code
stringlengths 2.5k
6.36M
| kind
stringclasses 2
values | parsed_code
stringlengths 0
404k
| quality_prob
float64 0
0.98
| learning_prob
float64 0.03
1
|
|---|---|---|---|---|
# Benchmarking
In this tutorial we will compare DaCe with other popular Python-accelerating libraries. The NumPy results should be a bit faster if an optimized version is installed (for example, compiled with Intel MKL).
**NOTE**: Running this notebook on a VM/cloud instance may run out of memory and crash the Jupyter kernel, due to inefficiency of the other frameworks. In that case, rerun the cells in [Dependencies](#Dependencies) and continue.
Table of Contents:
* [Dependencies](#Dependencies)
* [Simple programs](#Simple-programs-with-multiple-operators)
* [Loops](#Loops)
* [Varying sizes](#Varying-sizes)
* [Auto-parallelization](#Auto-parallelization)
* [Example: 3D Heat Diffusion](#3D-Heat-Diffusion)
* [Benchmarking and Instrumentation API](#Benchmarking-and-Instrumentation-API)
TL;DR DaCe is fast:

## Dependencies
First, let's make sure we have all the frameworks ready to go:
```
%pip install jax jaxlib
%pip install numba
%pip install pythran
# Your library here
# MKL for performance
%conda install mkl mkl-include mkl-devel
# matplotlib to draw the results
%pip install matplotlib
# Setup code for plotting
import matplotlib.pyplot as plt
def barplot(title, labels=False):
x = ['numpy'] + list(sorted(TIMES.keys() - {'numpy'}))
bars = [np.median(TIMES[key].timings) for key in x]
yerr = [np.std(TIMES[key].timings) for key in x]
color = [('#86add9' if 'dace' in key else 'salmon') for key in x]
p = plt.bar(x, bars, yerr=yerr, color=color)
plt.ylabel('Runtime [s]'); plt.xlabel('Implementation'); plt.title(title);
if labels:
plt.gca().bar_label(p)
pass
# Setup code for benchmarked frameworks
import numpy as np
import jax
import numba
import dace
# Pythran loads in a separate cell
%load_ext pythran.magic
```
## Simple programs with multiple operators
Let's start with a basic program with three different operations. This example program was taken from the [JAX README](https://github.com/google/jax#compilation-with-jit):
```
def slow_f(x):
return x * x + x * 2.0
```
First, let's measure the performance of NumPy as-is on this function:
```
a = np.random.rand(5000, 5000)
TIMES = {}
TIMES['numpy'] = %timeit -o slow_f(a)
```
Now we can construct Just-In-Time (JIT) compiled versions of this function, for each framework:
```
jax_f = jax.jit(slow_f)
numba_f = numba.jit(slow_f)
dace_f = dace.program(auto_optimize=True)(slow_f)
%%pythran
#pythran export pythran_f(float64[:,:])
def pythran_f(x):
return x * x + x * 2.0
```
Before we measure the time, we will run the functions first as a warmup, to allow compilers to run JIT compilation:
```
# On your marks...
%timeit -r 1 -n 1 jax_f(a).block_until_ready()
%timeit -r 1 -n 1 numba_f(a)
%timeit -r 1 -n 1 dace_f(a)
%timeit -r 1 -n 1 pythran_f(a)
pass
# ...get set...
# ...Go!
TIMES['jax'] = %timeit -o jax_f(a).block_until_ready()
TIMES['numba'] = %timeit -o numba_f(a)
TIMES['pythran'] = %timeit -o pythran_f(a)
TIMES['dace_jit'] = %timeit -o dace_f(a)
```
You could also precompile the program for faster runtimes (be aware that the return value is retained across calls!):
```
# Either provide type annotations on the `@dace.program`, or call `compile` with sample arguments
cprog = dace_f.compile(a)
TIMES['dace'] = %timeit -o cprog(a)
```
We can now plot the results:
```
barplot('Simple program, multiple operators')
```
## Loops
Here we test how interpreter overhead can be mitigated by the Python compiling frameworks. Let's take another application from Numba's [5 minute guide](https://numba.readthedocs.io/en/stable/user/5minguide.html):
```
def go_fast(a):
trace = 0.0
for i in range(a.shape[0]):
trace += np.tanh(a[i, i])
return a + trace
import numpy as np
b = np.random.rand(1000, 1000)
TIMES = {}
TIMES['numpy'] = %timeit -o go_fast(b)
numba_fast = numba.jit(go_fast)
import jax.numpy as jnp
@jax.jit
def jax_fast(a):
trace = 0.0
for i in range(a.shape[0]):
trace += jnp.tanh(a[i, i])
return a + trace
N = dace.symbol('N')
@dace.program(auto_optimize=True)
def dace_fast(a: dace.float64[N, N]):
trace = 0.0
for i in range(N):
trace += np.tanh(a[i, i])
return a + trace
%%pythran
from numpy import tanh
#pythran export pythran_fast(float64[:,:])
def pythran_fast(a):
trace = 0.0
for i in range(a.shape[0]):
trace += tanh(a[i, i])
return a + trace
import time
start = time.time()
csdfg = dace_fast.compile(b)
print('DaCe compilation time:', time.time() - start, 'seconds')
%timeit -r 1 -n 1 jax_fast(b).block_until_ready()
%timeit -r 1 -n 1 numba_fast(b)
%timeit -r 1 -n 1 pythran_fast(b)
```
Note that the slow JAX first run time is due to the inspector/executor model, in which the compilation time depends on the size of the array.
```
TIMES['jax'] = %timeit -o jax_fast(b).block_until_ready()
TIMES['numba'] = %timeit -o numba_fast(b)
TIMES['pythran'] = %timeit -o pythran_fast(b)
TIMES['dace'] = %timeit -o csdfg(b, N=b.shape[0])
barplot('Loops')
```
### Varying sizes
Since the DaCe program was defined symbolically, the input array size can be changed without recompilation:
```
sizes = [np.random.randint(700, 5000) for _ in range(10)]
arrays = [np.random.rand(n, n) for n in sizes]
def vary_size(call):
for a in arrays:
call(a)
def vary_size_dace(call):
for a, n in zip(arrays, sizes):
call(a, N=n)
def vary_size_jax(call):
for a in arrays:
call(a).block_until_ready()
TIMES = {}
TIMES['numpy'] = %timeit -o vary_size(go_fast)
TIMES['numba'] = %timeit -o vary_size(numba_fast)
TIMES['pythran'] = %timeit -o vary_size(pythran_fast)
TIMES['dace'] = %timeit -o vary_size_dace(csdfg)
TIMES['jax'] = %timeit -o vary_size_jax(jax_fast)
barplot('Loop - Varying sizes')
```
## Auto-parallelization
DaCe can use data-centric dependency analysis to not only track and reduce data movement, but also automatically extract parallel regions in code. Here we look at a simple program and how it is run in parallel. We use the `auto_optimize` flag in the `dace.program` decorator to automatically apply optimization heuristics.
```
def element_update(a):
return a * 5
def someforloop(A):
for i in range(A.shape[0]):
for j in range(A.shape[1]):
A[i, j] = element_update(A[i, j])
a = np.random.rand(1000, 1000)
daceloop = dace.program(auto_optimize=True)(someforloop)
```
Here it is compared with numpy and numba's similar capability:
```
numbaloop = numba.jit(parallel=True)(someforloop)
csdfg = daceloop.compile(a)
TIMES = {}
TIMES['numpy'] = %timeit -o someforloop(a)
TIMES['numba'] = %timeit -o numbaloop(a)
TIMES['dace'] = %timeit -o csdfg(a)
barplot('Automatic parallelization', labels=True)
```
As we can see, the nested call triggered the numba code to stay sequential, whereas the global data dependency analysis in DaCe allowed it to parallelize the code, yielding a performance of **549 µs** vs. 406 ms.
## 3D Heat Diffusion
As a more realistic application, the following program, `heat3d` is taken from the [NPBench numpy benchmark](https://github.com/spcl/npbench). It runs a three-dimensional stencil repeatedly to perform heat diffusion:
```
def heat3d(TSTEPS, A, B):
for t in range(1, TSTEPS):
B[1:-1, 1:-1,
1:-1] = (0.125 * (A[2:, 1:-1, 1:-1] - 2.0 * A[1:-1, 1:-1, 1:-1] +
A[:-2, 1:-1, 1:-1]) + 0.125 *
(A[1:-1, 2:, 1:-1] - 2.0 * A[1:-1, 1:-1, 1:-1] +
A[1:-1, :-2, 1:-1]) + 0.125 *
(A[1:-1, 1:-1, 2:] - 2.0 * A[1:-1, 1:-1, 1:-1] +
A[1:-1, 1:-1, 0:-2]) + A[1:-1, 1:-1, 1:-1])
A[1:-1, 1:-1,
1:-1] = (0.125 * (B[2:, 1:-1, 1:-1] - 2.0 * B[1:-1, 1:-1, 1:-1] +
B[:-2, 1:-1, 1:-1]) + 0.125 *
(B[1:-1, 2:, 1:-1] - 2.0 * B[1:-1, 1:-1, 1:-1] +
B[1:-1, :-2, 1:-1]) + 0.125 *
(B[1:-1, 1:-1, 2:] - 2.0 * B[1:-1, 1:-1, 1:-1] +
B[1:-1, 1:-1, 0:-2]) + B[1:-1, 1:-1, 1:-1])
# Using the "L" size
TSTEPS, N = 100, 70
A = np.fromfunction(lambda i, j, k: (i + j + (N - k)) * 10 / N, (N, N, N),
dtype=np.float64)
B = np.copy(A)
dace_heat3d = dace.program(auto_optimize=True)(heat3d)
numba_heat3d = numba.jit(nopython=True, parallel=True)(heat3d)
%%pythran
#pythran export pythran_heat3d(int, float64[:,:,:], float64[:,:,:])
def pythran_heat3d(TSTEPS, A, B):
for t in range(1, TSTEPS):
B[1:-1, 1:-1,
1:-1] = (0.125 * (A[2:, 1:-1, 1:-1] - 2.0 * A[1:-1, 1:-1, 1:-1] +
A[:-2, 1:-1, 1:-1]) + 0.125 *
(A[1:-1, 2:, 1:-1] - 2.0 * A[1:-1, 1:-1, 1:-1] +
A[1:-1, :-2, 1:-1]) + 0.125 *
(A[1:-1, 1:-1, 2:] - 2.0 * A[1:-1, 1:-1, 1:-1] +
A[1:-1, 1:-1, 0:-2]) + A[1:-1, 1:-1, 1:-1])
A[1:-1, 1:-1,
1:-1] = (0.125 * (B[2:, 1:-1, 1:-1] - 2.0 * B[1:-1, 1:-1, 1:-1] +
B[:-2, 1:-1, 1:-1]) + 0.125 *
(B[1:-1, 2:, 1:-1] - 2.0 * B[1:-1, 1:-1, 1:-1] +
B[1:-1, :-2, 1:-1]) + 0.125 *
(B[1:-1, 1:-1, 2:] - 2.0 * B[1:-1, 1:-1, 1:-1] +
B[1:-1, 1:-1, 0:-2]) + B[1:-1, 1:-1, 1:-1])
# Warmup
%timeit -r 1 -n 1 dace_heat3d(TSTEPS, A, B)
%timeit -r 1 -n 1 numba_heat3d(TSTEPS, A, B)
%timeit -r 1 -n 1 pythran_heat3d(TSTEPS, A, B)
TIMES = {}
TIMES['numpy'] = %timeit -o heat3d(TSTEPS, A, B)
TIMES['dace'] = %timeit -o dace_heat3d(TSTEPS, A, B)
TIMES['numba'] = %timeit -o numba_heat3d(TSTEPS, A, B)
TIMES['pythran'] = %timeit -o pythran_heat3d(TSTEPS, A, B)
barplot('3D Heat Diffusion', labels=True)
```
## Benchmarking and Instrumentation API
When optimizing programs in DaCe, it is useful to know the raw time the compiled program takes or any of its components. For this purpose, DaCe includes an instrumentation API, which allows you to time each SDFG, state, map, or tasklet directly from the code.
The instrumentation providers given in DaCe can measure different metrics: wall-clock time, GPU (CUDA/HIP) events, PAPI performance counters, and more (it's extensible).
Performance results are saved as report files in CSV format or the `chrome://tracing` JSON format for easy timeline view.
### Profiling API
First, we demonstrate the profiling API, which is a simple low-level timer that will run every called DaCe program a number of times and print out the median runtime.
```
# Setup some optional dependencies for viewing results and printing progress
%pip install pandas tqdm
# Temporarily set the DACE_profiling config to True
with dace.config.set_temporary('profiling', value=True):
# You can control the number of times a program is run with the treps configuration
with dace.config.set_temporary('treps', value=100):
daceloop(a)
```
This can also be controlled with environment variables. Setting `DACE_profiling=1` and `DACE_treps=100` achieves the same effect on the entire script.
The report is saved as a CSV file in the `.dacecache/<program>/profiling` folder, where `<program>` is the program or SDFG name.
```
import pandas as pd
df = pd.read_csv('.dacecache/someforloop/profiling/results-1644308750891.csv')
df.head(10)
```
### Instrumentation API
The Instrumentation API allows more fine-grained control over measuring program metrics. It creates a JSON report in `.dacecache/<program>/perf`, which can be obtained with the API or viewed with any Chrome Tracing capable viewer. More usage information and how to use the API to tune programs can be found in the [program tuning sample](https://github.com/spcl/dace/blob/master/samples/optimization/tuning.py).
```
@dace.program
def twomaps(A):
B = np.sin(A)
return B * 2.0
a = np.random.rand(1000, 1000)
sdfg = twomaps.to_sdfg(a)
sdfg
```
We will now instrument the each of the maps in the program separately, so see which one is a potential bottleneck:
```
# Get all maps
maps = [n for n, _ in sdfg.all_nodes_recursive() if isinstance(n, dace.nodes.MapEntry)]
# Instrument with wall-clock timer
for m in maps:
m.instrument = dace.InstrumentationType.Timer
# Run SDFG and create report
sdfg(a)
# Get the latest instrumentation report from .dacecache/twomaps/perf
report = sdfg.get_latest_report()
# Print report in a nicely readable format
print(report)
```
As we can see, the `np.sin` statement is more expensive than the multiplication statement.
These reports can also be loaded directly to the Visual Studio code plugin to overlay the information on the graph, as shown below:
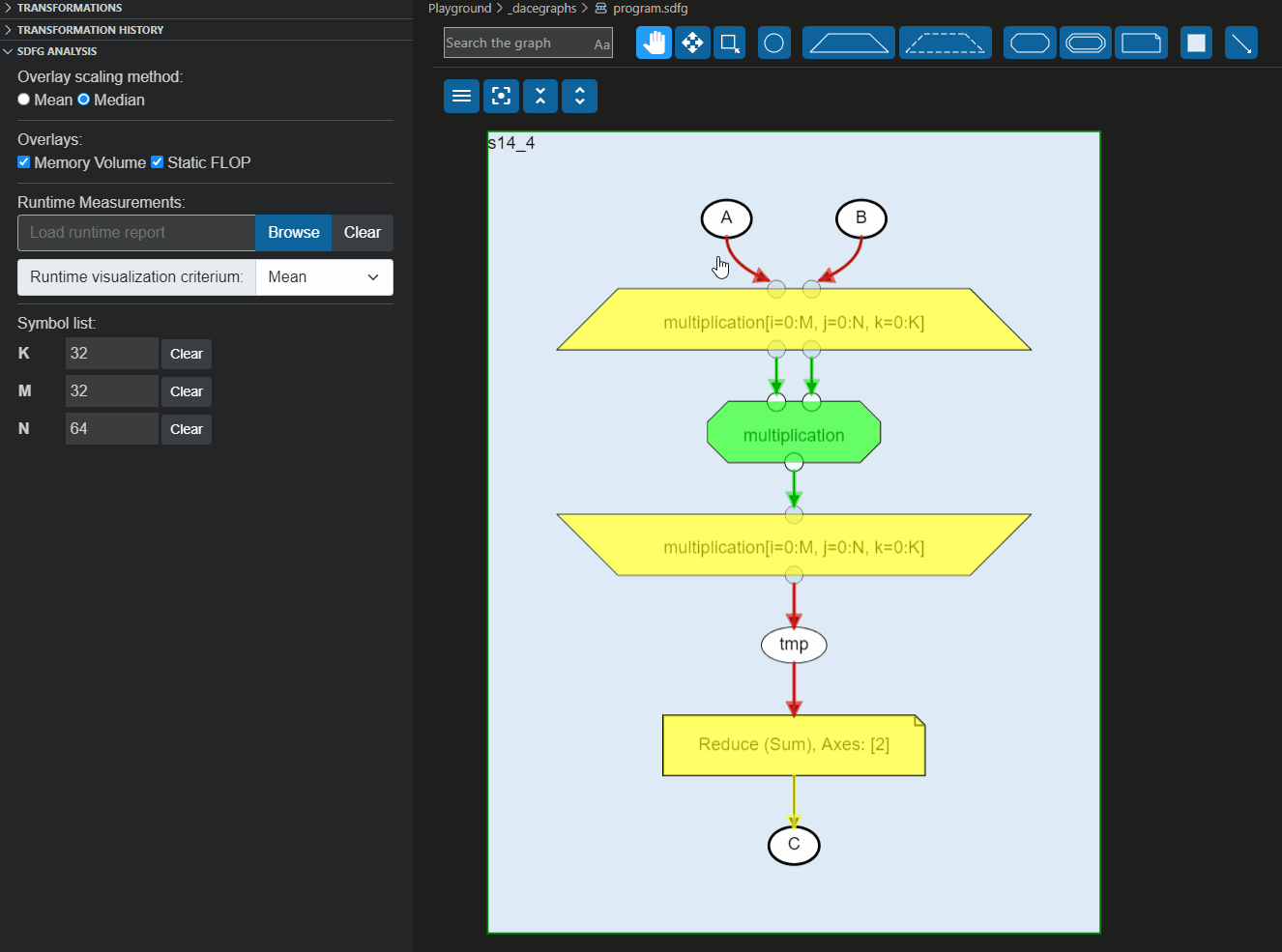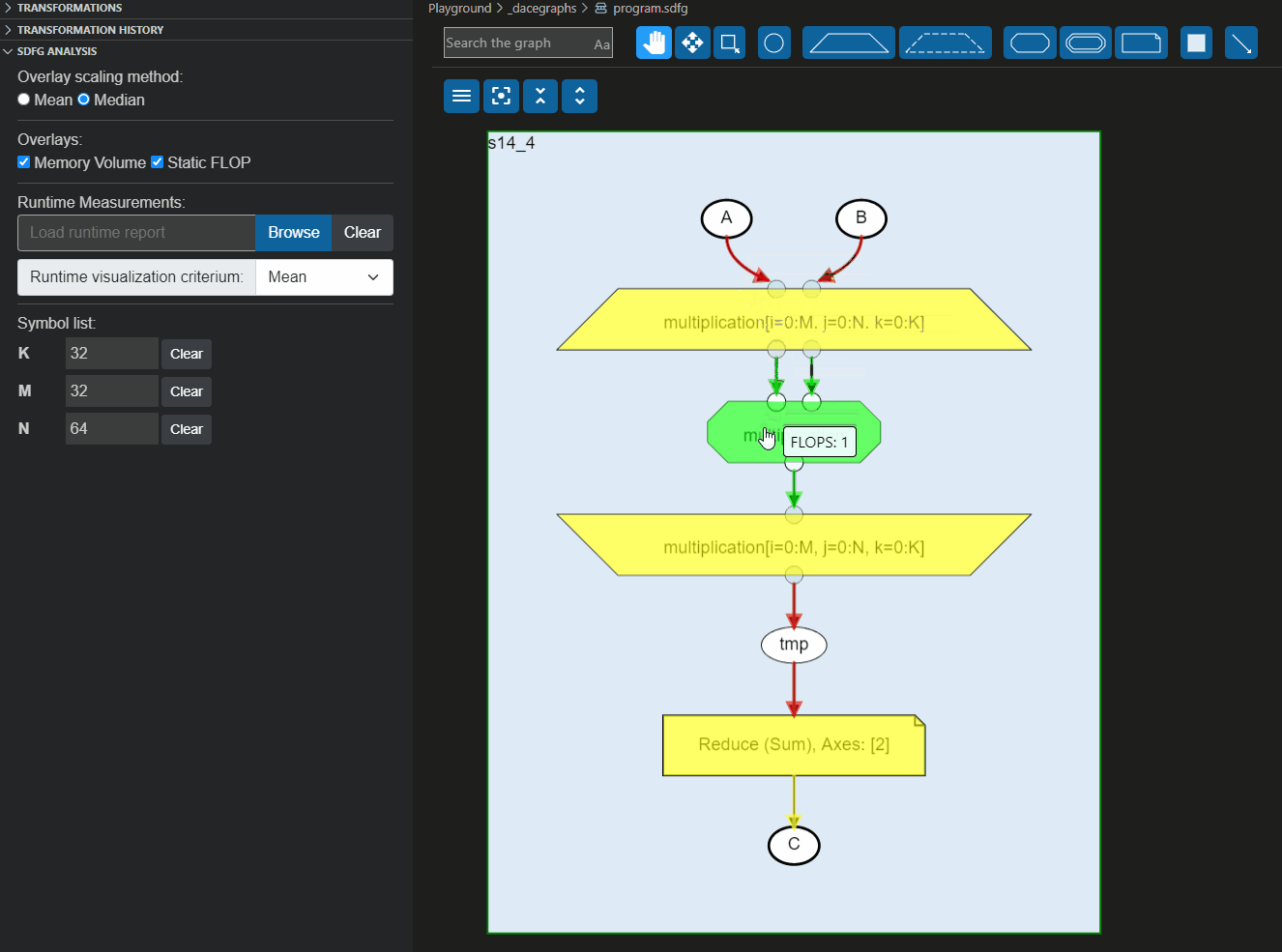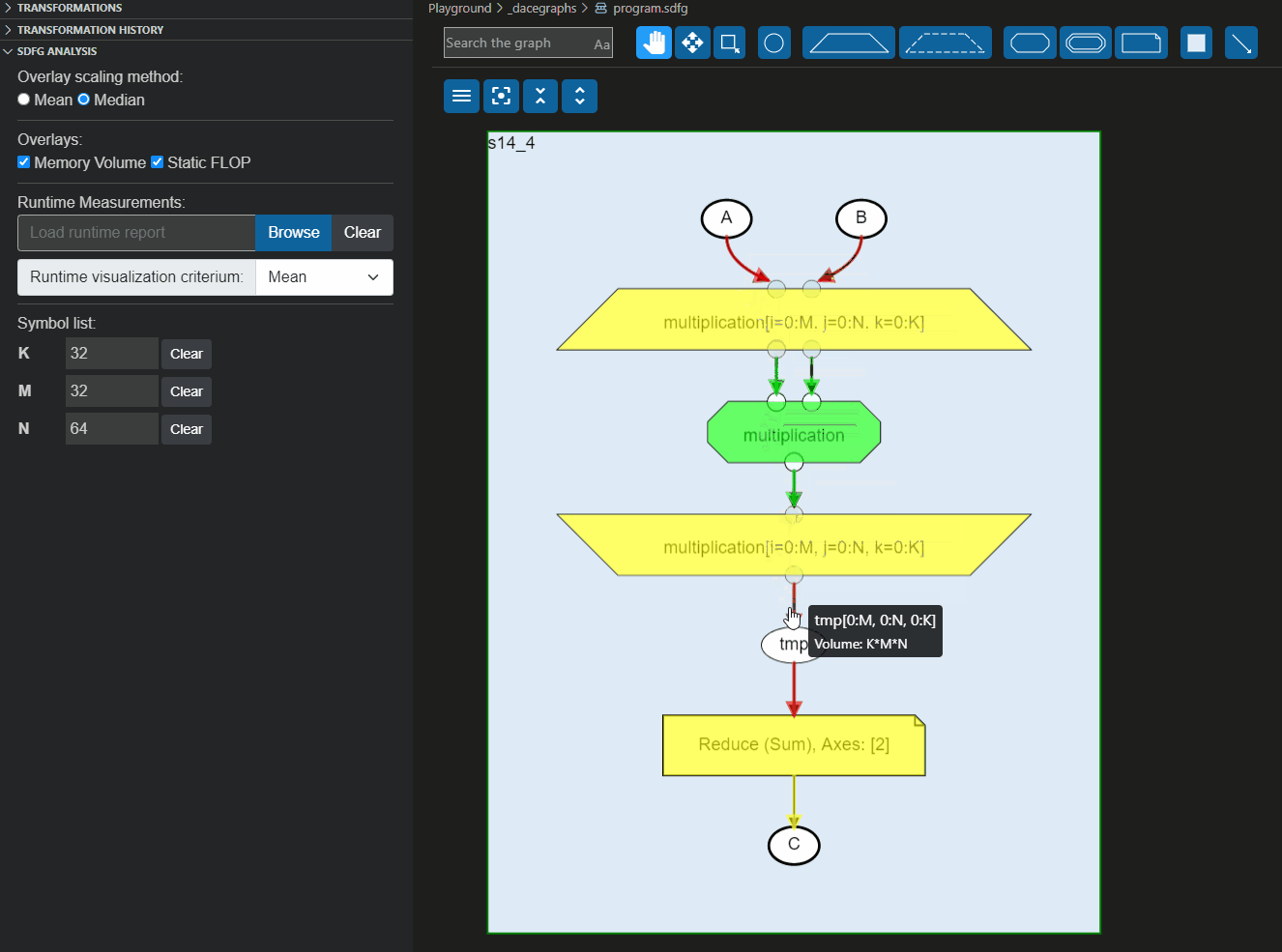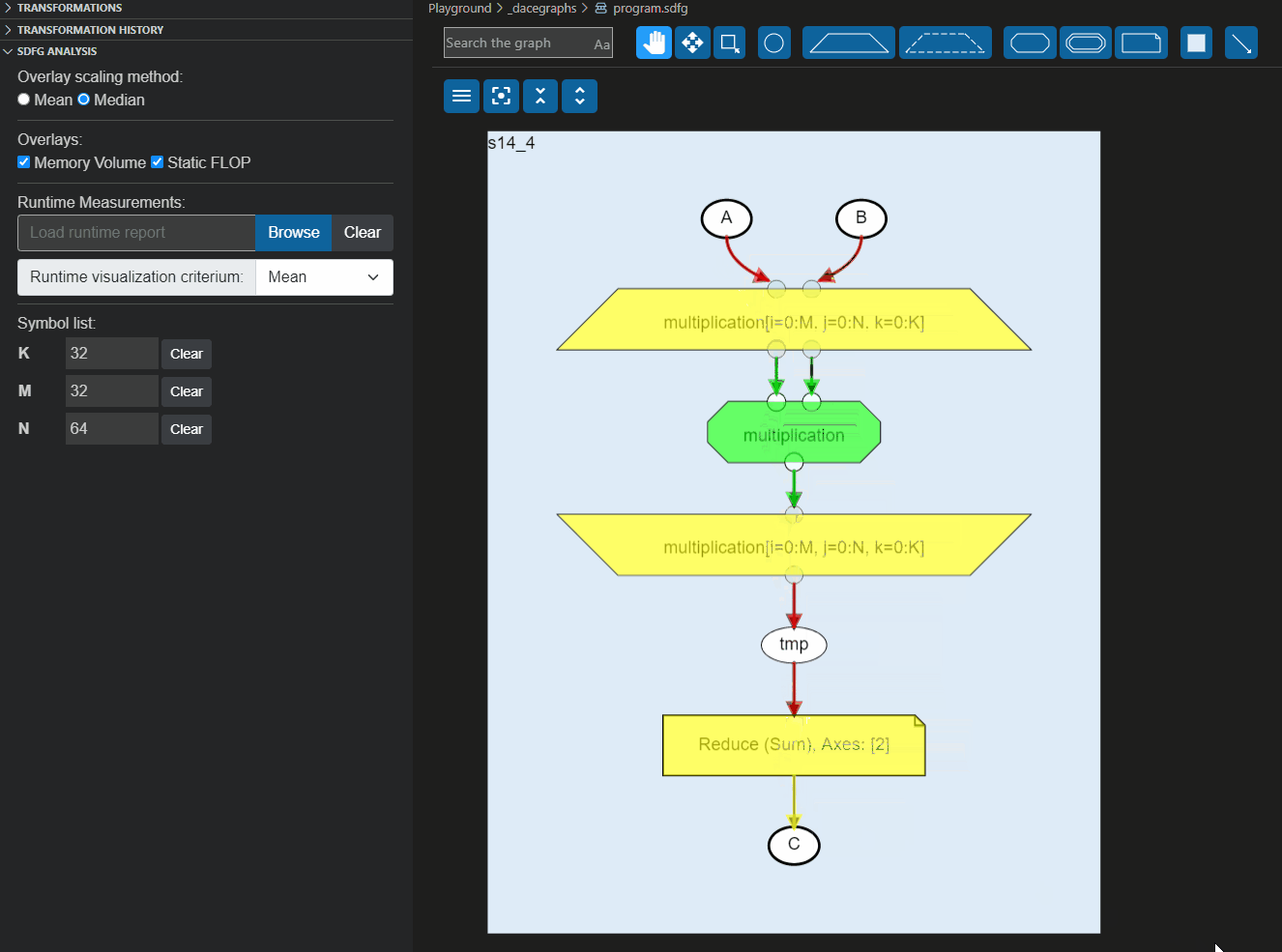
|
github_jupyter
|
%pip install jax jaxlib
%pip install numba
%pip install pythran
# Your library here
# MKL for performance
%conda install mkl mkl-include mkl-devel
# matplotlib to draw the results
%pip install matplotlib
# Setup code for plotting
import matplotlib.pyplot as plt
def barplot(title, labels=False):
x = ['numpy'] + list(sorted(TIMES.keys() - {'numpy'}))
bars = [np.median(TIMES[key].timings) for key in x]
yerr = [np.std(TIMES[key].timings) for key in x]
color = [('#86add9' if 'dace' in key else 'salmon') for key in x]
p = plt.bar(x, bars, yerr=yerr, color=color)
plt.ylabel('Runtime [s]'); plt.xlabel('Implementation'); plt.title(title);
if labels:
plt.gca().bar_label(p)
pass
# Setup code for benchmarked frameworks
import numpy as np
import jax
import numba
import dace
# Pythran loads in a separate cell
%load_ext pythran.magic
def slow_f(x):
return x * x + x * 2.0
a = np.random.rand(5000, 5000)
TIMES = {}
TIMES['numpy'] = %timeit -o slow_f(a)
jax_f = jax.jit(slow_f)
numba_f = numba.jit(slow_f)
dace_f = dace.program(auto_optimize=True)(slow_f)
%%pythran
#pythran export pythran_f(float64[:,:])
def pythran_f(x):
return x * x + x * 2.0
# On your marks...
%timeit -r 1 -n 1 jax_f(a).block_until_ready()
%timeit -r 1 -n 1 numba_f(a)
%timeit -r 1 -n 1 dace_f(a)
%timeit -r 1 -n 1 pythran_f(a)
pass
# ...get set...
# ...Go!
TIMES['jax'] = %timeit -o jax_f(a).block_until_ready()
TIMES['numba'] = %timeit -o numba_f(a)
TIMES['pythran'] = %timeit -o pythran_f(a)
TIMES['dace_jit'] = %timeit -o dace_f(a)
# Either provide type annotations on the `@dace.program`, or call `compile` with sample arguments
cprog = dace_f.compile(a)
TIMES['dace'] = %timeit -o cprog(a)
barplot('Simple program, multiple operators')
def go_fast(a):
trace = 0.0
for i in range(a.shape[0]):
trace += np.tanh(a[i, i])
return a + trace
import numpy as np
b = np.random.rand(1000, 1000)
TIMES = {}
TIMES['numpy'] = %timeit -o go_fast(b)
numba_fast = numba.jit(go_fast)
import jax.numpy as jnp
@jax.jit
def jax_fast(a):
trace = 0.0
for i in range(a.shape[0]):
trace += jnp.tanh(a[i, i])
return a + trace
N = dace.symbol('N')
@dace.program(auto_optimize=True)
def dace_fast(a: dace.float64[N, N]):
trace = 0.0
for i in range(N):
trace += np.tanh(a[i, i])
return a + trace
%%pythran
from numpy import tanh
#pythran export pythran_fast(float64[:,:])
def pythran_fast(a):
trace = 0.0
for i in range(a.shape[0]):
trace += tanh(a[i, i])
return a + trace
import time
start = time.time()
csdfg = dace_fast.compile(b)
print('DaCe compilation time:', time.time() - start, 'seconds')
%timeit -r 1 -n 1 jax_fast(b).block_until_ready()
%timeit -r 1 -n 1 numba_fast(b)
%timeit -r 1 -n 1 pythran_fast(b)
TIMES['jax'] = %timeit -o jax_fast(b).block_until_ready()
TIMES['numba'] = %timeit -o numba_fast(b)
TIMES['pythran'] = %timeit -o pythran_fast(b)
TIMES['dace'] = %timeit -o csdfg(b, N=b.shape[0])
barplot('Loops')
sizes = [np.random.randint(700, 5000) for _ in range(10)]
arrays = [np.random.rand(n, n) for n in sizes]
def vary_size(call):
for a in arrays:
call(a)
def vary_size_dace(call):
for a, n in zip(arrays, sizes):
call(a, N=n)
def vary_size_jax(call):
for a in arrays:
call(a).block_until_ready()
TIMES = {}
TIMES['numpy'] = %timeit -o vary_size(go_fast)
TIMES['numba'] = %timeit -o vary_size(numba_fast)
TIMES['pythran'] = %timeit -o vary_size(pythran_fast)
TIMES['dace'] = %timeit -o vary_size_dace(csdfg)
TIMES['jax'] = %timeit -o vary_size_jax(jax_fast)
barplot('Loop - Varying sizes')
def element_update(a):
return a * 5
def someforloop(A):
for i in range(A.shape[0]):
for j in range(A.shape[1]):
A[i, j] = element_update(A[i, j])
a = np.random.rand(1000, 1000)
daceloop = dace.program(auto_optimize=True)(someforloop)
numbaloop = numba.jit(parallel=True)(someforloop)
csdfg = daceloop.compile(a)
TIMES = {}
TIMES['numpy'] = %timeit -o someforloop(a)
TIMES['numba'] = %timeit -o numbaloop(a)
TIMES['dace'] = %timeit -o csdfg(a)
barplot('Automatic parallelization', labels=True)
def heat3d(TSTEPS, A, B):
for t in range(1, TSTEPS):
B[1:-1, 1:-1,
1:-1] = (0.125 * (A[2:, 1:-1, 1:-1] - 2.0 * A[1:-1, 1:-1, 1:-1] +
A[:-2, 1:-1, 1:-1]) + 0.125 *
(A[1:-1, 2:, 1:-1] - 2.0 * A[1:-1, 1:-1, 1:-1] +
A[1:-1, :-2, 1:-1]) + 0.125 *
(A[1:-1, 1:-1, 2:] - 2.0 * A[1:-1, 1:-1, 1:-1] +
A[1:-1, 1:-1, 0:-2]) + A[1:-1, 1:-1, 1:-1])
A[1:-1, 1:-1,
1:-1] = (0.125 * (B[2:, 1:-1, 1:-1] - 2.0 * B[1:-1, 1:-1, 1:-1] +
B[:-2, 1:-1, 1:-1]) + 0.125 *
(B[1:-1, 2:, 1:-1] - 2.0 * B[1:-1, 1:-1, 1:-1] +
B[1:-1, :-2, 1:-1]) + 0.125 *
(B[1:-1, 1:-1, 2:] - 2.0 * B[1:-1, 1:-1, 1:-1] +
B[1:-1, 1:-1, 0:-2]) + B[1:-1, 1:-1, 1:-1])
# Using the "L" size
TSTEPS, N = 100, 70
A = np.fromfunction(lambda i, j, k: (i + j + (N - k)) * 10 / N, (N, N, N),
dtype=np.float64)
B = np.copy(A)
dace_heat3d = dace.program(auto_optimize=True)(heat3d)
numba_heat3d = numba.jit(nopython=True, parallel=True)(heat3d)
%%pythran
#pythran export pythran_heat3d(int, float64[:,:,:], float64[:,:,:])
def pythran_heat3d(TSTEPS, A, B):
for t in range(1, TSTEPS):
B[1:-1, 1:-1,
1:-1] = (0.125 * (A[2:, 1:-1, 1:-1] - 2.0 * A[1:-1, 1:-1, 1:-1] +
A[:-2, 1:-1, 1:-1]) + 0.125 *
(A[1:-1, 2:, 1:-1] - 2.0 * A[1:-1, 1:-1, 1:-1] +
A[1:-1, :-2, 1:-1]) + 0.125 *
(A[1:-1, 1:-1, 2:] - 2.0 * A[1:-1, 1:-1, 1:-1] +
A[1:-1, 1:-1, 0:-2]) + A[1:-1, 1:-1, 1:-1])
A[1:-1, 1:-1,
1:-1] = (0.125 * (B[2:, 1:-1, 1:-1] - 2.0 * B[1:-1, 1:-1, 1:-1] +
B[:-2, 1:-1, 1:-1]) + 0.125 *
(B[1:-1, 2:, 1:-1] - 2.0 * B[1:-1, 1:-1, 1:-1] +
B[1:-1, :-2, 1:-1]) + 0.125 *
(B[1:-1, 1:-1, 2:] - 2.0 * B[1:-1, 1:-1, 1:-1] +
B[1:-1, 1:-1, 0:-2]) + B[1:-1, 1:-1, 1:-1])
# Warmup
%timeit -r 1 -n 1 dace_heat3d(TSTEPS, A, B)
%timeit -r 1 -n 1 numba_heat3d(TSTEPS, A, B)
%timeit -r 1 -n 1 pythran_heat3d(TSTEPS, A, B)
TIMES = {}
TIMES['numpy'] = %timeit -o heat3d(TSTEPS, A, B)
TIMES['dace'] = %timeit -o dace_heat3d(TSTEPS, A, B)
TIMES['numba'] = %timeit -o numba_heat3d(TSTEPS, A, B)
TIMES['pythran'] = %timeit -o pythran_heat3d(TSTEPS, A, B)
barplot('3D Heat Diffusion', labels=True)
# Setup some optional dependencies for viewing results and printing progress
%pip install pandas tqdm
# Temporarily set the DACE_profiling config to True
with dace.config.set_temporary('profiling', value=True):
# You can control the number of times a program is run with the treps configuration
with dace.config.set_temporary('treps', value=100):
daceloop(a)
import pandas as pd
df = pd.read_csv('.dacecache/someforloop/profiling/results-1644308750891.csv')
df.head(10)
@dace.program
def twomaps(A):
B = np.sin(A)
return B * 2.0
a = np.random.rand(1000, 1000)
sdfg = twomaps.to_sdfg(a)
sdfg
# Get all maps
maps = [n for n, _ in sdfg.all_nodes_recursive() if isinstance(n, dace.nodes.MapEntry)]
# Instrument with wall-clock timer
for m in maps:
m.instrument = dace.InstrumentationType.Timer
# Run SDFG and create report
sdfg(a)
# Get the latest instrumentation report from .dacecache/twomaps/perf
report = sdfg.get_latest_report()
# Print report in a nicely readable format
print(report)
| 0.428592 | 0.963506 |
# Work and drink
Imports and set magics:
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from matplotlib_venn import venn2 # `pip install matplotlib-venn`
from pandas_datareader import wb
import datetime
import pandas_datareader # install with `pip install pandas-datareader`
import pydst # install with `pip install git+https://github.com/elben10/pydst`
import statsmodels.api as sm
from matplotlib.ticker import StrMethodFormatter # to set decimals on axis
import ipywidgets as widgets # importing package for interactive figure
from IPython.display import display
```
# Importing data from the World Bank
For this data project we will use the package wb from pandas_datareader to import data from the World Bank. We will use data on alcohol consumption per capita in liters and GDP per capita. We predict alcohol consumption to increase with GDP per capita, because richer countries tend to have more spare time (We will not test this), and therefore have more time, where drinking is appropriate. For the analysis we first display the trend in each of alcohol consumption per capita and GDP per capita and afterwards display the correlation between the two. Alcohol per capita is measured in liters of pure alcohol consumped a year and GDP per capita is measured by USD PPP 2011. Lastly we estimate a simple OLS regression to see if there is a significant estimate.
```
# importing data on alcohol consumption
wb_alco = wb.download(indicator='SH.ALC.PCAP.LI', country = ['all'] ,start=1990, end=2020) #using the indicator to pull data from the world bank
wb_alco = wb_alco.rename(columns = {'SH.ALC.PCAP.LI': 'alcohol_per_capita'}) # renaming the alcohol row
wb_alco = wb_alco.reset_index() # resetting the index to ensure it is correct
wb_alco.year = wb_alco.year.astype(int) # convert year
wb_alco.country = wb_alco.country.astype('string') # convert country to the special pandas string type
#importing data on GDP per capita
wb_gdp = wb.download(indicator='NY.GDP.PCAP.KD', country=['all'], start=1990, end=2020)
wb_gdp = wb_gdp.rename(columns = {'NY.GDP.PCAP.KD':'GDP_per_capita'})
wb_gdp = wb_gdp.reset_index()
wb_gdp.year = wb_gdp.year.astype(int)
wb_gdp.country = wb_gdp.country.astype('string')
#importing data on unemployment
wb_mig = wb.download(indicator='SM.POP.TOTL.ZS', country=['all'], start=1990, end=2020)
wb_mig = wb_mig.rename(columns = {'SM.POP.TOTL.ZS':'migration'})
wb_mig = wb_mig.reset_index()
wb_mig.year = wb_mig.year.astype(int)
wb_mig.country = wb_mig.country.astype('string')
```
For the analysis we will use the years 2000, 2005, 2010, 2015 and 2018 as data on alcohol consumption per capita is only measured these years.
```
merged = pd.merge(wb_gdp, wb_alco, how = 'left', on = ['country', 'year']) #merging the the alcohol and GDP datsets
```
The dataset include "world", "Low income country" etc. in the "country" row, which we will not include in the cross-country analysis. The data is sorted such that after column 1519, it is only real countries. We use this sorting to choose only the countries.
```
merged = merged.iloc[1519:,:] #excluding the first 1519 values of merged.
merged = merged.dropna() #We delete every 'NaN'
merged.reset_index(inplace=True, drop = True) #resetting the index
merged #displaying data to see if all looks good
```
In the following we create "alco_income", which only include data on world and low-, middle-, and high income countries. We Will use this dataset to display the general evolution in the cinsumption and alcohol in the world and by income groups to see if they have the same trend.
```
merged_full = pd.merge(wb_gdp, wb_alco, how = 'left', on = ['country', 'year'])
merged_full = merged_full.dropna() #We delete every 'NaN'
merged_full.reset_index(inplace=True, drop = True)
alco_income = merged_full.loc[merged_full['country'].isin(['World', 'Low income', 'Middle income', 'High income'])]
World_years = alco_income.loc[alco_income['country'] == 'World', :]
```
# summary statistics
First we look at the summary statistics to get an idea of the range of the dataset. There is a large difference in income per capita with lowest being 262 USD a year and the higher 107.201 USd a year.
```
merged[['GDP_per_capita' , 'alcohol_per_capita']].describe()
def plot_1(df1, year):
I = df1['year'] == year
ax=df1.loc[I,:].hist('GDP_per_capita')
plt.title('distribution of GDP per capita')
plt.xlabel('USD')
plt.ylabel('Countries')
widgets.interact(plot_1,
df1 = widgets.fixed(merged),
year = widgets.IntSlider(description='year', min=2000, max=2018, steps=len(year.unique()),
continuous_update=True),
);
```
# Plot of the data
The two following figures the evolution of the average world consumption of alcohol per capita and the average GDP per capita. Both have increased in the period 2000-2018. GDP per capita have been steadily increasing, whereas the alcohol consumption per capita increase sharply from 2005 to 2010,and is more or less constant or even a bit downward sloping in the other periods. This does not support our hypothesis that GDP per capita and alcohol consumption per capita are positively correlated perfectly as we would expect them to have the same trend and kinks.
```
World_years.plot(x='year',y='GDP_per_capita',legend=True)
plt.gca().xaxis.set_major_formatter(StrMethodFormatter('{x:.0f}')) # Setting x-axis to not include decimals
plt.title('average World GDP per capita')
plt.ylabel('GDP per capita')
plt.xlabel('years')
World_years.plot(x='year',y='alcohol_per_capita',legend=True)
plt.gca().xaxis.set_major_formatter(StrMethodFormatter('{x:.0f}')) # Setting x-axis to not include decimals
plt.title('average World alcohol consumption per capita')
plt.ylabel('liters of pure alcohol')
plt.xlabel('years')
plt.tight_layout()
fig, (ax1, ax2) = plt.subplots(1,2)
ax1.plot(World_years['year'], World_years['GDP_per_capita'])
plt.gca().xaxis.set_major_formatter(StrMethodFormatter('{x:.0f}')) # Setting x-axis to not include decimals
ax1.set_title('World GDP per capita')
ax1.set_ylabel('GDP per capita')
ax1.set_xlabel('years')
ax2.plot(World_years['year'],World_years['alcohol_per_capita'])
plt.gca().xaxis.set_major_formatter(StrMethodFormatter('{x:.0f}')) # Setting x-axis to not include decimals
ax2.set_title('World alcohol consumption per capita')
ax2.set_ylabel('liters of pure alcohol')
ax2.set_xlabel('years')
plt.tight_layout()
```
To further investigate our dataset and the correlation between GDP per capita and alcohol consumption per capita, we divide the dataset by income groups. From the two figure below we see that high income countries drink more and low income countries drink less. We see that the shift to higher alcohol consumption 2010-2015 was driven by the middle income countries, whereas the high income countries have on average decreased their alcohol consumption per capita in the period, and the alcohol consumption of the low income countries have been constant.
```
alco_income.set_index('year', inplace=True)
alco_income.groupby('country')['alcohol_per_capita'].plot(legend=True)
plt.title('alcohol per capita per income and world')
plt.ylabel('liters of pure alcohol')
plt.gca().xaxis.set_major_formatter(StrMethodFormatter('{x:.0f}'))
alco_income.groupby('country')['GDP_per_capita'].plot(legend=True)
plt.title('GDP per capita per income and world')
plt.ylabel('USD')
plt.gca().xaxis.set_major_formatter(StrMethodFormatter('{x:.0f}'))
```
## Interactive figure
From the interactive figure below, we see that there is a positive correlation between GDP per capita and alohol consumption per capita in all years. From a graphical analysis of each year, it does not seem as the correlation between GDP per capita and alcohol consumtion per capita change. In all year there is a large cluster of countries, which drink 1-10 liters of alcohol every year and have very low GDP per capita.
```
def plot_2(df, year):
I = df['year'] == year
ax=df.loc[I,:].plot.scatter(x='GDP_per_capita', y='alcohol_per_capita', legend=False)
plt.title('Alcohol consumption and GDP per capita')
plt.xlabel('USD')
plt.ylabel('liters of pure alcohol')
x = merged.loc[:,'GDP_per_capita']
y = merged.loc[:,'alcohol_per_capita']
z = np.polyfit(x, y, 1)
p = np.poly1d(z)
plt.plot(x, p(x))
widgets.interact(plot_2,
df = widgets.fixed(merged),
year = widgets.Dropdown(description='year',
options=merged.year.unique(),
value=2000)
);
```
## OLS-regression
As we expect the causality to be from increased income to increased alcohol consumption, we estimate a simple OLS-regression with alcohol per capita as the dependent variable and GDP per capita as the independent variable. The estimate for GDP per capita is positive and very significant. From the estimate we see that a 1 USD increase in GDP per capita increase alcohol consumption with 0.00000774 liters of pure alcohol per year. This is equivalent to one unit, when income increases 2286 USD a year as there is approximately 0.0177 liters of pure alcohol to one unit. Even though the estimate is positive as we expected, the it should be interpret as a correlation.
```
x = merged.loc[:, 'GDP_per_capita' ]
y = merged.loc[:, 'alcohol_per_capita']
x = sm.add_constant(x)
model = sm.OLS(y, x).fit()
summary = model.summary()
print(summary)
```
'alcohol' is measured in liters of pure alcohol consumed. From the regression tabel we see that 1 additional liter of alcohol consumed increases GDP per capita by 1457 dollars. The estimate is highly significant with a p-value of 0.000.
# Conclusion
From this analysis we can conclude that alcohol and GDP per capita correlate positive.
|
github_jupyter
|
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from matplotlib_venn import venn2 # `pip install matplotlib-venn`
from pandas_datareader import wb
import datetime
import pandas_datareader # install with `pip install pandas-datareader`
import pydst # install with `pip install git+https://github.com/elben10/pydst`
import statsmodels.api as sm
from matplotlib.ticker import StrMethodFormatter # to set decimals on axis
import ipywidgets as widgets # importing package for interactive figure
from IPython.display import display
# importing data on alcohol consumption
wb_alco = wb.download(indicator='SH.ALC.PCAP.LI', country = ['all'] ,start=1990, end=2020) #using the indicator to pull data from the world bank
wb_alco = wb_alco.rename(columns = {'SH.ALC.PCAP.LI': 'alcohol_per_capita'}) # renaming the alcohol row
wb_alco = wb_alco.reset_index() # resetting the index to ensure it is correct
wb_alco.year = wb_alco.year.astype(int) # convert year
wb_alco.country = wb_alco.country.astype('string') # convert country to the special pandas string type
#importing data on GDP per capita
wb_gdp = wb.download(indicator='NY.GDP.PCAP.KD', country=['all'], start=1990, end=2020)
wb_gdp = wb_gdp.rename(columns = {'NY.GDP.PCAP.KD':'GDP_per_capita'})
wb_gdp = wb_gdp.reset_index()
wb_gdp.year = wb_gdp.year.astype(int)
wb_gdp.country = wb_gdp.country.astype('string')
#importing data on unemployment
wb_mig = wb.download(indicator='SM.POP.TOTL.ZS', country=['all'], start=1990, end=2020)
wb_mig = wb_mig.rename(columns = {'SM.POP.TOTL.ZS':'migration'})
wb_mig = wb_mig.reset_index()
wb_mig.year = wb_mig.year.astype(int)
wb_mig.country = wb_mig.country.astype('string')
merged = pd.merge(wb_gdp, wb_alco, how = 'left', on = ['country', 'year']) #merging the the alcohol and GDP datsets
merged = merged.iloc[1519:,:] #excluding the first 1519 values of merged.
merged = merged.dropna() #We delete every 'NaN'
merged.reset_index(inplace=True, drop = True) #resetting the index
merged #displaying data to see if all looks good
merged_full = pd.merge(wb_gdp, wb_alco, how = 'left', on = ['country', 'year'])
merged_full = merged_full.dropna() #We delete every 'NaN'
merged_full.reset_index(inplace=True, drop = True)
alco_income = merged_full.loc[merged_full['country'].isin(['World', 'Low income', 'Middle income', 'High income'])]
World_years = alco_income.loc[alco_income['country'] == 'World', :]
merged[['GDP_per_capita' , 'alcohol_per_capita']].describe()
def plot_1(df1, year):
I = df1['year'] == year
ax=df1.loc[I,:].hist('GDP_per_capita')
plt.title('distribution of GDP per capita')
plt.xlabel('USD')
plt.ylabel('Countries')
widgets.interact(plot_1,
df1 = widgets.fixed(merged),
year = widgets.IntSlider(description='year', min=2000, max=2018, steps=len(year.unique()),
continuous_update=True),
);
World_years.plot(x='year',y='GDP_per_capita',legend=True)
plt.gca().xaxis.set_major_formatter(StrMethodFormatter('{x:.0f}')) # Setting x-axis to not include decimals
plt.title('average World GDP per capita')
plt.ylabel('GDP per capita')
plt.xlabel('years')
World_years.plot(x='year',y='alcohol_per_capita',legend=True)
plt.gca().xaxis.set_major_formatter(StrMethodFormatter('{x:.0f}')) # Setting x-axis to not include decimals
plt.title('average World alcohol consumption per capita')
plt.ylabel('liters of pure alcohol')
plt.xlabel('years')
plt.tight_layout()
fig, (ax1, ax2) = plt.subplots(1,2)
ax1.plot(World_years['year'], World_years['GDP_per_capita'])
plt.gca().xaxis.set_major_formatter(StrMethodFormatter('{x:.0f}')) # Setting x-axis to not include decimals
ax1.set_title('World GDP per capita')
ax1.set_ylabel('GDP per capita')
ax1.set_xlabel('years')
ax2.plot(World_years['year'],World_years['alcohol_per_capita'])
plt.gca().xaxis.set_major_formatter(StrMethodFormatter('{x:.0f}')) # Setting x-axis to not include decimals
ax2.set_title('World alcohol consumption per capita')
ax2.set_ylabel('liters of pure alcohol')
ax2.set_xlabel('years')
plt.tight_layout()
alco_income.set_index('year', inplace=True)
alco_income.groupby('country')['alcohol_per_capita'].plot(legend=True)
plt.title('alcohol per capita per income and world')
plt.ylabel('liters of pure alcohol')
plt.gca().xaxis.set_major_formatter(StrMethodFormatter('{x:.0f}'))
alco_income.groupby('country')['GDP_per_capita'].plot(legend=True)
plt.title('GDP per capita per income and world')
plt.ylabel('USD')
plt.gca().xaxis.set_major_formatter(StrMethodFormatter('{x:.0f}'))
def plot_2(df, year):
I = df['year'] == year
ax=df.loc[I,:].plot.scatter(x='GDP_per_capita', y='alcohol_per_capita', legend=False)
plt.title('Alcohol consumption and GDP per capita')
plt.xlabel('USD')
plt.ylabel('liters of pure alcohol')
x = merged.loc[:,'GDP_per_capita']
y = merged.loc[:,'alcohol_per_capita']
z = np.polyfit(x, y, 1)
p = np.poly1d(z)
plt.plot(x, p(x))
widgets.interact(plot_2,
df = widgets.fixed(merged),
year = widgets.Dropdown(description='year',
options=merged.year.unique(),
value=2000)
);
x = merged.loc[:, 'GDP_per_capita' ]
y = merged.loc[:, 'alcohol_per_capita']
x = sm.add_constant(x)
model = sm.OLS(y, x).fit()
summary = model.summary()
print(summary)
| 0.467818 | 0.957675 |
<a href="https://colab.research.google.com/github/charleslien/wordle-greedy-strategies/blob/main/wordle.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
```
# Scraped from the wordle website
POSSIBLE_ANSWERS = ["cigar", "rebut", "sissy", "humph", "awake", "blush", "focal", "evade", "naval", "serve", "heath", "dwarf", "model", "karma", "stink", "grade", "quiet", "bench", "abate", "feign", "major", "death", "fresh", "crust", "stool", "colon", "abase", "marry", "react", "batty", "pride", "floss", "helix", "croak", "staff", "paper", "unfed", "whelp", "trawl", "outdo", "adobe", "crazy", "sower", "repay", "digit", "crate", "cluck", "spike", "mimic", "pound", "maxim", "linen", "unmet", "flesh", "booby", "forth", "first", "stand", "belly", "ivory", "seedy", "print", "yearn", "drain", "bribe", "stout", "panel", "crass", "flume", "offal", "agree", "error", "swirl", "argue", "bleed", "delta", "flick", "totem", "wooer", "front", "shrub", "parry", "biome", "lapel", "start", "greet", "goner", "golem", "lusty", "loopy", "round", "audit", "lying", "gamma", "labor", "islet", "civic", "forge", "corny", "moult", "basic", "salad", "agate", "spicy", "spray", "essay", "fjord", "spend", "kebab", "guild", "aback", "motor", "alone", "hatch", "hyper", "thumb", "dowry", "ought", "belch", "dutch", "pilot", "tweed", "comet", "jaunt", "enema", "steed", "abyss", "growl", "fling", "dozen", "boozy", "erode", "world", "gouge", "click", "briar", "great", "altar", "pulpy", "blurt", "coast", "duchy", "groin", "fixer", "group", "rogue", "badly", "smart", "pithy", "gaudy", "chill", "heron", "vodka", "finer", "surer", "radio", "rouge", "perch", "retch", "wrote", "clock", "tilde", "store", "prove", "bring", "solve", "cheat", "grime", "exult", "usher", "epoch", "triad", "break", "rhino", "viral", "conic", "masse", "sonic", "vital", "trace", "using", "peach", "champ", "baton", "brake", "pluck", "craze", "gripe", "weary", "picky", "acute", "ferry", "aside", "tapir", "troll", "unify", "rebus", "boost", "truss", "siege", "tiger", "banal", "slump", "crank", "gorge", "query", "drink", "favor", "abbey", "tangy", "panic", "solar", "shire", "proxy", "point", "robot", "prick", "wince", "crimp", "knoll", "sugar", "whack", "mount", "perky", "could", "wrung", "light", "those", "moist", "shard", "pleat", "aloft", "skill", "elder", "frame", "humor", "pause", "ulcer", "ultra", "robin", "cynic", "agora", "aroma", "caulk", "shake", "pupal", "dodge", "swill", "tacit", "other", "thorn", "trove", "bloke", "vivid", "spill", "chant", "choke", "rupee", "nasty", "mourn", "ahead", "brine", "cloth", "hoard", "sweet", "month", "lapse", "watch", "today", "focus", "smelt", "tease", "cater", "movie", "lynch", "saute", "allow", "renew", "their", "slosh", "purge", "chest", "depot", "epoxy", "nymph", "found", "shall", "harry", "stove", "lowly", "snout", "trope", "fewer", "shawl", "natal", "fibre", "comma", "foray", "scare", "stair", "black", "squad", "royal", "chunk", "mince", "slave", "shame", "cheek", "ample", "flair", "foyer", "cargo", "oxide", "plant", "olive", "inert", "askew", "heist", "shown", "zesty", "hasty", "trash", "fella", "larva", "forgo", "story", "hairy", "train", "homer", "badge", "midst", "canny", "fetus", "butch", "farce", "slung", "tipsy", "metal", "yield", "delve", "being", "scour", "glass", "gamer", "scrap", "money", "hinge", "album", "vouch", "asset", "tiara", "crept", "bayou", "atoll", "manor", "creak", "showy", "phase", "froth", "depth", "gloom", "flood", "trait", "girth", "piety", "payer", "goose", "float", "donor", "atone", "primo", "apron", "blown", "cacao", "loser", "input", "gloat", "awful", "brink", "smite", "beady", "rusty", "retro", "droll", "gawky", "hutch", "pinto", "gaily", "egret", "lilac", "sever", "field", "fluff", "hydro", "flack", "agape", "wench", "voice", "stead", "stalk", "berth", "madam", "night", "bland", "liver", "wedge", "augur", "roomy", "wacky", "flock", "angry", "bobby", "trite", "aphid", "tryst", "midge", "power", "elope", "cinch", "motto", "stomp", "upset", "bluff", "cramp", "quart", "coyly", "youth", "rhyme", "buggy", "alien", "smear", "unfit", "patty", "cling", "glean", "label", "hunky", "khaki", "poker", "gruel", "twice", "twang", "shrug", "treat", "unlit", "waste", "merit", "woven", "octal", "needy", "clown", "widow", "irony", "ruder", "gauze", "chief", "onset", "prize", "fungi", "charm", "gully", "inter", "whoop", "taunt", "leery", "class", "theme", "lofty", "tibia", "booze", "alpha", "thyme", "eclat", "doubt", "parer", "chute", "stick", "trice", "alike", "sooth", "recap", "saint", "liege", "glory", "grate", "admit", "brisk", "soggy", "usurp", "scald", "scorn", "leave", "twine", "sting", "bough", "marsh", "sloth", "dandy", "vigor", "howdy", "enjoy", "valid", "ionic", "equal", "unset", "floor", "catch", "spade", "stein", "exist", "quirk", "denim", "grove", "spiel", "mummy", "fault", "foggy", "flout", "carry", "sneak", "libel", "waltz", "aptly", "piney", "inept", "aloud", "photo", "dream", "stale", "vomit", "ombre", "fanny", "unite", "snarl", "baker", "there", "glyph", "pooch", "hippy", "spell", "folly", "louse", "gulch", "vault", "godly", "threw", "fleet", "grave", "inane", "shock", "crave", "spite", "valve", "skimp", "claim", "rainy", "musty", "pique", "daddy", "quasi", "arise", "aging", "valet", "opium", "avert", "stuck", "recut", "mulch", "genre", "plume", "rifle", "count", "incur", "total", "wrest", "mocha", "deter", "study", "lover", "safer", "rivet", "funny", "smoke", "mound", "undue", "sedan", "pagan", "swine", "guile", "gusty", "equip", "tough", "canoe", "chaos", "covet", "human", "udder", "lunch", "blast", "stray", "manga", "melee", "lefty", "quick", "paste", "given", "octet", "risen", "groan", "leaky", "grind", "carve", "loose", "sadly", "spilt", "apple", "slack", "honey", "final", "sheen", "eerie", "minty", "slick", "derby", "wharf", "spelt", "coach", "erupt", "singe", "price", "spawn", "fairy", "jiffy", "filmy", "stack", "chose", "sleep", "ardor", "nanny", "niece", "woozy", "handy", "grace", "ditto", "stank", "cream", "usual", "diode", "valor", "angle", "ninja", "muddy", "chase", "reply", "prone", "spoil", "heart", "shade", "diner", "arson", "onion", "sleet", "dowel", "couch", "palsy", "bowel", "smile", "evoke", "creek", "lance", "eagle", "idiot", "siren", "built", "embed", "award", "dross", "annul", "goody", "frown", "patio", "laden", "humid", "elite", "lymph", "edify", "might", "reset", "visit", "gusto", "purse", "vapor", "crock", "write", "sunny", "loath", "chaff", "slide", "queer", "venom", "stamp", "sorry", "still", "acorn", "aping", "pushy", "tamer", "hater", "mania", "awoke", "brawn", "swift", "exile", "birch", "lucky", "freer", "risky", "ghost", "plier", "lunar", "winch", "snare", "nurse", "house", "borax", "nicer", "lurch", "exalt", "about", "savvy", "toxin", "tunic", "pried", "inlay", "chump", "lanky", "cress", "eater", "elude", "cycle", "kitty", "boule", "moron", "tenet", "place", "lobby", "plush", "vigil", "index", "blink", "clung", "qualm", "croup", "clink", "juicy", "stage", "decay", "nerve", "flier", "shaft", "crook", "clean", "china", "ridge", "vowel", "gnome", "snuck", "icing", "spiny", "rigor", "snail", "flown", "rabid", "prose", "thank", "poppy", "budge", "fiber", "moldy", "dowdy", "kneel", "track", "caddy", "quell", "dumpy", "paler", "swore", "rebar", "scuba", "splat", "flyer", "horny", "mason", "doing", "ozone", "amply", "molar", "ovary", "beset", "queue", "cliff", "magic", "truce", "sport", "fritz", "edict", "twirl", "verse", "llama", "eaten", "range", "whisk", "hovel", "rehab", "macaw", "sigma", "spout", "verve", "sushi", "dying", "fetid", "brain", "buddy", "thump", "scion", "candy", "chord", "basin", "march", "crowd", "arbor", "gayly", "musky", "stain", "dally", "bless", "bravo", "stung", "title", "ruler", "kiosk", "blond", "ennui", "layer", "fluid", "tatty", "score", "cutie", "zebra", "barge", "matey", "bluer", "aider", "shook", "river", "privy", "betel", "frisk", "bongo", "begun", "azure", "weave", "genie", "sound", "glove", "braid", "scope", "wryly", "rover", "assay", "ocean", "bloom", "irate", "later", "woken", "silky", "wreck", "dwelt", "slate", "smack", "solid", "amaze", "hazel", "wrist", "jolly", "globe", "flint", "rouse", "civil", "vista", "relax", "cover", "alive", "beech", "jetty", "bliss", "vocal", "often", "dolly", "eight", "joker", "since", "event", "ensue", "shunt", "diver", "poser", "worst", "sweep", "alley", "creed", "anime", "leafy", "bosom", "dunce", "stare", "pudgy", "waive", "choir", "stood", "spoke", "outgo", "delay", "bilge", "ideal", "clasp", "seize", "hotly", "laugh", "sieve", "block", "meant", "grape", "noose", "hardy", "shied", "drawl", "daisy", "putty", "strut", "burnt", "tulip", "crick", "idyll", "vixen", "furor", "geeky", "cough", "naive", "shoal", "stork", "bathe", "aunty", "check", "prime", "brass", "outer", "furry", "razor", "elect", "evict", "imply", "demur", "quota", "haven", "cavil", "swear", "crump", "dough", "gavel", "wagon", "salon", "nudge", "harem", "pitch", "sworn", "pupil", "excel", "stony", "cabin", "unzip", "queen", "trout", "polyp", "earth", "storm", "until", "taper", "enter", "child", "adopt", "minor", "fatty", "husky", "brave", "filet", "slime", "glint", "tread", "steal", "regal", "guest", "every", "murky", "share", "spore", "hoist", "buxom", "inner", "otter", "dimly", "level", "sumac", "donut", "stilt", "arena", "sheet", "scrub", "fancy", "slimy", "pearl", "silly", "porch", "dingo", "sepia", "amble", "shady", "bread", "friar", "reign", "dairy", "quill", "cross", "brood", "tuber", "shear", "posit", "blank", "villa", "shank", "piggy", "freak", "which", "among", "fecal", "shell", "would", "algae", "large", "rabbi", "agony", "amuse", "bushy", "copse", "swoon", "knife", "pouch", "ascot", "plane", "crown", "urban", "snide", "relay", "abide", "viola", "rajah", "straw", "dilly", "crash", "amass", "third", "trick", "tutor", "woody", "blurb", "grief", "disco", "where", "sassy", "beach", "sauna", "comic", "clued", "creep", "caste", "graze", "snuff", "frock", "gonad", "drunk", "prong", "lurid", "steel", "halve", "buyer", "vinyl", "utile", "smell", "adage", "worry", "tasty", "local", "trade", "finch", "ashen", "modal", "gaunt", "clove", "enact", "adorn", "roast", "speck", "sheik", "missy", "grunt", "snoop", "party", "touch", "mafia", "emcee", "array", "south", "vapid", "jelly", "skulk", "angst", "tubal", "lower", "crest", "sweat", "cyber", "adore", "tardy", "swami", "notch", "groom", "roach", "hitch", "young", "align", "ready", "frond", "strap", "puree", "realm", "venue", "swarm", "offer", "seven", "dryer", "diary", "dryly", "drank", "acrid", "heady", "theta", "junto", "pixie", "quoth", "bonus", "shalt", "penne", "amend", "datum", "build", "piano", "shelf", "lodge", "suing", "rearm", "coral", "ramen", "worth", "psalm", "infer", "overt", "mayor", "ovoid", "glide", "usage", "poise", "randy", "chuck", "prank", "fishy", "tooth", "ether", "drove", "idler", "swath", "stint", "while", "begat", "apply", "slang", "tarot", "radar", "credo", "aware", "canon", "shift", "timer", "bylaw", "serum", "three", "steak", "iliac", "shirk", "blunt", "puppy", "penal", "joist", "bunny", "shape", "beget", "wheel", "adept", "stunt", "stole", "topaz", "chore", "fluke", "afoot", "bloat", "bully", "dense", "caper", "sneer", "boxer", "jumbo", "lunge", "space", "avail", "short", "slurp", "loyal", "flirt", "pizza", "conch", "tempo", "droop", "plate", "bible", "plunk", "afoul", "savoy", "steep", "agile", "stake", "dwell", "knave", "beard", "arose", "motif", "smash", "broil", "glare", "shove", "baggy", "mammy", "swamp", "along", "rugby", "wager", "quack", "squat", "snaky", "debit", "mange", "skate", "ninth", "joust", "tramp", "spurn", "medal", "micro", "rebel", "flank", "learn", "nadir", "maple", "comfy", "remit", "gruff", "ester", "least", "mogul", "fetch", "cause", "oaken", "aglow", "meaty", "gaffe", "shyly", "racer", "prowl", "thief", "stern", "poesy", "rocky", "tweet", "waist", "spire", "grope", "havoc", "patsy", "truly", "forty", "deity", "uncle", "swish", "giver", "preen", "bevel", "lemur", "draft", "slope", "annoy", "lingo", "bleak", "ditty", "curly", "cedar", "dirge", "grown", "horde", "drool", "shuck", "crypt", "cumin", "stock", "gravy", "locus", "wider", "breed", "quite", "chafe", "cache", "blimp", "deign", "fiend", "logic", "cheap", "elide", "rigid", "false", "renal", "pence", "rowdy", "shoot", "blaze", "envoy", "posse", "brief", "never", "abort", "mouse", "mucky", "sulky", "fiery", "media", "trunk", "yeast", "clear", "skunk", "scalp", "bitty", "cider", "koala", "duvet", "segue", "creme", "super", "grill", "after", "owner", "ember", "reach", "nobly", "empty", "speed", "gipsy", "recur", "smock", "dread", "merge", "burst", "kappa", "amity", "shaky", "hover", "carol", "snort", "synod", "faint", "haunt", "flour", "chair", "detox", "shrew", "tense", "plied", "quark", "burly", "novel", "waxen", "stoic", "jerky", "blitz", "beefy", "lyric", "hussy", "towel", "quilt", "below", "bingo", "wispy", "brash", "scone", "toast", "easel", "saucy", "value", "spice", "honor", "route", "sharp", "bawdy", "radii", "skull", "phony", "issue", "lager", "swell", "urine", "gassy", "trial", "flora", "upper", "latch", "wight", "brick", "retry", "holly", "decal", "grass", "shack", "dogma", "mover", "defer", "sober", "optic", "crier", "vying", "nomad", "flute", "hippo", "shark", "drier", "obese", "bugle", "tawny", "chalk", "feast", "ruddy", "pedal", "scarf", "cruel", "bleat", "tidal", "slush", "semen", "windy", "dusty", "sally", "igloo", "nerdy", "jewel", "shone", "whale", "hymen", "abuse", "fugue", "elbow", "crumb", "pansy", "welsh", "syrup", "terse", "suave", "gamut", "swung", "drake", "freed", "afire", "shirt", "grout", "oddly", "tithe", "plaid", "dummy", "broom", "blind", "torch", "enemy", "again", "tying", "pesky", "alter", "gazer", "noble", "ethos", "bride", "extol", "decor", "hobby", "beast", "idiom", "utter", "these", "sixth", "alarm", "erase", "elegy", "spunk", "piper", "scaly", "scold", "hefty", "chick", "sooty", "canal", "whiny", "slash", "quake", "joint", "swept", "prude", "heavy", "wield", "femme", "lasso", "maize", "shale", "screw", "spree", "smoky", "whiff", "scent", "glade", "spent", "prism", "stoke", "riper", "orbit", "cocoa", "guilt", "humus", "shush", "table", "smirk", "wrong", "noisy", "alert", "shiny", "elate", "resin", "whole", "hunch", "pixel", "polar", "hotel", "sword", "cleat", "mango", "rumba", "puffy", "filly", "billy", "leash", "clout", "dance", "ovate", "facet", "chili", "paint", "liner", "curio", "salty", "audio", "snake", "fable", "cloak", "navel", "spurt", "pesto", "balmy", "flash", "unwed", "early", "churn", "weedy", "stump", "lease", "witty", "wimpy", "spoof", "saner", "blend", "salsa", "thick", "warty", "manic", "blare", "squib", "spoon", "probe", "crepe", "knack", "force", "debut", "order", "haste", "teeth", "agent", "widen", "icily", "slice", "ingot", "clash", "juror", "blood", "abode", "throw", "unity", "pivot", "slept", "troop", "spare", "sewer", "parse", "morph", "cacti", "tacky", "spool", "demon", "moody", "annex", "begin", "fuzzy", "patch", "water", "lumpy", "admin", "omega", "limit", "tabby", "macho", "aisle", "skiff", "basis", "plank", "verge", "botch", "crawl", "lousy", "slain", "cubic", "raise", "wrack", "guide", "foist", "cameo", "under", "actor", "revue", "fraud", "harpy", "scoop", "climb", "refer", "olden", "clerk", "debar", "tally", "ethic", "cairn", "tulle", "ghoul", "hilly", "crude", "apart", "scale", "older", "plain", "sperm", "briny", "abbot", "rerun", "quest", "crisp", "bound", "befit", "drawn", "suite", "itchy", "cheer", "bagel", "guess", "broad", "axiom", "chard", "caput", "leant", "harsh", "curse", "proud", "swing", "opine", "taste", "lupus", "gumbo", "miner", "green", "chasm", "lipid", "topic", "armor", "brush", "crane", "mural", "abled", "habit", "bossy", "maker", "dusky", "dizzy", "lithe", "brook", "jazzy", "fifty", "sense", "giant", "surly", "legal", "fatal", "flunk", "began", "prune", "small", "slant", "scoff", "torus", "ninny", "covey", "viper", "taken", "moral", "vogue", "owing", "token", "entry", "booth", "voter", "chide", "elfin", "ebony", "neigh", "minim", "melon", "kneed", "decoy", "voila", "ankle", "arrow", "mushy", "tribe", "cease", "eager", "birth", "graph", "odder", "terra", "weird", "tried", "clack", "color", "rough", "weigh", "uncut", "ladle", "strip", "craft", "minus", "dicey", "titan", "lucid", "vicar", "dress", "ditch", "gypsy", "pasta", "taffy", "flame", "swoop", "aloof", "sight", "broke", "teary", "chart", "sixty", "wordy", "sheer", "leper", "nosey", "bulge", "savor", "clamp", "funky", "foamy", "toxic", "brand", "plumb", "dingy", "butte", "drill", "tripe", "bicep", "tenor", "krill", "worse", "drama", "hyena", "think", "ratio", "cobra", "basil", "scrum", "bused", "phone", "court", "camel", "proof", "heard", "angel", "petal", "pouty", "throb", "maybe", "fetal", "sprig", "spine", "shout", "cadet", "macro", "dodgy", "satyr", "rarer", "binge", "trend", "nutty", "leapt", "amiss", "split", "myrrh", "width", "sonar", "tower", "baron", "fever", "waver", "spark", "belie", "sloop", "expel", "smote", "baler", "above", "north", "wafer", "scant", "frill", "awash", "snack", "scowl", "frail", "drift", "limbo", "fence", "motel", "ounce", "wreak", "revel", "talon", "prior", "knelt", "cello", "flake", "debug", "anode", "crime", "salve", "scout", "imbue", "pinky", "stave", "vague", "chock", "fight", "video", "stone", "teach", "cleft", "frost", "prawn", "booty", "twist", "apnea", "stiff", "plaza", "ledge", "tweak", "board", "grant", "medic", "bacon", "cable", "brawl", "slunk", "raspy", "forum", "drone", "women", "mucus", "boast", "toddy", "coven", "tumor", "truer", "wrath", "stall", "steam", "axial", "purer", "daily", "trail", "niche", "mealy", "juice", "nylon", "plump", "merry", "flail", "papal", "wheat", "berry", "cower", "erect", "brute", "leggy", "snipe", "sinew", "skier", "penny", "jumpy", "rally", "umbra", "scary", "modem", "gross", "avian", "greed", "satin", "tonic", "parka", "sniff", "livid", "stark", "trump", "giddy", "reuse", "taboo", "avoid", "quote", "devil", "liken", "gloss", "gayer", "beret", "noise", "gland", "dealt", "sling", "rumor", "opera", "thigh", "tonga", "flare", "wound", "white", "bulky", "etude", "horse", "circa", "paddy", "inbox", "fizzy", "grain", "exert", "surge", "gleam", "belle", "salvo", "crush", "fruit", "sappy", "taker", "tract", "ovine", "spiky", "frank", "reedy", "filth", "spasm", "heave", "mambo", "right", "clank", "trust", "lumen", "borne", "spook", "sauce", "amber", "lathe", "carat", "corer", "dirty", "slyly", "affix", "alloy", "taint", "sheep", "kinky", "wooly", "mauve", "flung", "yacht", "fried", "quail", "brunt", "grimy", "curvy", "cagey", "rinse", "deuce", "state", "grasp", "milky", "bison", "graft", "sandy", "baste", "flask", "hedge", "girly", "swash", "boney", "coupe", "endow", "abhor", "welch", "blade", "tight", "geese", "miser", "mirth", "cloud", "cabal", "leech", "close", "tenth", "pecan", "droit", "grail", "clone", "guise", "ralph", "tango", "biddy", "smith", "mower", "payee", "serif", "drape", "fifth", "spank", "glaze", "allot", "truck", "kayak", "virus", "testy", "tepee", "fully", "zonal", "metro", "curry", "grand", "banjo", "axion", "bezel", "occur", "chain", "nasal", "gooey", "filer", "brace", "allay", "pubic", "raven", "plead", "gnash", "flaky", "munch", "dully", "eking", "thing", "slink", "hurry", "theft", "shorn", "pygmy", "ranch", "wring", "lemon", "shore", "mamma", "froze", "newer", "style", "moose", "antic", "drown", "vegan", "chess", "guppy", "union", "lever", "lorry", "image", "cabby", "druid", "exact", "truth", "dopey", "spear", "cried", "chime", "crony", "stunk", "timid", "batch", "gauge", "rotor", "crack", "curve", "latte", "witch", "bunch", "repel", "anvil", "soapy", "meter", "broth", "madly", "dried", "scene", "known", "magma", "roost", "woman", "thong", "punch", "pasty", "downy", "knead", "whirl", "rapid", "clang", "anger", "drive", "goofy", "email", "music", "stuff", "bleep", "rider", "mecca", "folio", "setup", "verso", "quash", "fauna", "gummy", "happy", "newly", "fussy", "relic", "guava", "ratty", "fudge", "femur", "chirp", "forte", "alibi", "whine", "petty", "golly", "plait", "fleck", "felon", "gourd", "brown", "thrum", "ficus", "stash", "decry", "wiser", "junta", "visor", "daunt", "scree", "impel", "await", "press", "whose", "turbo", "stoop", "speak", "mangy", "eying", "inlet", "crone", "pulse", "mossy", "staid", "hence", "pinch", "teddy", "sully", "snore", "ripen", "snowy", "attic", "going", "leach", "mouth", "hound", "clump", "tonal", "bigot", "peril", "piece", "blame", "haute", "spied", "undid", "intro", "basal", "shine", "gecko", "rodeo", "guard", "steer", "loamy", "scamp", "scram", "manly", "hello", "vaunt", "organ", "feral", "knock", "extra", "condo", "adapt", "willy", "polka", "rayon", "skirt", "faith", "torso", "match", "mercy", "tepid", "sleek", "riser", "twixt", "peace", "flush", "catty", "login", "eject", "roger", "rival", "untie", "refit", "aorta", "adult", "judge", "rower", "artsy", "rural", "shave"]
POSSIBLE_GUESSES = POSSIBLE_ANSWERS + ["aahed", "aalii", "aargh", "aarti", "abaca", "abaci", "abacs", "abaft", "abaka", "abamp", "aband", "abash", "abask", "abaya", "abbas", "abbed", "abbes", "abcee", "abeam", "abear", "abele", "abers", "abets", "abies", "abler", "ables", "ablet", "ablow", "abmho", "abohm", "aboil", "aboma", "aboon", "abord", "abore", "abram", "abray", "abrim", "abrin", "abris", "absey", "absit", "abuna", "abune", "abuts", "abuzz", "abyes", "abysm", "acais", "acari", "accas", "accoy", "acerb", "acers", "aceta", "achar", "ached", "aches", "achoo", "acids", "acidy", "acing", "acini", "ackee", "acker", "acmes", "acmic", "acned", "acnes", "acock", "acold", "acred", "acres", "acros", "acted", "actin", "acton", "acyls", "adaws", "adays", "adbot", "addax", "added", "adder", "addio", "addle", "adeem", "adhan", "adieu", "adios", "adits", "adman", "admen", "admix", "adobo", "adown", "adoze", "adrad", "adred", "adsum", "aduki", "adunc", "adust", "advew", "adyta", "adzed", "adzes", "aecia", "aedes", "aegis", "aeons", "aerie", "aeros", "aesir", "afald", "afara", "afars", "afear", "aflaj", "afore", "afrit", "afros", "agama", "agami", "agars", "agast", "agave", "agaze", "agene", "agers", "agger", "aggie", "aggri", "aggro", "aggry", "aghas", "agila", "agios", "agism", "agist", "agita", "aglee", "aglet", "agley", "agloo", "aglus", "agmas", "agoge", "agone", "agons", "agood", "agria", "agrin", "agros", "agued", "agues", "aguna", "aguti", "aheap", "ahent", "ahigh", "ahind", "ahing", "ahint", "ahold", "ahull", "ahuru", "aidas", "aided", "aides", "aidoi", "aidos", "aiery", "aigas", "aight", "ailed", "aimed", "aimer", "ainee", "ainga", "aioli", "aired", "airer", "airns", "airth", "airts", "aitch", "aitus", "aiver", "aiyee", "aizle", "ajies", "ajiva", "ajuga", "ajwan", "akees", "akela", "akene", "aking", "akita", "akkas", "alaap", "alack", "alamo", "aland", "alane", "alang", "alans", "alant", "alapa", "alaps", "alary", "alate", "alays", "albas", "albee", "alcid", "alcos", "aldea", "alder", "aldol", "aleck", "alecs", "alefs", "aleft", "aleph", "alews", "aleye", "alfas", "algal", "algas", "algid", "algin", "algor", "algum", "alias", "alifs", "aline", "alist", "aliya", "alkie", "alkos", "alkyd", "alkyl", "allee", "allel", "allis", "allod", "allyl", "almah", "almas", "almeh", "almes", "almud", "almug", "alods", "aloed", "aloes", "aloha", "aloin", "aloos", "alowe", "altho", "altos", "alula", "alums", "alure", "alvar", "alway", "amahs", "amain", "amate", "amaut", "amban", "ambit", "ambos", "ambry", "ameba", "ameer", "amene", "amens", "ament", "amias", "amice", "amici", "amide", "amido", "amids", "amies", "amiga", "amigo", "amine", "amino", "amins", "amirs", "amlas", "amman", "ammon", "ammos", "amnia", "amnic", "amnio", "amoks", "amole", "amort", "amour", "amove", "amowt", "amped", "ampul", "amrit", "amuck", "amyls", "anana", "anata", "ancho", "ancle", "ancon", "andro", "anear", "anele", "anent", "angas", "anglo", "anigh", "anile", "anils", "anima", "animi", "anion", "anise", "anker", "ankhs", "ankus", "anlas", "annal", "annas", "annat", "anoas", "anole", "anomy", "ansae", "antae", "antar", "antas", "anted", "antes", "antis", "antra", "antre", "antsy", "anura", "anyon", "apace", "apage", "apaid", "apayd", "apays", "apeak", "apeek", "apers", "apert", "apery", "apgar", "aphis", "apian", "apiol", "apish", "apism", "apode", "apods", "apoop", "aport", "appal", "appay", "appel", "appro", "appui", "appuy", "apres", "apses", "apsis", "apsos", "apted", "apter", "aquae", "aquas", "araba", "araks", "arame", "arars", "arbas", "arced", "archi", "arcos", "arcus", "ardeb", "ardri", "aread", "areae", "areal", "arear", "areas", "areca", "aredd", "arede", "arefy", "areic", "arene", "arepa", "arere", "arete", "arets", "arett", "argal", "argan", "argil", "argle", "argol", "argon", "argot", "argus", "arhat", "arias", "ariel", "ariki", "arils", "ariot", "arish", "arked", "arled", "arles", "armed", "armer", "armet", "armil", "arnas", "arnut", "aroba", "aroha", "aroid", "arpas", "arpen", "arrah", "arras", "arret", "arris", "arroz", "arsed", "arses", "arsey", "arsis", "artal", "artel", "artic", "artis", "aruhe", "arums", "arval", "arvee", "arvos", "aryls", "asana", "ascon", "ascus", "asdic", "ashed", "ashes", "ashet", "asked", "asker", "askoi", "askos", "aspen", "asper", "aspic", "aspie", "aspis", "aspro", "assai", "assam", "asses", "assez", "assot", "aster", "astir", "astun", "asura", "asway", "aswim", "asyla", "ataps", "ataxy", "atigi", "atilt", "atimy", "atlas", "atman", "atmas", "atmos", "atocs", "atoke", "atoks", "atoms", "atomy", "atony", "atopy", "atria", "atrip", "attap", "attar", "atuas", "audad", "auger", "aught", "aulas", "aulic", "auloi", "aulos", "aumil", "aunes", "aunts", "aurae", "aural", "aurar", "auras", "aurei", "aures", "auric", "auris", "aurum", "autos", "auxin", "avale", "avant", "avast", "avels", "avens", "avers", "avgas", "avine", "avion", "avise", "aviso", "avize", "avows", "avyze", "awarn", "awato", "awave", "aways", "awdls", "aweel", "aweto", "awing", "awmry", "awned", "awner", "awols", "awork", "axels", "axile", "axils", "axing", "axite", "axled", "axles", "axman", "axmen", "axoid", "axone", "axons", "ayahs", "ayaya", "ayelp", "aygre", "ayins", "ayont", "ayres", "ayrie", "azans", "azide", "azido", "azine", "azlon", "azoic", "azole", "azons", "azote", "azoth", "azuki", "azurn", "azury", "azygy", "azyme", "azyms", "baaed", "baals", "babas", "babel", "babes", "babka", "baboo", "babul", "babus", "bacca", "bacco", "baccy", "bacha", "bachs", "backs", "baddy", "baels", "baffs", "baffy", "bafts", "baghs", "bagie", "bahts", "bahus", "bahut", "bails", "bairn", "baisa", "baith", "baits", "baiza", "baize", "bajan", "bajra", "bajri", "bajus", "baked", "baken", "bakes", "bakra", "balas", "balds", "baldy", "baled", "bales", "balks", "balky", "balls", "bally", "balms", "baloo", "balsa", "balti", "balun", "balus", "bambi", "banak", "banco", "bancs", "banda", "bandh", "bands", "bandy", "baned", "banes", "bangs", "bania", "banks", "banns", "bants", "bantu", "banty", "banya", "bapus", "barbe", "barbs", "barby", "barca", "barde", "bardo", "bards", "bardy", "bared", "barer", "bares", "barfi", "barfs", "baric", "barks", "barky", "barms", "barmy", "barns", "barny", "barps", "barra", "barre", "barro", "barry", "barye", "basan", "based", "basen", "baser", "bases", "basho", "basij", "basks", "bason", "basse", "bassi", "basso", "bassy", "basta", "basti", "basto", "basts", "bated", "bates", "baths", "batik", "batta", "batts", "battu", "bauds", "bauks", "baulk", "baurs", "bavin", "bawds", "bawks", "bawls", "bawns", "bawrs", "bawty", "bayed", "bayer", "bayes", "bayle", "bayts", "bazar", "bazoo", "beads", "beaks", "beaky", "beals", "beams", "beamy", "beano", "beans", "beany", "beare", "bears", "beath", "beats", "beaty", "beaus", "beaut", "beaux", "bebop", "becap", "becke", "becks", "bedad", "bedel", "bedes", "bedew", "bedim", "bedye", "beedi", "beefs", "beeps", "beers", "beery", "beets", "befog", "begad", "begar", "begem", "begot", "begum", "beige", "beigy", "beins", "bekah", "belah", "belar", "belay", "belee", "belga", "bells", "belon", "belts", "bemad", "bemas", "bemix", "bemud", "bends", "bendy", "benes", "benet", "benga", "benis", "benne", "benni", "benny", "bento", "bents", "benty", "bepat", "beray", "beres", "bergs", "berko", "berks", "berme", "berms", "berob", "beryl", "besat", "besaw", "besee", "beses", "besit", "besom", "besot", "besti", "bests", "betas", "beted", "betes", "beths", "betid", "beton", "betta", "betty", "bever", "bevor", "bevue", "bevvy", "bewet", "bewig", "bezes", "bezil", "bezzy", "bhais", "bhaji", "bhang", "bhats", "bhels", "bhoot", "bhuna", "bhuts", "biach", "biali", "bialy", "bibbs", "bibes", "biccy", "bices", "bided", "bider", "bides", "bidet", "bidis", "bidon", "bield", "biers", "biffo", "biffs", "biffy", "bifid", "bigae", "biggs", "biggy", "bigha", "bight", "bigly", "bigos", "bijou", "biked", "biker", "bikes", "bikie", "bilbo", "bilby", "biled", "biles", "bilgy", "bilks", "bills", "bimah", "bimas", "bimbo", "binal", "bindi", "binds", "biner", "bines", "bings", "bingy", "binit", "binks", "bints", "biogs", "biont", "biota", "biped", "bipod", "birds", "birks", "birle", "birls", "biros", "birrs", "birse", "birsy", "bises", "bisks", "bisom", "bitch", "biter", "bites", "bitos", "bitou", "bitsy", "bitte", "bitts", "bivia", "bivvy", "bizes", "bizzo", "bizzy", "blabs", "blads", "blady", "blaer", "blaes", "blaff", "blags", "blahs", "blain", "blams", "blart", "blase", "blash", "blate", "blats", "blatt", "blaud", "blawn", "blaws", "blays", "blear", "blebs", "blech", "blees", "blent", "blert", "blest", "blets", "bleys", "blimy", "bling", "blini", "blins", "bliny", "blips", "blist", "blite", "blits", "blive", "blobs", "blocs", "blogs", "blook", "bloop", "blore", "blots", "blows", "blowy", "blubs", "blude", "bluds", "bludy", "blued", "blues", "bluet", "bluey", "bluid", "blume", "blunk", "blurs", "blype", "boabs", "boaks", "boars", "boart", "boats", "bobac", "bobak", "bobas", "bobol", "bobos", "bocca", "bocce", "bocci", "boche", "bocks", "boded", "bodes", "bodge", "bodhi", "bodle", "boeps", "boets", "boeuf", "boffo", "boffs", "bogan", "bogey", "boggy", "bogie", "bogle", "bogue", "bogus", "bohea", "bohos", "boils", "boing", "boink", "boite", "boked", "bokeh", "bokes", "bokos", "bolar", "bolas", "bolds", "boles", "bolix", "bolls", "bolos", "bolts", "bolus", "bomas", "bombe", "bombo", "bombs", "bonce", "bonds", "boned", "boner", "bones", "bongs", "bonie", "bonks", "bonne", "bonny", "bonza", "bonze", "booai", "booay", "boobs", "boody", "booed", "boofy", "boogy", "boohs", "books", "booky", "bools", "booms", "boomy", "boong", "boons", "boord", "boors", "boose", "boots", "boppy", "borak", "boral", "boras", "borde", "bords", "bored", "boree", "borel", "borer", "bores", "borgo", "boric", "borks", "borms", "borna", "boron", "borts", "borty", "bortz", "bosie", "bosks", "bosky", "boson", "bosun", "botas", "botel", "botes", "bothy", "botte", "botts", "botty", "bouge", "bouks", "boult", "bouns", "bourd", "bourg", "bourn", "bouse", "bousy", "bouts", "bovid", "bowat", "bowed", "bower", "bowes", "bowet", "bowie", "bowls", "bowne", "bowrs", "bowse", "boxed", "boxen", "boxes", "boxla", "boxty", "boyar", "boyau", "boyed", "boyfs", "boygs", "boyla", "boyos", "boysy", "bozos", "braai", "brach", "brack", "bract", "brads", "braes", "brags", "brail", "braks", "braky", "brame", "brane", "brank", "brans", "brant", "brast", "brats", "brava", "bravi", "braws", "braxy", "brays", "braza", "braze", "bream", "brede", "breds", "breem", "breer", "brees", "breid", "breis", "breme", "brens", "brent", "brere", "brers", "breve", "brews", "breys", "brier", "bries", "brigs", "briki", "briks", "brill", "brims", "brins", "brios", "brise", "briss", "brith", "brits", "britt", "brize", "broch", "brock", "brods", "brogh", "brogs", "brome", "bromo", "bronc", "brond", "brool", "broos", "brose", "brosy", "brows", "brugh", "bruin", "bruit", "brule", "brume", "brung", "brusk", "brust", "bruts", "buats", "buaze", "bubal", "bubas", "bubba", "bubbe", "bubby", "bubus", "buchu", "bucko", "bucks", "bucku", "budas", "budis", "budos", "buffa", "buffe", "buffi", "buffo", "buffs", "buffy", "bufos", "bufty", "buhls", "buhrs", "buiks", "buist", "bukes", "bulbs", "bulgy", "bulks", "bulla", "bulls", "bulse", "bumbo", "bumfs", "bumph", "bumps", "bumpy", "bunas", "bunce", "bunco", "bunde", "bundh", "bunds", "bundt", "bundu", "bundy", "bungs", "bungy", "bunia", "bunje", "bunjy", "bunko", "bunks", "bunns", "bunts", "bunty", "bunya", "buoys", "buppy", "buran", "buras", "burbs", "burds", "buret", "burfi", "burgh", "burgs", "burin", "burka", "burke", "burks", "burls", "burns", "buroo", "burps", "burqa", "burro", "burrs", "burry", "bursa", "burse", "busby", "buses", "busks", "busky", "bussu", "busti", "busts", "busty", "buteo", "butes", "butle", "butoh", "butts", "butty", "butut", "butyl", "buzzy", "bwana", "bwazi", "byded", "bydes", "byked", "bykes", "byres", "byrls", "byssi", "bytes", "byway", "caaed", "cabas", "caber", "cabob", "caboc", "cabre", "cacas", "cacks", "cacky", "cadee", "cades", "cadge", "cadgy", "cadie", "cadis", "cadre", "caeca", "caese", "cafes", "caffs", "caged", "cager", "cages", "cagot", "cahow", "caids", "cains", "caird", "cajon", "cajun", "caked", "cakes", "cakey", "calfs", "calid", "calif", "calix", "calks", "calla", "calls", "calms", "calmy", "calos", "calpa", "calps", "calve", "calyx", "caman", "camas", "cames", "camis", "camos", "campi", "campo", "camps", "campy", "camus", "caned", "caneh", "caner", "canes", "cangs", "canid", "canna", "canns", "canso", "canst", "canto", "cants", "canty", "capas", "caped", "capes", "capex", "caphs", "capiz", "caple", "capon", "capos", "capot", "capri", "capul", "carap", "carbo", "carbs", "carby", "cardi", "cards", "cardy", "cared", "carer", "cares", "caret", "carex", "carks", "carle", "carls", "carns", "carny", "carob", "carom", "caron", "carpi", "carps", "carrs", "carse", "carta", "carte", "carts", "carvy", "casas", "casco", "cased", "cases", "casks", "casky", "casts", "casus", "cates", "cauda", "cauks", "cauld", "cauls", "caums", "caups", "cauri", "causa", "cavas", "caved", "cavel", "caver", "caves", "cavie", "cawed", "cawks", "caxon", "ceaze", "cebid", "cecal", "cecum", "ceded", "ceder", "cedes", "cedis", "ceiba", "ceili", "ceils", "celeb", "cella", "celli", "cells", "celom", "celts", "cense", "cento", "cents", "centu", "ceorl", "cepes", "cerci", "cered", "ceres", "cerge", "ceria", "ceric", "cerne", "ceroc", "ceros", "certs", "certy", "cesse", "cesta", "cesti", "cetes", "cetyl", "cezve", "chace", "chack", "chaco", "chado", "chads", "chaft", "chais", "chals", "chams", "chana", "chang", "chank", "chape", "chaps", "chapt", "chara", "chare", "chark", "charr", "chars", "chary", "chats", "chave", "chavs", "chawk", "chaws", "chaya", "chays", "cheep", "chefs", "cheka", "chela", "chelp", "chemo", "chems", "chere", "chert", "cheth", "chevy", "chews", "chewy", "chiao", "chias", "chibs", "chica", "chich", "chico", "chics", "chiel", "chiks", "chile", "chimb", "chimo", "chimp", "chine", "ching", "chink", "chino", "chins", "chips", "chirk", "chirl", "chirm", "chiro", "chirr", "chirt", "chiru", "chits", "chive", "chivs", "chivy", "chizz", "choco", "chocs", "chode", "chogs", "choil", "choko", "choky", "chola", "choli", "cholo", "chomp", "chons", "choof", "chook", "choom", "choon", "chops", "chota", "chott", "chout", "choux", "chowk", "chows", "chubs", "chufa", "chuff", "chugs", "chums", "churl", "churr", "chuse", "chuts", "chyle", "chyme", "chynd", "cibol", "cided", "cides", "ciels", "ciggy", "cilia", "cills", "cimar", "cimex", "cinct", "cines", "cinqs", "cions", "cippi", "circs", "cires", "cirls", "cirri", "cisco", "cissy", "cists", "cital", "cited", "citer", "cites", "cives", "civet", "civie", "civvy", "clach", "clade", "clads", "claes", "clags", "clame", "clams", "clans", "claps", "clapt", "claro", "clart", "clary", "clast", "clats", "claut", "clave", "clavi", "claws", "clays", "cleck", "cleek", "cleep", "clefs", "clegs", "cleik", "clems", "clepe", "clept", "cleve", "clews", "clied", "clies", "clift", "clime", "cline", "clint", "clipe", "clips", "clipt", "clits", "cloam", "clods", "cloff", "clogs", "cloke", "clomb", "clomp", "clonk", "clons", "cloop", "cloot", "clops", "clote", "clots", "clour", "clous", "clows", "cloye", "cloys", "cloze", "clubs", "clues", "cluey", "clunk", "clype", "cnida", "coact", "coady", "coala", "coals", "coaly", "coapt", "coarb", "coate", "coati", "coats", "cobbs", "cobby", "cobia", "coble", "cobza", "cocas", "cocci", "cocco", "cocks", "cocky", "cocos", "codas", "codec", "coded", "coden", "coder", "codes", "codex", "codon", "coeds", "coffs", "cogie", "cogon", "cogue", "cohab", "cohen", "cohoe", "cohog", "cohos", "coifs", "coign", "coils", "coins", "coirs", "coits", "coked", "cokes", "colas", "colby", "colds", "coled", "coles", "coley", "colic", "colin", "colls", "colly", "colog", "colts", "colza", "comae", "comal", "comas", "combe", "combi", "combo", "combs", "comby", "comer", "comes", "comix", "commo", "comms", "commy", "compo", "comps", "compt", "comte", "comus", "coned", "cones", "coney", "confs", "conga", "conge", "congo", "conia", "conin", "conks", "conky", "conne", "conns", "conte", "conto", "conus", "convo", "cooch", "cooed", "cooee", "cooer", "cooey", "coofs", "cooks", "cooky", "cools", "cooly", "coomb", "cooms", "coomy", "coons", "coops", "coopt", "coost", "coots", "cooze", "copal", "copay", "coped", "copen", "coper", "copes", "coppy", "copra", "copsy", "coqui", "coram", "corbe", "corby", "cords", "cored", "cores", "corey", "corgi", "coria", "corks", "corky", "corms", "corni", "corno", "corns", "cornu", "corps", "corse", "corso", "cosec", "cosed", "coses", "coset", "cosey", "cosie", "costa", "coste", "costs", "cotan", "coted", "cotes", "coths", "cotta", "cotts", "coude", "coups", "courb", "courd", "coure", "cours", "couta", "couth", "coved", "coves", "covin", "cowal", "cowan", "cowed", "cowks", "cowls", "cowps", "cowry", "coxae", "coxal", "coxed", "coxes", "coxib", "coyau", "coyed", "coyer", "coypu", "cozed", "cozen", "cozes", "cozey", "cozie", "craal", "crabs", "crags", "craic", "craig", "crake", "crame", "crams", "crans", "crape", "craps", "crapy", "crare", "craws", "crays", "creds", "creel", "crees", "crems", "crena", "creps", "crepy", "crewe", "crews", "crias", "cribs", "cries", "crims", "crine", "crios", "cripe", "crips", "crise", "crith", "crits", "croci", "crocs", "croft", "crogs", "cromb", "crome", "cronk", "crons", "crool", "croon", "crops", "crore", "crost", "crout", "crows", "croze", "cruck", "crudo", "cruds", "crudy", "crues", "cruet", "cruft", "crunk", "cruor", "crura", "cruse", "crusy", "cruve", "crwth", "cryer", "ctene", "cubby", "cubeb", "cubed", "cuber", "cubes", "cubit", "cuddy", "cuffo", "cuffs", "cuifs", "cuing", "cuish", "cuits", "cukes", "culch", "culet", "culex", "culls", "cully", "culms", "culpa", "culti", "cults", "culty", "cumec", "cundy", "cunei", "cunit", "cunts", "cupel", "cupid", "cuppa", "cuppy", "curat", "curbs", "curch", "curds", "curdy", "cured", "curer", "cures", "curet", "curfs", "curia", "curie", "curli", "curls", "curns", "curny", "currs", "cursi", "curst", "cusec", "cushy", "cusks", "cusps", "cuspy", "cusso", "cusum", "cutch", "cuter", "cutes", "cutey", "cutin", "cutis", "cutto", "cutty", "cutup", "cuvee", "cuzes", "cwtch", "cyano", "cyans", "cycad", "cycas", "cyclo", "cyder", "cylix", "cymae", "cymar", "cymas", "cymes", "cymol", "cysts", "cytes", "cyton", "czars", "daals", "dabba", "daces", "dacha", "dacks", "dadah", "dadas", "dados", "daffs", "daffy", "dagga", "daggy", "dagos", "dahls", "daiko", "daine", "daint", "daker", "daled", "dales", "dalis", "dalle", "dalts", "daman", "damar", "dames", "damme", "damns", "damps", "dampy", "dancy", "dangs", "danio", "danks", "danny", "dants", "daraf", "darbs", "darcy", "dared", "darer", "dares", "darga", "dargs", "daric", "daris", "darks", "darky", "darns", "darre", "darts", "darzi", "dashi", "dashy", "datal", "dated", "dater", "dates", "datos", "datto", "daube", "daubs", "dauby", "dauds", "dault", "daurs", "dauts", "daven", "davit", "dawah", "dawds", "dawed", "dawen", "dawks", "dawns", "dawts", "dayan", "daych", "daynt", "dazed", "dazer", "dazes", "deads", "deair", "deals", "deans", "deare", "dearn", "dears", "deary", "deash", "deave", "deaws", "deawy", "debag", "debby", "debel", "debes", "debts", "debud", "debur", "debus", "debye", "decad", "decaf", "decan", "decko", "decks", "decos", "dedal", "deeds", "deedy", "deely", "deems", "deens", "deeps", "deere", "deers", "deets", "deeve", "deevs", "defat", "deffo", "defis", "defog", "degas", "degum", "degus", "deice", "deids", "deify", "deils", "deism", "deist", "deked", "dekes", "dekko", "deled", "deles", "delfs", "delft", "delis", "dells", "delly", "delos", "delph", "delts", "deman", "demes", "demic", "demit", "demob", "demoi", "demos", "dempt", "denar", "denay", "dench", "denes", "denet", "denis", "dents", "deoxy", "derat", "deray", "dered", "deres", "derig", "derma", "derms", "derns", "derny", "deros", "derro", "derry", "derth", "dervs", "desex", "deshi", "desis", "desks", "desse", "devas", "devel", "devis", "devon", "devos", "devot", "dewan", "dewar", "dewax", "dewed", "dexes", "dexie", "dhaba", "dhaks", "dhals", "dhikr", "dhobi", "dhole", "dholl", "dhols", "dhoti", "dhows", "dhuti", "diact", "dials", "diane", "diazo", "dibbs", "diced", "dicer", "dices", "dicht", "dicks", "dicky", "dicot", "dicta", "dicts", "dicty", "diddy", "didie", "didos", "didst", "diebs", "diels", "diene", "diets", "diffs", "dight", "dikas", "diked", "diker", "dikes", "dikey", "dildo", "dilli", "dills", "dimbo", "dimer", "dimes", "dimps", "dinar", "dined", "dines", "dinge", "dings", "dinic", "dinks", "dinky", "dinna", "dinos", "dints", "diols", "diota", "dippy", "dipso", "diram", "direr", "dirke", "dirks", "dirls", "dirts", "disas", "disci", "discs", "dishy", "disks", "disme", "dital", "ditas", "dited", "dites", "ditsy", "ditts", "ditzy", "divan", "divas", "dived", "dives", "divis", "divna", "divos", "divot", "divvy", "diwan", "dixie", "dixit", "diyas", "dizen", "djinn", "djins", "doabs", "doats", "dobby", "dobes", "dobie", "dobla", "dobra", "dobro", "docht", "docks", "docos", "docus", "doddy", "dodos", "doeks", "doers", "doest", "doeth", "doffs", "dogan", "doges", "dogey", "doggo", "doggy", "dogie", "dohyo", "doilt", "doily", "doits", "dojos", "dolce", "dolci", "doled", "doles", "dolia", "dolls", "dolma", "dolor", "dolos", "dolts", "domal", "domed", "domes", "domic", "donah", "donas", "donee", "doner", "donga", "dongs", "donko", "donna", "donne", "donny", "donsy", "doobs", "dooce", "doody", "dooks", "doole", "dools", "dooly", "dooms", "doomy", "doona", "doorn", "doors", "doozy", "dopas", "doped", "doper", "dopes", "dorad", "dorba", "dorbs", "doree", "dores", "doric", "doris", "dorks", "dorky", "dorms", "dormy", "dorps", "dorrs", "dorsa", "dorse", "dorts", "dorty", "dosai", "dosas", "dosed", "doseh", "doser", "doses", "dosha", "dotal", "doted", "doter", "dotes", "dotty", "douar", "douce", "doucs", "douks", "doula", "douma", "doums", "doups", "doura", "douse", "douts", "doved", "doven", "dover", "doves", "dovie", "dowar", "dowds", "dowed", "dower", "dowie", "dowle", "dowls", "dowly", "downa", "downs", "dowps", "dowse", "dowts", "doxed", "doxes", "doxie", "doyen", "doyly", "dozed", "dozer", "dozes", "drabs", "drack", "draco", "draff", "drags", "drail", "drams", "drant", "draps", "drats", "drave", "draws", "drays", "drear", "dreck", "dreed", "dreer", "drees", "dregs", "dreks", "drent", "drere", "drest", "dreys", "dribs", "drice", "dries", "drily", "drips", "dript", "droid", "droil", "droke", "drole", "drome", "drony", "droob", "droog", "drook", "drops", "dropt", "drouk", "drows", "drubs", "drugs", "drums", "drupe", "druse", "drusy", "druxy", "dryad", "dryas", "dsobo", "dsomo", "duads", "duals", "duans", "duars", "dubbo", "ducal", "ducat", "duces", "ducks", "ducky", "ducts", "duddy", "duded", "dudes", "duels", "duets", "duett", "duffs", "dufus", "duing", "duits", "dukas", "duked", "dukes", "dukka", "dulce", "dules", "dulia", "dulls", "dulse", "dumas", "dumbo", "dumbs", "dumka", "dumky", "dumps", "dunam", "dunch", "dunes", "dungs", "dungy", "dunks", "dunno", "dunny", "dunsh", "dunts", "duomi", "duomo", "duped", "duper", "dupes", "duple", "duply", "duppy", "dural", "duras", "dured", "dures", "durgy", "durns", "duroc", "duros", "duroy", "durra", "durrs", "durry", "durst", "durum", "durzi", "dusks", "dusts", "duxes", "dwaal", "dwale", "dwalm", "dwams", "dwang", "dwaum", "dweeb", "dwile", "dwine", "dyads", "dyers", "dyked", "dykes", "dykey", "dykon", "dynel", "dynes", "dzhos", "eagre", "ealed", "eales", "eaned", "eards", "eared", "earls", "earns", "earnt", "earst", "eased", "easer", "eases", "easle", "easts", "eathe", "eaved", "eaves", "ebbed", "ebbet", "ebons", "ebook", "ecads", "eched", "eches", "echos", "ecrus", "edema", "edged", "edger", "edges", "edile", "edits", "educe", "educt", "eejit", "eensy", "eeven", "eevns", "effed", "egads", "egers", "egest", "eggar", "egged", "egger", "egmas", "ehing", "eider", "eidos", "eigne", "eiked", "eikon", "eilds", "eisel", "ejido", "ekkas", "elain", "eland", "elans", "elchi", "eldin", "elemi", "elfed", "eliad", "elint", "elmen", "eloge", "elogy", "eloin", "elops", "elpee", "elsin", "elute", "elvan", "elven", "elver", "elves", "emacs", "embar", "embay", "embog", "embow", "embox", "embus", "emeer", "emend", "emerg", "emery", "emeus", "emics", "emirs", "emits", "emmas", "emmer", "emmet", "emmew", "emmys", "emoji", "emong", "emote", "emove", "empts", "emule", "emure", "emyde", "emyds", "enarm", "enate", "ended", "ender", "endew", "endue", "enews", "enfix", "eniac", "enlit", "enmew", "ennog", "enoki", "enols", "enorm", "enows", "enrol", "ensew", "ensky", "entia", "enure", "enurn", "envoi", "enzym", "eorls", "eosin", "epact", "epees", "ephah", "ephas", "ephod", "ephor", "epics", "epode", "epopt", "epris", "eques", "equid", "erbia", "erevs", "ergon", "ergos", "ergot", "erhus", "erica", "erick", "erics", "ering", "erned", "ernes", "erose", "erred", "erses", "eruct", "erugo", "eruvs", "erven", "ervil", "escar", "escot", "esile", "eskar", "esker", "esnes", "esses", "estoc", "estop", "estro", "etage", "etape", "etats", "etens", "ethal", "ethne", "ethyl", "etics", "etnas", "ettin", "ettle", "etuis", "etwee", "etyma", "eughs", "euked", "eupad", "euros", "eusol", "evens", "evert", "evets", "evhoe", "evils", "evite", "evohe", "ewers", "ewest", "ewhow", "ewked", "exams", "exeat", "execs", "exeem", "exeme", "exfil", "exies", "exine", "exing", "exits", "exode", "exome", "exons", "expat", "expos", "exude", "exuls", "exurb", "eyass", "eyers", "eyots", "eyras", "eyres", "eyrie", "eyrir", "ezine", "fabby", "faced", "facer", "faces", "facia", "facta", "facts", "faddy", "faded", "fader", "fades", "fadge", "fados", "faena", "faery", "faffs", "faffy", "faggy", "fagin", "fagot", "faiks", "fails", "faine", "fains", "fairs", "faked", "faker", "fakes", "fakey", "fakie", "fakir", "falaj", "falls", "famed", "fames", "fanal", "fands", "fanes", "fanga", "fango", "fangs", "fanks", "fanon", "fanos", "fanum", "faqir", "farad", "farci", "farcy", "fards", "fared", "farer", "fares", "farle", "farls", "farms", "faros", "farro", "farse", "farts", "fasci", "fasti", "fasts", "fated", "fates", "fatly", "fatso", "fatwa", "faugh", "fauld", "fauns", "faurd", "fauts", "fauve", "favas", "favel", "faver", "faves", "favus", "fawns", "fawny", "faxed", "faxes", "fayed", "fayer", "fayne", "fayre", "fazed", "fazes", "feals", "feare", "fears", "feart", "fease", "feats", "feaze", "feces", "fecht", "fecit", "fecks", "fedex", "feebs", "feeds", "feels", "feens", "feers", "feese", "feeze", "fehme", "feint", "feist", "felch", "felid", "fells", "felly", "felts", "felty", "femal", "femes", "femmy", "fends", "fendy", "fenis", "fenks", "fenny", "fents", "feods", "feoff", "ferer", "feres", "feria", "ferly", "fermi", "ferms", "ferns", "ferny", "fesse", "festa", "fests", "festy", "fetas", "feted", "fetes", "fetor", "fetta", "fetts", "fetwa", "feuar", "feuds", "feued", "feyed", "feyer", "feyly", "fezes", "fezzy", "fiars", "fiats", "fibro", "fices", "fiche", "fichu", "ficin", "ficos", "fides", "fidge", "fidos", "fiefs", "fient", "fiere", "fiers", "fiest", "fifed", "fifer", "fifes", "fifis", "figgy", "figos", "fiked", "fikes", "filar", "filch", "filed", "files", "filii", "filks", "fille", "fillo", "fills", "filmi", "films", "filos", "filum", "finca", "finds", "fined", "fines", "finis", "finks", "finny", "finos", "fiord", "fiqhs", "fique", "fired", "firer", "fires", "firie", "firks", "firms", "firns", "firry", "firth", "fiscs", "fisks", "fists", "fisty", "fitch", "fitly", "fitna", "fitte", "fitts", "fiver", "fives", "fixed", "fixes", "fixit", "fjeld", "flabs", "flaff", "flags", "flaks", "flamm", "flams", "flamy", "flane", "flans", "flaps", "flary", "flats", "flava", "flawn", "flaws", "flawy", "flaxy", "flays", "fleam", "fleas", "fleek", "fleer", "flees", "flegs", "fleme", "fleur", "flews", "flexi", "flexo", "fleys", "flics", "flied", "flies", "flimp", "flims", "flips", "flirs", "flisk", "flite", "flits", "flitt", "flobs", "flocs", "floes", "flogs", "flong", "flops", "flors", "flory", "flosh", "flota", "flote", "flows", "flubs", "flued", "flues", "fluey", "fluky", "flump", "fluor", "flurr", "fluty", "fluyt", "flyby", "flype", "flyte", "foals", "foams", "foehn", "fogey", "fogie", "fogle", "fogou", "fohns", "foids", "foils", "foins", "folds", "foley", "folia", "folic", "folie", "folks", "folky", "fomes", "fonda", "fonds", "fondu", "fones", "fonly", "fonts", "foods", "foody", "fools", "foots", "footy", "foram", "forbs", "forby", "fordo", "fords", "forel", "fores", "forex", "forks", "forky", "forme", "forms", "forts", "forza", "forze", "fossa", "fosse", "fouat", "fouds", "fouer", "fouet", "foule", "fouls", "fount", "fours", "fouth", "fovea", "fowls", "fowth", "foxed", "foxes", "foxie", "foyle", "foyne", "frabs", "frack", "fract", "frags", "fraim", "franc", "frape", "fraps", "frass", "frate", "frati", "frats", "fraus", "frays", "frees", "freet", "freit", "fremd", "frena", "freon", "frere", "frets", "fribs", "frier", "fries", "frigs", "frise", "frist", "frith", "frits", "fritt", "frize", "frizz", "froes", "frogs", "frons", "frore", "frorn", "frory", "frosh", "frows", "frowy", "frugs", "frump", "frush", "frust", "fryer", "fubar", "fubby", "fubsy", "fucks", "fucus", "fuddy", "fudgy", "fuels", "fuero", "fuffs", "fuffy", "fugal", "fuggy", "fugie", "fugio", "fugle", "fugly", "fugus", "fujis", "fulls", "fumed", "fumer", "fumes", "fumet", "fundi", "funds", "fundy", "fungo", "fungs", "funks", "fural", "furan", "furca", "furls", "furol", "furrs", "furth", "furze", "furzy", "fused", "fusee", "fusel", "fuses", "fusil", "fusks", "fusts", "fusty", "futon", "fuzed", "fuzee", "fuzes", "fuzil", "fyces", "fyked", "fykes", "fyles", "fyrds", "fytte", "gabba", "gabby", "gable", "gaddi", "gades", "gadge", "gadid", "gadis", "gadje", "gadjo", "gadso", "gaffs", "gaged", "gager", "gages", "gaids", "gains", "gairs", "gaita", "gaits", "gaitt", "gajos", "galah", "galas", "galax", "galea", "galed", "gales", "galls", "gally", "galop", "galut", "galvo", "gamas", "gamay", "gamba", "gambe", "gambo", "gambs", "gamed", "games", "gamey", "gamic", "gamin", "gamme", "gammy", "gamps", "ganch", "gandy", "ganef", "ganev", "gangs", "ganja", "ganof", "gants", "gaols", "gaped", "gaper", "gapes", "gapos", "gappy", "garbe", "garbo", "garbs", "garda", "gares", "garis", "garms", "garni", "garre", "garth", "garum", "gases", "gasps", "gaspy", "gasts", "gatch", "gated", "gater", "gates", "gaths", "gator", "gauch", "gaucy", "gauds", "gauje", "gault", "gaums", "gaumy", "gaups", "gaurs", "gauss", "gauzy", "gavot", "gawcy", "gawds", "gawks", "gawps", "gawsy", "gayal", "gazal", "gazar", "gazed", "gazes", "gazon", "gazoo", "geals", "geans", "geare", "gears", "geats", "gebur", "gecks", "geeks", "geeps", "geest", "geist", "geits", "gelds", "gelee", "gelid", "gelly", "gelts", "gemel", "gemma", "gemmy", "gemot", "genal", "genas", "genes", "genet", "genic", "genii", "genip", "genny", "genoa", "genom", "genro", "gents", "genty", "genua", "genus", "geode", "geoid", "gerah", "gerbe", "geres", "gerle", "germs", "germy", "gerne", "gesse", "gesso", "geste", "gests", "getas", "getup", "geums", "geyan", "geyer", "ghast", "ghats", "ghaut", "ghazi", "ghees", "ghest", "ghyll", "gibed", "gibel", "giber", "gibes", "gibli", "gibus", "gifts", "gigas", "gighe", "gigot", "gigue", "gilas", "gilds", "gilet", "gills", "gilly", "gilpy", "gilts", "gimel", "gimme", "gimps", "gimpy", "ginch", "ginge", "gings", "ginks", "ginny", "ginzo", "gipon", "gippo", "gippy", "girds", "girls", "girns", "giron", "giros", "girrs", "girsh", "girts", "gismo", "gisms", "gists", "gitch", "gites", "giust", "gived", "gives", "gizmo", "glace", "glads", "glady", "glaik", "glair", "glams", "glans", "glary", "glaum", "glaur", "glazy", "gleba", "glebe", "gleby", "glede", "gleds", "gleed", "gleek", "glees", "gleet", "gleis", "glens", "glent", "gleys", "glial", "glias", "glibs", "gliff", "glift", "glike", "glime", "glims", "glisk", "glits", "glitz", "gloam", "globi", "globs", "globy", "glode", "glogg", "gloms", "gloop", "glops", "glost", "glout", "glows", "gloze", "glued", "gluer", "glues", "gluey", "glugs", "glume", "glums", "gluon", "glute", "gluts", "gnarl", "gnarr", "gnars", "gnats", "gnawn", "gnaws", "gnows", "goads", "goafs", "goals", "goary", "goats", "goaty", "goban", "gobar", "gobbi", "gobbo", "gobby", "gobis", "gobos", "godet", "godso", "goels", "goers", "goest", "goeth", "goety", "gofer", "goffs", "gogga", "gogos", "goier", "gojis", "golds", "goldy", "goles", "golfs", "golpe", "golps", "gombo", "gomer", "gompa", "gonch", "gonef", "gongs", "gonia", "gonif", "gonks", "gonna", "gonof", "gonys", "gonzo", "gooby", "goods", "goofs", "googs", "gooks", "gooky", "goold", "gools", "gooly", "goons", "goony", "goops", "goopy", "goors", "goory", "goosy", "gopak", "gopik", "goral", "goras", "gored", "gores", "goris", "gorms", "gormy", "gorps", "gorse", "gorsy", "gosht", "gosse", "gotch", "goths", "gothy", "gotta", "gouch", "gouks", "goura", "gouts", "gouty", "gowan", "gowds", "gowfs", "gowks", "gowls", "gowns", "goxes", "goyim", "goyle", "graal", "grabs", "grads", "graff", "graip", "grama", "grame", "gramp", "grams", "grana", "grans", "grapy", "gravs", "grays", "grebe", "grebo", "grece", "greek", "grees", "grege", "grego", "grein", "grens", "grese", "greve", "grews", "greys", "grice", "gride", "grids", "griff", "grift", "grigs", "grike", "grins", "griot", "grips", "gript", "gripy", "grise", "grist", "grisy", "grith", "grits", "grize", "groat", "grody", "grogs", "groks", "groma", "grone", "groof", "grosz", "grots", "grouf", "grovy", "grows", "grrls", "grrrl", "grubs", "grued", "grues", "grufe", "grume", "grump", "grund", "gryce", "gryde", "gryke", "grype", "grypt", "guaco", "guana", "guano", "guans", "guars", "gucks", "gucky", "gudes", "guffs", "gugas", "guids", "guimp", "guiro", "gulag", "gular", "gulas", "gules", "gulet", "gulfs", "gulfy", "gulls", "gulph", "gulps", "gulpy", "gumma", "gummi", "gumps", "gundy", "gunge", "gungy", "gunks", "gunky", "gunny", "guqin", "gurdy", "gurge", "gurls", "gurly", "gurns", "gurry", "gursh", "gurus", "gushy", "gusla", "gusle", "gusli", "gussy", "gusts", "gutsy", "gutta", "gutty", "guyed", "guyle", "guyot", "guyse", "gwine", "gyals", "gyans", "gybed", "gybes", "gyeld", "gymps", "gynae", "gynie", "gynny", "gynos", "gyoza", "gypos", "gyppo", "gyppy", "gyral", "gyred", "gyres", "gyron", "gyros", "gyrus", "gytes", "gyved", "gyves", "haafs", "haars", "hable", "habus", "hacek", "hacks", "hadal", "haded", "hades", "hadji", "hadst", "haems", "haets", "haffs", "hafiz", "hafts", "haggs", "hahas", "haick", "haika", "haiks", "haiku", "hails", "haily", "hains", "haint", "hairs", "haith", "hajes", "hajis", "hajji", "hakam", "hakas", "hakea", "hakes", "hakim", "hakus", "halal", "haled", "haler", "hales", "halfa", "halfs", "halid", "hallo", "halls", "halma", "halms", "halon", "halos", "halse", "halts", "halva", "halwa", "hamal", "hamba", "hamed", "hames", "hammy", "hamza", "hanap", "hance", "hanch", "hands", "hangi", "hangs", "hanks", "hanky", "hansa", "hanse", "hants", "haole", "haoma", "hapax", "haply", "happi", "hapus", "haram", "hards", "hared", "hares", "harim", "harks", "harls", "harms", "harns", "haros", "harps", "harts", "hashy", "hasks", "hasps", "hasta", "hated", "hates", "hatha", "hauds", "haufs", "haugh", "hauld", "haulm", "hauls", "hault", "hauns", "hause", "haver", "haves", "hawed", "hawks", "hawms", "hawse", "hayed", "hayer", "hayey", "hayle", "hazan", "hazed", "hazer", "hazes", "heads", "heald", "heals", "heame", "heaps", "heapy", "heare", "hears", "heast", "heats", "heben", "hebes", "hecht", "hecks", "heder", "hedgy", "heeds", "heedy", "heels", "heeze", "hefte", "hefts", "heids", "heigh", "heils", "heirs", "hejab", "hejra", "heled", "heles", "helio", "hells", "helms", "helos", "helot", "helps", "helve", "hemal", "hemes", "hemic", "hemin", "hemps", "hempy", "hench", "hends", "henge", "henna", "henny", "henry", "hents", "hepar", "herbs", "herby", "herds", "heres", "herls", "herma", "herms", "herns", "heros", "herry", "herse", "hertz", "herye", "hesps", "hests", "hetes", "heths", "heuch", "heugh", "hevea", "hewed", "hewer", "hewgh", "hexad", "hexed", "hexer", "hexes", "hexyl", "heyed", "hiant", "hicks", "hided", "hider", "hides", "hiems", "highs", "hight", "hijab", "hijra", "hiked", "hiker", "hikes", "hikoi", "hilar", "hilch", "hillo", "hills", "hilts", "hilum", "hilus", "himbo", "hinau", "hinds", "hings", "hinky", "hinny", "hints", "hiois", "hiply", "hired", "hiree", "hirer", "hires", "hissy", "hists", "hithe", "hived", "hiver", "hives", "hizen", "hoaed", "hoagy", "hoars", "hoary", "hoast", "hobos", "hocks", "hocus", "hodad", "hodja", "hoers", "hogan", "hogen", "hoggs", "hoghs", "hohed", "hoick", "hoied", "hoiks", "hoing", "hoise", "hokas", "hoked", "hokes", "hokey", "hokis", "hokku", "hokum", "holds", "holed", "holes", "holey", "holks", "holla", "hollo", "holme", "holms", "holon", "holos", "holts", "homas", "homed", "homes", "homey", "homie", "homme", "homos", "honan", "honda", "honds", "honed", "honer", "hones", "hongi", "hongs", "honks", "honky", "hooch", "hoods", "hoody", "hooey", "hoofs", "hooka", "hooks", "hooky", "hooly", "hoons", "hoops", "hoord", "hoors", "hoosh", "hoots", "hooty", "hoove", "hopak", "hoped", "hoper", "hopes", "hoppy", "horah", "horal", "horas", "horis", "horks", "horme", "horns", "horst", "horsy", "hosed", "hosel", "hosen", "hoser", "hoses", "hosey", "hosta", "hosts", "hotch", "hoten", "hotty", "houff", "houfs", "hough", "houri", "hours", "houts", "hovea", "hoved", "hoven", "hoves", "howbe", "howes", "howff", "howfs", "howks", "howls", "howre", "howso", "hoxed", "hoxes", "hoyas", "hoyed", "hoyle", "hubby", "hucks", "hudna", "hudud", "huers", "huffs", "huffy", "huger", "huggy", "huhus", "huias", "hulas", "hules", "hulks", "hulky", "hullo", "hulls", "hully", "humas", "humfs", "humic", "humps", "humpy", "hunks", "hunts", "hurds", "hurls", "hurly", "hurra", "hurst", "hurts", "hushy", "husks", "husos", "hutia", "huzza", "huzzy", "hwyls", "hydra", "hyens", "hygge", "hying", "hykes", "hylas", "hyleg", "hyles", "hylic", "hymns", "hynde", "hyoid", "hyped", "hypes", "hypha", "hyphy", "hypos", "hyrax", "hyson", "hythe", "iambi", "iambs", "ibrik", "icers", "iched", "iches", "ichor", "icier", "icker", "ickle", "icons", "ictal", "ictic", "ictus", "idant", "ideas", "idees", "ident", "idled", "idles", "idola", "idols", "idyls", "iftar", "igapo", "igged", "iglus", "ihram", "ikans", "ikats", "ikons", "ileac", "ileal", "ileum", "ileus", "iliad", "ilial", "ilium", "iller", "illth", "imago", "imams", "imari", "imaum", "imbar", "imbed", "imide", "imido", "imids", "imine", "imino", "immew", "immit", "immix", "imped", "impis", "impot", "impro", "imshi", "imshy", "inapt", "inarm", "inbye", "incel", "incle", "incog", "incus", "incut", "indew", "india", "indie", "indol", "indow", "indri", "indue", "inerm", "infix", "infos", "infra", "ingan", "ingle", "inion", "inked", "inker", "inkle", "inned", "innit", "inorb", "inrun", "inset", "inspo", "intel", "intil", "intis", "intra", "inula", "inure", "inurn", "inust", "invar", "inwit", "iodic", "iodid", "iodin", "iotas", "ippon", "irade", "irids", "iring", "irked", "iroko", "irone", "irons", "isbas", "ishes", "isled", "isles", "isnae", "issei", "istle", "items", "ither", "ivied", "ivies", "ixias", "ixnay", "ixora", "ixtle", "izard", "izars", "izzat", "jaaps", "jabot", "jacal", "jacks", "jacky", "jaded", "jades", "jafas", "jaffa", "jagas", "jager", "jaggs", "jaggy", "jagir", "jagra", "jails", "jaker", "jakes", "jakey", "jalap", "jalop", "jambe", "jambo", "jambs", "jambu", "james", "jammy", "jamon", "janes", "janns", "janny", "janty", "japan", "japed", "japer", "japes", "jarks", "jarls", "jarps", "jarta", "jarul", "jasey", "jaspe", "jasps", "jatos", "jauks", "jaups", "javas", "javel", "jawan", "jawed", "jaxie", "jeans", "jeats", "jebel", "jedis", "jeels", "jeely", "jeeps", "jeers", "jeeze", "jefes", "jeffs", "jehad", "jehus", "jelab", "jello", "jells", "jembe", "jemmy", "jenny", "jeons", "jerid", "jerks", "jerry", "jesse", "jests", "jesus", "jetes", "jeton", "jeune", "jewed", "jewie", "jhala", "jiaos", "jibba", "jibbs", "jibed", "jiber", "jibes", "jiffs", "jiggy", "jigot", "jihad", "jills", "jilts", "jimmy", "jimpy", "jingo", "jinks", "jinne", "jinni", "jinns", "jirds", "jirga", "jirre", "jisms", "jived", "jiver", "jives", "jivey", "jnana", "jobed", "jobes", "jocko", "jocks", "jocky", "jocos", "jodel", "joeys", "johns", "joins", "joked", "jokes", "jokey", "jokol", "joled", "joles", "jolls", "jolts", "jolty", "jomon", "jomos", "jones", "jongs", "jonty", "jooks", "joram", "jorum", "jotas", "jotty", "jotun", "joual", "jougs", "jouks", "joule", "jours", "jowar", "jowed", "jowls", "jowly", "joyed", "jubas", "jubes", "jucos", "judas", "judgy", "judos", "jugal", "jugum", "jujus", "juked", "jukes", "jukus", "julep", "jumar", "jumby", "jumps", "junco", "junks", "junky", "jupes", "jupon", "jural", "jurat", "jurel", "jures", "justs", "jutes", "jutty", "juves", "juvie", "kaama", "kabab", "kabar", "kabob", "kacha", "kacks", "kadai", "kades", "kadis", "kafir", "kagos", "kagus", "kahal", "kaiak", "kaids", "kaies", "kaifs", "kaika", "kaiks", "kails", "kaims", "kaing", "kains", "kakas", "kakis", "kalam", "kales", "kalif", "kalis", "kalpa", "kamas", "kames", "kamik", "kamis", "kamme", "kanae", "kanas", "kandy", "kaneh", "kanes", "kanga", "kangs", "kanji", "kants", "kanzu", "kaons", "kapas", "kaphs", "kapok", "kapow", "kapus", "kaput", "karas", "karat", "karks", "karns", "karoo", "karos", "karri", "karst", "karsy", "karts", "karzy", "kasha", "kasme", "katal", "katas", "katis", "katti", "kaugh", "kauri", "kauru", "kaury", "kaval", "kavas", "kawas", "kawau", "kawed", "kayle", "kayos", "kazis", "kazoo", "kbars", "kebar", "kebob", "kecks", "kedge", "kedgy", "keech", "keefs", "keeks", "keels", "keema", "keeno", "keens", "keeps", "keets", "keeve", "kefir", "kehua", "keirs", "kelep", "kelim", "kells", "kelly", "kelps", "kelpy", "kelts", "kelty", "kembo", "kembs", "kemps", "kempt", "kempy", "kenaf", "kench", "kendo", "kenos", "kente", "kents", "kepis", "kerbs", "kerel", "kerfs", "kerky", "kerma", "kerne", "kerns", "keros", "kerry", "kerve", "kesar", "kests", "ketas", "ketch", "ketes", "ketol", "kevel", "kevil", "kexes", "keyed", "keyer", "khadi", "khafs", "khans", "khaph", "khats", "khaya", "khazi", "kheda", "kheth", "khets", "khoja", "khors", "khoum", "khuds", "kiaat", "kiack", "kiang", "kibbe", "kibbi", "kibei", "kibes", "kibla", "kicks", "kicky", "kiddo", "kiddy", "kidel", "kidge", "kiefs", "kiers", "kieve", "kievs", "kight", "kikes", "kikoi", "kiley", "kilim", "kills", "kilns", "kilos", "kilps", "kilts", "kilty", "kimbo", "kinas", "kinda", "kinds", "kindy", "kines", "kings", "kinin", "kinks", "kinos", "kiore", "kipes", "kippa", "kipps", "kirby", "kirks", "kirns", "kirri", "kisan", "kissy", "kists", "kited", "kiter", "kites", "kithe", "kiths", "kitul", "kivas", "kiwis", "klang", "klaps", "klett", "klick", "klieg", "kliks", "klong", "kloof", "kluge", "klutz", "knags", "knaps", "knarl", "knars", "knaur", "knawe", "knees", "knell", "knish", "knits", "knive", "knobs", "knops", "knosp", "knots", "knout", "knowe", "knows", "knubs", "knurl", "knurr", "knurs", "knuts", "koans", "koaps", "koban", "kobos", "koels", "koffs", "kofta", "kogal", "kohas", "kohen", "kohls", "koine", "kojis", "kokam", "kokas", "koker", "kokra", "kokum", "kolas", "kolos", "kombu", "konbu", "kondo", "konks", "kooks", "kooky", "koori", "kopek", "kophs", "kopje", "koppa", "korai", "koras", "korat", "kores", "korma", "koros", "korun", "korus", "koses", "kotch", "kotos", "kotow", "koura", "kraal", "krabs", "kraft", "krais", "krait", "krang", "krans", "kranz", "kraut", "krays", "kreep", "kreng", "krewe", "krona", "krone", "kroon", "krubi", "krunk", "ksars", "kubie", "kudos", "kudus", "kudzu", "kufis", "kugel", "kuias", "kukri", "kukus", "kulak", "kulan", "kulas", "kulfi", "kumis", "kumys", "kuris", "kurre", "kurta", "kurus", "kusso", "kutas", "kutch", "kutis", "kutus", "kuzus", "kvass", "kvell", "kwela", "kyack", "kyaks", "kyang", "kyars", "kyats", "kybos", "kydst", "kyles", "kylie", "kylin", "kylix", "kyloe", "kynde", "kynds", "kypes", "kyrie", "kytes", "kythe", "laari", "labda", "labia", "labis", "labra", "laced", "lacer", "laces", "lacet", "lacey", "lacks", "laddy", "laded", "lader", "lades", "laers", "laevo", "lagan", "lahal", "lahar", "laich", "laics", "laids", "laigh", "laika", "laiks", "laird", "lairs", "lairy", "laith", "laity", "laked", "laker", "lakes", "lakhs", "lakin", "laksa", "laldy", "lalls", "lamas", "lambs", "lamby", "lamed", "lamer", "lames", "lamia", "lammy", "lamps", "lanai", "lanas", "lanch", "lande", "lands", "lanes", "lanks", "lants", "lapin", "lapis", "lapje", "larch", "lards", "lardy", "laree", "lares", "largo", "laris", "larks", "larky", "larns", "larnt", "larum", "lased", "laser", "lases", "lassi", "lassu", "lassy", "lasts", "latah", "lated", "laten", "latex", "lathi", "laths", "lathy", "latke", "latus", "lauan", "lauch", "lauds", "laufs", "laund", "laura", "laval", "lavas", "laved", "laver", "laves", "lavra", "lavvy", "lawed", "lawer", "lawin", "lawks", "lawns", "lawny", "laxed", "laxer", "laxes", "laxly", "layed", "layin", "layup", "lazar", "lazed", "lazes", "lazos", "lazzi", "lazzo", "leads", "leady", "leafs", "leaks", "leams", "leans", "leany", "leaps", "leare", "lears", "leary", "leats", "leavy", "leaze", "leben", "leccy", "ledes", "ledgy", "ledum", "leear", "leeks", "leeps", "leers", "leese", "leets", "leeze", "lefte", "lefts", "leger", "leges", "legge", "leggo", "legit", "lehrs", "lehua", "leirs", "leish", "leman", "lemed", "lemel", "lemes", "lemma", "lemme", "lends", "lenes", "lengs", "lenis", "lenos", "lense", "lenti", "lento", "leone", "lepid", "lepra", "lepta", "lered", "leres", "lerps", "lesbo", "leses", "lests", "letch", "lethe", "letup", "leuch", "leuco", "leuds", "leugh", "levas", "levee", "leves", "levin", "levis", "lewis", "lexes", "lexis", "lezes", "lezza", "lezzy", "liana", "liane", "liang", "liard", "liars", "liart", "liber", "libra", "libri", "lichi", "licht", "licit", "licks", "lidar", "lidos", "liefs", "liens", "liers", "lieus", "lieve", "lifer", "lifes", "lifts", "ligan", "liger", "ligge", "ligne", "liked", "liker", "likes", "likin", "lills", "lilos", "lilts", "liman", "limas", "limax", "limba", "limbi", "limbs", "limby", "limed", "limen", "limes", "limey", "limma", "limns", "limos", "limpa", "limps", "linac", "linch", "linds", "lindy", "lined", "lines", "liney", "linga", "lings", "lingy", "linin", "links", "linky", "linns", "linny", "linos", "lints", "linty", "linum", "linux", "lions", "lipas", "lipes", "lipin", "lipos", "lippy", "liras", "lirks", "lirot", "lisks", "lisle", "lisps", "lists", "litai", "litas", "lited", "liter", "lites", "litho", "liths", "litre", "lived", "liven", "lives", "livor", "livre", "llano", "loach", "loads", "loafs", "loams", "loans", "loast", "loave", "lobar", "lobed", "lobes", "lobos", "lobus", "loche", "lochs", "locie", "locis", "locks", "locos", "locum", "loden", "lodes", "loess", "lofts", "logan", "loges", "loggy", "logia", "logie", "logoi", "logon", "logos", "lohan", "loids", "loins", "loipe", "loirs", "lokes", "lolls", "lolly", "lolog", "lomas", "lomed", "lomes", "loner", "longa", "longe", "longs", "looby", "looed", "looey", "loofa", "loofs", "looie", "looks", "looky", "looms", "loons", "loony", "loops", "loord", "loots", "loped", "loper", "lopes", "loppy", "loral", "loran", "lords", "lordy", "lorel", "lores", "loric", "loris", "losed", "losel", "losen", "loses", "lossy", "lotah", "lotas", "lotes", "lotic", "lotos", "lotsa", "lotta", "lotte", "lotto", "lotus", "loued", "lough", "louie", "louis", "louma", "lound", "louns", "loupe", "loups", "loure", "lours", "loury", "louts", "lovat", "loved", "loves", "lovey", "lovie", "lowan", "lowed", "lowes", "lownd", "lowne", "lowns", "lowps", "lowry", "lowse", "lowts", "loxed", "loxes", "lozen", "luach", "luaus", "lubed", "lubes", "lubra", "luces", "lucks", "lucre", "ludes", "ludic", "ludos", "luffa", "luffs", "luged", "luger", "luges", "lulls", "lulus", "lumas", "lumbi", "lumme", "lummy", "lumps", "lunas", "lunes", "lunet", "lungi", "lungs", "lunks", "lunts", "lupin", "lured", "lurer", "lures", "lurex", "lurgi", "lurgy", "lurks", "lurry", "lurve", "luser", "lushy", "lusks", "lusts", "lusus", "lutea", "luted", "luter", "lutes", "luvvy", "luxed", "luxer", "luxes", "lweis", "lyams", "lyard", "lyart", "lyase", "lycea", "lycee", "lycra", "lymes", "lynes", "lyres", "lysed", "lyses", "lysin", "lysis", "lysol", "lyssa", "lyted", "lytes", "lythe", "lytic", "lytta", "maaed", "maare", "maars", "mabes", "macas", "maced", "macer", "maces", "mache", "machi", "machs", "macks", "macle", "macon", "madge", "madid", "madre", "maerl", "mafic", "mages", "maggs", "magot", "magus", "mahoe", "mahua", "mahwa", "maids", "maiko", "maiks", "maile", "maill", "mails", "maims", "mains", "maire", "mairs", "maise", "maist", "makar", "makes", "makis", "makos", "malam", "malar", "malas", "malax", "males", "malic", "malik", "malis", "malls", "malms", "malmy", "malts", "malty", "malus", "malva", "malwa", "mamas", "mamba", "mamee", "mamey", "mamie", "manas", "manat", "mandi", "maneb", "maned", "maneh", "manes", "manet", "mangs", "manis", "manky", "manna", "manos", "manse", "manta", "manto", "manty", "manul", "manus", "mapau", "maqui", "marae", "marah", "maras", "marcs", "mardy", "mares", "marge", "margs", "maria", "marid", "marka", "marks", "marle", "marls", "marly", "marms", "maron", "maror", "marra", "marri", "marse", "marts", "marvy", "masas", "mased", "maser", "mases", "mashy", "masks", "massa", "massy", "masts", "masty", "masus", "matai", "mated", "mater", "mates", "maths", "matin", "matlo", "matte", "matts", "matza", "matzo", "mauby", "mauds", "mauls", "maund", "mauri", "mausy", "mauts", "mauzy", "maven", "mavie", "mavin", "mavis", "mawed", "mawks", "mawky", "mawns", "mawrs", "maxed", "maxes", "maxis", "mayan", "mayas", "mayed", "mayos", "mayst", "mazed", "mazer", "mazes", "mazey", "mazut", "mbira", "meads", "meals", "meane", "means", "meany", "meare", "mease", "meath", "meats", "mebos", "mechs", "mecks", "medii", "medle", "meeds", "meers", "meets", "meffs", "meins", "meint", "meiny", "meith", "mekka", "melas", "melba", "melds", "melic", "melik", "mells", "melts", "melty", "memes", "memos", "menad", "mends", "mened", "menes", "menge", "mengs", "mensa", "mense", "mensh", "menta", "mento", "menus", "meous", "meows", "merch", "mercs", "merde", "mered", "merel", "merer", "meres", "meril", "meris", "merks", "merle", "merls", "merse", "mesal", "mesas", "mesel", "meses", "meshy", "mesic", "mesne", "meson", "messy", "mesto", "meted", "metes", "metho", "meths", "metic", "metif", "metis", "metol", "metre", "meuse", "meved", "meves", "mewed", "mewls", "meynt", "mezes", "mezze", "mezzo", "mhorr", "miaou", "miaow", "miasm", "miaul", "micas", "miche", "micht", "micks", "micky", "micos", "micra", "middy", "midgy", "midis", "miens", "mieve", "miffs", "miffy", "mifty", "miggs", "mihas", "mihis", "miked", "mikes", "mikra", "mikva", "milch", "milds", "miler", "miles", "milfs", "milia", "milko", "milks", "mille", "mills", "milor", "milos", "milpa", "milts", "milty", "miltz", "mimed", "mimeo", "mimer", "mimes", "mimsy", "minae", "minar", "minas", "mincy", "minds", "mined", "mines", "minge", "mings", "mingy", "minis", "minke", "minks", "minny", "minos", "mints", "mired", "mires", "mirex", "mirid", "mirin", "mirks", "mirky", "mirly", "miros", "mirvs", "mirza", "misch", "misdo", "mises", "misgo", "misos", "missa", "mists", "misty", "mitch", "miter", "mites", "mitis", "mitre", "mitts", "mixed", "mixen", "mixer", "mixes", "mixte", "mixup", "mizen", "mizzy", "mneme", "moans", "moats", "mobby", "mobes", "mobey", "mobie", "moble", "mochi", "mochs", "mochy", "mocks", "moder", "modes", "modge", "modii", "modus", "moers", "mofos", "moggy", "mohel", "mohos", "mohrs", "mohua", "mohur", "moile", "moils", "moira", "moire", "moits", "mojos", "mokes", "mokis", "mokos", "molal", "molas", "molds", "moled", "moles", "molla", "molls", "molly", "molto", "molts", "molys", "momes", "momma", "mommy", "momus", "monad", "monal", "monas", "monde", "mondo", "moner", "mongo", "mongs", "monic", "monie", "monks", "monos", "monte", "monty", "moobs", "mooch", "moods", "mooed", "mooks", "moola", "mooli", "mools", "mooly", "moong", "moons", "moony", "moops", "moors", "moory", "moots", "moove", "moped", "moper", "mopes", "mopey", "moppy", "mopsy", "mopus", "morae", "moras", "morat", "moray", "morel", "mores", "moria", "morne", "morns", "morra", "morro", "morse", "morts", "mosed", "moses", "mosey", "mosks", "mosso", "moste", "mosts", "moted", "moten", "motes", "motet", "motey", "moths", "mothy", "motis", "motte", "motts", "motty", "motus", "motza", "mouch", "moues", "mould", "mouls", "moups", "moust", "mousy", "moved", "moves", "mowas", "mowed", "mowra", "moxas", "moxie", "moyas", "moyle", "moyls", "mozed", "mozes", "mozos", "mpret", "mucho", "mucic", "mucid", "mucin", "mucks", "mucor", "mucro", "mudge", "mudir", "mudra", "muffs", "mufti", "mugga", "muggs", "muggy", "muhly", "muids", "muils", "muirs", "muist", "mujik", "mulct", "muled", "mules", "muley", "mulga", "mulie", "mulla", "mulls", "mulse", "mulsh", "mumms", "mumps", "mumsy", "mumus", "munga", "munge", "mungo", "mungs", "munis", "munts", "muntu", "muons", "muras", "mured", "mures", "murex", "murid", "murks", "murls", "murly", "murra", "murre", "murri", "murrs", "murry", "murti", "murva", "musar", "musca", "mused", "muser", "muses", "muset", "musha", "musit", "musks", "musos", "musse", "mussy", "musth", "musts", "mutch", "muted", "muter", "mutes", "mutha", "mutis", "muton", "mutts", "muxed", "muxes", "muzak", "muzzy", "mvule", "myall", "mylar", "mynah", "mynas", "myoid", "myoma", "myope", "myops", "myopy", "mysid", "mythi", "myths", "mythy", "myxos", "mzees", "naams", "naans", "nabes", "nabis", "nabks", "nabla", "nabob", "nache", "nacho", "nacre", "nadas", "naeve", "naevi", "naffs", "nagas", "naggy", "nagor", "nahal", "naiad", "naifs", "naiks", "nails", "naira", "nairu", "naked", "naker", "nakfa", "nalas", "naled", "nalla", "named", "namer", "names", "namma", "namus", "nanas", "nance", "nancy", "nandu", "nanna", "nanos", "nanua", "napas", "naped", "napes", "napoo", "nappa", "nappe", "nappy", "naras", "narco", "narcs", "nards", "nares", "naric", "naris", "narks", "narky", "narre", "nashi", "natch", "nates", "natis", "natty", "nauch", "naunt", "navar", "naves", "navew", "navvy", "nawab", "nazes", "nazir", "nazis", "nduja", "neafe", "neals", "neaps", "nears", "neath", "neats", "nebek", "nebel", "necks", "neddy", "needs", "neeld", "neele", "neemb", "neems", "neeps", "neese", "neeze", "negro", "negus", "neifs", "neist", "neive", "nelis", "nelly", "nemas", "nemns", "nempt", "nenes", "neons", "neper", "nepit", "neral", "nerds", "nerka", "nerks", "nerol", "nerts", "nertz", "nervy", "nests", "netes", "netop", "netts", "netty", "neuks", "neume", "neums", "nevel", "neves", "nevus", "newbs", "newed", "newel", "newie", "newsy", "newts", "nexts", "nexus", "ngaio", "ngana", "ngati", "ngoma", "ngwee", "nicad", "nicht", "nicks", "nicol", "nidal", "nided", "nides", "nidor", "nidus", "niefs", "nieve", "nifes", "niffs", "niffy", "nifty", "niger", "nighs", "nihil", "nikab", "nikah", "nikau", "nills", "nimbi", "nimbs", "nimps", "niner", "nines", "ninon", "nipas", "nippy", "niqab", "nirls", "nirly", "nisei", "nisse", "nisus", "niter", "nites", "nitid", "niton", "nitre", "nitro", "nitry", "nitty", "nival", "nixed", "nixer", "nixes", "nixie", "nizam", "nkosi", "noahs", "nobby", "nocks", "nodal", "noddy", "nodes", "nodus", "noels", "noggs", "nohow", "noils", "noily", "noint", "noirs", "noles", "nolls", "nolos", "nomas", "nomen", "nomes", "nomic", "nomoi", "nomos", "nonas", "nonce", "nones", "nonet", "nongs", "nonis", "nonny", "nonyl", "noobs", "nooit", "nooks", "nooky", "noons", "noops", "nopal", "noria", "noris", "norks", "norma", "norms", "nosed", "noser", "noses", "notal", "noted", "noter", "notes", "notum", "nould", "noule", "nouls", "nouns", "nouny", "noups", "novae", "novas", "novum", "noway", "nowed", "nowls", "nowts", "nowty", "noxal", "noxes", "noyau", "noyed", "noyes", "nubby", "nubia", "nucha", "nuddy", "nuder", "nudes", "nudie", "nudzh", "nuffs", "nugae", "nuked", "nukes", "nulla", "nulls", "numbs", "numen", "nummy", "nunny", "nurds", "nurdy", "nurls", "nurrs", "nutso", "nutsy", "nyaff", "nyala", "nying", "nyssa", "oaked", "oaker", "oakum", "oared", "oases", "oasis", "oasts", "oaten", "oater", "oaths", "oaves", "obang", "obeah", "obeli", "obeys", "obias", "obied", "obiit", "obits", "objet", "oboes", "obole", "oboli", "obols", "occam", "ocher", "oches", "ochre", "ochry", "ocker", "ocrea", "octad", "octan", "octas", "octyl", "oculi", "odahs", "odals", "odeon", "odeum", "odism", "odist", "odium", "odors", "odour", "odyle", "odyls", "ofays", "offed", "offie", "oflag", "ofter", "ogams", "ogeed", "ogees", "oggin", "ogham", "ogive", "ogled", "ogler", "ogles", "ogmic", "ogres", "ohias", "ohing", "ohmic", "ohone", "oidia", "oiled", "oiler", "oinks", "oints", "ojime", "okapi", "okays", "okehs", "okras", "oktas", "oldie", "oleic", "olein", "olent", "oleos", "oleum", "olios", "ollas", "ollav", "oller", "ollie", "ology", "olpae", "olpes", "omasa", "omber", "ombus", "omens", "omers", "omits", "omlah", "omovs", "omrah", "oncer", "onces", "oncet", "oncus", "onely", "oners", "onery", "onium", "onkus", "onlay", "onned", "ontic", "oobit", "oohed", "oomph", "oonts", "ooped", "oorie", "ooses", "ootid", "oozed", "oozes", "opahs", "opals", "opens", "opepe", "oping", "oppos", "opsin", "opted", "opter", "orach", "oracy", "orals", "orang", "orant", "orate", "orbed", "orcas", "orcin", "ordos", "oread", "orfes", "orgia", "orgic", "orgue", "oribi", "oriel", "orixa", "orles", "orlon", "orlop", "ormer", "ornis", "orpin", "orris", "ortho", "orval", "orzos", "oscar", "oshac", "osier", "osmic", "osmol", "ossia", "ostia", "otaku", "otary", "ottar", "ottos", "oubit", "oucht", "ouens", "ouija", "oulks", "oumas", "oundy", "oupas", "ouped", "ouphe", "ouphs", "ourie", "ousel", "ousts", "outby", "outed", "outre", "outro", "outta", "ouzel", "ouzos", "ovals", "ovels", "ovens", "overs", "ovist", "ovoli", "ovolo", "ovule", "owche", "owies", "owled", "owler", "owlet", "owned", "owres", "owrie", "owsen", "oxbow", "oxers", "oxeye", "oxids", "oxies", "oxime", "oxims", "oxlip", "oxter", "oyers", "ozeki", "ozzie", "paals", "paans", "pacas", "paced", "pacer", "paces", "pacey", "pacha", "packs", "pacos", "pacta", "pacts", "padis", "padle", "padma", "padre", "padri", "paean", "paedo", "paeon", "paged", "pager", "pages", "pagle", "pagod", "pagri", "paiks", "pails", "pains", "paire", "pairs", "paisa", "paise", "pakka", "palas", "palay", "palea", "paled", "pales", "palet", "palis", "palki", "palla", "palls", "pally", "palms", "palmy", "palpi", "palps", "palsa", "pampa", "panax", "pance", "panda", "pands", "pandy", "paned", "panes", "panga", "pangs", "panim", "panko", "panne", "panni", "panto", "pants", "panty", "paoli", "paolo", "papas", "papaw", "papes", "pappi", "pappy", "parae", "paras", "parch", "pardi", "pards", "pardy", "pared", "paren", "pareo", "pares", "pareu", "parev", "parge", "pargo", "paris", "parki", "parks", "parky", "parle", "parly", "parma", "parol", "parps", "parra", "parrs", "parti", "parts", "parve", "parvo", "paseo", "pases", "pasha", "pashm", "paska", "paspy", "passe", "pasts", "pated", "paten", "pater", "pates", "paths", "patin", "patka", "patly", "patte", "patus", "pauas", "pauls", "pavan", "paved", "paven", "paver", "paves", "pavid", "pavin", "pavis", "pawas", "pawaw", "pawed", "pawer", "pawks", "pawky", "pawls", "pawns", "paxes", "payed", "payor", "paysd", "peage", "peags", "peaks", "peaky", "peals", "peans", "peare", "pears", "peart", "pease", "peats", "peaty", "peavy", "peaze", "pebas", "pechs", "pecke", "pecks", "pecky", "pedes", "pedis", "pedro", "peece", "peeks", "peels", "peens", "peeoy", "peepe", "peeps", "peers", "peery", "peeve", "peggy", "peghs", "peins", "peise", "peize", "pekan", "pekes", "pekin", "pekoe", "pelas", "pelau", "peles", "pelfs", "pells", "pelma", "pelon", "pelta", "pelts", "pends", "pendu", "pened", "penes", "pengo", "penie", "penis", "penks", "penna", "penni", "pents", "peons", "peony", "pepla", "pepos", "peppy", "pepsi", "perai", "perce", "percs", "perdu", "perdy", "perea", "peres", "peris", "perks", "perms", "perns", "perog", "perps", "perry", "perse", "perst", "perts", "perve", "pervo", "pervs", "pervy", "pesos", "pests", "pesty", "petar", "peter", "petit", "petre", "petri", "petti", "petto", "pewee", "pewit", "peyse", "phage", "phang", "phare", "pharm", "pheer", "phene", "pheon", "phese", "phial", "phish", "phizz", "phlox", "phoca", "phono", "phons", "phots", "phpht", "phuts", "phyla", "phyle", "piani", "pians", "pibal", "pical", "picas", "piccy", "picks", "picot", "picra", "picul", "piend", "piers", "piert", "pieta", "piets", "piezo", "pight", "pigmy", "piing", "pikas", "pikau", "piked", "piker", "pikes", "pikey", "pikis", "pikul", "pilae", "pilaf", "pilao", "pilar", "pilau", "pilaw", "pilch", "pilea", "piled", "pilei", "piler", "piles", "pilis", "pills", "pilow", "pilum", "pilus", "pimas", "pimps", "pinas", "pined", "pines", "pingo", "pings", "pinko", "pinks", "pinna", "pinny", "pinon", "pinot", "pinta", "pints", "pinup", "pions", "piony", "pious", "pioye", "pioys", "pipal", "pipas", "piped", "pipes", "pipet", "pipis", "pipit", "pippy", "pipul", "pirai", "pirls", "pirns", "pirog", "pisco", "pises", "pisky", "pisos", "pissy", "piste", "pitas", "piths", "piton", "pitot", "pitta", "piums", "pixes", "pized", "pizes", "plaas", "plack", "plage", "plans", "plaps", "plash", "plasm", "plast", "plats", "platt", "platy", "playa", "plays", "pleas", "plebe", "plebs", "plena", "pleon", "plesh", "plews", "plica", "plies", "plims", "pling", "plink", "ploat", "plods", "plong", "plonk", "plook", "plops", "plots", "plotz", "plouk", "plows", "ploye", "ploys", "plues", "pluff", "plugs", "plums", "plumy", "pluot", "pluto", "plyer", "poach", "poaka", "poake", "poboy", "pocks", "pocky", "podal", "poddy", "podex", "podge", "podgy", "podia", "poems", "poeps", "poets", "pogey", "pogge", "pogos", "pohed", "poilu", "poind", "pokal", "poked", "pokes", "pokey", "pokie", "poled", "poler", "poles", "poley", "polio", "polis", "polje", "polks", "polls", "polly", "polos", "polts", "polys", "pombe", "pomes", "pommy", "pomos", "pomps", "ponce", "poncy", "ponds", "pones", "poney", "ponga", "pongo", "pongs", "pongy", "ponks", "ponts", "ponty", "ponzu", "poods", "pooed", "poofs", "poofy", "poohs", "pooja", "pooka", "pooks", "pools", "poons", "poops", "poopy", "poori", "poort", "poots", "poove", "poovy", "popes", "poppa", "popsy", "porae", "poral", "pored", "porer", "pores", "porge", "porgy", "porin", "porks", "porky", "porno", "porns", "porny", "porta", "ports", "porty", "posed", "poses", "posey", "posho", "posts", "potae", "potch", "poted", "potes", "potin", "potoo", "potsy", "potto", "potts", "potty", "pouff", "poufs", "pouke", "pouks", "poule", "poulp", "poult", "poupe", "poupt", "pours", "pouts", "powan", "powin", "pownd", "powns", "powny", "powre", "poxed", "poxes", "poynt", "poyou", "poyse", "pozzy", "praam", "prads", "prahu", "prams", "prana", "prang", "praos", "prase", "prate", "prats", "pratt", "praty", "praus", "prays", "predy", "preed", "prees", "preif", "prems", "premy", "prent", "preon", "preop", "preps", "presa", "prese", "prest", "preve", "prexy", "preys", "prial", "pricy", "prief", "prier", "pries", "prigs", "prill", "prima", "primi", "primp", "prims", "primy", "prink", "prion", "prise", "priss", "proas", "probs", "prods", "proem", "profs", "progs", "proin", "proke", "prole", "proll", "promo", "proms", "pronk", "props", "prore", "proso", "pross", "prost", "prosy", "proto", "proul", "prows", "proyn", "prunt", "pruta", "pryer", "pryse", "pseud", "pshaw", "psion", "psoae", "psoai", "psoas", "psora", "psych", "psyop", "pubco", "pubes", "pubis", "pucan", "pucer", "puces", "pucka", "pucks", "puddy", "pudge", "pudic", "pudor", "pudsy", "pudus", "puers", "puffa", "puffs", "puggy", "pugil", "puhas", "pujah", "pujas", "pukas", "puked", "puker", "pukes", "pukey", "pukka", "pukus", "pulao", "pulas", "puled", "puler", "pules", "pulik", "pulis", "pulka", "pulks", "pulli", "pulls", "pully", "pulmo", "pulps", "pulus", "pumas", "pumie", "pumps", "punas", "punce", "punga", "pungs", "punji", "punka", "punks", "punky", "punny", "punto", "punts", "punty", "pupae", "pupas", "pupus", "purda", "pured", "pures", "purin", "puris", "purls", "purpy", "purrs", "pursy", "purty", "puses", "pusle", "pussy", "putid", "puton", "putti", "putto", "putts", "puzel", "pwned", "pyats", "pyets", "pygal", "pyins", "pylon", "pyned", "pynes", "pyoid", "pyots", "pyral", "pyran", "pyres", "pyrex", "pyric", "pyros", "pyxed", "pyxes", "pyxie", "pyxis", "pzazz", "qadis", "qaids", "qajaq", "qanat", "qapik", "qibla", "qophs", "qorma", "quads", "quaff", "quags", "quair", "quais", "quaky", "quale", "quant", "quare", "quass", "quate", "quats", "quayd", "quays", "qubit", "quean", "queme", "quena", "quern", "queyn", "queys", "quich", "quids", "quiff", "quims", "quina", "quine", "quino", "quins", "quint", "quipo", "quips", "quipu", "quire", "quirt", "quist", "quits", "quoad", "quods", "quoif", "quoin", "quoit", "quoll", "quonk", "quops", "qursh", "quyte", "rabat", "rabic", "rabis", "raced", "races", "rache", "racks", "racon", "radge", "radix", "radon", "raffs", "rafts", "ragas", "ragde", "raged", "ragee", "rager", "rages", "ragga", "raggs", "raggy", "ragis", "ragus", "rahed", "rahui", "raias", "raids", "raiks", "raile", "rails", "raine", "rains", "raird", "raita", "raits", "rajas", "rajes", "raked", "rakee", "raker", "rakes", "rakia", "rakis", "rakus", "rales", "ramal", "ramee", "ramet", "ramie", "ramin", "ramis", "rammy", "ramps", "ramus", "ranas", "rance", "rands", "ranee", "ranga", "rangi", "rangs", "rangy", "ranid", "ranis", "ranke", "ranks", "rants", "raped", "raper", "rapes", "raphe", "rappe", "rared", "raree", "rares", "rarks", "rased", "raser", "rases", "rasps", "rasse", "rasta", "ratal", "ratan", "ratas", "ratch", "rated", "ratel", "rater", "rates", "ratha", "rathe", "raths", "ratoo", "ratos", "ratus", "rauns", "raupo", "raved", "ravel", "raver", "raves", "ravey", "ravin", "rawer", "rawin", "rawly", "rawns", "raxed", "raxes", "rayah", "rayas", "rayed", "rayle", "rayne", "razed", "razee", "razer", "razes", "razoo", "readd", "reads", "reais", "reaks", "realo", "reals", "reame", "reams", "reamy", "reans", "reaps", "rears", "reast", "reata", "reate", "reave", "rebbe", "rebec", "rebid", "rebit", "rebop", "rebuy", "recal", "recce", "recco", "reccy", "recit", "recks", "recon", "recta", "recti", "recto", "redan", "redds", "reddy", "reded", "redes", "redia", "redid", "redip", "redly", "redon", "redos", "redox", "redry", "redub", "redux", "redye", "reech", "reede", "reeds", "reefs", "reefy", "reeks", "reeky", "reels", "reens", "reest", "reeve", "refed", "refel", "reffo", "refis", "refix", "refly", "refry", "regar", "reges", "reggo", "regie", "regma", "regna", "regos", "regur", "rehem", "reifs", "reify", "reiki", "reiks", "reink", "reins", "reird", "reist", "reive", "rejig", "rejon", "reked", "rekes", "rekey", "relet", "relie", "relit", "rello", "reman", "remap", "remen", "remet", "remex", "remix", "renay", "rends", "reney", "renga", "renig", "renin", "renne", "renos", "rente", "rents", "reoil", "reorg", "repeg", "repin", "repla", "repos", "repot", "repps", "repro", "reran", "rerig", "resat", "resaw", "resay", "resee", "reses", "resew", "resid", "resit", "resod", "resow", "resto", "rests", "resty", "resus", "retag", "retax", "retem", "retia", "retie", "retox", "revet", "revie", "rewan", "rewax", "rewed", "rewet", "rewin", "rewon", "rewth", "rexes", "rezes", "rheas", "rheme", "rheum", "rhies", "rhime", "rhine", "rhody", "rhomb", "rhone", "rhumb", "rhyne", "rhyta", "riads", "rials", "riant", "riata", "ribas", "ribby", "ribes", "riced", "ricer", "rices", "ricey", "richt", "ricin", "ricks", "rides", "ridgy", "ridic", "riels", "riems", "rieve", "rifer", "riffs", "rifte", "rifts", "rifty", "riggs", "rigol", "riled", "riles", "riley", "rille", "rills", "rimae", "rimed", "rimer", "rimes", "rimus", "rinds", "rindy", "rines", "rings", "rinks", "rioja", "riots", "riped", "ripes", "ripps", "rises", "rishi", "risks", "risps", "risus", "rites", "ritts", "ritzy", "rivas", "rived", "rivel", "riven", "rives", "riyal", "rizas", "roads", "roams", "roans", "roars", "roary", "roate", "robed", "robes", "roble", "rocks", "roded", "rodes", "roguy", "rohes", "roids", "roils", "roily", "roins", "roist", "rojak", "rojis", "roked", "roker", "rokes", "rolag", "roles", "rolfs", "rolls", "romal", "roman", "romeo", "romps", "ronde", "rondo", "roneo", "rones", "ronin", "ronne", "ronte", "ronts", "roods", "roofs", "roofy", "rooks", "rooky", "rooms", "roons", "roops", "roopy", "roosa", "roose", "roots", "rooty", "roped", "roper", "ropes", "ropey", "roque", "roral", "rores", "roric", "rorid", "rorie", "rorts", "rorty", "rosed", "roses", "roset", "roshi", "rosin", "rosit", "rosti", "rosts", "rotal", "rotan", "rotas", "rotch", "roted", "rotes", "rotis", "rotls", "roton", "rotos", "rotte", "rouen", "roues", "roule", "rouls", "roums", "roups", "roupy", "roust", "routh", "routs", "roved", "roven", "roves", "rowan", "rowed", "rowel", "rowen", "rowie", "rowme", "rownd", "rowth", "rowts", "royne", "royst", "rozet", "rozit", "ruana", "rubai", "rubby", "rubel", "rubes", "rubin", "ruble", "rubli", "rubus", "ruche", "rucks", "rudas", "rudds", "rudes", "rudie", "rudis", "rueda", "ruers", "ruffe", "ruffs", "rugae", "rugal", "ruggy", "ruing", "ruins", "rukhs", "ruled", "rules", "rumal", "rumbo", "rumen", "rumes", "rumly", "rummy", "rumpo", "rumps", "rumpy", "runch", "runds", "runed", "runes", "rungs", "runic", "runny", "runts", "runty", "rupia", "rurps", "rurus", "rusas", "ruses", "rushy", "rusks", "rusma", "russe", "rusts", "ruths", "rutin", "rutty", "ryals", "rybat", "ryked", "rykes", "rymme", "rynds", "ryots", "ryper", "saags", "sabal", "sabed", "saber", "sabes", "sabha", "sabin", "sabir", "sable", "sabot", "sabra", "sabre", "sacks", "sacra", "saddo", "sades", "sadhe", "sadhu", "sadis", "sados", "sadza", "safed", "safes", "sagas", "sager", "sages", "saggy", "sagos", "sagum", "saheb", "sahib", "saice", "saick", "saics", "saids", "saiga", "sails", "saims", "saine", "sains", "sairs", "saist", "saith", "sajou", "sakai", "saker", "sakes", "sakia", "sakis", "sakti", "salal", "salat", "salep", "sales", "salet", "salic", "salix", "salle", "salmi", "salol", "salop", "salpa", "salps", "salse", "salto", "salts", "salue", "salut", "saman", "samas", "samba", "sambo", "samek", "samel", "samen", "sames", "samey", "samfu", "sammy", "sampi", "samps", "sands", "saned", "sanes", "sanga", "sangh", "sango", "sangs", "sanko", "sansa", "santo", "sants", "saola", "sapan", "sapid", "sapor", "saran", "sards", "sared", "saree", "sarge", "sargo", "sarin", "saris", "sarks", "sarky", "sarod", "saros", "sarus", "saser", "sasin", "sasse", "satai", "satay", "sated", "satem", "sates", "satis", "sauba", "sauch", "saugh", "sauls", "sault", "saunt", "saury", "sauts", "saved", "saver", "saves", "savey", "savin", "sawah", "sawed", "sawer", "saxes", "sayed", "sayer", "sayid", "sayne", "sayon", "sayst", "sazes", "scabs", "scads", "scaff", "scags", "scail", "scala", "scall", "scams", "scand", "scans", "scapa", "scape", "scapi", "scarp", "scars", "scart", "scath", "scats", "scatt", "scaud", "scaup", "scaur", "scaws", "sceat", "scena", "scend", "schav", "schmo", "schul", "schwa", "sclim", "scody", "scogs", "scoog", "scoot", "scopa", "scops", "scots", "scoug", "scoup", "scowp", "scows", "scrab", "scrae", "scrag", "scran", "scrat", "scraw", "scray", "scrim", "scrip", "scrob", "scrod", "scrog", "scrow", "scudi", "scudo", "scuds", "scuff", "scuft", "scugs", "sculk", "scull", "sculp", "sculs", "scums", "scups", "scurf", "scurs", "scuse", "scuta", "scute", "scuts", "scuzz", "scyes", "sdayn", "sdein", "seals", "seame", "seams", "seamy", "seans", "seare", "sears", "sease", "seats", "seaze", "sebum", "secco", "sechs", "sects", "seder", "sedes", "sedge", "sedgy", "sedum", "seeds", "seeks", "seeld", "seels", "seely", "seems", "seeps", "seepy", "seers", "sefer", "segar", "segni", "segno", "segol", "segos", "sehri", "seifs", "seils", "seine", "seirs", "seise", "seism", "seity", "seiza", "sekos", "sekts", "selah", "seles", "selfs", "sella", "selle", "sells", "selva", "semee", "semes", "semie", "semis", "senas", "sends", "senes", "sengi", "senna", "senor", "sensa", "sensi", "sente", "senti", "sents", "senvy", "senza", "sepad", "sepal", "sepic", "sepoy", "septa", "septs", "serac", "serai", "seral", "sered", "serer", "seres", "serfs", "serge", "seric", "serin", "serks", "seron", "serow", "serra", "serre", "serrs", "serry", "servo", "sesey", "sessa", "setae", "setal", "seton", "setts", "sewan", "sewar", "sewed", "sewel", "sewen", "sewin", "sexed", "sexer", "sexes", "sexto", "sexts", "seyen", "shads", "shags", "shahs", "shako", "shakt", "shalm", "shaly", "shama", "shams", "shand", "shans", "shaps", "sharn", "shash", "shaul", "shawm", "shawn", "shaws", "shaya", "shays", "shchi", "sheaf", "sheal", "sheas", "sheds", "sheel", "shend", "shent", "sheol", "sherd", "shere", "shero", "shets", "sheva", "shewn", "shews", "shiai", "shiel", "shier", "shies", "shill", "shily", "shims", "shins", "ships", "shirr", "shirs", "shish", "shiso", "shist", "shite", "shits", "shiur", "shiva", "shive", "shivs", "shlep", "shlub", "shmek", "shmoe", "shoat", "shoed", "shoer", "shoes", "shogi", "shogs", "shoji", "shojo", "shola", "shool", "shoon", "shoos", "shope", "shops", "shorl", "shote", "shots", "shott", "showd", "shows", "shoyu", "shred", "shris", "shrow", "shtik", "shtum", "shtup", "shule", "shuln", "shuls", "shuns", "shura", "shute", "shuts", "shwas", "shyer", "sials", "sibbs", "sibyl", "sices", "sicht", "sicko", "sicks", "sicky", "sidas", "sided", "sider", "sides", "sidha", "sidhe", "sidle", "sield", "siens", "sient", "sieth", "sieur", "sifts", "sighs", "sigil", "sigla", "signa", "signs", "sijos", "sikas", "siker", "sikes", "silds", "siled", "silen", "siler", "siles", "silex", "silks", "sills", "silos", "silts", "silty", "silva", "simar", "simas", "simba", "simis", "simps", "simul", "sinds", "sined", "sines", "sings", "sinhs", "sinks", "sinky", "sinus", "siped", "sipes", "sippy", "sired", "siree", "sires", "sirih", "siris", "siroc", "sirra", "sirup", "sisal", "sises", "sista", "sists", "sitar", "sited", "sites", "sithe", "sitka", "situp", "situs", "siver", "sixer", "sixes", "sixmo", "sixte", "sizar", "sized", "sizel", "sizer", "sizes", "skags", "skail", "skald", "skank", "skart", "skats", "skatt", "skaws", "skean", "skear", "skeds", "skeed", "skeef", "skeen", "skeer", "skees", "skeet", "skegg", "skegs", "skein", "skelf", "skell", "skelm", "skelp", "skene", "skens", "skeos", "skeps", "skers", "skets", "skews", "skids", "skied", "skies", "skiey", "skimo", "skims", "skink", "skins", "skint", "skios", "skips", "skirl", "skirr", "skite", "skits", "skive", "skivy", "sklim", "skoal", "skody", "skoff", "skogs", "skols", "skool", "skort", "skosh", "skran", "skrik", "skuas", "skugs", "skyed", "skyer", "skyey", "skyfs", "skyre", "skyrs", "skyte", "slabs", "slade", "slaes", "slags", "slaid", "slake", "slams", "slane", "slank", "slaps", "slart", "slats", "slaty", "slaws", "slays", "slebs", "sleds", "sleer", "slews", "sleys", "slier", "slily", "slims", "slipe", "slips", "slipt", "slish", "slits", "slive", "sloan", "slobs", "sloes", "slogs", "sloid", "slojd", "slomo", "sloom", "sloot", "slops", "slopy", "slorm", "slots", "slove", "slows", "sloyd", "slubb", "slubs", "slued", "slues", "sluff", "slugs", "sluit", "slums", "slurb", "slurs", "sluse", "sluts", "slyer", "slype", "smaak", "smaik", "smalm", "smalt", "smarm", "smaze", "smeek", "smees", "smeik", "smeke", "smerk", "smews", "smirr", "smirs", "smits", "smogs", "smoko", "smolt", "smoor", "smoot", "smore", "smorg", "smout", "smowt", "smugs", "smurs", "smush", "smuts", "snabs", "snafu", "snags", "snaps", "snarf", "snark", "snars", "snary", "snash", "snath", "snaws", "snead", "sneap", "snebs", "sneck", "sneds", "sneed", "snees", "snell", "snibs", "snick", "snies", "snift", "snigs", "snips", "snipy", "snirt", "snits", "snobs", "snods", "snoek", "snoep", "snogs", "snoke", "snood", "snook", "snool", "snoot", "snots", "snowk", "snows", "snubs", "snugs", "snush", "snyes", "soaks", "soaps", "soare", "soars", "soave", "sobas", "socas", "soces", "socko", "socks", "socle", "sodas", "soddy", "sodic", "sodom", "sofar", "sofas", "softa", "softs", "softy", "soger", "sohur", "soils", "soily", "sojas", "sojus", "sokah", "soken", "sokes", "sokol", "solah", "solan", "solas", "solde", "soldi", "soldo", "solds", "soled", "solei", "soler", "soles", "solon", "solos", "solum", "solus", "soman", "somas", "sonce", "sonde", "sones", "songs", "sonly", "sonne", "sonny", "sonse", "sonsy", "sooey", "sooks", "sooky", "soole", "sools", "sooms", "soops", "soote", "soots", "sophs", "sophy", "sopor", "soppy", "sopra", "soral", "soras", "sorbo", "sorbs", "sorda", "sordo", "sords", "sored", "soree", "sorel", "sorer", "sores", "sorex", "sorgo", "sorns", "sorra", "sorta", "sorts", "sorus", "soths", "sotol", "souce", "souct", "sough", "souks", "souls", "soums", "soups", "soupy", "sours", "souse", "souts", "sowar", "sowce", "sowed", "sowff", "sowfs", "sowle", "sowls", "sowms", "sownd", "sowne", "sowps", "sowse", "sowth", "soyas", "soyle", "soyuz", "sozin", "spacy", "spado", "spaed", "spaer", "spaes", "spags", "spahi", "spail", "spain", "spait", "spake", "spald", "spale", "spall", "spalt", "spams", "spane", "spang", "spans", "spard", "spars", "spart", "spate", "spats", "spaul", "spawl", "spaws", "spayd", "spays", "spaza", "spazz", "speal", "spean", "speat", "specs", "spect", "speel", "speer", "speil", "speir", "speks", "speld", "spelk", "speos", "spets", "speug", "spews", "spewy", "spial", "spica", "spick", "spics", "spide", "spier", "spies", "spiff", "spifs", "spiks", "spile", "spims", "spina", "spink", "spins", "spirt", "spiry", "spits", "spitz", "spivs", "splay", "splog", "spode", "spods", "spoom", "spoor", "spoot", "spork", "sposh", "spots", "sprad", "sprag", "sprat", "spred", "sprew", "sprit", "sprod", "sprog", "sprue", "sprug", "spuds", "spued", "spuer", "spues", "spugs", "spule", "spume", "spumy", "spurs", "sputa", "spyal", "spyre", "squab", "squaw", "squeg", "squid", "squit", "squiz", "stabs", "stade", "stags", "stagy", "staig", "stane", "stang", "staph", "staps", "starn", "starr", "stars", "stats", "staun", "staws", "stays", "stean", "stear", "stedd", "stede", "steds", "steek", "steem", "steen", "steil", "stela", "stele", "stell", "steme", "stems", "stend", "steno", "stens", "stent", "steps", "stept", "stere", "stets", "stews", "stewy", "steys", "stich", "stied", "sties", "stilb", "stile", "stime", "stims", "stimy", "stipa", "stipe", "stire", "stirk", "stirp", "stirs", "stive", "stivy", "stoae", "stoai", "stoas", "stoat", "stobs", "stoep", "stogy", "stoit", "stoln", "stoma", "stond", "stong", "stonk", "stonn", "stook", "stoor", "stope", "stops", "stopt", "stoss", "stots", "stott", "stoun", "stoup", "stour", "stown", "stowp", "stows", "strad", "strae", "strag", "strak", "strep", "strew", "stria", "strig", "strim", "strop", "strow", "stroy", "strum", "stubs", "stude", "studs", "stull", "stulm", "stumm", "stums", "stuns", "stupa", "stupe", "sture", "sturt", "styed", "styes", "styli", "stylo", "styme", "stymy", "styre", "styte", "subah", "subas", "subby", "suber", "subha", "succi", "sucks", "sucky", "sucre", "sudds", "sudor", "sudsy", "suede", "suent", "suers", "suete", "suets", "suety", "sugan", "sughs", "sugos", "suhur", "suids", "suint", "suits", "sujee", "sukhs", "sukuk", "sulci", "sulfa", "sulfo", "sulks", "sulph", "sulus", "sumis", "summa", "sumos", "sumph", "sumps", "sunis", "sunks", "sunna", "sunns", "sunup", "supes", "supra", "surah", "sural", "suras", "surat", "surds", "sured", "sures", "surfs", "surfy", "surgy", "surra", "sused", "suses", "susus", "sutor", "sutra", "sutta", "swabs", "swack", "swads", "swage", "swags", "swail", "swain", "swale", "swaly", "swamy", "swang", "swank", "swans", "swaps", "swapt", "sward", "sware", "swarf", "swart", "swats", "swayl", "sways", "sweal", "swede", "sweed", "sweel", "sweer", "swees", "sweir", "swelt", "swerf", "sweys", "swies", "swigs", "swile", "swims", "swink", "swipe", "swire", "swiss", "swith", "swits", "swive", "swizz", "swobs", "swole", "swoln", "swops", "swopt", "swots", "swoun", "sybbe", "sybil", "syboe", "sybow", "sycee", "syces", "sycon", "syens", "syker", "sykes", "sylis", "sylph", "sylva", "symar", "synch", "syncs", "synds", "syned", "synes", "synth", "syped", "sypes", "syphs", "syrah", "syren", "sysop", "sythe", "syver", "taals", "taata", "taber", "tabes", "tabid", "tabis", "tabla", "tabor", "tabun", "tabus", "tacan", "taces", "tacet", "tache", "tacho", "tachs", "tacks", "tacos", "tacts", "taels", "tafia", "taggy", "tagma", "tahas", "tahrs", "taiga", "taigs", "taiko", "tails", "tains", "taira", "taish", "taits", "tajes", "takas", "takes", "takhi", "takin", "takis", "takky", "talak", "talaq", "talar", "talas", "talcs", "talcy", "talea", "taler", "tales", "talks", "talky", "talls", "talma", "talpa", "taluk", "talus", "tamal", "tamed", "tames", "tamin", "tamis", "tammy", "tamps", "tanas", "tanga", "tangi", "tangs", "tanhs", "tanka", "tanks", "tanky", "tanna", "tansy", "tanti", "tanto", "tanty", "tapas", "taped", "tapen", "tapes", "tapet", "tapis", "tappa", "tapus", "taras", "tardo", "tared", "tares", "targa", "targe", "tarns", "taroc", "tarok", "taros", "tarps", "tarre", "tarry", "tarsi", "tarts", "tarty", "tasar", "tased", "taser", "tases", "tasks", "tassa", "tasse", "tasso", "tatar", "tater", "tates", "taths", "tatie", "tatou", "tatts", "tatus", "taube", "tauld", "tauon", "taupe", "tauts", "tavah", "tavas", "taver", "tawai", "tawas", "tawed", "tawer", "tawie", "tawse", "tawts", "taxed", "taxer", "taxes", "taxis", "taxol", "taxon", "taxor", "taxus", "tayra", "tazza", "tazze", "teade", "teads", "teaed", "teaks", "teals", "teams", "tears", "teats", "teaze", "techs", "techy", "tecta", "teels", "teems", "teend", "teene", "teens", "teeny", "teers", "teffs", "teggs", "tegua", "tegus", "tehrs", "teiid", "teils", "teind", "teins", "telae", "telco", "teles", "telex", "telia", "telic", "tells", "telly", "teloi", "telos", "temed", "temes", "tempi", "temps", "tempt", "temse", "tench", "tends", "tendu", "tenes", "tenge", "tenia", "tenne", "tenno", "tenny", "tenon", "tents", "tenty", "tenue", "tepal", "tepas", "tepoy", "terai", "teras", "terce", "terek", "teres", "terfe", "terfs", "terga", "terms", "terne", "terns", "terry", "terts", "tesla", "testa", "teste", "tests", "tetes", "teths", "tetra", "tetri", "teuch", "teugh", "tewed", "tewel", "tewit", "texas", "texes", "texts", "thack", "thagi", "thaim", "thale", "thali", "thana", "thane", "thang", "thans", "thanx", "tharm", "thars", "thaws", "thawy", "thebe", "theca", "theed", "theek", "thees", "thegn", "theic", "thein", "thelf", "thema", "thens", "theow", "therm", "thesp", "thete", "thews", "thewy", "thigs", "thilk", "thill", "thine", "thins", "thiol", "thirl", "thoft", "thole", "tholi", "thoro", "thorp", "thous", "thowl", "thrae", "thraw", "thrid", "thrip", "throe", "thuds", "thugs", "thuja", "thunk", "thurl", "thuya", "thymi", "thymy", "tians", "tiars", "tical", "ticca", "ticed", "tices", "tichy", "ticks", "ticky", "tiddy", "tided", "tides", "tiers", "tiffs", "tifos", "tifts", "tiges", "tigon", "tikas", "tikes", "tikis", "tikka", "tilak", "tiled", "tiler", "tiles", "tills", "tilly", "tilth", "tilts", "timbo", "timed", "times", "timon", "timps", "tinas", "tinct", "tinds", "tinea", "tined", "tines", "tinge", "tings", "tinks", "tinny", "tints", "tinty", "tipis", "tippy", "tired", "tires", "tirls", "tiros", "tirrs", "titch", "titer", "titis", "titre", "titty", "titup", "tiyin", "tiyns", "tizes", "tizzy", "toads", "toady", "toaze", "tocks", "tocky", "tocos", "todde", "toeas", "toffs", "toffy", "tofts", "tofus", "togae", "togas", "toged", "toges", "togue", "tohos", "toile", "toils", "toing", "toise", "toits", "tokay", "toked", "toker", "tokes", "tokos", "tolan", "tolar", "tolas", "toled", "toles", "tolls", "tolly", "tolts", "tolus", "tolyl", "toman", "tombs", "tomes", "tomia", "tommy", "tomos", "tondi", "tondo", "toned", "toner", "tones", "toney", "tongs", "tonka", "tonks", "tonne", "tonus", "tools", "tooms", "toons", "toots", "toped", "topee", "topek", "toper", "topes", "tophe", "tophi", "tophs", "topis", "topoi", "topos", "toppy", "toque", "torah", "toran", "toras", "torcs", "tores", "toric", "torii", "toros", "torot", "torrs", "torse", "torsi", "torsk", "torta", "torte", "torts", "tosas", "tosed", "toses", "toshy", "tossy", "toted", "toter", "totes", "totty", "touks", "touns", "tours", "touse", "tousy", "touts", "touze", "touzy", "towed", "towie", "towns", "towny", "towse", "towsy", "towts", "towze", "towzy", "toyed", "toyer", "toyon", "toyos", "tozed", "tozes", "tozie", "trabs", "trads", "tragi", "traik", "trams", "trank", "tranq", "trans", "trant", "trape", "traps", "trapt", "trass", "trats", "tratt", "trave", "trayf", "trays", "treck", "treed", "treen", "trees", "trefa", "treif", "treks", "trema", "trems", "tress", "trest", "trets", "trews", "treyf", "treys", "triac", "tride", "trier", "tries", "triff", "trigo", "trigs", "trike", "trild", "trill", "trims", "trine", "trins", "triol", "trior", "trios", "trips", "tripy", "trist", "troad", "troak", "troat", "trock", "trode", "trods", "trogs", "trois", "troke", "tromp", "trona", "tronc", "trone", "tronk", "trons", "trooz", "troth", "trots", "trows", "troys", "trued", "trues", "trugo", "trugs", "trull", "tryer", "tryke", "tryma", "tryps", "tsade", "tsadi", "tsars", "tsked", "tsuba", "tsubo", "tuans", "tuart", "tuath", "tubae", "tubar", "tubas", "tubby", "tubed", "tubes", "tucks", "tufas", "tuffe", "tuffs", "tufts", "tufty", "tugra", "tuile", "tuina", "tuism", "tuktu", "tules", "tulpa", "tulsi", "tumid", "tummy", "tumps", "tumpy", "tunas", "tunds", "tuned", "tuner", "tunes", "tungs", "tunny", "tupek", "tupik", "tuple", "tuque", "turds", "turfs", "turfy", "turks", "turme", "turms", "turns", "turnt", "turps", "turrs", "tushy", "tusks", "tusky", "tutee", "tutti", "tutty", "tutus", "tuxes", "tuyer", "twaes", "twain", "twals", "twank", "twats", "tways", "tweel", "tween", "tweep", "tweer", "twerk", "twerp", "twier", "twigs", "twill", "twilt", "twink", "twins", "twiny", "twire", "twirp", "twite", "twits", "twoer", "twyer", "tyees", "tyers", "tyiyn", "tykes", "tyler", "tymps", "tynde", "tyned", "tynes", "typal", "typed", "types", "typey", "typic", "typos", "typps", "typto", "tyran", "tyred", "tyres", "tyros", "tythe", "tzars", "udals", "udons", "ugali", "ugged", "uhlan", "uhuru", "ukase", "ulama", "ulans", "ulema", "ulmin", "ulnad", "ulnae", "ulnar", "ulnas", "ulpan", "ulvas", "ulyie", "ulzie", "umami", "umbel", "umber", "umble", "umbos", "umbre", "umiac", "umiak", "umiaq", "ummah", "ummas", "ummed", "umped", "umphs", "umpie", "umpty", "umrah", "umras", "unais", "unapt", "unarm", "unary", "unaus", "unbag", "unban", "unbar", "unbed", "unbid", "unbox", "uncap", "unces", "uncia", "uncos", "uncoy", "uncus", "undam", "undee", "undos", "undug", "uneth", "unfix", "ungag", "unget", "ungod", "ungot", "ungum", "unhat", "unhip", "unica", "units", "unjam", "unked", "unket", "unkid", "unlaw", "unlay", "unled", "unlet", "unlid", "unman", "unmew", "unmix", "unpay", "unpeg", "unpen", "unpin", "unred", "unrid", "unrig", "unrip", "unsaw", "unsay", "unsee", "unsew", "unsex", "unsod", "untax", "untin", "unwet", "unwit", "unwon", "upbow", "upbye", "updos", "updry", "upend", "upjet", "uplay", "upled", "uplit", "upped", "upran", "uprun", "upsee", "upsey", "uptak", "upter", "uptie", "uraei", "urali", "uraos", "urare", "urari", "urase", "urate", "urbex", "urbia", "urdee", "ureal", "ureas", "uredo", "ureic", "urena", "urent", "urged", "urger", "urges", "urial", "urite", "urman", "urnal", "urned", "urped", "ursae", "ursid", "urson", "urubu", "urvas", "users", "usnea", "usque", "usure", "usury", "uteri", "uveal", "uveas", "uvula", "vacua", "vaded", "vades", "vagal", "vagus", "vails", "vaire", "vairs", "vairy", "vakas", "vakil", "vales", "valis", "valse", "vamps", "vampy", "vanda", "vaned", "vanes", "vangs", "vants", "vaped", "vaper", "vapes", "varan", "varas", "vardy", "varec", "vares", "varia", "varix", "varna", "varus", "varve", "vasal", "vases", "vasts", "vasty", "vatic", "vatus", "vauch", "vaute", "vauts", "vawte", "vaxes", "veale", "veals", "vealy", "veena", "veeps", "veers", "veery", "vegas", "veges", "vegie", "vegos", "vehme", "veils", "veily", "veins", "veiny", "velar", "velds", "veldt", "veles", "vells", "velum", "venae", "venal", "vends", "vendu", "veney", "venge", "venin", "vents", "venus", "verbs", "verra", "verry", "verst", "verts", "vertu", "vespa", "vesta", "vests", "vetch", "vexed", "vexer", "vexes", "vexil", "vezir", "vials", "viand", "vibes", "vibex", "vibey", "viced", "vices", "vichy", "viers", "views", "viewy", "vifda", "viffs", "vigas", "vigia", "vilde", "viler", "villi", "vills", "vimen", "vinal", "vinas", "vinca", "vined", "viner", "vines", "vinew", "vinic", "vinos", "vints", "viold", "viols", "vired", "vireo", "vires", "virga", "virge", "virid", "virls", "virtu", "visas", "vised", "vises", "visie", "visne", "vison", "visto", "vitae", "vitas", "vitex", "vitro", "vitta", "vivas", "vivat", "vivda", "viver", "vives", "vizir", "vizor", "vleis", "vlies", "vlogs", "voars", "vocab", "voces", "voddy", "vodou", "vodun", "voema", "vogie", "voids", "voile", "voips", "volae", "volar", "voled", "voles", "volet", "volks", "volta", "volte", "volti", "volts", "volva", "volve", "vomer", "voted", "votes", "vouge", "voulu", "vowed", "vower", "voxel", "vozhd", "vraic", "vrils", "vroom", "vrous", "vrouw", "vrows", "vuggs", "vuggy", "vughs", "vughy", "vulgo", "vulns", "vulva", "vutty", "waacs", "wacke", "wacko", "wacks", "wadds", "waddy", "waded", "wader", "wades", "wadge", "wadis", "wadts", "waffs", "wafts", "waged", "wages", "wagga", "wagyu", "wahoo", "waide", "waifs", "waift", "wails", "wains", "wairs", "waite", "waits", "wakas", "waked", "waken", "waker", "wakes", "wakfs", "waldo", "walds", "waled", "waler", "wales", "walie", "walis", "walks", "walla", "walls", "wally", "walty", "wamed", "wames", "wamus", "wands", "waned", "wanes", "waney", "wangs", "wanks", "wanky", "wanle", "wanly", "wanna", "wants", "wanty", "wanze", "waqfs", "warbs", "warby", "wards", "wared", "wares", "warez", "warks", "warms", "warns", "warps", "warre", "warst", "warts", "wases", "washy", "wasms", "wasps", "waspy", "wasts", "watap", "watts", "wauff", "waugh", "wauks", "waulk", "wauls", "waurs", "waved", "waves", "wavey", "wawas", "wawes", "wawls", "waxed", "waxer", "waxes", "wayed", "wazir", "wazoo", "weald", "weals", "weamb", "weans", "wears", "webby", "weber", "wecht", "wedel", "wedgy", "weeds", "weeke", "weeks", "weels", "weems", "weens", "weeny", "weeps", "weepy", "weest", "weete", "weets", "wefte", "wefts", "weids", "weils", "weirs", "weise", "weize", "wekas", "welds", "welke", "welks", "welkt", "wells", "welly", "welts", "wembs", "wends", "wenge", "wenny", "wents", "weros", "wersh", "wests", "wetas", "wetly", "wexed", "wexes", "whamo", "whams", "whang", "whaps", "whare", "whata", "whats", "whaup", "whaur", "wheal", "whear", "wheen", "wheep", "wheft", "whelk", "whelm", "whens", "whets", "whews", "wheys", "whids", "whift", "whigs", "whilk", "whims", "whins", "whios", "whips", "whipt", "whirr", "whirs", "whish", "whiss", "whist", "whits", "whity", "whizz", "whomp", "whoof", "whoot", "whops", "whore", "whorl", "whort", "whoso", "whows", "whump", "whups", "whyda", "wicca", "wicks", "wicky", "widdy", "wides", "wiels", "wifed", "wifes", "wifey", "wifie", "wifty", "wigan", "wigga", "wiggy", "wikis", "wilco", "wilds", "wiled", "wiles", "wilga", "wilis", "wilja", "wills", "wilts", "wimps", "winds", "wined", "wines", "winey", "winge", "wings", "wingy", "winks", "winna", "winns", "winos", "winze", "wiped", "wiper", "wipes", "wired", "wirer", "wires", "wirra", "wised", "wises", "wisha", "wisht", "wisps", "wists", "witan", "wited", "wites", "withe", "withs", "withy", "wived", "wiver", "wives", "wizen", "wizes", "woads", "woald", "wocks", "wodge", "woful", "wojus", "woker", "wokka", "wolds", "wolfs", "wolly", "wolve", "wombs", "womby", "womyn", "wonga", "wongi", "wonks", "wonky", "wonts", "woods", "wooed", "woofs", "woofy", "woold", "wools", "woons", "woops", "woopy", "woose", "woosh", "wootz", "words", "works", "worms", "wormy", "worts", "wowed", "wowee", "woxen", "wrang", "wraps", "wrapt", "wrast", "wrate", "wrawl", "wrens", "wrick", "wried", "wrier", "wries", "writs", "wroke", "wroot", "wroth", "wryer", "wuddy", "wudus", "wulls", "wurst", "wuses", "wushu", "wussy", "wuxia", "wyled", "wyles", "wynds", "wynns", "wyted", "wytes", "xebec", "xenia", "xenic", "xenon", "xeric", "xerox", "xerus", "xoana", "xrays", "xylan", "xylem", "xylic", "xylol", "xylyl", "xysti", "xysts", "yaars", "yabas", "yabba", "yabby", "yacca", "yacka", "yacks", "yaffs", "yager", "yages", "yagis", "yahoo", "yaird", "yakka", "yakow", "yales", "yamen", "yampy", "yamun", "yangs", "yanks", "yapok", "yapon", "yapps", "yappy", "yarak", "yarco", "yards", "yarer", "yarfa", "yarks", "yarns", "yarrs", "yarta", "yarto", "yates", "yauds", "yauld", "yaups", "yawed", "yawey", "yawls", "yawns", "yawny", "yawps", "ybore", "yclad", "ycled", "ycond", "ydrad", "ydred", "yeads", "yeahs", "yealm", "yeans", "yeard", "years", "yecch", "yechs", "yechy", "yedes", "yeeds", "yeesh", "yeggs", "yelks", "yells", "yelms", "yelps", "yelts", "yenta", "yente", "yerba", "yerds", "yerks", "yeses", "yesks", "yests", "yesty", "yetis", "yetts", "yeuks", "yeuky", "yeven", "yeves", "yewen", "yexed", "yexes", "yfere", "yiked", "yikes", "yills", "yince", "yipes", "yippy", "yirds", "yirks", "yirrs", "yirth", "yites", "yitie", "ylems", "ylike", "ylkes", "ymolt", "ympes", "yobbo", "yobby", "yocks", "yodel", "yodhs", "yodle", "yogas", "yogee", "yoghs", "yogic", "yogin", "yogis", "yoick", "yojan", "yoked", "yokel", "yoker", "yokes", "yokul", "yolks", "yolky", "yomim", "yomps", "yonic", "yonis", "yonks", "yoofs", "yoops", "yores", "yorks", "yorps", "youks", "yourn", "yours", "yourt", "youse", "yowed", "yowes", "yowie", "yowls", "yowza", "yrapt", "yrent", "yrivd", "yrneh", "ysame", "ytost", "yuans", "yucas", "yucca", "yucch", "yucko", "yucks", "yucky", "yufts", "yugas", "yuked", "yukes", "yukky", "yukos", "yulan", "yules", "yummo", "yummy", "yumps", "yupon", "yuppy", "yurta", "yurts", "yuzus", "zabra", "zacks", "zaida", "zaidy", "zaire", "zakat", "zaman", "zambo", "zamia", "zanja", "zante", "zanza", "zanze", "zappy", "zarfs", "zaris", "zatis", "zaxes", "zayin", "zazen", "zeals", "zebec", "zebub", "zebus", "zedas", "zeins", "zendo", "zerda", "zerks", "zeros", "zests", "zetas", "zexes", "zezes", "zhomo", "zibet", "ziffs", "zigan", "zilas", "zilch", "zilla", "zills", "zimbi", "zimbs", "zinco", "zincs", "zincy", "zineb", "zines", "zings", "zingy", "zinke", "zinky", "zippo", "zippy", "ziram", "zitis", "zizel", "zizit", "zlote", "zloty", "zoaea", "zobos", "zobus", "zocco", "zoeae", "zoeal", "zoeas", "zoism", "zoist", "zombi", "zonae", "zonda", "zoned", "zoner", "zones", "zonks", "zooea", "zooey", "zooid", "zooks", "zooms", "zoons", "zooty", "zoppa", "zoppo", "zoril", "zoris", "zorro", "zouks", "zowee", "zowie", "zulus", "zupan", "zupas", "zuppa", "zurfs", "zuzim", "zygal", "zygon", "zymes", "zymic"]
_RESPONSES_CACHE = {}
def getResponse(guess, answer):
optional_cached = _RESPONSES_CACHE.get((guess, answer), None)
if optional_cached is not None:
return optional_cached
answer_multiset = {}
greens_multiset = {}
for c_guess, c in zip(guess, answer):
answer_multiset[c] = answer_multiset.get(c, 0) + 1
if c_guess == c:
greens_multiset[c] = greens_multiset.get(c, 0) + 1
result = []
for c_answer, c in zip(answer, guess):
if c_answer == c:
result.append('g')
greens_multiset[c] -= 1
answer_multiset[c] -= 1
continue
if answer_multiset.get(c, 0) > greens_multiset.get(c, 0):
result.append('y')
answer_multiset[c] -= 1
continue
result.append('b')
joined = ''.join(result)
_RESPONSES_CACHE[(guess, answer)] = joined
return joined
assert(getResponse('puree', 'pleat') == 'gbbyb')
assert(getResponse('eejit', 'pleat') == 'ybbbg')
assert(getResponse('raise', 'pleat') == 'bybby')
assert(getResponse('llama', 'aloft') == 'bgybb')
import math
def getGreedyGuess(possible_guesses, possible_answers, metric_function):
best_guesses = None
best_guess_metric = float('inf')
for guess in possible_guesses:
response_counts = {}
for answer in possible_answers:
response = getResponse(guess, answer)
response_counts[response] = response_counts.get(response, 0) + 1
metric = metric_function(v for k, v in response_counts.items() if k != 'ggggg')
if best_guesses is None or best_guess_metric > metric:
best_guess_metric = metric
best_guesses = [guess]
elif best_guess_metric == metric:
best_guesses.append(guess)
return best_guesses
def getGreedyMinMaxBucket(possible_guesses, possible_answers):
return getGreedyGuess(possible_guesses,
possible_answers,
lambda vals: tuple(sorted(vals, reverse=True)))
def getGreedyExpectedBucket(possible_guesses, possible_answers):
return getGreedyGuess(possible_guesses,
possible_answers,
lambda vals: sum(val * val for val in vals))
def getGreedyNextWord(possible_guesses, possible_answers):
return getGreedyGuess(possible_guesses,
possible_answers,
lambda vals: -len(list(vals)))
def getGreedyInfiniteExponentialUtility(possible_guesses, possible_answers):
return getGreedyGuess(possible_answers,
possible_answers,
lambda vals: -len(list(vals)))
def _entropy(vals):
v = list(vals)
total = sum(v)
return 0 if (total == 0) else sum(val * math.log(val + 1) for val in v)
def getGreedyEntropy(possible_guesses, possible_answers):
return getGreedyGuess(possible_guesses,
possible_answers,
_entropy)
def getNewPossibleAnswers(possible_answers, guess, given_response):
return [answer for answer in possible_answers
if getResponse(guess, answer) == given_response]
print(f'Best guesses for min max: {getGreedyMinMaxBucket(POSSIBLE_GUESSES, POSSIBLE_ANSWERS)}')
print(f'Best guesses for EV: {getGreedyExpectedBucket(POSSIBLE_GUESSES, POSSIBLE_ANSWERS)}')
print(f'Best guesses for getting in 2: {getGreedyNextWord(POSSIBLE_GUESSES, POSSIBLE_ANSWERS)}')
print(f'Best guesses for getting for any cutoff: {getGreedyInfiniteExponentialUtility(POSSIBLE_GUESSES, POSSIBLE_ANSWERS)}')
print(f'Best guesses for maximizing information entropy: {getGreedyEntropy(POSSIBLE_GUESSES, POSSIBLE_ANSWERS)}')
```
##Trying the greedy min-max with the word of the day for 2/5/2022 (ALOFT)
```
new_possible_answers = POSSIBLE_ANSWERS
new_possible_answers = getNewPossibleAnswers(new_possible_answers, 'raise', 'bybbb')
print(new_possible_answers)
print(getGreedyMinMaxBucket(POSSIBLE_GUESSES, new_possible_answers))
new_possible_answers = getNewPossibleAnswers(new_possible_answers, 'cloak', 'bggyb')
print(new_possible_answers)
print(getGreedyMinMaxBucket(POSSIBLE_GUESSES, new_possible_answers))
new_possible_answers = getNewPossibleAnswers(new_possible_answers, 'dwarf', 'bbyby')
print(new_possible_answers)
print(getGreedyMinMaxBucket(POSSIBLE_GUESSES, new_possible_answers))
```
## Trying the greedy expected bucket size with ALOFT
```
new_possible_answers = POSSIBLE_ANSWERS
new_possible_answers = getNewPossibleAnswers(POSSIBLE_ANSWERS, 'roate', 'byyyb')
print(new_possible_answers)
print(getGreedyExpectedBucket(POSSIBLE_GUESSES, new_possible_answers))
new_possible_answers = getNewPossibleAnswers(POSSIBLE_ANSWERS, 'bloat', 'bggyg')
print(new_possible_answers)
print(getGreedyExpectedBucket(POSSIBLE_GUESSES, new_possible_answers))
```
## Viewing entire strategy trees
```
def printTree(possible_guesses, possible_answers, strategy, starting_guess=None, filename=None):
if filename is not None:
with open(filename, 'w') as f:
pass
return printTreeHelper(possible_guesses,
possible_answers,
strategy,
guess=starting_guess,
indent_level=0,
filename=filename) / len(possible_answers)
def printTreeHelper(possible_guesses, possible_answers, strategy, guess=None, indent_level=0, filename=None):
if guess is None:
guess = strategy(possible_guesses, possible_answers)[0]
responses = {}
for answer in possible_answers:
response = getResponse(guess, answer)
if response not in responses:
responses[response] = []
responses[response].append(answer)
def log(info):
if filename is None:
print(info)
else:
with open(filename, 'a') as f:
f.write(info)
f.write('\n')
guess_number = indent_level + 1
total_guesses = 0
items = sorted(responses.items(), key=lambda x: (x[0] != 'ggggg', len(x[1])))
log(f'{" " * indent_level}guess: {guess}')
for response, remaining in items:
if response == 'ggggg':
log(f'{" " * indent_level} took {guess_number} guesses')
total_guesses += indent_level + 1
continue
log(f'{" " * indent_level} {len(remaining)} remaining words for {response}: {remaining}')
total_guesses += printTreeHelper(possible_guesses, remaining, strategy, indent_level=guess_number, filename=filename)
return total_guesses
for starting_guess in ['raise', 'roate', 'trace', 'soare', 'crane', 'slate']:
print(f'Starting guess: {starting_guess}')
for strategy, name in [(getGreedyMinMaxBucket, 'minmax'), (getGreedyExpectedBucket, 'ev'), (getGreedyEntropy, 'entropy')]:
expected_guesses = printTree(POSSIBLE_GUESSES, POSSIBLE_ANSWERS, strategy, starting_guess=starting_guess, filename=f'{starting_guess}_{name}.txt')
print(f'Expected number of guesses for {name}: {expected_guesses}')
```
## Trying to get answer in 3 guesses
```
def getAverageNumBucketsAfterNextGuess(guess):
score = 0
responses = {}
for answer in POSSIBLE_ANSWERS:
response = getResponse(guess, answer)
if response not in responses:
responses[response] = [answer]
else:
responses[response].append(answer)
for response, possible_answers in responses.items():
score_2 = 0
for guess_2 in POSSIBLE_GUESSES:
responses_2 = set()
for answer in possible_answers:
responses_2.add(getResponse(guess_2, answer))
if len(responses_2) > score_2:
score_2 = len(responses_2)
score += score_2
return score
max_score = 1388
best_word = 'trace'
last_checkpoint = 'steep'
seen = False
for guess in POSSIBLE_GUESSES:
if not seen:
if guess == last_checkpoint:
seen = True
continue
score = getAverageNumBucketsAfterNextGuess(guess)
if score > max_score:
max_score = score
best_word = guess
print(f'Found new best guess: {guess} ({score})')
else:
print(f'Guess {guess} ({score}) could not beat {best_word} ({max_score})')
printTree(POSSIBLE_GUESSES, POSSIBLE_ANSWERS, getGreedyInfiniteExponentialUtility, filename='greedyexponential.txt')
```
## Lazy (human) strategy
Always type in the same first 2 words, minimizing maximum bucket size
```
min_guesses = []
min_max_bucket = float('inf')
for i, guess_1 in enumerate(POSSIBLE_GUESSES):
print(i, guess_1)
for j in range(i+1, len(POSSIBLE_GUESSES)):
guess_2 = POSSIBLE_GUESSES[j]
responses = {}
max_bucket = 0
for answer in POSSIBLE_ANSWERS:
response_1 = getResponse(guess_1, answer)
response_2 = getResponse(guess_2, answer)
combined = response_1 + response_2
if combined not in responses:
responses[combined] = []
responses[combined].append(answer)
max_bucket = max(max_bucket, len(responses[combined]))
if min_max_bucket > max_bucket:
min_max_bucket = max_bucket
min_guesses = [(guess_1, guess_2)]
print(f'Found best first 2 guesses ({max_bucket}): {guess_1}, {guess_2}')
elif min_max_bucket == max_bucket:
min_guesses.append((guess_1, guess_2))
print(f'Found best first 2 guesses ({max_bucket}): {guess_1}, {guess_2}')
print(f'Best first 2 guesses: {min_guesses}')
```
|
github_jupyter
|
# Scraped from the wordle website
POSSIBLE_ANSWERS = ["cigar", "rebut", "sissy", "humph", "awake", "blush", "focal", "evade", "naval", "serve", "heath", "dwarf", "model", "karma", "stink", "grade", "quiet", "bench", "abate", "feign", "major", "death", "fresh", "crust", "stool", "colon", "abase", "marry", "react", "batty", "pride", "floss", "helix", "croak", "staff", "paper", "unfed", "whelp", "trawl", "outdo", "adobe", "crazy", "sower", "repay", "digit", "crate", "cluck", "spike", "mimic", "pound", "maxim", "linen", "unmet", "flesh", "booby", "forth", "first", "stand", "belly", "ivory", "seedy", "print", "yearn", "drain", "bribe", "stout", "panel", "crass", "flume", "offal", "agree", "error", "swirl", "argue", "bleed", "delta", "flick", "totem", "wooer", "front", "shrub", "parry", "biome", "lapel", "start", "greet", "goner", "golem", "lusty", "loopy", "round", "audit", "lying", "gamma", "labor", "islet", "civic", "forge", "corny", "moult", "basic", "salad", "agate", "spicy", "spray", "essay", "fjord", "spend", "kebab", "guild", "aback", "motor", "alone", "hatch", "hyper", "thumb", "dowry", "ought", "belch", "dutch", "pilot", "tweed", "comet", "jaunt", "enema", "steed", "abyss", "growl", "fling", "dozen", "boozy", "erode", "world", "gouge", "click", "briar", "great", "altar", "pulpy", "blurt", "coast", "duchy", "groin", "fixer", "group", "rogue", "badly", "smart", "pithy", "gaudy", "chill", "heron", "vodka", "finer", "surer", "radio", "rouge", "perch", "retch", "wrote", "clock", "tilde", "store", "prove", "bring", "solve", "cheat", "grime", "exult", "usher", "epoch", "triad", "break", "rhino", "viral", "conic", "masse", "sonic", "vital", "trace", "using", "peach", "champ", "baton", "brake", "pluck", "craze", "gripe", "weary", "picky", "acute", "ferry", "aside", "tapir", "troll", "unify", "rebus", "boost", "truss", "siege", "tiger", "banal", "slump", "crank", "gorge", "query", "drink", "favor", "abbey", "tangy", "panic", "solar", "shire", "proxy", "point", "robot", "prick", "wince", "crimp", "knoll", "sugar", "whack", "mount", "perky", "could", "wrung", "light", "those", "moist", "shard", "pleat", "aloft", "skill", "elder", "frame", "humor", "pause", "ulcer", "ultra", "robin", "cynic", "agora", "aroma", "caulk", "shake", "pupal", "dodge", "swill", "tacit", "other", "thorn", "trove", "bloke", "vivid", "spill", "chant", "choke", "rupee", "nasty", "mourn", "ahead", "brine", "cloth", "hoard", "sweet", "month", "lapse", "watch", "today", "focus", "smelt", "tease", "cater", "movie", "lynch", "saute", "allow", "renew", "their", "slosh", "purge", "chest", "depot", "epoxy", "nymph", "found", "shall", "harry", "stove", "lowly", "snout", "trope", "fewer", "shawl", "natal", "fibre", "comma", "foray", "scare", "stair", "black", "squad", "royal", "chunk", "mince", "slave", "shame", "cheek", "ample", "flair", "foyer", "cargo", "oxide", "plant", "olive", "inert", "askew", "heist", "shown", "zesty", "hasty", "trash", "fella", "larva", "forgo", "story", "hairy", "train", "homer", "badge", "midst", "canny", "fetus", "butch", "farce", "slung", "tipsy", "metal", "yield", "delve", "being", "scour", "glass", "gamer", "scrap", "money", "hinge", "album", "vouch", "asset", "tiara", "crept", "bayou", "atoll", "manor", "creak", "showy", "phase", "froth", "depth", "gloom", "flood", "trait", "girth", "piety", "payer", "goose", "float", "donor", "atone", "primo", "apron", "blown", "cacao", "loser", "input", "gloat", "awful", "brink", "smite", "beady", "rusty", "retro", "droll", "gawky", "hutch", "pinto", "gaily", "egret", "lilac", "sever", "field", "fluff", "hydro", "flack", "agape", "wench", "voice", "stead", "stalk", "berth", "madam", "night", "bland", "liver", "wedge", "augur", "roomy", "wacky", "flock", "angry", "bobby", "trite", "aphid", "tryst", "midge", "power", "elope", "cinch", "motto", "stomp", "upset", "bluff", "cramp", "quart", "coyly", "youth", "rhyme", "buggy", "alien", "smear", "unfit", "patty", "cling", "glean", "label", "hunky", "khaki", "poker", "gruel", "twice", "twang", "shrug", "treat", "unlit", "waste", "merit", "woven", "octal", "needy", "clown", "widow", "irony", "ruder", "gauze", "chief", "onset", "prize", "fungi", "charm", "gully", "inter", "whoop", "taunt", "leery", "class", "theme", "lofty", "tibia", "booze", "alpha", "thyme", "eclat", "doubt", "parer", "chute", "stick", "trice", "alike", "sooth", "recap", "saint", "liege", "glory", "grate", "admit", "brisk", "soggy", "usurp", "scald", "scorn", "leave", "twine", "sting", "bough", "marsh", "sloth", "dandy", "vigor", "howdy", "enjoy", "valid", "ionic", "equal", "unset", "floor", "catch", "spade", "stein", "exist", "quirk", "denim", "grove", "spiel", "mummy", "fault", "foggy", "flout", "carry", "sneak", "libel", "waltz", "aptly", "piney", "inept", "aloud", "photo", "dream", "stale", "vomit", "ombre", "fanny", "unite", "snarl", "baker", "there", "glyph", "pooch", "hippy", "spell", "folly", "louse", "gulch", "vault", "godly", "threw", "fleet", "grave", "inane", "shock", "crave", "spite", "valve", "skimp", "claim", "rainy", "musty", "pique", "daddy", "quasi", "arise", "aging", "valet", "opium", "avert", "stuck", "recut", "mulch", "genre", "plume", "rifle", "count", "incur", "total", "wrest", "mocha", "deter", "study", "lover", "safer", "rivet", "funny", "smoke", "mound", "undue", "sedan", "pagan", "swine", "guile", "gusty", "equip", "tough", "canoe", "chaos", "covet", "human", "udder", "lunch", "blast", "stray", "manga", "melee", "lefty", "quick", "paste", "given", "octet", "risen", "groan", "leaky", "grind", "carve", "loose", "sadly", "spilt", "apple", "slack", "honey", "final", "sheen", "eerie", "minty", "slick", "derby", "wharf", "spelt", "coach", "erupt", "singe", "price", "spawn", "fairy", "jiffy", "filmy", "stack", "chose", "sleep", "ardor", "nanny", "niece", "woozy", "handy", "grace", "ditto", "stank", "cream", "usual", "diode", "valor", "angle", "ninja", "muddy", "chase", "reply", "prone", "spoil", "heart", "shade", "diner", "arson", "onion", "sleet", "dowel", "couch", "palsy", "bowel", "smile", "evoke", "creek", "lance", "eagle", "idiot", "siren", "built", "embed", "award", "dross", "annul", "goody", "frown", "patio", "laden", "humid", "elite", "lymph", "edify", "might", "reset", "visit", "gusto", "purse", "vapor", "crock", "write", "sunny", "loath", "chaff", "slide", "queer", "venom", "stamp", "sorry", "still", "acorn", "aping", "pushy", "tamer", "hater", "mania", "awoke", "brawn", "swift", "exile", "birch", "lucky", "freer", "risky", "ghost", "plier", "lunar", "winch", "snare", "nurse", "house", "borax", "nicer", "lurch", "exalt", "about", "savvy", "toxin", "tunic", "pried", "inlay", "chump", "lanky", "cress", "eater", "elude", "cycle", "kitty", "boule", "moron", "tenet", "place", "lobby", "plush", "vigil", "index", "blink", "clung", "qualm", "croup", "clink", "juicy", "stage", "decay", "nerve", "flier", "shaft", "crook", "clean", "china", "ridge", "vowel", "gnome", "snuck", "icing", "spiny", "rigor", "snail", "flown", "rabid", "prose", "thank", "poppy", "budge", "fiber", "moldy", "dowdy", "kneel", "track", "caddy", "quell", "dumpy", "paler", "swore", "rebar", "scuba", "splat", "flyer", "horny", "mason", "doing", "ozone", "amply", "molar", "ovary", "beset", "queue", "cliff", "magic", "truce", "sport", "fritz", "edict", "twirl", "verse", "llama", "eaten", "range", "whisk", "hovel", "rehab", "macaw", "sigma", "spout", "verve", "sushi", "dying", "fetid", "brain", "buddy", "thump", "scion", "candy", "chord", "basin", "march", "crowd", "arbor", "gayly", "musky", "stain", "dally", "bless", "bravo", "stung", "title", "ruler", "kiosk", "blond", "ennui", "layer", "fluid", "tatty", "score", "cutie", "zebra", "barge", "matey", "bluer", "aider", "shook", "river", "privy", "betel", "frisk", "bongo", "begun", "azure", "weave", "genie", "sound", "glove", "braid", "scope", "wryly", "rover", "assay", "ocean", "bloom", "irate", "later", "woken", "silky", "wreck", "dwelt", "slate", "smack", "solid", "amaze", "hazel", "wrist", "jolly", "globe", "flint", "rouse", "civil", "vista", "relax", "cover", "alive", "beech", "jetty", "bliss", "vocal", "often", "dolly", "eight", "joker", "since", "event", "ensue", "shunt", "diver", "poser", "worst", "sweep", "alley", "creed", "anime", "leafy", "bosom", "dunce", "stare", "pudgy", "waive", "choir", "stood", "spoke", "outgo", "delay", "bilge", "ideal", "clasp", "seize", "hotly", "laugh", "sieve", "block", "meant", "grape", "noose", "hardy", "shied", "drawl", "daisy", "putty", "strut", "burnt", "tulip", "crick", "idyll", "vixen", "furor", "geeky", "cough", "naive", "shoal", "stork", "bathe", "aunty", "check", "prime", "brass", "outer", "furry", "razor", "elect", "evict", "imply", "demur", "quota", "haven", "cavil", "swear", "crump", "dough", "gavel", "wagon", "salon", "nudge", "harem", "pitch", "sworn", "pupil", "excel", "stony", "cabin", "unzip", "queen", "trout", "polyp", "earth", "storm", "until", "taper", "enter", "child", "adopt", "minor", "fatty", "husky", "brave", "filet", "slime", "glint", "tread", "steal", "regal", "guest", "every", "murky", "share", "spore", "hoist", "buxom", "inner", "otter", "dimly", "level", "sumac", "donut", "stilt", "arena", "sheet", "scrub", "fancy", "slimy", "pearl", "silly", "porch", "dingo", "sepia", "amble", "shady", "bread", "friar", "reign", "dairy", "quill", "cross", "brood", "tuber", "shear", "posit", "blank", "villa", "shank", "piggy", "freak", "which", "among", "fecal", "shell", "would", "algae", "large", "rabbi", "agony", "amuse", "bushy", "copse", "swoon", "knife", "pouch", "ascot", "plane", "crown", "urban", "snide", "relay", "abide", "viola", "rajah", "straw", "dilly", "crash", "amass", "third", "trick", "tutor", "woody", "blurb", "grief", "disco", "where", "sassy", "beach", "sauna", "comic", "clued", "creep", "caste", "graze", "snuff", "frock", "gonad", "drunk", "prong", "lurid", "steel", "halve", "buyer", "vinyl", "utile", "smell", "adage", "worry", "tasty", "local", "trade", "finch", "ashen", "modal", "gaunt", "clove", "enact", "adorn", "roast", "speck", "sheik", "missy", "grunt", "snoop", "party", "touch", "mafia", "emcee", "array", "south", "vapid", "jelly", "skulk", "angst", "tubal", "lower", "crest", "sweat", "cyber", "adore", "tardy", "swami", "notch", "groom", "roach", "hitch", "young", "align", "ready", "frond", "strap", "puree", "realm", "venue", "swarm", "offer", "seven", "dryer", "diary", "dryly", "drank", "acrid", "heady", "theta", "junto", "pixie", "quoth", "bonus", "shalt", "penne", "amend", "datum", "build", "piano", "shelf", "lodge", "suing", "rearm", "coral", "ramen", "worth", "psalm", "infer", "overt", "mayor", "ovoid", "glide", "usage", "poise", "randy", "chuck", "prank", "fishy", "tooth", "ether", "drove", "idler", "swath", "stint", "while", "begat", "apply", "slang", "tarot", "radar", "credo", "aware", "canon", "shift", "timer", "bylaw", "serum", "three", "steak", "iliac", "shirk", "blunt", "puppy", "penal", "joist", "bunny", "shape", "beget", "wheel", "adept", "stunt", "stole", "topaz", "chore", "fluke", "afoot", "bloat", "bully", "dense", "caper", "sneer", "boxer", "jumbo", "lunge", "space", "avail", "short", "slurp", "loyal", "flirt", "pizza", "conch", "tempo", "droop", "plate", "bible", "plunk", "afoul", "savoy", "steep", "agile", "stake", "dwell", "knave", "beard", "arose", "motif", "smash", "broil", "glare", "shove", "baggy", "mammy", "swamp", "along", "rugby", "wager", "quack", "squat", "snaky", "debit", "mange", "skate", "ninth", "joust", "tramp", "spurn", "medal", "micro", "rebel", "flank", "learn", "nadir", "maple", "comfy", "remit", "gruff", "ester", "least", "mogul", "fetch", "cause", "oaken", "aglow", "meaty", "gaffe", "shyly", "racer", "prowl", "thief", "stern", "poesy", "rocky", "tweet", "waist", "spire", "grope", "havoc", "patsy", "truly", "forty", "deity", "uncle", "swish", "giver", "preen", "bevel", "lemur", "draft", "slope", "annoy", "lingo", "bleak", "ditty", "curly", "cedar", "dirge", "grown", "horde", "drool", "shuck", "crypt", "cumin", "stock", "gravy", "locus", "wider", "breed", "quite", "chafe", "cache", "blimp", "deign", "fiend", "logic", "cheap", "elide", "rigid", "false", "renal", "pence", "rowdy", "shoot", "blaze", "envoy", "posse", "brief", "never", "abort", "mouse", "mucky", "sulky", "fiery", "media", "trunk", "yeast", "clear", "skunk", "scalp", "bitty", "cider", "koala", "duvet", "segue", "creme", "super", "grill", "after", "owner", "ember", "reach", "nobly", "empty", "speed", "gipsy", "recur", "smock", "dread", "merge", "burst", "kappa", "amity", "shaky", "hover", "carol", "snort", "synod", "faint", "haunt", "flour", "chair", "detox", "shrew", "tense", "plied", "quark", "burly", "novel", "waxen", "stoic", "jerky", "blitz", "beefy", "lyric", "hussy", "towel", "quilt", "below", "bingo", "wispy", "brash", "scone", "toast", "easel", "saucy", "value", "spice", "honor", "route", "sharp", "bawdy", "radii", "skull", "phony", "issue", "lager", "swell", "urine", "gassy", "trial", "flora", "upper", "latch", "wight", "brick", "retry", "holly", "decal", "grass", "shack", "dogma", "mover", "defer", "sober", "optic", "crier", "vying", "nomad", "flute", "hippo", "shark", "drier", "obese", "bugle", "tawny", "chalk", "feast", "ruddy", "pedal", "scarf", "cruel", "bleat", "tidal", "slush", "semen", "windy", "dusty", "sally", "igloo", "nerdy", "jewel", "shone", "whale", "hymen", "abuse", "fugue", "elbow", "crumb", "pansy", "welsh", "syrup", "terse", "suave", "gamut", "swung", "drake", "freed", "afire", "shirt", "grout", "oddly", "tithe", "plaid", "dummy", "broom", "blind", "torch", "enemy", "again", "tying", "pesky", "alter", "gazer", "noble", "ethos", "bride", "extol", "decor", "hobby", "beast", "idiom", "utter", "these", "sixth", "alarm", "erase", "elegy", "spunk", "piper", "scaly", "scold", "hefty", "chick", "sooty", "canal", "whiny", "slash", "quake", "joint", "swept", "prude", "heavy", "wield", "femme", "lasso", "maize", "shale", "screw", "spree", "smoky", "whiff", "scent", "glade", "spent", "prism", "stoke", "riper", "orbit", "cocoa", "guilt", "humus", "shush", "table", "smirk", "wrong", "noisy", "alert", "shiny", "elate", "resin", "whole", "hunch", "pixel", "polar", "hotel", "sword", "cleat", "mango", "rumba", "puffy", "filly", "billy", "leash", "clout", "dance", "ovate", "facet", "chili", "paint", "liner", "curio", "salty", "audio", "snake", "fable", "cloak", "navel", "spurt", "pesto", "balmy", "flash", "unwed", "early", "churn", "weedy", "stump", "lease", "witty", "wimpy", "spoof", "saner", "blend", "salsa", "thick", "warty", "manic", "blare", "squib", "spoon", "probe", "crepe", "knack", "force", "debut", "order", "haste", "teeth", "agent", "widen", "icily", "slice", "ingot", "clash", "juror", "blood", "abode", "throw", "unity", "pivot", "slept", "troop", "spare", "sewer", "parse", "morph", "cacti", "tacky", "spool", "demon", "moody", "annex", "begin", "fuzzy", "patch", "water", "lumpy", "admin", "omega", "limit", "tabby", "macho", "aisle", "skiff", "basis", "plank", "verge", "botch", "crawl", "lousy", "slain", "cubic", "raise", "wrack", "guide", "foist", "cameo", "under", "actor", "revue", "fraud", "harpy", "scoop", "climb", "refer", "olden", "clerk", "debar", "tally", "ethic", "cairn", "tulle", "ghoul", "hilly", "crude", "apart", "scale", "older", "plain", "sperm", "briny", "abbot", "rerun", "quest", "crisp", "bound", "befit", "drawn", "suite", "itchy", "cheer", "bagel", "guess", "broad", "axiom", "chard", "caput", "leant", "harsh", "curse", "proud", "swing", "opine", "taste", "lupus", "gumbo", "miner", "green", "chasm", "lipid", "topic", "armor", "brush", "crane", "mural", "abled", "habit", "bossy", "maker", "dusky", "dizzy", "lithe", "brook", "jazzy", "fifty", "sense", "giant", "surly", "legal", "fatal", "flunk", "began", "prune", "small", "slant", "scoff", "torus", "ninny", "covey", "viper", "taken", "moral", "vogue", "owing", "token", "entry", "booth", "voter", "chide", "elfin", "ebony", "neigh", "minim", "melon", "kneed", "decoy", "voila", "ankle", "arrow", "mushy", "tribe", "cease", "eager", "birth", "graph", "odder", "terra", "weird", "tried", "clack", "color", "rough", "weigh", "uncut", "ladle", "strip", "craft", "minus", "dicey", "titan", "lucid", "vicar", "dress", "ditch", "gypsy", "pasta", "taffy", "flame", "swoop", "aloof", "sight", "broke", "teary", "chart", "sixty", "wordy", "sheer", "leper", "nosey", "bulge", "savor", "clamp", "funky", "foamy", "toxic", "brand", "plumb", "dingy", "butte", "drill", "tripe", "bicep", "tenor", "krill", "worse", "drama", "hyena", "think", "ratio", "cobra", "basil", "scrum", "bused", "phone", "court", "camel", "proof", "heard", "angel", "petal", "pouty", "throb", "maybe", "fetal", "sprig", "spine", "shout", "cadet", "macro", "dodgy", "satyr", "rarer", "binge", "trend", "nutty", "leapt", "amiss", "split", "myrrh", "width", "sonar", "tower", "baron", "fever", "waver", "spark", "belie", "sloop", "expel", "smote", "baler", "above", "north", "wafer", "scant", "frill", "awash", "snack", "scowl", "frail", "drift", "limbo", "fence", "motel", "ounce", "wreak", "revel", "talon", "prior", "knelt", "cello", "flake", "debug", "anode", "crime", "salve", "scout", "imbue", "pinky", "stave", "vague", "chock", "fight", "video", "stone", "teach", "cleft", "frost", "prawn", "booty", "twist", "apnea", "stiff", "plaza", "ledge", "tweak", "board", "grant", "medic", "bacon", "cable", "brawl", "slunk", "raspy", "forum", "drone", "women", "mucus", "boast", "toddy", "coven", "tumor", "truer", "wrath", "stall", "steam", "axial", "purer", "daily", "trail", "niche", "mealy", "juice", "nylon", "plump", "merry", "flail", "papal", "wheat", "berry", "cower", "erect", "brute", "leggy", "snipe", "sinew", "skier", "penny", "jumpy", "rally", "umbra", "scary", "modem", "gross", "avian", "greed", "satin", "tonic", "parka", "sniff", "livid", "stark", "trump", "giddy", "reuse", "taboo", "avoid", "quote", "devil", "liken", "gloss", "gayer", "beret", "noise", "gland", "dealt", "sling", "rumor", "opera", "thigh", "tonga", "flare", "wound", "white", "bulky", "etude", "horse", "circa", "paddy", "inbox", "fizzy", "grain", "exert", "surge", "gleam", "belle", "salvo", "crush", "fruit", "sappy", "taker", "tract", "ovine", "spiky", "frank", "reedy", "filth", "spasm", "heave", "mambo", "right", "clank", "trust", "lumen", "borne", "spook", "sauce", "amber", "lathe", "carat", "corer", "dirty", "slyly", "affix", "alloy", "taint", "sheep", "kinky", "wooly", "mauve", "flung", "yacht", "fried", "quail", "brunt", "grimy", "curvy", "cagey", "rinse", "deuce", "state", "grasp", "milky", "bison", "graft", "sandy", "baste", "flask", "hedge", "girly", "swash", "boney", "coupe", "endow", "abhor", "welch", "blade", "tight", "geese", "miser", "mirth", "cloud", "cabal", "leech", "close", "tenth", "pecan", "droit", "grail", "clone", "guise", "ralph", "tango", "biddy", "smith", "mower", "payee", "serif", "drape", "fifth", "spank", "glaze", "allot", "truck", "kayak", "virus", "testy", "tepee", "fully", "zonal", "metro", "curry", "grand", "banjo", "axion", "bezel", "occur", "chain", "nasal", "gooey", "filer", "brace", "allay", "pubic", "raven", "plead", "gnash", "flaky", "munch", "dully", "eking", "thing", "slink", "hurry", "theft", "shorn", "pygmy", "ranch", "wring", "lemon", "shore", "mamma", "froze", "newer", "style", "moose", "antic", "drown", "vegan", "chess", "guppy", "union", "lever", "lorry", "image", "cabby", "druid", "exact", "truth", "dopey", "spear", "cried", "chime", "crony", "stunk", "timid", "batch", "gauge", "rotor", "crack", "curve", "latte", "witch", "bunch", "repel", "anvil", "soapy", "meter", "broth", "madly", "dried", "scene", "known", "magma", "roost", "woman", "thong", "punch", "pasty", "downy", "knead", "whirl", "rapid", "clang", "anger", "drive", "goofy", "email", "music", "stuff", "bleep", "rider", "mecca", "folio", "setup", "verso", "quash", "fauna", "gummy", "happy", "newly", "fussy", "relic", "guava", "ratty", "fudge", "femur", "chirp", "forte", "alibi", "whine", "petty", "golly", "plait", "fleck", "felon", "gourd", "brown", "thrum", "ficus", "stash", "decry", "wiser", "junta", "visor", "daunt", "scree", "impel", "await", "press", "whose", "turbo", "stoop", "speak", "mangy", "eying", "inlet", "crone", "pulse", "mossy", "staid", "hence", "pinch", "teddy", "sully", "snore", "ripen", "snowy", "attic", "going", "leach", "mouth", "hound", "clump", "tonal", "bigot", "peril", "piece", "blame", "haute", "spied", "undid", "intro", "basal", "shine", "gecko", "rodeo", "guard", "steer", "loamy", "scamp", "scram", "manly", "hello", "vaunt", "organ", "feral", "knock", "extra", "condo", "adapt", "willy", "polka", "rayon", "skirt", "faith", "torso", "match", "mercy", "tepid", "sleek", "riser", "twixt", "peace", "flush", "catty", "login", "eject", "roger", "rival", "untie", "refit", "aorta", "adult", "judge", "rower", "artsy", "rural", "shave"]
