the hypotheses:
```
hypos = [6, 8, 12]
```
We can make the prior distribution like this:
```
prior = Pmf(1/3, hypos)
prior
```
As in the previous example, the prior probability gets broadcast across the hypotheses.
The `Pmf` object has two attributes:
* `qs` contains the quantities in the distribution;
* `ps` contains the corresponding probabilities.
```
prior.qs
prior.ps
```
Now we're ready to do the update.
Here's the likelihood of the data for each hypothesis.
```
likelihood1 = 1/6, 1/8, 1/12
```
And here's the update.
```
posterior = prior * likelihood1
posterior.normalize()
posterior
```
The posterior probability for the 6-sided die is $4/9$.
Now suppose I roll the same die again and get a 7.
Here are the likelihoods:
```
likelihood2 = 0, 1/8, 1/12
```
The likelihood for the 6-sided die is 0 because it is not possible to get a 7 on a 6-sided die.
The other two likelihoods are the same as in the previous update.
Here's the update:
```
posterior *= likelihood2
posterior.normalize()
posterior
```
After rolling a 1 and a 7, the posterior probability of the 8-sided die is about 69%.
## Updating dice
The following function is a more general version of the update in the previous section:
```
def update_dice(pmf, data):
"""Update pmf based on new data."""
hypos = pmf.qs
likelihood = 1 / hypos
impossible = (data > hypos)
likelihood[impossible] = 0
pmf *= likelihood
pmf.normalize()
```
The first parameter is a `Pmf` that represents the possible dice and their probabilities.
The second parameter is the outcome of rolling a die.
The first line selects quantities from the `Pmf` which represent the hypotheses.
Since the hypotheses are integers, we can use them to compute the likelihoods.
In general, if there are `n` sides on the die, the probability of any possible outcome is `1/n`.
However, we have to check for impossible outcomes!
If the outcome exceeds the hypothetical number of sides on the die, the probability of that outcome is 0.
`impossible` is a Boolean Series that is `True` for each impossible outcome.
I use it as an index into `likelihood` to set the corresponding probabilities to 0.
Finally, I multiply `pmf` by the likelihoods and normalize.
Here's how we can use this function to compute the updates in the previous section.
I start with a fresh copy of the prior distribution:
```
pmf = prior.copy()
```
And use `update_dice` to do the updates.
```
update_dice(pmf, 1)
update_dice(pmf, 7)
pmf
```
The result is the same.
## Summary
This chapter introduces the `empiricaldist` module, which provides `Pmf`, which we use to represent a set of hypotheses and their probabilities.
`empiricaldist` is based on Pandas; the `Pmf` class inherits from the Pandas `Series` class and provides additional features specific to probability mass functions.
We'll use `Pmf` and other classes from `empiricaldist` throughout the book because they simplify the code and make it more readable.
But it would be easy to do the same things directly with Pandas.
We use a `Pmf` to solve the cookie problem and the dice problem, which we saw in the previous chapter.
With a `Pmf` it is easy to perform sequential updates with multiple pieces of data.
We also solved a more general version of the cookie problem, with 101 bowls rather than two.
Then we computed the MAP, which is the quantity with the highest posterior probability.
In the next chapter, I'll introduce the Euro problem, and we will use the binomial distribution.
And, at last, we will make the leap from using Bayes's Theorem to doing Bayesian statistics.
But first you might want to work on the exercises.
## Exercises
**Exercise:** Suppose I have a box with a 6-sided die, an 8-sided die, and a 12-sided die.
I choose one of the dice at random, roll it four times, and get 1, 3, 5, and 7.
What is the probability that I chose the 8-sided die?
You can use the `update_dice` function or do the update yourself.
```
# Solution goes here
```
**Exercise:** In the previous version of the dice problem, the prior probabilities are the same because the box contains one of each die.
But suppose the box contains 1 die that is 4-sided, 2 dice that are 6-sided, 3 dice that are 8-sided, 4 dice that are 12-sided, and 5 dice that are 20-sided.
I choose a die, roll it, and get a 7.
What is the probability that I chose an 8-sided die?
Hint: To make the prior distribution, call `Pmf` with two parameters.
```
# Solution goes here
```
**Exercise:** Suppose I have two sock drawers.
One contains equal numbers of black and white socks.
The other contains equal numbers of red, green, and blue socks.
Suppose I choose a drawer at random, choose two socks at random, and I tell you that I got a matching pair.
What is the probability that the socks are white?
For simplicity, let's assume that there are so many socks in both drawers that removing one sock makes a negligible change to the proportions.
```
# Solution goes here
# Solution goes here
```
**Exercise:** Here's a problem from [Bayesian Data Analysis](http://www.stat.columbia.edu/~gelman/book/):
> Elvis Presley had a twin brother (who died at birth). What is the probability that Elvis was an identical twin?
Hint: In 1935, about 2/3 of twins were fraternal and 1/3 were identical.
```
# Solution goes here
# Solution goes here
# Solution goes here
```
|
github_jupyter
|
# If we're running on Colab, install empiricaldist
# https://pypi.org/project/empiricaldist/
import sys
IN_COLAB = 'google.colab' in sys.modules
if IN_COLAB:
!pip install empiricaldist
# Get utils.py
import os
if not os.path.exists('utils.py'):
!wget https://github.com/AllenDowney/ThinkBayes2/raw/master/soln/utils.py
from utils import set_pyplot_params
set_pyplot_params()
from empiricaldist import Pmf
!pip install empiricaldist
pip install empiricaldist
coin = Pmf()
coin['heads'] = 1/2
coin['tails'] = 1/2
coin
die = Pmf.from_seq([1,2,3,4,5,6])
die
letters = Pmf.from_seq(list('Mississippi'))
letters
letters['s']
try:
letters['t']
except KeyError as e:
print(type(e))
letters('s')
letters('t')
die([1,4,7])
prior = Pmf.from_seq(['Bowl 1', 'Bowl 2'])
prior
likelihood_vanilla = [0.75, 0.5]
posterior = prior * likelihood_vanilla
posterior
posterior.normalize()
posterior
posterior('Bowl 1')
posterior *= likelihood_vanilla
posterior.normalize()
posterior
likelihood_chocolate = [0.25, 0.5]
posterior *= likelihood_chocolate
posterior.normalize()
posterior
#id bowls101
import numpy as np
hypos = np.arange(101)
prior = Pmf(1, hypos)
prior.normalize()
likelihood_vanilla = hypos/100
posterior1 = prior * likelihood_vanilla
posterior1.normalize()
from utils import decorate
def decorate_bowls(title):
decorate(xlabel='Bowl #',
ylabel='PMF',
title=title)
prior.plot(label='prior', color='C5')
posterior1.plot(label='posterior', color='C4')
decorate_bowls('Posterior after one vanilla cookie')
posterior2 = posterior1 * likelihood_vanilla
posterior2.normalize()
posterior2.plot(label='posterior', color='C4')
decorate_bowls('Posterior after two vanilla cookies')
likelihood_chocolate = 1 - hypos/100
posterior3 = posterior2 * likelihood_chocolate
posterior3.normalize()
posterior3.plot(label='posterior', color='C4')
decorate_bowls('Posterior after 2 vanilla, 1 chocolate')
posterior3.idxmax()
posterior3.max_prob()
hypos = [6, 8, 12]
prior = Pmf(1/3, hypos)
prior
prior.qs
prior.ps
likelihood1 = 1/6, 1/8, 1/12
posterior = prior * likelihood1
posterior.normalize()
posterior
likelihood2 = 0, 1/8, 1/12
posterior *= likelihood2
posterior.normalize()
posterior
def update_dice(pmf, data):
"""Update pmf based on new data."""
hypos = pmf.qs
likelihood = 1 / hypos
impossible = (data > hypos)
likelihood[impossible] = 0
pmf *= likelihood
pmf.normalize()
pmf = prior.copy()
update_dice(pmf, 1)
update_dice(pmf, 7)
pmf
# Solution goes here
# Solution goes here
# Solution goes here
# Solution goes here
# Solution goes here
# Solution goes here
# Solution goes here
| 0.517815 | 0.980262 |
# chp02-Introduction(2013)
>因为我在做第三章的笔记时,才发现作者已经在2017推出了第二版,但难得写了这么多,就留下来了,所以这里的chp02-Introduction(2013)指的是2013年第一版的第二章,这一章的内容主要是直接使用pandas对一些数据集进行数据分析,可以说是实战篇。不过在2017年的第二版中,这一章的内容被移到了第十四章,内容为14.1,14.2,14.3。第十四章中把不同的数据集分成了不同的小节,阅读起来会更方便一些,而且有很多地方做了优化,推荐大家直接看第十四章的内容。
# 读取来自bit.ly的1.use.gov数据
这个数据集是bit.ly和美国政府网站use.gov合作,提供了一份从生成.gov或.mil短链接的用户那里手机来的匿名数据。
下面文中的各行格式为JSON,这里读取第一行看一看是什么东西
```
path = '../datasets/bitly_usagov/example.txt'
open(path).readline()
```
python有很多内置的模块能把JSON字符串转换成Python字典对象
```
import json
path = '../datasets/bitly_usagov/example.txt'
records = [json.loads(line) for line in open(path)]
```
上面这种方法叫做列表推导式, list comprehension, 在一组字符串上执行一条相同操作(比如这里的json.loads)
```
records[0]
records[0]['tz']
```
## 用纯python代码对时区进行计数
我们想知道数据集中出现在哪个时区(即tz字段)
```
time_zones = [rec['tz'] for rec in records]
```
看来并不是所有的记录都有时区字段。那么只需要在推导式的末尾加一个if 'tz' in rec判断即可
```
time_zones = [rec['tz'] for rec in records if 'tz' in rec]
time_zones[:10]
```
为了对时区进行计数,我们用两种方法:一个用python,比较麻烦。另一个用pandas,比较简单。
这里我们遍历时区的过程中将计数值保存在字典中:
```
def get_counts(sequence):
counts = {}
for x in sequence:
if x in counts:
counts[x] += 1
else:
counts[x] = 1
return counts
```
使用python标准库的话,能把代码写得更简洁一些:
```
from collections import defaultdict
def get_counts2(sequence):
counts = defaultdict(int) # 所有的值均会被初始化为0
for x in sequence:
counts[x] += 1
return counts
```
(译者:下面关于defaultdict的用法是我从Stack Overflow上找到的,英文比较多,简单的说就是通常如果一个字典里不存在一个key,调用的时候会报错,但是如果我们设置了了default,就不会被报错,而是会新建一个key,对应的value就是我们设置的int,这里int代表0)
**defaultdict** means that if a key is not found in the dictionary, then instead of a KeyError being thrown, a new entry is created. The type of this new entry is given by the argument of defaultdict.
```
somedict = {}
print(somedict[3]) # KeyError
someddict = defaultdict(int)
print(someddict[3]) # print int(), thus 0
```
Usually, a Python dictionary throws a KeyError if you try to get an item with a key that is not currently in the dictionary. The defaultdict in contrast will simply create any items that you try to access (provided of course they do not exist yet). To create such a "default" item, it calls the function object that you pass in the constructor (more precisely, it's an arbitrary "callable" object, which includes function and type objects). For the first example, default items are created using `int()`, which will return the integer object 0. For the second example, default items are created using `list()`, which returns a new empty list object.
```
someddict = defaultdict(int)
print(someddict[3])
someddict[3]
```
上面用函数的方式写出来是为了有更高的可用性。要对它进行时区处理,只需要将time_zones传入即可:
```
counts = get_counts(time_zones)
counts['America/New_York']
len(time_zones)
```
如何想要得到前10位的时区及其计数值,我们需要一些有关字典的处理技巧:
```
def top_counts(count_dict, n=10):
value_key_pairs = [(count, tz) for tz, count in count_dict.items()]
value_key_pairs.sort()
return value_key_pairs[-n:]
top_counts(counts)
```
如果用python标准库里的collections.Counter类,能让这个任务变得更简单
```
from collections import Counter
counts = Counter(time_zones)
counts.most_common(10)
```
## 用pandas对时区进行计数
DataFrame是pandas中最重要的数据结构,它用于将数据表示变为一个表格。从一组原始记录中创建DataFrame是很简单的:
```
from pandas import DataFrame, Series
import pandas as pd; import numpy as np
frame = DataFrame(records)
frame.head()
frame['tz'][:10]
```
这里frame的输出形式是summary view, 主要用于较大的dataframe对象。frame['tz']所返回的Series对象有一个value_counts方法,该方法可以让我们得到所需要的信息。
```
tz_counts = frame['tz'].value_counts()
tz_counts[:10]
```
我们想利用matplotlib为这段数据生成一张图片。所以,我们先给记录中未知或缺失的时区填上一个替代值。fillna函数可以替代缺失值(NA),而未知值(空字符串)则可以通过布尔型数组索引,加以替换:
```
clean_tz = frame['tz'].fillna('Missing')
clean_tz[clean_tz == ''] = 'Unknown'
tz_counts = clean_tz.value_counts()
tz_counts[:10]
```
利用counts对象的plot方法即可得到一张水平条形图:
```
%matplotlib inline
tz_counts[:10].plot(kind='barh', rot=0)
```
我们还可以对这种数据进行更多的处理。比如a字段含有执行URL操作的浏览器、设备、应用程序的相关信息:
```
frame['a'][1]
frame['a'][50]
frame['a'][51]
frame['a'][:5]
```
将这些USER_AGENT字符串中的所有信息都解析出来是一件挺郁闷的工作。不过只要掌握了Python内置的字符串函数和正则表达式,就方便了。比如,我们可以把字符串的第一节(与浏览器大致对应)分离出来得到另一份用户行为摘要:
```
results = Series([x.split()[0] for x in frame.a.dropna()])
results[:5]
results.value_counts()[:8]
```
现在,假设我们想按Windows和非Windows用户对时区统计信息进行分解。为了简单期间,我们假定只要agent字符串中含有“windows”就认为该用户是windows用户。由于有的agent缺失,所以先将他们从数据中移除:
```
cframe = frame[frame.a.notnull()]
```
其次根据a值计算出各行是否是windows:
```
operating_system = np.where(cframe['a'].str.contains('Windows'),
'Windows', 'Not Windows')
operating_system[:5]
```
接下来就可以根据时区和新得到的操作系统列表对数据进行分组了:
```
by_tz_os = cframe.groupby(['tz', operating_system])
by_tz_os.size()
```
然后通过size对分组结果进行计数(类似于上面的value_counts函数),并利用unstack对计数结果进行重塑:
```
agg_counts = by_tz_os.size().unstack().fillna(0)
agg_counts[:10]
```
最后,我们来选取最常出现的时区。为了达到这个目的,根据agg_counts中的行数构造了一个简洁索引数组:
```
indexer = agg_counts.sum(1)
indexer[:10]
indexer = agg_counts.sum(1).argsort()
indexer[:10]
```
然后我通过take按照这个顺序截取了最后10行:
```
count_subset = agg_counts.take(indexer)[-10:]
count_subset
```
这里也可以生成一张条形图。我们使用stacked=True来生成一张堆积条形图:
```
count_subset.plot(kind='barh', stacked=True)
```
由于这张图中不太容易看清楚较小分组中windows用户的相对比例,因此我们可以将各行规范化为“总计为1”并重新绘图:
```
normed_subset = count_subset.div(count_subset.sum(1), axis=0)
normed_subset.plot(kind='barh', stacked=True)
```
# MovieLens 1M数据集
这个数据集是电影评分数据:包括电影评分,电影元数据(风格类型,年代)以及关于用户的人口统计学数据(年龄,邮编,性别,职业等)。
MovieLens 1M数据集含有来自6000名用户对4000部电影的100万条评分数据。飞卫三个表:评分,用户信息,电影信息。这些数据都是dat文件格式,可以通过pandas.read_table将各个表分别读到一个pandas DataFrame对象中:
```
import pandas as pd
unames = ['user_id', 'gender', 'age', 'occupation', 'zip']
users = pd.read_table('../datasets/movielens/users.dat', sep='::', header=None, names=unames)
```
因为sep='::'有点像是正则表达式,于是有了上面的错误。在这个[帖子](https://stackoverflow.com/questions/27301477/python-file-path-failing-in-pycharm-regex-confusion)找到了解决方法。
Looks like on Python 2.7 Pandas just doesn't handle separators that look regexish. The initial "error" can be worked around by adding engine='python' as a named parameter in the call, as suggested in the warning.
```
users = pd.read_table('../datasets/movielens/users.dat', sep='::', header=None, names=unames, engine='python')
rnames = ['user_id', 'movie_id', 'rating', 'timestamp']
ratings = pd.read_table('../datasets/movielens/ratings.dat', sep='::', header=None, names=rnames, engine='python')
mnames = ['movie_id', 'title', 'genres']
movies = pd.read_table('../datasets/movielens/movies.dat', sep='::', header=None, names=mnames, engine='python')
```
加载前几行验证一下数据加载工作是否顺利
```
users[:5]
ratings[:5]
movies[:5]
```
注意,年龄和职业是以编码形式给出的,它们的具体含义请参考改数据集的REAMDE文件。分析散布在三个表中的数据不是一件轻松的事情。假设我们想要根据性别和年龄来计算某部电影的平均得分,如果将所有的数据都合并到一个表中的话,问题就简单多了。我们先用pandas的merge函数将ratings和users合并到一起,然后再将movies也合并进去。pandas会根据列名的重叠情况推断出哪些列是合并(或连接)键:
```
data = pd.merge(pd.merge(ratings, users), movies)
data.head()
data.ix[0]
```
现在,只要稍微熟悉一下pandas,就能轻松地根据任意个用户或电影属性对评分数据进行聚合操作了。为了按性别计算每部电影的平均得分,我们可以使用pivot_table方法:
```
mean_ratings = data.pivot_table('rating', index='title',
columns='gender', aggfunc='mean')
mean_ratings[:5]
```
该操作产生了另一个DataFrame,其内容为电影平均得分,行标为电影名称,列表为性别。现在,我们打算过滤掉评分数据不够250条的电影(这个数字可以自己设定)。为了达到这个目的,我们先对title进行分组,然后利用size()得到一个含有各电影分组大小的Series对象:
```
ratings_by_title = data.groupby('title').size()
ratings_by_title[:10]
active_titles = ratings_by_title.index[ratings_by_title >= 250]
active_titles
mean_ratings = mean_ratings.ix[active_titles]
mean_ratings.head()
top_female_ratings = mean_ratings.sort_values(by='F', ascending=False)
top_female_ratings[:10]
```
## 计算评分分歧
假设我们想要找出男性和女性观众分歧最大的电影。一个办法是给mean_ratings加上一个用于存放平均得分之差的列,并对其进行排序:
```
mean_ratings['diff'] = mean_ratings['M'] - mean_ratings['F']
```
按‘diff’排序即可得到分歧最大且女性观众更喜欢的电影:
```
sorted_by_diff = mean_ratings.sort_values(by='diff')
sorted_by_diff[:15]
```
对行反序,并取出前15行,得到的则是男性更喜欢的电影
```
sorted_by_diff[::-1][:15]
```
如果只是想要找出分歧最大的电影(不考虑性别因素),则可以计算得分数据的方差或标准差:
```
# 根据电影名称分组的得分数据的标准差
rating_std_by_title = data.groupby('title')['rating'].std()
# 根据active_titles进行过滤
rating_std_by_title = rating_std_by_title.ix[active_titles]
rating_std_by_title.sort_values(ascending=False)[:10]
movies[:5]
```
这里我们注意到,电影分类是以竖线`|`分割的字符串形式给出的。如果想对电影分类进行分析的话,就需要先将其转换成更有用的形式才行。我将在本书后续章节中讲到这种转换处理,到时候还会再用到这个数据。
# 1880-2010年间全美婴儿姓名
这个数据时从1880年到2010年婴儿名字频率数据。这个数据集可以用来做很多事,例如:
- 计算指定名字的年度比例
- 计算某个名字的相对排名
- 计算各年度最流行的名字,以及增长或减少最快的名字
- 分析名字趋势:元音、辅音、长度、总体多样性、拼写变化、首尾字母等
- 分析外源性趋势:圣经中的名字、名人、人口结构变化等
在babynames文件夹下可以看到这些文件是按年度来编号的
```
!head -n 10 ../datasets/babynames/yob1880.txt
```
由于这是一个非常标准的以逗号隔开的格式(即CSV文件),所以可以用pandas.read_csv将其加载到DataFrame中:
```
import pandas as pd
names1880 = pd.read_csv('../datasets/babynames/yob1880.txt', names=['names', 'sex', 'births'])
names1880.head()
```
这些文件中仅含有当年出现超过5次的名字。为了简单期间,我们可以用births列的sex分组小计,表示该年度的births总计:
```
names1880.groupby('sex').births.sum()
```
由于该数据集按年度被分割成了多个文件,所以第一件事情就是要将所有数据都组装到一个DataFrame里面,并加上一个year字段。使用pandas.concat可以做到:
```
# 2010是最后一个有效统计年度
years = range(1880, 2011)
pieces = []
columns = ['name', 'sex', 'births']
for year in years:
path = '../datasets/babynames/yob%d.txt' % year
frame = pd.read_csv(path, names=columns)
frame['year'] = year
pieces.append(frame)
# 将所有数据整合到单个DataFrame中
names = pd.concat(pieces, ignore_index=True)
```
这里要注意几件事。
- 第一,concat默认是按行将多个DataFrame组合到一起的;
- 第二,必须指定ignore_index=True,因为我们不希望保留read_csv所返回的原始行号。
现在我们得到了一个非常大的DataFrame,它含有全部的名字数据。现在names这个DataFrame看上去是:
```
names[:5]
```
有了这些数据后,我们就可以利用groupby或pivot_table在year和sex界别上对其进行聚合了:
```
total_births = names.pivot_table('births', index='year',
columns='sex', aggfunc=sum)
total_births.tail()
total_births.plot(title='Total births by sex and year')
```
下面我们来插入一个prop列,用于存放指定名字的婴儿数相对于总出生数的比列。prop值为0.02表示每100名婴儿中有2名去了当前这个名字。因此,我们先按year和sex分组,然后再将新列加到各个分组上:
```
def add_prop(group):
# 整数出发回向下圆整
# births = group.births.astype(float)
births = group.births
group['prop'] = births / births.sum()
return group
names = names.groupby(['year', 'sex']).apply(add_prop)
```
注意:由于births是整数,所以我们在计算分式时必须将分子或分母转换成浮点数(除非使用的是python3)。所以上面我们可以不加`births = group.births.astype(float)`这一句代码。
```
names[:5]
```
在执行这样的分组处理时,一般都应该做一些有效性检查,比如验证所有分组的prop的综合是否为1。由于这是一个浮点型数据,所以我们应该用np.allclose来检查这个分组总计值是否够近似于(可能不会精确等于)1:
```
np.allclose(names.groupby(['year', 'sex']).prop.sum(), 1)
```
这样就算完活了。为了便于实现进一步的分析,我们需要取出该数据的一个子集:每对sex/year组合的前1000个名字。这又是一个分组操作:
```
def get_top1000(group):
return group.sort_values(by='births', ascending=False)[:1000]
grouped = names.groupby(['year', 'sex'])
top1000 = grouped.apply(get_top1000)
```
如果喜欢DIY的话,也可以这样:
```
pieces =[]
for year, group in names.groupby(['year', 'sex']):
pieces.append(group.sort_values(by='births', ascending=False)[:1000])
top1000 = pd.concat(pieces, ignore_index=True)
top1000[:10]
```
接下来的数据分析工作就针对这个top1000数据集了
## 分析命名趋势
有了完整的数据集和刚才生成的top1000数据集,我们就可以开始分析各种命名趋势了。首先将前1000个名字分为男女两个部分:
```
boys = top1000[top1000.sex=='M']
girls = top1000[top1000.sex=='F']
```
这是两个简单的时间序列,只需要稍作整理即可绘制出相应的图标(比如每年叫做John和Mary的婴儿数)。我们先生成一张按year和name统计的总出生数透视表:
```
total_births = top1000.pivot_table('births', index='year',
columns='name', aggfunc=sum)
total_births[:5]
subset = total_births[['John', 'Harry', 'Mary', 'Marilyn']]
subset.plot(subplots=True, figsize=(12, 10), grid=False,
title="Number of births per year")
```
## 评价命名多样性的增长
上图反应的降低情况可能意味着父母愿意给小孩起常见的名字越来越少。这个假设可以从数据中得到验证。一个办法是计算最流行的1000个名字所占的比例,我按year和sex进行聚合并绘图:
```
table = top1000.pivot_table('prop', index='year',
columns='sex', aggfunc=sum)
table.plot(title='Sum of table1000.prop by year and sex',
yticks=np.linspace(0, 1.2, 13), xticks=range(1880, 2020, 10))
```
从图中可以看出,名字的多样性确实出现了增长(前1000项的比例降低)。另一个办法是计算占总出生人数前50%的不同名字的数量,这个数字不太好计算。我们只考虑2010年男孩的名字:
```
df = boys[boys.year == 2010]
df[:5]
```
对prop降序排列后,我们想知道前面多少个名字的人数加起来才够50%。虽然编写一个for循环也能达到目的,但NumPy有一种更聪明的矢量方式。先计算prop的累计和cumsum,,然后再通过searchsorted方法找出0.5应该被插入在哪个位置才能保证不破坏顺序:
```
prop_cumsum = df.sort_values(by='prop', ascending=False).prop.cumsum()
prop_cumsum[:10]
prop_cumsum.searchsorted(0.5)
```
由于数组索引是从0开始的,因此我们要给这个结果加1,即最终结果为117.拿1900年的数据来做个比较,这个数字要小得多:
```
df = boys[boys.year == 1900]
in1900 = df.sort_values(by='prop', ascending=False).prop.cumsum()
in1900[-10:]
in1900.searchsorted(0.5) + 1
```
现在就可以对所有year/sex组合执行这个计算了。按这两个字段进行groupby处理,然后用一个函数计算各分组的这个值:
```
def get_quantile_count(group, q=0.5):
group = group.sort_values(by='prop', ascending=False)
return group.prop.cumsum().searchsorted(q) + 1
diversity = top1000.groupby(['year', 'sex']).apply(get_quantile_count)
diversity = diversity.unstack('sex')
```
现在,这个diversity有两个时间序列(每个性别各一个,按年度索引)。通过IPython,可以看到其内容,还可以绘制图标
```
diversity.head()
diversity=diversity.astype(float)
diversity.plot(title='Number of popular names in top 50%')
diversity[:5]
```
> 注意,如果不加`diversity=diversity.astype(float)`的话,会报错显示,“no numeric data to plot” error。通过加上这句来更改数据类型,就能正常绘图了。
从图中可以看出,女孩名字的多样性总是比男孩高,而且还变得越来越高。我们可以自己分析一下具体是什么在驱动这个多样性(比如拼写形式的变化)。
## “最后一个字母”的变革
一位研究人员指出:近百年来,男孩名字在最后一个字母上的分布发生了显著的变化。为了了解具体的情况,我们首先将全部出生数据在年度、性别以及末字母上进行了聚合:
```
# 从name列中取出最后一个字母
get_last_letter = lambda x: x[-1]
last_letters = names.name.map(get_last_letter)
last_letters.name = 'last_letter'
table = names.pivot_table('births', index=last_letters,
columns=['sex', 'year'], aggfunc=sum)
print(type(last_letters))
print(last_letters[:5])
```
然后,我选出具有一个代表性的三年,并输出前几行:
```
subtable = table.reindex(columns=[1910, 1960, 2010], level='year')
subtable.head()
```
接下来我们需要安总出生数对该表进行规范化处理,一遍计算出个性别各末字母站总出生人数的比例:
```
subtable.sum()
letter_prop = subtable / subtable.sum()
letter_prop
```
有了这个字母比例数据后,就可以生成一张各年度各性别的条形图了:
```
import matplotlib.pyplot as plt
fig, axes = plt.subplots(2, 1, figsize=(10, 8))
letter_prop['M'].plot(kind='bar', rot=0, ax=axes[0], title='Male')
letter_prop['F'].plot(kind='bar', rot=0, ax=axes[1], title='Femal', legend=False)
```
从上图可以看出来,从20世纪60年代开始,以字母'n'结尾的男孩名字出现了显著的增长。回到之前创建的那个完整表,按年度和性别对其进行规范化处理,并在男孩名字中选取几个字母,最后进行转置以便将各个列做成一个时间序列:
```
# table = top1000.pivot_table('prop', index='year',
# columns='sex', aggfunc=sum)
letter_prop = table / table.sum()
letter_prop.head()
dny_ts = letter_prop.ix[['d', 'n', 'y'], 'M'].T
dny_ts.head()
```
有了这个时间序列的DataFrame后,就可以通过其plot方法绘制出一张趋势图:
```
dny_ts.plot()
```
## 变成女孩名字的男孩名字(以及相反的情况)
另一个有趣的趋势是,早年流行于男孩的名字近年来“变性了”,列入Lesley或Leslie。回到top1000数据集,找出其中以"lesl"开头的一组名字:
```
all_names = top1000.name.unique()
mask = np.array(['lesl' in x.lower() for x in all_names])
lesley_like = all_names[mask]
lesley_like
```
然后利用这个结果过滤其他的名字,并按名字分组计算出生数以查看相对频率:
```
filtered = top1000[top1000.name.isin(lesley_like)]
filtered.groupby('name').births.sum()
```
接下来,我们按性别和年度进行聚合,并按年度进行规范化处理:
```
table = filtered.pivot_table('births', index='year',
columns='sex', aggfunc='sum')
table[:5]
table = table.div(table.sum(1), axis=0)
table[:5]
table.tail()
```
现在,我们可以轻松绘制一张分性别的年度曲线图了:
```
table.plot(style={'M': 'k-', 'F': 'k--'})
```
|
github_jupyter
|
path = '../datasets/bitly_usagov/example.txt'
open(path).readline()
import json
path = '../datasets/bitly_usagov/example.txt'
records = [json.loads(line) for line in open(path)]
records[0]
records[0]['tz']
time_zones = [rec['tz'] for rec in records]
time_zones = [rec['tz'] for rec in records if 'tz' in rec]
time_zones[:10]
def get_counts(sequence):
counts = {}
for x in sequence:
if x in counts:
counts[x] += 1
else:
counts[x] = 1
return counts
from collections import defaultdict
def get_counts2(sequence):
counts = defaultdict(int) # 所有的值均会被初始化为0
for x in sequence:
counts[x] += 1
return counts
somedict = {}
print(somedict[3]) # KeyError
someddict = defaultdict(int)
print(someddict[3]) # print int(), thus 0
someddict = defaultdict(int)
print(someddict[3])
someddict[3]
counts = get_counts(time_zones)
counts['America/New_York']
len(time_zones)
def top_counts(count_dict, n=10):
value_key_pairs = [(count, tz) for tz, count in count_dict.items()]
value_key_pairs.sort()
return value_key_pairs[-n:]
top_counts(counts)
from collections import Counter
counts = Counter(time_zones)
counts.most_common(10)
from pandas import DataFrame, Series
import pandas as pd; import numpy as np
frame = DataFrame(records)
frame.head()
frame['tz'][:10]
tz_counts = frame['tz'].value_counts()
tz_counts[:10]
clean_tz = frame['tz'].fillna('Missing')
clean_tz[clean_tz == ''] = 'Unknown'
tz_counts = clean_tz.value_counts()
tz_counts[:10]
%matplotlib inline
tz_counts[:10].plot(kind='barh', rot=0)
frame['a'][1]
frame['a'][50]
frame['a'][51]
frame['a'][:5]
results = Series([x.split()[0] for x in frame.a.dropna()])
results[:5]
results.value_counts()[:8]
cframe = frame[frame.a.notnull()]
operating_system = np.where(cframe['a'].str.contains('Windows'),
'Windows', 'Not Windows')
operating_system[:5]
by_tz_os = cframe.groupby(['tz', operating_system])
by_tz_os.size()
agg_counts = by_tz_os.size().unstack().fillna(0)
agg_counts[:10]
indexer = agg_counts.sum(1)
indexer[:10]
indexer = agg_counts.sum(1).argsort()
indexer[:10]
count_subset = agg_counts.take(indexer)[-10:]
count_subset
count_subset.plot(kind='barh', stacked=True)
normed_subset = count_subset.div(count_subset.sum(1), axis=0)
normed_subset.plot(kind='barh', stacked=True)
import pandas as pd
unames = ['user_id', 'gender', 'age', 'occupation', 'zip']
users = pd.read_table('../datasets/movielens/users.dat', sep='::', header=None, names=unames)
users = pd.read_table('../datasets/movielens/users.dat', sep='::', header=None, names=unames, engine='python')
rnames = ['user_id', 'movie_id', 'rating', 'timestamp']
ratings = pd.read_table('../datasets/movielens/ratings.dat', sep='::', header=None, names=rnames, engine='python')
mnames = ['movie_id', 'title', 'genres']
movies = pd.read_table('../datasets/movielens/movies.dat', sep='::', header=None, names=mnames, engine='python')
users[:5]
ratings[:5]
movies[:5]
data = pd.merge(pd.merge(ratings, users), movies)
data.head()
data.ix[0]
mean_ratings = data.pivot_table('rating', index='title',
columns='gender', aggfunc='mean')
mean_ratings[:5]
ratings_by_title = data.groupby('title').size()
ratings_by_title[:10]
active_titles = ratings_by_title.index[ratings_by_title >= 250]
active_titles
mean_ratings = mean_ratings.ix[active_titles]
mean_ratings.head()
top_female_ratings = mean_ratings.sort_values(by='F', ascending=False)
top_female_ratings[:10]
mean_ratings['diff'] = mean_ratings['M'] - mean_ratings['F']
sorted_by_diff = mean_ratings.sort_values(by='diff')
sorted_by_diff[:15]
sorted_by_diff[::-1][:15]
# 根据电影名称分组的得分数据的标准差
rating_std_by_title = data.groupby('title')['rating'].std()
# 根据active_titles进行过滤
rating_std_by_title = rating_std_by_title.ix[active_titles]
rating_std_by_title.sort_values(ascending=False)[:10]
movies[:5]
!head -n 10 ../datasets/babynames/yob1880.txt
import pandas as pd
names1880 = pd.read_csv('../datasets/babynames/yob1880.txt', names=['names', 'sex', 'births'])
names1880.head()
names1880.groupby('sex').births.sum()
# 2010是最后一个有效统计年度
years = range(1880, 2011)
pieces = []
columns = ['name', 'sex', 'births']
for year in years:
path = '../datasets/babynames/yob%d.txt' % year
frame = pd.read_csv(path, names=columns)
frame['year'] = year
pieces.append(frame)
# 将所有数据整合到单个DataFrame中
names = pd.concat(pieces, ignore_index=True)
names[:5]
total_births = names.pivot_table('births', index='year',
columns='sex', aggfunc=sum)
total_births.tail()
total_births.plot(title='Total births by sex and year')
def add_prop(group):
# 整数出发回向下圆整
# births = group.births.astype(float)
births = group.births
group['prop'] = births / births.sum()
return group
names = names.groupby(['year', 'sex']).apply(add_prop)
names[:5]
np.allclose(names.groupby(['year', 'sex']).prop.sum(), 1)
def get_top1000(group):
return group.sort_values(by='births', ascending=False)[:1000]
grouped = names.groupby(['year', 'sex'])
top1000 = grouped.apply(get_top1000)
pieces =[]
for year, group in names.groupby(['year', 'sex']):
pieces.append(group.sort_values(by='births', ascending=False)[:1000])
top1000 = pd.concat(pieces, ignore_index=True)
top1000[:10]
boys = top1000[top1000.sex=='M']
girls = top1000[top1000.sex=='F']
total_births = top1000.pivot_table('births', index='year',
columns='name', aggfunc=sum)
total_births[:5]
subset = total_births[['John', 'Harry', 'Mary', 'Marilyn']]
subset.plot(subplots=True, figsize=(12, 10), grid=False,
title="Number of births per year")
table = top1000.pivot_table('prop', index='year',
columns='sex', aggfunc=sum)
table.plot(title='Sum of table1000.prop by year and sex',
yticks=np.linspace(0, 1.2, 13), xticks=range(1880, 2020, 10))
df = boys[boys.year == 2010]
df[:5]
prop_cumsum = df.sort_values(by='prop', ascending=False).prop.cumsum()
prop_cumsum[:10]
prop_cumsum.searchsorted(0.5)
df = boys[boys.year == 1900]
in1900 = df.sort_values(by='prop', ascending=False).prop.cumsum()
in1900[-10:]
in1900.searchsorted(0.5) + 1
def get_quantile_count(group, q=0.5):
group = group.sort_values(by='prop', ascending=False)
return group.prop.cumsum().searchsorted(q) + 1
diversity = top1000.groupby(['year', 'sex']).apply(get_quantile_count)
diversity = diversity.unstack('sex')
diversity.head()
diversity=diversity.astype(float)
diversity.plot(title='Number of popular names in top 50%')
diversity[:5]
# 从name列中取出最后一个字母
get_last_letter = lambda x: x[-1]
last_letters = names.name.map(get_last_letter)
last_letters.name = 'last_letter'
table = names.pivot_table('births', index=last_letters,
columns=['sex', 'year'], aggfunc=sum)
print(type(last_letters))
print(last_letters[:5])
subtable = table.reindex(columns=[1910, 1960, 2010], level='year')
subtable.head()
subtable.sum()
letter_prop = subtable / subtable.sum()
letter_prop
import matplotlib.pyplot as plt
fig, axes = plt.subplots(2, 1, figsize=(10, 8))
letter_prop['M'].plot(kind='bar', rot=0, ax=axes[0], title='Male')
letter_prop['F'].plot(kind='bar', rot=0, ax=axes[1], title='Femal', legend=False)
# table = top1000.pivot_table('prop', index='year',
# columns='sex', aggfunc=sum)
letter_prop = table / table.sum()
letter_prop.head()
dny_ts = letter_prop.ix[['d', 'n', 'y'], 'M'].T
dny_ts.head()
dny_ts.plot()
all_names = top1000.name.unique()
mask = np.array(['lesl' in x.lower() for x in all_names])
lesley_like = all_names[mask]
lesley_like
filtered = top1000[top1000.name.isin(lesley_like)]
filtered.groupby('name').births.sum()
table = filtered.pivot_table('births', index='year',
columns='sex', aggfunc='sum')
table[:5]
table = table.div(table.sum(1), axis=0)
table[:5]
table.tail()
table.plot(style={'M': 'k-', 'F': 'k--'})
| 0.260389 | 0.944022 |
# Monitor a Model
When you've deployed a model into production as a service, you'll want to monitor it to track usage and explore the requests it processes. You can use Azure Application Insights to monitor activity for a model service endpoint.
## Connect to your workspace
To get started, connect to your workspace.
**Note:** If you haven't already established an authenticated session with your Azure subscription, you'll be prompted to authenticate by clicking a link, entering an authentication code, and signing into Azure.
```
from azureml.core import Workspace
# Load the workspace from the saved config file
ws = Workspace.from_config()
print('Ready to work with', ws.name)
```
## Prepare a model for deployment
Now we need a model to deploy. Run the code below to:
1. Create and register a dataset.
2. Train a model using the dataset.
3. Register the model.
```
from azureml.core import Experiment
from azureml.core import Model
import pandas as pd
import numpy as np
import joblib
from sklearn.model_selection import train_test_split
from sklearn.tree import DecisionTreeClassifier
from sklearn.metrics import roc_auc_score, roc_curve
from azureml.core import Dataset
# Upload data files to the default datastore
default_ds = ws.get_default_datastore()
default_ds.upload_files(files=['./data/diabetes.csv', './data/diabetes2.csv'],
target_path='diabetes-data/',
overwrite=True,
show_progress=True)
#Create a tabular dataset from the path on the datastore
print('Creating dataset...')
data_set = Dataset.Tabular.from_delimited_files(path=(default_ds, 'diabetes-data/*.csv'))
# Register the tabular dataset
print('Registering dataset...')
try:
data_set = data_set.register(workspace=ws,
name='diabetes dataset',
description='diabetes data',
tags = {'format':'CSV'},
create_new_version=True)
except Exception as ex:
print(ex)
# Create an Azure ML experiment in your workspace
experiment = Experiment(workspace=ws, name='mslearn-train-diabetes')
run = experiment.start_logging()
print("Starting experiment:", experiment.name)
# load the diabetes dataset
print("Loading Data...")
diabetes = data_set.to_pandas_dataframe()
# Separate features and labels
X, y = diabetes[['Pregnancies','PlasmaGlucose','DiastolicBloodPressure','TricepsThickness','SerumInsulin','BMI','DiabetesPedigree','Age']].values, diabetes['Diabetic'].values
# Split data into training set and test set
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.30, random_state=0)
# Train a decision tree model
print('Training a decision tree model')
model = DecisionTreeClassifier().fit(X_train, y_train)
# calculate accuracy
y_hat = model.predict(X_test)
acc = np.average(y_hat == y_test)
print('Accuracy:', acc)
run.log('Accuracy', np.float(acc))
# calculate AUC
y_scores = model.predict_proba(X_test)
auc = roc_auc_score(y_test,y_scores[:,1])
print('AUC: ' + str(auc))
run.log('AUC', np.float(auc))
# Save the trained model
model_file = 'diabetes_model.pkl'
joblib.dump(value=model, filename=model_file)
run.upload_file(name = 'outputs/' + model_file, path_or_stream = './' + model_file)
# Complete the run
run.complete()
# Register the model
print('Registering model...')
run.register_model(model_path='outputs/diabetes_model.pkl', model_name='diabetes_model',
tags={'Training context':'Inline Training'},
properties={'AUC': run.get_metrics()['AUC'], 'Accuracy': run.get_metrics()['Accuracy']})
# Get the registered model
model = ws.models['diabetes_model']
print('Model trained and registered.')
```
### Deploy a model as a web service
Now you're ready to deploy the registered model as a web service.
First, create a folder for the deployment configuration files
```
import os
folder_name = 'diabetes_service'
# Create a folder for the web service files
experiment_folder = './' + folder_name
os.makedirs(experiment_folder, exist_ok=True)
print(folder_name, 'folder created.')
# Set path for scoring script
script_file = os.path.join(experiment_folder,"score_diabetes.py")
```
Now you need an entry script that the service will use to score new data.
```
%%writefile $script_file
import json
import joblib
import numpy as np
from azureml.core.model import Model
# Called when the service is loaded
def init():
global model
# Get the path to the deployed model file and load it
model_path = Model.get_model_path('diabetes_model')
model = joblib.load(model_path)
# Called when a request is received
def run(raw_data):
# Get the input data as a numpy array
data = json.loads(raw_data)['data']
np_data = np.array(data)
# Get a prediction from the model
predictions = model.predict(np_data)
# print the data and predictions (so they'll be logged!)
log_text = 'Data:' + str(data) + ' - Predictions:' + str(predictions)
print(log_text)
# Get the corresponding classname for each prediction (0 or 1)
classnames = ['not-diabetic', 'diabetic']
predicted_classes = []
for prediction in predictions:
predicted_classes.append(classnames[prediction])
# Return the predictions as JSON
return json.dumps(predicted_classes)
```
You'll also need a Conda configuration file for the service environment.
```
from azureml.core.conda_dependencies import CondaDependencies
# Add the dependencies for our model (AzureML defaults is already included)
myenv = CondaDependencies()
myenv.add_conda_package("scikit-learn")
# Save the environment config as a .yml file
env_file = folder_name + "/diabetes_env.yml"
with open(env_file,"w") as f:
f.write(myenv.serialize_to_string())
print("Saved dependency info in", env_file)
# Print the .yml file
with open(env_file,"r") as f:
print(f.read())
```
Now you can deploy the service (in this case, as an Azure Container Instance (ACI).
**Note**: This can take a few minutes - wait until the state is shown as **Healthy**.
```
from azureml.core.webservice import AciWebservice, Webservice
from azureml.core.model import Model
from azureml.core.model import InferenceConfig
# Configure the scoring environment
inference_config = InferenceConfig(runtime= "python",
entry_script=script_file,
conda_file=env_file)
service_name = "diabetes-service-app-insights"
deployment_config = AciWebservice.deploy_configuration(cpu_cores = 1, memory_gb = 1)
aci_service = Model.deploy(workspace=ws,
name= service_name,
models= [model],
inference_config= inference_config,
deployment_config=deployment_config)
aci_service.wait_for_deployment(show_output = True)
print(aci_service.state)
```
### Enable Application Insights
Next, you need to enable Application Insights for the service.
```
# Enable AppInsights
aci_service.update(enable_app_insights=True)
print('AppInsights enabled!')
```
### Use the web service
With the service deployed, now you can consume it from a client application.
First, determine the URL to which these applications must submit their requests.
```
endpoint = aci_service.scoring_uri
print(endpoint)
```
Now that you know the endpoint URI, an application can simply make an HTTP request, sending the patient data in JSON (or binary) format, and receive back the predicted class(es).
Tip: If an error occurs because the service endpoint isn't ready. Wait a few seconds and try again!
```
import requests
import json
# Create new data for inferencing
x_new = [[2,180,74,24,21,23.9091702,1.488172308,22],
[0,148,58,11,179,39.19207553,0.160829008,45]]
# Convert the array to a serializable list in a JSON document
input_json = json.dumps({"data": x_new})
# Set the content type
headers = { 'Content-Type':'application/json' }
# Get the predictions
predictions = requests.post(endpoint, input_json, headers = headers)
print(predictions.status_code)
if predictions.status_code == 200:
predicted_classes = json.loads(predictions.json())
for i in range(len(x_new)):
print ("Patient {}".format(x_new[i]), predicted_classes[i] )
```
Now you can view the data logged for the service endpoint:
1. In the [Azure portal](https://portal.azure.com/#home), open your Machine Learning workspace.
2. On the **Overview** page, click the link for the associated **Application Insights** resource.
3. In the Application Insights blade, click **Logs**.
**Note**: If this is the first time you've opened log analytics, you may need to click **Get Started** to open the query editor. If a tip explaining how to write a query is displayed, close it.
4. Paste the following query into the query editor and click **Run**
`traces`
`|where message == "STDOUT"`
`and customDimensions.["Service Name"] == "diabetes-service-app-insights"`
`|project timestamp, customDimensions.Content`
5. View the results. At first there may be none, because an ACI web service can take as long as five minutes to send the telemetry to Application Insights. Wait a few minutes and re-run the query until you see the logged data and predictions.
6. When you've reviewed the logged data, close the Application Insights query page.
### Delete the service
When you no longer need your service, you should delete it.
**Note**: If the service is in use, you may not be able to delete it immediately.
```
try:
aci_service.delete()
print('Service deleted.')
except Exception as ex:
print(ex.message)
```
For more information about using Application Insights to monitor a deployed service, see the [Azure Machine Learning documentation](https://docs.microsoft.com/azure/machine-learning/how-to-enable-app-insights).
|
github_jupyter
|
from azureml.core import Workspace
# Load the workspace from the saved config file
ws = Workspace.from_config()
print('Ready to work with', ws.name)
from azureml.core import Experiment
from azureml.core import Model
import pandas as pd
import numpy as np
import joblib
from sklearn.model_selection import train_test_split
from sklearn.tree import DecisionTreeClassifier
from sklearn.metrics import roc_auc_score, roc_curve
from azureml.core import Dataset
# Upload data files to the default datastore
default_ds = ws.get_default_datastore()
default_ds.upload_files(files=['./data/diabetes.csv', './data/diabetes2.csv'],
target_path='diabetes-data/',
overwrite=True,
show_progress=True)
#Create a tabular dataset from the path on the datastore
print('Creating dataset...')
data_set = Dataset.Tabular.from_delimited_files(path=(default_ds, 'diabetes-data/*.csv'))
# Register the tabular dataset
print('Registering dataset...')
try:
data_set = data_set.register(workspace=ws,
name='diabetes dataset',
description='diabetes data',
tags = {'format':'CSV'},
create_new_version=True)
except Exception as ex:
print(ex)
# Create an Azure ML experiment in your workspace
experiment = Experiment(workspace=ws, name='mslearn-train-diabetes')
run = experiment.start_logging()
print("Starting experiment:", experiment.name)
# load the diabetes dataset
print("Loading Data...")
diabetes = data_set.to_pandas_dataframe()
# Separate features and labels
X, y = diabetes[['Pregnancies','PlasmaGlucose','DiastolicBloodPressure','TricepsThickness','SerumInsulin','BMI','DiabetesPedigree','Age']].values, diabetes['Diabetic'].values
# Split data into training set and test set
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.30, random_state=0)
# Train a decision tree model
print('Training a decision tree model')
model = DecisionTreeClassifier().fit(X_train, y_train)
# calculate accuracy
y_hat = model.predict(X_test)
acc = np.average(y_hat == y_test)
print('Accuracy:', acc)
run.log('Accuracy', np.float(acc))
# calculate AUC
y_scores = model.predict_proba(X_test)
auc = roc_auc_score(y_test,y_scores[:,1])
print('AUC: ' + str(auc))
run.log('AUC', np.float(auc))
# Save the trained model
model_file = 'diabetes_model.pkl'
joblib.dump(value=model, filename=model_file)
run.upload_file(name = 'outputs/' + model_file, path_or_stream = './' + model_file)
# Complete the run
run.complete()
# Register the model
print('Registering model...')
run.register_model(model_path='outputs/diabetes_model.pkl', model_name='diabetes_model',
tags={'Training context':'Inline Training'},
properties={'AUC': run.get_metrics()['AUC'], 'Accuracy': run.get_metrics()['Accuracy']})
# Get the registered model
model = ws.models['diabetes_model']
print('Model trained and registered.')
import os
folder_name = 'diabetes_service'
# Create a folder for the web service files
experiment_folder = './' + folder_name
os.makedirs(experiment_folder, exist_ok=True)
print(folder_name, 'folder created.')
# Set path for scoring script
script_file = os.path.join(experiment_folder,"score_diabetes.py")
%%writefile $script_file
import json
import joblib
import numpy as np
from azureml.core.model import Model
# Called when the service is loaded
def init():
global model
# Get the path to the deployed model file and load it
model_path = Model.get_model_path('diabetes_model')
model = joblib.load(model_path)
# Called when a request is received
def run(raw_data):
# Get the input data as a numpy array
data = json.loads(raw_data)['data']
np_data = np.array(data)
# Get a prediction from the model
predictions = model.predict(np_data)
# print the data and predictions (so they'll be logged!)
log_text = 'Data:' + str(data) + ' - Predictions:' + str(predictions)
print(log_text)
# Get the corresponding classname for each prediction (0 or 1)
classnames = ['not-diabetic', 'diabetic']
predicted_classes = []
for prediction in predictions:
predicted_classes.append(classnames[prediction])
# Return the predictions as JSON
return json.dumps(predicted_classes)
from azureml.core.conda_dependencies import CondaDependencies
# Add the dependencies for our model (AzureML defaults is already included)
myenv = CondaDependencies()
myenv.add_conda_package("scikit-learn")
# Save the environment config as a .yml file
env_file = folder_name + "/diabetes_env.yml"
with open(env_file,"w") as f:
f.write(myenv.serialize_to_string())
print("Saved dependency info in", env_file)
# Print the .yml file
with open(env_file,"r") as f:
print(f.read())
from azureml.core.webservice import AciWebservice, Webservice
from azureml.core.model import Model
from azureml.core.model import InferenceConfig
# Configure the scoring environment
inference_config = InferenceConfig(runtime= "python",
entry_script=script_file,
conda_file=env_file)
service_name = "diabetes-service-app-insights"
deployment_config = AciWebservice.deploy_configuration(cpu_cores = 1, memory_gb = 1)
aci_service = Model.deploy(workspace=ws,
name= service_name,
models= [model],
inference_config= inference_config,
deployment_config=deployment_config)
aci_service.wait_for_deployment(show_output = True)
print(aci_service.state)
# Enable AppInsights
aci_service.update(enable_app_insights=True)
print('AppInsights enabled!')
endpoint = aci_service.scoring_uri
print(endpoint)
import requests
import json
# Create new data for inferencing
x_new = [[2,180,74,24,21,23.9091702,1.488172308,22],
[0,148,58,11,179,39.19207553,0.160829008,45]]
# Convert the array to a serializable list in a JSON document
input_json = json.dumps({"data": x_new})
# Set the content type
headers = { 'Content-Type':'application/json' }
# Get the predictions
predictions = requests.post(endpoint, input_json, headers = headers)
print(predictions.status_code)
if predictions.status_code == 200:
predicted_classes = json.loads(predictions.json())
for i in range(len(x_new)):
print ("Patient {}".format(x_new[i]), predicted_classes[i] )
try:
aci_service.delete()
print('Service deleted.')
except Exception as ex:
print(ex.message)
| 0.585575 | 0.907476 |
# SIT742: Modern Data Science
**(Assessment Task 02: Bank Marketing Data Analytics)**
---
- Materials in this module include resources collected from various open-source online repositories.
- You are free to use, change and distribute this package.
Prepared by **SIT742 Teaching Team**
---
**Project Group Information:**
- Names:
- Student IDs:
- Emails:
---
## 1. Import Spark
```
!apt-get install openjdk-8-jdk-headless -qq > /dev/null
!wget -q http://www-us.apache.org/dist/spark/spark-2.4.0/spark-2.4.0-bin-hadoop2.7.tgz
!tar xf spark-2.4.0-bin-hadoop2.7.tgz
!pip install -q findspark
import os
os.environ["JAVA_HOME"] = "/usr/lib/jvm/java-8-openjdk-amd64"
os.environ["SPARK_HOME"] = "/content/spark-2.4.0-bin-hadoop2.7"
import findspark
findspark.init()
from pyspark.sql import SparkSession
```
## 2. Read and check data
```
!pip install wget
import wget
link_to_data = 'https://github.com/tulip-lab/sit742/raw/master/Assessment/2019/data/bank.csv'
DataSet = wget.download(link_to_data)
!ls
spark = SparkSession.builder.appName('ml-bank').getOrCreate()
# Import the 'bank.csv' as a Spark dataframe and name it as df
df =
# check data distribution
# you may use printSchema()
```
## 3. Select features
```
# select features ('age', 'job', 'marital', 'education', 'default', 'balance', 'housing', 'loan', 'campaign', 'pdays', 'previous', 'poutcome', 'deposit') as df2
df2=
# remove invalid rows/records using spark.sql
# convert categorical features to numeric features using One hot encoding,
```
### 3.1 normalisation
```
# then apply Min-Max normalisation on each attribute using MinMaxScaler
from pyspark.ml.feature import MinMaxScaler
```
## 4. Unsupervised learning
### 4.1 K-means
```
# Perform unsupervised learning on df2 with k-means
# you can use whole df2 as both training and testing data,
# evaluate the clustering result using Accuracy.
from pyspark.ml.clustering import KMeans
from pyspark.ml.evaluation import ClusteringEvaluator
from pyspark.ml.evaluation import MulticlassClassificationEvaluator
from pyspark.ml.evaluation import BinaryClassificationEvaluator
```
### 4.2 PCA
```
# generate a scatter plot using the first two PCA components to investigate the data distribution.
from pyspark.ml.feature import PCA
from pyspark.ml.linalg import Vectors
```
## 5. Supervised learning
```
train, test = df2.randomSplit([0.7, 0.3], seed = 742)
print("Training Dataset Count: " + str(train.count()))
print("Test Dataset Count: " + str(test.count()))
```
### 5.1 LogisticRegression
```
# Logistic Regression
from pyspark.ml.classification import LogisticRegression
from pyspark.ml.evaluation import MulticlassClassificationEvaluator
from pyspark.ml.evaluation import BinaryClassificationEvaluator
# exam the coefficients
```
### 5.2 Decision tree
```
# Decision tree
from pyspark.ml.classification import DecisionTreeClassifier
```
### 5.3 NaiveBayes
```
# NaiveBayes
from pyspark.ml.classification import NaiveBayes
from pyspark.ml.evaluation import MulticlassClassificationEvaluator
```
|
github_jupyter
|
!apt-get install openjdk-8-jdk-headless -qq > /dev/null
!wget -q http://www-us.apache.org/dist/spark/spark-2.4.0/spark-2.4.0-bin-hadoop2.7.tgz
!tar xf spark-2.4.0-bin-hadoop2.7.tgz
!pip install -q findspark
import os
os.environ["JAVA_HOME"] = "/usr/lib/jvm/java-8-openjdk-amd64"
os.environ["SPARK_HOME"] = "/content/spark-2.4.0-bin-hadoop2.7"
import findspark
findspark.init()
from pyspark.sql import SparkSession
!pip install wget
import wget
link_to_data = 'https://github.com/tulip-lab/sit742/raw/master/Assessment/2019/data/bank.csv'
DataSet = wget.download(link_to_data)
!ls
spark = SparkSession.builder.appName('ml-bank').getOrCreate()
# Import the 'bank.csv' as a Spark dataframe and name it as df
df =
# check data distribution
# you may use printSchema()
# select features ('age', 'job', 'marital', 'education', 'default', 'balance', 'housing', 'loan', 'campaign', 'pdays', 'previous', 'poutcome', 'deposit') as df2