POSSIBLE_GUESSES = POSSIBLE_ANSWERS + ["aahed", "aalii", "aargh", "aarti", "abaca", "abaci", "abacs", "abaft", "abaka", "abamp", "aband", "abash", "abask", "abaya", "abbas", "abbed", "abbes", "abcee", "abeam", "abear", "abele", "abers", "abets", "abies", "abler", "ables", "ablet", "ablow", "abmho", "abohm", "aboil", "aboma", "aboon", "abord", "abore", "abram", "abray", "abrim", "abrin", "abris", "absey", "absit", "abuna", "abune", "abuts", "abuzz", "abyes", "abysm", "acais", "acari", "accas", "accoy", "acerb", "acers", "aceta", "achar", "ached", "aches", "achoo", "acids", "acidy", "acing", "acini", "ackee", "acker", "acmes", "acmic", "acned", "acnes", "acock", "acold", "acred", "acres", "acros", "acted", "actin", "acton", "acyls", "adaws", "adays", "adbot", "addax", "added", "adder", "addio", "addle", "adeem", "adhan", "adieu", "adios", "adits", "adman", "admen", "admix", "adobo", "adown", "adoze", "adrad", "adred", "adsum", "aduki", "adunc", "adust", "advew", "adyta", "adzed", "adzes", "aecia", "aedes", "aegis", "aeons", "aerie", "aeros", "aesir", "afald", "afara", "afars", "afear", "aflaj", "afore", "afrit", "afros", "agama", "agami", "agars", "agast", "agave", "agaze", "agene", "agers", "agger", "aggie", "aggri", "aggro", "aggry", "aghas", "agila", "agios", "agism", "agist", "agita", "aglee", "aglet", "agley", "agloo", "aglus", "agmas", "agoge", "agone", "agons", "agood", "agria", "agrin", "agros", "agued", "agues", "aguna", "aguti", "aheap", "ahent", "ahigh", "ahind", "ahing", "ahint", "ahold", "ahull", "ahuru", "aidas", "aided", "aides", "aidoi", "aidos", "aiery", "aigas", "aight", "ailed", "aimed", "aimer", "ainee", "ainga", "aioli", "aired", "airer", "airns", "airth", "airts", "aitch", "aitus", "aiver", "aiyee", "aizle", "ajies", "ajiva", "ajuga", "ajwan", "akees", "akela", "akene", "aking", "akita", "akkas", "alaap", "alack", "alamo", "aland", "alane", "alang", "alans", "alant", "alapa", "alaps", "alary", "alate", "alays", "albas", "albee", "alcid", "alcos", "aldea", "alder", "aldol", "aleck", "alecs", "alefs", "aleft", "aleph", "alews", "aleye", "alfas", "algal", "algas", "algid", "algin", "algor", "algum", "alias", "alifs", "aline", "alist", "aliya", "alkie", "alkos", "alkyd", "alkyl", "allee", "allel", "allis", "allod", "allyl", "almah", "almas", "almeh", "almes", "almud", "almug", "alods", "aloed", "aloes", "aloha", "aloin", "aloos", "alowe", "altho", "altos", "alula", "alums", "alure", "alvar", "alway", "amahs", "amain", "amate", "amaut", "amban", "ambit", "ambos", "ambry", "ameba", "ameer", "amene", "amens", "ament", "amias", "amice", "amici", "amide", "amido", "amids", "amies", "amiga", "amigo", "amine", "amino", "amins", "amirs", "amlas", "amman", "ammon", "ammos", "amnia", "amnic", "amnio", "amoks", "amole", "amort", "amour", "amove", "amowt", "amped", "ampul", "amrit", "amuck", "amyls", "anana", "anata", "ancho", "ancle", "ancon", "andro", "anear", "anele", "anent", "angas", "anglo", "anigh", "anile", "anils", "anima", "animi", "anion", "anise", "anker", "ankhs", "ankus", "anlas", "annal", "annas", "annat", "anoas", "anole", "anomy", "ansae", "antae", "antar", "antas", "anted", "antes", "antis", "antra", "antre", "antsy", "anura", "anyon", "apace", "apage", "apaid", "apayd", "apays", "apeak", "apeek", "apers", "apert", "apery", "apgar", "aphis", "apian", "apiol", "apish", "apism", "apode", "apods", "apoop", "aport", "appal", "appay", "appel", "appro", "appui", "appuy", "apres", "apses", "apsis", "apsos", "apted", "apter", "aquae", "aquas", "araba", "araks", "arame", "arars", "arbas", "arced", "archi", "arcos", "arcus", "ardeb", "ardri", "aread", "areae", "areal", "arear", "areas", "areca", "aredd", "arede", "arefy", "areic", "arene", "arepa", "arere", "arete", "arets", "arett", "argal", "argan", "argil", "argle", "argol", "argon", "argot", "argus", "arhat", "arias", "ariel", "ariki", "arils", "ariot", "arish", "arked", "arled", "arles", "armed", "armer", "armet", "armil", "arnas", "arnut", "aroba", "aroha", "aroid", "arpas", "arpen", "arrah", "arras", "arret", "arris", "arroz", "arsed", "arses", "arsey", "arsis", "artal", "artel", "artic", "artis", "aruhe", "arums", "arval", "arvee", "arvos", "aryls", "asana", "ascon", "ascus", "asdic", "ashed", "ashes", "ashet", "asked", "asker", "askoi", "askos", "aspen", "asper", "aspic", "aspie", "aspis", "aspro", "assai", "assam", "asses", "assez", "assot", "aster", "astir", "astun", "asura", "asway", "aswim", "asyla", "ataps", "ataxy", "atigi", "atilt", "atimy", "atlas", "atman", "atmas", "atmos", "atocs", "atoke", "atoks", "atoms", "atomy", "atony", "atopy", "atria", "atrip", "attap", "attar", "atuas", "audad", "auger", "aught", "aulas", "aulic", "auloi", "aulos", "aumil", "aunes", "aunts", "aurae", "aural", "aurar", "auras", "aurei", "aures", "auric", "auris", "aurum", "autos", "auxin", "avale", "avant", "avast", "avels", "avens", "avers", "avgas", "avine", "avion", "avise", "aviso", "avize", "avows", "avyze", "awarn", "awato", "awave", "aways", "awdls", "aweel", "aweto", "awing", "awmry", "awned", "awner", "awols", "awork", "axels", "axile", "axils", "axing", "axite", "axled", "axles", "axman", "axmen", "axoid", "axone", "axons", "ayahs", "ayaya", "ayelp", "aygre", "ayins", "ayont", "ayres", "ayrie", "azans", "azide", "azido", "azine", "azlon", "azoic", "azole", "azons", "azote", "azoth", "azuki", "azurn", "azury", "azygy", "azyme", "azyms", "baaed", "baals", "babas", "babel", "babes", "babka", "baboo", "babul", "babus", "bacca", "bacco", "baccy", "bacha", "bachs", "backs", "baddy", "baels", "baffs", "baffy", "bafts", "baghs", "bagie", "bahts", "bahus", "bahut", "bails", "bairn", "baisa", "baith", "baits", "baiza", "baize", "bajan", "bajra", "bajri", "bajus", "baked", "baken", "bakes", "bakra", "balas", "balds", "baldy", "baled", "bales", "balks", "balky", "balls", "bally", "balms", "baloo", "balsa", "balti", "balun", "balus", "bambi", "banak", "banco", "bancs", "banda", "bandh", "bands", "bandy", "baned", "banes", "bangs", "bania", "banks", "banns", "bants", "bantu", "banty", "banya", "bapus", "barbe", "barbs", "barby", "barca", "barde", "bardo", "bards", "bardy", "bared", "barer", "bares", "barfi", "barfs", "baric", "barks", "barky", "barms", "barmy", "barns", "barny", "barps", "barra", "barre", "barro", "barry", "barye", "basan", "based", "basen", "baser", "bases", "basho", "basij", "basks", "bason", "basse", "bassi", "basso", "bassy", "basta", "basti", "basto", "basts", "bated", "bates", "baths", "batik", "batta", "batts", "battu", "bauds", "bauks", "baulk", "baurs", "bavin", "bawds", "bawks", "bawls", "bawns", "bawrs", "bawty", "bayed", "bayer", "bayes", "bayle", "bayts", "bazar", "bazoo", "beads", "beaks", "beaky", "beals", "beams", "beamy", "beano", "beans", "beany", "beare", "bears", "beath", "beats", "beaty", "beaus", "beaut", "beaux", "bebop", "becap", "becke", "becks", "bedad", "bedel", "bedes", "bedew", "bedim", "bedye", "beedi", "beefs", "beeps", "beers", "beery", "beets", "befog", "begad", "begar", "begem", "begot", "begum", "beige", "beigy", "beins", "bekah", "belah", "belar", "belay", "belee", "belga", "bells", "belon", "belts", "bemad", "bemas", "bemix", "bemud", "bends", "bendy", "benes", "benet", "benga", "benis", "benne", "benni", "benny", "bento", "bents", "benty", "bepat", "beray", "beres", "bergs", "berko", "berks", "berme", "berms", "berob", "beryl", "besat", "besaw", "besee", "beses", "besit", "besom", "besot", "besti", "bests", "betas", "beted", "betes", "beths", "betid", "beton", "betta", "betty", "bever", "bevor", "bevue", "bevvy", "bewet", "bewig", "bezes", "bezil", "bezzy", "bhais", "bhaji", "bhang", "bhats", "bhels", "bhoot", "bhuna", "bhuts", "biach", "biali", "bialy", "bibbs", "bibes", "biccy", "bices", "bided", "bider", "bides", "bidet", "bidis", "bidon", "bield", "biers", "biffo", "biffs", "biffy", "bifid", "bigae", "biggs", "biggy", "bigha", "bight", "bigly", "bigos", "bijou", "biked", "biker", "bikes", "bikie", "bilbo", "bilby", "biled", "biles", "bilgy", "bilks", "bills", "bimah", "bimas", "bimbo", "binal", "bindi", "binds", "biner", "bines", "bings", "bingy", "binit", "binks", "bints", "biogs", "biont", "biota", "biped", "bipod", "birds", "birks", "birle", "birls", "biros", "birrs", "birse", "birsy", "bises", "bisks", "bisom", "bitch", "biter", "bites", "bitos", "bitou", "bitsy", "bitte", "bitts", "bivia", "bivvy", "bizes", "bizzo", "bizzy", "blabs", "blads", "blady", "blaer", "blaes", "blaff", "blags", "blahs", "blain", "blams", "blart", "blase", "blash", "blate", "blats", "blatt", "blaud", "blawn", "blaws", "blays", "blear", "blebs", "blech", "blees", "blent", "blert", "blest", "blets", "bleys", "blimy", "bling", "blini", "blins", "bliny", "blips", "blist", "blite", "blits", "blive", "blobs", "blocs", "blogs", "blook", "bloop", "blore", "blots", "blows", "blowy", "blubs", "blude", "bluds", "bludy", "blued", "blues", "bluet", "bluey", "bluid", "blume", "blunk", "blurs", "blype", "boabs", "boaks", "boars", "boart", "boats", "bobac", "bobak", "bobas", "bobol", "bobos", "bocca", "bocce", "bocci", "boche", "bocks", "boded", "bodes", "bodge", "bodhi", "bodle", "boeps", "boets", "boeuf", "boffo", "boffs", "bogan", "bogey", "boggy", "bogie", "bogle", "bogue", "bogus", "bohea", "bohos", "boils", "boing", "boink", "boite", "boked", "bokeh", "bokes", "bokos", "bolar", "bolas", "bolds", "boles", "bolix", "bolls", "bolos", "bolts", "bolus", "bomas", "bombe", "bombo", "bombs", "bonce", "bonds", "boned", "boner", "bones", "bongs", "bonie", "bonks", "bonne", "bonny", "bonza", "bonze", "booai", "booay", "boobs", "boody", "booed", "boofy", "boogy", "boohs", "books", "booky", "bools", "booms", "boomy", "boong", "boons", "boord", "boors", "boose", "boots", "boppy", "borak", "boral", "boras", "borde", "bords", "bored", "boree", "borel", "borer", "bores", "borgo", "boric", "borks", "borms", "borna", "boron", "borts", "borty", "bortz", "bosie", "bosks", "bosky", "boson", "bosun", "botas", "botel", "botes", "bothy", "botte", "botts", "botty", "bouge", "bouks", "boult", "bouns", "bourd", "bourg", "bourn", "bouse", "bousy", "bouts", "bovid", "bowat", "bowed", "bower", "bowes", "bowet", "bowie", "bowls", "bowne", "bowrs", "bowse", "boxed", "boxen", "boxes", "boxla", "boxty", "boyar", "boyau", "boyed", "boyfs", "boygs", "boyla", "boyos", "boysy", "bozos", "braai", "brach", "brack", "bract", "brads", "braes", "brags", "brail", "braks", "braky", "brame", "brane", "brank", "brans", "brant", "brast", "brats", "brava", "bravi", "braws", "braxy", "brays", "braza", "braze", "bream", "brede", "breds", "breem", "breer", "brees", "breid", "breis", "breme", "brens", "brent", "brere", "brers", "breve", "brews", "breys", "brier", "bries", "brigs", "briki", "briks", "brill", "brims", "brins", "brios", "brise", "briss", "brith", "brits", "britt", "brize", "broch", "brock", "brods", "brogh", "brogs", "brome", "bromo", "bronc", "brond", "brool", "broos", "brose", "brosy", "brows", "brugh", "bruin", "bruit", "brule", "brume", "brung", "brusk", "brust", "bruts", "buats", "buaze", "bubal", "bubas", "bubba", "bubbe", "bubby", "bubus", "buchu", "bucko", "bucks", "bucku", "budas", "budis", "budos", "buffa", "buffe", "buffi", "buffo", "buffs", "buffy", "bufos", "bufty", "buhls", "buhrs", "buiks", "buist", "bukes", "bulbs", "bulgy", "bulks", "bulla", "bulls", "bulse", "bumbo", "bumfs", "bumph", "bumps", "bumpy", "bunas", "bunce", "bunco", "bunde", "bundh", "bunds", "bundt", "bundu", "bundy", "bungs", "bungy", "bunia", "bunje", "bunjy", "bunko", "bunks", "bunns", "bunts", "bunty", "bunya", "buoys", "buppy", "buran", "buras", "burbs", "burds", "buret", "burfi", "burgh", "burgs", "burin", "burka", "burke", "burks", "burls", "burns", "buroo", "burps", "burqa", "burro", "burrs", "burry", "bursa", "burse", "busby", "buses", "busks", "busky", "bussu", "busti", "busts", "busty", "buteo", "butes", "butle", "butoh", "butts", "butty", "butut", "butyl", "buzzy", "bwana", "bwazi", "byded", "bydes", "byked", "bykes", "byres", "byrls", "byssi", "bytes", "byway", "caaed", "cabas", "caber", "cabob", "caboc", "cabre", "cacas", "cacks", "cacky", "cadee", "cades", "cadge", "cadgy", "cadie", "cadis", "cadre", "caeca", "caese", "cafes", "caffs", "caged", "cager", "cages", "cagot", "cahow", "caids", "cains", "caird", "cajon", "cajun", "caked", "cakes", "cakey", "calfs", "calid", "calif", "calix", "calks", "calla", "calls", "calms", "calmy", "calos", "calpa", "calps", "calve", "calyx", "caman", "camas", "cames", "camis", "camos", "campi", "campo", "camps", "campy", "camus", "caned", "caneh", "caner", "canes", "cangs", "canid", "canna", "canns", "canso", "canst", "canto", "cants", "canty", "capas", "caped", "capes", "capex", "caphs", "capiz", "caple", "capon", "capos", "capot", "capri", "capul", "carap", "carbo", "carbs", "carby", "cardi", "cards", "cardy", "cared", "carer", "cares", "caret", "carex", "carks", "carle", "carls", "carns", "carny", "carob", "carom", "caron", "carpi", "carps", "carrs", "carse", "carta", "carte", "carts", "carvy", "casas", "casco", "cased", "cases", "casks", "casky", "casts", "casus", "cates", "cauda", "cauks", "cauld", "cauls", "caums", "caups", "cauri", "causa", "cavas", "caved", "cavel", "caver", "caves", "cavie", "cawed", "cawks", "caxon", "ceaze", "cebid", "cecal", "cecum", "ceded", "ceder", "cedes", "cedis", "ceiba", "ceili", "ceils", "celeb", "cella", "celli", "cells", "celom", "celts", "cense", "cento", "cents", "centu", "ceorl", "cepes", "cerci", "cered", "ceres", "cerge", "ceria", "ceric", "cerne", "ceroc", "ceros", "certs", "certy", "cesse", "cesta", "cesti", "cetes", "cetyl", "cezve", "chace", "chack", "chaco", "chado", "chads", "chaft", "chais", "chals", "chams", "chana", "chang", "chank", "chape", "chaps", "chapt", "chara", "chare", "chark", "charr", "chars", "chary", "chats", "chave", "chavs", "chawk", "chaws", "chaya", "chays", "cheep", "chefs", "cheka", "chela", "chelp", "chemo", "chems", "chere", "chert", "cheth", "chevy", "chews", "chewy", "chiao", "chias", "chibs", "chica", "chich", "chico", "chics", "chiel", "chiks", "chile", "chimb", "chimo", "chimp", "chine", "ching", "chink", "chino", "chins", "chips", "chirk", "chirl", "chirm", "chiro", "chirr", "chirt", "chiru", "chits", "chive", "chivs", "chivy", "chizz", "choco", "chocs", "chode", "chogs", "choil", "choko", "choky", "chola", "choli", "cholo", "chomp", "chons", "choof", "chook", "choom", "choon", "chops", "chota", "chott", "chout", "choux", "chowk", "chows", "chubs", "chufa", "chuff", "chugs", "chums", "churl", "churr", "chuse", "chuts", "chyle", "chyme", "chynd", "cibol", "cided", "cides", "ciels", "ciggy", "cilia", "cills", "cimar", "cimex", "cinct", "cines", "cinqs", "cions", "cippi", "circs", "cires", "cirls", "cirri", "cisco", "cissy", "cists", "cital", "cited", "citer", "cites", "cives", "civet", "civie", "civvy", "clach", "clade", "clads", "claes", "clags", "clame", "clams", "clans", "claps", "clapt", "claro", "clart", "clary", "clast", "clats", "claut", "clave", "clavi", "claws", "clays", "cleck", "cleek", "cleep", "clefs", "clegs", "cleik", "clems", "clepe", "clept", "cleve", "clews", "clied", "clies", "clift", "clime", "cline", "clint", "clipe", "clips", "clipt", "clits", "cloam", "clods", "cloff", "clogs", "cloke", "clomb", "clomp", "clonk", "clons", "cloop", "cloot", "clops", "clote", "clots", "clour", "clous", "clows", "cloye", "cloys", "cloze", "clubs", "clues", "cluey", "clunk", "clype", "cnida", "coact", "coady", "coala", "coals", "coaly", "coapt", "coarb", "coate", "coati", "coats", "cobbs", "cobby", "cobia", "coble", "cobza", "cocas", "cocci", "cocco", "cocks", "cocky", "cocos", "codas", "codec", "coded", "coden", "coder", "codes", "codex", "codon", "coeds", "coffs", "cogie", "cogon", "cogue", "cohab", "cohen", "cohoe", "cohog", "cohos", "coifs", "coign", "coils", "coins", "coirs", "coits", "coked", "cokes", "colas", "colby", "colds", "coled", "coles", "coley", "colic", "colin", "colls", "colly", "colog", "colts", "colza", "comae", "comal", "comas", "combe", "combi", "combo", "combs", "comby", "comer", "comes", "comix", "commo", "comms", "commy", "compo", "comps", "compt", "comte", "comus", "coned", "cones", "coney", "confs", "conga", "conge", "congo", "conia", "conin", "conks", "conky", "conne", "conns", "conte", "conto", "conus", "convo", "cooch", "cooed", "cooee", "cooer", "cooey", "coofs", "cooks", "cooky", "cools", "cooly", "coomb", "cooms", "coomy", "coons", "coops", "coopt", "coost", "coots", "cooze", "copal", "copay", "coped", "copen", "coper", "copes", "coppy", "copra", "copsy", "coqui", "coram", "corbe", "corby", "cords", "cored", "cores", "corey", "corgi", "coria", "corks", "corky", "corms", "corni", "corno", "corns", "cornu", "corps", "corse", "corso", "cosec", "cosed", "coses", "coset", "cosey", "cosie", "costa", "coste", "costs", "cotan", "coted", "cotes", "coths", "cotta", "cotts", "coude", "coups", "courb", "courd", "coure", "cours", "couta", "couth", "coved", "coves", "covin", "cowal", "cowan", "cowed", "cowks", "cowls", "cowps", "cowry", "coxae", "coxal", "coxed", "coxes", "coxib", "coyau", "coyed", "coyer", "coypu", "cozed", "cozen", "cozes", "cozey", "cozie", "craal", "crabs", "crags", "craic", "craig", "crake", "crame", "crams", "crans", "crape", "craps", "crapy", "crare", "craws", "crays", "creds", "creel", "crees", "crems", "crena", "creps", "crepy", "crewe", "crews", "crias", "cribs", "cries", "crims", "crine", "crios", "cripe", "crips", "crise", "crith", "crits", "croci", "crocs", "croft", "crogs", "cromb", "crome", "cronk", "crons", "crool", "croon", "crops", "crore", "crost", "crout", "crows", "croze", "cruck", "crudo", "cruds", "crudy", "crues", "cruet", "cruft", "crunk", "cruor", "crura", "cruse", "crusy", "cruve", "crwth", "cryer", "ctene", "cubby", "cubeb", "cubed", "cuber", "cubes", "cubit", "cuddy", "cuffo", "cuffs", "cuifs", "cuing", "cuish", "cuits", "cukes", "culch", "culet", "culex", "culls", "cully", "culms", "culpa", "culti", "cults", "culty", "cumec", "cundy", "cunei", "cunit", "cunts", "cupel", "cupid", "cuppa", "cuppy", "curat", "curbs", "curch", "curds", "curdy", "cured", "curer", "cures", "curet", "curfs", "curia", "curie", "curli", "curls", "curns", "curny", "currs", "cursi", "curst", "cusec", "cushy", "cusks", "cusps", "cuspy", "cusso", "cusum", "cutch", "cuter", "cutes", "cutey", "cutin", "cutis", "cutto", "cutty", "cutup", "cuvee", "cuzes", "cwtch", "cyano", "cyans", "cycad", "cycas", "cyclo", "cyder", "cylix", "cymae", "cymar", "cymas", "cymes", "cymol", "cysts", "cytes", "cyton", "czars", "daals", "dabba", "daces", "dacha", "dacks", "dadah", "dadas", "dados", "daffs", "daffy", "dagga", "daggy", "dagos", "dahls", "daiko", "daine", "daint", "daker", "daled", "dales", "dalis", "dalle", "dalts", "daman", "damar", "dames", "damme", "damns", "damps", "dampy", "dancy", "dangs", "danio", "danks", "danny", "dants", "daraf", "darbs", "darcy", "dared", "darer", "dares", "darga", "dargs", "daric", "daris", "darks", "darky", "darns", "darre", "darts", "darzi", "dashi", "dashy", "datal", "dated", "dater", "dates", "datos", "datto", "daube", "daubs", "dauby", "dauds", "dault", "daurs", "dauts", "daven", "davit", "dawah", "dawds", "dawed", "dawen", "dawks", "dawns", "dawts", "dayan", "daych", "daynt", "dazed", "dazer", "dazes", "deads", "deair", "deals", "deans", "deare", "dearn", "dears", "deary", "deash", "deave", "deaws", "deawy", "debag", "debby", "debel", "debes", "debts", "debud", "debur", "debus", "debye", "decad", "decaf", "decan", "decko", "decks", "decos", "dedal", "deeds", "deedy", "deely", "deems", "deens", "deeps", "deere", "deers", "deets", "deeve", "deevs", "defat", "deffo", "defis", "defog", "degas", "degum", "degus", "deice", "deids", "deify", "deils", "deism", "deist", "deked", "dekes", "dekko", "deled", "deles", "delfs", "delft", "delis", "dells", "delly", "delos", "delph", "delts", "deman", "demes", "demic", "demit", "demob", "demoi", "demos", "dempt", "denar", "denay", "dench", "denes", "denet", "denis", "dents", "deoxy", "derat", "deray", "dered", "deres", "derig", "derma", "derms", "derns", "derny", "deros", "derro", "derry", "derth", "dervs", "desex", "deshi", "desis", "desks", "desse", "devas", "devel", "devis", "devon", "devos", "devot", "dewan", "dewar", "dewax", "dewed", "dexes", "dexie", "dhaba", "dhaks", "dhals", "dhikr", "dhobi", "dhole", "dholl", "dhols", "dhoti", "dhows", "dhuti", "diact", "dials", "diane", "diazo", "dibbs", "diced", "dicer", "dices", "dicht", "dicks", "dicky", "dicot", "dicta", "dicts", "dicty", "diddy", "didie", "didos", "didst", "diebs", "diels", "diene", "diets", "diffs", "dight", "dikas", "diked", "diker", "dikes", "dikey", "dildo", "dilli", "dills", "dimbo", "dimer", "dimes", "dimps", "dinar", "dined", "dines", "dinge", "dings", "dinic", "dinks", "dinky", "dinna", "dinos", "dints", "diols", "diota", "dippy", "dipso", "diram", "direr", "dirke", "dirks", "dirls", "dirts", "disas", "disci", "discs", "dishy", "disks", "disme", "dital", "ditas", "dited", "dites", "ditsy", "ditts", "ditzy", "divan", "divas", "dived", "dives", "divis", "divna", "divos", "divot", "divvy", "diwan", "dixie", "dixit", "diyas", "dizen", "djinn", "djins", "doabs", "doats", "dobby", "dobes", "dobie", "dobla", "dobra", "dobro", "docht", "docks", "docos", "docus", "doddy", "dodos", "doeks", "doers", "doest", "doeth", "doffs", "dogan", "doges", "dogey", "doggo", "doggy", "dogie", "dohyo", "doilt", "doily", "doits", "dojos", "dolce", "dolci", "doled", "doles", "dolia", "dolls", "dolma", "dolor", "dolos", "dolts", "domal", "domed", "domes", "domic", "donah", "donas", "donee", "doner", "donga", "dongs", "donko", "donna", "donne", "donny", "donsy", "doobs", "dooce", "doody", "dooks", "doole", "dools", "dooly", "dooms", "doomy", "doona", "doorn", "doors", "doozy", "dopas", "doped", "doper", "dopes", "dorad", "dorba", "dorbs", "doree", "dores", "doric", "doris", "dorks", "dorky", "dorms", "dormy", "dorps", "dorrs", "dorsa", "dorse", "dorts", "dorty", "dosai", "dosas", "dosed", "doseh", "doser", "doses", "dosha", "dotal", "doted", "doter", "dotes", "dotty", "douar", "douce", "doucs", "douks", "doula", "douma", "doums", "doups", "doura", "douse", "douts", "doved", "doven", "dover", "doves", "dovie", "dowar", "dowds", "dowed", "dower", "dowie", "dowle", "dowls", "dowly", "downa", "downs", "dowps", "dowse", "dowts", "doxed", "doxes", "doxie", "doyen", "doyly", "dozed", "dozer", "dozes", "drabs", "drack", "draco", "draff", "drags", "drail", "drams", "drant", "draps", "drats", "drave", "draws", "drays", "drear", "dreck", "dreed", "dreer", "drees", "dregs", "dreks", "drent", "drere", "drest", "dreys", "dribs", "drice", "dries", "drily", "drips", "dript", "droid", "droil", "droke", "drole", "drome", "drony", "droob", "droog", "drook", "drops", "dropt", "drouk", "drows", "drubs", "drugs", "drums", "drupe", "druse", "drusy", "druxy", "dryad", "dryas", "dsobo", "dsomo", "duads", "duals", "duans", "duars", "dubbo", "ducal", "ducat", "duces", "ducks", "ducky", "ducts", "duddy", "duded", "dudes", "duels", "duets", "duett", "duffs", "dufus", "duing", "duits", "dukas", "duked", "dukes", "dukka", "dulce", "dules", "dulia", "dulls", "dulse", "dumas", "dumbo", "dumbs", "dumka", "dumky", "dumps", "dunam", "dunch", "dunes", "dungs", "dungy", "dunks", "dunno", "dunny", "dunsh", "dunts", "duomi", "duomo", "duped", "duper", "dupes", "duple", "duply", "duppy", "dural", "duras", "dured", "dures", "durgy", "durns", "duroc", "duros", "duroy", "durra", "durrs", "durry", "durst", "durum", "durzi", "dusks", "dusts", "duxes", "dwaal", "dwale", "dwalm", "dwams", "dwang", "dwaum", "dweeb", "dwile", "dwine", "dyads", "dyers", "dyked", "dykes", "dykey", "dykon", "dynel", "dynes", "dzhos", "eagre", "ealed", "eales", "eaned", "eards", "eared", "earls", "earns", "earnt", "earst", "eased", "easer", "eases", "easle", "easts", "eathe", "eaved", "eaves", "ebbed", "ebbet", "ebons", "ebook", "ecads", "eched", "eches", "echos", "ecrus", "edema", "edged", "edger", "edges", "edile", "edits", "educe", "educt", "eejit", "eensy", "eeven", "eevns", "effed", "egads", "egers", "egest", "eggar", "egged", "egger", "egmas", "ehing", "eider", "eidos", "eigne", "eiked", "eikon", "eilds", "eisel", "ejido", "ekkas", "elain", "eland", "elans", "elchi", "eldin", "elemi", "elfed", "eliad", "elint", "elmen", "eloge", "elogy", "eloin", "elops", "elpee", "elsin", "elute", "elvan", "elven", "elver", "elves", "emacs", "embar", "embay", "embog", "embow", "embox", "embus", "emeer", "emend", "emerg", "emery", "emeus", "emics", "emirs", "emits", "emmas", "emmer", "emmet", "emmew", "emmys", "emoji", "emong", "emote", "emove", "empts", "emule", "emure", "emyde", "emyds", "enarm", "enate", "ended", "ender", "endew", "endue", "enews", "enfix", "eniac", "enlit", "enmew", "ennog", "enoki", "enols", "enorm", "enows", "enrol", "ensew", "ensky", "entia", "enure", "enurn", "envoi", "enzym", "eorls", "eosin", "epact", "epees", "ephah", "ephas", "ephod", "ephor", "epics", "epode", "epopt", "epris", "eques", "equid", "erbia", "erevs", "ergon", "ergos", "ergot", "erhus", "erica", "erick", "erics", "ering", "erned", "ernes", "erose", "erred", "erses", "eruct", "erugo", "eruvs", "erven", "ervil", "escar", "escot", "esile", "eskar", "esker", "esnes", "esses", "estoc", "estop", "estro", "etage", "etape", "etats", "etens", "ethal", "ethne", "ethyl", "etics", "etnas", "ettin", "ettle", "etuis", "etwee", "etyma", "eughs", "euked", "eupad", "euros", "eusol", "evens", "evert", "evets", "evhoe", "evils", "evite", "evohe", "ewers", "ewest", "ewhow", "ewked", "exams", "exeat", "execs", "exeem", "exeme", "exfil", "exies", "exine", "exing", "exits", "exode", "exome", "exons", "expat", "expos", "exude", "exuls", "exurb", "eyass", "eyers", "eyots", "eyras", "eyres", "eyrie", "eyrir", "ezine", "fabby", "faced", "facer", "faces", "facia", "facta", "facts", "faddy", "faded", "fader", "fades", "fadge", "fados", "faena", "faery", "faffs", "faffy", "faggy", "fagin", "fagot", "faiks", "fails", "faine", "fains", "fairs", "faked", "faker", "fakes", "fakey", "fakie", "fakir", "falaj", "falls", "famed", "fames", "fanal", "fands", "fanes", "fanga", "fango", "fangs", "fanks", "fanon", "fanos", "fanum", "faqir", "farad", "farci", "farcy", "fards", "fared", "farer", "fares", "farle", "farls", "farms", "faros", "farro", "farse", "farts", "fasci", "fasti", "fasts", "fated", "fates", "fatly", "fatso", "fatwa", "faugh", "fauld", "fauns", "faurd", "fauts", "fauve", "favas", "favel", "faver", "faves", "favus", "fawns", "fawny", "faxed", "faxes", "fayed", "fayer", "fayne", "fayre", "fazed", "fazes", "feals", "feare", "fears", "feart", "fease", "feats", "feaze", "feces", "fecht", "fecit", "fecks", "fedex", "feebs", "feeds", "feels", "feens", "feers", "feese", "feeze", "fehme", "feint", "feist", "felch", "felid", "fells", "felly", "felts", "felty", "femal", "femes", "femmy", "fends", "fendy", "fenis", "fenks", "fenny", "fents", "feods", "feoff", "ferer", "feres", "feria", "ferly", "fermi", "ferms", "ferns", "ferny", "fesse", "festa", "fests", "festy", "fetas", "feted", "fetes", "fetor", "fetta", "fetts", "fetwa", "feuar", "feuds", "feued", "feyed", "feyer", "feyly", "fezes", "fezzy", "fiars", "fiats", "fibro", "fices", "fiche", "fichu", "ficin", "ficos", "fides", "fidge", "fidos", "fiefs", "fient", "fiere", "fiers", "fiest", "fifed", "fifer", "fifes", "fifis", "figgy", "figos", "fiked", "fikes", "filar", "filch", "filed", "files", "filii", "filks", "fille", "fillo", "fills", "filmi", "films", "filos", "filum", "finca", "finds", "fined", "fines", "finis", "finks", "finny", "finos", "fiord", "fiqhs", "fique", "fired", "firer", "fires", "firie", "firks", "firms", "firns", "firry", "firth", "fiscs", "fisks", "fists", "fisty", "fitch", "fitly", "fitna", "fitte", "fitts", "fiver", "fives", "fixed", "fixes", "fixit", "fjeld", "flabs", "flaff", "flags", "flaks", "flamm", "flams", "flamy", "flane", "flans", "flaps", "flary", "flats", "flava", "flawn", "flaws", "flawy", "flaxy", "flays", "fleam", "fleas", "fleek", "fleer", "flees", "flegs", "fleme", "fleur", "flews", "flexi", "flexo", "fleys", "flics", "flied", "flies", "flimp", "flims", "flips", "flirs", "flisk", "flite", "flits", "flitt", "flobs", "flocs", "floes", "flogs", "flong", "flops", "flors", "flory", "flosh", "flota", "flote", "flows", "flubs", "flued", "flues", "fluey", "fluky", "flump", "fluor", "flurr", "fluty", "fluyt", "flyby", "flype", "flyte", "foals", "foams", "foehn", "fogey", "fogie", "fogle", "fogou", "fohns", "foids", "foils", "foins", "folds", "foley", "folia", "folic", "folie", "folks", "folky", "fomes", "fonda", "fonds", "fondu", "fones", "fonly", "fonts", "foods", "foody", "fools", "foots", "footy", "foram", "forbs", "forby", "fordo", "fords", "forel", "fores", "forex", "forks", "forky", "forme", "forms", "forts", "forza", "forze", "fossa", "fosse", "fouat", "fouds", "fouer", "fouet", "foule", "fouls", "fount", "fours", "fouth", "fovea", "fowls", "fowth", "foxed", "foxes", "foxie", "foyle", "foyne", "frabs", "frack", "fract", "frags", "fraim", "franc", "frape", "fraps", "frass", "frate", "frati", "frats", "fraus", "frays", "frees", "freet", "freit", "fremd", "frena", "freon", "frere", "frets", "fribs", "frier", "fries", "frigs", "frise", "frist", "frith", "frits", "fritt", "frize", "frizz", "froes", "frogs", "frons", "frore", "frorn", "frory", "frosh", "frows", "frowy", "frugs", "frump", "frush", "frust", "fryer", "fubar", "fubby", "fubsy", "fucks", "fucus", "fuddy", "fudgy", "fuels", "fuero", "fuffs", "fuffy", "fugal", "fuggy", "fugie", "fugio", "fugle", "fugly", "fugus", "fujis", "fulls", "fumed", "fumer", "fumes", "fumet", "fundi", "funds", "fundy", "fungo", "fungs", "funks", "fural", "furan", "furca", "furls", "furol", "furrs", "furth", "furze", "furzy", "fused", "fusee", "fusel", "fuses", "fusil", "fusks", "fusts", "fusty", "futon", "fuzed", "fuzee", "fuzes", "fuzil", "fyces", "fyked", "fykes", "fyles", "fyrds", "fytte", "gabba", "gabby", "gable", "gaddi", "gades", "gadge", "gadid", "gadis", "gadje", "gadjo", "gadso", "gaffs", "gaged", "gager", "gages", "gaids", "gains", "gairs", "gaita", "gaits", "gaitt", "gajos", "galah", "galas", "galax", "galea", "galed", "gales", "galls", "gally", "galop", "galut", "galvo", "gamas", "gamay", "gamba", "gambe", "gambo", "gambs", "gamed", "games", "gamey", "gamic", "gamin", "gamme", "gammy", "gamps", "ganch", "gandy", "ganef", "ganev", "gangs", "ganja", "ganof", "gants", "gaols", "gaped", "gaper", "gapes", "gapos", "gappy", "garbe", "garbo", "garbs", "garda", "gares", "garis", "garms", "garni", "garre", "garth", "garum", "gases", "gasps", "gaspy", "gasts", "gatch", "gated", "gater", "gates", "gaths", "gator", "gauch", "gaucy", "gauds", "gauje", "gault", "gaums", "gaumy", "gaups", "gaurs", "gauss", "gauzy", "gavot", "gawcy", "gawds", "gawks", "gawps", "gawsy", "gayal", "gazal", "gazar", "gazed", "gazes", "gazon", "gazoo", "geals", "geans", "geare", "gears", "geats", "gebur", "gecks", "geeks", "geeps", "geest", "geist", "geits", "gelds", "gelee", "gelid", "gelly", "gelts", "gemel", "gemma", "gemmy", "gemot", "genal", "genas", "genes", "genet", "genic", "genii", "genip", "genny", "genoa", "genom", "genro", "gents", "genty", "genua", "genus", "geode", "geoid", "gerah", "gerbe", "geres", "gerle", "germs", "germy", "gerne", "gesse", "gesso", "geste", "gests", "getas", "getup", "geums", "geyan", "geyer", "ghast", "ghats", "ghaut", "ghazi", "ghees", "ghest", "ghyll", "gibed", "gibel", "giber", "gibes", "gibli", "gibus", "gifts", "gigas", "gighe", "gigot", "gigue", "gilas", "gilds", "gilet", "gills", "gilly", "gilpy", "gilts", "gimel", "gimme", "gimps", "gimpy", "ginch", "ginge", "gings", "ginks", "ginny", "ginzo", "gipon", "gippo", "gippy", "girds", "girls", "girns", "giron", "giros", "girrs", "girsh", "girts", "gismo", "gisms", "gists", "gitch", "gites", "giust", "gived", "gives", "gizmo", "glace", "glads", "glady", "glaik", "glair", "glams", "glans", "glary", "glaum", "glaur", "glazy", "gleba", "glebe", "gleby", "glede", "gleds", "gleed", "gleek", "glees", "gleet", "gleis", "glens", "glent", "gleys", "glial", "glias", "glibs", "gliff", "glift", "glike", "glime", "glims", "glisk", "glits", "glitz", "gloam", "globi", "globs", "globy", "glode", "glogg", "gloms", "gloop", "glops", "glost", "glout", "glows", "gloze", "glued", "gluer", "glues", "gluey", "glugs", "glume", "glums", "gluon", "glute", "gluts", "gnarl", "gnarr", "gnars", "gnats", "gnawn", "gnaws", "gnows", "goads", "goafs", "goals", "goary", "goats", "goaty", "goban", "gobar", "gobbi", "gobbo", "gobby", "gobis", "gobos", "godet", "godso", "goels", "goers", "goest", "goeth", "goety", "gofer", "goffs", "gogga", "gogos", "goier", "gojis", "golds", "goldy", "goles", "golfs", "golpe", "golps", "gombo", "gomer", "gompa", "gonch", "gonef", "gongs", "gonia", "gonif", "gonks", "gonna", "gonof", "gonys", "gonzo", "gooby", "goods", "goofs", "googs", "gooks", "gooky", "goold", "gools", "gooly", "goons", "goony", "goops", "goopy", "goors", "goory", "goosy", "gopak", "gopik", "goral", "goras", "gored", "gores", "goris", "gorms", "gormy", "gorps", "gorse", "gorsy", "gosht", "gosse", "gotch", "goths", "gothy", "gotta", "gouch", "gouks", "goura", "gouts", "gouty", "gowan", "gowds", "gowfs", "gowks", "gowls", "gowns", "goxes", "goyim", "goyle", "graal", "grabs", "grads", "graff", "graip", "grama", "grame", "gramp", "grams", "grana", "grans", "grapy", "gravs", "grays", "grebe", "grebo", "grece", "greek", "grees", "grege", "grego", "grein", "grens", "grese", "greve", "grews", "greys", "grice", "gride", "grids", "griff", "grift", "grigs", "grike", "grins", "griot", "grips", "gript", "gripy", "grise", "grist", "grisy", "grith", "grits", "grize", "groat", "grody", "grogs", "groks", "groma", "grone", "groof", "grosz", "grots", "grouf", "grovy", "grows", "grrls", "grrrl", "grubs", "grued", "grues", "grufe", "grume", "grump", "grund", "gryce", "gryde", "gryke", "grype", "grypt", "guaco", "guana", "guano", "guans", "guars", "gucks", "gucky", "gudes", "guffs", "gugas", "guids", "guimp", "guiro", "gulag", "gular", "gulas", "gules", "gulet", "gulfs", "gulfy", "gulls", "gulph", "gulps", "gulpy", "gumma", "gummi", "gumps", "gundy", "gunge", "gungy", "gunks", "gunky", "gunny", "guqin", "gurdy", "gurge", "gurls", "gurly", "gurns", "gurry", "gursh", "gurus", "gushy", "gusla", "gusle", "gusli", "gussy", "gusts", "gutsy", "gutta", "gutty", "guyed", "guyle", "guyot", "guyse", "gwine", "gyals", "gyans", "gybed", "gybes", "gyeld", "gymps", "gynae", "gynie", "gynny", "gynos", "gyoza", "gypos", "gyppo", "gyppy", "gyral", "gyred", "gyres", "gyron", "gyros", "gyrus", "gytes", "gyved", "gyves", "haafs", "haars", "hable", "habus", "hacek", "hacks", "hadal", "haded", "hades", "hadji", "hadst", "haems", "haets", "haffs", "hafiz", "hafts", "haggs", "hahas", "haick", "haika", "haiks", "haiku", "hails", "haily", "hains", "haint", "hairs", "haith", "hajes", "hajis", "hajji", "hakam", "hakas", "hakea", "hakes", "hakim", "hakus", "halal", "haled", "haler", "hales", "halfa", "halfs", "halid", "hallo", "halls", "halma", "halms", "halon", "halos", "halse", "halts", "halva", "halwa", "hamal", "hamba", "hamed", "hames", "hammy", "hamza", "hanap", "hance", "hanch", "hands", "hangi", "hangs", "hanks", "hanky", "hansa", "hanse", "hants", "haole", "haoma", "hapax", "haply", "happi", "hapus", "haram", "hards", "hared", "hares", "harim", "harks", "harls", "harms", "harns", "haros", "harps", "harts", "hashy", "hasks", "hasps", "hasta", "hated", "hates", "hatha", "hauds", "haufs", "haugh", "hauld", "haulm", "hauls", "hault", "hauns", "hause", "haver", "haves", "hawed", "hawks", "hawms", "hawse", "hayed", "hayer", "hayey", "hayle", "hazan", "hazed", "hazer", "hazes", "heads", "heald", "heals", "heame", "heaps", "heapy", "heare", "hears", "heast", "heats", "heben", "hebes", "hecht", "hecks", "heder", "hedgy", "heeds", "heedy", "heels", "heeze", "hefte", "hefts", "heids", "heigh", "heils", "heirs", "hejab", "hejra", "heled", "heles", "helio", "hells", "helms", "helos", "helot", "helps", "helve", "hemal", "hemes", "hemic", "hemin", "hemps", "hempy", "hench", "hends", "henge", "henna", "henny", "henry", "hents", "hepar", "herbs", "herby", "herds", "heres", "herls", "herma", "herms", "herns", "heros", "herry", "herse", "hertz", "herye", "hesps", "hests", "hetes", "heths", "heuch", "heugh", "hevea", "hewed", "hewer", "hewgh", "hexad", "hexed", "hexer", "hexes", "hexyl", "heyed", "hiant", "hicks", "hided", "hider", "hides", "hiems", "highs", "hight", "hijab", "hijra", "hiked", "hiker", "hikes", "hikoi", "hilar", "hilch", "hillo", "hills", "hilts", "hilum", "hilus", "himbo", "hinau", "hinds", "hings", "hinky", "hinny", "hints", "hiois", "hiply", "hired", "hiree", "hirer", "hires", "hissy", "hists", "hithe", "hived", "hiver", "hives", "hizen", "hoaed", "hoagy", "hoars", "hoary", "hoast", "hobos", "hocks", "hocus", "hodad", "hodja", "hoers", "hogan", "hogen", "hoggs", "hoghs", "hohed", "hoick", "hoied", "hoiks", "hoing", "hoise", "hokas", "hoked", "hokes", "hokey", "hokis", "hokku", "hokum", "holds", "holed", "holes", "holey", "holks", "holla", "hollo", "holme", "holms", "holon", "holos", "holts", "homas", "homed", "homes", "homey", "homie", "homme", "homos", "honan", "honda", "honds", "honed", "honer", "hones", "hongi", "hongs", "honks", "honky", "hooch", "hoods", "hoody", "hooey", "hoofs", "hooka", "hooks", "hooky", "hooly", "hoons", "hoops", "hoord", "hoors", "hoosh", "hoots", "hooty", "hoove", "hopak", "hoped", "hoper", "hopes", "hoppy", "horah", "horal", "horas", "horis", "horks", "horme", "horns", "horst", "horsy", "hosed", "hosel", "hosen", "hoser", "hoses", "hosey", "hosta", "hosts", "hotch", "hoten", "hotty", "houff", "houfs", "hough", "houri", "hours", "houts", "hovea", "hoved", "hoven", "hoves", "howbe", "howes", "howff", "howfs", "howks", "howls", "howre", "howso", "hoxed", "hoxes", "hoyas", "hoyed", "hoyle", "hubby", "hucks", "hudna", "hudud", "huers", "huffs", "huffy", "huger", "huggy", "huhus", "huias", "hulas", "hules", "hulks", "hulky", "hullo", "hulls", "hully", "humas", "humfs", "humic", "humps", "humpy", "hunks", "hunts", "hurds", "hurls", "hurly", "hurra", "hurst", "hurts", "hushy", "husks", "husos", "hutia", "huzza", "huzzy", "hwyls", "hydra", "hyens", "hygge", "hying", "hykes", "hylas", "hyleg", "hyles", "hylic", "hymns", "hynde", "hyoid", "hyped", "hypes", "hypha", "hyphy", "hypos", "hyrax", "hyson", "hythe", "iambi", "iambs", "ibrik", "icers", "iched", "iches", "ichor", "icier", "icker", "ickle", "icons", "ictal", "ictic", "ictus", "idant", "ideas", "idees", "ident", "idled", "idles", "idola", "idols", "idyls", "iftar", "igapo", "igged", "iglus", "ihram", "ikans", "ikats", "ikons", "ileac", "ileal", "ileum", "ileus", "iliad", "ilial", "ilium", "iller", "illth", "imago", "imams", "imari", "imaum", "imbar", "imbed", "imide", "imido", "imids", "imine", "imino", "immew", "immit", "immix", "imped", "impis", "impot", "impro", "imshi", "imshy", "inapt", "inarm", "inbye", "incel", "incle", "incog", "incus", "incut", "indew", "india", "indie", "indol", "indow", "indri", "indue", "inerm", "infix", "infos", "infra", "ingan", "ingle", "inion", "inked", "inker", "inkle", "inned", "innit", "inorb", "inrun", "inset", "inspo", "intel", "intil", "intis", "intra", "inula", "inure", "inurn", "inust", "invar", "inwit", "iodic", "iodid", "iodin", "iotas", "ippon", "irade", "irids", "iring", "irked", "iroko", "irone", "irons", "isbas", "ishes", "isled", "isles", "isnae", "issei", "istle", "items", "ither", "ivied", "ivies", "ixias", "ixnay", "ixora", "ixtle", "izard", "izars", "izzat", "jaaps", "jabot", "jacal", "jacks", "jacky", "jaded", "jades", "jafas", "jaffa", "jagas", "jager", "jaggs", "jaggy", "jagir", "jagra", "jails", "jaker", "jakes", "jakey", "jalap", "jalop", "jambe", "jambo", "jambs", "jambu", "james", "jammy", "jamon", "janes", "janns", "janny", "janty", "japan", "japed", "japer", "japes", "jarks", "jarls", "jarps", "jarta", "jarul", "jasey", "jaspe", "jasps", "jatos", "jauks", "jaups", "javas", "javel", "jawan", "jawed", "jaxie", "jeans", "jeats", "jebel", "jedis", "jeels", "jeely", "jeeps", "jeers", "jeeze", "jefes", "jeffs", "jehad", "jehus", "jelab", "jello", "jells", "jembe", "jemmy", "jenny", "jeons", "jerid", "jerks", "jerry", "jesse", "jests", "jesus", "jetes", "jeton", "jeune", "jewed", "jewie", "jhala", "jiaos", "jibba", "jibbs", "jibed", "jiber", "jibes", "jiffs", "jiggy", "jigot", "jihad", "jills", "jilts", "jimmy", "jimpy", "jingo", "jinks", "jinne", "jinni", "jinns", "jirds", "jirga", "jirre", "jisms", "jived", "jiver", "jives", "jivey", "jnana", "jobed", "jobes", "jocko", "jocks", "jocky", "jocos", "jodel", "joeys", "johns", "joins", "joked", "jokes", "jokey", "jokol", "joled", "joles", "jolls", "jolts", "jolty", "jomon", "jomos", "jones", "jongs", "jonty", "jooks", "joram", "jorum", "jotas", "jotty", "jotun", "joual", "jougs", "jouks", "joule", "jours", "jowar", "jowed", "jowls", "jowly", "joyed", "jubas", "jubes", "jucos", "judas", "judgy", "judos", "jugal", "jugum", "jujus", "juked", "jukes", "jukus", "julep", "jumar", "jumby", "jumps", "junco", "junks", "junky", "jupes", "jupon", "jural", "jurat", "jurel", "jures", "justs", "jutes", "jutty", "juves", "juvie", "kaama", "kabab", "kabar", "kabob", "kacha", "kacks", "kadai", "kades", "kadis", "kafir", "kagos", "kagus", "kahal", "kaiak", "kaids", "kaies", "kaifs", "kaika", "kaiks", "kails", "kaims", "kaing", "kains", "kakas", "kakis", "kalam", "kales", "kalif", "kalis", "kalpa", "kamas", "kames", "kamik", "kamis", "kamme", "kanae", "kanas", "kandy", "kaneh", "kanes", "kanga", "kangs", "kanji", "kants", "kanzu", "kaons", "kapas", "kaphs", "kapok", "kapow", "kapus", "kaput", "karas", "karat", "karks", "karns", "karoo", "karos", "karri", "karst", "karsy", "karts", "karzy", "kasha", "kasme", "katal", "katas", "katis", "katti", "kaugh", "kauri", "kauru", "kaury", "kaval", "kavas", "kawas", "kawau", "kawed", "kayle", "kayos", "kazis", "kazoo", "kbars", "kebar", "kebob", "kecks", "kedge", "kedgy", "keech", "keefs", "keeks", "keels", "keema", "keeno", "keens", "keeps", "keets", "keeve", "kefir", "kehua", "keirs", "kelep", "kelim", "kells", "kelly", "kelps", "kelpy", "kelts", "kelty", "kembo", "kembs", "kemps", "kempt", "kempy", "kenaf", "kench", "kendo", "kenos", "kente", "kents", "kepis", "kerbs", "kerel", "kerfs", "kerky", "kerma", "kerne", "kerns", "keros", "kerry", "kerve", "kesar", "kests", "ketas", "ketch", "ketes", "ketol", "kevel", "kevil", "kexes", "keyed", "keyer", "khadi", "khafs", "khans", "khaph", "khats", "khaya", "khazi", "kheda", "kheth", "khets", "khoja", "khors", "khoum", "khuds", "kiaat", "kiack", "kiang", "kibbe", "kibbi", "kibei", "kibes", "kibla", "kicks", "kicky", "kiddo", "kiddy", "kidel", "kidge", "kiefs", "kiers", "kieve", "kievs", "kight", "kikes", "kikoi", "kiley", "kilim", "kills", "kilns", "kilos", "kilps", "kilts", "kilty", "kimbo", "kinas", "kinda", "kinds", "kindy", "kines", "kings", "kinin", "kinks", "kinos", "kiore", "kipes", "kippa", "kipps", "kirby", "kirks", "kirns", "kirri", "kisan", "kissy", "kists", "kited", "kiter", "kites", "kithe", "kiths", "kitul", "kivas", "kiwis", "klang", "klaps", "klett", "klick", "klieg", "kliks", "klong", "kloof", "kluge", "klutz", "knags", "knaps", "knarl", "knars", "knaur", "knawe", "knees", "knell", "knish", "knits", "knive", "knobs", "knops", "knosp", "knots", "knout", "knowe", "knows", "knubs", "knurl", "knurr", "knurs", "knuts", "koans", "koaps", "koban", "kobos", "koels", "koffs", "kofta", "kogal", "kohas", "kohen", "kohls", "koine", "kojis", "kokam", "kokas", "koker", "kokra", "kokum", "kolas", "kolos", "kombu", "konbu", "kondo", "konks", "kooks", "kooky", "koori", "kopek", "kophs", "kopje", "koppa", "korai", "koras", "korat", "kores", "korma", "koros", "korun", "korus", "koses", "kotch", "kotos", "kotow", "koura", "kraal", "krabs", "kraft", "krais", "krait", "krang", "krans", "kranz", "kraut", "krays", "kreep", "kreng", "krewe", "krona", "krone", "kroon", "krubi", "krunk", "ksars", "kubie", "kudos", "kudus", "kudzu", "kufis", "kugel", "kuias", "kukri", "kukus", "kulak", "kulan", "kulas", "kulfi", "kumis", "kumys", "kuris", "kurre", "kurta", "kurus", "kusso", "kutas", "kutch", "kutis", "kutus", "kuzus", "kvass", "kvell", "kwela", "kyack", "kyaks", "kyang", "kyars", "kyats", "kybos", "kydst", "kyles", "kylie", "kylin", "kylix", "kyloe", "kynde", "kynds", "kypes", "kyrie", "kytes", "kythe", "laari", "labda", "labia", "labis", "labra", "laced", "lacer", "laces", "lacet", "lacey", "lacks", "laddy", "laded", "lader", "lades", "laers", "laevo", "lagan", "lahal", "lahar", "laich", "laics", "laids", "laigh", "laika", "laiks", "laird", "lairs", "lairy", "laith", "laity", "laked", "laker", "lakes", "lakhs", "lakin", "laksa", "laldy", "lalls", "lamas", "lambs", "lamby", "lamed", "lamer", "lames", "lamia", "lammy", "lamps", "lanai", "lanas", "lanch", "lande", "lands", "lanes", "lanks", "lants", "lapin", "lapis", "lapje", "larch", "lards", "lardy", "laree", "lares", "largo", "laris", "larks", "larky", "larns", "larnt", "larum", "lased", "laser", "lases", "lassi", "lassu", "lassy", "lasts", "latah", "lated", "laten", "latex", "lathi", "laths", "lathy", "latke", "latus", "lauan", "lauch", "lauds", "laufs", "laund", "laura", "laval", "lavas", "laved", "laver", "laves", "lavra", "lavvy", "lawed", "lawer", "lawin", "lawks", "lawns", "lawny", "laxed", "laxer", "laxes", "laxly", "layed", "layin", "layup", "lazar", "lazed", "lazes", "lazos", "lazzi", "lazzo", "leads", "leady", "leafs", "leaks", "leams", "leans", "leany", "leaps", "leare", "lears", "leary", "leats", "leavy", "leaze", "leben", "leccy", "ledes", "ledgy", "ledum", "leear", "leeks", "leeps", "leers", "leese", "leets", "leeze", "lefte", "lefts", "leger", "leges", "legge", "leggo", "legit", "lehrs", "lehua", "leirs", "leish", "leman", "lemed", "lemel", "lemes", "lemma", "lemme", "lends", "lenes", "lengs", "lenis", "lenos", "lense", "lenti", "lento", "leone", "lepid", "lepra", "lepta", "lered", "leres", "lerps", "lesbo", "leses", "lests", "letch", "lethe", "letup", "leuch", "leuco", "leuds", "leugh", "levas", "levee", "leves", "levin", "levis", "lewis", "lexes", "lexis", "lezes", "lezza", "lezzy", "liana", "liane", "liang", "liard", "liars", "liart", "liber", "libra", "libri", "lichi", "licht", "licit", "licks", "lidar", "lidos", "liefs", "liens", "liers", "lieus", "lieve", "lifer", "lifes", "lifts", "ligan", "liger", "ligge", "ligne", "liked", "liker", "likes", "likin", "lills", "lilos", "lilts", "liman", "limas", "limax", "limba", "limbi", "limbs", "limby", "limed", "limen", "limes", "limey", "limma", "limns", "limos", "limpa", "limps", "linac", "linch", "linds", "lindy", "lined", "lines", "liney", "linga", "lings", "lingy", "linin", "links", "linky", "linns", "linny", "linos", "lints", "linty", "linum", "linux", "lions", "lipas", "lipes", "lipin", "lipos", "lippy", "liras", "lirks", "lirot", "lisks", "lisle", "lisps", "lists", "litai", "litas", "lited", "liter", "lites", "litho", "liths", "litre", "lived", "liven", "lives", "livor", "livre", "llano", "loach", "loads", "loafs", "loams", "loans", "loast", "loave", "lobar", "lobed", "lobes", "lobos", "lobus", "loche", "lochs", "locie", "locis", "locks", "locos", "locum", "loden", "lodes", "loess", "lofts", "logan", "loges", "loggy", "logia", "logie", "logoi", "logon", "logos", "lohan", "loids", "loins", "loipe", "loirs", "lokes", "lolls", "lolly", "lolog", "lomas", "lomed", "lomes", "loner", "longa", "longe", "longs", "looby", "looed", "looey", "loofa", "loofs", "looie", "looks", "looky", "looms", "loons", "loony", "loops", "loord", "loots", "loped", "loper", "lopes", "loppy", "loral", "loran", "lords", "lordy", "lorel", "lores", "loric", "loris", "losed", "losel", "losen", "loses", "lossy", "lotah", "lotas", "lotes", "lotic", "lotos", "lotsa", "lotta", "lotte", "lotto", "lotus", "loued", "lough", "louie", "louis", "louma", "lound", "louns", "loupe", "loups", "loure", "lours", "loury", "louts", "lovat", "loved", "loves", "lovey", "lovie", "lowan", "lowed", "lowes", "lownd", "lowne", "lowns", "lowps", "lowry", "lowse", "lowts", "loxed", "loxes", "lozen", "luach", "luaus", "lubed", "lubes", "lubra", "luces", "lucks", "lucre", "ludes", "ludic", "ludos", "luffa", "luffs", "luged", "luger", "luges", "lulls", "lulus", "lumas", "lumbi", "lumme", "lummy", "lumps", "lunas", "lunes", "lunet", "lungi", "lungs", "lunks", "lunts", "lupin", "lured", "lurer", "lures", "lurex", "lurgi", "lurgy", "lurks", "lurry", "lurve", "luser", "lushy", "lusks", "lusts", "lusus", "lutea", "luted", "luter", "lutes", "luvvy", "luxed", "luxer", "luxes", "lweis", "lyams", "lyard", "lyart", "lyase", "lycea", "lycee", "lycra", "lymes", "lynes", "lyres", "lysed", "lyses", "lysin", "lysis", "lysol", "lyssa", "lyted", "lytes", "lythe", "lytic", "lytta", "maaed", "maare", "maars", "mabes", "macas", "maced", "macer", "maces", "mache", "machi", "machs", "macks", "macle", "macon", "madge", "madid", "madre", "maerl", "mafic", "mages", "maggs", "magot", "magus", "mahoe", "mahua", "mahwa", "maids", "maiko", "maiks", "maile", "maill", "mails", "maims", "mains", "maire", "mairs", "maise", "maist", "makar", "makes", "makis", "makos", "malam", "malar", "malas", "malax", "males", "malic", "malik", "malis", "malls", "malms", "malmy", "malts", "malty", "malus", "malva", "malwa", "mamas", "mamba", "mamee", "mamey", "mamie", "manas", "manat", "mandi", "maneb", "maned", "maneh", "manes", "manet", "mangs", "manis", "manky", "manna", "manos", "manse", "manta", "manto", "manty", "manul", "manus", "mapau", "maqui", "marae", "marah", "maras", "marcs", "mardy", "mares", "marge", "margs", "maria", "marid", "marka", "marks", "marle", "marls", "marly", "marms", "maron", "maror", "marra", "marri", "marse", "marts", "marvy", "masas", "mased", "maser", "mases", "mashy", "masks", "massa", "massy", "masts", "masty", "masus", "matai", "mated", "mater", "mates", "maths", "matin", "matlo", "matte", "matts", "matza", "matzo", "mauby", "mauds", "mauls", "maund", "mauri", "mausy", "mauts", "mauzy", "maven", "mavie", "mavin", "mavis", "mawed", "mawks", "mawky", "mawns", "mawrs", "maxed", "maxes", "maxis", "mayan", "mayas", "mayed", "mayos", "mayst", "mazed", "mazer", "mazes", "mazey", "mazut", "mbira", "meads", "meals", "meane", "means", "meany", "meare", "mease", "meath", "meats", "mebos", "mechs", "mecks", "medii", "medle", "meeds", "meers", "meets", "meffs", "meins", "meint", "meiny", "meith", "mekka", "melas", "melba", "melds", "melic", "melik", "mells", "melts", "melty", "memes", "memos", "menad", "mends", "mened", "menes", "menge", "mengs", "mensa", "mense", "mensh", "menta", "mento", "menus", "meous", "meows", "merch", "mercs", "merde", "mered", "merel", "merer", "meres", "meril", "meris", "merks", "merle", "merls", "merse", "mesal", "mesas", "mesel", "meses", "meshy", "mesic", "mesne", "meson", "messy", "mesto", "meted", "metes", "metho", "meths", "metic", "metif", "metis", "metol", "metre", "meuse", "meved", "meves", "mewed", "mewls", "meynt", "mezes", "mezze", "mezzo", "mhorr", "miaou", "miaow", "miasm", "miaul", "micas", "miche", "micht", "micks", "micky", "micos", "micra", "middy", "midgy", "midis", "miens", "mieve", "miffs", "miffy", "mifty", "miggs", "mihas", "mihis", "miked", "mikes", "mikra", "mikva", "milch", "milds", "miler", "miles", "milfs", "milia", "milko", "milks", "mille", "mills", "milor", "milos", "milpa", "milts", "milty", "miltz", "mimed", "mimeo", "mimer", "mimes", "mimsy", "minae", "minar", "minas", "mincy", "minds", "mined", "mines", "minge", "mings", "mingy", "minis", "minke", "minks", "minny", "minos", "mints", "mired", "mires", "mirex", "mirid", "mirin", "mirks", "mirky", "mirly", "miros", "mirvs", "mirza", "misch", "misdo", "mises", "misgo", "misos", "missa", "mists", "misty", "mitch", "miter", "mites", "mitis", "mitre", "mitts", "mixed", "mixen", "mixer", "mixes", "mixte", "mixup", "mizen", "mizzy", "mneme", "moans", "moats", "mobby", "mobes", "mobey", "mobie", "moble", "mochi", "mochs", "mochy", "mocks", "moder", "modes", "modge", "modii", "modus", "moers", "mofos", "moggy", "mohel", "mohos", "mohrs", "mohua", "mohur", "moile", "moils", "moira", "moire", "moits", "mojos", "mokes", "mokis", "mokos", "molal", "molas", "molds", "moled", "moles", "molla", "molls", "molly", "molto", "molts", "molys", "momes", "momma", "mommy", "momus", "monad", "monal", "monas", "monde", "mondo", "moner", "mongo", "mongs", "monic", "monie", "monks", "monos", "monte", "monty", "moobs", "mooch", "moods", "mooed", "mooks", "moola", "mooli", "mools", "mooly", "moong", "moons", "moony", "moops", "moors", "moory", "moots", "moove", "moped", "moper", "mopes", "mopey", "moppy", "mopsy", "mopus", "morae", "moras", "morat", "moray", "morel", "mores", "moria", "morne", "morns", "morra", "morro", "morse", "morts", "mosed", "moses", "mosey", "mosks", "mosso", "moste", "mosts", "moted", "moten", "motes", "motet", "motey", "moths", "mothy", "motis", "motte", "motts", "motty", "motus", "motza", "mouch", "moues", "mould", "mouls", "moups", "moust", "mousy", "moved", "moves", "mowas", "mowed", "mowra", "moxas", "moxie", "moyas", "moyle", "moyls", "mozed", "mozes", "mozos", "mpret", "mucho", "mucic", "mucid", "mucin", "mucks", "mucor", "mucro", "mudge", "mudir", "mudra", "muffs", "mufti", "mugga", "muggs", "muggy", "muhly", "muids", "muils", "muirs", "muist", "mujik", "mulct", "muled", "mules", "muley", "mulga", "mulie", "mulla", "mulls", "mulse", "mulsh", "mumms", "mumps", "mumsy", "mumus", "munga", "munge", "mungo", "mungs", "munis", "munts", "muntu", "muons", "muras", "mured", "mures", "murex", "murid", "murks", "murls", "murly", "murra", "murre", "murri", "murrs", "murry", "murti", "murva", "musar", "musca", "mused", "muser", "muses", "muset", "musha", "musit", "musks", "musos", "musse", "mussy", "musth", "musts", "mutch", "muted", "muter", "mutes", "mutha", "mutis", "muton", "mutts", "muxed", "muxes", "muzak", "muzzy", "mvule", "myall", "mylar", "mynah", "mynas", "myoid", "myoma", "myope", "myops", "myopy", "mysid", "mythi", "myths", "mythy", "myxos", "mzees", "naams", "naans", "nabes", "nabis", "nabks", "nabla", "nabob", "nache", "nacho", "nacre", "nadas", "naeve", "naevi", "naffs", "nagas", "naggy", "nagor", "nahal", "naiad", "naifs", "naiks", "nails", "naira", "nairu", "naked", "naker", "nakfa", "nalas", "naled", "nalla", "named", "namer", "names", "namma", "namus", "nanas", "nance", "nancy", "nandu", "nanna", "nanos", "nanua", "napas", "naped", "napes", "napoo", "nappa", "nappe", "nappy", "naras", "narco", "narcs", "nards", "nares", "naric", "naris", "narks", "narky", "narre", "nashi", "natch", "nates", "natis", "natty", "nauch", "naunt", "navar", "naves", "navew", "navvy", "nawab", "nazes", "nazir", "nazis", "nduja", "neafe", "neals", "neaps", "nears", "neath", "neats", "nebek", "nebel", "necks", "neddy", "needs", "neeld", "neele", "neemb", "neems", "neeps", "neese", "neeze", "negro", "negus", "neifs", "neist", "neive", "nelis", "nelly", "nemas", "nemns", "nempt", "nenes", "neons", "neper", "nepit", "neral", "nerds", "nerka", "nerks", "nerol", "nerts", "nertz", "nervy", "nests", "netes", "netop", "netts", "netty", "neuks", "neume", "neums", "nevel", "neves", "nevus", "newbs", "newed", "newel", "newie", "newsy", "newts", "nexts", "nexus", "ngaio", "ngana", "ngati", "ngoma", "ngwee", "nicad", "nicht", "nicks", "nicol", "nidal", "nided", "nides", "nidor", "nidus", "niefs", "nieve", "nifes", "niffs", "niffy", "nifty", "niger", "nighs", "nihil", "nikab", "nikah", "nikau", "nills", "nimbi", "nimbs", "nimps", "niner", "nines", "ninon", "nipas", "nippy", "niqab", "nirls", "nirly", "nisei", "nisse", "nisus", "niter", "nites", "nitid", "niton", "nitre", "nitro", "nitry", "nitty", "nival", "nixed", "nixer", "nixes", "nixie", "nizam", "nkosi", "noahs", "nobby", "nocks", "nodal", "noddy", "nodes", "nodus", "noels", "noggs", "nohow", "noils", "noily", "noint", "noirs", "noles", "nolls", "nolos", "nomas", "nomen", "nomes", "nomic", "nomoi", "nomos", "nonas", "nonce", "nones", "nonet", "nongs", "nonis", "nonny", "nonyl", "noobs", "nooit", "nooks", "nooky", "noons", "noops", "nopal", "noria", "noris", "norks", "norma", "norms", "nosed", "noser", "noses", "notal", "noted", "noter", "notes", "notum", "nould", "noule", "nouls", "nouns", "nouny", "noups", "novae", "novas", "novum", "noway", "nowed", "nowls", "nowts", "nowty", "noxal", "noxes", "noyau", "noyed", "noyes", "nubby", "nubia", "nucha", "nuddy", "nuder", "nudes", "nudie", "nudzh", "nuffs", "nugae", "nuked", "nukes", "nulla", "nulls", "numbs", "numen", "nummy", "nunny", "nurds", "nurdy", "nurls", "nurrs", "nutso", "nutsy", "nyaff", "nyala", "nying", "nyssa", "oaked", "oaker", "oakum", "oared", "oases", "oasis", "oasts", "oaten", "oater", "oaths", "oaves", "obang", "obeah", "obeli", "obeys", "obias", "obied", "obiit", "obits", "objet", "oboes", "obole", "oboli", "obols", "occam", "ocher", "oches", "ochre", "ochry", "ocker", "ocrea", "octad", "octan", "octas", "octyl", "oculi", "odahs", "odals", "odeon", "odeum", "odism", "odist", "odium", "odors", "odour", "odyle", "odyls", "ofays", "offed", "offie", "oflag", "ofter", "ogams", "ogeed", "ogees", "oggin", "ogham", "ogive", "ogled", "ogler", "ogles", "ogmic", "ogres", "ohias", "ohing", "ohmic", "ohone", "oidia", "oiled", "oiler", "oinks", "oints", "ojime", "okapi", "okays", "okehs", "okras", "oktas", "oldie", "oleic", "olein", "olent", "oleos", "oleum", "olios", "ollas", "ollav", "oller", "ollie", "ology", "olpae", "olpes", "omasa", "omber", "ombus", "omens", "omers", "omits", "omlah", "omovs", "omrah", "oncer", "onces", "oncet", "oncus", "onely", "oners", "onery", "onium", "onkus", "onlay", "onned", "ontic", "oobit", "oohed", "oomph", "oonts", "ooped", "oorie", "ooses", "ootid", "oozed", "oozes", "opahs", "opals", "opens", "opepe", "oping", "oppos", "opsin", "opted", "opter", "orach", "oracy", "orals", "orang", "orant", "orate", "orbed", "orcas", "orcin", "ordos", "oread", "orfes", "orgia", "orgic", "orgue", "oribi", "oriel", "orixa", "orles", "orlon", "orlop", "ormer", "ornis", "orpin", "orris", "ortho", "orval", "orzos", "oscar", "oshac", "osier", "osmic", "osmol", "ossia", "ostia", "otaku", "otary", "ottar", "ottos", "oubit", "oucht", "ouens", "ouija", "oulks", "oumas", "oundy", "oupas", "ouped", "ouphe", "ouphs", "ourie", "ousel", "ousts", "outby", "outed", "outre", "outro", "outta", "ouzel", "ouzos", "ovals", "ovels", "ovens", "overs", "ovist", "ovoli", "ovolo", "ovule", "owche", "owies", "owled", "owler", "owlet", "owned", "owres", "owrie", "owsen", "oxbow", "oxers", "oxeye", "oxids", "oxies", "oxime", "oxims", "oxlip", "oxter", "oyers", "ozeki", "ozzie", "paals", "paans", "pacas", "paced", "pacer", "paces", "pacey", "pacha", "packs", "pacos", "pacta", "pacts", "padis", "padle", "padma", "padre", "padri", "paean", "paedo", "paeon", "paged", "pager", "pages", "pagle", "pagod", "pagri", "paiks", "pails", "pains", "paire", "pairs", "paisa", "paise", "pakka", "palas", "palay", "palea", "paled", "pales", "palet", "palis", "palki", "palla", "palls", "pally", "palms", "palmy", "palpi", "palps", "palsa", "pampa", "panax", "pance", "panda", "pands", "pandy", "paned", "panes", "panga", "pangs", "panim", "panko", "panne", "panni", "panto", "pants", "panty", "paoli", "paolo", "papas", "papaw", "papes", "pappi", "pappy", "parae", "paras", "parch", "pardi", "pards", "pardy", "pared", "paren", "pareo", "pares", "pareu", "parev", "parge", "pargo", "paris", "parki", "parks", "parky", "parle", "parly", "parma", "parol", "parps", "parra", "parrs", "parti", "parts", "parve", "parvo", "paseo", "pases", "pasha", "pashm", "paska", "paspy", "passe", "pasts", "pated", "paten", "pater", "pates", "paths", "patin", "patka", "patly", "patte", "patus", "pauas", "pauls", "pavan", "paved", "paven", "paver", "paves", "pavid", "pavin", "pavis", "pawas", "pawaw", "pawed", "pawer", "pawks", "pawky", "pawls", "pawns", "paxes", "payed", "payor", "paysd", "peage", "peags", "peaks", "peaky", "peals", "peans", "peare", "pears", "peart", "pease", "peats", "peaty", "peavy", "peaze", "pebas", "pechs", "pecke", "pecks", "pecky", "pedes", "pedis", "pedro", "peece", "peeks", "peels", "peens", "peeoy", "peepe", "peeps", "peers", "peery", "peeve", "peggy", "peghs", "peins", "peise", "peize", "pekan", "pekes", "pekin", "pekoe", "pelas", "pelau", "peles", "pelfs", "pells", "pelma", "pelon", "pelta", "pelts", "pends", "pendu", "pened", "penes", "pengo", "penie", "penis", "penks", "penna", "penni", "pents", "peons", "peony", "pepla", "pepos", "peppy", "pepsi", "perai", "perce", "percs", "perdu", "perdy", "perea", "peres", "peris", "perks", "perms", "perns", "perog", "perps", "perry", "perse", "perst", "perts", "perve", "pervo", "pervs", "pervy", "pesos", "pests", "pesty", "petar", "peter", "petit", "petre", "petri", "petti", "petto", "pewee", "pewit", "peyse", "phage", "phang", "phare", "pharm", "pheer", "phene", "pheon", "phese", "phial", "phish", "phizz", "phlox", "phoca", "phono", "phons", "phots", "phpht", "phuts", "phyla", "phyle", "piani", "pians", "pibal", "pical", "picas", "piccy", "picks", "picot", "picra", "picul", "piend", "piers", "piert", "pieta", "piets", "piezo", "pight", "pigmy", "piing", "pikas", "pikau", "piked", "piker", "pikes", "pikey", "pikis", "pikul", "pilae", "pilaf", "pilao", "pilar", "pilau", "pilaw", "pilch", "pilea", "piled", "pilei", "piler", "piles", "pilis", "pills", "pilow", "pilum", "pilus", "pimas", "pimps", "pinas", "pined", "pines", "pingo", "pings", "pinko", "pinks", "pinna", "pinny", "pinon", "pinot", "pinta", "pints", "pinup", "pions", "piony", "pious", "pioye", "pioys", "pipal", "pipas", "piped", "pipes", "pipet", "pipis", "pipit", "pippy", "pipul", "pirai", "pirls", "pirns", "pirog", "pisco", "pises", "pisky", "pisos", "pissy", "piste", "pitas", "piths", "piton", "pitot", "pitta", "piums", "pixes", "pized", "pizes", "plaas", "plack", "plage", "plans", "plaps", "plash", "plasm", "plast", "plats", "platt", "platy", "playa", "plays", "pleas", "plebe", "plebs", "plena", "pleon", "plesh", "plews", "plica", "plies", "plims", "pling", "plink", "ploat", "plods", "plong", "plonk", "plook", "plops", "plots", "plotz", "plouk", "plows", "ploye", "ploys", "plues", "pluff", "plugs", "plums", "plumy", "pluot", "pluto", "plyer", "poach", "poaka", "poake", "poboy", "pocks", "pocky", "podal", "poddy", "podex", "podge", "podgy", "podia", "poems", "poeps", "poets", "pogey", "pogge", "pogos", "pohed", "poilu", "poind", "pokal", "poked", "pokes", "pokey", "pokie", "poled", "poler", "poles", "poley", "polio", "polis", "polje", "polks", "polls", "polly", "polos", "polts", "polys", "pombe", "pomes", "pommy", "pomos", "pomps", "ponce", "poncy", "ponds", "pones", "poney", "ponga", "pongo", "pongs", "pongy", "ponks", "ponts", "ponty", "ponzu", "poods", "pooed", "poofs", "poofy", "poohs", "pooja", "pooka", "pooks", "pools", "poons", "poops", "poopy", "poori", "poort", "poots", "poove", "poovy", "popes", "poppa", "popsy", "porae", "poral", "pored", "porer", "pores", "porge", "porgy", "porin", "porks", "porky", "porno", "porns", "porny", "porta", "ports", "porty", "posed", "poses", "posey", "posho", "posts", "potae", "potch", "poted", "potes", "potin", "potoo", "potsy", "potto", "potts", "potty", "pouff", "poufs", "pouke", "pouks", "poule", "poulp", "poult", "poupe", "poupt", "pours", "pouts", "powan", "powin", "pownd", "powns", "powny", "powre", "poxed", "poxes", "poynt", "poyou", "poyse", "pozzy", "praam", "prads", "prahu", "prams", "prana", "prang", "praos", "prase", "prate", "prats", "pratt", "praty", "praus", "prays", "predy", "preed", "prees", "preif", "prems", "premy", "prent", "preon", "preop", "preps", "presa", "prese", "prest", "preve", "prexy", "preys", "prial", "pricy", "prief", "prier", "pries", "prigs", "prill", "prima", "primi", "primp", "prims", "primy", "prink", "prion", "prise", "priss", "proas", "probs", "prods", "proem", "profs", "progs", "proin", "proke", "prole", "proll", "promo", "proms", "pronk", "props", "prore", "proso", "pross", "prost", "prosy", "proto", "proul", "prows", "proyn", "prunt", "pruta", "pryer", "pryse", "pseud", "pshaw", "psion", "psoae", "psoai", "psoas", "psora", "psych", "psyop", "pubco", "pubes", "pubis", "pucan", "pucer", "puces", "pucka", "pucks", "puddy", "pudge", "pudic", "pudor", "pudsy", "pudus", "puers", "puffa", "puffs", "puggy", "pugil", "puhas", "pujah", "pujas", "pukas", "puked", "puker", "pukes", "pukey", "pukka", "pukus", "pulao", "pulas", "puled", "puler", "pules", "pulik", "pulis", "pulka", "pulks", "pulli", "pulls", "pully", "pulmo", "pulps", "pulus", "pumas", "pumie", "pumps", "punas", "punce", "punga", "pungs", "punji", "punka", "punks", "punky", "punny", "punto", "punts", "punty", "pupae", "pupas", "pupus", "purda", "pured", "pures", "purin", "puris", "purls", "purpy", "purrs", "pursy", "purty", "puses", "pusle", "pussy", "putid", "puton", "putti", "putto", "putts", "puzel", "pwned", "pyats", "pyets", "pygal", "pyins", "pylon", "pyned", "pynes", "pyoid", "pyots", "pyral", "pyran", "pyres", "pyrex", "pyric", "pyros", "pyxed", "pyxes", "pyxie", "pyxis", "pzazz", "qadis", "qaids", "qajaq", "qanat", "qapik", "qibla", "qophs", "qorma", "quads", "quaff", "quags", "quair", "quais", "quaky", "quale", "quant", "quare", "quass", "quate", "quats", "quayd", "quays", "qubit", "quean", "queme", "quena", "quern", "queyn", "queys", "quich", "quids", "quiff", "quims", "quina", "quine", "quino", "quins", "quint", "quipo", "quips", "quipu", "quire", "quirt", "quist", "quits", "quoad", "quods", "quoif", "quoin", "quoit", "quoll", "quonk", "quops", "qursh", "quyte", "rabat", "rabic", "rabis", "raced", "races", "rache", "racks", "racon", "radge", "radix", "radon", "raffs", "rafts", "ragas", "ragde", "raged", "ragee", "rager", "rages", "ragga", "raggs", "raggy", "ragis", "ragus", "rahed", "rahui", "raias", "raids", "raiks", "raile", "rails", "raine", "rains", "raird", "raita", "raits", "rajas", "rajes", "raked", "rakee", "raker", "rakes", "rakia", "rakis", "rakus", "rales", "ramal", "ramee", "ramet", "ramie", "ramin", "ramis", "rammy", "ramps", "ramus", "ranas", "rance", "rands", "ranee", "ranga", "rangi", "rangs", "rangy", "ranid", "ranis", "ranke", "ranks", "rants", "raped", "raper", "rapes", "raphe", "rappe", "rared", "raree", "rares", "rarks", "rased", "raser", "rases", "rasps", "rasse", "rasta", "ratal", "ratan", "ratas", "ratch", "rated", "ratel", "rater", "rates", "ratha", "rathe", "raths", "ratoo", "ratos", "ratus", "rauns", "raupo", "raved", "ravel", "raver", "raves", "ravey", "ravin", "rawer", "rawin", "rawly", "rawns", "raxed", "raxes", "rayah", "rayas", "rayed", "rayle", "rayne", "razed", "razee", "razer", "razes", "razoo", "readd", "reads", "reais", "reaks", "realo", "reals", "reame", "reams", "reamy", "reans", "reaps", "rears", "reast", "reata", "reate", "reave", "rebbe", "rebec", "rebid", "rebit", "rebop", "rebuy", "recal", "recce", "recco", "reccy", "recit", "recks", "recon", "recta", "recti", "recto", "redan", "redds", "reddy", "reded", "redes", "redia", "redid", "redip", "redly", "redon", "redos", "redox", "redry", "redub", "redux", "redye", "reech", "reede", "reeds", "reefs", "reefy", "reeks", "reeky", "reels", "reens", "reest", "reeve", "refed", "refel", "reffo", "refis", "refix", "refly", "refry", "regar", "reges", "reggo", "regie", "regma", "regna", "regos", "regur", "rehem", "reifs", "reify", "reiki", "reiks", "reink", "reins", "reird", "reist", "reive", "rejig", "rejon", "reked", "rekes", "rekey", "relet", "relie", "relit", "rello", "reman", "remap", "remen", "remet", "remex", "remix", "renay", "rends", "reney", "renga", "renig", "renin", "renne", "renos", "rente", "rents", "reoil", "reorg", "repeg", "repin", "repla", "repos", "repot", "repps", "repro", "reran", "rerig", "resat", "resaw", "resay", "resee", "reses", "resew", "resid", "resit", "resod", "resow", "resto", "rests", "resty", "resus", "retag", "retax", "retem", "retia", "retie", "retox", "revet", "revie", "rewan", "rewax", "rewed", "rewet", "rewin", "rewon", "rewth", "rexes", "rezes", "rheas", "rheme", "rheum", "rhies", "rhime", "rhine", "rhody", "rhomb", "rhone", "rhumb", "rhyne", "rhyta", "riads", "rials", "riant", "riata", "ribas", "ribby", "ribes", "riced", "ricer", "rices", "ricey", "richt", "ricin", "ricks", "rides", "ridgy", "ridic", "riels", "riems", "rieve", "rifer", "riffs", "rifte", "rifts", "rifty", "riggs", "rigol", "riled", "riles", "riley", "rille", "rills", "rimae", "rimed", "rimer", "rimes", "rimus", "rinds", "rindy", "rines", "rings", "rinks", "rioja", "riots", "riped", "ripes", "ripps", "rises", "rishi", "risks", "risps", "risus", "rites", "ritts", "ritzy", "rivas", "rived", "rivel", "riven", "rives", "riyal", "rizas", "roads", "roams", "roans", "roars", "roary", "roate", "robed", "robes", "roble", "rocks", "roded", "rodes", "roguy", "rohes", "roids", "roils", "roily", "roins", "roist", "rojak", "rojis", "roked", "roker", "rokes", "rolag", "roles", "rolfs", "rolls", "romal", "roman", "romeo", "romps", "ronde", "rondo", "roneo", "rones", "ronin", "ronne", "ronte", "ronts", "roods", "roofs", "roofy", "rooks", "rooky", "rooms", "roons", "roops", "roopy", "roosa", "roose", "roots", "rooty", "roped", "roper", "ropes", "ropey", "roque", "roral", "rores", "roric", "rorid", "rorie", "rorts", "rorty", "rosed", "roses", "roset", "roshi", "rosin", "rosit", "rosti", "rosts", "rotal", "rotan", "rotas", "rotch", "roted", "rotes", "rotis", "rotls", "roton", "rotos", "rotte", "rouen", "roues", "roule", "rouls", "roums", "roups", "roupy", "roust", "routh", "routs", "roved", "roven", "roves", "rowan", "rowed", "rowel", "rowen", "rowie", "rowme", "rownd", "rowth", "rowts", "royne", "royst", "rozet", "rozit", "ruana", "rubai", "rubby", "rubel", "rubes", "rubin", "ruble", "rubli", "rubus", "ruche", "rucks", "rudas", "rudds", "rudes", "rudie", "rudis", "rueda", "ruers", "ruffe", "ruffs", "rugae", "rugal", "ruggy", "ruing", "ruins", "rukhs", "ruled", "rules", "rumal", "rumbo", "rumen", "rumes", "rumly", "rummy", "rumpo", "rumps", "rumpy", "runch", "runds", "runed", "runes", "rungs", "runic", "runny", "runts", "runty", "rupia", "rurps", "rurus", "rusas", "ruses", "rushy", "rusks", "rusma", "russe", "rusts", "ruths", "rutin", "rutty", "ryals", "rybat", "ryked", "rykes", "rymme", "rynds", "ryots", "ryper", "saags", "sabal", "sabed", "saber", "sabes", "sabha", "sabin", "sabir", "sable", "sabot", "sabra", "sabre", "sacks", "sacra", "saddo", "sades", "sadhe", "sadhu", "sadis", "sados", "sadza", "safed", "safes", "sagas", "sager", "sages", "saggy", "sagos", "sagum", "saheb", "sahib", "saice", "saick", "saics", "saids", "saiga", "sails", "saims", "saine", "sains", "sairs", "saist", "saith", "sajou", "sakai", "saker", "sakes", "sakia", "sakis", "sakti", "salal", "salat", "salep", "sales", "salet", "salic", "salix", "salle", "salmi", "salol", "salop", "salpa", "salps", "salse", "salto", "salts", "salue", "salut", "saman", "samas", "samba", "sambo", "samek", "samel", "samen", "sames", "samey", "samfu", "sammy", "sampi", "samps", "sands", "saned", "sanes", "sanga", "sangh", "sango", "sangs", "sanko", "sansa", "santo", "sants", "saola", "sapan", "sapid", "sapor", "saran", "sards", "sared", "saree", "sarge", "sargo", "sarin", "saris", "sarks", "sarky", "sarod", "saros", "sarus", "saser", "sasin", "sasse", "satai", "satay", "sated", "satem", "sates", "satis", "sauba", "sauch", "saugh", "sauls", "sault", "saunt", "saury", "sauts", "saved", "saver", "saves", "savey", "savin", "sawah", "sawed", "sawer", "saxes", "sayed", "sayer", "sayid", "sayne", "sayon", "sayst", "sazes", "scabs", "scads", "scaff", "scags", "scail", "scala", "scall", "scams", "scand", "scans", "scapa", "scape", "scapi", "scarp", "scars", "scart", "scath", "scats", "scatt", "scaud", "scaup", "scaur", "scaws", "sceat", "scena", "scend", "schav", "schmo", "schul", "schwa", "sclim", "scody", "scogs", "scoog", "scoot", "scopa", "scops", "scots", "scoug", "scoup", "scowp", "scows", "scrab", "scrae", "scrag", "scran", "scrat", "scraw", "scray", "scrim", "scrip", "scrob", "scrod", "scrog", "scrow", "scudi", "scudo", "scuds", "scuff", "scuft", "scugs", "sculk", "scull", "sculp", "sculs", "scums", "scups", "scurf", "scurs", "scuse", "scuta", "scute", "scuts", "scuzz", "scyes", "sdayn", "sdein", "seals", "seame", "seams", "seamy", "seans", "seare", "sears", "sease", "seats", "seaze", "sebum", "secco", "sechs", "sects", "seder", "sedes", "sedge", "sedgy", "sedum", "seeds", "seeks", "seeld", "seels", "seely", "seems", "seeps", "seepy", "seers", "sefer", "segar", "segni", "segno", "segol", "segos", "sehri", "seifs", "seils", "seine", "seirs", "seise", "seism", "seity", "seiza", "sekos", "sekts", "selah", "seles", "selfs", "sella", "selle", "sells", "selva", "semee", "semes", "semie", "semis", "senas", "sends", "senes", "sengi", "senna", "senor", "sensa", "sensi", "sente", "senti", "sents", "senvy", "senza", "sepad", "sepal", "sepic", "sepoy", "septa", "septs", "serac", "serai", "seral", "sered", "serer", "seres", "serfs", "serge", "seric", "serin", "serks", "seron", "serow", "serra", "serre", "serrs", "serry", "servo", "sesey", "sessa", "setae", "setal", "seton", "setts", "sewan", "sewar", "sewed", "sewel", "sewen", "sewin", "sexed", "sexer", "sexes", "sexto", "sexts", "seyen", "shads", "shags", "shahs", "shako", "shakt", "shalm", "shaly", "shama", "shams", "shand", "shans", "shaps", "sharn", "shash", "shaul", "shawm", "shawn", "shaws", "shaya", "shays", "shchi", "sheaf", "sheal", "sheas", "sheds", "sheel", "shend", "shent", "sheol", "sherd", "shere", "shero", "shets", "sheva", "shewn", "shews", "shiai", "shiel", "shier", "shies", "shill", "shily", "shims", "shins", "ships", "shirr", "shirs", "shish", "shiso", "shist", "shite", "shits", "shiur", "shiva", "shive", "shivs", "shlep", "shlub", "shmek", "shmoe", "shoat", "shoed", "shoer", "shoes", "shogi", "shogs", "shoji", "shojo", "shola", "shool", "shoon", "shoos", "shope", "shops", "shorl", "shote", "shots", "shott", "showd", "shows", "shoyu", "shred", "shris", "shrow", "shtik", "shtum", "shtup", "shule", "shuln", "shuls", "shuns", "shura", "shute", "shuts", "shwas", "shyer", "sials", "sibbs", "sibyl", "sices", "sicht", "sicko", "sicks", "sicky", "sidas", "sided", "sider", "sides", "sidha", "sidhe", "sidle", "sield", "siens", "sient", "sieth", "sieur", "sifts", "sighs", "sigil", "sigla", "signa", "signs", "sijos", "sikas", "siker", "sikes", "silds", "siled", "silen", "siler", "siles", "silex", "silks", "sills", "silos", "silts", "silty", "silva", "simar", "simas", "simba", "simis", "simps", "simul", "sinds", "sined", "sines", "sings", "sinhs", "sinks", "sinky", "sinus", "siped", "sipes", "sippy", "sired", "siree", "sires", "sirih", "siris", "siroc", "sirra", "sirup", "sisal", "sises", "sista", "sists", "sitar", "sited", "sites", "sithe", "sitka", "situp", "situs", "siver", "sixer", "sixes", "sixmo", "sixte", "sizar", "sized", "sizel", "sizer", "sizes", "skags", "skail", "skald", "skank", "skart", "skats", "skatt", "skaws", "skean", "skear", "skeds", "skeed", "skeef", "skeen", "skeer", "skees", "skeet", "skegg", "skegs", "skein", "skelf", "skell", "skelm", "skelp", "skene", "skens", "skeos", "skeps", "skers", "skets", "skews", "skids", "skied", "skies", "skiey", "skimo", "skims", "skink", "skins", "skint", "skios", "skips", "skirl", "skirr", "skite", "skits", "skive", "skivy", "sklim", "skoal", "skody", "skoff", "skogs", "skols", "skool", "skort", "skosh", "skran", "skrik", "skuas", "skugs", "skyed", "skyer", "skyey", "skyfs", "skyre", "skyrs", "skyte", "slabs", "slade", "slaes", "slags", "slaid", "slake", "slams", "slane", "slank", "slaps", "slart", "slats", "slaty", "slaws", "slays", "slebs", "sleds", "sleer", "slews", "sleys", "slier", "slily", "slims", "slipe", "slips", "slipt", "slish", "slits", "slive", "sloan", "slobs", "sloes", "slogs", "sloid", "slojd", "slomo", "sloom", "sloot", "slops", "slopy", "slorm", "slots", "slove", "slows", "sloyd", "slubb", "slubs", "slued", "slues", "sluff", "slugs", "sluit", "slums", "slurb", "slurs", "sluse", "sluts", "slyer", "slype", "smaak", "smaik", "smalm", "smalt", "smarm", "smaze", "smeek", "smees", "smeik", "smeke", "smerk", "smews", "smirr", "smirs", "smits", "smogs", "smoko", "smolt", "smoor", "smoot", "smore", "smorg", "smout", "smowt", "smugs", "smurs", "smush", "smuts", "snabs", "snafu", "snags", "snaps", "snarf", "snark", "snars", "snary", "snash", "snath", "snaws", "snead", "sneap", "snebs", "sneck", "sneds", "sneed", "snees", "snell", "snibs", "snick", "snies", "snift", "snigs", "snips", "snipy", "snirt", "snits", "snobs", "snods", "snoek", "snoep", "snogs", "snoke", "snood", "snook", "snool", "snoot", "snots", "snowk", "snows", "snubs", "snugs", "snush", "snyes", "soaks", "soaps", "soare", "soars", "soave", "sobas", "socas", "soces", "socko", "socks", "socle", "sodas", "soddy", "sodic", "sodom", "sofar", "sofas", "softa", "softs", "softy", "soger", "sohur", "soils", "soily", "sojas", "sojus", "sokah", "soken", "sokes", "sokol", "solah", "solan", "solas", "solde", "soldi", "soldo", "solds", "soled", "solei", "soler", "soles", "solon", "solos", "solum", "solus", "soman", "somas", "sonce", "sonde", "sones", "songs", "sonly", "sonne", "sonny", "sonse", "sonsy", "sooey", "sooks", "sooky", "soole", "sools", "sooms", "soops", "soote", "soots", "sophs", "sophy", "sopor", "soppy", "sopra", "soral", "soras", "sorbo", "sorbs", "sorda", "sordo", "sords", "sored", "soree", "sorel", "sorer", "sores", "sorex", "sorgo", "sorns", "sorra", "sorta", "sorts", "sorus", "soths", "sotol", "souce", "souct", "sough", "souks", "souls", "soums", "soups", "soupy", "sours", "souse", "souts", "sowar", "sowce", "sowed", "sowff", "sowfs", "sowle", "sowls", "sowms", "sownd", "sowne", "sowps", "sowse", "sowth", "soyas", "soyle", "soyuz", "sozin", "spacy", "spado", "spaed", "spaer", "spaes", "spags", "spahi", "spail", "spain", "spait", "spake", "spald", "spale", "spall", "spalt", "spams", "spane", "spang", "spans", "spard", "spars", "spart", "spate", "spats", "spaul", "spawl", "spaws", "spayd", "spays", "spaza", "spazz", "speal", "spean", "speat", "specs", "spect", "speel", "speer", "speil", "speir", "speks", "speld", "spelk", "speos", "spets", "speug", "spews", "spewy", "spial", "spica", "spick", "spics", "spide", "spier", "spies", "spiff", "spifs", "spiks", "spile", "spims", "spina", "spink", "spins", "spirt", "spiry", "spits", "spitz", "spivs", "splay", "splog", "spode", "spods", "spoom", "spoor", "spoot", "spork", "sposh", "spots", "sprad", "sprag", "sprat", "spred", "sprew", "sprit", "sprod", "sprog", "sprue", "sprug", "spuds", "spued", "spuer", "spues", "spugs", "spule", "spume", "spumy", "spurs", "sputa", "spyal", "spyre", "squab", "squaw", "squeg", "squid", "squit", "squiz", "stabs", "stade", "stags", "stagy", "staig", "stane", "stang", "staph", "staps", "starn", "starr", "stars", "stats", "staun", "staws", "stays", "stean", "stear", "stedd", "stede", "steds", "steek", "steem", "steen", "steil", "stela", "stele", "stell", "steme", "stems", "stend", "steno", "stens", "stent", "steps", "stept", "stere", "stets", "stews", "stewy", "steys", "stich", "stied", "sties", "stilb", "stile", "stime", "stims", "stimy", "stipa", "stipe", "stire", "stirk", "stirp", "stirs", "stive", "stivy", "stoae", "stoai", "stoas", "stoat", "stobs", "stoep", "stogy", "stoit", "stoln", "stoma", "stond", "stong", "stonk", "stonn", "stook", "stoor", "stope", "stops", "stopt", "stoss", "stots", "stott", "stoun", "stoup", "stour", "stown", "stowp", "stows", "strad", "strae", "strag", "strak", "strep", "strew", "stria", "strig", "strim", "strop", "strow", "stroy", "strum", "stubs", "stude", "studs", "stull", "stulm", "stumm", "stums", "stuns", "stupa", "stupe", "sture", "sturt", "styed", "styes", "styli", "stylo", "styme", "stymy", "styre", "styte", "subah", "subas", "subby", "suber", "subha", "succi", "sucks", "sucky", "sucre", "sudds", "sudor", "sudsy", "suede", "suent", "suers", "suete", "suets", "suety", "sugan", "sughs", "sugos", "suhur", "suids", "suint", "suits", "sujee", "sukhs", "sukuk", "sulci", "sulfa", "sulfo", "sulks", "sulph", "sulus", "sumis", "summa", "sumos", "sumph", "sumps", "sunis", "sunks", "sunna", "sunns", "sunup", "supes", "supra", "surah", "sural", "suras", "surat", "surds", "sured", "sures", "surfs", "surfy", "surgy", "surra", "sused", "suses", "susus", "sutor", "sutra", "sutta", "swabs", "swack", "swads", "swage", "swags", "swail", "swain", "swale", "swaly", "swamy", "swang", "swank", "swans", "swaps", "swapt", "sward", "sware", "swarf", "swart", "swats", "swayl", "sways", "sweal", "swede", "sweed", "sweel", "sweer", "swees", "sweir", "swelt", "swerf", "sweys", "swies", "swigs", "swile", "swims", "swink", "swipe", "swire", "swiss", "swith", "swits", "swive", "swizz", "swobs", "swole", "swoln", "swops", "swopt", "swots", "swoun", "sybbe", "sybil", "syboe", "sybow", "sycee", "syces", "sycon", "syens", "syker", "sykes", "sylis", "sylph", "sylva", "symar", "synch", "syncs", "synds", "syned", "synes", "synth", "syped", "sypes", "syphs", "syrah", "syren", "sysop", "sythe", "syver", "taals", "taata", "taber", "tabes", "tabid", "tabis", "tabla", "tabor", "tabun", "tabus", "tacan", "taces", "tacet", "tache", "tacho", "tachs", "tacks", "tacos", "tacts", "taels", "tafia", "taggy", "tagma", "tahas", "tahrs", "taiga", "taigs", "taiko", "tails", "tains", "taira", "taish", "taits", "tajes", "takas", "takes", "takhi", "takin", "takis", "takky", "talak", "talaq", "talar", "talas", "talcs", "talcy", "talea", "taler", "tales", "talks", "talky", "talls", "talma", "talpa", "taluk", "talus", "tamal", "tamed", "tames", "tamin", "tamis", "tammy", "tamps", "tanas", "tanga", "tangi", "tangs", "tanhs", "tanka", "tanks", "tanky", "tanna", "tansy", "tanti", "tanto", "tanty", "tapas", "taped", "tapen", "tapes", "tapet", "tapis", "tappa", "tapus", "taras", "tardo", "tared", "tares", "targa", "targe", "tarns", "taroc", "tarok", "taros", "tarps", "tarre", "tarry", "tarsi", "tarts", "tarty", "tasar", "tased", "taser", "tases", "tasks", "tassa", "tasse", "tasso", "tatar", "tater", "tates", "taths", "tatie", "tatou", "tatts", "tatus", "taube", "tauld", "tauon", "taupe", "tauts", "tavah", "tavas", "taver", "tawai", "tawas", "tawed", "tawer", "tawie", "tawse", "tawts", "taxed", "taxer", "taxes", "taxis", "taxol", "taxon", "taxor", "taxus", "tayra", "tazza", "tazze", "teade", "teads", "teaed", "teaks", "teals", "teams", "tears", "teats", "teaze", "techs", "techy", "tecta", "teels", "teems", "teend", "teene", "teens", "teeny", "teers", "teffs", "teggs", "tegua", "tegus", "tehrs", "teiid", "teils", "teind", "teins", "telae", "telco", "teles", "telex", "telia", "telic", "tells", "telly", "teloi", "telos", "temed", "temes", "tempi", "temps", "tempt", "temse", "tench", "tends", "tendu", "tenes", "tenge", "tenia", "tenne", "tenno", "tenny", "tenon", "tents", "tenty", "tenue", "tepal", "tepas", "tepoy", "terai", "teras", "terce", "terek", "teres", "terfe", "terfs", "terga", "terms", "terne", "terns", "terry", "terts", "tesla", "testa", "teste", "tests", "tetes", "teths", "tetra", "tetri", "teuch", "teugh", "tewed", "tewel", "tewit", "texas", "texes", "texts", "thack", "thagi", "thaim", "thale", "thali", "thana", "thane", "thang", "thans", "thanx", "tharm", "thars", "thaws", "thawy", "thebe", "theca", "theed", "theek", "thees", "thegn", "theic", "thein", "thelf", "thema", "thens", "theow", "therm", "thesp", "thete", "thews", "thewy", "thigs", "thilk", "thill", "thine", "thins", "thiol", "thirl", "thoft", "thole", "tholi", "thoro", "thorp", "thous", "thowl", "thrae", "thraw", "thrid", "thrip", "throe", "thuds", "thugs", "thuja", "thunk", "thurl", "thuya", "thymi", "thymy", "tians", "tiars", "tical", "ticca", "ticed", "tices", "tichy", "ticks", "ticky", "tiddy", "tided", "tides", "tiers", "tiffs", "tifos", "tifts", "tiges", "tigon", "tikas", "tikes", "tikis", "tikka", "tilak", "tiled", "tiler", "tiles", "tills", "tilly", "tilth", "tilts", "timbo", "timed", "times", "timon", "timps", "tinas", "tinct", "tinds", "tinea", "tined", "tines", "tinge", "tings", "tinks", "tinny", "tints", "tinty", "tipis", "tippy", "tired", "tires", "tirls", "tiros", "tirrs", "titch", "titer", "titis", "titre", "titty", "titup", "tiyin", "tiyns", "tizes", "tizzy", "toads", "toady", "toaze", "tocks", "tocky", "tocos", "todde", "toeas", "toffs", "toffy", "tofts", "tofus", "togae", "togas", "toged", "toges", "togue", "tohos", "toile", "toils", "toing", "toise", "toits", "tokay", "toked", "toker", "tokes", "tokos", "tolan", "tolar", "tolas", "toled", "toles", "tolls", "tolly", "tolts", "tolus", "tolyl", "toman", "tombs", "tomes", "tomia", "tommy", "tomos", "tondi", "tondo", "toned", "toner", "tones", "toney", "tongs", "tonka", "tonks", "tonne", "tonus", "tools", "tooms", "toons", "toots", "toped", "topee", "topek", "toper", "topes", "tophe", "tophi", "tophs", "topis", "topoi", "topos", "toppy", "toque", "torah", "toran", "toras", "torcs", "tores", "toric", "torii", "toros", "torot", "torrs", "torse", "torsi", "torsk", "torta", "torte", "torts", "tosas", "tosed", "toses", "toshy", "tossy", "toted", "toter", "totes", "totty", "touks", "touns", "tours", "touse", "tousy", "touts", "touze", "touzy", "towed", "towie", "towns", "towny", "towse", "towsy", "towts", "towze", "towzy", "toyed", "toyer", "toyon", "toyos", "tozed", "tozes", "tozie", "trabs", "trads", "tragi", "traik", "trams", "trank", "tranq", "trans", "trant", "trape", "traps", "trapt", "trass", "trats", "tratt", "trave", "trayf", "trays", "treck", "treed", "treen", "trees", "trefa", "treif", "treks", "trema", "trems", "tress", "trest", "trets", "trews", "treyf", "treys", "triac", "tride", "trier", "tries", "triff", "trigo", "trigs", "trike", "trild", "trill", "trims", "trine", "trins", "triol", "trior", "trios", "trips", "tripy", "trist", "troad", "troak", "troat", "trock", "trode", "trods", "trogs", "trois", "troke", "tromp", "trona", "tronc", "trone", "tronk", "trons", "trooz", "troth", "trots", "trows", "troys", "trued", "trues", "trugo", "trugs", "trull", "tryer", "tryke", "tryma", "tryps", "tsade", "tsadi", "tsars", "tsked", "tsuba", "tsubo", "tuans", "tuart", "tuath", "tubae", "tubar", "tubas", "tubby", "tubed", "tubes", "tucks", "tufas", "tuffe", "tuffs", "tufts", "tufty", "tugra", "tuile", "tuina", "tuism", "tuktu", "tules", "tulpa", "tulsi", "tumid", "tummy", "tumps", "tumpy", "tunas", "tunds", "tuned", "tuner", "tunes", "tungs", "tunny", "tupek", "tupik", "tuple", "tuque", "turds", "turfs", "turfy", "turks", "turme", "turms", "turns", "turnt", "turps", "turrs", "tushy", "tusks", "tusky", "tutee", "tutti", "tutty", "tutus", "tuxes", "tuyer", "twaes", "twain", "twals", "twank", "twats", "tways", "tweel", "tween", "tweep", "tweer", "twerk", "twerp", "twier", "twigs", "twill", "twilt", "twink", "twins", "twiny", "twire", "twirp", "twite", "twits", "twoer", "twyer", "tyees", "tyers", "tyiyn", "tykes", "tyler", "tymps", "tynde", "tyned", "tynes", "typal", "typed", "types", "typey", "typic", "typos", "typps", "typto", "tyran", "tyred", "tyres", "tyros", "tythe", "tzars", "udals", "udons", "ugali", "ugged", "uhlan", "uhuru", "ukase", "ulama", "ulans", "ulema", "ulmin", "ulnad", "ulnae", "ulnar", "ulnas", "ulpan", "ulvas", "ulyie", "ulzie", "umami", "umbel", "umber", "umble", "umbos", "umbre", "umiac", "umiak", "umiaq", "ummah", "ummas", "ummed", "umped", "umphs", "umpie", "umpty", "umrah", "umras", "unais", "unapt", "unarm", "unary", "unaus", "unbag", "unban", "unbar", "unbed", "unbid", "unbox", "uncap", "unces", "uncia", "uncos", "uncoy", "uncus", "undam", "undee", "undos", "undug", "uneth", "unfix", "ungag", "unget", "ungod", "ungot", "ungum", "unhat", "unhip", "unica", "units", "unjam", "unked", "unket", "unkid", "unlaw", "unlay", "unled", "unlet", "unlid", "unman", "unmew", "unmix", "unpay", "unpeg", "unpen", "unpin", "unred", "unrid", "unrig", "unrip", "unsaw", "unsay", "unsee", "unsew", "unsex", "unsod", "untax", "untin", "unwet", "unwit", "unwon", "upbow", "upbye", "updos", "updry", "upend", "upjet", "uplay", "upled", "uplit", "upped", "upran", "uprun", "upsee", "upsey", "uptak", "upter", "uptie", "uraei", "urali", "uraos", "urare", "urari", "urase", "urate", "urbex", "urbia", "urdee", "ureal", "ureas", "uredo", "ureic", "urena", "urent", "urged", "urger", "urges", "urial", "urite", "urman", "urnal", "urned", "urped", "ursae", "ursid", "urson", "urubu", "urvas", "users", "usnea", "usque", "usure", "usury", "uteri", "uveal", "uveas", "uvula", "vacua", "vaded", "vades", "vagal", "vagus", "vails", "vaire", "vairs", "vairy", "vakas", "vakil", "vales", "valis", "valse", "vamps", "vampy", "vanda", "vaned", "vanes", "vangs", "vants", "vaped", "vaper", "vapes", "varan", "varas", "vardy", "varec", "vares", "varia", "varix", "varna", "varus", "varve", "vasal", "vases", "vasts", "vasty", "vatic", "vatus", "vauch", "vaute", "vauts", "vawte", "vaxes", "veale", "veals", "vealy", "veena", "veeps", "veers", "veery", "vegas", "veges", "vegie", "vegos", "vehme", "veils", "veily", "veins", "veiny", "velar", "velds", "veldt", "veles", "vells", "velum", "venae", "venal", "vends", "vendu", "veney", "venge", "venin", "vents", "venus", "verbs", "verra", "verry", "verst", "verts", "vertu", "vespa", "vesta", "vests", "vetch", "vexed", "vexer", "vexes", "vexil", "vezir", "vials", "viand", "vibes", "vibex", "vibey", "viced", "vices", "vichy", "viers", "views", "viewy", "vifda", "viffs", "vigas", "vigia", "vilde", "viler", "villi", "vills", "vimen", "vinal", "vinas", "vinca", "vined", "viner", "vines", "vinew", "vinic", "vinos", "vints", "viold", "viols", "vired", "vireo", "vires", "virga", "virge", "virid", "virls", "virtu", "visas", "vised", "vises", "visie", "visne", "vison", "visto", "vitae", "vitas", "vitex", "vitro", "vitta", "vivas", "vivat", "vivda", "viver", "vives", "vizir", "vizor", "vleis", "vlies", "vlogs", "voars", "vocab", "voces", "voddy", "vodou", "vodun", "voema", "vogie", "voids", "voile", "voips", "volae", "volar", "voled", "voles", "volet", "volks", "volta", "volte", "volti", "volts", "volva", "volve", "vomer", "voted", "votes", "vouge", "voulu", "vowed", "vower", "voxel", "vozhd", "vraic", "vrils", "vroom", "vrous", "vrouw", "vrows", "vuggs", "vuggy", "vughs", "vughy", "vulgo", "vulns", "vulva", "vutty", "waacs", "wacke", "wacko", "wacks", "wadds", "waddy", "waded", "wader", "wades", "wadge", "wadis", "wadts", "waffs", "wafts", "waged", "wages", "wagga", "wagyu", "wahoo", "waide", "waifs", "waift", "wails", "wains", "wairs", "waite", "waits", "wakas", "waked", "waken", "waker", "wakes", "wakfs", "waldo", "walds", "waled", "waler", "wales", "walie", "walis", "walks", "walla", "walls", "wally", "walty", "wamed", "wames", "wamus", "wands", "waned", "wanes", "waney", "wangs", "wanks", "wanky", "wanle", "wanly", "wanna", "wants", "wanty", "wanze", "waqfs", "warbs", "warby", "wards", "wared", "wares", "warez", "warks", "warms", "warns", "warps", "warre", "warst", "warts", "wases", "washy", "wasms", "wasps", "waspy", "wasts", "watap", "watts", "wauff", "waugh", "wauks", "waulk", "wauls", "waurs", "waved", "waves", "wavey", "wawas", "wawes", "wawls", "waxed", "waxer", "waxes", "wayed", "wazir", "wazoo", "weald", "weals", "weamb", "weans", "wears", "webby", "weber", "wecht", "wedel", "wedgy", "weeds", "weeke", "weeks", "weels", "weems", "weens", "weeny", "weeps", "weepy", "weest", "weete", "weets", "wefte", "wefts", "weids", "weils", "weirs", "weise", "weize", "wekas", "welds", "welke", "welks", "welkt", "wells", "welly", "welts", "wembs", "wends", "wenge", "wenny", "wents", "weros", "wersh", "wests", "wetas", "wetly", "wexed", "wexes", "whamo", "whams", "whang", "whaps", "whare", "whata", "whats", "whaup", "whaur", "wheal", "whear", "wheen", "wheep", "wheft", "whelk", "whelm", "whens", "whets", "whews", "wheys", "whids", "whift", "whigs", "whilk", "whims", "whins", "whios", "whips", "whipt", "whirr", "whirs", "whish", "whiss", "whist", "whits", "whity", "whizz", "whomp", "whoof", "whoot", "whops", "whore", "whorl", "whort", "whoso", "whows", "whump", "whups", "whyda", "wicca", "wicks", "wicky", "widdy", "wides", "wiels", "wifed", "wifes", "wifey", "wifie", "wifty", "wigan", "wigga", "wiggy", "wikis", "wilco", "wilds", "wiled", "wiles", "wilga", "wilis", "wilja", "wills", "wilts", "wimps", "winds", "wined", "wines", "winey", "winge", "wings", "wingy", "winks", "winna", "winns", "winos", "winze", "wiped", "wiper", "wipes", "wired", "wirer", "wires", "wirra", "wised", "wises", "wisha", "wisht", "wisps", "wists", "witan", "wited", "wites", "withe", "withs", "withy", "wived", "wiver", "wives", "wizen", "wizes", "woads", "woald", "wocks", "wodge", "woful", "wojus", "woker", "wokka", "wolds", "wolfs", "wolly", "wolve", "wombs", "womby", "womyn", "wonga", "wongi", "wonks", "wonky", "wonts", "woods", "wooed", "woofs", "woofy", "woold", "wools", "woons", "woops", "woopy", "woose", "woosh", "wootz", "words", "works", "worms", "wormy", "worts", "wowed", "wowee", "woxen", "wrang", "wraps", "wrapt", "wrast", "wrate", "wrawl", "wrens", "wrick", "wried", "wrier", "wries", "writs", "wroke", "wroot", "wroth", "wryer", "wuddy", "wudus", "wulls", "wurst", "wuses", "wushu", "wussy", "wuxia", "wyled", "wyles", "wynds", "wynns", "wyted", "wytes", "xebec", "xenia", "xenic", "xenon", "xeric", "xerox", "xerus", "xoana", "xrays", "xylan", "xylem", "xylic", "xylol", "xylyl", "xysti", "xysts", "yaars", "yabas", "yabba", "yabby", "yacca", "yacka", "yacks", "yaffs", "yager", "yages", "yagis", "yahoo", "yaird", "yakka", "yakow", "yales", "yamen", "yampy", "yamun", "yangs", "yanks", "yapok", "yapon", "yapps", "yappy", "yarak", "yarco", "yards", "yarer", "yarfa", "yarks", "yarns", "yarrs", "yarta", "yarto", "yates", "yauds", "yauld", "yaups", "yawed", "yawey", "yawls", "yawns", "yawny", "yawps", "ybore", "yclad", "ycled", "ycond", "ydrad", "ydred", "yeads", "yeahs", "yealm", "yeans", "yeard", "years", "yecch", "yechs", "yechy", "yedes", "yeeds", "yeesh", "yeggs", "yelks", "yells", "yelms", "yelps", "yelts", "yenta", "yente", "yerba", "yerds", "yerks", "yeses", "yesks", "yests", "yesty", "yetis", "yetts", "yeuks", "yeuky", "yeven", "yeves", "yewen", "yexed", "yexes", "yfere", "yiked", "yikes", "yills", "yince", "yipes", "yippy", "yirds", "yirks", "yirrs", "yirth", "yites", "yitie", "ylems", "ylike", "ylkes", "ymolt", "ympes", "yobbo", "yobby", "yocks", "yodel", "yodhs", "yodle", "yogas", "yogee", "yoghs", "yogic", "yogin", "yogis", "yoick", "yojan", "yoked", "yokel", "yoker", "yokes", "yokul", "yolks", "yolky", "yomim", "yomps", "yonic", "yonis", "yonks", "yoofs", "yoops", "yores", "yorks", "yorps", "youks", "yourn", "yours", "yourt", "youse", "yowed", "yowes", "yowie", "yowls", "yowza", "yrapt", "yrent", "yrivd", "yrneh", "ysame", "ytost", "yuans", "yucas", "yucca", "yucch", "yucko", "yucks", "yucky", "yufts", "yugas", "yuked", "yukes", "yukky", "yukos", "yulan", "yules", "yummo", "yummy", "yumps", "yupon", "yuppy", "yurta", "yurts", "yuzus", "zabra", "zacks", "zaida", "zaidy", "zaire", "zakat", "zaman", "zambo", "zamia", "zanja", "zante", "zanza", "zanze", "zappy", "zarfs", "zaris", "zatis", "zaxes", "zayin", "zazen", "zeals", "zebec", "zebub", "zebus", "zedas", "zeins", "zendo", "zerda", "zerks", "zeros", "zests", "zetas", "zexes", "zezes", "zhomo", "zibet", "ziffs", "zigan", "zilas", "zilch", "zilla", "zills", "zimbi", "zimbs", "zinco", "zincs", "zincy", "zineb", "zines", "zings", "zingy", "zinke", "zinky", "zippo", "zippy", "ziram", "zitis", "zizel", "zizit", "zlote", "zloty", "zoaea", "zobos", "zobus", "zocco", "zoeae", "zoeal", "zoeas", "zoism", "zoist", "zombi", "zonae", "zonda", "zoned", "zoner", "zones", "zonks", "zooea", "zooey", "zooid", "zooks", "zooms", "zoons", "zooty", "zoppa", "zoppo", "zoril", "zoris", "zorro", "zouks", "zowee", "zowie", "zulus", "zupan", "zupas", "zuppa", "zurfs", "zuzim", "zygal", "zygon", "zymes", "zymic"]
_RESPONSES_CACHE = {}
def getResponse(guess, answer):
optional_cached = _RESPONSES_CACHE.get((guess, answer), None)
if optional_cached is not None:
return optional_cached
answer_multiset = {}
greens_multiset = {}
for c_guess, c in zip(guess, answer):
answer_multiset[c] = answer_multiset.get(c, 0) + 1
if c_guess == c:
greens_multiset[c] = greens_multiset.get(c, 0) + 1
result = []
for c_answer, c in zip(answer, guess):
if c_answer == c:
result.append('g')
greens_multiset[c] -= 1
answer_multiset[c] -= 1
continue
if answer_multiset.get(c, 0) > greens_multiset.get(c, 0):
result.append('y')
answer_multiset[c] -= 1
continue
result.append('b')
joined = ''.join(result)
_RESPONSES_CACHE[(guess, answer)] = joined
return joined
assert(getResponse('puree', 'pleat') == 'gbbyb')
assert(getResponse('eejit', 'pleat') == 'ybbbg')
assert(getResponse('raise', 'pleat') == 'bybby')
assert(getResponse('llama', 'aloft') == 'bgybb')
import math
def getGreedyGuess(possible_guesses, possible_answers, metric_function):
best_guesses = None
best_guess_metric = float('inf')
for guess in possible_guesses:
response_counts = {}
for answer in possible_answers:
response = getResponse(guess, answer)
response_counts[response] = response_counts.get(response, 0) + 1
metric = metric_function(v for k, v in response_counts.items() if k != 'ggggg')
if best_guesses is None or best_guess_metric > metric:
best_guess_metric = metric
best_guesses = [guess]
elif best_guess_metric == metric:
best_guesses.append(guess)
return best_guesses
def getGreedyMinMaxBucket(possible_guesses, possible_answers):
return getGreedyGuess(possible_guesses,
possible_answers,
lambda vals: tuple(sorted(vals, reverse=True)))
def getGreedyExpectedBucket(possible_guesses, possible_answers):
return getGreedyGuess(possible_guesses,
possible_answers,
lambda vals: sum(val * val for val in vals))
def getGreedyNextWord(possible_guesses, possible_answers):
return getGreedyGuess(possible_guesses,
possible_answers,
lambda vals: -len(list(vals)))
def getGreedyInfiniteExponentialUtility(possible_guesses, possible_answers):
return getGreedyGuess(possible_answers,
possible_answers,
lambda vals: -len(list(vals)))
def _entropy(vals):
v = list(vals)
total = sum(v)
return 0 if (total == 0) else sum(val * math.log(val + 1) for val in v)
def getGreedyEntropy(possible_guesses, possible_answers):
return getGreedyGuess(possible_guesses,
possible_answers,
_entropy)
def getNewPossibleAnswers(possible_answers, guess, given_response):
return [answer for answer in possible_answers
if getResponse(guess, answer) == given_response]
print(f'Best guesses for min max: {getGreedyMinMaxBucket(POSSIBLE_GUESSES, POSSIBLE_ANSWERS)}')
print(f'Best guesses for EV: {getGreedyExpectedBucket(POSSIBLE_GUESSES, POSSIBLE_ANSWERS)}')
print(f'Best guesses for getting in 2: {getGreedyNextWord(POSSIBLE_GUESSES, POSSIBLE_ANSWERS)}')
print(f'Best guesses for getting for any cutoff: {getGreedyInfiniteExponentialUtility(POSSIBLE_GUESSES, POSSIBLE_ANSWERS)}')
print(f'Best guesses for maximizing information entropy: {getGreedyEntropy(POSSIBLE_GUESSES, POSSIBLE_ANSWERS)}')
new_possible_answers = POSSIBLE_ANSWERS
new_possible_answers = getNewPossibleAnswers(new_possible_answers, 'raise', 'bybbb')
print(new_possible_answers)
print(getGreedyMinMaxBucket(POSSIBLE_GUESSES, new_possible_answers))
new_possible_answers = getNewPossibleAnswers(new_possible_answers, 'cloak', 'bggyb')
print(new_possible_answers)
print(getGreedyMinMaxBucket(POSSIBLE_GUESSES, new_possible_answers))
new_possible_answers = getNewPossibleAnswers(new_possible_answers, 'dwarf', 'bbyby')
print(new_possible_answers)
print(getGreedyMinMaxBucket(POSSIBLE_GUESSES, new_possible_answers))
new_possible_answers = POSSIBLE_ANSWERS
new_possible_answers = getNewPossibleAnswers(POSSIBLE_ANSWERS, 'roate', 'byyyb')
print(new_possible_answers)
print(getGreedyExpectedBucket(POSSIBLE_GUESSES, new_possible_answers))
new_possible_answers = getNewPossibleAnswers(POSSIBLE_ANSWERS, 'bloat', 'bggyg')
print(new_possible_answers)
print(getGreedyExpectedBucket(POSSIBLE_GUESSES, new_possible_answers))
def printTree(possible_guesses, possible_answers, strategy, starting_guess=None, filename=None):
if filename is not None:
with open(filename, 'w') as f:
pass
return printTreeHelper(possible_guesses,
possible_answers,
strategy,
guess=starting_guess,
indent_level=0,
filename=filename) / len(possible_answers)
def printTreeHelper(possible_guesses, possible_answers, strategy, guess=None, indent_level=0, filename=None):
if guess is None:
guess = strategy(possible_guesses, possible_answers)[0]
responses = {}
for answer in possible_answers:
response = getResponse(guess, answer)
if response not in responses:
responses[response] = []
responses[response].append(answer)
def log(info):
if filename is None:
print(info)
else:
with open(filename, 'a') as f:
f.write(info)
f.write('\n')
guess_number = indent_level + 1
total_guesses = 0
items = sorted(responses.items(), key=lambda x: (x[0] != 'ggggg', len(x[1])))
log(f'{" " * indent_level}guess: {guess}')
for response, remaining in items:
if response == 'ggggg':
log(f'{" " * indent_level} took {guess_number} guesses')
total_guesses += indent_level + 1
continue
log(f'{" " * indent_level} {len(remaining)} remaining words for {response}: {remaining}')
total_guesses += printTreeHelper(possible_guesses, remaining, strategy, indent_level=guess_number, filename=filename)
return total_guesses
for starting_guess in ['raise', 'roate', 'trace', 'soare', 'crane', 'slate']:
print(f'Starting guess: {starting_guess}')
for strategy, name in [(getGreedyMinMaxBucket, 'minmax'), (getGreedyExpectedBucket, 'ev'), (getGreedyEntropy, 'entropy')]:
expected_guesses = printTree(POSSIBLE_GUESSES, POSSIBLE_ANSWERS, strategy, starting_guess=starting_guess, filename=f'{starting_guess}_{name}.txt')
print(f'Expected number of guesses for {name}: {expected_guesses}')
def getAverageNumBucketsAfterNextGuess(guess):
score = 0
responses = {}
for answer in POSSIBLE_ANSWERS:
response = getResponse(guess, answer)
if response not in responses:
responses[response] = [answer]
else:
responses[response].append(answer)
for response, possible_answers in responses.items():
score_2 = 0
for guess_2 in POSSIBLE_GUESSES:
responses_2 = set()
for answer in possible_answers:
responses_2.add(getResponse(guess_2, answer))
if len(responses_2) > score_2:
score_2 = len(responses_2)
score += score_2
return score
max_score = 1388
best_word = 'trace'
last_checkpoint = 'steep'
seen = False
for guess in POSSIBLE_GUESSES:
if not seen:
if guess == last_checkpoint:
seen = True
continue
score = getAverageNumBucketsAfterNextGuess(guess)
if score > max_score:
max_score = score
best_word = guess
print(f'Found new best guess: {guess} ({score})')
else:
print(f'Guess {guess} ({score}) could not beat {best_word} ({max_score})')
printTree(POSSIBLE_GUESSES, POSSIBLE_ANSWERS, getGreedyInfiniteExponentialUtility, filename='greedyexponential.txt')
min_guesses = []
min_max_bucket = float('inf')
for i, guess_1 in enumerate(POSSIBLE_GUESSES):
print(i, guess_1)
for j in range(i+1, len(POSSIBLE_GUESSES)):
guess_2 = POSSIBLE_GUESSES[j]
responses = {}
max_bucket = 0
for answer in POSSIBLE_ANSWERS:
response_1 = getResponse(guess_1, answer)
response_2 = getResponse(guess_2, answer)
combined = response_1 + response_2
if combined not in responses:
responses[combined] = []
responses[combined].append(answer)
max_bucket = max(max_bucket, len(responses[combined]))
if min_max_bucket > max_bucket:
min_max_bucket = max_bucket
min_guesses = [(guess_1, guess_2)]
print(f'Found best first 2 guesses ({max_bucket}): {guess_1}, {guess_2}')
elif min_max_bucket == max_bucket:
min_guesses.append((guess_1, guess_2))
print(f'Found best first 2 guesses ({max_bucket}): {guess_1}, {guess_2}')
print(f'Best first 2 guesses: {min_guesses}')
| 0.302082 | 0.706975 |
# Arrivlas and Departures
### Import Packages
```
import pandas as pd
import requests
import sqlalchemy
import os
from dotenv import load_dotenv, find_dotenv
from functools import wraps
import datetime as dt
import json
```
### Load variables from .env file
```
# load env data from .env file.
load_dotenv(find_dotenv(filename='.env'))
```
### Logging Wrapper
```
def log_step(func):
@wraps(func)
def wrapper(*args, **kwargs):
tic = dt.datetime.now()
result = func(*args, **kwargs)
time_taken = str(dt.datetime.now() - tic)
print(f"{func.__name__}:\n shape={result.shape} took {time_taken}s\n")
return result
return wrapper
```
## Get Airports from DB
```
schema="gans"
host="gans-aws.cs3d3b90junp.us-east-1.rds.amazonaws.com"
user="admin"
password = "pEjhiw-wygsy4-quhsos"
port=3306
con = f'mysql+pymysql://{user}:{password}@{host}:{port}/{schema}'
def get_airports_from_db(con):
sql = '''
SELECT * FROM airports
WHERE municipality_country = "Berlin,DE"
'''
airports = pd.read_sql(sql, con = con)
return airports
init_airports = get_airports_from_db(con)
def get_flight_schedules(airports):
responses_list = []
for index, airport_row in airports.iterrows():
url = "https://aerodatabox.p.rapidapi.com/flights/airports/icao/EDDB/2022-04-07T19:00/2022-04-08T07:00"
querystring = {"withLeg":"true","direction":"Both","withCancelled":"true","withCodeshared":"true","withCargo":"true","withPrivate":"true","withLocation":"true"}
headers = {
"X-RapidAPI-Host": "aerodatabox.p.rapidapi.com",
"X-RapidAPI-Key": "c1bc1a1acemsh99ced7306b1d2c9p1c10d7jsn078f60dc1a0e"
}
try:
# response = requests.request("GET", url, headers=headers, params=querystring).json()
# response["municipality_country"] = airport_row["municipality_country"]
# responses_list.append(response)
json_file = f = open("./response_list.json")
responses_list = json.load(f)
except Exception as e:
print("No data for:", airport_row["airport_ident"] )
raise e
continue
return responses_list
def unpack_responses(responses_list):
arrivals = pd.DataFrame()
departures = pd.DataFrame()
for response in responses_list:
city_arr =pd.json_normalize(response["departures"], sep="_")
city_dep = pd.json_normalize(response["arrivals"], sep="_")
arrivals = pd.concat([arrivals, city_arr])
departures = pd.concat([departures, city_dep])
return [arrivals, departures]
def clean_arrivals(df):
# rename_columns
df.rename(columns={
"number": "flight_number",
"call_sign": "flight_call_sign",
"status": "flight_status",
"is_cargo": "flight_is_cargo",
})
```
## Data Cleaning Pipeline
### Init Pipeline
```
@log_step
def init_pipeline(df):
return df.copy()
```
### Rename Columns
```
@log_step
def rename_columns(df):
return (
df.rename(columns={
"id": "city_id",
})
)
```
### Drop Columns
```
@log_step
def drop_columns(df):
return df.drop(columns=["city_state"])
```
### Add Columns
```
@log_step
def add_columns(df):
return (
df
.assign(municipality_country = lambda x: x["city_name"] + "," + x["city_country"])
.assign(created_at = dt.datetime.now())
)
```
### Adjust Datatypes
```
def adjust_datatypes(df):
# df["city_id"] = df["city_id"].astype("int64").astype("string")
return df
```
### Send to DB
```
def send_to_DB(df, table_name, if_exists="replace"):
con = f'mysql+pymysql://{os.environ["DB_USER"]}:{os.environ["DB_PASSWORD"]}@{os.environ["DB_HOST"]}:{os.environ["DB_PORT"]}/{os.environ["DB_SCHEMA"]}'
df.to_sql(
table_name,
con=con,
if_exists=if_exists,
index=False,
dtype={
'city_id': sqlalchemy.types.VARCHAR(length=30),
}
)
engine = sqlalchemy.create_engine(con)
with engine.connect() as engine:
engine.execute('ALTER TABLE `cities` ADD PRIMARY KEY (`municipality_country`);')
return df
```
## Lambda Handler
```
def lambda_handler():
airports = get_airports_from_db(con)
responses_list = get_flight_schedules(airports)
[arrivals, departures] = unpack_responses(responses_list)
arrivals = clean_arrivals(arrivals)
#departures = clean_arrivals(departures)
#send_to_DB([arrivals, departures])
return arrivals
arrivals = lambda_handler()
```
|
github_jupyter
|
import pandas as pd
import requests
import sqlalchemy
import os
from dotenv import load_dotenv, find_dotenv
from functools import wraps
import datetime as dt
import json
# load env data from .env file.
load_dotenv(find_dotenv(filename='.env'))
def log_step(func):
@wraps(func)
def wrapper(*args, **kwargs):
tic = dt.datetime.now()
result = func(*args, **kwargs)
time_taken = str(dt.datetime.now() - tic)
print(f"{func.__name__}:\n shape={result.shape} took {time_taken}s\n")
return result
return wrapper
schema="gans"
host="gans-aws.cs3d3b90junp.us-east-1.rds.amazonaws.com"
user="admin"
password = "pEjhiw-wygsy4-quhsos"
port=3306
con = f'mysql+pymysql://{user}:{password}@{host}:{port}/{schema}'
def get_airports_from_db(con):
sql = '''
SELECT * FROM airports
WHERE municipality_country = "Berlin,DE"
'''
airports = pd.read_sql(sql, con = con)
return airports
init_airports = get_airports_from_db(con)
def get_flight_schedules(airports):
responses_list = []
for index, airport_row in airports.iterrows():
url = "https://aerodatabox.p.rapidapi.com/flights/airports/icao/EDDB/2022-04-07T19:00/2022-04-08T07:00"
querystring = {"withLeg":"true","direction":"Both","withCancelled":"true","withCodeshared":"true","withCargo":"true","withPrivate":"true","withLocation":"true"}
headers = {
"X-RapidAPI-Host": "aerodatabox.p.rapidapi.com",
"X-RapidAPI-Key": "c1bc1a1acemsh99ced7306b1d2c9p1c10d7jsn078f60dc1a0e"
}
try:
# response = requests.request("GET", url, headers=headers, params=querystring).json()
# response["municipality_country"] = airport_row["municipality_country"]
# responses_list.append(response)
json_file = f = open("./response_list.json")
responses_list = json.load(f)
except Exception as e:
print("No data for:", airport_row["airport_ident"] )
raise e
continue
return responses_list
def unpack_responses(responses_list):
arrivals = pd.DataFrame()
departures = pd.DataFrame()
for response in responses_list:
city_arr =pd.json_normalize(response["departures"], sep="_")
city_dep = pd.json_normalize(response["arrivals"], sep="_")
arrivals = pd.concat([arrivals, city_arr])
departures = pd.concat([departures, city_dep])
return [arrivals, departures]
def clean_arrivals(df):
# rename_columns
df.rename(columns={
"number": "flight_number",
"call_sign": "flight_call_sign",
"status": "flight_status",
"is_cargo": "flight_is_cargo",
})
@log_step
def init_pipeline(df):
return df.copy()
@log_step
def rename_columns(df):
return (
df.rename(columns={
"id": "city_id",
})
)
@log_step
def drop_columns(df):
return df.drop(columns=["city_state"])
@log_step
def add_columns(df):
return (
df
.assign(municipality_country = lambda x: x["city_name"] + "," + x["city_country"])
.assign(created_at = dt.datetime.now())
)
def adjust_datatypes(df):
# df["city_id"] = df["city_id"].astype("int64").astype("string")
return df
def send_to_DB(df, table_name, if_exists="replace"):
con = f'mysql+pymysql://{os.environ["DB_USER"]}:{os.environ["DB_PASSWORD"]}@{os.environ["DB_HOST"]}:{os.environ["DB_PORT"]}/{os.environ["DB_SCHEMA"]}'
df.to_sql(
table_name,
con=con,
if_exists=if_exists,
index=False,
dtype={
'city_id': sqlalchemy.types.VARCHAR(length=30),
}
)
engine = sqlalchemy.create_engine(con)
with engine.connect() as engine:
engine.execute('ALTER TABLE `cities` ADD PRIMARY KEY (`municipality_country`);')
return df
def lambda_handler():
airports = get_airports_from_db(con)
responses_list = get_flight_schedules(airports)
[arrivals, departures] = unpack_responses(responses_list)
arrivals = clean_arrivals(arrivals)
#departures = clean_arrivals(departures)
#send_to_DB([arrivals, departures])
return arrivals
arrivals = lambda_handler()
| 0.308919 | 0.55254 |
# Machine Learning Engineer Nanodegree
## Supervised Learning
## Project: Finding Donors for *CharityML*
Welcome to the second project of the Machine Learning Engineer Nanodegree! In this notebook, some template code has already been provided for you, and it will be my job to implement the additional functionality necessary to successfully complete this project. Sections that begin with **'Implementation'** in the header indicate that the following block of code will require additional functionality which I must provide.
## Getting Started
In this project, I will employ several supervised algorithms of your choice to accurately model individuals' income using data collected from the 1994 U.S. Census. I will then choose the best candidate algorithm from preliminary results and further optimize this algorithm to best model the data. Your goal with this implementation is to construct a model that accurately predicts whether an individual makes more than $50,000. This sort of task can arise in a non-profit setting, where organizations survive on donations. Understanding an individual's income can help a non-profit better understand how large of a donation to request, or whether or not they should reach out to begin with. While it can be difficult to determine an individual's general income bracket directly from public sources, we can (as we will see) infer this value from other publically available features.
The dataset for this project originates from the [UCI Machine Learning Repository](https://archive.ics.uci.edu/ml/datasets/Census+Income). The datset was donated by Ron Kohavi and Barry Becker, after being published in the article _"Scaling Up the Accuracy of Naive-Bayes Classifiers: A Decision-Tree Hybrid"_. I can find the article by Ron Kohavi [online](https://www.aaai.org/Papers/KDD/1996/KDD96-033.pdf). The data we investigate here consists of small changes to the original dataset, such as removing the `'fnlwgt'` feature and records with missing or ill-formatted entries.
----
## Exploring the Data
Run the code cell below to load necessary Python libraries and load the census data. Note that the last column from this dataset, `'income'`, will be our target label (whether an individual makes more than, or at most, $50,000 annually). All other columns are features about each individual in the census database.
```
# Import libraries necessary for this project
import numpy as np
import pandas as pd
from time import time
from IPython.display import display # Allows the use of display() for DataFrames
# Import supplementary visualization code visuals.py
import visuals as vs
# Pretty display for notebooks
%matplotlib inline
# Load the Census dataset
data = pd.read_csv("census.csv")
# Success - Display the first record
display(data.head(n=10))
```
### Implementation: Data Exploration
A cursory investigation of the dataset will determine how many individuals fit into either group, and will tell us about the percentage of these individuals making more than \$50,000.
- The total number of records, `'n_records'`
- The number of individuals making more than \$50,000 annually, `'n_greater_50k'`.
- The number of individuals making at most \$50,000 annually, `'n_at_most_50k'`.
- The percentage of individuals making more than \$50,000 annually, `'greater_percent'`.
```
# Total number of records
n_records = data.shape[0]
# Number of records where individual's income is more than $50,000
n_greater_50k = data[(data.income == '>50K')].shape[0]
# Number of records where individual's income is at most $50,000
n_at_most_50k = data[(data.income == '<=50K')].shape[0]
# Percentage of individuals whose income is more than $50,000
greater_percent = (n_greater_50k/n_records)*100
# Print the results
print("Total number of records: {}".format(n_records))
print("Individuals making more than $50,000: {}".format(n_greater_50k))
print("Individuals making at most $50,000: {}".format(n_at_most_50k))
print("Percentage of individuals making more than $50,000: {}%".format(greater_percent))
```
** Featureset Exploration **
* **age**: continuous.
* **workclass**: Private, Self-emp-not-inc, Self-emp-inc, Federal-gov, Local-gov, State-gov, Without-pay, Never-worked.
* **education**: Bachelors, Some-college, 11th, HS-grad, Prof-school, Assoc-acdm, Assoc-voc, 9th, 7th-8th, 12th, Masters, 1st-4th, 10th, Doctorate, 5th-6th, Preschool.
* **education-num**: continuous.
* **marital-status**: Married-civ-spouse, Divorced, Never-married, Separated, Widowed, Married-spouse-absent, Married-AF-spouse.
* **occupation**: Tech-support, Craft-repair, Other-service, Sales, Exec-managerial, Prof-specialty, Handlers-cleaners, Machine-op-inspct, Adm-clerical, Farming-fishing, Transport-moving, Priv-house-serv, Protective-serv, Armed-Forces.
* **relationship**: Wife, Own-child, Husband, Not-in-family, Other-relative, Unmarried.
* **race**: Black, White, Asian-Pac-Islander, Amer-Indian-Eskimo, Other.
* **sex**: Female, Male.
* **capital-gain**: continuous.
* **capital-loss**: continuous.
* **hours-per-week**: continuous.
* **native-country**: United-States, Cambodia, England, Puerto-Rico, Canada, Germany, Outlying-US(Guam-USVI-etc), India, Japan, Greece, South, China, Cuba, Iran, Honduras, Philippines, Italy, Poland, Jamaica, Vietnam, Mexico, Portugal, Ireland, France, Dominican-Republic, Laos, Ecuador, Taiwan, Haiti, Columbia, Hungary, Guatemala, Nicaragua, Scotland, Thailand, Yugoslavia, El-Salvador, Trinadad&Tobago, Peru, Hong, Holand-Netherlands.
----
## Preparing the Data
Before data can be used as input for machine learning algorithms, it often must be cleaned, formatted, and restructured — this is typically known as **preprocessing**. Fortunately, for this dataset, there are no invalid or missing entries we must deal with, however, there are some qualities about certain features that must be adjusted. This preprocessing can help tremendously with the outcome and predictive power of nearly all learning algorithms.
### Transforming Skewed Continuous Features
A dataset may sometimes contain at least one feature whose values tend to lie near a single number, but will also have a non-trivial number of vastly larger or smaller values than that single number. Algorithms can be sensitive to such distributions of values and can underperform if the range is not properly normalized. With the census dataset two features fit this description: '`capital-gain'` and `'capital-loss'`.
Run the code cell below to plot a histogram of these two features. Note the range of the values present and how they are distributed.
```
# Split the data into features and target label
income_raw = data['income']
features_raw = data.drop('income', axis = 1)
# Visualize skewed continuous features of original data
vs.distribution(data)
```
For highly-skewed feature distributions such as `'capital-gain'` and `'capital-loss'`, it is common practice to apply a <a href="https://en.wikipedia.org/wiki/Data_transformation_(statistics)">logarithmic transformation</a> on the data so that the very large and very small values do not negatively affect the performance of a learning algorithm. Using a logarithmic transformation significantly reduces the range of values caused by outliers. Care must be taken when applying this transformation however: The logarithm of `0` is undefined, so we must translate the values by a small amount above `0` to apply the the logarithm successfully.
Run the code cell below to perform a transformation on the data and visualize the results. Again, note the range of values and how they are distributed.
```
# Log-transform the skewed features
skewed = ['capital-gain', 'capital-loss']
features_log_transformed = pd.DataFrame(data = features_raw)
features_log_transformed[skewed] = features_raw[skewed].apply(lambda x: np.log(x + 1))
# Visualize the new log distributions
vs.distribution(features_log_transformed, transformed = True)
```
### Normalizing Numerical Features
In addition to performing transformations on features that are highly skewed, it is often good practice to perform some type of scaling on numerical features. Applying a scaling to the data does not change the shape of each feature's distribution (such as `'capital-gain'` or `'capital-loss'` above); however, normalization ensures that each feature is treated equally when applying supervised learners. Note that once scaling is applied, observing the data in its raw form will no longer have the same original meaning, as exampled below.
Run the code cell below to normalize each numerical feature. We will use [`sklearn.preprocessing.MinMaxScaler`](http://scikit-learn.org/stable/modules/generated/sklearn.preprocessing.MinMaxScaler.html) for this.
```
# Import sklearn.preprocessing.StandardScaler
from sklearn.preprocessing import MinMaxScaler
# Initialize a scaler, then apply it to the features
scaler = MinMaxScaler() # default=(0, 1)
numerical = ['age', 'education-num', 'capital-gain', 'capital-loss', 'hours-per-week']
features_log_minmax_transform = pd.DataFrame(data = features_log_transformed)
features_log_minmax_transform[numerical] = scaler.fit_transform(features_log_transformed[numerical])
# Show an example of a record with scaling applied
display(features_log_minmax_transform.head(n = 5))
```
### Implementation: Data Preprocessing
From the table in **Exploring the Data** above, we can see there are several features for each record that are non-numeric. Typically, learning algorithms expect input to be numeric, which requires that non-numeric features (called *categorical variables*) be converted. One popular way to convert categorical variables is by using the **one-hot encoding** scheme. One-hot encoding creates a _"dummy"_ variable for each possible category of each non-numeric feature. For example, assume `someFeature` has three possible entries: `A`, `B`, or `C`. We then encode this feature into `someFeature_A`, `someFeature_B` and `someFeature_C`.
| | someFeature | | someFeature_A | someFeature_B | someFeature_C |
| :-: | :-: | | :-: | :-: | :-: |
| 0 | B | | 0 | 1 | 0 |
| 1 | C | ----> one-hot encode ----> | 0 | 0 | 1 |
| 2 | A | | 1 | 0 | 0 |
Additionally, as with the non-numeric features, we need to convert the non-numeric target label, `'income'` to numerical values for the learning algorithm to work. Since there are only two possible categories for this label ("<=50K" and ">50K"), we can avoid using one-hot encoding and simply encode these two categories as `0` and `1`, respectively. In code cell below, you will need to implement the following:
- Use [`pandas.get_dummies()`](http://pandas.pydata.org/pandas-docs/stable/generated/pandas.get_dummies.html?highlight=get_dummies#pandas.get_dummies) to perform one-hot encoding on the `'features_log_minmax_transform'` data.
- Convert the target label `'income_raw'` to numerical entries.
- Set records with "<=50K" to `0` and records with ">50K" to `1`.
```
# One-hot encode the 'features_log_minmax_transform' data using pandas.get_dummies()
features_final = pd.get_dummies(features_log_minmax_transform)
# Encode the 'income_raw' data to numerical values
income = income_raw.apply(lambda a: 0 if a == '<=50K' else 1)
# Print the number of features after one-hot encoding
encoded = list(features_final.columns)
print("{} total features after one-hot encoding.".format(len(encoded)))
# Uncomment the following line to see the encoded feature names
print (encoded)
```
### Shuffle and Split Data
Now all _categorical variables_ have been converted into numerical features, and all numerical features have been normalized. As always, we will now split the data (both features and their labels) into training and test sets. 80% of the data will be used for training and 20% for testing.
Run the code cell below to perform this split.
```
# Import train_test_split
from sklearn.cross_validation import train_test_split
# Split the 'features' and 'income' data into training and testing sets
X_train, X_test, y_train, y_test = train_test_split(features_final,
income,
test_size = 0.2,
random_state = 0)
# Show the results of the split
print("Training set has {} samples.".format(X_train.shape[0]))
print("Testing set has {} samples.".format(X_test.shape[0]))
```
----
## Evaluating Model Performance
In this section, we will investigate four different algorithms, and determine which is best at modeling the data. Three of these algorithms will be supervised learners of your choice, and the fourth algorithm is known as a *naive predictor*.
### Metrics and the Naive Predictor
*CharityML*, equipped with their research, knows individuals that make more than \$50,000 are most likely to donate to their charity. Because of this, *CharityML* is particularly interested in predicting who makes more than \$50,000 accurately. It would seem that using **accuracy** as a metric for evaluating a particular model's performace would be appropriate. Additionally, identifying someone that *does not* make more than \$50,000 as someone who does would be detrimental to *CharityML*, since they are looking to find individuals willing to donate. Therefore, a model's ability to precisely predict those that make more than \$50,000 is *more important* than the model's ability to **recall** those individuals. We can use **F-beta score** as a metric that considers both precision and recall:
$$ F_{\beta} = (1 + \beta^2) \cdot \frac{precision \cdot recall}{\left( \beta^2 \cdot precision \right) + recall} $$
In particular, when $\beta = 0.5$, more emphasis is placed on precision. This is called the **F$_{0.5}$ score** (or F-score for simplicity).
Looking at the distribution of classes (those who make at most \$50,000, and those who make more), it's clear most individuals do not make more than \$50,000. This can greatly affect **accuracy**, since we could simply say *"this person does not make more than \$50,000"* and generally be right, without ever looking at the data! Making such a statement would be called **naive**, since we have not considered any information to substantiate the claim. It is always important to consider the *naive prediction* for your data, to help establish a benchmark for whether a model is performing well. That been said, using that prediction would be pointless: If we predicted all people made less than \$50,000, *CharityML* would identify no one as donors.
#### Note: Recap of accuracy, precision, recall
** Accuracy ** measures how often the classifier makes the correct prediction. It’s the ratio of the number of correct predictions to the total number of predictions (the number of test data points).
** Precision ** tells us what proportion of messages we classified as spam, actually were spam.
It is a ratio of true positives(words classified as spam, and which are actually spam) to all positives(all words classified as spam, irrespective of whether that was the correct classificatio), in other words it is the ratio of
`[True Positives/(True Positives + False Positives)]`
** Recall(sensitivity)** tells us what proportion of messages that actually were spam were classified by us as spam.
It is a ratio of true positives(words classified as spam, and which are actually spam) to all the words that were actually spam, in other words it is the ratio of
`[True Positives/(True Positives + False Negatives)]`
For classification problems that are skewed in their classification distributions like in our case, for example if we had a 100 text messages and only 2 were spam and the rest 98 weren't, accuracy by itself is not a very good metric. We could classify 90 messages as not spam(including the 2 that were spam but we classify them as not spam, hence they would be false negatives) and 10 as spam(all 10 false positives) and still get a reasonably good accuracy score. For such cases, precision and recall come in very handy. These two metrics can be combined to get the F1 score, which is weighted average(harmonic mean) of the precision and recall scores. This score can range from 0 to 1, with 1 being the best possible F1 score(we take the harmonic mean as we are dealing with ratios).
### Question 1 - Naive Predictor Performace
* If we chose a model that always predicted an individual made more than $50,000, what would that model's accuracy and F-score be on this dataset? You must use the code cell below and assign your results to `'accuracy'` and `'fscore'` to be used later.
** Please note ** that the the purpose of generating a naive predictor is simply to show what a base model without any intelligence would look like. In the real world, ideally your base model would be either the results of a previous model or could be based on a research paper upon which you are looking to improve. When there is no benchmark model set, getting a result better than random choice is a place you could start from.
```
'''
TP = np.sum(income) # Counting the ones as this is the naive case. Note that 'income' is the 'income_raw' data
encoded to numerical values done in the data preprocessing step.
FP = income.count() - TP # Specific to the naive case
TN = 0 # No predicted negatives in the naive case
FN = 0 # No predicted negatives in the naive case
'''
# Calculate accuracy, precision and recall
accuracy = (n_greater_50k/n_records)
recall = 1 # Since we pick all the incomes >50K
precision = accuracy
# Calculate F-score using the formula above for beta = 0.5 and correct values for precision and recall.
fscore = (1+0.5**2)*(precision*recall)/((0.5**2 * precision)+recall)
# Print the results
print("Naive Predictor: [Accuracy score: {:.4f}, F-score: {:.4f}]".format(accuracy, fscore))
```
### Supervised Learning Models
**The following are some of the supervised learning models that are currently available in** [`scikit-learn`](http://scikit-learn.org/stable/supervised_learning.html)
- Gaussian Naive Bayes (GaussianNB)
- Decision Trees
- Ensemble Methods (Bagging, AdaBoost, Random Forest, Gradient Boosting)
- K-Nearest Neighbors (KNeighbors)
- Stochastic Gradient Descent Classifier (SGDC)
- Support Vector Machines (SVM)
- Logistic Regression
### Question 2 - Model Application
List three of the supervised learning models above that are appropriate for this problem that you will test on the census data. For each model chosen
- Describe one real-world application in industry where the model can be applied.
- What are the strengths of the model; when does it perform well?
- What are the weaknesses of the model; when does it perform poorly?
- What makes this model a good candidate for the problem, given what you know about the data?
** HINT: **
Structure your answer in the same format as above^, with 4 parts for each of the three models you pick. Please include references with your answer.
**Answer:**
KNN (K-Nearest Neighbours)
- The KNN model can be applied in the medical field to determine if a patient with a heart attack will have another heart attack or not based on the demographics, diet and clinical measurement of the patient.
- The KNN model performs well on a relatively small dataset with linear and non-linear classification boundaries. Also, the KNN model works well for the big datasets. The KNN model is simple and effective.
- The KNN model depends on the value K which is the number of the nearest neighbors. There is no straightforward way to determine the value of the K for a given model. For large datasets, the KNN model becomes computation intensive as there are many distance calculations.
- KNN model could be a good candidate for the problem because the problem dataset is relatively small; therefore, the computation cost will not be higher.
Referance: http://www.ijera.com/papers/Vol3_issue5/DI35605610.pdf
Support Vector Machines(SVMs)
- The SVM model is used for the classification of the protein and cancer based on the geans and biological problems of the patient.
- The advantages of the SVM model are the SVM model produces robust learning results, overfitting is not universal, works well with small datasets.
- The disadvantages of the SVM model are the model needs to be 2-class, and the training takes longer time for large datasets.
- The SVM model is a good candidate for the problem as it is a classification problem and the number of data points is relatively small. Also, SVM can help establish a reliable relationship between the output and each input variables.
Referance: http://www.cs.uky.edu/~jzhang/CS689/PPDM-Chapter2.pdf
Logistic Regression
- One real-life application of logistic regression is to detect if the given credit card transaction is fraud or not.
- The advantage of the logistic regression is the fact that it gives the probability of each outcome predicted by the model, unlike the linear model. Also, the logistic regression can be used for non-linear classification boundary.
- The primary disadvantage of the logistic regression is it requires a large dataset to produce a reliable model for the problem.
- The logistic regression is an excellent solution to the problem because we have big enough data set to produce a reliable model. The model will be robust against non-linear boundaries of the classification.
Referance: https://victorfang.wordpress.com/2011/05/10/advantages-and-disadvantages-of-logistic-regression/
### Implementation - Creating a Training and Predicting Pipeline
To properly evaluate the performance of each model you've chosen, it's important that you create a training and predicting pipeline that allows you to quickly and effectively train models using various sizes of training data and perform predictions on the testing data. Your implementation here will be used in the following section.
In the code block below, you will need to implement the following:
- Import `fbeta_score` and `accuracy_score` from [`sklearn.metrics`](http://scikit-learn.org/stable/modules/classes.html#sklearn-metrics-metrics).
- Fit the learner to the sampled training data and record the training time.
- Perform predictions on the test data `X_test`, and also on the first 300 training points `X_train[:300]`.
- Record the total prediction time.
- Calculate the accuracy score for both the training subset and testing set.
- Calculate the F-score for both the training subset and testing set.
- Make sure that you set the `beta` parameter!
```
#Import two metrics from sklearn - fbeta_score and accuracy_score
from sklearn.metrics import fbeta_score
from sklearn.metrics import accuracy_score
def train_predict(learner, sample_size, X_train, y_train, X_test, y_test):
'''
inputs:
- learner: the learning algorithm to be trained and predicted on
- sample_size: the size of samples (number) to be drawn from training set
- X_train: features training set
- y_train: income training set
- X_test: features testing set
- y_test: income testing set
'''
results = {}
# Fit the learner to the training data using slicing with 'sample_size' using .fit(training_features[:], training_labels[:])
start = time() # Get start time
learner = learner.fit(X_train[:sample_size], y_train[:sample_size])
end = time() # Get end time
# alculate the training time
results['train_time'] = end -start
# Get the predictions on the test set(X_test),
# then get predictions on the first 300 training samples(X_train) using .predict()
start = time() # Get start time
predictions_test = learner.predict( X_test )
predictions_train = learner.predict( X_train[:300] )
end = time() # Get end time
# Calculate the total prediction time
results['pred_time'] = end- start
# Compute accuracy on the first 300 training samples which is y_train[:300]
results['acc_train'] = accuracy_score( y_train[:300], predictions_train )
# Compute accuracy on test set using accuracy_score()
results['acc_test'] = accuracy_score( y_test, predictions_test )
# Compute F-score on the the first 300 training samples using fbeta_score()
results['f_train'] = fbeta_score( y_train[:300], predictions_train, 0.5 )
# Compute F-score on the test set which is y_test
results['f_test'] = fbeta_score( y_test, predictions_test, 0.5 )
# Success
print("{} trained on {} samples.".format(learner.__class__.__name__, sample_size))
# Return the results
return results
```
### Implementation: Initial Model Evaluation
In the code cell, you will need to implement the following:
- Import the three supervised learning models you've discussed in the previous section.
- Initialize the three models and store them in `'clf_A'`, `'clf_B'`, and `'clf_C'`.
- Use a `'random_state'` for each model you use, if provided.
- **Note:** Use the default settings for each model — you will tune one specific model in a later section.
- Calculate the number of records equal to 1%, 10%, and 100% of the training data.
- Store those values in `'samples_1'`, `'samples_10'`, and `'samples_100'` respectively.
**Note:** Depending on which algorithms you chose, the following implementation may take some time to run!
```
# Import the three supervised learning models from sklearn
from sklearn.neighbors import KNeighborsClassifier
from sklearn.svm import LinearSVC
from sklearn.linear_model import LogisticRegression
# Initialize the three models
clf_A = KNeighborsClassifier(n_neighbors=3)
clf_B = LinearSVC(random_state = 31)
clf_C = LogisticRegression(random_state = 31)
# Calculate the number of samples for 1%, 10%, and 100% of the training data
# samples_100 is the entire training set i.e. len(y_train)
# samples_10 is 10% of samples_100 (ensure to set the count of the values to be `int` and not `float`)
# samples_1 is 1% of samples_100 (ensure to set the count of the values to be `int` and not `float`)
samples_100 = len(X_train)
samples_10 = int(len(X_train)/10)
samples_1 = int(len(X_train)/100)
# Collect results on the learners
results = {}
for clf in [clf_A, clf_B, clf_C]:
clf_name = clf.__class__.__name__
results[clf_name] = {}
for i, samples in enumerate([samples_1, samples_10, samples_100]):
results[clf_name][i] = \
train_predict(clf, samples, X_train, y_train, X_test, y_test)
# Run metrics visualization for the three supervised learning models chosen
vs.evaluate(results, accuracy, fscore)
```
----
## Improving Results
In this final section, you will choose from the three supervised learning models the *best* model to use on the student data. You will then perform a grid search optimization for the model over the entire training set (`X_train` and `y_train`) by tuning at least one parameter to improve upon the untuned model's F-score.
### Question 3 - Choosing the Best Model
* Based on the evaluation you performed earlier, in one to two paragraphs, explain to *CharityML* which of the three models you believe to be most appropriate for the task of identifying individuals that make more than \$50,000.
**Answer:**
- From the graphs above, the logistic regression model is the best model to predict the individuals that make more than $50,000 since the F-score of the SVM model for 100 percent training data is better than KNN model and very close to the SVM model. Also, the SVM model takes about two seconds more for the model training than logistic regression; therefore, with little difference in F-score, the logistic regression delivers faster performance. With high accuracy, faster performance and good F-score, the logistic regression is the most suitable algorithm for the data. Since the linear SVC and the logistic regression produces linear boundaries to classify the data. On the other hand, KNN does not always produce a linear boundary to classify the data. Therefore, KNN tends to overfit the training data while the linear boundary usually generalizes the data well. By looking at the test data results, the logistic algorithm works well with the data since the linear classification boundary generalizes the data well.
### Question 4 - Describing the Model in Layman's Terms
* In one to two paragraphs, explain to *CharityML*, in layman's terms, how the final model chosen is supposed to work. Be sure that you are describing the major qualities of the model, such as how the model is trained and how the model makes a prediction. Avoid using advanced mathematical jargon, such as describing equations.
**Answer:**
- Logistic regression works best for the binary classification as in this case to classify the people with the income of more than 50,000 dollars. The linear regression predicts a value while the logistic regression produces a probability that a given input belongs to a class. Rather than telling the input belongs to class 1 or class 2, the logistic regression gives a probability like 0.8 for class 1 and 0.2 for class 2. Based on the probability value, the classification is determined. The training of the logistic model means finding the best line to separate two classes of data. The input data being either side of the line, the logistic regression value will be high for one class and low for another. If the input is close to the linear boundary, the logistic regression value will be close to 0.5 for both classes.
### Implementation: Model Tuning
Fine tune the chosen model. Use grid search (`GridSearchCV`) with at least one important parameter tuned with at least 3 different values. You will need to use the entire training set for this. In the code cell below, you will need to implement the following:
- Import [`sklearn.grid_search.GridSearchCV`](http://scikit-learn.org/0.17/modules/generated/sklearn.grid_search.GridSearchCV.html) and [`sklearn.metrics.make_scorer`](http://scikit-learn.org/stable/modules/generated/sklearn.metrics.make_scorer.html).
- Initialize the classifier you've chosen and store it in `clf`.
- Set a `random_state` if one is available to the same state you set before.
- Create a dictionary of parameters you wish to tune for the chosen model.
- Example: `parameters = {'parameter' : [list of values]}`.
- **Note:** Avoid tuning the `max_features` parameter of your learner if that parameter is available!
- Use `make_scorer` to create an `fbeta_score` scoring object (with $\beta = 0.5$).
- Perform grid search on the classifier `clf` using the `'scorer'`, and store it in `grid_obj`.
- Fit the grid search object to the training data (`X_train`, `y_train`), and store it in `grid_fit`.
**Note:** Depending on the algorithm chosen and the parameter list, the following implementation may take some time to run!
```
#Import 'GridSearchCV', 'make_scorer', and any other necessary libraries
from sklearn.grid_search import GridSearchCV
from sklearn.metrics import make_scorer
import random
#Initialize the classifier
clf = LogisticRegression(random_state = 31)
# Create the parameters list you wish to tune, using a dictionary if needed.
# parameters = {'parameter_1': [value1, value2], 'parameter_2': [value1, value2]}
parameters = { 'C' : [0.5, 1.0, 10.5], 'intercept_scaling' : [1.0, 10.5, 20.0]}
#''C' : [0.5, 1.0, 1.5], 'intercept_scaling' : [1.0, 1.5, 2.0],
# Make an fbeta_score scoring object using make_scorer()
scorer = make_scorer(fbeta_score, beta = 0.5)
#Perform grid search on the classifier using 'scorer' as the scoring method using GridSearchCV()
grid_obj = GridSearchCV(clf, param_grid = parameters, cv = 9, scoring = scorer)
#Fit the grid search object to the training data and find the optimal parameters using fit()
grid_fit = grid_obj.fit(X_train, y_train)
# Get the estimator
best_clf = grid_fit.best_estimator_
# Make predictions using the unoptimized and model
predictions = (clf.fit(X_train, y_train)).predict(X_test)
best_predictions = best_clf.predict(X_test)
# Report the before-and-afterscores
print("Unoptimized model\n------")
print("Accuracy score on testing data: {:.4f}".format(accuracy_score(y_test, predictions)))
print("F-score on testing data: {:.4f}".format(fbeta_score(y_test, predictions, beta = 0.5)))
print("\nOptimized Model\n------")
print("Final accuracy score on the testing data: {:.4f}".format(accuracy_score(y_test, best_predictions)))
print("Final F-score on the testing data: {:.4f}".format(fbeta_score(y_test, best_predictions, beta = 0.5)))
```
### Question 5 - Final Model Evaluation
* What is your optimized model's accuracy and F-score on the testing data?
* Are these scores better or worse than the unoptimized model?
* How do the results from your optimized model compare to the naive predictor benchmarks you found earlier in **Question 1**?_
**Note:** Fill in the table below with your results, and then provide discussion in the **Answer** box.
#### Results:
| Metric | Unoptimized Model | Optimized Model |
| :------------: | :---------------: | :-------------: |
| Accuracy Score | 0.8419 | 0.8419 |
| F-score | 0.6832 | 0.6832 |
**Answer:**
- The optimized model's accuracy and F-score on the testing data are 0.8419 and 0.6832 respectively. The accuracy and the F-score are the same for both optimized and unoptimized models. The naive predictor has the Accuracy score: 0.2478, F-score: 0.2917. Compare to the naive predictor; the optimized model is doing much better since both the accuracy and F-score is significantly higher for the optimized model.
----
## Feature Importance
An important task when performing supervised learning on a dataset like the census data we study here is determining which features provide the most predictive power. By focusing on the relationship between only a few crucial features and the target label we simplify our understanding of the phenomenon, which is most always a useful thing to do. In the case of this project, that means we wish to identify a small number of features that most strongly predict whether an individual makes at most or more than \$50,000.
Choose a scikit-learn classifier (e.g., adaboost, random forests) that has a `feature_importance_` attribute, which is a function that ranks the importance of features according to the chosen classifier. In the next python cell fit this classifier to training set and use this attribute to determine the top 5 most important features for the census dataset.
### Question 6 - Feature Relevance Observation
When **Exploring the Data**, it was shown there are thirteen available features for each individual on record in the census data. Of these thirteen records, which five features do you believe to be most important for prediction, and in what order would you rank them and why?
**Answer:**
- Hours-per-week, education_level, Age, capital-gain, capitol-loss are the essential features for the prediction in the order of high rank to low rank respectively. The hours-per-week features determine the hours of work for an individual. The more the hours, the more the salary in general. The higher the education level, the higher the income in most cases. Age is also crucial since the young people tend to work more and make more money while the same can not be said about children and old people. Capital gain and loss gives insight into the individual's overall income. If an individual has more than 50,000 dollars of income but has a significant capital loss, the individual is less likely to donate the money. Similarly, with income little, less than 50,000 dollars with significant capital gain could make an individual eligible to make donations.
### Implementation - Extracting Feature Importance
Choose a `scikit-learn` supervised learning algorithm that has a `feature_importance_` attribute availble for it. This attribute is a function that ranks the importance of each feature when making predictions based on the chosen algorithm.
In the code cell below, you will need to implement the following:
- Import a supervised learning model from sklearn if it is different from the three used earlier.
- Train the supervised model on the entire training set.
- Extract the feature importances using `'.feature_importances_'`.
```
#Import a supervised learning model that has 'feature_importances_'
from sklearn.ensemble import AdaBoostClassifier
#Train the supervised model on the training set using .fit(X_train, y_train)
model = AdaBoostClassifier(random_state = 31)
model.fit(X_train, y_train)
#Extract the feature importances using .feature_importances_
importances = model.feature_importances_
# Plot
vs.feature_plot(importances, X_train, y_train)
```
### Question 7 - Extracting Feature Importance
Observe the visualization created above which displays the five most relevant features for predicting if an individual makes at most or above \$50,000.
* How do these five features compare to the five features you discussed in **Question 6**?
* If you were close to the same answer, how does this visualization confirm your thoughts?
* If you were not close, why do you think these features are more relevant?
**Answer:**
- These five features are the five feature I predicted before but in different rank order. I predicted the capital-loss to be the fifth but it the first one in the graph because the capital-loss directly implies the loss in an individual's income; therefore, the less ability to donate. The age is in the second spot because the age plays a significant role in the income of an individual. Capital gain is the third feature which I predicted as fourth. Capitol-gain again is directly related to the income of an individual. The hours-per-week and the education-num follow the capital-gain as the more the value of the variables, the more the income and therefore, direct relationship with the ability to donate.
### Feature Selection
How does a model perform if we only use a subset of all the available features in the data? With less features required to train, the expectation is that training and prediction time is much lower — at the cost of performance metrics. From the visualization above, we see that the top five most important features contribute more than half of the importance of **all** features present in the data. This hints that we can attempt to *reduce the feature space* and simplify the information required for the model to learn. The code cell below will use the same optimized model you found earlier, and train it on the same training set *with only the top five important features*.
```
# Import functionality for cloning a model
from sklearn.base import clone
# Reduce the feature space
X_train_reduced = X_train[X_train.columns.values[(np.argsort(importances)[::-1])[:5]]]
X_test_reduced = X_test[X_test.columns.values[(np.argsort(importances)[::-1])[:5]]]
# Train on the "best" model found from grid search earlier
clf = (clone(best_clf)).fit(X_train_reduced, y_train)
# Make new predictions
reduced_predictions = clf.predict(X_test_reduced)
# Report scores from the final model using both versions of data
print("Final Model trained on full data\n------")
print("Accuracy on testing data: {:.4f}".format(accuracy_score(y_test, best_predictions)))
print("F-score on testing data: {:.4f}".format(fbeta_score(y_test, best_predictions, beta = 0.5)))
print("\nFinal Model trained on reduced data\n------")
print("Accuracy on testing data: {:.4f}".format(accuracy_score(y_test, reduced_predictions)))
print("F-score on testing data: {:.4f}".format(fbeta_score(y_test, reduced_predictions, beta = 0.5)))
```
### Question 8 - Effects of Feature Selection
* How does the final model's F-score and accuracy score on the reduced data using only five features compare to those same scores when all features are used?
* If training time was a factor, would you consider using the reduced data as your training set?
**Answer:**
- The reduced data model performed worst then the full data model. The reduced data model has both low accuracy and F-score than the full data model. Since the performance of the reduced data is significantly lower than the full data model, I would not consider using the reduced data as my training set.
|
github_jupyter
|
# Import libraries necessary for this project
import numpy as np
import pandas as pd
from time import time
from IPython.display import display # Allows the use of display() for DataFrames
# Import supplementary visualization code visuals.py
import visuals as vs
# Pretty display for notebooks
%matplotlib inline
# Load the Census dataset
data = pd.read_csv("census.csv")
# Success - Display the first record
display(data.head(n=10))
# Total number of records
n_records = data.shape[0]
# Number of records where individual's income is more than $50,000
n_greater_50k = data[(data.income == '>50K')].shape[0]
# Number of records where individual's income is at most $50,000
n_at_most_50k = data[(data.income == '<=50K')].shape[0]
# Percentage of individuals whose income is more than $50,000
greater_percent = (n_greater_50k/n_records)*100
# Print the results
print("Total number of records: {}".format(n_records))
print("Individuals making more than $50,000: {}".format(n_greater_50k))
print("Individuals making at most $50,000: {}".format(n_at_most_50k))
print("Percentage of individuals making more than $50,000: {}%".format(greater_percent))
# Split the data into features and target label
income_raw = data['income']
features_raw = data.drop('income', axis = 1)
# Visualize skewed continuous features of original data
vs.distribution(data)
# Log-transform the skewed features
skewed = ['capital-gain', 'capital-loss']
features_log_transformed = pd.DataFrame(data = features_raw)
features_log_transformed[skewed] = features_raw[skewed].apply(lambda x: np.log(x + 1))
# Visualize the new log distributions
vs.distribution(features_log_transformed, transformed = True)
# Import sklearn.preprocessing.StandardScaler
from sklearn.preprocessing import MinMaxScaler
# Initialize a scaler, then apply it to the features
scaler = MinMaxScaler() # default=(0, 1)
numerical = ['age', 'education-num', 'capital-gain', 'capital-loss', 'hours-per-week']
features_log_minmax_transform = pd.DataFrame(data = features_log_transformed)
features_log_minmax_transform[numerical] = scaler.fit_transform(features_log_transformed[numerical])
# Show an example of a record with scaling applied
display(features_log_minmax_transform.head(n = 5))
# One-hot encode the 'features_log_minmax_transform' data using pandas.get_dummies()
features_final = pd.get_dummies(features_log_minmax_transform)
# Encode the 'income_raw' data to numerical values
income = income_raw.apply(lambda a: 0 if a == '<=50K' else 1)
# Print the number of features after one-hot encoding
encoded = list(features_final.columns)
print("{} total features after one-hot encoding.".format(len(encoded)))
# Uncomment the following line to see the encoded feature names
print (encoded)
# Import train_test_split
from sklearn.cross_validation import train_test_split
# Split the 'features' and 'income' data into training and testing sets
X_train, X_test, y_train, y_test = train_test_split(features_final,
income,
test_size = 0.2,
random_state = 0)
# Show the results of the split
print("Training set has {} samples.".format(X_train.shape[0]))
print("Testing set has {} samples.".format(X_test.shape[0]))
'''
TP = np.sum(income) # Counting the ones as this is the naive case. Note that 'income' is the 'income_raw' data
encoded to numerical values done in the data preprocessing step.
FP = income.count() - TP # Specific to the naive case
TN = 0 # No predicted negatives in the naive case
FN = 0 # No predicted negatives in the naive case
'''
# Calculate accuracy, precision and recall
accuracy = (n_greater_50k/n_records)
recall = 1 # Since we pick all the incomes >50K
precision = accuracy
# Calculate F-score using the formula above for beta = 0.5 and correct values for precision and recall.
fscore = (1+0.5**2)*(precision*recall)/((0.5**2 * precision)+recall)
# Print the results
print("Naive Predictor: [Accuracy score: {:.4f}, F-score: {:.4f}]".format(accuracy, fscore))
#Import two metrics from sklearn - fbeta_score and accuracy_score
from sklearn.metrics import fbeta_score
from sklearn.metrics import accuracy_score
def train_predict(learner, sample_size, X_train, y_train, X_test, y_test):
'''
inputs:
- learner: the learning algorithm to be trained and predicted on
- sample_size: the size of samples (number) to be drawn from training set
- X_train: features training set
- y_train: income training set
- X_test: features testing set
- y_test: income testing set
'''
results = {}
# Fit the learner to the training data using slicing with 'sample_size' using .fit(training_features[:], training_labels[:])
start = time() # Get start time
learner = learner.fit(X_train[:sample_size], y_train[:sample_size])
end = time() # Get end time
# alculate the training time
results['train_time'] = end -start
# Get the predictions on the test set(X_test),
# then get predictions on the first 300 training samples(X_train) using .predict()
start = time() # Get start time
predictions_test = learner.predict( X_test )
predictions_train = learner.predict( X_train[:300] )
end = time() # Get end time
# Calculate the total prediction time
results['pred_time'] = end- start
# Compute accuracy on the first 300 training samples which is y_train[:300]
results['acc_train'] = accuracy_score( y_train[:300], predictions_train )
# Compute accuracy on test set using accuracy_score()
results['acc_test'] = accuracy_score( y_test, predictions_test )
# Compute F-score on the the first 300 training samples using fbeta_score()
results['f_train'] = fbeta_score( y_train[:300], predictions_train, 0.5 )
# Compute F-score on the test set which is y_test
results['f_test'] = fbeta_score( y_test, predictions_test, 0.5 )
# Success
print("{} trained on {} samples.".format(learner.__class__.__name__, sample_size))
# Return the results
return results
# Import the three supervised learning models from sklearn
from sklearn.neighbors import KNeighborsClassifier
from sklearn.svm import LinearSVC
from sklearn.linear_model import LogisticRegression
# Initialize the three models
clf_A = KNeighborsClassifier(n_neighbors=3)
clf_B = LinearSVC(random_state = 31)
clf_C = LogisticRegression(random_state = 31)
# Calculate the number of samples for 1%, 10%, and 100% of the training data
# samples_100 is the entire training set i.e. len(y_train)
# samples_10 is 10% of samples_100 (ensure to set the count of the values to be `int` and not `float`)
# samples_1 is 1% of samples_100 (ensure to set the count of the values to be `int` and not `float`)
samples_100 = len(X_train)
samples_10 = int(len(X_train)/10)
samples_1 = int(len(X_train)/100)
# Collect results on the learners
results = {}
for clf in [clf_A, clf_B, clf_C]:
clf_name = clf.__class__.__name__
results[clf_name] = {}
for i, samples in enumerate([samples_1, samples_10, samples_100]):
results[clf_name][i] = \
train_predict(clf, samples, X_train, y_train, X_test, y_test)
# Run metrics visualization for the three supervised learning models chosen
vs.evaluate(results, accuracy, fscore)
#Import 'GridSearchCV', 'make_scorer', and any other necessary libraries
from sklearn.grid_search import GridSearchCV
from sklearn.metrics import make_scorer
import random
#Initialize the classifier
clf = LogisticRegression(random_state = 31)
# Create the parameters list you wish to tune, using a dictionary if needed.
# parameters = {'parameter_1': [value1, value2], 'parameter_2': [value1, value2]}
parameters = { 'C' : [0.5, 1.0, 10.5], 'intercept_scaling' : [1.0, 10.5, 20.0]}
#''C' : [0.5, 1.0, 1.5], 'intercept_scaling' : [1.0, 1.5, 2.0],
# Make an fbeta_score scoring object using make_scorer()
scorer = make_scorer(fbeta_score, beta = 0.5)
#Perform grid search on the classifier using 'scorer' as the scoring method using GridSearchCV()
grid_obj = GridSearchCV(clf, param_grid = parameters, cv = 9, scoring = scorer)
#Fit the grid search object to the training data and find the optimal parameters using fit()
grid_fit = grid_obj.fit(X_train, y_train)
# Get the estimator
best_clf = grid_fit.best_estimator_
# Make predictions using the unoptimized and model
predictions = (clf.fit(X_train, y_train)).predict(X_test)
best_predictions = best_clf.predict(X_test)
# Report the before-and-afterscores
print("Unoptimized model\n------")
print("Accuracy score on testing data: {:.4f}".format(accuracy_score(y_test, predictions)))
print("F-score on testing data: {:.4f}".format(fbeta_score(y_test, predictions, beta = 0.5)))
print("\nOptimized Model\n------")
print("Final accuracy score on the testing data: {:.4f}".format(accuracy_score(y_test, best_predictions)))
print("Final F-score on the testing data: {:.4f}".format(fbeta_score(y_test, best_predictions, beta = 0.5)))
#Import a supervised learning model that has 'feature_importances_'
from sklearn.ensemble import AdaBoostClassifier
#Train the supervised model on the training set using .fit(X_train, y_train)
model = AdaBoostClassifier(random_state = 31)
model.fit(X_train, y_train)
#Extract the feature importances using .feature_importances_
importances = model.feature_importances_
# Plot
vs.feature_plot(importances, X_train, y_train)
# Import functionality for cloning a model
from sklearn.base import clone
# Reduce the feature space
X_train_reduced = X_train[X_train.columns.values[(np.argsort(importances)[::-1])[:5]]]
X_test_reduced = X_test[X_test.columns.values[(np.argsort(importances)[::-1])[:5]]]
# Train on the "best" model found from grid search earlier
clf = (clone(best_clf)).fit(X_train_reduced, y_train)
# Make new predictions
reduced_predictions = clf.predict(X_test_reduced)
# Report scores from the final model using both versions of data
print("Final Model trained on full data\n------")
print("Accuracy on testing data: {:.4f}".format(accuracy_score(y_test, best_predictions)))
print("F-score on testing data: {:.4f}".format(fbeta_score(y_test, best_predictions, beta = 0.5)))
print("\nFinal Model trained on reduced data\n------")
print("Accuracy on testing data: {:.4f}".format(accuracy_score(y_test, reduced_predictions)))
print("F-score on testing data: {:.4f}".format(fbeta_score(y_test, reduced_predictions, beta = 0.5)))
| 0.836087 | 0.98986 |
```
import os
import sys
import gc
import time
import json
import random
import math
import numpy as np
import torch
from torch.optim.adamw import AdamW
from torch.nn.utils import clip_grad_norm_
import torch.distributed as dist
from torch.utils.data.dataloader import DataLoader
from torchvision.utils import save_image
from wolf.data import load_datasets, get_batch, preprocess, postprocess
from wolf import WolfModel
from wolf.utils import total_grad_norm
from wolf.optim import ExponentialScheduler
from experiments.options import parse_args
from matplotlib import pyplot as plt
from tqdm import tqdm_notebook
import autoreload
%load_ext autoreload
%autoreload 2
def is_master(rank):
return rank <= 0
def is_distributed(rank):
return rank >= 0
def logging(info, logfile=None):
print(info)
if logfile is not None:
print(info, file=logfile)
logfile.flush()
def init_dataloader(args, train_data, val_data):
if is_distributed(args.rank):
train_sampler = torch.utils.data.distributed.DistributedSampler(train_data, rank=args.rank,
num_replicas=args.world_size,
shuffle=True)
else:
train_sampler = None
train_loader = DataLoader(train_data, batch_size=args.batch_size,
shuffle=(train_sampler is None), sampler=train_sampler,
num_workers=args.workers, pin_memory=True, drop_last=True)
if is_master(args.rank):
eval_batch = args.eval_batch_size
val_loader = DataLoader(val_data, batch_size=eval_batch, shuffle=False,
num_workers=args.workers, pin_memory=True)
else:
val_loader = None
return train_loader, train_sampler, val_loader
def setup(args):
def check_dataset():
if dataset == 'cifar10':
assert image_size == 32, 'CIFAR-10 expected image size 32 but got {}'.format(image_size)
elif dataset.startswith('lsun'):
assert image_size in [128, 256]
elif dataset == 'celeba':
assert image_size in [256, 512]
elif dataset == 'imagenet':
assert image_size in [64, 128, 256]
dataset = args.dataset
if args.category is not None:
dataset = dataset + '_' + args.category
image_size = args.image_size
check_dataset()
nc = 3
args.nx = image_size ** 2 * nc
n_bits = args.n_bits
args.n_bins = 2. ** n_bits
args.test_k = 1
model_path = args.model_path
args.checkpoint_name = os.path.join(model_path, 'checkpoint')
result_path = os.path.join(model_path, 'images')
args.result_path = result_path
data_path = args.data_path
if is_master(args.rank):
if not os.path.exists(model_path):
os.makedirs(model_path)
if not os.path.exists(result_path):
os.makedirs(result_path)
if args.recover < 0:
args.log = open(os.path.join(model_path, 'log.txt'), 'w')
else:
args.log = open(os.path.join(model_path, 'log.txt'), 'a')
else:
args.log = None
args.cuda = torch.cuda.is_available()
random_seed = args.seed + args.rank if args.rank >= 0 else args.seed
if args.recover >= 0:
random_seed += random.randint(0, 1024)
logging("Rank {}: random seed={}".format(args.rank, random_seed), logfile=args.log)
random.seed(random_seed)
np.random.seed(random_seed)
torch.manual_seed(random_seed)
device = torch.device('cuda', args.local_rank) if args.cuda else torch.device('cpu')
if args.cuda:
torch.cuda.set_device(device)
torch.cuda.manual_seed(random_seed)
torch.backends.cudnn.benchmark = True
args.world_size = int(os.environ["WORLD_SIZE"]) if is_distributed(args.rank) else 1
logging("Rank {}: ".format(args.rank) + str(args), args.log)
train_data, val_data = load_datasets(dataset, image_size, data_path=data_path)
train_index = np.arange(len(train_data))
np.random.shuffle(train_index)
val_index = np.arange(len(val_data))
if is_master(args.rank):
logging('Data size: training: {}, val: {}'.format(len(train_index), len(val_index)))
if args.recover >= 0:
params = json.load(open(os.path.join(model_path, 'config.json'), 'r'))
else:
params = json.load(open(args.config, 'r'))
json.dump(params, open(os.path.join(model_path, 'config.json'), 'w'), indent=2)
wolf = WolfModel.from_params(params)
wolf.to_device(device)
args.device = device
if args.recover >= 0:
wolf = WolfModel.load(args.model_path, args.device, 0)
# if args.recover >= 0:
# checkpoint_name = args.checkpoint_name + '{}.tar'.format(args.recover)
# print(f"Rank = {args.rank}, loading from checkpoint {checkpoint_name}")
# checkpoint = torch.load(checkpoint_name, map_location=args.device)
# start_epoch = checkpoint['epoch']
# last_step = checkpoint['step']
# wolf.load_state_dict(checkpoint['model'])
# best_epoch = checkpoint['best_epoch']
# best_nll = checkpoint['best_nll']
# best_bpd = checkpoint['best_bpd']
# best_nent = checkpoint['best_nent']
# best_nepd = checkpoint['best_nepd']
# best_kl = checkpoint['best_kl']
# del checkpoint
return args, (train_data, val_data), (train_index, val_index), wolf
debug=False
def eval(args, val_loader, wolf):
wolf.eval()
wolf.sync()
gnll = 0
nent = 0
kl = 0
num_insts = 0
device = args.device
n_bits = args.n_bits
n_bins = args.n_bins
nx = args.nx
test_k = args.test_k
results = []
for step, (data, y) in enumerate(val_loader):
batch_size = len(data)
data = data.to(device, non_blocking=True)
y = y.to(device, non_blocking=True)
with torch.no_grad():
attns = wolf.loss_attn(data, y=y, n_bits=n_bits, nsamples=test_k)
# print('shape is ', attns.shape)
# del attns
# gc.collect()
# size_bool = True
# for attn_idx in range(len(attns_)-1):
# size_bool = (size_bool and (attns_[attn_idx].shape == attns_[attn_idx].shape))
# if debug:
# if size_bool:
# print('the length of attentions is {}; the shape of attention is {}'.format(len(attns_), attns_[0].shape))
# else:
# print('Size not matched')
# gnll += loss_gen.sum().item()
# kl += loss_kl.sum().item()
# nent += loss_dequant.sum().item()
# num_insts += batch_size
results.append((data.to('cpu'), y.to('cpu'), attns.to('cpu')))
if step % 10 == 0:
print('Step: ', step)
torch.cuda.empty_cache()
# gnll = gnll / num_insts
# nent = nent / num_insts
# kl = kl / num_insts
# nll = gnll + kl + nent + np.log(n_bins / 2.) * nx
# bpd = nll / (nx * np.log(2.0))
# nepd = nent / (nx * np.log(2.0))
# logging('Avg NLL: {:.2f}, KL: {:.2f}, NENT: {:.2f}, BPD: {:.4f}, NEPD: {:.4f}'.format(
# nll, kl, nent, bpd, nepd), args.log)
return results
def main(args):
args, (train_data, val_data), (train_index, val_index), wolf = setup(args)
train_loader, train_sampler, val_loader = init_dataloader(args, train_data, val_data)
return eval(args, val_loader, wolf), wolf
args_dict = {'rank': -1,
'local_rank': 0,
'config': 'experiments/configs/cifar10/glow/glow-cat-uni.json',
'batch_size': 256,
'eval_batch_size': 512,
'batch_steps': 2,
'init_batch_size': 1024,
'epochs': 100,
'valid_epochs': 10,
'seed': 65537,
'train_k': 1,
'log_interval': 10,
'lr': 0.001,
'warmup_steps': 50,
'lr_decay': 0.999997,
'beta1': 0.9,
'beta2': 0.999,
'eps': 1e-08,
'weight_decay': 1e-06,
'amsgrad': False,
'grad_clip': 0.0,
'dataset': 'cifar10',
'category': None,
'image_size': 32,
'workers': 4,
'n_bits': 8,
'model_path': 'experiments/models/glow/cifar_linear_attn_model/',
'data_path': 'experiments/data/cifar_data1',
'recover': 1000,
}
from argparse import Namespace
args = Namespace(**args_dict)
results, wolf = main(args)
data_1, label_1, attn_1 = results[0]
plt.imshow(data_1[2].squeeze(0).permute(1, 2, 0))
plt.imshow(attn_1[2].permute(1, 2, 0))
# 40 samples, one row 4 pictures, 10 rows
f, axarr = plt.subplots(15, 8)
f.set_figheight(16)
f.set_figwidth(16)
for i in tqdm_notebook(range(60)):
idx_col = i // 15
idx_row = (i % 15)
axarr[idx_row, 2*idx_col].set_title(label_1[i].item())
axarr[idx_row, 2*idx_col].imshow(data_1[i].squeeze_(0).permute(1, 2, 0))
axarr[idx_row, 2*idx_col].axis('off')
# threshold = (torch.max(attn_1[i]) + torch.min(attn_1[i])) / 2
# axarr[i, 1].imshow((attn_1[i] > threshold).permute(1, 2, 0))
axarr[idx_row, 2*idx_col+1].imshow(attn_1[i].permute(1, 2, 0))
axarr[idx_row, 2*idx_col+1].axis('off')
def interpolation(args, img1, y1, img2, y2, model):
image_size = (3, args.image_size, args.image_size)
nsamples = 1
batch = 1
image_recons = []
num_recons = 5
img1 = img1.to(args.device)
img2 = img2.to(args.device)
y1 = y1.to(args.device)
y2 = y2.to(args.device)
with torch.no_grad():
_, eps1 = model.encode(img1, y=y1, n_bits=args.n_bits, nsamples=nsamples, random=True)
eps1 = eps1.view(batch * nsamples, *image_size)
z1 = model.encode_global(img1, y=y1, n_bits=args.n_bits)
z1 = z1.view(batch * nsamples, z1.size(2))
_, eps2 = model.encode(img2, y=y2, n_bits=args.n_bits, nsamples=nsamples, random=True)
eps2 = eps2.view(batch * nsamples, *image_size)
z2 = model.encode_global(img2, y=y2, n_bits=args.n_bits)
z2 = z2.view(batch * nsamples, z2.size(2))
fig, axs = plt.subplots(1, num_recons+2)
for i, delta in enumerate(torch.linspace(0., 1., steps=num_recons+2)):
new_z = z1 + (z2 - z1) * delta.item()
new_eps = eps1 + (eps2 - eps1) * delta.item()
img_recon = model.decode(new_eps, z=new_z, n_bits=args.n_bits).view(batch, nsamples, *image_size).cpu()
axs[i].imshow(img_recon[0][0].permute(1, 2, 0))
y_1_ = results[0][1]
interpolation(args, data_1[3].unsqueeze(0), y_1_[3].unsqueeze(0), data_1[7].unsqueeze(0), y_1_[7].unsqueeze(0), wolf)
def reconstruct(args, img, y, wolf):
print('reconstruct')
wolf.eval()
batch = 1
nsamples = 15
# index = np.arange(len(data))
# np.random.shuffle(index)
# img, y = get_batch(data, index[:batch])
img = img.to(args.device)
y = y.to(args.device)
with torch.no_grad():
image_size = (3, args.image_size, args.image_size)
_, epsilon = wolf.encode(img, y=y, n_bits=args.n_bits, nsamples=nsamples, random=True)
epsilon = epsilon.view(batch * nsamples, *image_size)
z = wolf.encode_global(img, y=y, n_bits=args.n_bits, nsamples=nsamples, random=True)
z = z.view(batch * nsamples, z.size(2))
# [batch, nsamples, c, h, w]
img_recon = wolf.decode(epsilon, z=z, n_bits=args.n_bits).view(batch, nsamples, *image_size)
# [batch, 1, c, h, w]
img = postprocess(preprocess(img, args.n_bits), args.n_bits).unsqueeze(1)
fig, axs = plt.subplots(1,2)
img_cpu = img.cpu()
img_recon_cpu = img_recon.cpu()
print(img_recon_cpu.shape)
axs[0].imshow(img_cpu[0][0].permute(1, 2, 0))
axs[1].imshow(img_recon_cpu[0][0].permute(1, 2, 0))
print(torch.norm(img_recon_cpu[0][0] - img_cpu[0][0]))
# [batch, nsamples + 1, c, h, w] -> [batch*(nsamples + 1), c, h, w]
comparison = torch.cat([img, img_recon], dim=1).view(-1, *image_size).cpu()
y_1_ = results[0][1]
reconstruct(args, data_1[9].unsqueeze(0), label_1[9].unsqueeze(0), wolf)
def synthesize_cat(args, epoch, wolf, label):
logging('sampling based on cat', args.log)
wolf.eval()
n = 64 if args.image_size > 128 else 256
nrow = int(math.sqrt(n))
taus = [0.7, 0.8, 0.9, 1.0]
image_size = (3, args.image_size, args.image_size)
device = args.device
# label = torch.Tensor(label).to(device).long()
label = label.to(device)
print(label)
for t in taus:
epsilon = torch.randn(n, *image_size, device=device)
epsilon = epsilon * t
z = wolf.encode_global(epsilon, label)
z = z.view(n, z.size(2))
imgs = wolf.decode(epsilon, z)
# imgs, attns = wolf.decode_with_attn(epsilon, z)
# print('img shape: ', imgs.shape)
# print('attn shape: ', attns.shape)
image_file = 'sample{}.t{:.1f}.png'.format(epoch, t)
attn_file = 'sample{}.t{:.1f}attn.png'.format(epoch, t)
save_image(imgs, os.path.join(args.result_path, image_file), nrow=nrow)
# save_image(attns, os.path.join(args.result_path, attn_file), nrow=nrow)
with torch.no_grad():
synthesize_cat(args, 224, wolf, label_1[1].unsqueeze(0))
label_1[1]
```
|
github_jupyter
|
import os
import sys
import gc
import time
import json
import random
import math
import numpy as np
import torch
from torch.optim.adamw import AdamW
from torch.nn.utils import clip_grad_norm_
import torch.distributed as dist
from torch.utils.data.dataloader import DataLoader
from torchvision.utils import save_image
from wolf.data import load_datasets, get_batch, preprocess, postprocess
from wolf import WolfModel
from wolf.utils import total_grad_norm
from wolf.optim import ExponentialScheduler
from experiments.options import parse_args
from matplotlib import pyplot as plt
from tqdm import tqdm_notebook
import autoreload
%load_ext autoreload
%autoreload 2
def is_master(rank):
return rank <= 0
def is_distributed(rank):
return rank >= 0
def logging(info, logfile=None):
print(info)
if logfile is not None:
print(info, file=logfile)
logfile.flush()
def init_dataloader(args, train_data, val_data):
if is_distributed(args.rank):
train_sampler = torch.utils.data.distributed.DistributedSampler(train_data, rank=args.rank,
num_replicas=args.world_size,
shuffle=True)
else:
train_sampler = None
train_loader = DataLoader(train_data, batch_size=args.batch_size,
shuffle=(train_sampler is None), sampler=train_sampler,
num_workers=args.workers, pin_memory=True, drop_last=True)
if is_master(args.rank):
eval_batch = args.eval_batch_size
val_loader = DataLoader(val_data, batch_size=eval_batch, shuffle=False,
num_workers=args.workers, pin_memory=True)
else:
val_loader = None
return train_loader, train_sampler, val_loader
def setup(args):
def check_dataset():
if dataset == 'cifar10':
assert image_size == 32, 'CIFAR-10 expected image size 32 but got {}'.format(image_size)
elif dataset.startswith('lsun'):
assert image_size in [128, 256]
elif dataset == 'celeba':
assert image_size in [256, 512]
elif dataset == 'imagenet':
assert image_size in [64, 128, 256]
dataset = args.dataset
if args.category is not None:
dataset = dataset + '_' + args.category
image_size = args.image_size
check_dataset()
nc = 3
args.nx = image_size ** 2 * nc
n_bits = args.n_bits
args.n_bins = 2. ** n_bits
args.test_k = 1
model_path = args.model_path
args.checkpoint_name = os.path.join(model_path, 'checkpoint')
result_path = os.path.join(model_path, 'images')
args.result_path = result_path
data_path = args.data_path
if is_master(args.rank):
if not os.path.exists(model_path):
os.makedirs(model_path)
if not os.path.exists(result_path):
os.makedirs(result_path)
if args.recover < 0:
args.log = open(os.path.join(model_path, 'log.txt'), 'w')
else:
args.log = open(os.path.join(model_path, 'log.txt'), 'a')
else:
args.log = None
args.cuda = torch.cuda.is_available()
random_seed = args.seed + args.rank if args.rank >= 0 else args.seed
if args.recover >= 0:
random_seed += random.randint(0, 1024)
logging("Rank {}: random seed={}".format(args.rank, random_seed), logfile=args.log)
random.seed(random_seed)
np.random.seed(random_seed)
torch.manual_seed(random_seed)
device = torch.device('cuda', args.local_rank) if args.cuda else torch.device('cpu')
if args.cuda:
torch.cuda.set_device(device)
torch.cuda.manual_seed(random_seed)
torch.backends.cudnn.benchmark = True
args.world_size = int(os.environ["WORLD_SIZE"]) if is_distributed(args.rank) else 1
logging("Rank {}: ".format(args.rank) + str(args), args.log)
train_data, val_data = load_datasets(dataset, image_size, data_path=data_path)
train_index = np.arange(len(train_data))
np.random.shuffle(train_index)
val_index = np.arange(len(val_data))
if is_master(args.rank):
logging('Data size: training: {}, val: {}'.format(len(train_index), len(val_index)))
if args.recover >= 0:
params = json.load(open(os.path.join(model_path, 'config.json'), 'r'))
else:
params = json.load(open(args.config, 'r'))
json.dump(params, open(os.path.join(model_path, 'config.json'), 'w'), indent=2)
wolf = WolfModel.from_params(params)
wolf.to_device(device)
args.device = device
if args.recover >= 0:
wolf = WolfModel.load(args.model_path, args.device, 0)
# if args.recover >= 0:
# checkpoint_name = args.checkpoint_name + '{}.tar'.format(args.recover)
# print(f"Rank = {args.rank}, loading from checkpoint {checkpoint_name}")
# checkpoint = torch.load(checkpoint_name, map_location=args.device)
# start_epoch = checkpoint['epoch']
# last_step = checkpoint['step']
# wolf.load_state_dict(checkpoint['model'])
# best_epoch = checkpoint['best_epoch']
# best_nll = checkpoint['best_nll']
# best_bpd = checkpoint['best_bpd']
# best_nent = checkpoint['best_nent']
# best_nepd = checkpoint['best_nepd']
# best_kl = checkpoint['best_kl']
# del checkpoint
return args, (train_data, val_data), (train_index, val_index), wolf
debug=False
def eval(args, val_loader, wolf):
wolf.eval()
wolf.sync()
gnll = 0
nent = 0
kl = 0
num_insts = 0
device = args.device
n_bits = args.n_bits
n_bins = args.n_bins
nx = args.nx
test_k = args.test_k
results = []
for step, (data, y) in enumerate(val_loader):
batch_size = len(data)
data = data.to(device, non_blocking=True)
y = y.to(device, non_blocking=True)
with torch.no_grad():
attns = wolf.loss_attn(data, y=y, n_bits=n_bits, nsamples=test_k)
# print('shape is ', attns.shape)
# del attns
# gc.collect()
# size_bool = True
# for attn_idx in range(len(attns_)-1):
# size_bool = (size_bool and (attns_[attn_idx].shape == attns_[attn_idx].shape))
# if debug:
# if size_bool:
# print('the length of attentions is {}; the shape of attention is {}'.format(len(attns_), attns_[0].shape))
# else:
# print('Size not matched')
# gnll += loss_gen.sum().item()
# kl += loss_kl.sum().item()
# nent += loss_dequant.sum().item()
# num_insts += batch_size
results.append((data.to('cpu'), y.to('cpu'), attns.to('cpu')))
if step % 10 == 0:
print('Step: ', step)
torch.cuda.empty_cache()
# gnll = gnll / num_insts
# nent = nent / num_insts
# kl = kl / num_insts
# nll = gnll + kl + nent + np.log(n_bins / 2.) * nx
# bpd = nll / (nx * np.log(2.0))
# nepd = nent / (nx * np.log(2.0))
# logging('Avg NLL: {:.2f}, KL: {:.2f}, NENT: {:.2f}, BPD: {:.4f}, NEPD: {:.4f}'.format(
# nll, kl, nent, bpd, nepd), args.log)
return results
def main(args):
args, (train_data, val_data), (train_index, val_index), wolf = setup(args)
train_loader, train_sampler, val_loader = init_dataloader(args, train_data, val_data)
return eval(args, val_loader, wolf), wolf
args_dict = {'rank': -1,
'local_rank': 0,
'config': 'experiments/configs/cifar10/glow/glow-cat-uni.json',
'batch_size': 256,
'eval_batch_size': 512,
'batch_steps': 2,
'init_batch_size': 1024,
'epochs': 100,
'valid_epochs': 10,
'seed': 65537,
'train_k': 1,
'log_interval': 10,
'lr': 0.001,
'warmup_steps': 50,
'lr_decay': 0.999997,
'beta1': 0.9,
'beta2': 0.999,
'eps': 1e-08,
'weight_decay': 1e-06,
'amsgrad': False,
'grad_clip': 0.0,
'dataset': 'cifar10',
'category': None,
'image_size': 32,
'workers': 4,
'n_bits': 8,
'model_path': 'experiments/models/glow/cifar_linear_attn_model/',
'data_path': 'experiments/data/cifar_data1',
'recover': 1000,
}
from argparse import Namespace
args = Namespace(**args_dict)
results, wolf = main(args)
data_1, label_1, attn_1 = results[0]
plt.imshow(data_1[2].squeeze(0).permute(1, 2, 0))
plt.imshow(attn_1[2].permute(1, 2, 0))
# 40 samples, one row 4 pictures, 10 rows
f, axarr = plt.subplots(15, 8)
f.set_figheight(16)
f.set_figwidth(16)
for i in tqdm_notebook(range(60)):
idx_col = i // 15
idx_row = (i % 15)
axarr[idx_row, 2*idx_col].set_title(label_1[i].item())
axarr[idx_row, 2*idx_col].imshow(data_1[i].squeeze_(0).permute(1, 2, 0))
axarr[idx_row, 2*idx_col].axis('off')
# threshold = (torch.max(attn_1[i]) + torch.min(attn_1[i])) / 2
# axarr[i, 1].imshow((attn_1[i] > threshold).permute(1, 2, 0))
axarr[idx_row, 2*idx_col+1].imshow(attn_1[i].permute(1, 2, 0))
axarr[idx_row, 2*idx_col+1].axis('off')
def interpolation(args, img1, y1, img2, y2, model):
image_size = (3, args.image_size, args.image_size)
nsamples = 1
batch = 1
image_recons = []
num_recons = 5
img1 = img1.to(args.device)
img2 = img2.to(args.device)
y1 = y1.to(args.device)
y2 = y2.to(args.device)
with torch.no_grad():
_, eps1 = model.encode(img1, y=y1, n_bits=args.n_bits, nsamples=nsamples, random=True)
eps1 = eps1.view(batch * nsamples, *image_size)
z1 = model.encode_global(img1, y=y1, n_bits=args.n_bits)
z1 = z1.view(batch * nsamples, z1.size(2))
_, eps2 = model.encode(img2, y=y2, n_bits=args.n_bits, nsamples=nsamples, random=True)
eps2 = eps2.view(batch * nsamples, *image_size)
z2 = model.encode_global(img2, y=y2, n_bits=args.n_bits)
z2 = z2.view(batch * nsamples, z2.size(2))
fig, axs = plt.subplots(1, num_recons+2)
for i, delta in enumerate(torch.linspace(0., 1., steps=num_recons+2)):
new_z = z1 + (z2 - z1) * delta.item()
new_eps = eps1 + (eps2 - eps1) * delta.item()
img_recon = model.decode(new_eps, z=new_z, n_bits=args.n_bits).view(batch, nsamples, *image_size).cpu()
axs[i].imshow(img_recon[0][0].permute(1, 2, 0))
y_1_ = results[0][1]
interpolation(args, data_1[3].unsqueeze(0), y_1_[3].unsqueeze(0), data_1[7].unsqueeze(0), y_1_[7].unsqueeze(0), wolf)
def reconstruct(args, img, y, wolf):
print('reconstruct')
wolf.eval()
batch = 1
nsamples = 15
# index = np.arange(len(data))
# np.random.shuffle(index)
# img, y = get_batch(data, index[:batch])
img = img.to(args.device)
y = y.to(args.device)
with torch.no_grad():
image_size = (3, args.image_size, args.image_size)
_, epsilon = wolf.encode(img, y=y, n_bits=args.n_bits, nsamples=nsamples, random=True)
epsilon = epsilon.view(batch * nsamples, *image_size)
z = wolf.encode_global(img, y=y, n_bits=args.n_bits, nsamples=nsamples, random=True)
z = z.view(batch * nsamples, z.size(2))
# [batch, nsamples, c, h, w]
img_recon = wolf.decode(epsilon, z=z, n_bits=args.n_bits).view(batch, nsamples, *image_size)
# [batch, 1, c, h, w]
img = postprocess(preprocess(img, args.n_bits), args.n_bits).unsqueeze(1)
fig, axs = plt.subplots(1,2)
img_cpu = img.cpu()
img_recon_cpu = img_recon.cpu()
print(img_recon_cpu.shape)
axs[0].imshow(img_cpu[0][0].permute(1, 2, 0))
axs[1].imshow(img_recon_cpu[0][0].permute(1, 2, 0))
print(torch.norm(img_recon_cpu[0][0] - img_cpu[0][0]))
# [batch, nsamples + 1, c, h, w] -> [batch*(nsamples + 1), c, h, w]
comparison = torch.cat([img, img_recon], dim=1).view(-1, *image_size).cpu()
y_1_ = results[0][1]
reconstruct(args, data_1[9].unsqueeze(0), label_1[9].unsqueeze(0), wolf)
def synthesize_cat(args, epoch, wolf, label):
logging('sampling based on cat', args.log)
wolf.eval()
n = 64 if args.image_size > 128 else 256
nrow = int(math.sqrt(n))
taus = [0.7, 0.8, 0.9, 1.0]
image_size = (3, args.image_size, args.image_size)
device = args.device
# label = torch.Tensor(label).to(device).long()
label = label.to(device)
print(label)
for t in taus:
epsilon = torch.randn(n, *image_size, device=device)
epsilon = epsilon * t
z = wolf.encode_global(epsilon, label)
z = z.view(n, z.size(2))
imgs = wolf.decode(epsilon, z)
# imgs, attns = wolf.decode_with_attn(epsilon, z)
# print('img shape: ', imgs.shape)
# print('attn shape: ', attns.shape)
image_file = 'sample{}.t{:.1f}.png'.format(epoch, t)
attn_file = 'sample{}.t{:.1f}attn.png'.format(epoch, t)
save_image(imgs, os.path.join(args.result_path, image_file), nrow=nrow)
# save_image(attns, os.path.join(args.result_path, attn_file), nrow=nrow)
with torch.no_grad():
synthesize_cat(args, 224, wolf, label_1[1].unsqueeze(0))
label_1[1]
| 0.295535 | 0.32888 |
# Deep Learning for Automatic Labeling of CT Images
## By: Ian Pan, MD.ai modified by Anouk Stein, MD.ai and Ross Filice MD, MedStar Georgetown University Hospital to predict chest, abdomen, or pelvic slices. Note lower chest/upper abdomen may have labels for both chest and abdomen.
```
!git clone https://github.com/rwfilice/bodypart.git
!pip install pydicom
from scipy.ndimage.interpolation import zoom
import matplotlib.pyplot as plt
import pydicom
import pandas as pd
import numpy as np
import glob
import os
import re
import json
from pathlib import Path
from keras.applications.imagenet_utils import preprocess_input
from keras.applications.mobilenet_v2 import MobileNetV2
from keras.callbacks import EarlyStopping, ReduceLROnPlateau
from keras import Model
from keras.layers import Dropout, Dense, GlobalAveragePooling2D
from keras import optimizers
from keras.models import model_from_json
import tensorflow as tf
# Set seed for reproducibility
tf.random.set_seed(88) ; np.random.seed(88)
# For data augmentation
from albumentations import (
Compose, OneOf, HorizontalFlip, Blur, RandomGamma, RandomContrast, RandomBrightness
)
testPath = Path('bodypart/testnpy')
testList = list(sorted(testPath.glob('**/*.npy'), key=lambda fn: int(re.search('-([0-9]*)', str(fn)).group(1))))
testList
def get_dicom_and_uid(path_to_npy):
'''
Given a filepath, return the npy file and corresponding SOPInstanceUID.
'''
path_to_npy = str(path_to_npy)
dicom_file = np.load(path_to_npy)
uid = path_to_npy.split('/')[-1].replace('.npy', '')
return dicom_file, uid
def convert_dicom_to_8bit(npy_file, width, level, imsize=(224.,224.), clip=True):
'''
Given a DICOM file, window specifications, and image size,
return the image as a Numpy array scaled to [0,255] of the specified size.
'''
array = npy_file.copy()
#array = array + int(dicom_file.RescaleIntercept) #we did this on preprocess
#array = array * int(dicom_file.RescaleSlope) #we did this on preprocess
array = np.clip(array, level - width / 2, level + width / 2)
# Rescale to [0, 255]
array -= np.min(array)
array /= np.max(array)
array *= 255.
array = array.astype('uint8')
if clip:
# Sometimes there is dead space around the images -- let's get rid of that
nonzeros = np.nonzero(array)
x1 = np.min(nonzeros[0]) ; x2 = np.max(nonzeros[0])
y1 = np.min(nonzeros[1]) ; y2 = np.max(nonzeros[1])
array = array[x1:x2,y1:y2]
# Resize image if necessary
resize_x = float(imsize[0]) / array.shape[0]
resize_y = float(imsize[1]) / array.shape[1]
if resize_x != 1. or resize_y != 1.:
array = zoom(array, [resize_x, resize_y], order=1, prefilter=False)
return np.expand_dims(array, axis=-1)
json_file = open('bodypart/model.json', 'r')
loaded_model_json = json_file.read()
json_file.close()
model = model_from_json(loaded_model_json)
model.load_weights('bodypart/tcga-mguh-multilabel.h5') #federated
#Inference
IMSIZE = 256
WINDOW_LEVEL, WINDOW_WIDTH = 50, 500
def predict(model, images, imsize):
'''
Small modifications to data generator to allow for prediction on test data.
'''
test_arrays = []
test_probas = []
test_uids = []
for im in images:
dicom_file, uid = get_dicom_and_uid(im)
try:
array = convert_dicom_to_8bit(dicom_file, WINDOW_WIDTH, WINDOW_LEVEL,
imsize=(imsize,imsize))
except:
continue
array = preprocess_input(array, mode='tf')
test_arrays.append(array)
test_probas.append(model.predict(np.expand_dims(array, axis=0)))
test_uids.append(uid)
return test_uids, test_arrays, test_probas
uids, X, y_prob = predict(model, testList, IMSIZE)
test_pred_df = pd.DataFrame({'uid': uids, 'X': X, 'y_prob': y_prob})
test_pred_df.apply(lambda row: row['y_prob'], axis=1)
chest = np.stack(test_pred_df['y_prob'])[:,0][:,0]
abd = np.stack(test_pred_df['y_prob'])[:,0][:,1]
pelv = np.stack(test_pred_df['y_prob'])[:,0][:,2]
plt.plot(chest)
plt.plot(abd)
plt.plot(pelv)
numaveslices = 5
avepreds = []
allpreds = np.stack(test_pred_df['y_prob'])[:,0]
for idx,arr in enumerate(allpreds):
low = int(max(0,idx-(numaveslices-1)/2))
high = int(min(len(allpreds),idx+(numaveslices+1)/2))
avepreds.append(np.mean(allpreds[low:high],axis=0))
chest = np.stack(avepreds)[:,0]
abd = np.stack(avepreds)[:,1]
pelv = np.stack(avepreds)[:,2]
#averaged over 5 slices
plt.plot(chest)
plt.plot(abd)
plt.plot(pelv)
def displayImages(imgs,labels):
numimgs = len(imgs)
plt.figure(figsize=(20,10))
for idx,img in enumerate(imgs):
dicom_file, uid = get_dicom_and_uid(img)
img = convert_dicom_to_8bit(dicom_file, WINDOW_WIDTH, WINDOW_LEVEL, clip=False)
plt.subplot("1%i%i" % (numimgs,idx+1))
plt.imshow(img[...,0],cmap='gray')
plt.title(labels[idx])
plt.axis('off')
#averaged over 5 slices
fig, ax1 = plt.subplots(figsize=(17,10))
ax1.set_xlabel("Slice Number", fontsize=20)
ax1.set_ylabel("Confidence", fontsize=20)
plt.xticks([0,30,60,90,120,150,180,210],fontsize=12)
plt.yticks(fontsize=12)
ax1.axvline(30,color='gray',ymax=0.1)
ax1.axvline(82,color='gray',ymax=0.1)
ax1.axvline(120,color='gray',ymax=0.1)
ax1.axvline(172,color='gray',ymax=0.1)
ax1.axvline(195,color='gray',ymax=0.1)
plt.plot(chest,linewidth=2,label="Chest")
plt.plot(abd,linewidth=2,label="Abdomen")
plt.plot(pelv,linewidth=2,label="Pelvis")
plt.legend(fontsize=16)
displayImages([testList[30],testList[82],testList[120],testList[172],testList[195]],[30,82,120,172,195])
```
|
github_jupyter
|
!git clone https://github.com/rwfilice/bodypart.git
!pip install pydicom
from scipy.ndimage.interpolation import zoom
import matplotlib.pyplot as plt
import pydicom
import pandas as pd
import numpy as np
import glob
import os
import re
import json
from pathlib import Path
from keras.applications.imagenet_utils import preprocess_input
from keras.applications.mobilenet_v2 import MobileNetV2
from keras.callbacks import EarlyStopping, ReduceLROnPlateau
from keras import Model
from keras.layers import Dropout, Dense, GlobalAveragePooling2D
from keras import optimizers
from keras.models import model_from_json
import tensorflow as tf
# Set seed for reproducibility
tf.random.set_seed(88) ; np.random.seed(88)
# For data augmentation
from albumentations import (
Compose, OneOf, HorizontalFlip, Blur, RandomGamma, RandomContrast, RandomBrightness
)
testPath = Path('bodypart/testnpy')
testList = list(sorted(testPath.glob('**/*.npy'), key=lambda fn: int(re.search('-([0-9]*)', str(fn)).group(1))))
testList
def get_dicom_and_uid(path_to_npy):
'''
Given a filepath, return the npy file and corresponding SOPInstanceUID.
'''
path_to_npy = str(path_to_npy)
dicom_file = np.load(path_to_npy)
uid = path_to_npy.split('/')[-1].replace('.npy', '')
return dicom_file, uid
def convert_dicom_to_8bit(npy_file, width, level, imsize=(224.,224.), clip=True):
'''
Given a DICOM file, window specifications, and image size,
return the image as a Numpy array scaled to [0,255] of the specified size.
'''
array = npy_file.copy()
#array = array + int(dicom_file.RescaleIntercept) #we did this on preprocess
#array = array * int(dicom_file.RescaleSlope) #we did this on preprocess
array = np.clip(array, level - width / 2, level + width / 2)
# Rescale to [0, 255]
array -= np.min(array)
array /= np.max(array)
array *= 255.
array = array.astype('uint8')
if clip:
# Sometimes there is dead space around the images -- let's get rid of that
nonzeros = np.nonzero(array)
x1 = np.min(nonzeros[0]) ; x2 = np.max(nonzeros[0])
y1 = np.min(nonzeros[1]) ; y2 = np.max(nonzeros[1])
array = array[x1:x2,y1:y2]
# Resize image if necessary
resize_x = float(imsize[0]) / array.shape[0]
resize_y = float(imsize[1]) / array.shape[1]
if resize_x != 1. or resize_y != 1.:
array = zoom(array, [resize_x, resize_y], order=1, prefilter=False)
return np.expand_dims(array, axis=-1)
json_file = open('bodypart/model.json', 'r')
loaded_model_json = json_file.read()
json_file.close()
model = model_from_json(loaded_model_json)
model.load_weights('bodypart/tcga-mguh-multilabel.h5') #federated
#Inference
IMSIZE = 256
WINDOW_LEVEL, WINDOW_WIDTH = 50, 500
def predict(model, images, imsize):
'''
Small modifications to data generator to allow for prediction on test data.
'''
test_arrays = []
test_probas = []
test_uids = []
for im in images:
dicom_file, uid = get_dicom_and_uid(im)
try:
array = convert_dicom_to_8bit(dicom_file, WINDOW_WIDTH, WINDOW_LEVEL,
imsize=(imsize,imsize))
except:
continue
array = preprocess_input(array, mode='tf')
test_arrays.append(array)
test_probas.append(model.predict(np.expand_dims(array, axis=0)))
test_uids.append(uid)
return test_uids, test_arrays, test_probas
uids, X, y_prob = predict(model, testList, IMSIZE)
test_pred_df = pd.DataFrame({'uid': uids, 'X': X, 'y_prob': y_prob})
test_pred_df.apply(lambda row: row['y_prob'], axis=1)
chest = np.stack(test_pred_df['y_prob'])[:,0][:,0]
abd = np.stack(test_pred_df['y_prob'])[:,0][:,1]
pelv = np.stack(test_pred_df['y_prob'])[:,0][:,2]
plt.plot(chest)
plt.plot(abd)
plt.plot(pelv)
numaveslices = 5
avepreds = []
allpreds = np.stack(test_pred_df['y_prob'])[:,0]
for idx,arr in enumerate(allpreds):
low = int(max(0,idx-(numaveslices-1)/2))
high = int(min(len(allpreds),idx+(numaveslices+1)/2))
avepreds.append(np.mean(allpreds[low:high],axis=0))
chest = np.stack(avepreds)[:,0]
abd = np.stack(avepreds)[:,1]
pelv = np.stack(avepreds)[:,2]
#averaged over 5 slices
plt.plot(chest)
plt.plot(abd)
plt.plot(pelv)
def displayImages(imgs,labels):
numimgs = len(imgs)
plt.figure(figsize=(20,10))
for idx,img in enumerate(imgs):
dicom_file, uid = get_dicom_and_uid(img)
img = convert_dicom_to_8bit(dicom_file, WINDOW_WIDTH, WINDOW_LEVEL, clip=False)
plt.subplot("1%i%i" % (numimgs,idx+1))
plt.imshow(img[...,0],cmap='gray')
plt.title(labels[idx])
plt.axis('off')
#averaged over 5 slices
fig, ax1 = plt.subplots(figsize=(17,10))
ax1.set_xlabel("Slice Number", fontsize=20)
ax1.set_ylabel("Confidence", fontsize=20)
plt.xticks([0,30,60,90,120,150,180,210],fontsize=12)
plt.yticks(fontsize=12)
ax1.axvline(30,color='gray',ymax=0.1)
ax1.axvline(82,color='gray',ymax=0.1)
ax1.axvline(120,color='gray',ymax=0.1)
ax1.axvline(172,color='gray',ymax=0.1)
ax1.axvline(195,color='gray',ymax=0.1)
plt.plot(chest,linewidth=2,label="Chest")
plt.plot(abd,linewidth=2,label="Abdomen")
plt.plot(pelv,linewidth=2,label="Pelvis")
plt.legend(fontsize=16)
displayImages([testList[30],testList[82],testList[120],testList[172],testList[195]],[30,82,120,172,195])
| 0.655557 | 0.730434 |
# Chapter 2: Single cell simulation with external feedfoward input (with BioNet)
In the previous tutorial we built a single cell and stimulated it with a current injection. In this example we will keep our single-cell network, but instead of stimulation by a step current, we'll set-up an external network that synapses onto our cell.
**Note** - scripts and files for running this tutorial can be found in the directory [sources/chapter02/](sources/chapter02)
**Requirements:**
* Python 2.7 or 3.6+
* bmtk
* NEURON 7.4+
## Step 1: Building the network.
Similar to the previous tutorial, we want to build and save a network consisting of a single biophysically detailed cell.
```
from bmtk.builder.networks import NetworkBuilder
cortex = NetworkBuilder('mcortex')
cortex.add_nodes(cell_name='Scnn1a_473845048',
potental='exc',
model_type='biophysical',
model_template='ctdb:Biophys1.hoc',
model_processing='aibs_perisomatic',
dynamics_params='472363762_fit.json',
morphology='Scnn1a_473845048_m.swc')
cortex.build()
cortex.save_nodes(output_dir='network')
```
But we will also want a collection of external spike-generating cells that will synapse onto our cell. To do this we create a second network which can represent thalamic input. We will call our network "mthalamus", and it will consist of 10 cells. These cells are not biophysical but instead "virtual" cells. Virtual cells don't have a morphology or the normal properties of a neuron, but rather act as spike generators.
```
thalamus = NetworkBuilder('mthalamus')
thalamus.add_nodes(N=10,
pop_name='tON',
potential='exc',
model_type='virtual')
```
Now that we built our nodes, we want to create a connection between our 10 thalamic cells onto our cortex cell. To do so we use the add_edges function like so:
```
thalamus.add_edges(source={'pop_name': 'tON'}, target=cortex.nodes(),
connection_rule=5,
syn_weight=0.001,
delay=2.0,
weight_function=None,
target_sections=['basal', 'apical'],
distance_range=[0.0, 150.0],
dynamics_params='AMPA_ExcToExc.json',
model_template='exp2syn')
```
Let us break down how this method call works:
```python
thalamus.add_edges(source={'pop_name': 'tON'}, target=cortex.nodes(),
```
* Here we specify which set of nodes to use as sources and targets. Our source/pre-synaptic cells are all thamalus cells with the property "pop_name=tON", which in this case is every thalmus cell (We could also use source=thalamus.nodes(), or source={'level_of_detail': 'filter'}). The target/post-synaptic is all cell(s) of the "cortex" network.
```python
connection_rule=5,
```
* The connection_rule parameter determines how many synapses exists between every source/target pair. In this very trivial case we are indicating that between every thamalic --> cortical cell connection, there are 5 synapatic connections. In future tutorials we will show how we can create more complex customized rules.
```python
syn_weight=0.001,
delay=2.0,
weight_function=None,
```
* Here we are specifying the connection weight. For every connection in this edge-type, there is a connection strenght of 5e-05 (units) and a connection dealy of 2 ms. The weight function is used to adjusted the weights before runtime. Later we will show how to create customized weight functions.
```python
target_sections=['basal', 'apical'],
distance_range=[0.0, 150.0],
```
* This is used by BioNet to determine where on the post-synaptic cell to place the synapse. By default placement is random within the given section and range.
```python
dynamics_params='AMPA_ExcToExc.json',
model_template='exp2syn')
```
* The params_file give the parameters of the synpase, including the time constant and potential. Here we are using an AMPA type synaptic model with an Excitatory connection. The set_params_function is used by BioNet to convert the model into a valid NEURON synaptic object.
Finally we are ready to build the model and save the thalamic nodes and edges.
```
thalamus.build()
thalamus.save_nodes(output_dir='network')
thalamus.save_edges(output_dir='network')
```
The network/ directory will contain multiple nodes and edges files. It should have nodes (and node-types) files for both the thalamus and cortex network. And edges (and edge-types) files for the thalamus --> cortex connections. Nodes and edges for different networks and their connections are spread out across different files which allows us in the future to rebuild, edit or replace part of setup in a piecemeal and efficent manner.
## Step 2: Setting up BioNet environment.
#### file structure.
Before running a simulation, we will need to create the runtime environment, including parameter files, run-script and configuration files. If using the tutorial these files will already be in place. Otherwise we can use a command-line:
```bash
$ python -m bmtk.utils.sim_setup -n network --membrane_report-vars v,cai --membrane_report-sections soma --tstop 2000.0 --dt 0.1 bionet
```
Also our cortex cell uses a Scnn1a model we can download from the Allen Cell-Types Database
```bash
$ wget http://celltypes.brain-map.org/neuronal_model/download/482934212
$ unzip 482934212
$ cp fit_parameters.json biophys_components/biophysical_neuron_templates/472363762_fit.json
$ cp reconstruction.swc biophys_components/morphologies/Scnn1a_473845048_m.swc
```
#### Spike Trains
We need to give our 10 thalamic cells spike trains. There are multiple ways to do this, but an easy way to use a csv file. The following function will create a file to provide the spikes for our 10 cells.
```
from bmtk.utils.spike_trains import SpikesGenerator
sg = SpikesGenerator(nodes='network/mthalamus_nodes.h5', t_max=3.0)
sg.set_rate(10.0)
sg.save_csv('thalamus_spikes.csv', in_ms=True)
```
The spikes file consists of 10 rows with 2 columns; the gid and a list of spike times (in milliseconds). Thus you can create your own if you want.
```
import pandas as pd
pd.read_csv('thalamus_spikes.csv', sep=' ')
```
The last thing that we need to do is to update the configuration file to read "thalamus_spikes.csv". To do so we open simulation_config.json in a text editor and add the following to the **input** section.
```json
"inputs": {
"lgn_spikes": {
"input_type": "spikes",
"module": "csv",
"input_file": "${BASE_DIR}/thalamus_spikes.csv",
"node_set": "mthalamus"
}
}
```
## 3. Running the simulation
Once our config file is setup we can run a simulation either through the command line:
```bash
$ python run_bionet.py config.json
```
or through the script
```
from bmtk.simulator import bionet
conf = bionet.Config.from_json('simulation_config.json')
conf.build_env()
net = bionet.BioNetwork.from_config(conf)
sim = bionet.BioSimulator.from_config(conf, network=net)
sim.run()
```
## 4. Analyzing the run
```
from bmtk.analyzer.spike_trains import to_dataframe
to_dataframe(config_file='simulation_config.json')
from bmtk.analyzer.cell_vars import plot_report
plot_report(config_file='simulation_config.json')
```
## 5. Additional things to do:
### Changing edge properties
When using the Network Builder add_edges method, we gave all the edges the same parameter values (delay, weight, target_section, etc.). All connection created by this method call constitute an single edge-type that share the same parameters, and are specified in the mthalamic_edge_type.csv file
```
import pandas as pd
pd.read_csv('network/mthalamus_mcortex_edge_types.csv', sep=' ')
```
(if in the build script we called add_edges multiple times, we'd have multiple edge-types).
Using a simple text-editor we can modify this file directly, change parameters before a simulation run without having to rebuild the entire network (although for a network this small it may not be beneficial).
#### weight_function
By default BioNet uses the value in syn_weight to set a synaptic weight, which is a constant stored in the network files. Often we will want to adjust the synaptic weight between simulations, but don't want to have to regenerate the network. BioNet allows us to specify custom synaptic weight functions that will calculate synaptic weight before each simulation.
To do so first we must set the value of 'weight_function' column. Either we can open up the file mthalamus_mcortex_edge_types.csv with a text-editor and change the column.
|edge_type_id | target_query | source_query | ... | weight_function |
|-------------|--------------|----------------|-----|-----------------|
|100 | * |pop_name=='tON' | ... |*adjusted_weight* |
or we can rebuild the edges
```python
thalamus.add_edges(source={'pop_name': 'tON'}, target=cortex.nodes(),
connection_rule=5,
syn_weight=0.001,
weight_function=adjusted_weight,
delay=2.0,
target_sections=['basal', 'apical'],
distance_range=[0.0, 150.0],
dynamics_params='AMPA_ExcToExc.json',
model_template='exp2syn')
```
Then we write a custom weight function. The weight functions will be called during the simulation when building each synapse, and requires three parameters - target_cell, source_cell, and edge_props. These three parameters are dictionaries which can be used to access properties of the source node, target node, and edge, respectively. The function must return a floating point number which will be used to set the synaptic weight
```python
def adjusted_weights(target_cell, source_cell, edge_props):
if target_cell['cell_name'] == 'Scnn1a':
return edge_prop["weight_max"]*0.5
elif target_cell['cell_name'] == 'Rorb'
return edge_prop["weight_max"]*1.5
else:
...
```
Finally we must tell BioNet where to access the function which we can do by using the add_weight_function.
```python
from bmtk.simulator import bionet
bionet.nrn.add_weight_function(adjusted_weights)
conf = bionet.Config.from_json('config.json')
...
```
### Using NWB for spike trains
Instead of using csv files to set the spike trains of our external network, we can also use nwb files. The typical setup would look like the following in the config file:
```json
"inputs": {
"LGN_spikes": {
"input_type": "spikes",
"module": "nwb",
"input_file": "$INPUT_DIR/lgn_spikes.nwb",
"node_set": "lgn",
"trial": "trial_0"
},
}
```
|
github_jupyter
|
from bmtk.builder.networks import NetworkBuilder
cortex = NetworkBuilder('mcortex')
cortex.add_nodes(cell_name='Scnn1a_473845048',
potental='exc',
model_type='biophysical',
model_template='ctdb:Biophys1.hoc',
model_processing='aibs_perisomatic',
dynamics_params='472363762_fit.json',
morphology='Scnn1a_473845048_m.swc')
cortex.build()
cortex.save_nodes(output_dir='network')
thalamus = NetworkBuilder('mthalamus')
thalamus.add_nodes(N=10,
pop_name='tON',
potential='exc',
model_type='virtual')
thalamus.add_edges(source={'pop_name': 'tON'}, target=cortex.nodes(),
connection_rule=5,
syn_weight=0.001,
delay=2.0,
weight_function=None,
target_sections=['basal', 'apical'],
distance_range=[0.0, 150.0],
dynamics_params='AMPA_ExcToExc.json',
model_template='exp2syn')
thalamus.add_edges(source={'pop_name': 'tON'}, target=cortex.nodes(),
connection_rule=5,
syn_weight=0.001,
delay=2.0,
weight_function=None,
target_sections=['basal', 'apical'],
distance_range=[0.0, 150.0],
dynamics_params='AMPA_ExcToExc.json',
model_template='exp2syn')
thalamus.build()
thalamus.save_nodes(output_dir='network')
thalamus.save_edges(output_dir='network')
$ python -m bmtk.utils.sim_setup -n network --membrane_report-vars v,cai --membrane_report-sections soma --tstop 2000.0 --dt 0.1 bionet
$ wget http://celltypes.brain-map.org/neuronal_model/download/482934212
$ unzip 482934212
$ cp fit_parameters.json biophys_components/biophysical_neuron_templates/472363762_fit.json
$ cp reconstruction.swc biophys_components/morphologies/Scnn1a_473845048_m.swc
from bmtk.utils.spike_trains import SpikesGenerator
sg = SpikesGenerator(nodes='network/mthalamus_nodes.h5', t_max=3.0)
sg.set_rate(10.0)
sg.save_csv('thalamus_spikes.csv', in_ms=True)
import pandas as pd
pd.read_csv('thalamus_spikes.csv', sep=' ')
"inputs": {
"lgn_spikes": {
"input_type": "spikes",
"module": "csv",
"input_file": "${BASE_DIR}/thalamus_spikes.csv",
"node_set": "mthalamus"
}
}
$ python run_bionet.py config.json
from bmtk.simulator import bionet
conf = bionet.Config.from_json('simulation_config.json')
conf.build_env()
net = bionet.BioNetwork.from_config(conf)
sim = bionet.BioSimulator.from_config(conf, network=net)
sim.run()
from bmtk.analyzer.spike_trains import to_dataframe
to_dataframe(config_file='simulation_config.json')
from bmtk.analyzer.cell_vars import plot_report
plot_report(config_file='simulation_config.json')
import pandas as pd
pd.read_csv('network/mthalamus_mcortex_edge_types.csv', sep=' ')
thalamus.add_edges(source={'pop_name': 'tON'}, target=cortex.nodes(),
connection_rule=5,
syn_weight=0.001,
weight_function=adjusted_weight,
delay=2.0,
target_sections=['basal', 'apical'],
distance_range=[0.0, 150.0],
dynamics_params='AMPA_ExcToExc.json',
model_template='exp2syn')
def adjusted_weights(target_cell, source_cell, edge_props):
if target_cell['cell_name'] == 'Scnn1a':
return edge_prop["weight_max"]*0.5
elif target_cell['cell_name'] == 'Rorb'
return edge_prop["weight_max"]*1.5
else:
...
from bmtk.simulator import bionet
bionet.nrn.add_weight_function(adjusted_weights)
conf = bionet.Config.from_json('config.json')
...