df2=
# remove invalid rows/records using spark.sql
# convert categorical features to numeric features using One hot encoding,
# then apply Min-Max normalisation on each attribute using MinMaxScaler
from pyspark.ml.feature import MinMaxScaler
# Perform unsupervised learning on df2 with k-means
# you can use whole df2 as both training and testing data,
# evaluate the clustering result using Accuracy.
from pyspark.ml.clustering import KMeans
from pyspark.ml.evaluation import ClusteringEvaluator
from pyspark.ml.evaluation import MulticlassClassificationEvaluator
from pyspark.ml.evaluation import BinaryClassificationEvaluator
# generate a scatter plot using the first two PCA components to investigate the data distribution.
from pyspark.ml.feature import PCA
from pyspark.ml.linalg import Vectors
train, test = df2.randomSplit([0.7, 0.3], seed = 742)
print("Training Dataset Count: " + str(train.count()))
print("Test Dataset Count: " + str(test.count()))
# Logistic Regression
from pyspark.ml.classification import LogisticRegression
from pyspark.ml.evaluation import MulticlassClassificationEvaluator
from pyspark.ml.evaluation import BinaryClassificationEvaluator
# exam the coefficients
# Decision tree
from pyspark.ml.classification import DecisionTreeClassifier
# NaiveBayes
from pyspark.ml.classification import NaiveBayes
from pyspark.ml.evaluation import MulticlassClassificationEvaluator
| 0.651244 | 0.826852 |
```
import pandas as pd
import numpy as np
import torch
ml1m_dir = '/data/vnkljukin/train.csv'
ml1m_rating = pd.read_csv(ml1m_dir)
ml1m_rating.rating += 1
ml1m_rating.rating //= 2
print('Range of userId is [{}, {}]'.format(ml1m_rating.userId.min(), ml1m_rating.userId.max()))
print('Range of itemId is [{}, {}]'.format(ml1m_rating.itemId.min(), ml1m_rating.itemId.max()))
# Test
test_dir = '/data/vnkljukin/solution.csv'
test_rating = pd.read_csv(test_dir)
test_rating.rating += 1
test_rating.rating //= 2
from bert_cnn import BertCNN
from utils import resume_checkpoint
config = {'alias': 'bert_conv_outer_simple_5',
'num_epoch': 200,
'batch_size': 2048,
'optimizer': 'adam',
'adam_lr': 1e-3,
'num_users': 42977,
'num_items': 328050,
'latent_dim': 64,
'l2_regularization': 0.,
'use_cuda': True,
'device_id': 0,
'pretrain': False,
'title_embeddings': '/data/vnkljukin/encoded_bert_128.npy',
'content_embeddings': None,
'model_dir': '/data/vnkljukin/checkpoints/{}_Epoch{}_HR{:.4f}_NDCG{:.4f}.model'
}
bert_cnn = BertCNN(config)
resume_checkpoint(bert_cnn,
'/data/vnkljukin/checkpoints/bert_conv_outer_simple_4_Epoch2_HR0.1862_NDCG0.1952.model',
config['device_id'])
ml1m_rating[ml1m_rating.rating == 0]
embedding_user = bert_cnn.embedding_user(torch.LongTensor([0]))[0]
embedding_user
embedding_item = bert_cnn.item_title(torch.LongTensor([93250]))[0]
embedding_item
bert_cnn.eval()
bert_cnn(torch.LongTensor([0]), torch.LongTensor([93250]))
import matplotlib.pyplot as plt
import seaborn as sns
sns.set()
plt.title('User embedding')
sns.heatmap(embedding_user.detach().numpy().reshape(-1, 1))
plt.show()
plt.title('Item embedding')
sns.heatmap(embedding_item.detach().numpy().reshape(-1, 1))
plt.show()
outer_map_pos = embedding_user.detach().numpy().reshape(-1, 1) @ embedding_item.detach().numpy().reshape(1, -1)
outer_map_pos.shape
plt.title('Positive interaction outer product map')
sns.heatmap(outer_map)
plt.show()
embedding_user = bert_cnn.embedding_user(torch.LongTensor([0]))[0]
embedding_item = bert_cnn.item_title(torch.LongTensor([221548]))[0]
outer_map_neg = embedding_user.detach().numpy().reshape(-1, 1) @ embedding_item.detach().numpy().reshape(1, -1)
vmin = min(outer_map_neg.min(), outer_map_pos.min())
vmax = max(outer_map_neg.max(), outer_map_pos.max())
plt.figure(figsize=(15, 5))
plt.suptitle('Interactions for one user and two different items')
plt.subplot(1, 2, 1)
plt.title('Positive interaction outer product map')
sns.heatmap(outer_map_pos, vmin=vmin, vmax=vmax, cbar=False)
plt.ylabel('user components')
plt.xlabel('item components')
plt.subplot(1, 2, 2)
plt.title('Negative interaction outer product map')
g2 = sns.heatmap(outer_map_neg, vmin=vmin, vmax=vmax)
g2.set_yticks([])
plt.xlabel('item components')
plt.savefig('/data/vnkljukin/outer_maps.png')
plt.show()
from tensorflow.python.summary.summary_iterator import summary_iterator
!ls runs
from collections import defaultdict
gmf_factor8_implicit = defaultdict(list)
for event in summary_iterator("runs/gmf_factor8-implict/events.out.tfevents.1586691393.almaren.velkerr.ru"):
for value in event.summary.value:
cause, item = value.tag.split('/')
if item in ('loss', 'HR', 'NDCG'):
gmf_factor8_implicit[item].append(value.simple_value)
gmf_factor8_implicit
mlp_factor8_bs512_reg_1e_7 = defaultdict(list)
for event in summary_iterator("runs/mlp_factor8_bs512_reg_1e-7/events.out.tfevents.1586781182.almaren.velkerr.ru"):
for value in event.summary.value:
cause, item = value.tag.split('/')
if item in ('loss', 'HR', 'NDCG'):
mlp_factor8_bs512_reg_1e_7[item].append(value.simple_value)
mlp_factor8_bs512_reg_1e_7
pretrain_neumf_factor16 = defaultdict(list)
for event in summary_iterator("runs/pretrain_neumf_factor16/events.out.tfevents.1586805539.almaren.velkerr.ru"):
for value in event.summary.value:
cause, item = value.tag.split('/')
if item in ('loss', 'HR', 'NDCG'):
pretrain_neumf_factor16[item].append(value.simple_value)
pretrain_neumf_factor16
w2v_neumf_title = defaultdict(list)
for event in summary_iterator("runs/w2v_neumf_title/events.out.tfevents.1586883854.almaren.velkerr.ru"):
for value in event.summary.value:
cause, item = value.tag.split('/')
if item in ('loss', 'HR', 'NDCG'):
w2v_neumf_title[item].append(value.simple_value)
w2v_neumf_title
!ls runs/bert_conv_outer_simple_5
bert_neumf_title = defaultdict(list)
for event in summary_iterator("runs/bert_neumf_title/events.out.tfevents.1586901360.imladris"):
for value in event.summary.value:
cause, item = value.tag.split('/')
if item in ('loss', 'HR', 'NDCG'):
bert_neumf_title[item].append(value.simple_value)
bert_neumf_title
bert_conv_outer_simple_all = defaultdict(list)
for event in summary_iterator("runs/bert_conv_outer_simple/events.out.tfevents.1586973914.almaren.velkerr.ru"):
for value in event.summary.value:
cause, item = value.tag.split('/')
if item in ('loss', 'HR', 'NDCG'):
bert_conv_outer_simple_all[item].append(value.simple_value)
bert_conv_outer_simple_all
bert_conv_outer_simple_5x = defaultdict(list)
for event in summary_iterator("runs/bert_conv_outer_simple_2/events.out.tfevents.1586984604.almaren.velkerr.ru"):
for value in event.summary.value:
cause, item = value.tag.split('/')
if item in ('loss', 'HR', 'NDCG'):
bert_conv_outer_simple_5x[item].append(value.simple_value)
bert_conv_outer_simple_5x
bert_conv_outer_simple_2x = defaultdict(list)
for event in summary_iterator("runs/bert_conv_outer_simple_3/events.out.tfevents.1587024598.almaren.velkerr.ru"):
for value in event.summary.value:
cause, item = value.tag.split('/')
if item in ('loss', 'HR', 'NDCG'):
bert_conv_outer_simple_2x[item].append(value.simple_value)
bert_conv_outer_simple_2x
bert_conv_outer_simple_1x = defaultdict(list)
for event in summary_iterator("runs/bert_conv_outer_simple_4/events.out.tfevents.1587031716.almaren.velkerr.ru"):
for value in event.summary.value:
cause, item = value.tag.split('/')
if item in ('loss', 'HR', 'NDCG'):
bert_conv_outer_simple_1x[item].append(value.simple_value)
bert_conv_outer_simple_1x
bert_conv_outer_simple_0_5x = defaultdict(list)
for event in summary_iterator("runs/bert_conv_outer_simple_5/events.out.tfevents.1587040797.almaren.velkerr.ru"):
for value in event.summary.value:
cause, item = value.tag.split('/')
if item in ('loss', 'HR', 'NDCG'):
bert_conv_outer_simple_0_5x[item].append(value.simple_value)
bert_conv_outer_simple_0_5x
plt.figure(figsize=(15, 4))
plt.subplot(1, 3, 1)
plt.title('Train loss')
plt.plot(gmf_factor8_implicit['loss'], label='GMF')
plt.plot(mlp_factor8_bs512_reg_1e_7['loss'], label='MLP')
plt.plot(pretrain_neumf_factor16['loss'], label='NMF')
plt.xticks(list(range(13)))
plt.xlabel('Epoch')
plt.legend()
plt.subplot(1, 3, 2)
plt.title('Hit Ratio@20')
plt.plot(gmf_factor8_implicit['HR'], label='GMF')
plt.plot(mlp_factor8_bs512_reg_1e_7['HR'], label='MLP')
plt.plot(pretrain_neumf_factor16['HR'], label='NMF')
plt.xticks(list(range(13)))
plt.xlabel('Epoch')
plt.legend()
plt.subplot(1, 3, 3)
plt.title('NDCG@20')
plt.plot(gmf_factor8_implicit['NDCG'], label='GMF')
plt.plot(mlp_factor8_bs512_reg_1e_7['NDCG'], label='MLP')
plt.plot(pretrain_neumf_factor16['NDCG'], label='NMF')
plt.xticks(list(range(13)))
plt.xlabel('Epoch')
plt.legend()
plt.savefig('/data/vnkljukin/baselines_recsys.png')
plt.show()
baseline_hr = max(pretrain_neumf_factor16['HR'])
baseline_ndcg = max(pretrain_neumf_factor16['NDCG'])
baseline_hr, baseline_ndcg
plt.figure(figsize=(15, 4))
plt.subplot(1, 3, 1)
plt.title('Train loss')
plt.plot(w2v_neumf_title['loss'], label='W2V MLP')
plt.plot(bert_neumf_title['loss'], label='BERT MLP')
plt.xticks(list(range(15)))
plt.xlabel('Epoch')
plt.legend()
plt.subplot(1, 3, 2)
plt.title('Hit Ratio@20')
plt.plot(w2v_neumf_title['HR'], label='W2V MLP')
plt.plot(bert_neumf_title['HR'], label='BERT MLP')
plt.axhline(baseline_hr, color='r', linestyle='dashed', label='basic baseline')
plt.xticks(list(range(15)))
plt.xlabel('Epoch')
plt.legend()
plt.subplot(1, 3, 3)
plt.title('NDCG@20')
plt.plot(w2v_neumf_title['NDCG'], label='W2V MLP')
plt.plot(bert_neumf_title['NDCG'], label='BERT MLP')
plt.axhline(baseline_ndcg, color='r', linestyle='dashed', label='basic baseline')
plt.xticks(list(range(15)))
plt.xlabel('Epoch')
plt.legend()
plt.savefig('/data/vnkljukin/bert_w2v_recsys.png')
plt.show()
plt.figure(figsize=(19, 4))
plt.subplot(1, 3, 1)
plt.title('Train loss')
plt.plot(bert_neumf_title['loss'], label='BERT MLP')
plt.plot(bert_conv_outer_simple_all['loss'], label='BERT CNN')
plt.plot(bert_conv_outer_simple_5x['loss'], label='BERT CNN 5x')
plt.plot(bert_conv_outer_simple_2x['loss'], label='BERT CNN 2x')
plt.plot(bert_conv_outer_simple_1x['loss'], label='BERT CNN 1x')
plt.plot(bert_conv_outer_simple_0_5x['loss'], label='BERT CNN 0.5x')
plt.xticks(list(range(15)))
plt.xlabel('Epoch')
plt.legend()
plt.subplot(1, 3, 2)
plt.title('Hit Ratio@20')
plt.plot(bert_neumf_title['HR'], label='BERT MLP')
plt.plot(bert_conv_outer_simple_all['HR'], label='BERT CNN')
plt.plot(bert_conv_outer_simple_5x['HR'], label='BERT CNN 5x')
plt.plot(bert_conv_outer_simple_2x['HR'], label='BERT CNN 2x')
plt.plot(bert_conv_outer_simple_1x['HR'], label='BERT CNN 1x')
plt.plot(bert_conv_outer_simple_0_5x['HR'], label='BERT CNN 0.5x')
plt.axhline(baseline_hr, color='r', linestyle='dashed', label='basic baseline')
plt.xticks(list(range(15)))
plt.xlabel('Epoch')
# plt.legend()
plt.subplot(1, 3, 3)
plt.title('NDCG@20')
plt.plot(bert_neumf_title['NDCG'], label='BERT MLP')
plt.plot(bert_conv_outer_simple_all['NDCG'], label='BERT CNN')
plt.plot(bert_conv_outer_simple_5x['NDCG'], label='BERT CNN 5x')
plt.plot(bert_conv_outer_simple_2x['NDCG'], label='BERT CNN 2x')
plt.plot(bert_conv_outer_simple_1x['NDCG'], label='BERT CNN 1x')
plt.plot(bert_conv_outer_simple_0_5x['NDCG'], label='BERT CNN 0.5x')
plt.axhline(baseline_ndcg, color='r', linestyle='dashed', label='basic baseline')
plt.xticks(list(range(15)))
plt.xlabel('Epoch')
plt.legend()
plt.savefig('/data/vnkljukin/bert_cnn_recsys.png')
plt.show()
```
|
github_jupyter
|
import pandas as pd
import numpy as np
import torch
ml1m_dir = '/data/vnkljukin/train.csv'
ml1m_rating = pd.read_csv(ml1m_dir)
ml1m_rating.rating += 1
ml1m_rating.rating //= 2
print('Range of userId is [{}, {}]'.format(ml1m_rating.userId.min(), ml1m_rating.userId.max()))
print('Range of itemId is [{}, {}]'.format(ml1m_rating.itemId.min(), ml1m_rating.itemId.max()))
# Test
test_dir = '/data/vnkljukin/solution.csv'
test_rating = pd.read_csv(test_dir)
test_rating.rating += 1
test_rating.rating //= 2
from bert_cnn import BertCNN
from utils import resume_checkpoint
config = {'alias': 'bert_conv_outer_simple_5',
'num_epoch': 200,
'batch_size': 2048,
'optimizer': 'adam',
'adam_lr': 1e-3,
'num_users': 42977,
'num_items': 328050,
'latent_dim': 64,
'l2_regularization': 0.,
'use_cuda': True,
'device_id': 0,
'pretrain': False,
'title_embeddings': '/data/vnkljukin/encoded_bert_128.npy',
'content_embeddings': None,
'model_dir': '/data/vnkljukin/checkpoints/{}_Epoch{}_HR{:.4f}_NDCG{:.4f}.model'
}
bert_cnn = BertCNN(config)
resume_checkpoint(bert_cnn,
'/data/vnkljukin/checkpoints/bert_conv_outer_simple_4_Epoch2_HR0.1862_NDCG0.1952.model',
config['device_id'])
ml1m_rating[ml1m_rating.rating == 0]
embedding_user = bert_cnn.embedding_user(torch.LongTensor([0]))[0]
embedding_user
embedding_item = bert_cnn.item_title(torch.LongTensor([93250]))[0]
embedding_item
bert_cnn.eval()
bert_cnn(torch.LongTensor([0]), torch.LongTensor([93250]))
import matplotlib.pyplot as plt
import seaborn as sns
sns.set()
plt.title('User embedding')
sns.heatmap(embedding_user.detach().numpy().reshape(-1, 1))
plt.show()
plt.title('Item embedding')
sns.heatmap(embedding_item.detach().numpy().reshape(-1, 1))
plt.show()
outer_map_pos = embedding_user.detach().numpy().reshape(-1, 1) @ embedding_item.detach().numpy().reshape(1, -1)
outer_map_pos.shape
plt.title('Positive interaction outer product map')
sns.heatmap(outer_map)
plt.show()
embedding_user = bert_cnn.embedding_user(torch.LongTensor([0]))[0]
embedding_item = bert_cnn.item_title(torch.LongTensor([221548]))[0]
outer_map_neg = embedding_user.detach().numpy().reshape(-1, 1) @ embedding_item.detach().numpy().reshape(1, -1)
vmin = min(outer_map_neg.min(), outer_map_pos.min())
vmax = max(outer_map_neg.max(), outer_map_pos.max())
plt.figure(figsize=(15, 5))
plt.suptitle('Interactions for one user and two different items')
plt.subplot(1, 2, 1)
plt.title('Positive interaction outer product map')
sns.heatmap(outer_map_pos, vmin=vmin, vmax=vmax, cbar=False)
plt.ylabel('user components')
plt.xlabel('item components')
plt.subplot(1, 2, 2)
plt.title('Negative interaction outer product map')
g2 = sns.heatmap(outer_map_neg, vmin=vmin, vmax=vmax)
g2.set_yticks([])
plt.xlabel('item components')
plt.savefig('/data/vnkljukin/outer_maps.png')
plt.show()
from tensorflow.python.summary.summary_iterator import summary_iterator
!ls runs
from collections import defaultdict
gmf_factor8_implicit = defaultdict(list)
for event in summary_iterator("runs/gmf_factor8-implict/events.out.tfevents.1586691393.almaren.velkerr.ru"):
for value in event.summary.value:
cause, item = value.tag.split('/')
if item in ('loss', 'HR', 'NDCG'):
gmf_factor8_implicit[item].append(value.simple_value)
gmf_factor8_implicit
mlp_factor8_bs512_reg_1e_7 = defaultdict(list)
for event in summary_iterator("runs/mlp_factor8_bs512_reg_1e-7/events.out.tfevents.1586781182.almaren.velkerr.ru"):
for value in event.summary.value:
cause, item = value.tag.split('/')
if item in ('loss', 'HR', 'NDCG'):
mlp_factor8_bs512_reg_1e_7[item].append(value.simple_value)
mlp_factor8_bs512_reg_1e_7
pretrain_neumf_factor16 = defaultdict(list)
for event in summary_iterator("runs/pretrain_neumf_factor16/events.out.tfevents.1586805539.almaren.velkerr.ru"):
for value in event.summary.value:
cause, item = value.tag.split('/')
if item in ('loss', 'HR', 'NDCG'):
pretrain_neumf_factor16[item].append(value.simple_value)
pretrain_neumf_factor16
w2v_neumf_title = defaultdict(list)
for event in summary_iterator("runs/w2v_neumf_title/events.out.tfevents.1586883854.almaren.velkerr.ru"):
for value in event.summary.value:
cause, item = value.tag.split('/')
if item in ('loss', 'HR', 'NDCG'):
w2v_neumf_title[item].append(value.simple_value)
w2v_neumf_title
!ls runs/bert_conv_outer_simple_5
bert_neumf_title = defaultdict(list)
for event in summary_iterator("runs/bert_neumf_title/events.out.tfevents.1586901360.imladris"):
for value in event.summary.value:
cause, item = value.tag.split('/')
if item in ('loss', 'HR', 'NDCG'):
bert_neumf_title[item].append(value.simple_value)
bert_neumf_title
bert_conv_outer_simple_all = defaultdict(list)
for event in summary_iterator("runs/bert_conv_outer_simple/events.out.tfevents.1586973914.almaren.velkerr.ru"):
for value in event.summary.value:
cause, item = value.tag.split('/')
if item in ('loss', 'HR', 'NDCG'):
bert_conv_outer_simple_all[item].append(value.simple_value)
bert_conv_outer_simple_all
bert_conv_outer_simple_5x = defaultdict(list)
for event in summary_iterator("runs/bert_conv_outer_simple_2/events.out.tfevents.1586984604.almaren.velkerr.ru"):
for value in event.summary.value:
cause, item = value.tag.split('/')
if item in ('loss', 'HR', 'NDCG'):
bert_conv_outer_simple_5x[item].append(value.simple_value)
bert_conv_outer_simple_5x
bert_conv_outer_simple_2x = defaultdict(list)
for event in summary_iterator("runs/bert_conv_outer_simple_3/events.out.tfevents.1587024598.almaren.velkerr.ru"):
for value in event.summary.value:
cause, item = value.tag.split('/')
if item in ('loss', 'HR', 'NDCG'):
bert_conv_outer_simple_2x[item].append(value.simple_value)
bert_conv_outer_simple_2x
bert_conv_outer_simple_1x = defaultdict(list)
for event in summary_iterator("runs/bert_conv_outer_simple_4/events.out.tfevents.1587031716.almaren.velkerr.ru"):
for value in event.summary.value:
cause, item = value.tag.split('/')
if item in ('loss', 'HR', 'NDCG'):
bert_conv_outer_simple_1x[item].append(value.simple_value)
bert_conv_outer_simple_1x
bert_conv_outer_simple_0_5x = defaultdict(list)
for event in summary_iterator("runs/bert_conv_outer_simple_5/events.out.tfevents.1587040797.almaren.velkerr.ru"):
for value in event.summary.value:
cause, item = value.tag.split('/')
if item in ('loss', 'HR', 'NDCG'):
bert_conv_outer_simple_0_5x[item].append(value.simple_value)
bert_conv_outer_simple_0_5x
plt.figure(figsize=(15, 4))
plt.subplot(1, 3, 1)
plt.title('Train loss')
plt.plot(gmf_factor8_implicit['loss'], label='GMF')
plt.plot(mlp_factor8_bs512_reg_1e_7['loss'], label='MLP')
plt.plot(pretrain_neumf_factor16['loss'], label='NMF')
plt.xticks(list(range(13)))
plt.xlabel('Epoch')
plt.legend()
plt.subplot(1, 3, 2)
plt.title('Hit Ratio@20')
plt.plot(gmf_factor8_implicit['HR'], label='GMF')
plt.plot(mlp_factor8_bs512_reg_1e_7['HR'], label='MLP')
plt.plot(pretrain_neumf_factor16['HR'], label='NMF')
plt.xticks(list(range(13)))
plt.xlabel('Epoch')
plt.legend()
plt.subplot(1, 3, 3)
plt.title('NDCG@20')
plt.plot(gmf_factor8_implicit['NDCG'], label='GMF')
plt.plot(mlp_factor8_bs512_reg_1e_7['NDCG'], label='MLP')
plt.plot(pretrain_neumf_factor16['NDCG'], label='NMF')
plt.xticks(list(range(13)))
plt.xlabel('Epoch')
plt.legend()
plt.savefig('/data/vnkljukin/baselines_recsys.png')
plt.show()
baseline_hr = max(pretrain_neumf_factor16['HR'])
baseline_ndcg = max(pretrain_neumf_factor16['NDCG'])
baseline_hr, baseline_ndcg
plt.figure(figsize=(15, 4))
plt.subplot(1, 3, 1)
plt.title('Train loss')
plt.plot(w2v_neumf_title['loss'], label='W2V MLP')
plt.plot(bert_neumf_title['loss'], label='BERT MLP')
plt.xticks(list(range(15)))
plt.xlabel('Epoch')
plt.legend()
plt.subplot(1, 3, 2)
plt.title('Hit Ratio@20')
plt.plot(w2v_neumf_title['HR'], label='W2V MLP')
plt.plot(bert_neumf_title['HR'], label='BERT MLP')
plt.axhline(baseline_hr, color='r', linestyle='dashed', label='basic baseline')
plt.xticks(list(range(15)))
plt.xlabel('Epoch')
plt.legend()
plt.subplot(1, 3, 3)
plt.title('NDCG@20')
plt.plot(w2v_neumf_title['NDCG'], label='W2V MLP')
plt.plot(bert_neumf_title['NDCG'], label='BERT MLP')
plt.axhline(baseline_ndcg, color='r', linestyle='dashed', label='basic baseline')
plt.xticks(list(range(15)))
plt.xlabel('Epoch')
plt.legend()
plt.savefig('/data/vnkljukin/bert_w2v_recsys.png')
plt.show()
plt.figure(figsize=(19, 4))
plt.subplot(1, 3, 1)
plt.title('Train loss')
plt.plot(bert_neumf_title['loss'], label='BERT MLP')
plt.plot(bert_conv_outer_simple_all['loss'], label='BERT CNN')
plt.plot(bert_conv_outer_simple_5x['loss'], label='BERT CNN 5x')
plt.plot(bert_conv_outer_simple_2x['loss'], label='BERT CNN 2x')
plt.plot(bert_conv_outer_simple_1x['loss'], label='BERT CNN 1x')
plt.plot(bert_conv_outer_simple_0_5x['loss'], label='BERT CNN 0.5x')
plt.xticks(list(range(15)))
plt.xlabel('Epoch')
plt.legend()
plt.subplot(1, 3, 2)
plt.title('Hit Ratio@20')
plt.plot(bert_neumf_title['HR'], label='BERT MLP')
plt.plot(bert_conv_outer_simple_all['HR'], label='BERT CNN')
plt.plot(bert_conv_outer_simple_5x['HR'], label='BERT CNN 5x')
plt.plot(bert_conv_outer_simple_2x['HR'], label='BERT CNN 2x')
plt.plot(bert_conv_outer_simple_1x['HR'], label='BERT CNN 1x')
plt.plot(bert_conv_outer_simple_0_5x['HR'], label='BERT CNN 0.5x')
plt.axhline(baseline_hr, color='r', linestyle='dashed', label='basic baseline')
plt.xticks(list(range(15)))
plt.xlabel('Epoch')
# plt.legend()
plt.subplot(1, 3, 3)
plt.title('NDCG@20')
plt.plot(bert_neumf_title['NDCG'], label='BERT MLP')
plt.plot(bert_conv_outer_simple_all['NDCG'], label='BERT CNN')
plt.plot(bert_conv_outer_simple_5x['NDCG'], label='BERT CNN 5x')
plt.plot(bert_conv_outer_simple_2x['NDCG'], label='BERT CNN 2x')
plt.plot(bert_conv_outer_simple_1x['NDCG'], label='BERT CNN 1x')
plt.plot(bert_conv_outer_simple_0_5x['NDCG'], label='BERT CNN 0.5x')
plt.axhline(baseline_ndcg, color='r', linestyle='dashed', label='basic baseline')
plt.xticks(list(range(15)))
plt.xlabel('Epoch')
plt.legend()
plt.savefig('/data/vnkljukin/bert_cnn_recsys.png')
plt.show()
| 0.436622 | 0.320914 |
[](https://colab.research.google.com/github/idealo/image-super-resolution/blob/master/notebooks/ISR_Training_Tutorial.ipynb)
# Install ISR
```
!pip install ISR
```
# Train
## Get the training data
Get your data to train the model. The div2k dataset linked here is for a scaling factor of 2. Beware of this later when training the model.
(for more options on how to get you data on Colab notebooks visit https://colab.research.google.com/notebooks/io.ipynb)
```
!wget http://data.vision.ee.ethz.ch/cvl/DIV2K/DIV2K_train_LR_bicubic_X2.zip
!wget http://data.vision.ee.ethz.ch/cvl/DIV2K/DIV2K_valid_LR_bicubic_X2.zip
!wget http://data.vision.ee.ethz.ch/cvl/DIV2K/DIV2K_train_HR.zip
!wget http://data.vision.ee.ethz.ch/cvl/DIV2K/DIV2K_valid_HR.zip
!mkdir div2k
!unzip -q DIV2K_valid_LR_bicubic_X2.zip -d div2k
!unzip -q DIV2K_train_LR_bicubic_X2.zip -d div2k
!unzip -q DIV2K_train_HR.zip -d div2k
!unzip -q DIV2K_valid_HR.zip -d div2k
```
## Create the models
Import the models from the ISR package and create
- a RRDN super scaling network
- a discriminator network for GANs training
- a VGG19 feature extractor to train with a perceptual loss function
Carefully select
- 'x': this is the upscaling factor (2 by default)
- 'layers_to_extract': these are the layers from the VGG19 that will be used in the perceptual loss (leave the default if you're not familiar with it)
- 'lr_patch_size': this is the size of the patches that will be extracted from the LR images and fed to the ISR network during training time
Play around with the other architecture parameters
```
from ISR.models import RRDN
from ISR.models import Discriminator
from ISR.models import Cut_VGG19
lr_train_patch_size = 40
layers_to_extract = [5, 9]
scale = 2
hr_train_patch_size = lr_train_patch_size * scale
rrdn = RRDN(arch_params={'C':4, 'D':3, 'G':64, 'G0':64, 'T':10, 'x':scale}, patch_size=lr_train_patch_size)
f_ext = Cut_VGG19(patch_size=hr_train_patch_size, layers_to_extract=layers_to_extract)
discr = Discriminator(patch_size=hr_train_patch_size, kernel_size=3)
```
## Give the models to the Trainer
The Trainer object will combine the networks, manage your training data and keep you up-to-date with the training progress through Tensorboard and the command line.
Here we do not use the pixel-wise MSE but only the perceptual loss by specifying the respective weights in `loss_weights`
```
from ISR.train import Trainer
loss_weights = {
'generator': 0.0,
'feature_extractor': 0.0833,
'discriminator': 0.01
}
losses = {
'generator': 'mae',
'feature_extractor': 'mse',
'discriminator': 'binary_crossentropy'
}
log_dirs = {'logs': './logs', 'weights': './weights'}
learning_rate = {'initial_value': 0.0004, 'decay_factor': 0.5, 'decay_frequency': 30}
flatness = {'min': 0.0, 'max': 0.15, 'increase': 0.01, 'increase_frequency': 5}
trainer = Trainer(
generator=rrdn,
discriminator=discr,
feature_extractor=f_ext,
lr_train_dir='div2k/DIV2K_train_LR_bicubic/X2/',
hr_train_dir='div2k/DIV2K_train_HR/',
lr_valid_dir='div2k/DIV2K_train_LR_bicubic/X2/',
hr_valid_dir='div2k/DIV2K_train_HR/',
loss_weights=loss_weights,
learning_rate=learning_rate,
flatness=flatness,
dataname='div2k',
log_dirs=log_dirs,
weights_generator=None,
weights_discriminator=None,
n_validation=40,
)
```
Choose epoch number, steps and batch size and start training
```
trainer.train(
epochs=1,
steps_per_epoch=20,
batch_size=4,
monitored_metrics={'val_generator_PSNR_Y': 'max'}
)
```
|
github_jupyter
|
!pip install ISR
!wget http://data.vision.ee.ethz.ch/cvl/DIV2K/DIV2K_train_LR_bicubic_X2.zip
!wget http://data.vision.ee.ethz.ch/cvl/DIV2K/DIV2K_valid_LR_bicubic_X2.zip
!wget http://data.vision.ee.ethz.ch/cvl/DIV2K/DIV2K_train_HR.zip
!wget http://data.vision.ee.ethz.ch/cvl/DIV2K/DIV2K_valid_HR.zip
!mkdir div2k
!unzip -q DIV2K_valid_LR_bicubic_X2.zip -d div2k
!unzip -q DIV2K_train_LR_bicubic_X2.zip -d div2k
!unzip -q DIV2K_train_HR.zip -d div2k
!unzip -q DIV2K_valid_HR.zip -d div2k
from ISR.models import RRDN
from ISR.models import Discriminator
from ISR.models import Cut_VGG19
lr_train_patch_size = 40
layers_to_extract = [5, 9]
scale = 2
hr_train_patch_size = lr_train_patch_size * scale
rrdn = RRDN(arch_params={'C':4, 'D':3, 'G':64, 'G0':64, 'T':10, 'x':scale}, patch_size=lr_train_patch_size)
f_ext = Cut_VGG19(patch_size=hr_train_patch_size, layers_to_extract=layers_to_extract)
discr = Discriminator(patch_size=hr_train_patch_size, kernel_size=3)
from ISR.train import Trainer
loss_weights = {
'generator': 0.0,
'feature_extractor': 0.0833,
'discriminator': 0.01
}
losses = {
'generator': 'mae',
'feature_extractor': 'mse',
'discriminator': 'binary_crossentropy'
}
log_dirs = {'logs': './logs', 'weights': './weights'}
learning_rate = {'initial_value': 0.0004, 'decay_factor': 0.5, 'decay_frequency': 30}
flatness = {'min': 0.0, 'max': 0.15, 'increase': 0.01, 'increase_frequency': 5}
trainer = Trainer(
generator=rrdn,
discriminator=discr,
feature_extractor=f_ext,
lr_train_dir='div2k/DIV2K_train_LR_bicubic/X2/',
hr_train_dir='div2k/DIV2K_train_HR/',
lr_valid_dir='div2k/DIV2K_train_LR_bicubic/X2/',
hr_valid_dir='div2k/DIV2K_train_HR/',
loss_weights=loss_weights,
learning_rate=learning_rate,
flatness=flatness,
dataname='div2k',
log_dirs=log_dirs,
weights_generator=None,
weights_discriminator=None,
n_validation=40,
)
trainer.train(
epochs=1,
steps_per_epoch=20,
batch_size=4,
monitored_metrics={'val_generator_PSNR_Y': 'max'}
)
| 0.527317 | 0.93744 |
```
from IPython.display import HTML
HTML('''<script>
code_show=true;
function code_toggle() {
if (code_show){
$('div.input').hide();
} else {
$('div.input').show();
}
code_show = !code_show
}
$( document ).ready(code_toggle);
</script>
The raw code for this IPython notebook is by default hidden for easier reading.
To toggle on/off the raw code, click <a href="javascript:code_toggle()">here</a>.''')
```

<h1 align='center'>Introduction to Python $\&$ the Jupyter Environment</h1>
<h4 align='center'>Workshop Facilitator: Laura Gutierrez Funderburk, Department of Mathematics at SFU, Callysto Developer, November 2019</h4>
<h2 align='center'>Introduction</h2>
In this workshop participants will have an opportunity to learn basic programming with Python while using the Jupyter environment to define, test and implement functions. At the end of the workshop, participants will be given resources and challenge exercises.
<h2 align='center'>Why Python</h2>
* Python is currently a "hot" programming language
* Python is (relatively) intuitive - there are lots of online resources available to learn and improve your skills
* Python is great for data analysis and visualization: a highly sought after skill in the current job market
<h2 align='center'>Workshop Overview</h2>
In this workshop we will cover basic for loops, if/else and while statements, importing libraries and 2D visualization.
Via a series of guided exercises, we will simulate an unfair probabilistic game with two players.
```
import matplotlib.patches as mpatches
from ipywidgets import interact, interact_manual, widgets
import random
import matplotlib.pyplot as plt
style = {'description_width': 'initial'}
def roll_dice():
positive_luck = random.choice([1,2,4,5,6])
negative_luck = random.choice([1,2,4,5,6])
if positive_luck - negative_luck >= 0:
return True
else:
return False
def simulate_unfair_game(points_A,points_C,number_of_games):
x_coord = []
y_coord_A = []
y_coord_C = []
value_A = points_A
value_C = points_C
turn_number = 1
while turn_number <= number_of_games:
if roll_dice():
x_coord.append(turn_number)
turn_number += 1
y_coord_A.append(value_A)
y_coord_C.append(value_C)
value_A = value_A + (value_C/2)
value_C = value_C - (value_C/2)
else:
x_coord.append(turn_number)
turn_number += 1
y_coord_A.append(value_A)
y_coord_C.append(value_C)
value_A = value_A - (value_A/2)
value_C = value_C + (value_A/2)
return [x_coord,y_coord_A,y_coord_C]
def plot_unfair_simulation(number_turns):
fig,ax = plt.subplots(figsize=(10,10))
ax.set_xlim([0,number_turns + 1])
ax.set_ylim([0,200])
ax.grid(True)
[x_co,y_co_A,y_co_C] = simulate_unfair_game(100,100,number_turns)
ax.plot(x_co,y_co_A,color='#8642f4',label="Alice")
ax.plot(x_co,y_co_C,color='#518900',label="Bob")
ax.set_ylabel("Number of points",fontsize=25)
ax.set_xlabel("Number of turns",fontsize=25)
ax.set_title("Positive vs Negative Luck Game Simulation",fontsize=25)
plt.legend(bbox_to_anchor=(1.05, 1), loc=2, borderaxespad=1.)
plt.show()
interact_manual(plot_unfair_simulation,number_turns=widgets.IntSlider(
value=10,
min=2,
max=100,
step=1,
description='Number of turns',
disabled=False,
continuous_update=False,
orientation='horizontal',
readout=True,
readout_format='d',
style =style
));
```
<h2 align='center'>Section 0. Setup</h2>
1. Go to http://sfu.syzygy.ca and press the Login icon 
2. Authenticate
3. Open a new terminal 
4. Enter the following command ```git clone https://github.com/lfunderburk/IntroToPython``` 
5. Press Enter
6. You can now access workshop material!
<h2 align='center'>Section 1. For loops, if/else and while statements</h2>
In this section we will go over basic syntax used in for loops, if/else and while statements.
```
# Basic for loop syntax
for i in range(1,5):
print(i*2)
# Basic if/else syntax
a_variable = True
if a_variable==True:
print("True that.")
else:
print("False that.")
# Basic while syntax
condition = 1
while condition<5:
print(condition*3)
condition = condition + 1
```
<h2 align='center'>Section 2. Defininign a Function</h2>
```
def my_function(number_repetitions):
for i in range(number_repetitions):
if number_repetitions%2==0:
return True
else:
return False
nu_rep = 10
my_function(nu_rep)
nu_rep = 5
my_function(nu_rep)
```
<h2 align='center'>Section 3. Importing Modules</h2>
```
import random
# To call a function from the random module use the syntax
# module.function()
# For example
dice_outcome = random.choice([1,2,3,4,5,6])
print(dice_outcome)
# Use a for loop
for i in range(5):
print(random.choice([1,2,3,4,5,6]))
```
<h2 align='center'>Setting up the Game</h2>
Let us begin to set up the game.
Let's suppose we have two dice P, N each with six faces. Let us use P to track the amount of "positive luck" and N the amount og "negative luck".
Let T denote total luck, where T is calculated by substracting the amount of negative luck from the amount of positive luck. If the number in P is higher than or equal to N, then we say we have "good luck". We say we have "bad luck" otherwise.
Complete the following function that simulates rolling the P and N dices. If we have good luck, it will return True. If we have bad luck it will return False.
A player is anyone who rolls P, N dice with the purpose of obtaining a good or a back luck outcome.
```
### Complete the blanks _________
def roll_dice():
positive_luck = random.choice([1,2,3,4,5,6])
negative_luck = random.choice([1,2,3,4,5,6])
# print(negative_luck)
# print(positive_luck)
if positive_luck - negative_luck >= 0:
return True
else:
return False
roll_dice()
```
We observe that the sample space in this game is given as follows:

From our definition of Good Luck, we see that we have a higher probability of obtaining a favourable outcome than a negative one. Therefore this is an unfair game.
Let us introduce two players Alice and Bob. Both Alice and Bob are given 100 points.
They roll the two dice once. If the outcome is Good luck Alice wins and takes half of Bob's points. If the outcome is Bad luck, Bob wins and takes half of Alice's points.
Players can reroll dice and take points from each other until one of the players decides to withdraw. Players can withdraw at any time during the game.
This is clearly a bad deal of Bob. Suppose both Bob and Alice are unaware that this is an unfair game.
Let us simulate what would happen if they chose to play.
Complete the function below.
```
def simulate_unfair_game(points_A,points_B,number_of_games):
# Store points in arrays
# We will plot these in the next exercise
# Number of games played
x_coord = []
# Alice's points
y_coord_A = []
# Bob's points
y_coord_B = []
# Set initial number of points
value_A = points_A
value_B = points_B
# Initialize number of turns to 1
turn_number = 1
#
while turn_number <= number_of_games:
# If outcome is Goog luck
if roll_dice():
# Add turn number to x_coord
x_coord.append(turn_number)
# Increase turn number
turn_number = turn_number + 1
# Add points for Alice and Bob
y_coord_A.append(value_A)
y_coord_B.append(value_B)
# Bob loses half of his points
value_B = value_B - (value_B/2)
# Since outcome is good luck, Alice gets half of Bob's points
value_A = value_A + value_B
# If outcome is Bad luck
else:
# Add turn number of x_cord
x_coord.append(turn_number)
# Increase number of turns
turn_number += 1
# Add points for Alice and Bob
y_coord_A.append(value_A)
y_coord_B.append(value_B)
# Since outcome is bad luck, Bob gets half of Alice's points
# Alice loses half of her points
value_A = value_A - value_A/2
value_B = value_B + value_A
return [x_coord,y_coord_A,y_coord_B]
[x_co,y_co_A,y_co_B] = simulate_unfair_game(100,100,10)
for item in [x_co,y_co_A,y_co_B]:
print(item)
```
<h2 align='center'>Section 4. 2D plotting with matplotlib</h2>
We begin by importing the pyplot function within the matplotlib module.
```
import matplotlib.pyplot as plt
# We begin by initializing our figure
fig,ax1 = plt.subplots(figsize=(10,10))
# Set x, y axis limits
ax1.set_xlim([0,10])
ax1.set_ylim([0,10])
# Plot a line
ax1.plot([2,8],[2,8])
# X, Y labels
ax1.set_ylabel("X axis",fontsize=25)
ax1.set_xlabel("Y axis",fontsize=25)
# Title
ax1.set_title("A Plot",fontsize=25)
# Show the figure
plt.show()
```
Print the outcome of our simulation
```
print([x_co,y_co_A,y_co_B])
```
The first array corresponds to the number of times they play.
The second array corresponds to the number of points Alice gets in each turn while the third array corresponds to the number of points Bob gets in each turn.
Let us plot the results.
```
def plotsimulation(initial_pointsa,initial_pointsb,number_turns):# Try different values in the function simulate_unfair_game() and rerun the cell
# Initialize figure
fig,ax = plt.subplots(figsize=(10,10))
# Adjust x, y axis
ax.set_xlim([0,number_turns + 1])
ax.set_ylim([0,200])
ax.grid(True)
# Begin simulation
[x_co,y_co_A,y_co_B] = simulate_unfair_game(initial_pointsa,initial_pointsb,number_turns)
# Plot Simulation
# Alice's points
ax.plot(x_co,y_co_A,color='#8642f4',label="Alice")
# Bob's points
ax.plot(x_co,y_co_B,color='#518900',label="Bob")
# Label the figure
ax.set_ylabel("Number of points",fontsize=25)
ax.set_xlabel("Number of turns",fontsize=25)
ax.set_title("Positive vs Negative Luck Game Simulation",fontsize=25)
plt.legend(bbox_to_anchor=(1.05, 1), loc=2, borderaxespad=1.)
# Show figure
plt.show()
```
#### Exercise
Turn the code above into a function whose parameters are total number of turns, and initial points for Bob and Alice.
What happens when each player is give a different number of points?
```
plotsimulation(150,60,10)
```
<h2 align='center'>BONUS: Widgets</h2>
We can create a nice user interface to run several simulations of the game.
The interact_manual and interact functions from the ipywidgets modules allows us to use sliders and buttons.
```
from ipywidgets import interact, interact_manual, widgets
def simple_function(x):
print(x)
style = {'description_width': 'initial'}
interact(simple_function,x=widgets.IntSlider(
value=10,
min=2,
max=100,
step=1,
description='Number of turns',
disabled=False,
continuous_update=False,
orientation='horizontal',
readout=True,
readout_format='d',
style =style
));
```
<h2 align='center'>Challenge</h2>
The cell below contains all functions we worked on during the workshop.
Substitute the plot_unfair_simulation(number_turns) for the function you defined plot_unfair_simulation(number_turns,points_bob,points_alicece)
Modify the interact_manual command so that widgets are added that would allow you to modify the initial number of points Bob and Alice are given.
```
from ipywidgets import interact, interact_manual, widgets
import random
import matplotlib.pyplot as plt
style = {'description_width': 'initial'}
def roll_dice():
positive_luck = random.choice([1,2,4,5,6])
negative_luck = random.choice([1,2,4,5,6])
if positive_luck - negative_luck >= 0:
return True
else:
return False
def simulate_unfair_game(points_A,points_B,number_of_games):
x_coord = []
y_coord_A = []
y_coord_B = []
value_A = points_A
value_B = points_B
turn_number = 1
while turn_number <= number_of_games:
if roll_dice():
x_coord.append(turn_number)
turn_number += 1
y_coord_A.append(value_A)
y_coord_B.append(value_B)
value_B = value_B - (value_B/2)
value_A = value_A + value_B
else:
x_coord.append(turn_number)
turn_number += 1
y_coord_A.append(value_A)
y_coord_B.append(value_B)
value_A = value_A - value_A/2
value_B = value_B + value_A
return [x_coord,y_coord_A,y_coord_B]
def plot_unfair_simulation(number_turns,points_bob,points_alice):
fig,ax = plt.subplots(figsize=(10,10))
ax.set_xlim([0,number_turns + 1])
ax.set_ylim([0,200])
ax.grid(True)
[x_co,y_co_A,y_co_B] = simulate_unfair_game(points_bob,points_alice,number_turns)
ax.plot(x_co,y_co_A,color='#8642f4',label="Alice")
ax.plot(x_co,y_co_B,color='#518900',label="Bob")
ax.set_ylabel("Number of points",fontsize=25)
ax.set_xlabel("Number of turns",fontsize=25)
ax.set_title("Positive vs Negative Luck Game Simulation",fontsize=25)
plt.legend(bbox_to_anchor=(1.05, 1), loc=2, borderaxespad=1.)
plt.show()
interact_manual(plot_unfair_simulation,number_turns=widgets.IntSlider(
value=10,
min=2,
max=100,
step=1,
description='Number of turns',
disabled=False,
continuous_update=False,
orientation='horizontal',
readout=True,
readout_format='d',
style =style
), points_bob=widgets.IntSlider(
value=100,
min=2,
max=100,
step=1,
description='Initial points for Bob',
disabled=False,
continuous_update=False,
orientation='horizontal',
readout=True,
readout_format='d',
style =style
), points_alice = widgets.IntSlider(
value=100,
min=2,
max=100,
step=1,
description='Initial points for Alice',
disabled=False,
continuous_update=False,
orientation='horizontal',
readout=True,
readout_format='d',
style =style));
```

|
github_jupyter
|
from IPython.display import HTML
HTML('''<script>
code_show=true;
function code_toggle() {
if (code_show){
$('div.input').hide();
} else {
$('div.input').show();
}
code_show = !code_show
}
$( document ).ready(code_toggle);
</script>
The raw code for this IPython notebook is by default hidden for easier reading.
To toggle on/off the raw code, click <a href="javascript:code_toggle()">here</a>.''')
import matplotlib.patches as mpatches
from ipywidgets import interact, interact_manual, widgets
import random
import matplotlib.pyplot as plt
style = {'description_width': 'initial'}
def roll_dice():
positive_luck = random.choice([1,2,4,5,6])
negative_luck = random.choice([1,2,4,5,6])
if positive_luck - negative_luck >= 0:
return True
else:
return False
def simulate_unfair_game(points_A,points_C,number_of_games):
x_coord = []
y_coord_A = []
y_coord_C = []
value_A = points_A
value_C = points_C
turn_number = 1
while turn_number <= number_of_games:
if roll_dice():
x_coord.append(turn_number)
turn_number += 1
y_coord_A.append(value_A)
y_coord_C.append(value_C)
value_A = value_A + (value_C/2)
value_C = value_C - (value_C/2)
else:
x_coord.append(turn_number)
turn_number += 1
y_coord_A.append(value_A)
y_coord_C.append(value_C)
value_A = value_A - (value_A/2)
value_C = value_C + (value_A/2)
return [x_coord,y_coord_A,y_coord_C]
def plot_unfair_simulation(number_turns):
fig,ax = plt.subplots(figsize=(10,10))
ax.set_xlim([0,number_turns + 1])
ax.set_ylim([0,200])
ax.grid(True)
[x_co,y_co_A,y_co_C] = simulate_unfair_game(100,100,number_turns)
ax.plot(x_co,y_co_A,color='#8642f4',label="Alice")
ax.plot(x_co,y_co_C,color='#518900',label="Bob")
ax.set_ylabel("Number of points",fontsize=25)
ax.set_xlabel("Number of turns",fontsize=25)
ax.set_title("Positive vs Negative Luck Game Simulation",fontsize=25)
plt.legend(bbox_to_anchor=(1.05, 1), loc=2, borderaxespad=1.)
plt.show()
interact_manual(plot_unfair_simulation,number_turns=widgets.IntSlider(
value=10,
min=2,
max=100,
step=1,
description='Number of turns',
disabled=False,
continuous_update=False,
orientation='horizontal',
readout=True,
readout_format='d',
style =style
));
# Basic for loop syntax
for i in range(1,5):
print(i*2)
# Basic if/else syntax
a_variable = True
if a_variable==True:
print("True that.")
else:
print("False that.")
# Basic while syntax
condition = 1
while condition<5:
print(condition*3)
condition = condition + 1
def my_function(number_repetitions):
for i in range(number_repetitions):
if number_repetitions%2==0:
return True
else:
return False
nu_rep = 10
my_function(nu_rep)
nu_rep = 5
my_function(nu_rep)
import random
# To call a function from the random module use the syntax
# module.function()
# For example
dice_outcome = random.choice([1,2,3,4,5,6])
print(dice_outcome)
# Use a for loop
for i in range(5):
print(random.choice([1,2,3,4,5,6]))
### Complete the blanks _________
def roll_dice():
positive_luck = random.choice([1,2,3,4,5,6])
negative_luck = random.choice([1,2,3,4,5,6])
# print(negative_luck)
# print(positive_luck)
if positive_luck - negative_luck >= 0:
return True
else:
return False
roll_dice()
def simulate_unfair_game(points_A,points_B,number_of_games):
# Store points in arrays
# We will plot these in the next exercise
# Number of games played
x_coord = []
# Alice's points
y_coord_A = []
# Bob's points
y_coord_B = []
# Set initial number of points
value_A = points_A
value_B = points_B
# Initialize number of turns to 1
turn_number = 1
#
while turn_number <= number_of_games:
# If outcome is Goog luck
if roll_dice():
# Add turn number to x_coord
x_coord.append(turn_number)
# Increase turn number
turn_number = turn_number + 1
# Add points for Alice and Bob
y_coord_A.append(value_A)
y_coord_B.append(value_B)
# Bob loses half of his points
value_B = value_B - (value_B/2)
# Since outcome is good luck, Alice gets half of Bob's points
value_A = value_A + value_B
# If outcome is Bad luck
else:
# Add turn number of x_cord
x_coord.append(turn_number)
# Increase number of turns
turn_number += 1
# Add points for Alice and Bob
y_coord_A.append(value_A)
y_coord_B.append(value_B)
# Since outcome is bad luck, Bob gets half of Alice's points
# Alice loses half of her points
value_A = value_A - value_A/2
value_B = value_B + value_A
return [x_coord,y_coord_A,y_coord_B]
[x_co,y_co_A,y_co_B] = simulate_unfair_game(100,100,10)
for item in [x_co,y_co_A,y_co_B]:
print(item)
import matplotlib.pyplot as plt
# We begin by initializing our figure
fig,ax1 = plt.subplots(figsize=(10,10))
# Set x, y axis limits
ax1.set_xlim([0,10])
ax1.set_ylim([0,10])
# Plot a line
ax1.plot([2,8],[2,8])
# X, Y labels
ax1.set_ylabel("X axis",fontsize=25)
ax1.set_xlabel("Y axis",fontsize=25)
# Title
ax1.set_title("A Plot",fontsize=25)
# Show the figure
plt.show()
print([x_co,y_co_A,y_co_B])
def plotsimulation(initial_pointsa,initial_pointsb,number_turns):# Try different values in the function simulate_unfair_game() and rerun the cell
# Initialize figure
fig,ax = plt.subplots(figsize=(10,10))
# Adjust x, y axis
ax.set_xlim([0,number_turns + 1])
ax.set_ylim([0,200])
ax.grid(True)
# Begin simulation
[x_co,y_co_A,y_co_B] = simulate_unfair_game(initial_pointsa,initial_pointsb,number_turns)
# Plot Simulation
# Alice's points
ax.plot(x_co,y_co_A,color='#8642f4',label="Alice")
# Bob's points
ax.plot(x_co,y_co_B,color='#518900',label="Bob")
# Label the figure
ax.set_ylabel("Number of points",fontsize=25)
ax.set_xlabel("Number of turns",fontsize=25)
ax.set_title("Positive vs Negative Luck Game Simulation",fontsize=25)
plt.legend(bbox_to_anchor=(1.05, 1), loc=2, borderaxespad=1.)
# Show figure
plt.show()
plotsimulation(150,60,10)
from ipywidgets import interact, interact_manual, widgets
def simple_function(x):
print(x)
style = {'description_width': 'initial'}
interact(simple_function,x=widgets.IntSlider(
value=10,
min=2,
max=100,
step=1,
description='Number of turns',
disabled=False,
continuous_update=False,
orientation='horizontal',
readout=True,
readout_format='d',
style =style
));
from ipywidgets import interact, interact_manual, widgets
import random
import matplotlib.pyplot as plt
style = {'description_width': 'initial'}
def roll_dice():
positive_luck = random.choice([1,2,4,5,6])
negative_luck = random.choice([1,2,4,5,6])
if positive_luck - negative_luck >= 0:
return True
else:
return False
def simulate_unfair_game(points_A,points_B,number_of_games):
x_coord = []
y_coord_A = []
y_coord_B = []
value_A = points_A
value_B = points_B
turn_number = 1
while turn_number <= number_of_games:
if roll_dice():
x_coord.append(turn_number)
turn_number += 1
y_coord_A.append(value_A)
y_coord_B.append(value_B)
value_B = value_B - (value_B/2)
value_A = value_A + value_B
else:
x_coord.append(turn_number)
turn_number += 1
y_coord_A.append(value_A)
y_coord_B.append(value_B)
value_A = value_A - value_A/2
value_B = value_B + value_A
return [x_coord,y_coord_A,y_coord_B]
def plot_unfair_simulation(number_turns,points_bob,points_alice):
fig,ax = plt.subplots(figsize=(10,10))
ax.set_xlim([0,number_turns + 1])
ax.set_ylim([0,200])
ax.grid(True)
[x_co,y_co_A,y_co_B] = simulate_unfair_game(points_bob,points_alice,number_turns)
ax.plot(x_co,y_co_A,color='#8642f4',label="Alice")
ax.plot(x_co,y_co_B,color='#518900',label="Bob")
ax.set_ylabel("Number of points",fontsize=25)
ax.set_xlabel("Number of turns",fontsize=25)
ax.set_title("Positive vs Negative Luck Game Simulation",fontsize=25)
plt.legend(bbox_to_anchor=(1.05, 1), loc=2, borderaxespad=1.)
plt.show()
interact_manual(plot_unfair_simulation,number_turns=widgets.IntSlider(
value=10,
min=2,
max=100,
step=1,
description='Number of turns',
disabled=False,
continuous_update=False,
orientation='horizontal',
readout=True,
readout_format='d',
style =style
), points_bob=widgets.IntSlider(
value=100,
min=2,
max=100,
step=1,
description='Initial points for Bob',
disabled=False,
continuous_update=False,
orientation='horizontal',
readout=True,
readout_format='d',
style =style
), points_alice = widgets.IntSlider(
value=100,
min=2,
max=100,
step=1,
description='Initial points for Alice',
disabled=False,
continuous_update=False,
orientation='horizontal',
readout=True,
readout_format='d',
style =style));
| 0.301156 | 0.909626 |
```
from IPython.core.interactiveshell import InteractiveShell
InteractiveShell.ast_node_interactivity = "all"
import pandas as pd
import numpy as np
from scipy.sparse import coo_matrix
import math as mt
from scipy.sparse.linalg import * #used for matrix multiplication
from scipy.sparse.linalg import svds
from scipy.sparse import csc_matrix
from sklearn.metrics import mean_squared_error
from math import sqrt
import time
def compute_svd(urm, K):
U, s, Vt = svds(urm, K) # 这里的s是一个个值,所以下面要转成矩阵的形式
dim = (len(s), len(s))
S = np.zeros(dim, dtype=np.float32)
for i in range(0, len(s)):
S[i, i] = mt.sqrt(s[i])
U = csc_matrix(U, dtype=np.float32)
S = csc_matrix(S, dtype=np.float32)
Vt = csc_matrix(Vt, dtype=np.float32)
return U, S, Vt
def user_item(test): # 得到键值对,用户:借过的书籍
ui = dict(test)
groups = test.groupby(['user_id'])
for item, group in groups:
ui[item] = set(group.loc[:, 'book_id'])
return ui
def Recommend(test_data,user,topk,svd_p_df):
items = list(set(list(test_data['book_id'])))
predictList = [svd_p_df.loc[user,item] for item in items]
series = pd.Series(predictList, index=items)
series = series.sort_values(ascending=False)[:topk]
return series
def recallAndPrecision(test,ui,svd_p_df): # 召回率和准确率
userID = set(test.loc[:, 'user_id'])
hit = 0
recall = 0
precision = 0
for userid in userID:
# trueItem = test[test.ix[:,0] == userid]
# trueItem= trueItem.ix[:,1]
trueItem = ui[userid]
preitem = Recommend(test_data,userid,5,svd_p_df)
for item in list(preitem.index):
if item in trueItem:
hit += 1
recall += len(trueItem)
precision += len(preitem)
return (hit / (recall * 1.0), hit / (precision * 1.0))
start=time.clock()
train_data = pd.read_csv('positive_negtive_data\positive_negtive_data_19_2VS3.csv')
test_data= pd.read_csv('user_book_score_time\\user_book_score_time_19_3VS2.csv',usecols=['user_id','book_id','final_score'])
ui = user_item(test_data)
n_users = train_data.user_id.nunique() # 用户、物品数去重统计
n_items = train_data.book_id.nunique()
print('用户个数为{},图书数目为{}'.format(n_users,n_items))
users = test_data.user_id.nunique() # 用户、物品数去重统计
items = test_data.book_id.nunique()
print('用户个数为{},图书数目为{}'.format(users,items))
train_data_array = train_data.final_score.values
train_row_array = train_data.user_id.values
train_col_array = train_data.book_id.values
test_data_array =test_data.final_score.values
test_row_array = test_data.user_id.values
test_col_array = test_data.book_id.values
train_data_sparse = coo_matrix((train_data_array, (train_row_array, train_col_array)),dtype=float)
test_data_sparse = coo_matrix((test_data_array, (test_row_array, test_col_array)),dtype=float)
n_users_list = train_data.user_id.unique()
print('用户列表为:{}'.format(n_users_list))
K=40
urm = train_data_sparse
MAX_PID = urm.shape[1]
MAX_UID = urm.shape[0]
U, S, Vt = compute_svd(urm, K)
svd_prediction = np.dot(np.dot(U, S), Vt)
svd_p_df = pd.DataFrame(svd_prediction.todense())
recall,precision=recallAndPrecision(test_data,ui,svd_p_df)
print('召回率为:{}'.format(recall))
print('准确率为:{}'.format(precision))
prediction_flatten = svd_prediction[train_data_sparse.todense().nonzero()]
train_data_matrix_flatten = train_data_sparse.todense()[train_data_sparse.todense().nonzero()]
sqrt_train_val=sqrt(mean_squared_error(prediction_flatten, train_data_matrix_flatten))
print('训练数据均方误差为:{}'.format(sqrt_train_val))
test_prediction_flatten = svd_prediction[test_data_sparse.todense().nonzero()]
test_data_matrix_flatten = test_data_sparse.todense()[test_data_sparse.todense().nonzero()]
sqrt_test_val=sqrt(mean_squared_error(test_prediction_flatten, test_data_matrix_flatten))
print('测试数据均方误差为:{}'.format(sqrt_test_val))
duration=time.clock()-start
print('耗费时间:{}'.format(duration))
```
|
github_jupyter
|
from IPython.core.interactiveshell import InteractiveShell
InteractiveShell.ast_node_interactivity = "all"
import pandas as pd
import numpy as np
from scipy.sparse import coo_matrix
import math as mt
from scipy.sparse.linalg import * #used for matrix multiplication
from scipy.sparse.linalg import svds
from scipy.sparse import csc_matrix
from sklearn.metrics import mean_squared_error
from math import sqrt
import time
def compute_svd(urm, K):
U, s, Vt = svds(urm, K) # 这里的s是一个个值,所以下面要转成矩阵的形式
dim = (len(s), len(s))
S = np.zeros(dim, dtype=np.float32)
for i in range(0, len(s)):
S[i, i] = mt.sqrt(s[i])
U = csc_matrix(U, dtype=np.float32)
S = csc_matrix(S, dtype=np.float32)
Vt = csc_matrix(Vt, dtype=np.float32)
return U, S, Vt
def user_item(test): # 得到键值对,用户:借过的书籍
ui = dict(test)
groups = test.groupby(['user_id'])
for item, group in groups:
ui[item] = set(group.loc[:, 'book_id'])
return ui
def Recommend(test_data,user,topk,svd_p_df):
items = list(set(list(test_data['book_id'])))
predictList = [svd_p_df.loc[user,item] for item in items]
series = pd.Series(predictList, index=items)
series = series.sort_values(ascending=False)[:topk]
return series
def recallAndPrecision(test,ui,svd_p_df): # 召回率和准确率
userID = set(test.loc[:, 'user_id'])
hit = 0
recall = 0
precision = 0
for userid in userID:
# trueItem = test[test.ix[:,0] == userid]
# trueItem= trueItem.ix[:,1]
trueItem = ui[userid]
preitem = Recommend(test_data,userid,5,svd_p_df)
for item in list(preitem.index):
if item in trueItem:
hit += 1
recall += len(trueItem)
precision += len(preitem)
return (hit / (recall * 1.0), hit / (precision * 1.0))
start=time.clock()
train_data = pd.read_csv('positive_negtive_data\positive_negtive_data_19_2VS3.csv')
test_data= pd.read_csv('user_book_score_time\\user_book_score_time_19_3VS2.csv',usecols=['user_id','book_id','final_score'])
ui = user_item(test_data)
n_users = train_data.user_id.nunique() # 用户、物品数去重统计
n_items = train_data.book_id.nunique()
print('用户个数为{},图书数目为{}'.format(n_users,n_items))
users = test_data.user_id.nunique() # 用户、物品数去重统计
items = test_data.book_id.nunique()
print('用户个数为{},图书数目为{}'.format(users,items))
train_data_array = train_data.final_score.values
train_row_array = train_data.user_id.values
train_col_array = train_data.book_id.values
test_data_array =test_data.final_score.values
test_row_array = test_data.user_id.values
test_col_array = test_data.book_id.values
train_data_sparse = coo_matrix((train_data_array, (train_row_array, train_col_array)),dtype=float)
test_data_sparse = coo_matrix((test_data_array, (test_row_array, test_col_array)),dtype=float)
n_users_list = train_data.user_id.unique()
print('用户列表为:{}'.format(n_users_list))
K=40
urm = train_data_sparse
MAX_PID = urm.shape[1]
MAX_UID = urm.shape[0]
U, S, Vt = compute_svd(urm, K)
svd_prediction = np.dot(np.dot(U, S), Vt)
svd_p_df = pd.DataFrame(svd_prediction.todense())
recall,precision=recallAndPrecision(test_data,ui,svd_p_df)
print('召回率为:{}'.format(recall))
print('准确率为:{}'.format(precision))
prediction_flatten = svd_prediction[train_data_sparse.todense().nonzero()]
train_data_matrix_flatten = train_data_sparse.todense()[train_data_sparse.todense().nonzero()]
sqrt_train_val=sqrt(mean_squared_error(prediction_flatten, train_data_matrix_flatten))
print('训练数据均方误差为:{}'.format(sqrt_train_val))
test_prediction_flatten = svd_prediction[test_data_sparse.todense().nonzero()]
test_data_matrix_flatten = test_data_sparse.todense()[test_data_sparse.todense().nonzero()]
sqrt_test_val=sqrt(mean_squared_error(test_prediction_flatten, test_data_matrix_flatten))
print('测试数据均方误差为:{}'.format(sqrt_test_val))
duration=time.clock()-start
print('耗费时间:{}'.format(duration))
| 0.347537 | 0.291612 |
```
import rustpycoils as rpc
import magpylib as mag3
import numpy as np
import math
import matplotlib.pyplot as plt
print("Version of magpylib compared : " + mag3.__version__)
```
# Comparsion to Magpylib
The magpylib library offers a wide range of magnetic field primitives such as geometric objects, dipoles and current primitives. The ideal current loop primitive his is the same moduled using a power series by R.H. Jackson and can therfore be directly compared for accuracy and speed in producing solenoidal fields.