"inputs": {
"LGN_spikes": {
"input_type": "spikes",
"module": "nwb",
"input_file": "$INPUT_DIR/lgn_spikes.nwb",
"node_set": "lgn",
"trial": "trial_0"
},
}
| 0.570571 | 0.97024 |
```
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
from astropy.table import Table
fast = np.loadtxt('data/epics_fast.txt')
slow = np.loadtxt('data/epics_slow.txt')
superfast = np.loadtxt('data/epics_superfast.txt')
from astropy.stats import mad_std
douglas = Table.read('data/douglas2017.vot')
douglas.add_index('EPIC')
from scipy.ndimage import gaussian_filter1d
from scipy.optimize import minimize
from scipy.stats import skew
from interpacf import interpolated_acf
plots = False
smoothed_amps_fast = dict()
newstat_fast = dict()
periods = dict()
baseline_flux_at_flares = []
smoothed_flux_dist = []
for i in range(len(fast)):
times, fluxes = np.load("data/{0}.npy".format(int(fast[i])))
clipped = ~np.isnan(fluxes)
times, fluxes = times[clipped], fluxes[clipped]
clip_flares = np.abs(fluxes - np.nanmedian(fluxes)) < 5*mad_std(fluxes, ignore_nan=True)
# Remove polynomial trend
fit = np.polyval(np.polyfit(times[clip_flares]-times.mean(), fluxes[clip_flares], 5), times-times.mean())
fluxes /= fit
period = douglas.loc[fast[i]]['Prot1']
phases = (times % period) / period
sort = np.argsort(phases)
sort_clipped = np.argsort(phases[clip_flares])
smoothed = gaussian_filter1d(fluxes[clip_flares][sort_clipped], 50, mode='nearest')
smoothed_sorted = smoothed[np.argsort(times[sort_clipped])]
interp_smoothed = np.interp(times, times[clip_flares], smoothed_sorted)
outliers = (fluxes - interp_smoothed) > 0.015 #np.percentile(fluxes, 95)
smoothed_amps_fast[fast[i]] = smoothed.max() - smoothed.min()
# fft = np.abs(np.fft.rfft(fluxes))**2
# freq = np.fft.rfftfreq(len(fluxes), times[1]-times[0])
newstat_fast[fast[i]] = douglas.loc[fast[i]]["Pw1"]#fft[np.abs(freq - 1/period).argmin()]
periods[fast[i]] = period
if np.count_nonzero(outliers) < 100:
baseline_flux_at_flares.append(interp_smoothed[outliers])#[np.argmax(fluxes[outliers])])
smoothed_flux_dist.append(smoothed)
if plots:
fig, ax = plt.subplots(1, 3, figsize=(16, 3))
ax[0].plot(times, fluxes)
ax[1].plot(phases, fluxes, '.', alpha=0.5)
ax[1].plot(phases[clip_flares][sort_clipped], smoothed, 'r')
ax[2].loglog(freq, fft)
ax[2].axhline(fft[1:].max())
ax[2].axvline(1/period)
# ax[2].axvline(freq[fft.argmax()])
# ax[2].plot(times[outliers], fluxes[outliers], '.', alpha=0.5)
# ax[2].plot(times[~outliers], fluxes[~outliers], '.', alpha=0.5)
# ax[2].plot(times, interp_smoothed, ',')
# ax[3].plot(interp_smoothed[outliers], fluxes[outliers], '.')
ax[1].axhline(0.99*smoothed.min())
ax[1].axhline(1.01*smoothed.max())
ax[1].set_ylim([smoothed.min(), smoothed.max()])
ax[1].axhline(np.median(fluxes[sort]), ls='--', color='k')
ax[1].axhline(np.mean(fluxes[sort]), ls='-.', color='gray')
ax[1].set_title("{0}".format(newstat_fast[fast[i]]))
plt.show()
plt.hist(np.hstack(smoothed_flux_dist), bins=100, density=True, lw=2, histtype='step');
plt.hist(np.hstack(baseline_flux_at_flares), bins=100, density=True, lw=2, histtype='step');
from scipy.stats import anderson_ksamp
print(anderson_ksamp([np.hstack(smoothed_flux_dist), np.hstack(baseline_flux_at_flares)]))
plots = False
smoothed_amps_slow = dict()
newstat_slow = dict()
baseline_flux_at_flares = []
smoothed_flux_dist = []
for i in range(len(slow)):
times, fluxes = np.load("data/{0}.npy".format(int(slow[i])))
if hasattr(times, "__len__"):
times, fluxes = np.load("data/{0}.npy".format(int(slow[i])))
clipped = ~np.isnan(fluxes)
times, fluxes = times[clipped], fluxes[clipped]
clip_flares = np.abs(fluxes - np.nanmedian(fluxes)) < 5*mad_std(fluxes, ignore_nan=True)
# Remove polynomial trend
fit = np.polyval(np.polyfit(times[clip_flares]-times.mean(), fluxes[clip_flares], 5), times-times.mean())
fluxes /= fit
period = douglas.loc[slow[i]]['Prot1']
phases = (times % period) / period
sort = np.argsort(phases)
sort_clipped = np.argsort(phases[clip_flares])
smoothed = gaussian_filter1d(fluxes[clip_flares][sort_clipped], 50, mode='nearest')
smoothed_sorted = smoothed[np.argsort(times[sort_clipped])]
interp_smoothed = np.interp(times, times[clip_flares], smoothed_sorted)
outliers = (fluxes - interp_smoothed) > 0.015 #np.percentile(fluxes, 95)
smoothed_amps_slow[slow[i]] = smoothed.max() - smoothed.min()
# fft = np.abs(np.fft.rfft(fluxes))**2
# freq = np.fft.rfftfreq(len(fluxes), times[1]-times[0])
if np.count_nonzero(outliers) < 100:
baseline_flux_at_flares.append(interp_smoothed[outliers])#[np.argmax(fluxes[outliers])])
smoothed_flux_dist.append(smoothed)
plt.hist(np.hstack(smoothed_flux_dist), bins=100, density=True, lw=2, histtype='step');
plt.hist(np.hstack(baseline_flux_at_flares), bins=100, density=True, lw=2, histtype='step');
print(anderson_ksamp([np.hstack(smoothed_flux_dist), np.hstack(baseline_flux_at_flares)]))
plots = False
smoothed_amps_superfast = dict()
newstat_superfast = dict()
baseline_flux_at_flares = []
smoothed_flux_dist = []
for i in range(len(superfast)):
times, fluxes = np.load("data/{0}.npy".format(int(superfast[i])))
if hasattr(times, "__len__"):
times, fluxes = np.load("data/{0}.npy".format(int(superfast[i])))
clipped = ~np.isnan(fluxes)
times, fluxes = times[clipped], fluxes[clipped]
clip_flares = np.abs(fluxes - np.nanmedian(fluxes)) < 5*mad_std(fluxes, ignore_nan=True)
# Remove polynomial trend
fit = np.polyval(np.polyfit(times[clip_flares]-times.mean(), fluxes[clip_flares], 5), times-times.mean())
fluxes /= fit
phases = (times % period) / period
sort = np.argsort(phases)
sort_clipped = np.argsort(phases[clip_flares])
smoothed = gaussian_filter1d(fluxes[clip_flares][sort_clipped], 50, mode='nearest')
smoothed_sorted = smoothed[np.argsort(times[sort_clipped])]
interp_smoothed = np.interp(times, times[clip_flares], smoothed_sorted)
outliers = (fluxes - interp_smoothed) > 0.015 #np.percentile(fluxes, 95)
smoothed_amps_superfast[superfast[i]] = smoothed.max() - smoothed.min()
# fft = np.abs(np.fft.rfft(fluxes))**2
# freq = np.fft.rfftfreq(len(fluxes), times[1]-times[0])
if np.count_nonzero(outliers) < 100:
baseline_flux_at_flares.append(interp_smoothed[outliers])#[np.argmax(fluxes[outliers])])
smoothed_flux_dist.append(smoothed)
plt.hist(np.hstack(smoothed_flux_dist), bins=100, density=True, lw=2, histtype='step');
plt.hist(np.hstack(baseline_flux_at_flares), bins=100, density=True, lw=2, histtype='step');
print(anderson_ksamp([np.hstack(smoothed_flux_dist), np.hstack(baseline_flux_at_flares)]))
```
|
github_jupyter
|
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
from astropy.table import Table
fast = np.loadtxt('data/epics_fast.txt')
slow = np.loadtxt('data/epics_slow.txt')
superfast = np.loadtxt('data/epics_superfast.txt')
from astropy.stats import mad_std
douglas = Table.read('data/douglas2017.vot')
douglas.add_index('EPIC')
from scipy.ndimage import gaussian_filter1d
from scipy.optimize import minimize
from scipy.stats import skew
from interpacf import interpolated_acf
plots = False
smoothed_amps_fast = dict()
newstat_fast = dict()
periods = dict()
baseline_flux_at_flares = []
smoothed_flux_dist = []
for i in range(len(fast)):
times, fluxes = np.load("data/{0}.npy".format(int(fast[i])))
clipped = ~np.isnan(fluxes)
times, fluxes = times[clipped], fluxes[clipped]
clip_flares = np.abs(fluxes - np.nanmedian(fluxes)) < 5*mad_std(fluxes, ignore_nan=True)
# Remove polynomial trend
fit = np.polyval(np.polyfit(times[clip_flares]-times.mean(), fluxes[clip_flares], 5), times-times.mean())
fluxes /= fit
period = douglas.loc[fast[i]]['Prot1']
phases = (times % period) / period
sort = np.argsort(phases)
sort_clipped = np.argsort(phases[clip_flares])
smoothed = gaussian_filter1d(fluxes[clip_flares][sort_clipped], 50, mode='nearest')
smoothed_sorted = smoothed[np.argsort(times[sort_clipped])]
interp_smoothed = np.interp(times, times[clip_flares], smoothed_sorted)
outliers = (fluxes - interp_smoothed) > 0.015 #np.percentile(fluxes, 95)
smoothed_amps_fast[fast[i]] = smoothed.max() - smoothed.min()
# fft = np.abs(np.fft.rfft(fluxes))**2
# freq = np.fft.rfftfreq(len(fluxes), times[1]-times[0])
newstat_fast[fast[i]] = douglas.loc[fast[i]]["Pw1"]#fft[np.abs(freq - 1/period).argmin()]
periods[fast[i]] = period
if np.count_nonzero(outliers) < 100:
baseline_flux_at_flares.append(interp_smoothed[outliers])#[np.argmax(fluxes[outliers])])
smoothed_flux_dist.append(smoothed)
if plots:
fig, ax = plt.subplots(1, 3, figsize=(16, 3))
ax[0].plot(times, fluxes)
ax[1].plot(phases, fluxes, '.', alpha=0.5)
ax[1].plot(phases[clip_flares][sort_clipped], smoothed, 'r')
ax[2].loglog(freq, fft)
ax[2].axhline(fft[1:].max())
ax[2].axvline(1/period)
# ax[2].axvline(freq[fft.argmax()])
# ax[2].plot(times[outliers], fluxes[outliers], '.', alpha=0.5)
# ax[2].plot(times[~outliers], fluxes[~outliers], '.', alpha=0.5)
# ax[2].plot(times, interp_smoothed, ',')
# ax[3].plot(interp_smoothed[outliers], fluxes[outliers], '.')
ax[1].axhline(0.99*smoothed.min())
ax[1].axhline(1.01*smoothed.max())
ax[1].set_ylim([smoothed.min(), smoothed.max()])
ax[1].axhline(np.median(fluxes[sort]), ls='--', color='k')
ax[1].axhline(np.mean(fluxes[sort]), ls='-.', color='gray')
ax[1].set_title("{0}".format(newstat_fast[fast[i]]))
plt.show()
plt.hist(np.hstack(smoothed_flux_dist), bins=100, density=True, lw=2, histtype='step');
plt.hist(np.hstack(baseline_flux_at_flares), bins=100, density=True, lw=2, histtype='step');
from scipy.stats import anderson_ksamp
print(anderson_ksamp([np.hstack(smoothed_flux_dist), np.hstack(baseline_flux_at_flares)]))
plots = False
smoothed_amps_slow = dict()
newstat_slow = dict()
baseline_flux_at_flares = []
smoothed_flux_dist = []
for i in range(len(slow)):
times, fluxes = np.load("data/{0}.npy".format(int(slow[i])))
if hasattr(times, "__len__"):
times, fluxes = np.load("data/{0}.npy".format(int(slow[i])))
clipped = ~np.isnan(fluxes)
times, fluxes = times[clipped], fluxes[clipped]
clip_flares = np.abs(fluxes - np.nanmedian(fluxes)) < 5*mad_std(fluxes, ignore_nan=True)
# Remove polynomial trend
fit = np.polyval(np.polyfit(times[clip_flares]-times.mean(), fluxes[clip_flares], 5), times-times.mean())
fluxes /= fit
period = douglas.loc[slow[i]]['Prot1']
phases = (times % period) / period
sort = np.argsort(phases)
sort_clipped = np.argsort(phases[clip_flares])
smoothed = gaussian_filter1d(fluxes[clip_flares][sort_clipped], 50, mode='nearest')
smoothed_sorted = smoothed[np.argsort(times[sort_clipped])]
interp_smoothed = np.interp(times, times[clip_flares], smoothed_sorted)
outliers = (fluxes - interp_smoothed) > 0.015 #np.percentile(fluxes, 95)
smoothed_amps_slow[slow[i]] = smoothed.max() - smoothed.min()
# fft = np.abs(np.fft.rfft(fluxes))**2
# freq = np.fft.rfftfreq(len(fluxes), times[1]-times[0])
if np.count_nonzero(outliers) < 100:
baseline_flux_at_flares.append(interp_smoothed[outliers])#[np.argmax(fluxes[outliers])])
smoothed_flux_dist.append(smoothed)
plt.hist(np.hstack(smoothed_flux_dist), bins=100, density=True, lw=2, histtype='step');
plt.hist(np.hstack(baseline_flux_at_flares), bins=100, density=True, lw=2, histtype='step');
print(anderson_ksamp([np.hstack(smoothed_flux_dist), np.hstack(baseline_flux_at_flares)]))
plots = False
smoothed_amps_superfast = dict()
newstat_superfast = dict()
baseline_flux_at_flares = []
smoothed_flux_dist = []
for i in range(len(superfast)):
times, fluxes = np.load("data/{0}.npy".format(int(superfast[i])))
if hasattr(times, "__len__"):
times, fluxes = np.load("data/{0}.npy".format(int(superfast[i])))
clipped = ~np.isnan(fluxes)
times, fluxes = times[clipped], fluxes[clipped]
clip_flares = np.abs(fluxes - np.nanmedian(fluxes)) < 5*mad_std(fluxes, ignore_nan=True)
# Remove polynomial trend
fit = np.polyval(np.polyfit(times[clip_flares]-times.mean(), fluxes[clip_flares], 5), times-times.mean())
fluxes /= fit
phases = (times % period) / period
sort = np.argsort(phases)
sort_clipped = np.argsort(phases[clip_flares])
smoothed = gaussian_filter1d(fluxes[clip_flares][sort_clipped], 50, mode='nearest')
smoothed_sorted = smoothed[np.argsort(times[sort_clipped])]
interp_smoothed = np.interp(times, times[clip_flares], smoothed_sorted)
outliers = (fluxes - interp_smoothed) > 0.015 #np.percentile(fluxes, 95)
smoothed_amps_superfast[superfast[i]] = smoothed.max() - smoothed.min()
# fft = np.abs(np.fft.rfft(fluxes))**2
# freq = np.fft.rfftfreq(len(fluxes), times[1]-times[0])
if np.count_nonzero(outliers) < 100:
baseline_flux_at_flares.append(interp_smoothed[outliers])#[np.argmax(fluxes[outliers])])
smoothed_flux_dist.append(smoothed)
plt.hist(np.hstack(smoothed_flux_dist), bins=100, density=True, lw=2, histtype='step');
plt.hist(np.hstack(baseline_flux_at_flares), bins=100, density=True, lw=2, histtype='step');
print(anderson_ksamp([np.hstack(smoothed_flux_dist), np.hstack(baseline_flux_at_flares)]))
| 0.457379 | 0.595493 |
```
# From here on, operate on train_set only.
# Later, we will automate all processing so it can be repeated on test_set.
# This page represents exploration of the ideas.
import pandas as pd
datapath="/Users/jasonmiller/Source/MachineLearning/datasets/housing/housing.csv"
all_data=pd.read_csv(datapath)
from sklearn.model_selection import train_test_split
train_set, test_set = train_test_split(all_data,test_size=0.2,random_state=42)
train_predictors = train_set.drop(["median_house_value"],axis=1)
train_labels = train_set["median_house_value"].copy()
train_set.head()
# Return to issue of missing data.
# Recall there were 207 null bedroom counts
# By chance, all of them ended up in test_set, but we will process train_set anyway.
# Generalize the fix to all columns since real data could have NaN anywhere.
# Choice 1 = remove whole rows i.e. data points: df.dropna(subset=["total_bedrooms"])
# Choice 2 = remove whole columns i.e. features: df.drop("total_bedrooms",axis=1)
# Choice 3 = change NaN to 3000 i.e. df["total_bedrooms"].fillna(3000,inplace=True)
# Choice 4 = use an imputer. Hits every column for us.
# Unfortunately, requires us to put aside non-numeric columns.
# Using median impute will perhaps have the least effect on outcome.
from sklearn.impute import SimpleImputer
def cleanse_NaN (df):
imputer=SimpleImputer(strategy="median")
numeric_only_df = df.drop("ocean_proximity",axis=1) # returns new data frame; original is not changed
imputer.fit(numeric_only_df)
numpy_array = imputer.transform(numeric_only_df) # replace NaN with column median in every column
transformed_df = pd.DataFrame(numpy_array,columns=numeric_only_df.columns,
index=numeric_only_df.index)
transformed_df["ocean_proximity"] = df["ocean_proximity"]
transformed_df.describe()
return transformed_df
train_set = cleanse_NaN(train_set)
train_set.head()
# Return to issue of categorical data.
# Seek to replace the quirky text in ocean_proximity.
train_set["ocean_proximity"].unique()
# This notation would extract column as pandas Series.
type(train_set["ocean_proximity"])
# In pandas, single brackets extracts a Series, double brackets extracts a DataFrame.
type(train_set[["ocean_proximity"]])
# First try: Convert text to ordinal.
import numpy as np
proximity = train_set[["ocean_proximity"]]
from sklearn.preprocessing import OrdinalEncoder
encoder = OrdinalEncoder()
numpy_array = encoder.fit_transform(proximity)
np.unique(numpy_array)
# This is bad since we don't want ML to think (0,1) are similar while (0,4) are different.
# Second try: One Hot Encoding.
from sklearn.preprocessing import OneHotEncoder
encoder = OneHotEncoder()
scipi_sparse_matrix = encoder.fit_transform(proximity)
scipi_sparse_matrix.toarray()
# This is good. Luckily, sklearn can deal with this data type.
# Return to isse of feature scaling.
# Most ML won't work with different ranges and different skew.
# First try: min-max scaling (i.e. normalization).
# All values mapped to range (0,1): new=(old-min)/(max-min).
# Requires all columns are numeric.
numeric_only_df = train_set.drop("ocean_proximity",axis=1)
from sklearn.preprocessing import MinMaxScaler
scaler = MinMaxScaler()
scaler.fit(numeric_only_df)
numpy_array = scaler.transform(numeric_only_df)
numpy_array
# Second try: standardization (i.e. z-scores with unit variance)
numeric_only_df = train_set.drop("ocean_proximity",axis=1)
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
scaler.fit(numeric_only_df)
numpy_array = scaler.transform(numeric_only_df)
numpy_array
# Done!
# Ready to make a transformation pipeline
```
|
github_jupyter
|
# From here on, operate on train_set only.
# Later, we will automate all processing so it can be repeated on test_set.
# This page represents exploration of the ideas.
import pandas as pd
datapath="/Users/jasonmiller/Source/MachineLearning/datasets/housing/housing.csv"
all_data=pd.read_csv(datapath)
from sklearn.model_selection import train_test_split
train_set, test_set = train_test_split(all_data,test_size=0.2,random_state=42)
train_predictors = train_set.drop(["median_house_value"],axis=1)
train_labels = train_set["median_house_value"].copy()
train_set.head()
# Return to issue of missing data.
# Recall there were 207 null bedroom counts
# By chance, all of them ended up in test_set, but we will process train_set anyway.
# Generalize the fix to all columns since real data could have NaN anywhere.
# Choice 1 = remove whole rows i.e. data points: df.dropna(subset=["total_bedrooms"])
# Choice 2 = remove whole columns i.e. features: df.drop("total_bedrooms",axis=1)
# Choice 3 = change NaN to 3000 i.e. df["total_bedrooms"].fillna(3000,inplace=True)
# Choice 4 = use an imputer. Hits every column for us.
# Unfortunately, requires us to put aside non-numeric columns.
# Using median impute will perhaps have the least effect on outcome.
from sklearn.impute import SimpleImputer
def cleanse_NaN (df):
imputer=SimpleImputer(strategy="median")
numeric_only_df = df.drop("ocean_proximity",axis=1) # returns new data frame; original is not changed
imputer.fit(numeric_only_df)
numpy_array = imputer.transform(numeric_only_df) # replace NaN with column median in every column
transformed_df = pd.DataFrame(numpy_array,columns=numeric_only_df.columns,
index=numeric_only_df.index)
transformed_df["ocean_proximity"] = df["ocean_proximity"]
transformed_df.describe()
return transformed_df
train_set = cleanse_NaN(train_set)
train_set.head()
# Return to issue of categorical data.
# Seek to replace the quirky text in ocean_proximity.
train_set["ocean_proximity"].unique()
# This notation would extract column as pandas Series.
type(train_set["ocean_proximity"])
# In pandas, single brackets extracts a Series, double brackets extracts a DataFrame.
type(train_set[["ocean_proximity"]])
# First try: Convert text to ordinal.
import numpy as np
proximity = train_set[["ocean_proximity"]]
from sklearn.preprocessing import OrdinalEncoder
encoder = OrdinalEncoder()
numpy_array = encoder.fit_transform(proximity)
np.unique(numpy_array)
# This is bad since we don't want ML to think (0,1) are similar while (0,4) are different.
# Second try: One Hot Encoding.
from sklearn.preprocessing import OneHotEncoder
encoder = OneHotEncoder()
scipi_sparse_matrix = encoder.fit_transform(proximity)
scipi_sparse_matrix.toarray()
# This is good. Luckily, sklearn can deal with this data type.
# Return to isse of feature scaling.
# Most ML won't work with different ranges and different skew.
# First try: min-max scaling (i.e. normalization).
# All values mapped to range (0,1): new=(old-min)/(max-min).
# Requires all columns are numeric.
numeric_only_df = train_set.drop("ocean_proximity",axis=1)
from sklearn.preprocessing import MinMaxScaler
scaler = MinMaxScaler()
scaler.fit(numeric_only_df)
numpy_array = scaler.transform(numeric_only_df)
numpy_array
# Second try: standardization (i.e. z-scores with unit variance)
numeric_only_df = train_set.drop("ocean_proximity",axis=1)
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
scaler.fit(numeric_only_df)
numpy_array = scaler.transform(numeric_only_df)
numpy_array
# Done!
# Ready to make a transformation pipeline
| 0.652906 | 0.593167 |
<a href="https://colab.research.google.com/github/nelslindahlx/NLP/blob/master/GCH_working_GPT2_corpus_example.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
This was working code and the corpus file was live on 6/8/2020.
> Please note that synthetic writing could be mean or otherwise terrible... My apologies in advance
# Working example of GPT-2 using gpt-2-simple
Want to know what GPU you have? Run the command below...
```
!nvidia-smi
```
The best method to use the OpenAI GPT-2 model is via gpt-2-simple. It works well enough. Some methods that I tried and that failed are commented out before it. The only method that seems to work is the gpt-2-simple
```
# !pip uninstall tensorflow -y
# !pip uninstall tensorflow-gpu -y
# !pip install tensorflow==1.14
# !pip install tensorflow-gpu==1.4.1
# !pip3 install tensorflow==1.12.0
!pip install gpt-2-simple
```
You can now use the 1x TensorFlow selection and check what GPU you have.
```
%tensorflow_version 1.x
```
This is our import of the GPT-2 we installed above. You get the warning that is the root of the GPT-2 problem noting that "The TensorFlow contrib module will not be included in TensorFlow 2.0.'
```
import gpt_2_simple as gpt2
import os
import requests
```
This snippet is using the 355M model. The other model numbers are commented out above for later adventure
```
# !python3 download_model.py 124M
# !python3 download_model.py 355M
# !python3 download_model.py 774M
# !python3 download_model.py 1558M
model_name = "355M"
if not os.path.isdir(os.path.join("models", model_name)):
print(f"Downloading {model_name} model...")
gpt2.download_gpt2(model_name=model_name) # model is saved into current directory under /content/models
```
# Getting the corpus file
To be able to go get files we need gdown
```
pip install gdown
```
We need to download the corpus file to do some training on it... This one gets the corpus I plan on using from a shareable Google Drive link
```
!gdown --id 1-VdKEgB-LpDSPtiIsI6iDfoDlN2BUAgz --output GCH.txt
```
Set the file name to my Graduation with Civic Honors corpus file GCH.txt...
```
file_name = "GCH.txt"
```
This will take 20 to 30 minutes to run...
```
sess = gpt2.start_tf_sess()
gpt2.finetune(sess,
dataset=file_name,
model_name='355M',
steps=2000,
restore_from='fresh',
run_name='run1',
print_every=10,
sample_every=200,
save_every=500
)
```
# Let's make some outputs
This prompt will generate text from the trained model
```
gpt2.generate(sess, run_name='run1')
```
You can change the prefix text, but it spews nonsense so far...
```
single_text = gpt2.generate(sess, prefix="Civil society will")
print(single_text)
```
|
github_jupyter
|
!nvidia-smi
# !pip uninstall tensorflow -y
# !pip uninstall tensorflow-gpu -y
# !pip install tensorflow==1.14
# !pip install tensorflow-gpu==1.4.1
# !pip3 install tensorflow==1.12.0
!pip install gpt-2-simple
%tensorflow_version 1.x
import gpt_2_simple as gpt2
import os
import requests
# !python3 download_model.py 124M
# !python3 download_model.py 355M
# !python3 download_model.py 774M
# !python3 download_model.py 1558M
model_name = "355M"
if not os.path.isdir(os.path.join("models", model_name)):
print(f"Downloading {model_name} model...")
gpt2.download_gpt2(model_name=model_name) # model is saved into current directory under /content/models
pip install gdown
!gdown --id 1-VdKEgB-LpDSPtiIsI6iDfoDlN2BUAgz --output GCH.txt
file_name = "GCH.txt"
sess = gpt2.start_tf_sess()
gpt2.finetune(sess,
dataset=file_name,
model_name='355M',
steps=2000,
restore_from='fresh',
run_name='run1',
print_every=10,
sample_every=200,
save_every=500
)
gpt2.generate(sess, run_name='run1')
single_text = gpt2.generate(sess, prefix="Civil society will")
print(single_text)
| 0.196865 | 0.929055 |
# Pyber Challenge
### 4.3 Loading and Reading CSV files
```
# Challenge psuedo-code.
# - Use groupby(), function with count() and sum()
# - Get Total number of rides,
# number of drivers,
# total fares,
# for each city type.
# Then, Calculate
# Avg. fare per ride,
# Avg. fare per driver,
# for each city type.
# End, Add New DataFrame:
# Format the columns.
# Add Matplotlib inline magic command
%matplotlib inline
# Dependencies and Setup
import matplotlib.pyplot as plt
import pandas as pd
# File to Load (Remember to change these)
city_data_to_load = "Resources/city_data.csv"
ride_data_to_load = "Resources/ride_data.csv"
# Read the City and Ride Data
city_data_df = pd.read_csv(city_data_to_load)
ride_data_df = pd.read_csv(ride_data_to_load)
```
### Merge the DataFrames
```
# Combine the data into a single dataset
pyber_data_df = pd.merge(ride_data_df, city_data_df, how="left", on=["city", "city"])
# Display the data table for preview
pyber_data_df.head()
```
## Deliverable 1: Get a Summary DataFrame
```
# 1. Get the total rides for each city type
total_rides_df = pyber_data_df.groupby(["type"]).count().ride_id
total_rides_df
# 2. Get the total drivers for each city type
total_drivers_df = city_data_df.groupby(["type"]).sum().driver_count
total_drivers_df
# 3. Get the total amount of fares for each city type
total_fares_df = pyber_data_df.groupby(["type"]).sum().fare
total_fares_df
# 4. Get the average fare per ride for each city type.
avg_fare_per_ride_df = total_fares_df / total_rides_df
avg_fare_per_ride_df
# 5. Get the average fare per driver for each city type.
avg_fare_per_driver_df = total_fares_df / total_drivers_df
avg_fare_per_driver_df
# 6. Create a PyBer summary DataFrame.
pyber_summary_df = pd.DataFrame(
{"Total Rides":total_rides_df,
"Total Drivers":total_drivers_df,
"Total Fares":total_fares_df,
"Average Fare per Ride":avg_fare_per_ride_df,
"Average Fare per Driver":avg_fare_per_driver_df}
)
pyber_summary_df
# 7. Cleaning up the DataFrame. Delete the index name
pyber_summary_df.index.name = None
pyber_summary_df
# 8. Format the columns.
pyber_summary_df["Total Rides"] = pyber_summary_df["Total Rides"].map("{:,}".format)
pyber_summary_df["Total Drivers"] = pyber_summary_df["Total Drivers"].map("{:,}".format)
pyber_summary_df["Total Fares"] = pyber_summary_df["Total Fares"].map("${:,.2f}".format)
pyber_summary_df["Average Fare per Ride"] = pyber_summary_df["Average Fare per Ride"].map("${:,.2f}".format)
pyber_summary_df["Average Fare per Driver"] = pyber_summary_df["Average Fare per Driver"].map("${:,.2f}".format)
pyber_summary_df
```
## Deliverable 2. Create a multiple line plot that shows the total weekly of the fares for each type of city.
```
# 1. Read the merged DataFrame
pyber_summary_df
# 2. Using groupby() to create a new DataFrame showing the sum of the fares
# for each date where the indices are the city type and date.
fare_by_city_date_df = pyber_data_df.groupby(['type','date']).sum()["fare"]
fare_by_city_date_df.head()
# 3. Reset the index on the DataFrame you created in #1. This is needed to use the 'pivot()' function.
# df = df.reset_index()
fare_by_city_date_df = fare_by_city_date_df.reset_index()
fare_by_city_date_df.head()
# (3.5)Reformat the DateTime because it's different than what is expected in the Homework Module.
fare_by_city_date_df['date'] = pd.to_datetime(fare_by_city_date_df['date'])
# 4. Create a pivot table with the 'date' as the index, the columns ='type', and values='fare'
# to get the total fares for each type of city by the date.
fare_by_city_date_df = fare_by_city_date_df.pivot(index='date', columns='type', values='fare')
fare_by_city_date_df.head()
# 5. Create a new DataFrame from the pivot table DataFrame using loc on the given dates, '2019-01-01':'2019-04-29'.
fare_dates_df = fare_by_city_date_df.loc['2019-01-01':'2019-04-29']
fare_dates_df.head()
# 6. Set the "date" index to datetime datatype. This is necessary to use the resample() method in Step 8.
# df.index = pd.to_datetime(df.index)
# This step was completed in step 3.5 in order to run .loc[] (above)- Because the info was in the incorrect format in the ride_data.csv.
# 7. Check that the datatype for the index is datetime using df.info()
fare_dates_df.info()
# 8. Create a new DataFrame using the "resample()" function by week 'W' and get the sum of the fares for each week.
fare_resample_df = fare_dates_df
fare_resample_df = fare_resample_df.resample('W').sum()
fare_resample_df.head()
# 8. Using the object-oriented interface method, plot the resample DataFrame using the df.plot() function.
# Import the style from Matplotlib.
from matplotlib import style
# Use the graph style fivethirtyeight.
style.use('fivethirtyeight')
fare_resample_df.plot(figsize=(18,5),)
plt.xlabel('')
plt.ylabel("Fare ($USD)")
plt.title("Total Fare by City Type")
plt.legend(loc = "best")
plt.savefig('analysis/Pyber_fare_summary.png');
```
|
github_jupyter
|
# Challenge psuedo-code.
# - Use groupby(), function with count() and sum()
# - Get Total number of rides,
# number of drivers,
# total fares,
# for each city type.
# Then, Calculate
# Avg. fare per ride,
# Avg. fare per driver,
# for each city type.
# End, Add New DataFrame:
# Format the columns.
# Add Matplotlib inline magic command
%matplotlib inline
# Dependencies and Setup
import matplotlib.pyplot as plt
import pandas as pd
# File to Load (Remember to change these)
city_data_to_load = "Resources/city_data.csv"
ride_data_to_load = "Resources/ride_data.csv"
# Read the City and Ride Data
city_data_df = pd.read_csv(city_data_to_load)
ride_data_df = pd.read_csv(ride_data_to_load)
# Combine the data into a single dataset
pyber_data_df = pd.merge(ride_data_df, city_data_df, how="left", on=["city", "city"])
# Display the data table for preview
pyber_data_df.head()
# 1. Get the total rides for each city type
total_rides_df = pyber_data_df.groupby(["type"]).count().ride_id
total_rides_df
# 2. Get the total drivers for each city type
total_drivers_df = city_data_df.groupby(["type"]).sum().driver_count
total_drivers_df
# 3. Get the total amount of fares for each city type
total_fares_df = pyber_data_df.groupby(["type"]).sum().fare
total_fares_df
# 4. Get the average fare per ride for each city type.
avg_fare_per_ride_df = total_fares_df / total_rides_df
avg_fare_per_ride_df
# 5. Get the average fare per driver for each city type.
avg_fare_per_driver_df = total_fares_df / total_drivers_df
avg_fare_per_driver_df
# 6. Create a PyBer summary DataFrame.
pyber_summary_df = pd.DataFrame(
{"Total Rides":total_rides_df,
"Total Drivers":total_drivers_df,
"Total Fares":total_fares_df,
"Average Fare per Ride":avg_fare_per_ride_df,
"Average Fare per Driver":avg_fare_per_driver_df}
)
pyber_summary_df
# 7. Cleaning up the DataFrame. Delete the index name
pyber_summary_df.index.name = None
pyber_summary_df
# 8. Format the columns.
pyber_summary_df["Total Rides"] = pyber_summary_df["Total Rides"].map("{:,}".format)
pyber_summary_df["Total Drivers"] = pyber_summary_df["Total Drivers"].map("{:,}".format)
pyber_summary_df["Total Fares"] = pyber_summary_df["Total Fares"].map("${:,.2f}".format)
pyber_summary_df["Average Fare per Ride"] = pyber_summary_df["Average Fare per Ride"].map("${:,.2f}".format)
pyber_summary_df["Average Fare per Driver"] = pyber_summary_df["Average Fare per Driver"].map("${:,.2f}".format)
pyber_summary_df
# 1. Read the merged DataFrame
pyber_summary_df
# 2. Using groupby() to create a new DataFrame showing the sum of the fares
# for each date where the indices are the city type and date.
fare_by_city_date_df = pyber_data_df.groupby(['type','date']).sum()["fare"]
fare_by_city_date_df.head()
# 3. Reset the index on the DataFrame you created in #1. This is needed to use the 'pivot()' function.
# df = df.reset_index()
fare_by_city_date_df = fare_by_city_date_df.reset_index()
fare_by_city_date_df.head()
# (3.5)Reformat the DateTime because it's different than what is expected in the Homework Module.
fare_by_city_date_df['date'] = pd.to_datetime(fare_by_city_date_df['date'])
# 4. Create a pivot table with the 'date' as the index, the columns ='type', and values='fare'
# to get the total fares for each type of city by the date.
fare_by_city_date_df = fare_by_city_date_df.pivot(index='date', columns='type', values='fare')
fare_by_city_date_df.head()
# 5. Create a new DataFrame from the pivot table DataFrame using loc on the given dates, '2019-01-01':'2019-04-29'.
fare_dates_df = fare_by_city_date_df.loc['2019-01-01':'2019-04-29']
fare_dates_df.head()
# 6. Set the "date" index to datetime datatype. This is necessary to use the resample() method in Step 8.
# df.index = pd.to_datetime(df.index)
# This step was completed in step 3.5 in order to run .loc[] (above)- Because the info was in the incorrect format in the ride_data.csv.
# 7. Check that the datatype for the index is datetime using df.info()
fare_dates_df.info()
# 8. Create a new DataFrame using the "resample()" function by week 'W' and get the sum of the fares for each week.
fare_resample_df = fare_dates_df
fare_resample_df = fare_resample_df.resample('W').sum()
fare_resample_df.head()
# 8. Using the object-oriented interface method, plot the resample DataFrame using the df.plot() function.
# Import the style from Matplotlib.
from matplotlib import style
# Use the graph style fivethirtyeight.
style.use('fivethirtyeight')
fare_resample_df.plot(figsize=(18,5),)
plt.xlabel('')
plt.ylabel("Fare ($USD)")
plt.title("Total Fare by City Type")
plt.legend(loc = "best")
plt.savefig('analysis/Pyber_fare_summary.png');
| 0.684686 | 0.855972 |
```
from cospar import reader, F, paramkeys, datakeys
datakeys = tuple(filter(lambda k: k not in {'t', 'dt'}, datakeys))
import math
import operator
import numpy as np
from functools import reduce
from everest import window
from everest.analysis import time_fourier
from everest.window.data import Data
from matplotlib.pyplot import get_cmap
%matplotlib inline
cut = reader[reduce(operator.__and__, (
F('f') == 1,
F('aspect') == 1,
F('temperatureField') == '_built_peaskauslu-thoesfthuec',
))]
tauRefs = reader[cut : 'tauRef']
datas, ts = reader[cut : datakeys], reader[cut : 't']
datasets = [
(tauRef, ts[k], dict(zip(datakeys, datas[k])))
for k, tauRef in tauRefs.items()
]
datasets.sort()
ffts = []
for tauRef, t, data in datasets:
Nu = data['Nu']
length = slice(-round(len(t) / math.sqrt(2)), None)
t, Nu = t[length], Nu[length]
freqs, trans = time_fourier(t, Nu, sampleFactor = 0.01, interpKind = 'quadratic')
ffts.append((tauRef, freqs, trans))
canvas = window.Canvas(size = (18, 9), facecolour = 'black')
ax = canvas.make_ax()
logTaus = [math.log10(tau) for tau in sorted(r[0] for r in datasets)]
normTau = lambda tau: (math.log10(tau) - min(logTaus)) / (max(logTaus) - min(logTaus))
# normTau = lambda tau: (math.log10(tau) - 5.4) / 0.3
cmap = get_cmap('plasma')
for tauRef, freqs, trans in ffts:
# if tauRef < 10 ** 5.4: continue
# if tauRef > 10 ** 5.7: continue
# if tauRef != 10 ** 5.55: continue
ax.line(
Data(freqs[1:], label = 'Occurrences per unit dimensionless time'),
Data(np.log10(trans[1:]), label = 'Nusselt number (log10)'),
c = cmap(normTau(tauRef)),
linewidth = 1.0,
marker = 'o',
markersize = 5,
)
ax.axes.colour = 'white'
ax.ticks.colour = 'white'
ax.grid.colour = 'grey'
ax.axes.title = 'Fourier transform of Nusselt number at steady-state\n{aspect = 1, curvature = 1}'
legendValues = logTaus
legendHandles = [r[0] for r in ax.collections]
legendLabels = [str(v) for v in legendValues]
handles, labels, _ = zip(*sorted(zip(legendHandles, legendLabels, legendValues), key = lambda r: r[-1]))
legend = ax.ax.legend(
handles,
labels,
loc = 'right',
framealpha = 0.,
labelcolor = 'white',
title = 'tau0 (10^n)',
bbox_to_anchor = (1.08, 0.5),
)
legend.properties()['title'].set_color('white')
canvas.show()
x, y1, y2 = [], [], []
for tauRef, freqs, trans in ffts:
x.append(math.log10(tauRef))
y1.append(np.mean(trans))
y2.append(freqs[list(trans).index(np.max(trans))])
canvas = window.Canvas(size = (12, 6), facecolour = 'black')
ax1 = canvas.make_ax()
ax2 = canvas.make_ax()
ax1.line(
Data(x, label = 'Yield strength (10^n)'),
Data(y1, label = 'Mean Fourier amplitude'),
c = get_cmap('rainbow')(0.25),
marker = 'o',
)
ax2.line(
Data(x, label = 'Yield strength (10^n)'),
Data(y2, label = 'Dominant frequency'),
c = get_cmap('rainbow')(0.75),
marker = 'o',
)
ax1.axes.colour = ax2.axes.colour = 'white'
ax1.ticks.x.colour = ax2.ticks.x.colour = 'white'
ax1.ticks.y.colour = get_cmap('rainbow')(0.25)
ax2.ticks.y.colour = get_cmap('rainbow')(0.75)
ax2.axes.y.swap()
ax1.grid.colour = ax2.grid.colour = 'grey'
ax2.grid.visible = False
ax2.axes.x.visible = False
ax1.axes.title = 'Fourier properties of the Nusselt profile as a function of yield strength\n{aspect = 1, curvature = 1}'
canvas.show()
1 / 2e-9
1 / 0.05
```
|
github_jupyter
|
from cospar import reader, F, paramkeys, datakeys
datakeys = tuple(filter(lambda k: k not in {'t', 'dt'}, datakeys))
import math
import operator
import numpy as np
from functools import reduce
from everest import window
from everest.analysis import time_fourier
from everest.window.data import Data
from matplotlib.pyplot import get_cmap
%matplotlib inline
cut = reader[reduce(operator.__and__, (
F('f') == 1,
F('aspect') == 1,
F('temperatureField') == '_built_peaskauslu-thoesfthuec',
))]
tauRefs = reader[cut : 'tauRef']
datas, ts = reader[cut : datakeys], reader[cut : 't']
datasets = [
(tauRef, ts[k], dict(zip(datakeys, datas[k])))
for k, tauRef in tauRefs.items()
]
datasets.sort()
ffts = []
for tauRef, t, data in datasets:
Nu = data['Nu']
length = slice(-round(len(t) / math.sqrt(2)), None)
t, Nu = t[length], Nu[length]
freqs, trans = time_fourier(t, Nu, sampleFactor = 0.01, interpKind = 'quadratic')
ffts.append((tauRef, freqs, trans))
canvas = window.Canvas(size = (18, 9), facecolour = 'black')
ax = canvas.make_ax()
logTaus = [math.log10(tau) for tau in sorted(r[0] for r in datasets)]
normTau = lambda tau: (math.log10(tau) - min(logTaus)) / (max(logTaus) - min(logTaus))
# normTau = lambda tau: (math.log10(tau) - 5.4) / 0.3
cmap = get_cmap('plasma')
for tauRef, freqs, trans in ffts:
# if tauRef < 10 ** 5.4: continue
# if tauRef > 10 ** 5.7: continue
# if tauRef != 10 ** 5.55: continue
ax.line(
Data(freqs[1:], label = 'Occurrences per unit dimensionless time'),
Data(np.log10(trans[1:]), label = 'Nusselt number (log10)'),
c = cmap(normTau(tauRef)),
linewidth = 1.0,
marker = 'o',
markersize = 5,
)
ax.axes.colour = 'white'
ax.ticks.colour = 'white'
ax.grid.colour = 'grey'
ax.axes.title = 'Fourier transform of Nusselt number at steady-state\n{aspect = 1, curvature = 1}'
legendValues = logTaus
legendHandles = [r[0] for r in ax.collections]
legendLabels = [str(v) for v in legendValues]
handles, labels, _ = zip(*sorted(zip(legendHandles, legendLabels, legendValues), key = lambda r: r[-1]))
legend = ax.ax.legend(
handles,
labels,
loc = 'right',
framealpha = 0.,
labelcolor = 'white',
title = 'tau0 (10^n)',
bbox_to_anchor = (1.08, 0.5),
)
legend.properties()['title'].set_color('white')
canvas.show()
x, y1, y2 = [], [], []
for tauRef, freqs, trans in ffts:
x.append(math.log10(tauRef))
y1.append(np.mean(trans))
y2.append(freqs[list(trans).index(np.max(trans))])
canvas = window.Canvas(size = (12, 6), facecolour = 'black')
ax1 = canvas.make_ax()
ax2 = canvas.make_ax()
ax1.line(
Data(x, label = 'Yield strength (10^n)'),
Data(y1, label = 'Mean Fourier amplitude'),
c = get_cmap('rainbow')(0.25),
marker = 'o',
)
ax2.line(
Data(x, label = 'Yield strength (10^n)'),
Data(y2, label = 'Dominant frequency'),
c = get_cmap('rainbow')(0.75),
marker = 'o',
)
ax1.axes.colour = ax2.axes.colour = 'white'
ax1.ticks.x.colour = ax2.ticks.x.colour = 'white'
ax1.ticks.y.colour = get_cmap('rainbow')(0.25)
ax2.ticks.y.colour = get_cmap('rainbow')(0.75)
ax2.axes.y.swap()
ax1.grid.colour = ax2.grid.colour = 'grey'
ax2.grid.visible = False
ax2.axes.x.visible = False
ax1.axes.title = 'Fourier properties of the Nusselt profile as a function of yield strength\n{aspect = 1, curvature = 1}'
canvas.show()
1 / 2e-9
1 / 0.05
| 0.449634 | 0.48249 |
## 1.0 Import Function
```
from META_TOOLBOX import *
from VIGA_PREFABRICADA_PROTENDIDA import *
```
## 2.0 Setup
```
N_REP = 10
N_ITER = 150
X_L = [0.20, 0.10, 0.10, 1/6.0]
X_U = [0.25, 0.15, 0.15, 1/3.5]
D = 4
M = 2
GAMMA = GAMMA_ASSEMBLY(X_L, X_U, D, M)
SETUP_FA = {
'N_REP': N_REP,
'N_ITER': N_ITER,
'N_POP': 10,
'D': D,
'X_L': X_L,
'X_U': X_U,
'BETA_0': 0.98,
'ALPHA_MIN': 0.25,
'ALPHA_MAX': 1.00,
'THETA': 0.95,
'GAMMA': GAMMA,
'NULL_DIC': None
}
# OBJ. Function
def OF_FUNCTION(X, NULL_DIC):
# Geometria da viga
VIGA = {
'H_W': X[0],
'B_W': X[1],
'B_FS': 0.30,
'B_FI': 0.30,
'H_FS': X[2],
'H_FI': X[2],
'H_SI': 0.07,
'H_II': 0.07,
'COB': 0.035,
'PHI_L': 12.5 / 1E3,
'A_BAR': 99 / 1000 ** 2,
'PHI_E': 10.0 / 1E3,
'L': 20,
'L_PISTA': 150,
'FATOR_SEC': 'I',
'DELTA_ANC': 6 / 1E3,
'TEMPO_CONC': [1.00, 15, 45, 50 * 365],
'TEMPO_ACO': [2.00, 16, 46, 51 * 365],
'TEMP': 20,
'U': 70,
'PERDA_INICIAL': 2.5,
'PERDA_TEMPO': 17.50,
'E_SCP': 200E6,
'PHO_S': 78,
'F_PK': 1900000,
'F_YK': 1710000,
'LAMBA_SIG': 1,
'TIPO_FIO_CORD_BAR': 'COR',
'TIPO_PROT': 'PRE',
'TIPO_ACO': 'RB',
'PHO_C': 25,
'F_CK': 40 * 1E3,
'CIMENTO': 'CP5',
'AGREGADO': 'GRA',
'ABAT': 0.09,
'G_2K': 1.55 + 0.70,
'Q_1K': 1.5,
'PSI_1': 0.40,
'PSI_2': 0.30,
'GAMMA_F1': 1.30,
'GAMMA_F2': 1.40,
'GAMMA_S':1.15,
'ETA_1':1.2,
'ETA_2':1.0,
'E_PPROPORCAO': X[3],
'IMPRESSÃO': False
}
G, A_C, A_SCP = VERIFICACAO_VIGA(VIGA)
PESO = VIGA['L'] * A_C * VIGA['PHO_C']
OF = PESO
for I_CONT in range(len(G)):
OF += (max(0, G[I_CONT]) ** 2) * 1E10
return OF
```
## 4.0 Example
```
[RESULTS_REP, BEST_REP, AVERAGE_REP, WORST_REP, STATUS] = FA_ALGORITHM_0001(OF_FUNCTION, SETUP_FA)
STATUS
BEST_REP_ID = STATUS[0]
BEST_REP_ID
BEST = BEST_REP[BEST_REP_ID]
AVERAGE = AVERAGE_REP[BEST_REP_ID]
WORST = WORST_REP[BEST_REP_ID]
PLOT_SETUP = {
'NAME': 'WANDER-OF',
'WIDTH': 0.40,
'HEIGHT': 0.20,
'DPI': 600,
'EXTENSION': '.svg',
'COLOR OF': '#000000',
'MARKER OF': 's',
'COLOR FIT': '#000000',
'MARKER FIT': 's',
'MARKER SIZE': 6,
'LINE WIDTH': 4,
'LINE STYLE': '--',
'OF AXIS LABEL': '$W (kN) $',
'X AXIS LABEL': 'Number of objective function evaluations',
'LABELS SIZE': 14,
'LABELS COLOR': '#000000',
'X AXIS SIZE': 14,
'Y AXIS SIZE': 14,
'AXISES COLOR': '#000000',
'ON GRID?': True,
'Y LOG': True,
'X LOG': True,
}
DATASET = {'X': BEST['NEOF'], 'OF': BEST['OF'], 'FIT': BEST['FIT']}
META_PLOT_001(DATASET, PLOT_SETUP)
PLOT_SETUP = {
'NAME': 'WANDER-OF',
'WIDTH': 0.40,
'HEIGHT': 0.20,
'DPI': 600,
'EXTENSION': '.svg',
'COLOR': '#00BFFF',
'MARKER': 's',
'MARKER SIZE': 6,
'LINE WIDTH': 4,
'LINE STYLE': '--',
'Y AXIS LABEL': '$Euller$',
'X AXIS LABEL': 'Number of objective function evaluations',
'LABELS SIZE': 14,
'LABELS COLOR': '#000000',
'X AXIS SIZE': 14,
'Y AXIS SIZE': 14,
'AXISES COLOR': '#000000',
'ON GRID?': True,
'Y LOG': True,
'X LOG': True,
}
DATASET = {'X': BEST['NEOF'], 'Y': BEST['OF']}
META_PLOT_002(DATASET, PLOT_SETUP)
PLOT_SETUP = {
'NAME': 'WANDER-OF',
'WIDTH': 0.40,
'HEIGHT': 0.20,
'DPI': 600,
'EXTENSION': '.svg',
'COLOR BEST': '#00008B',
'COLOR WORST': '#000000',
'COLOR AVERAGE': '#ffcbdb',
'MARKER': 's',
'MARKER SIZE': 6,
'LINE WIDTH': 4,
'LINE STYLE': '--',
'Y AXIS LABEL': '$W (kN) $',
'X AXIS LABEL': 'Number of objective function evaluations',
'LABELS SIZE': 14,
'LABELS COLOR': '#000000',
'X AXIS SIZE': 14,
'Y AXIS SIZE': 14,
'AXISES COLOR': '#000000',
'ON GRID?': True,
'LOC LEGEND': 'upper right',
'SIZE LEGEND': 12,
'Y LOG': True,
'X LOG': True
}
DATASET = {'X': BEST['NEOF'], 'BEST': BEST['OF'], 'AVERAGE': AVERAGE['OF'], 'WORST': WORST['OF']}
META_PLOT_003(DATASET, PLOT_SETUP)
PLOT_SETUP = {
'NAME': 'WANDER-OF',
'WIDTH': 0.40,
'HEIGHT': 0.20,
'DPI': 600,
'EXTENSION': '.svg',
'MARKER': 's',
'X AXIS LABEL': 'OF values',
'X AXIS SIZE': 14,
'Y AXIS SIZE': 14,
'LABELS SIZE': 14,
'LABELS COLOR': '#000000',
'COLOR': '#000000',
'AXISES COLOR': '#000000',
'BINS': 20,
'KDE': False,
}
DATASET = {'NUMBER OF REPETITIONS': N_REP, 'NUMBER OF ITERATIONS': N_ITER, 'OF OR FIT': 'OF', 'BEST': BEST_REP}
META_PLOT_004(DATASET, PLOT_SETUP)
BEST_REP[BEST_REP_ID]
X = BEST_REP[BEST_REP_ID]['X_POSITION'][N_ITER, :]
VIGA = {
'H_W': X[0],
'B_W': X[1],
'B_FS': 0.30,
'B_FI': 0.30,
'H_FS': X[2],
'H_FI': X[2],
'H_SI': 0.07,
'H_II': 0.07,
'COB': 0.035,
'PHI_L': 12.5 / 1E3,
'A_BAR': 99 / 1000 ** 2,
'PHI_E': 10.0 / 1E3,
'L': 20,
'L_PISTA': 150,
'FATOR_SEC': 'I',
'DELTA_ANC': 6 / 1E3,
'TEMPO_CONC': [1.00, 15, 45, 50 * 365],
'TEMPO_ACO': [2.00, 16, 46, 51 * 365],
'TEMP': 20,
'U': 70,
'PERDA_INICIAL': 2.5,
'PERDA_TEMPO': 17.50,
'E_SCP': 200E6,
'PHO_S': 78,
'F_PK': 1900000,
'F_YK': 1710000,
'LAMBA_SIG': 1,
'TIPO_FIO_CORD_BAR': 'COR',
'TIPO_PROT': 'PRE',
'TIPO_ACO': 'RB',
'PHO_C': 25,
'F_CK': 40 * 1E3,
'CIMENTO': 'CP5',
'AGREGADO': 'GRA',
'ABAT': 0.09,
'G_2K': 1.55 + 0.70,
'Q_1K': 1.5,
'PSI_1': 0.40,
'PSI_2': 0.30,
'GAMMA_F1': 1.30,
'GAMMA_F2': 1.40,
'GAMMA_S':1.15,
'ETA_1':1.2,
'ETA_2':1.0,
'E_PPROPORCAO': X[3],
'IMPRESSÃO': False
}
G, A_C, A_SCP = VERIFICACAO_VIGA(VIGA)
PESO = VIGA['L'] * A_C * VIGA['PHO_C']
OF = PESO
OF
G
```
|
github_jupyter
|
from META_TOOLBOX import *
from VIGA_PREFABRICADA_PROTENDIDA import *
N_REP = 10
N_ITER = 150
X_L = [0.20, 0.10, 0.10, 1/6.0]
X_U = [0.25, 0.15, 0.15, 1/3.5]
D = 4
M = 2
GAMMA = GAMMA_ASSEMBLY(X_L, X_U, D, M)
SETUP_FA = {
'N_REP': N_REP,
'N_ITER': N_ITER,
'N_POP': 10,
'D': D,
'X_L': X_L,
'X_U': X_U,
'BETA_0': 0.98,
'ALPHA_MIN': 0.25,
'ALPHA_MAX': 1.00,
'THETA': 0.95,
'GAMMA': GAMMA,
'NULL_DIC': None
}
# OBJ. Function
def OF_FUNCTION(X, NULL_DIC):
# Geometria da viga
VIGA = {
'H_W': X[0],
'B_W': X[1],
'B_FS': 0.30,
'B_FI': 0.30,
'H_FS': X[2],
'H_FI': X[2],
'H_SI': 0.07,
'H_II': 0.07,
'COB': 0.035,
'PHI_L': 12.5 / 1E3,
'A_BAR': 99 / 1000 ** 2,
'PHI_E': 10.0 / 1E3,
'L': 20,
'L_PISTA': 150,
'FATOR_SEC': 'I',
'DELTA_ANC': 6 / 1E3,
'TEMPO_CONC': [1.00, 15, 45, 50 * 365],
'TEMPO_ACO': [2.00, 16, 46, 51 * 365],
'TEMP': 20,
'U': 70,
'PERDA_INICIAL': 2.5,
'PERDA_TEMPO': 17.50,
'E_SCP': 200E6,
'PHO_S': 78,
'F_PK': 1900000,
'F_YK': 1710000,
'LAMBA_SIG': 1,
'TIPO_FIO_CORD_BAR': 'COR',
'TIPO_PROT': 'PRE',
'TIPO_ACO': 'RB',
'PHO_C': 25,
'F_CK': 40 * 1E3,
'CIMENTO': 'CP5',
'AGREGADO': 'GRA',
'ABAT': 0.09,
'G_2K': 1.55 + 0.70,
'Q_1K': 1.5,
'PSI_1': 0.40,
'PSI_2': 0.30,
'GAMMA_F1': 1.30,
'GAMMA_F2': 1.40,
'GAMMA_S':1.15,
'ETA_1':1.2,
'ETA_2':1.0,
'E_PPROPORCAO': X[3],
'IMPRESSÃO': False
}
G, A_C, A_SCP = VERIFICACAO_VIGA(VIGA)
PESO = VIGA['L'] * A_C * VIGA['PHO_C']
OF = PESO
for I_CONT in range(len(G)):
OF += (max(0, G[I_CONT]) ** 2) * 1E10
return OF
[RESULTS_REP, BEST_REP, AVERAGE_REP, WORST_REP, STATUS] = FA_ALGORITHM_0001(OF_FUNCTION, SETUP_FA)
STATUS
BEST_REP_ID = STATUS[0]
BEST_REP_ID
BEST = BEST_REP[BEST_REP_ID]
AVERAGE = AVERAGE_REP[BEST_REP_ID]
WORST = WORST_REP[BEST_REP_ID]
PLOT_SETUP = {
'NAME': 'WANDER-OF',
'WIDTH': 0.40,
'HEIGHT': 0.20,
'DPI': 600,
'EXTENSION': '.svg',
'COLOR OF': '#000000',
'MARKER OF': 's',
'COLOR FIT': '#000000',
'MARKER FIT': 's',
'MARKER SIZE': 6,
'LINE WIDTH': 4,
'LINE STYLE': '--',
'OF AXIS LABEL': '$W (kN) $',
'X AXIS LABEL': 'Number of objective function evaluations',
'LABELS SIZE': 14,
'LABELS COLOR': '#000000',
'X AXIS SIZE': 14,
'Y AXIS SIZE': 14,
'AXISES COLOR': '#000000',
'ON GRID?': True,
'Y LOG': True,
'X LOG': True,
}
DATASET = {'X': BEST['NEOF'], 'OF': BEST['OF'], 'FIT': BEST['FIT']}
META_PLOT_001(DATASET, PLOT_SETUP)
PLOT_SETUP = {
'NAME': 'WANDER-OF',
'WIDTH': 0.40,
'HEIGHT': 0.20,
'DPI': 600,
'EXTENSION': '.svg',
'COLOR': '#00BFFF',
'MARKER': 's',
'MARKER SIZE': 6,
'LINE WIDTH': 4,
'LINE STYLE': '--',
'Y AXIS LABEL': '$Euller$',
'X AXIS LABEL': 'Number of objective function evaluations',
'LABELS SIZE': 14,
'LABELS COLOR': '#000000',
'X AXIS SIZE': 14,
'Y AXIS SIZE': 14,
'AXISES COLOR': '#000000',
'ON GRID?': True,
'Y LOG': True,
'X LOG': True,
}
DATASET = {'X': BEST['NEOF'], 'Y': BEST['OF']}
META_PLOT_002(DATASET, PLOT_SETUP)
PLOT_SETUP = {
'NAME': 'WANDER-OF',
'WIDTH': 0.40,
'HEIGHT': 0.20,
'DPI': 600,
'EXTENSION': '.svg',
'COLOR BEST': '#00008B',
'COLOR WORST': '#000000',
'COLOR AVERAGE': '#ffcbdb',
'MARKER': 's',
'MARKER SIZE': 6,
'LINE WIDTH': 4,
'LINE STYLE': '--',
'Y AXIS LABEL': '$W (kN) $',
'X AXIS LABEL': 'Number of objective function evaluations',
'LABELS SIZE': 14,
'LABELS COLOR': '#000000',
'X AXIS SIZE': 14,
'Y AXIS SIZE': 14,
'AXISES COLOR': '#000000',
'ON GRID?': True,
'LOC LEGEND': 'upper right',
'SIZE LEGEND': 12,
'Y LOG': True,
'X LOG': True
}
DATASET = {'X': BEST['NEOF'], 'BEST': BEST['OF'], 'AVERAGE': AVERAGE['OF'], 'WORST': WORST['OF']}
META_PLOT_003(DATASET, PLOT_SETUP)
PLOT_SETUP = {
'NAME': 'WANDER-OF',
'WIDTH': 0.40,
'HEIGHT': 0.20,
'DPI': 600,
'EXTENSION': '.svg',
'MARKER': 's',
'X AXIS LABEL': 'OF values',
'X AXIS SIZE': 14,
'Y AXIS SIZE': 14,
'LABELS SIZE': 14,
'LABELS COLOR': '#000000',
'COLOR': '#000000',
'AXISES COLOR': '#000000',
'BINS': 20,
'KDE': False,
}
DATASET = {'NUMBER OF REPETITIONS': N_REP, 'NUMBER OF ITERATIONS': N_ITER, 'OF OR FIT': 'OF', 'BEST': BEST_REP}
META_PLOT_004(DATASET, PLOT_SETUP)
BEST_REP[BEST_REP_ID]
X = BEST_REP[BEST_REP_ID]['X_POSITION'][N_ITER, :]
VIGA = {
'H_W': X[0],
'B_W': X[1],
'B_FS': 0.30,
'B_FI': 0.30,
'H_FS': X[2],
'H_FI': X[2],
'H_SI': 0.07,
'H_II': 0.07,
'COB': 0.035,
'PHI_L': 12.5 / 1E3,
'A_BAR': 99 / 1000 ** 2,
'PHI_E': 10.0 / 1E3,
'L': 20,
'L_PISTA': 150,
'FATOR_SEC': 'I',
'DELTA_ANC': 6 / 1E3,
'TEMPO_CONC': [1.00, 15, 45, 50 * 365],
'TEMPO_ACO': [2.00, 16, 46, 51 * 365],
'TEMP': 20,
'U': 70,
'PERDA_INICIAL': 2.5,
'PERDA_TEMPO': 17.50,
'E_SCP': 200E6,
'PHO_S': 78,
'F_PK': 1900000,
'F_YK': 1710000,
'LAMBA_SIG': 1,
'TIPO_FIO_CORD_BAR': 'COR',
'TIPO_PROT': 'PRE',
'TIPO_ACO': 'RB',
'PHO_C': 25,
'F_CK': 40 * 1E3,
'CIMENTO': 'CP5',
'AGREGADO': 'GRA',
'ABAT': 0.09,
'G_2K': 1.55 + 0.70,
'Q_1K': 1.5,
'PSI_1': 0.40,
'PSI_2': 0.30,
'GAMMA_F1': 1.30,
'GAMMA_F2': 1.40,
'GAMMA_S':1.15,
'ETA_1':1.2,
'ETA_2':1.0,
'E_PPROPORCAO': X[3],
'IMPRESSÃO': False
}
G, A_C, A_SCP = VERIFICACAO_VIGA(VIGA)
PESO = VIGA['L'] * A_C * VIGA['PHO_C']
OF = PESO
OF
G
| 0.336985 | 0.666035 |
```
!pip install torchviz
from torch.utils.data.sampler import SubsetRandomSampler
from torch.autograd import Variable
from torchviz import make_dot
import torch
import torch.nn as nn
import torchvision
import torchvision.transforms as transforms
import torch.optim as optim
import matplotlib.pyplot as plt
import numpy as np # we always love numpy
import time
from sklearn.datasets import fetch_openml
# Load data from https://www.openml.org/d/554
X, y = fetch_openml('mnist_784', version=1, return_X_y=True)
print(X.shape)
print(y)
# Data set information
image_dims = 1, 28, 28
n_training_samples = 60000 # How many training images to use
n_test_samples = 10000 # How many test images to use
classes = tuple([str(x) for x in range(9+1)])
# Load the training set
train_set = torchvision.datasets.MNIST(
root='./cifardata', train=True, download=True, transform=transforms.ToTensor())
train_sampler = SubsetRandomSampler(np.arange(n_training_samples, dtype=np.int64))
#Load the test set
test_set = torchvision.datasets.MNIST(
root='./cifardata', train=False, download=True, transform=transforms.ToTensor())
test_sampler = SubsetRandomSampler(np.arange(n_test_samples, dtype=np.int64))
def disp_image(image, class_idx, predicted=None):
# need to reorder the tensor dimensions to work properly with imshow
plt.imshow(image, 'gray')
plt.axis('off')
if predicted:
plt.title("Actual: " + classes[class_idx] + " Predicted: " + classes[predicted])
else:
plt.title("Actual: " + classes[class_idx])
plt.show()
print("training set input data shape", train_set.data.shape)
print("Number of training outputs", len(train_set.targets))
x, y = train_set[1]
disp_image(x.reshape(28,28), y)
import seaborn as sns
sns.countplot(train_set.targets.numpy())
plt.xticks(ticks=range(10), labels=classes)
plt.show()
sns.countplot(test_set.targets.numpy())
plt.xticks(ticks=range(10), labels=classes)
plt.show()
class MyCNN(nn.Module):
# The init funciton in Pytorch classes is used to keep track of the parameters of the model
# specifically the ones we want to update with gradient descent + backprop
# So we need to make sure we keep track of all of them here
def __init__(self):
super(MyCNN, self).__init__()
# layers defined here
# Make sure you understand what this convolutional layer is doing.
# E.g., considering looking at help(nn.Conv2D). Draw a picture of what
# this layer does to the data.
# note: image_dims[0] will be 1 as there are 1 color channels (R, G, B)
num_kernels = 16
self.conv1 = nn.Conv2d(image_dims[0], num_kernels, kernel_size=3, stride=1, padding=1)
# Make sure you understand what this MaxPool2D layer is doing.
# E.g., considering looking at help(nn.MaxPool2d). Draw a picture of
# what this layer does to the data.
self.pool = nn.MaxPool2d(kernel_size=2, stride=2, padding=0)
# maxpool_output_size is the total amount of data coming out of that
# layer. We have an exercise that asks you to explain why the line of
# code below computes this quantity.
self.maxpool_output_size = int(num_kernels * (image_dims[1] / 2) * (image_dims[2] / 2))
# Add on a fully connected layer (like in our MLP)
# fc stands for fully connected
fc1_size = 64
self.fc1 = nn.Linear(self.maxpool_output_size, fc1_size)
# we'll use this activation function internally in the network
self.activation_func = torch.nn.ReLU()
# Convert our fully connected layer into outputs that we can compare to the result
fc2_size = len(classes)
self.fc2 = nn.Linear(fc1_size, fc2_size)
# Note: that the output will not represent the probability of the
# output being in each class. The loss function we will use
# `CrossEntropyLoss` will take care of convering these values to
# probabilities and then computing the log loss with respect to the
# true label. We could break this out into multiple steps, but it turns
# out that the algorithm will be more numerically stable if we do it in
# one go. We have included a cell to show you the documentation for
# `CrossEntropyLoss` if you'd like to check it out.
# The forward function in the class defines the operations performed on a given input to the model
# and returns the output of the model
def forward(self, x):
x = self.conv1(x)
x = self.pool(x)
x = self.activation_func(x)
# this code flattens the output of the convolution, max pool,
# activation sequence of steps into a vector
x = x.view(-1, self.maxpool_output_size)
x = self.fc1(x)
x = self.activation_func(x)
x = self.fc2(x)
return x
# The loss function (which we chose to include as a method of the class, but doesn't need to be)
# returns the loss and optimizer used by the model
def get_loss(self, learning_rate):
# Loss function
loss = nn.CrossEntropyLoss()
# Optimizer, self.parameters() returns all the Pytorch operations that are attributes of the class
optimizer = optim.Adam(self.parameters(), lr=learning_rate)
return loss, optimizer
def visualize_network(net):
# Visualize the architecture of the model
# We need to give the net a fake input for this library to visualize the architecture
fake_input = Variable(torch.zeros((1,image_dims[0], image_dims[1], image_dims[2]))).to(device)
outputs = net(fake_input)
# Plot the DAG (Directed Acyclic Graph) of the model
return make_dot(outputs, dict(net.named_parameters()))
# Define what device we want to use
device = 'cuda' # 'cpu' if we want to not use the gpu
# Initialize the model, loss, and optimization function
net = MyCNN()
# This tells our model to send all of the tensors and operations to the GPU (or keep them at the CPU if we're not using GPU)
net.to(device)
visualize_network(net)
# Define training parameters
batch_size = 32
learning_rate = 1e-2
n_epochs = 10
# Get our data into the mini batch size that we defined
train_loader = torch.utils.data.DataLoader(train_set, batch_size=batch_size,
sampler=train_sampler, num_workers=2)
test_loader = torch.utils.data.DataLoader(
test_set, batch_size=128, sampler=test_sampler, num_workers=2)
def train_model(net):
""" Train a the specified network.
Outputs a tuple with the following four elements
train_hist_x: the x-values (batch number) that the training set was
evaluated on.
train_loss_hist: the loss values for the training set corresponding to
the batch numbers returned in train_hist_x
test_hist_x: the x-values (batch number) that the test set was
evaluated on.
test_loss_hist: the loss values for the test set corresponding to
the batch numbers returned in test_hist_x
"""
loss, optimizer = net.get_loss(learning_rate)
# Define some parameters to keep track of metrics
print_every = 20
idx = 0
train_hist_x = []
train_loss_hist = []
test_hist_x = []
test_loss_hist = []
training_start_time = time.time()
# Loop for n_epochs
for epoch in range(n_epochs):
running_loss = 0.0
start_time = time.time()
for i, data in enumerate(train_loader, 0):
# Get inputs in right form
inputs, labels = data
inputs, labels = Variable(inputs).to(device), Variable(labels).to(device)
# In Pytorch, We need to always remember to set the optimizer gradients to 0 before we recompute the new gradients
optimizer.zero_grad()
# Forward pass
outputs = net(inputs)
# Compute the loss and find the loss with respect to each parameter of the model
loss_size = loss(outputs, labels)
loss_size.backward()
# Change each parameter with respect to the recently computed loss.
optimizer.step()
# Update statistics
running_loss += loss_size.data.item()
# Print every 20th batch of an epoch
if (i % print_every) == print_every-1:
print("Epoch {}, Iteration {}\t train_loss: {:.2f} took: {:.2f}s".format(
epoch + 1, i+1,running_loss / print_every, time.time() - start_time))
# Reset running loss and time
train_loss_hist.append(running_loss / print_every)
train_hist_x.append(idx)
running_loss = 0.0
start_time = time.time()
idx += 1
# At the end of the epoch, do a pass on the test set
total_test_loss = 0
for inputs, labels in test_loader:
# Wrap tensors in Variables
inputs, labels = Variable(inputs).to(device), Variable(labels).to(device)
# Forward pass
test_outputs = net(inputs)
test_loss_size = loss(test_outputs, labels)
total_test_loss += test_loss_size.data.item()
test_loss_hist.append(total_test_loss / len(test_loader))
test_hist_x.append(idx)
print("Validation loss = {:.2f}".format(
total_test_loss / len(test_loader)))
print("Training finished, took {:.2f}s".format(
time.time() - training_start_time))
return train_hist_x, train_loss_hist, test_hist_x, test_loss_hist
train_hist_x, train_loss_hist, test_hist_x, test_loss_hist = train_model(net)
plt.plot(train_hist_x,train_loss_hist)
plt.plot(test_hist_x,test_loss_hist)
plt.legend(['train loss', 'validation loss'])
plt.xlabel('Batch number')
plt.ylabel('Loss')
plt.show()
print(test_loss_hist[-1])
n_correct = 0
n_total = 0
for i, data in enumerate(test_loader, 0):
# Get inputs in right form
inputs, labels = data
inputs, labels = Variable(inputs).to(device), Variable(labels).to(device)
# Forward pass
outputs = net(inputs)
n_correct += np.sum(np.argmax(outputs.cpu().detach().numpy(), axis=1) == labels.cpu().numpy())
n_total += labels.shape[0]
print("Testing accuracy is", n_correct/n_total)
plt.subplots(4, 4)
for i in range(net.conv1.weight.shape[0]):
plt.subplot(4, 4, i+1)
kernel = net.conv1.weight[i].cpu().detach().numpy()
im = kernel.mean(axis=0)
plt.pcolor(im, cmap='gray')
plt.show()
```
|
github_jupyter
|
!pip install torchviz
from torch.utils.data.sampler import SubsetRandomSampler
from torch.autograd import Variable
from torchviz import make_dot
import torch
import torch.nn as nn
import torchvision
import torchvision.transforms as transforms
import torch.optim as optim
import matplotlib.pyplot as plt
import numpy as np # we always love numpy
import time
from sklearn.datasets import fetch_openml
# Load data from https://www.openml.org/d/554
X, y = fetch_openml('mnist_784', version=1, return_X_y=True)
print(X.shape)
print(y)
# Data set information
image_dims = 1, 28, 28
n_training_samples = 60000 # How many training images to use
n_test_samples = 10000 # How many test images to use
classes = tuple([str(x) for x in range(9+1)])
# Load the training set
train_set = torchvision.datasets.MNIST(
root='./cifardata', train=True, download=True, transform=transforms.ToTensor())
train_sampler = SubsetRandomSampler(np.arange(n_training_samples, dtype=np.int64))
#Load the test set
test_set = torchvision.datasets.MNIST(
root='./cifardata', train=False, download=True, transform=transforms.ToTensor())
test_sampler = SubsetRandomSampler(np.arange(n_test_samples, dtype=np.int64))
def disp_image(image, class_idx, predicted=None):
# need to reorder the tensor dimensions to work properly with imshow
plt.imshow(image, 'gray')
plt.axis('off')
if predicted:
plt.title("Actual: " + classes[class_idx] + " Predicted: " + classes[predicted])
else:
plt.title("Actual: " + classes[class_idx])
plt.show()
print("training set input data shape", train_set.data.shape)
print("Number of training outputs", len(train_set.targets))
x, y = train_set[1]
disp_image(x.reshape(28,28), y)
import seaborn as sns
sns.countplot(train_set.targets.numpy())
plt.xticks(ticks=range(10), labels=classes)
plt.show()
sns.countplot(test_set.targets.numpy())
plt.xticks(ticks=range(10), labels=classes)
plt.show()
class MyCNN(nn.Module):
# The init funciton in Pytorch classes is used to keep track of the parameters of the model
# specifically the ones we want to update with gradient descent + backprop
# So we need to make sure we keep track of all of them here
def __init__(self):
super(MyCNN, self).__init__()
# layers defined here
# Make sure you understand what this convolutional layer is doing.
# E.g., considering looking at help(nn.Conv2D). Draw a picture of what
# this layer does to the data.
# note: image_dims[0] will be 1 as there are 1 color channels (R, G, B)
num_kernels = 16
self.conv1 = nn.Conv2d(image_dims[0], num_kernels, kernel_size=3, stride=1, padding=1)
# Make sure you understand what this MaxPool2D layer is doing.
# E.g., considering looking at help(nn.MaxPool2d). Draw a picture of
# what this layer does to the data.
self.pool = nn.MaxPool2d(kernel_size=2, stride=2, padding=0)
# maxpool_output_size is the total amount of data coming out of that
# layer. We have an exercise that asks you to explain why the line of
# code below computes this quantity.
self.maxpool_output_size = int(num_kernels * (image_dims[1] / 2) * (image_dims[2] / 2))
# Add on a fully connected layer (like in our MLP)
# fc stands for fully connected
fc1_size = 64
self.fc1 = nn.Linear(self.maxpool_output_size, fc1_size)
# we'll use this activation function internally in the network
self.activation_func = torch.nn.ReLU()
# Convert our fully connected layer into outputs that we can compare to the result
fc2_size = len(classes)
self.fc2 = nn.Linear(fc1_size, fc2_size)
# Note: that the output will not represent the probability of the
# output being in each class. The loss function we will use
# `CrossEntropyLoss` will take care of convering these values to
# probabilities and then computing the log loss with respect to the
# true label. We could break this out into multiple steps, but it turns
# out that the algorithm will be more numerically stable if we do it in
# one go. We have included a cell to show you the documentation for
# `CrossEntropyLoss` if you'd like to check it out.
# The forward function in the class defines the operations performed on a given input to the model
# and returns the output of the model
def forward(self, x):
x = self.conv1(x)
x = self.pool(x)
x = self.activation_func(x)
# this code flattens the output of the convolution, max pool,
# activation sequence of steps into a vector
x = x.view(-1, self.maxpool_output_size)
x = self.fc1(x)
x = self.activation_func(x)
x = self.fc2(x)
return x
# The loss function (which we chose to include as a method of the class, but doesn't need to be)
# returns the loss and optimizer used by the model
def get_loss(self, learning_rate):
# Loss function
loss = nn.CrossEntropyLoss()
# Optimizer, self.parameters() returns all the Pytorch operations that are attributes of the class
optimizer = optim.Adam(self.parameters(), lr=learning_rate)
return loss, optimizer
def visualize_network(net):
# Visualize the architecture of the model
# We need to give the net a fake input for this library to visualize the architecture
fake_input = Variable(torch.zeros((1,image_dims[0], image_dims[1], image_dims[2]))).to(device)
outputs = net(fake_input)
# Plot the DAG (Directed Acyclic Graph) of the model
return make_dot(outputs, dict(net.named_parameters()))
# Define what device we want to use
device = 'cuda' # 'cpu' if we want to not use the gpu
# Initialize the model, loss, and optimization function
net = MyCNN()
# This tells our model to send all of the tensors and operations to the GPU (or keep them at the CPU if we're not using GPU)
net.to(device)
visualize_network(net)
# Define training parameters
batch_size = 32
learning_rate = 1e-2
n_epochs = 10
# Get our data into the mini batch size that we defined
train_loader = torch.utils.data.DataLoader(train_set, batch_size=batch_size,
sampler=train_sampler, num_workers=2)
test_loader = torch.utils.data.DataLoader(
test_set, batch_size=128, sampler=test_sampler, num_workers=2)
def train_model(net):
""" Train a the specified network.
Outputs a tuple with the following four elements
train_hist_x: the x-values (batch number) that the training set was
evaluated on.
train_loss_hist: the loss values for the training set corresponding to
the batch numbers returned in train_hist_x
test_hist_x: the x-values (batch number) that the test set was
evaluated on.
test_loss_hist: the loss values for the test set corresponding to
the batch numbers returned in test_hist_x
"""
loss, optimizer = net.get_loss(learning_rate)
# Define some parameters to keep track of metrics
print_every = 20
idx = 0
train_hist_x = []
train_loss_hist = []
test_hist_x = []
test_loss_hist = []
training_start_time = time.time()
# Loop for n_epochs
for epoch in range(n_epochs):
running_loss = 0.0
start_time = time.time()
for i, data in enumerate(train_loader, 0):
# Get inputs in right form
inputs, labels = data
inputs, labels = Variable(inputs).to(device), Variable(labels).to(device)
# In Pytorch, We need to always remember to set the optimizer gradients to 0 before we recompute the new gradients
optimizer.zero_grad()
# Forward pass
outputs = net(inputs)
# Compute the loss and find the loss with respect to each parameter of the model
loss_size = loss(outputs, labels)
loss_size.backward()
# Change each parameter with respect to the recently computed loss.
optimizer.step()
# Update statistics
running_loss += loss_size.data.item()
# Print every 20th batch of an epoch
if (i % print_every) == print_every-1:
print("Epoch {}, Iteration {}\t train_loss: {:.2f} took: {:.2f}s".format(
epoch + 1, i+1,running_loss / print_every, time.time() - start_time))
# Reset running loss and time
train_loss_hist.append(running_loss / print_every)
train_hist_x.append(idx)
running_loss = 0.0
start_time = time.time()
idx += 1
# At the end of the epoch, do a pass on the test set
total_test_loss = 0
for inputs, labels in test_loader:
# Wrap tensors in Variables
inputs, labels = Variable(inputs).to(device), Variable(labels).to(device)
# Forward pass
test_outputs = net(inputs)
test_loss_size = loss(test_outputs, labels)
total_test_loss += test_loss_size.data.item()
test_loss_hist.append(total_test_loss / len(test_loader))
test_hist_x.append(idx)
print("Validation loss = {:.2f}".format(
total_test_loss / len(test_loader)))
print("Training finished, took {:.2f}s".format(
time.time() - training_start_time))
return train_hist_x, train_loss_hist, test_hist_x, test_loss_hist
train_hist_x, train_loss_hist, test_hist_x, test_loss_hist = train_model(net)
plt.plot(train_hist_x,train_loss_hist)
plt.plot(test_hist_x,test_loss_hist)
plt.legend(['train loss', 'validation loss'])
plt.xlabel('Batch number')
plt.ylabel('Loss')
plt.show()
print(test_loss_hist[-1])
n_correct = 0
n_total = 0
for i, data in enumerate(test_loader, 0):
# Get inputs in right form
inputs, labels = data
inputs, labels = Variable(inputs).to(device), Variable(labels).to(device)
# Forward pass
outputs = net(inputs)
n_correct += np.sum(np.argmax(outputs.cpu().detach().numpy(), axis=1) == labels.cpu().numpy())
n_total += labels.shape[0]
print("Testing accuracy is", n_correct/n_total)
plt.subplots(4, 4)
for i in range(net.conv1.weight.shape[0]):
plt.subplot(4, 4, i+1)
kernel = net.conv1.weight[i].cpu().detach().numpy()
im = kernel.mean(axis=0)
plt.pcolor(im, cmap='gray')
plt.show()
| 0.957942 | 0.734358 |
<a href="https://colab.research.google.com/github/jeffheaton/t81_558_deep_learning/blob/master/t81_558_class_01_2_intro_python.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# T81-558: Applications of Deep Neural Networks
**Module 1: Python Preliminaries**
* Instructor: [Jeff Heaton](https://sites.wustl.edu/jeffheaton/), McKelvey School of Engineering, [Washington University in St. Louis](https://engineering.wustl.edu/Programs/Pages/default.aspx)
* For more information visit the [class website](https://sites.wustl.edu/jeffheaton/t81-558/).
# Module 1 Material
* Part 1.1: Course Overview [[Video]](https://www.youtube.com/watch?v=taxS7a-goNs&list=PLjy4p-07OYzulelvJ5KVaT2pDlxivl_BN) [[Notebook]](t81_558_class_01_1_overview.ipynb)
* **Part 1.2: Introduction to Python** [[Video]](https://www.youtube.com/watch?v=czq5d53vKvo&list=PLjy4p-07OYzulelvJ5KVaT2pDlxivl_BN) [[Notebook]](t81_558_class_01_2_intro_python.ipynb)
* Part 1.3: Python Lists, Dictionaries, Sets and JSON [[Video]](https://www.youtube.com/watch?v=kcGx2I5akSs&list=PLjy4p-07OYzulelvJ5KVaT2pDlxivl_BN) [[Notebook]](t81_558_class_01_3_python_collections.ipynb)
* Part 1.4: File Handling [[Video]](https://www.youtube.com/watch?v=FSuSLCMgCZc&list=PLjy4p-07OYzulelvJ5KVaT2pDlxivl_BN) [[Notebook]](t81_558_class_01_4_python_files.ipynb)
* Part 1.5: Functions, Lambdas, and Map/Reduce [[Video]](https://www.youtube.com/watch?v=jQH1ZCSj6Ng&list=PLjy4p-07OYzulelvJ5KVaT2pDlxivl_BN) [[Notebook]](t81_558_class_01_5_python_functional.ipynb)
# Google CoLab Instructions
The following code ensures that Google CoLab is running the correct version of TensorFlow.
```
try:
from google.colab import drive
%tensorflow_version 2.x
COLAB = True
print("Note: using Google CoLab")
except:
print("Note: not using Google CoLab")
COLAB = False
```
# Part 1.2: Introduction to Python
Python is an interpreted, high-level, general-purpose programming language. Created by Guido van Rossum and first released in 1991, Python's design philosophy emphasizes code readability with its notable use of significant whitespace. Its language constructs and object-oriented approach aim to help programmers write clear, logical code for small and large-scale projects. Python has become a common language for machine learning research and is the primary language for TensorFlow.
Python 3.0, released in 2008, was a significant revision of the language that is not entirely backward-compatible, and much Python 2 code does not run unmodified on Python 3. This course makes use of Python 3. Furthermore, TensorFlow is not compatible with versions of Python earlier than 3. A non-profit organization, the Python Software Foundation (PSF), manages and directs resources for Python development. On January 1, 2020, the PSF discontinued the Python 2 language and no longer provides security patches and other improvements. Python interpreters are available for many operating systems.
The first two modules of this course provide an introduction to some aspects of the Python programming language. However, entire books focus on Python. Two modules will not cover every detail of this language. The reader is encouraged to consult additional sources on the Python language.
Like most tutorials, we will begin by printing Hello World.
```
print("Hello World")
```
The above code passes a constant string, containing the text "hello world" to a function that is named print.
You can also leave comments in your code to explain what you are doing. Comments can begin anywhere in a line.
```
# Single line comment (this has no effect on your program)
print("Hello World") # Say hello
```
Strings are very versatile and allow your program to process textual information. Constant string, enclosed in quotes, define literal string values inside your program. Sometimes you may wish to define a larger amount of literal text inside of your program. This text might consist of multiple lines. The triple quote allows for multiple lines of text.
```
print("""Print
Multiple
Lines
""")
```
Like many languages uses single (') and double (") quotes interchangeably to denote literal string constants. The general convention is that double quotes should enclose actual text, such as words or sentences. Single quotes should enclose symbolic text, such as error codes. An example of an error code might be 'HTTP404'.
However, there is no difference between single and double quotes in Python, and you may use whichever you like. The following code makes use of a single quote.
```
print('Hello World')
```
In addition to strings, Python allows numbers as literal constants in programs. Python includes support for floating-point, integer, complex, and other types of numbers. This course will not make use of complex numbers. Unlike strings, quotes do not enclose numbers.
The presence of a decimal point differentiates floating-point and integer numbers. For example, the value 42 is an integer. Similarly, 42.5 is a floating-point number. If you wish to have a floating-point number, without a fraction part, you should specify a zero fraction. The value 42.0 is a floating-point number, although it has no fractional part. As an example, the following code prints two numbers.
```
print(42)
print(42.5)
```
So far, we have only seen how to define literal numeric and string values. These literal values are constant and do not change as your program runs. Variables allow your program to hold values that can change as the program runs. Variables have names that allow you to reference their values. The following code assigns an integer value to a variable named "a" and a string value to a variable named "b."
```
a = 10
b = "ten"
print(a)
print(b)
```
The key feature of variables is that they can change. The following code demonstrates how to change the values held by variables.
```
a = 10
print(a)
a = a + 1
print(a)
```
You can mix strings and variables for printing. This technique is called a formatted or interpolated string. The variables must be inside of the curly braces. In Python, this type of string is generally called an f-string. The f-string is denoted by placing an "f" just in front of the opening single or double quote that begins the string. The following code demonstrates the use of an f-string to mix several variables with a literal string.
```
a = 10
print(f'The value of a is {a}')
```
You can also use f-strings with math (called an expression). Curly braces can enclose any valid Python expression for printing. The following code demonstrates the use of an expression inside of the curly braces of an f-string.
```
a = 10
print(f'The value of a plus 5 is {a+5}')
```
Python has many ways to print numbers; these are all correct. However, for this course, we will use f-strings. The following code demonstrates some of the varied methods of printing numbers in Python.
```
a = 5
print(f'a is {a}') # Preferred method for this course.
print('a is {}'.format(a))
print('a is ' + str(a))
print('a is %d' % (a))
```
You can use if-statements to perform logic. Notice the indents? These if-statements are how Python defines blocks of code to execute together. A block usually begins after a colon and includes any lines at the same level of indent. Unlike many other programming languages, Python uses whitespace to define blocks of code. The fact that whitespace is significant to the meaning of program code is a frequent source of annoyance for new programmers of Python. Tabs and spaces are both used to define the scope in a Python program. Mixing both spaces and tabs in the same program is not recommended.
```
a = 5
if a>5:
print('The variable a is greater than 5.')
else:
print('The variable a is not greater than 5')
```
The following if-statement has multiple levels. It can be easy to indent these levels improperly, so be careful. This code contains a nested if-statement under the first "a==5" if-statement. Only if a is equal to 5 will the nested "b==6" if-statement be executed. Also, not that the "elif" command means "else if."
```
a = 5
b = 6
if a==5:
print('The variable a is 5')
if b==6:
print('The variable b is also 6')
elif a==6:
print('The variable a is 6')
```
It is also important to note that the double equal ("==") operator is used to test the equality of two expressions. The single equal ("=") operator is only used to assign values to variables in Python. The greater than (">"), less than ("<"), greater than or equal (">="), less than or equal ("<=") all perform as would generally be accepted. Testing for inequality is performed with the not equal ("!=") operator.
It is common in programming languages to loop over a range of numbers. Python accomplishes this through the use of the **range** operation. Here you can see a **for** loop and a **range** operation that causes the program to loop between 1 and 9.
```
for x in range(1, 10): # If you ever see xrange, you are in Python 2
print(x) # If you ever see print x (no parenthesis), you are in Python 2
```
This code illustrates some incompatibilities between Python 2 and Python 3. Before Python 3, it was acceptable to leave the parentheses off of a *print* function call. This method of invoking the *print* command is no longer allowed in Python 3. Similarly, it used to be a performance improvement to use the *xrange* command in place of range command at times. Python 3 incorporated all of the functionality of the *xrange* Python 2 command into the normal *range* command. As a result, the programmer should not use the *xrange* command in Python 3. If you see either of these constructs used in example code, then you are looking at an older Python 2 era example.
The *range* command is used in conjunction with loops to pass over a specific range of numbers. Cases, where you must loop over specific number ranges, are somewhat uncommon. Generally, programmers use loops on collections of items, rather than hard-coding numeric values into your code. Collections, as well as the operations that loops can perform on them, is covered later in this module.
The following is a further example of a looped printing of strings and numbers.
```
acc = 0
for x in range(1, 10):
acc += x
print(f"Adding {x}, sum so far is {acc}")
print(f"Final sum: {acc}")
```
|
github_jupyter
|
try:
from google.colab import drive
%tensorflow_version 2.x
COLAB = True
print("Note: using Google CoLab")
except:
print("Note: not using Google CoLab")
COLAB = False
print("Hello World")
# Single line comment (this has no effect on your program)
print("Hello World") # Say hello
print("""Print
Multiple
Lines
""")
print('Hello World')
print(42)
print(42.5)
a = 10
b = "ten"
print(a)
print(b)
a = 10
print(a)
a = a + 1
print(a)
a = 10
print(f'The value of a is {a}')
a = 10
print(f'The value of a plus 5 is {a+5}')
a = 5
print(f'a is {a}') # Preferred method for this course.
print('a is {}'.format(a))
print('a is ' + str(a))
print('a is %d' % (a))
a = 5
if a>5:
print('The variable a is greater than 5.')
else:
print('The variable a is not greater than 5')
a = 5
b = 6
if a==5:
print('The variable a is 5')
if b==6:
print('The variable b is also 6')
elif a==6:
print('The variable a is 6')
for x in range(1, 10): # If you ever see xrange, you are in Python 2
print(x) # If you ever see print x (no parenthesis), you are in Python 2
acc = 0
for x in range(1, 10):
acc += x
print(f"Adding {x}, sum so far is {acc}")
print(f"Final sum: {acc}")
| 0.18363 | 0.99294 |
```
import os,sys,inspect
currentdir = os.path.dirname(os.path.abspath(inspect.getfile(inspect.currentframe())))
parentdir = os.path.dirname(currentdir)
# Add parent dir to path, so that python finds the lenskit package
sys.path.insert(0,parentdir)
from lenskit.metrics import dataGenerator
from lenskit.metrics import topnFair
from lenskit import batch, topn, util, topnFair
from lenskit import crossfold as xf
from lenskit.algorithms import Recommender, als, user_knn as knn
import numpy as np
import pandas as pd
import math
%matplotlib inline
index=range(0,20)
#col_names1 = ["item", 'user']
col_names = ['item', 'user', 'rank', 'protected']
my_df = pd.DataFrame(index = index, columns = col_names)
my_df["item"] = np.arange(20)
my_df["user"] = [1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1]
my_df["rank"] = np.arange(20)
my_df["protected"] = [0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,1,1]
print(my_df)
#data = np.array([np.arange(10)], [np.arange(10)])
#data.head
res1= my_df.item.nunique()
print(res1)
res2 = my_df.loc[my_df['protected'] == 1]
print(res2.item.nunique())
rla = topnFair.FairRecListAnalysis(['user'])
rla.add_metric("rND")
rla.add_metric("rKL")
rla.add_metric("rRD")
results = rla.compute(my_df, my_df, 'protected')
results.head()
#test rND for my_df = [0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,1,1] 0/10 vs 2/20
dif = abs((0/10)-(2/20))
rnd = (dif/math.log(10+1,2)+(0/math.log(20+1,2)))
rnd
#test rND for my_df = [0,0,0,0,0,0,0,0,1,0,0,0,0,0,0,0,0,1,1,1] 1/10 vs 4/20
dif1 = abs((1/10)-(4/20))
rnd1 = (dif1/math.log(10+1,2)+(0/math.log(20+1,2)))
rnd1
#test rRD for my_df = [0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,1,1] 0/10 vs 2/20
ratio1 = 0/10
ratio2 = 2/18
dif = abs(ratio1-ratio2)
rrd = (dif/math.log(10+1,2)+(0/math.log(20+1,2)))
rrd
print(topnFair.getNormalizer(20,2,"rRD"))
nrRD = rrd/topnFair.getNormalizer(20,2,"rRD")
nrRD
#test rRD for my_df = [0,0,0,0,0,0,0,1,1,0,0,0,0,0,0,0,0,0,1,1] 2/10 vs 4/20
ratio1 = 2/8
ratio2 = 4/16
dif = abs(ratio1-ratio2)
rrd = (dif/math.log(10+1,2)+(0/math.log(20+1,2)))
rrd
#test rRD for my_df = [0,0,0,0,0,0,0,0,0,1,0,0,0,0,0,0,0,1,1,1] 1/10 vs 4/20
ratio1 = 1/9
ratio2 = 4/16
dif = abs(ratio1-ratio2)
rrd = (dif/math.log(10+1,2)+(0/math.log(20+1,2)))
rrd
#test rRD for my_df = 5/10 vs 10/20
ratio1 = 5/5
ratio2 = 10/10
dif = abs(ratio1-ratio2)
rrd1 = (dif/math.log(10+1,2)+(0/math.log(20+1,2)))
rrd1
print(topnFair.getNormalizer(20,10,"rRD"))
nrRD = rrd1/topnFair.getNormalizer(20,10,"rRD")
nrRD
#test rKL for my_df["protected"] = [0,0,0,0,0,0,0,0,0,1,0,0,0,0,0,0,0,1,1,1] 1/10 vs 4/20
px= 1/10
qx=4/20
rkl =(px*math.log(px/qx,2))+(1-px)*math.log((1-px)/(1-qx),2)
print(rkl)
print(rkl/math.log(11,2))
#test rKL for my_df["protected"] = [0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,1,1] 0/10 vs 2/20
# corner case -- px cant be 0 thus set manuale to 0,001
#px= 0/10 - px cant be 0 thus set manuale to 0,001
px = 0.001
qx=2/20
rkl2 =(px*math.log(px/qx,2))+(1-px)*math.log((1-px)/(1-qx),2)
print(rkl2)
print(rkl2/math.log(11,2))
```
|
github_jupyter
|
import os,sys,inspect
currentdir = os.path.dirname(os.path.abspath(inspect.getfile(inspect.currentframe())))
parentdir = os.path.dirname(currentdir)
# Add parent dir to path, so that python finds the lenskit package
sys.path.insert(0,parentdir)
from lenskit.metrics import dataGenerator
from lenskit.metrics import topnFair
from lenskit import batch, topn, util, topnFair
from lenskit import crossfold as xf
from lenskit.algorithms import Recommender, als, user_knn as knn
import numpy as np
import pandas as pd
import math
%matplotlib inline
index=range(0,20)
#col_names1 = ["item", 'user']
col_names = ['item', 'user', 'rank', 'protected']
my_df = pd.DataFrame(index = index, columns = col_names)
my_df["item"] = np.arange(20)
my_df["user"] = [1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1]
my_df["rank"] = np.arange(20)
my_df["protected"] = [0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,1,1]
print(my_df)
#data = np.array([np.arange(10)], [np.arange(10)])
#data.head
res1= my_df.item.nunique()
print(res1)
res2 = my_df.loc[my_df['protected'] == 1]
print(res2.item.nunique())
rla = topnFair.FairRecListAnalysis(['user'])
rla.add_metric("rND")
rla.add_metric("rKL")
rla.add_metric("rRD")
results = rla.compute(my_df, my_df, 'protected')
results.head()
#test rND for my_df = [0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,1,1] 0/10 vs 2/20
dif = abs((0/10)-(2/20))
rnd = (dif/math.log(10+1,2)+(0/math.log(20+1,2)))
rnd
#test rND for my_df = [0,0,0,0,0,0,0,0,1,0,0,0,0,0,0,0,0,1,1,1] 1/10 vs 4/20
dif1 = abs((1/10)-(4/20))
rnd1 = (dif1/math.log(10+1,2)+(0/math.log(20+1,2)))
rnd1
#test rRD for my_df = [0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,1,1] 0/10 vs 2/20
ratio1 = 0/10
ratio2 = 2/18
dif = abs(ratio1-ratio2)
rrd = (dif/math.log(10+1,2)+(0/math.log(20+1,2)))
rrd
print(topnFair.getNormalizer(20,2,"rRD"))
nrRD = rrd/topnFair.getNormalizer(20,2,"rRD")
nrRD
#test rRD for my_df = [0,0,0,0,0,0,0,1,1,0,0,0,0,0,0,0,0,0,1,1] 2/10 vs 4/20
ratio1 = 2/8
ratio2 = 4/16
dif = abs(ratio1-ratio2)
rrd = (dif/math.log(10+1,2)+(0/math.log(20+1,2)))
rrd
#test rRD for my_df = [0,0,0,0,0,0,0,0,0,1,0,0,0,0,0,0,0,1,1,1] 1/10 vs 4/20
ratio1 = 1/9
ratio2 = 4/16
dif = abs(ratio1-ratio2)
rrd = (dif/math.log(10+1,2)+(0/math.log(20+1,2)))
rrd
#test rRD for my_df = 5/10 vs 10/20
ratio1 = 5/5
ratio2 = 10/10
dif = abs(ratio1-ratio2)
rrd1 = (dif/math.log(10+1,2)+(0/math.log(20+1,2)))
rrd1
print(topnFair.getNormalizer(20,10,"rRD"))
nrRD = rrd1/topnFair.getNormalizer(20,10,"rRD")
nrRD
#test rKL for my_df["protected"] = [0,0,0,0,0,0,0,0,0,1,0,0,0,0,0,0,0,1,1,1] 1/10 vs 4/20
px= 1/10
qx=4/20
rkl =(px*math.log(px/qx,2))+(1-px)*math.log((1-px)/(1-qx),2)
print(rkl)
print(rkl/math.log(11,2))
#test rKL for my_df["protected"] = [0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,1,1] 0/10 vs 2/20
# corner case -- px cant be 0 thus set manuale to 0,001
#px= 0/10 - px cant be 0 thus set manuale to 0,001
px = 0.001
qx=2/20
rkl2 =(px*math.log(px/qx,2))+(1-px)*math.log((1-px)/(1-qx),2)
print(rkl2)
print(rkl2/math.log(11,2))
| 0.127748 | 0.191252 |
# Machine Learning Engineer Nanodegree
## Model Evaluation & Validation
## Project: Predicting Boston Housing Prices
Welcome to the first project of the Machine Learning Engineer Nanodegree! In this notebook, some template code has already been provided for you, and you will need to implement additional functionality to successfully complete this project. You will not need to modify the included code beyond what is requested. Sections that begin with **'Implementation'** in the header indicate that the following block of code will require additional functionality which you must provide. Instructions will be provided for each section and the specifics of the implementation are marked in the code block with a 'TODO' statement. Please be sure to read the instructions carefully!
In addition to implementing code, there will be questions that you must answer which relate to the project and your implementation. Each section where you will answer a question is preceded by a **'Question X'** header. Carefully read each question and provide thorough answers in the following text boxes that begin with **'Answer:'**. Your project submission will be evaluated based on your answers to each of the questions and the implementation you provide.
>**Note:** Code and Markdown cells can be executed using the **Shift + Enter** keyboard shortcut. In addition, Markdown cells can be edited by typically double-clicking the cell to enter edit mode.
## Getting Started
In this project, you will evaluate the performance and predictive power of a model that has been trained and tested on data collected from homes in suburbs of Boston, Massachusetts. A model trained on this data that is seen as a *good fit* could then be used to make certain predictions about a home — in particular, its monetary value. This model would prove to be invaluable for someone like a real estate agent who could make use of such information on a daily basis.
The dataset for this project originates from the [UCI Machine Learning Repository](https://archive.ics.uci.edu/ml/machine-learning-databases/housing/). The Boston housing data was collected in 1978 and each of the 506 entries represent aggregated data about 14 features for homes from various suburbs in Boston, Massachusetts. For the purposes of this project, the following preprocessing steps have been made to the dataset:
- 16 data points have an `'MEDV'` value of 50.0. These data points likely contain **missing or censored values** and have been removed.
- 1 data point has an `'RM'` value of 8.78. This data point can be considered an **outlier** and has been removed.
- The features `'RM'`, `'LSTAT'`, `'PTRATIO'`, and `'MEDV'` are essential. The remaining **non-relevant features** have been excluded.
- The feature `'MEDV'` has been **multiplicatively scaled** to account for 35 years of market inflation.
Run the code cell below to load the Boston housing dataset, along with a few of the necessary Python libraries required for this project. You will know the dataset loaded successfully if the size of the dataset is reported.
```
# Import libraries necessary for this project
import numpy as np
import pandas as pd
from sklearn.cross_validation import ShuffleSplit
# Import supplementary visualizations code visuals.py
import visuals as vs
# Pretty display for notebooks
%matplotlib inline
# Load the Boston housing dataset
data = pd.read_csv('housing.csv')
prices = data['MEDV']
features = data.drop('MEDV', axis = 1)
# Success
print("Boston housing dataset has {} data points with {} variables each.".format(*data.shape))
```
## Data Exploration
In this first section of this project, you will make a cursory investigation about the Boston housing data and provide your observations. Familiarizing yourself with the data through an explorative process is a fundamental practice to help you better understand and justify your results.
Since the main goal of this project is to construct a working model which has the capability of predicting the value of houses, we will need to separate the dataset into **features** and the **target variable**. The **features**, `'RM'`, `'LSTAT'`, and `'PTRATIO'`, give us quantitative information about each data point. The **target variable**, `'MEDV'`, will be the variable we seek to predict. These are stored in `features` and `prices`, respectively.
### Implementation: Calculate Statistics
For your very first coding implementation, you will calculate descriptive statistics about the Boston housing prices. Since `numpy` has already been imported for you, use this library to perform the necessary calculations. These statistics will be extremely important later on to analyze various prediction results from the constructed model.
In the code cell below, you will need to implement the following:
- Calculate the minimum, maximum, mean, median, and standard deviation of `'MEDV'`, which is stored in `prices`.
- Store each calculation in their respective variable.
```
# TODO: Minimum price of the data
minimum_price = np.min(prices)
# TODO: Maximum price of the data
maximum_price = np.max(prices)
# TODO: Mean price of the data
mean_price = np.mean(prices)
# TODO: Median price of the data
median_price = np.median(prices)
# TODO: Standard deviation of prices of the data
std_price = np.std(prices)
# Show the calculated statistics
print("Statistics for Boston housing dataset:\n")
print("Minimum price: ${}".format(minimum_price))
print("Maximum price: ${}".format(maximum_price))
print("Mean price: ${}".format(mean_price))
print("Median price ${}".format(median_price))
print("Standard deviation of prices: ${}".format(std_price))
```
### Question 1 - Feature Observation
As a reminder, we are using three features from the Boston housing dataset: `'RM'`, `'LSTAT'`, and `'PTRATIO'`. For each data point (neighborhood):
- `'RM'` is the average number of rooms among homes in the neighborhood.
- `'LSTAT'` is the percentage of homeowners in the neighborhood considered "lower class" (working poor).
- `'PTRATIO'` is the ratio of students to teachers in primary and secondary schools in the neighborhood.
** Using your intuition, for each of the three features above, do you think that an increase in the value of that feature would lead to an **increase** in the value of `'MEDV'` or a **decrease** in the value of `'MEDV'`? Justify your answer for each.**
**Hint:** This problem can phrased using examples like below.
* Would you expect a home that has an `'RM'` value(number of rooms) of 6 be worth more or less than a home that has an `'RM'` value of 7?
* Would you expect a neighborhood that has an `'LSTAT'` value(percent of lower class workers) of 15 have home prices be worth more or less than a neighborhood that has an `'LSTAT'` value of 20?
* Would you expect a neighborhood that has an `'PTRATIO'` value(ratio of students to teachers) of 10 have home prices be worth more or less than a neighborhood that has an `'PTRATIO'` value of 15?
**Answer:**
***RM***: I consider that if **the number of rooms, 'RM'**, within a house increases, the cost of the same will also. However, this may vary depending on the quality of the room, services and other factors. Therefore, **if the residence offers more availability of spaces, or rooms, then the 'MEDV', and the value of the house, will increase.**
***LSTAT***:Also, the cost of the house is also affected by the neighborhood in which it is located, 'LSTAT', because if the class in which it is found is medium-high, the price can influence positively (increases), whereas if the house is in a lower class zone, it can be affected in a negative way. In this way, if the house is located in a poor neighborhood, and it is a question of selling a house at a high cost, then the sale will fail, since no one will be able to pay for it. For this reason, the 'MEDV' depends intrinsically on LSTAT, although they are inversely proportional. **If the LSTAT increases, the 'MEDV' decreases.**
***PTRATIO***: If the residence is in an area where _more students live than teachers_, then the cost of the residence, **'MEDV', will be affected: it will decrease**. However, if _the majority of the population is a teacher_, people who are able to pay more for a house with certain luxuries, then **the value of the residence, MEDV, increases**. Finally, _if the percentage of student-teacher population is approximately equal_, then the most likely is that **the residence is cheap, and decrease.**
----
## Developing a Model
In this second section of the project, you will develop the tools and techniques necessary for a model to make a prediction. Being able to make accurate evaluations of each model's performance through the use of these tools and techniques helps to greatly reinforce the confidence in your predictions.
### Implementation: Define a Performance Metric
It is difficult to measure the quality of a given model without quantifying its performance over training and testing. This is typically done using some type of performance metric, whether it is through calculating some type of error, the goodness of fit, or some other useful measurement. For this project, you will be calculating the [*coefficient of determination*](http://stattrek.com/statistics/dictionary.aspx?definition=coefficient_of_determination), R<sup>2</sup>, to quantify your model's performance. The coefficient of determination for a model is a useful statistic in regression analysis, as it often describes how "good" that model is at making predictions.
The values for R<sup>2</sup> range from 0 to 1, which captures the percentage of squared correlation between the predicted and actual values of the **target variable**. A model with an R<sup>2</sup> of 0 is no better than a model that always predicts the *mean* of the target variable, whereas a model with an R<sup>2</sup> of 1 perfectly predicts the target variable. Any value between 0 and 1 indicates what percentage of the target variable, using this model, can be explained by the **features**. _A model can be given a negative R<sup>2</sup> as well, which indicates that the model is **arbitrarily worse** than one that always predicts the mean of the target variable._
For the `performance_metric` function in the code cell below, you will need to implement the following:
- Use `r2_score` from `sklearn.metrics` to perform a performance calculation between `y_true` and `y_predict`.
- Assign the performance score to the `score` variable.
```
# TODO: Import 'r2_score'
from sklearn.metrics import r2_score
def performance_metric(y_true, y_predict):
""" Calculates and returns the performance score between
true and predicted values based on the metric chosen. """
# TODO: Calculate the performance score between 'y_true' and 'y_predict'
score = r2_score(y_true,y_predict)
# Return the score
return score
```
### Question 2 - Goodness of Fit
Assume that a dataset contains five data points and a model made the following predictions for the target variable:
| True Value | Prediction |
| :-------------: | :--------: |
| 3.0 | 2.5 |
| -0.5 | 0.0 |
| 2.0 | 2.1 |
| 7.0 | 7.8 |
| 4.2 | 5.3 |
Run the code cell below to use the `performance_metric` function and calculate this model's coefficient of determination.
```
# Calculate the performance of this model
score = performance_metric([3, -0.5, 2, 7, 4.2], [2.5, 0.0, 2.1, 7.8, 5.3])
print("Model has a coefficient of determination, R^2, of {:.3f}.".format(score))
```
* Would you consider this model to have successfully captured the variation of the target variable?
* Why or why not?
** Hint: ** The R2 score is the proportion of the variance in the dependent variable that is predictable from the independent variable. In other words:
* R2 score of 0 means that the dependent variable cannot be predicted from the independent variable.
* R2 score of 1 means the dependent variable can be predicted from the independent variable.
* R2 score between 0 and 1 indicates the extent to which the dependent variable is predictable. An
* R2 score of 0.40 means that 40 percent of the variance in Y is predictable from X.
**Answer:**
**Yes, I consider this model have succesfully captured the variation of the target variable.**
Why? Well, we know that Coefficient of determination, R^2, determines the squared correlation between the actual and the predicted values, so considering that R^2 value is almost 1, we taking the borderline cases:
Given:
R^2 = 1 - alpha
If alpha ~ 1:
Both actual and predicted values are almost the same, so if R^2 ~ 0 then this is a bad model.
If alpha ~ 0:
the mean square error for the original model is smaller than the mean square error than the simple model, so if R^2 ~ 1 then this is a good model.
So, given that R^2 score is between 0 and 1, and alpha is approximately 0.0777, that indicates the dependent variable is predictable with a porcentage of 92.30%.
### Implementation: Shuffle and Split Data
Your next implementation requires that you take the Boston housing dataset and split the data into training and testing subsets. Typically, the data is also shuffled into a random order when creating the training and testing subsets to remove any bias in the ordering of the dataset.
For the code cell below, you will need to implement the following:
- Use `train_test_split` from `sklearn.cross_validation` to shuffle and split the `features` and `prices` data into training and testing sets.
- Split the data into 80% training and 20% testing.
- Set the `random_state` for `train_test_split` to a value of your choice. This ensures results are consistent.
- Assign the train and testing splits to `X_train`, `X_test`, `y_train`, and `y_test`.
```
# TODO: Import 'train_test_split'
from sklearn.cross_validation import train_test_split
# TODO: Shuffle and split the data into training and testing subsets
X_train, X_test, y_train, y_test = train_test_split(features, prices, test_size = 0.20, random_state = 42)
# Success
print("Training and testing split was successful.")
```
### Question 3 - Training and Testing
* What is the benefit to splitting a dataset into some ratio of training and testing subsets for a learning algorithm?
**Hint:** Think about how overfitting or underfitting is contingent upon how splits on data is done.
**Answer:**
The main idea when we divide a set of data into these two sets is to evaluate, validate and measure our model and its accuracy and take decisions to verify if the model chosen is the best one, using the training set. When we doing this, we can avoid common mistakes when selecting a model, because if we **simplify the problem too much**, an error of **underfitting** may arise , so the model has **high bias** and it could have the problem with training set and there is another problem if we over complicate the problem: **overfitting**, where model works very well with training set but not with testing set, it has a **high variance** and then **the model memorize instead of learn**.
These data sets help us to train our model, submit it under metrics, compare it with others possible models and later make decisions, either making use of cross-validation or we can see how the model behaves through learning curves or other metrics of "quality control" and accuracy. After training the model, we submit it to a test data that it has never seen before to evaluate its performance and if it works very well, so submit to production.
----
## Analyzing Model Performance
In this third section of the project, you'll take a look at several models' learning and testing performances on various subsets of training data. Additionally, you'll investigate one particular algorithm with an increasing `'max_depth'` parameter on the full training set to observe how model complexity affects performance. Graphing your model's performance based on varying criteria can be beneficial in the analysis process, such as visualizing behavior that may not have been apparent from the results alone.
### Learning Curves
The following code cell produces four graphs for a decision tree model with different maximum depths. Each graph visualizes the learning curves of the model for both training and testing as the size of the training set is increased. Note that the shaded region of a learning curve denotes the uncertainty of that curve (measured as the standard deviation). The model is scored on both the training and testing sets using R<sup>2</sup>, the coefficient of determination.
Run the code cell below and use these graphs to answer the following question.
```
# Produce learning curves for varying training set sizes and maximum depths
vs.ModelLearning(features, prices)
```
### Question 4 - Learning the Data
* Choose one of the graphs above and state the maximum depth for the model.
* What happens to the score of the training curve as more training points are added? What about the testing curve?
* Would having more training points benefit the model?
**Hint:** Are the learning curves converging to particular scores? Generally speaking, the more data you have, the better. But if your training and testing curves are converging with a score above your benchmark threshold, would this be necessary?
Think about the pros and cons of adding more training points based on if the training and testing curves are converging.
**Answer:**
_I have chosen the graph number 2_, when **max_depth = 3**
Considering that as the number of training points increases, the learning curve of training data decreases, while the test learning curve increases. However, it seems to be in there is a point, approximately between a number of training points of 300-350 begins to stabilize, that is, begins to converge. So, if we start adding more training points to the model it does not benefit the model at all. We can say that this is a good model.
In the graph number 1, **max_depth = 1**, we can see there is a underfitting problem because it has an underfitting problem since the function can converge at one point, but it will take too long. In the curve of the training data set: at the beginning it has a bad performance, but later it is stabilizing and the same thing happens with the curve of the test data set.
And the others graphs we can see there is a overfitting problem, because the learning curve of training data remains almost stable, so the model is memorizing and not learning.
### Complexity Curves
The following code cell produces a graph for a decision tree model that has been trained and validated on the training data using different maximum depths. The graph produces two complexity curves — one for training and one for validation. Similar to the **learning curves**, the shaded regions of both the complexity curves denote the uncertainty in those curves, and the model is scored on both the training and validation sets using the `performance_metric` function.
** Run the code cell below and use this graph to answer the following two questions Q5 and Q6. **
```
vs.ModelComplexity(X_train, y_train)
```
### Question 5 - Bias-Variance Tradeoff
* When the model is trained with a maximum depth of 1, does the model suffer from high bias or from high variance?
* How about when the model is trained with a maximum depth of 10? What visual cues in the graph justify your conclusions?
**Hint:** High bias is a sign of underfitting(model is not complex enough to pick up the nuances in the data) and high variance is a sign of overfitting(model is by-hearting the data and cannot generalize well). Think about which model(depth 1 or 10) aligns with which part of the tradeoff.
**Answer: **
When the model is trained with a **maximum depth of 1**, does the model suffer from **high bias (underfitting)** and when the model is trained with a **maximum depth of 10** suffer from **high variance (overfitting)**, both learning curves training and validation converge in different points.
We can also say that the model "remembers" training data too well that it fails to generalize to unseen data in testing set, when **max_depth = 10** and when **max_depth = 1** the curve of the training data set: at the beginning it has a bad performance, but later it is stabilizing and the same thing happens with the curve of the test data set.
### Question 6 - Best-Guess Optimal Model
* Which maximum depth do you think results in a model that best generalizes to unseen data?
* What intuition lead you to this answer?
** Hint: ** Look at the graph above Question 5 and see where the validation scores lie for the various depths that have been assigned to the model. Does it get better with increased depth? At what point do we get our best validation score without overcomplicating our model? And remember, Occams Razor states "Among competing hypotheses, the one with the fewest assumptions should be selected."
**Answer: **
I think that **maximum depth equals 3** results in a model that best generalizes to unseen data, because in the graph above of Q5, approximately in a score of 6-8, has better performance, and also in the Q4 I explain why it is better.
Since the behavior of the last graphs, with a maximum depth of 1 creates problems of underfitting and with 6-10 generates problems of overfitting, then it can be deduced that the ideal depth is in a range of 2 to 5.
Even if **we use an algorithm such as binary search**, you can probably find the best depth. In this case, based on the behavior of the learning cruvas of questions 4 (Decision Tree Regressor Learning Performances) and 5 (Decision Tree Regressor Complexity Performances), we see that the maximum depth of 3 turns out to have better performance.
-----
## Evaluating Model Performance
In this final section of the project, you will construct a model and make a prediction on the client's feature set using an optimized model from `fit_model`.
### Question 7 - Grid Search
* What is the grid search technique?
* How it can be applied to optimize a learning algorithm?
** Hint: ** When explaining the Grid Search technique, be sure to touch upon why it is used, what the 'grid' entails and what the end goal of this method is. To solidify your answer, you can also give an example of a parameter in a model that can be optimized using this approach.
**Answer:**
**Grid Search Technique**:
> It is the process of **adjusting parameters (hyper parameters)** to _determine the optimal values for a given model_. This process **scans the data to configure optimal parameters for a given model**.
>
> This algorithm is **very powerful because depending on the model used**, certain parameters will be varied. However, _it can become extremely expensive_.
>
> In general: **Build a model in each possible parameter combination and store the model, after that we choose the best model.**
Example:
Suppose you are going to train a Logistic Regression model. We have some candidates and we will train them with training dataset due to get their curves and coefficients.
- We are using cross-validation and their data to calculate F1 score of all models. After that, we'll choose the highest and finally we use the testing dataset to sure that it is the best model.
Hyper-parameters | Parameters | F1 score |
------- | -------------------|----------|
Degree = 1 | Linear polynomial | 0.5 |
Degree = 2 | Quadratic polynomial | 0.8 |
Degree = 3 | polynomial degree 3 | 0.4 |
Degree = 4 | polynomial degree 4 | 0.2 |
Where:
- **Parameters**: Coefficients of the polynomial.
- **Hyper-parameters/Model**: Degree of the polynomial.
### Question 8 - Cross-Validation
* What is the k-fold cross-validation training technique?
* What benefit does this technique provide for grid search when optimizing a model?
**Hint:** When explaining the k-fold cross validation technique, be sure to touch upon what 'k' is, how the dataset is split into different parts for training and testing and the number of times it is run based on the 'k' value.
When thinking about how k-fold cross validation helps grid search, think about the main drawbacks of grid search which are hinged upon **using a particular subset of data for training or testing** and how k-fold cv could help alleviate that. You can refer to the [docs](http://scikit-learn.org/stable/modules/cross_validation.html#cross-validation) for your answer.
**Answer:**
**K-fold Cross-validation:**
It is a technique used to evaluate the results of a statistical analysis and **ensure that they are independent of the partition between training and test data.**
> How does work it?
1. Take the original data and create from them **two separate sets**: _Training data set_ and _Cross validation data set_.
2. Training set will be divided into k buckets.
Example: K = 4:
* #b: Train blue
* #r: Train red
* ~b: Testing blue
* ~r: Testing red
K1 | K2 | K3 | K4 | Ki-train |
------- | -----------|--------|----------|---------------|
~b~r~r | #b#r#r| #b#r#r | #b#r#r | K1 train |
#b#r#r | ~b~r~r| #b#r#r | #b#r#r | K2 train |
#b#r#r | #b#r#r| ~b~r~r | #b#r#r | K3 train |
#b#r#r | #b#r#r| #b#r#r | ~b~r~r | K4 trian |
3. Train our model k-times.
- Everytime we train our model, we'll use a different buckets for our testing set and the remaining of them will be our training set.
4. We average the results to obtain a final model.
> Then, said model can be used on the validation set generated in the first part, since, it is assumed, it is this model that offered the best overall result during the training phase.
**Cross-validation therefore allows for a model to be trained and evaluated on a larger set of observations. This is ideal for a method like grid search, which as mentioned above will exhaustively fine tune and optimize a model’s hyperparameters**
### Implementation: Fitting a Model
Your final implementation requires that you bring everything together and train a model using the **decision tree algorithm**. To ensure that you are producing an optimized model, you will train the model using the grid search technique to optimize the `'max_depth'` parameter for the decision tree. The `'max_depth'` parameter can be thought of as how many questions the decision tree algorithm is allowed to ask about the data before making a prediction. Decision trees are part of a class of algorithms called *supervised learning algorithms*.
In addition, you will find your implementation is using `ShuffleSplit()` for an alternative form of cross-validation (see the `'cv_sets'` variable). While it is not the K-Fold cross-validation technique you describe in **Question 8**, this type of cross-validation technique is just as useful!. The `ShuffleSplit()` implementation below will create 10 (`'n_splits'`) shuffled sets, and for each shuffle, 20% (`'test_size'`) of the data will be used as the *validation set*. While you're working on your implementation, think about the contrasts and similarities it has to the K-fold cross-validation technique.
Please note that ShuffleSplit has different parameters in scikit-learn versions 0.17 and 0.18.
For the `fit_model` function in the code cell below, you will need to implement the following:
- Use [`DecisionTreeRegressor`](http://scikit-learn.org/stable/modules/generated/sklearn.tree.DecisionTreeRegressor.html) from `sklearn.tree` to create a decision tree regressor object.
- Assign this object to the `'regressor'` variable.
- Create a dictionary for `'max_depth'` with the values from 1 to 10, and assign this to the `'params'` variable.
- Use [`make_scorer`](http://scikit-learn.org/stable/modules/generated/sklearn.metrics.make_scorer.html) from `sklearn.metrics` to create a scoring function object.
- Pass the `performance_metric` function as a parameter to the object.
- Assign this scoring function to the `'scoring_fnc'` variable.
- Use [`GridSearchCV`](http://scikit-learn.org/0.17/modules/generated/sklearn.grid_search.GridSearchCV.html) from `sklearn.grid_search` to create a grid search object.
- Pass the variables `'regressor'`, `'params'`, `'scoring_fnc'`, and `'cv_sets'` as parameters to the object.
- Assign the `GridSearchCV` object to the `'grid'` variable.
```
# TODO: Import 'make_scorer', 'DecisionTreeRegressor', and 'GridSearchCV'
from sklearn.metrics import make_scorer
from sklearn.tree import DecisionTreeRegressor
from sklearn.model_selection import GridSearchCV
def fit_model(X, y):
""" Performs grid search over the 'max_depth' parameter for a
decision tree regressor trained on the input data [X, y]. """
# Create cross-validation sets from the training data
# sklearn version 0.18: ShuffleSplit(n_splits=10, test_size=0.1, train_size=None, random_state=None)
# sklearn versiin 0.17: ShuffleSplit(n, n_iter=10, test_size=0.1, train_size=None, random_state=None)
cv_sets = ShuffleSplit(X.shape[0], n_iter = 10, test_size = 0.20, random_state = 40)
# TODO: Create a decision tree regressor object
regressor = DecisionTreeRegressor(random_state=41)
# TODO: Create a dictionary for the parameter 'max_depth' with a range from 1 to 10
params = {"max_depth":range(1,11)}
# TODO: Transform 'performance_metric' into a scoring function using 'make_scorer'
scoring_fnc = make_scorer(performance_metric)
# TODO: Create the grid search cv object --> GridSearchCV()
# Make sure to include the right parameters in the object:
# (estimator, param_grid, scoring, cv) which have values 'regressor', 'params', 'scoring_fnc', and 'cv_sets' respectively.
grid = GridSearchCV(estimator = regressor, param_grid = params, scoring = scoring_fnc, cv=cv_sets)
# Fit the grid search object to the data to compute the optimal model
grid = grid.fit(X, y)
# Return the optimal model after fitting the data
return grid.best_estimator_
```
### Making Predictions
Once a model has been trained on a given set of data, it can now be used to make predictions on new sets of input data. In the case of a *decision tree regressor*, the model has learned *what the best questions to ask about the input data are*, and can respond with a prediction for the **target variable**. You can use these predictions to gain information about data where the value of the target variable is unknown — such as data the model was not trained on.
### Question 9 - Optimal Model
* What maximum depth does the optimal model have? How does this result compare to your guess in **Question 6**?
Run the code block below to fit the decision tree regressor to the training data and produce an optimal model.
```
# Fit the training data to the model using grid search
reg = fit_model(X_train, y_train)
# Produce the value for 'max_depth'
print("Parameter 'max_depth' is {} for the optimal model.".format(reg.get_params()['max_depth']))
```
** Hint: ** The answer comes from the output of the code snipped above.
**Answer:**
The maximum depth was 4 for the optimal model. I guess the best maximum depth was an value between 2 to 5 and I choose 3 like the best depth.
### Question 10 - Predicting Selling Prices
Imagine that you were a real estate agent in the Boston area looking to use this model to help price homes owned by your clients that they wish to sell. You have collected the following information from three of your clients:
| Feature | Client 1 | Client 2 | Client 3 |
| :---: | :---: | :---: | :---: |
| Total number of rooms in home | 5 rooms | 4 rooms | 8 rooms |
| Neighborhood poverty level (as %) | 17% | 32% | 3% |
| Student-teacher ratio of nearby schools | 15-to-1 | 22-to-1 | 12-to-1 |
* What price would you recommend each client sell his/her home at?
* Do these prices seem reasonable given the values for the respective features?
**Hint:** Use the statistics you calculated in the **Data Exploration** section to help justify your response. Of the three clients, client 3 has has the biggest house, in the best public school neighborhood with the lowest poverty level; while client 2 has the smallest house, in a neighborhood with a relatively high poverty rate and not the best public schools.
Run the code block below to have your optimized model make predictions for each client's home.
```
# Produce a matrix for client data
client_data = [[5, 17, 15], # Client 1
[4, 32, 22], # Client 2
[8, 3, 12]] # Client 3
# Show predictions
for i, price in enumerate(reg.predict(client_data)):
print("Predicted selling price for Client {}'s home: ${:,.2f}".format(i+1, price))
```
**Answer:**
Taking the following features:
- `'RM'` is the average number of rooms among homes in the neighborhood.
- `'LSTAT'` is the percentage of homeowners in the neighborhood considered "lower class" (working poor).
- `'PTRATIO'` is the ratio of students to teachers in primary and secondary schools in the neighborhood.
* **Client 1: 409,400.00** - With 5 rooms and a relatively low neighborhood poverty level and student-teacher ratio, this would be a good selling price. The price at which I recommend selling it is at $\$409,800.00$ so there is a little slack if someone tries to bargain, so I think it is a reasonable price.
* **Client 2: 229,682.35** - The house has 4 rooms are helping raise the price of their home, since both teacher-student ratio and provery levels are quite high both of which negatively impact the selling price. The price at which I recommend selling it is at $\$229,700.00$ so there is a little slack if someone tries to bargain, so I think it is a reasonable price.
* **Client 3: 938,053.85** - Finally, It is noticeable to see that the customer number 3 has a house with much more rooms, which makes 'MEDV' increase and that this is also located in a middle-upper class neighborhood and a low level ratio of students to teachers, which increases its cost even more. The total amount predicted is $\$931,636.36$, the price at which I recommend selling it is at $\$931, 800$ so there is a little slack if someone tries to bargain, so I think it is a reasonable price.
### Sensitivity
An optimal model is not necessarily a robust model. Sometimes, a model is either too complex or too simple to sufficiently generalize to new data. Sometimes, a model could use a learning algorithm that is not appropriate for the structure of the data given. Other times, the data itself could be too noisy or contain too few samples to allow a model to adequately capture the target variable — i.e., the model is underfitted.
**Run the code cell below to run the `fit_model` function ten times with different training and testing sets to see how the prediction for a specific client changes with respect to the data it's trained on.**
```
vs.PredictTrials(features, prices, fit_model, client_data)
```
### Question 11 - Applicability
* In a few sentences, discuss whether the constructed model should or should not be used in a real-world setting.
**Hint:** Take a look at the range in prices as calculated in the code snippet above. Some questions to answering:
- How relevant today is data that was collected from 1978? How important is inflation?
- Are the features present in the data sufficient to describe a home? Do you think factors like quality of apppliances in the home, square feet of the plot area, presence of pool or not etc should factor in?
- Is the model robust enough to make consistent predictions?
- Would data collected in an urban city like Boston be applicable in a rural city?
- Is it fair to judge the price of an individual home based on the characteristics of the entire neighborhood?
**Answer:**
1. **Relevancy**: I consider that the characteristics, although they are important, are not all. In addition, relevance matters, since this study was applied more than 40 years ago and most likely is not relevant to the current date (2019).
2. **Features**: Although the characteristics contemplated in this study helped determine a monetary value for property (in 1978), I consider that there are other variables that can be added that adapt to the current date, which very intrinsically depends on the place, some of these variables could be: proximity to transport systems, garden dimension (if it has), pool availability, etc.
3. **Application**: Considering that the study and model was explicitly intended for the city of Boston, trying to adapt this model to an urban area could be costly and time consuming, as well as erroneous in its predictions at first.
4. **Robustness**: It is not robust enough, since given that a greater number of characteristics are not contemplated, and that they are also current, the prediction of the model may be limited. In addition, it seems that the system is too sensitive and is not well generalized, since as it was seen in the above code, executing it several times for a specific client (as seen above) provides a wide variety of prices. For this reason, it would not be satistantial to apply it in the real world.
> **Note**: Once you have completed all of the code implementations and successfully answered each question above, you may finalize your work by exporting the iPython Notebook as an HTML document. You can do this by using the menu above and navigating to
**File -> Download as -> HTML (.html)**. Include the finished document along with this notebook as your submission.
|
github_jupyter
|
# Import libraries necessary for this project
import numpy as np
import pandas as pd
from sklearn.cross_validation import ShuffleSplit
# Import supplementary visualizations code visuals.py
import visuals as vs
# Pretty display for notebooks
%matplotlib inline
# Load the Boston housing dataset
data = pd.read_csv('housing.csv')
prices = data['MEDV']
features = data.drop('MEDV', axis = 1)
# Success
print("Boston housing dataset has {} data points with {} variables each.".format(*data.shape))
# TODO: Minimum price of the data
minimum_price = np.min(prices)
# TODO: Maximum price of the data
maximum_price = np.max(prices)
# TODO: Mean price of the data
mean_price = np.mean(prices)
# TODO: Median price of the data
median_price = np.median(prices)
# TODO: Standard deviation of prices of the data
std_price = np.std(prices)
# Show the calculated statistics
print("Statistics for Boston housing dataset:\n")
print("Minimum price: ${}".format(minimum_price))
print("Maximum price: ${}".format(maximum_price))
print("Mean price: ${}".format(mean_price))
print("Median price ${}".format(median_price))
print("Standard deviation of prices: ${}".format(std_price))
# TODO: Import 'r2_score'
from sklearn.metrics import r2_score
def performance_metric(y_true, y_predict):
""" Calculates and returns the performance score between
true and predicted values based on the metric chosen. """
# TODO: Calculate the performance score between 'y_true' and 'y_predict'
score = r2_score(y_true,y_predict)
# Return the score
return score
# Calculate the performance of this model
score = performance_metric([3, -0.5, 2, 7, 4.2], [2.5, 0.0, 2.1, 7.8, 5.3])
print("Model has a coefficient of determination, R^2, of {:.3f}.".format(score))
# TODO: Import 'train_test_split'
from sklearn.cross_validation import train_test_split
# TODO: Shuffle and split the data into training and testing subsets
X_train, X_test, y_train, y_test = train_test_split(features, prices, test_size = 0.20, random_state = 42)
# Success
print("Training and testing split was successful.")
# Produce learning curves for varying training set sizes and maximum depths
vs.ModelLearning(features, prices)
vs.ModelComplexity(X_train, y_train)
# TODO: Import 'make_scorer', 'DecisionTreeRegressor', and 'GridSearchCV'
from sklearn.metrics import make_scorer
from sklearn.tree import DecisionTreeRegressor
from sklearn.model_selection import GridSearchCV
def fit_model(X, y):
""" Performs grid search over the 'max_depth' parameter for a
decision tree regressor trained on the input data [X, y]. """
# Create cross-validation sets from the training data
# sklearn version 0.18: ShuffleSplit(n_splits=10, test_size=0.1, train_size=None, random_state=None)
# sklearn versiin 0.17: ShuffleSplit(n, n_iter=10, test_size=0.1, train_size=None, random_state=None)
cv_sets = ShuffleSplit(X.shape[0], n_iter = 10, test_size = 0.20, random_state = 40)
# TODO: Create a decision tree regressor object
regressor = DecisionTreeRegressor(random_state=41)
# TODO: Create a dictionary for the parameter 'max_depth' with a range from 1 to 10
params = {"max_depth":range(1,11)}
# TODO: Transform 'performance_metric' into a scoring function using 'make_scorer'
scoring_fnc = make_scorer(performance_metric)
# TODO: Create the grid search cv object --> GridSearchCV()
# Make sure to include the right parameters in the object:
# (estimator, param_grid, scoring, cv) which have values 'regressor', 'params', 'scoring_fnc', and 'cv_sets' respectively.
grid = GridSearchCV(estimator = regressor, param_grid = params, scoring = scoring_fnc, cv=cv_sets)
# Fit the grid search object to the data to compute the optimal model
grid = grid.fit(X, y)
# Return the optimal model after fitting the data
return grid.best_estimator_
# Fit the training data to the model using grid search
reg = fit_model(X_train, y_train)
# Produce the value for 'max_depth'
print("Parameter 'max_depth' is {} for the optimal model.".format(reg.get_params()['max_depth']))
# Produce a matrix for client data
client_data = [[5, 17, 15], # Client 1
[4, 32, 22], # Client 2
[8, 3, 12]] # Client 3
# Show predictions
for i, price in enumerate(reg.predict(client_data)):
print("Predicted selling price for Client {}'s home: ${:,.2f}".format(i+1, price))
vs.PredictTrials(features, prices, fit_model, client_data)
| 0.532425 | 0.994385 |
```
!curl localhost:8501/v1/models/face-mask-detection-serving
from tensorflow.keras.preprocessing.image import img_to_array
from tensorflow.keras.preprocessing.image import load_img
from imutils import paths
import numpy as np
import tensorflow as tf
from sklearn.metrics import accuracy_score, f1_score
import json
import requests
pathname = "testset"
imagePaths = list(paths.list_images(pathname))
MAP_CHARACTERS= {1: 'No Mask',
0: 'Mask'}
# Apply the same preprocessing as during training (resize and rescale)
image = tf.io.decode_image(open('testset/NoMask/frame_2021_12_21_16_10_05_0_1.jpg', 'rb').read(), channels=3)
image = tf.image.resize(image, [224, 224])
image = image/255.
# Convert the Tensor to a batch of Tensors and then to a list
image_tensor = tf.expand_dims(image, 0)
image_tensor = image_tensor.numpy().tolist()
# Define the endpoint with the format: http://localhost:8501/v1/models/MODEL_NAME:predict
endpoint = "http://localhost:8501/v1/models/face-mask-detection-serving:predict"
# Prepare the data that is going to be sent in the POST request
json_data = json.dumps({
"instances": image_tensor
})
# Send the request to the Prediction API
headers = {"content-type": "application/json"}
response = requests.post(url="http://localhost:8501/v1/models/face-mask-detection-serving:predict", data=json_data, headers=headers)
# Retrieve the highest probablity index of the Tensor (actual prediction)
prediction = tf.argmax(response.json()['predictions'], 1).numpy()[0]
# print(MAP_CHARACTERS[prediction.numpy()])
# >>> "homer_simpson"
MAP_CHARACTERS[prediction]
import grpc
from tensorflow_serving.apis import predict_pb2, prediction_service_pb2_grpc
# Apply the same preprocessing as during training (resize and rescale)
image = tf.io.decode_image(open('testset/NoMask/frame_2021_12_17_10_43_04_0.jpg', 'rb').read(), channels=3)
image = tf.image.resize(image, [224, 224])
image = image/255.
# Convert the Tensor to a batch of Tensors and then to a list
image_tensor = tf.expand_dims(image, 0)
image_tensor = image_tensor.numpy().tolist()
# Optional: define a custom message lenght in bytes
MAX_MESSAGE_LENGTH = 20000000
# Optional: define a request timeout in seconds
REQUEST_TIMEOUT = 5
# Open a gRPC insecure channel
channel = grpc.insecure_channel(
"localhost:8500",
options=[
("grpc.max_send_message_length", MAX_MESSAGE_LENGTH),
("grpc.max_receive_message_length", MAX_MESSAGE_LENGTH),
],
)
# Create the PredictionServiceStub
stub = prediction_service_pb2_grpc.PredictionServiceStub(channel)
# Create the PredictRequest and set its values
req = predict_pb2.PredictRequest()
req.model_spec.name = 'face-mask-detection-serving'
req.model_spec.signature_name = ''
# Convert to Tensor Proto and send the request
# Note that shape is in NHWC (num_samples x height x width x channels) format
tensor = tf.make_tensor_proto(image_tensor)
req.inputs["input_1"].CopyFrom(tensor) # Available at /metadata
# Send request
response = stub.Predict(req, REQUEST_TIMEOUT)
# Handle request's response
output_tensor_proto = response.outputs["dense_1"] # Available at /metadata
shape = tf.TensorShape(output_tensor_proto.tensor_shape)
result = tf.reshape(output_tensor_proto.float_val, shape)
result = tf.argmax(result, 1).numpy()[0]
print(MAP_CHARACTERS[result])
```
|
github_jupyter
|
!curl localhost:8501/v1/models/face-mask-detection-serving
from tensorflow.keras.preprocessing.image import img_to_array
from tensorflow.keras.preprocessing.image import load_img
from imutils import paths
import numpy as np
import tensorflow as tf
from sklearn.metrics import accuracy_score, f1_score
import json
import requests
pathname = "testset"
imagePaths = list(paths.list_images(pathname))
MAP_CHARACTERS= {1: 'No Mask',
0: 'Mask'}
# Apply the same preprocessing as during training (resize and rescale)
image = tf.io.decode_image(open('testset/NoMask/frame_2021_12_21_16_10_05_0_1.jpg', 'rb').read(), channels=3)
image = tf.image.resize(image, [224, 224])
image = image/255.
# Convert the Tensor to a batch of Tensors and then to a list
image_tensor = tf.expand_dims(image, 0)
image_tensor = image_tensor.numpy().tolist()
# Define the endpoint with the format: http://localhost:8501/v1/models/MODEL_NAME:predict
endpoint = "http://localhost:8501/v1/models/face-mask-detection-serving:predict"
# Prepare the data that is going to be sent in the POST request
json_data = json.dumps({
"instances": image_tensor
})
# Send the request to the Prediction API
headers = {"content-type": "application/json"}
response = requests.post(url="http://localhost:8501/v1/models/face-mask-detection-serving:predict", data=json_data, headers=headers)
# Retrieve the highest probablity index of the Tensor (actual prediction)
prediction = tf.argmax(response.json()['predictions'], 1).numpy()[0]
# print(MAP_CHARACTERS[prediction.numpy()])
# >>> "homer_simpson"
MAP_CHARACTERS[prediction]
import grpc
from tensorflow_serving.apis import predict_pb2, prediction_service_pb2_grpc
# Apply the same preprocessing as during training (resize and rescale)
image = tf.io.decode_image(open('testset/NoMask/frame_2021_12_17_10_43_04_0.jpg', 'rb').read(), channels=3)
image = tf.image.resize(image, [224, 224])
image = image/255.
# Convert the Tensor to a batch of Tensors and then to a list
image_tensor = tf.expand_dims(image, 0)
image_tensor = image_tensor.numpy().tolist()
# Optional: define a custom message lenght in bytes
MAX_MESSAGE_LENGTH = 20000000
# Optional: define a request timeout in seconds
REQUEST_TIMEOUT = 5
# Open a gRPC insecure channel
channel = grpc.insecure_channel(
"localhost:8500",
options=[
("grpc.max_send_message_length", MAX_MESSAGE_LENGTH),
("grpc.max_receive_message_length", MAX_MESSAGE_LENGTH),
],
)
# Create the PredictionServiceStub
stub = prediction_service_pb2_grpc.PredictionServiceStub(channel)
# Create the PredictRequest and set its values
req = predict_pb2.PredictRequest()
req.model_spec.name = 'face-mask-detection-serving'
req.model_spec.signature_name = ''
# Convert to Tensor Proto and send the request
# Note that shape is in NHWC (num_samples x height x width x channels) format
tensor = tf.make_tensor_proto(image_tensor)
req.inputs["input_1"].CopyFrom(tensor) # Available at /metadata
# Send request
response = stub.Predict(req, REQUEST_TIMEOUT)
# Handle request's response
output_tensor_proto = response.outputs["dense_1"] # Available at /metadata
shape = tf.TensorShape(output_tensor_proto.tensor_shape)
result = tf.reshape(output_tensor_proto.float_val, shape)
result = tf.argmax(result, 1).numpy()[0]
print(MAP_CHARACTERS[result])
| 0.737442 | 0.516717 |
### Python para Data Science
### Data Imput using pandas
### Edgar Acuna, Math Department, UPR-Mayaguez
### Agosto 20, 2019
### I. Reading data files using Pandas
```
import pandas as pd
#Leyendo de un archivo csv, cuya primera fila tiene los nombre de las variables
df = pd.read_csv('http://academic.uprm.edu/eacuna/loan.csv')
df.info()
print(df)
#Mostrando los primeros 5 elementos del dataframe df
df.head()
df.describe()
df.describe(include=['O'])
import matplotlib.pyplot as plt
%matplotlib inline
df.boxplot(column="AnosEmpleo", by="CasPropia")
#Mostrando los 5 ultimos elementos del dataframe df
df.tail()
#Leyendo los datos de la internet pero de tal manera que no tengan nombre las columnas
url= "http://academic.uprm.edu/eacuna/diabetes.dat"
names = ['preg', 'plas', 'pres', 'skin', 'test', 'mass', 'pedi', 'age', 'class']
data = pd.read_csv(url, names=names,header=None,sep='\t')
data.describe()
#Reading a data file without column names
url= "http://academic.uprm.edu/eacuna/diabetes.dat"
data = pd.read_csv(url,header=None,sep='\t')
data.columns = ['preg', 'plas', 'pres', 'skin', 'test', 'mass', 'pedi', 'age', 'class']
data.head()
data.shape
```
### Reading from the internet a data file containing missing values
```
breastdf=pd.read_csv("https://archive.ics.uci.edu/ml/machine-learning-databases/breast-cancer-wisconsin/breast-cancer-wisconsin.data",header=None, sep=",",na_values=['?'])
#breastdf.head()
breastdf.info()
```
### Reading a data file containing missing values
```
#Ejemplo1. Leyendo los datos de Titanic
titanic=pd.read_csv('http://academic.uprm.edu/eacuna/titanic.csv',header=0,na_values='')
titanic.info()
titanic.head()
#Hallando los missings values por columna
titanic.isnull().sum()
#Hallando el numero de filas con missing
titanic.isnull().T.any().sum()
#Hallando el numero de entradas de la tabla que son missing
titanic.isnull().sum().sum()
#Eliminando las filas que tienen missing values
dfclean=titanic.dropna()
#Size of the dataset after eliminating missings
dfclean.shape
```
## II. Subsetting
```
#este cojunto de datos esta disponible en kaggle.com
df=pd.read_csv("http://academic.uprm.edu/eacuna/student-por.csv",sep=",")
df.info()
df.head()
#consideranado las 4 primeras columnas
df1=df.iloc[1:3,2:4]
df1.head()
df2=df[['school','sex','age','address']]
df2.tail()
#considerando las 10 primeres filas solamente
df3=df.loc[0:10,]
df3
#considerando solo los estudiantes de la escuela GP
df4=df.query('school=="GP"')
df4.info()
#Otra manera
df[df['school']=="GP"]
pan=pd.read_csv("http://academic.uprm.edu/eacuna/PANyTANFPR.csv")
pan.head()
#Extrayendo solamente la informacion del PAN con fecha de emisison 201406 (Junio 2014)
pan1=pan.query("PROGRAMA=='PAN' & FECHA_EMISION==201406")
print(pan1.shape)
# Eliminando la informacion de algunos municipios que estan en el dtaframe pan1
pan2=pan1.query("MUNICIPIO!=['CASTAÑER','ANGELES','RIO PIEDRAS', 'RIO PIEDRAS 3','RIO PIEDRAS 4']")
print(pan2.shape)
#Haciendo ambos pasos a la vez
pan12= pan1.query("PROGRAMA=='PAN' & FECHA_EMISION==201406 & MUNICIPIO!=['CASTAÑER','ANGELES','RIO PIEDRAS', 'RIO PIEDRAS 3','RIO PIEDRAS 4']")
print(pan12.shape)
```
|
github_jupyter
|
import pandas as pd
#Leyendo de un archivo csv, cuya primera fila tiene los nombre de las variables
df = pd.read_csv('http://academic.uprm.edu/eacuna/loan.csv')
df.info()
print(df)
#Mostrando los primeros 5 elementos del dataframe df
df.head()
df.describe()
df.describe(include=['O'])
import matplotlib.pyplot as plt
%matplotlib inline
df.boxplot(column="AnosEmpleo", by="CasPropia")
#Mostrando los 5 ultimos elementos del dataframe df
df.tail()
#Leyendo los datos de la internet pero de tal manera que no tengan nombre las columnas
url= "http://academic.uprm.edu/eacuna/diabetes.dat"
names = ['preg', 'plas', 'pres', 'skin', 'test', 'mass', 'pedi', 'age', 'class']
data = pd.read_csv(url, names=names,header=None,sep='\t')
data.describe()
#Reading a data file without column names
url= "http://academic.uprm.edu/eacuna/diabetes.dat"
data = pd.read_csv(url,header=None,sep='\t')
data.columns = ['preg', 'plas', 'pres', 'skin', 'test', 'mass', 'pedi', 'age', 'class']
data.head()
data.shape
breastdf=pd.read_csv("https://archive.ics.uci.edu/ml/machine-learning-databases/breast-cancer-wisconsin/breast-cancer-wisconsin.data",header=None, sep=",",na_values=['?'])
#breastdf.head()
breastdf.info()
#Ejemplo1. Leyendo los datos de Titanic
titanic=pd.read_csv('http://academic.uprm.edu/eacuna/titanic.csv',header=0,na_values='')
titanic.info()
titanic.head()
#Hallando los missings values por columna
titanic.isnull().sum()
#Hallando el numero de filas con missing
titanic.isnull().T.any().sum()
#Hallando el numero de entradas de la tabla que son missing
titanic.isnull().sum().sum()
#Eliminando las filas que tienen missing values
dfclean=titanic.dropna()
#Size of the dataset after eliminating missings
dfclean.shape
#este cojunto de datos esta disponible en kaggle.com
df=pd.read_csv("http://academic.uprm.edu/eacuna/student-por.csv",sep=",")
df.info()
df.head()
#consideranado las 4 primeras columnas
df1=df.iloc[1:3,2:4]
df1.head()
df2=df[['school','sex','age','address']]
df2.tail()
#considerando las 10 primeres filas solamente
df3=df.loc[0:10,]
df3
#considerando solo los estudiantes de la escuela GP
df4=df.query('school=="GP"')
df4.info()
#Otra manera
df[df['school']=="GP"]
pan=pd.read_csv("http://academic.uprm.edu/eacuna/PANyTANFPR.csv")
pan.head()
#Extrayendo solamente la informacion del PAN con fecha de emisison 201406 (Junio 2014)
pan1=pan.query("PROGRAMA=='PAN' & FECHA_EMISION==201406")
print(pan1.shape)
# Eliminando la informacion de algunos municipios que estan en el dtaframe pan1
pan2=pan1.query("MUNICIPIO!=['CASTAÑER','ANGELES','RIO PIEDRAS', 'RIO PIEDRAS 3','RIO PIEDRAS 4']")
print(pan2.shape)
#Haciendo ambos pasos a la vez
pan12= pan1.query("PROGRAMA=='PAN' & FECHA_EMISION==201406 & MUNICIPIO!=['CASTAÑER','ANGELES','RIO PIEDRAS', 'RIO PIEDRAS 3','RIO PIEDRAS 4']")
print(pan12.shape)
| 0.27513 | 0.825836 |
# Object Detection: R-FCN and SSD-MobileNet
```
from __future__ import print_function
import os
import time
import random
import numpy as np
import tensorflow as tf
from PIL import Image
from object_detection.utils.visualization_utils import visualize_boxes_and_labels_on_image_array
%matplotlib inline
import matplotlib
from matplotlib import pyplot as plt
MODEL = 'rfcn' # Use 'rfcn' for R-FCN or 'ssdmobilenet' for SSD-MobileNet
PROTOCOL = 'grpc' # Use 'grpc' for GRPC or 'rest' for REST
IMAGES_PATH = '/home/<user>/coco/val/val2017' # Edit this to your COCO validation directory
if PROTOCOL == 'grpc':
import grpc
import tensorflow as tf
from tensorflow_serving.apis import predict_pb2
from tensorflow_serving.apis import prediction_service_pb2_grpc
SERVER_URL = 'localhost:8500'
elif PROTOCOL == 'rest':
import requests
SERVER_URL = 'http://localhost:8501/v1/models/{}:predict'.format(MODEL)
def get_random_image(image_dir):
image_path = os.path.join(image_dir, random.choice(os.listdir(image_dir)))
image = Image.open(image_path)
(im_width, im_height) = image.size
return np.array(image.getdata()).reshape((im_height, im_width, 3)).astype(np.uint8)
def visualize(output_dict, image_np):
new_dict = {}
if PROTOCOL == 'grpc':
new_dict['num_detections'] = int(output_dict['num_detections'].float_val[0])
new_dict['detection_classes'] = np.array(output_dict['detection_classes'].float_val).astype(np.uint8)
new_dict['detection_boxes'] = np.array(output_dict['detection_boxes'].float_val).reshape((-1,4))
new_dict['detection_scores'] = np.array(output_dict['detection_scores'].float_val)
new_dict['instance_masks'] = np.array(output_dict['instance_masks'].float_val)
elif PROTOCOL == 'rest':
new_dict['num_detections'] = int(output_dict['num_detections'])
new_dict['detection_classes'] = np.array(output_dict['detection_classes']).astype(np.uint8)
new_dict['detection_boxes'] = np.array(output_dict['detection_boxes'])
new_dict['detection_scores'] = np.array(output_dict['detection_scores'])
# Visualize the results of a detection
visualize_boxes_and_labels_on_image_array(
image_np,
new_dict['detection_boxes'],
new_dict['detection_classes'],
new_dict['detection_scores'],
{1: {'id': 1, 'name': 'object'}}, # Empty category index
instance_masks=None,
use_normalized_coordinates=True,
line_thickness=8)
plt.figure()
plt.imshow(image_np)
```
# Test Object Detection
```
batch_size = 1
np_image = get_random_image(IMAGES_PATH)
if PROTOCOL == 'grpc':
np_image = np.repeat(np.expand_dims(np_image, 0), batch_size, axis=0)
channel = grpc.insecure_channel(SERVER_URL)
stub = prediction_service_pb2_grpc.PredictionServiceStub(channel)
request = predict_pb2.PredictRequest()
request.model_spec.name = 'ssdmobilenet'
request.model_spec.signature_name = 'serving_default'
request.inputs['inputs'].CopyFrom(tf.contrib.util.make_tensor_proto(np_image))
result = stub.Predict(request)
visualize(result.outputs, np_image[0])
elif PROTOCOL == 'rest':
predict_request = '{"instances" : %s}' % np.expand_dims(np_image, 0).tolist()
result = requests.post(SERVER_URL, data=predict_request)
visualize(result.json()['predictions'][0], np_image)
```
# Measure Performance
```
def make_request(batch_size):
if PROTOCOL == 'rest':
np_images = np.repeat(np.expand_dims(get_random_image(IMAGES_PATH), 0).tolist(), batch_size, axis=0).tolist()
return '{"instances" : %s}' % np_images
elif PROTOCOL == 'grpc':
np_images = np.repeat(np.expand_dims(get_random_image(IMAGES_PATH), 0), batch_size, axis=0)
channel = grpc.insecure_channel(SERVER_URL)
stub = prediction_service_pb2_grpc.PredictionServiceStub(channel)
request = predict_pb2.PredictRequest()
request.model_spec.name = MODEL
request.model_spec.signature_name = 'serving_default'
request.inputs['inputs'].CopyFrom(tf.contrib.util.make_tensor_proto(np_images))
return (stub, request)
def send_request(predict_request):
if PROTOCOL == 'rest':
requests.post(SERVER_URL, data=predict_request)
elif PROTOCOL == 'grpc':
predict_request[0].Predict(predict_request[1])
def benchmark(batch_size=1, num_iteration=10, warm_up_iteration=2):
i = 0
total_time = 0
for _ in range(num_iteration):
i += 1
np_images = np.repeat(np.expand_dims(get_random_image(IMAGES_PATH), 0), batch_size, axis=0)
predict_request = make_request(batch_size)
start_time = time.time()
send_request(predict_request)
time_consume = time.time() - start_time
print('Iteration %d: %.3f sec' % (i, time_consume))
if i > warm_up_iteration:
total_time += time_consume
time_average = total_time / (num_iteration - warm_up_iteration)
print('Average time: %.3f sec' % (time_average))
print('Batch size = %d' % batch_size)
if batch_size == 1:
print('Latency: %.3f ms' % (time_average * 1000))
print('Throughput: %.3f images/sec' % (batch_size / time_average))
```
## Real-time Inference (latency, batch_size=1)
```
benchmark()
```
## Throughput (batch_size=128)
```
benchmark(batch_size=128)
```
|
github_jupyter
|
from __future__ import print_function
import os
import time
import random
import numpy as np
import tensorflow as tf
from PIL import Image
from object_detection.utils.visualization_utils import visualize_boxes_and_labels_on_image_array
%matplotlib inline
import matplotlib
from matplotlib import pyplot as plt
MODEL = 'rfcn' # Use 'rfcn' for R-FCN or 'ssdmobilenet' for SSD-MobileNet
PROTOCOL = 'grpc' # Use 'grpc' for GRPC or 'rest' for REST
IMAGES_PATH = '/home/<user>/coco/val/val2017' # Edit this to your COCO validation directory
if PROTOCOL == 'grpc':
import grpc
import tensorflow as tf
from tensorflow_serving.apis import predict_pb2
from tensorflow_serving.apis import prediction_service_pb2_grpc
SERVER_URL = 'localhost:8500'
elif PROTOCOL == 'rest':
import requests
SERVER_URL = 'http://localhost:8501/v1/models/{}:predict'.format(MODEL)
def get_random_image(image_dir):
image_path = os.path.join(image_dir, random.choice(os.listdir(image_dir)))
image = Image.open(image_path)
(im_width, im_height) = image.size
return np.array(image.getdata()).reshape((im_height, im_width, 3)).astype(np.uint8)
def visualize(output_dict, image_np):
new_dict = {}
if PROTOCOL == 'grpc':
new_dict['num_detections'] = int(output_dict['num_detections'].float_val[0])
new_dict['detection_classes'] = np.array(output_dict['detection_classes'].float_val).astype(np.uint8)
new_dict['detection_boxes'] = np.array(output_dict['detection_boxes'].float_val).reshape((-1,4))
new_dict['detection_scores'] = np.array(output_dict['detection_scores'].float_val)
new_dict['instance_masks'] = np.array(output_dict['instance_masks'].float_val)
elif PROTOCOL == 'rest':
new_dict['num_detections'] = int(output_dict['num_detections'])
new_dict['detection_classes'] = np.array(output_dict['detection_classes']).astype(np.uint8)
new_dict['detection_boxes'] = np.array(output_dict['detection_boxes'])
new_dict['detection_scores'] = np.array(output_dict['detection_scores'])
# Visualize the results of a detection
visualize_boxes_and_labels_on_image_array(
image_np,
new_dict['detection_boxes'],
new_dict['detection_classes'],
new_dict['detection_scores'],
{1: {'id': 1, 'name': 'object'}}, # Empty category index
instance_masks=None,
use_normalized_coordinates=True,
line_thickness=8)
plt.figure()
plt.imshow(image_np)
batch_size = 1
np_image = get_random_image(IMAGES_PATH)
if PROTOCOL == 'grpc':
np_image = np.repeat(np.expand_dims(np_image, 0), batch_size, axis=0)
channel = grpc.insecure_channel(SERVER_URL)
stub = prediction_service_pb2_grpc.PredictionServiceStub(channel)
request = predict_pb2.PredictRequest()
request.model_spec.name = 'ssdmobilenet'
request.model_spec.signature_name = 'serving_default'
request.inputs['inputs'].CopyFrom(tf.contrib.util.make_tensor_proto(np_image))
result = stub.Predict(request)
visualize(result.outputs, np_image[0])
elif PROTOCOL == 'rest':
predict_request = '{"instances" : %s}' % np.expand_dims(np_image, 0).tolist()
result = requests.post(SERVER_URL, data=predict_request)
visualize(result.json()['predictions'][0], np_image)
def make_request(batch_size):
if PROTOCOL == 'rest':
np_images = np.repeat(np.expand_dims(get_random_image(IMAGES_PATH), 0).tolist(), batch_size, axis=0).tolist()
return '{"instances" : %s}' % np_images
elif PROTOCOL == 'grpc':
np_images = np.repeat(np.expand_dims(get_random_image(IMAGES_PATH), 0), batch_size, axis=0)
channel = grpc.insecure_channel(SERVER_URL)
stub = prediction_service_pb2_grpc.PredictionServiceStub(channel)
request = predict_pb2.PredictRequest()
request.model_spec.name = MODEL
request.model_spec.signature_name = 'serving_default'
request.inputs['inputs'].CopyFrom(tf.contrib.util.make_tensor_proto(np_images))
return (stub, request)
def send_request(predict_request):
if PROTOCOL == 'rest':
requests.post(SERVER_URL, data=predict_request)
elif PROTOCOL == 'grpc':
predict_request[0].Predict(predict_request[1])
def benchmark(batch_size=1, num_iteration=10, warm_up_iteration=2):
i = 0
total_time = 0
for _ in range(num_iteration):
i += 1
np_images = np.repeat(np.expand_dims(get_random_image(IMAGES_PATH), 0), batch_size, axis=0)
predict_request = make_request(batch_size)
start_time = time.time()
send_request(predict_request)
time_consume = time.time() - start_time
print('Iteration %d: %.3f sec' % (i, time_consume))
if i > warm_up_iteration:
total_time += time_consume
time_average = total_time / (num_iteration - warm_up_iteration)
print('Average time: %.3f sec' % (time_average))
print('Batch size = %d' % batch_size)
if batch_size == 1:
print('Latency: %.3f ms' % (time_average * 1000))
print('Throughput: %.3f images/sec' % (batch_size / time_average))
benchmark()
benchmark(batch_size=128)
| 0.602062 | 0.645525 |
```
import math
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.linear_model import ElasticNet
from sklearn.datasets import make_regression
from sklearn import linear_model
from sklearn.model_selection import train_test_split
from sklearn.metrics import r2_score
from sklearn.preprocessing import StandardScaler, RobustScaler
from sklearn.linear_model import ElasticNetCV
df=pd.read_csv('New_Sample_Clean_latest.csv')
df.info()
df = df.drop(columns = ['comm_polulation'])
df
import pandas as pd
import numpy as np
import io
import json
import requests
def transformed_data():
url = 'https://raw.githubusercontent.com/shughestr/PIMS_2020_Real_Estate_data/master/sample_clean.csv'
df = pd.read_csv(url, error_bad_lines=False)
### Normalise absolute value of crime variables and income recipients by the respective population
for col in ['saf1','saf2','saf3','saf4','saf5','saf6','saf7','saf8','inc2']:
df[col] = 100 * df[col] / df['pop1']
### Adding vacancy rate column from other census dataset
data2 = requests.get('https://data.calgary.ca/resource/set9-futw.json')
df2 = pd.DataFrame(json.loads(data2.text))
df2 = df2.loc[df2['dwelling_type_code'].isin([str(x) for x in range(1,11)])]
df2 = df2.drop(labels = ['census_year', 'community','ward','dwelling_type','dwelling_type_code','dwelling_type_description'],axis=1)
df2[['dwelling_cnt', 'resident_cnt', 'ocpd_dwelling_cnt',
'vacant_dwelling_cnt', 'ocpd_ownership_cnt', 'renovation_dwelling_cnt',
'under_const_dwelling_cnt', 'inactive_cnt', 'other_purpose_cnt']] = df2[['dwelling_cnt', 'resident_cnt', 'ocpd_dwelling_cnt',
'vacant_dwelling_cnt', 'ocpd_ownership_cnt', 'renovation_dwelling_cnt',
'under_const_dwelling_cnt', 'inactive_cnt', 'other_purpose_cnt']].astype(int)
df2=df2.groupby(['code']).sum()
df2['vacancy_rate'] = df2['vacant_dwelling_cnt']/(df2['vacant_dwelling_cnt']+df2['ocpd_dwelling_cnt']) * 100
df2vac = df2['vacancy_rate']
vacdict = df2vac.to_dict()
df['vacancy_rate'] = 0
for x in vacdict:
df['vacancy_rate'] = np.where(df['COMM_CODE']==x, vacdict[x],df['vacancy_rate'])
return df
df_transformed = transformed_data()
df_transformed
df['vacancy_rate'] = df_transformed['vacancy_rate']
df.info()
df = df[df['pct_change']<0.5]
df
df = df[df['pct_change']>-0.5]
df_new = df
df_new
std_scaler = StandardScaler()
for column in ['pct_change', "YEAR_OF_CONSTRUCTION","saf1","saf2","saf3","saf4","saf5","saf6","saf7","saf8","mr5y","Inflation","pop1","pop2","pop3","pop4","pop5","pop6","lan1","lan2","inc1","inc2","inc3","inc4","own1","own2","own3","own4","lab1","lab2","lab3",'vacancy_rate','walk_score_comm','transit_score_comm','bike_score_comm']:
df_new[column] = std_scaler.fit_transform(df_new[column].values.reshape(-1,1))
df_new.dropna(inplace=True)
df_new['y']=df_new['pct_change']
train, test= train_test_split(df, test_size=0.2)
features=["YEAR_OF_CONSTRUCTION","saf1","saf2","saf3","saf4","saf5","saf6","saf7","saf8","mr5y","Inflation","pop1","pop2","pop3","pop4","pop5","pop6","lan1","lan2","inc1","inc2","inc3","inc4","own1","own2","own3","own4","lab1","lab2","lab3",'vacancy_rate','walk_score_comm','transit_score_comm','bike_score_comm']
features
x_train=train[features]
#y_train=np.sign(train["pct_change"])
y_train=train['y']
X_test=test[features]
#y_test=np.sign(test["pct_change"])
y_test=test['y']
encv = ElasticNetCV(alphas=(0.1, 0.01, 0.005, 0.0025, 0.001), l1_ratio=(0.1, 0.25, 0.5, 0.75, 0.8), normalize=True)
encv.fit(x_train, y_train)
print('ElasticNet optimal alpha: %.3f and L1 ratio: %.4f' % (encv.alpha_, encv.l1_ratio_))
enet = ElasticNet(alpha=0.001, l1_ratio=0.8)
enet.fit(x_train, y_train)
y_pred_enet=enet.predict(X_test)
r2_score_enet = r2_score(y_test, y_pred_enet)
print(enet)
print("r^2 on test data : %f" % r2_score_enet)
x_train, y_train = make_regression(n_features=34, random_state=0)
regr = ElasticNet(random_state=0)
regr.fit(x_train, y_train)
ElasticNet(random_state=0)
print(regr.coef_)
import matplotlib.pyplot as plt
import numpy as np
plt.rcdefaults()
fig, ax = plt.subplots(figsize=(15,20))
ax.barh(features, regr.coef_)
ax.set_yticks(features)
ax.set_yticklabels(features)
ax.invert_yaxis() # labels read top-to-bottom
ax.set_xlabel('coefficient')
ax.set_title('Elastic Net for coefficient and features')
plt.show()
```
### Conclusion from Elastic Net : Top 10 features(pop1, own3, own4,saf3,pop4,inc3,own1,inc4,mr5y,lab3)
|
github_jupyter
|
import math
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.linear_model import ElasticNet
from sklearn.datasets import make_regression
from sklearn import linear_model
from sklearn.model_selection import train_test_split
from sklearn.metrics import r2_score
from sklearn.preprocessing import StandardScaler, RobustScaler
from sklearn.linear_model import ElasticNetCV
df=pd.read_csv('New_Sample_Clean_latest.csv')
df.info()
df = df.drop(columns = ['comm_polulation'])
df
import pandas as pd
import numpy as np
import io
import json
import requests
def transformed_data():
url = 'https://raw.githubusercontent.com/shughestr/PIMS_2020_Real_Estate_data/master/sample_clean.csv'
df = pd.read_csv(url, error_bad_lines=False)
### Normalise absolute value of crime variables and income recipients by the respective population
for col in ['saf1','saf2','saf3','saf4','saf5','saf6','saf7','saf8','inc2']:
df[col] = 100 * df[col] / df['pop1']
### Adding vacancy rate column from other census dataset
data2 = requests.get('https://data.calgary.ca/resource/set9-futw.json')
df2 = pd.DataFrame(json.loads(data2.text))
df2 = df2.loc[df2['dwelling_type_code'].isin([str(x) for x in range(1,11)])]
df2 = df2.drop(labels = ['census_year', 'community','ward','dwelling_type','dwelling_type_code','dwelling_type_description'],axis=1)
df2[['dwelling_cnt', 'resident_cnt', 'ocpd_dwelling_cnt',
'vacant_dwelling_cnt', 'ocpd_ownership_cnt', 'renovation_dwelling_cnt',
'under_const_dwelling_cnt', 'inactive_cnt', 'other_purpose_cnt']] = df2[['dwelling_cnt', 'resident_cnt', 'ocpd_dwelling_cnt',
'vacant_dwelling_cnt', 'ocpd_ownership_cnt', 'renovation_dwelling_cnt',
'under_const_dwelling_cnt', 'inactive_cnt', 'other_purpose_cnt']].astype(int)
df2=df2.groupby(['code']).sum()
df2['vacancy_rate'] = df2['vacant_dwelling_cnt']/(df2['vacant_dwelling_cnt']+df2['ocpd_dwelling_cnt']) * 100
df2vac = df2['vacancy_rate']
vacdict = df2vac.to_dict()
df['vacancy_rate'] = 0
for x in vacdict:
df['vacancy_rate'] = np.where(df['COMM_CODE']==x, vacdict[x],df['vacancy_rate'])
return df
df_transformed = transformed_data()
df_transformed
df['vacancy_rate'] = df_transformed['vacancy_rate']
df.info()
df = df[df['pct_change']<0.5]
df
df = df[df['pct_change']>-0.5]
df_new = df
df_new
std_scaler = StandardScaler()
for column in ['pct_change', "YEAR_OF_CONSTRUCTION","saf1","saf2","saf3","saf4","saf5","saf6","saf7","saf8","mr5y","Inflation","pop1","pop2","pop3","pop4","pop5","pop6","lan1","lan2","inc1","inc2","inc3","inc4","own1","own2","own3","own4","lab1","lab2","lab3",'vacancy_rate','walk_score_comm','transit_score_comm','bike_score_comm']:
df_new[column] = std_scaler.fit_transform(df_new[column].values.reshape(-1,1))
df_new.dropna(inplace=True)
df_new['y']=df_new['pct_change']
train, test= train_test_split(df, test_size=0.2)
features=["YEAR_OF_CONSTRUCTION","saf1","saf2","saf3","saf4","saf5","saf6","saf7","saf8","mr5y","Inflation","pop1","pop2","pop3","pop4","pop5","pop6","lan1","lan2","inc1","inc2","inc3","inc4","own1","own2","own3","own4","lab1","lab2","lab3",'vacancy_rate','walk_score_comm','transit_score_comm','bike_score_comm']
features
x_train=train[features]
#y_train=np.sign(train["pct_change"])
y_train=train['y']
X_test=test[features]
#y_test=np.sign(test["pct_change"])
y_test=test['y']
encv = ElasticNetCV(alphas=(0.1, 0.01, 0.005, 0.0025, 0.001), l1_ratio=(0.1, 0.25, 0.5, 0.75, 0.8), normalize=True)
encv.fit(x_train, y_train)
print('ElasticNet optimal alpha: %.3f and L1 ratio: %.4f' % (encv.alpha_, encv.l1_ratio_))
enet = ElasticNet(alpha=0.001, l1_ratio=0.8)
enet.fit(x_train, y_train)
y_pred_enet=enet.predict(X_test)
r2_score_enet = r2_score(y_test, y_pred_enet)
print(enet)
print("r^2 on test data : %f" % r2_score_enet)
x_train, y_train = make_regression(n_features=34, random_state=0)
regr = ElasticNet(random_state=0)
regr.fit(x_train, y_train)
ElasticNet(random_state=0)
print(regr.coef_)
import matplotlib.pyplot as plt
import numpy as np
plt.rcdefaults()
fig, ax = plt.subplots(figsize=(15,20))
ax.barh(features, regr.coef_)
ax.set_yticks(features)
ax.set_yticklabels(features)
ax.invert_yaxis() # labels read top-to-bottom
ax.set_xlabel('coefficient')
ax.set_title('Elastic Net for coefficient and features')
plt.show()
| 0.447943 | 0.36869 |
# Using the PyTorch JIT Compiler with Pyro
This tutorial shows how to use the PyTorch [jit compiler](https://pytorch.org/docs/master/jit.html) in Pyro models.
#### Summary:
- You can use compiled functions in Pyro models.
- You cannot use pyro primitives inside compiled functions.
- If your model has static structure, you can use a `Jit*` version of an `ELBO` algorithm, e.g.
```diff
- Trace_ELBO()
+ JitTrace_ELBO()
```
- The [HMC](http://docs.pyro.ai/en/dev/mcmc.html#pyro.infer.mcmc.HMC) and [NUTS](http://docs.pyro.ai/en/dev/mcmc.html#pyro.infer.mcmc.NUTS) classes accept `jit_compile=True` kwarg.
- Models should input all tensors as `*args` and all non-tensors as `**kwargs`.
- Each different value of `**kwargs` triggers a separate compilation.
- Use `**kwargs` to specify all variation in structure (e.g. time series length).
- To ignore jit warnings in safe code blocks, use `with pyro.util.ignore_jit_warnings():`.
- To ignore all jit warnings in `HMC` or `NUTS`, pass `ignore_jit_warnings=True`.
#### Table of contents
- [Introduction](#Introduction)
- [A simple model](#A-simple-model)
- [Varying structure](#Varying-structure)
```
import os
import torch
import pyro
import pyro.distributions as dist
from torch.distributions import constraints
from pyro import poutine
from pyro.distributions.util import broadcast_shape
from pyro.infer import Trace_ELBO, JitTrace_ELBO, TraceEnum_ELBO, JitTraceEnum_ELBO, SVI
from pyro.infer.mcmc import MCMC, NUTS
from pyro.infer.autoguide import AutoDiagonalNormal
from pyro.optim import Adam
smoke_test = ('CI' in os.environ)
assert pyro.__version__.startswith('0.4.1')
pyro.enable_validation(True) # <---- This is always a good idea!
```
## Introduction
PyTorch 1.0 includes a [jit compiler](https://pytorch.org/docs/master/jit.html) to speed up models. You can think of compilation as a "static mode", whereas PyTorch usually operates in "eager mode".
Pyro supports the jit compiler in two ways. First you can use compiled functions inside Pyro models (but those functions cannot contain Pyro primitives). Second, you can use Pyro's jit inference algorithms to compile entire inference steps; in static models this can reduce the Python overhead of Pyro models and speed up inference.
The rest of this tutorial focuses on Pyro's jitted inference algorithms: [JitTrace_ELBO](http://docs.pyro.ai/en/dev/inference_algos.html#pyro.infer.trace_elbo.JitTrace_ELBO), [JitTraceGraph_ELBO](http://docs.pyro.ai/en/dev/inference_algos.html#pyro.infer.tracegraph_elbo.JitTraceGraph_ELBO), [JitTraceEnum_ELBO](http://docs.pyro.ai/en/dev/inference_algos.html#pyro.infer.traceenum_elbo.JitTraceEnum_ELBO), [JitMeanField_ELBO](http://docs.pyro.ai/en/dev/inference_algos.html#pyro.infer.trace_mean_field_elbo.JitTraceMeanField_ELBO), [HMC(jit_compile=True)](http://docs.pyro.ai/en/dev/mcmc.html#pyro.infer.mcmc.HMC), and [NUTS(jit_compile=True)](http://docs.pyro.ai/en/dev/mcmc.html#pyro.infer.mcmc.NUTS). For further reading, see the [examples/](https://github.com/uber/pyro/tree/dev/examples) directory, where most examples include a `--jit` option to run in compiled mode.
## A simple model
Let's start with a simple Gaussian model and an [autoguide](http://docs.pyro.ai/en/dev/infer.autoguide.html).
```
def model(data):
loc = pyro.sample("loc", dist.Normal(0., 10.))
scale = pyro.sample("scale", dist.LogNormal(0., 3.))
with pyro.plate("data", data.size(0)):
pyro.sample("obs", dist.Normal(loc, scale), obs=data)
guide = AutoDiagonalNormal(model)
data = dist.Normal(0.5, 2.).sample((100,))
```
First let's run as usual with an SVI object and `Trace_ELBO`.
```
%%time
pyro.clear_param_store()
elbo = Trace_ELBO()
svi = SVI(model, guide, Adam({'lr': 0.01}), elbo)
for i in range(2 if smoke_test else 1000):
svi.step(data)
```
Next to run with a jit compiled inference, we simply replace
```diff
- elbo = Trace_ELBO()
+ elbo = JitTrace_ELBO()
```
Also note that the `AutoDiagonalNormal` guide behaves a little differently on its first invocation (it runs the model to produce a prototype trace), and we don't want to record this warmup behavior when compiling. Thus we call the `guide(data)` once to initialize, then run the compiled SVI,
```
%%time
pyro.clear_param_store()
guide(data) # Do any lazy initialization before compiling.
elbo = JitTrace_ELBO()
svi = SVI(model, guide, Adam({'lr': 0.01}), elbo)
for i in range(2 if smoke_test else 1000):
svi.step(data)
```
Notice that we have a more than 2x speedup for this small model.
Let us now use the same model, but we will instead use MCMC to generate samples from the model's posterior. We will use the No-U-Turn(NUTS) sampler.
```
%%time
nuts_kernel = NUTS(model)
pyro.set_rng_seed(1)
mcmc_run = MCMC(nuts_kernel, num_samples=100).run(data)
```
We can compile the potential energy computation in NUTS using the `jit_compile=True` argument to the NUTS kernel. We also silence JIT warnings due to the presence of tensor constants in the model by using `ignore_jit_warnings=True`.
```
%%time
nuts_kernel = NUTS(model, jit_compile=True, ignore_jit_warnings=True)
pyro.set_rng_seed(1)
mcmc_run = MCMC(nuts_kernel, num_samples=100).run(data)
```
We notice a significant increase in sampling throughput when JIT compilation is enabled.
## Varying structure
Time series models often run on datasets of multiple time series with different lengths. To accomodate varying structure like this, Pyro requires models to separate all model inputs into tensors and non-tensors.$^\dagger$
- Non-tensor inputs should be passed as `**kwargs` to the model and guide. These can determine model structure, so that a model is compiled for each value of the passed `**kwargs`.
- Tensor inputs should be passed as `*args`. These must not determine model structure. However `len(args)` may determine model structure (as is used e.g. in semisupervised models).
To illustrate this with a time series model, we will pass in a sequence of observations as a tensor `arg` and the sequence length as a non-tensor `kwarg`:
```
def model(sequence, num_sequences, length, state_dim=16):
# This is a Gaussian HMM model.
with pyro.plate("states", state_dim):
trans = pyro.sample("trans", dist.Dirichlet(0.5 * torch.ones(state_dim)))
emit_loc = pyro.sample("emit_loc", dist.Normal(0., 10.))
emit_scale = pyro.sample("emit_scale", dist.LogNormal(0., 3.))
# We're doing manual data subsampling, so we need to scale to actual data size.
with poutine.scale(scale=num_sequences):
# We'll use enumeration inference over the hidden x.
x = 0
for t in pyro.markov(range(length)):
x = pyro.sample("x_{}".format(t), dist.Categorical(trans[x]),
infer={"enumerate": "parallel"})
pyro.sample("y_{}".format(t), dist.Normal(emit_loc[x], emit_scale),
obs=sequence[t])
guide = AutoDiagonalNormal(poutine.block(model, expose=["trans", "emit_scale", "emit_loc"]))
# This is fake data of different lengths.
lengths = [24] * 50 + [48] * 20 + [72] * 5
sequences = [torch.randn(length) for length in lengths]
```
Now lets' run SVI as usual.
```
%%time
pyro.clear_param_store()
elbo = TraceEnum_ELBO(max_plate_nesting=1)
svi = SVI(model, guide, Adam({'lr': 0.01}), elbo)
for i in range(1 if smoke_test else 10):
for sequence in sequences:
svi.step(sequence, # tensor args
num_sequences=len(sequences), length=len(sequence)) # non-tensor args
```
Again we'll simply swap in a `Jit*` implementation
```diff
- elbo = TraceEnum_ELBO(max_plate_nesting=1)
+ elbo = JitTraceEnum_ELBO(max_plate_nesting=1)
```
Note that we are manually specifying the `max_plate_nesting` arg. Usually Pyro can figure this out automatically by running the model once on the first invocation; however to avoid this extra work when we run the compiler on the first step, we pass this in manually.
```
%%time
pyro.clear_param_store()
# Do any lazy initialization before compiling.
guide(sequences[0], num_sequences=len(sequences), length=len(sequences[0]))
elbo = JitTraceEnum_ELBO(max_plate_nesting=1)
svi = SVI(model, guide, Adam({'lr': 0.01}), elbo)
for i in range(1 if smoke_test else 10):
for sequence in sequences:
svi.step(sequence, # tensor args
num_sequences=len(sequences), length=len(sequence)) # non-tensor args
```
Again we see more than 2x speedup. Note that since there were three different sequence lengths, compilation was triggered three times.
$^\dagger$ Note this section is only valid for SVI, and HMC/NUTS assume fixed model arguments.
|
github_jupyter
|
- Trace_ELBO()
+ JitTrace_ELBO()
```
- The [HMC](http://docs.pyro.ai/en/dev/mcmc.html#pyro.infer.mcmc.HMC) and [NUTS](http://docs.pyro.ai/en/dev/mcmc.html#pyro.infer.mcmc.NUTS) classes accept `jit_compile=True` kwarg.
- Models should input all tensors as `*args` and all non-tensors as `**kwargs`.
- Each different value of `**kwargs` triggers a separate compilation.
- Use `**kwargs` to specify all variation in structure (e.g. time series length).
- To ignore jit warnings in safe code blocks, use `with pyro.util.ignore_jit_warnings():`.
- To ignore all jit warnings in `HMC` or `NUTS`, pass `ignore_jit_warnings=True`.
#### Table of contents
- [Introduction](#Introduction)
- [A simple model](#A-simple-model)
- [Varying structure](#Varying-structure)
## Introduction
PyTorch 1.0 includes a [jit compiler](https://pytorch.org/docs/master/jit.html) to speed up models. You can think of compilation as a "static mode", whereas PyTorch usually operates in "eager mode".
Pyro supports the jit compiler in two ways. First you can use compiled functions inside Pyro models (but those functions cannot contain Pyro primitives). Second, you can use Pyro's jit inference algorithms to compile entire inference steps; in static models this can reduce the Python overhead of Pyro models and speed up inference.
The rest of this tutorial focuses on Pyro's jitted inference algorithms: [JitTrace_ELBO](http://docs.pyro.ai/en/dev/inference_algos.html#pyro.infer.trace_elbo.JitTrace_ELBO), [JitTraceGraph_ELBO](http://docs.pyro.ai/en/dev/inference_algos.html#pyro.infer.tracegraph_elbo.JitTraceGraph_ELBO), [JitTraceEnum_ELBO](http://docs.pyro.ai/en/dev/inference_algos.html#pyro.infer.traceenum_elbo.JitTraceEnum_ELBO), [JitMeanField_ELBO](http://docs.pyro.ai/en/dev/inference_algos.html#pyro.infer.trace_mean_field_elbo.JitTraceMeanField_ELBO), [HMC(jit_compile=True)](http://docs.pyro.ai/en/dev/mcmc.html#pyro.infer.mcmc.HMC), and [NUTS(jit_compile=True)](http://docs.pyro.ai/en/dev/mcmc.html#pyro.infer.mcmc.NUTS). For further reading, see the [examples/](https://github.com/uber/pyro/tree/dev/examples) directory, where most examples include a `--jit` option to run in compiled mode.
## A simple model
Let's start with a simple Gaussian model and an [autoguide](http://docs.pyro.ai/en/dev/infer.autoguide.html).
First let's run as usual with an SVI object and `Trace_ELBO`.
Next to run with a jit compiled inference, we simply replace
Also note that the `AutoDiagonalNormal` guide behaves a little differently on its first invocation (it runs the model to produce a prototype trace), and we don't want to record this warmup behavior when compiling. Thus we call the `guide(data)` once to initialize, then run the compiled SVI,
Notice that we have a more than 2x speedup for this small model.
Let us now use the same model, but we will instead use MCMC to generate samples from the model's posterior. We will use the No-U-Turn(NUTS) sampler.
We can compile the potential energy computation in NUTS using the `jit_compile=True` argument to the NUTS kernel. We also silence JIT warnings due to the presence of tensor constants in the model by using `ignore_jit_warnings=True`.
We notice a significant increase in sampling throughput when JIT compilation is enabled.
## Varying structure
Time series models often run on datasets of multiple time series with different lengths. To accomodate varying structure like this, Pyro requires models to separate all model inputs into tensors and non-tensors.$^\dagger$
- Non-tensor inputs should be passed as `**kwargs` to the model and guide. These can determine model structure, so that a model is compiled for each value of the passed `**kwargs`.
- Tensor inputs should be passed as `*args`. These must not determine model structure. However `len(args)` may determine model structure (as is used e.g. in semisupervised models).
To illustrate this with a time series model, we will pass in a sequence of observations as a tensor `arg` and the sequence length as a non-tensor `kwarg`:
Now lets' run SVI as usual.
Again we'll simply swap in a `Jit*` implementation
Note that we are manually specifying the `max_plate_nesting` arg. Usually Pyro can figure this out automatically by running the model once on the first invocation; however to avoid this extra work when we run the compiler on the first step, we pass this in manually.
| 0.900518 | 0.984927 |
```
# !wget https://storage.googleapis.com/bert_models/2018_10_18/uncased_L-12_H-768_A-12.zip
# !unzip uncased_L-12_H-768_A-12.zip
BERT_VOCAB = 'uncased_L-12_H-768_A-12/vocab.txt'
BERT_INIT_CHKPNT = 'uncased_L-12_H-768_A-12/bert_model.ckpt'
BERT_CONFIG = 'uncased_L-12_H-768_A-12/bert_config.json'
import bert
from bert import run_classifier
from bert import optimization
from bert import tokenization
from bert import modeling
import tensorflow as tf
tokenization.validate_case_matches_checkpoint(True,BERT_INIT_CHKPNT)
tokenizer = tokenization.FullTokenizer(
vocab_file=BERT_VOCAB, do_lower_case=True)
# !wget https://raw.githubusercontent.com/huseinzol05/NLP-Models-Tensorflow/master/neural-machine-translation/english-train
# !wget https://raw.githubusercontent.com/huseinzol05/NLP-Models-Tensorflow/master/neural-machine-translation/vietnam-train
import collections
def build_dataset(words, n_words, atleast=1):
count = [['PAD', 0], ['GO', 1], ['EOS', 2], ['UNK', 3]]
counter = collections.Counter(words).most_common(n_words)
counter = [i for i in counter if i[1] >= atleast]
count.extend(counter)
dictionary = dict()
for word, _ in count:
dictionary[word] = len(dictionary)
data = list()
unk_count = 0
for word in words:
index = dictionary.get(word, 0)
if index == 0:
unk_count += 1
data.append(index)
count[0][1] = unk_count
reversed_dictionary = dict(zip(dictionary.values(), dictionary.keys()))
return data, count, dictionary, reversed_dictionary
with open('english-train', 'r') as fopen:
text_from = fopen.read().lower().split('\n')[:-1]
with open('vietnam-train', 'r') as fopen:
text_to = fopen.read().lower().split('\n')[:-1]
print('len from: %d, len to: %d'%(len(text_from), len(text_to)))
concat_to = ' '.join(text_to).split()
vocabulary_size_to = len(list(set(concat_to)))
data_to, count_to, dictionary_to, rev_dictionary_to = build_dataset(concat_to, vocabulary_size_to)
print('vocab to size: %d'%(vocabulary_size_to))
print('Most common words', count_to[4:10])
print('Sample data', data_to[:10], [rev_dictionary_to[i] for i in data_to[:10]])
GO = dictionary_to['GO']
PAD = dictionary_to['PAD']
EOS = dictionary_to['EOS']
UNK = dictionary_to['UNK']
for i in range(len(text_to)):
text_to[i] += ' EOS'
MAX_SEQ_LENGTH = 200
from tqdm import tqdm
input_ids, input_masks, segment_ids = [], [], []
for text in tqdm(text_from):
tokens_a = tokenizer.tokenize(text)
if len(tokens_a) > MAX_SEQ_LENGTH - 2:
tokens_a = tokens_a[:(MAX_SEQ_LENGTH - 2)]
tokens = ["[CLS]"] + tokens_a + ["[SEP]"]
segment_id = [0] * len(tokens)
input_id = tokenizer.convert_tokens_to_ids(tokens)
input_mask = [1] * len(input_id)
padding = [0] * (MAX_SEQ_LENGTH - len(input_id))
input_id += padding
input_mask += padding
segment_id += padding
input_ids.append(input_id)
input_masks.append(input_mask)
segment_ids.append(segment_id)
bert_config = modeling.BertConfig.from_json_file(BERT_CONFIG)
epoch = 20
batch_size = 10
warmup_proportion = 0.1
num_train_steps = int(len(input_ids) / batch_size * epoch)
num_warmup_steps = int(num_train_steps * warmup_proportion)
class Chatbot:
def __init__(self, size_layer, num_layers, embedded_size,
to_dict_size, learning_rate, dropout = 0.5):
def gru_cell(reuse=False):
return tf.nn.rnn_cell.GRUCell(size_layer, reuse=reuse)
def attention(encoder_out, seq_len, reuse=False):
attention_mechanism = tf.contrib.seq2seq.LuongAttention(num_units = size_layer,
memory = encoder_out,
memory_sequence_length = seq_len)
return tf.contrib.seq2seq.AttentionWrapper(
cell = tf.nn.rnn_cell.MultiRNNCell([gru_cell(reuse) for _ in range(num_layers)]),
attention_mechanism = attention_mechanism,
attention_layer_size = size_layer)
self.X = tf.placeholder(tf.int32, [None, None])
self.segment_ids = tf.placeholder(tf.int32, [None, None])
self.input_masks = tf.placeholder(tf.int32, [None, None])
self.Y = tf.placeholder(tf.int32, [None, None])
self.X_seq_len = tf.count_nonzero(self.X, 1, dtype=tf.int32)
self.Y_seq_len = tf.count_nonzero(self.Y, 1, dtype=tf.int32)
batch_size = tf.shape(self.X)[0]
model = modeling.BertModel(
config=bert_config,
is_training=True,
input_ids=self.X,
input_mask=self.input_masks,
token_type_ids=self.segment_ids,
use_one_hot_embeddings=False)
self.encoder_out = model.get_sequence_output()
self.encoder_state = tf.layers.dense(model.get_pooled_output(), size_layer)
self.encoder_state = tuple(self.encoder_state for _ in range(num_layers))
main = tf.strided_slice(self.Y, [0, 0], [batch_size, -1], [1, 1])
decoder_input = tf.concat([tf.fill([batch_size, 1], GO), main], 1)
decoder_embeddings = tf.Variable(tf.random_uniform([to_dict_size, embedded_size], -1, 1))
decoder_cell = attention(self.encoder_out, self.X_seq_len)
dense_layer = tf.layers.Dense(to_dict_size)
training_helper = tf.contrib.seq2seq.TrainingHelper(
inputs = tf.nn.embedding_lookup(decoder_embeddings, decoder_input),
sequence_length = self.Y_seq_len,
time_major = False)
training_decoder = tf.contrib.seq2seq.BasicDecoder(
cell = decoder_cell,
helper = training_helper,
initial_state = decoder_cell.zero_state(batch_size, tf.float32).clone(cell_state=self.encoder_state),
output_layer = dense_layer)
training_decoder_output, _, _ = tf.contrib.seq2seq.dynamic_decode(
decoder = training_decoder,
impute_finished = True,
maximum_iterations = tf.reduce_max(self.Y_seq_len))
predicting_helper = tf.contrib.seq2seq.GreedyEmbeddingHelper(
embedding = decoder_embeddings,
start_tokens = tf.tile(tf.constant([GO], dtype=tf.int32), [batch_size]),
end_token = EOS)
predicting_decoder = tf.contrib.seq2seq.BasicDecoder(
cell = decoder_cell,
helper = predicting_helper,
initial_state = decoder_cell.zero_state(batch_size, tf.float32).clone(cell_state=self.encoder_state),
output_layer = dense_layer)
predicting_decoder_output, _, _ = tf.contrib.seq2seq.dynamic_decode(
decoder = predicting_decoder,
impute_finished = True,
maximum_iterations = 2 * tf.reduce_max(self.X_seq_len))
self.training_logits = training_decoder_output.rnn_output
self.predicting_ids = predicting_decoder_output.sample_id
masks = tf.sequence_mask(self.Y_seq_len, tf.reduce_max(self.Y_seq_len), dtype=tf.float32)
self.cost = tf.contrib.seq2seq.sequence_loss(logits = self.training_logits,
targets = self.Y,
weights = masks)
self.optimizer = optimization.create_optimizer(self.cost, learning_rate,
num_train_steps, num_warmup_steps, False)
y_t = tf.argmax(self.training_logits,axis=2)
y_t = tf.cast(y_t, tf.int32)
self.prediction = tf.boolean_mask(y_t, masks)
mask_label = tf.boolean_mask(self.Y, masks)
correct_pred = tf.equal(self.prediction, mask_label)
correct_index = tf.cast(correct_pred, tf.float32)
self.accuracy = tf.reduce_mean(tf.cast(correct_pred, tf.float32))
size_layer = 256
num_layers = 2
embedded_size = 128
learning_rate = 2e-5
tf.reset_default_graph()
sess = tf.InteractiveSession()
model = Chatbot(size_layer, num_layers, embedded_size,
len(dictionary_to), learning_rate)
sess.run(tf.global_variables_initializer())
sess.run(tf.global_variables_initializer())
var_lists = tf.get_collection(tf.GraphKeys.TRAINABLE_VARIABLES, scope = 'bert')
saver = tf.train.Saver(var_list = var_lists)
saver.restore(sess, BERT_INIT_CHKPNT)
def str_idx(corpus, dic):
X = []
for i in corpus:
ints = []
for k in i.split():
ints.append(dic.get(k, 2))
X.append(ints)
return X
def pad_sentence_batch(sentence_batch, pad_int):
padded_seqs = []
seq_lens = []
max_sentence_len = max([len(sentence) for sentence in sentence_batch])
for sentence in sentence_batch:
padded_seqs.append(sentence + [pad_int] * (max_sentence_len - len(sentence)))
seq_lens.append(len(sentence))
return padded_seqs, seq_lens
Y = str_idx(text_to, dictionary_to)
from tqdm import tqdm
import time
import numpy as np
for e in range(epoch):
accuracy, loss = 0, 0
pbar = tqdm(
range(0, len(input_ids), batch_size), desc = 'train minibatch loop'
)
for i in pbar:
index = min(i + batch_size, len(input_ids))
batch_x = input_ids[i: index]
batch_masks = input_masks[i: index]
batch_segment = segment_ids[i: index]
batch_y, seq_y = pad_sentence_batch(Y[i: index], PAD)
acc, cost, _ = sess.run(
[model.accuracy, model.cost, model.optimizer],
feed_dict = {
model.Y: batch_y,
model.X: batch_x,
model.segment_ids: batch_segment,
model.input_masks: batch_masks
},
)
assert not np.isnan(cost)
loss += cost
accuracy += acc
pbar.set_postfix(cost = cost, accuracy = acc)
loss /= len(input_ids) / batch_size
accuracy /= len(input_ids) / batch_size
print(
'epoch: %d, training loss: %f, training acc: %f\n'
% (e, loss, accuracy)
)
```
|
github_jupyter
|
# !wget https://storage.googleapis.com/bert_models/2018_10_18/uncased_L-12_H-768_A-12.zip
# !unzip uncased_L-12_H-768_A-12.zip
BERT_VOCAB = 'uncased_L-12_H-768_A-12/vocab.txt'
BERT_INIT_CHKPNT = 'uncased_L-12_H-768_A-12/bert_model.ckpt'
BERT_CONFIG = 'uncased_L-12_H-768_A-12/bert_config.json'
import bert
from bert import run_classifier
from bert import optimization
from bert import tokenization
from bert import modeling
import tensorflow as tf
tokenization.validate_case_matches_checkpoint(True,BERT_INIT_CHKPNT)
tokenizer = tokenization.FullTokenizer(
vocab_file=BERT_VOCAB, do_lower_case=True)
# !wget https://raw.githubusercontent.com/huseinzol05/NLP-Models-Tensorflow/master/neural-machine-translation/english-train
# !wget https://raw.githubusercontent.com/huseinzol05/NLP-Models-Tensorflow/master/neural-machine-translation/vietnam-train
import collections
def build_dataset(words, n_words, atleast=1):
count = [['PAD', 0], ['GO', 1], ['EOS', 2], ['UNK', 3]]
counter = collections.Counter(words).most_common(n_words)
counter = [i for i in counter if i[1] >= atleast]
count.extend(counter)
dictionary = dict()
for word, _ in count:
dictionary[word] = len(dictionary)
data = list()
unk_count = 0
for word in words:
index = dictionary.get(word, 0)
if index == 0:
unk_count += 1
data.append(index)
count[0][1] = unk_count
reversed_dictionary = dict(zip(dictionary.values(), dictionary.keys()))
return data, count, dictionary, reversed_dictionary
with open('english-train', 'r') as fopen:
text_from = fopen.read().lower().split('\n')[:-1]
with open('vietnam-train', 'r') as fopen:
text_to = fopen.read().lower().split('\n')[:-1]
print('len from: %d, len to: %d'%(len(text_from), len(text_to)))
concat_to = ' '.join(text_to).split()
vocabulary_size_to = len(list(set(concat_to)))
data_to, count_to, dictionary_to, rev_dictionary_to = build_dataset(concat_to, vocabulary_size_to)
print('vocab to size: %d'%(vocabulary_size_to))
print('Most common words', count_to[4:10])
print('Sample data', data_to[:10], [rev_dictionary_to[i] for i in data_to[:10]])
GO = dictionary_to['GO']
PAD = dictionary_to['PAD']
EOS = dictionary_to['EOS']
UNK = dictionary_to['UNK']
for i in range(len(text_to)):
text_to[i] += ' EOS'
MAX_SEQ_LENGTH = 200
from tqdm import tqdm
input_ids, input_masks, segment_ids = [], [], []
for text in tqdm(text_from):
tokens_a = tokenizer.tokenize(text)
if len(tokens_a) > MAX_SEQ_LENGTH - 2:
tokens_a = tokens_a[:(MAX_SEQ_LENGTH - 2)]
tokens = ["[CLS]"] + tokens_a + ["[SEP]"]
segment_id = [0] * len(tokens)
input_id = tokenizer.convert_tokens_to_ids(tokens)
input_mask = [1] * len(input_id)
padding = [0] * (MAX_SEQ_LENGTH - len(input_id))
input_id += padding
input_mask += padding
segment_id += padding
input_ids.append(input_id)
input_masks.append(input_mask)
segment_ids.append(segment_id)
bert_config = modeling.BertConfig.from_json_file(BERT_CONFIG)
epoch = 20
batch_size = 10
warmup_proportion = 0.1
num_train_steps = int(len(input_ids) / batch_size * epoch)
num_warmup_steps = int(num_train_steps * warmup_proportion)
class Chatbot:
def __init__(self, size_layer, num_layers, embedded_size,
to_dict_size, learning_rate, dropout = 0.5):
def gru_cell(reuse=False):
return tf.nn.rnn_cell.GRUCell(size_layer, reuse=reuse)
def attention(encoder_out, seq_len, reuse=False):
attention_mechanism = tf.contrib.seq2seq.LuongAttention(num_units = size_layer,
memory = encoder_out,
memory_sequence_length = seq_len)
return tf.contrib.seq2seq.AttentionWrapper(
cell = tf.nn.rnn_cell.MultiRNNCell([gru_cell(reuse) for _ in range(num_layers)]),
attention_mechanism = attention_mechanism,
attention_layer_size = size_layer)
self.X = tf.placeholder(tf.int32, [None, None])
self.segment_ids = tf.placeholder(tf.int32, [None, None])
self.input_masks = tf.placeholder(tf.int32, [None, None])
self.Y = tf.placeholder(tf.int32, [None, None])
self.X_seq_len = tf.count_nonzero(self.X, 1, dtype=tf.int32)
self.Y_seq_len = tf.count_nonzero(self.Y, 1, dtype=tf.int32)
batch_size = tf.shape(self.X)[0]
model = modeling.BertModel(
config=bert_config,
is_training=True,
input_ids=self.X,
input_mask=self.input_masks,
token_type_ids=self.segment_ids,
use_one_hot_embeddings=False)
self.encoder_out = model.get_sequence_output()
self.encoder_state = tf.layers.dense(model.get_pooled_output(), size_layer)
self.encoder_state = tuple(self.encoder_state for _ in range(num_layers))
main = tf.strided_slice(self.Y, [0, 0], [batch_size, -1], [1, 1])
decoder_input = tf.concat([tf.fill([batch_size, 1], GO), main], 1)
decoder_embeddings = tf.Variable(tf.random_uniform([to_dict_size, embedded_size], -1, 1))
decoder_cell = attention(self.encoder_out, self.X_seq_len)
dense_layer = tf.layers.Dense(to_dict_size)
training_helper = tf.contrib.seq2seq.TrainingHelper(
inputs = tf.nn.embedding_lookup(decoder_embeddings, decoder_input),
sequence_length = self.Y_seq_len,
time_major = False)
training_decoder = tf.contrib.seq2seq.BasicDecoder(
cell = decoder_cell,
helper = training_helper,
initial_state = decoder_cell.zero_state(batch_size, tf.float32).clone(cell_state=self.encoder_state),
output_layer = dense_layer)
training_decoder_output, _, _ = tf.contrib.seq2seq.dynamic_decode(
decoder = training_decoder,
impute_finished = True,
maximum_iterations = tf.reduce_max(self.Y_seq_len))
predicting_helper = tf.contrib.seq2seq.GreedyEmbeddingHelper(
embedding = decoder_embeddings,
start_tokens = tf.tile(tf.constant([GO], dtype=tf.int32), [batch_size]),
end_token = EOS)
predicting_decoder = tf.contrib.seq2seq.BasicDecoder(
cell = decoder_cell,
helper = predicting_helper,
initial_state = decoder_cell.zero_state(batch_size, tf.float32).clone(cell_state=self.encoder_state),
output_layer = dense_layer)
predicting_decoder_output, _, _ = tf.contrib.seq2seq.dynamic_decode(
decoder = predicting_decoder,
impute_finished = True,
maximum_iterations = 2 * tf.reduce_max(self.X_seq_len))
self.training_logits = training_decoder_output.rnn_output
self.predicting_ids = predicting_decoder_output.sample_id
masks = tf.sequence_mask(self.Y_seq_len, tf.reduce_max(self.Y_seq_len), dtype=tf.float32)
self.cost = tf.contrib.seq2seq.sequence_loss(logits = self.training_logits,
targets = self.Y,
weights = masks)
self.optimizer = optimization.create_optimizer(self.cost, learning_rate,
num_train_steps, num_warmup_steps, False)
y_t = tf.argmax(self.training_logits,axis=2)
y_t = tf.cast(y_t, tf.int32)
self.prediction = tf.boolean_mask(y_t, masks)
mask_label = tf.boolean_mask(self.Y, masks)
correct_pred = tf.equal(self.prediction, mask_label)
correct_index = tf.cast(correct_pred, tf.float32)
self.accuracy = tf.reduce_mean(tf.cast(correct_pred, tf.float32))
size_layer = 256
num_layers = 2
embedded_size = 128
learning_rate = 2e-5
tf.reset_default_graph()
sess = tf.InteractiveSession()
model = Chatbot(size_layer, num_layers, embedded_size,
len(dictionary_to), learning_rate)
sess.run(tf.global_variables_initializer())
sess.run(tf.global_variables_initializer())
var_lists = tf.get_collection(tf.GraphKeys.TRAINABLE_VARIABLES, scope = 'bert')
saver = tf.train.Saver(var_list = var_lists)
saver.restore(sess, BERT_INIT_CHKPNT)
def str_idx(corpus, dic):
X = []
for i in corpus:
ints = []
for k in i.split():
ints.append(dic.get(k, 2))
X.append(ints)
return X
def pad_sentence_batch(sentence_batch, pad_int):
padded_seqs = []
seq_lens = []
max_sentence_len = max([len(sentence) for sentence in sentence_batch])
for sentence in sentence_batch:
padded_seqs.append(sentence + [pad_int] * (max_sentence_len - len(sentence)))
seq_lens.append(len(sentence))
return padded_seqs, seq_lens
Y = str_idx(text_to, dictionary_to)
from tqdm import tqdm
import time
import numpy as np
for e in range(epoch):
accuracy, loss = 0, 0
pbar = tqdm(
range(0, len(input_ids), batch_size), desc = 'train minibatch loop'
)
for i in pbar:
index = min(i + batch_size, len(input_ids))
batch_x = input_ids[i: index]
batch_masks = input_masks[i: index]
batch_segment = segment_ids[i: index]
batch_y, seq_y = pad_sentence_batch(Y[i: index], PAD)
acc, cost, _ = sess.run(
[model.accuracy, model.cost, model.optimizer],
feed_dict = {
model.Y: batch_y,
model.X: batch_x,
model.segment_ids: batch_segment,
model.input_masks: batch_masks
},
)
assert not np.isnan(cost)
loss += cost
accuracy += acc
pbar.set_postfix(cost = cost, accuracy = acc)
loss /= len(input_ids) / batch_size
accuracy /= len(input_ids) / batch_size
print(
'epoch: %d, training loss: %f, training acc: %f\n'
% (e, loss, accuracy)
)
| 0.559049 | 0.426919 |
### Note
* Instructions have been included for each segment. You do not have to follow them exactly, but they are included to help you think through the steps.
```
# Dependencies and Setup
import pandas as pd
import numpy as np
# File to Load (Remember to Change These)
file_to_load = "Resources/purchase_data.csv"
# Read Purchasing File and store into Pandas data frame
Purchase_Data = pd.read_csv(file_to_load)
Purchase_Data.head()
```
## Player Count
* Display the total number of players
```
TotalPlayerCount=Purchase_Data["SN"].unique().size
TotalPlayerCount_df=pd.DataFrame([[TotalPlayerCount]])
TotalPlayerCount_df.columns=[['Total Players']]
TotalPlayerCount_df.head()
```
## Purchasing Analysis (Total)
* Run basic calculations to obtain number of unique items, average price, etc.
* Create a summary data frame to hold the results
* Optional: give the displayed data cleaner formatting
* Display the summary data frame
```
Average_Price = Purchase_Data ['Price'].mean()
Unique_items = len(Purchase_Data['Item ID'].unique())
Revenue = Purchase_Data ['Price'].sum()
Purchase_Count = Purchase_Data ['Purchase ID'].count()
Summary = pd.DataFrame({'Number of Unique Items': [Unique_items],'Average Price': ['${:,.2f}'.format(Average_Price)],
'Number of Purchases': [Purchase_Count],'Total Revenue': ['${:,.2f}'.format(Revenue)]})
Summary
```
## Gender Demographics
* Percentage and Count of Male Players
* Percentage and Count of Female Players
* Percentage and Count of Other / Non-Disclosed
```
Gender_Demo = Purchase_Data.groupby ("Gender")
Total_Count = Gender_Demo["SN"].nunique()
Gender_Demo.first()
Player_Perc = Total_Count / TotalPlayerCount *100
Demo_Gender = pd.DataFrame({'Total Count':Total_Count,'Percentage of Players': Player_Perc})
Demo_Gender["Percentage of Players"]=Demo_Gender["Percentage of Players"].map('{:.2f}%'.format)
Demo_Gender
type (Demo_Gender)
Demo_Gender
```
## Purchasing Analysis (Gender)
* Run basic calculations to obtain purchase count, avg. purchase price, avg. purchase total per person etc. by gender
* Create a summary data frame to hold the results
* Optional: give the displayed data cleaner formatting
* Display the summary data frame
```
Purch_Count = Gender_Demo["Purchase ID"].count()
Gender_Demo.first()
Avg_Purch = Gender_Demo["Price"].mean()
Total_Purch = Gender_Demo["Price"].sum()
Per_Person = Total_Purch / Total_Count
Analysis = pd.DataFrame({'Purchase Count':Purch_Count,'Average Purchase Price':(Avg_Purch),
'Total Purchase Value':Total_Purch,'Avg Total Purchase per Person':(Per_Person)})
Analysis
```
## Age Demographics
* Establish bins for ages
* Categorize the existing players using the age bins. Hint: use pd.cut()
* Calculate the numbers and percentages by age group
* Create a summary data frame to hold the results
* Optional: round the percentage column to two decimal points
* Display Age Demographics Table
```
Age_Bins = [0,9,14,19,24,29,34,39,41]
Bin_Names = ["<10","10-14","15-19","20-24","25-29","30-34","35-39","40+"]
Players_Age = Purchase_Data.loc[:,["Age","SN"]]
Purchase_Data["Age Group"] = pd.cut(Purchase_Data["Age"],Age_Bins, labels=Bin_Names)
Purchase_Data["Bin_Columns"] = pd.cut(Purchase_Data["Age"],Age_Bins,labels = Bin_Names)
Purchase_Data
Age_Group = Purchase_Data.groupby("Age Group")
Count_Age = Age_Group["SN"].nunique()
Age_Perc = (Count_Age/TotalPlayerCount) * 100
Age_Dem = pd.DataFrame({"Percentage of Players": Age_Perc, "Total Count": Count_Age})
Age_Dem.index.name = None
Age_Dem.style.format({"Percentage of Players":"{:,.2f}"})
Age_Dem
```
## Purchasing Analysis (Age)
* Bin the purchase_data data frame by age
* Run basic calculations to obtain purchase count, avg. purchase price, avg. purchase total per person etc. in the table below
* Create a summary data frame to hold the results
* Optional: give the displayed data cleaner formatting
* Display the summary data frame
```
Age_Bins = [0,9,14,19,24,29,34,39,41]
Bin_Names = ["<10","10-14","15-19","20-24","25-29","30-34","35-39","40+"]
Players_Age = Purchase_Data.loc[:,["Age","SN"]]
Purchase_Data["Age Group"] = pd.cut(Purchase_Data["Age"],Age_Bins, labels=Bin_Names)
Purchase_Data["Bin_Columns"] = pd.cut(Purchase_Data["Age"],Age_Bins,labels = Bin_Names)
TotalPurchValue = Age_Group["Price"].sum()
AvgPurchPerAge = TotalPurchValue/Count_Age
AvgPurchPriceAge = Age_Group["Price"].mean()
PurchCountAge = Age_Group["Purchase ID"].count()
Age_Graph = pd.DataFrame({"Purchase Count": PurchCountAge,
"Average Purchase Price": AvgPurchPriceAge,
"Total Purchase Value":TotalPurchValue,
"Average Purchase Total per Person": AvgPurchPerAge})
Age_Graph.index.name = None
Age_Graph.style.format({"Average Purchase Price":"${:,.2f}",
"Total Purchase Value":"${:,.2f}",
"Average Purchase Total per Person":"${:,.2f}"})
Age_Graph
```
## Top Spenders
* Run basic calculations to obtain the results in the table below
* Create a summary data frame to hold the results
* Sort the total purchase value column in descending order
* Optional: give the displayed data cleaner formatting
* Display a preview of the summary data frame
```
Spender = Purchase_Data.groupby("SN")
AvgPurchSpender = Spender["Price"].mean()
PurchTotalSpender = Spender["Price"].sum()
PurchCountSpender = Spender["Purchase ID"].count()
TopSpender = pd.DataFrame({"Purchase Count": PurchCountSpender,
"Average Purchase Price": AvgPurchSpender,
"Total Purchase Value":PurchTotalSpender})
SpenderFormat = TopSpender.sort_values(["Total Purchase Value"], ascending=False).head()
SpenderFormat.style.format({"Average Purchase Total":"${:,.2f}",
"Average Purchase Price":"${:,.2f}",
"Total Purchase Value":"${:,.2f}"})
```
## Most Popular Items
* Retrieve the Item ID, Item Name, and Item Price columns
* Group by Item ID and Item Name. Perform calculations to obtain purchase count, average item price, and total purchase value
* Create a summary data frame to hold the results
* Sort the purchase count column in descending order
* Optional: give the displayed data cleaner formatting
* Display a preview of the summary data frame
```
Items = Purchase_Data[["Item ID", "Item Name", "Price"]]
ItemStats = Items.groupby(["Item ID","Item Name"])
PurchaseItemCount = ItemStats["Price"].count()
PurchaseValue = (ItemStats["Price"].sum())
ItemPrc = PurchaseValue/PurchaseItemCount
MostPopularItem = pd.DataFrame({"Purchase Count": PurchaseItemCount,
"Item Price": ItemPrc,
"Total Purchase Value":PurchaseValue})
PopularFormat = MostPopularItem.sort_values(["Purchase Count"], ascending=False).head()
PopularFormat.style.format({"Item Price":"${:,.2f}",
"Total Purchase Value":"${:,.2f}"})
```
## Most Profitable Items
* Sort the above table by total purchase value in descending order
* Optional: give the displayed data cleaner formatting
* Display a preview of the data frame
```
PopularFormat = MostPopularItem.sort_values(["Total Purchase Value"], ascending=False).head()
PopularFormat.style.format({"Item Price":"${:,.2f}",
"Total Purchase Value":"${:,.2f}"})
```
|
github_jupyter
|
# Dependencies and Setup
import pandas as pd
import numpy as np
# File to Load (Remember to Change These)
file_to_load = "Resources/purchase_data.csv"
# Read Purchasing File and store into Pandas data frame
Purchase_Data = pd.read_csv(file_to_load)
Purchase_Data.head()
TotalPlayerCount=Purchase_Data["SN"].unique().size
TotalPlayerCount_df=pd.DataFrame([[TotalPlayerCount]])
TotalPlayerCount_df.columns=[['Total Players']]
TotalPlayerCount_df.head()
Average_Price = Purchase_Data ['Price'].mean()
Unique_items = len(Purchase_Data['Item ID'].unique())
Revenue = Purchase_Data ['Price'].sum()
Purchase_Count = Purchase_Data ['Purchase ID'].count()
Summary = pd.DataFrame({'Number of Unique Items': [Unique_items],'Average Price': ['${:,.2f}'.format(Average_Price)],
'Number of Purchases': [Purchase_Count],'Total Revenue': ['${:,.2f}'.format(Revenue)]})
Summary
Gender_Demo = Purchase_Data.groupby ("Gender")
Total_Count = Gender_Demo["SN"].nunique()
Gender_Demo.first()
Player_Perc = Total_Count / TotalPlayerCount *100
Demo_Gender = pd.DataFrame({'Total Count':Total_Count,'Percentage of Players': Player_Perc})
Demo_Gender["Percentage of Players"]=Demo_Gender["Percentage of Players"].map('{:.2f}%'.format)
Demo_Gender
type (Demo_Gender)
Demo_Gender
Purch_Count = Gender_Demo["Purchase ID"].count()
Gender_Demo.first()
Avg_Purch = Gender_Demo["Price"].mean()
Total_Purch = Gender_Demo["Price"].sum()
Per_Person = Total_Purch / Total_Count
Analysis = pd.DataFrame({'Purchase Count':Purch_Count,'Average Purchase Price':(Avg_Purch),
'Total Purchase Value':Total_Purch,'Avg Total Purchase per Person':(Per_Person)})
Analysis
Age_Bins = [0,9,14,19,24,29,34,39,41]
Bin_Names = ["<10","10-14","15-19","20-24","25-29","30-34","35-39","40+"]
Players_Age = Purchase_Data.loc[:,["Age","SN"]]
Purchase_Data["Age Group"] = pd.cut(Purchase_Data["Age"],Age_Bins, labels=Bin_Names)
Purchase_Data["Bin_Columns"] = pd.cut(Purchase_Data["Age"],Age_Bins,labels = Bin_Names)
Purchase_Data
Age_Group = Purchase_Data.groupby("Age Group")
Count_Age = Age_Group["SN"].nunique()
Age_Perc = (Count_Age/TotalPlayerCount) * 100
Age_Dem = pd.DataFrame({"Percentage of Players": Age_Perc, "Total Count": Count_Age})
Age_Dem.index.name = None
Age_Dem.style.format({"Percentage of Players":"{:,.2f}"})
Age_Dem
Age_Bins = [0,9,14,19,24,29,34,39,41]
Bin_Names = ["<10","10-14","15-19","20-24","25-29","30-34","35-39","40+"]
Players_Age = Purchase_Data.loc[:,["Age","SN"]]
Purchase_Data["Age Group"] = pd.cut(Purchase_Data["Age"],Age_Bins, labels=Bin_Names)
Purchase_Data["Bin_Columns"] = pd.cut(Purchase_Data["Age"],Age_Bins,labels = Bin_Names)
TotalPurchValue = Age_Group["Price"].sum()
AvgPurchPerAge = TotalPurchValue/Count_Age
AvgPurchPriceAge = Age_Group["Price"].mean()
PurchCountAge = Age_Group["Purchase ID"].count()
Age_Graph = pd.DataFrame({"Purchase Count": PurchCountAge,
"Average Purchase Price": AvgPurchPriceAge,
"Total Purchase Value":TotalPurchValue,
"Average Purchase Total per Person": AvgPurchPerAge})
Age_Graph.index.name = None
Age_Graph.style.format({"Average Purchase Price":"${:,.2f}",
"Total Purchase Value":"${:,.2f}",
"Average Purchase Total per Person":"${:,.2f}"})
Age_Graph
Spender = Purchase_Data.groupby("SN")
AvgPurchSpender = Spender["Price"].mean()
PurchTotalSpender = Spender["Price"].sum()
PurchCountSpender = Spender["Purchase ID"].count()
TopSpender = pd.DataFrame({"Purchase Count": PurchCountSpender,
"Average Purchase Price": AvgPurchSpender,
"Total Purchase Value":PurchTotalSpender})
SpenderFormat = TopSpender.sort_values(["Total Purchase Value"], ascending=False).head()
SpenderFormat.style.format({"Average Purchase Total":"${:,.2f}",
"Average Purchase Price":"${:,.2f}",
"Total Purchase Value":"${:,.2f}"})
Items = Purchase_Data[["Item ID", "Item Name", "Price"]]
ItemStats = Items.groupby(["Item ID","Item Name"])
PurchaseItemCount = ItemStats["Price"].count()
PurchaseValue = (ItemStats["Price"].sum())
ItemPrc = PurchaseValue/PurchaseItemCount
MostPopularItem = pd.DataFrame({"Purchase Count": PurchaseItemCount,
"Item Price": ItemPrc,
"Total Purchase Value":PurchaseValue})
PopularFormat = MostPopularItem.sort_values(["Purchase Count"], ascending=False).head()
PopularFormat.style.format({"Item Price":"${:,.2f}",
"Total Purchase Value":"${:,.2f}"})
PopularFormat = MostPopularItem.sort_values(["Total Purchase Value"], ascending=False).head()
PopularFormat.style.format({"Item Price":"${:,.2f}",
"Total Purchase Value":"${:,.2f}"})
| 0.404037 | 0.845688 |
# Function Practice Exercises
Problems are arranged in increasing difficulty:
* Warm up - these can be solved using basic comparisons and methods
* Level 1 - these may involve if/then conditional statements and simple methods
* Level 2 - these may require iterating over sequences, usually with some kind of loop
* Challenging - these will take some creativity to solve
## WARM UP SECTION:
#### LESSER OF TWO EVENS:
Write a function that returns the lesser of two given numbers *if* both numbers are even, but returns the greater if one or both numbers are odd
lesser_of_two_evens(2, 4) --> 2
lesser_of_two_evens(2, 5) --> 5
```
def lesser_of_two_evens(a, b):
if (a%2 == 0) and (b%2 == 0):
return min(a, b)
else:
return max(a, b)
lesser_of_two_evens(2, 4)
lesser_of_two_evens(7, 5)
```
#### ANIMAL CRACKERS:
Write a function takes a two-word string and returns True if both words begin with same letter
animal_crackers('Levelheaded Llama') --> True
animal_crackers('Crazy Kangaroo') --> False
```
animal_crackers = lambda s: s.split()[0][0].lower() == s.split()[1][0].lower()
animal_crackers('Levelheaded Llama')
animal_crackers('Crazy kangaroo')
```
#### MAKES TWENTY:
Given two integers, return True if the sum of the integers is 20 *or* if one of the integers is 20. If not, return False
makes_twenty(20, 10) --> True
makes_twenty(12, 8) --> True
makes_twenty(2 ,3) --> False
```
makes_twenty = lambda n1, n2: (n1 + n2 == 20) or (n1 == 20) or (n2 == 20)
makes_twenty(20, 10)
makes_twenty(2, 3)
makes_twenty(12, 8)
```
# LEVEL 1 PROBLEMS
#### OLD MACDONALD:
Write a function that capitalizes the first and fourth letters of a name
old_macdonald('macdonald') --> MacDonald
Note: `'macdonald'.capitalize()` returns `'Macdonald'`
```
old_macdonald = lambda name: name[:3].capitalize() + name[3:].capitalize()
old_macdonald('macdonald')
```
#### MASTER YODA:
Given a sentence, return a sentence with the words reversed
master_yoda('I am home') --> 'home am I'
master_yoda('We are ready') --> 'ready are We'
Note: The .join() method may be useful here. The .join() method allows you to join together strings in a list with some connector string. For example, some uses of the .join() method:
>>> "--".join(['a','b','c'])
>>> 'a--b--c'
This means if you had a list of words you wanted to turn back into a sentence, you could just join them with a single space string:
>>> " ".join(['Hello','world'])
>>> "Hello world"
```
master_yoda = lambda s: " ".join(s.split()[::-1])
master_yoda('I am home')
master_yoda('We are ready')
```
#### ALMOST THERE:
Given an integer n, return True if n is within 10 of either 100 or 200
almost_there(90) --> True
almost_there(104) --> True
almost_there(150) --> False
almost_there(209) --> True
NOTE: `abs(num)` returns the absolute value of a number
```
almost_there = lambda n: (abs(n - 100) <= 10) or (abs(n - 200) <= 10)
almost_there(104)
almost_there(150)
almost_there(210)
```
# LEVEL 2 PROBLEMS
#### FIND 33:
Given a list of integers, return True if the array contains a 3 next to a 3 somewhere.
has_33([1, 3, 3]) → True
has_33([1, 3, 1, 3]) → False
has_33([3, 1, 3]) → False
```
has_33 =lambda numbers: "33" in "".join(map(lambda numbers: str(numbers), numbers))
has_33([1, 3, 3])
has_33([1, 3, 1, 3])
has_33([3, 1, 3])
```
#### PAPER DOLL:
Given a string, return a string where for every character in the original there are three characters
paper_doll('Hello') --> 'HHHeeellllllooo'
paper_doll('Mississippi') --> 'MMMiiissssssiiippppppiii'
```
paper_doll = lambda s: "".join([char * 3 for char in s])
paper_doll('Hello')
paper_doll('Mississippi')
```
#### BLACKJACK:
Given three integers between 1 and 11, if their sum is less than or equal to 21, return their sum. If their sum exceeds 21 *and* there's an eleven, reduce the total sum by 10. Finally, if the sum (even after adjustment) exceeds 21, return 'BUST'
blackjack(5, 6, 7) --> 18
blackjack(9, 9, 9) --> 'BUST'
blackjack(9, 9, 11) --> 19
```
def blackjack(a, b, c):
if (a > 11) or (b > 11) or (c > 11):
return "The integers must be between 2 and 11!"
else:
sum_all = sum([a, b, c])
if sum_all <= 21:
return sum_all
else:
count_11 = [a, b, c].count(11)
while count_11 > 0:
sum_all -= 10
count_11 -= 1
if sum_all <= 21:
return sum_all
return "BUST"
# Check
blackjack(5, 6, 7)
# Check
blackjack(9, 9, 9)
# Check
blackjack(9, 9, 11)
blackjack(11, 11, 11)
```
#### SUMMER OF '69:
Return the sum of the numbers in the array, except ignore sections of numbers starting with a 6 and extending to the next 9 (every 6 will be followed by at least one 9). Return 0 for no numbers.
summer_69([1, 3, 5]) --> 9
summer_69([4, 5, 6, 7, 8, 9]) --> 9
summer_69([2, 1, 6, 9, 11]) --> 14
```
def summer_of_69(numbers):
index_6 = 0
index_9 = -1
if 6 in numbers and 9 in numbers:
index_6 = numbers.index(6)
index_9 = numbers.index(9)
return sum(numbers) - sum(numbers[index_6: index_9 + 1])
summer_of_69([1, 3, 5])
summer_of_69([4, 5, 6, 7, 8, 9])
summer_of_69([2, 1, 6, 9, 11])
```
# CHALLENGING PROBLEMS
#### SPY GAME:
Write a function that takes in a list of integers and returns True if it contains 007 in order
spy_game([1,2,4,0,0,7,5]) --> True
spy_game([1,0,2,4,0,5,7]) --> True
spy_game([1,7,2,0,4,5,0]) --> False
```
spy_game = lambda numbers: "007" in "".join(
map(lambda number: str(number), [num for num in numbers if num in [0, 7]]))
spy_game([1, 2, 4, 0, 0, 7, 5])
spy_game([1, 0, 2, 4, 0, 5, 7])
spy_game([1, 7, 2, 0, 4, 5, 0])
spy_game([0, 1, 7, 2, 0, 4, 5, 0])
```
#### COUNT PRIMES:
Write a function that returns the *number* of prime numbers that exist up to and including a given number
count_primes(100) --> 25
By convention, 0 and 1 are not prime.
```
def primes_up_to(upper_limit):
primes = [2]
for number in range(3, upper_limit + 1, 2):
for dividend in range(3, number//2):
if number % dividend == 0:
break
else:
primes.append(number)
return primes, len(primes)
primes_up_to(10)
primes_up_to(100)
```
### Just for fun:
#### PRINT BIG:
Write a function that takes in a single letter, and returns a 5x5 representation of that letter
print_big('a')
out: *
* *
*****
* *
* *
HINT: Consider making a dictionary of possible patterns, and mapping the alphabet to specific 5-line combinations of patterns. <br>For purposes of this exercise, it's ok if your dictionary stops at "E".