### Define the unit wire loop in each module
The 'unit' wire loop taken to be 1m radius with 1A current centered at the origin and aligned along the z axis
```
rustcoil = rpc.AxialSystem()
rustcoil.transform_z()
### Parameters
max_distance = 5
radius = 1
current = 1
length = 5 # As ratio of radius
off_axis = 0.70 # location in radial direction as percentage of radius
tol = 1e-20 #tolerance of rustypy series convergence
rustcoil.add_loop("loop",radius,0.0,current)
unit_loop = mag3.current.Circular(current=1, diameter=2000,position=(0.0,0.0,0.0)) ##magpylib defines in mm
get_magpy_fields = lambda positions: mag3.getB(sources=[unit_loop],observers=[positions])
```
#### Generate list of positions to compare in correct format
```
number_positions= 10000
off_axis_pos = np.full(number_positions,off_axis*radius)
z_positions = np.linspace(-length,length,number_positions)
positions = np.asarray(list(zip(off_axis_pos,np.zeros(number_positions),z_positions)))
```
### Compute magnetic fields
after finding the magnetic fields for each module find the absolute percentage difference in the magnetic fields.
Also computed the absolute percentage difference of the two common definitions of $\mu_0$, i.e. $4\pi \times 10^{-7}$H/m and $1.25663706212 \times 10^{-6}$ H/m which is $\approx -7.2$ in the log based representation plotted below.
```
fields_rusty = rustcoil.get_b(positions,tol)
fields_magpy = get_magpy_fields(positions*1000)*1e-3
difference_percentage_z = np.log10(100*abs(((fields_magpy-fields_rusty)/fields_magpy))) #absolute percentage difference
from scipy.constants import mu_0
mu_difference = np.log10(100*abs(((4*np.pi*1e-7) - (mu_0))/(1.25663706212*1e-6)))
fig, (ax1, ax2) = plt.subplots(1,2,figsize=(12,5),sharey=True,sharex=True)
def plot_error_lines(ax):
for i in range(2,13):
ax.axhline(y=math.log10(100*0.1**i),ls='--',c='orange')
ax.text(s= str(round(0.1**i*100,14))+ '%:',x = length*0.95,y=math.log10(100*0.1**i),backgroundcolor='white',ha='right')
ax1.axhline(y=mu_difference,c='green',label =r"$\mu_0$ error",lw=2,ls='--')
ax1.set_ylim(-11,1)
ax1.set_title('Axial Field')
ax1.plot(z_positions/radius,difference_percentage_z[:,2],color='purple',lw=2)
ax1.axhline(y=mu_difference,c='red')
ax1.set_xlabel(r'z/$r_0$')
ax1.set_ylabel('log(% error)')
plot_error_lines(ax1)
ax2.axhline(y=mu_difference,c='green',label =r"$\mu_0$ error",lw=2,ls='--')
plt.xlim(-length,length)
ax2.set_title('Radial Field')
ax2.plot(z_positions/radius,difference_percentage_z[:,0],color='purple',lw=2)
ax2.set_xlabel(r'z/$r_0$')
plt.legend(loc=3)
plot_error_lines(ax2)
fig.patch.set_facecolor('lightgrey')
plt.savefig('accuracy.png')
```
## Compare 2D
Now will compare the accuracy in a 2 plot with radial positions out to beyond the radius
```
number_grid_positions= 1000
z_positions = np.linspace(-5,5,number_grid_positions)
x_positions = np.linspace(0,2*radius,number_grid_positions)
g0,g1 = np.meshgrid(x_positions,z_positions)
grid_positions = np.asarray(list(zip(g0.ravel(),np.zeros(number_grid_positions**2) ,g1.ravel())))
grid_fields_rusty = rustcoil.get_b(grid_positions,tol)
grid_fields_rusty_radial = grid_fields_rusty[:,0]
grid_fields_rusty_axial = grid_fields_rusty[:,2]
grid_fields_magpy = get_magpy_fields(grid_positions*1000)*1e-3
grid_fields_magpy_radial = grid_fields_magpy[:,0]
grid_fields_magpy_axial = grid_fields_magpy[:,2]
radial_grid_abs_percentage_error=np.log10(100*abs(((grid_fields_rusty_radial-grid_fields_magpy_radial)/grid_fields_rusty_radial)))
axial_grid_abs_percentage_error=np.log10(100*abs(((grid_fields_rusty_axial-grid_fields_magpy_axial)/grid_fields_magpy_axial)))
from mpl_toolkits.axes_grid1 import make_axes_locatable
fig,(ax1,ax2) = plt.subplots(1,2,figsize=(12,5))
ax1.set_title('Axial Field Error')
contourf_ = ax1.contourf(g1,g0,axial_grid_abs_percentage_error.reshape(number_grid_positions,number_grid_positions), levels=range(-8,8),cmap='PuOr',vmax=2,vmin=-8)
divider = make_axes_locatable(ax1)
cax1 = divider.append_axes("right", size="5%", pad=0.05)
cbar = plt.colorbar(contourf_, cax=cax1)
ax1.axhline(y=1.0,c='black',label='radius')
ax1.legend()
ax1.set_ylabel('Radial distance /radius')
ax1.set_xlabel('Axial distance /radius')
ax2.set_title('Radial Field Error')
ax2.set_xlabel('Axial distance /radius')
contourf_2 = ax2.contourf(g1,g0,radial_grid_abs_percentage_error.reshape(number_grid_positions,number_grid_positions), levels=range(-8,8),cmap='PuOr',vmax=2,vmin=-8)
divider = make_axes_locatable(ax2)
cax2 = divider.append_axes("right", size="5%", pad=0.05)
cbar2 = plt.colorbar(contourf_2, cax=cax2)
ax2.axhline(y=1.0,c='black')
fig.patch.set_facecolor('lightgrey')
plt.savefig('accuracy2.png')
```
# Speed Comparisons
Rustycoils can give accurate magnetic field values for the ideal wire loop within a large percentage of the radius, It does not however give accurate field values for this primitive shape near to, or beyond, the radius unless the radial distance is much larger than the radius.
The speed of the algorithim is therefore an important aspect to determine if it is useful for a certain application. The radial positions are kept within 25% of the radius and the axial positions within the radius.
for these tests In this section perform some basic speed tests:
- Speed for single position with Ideal loop
- Speed for many simulataneous positions inside an ideal loop
- Speed for single position inside many Ideal loops (approximating a solenoid)
```
import timeit
def time_single():
import magpylib as mag3
### Parameters
max_distance = 5
radius = 1
current = 1
length = 1 # As ratio of radius how far from coil
off_axis_max = 0.25 # location in radial direction as percentage of radius
tol = 1e-20 # tolerance of rustypy series convergence
rustcoil = rpc.AxialSystem()
rustcoil.transform_z()
rustcoil.add_loop("loop",radius,0.0,current)
unit_loop = mag3.current.Circular(current=1, diameter=2000,position=(0.0,0.0,0.0)) ##magpylib defines in mm
get_magpy_fields = lambda positions: mag3.getB(sources=[unit_loop],observers=[positions]).sum(axis=0)
number_grid_positions= 1
z_positions = np.linspace(-length,length,number_grid_positions)
x_positions = np.linspace(0,off_axis_max*radius,number_grid_positions)
g0,g1 = np.meshgrid(x_positions,z_positions)
grid_positions = np.asarray([0.1,0.1,0.1]).reshape(-1,3)
grid_positions_magpy = np.asarray([100.0,100.0,100.0])
time_rusty = timeit.timeit("rustcoil.get_b(grid_positions,1e-8)",globals=locals(),number=100000)/100000
time_magpy = timeit.timeit("mag3.getB(sources=[unit_loop],observers=[grid_positions_magpy])",globals=locals(),number=100000)/100000
return time_rusty*1e6,time_magpy*1e6
time_rusty_single,time_magpy_single= time_single()
print(time_rusty_single,time_magpy_single)
def time_number_positions(n):
import magpylib as mag3
### Parameters
max_distance = 5
radius = 1
current = 1
length = 1 # As ratio of radius how far from coil
off_axis_max = 0.25 # location in radial direction as percentage of radius
tol = 1e-20 # tolerance of rustypy series convergence
rustcoil = rpc.AxialSystem()
rustcoil.transform_z()
rustcoil.add_loop("loop",radius,0.0,current)
unit_loop = mag3.current.Circular(current=1, diameter=2000,position=(0.0,0.0,0.0)) ##magpylib defines in mm
get_magpy_fields = lambda positions: mag3.getB(sources=[unit_loop],observers=[positions]).sum(axis=0)
number_grid_positions= int(np.sqrt(n))
z_positions = np.linspace(-length,length,number_grid_positions)
x_positions = np.linspace(0,off_axis_max*radius,number_grid_positions)
g0,g1 = np.meshgrid(x_positions,z_positions)
grid_positions = np.asarray(list(zip(g0.ravel(),np.zeros(number_grid_positions**2) ,g1.ravel())))
grid_positions_magpy = np.asarray(list(zip(g0.ravel(),np.zeros(number_grid_positions**2) ,g1.ravel())))*1000
time_rusty = timeit.timeit("rustcoil.get_b(grid_positions,1e-8)",globals=locals(),number=10000)/10000
time_magpy = timeit.timeit("mag3.getB(sources=[unit_loop],observers=[grid_positions_magpy])",globals=locals(),number=10000)/10000
return time_rusty*1e6,time_magpy*1e6
ns_many_pos = np.arange(1,8000,250)
times_rust_many_pos=[]
times_magpy_many_pos=[]
for n in ns_many_pos:
t_r,t_m = time_number_positions(n)
times_rust_many_pos.append(t_r)
times_magpy_many_pos.append(t_m)
plt.scatter(ns_many_pos,times_rust_many_pos,c='orange')
plt.scatter(ns_many_pos,times_magpy_many_pos,c='purple')
plt.plot(ns_many_pos,times_rust_many_pos,c='orange')
plt.plot(ns_many_pos,times_magpy_many_pos,c='purple')
def time_number_positions_more_coils(n,coils):
"""
"""
import magpylib as mag3
### Parameters
max_distance = 5
radius = 1
current = 1
length = 1 # As ratio of radius how far from coil
off_axis_max = 0.25 # location in radial direction as percentage of radius
tol = 1e-20 # tolerance of rustypy series convergence
rustcoil = rpc.AxialSystem()
rustcoil.transform_z()
mags=[]
for c in range(0,coils):
rustcoil.add_loop("loop"+str(c),radius,0.0,current)
mags.append(mag3.current.Circular(current=1, diameter=2000,position=(0.0,0.0,0.0))) ##magpylib defines in mm)
get_magpy_fields = lambda positions: mag3.getB(sources=mags,observers=[positions]).sum(axis=0)
number_grid_positions= int(np.sqrt(n))
z_positions = np.linspace(-length,length,number_grid_positions)
x_positions = np.linspace(0,off_axis_max*radius,number_grid_positions)
g0,g1 = np.meshgrid(x_positions,z_positions)
grid_positions = np.asarray(list(zip(g0.ravel(),np.zeros(number_grid_positions**2) ,g1.ravel())))
grid_positions_magpy = np.asarray(list(zip(g0.ravel(),np.zeros(number_grid_positions**2) ,g1.ravel())))*1000
time_rusty = timeit.timeit("rustcoil.get_b(grid_positions,1e-8)",globals=locals(),number=10000)/10000
time_magpy = timeit.timeit("mag3.getB(sources=mags,observers=[grid_positions_magpy])",globals=locals(),number=10000)/10000
return time_rusty*1e6,time_magpy*1e6
number_coils = np.arange(1,200,10)
times_rust_many_coils=[]
times_magpy_many_coils=[]
for n in number_coils:
t_r,t_m = time_number_positions_more_coils(1,n)
times_rust_many_coils.append(t_r)
times_magpy_many_coils.append(t_m)
plt.scatter(number_coils,times_rust_many_coils,c='orange')
plt.scatter(number_coils,times_magpy_many_coils,c='purple')
plt.plot(number_coils,times_rust_many_coils,c='orange')
plt.plot(number_coils,times_magpy_many_coils,c='purple')
fig,(ax2,ax1) = plt.subplots(1,2,figsize=(12,5))
ax1.plot(number_coils,times_magpy_many_coils,c='purple',label='magpylib')
ax1.scatter(number_coils,times_magpy_many_coils,c='purple')
ax1.plot(number_coils,times_rust_many_coils,c='orange',label='rustpycoils')
ax1.scatter(number_coils,times_rust_many_coils,c='orange')
ax1.legend()
ax1.set_title('Single Positions, Many Wire Loops')
ax1.set_ylabel(r'Time ($\mu s$)')
ax1.set_xlabel('Number of coils')
ax2.plot(ns_many_pos,times_magpy_many_pos,c='purple',label='magpylib')
ax2.scatter(ns_many_pos,times_magpy_many_pos,c='purple')
ax2.plot(ns_many_pos,times_rust_many_pos,c='orange',label='rustpycoils')
ax2.scatter(ns_many_pos,times_rust_many_pos,c='orange')
ax2.set_xlabel('Number of positions')
ax2.set_ylabel(r'Time ($\mu s$)')
ax2.set_title('Many Positions, Single Wire Loop')
ax2.text(100,2000,"rustpycoils: " + str(round(time_rusty_single,2)) + r' $\mu$s')
ax2.text(100,1800,"magpylib: " + str(round(time_magpy_single,2))+ r' $\mu$s')
ax2.arrow(2000, 1700, -2000, -1600)
ax2.arrow(0, 0, 0, 270)
## draw a bbox of the region of the inset axes in the parent axes and
## connecting lines between the bbox and the inset axes area
ax2.legend()
ax1.set_ylabel(r'Time ($\mu s$)')
fig.patch.set_facecolor('lightgrey')
plt.savefig('speed_comparison.png')
```
|
github_jupyter
|
import rustpycoils as rpc
import magpylib as mag3
import numpy as np
import math
import matplotlib.pyplot as plt
print("Version of magpylib compared : " + mag3.__version__)
rustcoil = rpc.AxialSystem()
rustcoil.transform_z()
### Parameters
max_distance = 5
radius = 1
current = 1
length = 5 # As ratio of radius
off_axis = 0.70 # location in radial direction as percentage of radius
tol = 1e-20 #tolerance of rustypy series convergence
rustcoil.add_loop("loop",radius,0.0,current)
unit_loop = mag3.current.Circular(current=1, diameter=2000,position=(0.0,0.0,0.0)) ##magpylib defines in mm
get_magpy_fields = lambda positions: mag3.getB(sources=[unit_loop],observers=[positions])
number_positions= 10000
off_axis_pos = np.full(number_positions,off_axis*radius)
z_positions = np.linspace(-length,length,number_positions)
positions = np.asarray(list(zip(off_axis_pos,np.zeros(number_positions),z_positions)))
fields_rusty = rustcoil.get_b(positions,tol)
fields_magpy = get_magpy_fields(positions*1000)*1e-3
difference_percentage_z = np.log10(100*abs(((fields_magpy-fields_rusty)/fields_magpy))) #absolute percentage difference
from scipy.constants import mu_0
mu_difference = np.log10(100*abs(((4*np.pi*1e-7) - (mu_0))/(1.25663706212*1e-6)))
fig, (ax1, ax2) = plt.subplots(1,2,figsize=(12,5),sharey=True,sharex=True)
def plot_error_lines(ax):
for i in range(2,13):
ax.axhline(y=math.log10(100*0.1**i),ls='--',c='orange')
ax.text(s= str(round(0.1**i*100,14))+ '%:',x = length*0.95,y=math.log10(100*0.1**i),backgroundcolor='white',ha='right')
ax1.axhline(y=mu_difference,c='green',label =r"$\mu_0$ error",lw=2,ls='--')
ax1.set_ylim(-11,1)
ax1.set_title('Axial Field')
ax1.plot(z_positions/radius,difference_percentage_z[:,2],color='purple',lw=2)
ax1.axhline(y=mu_difference,c='red')
ax1.set_xlabel(r'z/$r_0$')
ax1.set_ylabel('log(% error)')
plot_error_lines(ax1)
ax2.axhline(y=mu_difference,c='green',label =r"$\mu_0$ error",lw=2,ls='--')
plt.xlim(-length,length)
ax2.set_title('Radial Field')
ax2.plot(z_positions/radius,difference_percentage_z[:,0],color='purple',lw=2)
ax2.set_xlabel(r'z/$r_0$')
plt.legend(loc=3)
plot_error_lines(ax2)
fig.patch.set_facecolor('lightgrey')
plt.savefig('accuracy.png')
number_grid_positions= 1000
z_positions = np.linspace(-5,5,number_grid_positions)
x_positions = np.linspace(0,2*radius,number_grid_positions)
g0,g1 = np.meshgrid(x_positions,z_positions)
grid_positions = np.asarray(list(zip(g0.ravel(),np.zeros(number_grid_positions**2) ,g1.ravel())))
grid_fields_rusty = rustcoil.get_b(grid_positions,tol)
grid_fields_rusty_radial = grid_fields_rusty[:,0]
grid_fields_rusty_axial = grid_fields_rusty[:,2]
grid_fields_magpy = get_magpy_fields(grid_positions*1000)*1e-3
grid_fields_magpy_radial = grid_fields_magpy[:,0]
grid_fields_magpy_axial = grid_fields_magpy[:,2]
radial_grid_abs_percentage_error=np.log10(100*abs(((grid_fields_rusty_radial-grid_fields_magpy_radial)/grid_fields_rusty_radial)))
axial_grid_abs_percentage_error=np.log10(100*abs(((grid_fields_rusty_axial-grid_fields_magpy_axial)/grid_fields_magpy_axial)))
from mpl_toolkits.axes_grid1 import make_axes_locatable
fig,(ax1,ax2) = plt.subplots(1,2,figsize=(12,5))
ax1.set_title('Axial Field Error')
contourf_ = ax1.contourf(g1,g0,axial_grid_abs_percentage_error.reshape(number_grid_positions,number_grid_positions), levels=range(-8,8),cmap='PuOr',vmax=2,vmin=-8)
divider = make_axes_locatable(ax1)
cax1 = divider.append_axes("right", size="5%", pad=0.05)
cbar = plt.colorbar(contourf_, cax=cax1)
ax1.axhline(y=1.0,c='black',label='radius')
ax1.legend()
ax1.set_ylabel('Radial distance /radius')
ax1.set_xlabel('Axial distance /radius')
ax2.set_title('Radial Field Error')
ax2.set_xlabel('Axial distance /radius')
contourf_2 = ax2.contourf(g1,g0,radial_grid_abs_percentage_error.reshape(number_grid_positions,number_grid_positions), levels=range(-8,8),cmap='PuOr',vmax=2,vmin=-8)
divider = make_axes_locatable(ax2)
cax2 = divider.append_axes("right", size="5%", pad=0.05)
cbar2 = plt.colorbar(contourf_2, cax=cax2)
ax2.axhline(y=1.0,c='black')
fig.patch.set_facecolor('lightgrey')
plt.savefig('accuracy2.png')
import timeit
def time_single():
import magpylib as mag3
### Parameters
max_distance = 5
radius = 1
current = 1
length = 1 # As ratio of radius how far from coil
off_axis_max = 0.25 # location in radial direction as percentage of radius
tol = 1e-20 # tolerance of rustypy series convergence
rustcoil = rpc.AxialSystem()
rustcoil.transform_z()
rustcoil.add_loop("loop",radius,0.0,current)
unit_loop = mag3.current.Circular(current=1, diameter=2000,position=(0.0,0.0,0.0)) ##magpylib defines in mm
get_magpy_fields = lambda positions: mag3.getB(sources=[unit_loop],observers=[positions]).sum(axis=0)
number_grid_positions= 1
z_positions = np.linspace(-length,length,number_grid_positions)
x_positions = np.linspace(0,off_axis_max*radius,number_grid_positions)
g0,g1 = np.meshgrid(x_positions,z_positions)
grid_positions = np.asarray([0.1,0.1,0.1]).reshape(-1,3)
grid_positions_magpy = np.asarray([100.0,100.0,100.0])
time_rusty = timeit.timeit("rustcoil.get_b(grid_positions,1e-8)",globals=locals(),number=100000)/100000
time_magpy = timeit.timeit("mag3.getB(sources=[unit_loop],observers=[grid_positions_magpy])",globals=locals(),number=100000)/100000
return time_rusty*1e6,time_magpy*1e6
time_rusty_single,time_magpy_single= time_single()
print(time_rusty_single,time_magpy_single)
def time_number_positions(n):
import magpylib as mag3
### Parameters
max_distance = 5
radius = 1
current = 1
length = 1 # As ratio of radius how far from coil
off_axis_max = 0.25 # location in radial direction as percentage of radius
tol = 1e-20 # tolerance of rustypy series convergence
rustcoil = rpc.AxialSystem()
rustcoil.transform_z()
rustcoil.add_loop("loop",radius,0.0,current)
unit_loop = mag3.current.Circular(current=1, diameter=2000,position=(0.0,0.0,0.0)) ##magpylib defines in mm
get_magpy_fields = lambda positions: mag3.getB(sources=[unit_loop],observers=[positions]).sum(axis=0)
number_grid_positions= int(np.sqrt(n))
z_positions = np.linspace(-length,length,number_grid_positions)
x_positions = np.linspace(0,off_axis_max*radius,number_grid_positions)
g0,g1 = np.meshgrid(x_positions,z_positions)
grid_positions = np.asarray(list(zip(g0.ravel(),np.zeros(number_grid_positions**2) ,g1.ravel())))
grid_positions_magpy = np.asarray(list(zip(g0.ravel(),np.zeros(number_grid_positions**2) ,g1.ravel())))*1000
time_rusty = timeit.timeit("rustcoil.get_b(grid_positions,1e-8)",globals=locals(),number=10000)/10000
time_magpy = timeit.timeit("mag3.getB(sources=[unit_loop],observers=[grid_positions_magpy])",globals=locals(),number=10000)/10000
return time_rusty*1e6,time_magpy*1e6
ns_many_pos = np.arange(1,8000,250)
times_rust_many_pos=[]
times_magpy_many_pos=[]
for n in ns_many_pos:
t_r,t_m = time_number_positions(n)
times_rust_many_pos.append(t_r)
times_magpy_many_pos.append(t_m)
plt.scatter(ns_many_pos,times_rust_many_pos,c='orange')
plt.scatter(ns_many_pos,times_magpy_many_pos,c='purple')
plt.plot(ns_many_pos,times_rust_many_pos,c='orange')
plt.plot(ns_many_pos,times_magpy_many_pos,c='purple')
def time_number_positions_more_coils(n,coils):
"""
"""
import magpylib as mag3
### Parameters
max_distance = 5
radius = 1
current = 1
length = 1 # As ratio of radius how far from coil
off_axis_max = 0.25 # location in radial direction as percentage of radius
tol = 1e-20 # tolerance of rustypy series convergence
rustcoil = rpc.AxialSystem()
rustcoil.transform_z()
mags=[]
for c in range(0,coils):
rustcoil.add_loop("loop"+str(c),radius,0.0,current)
mags.append(mag3.current.Circular(current=1, diameter=2000,position=(0.0,0.0,0.0))) ##magpylib defines in mm)
get_magpy_fields = lambda positions: mag3.getB(sources=mags,observers=[positions]).sum(axis=0)
number_grid_positions= int(np.sqrt(n))
z_positions = np.linspace(-length,length,number_grid_positions)
x_positions = np.linspace(0,off_axis_max*radius,number_grid_positions)
g0,g1 = np.meshgrid(x_positions,z_positions)
grid_positions = np.asarray(list(zip(g0.ravel(),np.zeros(number_grid_positions**2) ,g1.ravel())))
grid_positions_magpy = np.asarray(list(zip(g0.ravel(),np.zeros(number_grid_positions**2) ,g1.ravel())))*1000
time_rusty = timeit.timeit("rustcoil.get_b(grid_positions,1e-8)",globals=locals(),number=10000)/10000
time_magpy = timeit.timeit("mag3.getB(sources=mags,observers=[grid_positions_magpy])",globals=locals(),number=10000)/10000
return time_rusty*1e6,time_magpy*1e6
number_coils = np.arange(1,200,10)
times_rust_many_coils=[]
times_magpy_many_coils=[]
for n in number_coils:
t_r,t_m = time_number_positions_more_coils(1,n)
times_rust_many_coils.append(t_r)
times_magpy_many_coils.append(t_m)
plt.scatter(number_coils,times_rust_many_coils,c='orange')
plt.scatter(number_coils,times_magpy_many_coils,c='purple')
plt.plot(number_coils,times_rust_many_coils,c='orange')
plt.plot(number_coils,times_magpy_many_coils,c='purple')
fig,(ax2,ax1) = plt.subplots(1,2,figsize=(12,5))
ax1.plot(number_coils,times_magpy_many_coils,c='purple',label='magpylib')
ax1.scatter(number_coils,times_magpy_many_coils,c='purple')
ax1.plot(number_coils,times_rust_many_coils,c='orange',label='rustpycoils')
ax1.scatter(number_coils,times_rust_many_coils,c='orange')
ax1.legend()
ax1.set_title('Single Positions, Many Wire Loops')
ax1.set_ylabel(r'Time ($\mu s$)')
ax1.set_xlabel('Number of coils')
ax2.plot(ns_many_pos,times_magpy_many_pos,c='purple',label='magpylib')
ax2.scatter(ns_many_pos,times_magpy_many_pos,c='purple')
ax2.plot(ns_many_pos,times_rust_many_pos,c='orange',label='rustpycoils')
ax2.scatter(ns_many_pos,times_rust_many_pos,c='orange')
ax2.set_xlabel('Number of positions')
ax2.set_ylabel(r'Time ($\mu s$)')
ax2.set_title('Many Positions, Single Wire Loop')
ax2.text(100,2000,"rustpycoils: " + str(round(time_rusty_single,2)) + r' $\mu$s')
ax2.text(100,1800,"magpylib: " + str(round(time_magpy_single,2))+ r' $\mu$s')
ax2.arrow(2000, 1700, -2000, -1600)
ax2.arrow(0, 0, 0, 270)
## draw a bbox of the region of the inset axes in the parent axes and
## connecting lines between the bbox and the inset axes area
ax2.legend()
ax1.set_ylabel(r'Time ($\mu s$)')
fig.patch.set_facecolor('lightgrey')
plt.savefig('speed_comparison.png')
| 0.479016 | 0.899696 |
# !!THIS IS UNDER PROGRESS!!
## Labeled Faces in the Wild (LFW) experiment
The face features are extracted using face_recognition/dlib library.