```
def print_big(letters):
patterns = {1: ' * ',
2: ' * * ',
3: '* *',
4: '*****',
5: '**** ',
6: ' * ',
7: ' * ',
8: '* * ',
9: '* '}
alphabet = {'A':[1, 2, 4, 3, 3],
'B':[5, 3, 5, 3, 5],
'C':[4, 9, 9, 9, 4],
'D':[5, 3, 3, 3, 5],
'E':[4, 9, 4, 9, 4]}
for char in list(letters.upper()):
for pattern in alphabet[char]:
print(patterns[pattern])
print("")
print_big("abcde")
```
|
github_jupyter
|
def lesser_of_two_evens(a, b):
if (a%2 == 0) and (b%2 == 0):
return min(a, b)
else:
return max(a, b)
lesser_of_two_evens(2, 4)
lesser_of_two_evens(7, 5)
animal_crackers = lambda s: s.split()[0][0].lower() == s.split()[1][0].lower()
animal_crackers('Levelheaded Llama')
animal_crackers('Crazy kangaroo')
makes_twenty = lambda n1, n2: (n1 + n2 == 20) or (n1 == 20) or (n2 == 20)
makes_twenty(20, 10)
makes_twenty(2, 3)
makes_twenty(12, 8)
old_macdonald = lambda name: name[:3].capitalize() + name[3:].capitalize()
old_macdonald('macdonald')
master_yoda = lambda s: " ".join(s.split()[::-1])
master_yoda('I am home')
master_yoda('We are ready')
almost_there = lambda n: (abs(n - 100) <= 10) or (abs(n - 200) <= 10)
almost_there(104)
almost_there(150)
almost_there(210)
has_33 =lambda numbers: "33" in "".join(map(lambda numbers: str(numbers), numbers))
has_33([1, 3, 3])
has_33([1, 3, 1, 3])
has_33([3, 1, 3])
paper_doll = lambda s: "".join([char * 3 for char in s])
paper_doll('Hello')
paper_doll('Mississippi')
def blackjack(a, b, c):
if (a > 11) or (b > 11) or (c > 11):
return "The integers must be between 2 and 11!"
else:
sum_all = sum([a, b, c])
if sum_all <= 21:
return sum_all
else:
count_11 = [a, b, c].count(11)
while count_11 > 0:
sum_all -= 10
count_11 -= 1
if sum_all <= 21:
return sum_all
return "BUST"
# Check
blackjack(5, 6, 7)
# Check
blackjack(9, 9, 9)
# Check
blackjack(9, 9, 11)
blackjack(11, 11, 11)
def summer_of_69(numbers):
index_6 = 0
index_9 = -1
if 6 in numbers and 9 in numbers:
index_6 = numbers.index(6)
index_9 = numbers.index(9)
return sum(numbers) - sum(numbers[index_6: index_9 + 1])
summer_of_69([1, 3, 5])
summer_of_69([4, 5, 6, 7, 8, 9])
summer_of_69([2, 1, 6, 9, 11])
spy_game = lambda numbers: "007" in "".join(
map(lambda number: str(number), [num for num in numbers if num in [0, 7]]))
spy_game([1, 2, 4, 0, 0, 7, 5])
spy_game([1, 0, 2, 4, 0, 5, 7])
spy_game([1, 7, 2, 0, 4, 5, 0])
spy_game([0, 1, 7, 2, 0, 4, 5, 0])
def primes_up_to(upper_limit):
primes = [2]
for number in range(3, upper_limit + 1, 2):
for dividend in range(3, number//2):
if number % dividend == 0:
break
else:
primes.append(number)
return primes, len(primes)
primes_up_to(10)
primes_up_to(100)
def print_big(letters):
patterns = {1: ' * ',
2: ' * * ',
3: '* *',
4: '*****',
5: '**** ',
6: ' * ',
7: ' * ',
8: '* * ',
9: '* '}
alphabet = {'A':[1, 2, 4, 3, 3],
'B':[5, 3, 5, 3, 5],
'C':[4, 9, 9, 9, 4],
'D':[5, 3, 3, 3, 5],
'E':[4, 9, 4, 9, 4]}
for char in list(letters.upper()):
for pattern in alphabet[char]:
print(patterns[pattern])
print("")
print_big("abcde")
| 0.326271 | 0.945399 |
```
import pandas as pd
from app import WinrateCollect2
wc = WinrateCollect2("heroes_page_list.csv")
wc.update_stat()
# prepare heros stats
df = pd.read_pickle("data")
df['hero'] = df['hero'].str.replace('%20',' ')
df = df.astype({'g': 'int32'})
df.head()
# prepare heros matchups
df_m = pd.read_pickle("matchups")
df_m['hero'] = df_m['hero'].str.replace('%20',' ')
df_m = df_m.astype({'w': 'int32'})
df_m = df_m.astype({'l': 'int32'})
df_m.head()
def get_matchup(enemy_heroes):
matchup = None
for e in enemy_heroes:
tmp = df_m.loc[(df_m['hero'] == e)][['matchup', 'w', 'l']].set_index('matchup')
if matchup is not None:
matchup = matchup.add(tmp, fill_value=0)
else:
matchup = tmp
matchup = matchup[~matchup.index.isin(enemy_heroes)]
center_g = 1
center_lr = 1
matchup = matchup.assign(g = matchup['w'] + matchup['l'])
matchup = matchup.assign(lr = matchup['l'] / (matchup['w'] + matchup['l']))
matchup = matchup.assign(g_qant = matchup['g'].rank(method='max', pct=True))
matchup = matchup.assign(lr_qant = matchup['lr'].rank(method='max', pct=True))
matchup = matchup.assign(qant_circle = (matchup['g_qant']-center_g)**2 + (matchup['lr_qant']-center_lr)**2)
matchup = matchup.assign(rank = 1 - matchup['qant_circle'])
df_top = matchup.sort_values(by=['qant_circle'],ascending=True)[['g', 'lr']]
df_top = df_top.loc[df_top['lr'] > 0.49]
return df_top
df_tmp = df.sort_values(['hero', 'g'], ascending=[True,False]).drop_duplicates(['hero'])
p1 = list(df_tmp.loc[(df['pos'] == 1)].hero)
p2 = list(df_tmp.loc[(df['pos'] == 2)].hero)
p3 = list(df_tmp.loc[(df['pos'] == 3)].hero)
p4 = list(df_tmp.loc[(df['pos'] == 4)].hero)
p5 = list(df_tmp.loc[(df['pos'] == 5)].hero)
# insert enemy heroes
enemies = ['Ancient Apparition', 'Hoodwink']
m = get_matchup(enemies)
# select pos
m[m.index.isin(p5)]
df_tmp = df.sort_values(['hero', 'g'], ascending=[True,False]).drop_duplicates(['hero'])
df_tmp
p1 = list(df_tmp.loc[(df['pos'] == 1)].hero)
pos = 1
# & (df['wr'] >= 52)
min_g = df.loc[(df['pos'] == pos)]['g'].max() / 100
df1 = df.loc[(df['pos'] == pos) & (df['g'] > min_g) ]
df_top3 = df1.sort_values(by=['wr'],ascending=False)
df_top3
'''
# old version
def tierS(pos):
min_g = df.loc[(df['pos'] == pos)]['g'].max() / 3
df1 = df.loc[(df['pos'] == pos) & (df['g'] > min_g) & (df['wr'] >= 52)]
df_top3 = df1.sort_values(by=['wr'],ascending=False)
return df_top3
def tierA(pos):
min_g = df.loc[(df['pos'] == pos)]['g'].max() / 4
df2 = tierS(pos)
df1 = df.loc[(df['pos'] == pos) & (df['g'] > min_g) & (df['wr'] >= 49)]
df1 = df1.sort_values(by=['wr'],ascending=False).head(14)
df_top8 = pd.concat([df1, df2]).drop_duplicates(keep=False)
return df_top8.head(11)
def topWR():
min_g = df['g'].max() / 40
df_top3 = df.sort_values(by=['wr'],ascending=False)
return df_top3.head(10)
def topTier():
min_g = df['g'].max() / 10
df1 = df.loc[(df['g'] > min_g) & (df['wr'] >= 49)]
df_top3 = df1.sort_values(by=['wr'],ascending=False)
return df_top3.head(15)
'''
# make games count less valuable
#center_g = 0.9
# make win rate less valuable
#center_wr = 0.9
center_g = 1
center_wr = 1
#.style.hide_index()
def tierS(pos):
# 5% - outliers
min_g = df.loc[(df['pos'] == pos)]['g'].max() / 20
df1 = df.loc[(df['pos'] == pos) & (df['g'] >= min_g)]
df1 = df1.assign(g_qant = df1['g'].rank(method='max', pct=True))
df1 = df1.assign(wr_qant = df1['wr'].rank(method='max', pct=True))
df1 = df1.assign(qant_circle = (df1['g_qant']-center_g)**2 + (df1['wr_qant']-center_wr)**2)
df1 = df1.assign(rank = 1 - df1['qant_circle'])
df2 = df1.loc[df1['qant_circle'] <= 0.0625]
df_top = df2.sort_values(by=['qant_circle'],ascending=True)[['hero', 'g', 'wr', 'rank']]
return df_top
def tierA(pos):
# 5% - outliers
min_g = df.loc[(df['pos'] == pos)]['g'].max() / 20
df1 = df.loc[(df['pos'] == pos) & (df['g'] >= min_g)]
df1 = df1.assign(g_qant = df1['g'].rank(method='max', pct=True))
df1 = df1.assign(wr_qant = df1['wr'].rank(method='max', pct=True))
df1 = df1.assign(qant_circle = (df1['g_qant']-center_g)**2 + (df1['wr_qant']-center_wr)**2)
df1 = df1.assign(rank = 1 - df1['qant_circle'])
df2 = df1.loc[(df1['qant_circle'] <= 0.25) & (df1['qant_circle'] > 0.0625)]
df_top = df2.sort_values(by=['qant_circle'],ascending=True)[['hero', 'g', 'wr', 'rank']]
return df_top
def tierB_plus(pos):
# 5% - outliers
min_g = df.loc[(df['pos'] == pos)]['g'].max() / 20
df1 = df.loc[(df['pos'] == pos) & (df['g'] >= min_g)]
df1 = df1.assign(g_qant = df1['g'].rank(method='max', pct=True))
df1 = df1.assign(wr_qant = df1['wr'].rank(method='max', pct=True))
df1 = df1.assign(qant_circle = (df1['g_qant']-center_g)**2 + (df1['wr_qant']-center_wr)**2)
df1 = df1.assign(rank = 1 - df1['qant_circle'])
df2 = df1.loc[(df1['qant_circle'] > 0.25)]
df_top = df2.sort_values(by=['qant_circle'],ascending=True)[['hero', 'g', 'wr', 'rank']]
return df_top
def topWR_lowpick():
df_top3 = df.sort_values(by=['wr'],ascending=False)
return df_top3.head(10)
def topHiddenImba_pos(pos):
# 5% - outliers
min_g = df.loc[(df['pos'] == pos)]['g'].max() / 20
df1 = df.loc[(df['pos'] == pos) & (df['g'] >= min_g)]
df1 = df1.assign(g_qant = df1['g'].rank(method='max', pct=True))
df1 = df1.assign(wr_qant = df1['wr'].rank(method='max', pct=True))
df1 = df1.assign(qant_circle = (df1['g_qant']-center_g/2)**2 + (df1['wr_qant']-center_wr)**2)
df1 = df1.assign(rank = 1 - df1['qant_circle'])
df2 = df1.loc[df1['qant_circle'] <= 0.25]
df_top = df2.sort_values(by=['qant_circle'],ascending=True)[['hero', 'g', 'wr', 'rank']]
return df_top.head(3)
def topWinrateOverallTiers():
min_g = df['g'].max() / 10
df1 = df.loc[(df['g'] > min_g) & (df['wr'] >= 49)]
df_top3 = df1.sort_values(by=['wr'],ascending=False)
return df_top3.head(10)
topWinrateOverallTiers()
topWR_lowpick()
tierS(1)
tierA(1)
tierB_plus(1)
tierS(2)
tierA(2)
tierS(3)
tierA(3)
tierS(4)
tierA(4)
topHiddenImba_pos(4)
tierS(5)
tierA(5)
topHiddenImba_pos(5)
import os
from datetime import datetime
fold = 'history/'+ datetime.today().strftime('%Y_%m_%d__%Hh_%Mm')
os.mkdir(fold)
import dataframe_image as dfi
tierS(1).dfi.export(fold + '/1_TierS.png')
tierA(1).dfi.export(fold + '/1_TierA.png')
tierB_plus(1).dfi.export(fold + '/1_TierBplus.png')
topWinrateOverallTiers().dfi.export(fold + '/topWinrate.png')
topWR_lowpick().dfi.export(fold + '/topLowpick.png')
tierS(2).dfi.export(fold + '/2_TierS.png')
tierA(2).dfi.export(fold + '/2_TierA.png')
tierS(3).dfi.export(fold + '/3_TierS.png')
tierA(3).dfi.export(fold + '/3_TierA.png')
tierS(4).dfi.export(fold + '/4_TierS.png')
tierA(4).dfi.export(fold + '/4_TierA.png')
topHiddenImba_pos(4).dfi.export(fold + '/4_HiddenImba.png')
tierS(5).dfi.export(fold + '/5_TierS.png')
tierA(5).dfi.export(fold + '/5_TierA.png')
topHiddenImba_pos(5).dfi.export(fold + '/5_HiddenImba.png')
from IPython.display import display, HTML
css = """
.output {
flex-direction: row;
}
"""
HTML('<style>{}</style>'.format(css))
display(tierS(1))
display(tierA(1))
#display(tierB_plus(1))
df1 = df.loc[(df['pos'] == 1)]
df1 = df1.sort_values(by=['g'],ascending=False)
df1
df1.describe()
# make games count less valuable
#center_g = 0.9
# make win rate less valuable
#center_wr = 0.9
center_g = 1
center_wr = 1
#.style.hide_index()
def tiers(pos):
# 5% - outliers
min_g = df.loc[(df['pos'] == pos)]['g'].max() / 20
df1 = df.loc[(df['pos'] == pos) & (df['g'] >= min_g)]
df1 = df1.assign(g_qant = df1['g'].rank(method='max', pct=True))
df1 = df1.assign(wr_qant = df1['wr'].rank(method='max', pct=True))
df1 = df1.assign(qant_circle = (df1['g_qant']-center_g)**2 + (df1['wr_qant']-center_wr)**2)
df1 = df1.assign(rank = 1 - df1['qant_circle'])
df1['tier'] = ''
df1['tier'] = df1['qant_circle'].apply(lambda x: 'S' if x <= 0.0625 else
('A' if (x <= 0.25) & (x > 0.0625) else
('B' if (x <= 0.5625) & (x > 0.25) else 'C+')))
df_top = df1.sort_values(by=['qant_circle'],ascending=True)[['hero', 'g', 'wr', 'rank', 'tier']]
df_top.index = range(1, len(df1.index) + 1)
return df_top
def topWR_lowpick():
df_top3 = df.sort_values(by=['wr'],ascending=False)
return df_top3.head(10)
def topHiddenImba_pos(pos):
# 5% - outliers
min_g = df.loc[(df['pos'] == pos)]['g'].max() / 20
df1 = df.loc[(df['pos'] == pos) & (df['g'] >= min_g)]
df1 = df1.assign(g_qant = df1['g'].rank(method='max', pct=True))
df1 = df1.assign(wr_qant = df1['wr'].rank(method='max', pct=True))
df1 = df1.assign(qant_circle = (df1['g_qant']-center_g/2)**2 + (df1['wr_qant']-center_wr)**2)
df1 = df1.assign(rank = 1 - df1['qant_circle'])
df2 = df1.loc[df1['qant_circle'] <= 0.25]
df_top = df2.sort_values(by=['qant_circle'],ascending=True)[['hero', 'g', 'wr', 'rank']]
df_top.index = range(1, len(df2.index) + 1)
return df_top.head(10)
def topWinrateOverallTiers():
min_g = df['g'].max() / 10
df1 = df.loc[(df['g'] > min_g) & (df['wr'] >= 49)]
df_top3 = df1.sort_values(by=['wr'],ascending=False)
return df_top3.head(10)
from IPython.display import display, HTML
css = """
.output {
flex-direction: row;
}
"""
HTML('<style>{}</style>'.format(css))
import seaborn as sns
cm = sns.light_palette("green", as_cmap=True)
subset = ['rank']
a = tiers(5)
a = a.style.background_gradient('RdYlGn', subset = subset)
a
display(tiers(4)),
display(topHiddenImba_pos(4))
```
|
github_jupyter
|
import pandas as pd
from app import WinrateCollect2
wc = WinrateCollect2("heroes_page_list.csv")
wc.update_stat()
# prepare heros stats
df = pd.read_pickle("data")
df['hero'] = df['hero'].str.replace('%20',' ')
df = df.astype({'g': 'int32'})
df.head()
# prepare heros matchups
df_m = pd.read_pickle("matchups")
df_m['hero'] = df_m['hero'].str.replace('%20',' ')
df_m = df_m.astype({'w': 'int32'})
df_m = df_m.astype({'l': 'int32'})
df_m.head()
def get_matchup(enemy_heroes):
matchup = None
for e in enemy_heroes:
tmp = df_m.loc[(df_m['hero'] == e)][['matchup', 'w', 'l']].set_index('matchup')
if matchup is not None:
matchup = matchup.add(tmp, fill_value=0)
else:
matchup = tmp
matchup = matchup[~matchup.index.isin(enemy_heroes)]
center_g = 1
center_lr = 1
matchup = matchup.assign(g = matchup['w'] + matchup['l'])
matchup = matchup.assign(lr = matchup['l'] / (matchup['w'] + matchup['l']))
matchup = matchup.assign(g_qant = matchup['g'].rank(method='max', pct=True))
matchup = matchup.assign(lr_qant = matchup['lr'].rank(method='max', pct=True))
matchup = matchup.assign(qant_circle = (matchup['g_qant']-center_g)**2 + (matchup['lr_qant']-center_lr)**2)
matchup = matchup.assign(rank = 1 - matchup['qant_circle'])
df_top = matchup.sort_values(by=['qant_circle'],ascending=True)[['g', 'lr']]
df_top = df_top.loc[df_top['lr'] > 0.49]
return df_top
df_tmp = df.sort_values(['hero', 'g'], ascending=[True,False]).drop_duplicates(['hero'])
p1 = list(df_tmp.loc[(df['pos'] == 1)].hero)
p2 = list(df_tmp.loc[(df['pos'] == 2)].hero)
p3 = list(df_tmp.loc[(df['pos'] == 3)].hero)
p4 = list(df_tmp.loc[(df['pos'] == 4)].hero)
p5 = list(df_tmp.loc[(df['pos'] == 5)].hero)
# insert enemy heroes
enemies = ['Ancient Apparition', 'Hoodwink']
m = get_matchup(enemies)
# select pos
m[m.index.isin(p5)]
df_tmp = df.sort_values(['hero', 'g'], ascending=[True,False]).drop_duplicates(['hero'])
df_tmp
p1 = list(df_tmp.loc[(df['pos'] == 1)].hero)
pos = 1
# & (df['wr'] >= 52)
min_g = df.loc[(df['pos'] == pos)]['g'].max() / 100
df1 = df.loc[(df['pos'] == pos) & (df['g'] > min_g) ]
df_top3 = df1.sort_values(by=['wr'],ascending=False)
df_top3
'''
# old version
def tierS(pos):
min_g = df.loc[(df['pos'] == pos)]['g'].max() / 3
df1 = df.loc[(df['pos'] == pos) & (df['g'] > min_g) & (df['wr'] >= 52)]
df_top3 = df1.sort_values(by=['wr'],ascending=False)
return df_top3
def tierA(pos):
min_g = df.loc[(df['pos'] == pos)]['g'].max() / 4
df2 = tierS(pos)
df1 = df.loc[(df['pos'] == pos) & (df['g'] > min_g) & (df['wr'] >= 49)]
df1 = df1.sort_values(by=['wr'],ascending=False).head(14)
df_top8 = pd.concat([df1, df2]).drop_duplicates(keep=False)
return df_top8.head(11)
def topWR():
min_g = df['g'].max() / 40
df_top3 = df.sort_values(by=['wr'],ascending=False)
return df_top3.head(10)
def topTier():
min_g = df['g'].max() / 10
df1 = df.loc[(df['g'] > min_g) & (df['wr'] >= 49)]
df_top3 = df1.sort_values(by=['wr'],ascending=False)
return df_top3.head(15)
'''
# make games count less valuable
#center_g = 0.9
# make win rate less valuable
#center_wr = 0.9
center_g = 1
center_wr = 1
#.style.hide_index()
def tierS(pos):
# 5% - outliers
min_g = df.loc[(df['pos'] == pos)]['g'].max() / 20
df1 = df.loc[(df['pos'] == pos) & (df['g'] >= min_g)]
df1 = df1.assign(g_qant = df1['g'].rank(method='max', pct=True))
df1 = df1.assign(wr_qant = df1['wr'].rank(method='max', pct=True))
df1 = df1.assign(qant_circle = (df1['g_qant']-center_g)**2 + (df1['wr_qant']-center_wr)**2)
df1 = df1.assign(rank = 1 - df1['qant_circle'])
df2 = df1.loc[df1['qant_circle'] <= 0.0625]
df_top = df2.sort_values(by=['qant_circle'],ascending=True)[['hero', 'g', 'wr', 'rank']]
return df_top
def tierA(pos):
# 5% - outliers
min_g = df.loc[(df['pos'] == pos)]['g'].max() / 20
df1 = df.loc[(df['pos'] == pos) & (df['g'] >= min_g)]
df1 = df1.assign(g_qant = df1['g'].rank(method='max', pct=True))
df1 = df1.assign(wr_qant = df1['wr'].rank(method='max', pct=True))
df1 = df1.assign(qant_circle = (df1['g_qant']-center_g)**2 + (df1['wr_qant']-center_wr)**2)
df1 = df1.assign(rank = 1 - df1['qant_circle'])
df2 = df1.loc[(df1['qant_circle'] <= 0.25) & (df1['qant_circle'] > 0.0625)]
df_top = df2.sort_values(by=['qant_circle'],ascending=True)[['hero', 'g', 'wr', 'rank']]
return df_top
def tierB_plus(pos):
# 5% - outliers
min_g = df.loc[(df['pos'] == pos)]['g'].max() / 20
df1 = df.loc[(df['pos'] == pos) & (df['g'] >= min_g)]
df1 = df1.assign(g_qant = df1['g'].rank(method='max', pct=True))
df1 = df1.assign(wr_qant = df1['wr'].rank(method='max', pct=True))
df1 = df1.assign(qant_circle = (df1['g_qant']-center_g)**2 + (df1['wr_qant']-center_wr)**2)
df1 = df1.assign(rank = 1 - df1['qant_circle'])
df2 = df1.loc[(df1['qant_circle'] > 0.25)]
df_top = df2.sort_values(by=['qant_circle'],ascending=True)[['hero', 'g', 'wr', 'rank']]
return df_top
def topWR_lowpick():
df_top3 = df.sort_values(by=['wr'],ascending=False)
return df_top3.head(10)
def topHiddenImba_pos(pos):
# 5% - outliers
min_g = df.loc[(df['pos'] == pos)]['g'].max() / 20
df1 = df.loc[(df['pos'] == pos) & (df['g'] >= min_g)]
df1 = df1.assign(g_qant = df1['g'].rank(method='max', pct=True))
df1 = df1.assign(wr_qant = df1['wr'].rank(method='max', pct=True))
df1 = df1.assign(qant_circle = (df1['g_qant']-center_g/2)**2 + (df1['wr_qant']-center_wr)**2)
df1 = df1.assign(rank = 1 - df1['qant_circle'])
df2 = df1.loc[df1['qant_circle'] <= 0.25]
df_top = df2.sort_values(by=['qant_circle'],ascending=True)[['hero', 'g', 'wr', 'rank']]
return df_top.head(3)
def topWinrateOverallTiers():
min_g = df['g'].max() / 10
df1 = df.loc[(df['g'] > min_g) & (df['wr'] >= 49)]
df_top3 = df1.sort_values(by=['wr'],ascending=False)
return df_top3.head(10)
topWinrateOverallTiers()
topWR_lowpick()
tierS(1)
tierA(1)
tierB_plus(1)
tierS(2)
tierA(2)
tierS(3)
tierA(3)
tierS(4)
tierA(4)
topHiddenImba_pos(4)
tierS(5)
tierA(5)
topHiddenImba_pos(5)
import os
from datetime import datetime
fold = 'history/'+ datetime.today().strftime('%Y_%m_%d__%Hh_%Mm')
os.mkdir(fold)
import dataframe_image as dfi
tierS(1).dfi.export(fold + '/1_TierS.png')
tierA(1).dfi.export(fold + '/1_TierA.png')
tierB_plus(1).dfi.export(fold + '/1_TierBplus.png')
topWinrateOverallTiers().dfi.export(fold + '/topWinrate.png')
topWR_lowpick().dfi.export(fold + '/topLowpick.png')
tierS(2).dfi.export(fold + '/2_TierS.png')
tierA(2).dfi.export(fold + '/2_TierA.png')
tierS(3).dfi.export(fold + '/3_TierS.png')
tierA(3).dfi.export(fold + '/3_TierA.png')
tierS(4).dfi.export(fold + '/4_TierS.png')
tierA(4).dfi.export(fold + '/4_TierA.png')
topHiddenImba_pos(4).dfi.export(fold + '/4_HiddenImba.png')
tierS(5).dfi.export(fold + '/5_TierS.png')
tierA(5).dfi.export(fold + '/5_TierA.png')
topHiddenImba_pos(5).dfi.export(fold + '/5_HiddenImba.png')
from IPython.display import display, HTML
css = """
.output {
flex-direction: row;
}
"""
HTML('<style>{}</style>'.format(css))
display(tierS(1))
display(tierA(1))
#display(tierB_plus(1))
df1 = df.loc[(df['pos'] == 1)]
df1 = df1.sort_values(by=['g'],ascending=False)
df1
df1.describe()
# make games count less valuable
#center_g = 0.9
# make win rate less valuable
#center_wr = 0.9
center_g = 1
center_wr = 1
#.style.hide_index()
def tiers(pos):
# 5% - outliers
min_g = df.loc[(df['pos'] == pos)]['g'].max() / 20
df1 = df.loc[(df['pos'] == pos) & (df['g'] >= min_g)]
df1 = df1.assign(g_qant = df1['g'].rank(method='max', pct=True))
df1 = df1.assign(wr_qant = df1['wr'].rank(method='max', pct=True))
df1 = df1.assign(qant_circle = (df1['g_qant']-center_g)**2 + (df1['wr_qant']-center_wr)**2)
df1 = df1.assign(rank = 1 - df1['qant_circle'])
df1['tier'] = ''
df1['tier'] = df1['qant_circle'].apply(lambda x: 'S' if x <= 0.0625 else
('A' if (x <= 0.25) & (x > 0.0625) else
('B' if (x <= 0.5625) & (x > 0.25) else 'C+')))
df_top = df1.sort_values(by=['qant_circle'],ascending=True)[['hero', 'g', 'wr', 'rank', 'tier']]
df_top.index = range(1, len(df1.index) + 1)
return df_top
def topWR_lowpick():
df_top3 = df.sort_values(by=['wr'],ascending=False)
return df_top3.head(10)
def topHiddenImba_pos(pos):
# 5% - outliers
min_g = df.loc[(df['pos'] == pos)]['g'].max() / 20
df1 = df.loc[(df['pos'] == pos) & (df['g'] >= min_g)]
df1 = df1.assign(g_qant = df1['g'].rank(method='max', pct=True))
df1 = df1.assign(wr_qant = df1['wr'].rank(method='max', pct=True))
df1 = df1.assign(qant_circle = (df1['g_qant']-center_g/2)**2 + (df1['wr_qant']-center_wr)**2)
df1 = df1.assign(rank = 1 - df1['qant_circle'])
df2 = df1.loc[df1['qant_circle'] <= 0.25]
df_top = df2.sort_values(by=['qant_circle'],ascending=True)[['hero', 'g', 'wr', 'rank']]
df_top.index = range(1, len(df2.index) + 1)
return df_top.head(10)
def topWinrateOverallTiers():
min_g = df['g'].max() / 10
df1 = df.loc[(df['g'] > min_g) & (df['wr'] >= 49)]
df_top3 = df1.sort_values(by=['wr'],ascending=False)
return df_top3.head(10)
from IPython.display import display, HTML
css = """
.output {
flex-direction: row;
}
"""
HTML('<style>{}</style>'.format(css))
import seaborn as sns
cm = sns.light_palette("green", as_cmap=True)
subset = ['rank']
a = tiers(5)
a = a.style.background_gradient('RdYlGn', subset = subset)
a
display(tiers(4)),
display(topHiddenImba_pos(4))
| 0.284576 | 0.259911 |
```
import numpy as np
%matplotlib inline
import matplotlib.pyplot as plt
import matplotlib.image as mimg
import os
BASE = '/Users/mchrusci/uj/shaper_data/adversarial/fixed'
def load(path):
adv_samples_path = os.path.join(BASE, path)
with np.load(adv_samples_path) as adv_samples:
X = adv_samples['X']
Y = adv_samples['Y']
pred = adv_samples['pred']
prob = adv_samples['prob']
return X, Y, pred, prob
def load_drawings(path):
with np.load(path) as data:
X = data['drawings']
Y = data['Y']
return X, Y
def plot_idxs(X, Y, pred, prob, targeted, idxs, name):
title = 'target = %d' if targeted else 'true = %d'
fig = plt.figure(figsize=(10,3))
fig.suptitle('%s adversarial samples' % name, fontsize=12)
def plot_one_sample(i):
plt.subplot(1, 5, i + 1)
plt.title(title % Y[idxs[i]] + '\npred = %d (%.2f)' % (pred[idxs[i]], prob[idxs[i]]))
plt.axis('off')
plt.imshow(X[idxs[i]].reshape(28, 28), cmap='gray')
for i in range(len(idxs)):
plot_one_sample(i)
plt.show()
def plot_random(X, Y, pred, prob, targeted):
idxs = np.random.randint(0, X.shape[0], 5)
plot_idxs(X, Y, pred, prob, targeted, idxs, 'Random')
def plot_random_drawings(X, n=None, accs=None):
idxs = np.random.randint(0, X.shape[0], 5)
fig = plt.figure(figsize=(10,3))
if n is None and accs is None:
pass
else:
fig.suptitle('Drawned mnist, n = %d, accurracy = %f' % (n, accs[n-1]), fontsize=12)
def plot_one_sample(i):
plt.subplot(1, 5, i + 1)
plt.axis('off')
plt.imshow(X[idxs[i]].reshape(28, 28), cmap='gray')
for i in range(len(idxs)):
plot_one_sample(i)
plt.show()
def plot_best(X, Y, pred, prob, targeted):
success_samples_idx = np.where(pred == Y) if targeted else np.where(pred != Y)
probs_sorted = prob.argsort()[::-1]
idxs = []
for i in probs_sorted:
if i in success_samples_idx[0]:
idxs.append(i)
if len(idxs) == 5:
break
plot_idxs(X, Y, pred, prob, targeted, idxs, 'Best')
def plot_drawing_defense(path, n):
seed = np.random.randint(0, 1000)
X, _ = load_drawings(os.path.join(BASE, path.split('.npz')[0] + f'-redrawned-{n}.npz'))
np.random.seed(seed)
plot_random_drawings(X)
def plot_defense_accs(accs, adv_acc, method):
plt.plot(accs)
plt.axhline(y=adv_acc, linestyle='--', c='C1')
plt.xlabel('n')
plt.ylabel('adversarial success')
plt.legend(['redrawn attack', 'attack'])
plt.title(method)
plt.show()
```
# Testowany model
accuracy = 0.9914
Model A (3,382,346 parameters): Conv(64, 5, 5) + Relu, Conv(64, 5, 5) + Relu,
Dropout(0.25), FC(128) + Relu, Dropout(0.5), FC + Softmax
### Accuracy na rysunkach
```
accs = [0.3267, 0.4862, 0.5953, 0.6875, 0.7613, 0.8129, 0.8528, 0.8829, 0.9054, 0.9241, 0.9355, 0.945, 0.9524, 0.9585, 0.9638, 0.9667, 0.9695, 0.9709, 0.9727, 0.9748, 0.9759, 0.9775, 0.9787, 0.9784, 0.98, 0.9794, 0.9809, 0.9817, 0.982, 0.9812, 0.9819, 0.9822, 0.9823, 0.9826, 0.9833, 0.9834, 0.9837, 0.9838, 0.9841, 0.9844, 0.9841, 0.9844, 0.9847, 0.9849, 0.9858, 0.9856, 0.9857, 0.9862, 0.9858, 0.9856, 0.9857, 0.9858, 0.9858, 0.986, 0.9859, 0.9856, 0.986, 0.9863, 0.9865, 0.9864, 0.9862, 0.9865, 0.9863, 0.9864, 0.9862, 0.9863, 0.9868, 0.9866, 0.9872, 0.9872, 0.9874, 0.9871, 0.9867, 0.9867, 0.9866, 0.9866, 0.9867, 0.9868, 0.9871, 0.9869, 0.9868, 0.9872, 0.9872, 0.9873, 0.9872, 0.9874, 0.987, 0.987, 0.9871, 0.9873, 0.9872, 0.9872, 0.9872, 0.9873, 0.987, 0.9869, 0.987, 0.987, 0.9871, 0.9871]
plt.plot(accs)
plt.xlabel('n')
plt.ylabel('accuracy')
plt.axhline(y=0.9914, linestyle='--', c='C1')
plt.legend(['drawings', 'originals'])
plt.show()
X, _ = load_drawings('/Users/mchrusci/uj/shaper_data/adversarial/drawned-mnist/test_drawings_npz/test-mnist-n-5.npz')
plot_random_drawings(X, 5, accs)
X, _ = load_drawings('/Users/mchrusci/uj/shaper_data/adversarial/drawned-mnist/test_drawings_npz/test-mnist-n-10.npz')
plot_random_drawings(X, 10, accs)
X, _ = load_drawings('/Users/mchrusci/uj/shaper_data/adversarial/drawned-mnist/test_drawings_npz/test-mnist-n-20.npz')
plot_random_drawings(X, 20, accs)
X, _ = load_drawings('/Users/mchrusci/uj/shaper_data/adversarial/drawned-mnist/test_drawings_npz/test-mnist-n-50.npz')
plot_random_drawings(X, 50, accs)
X, _ = load_drawings('/Users/mchrusci/uj/shaper_data/adversarial/drawned-mnist/test_drawings_npz/test-mnist-n-100.npz')
plot_random_drawings(X, 100, accs)
```
# Testowane ataki
norm = linf, eps = 0.3
* baseline
* DoM
* DoM-T
* Random
* transfer
* **FGS**
* **IFGS**
* Rand_FGS
* **Carlini**
### DoM
adversarial success before = 39.33%
average l2 perturbation = 5.63
```
X, Y, pred, prob = load('examples-to-draw/baseline-norm-linf-alpha-0.0-targeted-0-adv-samples.npz')
plot_random(X=X, Y=Y, pred=pred, prob=prob, targeted=False)
plot_best(X=X, Y=Y, pred=pred, prob=prob, targeted=False)
```
redrawned adversarial samples:
```
plot_drawing_defense('baseline-dom/baseline-norm-linf-alpha-0.0-targeted-0-adv-samples', n=100)
```
adversarial success after defense:
```
accs = [0.895, 0.8932, 0.8888, 0.8841, 0.8806, 0.8788, 0.8702, 0.8677, 0.8632, 0.8548, 0.8488, 0.8445, 0.8414, 0.8383, 0.8337, 0.8292, 0.8278, 0.826, 0.8221, 0.8187, 0.8147, 0.8129, 0.8088, 0.8025, 0.7971, 0.7899, 0.7882, 0.7853, 0.7768, 0.775, 0.7731, 0.7689, 0.7665, 0.7598, 0.7536, 0.7502, 0.746, 0.7456, 0.7394, 0.7386, 0.7354, 0.7324, 0.7294, 0.7267, 0.7262, 0.7221, 0.7192, 0.7169, 0.7164, 0.7135, 0.7126, 0.7105, 0.7059, 0.7038, 0.7017, 0.6995, 0.6979, 0.6959, 0.6953, 0.696, 0.6925, 0.6908, 0.6883, 0.6841, 0.6798, 0.6779, 0.6766, 0.6761, 0.6725, 0.6701, 0.6689, 0.6674, 0.6653, 0.6646, 0.6621, 0.6621, 0.6622, 0.6597, 0.6583, 0.6564, 0.6546, 0.6528, 0.6507, 0.648, 0.648, 0.6453, 0.6418, 0.6413, 0.6396, 0.6389, 0.6361, 0.6355, 0.6346, 0.6352, 0.6348, 0.6316, 0.6276, 0.6284, 0.6278, 0.6275]
plot_defense_accs(accs=accs, adv_acc=0.3933, method='DoM')
```
### DoM-T
adversarial success = 13.59%
average l2 perturbation = 5.56
```
X, Y, pred, prob = load('baseline-norm-linf-alpha-0.0-targeted-1-adv-samples.npz')
plot_random(X=X, Y=Y, pred=pred, prob=prob, targeted=True)
plot_best(X=X, Y=Y, pred=pred, prob=prob, targeted=True)
```
redrawned adversarial samples:
```
plot_drawing_defense('baseline-dom-targeted/baseline-norm-linf-alpha-0.0-targeted-1-adv-samples', n=100)
```
adversarial success after defense:
```
accs = [0.1521, 0.1807, 0.2014, 0.2183, 0.2281, 0.2417, 0.2527, 0.2562, 0.2608, 0.2617, 0.27, 0.272, 0.2746, 0.2796, 0.2751, 0.2751, 0.2773, 0.2759, 0.2789, 0.2768, 0.2776, 0.2749, 0.2752, 0.2739, 0.2725, 0.2732, 0.274, 0.2763, 0.277, 0.2755, 0.2728, 0.2725, 0.2686, 0.2687, 0.2633, 0.2627, 0.2604, 0.2616, 0.2605, 0.2593, 0.2575, 0.2562, 0.2578, 0.2561, 0.2538, 0.2518, 0.2508, 0.2519, 0.2492, 0.2499, 0.2496, 0.2498, 0.2479, 0.2485, 0.2475, 0.2467, 0.2478, 0.2461, 0.2438, 0.2428, 0.2428, 0.2421, 0.2416, 0.2395, 0.2377, 0.2366, 0.235, 0.2335, 0.2351, 0.2346, 0.236, 0.2356, 0.2338, 0.2336, 0.2338, 0.2343, 0.2319, 0.232, 0.2296, 0.2305, 0.2294, 0.2312, 0.2297, 0.2309, 0.2294, 0.2277, 0.2264, 0.2249, 0.2249, 0.2243, 0.2237, 0.2234, 0.2222, 0.2207, 0.2207, 0.2197, 0.2209, 0.2202, 0.2198, 0.2184]
plot_defense_accs(accs=accs, adv_acc=0.1359, method='DoM - targeted')
```
### Random
adversarial success = 7.59%
average l2 perturbation = 6.12
```
X, Y, pred, prob = load('baseline-norm-linf-alpha-0.6-targeted-0-adv-samples.npz')
plot_random(X=X, Y=Y, pred=pred, prob=prob, targeted=False)
plot_best(X=X, Y=Y, pred=pred, prob=prob, targeted=False)
```
redrawned adversarial samples:
```
plot_drawing_defense('baseline-random/baseline-norm-linf-alpha-0.6-targeted-0-adv-samples', n=50)
```
adversarial success after defense:
```
accs = [0.831, 0.7762, 0.7252, 0.6803, 0.6414, 0.6072, 0.5673, 0.5405, 0.5132, 0.4822, 0.461, 0.4358, 0.4162, 0.4025, 0.3849, 0.3719, 0.3608, 0.3449, 0.3351, 0.3261, 0.3158, 0.3065, 0.297, 0.2907, 0.2802, 0.273, 0.2668, 0.2604, 0.2573, 0.2518, 0.2446, 0.241, 0.2398, 0.2352, 0.2285, 0.2244, 0.2197, 0.2155, 0.2128, 0.2095, 0.2053, 0.2021, 0.1995, 0.1956, 0.1935, 0.1913, 0.1857, 0.1843, 0.1841, 0.1827, 0.1812, 0.1804, 0.1781, 0.1756, 0.1744, 0.173, 0.1706, 0.17, 0.1686, 0.1664, 0.1661, 0.1639, 0.1632, 0.1627, 0.1618, 0.1609, 0.1593, 0.1574, 0.1588, 0.1583, 0.1574, 0.1569, 0.1559, 0.156, 0.1541, 0.153, 0.1543, 0.1517, 0.15, 0.149, 0.1485, 0.1487, 0.1474, 0.1472, 0.1477, 0.1471, 0.1475, 0.1447, 0.1443, 0.1422, 0.1422, 0.1414, 0.1409, 0.1389, 0.1376, 0.1375, 0.1363, 0.1348, 0.1338, 0.1349]
plot_defense_accs(accs=accs, adv_acc=0.0759, method='Random')
```
### FGS
modelB $\rightarrow$ modelB: 89.0%
modelB $\rightarrow$ modelA: 66.3%
```
X, Y, pred, prob = load('fgs.npz')
plot_random(X=X, Y=Y, pred=pred, prob=prob, targeted=False)
plot_best(X=X, Y=Y, pred=pred, prob=prob, targeted=False)
```
redrawned adversarial samples:
```
plot_drawing_defense('fgs/fgs.npz', n=40)
```
adversarial success after defense:
```
accs = [0.8529, 0.8203, 0.7954, 0.7721, 0.7481, 0.7319, 0.7159, 0.6987, 0.6888, 0.6734, 0.6671, 0.6549, 0.6494, 0.647, 0.6374, 0.6331, 0.6292, 0.6217, 0.6158, 0.6129, 0.6075, 0.6031, 0.5997, 0.5973, 0.5942, 0.5949, 0.5939, 0.5881, 0.5877, 0.586, 0.5819, 0.582, 0.5797, 0.5754, 0.5742, 0.5715, 0.5726, 0.5728, 0.5717, 0.5707, 0.5714, 0.5703, 0.5695, 0.5696, 0.5687, 0.5692, 0.5693, 0.5711, 0.5676, 0.5672, 0.5673, 0.5682, 0.568, 0.5678, 0.5687, 0.5653, 0.5661, 0.5683, 0.5704, 0.5679, 0.5696, 0.5689, 0.5698, 0.5712, 0.5716, 0.5713, 0.5708, 0.5708, 0.5712, 0.5722, 0.5714, 0.5712, 0.5717, 0.572, 0.5715, 0.5703, 0.5695, 0.5697, 0.5712, 0.5695, 0.571, 0.5704, 0.5692, 0.5696, 0.5675, 0.5698, 0.5682, 0.5688, 0.5666, 0.5658, 0.567, 0.5656, 0.5683, 0.5685, 0.5675, 0.567, 0.5687, 0.5688, 0.5677, 0.5709]
plot_defense_accs(accs=accs, adv_acc=0.663, method='FGS')
```
### IFGS
modelB $\rightarrow$ modelB: 99.7%
modelB $\rightarrow$ modelA: 86.3%
```
X, Y, pred, prob = load('ifgs.npz')
plot_random(X=X, Y=Y, pred=pred, prob=prob, targeted=False)
plot_best(X=X, Y=Y, pred=pred, prob=prob, targeted=False)
```
redrawned adversarial samples:
```
plot_drawing_defense('ifgs/ifgs.npz', n=30)
```
adversarial success after defense:
```
accs = [0.8456, 0.8034, 0.7687, 0.7349, 0.7062, 0.6844, 0.6608, 0.6482, 0.635, 0.6165, 0.6072, 0.596, 0.595, 0.5848, 0.5712, 0.564, 0.5624, 0.5615, 0.559, 0.5542, 0.5508, 0.5474, 0.5459, 0.5474, 0.546, 0.5448, 0.5448, 0.5499, 0.5506, 0.5521, 0.5487, 0.5464, 0.5501, 0.5514, 0.5495, 0.5513, 0.5526, 0.5535, 0.5548, 0.5561, 0.5604, 0.561, 0.563, 0.5667, 0.566, 0.5657, 0.5673, 0.5697, 0.5705, 0.5721, 0.5724, 0.576, 0.5754, 0.5779, 0.578, 0.5783, 0.5797, 0.584, 0.5844, 0.5832, 0.5855, 0.5855, 0.5853, 0.587, 0.5865, 0.5886, 0.5908, 0.593, 0.5941, 0.594, 0.5959, 0.5954, 0.5971, 0.599, 0.599, 0.6025, 0.603, 0.6055, 0.6061, 0.6073, 0.6084, 0.609, 0.6097, 0.611, 0.6125, 0.6117, 0.6123, 0.6142, 0.6162, 0.6176, 0.6169, 0.6167, 0.6164, 0.6169, 0.6192, 0.6188, 0.6214, 0.6233, 0.6246, 0.6265]
plot_defense_accs(accs=accs, adv_acc=0.863, method='IFGS')
```
### RAND_FGS
modelB $\rightarrow$ modelB: 88.6%
modelB $\rightarrow$ modelA: 63.4%
```
X, Y, pred, prob = load('rand_fgs.npz')
plot_random(X=X, Y=Y, pred=pred, prob=prob, targeted=False)
plot_best(X=X, Y=Y, pred=pred, prob=prob, targeted=False)
```
redrawned adversarial samples:
```
plot_drawing_defense('rand_fgs/rand_fgs.npz', n=20)
```
adversarial success after defense:
```
accs = [0.8545, 0.8178, 0.7915, 0.767, 0.7427, 0.7191, 0.7018, 0.6817, 0.6682, 0.6558, 0.645, 0.6312, 0.6238, 0.6166, 0.6042, 0.5999, 0.5945, 0.5875, 0.5769, 0.5765, 0.5697, 0.5677, 0.5657, 0.5613, 0.5589, 0.553, 0.5488, 0.5463, 0.5434, 0.5398, 0.5384, 0.5378, 0.5374, 0.5363, 0.5364, 0.5357, 0.5322, 0.5322, 0.5296, 0.5306, 0.5298, 0.5265, 0.5246, 0.5245, 0.5246, 0.5264, 0.5241, 0.5232, 0.521, 0.5202, 0.5199, 0.5197, 0.5197, 0.5185, 0.5197, 0.5199, 0.5202, 0.5212, 0.5221, 0.5206, 0.5187, 0.5192, 0.5203, 0.519, 0.5192, 0.5167, 0.5151, 0.5181, 0.5189, 0.5183, 0.5188, 0.5169, 0.5172, 0.518, 0.5179, 0.519, 0.5164, 0.5147, 0.5159, 0.5158, 0.5161, 0.5169, 0.519, 0.5178, 0.5172, 0.519, 0.5209, 0.5189, 0.5164, 0.518, 0.5194, 0.5202, 0.521, 0.5191, 0.5206, 0.5184, 0.5188, 0.5185, 0.5193, 0.5206]
plot_defense_accs(accs=accs, adv_acc=0.634, method='RAND_FGS')
```
### Carlini
modelB $\rightarrow$ modelB: 100%
modelB $\rightarrow$ modelA: 85.9%
```
X, Y, pred, prob = load('CW.npz')
plot_random(X=X, Y=Y, pred=pred, prob=prob, targeted=False)
plot_best(X=X, Y=Y, pred=pred, prob=prob, targeted=False)
```
redrawned adversarial samples:
```
plot_drawing_defense('CW/CW.npz', n=20)
```
adversarial success after defense:
```
accs = [0.842, 0.792, 0.766, 0.714, 0.708, 0.672, 0.654, 0.636, 0.613, 0.601, 0.594, 0.594, 0.57, 0.56, 0.555, 0.549, 0.561, 0.526, 0.531, 0.525, 0.532, 0.53, 0.526, 0.526, 0.523, 0.536, 0.532, 0.528, 0.527, 0.533, 0.53, 0.526, 0.53, 0.531, 0.527, 0.527, 0.528, 0.531, 0.54, 0.542, 0.536, 0.541, 0.552, 0.553, 0.559, 0.558, 0.564, 0.557, 0.568, 0.56, 0.555, 0.557, 0.555, 0.556, 0.559, 0.55, 0.555, 0.55, 0.551, 0.552, 0.557, 0.56, 0.561, 0.569, 0.567, 0.566, 0.573, 0.579, 0.585, 0.582, 0.575, 0.58, 0.59, 0.589, 0.592, 0.588, 0.593, 0.591, 0.589, 0.589, 0.586, 0.585, 0.577, 0.58, 0.584, 0.582, 0.582, 0.588, 0.594, 0.592, 0.591, 0.596, 0.6, 0.603, 0.602, 0.604, 0.606, 0.606, 0.605, 0.609]
plot_defense_accs(accs=accs, adv_acc=0.859, method='Carlini')
```
|
github_jupyter
|
import numpy as np
%matplotlib inline
import matplotlib.pyplot as plt
import matplotlib.image as mimg
import os
BASE = '/Users/mchrusci/uj/shaper_data/adversarial/fixed'
def load(path):
adv_samples_path = os.path.join(BASE, path)
with np.load(adv_samples_path) as adv_samples:
X = adv_samples['X']
Y = adv_samples['Y']
pred = adv_samples['pred']
prob = adv_samples['prob']
return X, Y, pred, prob
def load_drawings(path):
with np.load(path) as data:
X = data['drawings']
Y = data['Y']
return X, Y
def plot_idxs(X, Y, pred, prob, targeted, idxs, name):
title = 'target = %d' if targeted else 'true = %d'
fig = plt.figure(figsize=(10,3))
fig.suptitle('%s adversarial samples' % name, fontsize=12)
def plot_one_sample(i):
plt.subplot(1, 5, i + 1)
plt.title(title % Y[idxs[i]] + '\npred = %d (%.2f)' % (pred[idxs[i]], prob[idxs[i]]))
plt.axis('off')
plt.imshow(X[idxs[i]].reshape(28, 28), cmap='gray')
for i in range(len(idxs)):
plot_one_sample(i)
plt.show()
def plot_random(X, Y, pred, prob, targeted):
idxs = np.random.randint(0, X.shape[0], 5)
plot_idxs(X, Y, pred, prob, targeted, idxs, 'Random')
def plot_random_drawings(X, n=None, accs=None):
idxs = np.random.randint(0, X.shape[0], 5)
fig = plt.figure(figsize=(10,3))
if n is None and accs is None:
pass
else:
fig.suptitle('Drawned mnist, n = %d, accurracy = %f' % (n, accs[n-1]), fontsize=12)
def plot_one_sample(i):
plt.subplot(1, 5, i + 1)
plt.axis('off')
plt.imshow(X[idxs[i]].reshape(28, 28), cmap='gray')
for i in range(len(idxs)):
plot_one_sample(i)
plt.show()
def plot_best(X, Y, pred, prob, targeted):
success_samples_idx = np.where(pred == Y) if targeted else np.where(pred != Y)
probs_sorted = prob.argsort()[::-1]
idxs = []
for i in probs_sorted:
if i in success_samples_idx[0]:
idxs.append(i)
if len(idxs) == 5:
break
plot_idxs(X, Y, pred, prob, targeted, idxs, 'Best')
def plot_drawing_defense(path, n):
seed = np.random.randint(0, 1000)
X, _ = load_drawings(os.path.join(BASE, path.split('.npz')[0] + f'-redrawned-{n}.npz'))
np.random.seed(seed)
plot_random_drawings(X)
def plot_defense_accs(accs, adv_acc, method):
plt.plot(accs)
plt.axhline(y=adv_acc, linestyle='--', c='C1')
plt.xlabel('n')
plt.ylabel('adversarial success')
plt.legend(['redrawn attack', 'attack'])
plt.title(method)
plt.show()
accs = [0.3267, 0.4862, 0.5953, 0.6875, 0.7613, 0.8129, 0.8528, 0.8829, 0.9054, 0.9241, 0.9355, 0.945, 0.9524, 0.9585, 0.9638, 0.9667, 0.9695, 0.9709, 0.9727, 0.9748, 0.9759, 0.9775, 0.9787, 0.9784, 0.98, 0.9794, 0.9809, 0.9817, 0.982, 0.9812, 0.9819, 0.9822, 0.9823, 0.9826, 0.9833, 0.9834, 0.9837, 0.9838, 0.9841, 0.9844, 0.9841, 0.9844, 0.9847, 0.9849, 0.9858, 0.9856, 0.9857, 0.9862, 0.9858, 0.9856, 0.9857, 0.9858, 0.9858, 0.986, 0.9859, 0.9856, 0.986, 0.9863, 0.9865, 0.9864, 0.9862, 0.9865, 0.9863, 0.9864, 0.9862, 0.9863, 0.9868, 0.9866, 0.9872, 0.9872, 0.9874, 0.9871, 0.9867, 0.9867, 0.9866, 0.9866, 0.9867, 0.9868, 0.9871, 0.9869, 0.9868, 0.9872, 0.9872, 0.9873, 0.9872, 0.9874, 0.987, 0.987, 0.9871, 0.9873, 0.9872, 0.9872, 0.9872, 0.9873, 0.987, 0.9869, 0.987, 0.987, 0.9871, 0.9871]
plt.plot(accs)
plt.xlabel('n')
plt.ylabel('accuracy')
plt.axhline(y=0.9914, linestyle='--', c='C1')
plt.legend(['drawings', 'originals'])
plt.show()
X, _ = load_drawings('/Users/mchrusci/uj/shaper_data/adversarial/drawned-mnist/test_drawings_npz/test-mnist-n-5.npz')
plot_random_drawings(X, 5, accs)
X, _ = load_drawings('/Users/mchrusci/uj/shaper_data/adversarial/drawned-mnist/test_drawings_npz/test-mnist-n-10.npz')
plot_random_drawings(X, 10, accs)
X, _ = load_drawings('/Users/mchrusci/uj/shaper_data/adversarial/drawned-mnist/test_drawings_npz/test-mnist-n-20.npz')
plot_random_drawings(X, 20, accs)
X, _ = load_drawings('/Users/mchrusci/uj/shaper_data/adversarial/drawned-mnist/test_drawings_npz/test-mnist-n-50.npz')
plot_random_drawings(X, 50, accs)
X, _ = load_drawings('/Users/mchrusci/uj/shaper_data/adversarial/drawned-mnist/test_drawings_npz/test-mnist-n-100.npz')
plot_random_drawings(X, 100, accs)
X, Y, pred, prob = load('examples-to-draw/baseline-norm-linf-alpha-0.0-targeted-0-adv-samples.npz')
plot_random(X=X, Y=Y, pred=pred, prob=prob, targeted=False)
plot_best(X=X, Y=Y, pred=pred, prob=prob, targeted=False)
plot_drawing_defense('baseline-dom/baseline-norm-linf-alpha-0.0-targeted-0-adv-samples', n=100)
accs = [0.895, 0.8932, 0.8888, 0.8841, 0.8806, 0.8788, 0.8702, 0.8677, 0.8632, 0.8548, 0.8488, 0.8445, 0.8414, 0.8383, 0.8337, 0.8292, 0.8278, 0.826, 0.8221, 0.8187, 0.8147, 0.8129, 0.8088, 0.8025, 0.7971, 0.7899, 0.7882, 0.7853, 0.7768, 0.775, 0.7731, 0.7689, 0.7665, 0.7598, 0.7536, 0.7502, 0.746, 0.7456, 0.7394, 0.7386, 0.7354, 0.7324, 0.7294, 0.7267, 0.7262, 0.7221, 0.7192, 0.7169, 0.7164, 0.7135, 0.7126, 0.7105, 0.7059, 0.7038, 0.7017, 0.6995, 0.6979, 0.6959, 0.6953, 0.696, 0.6925, 0.6908, 0.6883, 0.6841, 0.6798, 0.6779, 0.6766, 0.6761, 0.6725, 0.6701, 0.6689, 0.6674, 0.6653, 0.6646, 0.6621, 0.6621, 0.6622, 0.6597, 0.6583, 0.6564, 0.6546, 0.6528, 0.6507, 0.648, 0.648, 0.6453, 0.6418, 0.6413, 0.6396, 0.6389, 0.6361, 0.6355, 0.6346, 0.6352, 0.6348, 0.6316, 0.6276, 0.6284, 0.6278, 0.6275]
plot_defense_accs(accs=accs, adv_acc=0.3933, method='DoM')
X, Y, pred, prob = load('baseline-norm-linf-alpha-0.0-targeted-1-adv-samples.npz')
plot_random(X=X, Y=Y, pred=pred, prob=prob, targeted=True)
plot_best(X=X, Y=Y, pred=pred, prob=prob, targeted=True)
plot_drawing_defense('baseline-dom-targeted/baseline-norm-linf-alpha-0.0-targeted-1-adv-samples', n=100)
accs = [0.1521, 0.1807, 0.2014, 0.2183, 0.2281, 0.2417, 0.2527, 0.2562, 0.2608, 0.2617, 0.27, 0.272, 0.2746, 0.2796, 0.2751, 0.2751, 0.2773, 0.2759, 0.2789, 0.2768, 0.2776, 0.2749, 0.2752, 0.2739, 0.2725, 0.2732, 0.274, 0.2763, 0.277, 0.2755, 0.2728, 0.2725, 0.2686, 0.2687, 0.2633, 0.2627, 0.2604, 0.2616, 0.2605, 0.2593, 0.2575, 0.2562, 0.2578, 0.2561, 0.2538, 0.2518, 0.2508, 0.2519, 0.2492, 0.2499, 0.2496, 0.2498, 0.2479, 0.2485, 0.2475, 0.2467, 0.2478, 0.2461, 0.2438, 0.2428, 0.2428, 0.2421, 0.2416, 0.2395, 0.2377, 0.2366, 0.235, 0.2335, 0.2351, 0.2346, 0.236, 0.2356, 0.2338, 0.2336, 0.2338, 0.2343, 0.2319, 0.232, 0.2296, 0.2305, 0.2294, 0.2312, 0.2297, 0.2309, 0.2294, 0.2277, 0.2264, 0.2249, 0.2249, 0.2243, 0.2237, 0.2234, 0.2222, 0.2207, 0.2207, 0.2197, 0.2209, 0.2202, 0.2198, 0.2184]
plot_defense_accs(accs=accs, adv_acc=0.1359, method='DoM - targeted')
X, Y, pred, prob = load('baseline-norm-linf-alpha-0.6-targeted-0-adv-samples.npz')
plot_random(X=X, Y=Y, pred=pred, prob=prob, targeted=False)
plot_best(X=X, Y=Y, pred=pred, prob=prob, targeted=False)
plot_drawing_defense('baseline-random/baseline-norm-linf-alpha-0.6-targeted-0-adv-samples', n=50)
accs = [0.831, 0.7762, 0.7252, 0.6803, 0.6414, 0.6072, 0.5673, 0.5405, 0.5132, 0.4822, 0.461, 0.4358, 0.4162, 0.4025, 0.3849, 0.3719, 0.3608, 0.3449, 0.3351, 0.3261, 0.3158, 0.3065, 0.297, 0.2907, 0.2802, 0.273, 0.2668, 0.2604, 0.2573, 0.2518, 0.2446, 0.241, 0.2398, 0.2352, 0.2285, 0.2244, 0.2197, 0.2155, 0.2128, 0.2095, 0.2053, 0.2021, 0.1995, 0.1956, 0.1935, 0.1913, 0.1857, 0.1843, 0.1841, 0.1827, 0.1812, 0.1804, 0.1781, 0.1756, 0.1744, 0.173, 0.1706, 0.17, 0.1686, 0.1664, 0.1661, 0.1639, 0.1632, 0.1627, 0.1618, 0.1609, 0.1593, 0.1574, 0.1588, 0.1583, 0.1574, 0.1569, 0.1559, 0.156, 0.1541, 0.153, 0.1543, 0.1517, 0.15, 0.149, 0.1485, 0.1487, 0.1474, 0.1472, 0.1477, 0.1471, 0.1475, 0.1447, 0.1443, 0.1422, 0.1422, 0.1414, 0.1409, 0.1389, 0.1376, 0.1375, 0.1363, 0.1348, 0.1338, 0.1349]
plot_defense_accs(accs=accs, adv_acc=0.0759, method='Random')
X, Y, pred, prob = load('fgs.npz')
plot_random(X=X, Y=Y, pred=pred, prob=prob, targeted=False)
plot_best(X=X, Y=Y, pred=pred, prob=prob, targeted=False)
plot_drawing_defense('fgs/fgs.npz', n=40)
accs = [0.8529, 0.8203, 0.7954, 0.7721, 0.7481, 0.7319, 0.7159, 0.6987, 0.6888, 0.6734, 0.6671, 0.6549, 0.6494, 0.647, 0.6374, 0.6331, 0.6292, 0.6217, 0.6158, 0.6129, 0.6075, 0.6031, 0.5997, 0.5973, 0.5942, 0.5949, 0.5939, 0.5881, 0.5877, 0.586, 0.5819, 0.582, 0.5797, 0.5754, 0.5742, 0.5715, 0.5726, 0.5728, 0.5717, 0.5707, 0.5714, 0.5703, 0.5695, 0.5696, 0.5687, 0.5692, 0.5693, 0.5711, 0.5676, 0.5672, 0.5673, 0.5682, 0.568, 0.5678, 0.5687, 0.5653, 0.5661, 0.5683, 0.5704, 0.5679, 0.5696, 0.5689, 0.5698, 0.5712, 0.5716, 0.5713, 0.5708, 0.5708, 0.5712, 0.5722, 0.5714, 0.5712, 0.5717, 0.572, 0.5715, 0.5703, 0.5695, 0.5697, 0.5712, 0.5695, 0.571, 0.5704, 0.5692, 0.5696, 0.5675, 0.5698, 0.5682, 0.5688, 0.5666, 0.5658, 0.567, 0.5656, 0.5683, 0.5685, 0.5675, 0.567, 0.5687, 0.5688, 0.5677, 0.5709]
plot_defense_accs(accs=accs, adv_acc=0.663, method='FGS')
X, Y, pred, prob = load('ifgs.npz')
plot_random(X=X, Y=Y, pred=pred, prob=prob, targeted=False)
plot_best(X=X, Y=Y, pred=pred, prob=prob, targeted=False)
plot_drawing_defense('ifgs/ifgs.npz', n=30)
accs = [0.8456, 0.8034, 0.7687, 0.7349, 0.7062, 0.6844, 0.6608, 0.6482, 0.635, 0.6165, 0.6072, 0.596, 0.595, 0.5848, 0.5712, 0.564, 0.5624, 0.5615, 0.559, 0.5542, 0.5508, 0.5474, 0.5459, 0.5474, 0.546, 0.5448, 0.5448, 0.5499, 0.5506, 0.5521, 0.5487, 0.5464, 0.5501, 0.5514, 0.5495, 0.5513, 0.5526, 0.5535, 0.5548, 0.5561, 0.5604, 0.561, 0.563, 0.5667, 0.566, 0.5657, 0.5673, 0.5697, 0.5705, 0.5721, 0.5724, 0.576, 0.5754, 0.5779, 0.578, 0.5783, 0.5797, 0.584, 0.5844, 0.5832, 0.5855, 0.5855, 0.5853, 0.587, 0.5865, 0.5886, 0.5908, 0.593, 0.5941, 0.594, 0.5959, 0.5954, 0.5971, 0.599, 0.599, 0.6025, 0.603, 0.6055, 0.6061, 0.6073, 0.6084, 0.609, 0.6097, 0.611, 0.6125, 0.6117, 0.6123, 0.6142, 0.6162, 0.6176, 0.6169, 0.6167, 0.6164, 0.6169, 0.6192, 0.6188, 0.6214, 0.6233, 0.6246, 0.6265]
plot_defense_accs(accs=accs, adv_acc=0.863, method='IFGS')
X, Y, pred, prob = load('rand_fgs.npz')
plot_random(X=X, Y=Y, pred=pred, prob=prob, targeted=False)
plot_best(X=X, Y=Y, pred=pred, prob=prob, targeted=False)
plot_drawing_defense('rand_fgs/rand_fgs.npz', n=20)
accs = [0.8545, 0.8178, 0.7915, 0.767, 0.7427, 0.7191, 0.7018, 0.6817, 0.6682, 0.6558, 0.645, 0.6312, 0.6238, 0.6166, 0.6042, 0.5999, 0.5945, 0.5875, 0.5769, 0.5765, 0.5697, 0.5677, 0.5657, 0.5613, 0.5589, 0.553, 0.5488, 0.5463, 0.5434, 0.5398, 0.5384, 0.5378, 0.5374, 0.5363, 0.5364, 0.5357, 0.5322, 0.5322, 0.5296, 0.5306, 0.5298, 0.5265, 0.5246, 0.5245, 0.5246, 0.5264, 0.5241, 0.5232, 0.521, 0.5202, 0.5199, 0.5197, 0.5197, 0.5185, 0.5197, 0.5199, 0.5202, 0.5212, 0.5221, 0.5206, 0.5187, 0.5192, 0.5203, 0.519, 0.5192, 0.5167, 0.5151, 0.5181, 0.5189, 0.5183, 0.5188, 0.5169, 0.5172, 0.518, 0.5179, 0.519, 0.5164, 0.5147, 0.5159, 0.5158, 0.5161, 0.5169, 0.519, 0.5178, 0.5172, 0.519, 0.5209, 0.5189, 0.5164, 0.518, 0.5194, 0.5202, 0.521, 0.5191, 0.5206, 0.5184, 0.5188, 0.5185, 0.5193, 0.5206]
plot_defense_accs(accs=accs, adv_acc=0.634, method='RAND_FGS')
X, Y, pred, prob = load('CW.npz')
plot_random(X=X, Y=Y, pred=pred, prob=prob, targeted=False)
plot_best(X=X, Y=Y, pred=pred, prob=prob, targeted=False)
plot_drawing_defense('CW/CW.npz', n=20)
accs = [0.842, 0.792, 0.766, 0.714, 0.708, 0.672, 0.654, 0.636, 0.613, 0.601, 0.594, 0.594, 0.57, 0.56, 0.555, 0.549, 0.561, 0.526, 0.531, 0.525, 0.532, 0.53, 0.526, 0.526, 0.523, 0.536, 0.532, 0.528, 0.527, 0.533, 0.53, 0.526, 0.53, 0.531, 0.527, 0.527, 0.528, 0.531, 0.54, 0.542, 0.536, 0.541, 0.552, 0.553, 0.559, 0.558, 0.564, 0.557, 0.568, 0.56, 0.555, 0.557, 0.555, 0.556, 0.559, 0.55, 0.555, 0.55, 0.551, 0.552, 0.557, 0.56, 0.561, 0.569, 0.567, 0.566, 0.573, 0.579, 0.585, 0.582, 0.575, 0.58, 0.59, 0.589, 0.592, 0.588, 0.593, 0.591, 0.589, 0.589, 0.586, 0.585, 0.577, 0.58, 0.584, 0.582, 0.582, 0.588, 0.594, 0.592, 0.591, 0.596, 0.6, 0.603, 0.602, 0.604, 0.606, 0.606, 0.605, 0.609]
plot_defense_accs(accs=accs, adv_acc=0.859, method='Carlini')
| 0.211173 | 0.732059 |
# PROJECT DEFINITION
## Project Overview/Problem Statement
The project is to compare the performance of an autoencoder with one-class SVM model in detecting fraud in credit card transactions. The data has been anonymized due to privacy reason and it is publicly available here
https://www.kaggle.com/mlg-ulb/creditcardfraud
## Metrics
The metrics used to compare performance are **accuracy**, **recall** and **precision**. More emphasize will be given on recall in order to capture as many fraudulent cases as possible.
# DATA ANALYSIS AND EXPLORATION
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split, RandomizedSearchCV
from sklearn.svm import OneClassSVM
from sklearn.metrics import accuracy_score, precision_score, recall_score, f1_score, confusion_matrix, precision_recall_curve
df = pd.read_csv('creditcard.csv')
df.shape
display(df)
total_null = df.isnull().sum().sum()
print("Total null in the dataset is: {}".format(total_null))
df.groupby('Class')['Class'].count()
class_1 = df['Class'].mean()
class_0 = 1 - class_1
labels = 'Class 0', 'Class 1'
sizes = [class_0, class_1]
fig1, ax1 = plt.subplots()
ax1.pie(sizes, labels=labels, autopct='%1.4f%%', startangle=315)
ax1.axis('equal')
plt.show()
plt.scatter(df[df.Class==1]['Time'],df[df.Class==1]['Amount']);
plt.title("Fraud Amount vs Time");
plt.scatter(df[df.Class==0]['Time'],df[df.Class==0]['Amount']);
plt.title("Normal Amount vs Time");
plt.hist(df[df.Class==1]['Time']);
plt.title('Histogram of Time for Fraudulent case');
plt.hist(df[df.Class==0]['Time']);
plt.title('Histogram of Time for Normal case');
plt.hist(df[df.Class==1]['Amount'], bins=20);
plt.title('Histogram of Amount for Fraudulent case');
plt.xlabel('Amount');
plt.ylabel('Number of Transactions');
plt.hist(df[df.Class==0]['Amount'], bins=50);
plt.title('Histogram of Amount for Normal case');
plt.yscale('log')
plt.xlabel('Amount');
plt.ylabel('Number of Transactions (log)');
```
### Remarks:
- There are 31 columns in the dataframe with 28 PCA components, Time, Amount and Class.
- Based on the histograms above, the Amount does differ between fraudulent and normal transactions.
- Based on the scatter plots and histograms above, Time does not really matter in identifying Fraud. Time will not be included in the predictive modelling part.
- From the pie chart above, the dataset is highly inbalance with 492 frauds out of 284807 transactions.
- There is no nulls in the dataset, so further cleaning is not necessary.
# ONE-CLASS SVM
```
# Preparing data for modelling
def prep_data(df):
'''
INPUT:
df - input DataFrame
OUTPUT:
X_train - training input
X_test - testing input
y_test - testing ouput
'''
# Dropping Time as it does not matter to fraud detection
df = df.drop(['Time'], axis=1)
X_train, X_test = train_test_split(df, test_size=0.2, random_state=66)
X_train = X_train[X_train.Class == 0]
y_train = X_train.Class
X_train = X_train.drop(['Class'], axis=1)
y_test = X_test.Class
X_test = X_test.drop(['Class'], axis=1)
X_train = X_train.values
X_test = X_test.values
return X_train, X_test, y_test
df = pd.read_csv('creditcard.csv')
X_train, X_test, y_test = prep_data(df)
len(X_train)
# Training and testing the model
model = OneClassSVM(gamma='auto', nu=0.05)
model.fit(X_train[:50000]) #Not all train data is used because of long training time
y_pred = model.predict(X_test)
# y_pred = y_pred.apply(lambda x: 1 if x == -1 else 0)
for i in range(len(y_pred)):
y_pred[i] = 1 if y_pred[i] == -1 else 0
def print_metrics(actual, prediction):
'''
INPUT:
actual - expected output
prediction - predictted output
'''
accuracy = accuracy_score(actual, prediction)
recall = recall_score(actual, prediction)
precision = precision_score(actual, prediction)
f1 = f1_score(actual, prediction)
print('The accuracy score of the model = {}'.format(accuracy))
print('The recall score of the model = {}'.format(recall))
print('The precision score of the model = {}'.format(precision))
print('The f1 score of the model = {}'.format(f1))
print('\nConfusion Matrix:')
print(confusion_matrix(actual, prediction))
print_metrics(y_test, y_pred)
```
### Remarks:
- Not all train data is used to fit the one-class SVM model because it is taking a long time to complete.
- Bacause the dataset is highly unbalanced, the metric used to measure model performance is recall i.e., we would like to detect as many frauds as possible.
- Although recall is quite high (86%), the precision is very low i.e., we have wrongly identified 5542 normal transactions as fraudulent.
# AUTOENCODER
```
from sklearn.preprocessing import StandardScaler
from keras.models import Model, load_model
from keras.layers import Input, Dense
from keras.callbacks import ModelCheckpoint
# Preparing data for modelling
df = pd.read_csv('creditcard.csv')
df['Amount'] = StandardScaler().fit_transform(df['Amount'].values.reshape(-1, 1))
X_train, X_test, y_test = prep_data(df)
input_dim = X_train.shape[1]
input_layer = Input(shape=(input_dim, ))
encoder = Dense(32, activation="tanh")(input_layer)
encoder = Dense(16, activation="relu")(encoder)
decoder = Dense(16, activation='tanh')(encoder)
decoder = Dense(input_dim, activation='relu')(decoder)
autoencoder = Model(inputs=input_layer, outputs=decoder)
autoencoder.compile(optimizer='adam', loss='mean_squared_error', metrics=['accuracy'])
filepath = "autoencoder_model.h5"
checkpoint = ModelCheckpoint(filepath, monitor='val_acc', verbose=1, save_best_only=True)
history = autoencoder.fit(X_train, X_train, epochs=100, batch_size=32, shuffle=True,
validation_data=(X_test, X_test), callbacks=[checkpoint], verbose=1)
pd.DataFrame(history.history).to_csv('history.csv')
log = pd.read_csv('history.csv')
log.head()
plt.plot(log['loss'])
plt.plot(log['val_loss'])
plt.title('model loss')
plt.ylabel('loss')
plt.xlabel('epoch')
plt.legend(['train', 'test'], loc='upper right');
plt.plot(log['acc'])
plt.plot(log['val_acc'])
plt.title('model accuracy')
plt.ylabel('accuracy')
plt.xlabel('epoch')
plt.legend(['train', 'test'], loc='upper right');
```
### Remarks:
- An autoencoder reconstructs input signal as its output. Hence, the reconstruction error of the two classes, normal and fraud is to be compared in order to determine suitable threshold to detect a fraud.
```
autoencoder = load_model('autoencoder_model.h5')
pred = autoencoder.predict(X_test)
mse = np.mean(np.power(X_test - pred, 2), axis=1)
error_df = pd.DataFrame({'Reconstruction_Error': mse, 'Class': y_test})
display(error_df)
error_df_fraud = error_df[error_df.Class == 1]['Reconstruction_Error']
error_df_normal = error_df[error_df.Class == 0]['Reconstruction_Error']
error_df_fraud.describe()
error_df_normal.describe()
```
### Remarks:
- From the error distribution, all values for normal are lower than of for fraud which is intuitively correct, except for the max. The value for max is higher probably because of the highly unbalanced dataset.
- We have to select a threshold to make prediction i.e., whenever the error is bigger than the threshold, the transaction will be considered as a fraud.
```
threshold = 3
error_df['Prediction'] = error_df.Reconstruction_Error > threshold
error_df.Prediction = error_df.Prediction.apply(lambda x: 1 if x is True else 0)
print_metrics(error_df.Class, error_df.Prediction)
```
### Remarks:
- Although the recall of the autoencoder is lower than that of one-class SVM, the other metrics are higher.
- Since the metrics of the autoencoder depend on the threshold of the reconstruction error, let's see some plots.
```
precision, recall, threshold_error = precision_recall_curve(error_df.Class, error_df.Reconstruction_Error)
plt.plot(recall, precision, 'r')
plt.title('Precision vs. Recall')
plt.xlabel('Recall')
plt.ylabel('Precision')
plt.show()
```
### Remarks:
- Ideally we would like to have high precision and high recall.
```
plt.plot(threshold_error, recall[1:], 'b')
plt.plot(threshold_error, precision[1:], 'r')
plt.title('Precision and Recall vs. Error threshold')
plt.xlabel('Error Threshold')
plt.ylabel('Score')
plt.legend(['recall', 'precision'], loc='upper right');
plt.show()
```
### Remarks:
- We can see from the plot above that precision and recall depend on the error threshold.
- As the threshold increases, the recall decreases and precision increases.
- At a very low threshold, although recall is high, precision is low.
- At a very high threshold, although precision is high, recall is low.
### Can we make prediction using z-score of the error instead of the threshold?
```
fraud_mean = error_df_fraud.mean()
fraud_std = error_df_fraud.std()
normal_mean = error_df_normal.mean()
normal_std = error_df_normal.std()
error_df['fraud_z_score'] = (error_df.Reconstruction_Error - fraud_mean)/fraud_std
error_df['normal_z_score'] = (error_df.Reconstruction_Error - normal_mean)/normal_std
# It is considered a fraud when its z-score is smaller then normal's z-score
error_df['Prediction_by_z_score'] = np.abs(error_df.fraud_z_score) < np.abs(error_df.normal_z_score)
error_df.Prediction_by_z_score = error_df.Prediction_by_z_score.apply(lambda x: 1 if x is True else 0)
error_df
print_metrics(error_df.Class, error_df.Prediction_by_z_score)
```
### Remarks:
- The prediction scores obtained when using z-score are similar with the scores obtained using error threshold with threshold value equals 3.
- Using z-score is more systematic than manually selecting the threshold.
# RESULTS
```
results = {'Model': ['one-class SVM', 'autoencoder (threshold)', 'autoencoder (z-score)'],
'Accuracy': [90.2, 98.0, 97.7], 'Recall': [86.3, 82.1, 83.2], 'Precision': [1.5, 6.6, 5.8], 'F1-score': [2.9, 12.3, 10.9]}
results_df = pd.DataFrame(data=results)
results_df
```
# DISCUSSION
- From the table above, although autoencoders perform better than one-class SVM except for Recall score, their performance in general are similar.
- To make predictions, One-class SVM is more straightforward than antoencoder to use. This is because an autoencoder reconstructs the input as its output, hence decision has to be made how to use the reconstruction error to make predictions.
- Comparing the two antoencoder models, their performance is quite similar. It is interesting that the method using z-score of the reconstruction error has similar performance with manually selected threshold error method. This would eliminate the need for experimenting with the threshold values for making predictions.
- Note that not all data is used to train the one-class SVM model due to high computing power required whereas the entire training data was used to train the neural networks. It would be interesting to see if the performance of the one-class SVM would be different if we use all training data.
# CONCLUSION
## Reflection
We have explored credit card transactions data and use it to develop predictive model to detect fraudulent transactions. Two models have been investigated namely one-class SVM and antoencoder. The performance of the models have been compared and discussed. The relationship of Precision against Recall for the autoencoder and their dependencies on the reconstruction errors have been investigated.
The most interesting point from this project is that z-score of the reconstruction error can be used to make prediction for the autoencoder with similar performance compared with the method using a carefully and manually selected error threshold.
The most difficult part is getting both recall and precision scores high. I wonder if this is possible at all given the highly unbalanced dataset.
## Improvement
To improve the way to make predictions for the autoencoder, a method using z-score of the reconstruction errors has been implemented and discussed throughout the notebook.
|
github_jupyter
|
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split, RandomizedSearchCV
from sklearn.svm import OneClassSVM
from sklearn.metrics import accuracy_score, precision_score, recall_score, f1_score, confusion_matrix, precision_recall_curve
df = pd.read_csv('creditcard.csv')
df.shape
display(df)
total_null = df.isnull().sum().sum()
print("Total null in the dataset is: {}".format(total_null))
df.groupby('Class')['Class'].count()
class_1 = df['Class'].mean()
class_0 = 1 - class_1
labels = 'Class 0', 'Class 1'
sizes = [class_0, class_1]
fig1, ax1 = plt.subplots()
ax1.pie(sizes, labels=labels, autopct='%1.4f%%', startangle=315)
ax1.axis('equal')
plt.show()
plt.scatter(df[df.Class==1]['Time'],df[df.Class==1]['Amount']);
plt.title("Fraud Amount vs Time");
plt.scatter(df[df.Class==0]['Time'],df[df.Class==0]['Amount']);
plt.title("Normal Amount vs Time");
plt.hist(df[df.Class==1]['Time']);
plt.title('Histogram of Time for Fraudulent case');
plt.hist(df[df.Class==0]['Time']);
plt.title('Histogram of Time for Normal case');
plt.hist(df[df.Class==1]['Amount'], bins=20);
plt.title('Histogram of Amount for Fraudulent case');
plt.xlabel('Amount');
plt.ylabel('Number of Transactions');
plt.hist(df[df.Class==0]['Amount'], bins=50);
plt.title('Histogram of Amount for Normal case');
plt.yscale('log')
plt.xlabel('Amount');
plt.ylabel('Number of Transactions (log)');
# Preparing data for modelling
def prep_data(df):
'''
INPUT:
df - input DataFrame
OUTPUT:
X_train - training input
X_test - testing input
y_test - testing ouput
'''
# Dropping Time as it does not matter to fraud detection
df = df.drop(['Time'], axis=1)
X_train, X_test = train_test_split(df, test_size=0.2, random_state=66)
X_train = X_train[X_train.Class == 0]
y_train = X_train.Class
X_train = X_train.drop(['Class'], axis=1)
y_test = X_test.Class
X_test = X_test.drop(['Class'], axis=1)
X_train = X_train.values
X_test = X_test.values
return X_train, X_test, y_test
df = pd.read_csv('creditcard.csv')
X_train, X_test, y_test = prep_data(df)
len(X_train)
# Training and testing the model
model = OneClassSVM(gamma='auto', nu=0.05)
model.fit(X_train[:50000]) #Not all train data is used because of long training time
y_pred = model.predict(X_test)
# y_pred = y_pred.apply(lambda x: 1 if x == -1 else 0)
for i in range(len(y_pred)):
y_pred[i] = 1 if y_pred[i] == -1 else 0
def print_metrics(actual, prediction):
'''
INPUT:
actual - expected output
prediction - predictted output
'''
accuracy = accuracy_score(actual, prediction)
recall = recall_score(actual, prediction)
precision = precision_score(actual, prediction)
f1 = f1_score(actual, prediction)
print('The accuracy score of the model = {}'.format(accuracy))
print('The recall score of the model = {}'.format(recall))
print('The precision score of the model = {}'.format(precision))
print('The f1 score of the model = {}'.format(f1))
print('\nConfusion Matrix:')
print(confusion_matrix(actual, prediction))
print_metrics(y_test, y_pred)
from sklearn.preprocessing import StandardScaler
from keras.models import Model, load_model
from keras.layers import Input, Dense
from keras.callbacks import ModelCheckpoint
# Preparing data for modelling
df = pd.read_csv('creditcard.csv')
df['Amount'] = StandardScaler().fit_transform(df['Amount'].values.reshape(-1, 1))
X_train, X_test, y_test = prep_data(df)
input_dim = X_train.shape[1]
input_layer = Input(shape=(input_dim, ))
encoder = Dense(32, activation="tanh")(input_layer)
encoder = Dense(16, activation="relu")(encoder)
decoder = Dense(16, activation='tanh')(encoder)
decoder = Dense(input_dim, activation='relu')(decoder)
autoencoder = Model(inputs=input_layer, outputs=decoder)
autoencoder.compile(optimizer='adam', loss='mean_squared_error', metrics=['accuracy'])
filepath = "autoencoder_model.h5"
checkpoint = ModelCheckpoint(filepath, monitor='val_acc', verbose=1, save_best_only=True)
history = autoencoder.fit(X_train, X_train, epochs=100, batch_size=32, shuffle=True,
validation_data=(X_test, X_test), callbacks=[checkpoint], verbose=1)
pd.DataFrame(history.history).to_csv('history.csv')
log = pd.read_csv('history.csv')
log.head()
plt.plot(log['loss'])
plt.plot(log['val_loss'])
plt.title('model loss')
plt.ylabel('loss')
plt.xlabel('epoch')
plt.legend(['train', 'test'], loc='upper right');
plt.plot(log['acc'])
plt.plot(log['val_acc'])
plt.title('model accuracy')
plt.ylabel('accuracy')
plt.xlabel('epoch')
plt.legend(['train', 'test'], loc='upper right');
autoencoder = load_model('autoencoder_model.h5')
pred = autoencoder.predict(X_test)
mse = np.mean(np.power(X_test - pred, 2), axis=1)
error_df = pd.DataFrame({'Reconstruction_Error': mse, 'Class': y_test})
display(error_df)
error_df_fraud = error_df[error_df.Class == 1]['Reconstruction_Error']
error_df_normal = error_df[error_df.Class == 0]['Reconstruction_Error']
error_df_fraud.describe()
error_df_normal.describe()
threshold = 3
error_df['Prediction'] = error_df.Reconstruction_Error > threshold
error_df.Prediction = error_df.Prediction.apply(lambda x: 1 if x is True else 0)
print_metrics(error_df.Class, error_df.Prediction)
precision, recall, threshold_error = precision_recall_curve(error_df.Class, error_df.Reconstruction_Error)
plt.plot(recall, precision, 'r')
plt.title('Precision vs. Recall')
plt.xlabel('Recall')
plt.ylabel('Precision')
plt.show()
plt.plot(threshold_error, recall[1:], 'b')
plt.plot(threshold_error, precision[1:], 'r')
plt.title('Precision and Recall vs. Error threshold')
plt.xlabel('Error Threshold')
plt.ylabel('Score')
plt.legend(['recall', 'precision'], loc='upper right');
plt.show()
fraud_mean = error_df_fraud.mean()
fraud_std = error_df_fraud.std()
normal_mean = error_df_normal.mean()
normal_std = error_df_normal.std()
error_df['fraud_z_score'] = (error_df.Reconstruction_Error - fraud_mean)/fraud_std
error_df['normal_z_score'] = (error_df.Reconstruction_Error - normal_mean)/normal_std
# It is considered a fraud when its z-score is smaller then normal's z-score
error_df['Prediction_by_z_score'] = np.abs(error_df.fraud_z_score) < np.abs(error_df.normal_z_score)
error_df.Prediction_by_z_score = error_df.Prediction_by_z_score.apply(lambda x: 1 if x is True else 0)
error_df
print_metrics(error_df.Class, error_df.Prediction_by_z_score)
results = {'Model': ['one-class SVM', 'autoencoder (threshold)', 'autoencoder (z-score)'],
'Accuracy': [90.2, 98.0, 97.7], 'Recall': [86.3, 82.1, 83.2], 'Precision': [1.5, 6.6, 5.8], 'F1-score': [2.9, 12.3, 10.9]}
results_df = pd.DataFrame(data=results)
results_df
| 0.792865 | 0.92637 |
# Mount Drive
```
from google.colab import drive
drive.mount('/content/drive')
!pip install -U -q PyDrive
!pip install httplib2==0.15.0
import os
from pydrive.auth import GoogleAuth
from pydrive.drive import GoogleDrive
from pydrive.files import GoogleDriveFileList
from google.colab import auth
from oauth2client.client import GoogleCredentials
from getpass import getpass
import urllib
# 1. Authenticate and create the PyDrive client.
auth.authenticate_user()
gauth = GoogleAuth()
gauth.credentials = GoogleCredentials.get_application_default()
drive = GoogleDrive(gauth)
# Cloning CLIPPER to access modules.
if 'CLIPPER' not in os.listdir():
cmd_string = 'git clone https://github.com/PAL-ML/CLIPPER.git'
os.system(cmd_string)
```
# Installation
## Install multi label metrics dependencies
```
! pip install scikit-learn==0.24
```
## Install CLIP dependencies
```
import subprocess
CUDA_version = [s for s in subprocess.check_output(["nvcc", "--version"]).decode("UTF-8").split(", ") if s.startswith("release")][0].split(" ")[-1]
print("CUDA version:", CUDA_version)
if CUDA_version == "10.0":
torch_version_suffix = "+cu100"
elif CUDA_version == "10.1":
torch_version_suffix = "+cu101"
elif CUDA_version == "10.2":
torch_version_suffix = ""
else:
torch_version_suffix = "+cu110"
! pip install torch==1.7.1{torch_version_suffix} torchvision==0.8.2{torch_version_suffix} -f https://download.pytorch.org/whl/torch_stable.html ftfy regex
! pip install ftfy regex
! wget https://openaipublic.azureedge.net/clip/bpe_simple_vocab_16e6.txt.gz -O bpe_simple_vocab_16e6.txt.gz
!pip install git+https://github.com/Sri-vatsa/CLIP # using this fork because of visualization capabilities
```
## Install clustering dependencies
```
!pip -q install umap-learn>=0.3.7
```
## Install dataset manager dependencies
```
!pip install wget
```
# Imports
```
# ML Libraries
import tensorflow as tf
import tensorflow_hub as hub
import torch
import torch.nn as nn
import torchvision.models as models
import torchvision.transforms as transforms
import keras
# Data processing
import PIL
import base64
import imageio
import pandas as pd
import numpy as np
import json
from PIL import Image
import cv2
import imgaug.augmenters as iaa
# Plotting
import seaborn as sns
import matplotlib.pyplot as plt
from IPython.core.display import display, HTML
from matplotlib import cm
# Models
import clip
# Datasets
import tensorflow_datasets as tfds
# Misc
import progressbar
import logging
from abc import ABC, abstractmethod
import time
import urllib.request
import os
import itertools
from tqdm import tqdm
# Modules
from CLIPPER.code.ExperimentModules import embedding_models
from CLIPPER.code.ExperimentModules import simclr_data_augmentations
from CLIPPER.code.ExperimentModules.dataset_manager import DatasetManager
from CLIPPER.code.ExperimentModules.clip_few_shot import CLIPFewShotClassifier
from CLIPPER.code.ExperimentModules.utils import (save_npy, load_npy,
get_folder_id,
create_expt_dir,
save_to_drive,
load_all_from_drive_folder,
download_file_by_name,
delete_file_by_name)
logging.getLogger('googleapicliet.discovery_cache').setLevel(logging.ERROR)
```
# Initialization & Constants
**Edited**
```
dataset_name = 'ImagenetSketch'
folder_name = "ImagenetSketch-Embeddings-28-02-21"
# Change parentid to match that of experiments root folder in gdrive
parentid = '1bK72W-Um20EQDEyChNhNJthUNbmoSEjD'
# Filepaths
# train_labels_filename = "train_labels.npz"
val_labels_filename = "val_labels.npz"
# train_embeddings_filename_suffix = "_embeddings_train.npz"
val_embeddings_filename_suffix = "_embeddings_val.npz"
# Initialize sepcific experiment folder in drive
folderid = create_expt_dir(drive, parentid, folder_name)
```
# Load data
```
def get_ndarray_from_drive(drive, folderid, filename):
download_file_by_name(drive, folderid, filename)
return np.load(filename)['data']
# train_labels = get_ndarray_from_drive(drive, folderid, train_labels_filename)
val_labels = get_ndarray_from_drive(drive, folderid, val_labels_filename)
# test_labels = get_ndarray_from_drive(drive, folderid, test_labels_filename)
dm = DatasetManager()
test_data_generator = dm.load_dataset('imagenet_sketch', split='val')
classes = dm.get_class_names()
```
# Label dicts
```
!wget https://s3.amazonaws.com/deep-learning-models/image-models/imagenet_class_index.json
with open("imagenet_class_index.json") as f:
class_idx = json.load(f)
idx2label_dict = {str(k): class_idx[str(k)][1] for k in range(len(class_idx))}
class2id_dict = {class_idx[str(k)][0]: (str(k), class_idx[str(k)][1]) for k in range(len(class_idx))}
class_names = [class2id_dict[x][1].replace("_", " ") for x in classes]
```
# Create label dictionary
```
unique_labels = np.unique(val_labels)
print(len(unique_labels))
label_dictionary = {la:[] for la in unique_labels}
for i in range(len(val_labels)):
la = val_labels[i]
label_dictionary[la].append(i)
```
# CLIP zero shot eval
## Function definitions
```
def start_progress_bar(bar_len):
widgets = [
' [',
progressbar.Timer(format= 'elapsed time: %(elapsed)s'),
'] ',
progressbar.Bar('*'),' (',
progressbar.ETA(), ') ',
]
pbar = progressbar.ProgressBar(
max_value=bar_len, widgets=widgets
).start()
return pbar
def prepare_indices(
num_ways,
num_shot,
num_eval,
num_episodes,
label_dictionary,
test_labels,
shuffle=False
):
eval_indices = []
train_indices = []
wi_y = []
eval_y = []
label_dictionary = {la:label_dictionary[la] for la in label_dictionary if len(label_dictionary[la]) >= (num_shot+num_eval)}
unique_labels = list(label_dictionary.keys())
pbar = start_progress_bar(num_episodes)
for s in range(num_episodes):
# Setting random seed for replicability
np.random.seed(s)
_train_indices = []
_eval_indices = []
selected_labels = np.random.choice(unique_labels, size=num_ways, replace=False)
for la in selected_labels:
la_indices = label_dictionary[la]
select = np.random.choice(la_indices, size = num_shot+num_eval, replace=False)
tr_idx = list(select[:num_shot])
ev_idx = list(select[num_shot:])
_train_indices = _train_indices + tr_idx
_eval_indices = _eval_indices + ev_idx
if shuffle:
np.random.shuffle(_train_indices)
np.random.shuffle(_eval_indices)
train_indices.append(_train_indices)
eval_indices.append(_eval_indices)
_wi_y = test_labels[_train_indices]
_eval_y = test_labels[_eval_indices]
wi_y.append(_wi_y)
eval_y.append(_eval_y)
pbar.update(s+1)
return train_indices, eval_indices, wi_y, eval_y
def embed_images(
embedding_model,
train_indices,
num_augmentations,
trivial=False
):
def augment_image(image, num_augmentations, trivial):
""" Perform SimCLR augmentations on the image
"""
if np.max(image) > 1:
image = image/255
augmented_images = [image]
# augmentations = iaa.Sequential([
# iaa.Affine(
# translate_percent={'x':(-0.1, 0.1), 'y':(-0.1, 0.1)},
# rotate=(-15, 15),
# shear=(-15, 15),
# ),
# iaa.Fliplr(0.5)
# ])
def _run_filters(image):
width = image.shape[1]
height = image.shape[0]
image_aug = simclr_data_augmentations.random_crop_with_resize(
image,
height,
width
)
image_aug = tf.image.random_flip_left_right(image_aug)
image_aug = simclr_data_augmentations.random_color_jitter(image_aug)
image_aug = simclr_data_augmentations.random_blur(
image_aug,
height,
width
)
image_aug = tf.reshape(image_aug, [image.shape[0], image.shape[1], 3])
image_aug = tf.clip_by_value(image_aug, 0., 1.)
return image_aug.numpy()
for _ in range(num_augmentations):
# aug_image = augmentations(image=image)
if trivial:
aug_image = image
else:
aug_image = _run_filters(image)
augmented_images.append(aug_image)
augmented_images = np.stack(augmented_images)
return augmented_images
embedding_model.load_model()
unique_indices = np.unique(np.array(train_indices))
ds = dm.load_dataset('imagenet_sketch', split='val')
embeddings = []
IMAGE_IDX = 0
pbar = start_progress_bar(unique_indices.size+1)
num_done=0
for idx, item in enumerate(ds):
if idx in unique_indices:
image = item[IMAGE_IDX][0]
if num_augmentations > 0:
aug_images = augment_image(image, num_augmentations, trivial)
else:
aug_images = image
processed_images = embedding_model.preprocess_data(aug_images)
embedding = embedding_model.embed_images(processed_images)
embeddings.append(embedding)
num_done += 1
pbar.update(num_done+1)
if idx == unique_indices[-1]:
break
embeddings = np.stack(embeddings)
return unique_indices, embeddings
def evaluate_model_for_episode(
model,
eval_x,
eval_y,
label_mapping,
filtered_classes,
metrics=['hamming', 'jaccard', 'subset_accuracy', 'ap', 'map', 'c_f1', 'o_f1', 'c_precision', 'o_precision', 'c_recall', 'o_recall', 'top1_accuracy', 'top5_accuracy', 'classwise_accuracy', 'c_accuracy'],
multi_label=True
):
zs_weights = model.zeroshot_classifier(filtered_classes)
logits = model.predict_scores(eval_x, zs_weights).tolist()
pred_y = model.predict_label(eval_x, zs_weights)
pred_y = [label_mapping[l] for l in pred_y]
met = model.evaluate_single_label_metrics(
eval_x, eval_y, label_mapping, zs_weights, metrics
)
return pred_y, met, logits
def get_label_mapping_n_class_names(eval_y, class_names):
label_mapping = {}
unique_labels = np.unique(eval_y)
filtered_classes = [class_names[x] for x in unique_labels]
num_classes = len(unique_labels)
for c in range(num_classes):
label_mapping[c] = unique_labels[c]
return label_mapping, filtered_classes
# chenni change
def run_episode_through_model(
indices_and_embeddings,
train_indices,
eval_indices,
wi_y,
eval_y,
class_names,
num_augmentations=0,
train_epochs=None,
train_batch_size=5,
metrics=['hamming', 'jaccard', 'subset_accuracy', 'ap', 'map', 'c_f1', 'o_f1', 'c_precision', 'o_precision', 'c_recall', 'o_recall', 'top1_accuracy', 'top5_accuracy', 'classwise_accuracy', 'c_accuracy'],
embeddings=None,
multi_label=True
):
metrics_values = {m:[] for m in metrics}
indices_and_embeddings
eval_x = embeddings[eval_indices]
ep_logits = []
label_mapping, filtered_classes = get_label_mapping_n_class_names(eval_y, class_names)
clip_fs_parameters = {
"num_classes": num_ways,
"input_dims": eval_x.shape[-1],
"multi_label": multi_label
}
clip_fs_cls = CLIPFewShotClassifier(clip_fs_parameters)
pred_labels, metrics_values, logits = evaluate_model_for_episode(
clip_fs_cls,
eval_x,
eval_y,
label_mapping,
filtered_classes,
metrics=metrics,
multi_label=False
)
ep_logits = logits
#cc
return metrics_values, ep_logits
def run_evaluations(
indices_and_embeddings,
train_indices,
eval_indices,
wi_y,
eval_y,
num_episodes,
num_ways,
class_names,
verbose=True,
normalize=True,
train_epochs=None,
train_batch_size=5,
metrics=['accuracy', 'ap', 'map', 'c_f1', 'o_f1', 'c_precision', 'o_precision', 'c_recall', 'o_recall', 'top1_accuracy', 'top5_accuracy', 'classwise_accuracy', 'c_accuracy'],
embeddings=None,
num_augmentations=0,
multi_label=True
):
metrics_values = {m:[] for m in metrics}
all_logits = []
if verbose:
pbar = start_progress_bar(num_episodes)
for idx_ep in range(num_episodes):
_train_indices = train_indices[idx_ep]
_eval_indices = eval_indices[idx_ep]
_wi_y = [label for label in wi_y[idx_ep]]
_eval_y = [label for label in eval_y[idx_ep]]
met, ep_logits = run_episode_through_model(
indices_and_embeddings,
_train_indices,
_eval_indices,
_wi_y,
_eval_y,
class_names,
num_augmentations=num_augmentations,
train_epochs=train_epochs,
train_batch_size=train_batch_size,
embeddings=embeddings,
metrics=metrics,
multi_label=multi_label
)
all_logits.append(ep_logits)
for m in metrics:
metrics_values[m].append(met[m])
if verbose:
pbar.update(idx_ep+1)
return metrics_values, all_logits
def get_best_metric(mt, metric_name, optimal='max'):
if optimal=='max':
opt_value = np.max(np.mean(np.array(mt[metric_name]), axis=0))
if optimal=='min':
opt_value = np.min(np.mean(np.array(mt[metric_name]), axis=0))
return opt_value
```
# 5 way 5 shot
## Picking indices
```
num_ways = 5
num_shot = 5
num_eval = 15
shuffle = False
num_episodes = 100
train_indices, eval_indices, wi_y, eval_y = prepare_indices(
num_ways, num_shot, num_eval, num_episodes, label_dictionary, val_labels, shuffle
)
embedding_model = embedding_models.CLIPEmbeddingWrapper()
num_augmentations = 0
trivial=False
indices, embeddings = embed_images(
embedding_model,
train_indices,
num_augmentations,
trivial=trivial
)
```
## CLIP
```
clip_embeddings_test_fn = "clip" + val_embeddings_filename_suffix
clip_embeddings_test = get_ndarray_from_drive(drive, folderid, clip_embeddings_test_fn)
import warnings
warnings.filterwarnings('ignore')
if trivial:
#cc
results_filename = "new_metrics"+dataset_name+"_0t"+str(num_ways)+"w"+str(num_shot)+"s"+str(num_augmentations)+"a_trivial_clip_zs_metrics_with_logits.json"
else:
#cc
results_filename = "new_metrics"+dataset_name+"_0t"+str(num_ways)+"w"+str(num_shot)+"s"+str(num_augmentations)+"a_clip_zs_metrics_with_logits.json"
auth.authenticate_user()
gauth = GoogleAuth()
gauth.credentials = GoogleCredentials.get_application_default()
drive = GoogleDrive(gauth)
download_file_by_name(drive, folderid, results_filename)
if results_filename in os.listdir():
with open(results_filename, 'r') as f:
#cc
json_loaded = json.load(f)
#cc
clip_metrics_over_train_epochs = json_loaded['metrics']
#cc
logits_over_train_epochs = json_loaded["logits"]
else:
clip_metrics_over_train_epochs = []
logits_over_train_epochs = []
train_epochs_arr = [0]
multi_label=False
# metrics_vals = ['hamming', 'jaccard', 'f1_score'] # ['accuracy', 'f1_score']
for idx, train_epochs in enumerate(train_epochs_arr):
if idx < len(clip_metrics_over_train_epochs):
continue
print(train_epochs)
#cc
clip_metrics_thresholds, all_logits = run_evaluations(
(indices, embeddings),
train_indices,
eval_indices,
wi_y,
eval_y,
num_episodes,
num_ways,
class_names,
train_epochs=train_epochs,
num_augmentations=num_augmentations,
embeddings=clip_embeddings_test
)
clip_metrics_over_train_epochs.append(clip_metrics_thresholds)
#cc
logits_over_train_epochs.append(all_logits)
#cc
fin_list = []
#cc the whole for loop
for a1 in wi_y:
fin_a1_list = []
for a2 in a1:
new_val = str(a2)
fin_a1_list.append(new_val)
fin_list.append(fin_a1_list)
with open(results_filename, 'w') as f:
#cc
results = {'metrics': clip_metrics_over_train_epochs,
"logits": logits_over_train_epochs,
"true_labels": fin_list}
json.dump(results, f)
auth.authenticate_user()
gauth = GoogleAuth()
gauth.credentials = GoogleCredentials.get_application_default()
drive = GoogleDrive(gauth)
#delete_file_by_name(drive, folderid, results_filename)
#save_to_drive(drive, folderid, results_filename)
if trivial:
PLOT_DIR = "NewMetrics_clip_zs_Sigmoid_" + dataset_name + "_0t" + str(num_ways) + "w" + str(num_shot) + "s" + str(num_augmentations) + "a_trivial_plots"
else:
PLOT_DIR = "NewMetrics_clip_zs_Sigmoid_" + dataset_name + "_0t" + str(num_ways) + "w" + str(num_shot) + "s" + str(num_augmentations) + "a_plots"
os.mkdir(PLOT_DIR)
# chenni change whole block
all_metrics = ['accuracy', 'ap', 'map', 'c_f1', 'o_f1', 'c_precision', 'o_precision', 'c_recall', 'o_recall', 'top1_accuracy', 'top5_accuracy', 'classwise_accuracy', 'c_accuracy']
final_dict = {}
for ind_metric in all_metrics:
vals = []
final_array = []
for mt in clip_metrics_over_train_epochs:
ret_val = get_best_metric(mt,ind_metric,"max")
vals.append(ret_val)
final_array.append(vals)
final_dict[ind_metric] = final_array
if trivial:
graph_filename = "new_metrics"+dataset_name+"_0t"+str(num_ways)+"w"+str(num_shot)+"s"+str(num_augmentations)+"a_trivial_clip_zs_metrics_graphs.json"
else:
graph_filename = "new_metrics"+dataset_name+"_0t"+str(num_ways)+"w"+str(num_shot)+"s"+str(num_augmentations)+"a_clip_zs_metrics_graphs.json"
with open(graph_filename, 'w') as f:
json.dump(final_dict, f)
auth.authenticate_user()
gauth = GoogleAuth()
gauth.credentials = GoogleCredentials.get_application_default()
drive = GoogleDrive(gauth)
delete_file_by_name(drive, folderid, graph_filename)
save_to_drive(drive, folderid, graph_filename)
zip_dirname = PLOT_DIR + ".zip"
zip_source = PLOT_DIR
! zip -r $zip_dirname $zip_source
auth.authenticate_user()
gauth = GoogleAuth()
gauth.credentials = GoogleCredentials.get_application_default()
drive = GoogleDrive(gauth)
save_to_drive(drive, folderid, zip_dirname)
```
# 20 way 5 shot
## Picking indices
```
num_ways = 20
num_shot = 5
num_eval = 5
shuffle = False
num_episodes = 100
train_indices, eval_indices, wi_y, eval_y = prepare_indices(
num_ways, num_shot, num_eval, num_episodes, label_dictionary, val_labels, shuffle
)
embedding_model = embedding_models.CLIPEmbeddingWrapper()
num_augmentations = 0
trivial=False
indices, embeddings = embed_images(
embedding_model,
train_indices,
num_augmentations,
trivial=trivial
)
```
## CLIP
```
clip_embeddings_test_fn = "clip" + val_embeddings_filename_suffix
clip_embeddings_test = get_ndarray_from_drive(drive, folderid, clip_embeddings_test_fn)
import warnings
warnings.filterwarnings('ignore')
if trivial:
#cc
results_filename = "new_metrics"+dataset_name+"_0t"+str(num_ways)+"w"+str(num_shot)+"s"+str(num_augmentations)+"a_trivial_clip_zs_metrics_with_logits.json"
else:
#cc
results_filename = "new_metrics"+dataset_name+"_0t"+str(num_ways)+"w"+str(num_shot)+"s"+str(num_augmentations)+"a_metrics_clip_zs_with_logits.json"
auth.authenticate_user()
gauth = GoogleAuth()
gauth.credentials = GoogleCredentials.get_application_default()
drive = GoogleDrive(gauth)
download_file_by_name(drive, folderid, results_filename)
if results_filename in os.listdir():
with open(results_filename, 'r') as f:
#cc
json_loaded = json.load(f)
#cc
clip_metrics_over_train_epochs = json_loaded['metrics']
#cc
logits_over_train_epochs = json_loaded["logits"]
else:
clip_metrics_over_train_epochs = []
#cc
logits_over_train_epochs = []
train_epochs_arr = [0]
multi_label=False
# metrics_vals = ['hamming', 'jaccard', 'f1_score'] # ['accuracy', 'f1_score']
for idx, train_epochs in enumerate(train_epochs_arr):
if idx < len(clip_metrics_over_train_epochs):
continue
print(train_epochs)
#cc
clip_metrics_thresholds, all_logits = run_evaluations(
(indices, embeddings),
train_indices,
eval_indices,
wi_y,
eval_y,
num_episodes,
num_ways,
class_names,
train_epochs=train_epochs,
num_augmentations=num_augmentations,
embeddings=clip_embeddings_test
)
clip_metrics_over_train_epochs.append(clip_metrics_thresholds)
#cc
logits_over_train_epochs.append(all_logits)
#cc
fin_list = []
#cc the whole for loop
for a1 in wi_y:
fin_a1_list = []
for a2 in a1:
new_val = str(a2)
fin_a1_list.append(new_val)
fin_list.append(fin_a1_list)
with open(results_filename, 'w') as f:
#cc
results = {'metrics': clip_metrics_over_train_epochs,
"logits": logits_over_train_epochs,
"true_labels": fin_list}
json.dump(results, f)
auth.authenticate_user()
gauth = GoogleAuth()
gauth.credentials = GoogleCredentials.get_application_default()
drive = GoogleDrive(gauth)
delete_file_by_name(drive, folderid, results_filename)
save_to_drive(drive, folderid, results_filename)
if trivial:
PLOT_DIR = "NewMetrics_clip_zs_Sigmoid_" + dataset_name + "_0t" + str(num_ways) + "w" + str(num_shot) + "s" + str(num_augmentations) + "a_trivial_plots"
else:
PLOT_DIR = "NewMetrics_clip_zs_Sigmoid_" + dataset_name + "_0t" + str(num_ways) + "w" + str(num_shot) + "s" + str(num_augmentations) + "a_plots"
os.mkdir(PLOT_DIR)
all_metrics = ['accuracy', 'ap', 'map', 'c_f1', 'o_f1', 'c_precision', 'o_precision', 'c_recall', 'o_recall', 'top1_accuracy', 'top5_accuracy', 'classwise_accuracy', 'c_accuracy']
final_dict = {}
for ind_metric in all_metrics:
vals = []
final_array = []
for mt in clip_metrics_over_train_epochs:
ret_val = get_best_metric(mt,ind_metric,"max")
vals.append(ret_val)
final_array.append(vals)
final_dict[ind_metric] = final_array
if trivial:
graph_filename = "new_metrics"+dataset_name+"_0t"+str(num_ways)+"w"+str(num_shot)+"s"+str(num_augmentations)+"a_trivial_clip_zs_metrics_graphs.json"
else:
graph_filename = "new_metrics"+dataset_name+"_0t"+str(num_ways)+"w"+str(num_shot)+"s"+str(num_augmentations)+"a_clip_zs_metrics_graphs.json"
with open(graph_filename, 'w') as f:
json.dump(final_dict, f)
auth.authenticate_user()
gauth = GoogleAuth()
gauth.credentials = GoogleCredentials.get_application_default()
drive = GoogleDrive(gauth)
delete_file_by_name(drive, folderid, graph_filename)
save_to_drive(drive, folderid, graph_filename)
zip_dirname = PLOT_DIR + ".zip"
zip_source = PLOT_DIR
! zip -r $zip_dirname $zip_source
auth.authenticate_user()
gauth = GoogleAuth()
gauth.credentials = GoogleCredentials.get_application_default()
drive = GoogleDrive(gauth)
save_to_drive(drive, folderid, zip_dirname)
```
# 5 way 1 shot
## Picking indices
```
num_ways = 5
num_shot = 1
num_eval = 19
shuffle = False
num_episodes = 100
train_indices, eval_indices, wi_y, eval_y = prepare_indices(
num_ways, num_shot, num_eval, num_episodes, label_dictionary, val_labels, shuffle
)
embedding_model = embedding_models.CLIPEmbeddingWrapper()
num_augmentations = 0
trivial=False
indices, embeddings = embed_images(
embedding_model,
train_indices,
num_augmentations,
trivial=trivial
)
```
## CLIP
```
clip_embeddings_test_fn = "clip" + val_embeddings_filename_suffix
clip_embeddings_test = get_ndarray_from_drive(drive, folderid, clip_embeddings_test_fn)
import warnings
warnings.filterwarnings('ignore')
if trivial:
#cc
results_filename = "new_metrics"+dataset_name+"_0t"+str(num_ways)+"w"+str(num_shot)+"s"+str(num_augmentations)+"a_trivial_clip_zs_metrics_with_logits.json"
else:
#cc
results_filename = "new_metrics"+dataset_name+"_0t"+str(num_ways)+"w"+str(num_shot)+"s"+str(num_augmentations)+"a_clip_zs_metrics_with_logits.json"
auth.authenticate_user()
gauth = GoogleAuth()
gauth.credentials = GoogleCredentials.get_application_default()
drive = GoogleDrive(gauth)
download_file_by_name(drive, folderid, results_filename)
if results_filename in os.listdir():
with open(results_filename, 'r') as f:
#cc
json_loaded = json.load(f)
#cc
clip_metrics_over_train_epochs = json_loaded['metrics']
#cc
logits_over_train_epochs = json_loaded["logits"]
else:
clip_metrics_over_train_epochs = []
#cc
logits_over_train_epochs = []
train_epochs_arr = [0]
multi_label=False
# metrics_vals = ['hamming', 'jaccard', 'f1_score'] # ['accuracy', 'f1_score']
for idx, train_epochs in enumerate(train_epochs_arr):
if idx < len(clip_metrics_over_train_epochs):
continue
print(train_epochs)
#cc
clip_metrics_thresholds, all_logits = run_evaluations(
(indices, embeddings),
train_indices,
eval_indices,
wi_y,
eval_y,
num_episodes,
num_ways,
class_names,
train_epochs=train_epochs,
num_augmentations=num_augmentations,
embeddings=clip_embeddings_test
)
clip_metrics_over_train_epochs.append(clip_metrics_thresholds)
#cc
logits_over_train_epochs.append(all_logits)
#cc
fin_list = []
#cc the whole for loop
for a1 in wi_y:
fin_a1_list = []
for a2 in a1:
new_val = str(a2)
fin_a1_list.append(new_val)
fin_list.append(fin_a1_list)
with open(results_filename, 'w') as f:
#cc
results = {'metrics': clip_metrics_over_train_epochs,
"logits": logits_over_train_epochs,
"true_labels": fin_list}
json.dump(results, f)
auth.authenticate_user()
gauth = GoogleAuth()
gauth.credentials = GoogleCredentials.get_application_default()
drive = GoogleDrive(gauth)
delete_file_by_name(drive, folderid, results_filename)
save_to_drive(drive, folderid, results_filename)
if trivial:
PLOT_DIR = "NewMetrics_clip_zs_Sigmoid_" + dataset_name + "_0t" + str(num_ways) + "w" + str(num_shot) + "s" + str(num_augmentations) + "a_trivial_plots"
else:
PLOT_DIR = "NewMetrics_clip_zs_Sigmoid_" + dataset_name + "_0t" + str(num_ways) + "w" + str(num_shot) + "s" + str(num_augmentations) + "a_plots"
os.mkdir(PLOT_DIR)
all_metrics = ['accuracy', 'ap', 'map', 'c_f1', 'o_f1', 'c_precision', 'o_precision', 'c_recall', 'o_recall', 'top1_accuracy', 'top5_accuracy', 'classwise_accuracy', 'c_accuracy']
final_dict = {}
for ind_metric in all_metrics:
vals = []
final_array = []
for mt in clip_metrics_over_train_epochs:
ret_val = get_best_metric(mt,ind_metric,"max")
vals.append(ret_val)
final_array.append(vals)
final_dict[ind_metric] = final_array
if trivial:
graph_filename = "new_metrics"+dataset_name+"_0t"+str(num_ways)+"w"+str(num_shot)+"s"+str(num_augmentations)+"a_trivial_clip_zs_metrics_graphs.json"
else:
graph_filename = "new_metrics"+dataset_name+"_0t"+str(num_ways)+"w"+str(num_shot)+"s"+str(num_augmentations)+"a_clip_zs_metrics_graphs.json"
with open(graph_filename, 'w') as f:
json.dump(final_dict, f)
auth.authenticate_user()
gauth = GoogleAuth()
gauth.credentials = GoogleCredentials.get_application_default()
drive = GoogleDrive(gauth)
delete_file_by_name(drive, folderid, graph_filename)
save_to_drive(drive, folderid, graph_filename)
zip_dirname = PLOT_DIR + ".zip"
zip_source = PLOT_DIR
! zip -r $zip_dirname $zip_source
auth.authenticate_user()
gauth = GoogleAuth()
gauth.credentials = GoogleCredentials.get_application_default()
drive = GoogleDrive(gauth)
save_to_drive(drive, folderid, zip_dirname)
```
# 20 way 1 shot
## Picking indices
```
num_ways = 20
num_shot = 1
num_eval = 10
shuffle = False
num_episodes = 100
train_indices, eval_indices, wi_y, eval_y = prepare_indices(
num_ways, num_shot, num_eval, num_episodes, label_dictionary, val_labels, shuffle
)
embedding_model = embedding_models.CLIPEmbeddingWrapper()
num_augmentations = 0
trivial=False
indices, embeddings = embed_images(
embedding_model,
train_indices,
num_augmentations,
trivial=trivial
)
```
## CLIP
```
clip_embeddings_test_fn = "clip" + val_embeddings_filename_suffix
clip_embeddings_test = get_ndarray_from_drive(drive, folderid, clip_embeddings_test_fn)
import warnings
warnings.filterwarnings('ignore')
if trivial:
#cc
results_filename = "new_metrics"+dataset_name+"_0t"+str(num_ways)+"w"+str(num_shot)+"s"+str(num_augmentations)+"a_trivial_clip_zs_metrics_with_logits.json"
else:
#cc
results_filename = "new_metrics"+dataset_name+"_0t"+str(num_ways)+"w"+str(num_shot)+"s"+str(num_augmentations)+"a_clip_zs_metrics_with_logits.json"
auth.authenticate_user()
gauth = GoogleAuth()
gauth.credentials = GoogleCredentials.get_application_default()
drive = GoogleDrive(gauth)
download_file_by_name(drive, folderid, results_filename)
if results_filename in os.listdir():
with open(results_filename, 'r') as f:
#cc
json_loaded = json.load(f)
#cc
clip_metrics_over_train_epochs = json_loaded['metrics']
#cc
logits_over_train_epochs = json_loaded["logits"]
else:
clip_metrics_over_train_epochs = []
#cc
logits_over_train_epochs = []
train_epochs_arr = [0]
multi_label=False
# metrics_vals = ['hamming', 'jaccard', 'f1_score'] # ['accuracy', 'f1_score']
for idx, train_epochs in enumerate(train_epochs_arr):
if idx < len(clip_metrics_over_train_epochs):
continue
print(train_epochs)
#cc
clip_metrics_thresholds, all_logits = run_evaluations(
(indices, embeddings),
train_indices,
eval_indices,
wi_y,
eval_y,
num_episodes,
num_ways,
class_names,
train_epochs=train_epochs,
num_augmentations=num_augmentations,
embeddings=clip_embeddings_test
)
clip_metrics_over_train_epochs.append(clip_metrics_thresholds)
#cc
logits_over_train_epochs.append(all_logits)
#cc
fin_list = []
#cc the whole for loop
for a1 in wi_y:
fin_a1_list = []
for a2 in a1:
new_val = str(a2)
fin_a1_list.append(new_val)
fin_list.append(fin_a1_list)
with open(results_filename, 'w') as f:
#cc
results = {'metrics': clip_metrics_over_train_epochs,
"logits": logits_over_train_epochs,
"true_labels": fin_list}
json.dump(results, f)
auth.authenticate_user()
gauth = GoogleAuth()
gauth.credentials = GoogleCredentials.get_application_default()
drive = GoogleDrive(gauth)
delete_file_by_name(drive, folderid, results_filename)
save_to_drive(drive, folderid, results_filename)
if trivial:
PLOT_DIR = "NewMetrics_clip_zs_Sigmoid_" + dataset_name + "_0t" + str(num_ways) + "w" + str(num_shot) + "s" + str(num_augmentations) + "a_trivial_plots"
else:
PLOT_DIR = "NewMetrics_clip_zs_Sigmoid_" + dataset_name + "_0t" + str(num_ways) + "w" + str(num_shot) + "s" + str(num_augmentations) + "a_plots"
os.mkdir(PLOT_DIR)
all_metrics = ['accuracy', 'ap', 'map', 'c_f1', 'o_f1', 'c_precision', 'o_precision', 'c_recall', 'o_recall', 'top1_accuracy', 'top5_accuracy', 'classwise_accuracy', 'c_accuracy']
final_dict = {}
for ind_metric in all_metrics:
vals = []
final_array = []
for mt in clip_metrics_over_train_epochs:
ret_val = get_best_metric(mt,ind_metric,"max")
vals.append(ret_val)
final_array.append(vals)
final_dict[ind_metric] = final_array
if trivial:
graph_filename = "new_metrics"+dataset_name+"_0t"+str(num_ways)+"w"+str(num_shot)+"s"+str(num_augmentations)+"a_trivial_clip_zs_metrics_graphs.json"
else:
graph_filename = "new_metrics"+dataset_name+"_0t"+str(num_ways)+"w"+str(num_shot)+"s"+str(num_augmentations)+"a_clip_zs_metrics_graphs.json"
with open(graph_filename, 'w') as f:
json.dump(final_dict, f)
auth.authenticate_user()
gauth = GoogleAuth()
gauth.credentials = GoogleCredentials.get_application_default()
drive = GoogleDrive(gauth)
delete_file_by_name(drive, folderid, graph_filename)
save_to_drive(drive, folderid, graph_filename)
zip_dirname = PLOT_DIR + ".zip"
zip_source = PLOT_DIR
! zip -r $zip_dirname $zip_source
auth.authenticate_user()
gauth = GoogleAuth()
gauth.credentials = GoogleCredentials.get_application_default()
drive = GoogleDrive(gauth)
save_to_drive(drive, folderid, zip_dirname)
```
|
github_jupyter
|
from google.colab import drive
drive.mount('/content/drive')
!pip install -U -q PyDrive
!pip install httplib2==0.15.0
import os
from pydrive.auth import GoogleAuth
from pydrive.drive import GoogleDrive
from pydrive.files import GoogleDriveFileList
from google.colab import auth
from oauth2client.client import GoogleCredentials
from getpass import getpass
import urllib
# 1. Authenticate and create the PyDrive client.
auth.authenticate_user()
gauth = GoogleAuth()
gauth.credentials = GoogleCredentials.get_application_default()
drive = GoogleDrive(gauth)
# Cloning CLIPPER to access modules.
if 'CLIPPER' not in os.listdir():
cmd_string = 'git clone https://github.com/PAL-ML/CLIPPER.git'
os.system(cmd_string)
! pip install scikit-learn==0.24
import subprocess
CUDA_version = [s for s in subprocess.check_output(["nvcc", "--version"]).decode("UTF-8").split(", ") if s.startswith("release")][0].split(" ")[-1]
print("CUDA version:", CUDA_version)
if CUDA_version == "10.0":
torch_version_suffix = "+cu100"
elif CUDA_version == "10.1":
torch_version_suffix = "+cu101"
elif CUDA_version == "10.2":
torch_version_suffix = ""
else:
torch_version_suffix = "+cu110"
! pip install torch==1.7.1{torch_version_suffix} torchvision==0.8.2{torch_version_suffix} -f https://download.pytorch.org/whl/torch_stable.html ftfy regex
! pip install ftfy regex
! wget https://openaipublic.azureedge.net/clip/bpe_simple_vocab_16e6.txt.gz -O bpe_simple_vocab_16e6.txt.gz
!pip install git+https://github.com/Sri-vatsa/CLIP # using this fork because of visualization capabilities
!pip -q install umap-learn>=0.3.7
!pip install wget
# ML Libraries
import tensorflow as tf
import tensorflow_hub as hub
import torch
import torch.nn as nn
import torchvision.models as models
import torchvision.transforms as transforms
import keras
# Data processing
import PIL
import base64
import imageio
import pandas as pd
import numpy as np
import json
from PIL import Image
import cv2
import imgaug.augmenters as iaa
# Plotting
import seaborn as sns
import matplotlib.pyplot as plt
from IPython.core.display import display, HTML
from matplotlib import cm
# Models
import clip
# Datasets
import tensorflow_datasets as tfds
# Misc
import progressbar
import logging
from abc import ABC, abstractmethod
import time
import urllib.request
import os
import itertools
from tqdm import tqdm
# Modules
from CLIPPER.code.ExperimentModules import embedding_models
from CLIPPER.code.ExperimentModules import simclr_data_augmentations
from CLIPPER.code.ExperimentModules.dataset_manager import DatasetManager
from CLIPPER.code.ExperimentModules.clip_few_shot import CLIPFewShotClassifier
from CLIPPER.code.ExperimentModules.utils import (save_npy, load_npy,
get_folder_id,
create_expt_dir,
save_to_drive,
load_all_from_drive_folder,
download_file_by_name,
delete_file_by_name)
logging.getLogger('googleapicliet.discovery_cache').setLevel(logging.ERROR)
dataset_name = 'ImagenetSketch'
folder_name = "ImagenetSketch-Embeddings-28-02-21"
# Change parentid to match that of experiments root folder in gdrive
parentid = '1bK72W-Um20EQDEyChNhNJthUNbmoSEjD'
# Filepaths
# train_labels_filename = "train_labels.npz"
val_labels_filename = "val_labels.npz"
# train_embeddings_filename_suffix = "_embeddings_train.npz"
val_embeddings_filename_suffix = "_embeddings_val.npz"
# Initialize sepcific experiment folder in drive
folderid = create_expt_dir(drive, parentid, folder_name)
def get_ndarray_from_drive(drive, folderid, filename):
download_file_by_name(drive, folderid, filename)
return np.load(filename)['data']
# train_labels = get_ndarray_from_drive(drive, folderid, train_labels_filename)
val_labels = get_ndarray_from_drive(drive, folderid, val_labels_filename)
# test_labels = get_ndarray_from_drive(drive, folderid, test_labels_filename)
dm = DatasetManager()
test_data_generator = dm.load_dataset('imagenet_sketch', split='val')
classes = dm.get_class_names()
!wget https://s3.amazonaws.com/deep-learning-models/image-models/imagenet_class_index.json
with open("imagenet_class_index.json") as f:
class_idx = json.load(f)
idx2label_dict = {str(k): class_idx[str(k)][1] for k in range(len(class_idx))}
class2id_dict = {class_idx[str(k)][0]: (str(k), class_idx[str(k)][1]) for k in range(len(class_idx))}
class_names = [class2id_dict[x][1].replace("_", " ") for x in classes]
unique_labels = np.unique(val_labels)
print(len(unique_labels))
label_dictionary = {la:[] for la in unique_labels}
for i in range(len(val_labels)):
la = val_labels[i]
label_dictionary[la].append(i)
def start_progress_bar(bar_len):
widgets = [
' [',
progressbar.Timer(format= 'elapsed time: %(elapsed)s'),
'] ',
progressbar.Bar('*'),' (',
progressbar.ETA(), ') ',
]
pbar = progressbar.ProgressBar(
max_value=bar_len, widgets=widgets
).start()
return pbar
def prepare_indices(
num_ways,
num_shot,
num_eval,
num_episodes,
label_dictionary,
test_labels,
shuffle=False
):
eval_indices = []
train_indices = []
wi_y = []
eval_y = []
label_dictionary = {la:label_dictionary[la] for la in label_dictionary if len(label_dictionary[la]) >= (num_shot+num_eval)}
unique_labels = list(label_dictionary.keys())
pbar = start_progress_bar(num_episodes)
for s in range(num_episodes):
# Setting random seed for replicability
np.random.seed(s)
_train_indices = []
_eval_indices = []
selected_labels = np.random.choice(unique_labels, size=num_ways, replace=False)
for la in selected_labels:
la_indices = label_dictionary[la]
select = np.random.choice(la_indices, size = num_shot+num_eval, replace=False)
tr_idx = list(select[:num_shot])
ev_idx = list(select[num_shot:])
_train_indices = _train_indices + tr_idx
_eval_indices = _eval_indices + ev_idx
if shuffle:
np.random.shuffle(_train_indices)
np.random.shuffle(_eval_indices)
train_indices.append(_train_indices)
eval_indices.append(_eval_indices)
_wi_y = test_labels[_train_indices]
_eval_y = test_labels[_eval_indices]
wi_y.append(_wi_y)
eval_y.append(_eval_y)
pbar.update(s+1)
return train_indices, eval_indices, wi_y, eval_y
def embed_images(
embedding_model,
train_indices,
num_augmentations,
trivial=False
):
def augment_image(image, num_augmentations, trivial):
""" Perform SimCLR augmentations on the image
"""
if np.max(image) > 1:
image = image/255
augmented_images = [image]
# augmentations = iaa.Sequential([
# iaa.Affine(
# translate_percent={'x':(-0.1, 0.1), 'y':(-0.1, 0.1)},
# rotate=(-15, 15),
# shear=(-15, 15),
# ),
# iaa.Fliplr(0.5)
# ])
def _run_filters(image):
width = image.shape[1]
height = image.shape[0]
image_aug = simclr_data_augmentations.random_crop_with_resize(
image,
height,
width
)
image_aug = tf.image.random_flip_left_right(image_aug)
image_aug = simclr_data_augmentations.random_color_jitter(image_aug)
image_aug = simclr_data_augmentations.random_blur(
image_aug,
height,
width
)
image_aug = tf.reshape(image_aug, [image.shape[0], image.shape[1], 3])
image_aug = tf.clip_by_value(image_aug, 0., 1.)
return image_aug.numpy()
for _ in range(num_augmentations):
# aug_image = augmentations(image=image)
if trivial:
aug_image = image
else:
aug_image = _run_filters(image)
augmented_images.append(aug_image)
augmented_images = np.stack(augmented_images)
return augmented_images
embedding_model.load_model()
unique_indices = np.unique(np.array(train_indices))
ds = dm.load_dataset('imagenet_sketch', split='val')
embeddings = []
IMAGE_IDX = 0
pbar = start_progress_bar(unique_indices.size+1)
num_done=0
for idx, item in enumerate(ds):
if idx in unique_indices:
image = item[IMAGE_IDX][0]
if num_augmentations > 0:
aug_images = augment_image(image, num_augmentations, trivial)
else:
aug_images = image
processed_images = embedding_model.preprocess_data(aug_images)
embedding = embedding_model.embed_images(processed_images)
embeddings.append(embedding)
num_done += 1
pbar.update(num_done+1)
if idx == unique_indices[-1]:
break
embeddings = np.stack(embeddings)
return unique_indices, embeddings
def evaluate_model_for_episode(
model,
eval_x,
eval_y,
label_mapping,
filtered_classes,
metrics=['hamming', 'jaccard', 'subset_accuracy', 'ap', 'map', 'c_f1', 'o_f1', 'c_precision', 'o_precision', 'c_recall', 'o_recall', 'top1_accuracy', 'top5_accuracy', 'classwise_accuracy', 'c_accuracy'],
multi_label=True
):
zs_weights = model.zeroshot_classifier(filtered_classes)
logits = model.predict_scores(eval_x, zs_weights).tolist()
pred_y = model.predict_label(eval_x, zs_weights)
pred_y = [label_mapping[l] for l in pred_y]
met = model.evaluate_single_label_metrics(
eval_x, eval_y, label_mapping, zs_weights, metrics
)
return pred_y, met, logits
def get_label_mapping_n_class_names(eval_y, class_names):
label_mapping = {}
unique_labels = np.unique(eval_y)
filtered_classes = [class_names[x] for x in unique_labels]
num_classes = len(unique_labels)
for c in range(num_classes):
label_mapping[c] = unique_labels[c]
return label_mapping, filtered_classes
# chenni change
def run_episode_through_model(
indices_and_embeddings,
train_indices,
eval_indices,
wi_y,
eval_y,
class_names,
num_augmentations=0,
train_epochs=None,
train_batch_size=5,
metrics=['hamming', 'jaccard', 'subset_accuracy', 'ap', 'map', 'c_f1', 'o_f1', 'c_precision', 'o_precision', 'c_recall', 'o_recall', 'top1_accuracy', 'top5_accuracy', 'classwise_accuracy', 'c_accuracy'],
embeddings=None,
multi_label=True
):
metrics_values = {m:[] for m in metrics}
indices_and_embeddings
eval_x = embeddings[eval_indices]
ep_logits = []
label_mapping, filtered_classes = get_label_mapping_n_class_names(eval_y, class_names)
clip_fs_parameters = {
"num_classes": num_ways,
"input_dims": eval_x.shape[-1],
"multi_label": multi_label
}
clip_fs_cls = CLIPFewShotClassifier(clip_fs_parameters)
pred_labels, metrics_values, logits = evaluate_model_for_episode(
clip_fs_cls,
eval_x,
eval_y,
label_mapping,
filtered_classes,
metrics=metrics,
multi_label=False
)
ep_logits = logits
#cc
return metrics_values, ep_logits
def run_evaluations(
indices_and_embeddings,
train_indices,
eval_indices,
wi_y,
eval_y,
num_episodes,
num_ways,
class_names,
verbose=True,
normalize=True,
train_epochs=None,
train_batch_size=5,
metrics=['accuracy', 'ap', 'map', 'c_f1', 'o_f1', 'c_precision', 'o_precision', 'c_recall', 'o_recall', 'top1_accuracy', 'top5_accuracy', 'classwise_accuracy', 'c_accuracy'],
embeddings=None,
num_augmentations=0,
multi_label=True
):
metrics_values = {m:[] for m in metrics}
all_logits = []
if verbose:
pbar = start_progress_bar(num_episodes)
for idx_ep in range(num_episodes):
_train_indices = train_indices[idx_ep]
_eval_indices = eval_indices[idx_ep]
_wi_y = [label for label in wi_y[idx_ep]]
_eval_y = [label for label in eval_y[idx_ep]]
met, ep_logits = run_episode_through_model(
indices_and_embeddings,
_train_indices,
_eval_indices,
_wi_y,
_eval_y,
class_names,
num_augmentations=num_augmentations,
train_epochs=train_epochs,
train_batch_size=train_batch_size,
embeddings=embeddings,
metrics=metrics,
multi_label=multi_label
)
all_logits.append(ep_logits)
for m in metrics:
metrics_values[m].append(met[m])
if verbose:
pbar.update(idx_ep+1)
return metrics_values, all_logits
def get_best_metric(mt, metric_name, optimal='max'):
if optimal=='max':
opt_value = np.max(np.mean(np.array(mt[metric_name]), axis=0))
if optimal=='min':
opt_value = np.min(np.mean(np.array(mt[metric_name]), axis=0))
return opt_value
num_ways = 5
num_shot = 5
num_eval = 15
shuffle = False
num_episodes = 100
train_indices, eval_indices, wi_y, eval_y = prepare_indices(
num_ways, num_shot, num_eval, num_episodes, label_dictionary, val_labels, shuffle
)
embedding_model = embedding_models.CLIPEmbeddingWrapper()
num_augmentations = 0
trivial=False
indices, embeddings = embed_images(
embedding_model,
train_indices,
num_augmentations,
trivial=trivial
)
clip_embeddings_test_fn = "clip" + val_embeddings_filename_suffix
clip_embeddings_test = get_ndarray_from_drive(drive, folderid, clip_embeddings_test_fn)
import warnings
warnings.filterwarnings('ignore')
if trivial:
#cc
results_filename = "new_metrics"+dataset_name+"_0t"+str(num_ways)+"w"+str(num_shot)+"s"+str(num_augmentations)+"a_trivial_clip_zs_metrics_with_logits.json"
else:
#cc
results_filename = "new_metrics"+dataset_name+"_0t"+str(num_ways)+"w"+str(num_shot)+"s"+str(num_augmentations)+"a_clip_zs_metrics_with_logits.json"
auth.authenticate_user()
gauth = GoogleAuth()
gauth.credentials = GoogleCredentials.get_application_default()
drive = GoogleDrive(gauth)
download_file_by_name(drive, folderid, results_filename)
if results_filename in os.listdir():
with open(results_filename, 'r') as f:
#cc
json_loaded = json.load(f)
#cc
clip_metrics_over_train_epochs = json_loaded['metrics']
#cc
logits_over_train_epochs = json_loaded["logits"]
else:
clip_metrics_over_train_epochs = []
logits_over_train_epochs = []
train_epochs_arr = [0]
multi_label=False
# metrics_vals = ['hamming', 'jaccard', 'f1_score'] # ['accuracy', 'f1_score']
for idx, train_epochs in enumerate(train_epochs_arr):
if idx < len(clip_metrics_over_train_epochs):
continue
print(train_epochs)
#cc
clip_metrics_thresholds, all_logits = run_evaluations(
(indices, embeddings),
train_indices,
eval_indices,
wi_y,
eval_y,
num_episodes,
num_ways,
class_names,
train_epochs=train_epochs,
num_augmentations=num_augmentations,
embeddings=clip_embeddings_test
)
clip_metrics_over_train_epochs.append(clip_metrics_thresholds)
#cc
logits_over_train_epochs.append(all_logits)
#cc
fin_list = []
#cc the whole for loop
for a1 in wi_y:
fin_a1_list = []
for a2 in a1:
new_val = str(a2)
fin_a1_list.append(new_val)
fin_list.append(fin_a1_list)
with open(results_filename, 'w') as f:
#cc
results = {'metrics': clip_metrics_over_train_epochs,
"logits": logits_over_train_epochs,
"true_labels": fin_list}
json.dump(results, f)
auth.authenticate_user()
gauth = GoogleAuth()
gauth.credentials = GoogleCredentials.get_application_default()
drive = GoogleDrive(gauth)
#delete_file_by_name(drive, folderid, results_filename)
#save_to_drive(drive, folderid, results_filename)
if trivial:
PLOT_DIR = "NewMetrics_clip_zs_Sigmoid_" + dataset_name + "_0t" + str(num_ways) + "w" + str(num_shot) + "s" + str(num_augmentations) + "a_trivial_plots"
else:
PLOT_DIR = "NewMetrics_clip_zs_Sigmoid_" + dataset_name + "_0t" + str(num_ways) + "w" + str(num_shot) + "s" + str(num_augmentations) + "a_plots"
os.mkdir(PLOT_DIR)
# chenni change whole block
all_metrics = ['accuracy', 'ap', 'map', 'c_f1', 'o_f1', 'c_precision', 'o_precision', 'c_recall', 'o_recall', 'top1_accuracy', 'top5_accuracy', 'classwise_accuracy', 'c_accuracy']
final_dict = {}
for ind_metric in all_metrics:
vals = []
final_array = []
for mt in clip_metrics_over_train_epochs:
ret_val = get_best_metric(mt,ind_metric,"max")
vals.append(ret_val)
final_array.append(vals)
final_dict[ind_metric] = final_array
if trivial:
graph_filename = "new_metrics"+dataset_name+"_0t"+str(num_ways)+"w"+str(num_shot)+"s"+str(num_augmentations)+"a_trivial_clip_zs_metrics_graphs.json"
else:
graph_filename = "new_metrics"+dataset_name+"_0t"+str(num_ways)+"w"+str(num_shot)+"s"+str(num_augmentations)+"a_clip_zs_metrics_graphs.json"
with open(graph_filename, 'w') as f:
json.dump(final_dict, f)
auth.authenticate_user()
gauth = GoogleAuth()
gauth.credentials = GoogleCredentials.get_application_default()
drive = GoogleDrive(gauth)
delete_file_by_name(drive, folderid, graph_filename)
save_to_drive(drive, folderid, graph_filename)
zip_dirname = PLOT_DIR + ".zip"
zip_source = PLOT_DIR
! zip -r $zip_dirname $zip_source
auth.authenticate_user()
gauth = GoogleAuth()
gauth.credentials = GoogleCredentials.get_application_default()
drive = GoogleDrive(gauth)
save_to_drive(drive, folderid, zip_dirname)
num_ways = 20
num_shot = 5
num_eval = 5
shuffle = False
num_episodes = 100
train_indices, eval_indices, wi_y, eval_y = prepare_indices(
num_ways, num_shot, num_eval, num_episodes, label_dictionary, val_labels, shuffle
)
embedding_model = embedding_models.CLIPEmbeddingWrapper()
num_augmentations = 0
trivial=False
indices, embeddings = embed_images(
embedding_model,
train_indices,
num_augmentations,
trivial=trivial
)
clip_embeddings_test_fn = "clip" + val_embeddings_filename_suffix
clip_embeddings_test = get_ndarray_from_drive(drive, folderid, clip_embeddings_test_fn)
import warnings
warnings.filterwarnings('ignore')
if trivial:
#cc
results_filename = "new_metrics"+dataset_name+"_0t"+str(num_ways)+"w"+str(num_shot)+"s"+str(num_augmentations)+"a_trivial_clip_zs_metrics_with_logits.json"
else:
#cc
results_filename = "new_metrics"+dataset_name+"_0t"+str(num_ways)+"w"+str(num_shot)+"s"+str(num_augmentations)+"a_metrics_clip_zs_with_logits.json"
auth.authenticate_user()
gauth = GoogleAuth()
gauth.credentials = GoogleCredentials.get_application_default()
drive = GoogleDrive(gauth)
download_file_by_name(drive, folderid, results_filename)
if results_filename in os.listdir():
with open(results_filename, 'r') as f:
#cc
json_loaded = json.load(f)
#cc
clip_metrics_over_train_epochs = json_loaded['metrics']
#cc
logits_over_train_epochs = json_loaded["logits"]
else:
clip_metrics_over_train_epochs = []
#cc
logits_over_train_epochs = []
train_epochs_arr = [0]
multi_label=False
# metrics_vals = ['hamming', 'jaccard', 'f1_score'] # ['accuracy', 'f1_score']
for idx, train_epochs in enumerate(train_epochs_arr):
if idx < len(clip_metrics_over_train_epochs):
continue
print(train_epochs)
#cc
clip_metrics_thresholds, all_logits = run_evaluations(
(indices, embeddings),
train_indices,
eval_indices,
wi_y,
eval_y,
num_episodes,
num_ways,
class_names,
train_epochs=train_epochs,
num_augmentations=num_augmentations,
embeddings=clip_embeddings_test
)
clip_metrics_over_train_epochs.append(clip_metrics_thresholds)
#cc
logits_over_train_epochs.append(all_logits)
#cc
fin_list = []
#cc the whole for loop
for a1 in wi_y:
fin_a1_list = []
for a2 in a1:
new_val = str(a2)
fin_a1_list.append(new_val)
fin_list.append(fin_a1_list)
with open(results_filename, 'w') as f:
#cc
results = {'metrics': clip_metrics_over_train_epochs,
"logits": logits_over_train_epochs,
"true_labels": fin_list}
json.dump(results, f)
auth.authenticate_user()
gauth = GoogleAuth()
gauth.credentials = GoogleCredentials.get_application_default()
drive = GoogleDrive(gauth)
delete_file_by_name(drive, folderid, results_filename)
save_to_drive(drive, folderid, results_filename)
if trivial:
PLOT_DIR = "NewMetrics_clip_zs_Sigmoid_" + dataset_name + "_0t" + str(num_ways) + "w" + str(num_shot) + "s" + str(num_augmentations) + "a_trivial_plots"
else:
PLOT_DIR = "NewMetrics_clip_zs_Sigmoid_" + dataset_name + "_0t" + str(num_ways) + "w" + str(num_shot) + "s" + str(num_augmentations) + "a_plots"
os.mkdir(PLOT_DIR)
all_metrics = ['accuracy', 'ap', 'map', 'c_f1', 'o_f1', 'c_precision', 'o_precision', 'c_recall', 'o_recall', 'top1_accuracy', 'top5_accuracy', 'classwise_accuracy', 'c_accuracy']
final_dict = {}
for ind_metric in all_metrics:
vals = []
final_array = []
for mt in clip_metrics_over_train_epochs:
ret_val = get_best_metric(mt,ind_metric,"max")
vals.append(ret_val)
final_array.append(vals)
final_dict[ind_metric] = final_array
if trivial:
graph_filename = "new_metrics"+dataset_name+"_0t"+str(num_ways)+"w"+str(num_shot)+"s"+str(num_augmentations)+"a_trivial_clip_zs_metrics_graphs.json"
else:
graph_filename = "new_metrics"+dataset_name+"_0t"+str(num_ways)+"w"+str(num_shot)+"s"+str(num_augmentations)+"a_clip_zs_metrics_graphs.json"
with open(graph_filename, 'w') as f:
json.dump(final_dict, f)
auth.authenticate_user()
gauth = GoogleAuth()
gauth.credentials = GoogleCredentials.get_application_default()
drive = GoogleDrive(gauth)
delete_file_by_name(drive, folderid, graph_filename)
save_to_drive(drive, folderid, graph_filename)
zip_dirname = PLOT_DIR + ".zip"
zip_source = PLOT_DIR
! zip -r $zip_dirname $zip_source
auth.authenticate_user()
gauth = GoogleAuth()
gauth.credentials = GoogleCredentials.get_application_default()
drive = GoogleDrive(gauth)
save_to_drive(drive, folderid, zip_dirname)
num_ways = 5
num_shot = 1
num_eval = 19
shuffle = False
num_episodes = 100
train_indices, eval_indices, wi_y, eval_y = prepare_indices(
num_ways, num_shot, num_eval, num_episodes, label_dictionary, val_labels, shuffle
)
embedding_model = embedding_models.CLIPEmbeddingWrapper()
num_augmentations = 0
trivial=False
indices, embeddings = embed_images(
embedding_model,
train_indices,
num_augmentations,
trivial=trivial
)
clip_embeddings_test_fn = "clip" + val_embeddings_filename_suffix
clip_embeddings_test = get_ndarray_from_drive(drive, folderid, clip_embeddings_test_fn)
import warnings
warnings.filterwarnings('ignore')
if trivial:
#cc
results_filename = "new_metrics"+dataset_name+"_0t"+str(num_ways)+"w"+str(num_shot)+"s"+str(num_augmentations)+"a_trivial_clip_zs_metrics_with_logits.json"
else:
#cc
results_filename = "new_metrics"+dataset_name+"_0t"+str(num_ways)+"w"+str(num_shot)+"s"+str(num_augmentations)+"a_clip_zs_metrics_with_logits.json"
auth.authenticate_user()
gauth = GoogleAuth()
gauth.credentials = GoogleCredentials.get_application_default()
drive = GoogleDrive(gauth)
download_file_by_name(drive, folderid, results_filename)
if results_filename in os.listdir():
with open(results_filename, 'r') as f:
#cc
json_loaded = json.load(f)
#cc
clip_metrics_over_train_epochs = json_loaded['metrics']
#cc
logits_over_train_epochs = json_loaded["logits"]
else:
clip_metrics_over_train_epochs = []
#cc
logits_over_train_epochs = []
train_epochs_arr = [0]
multi_label=False
# metrics_vals = ['hamming', 'jaccard', 'f1_score'] # ['accuracy', 'f1_score']
for idx, train_epochs in enumerate(train_epochs_arr):
if idx < len(clip_metrics_over_train_epochs):
continue
print(train_epochs)
#cc
clip_metrics_thresholds, all_logits = run_evaluations(
(indices, embeddings),
train_indices,
eval_indices,
wi_y,
eval_y,
num_episodes,
num_ways,
class_names,
train_epochs=train_epochs,
num_augmentations=num_augmentations,
embeddings=clip_embeddings_test
)
clip_metrics_over_train_epochs.append(clip_metrics_thresholds)
#cc
logits_over_train_epochs.append(all_logits)
#cc
fin_list = []
#cc the whole for loop
for a1 in wi_y:
fin_a1_list = []
for a2 in a1:
new_val = str(a2)
fin_a1_list.append(new_val)
fin_list.append(fin_a1_list)
with open(results_filename, 'w') as f:
#cc
results = {'metrics': clip_metrics_over_train_epochs,
"logits": logits_over_train_epochs,
"true_labels": fin_list}
json.dump(results, f)
auth.authenticate_user()
gauth = GoogleAuth()
gauth.credentials = GoogleCredentials.get_application_default()
drive = GoogleDrive(gauth)
delete_file_by_name(drive, folderid, results_filename)
save_to_drive(drive, folderid, results_filename)
if trivial:
PLOT_DIR = "NewMetrics_clip_zs_Sigmoid_" + dataset_name + "_0t" + str(num_ways) + "w" + str(num_shot) + "s" + str(num_augmentations) + "a_trivial_plots"
else:
PLOT_DIR = "NewMetrics_clip_zs_Sigmoid_" + dataset_name + "_0t" + str(num_ways) + "w" + str(num_shot) + "s" + str(num_augmentations) + "a_plots"
os.mkdir(PLOT_DIR)
all_metrics = ['accuracy', 'ap', 'map', 'c_f1', 'o_f1', 'c_precision', 'o_precision', 'c_recall', 'o_recall', 'top1_accuracy', 'top5_accuracy', 'classwise_accuracy', 'c_accuracy']
final_dict = {}
for ind_metric in all_metrics:
vals = []
final_array = []
for mt in clip_metrics_over_train_epochs:
ret_val = get_best_metric(mt,ind_metric,"max")
vals.append(ret_val)
final_array.append(vals)
final_dict[ind_metric] = final_array
if trivial:
graph_filename = "new_metrics"+dataset_name+"_0t"+str(num_ways)+"w"+str(num_shot)+"s"+str(num_augmentations)+"a_trivial_clip_zs_metrics_graphs.json"
else:
graph_filename = "new_metrics"+dataset_name+"_0t"+str(num_ways)+"w"+str(num_shot)+"s"+str(num_augmentations)+"a_clip_zs_metrics_graphs.json"
with open(graph_filename, 'w') as f:
json.dump(final_dict, f)
auth.authenticate_user()
gauth = GoogleAuth()
gauth.credentials = GoogleCredentials.get_application_default()
drive = GoogleDrive(gauth)
delete_file_by_name(drive, folderid, graph_filename)
save_to_drive(drive, folderid, graph_filename)
zip_dirname = PLOT_DIR + ".zip"
zip_source = PLOT_DIR
! zip -r $zip_dirname $zip_source
auth.authenticate_user()
gauth = GoogleAuth()
gauth.credentials = GoogleCredentials.get_application_default()
drive = GoogleDrive(gauth)
save_to_drive(drive, folderid, zip_dirname)
num_ways = 20
num_shot = 1
num_eval = 10
shuffle = False
num_episodes = 100
train_indices, eval_indices, wi_y, eval_y = prepare_indices(
num_ways, num_shot, num_eval, num_episodes, label_dictionary, val_labels, shuffle
)
embedding_model = embedding_models.CLIPEmbeddingWrapper()
num_augmentations = 0
trivial=False
indices, embeddings = embed_images(
embedding_model,
train_indices,
num_augmentations,
trivial=trivial
)
clip_embeddings_test_fn = "clip" + val_embeddings_filename_suffix
clip_embeddings_test = get_ndarray_from_drive(drive, folderid, clip_embeddings_test_fn)
import warnings
warnings.filterwarnings('ignore')
if trivial:
#cc
results_filename = "new_metrics"+dataset_name+"_0t"+str(num_ways)+"w"+str(num_shot)+"s"+str(num_augmentations)+"a_trivial_clip_zs_metrics_with_logits.json"
else:
#cc
results_filename = "new_metrics"+dataset_name+"_0t"+str(num_ways)+"w"+str(num_shot)+"s"+str(num_augmentations)+"a_clip_zs_metrics_with_logits.json"
auth.authenticate_user()
gauth = GoogleAuth()
gauth.credentials = GoogleCredentials.get_application_default()
drive = GoogleDrive(gauth)
download_file_by_name(drive, folderid, results_filename)
if results_filename in os.listdir():
with open(results_filename, 'r') as f:
#cc
json_loaded = json.load(f)
#cc
clip_metrics_over_train_epochs = json_loaded['metrics']
#cc
logits_over_train_epochs = json_loaded["logits"]
else:
clip_metrics_over_train_epochs = []
#cc
logits_over_train_epochs = []
train_epochs_arr = [0]
multi_label=False
# metrics_vals = ['hamming', 'jaccard', 'f1_score'] # ['accuracy', 'f1_score']
for idx, train_epochs in enumerate(train_epochs_arr):
if idx < len(clip_metrics_over_train_epochs):
continue
print(train_epochs)
#cc
clip_metrics_thresholds, all_logits = run_evaluations(
(indices, embeddings),
train_indices,
eval_indices,
wi_y,
eval_y,
num_episodes,
num_ways,
class_names,
train_epochs=train_epochs,
num_augmentations=num_augmentations,
embeddings=clip_embeddings_test
)
clip_metrics_over_train_epochs.append(clip_metrics_thresholds)
#cc
logits_over_train_epochs.append(all_logits)
#cc
fin_list = []
#cc the whole for loop
for a1 in wi_y:
fin_a1_list = []
for a2 in a1:
new_val = str(a2)
fin_a1_list.append(new_val)
fin_list.append(fin_a1_list)
with open(results_filename, 'w') as f:
#cc
results = {'metrics': clip_metrics_over_train_epochs,
"logits": logits_over_train_epochs,
"true_labels": fin_list}
json.dump(results, f)
auth.authenticate_user()
gauth = GoogleAuth()
gauth.credentials = GoogleCredentials.get_application_default()
drive = GoogleDrive(gauth)
delete_file_by_name(drive, folderid, results_filename)
save_to_drive(drive, folderid, results_filename)
if trivial:
PLOT_DIR = "NewMetrics_clip_zs_Sigmoid_" + dataset_name + "_0t" + str(num_ways) + "w" + str(num_shot) + "s" + str(num_augmentations) + "a_trivial_plots"
else:
PLOT_DIR = "NewMetrics_clip_zs_Sigmoid_" + dataset_name + "_0t" + str(num_ways) + "w" + str(num_shot) + "s" + str(num_augmentations) + "a_plots"
os.mkdir(PLOT_DIR)
all_metrics = ['accuracy', 'ap', 'map', 'c_f1', 'o_f1', 'c_precision', 'o_precision', 'c_recall', 'o_recall', 'top1_accuracy', 'top5_accuracy', 'classwise_accuracy', 'c_accuracy']
final_dict = {}
for ind_metric in all_metrics:
vals = []
final_array = []
for mt in clip_metrics_over_train_epochs:
ret_val = get_best_metric(mt,ind_metric,"max")
vals.append(ret_val)
final_array.append(vals)
final_dict[ind_metric] = final_array
if trivial:
graph_filename = "new_metrics"+dataset_name+"_0t"+str(num_ways)+"w"+str(num_shot)+"s"+str(num_augmentations)+"a_trivial_clip_zs_metrics_graphs.json"
else:
graph_filename = "new_metrics"+dataset_name+"_0t"+str(num_ways)+"w"+str(num_shot)+"s"+str(num_augmentations)+"a_clip_zs_metrics_graphs.json"
with open(graph_filename, 'w') as f:
json.dump(final_dict, f)
auth.authenticate_user()
gauth = GoogleAuth()
gauth.credentials = GoogleCredentials.get_application_default()
drive = GoogleDrive(gauth)
delete_file_by_name(drive, folderid, graph_filename)
save_to_drive(drive, folderid, graph_filename)
zip_dirname = PLOT_DIR + ".zip"
zip_source = PLOT_DIR
! zip -r $zip_dirname $zip_source
auth.authenticate_user()
gauth = GoogleAuth()
gauth.credentials = GoogleCredentials.get_application_default()
drive = GoogleDrive(gauth)
save_to_drive(drive, folderid, zip_dirname)
| 0.426083 | 0.383555 |
# NYC Crash Data Analysis
This project studies the [Motor-Vehicle-Collisions-Crashes](https://data.cityofnewyork.us/Public-Safety/Motor-Vehicle-Collisions-Crashes/h9gi-nx95) data to find patterns in the data by visualizing it.
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import warnings
warnings.filterwarnings('ignore')
%matplotlib inline
import geoplot as gplt
import geopandas as gpd
import geoplot.crs as gcrs
# please proceed onlyafter changing root directory and input file name
# Config for the project
root_dir = 'G:\Projects\\NYC-Crash-Data-Analytics\\'
# Input filename
filename = 'G:\Projects\\NYC-Crash-Data-Analytics\Motor_Vehicle_Collisions_-_Crashes.csv'
# Cleaned filename
clean_filename = 'G:\Projects\\NYC-Crash-Data-Analytics\Motor_Vehicle_Collisions_-_Crashes_Cleaned.csv'
# NY Borough Boundaries shapefile
nybb_shapefile = root_dir + 'maps\nybb.shp'
# Index column
index_col = 'COLLISION_ID'
```
## Data Cleaning
The data cleaning involved the following processes:
* **Date Parsing:** The data has two fields for CRASH DATE and CRASH TIME. We combined these two fields into a DATETIME field to make filtering easier for the data investigation.
* **NA Value Filling:** The data contains NA values, to have a more consistent data, we filled the NA values depending on the type of data in the field. For example, the NA values in the BOROUGH field were filled with 'UNKOWN', the LATITUDE/LONGITUDE fields were set to 0.
```
# Data types for the columns in the data
dtypes = {
'CRASH DATE' : 'str',
'CRASH TIME' : 'str',
'BOROUGH' : 'str',
'ZIP CODE' : 'str',
'LATITUDE' : 'float64',
'LONGITUDE' : 'float64',
'LOCATION' : 'object',
'ON STREET NAME' : 'str',
'CROSS STREET NAME' : 'str',
'OFF STREET NAME' : 'str',
'NUMBER OF PERSONS INJURED' : 'float64',
'NUMBER OF PERSONS KILLED' : 'float64',
'NUMBER OF PEDESTRIANS INJURED' : 'float64',
'NUMBER OF PEDESTRIANS KILLED' : 'float64',
'NUMBER OF CYCLIST INJURED' : 'float64',
'NUMBER OF CYCLIST KILLED' : 'float64',
'NUMBER OF MOTORIST INJURED' : 'float64',
'NUMBER OF MOTORIST KILLED' : 'float64',
'CONTRIBUTING FACTOR VEHICLE 1' : 'str',
'CONTRIBUTING FACTOR VEHICLE 2' : 'str',
'CONTRIBUTING FACTOR VEHICLE 3' : 'str',
'CONTRIBUTING FACTOR VEHICLE 4' : 'str',
'CONTRIBUTING FACTOR VEHICLE 5' : 'str',
'COLLISION_ID' : 'int64',
'VEHICLE TYPE CODE 1' : 'category',
'VEHICLE TYPE CODE 2' : 'category',
'VEHICLE TYPE CODE 3' : 'category',
'VEHICLE TYPE CODE 4' : 'category',
'VEHICLE TYPE CODE 5' : 'category'
}
# Column-wise replacement values for NA
na_replace = {
'BOROUGH' : 'UNKNOWN',
'ZIP CODE' : 'UNKNOWN',
'LATITUDE' : 0,
'LONGITUDE' : 0,
'LOCATION' : '(0.0, 0.0)',
'ON STREET NAME' : '',
'CROSS STREET NAME' : '',
'OFF STREET NAME' : '',
'NUMBER OF PERSONS INJURED' : 0,
'NUMBER OF PERSONS KILLED' : 0,
'NUMBER OF PEDESTRIANS INJURED' : 0,
'NUMBER OF PEDESTRIANS KILLED' : 0,
'NUMBER OF CYCLIST INJURED' : 0,
'NUMBER OF CYCLIST KILLED' : 0,
'NUMBER OF MOTORIST INJURED' : 0,
'NUMBER OF MOTORIST KILLED' : 0,
'CONTRIBUTING FACTOR VEHICLE 1' : '',
'CONTRIBUTING FACTOR VEHICLE 2' : '',
'CONTRIBUTING FACTOR VEHICLE 3' : '',
'CONTRIBUTING FACTOR VEHICLE 4' : '',
'CONTRIBUTING FACTOR VEHICLE 5' : '',
'VEHICLE TYPE CODE 1' : '',
'VEHICLE TYPE CODE 2' : '',
'VEHICLE TYPE CODE 3' : '',
'VEHICLE TYPE CODE 4' : '',
'VEHICLE TYPE CODE 5' : ''
}
print('Reading CSV file %s ...' % filename)
crash_data = pd.read_csv(filename,
index_col=index_col,
dtype=dtypes,
parse_dates={'CRASH DATETIME' : ['CRASH DATE', 'CRASH TIME']},
infer_datetime_format=True
)
print('Filling NaN values ...')
for key, val in na_replace.items():
print('\t%s' % key)
crash_data[key] = crash_data[key].replace(np.nan, val)
print("Saving cleaned file to %s ..." % clean_filename)
crash_data.to_csv(clean_filename)
print("Cleaning complete.")
```
## Read Clean Data
```
crash_data = pd.read_csv(clean_filename,
index_col=index_col,
parse_dates=['CRASH DATETIME'])
before_20 = crash_data[crash_data['CRASH DATETIME'].dt.year < 2020]
during_20 = crash_data[crash_data['CRASH DATETIME'].dt.year == 2020]
crash_data.head()
```
## Q1
For the years before 2020, which boroughs had the most accidents? Did this distribution change during 2020?
```
accidents = pd.DataFrame()
# == Compute the distribution for accidents by Borough for years before 2020 ===
accidents['Before 2020'] = before_20['BOROUGH'].value_counts()
accidents['Before 2020'] = accidents['Before 2020'] / accidents['Before 2020'] \
.sum()
# ====== Compute the distribution for accidents by Borough for year 2020 =======
accidents['2020'] = during_20['BOROUGH'].value_counts()
accidents['2020'] = accidents['2020'] / accidents['2020'].sum()
# Plot distributions
accidents.plot.bar()
plt.show()
```
## Q2
For the years before 2020, which months had the most accidents? Students in the past have said they thought there were 10% fewer accidents in February then in January. Is this true or is this bogus? Did this distribution change during 2020?
```
accidents = pd.DataFrame()
# == Compute the distribution for accidents by Borough for years before 2020 ===
accidents['Before 2020'] = before_20['CRASH DATETIME'].dt.month.value_counts()
accidents['Before 2020'] = accidents['Before 2020'] / accidents['Before 2020'] \
.sum()
# ====== Compute the distribution for accidents by Borough for year 2020 =======
accidents['2020'] = during_20['CRASH DATETIME'].dt.month.value_counts()
accidents['2020'] = accidents['2020'] / accidents['2020'].sum()
print('February had %.2f%% fewer accidents than January.' \
% ((1 - (accidents['Before 2020'][2] / accidents['Before 2020'][1])) * 100))
# Stack the data
accidents = accidents.sort_index()
# Plot distributions
accidents.plot()
plt.show()
```
## Q3
For the years before 2020, which types of accidents were most prevalent? Did this distribution change during 2020?
```
accidents = pd.DataFrame()
# == Compute the distribution for accidents by Borough for years before 2020 ===
accidents['Before 2020'] = before_20['CONTRIBUTING FACTOR VEHICLE 1'] \
.value_counts()
accidents['Before 2020'] = accidents['Before 2020'] / accidents['Before 2020'] \
.sum()
# ====== Compute the distribution for accidents by Borough for year 2020 =======
accidents['2020'] = during_20['CONTRIBUTING FACTOR VEHICLE 1'].value_counts()
accidents['2020'] = accidents['2020'] / accidents['2020'].sum()
# Plot distributions
accidents.plot.bar(figsize=(22,8), fontsize=20)
plt.show()
```
## Q4
For the years before 2020, which days of the week had the most accidents? Did this distribution change during 2020?
```
# introduce a column for storing day the crash took place
before_20['DAY'] = before_20['CRASH DATETIME'].dt.day_name()
during_20['DAY'] = during_20['CRASH DATETIME'].dt.day_name()
# get the total number of crashes that happened on each day
crash_day_before_20 = before_20['DAY'].value_counts(ascending=True,normalize=True)*100
crash_day_during_20 = during_20['DAY'].value_counts(ascending=True,normalize=True)*100
# plot the total number of crashes to day it took place
print('---------------------')
print('\nNumber of crashes by day before 2020:\n\n', crash_day_before_20.to_string())
crash_day_before_20.plot(kind='barh', figsize=(12, 8))
plt.xlabel("No. of Crashes", labelpad=14)
plt.ylabel("Day of Week", labelpad=14)
plt.title("No. of Crashes by Day of Week for year before 2020", y=1.02)
plt.savefig('Results\crashes_by_day_bef_20.png')
print('\nSaved the graph for No. of Crashes by Day of Week for '
'year before 2020 to crashes_by_day_bef_20.png.')
plt.show()
# plt.clf()
# plot the total number of crashes to day it took place
print('---------------------')
print('\nNumber of crashes by day during 2020:\n\n', crash_day_during_20.to_string())
crash_day_during_20.plot(kind='barh', figsize=(12, 8))
plt.xlabel("No. of Crashes", labelpad=14)
plt.ylabel("Day of Week", labelpad=14)
plt.title("No. of Crashes by Day of Week for year during 2020", y=1.02)
plt.savefig('Results\crashes_by_day_in_20.png')
print('\nSaved the graph for No. of Crashes by Day of Week for '
'year during 2020 to crashes_by_day_in_20.png.')
plt.show()
print('---------------------')
# Stack the data
season_crashes = pd.concat([crash_day_before_20, crash_day_during_20], axis=1)
# Plot distributions
season_crashes.plot.bar(figsize=(12,9))
plt.ylabel("Percentage of Crashes", labelpad=6)
plt.title("Percentage of Crashes by Day of week", y=1.02)
plt.legend(['Before 2020','After 2020'])
plt.savefig('Results\crashes_by_day')
print('\nSaved the graph for No. of Crashes by Day of Week to crashes_by_day.png.')
```
## Q5
For a typical year before 2020, how given a seven day calendar that starts at 12:01 AM on Saturday and goes until 11:59 PM midnight on Sunday, when are accidents most likely to happen? Which day of the week is most likely to have an accident? What time of the day is most likely to have an accident. Does this change in 2020?
```
# introduce a column for storing day the crash took place
before_20['DAY'] = before_20['CRASH DATETIME'].dt.day_name()
during_20['DAY'] = during_20['CRASH DATETIME'].dt.day_name()
# introduce a column for storing day the crash took place
before_20['HOUR'] = before_20['CRASH DATETIME'].dt.hour
during_20['HOUR'] = during_20['CRASH DATETIME'].dt.hour
# group the data by day value
grp_by_day_before_20 = before_20.groupby(['DAY'])
grp_by_day_during_20 = during_20.groupby(['DAY'])
# ======== Retrieve and plot data for years before 2020 ========
plt.figure(figsize=(10, 6), dpi=80)
print('---------------------')
print('\nCrashes that happened every hour for each day before 2020:')
day_list = []
for entry in grp_by_day_before_20:
# get day of week
day_of_week = entry[0]
# sort the entries by hour
this_day_crashes = entry[1].sort_values(by=['HOUR'])
# get count of crashes that happened for every hour of day
crashes_by_hour = this_day_crashes['HOUR'].value_counts().sort_index(ascending=True)
print('\nFor ',day_of_week,'\nHOUR\tCrashes\n', crashes_by_hour)
day_list.append(day_of_week)
# plot the count of crashes that happened for every hour of day
crashes_by_hour.plot.line()
# give labels and add legend
plt.ylabel("No. of Crashes", labelpad=14)
plt.xlabel("Time of day", labelpad=14)
plt.legend(day_list)
plt.title("No. of Crashes by Time of day for years before 2020", y=1.02)
# save the graph
plt.savefig('Results\crashes_by_time_bef_20.png')
print("Saved No. of Crashes by Time of day for years before 2020 to crashes_by_time_bef_20.png")
plt.show()
# clear plot
# plt.cla()
print('---------------------')
# ======== Retrieve and plot data for year 2020 ========
print('\nCrashes that happened every hour for each day in 2020:')
day_list = []
plt.figure(figsize=(10, 6), dpi=80)
# give labels and add legend
plt.ylabel("No. of Crashes", labelpad=14)
plt.xlabel("Time of day", labelpad=14)
plt.title("No. of Crashes by Time of day during 2020", y=1.02)
for entry in grp_by_day_during_20:
# get day of week
day_of_week = entry[0]
# sort the entries by hour
this_day_crashes = entry[1].sort_values(by=['HOUR'])
# get count of crashes that happened for every hour of day
crashes_by_hour = this_day_crashes['HOUR'].value_counts().sort_index(ascending=True)
print('\nFor ',day_of_week,'\nHOUR\tCrashes\n', crashes_by_hour)
day_list.append(day_of_week)
# plot the count of crashes that happened for every hour of day
crashes_by_hour.plot.line()
plt.legend(day_list)
# save the graph
plt.savefig('Results\crashes_by_time_in_20.png')
print("Saved No. of Crashes by Time of day during 2020 to crashes_by_time_in_20.png")
plt.show()
print('---------------------')
```
## Q6
Does the timing of when accidents happen depend on the borough of NY City? Does the amount of change vary from year to year?
```
# group the data by day value
grp_by_borough_before_20 = before_20.groupby(['BOROUGH'])
grp_by_borough_during_20 = during_20.groupby(['BOROUGH'])
# ======== Retrieve and plot data for years before 2020 ========
plt.figure(figsize=(10, 6), dpi=80)
print('---------------------')
print('\nCrashes that happened every hour for each BOROUGH before 2020:')
day_list = []
for entry in grp_by_borough_before_20:
# get borough
day_of_week = entry[0]
# sort the entries by hour
this_day_crashes = entry[1].sort_values(by=['HOUR'])
# get count of crashes that happened for every hour of day in the borough
crashes_by_hour = this_day_crashes['HOUR'].value_counts().sort_index(ascending=True)
print('\nFor ',day_of_week,'\nHOUR\tCrashes\n', crashes_by_hour)
day_list.append(day_of_week)
# plot the count of crashes that happened for every hour in the borough
crashes_by_hour.plot.line()
# give labels and add legend
plt.ylabel("No. of Crashes", labelpad=14)
plt.xlabel("Time of day", labelpad=14)
plt.legend(day_list)
plt.title("No. of Crashes for every Borough by Time of day for years before 2020", y=1.02)
# save the graph
plt.savefig('Results\crashes_by_borough_bef_20.png')
print("Saved No. of Crashes by Time of day for years before 2020 to Results\crashes_by_borough_bef_20.png")
plt.show()
# clear plot
print('---------------------')
plt.figure(figsize=(10, 6), dpi=80)
# ======== Retrieve and plot data for year 2020 ========
print('\nCrashes that happened every hour for each BOROUGH in 2020:')
day_list = []
for entry in grp_by_borough_during_20:
# get borough
day_of_week = entry[0]
# sort the entries by hour
this_day_crashes = entry[1].sort_values(by=['HOUR'])
# get count of crashes that happened for every hour in the borough
crashes_by_hour = this_day_crashes['HOUR'].value_counts().sort_index(ascending=True)
print('\nFor ',day_of_week,'\nHOUR\tCrashes\n', crashes_by_hour)
day_list.append(day_of_week)
# plot the count of crashes that happened for every hour of day in the borough
crashes_by_hour.plot.line()
# give labels and add legend
plt.ylabel("No. of Crashes", labelpad=14)
plt.xlabel("Time of day", labelpad=14)
plt.legend(day_list)
plt.title("No. of Crashes for every Borough by Time of day during 2020", y=1.02)
# save the graph
plt.savefig('Results\crashes_by_borough_in_20.png')
print("Saved No. of Crashes by Time of day during 2020 to Results\crashes_by_borough_in_20.png")
plt.show()
print('---------------------')
```
## Q7
For the years before 2020, given the entire region of NY City, which regions are “hot spots,” or places where accidents are most likely to occur? You will need to do some Parzen density estimation on this. You will need to use the GPS coordinates. You may need to convert the GPS coordinates from degrees:minutes:seconds, to degrees.fractions of degrees (fractional degrees). You do not need to use the Haversine distance. Assume that longitude and latitude is Euclidean for NY City. Create a “heat map” of where accidents occur and overlay it on a map of the city.
Do the hot spot locations change in 2020?
```
nyc = gpd.read_file(
gplt.datasets.get_path('nyc_boroughs')
)
nyc.head()
ax = gplt.polyplot(nyc,
edgecolor="white",
facecolor="lightgray",
figsize=(12, 8))
accidents = crash_data[(crash_data['LATITUDE'] != 0) & (crash_data['LONGITUDE'] != 0)]
accidents = accidents[['LATITUDE', 'LONGITUDE']].head(4000)
gdf = gpd.GeoDataFrame(
accidents,
geometry=gpd.points_from_xy(accidents['LONGITUDE'], accidents['LATITUDE']))
# gdf = gpd.read_file(gplt.datasets.get_path("nyc_collision_factors"))
gplt.kdeplot(gdf, ax=ax)
```
## Q8
Compare the number of car-only accidents (car and car or car and obstacle) with car-pedestrian accidents (car and person or car and bicycle). Do these proportions change in 2020? Do they change in any particular location?
```
before_20_total = len(before_20)
before_20_car_ped = before_20[(before_20['NUMBER OF PEDESTRIANS INJURED'] > 0) \
| (before_20['NUMBER OF PEDESTRIANS KILLED'] > 0) \
| (before_20['NUMBER OF CYCLIST INJURED'] > 0) \
| (before_20['NUMBER OF CYCLIST KILLED'] > 0)]
before_20_car_car = pd.merge( \
before_20, \
before_20_car_ped, \
on=before_20.columns.tolist(), \
how='outer', \
indicator=True).query("_merge == 'left_only'").drop('_merge', 1)
during_20_total = len(during_20)
during_20_car_ped = during_20[(during_20['NUMBER OF PEDESTRIANS INJURED'] > 0) \
| (during_20['NUMBER OF PEDESTRIANS KILLED'] > 0) \
| (during_20['NUMBER OF CYCLIST INJURED'] > 0) \
| (during_20['NUMBER OF CYCLIST KILLED'] > 0)]
during_20_car_car = pd.merge( \
during_20, \
during_20_car_ped, \
on=during_20.columns.tolist(), \
how='outer', \
indicator=True).query("_merge == 'left_only'").drop('_merge', 1)
accidents = pd.DataFrame(
{'Before 2020': [len(before_20_car_ped), len(before_20_car_car)],
'2020': [len(during_20_car_ped), len(during_20_car_car)] },
index=['Car-Ped', 'Car-Car'])
accidents['Before 2020'] /= before_20_total
accidents['2020'] /= during_20_total
accidents.plot.bar()
plt.show()
nyc = gpd.read_file(
gplt.datasets.get_path('nyc_boroughs')
)
ax = gplt.polyplot(nyc,
edgecolor="white",
facecolor="lightgray",
figsize=(12, 8))
accidents = before_20_car_car[(before_20_car_car['LATITUDE'] != 0) & (before_20_car_car['LONGITUDE'] != 0)]
accidents = accidents[['LATITUDE', 'LONGITUDE']].head(4000)
gdf = gpd.GeoDataFrame(
accidents,
geometry=gpd.points_from_xy(accidents['LONGITUDE'], accidents['LATITUDE']))
# gdf = gpd.read_file(gplt.datasets.get_path("nyc_collision_factors"))
gplt.kdeplot(gdf, ax=ax)
ax = gplt.polyplot(nyc,
edgecolor="white",
facecolor="lightgray",
figsize=(12, 8))
accidents = before_20_car_ped[(before_20_car_ped['LATITUDE'] != 0) & (before_20_car_ped['LONGITUDE'] != 0)]
accidents = accidents[['LATITUDE', 'LONGITUDE']].head(4000)
gdf = gpd.GeoDataFrame(
accidents,
geometry=gpd.points_from_xy(accidents['LONGITUDE'], accidents['LATITUDE']))
# gdf = gpd.read_file(gplt.datasets.get_path("nyc_collision_factors"))
gplt.kdeplot(gdf, ax=ax)
```
## Q9
While working on the data, did you discover anything else you wanted to explore?
Some students in the past found relationships between weather and numbers of accidents.
One team identified the surge in the number of accidents that happened after the change to/from daylight savings time.
```
# introduce a column for storing month the crash took place
before_20['MONTH'] = before_20['CRASH DATETIME'].dt.month
during_20['MONTH'] = during_20['CRASH DATETIME'].dt.month
# introduce a column for storing season the crash took place
before_20['SEASON'] = before_20["MONTH"].apply(lambda month: ["Winter","Spring","Summer","Fall"][(month-1)//3])
during_20['SEASON'] = during_20["MONTH"].apply(lambda month: ["Winter","Spring","Summer","Fall"][(month-1)//3])
# get the total number of crashes that happened in each season
crash_season_before_20 = before_20['SEASON'].value_counts(normalize=True) * 100
crash_season_during_20 = during_20['SEASON'].value_counts(normalize=True) * 100
crash_season_before_20.sort_index(inplace=True)
crash_season_during_20.sort_index(inplace=True)
# group crashes by season
grp_by_season_before_20 = before_20.groupby(['SEASON'])
grp_by_season_in_20 = during_20.groupby(['SEASON'])
print('\nPercentage of crashes by Season before 2020:\n\n', crash_season_before_20.to_string())
print('\nPercentage of crashes by Season during 2020:\n\n', crash_season_during_20.to_string())
for season_data in grp_by_season_before_20:
# get season
season = season_data[0]
# sort the entries by month
this_season_crashes = season_data[1].sort_values(by=['MONTH'])
crash_by_boroughs = this_season_crashes['BOROUGH'].value_counts(normalize=True)*100
print('\nPercentage of Crashes during %s season:'%season)
print(crash_by_boroughs.to_string())
for season_data in grp_by_season_in_20:
# get season
season = season_data[0]
# sort the entries by month
this_season_crashes = season_data[1].sort_values(by=['MONTH'])
crash_by_boroughs = this_season_crashes['BOROUGH'].value_counts(normalize=True)*100
print('\nPercentage of Crashes during %s season:'%season)
print(crash_by_boroughs.to_string())
# ======== Retrieve and plot data ========
print('---------------------')
# Stack the data
season_crashes = pd.concat([crash_season_before_20, crash_season_during_20], axis=1)
# Plot distributions
season_crashes.plot.bar(figsize=(9,8))
plt.ylabel("No. of Crashes", labelpad=6)
plt.title("Percentage of Crashes by Season", y=1.02)
plt.legend(['Before 2020','After 2020'])
plt.savefig('Results\crashes_by_season.png')
print('\nSaved the graph for Percentage of Crashes by Season of Week for '
'year before 2020 to crashes_by_season.png.')
# plt.show()
print('---------------------\n')
```
## Q10
Suppose I tell you that there is an accident in one of these certain locations:\
a. Hope, or \
b. Hunts Point, or \
c. Central Brooklyn, \
d. Brairwood, or \
e. West Bronx
what else can you tell me about that accident just by the location – even before we dispatch emergency vehicles? How would you build a classifier for this? Is it likely to be a car-car, or car-pedestrian, or car-bicycle?
```
# a. Hope,
# b. Hunts Point 40.813°N 73.884°W
# c. Central Brooklyn 40.697°N 73.917°W
# d. Brairwood 40.71°N 73.81°W
# e. West Bronx 40.850°N 73.900°W
crash_data_total = len(crash_data)
crash_data_car_other = crash_data[(crash_data['NUMBER OF PEDESTRIANS INJURED'] > 0) \
| (crash_data['NUMBER OF PEDESTRIANS KILLED'] > 0)\
| (crash_data['NUMBER OF CYCLIST INJURED'] > 0) \
| (crash_data['NUMBER OF CYCLIST KILLED'] > 0)\
| (crash_data['NUMBER OF MOTORIST INJURED'] > 0)\
| (crash_data['NUMBER OF MOTORIST INJURED'] > 0)]
crash_data_car_ped = crash_data[(crash_data['NUMBER OF PEDESTRIANS INJURED'] > 0) \
| (crash_data['NUMBER OF PEDESTRIANS KILLED'] > 0)]
crash_data_car_bike = crash_data[(crash_data['NUMBER OF CYCLIST INJURED'] > 0) \
| (crash_data['NUMBER OF CYCLIST KILLED'] > 0)]
crash_data_car_motorist = crash_data[(crash_data['NUMBER OF MOTORIST INJURED'] > 0) \
| (crash_data['NUMBER OF MOTORIST KILLED'] > 0)]
crash_data_car_car = pd.merge( \
crash_data, \
crash_data_car_other, \
on=crash_data.columns.tolist(), \
how='outer', \
indicator=True).query("_merge == 'left_only'").drop('_merge', 1)
# get the percentage of car-car crashes happened in each borough
car_car_by_boroughs = crash_data_car_car['BOROUGH'].value_counts()
car_car_by_boroughs.sort_index(inplace=True)
print("Percentage of car-car crashes happened in each borough:\n",car_car_by_boroughs)
# get the percentage of car-ped crashes happened in each borough
car_ped_by_boroughs = crash_data_car_ped['BOROUGH'].value_counts()
car_ped_by_boroughs.sort_index(inplace=True)
print("Percentage of car-ped crashes happened in each borough:\n",car_ped_by_boroughs)
# get the percentage of car-bike crashes happened in each borough
car_bike_by_boroughs = crash_data_car_bike['BOROUGH'].value_counts()
car_bike_by_boroughs.sort_index(inplace=True)
print("Percentage of car-bike crashes happened in each borough:\n",car_bike_by_boroughs)
# get the percentage of car-bike crashes happened in each borough
car_motorist_by_boroughs = crash_data_car_motorist['BOROUGH'].value_counts()
car_motorist_by_boroughs.sort_index(inplace=True)
print("Percentage of car-motorist crashes happened in each borough:\n",car_motorist_by_boroughs)
def plot_type_of_crashes(crashes_by_borough,type):
# ======== Retrieve and plot data ========
plt.figure(figsize=(4, 4), dpi=80)
print('---------------------')
print("No. of "+type+" Crashes in every Borough:\n",crashes_by_borough)
crashes_by_borough.plot.barh()
# give labels and add legend
plt.xlabel("No. of Crashes", labelpad=4)
plt.yticks(rotation=45)
# plt.xlim(0,40)
# plt.ylabel("Borough", labelpad=4)
plt.title("No. of "+type+" Crashes for every Borough", y=1.02)
# save the graph
plt.savefig('Results\\'+type+"_by_borough.png")
print("\nSaved No. of "+type+" Crashes for every Borough to "+type+"_by_borough.png")
plt.show()
print('---------------------')
plot_type_of_crashes(car_car_by_boroughs,'car_car')
plot_type_of_crashes(car_ped_by_boroughs,'car_ped')
plot_type_of_crashes(car_bike_by_boroughs,'car_bike')
plot_type_of_crashes(car_motorist_by_boroughs,'car_motorist')
grp_by_borough = crash_data.groupby(['BOROUGH'])
cols = ['Car-Car','Car-Ped','Car-Bike','Car-Motorist']
for entry in grp_by_borough:
borough = entry[0]
data = entry[1]
car_other = data[ (data['NUMBER OF PEDESTRIANS INJURED'] > 0) \
| (data['NUMBER OF PEDESTRIANS KILLED'] > 0)\
| (data['NUMBER OF CYCLIST INJURED'] > 0) \
| (data['NUMBER OF CYCLIST KILLED'] > 0)\
| (data['NUMBER OF MOTORIST INJURED'] > 0)\
| (data['NUMBER OF MOTORIST INJURED'] > 0)]
car_ped = data[(data['NUMBER OF PEDESTRIANS INJURED'] > 0) \
| (data['NUMBER OF PEDESTRIANS KILLED'] > 0)]
car_bike = data[(data['NUMBER OF CYCLIST INJURED'] > 0) \
| (data['NUMBER OF CYCLIST KILLED'] > 0)]
car_motorist = data[(data['NUMBER OF MOTORIST INJURED'] > 0) \
| (data['NUMBER OF MOTORIST KILLED'] > 0)]
car_car = pd.merge( \
data, \
car_other, \
on=data.columns.tolist(), \
how='outer', \
indicator=True).query("_merge == 'left_only'").drop('_merge', 1)
print('---------------------')
types = [len(car_car),len(car_ped),len(car_bike),len(car_motorist)]
print(types)
print(cols)
# df = pd.DataFrame({'Count': types,'Crash Type': cols, })
df = pd.DataFrame(types, index = cols, columns = ['count'])
df.plot.barh()
print(borough)
print(df)
# plt.figure(figsize=(10, 6), dpi=80)
# # ======== Retrieve and plot data for year 2020 ========
# plt.plot
# # give labels and add legend
plt.xlabel("No. of Crashes", labelpad=2)
# plt.ylabel("Type of crashes", labelpad=2)
plt.title("No of types of crashes for "+borough, y=1.02)
# save the graph
plt.savefig('Results\\'+borough+'_crash_types.png')
print("Saved No. of Crashes by "+borough+"_crash_types.png")
plt.show()
print('---------------------')
```
|
github_jupyter
|
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import warnings
warnings.filterwarnings('ignore')
%matplotlib inline
import geoplot as gplt
import geopandas as gpd
import geoplot.crs as gcrs
# please proceed onlyafter changing root directory and input file name
# Config for the project
root_dir = 'G:\Projects\\NYC-Crash-Data-Analytics\\'
# Input filename
filename = 'G:\Projects\\NYC-Crash-Data-Analytics\Motor_Vehicle_Collisions_-_Crashes.csv'
# Cleaned filename
clean_filename = 'G:\Projects\\NYC-Crash-Data-Analytics\Motor_Vehicle_Collisions_-_Crashes_Cleaned.csv'
# NY Borough Boundaries shapefile
nybb_shapefile = root_dir + 'maps\nybb.shp'
# Index column
index_col = 'COLLISION_ID'
# Data types for the columns in the data
dtypes = {
'CRASH DATE' : 'str',
'CRASH TIME' : 'str',
'BOROUGH' : 'str',
'ZIP CODE' : 'str',
'LATITUDE' : 'float64',
'LONGITUDE' : 'float64',
'LOCATION' : 'object',
'ON STREET NAME' : 'str',
'CROSS STREET NAME' : 'str',
'OFF STREET NAME' : 'str',
'NUMBER OF PERSONS INJURED' : 'float64',
'NUMBER OF PERSONS KILLED' : 'float64',
'NUMBER OF PEDESTRIANS INJURED' : 'float64',
'NUMBER OF PEDESTRIANS KILLED' : 'float64',
'NUMBER OF CYCLIST INJURED' : 'float64',
'NUMBER OF CYCLIST KILLED' : 'float64',
'NUMBER OF MOTORIST INJURED' : 'float64',
'NUMBER OF MOTORIST KILLED' : 'float64',
'CONTRIBUTING FACTOR VEHICLE 1' : 'str',
'CONTRIBUTING FACTOR VEHICLE 2' : 'str',
'CONTRIBUTING FACTOR VEHICLE 3' : 'str',
'CONTRIBUTING FACTOR VEHICLE 4' : 'str',
'CONTRIBUTING FACTOR VEHICLE 5' : 'str',
'COLLISION_ID' : 'int64',
'VEHICLE TYPE CODE 1' : 'category',
'VEHICLE TYPE CODE 2' : 'category',
'VEHICLE TYPE CODE 3' : 'category',
'VEHICLE TYPE CODE 4' : 'category',
'VEHICLE TYPE CODE 5' : 'category'
}
# Column-wise replacement values for NA
na_replace = {
'BOROUGH' : 'UNKNOWN',
'ZIP CODE' : 'UNKNOWN',
'LATITUDE' : 0,
'LONGITUDE' : 0,
'LOCATION' : '(0.0, 0.0)',
'ON STREET NAME' : '',
'CROSS STREET NAME' : '',
'OFF STREET NAME' : '',
'NUMBER OF PERSONS INJURED' : 0,
'NUMBER OF PERSONS KILLED' : 0,
'NUMBER OF PEDESTRIANS INJURED' : 0,
'NUMBER OF PEDESTRIANS KILLED' : 0,
'NUMBER OF CYCLIST INJURED' : 0,
'NUMBER OF CYCLIST KILLED' : 0,
'NUMBER OF MOTORIST INJURED' : 0,
'NUMBER OF MOTORIST KILLED' : 0,
'CONTRIBUTING FACTOR VEHICLE 1' : '',
'CONTRIBUTING FACTOR VEHICLE 2' : '',
'CONTRIBUTING FACTOR VEHICLE 3' : '',
'CONTRIBUTING FACTOR VEHICLE 4' : '',
'CONTRIBUTING FACTOR VEHICLE 5' : '',
'VEHICLE TYPE CODE 1' : '',
'VEHICLE TYPE CODE 2' : '',
'VEHICLE TYPE CODE 3' : '',
'VEHICLE TYPE CODE 4' : '',
'VEHICLE TYPE CODE 5' : ''
}
print('Reading CSV file %s ...' % filename)
crash_data = pd.read_csv(filename,
index_col=index_col,
dtype=dtypes,
parse_dates={'CRASH DATETIME' : ['CRASH DATE', 'CRASH TIME']},
infer_datetime_format=True
)
print('Filling NaN values ...')
for key, val in na_replace.items():
print('\t%s' % key)
crash_data[key] = crash_data[key].replace(np.nan, val)
print("Saving cleaned file to %s ..." % clean_filename)
crash_data.to_csv(clean_filename)
print("Cleaning complete.")
crash_data = pd.read_csv(clean_filename,
index_col=index_col,
parse_dates=['CRASH DATETIME'])
before_20 = crash_data[crash_data['CRASH DATETIME'].dt.year < 2020]
during_20 = crash_data[crash_data['CRASH DATETIME'].dt.year == 2020]
crash_data.head()
accidents = pd.DataFrame()
# == Compute the distribution for accidents by Borough for years before 2020 ===
accidents['Before 2020'] = before_20['BOROUGH'].value_counts()
accidents['Before 2020'] = accidents['Before 2020'] / accidents['Before 2020'] \
.sum()
# ====== Compute the distribution for accidents by Borough for year 2020 =======
accidents['2020'] = during_20['BOROUGH'].value_counts()
accidents['2020'] = accidents['2020'] / accidents['2020'].sum()
# Plot distributions
accidents.plot.bar()
plt.show()
accidents = pd.DataFrame()
# == Compute the distribution for accidents by Borough for years before 2020 ===
accidents['Before 2020'] = before_20['CRASH DATETIME'].dt.month.value_counts()
accidents['Before 2020'] = accidents['Before 2020'] / accidents['Before 2020'] \
.sum()
# ====== Compute the distribution for accidents by Borough for year 2020 =======
accidents['2020'] = during_20['CRASH DATETIME'].dt.month.value_counts()
accidents['2020'] = accidents['2020'] / accidents['2020'].sum()
print('February had %.2f%% fewer accidents than January.' \
% ((1 - (accidents['Before 2020'][2] / accidents['Before 2020'][1])) * 100))
# Stack the data
accidents = accidents.sort_index()
# Plot distributions
accidents.plot()
plt.show()
accidents = pd.DataFrame()
# == Compute the distribution for accidents by Borough for years before 2020 ===
accidents['Before 2020'] = before_20['CONTRIBUTING FACTOR VEHICLE 1'] \
.value_counts()
accidents['Before 2020'] = accidents['Before 2020'] / accidents['Before 2020'] \
.sum()
# ====== Compute the distribution for accidents by Borough for year 2020 =======
accidents['2020'] = during_20['CONTRIBUTING FACTOR VEHICLE 1'].value_counts()
accidents['2020'] = accidents['2020'] / accidents['2020'].sum()
# Plot distributions
accidents.plot.bar(figsize=(22,8), fontsize=20)
plt.show()
# introduce a column for storing day the crash took place
before_20['DAY'] = before_20['CRASH DATETIME'].dt.day_name()
during_20['DAY'] = during_20['CRASH DATETIME'].dt.day_name()
# get the total number of crashes that happened on each day
crash_day_before_20 = before_20['DAY'].value_counts(ascending=True,normalize=True)*100
crash_day_during_20 = during_20['DAY'].value_counts(ascending=True,normalize=True)*100
# plot the total number of crashes to day it took place
print('---------------------')
print('\nNumber of crashes by day before 2020:\n\n', crash_day_before_20.to_string())
crash_day_before_20.plot(kind='barh', figsize=(12, 8))
plt.xlabel("No. of Crashes", labelpad=14)
plt.ylabel("Day of Week", labelpad=14)
plt.title("No. of Crashes by Day of Week for year before 2020", y=1.02)
plt.savefig('Results\crashes_by_day_bef_20.png')
print('\nSaved the graph for No. of Crashes by Day of Week for '
'year before 2020 to crashes_by_day_bef_20.png.')
plt.show()
# plt.clf()
# plot the total number of crashes to day it took place
print('---------------------')
print('\nNumber of crashes by day during 2020:\n\n', crash_day_during_20.to_string())
crash_day_during_20.plot(kind='barh', figsize=(12, 8))
plt.xlabel("No. of Crashes", labelpad=14)
plt.ylabel("Day of Week", labelpad=14)
plt.title("No. of Crashes by Day of Week for year during 2020", y=1.02)
plt.savefig('Results\crashes_by_day_in_20.png')
print('\nSaved the graph for No. of Crashes by Day of Week for '
'year during 2020 to crashes_by_day_in_20.png.')
plt.show()
print('---------------------')
# Stack the data
season_crashes = pd.concat([crash_day_before_20, crash_day_during_20], axis=1)
# Plot distributions
season_crashes.plot.bar(figsize=(12,9))
plt.ylabel("Percentage of Crashes", labelpad=6)
plt.title("Percentage of Crashes by Day of week", y=1.02)
plt.legend(['Before 2020','After 2020'])
plt.savefig('Results\crashes_by_day')
print('\nSaved the graph for No. of Crashes by Day of Week to crashes_by_day.png.')
# introduce a column for storing day the crash took place
before_20['DAY'] = before_20['CRASH DATETIME'].dt.day_name()
during_20['DAY'] = during_20['CRASH DATETIME'].dt.day_name()
# introduce a column for storing day the crash took place
before_20['HOUR'] = before_20['CRASH DATETIME'].dt.hour
during_20['HOUR'] = during_20['CRASH DATETIME'].dt.hour
# group the data by day value
grp_by_day_before_20 = before_20.groupby(['DAY'])
grp_by_day_during_20 = during_20.groupby(['DAY'])
# ======== Retrieve and plot data for years before 2020 ========
plt.figure(figsize=(10, 6), dpi=80)
print('---------------------')
print('\nCrashes that happened every hour for each day before 2020:')
day_list = []
for entry in grp_by_day_before_20:
# get day of week
day_of_week = entry[0]
# sort the entries by hour
this_day_crashes = entry[1].sort_values(by=['HOUR'])
# get count of crashes that happened for every hour of day
crashes_by_hour = this_day_crashes['HOUR'].value_counts().sort_index(ascending=True)
print('\nFor ',day_of_week,'\nHOUR\tCrashes\n', crashes_by_hour)
day_list.append(day_of_week)
# plot the count of crashes that happened for every hour of day
crashes_by_hour.plot.line()
# give labels and add legend
plt.ylabel("No. of Crashes", labelpad=14)
plt.xlabel("Time of day", labelpad=14)
plt.legend(day_list)
plt.title("No. of Crashes by Time of day for years before 2020", y=1.02)
# save the graph
plt.savefig('Results\crashes_by_time_bef_20.png')
print("Saved No. of Crashes by Time of day for years before 2020 to crashes_by_time_bef_20.png")
plt.show()
# clear plot
# plt.cla()
print('---------------------')
# ======== Retrieve and plot data for year 2020 ========
print('\nCrashes that happened every hour for each day in 2020:')
day_list = []
plt.figure(figsize=(10, 6), dpi=80)
# give labels and add legend
plt.ylabel("No. of Crashes", labelpad=14)
plt.xlabel("Time of day", labelpad=14)
plt.title("No. of Crashes by Time of day during 2020", y=1.02)
for entry in grp_by_day_during_20:
# get day of week
day_of_week = entry[0]
# sort the entries by hour
this_day_crashes = entry[1].sort_values(by=['HOUR'])
# get count of crashes that happened for every hour of day
crashes_by_hour = this_day_crashes['HOUR'].value_counts().sort_index(ascending=True)
print('\nFor ',day_of_week,'\nHOUR\tCrashes\n', crashes_by_hour)
day_list.append(day_of_week)
# plot the count of crashes that happened for every hour of day
crashes_by_hour.plot.line()
plt.legend(day_list)
# save the graph
plt.savefig('Results\crashes_by_time_in_20.png')
print("Saved No. of Crashes by Time of day during 2020 to crashes_by_time_in_20.png")
plt.show()
print('---------------------')
# group the data by day value
grp_by_borough_before_20 = before_20.groupby(['BOROUGH'])
grp_by_borough_during_20 = during_20.groupby(['BOROUGH'])
# ======== Retrieve and plot data for years before 2020 ========
plt.figure(figsize=(10, 6), dpi=80)
print('---------------------')
print('\nCrashes that happened every hour for each BOROUGH before 2020:')
day_list = []
for entry in grp_by_borough_before_20:
# get borough
day_of_week = entry[0]
# sort the entries by hour
this_day_crashes = entry[1].sort_values(by=['HOUR'])
# get count of crashes that happened for every hour of day in the borough
crashes_by_hour = this_day_crashes['HOUR'].value_counts().sort_index(ascending=True)
print('\nFor ',day_of_week,'\nHOUR\tCrashes\n', crashes_by_hour)
day_list.append(day_of_week)
# plot the count of crashes that happened for every hour in the borough
crashes_by_hour.plot.line()
# give labels and add legend
plt.ylabel("No. of Crashes", labelpad=14)
plt.xlabel("Time of day", labelpad=14)
plt.legend(day_list)
plt.title("No. of Crashes for every Borough by Time of day for years before 2020", y=1.02)
# save the graph
plt.savefig('Results\crashes_by_borough_bef_20.png')
print("Saved No. of Crashes by Time of day for years before 2020 to Results\crashes_by_borough_bef_20.png")
plt.show()
# clear plot
print('---------------------')
plt.figure(figsize=(10, 6), dpi=80)
# ======== Retrieve and plot data for year 2020 ========
print('\nCrashes that happened every hour for each BOROUGH in 2020:')
day_list = []
for entry in grp_by_borough_during_20:
# get borough
day_of_week = entry[0]
# sort the entries by hour
this_day_crashes = entry[1].sort_values(by=['HOUR'])
# get count of crashes that happened for every hour in the borough
crashes_by_hour = this_day_crashes['HOUR'].value_counts().sort_index(ascending=True)
print('\nFor ',day_of_week,'\nHOUR\tCrashes\n', crashes_by_hour)
day_list.append(day_of_week)
# plot the count of crashes that happened for every hour of day in the borough
crashes_by_hour.plot.line()
# give labels and add legend
plt.ylabel("No. of Crashes", labelpad=14)
plt.xlabel("Time of day", labelpad=14)
plt.legend(day_list)
plt.title("No. of Crashes for every Borough by Time of day during 2020", y=1.02)
# save the graph
plt.savefig('Results\crashes_by_borough_in_20.png')
print("Saved No. of Crashes by Time of day during 2020 to Results\crashes_by_borough_in_20.png")
plt.show()
print('---------------------')
nyc = gpd.read_file(
gplt.datasets.get_path('nyc_boroughs')
)
nyc.head()
ax = gplt.polyplot(nyc,
edgecolor="white",
facecolor="lightgray",
figsize=(12, 8))
accidents = crash_data[(crash_data['LATITUDE'] != 0) & (crash_data['LONGITUDE'] != 0)]
accidents = accidents[['LATITUDE', 'LONGITUDE']].head(4000)
gdf = gpd.GeoDataFrame(
accidents,
geometry=gpd.points_from_xy(accidents['LONGITUDE'], accidents['LATITUDE']))
# gdf = gpd.read_file(gplt.datasets.get_path("nyc_collision_factors"))
gplt.kdeplot(gdf, ax=ax)
before_20_total = len(before_20)
before_20_car_ped = before_20[(before_20['NUMBER OF PEDESTRIANS INJURED'] > 0) \
| (before_20['NUMBER OF PEDESTRIANS KILLED'] > 0) \
| (before_20['NUMBER OF CYCLIST INJURED'] > 0) \
| (before_20['NUMBER OF CYCLIST KILLED'] > 0)]
before_20_car_car = pd.merge( \
before_20, \
before_20_car_ped, \
on=before_20.columns.tolist(), \
how='outer', \
indicator=True).query("_merge == 'left_only'").drop('_merge', 1)
during_20_total = len(during_20)
during_20_car_ped = during_20[(during_20['NUMBER OF PEDESTRIANS INJURED'] > 0) \
| (during_20['NUMBER OF PEDESTRIANS KILLED'] > 0) \
| (during_20['NUMBER OF CYCLIST INJURED'] > 0) \
| (during_20['NUMBER OF CYCLIST KILLED'] > 0)]
during_20_car_car = pd.merge( \
during_20, \
during_20_car_ped, \
on=during_20.columns.tolist(), \
how='outer', \
indicator=True).query("_merge == 'left_only'").drop('_merge', 1)
accidents = pd.DataFrame(
{'Before 2020': [len(before_20_car_ped), len(before_20_car_car)],
'2020': [len(during_20_car_ped), len(during_20_car_car)] },
index=['Car-Ped', 'Car-Car'])
accidents['Before 2020'] /= before_20_total
accidents['2020'] /= during_20_total
accidents.plot.bar()
plt.show()
nyc = gpd.read_file(
gplt.datasets.get_path('nyc_boroughs')
)
ax = gplt.polyplot(nyc,
edgecolor="white",
facecolor="lightgray",
figsize=(12, 8))
accidents = before_20_car_car[(before_20_car_car['LATITUDE'] != 0) & (before_20_car_car['LONGITUDE'] != 0)]
accidents = accidents[['LATITUDE', 'LONGITUDE']].head(4000)
gdf = gpd.GeoDataFrame(
accidents,
geometry=gpd.points_from_xy(accidents['LONGITUDE'], accidents['LATITUDE']))
# gdf = gpd.read_file(gplt.datasets.get_path("nyc_collision_factors"))
gplt.kdeplot(gdf, ax=ax)
ax = gplt.polyplot(nyc,
edgecolor="white",
facecolor="lightgray",
figsize=(12, 8))
accidents = before_20_car_ped[(before_20_car_ped['LATITUDE'] != 0) & (before_20_car_ped['LONGITUDE'] != 0)]
accidents = accidents[['LATITUDE', 'LONGITUDE']].head(4000)
gdf = gpd.GeoDataFrame(
accidents,
geometry=gpd.points_from_xy(accidents['LONGITUDE'], accidents['LATITUDE']))
# gdf = gpd.read_file(gplt.datasets.get_path("nyc_collision_factors"))
gplt.kdeplot(gdf, ax=ax)
# introduce a column for storing month the crash took place
before_20['MONTH'] = before_20['CRASH DATETIME'].dt.month
during_20['MONTH'] = during_20['CRASH DATETIME'].dt.month
# introduce a column for storing season the crash took place
before_20['SEASON'] = before_20["MONTH"].apply(lambda month: ["Winter","Spring","Summer","Fall"][(month-1)//3])
during_20['SEASON'] = during_20["MONTH"].apply(lambda month: ["Winter","Spring","Summer","Fall"][(month-1)//3])
# get the total number of crashes that happened in each season
crash_season_before_20 = before_20['SEASON'].value_counts(normalize=True) * 100
crash_season_during_20 = during_20['SEASON'].value_counts(normalize=True) * 100
crash_season_before_20.sort_index(inplace=True)
crash_season_during_20.sort_index(inplace=True)
# group crashes by season
grp_by_season_before_20 = before_20.groupby(['SEASON'])
grp_by_season_in_20 = during_20.groupby(['SEASON'])
print('\nPercentage of crashes by Season before 2020:\n\n', crash_season_before_20.to_string())
print('\nPercentage of crashes by Season during 2020:\n\n', crash_season_during_20.to_string())
for season_data in grp_by_season_before_20:
# get season
season = season_data[0]
# sort the entries by month
this_season_crashes = season_data[1].sort_values(by=['MONTH'])
crash_by_boroughs = this_season_crashes['BOROUGH'].value_counts(normalize=True)*100
print('\nPercentage of Crashes during %s season:'%season)
print(crash_by_boroughs.to_string())
for season_data in grp_by_season_in_20:
# get season
season = season_data[0]
# sort the entries by month
this_season_crashes = season_data[1].sort_values(by=['MONTH'])
crash_by_boroughs = this_season_crashes['BOROUGH'].value_counts(normalize=True)*100
print('\nPercentage of Crashes during %s season:'%season)
print(crash_by_boroughs.to_string())
# ======== Retrieve and plot data ========
print('---------------------')
# Stack the data
season_crashes = pd.concat([crash_season_before_20, crash_season_during_20], axis=1)
# Plot distributions
season_crashes.plot.bar(figsize=(9,8))
plt.ylabel("No. of Crashes", labelpad=6)
plt.title("Percentage of Crashes by Season", y=1.02)
plt.legend(['Before 2020','After 2020'])
plt.savefig('Results\crashes_by_season.png')
print('\nSaved the graph for Percentage of Crashes by Season of Week for '
'year before 2020 to crashes_by_season.png.')
# plt.show()
print('---------------------\n')
# a. Hope,
# b. Hunts Point 40.813°N 73.884°W
# c. Central Brooklyn 40.697°N 73.917°W
# d. Brairwood 40.71°N 73.81°W
# e. West Bronx 40.850°N 73.900°W
crash_data_total = len(crash_data)
crash_data_car_other = crash_data[(crash_data['NUMBER OF PEDESTRIANS INJURED'] > 0) \
| (crash_data['NUMBER OF PEDESTRIANS KILLED'] > 0)\
| (crash_data['NUMBER OF CYCLIST INJURED'] > 0) \
| (crash_data['NUMBER OF CYCLIST KILLED'] > 0)\
| (crash_data['NUMBER OF MOTORIST INJURED'] > 0)\
| (crash_data['NUMBER OF MOTORIST INJURED'] > 0)]
crash_data_car_ped = crash_data[(crash_data['NUMBER OF PEDESTRIANS INJURED'] > 0) \
| (crash_data['NUMBER OF PEDESTRIANS KILLED'] > 0)]
crash_data_car_bike = crash_data[(crash_data['NUMBER OF CYCLIST INJURED'] > 0) \
| (crash_data['NUMBER OF CYCLIST KILLED'] > 0)]
crash_data_car_motorist = crash_data[(crash_data['NUMBER OF MOTORIST INJURED'] > 0) \
| (crash_data['NUMBER OF MOTORIST KILLED'] > 0)]
crash_data_car_car = pd.merge( \
crash_data, \
crash_data_car_other, \
on=crash_data.columns.tolist(), \
how='outer', \
indicator=True).query("_merge == 'left_only'").drop('_merge', 1)
# get the percentage of car-car crashes happened in each borough
car_car_by_boroughs = crash_data_car_car['BOROUGH'].value_counts()
car_car_by_boroughs.sort_index(inplace=True)
print("Percentage of car-car crashes happened in each borough:\n",car_car_by_boroughs)
# get the percentage of car-ped crashes happened in each borough
car_ped_by_boroughs = crash_data_car_ped['BOROUGH'].value_counts()
car_ped_by_boroughs.sort_index(inplace=True)
print("Percentage of car-ped crashes happened in each borough:\n",car_ped_by_boroughs)
# get the percentage of car-bike crashes happened in each borough
car_bike_by_boroughs = crash_data_car_bike['BOROUGH'].value_counts()
car_bike_by_boroughs.sort_index(inplace=True)
print("Percentage of car-bike crashes happened in each borough:\n",car_bike_by_boroughs)
# get the percentage of car-bike crashes happened in each borough
car_motorist_by_boroughs = crash_data_car_motorist['BOROUGH'].value_counts()
car_motorist_by_boroughs.sort_index(inplace=True)
print("Percentage of car-motorist crashes happened in each borough:\n",car_motorist_by_boroughs)
def plot_type_of_crashes(crashes_by_borough,type):
# ======== Retrieve and plot data ========
plt.figure(figsize=(4, 4), dpi=80)
print('---------------------')
print("No. of "+type+" Crashes in every Borough:\n",crashes_by_borough)
crashes_by_borough.plot.barh()
# give labels and add legend
plt.xlabel("No. of Crashes", labelpad=4)
plt.yticks(rotation=45)
# plt.xlim(0,40)
# plt.ylabel("Borough", labelpad=4)
plt.title("No. of "+type+" Crashes for every Borough", y=1.02)
# save the graph
plt.savefig('Results\\'+type+"_by_borough.png")
print("\nSaved No. of "+type+" Crashes for every Borough to "+type+"_by_borough.png")
plt.show()
print('---------------------')
plot_type_of_crashes(car_car_by_boroughs,'car_car')
plot_type_of_crashes(car_ped_by_boroughs,'car_ped')
plot_type_of_crashes(car_bike_by_boroughs,'car_bike')
plot_type_of_crashes(car_motorist_by_boroughs,'car_motorist')
grp_by_borough = crash_data.groupby(['BOROUGH'])
cols = ['Car-Car','Car-Ped','Car-Bike','Car-Motorist']
for entry in grp_by_borough:
borough = entry[0]
data = entry[1]
car_other = data[ (data['NUMBER OF PEDESTRIANS INJURED'] > 0) \
| (data['NUMBER OF PEDESTRIANS KILLED'] > 0)\
| (data['NUMBER OF CYCLIST INJURED'] > 0) \
| (data['NUMBER OF CYCLIST KILLED'] > 0)\
| (data['NUMBER OF MOTORIST INJURED'] > 0)\
| (data['NUMBER OF MOTORIST INJURED'] > 0)]
car_ped = data[(data['NUMBER OF PEDESTRIANS INJURED'] > 0) \
| (data['NUMBER OF PEDESTRIANS KILLED'] > 0)]
car_bike = data[(data['NUMBER OF CYCLIST INJURED'] > 0) \
| (data['NUMBER OF CYCLIST KILLED'] > 0)]
car_motorist = data[(data['NUMBER OF MOTORIST INJURED'] > 0) \
| (data['NUMBER OF MOTORIST KILLED'] > 0)]
car_car = pd.merge( \
data, \
car_other, \
on=data.columns.tolist(), \
how='outer', \
indicator=True).query("_merge == 'left_only'").drop('_merge', 1)
print('---------------------')
types = [len(car_car),len(car_ped),len(car_bike),len(car_motorist)]
print(types)
print(cols)
# df = pd.DataFrame({'Count': types,'Crash Type': cols, })
df = pd.DataFrame(types, index = cols, columns = ['count'])
df.plot.barh()
print(borough)
print(df)
# plt.figure(figsize=(10, 6), dpi=80)
# # ======== Retrieve and plot data for year 2020 ========
# plt.plot
# # give labels and add legend
plt.xlabel("No. of Crashes", labelpad=2)
# plt.ylabel("Type of crashes", labelpad=2)
plt.title("No of types of crashes for "+borough, y=1.02)
# save the graph
plt.savefig('Results\\'+borough+'_crash_types.png')
print("Saved No. of Crashes by "+borough+"_crash_types.png")
plt.show()
print('---------------------')
| 0.435902 | 0.814016 |
<a href="https://colab.research.google.com/github/Drake-HeleneVeenstra/workshops/blob/main/MLworkshop_clustering.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# **Klustering algoritmer för segmentering - från statistik till maskininlärning**
**1. Som förberedning för klusteringanalys behöver vi ladda in vissa libraries samt skapa data.**
Dessa libraries innehåller visualiseringsmöjligheter (matplotlib och seaborn) och de faktiska clusterings algoritmer (sklearn.cluster, hdbscan). Numpy underlättar datamanipulering, och sklearn.datasets ger möjlighet att skapa data (liknande 'load inline' in Qlik).
Sen skapar vi data och plottar den. Du kan ändra parametrar i moons och blobs för att ändra datans utseende.
```
# 1 - skapa dataset och scatterplot
# installation behövs för att kunna köra hdbscan
!pip install hdbscan
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import sklearn.cluster as cluster
import sklearn.datasets as data
import hdbscan
plot_kwds = {'alpha': 0.5, 's': 80, 'linewidths': 0}
# Här skapar vi data, i form av månar och blobs. Ändra n_samples, noise, centers, cluster_std efter behag
moons, _ = data.make_moons(n_samples=100, noise=0.05)
blobs, _ = data.make_blobs(n_samples=100, centers=[(1, 2.25), (-1.0, 1.9)], cluster_std=0.2)
datastack = np.vstack([moons, blobs])
# Plot:
plt.scatter(datastack.T[0], datastack.T[1], c='b', **plot_kwds)
plt.title('1 Scatterplot of generated data', fontsize=20)
frame = plt.gca()
frame.axes.get_xaxis().set_visible(False)
frame.axes.get_yaxis().set_visible(False)
```
**2a. Vår första algoritm blir en K-means klustering algoritm.** I kwargs-argumentet finns några utvalda hyperparameters definierad som påverkar utfall av klustering. 'n_clusters' bestämmer i hur många kluster datasetet blir uppdelat. 'random_state' bestämmer random seed som utgångspunkt. Att definiera denna betyder samma resultat när man skulle testa algoritmen igen nästa vecka. Testa att ändra antal clusters i variabeln 'kwargs' from 5 till något annat.
```
# 2a - K-means clustering on dataset
kwargs = {'n_clusters': 4, 'random_state': 38}
algorithm = cluster.KMeans
labels = algorithm(**kwargs).fit_predict(datastack)
palette = sns.color_palette('muted', np.unique(labels).max() + 1)
colors = [palette[x] if x >= 0 else (0.0, 0.0, 0.0) for x in labels]
plt.scatter(datastack.T[0], datastack.T[1], c=colors)
frame = plt.gca()
frame.axes.get_xaxis().set_visible(False)
frame.axes.get_yaxis().set_visible(False)
plt.title('2a Clustering method: {}'.format(str(algorithm.__name__)),
fontsize=20);
```
>K-means är en partitioning algoritm som har som antagandet att det finns globala partitions som är centroid-based. Denna algoritm behöver input om hur många klusters det finns att hitta i datasettet. Prestanda på denna dataset är dålig, den grupperar och segrererar tydlig felaktig, för den kan inte fånga en måna som en segment.
**2b. Vi tar en närmare titt på hur man kan modifiera andra hyperparametrar än antal kluster för att komma närmare verkligheten.** För det skapar vi ett andra dataset som består utav tre blobs, två nära varandra och en blob med stor spridning längre bort. I denna figur har vi inte än applicerat en klusteringalgoritm, färgerna i scatterplot visar bara hur de tre originalblobs är definierat.
```
# 2b - skapa ny dataset med spridda kluster, och plotta de tre blobs i olika färger
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import sklearn.cluster as cluster
import sklearn.datasets as data
plot_kwds = {'alpha': 0.5, 's': 80, 'linewidths': 0}
# Nu skapar vi bara blobs (inga moons), med större spridning
blobs_tmp_1, _ = data.make_blobs(n_samples=300, centers=[(1.2, 2.25)], cluster_std=0.4)
blobs_tmp_2, _ = data.make_blobs(n_samples=300, centers=[(2, 1)], cluster_std=0.4)
blobs_tmp_3, _ = data.make_blobs(n_samples=300, centers=[(20, 5)], cluster_std=5)
datastack_tmp = np.vstack([blobs_tmp_1, blobs_tmp_2, blobs_tmp_3])
# Plot:
plt.scatter(blobs_tmp_1.T[0], blobs_tmp_1.T[1], c='b', **plot_kwds)
plt.scatter(blobs_tmp_2.T[0], blobs_tmp_2.T[1], c='r', **plot_kwds)
plt.scatter(blobs_tmp_3.T[0], blobs_tmp_3.T[1], c='g', **plot_kwds)
plt.title('2b Scatterplot of generated data',fontsize=20)
frame = plt.gca()
frame.axes.get_xaxis().set_visible(False)
frame.axes.get_yaxis().set_visible(False)
```
**2c. Applicering av en K-means klustering likadant som i 2a**
```
# 2c - K-means algoritm
kwargs_tmp = {'n_clusters': 3, 'random_state': 38, 'init': 'random'}
algorithm = cluster.KMeans
labels = algorithm(**kwargs_tmp).fit_predict(datastack_tmp)
palette = sns.color_palette('muted', np.unique(labels).max() + 1)
colors = [palette[x] if x >= 0 else (0.0, 0.0, 0.0) for x in labels]
plt.scatter(datastack_tmp.T[0], datastack_tmp.T[1], c=colors)
frame = plt.gca()
frame.axes.get_xaxis().set_visible(False)
frame.axes.get_yaxis().set_visible(False)
plt.title('2c Clustering method: {}'.format(str(algorithm.__name__)),
fontsize=20);
```
**2d.** Det syns tydlig i figuren 2c ovan att klustering blir fel, stora spridda blobben är egentligen en blob och borde falla i en kluster/segment. De två små blobs till vänster blir sammanslagen i en och samma segment.
Dock har vi kunskap om vår data, vi vet ju vart blobbens centroid ligger. I riktig data har vi inte exakt information om det, men vi kan ha ungefärlig beskrivning av en centroid. **Vi kan lägga centroids som utgångsinformation för klusterdefinition i modellen:**
```
# 2d - vi definierar centroids som vi hade kunskap om som utgångspunkt av klusterdefinition ('init')
centers=np.asarray([[1, 2.25], [2, 1], [20, 5]])
kwargs_tmp = {'n_clusters': 3, 'random_state': 38, 'init': centers, 'n_init': 1}
algorithm = cluster.KMeans
labels = algorithm(**kwargs_tmp).fit_predict(datastack_tmp)
palette = sns.color_palette('muted', np.unique(labels).max() + 1)
colors = [palette[x] if x >= 0 else (0.0, 0.0, 0.0) for x in labels]
plt.scatter(datastack_tmp.T[0], datastack_tmp.T[1], c=colors)
frame = plt.gca()
frame.axes.get_xaxis().set_visible(False)
frame.axes.get_yaxis().set_visible(False)
plt.title('2d Clustering method: {}'.format(str(algorithm.__name__)),
fontsize=20);
```
>Det finns fler sätt att optimera K-means kluster. T.ex. kan du gå tillbaka till kod i 2c och testa att separera i fler än tre kluster. Blev det bättre resultat?
**3.** De månar i vårt originaldataset går inte att separera med en K-means klustering. Vi går vidare och testar flera algoritmer, och ändra några av deras hyperparametrar för att se huruvida dessa går att optimera. **Vi testar en Affinity Propagation algoritm näst**:
```
# 3 - Affinity Propagation
kwargs = {'preference': -6.0, 'damping': 0.85}
algorithm = cluster.AffinityPropagation
labels = algorithm(**kwargs).fit_predict(datastack)
palette = sns.color_palette('muted', np.unique(labels).max() + 1)
colors = [palette[x] if x >= 0 else (0.0, 0.0, 0.0) for x in labels]
plt.scatter(datastack.T[0], datastack.T[1], c=colors)
frame = plt.gca()
frame.axes.get_xaxis().set_visible(False)
frame.axes.get_yaxis().set_visible(False)
plt.title('3 Clustering method: {}'.format(str(algorithm.__name__)),
fontsize=20);
```
>Affinity Propagation är en graph-baserad point-voting system, följd av en centroid-baserad partitioning approach. Fördelen med den över K-means är att man inte behöver bestämma på förhand hur många segmenter det ska hittas. Nackdelen är att den har inte bättre performance än K-means i denna dataset. Dessutom är de hyperparametrar i denna modell inte lätt-optimerad, dessutom är algoritmen långsammare än K-means. Eftersom denna modell förbättrar inte vårt segmentering jämfört med K-means så diskuterar vi inte den i djupet men går vi vidare.
**4. Mean Shift clustering. Igen en centroid-baserad algoritm. Denna algoritm gör riktig klustering snarare än partitioning.** Skillnaden är att en partitioning algoritm fungerar som en top-down gruppering av alla datapunkter, vilket föredrar en mindre antal större grupper. Partitioning börjar istället bottom-up, och utgår från konnektiviteten mellan punkter. Om vi väljer 'cluster_all': False så visas att inte alla punkter rimligtvis tillhör någon segment. Ändra till True för att tvinga alla punkter att falla i en segment, och se att det försämras segmenteringen.
>
```
# 4 - Mean Shift clustering
kwargs = {'cluster_all': False}
algorithm = cluster.MeanShift
labels = algorithm(0.3, **kwargs).fit_predict(datastack)
palette = sns.color_palette('muted', np.unique(labels).max() + 1)
colors = [palette[x] if x >= 0 else (0.0, 0.0, 0.0) for x in labels]
plt.scatter(datastack.T[0], datastack.T[1], c=colors)
frame = plt.gca()
frame.axes.get_xaxis().set_visible(False)
frame.axes.get_yaxis().set_visible(False)
plt.title('4 Clustering method: {}'.format(str(algorithm.__name__)),
fontsize=20);
```
Vi har fortfarande inte sett en tydlig förbättring i prestanda av modellen. Det finns fler algoritmer att testa, dock de kommer inte att göra en förbättring för just den dataset. Om du vill kan du dock testa i kod 2a de följande algoritmer genom att substituera cluster.KMeans med cluster.AgglomerativeClustering eller cluster.SpectralClustering; välj antal kluster i kwargs med 'n_cluster' och ta bort 'random_state' parameter. Vi fortsätter här med någonting helt annorlunda:
5. DBSCAN står för density-based spatial clustering of application with noise. Som namnet antyder gör den en klustering baserat på density; hur tight punkter ligger tillsammans med sina grannar. DBSCAN visar ett tydlig överlägsen prestanda jämfört med de tidigare modeller. eps (epsilon) är en parameter som beskriver grannområdet från varje datapunkt modellen väljer som potentiell startpunkt för en kluster. min_samples står för minimal antal punkter som beskriver en 'dense region', alltså hur många punkter tight tillsammans är trovärdigt för dig som utgångspunkt (core point) för en kluster. Testa olika värden på dessa hyperparametrar i kwargs för att påverka klustering.
```
kwargs = {'eps': 0.25, 'min_samples': 5}
algorithm = cluster.DBSCAN
labels = algorithm(**kwargs).fit_predict(datastack)
palette = sns.color_palette('muted', np.unique(labels).max() + 1)
colors = [palette[x] if x >= 0 else (0.0, 0.0, 0.0) for x in labels]
plt.scatter(datastack.T[0], datastack.T[1], c=colors)
frame = plt.gca()
frame.axes.get_xaxis().set_visible(False)
frame.axes.get_yaxis().set_visible(False)
plt.title('Clustering method: {}'.format(str(algorithm.__name__)),
fontsize=20);
```
6. Vi har sparat det bästa för sist. HDBSCAN är en optimerad DBSCAN, h står för hierarchical. Alla punkter har en attribut vilket beskriver hur starkt membership till en viss kluster är. Så vi kan plotta det som varierande saturation, för att visa skillnad mellan core och ytterkanten av en kluster. Detta är direkt användbart om man tänker riktig data; för nu kan vi visa skillnaden mellan kunder / produkten som är typiskt för en segment jämfört med kunder/produkter som kan tillhöra en segment men har vissa avvikande kännetecken.
```
clusterer = hdbscan.HDBSCAN(algorithm='best', alpha=1.0,
approx_min_span_tree=True,
gen_min_span_tree=True, leaf_size=40,
metric='euclidean', min_cluster_size=6,
min_samples=None, p=None,
cluster_selection_method='eom')
clusterer = clusterer.fit(datastack)
palette = sns.color_palette('deep')
cluster_colors = [sns.desaturate(palette[col], sat)
if col >= 0 else (0.5, 0.5, 0.5) for col, sat in
zip(clusterer.labels_, clusterer.probabilities_)]
plt.scatter(datastack.T[0], datastack.T[1], c=cluster_colors, **plot_kwds)
frame = plt.gca()
frame.axes.get_xaxis().set_visible(False)
frame.axes.get_yaxis().set_visible(False)
plt.title('Clustering method: {} (leaf)'.format(str(hdbscan.HDBSCAN.__name__)),
fontsize=20);
```
Om du vill leka mer så är det följande att rekommendera:
* Gå till https://scikit-learn.org/stable/modules/classes.html#module-sklearn.cluster och hitta olika algoritmer, du kan ta en algoritm diskuterat här och lägga till fler hyperparametrar för finetuning.
* Ta datasettet från början och modulera den. Annorlunda dataset ger annorlunda prestanda av olika modeller. Om datasetet är enkelt så räcker det med enkel klustering. Dock du kommer att märka att (H)DBSCAN har överlägset prestanda och är ett bra klusteringval.
|
github_jupyter
|
# 1 - skapa dataset och scatterplot
# installation behövs för att kunna köra hdbscan
!pip install hdbscan
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import sklearn.cluster as cluster
import sklearn.datasets as data
import hdbscan
plot_kwds = {'alpha': 0.5, 's': 80, 'linewidths': 0}
# Här skapar vi data, i form av månar och blobs. Ändra n_samples, noise, centers, cluster_std efter behag
moons, _ = data.make_moons(n_samples=100, noise=0.05)
blobs, _ = data.make_blobs(n_samples=100, centers=[(1, 2.25), (-1.0, 1.9)], cluster_std=0.2)
datastack = np.vstack([moons, blobs])
# Plot:
plt.scatter(datastack.T[0], datastack.T[1], c='b', **plot_kwds)
plt.title('1 Scatterplot of generated data', fontsize=20)
frame = plt.gca()
frame.axes.get_xaxis().set_visible(False)
frame.axes.get_yaxis().set_visible(False)
# 2a - K-means clustering on dataset
kwargs = {'n_clusters': 4, 'random_state': 38}
algorithm = cluster.KMeans
labels = algorithm(**kwargs).fit_predict(datastack)
palette = sns.color_palette('muted', np.unique(labels).max() + 1)
colors = [palette[x] if x >= 0 else (0.0, 0.0, 0.0) for x in labels]
plt.scatter(datastack.T[0], datastack.T[1], c=colors)
frame = plt.gca()
frame.axes.get_xaxis().set_visible(False)
frame.axes.get_yaxis().set_visible(False)
plt.title('2a Clustering method: {}'.format(str(algorithm.__name__)),
fontsize=20);
# 2b - skapa ny dataset med spridda kluster, och plotta de tre blobs i olika färger
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import sklearn.cluster as cluster
import sklearn.datasets as data
plot_kwds = {'alpha': 0.5, 's': 80, 'linewidths': 0}
# Nu skapar vi bara blobs (inga moons), med större spridning
blobs_tmp_1, _ = data.make_blobs(n_samples=300, centers=[(1.2, 2.25)], cluster_std=0.4)
blobs_tmp_2, _ = data.make_blobs(n_samples=300, centers=[(2, 1)], cluster_std=0.4)
blobs_tmp_3, _ = data.make_blobs(n_samples=300, centers=[(20, 5)], cluster_std=5)
datastack_tmp = np.vstack([blobs_tmp_1, blobs_tmp_2, blobs_tmp_3])
# Plot:
plt.scatter(blobs_tmp_1.T[0], blobs_tmp_1.T[1], c='b', **plot_kwds)
plt.scatter(blobs_tmp_2.T[0], blobs_tmp_2.T[1], c='r', **plot_kwds)
plt.scatter(blobs_tmp_3.T[0], blobs_tmp_3.T[1], c='g', **plot_kwds)
plt.title('2b Scatterplot of generated data',fontsize=20)
frame = plt.gca()
frame.axes.get_xaxis().set_visible(False)
frame.axes.get_yaxis().set_visible(False)
# 2c - K-means algoritm
kwargs_tmp = {'n_clusters': 3, 'random_state': 38, 'init': 'random'}
algorithm = cluster.KMeans
labels = algorithm(**kwargs_tmp).fit_predict(datastack_tmp)
palette = sns.color_palette('muted', np.unique(labels).max() + 1)
colors = [palette[x] if x >= 0 else (0.0, 0.0, 0.0) for x in labels]
plt.scatter(datastack_tmp.T[0], datastack_tmp.T[1], c=colors)
frame = plt.gca()
frame.axes.get_xaxis().set_visible(False)
frame.axes.get_yaxis().set_visible(False)
plt.title('2c Clustering method: {}'.format(str(algorithm.__name__)),
fontsize=20);
# 2d - vi definierar centroids som vi hade kunskap om som utgångspunkt av klusterdefinition ('init')
centers=np.asarray([[1, 2.25], [2, 1], [20, 5]])
kwargs_tmp = {'n_clusters': 3, 'random_state': 38, 'init': centers, 'n_init': 1}
algorithm = cluster.KMeans
labels = algorithm(**kwargs_tmp).fit_predict(datastack_tmp)
palette = sns.color_palette('muted', np.unique(labels).max() + 1)
colors = [palette[x] if x >= 0 else (0.0, 0.0, 0.0) for x in labels]
plt.scatter(datastack_tmp.T[0], datastack_tmp.T[1], c=colors)
frame = plt.gca()
frame.axes.get_xaxis().set_visible(False)
frame.axes.get_yaxis().set_visible(False)
plt.title('2d Clustering method: {}'.format(str(algorithm.__name__)),
fontsize=20);
# 3 - Affinity Propagation
kwargs = {'preference': -6.0, 'damping': 0.85}
algorithm = cluster.AffinityPropagation
labels = algorithm(**kwargs).fit_predict(datastack)
palette = sns.color_palette('muted', np.unique(labels).max() + 1)
colors = [palette[x] if x >= 0 else (0.0, 0.0, 0.0) for x in labels]
plt.scatter(datastack.T[0], datastack.T[1], c=colors)
frame = plt.gca()
frame.axes.get_xaxis().set_visible(False)
frame.axes.get_yaxis().set_visible(False)
plt.title('3 Clustering method: {}'.format(str(algorithm.__name__)),
fontsize=20);
# 4 - Mean Shift clustering
kwargs = {'cluster_all': False}
algorithm = cluster.MeanShift
labels = algorithm(0.3, **kwargs).fit_predict(datastack)
palette = sns.color_palette('muted', np.unique(labels).max() + 1)
colors = [palette[x] if x >= 0 else (0.0, 0.0, 0.0) for x in labels]
plt.scatter(datastack.T[0], datastack.T[1], c=colors)
frame = plt.gca()
frame.axes.get_xaxis().set_visible(False)
frame.axes.get_yaxis().set_visible(False)
plt.title('4 Clustering method: {}'.format(str(algorithm.__name__)),
fontsize=20);
kwargs = {'eps': 0.25, 'min_samples': 5}
algorithm = cluster.DBSCAN
labels = algorithm(**kwargs).fit_predict(datastack)
palette = sns.color_palette('muted', np.unique(labels).max() + 1)
colors = [palette[x] if x >= 0 else (0.0, 0.0, 0.0) for x in labels]
plt.scatter(datastack.T[0], datastack.T[1], c=colors)
frame = plt.gca()
frame.axes.get_xaxis().set_visible(False)
frame.axes.get_yaxis().set_visible(False)
plt.title('Clustering method: {}'.format(str(algorithm.__name__)),
fontsize=20);
clusterer = hdbscan.HDBSCAN(algorithm='best', alpha=1.0,
approx_min_span_tree=True,
gen_min_span_tree=True, leaf_size=40,
metric='euclidean', min_cluster_size=6,
min_samples=None, p=None,
cluster_selection_method='eom')
clusterer = clusterer.fit(datastack)
palette = sns.color_palette('deep')
cluster_colors = [sns.desaturate(palette[col], sat)
if col >= 0 else (0.5, 0.5, 0.5) for col, sat in
zip(clusterer.labels_, clusterer.probabilities_)]
plt.scatter(datastack.T[0], datastack.T[1], c=cluster_colors, **plot_kwds)
frame = plt.gca()
frame.axes.get_xaxis().set_visible(False)
frame.axes.get_yaxis().set_visible(False)
plt.title('Clustering method: {} (leaf)'.format(str(hdbscan.HDBSCAN.__name__)),
fontsize=20);
| 0.48438 | 0.901531 |
## Applicazione del metodo del perceptron per la separazione lineare
```
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
import matplotlib.colors as mcolors
from matplotlib import cm
plt.style.use('fivethirtyeight')
plt.rcParams['font.family'] = 'sans-serif'
plt.rcParams['font.serif'] = 'Ubuntu'
plt.rcParams['font.monospace'] = 'Ubuntu Mono'
plt.rcParams['font.size'] = 10
plt.rcParams['axes.labelsize'] = 10
plt.rcParams['axes.labelweight'] = 'bold'
plt.rcParams['axes.titlesize'] = 10
plt.rcParams['xtick.labelsize'] = 8
plt.rcParams['ytick.labelsize'] = 8
plt.rcParams['legend.fontsize'] = 10
plt.rcParams['figure.titlesize'] = 12
plt.rcParams['image.cmap'] = 'jet'
plt.rcParams['image.interpolation'] = 'none'
plt.rcParams['figure.figsize'] = (16, 8)
plt.rcParams['lines.linewidth'] = 2
plt.rcParams['lines.markersize'] = 8
colors = ['xkcd:pale orange', 'xkcd:sea blue', 'xkcd:pale red', 'xkcd:sage green', 'xkcd:terra cotta', 'xkcd:dull purple', 'xkcd:teal', 'xkcd:goldenrod', 'xkcd:cadet blue',
'xkcd:scarlet']
cmap_big = cm.get_cmap('Spectral', 512)
cmap = mcolors.ListedColormap(cmap_big(np.linspace(0.7, 0.95, 256)))
```
normalizzazione dei valori delle features a media 0 e varianza 1
```
def normalizza(X):
mu = np.mean(X, axis=0)
sigma = np.std(X, axis=0, ddof=1)
return (X-mu)/sigma
```
produce e restituisce una serie di statistiche
```
def statistics(theta,X,t):
# applica il modello
y = np.dot(X,theta)
# attribuisce i punti alle due classi
y = np.where(y>0, 1, 0)
# costruisce la confusion matrix
confmat = np.zeros((2, 2))
for i in range(2):
for j in range(2):
confmat[i,j] = np.sum(np.where(y==i,1,0)*np.where(t==j,1,0))
return confmat
```
legge i dati in dataframe pandas
```
data = pd.read_csv("../dataset/ex2data1.txt", header=0, delimiter=',', names=['x1','x2','t'])
# calcola dimensione dei dati
n = len(data)
# calcola dimensionalità delle features
nfeatures = len(data.columns)-1
X = np.array(data[['x1','x2']])
t = np.array(data['t']).reshape(-1,1)
c=[colors[i] for i in np.nditer(t)]
deltax = max(X[:,0])-min(X[:,0])
deltay = max(X[:,1])-min(X[:,1])
minx = min(X[:,0])-deltax/10.0
maxx = max(X[:,0])+deltax/10.0
miny = min(X[:,1])-deltay/10.0
maxy = max(X[:,1])+deltay/10.0
fig = plt.figure(figsize=(10,10))
fig.patch.set_facecolor('white')
ax = fig.gca()
ax.scatter(X[:,0],X[:,1],s=40,c=c, marker='o', alpha=.7)
t1 = np.arange(minx, maxx,0.01)
plt.xlabel('x1', fontsize=10)
plt.ylabel('x2', fontsize=10)
plt.xticks(fontsize=10)
plt.yticks(fontsize=10)
plt.show()
X=normalizza(X)
```
aggiunge una colonna di valori -1 alla matrice delle features
```
X= np.column_stack((-np.ones(n), X))
# fissa il valore del parametro eta
eta = 0.25
# fissa una valore iniziale casuale per i coefficienti
theta = np.random.rand(nfeatures+1, 1) * 0.1 - 0.05
deltax = max(X[:,1])-min(X[:,1])
deltay = max(X[:,2])-min(X[:,2])
minx = min(X[:,1])-deltax/10.0
maxx = max(X[:,1])+deltax/10.0
miny = min(X[:,2])-deltay/10.0
maxy = max(X[:,2])+deltay/10.0
t1 = np.arange(minx, maxx, 0.01)
thetas = []
thetas.append(theta.copy())
for k in range(2*n+1):
# determina la classificazione effettuata applicando la soglia 0-1 ai valori forniti
# dalla combinazione lineare delle features e dei coefficienti
y = np.where(np.dot(X, theta)>0,1,0)
# somma o sottrae eta da tutti i coefficienti per ogni osservazione mal classificata
theta += eta * np.dot(X.T, t-y)
thetas.append(theta.copy())
k=30
theta = thetas[k]
cf=statistics(theta,X,t)
cost=np.trace(cf)/n
theta1=-theta[1]/theta[2]
theta0=theta[0]/theta[2]
t2 = theta0+theta1*t1
plt.figure(figsize=(12,12))
plt.scatter(X[:,1],X[:,2],s=30,c=c, marker='o', alpha=.9)
plt.plot(t1, theta0+theta1*t1, color=colors[3], linewidth=2, alpha=1)
plt.xlabel('x1', fontsize=10)
plt.ylabel('x2', fontsize=10)
plt.xticks(fontsize=10)
plt.yticks(fontsize=10)
plt.xlim(minx,maxx)
plt.ylim(miny,maxy)
plt.annotate('Step {0:d}, costo = {1:3.3f}'.format(k,cost), xy=(.03, .97), backgroundcolor='w', va='top',
xycoords='axes fraction', fontsize=12)
plt.show()
cf
```
|
github_jupyter
|
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
import matplotlib.colors as mcolors
from matplotlib import cm
plt.style.use('fivethirtyeight')
plt.rcParams['font.family'] = 'sans-serif'
plt.rcParams['font.serif'] = 'Ubuntu'
plt.rcParams['font.monospace'] = 'Ubuntu Mono'
plt.rcParams['font.size'] = 10
plt.rcParams['axes.labelsize'] = 10
plt.rcParams['axes.labelweight'] = 'bold'
plt.rcParams['axes.titlesize'] = 10
plt.rcParams['xtick.labelsize'] = 8
plt.rcParams['ytick.labelsize'] = 8
plt.rcParams['legend.fontsize'] = 10
plt.rcParams['figure.titlesize'] = 12
plt.rcParams['image.cmap'] = 'jet'
plt.rcParams['image.interpolation'] = 'none'
plt.rcParams['figure.figsize'] = (16, 8)
plt.rcParams['lines.linewidth'] = 2
plt.rcParams['lines.markersize'] = 8
colors = ['xkcd:pale orange', 'xkcd:sea blue', 'xkcd:pale red', 'xkcd:sage green', 'xkcd:terra cotta', 'xkcd:dull purple', 'xkcd:teal', 'xkcd:goldenrod', 'xkcd:cadet blue',
'xkcd:scarlet']
cmap_big = cm.get_cmap('Spectral', 512)
cmap = mcolors.ListedColormap(cmap_big(np.linspace(0.7, 0.95, 256)))
def normalizza(X):
mu = np.mean(X, axis=0)
sigma = np.std(X, axis=0, ddof=1)
return (X-mu)/sigma
def statistics(theta,X,t):
# applica il modello
y = np.dot(X,theta)
# attribuisce i punti alle due classi
y = np.where(y>0, 1, 0)
# costruisce la confusion matrix
confmat = np.zeros((2, 2))
for i in range(2):
for j in range(2):
confmat[i,j] = np.sum(np.where(y==i,1,0)*np.where(t==j,1,0))
return confmat
data = pd.read_csv("../dataset/ex2data1.txt", header=0, delimiter=',', names=['x1','x2','t'])
# calcola dimensione dei dati
n = len(data)
# calcola dimensionalità delle features
nfeatures = len(data.columns)-1
X = np.array(data[['x1','x2']])
t = np.array(data['t']).reshape(-1,1)
c=[colors[i] for i in np.nditer(t)]
deltax = max(X[:,0])-min(X[:,0])
deltay = max(X[:,1])-min(X[:,1])
minx = min(X[:,0])-deltax/10.0
maxx = max(X[:,0])+deltax/10.0
miny = min(X[:,1])-deltay/10.0
maxy = max(X[:,1])+deltay/10.0
fig = plt.figure(figsize=(10,10))
fig.patch.set_facecolor('white')
ax = fig.gca()
ax.scatter(X[:,0],X[:,1],s=40,c=c, marker='o', alpha=.7)
t1 = np.arange(minx, maxx,0.01)
plt.xlabel('x1', fontsize=10)
plt.ylabel('x2', fontsize=10)
plt.xticks(fontsize=10)
plt.yticks(fontsize=10)
plt.show()
X=normalizza(X)
X= np.column_stack((-np.ones(n), X))
# fissa il valore del parametro eta
eta = 0.25
# fissa una valore iniziale casuale per i coefficienti
theta = np.random.rand(nfeatures+1, 1) * 0.1 - 0.05
deltax = max(X[:,1])-min(X[:,1])
deltay = max(X[:,2])-min(X[:,2])
minx = min(X[:,1])-deltax/10.0
maxx = max(X[:,1])+deltax/10.0
miny = min(X[:,2])-deltay/10.0
maxy = max(X[:,2])+deltay/10.0
t1 = np.arange(minx, maxx, 0.01)
thetas = []
thetas.append(theta.copy())
for k in range(2*n+1):
# determina la classificazione effettuata applicando la soglia 0-1 ai valori forniti
# dalla combinazione lineare delle features e dei coefficienti
y = np.where(np.dot(X, theta)>0,1,0)
# somma o sottrae eta da tutti i coefficienti per ogni osservazione mal classificata
theta += eta * np.dot(X.T, t-y)
thetas.append(theta.copy())
k=30
theta = thetas[k]
cf=statistics(theta,X,t)
cost=np.trace(cf)/n
theta1=-theta[1]/theta[2]
theta0=theta[0]/theta[2]
t2 = theta0+theta1*t1
plt.figure(figsize=(12,12))
plt.scatter(X[:,1],X[:,2],s=30,c=c, marker='o', alpha=.9)
plt.plot(t1, theta0+theta1*t1, color=colors[3], linewidth=2, alpha=1)
plt.xlabel('x1', fontsize=10)
plt.ylabel('x2', fontsize=10)
plt.xticks(fontsize=10)
plt.yticks(fontsize=10)
plt.xlim(minx,maxx)
plt.ylim(miny,maxy)
plt.annotate('Step {0:d}, costo = {1:3.3f}'.format(k,cost), xy=(.03, .97), backgroundcolor='w', va='top',
xycoords='axes fraction', fontsize=12)
plt.show()
cf
| 0.216508 | 0.837354 |
# Introduction
<center><img src="https://i.imgur.com/9hLRsjZ.jpg" height=400></center>
This dataset was scraped from [nextspaceflight.com](https://nextspaceflight.com/launches/past/?page=1) and includes all the space missions since the beginning of Space Race between the USA and the Soviet Union in 1957!
### Install Package with Country Codes
```
%pip install iso3166
```
### Upgrade Plotly
Run the cell below if you are working with Google Colab.
```
%pip install --upgrade plotly
```
### Import Statements
```
import numpy as np
import pandas as pd
import plotly.express as px
import matplotlib.pyplot as plt
import seaborn as sns
# These might be helpful:
from iso3166 import countries
from datetime import datetime, timedelta
```
### Notebook Presentation
```
pd.options.display.float_format = '{:,.2f}'.format
```
### Load the Data
```
df_data = pd.read_csv('mission_launches.csv')
```
# Preliminary Data Exploration
* What is the shape of `df_data`?
* How many rows and columns does it have?
* What are the column names?
* Are there any NaN values or duplicates?
```
```
## Data Cleaning - Check for Missing Values and Duplicates
Consider removing columns containing junk data.
```
```
## Descriptive Statistics
```
```
# Number of Launches per Company
Create a chart that shows the number of space mission launches by organisation.
```
```
# Number of Active versus Retired Rockets
How many rockets are active compared to those that are decomissioned?
```
```
# Distribution of Mission Status
How many missions were successful?
How many missions failed?
```
```
# How Expensive are the Launches?
Create a histogram and visualise the distribution. The price column is given in USD millions (careful of missing values).
```
```
# Use a Choropleth Map to Show the Number of Launches by Country
* Create a choropleth map using [the plotly documentation](https://plotly.com/python/choropleth-maps/)
* Experiment with [plotly's available colours](https://plotly.com/python/builtin-colorscales/). I quite like the sequential colour `matter` on this map.
* You'll need to extract a `country` feature as well as change the country names that no longer exist.
Wrangle the Country Names
You'll need to use a 3 letter country code for each country. You might have to change some country names.
* Russia is the Russian Federation
* New Mexico should be USA
* Yellow Sea refers to China
* Shahrud Missile Test Site should be Iran
* Pacific Missile Range Facility should be USA
* Barents Sea should be Russian Federation
* Gran Canaria should be USA
You can use the iso3166 package to convert the country names to Alpha3 format.
```
```
# Use a Choropleth Map to Show the Number of Failures by Country
```
```
# Create a Plotly Sunburst Chart of the countries, organisations, and mission status.
```
```
# Analyse the Total Amount of Money Spent by Organisation on Space Missions
```
```
# Analyse the Amount of Money Spent by Organisation per Launch
```
```
# Chart the Number of Launches per Year
```
```
# Chart the Number of Launches Month-on-Month until the Present
Which month has seen the highest number of launches in all time? Superimpose a rolling average on the month on month time series chart.
```
```
# Launches per Month: Which months are most popular and least popular for launches?
Some months have better weather than others. Which time of year seems to be best for space missions?
```
```
# How has the Launch Price varied Over Time?
Create a line chart that shows the average price of rocket launches over time.
```
```
# Chart the Number of Launches over Time by the Top 10 Organisations.
How has the dominance of launches changed over time between the different players?
```
```
# Cold War Space Race: USA vs USSR
The cold war lasted from the start of the dataset up until 1991.
```
```
## Create a Plotly Pie Chart comparing the total number of launches of the USSR and the USA
Hint: Remember to include former Soviet Republics like Kazakhstan when analysing the total number of launches.