```
import requests
import cv2
import face_recognition
import matplotlib.pyplot as plt
import numpy as np
import math
import tarfile
import os
import pandas as pd
# optionally download and unpack Labeled Faces in the Wild (170MB)
face_archive = 'lfw.tgz'
face_dir = 'lfw'
download_faces = False
unpack_faces = False
if download_faces:
print ('Starting download ...')
url = "http://vis-www.cs.umass.edu/lfw/" + face_archive
response = requests.get(url)
if response.status_code == 200:
with open(face_archive, 'wb') as f:
f.write(response.content)
print ('Done.')
if unpack_faces:
print ('Unpacking faces...')
tf = tarfile.open(face_archive)
tf.extractall()
print ('Done.')
def extract_face_encodings(img_path):
# Read the image for face detection
img_for_recognition = face_recognition.load_image_file(img_path)
# Detect faces encodings / feature vector
face_encodings = face_recognition.face_encodings(img_for_recognition)
faces_count = len(face_encodings)
if (faces_count == 0):
print ('No face detected:', img_path)
return None
if faces_count == 1:
# OK
return face_encodings[0]
# print ('More faces detected:', faces_count, ', path:', img_path)
# Detect faces and use encodings for the face with the largest area
faces = face_recognition.face_locations(img_for_recognition)
selected_face_index = 0
max_face_area = 0
for i, face in enumerate(faces):
top, right, bottom, left = face
area = (top - bottom) * (right - left)
if area > max_face_area:
selected_face_index = i
max_face_area = area
return face_encodings[selected_face_index]
def draw_faces(img_path):
# Read the image for face detection
img_for_recognition = face_recognition.load_image_file(img_path)
# Detect faces
faces = face_recognition.face_locations(img_for_recognition)
for face in faces:
top, right, bottom, left = face
# Draw rectangle around face
cv2.rectangle(img_for_recognition, (left, top), (right, bottom), (0, 255, 255), 1)
return img_for_recognition
# e.g. 3 faces - in this case get the face with the largest area
img = draw_faces('lfw/George_Robertson/George_Robertson_0003.jpg')
# Draw original figure with rectangles
plt.figure(figsize=(5, 5))
plt.axis("off")
plt.imshow(cv2.cvtColor(img, cv2.COLOR_RGBA2RGB))
plt.show()
face_encodings = extract_face_encodings('lfw/George_Robertson/George_Robertson_0003.jpg')
# vector of 128 numbers serves as an input to NN
print ('face_encodings array length:', len(face_encodings))
# Process data
person_list = os.listdir(path = face_dir)
all_person_extracted_faces = []
image_count = 0
errors_count = 0
print ('Processing', len(person_list), 'images ...')
for person in person_list:
person_extracted_extracted_face_encodings = []
all_person_extracted_faces.append({'name' : person, 'faces' : person_extracted_extracted_face_encodings})
person_imgs = os.listdir(path = os.path.join(face_dir, person))
for person_img in person_imgs:
img_full_path = os.path.join(face_dir, person, person_img)
extracted_face_encodings = extract_face_encodings(img_full_path)
if extracted_face_encodings is not None:
person_extracted_extracted_face_encodings.append(extracted_face_encodings)
image_count += 1
if len(person_extracted_extracted_face_encodings) == 0:
errors_count += 1
print ('People found: ', len(person_list), ', images found:', image_count, ', errors:', errors_count)
# dlib extracts 128 features
input_dim = 128
print('Input dimension: ', input_dim)
# convert train output data into categories
num_classes = len(person_list)
num_samples = image_count
print('num_classes:', num_classes)
print('num_samples:', num_samples)
# prepare categories
y_train = np.zeros(shape=(num_samples, num_classes), dtype=np.float32)
row_pos = 0 # image
col_pos = 0 # category = person
for person_with_faces in all_person_extracted_faces:
faces = person_with_faces['faces']
faces_count = len(faces)
if faces_count > 0:
for j in range(0, faces_count):
y_train[row_pos, col_pos] = 1.0
row_pos += 1
# next person
col_pos += 1
# prepare train input data
x_train = []
for person_with_faces in all_person_extracted_faces:
face_encodings = person_with_faces['faces']
faces_count = len(faces)
if faces_count > 0:
x_train.extend(face_encodings)
x_train = np.array(x_train)
print('X_train.shape:', x_train.shape)
print('Y_train.shape:', y_train.shape)
from sklearn.model_selection import train_test_split
# split into train/test
X_train, X_test, Y_train, Y_test = train_test_split(x_train, y_train, test_size=0.1, random_state=42)
# Keras (from TensorFlow) imports for building of neural network
from tensorflow.keras import Sequential
from tensorflow.keras.layers import Dense, Dropout, Activation, Flatten
from tensorflow.keras.utils import to_categorical
from tensorflow.keras.models import load_model
from tensorflow.keras.losses import categorical_crossentropy
from tensorflow.keras.optimizers import Adam
from tensorflow.keras.regularizers import l2
# define CNN model for animal images classification
model = Sequential()
model.add(Dense(512, input_dim=input_dim))
model.add(Activation('relu'))
model.add(Dropout(0.2))
model.add(Dense(512))
model.add(Activation('relu'))
model.add(Dropout(0.2))
model.add(Dense(num_classes))
model.add(Activation('softmax'))
model.compile(loss=categorical_crossentropy,
optimizer=Adam(lr=1e-4),
metrics=['accuracy'])
model.summary()
#model.load_weights('lfw1.h5')
# learn NN - tried 100 epochs
epochs = 2
batch_size = 64
history = model.fit(
X_train,
Y_train,
epochs=epochs,
validation_data=(X_test, Y_test),
batch_size=batch_size,
verbose=1
)
model.save_weights('lfw1.h5')
history_dict = history.history
print (history_dict.keys())
loss_values = history_dict['loss']
val_loss_values = history_dict['val_loss']
epochs = range(1, len(loss_values) + 1)
plt.plot(epochs, loss_values, 'b', label='Training loss', color='blue')
plt.plot(epochs, val_loss_values, 'b', label='Validation loss', color='yellow')
plt.title('Training and validation loss')
plt.xlabel('Epochs')
plt.ylabel('Loss')
plt.legend()
plt.show()
accuracy = history_dict['accuracy']
val_accuracy = history_dict['val_accuracy']
plt.plot(epochs, accuracy, 'b', label='Training accuracy', color='red')
plt.plot(epochs, val_accuracy, 'b', label='Validation accuracy', color='green')
plt.title('Training and validation binary accuracy')
plt.xlabel('Epochs')
plt.ylabel('Accuracy')
plt.legend()
plt.show()
# using one of train images (images from other sources are not working is currently not demonstrative)
extracted_face = extract_face_encodings('lfw/Pamela_Anderson/Pamela_Anderson_0004.jpg')
X_single = np.array(extracted_face)
X_single = np.expand_dims(X_single, axis=0)
classes = model.predict_classes(X_single, batch_size=1, verbose=1)
predicted_person = all_person_extracted_faces[classes[0]]
predicted_person_name = predicted_person['name']
predicted_person_faces = predicted_person['faces']
print('It could be:', predicted_person_name)
```
|
github_jupyter
|
import requests
import cv2
import face_recognition
import matplotlib.pyplot as plt
import numpy as np
import math
import tarfile
import os
import pandas as pd
# optionally download and unpack Labeled Faces in the Wild (170MB)
face_archive = 'lfw.tgz'
face_dir = 'lfw'
download_faces = False
unpack_faces = False
if download_faces:
print ('Starting download ...')
url = "http://vis-www.cs.umass.edu/lfw/" + face_archive
response = requests.get(url)
if response.status_code == 200:
with open(face_archive, 'wb') as f:
f.write(response.content)
print ('Done.')
if unpack_faces:
print ('Unpacking faces...')
tf = tarfile.open(face_archive)
tf.extractall()
print ('Done.')
def extract_face_encodings(img_path):
# Read the image for face detection
img_for_recognition = face_recognition.load_image_file(img_path)
# Detect faces encodings / feature vector
face_encodings = face_recognition.face_encodings(img_for_recognition)
faces_count = len(face_encodings)
if (faces_count == 0):
print ('No face detected:', img_path)
return None
if faces_count == 1:
# OK
return face_encodings[0]
# print ('More faces detected:', faces_count, ', path:', img_path)
# Detect faces and use encodings for the face with the largest area
faces = face_recognition.face_locations(img_for_recognition)
selected_face_index = 0
max_face_area = 0
for i, face in enumerate(faces):
top, right, bottom, left = face
area = (top - bottom) * (right - left)
if area > max_face_area:
selected_face_index = i
max_face_area = area
return face_encodings[selected_face_index]
def draw_faces(img_path):
# Read the image for face detection
img_for_recognition = face_recognition.load_image_file(img_path)
# Detect faces
faces = face_recognition.face_locations(img_for_recognition)
for face in faces:
top, right, bottom, left = face
# Draw rectangle around face
cv2.rectangle(img_for_recognition, (left, top), (right, bottom), (0, 255, 255), 1)
return img_for_recognition
# e.g. 3 faces - in this case get the face with the largest area
img = draw_faces('lfw/George_Robertson/George_Robertson_0003.jpg')
# Draw original figure with rectangles
plt.figure(figsize=(5, 5))
plt.axis("off")
plt.imshow(cv2.cvtColor(img, cv2.COLOR_RGBA2RGB))
plt.show()
face_encodings = extract_face_encodings('lfw/George_Robertson/George_Robertson_0003.jpg')
# vector of 128 numbers serves as an input to NN
print ('face_encodings array length:', len(face_encodings))
# Process data
person_list = os.listdir(path = face_dir)
all_person_extracted_faces = []
image_count = 0
errors_count = 0
print ('Processing', len(person_list), 'images ...')
for person in person_list:
person_extracted_extracted_face_encodings = []
all_person_extracted_faces.append({'name' : person, 'faces' : person_extracted_extracted_face_encodings})
person_imgs = os.listdir(path = os.path.join(face_dir, person))
for person_img in person_imgs:
img_full_path = os.path.join(face_dir, person, person_img)
extracted_face_encodings = extract_face_encodings(img_full_path)
if extracted_face_encodings is not None:
person_extracted_extracted_face_encodings.append(extracted_face_encodings)
image_count += 1
if len(person_extracted_extracted_face_encodings) == 0:
errors_count += 1
print ('People found: ', len(person_list), ', images found:', image_count, ', errors:', errors_count)
# dlib extracts 128 features
input_dim = 128
print('Input dimension: ', input_dim)
# convert train output data into categories
num_classes = len(person_list)
num_samples = image_count
print('num_classes:', num_classes)
print('num_samples:', num_samples)
# prepare categories
y_train = np.zeros(shape=(num_samples, num_classes), dtype=np.float32)
row_pos = 0 # image
col_pos = 0 # category = person
for person_with_faces in all_person_extracted_faces:
faces = person_with_faces['faces']
faces_count = len(faces)
if faces_count > 0:
for j in range(0, faces_count):
y_train[row_pos, col_pos] = 1.0
row_pos += 1
# next person
col_pos += 1
# prepare train input data
x_train = []
for person_with_faces in all_person_extracted_faces:
face_encodings = person_with_faces['faces']
faces_count = len(faces)
if faces_count > 0:
x_train.extend(face_encodings)
x_train = np.array(x_train)
print('X_train.shape:', x_train.shape)
print('Y_train.shape:', y_train.shape)
from sklearn.model_selection import train_test_split
# split into train/test
X_train, X_test, Y_train, Y_test = train_test_split(x_train, y_train, test_size=0.1, random_state=42)
# Keras (from TensorFlow) imports for building of neural network
from tensorflow.keras import Sequential
from tensorflow.keras.layers import Dense, Dropout, Activation, Flatten
from tensorflow.keras.utils import to_categorical
from tensorflow.keras.models import load_model
from tensorflow.keras.losses import categorical_crossentropy
from tensorflow.keras.optimizers import Adam
from tensorflow.keras.regularizers import l2
# define CNN model for animal images classification
model = Sequential()
model.add(Dense(512, input_dim=input_dim))
model.add(Activation('relu'))
model.add(Dropout(0.2))
model.add(Dense(512))
model.add(Activation('relu'))
model.add(Dropout(0.2))
model.add(Dense(num_classes))
model.add(Activation('softmax'))
model.compile(loss=categorical_crossentropy,
optimizer=Adam(lr=1e-4),
metrics=['accuracy'])
model.summary()
#model.load_weights('lfw1.h5')
# learn NN - tried 100 epochs
epochs = 2
batch_size = 64
history = model.fit(
X_train,
Y_train,
epochs=epochs,
validation_data=(X_test, Y_test),
batch_size=batch_size,
verbose=1
)
model.save_weights('lfw1.h5')
history_dict = history.history
print (history_dict.keys())
loss_values = history_dict['loss']
val_loss_values = history_dict['val_loss']
epochs = range(1, len(loss_values) + 1)
plt.plot(epochs, loss_values, 'b', label='Training loss', color='blue')
plt.plot(epochs, val_loss_values, 'b', label='Validation loss', color='yellow')
plt.title('Training and validation loss')
plt.xlabel('Epochs')
plt.ylabel('Loss')
plt.legend()
plt.show()
accuracy = history_dict['accuracy']
val_accuracy = history_dict['val_accuracy']
plt.plot(epochs, accuracy, 'b', label='Training accuracy', color='red')
plt.plot(epochs, val_accuracy, 'b', label='Validation accuracy', color='green')
plt.title('Training and validation binary accuracy')
plt.xlabel('Epochs')
plt.ylabel('Accuracy')
plt.legend()
plt.show()
# using one of train images (images from other sources are not working is currently not demonstrative)
extracted_face = extract_face_encodings('lfw/Pamela_Anderson/Pamela_Anderson_0004.jpg')
X_single = np.array(extracted_face)
X_single = np.expand_dims(X_single, axis=0)
classes = model.predict_classes(X_single, batch_size=1, verbose=1)
predicted_person = all_person_extracted_faces[classes[0]]
predicted_person_name = predicted_person['name']
predicted_person_faces = predicted_person['faces']
print('It could be:', predicted_person_name)
| 0.535584 | 0.739728 |
```
import numpy as np
import matplotlib.pyplot as plt
import h5py
import time
import progressbar
def load_wf(filepath):
"""
Load a distinct energy levels
as a numpy array into memory
"""
file = h5py.File(filepath)
if "/numres" in file:
data = np.array(file["/numres"])
file.close()
return data
else:
file.close()
return -1
def load_timestep(filepath,nt,t0=0,):
"""
Load a timestep from a result file
and returns it as a complex numpy array.
"""
file = h5py.File(filepath)
rl = "/dset"+str(nt)+"real"
im = "dset"+str(nt)+"img"
if (rl in file) and (im in file):
imag = np.array(file[rl])
real = np.array(file[im])
res = real + 1j * imag
file.close()
return res
else:
file.close()
return -1
def get_coeff(filepath_stat, filepath_ev, t0, tmax, nt, toff,xmin, xmax, nx):
"""
Returns the absolute square of the evolution coefficients c_n = <psi_n | psi_x >
"""
# get the complex initial wavefunction
psin = load_wf(filepath_stat)*(1+1j*0)
cn = np.zeros(np.int32(nt/toff))
c1 = np.zeros(np.int32(nt/toff))
c2 = np.zeros(np.int32(nt/toff))
c3 = np.zeros(np.int32(nt/toff))
c4 = np.zeros(np.int32(nt/toff))
dx = (xmax-xmin)/nx
with progressbar.ProgressBar(max_value=int(nt)) as bar:
n1 = 1
n2 = 2
for i in range(0, int(nt),toff):
index = np.int32(i/toff)
psik = load_timestep(filepath_ev, i)
bar.update(i)
cn[index] = np.abs(np.trapz(np.conj(psik)*psin,dx=dx))**2
#cn = np.abs(psin)**2
return cn
fig = plt.figure(figsize=(14,10))
plt.subplot(221)
k4 = get_coeff("45_au_s2.h5","../../build/res.h5",0,5000,1e5,100,-45,45,1e5)
t = np.linspace(0, 5000,1e3)
plt.xlabel(r"time $(a.u.)$")
plt.ylabel(r"$|C_2(t)|^2$")
plt.plot(t,k4,label=r"$c_2(t)$")
plt.legend()
plt.subplot(222)
k3 = get_coeff("45_au_s3.h5","../../build/res.h5",0,5000,1e5,100,-45,45,1e5)
plt.xlabel(r"time $(a.u.)$")
plt.ylabel(r"$|C_n(t)|^2$")
plt.plot(t,k3,label=r"$c_3(t)$",c="green")
plt.legend()
plt.subplot(223)
k4 = get_coeff("45_au_s4.h5","../../build/res.h5",0,5000,1e5,100,-45,45,1e5)
plt.xlabel(r"time $(a.u.)$")
plt.ylabel(r"$|C_n(t)|^2$")
plt.plot(t,k4,label=r"$c_4(t)$",c="red")
plt.legend()
plt.subplot(224)
k4 = get_coeff("45_au_s5.h5","../../build/res.h5",0,5000,1e5,100,-45,45,1e5)
plt.xlabel(r"time $(a.u.)$")
plt.ylabel(r"$|C_n(t)|^2$")
plt.plot(t,k4,label=r"$c_5(t)$",c="purple")
plt.legend()
plt.show()
```
|
github_jupyter
|
import numpy as np
import matplotlib.pyplot as plt
import h5py
import time
import progressbar
def load_wf(filepath):
"""
Load a distinct energy levels
as a numpy array into memory
"""
file = h5py.File(filepath)
if "/numres" in file:
data = np.array(file["/numres"])
file.close()
return data
else:
file.close()
return -1
def load_timestep(filepath,nt,t0=0,):
"""
Load a timestep from a result file
and returns it as a complex numpy array.
"""
file = h5py.File(filepath)
rl = "/dset"+str(nt)+"real"
im = "dset"+str(nt)+"img"
if (rl in file) and (im in file):
imag = np.array(file[rl])
real = np.array(file[im])
res = real + 1j * imag
file.close()
return res
else:
file.close()
return -1
def get_coeff(filepath_stat, filepath_ev, t0, tmax, nt, toff,xmin, xmax, nx):
"""
Returns the absolute square of the evolution coefficients c_n = <psi_n | psi_x >
"""
# get the complex initial wavefunction
psin = load_wf(filepath_stat)*(1+1j*0)
cn = np.zeros(np.int32(nt/toff))
c1 = np.zeros(np.int32(nt/toff))
c2 = np.zeros(np.int32(nt/toff))
c3 = np.zeros(np.int32(nt/toff))
c4 = np.zeros(np.int32(nt/toff))
dx = (xmax-xmin)/nx
with progressbar.ProgressBar(max_value=int(nt)) as bar:
n1 = 1
n2 = 2
for i in range(0, int(nt),toff):
index = np.int32(i/toff)
psik = load_timestep(filepath_ev, i)
bar.update(i)
cn[index] = np.abs(np.trapz(np.conj(psik)*psin,dx=dx))**2
#cn = np.abs(psin)**2
return cn
fig = plt.figure(figsize=(14,10))
plt.subplot(221)
k4 = get_coeff("45_au_s2.h5","../../build/res.h5",0,5000,1e5,100,-45,45,1e5)
t = np.linspace(0, 5000,1e3)
plt.xlabel(r"time $(a.u.)$")
plt.ylabel(r"$|C_2(t)|^2$")
plt.plot(t,k4,label=r"$c_2(t)$")
plt.legend()
plt.subplot(222)
k3 = get_coeff("45_au_s3.h5","../../build/res.h5",0,5000,1e5,100,-45,45,1e5)
plt.xlabel(r"time $(a.u.)$")
plt.ylabel(r"$|C_n(t)|^2$")
plt.plot(t,k3,label=r"$c_3(t)$",c="green")
plt.legend()
plt.subplot(223)
k4 = get_coeff("45_au_s4.h5","../../build/res.h5",0,5000,1e5,100,-45,45,1e5)
plt.xlabel(r"time $(a.u.)$")
plt.ylabel(r"$|C_n(t)|^2$")
plt.plot(t,k4,label=r"$c_4(t)$",c="red")
plt.legend()
plt.subplot(224)
k4 = get_coeff("45_au_s5.h5","../../build/res.h5",0,5000,1e5,100,-45,45,1e5)
plt.xlabel(r"time $(a.u.)$")
plt.ylabel(r"$|C_n(t)|^2$")
plt.plot(t,k4,label=r"$c_5(t)$",c="purple")
plt.legend()
plt.show()
| 0.587943 | 0.599485 |
# Word Embeddings
**Learning Objectives**
You will learn:
1. How to use Embedding layer
1. How to create a classification model
1. Compile and train the model
1. How to retrieve the trained word embeddings, save them to disk and visualize it.
## Introduction
This notebook contains an introduction to word embeddings. You will train your own word embeddings using a simple Keras model for a sentiment classification task, and then visualize them in the [Embedding Projector](http://projector.tensorflow.org) (shown in the image below).
<img src="https://github.com/tensorflow/docs/blob/master/site/en/tutorials/text/images/embedding.jpg?raw=1" alt="Screenshot of the embedding projector" width="400"/>
## Representing text as numbers
Machine learning models take vectors (arrays of numbers) as input. When working with text, the first thing you must do is come up with a strategy to convert strings to numbers (or to "vectorize" the text) before feeding it to the model. In this section, you will look at three strategies for doing so.
### One-hot encodings
As a first idea, you might "one-hot" encode each word in your vocabulary. Consider the sentence "The cat sat on the mat". The vocabulary (or unique words) in this sentence is (cat, mat, on, sat, the). To represent each word, you will create a zero vector with length equal to the vocabulary, then place a one in the index that corresponds to the word. This approach is shown in the following diagram.
<img src="https://github.com/tensorflow/docs/blob/master/site/en/tutorials/text/images/one-hot.png?raw=1" alt="Diagram of one-hot encodings" width="400" />
To create a vector that contains the encoding of the sentence, you could then concatenate the one-hot vectors for each word.
Key point: This approach is inefficient. A one-hot encoded vector is sparse (meaning, most indices are zero). Imagine you have 10,000 words in the vocabulary. To one-hot encode each word, you would create a vector where 99.99% of the elements are zero.
### Encode each word with a unique number
A second approach you might try is to encode each word using a unique number. Continuing the example above, you could assign 1 to "cat", 2 to "mat", and so on. You could then encode the sentence "The cat sat on the mat" as a dense vector like [5, 1, 4, 3, 5, 2]. This appoach is efficient. Instead of a sparse vector, you now have a dense one (where all elements are full).
There are two downsides to this approach, however:
* The integer-encoding is arbitrary (it does not capture any relationship between words).
* An integer-encoding can be challenging for a model to interpret. A linear classifier, for example, learns a single weight for each feature. Because there is no relationship between the similarity of any two words and the similarity of their encodings, this feature-weight combination is not meaningful.
### Word embeddings
Word embeddings give us a way to use an efficient, dense representation in which similar words have a similar encoding. Importantly, you do not have to specify this encoding by hand. An embedding is a dense vector of floating point values (the length of the vector is a parameter you specify). Instead of specifying the values for the embedding manually, they are trainable parameters (weights learned by the model during training, in the same way a model learns weights for a dense layer). It is common to see word embeddings that are 8-dimensional (for small datasets), up to 1024-dimensions when working with large datasets. A higher dimensional embedding can capture fine-grained relationships between words, but takes more data to learn.
<img src="https://github.com/tensorflow/docs/blob/master/site/en/tutorials/text/images/embedding2.png?raw=1" alt="Diagram of an embedding" width="400"/>
Above is a diagram for a word embedding. Each word is represented as a 4-dimensional vector of floating point values. Another way to think of an embedding is as "lookup table". After these weights have been learned, you can encode each word by looking up the dense vector it corresponds to in the table.
Each learning objective will correspond to a __#TODO__ in the [student lab notebook](https://github.com/GoogleCloudPlatform/training-data-analyst/blob/master/courses/machine_learning/deepdive2/text_classification/labs/word_embeddings.ipynb) -- try to complete that notebook first before reviewing this solution notebook.
## Setup
```
# Use the chown command to change the ownership of repository to user.
!sudo chown -R jupyter:jupyter /home/jupyter/training-data-analyst
import io
import os
import re
import shutil
import string
import tensorflow as tf
from datetime import datetime
from tensorflow.keras import Model, Sequential
from tensorflow.keras.layers import Activation, Dense, Embedding, GlobalAveragePooling1D
from tensorflow.keras.layers.experimental.preprocessing import TextVectorization
```
This notebook uses TF2.x.
Please check your tensorflow version using the cell below.
```
# Show the currently installed version of TensorFlow
print("TensorFlow version: ",tf.version.VERSION)
```
### Download the IMDb Dataset
You will use the [Large Movie Review Dataset](http://ai.stanford.edu/~amaas/data/sentiment/) through the tutorial. You will train a sentiment classifier model on this dataset and in the process learn embeddings from scratch. To read more about loading a dataset from scratch, see the [Loading text tutorial](../load_data/text.ipynb).
Download the dataset using Keras file utility and take a look at the directories.
```
url = "https://ai.stanford.edu/~amaas/data/sentiment/aclImdb_v1.tar.gz"
dataset = tf.keras.utils.get_file("aclImdb_v1.tar.gz", url,
untar=True, cache_dir='.',
cache_subdir='')
dataset_dir = os.path.join(os.path.dirname(dataset), 'aclImdb')
os.listdir(dataset_dir)
```
Take a look at the `train/` directory. It has `pos` and `neg` folders with movie reviews labelled as positive and negative respectively. You will use reviews from `pos` and `neg` folders to train a binary classification model.
```
train_dir = os.path.join(dataset_dir, 'train')
os.listdir(train_dir)
```
The `train` directory also has additional folders which should be removed before creating training dataset.
```
remove_dir = os.path.join(train_dir, 'unsup')
shutil.rmtree(remove_dir)
```
Next, create a `tf.data.Dataset` using `tf.keras.preprocessing.text_dataset_from_directory`. You can read more about using this utility in this [text classification tutorial](https://www.tensorflow.org/tutorials/keras/text_classification).
Use the `train` directory to create both train and validation datasets with a split of 20% for validation.
```
batch_size = 1024
seed = 123
train_ds = tf.keras.preprocessing.text_dataset_from_directory(
'aclImdb/train', batch_size=batch_size, validation_split=0.2,
subset='training', seed=seed)
val_ds = tf.keras.preprocessing.text_dataset_from_directory(
'aclImdb/train', batch_size=batch_size, validation_split=0.2,
subset='validation', seed=seed)
```
Take a look at a few movie reviews and their labels `(1: positive, 0: negative)` from the train dataset.
```
for text_batch, label_batch in train_ds.take(1):
for i in range(5):
print(label_batch[i].numpy(), text_batch.numpy()[i])
```
### Configure the dataset for performance
These are two important methods you should use when loading data to make sure that I/O does not become blocking.
`.cache()` keeps data in memory after it's loaded off disk. This will ensure the dataset does not become a bottleneck while training your model. If your dataset is too large to fit into memory, you can also use this method to create a performant on-disk cache, which is more efficient to read than many small files.
`.prefetch()` overlaps data preprocessing and model execution while training.
You can learn more about both methods, as well as how to cache data to disk in the [data performance guide](https://www.tensorflow.org/guide/data_performance).
```
AUTOTUNE = tf.data.experimental.AUTOTUNE
train_ds = train_ds.cache().prefetch(buffer_size=AUTOTUNE)
val_ds = val_ds.cache().prefetch(buffer_size=AUTOTUNE)
```
## Using the Embedding layer
Keras makes it easy to use word embeddings. Take a look at the [Embedding](https://www.tensorflow.org/api_docs/python/tf/keras/layers/Embedding) layer.
The Embedding layer can be understood as a lookup table that maps from integer indices (which stand for specific words) to dense vectors (their embeddings). The dimensionality (or width) of the embedding is a parameter you can experiment with to see what works well for your problem, much in the same way you would experiment with the number of neurons in a Dense layer.
```
# Embed a 1,000 word vocabulary into 5 dimensions.
embedding_layer = tf.keras.layers.Embedding(1000, 5)
```
When you create an Embedding layer, the weights for the embedding are randomly initialized (just like any other layer). During training, they are gradually adjusted via backpropagation. Once trained, the learned word embeddings will roughly encode similarities between words (as they were learned for the specific problem your model is trained on).
If you pass an integer to an embedding layer, the result replaces each integer with the vector from the embedding table:
```
result = embedding_layer(tf.constant([1,2,3]))
result.numpy()
```
For text or sequence problems, the Embedding layer takes a 2D tensor of integers, of shape `(samples, sequence_length)`, where each entry is a sequence of integers. It can embed sequences of variable lengths. You could feed into the embedding layer above batches with shapes `(32, 10)` (batch of 32 sequences of length 10) or `(64, 15)` (batch of 64 sequences of length 15).
The returned tensor has one more axis than the input, the embedding vectors are aligned along the new last axis. Pass it a `(2, 3)` input batch and the output is `(2, 3, N)`
```
result = embedding_layer(tf.constant([[0,1,2],[3,4,5]]))
result.shape
```
When given a batch of sequences as input, an embedding layer returns a 3D floating point tensor, of shape `(samples, sequence_length, embedding_dimensionality)`. To convert from this sequence of variable length to a fixed representation there are a variety of standard approaches. You could use an RNN, Attention, or pooling layer before passing it to a Dense layer. This tutorial uses pooling because it's the simplest. The [Text Classification with an RNN](text_classification_rnn.ipynb) tutorial is a good next step.
## Text preprocessing
Next, define the dataset preprocessing steps required for your sentiment classification model. Initialize a TextVectorization layer with the desired parameters to vectorize movie reviews. You can learn more about using this layer in the [Text Classification](https://www.tensorflow.org/tutorials/keras/text_classification) tutorial.
```
# Create a custom standardization function to strip HTML break tags '<br />'.
def custom_standardization(input_data):
lowercase = tf.strings.lower(input_data)
stripped_html = tf.strings.regex_replace(lowercase, '<br />', ' ')
return tf.strings.regex_replace(stripped_html,
'[%s]' % re.escape(string.punctuation), '')
# Vocabulary size and number of words in a sequence.
vocab_size = 10000
sequence_length = 100
# Use the text vectorization layer to normalize, split, and map strings to
# integers. Note that the layer uses the custom standardization defined above.
# Set maximum_sequence length as all samples are not of the same length.
vectorize_layer = TextVectorization(
standardize=custom_standardization,
max_tokens=vocab_size,
output_mode='int',
output_sequence_length=sequence_length)
# Make a text-only dataset (no labels) and call adapt to build the vocabulary.
text_ds = train_ds.map(lambda x, y: x)
vectorize_layer.adapt(text_ds)
```
## Create a classification model
Use the [Keras Sequential API](../../guide/keras) to define the sentiment classification model. In this case it is a "Continuous bag of words" style model.
* The [`TextVectorization`](https://www.tensorflow.org/api_docs/python/tf/keras/layers/experimental/preprocessing/TextVectorization) layer transforms strings into vocabulary indices. You have already initialized `vectorize_layer` as a TextVectorization layer and built it's vocabulary by calling `adapt` on `text_ds`. Now vectorize_layer can be used as the first layer of your end-to-end classification model, feeding tranformed strings into the Embedding layer.
* The [`Embedding`](https://www.tensorflow.org/api_docs/python/tf/keras/layers/Embedding) layer takes the integer-encoded vocabulary and looks up the embedding vector for each word-index. These vectors are learned as the model trains. The vectors add a dimension to the output array. The resulting dimensions are: `(batch, sequence, embedding)`.
* The [`GlobalAveragePooling1D`](https://www.tensorflow.org/api_docs/python/tf/keras/layers/GlobalAveragePooling1D) layer returns a fixed-length output vector for each example by averaging over the sequence dimension. This allows the model to handle input of variable length, in the simplest way possible.
* The fixed-length output vector is piped through a fully-connected ([`Dense`](https://www.tensorflow.org/api_docs/python/tf/keras/layers/Dense)) layer with 16 hidden units.
* The last layer is densely connected with a single output node.
Caution: This model doesn't use masking, so the zero-padding is used as part of the input and hence the padding length may affect the output. To fix this, see the [masking and padding guide](../../guide/keras/masking_and_padding).
```
embedding_dim=16
model = Sequential([
vectorize_layer,
Embedding(vocab_size, embedding_dim, name="embedding"),
GlobalAveragePooling1D(),
Dense(16, activation='relu'),
Dense(1)
])
```
## Compile and train the model
Create a `tf.keras.callbacks.TensorBoard`.
```
tensorboard_callback = tf.keras.callbacks.TensorBoard(log_dir="logs")
```
Compile and train the model using the `Adam` optimizer and `BinaryCrossentropy` loss.
```
model.compile(optimizer='adam',
loss=tf.keras.losses.BinaryCrossentropy(from_logits=True),
metrics=['accuracy'])
model.fit(
train_ds,
validation_data=val_ds,
epochs=10,
callbacks=[tensorboard_callback])
```
With this approach the model reaches a validation accuracy of around 84% (note that the model is overfitting since training accuracy is higher).
Note: Your results may be a bit different, depending on how weights were randomly initialized before training the embedding layer.
You can look into the model summary to learn more about each layer of the model.
```
model.summary()
```
Visualize the model metrics in TensorBoard.
```
!tensorboard --bind_all --port=8081 --logdir logs
```
Run the following command in **Cloud Shell**:
<code>gcloud beta compute ssh --zone <instance-zone> <notebook-instance-name> --project <project-id> -- -L 8081:localhost:8081</code>
Make sure to replace <instance-zone>, <notebook-instance-name> and <project-id>.
In Cloud Shell, click **Web Preview** > **Change Port** and insert port number **8081**. Click **Change and Preview** to open the TensorBoard.

**To quit the TensorBoard, click Kernel > Interrupt kernel**.
## Retrieve the trained word embeddings and save them to disk
Next, retrieve the word embeddings learned during training. The embeddings are weights of the Embedding layer in the model. The weights matrix is of shape `(vocab_size, embedding_dimension)`.
Obtain the weights from the model using `get_layer()` and `get_weights()`. The `get_vocabulary()` function provides the vocabulary to build a metadata file with one token per line.
```
weights = model.get_layer('embedding').get_weights()[0]
vocab = vectorize_layer.get_vocabulary()
```
Write the weights to disk. To use the [Embedding Projector](http://projector.tensorflow.org), you will upload two files in tab separated format: a file of vectors (containing the embedding), and a file of meta data (containing the words).
```
out_v = io.open('vectors.tsv', 'w', encoding='utf-8')
out_m = io.open('metadata.tsv', 'w', encoding='utf-8')
for index, word in enumerate(vocab):
if index == 0: continue # skip 0, it's padding.
vec = weights[index]
out_v.write('\t'.join([str(x) for x in vec]) + "\n")
out_m.write(word + "\n")
out_v.close()
out_m.close()
```
Two files will created as `vectors.tsv` and `metadata.tsv`. Download both files.