```
```
## Create a Chart that Shows the Total Number of Launches Year-On-Year by the Two Superpowers
```
```
## Chart the Total Number of Mission Failures Year on Year.
```
```
## Chart the Percentage of Failures over Time
Did failures go up or down over time? Did the countries get better at minimising risk and improving their chances of success over time?
```
```
# For Every Year Show which Country was in the Lead in terms of Total Number of Launches up to and including including 2020)
Do the results change if we only look at the number of successful launches?
```
```
# Create a Year-on-Year Chart Showing the Organisation Doing the Most Number of Launches
Which organisation was dominant in the 1970s and 1980s? Which organisation was dominant in 2018, 2019 and 2020?
```
```
|
github_jupyter
|
%pip install iso3166
%pip install --upgrade plotly
import numpy as np
import pandas as pd
import plotly.express as px
import matplotlib.pyplot as plt
import seaborn as sns
# These might be helpful:
from iso3166 import countries
from datetime import datetime, timedelta
pd.options.display.float_format = '{:,.2f}'.format
df_data = pd.read_csv('mission_launches.csv')
```
## Data Cleaning - Check for Missing Values and Duplicates
Consider removing columns containing junk data.
## Descriptive Statistics
# Number of Launches per Company
Create a chart that shows the number of space mission launches by organisation.
# Number of Active versus Retired Rockets
How many rockets are active compared to those that are decomissioned?
# Distribution of Mission Status
How many missions were successful?
How many missions failed?
# How Expensive are the Launches?
Create a histogram and visualise the distribution. The price column is given in USD millions (careful of missing values).
# Use a Choropleth Map to Show the Number of Launches by Country
* Create a choropleth map using [the plotly documentation](https://plotly.com/python/choropleth-maps/)
* Experiment with [plotly's available colours](https://plotly.com/python/builtin-colorscales/). I quite like the sequential colour `matter` on this map.
* You'll need to extract a `country` feature as well as change the country names that no longer exist.
Wrangle the Country Names
You'll need to use a 3 letter country code for each country. You might have to change some country names.
* Russia is the Russian Federation
* New Mexico should be USA
* Yellow Sea refers to China
* Shahrud Missile Test Site should be Iran
* Pacific Missile Range Facility should be USA
* Barents Sea should be Russian Federation
* Gran Canaria should be USA
You can use the iso3166 package to convert the country names to Alpha3 format.
# Use a Choropleth Map to Show the Number of Failures by Country
# Create a Plotly Sunburst Chart of the countries, organisations, and mission status.
# Analyse the Total Amount of Money Spent by Organisation on Space Missions
# Analyse the Amount of Money Spent by Organisation per Launch
# Chart the Number of Launches per Year
# Chart the Number of Launches Month-on-Month until the Present
Which month has seen the highest number of launches in all time? Superimpose a rolling average on the month on month time series chart.
# Launches per Month: Which months are most popular and least popular for launches?
Some months have better weather than others. Which time of year seems to be best for space missions?
# How has the Launch Price varied Over Time?
Create a line chart that shows the average price of rocket launches over time.
# Chart the Number of Launches over Time by the Top 10 Organisations.
How has the dominance of launches changed over time between the different players?
# Cold War Space Race: USA vs USSR
The cold war lasted from the start of the dataset up until 1991.
## Create a Plotly Pie Chart comparing the total number of launches of the USSR and the USA
Hint: Remember to include former Soviet Republics like Kazakhstan when analysing the total number of launches.
## Create a Chart that Shows the Total Number of Launches Year-On-Year by the Two Superpowers
## Chart the Total Number of Mission Failures Year on Year.
## Chart the Percentage of Failures over Time
Did failures go up or down over time? Did the countries get better at minimising risk and improving their chances of success over time?
# For Every Year Show which Country was in the Lead in terms of Total Number of Launches up to and including including 2020)
Do the results change if we only look at the number of successful launches?
# Create a Year-on-Year Chart Showing the Organisation Doing the Most Number of Launches
Which organisation was dominant in the 1970s and 1980s? Which organisation was dominant in 2018, 2019 and 2020?
| 0.744099 | 0.989368 |
```
import pandas as pd
pd.set_option('display.max_columns', 100)
pd.set_option('display.max_rows', 200)
all_muts_df = pd.read_pickle("../data/4_10_with_uniq_midpts.pkl")
print(len(all_muts_df))
all_muts_df.head()
# df = all_muts_df[all_muts_df["carbon-source"].str.contains("glycerol")]
# df["exp ale"] = df.apply(lambda r: r["exp"] + " " + str(r["ale"]), axis=1)
# df
ops = ["cyaA", "glpFKX", "ptsHI-crr"]
op_df = pd.DataFrame()
for _, m in all_muts_df.iterrows():
for op in m["operons"]:
if op["name"] in ops:
op_df = op_df.append(m)
break
# display(len(op_df), op_df.head())
def get_sample_name(exp_name, ale, flask, isolate, tech_rep):
sample_name = exp_name + " " + str(int(ale)) + " " + str(int(flask)) + " " + str(int(isolate)) + " " + str(int(tech_rep))
return sample_name
op_df["sample"] = op_df.apply(lambda r: get_sample_name(r.exp, r.ale, r.flask, r.isolate, r.tech_rep), axis=1)
op_df.head()
op_df["Mutation Type"].unique()
feat_set = set()
for _, m in op_df.iterrows():
for f in m["genomic features"]:
feat_set.add(f["name"])
mat = pd.DataFrame(columns=op_df["sample"].unique(), index=feat_set)
mat = mat.fillna('')
for _, m in op_df.iterrows():
for f in m["genomic features"]:
curr_mut_type = mat.at[f["name"], m["sample"]]
if curr_mut_type != m["Mutation Type"]: # oncoprint doesn't double-count mutation types within the same feature+sample (it might; need to test, but not currently needed), therefore explicitely removing to remember this quirk
if curr_mut_type != '':
curr_mut_type += ';'
new_mut_type = m["Mutation Type"]
if new_mut_type == "AMP":
new_mut_type = "CNV"
curr_mut_type += new_mut_type
mat.at[f["name"], m["sample"]] = curr_mut_type
mat
# removing pdxK entry since it's a DEL to the ptsHI-crr terminator that overlaps with the pdkX encoded on the opposite strand.
mat = mat[mat.index!="pdxK"]
mat
mat.to_csv("./glpK_cyaA_crr_op_oncoprint_mat_gly_genes.csv")
op_df.exp.unique()
exp_id_d = {
"GLU": "GLU",
"GYD": "GYD",
"MG1655-M9-NC_000913_3gb-stationary-37-m-tartrate2": "m-tartrate2",
"PGI": "pgiKO_1",
"pgi": 'pgiKO_2',
'SSW_GLU_GLY': 'SSW_GLU_GLY',
'SSW_GLU_XYL': 'SSW_GLU_XYL',
'SSW_GLY': 'SSW_GLY',
'TOL_hexamethylenediamine': 'TOL_hexamethylenediamine',
'pgiBME':'pgiBME',
'pgiHSA':'pgiHSA',
'pgiPAE':'pgiPAE',
'pts':'ptsHI-crrKO',
"wt": "gentamycin",
}
op_df["exp id"] = op_df["exp"].apply(lambda exp_name: exp_id_d[exp_name])
op_df.head()
cond_cols = {
'base-media',
'calcium-source',
'carbon-source',
'nitrogen-source',
'phosphorous-source',
'strain-description',
'sulfur-source',
'supplement',
'temperature',
'exp id'
}
cond_mat = pd.DataFrame(
columns=op_df["sample"].unique(),
index=cond_cols
)
cond_mat = cond_mat.fillna('')
for _, m in op_df.iterrows():
for cond_col in cond_cols:
cond_mat.at[cond_col, m['sample']] = m[cond_col]
# Serine's default supplement annotation is too large.
cond_mat = cond_mat.T
cond_mat["supplement"] = cond_mat["supplement"].apply(lambda x: "glycine(2mM) & L-serine" if x == "glycine(2mM) L-Serine(varying) Wolfe's vitamin solution trace elements(X1)" else x)
cond_mat["supplement"] = cond_mat["supplement"].apply(lambda x: "NaCl(0.5) trace elements" if x == "NaCl(0.5g/L) trace elements" else x)
cond_mat["temperature"] = cond_mat["temperature"].apply(lambda x: x.replace("celsius", "Celsius"))
cond_mat = cond_mat.T
cond_mat.to_csv("./glyK_cyaA_crr_op_oncoprint_mat_conditions.csv")
cond_mat
```
|
github_jupyter
|
import pandas as pd
pd.set_option('display.max_columns', 100)
pd.set_option('display.max_rows', 200)
all_muts_df = pd.read_pickle("../data/4_10_with_uniq_midpts.pkl")
print(len(all_muts_df))
all_muts_df.head()
# df = all_muts_df[all_muts_df["carbon-source"].str.contains("glycerol")]
# df["exp ale"] = df.apply(lambda r: r["exp"] + " " + str(r["ale"]), axis=1)
# df
ops = ["cyaA", "glpFKX", "ptsHI-crr"]
op_df = pd.DataFrame()
for _, m in all_muts_df.iterrows():
for op in m["operons"]:
if op["name"] in ops:
op_df = op_df.append(m)
break
# display(len(op_df), op_df.head())
def get_sample_name(exp_name, ale, flask, isolate, tech_rep):
sample_name = exp_name + " " + str(int(ale)) + " " + str(int(flask)) + " " + str(int(isolate)) + " " + str(int(tech_rep))
return sample_name
op_df["sample"] = op_df.apply(lambda r: get_sample_name(r.exp, r.ale, r.flask, r.isolate, r.tech_rep), axis=1)
op_df.head()
op_df["Mutation Type"].unique()
feat_set = set()
for _, m in op_df.iterrows():
for f in m["genomic features"]:
feat_set.add(f["name"])
mat = pd.DataFrame(columns=op_df["sample"].unique(), index=feat_set)
mat = mat.fillna('')
for _, m in op_df.iterrows():
for f in m["genomic features"]:
curr_mut_type = mat.at[f["name"], m["sample"]]
if curr_mut_type != m["Mutation Type"]: # oncoprint doesn't double-count mutation types within the same feature+sample (it might; need to test, but not currently needed), therefore explicitely removing to remember this quirk
if curr_mut_type != '':
curr_mut_type += ';'
new_mut_type = m["Mutation Type"]
if new_mut_type == "AMP":
new_mut_type = "CNV"
curr_mut_type += new_mut_type
mat.at[f["name"], m["sample"]] = curr_mut_type
mat
# removing pdxK entry since it's a DEL to the ptsHI-crr terminator that overlaps with the pdkX encoded on the opposite strand.
mat = mat[mat.index!="pdxK"]
mat
mat.to_csv("./glpK_cyaA_crr_op_oncoprint_mat_gly_genes.csv")
op_df.exp.unique()
exp_id_d = {
"GLU": "GLU",
"GYD": "GYD",
"MG1655-M9-NC_000913_3gb-stationary-37-m-tartrate2": "m-tartrate2",
"PGI": "pgiKO_1",
"pgi": 'pgiKO_2',
'SSW_GLU_GLY': 'SSW_GLU_GLY',
'SSW_GLU_XYL': 'SSW_GLU_XYL',
'SSW_GLY': 'SSW_GLY',
'TOL_hexamethylenediamine': 'TOL_hexamethylenediamine',
'pgiBME':'pgiBME',
'pgiHSA':'pgiHSA',
'pgiPAE':'pgiPAE',
'pts':'ptsHI-crrKO',
"wt": "gentamycin",
}
op_df["exp id"] = op_df["exp"].apply(lambda exp_name: exp_id_d[exp_name])
op_df.head()
cond_cols = {
'base-media',
'calcium-source',
'carbon-source',
'nitrogen-source',
'phosphorous-source',
'strain-description',
'sulfur-source',
'supplement',
'temperature',
'exp id'
}
cond_mat = pd.DataFrame(
columns=op_df["sample"].unique(),
index=cond_cols
)
cond_mat = cond_mat.fillna('')
for _, m in op_df.iterrows():
for cond_col in cond_cols:
cond_mat.at[cond_col, m['sample']] = m[cond_col]
# Serine's default supplement annotation is too large.
cond_mat = cond_mat.T
cond_mat["supplement"] = cond_mat["supplement"].apply(lambda x: "glycine(2mM) & L-serine" if x == "glycine(2mM) L-Serine(varying) Wolfe's vitamin solution trace elements(X1)" else x)
cond_mat["supplement"] = cond_mat["supplement"].apply(lambda x: "NaCl(0.5) trace elements" if x == "NaCl(0.5g/L) trace elements" else x)
cond_mat["temperature"] = cond_mat["temperature"].apply(lambda x: x.replace("celsius", "Celsius"))
cond_mat = cond_mat.T
cond_mat.to_csv("./glyK_cyaA_crr_op_oncoprint_mat_conditions.csv")
cond_mat
| 0.249722 | 0.212784 |
# Datasets
The preferred way to ingest data into TensorFlow estimators is by using the `tf.data.Dataset` class. There are a couple reasons for this:
1. Datasets automatically manage memory and resources
2. They separate data ingestion from modeling. This means that modeling steps can be ran concurrently with data i/o operations, speeding up training
3. They make batching/randomization from giant datasets split up over multiple files easy.
**Note:** There are lots of examples online using things like `QueueRunner`s (check what the thing actually is) that were popular before `Dataset`s were introduced.
## 1. The data
We need some data to work with. To start, we will just use a few .csv files. Later on we'll talk about .tfrecords, which is the preferred data format for TensorFlow.
Let's load up the Boston data set for the test data. To make things more interesting, we'll make the columns have some different data types and split it into several .csv files. (Clearly this entirely unnecessary to do with these data, but it will put us in a situation closer to reality.)
```
import pathlib
import numpy as np
import pandas as pd
import tensorflow as tf
from matplotlib import pyplot as plt
plt.style.use('seaborn')
from sklearn.datasets import load_boston
boston = load_boston()
features, labels = boston.data, boston.target
columns = [c.lower() for c in boston.feature_names]
df = pd.DataFrame(features, columns=columns)
df['chas'] = df['chas'].map({0.: 'Y', 1.: 'N'})
df['rad'] = df['rad'].astype(np.int64)
df['target'] = labels
df.head()
# Split into multiple files
n_shards = 5
shard_size = len(df) // n_shards
data_dir = pathlib.Path('../data/sharded_data')
if not data_dir.exists(parents=True):
data_dir.mkdir()
df = df.sample(frac=1)
for i in range(n_shards):
idx_start = i * shard_size
idx_end = (i + 1) * shard_size
df.iloc[idx_start:idx_end].to_csv(data_dir / 'boston-{0}.csv'.format(i), index=False)
```
## 2. Reading in data from a file
The general way we get in data with a `Dataset` is by instantiating a Dataset object, converting it to an iterator using the `make_one_shot_iterator` method, and get a batch of data with the iterator's `get_next` method. The `get_next` method returns an op (*verify this...*) which is why it is called only once (instead of each time we want to get the next batch of data).
Since we are reading in a single .csv, we use `TextLineDataset`, which reads in plain text files and returns a dataset where the rows of the text document are the records of the dataset.
```
# Read a single file as text
file = (data_dir / 'boston-0.csv').as_posix()
dataset = tf.data.TextLineDataset(file)
iterator = dataset.make_one_shot_iterator()
batch = iterator.get_next()
with tf.Session() as sess:
batch1 = sess.run(batch)
batch2 = sess.run(batch)
batch3 = sess.run(batch)
for b in (batch1, batch2, batch3):
print(b)
```
Note that the three batches we got are each just a single string, and we also got the text of the header. We don't want to be including the header at all in the data, and we want an array for each row, not just a single string.
The way datasets are modified is by chaining on methods which change the behavior of the dataset. Dealing with the header is straight forward; we can just use the dataset's `skip` method to skip the first row. To parse the rows as arrays and not a single string, we use the `map` method, which will apply the same function to every row of the dataset. TensorFlow provides a `decode_csv` function which converts a string tensor representing a row of a csv file into a tuple of tensors for each field of the csv.
```
# Decode csv requires a list of default values to use for each tensor
# produced. The defaults are passed as a list of lists.
DEFAULT_VALUES = [[0.0]] * 14
DEFAULT_VALUES[3] = ['_UNKNOWN']; DEFAULT_VALUES[8] = 0
def parse_row(row):
return tf.decode_csv(row, record_defaults=DEFAULT_VALUES)
dataset = tf.data.TextLineDataset(file)
dataset = dataset.skip(1) # skip the header
dataset = dataset.map(parse_row) # convert string to array
iterator = dataset.make_one_shot_iterator()
batch = iterator.get_next()
with tf.Session() as sess:
batch1 = sess.run(batch)
batch2 = sess.run(batch)
batch3 = sess.run(batch)
for b in (batch1, batch2, batch3):
print(b)
```
*Aside:* Since the `batch` op now produces a tuple of tensors instead of a single tensor, we're using `sess.run` instead of `batch.eval`.
If all of our data is in a single file, that is it. We have our data input pipeline. We can apply additonal methods to spruce up our Dataset by shuffling the data, taking batches of more than just one element, improving memory management, and so on.
## 3. Reading in data from multiple files
Now that we've successfully read data from a single file, let's do multiple. The general idea is to first make a dataset of file names, and use the map method to make a dataset of datasets. This doesn't literally work: The iterator made from a dataset returns tensors, so has to have one of the allowable tensor datatypes. Hence, it can't return a dataset itself. However, there is a `flat_map` method which applies a function to the rows of all of the would-be datasets while simultaneously flattening them into a single dataset. This avoids ever actually having a dataset who returns tensors of type "dataset".
```
# Can use wildcards for data with similar names
file = (data_dir / 'boston-*.csv').as_posix()
dataset = tf.data.Dataset.list_files(file)
iterator = dataset.make_one_shot_iterator()
batch = iterator.get_next()
with tf.Session() as sess:
batch1 = sess.run(batch)
batch2 = sess.run(batch)
batch3 = sess.run(batch)
# Just getting a dataset of individual file names
for b in (batch1, batch2, batch3):
print(b)
# Convert each file name into a dataset and flat_map
# Get dataset of file names
dataset = tf.data.Dataset.list_files(file)
# Combine all files into a single text dataset (without headers)
dataset = dataset.flat_map(lambda f: tf.data.TextLineDataset(f).skip(1))
# Convert each row into a tuple
dataset = dataset.map(parse_row)
iterator = dataset.make_one_shot_iterator()
batch = iterator.get_next()
with tf.Session() as sess:
batch1 = sess.run(batch)
batch2 = sess.run(batch)
batch3 = sess.run(batch)
# Just getting a dataset of individual file names
for b in (batch1, batch2, batch3):
print(b)
```
## 4. Some handy methods
While actually training a model, there are a few things we want to do:
1. Shuffle the data
2. Repeat the dataset for training over multiple epochs
3. Get batches of data
4. Preload the next batch of data while training...
(We also want to feed data into the `Estimator` during training as a tuple consisting of a dict of features and a label. This can be done in the `parse_row` function we wrote above. We'll go into this in more detail when we talk about `Estimators`.)
```
# Training parameters
n_epochs = 5
batch_size = 2
# Build data set
dataset = tf.data.Dataset.list_files(file)
dataset = dataset.flat_map(lambda f: tf.data.TextLineDataset(f).skip(1))
dataset = dataset.map(parse_row)
# Repeat the dataset
dataset = dataset.repeat(n_epochs)
# Shuffle data
dataset = dataset.shuffle(buffer_size=1024)
# Get a batch of data
dataset = dataset.batch(batch_size)
# Preload next batch to speed up training
dataset = dataset.prefetch(buffer_size=batch_size)
iterator = dataset.make_one_shot_iterator()
batch = iterator.get_next()
with tf.Session() as sess:
batch1 = sess.run(batch)
print(batch1)
```
A couple of remarks:
1. The number of repeats can be set to `None` in which case (according to the TensorFlow documentation) the model being fed by the dataset will train indefinitely. I am not sure how long indefinitely actually is?
2. When shuffling, the `buffer_size` parameter specifies how many records to read into memory and then shuffle. The smaller this number is, the less randomized the data will actually be; the larger it is the more memory is used. Here I am only reading in a KB of data into memory at a time. In real life you'd want to use several MB or a few GB if you got the ram for it. *I should check if buffer_size refers to the number of records or the max memory footprint of the loaded data...*
3. For prefetching, `buffer_size` specifies how many records to load into memory in advance. This is useful for speeding up training by allowing the dataset to load and process the next batch of training data while the previous batch is being consumed by the model.
There are a lot of other things that can be done to improve the efficiency of this bad boy, such as using "fused ops" which do several of these steps at once. For more information check out https://www.tensorflow.org/guide/performance/datasets
|
github_jupyter
|
import pathlib
import numpy as np
import pandas as pd
import tensorflow as tf
from matplotlib import pyplot as plt
plt.style.use('seaborn')
from sklearn.datasets import load_boston
boston = load_boston()
features, labels = boston.data, boston.target
columns = [c.lower() for c in boston.feature_names]
df = pd.DataFrame(features, columns=columns)
df['chas'] = df['chas'].map({0.: 'Y', 1.: 'N'})
df['rad'] = df['rad'].astype(np.int64)
df['target'] = labels
df.head()
# Split into multiple files
n_shards = 5
shard_size = len(df) // n_shards
data_dir = pathlib.Path('../data/sharded_data')
if not data_dir.exists(parents=True):
data_dir.mkdir()
df = df.sample(frac=1)
for i in range(n_shards):
idx_start = i * shard_size
idx_end = (i + 1) * shard_size
df.iloc[idx_start:idx_end].to_csv(data_dir / 'boston-{0}.csv'.format(i), index=False)
# Read a single file as text
file = (data_dir / 'boston-0.csv').as_posix()
dataset = tf.data.TextLineDataset(file)
iterator = dataset.make_one_shot_iterator()
batch = iterator.get_next()
with tf.Session() as sess:
batch1 = sess.run(batch)
batch2 = sess.run(batch)
batch3 = sess.run(batch)
for b in (batch1, batch2, batch3):
print(b)
# Decode csv requires a list of default values to use for each tensor
# produced. The defaults are passed as a list of lists.
DEFAULT_VALUES = [[0.0]] * 14
DEFAULT_VALUES[3] = ['_UNKNOWN']; DEFAULT_VALUES[8] = 0
def parse_row(row):
return tf.decode_csv(row, record_defaults=DEFAULT_VALUES)
dataset = tf.data.TextLineDataset(file)
dataset = dataset.skip(1) # skip the header
dataset = dataset.map(parse_row) # convert string to array
iterator = dataset.make_one_shot_iterator()
batch = iterator.get_next()
with tf.Session() as sess:
batch1 = sess.run(batch)
batch2 = sess.run(batch)
batch3 = sess.run(batch)
for b in (batch1, batch2, batch3):
print(b)
# Can use wildcards for data with similar names
file = (data_dir / 'boston-*.csv').as_posix()
dataset = tf.data.Dataset.list_files(file)
iterator = dataset.make_one_shot_iterator()
batch = iterator.get_next()
with tf.Session() as sess:
batch1 = sess.run(batch)
batch2 = sess.run(batch)
batch3 = sess.run(batch)
# Just getting a dataset of individual file names
for b in (batch1, batch2, batch3):
print(b)
# Convert each file name into a dataset and flat_map
# Get dataset of file names
dataset = tf.data.Dataset.list_files(file)
# Combine all files into a single text dataset (without headers)
dataset = dataset.flat_map(lambda f: tf.data.TextLineDataset(f).skip(1))
# Convert each row into a tuple
dataset = dataset.map(parse_row)
iterator = dataset.make_one_shot_iterator()
batch = iterator.get_next()
with tf.Session() as sess:
batch1 = sess.run(batch)
batch2 = sess.run(batch)
batch3 = sess.run(batch)
# Just getting a dataset of individual file names
for b in (batch1, batch2, batch3):
print(b)
# Training parameters
n_epochs = 5
batch_size = 2
# Build data set
dataset = tf.data.Dataset.list_files(file)
dataset = dataset.flat_map(lambda f: tf.data.TextLineDataset(f).skip(1))
dataset = dataset.map(parse_row)
# Repeat the dataset
dataset = dataset.repeat(n_epochs)
# Shuffle data
dataset = dataset.shuffle(buffer_size=1024)
# Get a batch of data
dataset = dataset.batch(batch_size)
# Preload next batch to speed up training
dataset = dataset.prefetch(buffer_size=batch_size)
iterator = dataset.make_one_shot_iterator()
batch = iterator.get_next()
with tf.Session() as sess:
batch1 = sess.run(batch)
print(batch1)
| 0.628635 | 0.984381 |
```
# We're going to install this module to help us parse datetimes from the raw dataset
!pip install dateparser
#InsertOne - inserts a document
#UpdateOne - updates a document
from pymongo import MongoClient, InsertOne, UpdateOne
import pprint
import dateparser
from bson.json_util import loads
# Replace XXXX with your connection URI from the Atlas UI
client = MongoClient('mongodb+srv://dbAdmin:pa55word@mflix.phy3v.mongodb.net/mflix_db?retryWrites=true&w=majority')
#cleansing json file
people_raw = client.cleansing['people-raw']
batch_size = 1000
inserts = []
count = 0
# There are over 50,000 lines, so this might take a while...
# Make sure to wait until the cell finishes executing before moving on (the * will turn into a number)
with open("./people-raw.json") as dataset:
for line in dataset:
#loads a document to the inserts list
inserts.append(InsertOne(loads(line)))
count += 1
if count == batch_size:
#write 1000 documents at a time to the inserts list
people_raw.bulk_write(inserts)
inserts = []
count = 0
if inserts:
people_raw.bulk_write(inserts)
count = 0
# Confirm that 50,474 documents are in your collection before moving on
people_raw.count_documents({})
people_raw.find_one()
# Replace YYYY with a query on the people-raw collection that will return a cursor with only
# documents where the birthday field is a string
people_with_string_birthdays = people_raw.find({"birthday":{"$type": "string"}})
# This is the answer to verify you completed the lab
#print2 = pprint.pprint(list(people_with_string_birthdays))
people_with_string_birthdays.count()
updates = []
# Again, we're updating several thousand documents, so this will take a little while
for person in people_with_string_birthdays:
# Pymongo converts datetime objects into BSON Dates. The dateparser.parse function returns a
# datetime object, so we can simply do the following to update the field properly.
# Replace ZZZZ with the correct update operator
updates.append(UpdateOne({ "_id": person["_id"]}, { "$set": {"birthday": dateparser.parse(person["birthday"])}}))
count += 1
if count == batch_size:
people_raw.bulk_write(updates)
updates = []
count = 0
if updates:
people_raw.bulk_write(updates)
count = 0
# If everything went well this should be zero
people_with_string_birthdays.count()
```
|
github_jupyter
|
# We're going to install this module to help us parse datetimes from the raw dataset
!pip install dateparser
#InsertOne - inserts a document
#UpdateOne - updates a document
from pymongo import MongoClient, InsertOne, UpdateOne
import pprint
import dateparser
from bson.json_util import loads
# Replace XXXX with your connection URI from the Atlas UI
client = MongoClient('mongodb+srv://dbAdmin:pa55word@mflix.phy3v.mongodb.net/mflix_db?retryWrites=true&w=majority')
#cleansing json file
people_raw = client.cleansing['people-raw']
batch_size = 1000
inserts = []
count = 0
# There are over 50,000 lines, so this might take a while...
# Make sure to wait until the cell finishes executing before moving on (the * will turn into a number)
with open("./people-raw.json") as dataset:
for line in dataset:
#loads a document to the inserts list
inserts.append(InsertOne(loads(line)))
count += 1
if count == batch_size:
#write 1000 documents at a time to the inserts list
people_raw.bulk_write(inserts)
inserts = []
count = 0
if inserts:
people_raw.bulk_write(inserts)
count = 0
# Confirm that 50,474 documents are in your collection before moving on
people_raw.count_documents({})
people_raw.find_one()
# Replace YYYY with a query on the people-raw collection that will return a cursor with only
# documents where the birthday field is a string
people_with_string_birthdays = people_raw.find({"birthday":{"$type": "string"}})
# This is the answer to verify you completed the lab
#print2 = pprint.pprint(list(people_with_string_birthdays))
people_with_string_birthdays.count()
updates = []
# Again, we're updating several thousand documents, so this will take a little while
for person in people_with_string_birthdays:
# Pymongo converts datetime objects into BSON Dates. The dateparser.parse function returns a
# datetime object, so we can simply do the following to update the field properly.
# Replace ZZZZ with the correct update operator
updates.append(UpdateOne({ "_id": person["_id"]}, { "$set": {"birthday": dateparser.parse(person["birthday"])}}))
count += 1
if count == batch_size:
people_raw.bulk_write(updates)
updates = []
count = 0
if updates:
people_raw.bulk_write(updates)
count = 0
# If everything went well this should be zero
people_with_string_birthdays.count()
| 0.313 | 0.301706 |
```
!curl -O https://download.pytorch.org/tutorial/data.zip
!unzip data.zip
import glob
import unicodedata
import string
import os
import torch
import torch.nn as nn
import torch.optim as optim
import random
import time
import math
import matplotlib.pyplot as plt
import matplotlib.ticker as ticker
if torch.cuda.is_available():
device = torch.device('cuda')
else:
device = torch.device('cpu')
device
filenames = glob.glob('data/names/*.txt')
all_letters = string.ascii_letters + " .,;'"
n_letters = len(all_letters)
def unicode_to_ascii(s):
return ''.join(
c for c in unicodedata.normalize('NFD', s)
if unicodedata.category(c) != 'Mn'
and c in all_letters
)
def readlines(fname):
lines = open(fname, encoding='utf-8').read().strip().split('\n')
return list(set([unicode_to_ascii(line) for line in lines]))
categories=[]
category_dict = {}
for fname in filenames:
cat=os.path.splitext(os.path.basename(fname))[0]
categories.append(cat)
category_dict[cat] = readlines(fname)
n_categories = len(categories)
def letter_to_index(l):
return all_letters.find(l)
def letter_to_tensor(l):
ten = torch.zeros(1, n_letters)
ten[0,letter_to_index(l)] = 1.0
return ten
def string_to_tensor(s):
ten = torch.zeros(len(s),1,n_letters,device=device)
for i, l in enumerate(s):
ten[i][0][letter_to_index(l)] = 1.0
return ten
[letter_to_tensor(l) for l in category_dict['German'][0]]
print(string_to_tensor(category_dict['German'][0]).size())
class Rnn(nn.Module):
def __init__(self, input_size, hidden_size, output_size):
super(Rnn, self).__init__()
self.hidden_size = hidden_size
self.i2h = nn.Linear(input_size + hidden_size, hidden_size)
self.i2o = nn.Linear(input_size + hidden_size, output_size)
self.softmax = nn.LogSoftmax(dim=1)
def forward(self, input, hidden):
combined = torch.cat((input, hidden), 1)
hidden = self.i2h(combined)
output = self.i2o(combined)
output = self.softmax(output)
return output, hidden
def initHidden(self):
return torch.zeros(1, self.hidden_size,device=device)
n_hidden = 128
def categoryFromOutput(out):
top_n, top_i = out.topk(1)
cat_i = top_i[0].item()
return categories[cat_i], cat_i, top_n[0].item()
def randomChoice(l):
return l[random.randint(0,len(l)-1)]
def randomTrainingExample():
cat = randomChoice(categories)
line = randomChoice(category_dict[cat])
cat_tensor = torch.tensor([categories.index(cat)], dtype=torch.long,device=device)
line_tensor = string_to_tensor(line)
return cat, line, cat_tensor, line_tensor
rnn = Rnn(n_letters, n_hidden, n_categories)
rnn.cuda()
criterion = nn.NLLLoss()
learning_rate = 0.005
optimizer = optim.SGD(rnn.parameters(), lr=learning_rate)
def train(category_tensor, line_tesor):
hidden = rnn.initHidden()
optimizer.zero_grad()
for i in range(line_tensor.size()[0]):
output, hidden = rnn(line_tensor[i], hidden)
loss = criterion(output, category_tensor)
loss.backward()
optimizer.step()
return output, loss.item()
n_iters = 1000000
print_every = 5000
plot_every = 1000
def timeSince(t):
now = time.time()
s = now - t
m = math.floor(s/60)
s -= m*60
return '{}:{}'.format(m,s)
current_loss = 0
all_losses = []
start = time.time()
for iter in range(1, n_iters+1):
category, line, category_tensor, line_tensor = randomTrainingExample()
output, loss = train(category_tensor, line_tensor)
current_loss += loss
if iter % print_every == 0:
guess, guess_i, _ = categoryFromOutput(output)
correct = '✓' if guess == category else '✗ ({})'.format(category)
print('{} {}% ({}) {} {} / {} {}'.format(iter, iter / n_iters * 100,
timeSince(start), loss, line,
guess, correct))
if iter % plot_every == 0:
all_losses.append(current_loss / plot_every)
current_loss = 0
plt.figure()
plt.plot(all_losses)
def evaluate(line_tensor):
hidden = rnn.initHidden()
for i in range(line_tensor.size()[0]):
output, hidden = rnn(line_tensor[i], hidden)
return output
confusion = torch.zeros(n_categories, n_categories, device=device)
n_confusion = 10000
for i in range(n_confusion):
category, line, category_tensor, line_tensor = randomTrainingExample()
output = evaluate(line_tensor)
guess, guess_i, _ = categoryFromOutput(output)
category_i = categories.index(category)
confusion[category_i][guess_i] += 1
for i in range(n_categories):
confusion[i] = confusion[i] / confusion[i].sum()
fig = plt.figure(figsize=(12,12))
ax = fig.add_subplot(111)
cax = ax.matshow(confusion.cpu().numpy())
fig.colorbar(cax)
ax.set_xticklabels(['']+categories,rotation=90)
ax.set_yticklabels(['']+categories)
ax.xaxis.set_major_locator(ticker.MultipleLocator(1))
ax.yaxis.set_major_locator(ticker.MultipleLocator(1))
plt.show()
```
|
github_jupyter
|
!curl -O https://download.pytorch.org/tutorial/data.zip
!unzip data.zip
import glob
import unicodedata
import string
import os
import torch
import torch.nn as nn
import torch.optim as optim
import random
import time
import math
import matplotlib.pyplot as plt
import matplotlib.ticker as ticker
if torch.cuda.is_available():
device = torch.device('cuda')
else:
device = torch.device('cpu')
device
filenames = glob.glob('data/names/*.txt')
all_letters = string.ascii_letters + " .,;'"
n_letters = len(all_letters)
def unicode_to_ascii(s):
return ''.join(
c for c in unicodedata.normalize('NFD', s)
if unicodedata.category(c) != 'Mn'
and c in all_letters
)
def readlines(fname):
lines = open(fname, encoding='utf-8').read().strip().split('\n')
return list(set([unicode_to_ascii(line) for line in lines]))
categories=[]
category_dict = {}
for fname in filenames:
cat=os.path.splitext(os.path.basename(fname))[0]
categories.append(cat)
category_dict[cat] = readlines(fname)
n_categories = len(categories)
def letter_to_index(l):
return all_letters.find(l)
def letter_to_tensor(l):
ten = torch.zeros(1, n_letters)
ten[0,letter_to_index(l)] = 1.0
return ten
def string_to_tensor(s):
ten = torch.zeros(len(s),1,n_letters,device=device)
for i, l in enumerate(s):
ten[i][0][letter_to_index(l)] = 1.0
return ten
[letter_to_tensor(l) for l in category_dict['German'][0]]
print(string_to_tensor(category_dict['German'][0]).size())
class Rnn(nn.Module):
def __init__(self, input_size, hidden_size, output_size):
super(Rnn, self).__init__()
self.hidden_size = hidden_size
self.i2h = nn.Linear(input_size + hidden_size, hidden_size)
self.i2o = nn.Linear(input_size + hidden_size, output_size)
self.softmax = nn.LogSoftmax(dim=1)
def forward(self, input, hidden):
combined = torch.cat((input, hidden), 1)
hidden = self.i2h(combined)
output = self.i2o(combined)
output = self.softmax(output)
return output, hidden
def initHidden(self):
return torch.zeros(1, self.hidden_size,device=device)
n_hidden = 128
def categoryFromOutput(out):
top_n, top_i = out.topk(1)
cat_i = top_i[0].item()
return categories[cat_i], cat_i, top_n[0].item()
def randomChoice(l):
return l[random.randint(0,len(l)-1)]
def randomTrainingExample():
cat = randomChoice(categories)
line = randomChoice(category_dict[cat])
cat_tensor = torch.tensor([categories.index(cat)], dtype=torch.long,device=device)
line_tensor = string_to_tensor(line)
return cat, line, cat_tensor, line_tensor
rnn = Rnn(n_letters, n_hidden, n_categories)
rnn.cuda()
criterion = nn.NLLLoss()
learning_rate = 0.005
optimizer = optim.SGD(rnn.parameters(), lr=learning_rate)
def train(category_tensor, line_tesor):
hidden = rnn.initHidden()
optimizer.zero_grad()
for i in range(line_tensor.size()[0]):
output, hidden = rnn(line_tensor[i], hidden)
loss = criterion(output, category_tensor)
loss.backward()
optimizer.step()
return output, loss.item()
n_iters = 1000000
print_every = 5000
plot_every = 1000
def timeSince(t):
now = time.time()
s = now - t
m = math.floor(s/60)
s -= m*60
return '{}:{}'.format(m,s)
current_loss = 0
all_losses = []
start = time.time()
for iter in range(1, n_iters+1):
category, line, category_tensor, line_tensor = randomTrainingExample()
output, loss = train(category_tensor, line_tensor)
current_loss += loss
if iter % print_every == 0:
guess, guess_i, _ = categoryFromOutput(output)
correct = '✓' if guess == category else '✗ ({})'.format(category)
print('{} {}% ({}) {} {} / {} {}'.format(iter, iter / n_iters * 100,
timeSince(start), loss, line,
guess, correct))
if iter % plot_every == 0:
all_losses.append(current_loss / plot_every)
current_loss = 0
plt.figure()
plt.plot(all_losses)
def evaluate(line_tensor):
hidden = rnn.initHidden()
for i in range(line_tensor.size()[0]):
output, hidden = rnn(line_tensor[i], hidden)
return output
confusion = torch.zeros(n_categories, n_categories, device=device)
n_confusion = 10000
for i in range(n_confusion):
category, line, category_tensor, line_tensor = randomTrainingExample()
output = evaluate(line_tensor)
guess, guess_i, _ = categoryFromOutput(output)
category_i = categories.index(category)
confusion[category_i][guess_i] += 1
for i in range(n_categories):
confusion[i] = confusion[i] / confusion[i].sum()
fig = plt.figure(figsize=(12,12))
ax = fig.add_subplot(111)
cax = ax.matshow(confusion.cpu().numpy())
fig.colorbar(cax)
ax.set_xticklabels(['']+categories,rotation=90)
ax.set_yticklabels(['']+categories)
ax.xaxis.set_major_locator(ticker.MultipleLocator(1))
ax.yaxis.set_major_locator(ticker.MultipleLocator(1))
plt.show()
| 0.811041 | 0.460774 |
# Map extraction - Example with two maps
This notebook provides an overview of the work done on maps in order to obtain the whole processed dataset announced in milestone 1. It shows how we extract polygons from raw maps (gifs or pdf). This process has been automated with a script for all the directories.
```
from pylab import contour
import matplotlib.pyplot as plt
from PIL import ImageFilter, Image, ImageDraw
from datetime import date, timedelta
import numpy as np
from PIL import Image
import cv2
from skimage import measure
import os
import pandas as pd
from scipy.spatial import distance
import json
import visvalingamwyatt as vw
import folium
%matplotlib inline
```
First, we define the colors and the arrays of colors we will need.
```
black = np.array([0, 0, 0])
white = np.array([255, 255, 255])
green = np.array([204, 255, 102])
yellow = np.array([255, 255, 0])
orange = np.array([255, 153, 0])
red = np.array([255, 0, 0])
danger_colors_code = ['#ccff66', '#ffff00', '#ff9900', '#ff0000']
shades_danger = [green, yellow, orange, red]
danger_image_shades = [green, yellow, orange, red, white]
light_blue = np.array([213, 252, 252])
light_medium_blue = np.array([168, 217, 241])
medium_blue = np.array([121, 161, 229])
dark_medium_blue = np.array([68, 89, 215])
dark_blue = np.array([47, 36, 162])
purple = np.array([91, 32, 196])
snow_color_code = ['#d5fcfc', '#a8d9f1', '#79a1e5', '#4459d7', '#2f24a2', '#5b20c4']
shades_snow = [light_blue, light_medium_blue, medium_blue, dark_medium_blue, dark_blue, purple]
shades_grey = [np.array([c,c,c]) for c in range(255)]
snow_image_shades = [light_blue, light_medium_blue, medium_blue, dark_medium_blue, dark_blue, purple, white]
raw_red = np.array([255, 0, 0])
raw_green = np.array([0, 255, 0])
raw_blue = np.array([0, 0, 255])
raw_pink = np.array([255, 0, 255])
raw_pink = np.array([255, 0, 255])
raw_cyan = np.array([0, 255, 255])
raw_yellow = np.array([255, 255, 0])
```
The defined functions below will be using those set of colors.
```
def keep_colors(img, colors, replace_with=white):
"""return a new image with only the `colors` selected, other pixel are `replace_with`"""
keep = np.zeros(img.shape[:2], dtype=bool)
for c in colors:
keep = keep | (c == img).all(axis=-1)
new_img = img.copy()
new_img[~keep] = replace_with
return new_img
def remove_colors(img, colors, replace_with=white):
"""return a new image without the `colors` selected which will be replaced by `replace_with`"""
keep = np.zeros(img.shape[:2], dtype=bool)
for c in colors:
keep = keep | (c == img).all(axis=-1)
new_img = img.copy()
new_img[keep] = replace_with
return new_img
def replace_color(img, color_map):
"""return a new image replacing the image colors which will be mapped to their corresponding colors in `color_map` (df)"""
new_img = img.copy()
for _, (source, target) in color_map.iterrows():
new_img[(img == source).all(axis=-1)] = target
return new_img
def build_color_map(img_arr, image_shades):
"""return colormap as dataframe"""
im_df = pd.DataFrame([img_arr[i,j,:] for i,j in np.ndindex(img_arr.shape[0],img_arr.shape[1])])
im_df = im_df.drop_duplicates()
image_colors = im_df.as_matrix()
colors = np.zeros(image_colors.shape)
dist = distance.cdist(image_colors, image_shades, 'sqeuclidean')
for j in range(dist.shape[0]):
distances = dist[j,:]
colors[j, :] = image_shades[distances.argmin()]
color_map = pd.DataFrame(
{'source': image_colors.tolist(),
'target': colors.tolist()
})
return color_map
```
Here are the two images we will process.
```
danger_path = '../data/slf/2001/nbk/de/gif/20001230_nbk_de_c.gif'
snow_path = '../data/slf/2010/hstop/en/gif/20100103_hstop_en_c.gif'
danger_img = Image.open(danger_path)
danger_img = danger_img.convert('RGB')
danger_img_arr = np.array(danger_img)
snow_img = Image.open(snow_path)
snow_img = snow_img.convert('RGB')
snow_img_arr = np.array(snow_img)
fig, axes = plt.subplots(1, 2, figsize=(14,10))
# original danger image
axes[0].imshow(danger_img_arr);
axes[0].set_title('Original danger image');
# original snow image
axes[1].imshow(snow_img_arr);
axes[1].set_title('Original snow image');
def numpify(o):
if not isinstance(o, np.ndarray):
o = np.array(o)
return o
def coord_color(img, color):
return np.array(list(zip(*(img == color).all(-1).nonzero())))
def open_mask(height, width):
masks_path = '../map-masks/'
mask_name = '{}x{}.gif'.format(height, width)
mask_path = os.path.join(masks_path, mask_name)
mask = Image.open(mask_path)
mask = mask.convert('RGB')
mask = np.array(mask)
landmarks_pix = {
geo_point: (width, height)
for geo_point, color in landmarks_colors.items()
for height, width in coord_color(mask, color)
}
binary_mask = (mask != 255).any(-1) # different of white
return binary_mask, landmarks_pix
# remove contours areas that have more than 30% of white
WHITE_RATIO_THRESHOLD = .3
def color_contours(img, color):
img = numpify(img)
color = numpify(color)
mask = (img == color[:3]).all(axis=-1)
monocholor = img.copy()
monocholor[~mask] = 255
contours = measure.find_contours(mask, 0.5)
# heuristic filter for contours
filter_contours = []
for c in contours:
region = Image.new("L", [img.shape[1], img.shape[0]], 0)
ImageDraw.Draw(region).polygon(list(map(lambda t: (t[1],t[0]), c)), fill=1)
region = np.array(region).astype(bool)
white_ratio = (monocholor == 255).all(axis=-1)[region].mean()
if white_ratio <= WHITE_RATIO_THRESHOLD:
filter_contours.append(c)
return filter_contours
```
We will use the following two binary masks to clip our images in order to extract the useful information only and therefore remove the legends along with the logos and titles.
```
# load mask of this size
leman_west = (6.148131, 46.206042)
quatre_canton_north = (8.435177, 47.082150)
majeur_east = (8.856851, 46.151857)
east_end = (10.472221, 46.544303)
constance_nw = (9.035247, 47.812716)
jura = (6.879290, 47.352935)
landmarks_colors = {
leman_west: raw_red,
quatre_canton_north: raw_green,
majeur_east: raw_blue,
constance_nw: raw_pink,
east_end: raw_yellow,
jura: raw_cyan
}
d_binary_mask, d_landmarks_pix = open_mask(*danger_img_arr.shape[:2])
s_binary_mask, s_landmarks_pix = open_mask(*snow_img_arr.shape[:2])
#display binary masks
fig, axes = plt.subplots(1, 2, figsize=(14,10))
# mask corresponding to danger image
axes[0].imshow(d_binary_mask);
widths, heights = list(zip(*d_landmarks_pix.values()))
axes[0].scatter(widths, heights);
axes[0].set_title('Mask informations (danger)');
# mask corresponding to danger image
axes[1].imshow(s_binary_mask);
widths, heights = list(zip(*s_landmarks_pix.values()))
axes[1].scatter(widths, heights);
axes[1].set_title('Mask informations (snow)');
fig, axes = plt.subplots(4, 2, figsize= (14,20))
# -------------------------------------------
# DANGER IMAGE
# -------------------------------------------
# original image
axes[0][0].imshow(danger_img_arr);
axes[0][0].set_title('Original image (danger)');
# keep useful colors
d_regions_only = keep_colors(danger_img_arr, shades_danger)
axes[0][1].imshow(d_regions_only);
axes[0][1].set_title('Keep only danger colors');
# clip the binary mask to remove color key
d_regions_only[~d_binary_mask] = 255
d_regions_only = Image.fromarray(d_regions_only).convert('RGB')
d_smoothed = d_regions_only.filter(ImageFilter.MedianFilter(7))
axes[1][0].imshow(d_smoothed);
axes[1][0].set_title('Smoothed with median filter (danger)');
# extract contours
axes[1][1].set_xlim([0, danger_img_arr.shape[1]])
axes[1][1].set_ylim([0, danger_img_arr.shape[0]])
axes[1][1].invert_yaxis()
axes[1][1].set_title('Regions contours')
for color in shades_danger:
contours = color_contours(d_smoothed, color)
for contour in contours:
axes[1][1].plot(contour[:, 1], contour[:, 0], linewidth=2, c=[x / 255 for x in color])
# -------------------------------------------
# SNOW IMAGE
# -------------------------------------------
# original image
axes[2][0].imshow(snow_img_arr);
axes[2][0].set_title('Original image (snow)');
#preprocessing to remove most of the noise
#remove grey colors
nogrey_img_arr = remove_colors(snow_img_arr, shades_grey)
#build colormap
color_map = build_color_map(nogrey_img_arr, snow_image_shades)
#map image colors to registered shades
new_img_arr = replace_color(nogrey_img_arr, color_map=color_map)
# keep useful colors
s_regions_only = keep_colors(new_img_arr, shades_snow)
axes[2][1].imshow(s_regions_only);
axes[2][1].set_title('Keep only snow colors');
# clip the binary mask to remove color key
s_regions_only[~s_binary_mask] = 255
s_regions_only = Image.fromarray(s_regions_only).convert('RGB')
s_smoothed = s_regions_only.filter(ImageFilter.MedianFilter(7))
axes[3][0].imshow(s_smoothed);
axes[3][0].set_title('Smoothed with median filter (danger)');
# extract contours
axes[3][1].set_xlim([0, snow_img_arr.shape[1]])
axes[3][1].set_ylim([0, snow_img_arr.shape[0]])
axes[3][1].invert_yaxis()
axes[3][1].set_title('Regions contours')
for color in shades_snow:
contours = color_contours(s_smoothed, color)
for contour in contours:
axes[3][1].plot(contour[:, 1], contour[:, 0], linewidth=2, c=[x / 255 for x in color])
```
# Contours to map polygon
Once we have contours we want to transform it into geographic coordinates and simplify the polygons.
To do this transformation, we use 5 points on the map to learn a transformation matrix $T$ that maps a pixel of the image to a geolocation. We could use only 3 points to have a valid transformation, but to dicrease the error we use 6 and solve a leastquare problem.
|**Location **| **Color**|
|---|---|
|Leman W| red|
|Quatre-cantons N| green|
|Lac majeur E| blue|
|Lac Constance NW| pink|
|Swiss E| yellow|
|Jura| cyan|
```
d_pix = np.array(list(map(numpify, d_landmarks_pix.values())))
d_coord = np.array(list(map(numpify, d_landmarks_pix.keys())))
# add 1 bias raw
d_pix_ext = np.vstack([np.ones((1,d_pix.shape[0])), d_pix.T])
d_coord_ext = np.vstack([np.ones((1,d_pix.shape[0])), d_coord.T])
# T = np.linalg.solve(
T = np.linalg.lstsq(d_pix_ext.T, d_coord_ext.T)[0]
s_pix = np.array(list(map(numpify, s_landmarks_pix.values())))
s_coord = np.array(list(map(numpify, s_landmarks_pix.keys())))
# add 1 bias raw
s_pix_ext = np.vstack([np.ones((1,s_pix.shape[0])), s_pix.T])
s_coord_ext = np.vstack([np.ones((1,s_pix.shape[0])), s_coord.T])
# T = np.linalg.solve(
T = np.linalg.lstsq(s_pix_ext.T, s_coord_ext.T)[0]
def transform_pix2map(points):
"""n x 2 array"""
points_ext = np.hstack([np.ones((points.shape[0], 1)), points])
points_map = points_ext.dot(T)
return points_map[:, 1:]
```
Obtained danger GeoJSON:
```
SMOOTHING_THRESHOLD = 0.0001
geo_json = {
"type": "FeatureCollection",
"features": []
}
for danger_level, color in enumerate(shades_danger):
for contour in color_contours(d_smoothed, color):
contour_right = contour.copy()
contour_right[:,0] = contour[:,1]
contour_right[:,1] = contour[:,0]
contour_right = transform_pix2map(contour_right)
simplifier = vw.Simplifier(contour_right)
contour_right = simplifier.simplify(threshold=SMOOTHING_THRESHOLD)
geo_json['features'].append({
"type": "Feature",
"properties": {
"date": "TODO",
"danger_level": danger_level + 1
},
"geometry": {
"type": "Polygon",
"coordinates": [ list(reversed(contour_right.tolist())) ]
}
})
switzerland = (46.875893, 8.289321)
tiles = 'https://server.arcgisonline.com/ArcGIS/rest/services/World_Topo_Map/MapServer/tile/{z}/{y}/{x}'
attr = 'Tiles © Esri — Esri, DeLorme, NAVTEQ, TomTom, Intermap, iPC, USGS, FAO, NPS, NRCAN, GeoBase, Kadaster NL, Ordnance Survey, Esri Japan, METI, Esri China (Hong Kong), and the GIS User Community'
m = folium.Map(location=switzerland, zoom_start=8, tiles=tiles, attr=attr)
colors = danger_colors_code
def style_function(risk_region):
level = risk_region['properties']['danger_level']
color = colors[level - 1]
return {
'fillOpacity': .5,
'weight': 0,
'fillColor': color,
'color': 'white',
}
folium.GeoJson(
geo_json,
name='geojson',
style_function=style_function
).add_to(m)
m
```
Obtained snow GeoJSON:
```
SMOOTHING_THRESHOLD = 0.0001
geo_json = {
"type": "FeatureCollection",
"features": []
}
for snow_level, color in enumerate(shades_snow):
for contour in color_contours(s_smoothed, color):
contour_right = contour.copy()
contour_right[:,0] = contour[:,1]
contour_right[:,1] = contour[:,0]
contour_right = transform_pix2map(contour_right)
simplifier = vw.Simplifier(contour_right)
contour_right = simplifier.simplify(threshold=SMOOTHING_THRESHOLD)
geo_json['features'].append({
"type": "Feature",
"properties": {
"date": "TODO",
"snow_level": snow_level + 1
},
"geometry": {
"type": "Polygon",
"coordinates": [ list(reversed(contour_right.tolist())) ]
}
})
switzerland = (46.875893, 8.289321)
tiles = 'https://server.arcgisonline.com/ArcGIS/rest/services/World_Topo_Map/MapServer/tile/{z}/{y}/{x}'
attr = 'Tiles © Esri — Esri, DeLorme, NAVTEQ, TomTom, Intermap, iPC, USGS, FAO, NPS, NRCAN, GeoBase, Kadaster NL, Ordnance Survey, Esri Japan, METI, Esri China (Hong Kong), and the GIS User Community'
m = folium.Map(location=switzerland, zoom_start=8, tiles=tiles, attr=attr)
colors = snow_color_code
def style_function(risk_region):
level = risk_region['properties']['danger_level']
color = colors[level - 1]
return {
'fillOpacity': .5,
'weight': 0,
'fillColor': color,
'color': 'white',
}
folium.GeoJson(
geo_json,
name='geojson',
style_function=style_function
).add_to(m)
m
```
|
github_jupyter
|
from pylab import contour
import matplotlib.pyplot as plt
from PIL import ImageFilter, Image, ImageDraw
from datetime import date, timedelta
import numpy as np
from PIL import Image
import cv2
from skimage import measure
import os
import pandas as pd
from scipy.spatial import distance
import json
import visvalingamwyatt as vw
import folium
%matplotlib inline
black = np.array([0, 0, 0])
white = np.array([255, 255, 255])
green = np.array([204, 255, 102])
yellow = np.array([255, 255, 0])
orange = np.array([255, 153, 0])
red = np.array([255, 0, 0])
danger_colors_code = ['#ccff66', '#ffff00', '#ff9900', '#ff0000']
shades_danger = [green, yellow, orange, red]
danger_image_shades = [green, yellow, orange, red, white]
light_blue = np.array([213, 252, 252])
light_medium_blue = np.array([168, 217, 241])
medium_blue = np.array([121, 161, 229])
dark_medium_blue = np.array([68, 89, 215])
dark_blue = np.array([47, 36, 162])
purple = np.array([91, 32, 196])
snow_color_code = ['#d5fcfc', '#a8d9f1', '#79a1e5', '#4459d7', '#2f24a2', '#5b20c4']
shades_snow = [light_blue, light_medium_blue, medium_blue, dark_medium_blue, dark_blue, purple]
shades_grey = [np.array([c,c,c]) for c in range(255)]
snow_image_shades = [light_blue, light_medium_blue, medium_blue, dark_medium_blue, dark_blue, purple, white]
raw_red = np.array([255, 0, 0])
raw_green = np.array([0, 255, 0])
raw_blue = np.array([0, 0, 255])
raw_pink = np.array([255, 0, 255])
raw_pink = np.array([255, 0, 255])
raw_cyan = np.array([0, 255, 255])
raw_yellow = np.array([255, 255, 0])
def keep_colors(img, colors, replace_with=white):
"""return a new image with only the `colors` selected, other pixel are `replace_with`"""
keep = np.zeros(img.shape[:2], dtype=bool)
for c in colors:
keep = keep | (c == img).all(axis=-1)
new_img = img.copy()
new_img[~keep] = replace_with
return new_img
def remove_colors(img, colors, replace_with=white):
"""return a new image without the `colors` selected which will be replaced by `replace_with`"""
keep = np.zeros(img.shape[:2], dtype=bool)
for c in colors:
keep = keep | (c == img).all(axis=-1)
new_img = img.copy()
new_img[keep] = replace_with
return new_img
def replace_color(img, color_map):
"""return a new image replacing the image colors which will be mapped to their corresponding colors in `color_map` (df)"""
new_img = img.copy()
for _, (source, target) in color_map.iterrows():
new_img[(img == source).all(axis=-1)] = target
return new_img
def build_color_map(img_arr, image_shades):
"""return colormap as dataframe"""
im_df = pd.DataFrame([img_arr[i,j,:] for i,j in np.ndindex(img_arr.shape[0],img_arr.shape[1])])
im_df = im_df.drop_duplicates()
image_colors = im_df.as_matrix()
colors = np.zeros(image_colors.shape)
dist = distance.cdist(image_colors, image_shades, 'sqeuclidean')
for j in range(dist.shape[0]):
distances = dist[j,:]
colors[j, :] = image_shades[distances.argmin()]
color_map = pd.DataFrame(
{'source': image_colors.tolist(),
'target': colors.tolist()
})
return color_map
danger_path = '../data/slf/2001/nbk/de/gif/20001230_nbk_de_c.gif'
snow_path = '../data/slf/2010/hstop/en/gif/20100103_hstop_en_c.gif'
danger_img = Image.open(danger_path)
danger_img = danger_img.convert('RGB')
danger_img_arr = np.array(danger_img)
snow_img = Image.open(snow_path)
snow_img = snow_img.convert('RGB')
snow_img_arr = np.array(snow_img)
fig, axes = plt.subplots(1, 2, figsize=(14,10))
# original danger image
axes[0].imshow(danger_img_arr);
axes[0].set_title('Original danger image');
# original snow image
axes[1].imshow(snow_img_arr);
axes[1].set_title('Original snow image');
def numpify(o):
if not isinstance(o, np.ndarray):
o = np.array(o)
return o
def coord_color(img, color):
return np.array(list(zip(*(img == color).all(-1).nonzero())))
def open_mask(height, width):
masks_path = '../map-masks/'
mask_name = '{}x{}.gif'.format(height, width)
mask_path = os.path.join(masks_path, mask_name)
mask = Image.open(mask_path)
mask = mask.convert('RGB')
mask = np.array(mask)
landmarks_pix = {
geo_point: (width, height)
for geo_point, color in landmarks_colors.items()
for height, width in coord_color(mask, color)
}
binary_mask = (mask != 255).any(-1) # different of white
return binary_mask, landmarks_pix
# remove contours areas that have more than 30% of white
WHITE_RATIO_THRESHOLD = .3
def color_contours(img, color):
img = numpify(img)
color = numpify(color)
mask = (img == color[:3]).all(axis=-1)
monocholor = img.copy()
monocholor[~mask] = 255
contours = measure.find_contours(mask, 0.5)
# heuristic filter for contours
filter_contours = []
for c in contours:
region = Image.new("L", [img.shape[1], img.shape[0]], 0)
ImageDraw.Draw(region).polygon(list(map(lambda t: (t[1],t[0]), c)), fill=1)
region = np.array(region).astype(bool)
white_ratio = (monocholor == 255).all(axis=-1)[region].mean()
if white_ratio <= WHITE_RATIO_THRESHOLD:
filter_contours.append(c)
return filter_contours
# load mask of this size
leman_west = (6.148131, 46.206042)
quatre_canton_north = (8.435177, 47.082150)
majeur_east = (8.856851, 46.151857)
east_end = (10.472221, 46.544303)
constance_nw = (9.035247, 47.812716)
jura = (6.879290, 47.352935)
landmarks_colors = {
leman_west: raw_red,
quatre_canton_north: raw_green,
majeur_east: raw_blue,
constance_nw: raw_pink,
east_end: raw_yellow,
jura: raw_cyan
}
d_binary_mask, d_landmarks_pix = open_mask(*danger_img_arr.shape[:2])
s_binary_mask, s_landmarks_pix = open_mask(*snow_img_arr.shape[:2])
#display binary masks
fig, axes = plt.subplots(1, 2, figsize=(14,10))
# mask corresponding to danger image
axes[0].imshow(d_binary_mask);
widths, heights = list(zip(*d_landmarks_pix.values()))
axes[0].scatter(widths, heights);
axes[0].set_title('Mask informations (danger)');
# mask corresponding to danger image
axes[1].imshow(s_binary_mask);
widths, heights = list(zip(*s_landmarks_pix.values()))
axes[1].scatter(widths, heights);
axes[1].set_title('Mask informations (snow)');
fig, axes = plt.subplots(4, 2, figsize= (14,20))
# -------------------------------------------
# DANGER IMAGE
# -------------------------------------------
# original image
axes[0][0].imshow(danger_img_arr);
axes[0][0].set_title('Original image (danger)');
# keep useful colors
d_regions_only = keep_colors(danger_img_arr, shades_danger)
axes[0][1].imshow(d_regions_only);
axes[0][1].set_title('Keep only danger colors');
# clip the binary mask to remove color key
d_regions_only[~d_binary_mask] = 255
d_regions_only = Image.fromarray(d_regions_only).convert('RGB')
d_smoothed = d_regions_only.filter(ImageFilter.MedianFilter(7))
axes[1][0].imshow(d_smoothed);
axes[1][0].set_title('Smoothed with median filter (danger)');
# extract contours
axes[1][1].set_xlim([0, danger_img_arr.shape[1]])
axes[1][1].set_ylim([0, danger_img_arr.shape[0]])
axes[1][1].invert_yaxis()
axes[1][1].set_title('Regions contours')
for color in shades_danger:
contours = color_contours(d_smoothed, color)
for contour in contours:
axes[1][1].plot(contour[:, 1], contour[:, 0], linewidth=2, c=[x / 255 for x in color])
# -------------------------------------------
# SNOW IMAGE
# -------------------------------------------
# original image
axes[2][0].imshow(snow_img_arr);
axes[2][0].set_title('Original image (snow)');
#preprocessing to remove most of the noise
#remove grey colors
nogrey_img_arr = remove_colors(snow_img_arr, shades_grey)
#build colormap
color_map = build_color_map(nogrey_img_arr, snow_image_shades)
#map image colors to registered shades
new_img_arr = replace_color(nogrey_img_arr, color_map=color_map)
# keep useful colors
s_regions_only = keep_colors(new_img_arr, shades_snow)
axes[2][1].imshow(s_regions_only);
axes[2][1].set_title('Keep only snow colors');
# clip the binary mask to remove color key
s_regions_only[~s_binary_mask] = 255
s_regions_only = Image.fromarray(s_regions_only).convert('RGB')
s_smoothed = s_regions_only.filter(ImageFilter.MedianFilter(7))
axes[3][0].imshow(s_smoothed);
axes[3][0].set_title('Smoothed with median filter (danger)');
# extract contours
axes[3][1].set_xlim([0, snow_img_arr.shape[1]])
axes[3][1].set_ylim([0, snow_img_arr.shape[0]])
axes[3][1].invert_yaxis()
axes[3][1].set_title('Regions contours')
for color in shades_snow:
contours = color_contours(s_smoothed, color)
for contour in contours:
axes[3][1].plot(contour[:, 1], contour[:, 0], linewidth=2, c=[x / 255 for x in color])
d_pix = np.array(list(map(numpify, d_landmarks_pix.values())))
d_coord = np.array(list(map(numpify, d_landmarks_pix.keys())))
# add 1 bias raw
d_pix_ext = np.vstack([np.ones((1,d_pix.shape[0])), d_pix.T])
d_coord_ext = np.vstack([np.ones((1,d_pix.shape[0])), d_coord.T])
# T = np.linalg.solve(
T = np.linalg.lstsq(d_pix_ext.T, d_coord_ext.T)[0]
s_pix = np.array(list(map(numpify, s_landmarks_pix.values())))
s_coord = np.array(list(map(numpify, s_landmarks_pix.keys())))
# add 1 bias raw
s_pix_ext = np.vstack([np.ones((1,s_pix.shape[0])), s_pix.T])
s_coord_ext = np.vstack([np.ones((1,s_pix.shape[0])), s_coord.T])
# T = np.linalg.solve(
T = np.linalg.lstsq(s_pix_ext.T, s_coord_ext.T)[0]
def transform_pix2map(points):
"""n x 2 array"""
points_ext = np.hstack([np.ones((points.shape[0], 1)), points])
points_map = points_ext.dot(T)
return points_map[:, 1:]
SMOOTHING_THRESHOLD = 0.0001
geo_json = {
"type": "FeatureCollection",
"features": []
}
for danger_level, color in enumerate(shades_danger):
for contour in color_contours(d_smoothed, color):
contour_right = contour.copy()
contour_right[:,0] = contour[:,1]
contour_right[:,1] = contour[:,0]
contour_right = transform_pix2map(contour_right)
simplifier = vw.Simplifier(contour_right)
contour_right = simplifier.simplify(threshold=SMOOTHING_THRESHOLD)
geo_json['features'].append({
"type": "Feature",
"properties": {
"date": "TODO",
"danger_level": danger_level + 1
},
"geometry": {
"type": "Polygon",
"coordinates": [ list(reversed(contour_right.tolist())) ]
}
})
switzerland = (46.875893, 8.289321)
tiles = 'https://server.arcgisonline.com/ArcGIS/rest/services/World_Topo_Map/MapServer/tile/{z}/{y}/{x}'
attr = 'Tiles © Esri — Esri, DeLorme, NAVTEQ, TomTom, Intermap, iPC, USGS, FAO, NPS, NRCAN, GeoBase, Kadaster NL, Ordnance Survey, Esri Japan, METI, Esri China (Hong Kong), and the GIS User Community'
m = folium.Map(location=switzerland, zoom_start=8, tiles=tiles, attr=attr)
colors = danger_colors_code
def style_function(risk_region):
level = risk_region['properties']['danger_level']
color = colors[level - 1]
return {
'fillOpacity': .5,
'weight': 0,
'fillColor': color,
'color': 'white',
}
folium.GeoJson(
geo_json,
name='geojson',
style_function=style_function
).add_to(m)
m
SMOOTHING_THRESHOLD = 0.0001
geo_json = {
"type": "FeatureCollection",
"features": []
}
for snow_level, color in enumerate(shades_snow):
for contour in color_contours(s_smoothed, color):
contour_right = contour.copy()
contour_right[:,0] = contour[:,1]
contour_right[:,1] = contour[:,0]
contour_right = transform_pix2map(contour_right)
simplifier = vw.Simplifier(contour_right)
contour_right = simplifier.simplify(threshold=SMOOTHING_THRESHOLD)
geo_json['features'].append({
"type": "Feature",
"properties": {
"date": "TODO",
"snow_level": snow_level + 1
},
"geometry": {
"type": "Polygon",
"coordinates": [ list(reversed(contour_right.tolist())) ]
}
})
switzerland = (46.875893, 8.289321)
tiles = 'https://server.arcgisonline.com/ArcGIS/rest/services/World_Topo_Map/MapServer/tile/{z}/{y}/{x}'
attr = 'Tiles © Esri — Esri, DeLorme, NAVTEQ, TomTom, Intermap, iPC, USGS, FAO, NPS, NRCAN, GeoBase, Kadaster NL, Ordnance Survey, Esri Japan, METI, Esri China (Hong Kong), and the GIS User Community'
m = folium.Map(location=switzerland, zoom_start=8, tiles=tiles, attr=attr)
colors = snow_color_code
def style_function(risk_region):
level = risk_region['properties']['danger_level']
color = colors[level - 1]
return {
'fillOpacity': .5,
'weight': 0,
'fillColor': color,
'color': 'white',
}
folium.GeoJson(
geo_json,
name='geojson',
style_function=style_function
).add_to(m)
m
| 0.714728 | 0.903975 |
<a href="https://colab.research.google.com/github/partha1189/machine_learning/blob/master/Common_patterns_in_time_series.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
```
import numpy as np
import matplotlib.pyplot as plt
def plot_series(time, series, format='-', start=0, end=None, label=None):
plt.plot(time[start:end], series[start:end], format, label = label)
plt.xlabel('Time')
plt.ylabel('Value')
if label:
plt.legend(fontsize=14)
plt.grid(True)
```
Trend & Seasonality
```
def trend(time, slope=0):
return slope * time
time = np.arange( 4 * 365 + 1)
time
baseline = 10
series = baseline + trend(time, 0.1)
plt.figure(figsize=(10, 6))
plot_series(time, series)
plt.show()
series
def seasonal_pattern(season_time):
return np.where(season_time < 0.4,
np.cos(season_time * 2 * np.pi),
1 / np.exp(3 * season_time))
def seasonality(time, period, amplitude=1, phase=0):
season_time = ((time + phase) % period) / period
return amplitude * seasonal_pattern(season_time)
amplitude = 40
series = seasonality(time, period=365, amplitude=amplitude)
plt.figure(figsize=(10, 6))
plot_series(time, series)
plt.show()
amplitude = 20
series = seasonality(time, period = 365, amplitude=amplitude, phase=5)
series = series + trend(time, slope=0.1)
plt.figure(figsize=(10, 6))
plot_series(time, series)
plt.show()
```
Noise
```
def white_noise(time, noise_level =1, seed=None):
rnd = np.random.RandomState(seed)
return rnd.randn(len(time)) * noise_level
noise_level = 5
noise = white_noise(time, noise_level, seed = 42)
plt.figure(figsize=(10, 6))
plot_series(time, noise)
plt.show()
series += noise
plt.figure(figsize=(10, 6))
plot_series(time, series)
plt.show()
time = np.arange(4 * 365 + 1)
slope = 0.05
baseline = 10
amplitude = 40
series = baseline + trend(time, slope) + seasonality(time, period=365, amplitude=amplitude)
noise_level = 5
noise = white_noise(time, noise_level, seed=42)
series += noise
plt.figure(figsize=(10, 6))
plot_series(time, series)
plt.show()
split_time = 1000
time_train = time[:split_time]
x_train = series[:split_time]
time_valid = time[split_time:]
x_valid = series[split_time:]
```
NAIVE FORECAST
```
naive_forecast = series[split_time - 1: -1]
plt.figure(figsize=(10, 6))
plot_series(time_valid, x_valid, label="Series")
plot_series(time_valid, naive_forecast, label="Forecast")
plt.figure(figsize=(10, 6))
plot_series(time_valid, x_valid, start=0, end=150, label="Series")
plot_series(time_valid, naive_forecast, start=1, end=151, label="Forecast")
errors = naive_forecast - x_valid
abs_errors = np.abs(errors)
abs_errors
mae = np.mean(abs_errors)
mae
import tensorflow as tf
tf.keras.losses.mean_absolute_error(x_valid, naive_forecast).numpy()
```
|
github_jupyter
|
import numpy as np
import matplotlib.pyplot as plt
def plot_series(time, series, format='-', start=0, end=None, label=None):
plt.plot(time[start:end], series[start:end], format, label = label)
plt.xlabel('Time')
plt.ylabel('Value')
if label:
plt.legend(fontsize=14)
plt.grid(True)
def trend(time, slope=0):
return slope * time
time = np.arange( 4 * 365 + 1)
time
baseline = 10
series = baseline + trend(time, 0.1)
plt.figure(figsize=(10, 6))
plot_series(time, series)
plt.show()
series
def seasonal_pattern(season_time):
return np.where(season_time < 0.4,
np.cos(season_time * 2 * np.pi),
1 / np.exp(3 * season_time))
def seasonality(time, period, amplitude=1, phase=0):
season_time = ((time + phase) % period) / period
return amplitude * seasonal_pattern(season_time)
amplitude = 40
series = seasonality(time, period=365, amplitude=amplitude)
plt.figure(figsize=(10, 6))
plot_series(time, series)
plt.show()
amplitude = 20
series = seasonality(time, period = 365, amplitude=amplitude, phase=5)
series = series + trend(time, slope=0.1)
plt.figure(figsize=(10, 6))
plot_series(time, series)
plt.show()
def white_noise(time, noise_level =1, seed=None):
rnd = np.random.RandomState(seed)
return rnd.randn(len(time)) * noise_level
noise_level = 5
noise = white_noise(time, noise_level, seed = 42)
plt.figure(figsize=(10, 6))
plot_series(time, noise)
plt.show()
series += noise
plt.figure(figsize=(10, 6))
plot_series(time, series)
plt.show()
time = np.arange(4 * 365 + 1)
slope = 0.05
baseline = 10
amplitude = 40
series = baseline + trend(time, slope) + seasonality(time, period=365, amplitude=amplitude)
noise_level = 5
noise = white_noise(time, noise_level, seed=42)
series += noise
plt.figure(figsize=(10, 6))
plot_series(time, series)
plt.show()
split_time = 1000
time_train = time[:split_time]
x_train = series[:split_time]
time_valid = time[split_time:]
x_valid = series[split_time:]
naive_forecast = series[split_time - 1: -1]
plt.figure(figsize=(10, 6))
plot_series(time_valid, x_valid, label="Series")
plot_series(time_valid, naive_forecast, label="Forecast")
plt.figure(figsize=(10, 6))
plot_series(time_valid, x_valid, start=0, end=150, label="Series")
plot_series(time_valid, naive_forecast, start=1, end=151, label="Forecast")
errors = naive_forecast - x_valid
abs_errors = np.abs(errors)
abs_errors
mae = np.mean(abs_errors)
mae
import tensorflow as tf
tf.keras.losses.mean_absolute_error(x_valid, naive_forecast).numpy()
| 0.776962 | 0.984884 |
# Vegetarian trends at food.com
The interest in vegetarian and vegan food has grown steadily over the past two decades.
The number of searches for 'veganism' in the UK has increased 900% from 2009 to 2019.[1]
And although in 2015, just 3.4% of all Americans said they were vegetarian, fully a quarter of 25- to 34-year-olds identified as such.[2]
Below, I will explore whether this vegetarian trend extends to the users of [food.com](https://www.food.com/), one of the leading online recipe websites.
[1] [Veganism: Why are vegan diets on the rise?](https://www.bbc.com/news/business-44488051)
[2] [The year of the vegan, The Economist](https://worldin2019.economist.com/theyearofthevegan)
## Questions
1. Is there a positive trend in the number of vegetarian recipes posted on food.com between 2008 and 2017?
2. Is there a positive trend in the number of interactions with vegetarian recipes on Food.com between 2008 and 2017?
2. Is there a difference in the ratings vegetarian recipes received compared to non-vegetarian recipes between 2008 and 2017?
## Dataset
I will be using the [food.com dataset](https://www.kaggle.com/shuyangli94/food-com-recipes-and-user-interactions) by Bodhisattwa Prasad Majumder, Shuyang Li, Jianmo Ni, and Julian McAuley.
The dataset consists of 180K+ recipes and 700K+ recipe reviews covering 18 years of user interactions and uploads on Food.com (formerly GeniusKitchen).
For the purpose of this exploration study, I will look at data between 2008 and 2017.
## Imports
```
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from matplotlib_venn import venn2
import seaborn as sns
from ast import literal_eval
%matplotlib inline
```
## Palettes
```
veg_meat = ["#454d66", "#b7e778", "#1fab89"]
sns.set_palette(veg_meat)
```
## Data import & summary
```
recipes = pd.read_csv('/kaggle/input/food-com-recipes-and-user-interactions/RAW_recipes.csv')
interactions = pd.read_csv('/kaggle/input/food-com-recipes-and-user-interactions/RAW_interactions.csv')
recipes.head()
print(recipes.info())
recipes.describe()
```
2,147,484,000 minutes sounds like a very long time to cook a recipe. Let's take a closer look at the `minutes` column later on.
```
recipes[['minutes', 'n_steps', 'n_ingredients']].hist()
interactions.head()
print(interactions.info())
interactions.describe()
interactions['rating'].hist()
```
Users tend to rate recipes very highly.
This is unlikely to mean that they are universally satisfied with recipes on food.com. More likely, users who don't like the results of a recipe either don't bother to rate, or blame their own cooking skills, giving the recipe author a benefit of the doubt.
## Data wrangling
Let's start by filtering the dataset down to our chosen 10-year interval.
```
from_year, to_year = '2008-01-01','2017-12-31'
recipes['submitted'] = pd.to_datetime(recipes['submitted'])
recipes['submitted'] = recipes['submitted'].apply(lambda x: x.tz_localize(None))
recipes_l0y = recipes.loc[recipes['submitted'].between(from_year, to_year, inclusive=False)]
interactions['date'] = pd.to_datetime(interactions['date'])
interactions['date'] = interactions['date'].apply(lambda x: x.tz_localize(None))
interactions_l0y = interactions.loc[interactions['date'].between(from_year, to_year, inclusive=False)]
print(recipes_l0y.shape)
print(interactions_l0y.shape)
```
### Remove outliers
```
sns.boxplot(x=recipes_l0y["minutes"])
```
There is clearly at least one extreme outlier in the data set. 694 days is too long of a preparation time for even the tastiest of recipes!
```
# calculate the first quartile, third quartile and the interquartile range
Q1 = recipes_l0y['minutes'].quantile(0.25)
Q3 = recipes_l0y['minutes'].quantile(0.75)
IQR = Q3 - Q1
# calculate the maximum value and minimum values according to the Tukey rule
max_value = Q3 + 1.5 * IQR
min_value = Q1 - 1.5 * IQR
# filter the data for values that are greater than max_value or less than min_value
minutes_outliers = recipes_l0y[(recipes_l0y['minutes'] > max_value) | (recipes_l0y['minutes'] < min_value)]
minutes_outliers.sort_values('minutes')
```
As we can see above, the Tukey method filters out many reasonable recipes as outliers. Some recipes, such as pickles, extracts and liqueurs can take many days to prepare, and should not be excluded.
The one extreme outlier at 1051200 minutes is the [How to Preserve a Husband](https://www.food.com/recipe/how-to-preserve-a-husband-447963) recipe. Although it is no doubt very valuable, I will exclude it from the rest of this exploration.
### Exclude How to Preserve a Husband recipe
```
# filter out recipes that take longer than 730 days as outliers
recipes_l0y = recipes_l0y.query('minutes < 1051200')
```
### Rating count and average by recipe and year
```
recipes_l0y['year'] = recipes_l0y['submitted'].dt.year
interactions_l0y['year'] = interactions_l0y['date'].dt.year
ratings_by_recipe = interactions_l0y.groupby(['recipe_id', 'year']).agg(
rating_cnt = ('rating', 'count'),
rating_avg = ('rating', 'mean'),
)
ratings_by_recipe.head()
```
### Merge recipes and ratings
```
recipes_and_ratings = recipes_l0y.merge(ratings_by_recipe, left_on='id', right_on='recipe_id')
recipes_and_ratings.head(2)
```
### Tags to lists
```
# convert the tags column to list format
recipes_and_ratings['tags'] = recipes_and_ratings['tags'].apply(lambda x: literal_eval(str(x)))
```
### Add vegan and vegetarian columns, check overlap
```
# add vegetarian and vegan boolean columns
recipes_and_ratings['vegetarian'] = ['vegetarian' in tag for tag in recipes_and_ratings['tags']]
recipes_and_ratings['vegan'] = ['vegan' in tag for tag in recipes_and_ratings['tags']]
recipes_and_ratings = recipes_and_ratings.drop(columns=['name', 'tags', 'nutrition', 'steps', 'description', 'ingredients'])
recipes_and_ratings.head(2)
#plot a venn diagram of vegetarian and vegan recipe counts
vegetarian_cnt = len(recipes_and_ratings.query('vegetarian == True'))
vegan_cnt = len(recipes_and_ratings.query('vegan == True'))
intersect_cnt = len(recipes_and_ratings.query('vegetarian == True and vegan == True'))
venn2(subsets = (vegetarian_cnt, vegan_cnt-intersect_cnt, intersect_cnt), set_labels = ('Vegetarian', 'Vegan'), set_colors=('#b7e778', '#031c16', '#031c16'), alpha = 1)
```
As expected, we can se that the `vegetarian` tag is a superset of the `vegan` category, so we don't need to preprocess the tags any further.
Given the very high likelyhood of users forgetting to tag vegan recipes as vegetarian, we can assume that the tags were automatically generated by the system.
## Exploration
### New recipes by year
```
df = recipes_and_ratings.groupby(['year', 'vegetarian']).agg(
recipe_cnt = ('id', 'count')
).reset_index()
plt.figure(figsize=(12,6))
ax = sns.lineplot(data=df, x='year', y='recipe_cnt', hue='vegetarian', linewidth=2.5)
ax.set(ylim=(0, None))
ax.set_title('Number of new recipes by year')
ax
```
We can see that there has been a rapid decline in the number of new recipes posted on food.com over the past decade.
Assuming that the data set is not missing information for some of the more recent years,this decline is somewhat perplexing, as food.com is the second largest recipe website, and internet usage overall has [increased by 10 percentage points](https://www.pewresearch.org/fact-tank/2018/09/28/internet-social-media-use-and-device-ownership-in-u-s-have-plateaued-after-years-of-growth/) in the last decade.
Nevertheless, there could be a number of explanations, which we unfortunately won't have the opportunity to answer in this study:
* There is a saturation of recipes on the website. Everything there is to cook is already covered.
* Related to the preciding point, Food.com might have stopped investing in attracting new contributors.
* Users have shifted their recipe consumption to mobile apps, made by other providers.
* Most prolific recipe authors launched their own recipe blogs, or moved to social media.
Whatever the reason for the decline, let's see whether it has had an equal impact on vegetarian and non-vegetarian recipes.
```
df = recipes_and_ratings.groupby(['year']).agg(
total_cnt = ('id', 'count'),
vegetarian_cnt = ('vegetarian', 'sum'),
vegan_cnt = ('vegan', 'sum'),
).reset_index()
df['vegetarian_pct'] = df['vegetarian_cnt'] / df['total_cnt'] * 100
df['vegan_pct'] = df['vegan_cnt'] / df['total_cnt'] * 100
plt.figure(figsize=(12,6))
ax = sns.lineplot(data=pd.melt(df[['year', 'vegetarian_pct', 'vegan_pct']], ['year']), x='year', y='value', palette=veg_meat[1:], hue='variable', linewidth=2.5)
ax.set(ylim=(0, 100))
ax.set_title('Percent of vegetarian recipes by year')
ax
```
The vegetarian category has declined at the same rate as the non-vegetarian portion of food.com, and did even worse between 2014–2017.
### Ratings by year
```
ratings_by_recipe = interactions_l0y.groupby(['recipe_id', 'year']).agg(
rating_cnt = ('rating', 'count'),
rating_avg = ('rating', 'mean'),
).reset_index()
ratings_by_recipe = ratings_by_recipe.merge(recipes_and_ratings[['id', 'vegetarian', 'vegan']], left_on='recipe_id', right_on='id')
df = ratings_by_recipe.groupby(['year', 'vegetarian']).agg(
rating_cnt = ('rating_cnt', 'sum'),
rating_avg = ('rating_avg', 'mean'),
).reset_index()
plt.figure(figsize=(12,6))
ax = sns.lineplot(data=df, x='year', y='rating_cnt', hue='vegetarian', linewidth=2.5)
ax.set_title('Recipe ratings by year')
ax
```
We can see that there has been a similar decline in the number of interactions (reviews) on the recipes.
The decline started one year later, which could probably be explained by a lag between new recipe postings and ratings. That is, the spike in new recipe postings in 2008 would only convert into interactions in the following year.
Again, let's see whether the decline has had an equal impact on vegetarian and non-vegetarian recipes.
```
interactions_by_recipe_and_year = interactions_l0y.reset_index().groupby(['recipe_id', 'year']).agg(
rating_cnt = ('index', 'count'),
rating_avg = ('rating', 'mean'),
).reset_index()
interactions_and_recipes = interactions_by_recipe_and_year[['recipe_id', 'year', 'rating_cnt', 'rating_avg']].merge(recipes_and_ratings[['id', 'vegetarian', 'vegan']], left_on='recipe_id', right_on='id')
interactions_and_recipes['vegetarian_rating_cnt'] = np.where(interactions_and_recipes['vegetarian'] == True, interactions_and_recipes['rating_cnt'], 0)
interactions_and_recipes['vegan_rating_cnt'] = np.where(interactions_and_recipes['vegan'] == True, interactions_and_recipes['rating_cnt'], 0)
df = interactions_and_recipes.groupby(['year']).agg(
total_cnt = ('rating_cnt', 'sum'),
vegetarian_cnt = ('vegetarian_rating_cnt', 'sum'),
vegan_cnt = ('vegan_rating_cnt', 'sum'),
).reset_index()
df['vegetarian_pct'] = df['vegetarian_cnt'] / df['total_cnt'] * 100
df['vegan_pct'] = df['vegan_cnt'] / df['total_cnt'] * 100
plt.figure(figsize=(12,6))
ax = sns.lineplot(data=pd.melt(df[['year', 'vegetarian_pct', 'vegan_pct']], ['year']), x='year', y='value', palette=veg_meat[1:], hue='variable', linewidth=2.5)
ax.set(ylim=(0, 100))
ax.set_title('Percent of votes on vegetarian recipes by year')
ax
```
The share of ratings posted on vegetarian and vegan recipes has remained flat through the 10-year period. This time, we don't even see a decline in the period between 2014 and 2017.
This may suggest that although the number of vegetarian contributors has declined at a faster rate than that of non-vegetarian authors, the reader composition remained roughly the same.
```
df = ratings_by_recipe.groupby(['year', 'vegetarian']).agg(
rating_avg = ('rating_avg', 'mean')
).reset_index()
plt.figure(figsize=(12,6))
ax = sns.lineplot(data=df, x='year', y='rating_avg', hue='vegetarian', linewidth=2.5)
ax.set(ylim=(0, 5))
ax.set_title('Average recipe rating by year')
ax
```
The average rating for vegetarian recipes was roughly the same until 2013, but has since grown to ~0.2 points above that of non-vegetarian recipes.
## Cohort analysis
I will next conduct a cohort retention analysis to confirm that the number of vegetarian contributors has indeed declined at a faster rate than that of non-vegetarian authors.
Some of the code below was taken from: http://www.gregreda.com/2015/08/23/cohort-analysis-with-python/
### Add submission year column
```
recipes_and_cohorts = recipes_and_ratings.copy()
recipes_and_cohorts['submitted_year'] = recipes_and_cohorts['submitted'].apply(lambda x: x.strftime('%Y'))
```
### Divide users into cohorts
```
# add cohort column — the year of the user's first recipe submission
recipes_and_cohorts.set_index('contributor_id', inplace=True)
recipes_and_cohorts['contributor_cohort'] = recipes_and_cohorts.groupby(level=0)['submitted'].min().apply(lambda x: x.strftime('%Y'))
recipes_and_cohorts.reset_index(inplace=True)
recipes_and_cohorts.head()
def add_cohort_periods(df):
"""
Creates a `cohort_period` column, which is the Nth period based on the contributor's first recipe.
"""
df['cohort_period'] = np.arange(len(df)) + 1
return df
def group_into_cohorts(df):
"""
Aggregates contributor count, recipe count and cohort period by contributor cohort and year of submission.
"""
df = df.groupby(['contributor_cohort', 'submitted_year']).agg(
contributor_cnt = ('contributor_id', 'nunique'),
recipe_cnt = ('id', 'nunique'),
)
df = df.groupby('contributor_cohort').apply(add_cohort_periods)
return df
# non-vegetarian cohorts
cohorts_nonveg = group_into_cohorts(recipes_and_cohorts[recipes_and_cohorts['vegetarian'] == False])
# vegetarian cohorts
cohorts_veg = group_into_cohorts(recipes_and_cohorts[recipes_and_cohorts['vegetarian'] == True])
cohorts_veg.head()
```
### User retention by cohort group
```
def calculate_cohort_sizes(df):
"""
Calculates cohort sizes.
"""
df.reset_index(inplace=True)
df.set_index(['contributor_cohort', 'cohort_period'], inplace=True)
return df['contributor_cnt'].groupby('contributor_cohort').first()
# calculate cohort sizes
cohort_sizes_nonveg = calculate_cohort_sizes(cohorts_nonveg)
cohort_sizes_veg = calculate_cohort_sizes(cohorts_veg)
cohort_sizes_veg.head()
def convert_cohort_counts_to_pct(df, cohort_sizes):
"""
Converts cohort period contributor counts to percentages.
"""
df = df.unstack(0).divide(cohort_sizes, axis=1)
df.reset_index(inplace=True)
return df
# convert cohort period contributor counts to percentages
contributor_retention_nonveg = convert_cohort_counts_to_pct(cohorts_nonveg['contributor_cnt'], cohort_sizes_nonveg)
contributor_retention_veg = convert_cohort_counts_to_pct(cohorts_veg['contributor_cnt'], cohort_sizes_veg)
contributor_retention_veg
def plot_retention_curves(df, cohorts, title, position):
"""
Plots retention curves for cohorts.
"""
plot = sns.lineplot(
data=pd.melt(contributor_retention_nonveg[['cohort_period'] + cohorts], ['cohort_period']),
x='cohort_period',
y='value',
palette='rocket_r',
hue='contributor_cohort',
linewidth=2.5,
ax=ax[position])
plot.set(xlim=(0, 8))
plot.set(ylim=(0, 1))
plot.set(xlabel='Cohort period')
plot.set(ylabel='Active contributors')
plot.set_title('Contributor retention by cohort: ' + title)
return
# plot contributor retention curves
fig, ax = plt.subplots(1, 2, figsize=(12,6))
cohorts_to_display = ['2008', '2009', '2010', '2011']
plot_retention_curves(contributor_retention_nonveg, cohorts_to_display, 'Non-vegetarian', 0)
plot_retention_curves(contributor_retention_veg, cohorts_to_display, 'Vegetarian', 1)
fig.show()
```
We can see that contributor retention at food.com has deteriorated significantly over the years.
The difference between the churn of vegetarian and non-vegetarian contributors appears very mild, however, suggesting that higher atrition rate is not the reason for the decline in the proportion of new vegetarian recipes on the site.
## Contributor acquisition
The alternative explanation for the drop in the share of new vegetarian recipes is that food.com acquires non-vegetarian contributors at a faster rate than vegetarian authors.
```
# get first recipe by contributor
df = recipes_and_cohorts.groupby('contributor_id').agg(
vegetarian = ('vegetarian', 'mean'),
contributor_cohort = ('contributor_cohort', 'min'),
)
# counting contributors with >50% of vegetarian contibutions as vegetarians
df.reset_index(inplace=True)
df = df.round(0)
# get first recipe by contributor
df = df.groupby(['contributor_cohort', 'vegetarian']).agg(
contributor_cnt = ('contributor_id', 'count'),
)
# counting contributors with >50% of vegetarian contibutions as vegetarians
df.reset_index(inplace=True)
df['vegetarian'] = df['vegetarian'].astype(bool)
plt.figure(figsize=(12,6))
ax = sns.lineplot(data=df, x='contributor_cohort', y='contributor_cnt', palette=veg_meat[:2], hue='vegetarian', linewidth=2.5)
ax.set(xlabel='New contributors')
ax.set(ylabel='Year')
ax.set_title('New contributors by year')
```
Let's try the same with a logarithmic yscale.
```
plt.figure(figsize=(12,6))
ax = sns.lineplot(data=df, x='contributor_cohort', y='contributor_cnt', palette=veg_meat[:2], hue='vegetarian', linewidth=2.5)
ax.set(yscale="log")
ax.set(xlabel='New contributors')
ax.set(ylabel='Year')
ax.set_title('New contributors by year (log)')
```
Indeed, we can see that from 2014 to 2017, the number of vegetarian contributors has dropped at a faster rate than that of non-vegetarian contributors.
## Conclusion
**Is there a positive trend in the number of vegetarian recipes posted on food.com between 2008 and 2017?**
There is no positive trand. Between 2008 and 2013, the number of new vegetarian recipes has fallen year-over-year at the same rate as the number of new non-vegetarian recipes. It then started to fall at an even faster rate than for non-vegetarian recipes between 2014 and 2017.
**Is there a positive trend in the number of interactions with vegetarian recipes on Food.com between 2008 and 2017?**
There is no positive trand. The share of reviews posted on vegetarian and non-vegetarian recipes hsa remained unchanged over the 10-year period.
**Is there a difference in the ratings vegetarian recipes received compared to non-vegetarian recipes between 2008 and 2017?**
Yes, there is a slight positive trend. Vegetarian recipes were rated the same between 2008 and 2013, but started to attract higher ratings from there on. As of 2018, vegetarian recipe average ratings are 0.2 points higher than those of non-vegetarian recipes.
### Business implications
We can conclude that the growing vegetarian trend has had no positive impact on food.com, and might have even had a negative impact on our contributor acqusition.
|
github_jupyter
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from matplotlib_venn import venn2
import seaborn as sns
from ast import literal_eval
%matplotlib inline
veg_meat = ["#454d66", "#b7e778", "#1fab89"]
sns.set_palette(veg_meat)
recipes = pd.read_csv('/kaggle/input/food-com-recipes-and-user-interactions/RAW_recipes.csv')
interactions = pd.read_csv('/kaggle/input/food-com-recipes-and-user-interactions/RAW_interactions.csv')
recipes.head()
print(recipes.info())
recipes.describe()
recipes[['minutes', 'n_steps', 'n_ingredients']].hist()
interactions.head()
print(interactions.info())
interactions.describe()
interactions['rating'].hist()
from_year, to_year = '2008-01-01','2017-12-31'
recipes['submitted'] = pd.to_datetime(recipes['submitted'])
recipes['submitted'] = recipes['submitted'].apply(lambda x: x.tz_localize(None))
recipes_l0y = recipes.loc[recipes['submitted'].between(from_year, to_year, inclusive=False)]
interactions['date'] = pd.to_datetime(interactions['date'])
interactions['date'] = interactions['date'].apply(lambda x: x.tz_localize(None))
interactions_l0y = interactions.loc[interactions['date'].between(from_year, to_year, inclusive=False)]
print(recipes_l0y.shape)
print(interactions_l0y.shape)
sns.boxplot(x=recipes_l0y["minutes"])
# calculate the first quartile, third quartile and the interquartile range
Q1 = recipes_l0y['minutes'].quantile(0.25)
Q3 = recipes_l0y['minutes'].quantile(0.75)
IQR = Q3 - Q1
# calculate the maximum value and minimum values according to the Tukey rule
max_value = Q3 + 1.5 * IQR
min_value = Q1 - 1.5 * IQR
# filter the data for values that are greater than max_value or less than min_value
minutes_outliers = recipes_l0y[(recipes_l0y['minutes'] > max_value) | (recipes_l0y['minutes'] < min_value)]
minutes_outliers.sort_values('minutes')
# filter out recipes that take longer than 730 days as outliers
recipes_l0y = recipes_l0y.query('minutes < 1051200')
recipes_l0y['year'] = recipes_l0y['submitted'].dt.year
interactions_l0y['year'] = interactions_l0y['date'].dt.year
ratings_by_recipe = interactions_l0y.groupby(['recipe_id', 'year']).agg(
rating_cnt = ('rating', 'count'),
rating_avg = ('rating', 'mean'),
)
ratings_by_recipe.head()
recipes_and_ratings = recipes_l0y.merge(ratings_by_recipe, left_on='id', right_on='recipe_id')
recipes_and_ratings.head(2)
# convert the tags column to list format
recipes_and_ratings['tags'] = recipes_and_ratings['tags'].apply(lambda x: literal_eval(str(x)))
# add vegetarian and vegan boolean columns
recipes_and_ratings['vegetarian'] = ['vegetarian' in tag for tag in recipes_and_ratings['tags']]
recipes_and_ratings['vegan'] = ['vegan' in tag for tag in recipes_and_ratings['tags']]
recipes_and_ratings = recipes_and_ratings.drop(columns=['name', 'tags', 'nutrition', 'steps', 'description', 'ingredients'])
recipes_and_ratings.head(2)
#plot a venn diagram of vegetarian and vegan recipe counts
vegetarian_cnt = len(recipes_and_ratings.query('vegetarian == True'))
vegan_cnt = len(recipes_and_ratings.query('vegan == True'))
intersect_cnt = len(recipes_and_ratings.query('vegetarian == True and vegan == True'))
venn2(subsets = (vegetarian_cnt, vegan_cnt-intersect_cnt, intersect_cnt), set_labels = ('Vegetarian', 'Vegan'), set_colors=('#b7e778', '#031c16', '#031c16'), alpha = 1)
df = recipes_and_ratings.groupby(['year', 'vegetarian']).agg(
recipe_cnt = ('id', 'count')
).reset_index()
plt.figure(figsize=(12,6))
ax = sns.lineplot(data=df, x='year', y='recipe_cnt', hue='vegetarian', linewidth=2.5)
ax.set(ylim=(0, None))
ax.set_title('Number of new recipes by year')
ax
df = recipes_and_ratings.groupby(['year']).agg(
total_cnt = ('id', 'count'),
vegetarian_cnt = ('vegetarian', 'sum'),
vegan_cnt = ('vegan', 'sum'),
).reset_index()
df['vegetarian_pct'] = df['vegetarian_cnt'] / df['total_cnt'] * 100
df['vegan_pct'] = df['vegan_cnt'] / df['total_cnt'] * 100
plt.figure(figsize=(12,6))
ax = sns.lineplot(data=pd.melt(df[['year', 'vegetarian_pct', 'vegan_pct']], ['year']), x='year', y='value', palette=veg_meat[1:], hue='variable', linewidth=2.5)
ax.set(ylim=(0, 100))
ax.set_title('Percent of vegetarian recipes by year')
ax
ratings_by_recipe = interactions_l0y.groupby(['recipe_id', 'year']).agg(
rating_cnt = ('rating', 'count'),
rating_avg = ('rating', 'mean'),
).reset_index()
ratings_by_recipe = ratings_by_recipe.merge(recipes_and_ratings[['id', 'vegetarian', 'vegan']], left_on='recipe_id', right_on='id')
df = ratings_by_recipe.groupby(['year', 'vegetarian']).agg(
rating_cnt = ('rating_cnt', 'sum'),
rating_avg = ('rating_avg', 'mean'),
).reset_index()
plt.figure(figsize=(12,6))
ax = sns.lineplot(data=df, x='year', y='rating_cnt', hue='vegetarian', linewidth=2.5)
ax.set_title('Recipe ratings by year')
ax
interactions_by_recipe_and_year = interactions_l0y.reset_index().groupby(['recipe_id', 'year']).agg(
rating_cnt = ('index', 'count'),
rating_avg = ('rating', 'mean'),
).reset_index()
interactions_and_recipes = interactions_by_recipe_and_year[['recipe_id', 'year', 'rating_cnt', 'rating_avg']].merge(recipes_and_ratings[['id', 'vegetarian', 'vegan']], left_on='recipe_id', right_on='id')
interactions_and_recipes['vegetarian_rating_cnt'] = np.where(interactions_and_recipes['vegetarian'] == True, interactions_and_recipes['rating_cnt'], 0)
interactions_and_recipes['vegan_rating_cnt'] = np.where(interactions_and_recipes['vegan'] == True, interactions_and_recipes['rating_cnt'], 0)
df = interactions_and_recipes.groupby(['year']).agg(
total_cnt = ('rating_cnt', 'sum'),
vegetarian_cnt = ('vegetarian_rating_cnt', 'sum'),
vegan_cnt = ('vegan_rating_cnt', 'sum'),
).reset_index()
df['vegetarian_pct'] = df['vegetarian_cnt'] / df['total_cnt'] * 100
df['vegan_pct'] = df['vegan_cnt'] / df['total_cnt'] * 100
plt.figure(figsize=(12,6))
ax = sns.lineplot(data=pd.melt(df[['year', 'vegetarian_pct', 'vegan_pct']], ['year']), x='year', y='value', palette=veg_meat[1:], hue='variable', linewidth=2.5)
ax.set(ylim=(0, 100))
ax.set_title('Percent of votes on vegetarian recipes by year')
ax
df = ratings_by_recipe.groupby(['year', 'vegetarian']).agg(
rating_avg = ('rating_avg', 'mean')
).reset_index()
plt.figure(figsize=(12,6))
ax = sns.lineplot(data=df, x='year', y='rating_avg', hue='vegetarian', linewidth=2.5)
ax.set(ylim=(0, 5))
ax.set_title('Average recipe rating by year')
ax
recipes_and_cohorts = recipes_and_ratings.copy()
recipes_and_cohorts['submitted_year'] = recipes_and_cohorts['submitted'].apply(lambda x: x.strftime('%Y'))
# add cohort column — the year of the user's first recipe submission
recipes_and_cohorts.set_index('contributor_id', inplace=True)
recipes_and_cohorts['contributor_cohort'] = recipes_and_cohorts.groupby(level=0)['submitted'].min().apply(lambda x: x.strftime('%Y'))
recipes_and_cohorts.reset_index(inplace=True)
recipes_and_cohorts.head()
def add_cohort_periods(df):
"""
Creates a `cohort_period` column, which is the Nth period based on the contributor's first recipe.
"""
df['cohort_period'] = np.arange(len(df)) + 1
return df
def group_into_cohorts(df):
"""
Aggregates contributor count, recipe count and cohort period by contributor cohort and year of submission.
"""
df = df.groupby(['contributor_cohort', 'submitted_year']).agg(
contributor_cnt = ('contributor_id', 'nunique'),
recipe_cnt = ('id', 'nunique'),
)
df = df.groupby('contributor_cohort').apply(add_cohort_periods)
return df
# non-vegetarian cohorts
cohorts_nonveg = group_into_cohorts(recipes_and_cohorts[recipes_and_cohorts['vegetarian'] == False])
# vegetarian cohorts
cohorts_veg = group_into_cohorts(recipes_and_cohorts[recipes_and_cohorts['vegetarian'] == True])
cohorts_veg.head()
def calculate_cohort_sizes(df):
"""
Calculates cohort sizes.
"""
df.reset_index(inplace=True)
df.set_index(['contributor_cohort', 'cohort_period'], inplace=True)
return df['contributor_cnt'].groupby('contributor_cohort').first()
# calculate cohort sizes
cohort_sizes_nonveg = calculate_cohort_sizes(cohorts_nonveg)
cohort_sizes_veg = calculate_cohort_sizes(cohorts_veg)
cohort_sizes_veg.head()
def convert_cohort_counts_to_pct(df, cohort_sizes):
"""
Converts cohort period contributor counts to percentages.
"""
df = df.unstack(0).divide(cohort_sizes, axis=1)
df.reset_index(inplace=True)
return df
# convert cohort period contributor counts to percentages
contributor_retention_nonveg = convert_cohort_counts_to_pct(cohorts_nonveg['contributor_cnt'], cohort_sizes_nonveg)
contributor_retention_veg = convert_cohort_counts_to_pct(cohorts_veg['contributor_cnt'], cohort_sizes_veg)
contributor_retention_veg
def plot_retention_curves(df, cohorts, title, position):
"""
Plots retention curves for cohorts.
"""
plot = sns.lineplot(
data=pd.melt(contributor_retention_nonveg[['cohort_period'] + cohorts], ['cohort_period']),
x='cohort_period',
y='value',
palette='rocket_r',
hue='contributor_cohort',
linewidth=2.5,
ax=ax[position])
plot.set(xlim=(0, 8))
plot.set(ylim=(0, 1))
plot.set(xlabel='Cohort period')
plot.set(ylabel='Active contributors')
plot.set_title('Contributor retention by cohort: ' + title)
return
# plot contributor retention curves
fig, ax = plt.subplots(1, 2, figsize=(12,6))
cohorts_to_display = ['2008', '2009', '2010', '2011']
plot_retention_curves(contributor_retention_nonveg, cohorts_to_display, 'Non-vegetarian', 0)
plot_retention_curves(contributor_retention_veg, cohorts_to_display, 'Vegetarian', 1)
fig.show()
# get first recipe by contributor
df = recipes_and_cohorts.groupby('contributor_id').agg(
vegetarian = ('vegetarian', 'mean'),
contributor_cohort = ('contributor_cohort', 'min'),
)
# counting contributors with >50% of vegetarian contibutions as vegetarians
df.reset_index(inplace=True)
df = df.round(0)
# get first recipe by contributor
df = df.groupby(['contributor_cohort', 'vegetarian']).agg(
contributor_cnt = ('contributor_id', 'count'),
)
# counting contributors with >50% of vegetarian contibutions as vegetarians
df.reset_index(inplace=True)
df['vegetarian'] = df['vegetarian'].astype(bool)
plt.figure(figsize=(12,6))
ax = sns.lineplot(data=df, x='contributor_cohort', y='contributor_cnt', palette=veg_meat[:2], hue='vegetarian', linewidth=2.5)
ax.set(xlabel='New contributors')
ax.set(ylabel='Year')
ax.set_title('New contributors by year')
plt.figure(figsize=(12,6))
ax = sns.lineplot(data=df, x='contributor_cohort', y='contributor_cnt', palette=veg_meat[:2], hue='vegetarian', linewidth=2.5)
ax.set(yscale="log")
ax.set(xlabel='New contributors')
ax.set(ylabel='Year')
ax.set_title('New contributors by year (log)')
| 0.462959 | 0.950641 |
```
from sklearn.metrics import mean_absolute_error,mean_squared_error
from sklearn.model_selection import KFold,train_test_split
import matplotlib.pyplot as plt
import seaborn as sns
import lightgbm as lgb
import pandas as pd
import numpy as np
from math import pi
from featexp import *
import gc
import os
path_train='D:/data_mywork/math_modeling/train_set/'
path_test='D:/data_mywork/math_modeling/test_set/'
path_dir=os.listdir(path_train)
path_dir1=os.listdir(path_test)
def feat(data):
feat=['Frequency Band','RS Power','Cell Clutter Index','Clutter Index']
train_x=data.loc[:,feat]
train_x['d']=np.sqrt((data['Cell X']-data['X'])**2+(data['Cell Y']-data['Y'])**2)
train_x['Hb']=data['Cell Altitude']+data['Cell Building Height']+data['Altitude']
train_x['Hue']=data['Altitude']+data['Building Height']
train_x['thetaM']=np.radians(data['Mechanical Downtilt'])
train_x['thetaE']=np.radians(data['Electrical Downtilt'])
train_x['delta_Hv']=train_x['Hb']-train_x['d']*np.tan(train_x['thetaM']+train_x['thetaE'])
train_x['theta_uc']=np.arctan(abs(train_x['Hb']-train_x['Hue'])/train_x['d'])
#train_x['log_freq']=np.log10(data['Frequency Band'])
train_x['log_Hb']=np.log10(train_x['Hb']+1)
train_x['log_Hue']=np.log10(train_x['Hue']+1)
train_x['log_d']=np.log10(train_x['d']+1)
train_x['dmulHue_log']=train_x['log_Hue']*train_x['log_d']
train_x['Azimuth_rad']=np.radians(data['Azimuth'])
train_x['dX']=data['X']-data['Cell X']
train_x['dY']=data['Y']-data['Cell Y']
train_x['theta_XY']=np.arctan2(train_x['dX'],train_x['dY'])
train_x['theta_XY'].loc[train_x['dY']<0]=train_x['theta_XY'].loc[train_x['dY']<0]+pi
train_x['theta_XY'].loc[(train_x['dY']>=0)&(train_x['dX']<0)]=train_x['theta_XY'].loc[(train_x['dY']>=0)&(train_x['dX']<0)]+2*pi
train_x['theta_XY_A']=train_x['theta_XY']-train_x['Azimuth_rad']
train_x=train_x.drop(['Azimuth_rad','theta_XY','dX','dY'],axis=1)
return train_x
#读取训练数据
train_data=[]
for f in path_dir:
train_data.append(pd.read_csv(path_train+f))
train_data=pd.concat(train_data).reset_index(drop=True)
#读取测试数据
test_data=[]
for f in path_dir1:
test_data.append(pd.read_csv(path_test+f))
test_data=pd.concat(test_data).reset_index(drop=True)
CellIndex=test_data['Cell Index']
#训练数据转化
train_set=[]
chunk=1000000
for i in range(1,14):
train_x=feat(train_data[(i-1)*chunk:i*chunk])
train_set.append(train_x)
train_set=pd.concat(train_set)
train_set['label']=train_data['RSRP']
#测试数据转化
test_set=feat(test_data)
del train_data
#按Frequency Band分3类,做成dict
def DataDivison(data_set):
groups=data_set.groupby('Frequency Band')
data_sets={}
for key,group in groups:
group=group.drop(['Frequency Band'],axis=1)
data_sets[key]=group
return data_sets
#分别建模
train_sets=DataDivison(train_set)
test_sets=DataDivison(test_set)
dataSetKey=[2585.0, 2604.8, 2624.6]
#trainSet1,ValSet1=train_test_split(train_sets,test_size=0.33, random_state=42)
#trainSet2,ValSet2=train_test_split(train_sets,test_size=0.33, random_state=42)
#trainSet3,ValSet3=train_test_split(train_sets,test_size=0.33, random_state=42)
#gc.collect()
#para
def display_importances(feature_importance_df_):
cols = feature_importance_df_[["Feature", "importance"]].groupby("Feature").mean().sort_values(by="importance", ascending=False)[:40].index
best_features = feature_importance_df_.loc[feature_importance_df_.Feature.isin(cols)]
plt.figure(figsize=(8, 10))
sns.barplot(x="importance", y="Feature", data=best_features.sort_values(by="importance", ascending=False))
plt.title('LightGBM Features (avg over folds)')
plt.tight_layout()
plt.show()
params = {
'learning_rate': 0.08,
'boosting_type': 'gbdt',
'objective': 'regression_l1',
'metric': 'mae',
'feature_fraction': 0.6,
'bagging_fraction': 0.8,
'bagging_freq': 5,
'num_leaves': 100,
'verbose': -1,
'max_depth': -1,
'reg_alpha':2.2,
'reg_lambda':1.4,
'nthread': 8
}
def lgb_reg(params,train,targets,test):
features=train.columns
folds = KFold(n_splits=5, shuffle=True, random_state=1420)
oof = np.zeros(len(train))
predictions = np.zeros(len(test))
feature_importance_df = pd.DataFrame()
for fold_, (trn_idx, val_idx) in enumerate(folds.split(train.values, targets.values)):
print("Fold {}".format(fold_))
#n=len(trn_idx)
trn_data = lgb.Dataset(train.iloc[trn_idx][features],label=targets.iloc[trn_idx])
val_data = lgb.Dataset(train.iloc[val_idx][features],label=targets.iloc[val_idx])
num_round = 10000
clf = lgb.train(params, trn_data, num_round, valid_sets = [trn_data, val_data], verbose_eval=1000, early_stopping_rounds = 200)
oof[val_idx] = clf.predict(train.iloc[val_idx][features], num_iteration=clf.best_iteration)
fold_importance_df = pd.DataFrame()
fold_importance_df["Feature"] = features
fold_importance_df["importance"] = clf.feature_importance()
fold_importance_df["fold"] = fold_ + 1
feature_importance_df = pd.concat([feature_importance_df, fold_importance_df], axis=0)
predictions += clf.predict(test[features], num_iteration=clf.best_iteration) / folds.n_splits
Score=mean_absolute_error(targets, oof)
print("CV score: {:<8.5f}".format(Score))
return oof,predictions,feature_importance_df
features=['RS Power', 'Cell Clutter Index', 'Clutter Index',
'd', 'Hb', 'Hue', 'thetaM', 'thetaE', 'delta_Hv', 'theta_uc', 'log_Hb',
'log_Hue', 'log_d', 'dmulHue_log', 'theta_XY_A']
#oof,predictions,feature_importance_df=lgb_reg(params,trainSet1[features][:100000],trainSet1['target'][:100000],testSet1[features][100000:120000])
#按类别训练并预测
result=pd.DataFrame()
result['CellIndex']=CellIndex
for key in test_sets.keys():
oof,predictions,feature_importance_df=lgb_reg(params,train_sets[key][features],train_set[key]['label'],test_sets[key][features])
result['predict'].loc[test_sets[key].index]=predictions
#result.to_csv(path_result+'result.csv')
plt.scatter(testSet1['target'][100000:120000],predictions)
display_importances(feature_importance_df)
```
|
github_jupyter
|
from sklearn.metrics import mean_absolute_error,mean_squared_error
from sklearn.model_selection import KFold,train_test_split
import matplotlib.pyplot as plt
import seaborn as sns
import lightgbm as lgb
import pandas as pd
import numpy as np
from math import pi
from featexp import *
import gc
import os
path_train='D:/data_mywork/math_modeling/train_set/'
path_test='D:/data_mywork/math_modeling/test_set/'
path_dir=os.listdir(path_train)
path_dir1=os.listdir(path_test)
def feat(data):
feat=['Frequency Band','RS Power','Cell Clutter Index','Clutter Index']
train_x=data.loc[:,feat]
train_x['d']=np.sqrt((data['Cell X']-data['X'])**2+(data['Cell Y']-data['Y'])**2)
train_x['Hb']=data['Cell Altitude']+data['Cell Building Height']+data['Altitude']
train_x['Hue']=data['Altitude']+data['Building Height']
train_x['thetaM']=np.radians(data['Mechanical Downtilt'])
train_x['thetaE']=np.radians(data['Electrical Downtilt'])
train_x['delta_Hv']=train_x['Hb']-train_x['d']*np.tan(train_x['thetaM']+train_x['thetaE'])
train_x['theta_uc']=np.arctan(abs(train_x['Hb']-train_x['Hue'])/train_x['d'])
#train_x['log_freq']=np.log10(data['Frequency Band'])
train_x['log_Hb']=np.log10(train_x['Hb']+1)
train_x['log_Hue']=np.log10(train_x['Hue']+1)
train_x['log_d']=np.log10(train_x['d']+1)
train_x['dmulHue_log']=train_x['log_Hue']*train_x['log_d']
train_x['Azimuth_rad']=np.radians(data['Azimuth'])
train_x['dX']=data['X']-data['Cell X']
train_x['dY']=data['Y']-data['Cell Y']
train_x['theta_XY']=np.arctan2(train_x['dX'],train_x['dY'])
train_x['theta_XY'].loc[train_x['dY']<0]=train_x['theta_XY'].loc[train_x['dY']<0]+pi
train_x['theta_XY'].loc[(train_x['dY']>=0)&(train_x['dX']<0)]=train_x['theta_XY'].loc[(train_x['dY']>=0)&(train_x['dX']<0)]+2*pi
train_x['theta_XY_A']=train_x['theta_XY']-train_x['Azimuth_rad']
train_x=train_x.drop(['Azimuth_rad','theta_XY','dX','dY'],axis=1)
return train_x
#读取训练数据
train_data=[]
for f in path_dir:
train_data.append(pd.read_csv(path_train+f))
train_data=pd.concat(train_data).reset_index(drop=True)
#读取测试数据
test_data=[]
for f in path_dir1:
test_data.append(pd.read_csv(path_test+f))
test_data=pd.concat(test_data).reset_index(drop=True)
CellIndex=test_data['Cell Index']
#训练数据转化
train_set=[]
chunk=1000000
for i in range(1,14):
train_x=feat(train_data[(i-1)*chunk:i*chunk])
train_set.append(train_x)
train_set=pd.concat(train_set)
train_set['label']=train_data['RSRP']
#测试数据转化
test_set=feat(test_data)
del train_data
#按Frequency Band分3类,做成dict
def DataDivison(data_set):
groups=data_set.groupby('Frequency Band')
data_sets={}
for key,group in groups:
group=group.drop(['Frequency Band'],axis=1)
data_sets[key]=group
return data_sets
#分别建模
train_sets=DataDivison(train_set)
test_sets=DataDivison(test_set)
dataSetKey=[2585.0, 2604.8, 2624.6]
#trainSet1,ValSet1=train_test_split(train_sets,test_size=0.33, random_state=42)
#trainSet2,ValSet2=train_test_split(train_sets,test_size=0.33, random_state=42)
#trainSet3,ValSet3=train_test_split(train_sets,test_size=0.33, random_state=42)
#gc.collect()
#para
def display_importances(feature_importance_df_):
cols = feature_importance_df_[["Feature", "importance"]].groupby("Feature").mean().sort_values(by="importance", ascending=False)[:40].index
best_features = feature_importance_df_.loc[feature_importance_df_.Feature.isin(cols)]
plt.figure(figsize=(8, 10))
sns.barplot(x="importance", y="Feature", data=best_features.sort_values(by="importance", ascending=False))
plt.title('LightGBM Features (avg over folds)')
plt.tight_layout()
plt.show()
params = {
'learning_rate': 0.08,
'boosting_type': 'gbdt',
'objective': 'regression_l1',
'metric': 'mae',
'feature_fraction': 0.6,
'bagging_fraction': 0.8,
'bagging_freq': 5,
'num_leaves': 100,
'verbose': -1,
'max_depth': -1,
'reg_alpha':2.2,
'reg_lambda':1.4,
'nthread': 8
}
def lgb_reg(params,train,targets,test):
features=train.columns
folds = KFold(n_splits=5, shuffle=True, random_state=1420)
oof = np.zeros(len(train))
predictions = np.zeros(len(test))
feature_importance_df = pd.DataFrame()
for fold_, (trn_idx, val_idx) in enumerate(folds.split(train.values, targets.values)):
print("Fold {}".format(fold_))
#n=len(trn_idx)
trn_data = lgb.Dataset(train.iloc[trn_idx][features],label=targets.iloc[trn_idx])
val_data = lgb.Dataset(train.iloc[val_idx][features],label=targets.iloc[val_idx])
num_round = 10000
clf = lgb.train(params, trn_data, num_round, valid_sets = [trn_data, val_data], verbose_eval=1000, early_stopping_rounds = 200)
oof[val_idx] = clf.predict(train.iloc[val_idx][features], num_iteration=clf.best_iteration)
fold_importance_df = pd.DataFrame()
fold_importance_df["Feature"] = features
fold_importance_df["importance"] = clf.feature_importance()
fold_importance_df["fold"] = fold_ + 1
feature_importance_df = pd.concat([feature_importance_df, fold_importance_df], axis=0)
predictions += clf.predict(test[features], num_iteration=clf.best_iteration) / folds.n_splits
Score=mean_absolute_error(targets, oof)
print("CV score: {:<8.5f}".format(Score))
return oof,predictions,feature_importance_df
features=['RS Power', 'Cell Clutter Index', 'Clutter Index',
'd', 'Hb', 'Hue', 'thetaM', 'thetaE', 'delta_Hv', 'theta_uc', 'log_Hb',
'log_Hue', 'log_d', 'dmulHue_log', 'theta_XY_A']
#oof,predictions,feature_importance_df=lgb_reg(params,trainSet1[features][:100000],trainSet1['target'][:100000],testSet1[features][100000:120000])
#按类别训练并预测
result=pd.DataFrame()
result['CellIndex']=CellIndex
for key in test_sets.keys():
oof,predictions,feature_importance_df=lgb_reg(params,train_sets[key][features],train_set[key]['label'],test_sets[key][features])
result['predict'].loc[test_sets[key].index]=predictions
#result.to_csv(path_result+'result.csv')
plt.scatter(testSet1['target'][100000:120000],predictions)
display_importances(feature_importance_df)
| 0.311113 | 0.326406 |
# Using Tensorflow DALI plugin: DALI and tf.data
### Overview
DALI offers integration with [tf.data API](https://www.tensorflow.org/guide/data). Using this approach you can easily connect DALI pipeline with various TensorFlow APIs and use it as a data source for your model. This tutorial shows how to do it using well known [MNIST](http://yann.lecun.com/exdb/mnist/) converted to LMDB format. You can find it in [DALI_extra](https://github.com/NVIDIA/DALI_extra) - DALI test data repository.
We start with creating a DALI pipeline to read, decode and normalize MNIST images and read corresponding labels.
`DALI_EXTRA_PATH` environment variable should point to the place where data from [DALI extra repository](https://github.com/NVIDIA/DALI_extra) is downloaded. Please make sure that the proper release tag is checked out.
```
from nvidia.dali import pipeline_def, Pipeline
import nvidia.dali.fn as fn
import nvidia.dali.types as types
import os
BATCH_SIZE = 64
DROPOUT = 0.2
IMAGE_SIZE = 28
NUM_CLASSES = 10
HIDDEN_SIZE = 128
EPOCHS = 5
ITERATIONS_PER_EPOCH = 100
# Path to MNIST dataset
data_path = os.path.join(os.environ['DALI_EXTRA_PATH'], 'db/MNIST/training/')
@pipeline_def(device_id=0, batch_size=BATCH_SIZE)
def mnist_pipeline(device):
jpegs, labels = fn.readers.caffe2(path=data_path, random_shuffle=True)
images = fn.decoders.image(
jpegs, device='mixed' if device == 'gpu' else 'cpu', output_type=types.GRAY)
images = fn.crop_mirror_normalize(
images, device=device, dtype=types.FLOAT, std=[255.], output_layout="CHW")
if device == 'gpu':
labels = labels.gpu()
return images, labels
```
Next step is to wrap an instance of `MnistPipeline` with a `DALIDataset` object from DALI TensorFlow plugin. This class is compatible with `tf.data.Dataset`. Other parameters are shapes and types of the outputs of the pipeline. Here we return images and labels. It means we have two outputs one of type `tf.float32` for images and on of type `tf.int32` for labels.
```
import nvidia.dali.plugin.tf as dali_tf
import tensorflow as tf
import tensorflow.compat.v1 as tf_v1
import logging
tf.get_logger().setLevel(logging.ERROR)
# Create pipeline
pipeline = mnist_pipeline(device='cpu')
# Define shapes and types of the outputs
shapes = (
(BATCH_SIZE, IMAGE_SIZE, IMAGE_SIZE),
(BATCH_SIZE))
dtypes = (
tf.float32,
tf.int32)
# Create dataset
with tf.device('/cpu:0'):
mnist_set = dali_tf.DALIDataset(
pipeline=pipeline,
batch_size=BATCH_SIZE,
output_shapes=shapes,
output_dtypes=dtypes,
device_id=0)
```
We are ready to start the training. Following sections show how to do it with different APIs availible in TensorFlow.
### Keras
First, we pass `mnist_set` to model created with `tf.keras` and use `model.fit` method to train it.
```
# Create the model
model = tf.keras.models.Sequential([
tf.keras.layers.Input(shape=(IMAGE_SIZE, IMAGE_SIZE), name='images'),
tf.keras.layers.Flatten(input_shape=(IMAGE_SIZE, IMAGE_SIZE)),
tf.keras.layers.Dense(HIDDEN_SIZE, activation='relu'),
tf.keras.layers.Dropout(DROPOUT),
tf.keras.layers.Dense(NUM_CLASSES, activation='softmax')])
model.compile(
optimizer='adam',
loss='sparse_categorical_crossentropy',
metrics=['accuracy'])
# Train using DALI dataset
model.fit(
mnist_set,
epochs=EPOCHS,
steps_per_epoch=ITERATIONS_PER_EPOCH)
```
As you can see, it was very easy to integrate DALI pipeline with `tf.keras` API.
The code above performed the training using the CPU. Both the DALI pipeline and the model were using the CPU.
We can easily move the whole processing to the GPU. First, we create a pipeline that uses the GPU with ID = 0. Next we place both the DALI dataset and the model on the same GPU.
```
# Define the model and place it on the GPU
with tf.device('/gpu:0'):
mnist_set = dali_tf.DALIDataset(
pipeline=mnist_pipeline(device='gpu'),
batch_size=BATCH_SIZE,
output_shapes=shapes,
output_dtypes=dtypes,
device_id=0)
model = tf.keras.models.Sequential([
tf.keras.layers.Input(shape=(IMAGE_SIZE, IMAGE_SIZE), name='images'),
tf.keras.layers.Flatten(input_shape=(IMAGE_SIZE, IMAGE_SIZE)),
tf.keras.layers.Dense(HIDDEN_SIZE, activation='relu'),
tf.keras.layers.Dropout(DROPOUT),
tf.keras.layers.Dense(NUM_CLASSES, activation='softmax')])
model.compile(
optimizer='adam',
loss='sparse_categorical_crossentropy',
metrics=['accuracy'])
```
We move the training to the GPU as well. This allows TensorFlow to pick up GPU instance of DALI dataset.
```
# Train on the GPU
with tf.device('/gpu:0'):
model.fit(
mnist_set,
epochs=EPOCHS,
steps_per_epoch=ITERATIONS_PER_EPOCH)
```
It is important to note here, that there is no intermediate CPU buffer between DALI and TensorFlow in the execution above. DALI GPU outputs are copied straight to TF GPU Tensors used by the model.
In this particular toy example performance of the GPU variant is lower than the CPU one. The MNIST images are small and nvJPEG decoder used in the GPU DALI pipeline to decode them is not well suited for such circumstance. We use it here to show how to integrate it properly in the real life case.
### Estimators
Another popular TensorFlow API is `tf.estimator` API. This section shows how to use DALI dataset as a data source for model based on this API.
First we create the model.
```
# Define the feature columns for Estimator
feature_columns = [tf.feature_column.numeric_column(
"images", shape=[IMAGE_SIZE, IMAGE_SIZE])]
# And the run config
run_config = tf.estimator.RunConfig(
model_dir='/tmp/tensorflow-checkpoints',
device_fn=lambda op: '/gpu:0')
# Finally create the model based on `DNNClassifier`
model = tf.estimator.DNNClassifier(
feature_columns=feature_columns,
hidden_units=[HIDDEN_SIZE],
n_classes=NUM_CLASSES,
dropout=DROPOUT,
config=run_config,
optimizer='Adam')
```
In `tf.estimator` API data is passed to the model with the function returning the dataset. We define this function to return DALI dataset placed on the GPU.
```
def train_data_fn():
with tf.device('/gpu:0'):
mnist_set = dali_tf.DALIDataset(
fail_on_device_mismatch=False,
pipeline=mnist_pipeline(device='gpu'),
batch_size=BATCH_SIZE,
output_shapes=shapes,
output_dtypes=dtypes,
device_id=0)
mnist_set = mnist_set.map(
lambda features, labels: ({'images': features}, labels))
return mnist_set
```
With everything set up we are ready to run the training.
```
# Running the training on the GPU
model.train(input_fn=train_data_fn, steps=EPOCHS * ITERATIONS_PER_EPOCH)
def test_data_fn():
with tf.device('/cpu:0'):
mnist_set = dali_tf.DALIDataset(
fail_on_device_mismatch=False,
pipeline=mnist_pipeline(device='cpu'),
batch_size=BATCH_SIZE,
output_shapes=shapes,
output_dtypes=dtypes,
device_id=0)
mnist_set = mnist_set.map(
lambda features, labels: ({'images': features}, labels))
return mnist_set
model.evaluate(input_fn=test_data_fn, steps=ITERATIONS_PER_EPOCH)
```
### Custom Models and Training Loops
Finally, the last part of this tutorial focuses on integrating DALI dataset with custom models and training loops. A complete example below shows from start to finish how to use DALI dataset with native TensorFlow model and run training using `tf.Session`.
First step is to define the model and the dataset and place both on the GPU.
```
tf.compat.v1.disable_eager_execution()
tf_v1.reset_default_graph()
with tf.device('/gpu:0'):
mnist_set = dali_tf.DALIDataset(
pipeline=mnist_pipeline(device='gpu'),
batch_size=BATCH_SIZE,
output_shapes=shapes,
output_dtypes=dtypes,
device_id=0)
iterator = tf_v1.data.make_initializable_iterator(mnist_set)
images, labels = iterator.get_next()
labels = tf_v1.reshape(
tf_v1.one_hot(labels, NUM_CLASSES),
[BATCH_SIZE, NUM_CLASSES])
with tf_v1.variable_scope('mnist_net', reuse=False):
images = tf_v1.layers.flatten(images)
images = tf_v1.layers.dense(images, HIDDEN_SIZE, activation=tf_v1.nn.relu)
images = tf_v1.layers.dropout(images, rate=DROPOUT, training=True)
images = tf_v1.layers.dense(images, NUM_CLASSES, activation=tf_v1.nn.softmax)
logits_train = images
loss_op = tf_v1.reduce_mean(tf.nn.softmax_cross_entropy_with_logits(
logits=logits_train, labels=labels))
train_step = tf_v1.train.AdamOptimizer().minimize(loss_op)
correct_pred = tf_v1.equal(
tf_v1.argmax(logits_train, 1), tf_v1.argmax(labels, 1))
accuracy = tf_v1.reduce_mean(tf_v1.cast(correct_pred, tf_v1.float32))
```
With `tf.Session` we can run this model and train it on the GPU.
```
with tf_v1.Session() as sess:
sess.run(tf_v1.global_variables_initializer())
sess.run(iterator.initializer)
for i in range(EPOCHS * ITERATIONS_PER_EPOCH):
sess.run(train_step)
if i % ITERATIONS_PER_EPOCH == 0:
train_accuracy = sess.run(accuracy)
print("Step %d, accuracy: %g" % (i, train_accuracy))
final_accuracy = 0
for _ in range(ITERATIONS_PER_EPOCH):
final_accuracy = final_accuracy + sess.run(accuracy)
final_accuracy = final_accuracy / ITERATIONS_PER_EPOCH
print('Final accuracy: ', final_accuracy)
```
|
github_jupyter
|
from nvidia.dali import pipeline_def, Pipeline
import nvidia.dali.fn as fn
import nvidia.dali.types as types
import os
BATCH_SIZE = 64
DROPOUT = 0.2
IMAGE_SIZE = 28
NUM_CLASSES = 10
HIDDEN_SIZE = 128
EPOCHS = 5
ITERATIONS_PER_EPOCH = 100
# Path to MNIST dataset
data_path = os.path.join(os.environ['DALI_EXTRA_PATH'], 'db/MNIST/training/')
@pipeline_def(device_id=0, batch_size=BATCH_SIZE)
def mnist_pipeline(device):
jpegs, labels = fn.readers.caffe2(path=data_path, random_shuffle=True)
images = fn.decoders.image(
jpegs, device='mixed' if device == 'gpu' else 'cpu', output_type=types.GRAY)
images = fn.crop_mirror_normalize(
images, device=device, dtype=types.FLOAT, std=[255.], output_layout="CHW")
if device == 'gpu':
labels = labels.gpu()
return images, labels
import nvidia.dali.plugin.tf as dali_tf
import tensorflow as tf
import tensorflow.compat.v1 as tf_v1
import logging
tf.get_logger().setLevel(logging.ERROR)
# Create pipeline
pipeline = mnist_pipeline(device='cpu')
# Define shapes and types of the outputs
shapes = (
(BATCH_SIZE, IMAGE_SIZE, IMAGE_SIZE),
(BATCH_SIZE))
dtypes = (
tf.float32,
tf.int32)
# Create dataset
with tf.device('/cpu:0'):
mnist_set = dali_tf.DALIDataset(
pipeline=pipeline,
batch_size=BATCH_SIZE,
output_shapes=shapes,
output_dtypes=dtypes,
device_id=0)
# Create the model
model = tf.keras.models.Sequential([
tf.keras.layers.Input(shape=(IMAGE_SIZE, IMAGE_SIZE), name='images'),
tf.keras.layers.Flatten(input_shape=(IMAGE_SIZE, IMAGE_SIZE)),
tf.keras.layers.Dense(HIDDEN_SIZE, activation='relu'),
tf.keras.layers.Dropout(DROPOUT),
tf.keras.layers.Dense(NUM_CLASSES, activation='softmax')])
model.compile(
optimizer='adam',
loss='sparse_categorical_crossentropy',
metrics=['accuracy'])
# Train using DALI dataset
model.fit(
mnist_set,
epochs=EPOCHS,
steps_per_epoch=ITERATIONS_PER_EPOCH)
# Define the model and place it on the GPU
with tf.device('/gpu:0'):
mnist_set = dali_tf.DALIDataset(
pipeline=mnist_pipeline(device='gpu'),
batch_size=BATCH_SIZE,
output_shapes=shapes,
output_dtypes=dtypes,
device_id=0)
model = tf.keras.models.Sequential([
tf.keras.layers.Input(shape=(IMAGE_SIZE, IMAGE_SIZE), name='images'),
tf.keras.layers.Flatten(input_shape=(IMAGE_SIZE, IMAGE_SIZE)),
tf.keras.layers.Dense(HIDDEN_SIZE, activation='relu'),
tf.keras.layers.Dropout(DROPOUT),
tf.keras.layers.Dense(NUM_CLASSES, activation='softmax')])
model.compile(
optimizer='adam',
loss='sparse_categorical_crossentropy',
metrics=['accuracy'])
# Train on the GPU
with tf.device('/gpu:0'):
model.fit(
mnist_set,
epochs=EPOCHS,
steps_per_epoch=ITERATIONS_PER_EPOCH)
# Define the feature columns for Estimator
feature_columns = [tf.feature_column.numeric_column(
"images", shape=[IMAGE_SIZE, IMAGE_SIZE])]
# And the run config
run_config = tf.estimator.RunConfig(
model_dir='/tmp/tensorflow-checkpoints',
device_fn=lambda op: '/gpu:0')
# Finally create the model based on `DNNClassifier`
model = tf.estimator.DNNClassifier(
feature_columns=feature_columns,
hidden_units=[HIDDEN_SIZE],
n_classes=NUM_CLASSES,
dropout=DROPOUT,
config=run_config,
optimizer='Adam')
def train_data_fn():
with tf.device('/gpu:0'):
mnist_set = dali_tf.DALIDataset(
fail_on_device_mismatch=False,
pipeline=mnist_pipeline(device='gpu'),
batch_size=BATCH_SIZE,
output_shapes=shapes,
output_dtypes=dtypes,
device_id=0)
mnist_set = mnist_set.map(
lambda features, labels: ({'images': features}, labels))
return mnist_set
# Running the training on the GPU
model.train(input_fn=train_data_fn, steps=EPOCHS * ITERATIONS_PER_EPOCH)
def test_data_fn():
with tf.device('/cpu:0'):
mnist_set = dali_tf.DALIDataset(
fail_on_device_mismatch=False,
pipeline=mnist_pipeline(device='cpu'),
batch_size=BATCH_SIZE,
output_shapes=shapes,
output_dtypes=dtypes,
device_id=0)
mnist_set = mnist_set.map(
lambda features, labels: ({'images': features}, labels))
return mnist_set
model.evaluate(input_fn=test_data_fn, steps=ITERATIONS_PER_EPOCH)
tf.compat.v1.disable_eager_execution()
tf_v1.reset_default_graph()
with tf.device('/gpu:0'):
mnist_set = dali_tf.DALIDataset(
pipeline=mnist_pipeline(device='gpu'),
batch_size=BATCH_SIZE,
output_shapes=shapes,
output_dtypes=dtypes,
device_id=0)
iterator = tf_v1.data.make_initializable_iterator(mnist_set)
images, labels = iterator.get_next()
labels = tf_v1.reshape(
tf_v1.one_hot(labels, NUM_CLASSES),
[BATCH_SIZE, NUM_CLASSES])
with tf_v1.variable_scope('mnist_net', reuse=False):
images = tf_v1.layers.flatten(images)
images = tf_v1.layers.dense(images, HIDDEN_SIZE, activation=tf_v1.nn.relu)
images = tf_v1.layers.dropout(images, rate=DROPOUT, training=True)
images = tf_v1.layers.dense(images, NUM_CLASSES, activation=tf_v1.nn.softmax)
logits_train = images
loss_op = tf_v1.reduce_mean(tf.nn.softmax_cross_entropy_with_logits(
logits=logits_train, labels=labels))
train_step = tf_v1.train.AdamOptimizer().minimize(loss_op)
correct_pred = tf_v1.equal(
tf_v1.argmax(logits_train, 1), tf_v1.argmax(labels, 1))
accuracy = tf_v1.reduce_mean(tf_v1.cast(correct_pred, tf_v1.float32))
with tf_v1.Session() as sess:
sess.run(tf_v1.global_variables_initializer())
sess.run(iterator.initializer)
for i in range(EPOCHS * ITERATIONS_PER_EPOCH):
sess.run(train_step)
if i % ITERATIONS_PER_EPOCH == 0:
train_accuracy = sess.run(accuracy)
print("Step %d, accuracy: %g" % (i, train_accuracy))
final_accuracy = 0
for _ in range(ITERATIONS_PER_EPOCH):
final_accuracy = final_accuracy + sess.run(accuracy)
final_accuracy = final_accuracy / ITERATIONS_PER_EPOCH
print('Final accuracy: ', final_accuracy)
| 0.744192 | 0.944842 |
# Randomised Iterative Improvement on MAXSAT
Set classpath to the pre-compiled jar
```
@file:DependsOn("../build/libs/kglsm.jar")
@file:DependsOn("ch.qos.logback:logback-classic:1.2.3")
@file:DependsOn("ch.qos.logback:logback-core:1.2.3")
@file:DependsOn("io.github.microutils:kotlin-logging-jvm:2.0.3")
```
Define imports
```
import com.sihvi.glsm.problem.MAXSAT
import com.sihvi.glsm.sls.GLSMBuilder
import com.sihvi.glsm.sls.StateMachineTransition
import com.sihvi.glsm.space.BooleanSearchSpace
import com.sihvi.glsm.space.SearchSpace
import com.sihvi.glsm.strategy.IIMode
import com.sihvi.glsm.strategy.IterativeImprovementStrategy
import com.sihvi.glsm.strategy.RandomWalkStrategy
import com.sihvi.glsm.transitionpredicate.NoImprovementPredicate
import com.sihvi.glsm.transitionpredicate.NotPredicate
import com.sihvi.glsm.transitionpredicate.ProbabilisticPredicate
import com.sihvi.glsm.memory.Memory
import com.sihvi.glsm.memory.BasicMemory
import com.sihvi.glsm.memory.BasicSolution
```
## Problem
We are using MAXSAT problem as an example. Get an instance of the problem.
```
val noVariables = 20
val problemInstance = MAXSAT.getRandomInstance(noVariables,50,2)
println(problemInstance)
```
## Search space
Boolean search space defines two methods on an array of booleans: to get a neighbourhood and a random neighbour.
In this case, the members of the neighbourhood are exactly Hamming distance of 1 away from the current solution (i.e. one boolean value flipped)
```
val space = BooleanSearchSpace(noVariables)
```
## Memory
Basic memory that holds:
* Current solution and its cost
* Best solution and best cost
* Number of steps performed
* Number of steps performed without improvement (when it was expected)
```
val initialSolution = space.getInitial()
val memory = BasicMemory(BasicSolution(initialSolution, problemInstance.evaluate(initialSolution)))
```
## GLSM and Strategies
Randomised Iterative Improvement consists of two strategies that are flipped probabilistically
* Iterative Best Improvement Strategy -- picks a solution from a neighbourhood that gives the best improvement
* Random Walk Strategy -- randomly picks a solution from a neighbourhood
The termination predicate of choice here is No Improvement Predicate, i.e. we terminate the search if there were n steps without improvement (in this case 10)
```
val terminationPredicate = NoImprovementPredicate(10)
val wp = 0.1
val toRandomPredicate = ProbabilisticPredicate(to = wp)
val toIIPredicate = NotPredicate(toRandomPredicate)
val walk = RandomWalkStrategy<Boolean>()
val iterativeImprovement = IterativeImprovementStrategy<Boolean>(IIMode.BEST, true)
```
As all the components are defined, we can now build the GLSM with strategies and transitions between them
```
val glsm = GLSMBuilder<Boolean, BasicSolution<Boolean>>()
.addStrategy(iterativeImprovement)
.addStrategy(walk)
.addTransition(StateMachineTransition(0, 1, toRandomPredicate))
.addTransition(StateMachineTransition(1, 0, toIIPredicate))
.addTransition(StateMachineTransition(0, -1, terminationPredicate))
.build()
glsm
glsm.toASCII()
```
# Solve
With everything ready we can run GLSM on our problem instance
```
val finalSolution = glsm.solve(memory, space, problemInstance::evaluate)
println("Steps taken: " + memory.stepCount)
println("Solution: " + finalSolution.solution.joinToString(", "))
println("Cost: " + finalSolution.cost)
```
|
github_jupyter
|
@file:DependsOn("../build/libs/kglsm.jar")
@file:DependsOn("ch.qos.logback:logback-classic:1.2.3")
@file:DependsOn("ch.qos.logback:logback-core:1.2.3")
@file:DependsOn("io.github.microutils:kotlin-logging-jvm:2.0.3")
import com.sihvi.glsm.problem.MAXSAT
import com.sihvi.glsm.sls.GLSMBuilder
import com.sihvi.glsm.sls.StateMachineTransition
import com.sihvi.glsm.space.BooleanSearchSpace
import com.sihvi.glsm.space.SearchSpace
import com.sihvi.glsm.strategy.IIMode
import com.sihvi.glsm.strategy.IterativeImprovementStrategy
import com.sihvi.glsm.strategy.RandomWalkStrategy
import com.sihvi.glsm.transitionpredicate.NoImprovementPredicate
import com.sihvi.glsm.transitionpredicate.NotPredicate
import com.sihvi.glsm.transitionpredicate.ProbabilisticPredicate
import com.sihvi.glsm.memory.Memory
import com.sihvi.glsm.memory.BasicMemory
import com.sihvi.glsm.memory.BasicSolution
val noVariables = 20
val problemInstance = MAXSAT.getRandomInstance(noVariables,50,2)
println(problemInstance)
val space = BooleanSearchSpace(noVariables)
val initialSolution = space.getInitial()
val memory = BasicMemory(BasicSolution(initialSolution, problemInstance.evaluate(initialSolution)))
val terminationPredicate = NoImprovementPredicate(10)
val wp = 0.1
val toRandomPredicate = ProbabilisticPredicate(to = wp)
val toIIPredicate = NotPredicate(toRandomPredicate)
val walk = RandomWalkStrategy<Boolean>()
val iterativeImprovement = IterativeImprovementStrategy<Boolean>(IIMode.BEST, true)
val glsm = GLSMBuilder<Boolean, BasicSolution<Boolean>>()
.addStrategy(iterativeImprovement)
.addStrategy(walk)
.addTransition(StateMachineTransition(0, 1, toRandomPredicate))
.addTransition(StateMachineTransition(1, 0, toIIPredicate))
.addTransition(StateMachineTransition(0, -1, terminationPredicate))
.build()
glsm
glsm.toASCII()
val finalSolution = glsm.solve(memory, space, problemInstance::evaluate)
println("Steps taken: " + memory.stepCount)
println("Solution: " + finalSolution.solution.joinToString(", "))
println("Cost: " + finalSolution.cost)
| 0.378574 | 0.703027 |
#1. Install Dependencies
First install the libraries needed to execute recipes, this only needs to be done once, then click play.
```
!pip install git+https://github.com/google/starthinker
```
#2. Get Cloud Project ID
To run this recipe [requires a Google Cloud Project](https://github.com/google/starthinker/blob/master/tutorials/cloud_project.md), this only needs to be done once, then click play.
```
CLOUD_PROJECT = 'PASTE PROJECT ID HERE'
print("Cloud Project Set To: %s" % CLOUD_PROJECT)
```
#3. Get Client Credentials
To read and write to various endpoints requires [downloading client credentials](https://github.com/google/starthinker/blob/master/tutorials/cloud_client_installed.md), this only needs to be done once, then click play.
```
CLIENT_CREDENTIALS = 'PASTE CREDENTIALS HERE'
print("Client Credentials Set To: %s" % CLIENT_CREDENTIALS)
```
#4. Enter Trends Places To BigQuery Via Values Parameters
Move using hard coded WOEID values.
1. Provide <a href='https://apps.twitter.com/' target='_blank'>Twitter credentials</a>.
1. Provide a comma delimited list of WOEIDs.
1. Specify BigQuery dataset and table to write API call results to.
1. Writes: WOEID, Name, Url, Promoted_Content, Query, Tweet_Volume
1. Note Twitter API is rate limited to 15 requests per 15 minutes. So keep WOEID lists short.
Modify the values below for your use case, can be done multiple times, then click play.
```
FIELDS = {
'auth_write': 'service', # Credentials used for writing data.
'secret': '',
'key': '',
'woeids': [],
'destination_dataset': '',
'destination_table': '',
}
print("Parameters Set To: %s" % FIELDS)
```
#5. Execute Trends Places To BigQuery Via Values
This does NOT need to be modified unles you are changing the recipe, click play.
```
from starthinker.util.project import project
from starthinker.script.parse import json_set_fields, json_expand_includes
USER_CREDENTIALS = '/content/user.json'
TASKS = [
{
'twitter': {
'auth': 'user',
'secret': {'field': {'name': 'secret','kind': 'string','order': 1,'default': ''}},
'key': {'field': {'name': 'key','kind': 'string','order': 2,'default': ''}},
'trends': {
'places': {
'single_cell': True,
'values': {'field': {'name': 'woeids','kind': 'integer_list','order': 3,'default': []}}
}
},
'out': {
'bigquery': {
'dataset': {'field': {'name': 'destination_dataset','kind': 'string','order': 6,'default': ''}},
'table': {'field': {'name': 'destination_table','kind': 'string','order': 7,'default': ''}}
}
}
}
}
]
json_set_fields(TASKS, FIELDS)
json_expand_includes(TASKS)
project.initialize(_recipe={ 'tasks':TASKS }, _project=CLOUD_PROJECT, _user=USER_CREDENTIALS, _client=CLIENT_CREDENTIALS, _verbose=True)
project.execute()
```
|
github_jupyter
|
!pip install git+https://github.com/google/starthinker
CLOUD_PROJECT = 'PASTE PROJECT ID HERE'
print("Cloud Project Set To: %s" % CLOUD_PROJECT)
CLIENT_CREDENTIALS = 'PASTE CREDENTIALS HERE'
print("Client Credentials Set To: %s" % CLIENT_CREDENTIALS)
FIELDS = {
'auth_write': 'service', # Credentials used for writing data.
'secret': '',
'key': '',
'woeids': [],
'destination_dataset': '',
'destination_table': '',
}
print("Parameters Set To: %s" % FIELDS)
from starthinker.util.project import project
from starthinker.script.parse import json_set_fields, json_expand_includes
USER_CREDENTIALS = '/content/user.json'
TASKS = [
{
'twitter': {
'auth': 'user',
'secret': {'field': {'name': 'secret','kind': 'string','order': 1,'default': ''}},
'key': {'field': {'name': 'key','kind': 'string','order': 2,'default': ''}},
'trends': {
'places': {
'single_cell': True,
'values': {'field': {'name': 'woeids','kind': 'integer_list','order': 3,'default': []}}
}
},
'out': {
'bigquery': {
'dataset': {'field': {'name': 'destination_dataset','kind': 'string','order': 6,'default': ''}},
'table': {'field': {'name': 'destination_table','kind': 'string','order': 7,'default': ''}}
}
}
}
}
]
json_set_fields(TASKS, FIELDS)
json_expand_includes(TASKS)
project.initialize(_recipe={ 'tasks':TASKS }, _project=CLOUD_PROJECT, _user=USER_CREDENTIALS, _client=CLIENT_CREDENTIALS, _verbose=True)
project.execute()
| 0.259263 | 0.72309 |
# Idealized entrainment
This notebook runs [GOTM](https://gotm.net/) using the [entrainment](https://gotm.net/cases/entrainment/) test case, an idealized wind stress-driven entrainment case with no rotation, in which the mixed layer gradually entrains into an underlying non-turbulent region with constant stable stratification.
Four turbulence closure schemes are used:
- GLS-C01A: The generic length scale ([Umlauf and Burchard, 2003](https://doi.org/10.1357/002224003322005087)) model in the $k$-$\epsilon$ formulation with the weak-equilibrium stability function by [Canuto et al., 2001](https://doi.org/10.1175/1520-0485(2001)031%3C1413:OTPIOP%3E2.0.CO;2) (C01A).
- Three variants of KPP via [CVMix](http://cvmix.github.io):
- KPP-CVMix ([Large et al., 1994](https://doi.org/10.1029/94RG01872), [Griffies et al., 2015](https://github.com/CVMix/CVMix-description/raw/master/cvmix.pdf))
- KPPLT-VR12 ([Li et al., 2016](https://doi.org/10.1016%2Fj.ocemod.2015.07.020))
- KPPLT-LF17 ([Li and Fox-Kemper, 2017](https://doi.org/10.1175%2FJPO-D-17-0085.1))
Run the case with a combination of vertical resolution of [0.2, 1., 5.] m and time step of [6, 60, 600] s.
```
import sys
import copy
import numpy as np
import matplotlib.pyplot as plt
# add the path of gotmtool
sys.path.append("../gotmtool")
from gotmtool import *
```
## Create a model
Create a model with environment file `../../.gotm_env.yaml`, which is created by `gotm_env_init.py`.
```
m = Model(name='Entrainment', environ='../gotmtool/.gotm_env.yaml')
```
Take a look at what are defined in the environment file.
```
for key in m.environ:
print('{:>15s}: {}'.format(key, m.environ[key]) )
```
## Build the model
```
%%time
m.build()
```
## Configuration
Initialize the GOTM configuration
```
cfg = m.init_config()
```
Update the configuration
```
# setup
title = 'Shear-driven Entrainment'
depth = 50.0
cfg['title'] = title
cfg['location']['name'] = 'equator'
cfg['location']['latitude'] = 0.0
cfg['location']['longitude'] = 0.0
cfg['location']['depth'] = depth
cfg['time']['start'] = '2005-01-01 00:00:00'
cfg['time']['stop'] = '2005-01-02 06:00:00'
# output
cfg['output'] = {}
cfg['output']['gotm_out'] = {}
cfg['output']['gotm_out']['use'] = True
cfg['output']['gotm_out']['title'] = title
cfg['output']['gotm_out']['time_unit'] = 'second'
cfg['output']['gotm_out']['time_step'] = 1800
cfg['output']['gotm_out']['variables'] = [{}]
cfg['output']['gotm_out']['variables'][0]['source'] = '*'
# forcing
cfg['temperature']['method'] = 'buoyancy'
cfg['temperature']['two_layer']['t_s'] = 20.0
cfg['temperature']['NN'] = 1e-4
cfg['salinity']['method'] = 'constant'
cfg['salinity']['constant_value'] = 35.0
cfg['surface']['fluxes']['method'] = 'off'
cfg['surface']['fluxes']['heat']['method'] = 'constant'
# since KPPLT-LF17 only use the Langmuir enhanced entrainment
# under destabilizing conditions, use a small destabilizing heat
# flux to activate it
cfg['surface']['fluxes']['heat']['constant_value'] = -1.0e-12
cfg['surface']['fluxes']['tx']['method'] = 'constant'
cfg['surface']['fluxes']['tx']['constant_value'] = 1.0e-1
cfg['surface']['fluxes']['ty']['method'] = 'constant'
cfg['surface']['fluxes']['ty']['constant_value'] = 0.0
cfg['waves']['stokes_drift']['us']['method'] = 'exponential'
cfg['waves']['stokes_drift']['vs']['method'] = 'exponential'
cfg['waves']['stokes_drift']['exponential']['us0']['method'] = 'constant'
cfg['waves']['stokes_drift']['exponential']['us0']['constant_value'] = 0.111
cfg['waves']['stokes_drift']['exponential']['vs0']['method'] = 'constant'
cfg['waves']['stokes_drift']['exponential']['vs0']['constant_value'] = 0.0
cfg['waves']['stokes_drift']['exponential']['ds']['method'] = 'constant'
cfg['waves']['stokes_drift']['exponential']['ds']['constant_value'] = 5.0
# EOS
cfg['eq_state']['method'] = 'unesco'
cfg['eq_state']['form'] = 'full-pot'
# buoyancy
cfg['buoyancy']['NN_ini'] = 1.0e-4
# configure GLS-C01A
cfg['turbulence']['turb_method'] = 'second_order'
cfg['turbulence']['tke_method'] = 'tke'
cfg['turbulence']['len_scale_method'] = 'gls'
cfg['turbulence']['scnd']['method'] = 'weak_eq_kb_eq'
cfg['turbulence']['scnd']['scnd_coeff'] = 'canuto-a'
cfg['turbulence']['turb_param']['length_lim'] = 'false'
cfg['turbulence']['turb_param']['compute_c3'] = 'true'
cfg['turbulence']['turb_param']['Ri_st'] = 0.25
cfg['turbulence']['generic']['gen_m'] = 1.5
cfg['turbulence']['generic']['gen_n'] = -1.0
cfg['turbulence']['generic']['gen_p'] = 3.0
cfg['turbulence']['generic']['cpsi1'] = 1.44
cfg['turbulence']['generic']['cpsi2'] = 1.92
cfg['turbulence']['generic']['cpsi3minus'] = -0.63
cfg['turbulence']['generic']['cpsi3plus'] = 1.0
cfg['turbulence']['generic']['sig_kpsi'] = 1.0
cfg['turbulence']['generic']['sig_psi'] = 1.3
```
## Run the model
Set the configurations and labels for each run
```
cfgs = []
labels = []
for dz in [0.2, 1., 5.]:
nlev = int(depth/dz)
cfg['grid']['nlev'] = nlev
cfg['output']['gotm_out']['k1_stop'] = nlev+1
cfg['output']['gotm_out']['k_stop'] = nlev
for dt in [6.0, 60.0, 600.0]:
nt = int(30.*3600/dt)
cfg['time']['dt'] = dt
label = '_Nz{:d}_Nt{:d}'.format(nlev, nt)
cfg['turbulence']['turb_method'] = 'second_order'
cfgs.append(copy.deepcopy(cfg))
labels.append('GLS-C01A'+label)
cfg['turbulence']['turb_method'] = 'cvmix'
cfg['cvmix']['surface_layer']['kpp']['langmuir_method'] = 'none'
cfgs.append(copy.deepcopy(cfg))
labels.append('KPP-CVMix'+label)
cfg['cvmix']['surface_layer']['kpp']['langmuir_method'] = 'lwf16'
cfgs.append(copy.deepcopy(cfg))
labels.append('KPPLT-VR12'+label)
cfg['cvmix']['surface_layer']['kpp']['langmuir_method'] = 'lf17'
cfgs.append(copy.deepcopy(cfg))
labels.append('KPPLT-LF17'+label)
```
Run the cases in parallel with 2 processes
```
%%time
sims = m.run_batch(configs=cfgs, labels=labels, nproc=2)
```
|
github_jupyter
|
import sys
import copy
import numpy as np
import matplotlib.pyplot as plt
# add the path of gotmtool
sys.path.append("../gotmtool")
from gotmtool import *
m = Model(name='Entrainment', environ='../gotmtool/.gotm_env.yaml')
for key in m.environ:
print('{:>15s}: {}'.format(key, m.environ[key]) )
%%time
m.build()
cfg = m.init_config()
# setup
title = 'Shear-driven Entrainment'
depth = 50.0
cfg['title'] = title
cfg['location']['name'] = 'equator'
cfg['location']['latitude'] = 0.0
cfg['location']['longitude'] = 0.0
cfg['location']['depth'] = depth
cfg['time']['start'] = '2005-01-01 00:00:00'
cfg['time']['stop'] = '2005-01-02 06:00:00'
# output
cfg['output'] = {}
cfg['output']['gotm_out'] = {}
cfg['output']['gotm_out']['use'] = True
cfg['output']['gotm_out']['title'] = title
cfg['output']['gotm_out']['time_unit'] = 'second'
cfg['output']['gotm_out']['time_step'] = 1800
cfg['output']['gotm_out']['variables'] = [{}]
cfg['output']['gotm_out']['variables'][0]['source'] = '*'
# forcing
cfg['temperature']['method'] = 'buoyancy'
cfg['temperature']['two_layer']['t_s'] = 20.0
cfg['temperature']['NN'] = 1e-4
cfg['salinity']['method'] = 'constant'
cfg['salinity']['constant_value'] = 35.0
cfg['surface']['fluxes']['method'] = 'off'
cfg['surface']['fluxes']['heat']['method'] = 'constant'
# since KPPLT-LF17 only use the Langmuir enhanced entrainment
# under destabilizing conditions, use a small destabilizing heat
# flux to activate it
cfg['surface']['fluxes']['heat']['constant_value'] = -1.0e-12
cfg['surface']['fluxes']['tx']['method'] = 'constant'
cfg['surface']['fluxes']['tx']['constant_value'] = 1.0e-1
cfg['surface']['fluxes']['ty']['method'] = 'constant'
cfg['surface']['fluxes']['ty']['constant_value'] = 0.0
cfg['waves']['stokes_drift']['us']['method'] = 'exponential'
cfg['waves']['stokes_drift']['vs']['method'] = 'exponential'
cfg['waves']['stokes_drift']['exponential']['us0']['method'] = 'constant'
cfg['waves']['stokes_drift']['exponential']['us0']['constant_value'] = 0.111
cfg['waves']['stokes_drift']['exponential']['vs0']['method'] = 'constant'
cfg['waves']['stokes_drift']['exponential']['vs0']['constant_value'] = 0.0
cfg['waves']['stokes_drift']['exponential']['ds']['method'] = 'constant'
cfg['waves']['stokes_drift']['exponential']['ds']['constant_value'] = 5.0
# EOS
cfg['eq_state']['method'] = 'unesco'
cfg['eq_state']['form'] = 'full-pot'
# buoyancy
cfg['buoyancy']['NN_ini'] = 1.0e-4
# configure GLS-C01A
cfg['turbulence']['turb_method'] = 'second_order'
cfg['turbulence']['tke_method'] = 'tke'
cfg['turbulence']['len_scale_method'] = 'gls'
cfg['turbulence']['scnd']['method'] = 'weak_eq_kb_eq'
cfg['turbulence']['scnd']['scnd_coeff'] = 'canuto-a'
cfg['turbulence']['turb_param']['length_lim'] = 'false'
cfg['turbulence']['turb_param']['compute_c3'] = 'true'
cfg['turbulence']['turb_param']['Ri_st'] = 0.25
cfg['turbulence']['generic']['gen_m'] = 1.5
cfg['turbulence']['generic']['gen_n'] = -1.0
cfg['turbulence']['generic']['gen_p'] = 3.0
cfg['turbulence']['generic']['cpsi1'] = 1.44
cfg['turbulence']['generic']['cpsi2'] = 1.92
cfg['turbulence']['generic']['cpsi3minus'] = -0.63
cfg['turbulence']['generic']['cpsi3plus'] = 1.0
cfg['turbulence']['generic']['sig_kpsi'] = 1.0
cfg['turbulence']['generic']['sig_psi'] = 1.3
cfgs = []
labels = []
for dz in [0.2, 1., 5.]:
nlev = int(depth/dz)
cfg['grid']['nlev'] = nlev
cfg['output']['gotm_out']['k1_stop'] = nlev+1
cfg['output']['gotm_out']['k_stop'] = nlev
for dt in [6.0, 60.0, 600.0]:
nt = int(30.*3600/dt)
cfg['time']['dt'] = dt
label = '_Nz{:d}_Nt{:d}'.format(nlev, nt)
cfg['turbulence']['turb_method'] = 'second_order'
cfgs.append(copy.deepcopy(cfg))
labels.append('GLS-C01A'+label)
cfg['turbulence']['turb_method'] = 'cvmix'
cfg['cvmix']['surface_layer']['kpp']['langmuir_method'] = 'none'
cfgs.append(copy.deepcopy(cfg))
labels.append('KPP-CVMix'+label)
cfg['cvmix']['surface_layer']['kpp']['langmuir_method'] = 'lwf16'
cfgs.append(copy.deepcopy(cfg))
labels.append('KPPLT-VR12'+label)
cfg['cvmix']['surface_layer']['kpp']['langmuir_method'] = 'lf17'
cfgs.append(copy.deepcopy(cfg))
labels.append('KPPLT-LF17'+label)
%%time
sims = m.run_batch(configs=cfgs, labels=labels, nproc=2)
| 0.114554 | 0.920932 |
# Generating Random Bayesian Network
This notebook shows how to generate singly- and multi-connected Bayesian Belief Networks (BBNs). The algorithms are taken directly from [here](https://pdfs.semanticscholar.org/5273/2fb57129443592024b0e7e46c2a1ec36639c.pdf). When generating a BBN, you have to generate
* the structure, which is a directed acyclic graph (DAG), and
* the parameters, which are local probability models.
In this notebook, the parameters are assumed to take on the [Dirichlet-Multinomial](https://en.wikipedia.org/wiki/Dirichlet-multinomial_distribution) distribution. If you are wondering, a singly-connected BBN is one when, ignoring the directions of the edges in the DAG, there is at most one path between any two nodes. A multi-connected BBN is one that is not singly-connected (it is defined by the negation of a singly-connected BBN). The BBNs generated using this approach gaurantees the distribution of the BBNs are uniformly distributed (look at the paper for proofs, details, and benefits of having this property).
## Generate the structure
Here, we generate the DAGs of a singly- and multi-connected BBNs. Note that when we visualize the DAGs, we do so by converting it to an undirected graph only because the layout algorithm is more aesthetically pleasing.
```
import warnings
import networkx as nx
import numpy as np
import json
import matplotlib.pyplot as plt
from networkx.algorithms.dag import is_directed_acyclic_graph
from networkx.algorithms.shortest_paths.generic import shortest_path
np.random.seed(37)
def get_simple_ordered_tree(n):
"""
Generates a simple-ordered tree. The tree is just a
directed acyclic graph of n nodes with the structure
0 --> 1 --> .... --> n.
"""
g = nx.DiGraph()
for i in range(n):
g.add_node(i)
for i in range(n - 1):
g.add_edges_from([(i, i+1, {})])
return g
def convert_to_undirected_graph(g):
"""
Converts a directed acyclic graph (DAG) to an undirected graph.
We need to convert a DAG to an undirected one to use
some API calls to operate over the undirected graph. For example,
in checking for connectedness of a graph, the API has a method
to check for connectedness of an undirected graph, but not a
DAG.
"""
u = nx.Graph()
for n in g.nodes:
u.add_node(n)
for e in g.edges:
u.add_edges_from([(e[0], e[1], {})])
return u
def is_connected(g):
"""
Checks if a the directed acyclic graph is connected.
"""
u = convert_to_undirected_graph(g)
return nx.is_connected(u)
def get_random_node_pair(n):
"""
Randomly generates a pair of nodes.
"""
i = np.random.randint(0, n)
j = i
while j == i:
j = np.random.randint(0, n)
return i, j
def edge_exists(i, j, g):
"""
Checks if the edge i --> j exists in the graph, g.
"""
return j in list(g.successors(i))
def del_edge(i, j, g):
"""
Deletes the edge i --> j in the graph, g. The edge is only
deleted if this removal does NOT cause the graph to be
disconnected.
"""
if g.has_edge(i, j) is True:
g.remove_edge(i, j)
if is_connected(g) is False:
g.add_edges_from([(i, j, {})])
def add_edge(i, j, g):
"""
Adds an edge i --> j to the graph, g. The edge is only
added if this addition does NOT cause the graph to have
cycles.
"""
g.add_edges_from([(i, j, {})])
if is_directed_acyclic_graph(g) is False:
g.remove_edge(i, j)
def find_predecessor(i, j, g):
"""
Finds a predecessor, k, in the path between two nodes, i and j,
in the graph, g. We assume g is connected, and there is a
path between i and j (ignoring the direction of the edges).
We want to find a k, that is a parent of j, that is in
the path between i and j. In some cases, we may not find
such a k.
"""
parents = list(g.predecessors(j))
u = convert_to_undirected_graph(g)
for pa in parents:
try:
path = shortest_path(u, pa, i)
return pa
except:
pass
return None
def generate_multi_connected_structure(n, max_iter=10):
"""
Generates a multi-connected directed acyclic graph.
"""
g = get_simple_ordered_tree(n)
for it in range(max_iter):
i, j = get_random_node_pair(n)
if g.has_edge(i, j) is True:
del_edge(i, j, g)
else:
add_edge(i, j, g)
return g
def generate_singly_structure(n, max_iter=10):
"""
Generates a singly-connected directed acyclic graph.
"""
g = get_simple_ordered_tree(n)
counter = 0
for it in range(max_iter):
i, j = get_random_node_pair(n)
if g.has_edge(i, j) is True or g.has_edge(j, i) is True:
pass
else:
p = np.random.random()
k = find_predecessor(i, j, g)
if k is not None:
g.remove_edge(k, j)
if p < 0.5:
g.add_edges_from([(j, i, {})])
else:
g.add_edges_from([(i, j, {})])
if is_connected(g) is False:
g.add_edges_from([(k, j, {})])
if p < 0.5:
g.remove_edge(j, i)
else:
g.remove_edge(i, j)
return g
```
### Generate DAG for singly-connected BBN
```
with warnings.catch_warnings(record=True):
s = generate_singly_structure(5, 1000)
plt.figure(figsize=(10, 5))
plt.subplot(111)
nx.draw(convert_to_undirected_graph(s), with_labels=True, font_weight='bold')
```
### Generate DAG for multi-connected BBN
```
with warnings.catch_warnings(record=True):
m = generate_multi_connected_structure(5, 10)
plt.figure(figsize=(10, 5))
plt.subplot(111)
nx.draw(convert_to_undirected_graph(m), with_labels=True, font_weight='bold')
```
## Generate the parameters
Here, we generate parameters for the BBNs.
```
from scipy.stats import dirichlet, multinomial
def generate_num_values(n, max_values=2):
"""
For each node, i, in the nodes, n, determine the number of values
the node (or equivalently, variable) has. Every node/variable in a
Bayesian Network should have 2 or more values. This generates
the number of values each variable will have. Each number will be
sampled uniformly.
"""
return np.array([max(np.random.randint(0, max_values) + 1, 2) for _ in range(n)])
def generate_alphas(n, max_alpha=10):
"""
Generate random number for the alpha's (the hyperparameters).
Each number will be in the range [1, max_alpha]. Each number will
be sampled uniformly.
"""
return [np.random.randint(1, max_alpha + 1) for i in range(n)]
def sample_dirichlet(n, max_alpha=10):
"""
Samples from the Dirichlet distribution to a produce
a probability vector of length n. The sum of each probability
in the probability vector should sum to 1.
"""
return np.array(dirichlet.rvs(generate_alphas(n, max_alpha))[0])
def get_num_parent_instantiations(parents, num_values):
num_pa_instantiations = 1
for pa in parents:
num_pa_values = num_values[pa]
num_pa_instantiations *= num_pa_values
return num_pa_instantiations
def generate_dirichlet_parameters(i, parents, num_values, max_alpha=10):
"""
Randomly and uniformly generate parameters for a node i. A matrix
of parameters will be returned. The matrix will represent the
condtional probability table of the node i. The matrix will have
the dimensions m (rows) by n (columns), m x n, where m is the
product of the domain sizes of the parents, and n is the domain
size of the node. The domain size is just the number of values
that a node (variable) has, which should always be greater than
or equal to 2.
"""
num_pa_instantiations = get_num_parent_instantiations(parents, num_values)
n = num_values[i]
cpt = []
for pa_instantiation in range(num_pa_instantiations):
probs = sample_dirichlet(n, max_alpha)
cpt.append(probs)
return np.array(cpt)
def generate_parameters(g, max_values=2, max_alpha=10):
"""
Generates parameters for each node in the graph, g.
A dictionary indexed by the node's id will give its
(sampled) parameters and its parents.
"""
num_nodes = len(list(g.nodes))
num_values = generate_num_values(num_nodes, max_values)
g_params = {}
for i in g.nodes:
parents = list(g.predecessors(i))
params = generate_dirichlet_parameters(i, parents, num_values, max_alpha)
g_params[i] = {
'parents': parents,
'params': params,
'shape': [get_num_parent_instantiations(parents, num_values), num_values[i]]
}
return g_params
```
### Generate parameters for singly-connected BBN
```
s_params = generate_parameters(s)
print(s_params)
```
### Generate parameters for muti-connected BBN
```
m_params = generate_parameters(m)
print(m_params)
```
## Persist (save) the Bayesian Belief Network
Here, we show how to save the BBN (the DAG and parameters). Note that we save it to a JSON file format. There are simply too many formats for BBNs, but the JSON format here has all the information you need to convert it to any other format.
```
def to_json(g, params, pretty=True):
to_int_arr = lambda arr: [int(item) for item in arr]
j = {}
j['nodes'] = list(g.nodes)
j['edges'] = [{'pa': e[0], 'ch': e[1]} for e in g.edges]
j['parameters'] = [{'node': k,
'params': list(v['params'].flatten()),
'shape': to_int_arr(v['shape'])}
for k, v in params.items()]
if pretty:
return json.dumps(j, indent=2, sort_keys=False)
return json.dumps(j)
```
### Persist singly-connected BBN
```
s_json = to_json(s, s_params)
print(s_json)
with open('./output/singly-connected.json', 'w') as fhandle:
fhandle.write(to_json(s, s_params, pretty=True))
```
### Persist multi-connected BBN
```
m_json = to_json(m, m_params)
print(m_json)
with open('./output/multi-connected.json', 'w') as fhandle:
fhandle.write(to_json(m, m_params, pretty=True))
```
## All-in-one (AIO) example
Here's a simple AIO example of generating a singly-connected BBN and its corresponding JSON.
```
g = generate_singly_structure(5, 1000)
p = generate_parameters(g)
j = to_json(g, p)
```
|
github_jupyter
|
import warnings
import networkx as nx
import numpy as np
import json
import matplotlib.pyplot as plt
from networkx.algorithms.dag import is_directed_acyclic_graph
from networkx.algorithms.shortest_paths.generic import shortest_path
np.random.seed(37)
def get_simple_ordered_tree(n):
"""
Generates a simple-ordered tree. The tree is just a
directed acyclic graph of n nodes with the structure
0 --> 1 --> .... --> n.
"""
g = nx.DiGraph()
for i in range(n):
g.add_node(i)
for i in range(n - 1):
g.add_edges_from([(i, i+1, {})])
return g
def convert_to_undirected_graph(g):
"""
Converts a directed acyclic graph (DAG) to an undirected graph.
We need to convert a DAG to an undirected one to use
some API calls to operate over the undirected graph. For example,
in checking for connectedness of a graph, the API has a method
to check for connectedness of an undirected graph, but not a
DAG.
"""
u = nx.Graph()
for n in g.nodes:
u.add_node(n)
for e in g.edges:
u.add_edges_from([(e[0], e[1], {})])
return u
def is_connected(g):
"""
Checks if a the directed acyclic graph is connected.
"""
u = convert_to_undirected_graph(g)
return nx.is_connected(u)
def get_random_node_pair(n):
"""
Randomly generates a pair of nodes.
"""
i = np.random.randint(0, n)
j = i
while j == i:
j = np.random.randint(0, n)
return i, j
def edge_exists(i, j, g):
"""
Checks if the edge i --> j exists in the graph, g.
"""
return j in list(g.successors(i))
def del_edge(i, j, g):
"""
Deletes the edge i --> j in the graph, g. The edge is only
deleted if this removal does NOT cause the graph to be
disconnected.
"""
if g.has_edge(i, j) is True:
g.remove_edge(i, j)
if is_connected(g) is False:
g.add_edges_from([(i, j, {})])
def add_edge(i, j, g):
"""
Adds an edge i --> j to the graph, g. The edge is only
added if this addition does NOT cause the graph to have
cycles.
"""
g.add_edges_from([(i, j, {})])
if is_directed_acyclic_graph(g) is False:
g.remove_edge(i, j)
def find_predecessor(i, j, g):
"""
Finds a predecessor, k, in the path between two nodes, i and j,
in the graph, g. We assume g is connected, and there is a
path between i and j (ignoring the direction of the edges).
We want to find a k, that is a parent of j, that is in
the path between i and j. In some cases, we may not find
such a k.
"""
parents = list(g.predecessors(j))
u = convert_to_undirected_graph(g)
for pa in parents:
try:
path = shortest_path(u, pa, i)
return pa
except:
pass
return None
def generate_multi_connected_structure(n, max_iter=10):
"""
Generates a multi-connected directed acyclic graph.
"""
g = get_simple_ordered_tree(n)
for it in range(max_iter):
i, j = get_random_node_pair(n)
if g.has_edge(i, j) is True:
del_edge(i, j, g)
else:
add_edge(i, j, g)
return g
def generate_singly_structure(n, max_iter=10):
"""
Generates a singly-connected directed acyclic graph.
"""
g = get_simple_ordered_tree(n)
counter = 0
for it in range(max_iter):
i, j = get_random_node_pair(n)
if g.has_edge(i, j) is True or g.has_edge(j, i) is True:
pass
else:
p = np.random.random()
k = find_predecessor(i, j, g)
if k is not None:
g.remove_edge(k, j)
if p < 0.5:
g.add_edges_from([(j, i, {})])
else:
g.add_edges_from([(i, j, {})])
if is_connected(g) is False:
g.add_edges_from([(k, j, {})])
if p < 0.5:
g.remove_edge(j, i)
else:
g.remove_edge(i, j)
return g
with warnings.catch_warnings(record=True):
s = generate_singly_structure(5, 1000)
plt.figure(figsize=(10, 5))
plt.subplot(111)
nx.draw(convert_to_undirected_graph(s), with_labels=True, font_weight='bold')
with warnings.catch_warnings(record=True):
m = generate_multi_connected_structure(5, 10)
plt.figure(figsize=(10, 5))
plt.subplot(111)
nx.draw(convert_to_undirected_graph(m), with_labels=True, font_weight='bold')
from scipy.stats import dirichlet, multinomial
def generate_num_values(n, max_values=2):
"""
For each node, i, in the nodes, n, determine the number of values
the node (or equivalently, variable) has. Every node/variable in a
Bayesian Network should have 2 or more values. This generates
the number of values each variable will have. Each number will be
sampled uniformly.
"""
return np.array([max(np.random.randint(0, max_values) + 1, 2) for _ in range(n)])
def generate_alphas(n, max_alpha=10):
"""
Generate random number for the alpha's (the hyperparameters).
Each number will be in the range [1, max_alpha]. Each number will
be sampled uniformly.
"""
return [np.random.randint(1, max_alpha + 1) for i in range(n)]
def sample_dirichlet(n, max_alpha=10):
"""
Samples from the Dirichlet distribution to a produce
a probability vector of length n. The sum of each probability
in the probability vector should sum to 1.
"""
return np.array(dirichlet.rvs(generate_alphas(n, max_alpha))[0])
def get_num_parent_instantiations(parents, num_values):
num_pa_instantiations = 1
for pa in parents:
num_pa_values = num_values[pa]
num_pa_instantiations *= num_pa_values
return num_pa_instantiations
def generate_dirichlet_parameters(i, parents, num_values, max_alpha=10):
"""
Randomly and uniformly generate parameters for a node i. A matrix
of parameters will be returned. The matrix will represent the
condtional probability table of the node i. The matrix will have
the dimensions m (rows) by n (columns), m x n, where m is the
product of the domain sizes of the parents, and n is the domain
size of the node. The domain size is just the number of values
that a node (variable) has, which should always be greater than
or equal to 2.
"""
num_pa_instantiations = get_num_parent_instantiations(parents, num_values)
n = num_values[i]
cpt = []
for pa_instantiation in range(num_pa_instantiations):
probs = sample_dirichlet(n, max_alpha)
cpt.append(probs)
return np.array(cpt)
def generate_parameters(g, max_values=2, max_alpha=10):
"""
Generates parameters for each node in the graph, g.
A dictionary indexed by the node's id will give its
(sampled) parameters and its parents.
"""
num_nodes = len(list(g.nodes))
num_values = generate_num_values(num_nodes, max_values)
g_params = {}
for i in g.nodes:
parents = list(g.predecessors(i))
params = generate_dirichlet_parameters(i, parents, num_values, max_alpha)
g_params[i] = {
'parents': parents,
'params': params,
'shape': [get_num_parent_instantiations(parents, num_values), num_values[i]]
}
return g_params
s_params = generate_parameters(s)
print(s_params)
m_params = generate_parameters(m)
print(m_params)
def to_json(g, params, pretty=True):
to_int_arr = lambda arr: [int(item) for item in arr]
j = {}
j['nodes'] = list(g.nodes)
j['edges'] = [{'pa': e[0], 'ch': e[1]} for e in g.edges]
j['parameters'] = [{'node': k,
'params': list(v['params'].flatten()),
'shape': to_int_arr(v['shape'])}
for k, v in params.items()]
if pretty:
return json.dumps(j, indent=2, sort_keys=False)
return json.dumps(j)
s_json = to_json(s, s_params)
print(s_json)
with open('./output/singly-connected.json', 'w') as fhandle:
fhandle.write(to_json(s, s_params, pretty=True))
m_json = to_json(m, m_params)
print(m_json)
with open('./output/multi-connected.json', 'w') as fhandle:
fhandle.write(to_json(m, m_params, pretty=True))
g = generate_singly_structure(5, 1000)
p = generate_parameters(g)
j = to_json(g, p)
| 0.632389 | 0.976197 |
```
from sklearn.metrics.pairwise import cosine_similarity
"""
paper 2 - ALTERNATIVE SIGNATURE-BASED ('VECTOR SPACE') APPROACH
Clustering GP practices according to similarities in signature paths
Given data are available up to mid December 2017, models are trained to predict feedback for September
and October 2017. Model testing takes place for feedback in November and December 2017. Models are trained and
tested for GP practices which:
- have at least 2 reviews from period preceding September 2017
- received some feedback in November and December 2017
"""
def cluster_pred(csv_file, depvar='q9', ignore_nan=True, train = None, test = None, day_min=735008,log=False,degs=2, sigs_with_depvar=True, cutoff = 10):
"""
To do:
- can add options for different distance measures between pairs of vectors
"""
#preliminary tests
if test == None and test == train:
test = list(range(736621,736681))
train = list(range(day_min,736621))
elif type(test) == list and type(train) == list:
if len(test) == 0 or len(train) == 0:
print('TypeError: test and train should be non-empty lists of day ID numbers')
return()
else:
print("TypeError: define test and train attributes or leave default values, i.e. None")
return()
# test for overlap of train_range and test_range
if min(test) <= max(train):
print("AttributeError: select non-overlapping ranges of days for test and train. Also, days included in train should pre-date days included in test")
return()
# test whether day_min is prior to day ranges defined in y_train and y_test
if day_min >= min(test) or day_min > min(train):
print("AttributeError: select ranges of days for test and train which post-date day_min")
return()
window = max(test) - min(test) + 1
# generate data paths and signatures for each GP practice - "train" period. Retain only data points where dependent variable data are available
x_train = make_paths(csv_file, depvar=depvar, min_day=day_min, max_day=max(train)-window, ignore_nan = ignore_nan)
x_train = make_sigs(x_train, degs, 'sigs_xtrain.csv', log=log, include_depvar=sigs_with_depvar)
y_train = make_depvar(csv_file,depvar, list(range(max(train)-window+1,max(train)+1)))
y_train = pd.DataFrame(y_train)
y_tmp = [str(x) for x in list(y_train.columns)]
x_tmp = [str(x) for x in list(x_train.columns)]
y_train.columns = y_tmp
x_train.columns = x_tmp
train_summary = pd.merge(left=y_train,right=x_train, left_on='0', right_on='0', how='inner')
train_summary.rename(columns={'1_x': 'y_train', '1_y': '1'}, inplace=True)
# generate data paths and signatures for each GP practice - "test" period. Retain only data points where dependent variable data are available
x_test = make_paths(csv_file, depvar=depvar, min_day=day_min+window, max_day=min(test)-1, ignore_nan = ignore_nan)
x_test = make_sigs(x_test, degs, 'sigs_xtest.csv', log=log, include_depvar=sigs_with_depvar)
y_test = make_depvar(csv_file,depvar, list(range(min(test),max(test)+1)))
y_test = pd.DataFrame(y_test)
y_tmp = [str(x) for x in list(y_test.columns)]
x_tmp = [str(x) for x in list(x_test.columns)]
y_test.columns = y_tmp
x_test.columns = x_tmp
test_summary = pd.merge(left=y_test,right=x_test, left_on='0', right_on='0', how='inner')
test_summary.rename(columns={'1_x': 'y_test', '1_y': '1'}, inplace=True)
# predict train dates (with variable "n" parameter which corresponds to the number of nearest vectors considered)
train_summary = train_summary.values.tolist()
# compute cosine similarities for each GP practice
x = []
y = []
depvar_vals = []
for i,v in enumerate(train_summary):
if i < 10000000000: # sample size can be modified for exercise
x.append(v[2:])
y.append(v[2:])
depvar_vals.append(int(v[1])) #depvar values get rounded to the nearest integer
r = cosine_similarity(np.array(x), np.array(y)) #computes cosine similarity score of every vector in 'y' to each vector in 'x'
# associate predictions with gp_ids and depvars rounded to nearest integer
probs = {}
for gp, row in enumerate(r): #every 'row' contains similarities of all vectors to a given vector of 'x'
i = []
ii = []
iii = []
iv = []
v = []
for ind, element in enumerate(row):
if depvar_vals[ind] == 1 and gp != ind:
i.append(element)
elif depvar_vals[ind] == 2 and gp != ind:
ii.append(element)
elif depvar_vals[ind] == 3 and gp != ind:
iii.append(element)
elif depvar_vals[ind] == 4 and gp != ind:
iv.append(element)
elif depvar_vals[ind] == 5 and gp != ind:
v.append(element)
elif gp == ind:
pass
else:
print('something went wrong with prediction calculations')
i = sorted(i, reverse=True)[:cutoff]
i = [xx for xx in i if xx > 0]
ii = sorted(ii, reverse=True)[:cutoff]
ii = [xx for xx in ii if xx > 0]
iii = sorted(iii, reverse=True)[:cutoff]
iii = [xx for xx in iii if xx > 0]
iv = sorted(iv, reverse=True)[:cutoff]
iv = [xx for xx in iv if xx > 0]
v = sorted(v, reverse=True)[:cutoff]
v = [xx for xx in v if xx > 0]
overall = i[:]
overall.extend(ii)
overall.extend(iii)
overall.extend(iv)
overall.extend(v)
overall = sum(overall)
tmp = []
for xx in [i,ii,iii,iv,v]:
prob = sum(xx) / overall
tmp.append(prob)
probs[train_summary[gp][0]] = tmp
#populate the train object to return the predictions and actual values
train = []
for i, xx in enumerate(probs):
tmp = [str(int(xx))]
tmp.extend(probs[xx])
tmp.extend([probs[xx].index(max(probs[xx]))+1 , int(train_summary[i][1])])
train.append(tmp)
train = pd.DataFrame(train, columns=['gp_id', "prob1", "prob2", "prob3", "prob4", "prob5", "yhat_train", "y_train"])
# predict values for the test period (last 60 days of feedback) by looking at the similarity between pairs of signatures
# do it the bayesian way (select the highest probability result, and then calculate test MSE error)
test_summary = test_summary.values.tolist()
x = []
# test_vals = []
for i,v in enumerate(test_summary):
if i < 10000000000: # sample size can be modified for exercise
x.append(v[2:])
# test_vals.append(int(v[1])) #depvar values get rounded to the nearest integer
r = cosine_similarity(np.array(x), np.array(y)) #computes cosine similarity score of every vector in 'y' to each vector in 'x'
# associate predictions with gp_ids and depvars rounded to nearest integer
probs = {}
for gp, row in enumerate(r): #every 'row' contains similarities of all vectors to a given vector of 'x'
i = []
ii = []
iii = []
iv = []
v = []
for ind, element in enumerate(row):
if depvar_vals[ind] == 1:
i.append(element)
elif depvar_vals[ind] == 2:
ii.append(element)
elif depvar_vals[ind] == 3:
iii.append(element)
elif depvar_vals[ind] == 4:
iv.append(element)
elif depvar_vals[ind] == 5:
v.append(element)
else:
print('something went wrong with prediction calculations')
i = sorted(i, reverse=True)[:cutoff]
i = [xx for xx in i if xx > 0]
ii = sorted(ii, reverse=True)[:cutoff]
ii = [xx for xx in ii if xx > 0]
iii = sorted(iii, reverse=True)[:cutoff]
iii = [xx for xx in iii if xx > 0]
iv = sorted(iv, reverse=True)[:cutoff]
iv = [xx for xx in iv if xx > 0]
v = sorted(v, reverse=True)[:cutoff]
v = [xx for xx in v if xx > 0]
overall = i[:]
overall.extend(ii)
overall.extend(iii)
overall.extend(iv)
overall.extend(v)
overall = sum(overall)
tmp = []
for xx in [i,ii,iii,iv,v]:
prob = sum(xx) / overall
tmp.append(prob)
probs[test_summary[gp][0]] = tmp
#populate the train object to return the predictions and actual values
test = []
for i, xx in enumerate(probs):
tmp = [str(int(xx))]
tmp.extend(probs[xx])
tmp.extend([probs[xx].index(max(probs[xx]))+1 , int(test_summary[i][1])])
test.append(tmp)
test = pd.DataFrame(test, columns=['gp_id', "prob1", "prob2", "prob3", "prob4", "prob5", "yhat_test", "y_test"])
results = [train,test]
return results
# clus_res = cluster_pred('r_output.csv',depvar='q9', cutoff = 2)
print('ok')
def clus_mse_calculator (chosen_cutoffs):
"""
Returns list of train and test MSE errors for predictions for a list of chosen 'cutoff' values.
The 'cutoff' parameter is used in cluster_pred() function
"""
clus_res_combo = []
for c in chosen_cutoffs:
print('compute results for cutoff=' +str(c))
clus_res = cluster_pred('r_output.csv',depvar='q9', cutoff = c)
train_mse = mean_squared_error(np.array(clus_res[0]['yhat_train']),np.array(clus_res[0]['y_train']))
test_mse = mean_squared_error(np.array(clus_res[1]['yhat_test']),np.array(clus_res[1]['y_test']))
clus_res_combo.append([c, train_mse, test_mse])
print([c, train_mse, test_mse])
print('all candidate models completed')
# clus_res_combo = clus_mse_calculator(list(range(1,31)))
# clus_res_combo1001to1030 = clus_mse_calculator(list(range(1001,1031)))
# clus_res_combo101to130 = clus_mse_calculator(list(range(101,131)))
for x in clus_res_combo:
print(x)
x = pd.DataFrame(t_results, columns = ['test_err','train_err'])
x['t_val'] = t_candidates
ns = [x[0] for x in clus_res_combo]
tr = [x[1] for x in clus_res_combo]
te = [x[2] for x in clus_res_combo]
print("Prediction errors in model training")
plt.plot(ns, tr, 'ro')
plt.xlabel('Number of top "n" models used to identify the most probable Likert-scale response')
plt.ylabel('average mean squared error')
plt.show()
print("Prediction errors in model testing")
plt.plot(ns, te, 'ro')
plt.xlabel('Number of top "n" models used to identify the most probable Likert-scale response')
plt.ylabel('average mean squared error')
plt.show()
# best clustering-based model
clus_res = cluster_pred('r_output.csv',depvar='q9', cutoff = 22)
mean_squared_error(np.array(clus_res[0]['yhat_train']),np.array(clus_res[0]['y_train']))
```
|
github_jupyter
|
from sklearn.metrics.pairwise import cosine_similarity
"""
paper 2 - ALTERNATIVE SIGNATURE-BASED ('VECTOR SPACE') APPROACH
Clustering GP practices according to similarities in signature paths
Given data are available up to mid December 2017, models are trained to predict feedback for September
and October 2017. Model testing takes place for feedback in November and December 2017. Models are trained and
tested for GP practices which:
- have at least 2 reviews from period preceding September 2017
- received some feedback in November and December 2017
"""
def cluster_pred(csv_file, depvar='q9', ignore_nan=True, train = None, test = None, day_min=735008,log=False,degs=2, sigs_with_depvar=True, cutoff = 10):
"""
To do:
- can add options for different distance measures between pairs of vectors
"""
#preliminary tests
if test == None and test == train:
test = list(range(736621,736681))
train = list(range(day_min,736621))
elif type(test) == list and type(train) == list:
if len(test) == 0 or len(train) == 0:
print('TypeError: test and train should be non-empty lists of day ID numbers')
return()
else:
print("TypeError: define test and train attributes or leave default values, i.e. None")
return()
# test for overlap of train_range and test_range
if min(test) <= max(train):
print("AttributeError: select non-overlapping ranges of days for test and train. Also, days included in train should pre-date days included in test")
return()
# test whether day_min is prior to day ranges defined in y_train and y_test
if day_min >= min(test) or day_min > min(train):
print("AttributeError: select ranges of days for test and train which post-date day_min")
return()
window = max(test) - min(test) + 1
# generate data paths and signatures for each GP practice - "train" period. Retain only data points where dependent variable data are available
x_train = make_paths(csv_file, depvar=depvar, min_day=day_min, max_day=max(train)-window, ignore_nan = ignore_nan)
x_train = make_sigs(x_train, degs, 'sigs_xtrain.csv', log=log, include_depvar=sigs_with_depvar)
y_train = make_depvar(csv_file,depvar, list(range(max(train)-window+1,max(train)+1)))
y_train = pd.DataFrame(y_train)
y_tmp = [str(x) for x in list(y_train.columns)]
x_tmp = [str(x) for x in list(x_train.columns)]
y_train.columns = y_tmp
x_train.columns = x_tmp
train_summary = pd.merge(left=y_train,right=x_train, left_on='0', right_on='0', how='inner')
train_summary.rename(columns={'1_x': 'y_train', '1_y': '1'}, inplace=True)
# generate data paths and signatures for each GP practice - "test" period. Retain only data points where dependent variable data are available
x_test = make_paths(csv_file, depvar=depvar, min_day=day_min+window, max_day=min(test)-1, ignore_nan = ignore_nan)
x_test = make_sigs(x_test, degs, 'sigs_xtest.csv', log=log, include_depvar=sigs_with_depvar)
y_test = make_depvar(csv_file,depvar, list(range(min(test),max(test)+1)))
y_test = pd.DataFrame(y_test)
y_tmp = [str(x) for x in list(y_test.columns)]
x_tmp = [str(x) for x in list(x_test.columns)]
y_test.columns = y_tmp
x_test.columns = x_tmp
test_summary = pd.merge(left=y_test,right=x_test, left_on='0', right_on='0', how='inner')
test_summary.rename(columns={'1_x': 'y_test', '1_y': '1'}, inplace=True)
# predict train dates (with variable "n" parameter which corresponds to the number of nearest vectors considered)
train_summary = train_summary.values.tolist()
# compute cosine similarities for each GP practice
x = []
y = []
depvar_vals = []
for i,v in enumerate(train_summary):
if i < 10000000000: # sample size can be modified for exercise
x.append(v[2:])
y.append(v[2:])
depvar_vals.append(int(v[1])) #depvar values get rounded to the nearest integer
r = cosine_similarity(np.array(x), np.array(y)) #computes cosine similarity score of every vector in 'y' to each vector in 'x'
# associate predictions with gp_ids and depvars rounded to nearest integer
probs = {}
for gp, row in enumerate(r): #every 'row' contains similarities of all vectors to a given vector of 'x'
i = []
ii = []
iii = []
iv = []
v = []
for ind, element in enumerate(row):
if depvar_vals[ind] == 1 and gp != ind:
i.append(element)
elif depvar_vals[ind] == 2 and gp != ind:
ii.append(element)
elif depvar_vals[ind] == 3 and gp != ind:
iii.append(element)
elif depvar_vals[ind] == 4 and gp != ind:
iv.append(element)
elif depvar_vals[ind] == 5 and gp != ind:
v.append(element)
elif gp == ind:
pass
else:
print('something went wrong with prediction calculations')
i = sorted(i, reverse=True)[:cutoff]
i = [xx for xx in i if xx > 0]
ii = sorted(ii, reverse=True)[:cutoff]
ii = [xx for xx in ii if xx > 0]
iii = sorted(iii, reverse=True)[:cutoff]
iii = [xx for xx in iii if xx > 0]
iv = sorted(iv, reverse=True)[:cutoff]
iv = [xx for xx in iv if xx > 0]
v = sorted(v, reverse=True)[:cutoff]
v = [xx for xx in v if xx > 0]
overall = i[:]
overall.extend(ii)
overall.extend(iii)
overall.extend(iv)
overall.extend(v)
overall = sum(overall)
tmp = []
for xx in [i,ii,iii,iv,v]:
prob = sum(xx) / overall
tmp.append(prob)
probs[train_summary[gp][0]] = tmp
#populate the train object to return the predictions and actual values
train = []
for i, xx in enumerate(probs):
tmp = [str(int(xx))]
tmp.extend(probs[xx])
tmp.extend([probs[xx].index(max(probs[xx]))+1 , int(train_summary[i][1])])
train.append(tmp)
train = pd.DataFrame(train, columns=['gp_id', "prob1", "prob2", "prob3", "prob4", "prob5", "yhat_train", "y_train"])
# predict values for the test period (last 60 days of feedback) by looking at the similarity between pairs of signatures
# do it the bayesian way (select the highest probability result, and then calculate test MSE error)
test_summary = test_summary.values.tolist()
x = []
# test_vals = []
for i,v in enumerate(test_summary):
if i < 10000000000: # sample size can be modified for exercise
x.append(v[2:])
# test_vals.append(int(v[1])) #depvar values get rounded to the nearest integer
r = cosine_similarity(np.array(x), np.array(y)) #computes cosine similarity score of every vector in 'y' to each vector in 'x'
# associate predictions with gp_ids and depvars rounded to nearest integer
probs = {}
for gp, row in enumerate(r): #every 'row' contains similarities of all vectors to a given vector of 'x'
i = []
ii = []
iii = []
iv = []
v = []
for ind, element in enumerate(row):
if depvar_vals[ind] == 1:
i.append(element)
elif depvar_vals[ind] == 2:
ii.append(element)
elif depvar_vals[ind] == 3:
iii.append(element)
elif depvar_vals[ind] == 4:
iv.append(element)
elif depvar_vals[ind] == 5:
v.append(element)
else:
print('something went wrong with prediction calculations')
i = sorted(i, reverse=True)[:cutoff]
i = [xx for xx in i if xx > 0]
ii = sorted(ii, reverse=True)[:cutoff]
ii = [xx for xx in ii if xx > 0]
iii = sorted(iii, reverse=True)[:cutoff]
iii = [xx for xx in iii if xx > 0]
iv = sorted(iv, reverse=True)[:cutoff]
iv = [xx for xx in iv if xx > 0]
v = sorted(v, reverse=True)[:cutoff]
v = [xx for xx in v if xx > 0]
overall = i[:]
overall.extend(ii)
overall.extend(iii)
overall.extend(iv)
overall.extend(v)
overall = sum(overall)
tmp = []
for xx in [i,ii,iii,iv,v]:
prob = sum(xx) / overall
tmp.append(prob)
probs[test_summary[gp][0]] = tmp
#populate the train object to return the predictions and actual values
test = []
for i, xx in enumerate(probs):
tmp = [str(int(xx))]
tmp.extend(probs[xx])
tmp.extend([probs[xx].index(max(probs[xx]))+1 , int(test_summary[i][1])])
test.append(tmp)
test = pd.DataFrame(test, columns=['gp_id', "prob1", "prob2", "prob3", "prob4", "prob5", "yhat_test", "y_test"])
results = [train,test]
return results
# clus_res = cluster_pred('r_output.csv',depvar='q9', cutoff = 2)
print('ok')
def clus_mse_calculator (chosen_cutoffs):
"""
Returns list of train and test MSE errors for predictions for a list of chosen 'cutoff' values.
The 'cutoff' parameter is used in cluster_pred() function
"""
clus_res_combo = []
for c in chosen_cutoffs:
print('compute results for cutoff=' +str(c))
clus_res = cluster_pred('r_output.csv',depvar='q9', cutoff = c)
train_mse = mean_squared_error(np.array(clus_res[0]['yhat_train']),np.array(clus_res[0]['y_train']))
test_mse = mean_squared_error(np.array(clus_res[1]['yhat_test']),np.array(clus_res[1]['y_test']))
clus_res_combo.append([c, train_mse, test_mse])
print([c, train_mse, test_mse])
print('all candidate models completed')
# clus_res_combo = clus_mse_calculator(list(range(1,31)))
# clus_res_combo1001to1030 = clus_mse_calculator(list(range(1001,1031)))
# clus_res_combo101to130 = clus_mse_calculator(list(range(101,131)))
for x in clus_res_combo:
print(x)
x = pd.DataFrame(t_results, columns = ['test_err','train_err'])
x['t_val'] = t_candidates
ns = [x[0] for x in clus_res_combo]
tr = [x[1] for x in clus_res_combo]
te = [x[2] for x in clus_res_combo]
print("Prediction errors in model training")
plt.plot(ns, tr, 'ro')
plt.xlabel('Number of top "n" models used to identify the most probable Likert-scale response')
plt.ylabel('average mean squared error')
plt.show()
print("Prediction errors in model testing")
plt.plot(ns, te, 'ro')
plt.xlabel('Number of top "n" models used to identify the most probable Likert-scale response')
plt.ylabel('average mean squared error')
plt.show()
# best clustering-based model
clus_res = cluster_pred('r_output.csv',depvar='q9', cutoff = 22)
mean_squared_error(np.array(clus_res[0]['yhat_train']),np.array(clus_res[0]['y_train']))
| 0.58522 | 0.604778 |
# Brain tumor 3D segmentation with MONAI
This tutorial shows how to construct a training workflow of multi-labels segmentation task.
And it contains below features:
1. Transforms for dictionary format data.
1. Define a new transform according to MONAI transform API.
1. Load Nifti image with metadata, load a list of images and stack them.
1. Randomly adjust intensity for data augmentation.
1. Cache IO and transforms to accelerate training and validation.
1. 3D UNet model, Dice loss function, Mean Dice metric for 3D segmentation task.
1. Deterministic training for reproducibility.
The dataset comes from http://medicaldecathlon.com/.
Target: Gliomas segmentation necrotic/active tumour and oedema
Modality: Multimodal multisite MRI data (FLAIR, T1w, T1gd,T2w)
Size: 750 4D volumes (484 Training + 266 Testing)
Source: BRATS 2016 and 2017 datasets.
Challenge: Complex and heterogeneously-located targets
Below figure shows image patches with the tumor sub-regions that are annotated in the different modalities (top left) and the final labels for the whole dataset (right).
(Figure taken from the [BraTS IEEE TMI paper](https://ieeexplore.ieee.org/document/6975210/))

The image patches show from left to right:
1. the whole tumor (yellow) visible in T2-FLAIR (Fig.A).
1. the tumor core (red) visible in T2 (Fig.B).
1. the enhancing tumor structures (light blue) visible in T1Gd, surrounding the cystic/necrotic components of the core (green) (Fig. C).
1. The segmentations are combined to generate the final labels of the tumor sub-regions (Fig.D): edema (yellow), non-enhancing solid core (red), necrotic/cystic core (green), enhancing core (blue).
[](https://colab.research.google.com/github/Project-MONAI/tutorials/blob/master/3d_segmentation/brats_segmentation_3d.ipynb)
## Setup environment
```
!python -c "import monai" || pip install -q "monai-weekly[nibabel, tqdm]"
!python -c "import matplotlib" || pip install -q matplotlib
%matplotlib inline
```
## Setup imports
```
# Copyright 2020 MONAI Consortium
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
# http://www.apache.org/licenses/LICENSE-2.0
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
import os
import shutil
import tempfile
import matplotlib.pyplot as plt
import numpy as np
from monai.apps import DecathlonDataset
from monai.config import print_config
from monai.data import DataLoader
from monai.losses import DiceLoss
from monai.metrics import DiceMetric
from monai.networks.nets import UNet
from monai.transforms import (
Activations,
AsChannelFirstd,
AsDiscrete,
CenterSpatialCropd,
Compose,
LoadImaged,
MapTransform,
NormalizeIntensityd,
Orientationd,
RandFlipd,
RandScaleIntensityd,
RandShiftIntensityd,
RandSpatialCropd,
Spacingd,
ToTensord,
)
from monai.utils import set_determinism
import torch
print_config()
```
## Setup data directory
You can specify a directory with the `MONAI_DATA_DIRECTORY` environment variable.
This allows you to save results and reuse downloads.
If not specified a temporary directory will be used.
```
directory = os.environ.get("MONAI_DATA_DIRECTORY")
root_dir = tempfile.mkdtemp() if directory is None else directory
print(root_dir)
```
## Set deterministic training for reproducibility
```
set_determinism(seed=0)
```
## Define a new transform to convert brain tumor labels
Here we convert the multi-classes labels into multi-labels segmentation task in One-Hot format.
```
class ConvertToMultiChannelBasedOnBratsClassesd(MapTransform):
"""
Convert labels to multi channels based on brats classes:
label 1 is the peritumoral edema
label 2 is the GD-enhancing tumor
label 3 is the necrotic and non-enhancing tumor core
The possible classes are TC (Tumor core), WT (Whole tumor)
and ET (Enhancing tumor).
"""
def __call__(self, data):
d = dict(data)
for key in self.keys:
result = []
# merge label 2 and label 3 to construct TC
result.append(np.logical_or(d[key] == 2, d[key] == 3))
# merge labels 1, 2 and 3 to construct WT
result.append(
np.logical_or(
np.logical_or(d[key] == 2, d[key] == 3), d[key] == 1
)
)
# label 2 is ET
result.append(d[key] == 2)
d[key] = np.stack(result, axis=0).astype(np.float32)
return d
```
## Setup transforms for training and validation
```
train_transform = Compose(
[
# load 4 Nifti images and stack them together
LoadImaged(keys=["image", "label"]),
AsChannelFirstd(keys="image"),
ConvertToMultiChannelBasedOnBratsClassesd(keys="label"),
Spacingd(
keys=["image", "label"],
pixdim=(1.5, 1.5, 2.0),
mode=("bilinear", "nearest"),
),
Orientationd(keys=["image", "label"], axcodes="RAS"),
RandSpatialCropd(
keys=["image", "label"], roi_size=[128, 128, 64], random_size=False
),
RandFlipd(keys=["image", "label"], prob=0.5, spatial_axis=0),
NormalizeIntensityd(keys="image", nonzero=True, channel_wise=True),
RandScaleIntensityd(keys="image", factors=0.1, prob=0.5),
RandShiftIntensityd(keys="image", offsets=0.1, prob=0.5),
ToTensord(keys=["image", "label"]),
]
)
val_transform = Compose(
[
LoadImaged(keys=["image", "label"]),
AsChannelFirstd(keys="image"),
ConvertToMultiChannelBasedOnBratsClassesd(keys="label"),
Spacingd(
keys=["image", "label"],
pixdim=(1.5, 1.5, 2.0),
mode=("bilinear", "nearest"),
),
Orientationd(keys=["image", "label"], axcodes="RAS"),
CenterSpatialCropd(keys=["image", "label"], roi_size=[128, 128, 64]),
NormalizeIntensityd(keys="image", nonzero=True, channel_wise=True),
ToTensord(keys=["image", "label"]),
]
)
```
## Quickly load data with DecathlonDataset
Here we use `DecathlonDataset` to automatically download and extract the dataset.
It inherits MONAI `CacheDataset`, so we set `cache_num=100` to cache 100 items for training and use the defaut args to cache all the items for validation.
```
train_ds = DecathlonDataset(
root_dir=root_dir,
task="Task01_BrainTumour",
transform=train_transform,
section="training",
download=True,
num_workers=4,
cache_num=100,
)
train_loader = DataLoader(train_ds, batch_size=2, shuffle=True, num_workers=4)
val_ds = DecathlonDataset(
root_dir=root_dir,
task="Task01_BrainTumour",
transform=val_transform,
section="validation",
download=False,
num_workers=4,
)
val_loader = DataLoader(val_ds, batch_size=2, shuffle=False, num_workers=4)
```
## Check data shape and visualize
```
# pick one image from DecathlonDataset to visualize and check the 4 channels
print(f"image shape: {val_ds[2]['image'].shape}")
plt.figure("image", (24, 6))
for i in range(4):
plt.subplot(1, 4, i + 1)
plt.title(f"image channel {i}")
plt.imshow(val_ds[2]["image"][i, :, :, 20].detach().cpu(), cmap="gray")
plt.show()
# also visualize the 3 channels label corresponding to this image
print(f"label shape: {val_ds[2]['label'].shape}")
plt.figure("label", (18, 6))
for i in range(3):
plt.subplot(1, 3, i + 1)
plt.title(f"label channel {i}")
plt.imshow(val_ds[2]["label"][i, :, :, 20].detach().cpu())
plt.show()
```
## Create Model, Loss, Optimizer
```
# standard PyTorch program style: create UNet, DiceLoss and Adam optimizer
device = torch.device("cuda:0")
model = UNet(
dimensions=3,
in_channels=4,
out_channels=3,
channels=(16, 32, 64, 128, 256),
strides=(2, 2, 2, 2),
num_res_units=2,
).to(device)
loss_function = DiceLoss(to_onehot_y=False, sigmoid=True, squared_pred=True)
optimizer = torch.optim.Adam(
model.parameters(), 1e-4, weight_decay=1e-5, amsgrad=True
)
```
## Execute a typical PyTorch training process
```
max_epochs = 180
val_interval = 2
best_metric = -1
best_metric_epoch = -1
epoch_loss_values = []
metric_values = []
metric_values_tc = []
metric_values_wt = []
metric_values_et = []
for epoch in range(max_epochs):
print("-" * 10)
print(f"epoch {epoch + 1}/{max_epochs}")
model.train()
epoch_loss = 0
step = 0
for batch_data in train_loader:
step += 1
inputs, labels = (
batch_data["image"].to(device),
batch_data["label"].to(device),
)
optimizer.zero_grad()
outputs = model(inputs)
loss = loss_function(outputs, labels)
loss.backward()
optimizer.step()
epoch_loss += loss.item()
print(
f"{step}/{len(train_ds) // train_loader.batch_size}"
f", train_loss: {loss.item():.4f}"
)
epoch_loss /= step
epoch_loss_values.append(epoch_loss)
print(f"epoch {epoch + 1} average loss: {epoch_loss:.4f}")
if (epoch + 1) % val_interval == 0:
model.eval()
with torch.no_grad():
dice_metric = DiceMetric(include_background=True, reduction="mean")
post_trans = Compose(
[Activations(sigmoid=True), AsDiscrete(threshold_values=True)]
)
metric_sum = metric_sum_tc = metric_sum_wt = metric_sum_et = 0.0
metric_count = (
metric_count_tc
) = metric_count_wt = metric_count_et = 0
for val_data in val_loader:
val_inputs, val_labels = (
val_data["image"].to(device),
val_data["label"].to(device),
)
val_outputs = model(val_inputs)
val_outputs = post_trans(val_outputs)
# compute overall mean dice
value, not_nans = dice_metric(y_pred=val_outputs, y=val_labels)
not_nans = not_nans.item()
metric_count += not_nans
metric_sum += value.item() * not_nans
# compute mean dice for TC
value_tc, not_nans = dice_metric(
y_pred=val_outputs[:, 0:1], y=val_labels[:, 0:1]
)
not_nans = not_nans.item()
metric_count_tc += not_nans
metric_sum_tc += value_tc.item() * not_nans
# compute mean dice for WT
value_wt, not_nans = dice_metric(
y_pred=val_outputs[:, 1:2], y=val_labels[:, 1:2]
)
not_nans = not_nans.item()
metric_count_wt += not_nans
metric_sum_wt += value_wt.item() * not_nans
# compute mean dice for ET
value_et, not_nans = dice_metric(
y_pred=val_outputs[:, 2:3], y=val_labels[:, 2:3]
)
not_nans = not_nans.item()
metric_count_et += not_nans
metric_sum_et += value_et.item() * not_nans
metric = metric_sum / metric_count
metric_values.append(metric)
metric_tc = metric_sum_tc / metric_count_tc
metric_values_tc.append(metric_tc)
metric_wt = metric_sum_wt / metric_count_wt
metric_values_wt.append(metric_wt)
metric_et = metric_sum_et / metric_count_et
metric_values_et.append(metric_et)
if metric > best_metric:
best_metric = metric
best_metric_epoch = epoch + 1
torch.save(
model.state_dict(),
os.path.join(root_dir, "best_metric_model.pth"),
)
print("saved new best metric model")
print(
f"current epoch: {epoch + 1} current mean dice: {metric:.4f}"
f" tc: {metric_tc:.4f} wt: {metric_wt:.4f} et: {metric_et:.4f}"
f"\nbest mean dice: {best_metric:.4f}"
f" at epoch: {best_metric_epoch}"
)
print(
f"train completed, best_metric: {best_metric:.4f}"
f" at epoch: {best_metric_epoch}"
)
```
## Plot the loss and metric
```
plt.figure("train", (12, 6))
plt.subplot(1, 2, 1)
plt.title("Epoch Average Loss")
x = [i + 1 for i in range(len(epoch_loss_values))]
y = epoch_loss_values
plt.xlabel("epoch")
plt.plot(x, y, color="red")
plt.subplot(1, 2, 2)
plt.title("Val Mean Dice")
x = [val_interval * (i + 1) for i in range(len(metric_values))]
y = metric_values
plt.xlabel("epoch")
plt.plot(x, y, color="green")
plt.show()
plt.figure("train", (18, 6))
plt.subplot(1, 3, 1)
plt.title("Val Mean Dice TC")
x = [val_interval * (i + 1) for i in range(len(metric_values_tc))]
y = metric_values_tc
plt.xlabel("epoch")
plt.plot(x, y, color="blue")
plt.subplot(1, 3, 2)
plt.title("Val Mean Dice WT")
x = [val_interval * (i + 1) for i in range(len(metric_values_wt))]
y = metric_values_wt
plt.xlabel("epoch")
plt.plot(x, y, color="brown")
plt.subplot(1, 3, 3)
plt.title("Val Mean Dice ET")
x = [val_interval * (i + 1) for i in range(len(metric_values_et))]
y = metric_values_et
plt.xlabel("epoch")
plt.plot(x, y, color="purple")
plt.show()
```
## Check best model output with the input image and label
```
model.load_state_dict(
torch.load(os.path.join(root_dir, "best_metric_model.pth"))
)
model.eval()
with torch.no_grad():
# select one image to evaluate and visualize the model output
val_input = val_ds[6]["image"].unsqueeze(0).to(device)
val_output = model(val_input)
plt.figure("image", (24, 6))
for i in range(4):
plt.subplot(1, 4, i + 1)
plt.title(f"image channel {i}")
plt.imshow(val_ds[6]["image"][i, :, :, 20].detach().cpu(), cmap="gray")
plt.show()
# visualize the 3 channels label corresponding to this image
plt.figure("label", (18, 6))
for i in range(3):
plt.subplot(1, 3, i + 1)
plt.title(f"label channel {i}")
plt.imshow(val_ds[6]["label"][i, :, :, 20].detach().cpu())
plt.show()
# visualize the 3 channels model output corresponding to this image
plt.figure("output", (18, 6))
for i in range(3):
plt.subplot(1, 3, i + 1)
plt.title(f"output channel {i}")
plt.imshow(val_output[0, i, :, :, 20].detach().cpu())
plt.show()
```
## Cleanup data directory
Remove directory if a temporary was used.
```
if directory is None:
shutil.rmtree(root_dir)
```
|
github_jupyter
|
!python -c "import monai" || pip install -q "monai-weekly[nibabel, tqdm]"
!python -c "import matplotlib" || pip install -q matplotlib
%matplotlib inline
# Copyright 2020 MONAI Consortium
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
# http://www.apache.org/licenses/LICENSE-2.0
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
import os
import shutil
import tempfile
import matplotlib.pyplot as plt
import numpy as np
from monai.apps import DecathlonDataset
from monai.config import print_config
from monai.data import DataLoader
from monai.losses import DiceLoss
from monai.metrics import DiceMetric
from monai.networks.nets import UNet
from monai.transforms import (
Activations,
AsChannelFirstd,
AsDiscrete,
CenterSpatialCropd,
Compose,
LoadImaged,
MapTransform,
NormalizeIntensityd,
Orientationd,
RandFlipd,
RandScaleIntensityd,
RandShiftIntensityd,
RandSpatialCropd,
Spacingd,
ToTensord,
)
from monai.utils import set_determinism
import torch
print_config()
directory = os.environ.get("MONAI_DATA_DIRECTORY")
root_dir = tempfile.mkdtemp() if directory is None else directory
print(root_dir)
set_determinism(seed=0)
class ConvertToMultiChannelBasedOnBratsClassesd(MapTransform):
"""
Convert labels to multi channels based on brats classes:
label 1 is the peritumoral edema
label 2 is the GD-enhancing tumor
label 3 is the necrotic and non-enhancing tumor core
The possible classes are TC (Tumor core), WT (Whole tumor)
and ET (Enhancing tumor).
"""
def __call__(self, data):
d = dict(data)
for key in self.keys:
result = []
# merge label 2 and label 3 to construct TC
result.append(np.logical_or(d[key] == 2, d[key] == 3))
# merge labels 1, 2 and 3 to construct WT
result.append(
np.logical_or(
np.logical_or(d[key] == 2, d[key] == 3), d[key] == 1
)
)
# label 2 is ET
result.append(d[key] == 2)
d[key] = np.stack(result, axis=0).astype(np.float32)
return d
train_transform = Compose(
[
# load 4 Nifti images and stack them together
LoadImaged(keys=["image", "label"]),
AsChannelFirstd(keys="image"),
ConvertToMultiChannelBasedOnBratsClassesd(keys="label"),
Spacingd(
keys=["image", "label"],
pixdim=(1.5, 1.5, 2.0),
mode=("bilinear", "nearest"),
),
Orientationd(keys=["image", "label"], axcodes="RAS"),
RandSpatialCropd(
keys=["image", "label"], roi_size=[128, 128, 64], random_size=False
),
RandFlipd(keys=["image", "label"], prob=0.5, spatial_axis=0),
NormalizeIntensityd(keys="image", nonzero=True, channel_wise=True),
RandScaleIntensityd(keys="image", factors=0.1, prob=0.5),
RandShiftIntensityd(keys="image", offsets=0.1, prob=0.5),
ToTensord(keys=["image", "label"]),
]
)
val_transform = Compose(
[
LoadImaged(keys=["image", "label"]),
AsChannelFirstd(keys="image"),
ConvertToMultiChannelBasedOnBratsClassesd(keys="label"),
Spacingd(
keys=["image", "label"],
pixdim=(1.5, 1.5, 2.0),
mode=("bilinear", "nearest"),
),
Orientationd(keys=["image", "label"], axcodes="RAS"),
CenterSpatialCropd(keys=["image", "label"], roi_size=[128, 128, 64]),
NormalizeIntensityd(keys="image", nonzero=True, channel_wise=True),
ToTensord(keys=["image", "label"]),
]
)
train_ds = DecathlonDataset(
root_dir=root_dir,
task="Task01_BrainTumour",
transform=train_transform,
section="training",
download=True,
num_workers=4,
cache_num=100,
)
train_loader = DataLoader(train_ds, batch_size=2, shuffle=True, num_workers=4)
val_ds = DecathlonDataset(
root_dir=root_dir,
task="Task01_BrainTumour",
transform=val_transform,
section="validation",
download=False,
num_workers=4,
)
val_loader = DataLoader(val_ds, batch_size=2, shuffle=False, num_workers=4)
# pick one image from DecathlonDataset to visualize and check the 4 channels
print(f"image shape: {val_ds[2]['image'].shape}")
plt.figure("image", (24, 6))
for i in range(4):
plt.subplot(1, 4, i + 1)
plt.title(f"image channel {i}")
plt.imshow(val_ds[2]["image"][i, :, :, 20].detach().cpu(), cmap="gray")
plt.show()
# also visualize the 3 channels label corresponding to this image
print(f"label shape: {val_ds[2]['label'].shape}")
plt.figure("label", (18, 6))
for i in range(3):
plt.subplot(1, 3, i + 1)
plt.title(f"label channel {i}")
plt.imshow(val_ds[2]["label"][i, :, :, 20].detach().cpu())
plt.show()
# standard PyTorch program style: create UNet, DiceLoss and Adam optimizer
device = torch.device("cuda:0")
model = UNet(
dimensions=3,
in_channels=4,
out_channels=3,
channels=(16, 32, 64, 128, 256),
strides=(2, 2, 2, 2),
num_res_units=2,
).to(device)
loss_function = DiceLoss(to_onehot_y=False, sigmoid=True, squared_pred=True)
optimizer = torch.optim.Adam(
model.parameters(), 1e-4, weight_decay=1e-5, amsgrad=True
)
max_epochs = 180
val_interval = 2
best_metric = -1
best_metric_epoch = -1
epoch_loss_values = []
metric_values = []
metric_values_tc = []
metric_values_wt = []
metric_values_et = []
for epoch in range(max_epochs):
print("-" * 10)
print(f"epoch {epoch + 1}/{max_epochs}")
model.train()
epoch_loss = 0
step = 0
for batch_data in train_loader:
step += 1
inputs, labels = (
batch_data["image"].to(device),
batch_data["label"].to(device),
)
optimizer.zero_grad()
outputs = model(inputs)
loss = loss_function(outputs, labels)
loss.backward()
optimizer.step()
epoch_loss += loss.item()
print(
f"{step}/{len(train_ds) // train_loader.batch_size}"
f", train_loss: {loss.item():.4f}"
)
epoch_loss /= step
epoch_loss_values.append(epoch_loss)
print(f"epoch {epoch + 1} average loss: {epoch_loss:.4f}")
if (epoch + 1) % val_interval == 0:
model.eval()
with torch.no_grad():
dice_metric = DiceMetric(include_background=True, reduction="mean")
post_trans = Compose(
[Activations(sigmoid=True), AsDiscrete(threshold_values=True)]
)
metric_sum = metric_sum_tc = metric_sum_wt = metric_sum_et = 0.0
metric_count = (
metric_count_tc
) = metric_count_wt = metric_count_et = 0
for val_data in val_loader:
val_inputs, val_labels = (
val_data["image"].to(device),
val_data["label"].to(device),
)
val_outputs = model(val_inputs)
val_outputs = post_trans(val_outputs)
# compute overall mean dice
value, not_nans = dice_metric(y_pred=val_outputs, y=val_labels)
not_nans = not_nans.item()
metric_count += not_nans
metric_sum += value.item() * not_nans
# compute mean dice for TC
value_tc, not_nans = dice_metric(
y_pred=val_outputs[:, 0:1], y=val_labels[:, 0:1]
)
not_nans = not_nans.item()
metric_count_tc += not_nans
metric_sum_tc += value_tc.item() * not_nans
# compute mean dice for WT
value_wt, not_nans = dice_metric(
y_pred=val_outputs[:, 1:2], y=val_labels[:, 1:2]
)
not_nans = not_nans.item()
metric_count_wt += not_nans
metric_sum_wt += value_wt.item() * not_nans
# compute mean dice for ET
value_et, not_nans = dice_metric(
y_pred=val_outputs[:, 2:3], y=val_labels[:, 2:3]
)
not_nans = not_nans.item()
metric_count_et += not_nans
metric_sum_et += value_et.item() * not_nans
metric = metric_sum / metric_count
metric_values.append(metric)
metric_tc = metric_sum_tc / metric_count_tc
metric_values_tc.append(metric_tc)
metric_wt = metric_sum_wt / metric_count_wt
metric_values_wt.append(metric_wt)
metric_et = metric_sum_et / metric_count_et
metric_values_et.append(metric_et)
if metric > best_metric:
best_metric = metric
best_metric_epoch = epoch + 1
torch.save(
model.state_dict(),
os.path.join(root_dir, "best_metric_model.pth"),
)
print("saved new best metric model")
print(
f"current epoch: {epoch + 1} current mean dice: {metric:.4f}"
f" tc: {metric_tc:.4f} wt: {metric_wt:.4f} et: {metric_et:.4f}"
f"\nbest mean dice: {best_metric:.4f}"
f" at epoch: {best_metric_epoch}"
)
print(
f"train completed, best_metric: {best_metric:.4f}"
f" at epoch: {best_metric_epoch}"
)
plt.figure("train", (12, 6))
plt.subplot(1, 2, 1)
plt.title("Epoch Average Loss")
x = [i + 1 for i in range(len(epoch_loss_values))]
y = epoch_loss_values
plt.xlabel("epoch")
plt.plot(x, y, color="red")
plt.subplot(1, 2, 2)
plt.title("Val Mean Dice")
x = [val_interval * (i + 1) for i in range(len(metric_values))]
y = metric_values
plt.xlabel("epoch")
plt.plot(x, y, color="green")
plt.show()
plt.figure("train", (18, 6))
plt.subplot(1, 3, 1)
plt.title("Val Mean Dice TC")
x = [val_interval * (i + 1) for i in range(len(metric_values_tc))]
y = metric_values_tc
plt.xlabel("epoch")
plt.plot(x, y, color="blue")
plt.subplot(1, 3, 2)
plt.title("Val Mean Dice WT")
x = [val_interval * (i + 1) for i in range(len(metric_values_wt))]
y = metric_values_wt
plt.xlabel("epoch")
plt.plot(x, y, color="brown")
plt.subplot(1, 3, 3)
plt.title("Val Mean Dice ET")
x = [val_interval * (i + 1) for i in range(len(metric_values_et))]
y = metric_values_et
plt.xlabel("epoch")
plt.plot(x, y, color="purple")
plt.show()
model.load_state_dict(
torch.load(os.path.join(root_dir, "best_metric_model.pth"))
)
model.eval()
with torch.no_grad():
# select one image to evaluate and visualize the model output
val_input = val_ds[6]["image"].unsqueeze(0).to(device)
val_output = model(val_input)
plt.figure("image", (24, 6))
for i in range(4):
plt.subplot(1, 4, i + 1)
plt.title(f"image channel {i}")
plt.imshow(val_ds[6]["image"][i, :, :, 20].detach().cpu(), cmap="gray")
plt.show()
# visualize the 3 channels label corresponding to this image
plt.figure("label", (18, 6))
for i in range(3):
plt.subplot(1, 3, i + 1)
plt.title(f"label channel {i}")
plt.imshow(val_ds[6]["label"][i, :, :, 20].detach().cpu())
plt.show()
# visualize the 3 channels model output corresponding to this image
plt.figure("output", (18, 6))
for i in range(3):
plt.subplot(1, 3, i + 1)
plt.title(f"output channel {i}")
plt.imshow(val_output[0, i, :, :, 20].detach().cpu())
plt.show()
if directory is None:
shutil.rmtree(root_dir)
| 0.748904 | 0.985029 |
```
%matplotlib inline
%reload_ext autoreload
%autoreload 2
import os
import csv
import gzip
import numpy as np
import urllib.request
from scipy.misc import imsave
path = 'datasets/MNIST/'
# loading MNIST data
def get_data_dict(output_path):
"""
Downloading and reading MNIST dataset
Returns dict of train and val images (here called t10k)
:param ouput_path: target directory
"""
# If not already there, downloading MNIST data:
files = ['train-images-idx3-ubyte.gz',
'train-labels-idx1-ubyte.gz',
't10k-images-idx3-ubyte.gz',
't10k-labels-idx1-ubyte.gz']
if not os.path.isdir(output_path):
print('creating MNIST directory')
os.mkdir(output_path)
if not len(os.listdir(output_path)) != 0:
for f in files:
URL = 'http://yann.lecun.com/exdb/mnist/'
if not os.path.isfile(output_path + f):
print(f)
print(f"Downloading MNIST {f} dataset")
fname, _ = urllib.request.urlretrieve(URL + f, output_path + f)
fnames.append(fname)
print('Done')
# Reading and loading data from directory
data = {'train': [[],[]], 't10k': [[],[]]}
for d in os.listdir(output_path):
with gzip.open(output_path + d, 'rb') as f:
ds = d.split('-')[0]
if 'labels' in d:
data[ds][1] = np.frombuffer(f.read(), np.uint8, offset = 8)
else:
data[ds][0] = np.frombuffer(f.read(), np.uint8, offset = 16).reshape(-1, 28, 28)
print('data loaded')
return data
data_dict = get_data_dict(path)
train_mean = ((data_dict['train'][0]) / 255).mean()
train_stdv = ((data_dict['train'][0]) / 255).std()
print(data_dict['train'][0].shape)
print(train_mean, train_stdv)
# Saving MNIST dataset as images and csv's
def save_as_csv_and_jpgs(data_dict, out_path):
"""
Saving images as .jpg and labels in .csv file
TODO: add tqdm to track progress
:param path: target directory
"""
for key in data_dict.keys():
full_path = out_path + key
if not os.path.isdir(full_path):
os.mkdir(full_path)
with open(full_path + "/labels.csv", 'w', newline='') as csvFile:
writer = csv.writer(csvFile, delimiter=',', quotechar='"')
for i in range(len(data_dict[key][0])):
imsave(full_path + '/' + str(i) + ".jpg", data_dict[key][0][i])
writer.writerow([str(i) + ".jpg", data_dict[key][1][i]])
save_as_csv_and_jpgs(data_dict, path)
```
|
github_jupyter
|
%matplotlib inline
%reload_ext autoreload
%autoreload 2
import os
import csv
import gzip
import numpy as np
import urllib.request
from scipy.misc import imsave
path = 'datasets/MNIST/'
# loading MNIST data
def get_data_dict(output_path):
"""
Downloading and reading MNIST dataset
Returns dict of train and val images (here called t10k)
:param ouput_path: target directory
"""
# If not already there, downloading MNIST data:
files = ['train-images-idx3-ubyte.gz',
'train-labels-idx1-ubyte.gz',
't10k-images-idx3-ubyte.gz',
't10k-labels-idx1-ubyte.gz']
if not os.path.isdir(output_path):
print('creating MNIST directory')
os.mkdir(output_path)
if not len(os.listdir(output_path)) != 0:
for f in files:
URL = 'http://yann.lecun.com/exdb/mnist/'
if not os.path.isfile(output_path + f):
print(f)
print(f"Downloading MNIST {f} dataset")
fname, _ = urllib.request.urlretrieve(URL + f, output_path + f)
fnames.append(fname)
print('Done')
# Reading and loading data from directory
data = {'train': [[],[]], 't10k': [[],[]]}
for d in os.listdir(output_path):
with gzip.open(output_path + d, 'rb') as f:
ds = d.split('-')[0]
if 'labels' in d:
data[ds][1] = np.frombuffer(f.read(), np.uint8, offset = 8)
else:
data[ds][0] = np.frombuffer(f.read(), np.uint8, offset = 16).reshape(-1, 28, 28)
print('data loaded')
return data
data_dict = get_data_dict(path)
train_mean = ((data_dict['train'][0]) / 255).mean()
train_stdv = ((data_dict['train'][0]) / 255).std()
print(data_dict['train'][0].shape)
print(train_mean, train_stdv)
# Saving MNIST dataset as images and csv's
def save_as_csv_and_jpgs(data_dict, out_path):
"""
Saving images as .jpg and labels in .csv file
TODO: add tqdm to track progress
:param path: target directory
"""
for key in data_dict.keys():
full_path = out_path + key
if not os.path.isdir(full_path):
os.mkdir(full_path)
with open(full_path + "/labels.csv", 'w', newline='') as csvFile:
writer = csv.writer(csvFile, delimiter=',', quotechar='"')
for i in range(len(data_dict[key][0])):
imsave(full_path + '/' + str(i) + ".jpg", data_dict[key][0][i])
writer.writerow([str(i) + ".jpg", data_dict[key][1][i]])
save_as_csv_and_jpgs(data_dict, path)
| 0.229881 | 0.306281 |
```
import re
from gensim import models, corpora
import nltk
from nltk import word_tokenize
from nltk.corpus import stopwords
from nltk.stem import WordNetLemmatizer
import spacy
import gensim
import pandas as pd
from gensim.models.coherencemodel import CoherenceModel
import pickle
#nltk.download('stopwords')
#nltk.download('punkt')
start_year = 1981
end_year = 2020
# Combine data from individual files into only giant list
data = []
for k in range(end_year - start_year):
try:
yearly_articles = (pd.read_csv(r'C:\Users\$ubhajit\Downloads\Technocolabs Project\ExchangeRateForecast\data\news\articles/articles_{}.csv'.format(start_year+k)))['article']
yearly_articles = [re.sub('\s+', ' ', str(sent)) for sent in yearly_articles]
yearly_articles = [re.sub("\'", "", str(sent)) for sent in yearly_articles]
for article in yearly_articles:
data.append(article)
except:
continue
NO_DOCUMENTS = len(data)
print(NO_DOCUMENTS)
NUM_TOPICS = 7
STOPWORDS = stopwords.words('english')
STOPWORDS.extend(['new', 'inc', 'like', 'one', 'two', 'inc.', 'nan'])
# ps = PorterStemmer()
lemm = WordNetLemmatizer()
nlp = spacy.load('en_core_web_sm', disable=['parser', 'ner'])
def lemmatization(texts, allowed_postags=['NOUN', 'ADJ', 'VERB', 'ADV']):
texts_out = []
for sent in texts:
doc = nlp(" ".join(sent))
texts_out.append([token.lemma_ for token in doc if token.pos_ in allowed_postags])
return texts_out
def clean_text(text):
tokenized_text = word_tokenize(text.lower())
cleaned_text = [t for t in tokenized_text if t not in STOPWORDS and re.match('[a-zA-Z\-][a-zA-Z\-]{2,}', t)]
return cleaned_text
tokenized_data = [clean_text(document) for document in data]
data_lemmatized = lemmatization(tokenized_data, allowed_postags=['NOUN', 'ADJ'])
# Build a Dictionary - association word to numeric id
dictionary = corpora.Dictionary(data_lemmatized)
dictionary.save(r"C:\Users\$ubhajit\Downloads\Technocolabs Project\ExchangeRateForecast\models\saved_model_data\lemmatized_dictionary.dic")
# Transform the collection of texts to a numerical form
corpus = [dictionary.doc2bow(text) for text in data_lemmatized]
corpora.MmCorpus.serialize(r"C:\Users\$ubhajit\Downloads\Technocolabs Project\ExchangeRateForecast\models\saved_model_data\serialised_corpus.mm", corpus)
# Build the LDA model
dictionary = corpora.Dictionary.load(r"C:\Users\$ubhajit\Downloads\Technocolabs Project\ExchangeRateForecast\models\saved_model_data\lemmatized_dictionary.dic")
mm = corpora.MmCorpus(r"C:\Users\$ubhajit\Downloads\Technocolabs Project\ExchangeRateForecast\models\saved_model_data\serialised_corpus.mm")
lda_model = models.ldamulticore.LdaMulticore(corpus=mm, random_state=100, num_topics=NUM_TOPICS, id2word=dictionary, workers=2)
lda_model.save(r'C:\Users\$ubhajit\Downloads\Technocolabs Project\ExchangeRateForecast\models\saved_model_data/lda_{}.model'.format(NUM_TOPICS))
lda_model = models.LdaModel.load(r'C:\Users\$ubhajit\Downloads\Technocolabs Project\ExchangeRateForecast\models\saved_model_data/lda_{}.model'.format(NUM_TOPICS))
def doc_topics(mapping, num_topics):
doc_topic_mapping = []
for index, doc in enumerate(mapping):
obj = {}
for i in range(num_topics):
obj['news_topic#{}'.format(i)] = 0
for topic in doc:
obj['news_topic#{}'.format(topic[0])] = 1
doc_topic_mapping.append(obj)
return pd.DataFrame(doc_topic_mapping)
document_topics = doc_topics([lda_model.get_document_topics(item) for item in mm], NUM_TOPICS)
documents = pd.DataFrame()
for year in range(end_year - start_year):
filename = r'C:\Users\$ubhajit\Downloads\Technocolabs Project\ExchangeRateForecast\data\news\articles/articles_{}.csv'.format(start_year+year)
if documents.empty:
documents = pd.read_csv(filename)
else:
documents = pd.concat([documents, pd.read_csv(filename)], sort=False)
documents = documents.reset_index(drop=True)
combined_data = pd.concat([documents,document_topics], axis=1, sort=False).reset_index(drop=True)
combined_data.to_csv('documents_to_topics_{}.csv'.format(NUM_TOPICS), index = False)
print("LDA Model:")
for idx in range(NUM_TOPICS):
# Print the first 10 most representative topics
print("Topic #%s:" % idx, re.sub('[\"\+\s\d\.*]+', ' ', lda_model.print_topic(idx, 40)))
print("- " * 20)
```
|
github_jupyter
|
import re
from gensim import models, corpora
import nltk
from nltk import word_tokenize
from nltk.corpus import stopwords
from nltk.stem import WordNetLemmatizer
import spacy
import gensim
import pandas as pd
from gensim.models.coherencemodel import CoherenceModel
import pickle
#nltk.download('stopwords')
#nltk.download('punkt')
start_year = 1981
end_year = 2020
# Combine data from individual files into only giant list
data = []
for k in range(end_year - start_year):
try:
yearly_articles = (pd.read_csv(r'C:\Users\$ubhajit\Downloads\Technocolabs Project\ExchangeRateForecast\data\news\articles/articles_{}.csv'.format(start_year+k)))['article']
yearly_articles = [re.sub('\s+', ' ', str(sent)) for sent in yearly_articles]
yearly_articles = [re.sub("\'", "", str(sent)) for sent in yearly_articles]
for article in yearly_articles:
data.append(article)
except:
continue
NO_DOCUMENTS = len(data)
print(NO_DOCUMENTS)
NUM_TOPICS = 7
STOPWORDS = stopwords.words('english')
STOPWORDS.extend(['new', 'inc', 'like', 'one', 'two', 'inc.', 'nan'])
# ps = PorterStemmer()
lemm = WordNetLemmatizer()
nlp = spacy.load('en_core_web_sm', disable=['parser', 'ner'])
def lemmatization(texts, allowed_postags=['NOUN', 'ADJ', 'VERB', 'ADV']):
texts_out = []
for sent in texts:
doc = nlp(" ".join(sent))
texts_out.append([token.lemma_ for token in doc if token.pos_ in allowed_postags])
return texts_out
def clean_text(text):
tokenized_text = word_tokenize(text.lower())
cleaned_text = [t for t in tokenized_text if t not in STOPWORDS and re.match('[a-zA-Z\-][a-zA-Z\-]{2,}', t)]
return cleaned_text
tokenized_data = [clean_text(document) for document in data]
data_lemmatized = lemmatization(tokenized_data, allowed_postags=['NOUN', 'ADJ'])
# Build a Dictionary - association word to numeric id
dictionary = corpora.Dictionary(data_lemmatized)
dictionary.save(r"C:\Users\$ubhajit\Downloads\Technocolabs Project\ExchangeRateForecast\models\saved_model_data\lemmatized_dictionary.dic")
# Transform the collection of texts to a numerical form
corpus = [dictionary.doc2bow(text) for text in data_lemmatized]
corpora.MmCorpus.serialize(r"C:\Users\$ubhajit\Downloads\Technocolabs Project\ExchangeRateForecast\models\saved_model_data\serialised_corpus.mm", corpus)
# Build the LDA model
dictionary = corpora.Dictionary.load(r"C:\Users\$ubhajit\Downloads\Technocolabs Project\ExchangeRateForecast\models\saved_model_data\lemmatized_dictionary.dic")
mm = corpora.MmCorpus(r"C:\Users\$ubhajit\Downloads\Technocolabs Project\ExchangeRateForecast\models\saved_model_data\serialised_corpus.mm")
lda_model = models.ldamulticore.LdaMulticore(corpus=mm, random_state=100, num_topics=NUM_TOPICS, id2word=dictionary, workers=2)
lda_model.save(r'C:\Users\$ubhajit\Downloads\Technocolabs Project\ExchangeRateForecast\models\saved_model_data/lda_{}.model'.format(NUM_TOPICS))
lda_model = models.LdaModel.load(r'C:\Users\$ubhajit\Downloads\Technocolabs Project\ExchangeRateForecast\models\saved_model_data/lda_{}.model'.format(NUM_TOPICS))
def doc_topics(mapping, num_topics):
doc_topic_mapping = []
for index, doc in enumerate(mapping):
obj = {}
for i in range(num_topics):
obj['news_topic#{}'.format(i)] = 0
for topic in doc:
obj['news_topic#{}'.format(topic[0])] = 1
doc_topic_mapping.append(obj)
return pd.DataFrame(doc_topic_mapping)
document_topics = doc_topics([lda_model.get_document_topics(item) for item in mm], NUM_TOPICS)
documents = pd.DataFrame()
for year in range(end_year - start_year):
filename = r'C:\Users\$ubhajit\Downloads\Technocolabs Project\ExchangeRateForecast\data\news\articles/articles_{}.csv'.format(start_year+year)
if documents.empty:
documents = pd.read_csv(filename)
else:
documents = pd.concat([documents, pd.read_csv(filename)], sort=False)
documents = documents.reset_index(drop=True)
combined_data = pd.concat([documents,document_topics], axis=1, sort=False).reset_index(drop=True)
combined_data.to_csv('documents_to_topics_{}.csv'.format(NUM_TOPICS), index = False)
print("LDA Model:")
for idx in range(NUM_TOPICS):
# Print the first 10 most representative topics
print("Topic #%s:" % idx, re.sub('[\"\+\s\d\.*]+', ' ', lda_model.print_topic(idx, 40)))
print("- " * 20)
| 0.247169 | 0.156008 |
## A quick introduction to HDF5 files for seismic
We will read a preformed dataset from a NumPy file, then save the dataset as an HDF5 file.