```
try:
from google.colab import files
files.download('vectors.tsv')
files.download('metadata.tsv')
except Exception as e:
pass
```
## Visualize the embeddings
To visualize the embeddings, upload them to the embedding projector.
Open the [Embedding Projector](http://projector.tensorflow.org/).
* Click on "Load".
* Upload the two files you created above: `vecs.tsv` and `meta.tsv`.
The embeddings you have trained will now be displayed. You can search for words to find their closest neighbors. For example, try searching for "beautiful". You may see neighbors like "wonderful".
Note: Experimentally, you may be able to produce more interpretable embeddings by using a simpler model. Try deleting the `Dense(16)` layer, retraining the model, and visualizing the embeddings again.
Note: Typically, a much larger dataset is needed to train more interpretable word embeddings. This tutorial uses a small IMDb dataset for the purpose of demonstration.
## Next Steps
This tutorial has shown you how to train and visualize word embeddings from scratch on a small dataset.
* To train word embeddings using Word2Vec algorithm, try the [Word2Vec](https://www.tensorflow.org/tutorials/text/word2vec) tutorial.
* To learn more about advanced text processing, read the [Transformer model for language understanding](https://www.tensorflow.org/tutorials/text/transformer).
|
github_jupyter
|
# Use the chown command to change the ownership of repository to user.
!sudo chown -R jupyter:jupyter /home/jupyter/training-data-analyst
import io
import os
import re
import shutil
import string
import tensorflow as tf
from datetime import datetime
from tensorflow.keras import Model, Sequential
from tensorflow.keras.layers import Activation, Dense, Embedding, GlobalAveragePooling1D
from tensorflow.keras.layers.experimental.preprocessing import TextVectorization
# Show the currently installed version of TensorFlow
print("TensorFlow version: ",tf.version.VERSION)
url = "https://ai.stanford.edu/~amaas/data/sentiment/aclImdb_v1.tar.gz"
dataset = tf.keras.utils.get_file("aclImdb_v1.tar.gz", url,
untar=True, cache_dir='.',
cache_subdir='')
dataset_dir = os.path.join(os.path.dirname(dataset), 'aclImdb')
os.listdir(dataset_dir)
train_dir = os.path.join(dataset_dir, 'train')
os.listdir(train_dir)
remove_dir = os.path.join(train_dir, 'unsup')
shutil.rmtree(remove_dir)
batch_size = 1024
seed = 123
train_ds = tf.keras.preprocessing.text_dataset_from_directory(
'aclImdb/train', batch_size=batch_size, validation_split=0.2,
subset='training', seed=seed)
val_ds = tf.keras.preprocessing.text_dataset_from_directory(
'aclImdb/train', batch_size=batch_size, validation_split=0.2,
subset='validation', seed=seed)
for text_batch, label_batch in train_ds.take(1):
for i in range(5):
print(label_batch[i].numpy(), text_batch.numpy()[i])
AUTOTUNE = tf.data.experimental.AUTOTUNE
train_ds = train_ds.cache().prefetch(buffer_size=AUTOTUNE)
val_ds = val_ds.cache().prefetch(buffer_size=AUTOTUNE)
# Embed a 1,000 word vocabulary into 5 dimensions.
embedding_layer = tf.keras.layers.Embedding(1000, 5)
result = embedding_layer(tf.constant([1,2,3]))
result.numpy()
result = embedding_layer(tf.constant([[0,1,2],[3,4,5]]))
result.shape
# Create a custom standardization function to strip HTML break tags '<br />'.
def custom_standardization(input_data):
lowercase = tf.strings.lower(input_data)
stripped_html = tf.strings.regex_replace(lowercase, '<br />', ' ')
return tf.strings.regex_replace(stripped_html,
'[%s]' % re.escape(string.punctuation), '')
# Vocabulary size and number of words in a sequence.
vocab_size = 10000
sequence_length = 100
# Use the text vectorization layer to normalize, split, and map strings to
# integers. Note that the layer uses the custom standardization defined above.
# Set maximum_sequence length as all samples are not of the same length.
vectorize_layer = TextVectorization(
standardize=custom_standardization,
max_tokens=vocab_size,
output_mode='int',
output_sequence_length=sequence_length)
# Make a text-only dataset (no labels) and call adapt to build the vocabulary.
text_ds = train_ds.map(lambda x, y: x)
vectorize_layer.adapt(text_ds)
embedding_dim=16
model = Sequential([
vectorize_layer,
Embedding(vocab_size, embedding_dim, name="embedding"),
GlobalAveragePooling1D(),
Dense(16, activation='relu'),
Dense(1)
])
tensorboard_callback = tf.keras.callbacks.TensorBoard(log_dir="logs")
model.compile(optimizer='adam',
loss=tf.keras.losses.BinaryCrossentropy(from_logits=True),
metrics=['accuracy'])
model.fit(
train_ds,
validation_data=val_ds,
epochs=10,
callbacks=[tensorboard_callback])
model.summary()
!tensorboard --bind_all --port=8081 --logdir logs
weights = model.get_layer('embedding').get_weights()[0]
vocab = vectorize_layer.get_vocabulary()
out_v = io.open('vectors.tsv', 'w', encoding='utf-8')
out_m = io.open('metadata.tsv', 'w', encoding='utf-8')
for index, word in enumerate(vocab):
if index == 0: continue # skip 0, it's padding.
vec = weights[index]
out_v.write('\t'.join([str(x) for x in vec]) + "\n")
out_m.write(word + "\n")
out_v.close()
out_m.close()
try:
from google.colab import files
files.download('vectors.tsv')
files.download('metadata.tsv')
except Exception as e:
pass
| 0.660063 | 0.990064 |
# DSCI 525 - Web and Cloud Computing
## Milestone 1: Tackling big data on your laptop
### Group #4
### Members: Heidi Ye, Junting He, Kamal MoravejJahromi, Tanmay Sharma
### GitHub Repo: **https://github.com/UBC-MDS/group4-525**
## Loading the libraries
```
import re
import os
import glob
import zipfile
import requests
from urllib.request import urlretrieve
import json
import pandas as pd
from memory_profiler import memory_usage
import dask.dataframe as dd
import pyarrow.feather as feather
import pyarrow.dataset as ds
%load_ext rpy2.ipython
%load_ext memory_profiler
```
#### Note: Code across this lab has been adapted from the DSCI-525 lectures
## 1. Downloading the data
```
# Necessary metadata
article_id = 14096681
url = f"https://api.figshare.com/v2/articles/{article_id}"
headers = {"Content-Type": "application/json"}
output_directory = "figsharerainfall/"
response = requests.request("GET", url, headers=headers)
data = json.loads(response.text)
files = data["files"]
files
```
## 2. Unzipping Data
```
%%time
files_to_dl = ["data.zip"]
for file in files:
if file["name"] in files_to_dl:
os.makedirs(output_directory, exist_ok=True)
urlretrieve(file["download_url"], output_directory + file["name"])
%%time
with zipfile.ZipFile(os.path.join(output_directory, "data.zip"), 'r') as f:
f.extractall(output_directory)
```
## 3. Combining data CSVs
```
df = pd.read_csv("./figsharerainfall/ACCESS-CM2_daily_rainfall_NSW.csv")
df
%%time
%memit
#Merging all the csv files
files = glob.glob('figsharerainfall/*NSW.csv')
df = pd.concat((pd.read_csv(file, index_col=0)
.assign(model=re.findall(r'/([^_]*)', file)[0])
for file in files)
)
df.to_csv("figsharerainfall/combined_data.csv")
```
The following table summerizes the `cpu times` and `Wall time` of combining csv files for all team members.
| | Memory Usage | CPU Time | Wall Time | OS | Memory (RAM) | CPUs |
|---|---|---|---|---|---|---|
| Heidi | 0.04 MiB | 6min 45s | 7min 10s | mac OS Catalina | 16 GB 3733 MHz LPDDR4X | 2 GHz Quad-Core Intel Core i5 |
| Junting | 0.05 MiB | 15min 8s | 15min 36s | mac OS Catalina |16GB of 2400MHz DDR4 | 2.6GHz 6-core Intel Core i7 |
| Kamal | 0.79 MiB | 9min 20s | 9min 43s | Windows 10 Pro | 8 GB DDR4 SDRAM | Intel Core i7-8650U Quad-Core |
| Tanmay | 0.04 MiB | 5min 23s | 5min 28s | mac OS Big Sur | 16 GB 2667 MHz DDR4 | 2.6 GHz 6-Core Intel Core i7 |
- We were able to combine the CSV files using Pandas concat method on both macOS and Windows operating systems and on the machines of all the 4 team-members.
- `Memory usage` ranged from `0.04 MiB` to `0.79 MiB`, `CPU time` from `5min 23s` to `15min 8s` and Wall time from `5min 28s` to `15min 36s`.
- Memory usage and CPU processing times are also impacted by background processes on the individual machines.
```
%%sh
du -sh figsharerainfall/combined_data.csv
```
## 4. Load the combined CSV to memory and perform a simple EDA
```
%%time
%%memit
#loading the entire data to the memory using Pandas
df = pd.read_csv("figsharerainfall/combined_data.csv")
print(df["model"].value_counts())
```
| | Memory Usage | CPU Time | Wall Time | OS | Memory (RAM) | CPUs |
|---|---|---|---|---|---|---|
| Heidi | 2933.94 MiB | 54.9 s | 1min 20s | mac OS Catalina | 16 GB 3733 MHz LPDDR4X | 2 GHz Quad-Core Intel Core i5 |
| Junting | 4149.87 MiB | 2min 54s | 3min 11s | mac OS Catalina |16GB of 2400MHz DDR4 | 2.6GHz 6-core Intel Core i7 |
| Kamal | 1084.17 MiB | 4min 20s | 4min 46s | Windows 10 Pro | 8 GB DDR4 SDRAM | Intel Core i7-8650U Quad-Core |
| Tanmay | 3921.75 MiB | 1min 1s | 1min 4s | mac OS Big Sur | 16 GB 2667 MHz DDR4 | 2.6 GHz 6-Core Intel Core i7 |
- We were able to load the combined CSV files using Pandas read_csv method on both macOS and Windows operating systems and on the machines of all the 4 team-members.
- `Memory usage` ranged from `1084.17 MiB` to `4149.87 MiB`, `CPU time` from `54.9 s` to `4min 20s` and Wall time from `1min 4s` to `4min 46s`.
- Memory usage and CPU processing times are also impacted by background processes on the individual machines.
```
df.head()
#checking datatypes for columns
df.dtypes
```
### 4.1. Investigate approaches to reduce memory usage while performing the EDA
### 4.1.1. Changing dtype of the data and loading just the columns we want
```
print(f"Memory usage with float64: {df[['lat_min','lat_max','rain (mm/day)']].memory_usage().sum() / 1e6:.2f} MB")
print(f"Memory usage with float32: {df[['lat_min','lat_max','rain (mm/day)']].astype('float32', errors='ignore').memory_usage().sum() / 1e6:.2f} MB")
```
### 4.1.2. Loading data in chunks using Pandas
```
%%time
%%memit
counts = pd.Series(dtype=int)
for chunk in pd.read_csv("figsharerainfall/combined_data.csv", chunksize=10_000_000):
counts = counts.add(chunk["model"].value_counts(), fill_value=0)
print(counts.astype(int))
```
| | Memory Usage | CPU Time | Wall Time | OS | Memory (RAM) | CPUs |
|---|---|---|---|---|---|---|
| Heidi | 5967.77 MiB | 53 s | 1min 2s | mac OS Catalina | 16 GB 3733 MHz LPDDR4X | 2 GHz Quad-Core Intel Core i5 |
| Junting |2163.42 | 2min 18s | 2min 19s | mac OS Catalina |16GB of 2400MHz DDR4 | 2.6GHz 6-core Intel Core i7 |
| Kamal | 1619.86 MiB | 1min 54s | 1min 59s | Windows 10 Pro | 8 GB DDR4 SDRAM | Intel Core i7-8650U Quad-Core |
| Tanmay | 1271.02 MiB | 55.6 s | 56.5 s | mac OS Big Sur | 16 GB 2667 MHz DDR4 | 2.6 GHz 6-Core Intel Core i7 |
- We were able to load data in chunks using Pandas on both macOS and Windows operating systems and on the machines of all the 4 team-members..
- `Memory usage` ranged from `1271.02 MiB` to `5967.77 MiB`, `CPU time` from `53 s` to `2min 18s` and Wall time from `56.5 s` to `2min 19s`.
- Memory usage and CPU processing times are also impacted by background processes on the individual machines.
### 4.1.2. Loading data using Dask
```
%%time
%%memit
# Using dask
ddf = dd.read_csv('figsharerainfall/combined_data.csv')
print(ddf["model"].value_counts().compute())
```
| | Memory Usage | CPU Time | Wall Time | OS | Memory (RAM) | CPUs |
|---|---|---|---|---|---|---|
| Heidi | 4714.38 MiB | 1min 32s | 42.6 s | mac OS Catalina | 16 GB 3733 MHz LPDDR4X | 2 GHz Quad-Core Intel Core i5 |
| Junting | 1817.48 MiB | 3min 43s | 1min 19s | mac OS Catalina |16GB of 2400MHz DDR4 | 2.6GHz 6-core Intel Core i7 |
| Kamal | 1690.57 MiB| 2min 24s | 1min 2s | Windows 10 Pro | 8 GB DDR4 SDRAM | Intel Core i7-8650U Quad-Core |
| Tanmay | 1797.21 MiB | 1min 31s | 34.3 s | mac OS Big Sur | 16 GB 2667 MHz DDR4 | 2.6 GHz 6-Core Intel Core i7 |
- We were able to load data using Dask on both macOS and Windows operating systems and on the machines of all the 4 team-members..
- `Memory usage` ranged from `1690.57 MiB` to `4714.38 MiB`, `CPU time` from `1min 31s` to `3min 43s` and Wall time from `34.3 s` to `1min 19s`.
- Memory usage and CPU processing times are also impacted by background processes on the individual machines.
### 4.2. Discuss your observations.
We tried the following approaches to reduce the memory usage while performing the EDA:
1. Changing dtype of the data and loading just the columns we want:
- We loaded only 3 out of the 5 attributes, namely, 'lat_min','lat_max', and 'rain (mm/day)'.
- We changed the data type of these attributes to float32 from the orignial float64.
- Memory usage with float32: 750.17 MB was almost half of the memory usage with float64: 1500.33 MB.
- This validates the hypothesis that using lower data types(float32 vs 64 in this case) leads to more efficient memory usage.
2. Loading data in chunks using Pandas
- We loaded the combined csv file using a chunksize=10_000_000 while performing the EDA.
- We observed a decline in peak memory usage from 8403.29 MiB to 6374.36 MiB.
- Wall Time decreased from 1min 22s to 1min 15s.
- We do not see a significant change in the Wall and CPU times.
- We hypothesize that this impact would be more pronoucned when doing more memory intensive operations in EDA and using smaller chunk sizes would further reduce the memory usage.
3. Loading data using Dask
- We next loaded the combined CSV using a dask object.
- We observed a decline in peak memory usage from 8403.29 MiB to 6610.67 MiB.
- Wall Time decreased significantly from 1min 22s to 46 s.
- We also notice that the CPU time was higher than the wall time (1min 33s vs 46s) suggesting that the CPU was performing operations in parallel.
In summary, loading the entire data (combined_csv) to memory at once has the longest wall time and the highest memory usage as expected. We have looked at three different approaches to load the data more efficiently i.e. loading the entire data using pandas, loading the data in chunks, and loading data using Dask. We conclude that if we want to reduce the memory usage and the processing time, loading with Dask is the best option.
### 5. Perform a simple EDA in R
```
%%time
%%memit
dataset = ds.dataset("figsharerainfall/combined_data.csv", format="csv")
table = dataset.to_table()
%%time
# writing in feather format
feather.write_feather(table, 'figsharerainfall/combined_data.feather')
%%time
%%R
library(arrow)
start_time <- Sys.time()
r_table <- arrow::read_feather("figsharerainfall/combined_data.feather")
print(class(r_table))
library(dplyr)
result <- r_table %>% count(model)
end_time <- Sys.time()
print(result)
print(end_time - start_time)
%%R
library(tidyverse)
r_table <- r_table %>% rename(rain_mmperday = `rain (mm/day)`)
summary_table <- r_table %>%
drop_na() %>%
summarise(median_lat_min = median(lat_min),
median_lat_max = median(lat_max),
median_lon_min = median(lon_min),
median_lon_max = median(lon_max),
median_rain = median(rain_mmperday))
summary_table
```
### 5.1 Discuss why you chose this approach over others
### 5.1.1. Reasons to choose feather
We chose `feather file format` based on the following reasons:
- The `feather` file format is **faster** compared with the `parquet` file and `arrow exchange` while writing files. It writes data with lesser serialization and deserialization that would result in a higher input/output speed. As we can see in our case, the Wall time for writing the feather file is almost half of the parquet file's wall time.
- The `feather` file format can effectively **transfer between python and R programming languages** due to the embedded API that would result in faster reading and writing data using R.
- `Feather` also takes **fewer memories** compared with the CSV file format. We can see that the CSV file takes `5.7 GB` while the feather file format takes `1.1 GB` space.
- We observed that the `partitioned.parquet` and `parquet files` take less space than the `feather` file format. However, the higher speed of writing and reading feather speeds the data queries and analysis.
- `Arrow` only support some operations. The `feather` does not have this limitation.
In summary, feather was selected over Parquet, Pandas Exchange, and Arrow Exchange for its comparatively high I/O speed, minimal memory on disk, and the fact that unpacking is not necessary for the data to be loaded back into RAM. Additionally, feather is relatively easy to use and is a suitable choice since the intent is not term storage.
### 5.1.2. Challenges and discussion
One of biggest challenges associated with this size of data was that computational speed became extremely slow. It was not uncommon for simple tasks to take upwards of 15 minutes. In addition, even after the data was read in, the manipulation of data was still fairly slow. One approach was to read in the data in chunks to minimize the amount of data available at one time. Although this approach may work in some use cases, it is not without its limitations. For example, there may be instances where we need full access to all the data and chunking could result in sampling the data incorrectly. Other alternatives explored in this milestone include changing the dtype of the data as well as loading in data via Dask. Again, this provided some computational savings but likely would not scale well for even larger datasets. To tackle the insufficient memory challenge certain team members ended up deleting certain files and terminating applications to make more memory available.
Another challenge was that runtime did vary from machine to machine. There were instances where the same code could take three or four times longer to run depending on the system being used. This type of inconsistency makes working in this environment fairly unpredictable under tight deadlines. In this milestone, there was no apparent method of overcoming this issue.
|
github_jupyter
|
import re
import os
import glob
import zipfile
import requests
from urllib.request import urlretrieve
import json
import pandas as pd
from memory_profiler import memory_usage
import dask.dataframe as dd
import pyarrow.feather as feather
import pyarrow.dataset as ds
%load_ext rpy2.ipython
%load_ext memory_profiler
# Necessary metadata
article_id = 14096681
url = f"https://api.figshare.com/v2/articles/{article_id}"
headers = {"Content-Type": "application/json"}
output_directory = "figsharerainfall/"
response = requests.request("GET", url, headers=headers)
data = json.loads(response.text)
files = data["files"]
files
%%time
files_to_dl = ["data.zip"]
for file in files:
if file["name"] in files_to_dl:
os.makedirs(output_directory, exist_ok=True)
urlretrieve(file["download_url"], output_directory + file["name"])
%%time
with zipfile.ZipFile(os.path.join(output_directory, "data.zip"), 'r') as f:
f.extractall(output_directory)
df = pd.read_csv("./figsharerainfall/ACCESS-CM2_daily_rainfall_NSW.csv")
df
%%time
%memit
#Merging all the csv files
files = glob.glob('figsharerainfall/*NSW.csv')
df = pd.concat((pd.read_csv(file, index_col=0)
.assign(model=re.findall(r'/([^_]*)', file)[0])
for file in files)
)
df.to_csv("figsharerainfall/combined_data.csv")
%%sh
du -sh figsharerainfall/combined_data.csv
%%time
%%memit
#loading the entire data to the memory using Pandas
df = pd.read_csv("figsharerainfall/combined_data.csv")
print(df["model"].value_counts())
df.head()
#checking datatypes for columns
df.dtypes
print(f"Memory usage with float64: {df[['lat_min','lat_max','rain (mm/day)']].memory_usage().sum() / 1e6:.2f} MB")
print(f"Memory usage with float32: {df[['lat_min','lat_max','rain (mm/day)']].astype('float32', errors='ignore').memory_usage().sum() / 1e6:.2f} MB")
%%time
%%memit
counts = pd.Series(dtype=int)
for chunk in pd.read_csv("figsharerainfall/combined_data.csv", chunksize=10_000_000):
counts = counts.add(chunk["model"].value_counts(), fill_value=0)
print(counts.astype(int))
%%time
%%memit
# Using dask
ddf = dd.read_csv('figsharerainfall/combined_data.csv')
print(ddf["model"].value_counts().compute())
%%time
%%memit
dataset = ds.dataset("figsharerainfall/combined_data.csv", format="csv")
table = dataset.to_table()
%%time
# writing in feather format
feather.write_feather(table, 'figsharerainfall/combined_data.feather')
%%time
%%R
library(arrow)
start_time <- Sys.time()
r_table <- arrow::read_feather("figsharerainfall/combined_data.feather")
print(class(r_table))
library(dplyr)
result <- r_table %>% count(model)
end_time <- Sys.time()
print(result)
print(end_time - start_time)
%%R
library(tidyverse)
r_table <- r_table %>% rename(rain_mmperday = `rain (mm/day)`)
summary_table <- r_table %>%
drop_na() %>%
summarise(median_lat_min = median(lat_min),
median_lat_max = median(lat_max),
median_lon_min = median(lon_min),
median_lon_max = median(lon_max),
median_rain = median(rain_mmperday))
summary_table
| 0.23634 | 0.809916 |
<table class="ee-notebook-buttons" align="left">
<td><a target="_blank" href="https://github.com/giswqs/earthengine-py-notebooks/tree/master/AssetManagement/export_vector.ipynb"><img width=32px src="https://www.tensorflow.org/images/GitHub-Mark-32px.png" /> View source on GitHub</a></td>
<td><a target="_blank" href="https://nbviewer.jupyter.org/github/giswqs/earthengine-py-notebooks/blob/master/AssetManagement/export_vector.ipynb"><img width=26px src="https://upload.wikimedia.org/wikipedia/commons/thumb/3/38/Jupyter_logo.svg/883px-Jupyter_logo.svg.png" />Notebook Viewer</a></td>
<td><a target="_blank" href="https://colab.research.google.com/github/giswqs/earthengine-py-notebooks/blob/master/AssetManagement/export_vector.ipynb"><img src="https://www.tensorflow.org/images/colab_logo_32px.png" /> Run in Google Colab</a></td>
</table>
## Install Earth Engine API and geemap
Install the [Earth Engine Python API](https://developers.google.com/earth-engine/python_install) and [geemap](https://github.com/giswqs/geemap). The **geemap** Python package is built upon the [ipyleaflet](https://github.com/jupyter-widgets/ipyleaflet) and [folium](https://github.com/python-visualization/folium) packages and implements several methods for interacting with Earth Engine data layers, such as `Map.addLayer()`, `Map.setCenter()`, and `Map.centerObject()`.
The following script checks if the geemap package has been installed. If not, it will install geemap, which automatically installs its [dependencies](https://github.com/giswqs/geemap#dependencies), including earthengine-api, folium, and ipyleaflet.
**Important note**: A key difference between folium and ipyleaflet is that ipyleaflet is built upon ipywidgets and allows bidirectional communication between the front-end and the backend enabling the use of the map to capture user input, while folium is meant for displaying static data only ([source](https://blog.jupyter.org/interactive-gis-in-jupyter-with-ipyleaflet-52f9657fa7a)). Note that [Google Colab](https://colab.research.google.com/) currently does not support ipyleaflet ([source](https://github.com/googlecolab/colabtools/issues/60#issuecomment-596225619)). Therefore, if you are using geemap with Google Colab, you should use [`import geemap.eefolium`](https://github.com/giswqs/geemap/blob/master/geemap/eefolium.py). If you are using geemap with [binder](https://mybinder.org/) or a local Jupyter notebook server, you can use [`import geemap`](https://github.com/giswqs/geemap/blob/master/geemap/geemap.py), which provides more functionalities for capturing user input (e.g., mouse-clicking and moving).
```
# Installs geemap package
import subprocess
try:
import geemap
except ImportError:
print('geemap package not installed. Installing ...')
subprocess.check_call(["python", '-m', 'pip', 'install', 'geemap'])
# Checks whether this notebook is running on Google Colab
try:
import google.colab
import geemap.eefolium as geemap
except:
import geemap
# Authenticates and initializes Earth Engine
import ee
try:
ee.Initialize()
except Exception as e:
ee.Authenticate()
ee.Initialize()
```
## Create an interactive map
The default basemap is `Google Maps`. [Additional basemaps](https://github.com/giswqs/geemap/blob/master/geemap/basemaps.py) can be added using the `Map.add_basemap()` function.
```
Map = geemap.Map(center=[40,-100], zoom=4)
Map
```
## Add Earth Engine Python script
```
# Add Earth Engine dataset
```
## Display Earth Engine data layers
```
Map.addLayerControl() # This line is not needed for ipyleaflet-based Map.
Map
```
|
github_jupyter
|
# Installs geemap package
import subprocess
try:
import geemap
except ImportError:
print('geemap package not installed. Installing ...')
subprocess.check_call(["python", '-m', 'pip', 'install', 'geemap'])
# Checks whether this notebook is running on Google Colab
try:
import google.colab
import geemap.eefolium as geemap
except:
import geemap
# Authenticates and initializes Earth Engine
import ee
try:
ee.Initialize()
except Exception as e:
ee.Authenticate()
ee.Initialize()
Map = geemap.Map(center=[40,-100], zoom=4)
Map
# Add Earth Engine dataset
Map.addLayerControl() # This line is not needed for ipyleaflet-based Map.
Map
| 0.5083 | 0.958654 |
```
import time
import os
import pyautogui
sia = str(input("Digite o ano e o mês 'AAMM': ") )
ab = input("Digite 'apac' ou 'bpa': ")
cnes = input("Digite o CNES do estabelecimento: ")
user = "MESTRE"
senha = "TRONCO"
sia2 = str('SIA32'+sia)
cnes2 = str(cnes+'APAC')
os.startfile(sia2)
time.sleep(2)
pyautogui.write(user)
time.sleep(0.2)
pyautogui.press('Enter')
time.sleep(0.2)
pyautogui.write(senha)
time.sleep(0.2)
pyautogui.press('Enter')
if ab == 'bpa':
time.sleep(0.2)
pyautogui.press([ 'Right','Right','Right','Right' ])
time.sleep(0.2)
pyautogui.press('Enter')
time.sleep(0.2)
pyautogui.press(['Down','Down'])
time.sleep(0.5)
pyautogui.press('Enter')
time.sleep(0.2)
pyautogui.press(['Down','Down'])
time.sleep(0.5)
pyautogui.press('Enter')
time.sleep(0.2)
pyautogui.write(cnes)
time.sleep(1)
pyautogui.press('Right')
time.sleep(0.2)
pyautogui.press('Enter')
time.sleep(3)
pyautogui.press(['Delete','Delete','Delete','Delete'])
time.sleep(0.2)
pyautogui.write(cnes)
time.sleep(0.2)
pyautogui.press('Enter')
time.sleep(2)
pyautogui.press(['Esc','Esc','Esc','Esc'])
elif ab == 'apac':
if cnes == '9734872' or cnes == '2019345' or cnes == '5257158' or cnes == '2012677' or cnes == '2019434' or cnes == '2012685' or cnes == '2018098':
time.sleep(0.2)
pyautogui.press([ 'Right','Right','Right','Right' ])
time.sleep(0.2)
pyautogui.press('Enter')
time.sleep(0.2)
pyautogui.press(['Down','Down','Down'])
time.sleep(0.5)
pyautogui.press('Enter')
time.sleep(0.2)
pyautogui.press(['Down'])
time.sleep(0.5)
pyautogui.press('Enter')
time.sleep(0.2)
pyautogui.write(cnes)
time.sleep(1)
pyautogui.press('Right')
time.sleep(0.2)
pyautogui.press('Enter')
time.sleep(3)
pyautogui.press(['Delete','Delete','Delete','Delete'])
time.sleep(0.2)
pyautogui.write(cnes2)
time.sleep(0.2)
pyautogui.press('Enter')
time.sleep(10)
pyautogui.press(['Esc','Esc','Esc','Esc'])
else:
time.sleep(0.2)
pyautogui.press([ 'Right','Right','Right','Right' ])
time.sleep(0.2)
pyautogui.press('Enter')
time.sleep(0.2)
pyautogui.press(['Down','Down','Down'])
time.sleep(0.5)
pyautogui.press('Enter')
time.sleep(0.2)
pyautogui.press(['Down'])
time.sleep(0.5)
pyautogui.press('Enter')
time.sleep(0.2)
pyautogui.write(cnes)
time.sleep(1)
pyautogui.press('Right')
time.sleep(0.2)
pyautogui.press('Enter')
time.sleep(3)
pyautogui.press(['Delete','Delete','Delete','Delete'])
time.sleep(0.2)
pyautogui.write(cnes2)
time.sleep(0.2)
pyautogui.press('Enter')
time.sleep(3)
pyautogui.press(['Esc','Esc','Esc','Esc'])
print("Será que fui feliz?")