This notebook follows on from `Intro_to_seismic.ipynb`.
## What are HDF5 files?
'HDF' stands for _hierarchical data format_.
An HDF5 **File** can contain multiple **Group** and **Dataset** items.
- A **Group** is a bit like a file system directory, and a bit like a Python dictionary. Groups can be arbitrarily nested (hence the _H_ in HDF). They can contain Datasets, or other Groups. The **File** is, for all intents and purposes, a root-level group.
- A **Dataset** is a lot like a NumPy array. It's an n-dimensional (hyper-)rectangular data object, containing elements of homogenous type.
Both Groups and Datasets can have **Attributes**: a dictionary-like `attrs` object attached to them, which holds metadata.
HDF5 files support compression, error detection, metadata, and other useful things. They also support chunking, which can dramatically speed up data access on large files ([more about this](http://geology.beer/2015/02/10/hdf-for-large-arrays/)).
```
import numpy as np
%matplotlib inline
import matplotlib.pyplot as plt
volume = np.load('../data/F3_volume_3x3_16bit.npy')
volume.shape
```
## Saving to HDF5
We'll be using the HDF5 library, via [the `h5py` Python library](http://docs.h5py.org/en/stable/index.html).
If you need to, you can install `h5py` from the Notebook with: `!conda install h5py`.
```
import h5py
```
The `h5py.File` object is a context manager, so we will use it that way. Let's write our seismic volume to an HDF5 file:
```
with h5py.File('../data/f3_seismic.hdf5', 'w') as f:
dset = f.create_dataset("volume", data=volume)
```
That has saved a file:
```
%ls -l ../data/f3_seismic.hdf5
```
This is a bit bigger than the array:
```
volume.nbytes
```
So there's a little bit more overhead than a NumPy binary file:
```
%ls -l ../data/F3_volume_3x3_16bit.npy
```
We can compress the dataset (there's not a lot you can do on this data, but it does squeeze down a bit) — this reduces the file size, but takes a little time.
```
with h5py.File('../data/f3_seismic.hdf5', 'w') as f:
dset = f.create_dataset("volume", data=volume, compression='gzip')
%ls -l ../data/f3_seismic.hdf5
```
Let's add a bit of metadata:
```
with h5py.File('../data/f3_seismic.hdf5', 'w') as f:
grp = f.create_group('amplitude_group')
dset = f.create_dataset("amplitude_group/timeseries_dataset", data=volume)
# Let's add some attributes (metadata) to the root-level File object...
f.attrs['survey'] = 'F3'
f.attrs['location'] = 'Netherlands'
f.attrs['owner'] = 'NAM/NLOG/TNO/dGB'
f.attrs['licence'] = 'CC-BY-SA'
# ...and to the group...
grp.attrs['kind'] = 'Raw seismic amplitude, no units'
# ...and to the dataset.
dset.attrs['domain'] = 'time'
dset.attrs['dt'] = '0.004'
```
## Reading HDF5
You will need to poke around a bt to figure out what the paths and datasets are. At first, it's best just to read the metadata, unless you know what you're expecting to find.
```
with h5py.File('../data/f3_seismic.hdf5', 'r') as f:
print([k for k in f.keys()])
print([k for k in f.attrs])
with h5py.File('../data/f3_seismic.hdf5', 'r') as f:
print([k for k in f['amplitude_group'].keys()])
print([k for k in f['amplitude_group'].attrs])
```
Once you've figured out what you want, you can read the data:
```
with h5py.File('../data/f3_seismic.hdf5', 'r') as f:
data = f['amplitude_group/timeseries_dataset'][:]
data.shape
```
But we can also read slices from the volume without loading it all into memory:
```
with h5py.File('../data/f3_seismic.hdf5', 'r') as f:
data = f['amplitude_group/timeseries_dataset'][..., 200]
plt.imshow(data)
```
## Adding a new dataset to an HDF5 file
Let's add a frequency cube with dimensions inlines, crosslines, frequency.
```
import scipy.signal
freqs, Pxx_den = scipy.signal.welch(volume, fs=250)
plt.imshow(Pxx_den[:, :, 20])
Pxx_den.shape
with h5py.File('../data/f3_seismic.hdf5', 'r+') as f:
dset = f.create_dataset("amplitude_group/frequency_dataset", data=Pxx_den)
# This time we only want to add metadata to the dataset.
dset.attrs['domain'] = 'frequency'
dset.attrs['df'] = str(125 / 128)
```
The HDF5 file has grown commensurately:
```
%ls -l ../data/f3_seismic.hdf5
with h5py.File('../data/f3_seismic.hdf5', 'r') as f:
print([k for k in f['amplitude_group'].keys()])
print([k for k in f['amplitude_group/frequency_dataset'].attrs])
with h5py.File('../data/f3_seismic.hdf5', 'r') as f:
print('domain =', f['amplitude_group/frequency_dataset'].attrs['domain'])
print('df =', f['amplitude_group/frequency_dataset'].attrs['df'])
with h5py.File('../data/f3_seismic.hdf5', 'r') as f:
print(np.mean(f['amplitude_group/frequency_dataset']))
```
## Modifying an existing dataset
You can't point a name at a new dataset, but you can change the values inside a dataset — as long as it doen't change shape.
```
with h5py.File('../data/f3_seismic.hdf5', 'r+') as f:
data = f["amplitude_group/frequency_dataset"]
data[...] = np.sqrt(Pxx_den)
with h5py.File('../data/f3_seismic.hdf5', 'r') as f:
print(np.mean(f['amplitude_group/frequency_dataset']))
```
If it changes shape, you'll have to delete it and add it again:
```
data = np.mean(Pxx_den, axis=1)
data.shape
with h5py.File('../data/f3_seismic.hdf5', 'r+') as f:
del(f["amplitude_group/frequency_dataset"])
dset = f.create_dataset("amplitude_group/frequency_dataset", data=data)
# This time we only want to add metadata to the dataset.
dset.attrs['domain'] = 'frequency'
dset.attrs['df'] = str(125 / 128)
with h5py.File('../data/f3_seismic.hdf5', 'r') as f:
print(f['amplitude_group/frequency_dataset'].shape)
```
<hr />
<div>
<img src="https://avatars1.githubusercontent.com/u/1692321?s=50"><p style="text-align:center">© Agile Geoscience 2018</p>
</div>
|
github_jupyter
|
import numpy as np
%matplotlib inline
import matplotlib.pyplot as plt
volume = np.load('../data/F3_volume_3x3_16bit.npy')
volume.shape
import h5py
with h5py.File('../data/f3_seismic.hdf5', 'w') as f:
dset = f.create_dataset("volume", data=volume)
%ls -l ../data/f3_seismic.hdf5
volume.nbytes
%ls -l ../data/F3_volume_3x3_16bit.npy
with h5py.File('../data/f3_seismic.hdf5', 'w') as f:
dset = f.create_dataset("volume", data=volume, compression='gzip')
%ls -l ../data/f3_seismic.hdf5
with h5py.File('../data/f3_seismic.hdf5', 'w') as f:
grp = f.create_group('amplitude_group')
dset = f.create_dataset("amplitude_group/timeseries_dataset", data=volume)
# Let's add some attributes (metadata) to the root-level File object...
f.attrs['survey'] = 'F3'
f.attrs['location'] = 'Netherlands'
f.attrs['owner'] = 'NAM/NLOG/TNO/dGB'
f.attrs['licence'] = 'CC-BY-SA'
# ...and to the group...
grp.attrs['kind'] = 'Raw seismic amplitude, no units'
# ...and to the dataset.
dset.attrs['domain'] = 'time'
dset.attrs['dt'] = '0.004'
with h5py.File('../data/f3_seismic.hdf5', 'r') as f:
print([k for k in f.keys()])
print([k for k in f.attrs])
with h5py.File('../data/f3_seismic.hdf5', 'r') as f:
print([k for k in f['amplitude_group'].keys()])
print([k for k in f['amplitude_group'].attrs])
with h5py.File('../data/f3_seismic.hdf5', 'r') as f:
data = f['amplitude_group/timeseries_dataset'][:]
data.shape
with h5py.File('../data/f3_seismic.hdf5', 'r') as f:
data = f['amplitude_group/timeseries_dataset'][..., 200]
plt.imshow(data)
import scipy.signal
freqs, Pxx_den = scipy.signal.welch(volume, fs=250)
plt.imshow(Pxx_den[:, :, 20])
Pxx_den.shape
with h5py.File('../data/f3_seismic.hdf5', 'r+') as f:
dset = f.create_dataset("amplitude_group/frequency_dataset", data=Pxx_den)
# This time we only want to add metadata to the dataset.
dset.attrs['domain'] = 'frequency'
dset.attrs['df'] = str(125 / 128)
%ls -l ../data/f3_seismic.hdf5
with h5py.File('../data/f3_seismic.hdf5', 'r') as f:
print([k for k in f['amplitude_group'].keys()])
print([k for k in f['amplitude_group/frequency_dataset'].attrs])
with h5py.File('../data/f3_seismic.hdf5', 'r') as f:
print('domain =', f['amplitude_group/frequency_dataset'].attrs['domain'])
print('df =', f['amplitude_group/frequency_dataset'].attrs['df'])
with h5py.File('../data/f3_seismic.hdf5', 'r') as f:
print(np.mean(f['amplitude_group/frequency_dataset']))
with h5py.File('../data/f3_seismic.hdf5', 'r+') as f:
data = f["amplitude_group/frequency_dataset"]
data[...] = np.sqrt(Pxx_den)
with h5py.File('../data/f3_seismic.hdf5', 'r') as f:
print(np.mean(f['amplitude_group/frequency_dataset']))
data = np.mean(Pxx_den, axis=1)
data.shape
with h5py.File('../data/f3_seismic.hdf5', 'r+') as f:
del(f["amplitude_group/frequency_dataset"])
dset = f.create_dataset("amplitude_group/frequency_dataset", data=data)
# This time we only want to add metadata to the dataset.
dset.attrs['domain'] = 'frequency'
dset.attrs['df'] = str(125 / 128)
with h5py.File('../data/f3_seismic.hdf5', 'r') as f:
print(f['amplitude_group/frequency_dataset'].shape)
| 0.295636 | 0.937153 |
```
from fractions import Fraction
import numpy as np
def answer(m):
""" Calculates the probabilities of reaching the terminal states"""
# Get number of states.
no_states = len(m)
# Edge case 0: empty matrix.
if (no_states == 0):
print("Input matrix is empty")
return []
# Edge case 1: 1d matrix - Test 4 passed.
if (no_states == 1):
print("Input matrix is 1d")
return [1, 1] # 0th state is final state for sure;)
# Calculate tmp variable - sums of rows
row_sums = [sum(i) for i in m]
#print("row_sums=", row_sums)
# Get absorbing states.
absorbing_states = []
not_absorbing_states = []
# Warning - assuming that m is square matrix
transition_matrix = np.matrix(m, dtype=float)
print("transition_matrix=",transition_matrix)
for i in range(no_states):
# If there are no outputs.
if (row_sums[i] == 0):
absorbing_states.append(i)
transition_matrix[i,i] = 1
# Or all outputs lead to the same node (diagonal):
elif (row_sums[i] == transition_matrix[i,i]) :
absorbing_states.append(i)
transition_matrix[i,i] = 1
else:
not_absorbing_states.append(i)
transition_matrix[i,:] = [float(el) / row_sums[i] for el in m[i]]
print("absorbing states ({}) = {}".format(len(absorbing_states), absorbing_states))
print("not absorbing states ({}) = {}".format(len(not_absorbing_states), not_absorbing_states))
print("transition_matrix=",transition_matrix)
# Edge case 2: no terminal states (task states clearly that this cannot happen, but just in case...)
if (len(absorbing_states) == 0):
print("There are no absorbing states!")
return []
# The task clearly states that it is an absorbing Markov chain.
# Edge case 3: all states are terminal states - which means that there are no transitions!
# Edge case 1 is a special case of this edge case.
if (len(not_absorbing_states) == 0):
print("All states are absorbing!")
res = [1] # 0-th state is the one where we will always finish
for _ in range(len(not_absorbing_states)-1):
res.append(0)
res.append(1) # denominator
return res
# Change absorbing transition matrix into "standard form".
# Swap cols and rows using advanced indexing.
transition_matrix[:, :] = transition_matrix [: , absorbing_states + not_absorbing_states]
transition_matrix[:, :] = transition_matrix [absorbing_states + not_absorbing_states, :]
print("P =\n",transition_matrix)
# Get R submatrix - transitions from not absorbing to absorbing states.
R = transition_matrix[len(absorbing_states):, :len(absorbing_states)]
print("R =\n",R)
# Get Q submatrix - transitions from not absorbing to not absorbing states.
Q = transition_matrix[len(absorbing_states):, len(absorbing_states):]
print("Q =\n",Q)
# Calculate the fundamental matrix F.
F = (np.eye(len(not_absorbing_states)) - Q).I
print("F =\n",F)
# Finally, calculate the limiting matrix - we can skip that at all.
#P_limit = np.concatenate([np.concatenate( [np.eye(len(absorbing_states)),
# np.zeros(shape=(len(absorbing_states), len(not_absorbing_states)))], axis=1),
# np.concatenate( [F * R,
# np.zeros(shape=(len(not_absorbing_states), len(not_absorbing_states)))], axis=1)],
# axis =0)
#print("P limit =\n",P_limit)
# Only FxR part is interesting.
FxR_limit = F * R
print("FxR_limit =\n",FxR_limit)
# Get probabilities of starting from state 0 to final.
# As we already fixed the case of s0 being terminal, now we are sure that s0 is not terminal,
# thus it is related to the first vector of FxR part of limiting matrix.
absorbing_state_probabilities = FxR_limit[0,:].tolist()[0]
print("absorbing_state_probabilities =\n", absorbing_state_probabilities)
numerators = []
denominators = []
fractions = [ Fraction(prob).limit_denominator() for prob in absorbing_state_probabilities]
#print("Fractions: {}".format(fractions))
# Handle separatelly numerators and denominators.
for frac in fractions:
numerators.append(frac.numerator)
denominators.append(frac.denominator)
print("numerators: {}".format(numerators))
print("denominators: {}".format(denominators))
# Calculate factors
max_den = max(denominators)
factors = [max_den // den for den in denominators]
print("factors: {}".format(factors))
# Bring to common denominator.
final_numerators = [num * fac for num, fac in zip(numerators, factors)]
print("final_numerators: {}".format(final_numerators))
# Sanity check
if (sum(final_numerators) != max_den ):
print("Error! Numerators do not sum to denominator!")
# Format output
output = []
output = [int(el) for el in final_numerators]
output.append(max_den)
return output
if __name__ == "__main__":
ore_trans_mat = [
[0,1,0,0,0,1], # s0, the initial state, goes to s1 and s5 with equal probability
[4,0,0,3,2,0], # s1 can become s0, s3, or s4, but with different probabilities
[0,0,0,0,0,0], # s2 is terminal, and unreachable (never observed in practice)
[0,0,0,0,0,0], # s3 is terminalnumerators
[0,0,0,0,0,0], # s4 is terminal
[0,0,0,0,0,0], # s5 is terminal
]
#ore_trans_mat = [
# [1, 0, 0, 0],
# [0, 1, 0, 0],
# [0, 0, 1, 0],
# [0, 0, 0, 1]
#]
#ore_trans_mat = [
# [1000, 2000, 3000, 4000],
# [0, 1000, 0, 0],
# [0, 0, 10001, 0],
# [0, 0, 0, 16000]
#]
#ore_trans_mat = [[0, 2, 1, 0, 0], [0, 0, 0, 3, 4], [0, 0, 0, 0, 0], [0, 0, 0, 0, 0], [0, 0, 0, 0, 0]]
#ore_trans_mat = [[0, 1, 0, 0, 0, 1], [4, 0, 0, 3, 2, 0], [0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0]]
# Tricky cases!
#ore_trans_mat = [[], []]
#ore_trans_mat = [[0, 1, 0], [0, 1, 0], [0, 1, 0]]
#ore_trans_mat = [[0, 2, 3, 4]]
#ore_trans_mat = [[0, 2], [1], [0], [0, 0]]
#ore_trans_mat = [[1]]
#ore_trans_mat = [[0, 0], [0, 1]]
#ore_trans_mat = [[0,1,0,1], [1, 0, 0, 1], [0, 0, 0, 0], [0, 1, 1, 0]]
#ore_trans_mat
#ore_trans_mat = [[0, .3, .3, .4],
# [0, 0, 0, 0],
# [0, 0, 1, 0],
# [.8, .1, .1, 0]]
#ore_trans_mat = [[1, 0, 0, 0],
# [0, 1, 0, 0],
# [.1, 0, .8, .1],
# [.1, .1, .4, .4]]
print("ore_trans_mat=",ore_trans_mat)
print("answer =",answer(ore_trans_mat))
```
|
github_jupyter
|
from fractions import Fraction
import numpy as np
def answer(m):
""" Calculates the probabilities of reaching the terminal states"""
# Get number of states.
no_states = len(m)
# Edge case 0: empty matrix.
if (no_states == 0):
print("Input matrix is empty")
return []
# Edge case 1: 1d matrix - Test 4 passed.
if (no_states == 1):
print("Input matrix is 1d")
return [1, 1] # 0th state is final state for sure;)
# Calculate tmp variable - sums of rows
row_sums = [sum(i) for i in m]
#print("row_sums=", row_sums)
# Get absorbing states.
absorbing_states = []
not_absorbing_states = []
# Warning - assuming that m is square matrix
transition_matrix = np.matrix(m, dtype=float)
print("transition_matrix=",transition_matrix)
for i in range(no_states):
# If there are no outputs.
if (row_sums[i] == 0):
absorbing_states.append(i)
transition_matrix[i,i] = 1
# Or all outputs lead to the same node (diagonal):
elif (row_sums[i] == transition_matrix[i,i]) :
absorbing_states.append(i)
transition_matrix[i,i] = 1
else:
not_absorbing_states.append(i)
transition_matrix[i,:] = [float(el) / row_sums[i] for el in m[i]]
print("absorbing states ({}) = {}".format(len(absorbing_states), absorbing_states))
print("not absorbing states ({}) = {}".format(len(not_absorbing_states), not_absorbing_states))
print("transition_matrix=",transition_matrix)
# Edge case 2: no terminal states (task states clearly that this cannot happen, but just in case...)
if (len(absorbing_states) == 0):
print("There are no absorbing states!")
return []
# The task clearly states that it is an absorbing Markov chain.
# Edge case 3: all states are terminal states - which means that there are no transitions!
# Edge case 1 is a special case of this edge case.
if (len(not_absorbing_states) == 0):
print("All states are absorbing!")
res = [1] # 0-th state is the one where we will always finish
for _ in range(len(not_absorbing_states)-1):
res.append(0)
res.append(1) # denominator
return res
# Change absorbing transition matrix into "standard form".
# Swap cols and rows using advanced indexing.
transition_matrix[:, :] = transition_matrix [: , absorbing_states + not_absorbing_states]
transition_matrix[:, :] = transition_matrix [absorbing_states + not_absorbing_states, :]
print("P =\n",transition_matrix)
# Get R submatrix - transitions from not absorbing to absorbing states.
R = transition_matrix[len(absorbing_states):, :len(absorbing_states)]
print("R =\n",R)
# Get Q submatrix - transitions from not absorbing to not absorbing states.
Q = transition_matrix[len(absorbing_states):, len(absorbing_states):]
print("Q =\n",Q)
# Calculate the fundamental matrix F.
F = (np.eye(len(not_absorbing_states)) - Q).I
print("F =\n",F)
# Finally, calculate the limiting matrix - we can skip that at all.
#P_limit = np.concatenate([np.concatenate( [np.eye(len(absorbing_states)),
# np.zeros(shape=(len(absorbing_states), len(not_absorbing_states)))], axis=1),
# np.concatenate( [F * R,
# np.zeros(shape=(len(not_absorbing_states), len(not_absorbing_states)))], axis=1)],
# axis =0)
#print("P limit =\n",P_limit)
# Only FxR part is interesting.
FxR_limit = F * R
print("FxR_limit =\n",FxR_limit)
# Get probabilities of starting from state 0 to final.
# As we already fixed the case of s0 being terminal, now we are sure that s0 is not terminal,
# thus it is related to the first vector of FxR part of limiting matrix.
absorbing_state_probabilities = FxR_limit[0,:].tolist()[0]
print("absorbing_state_probabilities =\n", absorbing_state_probabilities)
numerators = []
denominators = []
fractions = [ Fraction(prob).limit_denominator() for prob in absorbing_state_probabilities]
#print("Fractions: {}".format(fractions))
# Handle separatelly numerators and denominators.
for frac in fractions:
numerators.append(frac.numerator)
denominators.append(frac.denominator)
print("numerators: {}".format(numerators))
print("denominators: {}".format(denominators))
# Calculate factors
max_den = max(denominators)
factors = [max_den // den for den in denominators]
print("factors: {}".format(factors))
# Bring to common denominator.
final_numerators = [num * fac for num, fac in zip(numerators, factors)]
print("final_numerators: {}".format(final_numerators))
# Sanity check
if (sum(final_numerators) != max_den ):
print("Error! Numerators do not sum to denominator!")
# Format output
output = []
output = [int(el) for el in final_numerators]
output.append(max_den)
return output
if __name__ == "__main__":
ore_trans_mat = [
[0,1,0,0,0,1], # s0, the initial state, goes to s1 and s5 with equal probability
[4,0,0,3,2,0], # s1 can become s0, s3, or s4, but with different probabilities
[0,0,0,0,0,0], # s2 is terminal, and unreachable (never observed in practice)
[0,0,0,0,0,0], # s3 is terminalnumerators
[0,0,0,0,0,0], # s4 is terminal
[0,0,0,0,0,0], # s5 is terminal
]
#ore_trans_mat = [
# [1, 0, 0, 0],
# [0, 1, 0, 0],
# [0, 0, 1, 0],
# [0, 0, 0, 1]
#]
#ore_trans_mat = [
# [1000, 2000, 3000, 4000],
# [0, 1000, 0, 0],
# [0, 0, 10001, 0],
# [0, 0, 0, 16000]
#]
#ore_trans_mat = [[0, 2, 1, 0, 0], [0, 0, 0, 3, 4], [0, 0, 0, 0, 0], [0, 0, 0, 0, 0], [0, 0, 0, 0, 0]]
#ore_trans_mat = [[0, 1, 0, 0, 0, 1], [4, 0, 0, 3, 2, 0], [0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0]]
# Tricky cases!
#ore_trans_mat = [[], []]
#ore_trans_mat = [[0, 1, 0], [0, 1, 0], [0, 1, 0]]
#ore_trans_mat = [[0, 2, 3, 4]]
#ore_trans_mat = [[0, 2], [1], [0], [0, 0]]
#ore_trans_mat = [[1]]
#ore_trans_mat = [[0, 0], [0, 1]]
#ore_trans_mat = [[0,1,0,1], [1, 0, 0, 1], [0, 0, 0, 0], [0, 1, 1, 0]]
#ore_trans_mat
#ore_trans_mat = [[0, .3, .3, .4],
# [0, 0, 0, 0],
# [0, 0, 1, 0],
# [.8, .1, .1, 0]]
#ore_trans_mat = [[1, 0, 0, 0],
# [0, 1, 0, 0],
# [.1, 0, .8, .1],
# [.1, .1, .4, .4]]
print("ore_trans_mat=",ore_trans_mat)
print("answer =",answer(ore_trans_mat))
| 0.483161 | 0.734691 |
## Quantum Computing - Exploration
```
import numpy as np
from qiskit import *
```
### First, create a quantum circuit with 3 qubits.
```
circ = QuantumCircuit(3)
```
This circuit provides the basis for the quantum calculation's we'll do later. Essentially, this is the statevector. Let's perform some gate operations on this circuit to create a GHZ statevector.
```
circ.h(0)
circ.cx(0,1)
circ.cx(0,2)
```
Awesome! Now let's visualize this state circuit.
```
circ.draw()
```
Now that we have a quantum circuit set up, let's use a package to simulate it. This package is Qiskit Aer. To start the simulation, we need to choose a backend (section of Aer we want to use).
```
backend = Aer.get_backend('statevector_simulator')
```
Below I go through the commands to get the quantum state from a given circuit.
1. execute a circuit on a backend simulation, which returns a job object
2. take the result of that given job object and pul the statevector from that result.
This returns a statevector of dimension $2^n$, where n is the number of qubits.
```
job = execute(circ, backend)
result = job.result()
outputstate = result.get_statevector(circ,decimals=3)
print(outputstate)
```
Let's visualize this state.
```
from qiskit.visualization import plot_state_city
plot_state_city(outputstate)
```
Interpreting this output can be pretty difficult. These are the real and imaginary state density matrices, from which the probability of any measurment can be asertained.
### Next up: Unitary Simulations
Aer also has a unitary simulator for use in calculations where all the elements in the circuit are unitary operations. The unitary simulator returns a $2^n x 2^n$ matrix.
```
backend = Aer.get_backend('unitary_simulator')
job = execute(circ, backend)
result = job.result()
print(result.get_unitary(circ,decimals=3))
plot_state_city(result.get_unitary(circ,decimals=3))
```
### Measurment Simulations
Yet another Aer backend is available to simulate actual measurments, not simply ideal/theoretical information on the state that the previous backends gave us.
```
meas = QuantumCircuit(3,3)
meas.barrier(range(3))
#This next line maps the quantum measurment to classical bits
meas.measure(range(3),range(3))
qc = circ+meas
qc.draw()
```
To simulate the circuit, we use the qasm simulator in Aer. Each run of this GHZ circuit will yield either 000 or 111. We can use the execute command to build up statistics on the distribution of these results.
```
backend_sim = Aer.get_backend('qasm_simulator')
job_sim = execute(qc,backend_sim,shots=1024)
result_sim = job_sim.result()
counts = result_sim.get_counts(qc)
print(counts)
from qiskit.visualization import plot_histogram
plot_histogram(counts)
```
### IBM Q
Qiskit is supported by IBM, and we can use IBM's public quantum computers and API to test our code!
```
from qiskit import IBMQ
IBMQ.save_account('caa130bd16fc8c10d09c1e18bf99e63d1285cbe215c8b82ed6c30cc5997077238a2908271e30cc407577d4d34bd400fbabe66a90c959be45f8269f891cc645c9', overwrite = True)
IBMQ.load_account()
IBMQ.providers()
provider = IBMQ.get_provider(group='open')
provider.backends()
backend = provider.get_backend('ibmqx2')
from qiskit.tools.monitor import job_monitor
job_exp = execute(qc,backend=backend)
job_monitor(job_exp)
result_exp = job_exp.result()
counts_exp = result_exp.get_counts(qc)
plot_histogram([counts_exp,counts], legend=['Device','Simulator'])
```
## Part 4 - Characteristics of Quantum Circuits
```
#create a 12 qubit circuit
qc = QuantumCircuit(12)
#run a hadamard gate on each of the first five, and put each of the results of these into a bell state (through a c-not gate) with the subsequent 5 qubits.
for idx in range(5):
qc.h(idx)
qc.cx(idx, idx+5)
#q1 is also in a bell state to q7
qc.cx(1, 7)
#simply NOT q8
qc.x(8)
#etc
qc.cx(1, 9)
qc.x(7)
qc.cx(1, 11)
qc.swap(6, 11)
qc.swap(6, 9)
qc.swap(6, 10)
qc.x(6)
qc.draw()
#we can get the number of gates or operations performed on the qubits by using:
print(qc.count_ops())
#or, for total:
print(qc.size())
```
The performance of a quantum circuit can roughly trace back to the number of layers it has, or its "depth". This is generally the number of operations the computer needs to perform in sequence. This doesn't add to the count any operations that can be performed in parallel.
```
qc.depth()
```
We can also get the maximum number of individual circuits this calculation could be broken down to. This is especially useful for simplifying complicated circuits.
```
qc.num_unitary_factors()
```
Let's take the circuit above and add classical registers!
```
qc2 = QuantumCircuit(12, 12)
for idx in range(5):
qc2.h(idx)
qc2.cx(idx, idx+5)
qc2.cx(1, 7)
qc2.x(8)
qc2.cx(1, 9)
qc2.x(7)
qc2.cx(1, 11)
qc2.swap(6, 11)
qc2.swap(6, 9)
qc2.swap(6, 10)
qc2.x(6)
qc2.barrier()
qc2.measure(range(12), range(12))
qc2.draw()
```
It's also useful to see that some gates, such as the SWAP gate, is not actually an atomic operation. This can be seen using the decompose function:
```
qc3 = QuantumCircuit(2)
qc3.swap(0,1)
qc3.decompose().draw() # This decomposes the swap gate into the gates we want
```
## Part 5 - Let's introduce the Transpiler
```
from qiskit.visualization import plot_gate_map, plot_circuit_layout
import matplotlib.pyplot as plt
```
Consider this 5 qubit GHZ state circuit:
```
ghz = QuantumCircuit(5, 5)
ghz.h(0)
for idx in range(1,5):
ghz.cx(0,idx)
ghz.barrier(range(5))
ghz.measure(range(5), range(5))
ghz.draw()
```
Before we go further into learning how transpile() affects this circuit, let's introduce a way to understand some of the obscure quantum computing functions.
```
import inspect
#This shows all the default parameters for the transpile function.
inspect.signature(transpile)
inspect.signature(QuantumCircuit)
qc = QuantumCircuit(2, 1)
qc.h(0)
qc.x(1)
qc.cu1(np.pi/4, 0, 1)
qc.h(0)
qc.measure([0], [0])
qc.draw()
```
In the above circuit, we see we have an H, X, and U1 gate. None of these are basis gates. Let's decompose the circuit:
```
qc_basis = qc.decompose()
qc_basis.draw()
```
Let's look at another example: the Tofolli gate, a controlled-controlled-not gate. This is a 3 qubit gate!
```
ccx_circ = QuantumCircuit(3)
ccx_circ.ccx(0, 1, 2)
ccx_circ.draw()
ccx_circ.decompose().draw()
```
Wow! This is a big decomposition.
We also want to be able to map the circuit, which is entirely virtual, to physical qubits on specific providers. This is done with the plot_gate_map tool
```
backend = provider.get_backend('ibmq_16_melbourne')
plot_gate_map(backend, plot_directed=True)
backend = provider.get_backend('ibmq_16_melbourne')
new_circ_lv0 = transpile(ghz, backend=backend, optimization_level=0)
plot_circuit_layout(new_circ_lv0, backend)
backend = provider.get_backend('ibmq_16_melbourne')
new_circ_lv3 = transpile(ghz, backend=backend, optimization_level=3)
print('Depth:', new_circ_lv3.depth())
plot_circuit_layout(new_circ_lv3, backend)
```
With these three layouts, we can choose between them and execute one. This step of transpiling is typically done within the execute function.
```
from qiskit.tools.monitor import job_monitor
job1 = execute(new_circ_lv3, backend)
job_monitor(job1)
```
From the initial chart, we can choose our own initial mapping if we want. Here's two examples of using this level of control:
```
# Virtual -> physical
# 0 -> 11
# 1 -> 12
# 2 -> 10
# 3 -> 2
# 4 -> 4
good_ghz = transpile(ghz, backend, initial_layout=[11,12,10,2,4])
print('Depth:', good_ghz.depth())
plot_circuit_layout(good_ghz, backend)
job2 = execute(good_ghz, backend)
job_monitor(job2)
# Virtual -> physical
# 0 -> 0
# 1 -> 6
# 2 -> 10
# 3 -> 13
# 4 -> 7
bad_ghz = transpile(ghz, backend, initial_layout=[0,6,10,13,7])
print('Depth:', bad_ghz.depth())
plot_circuit_layout(bad_ghz, backend)
job3 = execute(bad_ghz, backend)
job_monitor(job3)
counts1 = job1.result().get_counts()
counts2 = job2.result().get_counts()
counts3 = job3.result().get_counts()
plot_histogram([counts1, counts2, counts3],
figsize=(15,6),
legend=['level3', 'good', 'bad'])
```
Different provider devices also change the topology of the circuit depending on the entanglement they support! This can be further explored in the transpiling tutorial; but I'm gunna move on.
## Intro to Qiskit Terra:
```
from math import pi
from qiskit import QuantumCircuit, ClassicalRegister, QuantumRegister, execute
from qiskit.tools.visualization import circuit_drawer
from qiskit.quantum_info import state_fidelity
from qiskit import BasicAer
backend = BasicAer.get_backend('unitary_simulator')
```
## Single Qubit Quantum states
A single qubit quantum state can be written as
$$\left|\psi\right\rangle = \alpha\left|0\right\rangle + \beta \left|1\right\rangle$$
where $\alpha$ and $\beta$ are complex numbers. In a measurement the probability of the bit being in $\left|0\right\rangle$ is $|\alpha|^2$ and $\left|1\right\rangle$ is $|\beta|^2$. As a vector this is
$$
\left|\psi\right\rangle =
\begin{pmatrix}
\alpha \\
\beta
\end{pmatrix}.
$$
Note due to conservation probability $|\alpha|^2+ |\beta|^2 = 1$ and since global phase is undetectable $\left|\psi\right\rangle := e^{i\delta} \left|\psi\right\rangle$ we only requires two real numbers to describe a single qubit quantum state.
A convenient representation is
$$\left|\psi\right\rangle = \cos(\theta/2)\left|0\right\rangle + \sin(\theta/2)e^{i\phi}\left|1\right\rangle$$
where $0\leq \phi < 2\pi$, and $0\leq \theta \leq \pi$. From this it is clear that there is a one-to-one correspondence between qubit states ($\mathbb{C}^2$) and the points on the surface of a unit sphere ($\mathbb{R}^3$). This is called the Bloch sphere representation of a qubit state.
Quantum gates/operations are usually represented as matrices. A gate which acts on a qubit is represented by a $2\times 2$ unitary matrix $U$. The action of the quantum gate is found by multiplying the matrix representing the gate with the vector which represents the quantum state.
$$\left|\psi'\right\rangle = U\left|\psi\right\rangle$$
A general unitary must be able to take the $\left|0\right\rangle$ to the above state. That is
$$
U = \begin{pmatrix}
\cos(\theta/2) & a \\
e^{i\phi}\sin(\theta/2) & b
\end{pmatrix}
$$
where $a$ and $b$ are complex numbers constrained such that $U^\dagger U = I$ for all $0\leq\theta\leq\pi$ and $0\leq \phi<2\pi$. This gives 3 constraints and as such $a\rightarrow -e^{i\lambda}\sin(\theta/2)$ and $b\rightarrow e^{i\lambda+i\phi}\cos(\theta/2)$ where $0\leq \lambda<2\pi$ giving
$$
U = \begin{pmatrix}
\cos(\theta/2) & -e^{i\lambda}\sin(\theta/2) \\
e^{i\phi}\sin(\theta/2) & e^{i\lambda+i\phi}\cos(\theta/2)
\end{pmatrix}.
$$
This is the most general form of a single qubit unitary.
## Single-Qubit Gates
The single-qubit gates available are:
- u gates
- Identity gate
- Pauli gates
- Clifford gates
- $C3$ gates
- Standard rotation gates
We have provided a backend: `unitary_simulator` to allow you to calculate the unitary matrices.
```
q = QuantumRegister(1)
qc = QuantumCircuit(q)
qc.u3(pi/2,pi/2,pi/2,q)
qc.draw()
backend = provider.get_backend('ibmqx2')
job = execute(qc,backend)
job.result().get_unitary(qc,decimals=3)
```
### u gates
In Qiskit we give you access to the general unitary using the $u3$ gate
$$
u3(\theta, \phi, \lambda) = U(\theta, \phi, \lambda)
$$
The $u2(\phi, \lambda) =u3(\pi/2, \phi, \lambda)$ has the matrix form
$$
u2(\phi, \lambda) =
\frac{1}{\sqrt{2}} \begin{pmatrix}
1 & -e^{i\lambda} \\
e^{i\phi} & e^{i(\phi + \lambda)}
\end{pmatrix}.
$$
This is a useful gate as it allows us to create superpositions
The $u1(\lambda)= u3(0, 0, \lambda)$ gate has the matrix form
$$
u1(\lambda) =
\begin{pmatrix}
1 & 0 \\
0 & e^{i \lambda}
\end{pmatrix},
$$
which is a useful as it allows us to apply a quantum phase.
### Pauli gates
#### $X$: bit-flip gate
The bit-flip gate $X$ is defined as:
$$
X =
\begin{pmatrix}
0 & 1\\
1 & 0
\end{pmatrix}= u3(\pi,0,\pi)
$$
```
qc = QuantumCircuit(q)
qc.x(q)
qc.draw()
```
#### $Y$: bit- and phase-flip gate
The $Y$ gate is defined as:
$$
Y =
\begin{pmatrix}
0 & -i\\
i & 0
\end{pmatrix}=u3(\pi,\pi/2,\pi/2)
$$
#### $Z$: phase-flip gate
The phase flip gate $Z$ is defined as:
$$
Z =
\begin{pmatrix}
1 & 0\\
0 & -1
\end{pmatrix}=u1(\pi)
$$
### Clifford gates
#### Hadamard gate
$$
H =
\frac{1}{\sqrt{2}}
\begin{pmatrix}
1 & 1\\
1 & -1
\end{pmatrix}= u2(0,\pi)
$$
The Hadamard gate acts on a single qubit. It maps the basis state $|0>$ to $\frac{1}{\sqrt 2}(|0> + |1>)$ and $|1>$ to $\frac{1}{\sqrt 2}(|0> - |1>)$, thus it creates a superposition state within a single qubit.
## Multi-Qubit Gates
### Mathematical Preliminaries
The space of quantum computer grows exponential with the number of qubits. For $n$ qubits the complex vector space has dimensions $d=2^n$. To describe states of a multi-qubit system, the tensor product is used to "glue together" operators and basis vectors.
Let's start by considering a 2-qubit system. Given two operators $A$ and $B$ that each act on one qubit, the joint operator $A \otimes B$ acting on two qubits is
$$\begin{equation}
A\otimes B =
\begin{pmatrix}
A_{00} \begin{pmatrix}
B_{00} & B_{01} \\
B_{10} & B_{11}
\end{pmatrix} & A_{01} \begin{pmatrix}
B_{00} & B_{01} \\
B_{10} & B_{11}
\end{pmatrix} \\
A_{10} \begin{pmatrix}
B_{00} & B_{01} \\
B_{10} & B_{11}
\end{pmatrix} & A_{11} \begin{pmatrix}
B_{00} & B_{01} \\
B_{10} & B_{11}
\end{pmatrix}
\end{pmatrix},
\end{equation}$$
where $A_{jk}$ and $B_{lm}$ are the matrix elements of $A$ and $B$, respectively.
Analogously, the basis vectors for the 2-qubit system are formed using the tensor product of basis vectors for a single qubit:
$$\begin{equation}\begin{split}
\left|{00}\right\rangle &= \begin{pmatrix}
1 \begin{pmatrix}
1 \\
0
\end{pmatrix} \\
0 \begin{pmatrix}
1 \\
0
\end{pmatrix}
\end{pmatrix} = \begin{pmatrix} 1 \\ 0 \\ 0 \\0 \end{pmatrix}~~~\left|{01}\right\rangle = \begin{pmatrix}
1 \begin{pmatrix}
0 \\
1
\end{pmatrix} \\
0 \begin{pmatrix}
0 \\
1
\end{pmatrix}
\end{pmatrix} = \begin{pmatrix}0 \\ 1 \\ 0 \\ 0 \end{pmatrix}\end{split}
\end{equation}$$
$$\begin{equation}\begin{split}\left|{10}\right\rangle = \begin{pmatrix}
0\begin{pmatrix}
1 \\
0
\end{pmatrix} \\
1\begin{pmatrix}
1 \\
0
\end{pmatrix}
\end{pmatrix} = \begin{pmatrix} 0 \\ 0 \\ 1 \\ 0 \end{pmatrix}~~~ \left|{11}\right\rangle = \begin{pmatrix}
0 \begin{pmatrix}
0 \\
1
\end{pmatrix} \\
1\begin{pmatrix}
0 \\
1
\end{pmatrix}
\end{pmatrix} = \begin{pmatrix} 0 \\ 0 \\ 0 \\1 \end{pmatrix}\end{split}
\end{equation}.$$
Note we've introduced a shorthand for the tensor product of basis vectors, wherein $\left|0\right\rangle \otimes \left|0\right\rangle$ is written as $\left|00\right\rangle$. The state of an $n$-qubit system can described using the $n$-fold tensor product of single-qubit basis vectors. Notice that the basis vectors for a 2-qubit system are 4-dimensional; in general, the basis vectors of an $n$-qubit sytsem are $2^{n}$-dimensional, as noted earlier.
### Basis vector ordering in Qiskit
Within the physics community, the qubits of a multi-qubit systems are typically ordered with the first qubit on the left-most side of the tensor product and the last qubit on the right-most side. For instance, if the first qubit is in state $\left|0\right\rangle$ and second is in state $\left|1\right\rangle$, their joint state would be $\left|01\right\rangle$. Qiskit uses a slightly different ordering of the qubits, in which the qubits are represented from the most significant bit (MSB) on the left to the least significant bit (LSB) on the right (big-endian). This is similar to bitstring representation on classical computers, and enables easy conversion from bitstrings to integers after measurements are performed. For the example just given, the joint state would be represented as $\left|10\right\rangle$. Importantly, _this change in the representation of multi-qubit states affects the way multi-qubit gates are represented in Qiskit_, as discussed below.
The representation used in Qiskit enumerates the basis vectors in increasing order of the integers they represent. For instance, the basis vectors for a 2-qubit system would be ordered as $\left|00\right\rangle$, $\left|01\right\rangle$, $\left|10\right\rangle$, and $\left|11\right\rangle$. Thinking of the basis vectors as bit strings, they encode the integers 0,1,2 and 3, respectively.
### Controlled operations on qubits
A common multi-qubit gate involves the application of a gate to one qubit, conditioned on the state of another qubit. For instance, we might want to flip the state of the second qubit when the first qubit is in $\left|0\right\rangle$. Such gates are known as _controlled gates_. The standard multi-qubit gates consist of two-qubit gates and three-qubit gates. The two-qubit gates are:
- controlled Pauli gates
- controlled Hadamard gate
- controlled rotation gates
- controlled phase gate
- controlled u3 gate
- swap gate
The three-qubit gates are:
- Toffoli gate
- Fredkin gate
|
github_jupyter
|
import numpy as np
from qiskit import *
circ = QuantumCircuit(3)
circ.h(0)
circ.cx(0,1)
circ.cx(0,2)
circ.draw()
backend = Aer.get_backend('statevector_simulator')
job = execute(circ, backend)
result = job.result()
outputstate = result.get_statevector(circ,decimals=3)
print(outputstate)
from qiskit.visualization import plot_state_city
plot_state_city(outputstate)
backend = Aer.get_backend('unitary_simulator')
job = execute(circ, backend)
result = job.result()
print(result.get_unitary(circ,decimals=3))
plot_state_city(result.get_unitary(circ,decimals=3))
meas = QuantumCircuit(3,3)
meas.barrier(range(3))
#This next line maps the quantum measurment to classical bits
meas.measure(range(3),range(3))
qc = circ+meas
qc.draw()
backend_sim = Aer.get_backend('qasm_simulator')
job_sim = execute(qc,backend_sim,shots=1024)
result_sim = job_sim.result()
counts = result_sim.get_counts(qc)
print(counts)
from qiskit.visualization import plot_histogram
plot_histogram(counts)
from qiskit import IBMQ
IBMQ.save_account('caa130bd16fc8c10d09c1e18bf99e63d1285cbe215c8b82ed6c30cc5997077238a2908271e30cc407577d4d34bd400fbabe66a90c959be45f8269f891cc645c9', overwrite = True)
IBMQ.load_account()
IBMQ.providers()
provider = IBMQ.get_provider(group='open')
provider.backends()
backend = provider.get_backend('ibmqx2')
from qiskit.tools.monitor import job_monitor
job_exp = execute(qc,backend=backend)
job_monitor(job_exp)
result_exp = job_exp.result()
counts_exp = result_exp.get_counts(qc)
plot_histogram([counts_exp,counts], legend=['Device','Simulator'])
#create a 12 qubit circuit
qc = QuantumCircuit(12)
#run a hadamard gate on each of the first five, and put each of the results of these into a bell state (through a c-not gate) with the subsequent 5 qubits.
for idx in range(5):
qc.h(idx)
qc.cx(idx, idx+5)
#q1 is also in a bell state to q7
qc.cx(1, 7)
#simply NOT q8
qc.x(8)
#etc
qc.cx(1, 9)
qc.x(7)
qc.cx(1, 11)
qc.swap(6, 11)
qc.swap(6, 9)
qc.swap(6, 10)
qc.x(6)
qc.draw()
#we can get the number of gates or operations performed on the qubits by using:
print(qc.count_ops())
#or, for total:
print(qc.size())
qc.depth()
qc.num_unitary_factors()
qc2 = QuantumCircuit(12, 12)
for idx in range(5):
qc2.h(idx)
qc2.cx(idx, idx+5)
qc2.cx(1, 7)
qc2.x(8)
qc2.cx(1, 9)
qc2.x(7)
qc2.cx(1, 11)
qc2.swap(6, 11)
qc2.swap(6, 9)
qc2.swap(6, 10)
qc2.x(6)
qc2.barrier()
qc2.measure(range(12), range(12))
qc2.draw()
qc3 = QuantumCircuit(2)
qc3.swap(0,1)
qc3.decompose().draw() # This decomposes the swap gate into the gates we want
from qiskit.visualization import plot_gate_map, plot_circuit_layout
import matplotlib.pyplot as plt
ghz = QuantumCircuit(5, 5)
ghz.h(0)
for idx in range(1,5):
ghz.cx(0,idx)
ghz.barrier(range(5))
ghz.measure(range(5), range(5))
ghz.draw()
import inspect
#This shows all the default parameters for the transpile function.
inspect.signature(transpile)
inspect.signature(QuantumCircuit)
qc = QuantumCircuit(2, 1)
qc.h(0)
qc.x(1)
qc.cu1(np.pi/4, 0, 1)
qc.h(0)
qc.measure([0], [0])
qc.draw()
qc_basis = qc.decompose()
qc_basis.draw()
ccx_circ = QuantumCircuit(3)
ccx_circ.ccx(0, 1, 2)
ccx_circ.draw()
ccx_circ.decompose().draw()
backend = provider.get_backend('ibmq_16_melbourne')
plot_gate_map(backend, plot_directed=True)
backend = provider.get_backend('ibmq_16_melbourne')
new_circ_lv0 = transpile(ghz, backend=backend, optimization_level=0)
plot_circuit_layout(new_circ_lv0, backend)
backend = provider.get_backend('ibmq_16_melbourne')
new_circ_lv3 = transpile(ghz, backend=backend, optimization_level=3)
print('Depth:', new_circ_lv3.depth())
plot_circuit_layout(new_circ_lv3, backend)
from qiskit.tools.monitor import job_monitor
job1 = execute(new_circ_lv3, backend)
job_monitor(job1)
# Virtual -> physical
# 0 -> 11
# 1 -> 12
# 2 -> 10
# 3 -> 2
# 4 -> 4
good_ghz = transpile(ghz, backend, initial_layout=[11,12,10,2,4])
print('Depth:', good_ghz.depth())
plot_circuit_layout(good_ghz, backend)
job2 = execute(good_ghz, backend)
job_monitor(job2)
# Virtual -> physical
# 0 -> 0
# 1 -> 6
# 2 -> 10
# 3 -> 13
# 4 -> 7
bad_ghz = transpile(ghz, backend, initial_layout=[0,6,10,13,7])
print('Depth:', bad_ghz.depth())
plot_circuit_layout(bad_ghz, backend)
job3 = execute(bad_ghz, backend)
job_monitor(job3)
counts1 = job1.result().get_counts()
counts2 = job2.result().get_counts()
counts3 = job3.result().get_counts()
plot_histogram([counts1, counts2, counts3],
figsize=(15,6),
legend=['level3', 'good', 'bad'])
from math import pi
from qiskit import QuantumCircuit, ClassicalRegister, QuantumRegister, execute
from qiskit.tools.visualization import circuit_drawer
from qiskit.quantum_info import state_fidelity
from qiskit import BasicAer
backend = BasicAer.get_backend('unitary_simulator')
q = QuantumRegister(1)
qc = QuantumCircuit(q)
qc.u3(pi/2,pi/2,pi/2,q)
qc.draw()
backend = provider.get_backend('ibmqx2')
job = execute(qc,backend)
job.result().get_unitary(qc,decimals=3)
qc = QuantumCircuit(q)
qc.x(q)
qc.draw()
| 0.534127 | 0.987888 |
```
# Python ≥3.5 is required
import sys
assert sys.version_info >= (3, 5)
# Scikit-Learn ≥0.20 is required
import sklearn
assert sklearn.__version__ >= "0.20"
# Common imports
import numpy as np
import os
# to make this notebook's output stable across runs
np.random.seed(42)
# To plot pretty figures
%matplotlib inline
import matplotlib as mpl
import matplotlib.pyplot as plt
mpl.rc('axes', labelsize=14)
mpl.rc('xtick', labelsize=12)
mpl.rc('ytick', labelsize=12)
# Where to save the figures
PROJECT_ROOT_DIR = "."
CHAPTER_ID = "training_linear_models"
IMAGES_PATH = os.path.join(PROJECT_ROOT_DIR, "images", CHAPTER_ID)
os.makedirs(IMAGES_PATH, exist_ok=True)
def save_fig(fig_id, tight_layout=True, fig_extension="png", resolution=300):
path = os.path.join(IMAGES_PATH, fig_id + "." + fig_extension)
print("Saving figure", fig_id)
if tight_layout:
plt.tight_layout()
plt.savefig(path, format=fig_extension, dpi=resolution)
# Ignore useless warnings (see SciPy issue #5998)
import warnings
warnings.filterwarnings(action="ignore", message="^internal gelsd")
from sklearn import datasets
iris = datasets.load_iris()
list(iris.keys())
print(iris.DESCR)
X = iris["data"][:, 3:] # petal width
y = (iris["target"] == 2).astype(np.int) # 1 if Iris virginica, else 0
from sklearn.linear_model import LogisticRegression
log_reg = LogisticRegression(solver="lbfgs", random_state=42)
log_reg.fit(X, y)
X_new = np.linspace(0, 3, 1000).reshape(-1, 1)
y_proba = log_reg.predict_proba(X_new)
plt.plot(X_new, y_proba[:, 1], "g-", linewidth=2, label="Iris virginica")
plt.plot(X_new, y_proba[:, 0], "b--", linewidth=2, label="Not Iris virginica")
X_new = np.linspace(0, 3, 1000).reshape(-1, 1)
y_proba = log_reg.predict_proba(X_new)
decision_boundary = X_new[y_proba[:, 1] >= 0.5][0]
plt.figure(figsize=(8, 3))
plt.plot(X[y==0], y[y==0], "bs")
plt.plot(X[y==1], y[y==1], "g^")
plt.plot([decision_boundary, decision_boundary], [-1, 2], "k:", linewidth=2)
plt.plot(X_new, y_proba[:, 1], "g-", linewidth=2, label="Iris virginica")
plt.plot(X_new, y_proba[:, 0], "b--", linewidth=2, label="Ikke Iris virginica")
plt.text(decision_boundary+0.02, 0.15, "Decision boundary", fontsize=14, color="k", ha="center")
plt.arrow(decision_boundary, 0.08, -0.3, 0, head_width=0.05, head_length=0.1, fc='b', ec='b')
plt.arrow(decision_boundary, 0.92, 0.3, 0, head_width=0.05, head_length=0.1, fc='g', ec='g')
plt.xlabel("Kronbladets bredde (cm)", fontsize=14)
plt.ylabel("Sandsynlighed", fontsize=14)
plt.legend(loc="center left", fontsize=14)
plt.axis([0, 3, -0.02, 1.02])
save_fig("logistic_regression_plot_folkeskolendk")
plt.show()
decision_boundary
log_reg.predict([[1.7], [1.5]])
from sklearn.linear_model import LogisticRegression
X = iris["data"][:, (2, 3)] # petal length, petal width
y = (iris["target"] == 2).astype(np.int)
log_reg = LogisticRegression(solver="lbfgs", C=10**10, random_state=42)
log_reg.fit(X, y)
x0, x1 = np.meshgrid(
np.linspace(2.9, 7, 500).reshape(-1, 1),
np.linspace(0.8, 2.7, 200).reshape(-1, 1),
)
X_new = np.c_[x0.ravel(), x1.ravel()]
y_proba = log_reg.predict_proba(X_new)
plt.figure(figsize=(10, 4))
plt.plot(X[y==0, 0], X[y==0, 1], "bs")
plt.plot(X[y==1, 0], X[y==1, 1], "g^")
zz = y_proba[:, 1].reshape(x0.shape)
contour = plt.contour(x0, x1, zz, cmap=plt.cm.brg)
left_right = np.array([2.9, 7])
boundary = -(log_reg.coef_[0][0] * left_right + log_reg.intercept_[0]) / log_reg.coef_[0][1]
plt.clabel(contour, inline=1, fontsize=12)
plt.plot(left_right, boundary, "k--", linewidth=3)
plt.text(3.5, 1.5, "IKKE Iris virginica", fontsize=14, color="b", ha="center")
plt.text(6.5, 2.3, "Iris virginica", fontsize=14, color="g", ha="center")
plt.xlabel("Kronbladets længde", fontsize=14)
plt.ylabel("Kronbladets bredde", fontsize=14)
plt.axis([2.9, 7, 0.8, 2.7])
save_fig("logistic_regression_contour_plot")
plt.show()
X = iris["data"][:, (2, 3)] # petal length, petal width
y = iris["target"]
softmax_reg = LogisticRegression(multi_class="multinomial",solver="lbfgs", C=10, random_state=42)
softmax_reg.fit(X, y)
X = iris["data"][:, (2, 3)] # petal length, petal width
y = iris["target"]
softmax_reg = LogisticRegression(multi_class="multinomial",solver="lbfgs", C=10, random_state=42)
softmax_reg.fit(X, y)
x0, x1 = np.meshgrid(
np.linspace(0, 8, 500).reshape(-1, 1),
np.linspace(0, 3.5, 200).reshape(-1, 1),
)
X_new = np.c_[x0.ravel(), x1.ravel()]
y_proba = softmax_reg.predict_proba(X_new)
y_predict = softmax_reg.predict(X_new)
zz1 = y_proba[:, 1].reshape(x0.shape)
zz = y_predict.reshape(x0.shape)
plt.figure(figsize=(10, 4))
plt.plot(X[y==2, 0], X[y==2, 1], "g^", label="Iris virginica")
plt.plot(X[y==1, 0], X[y==1, 1], "bs", label="Iris versicolor")
plt.plot(X[y==0, 0], X[y==0, 1], "yo", label="Iris setosa")
from matplotlib.colors import ListedColormap
custom_cmap = ListedColormap(['#fafab0','#9898ff','#a0faa0'])
plt.contourf(x0, x1, zz, cmap=custom_cmap)
contour = plt.contour(x0, x1, zz1, cmap=plt.cm.brg)
plt.clabel(contour, inline=1, fontsize=12)
plt.xlabel("Kronbladets længde", fontsize=14)
plt.ylabel("Kronbladet bredde", fontsize=14)
plt.legend(loc="center left", fontsize=14)
plt.axis([0, 7, 0, 3.5])
save_fig("softmax_regression_contour_plot_folkeskolendk")
plt.show()
softmax_reg.predict([[5, 2]])
softmax_reg.predict_proba([[5, 2]])
```
|
github_jupyter
|
# Python ≥3.5 is required
import sys
assert sys.version_info >= (3, 5)
# Scikit-Learn ≥0.20 is required
import sklearn
assert sklearn.__version__ >= "0.20"
# Common imports
import numpy as np
import os
# to make this notebook's output stable across runs
np.random.seed(42)
# To plot pretty figures
%matplotlib inline
import matplotlib as mpl
import matplotlib.pyplot as plt
mpl.rc('axes', labelsize=14)
mpl.rc('xtick', labelsize=12)
mpl.rc('ytick', labelsize=12)
# Where to save the figures
PROJECT_ROOT_DIR = "."
CHAPTER_ID = "training_linear_models"
IMAGES_PATH = os.path.join(PROJECT_ROOT_DIR, "images", CHAPTER_ID)
os.makedirs(IMAGES_PATH, exist_ok=True)
def save_fig(fig_id, tight_layout=True, fig_extension="png", resolution=300):
path = os.path.join(IMAGES_PATH, fig_id + "." + fig_extension)
print("Saving figure", fig_id)
if tight_layout:
plt.tight_layout()
plt.savefig(path, format=fig_extension, dpi=resolution)
# Ignore useless warnings (see SciPy issue #5998)
import warnings
warnings.filterwarnings(action="ignore", message="^internal gelsd")
from sklearn import datasets
iris = datasets.load_iris()
list(iris.keys())
print(iris.DESCR)
X = iris["data"][:, 3:] # petal width
y = (iris["target"] == 2).astype(np.int) # 1 if Iris virginica, else 0
from sklearn.linear_model import LogisticRegression
log_reg = LogisticRegression(solver="lbfgs", random_state=42)
log_reg.fit(X, y)
X_new = np.linspace(0, 3, 1000).reshape(-1, 1)
y_proba = log_reg.predict_proba(X_new)
plt.plot(X_new, y_proba[:, 1], "g-", linewidth=2, label="Iris virginica")
plt.plot(X_new, y_proba[:, 0], "b--", linewidth=2, label="Not Iris virginica")
X_new = np.linspace(0, 3, 1000).reshape(-1, 1)
y_proba = log_reg.predict_proba(X_new)
decision_boundary = X_new[y_proba[:, 1] >= 0.5][0]
plt.figure(figsize=(8, 3))
plt.plot(X[y==0], y[y==0], "bs")
plt.plot(X[y==1], y[y==1], "g^")
plt.plot([decision_boundary, decision_boundary], [-1, 2], "k:", linewidth=2)
plt.plot(X_new, y_proba[:, 1], "g-", linewidth=2, label="Iris virginica")
plt.plot(X_new, y_proba[:, 0], "b--", linewidth=2, label="Ikke Iris virginica")
plt.text(decision_boundary+0.02, 0.15, "Decision boundary", fontsize=14, color="k", ha="center")
plt.arrow(decision_boundary, 0.08, -0.3, 0, head_width=0.05, head_length=0.1, fc='b', ec='b')
plt.arrow(decision_boundary, 0.92, 0.3, 0, head_width=0.05, head_length=0.1, fc='g', ec='g')
plt.xlabel("Kronbladets bredde (cm)", fontsize=14)
plt.ylabel("Sandsynlighed", fontsize=14)
plt.legend(loc="center left", fontsize=14)
plt.axis([0, 3, -0.02, 1.02])
save_fig("logistic_regression_plot_folkeskolendk")
plt.show()
decision_boundary
log_reg.predict([[1.7], [1.5]])
from sklearn.linear_model import LogisticRegression
X = iris["data"][:, (2, 3)] # petal length, petal width
y = (iris["target"] == 2).astype(np.int)
log_reg = LogisticRegression(solver="lbfgs", C=10**10, random_state=42)
log_reg.fit(X, y)
x0, x1 = np.meshgrid(
np.linspace(2.9, 7, 500).reshape(-1, 1),
np.linspace(0.8, 2.7, 200).reshape(-1, 1),
)
X_new = np.c_[x0.ravel(), x1.ravel()]
y_proba = log_reg.predict_proba(X_new)
plt.figure(figsize=(10, 4))
plt.plot(X[y==0, 0], X[y==0, 1], "bs")
plt.plot(X[y==1, 0], X[y==1, 1], "g^")
zz = y_proba[:, 1].reshape(x0.shape)
contour = plt.contour(x0, x1, zz, cmap=plt.cm.brg)
left_right = np.array([2.9, 7])
boundary = -(log_reg.coef_[0][0] * left_right + log_reg.intercept_[0]) / log_reg.coef_[0][1]
plt.clabel(contour, inline=1, fontsize=12)
plt.plot(left_right, boundary, "k--", linewidth=3)
plt.text(3.5, 1.5, "IKKE Iris virginica", fontsize=14, color="b", ha="center")
plt.text(6.5, 2.3, "Iris virginica", fontsize=14, color="g", ha="center")
plt.xlabel("Kronbladets længde", fontsize=14)
plt.ylabel("Kronbladets bredde", fontsize=14)
plt.axis([2.9, 7, 0.8, 2.7])
save_fig("logistic_regression_contour_plot")
plt.show()
X = iris["data"][:, (2, 3)] # petal length, petal width
y = iris["target"]
softmax_reg = LogisticRegression(multi_class="multinomial",solver="lbfgs", C=10, random_state=42)
softmax_reg.fit(X, y)
X = iris["data"][:, (2, 3)] # petal length, petal width
y = iris["target"]
softmax_reg = LogisticRegression(multi_class="multinomial",solver="lbfgs", C=10, random_state=42)
softmax_reg.fit(X, y)
x0, x1 = np.meshgrid(
np.linspace(0, 8, 500).reshape(-1, 1),
np.linspace(0, 3.5, 200).reshape(-1, 1),
)
X_new = np.c_[x0.ravel(), x1.ravel()]
y_proba = softmax_reg.predict_proba(X_new)
y_predict = softmax_reg.predict(X_new)
zz1 = y_proba[:, 1].reshape(x0.shape)
zz = y_predict.reshape(x0.shape)
plt.figure(figsize=(10, 4))
plt.plot(X[y==2, 0], X[y==2, 1], "g^", label="Iris virginica")
plt.plot(X[y==1, 0], X[y==1, 1], "bs", label="Iris versicolor")
plt.plot(X[y==0, 0], X[y==0, 1], "yo", label="Iris setosa")
from matplotlib.colors import ListedColormap
custom_cmap = ListedColormap(['#fafab0','#9898ff','#a0faa0'])
plt.contourf(x0, x1, zz, cmap=custom_cmap)
contour = plt.contour(x0, x1, zz1, cmap=plt.cm.brg)
plt.clabel(contour, inline=1, fontsize=12)
plt.xlabel("Kronbladets længde", fontsize=14)
plt.ylabel("Kronbladet bredde", fontsize=14)
plt.legend(loc="center left", fontsize=14)
plt.axis([0, 7, 0, 3.5])
save_fig("softmax_regression_contour_plot_folkeskolendk")
plt.show()
softmax_reg.predict([[5, 2]])
softmax_reg.predict_proba([[5, 2]])
| 0.45641 | 0.682716 |
# Import buildings data to the 3DCityDB
This notebook shows how the **IntegrCiTy Data Access Layer** (DAL) can be used to import data to the 3DCityDB.
In this specific example, **geometric data** (2D footprints) and **energy data** (gas consumption) of buildings is **extracted** from separate sources (shapefiles, CSV data), **consolidated** and **stored** in the database.
## Extracting the data
In real life, collecting data is often a long and laborious task.
And once you have all the data you need, it is typically fragmented over various sources with various formats.
In this example, the data is spread over several files:
1. **Shapefile data**: A common format for GIS-related data is the shapefile format. In this example, the shapefile contains the 2D footprints of buildings and information which type of energy carrier they use for heating.
2. **CSV data**: Files containing comma-separated values (CSV) are probably the most common format for data exchange. In this example, the CSV data file contains gas consumption profiles of buildings.
In this example, both data sets use the same ID to refer to the same building (e.g., '*building175*'), which makes it possible to easily link the data from both sets.
In real-life applications it might take another pre-processing step to link the available data.
### Extracting shapefile data
The image below visualizes the buildings data from the shapefile (with the help of [QGIS](https://www.qgis.org/)).
<img src="./img/buildings_sf.png" style="height:12cm">
For the purpose of this example, the [Python Shapefile Library (PyShp)](https://pypi.org/project/pyshp/) is used to exatract this data:
```
import shapefile, os
file_path = os.path.join( os.getcwd(), '..', '1_data', 'shapefiles', 'buildings' )
buildings_sf = shapefile.Reader( file_path )
```
Take a look which data attributes are available (compare with figure above):
```
buildings_sf.fields
```
### Extracting CSV data
The image below visualizes a subset of the buildings data from the CSV file.
<img src="./img/buildings_df.png" style="height:9cm">
For the purpose of this example, the [pandas library](https://pandas.pydata.org/) is used to extract this data:
```
import pandas as pd
file_path = os.path.join( os.getcwd(), '..', '1_data', 'profiles', 'gas_consumption.csv' )
buildings_df = pd.read_csv( file_path )
```
Take a look at the imported data:
```
buildings_df
```
## Accessing the 3DCityDB through the IntegrCiTy DAL
The IntegrCiTy DAL is implemented in [Python package dblayer](https://github.com/IntegrCiTy/dblayer).
The following lines import the core of the package (*dblayer*) and additional wrappers for 3DCityDB SQL functions (*dblayer.func*), which will be used further down in this notebook.
```
from dblayer import *
from dblayer.func.func_citydb_view_nrg import *
from dblayer.func.func_citydb_pkg import *
from dblayer.func.func_postgis_geom import *
```
Provide the connection parameters for the 3DCityDB instance with the help of class *PostgreSQLConnectionInfo*:
```
connect = PostgreSQLConnectionInfo(
user = 'postgres',
pwd = 'postgres',
host = 'localhost',
port = '5432',
dbname = 'citydb'
)
```
Connect to the 3DCityDB with the help of class *DBAccess*. This starts an [SQLAlchemy session](https://docs.sqlalchemy.org/en/13/orm/session.html) in the background, which allows to interact with the database.
```
db_access = DBAccess()
db_access.connect_to_citydb( connect )
```
This tutorial is intended to work with a clean and empty 3DCityDB instance.
Hence, the next lines delete all content from the database for this tutorial.
If you already have a 3DCityDB instance filled with data, you should create a new, empty instance to work with!
**ATTENTION: The next two lines delete all content from the connected 3DCityDB instance!**
```
db_access.cleanup_citydb_schema()
db_access.cleanup_simpkg_schema()
```
Specify the *spatial reference identifier* (SRID) used by the 3DCityDB instance.
If you have used the [setup scripts](https://github.com/IntegrCiTy/dblayer/tree/master/scripts) for installing the extended 3DCityDB provided as part of package *dblayer*, then the default SRID is [4326](https://epsg.io/4326).
For this tutorial, this choice does note make much sense from a geographical point of view, but it is good enough to show you how the IntegrCiTy toolchain works.
If you have a 3DCityDB instance running that has been configured with a different SRID, then change it accordingly (here and in all subsequent notebooks).
```
srid=4326
```
## Consolidating the data
The next step is to merge the data from the two sources and add it to the 3DCityDB.
Since both data sources use the same identifier for buildings, the merging is rather straight forward.
For adding the appropriate CityGML and ADE objects to the 3DCityDB, the IntegrCiTy DAL provides easy access to the SQL functions defined in the respective schemas.
They are executed by calling the DBAccess function *add_citydb_object* with the SQL function name as first parameter, followed by a list of its arguments.
The following lines iterate through the extracted data and then add new objects to the 3DCityDB:
```
import datetime
# Iterate through all entries from the shapefile.
for data in buildings_sf:
# Retrieve the attribute 'name' and 'e_carrier' from this entry:
# - 'name' is the identifier that is also used in CSV data to refer to a specific building
# - 'e_carrier' specifies which energy carrier the building uses for heating
building_name = data.record['name']
building_energy_carrier = data.record['e_carrier']
# Convert the shape points defining the buildings's 2D footprint to a list
# of instances of class Point2D (class provided by package dblayer).
geom_2d_points = [ Point2D( p[0], p[1] ) for p in data.shape.points ]
# Execute function 'geom_from_2dpolygon' to convert the list of points to a
# 3DCityDB geometry.
geom = db_access.execute_function(
geom_from_2dpolygon( geom_2d_points, srid )
)
# Add the geometry as city object to the 3DCityDB with the help of SQL function
# 'insert_surface_geometry' (SQL function defined in database schema 'citydb_pkg').
geom_id = db_access.add_citydb_object(
insert_surface_geometry,
geometry = geom
)
# Add the building as city object to the 3DCityDB with the help of SQL function
# 'insert_building' (SQL function defined in database schema 'citydb_pkg').
building_id = db_access.add_citydb_object(
insert_building,
name = building_name,
lod0_footprint_id = geom_id,
)
if building_energy_carrier == 'gas':
# Retrieve the gas demand profile for this building.
gas_consumption_profile = buildings_df[building_name].tolist()
# Add the profile as city object to the 3DCityDB with the help of SQL function
# 'nrg8_insert_regular_time_series' (SQL function defined in database schema 'citydb_view').
time_series_id = db_access.add_citydb_object(
insert_regular_time_series,
name = 'ts_gas_consumption_{}'.format( building_name ),
description = 'gas consumption profile of {}'.format( building_name ),
acquisition_method = 'Simulation',
interpolation_type = 'AverageInSucceedingInterval',
values_array = gas_consumption_profile,
values_unit = 'kW',
temporal_extent_begin = datetime.datetime( 2020, 1, 1, 0, 0, 0 ),
temporal_extent_end = datetime.datetime( 2020, 1, 1, 23, 0, 0 ),
time_interval = 1,
time_interval_unit = 'h'
)
# Add a final energy object to the 3DCityDB and link it to the time series, with
# the help of SQL function 'nrg8_insert_final_energy' (SQL function defined in
# database schema 'citydb_view').
final_energy_id = db_access.add_citydb_object(
insert_final_energy,
name = 'final_energy_{}'.format( building_name ),
description = 'gas consumption of {}'.format( building_name ),
nrg_car_type = 'NaturalGas',
time_series_id = time_series_id
)
# Add a boiler object to the 3DCityDB and link it to the building, with
# the help of SQL function 'nrg8_insert_boiler' (SQL function defined in
# database schema 'citydb_view').
boiler_id = db_access.add_citydb_object(
insert_boiler,
name = 'gas_boiler_{}'.format( building_name ),
description = 'gas boiler of {}'.format( building_name ),
inst_in_ctyobj_id = building_id
)
# Add an energy conversion system object to the 3DCityDB, in order to link
# the boiler to the final energy use. This is done with the help of SQL function
# 'nrg8_insert_boiler' (SQL function defined in database schema 'citydb_view').
db_access.add_citydb_object(
insert_conv_sys_to_final_nrg,
conv_system_id = boiler_id,
final_nrg_id = final_energy_id,
role = 'consumption'
)
```
## Storing the data to the 3DCityDB
Above, the data was *added* to the database session. In order to make it persistent, i.e., to store it permanently in the database, it has to be *committed* to the 3DCityDB.
This is done via *commit_citydb_session*:
```
db_access.commit_citydb_session()
```
Finally, delete the instance of class DBAccess to close the session.
```
del db_access
```
Next up is notebook [2b_gas_network.ipynb](./2b_gas_network.ipynb), which demonstrates how to use the IntegrCiTy DAL to store a gas network to the 3DCityDB.
|
github_jupyter
|
import shapefile, os
file_path = os.path.join( os.getcwd(), '..', '1_data', 'shapefiles', 'buildings' )
buildings_sf = shapefile.Reader( file_path )
buildings_sf.fields
import pandas as pd
file_path = os.path.join( os.getcwd(), '..', '1_data', 'profiles', 'gas_consumption.csv' )
buildings_df = pd.read_csv( file_path )
buildings_df
from dblayer import *
from dblayer.func.func_citydb_view_nrg import *
from dblayer.func.func_citydb_pkg import *
from dblayer.func.func_postgis_geom import *
connect = PostgreSQLConnectionInfo(
user = 'postgres',
pwd = 'postgres',
host = 'localhost',
port = '5432',
dbname = 'citydb'
)
db_access = DBAccess()
db_access.connect_to_citydb( connect )
db_access.cleanup_citydb_schema()
db_access.cleanup_simpkg_schema()
srid=4326
import datetime
# Iterate through all entries from the shapefile.
for data in buildings_sf:
# Retrieve the attribute 'name' and 'e_carrier' from this entry:
# - 'name' is the identifier that is also used in CSV data to refer to a specific building
# - 'e_carrier' specifies which energy carrier the building uses for heating
building_name = data.record['name']
building_energy_carrier = data.record['e_carrier']
# Convert the shape points defining the buildings's 2D footprint to a list
# of instances of class Point2D (class provided by package dblayer).
geom_2d_points = [ Point2D( p[0], p[1] ) for p in data.shape.points ]
# Execute function 'geom_from_2dpolygon' to convert the list of points to a
# 3DCityDB geometry.
geom = db_access.execute_function(
geom_from_2dpolygon( geom_2d_points, srid )
)
# Add the geometry as city object to the 3DCityDB with the help of SQL function
# 'insert_surface_geometry' (SQL function defined in database schema 'citydb_pkg').
geom_id = db_access.add_citydb_object(
insert_surface_geometry,
geometry = geom
)
# Add the building as city object to the 3DCityDB with the help of SQL function
# 'insert_building' (SQL function defined in database schema 'citydb_pkg').
building_id = db_access.add_citydb_object(
insert_building,
name = building_name,
lod0_footprint_id = geom_id,
)
if building_energy_carrier == 'gas':
# Retrieve the gas demand profile for this building.
gas_consumption_profile = buildings_df[building_name].tolist()
# Add the profile as city object to the 3DCityDB with the help of SQL function
# 'nrg8_insert_regular_time_series' (SQL function defined in database schema 'citydb_view').
time_series_id = db_access.add_citydb_object(
insert_regular_time_series,
name = 'ts_gas_consumption_{}'.format( building_name ),
description = 'gas consumption profile of {}'.format( building_name ),
acquisition_method = 'Simulation',
interpolation_type = 'AverageInSucceedingInterval',
values_array = gas_consumption_profile,
values_unit = 'kW',
temporal_extent_begin = datetime.datetime( 2020, 1, 1, 0, 0, 0 ),
temporal_extent_end = datetime.datetime( 2020, 1, 1, 23, 0, 0 ),
time_interval = 1,
time_interval_unit = 'h'
)
# Add a final energy object to the 3DCityDB and link it to the time series, with
# the help of SQL function 'nrg8_insert_final_energy' (SQL function defined in
# database schema 'citydb_view').
final_energy_id = db_access.add_citydb_object(
insert_final_energy,
name = 'final_energy_{}'.format( building_name ),
description = 'gas consumption of {}'.format( building_name ),
nrg_car_type = 'NaturalGas',
time_series_id = time_series_id
)
# Add a boiler object to the 3DCityDB and link it to the building, with
# the help of SQL function 'nrg8_insert_boiler' (SQL function defined in
# database schema 'citydb_view').
boiler_id = db_access.add_citydb_object(
insert_boiler,
name = 'gas_boiler_{}'.format( building_name ),
description = 'gas boiler of {}'.format( building_name ),
inst_in_ctyobj_id = building_id
)
# Add an energy conversion system object to the 3DCityDB, in order to link
# the boiler to the final energy use. This is done with the help of SQL function
# 'nrg8_insert_boiler' (SQL function defined in database schema 'citydb_view').
db_access.add_citydb_object(
insert_conv_sys_to_final_nrg,
conv_system_id = boiler_id,
final_nrg_id = final_energy_id,
role = 'consumption'
)
db_access.commit_citydb_session()
del db_access
| 0.499268 | 0.967349 |
```
import asyncio
import time
from indy import anoncreds, crypto, did, ledger, pool, wallet
import json
from typing import Optional
async def run():
print("Getting started -> started")
print("Open Pool Ledger")
# Set protocol version 2 to work with Indy Node 1.4
await pool.set_protocol_version(2)
pool_name = 'pool1'
pool_config = json.dumps({"genesis_txn": '/home/indy/sandbox/pool_transactions_genesis'})
await pool.create_pool_ledger_config(pool_name, pool_config)
pool_handle = await pool.open_pool_ledger(pool_name, None)
print("==============================")
print("=== Getting Trust Anchor credentials for Faber, Acme, Thrift and Government ==")
print("------------------------------")
print("\"Sovrin Steward\" -> Create wallet")
steward_wallet_config = json.dumps({"id": "sovrin_steward_wallet"})
steward_wallet_credentials = json.dumps({"key": "steward_wallet_key"})
try:
await wallet.create_wallet(steward_wallet_config, steward_wallet_credentials)
except IndyError as ex:
if ex.error_code == ErrorCode.WalletAlreadyExistsError:
pass
steward_wallet = await wallet.open_wallet(steward_wallet_config, steward_wallet_credentials)
print("\"Sovrin Steward\" -> Create and store in Wallet DID from seed")
steward_did_info = {'seed': '000000000000000000000000Steward1'}
(steward_did, steward_key) = await did.create_and_store_my_did(steward_wallet, json.dumps(steward_did_info))
print("==============================")
print("== Getting Trust Anchor credentials - Government Onboarding ==")
print("------------------------------")
government_wallet_config = json.dumps({"id": "government_wallet"})
government_wallet_credentials = json.dumps({"key": "government_wallet_key"})
government_wallet, steward_government_key, government_steward_did, government_steward_key, _ \
= await onboarding(pool_handle, "Sovrin Steward", steward_wallet, steward_did, "Government", None,
government_wallet_config, government_wallet_credentials)
print("==============================")
print("== Getting Trust Anchor credentials - Government getting Verinym ==")
print("------------------------------")
government_did = await get_verinym(pool_handle, "Sovrin Steward", steward_wallet, steward_did,
steward_government_key, "Government", government_wallet, government_steward_did,
government_steward_key, 'TRUST_ANCHOR')
print("==============================")
print("== Getting Trust Anchor credentials - Faber Onboarding ==")
print("------------------------------")
faber_wallet_config = json.dumps({"id": "faber_wallet"})
faber_wallet_credentials = json.dumps({"key": "faber_wallet_key"})
faber_wallet, steward_faber_key, faber_steward_did, faber_steward_key, _ = \
await onboarding(pool_handle, "Sovrin Steward", steward_wallet, steward_did, "Faber", None,
faber_wallet_config, faber_wallet_credentials)
print("==============================")
print("== Getting Trust Anchor credentials - Faber getting Verinym ==")
print("------------------------------")
faber_did = await get_verinym(pool_handle, "Sovrin Steward", steward_wallet, steward_did, steward_faber_key,
"Faber", faber_wallet, faber_steward_did, faber_steward_key, 'TRUST_ANCHOR')
print("==============================")
print("== Getting Trust Anchor credentials - Acme Onboarding ==")
print("------------------------------")
acme_wallet_config = json.dumps({"id": "acme_wallet"})
acme_wallet_credentials = json.dumps({"key": "acme_wallet_key"})
acme_wallet, steward_acme_key, acme_steward_did, acme_steward_key, _ = \
await onboarding(pool_handle, "Sovrin Steward", steward_wallet, steward_did, "Acme", None,
acme_wallet_config, acme_wallet_credentials)
print("==============================")
print("== Getting Trust Anchor credentials - Acme getting Verinym ==")
print("------------------------------")
acme_did = await get_verinym(pool_handle, "Sovrin Steward", steward_wallet, steward_did, steward_acme_key,
"Acme", acme_wallet, acme_steward_did, acme_steward_key, 'TRUST_ANCHOR')
print("==============================")
print("== Getting Trust Anchor credentials - Thrift Onboarding ==")
print("------------------------------")
thrift_wallet_config = json.dumps({"id": "thrift_wallet"})
thrift_wallet_credentials = json.dumps({"key": "thrift_wallet_key"})
thrift_wallet, steward_thrift_key, thrift_steward_did, thrift_steward_key, _ = \
await onboarding(pool_handle, "Sovrin Steward", steward_wallet, steward_did, "Thrift", None,
thrift_wallet_config, thrift_wallet_credentials)
print("==============================")
print("== Getting Trust Anchor credentials - Thrift getting Verinym ==")
print("------------------------------")
thrift_did = await get_verinym(pool_handle, "Sovrin Steward", steward_wallet, steward_did, steward_thrift_key,
"Thrift", thrift_wallet, thrift_steward_did, thrift_steward_key, 'TRUST_ANCHOR')
print("==============================")
print("=== Credential Schemas Setup ==")
print("------------------------------")
print("\"Government\" -> Create \"Job-Certificate\" Schema")
(job_certificate_schema_id, job_certificate_schema) = \
await anoncreds.issuer_create_schema(government_did, 'Job-Certificate', '0.2',
json.dumps(['first_name', 'last_name', 'salary', 'employee_status',
'experience']))
print("\"Government\" -> Send \"Job-Certificate\" Schema to Ledger")
await send_schema(pool_handle, government_wallet, government_did, job_certificate_schema)
print("\"Government\" -> Create \"Transcript\" Schema")
(transcript_schema_id, transcript_schema) = \
await anoncreds.issuer_create_schema(government_did, 'Transcript', '1.2',
json.dumps(['first_name', 'last_name', 'degree', 'status',
'year', 'average', 'ssn']))
print("\"Government\" -> Send \"Transcript\" Schema to Ledger")
await send_schema(pool_handle, government_wallet, government_did, transcript_schema)
print("==============================")
print("=== Faber Credential Definition Setup ==")
print("------------------------------")
print("\"Faber\" -> Get \"Transcript\" Schema from Ledger")
(_, transcript_schema) = await get_schema(pool_handle, faber_did, transcript_schema_id)
print("\"Faber\" -> Create and store in Wallet \"Faber Transcript\" Credential Definition")
(faber_transcript_cred_def_id, faber_transcript_cred_def_json) = \
await anoncreds.issuer_create_and_store_credential_def(faber_wallet, faber_did, transcript_schema,
'TAG1', 'CL', '{"support_revocation": false}')
print("\"Faber\" -> Send \"Faber Transcript\" Credential Definition to Ledger")
await send_cred_def(pool_handle, faber_wallet, faber_did, faber_transcript_cred_def_json)
print("==============================")
print("=== Acme Credential Definition Setup ==")
print("------------------------------")
print("\"Acme\" -> Get from Ledger \"Job-Certificate\" Schema")
(_, job_certificate_schema) = await get_schema(pool_handle, acme_did, job_certificate_schema_id)
print("\"Acme\" -> Create and store in Wallet \"Acme Job-Certificate\" Credential Definition")
(acme_job_certificate_cred_def_id, acme_job_certificate_cred_def_json) = \
await anoncreds.issuer_create_and_store_credential_def(acme_wallet, acme_did, job_certificate_schema,
'TAG1', 'CL', '{"support_revocation": false}')
print("\"Acme\" -> Send \"Acme Job-Certificate\" Credential Definition to Ledger")
await send_cred_def(pool_handle, acme_wallet, acme_did, acme_job_certificate_cred_def_json)
print("==============================")
print("=== Getting Transcript with Faber ==")
print("==============================")
print("== Getting Transcript with Faber - Onboarding ==")
print("------------------------------")
alice_wallet_config = json.dumps({"id": "alice_wallet"})
alice_wallet_credentials = json.dumps({"key": "alice_wallet_key"})
alice_wallet, faber_alice_key, alice_faber_did, alice_faber_key, faber_alice_connection_response \
= await onboarding(pool_handle, "Faber", faber_wallet, faber_did, "Alice", None,
alice_wallet_config, alice_wallet_credentials)
time.sleep(1)
print("==============================")
print("== Getting Transcript with Faber - Getting Transcript Credential ==")
print("------------------------------")
print("\"Faber\" -> Create \"Transcript\" Credential Offer for Alice")
transcript_cred_offer_json = \
await anoncreds.issuer_create_credential_offer(faber_wallet, faber_transcript_cred_def_id)
print("\"Faber\" -> Get key for Alice did")
alice_faber_verkey = await did.key_for_did(pool_handle, acme_wallet, faber_alice_connection_response['did'])
print("\"Faber\" -> Authcrypt \"Transcript\" Credential Offer for Alice")
authcrypted_transcript_cred_offer = await crypto.auth_crypt(faber_wallet, faber_alice_key, alice_faber_verkey,
transcript_cred_offer_json.encode('utf-8'))
print("\"Faber\" -> Send authcrypted \"Transcript\" Credential Offer to Alice")
print("\"Alice\" -> Authdecrypted \"Transcript\" Credential Offer from Faber")
faber_alice_verkey, authdecrypted_transcript_cred_offer_json, authdecrypted_transcript_cred_offer = \
await auth_decrypt(alice_wallet, alice_faber_key, authcrypted_transcript_cred_offer)
print("\"Alice\" -> Create and store \"Alice\" Master Secret in Wallet")
alice_master_secret_id = await anoncreds.prover_create_master_secret(alice_wallet, None)
print("\"Alice\" -> Get \"Faber Transcript\" Credential Definition from Ledger")
(faber_transcript_cred_def_id, faber_transcript_cred_def) = \
await get_cred_def(pool_handle, alice_faber_did, authdecrypted_transcript_cred_offer['cred_def_id'])
print("\"Alice\" -> Create \"Transcript\" Credential Request for Faber")
(transcript_cred_request_json, transcript_cred_request_metadata_json) = \
await anoncreds.prover_create_credential_req(alice_wallet, alice_faber_did,
authdecrypted_transcript_cred_offer_json,
faber_transcript_cred_def, alice_master_secret_id)
print("\"Alice\" -> Authcrypt \"Transcript\" Credential Request for Faber")
authcrypted_transcript_cred_request = await crypto.auth_crypt(alice_wallet, alice_faber_key, faber_alice_verkey,
transcript_cred_request_json.encode('utf-8'))
print("\"Alice\" -> Send authcrypted \"Transcript\" Credential Request to Faber")
print("\"Faber\" -> Authdecrypt \"Transcript\" Credential Request from Alice")
alice_faber_verkey, authdecrypted_transcript_cred_request_json, _ = \
await auth_decrypt(faber_wallet, faber_alice_key, authcrypted_transcript_cred_request)
print("\"Faber\" -> Create \"Transcript\" Credential for Alice")
transcript_cred_values = json.dumps({
"first_name": {"raw": "Alice", "encoded": "1139481716457488690172217916278103335"},
"last_name": {"raw": "Garcia", "encoded": "5321642780241790123587902456789123452"},
"degree": {"raw": "Bachelor of Science, Marketing", "encoded": "12434523576212321"},
"status": {"raw": "graduated", "encoded": "2213454313412354"},
"ssn": {"raw": "123-45-6789", "encoded": "3124141231422543541"},
"year": {"raw": "2015", "encoded": "2015"},
"average": {"raw": "5", "encoded": "5"}
})
transcript_cred_json, _, _ = \
await anoncreds.issuer_create_credential(faber_wallet, transcript_cred_offer_json,
authdecrypted_transcript_cred_request_json,
transcript_cred_values, None, None)
print("\"Faber\" -> Authcrypt \"Transcript\" Credential for Alice")
authcrypted_transcript_cred_json = await crypto.auth_crypt(faber_wallet, faber_alice_key, alice_faber_verkey,
transcript_cred_json.encode('utf-8'))
print("\"Faber\" -> Send authcrypted \"Transcript\" Credential to Alice")
print("\"Alice\" -> Authdecrypted \"Transcript\" Credential from Faber")
_, authdecrypted_transcript_cred_json, _ = \
await auth_decrypt(alice_wallet, alice_faber_key, authcrypted_transcript_cred_json)
print("\"Alice\" -> Store \"Transcript\" Credential from Faber")
await anoncreds.prover_store_credential(alice_wallet, None, transcript_cred_request_metadata_json,
authdecrypted_transcript_cred_json, faber_transcript_cred_def, None)
print("==============================")
print("=== Apply for the job with Acme ==")
print("==============================")
print("== Apply for the job with Acme - Onboarding ==")
print("------------------------------")
alice_wallet, acme_alice_key, alice_acme_did, alice_acme_key, acme_alice_connection_response = \
await onboarding(pool_handle, "Acme", acme_wallet, acme_did, "Alice", alice_wallet,
alice_wallet_config, alice_wallet_credentials)
print("==============================")
print("== Apply for the job with Acme - Transcript proving ==")
print("------------------------------")
print("\"Acme\" -> Create \"Job-Application\" Proof Request")
job_application_proof_request_json = json.dumps({
'nonce': '1432422343242122312411212',
'name': 'Job-Application',
'version': '0.1',
'requested_attributes': {
'attr1_referent': {
'name': 'first_name'
},
'attr2_referent': {
'name': 'last_name'
},
'attr3_referent': {
'name': 'degree',
'restrictions': [{'cred_def_id': faber_transcript_cred_def_id}]
},
'attr4_referent': {
'name': 'status',
'restrictions': [{'cred_def_id': faber_transcript_cred_def_id}]
},
'attr5_referent': {
'name': 'ssn',
'restrictions': [{'cred_def_id': faber_transcript_cred_def_id}]
},
'attr6_referent': {
'name': 'phone_number'
}
},
'requested_predicates': {
'predicate1_referent': {
'name': 'average',
'p_type': '>=',
'p_value': 4,
'restrictions': [{'cred_def_id': faber_transcript_cred_def_id}]
}
}
})
print("\"Acme\" -> Get key for Alice did")
alice_acme_verkey = await did.key_for_did(pool_handle, acme_wallet, acme_alice_connection_response['did'])
print("\"Acme\" -> Authcrypt \"Job-Application\" Proof Request for Alice")
authcrypted_job_application_proof_request_json = \
await crypto.auth_crypt(acme_wallet, acme_alice_key, alice_acme_verkey,
job_application_proof_request_json.encode('utf-8'))
print("\"Acme\" -> Send authcrypted \"Job-Application\" Proof Request to Alice")
print("\"Alice\" -> Authdecrypt \"Job-Application\" Proof Request from Acme")
acme_alice_verkey, authdecrypted_job_application_proof_request_json, _ = \
await auth_decrypt(alice_wallet, alice_acme_key, authcrypted_job_application_proof_request_json)
print("\"Alice\" -> Get credentials for \"Job-Application\" Proof Request")
search_for_job_application_proof_request = \
await anoncreds.prover_search_credentials_for_proof_req(alice_wallet,
authdecrypted_job_application_proof_request_json, None)
cred_for_attr1 = await get_credential_for_referent(search_for_job_application_proof_request, 'attr1_referent')
cred_for_attr2 = await get_credential_for_referent(search_for_job_application_proof_request, 'attr2_referent')
cred_for_attr3 = await get_credential_for_referent(search_for_job_application_proof_request, 'attr3_referent')
cred_for_attr4 = await get_credential_for_referent(search_for_job_application_proof_request, 'attr4_referent')
cred_for_attr5 = await get_credential_for_referent(search_for_job_application_proof_request, 'attr5_referent')
cred_for_predicate1 = \
await get_credential_for_referent(search_for_job_application_proof_request, 'predicate1_referent')
await anoncreds.prover_close_credentials_search_for_proof_req(search_for_job_application_proof_request)
creds_for_job_application_proof = {cred_for_attr1['referent']: cred_for_attr1,
cred_for_attr2['referent']: cred_for_attr2,
cred_for_attr3['referent']: cred_for_attr3,
cred_for_attr4['referent']: cred_for_attr4,
cred_for_attr5['referent']: cred_for_attr5,
cred_for_predicate1['referent']: cred_for_predicate1}
schemas_json, cred_defs_json, revoc_states_json = \
await prover_get_entities_from_ledger(pool_handle, alice_faber_did, creds_for_job_application_proof, 'Alice')
print("\"Alice\" -> Create \"Job-Application\" Proof")
job_application_requested_creds_json = json.dumps({
'self_attested_attributes': {
'attr1_referent': 'Alice',
'attr2_referent': 'Garcia',
'attr6_referent': '123-45-6789'
},
'requested_attributes': {
'attr3_referent': {'cred_id': cred_for_attr3['referent'], 'revealed': True},
'attr4_referent': {'cred_id': cred_for_attr4['referent'], 'revealed': True},
'attr5_referent': {'cred_id': cred_for_attr5['referent'], 'revealed': True},
},
'requested_predicates': {'predicate1_referent': {'cred_id': cred_for_predicate1['referent']}}
})
job_application_proof_json = \
await anoncreds.prover_create_proof(alice_wallet, authdecrypted_job_application_proof_request_json,
job_application_requested_creds_json, alice_master_secret_id,
schemas_json, cred_defs_json, revoc_states_json)
print("\"Alice\" -> Authcrypt \"Job-Application\" Proof for Acme")
authcrypted_job_application_proof_json = await crypto.auth_crypt(alice_wallet, alice_acme_key, acme_alice_verkey,
job_application_proof_json.encode('utf-8'))
print("\"Alice\" -> Send authcrypted \"Job-Application\" Proof to Acme")
print("\"Acme\" -> Authdecrypted \"Job-Application\" Proof from Alice")
_, decrypted_job_application_proof_json, decrypted_job_application_proof = \
await auth_decrypt(acme_wallet, acme_alice_key, authcrypted_job_application_proof_json)
schemas_json, cred_defs_json, revoc_ref_defs_json, revoc_regs_json = \
await verifier_get_entities_from_ledger(pool_handle, acme_did,
decrypted_job_application_proof['identifiers'], 'Acme')
print("\"Acme\" -> Verify \"Job-Application\" Proof from Alice")
assert 'Bachelor of Science, Marketing' == \
decrypted_job_application_proof['requested_proof']['revealed_attrs']['attr3_referent']['raw']
assert 'graduated' == \
decrypted_job_application_proof['requested_proof']['revealed_attrs']['attr4_referent']['raw']
assert '123-45-6789' == \
decrypted_job_application_proof['requested_proof']['revealed_attrs']['attr5_referent']['raw']
assert 'Alice' == decrypted_job_application_proof['requested_proof']['self_attested_attrs']['attr1_referent']
assert 'Garcia' == decrypted_job_application_proof['requested_proof']['self_attested_attrs']['attr2_referent']
assert '123-45-6789' == decrypted_job_application_proof['requested_proof']['self_attested_attrs']['attr6_referent']
assert await anoncreds.verifier_verify_proof(job_application_proof_request_json,
decrypted_job_application_proof_json,
schemas_json, cred_defs_json, revoc_ref_defs_json, revoc_regs_json)
print("==============================")
print("== Apply for the job with Acme - Getting Job-Certificate Credential ==")
print("------------------------------")
print("\"Acme\" -> Create \"Job-Certificate\" Credential Offer for Alice")
job_certificate_cred_offer_json = \
await anoncreds.issuer_create_credential_offer(acme_wallet, acme_job_certificate_cred_def_id)
print("\"Acme\" -> Get key for Alice did")
alice_acme_verkey = await did.key_for_did(pool_handle, acme_wallet, acme_alice_connection_response['did'])
print("\"Acme\" -> Authcrypt \"Job-Certificate\" Credential Offer for Alice")
authcrypted_job_certificate_cred_offer = await crypto.auth_crypt(acme_wallet, acme_alice_key, alice_acme_verkey,
job_certificate_cred_offer_json.encode('utf-8'))
print("\"Acme\" -> Send authcrypted \"Job-Certificate\" Credential Offer to Alice")
print("\"Alice\" -> Authdecrypted \"Job-Certificate\" Credential Offer from Acme")
acme_alice_verkey, authdecrypted_job_certificate_cred_offer_json, authdecrypted_job_certificate_cred_offer = \
await auth_decrypt(alice_wallet, alice_acme_key, authcrypted_job_certificate_cred_offer)
print("\"Alice\" -> Get \"Acme Job-Certificate\" Credential Definition from Ledger")
(_, acme_job_certificate_cred_def) = \
await get_cred_def(pool_handle, alice_acme_did, authdecrypted_job_certificate_cred_offer['cred_def_id'])
print("\"Alice\" -> Create and store in Wallet \"Job-Certificate\" Credential Request for Acme")
(job_certificate_cred_request_json, job_certificate_cred_request_metadata_json) = \
await anoncreds.prover_create_credential_req(alice_wallet, alice_acme_did,
authdecrypted_job_certificate_cred_offer_json,
acme_job_certificate_cred_def, alice_master_secret_id)
print("\"Alice\" -> Authcrypt \"Job-Certificate\" Credential Request for Acme")
authcrypted_job_certificate_cred_request_json = \
await crypto.auth_crypt(alice_wallet, alice_acme_key, acme_alice_verkey,
job_certificate_cred_request_json.encode('utf-8'))
print("\"Alice\" -> Send authcrypted \"Job-Certificate\" Credential Request to Acme")
print("\"Acme\" -> Authdecrypt \"Job-Certificate\" Credential Request from Alice")
alice_acme_verkey, authdecrypted_job_certificate_cred_request_json, _ = \
await auth_decrypt(acme_wallet, acme_alice_key, authcrypted_job_certificate_cred_request_json)
print("\"Acme\" -> Create \"Job-Certificate\" Credential for Alice")
alice_job_certificate_cred_values_json = json.dumps({
"first_name": {"raw": "Alice", "encoded": "245712572474217942457235975012103335"},
"last_name": {"raw": "Garcia", "encoded": "312643218496194691632153761283356127"},
"employee_status": {"raw": "Permanent", "encoded": "2143135425425143112321314321"},
"salary": {"raw": "2400", "encoded": "2400"},
"experience": {"raw": "10", "encoded": "10"}
})
job_certificate_cred_json, _, _ = \
await anoncreds.issuer_create_credential(acme_wallet, job_certificate_cred_offer_json,
authdecrypted_job_certificate_cred_request_json,
alice_job_certificate_cred_values_json, None, None)
print("\"Acme\" -> Authcrypt \"Job-Certificate\" Credential for Alice")
authcrypted_job_certificate_cred_json = \
await crypto.auth_crypt(acme_wallet, acme_alice_key, alice_acme_verkey,
job_certificate_cred_json.encode('utf-8'))
print("\"Acme\" -> Send authcrypted \"Job-Certificate\" Credential to Alice")
print("\"Alice\" -> Authdecrypted \"Job-Certificate\" Credential from Acme")
_, authdecrypted_job_certificate_cred_json, _ = \
await auth_decrypt(alice_wallet, alice_acme_key, authcrypted_job_certificate_cred_json)
print("\"Alice\" -> Store \"Job-Certificate\" Credential")
await anoncreds.prover_store_credential(alice_wallet, None, job_certificate_cred_request_metadata_json,
authdecrypted_job_certificate_cred_json,
acme_job_certificate_cred_def_json, None)
print("==============================")
print("=== Apply for the loan with Thrift ==")
print("==============================")
print("== Apply for the loan with Thrift - Onboarding ==")
print("------------------------------")
_, thrift_alice_key, alice_thrift_did, alice_thrift_key, \
thrift_alice_connection_response = await onboarding(pool_handle, "Thrift", thrift_wallet, thrift_did, "Alice",
alice_wallet, alice_wallet_config, alice_wallet_credentials)
print("==============================")
print("== Apply for the loan with Thrift - Job-Certificate proving ==")
print("------------------------------")
print("\"Thrift\" -> Create \"Loan-Application-Basic\" Proof Request")
apply_loan_proof_request_json = json.dumps({
'nonce': '123432421212',
'name': 'Loan-Application-Basic',
'version': '0.1',
'requested_attributes': {
'attr1_referent': {
'name': 'employee_status',
'restrictions': [{'cred_def_id': acme_job_certificate_cred_def_id}]
}
},
'requested_predicates': {
'predicate1_referent': {
'name': 'salary',
'p_type': '>=',
'p_value': 2000,
'restrictions': [{'cred_def_id': acme_job_certificate_cred_def_id}]
},
'predicate2_referent': {
'name': 'experience',
'p_type': '>=',
'p_value': 1,
'restrictions': [{'cred_def_id': acme_job_certificate_cred_def_id}]
}
}
})
print("\"Thrift\" -> Get key for Alice did")
alice_thrift_verkey = await did.key_for_did(pool_handle, thrift_wallet, thrift_alice_connection_response['did'])
print("\"Thrift\" -> Authcrypt \"Loan-Application-Basic\" Proof Request for Alice")
authcrypted_apply_loan_proof_request_json = \
await crypto.auth_crypt(thrift_wallet, thrift_alice_key, alice_thrift_verkey,
apply_loan_proof_request_json.encode('utf-8'))
print("\"Thrift\" -> Send authcrypted \"Loan-Application-Basic\" Proof Request to Alice")
print("\"Alice\" -> Authdecrypt \"Loan-Application-Basic\" Proof Request from Thrift")
thrift_alice_verkey, authdecrypted_apply_loan_proof_request_json, _ = \
await auth_decrypt(alice_wallet, alice_thrift_key, authcrypted_apply_loan_proof_request_json)
print("\"Alice\" -> Get credentials for \"Loan-Application-Basic\" Proof Request")
search_for_apply_loan_proof_request = \
await anoncreds.prover_search_credentials_for_proof_req(alice_wallet,
authdecrypted_apply_loan_proof_request_json, None)
cred_for_attr1 = await get_credential_for_referent(search_for_apply_loan_proof_request, 'attr1_referent')
cred_for_predicate1 = await get_credential_for_referent(search_for_apply_loan_proof_request, 'predicate1_referent')
cred_for_predicate2 = await get_credential_for_referent(search_for_apply_loan_proof_request, 'predicate2_referent')
await anoncreds.prover_close_credentials_search_for_proof_req(search_for_apply_loan_proof_request)
creds_for_apply_loan_proof = {cred_for_attr1['referent']: cred_for_attr1,
cred_for_predicate1['referent']: cred_for_predicate1,
cred_for_predicate2['referent']: cred_for_predicate2}
schemas_json, cred_defs_json, revoc_states_json = \
await prover_get_entities_from_ledger(pool_handle, alice_thrift_did, creds_for_apply_loan_proof, 'Alice')
print("\"Alice\" -> Create \"Loan-Application-Basic\" Proof")
apply_loan_requested_creds_json = json.dumps({
'self_attested_attributes': {},
'requested_attributes': {
'attr1_referent': {'cred_id': cred_for_attr1['referent'], 'revealed': True}
},
'requested_predicates': {
'predicate1_referent': {'cred_id': cred_for_predicate1['referent']},
'predicate2_referent': {'cred_id': cred_for_predicate2['referent']}
}
})
alice_apply_loan_proof_json = \
await anoncreds.prover_create_proof(alice_wallet, authdecrypted_apply_loan_proof_request_json,
apply_loan_requested_creds_json, alice_master_secret_id, schemas_json,
cred_defs_json, revoc_states_json)
print("\"Alice\" -> Authcrypt \"Loan-Application-Basic\" Proof for Thrift")
authcrypted_alice_apply_loan_proof_json = \
await crypto.auth_crypt(alice_wallet, alice_thrift_key, thrift_alice_verkey,
alice_apply_loan_proof_json.encode('utf-8'))
print("\"Alice\" -> Send authcrypted \"Loan-Application-Basic\" Proof to Thrift")
print("\"Thrift\" -> Authdecrypted \"Loan-Application-Basic\" Proof from Alice")
_, authdecrypted_alice_apply_loan_proof_json, authdecrypted_alice_apply_loan_proof = \
await auth_decrypt(thrift_wallet, thrift_alice_key, authcrypted_alice_apply_loan_proof_json)
print("\"Thrift\" -> Get Schemas, Credential Definitions and Revocation Registries from Ledger"
" required for Proof verifying")
schemas_json, cred_defs_json, revoc_defs_json, revoc_regs_json = \
await verifier_get_entities_from_ledger(pool_handle, thrift_did,
authdecrypted_alice_apply_loan_proof['identifiers'], 'Thrift')
print("\"Thrift\" -> Verify \"Loan-Application-Basic\" Proof from Alice")
assert 'Permanent' == \
authdecrypted_alice_apply_loan_proof['requested_proof']['revealed_attrs']['attr1_referent']['raw']
assert await anoncreds.verifier_verify_proof(apply_loan_proof_request_json,
authdecrypted_alice_apply_loan_proof_json,
schemas_json, cred_defs_json, revoc_defs_json, revoc_regs_json)
print("==============================")
print("==============================")
print("== Apply for the loan with Thrift - Transcript and Job-Certificate proving ==")
print("------------------------------")
print("\"Thrift\" -> Create \"Loan-Application-KYC\" Proof Request")
apply_loan_kyc_proof_request_json = json.dumps({
'nonce': '123432421212',
'name': 'Loan-Application-KYC',
'version': '0.1',
'requested_attributes': {
'attr1_referent': {'name': 'first_name'},
'attr2_referent': {'name': 'last_name'},
'attr3_referent': {'name': 'ssn'}
},
'requested_predicates': {}
})
print("\"Thrift\" -> Get key for Alice did")
alice_thrift_verkey = await did.key_for_did(pool_handle, thrift_wallet, thrift_alice_connection_response['did'])
print("\"Thrift\" -> Authcrypt \"Loan-Application-KYC\" Proof Request for Alice")
authcrypted_apply_loan_kyc_proof_request_json = \
await crypto.auth_crypt(thrift_wallet, thrift_alice_key, alice_thrift_verkey,
apply_loan_kyc_proof_request_json.encode('utf-8'))
print("\"Thrift\" -> Send authcrypted \"Loan-Application-KYC\" Proof Request to Alice")
print("\"Alice\" -> Authdecrypt \"Loan-Application-KYC\" Proof Request from Thrift")
thrift_alice_verkey, authdecrypted_apply_loan_kyc_proof_request_json, _ = \
await auth_decrypt(alice_wallet, alice_thrift_key, authcrypted_apply_loan_kyc_proof_request_json)
print("\"Alice\" -> Get credentials for \"Loan-Application-KYC\" Proof Request")
search_for_apply_loan_kyc_proof_request = \
await anoncreds.prover_search_credentials_for_proof_req(alice_wallet,
authdecrypted_apply_loan_kyc_proof_request_json, None)
cred_for_attr1 = await get_credential_for_referent(search_for_apply_loan_kyc_proof_request, 'attr1_referent')
cred_for_attr2 = await get_credential_for_referent(search_for_apply_loan_kyc_proof_request, 'attr2_referent')
cred_for_attr3 = await get_credential_for_referent(search_for_apply_loan_kyc_proof_request, 'attr3_referent')
await anoncreds.prover_close_credentials_search_for_proof_req(search_for_apply_loan_kyc_proof_request)
creds_for_apply_loan_kyc_proof = {cred_for_attr1['referent']: cred_for_attr1,
cred_for_attr2['referent']: cred_for_attr2,
cred_for_attr3['referent']: cred_for_attr3}
schemas_json, cred_defs_json, revoc_states_json = \
await prover_get_entities_from_ledger(pool_handle, alice_thrift_did, creds_for_apply_loan_kyc_proof, 'Alice')
print("\"Alice\" -> Create \"Loan-Application-KYC\" Proof")
apply_loan_kyc_requested_creds_json = json.dumps({
'self_attested_attributes': {},
'requested_attributes': {
'attr1_referent': {'cred_id': cred_for_attr1['referent'], 'revealed': True},
'attr2_referent': {'cred_id': cred_for_attr2['referent'], 'revealed': True},
'attr3_referent': {'cred_id': cred_for_attr3['referent'], 'revealed': True}
},
'requested_predicates': {}
})
alice_apply_loan_kyc_proof_json = \
await anoncreds.prover_create_proof(alice_wallet, authdecrypted_apply_loan_kyc_proof_request_json,
apply_loan_kyc_requested_creds_json, alice_master_secret_id,
schemas_json, cred_defs_json, revoc_states_json)
print("\"Alice\" -> Authcrypt \"Loan-Application-KYC\" Proof for Thrift")
authcrypted_alice_apply_loan_kyc_proof_json = \
await crypto.auth_crypt(alice_wallet, alice_thrift_key, thrift_alice_verkey,
alice_apply_loan_kyc_proof_json.encode('utf-8'))
print("\"Alice\" -> Send authcrypted \"Loan-Application-KYC\" Proof to Thrift")
print("\"Thrift\" -> Authdecrypted \"Loan-Application-KYC\" Proof from Alice")
_, authdecrypted_alice_apply_loan_kyc_proof_json, authdecrypted_alice_apply_loan_kyc_proof = \
await auth_decrypt(thrift_wallet, thrift_alice_key, authcrypted_alice_apply_loan_kyc_proof_json)
print("\"Thrift\" -> Get Schemas, Credential Definitions and Revocation Registries from Ledger"
" required for Proof verifying")
schemas_json, cred_defs_json, revoc_defs_json, revoc_regs_json = \
await verifier_get_entities_from_ledger(pool_handle, thrift_did,
authdecrypted_alice_apply_loan_kyc_proof['identifiers'], 'Thrift')
print("\"Thrift\" -> Verify \"Loan-Application-KYC\" Proof from Alice")
assert 'Alice' == \
authdecrypted_alice_apply_loan_kyc_proof['requested_proof']['revealed_attrs']['attr1_referent']['raw']
assert 'Garcia' == \
authdecrypted_alice_apply_loan_kyc_proof['requested_proof']['revealed_attrs']['attr2_referent']['raw']
assert '123-45-6789' == \
authdecrypted_alice_apply_loan_kyc_proof['requested_proof']['revealed_attrs']['attr3_referent']['raw']
assert await anoncreds.verifier_verify_proof(apply_loan_kyc_proof_request_json,
authdecrypted_alice_apply_loan_kyc_proof_json,
schemas_json, cred_defs_json, revoc_defs_json, revoc_regs_json)
print("==============================")
print(" \"Sovrin Steward\" -> Close and Delete wallet")
await wallet.close_wallet(steward_wallet)
await wallet.delete_wallet(steward_wallet_config, steward_wallet_credentials)
print("\"Government\" -> Close and Delete wallet")
await wallet.close_wallet(government_wallet)
await wallet.delete_wallet(government_wallet_config, government_wallet_credentials)
print("\"Faber\" -> Close and Delete wallet")
await wallet.close_wallet(faber_wallet)
await wallet.delete_wallet(faber_wallet_config, faber_wallet_credentials)
print("\"Acme\" -> Close and Delete wallet")
await wallet.close_wallet(acme_wallet)
await wallet.delete_wallet(acme_wallet_config, acme_wallet_credentials)
print("\"Thrift\" -> Close and Delete wallet")
await wallet.close_wallet(thrift_wallet)
await wallet.delete_wallet(thrift_wallet_config, thrift_wallet_credentials)
print("\"Alice\" -> Close and Delete wallet")
await wallet.close_wallet(alice_wallet)
await wallet.delete_wallet(alice_wallet_config, alice_wallet_credentials)
print("Close and Delete pool")
await pool.close_pool_ledger(pool_handle)
await pool.delete_pool_ledger_config(pool_name)
print("Getting started -> done")
async def onboarding(pool_handle, _from, from_wallet, from_did, to,
to_wallet: Optional[str], to_wallet_config: str, to_wallet_credentials: str):
print("\"{}\" -> Create and store in Wallet \"{} {}\" DID".format(_from, _from, to))
(from_to_did, from_to_key) = await did.create_and_store_my_did(from_wallet, "{}")
print("\"{}\" -> Send Nym to Ledger for \"{} {}\" DID".format(_from, _from, to))
await send_nym(pool_handle, from_wallet, from_did, from_to_did, from_to_key, None)
print("\"{}\" -> Send connection request to {} with \"{} {}\" DID and nonce".format(_from, to, _from, to))
connection_request = {
'did': from_to_did,
'nonce': 123456789
}
if not to_wallet:
print("\"{}\" -> Create wallet".format(to))
try:
await wallet.create_wallet(to_wallet_config, to_wallet_credentials)
except IndyError as ex:
if ex.error_code == ErrorCode.PoolLedgerConfigAlreadyExistsError:
pass
to_wallet = await wallet.open_wallet(to_wallet_config, to_wallet_credentials)
print("\"{}\" -> Create and store in Wallet \"{} {}\" DID".format(to, to, _from))
(to_from_did, to_from_key) = await did.create_and_store_my_did(to_wallet, "{}")
print("\"{}\" -> Get key for did from \"{}\" connection request".format(to, _from))
from_to_verkey = await did.key_for_did(pool_handle, to_wallet, connection_request['did'])
print("\"{}\" -> Anoncrypt connection response for \"{}\" with \"{} {}\" DID, verkey and nonce"
.format(to, _from, to, _from))
connection_response = json.dumps({
'did': to_from_did,
'verkey': to_from_key,
'nonce': connection_request['nonce']
})
anoncrypted_connection_response = await crypto.anon_crypt(from_to_verkey, connection_response.encode('utf-8'))
print("\"{}\" -> Send anoncrypted connection response to \"{}\"".format(to, _from))
print("\"{}\" -> Anondecrypt connection response from \"{}\"".format(_from, to))
decrypted_connection_response = \
json.loads((await crypto.anon_decrypt(from_wallet, from_to_key,
anoncrypted_connection_response)).decode("utf-8"))
print("\"{}\" -> Authenticates \"{}\" by comparision of Nonce".format(_from, to))
assert connection_request['nonce'] == decrypted_connection_response['nonce']
print("\"{}\" -> Send Nym to Ledger for \"{} {}\" DID".format(_from, to, _from))
await send_nym(pool_handle, from_wallet, from_did, to_from_did, to_from_key, None)
return to_wallet, from_to_key, to_from_did, to_from_key, decrypted_connection_response
async def get_verinym(pool_handle, _from, from_wallet, from_did, from_to_key,
to, to_wallet, to_from_did, to_from_key, role):
print("\"{}\" -> Create and store in Wallet \"{}\" new DID".format(to, to))
(to_did, to_key) = await did.create_and_store_my_did(to_wallet, "{}")
print("\"{}\" -> Authcrypt \"{} DID info\" for \"{}\"".format(to, to, _from))
did_info_json = json.dumps({
'did': to_did,
'verkey': to_key
})
authcrypted_did_info_json = \
await crypto.auth_crypt(to_wallet, to_from_key, from_to_key, did_info_json.encode('utf-8'))
print("\"{}\" -> Send authcrypted \"{} DID info\" to {}".format(to, to, _from))
print("\"{}\" -> Authdecrypted \"{} DID info\" from {}".format(_from, to, to))
sender_verkey, authdecrypted_did_info_json, authdecrypted_did_info = \
await auth_decrypt(from_wallet, from_to_key, authcrypted_did_info_json)
print("\"{}\" -> Authenticate {} by comparision of Verkeys".format(_from, to, ))
assert sender_verkey == await did.key_for_did(pool_handle, from_wallet, to_from_did)
print("\"{}\" -> Send Nym to Ledger for \"{} DID\" with {} Role".format(_from, to, role))
await send_nym(pool_handle, from_wallet, from_did, authdecrypted_did_info['did'],
authdecrypted_did_info['verkey'], role)
return to_did
async def send_nym(pool_handle, wallet_handle, _did, new_did, new_key, role):
nym_request = await ledger.build_nym_request(_did, new_did, new_key, None, role)
await ledger.sign_and_submit_request(pool_handle, wallet_handle, _did, nym_request)
async def send_schema(pool_handle, wallet_handle, _did, schema):
schema_request = await ledger.build_schema_request(_did, schema)
await ledger.sign_and_submit_request(pool_handle, wallet_handle, _did, schema_request)
async def send_cred_def(pool_handle, wallet_handle, _did, cred_def_json):
cred_def_request = await ledger.build_cred_def_request(_did, cred_def_json)
await ledger.sign_and_submit_request(pool_handle, wallet_handle, _did, cred_def_request)
async def get_schema(pool_handle, _did, schema_id):
get_schema_request = await ledger.build_get_schema_request(_did, schema_id)
get_schema_response = await ledger.submit_request(pool_handle, get_schema_request)
return await ledger.parse_get_schema_response(get_schema_response)
async def get_cred_def(pool_handle, _did, schema_id):
get_cred_def_request = await ledger.build_get_cred_def_request(_did, schema_id)
get_cred_def_response = await ledger.submit_request(pool_handle, get_cred_def_request)
return await ledger.parse_get_cred_def_response(get_cred_def_response)
async def get_credential_for_referent(search_handle, referent):
credentials = json.loads(
await anoncreds.prover_fetch_credentials_for_proof_req(search_handle, referent, 10))
return credentials[0]['cred_info']
async def prover_get_entities_from_ledger(pool_handle, _did, identifiers, actor):
schemas = {}
cred_defs = {}
rev_states = {}
for item in identifiers.values():
print("\"{}\" -> Get Schema from Ledger".format(actor))
(received_schema_id, received_schema) = await get_schema(pool_handle, _did, item['schema_id'])
schemas[received_schema_id] = json.loads(received_schema)
print("\"{}\" -> Get Credential Definition from Ledger".format(actor))
(received_cred_def_id, received_cred_def) = await get_cred_def(pool_handle, _did, item['cred_def_id'])
cred_defs[received_cred_def_id] = json.loads(received_cred_def)
if 'rev_reg_seq_no' in item:
pass # TODO Create Revocation States
return json.dumps(schemas), json.dumps(cred_defs), json.dumps(rev_states)
async def verifier_get_entities_from_ledger(pool_handle, _did, identifiers, actor):
schemas = {}
cred_defs = {}
rev_reg_defs = {}
rev_regs = {}
for item in identifiers:
print("\"{}\" -> Get Schema from Ledger".format(actor))
(received_schema_id, received_schema) = await get_schema(pool_handle, _did, item['schema_id'])
schemas[received_schema_id] = json.loads(received_schema)
print("\"{}\" -> Get Credential Definition from Ledger".format(actor))
(received_cred_def_id, received_cred_def) = await get_cred_def(pool_handle, _did, item['cred_def_id'])
cred_defs[received_cred_def_id] = json.loads(received_cred_def)
if 'rev_reg_seq_no' in item:
pass # TODO Get Revocation Definitions and Revocation Registries
return json.dumps(schemas), json.dumps(cred_defs), json.dumps(rev_reg_defs), json.dumps(rev_regs)
async def auth_decrypt(wallet_handle, key, message):
from_verkey, decrypted_message_json = await crypto.auth_decrypt(wallet_handle, key, message)
decrypted_message_json = decrypted_message_json.decode("utf-8")
decrypted_message = json.loads(decrypted_message_json)
return from_verkey, decrypted_message_json, decrypted_message
if __name__ == '__main__':
loop = asyncio.new_event_loop()
asyncio.set_event_loop(loop)
loop.run_until_complete(run())
time.sleep(1) # FIXME waiting for libindy thread complete
```
|
github_jupyter
|
import asyncio
import time
from indy import anoncreds, crypto, did, ledger, pool, wallet
import json
from typing import Optional
async def run():
print("Getting started -> started")
print("Open Pool Ledger")
# Set protocol version 2 to work with Indy Node 1.4
await pool.set_protocol_version(2)
pool_name = 'pool1'
pool_config = json.dumps({"genesis_txn": '/home/indy/sandbox/pool_transactions_genesis'})
await pool.create_pool_ledger_config(pool_name, pool_config)
pool_handle = await pool.open_pool_ledger(pool_name, None)
print("==============================")
print("=== Getting Trust Anchor credentials for Faber, Acme, Thrift and Government ==")
print("------------------------------")
print("\"Sovrin Steward\" -> Create wallet")
steward_wallet_config = json.dumps({"id": "sovrin_steward_wallet"})
steward_wallet_credentials = json.dumps({"key": "steward_wallet_key"})
try:
await wallet.create_wallet(steward_wallet_config, steward_wallet_credentials)
except IndyError as ex:
if ex.error_code == ErrorCode.WalletAlreadyExistsError:
pass
steward_wallet = await wallet.open_wallet(steward_wallet_config, steward_wallet_credentials)
print("\"Sovrin Steward\" -> Create and store in Wallet DID from seed")
steward_did_info = {'seed': '000000000000000000000000Steward1'}
(steward_did, steward_key) = await did.create_and_store_my_did(steward_wallet, json.dumps(steward_did_info))
print("==============================")
print("== Getting Trust Anchor credentials - Government Onboarding ==")
print("------------------------------")
government_wallet_config = json.dumps({"id": "government_wallet"})
government_wallet_credentials = json.dumps({"key": "government_wallet_key"})
government_wallet, steward_government_key, government_steward_did, government_steward_key, _ \
= await onboarding(pool_handle, "Sovrin Steward", steward_wallet, steward_did, "Government", None,
government_wallet_config, government_wallet_credentials)
print("==============================")
print("== Getting Trust Anchor credentials - Government getting Verinym ==")
print("------------------------------")
government_did = await get_verinym(pool_handle, "Sovrin Steward", steward_wallet, steward_did,
steward_government_key, "Government", government_wallet, government_steward_did,
government_steward_key, 'TRUST_ANCHOR')
print("==============================")
print("== Getting Trust Anchor credentials - Faber Onboarding ==")
print("------------------------------")
faber_wallet_config = json.dumps({"id": "faber_wallet"})
faber_wallet_credentials = json.dumps({"key": "faber_wallet_key"})
faber_wallet, steward_faber_key, faber_steward_did, faber_steward_key, _ = \
await onboarding(pool_handle, "Sovrin Steward", steward_wallet, steward_did, "Faber", None,
faber_wallet_config, faber_wallet_credentials)
print("==============================")
print("== Getting Trust Anchor credentials - Faber getting Verinym ==")
print("------------------------------")
faber_did = await get_verinym(pool_handle, "Sovrin Steward", steward_wallet, steward_did, steward_faber_key,
"Faber", faber_wallet, faber_steward_did, faber_steward_key, 'TRUST_ANCHOR')
print("==============================")
print("== Getting Trust Anchor credentials - Acme Onboarding ==")
print("------------------------------")
acme_wallet_config = json.dumps({"id": "acme_wallet"})
acme_wallet_credentials = json.dumps({"key": "acme_wallet_key"})
acme_wallet, steward_acme_key, acme_steward_did, acme_steward_key, _ = \
await onboarding(pool_handle, "Sovrin Steward", steward_wallet, steward_did, "Acme", None,
acme_wallet_config, acme_wallet_credentials)
print("==============================")
print("== Getting Trust Anchor credentials - Acme getting Verinym ==")
print("------------------------------")
acme_did = await get_verinym(pool_handle, "Sovrin Steward", steward_wallet, steward_did, steward_acme_key,
"Acme", acme_wallet, acme_steward_did, acme_steward_key, 'TRUST_ANCHOR')
print("==============================")
print("== Getting Trust Anchor credentials - Thrift Onboarding ==")
print("------------------------------")
thrift_wallet_config = json.dumps({"id": "thrift_wallet"})
thrift_wallet_credentials = json.dumps({"key": "thrift_wallet_key"})
thrift_wallet, steward_thrift_key, thrift_steward_did, thrift_steward_key, _ = \
await onboarding(pool_handle, "Sovrin Steward", steward_wallet, steward_did, "Thrift", None,
thrift_wallet_config, thrift_wallet_credentials)
print("==============================")
print("== Getting Trust Anchor credentials - Thrift getting Verinym ==")
print("------------------------------")
thrift_did = await get_verinym(pool_handle, "Sovrin Steward", steward_wallet, steward_did, steward_thrift_key,
"Thrift", thrift_wallet, thrift_steward_did, thrift_steward_key, 'TRUST_ANCHOR')
print("==============================")
print("=== Credential Schemas Setup ==")
print("------------------------------")
print("\"Government\" -> Create \"Job-Certificate\" Schema")
(job_certificate_schema_id, job_certificate_schema) = \
await anoncreds.issuer_create_schema(government_did, 'Job-Certificate', '0.2',
json.dumps(['first_name', 'last_name', 'salary', 'employee_status',
'experience']))
print("\"Government\" -> Send \"Job-Certificate\" Schema to Ledger")
await send_schema(pool_handle, government_wallet, government_did, job_certificate_schema)
print("\"Government\" -> Create \"Transcript\" Schema")
(transcript_schema_id, transcript_schema) = \
await anoncreds.issuer_create_schema(government_did, 'Transcript', '1.2',
json.dumps(['first_name', 'last_name', 'degree', 'status',
'year', 'average', 'ssn']))
print("\"Government\" -> Send \"Transcript\" Schema to Ledger")
await send_schema(pool_handle, government_wallet, government_did, transcript_schema)
print("==============================")
print("=== Faber Credential Definition Setup ==")
print("------------------------------")
print("\"Faber\" -> Get \"Transcript\" Schema from Ledger")
(_, transcript_schema) = await get_schema(pool_handle, faber_did, transcript_schema_id)
print("\"Faber\" -> Create and store in Wallet \"Faber Transcript\" Credential Definition")
(faber_transcript_cred_def_id, faber_transcript_cred_def_json) = \
await anoncreds.issuer_create_and_store_credential_def(faber_wallet, faber_did, transcript_schema,
'TAG1', 'CL', '{"support_revocation": false}')
print("\"Faber\" -> Send \"Faber Transcript\" Credential Definition to Ledger")
await send_cred_def(pool_handle, faber_wallet, faber_did, faber_transcript_cred_def_json)
print("==============================")
print("=== Acme Credential Definition Setup ==")
print("------------------------------")
print("\"Acme\" -> Get from Ledger \"Job-Certificate\" Schema")
(_, job_certificate_schema) = await get_schema(pool_handle, acme_did, job_certificate_schema_id)
print("\"Acme\" -> Create and store in Wallet \"Acme Job-Certificate\" Credential Definition")
(acme_job_certificate_cred_def_id, acme_job_certificate_cred_def_json) = \
await anoncreds.issuer_create_and_store_credential_def(acme_wallet, acme_did, job_certificate_schema,
'TAG1', 'CL', '{"support_revocation": false}')
print("\"Acme\" -> Send \"Acme Job-Certificate\" Credential Definition to Ledger")
await send_cred_def(pool_handle, acme_wallet, acme_did, acme_job_certificate_cred_def_json)
print("==============================")
print("=== Getting Transcript with Faber ==")
print("==============================")
print("== Getting Transcript with Faber - Onboarding ==")
print("------------------------------")
alice_wallet_config = json.dumps({"id": "alice_wallet"})
alice_wallet_credentials = json.dumps({"key": "alice_wallet_key"})
alice_wallet, faber_alice_key, alice_faber_did, alice_faber_key, faber_alice_connection_response \
= await onboarding(pool_handle, "Faber", faber_wallet, faber_did, "Alice", None,
alice_wallet_config, alice_wallet_credentials)
time.sleep(1)
print("==============================")
print("== Getting Transcript with Faber - Getting Transcript Credential ==")
print("------------------------------")
print("\"Faber\" -> Create \"Transcript\" Credential Offer for Alice")
transcript_cred_offer_json = \
await anoncreds.issuer_create_credential_offer(faber_wallet, faber_transcript_cred_def_id)
print("\"Faber\" -> Get key for Alice did")
alice_faber_verkey = await did.key_for_did(pool_handle, acme_wallet, faber_alice_connection_response['did'])
print("\"Faber\" -> Authcrypt \"Transcript\" Credential Offer for Alice")
authcrypted_transcript_cred_offer = await crypto.auth_crypt(faber_wallet, faber_alice_key, alice_faber_verkey,
transcript_cred_offer_json.encode('utf-8'))
print("\"Faber\" -> Send authcrypted \"Transcript\" Credential Offer to Alice")
print("\"Alice\" -> Authdecrypted \"Transcript\" Credential Offer from Faber")
faber_alice_verkey, authdecrypted_transcript_cred_offer_json, authdecrypted_transcript_cred_offer = \
await auth_decrypt(alice_wallet, alice_faber_key, authcrypted_transcript_cred_offer)
print("\"Alice\" -> Create and store \"Alice\" Master Secret in Wallet")
alice_master_secret_id = await anoncreds.prover_create_master_secret(alice_wallet, None)
print("\"Alice\" -> Get \"Faber Transcript\" Credential Definition from Ledger")
(faber_transcript_cred_def_id, faber_transcript_cred_def) = \
await get_cred_def(pool_handle, alice_faber_did, authdecrypted_transcript_cred_offer['cred_def_id'])
print("\"Alice\" -> Create \"Transcript\" Credential Request for Faber")
(transcript_cred_request_json, transcript_cred_request_metadata_json) = \
await anoncreds.prover_create_credential_req(alice_wallet, alice_faber_did,
authdecrypted_transcript_cred_offer_json,
faber_transcript_cred_def, alice_master_secret_id)
print("\"Alice\" -> Authcrypt \"Transcript\" Credential Request for Faber")
authcrypted_transcript_cred_request = await crypto.auth_crypt(alice_wallet, alice_faber_key, faber_alice_verkey,
transcript_cred_request_json.encode('utf-8'))
print("\"Alice\" -> Send authcrypted \"Transcript\" Credential Request to Faber")
print("\"Faber\" -> Authdecrypt \"Transcript\" Credential Request from Alice")
alice_faber_verkey, authdecrypted_transcript_cred_request_json, _ = \
await auth_decrypt(faber_wallet, faber_alice_key, authcrypted_transcript_cred_request)
print("\"Faber\" -> Create \"Transcript\" Credential for Alice")
transcript_cred_values = json.dumps({
"first_name": {"raw": "Alice", "encoded": "1139481716457488690172217916278103335"},
"last_name": {"raw": "Garcia", "encoded": "5321642780241790123587902456789123452"},
"degree": {"raw": "Bachelor of Science, Marketing", "encoded": "12434523576212321"},
"status": {"raw": "graduated", "encoded": "2213454313412354"},
"ssn": {"raw": "123-45-6789", "encoded": "3124141231422543541"},
"year": {"raw": "2015", "encoded": "2015"},
"average": {"raw": "5", "encoded": "5"}
})
transcript_cred_json, _, _ = \
await anoncreds.issuer_create_credential(faber_wallet, transcript_cred_offer_json,
authdecrypted_transcript_cred_request_json,
transcript_cred_values, None, None)
print("\"Faber\" -> Authcrypt \"Transcript\" Credential for Alice")
authcrypted_transcript_cred_json = await crypto.auth_crypt(faber_wallet, faber_alice_key, alice_faber_verkey,
transcript_cred_json.encode('utf-8'))
print("\"Faber\" -> Send authcrypted \"Transcript\" Credential to Alice")
print("\"Alice\" -> Authdecrypted \"Transcript\" Credential from Faber")
_, authdecrypted_transcript_cred_json, _ = \
await auth_decrypt(alice_wallet, alice_faber_key, authcrypted_transcript_cred_json)
print("\"Alice\" -> Store \"Transcript\" Credential from Faber")
await anoncreds.prover_store_credential(alice_wallet, None, transcript_cred_request_metadata_json,
authdecrypted_transcript_cred_json, faber_transcript_cred_def, None)
print("==============================")
print("=== Apply for the job with Acme ==")
print("==============================")
print("== Apply for the job with Acme - Onboarding ==")
print("------------------------------")
alice_wallet, acme_alice_key, alice_acme_did, alice_acme_key, acme_alice_connection_response = \
await onboarding(pool_handle, "Acme", acme_wallet, acme_did, "Alice", alice_wallet,
alice_wallet_config, alice_wallet_credentials)
print("==============================")
print("== Apply for the job with Acme - Transcript proving ==")
print("------------------------------")
print("\"Acme\" -> Create \"Job-Application\" Proof Request")
job_application_proof_request_json = json.dumps({
'nonce': '1432422343242122312411212',
'name': 'Job-Application',
'version': '0.1',
'requested_attributes': {
'attr1_referent': {
'name': 'first_name'
},
'attr2_referent': {
'name': 'last_name'
},
'attr3_referent': {
'name': 'degree',
'restrictions': [{'cred_def_id': faber_transcript_cred_def_id}]
},
'attr4_referent': {
'name': 'status',
'restrictions': [{'cred_def_id': faber_transcript_cred_def_id}]
},
'attr5_referent': {
'name': 'ssn',
'restrictions': [{'cred_def_id': faber_transcript_cred_def_id}]
},
'attr6_referent': {
'name': 'phone_number'
}
},
'requested_predicates': {
'predicate1_referent': {
'name': 'average',
'p_type': '>=',
'p_value': 4,
'restrictions': [{'cred_def_id': faber_transcript_cred_def_id}]
}
}
})
print("\"Acme\" -> Get key for Alice did")
alice_acme_verkey = await did.key_for_did(pool_handle, acme_wallet, acme_alice_connection_response['did'])
print("\"Acme\" -> Authcrypt \"Job-Application\" Proof Request for Alice")
authcrypted_job_application_proof_request_json = \
await crypto.auth_crypt(acme_wallet, acme_alice_key, alice_acme_verkey,
job_application_proof_request_json.encode('utf-8'))
print("\"Acme\" -> Send authcrypted \"Job-Application\" Proof Request to Alice")
print("\"Alice\" -> Authdecrypt \"Job-Application\" Proof Request from Acme")
acme_alice_verkey, authdecrypted_job_application_proof_request_json, _ = \
await auth_decrypt(alice_wallet, alice_acme_key, authcrypted_job_application_proof_request_json)
print("\"Alice\" -> Get credentials for \"Job-Application\" Proof Request")
search_for_job_application_proof_request = \
await anoncreds.prover_search_credentials_for_proof_req(alice_wallet,
authdecrypted_job_application_proof_request_json, None)
cred_for_attr1 = await get_credential_for_referent(search_for_job_application_proof_request, 'attr1_referent')
cred_for_attr2 = await get_credential_for_referent(search_for_job_application_proof_request, 'attr2_referent')
cred_for_attr3 = await get_credential_for_referent(search_for_job_application_proof_request, 'attr3_referent')
cred_for_attr4 = await get_credential_for_referent(search_for_job_application_proof_request, 'attr4_referent')
cred_for_attr5 = await get_credential_for_referent(search_for_job_application_proof_request, 'attr5_referent')
cred_for_predicate1 = \
await get_credential_for_referent(search_for_job_application_proof_request, 'predicate1_referent')
await anoncreds.prover_close_credentials_search_for_proof_req(search_for_job_application_proof_request)
creds_for_job_application_proof = {cred_for_attr1['referent']: cred_for_attr1,
cred_for_attr2['referent']: cred_for_attr2,
cred_for_attr3['referent']: cred_for_attr3,
cred_for_attr4['referent']: cred_for_attr4,
cred_for_attr5['referent']: cred_for_attr5,
cred_for_predicate1['referent']: cred_for_predicate1}
schemas_json, cred_defs_json, revoc_states_json = \
await prover_get_entities_from_ledger(pool_handle, alice_faber_did, creds_for_job_application_proof, 'Alice')
print("\"Alice\" -> Create \"Job-Application\" Proof")
job_application_requested_creds_json = json.dumps({
'self_attested_attributes': {
'attr1_referent': 'Alice',
'attr2_referent': 'Garcia',
'attr6_referent': '123-45-6789'
},
'requested_attributes': {
'attr3_referent': {'cred_id': cred_for_attr3['referent'], 'revealed': True},
'attr4_referent': {'cred_id': cred_for_attr4['referent'], 'revealed': True},
'attr5_referent': {'cred_id': cred_for_attr5['referent'], 'revealed': True},
},
'requested_predicates': {'predicate1_referent': {'cred_id': cred_for_predicate1['referent']}}
})
job_application_proof_json = \
await anoncreds.prover_create_proof(alice_wallet, authdecrypted_job_application_proof_request_json,
job_application_requested_creds_json, alice_master_secret_id,
schemas_json, cred_defs_json, revoc_states_json)
print("\"Alice\" -> Authcrypt \"Job-Application\" Proof for Acme")
authcrypted_job_application_proof_json = await crypto.auth_crypt(alice_wallet, alice_acme_key, acme_alice_verkey,
job_application_proof_json.encode('utf-8'))
print("\"Alice\" -> Send authcrypted \"Job-Application\" Proof to Acme")
print("\"Acme\" -> Authdecrypted \"Job-Application\" Proof from Alice")
_, decrypted_job_application_proof_json, decrypted_job_application_proof = \
await auth_decrypt(acme_wallet, acme_alice_key, authcrypted_job_application_proof_json)
schemas_json, cred_defs_json, revoc_ref_defs_json, revoc_regs_json = \
await verifier_get_entities_from_ledger(pool_handle, acme_did,
decrypted_job_application_proof['identifiers'], 'Acme')
print("\"Acme\" -> Verify \"Job-Application\" Proof from Alice")
assert 'Bachelor of Science, Marketing' == \
decrypted_job_application_proof['requested_proof']['revealed_attrs']['attr3_referent']['raw']
assert 'graduated' == \
decrypted_job_application_proof['requested_proof']['revealed_attrs']['attr4_referent']['raw']
assert '123-45-6789' == \
decrypted_job_application_proof['requested_proof']['revealed_attrs']['attr5_referent']['raw']
assert 'Alice' == decrypted_job_application_proof['requested_proof']['self_attested_attrs']['attr1_referent']
assert 'Garcia' == decrypted_job_application_proof['requested_proof']['self_attested_attrs']['attr2_referent']
assert '123-45-6789' == decrypted_job_application_proof['requested_proof']['self_attested_attrs']['attr6_referent']
assert await anoncreds.verifier_verify_proof(job_application_proof_request_json,
decrypted_job_application_proof_json,
schemas_json, cred_defs_json, revoc_ref_defs_json, revoc_regs_json)
print("==============================")
print("== Apply for the job with Acme - Getting Job-Certificate Credential ==")
print("------------------------------")
print("\"Acme\" -> Create \"Job-Certificate\" Credential Offer for Alice")
job_certificate_cred_offer_json = \
await anoncreds.issuer_create_credential_offer(acme_wallet, acme_job_certificate_cred_def_id)
print("\"Acme\" -> Get key for Alice did")
alice_acme_verkey = await did.key_for_did(pool_handle, acme_wallet, acme_alice_connection_response['did'])
print("\"Acme\" -> Authcrypt \"Job-Certificate\" Credential Offer for Alice")
authcrypted_job_certificate_cred_offer = await crypto.auth_crypt(acme_wallet, acme_alice_key, alice_acme_verkey,
job_certificate_cred_offer_json.encode('utf-8'))
print("\"Acme\" -> Send authcrypted \"Job-Certificate\" Credential Offer to Alice")
print("\"Alice\" -> Authdecrypted \"Job-Certificate\" Credential Offer from Acme")
acme_alice_verkey, authdecrypted_job_certificate_cred_offer_json, authdecrypted_job_certificate_cred_offer = \
await auth_decrypt(alice_wallet, alice_acme_key, authcrypted_job_certificate_cred_offer)
print("\"Alice\" -> Get \"Acme Job-Certificate\" Credential Definition from Ledger")
(_, acme_job_certificate_cred_def) = \
await get_cred_def(pool_handle, alice_acme_did, authdecrypted_job_certificate_cred_offer['cred_def_id'])
print("\"Alice\" -> Create and store in Wallet \"Job-Certificate\" Credential Request for Acme")
(job_certificate_cred_request_json, job_certificate_cred_request_metadata_json) = \
await anoncreds.prover_create_credential_req(alice_wallet, alice_acme_did,
authdecrypted_job_certificate_cred_offer_json,
acme_job_certificate_cred_def, alice_master_secret_id)
print("\"Alice\" -> Authcrypt \"Job-Certificate\" Credential Request for Acme")
authcrypted_job_certificate_cred_request_json = \
await crypto.auth_crypt(alice_wallet, alice_acme_key, acme_alice_verkey,
job_certificate_cred_request_json.encode('utf-8'))
print("\"Alice\" -> Send authcrypted \"Job-Certificate\" Credential Request to Acme")
print("\"Acme\" -> Authdecrypt \"Job-Certificate\" Credential Request from Alice")
alice_acme_verkey, authdecrypted_job_certificate_cred_request_json, _ = \
await auth_decrypt(acme_wallet, acme_alice_key, authcrypted_job_certificate_cred_request_json)
print("\"Acme\" -> Create \"Job-Certificate\" Credential for Alice")
alice_job_certificate_cred_values_json = json.dumps({
"first_name": {"raw": "Alice", "encoded": "245712572474217942457235975012103335"},
"last_name": {"raw": "Garcia", "encoded": "312643218496194691632153761283356127"},
"employee_status": {"raw": "Permanent", "encoded": "2143135425425143112321314321"},
"salary": {"raw": "2400", "encoded": "2400"},
"experience": {"raw": "10", "encoded": "10"}
})
job_certificate_cred_json, _, _ = \
await anoncreds.issuer_create_credential(acme_wallet, job_certificate_cred_offer_json,
authdecrypted_job_certificate_cred_request_json,
alice_job_certificate_cred_values_json, None, None)
print("\"Acme\" -> Authcrypt \"Job-Certificate\" Credential for Alice")
authcrypted_job_certificate_cred_json = \
await crypto.auth_crypt(acme_wallet, acme_alice_key, alice_acme_verkey,
job_certificate_cred_json.encode('utf-8'))
print("\"Acme\" -> Send authcrypted \"Job-Certificate\" Credential to Alice")
print("\"Alice\" -> Authdecrypted \"Job-Certificate\" Credential from Acme")
_, authdecrypted_job_certificate_cred_json, _ = \
await auth_decrypt(alice_wallet, alice_acme_key, authcrypted_job_certificate_cred_json)
print("\"Alice\" -> Store \"Job-Certificate\" Credential")
await anoncreds.prover_store_credential(alice_wallet, None, job_certificate_cred_request_metadata_json,
authdecrypted_job_certificate_cred_json,
acme_job_certificate_cred_def_json, None)
print("==============================")
print("=== Apply for the loan with Thrift ==")
print("==============================")
print("== Apply for the loan with Thrift - Onboarding ==")
print("------------------------------")
_, thrift_alice_key, alice_thrift_did, alice_thrift_key, \
thrift_alice_connection_response = await onboarding(pool_handle, "Thrift", thrift_wallet, thrift_did, "Alice",
alice_wallet, alice_wallet_config, alice_wallet_credentials)
print("==============================")
print("== Apply for the loan with Thrift - Job-Certificate proving ==")
print("------------------------------")
print("\"Thrift\" -> Create \"Loan-Application-Basic\" Proof Request")
apply_loan_proof_request_json = json.dumps({
'nonce': '123432421212',
'name': 'Loan-Application-Basic',
'version': '0.1',
'requested_attributes': {
'attr1_referent': {
'name': 'employee_status',
'restrictions': [{'cred_def_id': acme_job_certificate_cred_def_id}]
}
},
'requested_predicates': {
'predicate1_referent': {
'name': 'salary',
'p_type': '>=',
'p_value': 2000,
'restrictions': [{'cred_def_id': acme_job_certificate_cred_def_id}]
},
'predicate2_referent': {
'name': 'experience',
'p_type': '>=',
'p_value': 1,
'restrictions': [{'cred_def_id': acme_job_certificate_cred_def_id}]
}
}
})
print("\"Thrift\" -> Get key for Alice did")
alice_thrift_verkey = await did.key_for_did(pool_handle, thrift_wallet, thrift_alice_connection_response['did'])
print("\"Thrift\" -> Authcrypt \"Loan-Application-Basic\" Proof Request for Alice")
authcrypted_apply_loan_proof_request_json = \
await crypto.auth_crypt(thrift_wallet, thrift_alice_key, alice_thrift_verkey,
apply_loan_proof_request_json.encode('utf-8'))
print("\"Thrift\" -> Send authcrypted \"Loan-Application-Basic\" Proof Request to Alice")
print("\"Alice\" -> Authdecrypt \"Loan-Application-Basic\" Proof Request from Thrift")
thrift_alice_verkey, authdecrypted_apply_loan_proof_request_json, _ = \
await auth_decrypt(alice_wallet, alice_thrift_key, authcrypted_apply_loan_proof_request_json)
print("\"Alice\" -> Get credentials for \"Loan-Application-Basic\" Proof Request")
search_for_apply_loan_proof_request = \
await anoncreds.prover_search_credentials_for_proof_req(alice_wallet,
authdecrypted_apply_loan_proof_request_json, None)
cred_for_attr1 = await get_credential_for_referent(search_for_apply_loan_proof_request, 'attr1_referent')
cred_for_predicate1 = await get_credential_for_referent(search_for_apply_loan_proof_request, 'predicate1_referent')
cred_for_predicate2 = await get_credential_for_referent(search_for_apply_loan_proof_request, 'predicate2_referent')
await anoncreds.prover_close_credentials_search_for_proof_req(search_for_apply_loan_proof_request)
creds_for_apply_loan_proof = {cred_for_attr1['referent']: cred_for_attr1,
cred_for_predicate1['referent']: cred_for_predicate1,
cred_for_predicate2['referent']: cred_for_predicate2}
schemas_json, cred_defs_json, revoc_states_json = \
await prover_get_entities_from_ledger(pool_handle, alice_thrift_did, creds_for_apply_loan_proof, 'Alice')
print("\"Alice\" -> Create \"Loan-Application-Basic\" Proof")
apply_loan_requested_creds_json = json.dumps({
'self_attested_attributes': {},
'requested_attributes': {
'attr1_referent': {'cred_id': cred_for_attr1['referent'], 'revealed': True}
},
'requested_predicates': {
'predicate1_referent': {'cred_id': cred_for_predicate1['referent']},
'predicate2_referent': {'cred_id': cred_for_predicate2['referent']}
}
})
alice_apply_loan_proof_json = \
await anoncreds.prover_create_proof(alice_wallet, authdecrypted_apply_loan_proof_request_json,
apply_loan_requested_creds_json, alice_master_secret_id, schemas_json,
cred_defs_json, revoc_states_json)
print("\"Alice\" -> Authcrypt \"Loan-Application-Basic\" Proof for Thrift")
authcrypted_alice_apply_loan_proof_json = \
await crypto.auth_crypt(alice_wallet, alice_thrift_key, thrift_alice_verkey,
alice_apply_loan_proof_json.encode('utf-8'))
print("\"Alice\" -> Send authcrypted \"Loan-Application-Basic\" Proof to Thrift")
print("\"Thrift\" -> Authdecrypted \"Loan-Application-Basic\" Proof from Alice")
_, authdecrypted_alice_apply_loan_proof_json, authdecrypted_alice_apply_loan_proof = \
await auth_decrypt(thrift_wallet, thrift_alice_key, authcrypted_alice_apply_loan_proof_json)
print("\"Thrift\" -> Get Schemas, Credential Definitions and Revocation Registries from Ledger"
" required for Proof verifying")
schemas_json, cred_defs_json, revoc_defs_json, revoc_regs_json = \
await verifier_get_entities_from_ledger(pool_handle, thrift_did,
authdecrypted_alice_apply_loan_proof['identifiers'], 'Thrift')
print("\"Thrift\" -> Verify \"Loan-Application-Basic\" Proof from Alice")
assert 'Permanent' == \
authdecrypted_alice_apply_loan_proof['requested_proof']['revealed_attrs']['attr1_referent']['raw']
assert await anoncreds.verifier_verify_proof(apply_loan_proof_request_json,
authdecrypted_alice_apply_loan_proof_json,
schemas_json, cred_defs_json, revoc_defs_json, revoc_regs_json)
print("==============================")
print("==============================")
print("== Apply for the loan with Thrift - Transcript and Job-Certificate proving ==")
print("------------------------------")
print("\"Thrift\" -> Create \"Loan-Application-KYC\" Proof Request")
apply_loan_kyc_proof_request_json = json.dumps({
'nonce': '123432421212',
'name': 'Loan-Application-KYC',
'version': '0.1',
'requested_attributes': {
'attr1_referent': {'name': 'first_name'},
'attr2_referent': {'name': 'last_name'},
'attr3_referent': {'name': 'ssn'}
},
'requested_predicates': {}
})
print("\"Thrift\" -> Get key for Alice did")
alice_thrift_verkey = await did.key_for_did(pool_handle, thrift_wallet, thrift_alice_connection_response['did'])
print("\"Thrift\" -> Authcrypt \"Loan-Application-KYC\" Proof Request for Alice")
authcrypted_apply_loan_kyc_proof_request_json = \
await crypto.auth_crypt(thrift_wallet, thrift_alice_key, alice_thrift_verkey,
apply_loan_kyc_proof_request_json.encode('utf-8'))
print("\"Thrift\" -> Send authcrypted \"Loan-Application-KYC\" Proof Request to Alice")
print("\"Alice\" -> Authdecrypt \"Loan-Application-KYC\" Proof Request from Thrift")
thrift_alice_verkey, authdecrypted_apply_loan_kyc_proof_request_json, _ = \
await auth_decrypt(alice_wallet, alice_thrift_key, authcrypted_apply_loan_kyc_proof_request_json)
print("\"Alice\" -> Get credentials for \"Loan-Application-KYC\" Proof Request")
search_for_apply_loan_kyc_proof_request = \
await anoncreds.prover_search_credentials_for_proof_req(alice_wallet,
authdecrypted_apply_loan_kyc_proof_request_json, None)
cred_for_attr1 = await get_credential_for_referent(search_for_apply_loan_kyc_proof_request, 'attr1_referent')
cred_for_attr2 = await get_credential_for_referent(search_for_apply_loan_kyc_proof_request, 'attr2_referent')
cred_for_attr3 = await get_credential_for_referent(search_for_apply_loan_kyc_proof_request, 'attr3_referent')
await anoncreds.prover_close_credentials_search_for_proof_req(search_for_apply_loan_kyc_proof_request)
creds_for_apply_loan_kyc_proof = {cred_for_attr1['referent']: cred_for_attr1,
cred_for_attr2['referent']: cred_for_attr2,
cred_for_attr3['referent']: cred_for_attr3}
schemas_json, cred_defs_json, revoc_states_json = \
await prover_get_entities_from_ledger(pool_handle, alice_thrift_did, creds_for_apply_loan_kyc_proof, 'Alice')
print("\"Alice\" -> Create \"Loan-Application-KYC\" Proof")
apply_loan_kyc_requested_creds_json = json.dumps({
'self_attested_attributes': {},
'requested_attributes': {
'attr1_referent': {'cred_id': cred_for_attr1['referent'], 'revealed': True},
'attr2_referent': {'cred_id': cred_for_attr2['referent'], 'revealed': True},
'attr3_referent': {'cred_id': cred_for_attr3['referent'], 'revealed': True}
},
'requested_predicates': {}
})
alice_apply_loan_kyc_proof_json = \
await anoncreds.prover_create_proof(alice_wallet, authdecrypted_apply_loan_kyc_proof_request_json,
apply_loan_kyc_requested_creds_json, alice_master_secret_id,
schemas_json, cred_defs_json, revoc_states_json)
print("\"Alice\" -> Authcrypt \"Loan-Application-KYC\" Proof for Thrift")
authcrypted_alice_apply_loan_kyc_proof_json = \
await crypto.auth_crypt(alice_wallet, alice_thrift_key, thrift_alice_verkey,
alice_apply_loan_kyc_proof_json.encode('utf-8'))
print("\"Alice\" -> Send authcrypted \"Loan-Application-KYC\" Proof to Thrift")
print("\"Thrift\" -> Authdecrypted \"Loan-Application-KYC\" Proof from Alice")
_, authdecrypted_alice_apply_loan_kyc_proof_json, authdecrypted_alice_apply_loan_kyc_proof = \
await auth_decrypt(thrift_wallet, thrift_alice_key, authcrypted_alice_apply_loan_kyc_proof_json)
print("\"Thrift\" -> Get Schemas, Credential Definitions and Revocation Registries from Ledger"
" required for Proof verifying")
schemas_json, cred_defs_json, revoc_defs_json, revoc_regs_json = \
await verifier_get_entities_from_ledger(pool_handle, thrift_did,
authdecrypted_alice_apply_loan_kyc_proof['identifiers'], 'Thrift')
print("\"Thrift\" -> Verify \"Loan-Application-KYC\" Proof from Alice")
assert 'Alice' == \
authdecrypted_alice_apply_loan_kyc_proof['requested_proof']['revealed_attrs']['attr1_referent']['raw']
assert 'Garcia' == \
authdecrypted_alice_apply_loan_kyc_proof['requested_proof']['revealed_attrs']['attr2_referent']['raw']
assert '123-45-6789' == \
authdecrypted_alice_apply_loan_kyc_proof['requested_proof']['revealed_attrs']['attr3_referent']['raw']
assert await anoncreds.verifier_verify_proof(apply_loan_kyc_proof_request_json,
authdecrypted_alice_apply_loan_kyc_proof_json,
schemas_json, cred_defs_json, revoc_defs_json, revoc_regs_json)
print("==============================")
print(" \"Sovrin Steward\" -> Close and Delete wallet")
await wallet.close_wallet(steward_wallet)
await wallet.delete_wallet(steward_wallet_config, steward_wallet_credentials)
print("\"Government\" -> Close and Delete wallet")
await wallet.close_wallet(government_wallet)
await wallet.delete_wallet(government_wallet_config, government_wallet_credentials)
print("\"Faber\" -> Close and Delete wallet")
await wallet.close_wallet(faber_wallet)
await wallet.delete_wallet(faber_wallet_config, faber_wallet_credentials)
print("\"Acme\" -> Close and Delete wallet")
await wallet.close_wallet(acme_wallet)
await wallet.delete_wallet(acme_wallet_config, acme_wallet_credentials)
print("\"Thrift\" -> Close and Delete wallet")
await wallet.close_wallet(thrift_wallet)
await wallet.delete_wallet(thrift_wallet_config, thrift_wallet_credentials)
print("\"Alice\" -> Close and Delete wallet")
await wallet.close_wallet(alice_wallet)
await wallet.delete_wallet(alice_wallet_config, alice_wallet_credentials)
print("Close and Delete pool")
await pool.close_pool_ledger(pool_handle)
await pool.delete_pool_ledger_config(pool_name)
print("Getting started -> done")
async def onboarding(pool_handle, _from, from_wallet, from_did, to,
to_wallet: Optional[str], to_wallet_config: str, to_wallet_credentials: str):
print("\"{}\" -> Create and store in Wallet \"{} {}\" DID".format(_from, _from, to))
(from_to_did, from_to_key) = await did.create_and_store_my_did(from_wallet, "{}")
print("\"{}\" -> Send Nym to Ledger for \"{} {}\" DID".format(_from, _from, to))
await send_nym(pool_handle, from_wallet, from_did, from_to_did, from_to_key, None)
print("\"{}\" -> Send connection request to {} with \"{} {}\" DID and nonce".format(_from, to, _from, to))
connection_request = {
'did': from_to_did,
'nonce': 123456789
}
if not to_wallet:
print("\"{}\" -> Create wallet".format(to))
try:
await wallet.create_wallet(to_wallet_config, to_wallet_credentials)
except IndyError as ex:
if ex.error_code == ErrorCode.PoolLedgerConfigAlreadyExistsError:
pass
to_wallet = await wallet.open_wallet(to_wallet_config, to_wallet_credentials)
print("\"{}\" -> Create and store in Wallet \"{} {}\" DID".format(to, to, _from))
(to_from_did, to_from_key) = await did.create_and_store_my_did(to_wallet, "{}")
print("\"{}\" -> Get key for did from \"{}\" connection request".format(to, _from))
from_to_verkey = await did.key_for_did(pool_handle, to_wallet, connection_request['did'])
print("\"{}\" -> Anoncrypt connection response for \"{}\" with \"{} {}\" DID, verkey and nonce"
.format(to, _from, to, _from))
connection_response = json.dumps({
'did': to_from_did,
'verkey': to_from_key,
'nonce': connection_request['nonce']
})
anoncrypted_connection_response = await crypto.anon_crypt(from_to_verkey, connection_response.encode('utf-8'))
print("\"{}\" -> Send anoncrypted connection response to \"{}\"".format(to, _from))
print("\"{}\" -> Anondecrypt connection response from \"{}\"".format(_from, to))
decrypted_connection_response = \
json.loads((await crypto.anon_decrypt(from_wallet, from_to_key,
anoncrypted_connection_response)).decode("utf-8"))
print("\"{}\" -> Authenticates \"{}\" by comparision of Nonce".format(_from, to))
assert connection_request['nonce'] == decrypted_connection_response['nonce']
print("\"{}\" -> Send Nym to Ledger for \"{} {}\" DID".format(_from, to, _from))
await send_nym(pool_handle, from_wallet, from_did, to_from_did, to_from_key, None)
return to_wallet, from_to_key, to_from_did, to_from_key, decrypted_connection_response
async def get_verinym(pool_handle, _from, from_wallet, from_did, from_to_key,
to, to_wallet, to_from_did, to_from_key, role):
print("\"{}\" -> Create and store in Wallet \"{}\" new DID".format(to, to))
(to_did, to_key) = await did.create_and_store_my_did(to_wallet, "{}")
print("\"{}\" -> Authcrypt \"{} DID info\" for \"{}\"".format(to, to, _from))
did_info_json = json.dumps({
'did': to_did,
'verkey': to_key
})
authcrypted_did_info_json = \
await crypto.auth_crypt(to_wallet, to_from_key, from_to_key, did_info_json.encode('utf-8'))
print("\"{}\" -> Send authcrypted \"{} DID info\" to {}".format(to, to, _from))
print("\"{}\" -> Authdecrypted \"{} DID info\" from {}".format(_from, to, to))
sender_verkey, authdecrypted_did_info_json, authdecrypted_did_info = \
await auth_decrypt(from_wallet, from_to_key, authcrypted_did_info_json)
print("\"{}\" -> Authenticate {} by comparision of Verkeys".format(_from, to, ))
assert sender_verkey == await did.key_for_did(pool_handle, from_wallet, to_from_did)
print("\"{}\" -> Send Nym to Ledger for \"{} DID\" with {} Role".format(_from, to, role))
await send_nym(pool_handle, from_wallet, from_did, authdecrypted_did_info['did'],
authdecrypted_did_info['verkey'], role)
return to_did
async def send_nym(pool_handle, wallet_handle, _did, new_did, new_key, role):
nym_request = await ledger.build_nym_request(_did, new_did, new_key, None, role)
await ledger.sign_and_submit_request(pool_handle, wallet_handle, _did, nym_request)
async def send_schema(pool_handle, wallet_handle, _did, schema):
schema_request = await ledger.build_schema_request(_did, schema)
await ledger.sign_and_submit_request(pool_handle, wallet_handle, _did, schema_request)
async def send_cred_def(pool_handle, wallet_handle, _did, cred_def_json):
cred_def_request = await ledger.build_cred_def_request(_did, cred_def_json)
await ledger.sign_and_submit_request(pool_handle, wallet_handle, _did, cred_def_request)
async def get_schema(pool_handle, _did, schema_id):
get_schema_request = await ledger.build_get_schema_request(_did, schema_id)
get_schema_response = await ledger.submit_request(pool_handle, get_schema_request)
return await ledger.parse_get_schema_response(get_schema_response)
async def get_cred_def(pool_handle, _did, schema_id):
get_cred_def_request = await ledger.build_get_cred_def_request(_did, schema_id)
get_cred_def_response = await ledger.submit_request(pool_handle, get_cred_def_request)
return await ledger.parse_get_cred_def_response(get_cred_def_response)
async def get_credential_for_referent(search_handle, referent):
credentials = json.loads(
await anoncreds.prover_fetch_credentials_for_proof_req(search_handle, referent, 10))
return credentials[0]['cred_info']
async def prover_get_entities_from_ledger(pool_handle, _did, identifiers, actor):
schemas = {}
cred_defs = {}
rev_states = {}
for item in identifiers.values():
print("\"{}\" -> Get Schema from Ledger".format(actor))
(received_schema_id, received_schema) = await get_schema(pool_handle, _did, item['schema_id'])
schemas[received_schema_id] = json.loads(received_schema)
print("\"{}\" -> Get Credential Definition from Ledger".format(actor))
(received_cred_def_id, received_cred_def) = await get_cred_def(pool_handle, _did, item['cred_def_id'])
cred_defs[received_cred_def_id] = json.loads(received_cred_def)
if 'rev_reg_seq_no' in item:
pass # TODO Create Revocation States
return json.dumps(schemas), json.dumps(cred_defs), json.dumps(rev_states)
async def verifier_get_entities_from_ledger(pool_handle, _did, identifiers, actor):
schemas = {}
cred_defs = {}
rev_reg_defs = {}
rev_regs = {}
for item in identifiers:
print("\"{}\" -> Get Schema from Ledger".format(actor))
(received_schema_id, received_schema) = await get_schema(pool_handle, _did, item['schema_id'])
schemas[received_schema_id] = json.loads(received_schema)
print("\"{}\" -> Get Credential Definition from Ledger".format(actor))
(received_cred_def_id, received_cred_def) = await get_cred_def(pool_handle, _did, item['cred_def_id'])
cred_defs[received_cred_def_id] = json.loads(received_cred_def)
if 'rev_reg_seq_no' in item:
pass # TODO Get Revocation Definitions and Revocation Registries
return json.dumps(schemas), json.dumps(cred_defs), json.dumps(rev_reg_defs), json.dumps(rev_regs)
async def auth_decrypt(wallet_handle, key, message):
from_verkey, decrypted_message_json = await crypto.auth_decrypt(wallet_handle, key, message)
decrypted_message_json = decrypted_message_json.decode("utf-8")
decrypted_message = json.loads(decrypted_message_json)
return from_verkey, decrypted_message_json, decrypted_message
if __name__ == '__main__':
loop = asyncio.new_event_loop()
asyncio.set_event_loop(loop)
loop.run_until_complete(run())
time.sleep(1) # FIXME waiting for libindy thread complete
| 0.296858 | 0.222236 |
# 5.5 卷积神经网络(LeNet)
```
import os
import time
import torch
from torch import nn, optim
import sys
sys.path.append("..")
import d2lzh_pytorch as d2l
os.environ["CUDA_VISIBLE_DEVICES"] = "0"
device = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
print(torch.__version__)
print(device)
```
## 5.5.1 LeNet模型
```
class LeNet(nn.Module):
def __init__(self):
super(LeNet, self).__init__()
self.conv = nn.Sequential(
nn.Conv2d(1, 6, 5), # in_channels, out_channels, kernel_size
nn.Sigmoid(),
nn.MaxPool2d(2, 2), # kernel_size, stride
nn.Conv2d(6, 16, 5),
nn.Sigmoid(),
nn.MaxPool2d(2, 2)
)
self.fc = nn.Sequential(
nn.Linear(16*5*5, 120),
nn.Sigmoid(),
nn.Linear(120, 84),
nn.Sigmoid(),
nn.Linear(84, 10)
)
def forward(self, img):
feature = self.conv(img)
output = self.fc(feature.view(img.shape[0], -1))
return output
net = LeNet()
print(net)
```
## 5.5.2 获取数据和训练模型
```
batch_size = 256
train_iter, test_iter = d2l.load_data_fashion_mnist(batch_size=batch_size)
# 本函数已保存在d2lzh_pytorch包中方便以后使用。该函数将被逐步改进:它的完整实现将在“图像增广”一节中描述
def evaluate_accuracy(data_iter, net, device=None):
if device is None and isinstance(net, torch.nn.Module):
# 如果没指定device就使用net的device
device = list(net.parameters())[0].device
acc_sum, n = 0.0, 0
with torch.no_grad():
for X, y in data_iter:
if isinstance(net, torch.nn.Module):
net.eval() # 评估模式, 这会关闭dropout
acc_sum += (net(X.to(device)).argmax(dim=1) == y.to(device)).float().sum().cpu().item()
net.train() # 改回训练模式
else: # 自定义的模型, 3.13节之后不会用到, 不考虑GPU
if('is_training' in net.__code__.co_varnames): # 如果有is_training这个参数
# 将is_training设置成False
acc_sum += (net(X, is_training=False).argmax(dim=1) == y).float().sum().item()
else:
acc_sum += (net(X).argmax(dim=1) == y).float().sum().item()
n += y.shape[0]
return acc_sum / n
# 本函数已保存在d2lzh_pytorch包中方便以后使用
def train_ch5(net, train_iter, test_iter, batch_size, optimizer, device, num_epochs):
net = net.to(device)
print("training on ", device)
loss = torch.nn.CrossEntropyLoss()
batch_count = 0
for epoch in range(num_epochs):
train_l_sum, train_acc_sum, n, start = 0.0, 0.0, 0, time.time()
for X, y in train_iter:
X = X.to(device)
y = y.to(device)
y_hat = net(X)
l = loss(y_hat, y)
optimizer.zero_grad()
l.backward()
optimizer.step()
train_l_sum += l.cpu().item()
train_acc_sum += (y_hat.argmax(dim=1) == y).sum().cpu().item()
n += y.shape[0]
batch_count += 1
test_acc = evaluate_accuracy(test_iter, net)
print('epoch %d, loss %.4f, train acc %.3f, test acc %.3f, time %.1f sec'
% (epoch + 1, train_l_sum / batch_count, train_acc_sum / n, test_acc, time.time() - start))
lr, num_epochs = 0.001, 5
optimizer = torch.optim.Adam(net.parameters(), lr=lr)
train_ch5(net, train_iter, test_iter, batch_size, optimizer, device, num_epochs)
```
|
github_jupyter
|
import os
import time
import torch
from torch import nn, optim
import sys
sys.path.append("..")
import d2lzh_pytorch as d2l
os.environ["CUDA_VISIBLE_DEVICES"] = "0"
device = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
print(torch.__version__)
print(device)
class LeNet(nn.Module):
def __init__(self):
super(LeNet, self).__init__()
self.conv = nn.Sequential(
nn.Conv2d(1, 6, 5), # in_channels, out_channels, kernel_size
nn.Sigmoid(),
nn.MaxPool2d(2, 2), # kernel_size, stride
nn.Conv2d(6, 16, 5),
nn.Sigmoid(),
nn.MaxPool2d(2, 2)
)
self.fc = nn.Sequential(
nn.Linear(16*5*5, 120),
nn.Sigmoid(),
nn.Linear(120, 84),
nn.Sigmoid(),
nn.Linear(84, 10)
)
def forward(self, img):
feature = self.conv(img)
output = self.fc(feature.view(img.shape[0], -1))
return output
net = LeNet()
print(net)
batch_size = 256
train_iter, test_iter = d2l.load_data_fashion_mnist(batch_size=batch_size)
# 本函数已保存在d2lzh_pytorch包中方便以后使用。该函数将被逐步改进:它的完整实现将在“图像增广”一节中描述
def evaluate_accuracy(data_iter, net, device=None):
if device is None and isinstance(net, torch.nn.Module):
# 如果没指定device就使用net的device
device = list(net.parameters())[0].device
acc_sum, n = 0.0, 0
with torch.no_grad():
for X, y in data_iter:
if isinstance(net, torch.nn.Module):
net.eval() # 评估模式, 这会关闭dropout
acc_sum += (net(X.to(device)).argmax(dim=1) == y.to(device)).float().sum().cpu().item()
net.train() # 改回训练模式
else: # 自定义的模型, 3.13节之后不会用到, 不考虑GPU
if('is_training' in net.__code__.co_varnames): # 如果有is_training这个参数
# 将is_training设置成False
acc_sum += (net(X, is_training=False).argmax(dim=1) == y).float().sum().item()
else:
acc_sum += (net(X).argmax(dim=1) == y).float().sum().item()
n += y.shape[0]
return acc_sum / n
# 本函数已保存在d2lzh_pytorch包中方便以后使用
def train_ch5(net, train_iter, test_iter, batch_size, optimizer, device, num_epochs):
net = net.to(device)
print("training on ", device)
loss = torch.nn.CrossEntropyLoss()
batch_count = 0
for epoch in range(num_epochs):
train_l_sum, train_acc_sum, n, start = 0.0, 0.0, 0, time.time()
for X, y in train_iter:
X = X.to(device)
y = y.to(device)
y_hat = net(X)
l = loss(y_hat, y)
optimizer.zero_grad()
l.backward()
optimizer.step()
train_l_sum += l.cpu().item()
train_acc_sum += (y_hat.argmax(dim=1) == y).sum().cpu().item()
n += y.shape[0]
batch_count += 1
test_acc = evaluate_accuracy(test_iter, net)
print('epoch %d, loss %.4f, train acc %.3f, test acc %.3f, time %.1f sec'
% (epoch + 1, train_l_sum / batch_count, train_acc_sum / n, test_acc, time.time() - start))
lr, num_epochs = 0.001, 5
optimizer = torch.optim.Adam(net.parameters(), lr=lr)
train_ch5(net, train_iter, test_iter, batch_size, optimizer, device, num_epochs)
| 0.63624 | 0.820793 |
```
import graphlab as gl
import tensorflow as tf
SF = gl.SFrame.read_csv('ISCX_Botnet-Testing.csv',verbose=False)
print "Done reading"
#SF.head(5)
#SF.column_names()
SF = SF[(SF['Source Port']!='')|(SF['Destination Port']!='')]
len(SF)
SF.save('ISCX_Botnet-Testing_PortsOnly.csv')
SF = gl.SFrame.read_csv('ISCX_Botnet-Testing_Ports_Only.csv',verbose=False)
print 'Done Reading'
#type(SF['tcp_Flags'][0])
#SF['tcp_Flags'].unique()
#SF['IP_Flags'].unique()
SF['tcp_Flags'] = SF['tcp_Flags'].apply(lambda x:int(x,16) if x!='' else 0)
len(SF[SF['tcp_Flags']==0])
len(SF[SF['Protocol']=='TCP'])
##Testing code for time comparison in iteration
"""import time
start = time.time()
for i in SF:
i['tcp_Flags']
print time.time()-start"""
len(SF)
len(SF[SF['tcp_Flags']!=0])
print len(SF[SF['Protocol']=='UDP']),len(SF[(SF['tcp_Flags']==0)&(SF['Protocol']=='UDP')])
print len(SF[(SF['tcp_Flags']==0)]),len(SF[(SF['tcp_Flags']==0)&(SF['Protocol']!='TCP')])
Protocols = sorted(SF['Protocol'].unique())
#print Protocols
```
## Some analysis for number of packets of each protocol type
```
"""lt = []
gt = []
for x in Protocols:
print x
temp = len(SF[SF['Protocol']==x])
if temp<1000:
lt.append(x)
else:
gt.append(x)"""
#print lt,"\n\n",gt
#print len(lt),len(gt)
SF[SF['Protocol']=='ICMP'].head(3)
len(SF[SF['Protocol']=='ICMP'])
```
IRC
"192.168.2.112",
"131.202.243.84",
"192.168.5.122",
"198.164.30.2",
"192.168.2.110",
"192.168.5.122",
"192.168.4.118",
"192.168.5.122",
"192.168.2.113",
"192.168.5.122",
"192.168.1.103",
"192.168.5.122",
"192.168.4.120",
"192.168.5.122",
"192.168.2.112",
"192.168.2.110",
"192.168.2.112",
"192.168.4.120",
"192.168.2.112",
"192.168.1.103",
"192.168.2.112",
"192.168.2.113",
"192.168.2.112",
"192.168.4.118",
"192.168.2.112",
"192.168.2.109",
"192.168.2.112",
"192.168.2.105",
"192.168.1.105",
"192.168.5.122",
```
iplist = [
"147.32.84.180",
"147.32.84.170",
"147.32.84.150",
"147.32.84.140",
"147.32.84.130",
"147.32.84.160",
"10.0.2.15",
"192.168.106.141",
"192.168.106.131",
"172.16.253.130",
"172.16.253.131",
"172.16.253.129",
"172.16.253.240",
"74.78.117.238",
"158.65.110.24",
"192.168.3.35",
"192.168.3.25",
"192.168.3.65",
"172.29.0.116",
"172.29.0.109",
"172.16.253.132",
"192.168.248.165",
"10.37.130.4"]
MasterBot = [ ("192.168.2.112", "131.202.243.84"), ("192.168.5.122", "198.164.30.2"), ("192.168.2.110", "192.168.5.122"),( "192.168.4.118", "192.168.5.122"), ("192.168.2.113", "192.168.5.122"), ("192.168.1.103", "192.168.5.122"), ("192.168.4.120", "192.168.5.122"), ("192.168.2.112", "192.168.2.110"), ("192.168.2.112", "192.168.4.120"), ("192.168.2.112", "192.168.1.103"), ("192.168.2.112", "192.168.2.113"), ("192.168.2.112", "192.168.4.118"), ("192.168.2.112", "192.168.2.109"), ("192.168.2.112", "192.168.2.105"), ("192.168.1.105", "192.168.5.122")]
iplist = gl.SArray(iplist)
iplist = iplist.unique()
iplist = list(iplist)
def fcheckIP(x):
if x['Source'] in iplist:
return 1
else:
if ((x['Source'],x['Destination']) in MasterBot ) or ((x['Destination'],x['Source']) in MasterBot ) :
return 1
else:
return 0
SF['isBot'] = SF[['Source','Destination']].apply(lambda x: fcheckIP(x))
SF[(SF['Source']=='192.168.2.112') & (SF['isBot']==1)]
```
Botnet Labelled packets and their percentage.
```
temp = len(SF[SF['isBot']==1])
print temp, (temp*1.0)/len(SF)*100
SF.head()
iplist = [
"192.168.2.112",
"131.202.243.84",
"192.168.5.122",
"198.164.30.2",
"192.168.2.110",
"192.168.5.122",
"192.168.4.118",
"192.168.5.122",
"192.168.2.113",
"192.168.5.122",
"192.168.1.103",
"192.168.5.122",
"192.168.4.120",
"192.168.5.122",
"192.168.2.112",
"192.168.2.110",
"192.168.2.112",
"192.168.4.120",
"192.168.2.112",
"192.168.1.103",
"192.168.2.112",
"192.168.2.113",
"192.168.2.112",
"192.168.4.118",
"192.168.2.112",
"192.168.2.109",
"192.168.2.112",
"192.168.2.105",
"192.168.1.105",
"192.168.5.122",
"147.32.84.180",
"147.32.84.170",
"147.32.84.150",
"147.32.84.140",
"147.32.84.130",
"147.32.84.160",
"10.0.2.15",
"192.168.106.141",
"192.168.106.131",
"172.16.253.130",
"172.16.253.131",
"172.16.253.129",
"172.16.253.240",
"74.78.117.238",
"158.65.110.24",
"192.168.3.35",
"192.168.3.25",
"192.168.3.65",
"172.29.0.116",
"172.29.0.109",
"172.16.253.132",
"192.168.248.165",
"10.37.130.4"]
iplist = gl.SArray(iplist)
iplist = iplist.unique()
iplist = list(iplist)
temp = SF[SF['Protocol']=='ICMP']
for x in iplist:
print x, " Destination : ",len(temp[temp['Destination']==x])," Source : ",len(temp[temp['Source']==x])
dest = temp.groupby('Destination',{
'Count':gl.aggregate.COUNT()
})
dest = dest.sort('Count',ascending=False)
dest.head()
src = temp.groupby('Source',{
'Count':gl.aggregate.COUNT()
})
src = src.sort('Count',ascending=False)
src.head()
len(SF[(SF['Source']=='10.0.0.254')&(SF['Protocol']!='ICMP')&(SF['Protocol']!='DNS')])
#sorted(iplist)
## To check whether No. field is primary key or not
print len(SF['No.'].unique()),len(SF)
```
# Sorting will help in identifying flow effectively
```
sorting_features = ['Source','Destination','Source Port','Destination Port','Protocol','Time']
#STD = SF[['No.','Time','Source','Destination','Source Port','Destination Port','Protocol','tcp_Flags']]
#STD = STD.sort(sorting_features)
#print 'Done sorting STD'
SF = SF.sort(sorting_features)
print 'Done sorting SF'
#STD.save('ISCX_Botnet-Testing_Ports_Only_Sorted_Flow.csv')
#print 'Done saving STD'
#STD = None
SF.save('ISCX_Botnet-Testing_Ports_Only_Sorted.csv')
print 'Done saving SF'
#STD = None
SF.head(3)
type(SF['Time'][0])
SF['tcp_Flags'].unique()
```
### Checking if No. is the unique primary key for data for
```
print len(SF),len(SF['No.'].unique())
```
## Back to flow identification Unidirectional
```
## function for comparing two different flows based on columns
def compare(x,y,columns):
try:
val = True
for column in columns:
if x[column]!=y[column]:
val = False
break;
return val
except KeyError:
print "Column does not exist in the data, check again!"
except:
print "Some unknown error"
##Code for logic of flow identification
import pickle
FlowCols = ['Source','Destination','Source Port','Destination Port','Protocol']
SF['FlowNo.'] = None
FlowNo = 0 ##Unique Flow Number for each flow
#FFlow = []
prev = None
Flow = [] ##Stores all flows in form of list of dictionary
#cFlow = [] ##Store the current flow (all details)
count = 0
fc = 0
startTime = None ##Start Time of each flow to implement timeout
for x in SF:
if count%500000 == 0:
#fName = 'Flow'+str(fc)+'.pkl'
#print 'Saving : ',fName
#pickle.dump(Flow,open(fName,'w'))
#print 'Saved'
print 'Running '+str(count)+' Done !'
#c = fc + 1
#Flow = []
count = count+1
if prev is None:
startTime = x['Time']
Flow.append(FlowNo)
#cFlow.append(x['No.'])
prev = x
elif compare(x,prev,FlowCols):
fc=fc+1
if x['tcp_Flags']&1 or x['tcp_Flags']&4 or x['Time']-startTime>=120:
Flow.append(FlowNo)
prev = None
FlowNo = FlowNo + 1
#cFlow.append(x['No.'])
#FFlow.append(cFlow)
#cFlow = []
else:
#cFlow.append(x['No.'])
Flow.append(FlowNo)
prev = x
else:
FlowNo = FlowNo + 1
Flow.append(FlowNo)
#FFlow.append(cFlow)
#cFlow = []
#cFlow.append(x['No.'])
prev = x
startTime = x['Time']
print len(gl.SArray(Flow).unique())
```
## Flow Identification Bidirectional
```
def flow_id(x):
if x['Source']>x['Destination']:
return x['Source']+'-'+x['Destination']+'-'+str(x['Source Port'])+'-'+str(x['Destination Port'])+'-'+x['Protocol']
else:
return x['Destination']+'-'+x['Source']+'-'+str(x['Destination Port'])+'-'+str(x['Source Port'])+'-'+x['Protocol']
SF['UFid'] = SF.apply(lambda x:flow_id(x))
## function for comparing two different flows based on columns
def compareUF(x,y):
if x!=y:
return False
return True
##Code for logic of Bidirectional flow identification
import pickle
FlowNo = 0 ##Unique Flow Number for each flow
prev = None
Flow = [] ##Stores all flows in form of list of dictionary
#cFlow = [] ##Store the current flow (all details)
count = 0
fc = 0
startTime = None ##Start Time of each flow to implement timeout
SF = SF.sort(['UFid','Time'])
print 'Done Sorting'
for x in SF:
if count%500000 == 0:
print 'Running '+str(count)+' Done !'
count = count+1
if prev is None:
if startTime is None:
startTime = x['Time']
Flow.append(FlowNo)
prev = x['UFid']
elif compareUF(x['UFid'],prev):
if x['tcp_Flags']&1:
Flow.append(FlowNo)
prev = None
startTime = None
FlowNo = FlowNo + 1
elif x['Time']-startTime>=120:
FlowNo = FlowNo + 1
Flow.append(FlowNo)
prev = None
startTime = x['Time']
else:
Flow.append(FlowNo)
prev = x['UFid']
else:
FlowNo = FlowNo + 1
Flow.append(FlowNo)
prev = x['UFid']
startTime = x['Time']
print len(gl.SArray(Flow).unique())
SF['Flow'] = gl.SArray(Flow)
temp = SF.groupby('Flow',{
'Count':gl.aggregate.COUNT()
})
len(temp[temp['Count']>1])
temp = SF.groupby('Flow',{
'NumBots' : gl.aggregate.SUM('isBot')
})
NumBotFlows = len(temp[temp['NumBots']>1])
print NumBotFlows, NumBotFlows*1.0/len(SF['Flow'].unique())
len(Flow)
len(SF)
SF['FlowNo.'] = gl.SArray(Flow)
##Code for checking authenticity of flow logic
#STD[(STD['Source']=='0.0.0.0')&(STD['Destination']=='255.255.255.255')&(STD['Source Port']=='68')&(STD['Destination Port']=='67')].sort('Time')
## Code to check if in any flows there are some No.s which are in decreasing order (Indicative or Decrepancies)
## UPDATE: No. does not indicate same relation in time, so Data collected is right !
"""count = 0
for li in Flow:
for i in range(1,len(li)):
if li[i]<li[i-1]:
#print li
count = count+1
break;
print count"""
import pickle
pickle.dump(Flow,open('Flow.pkl','w'))
SF.save('ISCX_Botnet-Testing_Ports_Only_Sorted_Flow.csv')
SF.head(3)
## First Packet Length
FlowFeatures = ['Source','Destination','Source Port','Destination Port','Protocol']
FPL = SF.groupby(['FlowNo.'],{
'Time':gl.aggregate.MIN('Time')
})
print len(FPL)
FPL = FPL.join(SF,on =['FlowNo.','Time'])[['FlowNo.','Length']].unique()
FPL = FPL.groupby(['FlowNo.'],{
'Length':gl.aggregate.AVG('Length')
})
print len(FPL)
FPL.save('FirstPacketLength.csv')
```
## 18/10/2016
```
import graphlab as gl
import tensorflow as tf
SF = gl.SFrame.read_csv('ISCX_Botnet-Testing_Ports_Only_Sorted_Flow.csv',verbose=False)
## Number of packets per flow
temp = SF.groupby(['FlowNo.'],{
'NumPackets':gl.aggregate.COUNT()
})
print temp.head(3)
temp.save('NumPacketsPerFlow.csv')
## Number of bytes exchanged
temp = SF.groupby(['FlowNo.'],{
'BytesEx':gl.aggregate.SUM('Length')
})
temp.save('BytexExchangedPerFlow.csv')
temp.head(3)
## Standard deviation of packet length
temp = SF.groupby(['FlowNo.'],{
'StdDevLen':gl.aggregate.STDV('Length')
})
temp.save('StdDevLenPerFlow.csv')
temp.sort('StdDevLen')[-10:]
## Same length packet ratio
temp2 = SF.groupby(['FlowNo.'],{
'SameLenPktRatio':gl.aggregate.COUNT_DISTINCT('Length')
})
##temp from number of packets computation
temp = SF.groupby(['FlowNo.'],{
'NumPackets':gl.aggregate.COUNT()
})
temp = temp.join(temp2,on='FlowNo.')
temp['SameLenPktRatio'] = temp['SameLenPktRatio']*1.0/temp['NumPackets']
temp2 = None
temp = temp[['FlowNo.','SameLenPktRatio']]
temp.save('SameLenPktRatio.csv')
temp.head(3)
def tfn(x):
if 'udp' in x.split(':'):
return 1
return 0
SF['hasUDP'] = SF['Protocols in frame'].apply(lambda x:tfn(x))
test = SF[SF['hasUDP']==1]
test['colDiff'] = test['Length'] - test['udp_Length']
test[test['colDiff']==-1].head(3)
test[test['colDiff']<-20].head(3)
## Duration of flow
timeF = SF.groupby(['FlowNo.'],{
'startTime':gl.aggregate.MIN('Time'),
'endTime':gl.aggregate.MAX('Time')
})
timeF['Duration'] = timeF['endTime'] - timeF['startTime']
timeF[['FlowNo.','Duration']].save('DurationFlow.csv')
timeF = timeF[['FlowNo.','Duration']]
## Average packets per second
temp = gl.SFrame.read_csv('NumPacketsPerFlow.csv',verbose=False)
temp = temp.join(timeF,on=['FlowNo.'])
temp['AvgPktPerSec'] = temp.apply(lambda x:0.0 if x['Duration'] == 0.0 else x['NumPackets']*1.0/x['Duration'])
temp = temp[['FlowNo.','AvgPktPerSec']]
temp.save('AvgPktPerSecFlow.csv')
temp.sort('AvgPktPerSec')[-10:]
##Average Bits Per Second
temp = gl.SFrame.read_csv('BytexExchangedPerFlow.csv',verbose=False)
temp = temp.join(timeF,on=['FlowNo.'])
temp['BitsPerSec'] = temp.apply(lambda x:0.0 if x['Duration'] == 0.0 else x['BytesEx']*8.0/x['Duration'])
temp = temp[['FlowNo.','BitsPerSec']]
temp.save('BitsPerSecPerFlow.csv')
temp.sort('BitsPerSec')[-5:]
## Average Packet Lentgth
temp = SF.groupby(['FlowNo.'],{
'APL':gl.aggregate.AVG('Length')
})
temp.save('AveragePktLengthFlow.csv')
test = SF[SF['hasUDP']==1]
test['colDiff'] = test['Length'] - test['udp_Length']
len(test[test['colDiff']<0])
len(test)
## Number of Reconnects, sort FlowNo, SeqNo
def tfn(x):
if 'udp' in x.split(':') or 'tcp' in x.split(':'):
return 1
return 0
temp = list(SF['Protocols in frame'].apply(lambda x:tfn(x)))
len(temp)
sum(temp)
SF.head(1)
type(SF['TCP Segment Len'][0])
type(SF['udp_Length'][0])
len(SF[(SF['udp_Length'] == None)&(SF['TCP Segment Len'] == '')])
SF[(SF['udp_Length'] == None)&(SF['TCP Segment Len'] == '')]['Protocols in frame'].unique()
print len(SF[SF['Protocols in frame']=='eth:ethertype:ip:icmp:ip:tcp:http:urlencoded-form']),len(SF[SF['Protocols in frame']=='eth:ethertype:ip:icmp:ip:tcp']),len(SF[SF['Protocols in frame']=='eth:ethertype:ip:icmp:ip:tcp:http:data'])
## Inter arrival time
SF['IAT'] = 0
SF = SF.sort(['FlowNo.','Time'])
prev = None
prevT = None
li = []
for x in SF:
if prev is None or x['FlowNo.']!= prev:
li.append(0)
else:
li.append(x['Time']-prevT)
prev = x['FlowNo.']
prevT = x['Time']
SF['IAT'] = gl.SArray(li)
SF.save('ISCX_Botnet-Testing_Ports_Only_Sorted_Flow_IAT.csv')
SF.head(3)
#len(SF[(SF['udp_Length']>=8)&(SF['udp_Length']<=16)])
#print len(SF[SF['udp_Length']==8])
#print len(SF[SF['udp_Length']==16])
#SF[SF['Protocols in frame']=='eth:llc:ipx'].head(3)
#SF[SF['Protocols in frame']=='eth:ethertype:ipx'].head(3)
#print len(SF[SF['Protocols in frame']=='eth:ipx'])
#SF[SF['udp_Length']==0]
print len(SF[SF['Protocols in frame']=='eth:ipx'])
SF[SF['hasUDP']==1]['Protocols in frame'].unique()
```
# Is Null feature
### Number of TCP Null packets
```
len(SF[SF['Protocol']=='TCP'])
```
### Number of UDP NUll Packets
```
len(SF[SF['udp_Length']==8]), len(SF[SF['Protocol']=='UDP'])
SF[SF['TCP Segment Len']=='0']['Protocols in frame'].unique()
```
### Null Packets exchanged,
TCP -> tcp segment len =0,
UDP -> udp len = 8,
non tcp udp -> length - individual headers
```
tt = SF[(SF['TCP Segment Len']!='0')&(SF['udp_Length']!=8 )]
len(tt)
## Null Packets handling
def checkNull(x):
if(x['TCP Segment Len']=='0' or x['udp_Length']==8 ):
return 1
elif('ipx' in x['Protocols in frame'].split(':')):
l = x['Length'] - 30
if('eth' in x['Protocols in frame'].split(':')):
l = l - 14
if('ethtype' in x['Protocols in frame'].split(':')):
l = l - 2
if('llc' in x['Protocols in frame'].split(':')):
l = l - 8
if(l==0 or l==-1):
return 1
return 0
SF['isNull'] = SF.apply(lambda x:checkNull(x))
len(SF[SF['isNull']==1])
NPEx = SF.groupby(['FlowNo.'],{
'NPEx':gl.aggregate.SUM('isNull')
})
NPEx.save('NumberNullPacketsEXc.csv')
```
### Number of Reconnects - considering only TCP reconnects, using sequence number
```
type(SF['Sequence number'][0])
recon = SF[SF['Sequence number']!=''].groupby(['FlowNo.'],{
'total_seq_no.' : gl.aggregate.COUNT('Sequence number'),
'distinct_seq_no.' : gl.aggregate.COUNT_DISTINCT('Sequence number')
})
recon['reconnects'] = recon['total_seq_no.'] - recon['distinct_seq_no.']
recon.head()
recon[['FlowNo.','reconnects']].save('ReconnectsFlow.csv')
#SF[SF['FlowNo.']==79732]['Info']
```
A lot of these reconnects can be simple retransmissions - due to out of order/timeout etcb
```
## Ratio of incoming to outgoing packets
convo = SF.groupby(['FlowNo.'],{
'start_time': gl.aggregate.MIN('Time'),
'protocol':gl.aggregate.SELECT_ONE('Protocol'),
'src_ip': gl.aggregate.SELECT_ONE('Source'),
'dst_ip': gl.aggregate.SELECT_ONE('Destination'),
'src_port': gl.aggregate.SELECT_ONE('Source Port'),
'dst_port': gl.aggregate.SELECT_ONE('Destination Port')
})
convo.sort('start_time')
convo['rev_flow_no.'] = -1
convo.head()
```
for x in convo:
if x['rev_flow_no.']==-1:
for y in convo:
if y['rev_flow_no.']==-1 & (x['src_ip']==y['dst_ip']) & (x['src_port']==y['dst_port']) & (y['src_ip']==x['dst_ip']) &(y['src_port']==x['dst_port']) & (x['protocol']==y['protocol']) :
x['rev_flow_no.'] = y['FlowNo.']
y['rev_flow_no.'] = x['FlowNo.']
break
if y['start_time']-x['start_time'] > 100 :
break
```
temp1 = convo
temp1.rename({'src_ip':'dst_ip1', 'src_port':'dst_port1'})
temp1.rename({'dst_ip':'src_ip','dst_port':'src_port'})
temp1.rename({'dst_ip1':'dst_ip', 'dst_port1':'dst_port'})
temp1.rename({'start_time':'return_time'})
temp1.rename({'FlowNo.':'rev_flow'})
temp1 = temp1['src_ip','dst_ip','src_port','dst_port','protocol','return_time','rev_flow']
convo = SF.groupby(['FlowNo.'],{
'start_time': gl.aggregate.MIN('Time'),
'protocol':gl.aggregate.SELECT_ONE('Protocol'),
'src_ip': gl.aggregate.SELECT_ONE('Source'),
'dst_ip': gl.aggregate.SELECT_ONE('Destination'),
'src_port': gl.aggregate.SELECT_ONE('Source Port'),
'dst_port': gl.aggregate.SELECT_ONE('Destination Port')
})
convo.sort('start_time')
temp2 = temp1.join(convo,on=['src_ip','dst_ip','src_port','dst_port','protocol'])
temp2
convo
temp1[(temp1['src_ip']=='66.249.73.56') & (temp1['src_port']==52954)]
temp2['reply_time'] = temp2['return_time'] - temp2['start_time']
temp2.head()
temp2['reply_time'].unique()
len(temp2[(temp2['reply_time']<100) & (temp2['reply_time']>-100)])
temp3 = temp2[(temp2['reply_time']<100) & (temp2['reply_time']>-100)]
temp4 = temp3.groupby(['src_ip','dst_ip','src_port','dst_port','protocol','start_time','FlowNo.'],{
'rep_time': gl.aggregate.MIN('reply_time'),
'rev_flow_no.' : gl.aggregate.ARGMIN('reply_time','rev_flow')
})
temp4.head()
temp = gl.SFrame.read_csv('NumPacketsPerFlow.csv',verbose=False)
temp2 = temp.join(temp4,on=['FlowNo.'])
temp.rename({'FlowNo.':'rev_flow_no.'})
temp2 = temp.join(temp2,on=['rev_flow_no.'])
temp2
temp2['IOPR'] = temp2.apply(lambda x:0.0 if x['NumPackets'] == 0 else x['NumPackets.1']/x['NumPackets'])
temp2 = temp2[['FlowNo.','IOPR']]
temp2.save('IOPR.csv')
temp2.sort('IOPR')[-10:]
SF.save('ISCX_Botnet-Testing_Ports_Only_Sorted_Flow_IAT_Null.csv')
```
### 22-10-2016
```
import graphlab as gl
import tensorflow as tf
SF = gl.SFrame.read_csv('ISCX_Botnet-Testing_Ports_Only_Sorted_Flow_IAT_Null.csv',verbose=False)
SF.head(3)
len(SF[SF['Source']=='10.37.130.4'])
len(SF[SF['Destination']=='10.37.130.4'])
len(SF[SF['Source']=='10.37.130.4']['FlowNo.'].unique())
len(SF['FlowNo.'].unique())
len(SF[SF['Source']=='147.32.84.140']['FlowNo.'].unique())
SF.head()
Flows = gl.SFrame.read_csv('DurationFlow.csv')
Flows['Duration'].unique()
Flows.groupby(['Duration'],{
'count':gl.aggregate.COUNT()
})
print min(SF['Time']),max(SF['Time'])
import graphlab as gl
tp = gl.SFrame.read_csv('ISCX_Botnet-Testing.csv')
tp = tp[(tp['Source Port']=='')|(tp['Destination Port']=='')]
for x in sorted(tp['Protocol'].unique()):
print x,len(tp[tp['Protocol']==x])
tp = tp[(tp['Source Port']=='')]
for x in sorted(tp['Protocol'].unique()):
print x,len(tp[tp['Protocol']==x])
tp = tp[(tp['Destination Port']=='')]
for x in sorted(tp['Protocol'].unique()):
print x,len(tp[tp['Protocol']==x])
len(tp)
for x in sorted(tp['Protocol'].unique()):
print len(tp[tp['Protocol']==x]),',',
import graphlab as gl
import tensorflow as tf
SF = gl.SFrame.read_csv('ISCX_Botnet-Testing.csv')
SF[SF['No.']==1944398]
```
# 28/10/16
Undirectional Flowscombining all features and labels
```
SF = gl.SFrame.read_csv('ISCX_Botnet-Testing_Ports_Only_Sorted_Flow_IAT_Null.csv',verbose=False)
SF.head()
# Avg packet length
AvgpackLen = gl.SFrame.read_csv('AveragePktLengthFlow.csv')
SF = SF.join(AvgpackLen)
SamePackLen = gl.SFrame.read_csv('SameLenPktRatio.csv')
SF = SF.join(SamePackLen,on = 'FlowNo.')
BytesExch = gl.SFrame.read_csv('BytexExchangedPerFlow.csv')
SF = SF.join(BytesExch,on = 'FlowNo.')
FrstPkl = gl.SFrame.read_csv('FirstPacketLength.csv')
SF = SF.join(FrstPkl,on = 'FlowNo.')
STDVLen = gl.SFrame.read_csv('StdDevLenPerFlow.csv')
SF = SF.join(STDVLen ,on = 'FlowNo.')
Avgpkpersec = gl.SFrame.read_csv('AvgPktPerSecFlow.csv')
SF = SF.join(Avgpkpersec ,on = 'FlowNo.')
Bitsperse = gl.SFrame.read_csv('BitsPerSecPerFlow.csv')
SF = SF.join(Bitsperse ,on = 'FlowNo.')
DurFlow = gl.SFrame.read_csv('DurationFlow.csv')
SF = SF.join(DurFlow ,on = 'FlowNo.')
NumPks = gl.SFrame.read_csv('NumPacketsPerFlow.csv')
SF = SF.join(NumPks, on='FlowNo.')
NullPks = gl.SFrame().read_csv('NumberNullPacketsEXc.csv')
SF = SF.join(NullPks, on='FlowNo.')
Recons = gl.SFrame.read_csv('ReconnectsFlow.csv')
SF = SF.join(Recons, on = ['FlowNo.'])
SF.column_names
SF.save('Botnet_Testing_all_features.csv')
#sf_valid_train, sf_test = SF.random_split(.8, seed=5)
#sf_valid, sf_train = sf_valid_train.random_split(.2, seed=5)
#X_train, y_train = sf_train.drop('isBot'), sf_train['isBot']
#X_valid, y_valid = sf_valid_train
#X_train_valid, y_train_valid = X[:3475000], y[:3475000]
#X_test, y_test = X[3475000:], y[3475000:]
```
#Train uncalibrated random forest classifier on whole train and validation
#data and evaluate on test data
clf = RandomForestClassifier(n_estimators=25)
clf.fit(X_train_valid, y_train_valid)
clf_probs = clf.predict_proba(X_test)
score = log_loss(y_test, clf_probs)
#Train random forest classifier, calibrate on validation data and evaluate
#on test data
clf = RandomForestClassifier(n_estimators=25)
clf.fit(X_train, y_train)
clf_probs = clf.predict_proba(X_test)
sig_clf = CalibratedClassifierCV(clf, method="sigmoid", cv="prefit")
sig_clf.fit(X_valid, y_valid)
sig_clf_probs = sig_clf.predict_proba(X_test)
sig_score = log_loss(y_test, sig_clf_probs)
|
github_jupyter
|
import graphlab as gl
import tensorflow as tf
SF = gl.SFrame.read_csv('ISCX_Botnet-Testing.csv',verbose=False)
print "Done reading"
#SF.head(5)
#SF.column_names()
SF = SF[(SF['Source Port']!='')|(SF['Destination Port']!='')]
len(SF)
SF.save('ISCX_Botnet-Testing_PortsOnly.csv')
SF = gl.SFrame.read_csv('ISCX_Botnet-Testing_Ports_Only.csv',verbose=False)
print 'Done Reading'
#type(SF['tcp_Flags'][0])
#SF['tcp_Flags'].unique()
#SF['IP_Flags'].unique()
SF['tcp_Flags'] = SF['tcp_Flags'].apply(lambda x:int(x,16) if x!='' else 0)
len(SF[SF['tcp_Flags']==0])
len(SF[SF['Protocol']=='TCP'])
##Testing code for time comparison in iteration
"""import time
start = time.time()
for i in SF:
i['tcp_Flags']
print time.time()-start"""
len(SF)
len(SF[SF['tcp_Flags']!=0])
print len(SF[SF['Protocol']=='UDP']),len(SF[(SF['tcp_Flags']==0)&(SF['Protocol']=='UDP')])
print len(SF[(SF['tcp_Flags']==0)]),len(SF[(SF['tcp_Flags']==0)&(SF['Protocol']!='TCP')])
Protocols = sorted(SF['Protocol'].unique())
#print Protocols
"""lt = []
gt = []
for x in Protocols:
print x
temp = len(SF[SF['Protocol']==x])
if temp<1000:
lt.append(x)
else:
gt.append(x)"""
#print lt,"\n\n",gt
#print len(lt),len(gt)
SF[SF['Protocol']=='ICMP'].head(3)
len(SF[SF['Protocol']=='ICMP'])
iplist = [
"147.32.84.180",
"147.32.84.170",
"147.32.84.150",
"147.32.84.140",
"147.32.84.130",
"147.32.84.160",
"10.0.2.15",
"192.168.106.141",
"192.168.106.131",
"172.16.253.130",
"172.16.253.131",
"172.16.253.129",
"172.16.253.240",
"74.78.117.238",
"158.65.110.24",
"192.168.3.35",
"192.168.3.25",
"192.168.3.65",
"172.29.0.116",
"172.29.0.109",
"172.16.253.132",
"192.168.248.165",
"10.37.130.4"]
MasterBot = [ ("192.168.2.112", "131.202.243.84"), ("192.168.5.122", "198.164.30.2"), ("192.168.2.110", "192.168.5.122"),( "192.168.4.118", "192.168.5.122"), ("192.168.2.113", "192.168.5.122"), ("192.168.1.103", "192.168.5.122"), ("192.168.4.120", "192.168.5.122"), ("192.168.2.112", "192.168.2.110"), ("192.168.2.112", "192.168.4.120"), ("192.168.2.112", "192.168.1.103"), ("192.168.2.112", "192.168.2.113"), ("192.168.2.112", "192.168.4.118"), ("192.168.2.112", "192.168.2.109"), ("192.168.2.112", "192.168.2.105"), ("192.168.1.105", "192.168.5.122")]
iplist = gl.SArray(iplist)
iplist = iplist.unique()
iplist = list(iplist)
def fcheckIP(x):
if x['Source'] in iplist:
return 1
else:
if ((x['Source'],x['Destination']) in MasterBot ) or ((x['Destination'],x['Source']) in MasterBot ) :
return 1
else:
return 0
SF['isBot'] = SF[['Source','Destination']].apply(lambda x: fcheckIP(x))
SF[(SF['Source']=='192.168.2.112') & (SF['isBot']==1)]
temp = len(SF[SF['isBot']==1])
print temp, (temp*1.0)/len(SF)*100
SF.head()
iplist = [
"192.168.2.112",
"131.202.243.84",
"192.168.5.122",
"198.164.30.2",
"192.168.2.110",
"192.168.5.122",
"192.168.4.118",
"192.168.5.122",
"192.168.2.113",
"192.168.5.122",
"192.168.1.103",
"192.168.5.122",
"192.168.4.120",
"192.168.5.122",
"192.168.2.112",
"192.168.2.110",
"192.168.2.112",
"192.168.4.120",
"192.168.2.112",
"192.168.1.103",
"192.168.2.112",
"192.168.2.113",
"192.168.2.112",
"192.168.4.118",
"192.168.2.112",
"192.168.2.109",
"192.168.2.112",
"192.168.2.105",
"192.168.1.105",
"192.168.5.122",
"147.32.84.180",
"147.32.84.170",
"147.32.84.150",
"147.32.84.140",
"147.32.84.130",
"147.32.84.160",
"10.0.2.15",
"192.168.106.141",
"192.168.106.131",
"172.16.253.130",
"172.16.253.131",
"172.16.253.129",
"172.16.253.240",
"74.78.117.238",
"158.65.110.24",
"192.168.3.35",
"192.168.3.25",
"192.168.3.65",
"172.29.0.116",
"172.29.0.109",
"172.16.253.132",
"192.168.248.165",
"10.37.130.4"]
iplist = gl.SArray(iplist)
iplist = iplist.unique()
iplist = list(iplist)
temp = SF[SF['Protocol']=='ICMP']
for x in iplist:
print x, " Destination : ",len(temp[temp['Destination']==x])," Source : ",len(temp[temp['Source']==x])
dest = temp.groupby('Destination',{
'Count':gl.aggregate.COUNT()
})
dest = dest.sort('Count',ascending=False)
dest.head()
src = temp.groupby('Source',{
'Count':gl.aggregate.COUNT()
})
src = src.sort('Count',ascending=False)
src.head()
len(SF[(SF['Source']=='10.0.0.254')&(SF['Protocol']!='ICMP')&(SF['Protocol']!='DNS')])
#sorted(iplist)
## To check whether No. field is primary key or not
print len(SF['No.'].unique()),len(SF)
sorting_features = ['Source','Destination','Source Port','Destination Port','Protocol','Time']
#STD = SF[['No.','Time','Source','Destination','Source Port','Destination Port','Protocol','tcp_Flags']]
#STD = STD.sort(sorting_features)
#print 'Done sorting STD'
SF = SF.sort(sorting_features)
print 'Done sorting SF'
#STD.save('ISCX_Botnet-Testing_Ports_Only_Sorted_Flow.csv')
#print 'Done saving STD'
#STD = None
SF.save('ISCX_Botnet-Testing_Ports_Only_Sorted.csv')
print 'Done saving SF'
#STD = None
SF.head(3)
type(SF['Time'][0])
SF['tcp_Flags'].unique()
print len(SF),len(SF['No.'].unique())
## function for comparing two different flows based on columns
def compare(x,y,columns):
try:
val = True
for column in columns:
if x[column]!=y[column]:
val = False
break;
return val
except KeyError:
print "Column does not exist in the data, check again!"
except:
print "Some unknown error"
##Code for logic of flow identification
import pickle
FlowCols = ['Source','Destination','Source Port','Destination Port','Protocol']
SF['FlowNo.'] = None
FlowNo = 0 ##Unique Flow Number for each flow
#FFlow = []
prev = None
Flow = [] ##Stores all flows in form of list of dictionary
#cFlow = [] ##Store the current flow (all details)
count = 0
fc = 0
startTime = None ##Start Time of each flow to implement timeout
for x in SF:
if count%500000 == 0:
#fName = 'Flow'+str(fc)+'.pkl'
#print 'Saving : ',fName
#pickle.dump(Flow,open(fName,'w'))
#print 'Saved'
print 'Running '+str(count)+' Done !'
#c = fc + 1
#Flow = []
count = count+1
if prev is None:
startTime = x['Time']
Flow.append(FlowNo)
#cFlow.append(x['No.'])
prev = x
elif compare(x,prev,FlowCols):
fc=fc+1
if x['tcp_Flags']&1 or x['tcp_Flags']&4 or x['Time']-startTime>=120:
Flow.append(FlowNo)
prev = None
FlowNo = FlowNo + 1
#cFlow.append(x['No.'])
#FFlow.append(cFlow)
#cFlow = []
else:
#cFlow.append(x['No.'])
Flow.append(FlowNo)
prev = x
else:
FlowNo = FlowNo + 1
Flow.append(FlowNo)
#FFlow.append(cFlow)
#cFlow = []
#cFlow.append(x['No.'])
prev = x
startTime = x['Time']
print len(gl.SArray(Flow).unique())
def flow_id(x):
if x['Source']>x['Destination']:
return x['Source']+'-'+x['Destination']+'-'+str(x['Source Port'])+'-'+str(x['Destination Port'])+'-'+x['Protocol']
else:
return x['Destination']+'-'+x['Source']+'-'+str(x['Destination Port'])+'-'+str(x['Source Port'])+'-'+x['Protocol']
SF['UFid'] = SF.apply(lambda x:flow_id(x))
## function for comparing two different flows based on columns
def compareUF(x,y):
if x!=y:
return False
return True
##Code for logic of Bidirectional flow identification
import pickle
FlowNo = 0 ##Unique Flow Number for each flow
prev = None
Flow = [] ##Stores all flows in form of list of dictionary
#cFlow = [] ##Store the current flow (all details)
count = 0
fc = 0
startTime = None ##Start Time of each flow to implement timeout
SF = SF.sort(['UFid','Time'])
print 'Done Sorting'
for x in SF:
if count%500000 == 0:
print 'Running '+str(count)+' Done !'
count = count+1
if prev is None:
if startTime is None:
startTime = x['Time']
Flow.append(FlowNo)
prev = x['UFid']
elif compareUF(x['UFid'],prev):
if x['tcp_Flags']&1:
Flow.append(FlowNo)
prev = None
startTime = None
FlowNo = FlowNo + 1
elif x['Time']-startTime>=120:
FlowNo = FlowNo + 1
Flow.append(FlowNo)
prev = None
startTime = x['Time']
else:
Flow.append(FlowNo)
prev = x['UFid']
else:
FlowNo = FlowNo + 1
Flow.append(FlowNo)
prev = x['UFid']
startTime = x['Time']
print len(gl.SArray(Flow).unique())
SF['Flow'] = gl.SArray(Flow)
temp = SF.groupby('Flow',{
'Count':gl.aggregate.COUNT()
})
len(temp[temp['Count']>1])
temp = SF.groupby('Flow',{
'NumBots' : gl.aggregate.SUM('isBot')
})
NumBotFlows = len(temp[temp['NumBots']>1])
print NumBotFlows, NumBotFlows*1.0/len(SF['Flow'].unique())
len(Flow)
len(SF)
SF['FlowNo.'] = gl.SArray(Flow)
##Code for checking authenticity of flow logic
#STD[(STD['Source']=='0.0.0.0')&(STD['Destination']=='255.255.255.255')&(STD['Source Port']=='68')&(STD['Destination Port']=='67')].sort('Time')
## Code to check if in any flows there are some No.s which are in decreasing order (Indicative or Decrepancies)
## UPDATE: No. does not indicate same relation in time, so Data collected is right !
"""count = 0
for li in Flow:
for i in range(1,len(li)):
if li[i]<li[i-1]:
#print li
count = count+1
break;
print count"""
import pickle
pickle.dump(Flow,open('Flow.pkl','w'))
SF.save('ISCX_Botnet-Testing_Ports_Only_Sorted_Flow.csv')
SF.head(3)
## First Packet Length
FlowFeatures = ['Source','Destination','Source Port','Destination Port','Protocol']
FPL = SF.groupby(['FlowNo.'],{
'Time':gl.aggregate.MIN('Time')
})
print len(FPL)
FPL = FPL.join(SF,on =['FlowNo.','Time'])[['FlowNo.','Length']].unique()
FPL = FPL.groupby(['FlowNo.'],{
'Length':gl.aggregate.AVG('Length')
})
print len(FPL)
FPL.save('FirstPacketLength.csv')
import graphlab as gl
import tensorflow as tf
SF = gl.SFrame.read_csv('ISCX_Botnet-Testing_Ports_Only_Sorted_Flow.csv',verbose=False)
## Number of packets per flow
temp = SF.groupby(['FlowNo.'],{
'NumPackets':gl.aggregate.COUNT()
})
print temp.head(3)
temp.save('NumPacketsPerFlow.csv')
## Number of bytes exchanged
temp = SF.groupby(['FlowNo.'],{
'BytesEx':gl.aggregate.SUM('Length')
})
temp.save('BytexExchangedPerFlow.csv')
temp.head(3)
## Standard deviation of packet length
temp = SF.groupby(['FlowNo.'],{
'StdDevLen':gl.aggregate.STDV('Length')
})
temp.save('StdDevLenPerFlow.csv')
temp.sort('StdDevLen')[-10:]
## Same length packet ratio
temp2 = SF.groupby(['FlowNo.'],{
'SameLenPktRatio':gl.aggregate.COUNT_DISTINCT('Length')
})
##temp from number of packets computation
temp = SF.groupby(['FlowNo.'],{
'NumPackets':gl.aggregate.COUNT()
})
temp = temp.join(temp2,on='FlowNo.')
temp['SameLenPktRatio'] = temp['SameLenPktRatio']*1.0/temp['NumPackets']
temp2 = None
temp = temp[['FlowNo.','SameLenPktRatio']]
temp.save('SameLenPktRatio.csv')
temp.head(3)
def tfn(x):
if 'udp' in x.split(':'):
return 1
return 0
SF['hasUDP'] = SF['Protocols in frame'].apply(lambda x:tfn(x))
test = SF[SF['hasUDP']==1]
test['colDiff'] = test['Length'] - test['udp_Length']
test[test['colDiff']==-1].head(3)
test[test['colDiff']<-20].head(3)
## Duration of flow
timeF = SF.groupby(['FlowNo.'],{
'startTime':gl.aggregate.MIN('Time'),
'endTime':gl.aggregate.MAX('Time')
})
timeF['Duration'] = timeF['endTime'] - timeF['startTime']
timeF[['FlowNo.','Duration']].save('DurationFlow.csv')
timeF = timeF[['FlowNo.','Duration']]
## Average packets per second
temp = gl.SFrame.read_csv('NumPacketsPerFlow.csv',verbose=False)
temp = temp.join(timeF,on=['FlowNo.'])
temp['AvgPktPerSec'] = temp.apply(lambda x:0.0 if x['Duration'] == 0.0 else x['NumPackets']*1.0/x['Duration'])
temp = temp[['FlowNo.','AvgPktPerSec']]
temp.save('AvgPktPerSecFlow.csv')
temp.sort('AvgPktPerSec')[-10:]
##Average Bits Per Second
temp = gl.SFrame.read_csv('BytexExchangedPerFlow.csv',verbose=False)
temp = temp.join(timeF,on=['FlowNo.'])
temp['BitsPerSec'] = temp.apply(lambda x:0.0 if x['Duration'] == 0.0 else x['BytesEx']*8.0/x['Duration'])
temp = temp[['FlowNo.','BitsPerSec']]
temp.save('BitsPerSecPerFlow.csv')
temp.sort('BitsPerSec')[-5:]
## Average Packet Lentgth
temp = SF.groupby(['FlowNo.'],{
'APL':gl.aggregate.AVG('Length')
})
temp.save('AveragePktLengthFlow.csv')
test = SF[SF['hasUDP']==1]
test['colDiff'] = test['Length'] - test['udp_Length']
len(test[test['colDiff']<0])
len(test)
## Number of Reconnects, sort FlowNo, SeqNo
def tfn(x):
if 'udp' in x.split(':') or 'tcp' in x.split(':'):
return 1
return 0
temp = list(SF['Protocols in frame'].apply(lambda x:tfn(x)))
len(temp)
sum(temp)
SF.head(1)
type(SF['TCP Segment Len'][0])
type(SF['udp_Length'][0])
len(SF[(SF['udp_Length'] == None)&(SF['TCP Segment Len'] == '')])
SF[(SF['udp_Length'] == None)&(SF['TCP Segment Len'] == '')]['Protocols in frame'].unique()
print len(SF[SF['Protocols in frame']=='eth:ethertype:ip:icmp:ip:tcp:http:urlencoded-form']),len(SF[SF['Protocols in frame']=='eth:ethertype:ip:icmp:ip:tcp']),len(SF[SF['Protocols in frame']=='eth:ethertype:ip:icmp:ip:tcp:http:data'])
## Inter arrival time
SF['IAT'] = 0
SF = SF.sort(['FlowNo.','Time'])
prev = None
prevT = None
li = []
for x in SF:
if prev is None or x['FlowNo.']!= prev:
li.append(0)
else:
li.append(x['Time']-prevT)
prev = x['FlowNo.']
prevT = x['Time']
SF['IAT'] = gl.SArray(li)
SF.save('ISCX_Botnet-Testing_Ports_Only_Sorted_Flow_IAT.csv')
SF.head(3)
#len(SF[(SF['udp_Length']>=8)&(SF['udp_Length']<=16)])
#print len(SF[SF['udp_Length']==8])
#print len(SF[SF['udp_Length']==16])
#SF[SF['Protocols in frame']=='eth:llc:ipx'].head(3)
#SF[SF['Protocols in frame']=='eth:ethertype:ipx'].head(3)
#print len(SF[SF['Protocols in frame']=='eth:ipx'])
#SF[SF['udp_Length']==0]
print len(SF[SF['Protocols in frame']=='eth:ipx'])
SF[SF['hasUDP']==1]['Protocols in frame'].unique()
len(SF[SF['Protocol']=='TCP'])
len(SF[SF['udp_Length']==8]), len(SF[SF['Protocol']=='UDP'])
SF[SF['TCP Segment Len']=='0']['Protocols in frame'].unique()
tt = SF[(SF['TCP Segment Len']!='0')&(SF['udp_Length']!=8 )]
len(tt)
## Null Packets handling
def checkNull(x):
if(x['TCP Segment Len']=='0' or x['udp_Length']==8 ):
return 1
elif('ipx' in x['Protocols in frame'].split(':')):
l = x['Length'] - 30
if('eth' in x['Protocols in frame'].split(':')):
l = l - 14
if('ethtype' in x['Protocols in frame'].split(':')):
l = l - 2
if('llc' in x['Protocols in frame'].split(':')):
l = l - 8
if(l==0 or l==-1):
return 1
return 0
SF['isNull'] = SF.apply(lambda x:checkNull(x))
len(SF[SF['isNull']==1])
NPEx = SF.groupby(['FlowNo.'],{
'NPEx':gl.aggregate.SUM('isNull')
})
NPEx.save('NumberNullPacketsEXc.csv')
type(SF['Sequence number'][0])
recon = SF[SF['Sequence number']!=''].groupby(['FlowNo.'],{
'total_seq_no.' : gl.aggregate.COUNT('Sequence number'),
'distinct_seq_no.' : gl.aggregate.COUNT_DISTINCT('Sequence number')
})
recon['reconnects'] = recon['total_seq_no.'] - recon['distinct_seq_no.']
recon.head()
recon[['FlowNo.','reconnects']].save('ReconnectsFlow.csv')
#SF[SF['FlowNo.']==79732]['Info']
## Ratio of incoming to outgoing packets
convo = SF.groupby(['FlowNo.'],{
'start_time': gl.aggregate.MIN('Time'),
'protocol':gl.aggregate.SELECT_ONE('Protocol'),
'src_ip': gl.aggregate.SELECT_ONE('Source'),
'dst_ip': gl.aggregate.SELECT_ONE('Destination'),
'src_port': gl.aggregate.SELECT_ONE('Source Port'),
'dst_port': gl.aggregate.SELECT_ONE('Destination Port')
})
convo.sort('start_time')
convo['rev_flow_no.'] = -1
convo.head()
temp1 = convo
temp1.rename({'src_ip':'dst_ip1', 'src_port':'dst_port1'})
temp1.rename({'dst_ip':'src_ip','dst_port':'src_port'})
temp1.rename({'dst_ip1':'dst_ip', 'dst_port1':'dst_port'})
temp1.rename({'start_time':'return_time'})
temp1.rename({'FlowNo.':'rev_flow'})
temp1 = temp1['src_ip','dst_ip','src_port','dst_port','protocol','return_time','rev_flow']
convo = SF.groupby(['FlowNo.'],{
'start_time': gl.aggregate.MIN('Time'),
'protocol':gl.aggregate.SELECT_ONE('Protocol'),
'src_ip': gl.aggregate.SELECT_ONE('Source'),
'dst_ip': gl.aggregate.SELECT_ONE('Destination'),
'src_port': gl.aggregate.SELECT_ONE('Source Port'),
'dst_port': gl.aggregate.SELECT_ONE('Destination Port')
})
convo.sort('start_time')
temp2 = temp1.join(convo,on=['src_ip','dst_ip','src_port','dst_port','protocol'])
temp2
convo
temp1[(temp1['src_ip']=='66.249.73.56') & (temp1['src_port']==52954)]
temp2['reply_time'] = temp2['return_time'] - temp2['start_time']
temp2.head()
temp2['reply_time'].unique()
len(temp2[(temp2['reply_time']<100) & (temp2['reply_time']>-100)])
temp3 = temp2[(temp2['reply_time']<100) & (temp2['reply_time']>-100)]
temp4 = temp3.groupby(['src_ip','dst_ip','src_port','dst_port','protocol','start_time','FlowNo.'],{
'rep_time': gl.aggregate.MIN('reply_time'),
'rev_flow_no.' : gl.aggregate.ARGMIN('reply_time','rev_flow')
})
temp4.head()
temp = gl.SFrame.read_csv('NumPacketsPerFlow.csv',verbose=False)
temp2 = temp.join(temp4,on=['FlowNo.'])
temp.rename({'FlowNo.':'rev_flow_no.'})
temp2 = temp.join(temp2,on=['rev_flow_no.'])
temp2
temp2['IOPR'] = temp2.apply(lambda x:0.0 if x['NumPackets'] == 0 else x['NumPackets.1']/x['NumPackets'])
temp2 = temp2[['FlowNo.','IOPR']]
temp2.save('IOPR.csv')
temp2.sort('IOPR')[-10:]
SF.save('ISCX_Botnet-Testing_Ports_Only_Sorted_Flow_IAT_Null.csv')
import graphlab as gl
import tensorflow as tf
SF = gl.SFrame.read_csv('ISCX_Botnet-Testing_Ports_Only_Sorted_Flow_IAT_Null.csv',verbose=False)
SF.head(3)
len(SF[SF['Source']=='10.37.130.4'])
len(SF[SF['Destination']=='10.37.130.4'])
len(SF[SF['Source']=='10.37.130.4']['FlowNo.'].unique())
len(SF['FlowNo.'].unique())
len(SF[SF['Source']=='147.32.84.140']['FlowNo.'].unique())
SF.head()
Flows = gl.SFrame.read_csv('DurationFlow.csv')
Flows['Duration'].unique()
Flows.groupby(['Duration'],{
'count':gl.aggregate.COUNT()
})
print min(SF['Time']),max(SF['Time'])
import graphlab as gl
tp = gl.SFrame.read_csv('ISCX_Botnet-Testing.csv')
tp = tp[(tp['Source Port']=='')|(tp['Destination Port']=='')]
for x in sorted(tp['Protocol'].unique()):
print x,len(tp[tp['Protocol']==x])
tp = tp[(tp['Source Port']=='')]
for x in sorted(tp['Protocol'].unique()):
print x,len(tp[tp['Protocol']==x])
tp = tp[(tp['Destination Port']=='')]
for x in sorted(tp['Protocol'].unique()):
print x,len(tp[tp['Protocol']==x])
len(tp)
for x in sorted(tp['Protocol'].unique()):
print len(tp[tp['Protocol']==x]),',',
import graphlab as gl
import tensorflow as tf
SF = gl.SFrame.read_csv('ISCX_Botnet-Testing.csv')
SF[SF['No.']==1944398]
SF = gl.SFrame.read_csv('ISCX_Botnet-Testing_Ports_Only_Sorted_Flow_IAT_Null.csv',verbose=False)
SF.head()
# Avg packet length
AvgpackLen = gl.SFrame.read_csv('AveragePktLengthFlow.csv')
SF = SF.join(AvgpackLen)
SamePackLen = gl.SFrame.read_csv('SameLenPktRatio.csv')
SF = SF.join(SamePackLen,on = 'FlowNo.')
BytesExch = gl.SFrame.read_csv('BytexExchangedPerFlow.csv')
SF = SF.join(BytesExch,on = 'FlowNo.')
FrstPkl = gl.SFrame.read_csv('FirstPacketLength.csv')
SF = SF.join(FrstPkl,on = 'FlowNo.')
STDVLen = gl.SFrame.read_csv('StdDevLenPerFlow.csv')
SF = SF.join(STDVLen ,on = 'FlowNo.')
Avgpkpersec = gl.SFrame.read_csv('AvgPktPerSecFlow.csv')
SF = SF.join(Avgpkpersec ,on = 'FlowNo.')
Bitsperse = gl.SFrame.read_csv('BitsPerSecPerFlow.csv')
SF = SF.join(Bitsperse ,on = 'FlowNo.')
DurFlow = gl.SFrame.read_csv('DurationFlow.csv')
SF = SF.join(DurFlow ,on = 'FlowNo.')
NumPks = gl.SFrame.read_csv('NumPacketsPerFlow.csv')
SF = SF.join(NumPks, on='FlowNo.')
NullPks = gl.SFrame().read_csv('NumberNullPacketsEXc.csv')
SF = SF.join(NullPks, on='FlowNo.')
Recons = gl.SFrame.read_csv('ReconnectsFlow.csv')
SF = SF.join(Recons, on = ['FlowNo.'])
SF.column_names
SF.save('Botnet_Testing_all_features.csv')
#sf_valid_train, sf_test = SF.random_split(.8, seed=5)
#sf_valid, sf_train = sf_valid_train.random_split(.2, seed=5)
#X_train, y_train = sf_train.drop('isBot'), sf_train['isBot']
#X_valid, y_valid = sf_valid_train
#X_train_valid, y_train_valid = X[:3475000], y[:3475000]
#X_test, y_test = X[3475000:], y[3475000:]
| 0.056993 | 0.370083 |
# Deep Q-Network (DQN)
---
In this notebook, you will implement a DQN agent with OpenAI Gym's LunarLander-v2 environment.
### 1. Import the Necessary Packages
```
import gym
import random
import torch
import numpy as np
from collections import deque
import matplotlib.pyplot as plt
%matplotlib inline
```
### 2. Instantiate the Environment and Agent
Initialize the environment in the code cell below.
```
env = gym.make('LunarLander-v2')
env.seed(0)
print('State shape: ', env.observation_space.shape)
print('Number of actions: ', env.action_space.n)
```
Please refer to the instructions in `Deep_Q_Network.ipynb` if you would like to write your own DQN agent. Otherwise, run the code cell below to load the solution files.
```
agent = Agent(state_size=8, action_size=4, seed=0)
# watch an untrained agent
state = env.reset()
for j in range(200):
action = agent.act(state)
env.render()
state, reward, done, _ = env.step(action)
if done:
break
env.close()
```
### 3. Train the Agent with DQN
Run the code cell below to train the agent from scratch. You are welcome to amend the supplied values of the parameters in the function, to try to see if you can get better performance!
Alternatively, you can skip to the next step below (**4. Watch a Smart Agent!**), to load the saved model weights from a pre-trained agent.
```
def dqn(n_episodes=2000, max_t=1000, eps_start=1.0, eps_end=0.01, eps_decay=0.995):
"""Deep Q-Learning.
Params
======
n_episodes (int): maximum number of training episodes
max_t (int): maximum number of timesteps per episode
eps_start (float): starting value of epsilon, for epsilon-greedy action selection
eps_end (float): minimum value of epsilon
eps_decay (float): multiplicative factor (per episode) for decreasing epsilon
"""
scores = [] # list containing scores from each episode
scores_window = deque(maxlen=100) # last 100 scores
eps = eps_start # initialize epsilon
for i_episode in range(1, n_episodes+1):
state = env.reset()
score = 0
for t in range(max_t):
action = agent.act(state, eps)
next_state, reward, done, _ = env.step(action)
agent.step(state, action, reward, next_state, done)
state = next_state
score += reward
if done:
break
scores_window.append(score) # save most recent score
scores.append(score) # save most recent score
eps = max(eps_end, eps_decay*eps) # decrease epsilon
print('\rEpisode {}\tAverage Score: {:.2f}'.format(i_episode, np.mean(scores_window)), end="")
if i_episode % 100 == 0:
print('\rEpisode {}\tAverage Score: {:.2f}'.format(i_episode, np.mean(scores_window)))
if np.mean(scores_window)>=200.0:
print('\nEnvironment solved in {:d} episodes!\tAverage Score: {:.2f}'.format(i_episode-100, np.mean(scores_window)))
torch.save(agent.qnetwork_local.state_dict(), 'checkpoint.pth')
break
return scores
scores = dqn()
# plot the scores
fig = plt.figure()
ax = fig.add_subplot(111)
plt.plot(np.arange(len(scores)), scores)
plt.ylabel('Score')
plt.xlabel('Episode #')
plt.show()
```
### 4. Watch a Smart Agent!
In the next code cell, you will load the trained weights from file to watch a smart agent!
```
# load the weights from file
agent.qnetwork_local.load_state_dict(torch.load('checkpoint.pth'))
for i in range(3):
state = env.reset()
for j in range(200):
action = agent.act(state)
env.render()
state, reward, done, _ = env.step(action)
if done:
break
env.close()
```
### 5. Explore
In this exercise, you have implemented a DQN agent and demonstrated how to use it to solve an OpenAI Gym environment. To continue your learning, you are encouraged to complete any (or all!) of the following tasks:
- Amend the various hyperparameters and network architecture to see if you can get your agent to solve the environment faster. Once you build intuition for the hyperparameters that work well with this environment, try solving a different OpenAI Gym task with discrete actions!
- You may like to implement some improvements such as prioritized experience replay, Double DQN, or Dueling DQN!
- Write a blog post explaining the intuition behind the DQN algorithm and demonstrating how to use it to solve an RL environment of your choosing.
|
github_jupyter
|
import gym
import random
import torch
import numpy as np
from collections import deque
import matplotlib.pyplot as plt
%matplotlib inline
env = gym.make('LunarLander-v2')
env.seed(0)
print('State shape: ', env.observation_space.shape)
print('Number of actions: ', env.action_space.n)
agent = Agent(state_size=8, action_size=4, seed=0)
# watch an untrained agent
state = env.reset()
for j in range(200):
action = agent.act(state)
env.render()
state, reward, done, _ = env.step(action)
if done:
break
env.close()
def dqn(n_episodes=2000, max_t=1000, eps_start=1.0, eps_end=0.01, eps_decay=0.995):
"""Deep Q-Learning.