```
TRONCO
## o
s.s
tart(sia)
2012030
time.sleep(1)
pyautogui.press('Right')
time.sleep(0.2)
pyautogui.press('Enter')
time.sleep(5)
pyautogui.press(['Delete','Delete','Delete','Delete'])
time.sleep(0.2)
pyautogui.write(cnes)
time.sleep(0.2)
pyautogui.press('Enter')
|
github_jupyter
|
import time
import os
import pyautogui
sia = str(input("Digite o ano e o mês 'AAMM': ") )
ab = input("Digite 'apac' ou 'bpa': ")
cnes = input("Digite o CNES do estabelecimento: ")
user = "MESTRE"
senha = "TRONCO"
sia2 = str('SIA32'+sia)
cnes2 = str(cnes+'APAC')
os.startfile(sia2)
time.sleep(2)
pyautogui.write(user)
time.sleep(0.2)
pyautogui.press('Enter')
time.sleep(0.2)
pyautogui.write(senha)
time.sleep(0.2)
pyautogui.press('Enter')
if ab == 'bpa':
time.sleep(0.2)
pyautogui.press([ 'Right','Right','Right','Right' ])
time.sleep(0.2)
pyautogui.press('Enter')
time.sleep(0.2)
pyautogui.press(['Down','Down'])
time.sleep(0.5)
pyautogui.press('Enter')
time.sleep(0.2)
pyautogui.press(['Down','Down'])
time.sleep(0.5)
pyautogui.press('Enter')
time.sleep(0.2)
pyautogui.write(cnes)
time.sleep(1)
pyautogui.press('Right')
time.sleep(0.2)
pyautogui.press('Enter')
time.sleep(3)
pyautogui.press(['Delete','Delete','Delete','Delete'])
time.sleep(0.2)
pyautogui.write(cnes)
time.sleep(0.2)
pyautogui.press('Enter')
time.sleep(2)
pyautogui.press(['Esc','Esc','Esc','Esc'])
elif ab == 'apac':
if cnes == '9734872' or cnes == '2019345' or cnes == '5257158' or cnes == '2012677' or cnes == '2019434' or cnes == '2012685' or cnes == '2018098':
time.sleep(0.2)
pyautogui.press([ 'Right','Right','Right','Right' ])
time.sleep(0.2)
pyautogui.press('Enter')
time.sleep(0.2)
pyautogui.press(['Down','Down','Down'])
time.sleep(0.5)
pyautogui.press('Enter')
time.sleep(0.2)
pyautogui.press(['Down'])
time.sleep(0.5)
pyautogui.press('Enter')
time.sleep(0.2)
pyautogui.write(cnes)
time.sleep(1)
pyautogui.press('Right')
time.sleep(0.2)
pyautogui.press('Enter')
time.sleep(3)
pyautogui.press(['Delete','Delete','Delete','Delete'])
time.sleep(0.2)
pyautogui.write(cnes2)
time.sleep(0.2)
pyautogui.press('Enter')
time.sleep(10)
pyautogui.press(['Esc','Esc','Esc','Esc'])
else:
time.sleep(0.2)
pyautogui.press([ 'Right','Right','Right','Right' ])
time.sleep(0.2)
pyautogui.press('Enter')
time.sleep(0.2)
pyautogui.press(['Down','Down','Down'])
time.sleep(0.5)
pyautogui.press('Enter')
time.sleep(0.2)
pyautogui.press(['Down'])
time.sleep(0.5)
pyautogui.press('Enter')
time.sleep(0.2)
pyautogui.write(cnes)
time.sleep(1)
pyautogui.press('Right')
time.sleep(0.2)
pyautogui.press('Enter')
time.sleep(3)
pyautogui.press(['Delete','Delete','Delete','Delete'])
time.sleep(0.2)
pyautogui.write(cnes2)
time.sleep(0.2)
pyautogui.press('Enter')
time.sleep(3)
pyautogui.press(['Esc','Esc','Esc','Esc'])
print("Será que fui feliz?")
| 0.078913 | 0.2438 |
# AWS Marketplace Product Usage Demonstration - Model Packages
## Using Model Package ARN with Amazon SageMaker APIs
This sample notebook demonstrates two new functionalities added to Amazon SageMaker:
1. Using a Model Package ARN for inference via Batch Transform jobs / Live Endpoints
2. Using a Marketplace Model Package ARN - we will use [Scikit Decision Trees - Pretrained Model](https://aws.amazon.com/marketplace/pp/prodview-7qop4x5ahrdhe?qid=1543169069960&sr=0-2&ref_=srh_res_product_title)
## Overall flow diagram
<img src="images/ModelPackageE2EFlow.jpg">
## Compatibility
This notebook is compatible only with [Scikit Decision Trees - Pretrained Model](https://aws.amazon.com/marketplace/pp/prodview-7qop4x5ahrdhe?qid=1543169069960&sr=0-2&ref_=srh_res_product_title) sample model that is published to AWS Marketplace
## Set up the environment
```
import sagemaker as sage
from sagemaker import get_execution_role
from sagemaker.serializers import CSVSerializer
role = get_execution_role()
# S3 prefixes
common_prefix = "DEMO-scikit-byo-iris"
batch_inference_input_prefix = common_prefix + "/batch-inference-input-data"
from sagemaker.predictor import Predictor
```
### Create the session
The session remembers our connection parameters to Amazon SageMaker. We'll use it to perform all of our Amazon SageMaker operations.
```
sagemaker_session = sage.Session()
```
## Create Model
Now we use the above Model Package to create a model
```
from src.scikit_product_arns import ScikitArnProvider
modelpackage_arn = ScikitArnProvider.get_model_package_arn(sagemaker_session.boto_region_name)
print("Using model package arn " + modelpackage_arn)
from sagemaker import ModelPackage
model = ModelPackage(
role=role, model_package_arn=modelpackage_arn, sagemaker_session=sagemaker_session
)
```
## Batch Transform Job
Now let's use the model built to run a batch inference job and verify it works.
### Batch Transform Input Preparation
The snippet below is removing the "label" column (column indexed at 0) and retaining the rest to be batch transform's input.
NOTE: This is the same training data, which is a no-no from a statistical/ML science perspective. But the aim of this notebook is to demonstrate how things work end-to-end.
```
import pandas as pd
## Remove first column that contains the label
shape = pd.read_csv("data/training/iris.csv", header=None).drop([0], axis=1)
TRANSFORM_WORKDIR = "data/transform"
shape.to_csv(TRANSFORM_WORKDIR + "/batchtransform_test.csv", index=False, header=False)
transform_input = (
sagemaker_session.upload_data(TRANSFORM_WORKDIR, key_prefix=batch_inference_input_prefix)
+ "/batchtransform_test.csv"
)
print("Transform input uploaded to " + transform_input)
import json
import uuid
transformer = model.transformer(1, "ml.m4.xlarge")
transformer.transform(transform_input, content_type="text/csv")
transformer.wait()
print("Batch Transform output saved to " + transformer.output_path)
```
#### Inspect the Batch Transform Output in S3
```
from urllib.parse import urlparse
parsed_url = urlparse(transformer.output_path)
bucket_name = parsed_url.netloc
file_key = "{}/{}.out".format(parsed_url.path[1:], "batchtransform_test.csv")
s3_client = sagemaker_session.boto_session.client("s3")
response = s3_client.get_object(Bucket=sagemaker_session.default_bucket(), Key=file_key)
response_bytes = response["Body"].read().decode("utf-8")
print(response_bytes)
```
## Live Inference Endpoint
Now we demonstrate the creation of an endpoint for live inference
```
endpoint_name = "scikit-model"
predictor = model.deploy(1, "ml.m4.xlarge", endpoint_name=endpoint_name)
```
### Choose some data and use it for a prediction
In order to do some predictions, we'll extract some of the data we used for training and do predictions against it. This is, of course, bad statistical practice, but a good way to see how the mechanism works.
```
TRAINING_WORKDIR = "data/training"
shape = pd.read_csv(TRAINING_WORKDIR + "/iris.csv", header=None)
import itertools
a = [50 * i for i in range(3)]
b = [40 + i for i in range(10)]
indices = [i + j for i, j in itertools.product(a, b)]
test_data = shape.iloc[indices[:-1]]
test_X = test_data.iloc[:, 1:]
test_y = test_data.iloc[:, 0]
predictor = Predictor(
endpoint_name=endpoint_name, sagemaker_session=None, serializer=CSVSerializer()
)
print(predictor.predict(test_X.values).decode("utf-8"))
```
### Cleanup endpoint
```
model.sagemaker_session.delete_endpoint(endpoint_name)
model.sagemaker_session.delete_endpoint_config(endpoint_name)
model.delete_model()
```
|
github_jupyter
|
import sagemaker as sage
from sagemaker import get_execution_role
from sagemaker.serializers import CSVSerializer
role = get_execution_role()
# S3 prefixes
common_prefix = "DEMO-scikit-byo-iris"
batch_inference_input_prefix = common_prefix + "/batch-inference-input-data"
from sagemaker.predictor import Predictor
sagemaker_session = sage.Session()
from src.scikit_product_arns import ScikitArnProvider
modelpackage_arn = ScikitArnProvider.get_model_package_arn(sagemaker_session.boto_region_name)
print("Using model package arn " + modelpackage_arn)
from sagemaker import ModelPackage
model = ModelPackage(
role=role, model_package_arn=modelpackage_arn, sagemaker_session=sagemaker_session
)
import pandas as pd
## Remove first column that contains the label
shape = pd.read_csv("data/training/iris.csv", header=None).drop([0], axis=1)
TRANSFORM_WORKDIR = "data/transform"
shape.to_csv(TRANSFORM_WORKDIR + "/batchtransform_test.csv", index=False, header=False)
transform_input = (
sagemaker_session.upload_data(TRANSFORM_WORKDIR, key_prefix=batch_inference_input_prefix)
+ "/batchtransform_test.csv"
)
print("Transform input uploaded to " + transform_input)
import json
import uuid
transformer = model.transformer(1, "ml.m4.xlarge")
transformer.transform(transform_input, content_type="text/csv")
transformer.wait()
print("Batch Transform output saved to " + transformer.output_path)
from urllib.parse import urlparse
parsed_url = urlparse(transformer.output_path)
bucket_name = parsed_url.netloc
file_key = "{}/{}.out".format(parsed_url.path[1:], "batchtransform_test.csv")
s3_client = sagemaker_session.boto_session.client("s3")
response = s3_client.get_object(Bucket=sagemaker_session.default_bucket(), Key=file_key)
response_bytes = response["Body"].read().decode("utf-8")
print(response_bytes)
endpoint_name = "scikit-model"
predictor = model.deploy(1, "ml.m4.xlarge", endpoint_name=endpoint_name)
TRAINING_WORKDIR = "data/training"
shape = pd.read_csv(TRAINING_WORKDIR + "/iris.csv", header=None)
import itertools
a = [50 * i for i in range(3)]
b = [40 + i for i in range(10)]
indices = [i + j for i, j in itertools.product(a, b)]
test_data = shape.iloc[indices[:-1]]
test_X = test_data.iloc[:, 1:]
test_y = test_data.iloc[:, 0]
predictor = Predictor(
endpoint_name=endpoint_name, sagemaker_session=None, serializer=CSVSerializer()
)
print(predictor.predict(test_X.values).decode("utf-8"))
model.sagemaker_session.delete_endpoint(endpoint_name)
model.sagemaker_session.delete_endpoint_config(endpoint_name)
model.delete_model()
| 0.305386 | 0.911731 |
```
import sklearn
import sklearn
from sklearn.tree import LinearDecisionTreeRegressor as ldtr
from sklearn.linear_model import LinearRegression
from sklearn.svm import LinearSVR
from sklearn.svm import SVR
from sklearn.tree import DecisionTreeRegressor
from sklearn.linear_model import SGDRegressor
from sklearn.ensemble import RandomForestRegressor
from sklearn.feature_selection import SelectKBest, f_regression
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.metrics import mean_squared_error
from sklearn import datasets
from sklearn.pipeline import make_pipeline
from sklearn.preprocessing import StandardScaler
from sklearn.datasets import make_regression
from sklearn.model_selection import train_test_split
X,y = datasets.load_diabetes(return_X_y=True)
X = X[:, 2].reshape(-1, 1)
X_train = X[:-20]
X_test = X[-20:]
y_train = y[:-20]
y_test = y[-20:]
data= pd.DataFrame(X)
data['10'] = y
data.head()
data.corr()
X, y = make_regression(n_features=4)
X_train, X_test, y_train, y_test = train_test_split(X,y,test_size = 0.2)
m = 150
#np.random.seed(42)
X = np.random.rand(m, 3) * 2
X= np.sort(X, axis= 0)
y = 4 * (X[:, 0] - 0.5) ** 2 + 6 * (X[:, 1] - 0.5)**2 + 2 * (X[:, 2] - 0.5)**5
y = y + np.random.randn(m,) / 10
X_train, X_test, y_train, y_test = train_test_split(X,y,test_size = 0.2)
X_train.shape
reg1 = ldtr(max_depth= 2)
reg2 = LinearRegression(n_jobs = -1)
reg3 = LinearSVR()
reg4 = SVR(kernel = 'rbf')
reg5 = DecisionTreeRegressor(max_depth = 2, min_samples_leaf= 3, min_samples_split= 6)
reg6 = SGDRegressor(n_iter_no_change=250, penalty=None, eta0=0.0001, max_iter=100000)
reg7 = RandomForestRegressor(max_depth = 2,n_jobs = -1)
reg1.fit(X_train, y_train)
reg2.fit(X_train, y_train)
reg3.fit(X_train, y_train)
reg4.fit(X_train, y_train)
reg5.fit(X_train, y_train)
reg6.fit(X_train, y_train)
reg7.fit(X_train, y_train)
y_pred1 = reg1.predict(X_test)
y_pred2 = reg2.predict(X_test)
y_pred3 = reg3.predict(X_test)
y_pred4 = reg4.predict(X_test)
y_pred5 = reg5.predict(X_test)
y_pred6 = reg6.predict(X_test)
y_pred7 = reg7.predict(X_test)
print('Mean squared error: %.2f'% mean_squared_error(y_test, y_pred1))
print('Mean squared error: %.2f'% mean_squared_error(y_test, y_pred2))
print('Mean squared error: %.2f'% mean_squared_error(y_test, y_pred3))
print('Mean squared error: %.2f'% mean_squared_error(y_test, y_pred4))
print('Mean squared error: %.2f'% mean_squared_error(y_test, y_pred5))
print('Mean squared error: %.2f'% mean_squared_error(y_test, y_pred6))
print('Mean squared error: %.2f'% mean_squared_error(y_test, y_pred7))
for i in range(X_train.shape[1]):
plt.scatter(X_train[:, i], y_train, color='black')
plt.scatter(X_train[:, i], reg1.predict(X_train), color='blue')
plt.scatter(X_train[:, i], reg7.predict(X_train), color='green')
plt.show()
for i in range(X_test.shape[1]):
plt.scatter(X_test[:, i], y_test, color='black')
plt.scatter(X_test[:, i], y_pred1, color='blue')
plt.show()
plt.scatter(X_test[:, i], y_test, color='black')
plt.scatter(X_test[:, i],y_pred2, color='orange')
plt.show()
plt.scatter(X_test[:, i], y_test, color='black')
plt.scatter(X_test[:, i],y_pred3, color='red')
plt.show()
plt.scatter(X_test[:, i], y_test, color='black')
plt.scatter(X_test[:, i], y_pred4, color='green')
plt.show()
plt.scatter(X_test[:, i], y_test, color='black')
plt.scatter(X_test[:, i], y_pred5, color='purple')
plt.show()
```
|
github_jupyter
|
import sklearn
import sklearn
from sklearn.tree import LinearDecisionTreeRegressor as ldtr
from sklearn.linear_model import LinearRegression
from sklearn.svm import LinearSVR
from sklearn.svm import SVR
from sklearn.tree import DecisionTreeRegressor
from sklearn.linear_model import SGDRegressor
from sklearn.ensemble import RandomForestRegressor
from sklearn.feature_selection import SelectKBest, f_regression
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.metrics import mean_squared_error
from sklearn import datasets
from sklearn.pipeline import make_pipeline
from sklearn.preprocessing import StandardScaler
from sklearn.datasets import make_regression
from sklearn.model_selection import train_test_split
X,y = datasets.load_diabetes(return_X_y=True)
X = X[:, 2].reshape(-1, 1)
X_train = X[:-20]
X_test = X[-20:]
y_train = y[:-20]
y_test = y[-20:]
data= pd.DataFrame(X)
data['10'] = y
data.head()
data.corr()
X, y = make_regression(n_features=4)
X_train, X_test, y_train, y_test = train_test_split(X,y,test_size = 0.2)
m = 150
#np.random.seed(42)
X = np.random.rand(m, 3) * 2
X= np.sort(X, axis= 0)
y = 4 * (X[:, 0] - 0.5) ** 2 + 6 * (X[:, 1] - 0.5)**2 + 2 * (X[:, 2] - 0.5)**5
y = y + np.random.randn(m,) / 10
X_train, X_test, y_train, y_test = train_test_split(X,y,test_size = 0.2)
X_train.shape
reg1 = ldtr(max_depth= 2)
reg2 = LinearRegression(n_jobs = -1)
reg3 = LinearSVR()
reg4 = SVR(kernel = 'rbf')
reg5 = DecisionTreeRegressor(max_depth = 2, min_samples_leaf= 3, min_samples_split= 6)
reg6 = SGDRegressor(n_iter_no_change=250, penalty=None, eta0=0.0001, max_iter=100000)
reg7 = RandomForestRegressor(max_depth = 2,n_jobs = -1)
reg1.fit(X_train, y_train)
reg2.fit(X_train, y_train)
reg3.fit(X_train, y_train)
reg4.fit(X_train, y_train)
reg5.fit(X_train, y_train)
reg6.fit(X_train, y_train)
reg7.fit(X_train, y_train)
y_pred1 = reg1.predict(X_test)
y_pred2 = reg2.predict(X_test)
y_pred3 = reg3.predict(X_test)
y_pred4 = reg4.predict(X_test)
y_pred5 = reg5.predict(X_test)
y_pred6 = reg6.predict(X_test)
y_pred7 = reg7.predict(X_test)
print('Mean squared error: %.2f'% mean_squared_error(y_test, y_pred1))
print('Mean squared error: %.2f'% mean_squared_error(y_test, y_pred2))
print('Mean squared error: %.2f'% mean_squared_error(y_test, y_pred3))
print('Mean squared error: %.2f'% mean_squared_error(y_test, y_pred4))
print('Mean squared error: %.2f'% mean_squared_error(y_test, y_pred5))
print('Mean squared error: %.2f'% mean_squared_error(y_test, y_pred6))
print('Mean squared error: %.2f'% mean_squared_error(y_test, y_pred7))
for i in range(X_train.shape[1]):
plt.scatter(X_train[:, i], y_train, color='black')
plt.scatter(X_train[:, i], reg1.predict(X_train), color='blue')
plt.scatter(X_train[:, i], reg7.predict(X_train), color='green')
plt.show()
for i in range(X_test.shape[1]):
plt.scatter(X_test[:, i], y_test, color='black')
plt.scatter(X_test[:, i], y_pred1, color='blue')
plt.show()
plt.scatter(X_test[:, i], y_test, color='black')
plt.scatter(X_test[:, i],y_pred2, color='orange')
plt.show()
plt.scatter(X_test[:, i], y_test, color='black')
plt.scatter(X_test[:, i],y_pred3, color='red')
plt.show()
plt.scatter(X_test[:, i], y_test, color='black')
plt.scatter(X_test[:, i], y_pred4, color='green')
plt.show()
plt.scatter(X_test[:, i], y_test, color='black')
plt.scatter(X_test[:, i], y_pred5, color='purple')
plt.show()
| 0.824356 | 0.762247 |
# VacationPy
----
#### Note
* Keep an eye on your API usage. Use https://developers.google.com/maps/reporting/gmp-reporting as reference for how to monitor your usage and billing.
* Instructions have been included for each segment. You do not have to follow them exactly, but they are included to help you think through the steps.
```
# Dependencies and Setup
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
import requests
import gmaps
import os
# Import API key
from api_keys import g_key
```
### Store Part I results into DataFrame
* Load the csv exported in Part I to a DataFrame
```
csv_file = os.path.join('cityweather.csv')
city_df = pd.read_csv(csv_file)
city_df
```
### Humidity Heatmap
* Configure gmaps.
* Use the Lat and Lng as locations and Humidity as the weight.
* Add Heatmap layer to map.
```
gmaps.configure(api_key=g_key)
cities = city_df[['Lat', 'Lng']]
humidity = city_df['Humidity']
heat_map = gmaps.figure(center=(46.0, -5.0), zoom_level=2)
heat_layer = gmaps.heatmap_layer(cities, weights=humidity, dissipating=False)
heat_layer.max_intensity = 100
heat_layer.point_radius=5
heat_map.add_layer(heat_layer)
heat_map
```
### Create new DataFrame fitting weather criteria
* Narrow down the cities to fit weather conditions.
* Drop any rows will null values.
```
ideal_cities_df = city_df[(city_df['Temperature'] < 80) & (city_df['Temperature'] < 70)]
ideal_cities_df = ideal_cities_df[ideal_cities_df['Wind Speed'] < 10]
ideal_cities_df = ideal_cities_df[ideal_cities_df['Clouds'] == 0]
hotel_df = ideal_cities_df
hotel_df
```
### Hotel Map
* Store into variable named `hotel_df`.
* Add a "Hotel Name" column to the DataFrame.
* Set parameters to search for hotels with 5000 meters.
* Hit the Google Places API for each city's coordinates.
* Store the first Hotel result into the DataFrame.
* Plot markers on top of the heatmap.
```
hotel_df['Hotel Name'] = ""
hotel_df
for index, row in hotel_df.iterrows():
try:
base_url = "https://maps.googleapis.com/maps/api/place/nearbysearch/json"
params = {
"keyword": "hotel",
"radius": 5000,
"key": g_key,
}
lat = row['Lat']
lng = row['Lng']
params['location'] = f"{lat}, {lng}"
hotel_data = requests.get(base_url, params=params).json()
hotel_df.loc[index, "Hotel Name"] = hotel_data["results"][0]["name"]
except IndexError:
hotel_df.loc[index, "Hotel Name"] = "NaN"
hotel_df
# NOTE: Do not change any of the code in this cell
# Using the template add the hotel marks to the heatmap
info_box_template = """
<dl>
<dt>Name</dt><dd>{Hotel Name}</dd>
<dt>City</dt><dd>{City}</dd>
<dt>Country</dt><dd>{Country}</dd>
</dl>
"""
# Store the DataFrame Row
# NOTE: be sure to update with your DataFrame name
hotel_info = [info_box_template.format(**row) for index, row in hotel_df.iterrows()]
locations = hotel_df[["Lat", "Lng"]]
# Add marker layer ontop of heat map
markers = gmaps.marker_layer(locations, info_box_content = hotel_info)
heat_map.add_layer(markers)
heat_map
# Display figure
```
|
github_jupyter
|
# Dependencies and Setup
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
import requests
import gmaps
import os
# Import API key
from api_keys import g_key
csv_file = os.path.join('cityweather.csv')
city_df = pd.read_csv(csv_file)
city_df
gmaps.configure(api_key=g_key)
cities = city_df[['Lat', 'Lng']]
humidity = city_df['Humidity']
heat_map = gmaps.figure(center=(46.0, -5.0), zoom_level=2)
heat_layer = gmaps.heatmap_layer(cities, weights=humidity, dissipating=False)
heat_layer.max_intensity = 100
heat_layer.point_radius=5
heat_map.add_layer(heat_layer)
heat_map
ideal_cities_df = city_df[(city_df['Temperature'] < 80) & (city_df['Temperature'] < 70)]
ideal_cities_df = ideal_cities_df[ideal_cities_df['Wind Speed'] < 10]
ideal_cities_df = ideal_cities_df[ideal_cities_df['Clouds'] == 0]
hotel_df = ideal_cities_df
hotel_df
hotel_df['Hotel Name'] = ""
hotel_df
for index, row in hotel_df.iterrows():
try:
base_url = "https://maps.googleapis.com/maps/api/place/nearbysearch/json"
params = {
"keyword": "hotel",
"radius": 5000,
"key": g_key,
}
lat = row['Lat']
lng = row['Lng']
params['location'] = f"{lat}, {lng}"
hotel_data = requests.get(base_url, params=params).json()
hotel_df.loc[index, "Hotel Name"] = hotel_data["results"][0]["name"]
except IndexError:
hotel_df.loc[index, "Hotel Name"] = "NaN"
hotel_df
# NOTE: Do not change any of the code in this cell
# Using the template add the hotel marks to the heatmap
info_box_template = """
<dl>
<dt>Name</dt><dd>{Hotel Name}</dd>
<dt>City</dt><dd>{City}</dd>
<dt>Country</dt><dd>{Country}</dd>
</dl>
"""
# Store the DataFrame Row
# NOTE: be sure to update with your DataFrame name
hotel_info = [info_box_template.format(**row) for index, row in hotel_df.iterrows()]
locations = hotel_df[["Lat", "Lng"]]
# Add marker layer ontop of heat map
markers = gmaps.marker_layer(locations, info_box_content = hotel_info)
heat_map.add_layer(markers)
heat_map
# Display figure
| 0.316475 | 0.855851 |
# Numerical Analysis - 7
###### Rafael Barsotti
#### 1) Implemente o algoritmo dado em sala para obter uma sequência de 1000 números pseudoaleatórios no interval [0, 1]. Plote o resultado obtido e compare com o gráfico obtido pra 1000 pontos uniformes gerados pelo python.
```
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import warnings
warnings.filterwarnings('ignore')
low = 0
high = 1
size = 1000
seed = 3
#Função Random do Python
def rand_py():
x = np.random.uniform(low,high,size)
sns.distplot(x, rug=True, rug_kws={"color": "b"}, kde_kws={"color": "k", "lw": 3, "label": "Python Random"},
hist_kws={"histtype": "step", "linewidth": 3,
"alpha": 1, "color": "b"})
plt.show()
#Função Random da Aula
def rand_aula():
li = seed
x = []
a = 7**5
b = (2**31)-1
for i in range(size):
li = a*li%b
x.append(li/b)
sns.distplot(x, rug=True, rug_kws={"color": "g"}, kde_kws={"color": "k", "lw": 3, "label": "Aula Random"},
hist_kws={"histtype": "step", "linewidth": 3,
"alpha": 1, "color": "g"})
plt.show()
rand_py()
rand_aula()
```
#### 2a) Implementando o método da quadratura de Gauss com três nós, obtenha uma aproximação para $\int_{0}^{2}e^{x^2}$ .Estime o erro da aproximação e compare com o erro obtido usando a integral do polinômio interpolador de grau 2.
```
import math as m
import scipy.integrate as integrate
import scipy.special as special
from scipy.special import erf
def f(x):
return m.e**(x**2)
def Quadratura_Gauss_3():
g = 0
x = [0.5+1/12*m.sqrt(10/3),0.5-1/12*m.sqrt(10/3),0.5+1/12*m.sqrt(10/3),0.5-1/12*m.sqrt(10/3)]
y = [(1-m.sqrt(1/7*(3-4*m.sqrt(0.3)))),(1-m.sqrt(1/7*(3+4*m.sqrt(0.3)))),(1+m.sqrt(1/7*(3-4*m.sqrt(0.3)))),
(1+m.sqrt(1/7*(3+4*m.sqrt(0.3))))]
for i in range(4):
g += m.e**-(y[i]**2) * x[i]
return(g)
# ???
def Poli2():
p2 = erf(2)*m.sqrt(m.pi)/2
return(p2)
def Error_Gauss():
a = Poli2()
b = Quadratura_Gauss_3()
e = abs(a-b)
return e
e = Error_Gauss()
print('O Erro estimado da Quadratura de Gauss foi {}.'.format(e))
```
#### 2b) Implementando o MCMC obtenha uma aproximação para a integral do item anterior. Qual é o valor de $n$ necessário para obter o mesmo erro obtido pela regra de Simpson? Qual é o valorde $n$ necessário para obter o mesmo erro obtido pelo método de Gauss?
```
def MCMC_N_to_Gauss():
int_real = Poli2()
n = 1
e_MCMC = 100
while > e:
x = np.linspace(0, 2, n)
y = f(x)
a = y * 2/n
int_MCMC = sum(a)
e_MCMC = abs(int_real - int_MCMC)
n += 1
print('N de MCMC para error menor ou igual ao Gauss {}.'.format(e_MC))
MCMC_N_to_Gauss()
def MCMC_N_to_Simpson():
n = 1
e_MCMC = 100
while erroMC2 > 0.13333333:
x = np.linspace(0, 2, n)
y = f(x)
a = y * 2/n
int_MCMC = sum(a)
e_MCMC = abs(int_real - int_MCMC)
n += 1
print('N de MCMC para error menor ou igual ao Gauss {}.'.format(e_MC))
```
|
github_jupyter
|
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import warnings
warnings.filterwarnings('ignore')
low = 0
high = 1
size = 1000
seed = 3
#Função Random do Python
def rand_py():
x = np.random.uniform(low,high,size)
sns.distplot(x, rug=True, rug_kws={"color": "b"}, kde_kws={"color": "k", "lw": 3, "label": "Python Random"},
hist_kws={"histtype": "step", "linewidth": 3,
"alpha": 1, "color": "b"})
plt.show()
#Função Random da Aula
def rand_aula():
li = seed
x = []
a = 7**5
b = (2**31)-1
for i in range(size):
li = a*li%b
x.append(li/b)
sns.distplot(x, rug=True, rug_kws={"color": "g"}, kde_kws={"color": "k", "lw": 3, "label": "Aula Random"},
hist_kws={"histtype": "step", "linewidth": 3,
"alpha": 1, "color": "g"})
plt.show()
rand_py()
rand_aula()
import math as m
import scipy.integrate as integrate
import scipy.special as special
from scipy.special import erf
def f(x):
return m.e**(x**2)
def Quadratura_Gauss_3():
g = 0
x = [0.5+1/12*m.sqrt(10/3),0.5-1/12*m.sqrt(10/3),0.5+1/12*m.sqrt(10/3),0.5-1/12*m.sqrt(10/3)]
y = [(1-m.sqrt(1/7*(3-4*m.sqrt(0.3)))),(1-m.sqrt(1/7*(3+4*m.sqrt(0.3)))),(1+m.sqrt(1/7*(3-4*m.sqrt(0.3)))),
(1+m.sqrt(1/7*(3+4*m.sqrt(0.3))))]
for i in range(4):
g += m.e**-(y[i]**2) * x[i]
return(g)
# ???
def Poli2():
p2 = erf(2)*m.sqrt(m.pi)/2
return(p2)
def Error_Gauss():
a = Poli2()
b = Quadratura_Gauss_3()
e = abs(a-b)
return e
e = Error_Gauss()
print('O Erro estimado da Quadratura de Gauss foi {}.'.format(e))
def MCMC_N_to_Gauss():
int_real = Poli2()
n = 1
e_MCMC = 100
while > e:
x = np.linspace(0, 2, n)
y = f(x)
a = y * 2/n
int_MCMC = sum(a)
e_MCMC = abs(int_real - int_MCMC)
n += 1
print('N de MCMC para error menor ou igual ao Gauss {}.'.format(e_MC))
MCMC_N_to_Gauss()
def MCMC_N_to_Simpson():
n = 1
e_MCMC = 100
while erroMC2 > 0.13333333:
x = np.linspace(0, 2, n)
y = f(x)
a = y * 2/n
int_MCMC = sum(a)
e_MCMC = abs(int_real - int_MCMC)
n += 1
print('N de MCMC para error menor ou igual ao Gauss {}.'.format(e_MC))
| 0.306942 | 0.920469 |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.