Params
======
n_episodes (int): maximum number of training episodes
max_t (int): maximum number of timesteps per episode
eps_start (float): starting value of epsilon, for epsilon-greedy action selection
eps_end (float): minimum value of epsilon
eps_decay (float): multiplicative factor (per episode) for decreasing epsilon
"""
scores = [] # list containing scores from each episode
scores_window = deque(maxlen=100) # last 100 scores
eps = eps_start # initialize epsilon
for i_episode in range(1, n_episodes+1):
state = env.reset()
score = 0
for t in range(max_t):
action = agent.act(state, eps)
next_state, reward, done, _ = env.step(action)
agent.step(state, action, reward, next_state, done)
state = next_state
score += reward
if done:
break
scores_window.append(score) # save most recent score
scores.append(score) # save most recent score
eps = max(eps_end, eps_decay*eps) # decrease epsilon
print('\rEpisode {}\tAverage Score: {:.2f}'.format(i_episode, np.mean(scores_window)), end="")
if i_episode % 100 == 0:
print('\rEpisode {}\tAverage Score: {:.2f}'.format(i_episode, np.mean(scores_window)))
if np.mean(scores_window)>=200.0:
print('\nEnvironment solved in {:d} episodes!\tAverage Score: {:.2f}'.format(i_episode-100, np.mean(scores_window)))
torch.save(agent.qnetwork_local.state_dict(), 'checkpoint.pth')
break
return scores
scores = dqn()
# plot the scores
fig = plt.figure()
ax = fig.add_subplot(111)
plt.plot(np.arange(len(scores)), scores)
plt.ylabel('Score')
plt.xlabel('Episode #')
plt.show()
# load the weights from file
agent.qnetwork_local.load_state_dict(torch.load('checkpoint.pth'))
for i in range(3):
state = env.reset()
for j in range(200):
action = agent.act(state)
env.render()
state, reward, done, _ = env.step(action)
if done:
break
env.close()
| 0.605916 | 0.956268 |
# Dokumentation der Tags
## s[]s Stichwort
## p[]p Person
Für die Ansetzung von Personennamen kommen folgende Stichwortteile bzw. Ordnungshilfen in Frage:
1. Name,
2. Vorname(n),
3. Adelstitel/Herrschaftsbezeichnung,
4. Akademischer Grad,
5. Beruf,
6. Ort
7. Sonstiges
8. Präfixe vor Personennamen
9. Pseudonym / angenommene Namen
Beispiel:
- der Aktentitel enthält:
... der Prager Buchhändler J. Gelder ...
- in Kat. 40 wird aufgenommen:
... der p[6{Prag}er 5{Buchhändler} 2{J}.{<ohannes>} 1{Gelder}]p ...
Im Personenregister entsteht daraus:
Gelder, Johannes <Buchhändler in Prag>
Zusätzlich werden Berufe und Orte indiziert, d.h. verklammerte Indizierung:
der p[6{o[Prag]o}er 5{s[Buchhändler]s} 2{J<ohannes>}. 1{Gelder}]p
Namen von Adeligen/Königen/Herzögen usw. werden nur unter dem Vornamen in Klammer 2 indiziert. Bekannte Zusätze werden in <> eingefügt.
## k[]k Körperschaft
## o[]o Ortsregister
## t[]t Titel
## z[]z Zeit
```
import json
import re
import csv
import glob
jsonpath = '../json'
csvpath = '../csv'
docpath = '../docx'
with open(f"{jsonpath}/2021-03-16-inventar-gesamt.json", 'r') as f:
db = json.load(f)
# logik zum parsen der lemmata aus den kategorien-strings, gibt eine liste aller lemmata einer kategorie aus.
def re_parse(type, string):
# print(f"type {type} und string {string}\n")
ergebnis_liste = list()
if substring_re_ergebnisse := re.finditer(r"([a-z])\[(.+)\]\1", string):
for substring_match in substring_re_ergebnisse:
ergebnis_liste.extend(re_parse(substring_match.group(1), substring_match.group(2)))
match type:
case "p":
person = dict()
string = re.sub(r"([tksop]\[)|(\][tksop])","",string)
if re_matches := re.findall(r"([1-9])\{([^\{]+)}", string):
for re_match in re_matches:
match re_match[0]:
case "1":
person["name"] = re_match[1]
case "2":
person["vorname"] = re_match[1]
case "3":
person["adelstitel"] = re_match[1]
case "4":
person["akademischergrad"] = re_match[1]
case "5":
person["beruf"] = re_match[1]
case "6":
person["ort"] = re_match[1]
case "7":
person["sonstiges"] = re_match[1]
case "8":
person["praefix"] = re_match[1]
case "9":
person["pseudonym"] = re_match[1]
else:
person["name"] = string
ergebnis_liste.append({'p':person})
return ergebnis_liste
case "s":
newstring = re.sub(r"([tksopz]\[)|(\][tksopz])","",string)
if re_matches := re.findall(r"([1-9])\{([^\{]+)}", newstring):
schlagwort = dict()
for re_match in re_matches:
match re_match[0]:
case "1":
schlagwort["1"] = re_match[1]
case "2":
schlagwort["2"] = re_match[1]
case "3":
schlagwort["3"] = re_match[1]
else:
schlagwort = string
ergebnis_liste.append({'s':schlagwort})
return ergebnis_liste
case "k":
string = re.sub(r"([tksopt]\[)|(\][tksopt])","",string)
string = re.sub(r"([1-9]\{)|(\})", "", string)
if re_matches := re.findall(r"([1-9])\{([^\{]+)}", string):
koerperschaft = dict()
for re_match in re_matches:
match re_match[0]:
case "1":
koerperschaft["1"] = re_match[1]
case "2":
koerperschaft["2"] = re_match[1]
else:
koerperschaft = string
ergebnis_liste.append({'k':koerperschaft})
return ergebnis_liste
case "o":
if re_matches := re.findall(r"([1-9])\{([^\{]+)}", string):
ort = dict()
for re_match in re_matches:
match re_match[0]:
case "1":
ort["1"] = re_match[1]
case "2":
ort["2"] = re_match[1]
case "3":
ort["3"] = re_match[1]
else:
ort = string
ergebnis_liste.append({'o': ort})
return ergebnis_liste
case "t":
titel = re.sub(r"([tksopz]\[)|(\][tksopz])","",string)
titel = re.sub(r"([1-9]\{)|(\})", "", titel)
ergebnis_liste.append({'t':titel})
return ergebnis_liste
case "z":
return ergebnis_liste
# nimmt eine Liste aller lemmata eines faszikels entgegen, formatiert diese und schreibt sie in die passende csv-datei
def write_faszikellemmata(sig, faszikel_lemmata):
for lemma in faszikel_lemmata:
match list(lemma.keys())[0]:
case 'p':
person_format = ""
if "name" in lemma['p']:
person_format = lemma['p']["name"]
if "vorname" in lemma['p']:
person_format += f', {lemma["p"]["vorname"]}'
if "praefix" in lemma['p']:
person_format += f' {lemma["p"]["praefix"]}'
if "akademischergrad" in lemma['p']:
person_format += f' ({lemma["p"]["akademischergrad"]})'
if "adelstitel" in lemma['p'] and "beruf" not in lemma['p']:
if "ort" in lemma['p']:
person_format += f'{lemma["p"]["ort"]}, '
person_format += f'{lemma["p"]["adelstitel"]} '
if "vorname" in lemma['p']:
person_format += f'{lemma["p"]["vorname"]} '
if "name" in lemma['p']:
person_format += f'{lemma["p"]["name"] } '
if "praefix" in lemma['p']:
person_format += f'{lemma["p"]["praefix"]}'
elif "beruf" in lemma['p'] or "ort" in lemma['p'] or "pseudonym" in lemma['p'] or "sonstiges" in lemma['p']:
person_format += ' <'
if "beruf" in lemma['p'] and "ort" in lemma['p']:
person_format += f'{lemma["p"]["beruf"]} in {lemma["p"]["ort"]}'
elif "beruf" in lemma['p'] and "ort" not in lemma['p']:
person_format += f"{lemma['p']['beruf']}"
elif "ort" in lemma['p'] and "beruf" not in lemma['p']:
person_format += lemma['p']["ort"]
if "adelstitel" in lemma['p']:
person_format += lemma['p']['adelstitel']
if "sonstiges" in lemma['p']:
person_format += f" {lemma['p']['sonstiges']}"
person_format += '>'
with open(f"{csvpath}/personen.csv","a", newline='') as p:
csvwriter = csv.writer(p, delimiter=',', quoting=csv.QUOTE_ALL, escapechar='\\', quotechar='"')
csvwriter.writerow([person_format,sig])
case 't':
with open(f'{csvpath}/titel.csv', 'a', newline='') as t:
csvwriter = csv.writer(t, delimiter=',', quoting=csv.QUOTE_ALL, escapechar='\\', quotechar='"')
csvwriter.writerow([lemma["t"],sig])
case 's':
if isinstance(lemma['s'], dict):
schlagwort_format = str()
if "1" in lemma['s']:
schlagwort_format += lemma['s']['1']
if "2" in lemma['s']:
schlagwort_format += f", {lemma['s']['2']}"
if "3" in lemma['s']:
schlagwort_format += f", {lemma['s']['3']}"
else:
schlagwort_format = lemma['s']
with open(f'{csvpath}/schlagworte.csv', 'a', newline='') as s:
csvwriter = csv.writer(s, delimiter=',', quoting=csv.QUOTE_ALL, escapechar='\\', quotechar='"')
csvwriter.writerow([schlagwort_format,sig])
case 'o':
if isinstance(lemma['o'], dict):
ort_format = str()
if "1" in lemma['o']:
ort_format = lemma['o']['1']
if "2" in lemma['o']:
ort_format += f", {lemma['o']['2']}"
if "3" in lemma['o']:
ort_format += f", {lemma['o']['3']}"
else:
ort_format = lemma['o']
with open(f'{csvpath}/orte.csv', 'a', newline='') as o:
csvwriter = csv.writer(o, delimiter=',', quoting=csv.QUOTE_ALL, escapechar='\\', quotechar='"')
csvwriter.writerow([ort_format,sig])
case 'k':
if isinstance(lemma['k'], dict):
koerperschaft_format = str()
if "1" in lemma['k']:
koerperschaft_format = lemma['k']['1']
if "2" in lemma['k']:
koerperschaft_format += f", {lemma['k']['2']}"
else:
koerperschaft_format = lemma['k']
with open(f'{csvpath}/koerperschaften.csv', 'a', newline='') as k:
csvwriter = csv.writer(k, delimiter=',', quoting=csv.QUOTE_ALL, escapechar='\\', quotechar='"')
csvwriter.writerow([koerperschaft_format,sig])
#main
# print(f'{len(db["archive"])} Archive')
lemmata = list()
for archiv in db["archive"].items():
# print(archiv)
for bestand in archiv[1]["bestaende"].items():
# print(bestand)
if "unterbestaende" in bestand[1]:
for unterbestand in bestand[1]["unterbestaende"].items():
if "faszikel" in unterbestand[1]:
for faszikel in unterbestand[1]["faszikel"].items():
faszikel_lemmata = list()
sig = f"{archiv[0]} / {bestand[0]} / {unterbestand[0]} / {faszikel[0]}"
for kategorie in faszikel[1].items():
if re_ergebnisse := re.finditer(r"([a-z])\[(.+)\]\1", kategorie[1]):
for match in re_ergebnisse:
faszikel_lemmata.extend(re_parse(match.group(1), match.group(2)))
write_faszikellemmata(sig, faszikel_lemmata)
elif "faszikel" in bestand[1]:
for faszikel in bestand[1]["faszikel"].items():
faszikel_lemmata = list()
sig = f"{archiv[0]} / {bestand[0]} / {faszikel[0]}"
for kategorie in faszikel[1].items():
if re_ergebnisse := re.finditer(r"([a-z])\[(.+)\]\1", kategorie[1]):
for match in re_ergebnisse:
faszikel_lemmata.extend(re_parse(match.group(1), match.group(2)))
write_faszikellemmata(sig, faszikel_lemmata)
```
# CSV-Dateien sortieren
```
file_list = glob.glob(f'{csvpath}/*.csv')
for file in file_list:
with open(file, 'r') as f:
reader = csv.reader(f, delimiter=",", quoting=csv.QUOTE_ALL, escapechar='\\', quotechar='"')
sortedlist = sorted(reader, key=lambda row: row[0])
with open(file, 'w') as f:
writer = csv.writer(f, delimiter=',', quoting=csv.QUOTE_ALL, escapechar='\\', quotechar='"')
for item in sortedlist:
writer.writerow(item)
```
|
github_jupyter
|
import json
import re
import csv
import glob
jsonpath = '../json'
csvpath = '../csv'
docpath = '../docx'
with open(f"{jsonpath}/2021-03-16-inventar-gesamt.json", 'r') as f:
db = json.load(f)
# logik zum parsen der lemmata aus den kategorien-strings, gibt eine liste aller lemmata einer kategorie aus.
def re_parse(type, string):
# print(f"type {type} und string {string}\n")
ergebnis_liste = list()
if substring_re_ergebnisse := re.finditer(r"([a-z])\[(.+)\]\1", string):
for substring_match in substring_re_ergebnisse:
ergebnis_liste.extend(re_parse(substring_match.group(1), substring_match.group(2)))
match type:
case "p":
person = dict()
string = re.sub(r"([tksop]\[)|(\][tksop])","",string)
if re_matches := re.findall(r"([1-9])\{([^\{]+)}", string):
for re_match in re_matches:
match re_match[0]:
case "1":
person["name"] = re_match[1]
case "2":
person["vorname"] = re_match[1]
case "3":
person["adelstitel"] = re_match[1]
case "4":
person["akademischergrad"] = re_match[1]
case "5":
person["beruf"] = re_match[1]
case "6":
person["ort"] = re_match[1]
case "7":
person["sonstiges"] = re_match[1]
case "8":
person["praefix"] = re_match[1]
case "9":
person["pseudonym"] = re_match[1]
else:
person["name"] = string
ergebnis_liste.append({'p':person})
return ergebnis_liste
case "s":
newstring = re.sub(r"([tksopz]\[)|(\][tksopz])","",string)
if re_matches := re.findall(r"([1-9])\{([^\{]+)}", newstring):
schlagwort = dict()
for re_match in re_matches:
match re_match[0]:
case "1":
schlagwort["1"] = re_match[1]
case "2":
schlagwort["2"] = re_match[1]
case "3":
schlagwort["3"] = re_match[1]
else:
schlagwort = string
ergebnis_liste.append({'s':schlagwort})
return ergebnis_liste
case "k":
string = re.sub(r"([tksopt]\[)|(\][tksopt])","",string)
string = re.sub(r"([1-9]\{)|(\})", "", string)
if re_matches := re.findall(r"([1-9])\{([^\{]+)}", string):
koerperschaft = dict()
for re_match in re_matches:
match re_match[0]:
case "1":
koerperschaft["1"] = re_match[1]
case "2":
koerperschaft["2"] = re_match[1]
else:
koerperschaft = string
ergebnis_liste.append({'k':koerperschaft})
return ergebnis_liste
case "o":
if re_matches := re.findall(r"([1-9])\{([^\{]+)}", string):
ort = dict()
for re_match in re_matches:
match re_match[0]:
case "1":
ort["1"] = re_match[1]
case "2":
ort["2"] = re_match[1]
case "3":
ort["3"] = re_match[1]
else:
ort = string
ergebnis_liste.append({'o': ort})
return ergebnis_liste
case "t":
titel = re.sub(r"([tksopz]\[)|(\][tksopz])","",string)
titel = re.sub(r"([1-9]\{)|(\})", "", titel)
ergebnis_liste.append({'t':titel})
return ergebnis_liste
case "z":
return ergebnis_liste
# nimmt eine Liste aller lemmata eines faszikels entgegen, formatiert diese und schreibt sie in die passende csv-datei
def write_faszikellemmata(sig, faszikel_lemmata):
for lemma in faszikel_lemmata:
match list(lemma.keys())[0]:
case 'p':
person_format = ""
if "name" in lemma['p']:
person_format = lemma['p']["name"]
if "vorname" in lemma['p']:
person_format += f', {lemma["p"]["vorname"]}'
if "praefix" in lemma['p']:
person_format += f' {lemma["p"]["praefix"]}'
if "akademischergrad" in lemma['p']:
person_format += f' ({lemma["p"]["akademischergrad"]})'
if "adelstitel" in lemma['p'] and "beruf" not in lemma['p']:
if "ort" in lemma['p']:
person_format += f'{lemma["p"]["ort"]}, '
person_format += f'{lemma["p"]["adelstitel"]} '
if "vorname" in lemma['p']:
person_format += f'{lemma["p"]["vorname"]} '
if "name" in lemma['p']:
person_format += f'{lemma["p"]["name"] } '
if "praefix" in lemma['p']:
person_format += f'{lemma["p"]["praefix"]}'
elif "beruf" in lemma['p'] or "ort" in lemma['p'] or "pseudonym" in lemma['p'] or "sonstiges" in lemma['p']:
person_format += ' <'
if "beruf" in lemma['p'] and "ort" in lemma['p']:
person_format += f'{lemma["p"]["beruf"]} in {lemma["p"]["ort"]}'
elif "beruf" in lemma['p'] and "ort" not in lemma['p']:
person_format += f"{lemma['p']['beruf']}"
elif "ort" in lemma['p'] and "beruf" not in lemma['p']:
person_format += lemma['p']["ort"]
if "adelstitel" in lemma['p']:
person_format += lemma['p']['adelstitel']
if "sonstiges" in lemma['p']:
person_format += f" {lemma['p']['sonstiges']}"
person_format += '>'
with open(f"{csvpath}/personen.csv","a", newline='') as p:
csvwriter = csv.writer(p, delimiter=',', quoting=csv.QUOTE_ALL, escapechar='\\', quotechar='"')
csvwriter.writerow([person_format,sig])
case 't':
with open(f'{csvpath}/titel.csv', 'a', newline='') as t:
csvwriter = csv.writer(t, delimiter=',', quoting=csv.QUOTE_ALL, escapechar='\\', quotechar='"')
csvwriter.writerow([lemma["t"],sig])
case 's':
if isinstance(lemma['s'], dict):
schlagwort_format = str()
if "1" in lemma['s']:
schlagwort_format += lemma['s']['1']
if "2" in lemma['s']:
schlagwort_format += f", {lemma['s']['2']}"
if "3" in lemma['s']:
schlagwort_format += f", {lemma['s']['3']}"
else:
schlagwort_format = lemma['s']
with open(f'{csvpath}/schlagworte.csv', 'a', newline='') as s:
csvwriter = csv.writer(s, delimiter=',', quoting=csv.QUOTE_ALL, escapechar='\\', quotechar='"')
csvwriter.writerow([schlagwort_format,sig])
case 'o':
if isinstance(lemma['o'], dict):
ort_format = str()
if "1" in lemma['o']:
ort_format = lemma['o']['1']
if "2" in lemma['o']:
ort_format += f", {lemma['o']['2']}"
if "3" in lemma['o']:
ort_format += f", {lemma['o']['3']}"
else:
ort_format = lemma['o']
with open(f'{csvpath}/orte.csv', 'a', newline='') as o:
csvwriter = csv.writer(o, delimiter=',', quoting=csv.QUOTE_ALL, escapechar='\\', quotechar='"')
csvwriter.writerow([ort_format,sig])
case 'k':
if isinstance(lemma['k'], dict):
koerperschaft_format = str()
if "1" in lemma['k']:
koerperschaft_format = lemma['k']['1']
if "2" in lemma['k']:
koerperschaft_format += f", {lemma['k']['2']}"
else:
koerperschaft_format = lemma['k']
with open(f'{csvpath}/koerperschaften.csv', 'a', newline='') as k:
csvwriter = csv.writer(k, delimiter=',', quoting=csv.QUOTE_ALL, escapechar='\\', quotechar='"')
csvwriter.writerow([koerperschaft_format,sig])
#main
# print(f'{len(db["archive"])} Archive')
lemmata = list()
for archiv in db["archive"].items():
# print(archiv)
for bestand in archiv[1]["bestaende"].items():
# print(bestand)
if "unterbestaende" in bestand[1]:
for unterbestand in bestand[1]["unterbestaende"].items():
if "faszikel" in unterbestand[1]:
for faszikel in unterbestand[1]["faszikel"].items():
faszikel_lemmata = list()
sig = f"{archiv[0]} / {bestand[0]} / {unterbestand[0]} / {faszikel[0]}"
for kategorie in faszikel[1].items():
if re_ergebnisse := re.finditer(r"([a-z])\[(.+)\]\1", kategorie[1]):
for match in re_ergebnisse:
faszikel_lemmata.extend(re_parse(match.group(1), match.group(2)))
write_faszikellemmata(sig, faszikel_lemmata)
elif "faszikel" in bestand[1]:
for faszikel in bestand[1]["faszikel"].items():
faszikel_lemmata = list()
sig = f"{archiv[0]} / {bestand[0]} / {faszikel[0]}"
for kategorie in faszikel[1].items():
if re_ergebnisse := re.finditer(r"([a-z])\[(.+)\]\1", kategorie[1]):
for match in re_ergebnisse:
faszikel_lemmata.extend(re_parse(match.group(1), match.group(2)))
write_faszikellemmata(sig, faszikel_lemmata)
file_list = glob.glob(f'{csvpath}/*.csv')
for file in file_list:
with open(file, 'r') as f:
reader = csv.reader(f, delimiter=",", quoting=csv.QUOTE_ALL, escapechar='\\', quotechar='"')
sortedlist = sorted(reader, key=lambda row: row[0])
with open(file, 'w') as f:
writer = csv.writer(f, delimiter=',', quoting=csv.QUOTE_ALL, escapechar='\\', quotechar='"')
for item in sortedlist:
writer.writerow(item)
| 0.074744 | 0.520314 |
# Naive Bayes Model for Newsgroups Data
For an explanation of the Naive Bayes model, see [our course notes](https://jennselby.github.io/MachineLearningCourseNotes/#naive-bayes).
This notebook uses code from http://scikit-learn.org/stable/tutorial/text_analytics/working_with_text_data.html.
## Instructions
0. If you haven't already, follow [the setup instructions here](https://jennselby.github.io/MachineLearningCourseNotes/#setting-up-python3) to get all necessary software installed.
0. Read through the code in the following sections:
* [Newgroups Data](#Newgroups-Data)
* [Model Training](#Model-Training)
* [Prediction](#Prediction)
0. Complete at least one of the following exercises:
* [Exercise Option #1 - Standard Difficulty](#Exercise-Option-#1---Standard-Difficulty)
* [Exercise Option #2 - Advanced Difficulty](#Exercise-Option-#2---Advanced-Difficulty)
```
from sklearn.datasets import fetch_20newsgroups # the 20 newgroups set is included in scikit-learn
from sklearn.naive_bayes import MultinomialNB # we need this for our Naive Bayes model
# These next two are about processing the data. We'll look into this more later in the semester.
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.feature_extraction.text import TfidfTransformer
```
## Newgroups Data
Back in the day, [Usenet](https://en.wikipedia.org/wiki/Usenet_newsgroup) was a popular discussion system where people could discuss topics in relevant newsgroups (think Slack channel or subreddit). At some point, someone pulled together messages sent to 20 different newsgroups, to use as [a dataset for doing text processing](http://qwone.com/~jason/20Newsgroups/).
We are going to pull out messages from just a few different groups to try out a Naive Bayes model.
Examine the newsgroups dictionary, to make sure you understand the dataset.
**Note**: If you get an error about SSL certificates, you can fix this with the following:
1. In Finder, click on Applications in the list on the left panel
1. Double click to go into the Python folder (it will be called something like Python 3.7)
1. Double click on the Install Certificates command in that folder
```
# which newsgroups we want to download
newsgroup_names = ['comp.graphics', 'rec.sport.hockey', 'sci.electronics', 'sci.space']
# get the newsgroup data (organized much like the iris data)
newsgroups = fetch_20newsgroups(categories=newsgroup_names, shuffle=True, random_state=265)
newsgroups.keys()
```
This next part does some processing of the data, because the scikit-learn Naive Bayes module is expecting numerical data rather than text data. We will talk more about what this code is doing later in the semester. For now, you can ignore it.
```
# Convert the text into numbers that represent each word (bag of words method)
word_vector = CountVectorizer()
word_vector_counts = word_vector.fit_transform(newsgroups.data)
# Account for the length of the documents:
# get the frequency with which the word occurs instead of the raw number of times
term_freq_transformer = TfidfTransformer()
term_freq = term_freq_transformer.fit_transform(word_vector_counts)
```
## Model Training
Now we fit the Naive Bayes model to the subset of the 20 newsgroups data that we've pulled out.
```
# Train the Naive Bayes model
model = MultinomialNB().fit(term_freq, newsgroups.target)
```
## Prediction
Let's see how the model does on some (very short) documents that we made up to fit into the specific categories our model is trained on.
```
# Predict some new fake documents
fake_docs = [
'That GPU has amazing performance with a lot of shaders',
'The player had a wicked slap shot',
'I spent all day yesterday soldering banks of capacitors',
'Today I have to solder a bank of capacitors',
'NASA has rovers on Mars']
fake_counts = word_vector.transform(fake_docs)
fake_term_freq = term_freq_transformer.transform(fake_counts)
predicted = model.predict(fake_term_freq)
print('Predictions:')
for doc, group in zip(fake_docs, predicted):
print('\t{0} => {1}'.format(doc, newsgroups.target_names[group]))
probabilities = model.predict_proba(fake_term_freq)
print('Probabilities:')
print(''.join(['{:17}'.format(name) for name in newsgroups.target_names]))
for probs in probabilities:
print(''.join(['{:<17.8}'.format(prob) for prob in probs]))
```
# Exercise Option #1 - Standard Difficulty
Modify the fake documents and add some new documents of your own.
What words in your documents have particularly large effects on the model probabilities? Note that we're not looking for documents that consist of a single word, but for words that, when included or excluded from a document, tend to change the model's output.
# Exercise Option #2 - Advanced Difficulty
Write some code to count up how often the words you found in the exercise above appear in each category in the training dataset. Does this match up with your intuition?
|
github_jupyter
|
from sklearn.datasets import fetch_20newsgroups # the 20 newgroups set is included in scikit-learn
from sklearn.naive_bayes import MultinomialNB # we need this for our Naive Bayes model
# These next two are about processing the data. We'll look into this more later in the semester.
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.feature_extraction.text import TfidfTransformer
# which newsgroups we want to download
newsgroup_names = ['comp.graphics', 'rec.sport.hockey', 'sci.electronics', 'sci.space']
# get the newsgroup data (organized much like the iris data)
newsgroups = fetch_20newsgroups(categories=newsgroup_names, shuffle=True, random_state=265)
newsgroups.keys()
# Convert the text into numbers that represent each word (bag of words method)
word_vector = CountVectorizer()
word_vector_counts = word_vector.fit_transform(newsgroups.data)
# Account for the length of the documents:
# get the frequency with which the word occurs instead of the raw number of times
term_freq_transformer = TfidfTransformer()
term_freq = term_freq_transformer.fit_transform(word_vector_counts)
# Train the Naive Bayes model
model = MultinomialNB().fit(term_freq, newsgroups.target)
# Predict some new fake documents
fake_docs = [
'That GPU has amazing performance with a lot of shaders',
'The player had a wicked slap shot',
'I spent all day yesterday soldering banks of capacitors',
'Today I have to solder a bank of capacitors',
'NASA has rovers on Mars']
fake_counts = word_vector.transform(fake_docs)
fake_term_freq = term_freq_transformer.transform(fake_counts)
predicted = model.predict(fake_term_freq)
print('Predictions:')
for doc, group in zip(fake_docs, predicted):
print('\t{0} => {1}'.format(doc, newsgroups.target_names[group]))
probabilities = model.predict_proba(fake_term_freq)
print('Probabilities:')
print(''.join(['{:17}'.format(name) for name in newsgroups.target_names]))
for probs in probabilities:
print(''.join(['{:<17.8}'.format(prob) for prob in probs]))
| 0.686265 | 0.989918 |
```
from pyFrame import Frame, Beam
import ipywidgets as widgets
import copy
BedFrame = Frame.Frame()
```
### Define Materials
```
#C24 Norm for German Construction wood in mm
E = 11000
G = 690
KVH = Beam.Material(E,G)
KVH.tensile_yield_strength = 14
KVH.compressive_yield_strength = 21
KVH.flexural_yield_strength = 24
KVH.shear_strength = 4
```
### Define Crosssections
```
def rect_crosssection(width, height):
return Beam.Crosssection([(0,0),(0,height),(width,height),(width,0)])
T60100 = rect_crosssection(60,100)
T60120 = rect_crosssection(60,120)
T80100 = rect_crosssection(80,100)
T80140 = rect_crosssection(80,140)
T100100 = rect_crosssection(100,100)
T10060 = rect_crosssection(100, 60)
T12060 = rect_crosssection(120, 60)
T10080 = rect_crosssection(100, 80)
T14080 = rect_crosssection(140, 80)
T60120.plot()
T80140.plot()
T100100.plot()
```
## Create Base Frame
### Define Nodes
```
#addding nodes to Frame
xDim = 2200
yDim = 2600
zDim = 3000
BedFrame.addNode('G1', 0,0,0)
BedFrame.addNode('G2',xDim,0,0)
BedFrame.addNode('G3', 0,yDim,0)
BedFrame.addNode('G4',xDim,yDim,0)
BedFrame.addNode('G5', 0,1000,0)
BedFrame.addNode('T1', 0,1000,zDim)
BedFrame.addNode('T2',xDim,1000,zDim)
BedFrame.addNode('T3', 0,yDim,zDim)
BedFrame.addNode('T4',xDim,yDim,zDim)
BedFrame.addNode('P1', 0,0,zDim - 1000)
BedFrame.addNode('P2',xDim,0,zDim - 1000)
BedFrame.addNode('P3', 0,1000,zDim - 1000)
BedFrame.addNode('P4',xDim,1000,zDim - 1000)
```
### Add Members
```
#add Members
BedFrame.addMember('col1', 'G1','P1', KVH, T14080)
BedFrame.addMember('col2', 'G2','P2', KVH, T14080)
BedFrame.addMember('col3', 'G3','T3', KVH, T14080)
BedFrame.addMember('col4', 'G4','T4', KVH, T14080)
BedFrame.addMember('col5', 'G5','P3', KVH, T14080)
BedFrame.addMember('TopBeam1', 'T1','T2', KVH, T60100)
BedFrame.addMember('TopBeam2', 'T3','T4', KVH, T60100)
BedFrame.addMember('TopBeam3', 'T1','T3', KVH, T80140)
BedFrame.addMember('TopBeam4', 'T2','T4', KVH, T80140)
BedFrame.addMember('PlattFormBeam1', 'P1','P2', KVH, T60100)
BedFrame.addMember('PlattFormBeam2', 'P3','P4', KVH, T60100)
BedFrame.addMember('PlattFormBeam3', 'P1','P3', KVH, T60120)
BedFrame.addMember('PlattFormBeam4', 'P2','P4', KVH, T60120)
BedFrame.addMember('Diagonal2', 'P4','T2', KVH, T60120)
BedFrame.addMember('Diagonal1', 'P3','T1', KVH, T60120)
BedFrame.addMember('Diagonal3', 'P1','T1', KVH, T60100)
BedFrame.addMember('Diagonal4', 'P2','T2', KVH, T60100)
BedFrame.plot(deformed=False, showMemberName= False, showNodeName = False)
#create supports
BedFrame.makeSupport('G1', Ux=0,Uy=0,Uz=0,Rx=0,Ry=0,Rz=0)
BedFrame.makeSupport('G2', Ux=0,Uy=0,Uz=0,Rx=0,Ry=0,Rz=0)
BedFrame.makeSupport('G3', Ux=0,Uy=0,Uz=0,Rx=0,Ry=0,Rz=0)
BedFrame.makeSupport('G4', Ux=0,Uy=0,Uz=0,Rx=0,Ry=0,Rz=0)
BedFrame.makeSupport('G5', Ux=0,Uy=0,Uz=0,Rx=0,Ry=0,Rz=0)
#BedFrame.addNodeLoad('5', Fy=lol*1000,Fx=lol*1000)
#set member loads
BedLoad = 10000
x1 = 100
x2 = 1500
a = abs(BedFrame.Nodes['T2'].x- BedFrame.Nodes['T3'].x)
b = abs(BedFrame.Nodes['T2'].y- BedFrame.Nodes['T3'].y)
BedFrame.addMemberPtForce('TopBeam3', x=x1,Fz=-BedLoad/4)
BedFrame.addMemberPtForce('TopBeam3', x=x1,Fx = BedLoad/4)
BedFrame.addMemberPtForce('TopBeam3', x=x2,Fz=-BedLoad)
BedFrame.addMemberPtForce('TopBeam3', x=x2,Fy=-BedLoad/4)
BedFrame.addMemberPtForce('TopBeam3', x=x2,Fx=-BedLoad/4*a/((a**2+b**2)**(0.5)), Fy=-BedLoad/4*b/((a**2+b**2)**(0.5)))
BedFrame.addMemberPtForce('TopBeam3', x=x2,Fx=-BedLoad/4)
BedFrame.addMemberPtForce('TopBeam4', x=x1,Fz=-BedLoad/4)
BedFrame.addMemberPtForce('TopBeam4', x=x1,Fx=BedLoad/4*a/((a**2+b**2)**(0.5)), Fy=BedLoad/4*b/((a**2+b**2)**(0.5)))
BedFrame.addMemberPtForce('TopBeam4', x=x2,Fz=-BedLoad/4)
BedFrame.addMemberPtForce('TopBeam4', x=x2,Fy=BedLoad/4)
BedFrame.analyze()
#BedFrame.plot()
widgets.interact(BedFrame.plot,
label_offset=(0.01, 0.1, 0.01),
xMargin=(0.25, 3, 0.25),
yMargin=(0.25, 3, 0.25),
zMargin=(0.5, 3, 0.25),
elevation=(0,360,10),
rotation=(0,360,10),
xFac=(1,50,1))
```
### Compute minimum Margin of Safeties
```
BedFrame.MoS()
widgets.interact(BedFrame.Members['col4'].plot,
label_offset=(0.01, 0.1, 0.01),
xMargin=(0.25, 3, 0.25),
yMargin=(0.25, 3, 0.25),
zMargin=(0.5, 3, 0.25),
elevation=(0,360,10),
rotation=(0,360,10),
xFac=(1,20,1))
```
# Due to Material Constraints Need to Reiterate
### At Bauhaus, I noticed that it is cheaper to buy the same kind of wood to get discounts. Also, they ran out of 80x140 beams thus I had buy 160x40 beams
```
BedFrame2 = Frame.Frame()
```
### New Beams
```
T80160 = rect_crosssection(80,160)
T16080 = rect_crosssection(160,80)
T80160.plot()
T16080.plot()
```
## Create Base Frame
### Define Nodes
```
#addding nodes to Frame
xDim = 2200
yDim = 2600
zDim = 3000
BedFrame2.addNode('G1', 0,0,0)
BedFrame2.addNode('G2',xDim,0,0)
BedFrame2.addNode('G3', 0,yDim,0)
BedFrame2.addNode('G4',xDim,yDim,0)
BedFrame2.addNode('G5', 0,1000,0)
BedFrame2.addNode('T1', 0,1000,zDim)
BedFrame2.addNode('T2',xDim,1000,zDim)
BedFrame2.addNode('T3', 0,yDim,zDim)
BedFrame2.addNode('T4',xDim,yDim,zDim)
BedFrame2.addNode('P1', 0,0,zDim - 1000)
BedFrame2.addNode('P1.1', xDim/4,0,zDim - 1000)
BedFrame2.addNode('P1.2', 2*xDim/4,0,zDim - 1000)
BedFrame2.addNode('P1.3', 3*xDim/4,0,zDim - 1000)
BedFrame2.addNode('P2',xDim,0,zDim - 1000)
BedFrame2.addNode('P3', 0,1000,zDim - 1000)
BedFrame2.addNode('P3.1', xDim/4,1000,zDim - 1000)
BedFrame2.addNode('P3.2', 2*xDim/4,1000,zDim - 1000)
BedFrame2.addNode('P3.3', 3*xDim/4,1000,zDim - 1000)
BedFrame2.addNode('P4',xDim,1000,zDim - 1000)
BedFrame2.addNode('P5', 0,yDim,zDim - 1000)
BedFrame2.addNode('P6',xDim,yDim,zDim - 1000)
```
### Add Members
```
#add Members
BedFrame2.addMember('col1', 'G1','P1', KVH, T16080)
BedFrame2.addMember('col2', 'G2','P2', KVH, T16080)
BedFrame2.addMember('col3A', 'G3','P5', KVH, T16080)
BedFrame2.addMember('col4A', 'G4','P6', KVH, T16080)
BedFrame2.addMember('col3B', 'P5', 'T3', KVH, T16080)
BedFrame2.addMember('col4B', 'P6', 'T4', KVH, T16080)
BedFrame2.addMember('col5', 'G5','P3', KVH, T16080)
BedFrame2.addMember('TopBeam1', 'T1','T2', KVH, T80160)
BedFrame2.addMember('TopBeam2', 'T3','T4', KVH, T80160)
BedFrame2.addMember('TopBeam3', 'T1','T3', KVH, T80160)
BedFrame2.addMember('TopBeam4', 'T2','T4', KVH, T80160)
BedFrame2.addMember('PlattFormBeam1.1', 'P1','P1.1', KVH, T80160)
BedFrame2.addMember('PlattFormBeam1.2', 'P1.1','P1.2', KVH, T80160)
BedFrame2.addMember('PlattFormBeam1.3', 'P1.2','P1.3', KVH, T80160)
BedFrame2.addMember('PlattFormBeam1.4', 'P1.3','P2', KVH, T80160)
BedFrame2.addMember('PlattFormBeam2.1', 'P3','P3.1', KVH, T80160)
BedFrame2.addMember('PlattFormBeam2.2', 'P3.1','P3.2', KVH, T80160)
BedFrame2.addMember('PlattFormBeam2.3', 'P3.2','P3.3', KVH, T80160)
BedFrame2.addMember('PlattFormBeam2.4', 'P3.3','P4', KVH, T80160)
BedFrame2.addMember('PlattFormBeam3', 'P1','P3', KVH, T80160)
BedFrame2.addMember('PlattFormBeam4', 'P2','P4', KVH, T80160)
BedFrame2.addMember('PlattFormBeam5.1', 'P1.1','P3.1', KVH, T80160)
BedFrame2.addMember('PlattFormBeam5.2', 'P1.2','P3.2', KVH, T80160)
BedFrame2.addMember('PlattFormBeam5.3', 'P1.3','P3.3', KVH, T80160)
BedFrame2.addMember('PlattFormBeam6', 'P5','P6', KVH, T80160)
BedFrame2.addMember('Diagonal2', 'P4','T2', KVH, T80160)
BedFrame2.addMember('Diagonal1', 'P3','T1', KVH, T80160)
BedFrame2.addMember('Diagonal3', 'P1','T1', KVH, T80160)
BedFrame2.addMember('Diagonal4', 'P2','T2', KVH, T80160)
BedFrame2.plot(deformed=False, showMemberName= False, showNodeName = False)
#create supports
BedFrame2.makeSupport('G1', Ux=0,Uy=0,Uz=0,Rx=0,Ry=0,Rz=0)
BedFrame2.makeSupport('G2', Ux=0,Uy=0,Uz=0,Rx=0,Ry=0,Rz=0)
BedFrame2.makeSupport('G3', Ux=0,Uy=0,Uz=0,Rx=0,Ry=0,Rz=0)
BedFrame2.makeSupport('G4', Ux=0,Uy=0,Uz=0,Rx=0,Ry=0,Rz=0)
BedFrame2.makeSupport('G5', Ux=0,Uy=0,Uz=0,Rx=0,Ry=0,Rz=0)
#set member loads
BedLoad = 10000
x1 = 100
x2 = 1500
a = abs(BedFrame2.Nodes['T2'].x- BedFrame2.Nodes['T3'].x)
b = abs(BedFrame2.Nodes['T2'].y- BedFrame2.Nodes['T3'].y)
BedFrame2.addMemberPtForce('TopBeam3', x=x1,Fz=-BedLoad/4)
BedFrame2.addMemberPtForce('TopBeam3', x=x1,Fx = BedLoad/4)
BedFrame2.addMemberPtForce('TopBeam3', x=x2,Fz=-BedLoad)
BedFrame2.addMemberPtForce('TopBeam3', x=x2,Fy=-BedLoad/4)
BedFrame2.addMemberPtForce('TopBeam3', x=x2,Fx=-BedLoad/4*a/((a**2+b**2)**(0.5)), Fy=-BedLoad/4*b/((a**2+b**2)**(0.5)))
BedFrame2.addMemberPtForce('TopBeam3', x=x2,Fx=-BedLoad/4)
BedFrame2.addMemberPtForce('TopBeam4', x=x1,Fz=-BedLoad/4)
BedFrame2.addMemberPtForce('TopBeam4', x=x1,Fx=BedLoad/4*a/((a**2+b**2)**(0.5)), Fy=BedLoad/4*b/((a**2+b**2)**(0.5)))
BedFrame2.addMemberPtForce('TopBeam4', x=x2,Fz=-BedLoad/4)
BedFrame2.addMemberPtForce('TopBeam4', x=x2,Fy=BedLoad/4)
U = BedFrame2.analyze()
#BedFrame.plot()
widgets.interact(BedFrame2.plot,
label_offset=(0.01, 0.1, 0.01),
xMargin=(0.25, 3, 0.25),
yMargin=(0.25, 3, 0.25),
zMargin=(0.5, 3, 0.25),
elevation=(0,360,10),
rotation=(0,360,10),
xFac=(1,200,10))
```
### Compute minimum Margin of Safeties
```
BedFrame2.MoS()
```
## Evaluation
With the new wood, we notice that our minimum MoS is much larger then the original Frame.
If we play with the Loads, we find that the Frame Fails at about 26 kN !!
Thus, we expect this frame to carry 10 kN with a MoS of 1.6 at the TopBeam3
|
github_jupyter
|
from pyFrame import Frame, Beam
import ipywidgets as widgets
import copy
BedFrame = Frame.Frame()
#C24 Norm for German Construction wood in mm
E = 11000
G = 690
KVH = Beam.Material(E,G)
KVH.tensile_yield_strength = 14
KVH.compressive_yield_strength = 21
KVH.flexural_yield_strength = 24
KVH.shear_strength = 4
def rect_crosssection(width, height):
return Beam.Crosssection([(0,0),(0,height),(width,height),(width,0)])
T60100 = rect_crosssection(60,100)
T60120 = rect_crosssection(60,120)
T80100 = rect_crosssection(80,100)
T80140 = rect_crosssection(80,140)
T100100 = rect_crosssection(100,100)
T10060 = rect_crosssection(100, 60)
T12060 = rect_crosssection(120, 60)
T10080 = rect_crosssection(100, 80)
T14080 = rect_crosssection(140, 80)
T60120.plot()
T80140.plot()
T100100.plot()
#addding nodes to Frame
xDim = 2200
yDim = 2600
zDim = 3000
BedFrame.addNode('G1', 0,0,0)
BedFrame.addNode('G2',xDim,0,0)
BedFrame.addNode('G3', 0,yDim,0)
BedFrame.addNode('G4',xDim,yDim,0)
BedFrame.addNode('G5', 0,1000,0)
BedFrame.addNode('T1', 0,1000,zDim)
BedFrame.addNode('T2',xDim,1000,zDim)
BedFrame.addNode('T3', 0,yDim,zDim)
BedFrame.addNode('T4',xDim,yDim,zDim)
BedFrame.addNode('P1', 0,0,zDim - 1000)
BedFrame.addNode('P2',xDim,0,zDim - 1000)
BedFrame.addNode('P3', 0,1000,zDim - 1000)
BedFrame.addNode('P4',xDim,1000,zDim - 1000)
#add Members
BedFrame.addMember('col1', 'G1','P1', KVH, T14080)
BedFrame.addMember('col2', 'G2','P2', KVH, T14080)
BedFrame.addMember('col3', 'G3','T3', KVH, T14080)
BedFrame.addMember('col4', 'G4','T4', KVH, T14080)
BedFrame.addMember('col5', 'G5','P3', KVH, T14080)
BedFrame.addMember('TopBeam1', 'T1','T2', KVH, T60100)
BedFrame.addMember('TopBeam2', 'T3','T4', KVH, T60100)
BedFrame.addMember('TopBeam3', 'T1','T3', KVH, T80140)
BedFrame.addMember('TopBeam4', 'T2','T4', KVH, T80140)
BedFrame.addMember('PlattFormBeam1', 'P1','P2', KVH, T60100)
BedFrame.addMember('PlattFormBeam2', 'P3','P4', KVH, T60100)
BedFrame.addMember('PlattFormBeam3', 'P1','P3', KVH, T60120)
BedFrame.addMember('PlattFormBeam4', 'P2','P4', KVH, T60120)
BedFrame.addMember('Diagonal2', 'P4','T2', KVH, T60120)
BedFrame.addMember('Diagonal1', 'P3','T1', KVH, T60120)
BedFrame.addMember('Diagonal3', 'P1','T1', KVH, T60100)
BedFrame.addMember('Diagonal4', 'P2','T2', KVH, T60100)
BedFrame.plot(deformed=False, showMemberName= False, showNodeName = False)
#create supports
BedFrame.makeSupport('G1', Ux=0,Uy=0,Uz=0,Rx=0,Ry=0,Rz=0)
BedFrame.makeSupport('G2', Ux=0,Uy=0,Uz=0,Rx=0,Ry=0,Rz=0)
BedFrame.makeSupport('G3', Ux=0,Uy=0,Uz=0,Rx=0,Ry=0,Rz=0)
BedFrame.makeSupport('G4', Ux=0,Uy=0,Uz=0,Rx=0,Ry=0,Rz=0)
BedFrame.makeSupport('G5', Ux=0,Uy=0,Uz=0,Rx=0,Ry=0,Rz=0)
#BedFrame.addNodeLoad('5', Fy=lol*1000,Fx=lol*1000)
#set member loads
BedLoad = 10000
x1 = 100
x2 = 1500
a = abs(BedFrame.Nodes['T2'].x- BedFrame.Nodes['T3'].x)
b = abs(BedFrame.Nodes['T2'].y- BedFrame.Nodes['T3'].y)
BedFrame.addMemberPtForce('TopBeam3', x=x1,Fz=-BedLoad/4)
BedFrame.addMemberPtForce('TopBeam3', x=x1,Fx = BedLoad/4)
BedFrame.addMemberPtForce('TopBeam3', x=x2,Fz=-BedLoad)
BedFrame.addMemberPtForce('TopBeam3', x=x2,Fy=-BedLoad/4)
BedFrame.addMemberPtForce('TopBeam3', x=x2,Fx=-BedLoad/4*a/((a**2+b**2)**(0.5)), Fy=-BedLoad/4*b/((a**2+b**2)**(0.5)))
BedFrame.addMemberPtForce('TopBeam3', x=x2,Fx=-BedLoad/4)
BedFrame.addMemberPtForce('TopBeam4', x=x1,Fz=-BedLoad/4)
BedFrame.addMemberPtForce('TopBeam4', x=x1,Fx=BedLoad/4*a/((a**2+b**2)**(0.5)), Fy=BedLoad/4*b/((a**2+b**2)**(0.5)))
BedFrame.addMemberPtForce('TopBeam4', x=x2,Fz=-BedLoad/4)
BedFrame.addMemberPtForce('TopBeam4', x=x2,Fy=BedLoad/4)
BedFrame.analyze()
#BedFrame.plot()
widgets.interact(BedFrame.plot,
label_offset=(0.01, 0.1, 0.01),
xMargin=(0.25, 3, 0.25),
yMargin=(0.25, 3, 0.25),
zMargin=(0.5, 3, 0.25),
elevation=(0,360,10),
rotation=(0,360,10),
xFac=(1,50,1))
BedFrame.MoS()
widgets.interact(BedFrame.Members['col4'].plot,
label_offset=(0.01, 0.1, 0.01),
xMargin=(0.25, 3, 0.25),
yMargin=(0.25, 3, 0.25),
zMargin=(0.5, 3, 0.25),
elevation=(0,360,10),
rotation=(0,360,10),
xFac=(1,20,1))
BedFrame2 = Frame.Frame()
T80160 = rect_crosssection(80,160)
T16080 = rect_crosssection(160,80)
T80160.plot()
T16080.plot()
#addding nodes to Frame
xDim = 2200
yDim = 2600
zDim = 3000
BedFrame2.addNode('G1', 0,0,0)
BedFrame2.addNode('G2',xDim,0,0)
BedFrame2.addNode('G3', 0,yDim,0)
BedFrame2.addNode('G4',xDim,yDim,0)
BedFrame2.addNode('G5', 0,1000,0)
BedFrame2.addNode('T1', 0,1000,zDim)
BedFrame2.addNode('T2',xDim,1000,zDim)
BedFrame2.addNode('T3', 0,yDim,zDim)
BedFrame2.addNode('T4',xDim,yDim,zDim)
BedFrame2.addNode('P1', 0,0,zDim - 1000)
BedFrame2.addNode('P1.1', xDim/4,0,zDim - 1000)
BedFrame2.addNode('P1.2', 2*xDim/4,0,zDim - 1000)
BedFrame2.addNode('P1.3', 3*xDim/4,0,zDim - 1000)
BedFrame2.addNode('P2',xDim,0,zDim - 1000)
BedFrame2.addNode('P3', 0,1000,zDim - 1000)
BedFrame2.addNode('P3.1', xDim/4,1000,zDim - 1000)
BedFrame2.addNode('P3.2', 2*xDim/4,1000,zDim - 1000)
BedFrame2.addNode('P3.3', 3*xDim/4,1000,zDim - 1000)
BedFrame2.addNode('P4',xDim,1000,zDim - 1000)
BedFrame2.addNode('P5', 0,yDim,zDim - 1000)
BedFrame2.addNode('P6',xDim,yDim,zDim - 1000)
#add Members
BedFrame2.addMember('col1', 'G1','P1', KVH, T16080)
BedFrame2.addMember('col2', 'G2','P2', KVH, T16080)
BedFrame2.addMember('col3A', 'G3','P5', KVH, T16080)
BedFrame2.addMember('col4A', 'G4','P6', KVH, T16080)
BedFrame2.addMember('col3B', 'P5', 'T3', KVH, T16080)
BedFrame2.addMember('col4B', 'P6', 'T4', KVH, T16080)
BedFrame2.addMember('col5', 'G5','P3', KVH, T16080)
BedFrame2.addMember('TopBeam1', 'T1','T2', KVH, T80160)
BedFrame2.addMember('TopBeam2', 'T3','T4', KVH, T80160)
BedFrame2.addMember('TopBeam3', 'T1','T3', KVH, T80160)
BedFrame2.addMember('TopBeam4', 'T2','T4', KVH, T80160)
BedFrame2.addMember('PlattFormBeam1.1', 'P1','P1.1', KVH, T80160)
BedFrame2.addMember('PlattFormBeam1.2', 'P1.1','P1.2', KVH, T80160)
BedFrame2.addMember('PlattFormBeam1.3', 'P1.2','P1.3', KVH, T80160)
BedFrame2.addMember('PlattFormBeam1.4', 'P1.3','P2', KVH, T80160)
BedFrame2.addMember('PlattFormBeam2.1', 'P3','P3.1', KVH, T80160)
BedFrame2.addMember('PlattFormBeam2.2', 'P3.1','P3.2', KVH, T80160)
BedFrame2.addMember('PlattFormBeam2.3', 'P3.2','P3.3', KVH, T80160)
BedFrame2.addMember('PlattFormBeam2.4', 'P3.3','P4', KVH, T80160)
BedFrame2.addMember('PlattFormBeam3', 'P1','P3', KVH, T80160)
BedFrame2.addMember('PlattFormBeam4', 'P2','P4', KVH, T80160)
BedFrame2.addMember('PlattFormBeam5.1', 'P1.1','P3.1', KVH, T80160)
BedFrame2.addMember('PlattFormBeam5.2', 'P1.2','P3.2', KVH, T80160)
BedFrame2.addMember('PlattFormBeam5.3', 'P1.3','P3.3', KVH, T80160)
BedFrame2.addMember('PlattFormBeam6', 'P5','P6', KVH, T80160)
BedFrame2.addMember('Diagonal2', 'P4','T2', KVH, T80160)
BedFrame2.addMember('Diagonal1', 'P3','T1', KVH, T80160)
BedFrame2.addMember('Diagonal3', 'P1','T1', KVH, T80160)
BedFrame2.addMember('Diagonal4', 'P2','T2', KVH, T80160)
BedFrame2.plot(deformed=False, showMemberName= False, showNodeName = False)
#create supports
BedFrame2.makeSupport('G1', Ux=0,Uy=0,Uz=0,Rx=0,Ry=0,Rz=0)
BedFrame2.makeSupport('G2', Ux=0,Uy=0,Uz=0,Rx=0,Ry=0,Rz=0)
BedFrame2.makeSupport('G3', Ux=0,Uy=0,Uz=0,Rx=0,Ry=0,Rz=0)
BedFrame2.makeSupport('G4', Ux=0,Uy=0,Uz=0,Rx=0,Ry=0,Rz=0)
BedFrame2.makeSupport('G5', Ux=0,Uy=0,Uz=0,Rx=0,Ry=0,Rz=0)
#set member loads
BedLoad = 10000
x1 = 100
x2 = 1500
a = abs(BedFrame2.Nodes['T2'].x- BedFrame2.Nodes['T3'].x)
b = abs(BedFrame2.Nodes['T2'].y- BedFrame2.Nodes['T3'].y)
BedFrame2.addMemberPtForce('TopBeam3', x=x1,Fz=-BedLoad/4)
BedFrame2.addMemberPtForce('TopBeam3', x=x1,Fx = BedLoad/4)
BedFrame2.addMemberPtForce('TopBeam3', x=x2,Fz=-BedLoad)
BedFrame2.addMemberPtForce('TopBeam3', x=x2,Fy=-BedLoad/4)
BedFrame2.addMemberPtForce('TopBeam3', x=x2,Fx=-BedLoad/4*a/((a**2+b**2)**(0.5)), Fy=-BedLoad/4*b/((a**2+b**2)**(0.5)))
BedFrame2.addMemberPtForce('TopBeam3', x=x2,Fx=-BedLoad/4)
BedFrame2.addMemberPtForce('TopBeam4', x=x1,Fz=-BedLoad/4)
BedFrame2.addMemberPtForce('TopBeam4', x=x1,Fx=BedLoad/4*a/((a**2+b**2)**(0.5)), Fy=BedLoad/4*b/((a**2+b**2)**(0.5)))
BedFrame2.addMemberPtForce('TopBeam4', x=x2,Fz=-BedLoad/4)
BedFrame2.addMemberPtForce('TopBeam4', x=x2,Fy=BedLoad/4)
U = BedFrame2.analyze()
#BedFrame.plot()
widgets.interact(BedFrame2.plot,
label_offset=(0.01, 0.1, 0.01),
xMargin=(0.25, 3, 0.25),
yMargin=(0.25, 3, 0.25),
zMargin=(0.5, 3, 0.25),
elevation=(0,360,10),
rotation=(0,360,10),
xFac=(1,200,10))
BedFrame2.MoS()
| 0.384797 | 0.797439 |
# Imports
```
import csv
import io
import matplotlib.pyplot as plt
import tensorflow as tf
import numpy as np
from tensorflow.keras.preprocessing.text import Tokenizer
from tensorflow.keras.preprocessing.sequence import pad_sequences
from tensorflow.keras.utils import to_categorical
from google.colab import files
```
# Hyper Parameters
```
vocab_size = 8000
embedding_dim = 64
max_length = 120
trunc_type='post'
padding_type='post'
oov_tok = "<OOV>"
training_portion = .8
num_epochs = 20
```
# Dataset Loading and Preprocessing
```
!wget --no-check-certificate \
https://storage.googleapis.com/laurencemoroney-blog.appspot.com/bbc-text.csv \
-O /tmp/bbc-text.csv
sentences = []
labels = []
stopwords = [ "a", "about", "above", "after", "again", "against", "all", "am", "an", "and", "any", "are", "as", "at", "be", "because", "been", "before", "being", "below", "between", "both", "but", "by", "could", "did", "do", "does", "doing", "down", "during", "each", "few", "for", "from", "further", "had", "has", "have", "having", "he", "he'd", "he'll", "he's", "her", "here", "here's", "hers", "herself", "him", "himself", "his", "how", "how's", "i", "i'd", "i'll", "i'm", "i've", "if", "in", "into", "is", "it", "it's", "its", "itself", "let's", "me", "more", "most", "my", "myself", "nor", "of", "on", "once", "only", "or", "other", "ought", "our", "ours", "ourselves", "out", "over", "own", "same", "she", "she'd", "she'll", "she's", "should", "so", "some", "such", "than", "that", "that's", "the", "their", "theirs", "them", "themselves", "then", "there", "there's", "these", "they", "they'd", "they'll", "they're", "they've", "this", "those", "through", "to", "too", "under", "until", "up", "very", "was", "we", "we'd", "we'll", "we're", "we've", "were", "what", "what's", "when", "when's", "where", "where's", "which", "while", "who", "who's", "whom", "why", "why's", "with", "would", "you", "you'd", "you'll", "you're", "you've", "your", "yours", "yourself", "yourselves" ]
with open("/tmp/bbc-text.csv", 'r') as csvfile:
reader = csv.reader(csvfile, delimiter=',')
next(reader)
for row in reader:
labels.append(row[0])
sentence = row[1]
for word in stopwords:
token = " " + word + " "
sentence = sentence.replace(token, " ")
sentences.append(sentence)
print(len(labels))
print(len(sentences))
train_size = int(len(sentences) * training_portion)
train_sentences = sentences[:train_size]
train_labels = labels[:train_size]
validation_sentences = sentences[train_size:]
validation_labels = labels[train_size:]
print(train_size)
print(len(train_sentences))
print(len(train_labels))
print(len(validation_sentences))
print(len(validation_labels))
tokenizer = Tokenizer(num_words = vocab_size, oov_token=oov_tok)
tokenizer.fit_on_texts(train_sentences)
word_index = tokenizer.word_index
train_sequences = tokenizer.texts_to_sequences(train_sentences)
train_padded = pad_sequences(train_sequences, padding=padding_type, maxlen=max_length)
validation_sequences = tokenizer.texts_to_sequences(validation_sentences)
validation_padded = pad_sequences(validation_sequences, padding=padding_type, maxlen=max_length)
label_tokenizer = Tokenizer()
label_tokenizer.fit_on_texts(labels)
training_label_seq = to_categorical(np.array(label_tokenizer.texts_to_sequences(train_labels)), num_classes = 6)
validation_label_seq = to_categorical(np.array(label_tokenizer.texts_to_sequences(validation_labels)), num_classes = 6)
print(training_label_seq.shape)
print(validation_label_seq.shape)
```
# Model
```
model = tf.keras.Sequential([tf.keras.layers.Embedding(vocab_size, embedding_dim, input_length = max_length),
tf.keras.layers.LSTM(64 ,return_state = False),
tf.keras.layers.Dense(256, activation='relu'),
tf.keras.layers.Dropout(0.2),
tf.keras.layers.Dense(6, activation='softmax')
])
model.compile(loss='categorical_crossentropy', optimizer='adam', metrics=['accuracy'])
model.summary()
```
# Training and Validation
```
history = model.fit(train_padded, training_label_seq, epochs=num_epochs, validation_data=(validation_padded, validation_label_seq), verbose=1)
```
# Accuracy and Loss Plot
```
def plot_graphs(history, string):
plt.plot(history.history[string])
plt.plot(history.history['val_'+string])
plt.xlabel("Epochs")
plt.ylabel(string)
plt.legend([string, 'val_'+string])
plt.show()
plot_graphs(history, "accuracy")
plot_graphs(history, "loss")
```
# Word Embedding Visualisation
```
reverse_word_index = dict([(value, key) for (key, value) in word_index.items()])
def decode_sentence(text):
return ' '.join([reverse_word_index.get(i, '?') for i in text])
e = model.layers[0]
weights = e.get_weights()[0]
print(weights.shape) # shape: (vocab_size, embedding_dim)
out_v = io.open('vecs.tsv', 'w', encoding='utf-8')
out_m = io.open('meta.tsv', 'w', encoding='utf-8')
for word_num in range(1, vocab_size):
word = reverse_word_index[word_num]
embeddings = weights[word_num]
out_m.write(word + "\n")
out_v.write('\t'.join([str(x) for x in embeddings]) + "\n")
out_v.close()
out_m.close()
files.download('vecs.tsv')
files.download('meta.tsv')
```
|
github_jupyter
|
import csv
import io
import matplotlib.pyplot as plt
import tensorflow as tf
import numpy as np
from tensorflow.keras.preprocessing.text import Tokenizer
from tensorflow.keras.preprocessing.sequence import pad_sequences
from tensorflow.keras.utils import to_categorical
from google.colab import files
vocab_size = 8000
embedding_dim = 64
max_length = 120
trunc_type='post'
padding_type='post'
oov_tok = "<OOV>"
training_portion = .8
num_epochs = 20
!wget --no-check-certificate \
https://storage.googleapis.com/laurencemoroney-blog.appspot.com/bbc-text.csv \
-O /tmp/bbc-text.csv
sentences = []
labels = []
stopwords = [ "a", "about", "above", "after", "again", "against", "all", "am", "an", "and", "any", "are", "as", "at", "be", "because", "been", "before", "being", "below", "between", "both", "but", "by", "could", "did", "do", "does", "doing", "down", "during", "each", "few", "for", "from", "further", "had", "has", "have", "having", "he", "he'd", "he'll", "he's", "her", "here", "here's", "hers", "herself", "him", "himself", "his", "how", "how's", "i", "i'd", "i'll", "i'm", "i've", "if", "in", "into", "is", "it", "it's", "its", "itself", "let's", "me", "more", "most", "my", "myself", "nor", "of", "on", "once", "only", "or", "other", "ought", "our", "ours", "ourselves", "out", "over", "own", "same", "she", "she'd", "she'll", "she's", "should", "so", "some", "such", "than", "that", "that's", "the", "their", "theirs", "them", "themselves", "then", "there", "there's", "these", "they", "they'd", "they'll", "they're", "they've", "this", "those", "through", "to", "too", "under", "until", "up", "very", "was", "we", "we'd", "we'll", "we're", "we've", "were", "what", "what's", "when", "when's", "where", "where's", "which", "while", "who", "who's", "whom", "why", "why's", "with", "would", "you", "you'd", "you'll", "you're", "you've", "your", "yours", "yourself", "yourselves" ]
with open("/tmp/bbc-text.csv", 'r') as csvfile:
reader = csv.reader(csvfile, delimiter=',')
next(reader)
for row in reader:
labels.append(row[0])
sentence = row[1]
for word in stopwords:
token = " " + word + " "
sentence = sentence.replace(token, " ")
sentences.append(sentence)
print(len(labels))
print(len(sentences))
train_size = int(len(sentences) * training_portion)
train_sentences = sentences[:train_size]
train_labels = labels[:train_size]
validation_sentences = sentences[train_size:]
validation_labels = labels[train_size:]
print(train_size)
print(len(train_sentences))
print(len(train_labels))
print(len(validation_sentences))
print(len(validation_labels))
tokenizer = Tokenizer(num_words = vocab_size, oov_token=oov_tok)
tokenizer.fit_on_texts(train_sentences)
word_index = tokenizer.word_index
train_sequences = tokenizer.texts_to_sequences(train_sentences)
train_padded = pad_sequences(train_sequences, padding=padding_type, maxlen=max_length)
validation_sequences = tokenizer.texts_to_sequences(validation_sentences)
validation_padded = pad_sequences(validation_sequences, padding=padding_type, maxlen=max_length)
label_tokenizer = Tokenizer()
label_tokenizer.fit_on_texts(labels)
training_label_seq = to_categorical(np.array(label_tokenizer.texts_to_sequences(train_labels)), num_classes = 6)
validation_label_seq = to_categorical(np.array(label_tokenizer.texts_to_sequences(validation_labels)), num_classes = 6)
print(training_label_seq.shape)
print(validation_label_seq.shape)
model = tf.keras.Sequential([tf.keras.layers.Embedding(vocab_size, embedding_dim, input_length = max_length),
tf.keras.layers.LSTM(64 ,return_state = False),
tf.keras.layers.Dense(256, activation='relu'),
tf.keras.layers.Dropout(0.2),
tf.keras.layers.Dense(6, activation='softmax')
])
model.compile(loss='categorical_crossentropy', optimizer='adam', metrics=['accuracy'])
model.summary()
history = model.fit(train_padded, training_label_seq, epochs=num_epochs, validation_data=(validation_padded, validation_label_seq), verbose=1)
def plot_graphs(history, string):
plt.plot(history.history[string])
plt.plot(history.history['val_'+string])
plt.xlabel("Epochs")
plt.ylabel(string)
plt.legend([string, 'val_'+string])
plt.show()
plot_graphs(history, "accuracy")
plot_graphs(history, "loss")
reverse_word_index = dict([(value, key) for (key, value) in word_index.items()])
def decode_sentence(text):
return ' '.join([reverse_word_index.get(i, '?') for i in text])
e = model.layers[0]
weights = e.get_weights()[0]
print(weights.shape) # shape: (vocab_size, embedding_dim)
out_v = io.open('vecs.tsv', 'w', encoding='utf-8')
out_m = io.open('meta.tsv', 'w', encoding='utf-8')
for word_num in range(1, vocab_size):
word = reverse_word_index[word_num]
embeddings = weights[word_num]
out_m.write(word + "\n")
out_v.write('\t'.join([str(x) for x in embeddings]) + "\n")
out_v.close()
out_m.close()
files.download('vecs.tsv')
files.download('meta.tsv')
| 0.562657 | 0.733643 |
# Prostate Cancer
In this notebook, we use blocked Gibbs sampling to examine the relationship between a prostate specific antigen and cancer volume.
```
import collections
import copy
import functools
from typing import Callable, NamedTuple
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
from scipy import stats
import seaborn as sns
import tensorflow as tf
import tensorflow_probability as tfp
import prostate
from stat570.linear_model import linear_regression
from stat570.mcmc import gibbs_sampling
np.set_printoptions(suppress=True)
prostate_data = prostate.load_data()
prostate_data.head()
```
## TensorFlow `input_fn`
We carry out our computations in TensorFlow, so we'll convert our data into tensors.
```
def prostate_input_fn():
features = prostate_data.to_dict('list')
labels = features.pop('lpsa')
return tf.data.Dataset.from_tensors((features, labels))
```
## Sampling Functions
In Gibbs sampling, we sample from the posterior conditional distributions. The inverse error variance (also known as precision) is gamma-distributed, and the coefficients are normally distributed. It's *blocked* Gibbs sampling since we draw both coefficients at once.
```
def make_inverse_error_variance_dist(
prior_concentration, prior_rate, features, labels, beta):
"""Makes the the posterior distribution for inverse error variance."""
with tf.name_scope('make_inverse_error_variance_dist',
values=[
prior_concentration,
prior_rate,
features,
labels,
beta,
]):
posterior_concentration = (
prior_concentration +
tf.divide(tf.cast(tf.shape(features)[0], tf.float32), 2.))
posterior_rate = (prior_rate +
tf.nn.l2_loss(labels - tf.tensordot(features, beta, 1)))
return tf.distributions.Gamma(
concentration=posterior_concentration, rate=posterior_rate,
name='posterior_inverse_error_variance')
def make_beta_dist(prior_mean, prior_variance, features, labels, inverse_error_variance):
"""Makes the posterior distribution for model coefficients."""
shape = int(prior_mean.shape[0])
with tf.name_scope('make_beta_dist',
values=[
inverse_error_variance,
features,
labels,
prior_mean,
prior_variance,
]):
transposed_features = tf.transpose(features)
gramian_matrix = tf.matmul(transposed_features, features)
mle_mean = tf.squeeze(tf.linalg.cholesky_solve(
tf.linalg.cholesky(gramian_matrix),
tf.matmul(transposed_features, tf.expand_dims(labels, -1))))
mle_precision = gramian_matrix*inverse_error_variance
posterior_precision = mle_precision + tf.eye(shape)/prior_variance
posterior_covariance = tf.linalg.cholesky_solve(
tf.linalg.cholesky(posterior_precision), tf.eye(shape))
posterior_mean = tf.tensordot(
tf.matmul(posterior_covariance, mle_precision),
mle_mean - prior_mean, axes=1) + prior_mean
return tfp.distributions.MultivariateNormalFullCovariance(
loc=posterior_mean, covariance_matrix=posterior_covariance,
name='posterior_beta')
```
## Model
Now, we specify the model. The generative process is specified in `forward`. We build our conditional disributions based on the data and use them to construct the transition kernel for Markov Chain Monte Carlo (MCMC) sampling.
The code for [`gibbs_sampling.GibbsSamplingKernel`](https://github.com/ppham27/stat570/blob/master/stat570/mcmc/gibbs_sampling.py) can be on my [GitHub](https://github.com/ppham27/stat570/blob/master/stat570/mcmc/gibbs_sampling.py).
```
def model_fn(features, labels, mode, params, config):
del config
prior_inverse_error_variance_concentration = (
params['prior']['inverse_error_variance']['concentration'])
prior_inverse_error_variance_rate = (
params['prior']['inverse_error_variance']['rate'])
prior_beta_mean = tf.constant(params['prior']['beta']['mean'],
dtype=tf.float32)
prior_beta_variance = tf.constant(params['prior']['beta']['variance'],
dtype=tf.float32)
def forward(features):
inverse_error_variance = tfp.edward2.Gamma(
concentration=prior_inverse_error_variance_concentration,
rate=prior_inverse_error_variance_rate,
name='inverse_error_variance')
beta = tfp.edward2.MultivariateNormalDiag(
loc=prior_beta_mean,
scale_identity_multiplier=tf.sqrt(prior_beta_variance), name='beta')
return tfp.edward2.Normal(
loc=tf.tensordot(features, beta, axes=1), scale=1/tf.sqrt(inverse_error_variance),
name='labels')
features = tf.feature_column.input_layer(
features, [tf.feature_column.numeric_column('lcavol')])
features = tf.concat((tf.ones_like(features), features), axis=-1)
if mode == tf.estimator.ModeKeys.PREDICT:
return forward(features).value
log_joint_fn = functools.partial(
tfp.edward2.make_log_joint_fn(lambda: forward(features)),
labels=labels)
if mode == tf.estimator.ModeKeys.EVAL:
return log_joint_fn(labels=labels) # currently will error
samplers = [
functools.partial(
make_inverse_error_variance_dist,
prior_inverse_error_variance_concentration,
prior_inverse_error_variance_rate,
features, labels),
functools.partial(
make_beta_dist, prior_beta_mean, prior_beta_variance, features, labels),
]
kernel = tfp.mcmc.MetropolisHastings(
inner_kernel=gibbs_sampling.GibbsSamplingKernel(
samplers=samplers,
target_log_prob_fn=lambda inverse_error_variance, beta: log_joint_fn(
inverse_error_variance=inverse_error_variance,
beta=beta)))
samples, _ = tfp.mcmc.sample_chain(
num_results=params['mcmc']['num_results'],
current_state=(
tf.convert_to_tensor(
params['mcmc']['initial_state']['inverse_error_variance'], tf.float32),
tf.convert_to_tensor(
params['mcmc']['initial_state']['beta'], tf.float32)),
kernel=kernel,
num_burnin_steps=params['mcmc']['num_burnin_steps'],
num_steps_between_results=len(samplers) - 1,
parallel_iterations=1)
return samples
```
## Running MCMC
Prior parameters are taken from the homework. A gamma distribution with $0$ shape and $0$ rate is improper so very small values were used.
```
_DEFAULT_PARAMS = {
'prior': {
'inverse_error_variance': {
'concentration': 0.1, # Also called shape and denoted alpha
'rate': 0.1, # Usually denoted by beta.
},
'beta': {
'mean': [0., 0.],
'variance': 2., # Enforce equal variance and no covariance.
},
},
'mcmc': {
'num_burnin_steps': 0,
'num_results': 128,
'initial_state': {
'inverse_error_variance': 1.,
'beta': [0., 0.],
},
},
}
def get_mle_params():
mle_params = copy.deepcopy(_DEFAULT_PARAMS)
mle_model = linear_regression.LinearRegression.from_data_frame(
prostate_data, ['lcavol'], 'lpsa')
mle_params['mcmc']['initial_state']['inverse_error_variance'] = (
1./mle_model.residual_variance_)
mle_params['mcmc']['initial_state']['beta'] = mle_model.coefficients_['estimate'].values
return mle_params
def get_prior_params(params=_DEFAULT_PARAMS):
prior_params = copy.deepcopy(params)
prior_params['mcmc']['initial_state']['inverse_error_variance'] = stats.gamma.rvs(
params['prior']['inverse_error_variance']['concentration'],
scale=1/params['prior']['inverse_error_variance']['rate'])
prior_params['mcmc']['initial_state']['beta'] = stats.multivariate_normal.rvs(
mean=params['prior']['beta']['mean'],
cov=params['prior']['beta']['variance']*np.eye(len(params['prior']['beta']['mean'])))
return prior_params
```
### TensorFlow
We start a TensorFlow session to run the two chains.
```
def run_chains(params_dict):
graph = tf.Graph()
with graph.as_default():
features, labels = prostate_input_fn().repeat().make_one_shot_iterator().get_next()
chain_ops = {
key: model_fn(features, labels, tf.estimator.ModeKeys.TRAIN,
params, tf.estimator.RunConfig())
for key, params in params_dict.items()
}
init_op = tf.group(tf.global_variables_initializer())
graph.finalize()
with graph.as_default(), tf.Session() as sess:
sess.run(init_op)
return sess.run(chain_ops)
states = run_chains({'mle': get_mle_params(), 'prior': get_prior_params()})
```
## Trace Plots
To examine how many burn-in steps are needed, we qualitatively examine when the trace becomes stationary.
```
TracePlotOptions = NamedTuple('TracePlotOptions', [
('label', str),
('transform', Callable[[np.array], np.array]),
])
def plot_traces(states_dict, options, title):
def plot_trace(ax, states_dict, label):
for key, states in states_dict.items():
ax.plot(np.arange(len(states)) + 1, states, label=key)
ax.grid(True)
ax.set_ylabel(label)
states_dict = {
key: np.hstack([value[:,np.newaxis] if value.ndim == 1 else value for value in values])
for key, values in states_dict.items()
}
assert np.all(
[value.shape[1] == len(options)
for value in states_dict.values()]),'Options must be provided for each parameter.'
fig, axes = plt.subplots(figsize=(6, 6), nrows=len(options))
for i, (ax, option) in enumerate(zip(axes, options)):
plot_trace(
ax,
{key: option.transform(value[:,i]) for key, value in states_dict.items()},
option.label)
axes[0].legend()
axes[-1].set_xlabel('Result Number')
fig.suptitle(title)
fig.tight_layout()
fig.subplots_adjust(top=0.94)
return fig
TRACE_PLOT_OPTIONS = [
TracePlotOptions(label='$\\log\\sigma^2$', transform=lambda x: np.log(1/x)),
TracePlotOptions(label='$\\beta_0$', transform=lambda x: x),
TracePlotOptions(label='$\\beta_1$', transform=lambda x: x),
]
fig = plot_traces(states, TRACE_PLOT_OPTIONS, 'MCMC Trace Plots')
fig.savefig('p1_trace.pdf', bbox_inches='tight')
```
## Running Full Chain
Not much burn-in is needed, so we can run the full chain now.
```
def merge_params(params, updates):
for k, v in updates.items():
if isinstance(v, collections.Mapping):
params[k] = merge_params(params.get(k, {}), v)
else:
params[k] = v
return params
_MCMC_PARAMS = {
'mcmc': {
'num_results': 2048,
'num_burnin_steps': 128,
},
}
states = run_chains({
'mle': merge_params(get_mle_params(), _MCMC_PARAMS),
'prior': merge_params(get_prior_params(), _MCMC_PARAMS),
})
states_data_frame = pd.DataFrame(collections.OrderedDict([
('$\\sigma$', np.sqrt(1/np.concatenate([v[0] for v in states.values()]))),
('$\\beta_0$', np.concatenate([v[1][:,0] for v in states.values()])),
('$\\beta_1$', np.concatenate([v[1][:,1] for v in states.values()]))
]))
states_summary = pd.DataFrame(collections.OrderedDict([
('Posterior mean', states_data_frame.mean()),
('Standard deviation', states_data_frame.std(ddof=1)),
('10% quantile', states_data_frame.quantile(.1)),
('50% quantile', states_data_frame.quantile(.5)),
('90% quantile', states_data_frame.quantile(.9)),
]))
with open('p1_summary.tex', 'w') as f:
f.write(states_summary.to_latex(escape=False).replace('%', '\\%'))
states_summary
fig, axes = plt.subplots(figsize=(6,3), ncols=len(states_data_frame.columns))
for ax, (column_name, samples) in zip(axes, states_data_frame.items()):
sns.distplot(samples, kde=False, ax=ax)
ax.grid(True)
fig.suptitle('Posterior Univariate Marginal Distributions')
fig.tight_layout()
fig.subplots_adjust(top=0.90)
fig.savefig('p1_univariate_marginals.pdf', bbox_inches='tight')
_BIVARIATE_PAIRS = {
'beta0_beta1': ('$\\beta_0$', '$\\beta_1$'),
'beta0_sigma': ('$\\beta_0$', '$\\sigma$'),
'beta1_sigma': ('$\\beta_1$', '$\\sigma$'),
}
for label, (x, y) in _BIVARIATE_PAIRS.items():
joint_grid = sns.jointplot(x, y, data=states_data_frame, alpha=1/8)
joint_grid.ax_joint.grid(True)
joint_grid.fig.set_figwidth(5)
joint_grid.fig.set_figheight(5)
joint_grid.fig.tight_layout()
joint_grid.fig.savefig('p1_{}.pdf'.format(label), bbox_inches='tight')
```
## Empirical Estimate for $\mathbb{P}\left(\beta_1 > 0.5\right)$
We can estimate how significant the relationship between the prostate specific antigen and cancer volume by computing the emprical estimate for probability that the coefficient is greater than $0.5$.
```
np.mean(states_data_frame['$\\beta_1$'] > 0.5)
```
|
github_jupyter
|
import collections
import copy
import functools
from typing import Callable, NamedTuple
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
from scipy import stats
import seaborn as sns
import tensorflow as tf
import tensorflow_probability as tfp
import prostate
from stat570.linear_model import linear_regression
from stat570.mcmc import gibbs_sampling
np.set_printoptions(suppress=True)
prostate_data = prostate.load_data()
prostate_data.head()
def prostate_input_fn():
features = prostate_data.to_dict('list')
labels = features.pop('lpsa')
return tf.data.Dataset.from_tensors((features, labels))
def make_inverse_error_variance_dist(
prior_concentration, prior_rate, features, labels, beta):
"""Makes the the posterior distribution for inverse error variance."""
with tf.name_scope('make_inverse_error_variance_dist',
values=[
prior_concentration,
prior_rate,
features,
labels,
beta,
]):
posterior_concentration = (
prior_concentration +
tf.divide(tf.cast(tf.shape(features)[0], tf.float32), 2.))
posterior_rate = (prior_rate +
tf.nn.l2_loss(labels - tf.tensordot(features, beta, 1)))
return tf.distributions.Gamma(
concentration=posterior_concentration, rate=posterior_rate,
name='posterior_inverse_error_variance')
def make_beta_dist(prior_mean, prior_variance, features, labels, inverse_error_variance):
"""Makes the posterior distribution for model coefficients."""
shape = int(prior_mean.shape[0])
with tf.name_scope('make_beta_dist',
values=[
inverse_error_variance,
features,
labels,
prior_mean,
prior_variance,
]):
transposed_features = tf.transpose(features)
gramian_matrix = tf.matmul(transposed_features, features)
mle_mean = tf.squeeze(tf.linalg.cholesky_solve(
tf.linalg.cholesky(gramian_matrix),
tf.matmul(transposed_features, tf.expand_dims(labels, -1))))
mle_precision = gramian_matrix*inverse_error_variance
posterior_precision = mle_precision + tf.eye(shape)/prior_variance
posterior_covariance = tf.linalg.cholesky_solve(
tf.linalg.cholesky(posterior_precision), tf.eye(shape))
posterior_mean = tf.tensordot(
tf.matmul(posterior_covariance, mle_precision),
mle_mean - prior_mean, axes=1) + prior_mean
return tfp.distributions.MultivariateNormalFullCovariance(
loc=posterior_mean, covariance_matrix=posterior_covariance,
name='posterior_beta')
def model_fn(features, labels, mode, params, config):
del config
prior_inverse_error_variance_concentration = (
params['prior']['inverse_error_variance']['concentration'])
prior_inverse_error_variance_rate = (
params['prior']['inverse_error_variance']['rate'])
prior_beta_mean = tf.constant(params['prior']['beta']['mean'],
dtype=tf.float32)
prior_beta_variance = tf.constant(params['prior']['beta']['variance'],
dtype=tf.float32)
def forward(features):
inverse_error_variance = tfp.edward2.Gamma(
concentration=prior_inverse_error_variance_concentration,
rate=prior_inverse_error_variance_rate,
name='inverse_error_variance')
beta = tfp.edward2.MultivariateNormalDiag(
loc=prior_beta_mean,
scale_identity_multiplier=tf.sqrt(prior_beta_variance), name='beta')
return tfp.edward2.Normal(
loc=tf.tensordot(features, beta, axes=1), scale=1/tf.sqrt(inverse_error_variance),
name='labels')
features = tf.feature_column.input_layer(
features, [tf.feature_column.numeric_column('lcavol')])
features = tf.concat((tf.ones_like(features), features), axis=-1)
if mode == tf.estimator.ModeKeys.PREDICT:
return forward(features).value
log_joint_fn = functools.partial(
tfp.edward2.make_log_joint_fn(lambda: forward(features)),
labels=labels)
if mode == tf.estimator.ModeKeys.EVAL:
return log_joint_fn(labels=labels) # currently will error
samplers = [
functools.partial(
make_inverse_error_variance_dist,
prior_inverse_error_variance_concentration,
prior_inverse_error_variance_rate,
features, labels),
functools.partial(
make_beta_dist, prior_beta_mean, prior_beta_variance, features, labels),
]
kernel = tfp.mcmc.MetropolisHastings(
inner_kernel=gibbs_sampling.GibbsSamplingKernel(
samplers=samplers,
target_log_prob_fn=lambda inverse_error_variance, beta: log_joint_fn(
inverse_error_variance=inverse_error_variance,
beta=beta)))
samples, _ = tfp.mcmc.sample_chain(
num_results=params['mcmc']['num_results'],
current_state=(
tf.convert_to_tensor(
params['mcmc']['initial_state']['inverse_error_variance'], tf.float32),
tf.convert_to_tensor(
params['mcmc']['initial_state']['beta'], tf.float32)),
kernel=kernel,
num_burnin_steps=params['mcmc']['num_burnin_steps'],
num_steps_between_results=len(samplers) - 1,
parallel_iterations=1)
return samples
_DEFAULT_PARAMS = {
'prior': {
'inverse_error_variance': {
'concentration': 0.1, # Also called shape and denoted alpha
'rate': 0.1, # Usually denoted by beta.
},
'beta': {
'mean': [0., 0.],
'variance': 2., # Enforce equal variance and no covariance.
},
},
'mcmc': {
'num_burnin_steps': 0,
'num_results': 128,
'initial_state': {
'inverse_error_variance': 1.,
'beta': [0., 0.],
},
},
}
def get_mle_params():
mle_params = copy.deepcopy(_DEFAULT_PARAMS)
mle_model = linear_regression.LinearRegression.from_data_frame(
prostate_data, ['lcavol'], 'lpsa')
mle_params['mcmc']['initial_state']['inverse_error_variance'] = (
1./mle_model.residual_variance_)
mle_params['mcmc']['initial_state']['beta'] = mle_model.coefficients_['estimate'].values
return mle_params
def get_prior_params(params=_DEFAULT_PARAMS):
prior_params = copy.deepcopy(params)
prior_params['mcmc']['initial_state']['inverse_error_variance'] = stats.gamma.rvs(
params['prior']['inverse_error_variance']['concentration'],
scale=1/params['prior']['inverse_error_variance']['rate'])
prior_params['mcmc']['initial_state']['beta'] = stats.multivariate_normal.rvs(
mean=params['prior']['beta']['mean'],
cov=params['prior']['beta']['variance']*np.eye(len(params['prior']['beta']['mean'])))
return prior_params
def run_chains(params_dict):
graph = tf.Graph()
with graph.as_default():
features, labels = prostate_input_fn().repeat().make_one_shot_iterator().get_next()
chain_ops = {
key: model_fn(features, labels, tf.estimator.ModeKeys.TRAIN,
params, tf.estimator.RunConfig())
for key, params in params_dict.items()
}
init_op = tf.group(tf.global_variables_initializer())
graph.finalize()
with graph.as_default(), tf.Session() as sess:
sess.run(init_op)
return sess.run(chain_ops)
states = run_chains({'mle': get_mle_params(), 'prior': get_prior_params()})
TracePlotOptions = NamedTuple('TracePlotOptions', [
('label', str),
('transform', Callable[[np.array], np.array]),
])
def plot_traces(states_dict, options, title):
def plot_trace(ax, states_dict, label):
for key, states in states_dict.items():
ax.plot(np.arange(len(states)) + 1, states, label=key)
ax.grid(True)
ax.set_ylabel(label)
states_dict = {
key: np.hstack([value[:,np.newaxis] if value.ndim == 1 else value for value in values])
for key, values in states_dict.items()
}
assert np.all(
[value.shape[1] == len(options)
for value in states_dict.values()]),'Options must be provided for each parameter.'
fig, axes = plt.subplots(figsize=(6, 6), nrows=len(options))
for i, (ax, option) in enumerate(zip(axes, options)):
plot_trace(
ax,
{key: option.transform(value[:,i]) for key, value in states_dict.items()},
option.label)
axes[0].legend()
axes[-1].set_xlabel('Result Number')
fig.suptitle(title)
fig.tight_layout()
fig.subplots_adjust(top=0.94)
return fig
TRACE_PLOT_OPTIONS = [
TracePlotOptions(label='$\\log\\sigma^2$', transform=lambda x: np.log(1/x)),
TracePlotOptions(label='$\\beta_0$', transform=lambda x: x),
TracePlotOptions(label='$\\beta_1$', transform=lambda x: x),
]
fig = plot_traces(states, TRACE_PLOT_OPTIONS, 'MCMC Trace Plots')
fig.savefig('p1_trace.pdf', bbox_inches='tight')
def merge_params(params, updates):
for k, v in updates.items():
if isinstance(v, collections.Mapping):
params[k] = merge_params(params.get(k, {}), v)
else:
params[k] = v
return params
_MCMC_PARAMS = {
'mcmc': {
'num_results': 2048,
'num_burnin_steps': 128,
},
}
states = run_chains({
'mle': merge_params(get_mle_params(), _MCMC_PARAMS),
'prior': merge_params(get_prior_params(), _MCMC_PARAMS),
})
states_data_frame = pd.DataFrame(collections.OrderedDict([
('$\\sigma$', np.sqrt(1/np.concatenate([v[0] for v in states.values()]))),
('$\\beta_0$', np.concatenate([v[1][:,0] for v in states.values()])),
('$\\beta_1$', np.concatenate([v[1][:,1] for v in states.values()]))
]))
states_summary = pd.DataFrame(collections.OrderedDict([
('Posterior mean', states_data_frame.mean()),
('Standard deviation', states_data_frame.std(ddof=1)),
('10% quantile', states_data_frame.quantile(.1)),
('50% quantile', states_data_frame.quantile(.5)),
('90% quantile', states_data_frame.quantile(.9)),
]))
with open('p1_summary.tex', 'w') as f:
f.write(states_summary.to_latex(escape=False).replace('%', '\\%'))
states_summary
fig, axes = plt.subplots(figsize=(6,3), ncols=len(states_data_frame.columns))
for ax, (column_name, samples) in zip(axes, states_data_frame.items()):
sns.distplot(samples, kde=False, ax=ax)
ax.grid(True)
fig.suptitle('Posterior Univariate Marginal Distributions')
fig.tight_layout()
fig.subplots_adjust(top=0.90)
fig.savefig('p1_univariate_marginals.pdf', bbox_inches='tight')
_BIVARIATE_PAIRS = {
'beta0_beta1': ('$\\beta_0$', '$\\beta_1$'),
'beta0_sigma': ('$\\beta_0$', '$\\sigma$'),
'beta1_sigma': ('$\\beta_1$', '$\\sigma$'),
}
for label, (x, y) in _BIVARIATE_PAIRS.items():
joint_grid = sns.jointplot(x, y, data=states_data_frame, alpha=1/8)
joint_grid.ax_joint.grid(True)
joint_grid.fig.set_figwidth(5)
joint_grid.fig.set_figheight(5)
joint_grid.fig.tight_layout()
joint_grid.fig.savefig('p1_{}.pdf'.format(label), bbox_inches='tight')
np.mean(states_data_frame['$\\beta_1$'] > 0.5)
| 0.839405 | 0.928959 |
```
%pwd
%cd ../..
import argparse
import sys
from datetime import datetime
from pathlib import Path
import numpy as np
import tensorflow as tf
import tensorflow_addons as tfa
import tensorflow.keras.backend as K
from dotenv import load_dotenv, find_dotenv
from tensorflow.keras.callbacks import TensorBoard, ModelCheckpoint, ReduceLROnPlateau, EarlyStopping, TerminateOnNaN
from tensorflow.keras.optimizers import Adam
from tensorflow.python.keras.preprocessing.image import ImageDataGenerator
from models import build_simple_conv_net
BASE_DIR = Path().resolve()
LOGS_DIR = BASE_DIR.joinpath('logs', f'{datetime.now().strftime("%m-%d-%Y_%H:%M:%S")}')
WEIGHTS_OUTPUT_PATH = LOGS_DIR.joinpath('ep{epoch:03d}-loss{loss:.4f}-val_loss{val_loss:.4f}.h5')
MODEL_OUTPUT_PATH = LOGS_DIR.joinpath('trained_model.h5')
ENV_FILE = find_dotenv()
if ENV_FILE:
load_dotenv(ENV_FILE)
input_shape = np.asarray([300, 300, 3])
checkpoint = ModelCheckpoint(str(WEIGHTS_OUTPUT_PATH),
monitor='val_loss',
verbose=1,
save_weights_only=True,
save_best_only=True)
logging = TensorBoard(log_dir=str(LOGS_DIR))
early_stopping = EarlyStopping(monitor='val_loss', min_delta=0, patience=50, verbose=1)
terminate_on_nan = TerminateOnNaN()
callbacks = [logging, checkpoint, early_stopping, terminate_on_nan]
optimizer = Adam()
model = build_simple_conv_net(input_shape)
model.compile(loss='binary_crossentropy', optimizer=optimizer, metrics=['accuracy', tfa.metrics.F1Score(num_classes=2,average="micro")])
epochs = 100
train_datagen = ImageDataGenerator(rescale=1 / 255)
val_datagen = ImageDataGenerator(rescale=1 / 255)
train_generator = train_datagen.flow_from_directory(
'/var/data/Downloads/archive/chest_xray/chest_xray/train/',
target_size=input_shape[:2],
batch_size=16,
class_mode='binary'
)
validation_generator = val_datagen.flow_from_directory(
'/var/data/Downloads/archive/chest_xray/chest_xray/val/',
target_size=input_shape[:2],
batch_size=16,
class_mode='binary'
)
history = model.fit(
train_generator,
epochs=epochs,
validation_data=validation_generator,
callbacks=callbacks
)
test_datagen = ImageDataGenerator(rescale = 1/255)
test_generator = test_datagen.flow_from_directory(
'/var/data/Downloads/archive/chest_xray/chest_xray/test/',
target_size=input_shape[:2],
batch_size = 156,
class_mode = 'binary'
)
eval_result = model.evaluate(test_generator)
print(f'test loss: {eval_result[0]}')
print(f'test accuracy: {eval_result[1]}')
model.save(MODEL_OUTPUT_PATH)
```
|
github_jupyter
|
%pwd
%cd ../..
import argparse
import sys
from datetime import datetime
from pathlib import Path
import numpy as np
import tensorflow as tf
import tensorflow_addons as tfa
import tensorflow.keras.backend as K
from dotenv import load_dotenv, find_dotenv
from tensorflow.keras.callbacks import TensorBoard, ModelCheckpoint, ReduceLROnPlateau, EarlyStopping, TerminateOnNaN
from tensorflow.keras.optimizers import Adam
from tensorflow.python.keras.preprocessing.image import ImageDataGenerator
from models import build_simple_conv_net
BASE_DIR = Path().resolve()
LOGS_DIR = BASE_DIR.joinpath('logs', f'{datetime.now().strftime("%m-%d-%Y_%H:%M:%S")}')
WEIGHTS_OUTPUT_PATH = LOGS_DIR.joinpath('ep{epoch:03d}-loss{loss:.4f}-val_loss{val_loss:.4f}.h5')
MODEL_OUTPUT_PATH = LOGS_DIR.joinpath('trained_model.h5')
ENV_FILE = find_dotenv()
if ENV_FILE:
load_dotenv(ENV_FILE)
input_shape = np.asarray([300, 300, 3])
checkpoint = ModelCheckpoint(str(WEIGHTS_OUTPUT_PATH),
monitor='val_loss',
verbose=1,
save_weights_only=True,
save_best_only=True)
logging = TensorBoard(log_dir=str(LOGS_DIR))
early_stopping = EarlyStopping(monitor='val_loss', min_delta=0, patience=50, verbose=1)
terminate_on_nan = TerminateOnNaN()
callbacks = [logging, checkpoint, early_stopping, terminate_on_nan]
optimizer = Adam()
model = build_simple_conv_net(input_shape)
model.compile(loss='binary_crossentropy', optimizer=optimizer, metrics=['accuracy', tfa.metrics.F1Score(num_classes=2,average="micro")])
epochs = 100
train_datagen = ImageDataGenerator(rescale=1 / 255)
val_datagen = ImageDataGenerator(rescale=1 / 255)
train_generator = train_datagen.flow_from_directory(
'/var/data/Downloads/archive/chest_xray/chest_xray/train/',
target_size=input_shape[:2],
batch_size=16,
class_mode='binary'
)
validation_generator = val_datagen.flow_from_directory(
'/var/data/Downloads/archive/chest_xray/chest_xray/val/',
target_size=input_shape[:2],
batch_size=16,
class_mode='binary'
)
history = model.fit(
train_generator,
epochs=epochs,
validation_data=validation_generator,
callbacks=callbacks
)
test_datagen = ImageDataGenerator(rescale = 1/255)
test_generator = test_datagen.flow_from_directory(
'/var/data/Downloads/archive/chest_xray/chest_xray/test/',
target_size=input_shape[:2],
batch_size = 156,
class_mode = 'binary'
)
eval_result = model.evaluate(test_generator)
print(f'test loss: {eval_result[0]}')
print(f'test accuracy: {eval_result[1]}')
model.save(MODEL_OUTPUT_PATH)
| 0.458106 | 0.172764 |
# Importing relevant libraries
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import os, re
import nltk
from nltk.tokenize import word_tokenize
from nltk.corpus import stopwords
from nltk.tag import pos_tag
from nltk.stem import WordNetLemmatizer
from nltk import FreqDist
```
# Importing data from Google Drive
```
from google.colab import drive
drive.mount('/content/drive')
train_reviews = []
for line in open('/content/drive/My Drive/movie_data/full_train.txt', 'r', encoding='utf-8'):
train_reviews.append(line.strip())
test_reviews = []
for line in open('/content/drive/My Drive/movie_data/full_test.txt', 'r', encoding='utf-8'):
test_reviews.append(line.strip())
```
# Cleaning and Preprocessing
### Remove punctuations, HTML tags, etc.
```
REPLACE_NO_SPACE = re.compile("[.;:!\'?,\"()\[\]]")
REPLACE_WITH_SPACE = re.compile("(<br\s*/><br\s*/>)|(\-)|(\/)")
def preprocess_reviews(reviews):
reviews = [REPLACE_NO_SPACE.sub("", line.lower()) for line in reviews]
reviews = [REPLACE_WITH_SPACE.sub(" ", line) for line in reviews]
return reviews
train_reviews_clean = preprocess_reviews(train_reviews)
test_reviews_clean = preprocess_reviews(test_reviews)
```
### Tokenizing the data
```
nltk.download('punkt')
train_reviews_clean_tokens = []
for review in train_reviews_clean:
train_reviews_clean_tokens.append(word_tokenize(review))
test_reviews_clean_tokens = []
for review in test_reviews_clean:
test_reviews_clean_tokens.append(word_tokenize(review))
```
## Removing stopwords
```
def remove_noise(review_tokens, stop_words = ()):
return [token for token in review_tokens if token not in stop_words]
nltk.download('stopwords')
stop_words = stopwords.words('english')
cleaned_train_reviews = []
cleaned_test_reviews = []
for tokens in train_reviews_clean_tokens:
cleaned_train_reviews.append(remove_noise(tokens, stop_words))
for tokens in test_reviews_clean_tokens:
cleaned_test_reviews.append(remove_noise(tokens, stop_words))
```
## Normalizing the data - Lemmatization
```
def lemmatizeReview(review):
lemmatizer = WordNetLemmatizer()
lemmatizedReviews = []
for token, tag in pos_tag(review):
if tag.startswith('NN'):
pos = 'n'
elif tag.startswith('VB'):
pos = 'v'
else:
pos = 'a'
token = lemmatizer.lemmatize(token, pos)
lemmatizedReviews.append(token.lower())
return lemmatizedReviews
nltk.download('wordnet') # used for stemming pupose inside WordNetLemmatizer
nltk.download('averaged_perceptron_tagger') # User for tagging pupose inside pos_tag
train_reviews_normalized = [lemmatizeReview(review) for review in cleaned_train_reviews]
test_reviews_normalized = [lemmatizeReview(review) for review in cleaned_test_reviews]
```
## Let's have a look at the word count
Instead of simply noting whether a word appears in the review or not, we can include the number of times a given word appears. This can give our sentiment classifier a lot more predictive power. For example, if a movie reviewer says ‘amazing’ or ‘terrible’ multiple times in a review it is considerably more probable that the review is positive or negative, respectively.
```
# get a single list of tokens - all tokens in a single list
def get_all_words(tokens):
all_tokens = []
for token_list in tokens:
for token in token_list:
all_tokens.append(token)
return all_tokens
all_train_tokens_list = get_all_words(train_reviews_normalized)
all_test_tokens_list = get_all_words(test_reviews_normalized)
train_token_frequency = FreqDist(all_train_tokens_list)
test_token_frequency = FreqDist(all_test_tokens_list)
print(train_token_frequency.most_common(10))
print(test_token_frequency.most_common(10))
```
Most common words are same in both training and testing set
# Building the classification model
## Preparing the data for training and testing
```
def get_train_reviews_tokes(review_normalized):
for token_list in review_normalized:
yield dict([token, True] for token in token_list)
train_reviews_for_model = get_train_reviews_tokes(train_reviews_normalized)
test_reviews_for_model = get_train_reviews_tokes(test_reviews_normalized)
COUNT = 1
train_dataset = []
for review_dict in train_reviews_for_model:
if COUNT <= 12500:
train_dataset.append((review_dict, 1))
else:
train_dataset.append((review_dict, 0))
COUNT += 1
COUNT = 1
test_dataset = []
for review_dict in test_reviews_for_model:
if COUNT <= 12500:
test_dataset.append((review_dict, 1))
else:
test_dataset.append((review_dict, 0))
COUNT += 1
import random
random.shuffle(train_dataset)
train_dataset[24500]
occurence = []
for i in range(1000):
for tup in train_dataset:
if tup[1] == 0:
occurence.append(i)
break
len(occurence)
```
|
github_jupyter
|
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import os, re
import nltk
from nltk.tokenize import word_tokenize
from nltk.corpus import stopwords
from nltk.tag import pos_tag
from nltk.stem import WordNetLemmatizer
from nltk import FreqDist
from google.colab import drive
drive.mount('/content/drive')
train_reviews = []
for line in open('/content/drive/My Drive/movie_data/full_train.txt', 'r', encoding='utf-8'):
train_reviews.append(line.strip())
test_reviews = []
for line in open('/content/drive/My Drive/movie_data/full_test.txt', 'r', encoding='utf-8'):
test_reviews.append(line.strip())
REPLACE_NO_SPACE = re.compile("[.;:!\'?,\"()\[\]]")
REPLACE_WITH_SPACE = re.compile("(<br\s*/><br\s*/>)|(\-)|(\/)")
def preprocess_reviews(reviews):
reviews = [REPLACE_NO_SPACE.sub("", line.lower()) for line in reviews]
reviews = [REPLACE_WITH_SPACE.sub(" ", line) for line in reviews]
return reviews
train_reviews_clean = preprocess_reviews(train_reviews)
test_reviews_clean = preprocess_reviews(test_reviews)
nltk.download('punkt')
train_reviews_clean_tokens = []
for review in train_reviews_clean:
train_reviews_clean_tokens.append(word_tokenize(review))
test_reviews_clean_tokens = []
for review in test_reviews_clean:
test_reviews_clean_tokens.append(word_tokenize(review))
def remove_noise(review_tokens, stop_words = ()):
return [token for token in review_tokens if token not in stop_words]
nltk.download('stopwords')
stop_words = stopwords.words('english')
cleaned_train_reviews = []
cleaned_test_reviews = []
for tokens in train_reviews_clean_tokens:
cleaned_train_reviews.append(remove_noise(tokens, stop_words))
for tokens in test_reviews_clean_tokens:
cleaned_test_reviews.append(remove_noise(tokens, stop_words))
def lemmatizeReview(review):
lemmatizer = WordNetLemmatizer()
lemmatizedReviews = []
for token, tag in pos_tag(review):
if tag.startswith('NN'):
pos = 'n'
elif tag.startswith('VB'):
pos = 'v'
else:
pos = 'a'
token = lemmatizer.lemmatize(token, pos)
lemmatizedReviews.append(token.lower())
return lemmatizedReviews
nltk.download('wordnet') # used for stemming pupose inside WordNetLemmatizer
nltk.download('averaged_perceptron_tagger') # User for tagging pupose inside pos_tag
train_reviews_normalized = [lemmatizeReview(review) for review in cleaned_train_reviews]
test_reviews_normalized = [lemmatizeReview(review) for review in cleaned_test_reviews]
# get a single list of tokens - all tokens in a single list
def get_all_words(tokens):
all_tokens = []
for token_list in tokens:
for token in token_list:
all_tokens.append(token)
return all_tokens
all_train_tokens_list = get_all_words(train_reviews_normalized)
all_test_tokens_list = get_all_words(test_reviews_normalized)
train_token_frequency = FreqDist(all_train_tokens_list)
test_token_frequency = FreqDist(all_test_tokens_list)
print(train_token_frequency.most_common(10))
print(test_token_frequency.most_common(10))
def get_train_reviews_tokes(review_normalized):
for token_list in review_normalized:
yield dict([token, True] for token in token_list)
train_reviews_for_model = get_train_reviews_tokes(train_reviews_normalized)
test_reviews_for_model = get_train_reviews_tokes(test_reviews_normalized)
COUNT = 1
train_dataset = []
for review_dict in train_reviews_for_model:
if COUNT <= 12500:
train_dataset.append((review_dict, 1))
else:
train_dataset.append((review_dict, 0))
COUNT += 1
COUNT = 1
test_dataset = []
for review_dict in test_reviews_for_model:
if COUNT <= 12500:
test_dataset.append((review_dict, 1))
else:
test_dataset.append((review_dict, 0))
COUNT += 1
import random
random.shuffle(train_dataset)
train_dataset[24500]
occurence = []
for i in range(1000):
for tup in train_dataset:
if tup[1] == 0:
occurence.append(i)
break
len(occurence)
| 0.154951 | 0.699819 |
<a href="https://colab.research.google.com/github/zangell44/DS-Sprint-03-Creating-Professional-Portfolios/blob/master/DS_Unit_1_Sprint_Challenge_3.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# Data Science Unit 1 Sprint Challenge 3
# Creating Professional Portfolios
For your Sprint Challenge, you will **write about your upcoming [data storytelling portfolio project](https://learn.lambdaschool.com/ds/module/recedjanlbpqxic2r)**.
(Don't worry, you don't have to choose your final idea now. For this challenge, you can write about any idea you're considering.)
# Part 1
**Describe an idea** you could work on for your upcoming data storytelling project. What's your hypothesis?
#### Write a [lede](https://www.thoughtco.com/how-to-write-a-great-lede-2074346) (first paragraph)
- Put the bottom line up front.
- Use 60 words or fewer. (The [Hemingway App](http://www.hemingwayapp.com/) gives you word count.)
[This is hard](https://quoteinvestigator.com/2012/04/28/shorter-letter/), but you can do it!
#### Stretch goals
- Write more about your idea. Tell us what the story's about. Show us why it's interesting. Continue to follow the inverted pyramid structure.
- Improve your readability. Post your "before & after" scores from the Hemingway App.
**Idea - The Basics**
I would like to delve further into wealth/income inequality in different countries. Many analyses/metrics are *static* measures of inequality in outcomes.
For example, the [Gini Index](https://www.investopedia.com/terms/g/gini-index.asp) is a common metric for reporting inequality, measuring how incomes deviate from perfect equality.
I would argue it's more complicated. Equality of *opportunity* is at least as important as equality of outcomes. I suspect that opportuntity equality differs substantially in countries where outcome equality is similar.
One way to test this is by comparing correlations between income quintiles (or other groups). I would pose the question - "If you're born into the lowest income quintile in the US how likely are you to land in the lowest income quintile when you're older? the second quinitle? third? fourth? fifth?"
**Lede **
Across the world, [Standard measures of inequality](http://www.pewresearch.org/fact-tank/2015/09/22/the-many-ways-to-measure-economic-inequality/ "Inequality in the US") exhibit concerning trends. If trends persist, history suggests *at best * civil unrest will ensue. To better understand inequality, we must also examine dynamic inequality measures.
Dynamic equality, the American Dream, is equality of opportunity. Has inequality of opportunity exhibited a parallel increase in past decades?
# Part 2
#### Find sources
- Link to at least 2 relevant sources for your topic. Sources could include any data or writing about your topic.
- Use [Markdown](https://commonmark.org/help/) to format your links.
- Summarize each source in 1-2 sentences.
#### Stretch goals
- Find more sources.
- Use Markdown to add images from your sources.
- Critically evaluate your sources in writing.
**Sources**
[OECD - Inequality and Poverty](http://www.oecd.org/social/inequality-and-poverty.htm)
The OECD has extensive data on OECD nations. Further investigation is necessary to determine if the site has the data I'm looking for.
[Opportunity Insights](https://opportunityinsights.org/data/)
Opportunity Inights, run by a team of researchers at Harvard University, aims to analyze data on inequality and opportunity to create policies generating better outcomes. The organization appears to have a good quantity of open source data and publishes research regularly (including data, replication code, etc).
The downside is the data appears to be mostly, if not exclusively, US based. This represents a good starting point, but eventually I would like to compare across countries.
Given the academic background of the organization, this site's data quality should be high. I would feel comfortable using it with limited data validation.
[World Bank Global Database on Intergenerational Mobility](https://www.worldbank.org/en/topic/poverty/brief/what-is-the-global-database-on-intergenerational-mobility-gdim)
Per the World Bank this database "estimates of intergenerational mobility covering 148 economies for cohorts between 1940 and 1989. This translates to a world population coverage of 96 percent".
The dataset is extensively documented, but both usefulness and quality of the data require further investigation. Some of the data is based on surveys, which are notoriously unreliable.
# Part 3
#### Plan your next steps
- Describe at least 2 actions you'd take to get started with your project.
- Use Markdown headings and lists to organize your plan.
#### Stretch goals
- Add detail to your plan.
- Publish your project proposal on your GitHub Pages site.
## Next Steps
### Find Data on Income/Wealth Outcomes by Income/Wealth of Family at Birth
I can't do much without data to examine the problem. It remains to be seen how difficult it is to.
#### US Data
Analyze US Data, starting with [Opportunity Insights](https://opportunityinsights.org/data/). Data for only the US should be easy enough to find.
#### World Data
My ideal data would look something like this -
For each country, a table in the form of:
PersonID | Birth Year | Parent's Income at Birth | Person's Current Income
--- | --- | --- | ---
1 | 1980 | 20,000 | 30,000
... | ... | ... | ...
Income data could be either the income amount, or the percentile. Either would suffice for analysis.
Summary statistics pre-compiled would do for the initial analysis. Additional factors would be useful for further granular analysis of structural factors.
Promising sources include:
* [World Bank Global Database on Intergenerational Mobility](https://www.worldbank.org/en/topic/poverty/brief/what-is-the-global-database-on-intergenerational-mobility-gdim)
### Examine Existing Academic Research on Dynamic Inequality Measures
Lots has been written about inequality, from Thomas Piketty to the World Bank.
I read (most of) Capital in the 21st Century, and the rest is still sitting on my book shelf. I'd like to go back and examine some of the factors Piketty concludes are responsible for wealth inequality.
Plenty of other academic reserach on both wealth and income inequality has been published. This often is found alongisde data sources, including several of those cited above.
I am particularly interested in (1) Existing research on dynamic inequality measures (2) how dynamic inequality measures compare to static ones.
### Using Data and Existing Research, Quantify Dynamic Inequality
Visuals, analysis, etc. is great, but to meaningfully communicate about social mobility, I'll need some statistic. The Gini coefficient communicates a great deal about static inequality in one number.
If there is not an equivialent for dynamic inequality, I'll have to create one.
### Visualize Dynamic Inequality
Create a sweet world map to visualize how dynamic inequality varies.
```
```
|
github_jupyter
| 0.096376 | 0.905615 |
|
# In Class: ggplot2 - II
## Loading packages and data
In this session, we will continue our analysis of COVID-19 data in the US, but this time look more locally at individual case data from Montgomery county in Pennsylvania (Data which one of your course directors may or may not have been obsessively analyzing since March 2020).
We will look at the number of cases across townships, the age distribution of these cases and how outcomes are associated with age.
Unlike the previous session, where we used mostly geoms with 'identity' stats (e.g. geom_point), here we will use functions that calculate different stats from the data including counts, bins and density.
We have already downloaded the data for you. As before, load the data using the code below and take a look at it so you can get an idea of how it is structured.
Execute the following code below to get started:
```
library(tidyverse)
options(repr.plot.width=10, repr.plot.height=3) #set size for plots in this notebook
data <- as_tibble(read.csv('data.csv'))
head(data)
```
As before, run the code below to convert the column DateReported to Class "Date".
```
data$DateReported = as.Date(data$DateReported,format = '%Y-%m-%d')
```
In this dataset, every line is a single confirmed COVID-19 case.
First, let us plot the number of cases in the county as a function of time. In the last in class session, we had the total number cases reportd each day as a variable, so it was very intutive to plot that number over the date using geom_point.
Here, we would need to summarize the number of rows reported each day. You have already done this two classes ago! (Hint: you would like to make a bar plot)
**Q1.** Write the ggplot code that:
- creates a barplot for the number of cases reported per day.
Provide and execute your code below:
**Q2.** What statistic is being used as a summary in this plot? *(Hint: check the documentation for geom_bar)*
Next, let us investigate how many cases have been reported in each township (indicated in the column labelled *Name*).
To do that, we need to produce a new table (call it **count_data**) with two columns:
the column "Name" that contains the name of each township
and the column "n" that contains the number of cases (which is the number of rows reported by each township).
Remember back in R-III how we used different functions in dplyr (tidyverse) to summarize and group data? If you don't recall, look back at your R-III prelab or inclass assignment to help you answer this next question:
**Q3.** Use dplyr's **group_by()** and **summarize()** function, make a new table that
- groups the data by *Name*
- summarizes the total count (name this "n") for each Name (hint: n() might be helpful here)
- stores the output in a new variable called **count_data**.
- check the output of **count_data** using the **head()** function
Provide and Execute your code below:
Now let's add an additional piece of data. It would be interesting to compare the number of confirmed cases with the population size in each township and calculate the percent of population that had a confirmed infection.
For that, we will need to add a column to our **count_data** with the population size for each township, and another column with the ratio of these numbers.
Execute the code below to load a table with population size.
```
pop_size <- as_tibble(read.csv('montco_2010_census.csv'))
head(pop_size)
```
(This data is from the 2010 census, but not much happens in the suburbs....)
Now you will have to *join* the two tables and add another column - *pct_infected* - with the ratio.
**Q4.** Modify the code below (to the contents of the mutate function) and add the missing part to calculate the ratio.
Execute the code after you have modified it.
```
count_data <- count_data %>% inner_join(pop_size) %>% mutate(___________________)
head(count_data)
```
(Note the variable that was used to join the two tables. This time it was easy because both tables had a single shared variable, but in other cases you might need to specify which variable to use for the join.)
Now, let's compare the size of each township with the percent of confirmed infections.
**Q5.** Produce a ggplot2 dot plot to compare these two variables.
Provide and Execute the code below:
Looking at this plot, do you think there is a clear relationship? Why or why not?
Next, let's choose the top four townships with the highest number of infections for further analysis.
Execute the code below to sort **count_data** by the total number of confirmed infections.
```
count_data <- count_data %>% arrange(n)
head(count_data)
```
ooops... the code sorted the column n by an ascending order!
**Q6.** Modify the code below (to the contents of arrange) such that the order would be reversed. (hint: is there a descending function in tidyverse?)
Execeute the code below after you modify.
```
count_data <- count_data %>% arrange(______)
head(count_data)
```
Great! Now that we know which four townships have the highest number of infections, we will focus on them for addtional analysis.
**Q7.** Reuse the code from **Q1** and replot these four townships, such that each will have a seperate plot:
- Filter the **data** variable to include only these four townships
- Use **facet_wrap()** to plot the data for the four townships separatly
Remember that you can use %>% to input data directly into ggplot, try to write this in one command without saving any addtional variables.
Provide and execute the code below:
Each bar in our plot contains all the cases reported on that specific day, we can easily add information to these bars by dividing them according to an addtional categorical variable. For example, say we would like to know which age groups the cases reported on each day belong to.
**Q8.** Build on the code in **Q7** to:
- add one additional aesthetic mapping using the paramater "fill", and assigning the variable *Age_Range* to it.
Provide and execute the code below:
**Q9.** Look closely at these plots.
Do you see a difference in age composition between the first and second wave in Lower Merion?
Finally, we all know that there is a strong relationship between age and outcome of Sars-CoV2 infection. So let's check if this is reflected in the data.
In the original dataset from montoco (**data**), there is a column called *Hospitalized* which contains some outcome data.
Execute the code below to make a density plot of the age distribution for the different outcomes.
```
ggplot(data) + geom_density(aes(x=Age,fill=Hospitalized))
```
This is nice, but it is a bit hard to see the distribution of cases that were not hospitalized.
However, we can add a parameter to 'geom' which will make the fill semi-transparent (hint: read about potential arguements you can provide to the **geom_density()** function).
**Q10.** Modify the code from the plot above to add some transparency, and replot it here.
Provide and execute the code below:
# Homework
For homework, let's look at the rate of vaccination across different US states.
Load the data from the file
**us-daily-covid-vaccine-doses-administered.csv**
This file contains the number of reported shots per day for each US state.
**Q11.** Plot the number of shots per day over time for four of your favorite states.
Provide and Execute your code below:
**Q12.** Assuming:
- That the vaccination rate estimated by the average in the last 7 recorded days persists until the end of the pandemic
- and that everyone who is offered a vaccine accepts one
How many days will it take for each of these states reach 75% vaccination, a level close to hopefully a level of 'herd immunity'? And what precise day is that? (Do note that the rates of vaccination are actually increasing as we go along. So the guess you are making here is in this way a very conservative one!)
|
github_jupyter
|
library(tidyverse)
options(repr.plot.width=10, repr.plot.height=3) #set size for plots in this notebook
data <- as_tibble(read.csv('data.csv'))
head(data)
data$DateReported = as.Date(data$DateReported,format = '%Y-%m-%d')
pop_size <- as_tibble(read.csv('montco_2010_census.csv'))
head(pop_size)
count_data <- count_data %>% inner_join(pop_size) %>% mutate(___________________)
head(count_data)
count_data <- count_data %>% arrange(n)
head(count_data)
count_data <- count_data %>% arrange(______)
head(count_data)
ggplot(data) + geom_density(aes(x=Age,fill=Hospitalized))
| 0.423458 | 0.98746 |
```
!pip install python-dotenv
from dotenv import load_dotenv, find_dotenv
#find .env automatically by walking up directories until it's found
dotenv_path = find_dotenv()
#load up the entries as environment variables
load_dotenv(dotenv_path)
KAGGLE_USERNAME = os.environ.get("KAGGLE_USERNAME")
print(KAGGLE_USERNAME)
KAGGLE_PASSWORD = os.environ.get("KAGGLE_PASSWORD")
print(KAGGLE_PASSWORD)
import requests
from requests import session
import os
from dotenv import load_dotenv, find_dotenv
from contextlib import closing
import csv
import logging
import os
payload={
'eamil': os.environ.get("KAGGLE_USERNAME"),
'password': os.environ.get("KAGGLE_PASSWORD"),
'X-XSRF-TOKEN': 'CfDJ8LdUzqlsSWBPr4Ce3rb9VL_y_NbFaUd3UvKyA87g-6f_duSDyTbcvnSdqgsUUd6f9nZZgzvwtknzZy7kB1hVo4NezWLbcDUCa0dwX2diSkdFx6_nXb0oy0eNClalAHBUBJB0YPXmzvED6srSMEKcLjY'
}
headers = {'cookie': 'ka_sessionid=de5a7b94fc1b8190d230a660db2eeabe; CSRF-TOKEN=CfDJ8LdUzqlsSWBPr4Ce3rb9VL8HNVA6MY5KS81DxnibiALk1rubVNl8mWVI9mGKxhxTn2_7e7VSCEgiYx_VcshnUQGIsyVOqjQBNSFrsVIOBMRlPAIcSB0BYU56cS4PtrBFDJau8LGVGyHqndtWrG7gNW4; GCLB=CLz3y_uEmtrGQA; _ga=GA1.2.1051186253.1587541272; _gid=GA1.2.965433564.1587541272; searchToken=1f4bc0e6-d0f4-4aa5-8d77-3d936b25bf7c; intercom-session-koj6gxx6=dzNYQWdZWjQxQWVwa3hzVEFSTkE1U1pZMUQwaUxPVXZueXR1bnBkOE5GUTVPOWVreGRKSnR3Njd5TUFnNmIwTC0tWTZ2aDlpaFBuRFpGVm1RR2RCaWFwQT09--1b2853c28de7ff83fc33352e16ec8d350fa54b01; XSRF-TOKEN=CfDJ8LdUzqlsSWBPr4Ce3rb9VL_y_NbFaUd3UvKyA87g-6f_duSDyTbcvnSdqgsUUd6f9nZZgzvwtknzZy7kB1hVo4NezWLbcDUCa0dwX2diSkdFx6_nXb0oy0eNClalAHBUBJB0YPXmzvED6srSMEKcLjY; CLIENT-TOKEN=eyJhbGciOiJub25lIiwidHlwIjoiSldUIn0.eyJpc3MiOiJrYWdnbGUiLCJhdWQiOiJjbGllbnQiLCJzdWIiOm51bGwsIm5idCI6IjIwMjAtMDQtMjJUMTA6MTU6NDguNjg3MjAyMloiLCJpYXQiOiIyMDIwLTA0LTIyVDEwOjE1OjQ4LjY4NzIwMjJaIiwianRpIjoiZTE3NzIxMzAtMjZjNC00MjIwLWI4NWItMDczNTZjMzAxN2RmIiwiZXhwIjoiMjAyMC0wNS0yMlQxMDoxNTo0OC42ODcyMDIyWiIsImFub24iOnRydWUsImZmIjpbIkZsZXhpYmxlR3B1IiwiS2VybmVsc0ludGVybmV0IiwiRGF0YUV4cGxvcmVyVjIiLCJEYXRhU291cmNlU2VsZWN0b3JWMiIsIktlcm5lbHNWaWV3ZXJJbm5lclRhYmxlT2ZDb250ZW50cyIsIkZvcnVtV2F0Y2hEZXByZWNhdGVkIiwiTmV3S2VybmVsV2VsY29tZSIsIk1kZUltYWdlVXBsb2FkZXIiLCJLZXJuZWxzUXVpY2tWZXJzaW9ucyIsIkRpc2FibGVDdXN0b21QYWNrYWdlcyIsIlBpbk9yaWdpbmFsRG9ja2VyVmVyc2lvbiIsIlBob25lVmVyaWZ5Rm9yR3B1IiwiQ2xvdWRTZXJ2aWNlc0tlcm5lbEludGVnIiwiVXNlclNlY3JldHNLZXJuZWxJbnRlZyIsIk5hdmlnYXRpb25SZWRlc2lnbiIsIktlcm5lbHNTbmlwcGV0cyIsIktlcm5lbFdlbGNvbWVMb2FkRnJvbVVybCIsIlRwdUtlcm5lbEludGVnIiwiS2VybmVsc0ZpcmViYXNlTG9uZ1BvbGxpbmciLCJEYXRhc2V0TGl2ZU1vdW50IiwiRGF0YXNldHNUYXNrT25Ob3RlYm9va0xpc3RpbmciXX0.; _gat_gtag_UA_12629138_1=1'}
url ='https://www.kaggle.com/c/titanic/download/GQf0y8ebHO0C4JXscPPp%2Fversions%2FXkNkvXwqPPVG0Qt3MtQT%2Ffiles%2Ftrain.csv'
url2 = 'https://www.kaggle.com/c/titanic/notebooks'
#可以成功登入,但是下載csv時還是被擋住了
with session() as c:
#post request
response = c.get(url)
response=c.post('https://www.kaggle.com/account/email-signin?returnUrl=https%3A%2F%2Fwww.kaggle.com%2Fc%2Ftitanic%2Fdata',
data=payload,headers=headers)
print(response)
for chunk in r.iter_content(chunk_size=(512*1024)):
if chunk:
print(chunk)
# with closing(c.get(url)) as r:
# print(r)
# lines = (line.decode('utf-8') for line in r.iter_lines())
# for row in csv.reader(lines):
# print(row)
#kaggle登入,下載目前失敗
def extract_data(url,file_path):
with session() as c:
c.post('https://www.kaggle.com/account/email-signin?returnUrl=https%3A%2F%2Fwww.kaggle.com%2Fc%2Ftitanic%2Fdata'
data=payload,headers=headers)
with open(file_path,'w') as handle:
response = c.get(url,stream=True)
for block in response.iter_content(1024)
handle.write(block)
#測試下載csv檔案,可以成功
url='https://www.stats.govt.nz/assets/Uploads/Annual-enterprise-survey/Annual-enterprise-survey-2018-financial-year-provisional/Download-data/annual-enterprise-survey-2018-financial-year-provisional-csv.csv'
with session() as c:
with closing(c.get(url)) as r:
lines = (line.decode('utf-8') for line in r.iter_lines())
for row in csv.reader(lines):
print(row)
def main(project_dir):
logger= logger.getLogger(__name__)
logger.info('getting raw date')
train_url = 'https://www.kaggle.com/c/titanic/download/GQf0y8ebHO0C4JXscPPp%2Fversions%2FXkNkvXwqPPVG0Qt3MtQT%2Ffiles%2Ftrain.csv'
test_url = 'http'
```
|
github_jupyter
|
!pip install python-dotenv
from dotenv import load_dotenv, find_dotenv
#find .env automatically by walking up directories until it's found
dotenv_path = find_dotenv()
#load up the entries as environment variables
load_dotenv(dotenv_path)
KAGGLE_USERNAME = os.environ.get("KAGGLE_USERNAME")
print(KAGGLE_USERNAME)
KAGGLE_PASSWORD = os.environ.get("KAGGLE_PASSWORD")
print(KAGGLE_PASSWORD)
import requests
from requests import session
import os
from dotenv import load_dotenv, find_dotenv
from contextlib import closing
import csv
import logging
import os
payload={
'eamil': os.environ.get("KAGGLE_USERNAME"),
'password': os.environ.get("KAGGLE_PASSWORD"),
'X-XSRF-TOKEN': 'CfDJ8LdUzqlsSWBPr4Ce3rb9VL_y_NbFaUd3UvKyA87g-6f_duSDyTbcvnSdqgsUUd6f9nZZgzvwtknzZy7kB1hVo4NezWLbcDUCa0dwX2diSkdFx6_nXb0oy0eNClalAHBUBJB0YPXmzvED6srSMEKcLjY'
}
headers = {'cookie': 'ka_sessionid=de5a7b94fc1b8190d230a660db2eeabe; CSRF-TOKEN=CfDJ8LdUzqlsSWBPr4Ce3rb9VL8HNVA6MY5KS81DxnibiALk1rubVNl8mWVI9mGKxhxTn2_7e7VSCEgiYx_VcshnUQGIsyVOqjQBNSFrsVIOBMRlPAIcSB0BYU56cS4PtrBFDJau8LGVGyHqndtWrG7gNW4; GCLB=CLz3y_uEmtrGQA; _ga=GA1.2.1051186253.1587541272; _gid=GA1.2.965433564.1587541272; searchToken=1f4bc0e6-d0f4-4aa5-8d77-3d936b25bf7c; intercom-session-koj6gxx6=dzNYQWdZWjQxQWVwa3hzVEFSTkE1U1pZMUQwaUxPVXZueXR1bnBkOE5GUTVPOWVreGRKSnR3Njd5TUFnNmIwTC0tWTZ2aDlpaFBuRFpGVm1RR2RCaWFwQT09--1b2853c28de7ff83fc33352e16ec8d350fa54b01; XSRF-TOKEN=CfDJ8LdUzqlsSWBPr4Ce3rb9VL_y_NbFaUd3UvKyA87g-6f_duSDyTbcvnSdqgsUUd6f9nZZgzvwtknzZy7kB1hVo4NezWLbcDUCa0dwX2diSkdFx6_nXb0oy0eNClalAHBUBJB0YPXmzvED6srSMEKcLjY; CLIENT-TOKEN=eyJhbGciOiJub25lIiwidHlwIjoiSldUIn0.eyJpc3MiOiJrYWdnbGUiLCJhdWQiOiJjbGllbnQiLCJzdWIiOm51bGwsIm5idCI6IjIwMjAtMDQtMjJUMTA6MTU6NDguNjg3MjAyMloiLCJpYXQiOiIyMDIwLTA0LTIyVDEwOjE1OjQ4LjY4NzIwMjJaIiwianRpIjoiZTE3NzIxMzAtMjZjNC00MjIwLWI4NWItMDczNTZjMzAxN2RmIiwiZXhwIjoiMjAyMC0wNS0yMlQxMDoxNTo0OC42ODcyMDIyWiIsImFub24iOnRydWUsImZmIjpbIkZsZXhpYmxlR3B1IiwiS2VybmVsc0ludGVybmV0IiwiRGF0YUV4cGxvcmVyVjIiLCJEYXRhU291cmNlU2VsZWN0b3JWMiIsIktlcm5lbHNWaWV3ZXJJbm5lclRhYmxlT2ZDb250ZW50cyIsIkZvcnVtV2F0Y2hEZXByZWNhdGVkIiwiTmV3S2VybmVsV2VsY29tZSIsIk1kZUltYWdlVXBsb2FkZXIiLCJLZXJuZWxzUXVpY2tWZXJzaW9ucyIsIkRpc2FibGVDdXN0b21QYWNrYWdlcyIsIlBpbk9yaWdpbmFsRG9ja2VyVmVyc2lvbiIsIlBob25lVmVyaWZ5Rm9yR3B1IiwiQ2xvdWRTZXJ2aWNlc0tlcm5lbEludGVnIiwiVXNlclNlY3JldHNLZXJuZWxJbnRlZyIsIk5hdmlnYXRpb25SZWRlc2lnbiIsIktlcm5lbHNTbmlwcGV0cyIsIktlcm5lbFdlbGNvbWVMb2FkRnJvbVVybCIsIlRwdUtlcm5lbEludGVnIiwiS2VybmVsc0ZpcmViYXNlTG9uZ1BvbGxpbmciLCJEYXRhc2V0TGl2ZU1vdW50IiwiRGF0YXNldHNUYXNrT25Ob3RlYm9va0xpc3RpbmciXX0.; _gat_gtag_UA_12629138_1=1'}
url ='https://www.kaggle.com/c/titanic/download/GQf0y8ebHO0C4JXscPPp%2Fversions%2FXkNkvXwqPPVG0Qt3MtQT%2Ffiles%2Ftrain.csv'
url2 = 'https://www.kaggle.com/c/titanic/notebooks'
#可以成功登入,但是下載csv時還是被擋住了
with session() as c:
#post request
response = c.get(url)
response=c.post('https://www.kaggle.com/account/email-signin?returnUrl=https%3A%2F%2Fwww.kaggle.com%2Fc%2Ftitanic%2Fdata',
data=payload,headers=headers)
print(response)
for chunk in r.iter_content(chunk_size=(512*1024)):
if chunk:
print(chunk)
# with closing(c.get(url)) as r:
# print(r)
# lines = (line.decode('utf-8') for line in r.iter_lines())
# for row in csv.reader(lines):
# print(row)
#kaggle登入,下載目前失敗
def extract_data(url,file_path):
with session() as c:
c.post('https://www.kaggle.com/account/email-signin?returnUrl=https%3A%2F%2Fwww.kaggle.com%2Fc%2Ftitanic%2Fdata'
data=payload,headers=headers)
with open(file_path,'w') as handle:
response = c.get(url,stream=True)
for block in response.iter_content(1024)
handle.write(block)
#測試下載csv檔案,可以成功
url='https://www.stats.govt.nz/assets/Uploads/Annual-enterprise-survey/Annual-enterprise-survey-2018-financial-year-provisional/Download-data/annual-enterprise-survey-2018-financial-year-provisional-csv.csv'
with session() as c:
with closing(c.get(url)) as r:
lines = (line.decode('utf-8') for line in r.iter_lines())
for row in csv.reader(lines):
print(row)
def main(project_dir):
logger= logger.getLogger(__name__)
logger.info('getting raw date')
train_url = 'https://www.kaggle.com/c/titanic/download/GQf0y8ebHO0C4JXscPPp%2Fversions%2FXkNkvXwqPPVG0Qt3MtQT%2Ffiles%2Ftrain.csv'
test_url = 'http'
| 0.089095 | 0.088033 |
This Notebook presents all experimental results, with the code used to compute them.
Results are based on two-fold cross-validation, by splitting the RWC Pop dataset between odd and even songs.
Results are then averaged on both test subsets.
```
import numpy as np
import musicntd.scripts.final_tests as tests # Paths are defined in scripts/default_paths.py, and you should modify them in order for the Notebook to work.
penalty_weight = 1
penalty_range = [1]
core_dimensions_list = [8,16,24,32,40]
```
# Baseline (HALS on chromagram)
```
core_dimensions_chromas = [12,16,20,24,28,32,36,40,44,48]
# Note that the baseline with chromagrams is based on much more dimensions than Mel/Log Mel ones.
odd_params, odd_zero_five, odd_three = tests.several_ranks_with_cross_validation_of_param_RWC("even_songs", "odd_songs", "pcp", init = "chromas",
ranks_frequency = [12], ranks_rhythm = core_dimensions_chromas, ranks_pattern = core_dimensions_chromas,
penalty_range = penalty_range, update_rule = "hals", annotations_type = "MIREX10", penalty_func = "modulo8", convolution_type = "mixed")
even_params, even_zero_five, even_three = tests.several_ranks_with_cross_validation_of_param_RWC("odd_songs", "even_songs", "pcp", init = "chromas",
ranks_frequency = [12], ranks_rhythm = core_dimensions_chromas, ranks_pattern = core_dimensions_chromas,
penalty_range = penalty_range, update_rule = "hals", annotations_type = "MIREX10", penalty_func = "modulo8", convolution_type = "mixed")
print(f"0.5 seconds: {[(odd_zero_five[i] + even_zero_five[i])/2 for i in range(3,6)]}")
print(f"3 seconds: {[(odd_three[i] + even_three[i])/2 for i in range(3,6)]}")
```
# Mel Spectrogram
```
feature = "mel_grill"
```
## Feature
```
zero_five_feature, three_feature = tests.fixed_conditions_feature("full", feature, penalty_weight = penalty_weight, annotations_type = "MIREX10", penalty_func = "modulo8", convolution_type = "mixed")
```
## HALS
```
odd_params, odd_zero_five, odd_three = tests.several_ranks_with_cross_validation_of_param_RWC("even_songs", "odd_songs", feature,
ranks_frequency = core_dimensions_list, ranks_rhythm = core_dimensions_list, ranks_pattern = core_dimensions_list,
penalty_range = penalty_range, update_rule = "hals", annotations_type = "MIREX10", penalty_func = "modulo8", convolution_type = "mixed")
even_params, even_zero_five, even_three = tests.several_ranks_with_cross_validation_of_param_RWC("odd_songs", "even_songs", feature,
ranks_frequency = core_dimensions_list, ranks_rhythm = core_dimensions_list, ranks_pattern = core_dimensions_list,
penalty_range = penalty_range, update_rule = "hals", annotations_type = "MIREX10", penalty_func = "modulo8", convolution_type = "mixed")
print(f"0.5 seconds: {[(odd_zero_five[i] + even_zero_five[i])/2 for i in range(3,6)]}")
print(f"3 seconds: {[(odd_three[i] + even_three[i])/2 for i in range(3,6)]}")
```
## MU
### Beta = 1
```
odd_params, odd_zero_five, odd_three = tests.several_ranks_with_cross_validation_of_param_RWC("even_songs", "odd_songs", feature,
ranks_frequency = core_dimensions_list, ranks_rhythm = core_dimensions_list, ranks_pattern = core_dimensions_list, penalty_range = penalty_range,
update_rule = "mu", beta = 1, annotations_type = "MIREX10", penalty_func = "modulo8", convolution_type = "mixed")
even_params, even_zero_five, even_three = tests.several_ranks_with_cross_validation_of_param_RWC("odd_songs", "even_songs", feature,
ranks_frequency = core_dimensions_list, ranks_rhythm = core_dimensions_list, ranks_pattern = core_dimensions_list, penalty_range = penalty_range,
update_rule = "mu", beta = 1, annotations_type = "MIREX10", penalty_func = "modulo8", convolution_type = "mixed")
print(f"0.5 seconds: {[(odd_zero_five[i] + even_zero_five[i])/2 for i in range(3,6)]}")
print(f"3 seconds: {[(odd_three[i] + even_three[i])/2 for i in range(3,6)]}")
```
### Beta = 0
```
odd_params, odd_zero_five, odd_three = tests.several_ranks_with_cross_validation_of_param_RWC("even_songs", "odd_songs", feature,
ranks_frequency = core_dimensions_list, ranks_rhythm = core_dimensions_list, ranks_pattern = core_dimensions_list, penalty_range = penalty_range,
update_rule = "mu", beta = 0, annotations_type = "MIREX10", penalty_func = "modulo8", convolution_type = "mixed")
even_params, even_zero_five, even_three = tests.several_ranks_with_cross_validation_of_param_RWC("odd_songs", "even_songs", feature,
ranks_frequency = core_dimensions_list, ranks_rhythm = core_dimensions_list, ranks_pattern = core_dimensions_list, penalty_range = penalty_range,
update_rule = "mu", beta = 0, annotations_type = "MIREX10", penalty_func = "modulo8", convolution_type = "mixed")
print(f"0.5 seconds: {[(odd_zero_five[i] + even_zero_five[i])/2 for i in range(3,6)]}")
print(f"3 seconds: {[(odd_three[i] + even_three[i])/2 for i in range(3,6)]}")
```
# Nonnegative Log Mel Spectrogram
```
feature = "nn_log_mel_grill"
```
## Feature
```
zero_five_feature, three_feature = tests.fixed_conditions_feature("full", feature, penalty_weight = penalty_weight, annotations_type = "MIREX10", penalty_func = "modulo8", convolution_type = "mixed")
```
## HALS
```
odd_params, odd_zero_five, odd_three = tests.several_ranks_with_cross_validation_of_param_RWC("even_songs", "odd_songs", feature,
ranks_frequency = core_dimensions_list, ranks_rhythm = core_dimensions_list, ranks_pattern = core_dimensions_list,
penalty_range = penalty_range, update_rule = "hals", annotations_type = "MIREX10", penalty_func = "modulo8", convolution_type = "mixed")
even_params, even_zero_five, even_three = tests.several_ranks_with_cross_validation_of_param_RWC("odd_songs", "even_songs", feature,
ranks_frequency = core_dimensions_list, ranks_rhythm = core_dimensions_list, ranks_pattern = core_dimensions_list,
penalty_range = penalty_range, update_rule = "hals", annotations_type = "MIREX10", penalty_func = "modulo8", convolution_type = "mixed")
print(f"0.5 seconds: {[(odd_zero_five[i] + even_zero_five[i])/2 for i in range(3,6)]}")
print(f"3 seconds: {[(odd_three[i] + even_three[i])/2 for i in range(3,6)]}")
```
## MU
### Beta = 1
```
odd_params, odd_zero_five, odd_three = tests.several_ranks_with_cross_validation_of_param_RWC("even_songs", "odd_songs", feature,
ranks_frequency = core_dimensions_list, ranks_rhythm = core_dimensions_list, ranks_pattern = core_dimensions_list, penalty_range = penalty_range,
update_rule = "mu", beta = 1, annotations_type = "MIREX10", penalty_func = "modulo8", convolution_type = "mixed")
even_params, even_zero_five, even_three = tests.several_ranks_with_cross_validation_of_param_RWC("odd_songs", "even_songs", feature,
ranks_frequency = core_dimensions_list, ranks_rhythm = core_dimensions_list, ranks_pattern = core_dimensions_list, penalty_range = penalty_range,
update_rule = "mu", beta = 1, annotations_type = "MIREX10", penalty_func = "modulo8", convolution_type = "mixed")
print(f"0.5 seconds: {[(odd_zero_five[i] + even_zero_five[i])/2 for i in range(3,6)]}")
print(f"3 seconds: {[(odd_three[i] + even_three[i])/2 for i in range(3,6)]}")
```
### Beta = 0
```
odd_params, odd_zero_five, odd_three = tests.several_ranks_with_cross_validation_of_param_RWC("even_songs", "odd_songs", feature,
ranks_frequency = core_dimensions_list, ranks_rhythm = core_dimensions_list, ranks_pattern = core_dimensions_list, penalty_range = penalty_range,
update_rule = "mu", beta = 0, annotations_type = "MIREX10", penalty_func = "modulo8", convolution_type = "mixed")
even_params, even_zero_five, even_three = tests.several_ranks_with_cross_validation_of_param_RWC("odd_songs", "even_songs", feature,
ranks_frequency = core_dimensions_list, ranks_rhythm = core_dimensions_list, ranks_pattern = core_dimensions_list, penalty_range = penalty_range,
update_rule = "mu", beta = 0, annotations_type = "MIREX10", penalty_func = "modulo8", convolution_type = "mixed")
print(f"0.5 seconds: {[(odd_zero_five[i] + even_zero_five[i])/2 for i in range(3,6)]}")
print(f"3 seconds: {[(odd_three[i] + even_three[i])/2 for i in range(3,6)]}")
```
|
github_jupyter
|
import numpy as np
import musicntd.scripts.final_tests as tests # Paths are defined in scripts/default_paths.py, and you should modify them in order for the Notebook to work.
penalty_weight = 1
penalty_range = [1]
core_dimensions_list = [8,16,24,32,40]
core_dimensions_chromas = [12,16,20,24,28,32,36,40,44,48]
# Note that the baseline with chromagrams is based on much more dimensions than Mel/Log Mel ones.
odd_params, odd_zero_five, odd_three = tests.several_ranks_with_cross_validation_of_param_RWC("even_songs", "odd_songs", "pcp", init = "chromas",
ranks_frequency = [12], ranks_rhythm = core_dimensions_chromas, ranks_pattern = core_dimensions_chromas,
penalty_range = penalty_range, update_rule = "hals", annotations_type = "MIREX10", penalty_func = "modulo8", convolution_type = "mixed")
even_params, even_zero_five, even_three = tests.several_ranks_with_cross_validation_of_param_RWC("odd_songs", "even_songs", "pcp", init = "chromas",
ranks_frequency = [12], ranks_rhythm = core_dimensions_chromas, ranks_pattern = core_dimensions_chromas,
penalty_range = penalty_range, update_rule = "hals", annotations_type = "MIREX10", penalty_func = "modulo8", convolution_type = "mixed")
print(f"0.5 seconds: {[(odd_zero_five[i] + even_zero_five[i])/2 for i in range(3,6)]}")
print(f"3 seconds: {[(odd_three[i] + even_three[i])/2 for i in range(3,6)]}")
feature = "mel_grill"
zero_five_feature, three_feature = tests.fixed_conditions_feature("full", feature, penalty_weight = penalty_weight, annotations_type = "MIREX10", penalty_func = "modulo8", convolution_type = "mixed")
odd_params, odd_zero_five, odd_three = tests.several_ranks_with_cross_validation_of_param_RWC("even_songs", "odd_songs", feature,
ranks_frequency = core_dimensions_list, ranks_rhythm = core_dimensions_list, ranks_pattern = core_dimensions_list,
penalty_range = penalty_range, update_rule = "hals", annotations_type = "MIREX10", penalty_func = "modulo8", convolution_type = "mixed")
even_params, even_zero_five, even_three = tests.several_ranks_with_cross_validation_of_param_RWC("odd_songs", "even_songs", feature,
ranks_frequency = core_dimensions_list, ranks_rhythm = core_dimensions_list, ranks_pattern = core_dimensions_list,
penalty_range = penalty_range, update_rule = "hals", annotations_type = "MIREX10", penalty_func = "modulo8", convolution_type = "mixed")
print(f"0.5 seconds: {[(odd_zero_five[i] + even_zero_five[i])/2 for i in range(3,6)]}")
print(f"3 seconds: {[(odd_three[i] + even_three[i])/2 for i in range(3,6)]}")
odd_params, odd_zero_five, odd_three = tests.several_ranks_with_cross_validation_of_param_RWC("even_songs", "odd_songs", feature,
ranks_frequency = core_dimensions_list, ranks_rhythm = core_dimensions_list, ranks_pattern = core_dimensions_list, penalty_range = penalty_range,
update_rule = "mu", beta = 1, annotations_type = "MIREX10", penalty_func = "modulo8", convolution_type = "mixed")
even_params, even_zero_five, even_three = tests.several_ranks_with_cross_validation_of_param_RWC("odd_songs", "even_songs", feature,
ranks_frequency = core_dimensions_list, ranks_rhythm = core_dimensions_list, ranks_pattern = core_dimensions_list, penalty_range = penalty_range,
update_rule = "mu", beta = 1, annotations_type = "MIREX10", penalty_func = "modulo8", convolution_type = "mixed")
print(f"0.5 seconds: {[(odd_zero_five[i] + even_zero_five[i])/2 for i in range(3,6)]}")
print(f"3 seconds: {[(odd_three[i] + even_three[i])/2 for i in range(3,6)]}")
odd_params, odd_zero_five, odd_three = tests.several_ranks_with_cross_validation_of_param_RWC("even_songs", "odd_songs", feature,
ranks_frequency = core_dimensions_list, ranks_rhythm = core_dimensions_list, ranks_pattern = core_dimensions_list, penalty_range = penalty_range,
update_rule = "mu", beta = 0, annotations_type = "MIREX10", penalty_func = "modulo8", convolution_type = "mixed")
even_params, even_zero_five, even_three = tests.several_ranks_with_cross_validation_of_param_RWC("odd_songs", "even_songs", feature,
ranks_frequency = core_dimensions_list, ranks_rhythm = core_dimensions_list, ranks_pattern = core_dimensions_list, penalty_range = penalty_range,
update_rule = "mu", beta = 0, annotations_type = "MIREX10", penalty_func = "modulo8", convolution_type = "mixed")
print(f"0.5 seconds: {[(odd_zero_five[i] + even_zero_five[i])/2 for i in range(3,6)]}")
print(f"3 seconds: {[(odd_three[i] + even_three[i])/2 for i in range(3,6)]}")
feature = "nn_log_mel_grill"
zero_five_feature, three_feature = tests.fixed_conditions_feature("full", feature, penalty_weight = penalty_weight, annotations_type = "MIREX10", penalty_func = "modulo8", convolution_type = "mixed")
odd_params, odd_zero_five, odd_three = tests.several_ranks_with_cross_validation_of_param_RWC("even_songs", "odd_songs", feature,
ranks_frequency = core_dimensions_list, ranks_rhythm = core_dimensions_list, ranks_pattern = core_dimensions_list,
penalty_range = penalty_range, update_rule = "hals", annotations_type = "MIREX10", penalty_func = "modulo8", convolution_type = "mixed")
even_params, even_zero_five, even_three = tests.several_ranks_with_cross_validation_of_param_RWC("odd_songs", "even_songs", feature,
ranks_frequency = core_dimensions_list, ranks_rhythm = core_dimensions_list, ranks_pattern = core_dimensions_list,
penalty_range = penalty_range, update_rule = "hals", annotations_type = "MIREX10", penalty_func = "modulo8", convolution_type = "mixed")
print(f"0.5 seconds: {[(odd_zero_five[i] + even_zero_five[i])/2 for i in range(3,6)]}")
print(f"3 seconds: {[(odd_three[i] + even_three[i])/2 for i in range(3,6)]}")
odd_params, odd_zero_five, odd_three = tests.several_ranks_with_cross_validation_of_param_RWC("even_songs", "odd_songs", feature,
ranks_frequency = core_dimensions_list, ranks_rhythm = core_dimensions_list, ranks_pattern = core_dimensions_list, penalty_range = penalty_range,
update_rule = "mu", beta = 1, annotations_type = "MIREX10", penalty_func = "modulo8", convolution_type = "mixed")
even_params, even_zero_five, even_three = tests.several_ranks_with_cross_validation_of_param_RWC("odd_songs", "even_songs", feature,
ranks_frequency = core_dimensions_list, ranks_rhythm = core_dimensions_list, ranks_pattern = core_dimensions_list, penalty_range = penalty_range,
update_rule = "mu", beta = 1, annotations_type = "MIREX10", penalty_func = "modulo8", convolution_type = "mixed")
print(f"0.5 seconds: {[(odd_zero_five[i] + even_zero_five[i])/2 for i in range(3,6)]}")
print(f"3 seconds: {[(odd_three[i] + even_three[i])/2 for i in range(3,6)]}")
odd_params, odd_zero_five, odd_three = tests.several_ranks_with_cross_validation_of_param_RWC("even_songs", "odd_songs", feature,
ranks_frequency = core_dimensions_list, ranks_rhythm = core_dimensions_list, ranks_pattern = core_dimensions_list, penalty_range = penalty_range,
update_rule = "mu", beta = 0, annotations_type = "MIREX10", penalty_func = "modulo8", convolution_type = "mixed")
even_params, even_zero_five, even_three = tests.several_ranks_with_cross_validation_of_param_RWC("odd_songs", "even_songs", feature,
ranks_frequency = core_dimensions_list, ranks_rhythm = core_dimensions_list, ranks_pattern = core_dimensions_list, penalty_range = penalty_range,
update_rule = "mu", beta = 0, annotations_type = "MIREX10", penalty_func = "modulo8", convolution_type = "mixed")
print(f"0.5 seconds: {[(odd_zero_five[i] + even_zero_five[i])/2 for i in range(3,6)]}")
print(f"3 seconds: {[(odd_three[i] + even_three[i])/2 for i in range(3,6)]}")
| 0.493164 | 0.847084 |
<img src="http://akhavanpour.ir/notebook/images/srttu.gif" alt="SRTTU" style="width: 150px;"/>
[](https://notebooks.azure.com/import/gh/Alireza-Akhavan/class.vision)
# Deep Convolutional GAN (DCGAN)
```
%matplotlib inline
from keras.datasets import mnist
from keras.layers import Input, Dense, Reshape, Flatten, Dropout
from keras.layers import BatchNormalization, Activation, ZeroPadding2D
from keras.layers.advanced_activations import LeakyReLU
from keras.layers.convolutional import Conv2D, Conv2DTranspose
from keras.models import Sequential, Model
from keras.optimizers import Adam
import matplotlib.pyplot as plt
import numpy as np
img_rows = 28
img_cols = 28
channels = 1
img_shape = (img_rows, img_cols, channels)
z_dim = 100
```
## Generator
```
def generator(img_shape, z_dim):
model = Sequential()
# Reshape input into 7x7x256 tensor via a fully connected layer
model.add(Dense(256 * 7 * 7, input_dim=z_dim))
model.add(Reshape((7, 7, 256)))
# Transposed convolution layer, from 7x7x256 into 14x14x128 tensor
model.add(Conv2DTranspose(
128, kernel_size=3, strides=2, padding='same'))
# Batch normalization
model.add(BatchNormalization())
# Leaky ReLU
model.add(LeakyReLU(alpha=0.01))
# Transposed convolution layer, from 14x14x128 to 14x14x64 tensor
model.add(Conv2DTranspose(
64, kernel_size=3, strides=1, padding='same'))
# Batch normalization
model.add(BatchNormalization())
# Leaky ReLU
model.add(LeakyReLU(alpha=0.01))
# Transposed convolution layer, from 14x14x64 to 28x28x1 tensor
model.add(Conv2DTranspose(
1, kernel_size=3, strides=2, padding='same'))
# Tanh activation
model.add(Activation('tanh'))
z = Input(shape=(z_dim,))
img = model(z)
return Model(z, img)
```
## Discriminator
```
def discriminator(img_shape):
model = Sequential()
# Convolutional layer, from 28x28x1 into 14x14x32 tensor
model.add(Conv2D(32, kernel_size=3, strides=2,
input_shape=img_shape, padding='same'))
# Leaky ReLU
model.add(LeakyReLU(alpha=0.01))
# Convolutional layer, from 14x14x32 into 7x7x64 tensor
model.add(Conv2D(64, kernel_size=3, strides=2,
input_shape=img_shape, padding='same'))
# Batch normalization
model.add(BatchNormalization())
# Leaky ReLU
model.add(LeakyReLU(alpha=0.01))
# Convolutional layer, from 7x7x64 tensor into 3x3x128 tensor
model.add(Conv2D(128, kernel_size=3, strides=2,
input_shape=img_shape, padding='same'))
# Batch normalization
model.add(BatchNormalization())
# Leaky ReLU
model.add(LeakyReLU(alpha=0.01))
# Flatten the tensor and apply sigmoid activation function
model.add(Flatten())
model.add(Dense(1, activation='sigmoid'))
img = Input(shape=img_shape)
prediction = model(img)
return Model(img, prediction)
```
## Build the Model
```
# Build and compile the Discriminator
discriminator = discriminator(img_shape)
discriminator.compile(loss='binary_crossentropy',
optimizer=Adam(), metrics=['accuracy'])
# Build the Generator
generator = generator(img_shape, z_dim)
# Generated image to be used as input
z = Input(shape=(100,))
img = generator(z)
# Keep Discriminator’s parameters constant during Generator training
discriminator.trainable = False
# The Discriminator’s prediction
prediction = discriminator(img)
# Combined GAN model to train the Generator
combined = Model(z, prediction)
combined.compile(loss='binary_crossentropy', optimizer=Adam())
```
## Training
```
from dataset import load_hoda
def hoda_dataset():
X_train, _, _, _ = load_hoda(training_sample_size=60000,
test_sample_size=400,size=28)
X_train = np.array(X_train)
X_train = X_train.reshape(60000, 28, 28)
return X_train
losses = []
accuracies = []
def train(iterations, batch_size, sample_interval):
# Load the MNIST dataset
#(X_train, _), (_, _) = mnist.load_data()
# Load the HODA dataset
X_train = hoda_dataset()
# Rescale -1 to 1
X_train = X_train / 127.5 - 1.
X_train = np.expand_dims(X_train, axis=3)
# Labels for real and fake examples
real = np.ones((batch_size, 1))
fake = np.zeros((batch_size, 1))
for iteration in range(iterations):
# -------------------------
# Train the Discriminator
# -------------------------
# Select a random batch of real images
idx = np.random.randint(0, X_train.shape[0], batch_size)
imgs = X_train[idx]
# Generate a batch of fake images
z = np.random.normal(0, 1, (batch_size, 100))
gen_imgs = generator.predict(z)
# Discriminator loss
d_loss_real = discriminator.train_on_batch(imgs, real)
d_loss_fake = discriminator.train_on_batch(gen_imgs, fake)
d_loss = 0.5 * np.add(d_loss_real, d_loss_fake)
# ---------------------
# Train the Generator
# ---------------------
# Generate a batch of fake images
z = np.random.normal(0, 1, (batch_size, 100))
gen_imgs = generator.predict(z)
# Generator loss
g_loss = combined.train_on_batch(z, real)
if iteration % sample_interval == 0:
# Output training progress
print ("%d [D loss: %f, acc.: %.2f%%] [G loss: %f]" %
(iteration, d_loss[0], 100*d_loss[1], g_loss))
# Save losses and accuracies so they can be plotted after training
losses.append((d_loss[0], g_loss))
accuracies.append(100*d_loss[1])
# Output generated image samples
sample_images(iteration)
def sample_images(iteration, image_grid_rows=4, image_grid_columns=4):
# Sample random noise
z = np.random.normal(0, 1,
(image_grid_rows * image_grid_columns, z_dim))
# Generate images from random noise
gen_imgs = generator.predict(z)
# Rescale images to 0-1
gen_imgs = 0.5 * gen_imgs + 0.5
# Set image grid
fig, axs = plt.subplots(image_grid_rows, image_grid_columns,
figsize=(4,4), sharey=True, sharex=True)
cnt = 0
for i in range(image_grid_rows):
for j in range(image_grid_columns):
# Output image grid
axs[i,j].imshow(gen_imgs[cnt, :,:,0], cmap='gray')
axs[i,j].axis('off')
cnt += 1
```
## Train the Model and Inspect Output
```
# Suppress warnings because the warning Keras gives us about non-trainable parameters is by design:
# The Generator trainable parameters are intentionally held constant during Discriminator training and vice versa
import warnings; warnings.simplefilter('ignore')
iterations = 20000
batch_size = 128
sample_interval = 1000
# Train the GAN for the specified number of iterations
train(iterations, batch_size, sample_interval)
losses = np.array(losses)
# Plot training losses for Discriminator and Generator
plt.figure(figsize=(10,5))
plt.plot(losses.T[0], label="Discriminator Loss")
plt.plot(losses.T[1], label="Generator Loss")
plt.title("Training Losses")
plt.legend()
accuracies = np.array(accuracies)
# Plot Discriminator accuracy
plt.figure(figsize=(10,5))
plt.plot(accuracies, label="Discriminator Accuracy")
plt.title("Discriminator Accuracy")
plt.legend()
```
### <div style="direction:rtl;text-align:right;font-family:Tahoma">کدها برگرفته از:
</div>
[https://github.com/GANs-in-Action/gans-in-action/blob/master/chapter-4/Chapter_4_DCGAN.ipynb](https://github.com/GANs-in-Action/gans-in-action/blob/master/chapter-4/Chapter_4_DCGAN.ipynb)
<div class="alert alert-block alert-info">
<div style="direction:rtl;text-align:right;font-family:B Lotus, B Nazanin, Tahoma"> دانشگاه تربیت دبیر شهید رجایی<br>مباحث ویژه 2 - یادگیری عمیق پیشرفته<br>علیرضا اخوان پور<br>97-98<br>
</div>
<a href="https://www.srttu.edu/">SRTTU.edu</a> - <a href="http://class.vision">Class.Vision</a> - <a href="http://AkhavanPour.ir">AkhavanPour.ir</a>
</div>
|
github_jupyter
|
%matplotlib inline
from keras.datasets import mnist
from keras.layers import Input, Dense, Reshape, Flatten, Dropout
from keras.layers import BatchNormalization, Activation, ZeroPadding2D
from keras.layers.advanced_activations import LeakyReLU
from keras.layers.convolutional import Conv2D, Conv2DTranspose
from keras.models import Sequential, Model
from keras.optimizers import Adam
import matplotlib.pyplot as plt
import numpy as np
img_rows = 28
img_cols = 28
channels = 1
img_shape = (img_rows, img_cols, channels)
z_dim = 100
def generator(img_shape, z_dim):
model = Sequential()
# Reshape input into 7x7x256 tensor via a fully connected layer
model.add(Dense(256 * 7 * 7, input_dim=z_dim))
model.add(Reshape((7, 7, 256)))
# Transposed convolution layer, from 7x7x256 into 14x14x128 tensor
model.add(Conv2DTranspose(
128, kernel_size=3, strides=2, padding='same'))
# Batch normalization
model.add(BatchNormalization())
# Leaky ReLU
model.add(LeakyReLU(alpha=0.01))
# Transposed convolution layer, from 14x14x128 to 14x14x64 tensor
model.add(Conv2DTranspose(
64, kernel_size=3, strides=1, padding='same'))
# Batch normalization
model.add(BatchNormalization())
# Leaky ReLU
model.add(LeakyReLU(alpha=0.01))
# Transposed convolution layer, from 14x14x64 to 28x28x1 tensor
model.add(Conv2DTranspose(
1, kernel_size=3, strides=2, padding='same'))
# Tanh activation
model.add(Activation('tanh'))
z = Input(shape=(z_dim,))
img = model(z)
return Model(z, img)
def discriminator(img_shape):
model = Sequential()
# Convolutional layer, from 28x28x1 into 14x14x32 tensor
model.add(Conv2D(32, kernel_size=3, strides=2,
input_shape=img_shape, padding='same'))
# Leaky ReLU
model.add(LeakyReLU(alpha=0.01))
# Convolutional layer, from 14x14x32 into 7x7x64 tensor
model.add(Conv2D(64, kernel_size=3, strides=2,
input_shape=img_shape, padding='same'))
# Batch normalization
model.add(BatchNormalization())
# Leaky ReLU
model.add(LeakyReLU(alpha=0.01))
# Convolutional layer, from 7x7x64 tensor into 3x3x128 tensor
model.add(Conv2D(128, kernel_size=3, strides=2,
input_shape=img_shape, padding='same'))
# Batch normalization
model.add(BatchNormalization())
# Leaky ReLU
model.add(LeakyReLU(alpha=0.01))
# Flatten the tensor and apply sigmoid activation function
model.add(Flatten())
model.add(Dense(1, activation='sigmoid'))
img = Input(shape=img_shape)
prediction = model(img)
return Model(img, prediction)
# Build and compile the Discriminator
discriminator = discriminator(img_shape)
discriminator.compile(loss='binary_crossentropy',
optimizer=Adam(), metrics=['accuracy'])
# Build the Generator
generator = generator(img_shape, z_dim)
# Generated image to be used as input
z = Input(shape=(100,))
img = generator(z)
# Keep Discriminator’s parameters constant during Generator training
discriminator.trainable = False
# The Discriminator’s prediction
prediction = discriminator(img)
# Combined GAN model to train the Generator
combined = Model(z, prediction)
combined.compile(loss='binary_crossentropy', optimizer=Adam())
from dataset import load_hoda
def hoda_dataset():
X_train, _, _, _ = load_hoda(training_sample_size=60000,
test_sample_size=400,size=28)
X_train = np.array(X_train)
X_train = X_train.reshape(60000, 28, 28)
return X_train
losses = []
accuracies = []
def train(iterations, batch_size, sample_interval):
# Load the MNIST dataset
#(X_train, _), (_, _) = mnist.load_data()
# Load the HODA dataset
X_train = hoda_dataset()
# Rescale -1 to 1
X_train = X_train / 127.5 - 1.
X_train = np.expand_dims(X_train, axis=3)
# Labels for real and fake examples
real = np.ones((batch_size, 1))
fake = np.zeros((batch_size, 1))
for iteration in range(iterations):
# -------------------------
# Train the Discriminator
# -------------------------
# Select a random batch of real images
idx = np.random.randint(0, X_train.shape[0], batch_size)
imgs = X_train[idx]
# Generate a batch of fake images
z = np.random.normal(0, 1, (batch_size, 100))
gen_imgs = generator.predict(z)
# Discriminator loss
d_loss_real = discriminator.train_on_batch(imgs, real)
d_loss_fake = discriminator.train_on_batch(gen_imgs, fake)
d_loss = 0.5 * np.add(d_loss_real, d_loss_fake)
# ---------------------
# Train the Generator
# ---------------------
# Generate a batch of fake images
z = np.random.normal(0, 1, (batch_size, 100))
gen_imgs = generator.predict(z)
# Generator loss
g_loss = combined.train_on_batch(z, real)
if iteration % sample_interval == 0:
# Output training progress
print ("%d [D loss: %f, acc.: %.2f%%] [G loss: %f]" %
(iteration, d_loss[0], 100*d_loss[1], g_loss))
# Save losses and accuracies so they can be plotted after training
losses.append((d_loss[0], g_loss))
accuracies.append(100*d_loss[1])
# Output generated image samples
sample_images(iteration)
def sample_images(iteration, image_grid_rows=4, image_grid_columns=4):
# Sample random noise
z = np.random.normal(0, 1,
(image_grid_rows * image_grid_columns, z_dim))
# Generate images from random noise
gen_imgs = generator.predict(z)
# Rescale images to 0-1
gen_imgs = 0.5 * gen_imgs + 0.5
# Set image grid
fig, axs = plt.subplots(image_grid_rows, image_grid_columns,
figsize=(4,4), sharey=True, sharex=True)
cnt = 0
for i in range(image_grid_rows):
for j in range(image_grid_columns):
# Output image grid
axs[i,j].imshow(gen_imgs[cnt, :,:,0], cmap='gray')
axs[i,j].axis('off')
cnt += 1
# Suppress warnings because the warning Keras gives us about non-trainable parameters is by design:
# The Generator trainable parameters are intentionally held constant during Discriminator training and vice versa
import warnings; warnings.simplefilter('ignore')
iterations = 20000
batch_size = 128
sample_interval = 1000
# Train the GAN for the specified number of iterations
train(iterations, batch_size, sample_interval)
losses = np.array(losses)
# Plot training losses for Discriminator and Generator
plt.figure(figsize=(10,5))
plt.plot(losses.T[0], label="Discriminator Loss")
plt.plot(losses.T[1], label="Generator Loss")
plt.title("Training Losses")
plt.legend()
accuracies = np.array(accuracies)
# Plot Discriminator accuracy
plt.figure(figsize=(10,5))
plt.plot(accuracies, label="Discriminator Accuracy")
plt.title("Discriminator Accuracy")
plt.legend()
| 0.949248 | 0.955152 |
Objetivos:
1. Identificar cada una de las variables si es:
* cualitativa
* cuantitativa discreta
* cuantitativa continua.
2. Histograma qué distribución sigue?
```
import pandas as pd
data = pd.read_csv('/content/conjunto_de_datos_concentradohogar_enigh_2018_ns.csv')
data.sample(n=25)
#procedemos a ver los nobres de las columas
data.info(verbose = True)
# seleccionamos las columnas que son de tipo cualitativo y cuantiativo
variables_cuali = data.iloc[:,0:12]
variables_cuanti = data.iloc[:,13:]
var_cuanti = variables_cuanti.columns
# Añdimos las columnas cuantitativas que hacen falta
variables_cuanti_2 = data[['est_dis','upm','factor']]
var_cuantitativa = pd.concat([variables_cuanti,variables_cuanti_2],axis='columns')
# eliminamos algunas columnas cuantiativas dentro de las cualitativas
variables_cuali.drop(['est_dis','upm','factor'], axis='columns',inplace=True)
var_cuali = variables_cuali.columns
```
Procedemos a identificar que variables son cualitativas y que variables son cuantitativas, maas adelante identificaremos las discretas y las continuas.
```
# imprimimos las columnas cualitativas
for i in var_cuali:
print("La columna {} es: cualitativa ".format(i))
for i in var_cuantitativa:
print("La columna {} es: cuantitativa".format(i))
```
Procedemos a identificar el tipo de variable cuantitativa:
* discreta - Son aquellas que obedecen a una cantidad numérica exacta, que puede tomar **únicamente valores enteros y que sólo puede tomar valores dentro de un conjunto definido.**
* continua - Son variables numéricas que permiten **expresiones decimales porque pueden tomar cualquier valor real dentro de un intervalo** (es decir, hay infinitas posibilidades).
```
variables_cuanti.head()
# Separamos las culumnas por discretas y continuas
var_cuanti_discret = ['hombres','mujeres','mayores','menores','p12_64','p65mas','ocupados','percep_ing','perc_ocupa']
for i in var_cuanti_discret:
print("La columna {} es una vaiable continua discreta".format(i))
var_cuanti_conti = variables_cuanti.drop(var_cuanti_discret , axis=1)
cuanti_cont_list = var_cuanti_conti.columns
for i in cuanti_cont_list:
print("La variiable {} es cuantitativa continua.".format(i))
```
---
## Gráficos de las frecuencias absolutas de las variables categoricas.
```
import matplotlib.pyplot as plt
import seaborn as sns
var_cuali_l =['tam_loc',
'est_socio',
'clase_hog',
'sexo_jefe',
'educa_jefe']
var_cuali_l
fig, ax = plt.subplots(2, 3, figsize=(20,10))
for variable, subplot in zip(var_cuali_l, ax.flatten()):
sns.countplot(data[variable], ax=subplot)
for label in subplot.get_xticklabels():
label.set_rotation(90)
var_cuali
def make_autopct(values):
def my_autopct(pct):
total = sum(values)
val = int(round(pct*total/100.0))
return '{p:.2f}% ({v:d})'.format(p=pct,v=val)
return my_autopct
colors = ['#C70039','#FF5733','#FFC300','#DAF7A6']
plot = data['est_socio'].value_counts().plot(kind='pie', autopct=make_autopct(data['est_socio']),
figsize=(7, 7),colors=colors)
_ = plt.title('Clasificacion de viviendas')
plot = data['educa_jefe'].value_counts().plot(kind='pie', autopct=make_autopct(data['educa_jefe']),
figsize=(9, 9))
_ = plt.title('Educacion del jefe del hogar')
plt.show()
plot = data['sexo_jefe'].value_counts().plot(kind='pie', autopct=make_autopct(data['sexo_jefe']),
figsize=(7, 7),colors=['green','pink'])
_ = plt.title('Sexo del jefe del hogar')
plt.show()
# plot = data['tam_loc'].value_counts().plot(kind='pie', autopct=make_autopct(data['tam_loc']),
figsize=(7, 7),colors=colors)
# _ = plt.title('Clasificacion de viviendas')
```
---
## Variables Cuantitativas.
## **Variables Cuantitativas Discretas:**
```
sns.set()
data[var_cuanti_discret].hist(bins=15, figsize=(15, 6), layout=(2, 5));
```
## **Variables Cuantitativas Continuas**
```
data[var_cuanti_conti]
grafica_1_cont = ['ing_cor'
,'ingtrab'
,'trabajo'
,'sueldos'
,'horas_extr'
,'comisiones'
,'aguinaldo'
,'indemtrab'
,'otra_rem'
,'remu_espec']
grafica_2_cont = ['negocio',
'noagrop'
,'industria'
,'comercio'
,'servicios'
,'agrope',
'agricolas'
,'pecuarios'
,'reproducc'
,'pesca'
]
grafica_3_cont = ['otros_trab'
,'rentas'
,'utilidad'
,'arrenda'
,'transfer'
,'jubilacion'
,'becas'
,'donativos'
,'remesas'
,'bene_gob']
grafica_4_cont = ['transf_gas',
'percep_tot',
'retiro_inv'
,'prestamos'
,'otras_perc'
,'ero_nm_viv'
,'ero_nm_hog'
,'erogac_tot'
,'cuota_viv'
,'mater_serv']
fig, ax = plt.subplots(2, 5, figsize=(20,10))
for variable, subplot in zip(grafica_1_cont, ax.flatten()):
sns.kdeplot(data[variable], ax=subplot,color='orange')
for label in subplot.get_xticklabels():
label.set_rotation(90)
ig, ax = plt.subplots(2, 5, figsize=(20,10))
for variable, subplot in zip(grafica_2_cont, ax.flatten()):
sns.kdeplot(data[variable], ax=subplot,color='red')
for label in subplot.get_xticklabels():
label.set_rotation(90)
ig, ax = plt.subplots(2, 5, figsize=(20,10))
for variable, subplot in zip(grafica_3_cont, ax.flatten()):
sns.kdeplot(data[variable], ax=subplot,color='blue')
for label in subplot.get_xticklabels():
label.set_rotation(90)
ig, ax = plt.subplots(2, 5, figsize=(20,10))
for variable, subplot in zip(grafica_4_cont, ax.flatten()):
sns.kdeplot(data[variable], ax=subplot,color='purple')
for label in subplot.get_xticklabels():
label.set_rotation(90)
```
## RElaciones entre categoricos y numericos
```
fig, ax = plt.subplots(2, 3, figsize=(25, 15))
for var, subplot in zip(var_cuali_l, ax.flatten()):
sns.boxplot(x=var, y='sueldos', data=data, ax=subplot)
```
|
github_jupyter
|
import pandas as pd
data = pd.read_csv('/content/conjunto_de_datos_concentradohogar_enigh_2018_ns.csv')
data.sample(n=25)
#procedemos a ver los nobres de las columas
data.info(verbose = True)
# seleccionamos las columnas que son de tipo cualitativo y cuantiativo
variables_cuali = data.iloc[:,0:12]
variables_cuanti = data.iloc[:,13:]
var_cuanti = variables_cuanti.columns
# Añdimos las columnas cuantitativas que hacen falta
variables_cuanti_2 = data[['est_dis','upm','factor']]
var_cuantitativa = pd.concat([variables_cuanti,variables_cuanti_2],axis='columns')
# eliminamos algunas columnas cuantiativas dentro de las cualitativas
variables_cuali.drop(['est_dis','upm','factor'], axis='columns',inplace=True)
var_cuali = variables_cuali.columns
# imprimimos las columnas cualitativas
for i in var_cuali:
print("La columna {} es: cualitativa ".format(i))
for i in var_cuantitativa:
print("La columna {} es: cuantitativa".format(i))
variables_cuanti.head()
# Separamos las culumnas por discretas y continuas
var_cuanti_discret = ['hombres','mujeres','mayores','menores','p12_64','p65mas','ocupados','percep_ing','perc_ocupa']
for i in var_cuanti_discret:
print("La columna {} es una vaiable continua discreta".format(i))
var_cuanti_conti = variables_cuanti.drop(var_cuanti_discret , axis=1)
cuanti_cont_list = var_cuanti_conti.columns
for i in cuanti_cont_list:
print("La variiable {} es cuantitativa continua.".format(i))
import matplotlib.pyplot as plt
import seaborn as sns
var_cuali_l =['tam_loc',
'est_socio',
'clase_hog',
'sexo_jefe',
'educa_jefe']
var_cuali_l
fig, ax = plt.subplots(2, 3, figsize=(20,10))
for variable, subplot in zip(var_cuali_l, ax.flatten()):
sns.countplot(data[variable], ax=subplot)
for label in subplot.get_xticklabels():
label.set_rotation(90)
var_cuali
def make_autopct(values):
def my_autopct(pct):
total = sum(values)
val = int(round(pct*total/100.0))
return '{p:.2f}% ({v:d})'.format(p=pct,v=val)
return my_autopct
colors = ['#C70039','#FF5733','#FFC300','#DAF7A6']
plot = data['est_socio'].value_counts().plot(kind='pie', autopct=make_autopct(data['est_socio']),
figsize=(7, 7),colors=colors)
_ = plt.title('Clasificacion de viviendas')
plot = data['educa_jefe'].value_counts().plot(kind='pie', autopct=make_autopct(data['educa_jefe']),
figsize=(9, 9))
_ = plt.title('Educacion del jefe del hogar')
plt.show()
plot = data['sexo_jefe'].value_counts().plot(kind='pie', autopct=make_autopct(data['sexo_jefe']),
figsize=(7, 7),colors=['green','pink'])
_ = plt.title('Sexo del jefe del hogar')
plt.show()
# plot = data['tam_loc'].value_counts().plot(kind='pie', autopct=make_autopct(data['tam_loc']),
figsize=(7, 7),colors=colors)
# _ = plt.title('Clasificacion de viviendas')
sns.set()
data[var_cuanti_discret].hist(bins=15, figsize=(15, 6), layout=(2, 5));
data[var_cuanti_conti]
grafica_1_cont = ['ing_cor'
,'ingtrab'
,'trabajo'
,'sueldos'
,'horas_extr'
,'comisiones'
,'aguinaldo'
,'indemtrab'
,'otra_rem'
,'remu_espec']
grafica_2_cont = ['negocio',
'noagrop'
,'industria'
,'comercio'
,'servicios'
,'agrope',
'agricolas'
,'pecuarios'
,'reproducc'
,'pesca'
]
grafica_3_cont = ['otros_trab'
,'rentas'
,'utilidad'
,'arrenda'
,'transfer'
,'jubilacion'
,'becas'
,'donativos'
,'remesas'
,'bene_gob']
grafica_4_cont = ['transf_gas',
'percep_tot',
'retiro_inv'
,'prestamos'
,'otras_perc'
,'ero_nm_viv'
,'ero_nm_hog'
,'erogac_tot'
,'cuota_viv'
,'mater_serv']
fig, ax = plt.subplots(2, 5, figsize=(20,10))
for variable, subplot in zip(grafica_1_cont, ax.flatten()):
sns.kdeplot(data[variable], ax=subplot,color='orange')
for label in subplot.get_xticklabels():
label.set_rotation(90)
ig, ax = plt.subplots(2, 5, figsize=(20,10))
for variable, subplot in zip(grafica_2_cont, ax.flatten()):
sns.kdeplot(data[variable], ax=subplot,color='red')
for label in subplot.get_xticklabels():
label.set_rotation(90)
ig, ax = plt.subplots(2, 5, figsize=(20,10))
for variable, subplot in zip(grafica_3_cont, ax.flatten()):
sns.kdeplot(data[variable], ax=subplot,color='blue')
for label in subplot.get_xticklabels():
label.set_rotation(90)
ig, ax = plt.subplots(2, 5, figsize=(20,10))
for variable, subplot in zip(grafica_4_cont, ax.flatten()):
sns.kdeplot(data[variable], ax=subplot,color='purple')
for label in subplot.get_xticklabels():
label.set_rotation(90)
fig, ax = plt.subplots(2, 3, figsize=(25, 15))
for var, subplot in zip(var_cuali_l, ax.flatten()):
sns.boxplot(x=var, y='sueldos', data=data, ax=subplot)
| 0.253214 | 0.88573 |
# Multiple models comparison
This notebook will run three forecasting algorithms on the same dataset and compare their performances.
The algorithms are:
- Prophet
- ETS
- DeepAR+
## Setup
```
import boto3
from time import sleep
import pandas as pd
import seaborn as sns
import pprint
pp = pprint.PrettyPrinter(indent=2) # Better display for dictionaries
```
The line below will retrieve your shared variables from the first notebook.
```
%store -r
```
The last part of the setup process is to validate that your account can communicate with Amazon Forecast, the cell below does just that.
```
forecast = boto3.client(service_name='forecast')
forecastquery = boto3.client(service_name='forecastquery')
```
## Create the predictors
The first step is to create a dictionary where to store useful information about the algorithms: their name, ARN and eventually their performance metrics.
```
algos = ['Prophet', 'ETS', 'Deep_AR_Plus']
predictors = {a:{} for a in algos}
for p in predictors:
predictors[p]['predictor_name'] = project + '_' + p + '_algo'
predictors[p]['algorithm_arn'] = 'arn:aws:forecast:::algorithm/' + p
pp.pprint(predictors)
```
Here we also define our forecast horizon: the number of time points to be predicted in the future. For weekly data, a value of 12 means 12 weeks. Our example is hourly data, we try forecast the next day, so we can set to 24.
```
forecastHorizon = 24
```
The following function actually creates the predictor as specified by several parameters. We will call this function once for each of the 3 algorithms.
```
def create_predictor_response(pred_name, algo_arn, forecast_horizon):
response=forecast.create_predictor(PredictorName=pred_name,
AlgorithmArn=algo_arn,
ForecastHorizon=forecast_horizon,
PerformAutoML= False,
PerformHPO=False,
EvaluationParameters= {"NumberOfBacktestWindows": 1,
"BackTestWindowOffset": 24},
InputDataConfig= {"DatasetGroupArn": datasetGroupArn},
FeaturizationConfig= {"ForecastFrequency": "H",
"Featurizations":
[
{"AttributeName": "target_value",
"FeaturizationPipeline":
[
{"FeaturizationMethodName": "filling",
"FeaturizationMethodParameters":
{"frontfill": "none",
"middlefill": "zero",
"backfill": "zero"}
}
]
}
]
}
)
return response
```
For all 3 algorithms, we invoke their creation and wait until they are complete. We also store their performance in our dictionary.
```
for p in predictors.keys():
predictor_response = create_predictor_response(predictors[p]['predictor_name'], predictors[p]['algorithm_arn'], forecastHorizon)
predictorArn=predictor_response['PredictorArn']
# wait for the predictor to be actually created
print('------------------ Creating ' + p)
while True:
predictorStatus = forecast.describe_predictor(PredictorArn=predictorArn)['Status']
print(predictorStatus)
if predictorStatus != 'ACTIVE' and predictorStatus != 'CREATE_FAILED':
sleep(30)
else:
predictors[p]['predictor_arn'] = predictorArn # save it, just for reference
break
# compute and store performance metrics, then proceed with the next algorithm
predictors[p]['accuracy'] = forecast.get_accuracy_metrics(PredictorArn=predictorArn)
```
**TODO:** (Bar?)plot RMSE, 0.9-, 0.5- and 0.1-quantile LossValues for each algorithm
This is what we stored so far for DeepAR+:
```
pp.pprint(predictors['Deep_AR_Plus'])
```
## Visualize results
We use `seaborn` as it interacts well with `pandas` DataFrames.
Looping over our dictionary, we can retrieve the Root Mean Square Error (RMSE) for each predictor and plot it as a bar plot.
```
scores = pd.DataFrame(columns=['predictor', 'RMSE'])
for p in predictors:
score = predictors[p]['accuracy']['PredictorEvaluationResults'][0]['TestWindows'][0]['Metrics']['RMSE']
scores = scores.append(pd.DataFrame({'predictor':[p], 'RMSE':[score]}), ignore_index=True)
fig = sns.barplot(data=scores, x='predictor', y='RMSE').set_title('Root Mean Square Error')
```
|
github_jupyter
|
import boto3
from time import sleep
import pandas as pd
import seaborn as sns
import pprint
pp = pprint.PrettyPrinter(indent=2) # Better display for dictionaries
%store -r
forecast = boto3.client(service_name='forecast')
forecastquery = boto3.client(service_name='forecastquery')
algos = ['Prophet', 'ETS', 'Deep_AR_Plus']
predictors = {a:{} for a in algos}
for p in predictors:
predictors[p]['predictor_name'] = project + '_' + p + '_algo'
predictors[p]['algorithm_arn'] = 'arn:aws:forecast:::algorithm/' + p
pp.pprint(predictors)
forecastHorizon = 24
def create_predictor_response(pred_name, algo_arn, forecast_horizon):
response=forecast.create_predictor(PredictorName=pred_name,
AlgorithmArn=algo_arn,
ForecastHorizon=forecast_horizon,
PerformAutoML= False,
PerformHPO=False,
EvaluationParameters= {"NumberOfBacktestWindows": 1,
"BackTestWindowOffset": 24},
InputDataConfig= {"DatasetGroupArn": datasetGroupArn},
FeaturizationConfig= {"ForecastFrequency": "H",
"Featurizations":
[
{"AttributeName": "target_value",
"FeaturizationPipeline":
[
{"FeaturizationMethodName": "filling",
"FeaturizationMethodParameters":
{"frontfill": "none",
"middlefill": "zero",
"backfill": "zero"}
}
]
}
]
}
)
return response
for p in predictors.keys():
predictor_response = create_predictor_response(predictors[p]['predictor_name'], predictors[p]['algorithm_arn'], forecastHorizon)
predictorArn=predictor_response['PredictorArn']
# wait for the predictor to be actually created
print('------------------ Creating ' + p)
while True:
predictorStatus = forecast.describe_predictor(PredictorArn=predictorArn)['Status']
print(predictorStatus)
if predictorStatus != 'ACTIVE' and predictorStatus != 'CREATE_FAILED':
sleep(30)
else:
predictors[p]['predictor_arn'] = predictorArn # save it, just for reference
break
# compute and store performance metrics, then proceed with the next algorithm
predictors[p]['accuracy'] = forecast.get_accuracy_metrics(PredictorArn=predictorArn)
pp.pprint(predictors['Deep_AR_Plus'])
scores = pd.DataFrame(columns=['predictor', 'RMSE'])
for p in predictors:
score = predictors[p]['accuracy']['PredictorEvaluationResults'][0]['TestWindows'][0]['Metrics']['RMSE']
scores = scores.append(pd.DataFrame({'predictor':[p], 'RMSE':[score]}), ignore_index=True)
fig = sns.barplot(data=scores, x='predictor', y='RMSE').set_title('Root Mean Square Error')
| 0.448668 | 0.926103 |
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import pickle
import math
from pyFTS.benchmarks import Measures
```
## Auxiliary Functions
```
def normalize(df):
mindf = df.min()
maxdf = df.max()
return (df-mindf)/(maxdf-mindf)
def denormalize(norm, _min, _max):
return [(n * (_max-_min)) + _min for n in norm]
def save_obj(obj, name ):
with open('results/'+ name + '.pkl', 'wb') as f:
pickle.dump(obj, f, pickle.HIGHEST_PROTOCOL)
def load_obj(name ):
with open('results/' + name + '.pkl', 'rb') as f:
return pickle.load(f)
```
## Load Dataset
```
#Set target and input variables
target_station = 'DHHL_3'
#All neighbor stations with residual correlation greater than .90
neighbor_stations_90 = ['DHHL_3', 'DHHL_4','DHHL_5','DHHL_10','DHHL_11','DHHL_9','DHHL_2', 'DHHL_6','DHHL_7','DHHL_8']
df = pd.read_pickle("df_oahu.pkl")
## Remove columns with many corrupted or missing values
df.drop(columns=['AP_1', 'AP_7'], inplace=True)
# Get data form the interval of interest
interval = ((df.index >= '2010-06') & (df.index < '2010-08'))
df = df.loc[interval]
#Normalize Data
# Save Min-Max for Denorm
min_raw = df[target_station].min()
max_raw = df[target_station].max()
# Perform Normalization
norm_df = normalize(df)
def get_final_forecast(forecast_list):
forecast_final = []
for fcst in forecast_list:
forecast_final.append(denormalize(fcst, min_raw, max_raw))
return forecast_final
```
## Create Rolling window indexes
- Training: 4 weeks
- Validation: 1 week
- Test: 1 week
Roliing daily
```
import datetime
def getRollingWindow(index):
pivot = index
train_start = pivot.strftime('%Y-%m-%d')
pivot = pivot + datetime.timedelta(days=27)
train_end = pivot.strftime('%Y-%m-%d')
pivot = pivot + datetime.timedelta(days=1)
validation_start = pivot.strftime('%Y-%m-%d')
pivot = pivot + datetime.timedelta(days=6)
validation_end = pivot.strftime('%Y-%m-%d')
pivot = pivot + datetime.timedelta(days=1)
test_start = pivot.strftime('%Y-%m-%d')
pivot = pivot + datetime.timedelta(days=6)
test_end = pivot.strftime('%Y-%m-%d')
return train_start, train_end, validation_start, validation_end, test_start, test_end
def calculate_rolling_error(cv_name, df, forecasts, order_list):
cv_results = pd.DataFrame(columns=['Split', 'RMSE', 'SMAPE', 'U'])
limit = df.index[-1].strftime('%Y-%m-%d')
test_end = ""
index = df.index[0]
for i in np.arange(len(forecasts)):
train_start, train_end, validation_start, validation_end, test_start, test_end = getRollingWindow(index)
test = df[test_start : test_end]
yhat = forecasts[i]
order = order_list[i]
rmse = Measures.rmse(test[target_station].iloc[order:], yhat[:-1])
smape = Measures.smape(test[target_station].iloc[order:], yhat[:-1])
u = Measures.UStatistic(test[target_station].iloc[order:], yhat[:-1])
res = {'Split' : index.strftime('%Y-%m-%d') ,'RMSE' : rmse, 'SMAPE' : smape, 'U' : u}
cv_results = cv_results.append(res, ignore_index=True)
cv_results.to_csv(cv_name+".csv")
index = index + datetime.timedelta(days=7)
```
## Persistence
```
def persistence_forecast(train, test, step):
predictions = []
for t in np.arange(0,len(test), step):
yhat = [test.iloc[t]] * step
predictions.extend(yhat)
return predictions
def rolling_cv_persistence(df, step):
forecasts = []
lags_list = []
limit = df.index[-1].strftime('%Y-%m-%d')
test_end = ""
index = df.index[0]
while test_end < limit :
print("Index: ", index.strftime('%Y-%m-%d'))
train_start, train_end, validation_start, validation_end, test_start, test_end = getRollingWindow(index)
index = index + datetime.timedelta(days=7)
train = df[train_start : train_end]
validation = df[validation_start : validation_end]
test = df[test_start : test_end]
# Concat train & validation for test
train = train.append(validation)
yhat = persistence_forecast(train[target_station], test[target_station], step)
lags_list.append(1)
forecasts.append(yhat)
return forecasts, lags_list
forecasts, order_list = rolling_cv_persistence(norm_df, 1)
forecasts_final = get_final_forecast(forecasts)
calculate_rolling_error("rolling_cv_oahu_raw_persistence", df, forecasts_final, order_list)
```
## SARIMA
```
from statsmodels.tsa.statespace.sarimax import SARIMAX
from itertools import product
import sys
def evaluate_SARIMA_models(test_name, train, validation, parameters_list, period_length):
sarima_results = pd.DataFrame(columns=['Order','RMSE'])
best_score, best_cfg = float("inf"), None
for param in parameters_list:
arima_order = (param[0],param[1],param[2])
sarima_order = (param[3],param[4],param[5],period_length)
print('Testing SARIMA%s %s ' % (str(arima_order),str(sarima_order)))
try:
fcst = sarima_forecast(train, validation, arima_order, sarima_order)
rmse = Measures.rmse(validation.values, fcst)
if rmse < best_score:
best_score, best_cfg = rmse, (arima_order, sarima_order)
res = {'Parameters' : str(param) ,'RMSE' : rmse}
print('SARIMA%s %s RMSE=%.3f' % (str(arima_order),str(sarima_order),rmse))
sarima_results = sarima_results.append(res, ignore_index=True)
sarima_results.to_csv(test_name+".csv")
except:
print(sys.exc_info())
print('Invalid model%s %s ' % (str(arima_order),str(sarima_order)))
continue
print('Best SARIMA(%s) RMSE=%.3f' % (best_cfg, best_score))
return best_cfg
def sarima_forecast(train, test, arima_order, sarima_order):
predictions = []
window_size = sarima_order[3] * 5
history = list(train.iloc[-window_size:])
model = SARIMAX(history, order=arima_order, seasonal_order=sarima_order,enforce_invertibility=False,enforce_stationarity=False)
model_fit = model.fit(disp=True,enforce_invertibility=False, maxiter=10)
#save the state parameter
est_params = model_fit.params
est_state = model_fit.predicted_state[:, -1]
est_state_cov = model_fit.predicted_state_cov[:, :, -1]
st = 0
for t in np.arange(1,len(test)+1,1):
obs = test.iloc[st:t].values
history.extend(obs)
history = history[-window_size:]
mod_updated = SARIMAX(history, order=arima_order, seasonal_order=sarima_order,enforce_invertibility=False,enforce_stationarity=False)
mod_updated.initialize_known(est_state, est_state_cov)
mod_frcst = mod_updated.smooth(est_params)
yhat = mod_frcst.forecast(1)
predictions.extend(yhat)
est_params = mod_frcst.params
est_state = mod_frcst.predicted_state[:, -1]
est_state_cov = mod_frcst.predicted_state_cov[:, :, -1]
st = t
return predictions
def rolling_cv_SARIMA(df, step):
# p_values = [0,1,2]
# d_values = [0,1]
# q_values = [0,1,2]
# P_values = [0,1]
# D_Values = [0,1]
# Q_Values = [0,1]
p_values = [2]
d_values = [1]
q_values = [2]
P_values = [1]
D_Values = [1]
Q_Values = [1]
parameters = product(p_values, d_values, q_values, P_values, D_Values, Q_Values)
parameters_list = list(parameters)
period_length = 61 #de 5:00 as 20:00
forecasts = []
lags_list = []
limit = df.index[-1].strftime('%Y-%m-%d')
test_end = ""
index = df.index[0]
while test_end < limit :
print("Index: ", index.strftime('%Y-%m-%d'))
train_start, train_end, validation_start, validation_end, test_start, test_end = getRollingWindow(index)
index = index + datetime.timedelta(days=7)
train = df[train_start : train_end]
validation = df[validation_start : validation_end]
test = df[test_start : test_end]
# Perform grid search
#(arima_params, sarima_params) = evaluate_SARIMA_models("nested_test_sarima_oahu", train[target_station], validation[target_station], parameters_list, period_length)
arima_params = (2, 1, 2)
sarima_params = (1, 1, 1, 61)
# Concat train & validation for test
train = train.append(validation)
yhat = sarima_forecast(train[target_station], test[target_station], arima_params, sarima_params)
lags_list.append(1)
forecasts.append(yhat)
return forecasts, lags_list
forecasts, order_list = rolling_cv_SARIMA(norm_df, 1)
forecasts_final, order_list = get_final_forecast(forecasts)
calculate_rolling_error("rolling_cv_oahu_raw_sarima", df, forecasts_final, order_list)
```
## Vector Autoregressive
```
from statsmodels.tsa.api import VAR, DynamicVAR
def evaluate_VAR_models(test_name, train, validation,target, maxlags_list):
var_results = pd.DataFrame(columns=['Order','RMSE'])
best_score, best_cfg, best_model = float("inf"), None, None
for lgs in maxlags_list:
model = VAR(train)
results = model.fit(maxlags=lgs, ic='aic')
order = results.k_ar
forecast = []
for i in range(len(validation)-order) :
forecast.extend(results.forecast(validation.values[i:i+order],1))
forecast_df = pd.DataFrame(columns=validation.columns, data=forecast)
rmse = Measures.rmse(validation[target].iloc[order:], forecast_df[target].values)
if rmse < best_score:
best_score, best_cfg, best_model = rmse, order, results
res = {'Order' : str(order) ,'RMSE' : rmse}
print('VAR (%s) RMSE=%.3f' % (str(order),rmse))
var_results = var_results.append(res, ignore_index=True)
var_results.to_csv(test_name+".csv")
print('Best VAR(%s) RMSE=%.3f' % (best_cfg, best_score))
return best_model
def var_forecast(train, test, target, order, step):
model = VAR(train.values)
results = model.fit(maxlags=order)
lag_order = results.k_ar
print("Lag order:" + str(lag_order))
forecast = []
for i in np.arange(0,len(test)-lag_order+1,step) :
forecast.extend(results.forecast(test.values[i:i+lag_order],step))
forecast_df = pd.DataFrame(columns=test.columns, data=forecast)
return forecast_df[target].values
def nested_cv_var(df, step):
maxlags_list = [1,2,4,6,8,10,20,40]
forecasts = []
order_list = []
for i in np.arange(n_folds):
train = getNestedData(df, train_inds[i])
validation = getNestedData(df, val_inds[i])
test = getNestedData(df, test_inds[i])
# Perform grid search
best_model = evaluate_VAR_models("nested_test_var_oahu", train[neighbor_stations_90], validation[neighbor_stations_90],target_station, maxlags_list)
# Concat train & validation for test
train = train.append(validation)
order = best_model.k_ar
yhat = var_forecast(train[neighbor_stations_90], test[neighbor_stations_90], target_station, order, step)
order_list.append(order)
forecasts.append(yhat)
return forecasts, order_list
def rolling_cv_var(df, step):
maxlags_list = [1,2,4,6,8,10,20,40]
forecasts = []
order_list = []
limit = df.index[-1].strftime('%Y-%m-%d')
test_end = ""
index = df.index[0]
while test_end < limit :
print("Index: ", index.strftime('%Y-%m-%d'))
train_start, train_end, validation_start, validation_end, test_start, test_end = getRollingWindow(index)
index = index + datetime.timedelta(days=7)
train = df[train_start : train_end]
validation = df[validation_start : validation_end]
test = df[test_start : test_end]
# Perform grid search
best_model = evaluate_VAR_models("nested_test_var_oahu", train[neighbor_stations_90], validation[neighbor_stations_90],target_station, maxlags_list)
# Concat train & validation for test
train = train.append(validation)
order = best_model.k_ar
yhat = var_forecast(train[neighbor_stations_90], test[neighbor_stations_90], target_station, order, step)
order_list.append(order)
forecasts.append(yhat)
return forecasts, order_list
forecasts, order_list = rolling_cv_var(norm_df, 1)
forecasts_final = get_final_forecast(forecasts)
calculate_rolling_error("rolling_cv_oahu_raw_var", df, forecasts_final, order_list)
```
## High Order FTS
```
from pyFTS.partitioners import Grid, Entropy, Util as pUtil
from pyFTS.models import hofts
def evaluate_hofts_models(test_name, train, validation, partitioners_list, order_list, partitions_list):
hofts_results = pd.DataFrame(columns=['Partitioner','Partitions','Order','RMSE'])
best_score, best_cfg, best_model = float("inf"), None, None
for _partitioner in partitioners_list:
for _order in order_list:
for npartitions in partitions_list:
fuzzy_sets = _partitioner(data=train.values, npart=npartitions)
model_simple_hofts = hofts.HighOrderFTS(order=_order)
model_simple_hofts.fit(train.values, order=_order, partitioner=fuzzy_sets)
forecast = model_simple_hofts.predict(validation.values)
rmse = Measures.rmse(validation.iloc[_order:], forecast[:-1])
if rmse < best_score:
best_score, best_cfg = rmse, (_order,npartitions,_partitioner)
best_model = model_simple_hofts
res = {'Partitioner':str(_partitioner), 'Partitions':npartitions, 'Order' : str(_order) ,'RMSE' : rmse}
print('HOFTS %s - %s - %s RMSE=%.3f' % (str(_partitioner), npartitions, str(_order),rmse))
hofts_results = hofts_results.append(res, ignore_index=True)
hofts_results.to_csv(test_name+".csv")
print('Best HOFTS(%s) RMSE=%.3f' % (best_cfg, best_score))
return best_cfg
def hofts_forecast(train_df, test_df, _order, _partitioner, _npartitions):
fuzzy_sets = _partitioner(data=train_df.values, npart=_npartitions)
model_simple_hofts = hofts.HighOrderFTS()
model_simple_hofts.fit(train_df.values, order=_order, partitioner=fuzzy_sets)
forecast = model_simple_hofts.predict(test_df.values)
return forecast
def rolling_cv_hofts(df, step):
partitioners_list = [Grid.GridPartitioner, Entropy.EntropyPartitioner]
eval_order_list = np.arange(1,3)
partitions_list = np.arange(80,100,10)
forecasts = []
order_list = []
limit = df.index[-1].strftime('%Y-%m-%d')
test_end = ""
index = df.index[0]
while test_end < limit :
print("Index: ", index.strftime('%Y-%m-%d'))
train_start, train_end, validation_start, validation_end, test_start, test_end = getRollingWindow(index)
index = index + datetime.timedelta(days=1)
train = df[train_start : train_end]
validation = df[validation_start : validation_end]
test = df[test_start : test_end]
# Perform grid search
(order,nparts,partitioner) = evaluate_hofts_models("nested_eval_hofts_oahu", train[target_station], validation[target_station], partitioners_list, eval_order_list, partitions_list)
# Concat train & validation for test
train = train.append(validation)
# Perform forecast
yhat = hofts_forecast(train[target_station], test[target_station], order, partitioner, nparts)
order_list.append(order)
forecasts.append(yhat)
return forecasts, order_list
forecasts, order_list = rolling_cv_hofts(norm_df, 1)
forecasts_final = get_final_forecast(forecasts)
calculate_rolling_error("rolling_cv_oahu_raw_hofts", df, forecasts_final, order_list)
```
## Conditional Variance FTS
```
from pyFTS.models.nonstationary import cvfts
from pyFTS.models.nonstationary import partitioners as nspartitioners
def evaluate_cvfts_models(test_name, train, validation, partitions_list):
cvfts_results = pd.DataFrame(columns=['Partitions','RMSE'])
best_score, best_cfg, best_model = float("inf"), None, None
for npartitions in partitions_list:
fuzzy_sets = nspartitioners.PolynomialNonStationaryPartitioner(data=train.values, part=Grid.GridPartitioner(data=train.values, npart=npartitions), degree=2)
model_cvfts = cvfts.ConditionalVarianceFTS()
model_cvfts.fit(train.values, parameters=1, partitioner=fuzzy_sets, num_batches=1000)
forecast = model_cvfts.predict(validation.values)
rmse = Measures.rmse(validation.iloc[1:], forecast[:-1])
if rmse < best_score:
best_score, best_cfg = rmse, npartitions
best_model = model_cvfts
res = {'Partitions':npartitions, 'RMSE' : rmse}
print('CVFTS %s - RMSE=%.3f' % (npartitions, rmse))
cvfts_results = cvfts_results.append(res, ignore_index=True)
cvfts_results.to_csv(test_name+".csv")
print('Best CVFTS(%s) RMSE=%.3f' % (best_cfg, best_score))
return best_cfg
def cvfts_forecast(train, test, _partitions):
fuzzy_sets = nspartitioners.PolynomialNonStationaryPartitioner(data=train.values, part=Grid.GridPartitioner(data=train.values, npart=_partitions), degree=2)
model_cvfts = cvfts.ConditionalVarianceFTS()
model_cvfts.fit(train.values, parameters=1, partitioner=fuzzy_sets, num_batches=1000)
forecast = model_cvfts.predict(test.values)
return forecast
def rolling_cv_cvfts(df, step):
partitions_list = np.arange(80,100,10)
forecasts = []
limit = df.index[-1].strftime('%Y-%m-%d')
test_end = ""
index = df.index[0]
while test_end < limit :
print("Index: ", index.strftime('%Y-%m-%d'))
train_start, train_end, validation_start, validation_end, test_start, test_end = getRollingWindow(index)
index = index + datetime.timedelta(days=7)
train = df[train_start : train_end]
validation = df[validation_start : validation_end]
test = df[test_start : test_end]
# Perform grid search
nparts = evaluate_cvfts_models("nested_eval_cvfts_oahu", train[target_station], validation[target_station], partitions_list)
# Concat train & validation for test
train = train.append(validation)
# Perform forecast
yhat = cvfts_forecast(train[target_station], test[target_station],nparts)
order_list.append(1)
forecasts.append(yhat)
return forecasts, order_list
forecasts, order_list = rolling_cv_cvfts(norm_df, 1)
forecasts_final = get_final_forecast(forecasts)
calculate_rolling_error("rolling_cv_oahu_raw_cvfts", df, forecasts_final, order_list)
```
## Clustered Multivariate FTS
```
!pip install -U git+https://github.com/cseveriano/spatio-temporal-forecasting
from models import KMeansPartitioner
from models import sthofts
def cmvfts_forecast(train_df, test_df, target, _order, npartitions):
_partitioner = KMeansPartitioner.KMeansPartitioner(data=train_df.values, npart=npartitions, batch_size=1000, init_size=npartitions*3)
model_sthofts = sthofts.SpatioTemporalHighOrderFTS(membership_threshold=0.6)
model_sthofts.fit(train_df.values, num_batches=100, order=_order, partitioner=_partitioner)
forecast = model_sthofts.predict(test_df.values)
forecast_df = pd.DataFrame(data=forecast, columns=test_df.columns)
return forecast_df[target].values
def evaluate_cmvfts_models(test_name, train, validation, order_list, partitions_list):
cmvfts_results = pd.DataFrame(columns=['Partitions','Order','RMSE'])
best_score, best_cfg = float("inf"), None
for _order in order_list:
for npartitions in partitions_list:
forecast = cmvfts_forecast(train, validation, target_station, _order, npartitions)
rmse = Measures.rmse(validation[target_station].iloc[_order:], forecast[:-1])
if rmse < best_score:
best_score, best_cfg = rmse, (_order,npartitions)
res = {'Partitions':npartitions, 'Order' : str(_order) ,'RMSE' : rmse}
print('CMVFTS %s - %s RMSE=%.3f' % (npartitions, str(_order),rmse))
cmvfts_results = cmvfts_results.append(res, ignore_index=True)
cmvfts_results.to_csv(test_name+".csv")
print('Best CMVFTS(%s) RMSE=%.3f' % (best_cfg, best_score))
return best_cfg
def rolling_cv_cmvfts(df, step):
eval_order_list = np.arange(1,3)
partitions_list = np.arange(10,100,10)
forecasts = []
order_list = []
limit = df.index[-1].strftime('%Y-%m-%d')
test_end = ""
index = df.index[0]
while test_end < limit :
print("Index: ", index.strftime('%Y-%m-%d'))
train_start, train_end, validation_start, validation_end, test_start, test_end = getRollingWindow(index)
index = index + datetime.timedelta(days=7)
train = df[train_start : train_end]
validation = df[validation_start : validation_end]
test = df[test_start : test_end]
# Perform grid search
(order,nparts) = evaluate_cmvfts_models("nested_eval_cmvfts_oahu", train[neighbor_stations_90], validation[neighbor_stations_90], eval_order_list, partitions_list)
# Concat train & validation for test
train = train.append(validation)
# Perform forecast
yhat = cmvfts_forecast(train[neighbor_stations_90], test[neighbor_stations_90], order, nparts)
order_list.append(order)
forecasts.append(yhat)
return forecasts, order_list
forecasts, order_list = rolling_cv_cmvfts(norm_df, 1)
forecasts_final = get_final_forecast(forecasts)
calculate_rolling_error("rolling_cv_oahu_raw_cmvfts", df, forecasts_final, order_list)
```
## LSTM - Multivariate
```
from keras.models import Sequential
from keras.layers import Dense
from keras.layers import LSTM
from keras.layers import Dropout
from keras.constraints import maxnorm
# convert series to supervised learning
def series_to_supervised(data, n_in=1, n_out=1, dropnan=True):
n_vars = 1 if type(data) is list else data.shape[1]
df = pd.DataFrame(data)
cols, names = list(), list()
# input sequence (t-n, ... t-1)
for i in range(n_in, 0, -1):
cols.append(df.shift(i))
names += [('var%d(t-%d)' % (j+1, i)) for j in range(n_vars)]
# forecast sequence ( t, t+1, ... t+n)
for i in range(0, n_out):
cols.append(df.shift(-i))
if i == 0:
names += [('var%d(t)' % (j+1)) for j in range(n_vars)]
else:
names += [('var%d(t+%d)' % (j+1, i)) for j in range(n_vars)]
# put it all together
agg = pd.concat(cols, axis=1)
agg.columns = names
# drop rows with NaN values
if dropnan:
agg.dropna(inplace=True)
return agg
def lstm_multi_forecast(train_df, test_df, _order, _steps, _neurons, _epochs):
nfeat = len(train_df.columns)
nlags = _order
nsteps = _steps
nobs = nlags * nfeat
train_reshaped_df = series_to_supervised(train_df, n_in=nlags, n_out=nsteps)
train_X, train_Y = train_reshaped_df.iloc[:,:nobs].values, train_reshaped_df.iloc[:,-nfeat].values
train_X = train_X.reshape((train_X.shape[0], nlags, nfeat))
test_reshaped_df = series_to_supervised(test_df, n_in=nlags, n_out=nsteps)
test_X, test_Y = test_reshaped_df.iloc[:,:nobs].values, test_reshaped_df.iloc[:,-nfeat].values
test_X = test_X.reshape((test_X.shape[0], nlags, nfeat))
# design network
model = Sequential()
model.add(LSTM(_neurons, input_shape=(train_X.shape[1], train_X.shape[2])))
model.add(Dense(1))
model.compile(loss='mae', optimizer='adam')
# fit network
model.fit(train_X, train_Y, epochs=_epochs, batch_size=1000, verbose=False, shuffle=False)
forecast = model.predict(test_X)
fcst = [f[0] for f in forecast]
return fcst
def evaluate_multivariate_lstm_models(test_name, train_df, validation_df, neurons_list, order_list, epochs_list):
lstm_results = pd.DataFrame(columns=['Neurons','Order','Epochs','RMSE'])
best_score, best_cfg = float("inf"), None
nfeat = len(train_df.columns)
nsteps = 1
for _neurons in neurons_list:
for _order in order_list:
for epochs in epochs_list:
nobs = nfeat * _order
train_reshaped_df = series_to_supervised(train_df, n_in=_order, n_out=nsteps)
train_X, train_Y = train_reshaped_df.iloc[:,:nobs].values, train_reshaped_df.iloc[:,-nfeat].values
train_X = train_X.reshape((train_X.shape[0], _order, nfeat))
val_reshaped_df = series_to_supervised(validation_df, n_in=_order, n_out=nsteps)
validation_X, validation_Y = val_reshaped_df.iloc[:,:nobs].values, val_reshaped_df.iloc[:,-nfeat].values
validation_X = validation_X.reshape((validation_X.shape[0], _order, nfeat))
# design network
model = Sequential()
model.add(LSTM(_neurons, input_shape=(train_X.shape[1], train_X.shape[2])))
model.add(Dense(1))
model.compile(loss='mae', optimizer='adam')
# fit network
history = model.fit(train_X, train_Y, epochs=epochs, batch_size=1000, verbose=False, shuffle=False)
forecast = model.predict(validation_X)
fcst = [f[0] for f in forecast]
rmse = Measures.rmse(validation_Y, fcst)
#rmse = math.sqrt(mean_squared_error(validation_Y, forecast))
params = (_neurons, _order,epochs)
if rmse < best_score:
best_score, best_cfg = rmse, params
res = {'Neurons':_neurons, 'Order':_order, 'Epochs' : epochs ,'RMSE' : rmse}
print('LSTM %s RMSE=%.3f' % (params,rmse))
lstm_results = lstm_results.append(res, ignore_index=True)
lstm_results.to_csv(test_name+".csv")
print('Best LSTM(%s) RMSE=%.3f' % (best_cfg, best_score))
return best_cfg
def rolling_cv_lstm_multi(df, step):
neurons_list = np.arange(50,110,50)
order_list = np.arange(2,4)
epochs_list = [100]
neurons_list = [5]
order_list = [2]
epochs_list = [1]
lags_list = []
forecasts = []
limit = df.index[-1].strftime('%Y-%m-%d')
test_end = ""
index = df.index[0]
while test_end < limit :
print("Index: ", index.strftime('%Y-%m-%d'))
train_start, train_end, validation_start, validation_end, test_start, test_end = getRollingWindow(index)
index = index + datetime.timedelta(days=7)
train = df[train_start : train_end]
validation = df[validation_start : validation_end]
test = df[test_start : test_end]
# Perform grid search
(_neurons, _order,epochs) = evaluate_multivariate_lstm_models("nested_eval_lstm_multi_oahu", train[neighbor_stations_90], validation[neighbor_stations_90], neurons_list, order_list, epochs_list)
# Concat train & validation for test
train = train.append(validation)
# Perform forecast
yhat = lstm_multi_forecast(train[neighbor_stations_90], test[neighbor_stations_90], _order, 1, _neurons,epochs)
yhat.append(0) #para manter o formato do vetor de metricas
lags_list.append(_order)
forecasts.append(yhat)
return forecasts, lags_list
forecasts, order_list = rolling_cv_lstm_multi(norm_df, 1)
forecasts_final = get_final_forecast(forecasts)
calculate_rolling_error("rolling_cv_oahu_raw_lstm_multi", df, forecasts_final, order_list)
```
## LSTM Univariate
```
def rolling_cv_lstm_uni(df, step):
neurons_list = np.arange(50,110,50)
order_list = np.arange(2,4)
epochs_list = [100]
neurons_list = [5]
order_list = [2]
epochs_list = [1]
lags_list = []
forecasts = []
limit = df.index[-1].strftime('%Y-%m-%d')
test_end = ""
index = df.index[0]
while test_end < limit :
print("Index: ", index.strftime('%Y-%m-%d'))
train_start, train_end, validation_start, validation_end, test_start, test_end = getRollingWindow(index)
index = index + datetime.timedelta(days=7)
train = df[train_start : train_end]
validation = df[validation_start : validation_end]
test = df[test_start : test_end]
# Perform grid search
(_neurons, _order,epochs) = evaluate_multivariate_lstm_models("nested_eval_lstm_multi_oahu", train[[target_station]], validation[[target_station]], neurons_list, order_list, epochs_list)
# Concat train & validation for test
train = train.append(validation)
# Perform forecast
yhat = lstm_multi_forecast(train[[target_station]], test[[target_station]], _order, 1, _neurons,epochs)
yhat.append(0) #para manter o formato do vetor de metricas
lags_list.append(_order)
forecasts.append(yhat)
return forecasts, lags_list
forecasts, order_list = rolling_cv_lstm_uni(norm_df, 1)
forecasts_final = get_final_forecast(forecasts)
calculate_rolling_error("rolling_cv_oahu_raw_lstm_uni", df, forecasts_final, order_list)
```
## MLP Multivariate
```
def mlp_multi_forecast(train_df, test_df, _order, _steps, _neurons, _epochs):
nfeat = len(train_df.columns)
nlags = _order
nsteps = _steps
nobs = nlags * nfeat
train_reshaped_df = series_to_supervised(train_df, n_in=nlags, n_out=nsteps)
train_X, train_Y = train_reshaped_df.iloc[:,:nobs].values, train_reshaped_df.iloc[:,-nfeat].values
test_reshaped_df = series_to_supervised(test_df, n_in=nlags, n_out=nsteps)
test_X, test_Y = test_reshaped_df.iloc[:,:nobs].values, test_reshaped_df.iloc[:,-nfeat].values
# design network
model = designMLPNetwork(_neurons,train_X.shape[1])
# fit network
model.fit(train_X, train_Y, epochs=_epochs, batch_size=1000, verbose=False, shuffle=False)
forecast = model.predict(test_X)
fcst = [f[0] for f in forecast]
return fcst
def evaluate_multivariate_mlp_models(test_name, train_df, validation_df, neurons_list, order_list, epochs_list):
lstm_results = pd.DataFrame(columns=['Neurons','Order','Epochs','RMSE'])
best_score, best_cfg = float("inf"), None
nfeat = len(train_df.columns)
nsteps = 1
for _neurons in neurons_list:
for _order in order_list:
for epochs in epochs_list:
nobs = nfeat * _order
train_reshaped_df = series_to_supervised(train_df, n_in=_order, n_out=nsteps)
train_X, train_Y = train_reshaped_df.iloc[:,:nobs].values, train_reshaped_df.iloc[:,-nfeat].values
val_reshaped_df = series_to_supervised(validation_df, n_in=_order, n_out=nsteps)
validation_X, validation_Y = val_reshaped_df.iloc[:,:nobs].values, val_reshaped_df.iloc[:,-nfeat].values
model = designMLPNetwork(_neurons,train_X.shape[1])
# fit network
history = model.fit(train_X, train_Y, epochs=epochs, batch_size=1000, verbose=False, shuffle=False)
forecast = model.predict(validation_X)
fcst = [f[0] for f in forecast]
rmse = Measures.rmse(validation_Y, fcst)
#rmse = math.sqrt(mean_squared_error(validation_Y, forecast))
params = (_neurons, _order,epochs)
if rmse < best_score:
best_score, best_cfg = rmse, params
res = {'Neurons':_neurons, 'Order':_order, 'Epochs' : epochs ,'RMSE' : rmse}
print('LSTM %s RMSE=%.3f' % (params,rmse))
lstm_results = lstm_results.append(res, ignore_index=True)
lstm_results.to_csv(test_name+".csv")
print('Best MLP(%s) RMSE=%.3f' % (best_cfg, best_score))
return best_cfg
def designMLPNetwork(neurons, shape):
model = Sequential()
model.add(Dense(neurons, activation='relu', input_dim=shape))
model.add(Dense(neurons, activation='relu'))
model.add(Dense(neurons, activation='relu'))
model.add(Dense(1))
model.compile(loss='mean_squared_error', optimizer='adam')
return model
def rolling_cv_mlp_multi(df, step):
neurons_list = np.arange(50,110,50)
order_list = np.arange(2,4)
epochs_list = [100]
neurons_list = [50]
order_list = [4]
epochs_list = [500]
lags_list = []
forecasts = []
limit = df.index[-1].strftime('%Y-%m-%d')
test_end = ""
index = df.index[0]
while test_end < limit :
print("Index: ", index.strftime('%Y-%m-%d'))
train_start, train_end, validation_start, validation_end, test_start, test_end = getRollingWindow(index)
index = index + datetime.timedelta(days=7)
train = df[train_start : train_end]
validation = df[validation_start : validation_end]
test = df[test_start : test_end]
# Perform grid search
(_neurons, _order,epochs) = evaluate_multivariate_mlp_models("nested_eval_mlp_multi_oahu", train[neighbor_stations_90], validation[neighbor_stations_90], neurons_list, order_list, epochs_list)
# Concat train & validation for test
train = train.append(validation)
# Perform forecast
yhat = mlp_multi_forecast(train[neighbor_stations_90], test[neighbor_stations_90], _order, 1, _neurons,epochs)
yhat.append(0) #para manter o formato do vetor de metricas
lags_list.append(_order)
forecasts.append(yhat)
return forecasts, lags_list
forecasts, order_list = rolling_cv_mlp_multi(norm_df, 1)
forecasts_final = get_final_forecast(forecasts)
calculate_rolling_error("rolling_cv_oahu_raw_mlp_multi", df, forecasts_final, order_list)
```
## MLP Univariate
```
def rolling_cv_mlp_uni(df, step):
neurons_list = [50]
order_list = [4]
epochs_list = [500]
lags_list = []
forecasts = []
limit = df.index[-1].strftime('%Y-%m-%d')
test_end = ""
index = df.index[0]
while test_end < limit :
print("Index: ", index.strftime('%Y-%m-%d'))
train_start, train_end, validation_start, validation_end, test_start, test_end = getRollingWindow(index)
index = index + datetime.timedelta(days=7)
train = df[train_start : train_end]
validation = df[validation_start : validation_end]
test = df[test_start : test_end]
# Perform grid search
(_neurons, _order,epochs) = evaluate_multivariate_mlp_models("nested_eval_mlp_uni_oahu", train[[target_station]], validation[[target_station]], neurons_list, order_list, epochs_list)
# Concat train & validation for test
train = train.append(validation)
# Perform forecast
yhat = mlp_multi_forecast(train[[target_station]], test[[target_station]], _order, 1, _neurons,epochs)
yhat.append(0) #para manter o formato do vetor de metricas
lags_list.append(_order)
forecasts.append(yhat)
return forecasts, lags_list
forecasts, order_list = rolling_cv_mlp_uni(norm_df, 1)
forecasts_final = get_final_forecast(forecasts)
calculate_rolling_error("rolling_cv_oahu_raw_mlp_uni", df, forecasts_final, order_list)
```
|
github_jupyter
|
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import pickle
import math
from pyFTS.benchmarks import Measures
def normalize(df):
mindf = df.min()
maxdf = df.max()
return (df-mindf)/(maxdf-mindf)
def denormalize(norm, _min, _max):
return [(n * (_max-_min)) + _min for n in norm]
def save_obj(obj, name ):
with open('results/'+ name + '.pkl', 'wb') as f:
pickle.dump(obj, f, pickle.HIGHEST_PROTOCOL)
def load_obj(name ):
with open('results/' + name + '.pkl', 'rb') as f:
return pickle.load(f)
#Set target and input variables
target_station = 'DHHL_3'
#All neighbor stations with residual correlation greater than .90
neighbor_stations_90 = ['DHHL_3', 'DHHL_4','DHHL_5','DHHL_10','DHHL_11','DHHL_9','DHHL_2', 'DHHL_6','DHHL_7','DHHL_8']
df = pd.read_pickle("df_oahu.pkl")
## Remove columns with many corrupted or missing values
df.drop(columns=['AP_1', 'AP_7'], inplace=True)
# Get data form the interval of interest
interval = ((df.index >= '2010-06') & (df.index < '2010-08'))
df = df.loc[interval]
#Normalize Data
# Save Min-Max for Denorm
min_raw = df[target_station].min()
max_raw = df[target_station].max()
# Perform Normalization
norm_df = normalize(df)
def get_final_forecast(forecast_list):
forecast_final = []
for fcst in forecast_list:
forecast_final.append(denormalize(fcst, min_raw, max_raw))
return forecast_final
import datetime
def getRollingWindow(index):
pivot = index
train_start = pivot.strftime('%Y-%m-%d')
pivot = pivot + datetime.timedelta(days=27)
train_end = pivot.strftime('%Y-%m-%d')
pivot = pivot + datetime.timedelta(days=1)
validation_start = pivot.strftime('%Y-%m-%d')
pivot = pivot + datetime.timedelta(days=6)
validation_end = pivot.strftime('%Y-%m-%d')
pivot = pivot + datetime.timedelta(days=1)
test_start = pivot.strftime('%Y-%m-%d')
pivot = pivot + datetime.timedelta(days=6)
test_end = pivot.strftime('%Y-%m-%d')
return train_start, train_end, validation_start, validation_end, test_start, test_end
def calculate_rolling_error(cv_name, df, forecasts, order_list):
cv_results = pd.DataFrame(columns=['Split', 'RMSE', 'SMAPE', 'U'])
limit = df.index[-1].strftime('%Y-%m-%d')
test_end = ""
index = df.index[0]
for i in np.arange(len(forecasts)):
train_start, train_end, validation_start, validation_end, test_start, test_end = getRollingWindow(index)
test = df[test_start : test_end]
yhat = forecasts[i]
order = order_list[i]
rmse = Measures.rmse(test[target_station].iloc[order:], yhat[:-1])
smape = Measures.smape(test[target_station].iloc[order:], yhat[:-1])
u = Measures.UStatistic(test[target_station].iloc[order:], yhat[:-1])
res = {'Split' : index.strftime('%Y-%m-%d') ,'RMSE' : rmse, 'SMAPE' : smape, 'U' : u}
cv_results = cv_results.append(res, ignore_index=True)
cv_results.to_csv(cv_name+".csv")
index = index + datetime.timedelta(days=7)
def persistence_forecast(train, test, step):
predictions = []
for t in np.arange(0,len(test), step):
yhat = [test.iloc[t]] * step
predictions.extend(yhat)
return predictions
def rolling_cv_persistence(df, step):
forecasts = []
lags_list = []
limit = df.index[-1].strftime('%Y-%m-%d')
test_end = ""
index = df.index[0]
while test_end < limit :
print("Index: ", index.strftime('%Y-%m-%d'))
train_start, train_end, validation_start, validation_end, test_start, test_end = getRollingWindow(index)
index = index + datetime.timedelta(days=7)
train = df[train_start : train_end]
validation = df[validation_start : validation_end]
test = df[test_start : test_end]
# Concat train & validation for test
train = train.append(validation)
yhat = persistence_forecast(train[target_station], test[target_station], step)
lags_list.append(1)
forecasts.append(yhat)
return forecasts, lags_list
forecasts, order_list = rolling_cv_persistence(norm_df, 1)
forecasts_final = get_final_forecast(forecasts)
calculate_rolling_error("rolling_cv_oahu_raw_persistence", df, forecasts_final, order_list)
from statsmodels.tsa.statespace.sarimax import SARIMAX
from itertools import product
import sys
def evaluate_SARIMA_models(test_name, train, validation, parameters_list, period_length):
sarima_results = pd.DataFrame(columns=['Order','RMSE'])
best_score, best_cfg = float("inf"), None
for param in parameters_list:
arima_order = (param[0],param[1],param[2])
sarima_order = (param[3],param[4],param[5],period_length)
print('Testing SARIMA%s %s ' % (str(arima_order),str(sarima_order)))
try:
fcst = sarima_forecast(train, validation, arima_order, sarima_order)
rmse = Measures.rmse(validation.values, fcst)
if rmse < best_score:
best_score, best_cfg = rmse, (arima_order, sarima_order)
res = {'Parameters' : str(param) ,'RMSE' : rmse}
print('SARIMA%s %s RMSE=%.3f' % (str(arima_order),str(sarima_order),rmse))
sarima_results = sarima_results.append(res, ignore_index=True)
sarima_results.to_csv(test_name+".csv")
except:
print(sys.exc_info())
print('Invalid model%s %s ' % (str(arima_order),str(sarima_order)))
continue
print('Best SARIMA(%s) RMSE=%.3f' % (best_cfg, best_score))
return best_cfg
def sarima_forecast(train, test, arima_order, sarima_order):
predictions = []
window_size = sarima_order[3] * 5
history = list(train.iloc[-window_size:])
model = SARIMAX(history, order=arima_order, seasonal_order=sarima_order,enforce_invertibility=False,enforce_stationarity=False)
model_fit = model.fit(disp=True,enforce_invertibility=False, maxiter=10)
#save the state parameter
est_params = model_fit.params
est_state = model_fit.predicted_state[:, -1]
est_state_cov = model_fit.predicted_state_cov[:, :, -1]
st = 0
for t in np.arange(1,len(test)+1,1):
obs = test.iloc[st:t].values
history.extend(obs)
history = history[-window_size:]
mod_updated = SARIMAX(history, order=arima_order, seasonal_order=sarima_order,enforce_invertibility=False,enforce_stationarity=False)
mod_updated.initialize_known(est_state, est_state_cov)
mod_frcst = mod_updated.smooth(est_params)
yhat = mod_frcst.forecast(1)
predictions.extend(yhat)
est_params = mod_frcst.params
est_state = mod_frcst.predicted_state[:, -1]
est_state_cov = mod_frcst.predicted_state_cov[:, :, -1]
st = t
return predictions
def rolling_cv_SARIMA(df, step):
# p_values = [0,1,2]
# d_values = [0,1]
# q_values = [0,1,2]
# P_values = [0,1]
# D_Values = [0,1]
# Q_Values = [0,1]
p_values = [2]
d_values = [1]
q_values = [2]
P_values = [1]
D_Values = [1]
Q_Values = [1]
parameters = product(p_values, d_values, q_values, P_values, D_Values, Q_Values)
parameters_list = list(parameters)
period_length = 61 #de 5:00 as 20:00
forecasts = []
lags_list = []
limit = df.index[-1].strftime('%Y-%m-%d')
test_end = ""
index = df.index[0]
while test_end < limit :
print("Index: ", index.strftime('%Y-%m-%d'))
train_start, train_end, validation_start, validation_end, test_start, test_end = getRollingWindow(index)
index = index + datetime.timedelta(days=7)
train = df[train_start : train_end]
validation = df[validation_start : validation_end]
test = df[test_start : test_end]
# Perform grid search
#(arima_params, sarima_params) = evaluate_SARIMA_models("nested_test_sarima_oahu", train[target_station], validation[target_station], parameters_list, period_length)
arima_params = (2, 1, 2)
sarima_params = (1, 1, 1, 61)
# Concat train & validation for test
train = train.append(validation)
yhat = sarima_forecast(train[target_station], test[target_station], arima_params, sarima_params)
lags_list.append(1)
forecasts.append(yhat)
return forecasts, lags_list
forecasts, order_list = rolling_cv_SARIMA(norm_df, 1)
forecasts_final, order_list = get_final_forecast(forecasts)
calculate_rolling_error("rolling_cv_oahu_raw_sarima", df, forecasts_final, order_list)
from statsmodels.tsa.api import VAR, DynamicVAR
def evaluate_VAR_models(test_name, train, validation,target, maxlags_list):
var_results = pd.DataFrame(columns=['Order','RMSE'])
best_score, best_cfg, best_model = float("inf"), None, None
for lgs in maxlags_list:
model = VAR(train)
results = model.fit(maxlags=lgs, ic='aic')
order = results.k_ar
forecast = []
for i in range(len(validation)-order) :
forecast.extend(results.forecast(validation.values[i:i+order],1))
forecast_df = pd.DataFrame(columns=validation.columns, data=forecast)
rmse = Measures.rmse(validation[target].iloc[order:], forecast_df[target].values)
if rmse < best_score:
best_score, best_cfg, best_model = rmse, order, results
res = {'Order' : str(order) ,'RMSE' : rmse}
print('VAR (%s) RMSE=%.3f' % (str(order),rmse))
var_results = var_results.append(res, ignore_index=True)
var_results.to_csv(test_name+".csv")
print('Best VAR(%s) RMSE=%.3f' % (best_cfg, best_score))
return best_model
def var_forecast(train, test, target, order, step):
model = VAR(train.values)
results = model.fit(maxlags=order)
lag_order = results.k_ar
print("Lag order:" + str(lag_order))
forecast = []
for i in np.arange(0,len(test)-lag_order+1,step) :
forecast.extend(results.forecast(test.values[i:i+lag_order],step))
forecast_df = pd.DataFrame(columns=test.columns, data=forecast)
return forecast_df[target].values
def nested_cv_var(df, step):
maxlags_list = [1,2,4,6,8,10,20,40]
forecasts = []
order_list = []
for i in np.arange(n_folds):
train = getNestedData(df, train_inds[i])
validation = getNestedData(df, val_inds[i])
test = getNestedData(df, test_inds[i])
# Perform grid search
best_model = evaluate_VAR_models("nested_test_var_oahu", train[neighbor_stations_90], validation[neighbor_stations_90],target_station, maxlags_list)
# Concat train & validation for test
train = train.append(validation)
order = best_model.k_ar
yhat = var_forecast(train[neighbor_stations_90], test[neighbor_stations_90], target_station, order, step)
order_list.append(order)
forecasts.append(yhat)
return forecasts, order_list
def rolling_cv_var(df, step):
maxlags_list = [1,2,4,6,8,10,20,40]
forecasts = []
order_list = []
limit = df.index[-1].strftime('%Y-%m-%d')
test_end = ""
index = df.index[0]
while test_end < limit :
print("Index: ", index.strftime('%Y-%m-%d'))
train_start, train_end, validation_start, validation_end, test_start, test_end = getRollingWindow(index)
index = index + datetime.timedelta(days=7)
train = df[train_start : train_end]
validation = df[validation_start : validation_end]
test = df[test_start : test_end]
# Perform grid search
best_model = evaluate_VAR_models("nested_test_var_oahu", train[neighbor_stations_90], validation[neighbor_stations_90],target_station, maxlags_list)
# Concat train & validation for test
train = train.append(validation)
order = best_model.k_ar
yhat = var_forecast(train[neighbor_stations_90], test[neighbor_stations_90], target_station, order, step)
order_list.append(order)
forecasts.append(yhat)
return forecasts, order_list
forecasts, order_list = rolling_cv_var(norm_df, 1)
forecasts_final = get_final_forecast(forecasts)
calculate_rolling_error("rolling_cv_oahu_raw_var", df, forecasts_final, order_list)
from pyFTS.partitioners import Grid, Entropy, Util as pUtil
from pyFTS.models import hofts
def evaluate_hofts_models(test_name, train, validation, partitioners_list, order_list, partitions_list):
hofts_results = pd.DataFrame(columns=['Partitioner','Partitions','Order','RMSE'])
best_score, best_cfg, best_model = float("inf"), None, None
for _partitioner in partitioners_list:
for _order in order_list:
for npartitions in partitions_list:
fuzzy_sets = _partitioner(data=train.values, npart=npartitions)
model_simple_hofts = hofts.HighOrderFTS(order=_order)
model_simple_hofts.fit(train.values, order=_order, partitioner=fuzzy_sets)
forecast = model_simple_hofts.predict(validation.values)
rmse = Measures.rmse(validation.iloc[_order:], forecast[:-1])
if rmse < best_score:
best_score, best_cfg = rmse, (_order,npartitions,_partitioner)
best_model = model_simple_hofts
res = {'Partitioner':str(_partitioner), 'Partitions':npartitions, 'Order' : str(_order) ,'RMSE' : rmse}
print('HOFTS %s - %s - %s RMSE=%.3f' % (str(_partitioner), npartitions, str(_order),rmse))
hofts_results = hofts_results.append(res, ignore_index=True)
hofts_results.to_csv(test_name+".csv")
print('Best HOFTS(%s) RMSE=%.3f' % (best_cfg, best_score))
return best_cfg
def hofts_forecast(train_df, test_df, _order, _partitioner, _npartitions):
fuzzy_sets = _partitioner(data=train_df.values, npart=_npartitions)
model_simple_hofts = hofts.HighOrderFTS()
model_simple_hofts.fit(train_df.values, order=_order, partitioner=fuzzy_sets)
forecast = model_simple_hofts.predict(test_df.values)
return forecast
def rolling_cv_hofts(df, step):
partitioners_list = [Grid.GridPartitioner, Entropy.EntropyPartitioner]
eval_order_list = np.arange(1,3)
partitions_list = np.arange(80,100,10)
forecasts = []
order_list = []
limit = df.index[-1].strftime('%Y-%m-%d')
test_end = ""
index = df.index[0]
while test_end < limit :
print("Index: ", index.strftime('%Y-%m-%d'))
train_start, train_end, validation_start, validation_end, test_start, test_end = getRollingWindow(index)
index = index + datetime.timedelta(days=1)
train = df[train_start : train_end]
validation = df[validation_start : validation_end]
test = df[test_start : test_end]
# Perform grid search
(order,nparts,partitioner) = evaluate_hofts_models("nested_eval_hofts_oahu", train[target_station], validation[target_station], partitioners_list, eval_order_list, partitions_list)
# Concat train & validation for test
train = train.append(validation)
# Perform forecast
yhat = hofts_forecast(train[target_station], test[target_station], order, partitioner, nparts)
order_list.append(order)
forecasts.append(yhat)
return forecasts, order_list
forecasts, order_list = rolling_cv_hofts(norm_df, 1)
forecasts_final = get_final_forecast(forecasts)
calculate_rolling_error("rolling_cv_oahu_raw_hofts", df, forecasts_final, order_list)
from pyFTS.models.nonstationary import cvfts
from pyFTS.models.nonstationary import partitioners as nspartitioners
def evaluate_cvfts_models(test_name, train, validation, partitions_list):
cvfts_results = pd.DataFrame(columns=['Partitions','RMSE'])
best_score, best_cfg, best_model = float("inf"), None, None
for npartitions in partitions_list:
fuzzy_sets = nspartitioners.PolynomialNonStationaryPartitioner(data=train.values, part=Grid.GridPartitioner(data=train.values, npart=npartitions), degree=2)
model_cvfts = cvfts.ConditionalVarianceFTS()
model_cvfts.fit(train.values, parameters=1, partitioner=fuzzy_sets, num_batches=1000)
forecast = model_cvfts.predict(validation.values)
rmse = Measures.rmse(validation.iloc[1:], forecast[:-1])
if rmse < best_score:
best_score, best_cfg = rmse, npartitions
best_model = model_cvfts
res = {'Partitions':npartitions, 'RMSE' : rmse}
print('CVFTS %s - RMSE=%.3f' % (npartitions, rmse))
cvfts_results = cvfts_results.append(res, ignore_index=True)
cvfts_results.to_csv(test_name+".csv")
print('Best CVFTS(%s) RMSE=%.3f' % (best_cfg, best_score))
return best_cfg
def cvfts_forecast(train, test, _partitions):
fuzzy_sets = nspartitioners.PolynomialNonStationaryPartitioner(data=train.values, part=Grid.GridPartitioner(data=train.values, npart=_partitions), degree=2)
model_cvfts = cvfts.ConditionalVarianceFTS()
model_cvfts.fit(train.values, parameters=1, partitioner=fuzzy_sets, num_batches=1000)
forecast = model_cvfts.predict(test.values)
return forecast
def rolling_cv_cvfts(df, step):
partitions_list = np.arange(80,100,10)
forecasts = []
limit = df.index[-1].strftime('%Y-%m-%d')
test_end = ""
index = df.index[0]
while test_end < limit :
print("Index: ", index.strftime('%Y-%m-%d'))
train_start, train_end, validation_start, validation_end, test_start, test_end = getRollingWindow(index)
index = index + datetime.timedelta(days=7)
train = df[train_start : train_end]
validation = df[validation_start : validation_end]
test = df[test_start : test_end]
# Perform grid search
nparts = evaluate_cvfts_models("nested_eval_cvfts_oahu", train[target_station], validation[target_station], partitions_list)
# Concat train & validation for test
train = train.append(validation)
# Perform forecast
yhat = cvfts_forecast(train[target_station], test[target_station],nparts)
order_list.append(1)
forecasts.append(yhat)
return forecasts, order_list
forecasts, order_list = rolling_cv_cvfts(norm_df, 1)
forecasts_final = get_final_forecast(forecasts)
calculate_rolling_error("rolling_cv_oahu_raw_cvfts", df, forecasts_final, order_list)
!pip install -U git+https://github.com/cseveriano/spatio-temporal-forecasting
from models import KMeansPartitioner
from models import sthofts
def cmvfts_forecast(train_df, test_df, target, _order, npartitions):
_partitioner = KMeansPartitioner.KMeansPartitioner(data=train_df.values, npart=npartitions, batch_size=1000, init_size=npartitions*3)
model_sthofts = sthofts.SpatioTemporalHighOrderFTS(membership_threshold=0.6)
model_sthofts.fit(train_df.values, num_batches=100, order=_order, partitioner=_partitioner)
forecast = model_sthofts.predict(test_df.values)
forecast_df = pd.DataFrame(data=forecast, columns=test_df.columns)
return forecast_df[target].values
def evaluate_cmvfts_models(test_name, train, validation, order_list, partitions_list):
cmvfts_results = pd.DataFrame(columns=['Partitions','Order','RMSE'])
best_score, best_cfg = float("inf"), None
for _order in order_list:
for npartitions in partitions_list:
forecast = cmvfts_forecast(train, validation, target_station, _order, npartitions)
rmse = Measures.rmse(validation[target_station].iloc[_order:], forecast[:-1])
if rmse < best_score:
best_score, best_cfg = rmse, (_order,npartitions)
res = {'Partitions':npartitions, 'Order' : str(_order) ,'RMSE' : rmse}
print('CMVFTS %s - %s RMSE=%.3f' % (npartitions, str(_order),rmse))
cmvfts_results = cmvfts_results.append(res, ignore_index=True)
cmvfts_results.to_csv(test_name+".csv")
print('Best CMVFTS(%s) RMSE=%.3f' % (best_cfg, best_score))
return best_cfg
def rolling_cv_cmvfts(df, step):
eval_order_list = np.arange(1,3)
partitions_list = np.arange(10,100,10)
forecasts = []
order_list = []
limit = df.index[-1].strftime('%Y-%m-%d')
test_end = ""
index = df.index[0]
while test_end < limit :
print("Index: ", index.strftime('%Y-%m-%d'))
train_start, train_end, validation_start, validation_end, test_start, test_end = getRollingWindow(index)
index = index + datetime.timedelta(days=7)
train = df[train_start : train_end]
validation = df[validation_start : validation_end]
test = df[test_start : test_end]
# Perform grid search
(order,nparts) = evaluate_cmvfts_models("nested_eval_cmvfts_oahu", train[neighbor_stations_90], validation[neighbor_stations_90], eval_order_list, partitions_list)
# Concat train & validation for test
train = train.append(validation)
# Perform forecast
yhat = cmvfts_forecast(train[neighbor_stations_90], test[neighbor_stations_90], order, nparts)
order_list.append(order)
forecasts.append(yhat)
return forecasts, order_list
forecasts, order_list = rolling_cv_cmvfts(norm_df, 1)
forecasts_final = get_final_forecast(forecasts)
calculate_rolling_error("rolling_cv_oahu_raw_cmvfts", df, forecasts_final, order_list)
from keras.models import Sequential
from keras.layers import Dense
from keras.layers import LSTM
from keras.layers import Dropout
from keras.constraints import maxnorm
# convert series to supervised learning
def series_to_supervised(data, n_in=1, n_out=1, dropnan=True):
n_vars = 1 if type(data) is list else data.shape[1]
df = pd.DataFrame(data)
cols, names = list(), list()
# input sequence (t-n, ... t-1)
for i in range(n_in, 0, -1):
cols.append(df.shift(i))
names += [('var%d(t-%d)' % (j+1, i)) for j in range(n_vars)]
# forecast sequence ( t, t+1, ... t+n)
for i in range(0, n_out):
cols.append(df.shift(-i))
if i == 0:
names += [('var%d(t)' % (j+1)) for j in range(n_vars)]
else:
names += [('var%d(t+%d)' % (j+1, i)) for j in range(n_vars)]
# put it all together
agg = pd.concat(cols, axis=1)
agg.columns = names
# drop rows with NaN values
if dropnan:
agg.dropna(inplace=True)
return agg
def lstm_multi_forecast(train_df, test_df, _order, _steps, _neurons, _epochs):
nfeat = len(train_df.columns)
nlags = _order
nsteps = _steps
nobs = nlags * nfeat
train_reshaped_df = series_to_supervised(train_df, n_in=nlags, n_out=nsteps)
train_X, train_Y = train_reshaped_df.iloc[:,:nobs].values, train_reshaped_df.iloc[:,-nfeat].values
train_X = train_X.reshape((train_X.shape[0], nlags, nfeat))
test_reshaped_df = series_to_supervised(test_df, n_in=nlags, n_out=nsteps)
test_X, test_Y = test_reshaped_df.iloc[:,:nobs].values, test_reshaped_df.iloc[:,-nfeat].values
test_X = test_X.reshape((test_X.shape[0], nlags, nfeat))
# design network
model = Sequential()
model.add(LSTM(_neurons, input_shape=(train_X.shape[1], train_X.shape[2])))
model.add(Dense(1))
model.compile(loss='mae', optimizer='adam')
# fit network
model.fit(train_X, train_Y, epochs=_epochs, batch_size=1000, verbose=False, shuffle=False)
forecast = model.predict(test_X)
fcst = [f[0] for f in forecast]
return fcst
def evaluate_multivariate_lstm_models(test_name, train_df, validation_df, neurons_list, order_list, epochs_list):
lstm_results = pd.DataFrame(columns=['Neurons','Order','Epochs','RMSE'])
best_score, best_cfg = float("inf"), None
nfeat = len(train_df.columns)
nsteps = 1
for _neurons in neurons_list:
for _order in order_list:
for epochs in epochs_list:
nobs = nfeat * _order
train_reshaped_df = series_to_supervised(train_df, n_in=_order, n_out=nsteps)
train_X, train_Y = train_reshaped_df.iloc[:,:nobs].values, train_reshaped_df.iloc[:,-nfeat].values
train_X = train_X.reshape((train_X.shape[0], _order, nfeat))
val_reshaped_df = series_to_supervised(validation_df, n_in=_order, n_out=nsteps)
validation_X, validation_Y = val_reshaped_df.iloc[:,:nobs].values, val_reshaped_df.iloc[:,-nfeat].values
validation_X = validation_X.reshape((validation_X.shape[0], _order, nfeat))
# design network
model = Sequential()
model.add(LSTM(_neurons, input_shape=(train_X.shape[1], train_X.shape[2])))
model.add(Dense(1))
model.compile(loss='mae', optimizer='adam')
# fit network
history = model.fit(train_X, train_Y, epochs=epochs, batch_size=1000, verbose=False, shuffle=False)
forecast = model.predict(validation_X)
fcst = [f[0] for f in forecast]
rmse = Measures.rmse(validation_Y, fcst)
#rmse = math.sqrt(mean_squared_error(validation_Y, forecast))
params = (_neurons, _order,epochs)
if rmse < best_score:
best_score, best_cfg = rmse, params
res = {'Neurons':_neurons, 'Order':_order, 'Epochs' : epochs ,'RMSE' : rmse}
print('LSTM %s RMSE=%.3f' % (params,rmse))
lstm_results = lstm_results.append(res, ignore_index=True)
lstm_results.to_csv(test_name+".csv")
print('Best LSTM(%s) RMSE=%.3f' % (best_cfg, best_score))
return best_cfg
def rolling_cv_lstm_multi(df, step):
neurons_list = np.arange(50,110,50)
order_list = np.arange(2,4)
epochs_list = [100]
neurons_list = [5]
order_list = [2]
epochs_list = [1]
lags_list = []
forecasts = []
limit = df.index[-1].strftime('%Y-%m-%d')
test_end = ""
index = df.index[0]
while test_end < limit :
print("Index: ", index.strftime('%Y-%m-%d'))
train_start, train_end, validation_start, validation_end, test_start, test_end = getRollingWindow(index)
index = index + datetime.timedelta(days=7)
train = df[train_start : train_end]
validation = df[validation_start : validation_end]
test = df[test_start : test_end]
# Perform grid search
(_neurons, _order,epochs) = evaluate_multivariate_lstm_models("nested_eval_lstm_multi_oahu", train[neighbor_stations_90], validation[neighbor_stations_90], neurons_list, order_list, epochs_list)
# Concat train & validation for test
train = train.append(validation)
# Perform forecast
yhat = lstm_multi_forecast(train[neighbor_stations_90], test[neighbor_stations_90], _order, 1, _neurons,epochs)
yhat.append(0) #para manter o formato do vetor de metricas
lags_list.append(_order)
forecasts.append(yhat)
return forecasts, lags_list
forecasts, order_list = rolling_cv_lstm_multi(norm_df, 1)
forecasts_final = get_final_forecast(forecasts)
calculate_rolling_error("rolling_cv_oahu_raw_lstm_multi", df, forecasts_final, order_list)
def rolling_cv_lstm_uni(df, step):
neurons_list = np.arange(50,110,50)
order_list = np.arange(2,4)
epochs_list = [100]
neurons_list = [5]
order_list = [2]
epochs_list = [1]
lags_list = []
forecasts = []
limit = df.index[-1].strftime('%Y-%m-%d')
test_end = ""
index = df.index[0]
while test_end < limit :
print("Index: ", index.strftime('%Y-%m-%d'))
train_start, train_end, validation_start, validation_end, test_start, test_end = getRollingWindow(index)
index = index + datetime.timedelta(days=7)
train = df[train_start : train_end]
validation = df[validation_start : validation_end]
test = df[test_start : test_end]
# Perform grid search
(_neurons, _order,epochs) = evaluate_multivariate_lstm_models("nested_eval_lstm_multi_oahu", train[[target_station]], validation[[target_station]], neurons_list, order_list, epochs_list)
# Concat train & validation for test
train = train.append(validation)
# Perform forecast
yhat = lstm_multi_forecast(train[[target_station]], test[[target_station]], _order, 1, _neurons,epochs)
yhat.append(0) #para manter o formato do vetor de metricas
lags_list.append(_order)
forecasts.append(yhat)
return forecasts, lags_list
forecasts, order_list = rolling_cv_lstm_uni(norm_df, 1)
forecasts_final = get_final_forecast(forecasts)
calculate_rolling_error("rolling_cv_oahu_raw_lstm_uni", df, forecasts_final, order_list)
def mlp_multi_forecast(train_df, test_df, _order, _steps, _neurons, _epochs):
nfeat = len(train_df.columns)
nlags = _order
nsteps = _steps
nobs = nlags * nfeat
train_reshaped_df = series_to_supervised(train_df, n_in=nlags, n_out=nsteps)
train_X, train_Y = train_reshaped_df.iloc[:,:nobs].values, train_reshaped_df.iloc[:,-nfeat].values
test_reshaped_df = series_to_supervised(test_df, n_in=nlags, n_out=nsteps)
test_X, test_Y = test_reshaped_df.iloc[:,:nobs].values, test_reshaped_df.iloc[:,-nfeat].values
# design network
model = designMLPNetwork(_neurons,train_X.shape[1])
# fit network
model.fit(train_X, train_Y, epochs=_epochs, batch_size=1000, verbose=False, shuffle=False)
forecast = model.predict(test_X)
fcst = [f[0] for f in forecast]
return fcst
def evaluate_multivariate_mlp_models(test_name, train_df, validation_df, neurons_list, order_list, epochs_list):
lstm_results = pd.DataFrame(columns=['Neurons','Order','Epochs','RMSE'])
best_score, best_cfg = float("inf"), None
nfeat = len(train_df.columns)
nsteps = 1
for _neurons in neurons_list:
for _order in order_list:
for epochs in epochs_list:
nobs = nfeat * _order
train_reshaped_df = series_to_supervised(train_df, n_in=_order, n_out=nsteps)
train_X, train_Y = train_reshaped_df.iloc[:,:nobs].values, train_reshaped_df.iloc[:,-nfeat].values
val_reshaped_df = series_to_supervised(validation_df, n_in=_order, n_out=nsteps)
validation_X, validation_Y = val_reshaped_df.iloc[:,:nobs].values, val_reshaped_df.iloc[:,-nfeat].values
model = designMLPNetwork(_neurons,train_X.shape[1])
# fit network
history = model.fit(train_X, train_Y, epochs=epochs, batch_size=1000, verbose=False, shuffle=False)
forecast = model.predict(validation_X)
fcst = [f[0] for f in forecast]
rmse = Measures.rmse(validation_Y, fcst)
#rmse = math.sqrt(mean_squared_error(validation_Y, forecast))
params = (_neurons, _order,epochs)
if rmse < best_score:
best_score, best_cfg = rmse, params
res = {'Neurons':_neurons, 'Order':_order, 'Epochs' : epochs ,'RMSE' : rmse}
print('LSTM %s RMSE=%.3f' % (params,rmse))
lstm_results = lstm_results.append(res, ignore_index=True)
lstm_results.to_csv(test_name+".csv")
print('Best MLP(%s) RMSE=%.3f' % (best_cfg, best_score))
return best_cfg
def designMLPNetwork(neurons, shape):
model = Sequential()
model.add(Dense(neurons, activation='relu', input_dim=shape))
model.add(Dense(neurons, activation='relu'))
model.add(Dense(neurons, activation='relu'))
model.add(Dense(1))
model.compile(loss='mean_squared_error', optimizer='adam')
return model
def rolling_cv_mlp_multi(df, step):
neurons_list = np.arange(50,110,50)
order_list = np.arange(2,4)
epochs_list = [100]
neurons_list = [50]
order_list = [4]
epochs_list = [500]
lags_list = []
forecasts = []
limit = df.index[-1].strftime('%Y-%m-%d')
test_end = ""
index = df.index[0]
while test_end < limit :
print("Index: ", index.strftime('%Y-%m-%d'))
train_start, train_end, validation_start, validation_end, test_start, test_end = getRollingWindow(index)
index = index + datetime.timedelta(days=7)
train = df[train_start : train_end]
validation = df[validation_start : validation_end]
test = df[test_start : test_end]
# Perform grid search
(_neurons, _order,epochs) = evaluate_multivariate_mlp_models("nested_eval_mlp_multi_oahu", train[neighbor_stations_90], validation[neighbor_stations_90], neurons_list, order_list, epochs_list)
# Concat train & validation for test
train = train.append(validation)
# Perform forecast
yhat = mlp_multi_forecast(train[neighbor_stations_90], test[neighbor_stations_90], _order, 1, _neurons,epochs)
yhat.append(0) #para manter o formato do vetor de metricas
lags_list.append(_order)
forecasts.append(yhat)
return forecasts, lags_list
forecasts, order_list = rolling_cv_mlp_multi(norm_df, 1)
forecasts_final = get_final_forecast(forecasts)
calculate_rolling_error("rolling_cv_oahu_raw_mlp_multi", df, forecasts_final, order_list)
def rolling_cv_mlp_uni(df, step):
neurons_list = [50]
order_list = [4]
epochs_list = [500]
lags_list = []
forecasts = []
limit = df.index[-1].strftime('%Y-%m-%d')
test_end = ""
index = df.index[0]
while test_end < limit :
print("Index: ", index.strftime('%Y-%m-%d'))
train_start, train_end, validation_start, validation_end, test_start, test_end = getRollingWindow(index)
index = index + datetime.timedelta(days=7)
train = df[train_start : train_end]
validation = df[validation_start : validation_end]
test = df[test_start : test_end]
# Perform grid search
(_neurons, _order,epochs) = evaluate_multivariate_mlp_models("nested_eval_mlp_uni_oahu", train[[target_station]], validation[[target_station]], neurons_list, order_list, epochs_list)
# Concat train & validation for test
train = train.append(validation)
# Perform forecast
yhat = mlp_multi_forecast(train[[target_station]], test[[target_station]], _order, 1, _neurons,epochs)
yhat.append(0) #para manter o formato do vetor de metricas
lags_list.append(_order)
forecasts.append(yhat)
return forecasts, lags_list
forecasts, order_list = rolling_cv_mlp_uni(norm_df, 1)
forecasts_final = get_final_forecast(forecasts)
calculate_rolling_error("rolling_cv_oahu_raw_mlp_uni", df, forecasts_final, order_list)
| 0.344333 | 0.611063 |
```
import torch
import torch.nn as nn
import torch.distributions as td
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.animation as animation
```
## Environment
Here, we are going to use a simple environment to test our model. Suppose we control a particle (represented by white rectangular) that moves in a 2D plane. We can directly control the velocity of the particle along $x$ and $y$ axis. The task is to control the particle moving towards a goal position (represented by red rectangular) at the center of the plane (see video below).
<img src="./particle_sys_gif.gif" width=200 height=200 />
Now, assume we can not directly observe the position of the particle and the goal, but we can access to some visual observation that is represented as images (see figure below).
<img src="./particle_sys.png" width=150 height=150 />
Now we are going to learn a state space model of this environment. Given the learnt model, we will be able to forward simulate trajectoris in latent space and plan some actions that minimize the distance to the goal position.
```
class ParticleEnv:
def __init__(self):
self.state = np.array([0, 0])
self.goal_state = np.array([0.5, 0.5])
self.action_scale = 0.025
self.step_count = 0
def reset(self):
self.step_count = 0
self.state = 0.8 * (np.random.rand(2) - 0.5) + 0.5
reward = -np.sqrt(np.square(self.state - self.goal_state).sum())
state_image = self.generate_image()
# potential_goal_states = [[0.0, 0.0], [0.9, 0.9], [0.9, 0.0], [0.0, 0.9]]
# self.goal_state = np.array(potential_goal_states[random.choice([0, 1, 2, 3])])
return self.state.copy(), state_image, reward
def step(self, action):
self.state += action * self.action_scale
self.state = np.clip(self.state, a_min=0.0, a_max=1.0)
reward = -np.sqrt(np.square(self.state - self.goal_state).sum())
if np.sqrt(np.square(self.state - self.goal_state).sum()) < 0.01:
done = True
else:
done = False
state_image = self.generate_image()
self.step_count += 1
if self.step_count > 10:
done = True
return self.state.copy(), state_image, reward, done
def generate_image(self):
resolution = 32
radius = 3
image_canvas = np.zeros(shape=[3, resolution, resolution])
pixel_x = int(self.state[0].item() * (resolution - 1))
pixel_y = int(self.state[1].item() * (resolution - 1))
for i in range(radius):
for j in range(radius):
image_canvas[:, np.clip(pixel_x + i, 0, resolution - 1), np.clip(pixel_y + j, 0, resolution - 1)] = 1.0
image_canvas[:, np.clip(pixel_x - i, 0, resolution - 1), np.clip(pixel_y - j, 0, resolution - 1)] = 1.0
image_canvas[:, np.clip(pixel_x + i, 0, resolution - 1), np.clip(pixel_y - j, 0, resolution - 1)] = 1.0
image_canvas[:, np.clip(pixel_x - i, 0, resolution - 1), np.clip(pixel_y + j, 0, resolution - 1)] = 1.0
pixel_x = int(self.goal_state[0].item() * (resolution - 1))
pixel_y = int(self.goal_state[1].item() * (resolution - 1))
for i in range(radius):
for j in range(radius):
image_canvas[0, np.clip(pixel_x + i, 0, resolution - 1), np.clip(pixel_y + j, 0, resolution - 1)] = 1.0
image_canvas[0, np.clip(pixel_x - i, 0, resolution - 1), np.clip(pixel_y - j, 0, resolution - 1)] = 1.0
image_canvas[0, np.clip(pixel_x + i, 0, resolution - 1), np.clip(pixel_y - j, 0, resolution - 1)] = 1.0
image_canvas[0, np.clip(pixel_x - i, 0, resolution - 1), np.clip(pixel_y + j, 0, resolution - 1)] = 1.0
return image_canvas
```
Here, we define a ReplayBuffer to collect data when interacting with the environment. The ReplayBuffer will record the visual observations (images), actions, reward (negative distance to the goal) and terminal flag at each time step. We also define some neural network template that will be used later.
```
class ReplayBuffer(object):
"""Buffer to store and replay environment transitions."""
def __init__(self, obs_shape, action_shape, reward_shape, capacity, batch_size, length, device='cpu'):
self.capacity = capacity
self.batch_size = batch_size
self.length = length
self.device = device
# Initialize all the buffers
self.obs_buffer = np.empty(shape=(capacity, *obs_shape), dtype=np.float32)
self.action_buffer = np.empty(shape=(capacity, *action_shape), dtype=np.float32)
self.reward_buffer = np.empty(shape=(capacity, *reward_shape), dtype=np.float32)
self.done_buffer = np.empty(shape=(capacity, *reward_shape), dtype=np.float32)
self.idx = 0
def add(self, obs, action, reward, done):
if self.idx < self.capacity:
self.obs_buffer[self.idx] = obs
self.action_buffer[self.idx] = action
self.reward_buffer[self.idx] = reward
self.done_buffer[self.idx] = done
self.idx += 1
else:
self.obs_buffer = self.obs_buffer[1:]
self.obs_buffer = np.append(self.obs_buffer,
obs.reshape((1, obs.shape[0], obs.shape[1], obs.shape[2])),
axis=0)
self.action_buffer = self.action_buffer[1:]
self.action_buffer = np.append(self.action_buffer,
action.reshape((1, action.shape[0])),
axis=0)
self.reward_buffer = self.reward_buffer[1:]
self.reward_buffer = np.append(self.reward_buffer,
reward.reshape((1, 1)),
axis=0)
self.done_buffer = self.done_buffer[1:]
self.done_buffer = np.append(self.done_buffer,
done.reshape((1, done.shape[0])),
axis=0)
def sample(self):
idxs = np.random.randint(
0, self.capacity - self.length + 1 if self.idx == self.capacity else self.idx - self.length + 1,
size=self.batch_size)
obses = torch.as_tensor(self.obs_buffer[idxs], device=self.device).unsqueeze(1).float()
actions = torch.as_tensor(self.action_buffer[idxs], device=self.device).unsqueeze(1).float()
rewards = torch.as_tensor(self.reward_buffer[idxs], device=self.device).unsqueeze(1).float()
dones = torch.as_tensor(self.done_buffer[idxs], device=self.device).unsqueeze(1).float()
for i in range(1, self.length):
next_obses = torch.as_tensor(self.obs_buffer[idxs + i], device=self.device).unsqueeze(1).float()
next_actions = torch.as_tensor(self.action_buffer[idxs + i], device=self.device).unsqueeze(1).float()
next_rewards = torch.as_tensor(self.reward_buffer[idxs + i], device=self.device).unsqueeze(1).float()
next_dones = torch.as_tensor(self.done_buffer[idxs + i], device=self.device).unsqueeze(1).float()
obses = torch.cat((obses, next_obses), 1)
actions = torch.cat((actions, next_actions), 1)
rewards = torch.cat((rewards, next_rewards), 1)
dones = torch.cat((dones, next_dones), 1)
return obses, actions, rewards, dones
class CNNDenseModel(nn.Module):
def __init__(self, embed_dim: int, layers: int, h_dim: int,
activation=nn.ReLU, min=1e-4, max=10.0):
super().__init__()
self._embed_size = embed_dim
self._layers = layers
self._hidden_size = h_dim
self.activation = activation
self.model = self.build_model()
self.soft_plus = nn.Softplus()
self._min = min
self._max = max
self.conv_channels = 4
self.conv_in = nn.Sequential(torch.nn.Conv2d(in_channels=3,
out_channels=self.conv_channels,
kernel_size=3,
stride=3))
self.fc_out = self.build_model()
def build_model(self):
model = [nn.Linear(400, self._hidden_size)]
model += [self.activation()]
for i in range(self._layers - 1):
model += [nn.Linear(self._hidden_size, self._hidden_size)]
model += [self.activation()]
model += [nn.Linear(self._hidden_size, self._embed_size)]
return nn.Sequential(*model)
def forward(self, obs_visual):
x_visual = self.conv_in(obs_visual)
x_visual = x_visual.contiguous()
x_visual = x_visual.view(-1, self.conv_channels * 10 * 10)
x = self.fc_out(x_visual)
return x
class CNNDecoder(torch.nn.Module):
def __init__(self, z_dim=10, h_dim=64):
super().__init__()
self.conv_channels = 4
self.fc = nn.Sequential(torch.nn.Linear(z_dim, h_dim),
torch.nn.ReLU(),
torch.nn.Linear(h_dim, self.conv_channels * 10 * 10))
self.deconv = nn.Sequential(
torch.nn.ConvTranspose2d(in_channels=self.conv_channels, out_channels=3, kernel_size=5, stride=3))
def forward(self, x):
x = self.fc(x)
h = x.view(-1, self.conv_channels, 10, 10)
h = self.deconv(h) # , output_size=(x.size(0), 3, 28, 28))
return h
class DenseModelNormal(nn.Module):
def __init__(self, feature_dim: int, output_shape: tuple, layers: int, h_dim: int, activation=nn.ELU,
min=1e-4, max=10.0):
super().__init__()
self._output_shape = output_shape
self._layers = layers
self._hidden_size = h_dim
self.activation = activation
# For adjusting pytorch to tensorflow
self._feature_size = feature_dim
# Defining the structure of the NN
self.model = self.build_model()
self.soft_plus = nn.Softplus()
self._min = min
self._max = max
def build_model(self):
model = [nn.Linear(self._feature_size, self._hidden_size)]
model += [self.activation()]
for i in range(self._layers - 1):
model += [nn.Linear(self._hidden_size, self._hidden_size)]
model += [self.activation()]
model += [nn.Linear(self._hidden_size, 2 * int(np.prod(self._output_shape)))]
return nn.Sequential(*model)
def forward(self, features):
dist_inputs = self.model(features)
reshaped_inputs_mean = torch.reshape(dist_inputs[..., :np.prod(self._output_shape)],
features.shape[:-1] + self._output_shape)
reshaped_inputs_std = torch.reshape(dist_inputs[..., np.prod(self._output_shape):],
features.shape[:-1] + self._output_shape)
reshaped_inputs_std = torch.clamp(self.soft_plus(reshaped_inputs_std), min=self._min, max=self._max)
return td.independent.Independent(td.Normal(reshaped_inputs_mean, reshaped_inputs_std), len(self._output_shape))
```
## State Space Model
Here, we define our State Space Model (SSM). Intuitively, the SSM models an agent that is sequentially taking actions in a world and receiving rewards and visual observations. The observation $x_t$ at time $t$ visual observations are generated from the latent state $z_t$. The model assumes Markovian transitions where the next state is conditioned upon the current state and the action $a_t$ taken by the agent. Upon taking an action, the agent receives reward $r_t$. Given the graphical structure in the figure below, the SSM's joint distribution factorizes as:
<img src="./pgm.png" width=400 height=400 />
\begin{align}
&p_\theta(x_{1:T},r_{1:T},z_{0:T},a_{1:T-1}) = \prod_{t=1}^{T} p_\theta(x_{t}|z_{t})p_\theta(r_{t}|z_{t})p_\theta(z_{t}|z_{t-1},a_{t-1})p_\psi(a_{t-1}|z_{t-1})p(z_0)
\end{align}
Each of the factorized distributions are modelled using nonlinear functions:
* Transitions: $p_\theta(z_{t}|z_{t-1},a_{t-1}) = p(z_{t}| f_\theta(z_{t-1},a_{t-1}))$
* Observations: $p_\theta(x_{t}|z_{t}) = p(x_{t}| d_\theta(z_{t})) $
* Rewards: $p_\theta(r_{t}|z_{t}) = p(r_{t}| r_\theta(z_{t})) $
where $f_\theta$, $d^m_\theta$, $r_\theta$, and $\pi_\psi$ are neural networks parameterized by $\theta$. Given nonlinearity of these factorized distributions, the posterior distribution $p(z_{1:T}|x_{1:T},a_{1:T-1})$ is intractable. Thus, we approximate it by
$$p(z_{1:T}|x_{1:T},a_{1:T-1})\approx\hat{q}_{\phi}(z_{1:T}|x_{1:T},a_{1:T-1})=\prod_{t=2}^T q_\phi(z_{t}|g_\phi(x_{t}, z_{t-1}, a_{t-1}))p(z_1)$$
where $q_\phi$ is modeled as a Gaussian distribution and $g_\theta(x_{t}, z_{t-1}, a_{t-1})$ is a neural network parameterized by $\phi$, which is typically called inference network.
Given all these distributions and trajectories of the form $\tau = \left\{(x_{t}, a_{t}, r_{t})\right\}_{t=1}^{T}$ that samples from the ReplayBuffer, we seek to learn the parameters $\theta$ and $\phi$. Because maximum likelihood estimation is intractable in this setting, we optimize the evidence lower bound (ELBO) under the data distribution $p_d$ using a variational distribution $q$ over the latent state variables $z_t$
\begin{align}
\mathbb{E}_{p_d}[\mathrm{ELBO}] \leq \mathbb{E}_{p_d}[\log p_\theta(x_{1:T},r_{1:T}|a_{1:T-1})]
\end{align}
where
\begin{align}
\mathrm{ELBO} = & \sum_{t=1}^{T}\Big(\displaystyle \mathop{\mathbb{E}}_{\hat{q}_{\phi}(z_{t})}\left[\log p_{\theta}(x_{t}|z_{t})\right] +\displaystyle\mathop{\mathbb{E}}_{\hat{q}_{\phi}(z_{t})}\left[\log p_{\theta}(r_{t}|z_{t})\right] \nonumber\\
&- \displaystyle\mathop{\mathbb{E}}_{\hat{q}_{\phi}(z_{t-1})}\left[\mathrm{KL}\left[\hat{q}_{\phi}(z_{t}) \| p_{\theta}(z_{t}|z_{t-1},a_{t-1})\right]\right]\Big)
\end{align}
```
class SequentialVAE(nn.Module):
def __init__(self, z_dim=8, action_dim=2, embed_dim=32):
super().__init__()
self.image_embed_net = CNNDenseModel(embed_dim=embed_dim, h_dim=64, layers=2)
self.inference_net = DenseModelNormal(feature_dim=embed_dim + z_dim + action_dim,
output_shape=(z_dim,),
layers=2,
h_dim=32)
self.transition_net = DenseModelNormal(feature_dim=z_dim + action_dim,
output_shape=(z_dim,),
layers=2,
h_dim=32)
self.emission_net = CNNDecoder(z_dim=z_dim, h_dim=32)
self.reward_net = DenseModelNormal(feature_dim=z_dim,
output_shape=(1,),
layers=2,
h_dim=32)
self.z_dim = z_dim
self.action_dim = action_dim
self.optimizer = torch.optim.Adam(params=self.parameters(), lr=0.002)
self.anneal_factor = 2.0
def get_z_post_dist(self, obs, z=None, action=None):
obs_embed = self.image_embed_net(obs)
if len(obs.shape) == 3:
obs_embed = obs_embed[0]
if z is None:
z = torch.zeros(obs_embed.shape[:-1] + (self.z_dim,))
if action is None:
action = torch.zeros(obs_embed.shape[:-1] + (self.action_dim,))
z_post_dist = self.inference_net(torch.cat([obs_embed, z, action], dim=-1))
return z_post_dist
def cross_entropy_planning(self, z):
horizon = 4
sample_size = 100
discount_factor = 0.8
action_samples = torch.randint(low=-1, high=2, size=(sample_size, horizon, self.action_dim))
accumulated_reward = torch.zeros((1, 1))
z_temp = z.clone().unsqueeze(0).repeat(sample_size, 1)
for t in range(horizon):
z_temp_dist = self.transition_net(torch.cat([z_temp, action_samples[:, t]], dim=-1))
z_temp = z_temp_dist.mean
reward = self.reward_net(z_temp).mean
accumulated_reward = accumulated_reward + reward * (discount_factor ** t)
opti_id = torch.argmax(accumulated_reward, dim=0)
opti_action_sample = action_samples[opti_id.item()]
return opti_action_sample[0]
def learn(self, replay_buffer, episode_count):
max_epoch = 200
for i in range(max_epoch):
obs, action, reward, done = replay_buffer.sample()
batch_t = obs.shape[1]
# get latent_state
z_post_rsample = [[]] * batch_t
z_post_mean = [[]] * batch_t
z_post_std = [[]] * batch_t
for t in range(batch_t):
if t == 0:
z_post_dist = self.get_z_post_dist(obs[:, t])
else:
z_post_dist = self.get_z_post_dist(obs[:, t], z_post_rsample[t - 1], action[:, t - 1])
z_post_rsample[t] = z_post_dist.rsample()
z_post_mean[t] = z_post_dist.mean
z_post_std[t] = z_post_dist.stddev
z_post_rsample = torch.stack(z_post_rsample, dim=1)
z_post_mean = torch.stack(z_post_mean, dim=1)
z_post_std = torch.stack(z_post_std, dim=1)
z_trans_mean = [[]] * batch_t
z_trans_std = [[]] * batch_t
for t in range(batch_t):
if t == 0:
z_trans_mean[t] = z_post_mean[:, t]
z_trans_std[t] = z_post_std[:, t]
else:
z_trans_dist = self.transition_net(torch.cat([z_post_rsample[:, t - 1], action[:, t]], dim=-1))
z_trans_mean[t] = z_trans_dist.mean
z_trans_std[t] = z_trans_dist.stddev
z_trans_mean = torch.stack(z_trans_mean, dim=1)
z_trans_std = torch.stack(z_trans_std, dim=1)
obs_rec = self.emission_net(z_post_rsample)
obs_rec_loss = 100 * torch.square(obs_rec - obs.view(-1, 3, 32, 32)).mean()
reward_dist = self.reward_net(z_post_mean.detach())
reward_rec_loss = -reward_dist.log_prob(reward).mean()
kl_loss = (z_trans_std.log() - z_post_std.log()
+ (z_post_std.pow(2) + (z_trans_mean - z_post_mean).pow(2))
/ (2 * z_trans_std.pow(2) + 1e-5) - 0.5).mean()
self.optimizer.zero_grad()
loss = obs_rec_loss + reward_rec_loss + 1.0 * torch.max(kl_loss, torch.ones(1) * self.anneal_factor)
loss.backward()
self.optimizer.step()
self.anneal_factor = np.clip(self.anneal_factor * 0.9, 0.1, 10.0)
print(
f'{episode_count}: obs_loss:{obs_rec_loss.item()}, reward_loss:{reward_rec_loss.item()}, kl_loss:{kl_loss.item()}')
```
## Main Loop
Without loss of generality, we begin at time-step $t-1$ where we have a sample of $z_{t-1}$ and taken a action $a_{t-1}$. At time step $t$, we first recieve an visual observation $x_t$ and reward $r_t$. Then,we pass $z_{t-1}$, $a_{t-1}$ and $x_t$ into the variational distribution $q_\phi(z_t|z_{t-1}, a_{t-1}, x_t)$ and sample a $z_t$ from $q_\phi$. Given the sample $z_t$, we optimize the following objective:
\begin{align}
\mathrm{argmax}_{a_{t:t+H-1}} \ \ J = \mathrm{E}_{p(z_{t+1:t+H}\ \ \ \ |a_{t:t+H-1}\ \ \ \ ,z_t)}\left[\sum_{k=t+1}^{t+H} {\gamma^tr(z_t)} \right]
\end{align}
where $p(z_{t+1:t+H}|a_{t:t+H-1},z_t)=\prod_{k=t+1}^{t+H}p_\theta(z_{k}|f_\theta(z_{k-1},a_{k-1}))$ We choose action $a_t$ to execute and replan at each time step. After that,environment fowards to the $t+1$.
```
def main(test=False):
env = ParticleEnv()
replay_buffer = ReplayBuffer(obs_shape=(3, 32, 32),
action_shape=(2,),
reward_shape=(1,),
capacity=1000,
batch_size=50,
length=10)
model = SequentialVAE(z_dim=5, action_dim=2)
if test:
model.load_state_dict(torch.load('./model.pt'))
model.eval()
_, obs, reward = env.reset()
z_post_mean = None
action = None
max_episode = 100
episode_count = 0
episode_data_size = 200
visualize_freq = 5
ims = []
fig, ax = plt.subplots()
while True:
avg_reward = 0.0
for i in range(episode_data_size):
z_post_mean = model.get_z_post_dist(torch.as_tensor(obs).float(), z_post_mean, action).mean
action = model.cross_entropy_planning(z_post_mean)
_, obs, reward, done = env.step(action.detach().numpy())
avg_reward += reward
visualize_image = obs.transpose((1, 2, 0))
if episode_count % visualize_freq == 0:
im = ax.imshow(visualize_image, animated=True)
ims.append([im])
replay_buffer.add(obs, action, reward, np.array([done]))
if done:
_, obs, reward = env.reset()
z_post_mean = None
action = None
avg_reward /= episode_data_size
print(f'avg_reward:{avg_reward}')
if episode_count % visualize_freq == 0:
ani = animation.ArtistAnimation(fig, ims, interval=50, blit=True,
repeat_delay=1000)
writergif = animation.PillowWriter(fps=30)
if test:
file_name = f'./test_episode_{episode_count}.gif'
else:
file_name = f'./episode_{episode_count}.gif'
ani.save(file_name, writer=writergif)
ims.clear()
if not test:
# train model using collected data
model.train()
model.learn(replay_buffer, episode_count)
model.eval()
torch.save(model.state_dict(), './model.pt')
episode_count += 1
if episode_count > max_episode:
break
main(False)
```
|
github_jupyter
|
import torch
import torch.nn as nn
import torch.distributions as td
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.animation as animation
class ParticleEnv:
def __init__(self):
self.state = np.array([0, 0])
self.goal_state = np.array([0.5, 0.5])
self.action_scale = 0.025
self.step_count = 0
def reset(self):
self.step_count = 0
self.state = 0.8 * (np.random.rand(2) - 0.5) + 0.5
reward = -np.sqrt(np.square(self.state - self.goal_state).sum())
state_image = self.generate_image()
# potential_goal_states = [[0.0, 0.0], [0.9, 0.9], [0.9, 0.0], [0.0, 0.9]]
# self.goal_state = np.array(potential_goal_states[random.choice([0, 1, 2, 3])])
return self.state.copy(), state_image, reward
def step(self, action):
self.state += action * self.action_scale
self.state = np.clip(self.state, a_min=0.0, a_max=1.0)
reward = -np.sqrt(np.square(self.state - self.goal_state).sum())
if np.sqrt(np.square(self.state - self.goal_state).sum()) < 0.01:
done = True
else:
done = False
state_image = self.generate_image()
self.step_count += 1
if self.step_count > 10:
done = True
return self.state.copy(), state_image, reward, done
def generate_image(self):
resolution = 32
radius = 3
image_canvas = np.zeros(shape=[3, resolution, resolution])
pixel_x = int(self.state[0].item() * (resolution - 1))
pixel_y = int(self.state[1].item() * (resolution - 1))
for i in range(radius):
for j in range(radius):
image_canvas[:, np.clip(pixel_x + i, 0, resolution - 1), np.clip(pixel_y + j, 0, resolution - 1)] = 1.0
image_canvas[:, np.clip(pixel_x - i, 0, resolution - 1), np.clip(pixel_y - j, 0, resolution - 1)] = 1.0
image_canvas[:, np.clip(pixel_x + i, 0, resolution - 1), np.clip(pixel_y - j, 0, resolution - 1)] = 1.0
image_canvas[:, np.clip(pixel_x - i, 0, resolution - 1), np.clip(pixel_y + j, 0, resolution - 1)] = 1.0
pixel_x = int(self.goal_state[0].item() * (resolution - 1))
pixel_y = int(self.goal_state[1].item() * (resolution - 1))
for i in range(radius):
for j in range(radius):
image_canvas[0, np.clip(pixel_x + i, 0, resolution - 1), np.clip(pixel_y + j, 0, resolution - 1)] = 1.0
image_canvas[0, np.clip(pixel_x - i, 0, resolution - 1), np.clip(pixel_y - j, 0, resolution - 1)] = 1.0
image_canvas[0, np.clip(pixel_x + i, 0, resolution - 1), np.clip(pixel_y - j, 0, resolution - 1)] = 1.0
image_canvas[0, np.clip(pixel_x - i, 0, resolution - 1), np.clip(pixel_y + j, 0, resolution - 1)] = 1.0
return image_canvas
class ReplayBuffer(object):
"""Buffer to store and replay environment transitions."""
def __init__(self, obs_shape, action_shape, reward_shape, capacity, batch_size, length, device='cpu'):
self.capacity = capacity
self.batch_size = batch_size
self.length = length
self.device = device
# Initialize all the buffers
self.obs_buffer = np.empty(shape=(capacity, *obs_shape), dtype=np.float32)
self.action_buffer = np.empty(shape=(capacity, *action_shape), dtype=np.float32)
self.reward_buffer = np.empty(shape=(capacity, *reward_shape), dtype=np.float32)
self.done_buffer = np.empty(shape=(capacity, *reward_shape), dtype=np.float32)
self.idx = 0
def add(self, obs, action, reward, done):
if self.idx < self.capacity:
self.obs_buffer[self.idx] = obs
self.action_buffer[self.idx] = action
self.reward_buffer[self.idx] = reward
self.done_buffer[self.idx] = done
self.idx += 1
else:
self.obs_buffer = self.obs_buffer[1:]
self.obs_buffer = np.append(self.obs_buffer,
obs.reshape((1, obs.shape[0], obs.shape[1], obs.shape[2])),
axis=0)
self.action_buffer = self.action_buffer[1:]
self.action_buffer = np.append(self.action_buffer,
action.reshape((1, action.shape[0])),
axis=0)
self.reward_buffer = self.reward_buffer[1:]
self.reward_buffer = np.append(self.reward_buffer,
reward.reshape((1, 1)),
axis=0)
self.done_buffer = self.done_buffer[1:]
self.done_buffer = np.append(self.done_buffer,
done.reshape((1, done.shape[0])),
axis=0)
def sample(self):
idxs = np.random.randint(
0, self.capacity - self.length + 1 if self.idx == self.capacity else self.idx - self.length + 1,
size=self.batch_size)
obses = torch.as_tensor(self.obs_buffer[idxs], device=self.device).unsqueeze(1).float()
actions = torch.as_tensor(self.action_buffer[idxs], device=self.device).unsqueeze(1).float()
rewards = torch.as_tensor(self.reward_buffer[idxs], device=self.device).unsqueeze(1).float()
dones = torch.as_tensor(self.done_buffer[idxs], device=self.device).unsqueeze(1).float()
for i in range(1, self.length):
next_obses = torch.as_tensor(self.obs_buffer[idxs + i], device=self.device).unsqueeze(1).float()
next_actions = torch.as_tensor(self.action_buffer[idxs + i], device=self.device).unsqueeze(1).float()
next_rewards = torch.as_tensor(self.reward_buffer[idxs + i], device=self.device).unsqueeze(1).float()
next_dones = torch.as_tensor(self.done_buffer[idxs + i], device=self.device).unsqueeze(1).float()
obses = torch.cat((obses, next_obses), 1)
actions = torch.cat((actions, next_actions), 1)
rewards = torch.cat((rewards, next_rewards), 1)
dones = torch.cat((dones, next_dones), 1)
return obses, actions, rewards, dones
class CNNDenseModel(nn.Module):
def __init__(self, embed_dim: int, layers: int, h_dim: int,
activation=nn.ReLU, min=1e-4, max=10.0):
super().__init__()
self._embed_size = embed_dim
self._layers = layers
self._hidden_size = h_dim
self.activation = activation
self.model = self.build_model()
self.soft_plus = nn.Softplus()
self._min = min
self._max = max
self.conv_channels = 4
self.conv_in = nn.Sequential(torch.nn.Conv2d(in_channels=3,
out_channels=self.conv_channels,
kernel_size=3,
stride=3))
self.fc_out = self.build_model()
def build_model(self):
model = [nn.Linear(400, self._hidden_size)]
model += [self.activation()]
for i in range(self._layers - 1):
model += [nn.Linear(self._hidden_size, self._hidden_size)]
model += [self.activation()]
model += [nn.Linear(self._hidden_size, self._embed_size)]
return nn.Sequential(*model)
def forward(self, obs_visual):
x_visual = self.conv_in(obs_visual)
x_visual = x_visual.contiguous()
x_visual = x_visual.view(-1, self.conv_channels * 10 * 10)
x = self.fc_out(x_visual)
return x
class CNNDecoder(torch.nn.Module):
def __init__(self, z_dim=10, h_dim=64):
super().__init__()
self.conv_channels = 4
self.fc = nn.Sequential(torch.nn.Linear(z_dim, h_dim),
torch.nn.ReLU(),
torch.nn.Linear(h_dim, self.conv_channels * 10 * 10))
self.deconv = nn.Sequential(
torch.nn.ConvTranspose2d(in_channels=self.conv_channels, out_channels=3, kernel_size=5, stride=3))
def forward(self, x):
x = self.fc(x)
h = x.view(-1, self.conv_channels, 10, 10)
h = self.deconv(h) # , output_size=(x.size(0), 3, 28, 28))
return h
class DenseModelNormal(nn.Module):
def __init__(self, feature_dim: int, output_shape: tuple, layers: int, h_dim: int, activation=nn.ELU,
min=1e-4, max=10.0):
super().__init__()
self._output_shape = output_shape
self._layers = layers
self._hidden_size = h_dim
self.activation = activation
# For adjusting pytorch to tensorflow
self._feature_size = feature_dim
# Defining the structure of the NN
self.model = self.build_model()
self.soft_plus = nn.Softplus()
self._min = min
self._max = max
def build_model(self):
model = [nn.Linear(self._feature_size, self._hidden_size)]
model += [self.activation()]
for i in range(self._layers - 1):
model += [nn.Linear(self._hidden_size, self._hidden_size)]
model += [self.activation()]
model += [nn.Linear(self._hidden_size, 2 * int(np.prod(self._output_shape)))]
return nn.Sequential(*model)
def forward(self, features):
dist_inputs = self.model(features)
reshaped_inputs_mean = torch.reshape(dist_inputs[..., :np.prod(self._output_shape)],
features.shape[:-1] + self._output_shape)
reshaped_inputs_std = torch.reshape(dist_inputs[..., np.prod(self._output_shape):],
features.shape[:-1] + self._output_shape)
reshaped_inputs_std = torch.clamp(self.soft_plus(reshaped_inputs_std), min=self._min, max=self._max)
return td.independent.Independent(td.Normal(reshaped_inputs_mean, reshaped_inputs_std), len(self._output_shape))
class SequentialVAE(nn.Module):
def __init__(self, z_dim=8, action_dim=2, embed_dim=32):
super().__init__()
self.image_embed_net = CNNDenseModel(embed_dim=embed_dim, h_dim=64, layers=2)
self.inference_net = DenseModelNormal(feature_dim=embed_dim + z_dim + action_dim,
output_shape=(z_dim,),
layers=2,
h_dim=32)
self.transition_net = DenseModelNormal(feature_dim=z_dim + action_dim,
output_shape=(z_dim,),
layers=2,
h_dim=32)
self.emission_net = CNNDecoder(z_dim=z_dim, h_dim=32)
self.reward_net = DenseModelNormal(feature_dim=z_dim,
output_shape=(1,),
layers=2,
h_dim=32)
self.z_dim = z_dim
self.action_dim = action_dim
self.optimizer = torch.optim.Adam(params=self.parameters(), lr=0.002)
self.anneal_factor = 2.0
def get_z_post_dist(self, obs, z=None, action=None):
obs_embed = self.image_embed_net(obs)
if len(obs.shape) == 3:
obs_embed = obs_embed[0]
if z is None:
z = torch.zeros(obs_embed.shape[:-1] + (self.z_dim,))
if action is None:
action = torch.zeros(obs_embed.shape[:-1] + (self.action_dim,))
z_post_dist = self.inference_net(torch.cat([obs_embed, z, action], dim=-1))
return z_post_dist
def cross_entropy_planning(self, z):
horizon = 4
sample_size = 100
discount_factor = 0.8
action_samples = torch.randint(low=-1, high=2, size=(sample_size, horizon, self.action_dim))
accumulated_reward = torch.zeros((1, 1))
z_temp = z.clone().unsqueeze(0).repeat(sample_size, 1)
for t in range(horizon):
z_temp_dist = self.transition_net(torch.cat([z_temp, action_samples[:, t]], dim=-1))
z_temp = z_temp_dist.mean
reward = self.reward_net(z_temp).mean
accumulated_reward = accumulated_reward + reward * (discount_factor ** t)
opti_id = torch.argmax(accumulated_reward, dim=0)
opti_action_sample = action_samples[opti_id.item()]
return opti_action_sample[0]
def learn(self, replay_buffer, episode_count):
max_epoch = 200
for i in range(max_epoch):
obs, action, reward, done = replay_buffer.sample()
batch_t = obs.shape[1]
# get latent_state
z_post_rsample = [[]] * batch_t
z_post_mean = [[]] * batch_t
z_post_std = [[]] * batch_t
for t in range(batch_t):
if t == 0:
z_post_dist = self.get_z_post_dist(obs[:, t])
else:
z_post_dist = self.get_z_post_dist(obs[:, t], z_post_rsample[t - 1], action[:, t - 1])
z_post_rsample[t] = z_post_dist.rsample()
z_post_mean[t] = z_post_dist.mean
z_post_std[t] = z_post_dist.stddev
z_post_rsample = torch.stack(z_post_rsample, dim=1)
z_post_mean = torch.stack(z_post_mean, dim=1)
z_post_std = torch.stack(z_post_std, dim=1)
z_trans_mean = [[]] * batch_t
z_trans_std = [[]] * batch_t
for t in range(batch_t):
if t == 0:
z_trans_mean[t] = z_post_mean[:, t]
z_trans_std[t] = z_post_std[:, t]
else:
z_trans_dist = self.transition_net(torch.cat([z_post_rsample[:, t - 1], action[:, t]], dim=-1))
z_trans_mean[t] = z_trans_dist.mean
z_trans_std[t] = z_trans_dist.stddev
z_trans_mean = torch.stack(z_trans_mean, dim=1)
z_trans_std = torch.stack(z_trans_std, dim=1)
obs_rec = self.emission_net(z_post_rsample)
obs_rec_loss = 100 * torch.square(obs_rec - obs.view(-1, 3, 32, 32)).mean()
reward_dist = self.reward_net(z_post_mean.detach())
reward_rec_loss = -reward_dist.log_prob(reward).mean()
kl_loss = (z_trans_std.log() - z_post_std.log()
+ (z_post_std.pow(2) + (z_trans_mean - z_post_mean).pow(2))
/ (2 * z_trans_std.pow(2) + 1e-5) - 0.5).mean()
self.optimizer.zero_grad()
loss = obs_rec_loss + reward_rec_loss + 1.0 * torch.max(kl_loss, torch.ones(1) * self.anneal_factor)
loss.backward()
self.optimizer.step()
self.anneal_factor = np.clip(self.anneal_factor * 0.9, 0.1, 10.0)
print(
f'{episode_count}: obs_loss:{obs_rec_loss.item()}, reward_loss:{reward_rec_loss.item()}, kl_loss:{kl_loss.item()}')
def main(test=False):
env = ParticleEnv()
replay_buffer = ReplayBuffer(obs_shape=(3, 32, 32),
action_shape=(2,),
reward_shape=(1,),
capacity=1000,
batch_size=50,
length=10)
model = SequentialVAE(z_dim=5, action_dim=2)
if test:
model.load_state_dict(torch.load('./model.pt'))
model.eval()
_, obs, reward = env.reset()
z_post_mean = None
action = None
max_episode = 100
episode_count = 0
episode_data_size = 200
visualize_freq = 5
ims = []
fig, ax = plt.subplots()
while True:
avg_reward = 0.0
for i in range(episode_data_size):
z_post_mean = model.get_z_post_dist(torch.as_tensor(obs).float(), z_post_mean, action).mean
action = model.cross_entropy_planning(z_post_mean)
_, obs, reward, done = env.step(action.detach().numpy())
avg_reward += reward
visualize_image = obs.transpose((1, 2, 0))
if episode_count % visualize_freq == 0:
im = ax.imshow(visualize_image, animated=True)
ims.append([im])
replay_buffer.add(obs, action, reward, np.array([done]))
if done:
_, obs, reward = env.reset()
z_post_mean = None
action = None
avg_reward /= episode_data_size
print(f'avg_reward:{avg_reward}')
if episode_count % visualize_freq == 0:
ani = animation.ArtistAnimation(fig, ims, interval=50, blit=True,
repeat_delay=1000)
writergif = animation.PillowWriter(fps=30)
if test:
file_name = f'./test_episode_{episode_count}.gif'
else:
file_name = f'./episode_{episode_count}.gif'
ani.save(file_name, writer=writergif)
ims.clear()
if not test:
# train model using collected data
model.train()
model.learn(replay_buffer, episode_count)
model.eval()
torch.save(model.state_dict(), './model.pt')
episode_count += 1
if episode_count > max_episode:
break
main(False)
| 0.853394 | 0.923454 |
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import warnings
warnings.filterwarnings("ignore")
%matplotlib inline
sns.set_style('whitegrid')
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import classification_report
from sklearn.metrics import confusion_matrix
from sklearn.model_selection import cross_val_predict,cross_val_score
from sklearn.metrics import roc_auc_score,roc_curve
from sklearn.model_selection import train_test_split
from sklearn import metrics
from sklearn.metrics import mean_squared_error
training_data = pd.read_csv("../Data/aps_failure_training_set.csv",na_values="na")
training_data.head()
```
# Preprocessing
```
plt.figure(figsize=(20,12))
sns.heatmap(training_data.isnull(),yticklabels=False,cbar=False,cmap = 'viridis')
```
# Missing value handling
We are going to use different approches with missing values:
1. Removing the column having 80% missing values (**Self intuition)
2. Keeping all the features
3. Later, we will try to implement some feature engineering
**For the rest of the missing values, we are replacing them with their mean() for now (**Ref)
<big><b>Second Approach</b>
```
sample_training_data = training_data
sample_training_data.fillna(sample_training_data.mean(),inplace=True)
#after replacing with mean()
plt.figure(figsize=(20,12))
sns.heatmap(sample_training_data.isnull(),yticklabels=False,cbar=False,cmap='viridis')
#as all the other values are numerical except Class column so we can replace them with 1 and 0
sample_training_data = sample_training_data.replace('neg',0)
sample_training_data = sample_training_data.replace('pos',1)
sample_training_data.head()
```
# Testing Data preprocessing
```
testing_data = pd.read_csv("../Data/aps_failure_test_set.csv",na_values="na")
testing_data.head()
sample_testing_data = testing_data
sample_testing_data.fillna(sample_testing_data.mean(),inplace=True)
#after replacing with mean()
plt.figure(figsize=(20,12))
sns.heatmap(sample_testing_data.isnull(),yticklabels=False,cbar=False,cmap='viridis')
#as all the other values are numerical except Class column so we can replace them with 1 and 0
sample_testing_data = sample_testing_data.replace('neg',0)
sample_testing_data = sample_testing_data.replace('pos',1)
sample_testing_data.head()
```
# Model implementation with Cross validation
```
X = sample_training_data.drop('class',axis=1)
y = sample_training_data['class']
CV_prediction = cross_val_predict(LogisticRegression(),X,y,cv = 5)
CV_score = cross_val_score(LogisticRegression(),X,y,cv = 5)
#mean cross validation score
np.mean(CV_score)
print(classification_report(y,CV_prediction))
tn, fp, fn, tp = confusion_matrix(y, CV_prediction).ravel()
confusionData = [[tn,fp],[fn,tp]]
pd.DataFrame(confusionData,columns=['FN','FP'],index=['TN','TP'])
cost = 10*fp+500*fn
values = {'Score':[cost],'Number of Type 1 faults':[fp],'Number of Type 2 faults':[fn]}
pd.DataFrame(values)
```
<b> Improved from approach 1 | type 2 fault decreases </b>
```
print(metrics.accuracy_score(y, CV_prediction)) #almost same
print(metrics.r2_score(y, CV_prediction)) # improved
print(metrics.f1_score(y, CV_prediction)) #improved
print(mean_squared_error(y,CV_prediction))
CV_prediction = cross_val_predict(LogisticRegression(),X,y,cv = 10)
CV_score = cross_val_score(LogisticRegression(),X,y,cv = 10)
print(classification_report(y,CV_prediction))
tn, fp, fn, tp = confusion_matrix(y, CV_prediction).ravel()
cost = 10*fp+500*fn
values = {'Score':[cost],'Number of Type 1 faults':[fp],'Number of Type 2 faults':[fn]}
pd.DataFrame(values)
print(metrics.accuracy_score(y, CV_prediction)) #almost same
print(metrics.r2_score(y, CV_prediction)) # improved
print(metrics.f1_score(y, CV_prediction)) #improved
print(mean_squared_error(y,CV_prediction))
```
# Try with test train split
```
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state=101)
logmodel = LogisticRegression()
logmodel.fit(X_train,y_train)
regularPrediction = logmodel.predict(X_test)
print(classification_report(y_test,regularPrediction))
print(metrics.accuracy_score(y_test, regularPrediction))
print(metrics.r2_score(y_test, regularPrediction)) # as the ans is negative , so reverese correlation
print(metrics.f1_score(y_test, regularPrediction))
#testing error
print(metrics.mean_squared_error(y_test, regularPrediction))
#Training error
temp = logmodel.predict(X_train)
mean_squared_error(y_train,temp)
#confusion matrix
print(confusion_matrix(y_test,regularPrediction))
tn, fp, fn, tp = confusion_matrix(y_test,regularPrediction).ravel()
confusionData = [[tn,fp],[fn,tp]]
pd.DataFrame(confusionData,columns=['FN','FP'],index=['TN','TP'])
#without modified threshold
cost = 10*fp+500*fn
values = {'Score':[cost],'Number of Type 1 faults':[fp],'Number of Type 2 faults':[fn]}
pd.DataFrame(values)
logit_roc_auc = roc_auc_score(y_test, logmodel.predict_proba(X_test)[:,1])
fpr, tpr, thresholds = roc_curve(y_test,logmodel.predict_proba(X_test)[:,1])
plt.figure()
plt.plot(fpr, tpr, label='Logistic Regression (area = %0.2f)' % logit_roc_auc)
plt.plot([0, 1], [0, 1],'r--')
plt.xlim([0.0, 1.0])
plt.ylim([0.0, 1.0])
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
plt.title('Receiver operating characteristic')
plt.legend(loc="upper center")
plt.savefig('Log_ROC')
# create the axis of thresholds (scores)
ax2 = plt.gca().twinx()
ax2.plot(fpr, thresholds, markeredgecolor='g',linestyle='dashed', color='g',label = 'Threshold')
ax2.set_ylabel('Threshold',color='g')
ax2.set_ylim([thresholds[-1],thresholds[0]])
ax2.set_xlim([fpr[0],fpr[-1]])
plt.legend(loc="lower right")
plt.savefig('roc_and_threshold.png')
plt.show()
#wtih different threshold
THRESHOLD = 0.253 #optimal one chosen manually
thresholdPrediction = np.where(logmodel.predict_proba(X_test)[:,1] > THRESHOLD, 1,0)
tn, fp, fn, tp = confusion_matrix(y_test,thresholdPrediction).ravel()
cost = 10*fp+500*fn
values = {'Score':[cost],'Number of Type 1 faults':[fp],'Number of Type 2 faults':[fn]}
pd.DataFrame(values)
```
# Testing Data implementation
```
logmodel.fit(X,y)
testData_X = sample_testing_data.drop('class',axis=1)
testData_y = sample_testing_data['class']
#as the logmodel is already fitted with training data X_train and y_train so,
testDataPrediction = logmodel.predict(testData_X)
print(classification_report(testData_y,testDataPrediction))
print(metrics.accuracy_score(testData_y, testDataPrediction)) #improved
print(metrics.r2_score(testData_y, testDataPrediction)) # improved
print(metrics.f1_score(testData_y, testDataPrediction)) #improved
#testing error
print(metrics.mean_squared_error(testData_y, testDataPrediction)) #testing error increased
#Training error
temp = logmodel.predict(X)
mean_squared_error(y,temp)
#confusion matrix
print(confusion_matrix(testData_y, testDataPrediction))
tn, fp, fn, tp = confusion_matrix(testData_y, testDataPrediction).ravel()
confusionData = [[tn,fp],[fn,tp]]
pd.DataFrame(confusionData,columns=['FN','FP'],index=['TN','TP'])
#without modified threshold
cost = 10*fp+500*fn
values = {'Score':[cost],'Number of Type 1 faults':[fp],'Number of Type 2 faults':[fn]}
pd.DataFrame(values)
logit_roc_auc = roc_auc_score(testData_y, logmodel.predict_proba(testData_X)[:,1])
fpr, tpr, thresholds = roc_curve(testData_y,logmodel.predict_proba(testData_X)[:,1])
plt.figure()
plt.plot(fpr, tpr, label='Logistic Regression (area = %0.2f)' % logit_roc_auc)
plt.plot([0, 1], [0, 1],'r--')
plt.xlim([0.0, 1.0])
plt.ylim([0.0, 1.0])
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
plt.title('Receiver operating characteristic')
plt.legend(loc="lower right")
plt.savefig('Log_ROC')
# create the axis of thresholds (scores)
ax2 = plt.gca().twinx()
ax2.plot(fpr, thresholds, markeredgecolor='g',linestyle='dashed', color='g',label = 'Threshold')
ax2.set_ylabel('Threshold',color='g')
ax2.set_ylim([thresholds[-1],thresholds[0]])
ax2.set_xlim([fpr[0],fpr[-1]])
plt.legend(loc="lower right")
plt.savefig('roc_and_threshold.png')
plt.show()
#wtih different threshold
THRESHOLD = 0.253 #optimal one chosen manually with test train split
thresholdTestingPrediction = np.where(logmodel.predict_proba(testData_X)[:,1] > THRESHOLD, 1,0)
testing_tn, testing_fp, testing_fn, testing_tp = confusion_matrix(testData_y,thresholdTestingPrediction).ravel()
testing_cost = 10*testing_fp+500*testing_fn
testing_values = {'Score':[testing_cost],'Number of Type 1 faults':[testing_fp],'Number of Type 2 faults':[testing_fn]}
pd.DataFrame(testing_values)
```
# Final Score is 47860
```
len(thresholdTestingPrediction)
print(metrics.accuracy_score(testData_y, thresholdTestingPrediction)) #improved
print(metrics.r2_score(testData_y, thresholdTestingPrediction)) # improved
print(metrics.f1_score(testData_y, thresholdTestingPrediction)) #improved
#testing error
print(metrics.mean_squared_error(testData_y, thresholdTestingPrediction)) #testing error increased
print(metrics.recall_score(testData_y, thresholdTestingPrediction))
```
|
github_jupyter
|
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import warnings
warnings.filterwarnings("ignore")
%matplotlib inline
sns.set_style('whitegrid')
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import classification_report
from sklearn.metrics import confusion_matrix
from sklearn.model_selection import cross_val_predict,cross_val_score
from sklearn.metrics import roc_auc_score,roc_curve
from sklearn.model_selection import train_test_split
from sklearn import metrics
from sklearn.metrics import mean_squared_error
training_data = pd.read_csv("../Data/aps_failure_training_set.csv",na_values="na")
training_data.head()
plt.figure(figsize=(20,12))
sns.heatmap(training_data.isnull(),yticklabels=False,cbar=False,cmap = 'viridis')
sample_training_data = training_data
sample_training_data.fillna(sample_training_data.mean(),inplace=True)
#after replacing with mean()
plt.figure(figsize=(20,12))
sns.heatmap(sample_training_data.isnull(),yticklabels=False,cbar=False,cmap='viridis')
#as all the other values are numerical except Class column so we can replace them with 1 and 0
sample_training_data = sample_training_data.replace('neg',0)
sample_training_data = sample_training_data.replace('pos',1)
sample_training_data.head()
testing_data = pd.read_csv("../Data/aps_failure_test_set.csv",na_values="na")
testing_data.head()
sample_testing_data = testing_data
sample_testing_data.fillna(sample_testing_data.mean(),inplace=True)
#after replacing with mean()
plt.figure(figsize=(20,12))
sns.heatmap(sample_testing_data.isnull(),yticklabels=False,cbar=False,cmap='viridis')
#as all the other values are numerical except Class column so we can replace them with 1 and 0
sample_testing_data = sample_testing_data.replace('neg',0)
sample_testing_data = sample_testing_data.replace('pos',1)
sample_testing_data.head()
X = sample_training_data.drop('class',axis=1)
y = sample_training_data['class']
CV_prediction = cross_val_predict(LogisticRegression(),X,y,cv = 5)
CV_score = cross_val_score(LogisticRegression(),X,y,cv = 5)
#mean cross validation score
np.mean(CV_score)
print(classification_report(y,CV_prediction))
tn, fp, fn, tp = confusion_matrix(y, CV_prediction).ravel()
confusionData = [[tn,fp],[fn,tp]]
pd.DataFrame(confusionData,columns=['FN','FP'],index=['TN','TP'])
cost = 10*fp+500*fn
values = {'Score':[cost],'Number of Type 1 faults':[fp],'Number of Type 2 faults':[fn]}
pd.DataFrame(values)
print(metrics.accuracy_score(y, CV_prediction)) #almost same
print(metrics.r2_score(y, CV_prediction)) # improved
print(metrics.f1_score(y, CV_prediction)) #improved
print(mean_squared_error(y,CV_prediction))
CV_prediction = cross_val_predict(LogisticRegression(),X,y,cv = 10)
CV_score = cross_val_score(LogisticRegression(),X,y,cv = 10)
print(classification_report(y,CV_prediction))
tn, fp, fn, tp = confusion_matrix(y, CV_prediction).ravel()
cost = 10*fp+500*fn
values = {'Score':[cost],'Number of Type 1 faults':[fp],'Number of Type 2 faults':[fn]}
pd.DataFrame(values)
print(metrics.accuracy_score(y, CV_prediction)) #almost same
print(metrics.r2_score(y, CV_prediction)) # improved
print(metrics.f1_score(y, CV_prediction)) #improved
print(mean_squared_error(y,CV_prediction))
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state=101)
logmodel = LogisticRegression()
logmodel.fit(X_train,y_train)
regularPrediction = logmodel.predict(X_test)
print(classification_report(y_test,regularPrediction))
print(metrics.accuracy_score(y_test, regularPrediction))
print(metrics.r2_score(y_test, regularPrediction)) # as the ans is negative , so reverese correlation
print(metrics.f1_score(y_test, regularPrediction))
#testing error
print(metrics.mean_squared_error(y_test, regularPrediction))
#Training error
temp = logmodel.predict(X_train)
mean_squared_error(y_train,temp)
#confusion matrix
print(confusion_matrix(y_test,regularPrediction))
tn, fp, fn, tp = confusion_matrix(y_test,regularPrediction).ravel()
confusionData = [[tn,fp],[fn,tp]]
pd.DataFrame(confusionData,columns=['FN','FP'],index=['TN','TP'])
#without modified threshold
cost = 10*fp+500*fn
values = {'Score':[cost],'Number of Type 1 faults':[fp],'Number of Type 2 faults':[fn]}
pd.DataFrame(values)
logit_roc_auc = roc_auc_score(y_test, logmodel.predict_proba(X_test)[:,1])
fpr, tpr, thresholds = roc_curve(y_test,logmodel.predict_proba(X_test)[:,1])
plt.figure()
plt.plot(fpr, tpr, label='Logistic Regression (area = %0.2f)' % logit_roc_auc)
plt.plot([0, 1], [0, 1],'r--')
plt.xlim([0.0, 1.0])
plt.ylim([0.0, 1.0])
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
plt.title('Receiver operating characteristic')
plt.legend(loc="upper center")
plt.savefig('Log_ROC')
# create the axis of thresholds (scores)
ax2 = plt.gca().twinx()
ax2.plot(fpr, thresholds, markeredgecolor='g',linestyle='dashed', color='g',label = 'Threshold')
ax2.set_ylabel('Threshold',color='g')
ax2.set_ylim([thresholds[-1],thresholds[0]])
ax2.set_xlim([fpr[0],fpr[-1]])
plt.legend(loc="lower right")
plt.savefig('roc_and_threshold.png')
plt.show()
#wtih different threshold
THRESHOLD = 0.253 #optimal one chosen manually
thresholdPrediction = np.where(logmodel.predict_proba(X_test)[:,1] > THRESHOLD, 1,0)
tn, fp, fn, tp = confusion_matrix(y_test,thresholdPrediction).ravel()
cost = 10*fp+500*fn
values = {'Score':[cost],'Number of Type 1 faults':[fp],'Number of Type 2 faults':[fn]}
pd.DataFrame(values)
logmodel.fit(X,y)
testData_X = sample_testing_data.drop('class',axis=1)
testData_y = sample_testing_data['class']
#as the logmodel is already fitted with training data X_train and y_train so,
testDataPrediction = logmodel.predict(testData_X)
print(classification_report(testData_y,testDataPrediction))
print(metrics.accuracy_score(testData_y, testDataPrediction)) #improved
print(metrics.r2_score(testData_y, testDataPrediction)) # improved
print(metrics.f1_score(testData_y, testDataPrediction)) #improved
#testing error
print(metrics.mean_squared_error(testData_y, testDataPrediction)) #testing error increased
#Training error
temp = logmodel.predict(X)
mean_squared_error(y,temp)
#confusion matrix
print(confusion_matrix(testData_y, testDataPrediction))
tn, fp, fn, tp = confusion_matrix(testData_y, testDataPrediction).ravel()
confusionData = [[tn,fp],[fn,tp]]
pd.DataFrame(confusionData,columns=['FN','FP'],index=['TN','TP'])
#without modified threshold
cost = 10*fp+500*fn
values = {'Score':[cost],'Number of Type 1 faults':[fp],'Number of Type 2 faults':[fn]}
pd.DataFrame(values)
logit_roc_auc = roc_auc_score(testData_y, logmodel.predict_proba(testData_X)[:,1])
fpr, tpr, thresholds = roc_curve(testData_y,logmodel.predict_proba(testData_X)[:,1])
plt.figure()
plt.plot(fpr, tpr, label='Logistic Regression (area = %0.2f)' % logit_roc_auc)
plt.plot([0, 1], [0, 1],'r--')
plt.xlim([0.0, 1.0])
plt.ylim([0.0, 1.0])
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
plt.title('Receiver operating characteristic')
plt.legend(loc="lower right")
plt.savefig('Log_ROC')
# create the axis of thresholds (scores)
ax2 = plt.gca().twinx()
ax2.plot(fpr, thresholds, markeredgecolor='g',linestyle='dashed', color='g',label = 'Threshold')
ax2.set_ylabel('Threshold',color='g')
ax2.set_ylim([thresholds[-1],thresholds[0]])
ax2.set_xlim([fpr[0],fpr[-1]])
plt.legend(loc="lower right")
plt.savefig('roc_and_threshold.png')
plt.show()
#wtih different threshold
THRESHOLD = 0.253 #optimal one chosen manually with test train split
thresholdTestingPrediction = np.where(logmodel.predict_proba(testData_X)[:,1] > THRESHOLD, 1,0)
testing_tn, testing_fp, testing_fn, testing_tp = confusion_matrix(testData_y,thresholdTestingPrediction).ravel()
testing_cost = 10*testing_fp+500*testing_fn
testing_values = {'Score':[testing_cost],'Number of Type 1 faults':[testing_fp],'Number of Type 2 faults':[testing_fn]}
pd.DataFrame(testing_values)
len(thresholdTestingPrediction)
print(metrics.accuracy_score(testData_y, thresholdTestingPrediction)) #improved
print(metrics.r2_score(testData_y, thresholdTestingPrediction)) # improved
print(metrics.f1_score(testData_y, thresholdTestingPrediction)) #improved
#testing error
print(metrics.mean_squared_error(testData_y, thresholdTestingPrediction)) #testing error increased
print(metrics.recall_score(testData_y, thresholdTestingPrediction))
| 0.495117 | 0.8586 |
# Anna KaRNNa
In this notebook, I'll build a character-wise RNN trained on Anna Karenina, one of my all-time favorite books. It'll be able to generate new text based on the text from the book.
This network is based off of Andrej Karpathy's [post on RNNs](http://karpathy.github.io/2015/05/21/rnn-effectiveness/) and [implementation in Torch](https://github.com/karpathy/char-rnn). Also, some information [here at r2rt](http://r2rt.com/recurrent-neural-networks-in-tensorflow-ii.html) and from [Sherjil Ozair](https://github.com/sherjilozair/char-rnn-tensorflow) on GitHub. Below is the general architecture of the character-wise RNN.
<img src="assets/charseq.jpeg" width="500">
```
import time
from collections import namedtuple
import numpy as np
import tensorflow as tf
```
First we'll load the text file and convert it into integers for our network to use. Here I'm creating a couple dictionaries to convert the characters to and from integers. Encoding the characters as integers makes it easier to use as input in the network.
```
with open('anna.txt', 'r') as f:
text=f.read()
vocab = sorted(set(text))
vocab_to_int = {c: i for i, c in enumerate(vocab)}
int_to_vocab = dict(enumerate(vocab))
encoded = np.array([vocab_to_int[c] for c in text], dtype=np.int32)
```
Let's check out the first 100 characters, make sure everything is peachy. According to the [American Book Review](http://americanbookreview.org/100bestlines.asp), this is the 6th best first line of a book ever.
```
text[:100]
```
And we can see the characters encoded as integers.
```
encoded[:100]
```
Since the network is working with individual characters, it's similar to a classification problem in which we are trying to predict the next character from the previous text. Here's how many 'classes' our network has to pick from.
```
len(vocab)
```
## Making training mini-batches
Here is where we'll make our mini-batches for training. Remember that we want our batches to be multiple sequences of some desired number of sequence steps. Considering a simple example, our batches would look like this:
<img src="assets/sequence_batching@1x.png" width=500px>
<br>
We have our text encoded as integers as one long array in `encoded`. Let's create a function that will give us an iterator for our batches. I like using [generator functions](https://jeffknupp.com/blog/2013/04/07/improve-your-python-yield-and-generators-explained/) to do this. Then we can pass `encoded` into this function and get our batch generator.
The first thing we need to do is discard some of the text so we only have completely full batches. Each batch contains $N \times M$ characters, where $N$ is the batch size (the number of sequences) and $M$ is the number of steps. Then, to get the number of batches we can make from some array `arr`, you divide the length of `arr` by the batch size. Once you know the number of batches and the batch size, you can get the total number of characters to keep.
After that, we need to split `arr` into $N$ sequences. You can do this using `arr.reshape(size)` where `size` is a tuple containing the dimensions sizes of the reshaped array. We know we want $N$ sequences (`n_seqs` below), let's make that the size of the first dimension. For the second dimension, you can use `-1` as a placeholder in the size, it'll fill up the array with the appropriate data for you. After this, you should have an array that is $N \times (M * K)$ where $K$ is the number of batches.
Now that we have this array, we can iterate through it to get our batches. The idea is each batch is a $N \times M$ window on the array. For each subsequent batch, the window moves over by `n_steps`. We also want to create both the input and target arrays. Remember that the targets are the inputs shifted over one character. You'll usually see the first input character used as the last target character, so something like this:
```python
y[:, :-1], y[:, -1] = x[:, 1:], x[:, 0]
```
where `x` is the input batch and `y` is the target batch.
The way I like to do this window is use `range` to take steps of size `n_steps` from $0$ to `arr.shape[1]`, the total number of steps in each sequence. That way, the integers you get from `range` always point to the start of a batch, and each window is `n_steps` wide.
```
def get_batches(arr, n_seqs, n_steps):
'''Create a generator that returns batches of size
n_seqs x n_steps from arr.
Arguments
---------
arr: Array you want to make batches from
n_seqs: Batch size, the number of sequences per batch
n_steps: Number of sequence steps per batch
'''
# Get the number of characters per batch and number of batches we can make
characters_per_batch = n_seqs * n_steps
n_batches = len(arr)//characters_per_batch
# Keep only enough characters to make full batches
arr = arr[:n_batches * characters_per_batch]
# Reshape into n_seqs rows
arr = arr.reshape((n_seqs, -1))
for n in range(0, arr.shape[1], n_steps):
# The features
x = arr[:, n:n+n_steps]
# The targets, shifted by one
y = np.zeros_like(x)
y[:, :-1], y[:, -1] = x[:, 1:], x[:, 0]
yield x, y
```
Now I'll make my data sets and we can check out what's going on here. Here I'm going to use a batch size of 10 and 50 sequence steps.
```
batches = get_batches(encoded, 10, 50)
x, y = next(batches)
print('x\n', x[:10, :10])
print('\ny\n', y[:10, :10])
```
If you implemented `get_batches` correctly, the above output should look something like
```
x
[[55 63 69 22 6 76 45 5 16 35]
[ 5 69 1 5 12 52 6 5 56 52]
[48 29 12 61 35 35 8 64 76 78]
[12 5 24 39 45 29 12 56 5 63]
[ 5 29 6 5 29 78 28 5 78 29]
[ 5 13 6 5 36 69 78 35 52 12]
[63 76 12 5 18 52 1 76 5 58]
[34 5 73 39 6 5 12 52 36 5]
[ 6 5 29 78 12 79 6 61 5 59]
[ 5 78 69 29 24 5 6 52 5 63]]
y
[[63 69 22 6 76 45 5 16 35 35]
[69 1 5 12 52 6 5 56 52 29]
[29 12 61 35 35 8 64 76 78 28]
[ 5 24 39 45 29 12 56 5 63 29]
[29 6 5 29 78 28 5 78 29 45]
[13 6 5 36 69 78 35 52 12 43]
[76 12 5 18 52 1 76 5 58 52]
[ 5 73 39 6 5 12 52 36 5 78]
[ 5 29 78 12 79 6 61 5 59 63]
[78 69 29 24 5 6 52 5 63 76]]
```
although the exact numbers will be different. Check to make sure the data is shifted over one step for `y`.
## Building the model
Below is where you'll build the network. We'll break it up into parts so it's easier to reason about each bit. Then we can connect them up into the whole network.
<img src="assets/charRNN.png" width=500px>
### Inputs
First off we'll create our input placeholders. As usual we need placeholders for the training data and the targets. We'll also create a placeholder for dropout layers called `keep_prob`.
```
def build_inputs(batch_size, num_steps):
''' Define placeholders for inputs, targets, and dropout
Arguments
---------
batch_size: Batch size, number of sequences per batch
num_steps: Number of sequence steps in a batch
'''
# Declare placeholders we'll feed into the graph
inputs = tf.placeholder(tf.int32, [batch_size, num_steps], name='inputs')
targets = tf.placeholder(tf.int32, [batch_size, num_steps], name='targets')
# Keep probability placeholder for drop out layers
keep_prob = tf.placeholder(tf.float32, name='keep_prob')
return inputs, targets, keep_prob
```
### LSTM Cell
Here we will create the LSTM cell we'll use in the hidden layer. We'll use this cell as a building block for the RNN. So we aren't actually defining the RNN here, just the type of cell we'll use in the hidden layer.
We first create a basic LSTM cell with
```python
lstm = tf.contrib.rnn.BasicLSTMCell(num_units)
```
where `num_units` is the number of units in the hidden layers in the cell. Then we can add dropout by wrapping it with
```python
tf.contrib.rnn.DropoutWrapper(lstm, output_keep_prob=keep_prob)
```
You pass in a cell and it will automatically add dropout to the inputs or outputs. Finally, we can stack up the LSTM cells into layers with [`tf.contrib.rnn.MultiRNNCell`](https://www.tensorflow.org/versions/r1.0/api_docs/python/tf/contrib/rnn/MultiRNNCell). With this, you pass in a list of cells and it will send the output of one cell into the next cell. Previously with TensorFlow 1.0, you could do this
```python
tf.contrib.rnn.MultiRNNCell([cell]*num_layers)
```
This might look a little weird if you know Python well because this will create a list of the same `cell` object. However, TensorFlow 1.0 will create different weight matrices for all `cell` objects. But, starting with TensorFlow 1.1 you actually need to create new cell objects in the list. To get it to work in TensorFlow 1.1, it should look like
```python
def build_cell(num_units, keep_prob):
lstm = tf.contrib.rnn.BasicLSTMCell(num_units)
drop = tf.contrib.rnn.DropoutWrapper(lstm, output_keep_prob=keep_prob)
return drop
tf.contrib.rnn.MultiRNNCell([build_cell(num_units, keep_prob) for _ in range(num_layers)])
```
Even though this is actually multiple LSTM cells stacked on each other, you can treat the multiple layers as one cell.
We also need to create an initial cell state of all zeros. This can be done like so
```python
initial_state = cell.zero_state(batch_size, tf.float32)
```
Below, we implement the `build_lstm` function to create these LSTM cells and the initial state.
```
def build_lstm(lstm_size, num_layers, batch_size, keep_prob):
''' Build LSTM cell.
Arguments
---------
keep_prob: Scalar tensor (tf.placeholder) for the dropout keep probability
lstm_size: Size of the hidden layers in the LSTM cells
num_layers: Number of LSTM layers
batch_size: Batch size
'''
### Build the LSTM Cell
def build_cell(lstm_size, keep_prob):
# Use a basic LSTM cell
lstm = tf.contrib.rnn.BasicLSTMCell(lstm_size)
# Add dropout to the cell
drop = tf.contrib.rnn.DropoutWrapper(lstm, output_keep_prob=keep_prob)
return drop
# Stack up multiple LSTM layers, for deep learning
cell = tf.contrib.rnn.MultiRNNCell([build_cell(lstm_size, keep_prob) for _ in range(num_layers)])
initial_state = cell.zero_state(batch_size, tf.float32)
return cell, initial_state
```
### RNN Output
Here we'll create the output layer. We need to connect the output of the RNN cells to a full connected layer with a softmax output. The softmax output gives us a probability distribution we can use to predict the next character.
If our input has batch size $N$, number of steps $M$, and the hidden layer has $L$ hidden units, then the output is a 3D tensor with size $N \times M \times L$. The output of each LSTM cell has size $L$, we have $M$ of them, one for each sequence step, and we have $N$ sequences. So the total size is $N \times M \times L$.
We are using the same fully connected layer, the same weights, for each of the outputs. Then, to make things easier, we should reshape the outputs into a 2D tensor with shape $(M * N) \times L$. That is, one row for each sequence and step, where the values of each row are the output from the LSTM cells.
One we have the outputs reshaped, we can do the matrix multiplication with the weights. We need to wrap the weight and bias variables in a variable scope with `tf.variable_scope(scope_name)` because there are weights being created in the LSTM cells. TensorFlow will throw an error if the weights created here have the same names as the weights created in the LSTM cells, which they will be default. To avoid this, we wrap the variables in a variable scope so we can give them unique names.
```
def build_output(lstm_output, in_size, out_size):
''' Build a softmax layer, return the softmax output and logits.
Arguments
---------
x: Input tensor
in_size: Size of the input tensor, for example, size of the LSTM cells
out_size: Size of this softmax layer
'''
# Reshape output so it's a bunch of rows, one row for each step for each sequence.
# That is, the shape should be batch_size*num_steps rows by lstm_size columns
seq_output = tf.concat(lstm_output, axis=1)
x = tf.reshape(seq_output, [-1, in_size])
# Connect the RNN outputs to a softmax layer
with tf.variable_scope('softmax'):
softmax_w = tf.Variable(tf.truncated_normal((in_size, out_size), stddev=0.1))
softmax_b = tf.Variable(tf.zeros(out_size))
# Since output is a bunch of rows of RNN cell outputs, logits will be a bunch
# of rows of logit outputs, one for each step and sequence
logits = tf.matmul(x, softmax_w) + softmax_b
# Use softmax to get the probabilities for predicted characters
out = tf.nn.softmax(logits, name='predictions')
return out, logits
```
### Training loss
Next up is the training loss. We get the logits and targets and calculate the softmax cross-entropy loss. First we need to one-hot encode the targets, we're getting them as encoded characters. Then, reshape the one-hot targets so it's a 2D tensor with size $(M*N) \times C$ where $C$ is the number of classes/characters we have. Remember that we reshaped the LSTM outputs and ran them through a fully connected layer with $C$ units. So our logits will also have size $(M*N) \times C$.
Then we run the logits and targets through `tf.nn.softmax_cross_entropy_with_logits` and find the mean to get the loss.
```
def build_loss(logits, targets, lstm_size, num_classes):
''' Calculate the loss from the logits and the targets.
Arguments
---------
logits: Logits from final fully connected layer
targets: Targets for supervised learning
lstm_size: Number of LSTM hidden units
num_classes: Number of classes in targets
'''
# One-hot encode targets and reshape to match logits, one row per batch_size per step
y_one_hot = tf.one_hot(targets, num_classes)
y_reshaped = tf.reshape(y_one_hot, logits.get_shape())
# Softmax cross entropy loss
loss = tf.nn.softmax_cross_entropy_with_logits(logits=logits, labels=y_reshaped)
loss = tf.reduce_mean(loss)
return loss
```
### Optimizer
Here we build the optimizer. Normal RNNs have have issues gradients exploding and disappearing. LSTMs fix the disappearance problem, but the gradients can still grow without bound. To fix this, we can clip the gradients above some threshold. That is, if a gradient is larger than that threshold, we set it to the threshold. This will ensure the gradients never grow overly large. Then we use an AdamOptimizer for the learning step.
```
def build_optimizer(loss, learning_rate, grad_clip):
''' Build optmizer for training, using gradient clipping.
Arguments:
loss: Network loss
learning_rate: Learning rate for optimizer
'''
# Optimizer for training, using gradient clipping to control exploding gradients
tvars = tf.trainable_variables()
grads, _ = tf.clip_by_global_norm(tf.gradients(loss, tvars), grad_clip)
train_op = tf.train.AdamOptimizer(learning_rate)
optimizer = train_op.apply_gradients(zip(grads, tvars))
return optimizer
```
### Build the network
Now we can put all the pieces together and build a class for the network. To actually run data through the LSTM cells, we will use [`tf.nn.dynamic_rnn`](https://www.tensorflow.org/versions/r1.0/api_docs/python/tf/nn/dynamic_rnn). This function will pass the hidden and cell states across LSTM cells appropriately for us. It returns the outputs for each LSTM cell at each step for each sequence in the mini-batch. It also gives us the final LSTM state. We want to save this state as `final_state` so we can pass it to the first LSTM cell in the the next mini-batch run. For `tf.nn.dynamic_rnn`, we pass in the cell and initial state we get from `build_lstm`, as well as our input sequences. Also, we need to one-hot encode the inputs before going into the RNN.
```
class CharRNN:
def __init__(self, num_classes, batch_size=64, num_steps=50,
lstm_size=128, num_layers=2, learning_rate=0.001,
grad_clip=5, sampling=False):
# When we're using this network for sampling later, we'll be passing in
# one character at a time, so providing an option for that
if sampling == True:
batch_size, num_steps = 1, 1
else:
batch_size, num_steps = batch_size, num_steps
tf.reset_default_graph()
# Build the input placeholder tensors
self.inputs, self.targets, self.keep_prob = build_inputs(batch_size, num_steps)
# Build the LSTM cell
cell, self.initial_state = build_lstm(lstm_size, num_layers, batch_size, self.keep_prob)
### Run the data through the RNN layers
# First, one-hot encode the input tokens
x_one_hot = tf.one_hot(self.inputs, num_classes)
# Run each sequence step through the RNN and collect the outputs
outputs, state = tf.nn.dynamic_rnn(cell, x_one_hot, initial_state=self.initial_state)
self.final_state = state
# Get softmax predictions and logits
self.prediction, self.logits = build_output(outputs, lstm_size, num_classes)
# Loss and optimizer (with gradient clipping)
self.loss = build_loss(self.logits, self.targets, lstm_size, num_classes)
self.optimizer = build_optimizer(self.loss, learning_rate, grad_clip)
```
## Hyperparameters
Here I'm defining the hyperparameters for the network.
* `batch_size` - Number of sequences running through the network in one pass.
* `num_steps` - Number of characters in the sequence the network is trained on. Larger is better typically, the network will learn more long range dependencies. But it takes longer to train. 100 is typically a good number here.
* `lstm_size` - The number of units in the hidden layers.
* `num_layers` - Number of hidden LSTM layers to use
* `learning_rate` - Learning rate for training
* `keep_prob` - The dropout keep probability when training. If you're network is overfitting, try decreasing this.
Here's some good advice from Andrej Karpathy on training the network. I'm going to copy it in here for your benefit, but also link to [where it originally came from](https://github.com/karpathy/char-rnn#tips-and-tricks).
> ## Tips and Tricks
>### Monitoring Validation Loss vs. Training Loss
>If you're somewhat new to Machine Learning or Neural Networks it can take a bit of expertise to get good models. The most important quantity to keep track of is the difference between your training loss (printed during training) and the validation loss (printed once in a while when the RNN is run on the validation data (by default every 1000 iterations)). In particular:
> - If your training loss is much lower than validation loss then this means the network might be **overfitting**. Solutions to this are to decrease your network size, or to increase dropout. For example you could try dropout of 0.5 and so on.
> - If your training/validation loss are about equal then your model is **underfitting**. Increase the size of your model (either number of layers or the raw number of neurons per layer)
> ### Approximate number of parameters
> The two most important parameters that control the model are `lstm_size` and `num_layers`. I would advise that you always use `num_layers` of either 2/3. The `lstm_size` can be adjusted based on how much data you have. The two important quantities to keep track of here are:
> - The number of parameters in your model. This is printed when you start training.
> - The size of your dataset. 1MB file is approximately 1 million characters.
>These two should be about the same order of magnitude. It's a little tricky to tell. Here are some examples:
> - I have a 100MB dataset and I'm using the default parameter settings (which currently print 150K parameters). My data size is significantly larger (100 mil >> 0.15 mil), so I expect to heavily underfit. I am thinking I can comfortably afford to make `lstm_size` larger.
> - I have a 10MB dataset and running a 10 million parameter model. I'm slightly nervous and I'm carefully monitoring my validation loss. If it's larger than my training loss then I may want to try to increase dropout a bit and see if that helps the validation loss.
> ### Best models strategy
>The winning strategy to obtaining very good models (if you have the compute time) is to always err on making the network larger (as large as you're willing to wait for it to compute) and then try different dropout values (between 0,1). Whatever model has the best validation performance (the loss, written in the checkpoint filename, low is good) is the one you should use in the end.
>It is very common in deep learning to run many different models with many different hyperparameter settings, and in the end take whatever checkpoint gave the best validation performance.
>By the way, the size of your training and validation splits are also parameters. Make sure you have a decent amount of data in your validation set or otherwise the validation performance will be noisy and not very informative.
```
batch_size = 100 # Sequences per batch
num_steps = 100 # Number of sequence steps per batch
lstm_size = 512 # Size of hidden layers in LSTMs
num_layers = 2 # Number of LSTM layers
learning_rate = 0.001 # Learning rate
keep_prob = 0.5 # Dropout keep probability
```
## Time for training
This is typical training code, passing inputs and targets into the network, then running the optimizer. Here we also get back the final LSTM state for the mini-batch. Then, we pass that state back into the network so the next batch can continue the state from the previous batch. And every so often (set by `save_every_n`) I save a checkpoint.
Here I'm saving checkpoints with the format
`i{iteration number}_l{# hidden layer units}.ckpt`
```
epochs = 20
# Save every N iterations
save_every_n = 200
model = CharRNN(len(vocab), batch_size=batch_size, num_steps=num_steps,
lstm_size=lstm_size, num_layers=num_layers,
learning_rate=learning_rate)
saver = tf.train.Saver(max_to_keep=100)
with tf.Session() as sess:
sess.run(tf.global_variables_initializer())
# Use the line below to load a checkpoint and resume training
#saver.restore(sess, 'checkpoints/______.ckpt')
counter = 0
for e in range(epochs):
# Train network
new_state = sess.run(model.initial_state)
loss = 0
for x, y in get_batches(encoded, batch_size, num_steps):
counter += 1
start = time.time()
feed = {model.inputs: x,
model.targets: y,
model.keep_prob: keep_prob,
model.initial_state: new_state}
batch_loss, new_state, _ = sess.run([model.loss,
model.final_state,
model.optimizer],
feed_dict=feed)
end = time.time()
print('Epoch: {}/{}... '.format(e+1, epochs),
'Training Step: {}... '.format(counter),
'Training loss: {:.4f}... '.format(batch_loss),
'{:.4f} sec/batch'.format((end-start)))
if (counter % save_every_n == 0):
saver.save(sess, "checkpoints/i{}_l{}.ckpt".format(counter, lstm_size))
saver.save(sess, "checkpoints/i{}_l{}.ckpt".format(counter, lstm_size))
```
#### Saved checkpoints
Read up on saving and loading checkpoints here: https://www.tensorflow.org/programmers_guide/variables
```
tf.train.get_checkpoint_state('checkpoints')
```
## Sampling
Now that the network is trained, we'll can use it to generate new text. The idea is that we pass in a character, then the network will predict the next character. We can use the new one, to predict the next one. And we keep doing this to generate all new text. I also included some functionality to prime the network with some text by passing in a string and building up a state from that.
The network gives us predictions for each character. To reduce noise and make things a little less random, I'm going to only choose a new character from the top N most likely characters.
```
def pick_top_n(preds, vocab_size, top_n=5):
p = np.squeeze(preds)
p[np.argsort(p)[:-top_n]] = 0
p = p / np.sum(p)
c = np.random.choice(vocab_size, 1, p=p)[0]
return c
def sample(checkpoint, n_samples, lstm_size, vocab_size, prime="The "):
samples = [c for c in prime]
model = CharRNN(len(vocab), lstm_size=lstm_size, sampling=True)
saver = tf.train.Saver()
with tf.Session() as sess:
saver.restore(sess, checkpoint)
new_state = sess.run(model.initial_state)
for c in prime:
x = np.zeros((1, 1))
x[0,0] = vocab_to_int[c]
feed = {model.inputs: x,
model.keep_prob: 1.,
model.initial_state: new_state}
preds, new_state = sess.run([model.prediction, model.final_state],
feed_dict=feed)
c = pick_top_n(preds, len(vocab))
samples.append(int_to_vocab[c])
for i in range(n_samples):
x[0,0] = c
feed = {model.inputs: x,
model.keep_prob: 1.,
model.initial_state: new_state}
preds, new_state = sess.run([model.prediction, model.final_state],
feed_dict=feed)
c = pick_top_n(preds, len(vocab))
samples.append(int_to_vocab[c])
return ''.join(samples)
```
Here, pass in the path to a checkpoint and sample from the network.
```
tf.train.latest_checkpoint('checkpoints')
checkpoint = tf.train.latest_checkpoint('checkpoints')
samp = sample(checkpoint, 2000, lstm_size, len(vocab), prime="Far")
print(samp)
checkpoint = 'checkpoints/i200_l512.ckpt'
samp = sample(checkpoint, 1000, lstm_size, len(vocab), prime="Far")
print(samp)
checkpoint = 'checkpoints/i600_l512.ckpt'
samp = sample(checkpoint, 1000, lstm_size, len(vocab), prime="Far")
print(samp)
checkpoint = 'checkpoints/i1200_l512.ckpt'
samp = sample(checkpoint, 1000, lstm_size, len(vocab), prime="Far")
print(samp)
```
|
github_jupyter
|
import time
from collections import namedtuple
import numpy as np
import tensorflow as tf
with open('anna.txt', 'r') as f:
text=f.read()
vocab = sorted(set(text))
vocab_to_int = {c: i for i, c in enumerate(vocab)}
int_to_vocab = dict(enumerate(vocab))
encoded = np.array([vocab_to_int[c] for c in text], dtype=np.int32)
text[:100]
encoded[:100]
len(vocab)
y[:, :-1], y[:, -1] = x[:, 1:], x[:, 0]
def get_batches(arr, n_seqs, n_steps):
'''Create a generator that returns batches of size
n_seqs x n_steps from arr.
Arguments
---------
arr: Array you want to make batches from
n_seqs: Batch size, the number of sequences per batch
n_steps: Number of sequence steps per batch
'''
# Get the number of characters per batch and number of batches we can make
characters_per_batch = n_seqs * n_steps
n_batches = len(arr)//characters_per_batch
# Keep only enough characters to make full batches
arr = arr[:n_batches * characters_per_batch]
# Reshape into n_seqs rows
arr = arr.reshape((n_seqs, -1))
for n in range(0, arr.shape[1], n_steps):
# The features
x = arr[:, n:n+n_steps]
# The targets, shifted by one
y = np.zeros_like(x)
y[:, :-1], y[:, -1] = x[:, 1:], x[:, 0]
yield x, y
batches = get_batches(encoded, 10, 50)
x, y = next(batches)
print('x\n', x[:10, :10])
print('\ny\n', y[:10, :10])
x
[[55 63 69 22 6 76 45 5 16 35]
[ 5 69 1 5 12 52 6 5 56 52]
[48 29 12 61 35 35 8 64 76 78]
[12 5 24 39 45 29 12 56 5 63]
[ 5 29 6 5 29 78 28 5 78 29]
[ 5 13 6 5 36 69 78 35 52 12]
[63 76 12 5 18 52 1 76 5 58]
[34 5 73 39 6 5 12 52 36 5]
[ 6 5 29 78 12 79 6 61 5 59]
[ 5 78 69 29 24 5 6 52 5 63]]
y
[[63 69 22 6 76 45 5 16 35 35]
[69 1 5 12 52 6 5 56 52 29]
[29 12 61 35 35 8 64 76 78 28]
[ 5 24 39 45 29 12 56 5 63 29]
[29 6 5 29 78 28 5 78 29 45]
[13 6 5 36 69 78 35 52 12 43]
[76 12 5 18 52 1 76 5 58 52]
[ 5 73 39 6 5 12 52 36 5 78]
[ 5 29 78 12 79 6 61 5 59 63]
[78 69 29 24 5 6 52 5 63 76]]
```
although the exact numbers will be different. Check to make sure the data is shifted over one step for `y`.
## Building the model
Below is where you'll build the network. We'll break it up into parts so it's easier to reason about each bit. Then we can connect them up into the whole network.
<img src="assets/charRNN.png" width=500px>
### Inputs
First off we'll create our input placeholders. As usual we need placeholders for the training data and the targets. We'll also create a placeholder for dropout layers called `keep_prob`.
### LSTM Cell
Here we will create the LSTM cell we'll use in the hidden layer. We'll use this cell as a building block for the RNN. So we aren't actually defining the RNN here, just the type of cell we'll use in the hidden layer.
We first create a basic LSTM cell with
where `num_units` is the number of units in the hidden layers in the cell. Then we can add dropout by wrapping it with
You pass in a cell and it will automatically add dropout to the inputs or outputs. Finally, we can stack up the LSTM cells into layers with [`tf.contrib.rnn.MultiRNNCell`](https://www.tensorflow.org/versions/r1.0/api_docs/python/tf/contrib/rnn/MultiRNNCell). With this, you pass in a list of cells and it will send the output of one cell into the next cell. Previously with TensorFlow 1.0, you could do this
This might look a little weird if you know Python well because this will create a list of the same `cell` object. However, TensorFlow 1.0 will create different weight matrices for all `cell` objects. But, starting with TensorFlow 1.1 you actually need to create new cell objects in the list. To get it to work in TensorFlow 1.1, it should look like
Even though this is actually multiple LSTM cells stacked on each other, you can treat the multiple layers as one cell.
We also need to create an initial cell state of all zeros. This can be done like so
Below, we implement the `build_lstm` function to create these LSTM cells and the initial state.
### RNN Output
Here we'll create the output layer. We need to connect the output of the RNN cells to a full connected layer with a softmax output. The softmax output gives us a probability distribution we can use to predict the next character.
If our input has batch size $N$, number of steps $M$, and the hidden layer has $L$ hidden units, then the output is a 3D tensor with size $N \times M \times L$. The output of each LSTM cell has size $L$, we have $M$ of them, one for each sequence step, and we have $N$ sequences. So the total size is $N \times M \times L$.
We are using the same fully connected layer, the same weights, for each of the outputs. Then, to make things easier, we should reshape the outputs into a 2D tensor with shape $(M * N) \times L$. That is, one row for each sequence and step, where the values of each row are the output from the LSTM cells.
One we have the outputs reshaped, we can do the matrix multiplication with the weights. We need to wrap the weight and bias variables in a variable scope with `tf.variable_scope(scope_name)` because there are weights being created in the LSTM cells. TensorFlow will throw an error if the weights created here have the same names as the weights created in the LSTM cells, which they will be default. To avoid this, we wrap the variables in a variable scope so we can give them unique names.
### Training loss
Next up is the training loss. We get the logits and targets and calculate the softmax cross-entropy loss. First we need to one-hot encode the targets, we're getting them as encoded characters. Then, reshape the one-hot targets so it's a 2D tensor with size $(M*N) \times C$ where $C$ is the number of classes/characters we have. Remember that we reshaped the LSTM outputs and ran them through a fully connected layer with $C$ units. So our logits will also have size $(M*N) \times C$.
Then we run the logits and targets through `tf.nn.softmax_cross_entropy_with_logits` and find the mean to get the loss.
### Optimizer
Here we build the optimizer. Normal RNNs have have issues gradients exploding and disappearing. LSTMs fix the disappearance problem, but the gradients can still grow without bound. To fix this, we can clip the gradients above some threshold. That is, if a gradient is larger than that threshold, we set it to the threshold. This will ensure the gradients never grow overly large. Then we use an AdamOptimizer for the learning step.
### Build the network
Now we can put all the pieces together and build a class for the network. To actually run data through the LSTM cells, we will use [`tf.nn.dynamic_rnn`](https://www.tensorflow.org/versions/r1.0/api_docs/python/tf/nn/dynamic_rnn). This function will pass the hidden and cell states across LSTM cells appropriately for us. It returns the outputs for each LSTM cell at each step for each sequence in the mini-batch. It also gives us the final LSTM state. We want to save this state as `final_state` so we can pass it to the first LSTM cell in the the next mini-batch run. For `tf.nn.dynamic_rnn`, we pass in the cell and initial state we get from `build_lstm`, as well as our input sequences. Also, we need to one-hot encode the inputs before going into the RNN.
## Hyperparameters
Here I'm defining the hyperparameters for the network.
* `batch_size` - Number of sequences running through the network in one pass.
* `num_steps` - Number of characters in the sequence the network is trained on. Larger is better typically, the network will learn more long range dependencies. But it takes longer to train. 100 is typically a good number here.
* `lstm_size` - The number of units in the hidden layers.
* `num_layers` - Number of hidden LSTM layers to use
* `learning_rate` - Learning rate for training
* `keep_prob` - The dropout keep probability when training. If you're network is overfitting, try decreasing this.
Here's some good advice from Andrej Karpathy on training the network. I'm going to copy it in here for your benefit, but also link to [where it originally came from](https://github.com/karpathy/char-rnn#tips-and-tricks).
> ## Tips and Tricks
>### Monitoring Validation Loss vs. Training Loss
>If you're somewhat new to Machine Learning or Neural Networks it can take a bit of expertise to get good models. The most important quantity to keep track of is the difference between your training loss (printed during training) and the validation loss (printed once in a while when the RNN is run on the validation data (by default every 1000 iterations)). In particular:
> - If your training loss is much lower than validation loss then this means the network might be **overfitting**. Solutions to this are to decrease your network size, or to increase dropout. For example you could try dropout of 0.5 and so on.
> - If your training/validation loss are about equal then your model is **underfitting**. Increase the size of your model (either number of layers or the raw number of neurons per layer)
> ### Approximate number of parameters
> The two most important parameters that control the model are `lstm_size` and `num_layers`. I would advise that you always use `num_layers` of either 2/3. The `lstm_size` can be adjusted based on how much data you have. The two important quantities to keep track of here are:
> - The number of parameters in your model. This is printed when you start training.
> - The size of your dataset. 1MB file is approximately 1 million characters.
>These two should be about the same order of magnitude. It's a little tricky to tell. Here are some examples:
> - I have a 100MB dataset and I'm using the default parameter settings (which currently print 150K parameters). My data size is significantly larger (100 mil >> 0.15 mil), so I expect to heavily underfit. I am thinking I can comfortably afford to make `lstm_size` larger.
> - I have a 10MB dataset and running a 10 million parameter model. I'm slightly nervous and I'm carefully monitoring my validation loss. If it's larger than my training loss then I may want to try to increase dropout a bit and see if that helps the validation loss.
> ### Best models strategy
>The winning strategy to obtaining very good models (if you have the compute time) is to always err on making the network larger (as large as you're willing to wait for it to compute) and then try different dropout values (between 0,1). Whatever model has the best validation performance (the loss, written in the checkpoint filename, low is good) is the one you should use in the end.
>It is very common in deep learning to run many different models with many different hyperparameter settings, and in the end take whatever checkpoint gave the best validation performance.
>By the way, the size of your training and validation splits are also parameters. Make sure you have a decent amount of data in your validation set or otherwise the validation performance will be noisy and not very informative.
## Time for training
This is typical training code, passing inputs and targets into the network, then running the optimizer. Here we also get back the final LSTM state for the mini-batch. Then, we pass that state back into the network so the next batch can continue the state from the previous batch. And every so often (set by `save_every_n`) I save a checkpoint.
Here I'm saving checkpoints with the format
`i{iteration number}_l{# hidden layer units}.ckpt`
#### Saved checkpoints
Read up on saving and loading checkpoints here: https://www.tensorflow.org/programmers_guide/variables
## Sampling
Now that the network is trained, we'll can use it to generate new text. The idea is that we pass in a character, then the network will predict the next character. We can use the new one, to predict the next one. And we keep doing this to generate all new text. I also included some functionality to prime the network with some text by passing in a string and building up a state from that.
The network gives us predictions for each character. To reduce noise and make things a little less random, I'm going to only choose a new character from the top N most likely characters.
Here, pass in the path to a checkpoint and sample from the network.
| 0.789518 | 0.992108 |
## Tutorial for contextual-AI with H2O Model
This tutorial is to show how to create a wrapper for H2O model and make it compatible with contextual-ai library function calls.
We will train a simple `H2ORandomForestEstimator` model to classify the iris dataset, in the end we want to generate a model interpretation reports including the following:
- Feature importance globally from shap values
- Model per-class feature importance by aggregating explanations
Note: please install h2o library in order to run this tutorial.
```
import sys
import json
import sklearn
import sklearn.datasets
import sklearn.ensemble
import pandas as pd
import numpy as np
import h2o
from h2o.estimators.random_forest import H2ORandomForestEstimator
np.random.seed(1)
```
We will start the demo by training a H2O model with iris data.
### Initialize the H2O virtual cluster
```
h2o.init(max_mem_size = "500M", nthreads = 6)
# h2o.remove_all()
```
### Data preparation for training a H2O model
1. Load the dataset and split the data into train and test
```
iris = sklearn.datasets.load_iris()
feature_names = iris.feature_names
class_labels = 'species'
train, test, labels_train, labels_test = sklearn.model_selection.train_test_split(iris.data, iris.target, train_size=0.80)
```
2. Create the H2O Dataframe with column names and target column name
```
train_h2o_df = h2o.H2OFrame(train)
train_h2o_df.set_names(iris.feature_names)
train_h2o_df['species'] = h2o.H2OFrame(iris.target_names[labels_train])
train_h2o_df['species'] = train_h2o_df['species'].asfactor()
test_h2o_df = h2o.H2OFrame(test)
test_h2o_df.set_names(iris.feature_names)
test_h2o_df['species'] = h2o.H2OFrame(iris.target_names[labels_test])
test_h2o_df['species'] = test_h2o_df['species'].asfactor()
```
### Train a H2O Model
```
iris_h2o_model = H2ORandomForestEstimator(
model_id="iris_random_forest",
ntrees=500,
stopping_rounds=2,
score_each_iteration=True,
seed=1000000,
balance_classes=False,
histogram_type="AUTO")
# train
iris_h2o_model.train(x=feature_names, y='species', training_frame=train_h2o_df)
# evaluate
iris_h2o_model.model_performance(test_h2o_df)
```
## Wrapper class for H2O model
Next, we will need a wrapper class to make the H2O model behave like a scikit-learn model, which contextual-AI current supports. The wrapper mainly needs a predict function to fulfill the requirements that takes in a numpy array / pandas DataFrame and output an numpy array as prediction probability, while inside the wrapper, the input data is formatted to H2O DataFrame and feed to H2O model.
```
class h2o_predict_proba_wrapper:
def __init__(self,model,column_names):
"""
model: H2O model object
column_names: feature names from the training data frame
"""
self.model = model
self.column_names = column_names
def predict_proba(self,data_array):
'''
data_array: can be both numpy array type or pandas data frame
'''
shape_tuple = np.shape(data_array)
if len(shape_tuple) == 1:
data_array = data_array.reshape(1, -1)
# Convert a numpy array into H2O data frame
pandas_df = pd.DataFrame(data = data_array,columns = self.column_names)
h2o_df = h2o.H2OFrame(pandas_df)
# Predict with the H2O model
predictions = self.model.predict(h2o_df).as_data_frame()
# the first column is the class labels, the rest are probabilities for each class
predictions = predictions.iloc[:,1:].as_matrix()
return predictions
```
### Simple test on the wrapper
```
# initialize the wrapper
h2o_drf_wrapper = h2o_predict_proba_wrapper(iris_h2o_model,feature_names)
# convert H2O dataframe to pandas dataframe
test_pandas_df = test_h2o_df[feature_names].as_data_frame()
# call predict_proba from the wrapper to get the prediction probability
h2o_drf_wrapper.predict_proba(test_pandas_df)
```
## Contextual-AI Report Demo
1. Define the global virables
```
# convert data into pandas dataframe
train_pandas_df = train_h2o_df[feature_names].as_data_frame()
# initialize the h2o wrapper
h2o_drf_wrapper = h2o_predict_proba_wrapper(iris_h2o_model,feature_names)
# define the global variables used in the config file
X_train = train_pandas_df
target_names_list = ["versicolor","virginica","setosa"]
clf = h2o_drf_wrapper
clf_fn = h2o_drf_wrapper.predict_proba
feature_names = train_pandas_df.columns.tolist()
```
2. Define the config json file used for compiler
```
import json
json_config = 'iris_config.json'
with open(json_config) as file:
config = json.load(file)
config
```
3. Import the corresponding module and render the report
```
sys.path.append('../../..')
from xai.compiler.base import Configuration, Controller
controller = Controller(config=Configuration(config, locals()))
controller.render()
```
|
github_jupyter
|
import sys
import json
import sklearn
import sklearn.datasets
import sklearn.ensemble
import pandas as pd
import numpy as np
import h2o
from h2o.estimators.random_forest import H2ORandomForestEstimator
np.random.seed(1)
h2o.init(max_mem_size = "500M", nthreads = 6)
# h2o.remove_all()
iris = sklearn.datasets.load_iris()
feature_names = iris.feature_names
class_labels = 'species'
train, test, labels_train, labels_test = sklearn.model_selection.train_test_split(iris.data, iris.target, train_size=0.80)
train_h2o_df = h2o.H2OFrame(train)
train_h2o_df.set_names(iris.feature_names)
train_h2o_df['species'] = h2o.H2OFrame(iris.target_names[labels_train])
train_h2o_df['species'] = train_h2o_df['species'].asfactor()
test_h2o_df = h2o.H2OFrame(test)
test_h2o_df.set_names(iris.feature_names)
test_h2o_df['species'] = h2o.H2OFrame(iris.target_names[labels_test])
test_h2o_df['species'] = test_h2o_df['species'].asfactor()
iris_h2o_model = H2ORandomForestEstimator(
model_id="iris_random_forest",
ntrees=500,
stopping_rounds=2,
score_each_iteration=True,
seed=1000000,
balance_classes=False,
histogram_type="AUTO")
# train
iris_h2o_model.train(x=feature_names, y='species', training_frame=train_h2o_df)
# evaluate
iris_h2o_model.model_performance(test_h2o_df)
class h2o_predict_proba_wrapper:
def __init__(self,model,column_names):
"""
model: H2O model object
column_names: feature names from the training data frame
"""
self.model = model
self.column_names = column_names
def predict_proba(self,data_array):
'''
data_array: can be both numpy array type or pandas data frame
'''
shape_tuple = np.shape(data_array)
if len(shape_tuple) == 1:
data_array = data_array.reshape(1, -1)
# Convert a numpy array into H2O data frame
pandas_df = pd.DataFrame(data = data_array,columns = self.column_names)
h2o_df = h2o.H2OFrame(pandas_df)
# Predict with the H2O model
predictions = self.model.predict(h2o_df).as_data_frame()
# the first column is the class labels, the rest are probabilities for each class
predictions = predictions.iloc[:,1:].as_matrix()
return predictions
# initialize the wrapper
h2o_drf_wrapper = h2o_predict_proba_wrapper(iris_h2o_model,feature_names)
# convert H2O dataframe to pandas dataframe
test_pandas_df = test_h2o_df[feature_names].as_data_frame()
# call predict_proba from the wrapper to get the prediction probability
h2o_drf_wrapper.predict_proba(test_pandas_df)
# convert data into pandas dataframe
train_pandas_df = train_h2o_df[feature_names].as_data_frame()
# initialize the h2o wrapper
h2o_drf_wrapper = h2o_predict_proba_wrapper(iris_h2o_model,feature_names)
# define the global variables used in the config file
X_train = train_pandas_df
target_names_list = ["versicolor","virginica","setosa"]
clf = h2o_drf_wrapper
clf_fn = h2o_drf_wrapper.predict_proba
feature_names = train_pandas_df.columns.tolist()
import json
json_config = 'iris_config.json'
with open(json_config) as file:
config = json.load(file)
config
sys.path.append('../../..')
from xai.compiler.base import Configuration, Controller
controller = Controller(config=Configuration(config, locals()))
controller.render()
| 0.389198 | 0.976378 |
# Degree Planner Backend Algorithm
## TODO:
* In class Graph, use dict for nodes
* In class Graph, finish isCompleted()
* Add all components to Node class (full name, description, etc.)
* In Graph, nodesToRemove(), return 2 lists: required, and possible to take
* Finish God
```
class Node(object):
name = ""
credits = None
prereq = []
required = False
completed = False
description = ""
fullName = ""
priorityNumber = 0 # Number of edges to that node
offeredF = False
offeredS = False
offeredSu = False
offeredEvenYear = False
offeredOddYear = False
def __init__(self, n, p, c, r):
self.name = n
if p != None:
self.prereq = p
self.required = r
def __str__(self):
if len(self.prereq) == 0:
return str(self.name) + ' [] ' + str(self.required)
retVal = self.name
retVal += ' ['
for item in self.prereq:
retVal += (str(item.name) + ', ')
retVal = retVal[:-2]
retVal += '] '
retVal += str(self.required)
return retVal
def canComplete(self):
if self.completed == True:
return False
for pr in self.prereq:
if pr.completed == False:
return False
return True
class Graph(object):
nodes = {}
def __init__(self, nodesDict):
self.nodes = nodesDict
def __str__(self):
return self.nodes
def nodesToRemove(self):
retList = []
for key, val in self.nodes.items():
#print(str(node))
if(val.canComplete()):
retList.append(val)
retList.sort(key=lambda x: x.priorityNumber, reverse=True)
return retList
def isCompleted(self): #Sets degree requirements
for node in self.nodes:
#print(str(node))
if(not node.completed):
return False
return True
def completeCourses(listOfCourses):
for course in listOfCourses:
for catalogCourse in nodes:
if course.name == catalogCourse.name:
catalogCourse.completed = True
listOfCourses.remove(course)
for course in listOfCourses:
nodes.append(course)
class God(object): #God algorithm!
catalogYear = None
coursesComplete = []
allCourses = []
terms = []
termCredits = []
graph = None
def __init__(self, catalogY, coursesComplete):
self.catalogYear = catalogY
self.coursesComplete = coursesComplete
letThereBeLight()
def getTerms(self):
return None
def letThereBeLight(self):
# get courses from a specific year catalog
self.allCourses = testNodes
self.graph = Graph(self.allCourses)
self.graph.completeCourses(self.coursesComplete)
genNumber = 0
while(not graph.isCompleted()):
gen = createGeneration(genNumber)
self.terms.append(gen)
genNumber += 1
# Choose high priority classes, then required, then possible to take
def createGeneration(self, genNum):
credits = 0
while(self.termCredits >= credits):
classToAdd = GetClass(self.termCredits-credits)
def GetClass(credits):
pass
A = Node('A', None, 3, True)
B = Node('B', [A], 3, True)
C = Node('C', None, 3, False)
D = Node('B', [B], 3, True)
E = Node('B', [B], 3, True)
F = Node('B', [B], 3, True)
A.completed = True
testNodes = {'A':A, 'B':B, 'C':C, 'D':D, 'E':E, 'F':F}
str(B)
catalogTest = Graph(testNodes)
catalogTest.nodes
catalogTest.nodesToRemove()
str(catalogTest.nodesToRemove()[0])
str(catalogTest.nodesToRemove()[1])
```
|
github_jupyter
|
class Node(object):
name = ""
credits = None
prereq = []
required = False
completed = False
description = ""
fullName = ""
priorityNumber = 0 # Number of edges to that node
offeredF = False
offeredS = False
offeredSu = False
offeredEvenYear = False
offeredOddYear = False
def __init__(self, n, p, c, r):
self.name = n
if p != None:
self.prereq = p
self.required = r
def __str__(self):
if len(self.prereq) == 0:
return str(self.name) + ' [] ' + str(self.required)
retVal = self.name
retVal += ' ['
for item in self.prereq:
retVal += (str(item.name) + ', ')
retVal = retVal[:-2]
retVal += '] '
retVal += str(self.required)
return retVal
def canComplete(self):
if self.completed == True:
return False
for pr in self.prereq:
if pr.completed == False:
return False
return True
class Graph(object):
nodes = {}
def __init__(self, nodesDict):
self.nodes = nodesDict
def __str__(self):
return self.nodes
def nodesToRemove(self):
retList = []
for key, val in self.nodes.items():
#print(str(node))
if(val.canComplete()):
retList.append(val)
retList.sort(key=lambda x: x.priorityNumber, reverse=True)
return retList
def isCompleted(self): #Sets degree requirements
for node in self.nodes:
#print(str(node))
if(not node.completed):
return False
return True
def completeCourses(listOfCourses):
for course in listOfCourses:
for catalogCourse in nodes:
if course.name == catalogCourse.name:
catalogCourse.completed = True
listOfCourses.remove(course)
for course in listOfCourses:
nodes.append(course)
class God(object): #God algorithm!
catalogYear = None
coursesComplete = []
allCourses = []
terms = []
termCredits = []
graph = None
def __init__(self, catalogY, coursesComplete):
self.catalogYear = catalogY
self.coursesComplete = coursesComplete
letThereBeLight()
def getTerms(self):
return None
def letThereBeLight(self):
# get courses from a specific year catalog
self.allCourses = testNodes
self.graph = Graph(self.allCourses)
self.graph.completeCourses(self.coursesComplete)
genNumber = 0
while(not graph.isCompleted()):
gen = createGeneration(genNumber)
self.terms.append(gen)
genNumber += 1
# Choose high priority classes, then required, then possible to take
def createGeneration(self, genNum):
credits = 0
while(self.termCredits >= credits):
classToAdd = GetClass(self.termCredits-credits)
def GetClass(credits):
pass
A = Node('A', None, 3, True)
B = Node('B', [A], 3, True)
C = Node('C', None, 3, False)
D = Node('B', [B], 3, True)
E = Node('B', [B], 3, True)
F = Node('B', [B], 3, True)
A.completed = True
testNodes = {'A':A, 'B':B, 'C':C, 'D':D, 'E':E, 'F':F}
str(B)
catalogTest = Graph(testNodes)
catalogTest.nodes
catalogTest.nodesToRemove()
str(catalogTest.nodesToRemove()[0])
str(catalogTest.nodesToRemove()[1])
| 0.247169 | 0.749294 |
<center>
<img src="https://cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud/IBMDeveloperSkillsNetwork-PY0101EN-SkillsNetwork/IDSNlogo.png" width="300" alt="cognitiveclass.ai logo" />
</center>
# Reading Files Python
Estimated time needed: **40** minutes
## Objectives
After completing this lab you will be able to:
- Read text files using Python libraries
<h2>Table of Contents</h2>
<div class="alert alert-block alert-info" style="margin-top: 20px">
<ul>
<li><a href="download">Download Data</a></li>
<li><a href="read">Reading Text Files</a></li>
<li><a href="better">A Better Way to Open a File</a></li>
</ul>
</div>
<hr>
<h2 id="download">Download Data</h2>
```
import urllib.request
url = 'https://cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud/IBMDeveloperSkillsNetwork-PY0101EN-SkillsNetwork/labs/Module%204/data/example1.txt'
filename = 'Example1.txt'
urllib.request.urlretrieve(url, filename)
# Download Example file
!wget -O /resources/data/Example1.txt https://cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud/IBMDeveloperSkillsNetwork-PY0101EN-SkillsNetwork/labs/Module%204/data/example1.txt
```
<hr>
<h2 id="read">Reading Text Files</h2>
One way to read or write a file in Python is to use the built-in <code>open</code> function. The <code>open</code> function provides a <b>File object</b> that contains the methods and attributes you need in order to read, save, and manipulate the file. In this notebook, we will only cover <b>.txt</b> files. The first parameter you need is the file path and the file name. An example is shown as follow:
<img src="https://cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud/IBMDeveloperSkillsNetwork-PY0101EN-SkillsNetwork/labs/Module%204/images/ReadOpen.png" width="500" />
The mode argument is optional and the default value is <b>r</b>. In this notebook we only cover two modes:
<ul>
<li><b>r</b> Read mode for reading files </li>
<li><b>w</b> Write mode for writing files</li>
</ul>
For the next example, we will use the text file <b>Example1.txt</b>. The file is shown as follow:
<img src="https://cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud/IBMDeveloperSkillsNetwork-PY0101EN-SkillsNetwork/labs/Module%204/images/ReadFile.png" width="100" />
We read the file:
```
# Read the Example1.txt
example1 = "Example1.txt"
file1 = open(example1, "r")
```
We can view the attributes of the file.
The name of the file:
```
# Print the path of file
file1.name
```
The mode the file object is in:
```
# Print the mode of file, either 'r' or 'w'
file1.mode
```
We can read the file and assign it to a variable :
```
# Read the file
FileContent = file1.read()
FileContent
```
The <b>/n</b> means that there is a new line.
We can print the file:
```
# Print the file with '\n' as a new line
print(FileContent)
```
The file is of type string:
```
# Type of file content
type(FileContent)
```
It is very important that the file is closed in the end. This frees up resources and ensures consistency across different python versions.
```
# Close file after finish
file1.close()
```
<hr>
<h2 id="better">A Better Way to Open a File</h2>
Using the <code>with</code> statement is better practice, it automatically closes the file even if the code encounters an exception. The code will run everything in the indent block then close the file object.
```
# Open file using with
with open(example1, "r") as file1:
FileContent = file1.read()
print(FileContent)
```
The file object is closed, you can verify it by running the following cell:
```
# Verify if the file is closed
file1.closed
```
We can see the info in the file:
```
# See the content of file
print(FileContent)
```
The syntax is a little confusing as the file object is after the <code>as</code> statement. We also don’t explicitly close the file. Therefore we summarize the steps in a figure:
<img src="https://cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud/IBMDeveloperSkillsNetwork-PY0101EN-SkillsNetwork/labs/Module%204/images/ReadWith.png" width="500" />
We don’t have to read the entire file, for example, we can read the first 4 characters by entering three as a parameter to the method **.read()**:
```
# Read first four characters
with open(example1, "r") as file1:
print(file1.read(4))
```
Once the method <code>.read(4)</code> is called the first 4 characters are called. If we call the method again, the next 4 characters are called. The output for the following cell will demonstrate the process for different inputs to the method <code>read()</code>:
```
# Read certain amount of characters
with open(example1, "r") as file1:
print(file1.read(4))
print(file1.read(4))
print(file1.read(7))
print(file1.read(15))
```
The process is illustrated in the below figure, and each color represents the part of the file read after the method <code>read()</code> is called:
<img src="https://cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud/IBMDeveloperSkillsNetwork-PY0101EN-SkillsNetwork/labs/Module%204/images/read.png" width="500" />
Here is an example using the same file, but instead we read 16, 5, and then 9 characters at a time:
```
# Read certain amount of characters
with open(example1, "r") as file1:
print(file1.read(16))
print(file1.read(5))
print(file1.read(9))
```
We can also read one line of the file at a time using the method <code>readline()</code>:
```
# Read one line
with open(example1, "r") as file1:
print("first line: " + file1.readline())
```
We can also pass an argument to <code> readline() </code> to specify the number of charecters we want to read. However, unlike <code> read()</code>, <code> readline()</code> can only read one line at most.
```
with open(example1, "r") as file1:
print(file1.readline(20)) # does not read past the end of line
print(file1.read(20)) # Returns the next 20 chars
```
We can use a loop to iterate through each line:
```
# Iterate through the lines
with open(example1,"r") as file1:
i = 0;
for line in file1:
print("Iteration", str(i), ": ", line)
i = i + 1
```
We can use the method <code>readlines()</code> to save the text file to a list:
```
# Read all lines and save as a list
with open(example1, "r") as file1:
FileasList = file1.readlines()
```
Each element of the list corresponds to a line of text:
```
# Print the first line
FileasList[0]
```
# Print the second line
FileasList[1]
```
# Print the third line
FileasList[2]
```
<hr>
<h2>The last exercise!</h2>
<p>Congratulations, you have completed your first lesson and hands-on lab in Python. However, there is one more thing you need to do. The Data Science community encourages sharing work. The best way to share and showcase your work is to share it on GitHub. By sharing your notebook on GitHub you are not only building your reputation with fellow data scientists, but you can also show it off when applying for a job. Even though this was your first piece of work, it is never too early to start building good habits. So, please read and follow <a href="https://cognitiveclass.ai/blog/data-scientists-stand-out-by-sharing-your-notebooks/" target="_blank">this article</a> to learn how to share your work.
<hr>
## Author
<a href="https://www.linkedin.com/in/joseph-s-50398b136/" target="_blank">Joseph Santarcangelo</a>
## Other contributors
<a href="www.linkedin.com/in/jiahui-mavis-zhou-a4537814a">Mavis Zhou</a>
## Change Log
| Date (YYYY-MM-DD) | Version | Changed By | Change Description |
| ----------------- | ------- | ------------- | --------------------------------------------------------- |
| 2020-09-30 | 1.3 | Malika | Deleted exericse "Weather Data" |
| 2020-09-30 | 1.2 | Malika Singla | Weather Data dataset link added |
| 2020-09-30 | 1.1 | Arjun Swani | Added exericse "Weather Data" |
| 2020-09-30 | 1.0 | Arjun Swani | Added blurbs about closing files and read() vs readline() |
| 2020-08-26 | 0.2 | Lavanya | Moved lab to course repo in GitLab |
| | | | |
| | | | |
<hr/>
## <h3 align="center"> © IBM Corporation 2020. All rights reserved. <h3/>
|
github_jupyter
|
import urllib.request
url = 'https://cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud/IBMDeveloperSkillsNetwork-PY0101EN-SkillsNetwork/labs/Module%204/data/example1.txt'
filename = 'Example1.txt'
urllib.request.urlretrieve(url, filename)
# Download Example file
!wget -O /resources/data/Example1.txt https://cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud/IBMDeveloperSkillsNetwork-PY0101EN-SkillsNetwork/labs/Module%204/data/example1.txt
# Read the Example1.txt
example1 = "Example1.txt"
file1 = open(example1, "r")
# Print the path of file
file1.name
# Print the mode of file, either 'r' or 'w'
file1.mode
# Read the file
FileContent = file1.read()
FileContent
# Print the file with '\n' as a new line
print(FileContent)
# Type of file content
type(FileContent)
# Close file after finish
file1.close()
# Open file using with
with open(example1, "r") as file1:
FileContent = file1.read()
print(FileContent)
# Verify if the file is closed
file1.closed
# See the content of file
print(FileContent)
# Read first four characters
with open(example1, "r") as file1:
print(file1.read(4))
# Read certain amount of characters
with open(example1, "r") as file1:
print(file1.read(4))
print(file1.read(4))
print(file1.read(7))
print(file1.read(15))
# Read certain amount of characters
with open(example1, "r") as file1:
print(file1.read(16))
print(file1.read(5))
print(file1.read(9))
# Read one line
with open(example1, "r") as file1:
print("first line: " + file1.readline())
with open(example1, "r") as file1:
print(file1.readline(20)) # does not read past the end of line
print(file1.read(20)) # Returns the next 20 chars
# Iterate through the lines
with open(example1,"r") as file1:
i = 0;
for line in file1:
print("Iteration", str(i), ": ", line)
i = i + 1
# Read all lines and save as a list
with open(example1, "r") as file1:
FileasList = file1.readlines()
# Print the first line
FileasList[0]
# Print the third line
FileasList[2]
| 0.346431 | 0.949949 |
# Kevin Daum - Ch. 1 HW
Problems: 1-4, 6, 8, 10-12, 14, 16, 17,18,20,22,24,26,29-31 for all students
33,35,42 for grad students
```
%matplotlib inline
import numpy as np
import scipy.stats as st
import matplotlib.pyplot as plt
import pandas as pd
import statsmodels.formula.api as sm
from sklearn.linear_model import LinearRegression
import seaborn as sns
from IPython.display import Image
```
## 1.
If the clerical errors are made _after_ charging the customers, e.g. when recording the sales amounts, I would argue that the actual relationship between units sold and dollar sales is still functional. However, if the errors are made _before_ charging customers, so that there are actually inconsistencies in dollars made for sales, then **no, the relation is no longer functional**, since it can no longer be expressed as a mathematical formula.
## 2.
$Y = 2X + 300$. It's a functional relation.
## 3.
I tend to agree with the objection; however, it's not a perfect world and I think that a stakeholder wanting to control production processes should do their due diligence by taking samples and doing a regression analysis to test whether the relation is actually mathematical.
## 4.
The random error term $\epsilon_i$
## 8.
Yes, $E\{Y\}$ would still be 104. No, the Y value for this new observation would probably not be 108.
## 10.
No, because all the observations were made at the same point in time. It is likely that each of the programmers' individual salaries will continually increase during the duration of their tenure at the company. The curvilinear relationship is probably an indication of some other factor(s), perhaps that programmers older than 47 tend to work less hours or were originally hired at a time when salaries were significantly lower or when the position was less skilled. We don't know, but we do know that the study tells us nothing about what happens to an individual programmer's salary over time.
# 11.
```
def before_training(x):
return x
def after_training(x):
return 20 + 0.95*x
xmin, xmax = 40, 100
# plot production output before training in blue
plt.plot([xmin, xmax], [before_training(xmin), before_training(xmax)], 'b-')
# plot production output after training in red
plt.plot([xmin, xmax], [after_training(xmin), after_training(xmax)], 'r-')
plt.show()
```
As you can see, production after training is higher than production before training over the entire possible range of X values.
## 12.
a. observational
b. Because the investigator used observational data rather than a controlled experiment, there is not adequate information to claim and cause-and-effect relationship between exercise and number of colds.
c. weather, ability, proximity to other people
d. The investigator could organize a designed experiment with a cohort of participants for which all other relevant variables are roughly equal. Participants would be randomly assigned amounts of weekly exercise.
## 14.
```
treatments = list(range(1,5))
print(treatments)
batches = list(range(1,17))
print(batches)
print()
assignments = {}
for t in treatments:
assignments[t] = []
for i in range(4):
assignments[t].append(batches.pop(np.random.random_integers(0,len(batches)-1)))
print(assignments)
```
## 16.
$Y(X)$ will not be normally distributed; we're hoping that $Y(X)$ will have a linear relationship. However, I think it is true that $Y_i$ is normally distributed, since we expect values of $Y_i$ to be centered around $\beta_0 + \beta_1X_i$.
## 17.
We don't estimate $b_0$ and $b_1$; $b_0$ and $b_1$ _are_ the estimates of $\beta_0$ and $\beta_1$ in (1.1).
## 18.
Yes, because when regression model (1.1) is fitted to a set of $n$ cases by the method of least squares, I think $\epsilon_i = e_i$. And even outside of a least squares fit, by definition in (1.1), $E(\epsilon_i) = 0$, which I think would mean that $\sum \epsilon_i = 0$ no matter the method of fit.
## 20.
a.
$\hat Y = − 0.580 + 15.0352X$
b.
```
Image(filename='/Users/kevin/Dropbox/School/STA-580/ch1hw/plot20.png')
```
It's a good fit. The Pearson correlation coefficient is 0.979.
c. $b_0$ would be the time taken to service 0 copiers, which is meaningless.
d.
```
-0.580+15.0352*5
```
$\hat Y(5)$ = 74.60 minutes
## 22.
a.
$\hat Y = 168.600 + 2.03438X$
```
Image(filename='/Users/kevin/Dropbox/School/STA-580/ch1hw/plot22.png')
```
Yes, it's a good fit. The Pearson correlation coefficient is 0.986.
b.
```
168.600+2.03438*40
```
$\hat Y(40) =$ 249.98 Brinell units
c. 2.034 Brinell units
## 24.
a.
I'm following along with http://connor-johnson.com/2014/02/18/linear-regression-with-python/.
```
df24 = pd.read_table('/Users/kevin/Dropbox/School/STA-580/ch1hw/Ch1-20.csv', sep=',', index_col=False)
df24['Eins'] = np.ones(( len(df24), ))
df24.head() # show first five rows
y24 = df24.time
x24 = df24[['copiers', 'Eins']]
result24 = sm.OLS(y24, x24).fit()
result24.summary()
# residuals
result24.resid
# sum of squared residuals
result24.ssr
```
_I just realized that I'm not supposed to turn these problems in until the group project, so I'll move on to the next one..._
## 26.
a.
```
df26 = pd.read_table('/Users/kevin/Dropbox/School/STA-580/ch1hw/Ch1-22.csv', sep=',', index_col=False)
df26['Eins'] = np.ones(( len(df26), ))
df26
# let's see what the data looks like
sns.lmplot('time', 'hardness', df26)
# perform the least-squares linear regression
y26 = df26.hardness
x26 = df26[['time', 'Eins']]
result26 = sm.OLS(y26, x26).fit()
# residuals
result26.resid
result26.resid.sum()
```
The residuals sure do sum to zero!
b.
```
# variance
result26.resid.var()
# standard deviation
result26.resid.std()
```
$\sigma$ is expressed in the same units as $Y$, Brinell (hardness) units.
## 29.
It just means that the y-intercept will be zero, so the regression function will go through the point (0,0).
## 30.
It means that the regression function has a slope of zero, meaning it will plot on a graph as a horizontal line.
## 31.
No, it would not. I would expect the error terms to be much smaller in this case. No, I would expect that the error terms _would_ be correlated since they are coming from the same experimental unit. For example, if the item has a lower-than-average hardness at the first measurement, it is not likely to suddenly start hardening quicker and have a higher-than-average hardness in another measurement; it will likely have lower-than-average hardness values for all of the subsequent measurements.
## 33.
It's been ten years since I've done any calculus, but I think I can follow the example on p. 17 of the text. Our regression model is
$$Y_i = \beta_0 + \epsilon_i$$
Substituting $\beta_1=0$ into (1.8) we have
$$Q = \sum_{i=1}^n (Y_i-\beta_0)^2 \tag{33.1}$$
We want to minimize Q. Differentiating (33.1) with respect to $\beta_0$ gives
$$\frac{\partial Q}{\partial \beta_0} = -2\sum_{i=1}^n (Y_i-\beta_0) \tag {33.2}$$
We set (33.2) equal to zero and use $b_0$ as the value of $\beta_0$ that minimizes Q:
$$ -2\sum_{i=1}^n (Y_i-\beta_0) = 0 $$
Expanding, we have
$$ \sum Y_i - nb_0 = 0 $$
Solving for $b_0$, we end up with
$$ b_0 = \frac{\sum Y_i}{n} = \bar Y$$
## 35.
(1.18) follows easily from (1.17):
$$\sum_{i=1}^n e_i = 0 \tag{1.17}$$
Substituting for $e_i$ from (1.16), we have:
$$\sum_{i=1}^n Y_i - \hat Y_i = 0$$
Adding $\sum_{i=1}^n \hat Y_i$ to both sides, we end up with (1.18):
$$\sum_{i=1}^n Y_i = \sum_{i=1}^n \hat Y_i \tag{1.18}$$
## 42.
a.
Substituting $\beta_0 = 0$, $n=6$ and $\sigma^2 = 16$ into (1.26), we have:
$$L(\beta_1) = \frac{1}{(32\pi)^3}exp\left[-\frac{1}{32}\sum_{i=1}^n(Y_i-\beta_{1}X_i)^2\right]$$
b.
```
data42 = {7:128, 12:213, 4:75, 14:250, 25:446, 30:540}
def galley_likelihood(b1, data):
summation = sum([(y-b1*x)**2 for (x,y) in data.items()])
return 1/((32*np.pi)**3)*np.exp(-1/32*summation)
b1_values = range(17,20)
for b in b1_values:
print ('L({b}) = {L}'.format(b=b, L=galley_likelihood(b,data42)))
```
The likelihood function is largest for $\beta_1 = 18$.
c.
```
sum([x*y for x,y in data42.items()])/sum([x**2 for x in data42.keys()])
```
Yes, this is consistent with my results in (b).
d.
```
likelihood_data = {'b1': [], 'likelihood': []}
for b in b1_values:
likelihood_data['b1'].append(b)
likelihood_data['likelihood'].append(galley_likelihood(b,data42))
likelihood_data
df42_likelihood = pd.DataFrame(likelihood_data)
df42_likelihood
sns.jointplot(x='b1', y='likelihood', data=df42_likelihood, ylim=(-1e-08,5e-07))
```
I'm just plotting the values of $L(\beta_1)$ for the three given values of $\beta_1$ that I found in (b). I'm not sure if that's the likelihood function the question is asking me to plot. Of course this agrees with my result in (c).
|
github_jupyter
|
%matplotlib inline
import numpy as np
import scipy.stats as st
import matplotlib.pyplot as plt
import pandas as pd
import statsmodels.formula.api as sm
from sklearn.linear_model import LinearRegression
import seaborn as sns
from IPython.display import Image
def before_training(x):
return x
def after_training(x):
return 20 + 0.95*x
xmin, xmax = 40, 100
# plot production output before training in blue
plt.plot([xmin, xmax], [before_training(xmin), before_training(xmax)], 'b-')
# plot production output after training in red
plt.plot([xmin, xmax], [after_training(xmin), after_training(xmax)], 'r-')
plt.show()
treatments = list(range(1,5))
print(treatments)
batches = list(range(1,17))
print(batches)
print()
assignments = {}
for t in treatments:
assignments[t] = []
for i in range(4):
assignments[t].append(batches.pop(np.random.random_integers(0,len(batches)-1)))
print(assignments)
Image(filename='/Users/kevin/Dropbox/School/STA-580/ch1hw/plot20.png')
-0.580+15.0352*5
Image(filename='/Users/kevin/Dropbox/School/STA-580/ch1hw/plot22.png')
168.600+2.03438*40
df24 = pd.read_table('/Users/kevin/Dropbox/School/STA-580/ch1hw/Ch1-20.csv', sep=',', index_col=False)
df24['Eins'] = np.ones(( len(df24), ))
df24.head() # show first five rows
y24 = df24.time
x24 = df24[['copiers', 'Eins']]
result24 = sm.OLS(y24, x24).fit()
result24.summary()
# residuals
result24.resid
# sum of squared residuals
result24.ssr
df26 = pd.read_table('/Users/kevin/Dropbox/School/STA-580/ch1hw/Ch1-22.csv', sep=',', index_col=False)
df26['Eins'] = np.ones(( len(df26), ))
df26
# let's see what the data looks like
sns.lmplot('time', 'hardness', df26)
# perform the least-squares linear regression
y26 = df26.hardness
x26 = df26[['time', 'Eins']]
result26 = sm.OLS(y26, x26).fit()
# residuals
result26.resid
result26.resid.sum()
# variance
result26.resid.var()
# standard deviation
result26.resid.std()
data42 = {7:128, 12:213, 4:75, 14:250, 25:446, 30:540}
def galley_likelihood(b1, data):
summation = sum([(y-b1*x)**2 for (x,y) in data.items()])
return 1/((32*np.pi)**3)*np.exp(-1/32*summation)
b1_values = range(17,20)
for b in b1_values:
print ('L({b}) = {L}'.format(b=b, L=galley_likelihood(b,data42)))
sum([x*y for x,y in data42.items()])/sum([x**2 for x in data42.keys()])
likelihood_data = {'b1': [], 'likelihood': []}
for b in b1_values:
likelihood_data['b1'].append(b)
likelihood_data['likelihood'].append(galley_likelihood(b,data42))
likelihood_data
df42_likelihood = pd.DataFrame(likelihood_data)
df42_likelihood
sns.jointplot(x='b1', y='likelihood', data=df42_likelihood, ylim=(-1e-08,5e-07))
| 0.566858 | 0.88996 |
```
import os, json
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
import seaborn as sns
import string, re
import sys, zipfile
from collections import Counter
from keras.callbacks import EarlyStopping, ModelCheckpoint, ReduceLROnPlateau
from keras.initializers import Constant
from keras.layers import Dense, Input, GlobalMaxPooling1D
from keras.layers import Conv1D, MaxPooling1D, Embedding
from keras.layers import Activation, Dense, Dropout, SpatialDropout1D
from keras.layers import Bidirectional, CuDNNLSTM
from keras.models import Sequential, Model
from keras.preprocessing.text import Tokenizer
from keras.preprocessing.sequence import pad_sequences
from keras.utils import to_categorical
from nltk.corpus import names, stopwords, words
from nltk.stem import WordNetLemmatizer
from scipy import stats
from sklearn.metrics import f1_score, make_scorer, classification_report, confusion_matrix, roc_curve, roc_auc_score
from sklearn.model_selection import train_test_split
sys.path.insert(0, './code/')
from text_cleaning import TextClean
%matplotlib inline
glove = "../glove/glove.840B.300d.zip"
embedding_dimension = 300
max_words = 20000
val_split = 0.1
a = TextClean('.', app_name='temple_')
df = pd.read_csv('app_ratings.csv')
df.dropna(subset=['cleaned_text'], how='all', inplace = True)
df_p = df.loc[(df['stars'] > 3) & (df['polsum'] > 0.1)]
df_l = df.loc[(df['stars'] < 3) & (df['polsum'] < 0.1)]
X = list(df_p['cleaned_text']) + list(df_l['cleaned_text'])
y = list(df_p['stars']) + list(df_l['stars'])
X_, y_ = a.even_sample(X, y)
x_a = pd.DataFrame(X_)
x_a['y'] = y_
x_a = x_a.loc[x_a['y']!=3]
x_a['label'] = [1 if x > 3 else 0 for x in list(x_a['y'])]
X_ = list(x_a[0])
y = list(x_a['label'])
sequence_length = max([len(x) for x in X_]) + 1
tokenizer = Tokenizer(num_words=max_words, split=' ', oov_token='<unk>', filters=' ')
tokenizer.fit_on_texts(X_)
X = tokenizer.texts_to_sequences(X_)
X = pad_sequences(X, sequence_length)
y = np.array(y)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.1)
print("test set size " + str(len(X_test)))
vocab_size = min(max_words, len(tokenizer.word_index)) + 1
def glove(glove_dir, wordindex):
embedding_weights = {}
count_all_words = 0
with zipfile.ZipFile(glove_dir) as z:
with z.open("glove.840B.300d.txt") as f:
for line in f:
vals = line.split()
word = str(vals[0].decode("utf-8"))
if word in wordindex:
count_all_words+=1
coefs = np.asarray(vals[1:], dtype='float32')
coefs/=np.linalg.norm(coefs)
embedding_weights[word] = coefs
if count_all_words==len(wordindex) - 1:
break
return embedding_weights
embeddings_index = glove("./glove/glove.840B.300d.zip", tokenizer.word_index)
embedding_matrix = np.zeros((vocab_size, 300))
for word, i in tokenizer.word_index.items():
embedding_vector = embeddings_index.get(word)
if embedding_vector is not None:
embedding_matrix[i] = embedding_vector
else:
# doesn't exist, assign a random vector
embedding_matrix[i] = np.random.randn(300)
model = Sequential([
Embedding(vocab_size, embedding_dimension, embeddings_initializer=Constant(embedding_matrix),
input_length=sequence_length,trainable=False),
SpatialDropout1D(0.5),
Bidirectional(CuDNNLSTM(64, return_sequences=True)),
Dropout(0.5),
Bidirectional(CuDNNLSTM(64)),
Dropout(0.5),
Dense(units=1, activation='sigmoid')
])
model.compile(loss = 'binary_crossentropy', optimizer='adam', metrics = ['accuracy'])
print(model.summary())
batch_size = 64
early_stopping = EarlyStopping(monitor='val_loss', patience=5, mode='min')
save_best = ModelCheckpoint('reviews_binary_lstm.hdf', save_best_only=True,
monitor='val_loss', mode='min')
history = model.fit(X_train, y_train,
epochs=20,
batch_size=batch_size,
callbacks=[early_stopping,save_best],
verbose=1, validation_split=0.1)
pd.DataFrame(history.history).plot(figsize=(8, 5))
plt.axis([0, 20, 0, 1.0])
plt.grid(True)
plt.show()
preds = model.predict_classes(X_test)
matrix = confusion_matrix(y_test, preds)
sns.heatmap(matrix, square=True, annot=True, fmt='d', cbar=False,
xticklabels=[0,1],yticklabels=[0,1])
plt.xlabel('predicted label')
plt.ylabel('label actual')
print(classification_report(y_test, preds))
import pickle
with open('binary_tokenizer.pickle', 'wb') as handle:
pickle.dump(tokenizer, handle, protocol=pickle.HIGHEST_PROTOCOL)
info = {
'tokenizer':'binary_tokenizer.pickle',
'model':'reviews_binary_lstm.hdf',
'max_sequence':sequence_length,
'max_words':max_words,
'vocab_size':vocab_size
}
import json
with open('binary_project_info.json', 'w') as outfile:
json.dump(info, outfile)
```
|
github_jupyter
|
import os, json
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
import seaborn as sns
import string, re
import sys, zipfile
from collections import Counter
from keras.callbacks import EarlyStopping, ModelCheckpoint, ReduceLROnPlateau
from keras.initializers import Constant
from keras.layers import Dense, Input, GlobalMaxPooling1D
from keras.layers import Conv1D, MaxPooling1D, Embedding
from keras.layers import Activation, Dense, Dropout, SpatialDropout1D
from keras.layers import Bidirectional, CuDNNLSTM
from keras.models import Sequential, Model
from keras.preprocessing.text import Tokenizer
from keras.preprocessing.sequence import pad_sequences
from keras.utils import to_categorical
from nltk.corpus import names, stopwords, words
from nltk.stem import WordNetLemmatizer
from scipy import stats
from sklearn.metrics import f1_score, make_scorer, classification_report, confusion_matrix, roc_curve, roc_auc_score
from sklearn.model_selection import train_test_split
sys.path.insert(0, './code/')
from text_cleaning import TextClean
%matplotlib inline
glove = "../glove/glove.840B.300d.zip"
embedding_dimension = 300
max_words = 20000
val_split = 0.1
a = TextClean('.', app_name='temple_')
df = pd.read_csv('app_ratings.csv')
df.dropna(subset=['cleaned_text'], how='all', inplace = True)
df_p = df.loc[(df['stars'] > 3) & (df['polsum'] > 0.1)]
df_l = df.loc[(df['stars'] < 3) & (df['polsum'] < 0.1)]
X = list(df_p['cleaned_text']) + list(df_l['cleaned_text'])
y = list(df_p['stars']) + list(df_l['stars'])
X_, y_ = a.even_sample(X, y)
x_a = pd.DataFrame(X_)
x_a['y'] = y_
x_a = x_a.loc[x_a['y']!=3]
x_a['label'] = [1 if x > 3 else 0 for x in list(x_a['y'])]
X_ = list(x_a[0])
y = list(x_a['label'])
sequence_length = max([len(x) for x in X_]) + 1
tokenizer = Tokenizer(num_words=max_words, split=' ', oov_token='<unk>', filters=' ')
tokenizer.fit_on_texts(X_)
X = tokenizer.texts_to_sequences(X_)
X = pad_sequences(X, sequence_length)
y = np.array(y)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.1)
print("test set size " + str(len(X_test)))
vocab_size = min(max_words, len(tokenizer.word_index)) + 1
def glove(glove_dir, wordindex):
embedding_weights = {}
count_all_words = 0
with zipfile.ZipFile(glove_dir) as z:
with z.open("glove.840B.300d.txt") as f:
for line in f:
vals = line.split()
word = str(vals[0].decode("utf-8"))
if word in wordindex:
count_all_words+=1
coefs = np.asarray(vals[1:], dtype='float32')
coefs/=np.linalg.norm(coefs)
embedding_weights[word] = coefs
if count_all_words==len(wordindex) - 1:
break
return embedding_weights
embeddings_index = glove("./glove/glove.840B.300d.zip", tokenizer.word_index)
embedding_matrix = np.zeros((vocab_size, 300))
for word, i in tokenizer.word_index.items():
embedding_vector = embeddings_index.get(word)
if embedding_vector is not None:
embedding_matrix[i] = embedding_vector
else:
# doesn't exist, assign a random vector
embedding_matrix[i] = np.random.randn(300)
model = Sequential([
Embedding(vocab_size, embedding_dimension, embeddings_initializer=Constant(embedding_matrix),
input_length=sequence_length,trainable=False),
SpatialDropout1D(0.5),
Bidirectional(CuDNNLSTM(64, return_sequences=True)),
Dropout(0.5),
Bidirectional(CuDNNLSTM(64)),
Dropout(0.5),
Dense(units=1, activation='sigmoid')
])
model.compile(loss = 'binary_crossentropy', optimizer='adam', metrics = ['accuracy'])
print(model.summary())
batch_size = 64
early_stopping = EarlyStopping(monitor='val_loss', patience=5, mode='min')
save_best = ModelCheckpoint('reviews_binary_lstm.hdf', save_best_only=True,
monitor='val_loss', mode='min')
history = model.fit(X_train, y_train,
epochs=20,
batch_size=batch_size,
callbacks=[early_stopping,save_best],
verbose=1, validation_split=0.1)
pd.DataFrame(history.history).plot(figsize=(8, 5))
plt.axis([0, 20, 0, 1.0])
plt.grid(True)
plt.show()
preds = model.predict_classes(X_test)
matrix = confusion_matrix(y_test, preds)
sns.heatmap(matrix, square=True, annot=True, fmt='d', cbar=False,
xticklabels=[0,1],yticklabels=[0,1])
plt.xlabel('predicted label')
plt.ylabel('label actual')
print(classification_report(y_test, preds))
import pickle
with open('binary_tokenizer.pickle', 'wb') as handle:
pickle.dump(tokenizer, handle, protocol=pickle.HIGHEST_PROTOCOL)
info = {
'tokenizer':'binary_tokenizer.pickle',
'model':'reviews_binary_lstm.hdf',
'max_sequence':sequence_length,
'max_words':max_words,
'vocab_size':vocab_size
}
import json
with open('binary_project_info.json', 'w') as outfile:
json.dump(info, outfile)
| 0.447702 | 0.253237 |
```
import pandas as pd
import numpy as np
from sklearn.preprocessing import MinMaxScaler
from sklearn.preprocessing import StandardScaler
from sklearn import ensemble
from sklearn.utils import shuffle
from sklearn.metrics import mean_squared_error
from sklearn.metrics import r2_score
from sklearn.model_selection import train_test_split
from sklearn.multioutput import MultiOutputRegressor
from sklearn.ensemble import RandomForestRegressor
from matplotlib import pyplot as plt
def find_nearest(array,value):
idx = (np.abs(array-value)).argmin()
return array[idx]
def refine_data(arr1,arr2,arr3):
refined_data = []
for i in range(arr1.shape[0]):
rt = find_nearest(arr1[i],arr3[i])
refined_data.append(rt.round(3))
refined_data = np.array(refined_data)
return refined_data
def get_metrics(arr1,arr2):
mse = mean_squared_error(arr1,arr2)
r2 = r2_score(arr1,arr2)
return mse,r2
X = pd.read_csv('X_data_t3',sep = ',')
y = pd.read_csv('y_data',sep = ',')
time = np.array(pd.read_csv('time',sep = ',',header = None).dropna(axis = 'columns'))
abundance = np.array(pd.read_csv('abundance',sep = ',',header = None).dropna(axis = 'columns'))
baseline = np.array(pd.read_csv('baseline',sep = ',',header = None).dropna(axis = 'columns'))
X,y,time,abundance,baseline = shuffle(X,y,time,abundance,baseline)
Xleft = np.array(X[['maxRT_t','x_start_t','diff_start']])
Xright = np.array(X[['maxRT_t','x_end_t','diff_end']])
#Xleft = np.array(X[['maxRT_t','maxRT_ab','maxRT_baseline','x_start_t','x_start_ab']])
#Xright = np.array(X[['maxRT_t','maxRT_ab','maxRT_baseline','x_end_t','x_end_ab']])
#Xleft = np.array(X[['maxRT_t','maxRT_ab','x_start_t','x_start_ab','diff_start']])
#Xright = np.array(X[['maxRT_t','maxRT_ab','x_end_t','x_end_ab','diff_end']])
yleft = np.array(y['y_left_t'])
yright = np.array(y['y_right_t'])
scaler_left = MinMaxScaler(feature_range = (0,1))
Xleft = scaler_left.fit_transform(Xleft)
scaler_right = MinMaxScaler(feature_range = (0,1))
Xright = scaler_right.fit_transform(Xright)
Xright
test_size = 0.05
random_state = 42
Xleft_train,Xleft_test,yleft_train,yleft_test,tleft_train,tleft_test,aleft_train,aleft_test = train_test_split(Xleft,yleft,time,abundance,test_size=test_size,random_state=random_state)
Xright_train,Xright_test,yright_train,yright_test,tright_train,tright_test,aright_train,aright_test = train_test_split(Xright,yright,time,abundance,test_size=test_size,random_state=random_state)
n_estimators = np.linspace(1000000,2000000,2,dtype=int)
max_depth = np.linspace(100000,200000,2,dtype=int)
mse_left,r2_left = [[] for i in range(len(n_estimators))], [[] for i in range(len(n_estimators))]
mse_right,r2_right = [[] for i in range(len(n_estimators))], [[] for i in range(len(n_estimators))]
yleft_, yright_ = [[] for i in range(len(n_estimators))], [[] for i in range(len(n_estimators))]
for i in range(len(n_estimators)):
for j in range(len(max_depth)):
params = {'n_estimators': n_estimators[i], 'max_depth': max_depth[j], 'min_samples_split': 5,
'learning_rate': 0.01, 'loss': 'ls'}
clf_left = ensemble.GradientBoostingRegressor(**params)
clf_right = ensemble.GradientBoostingRegressor(**params)
clf_left.fit(Xleft_train,yleft_train)
clf_right.fit(Xright_train,yright_train)
yleft_predict = clf_left.predict(Xleft_test).round(3)
yright_predict = clf_right.predict(Xright_test).round(3)
yleft_refined = refine_data(tleft_test,aleft_test,yleft_predict)
yright_refined = refine_data(tright_test,aright_test,yright_predict)
mse_1,r2_1 = get_metrics(yleft_test,yleft_predict)
mse_2,r2_2 = get_metrics(yright_test,yright_predict)
mse_left[i].append(mse_1)
r2_left[i].append(r2_1)
mse_right[i].append(mse_2)
r2_right[i].append(r2_2)
yleft_[i].append(yleft_refined)
yright_[i].append(yright_refined)
yleft_ = np.array(yleft_)
yright_ = np.array(yright_)
mse_left = np.array(mse_left)
mse_right = np.array(mse_right)
r2_left = np.array(r2_left)
r2_right = np.array(r2_right)
r2_left
r2_right
mse_left
mse_right
yleft_[1][1]
yleft_test
```
|
github_jupyter
|
import pandas as pd
import numpy as np
from sklearn.preprocessing import MinMaxScaler
from sklearn.preprocessing import StandardScaler
from sklearn import ensemble
from sklearn.utils import shuffle
from sklearn.metrics import mean_squared_error
from sklearn.metrics import r2_score
from sklearn.model_selection import train_test_split
from sklearn.multioutput import MultiOutputRegressor
from sklearn.ensemble import RandomForestRegressor
from matplotlib import pyplot as plt
def find_nearest(array,value):
idx = (np.abs(array-value)).argmin()
return array[idx]
def refine_data(arr1,arr2,arr3):
refined_data = []
for i in range(arr1.shape[0]):
rt = find_nearest(arr1[i],arr3[i])
refined_data.append(rt.round(3))
refined_data = np.array(refined_data)
return refined_data
def get_metrics(arr1,arr2):
mse = mean_squared_error(arr1,arr2)
r2 = r2_score(arr1,arr2)
return mse,r2
X = pd.read_csv('X_data_t3',sep = ',')
y = pd.read_csv('y_data',sep = ',')
time = np.array(pd.read_csv('time',sep = ',',header = None).dropna(axis = 'columns'))
abundance = np.array(pd.read_csv('abundance',sep = ',',header = None).dropna(axis = 'columns'))
baseline = np.array(pd.read_csv('baseline',sep = ',',header = None).dropna(axis = 'columns'))
X,y,time,abundance,baseline = shuffle(X,y,time,abundance,baseline)
Xleft = np.array(X[['maxRT_t','x_start_t','diff_start']])
Xright = np.array(X[['maxRT_t','x_end_t','diff_end']])
#Xleft = np.array(X[['maxRT_t','maxRT_ab','maxRT_baseline','x_start_t','x_start_ab']])
#Xright = np.array(X[['maxRT_t','maxRT_ab','maxRT_baseline','x_end_t','x_end_ab']])
#Xleft = np.array(X[['maxRT_t','maxRT_ab','x_start_t','x_start_ab','diff_start']])
#Xright = np.array(X[['maxRT_t','maxRT_ab','x_end_t','x_end_ab','diff_end']])
yleft = np.array(y['y_left_t'])
yright = np.array(y['y_right_t'])
scaler_left = MinMaxScaler(feature_range = (0,1))
Xleft = scaler_left.fit_transform(Xleft)
scaler_right = MinMaxScaler(feature_range = (0,1))
Xright = scaler_right.fit_transform(Xright)
Xright
test_size = 0.05
random_state = 42
Xleft_train,Xleft_test,yleft_train,yleft_test,tleft_train,tleft_test,aleft_train,aleft_test = train_test_split(Xleft,yleft,time,abundance,test_size=test_size,random_state=random_state)
Xright_train,Xright_test,yright_train,yright_test,tright_train,tright_test,aright_train,aright_test = train_test_split(Xright,yright,time,abundance,test_size=test_size,random_state=random_state)
n_estimators = np.linspace(1000000,2000000,2,dtype=int)
max_depth = np.linspace(100000,200000,2,dtype=int)
mse_left,r2_left = [[] for i in range(len(n_estimators))], [[] for i in range(len(n_estimators))]
mse_right,r2_right = [[] for i in range(len(n_estimators))], [[] for i in range(len(n_estimators))]
yleft_, yright_ = [[] for i in range(len(n_estimators))], [[] for i in range(len(n_estimators))]
for i in range(len(n_estimators)):
for j in range(len(max_depth)):
params = {'n_estimators': n_estimators[i], 'max_depth': max_depth[j], 'min_samples_split': 5,
'learning_rate': 0.01, 'loss': 'ls'}
clf_left = ensemble.GradientBoostingRegressor(**params)
clf_right = ensemble.GradientBoostingRegressor(**params)
clf_left.fit(Xleft_train,yleft_train)
clf_right.fit(Xright_train,yright_train)
yleft_predict = clf_left.predict(Xleft_test).round(3)
yright_predict = clf_right.predict(Xright_test).round(3)
yleft_refined = refine_data(tleft_test,aleft_test,yleft_predict)
yright_refined = refine_data(tright_test,aright_test,yright_predict)
mse_1,r2_1 = get_metrics(yleft_test,yleft_predict)
mse_2,r2_2 = get_metrics(yright_test,yright_predict)
mse_left[i].append(mse_1)
r2_left[i].append(r2_1)
mse_right[i].append(mse_2)
r2_right[i].append(r2_2)
yleft_[i].append(yleft_refined)
yright_[i].append(yright_refined)
yleft_ = np.array(yleft_)
yright_ = np.array(yright_)
mse_left = np.array(mse_left)
mse_right = np.array(mse_right)
r2_left = np.array(r2_left)
r2_right = np.array(r2_right)
r2_left
r2_right
mse_left
mse_right
yleft_[1][1]
yleft_test
| 0.521715 | 0.607489 |
## First Last - Homework \#3
```
import numpy as np
from astropy.table import QTable
```
#### The columns for the dataset 'MainBelt.csv' are:
* `Col 1: Asteroid Name and Number (string)`
* `Col 2: Asteroid Absolute Magnitude (float)`
* `Col 3: Asteroid Albedo (float)`
### Read in the datafile `MainBelt.csv`
### Write a function to compute the Diameters of the MBAs
### Write a function to compute the Masses of the MBAs. Assume $\rho$ = 3,000 kg/m$^3$
### List the names of the 5 most massive MBAs
### List the names of the 5 least massive MBAs
### Determine the total mass of all of the MBAs
### Determine the fraction of the total mass contained in the 5 largest MBAs
### How does the total mass of the MBAs compare to the mass of the Moon ($7.35\ \times\ 10^{22}$ kg)?
### How many MBAs does it take to get 90% of the total mass of the MBAs (start from the most massive)
### Due Tue Jan 24 - 5pm
- `Make sure to change the filename to your name!`
- `Make sure to change the Title to your name!`
- `File -> Download as -> HTML (.html)`
- `upload your .html and .ipynb file to the class Canvas page`
***
## Absolute Magnitude, Size and Mass
The absolute magnitude **H** of a solar system object is the magnitude that it would have if it was 1 AU from the Earth and 1 AU from the Sun while having a phase angle of 0 degrees. Remember, astronomical magnitudes are "backwards". The magnitude of a bright object is *less* than the magnitude of a dimmer object.
The phase angle **$\theta$** in astronomical observations is the angle between the light incident onto an observed object and the light reflected from the object. In the context of astronomical observations, this is usually the angle $\angle$ Sun-object-observer.

It is actually impossible for an object to be 1 AU from the Earth and 1 AU from the Sun while having a phase angle of 0 degrees. The Sun and the Earth would occupy the same point. However, this is the definition of **H**. Please note, that the definition of absolute magnitude for solar system objects is *very* different from the definition of absolute magnitude used for stars.
The Absolute magnitude of an object is related to the physical size of the object via the equation:
$$\large D\ (\textrm{in km}) = \frac{1329}{\sqrt{A}}\ 10^{-0.2H}$$
Where **D** is the diameter of the object in kilometers and **A** is the geometric albedo of the object.
The geometric albedo **A** of an astronomical body is the ratio of its actual brightness at zero phase angle (i.e., as seen from the light source) to that of an idealized flat, fully reflecting, diffusively scattering disk with the same cross-section.
Perfect reflectors of light have A = 1 and perfect absorbers have **A** = 0. Typical asteroids have a wide range of values: 0.02 $<$ A $<$ 0.8.
Once we have the diameter of the object, we can determine the mass if we know the density ($\rho$).
$ \large \mathrm{Mass} = \rho \cdot \mathrm{Volume} = \rho \cdot \frac{4}{3} \pi r^3 \ = \ \rho \cdot \frac{1}{6} \pi D^3 $
This assumes that the object is spherical (this is **not** a very good assumption for asteroids).
|
github_jupyter
|
import numpy as np
from astropy.table import QTable
| 0.380299 | 0.968974 |
## Pandas
### Instructions
This assignment will be done completely inside this Jupyter notebook with answers placed in the cell provided.
All python imports that are needed shown.
Follow all the instructions in this notebook to complete these tasks.
Make sure the CSV data files is in the same folder as this notebook - alumni.csv, groceries.csv
```
from google.colab import drive
drive.mount('/content/gdrive', force_remount= True)
import sys
sys.path.append('/content/gdrive/My Drive/DataScienceSchool/ADS-Assignment-1')
# Imports needed to complete this assignment
import pandas as pd
import matplotlib.pyplot as plt
%matplotlib inline
import seaborn as sns
```
### Question 1 : Import CSV file (1 Mark)
Write code to load the alumni csv dataset into a Pandas DataFrame called 'alumni'.
```
#q1 (1)
alumni_df = pd.read_csv("/content/gdrive/My Drive/DataScienceSchool/ADS-Assignment-1/alumni.csv")
alumni_df.head()
```
### Question 2 : Understand the data set (5 Marks)
Use the following pandas commands to understand the data set: a) head, b) tail, c) dtypes, d) info, e) describe
```
#a) (1)
alumni_df.head()
#b) (1)
alumni_df.tail()
#c) (1)
alumni_df.dtypes
#d) (1)
alumni_df.info
#e) (1)
alumni_df.describe
```
### Question 3 : Cleaning the data set - part A (3 Marks)
a) Use clean_currency method below to strip out commas and dollar signs from Savings ($) column and put into a new column called 'Savings'.
```
def clean_currency(curr):
return float(curr.replace(",", "").replace("$", ""))
clean_currency("$66,000")
#a) (2)
alumni_df['Savings'] = alumni_df['Savings ($)'].apply(clean_currency)
alumni_df
```
b) Uncomment 'alumni.dtypes.Savings' to check that the type change has occurred
```
#b) (1)
alumni_df.dtypes.Savings
```
### Question 4 : Cleaning the data set - part B (5 Marks)
a) Run the 'alumni["Gender"].value_counts()' to see the incorrect 'M' fields that need to be converted to 'Male'
```
# a) (1)
alumni_df["Gender"].value_counts()
```
b) Now use a '.str.replace' on the 'Gender' column to covert the incorrect 'M' fields. Hint: We must use ^...$ to restrict the pattern to match the whole string.
```
# b) (1)
alumni_df["Gender"].str.replace("M","Male")
# b) (1)
alumni_df["Gender"].str.replace("^[M]$","Male")
```
c) That didn't the set alumni["Gender"] column however. You will need to update the column when using the replace command 'alumni["Gender"]=<replace command>', show how this is done below
```
# c) (1)
alumni_df["Gender"] = alumni_df["Gender"].str.replace("^[M]$","Male")
alumni_df["Gender"]
```
d) You can set it directly by using the df.loc command, show how this can be done by using the 'df.loc[row_indexer,col_indexer] = value' command to convert the 'M' to 'Male'
```
# d) (1)
alumni_df.loc[[1, 2,3], "Gender"] = "Male"
alumni_df["Gender"]
```
e) Now run the 'value_counts' for Gender again to see the correct columns - 'Male' and 'Female'
```
# e) (1)
alumni_df.value_counts
```
### Question 5 : Working with the data set (4)
a) get the median, b) mean and c) standard deviation for the 'Salary' column
```
# a)(1)
alumni_df.median
# b)(1)
alumni_df.mean
# c)(1)
alumni_df.std
```
d) identify which alumni paid more than $15000 in fees, using the 'Fee' column
```
# d) (1)
alumni_df[alumni_df.Fee >15000]
```
### Question 6 : Visualise the data set (4 Marks)
a) Using the 'Diploma Type' column, plot a bar chart and show its value counts.
```
#a) (1)
alumni_df["Diploma Type"].value_counts()
```
b) Now create a box plot comparison between 'Savings' and 'Salary' columns
```
#b) (1)
alumni_df["Diploma Type"].value_counts().plot(kind = "bar")
```
c) Generate a histogram with the 'Salary' column and use 12 bins.
```
#c) (1)
alumni_df["Salary"].plot(kind = "hist" , bins = 10)
```
d) Generate a scatter plot comparing 'Salary' and 'Savings' columns.
```
#d) (1)
alumni_df.plot(kind = "scatter", x = "Savings", y = "Salary")
```
### Question 7 : Contingency Table (2 Marks)
Using both the 'Martial Status' and 'Defaulted' create a contingency table. Hint: crosstab
```
# Q7 (2)
alumni_df["Marital Status"].value_counts()
alumni_df["Defaulted"].value_counts()
pd.crosstab(index=alumni_df["Marital Status"], columns=alumni_df["Defaulted"])
```
|
github_jupyter
|
from google.colab import drive
drive.mount('/content/gdrive', force_remount= True)
import sys
sys.path.append('/content/gdrive/My Drive/DataScienceSchool/ADS-Assignment-1')
# Imports needed to complete this assignment
import pandas as pd
import matplotlib.pyplot as plt
%matplotlib inline
import seaborn as sns
#q1 (1)
alumni_df = pd.read_csv("/content/gdrive/My Drive/DataScienceSchool/ADS-Assignment-1/alumni.csv")
alumni_df.head()
#a) (1)
alumni_df.head()
#b) (1)
alumni_df.tail()
#c) (1)
alumni_df.dtypes
#d) (1)
alumni_df.info
#e) (1)
alumni_df.describe
def clean_currency(curr):
return float(curr.replace(",", "").replace("$", ""))
clean_currency("$66,000")
#a) (2)
alumni_df['Savings'] = alumni_df['Savings ($)'].apply(clean_currency)
alumni_df
#b) (1)
alumni_df.dtypes.Savings
# a) (1)
alumni_df["Gender"].value_counts()
# b) (1)
alumni_df["Gender"].str.replace("M","Male")
# b) (1)
alumni_df["Gender"].str.replace("^[M]$","Male")
# c) (1)
alumni_df["Gender"] = alumni_df["Gender"].str.replace("^[M]$","Male")
alumni_df["Gender"]
# d) (1)
alumni_df.loc[[1, 2,3], "Gender"] = "Male"
alumni_df["Gender"]
# e) (1)
alumni_df.value_counts
# a)(1)
alumni_df.median
# b)(1)
alumni_df.mean
# c)(1)
alumni_df.std
# d) (1)
alumni_df[alumni_df.Fee >15000]
#a) (1)
alumni_df["Diploma Type"].value_counts()
#b) (1)
alumni_df["Diploma Type"].value_counts().plot(kind = "bar")
#c) (1)
alumni_df["Salary"].plot(kind = "hist" , bins = 10)
#d) (1)
alumni_df.plot(kind = "scatter", x = "Savings", y = "Salary")
# Q7 (2)
alumni_df["Marital Status"].value_counts()
alumni_df["Defaulted"].value_counts()
pd.crosstab(index=alumni_df["Marital Status"], columns=alumni_df["Defaulted"])
| 0.207375 | 0.870046 |
This notebook is part of the `nbsphinx` documentation: https://nbsphinx.readthedocs.io/.
# Markdown Cells
We can use *emphasis*, **boldface**, `preformatted text`.
> It looks like strike-out text is not supported: ~~strikethrough~~.
* Red
* Green
* Blue
Note: JupyterLab and JupyterNotebook uses a different Markdown parser than nbsphinx (which currently uses Pandoc).
In case that your Bulletpoints do render in the notebook and do not render with nbsphinx, please add one blank line before the bulletpoints.
***
1. One
1. Two
1. Three
Arbitrary Unicode characters should be supported, e.g. łßō.
Note, however, that this only works if your HTML browser and your LaTeX processor provide the appropriate fonts.
## Equations
Inline equations like $\text{e}^{i\pi} = -1$
can be created by putting a LaTeX expression between two dollar signs, like this:
`$\text{e}^{i\pi} = -1$`.
<div class="alert alert-info">
Note
Avoid leading and trailing spaces around math expressions, otherwise errors like the following will occur when Sphinx is running:
ERROR: Unknown interpreted text role "raw-latex".
See also the [pandoc docs](https://pandoc.org/MANUAL.html#math):
> Anything between two `$` characters will be treated as TeX math. The opening `$` must have a non-space character immediately to its right, while the closing `$` must have a non-space character immediately to its left, and must not be followed immediately by a digit.
</div>
Equations can also be displayed on their own line like this:
\begin{equation}
\int\limits_{-\infty}^\infty f(x) \delta(x - x_0) dx = f(x_0).
\end{equation}
This can be done by simply using one of the LaTeX math environments, like so:
```
\begin{equation}
\int\limits_{-\infty}^\infty f(x) \delta(x - x_0) dx = f(x_0)
\end{equation}
```
<div class="alert alert-info">
Note
For equations to be shown in HTML output,
you have to specify a
[math extension](https://www.sphinx-doc.org/en/master/usage/extensions/math.html)
in your [extensions](usage.ipynb#extensions) setting, e.g.:
```python
extensions = [
'nbsphinx',
'sphinx.ext.mathjax',
# ... other useful extensions ...
]
```
</div>
### Automatic Equation Numbering
This is not automatically enabled in Jupyter notebooks,
but you can install a notebook extension in order to enable equation numbering:
https://jupyter-contrib-nbextensions.readthedocs.io/en/latest/nbextensions/equation-numbering/readme.html.
Automatic Equation Numbering is enabled on https://nbviewer.jupyter.org/,
see e.g. the latest version of this very notebook at the link https://nbviewer.jupyter.org/github/spatialaudio/nbsphinx/blob/master/doc/markdown-cells.ipynb#Automatic-Equation-Numbering.
When using `nbsphinx`, you can use the following `mathjax3_config` setting in your `conf.py` file
to enable automatic equation numbering in HTML output.
```python
mathjax3_config = {
'tex': {'tags': 'ams', 'useLabelIds': True},
}
```
This works for Sphinx version 4 (and higher), which uses MathJax version 3.
For older Sphinx versions, the corresponding configuration looks like this:
```python
mathjax_config = {
'TeX': {'equationNumbers': {'autoNumber': 'AMS', 'useLabelIds': True}},
}
```
In LaTeX output, the equations are numbered by default.
You can use `\label{...}` to give a unique label to an equation:
\begin{equation}
\phi = \frac{1 + \sqrt{5}}{2}
\label{golden-mean}
\end{equation}
```
\begin{equation}
\phi = \frac{1 + \sqrt{5}}{2}
\label{golden-mean}
\end{equation}
```
If automatic equation numbering is enabled,
you can later reference that equation using its label.
You can use `\eqref{golden-mean}` for a reference with parentheses: \eqref{golden-mean},
or `\ref{golden-mean}` for a reference without them: \ref{golden-mean}.
In HTML output, these equation references only work for equations within a single HTML page.
In LaTeX output, equations from other notebooks can be referenced, e.g. \eqref{fibonacci-recurrence}.
### Manual Equation Numbering
If you prefer to assign equation numbers (or some kind of names) manually,
you can do so with `\tag{...}`:
\begin{equation}
a^2 + b^2 = c^2
\tag{99.4}
\label{pythagoras}
\end{equation}
```
\begin{equation}
a^2 + b^2 = c^2
\tag{99.4}
\label{pythagoras}
\end{equation}
```
The above equation has the number \ref{pythagoras}.
## Citations
According to https://nbconvert.readthedocs.io/en/latest/latex_citations.html,
`nbconvert` supports citations using a special HTML-based syntax.
`nbsphinx` supports the same syntax.
Example: <cite data-cite="kluyver2016jupyter">Kluyver et al. (2016)</cite>.
```html
<cite data-cite="kluyver2016jupyter">Kluyver et al. (2016)</cite>
```
You don't actually have to use `<cite>`,
any inline HTML tag can be used, e.g. `<strong>`:
<strong data-cite="perez2011python">Python: An Ecosystem for Scientific Computing</strong>.
```html
<strong data-cite="perez2011python">Python: An Ecosystem for Scientific Computing</strong>
```
You'll also have to define a list of references,
see [the section about references](a-normal-rst-file.rst#references).
There is also a Notebook extension which may or may not be useful: https://github.com/takluyver/cite2c.
### Footnote citations
Since version `2.0.0` of sphinxcontrib-bibtex, [footnote citations](https://sphinxcontrib-bibtex.readthedocs.io/en/latest/usage.html#role-footcite) are possible. This generates footnotes for all foot-citations up to the point of the [bibliography directive](a-normal-rst-file.rst#footnote-citations), which is typically placed at the end of the source file.
Depending on whether the documentation is rendered into HTML or into LaTeX/PDF, the citations are either placed into a bibliography as ordinary citations (HTML output) or placed into the footnotes of the citation's respective page (PDF).
Example: <cite data-footcite="perez2011python">Pérez et al. (2011)</cite>.
```html
<cite data-footcite="perez2011python">Pérez et al. (2011)</cite>
```
As footnote references are restricted to their own Jupyter notebook or other source file, a raw nbconvert cell of reST format (see [the section about raw cells](raw-cells.ipynb)) can be added to the notebook, containing the
```rst
.. footbibliography::
```
directive.
Alternatively, one can use the [nbsphinx epilog](prolog-and-epilog.ipynb) by setting it to, e.g.,
```python
nbsphinx_epilog = r"""
.. footbibliography::
"""
```
## Code
We can also write code with nice syntax highlighting:
```python3
print("Hello, world!")
```
## Tables
A | B | A and B
------|-------|--------
False | False | False
True | False | False
False | True | False
True | True | True
## Images
Local image: 

Remote image: 

### Using the HTML `<img>` tag
The aforementioned Markdown syntax for including images
doesn't allow specifying the image size.
If you want to control the size of the included image,
you can use the HTML
[\<img\>](https://www.w3.org/TR/html52/semantics-embedded-content.html#the-img-element)
element with the `width` attribute like this:
```html
<img src="images/notebook_icon.png" alt="Jupyter notebook icon" width="300">
```
<img src="images/notebook_icon.png" alt="Jupyter notebook icon" width="300">
In addition to the `src`, `alt`, `width` and `height` attributes,
you can also use the `class` attribute,
which is simply forwarded to the HTML output (and ignored in LaTeX output).
All other attributes are ignored.
### SVG support for LaTeX
LaTeX doesn't support SVG images, but there are Sphinx extensions that can be used for automatically converting SVG images for inclusion in LaTeX output.
Just include one of the following options in the list of
[extensions](usage.ipynb#extensions)
in your `conf.py` file.
* `'sphinxcontrib.inkscapeconverter'` or `'sphinxcontrib.rsvgconverter'`:
See https://github.com/missinglinkelectronics/sphinxcontrib-svg2pdfconverter
for installation instructions.
The external programs `inkscape` or `rsvg-convert`
(Debian/Ubuntu package `librsvg2-bin`; `conda` package `librsvg`)
are needed, respectively.
* `'sphinx.ext.imgconverter'`:
This is a built-in Sphinx extension, see
https://www.sphinx-doc.org/en/master/usage/extensions/imgconverter.html.
This needs the external program `convert` from *ImageMagick*.
The disadvantage of this extension is that SVGs are converted to bitmap images.
If one of those extensions is installed, SVG images can be used even for LaTeX output:


Remote SVG images can also be used (and will be shown in the LaTeX output):


## Cell Attachments
Images can also be embedded in the notebook itself. Just drag an image file into the Markdown cell you are just editing or copy and paste some image data from an image editor/viewer.
The generated Markdown code will look just like a "normal" image link, except that it will have an `attachment:` prefix:

These are cell attachments: 

In the Jupyter Notebook, there is a speciall "Attachments" cell toolbar which you can use to see all attachments of a cell and delete them, if needed.
## HTML Elements (HTML only)
It is allowed to use plain HTML elements within Markdown cells.
Those elements are passed through to the HTML output and are ignored for the LaTeX output.
Below are a few examples.
HTML5 [audio](https://developer.mozilla.org/en-US/docs/Web/HTML/Element/audio) elements can be created like this:
```html
<audio src="https://example.org/audio.ogg" controls>alternative text</audio>
```
Example:
<audio src="https://upload.wikimedia.org/wikipedia/commons/6/61/DescenteInfinie.ogg" controls>The HTML audio element is not supported!</audio>
HTML5 [video](https://developer.mozilla.org/en-US/docs/Web/HTML/Element/video) elements can be created like this:
```html
<video src="https://example.org/video.ogv" controls>alternative text</video>
```
Example:
<video src="https://upload.wikimedia.org/wikipedia/commons/4/42/Shepard_Calais_1906_FrenchGP.ogv" controls autoplay loop>The HTML video element is not supported!</video>
The alternative text is shown in browsers that don't support those elements. The same text is also shown in Sphinx's LaTeX output.
<div class="alert alert-info">
**Note:** You can also use local files for the `<audio>` and `<video>` elements, but you have to create a link to the source file somewhere, because only then are the local files copied to the HTML output directory!
You should do that anyway to make the audio/video file accessible to browsers that don't support the `<audio>` and `<video>` elements.
</div>
## Info/Warning Boxes
<div class="alert alert-warning">
Warning
This is an *experimental feature*!
Its usage will probably change in the future or it might be removed completely!
</div>
Until there is an info/warning extension for Markdown/CommonMark (see [this issue](https://github.com/jupyter/notebook/issues/1292)), such boxes can be created by using HTML `<div>` elements like this:
```html
<div class="alert alert-info">
Note
This is a note!
</div>
```
For this to work reliably, you should obey the following guidelines:
* The `class` attribute has to be either `"alert alert-info"` or `"alert alert-warning"`, other values will not be converted correctly.
* No further attributes are allowed.
* For compatibility with CommonMark, you should add an empty line between the `<div>` start tag and the beginning of the content.
<div class="alert alert-warning">
Warning
While this works nicely with `nbsphinx`, JupyterLab and the Classic Jupyter Notebook,
This doesn't work correctly in `nbconvert`
and by extension on https://nbviewer.jupyter.org/ and Github's notebook preview.
See https://github.com/jupyter/nbconvert/issues/1125.
</div>
<div class="alert alert-info">
Note
The text can contain further Markdown formatting.
It is even possible to have nested boxes:
<div class="alert alert-warning">
... but please don't *overuse* this!
</div>
</div>
## Links to Other Notebooks
Relative links to local notebooks can be used:
[a link to a notebook in a subdirectory](subdir/a-notebook-in-a-subdir.ipynb),
[a link to an orphan notebook](orphan.ipynb)
(latter won't work in LaTeX output, because orphan pages are not included there).
This is how a link is created in Markdown:
```
[a link to a notebook in a subdirectory](subdir/a-notebook-in-a-subdir.ipynb)
```
Markdown also supports *reference-style* links:
[a reference-style link][mylink],
[another version of the same link][mylink].
[mylink]: subdir/a-notebook-in-a-subdir.ipynb
These can be created with this syntax:
```
[a reference-style link][mylink]
[mylink]: subdir/a-notebook-in-a-subdir.ipynb
```
Links to sub-sections are also possible, e.g.
[this subsection](subdir/a-notebook-in-a-subdir.ipynb#A-Sub-Section).
This link was created with:
```
[this subsection](subdir/a-notebook-in-a-subdir.ipynb#A-Sub-Section)
```
You just have to remember to replace spaces with hyphens!
BTW, links to sections of the current notebook work, too, e.g.
[beginning of this section](#Links-to-Other-Notebooks).
This can be done, as expected, like this:
```
[beginning of this section](#Links-to-Other-Notebooks)
```
It's also possible to create a
[link to the beginning of the current page](#),
by simply using a `#` character:
```
[link to the beginning of the current page](#)
```
## Links to `*.rst` Files (and Other Sphinx Source Files)
Links to files whose extension is in the configuration value [source_suffix](https://www.sphinx-doc.org/en/master/config.html#confval-source_suffix), will be converted to links to the generated HTML/LaTeX pages. Example: [A reStructuredText file](a-normal-rst-file.rst).
This was created with:
```
[A reStructuredText file](a-normal-rst-file.rst)
```
Links to sub-sections are also possible. Example: [Sphinx Directives](a-normal-rst-file.rst#sphinx-directives-for-info-warning-boxes).
This was created with:
```
[Sphinx Directives](a-normal-rst-file.rst#sphinx-directives-for-info-warning-boxes)
```
<div class="alert alert-info">
Note
Sphinx section anchors are different from Jupyter section anchors!
To create a link to a subsection in an `.rst` file (or another non-notebook source file), you not only have to replace spaces with hyphens, but also slashes and some other characters.
In case of doubt, just check the target HTML page generated by Sphinx.
</div>
## Links to Local Files
Links to local files (other than Jupyter notebooks and other Sphinx source files) are also possible, e.g. [requirements.txt](requirements.txt).
This was simply created with:
```
[requirements.txt](requirements.txt)
```
The linked files are automatically copied to the HTML output directory.
For LaTeX output, links are created,
but the files are not copied to the target directory.
## Links to Domain Objects
Links to [Sphinx domain objects](https://www.sphinx-doc.org/en/master/usage/restructuredtext/domains.html) (such as a Python class or JavaScript function) are also possible. For example:
[example_python_function()](a-normal-rst-file.rst#example_python_function).
This was created with:
```
[example_python_function()](a-normal-rst-file.rst#example_python_function)
```
This is especially useful for use with the Sphinx [autodoc](https://www.sphinx-doc.org/en/master/ext/autodoc.html) extension!
|
github_jupyter
|
\begin{equation}
\int\limits_{-\infty}^\infty f(x) \delta(x - x_0) dx = f(x_0)
\end{equation}
extensions = [
'nbsphinx',
'sphinx.ext.mathjax',
# ... other useful extensions ...
]
mathjax3_config = {
'tex': {'tags': 'ams', 'useLabelIds': True},
}
mathjax_config = {
'TeX': {'equationNumbers': {'autoNumber': 'AMS', 'useLabelIds': True}},
}
\begin{equation}
\phi = \frac{1 + \sqrt{5}}{2}
\label{golden-mean}
\end{equation}
\begin{equation}
a^2 + b^2 = c^2
\tag{99.4}
\label{pythagoras}
\end{equation}
<cite data-cite="kluyver2016jupyter">Kluyver et al. (2016)</cite>
<strong data-cite="perez2011python">Python: An Ecosystem for Scientific Computing</strong>
<cite data-footcite="perez2011python">Pérez et al. (2011)</cite>
.. footbibliography::
nbsphinx_epilog = r"""
.. footbibliography::
"""
## Tables
A | B | A and B
------|-------|--------
False | False | False
True | False | False
False | True | False
True | True | True
## Images
Local image: 

Remote image: 

### Using the HTML `<img>` tag
The aforementioned Markdown syntax for including images
doesn't allow specifying the image size.
If you want to control the size of the included image,
you can use the HTML
[\<img\>](https://www.w3.org/TR/html52/semantics-embedded-content.html#the-img-element)
element with the `width` attribute like this:
<img src="images/notebook_icon.png" alt="Jupyter notebook icon" width="300">
In addition to the `src`, `alt`, `width` and `height` attributes,
you can also use the `class` attribute,
which is simply forwarded to the HTML output (and ignored in LaTeX output).
All other attributes are ignored.
### SVG support for LaTeX
LaTeX doesn't support SVG images, but there are Sphinx extensions that can be used for automatically converting SVG images for inclusion in LaTeX output.
Just include one of the following options in the list of
[extensions](usage.ipynb#extensions)
in your `conf.py` file.
* `'sphinxcontrib.inkscapeconverter'` or `'sphinxcontrib.rsvgconverter'`:
See https://github.com/missinglinkelectronics/sphinxcontrib-svg2pdfconverter
for installation instructions.
The external programs `inkscape` or `rsvg-convert`
(Debian/Ubuntu package `librsvg2-bin`; `conda` package `librsvg`)
are needed, respectively.
* `'sphinx.ext.imgconverter'`:
This is a built-in Sphinx extension, see
https://www.sphinx-doc.org/en/master/usage/extensions/imgconverter.html.
This needs the external program `convert` from *ImageMagick*.
The disadvantage of this extension is that SVGs are converted to bitmap images.
If one of those extensions is installed, SVG images can be used even for LaTeX output:


Remote SVG images can also be used (and will be shown in the LaTeX output):


## Cell Attachments
Images can also be embedded in the notebook itself. Just drag an image file into the Markdown cell you are just editing or copy and paste some image data from an image editor/viewer.
The generated Markdown code will look just like a "normal" image link, except that it will have an `attachment:` prefix:

These are cell attachments: 

In the Jupyter Notebook, there is a speciall "Attachments" cell toolbar which you can use to see all attachments of a cell and delete them, if needed.
## HTML Elements (HTML only)
It is allowed to use plain HTML elements within Markdown cells.
Those elements are passed through to the HTML output and are ignored for the LaTeX output.
Below are a few examples.
HTML5 [audio](https://developer.mozilla.org/en-US/docs/Web/HTML/Element/audio) elements can be created like this:
Example:
<audio src="https://upload.wikimedia.org/wikipedia/commons/6/61/DescenteInfinie.ogg" controls>The HTML audio element is not supported!</audio>
HTML5 [video](https://developer.mozilla.org/en-US/docs/Web/HTML/Element/video) elements can be created like this:
Example:
<video src="https://upload.wikimedia.org/wikipedia/commons/4/42/Shepard_Calais_1906_FrenchGP.ogv" controls autoplay loop>The HTML video element is not supported!</video>
The alternative text is shown in browsers that don't support those elements. The same text is also shown in Sphinx's LaTeX output.
<div class="alert alert-info">
**Note:** You can also use local files for the `<audio>` and `<video>` elements, but you have to create a link to the source file somewhere, because only then are the local files copied to the HTML output directory!
You should do that anyway to make the audio/video file accessible to browsers that don't support the `<audio>` and `<video>` elements.
</div>
## Info/Warning Boxes
<div class="alert alert-warning">
Warning
This is an *experimental feature*!
Its usage will probably change in the future or it might be removed completely!
</div>
Until there is an info/warning extension for Markdown/CommonMark (see [this issue](https://github.com/jupyter/notebook/issues/1292)), such boxes can be created by using HTML `<div>` elements like this:
For this to work reliably, you should obey the following guidelines:
* The `class` attribute has to be either `"alert alert-info"` or `"alert alert-warning"`, other values will not be converted correctly.
* No further attributes are allowed.
* For compatibility with CommonMark, you should add an empty line between the `<div>` start tag and the beginning of the content.
<div class="alert alert-warning">
Warning
While this works nicely with `nbsphinx`, JupyterLab and the Classic Jupyter Notebook,
This doesn't work correctly in `nbconvert`
and by extension on https://nbviewer.jupyter.org/ and Github's notebook preview.
See https://github.com/jupyter/nbconvert/issues/1125.
</div>
<div class="alert alert-info">
Note
The text can contain further Markdown formatting.
It is even possible to have nested boxes:
<div class="alert alert-warning">
... but please don't *overuse* this!
</div>
</div>
## Links to Other Notebooks
Relative links to local notebooks can be used:
[a link to a notebook in a subdirectory](subdir/a-notebook-in-a-subdir.ipynb),
[a link to an orphan notebook](orphan.ipynb)
(latter won't work in LaTeX output, because orphan pages are not included there).
This is how a link is created in Markdown:
Markdown also supports *reference-style* links:
[a reference-style link][mylink],
[another version of the same link][mylink].
[mylink]: subdir/a-notebook-in-a-subdir.ipynb
These can be created with this syntax:
Links to sub-sections are also possible, e.g.
[this subsection](subdir/a-notebook-in-a-subdir.ipynb#A-Sub-Section).
This link was created with:
You just have to remember to replace spaces with hyphens!
BTW, links to sections of the current notebook work, too, e.g.
[beginning of this section](#Links-to-Other-Notebooks).
This can be done, as expected, like this:
It's also possible to create a
[link to the beginning of the current page](#),
by simply using a `#` character:
## Links to `*.rst` Files (and Other Sphinx Source Files)
Links to files whose extension is in the configuration value [source_suffix](https://www.sphinx-doc.org/en/master/config.html#confval-source_suffix), will be converted to links to the generated HTML/LaTeX pages. Example: [A reStructuredText file](a-normal-rst-file.rst).
This was created with:
Links to sub-sections are also possible. Example: [Sphinx Directives](a-normal-rst-file.rst#sphinx-directives-for-info-warning-boxes).
This was created with:
<div class="alert alert-info">
Note
Sphinx section anchors are different from Jupyter section anchors!
To create a link to a subsection in an `.rst` file (or another non-notebook source file), you not only have to replace spaces with hyphens, but also slashes and some other characters.
In case of doubt, just check the target HTML page generated by Sphinx.
</div>
## Links to Local Files
Links to local files (other than Jupyter notebooks and other Sphinx source files) are also possible, e.g. [requirements.txt](requirements.txt).
This was simply created with:
The linked files are automatically copied to the HTML output directory.
For LaTeX output, links are created,
but the files are not copied to the target directory.
## Links to Domain Objects
Links to [Sphinx domain objects](https://www.sphinx-doc.org/en/master/usage/restructuredtext/domains.html) (such as a Python class or JavaScript function) are also possible. For example:
[example_python_function()](a-normal-rst-file.rst#example_python_function).
This was created with:
| 0.799716 | 0.905615 |
```
%pylab inline
import pandas as pd
from sklearn.model_selection import StratifiedShuffleSplit
from sklearn.model_selection import train_test_split
normal_df = pd.read_table('/mnt/disks/data/histopath_data/pyvirchow-out-normal-patches/master_df.tsv')
normal_df['slide_type'] = 'normal'
normal_df['is_tumor'] = False
tumor_df = pd.read_table('/mnt/disks/data/histopath_data/pyvirchow-out-tumor-patches//master_df.tsv')
tumor_df['slide_type'] = 'tumor'
master_df = pd.concat([normal_df, tumor_df])
```
# Strategy
We will work with 200k patches for training and 30k patches for validation. The procedure is as:
1. Generate 'normal' or 'tumor' with probability = 0.5
2. Select a sample belonging to that category
If tumor : Sample x tumor patches from the tumor such that, x/(total_tumor_patches) =
```
split = StratifiedShuffleSplit(n_splits=1, test_size=0.2, random_state=42)
for train_index, test_index in split.split(tumor_df, tumor_df['is_tumor']):
train_samples = tumor_df.loc[train_index]
validation_samples = tumor_df.loc[test_index]
len(train_samples[train_samples.is_tumor==True].index)
normal_slides_that_are_tumors = ['normal_086', 'normal_144']
# The following slides should not be used for generating
# normal patches
# See "Detecting Cancer Metastases on Gigapixel Pathology Images"
tumor_slides_with_nonexhaustive_tumor_annotation = ['tumor_{}'.format(x) for x in ['018', '020', '029',
'033', '044', '046',
'051', '054', '055',
'079', '092', '095',
'010', '025', '034',
'056', '067', '085',
'110']]
df = tumor_df['uid'].groupby(tumor_df['is_tumor']).value_counts().rename('counts')#.reset_index(drop=True)#.groupby('is_tumor')
df = pd.DataFrame(df).reset_index()
all_tumor_slides = sorted(tumor_df['uid'].unique())
all_normal_slides = sorted(normal_df['uid'].unique())
# Remove normal slides that are tumors
all_normal_slides = [x for x in all_normal_slides if x not in normal_slides_that_are_tumors]
# Keep 10 slides for validation
tumor_train_slides, tumor_validate_slides = train_test_split(all_tumor_slides, test_size=0.08, random_state=42)
normal_train_slides, normal_validate_slides = train_test_split(all_normal_slides, test_size=0.08, random_state=42)
tumor_train_slides = sorted(tumor_train_slides)
tumor_validate_slides = sorted(tumor_validate_slides)
normal_train_slides = sorted(normal_train_slides)
normal_validate_slides = sorted(normal_validate_slides)
tumor_validate_slides
```
# How many patches in train/test dataset?
```
tumor_train_df = tumor_df[tumor_df.uid.isin(tumor_train_slides)]
tumor_validate_df = tumor_df[tumor_df.uid.isin(tumor_validate_slides)]
tumor_train_df.reset_index().to_csv('/mnt/disks/data/histopath_data/pyvirchow-out-df/tumor_train_df.tsv',
sep='\t',
index=False,
header=True)
tumor_validate_df.reset_index().to_csv('/mnt/disks/data/histopath_data/pyvirchow-out-df/tumor_validate_df.tsv',
sep='\t',
index=False,
header=True)
tumor_train_df_summary = tumor_train_df['uid'].groupby(tumor_train_df['is_tumor']).value_counts().groupby('is_tumor').describe()
tumor_train_df_summary['total'] = tumor_train_df_summary['mean'] * tumor_train_df_summary['count']
tumor_train_df_summary['cumprop']= tumor_train_df_summary.total.cumsum()/tumor_train_df_summary['total'].sum()
total_tumor_train_patches = tumor_train_df_summary.loc[True]['total']
tumor_train_df_summary.reset_index().to_csv('/mnt/disks/data/histopath_data/pyvirchow-out-df//tumor_train_df_summary.tsv',
sep='\t',
index=False,
header=True)
tumor_train_df_summary
total_tumor_train_patches
tumor_validate_df_summary = tumor_validate_df['uid'].groupby(tumor_validate_df['is_tumor']).value_counts().groupby('is_tumor').describe()
tumor_validate_df_summary['total'] = tumor_validate_df_summary['mean'] * tumor_validate_df_summary['count']
tumor_validate_df_summary['cumprop']= tumor_validate_df_summary.total.cumsum()/tumor_validate_df_summary['total'].sum()
total_tumor_validate_patches = tumor_validate_df_summary.loc[True]['total']
tumor_validate_df_summary.reset_index().to_csv('/mnt/disks/data/histopath_data/pyvirchow-out-df//tumor_validate_df_summary.tsv',
sep='\t',
index=False,
header=True)
tumor_validate_df_summary
normal_train_df = normal_df[normal_df.uid.isin(normal_train_slides)]
normal_validate_df = normal_df[normal_df.uid.isin(normal_validate_slides)]
normal_train_df.reset_index().to_csv('/mnt/disks/data/histopath_data/pyvirchow-out-df//normal_train_df.tsv',
sep='\t',
index=False,
header=True)
normal_validate_df.reset_index().to_csv('/mnt/disks/data/histopath_data/pyvirchow-out-df//normal_validate_df.tsv',
sep='\t',
index=False,
header=True)
normal_train_df_summary = normal_train_df['uid'].value_counts().describe()
normal_train_df_summary['total'] = normal_train_df_summary['mean'] * normal_train_df_summary['count']
normal_train_df_summary = pd.DataFrame(normal_train_df_summary).T
total_normal_train_patches = normal_train_df_summary['total']
normal_train_df_summary.to_csv('/mnt/disks/data/histopath_data/pyvirchow-out-df/normal_train_df_summary.tsv',
sep='\t',
index=False,
header=True)
normal_train_df_summary
normal_validate_df_summary = normal_validate_df['uid'].value_counts().describe()
normal_validate_df_summary['total'] = normal_validate_df_summary['mean'] * normal_validate_df_summary['count']
normal_validate_df_summary = pd.DataFrame(normal_validate_df_summary).T
total_normal_validate_patches = normal_validate_df_summary['total']
normal_validate_df_summary.to_csv('/mnt/disks/data/histopath_data/pyvirchow-out-df/normal_validate_df_summary.tsv',
sep='\t',
index=False,
header=True)
normal_validate_df_summary
```
So we have a lot of normal patches and hence that will require subsampling.
We will subsample so that we subsample as many normal patches from tumor samples such that they match up
the counts of tumor patches (close to 225k)
We start off training with an equal number of normal patches. Then given the tumor samples, we generate half of these normal patches from the valid tumor samples and the other half from the actual normal samples.
```
normal_train_df_finalized = pd.DataFrame()
relevant_tumor_normal_patches = tumor_train_df[(tumor_train_df.is_tumor==False) & (~tumor_train_df.uid.isin(tumor_slides_with_nonexhaustive_tumor_annotation))]
patch_frac = len(relevant_tumor_normal_patches.index)/(total_tumor_train_patches/2)
patch_frac = 1/patch_frac
for seed, tumor_uid in enumerate(tumor_train_slides):
if tumor_uid in tumor_slides_with_nonexhaustive_tumor_annotation:
# Should not use these slides for generating normal patches
continue
# How many normal patches does this uid have?
tumor_train_uid_df = tumor_train_df[tumor_train_df.uid==tumor_uid]
tumor_train_uid_df_normal = tumor_train_uid_df[tumor_train_uid_df.is_tumor==False]
normal_patches_in_uid = len(tumor_train_uid_df_normal.index)
tumor_patches_in_uid = len(tumor_train_uid_df[tumor_train_uid_df.is_tumor==True].index)
# subsample these proprotion of
selected_patches = tumor_train_uid_df_normal.sample(frac=patch_frac, random_state=seed+42)
normal_train_df_finalized = pd.concat([normal_train_df_finalized, selected_patches])
patch_frac
len(normal_train_df_finalized.index)
# Select the remaining half from the actual normal slides.
patch_frac = len(normal_train_df.index)/(total_tumor_train_patches)
patch_frac = 1/patch_frac
for seed, normal_uid in enumerate(normal_train_slides):
# How many normal patches does this uid have?
normal_train_uid_df = normal_train_df[normal_train_df.uid==normal_uid]
normal_patches_in_uid = len(normal_train_uid_df.index)
# subsample these proprotion of
selected_patches = normal_train_uid_df.sample(frac=patch_frac, random_state=seed+42)
normal_train_df_finalized = pd.concat([normal_train_df_finalized, selected_patches])
len(normal_train_df_finalized.index)
total_tumor_train_patches
total_tumor_validate_patches
# Do the same thing for the validation
normal_validate_df_finalized = pd.DataFrame()
relevant_tumor_normal_patches = tumor_validate_df[(tumor_validate_df.is_tumor==False) & (~tumor_validate_df.uid.isin(tumor_slides_with_nonexhaustive_tumor_annotation))]
patch_frac = len(relevant_tumor_normal_patches.index)/(total_tumor_validate_patches/2)
patch_frac = 1/patch_frac
for seed, tumor_uid in enumerate(tumor_validate_slides):
if tumor_uid in tumor_slides_with_nonexhaustive_tumor_annotation:
# Should not use these slides for generating normal patches
continue
# How many normal patches does this uid have?
tumor_validate_uid_df = tumor_validate_df[tumor_validate_df.uid==tumor_uid]
tumor_validate_uid_df_normal = tumor_validate_uid_df[tumor_validate_uid_df.is_tumor==False]
normal_patches_in_uid = len(tumor_validate_uid_df_normal.index)
tumor_patches_in_uid = len(tumor_validate_uid_df[tumor_validate_uid_df.is_tumor==True].index)
# subsample these proprotion of
selected_patches = tumor_validate_uid_df_normal.sample(frac=patch_frac, random_state=seed+42)
normal_validate_df_finalized = pd.concat([normal_validate_df_finalized, selected_patches])
print(len(normal_validate_df_finalized.index))
patch_frac = len(relevant_tumor_normal_patches.index)/(total_tumor_validate_patches)
patch_frac = 1/patch_frac
patch_frac
total_tumor_validate_patches
# Select the remaining half from the actual normal slides.
patch_frac = len(normal_validate_df.index)/(total_tumor_validate_patches/2)
patch_frac = 1/patch_frac
for seed, normal_uid in enumerate(normal_validate_slides):
# How many normal patches does this uid have?
normal_validate_uid_df = normal_validate_df[normal_validate_df.uid==normal_uid]
normal_patches_in_uid = len(normal_validate_uid_df.index)
# subsample these proprotion of
selected_patches = normal_validate_uid_df.sample(frac=patch_frac, random_state=seed+42)
normal_validate_df_finalized = pd.concat([normal_validate_df_finalized, selected_patches])
print(len(normal_validate_df_finalized.index))
print(len(tumor_validate_df.index))
normal_train_df_finalized.to_csv('/mnt/disks/data/histopath_data/pyvirchow-out-df//normal_train_df_finalized.tsv',
sep='\t',
index=False,
header=True)
normal_validate_df_finalized.to_csv('/mnt/disks/data/histopath_data/pyvirchow-out-df//normal_validate_df_finalized.tsv',
sep='\t',
index=False,
header=True)
# Let's just merge the datasets for training
tumor_train_df_tumor = tumor_train_df[tumor_train_df.is_tumor==True]
normal_train_df_finalized['is_tumor'] = False
tumor_validate_df_tumor = tumor_validate_df[tumor_validate_df.is_tumor==True]
normal_validate_df_finalized['is_tumor'] = False
train_df = pd.concat([tumor_train_df_tumor, normal_train_df_finalized])
# Shuffle before saving so that we do not have things together
train_df = train_df.sample(frac=1, random_state=42)
validate_df = pd.concat([tumor_validate_df_tumor, normal_validate_df_finalized])
# Shuffle before saving so that we do not have things together
validate_df = validate_df.sample(frac=1, random_state=42)
train_df.to_csv('/mnt/disks/data/histopath_data/pyvirchow-out-df/train_df.tsv',
sep='\t',
index=False,
header=True)
validate_df.to_csv('//mnt/disks/data/histopath_data/pyvirchow-out-df/validate_df.tsv',
sep='\t',
index=False,
header=True)
len(train_df.index)
len(tumor_validate_df.index)
len(normal_validate_df_finalized.index)
patch_frac = len(normal_validate_df.index)/(total_tumor_validate_patches/2)
patch_frac = 1/patch_frac
patch_frac
len(validate_df.index)
total_tumor_train_patches
```
# Full fraction
```
normal_train_df_finalized = pd.DataFrame()
relevant_tumor_normal_patches = tumor_train_df[(tumor_train_df.is_tumor==False) & (~tumor_train_df.uid.isin(tumor_slides_with_nonexhaustive_tumor_annotation))]
patch_frac = len(relevant_tumor_normal_patches.index)/(total_tumor_train_patches/2)
patch_frac = 1/patch_frac
for seed, tumor_uid in enumerate(tumor_train_slides):
if tumor_uid in tumor_slides_with_nonexhaustive_tumor_annotation:
# Should not use these slides for generating normal patches
continue
# How many normal patches does this uid have?
tumor_train_uid_df = tumor_train_df[tumor_train_df.uid==tumor_uid]
tumor_train_uid_df_normal = tumor_train_uid_df[tumor_train_uid_df.is_tumor==False]
normal_patches_in_uid = len(tumor_train_uid_df_normal.index)
tumor_patches_in_uid = len(tumor_train_uid_df[tumor_train_uid_df.is_tumor==True].index)
# subsample these proprotion of
selected_patches = tumor_train_uid_df_normal.sample(frac=patch_frac, random_state=seed+42)
normal_train_df_finalized = pd.concat([normal_train_df_finalized, selected_patches])
len(tumor_train_uid_df_normal.index)
patch_frac
patch_frac = len(normal_train_df.index)/(total_tumor_train_patches)
patch_frac = 1/patch_frac
patch_frac
len(normal_train_df_finalized.index)
# Select the remaining half from the actual normal slides.
patch_frac = len(normal_train_df.index)/(total_tumor_train_patches/2)
patch_frac = 1/patch_frac
for seed, normal_uid in enumerate(normal_train_slides):
# How many normal patches does this uid have?
normal_train_uid_df = normal_train_df[normal_train_df.uid==normal_uid]
normal_patches_in_uid = len(normal_train_uid_df.index)
# subsample these proprotion of
selected_patches = normal_train_uid_df.sample(frac=patch_frac, random_state=seed+42)
normal_train_df_finalized = pd.concat([normal_train_df_finalized, selected_patches])
len(normal_train_df_finalized.index)
total_tumor_train_patches
total_tumor_validate_patches
# Do the same thing for the validation
normal_validate_df_finalized = pd.DataFrame()
relevant_tumor_normal_patches = tumor_validate_df[(tumor_validate_df.is_tumor==False) & (~tumor_validate_df.uid.isin(tumor_slides_with_nonexhaustive_tumor_annotation))]
patch_frac = len(relevant_tumor_normal_patches.index)/(total_tumor_validate_patches)
patch_frac = 1/patch_frac
for seed, tumor_uid in enumerate(tumor_validate_slides):
if tumor_uid in tumor_slides_with_nonexhaustive_tumor_annotation:
# Should not use these slides for generating normal patches
continue
# How many normal patches does this uid have?
tumor_validate_uid_df = tumor_validate_df[tumor_validate_df.uid==tumor_uid]
tumor_validate_uid_df_normal = tumor_validate_uid_df[tumor_validate_uid_df.is_tumor==False]
normal_patches_in_uid = len(tumor_validate_uid_df_normal.index)
tumor_patches_in_uid = len(tumor_validate_uid_df[tumor_validate_uid_df.is_tumor==True].index)
# subsample these proprotion of
selected_patches = tumor_validate_uid_df_normal.sample(frac=patch_frac, random_state=seed+42)
normal_validate_df_finalized = pd.concat([normal_validate_df_finalized, selected_patches])
print(len(normal_validate_df_finalized.index))
patch_frac = len(relevant_tumor_normal_patches.index)/(total_tumor_validate_patches)
patch_frac = 1/patch_frac
patch_frac
total_tumor_validate_patches
# Select the remaining half from the actual normal slides.
patch_frac = len(normal_validate_df.index)/(total_tumor_validate_patches)
patch_frac = 1/patch_frac
for seed, normal_uid in enumerate(normal_validate_slides):
# How many normal patches does this uid have?
normal_validate_uid_df = normal_validate_df[normal_validate_df.uid==normal_uid]
normal_patches_in_uid = len(normal_validate_uid_df.index)
# subsample these proprotion of
selected_patches = normal_validate_uid_df.sample(frac=patch_frac, random_state=seed+42)
normal_validate_df_finalized = pd.concat([normal_validate_df_finalized, selected_patches])
print(len(normal_validate_df_finalized.index))
print(len(tumor_validate_df.index))
normal_train_df_finalized.to_csv('/mnt/disks/data/histopath_data/pyvirchow-out-df//normal_train_df_finalized_fullfrac.tsv',
sep='\t',
index=False,
header=True)
normal_validate_df_finalized.to_csv('/mnt/disks/data/histopath_data/pyvirchow-out-df//normal_validate_df_finalized_fullfrac.tsv',
sep='\t',
index=False,
header=True)
# Let's just merge the datasets for training
tumor_train_df_tumor = tumor_train_df[tumor_train_df.is_tumor==True]
normal_train_df_finalized['is_tumor'] = False
tumor_validate_df_tumor = tumor_validate_df[tumor_validate_df.is_tumor==True]
normal_validate_df_finalized['is_tumor'] = False
train_df = pd.concat([tumor_train_df_tumor, normal_train_df_finalized])
# Shuffle before saving so that we do not have things together
train_df = train_df.sample(frac=1, random_state=42)
validate_df = pd.concat([tumor_validate_df_tumor, normal_validate_df_finalized])
# Shuffle before saving so that we do not have things together
validate_df = validate_df.sample(frac=1, random_state=42)
train_df.to_csv('/mnt/disks/data/histopath_data/pyvirchow-out-df/train_df_fullfrac.tsv',
sep='\t',
index=False,
header=True)
validate_df.to_csv('//mnt/disks/data/histopath_data/pyvirchow-out-df/validate_df_fullfrac.tsv',
sep='\t',
index=False,
header=True)
len(train_df.index)
len(validate_df.index)
len(train_df[train_df.is_tumor==True].index)
len(train_df[train_df.is_tumor==False].index)
```
|
github_jupyter
|
%pylab inline
import pandas as pd
from sklearn.model_selection import StratifiedShuffleSplit
from sklearn.model_selection import train_test_split
normal_df = pd.read_table('/mnt/disks/data/histopath_data/pyvirchow-out-normal-patches/master_df.tsv')
normal_df['slide_type'] = 'normal'
normal_df['is_tumor'] = False
tumor_df = pd.read_table('/mnt/disks/data/histopath_data/pyvirchow-out-tumor-patches//master_df.tsv')
tumor_df['slide_type'] = 'tumor'
master_df = pd.concat([normal_df, tumor_df])
split = StratifiedShuffleSplit(n_splits=1, test_size=0.2, random_state=42)
for train_index, test_index in split.split(tumor_df, tumor_df['is_tumor']):
train_samples = tumor_df.loc[train_index]
validation_samples = tumor_df.loc[test_index]
len(train_samples[train_samples.is_tumor==True].index)
normal_slides_that_are_tumors = ['normal_086', 'normal_144']
# The following slides should not be used for generating
# normal patches
# See "Detecting Cancer Metastases on Gigapixel Pathology Images"
tumor_slides_with_nonexhaustive_tumor_annotation = ['tumor_{}'.format(x) for x in ['018', '020', '029',
'033', '044', '046',
'051', '054', '055',
'079', '092', '095',
'010', '025', '034',
'056', '067', '085',
'110']]
df = tumor_df['uid'].groupby(tumor_df['is_tumor']).value_counts().rename('counts')#.reset_index(drop=True)#.groupby('is_tumor')
df = pd.DataFrame(df).reset_index()
all_tumor_slides = sorted(tumor_df['uid'].unique())
all_normal_slides = sorted(normal_df['uid'].unique())
# Remove normal slides that are tumors
all_normal_slides = [x for x in all_normal_slides if x not in normal_slides_that_are_tumors]
# Keep 10 slides for validation
tumor_train_slides, tumor_validate_slides = train_test_split(all_tumor_slides, test_size=0.08, random_state=42)
normal_train_slides, normal_validate_slides = train_test_split(all_normal_slides, test_size=0.08, random_state=42)
tumor_train_slides = sorted(tumor_train_slides)
tumor_validate_slides = sorted(tumor_validate_slides)
normal_train_slides = sorted(normal_train_slides)
normal_validate_slides = sorted(normal_validate_slides)
tumor_validate_slides
tumor_train_df = tumor_df[tumor_df.uid.isin(tumor_train_slides)]
tumor_validate_df = tumor_df[tumor_df.uid.isin(tumor_validate_slides)]
tumor_train_df.reset_index().to_csv('/mnt/disks/data/histopath_data/pyvirchow-out-df/tumor_train_df.tsv',
sep='\t',
index=False,
header=True)
tumor_validate_df.reset_index().to_csv('/mnt/disks/data/histopath_data/pyvirchow-out-df/tumor_validate_df.tsv',
sep='\t',
index=False,
header=True)
tumor_train_df_summary = tumor_train_df['uid'].groupby(tumor_train_df['is_tumor']).value_counts().groupby('is_tumor').describe()
tumor_train_df_summary['total'] = tumor_train_df_summary['mean'] * tumor_train_df_summary['count']
tumor_train_df_summary['cumprop']= tumor_train_df_summary.total.cumsum()/tumor_train_df_summary['total'].sum()
total_tumor_train_patches = tumor_train_df_summary.loc[True]['total']
tumor_train_df_summary.reset_index().to_csv('/mnt/disks/data/histopath_data/pyvirchow-out-df//tumor_train_df_summary.tsv',
sep='\t',
index=False,
header=True)
tumor_train_df_summary
total_tumor_train_patches
tumor_validate_df_summary = tumor_validate_df['uid'].groupby(tumor_validate_df['is_tumor']).value_counts().groupby('is_tumor').describe()
tumor_validate_df_summary['total'] = tumor_validate_df_summary['mean'] * tumor_validate_df_summary['count']
tumor_validate_df_summary['cumprop']= tumor_validate_df_summary.total.cumsum()/tumor_validate_df_summary['total'].sum()
total_tumor_validate_patches = tumor_validate_df_summary.loc[True]['total']
tumor_validate_df_summary.reset_index().to_csv('/mnt/disks/data/histopath_data/pyvirchow-out-df//tumor_validate_df_summary.tsv',
sep='\t',
index=False,
header=True)
tumor_validate_df_summary
normal_train_df = normal_df[normal_df.uid.isin(normal_train_slides)]
normal_validate_df = normal_df[normal_df.uid.isin(normal_validate_slides)]
normal_train_df.reset_index().to_csv('/mnt/disks/data/histopath_data/pyvirchow-out-df//normal_train_df.tsv',
sep='\t',
index=False,
header=True)
normal_validate_df.reset_index().to_csv('/mnt/disks/data/histopath_data/pyvirchow-out-df//normal_validate_df.tsv',
sep='\t',
index=False,
header=True)
normal_train_df_summary = normal_train_df['uid'].value_counts().describe()
normal_train_df_summary['total'] = normal_train_df_summary['mean'] * normal_train_df_summary['count']
normal_train_df_summary = pd.DataFrame(normal_train_df_summary).T
total_normal_train_patches = normal_train_df_summary['total']
normal_train_df_summary.to_csv('/mnt/disks/data/histopath_data/pyvirchow-out-df/normal_train_df_summary.tsv',
sep='\t',
index=False,
header=True)
normal_train_df_summary
normal_validate_df_summary = normal_validate_df['uid'].value_counts().describe()
normal_validate_df_summary['total'] = normal_validate_df_summary['mean'] * normal_validate_df_summary['count']
normal_validate_df_summary = pd.DataFrame(normal_validate_df_summary).T
total_normal_validate_patches = normal_validate_df_summary['total']
normal_validate_df_summary.to_csv('/mnt/disks/data/histopath_data/pyvirchow-out-df/normal_validate_df_summary.tsv',
sep='\t',
index=False,
header=True)
normal_validate_df_summary
normal_train_df_finalized = pd.DataFrame()
relevant_tumor_normal_patches = tumor_train_df[(tumor_train_df.is_tumor==False) & (~tumor_train_df.uid.isin(tumor_slides_with_nonexhaustive_tumor_annotation))]
patch_frac = len(relevant_tumor_normal_patches.index)/(total_tumor_train_patches/2)
patch_frac = 1/patch_frac
for seed, tumor_uid in enumerate(tumor_train_slides):
if tumor_uid in tumor_slides_with_nonexhaustive_tumor_annotation:
# Should not use these slides for generating normal patches
continue
# How many normal patches does this uid have?
tumor_train_uid_df = tumor_train_df[tumor_train_df.uid==tumor_uid]
tumor_train_uid_df_normal = tumor_train_uid_df[tumor_train_uid_df.is_tumor==False]
normal_patches_in_uid = len(tumor_train_uid_df_normal.index)
tumor_patches_in_uid = len(tumor_train_uid_df[tumor_train_uid_df.is_tumor==True].index)
# subsample these proprotion of
selected_patches = tumor_train_uid_df_normal.sample(frac=patch_frac, random_state=seed+42)
normal_train_df_finalized = pd.concat([normal_train_df_finalized, selected_patches])
patch_frac
len(normal_train_df_finalized.index)
# Select the remaining half from the actual normal slides.
patch_frac = len(normal_train_df.index)/(total_tumor_train_patches)
patch_frac = 1/patch_frac
for seed, normal_uid in enumerate(normal_train_slides):
# How many normal patches does this uid have?
normal_train_uid_df = normal_train_df[normal_train_df.uid==normal_uid]
normal_patches_in_uid = len(normal_train_uid_df.index)
# subsample these proprotion of
selected_patches = normal_train_uid_df.sample(frac=patch_frac, random_state=seed+42)
normal_train_df_finalized = pd.concat([normal_train_df_finalized, selected_patches])
len(normal_train_df_finalized.index)
total_tumor_train_patches
total_tumor_validate_patches
# Do the same thing for the validation
normal_validate_df_finalized = pd.DataFrame()
relevant_tumor_normal_patches = tumor_validate_df[(tumor_validate_df.is_tumor==False) & (~tumor_validate_df.uid.isin(tumor_slides_with_nonexhaustive_tumor_annotation))]
patch_frac = len(relevant_tumor_normal_patches.index)/(total_tumor_validate_patches/2)
patch_frac = 1/patch_frac
for seed, tumor_uid in enumerate(tumor_validate_slides):
if tumor_uid in tumor_slides_with_nonexhaustive_tumor_annotation:
# Should not use these slides for generating normal patches
continue
# How many normal patches does this uid have?
tumor_validate_uid_df = tumor_validate_df[tumor_validate_df.uid==tumor_uid]
tumor_validate_uid_df_normal = tumor_validate_uid_df[tumor_validate_uid_df.is_tumor==False]
normal_patches_in_uid = len(tumor_validate_uid_df_normal.index)
tumor_patches_in_uid = len(tumor_validate_uid_df[tumor_validate_uid_df.is_tumor==True].index)
# subsample these proprotion of
selected_patches = tumor_validate_uid_df_normal.sample(frac=patch_frac, random_state=seed+42)
normal_validate_df_finalized = pd.concat([normal_validate_df_finalized, selected_patches])
print(len(normal_validate_df_finalized.index))
patch_frac = len(relevant_tumor_normal_patches.index)/(total_tumor_validate_patches)
patch_frac = 1/patch_frac
patch_frac
total_tumor_validate_patches
# Select the remaining half from the actual normal slides.
patch_frac = len(normal_validate_df.index)/(total_tumor_validate_patches/2)
patch_frac = 1/patch_frac
for seed, normal_uid in enumerate(normal_validate_slides):
# How many normal patches does this uid have?
normal_validate_uid_df = normal_validate_df[normal_validate_df.uid==normal_uid]
normal_patches_in_uid = len(normal_validate_uid_df.index)
# subsample these proprotion of
selected_patches = normal_validate_uid_df.sample(frac=patch_frac, random_state=seed+42)
normal_validate_df_finalized = pd.concat([normal_validate_df_finalized, selected_patches])
print(len(normal_validate_df_finalized.index))
print(len(tumor_validate_df.index))
normal_train_df_finalized.to_csv('/mnt/disks/data/histopath_data/pyvirchow-out-df//normal_train_df_finalized.tsv',
sep='\t',
index=False,
header=True)
normal_validate_df_finalized.to_csv('/mnt/disks/data/histopath_data/pyvirchow-out-df//normal_validate_df_finalized.tsv',
sep='\t',
index=False,
header=True)
# Let's just merge the datasets for training
tumor_train_df_tumor = tumor_train_df[tumor_train_df.is_tumor==True]
normal_train_df_finalized['is_tumor'] = False
tumor_validate_df_tumor = tumor_validate_df[tumor_validate_df.is_tumor==True]
normal_validate_df_finalized['is_tumor'] = False
train_df = pd.concat([tumor_train_df_tumor, normal_train_df_finalized])
# Shuffle before saving so that we do not have things together
train_df = train_df.sample(frac=1, random_state=42)
validate_df = pd.concat([tumor_validate_df_tumor, normal_validate_df_finalized])
# Shuffle before saving so that we do not have things together
validate_df = validate_df.sample(frac=1, random_state=42)
train_df.to_csv('/mnt/disks/data/histopath_data/pyvirchow-out-df/train_df.tsv',
sep='\t',
index=False,
header=True)
validate_df.to_csv('//mnt/disks/data/histopath_data/pyvirchow-out-df/validate_df.tsv',
sep='\t',
index=False,
header=True)
len(train_df.index)
len(tumor_validate_df.index)
len(normal_validate_df_finalized.index)
patch_frac = len(normal_validate_df.index)/(total_tumor_validate_patches/2)
patch_frac = 1/patch_frac
patch_frac
len(validate_df.index)
total_tumor_train_patches
normal_train_df_finalized = pd.DataFrame()
relevant_tumor_normal_patches = tumor_train_df[(tumor_train_df.is_tumor==False) & (~tumor_train_df.uid.isin(tumor_slides_with_nonexhaustive_tumor_annotation))]
patch_frac = len(relevant_tumor_normal_patches.index)/(total_tumor_train_patches/2)
patch_frac = 1/patch_frac
for seed, tumor_uid in enumerate(tumor_train_slides):
if tumor_uid in tumor_slides_with_nonexhaustive_tumor_annotation:
# Should not use these slides for generating normal patches
continue
# How many normal patches does this uid have?
tumor_train_uid_df = tumor_train_df[tumor_train_df.uid==tumor_uid]
tumor_train_uid_df_normal = tumor_train_uid_df[tumor_train_uid_df.is_tumor==False]
normal_patches_in_uid = len(tumor_train_uid_df_normal.index)
tumor_patches_in_uid = len(tumor_train_uid_df[tumor_train_uid_df.is_tumor==True].index)
# subsample these proprotion of
selected_patches = tumor_train_uid_df_normal.sample(frac=patch_frac, random_state=seed+42)
normal_train_df_finalized = pd.concat([normal_train_df_finalized, selected_patches])
len(tumor_train_uid_df_normal.index)
patch_frac
patch_frac = len(normal_train_df.index)/(total_tumor_train_patches)
patch_frac = 1/patch_frac
patch_frac
len(normal_train_df_finalized.index)
# Select the remaining half from the actual normal slides.
patch_frac = len(normal_train_df.index)/(total_tumor_train_patches/2)
patch_frac = 1/patch_frac
for seed, normal_uid in enumerate(normal_train_slides):
# How many normal patches does this uid have?
normal_train_uid_df = normal_train_df[normal_train_df.uid==normal_uid]
normal_patches_in_uid = len(normal_train_uid_df.index)
# subsample these proprotion of
selected_patches = normal_train_uid_df.sample(frac=patch_frac, random_state=seed+42)
normal_train_df_finalized = pd.concat([normal_train_df_finalized, selected_patches])
len(normal_train_df_finalized.index)
total_tumor_train_patches
total_tumor_validate_patches
# Do the same thing for the validation
normal_validate_df_finalized = pd.DataFrame()
relevant_tumor_normal_patches = tumor_validate_df[(tumor_validate_df.is_tumor==False) & (~tumor_validate_df.uid.isin(tumor_slides_with_nonexhaustive_tumor_annotation))]
patch_frac = len(relevant_tumor_normal_patches.index)/(total_tumor_validate_patches)
patch_frac = 1/patch_frac
for seed, tumor_uid in enumerate(tumor_validate_slides):
if tumor_uid in tumor_slides_with_nonexhaustive_tumor_annotation:
# Should not use these slides for generating normal patches
continue
# How many normal patches does this uid have?
tumor_validate_uid_df = tumor_validate_df[tumor_validate_df.uid==tumor_uid]
tumor_validate_uid_df_normal = tumor_validate_uid_df[tumor_validate_uid_df.is_tumor==False]
normal_patches_in_uid = len(tumor_validate_uid_df_normal.index)
tumor_patches_in_uid = len(tumor_validate_uid_df[tumor_validate_uid_df.is_tumor==True].index)
# subsample these proprotion of
selected_patches = tumor_validate_uid_df_normal.sample(frac=patch_frac, random_state=seed+42)
normal_validate_df_finalized = pd.concat([normal_validate_df_finalized, selected_patches])
print(len(normal_validate_df_finalized.index))
patch_frac = len(relevant_tumor_normal_patches.index)/(total_tumor_validate_patches)
patch_frac = 1/patch_frac
patch_frac
total_tumor_validate_patches
# Select the remaining half from the actual normal slides.
patch_frac = len(normal_validate_df.index)/(total_tumor_validate_patches)
patch_frac = 1/patch_frac
for seed, normal_uid in enumerate(normal_validate_slides):
# How many normal patches does this uid have?
normal_validate_uid_df = normal_validate_df[normal_validate_df.uid==normal_uid]
normal_patches_in_uid = len(normal_validate_uid_df.index)
# subsample these proprotion of
selected_patches = normal_validate_uid_df.sample(frac=patch_frac, random_state=seed+42)
normal_validate_df_finalized = pd.concat([normal_validate_df_finalized, selected_patches])
print(len(normal_validate_df_finalized.index))
print(len(tumor_validate_df.index))
normal_train_df_finalized.to_csv('/mnt/disks/data/histopath_data/pyvirchow-out-df//normal_train_df_finalized_fullfrac.tsv',
sep='\t',
index=False,
header=True)
normal_validate_df_finalized.to_csv('/mnt/disks/data/histopath_data/pyvirchow-out-df//normal_validate_df_finalized_fullfrac.tsv',
sep='\t',
index=False,
header=True)
# Let's just merge the datasets for training
tumor_train_df_tumor = tumor_train_df[tumor_train_df.is_tumor==True]
normal_train_df_finalized['is_tumor'] = False
tumor_validate_df_tumor = tumor_validate_df[tumor_validate_df.is_tumor==True]
normal_validate_df_finalized['is_tumor'] = False
train_df = pd.concat([tumor_train_df_tumor, normal_train_df_finalized])
# Shuffle before saving so that we do not have things together
train_df = train_df.sample(frac=1, random_state=42)
validate_df = pd.concat([tumor_validate_df_tumor, normal_validate_df_finalized])
# Shuffle before saving so that we do not have things together
validate_df = validate_df.sample(frac=1, random_state=42)
train_df.to_csv('/mnt/disks/data/histopath_data/pyvirchow-out-df/train_df_fullfrac.tsv',
sep='\t',
index=False,
header=True)
validate_df.to_csv('//mnt/disks/data/histopath_data/pyvirchow-out-df/validate_df_fullfrac.tsv',
sep='\t',
index=False,
header=True)
len(train_df.index)
len(validate_df.index)
len(train_df[train_df.is_tumor==True].index)
len(train_df[train_df.is_tumor==False].index)
| 0.401453 | 0.763528 |
```
import cadquery as cq
from jupyter_cadquery.cadquery import show
from jupyter_cadquery import set_defaults
set_defaults(display='cell', axes=False, axes0=True, grid=True,
default_color='lightgrey', tools=False)
from cq_gears import SpurGear, HerringboneGear
```
## Spur Gear
```
spur_gear = SpurGear(module=1.0, teeth_number=19, width=5.0, bore_d=5.0)
wp = cq.Workplane('XY').gear(spur_gear)
show(wp)
```
## Spur gear train
```
spur_gear = SpurGear(module=1.0, teeth_number=13, width=5.0, bore_d=5.0)
wp = (cq.Workplane('XY')
# Pushing an array of 4 points with spacing equal to the gear's pitch diameter
.rarray(xSpacing=spur_gear.r0 * 2.0,
ySpacing=1.0, xCount=4, yCount=1, center=False)
# Create 4 meshing gears
.gear(spur_gear)
.moveTo(spur_gear.r0 * 2 * 4, 0.0)
# Create an additional gear with the same profile but with different
# bore diameter and hub
.addGear(spur_gear, bore_d=3.0, hub_d=8.0, hub_length=4.0))
show(wp)
```
## Helical Gears
```
# Create two meshing helical gears with different tooth count and face width
helical_gear1 = SpurGear(module=1.0, teeth_number=29, width=16.0,
# the sign of helix_angle determines direction:
# positive=clockwise, negative=counterclockwise
helix_angle=40.0,
bore_d=20.0)
helical_gear2 = SpurGear(module=1.0, teeth_number=17, width=8.0,
helix_angle=-40.0,
bore_d=10.0)
wp = (cq.Workplane('XY')
.gear(helical_gear1)
.moveTo(helical_gear1.r0 + helical_gear2.r0, 0.0)
.addGear(helical_gear2))
show(wp)
```
## Herringbone Gear
```
# Herringbone helical gear variation
hb_gear = HerringboneGear(module=1.0, teeth_number=42, width=10.0,
helix_angle=-20.0, bore_d=28.0)
wp = cq.Workplane('XY').gear(hb_gear)
show(wp)
```
## Beaten up Spur Gear
```
# Build a gear with some missing teeth
beaten_spur_gear = SpurGear(module=1.0, teeth_number=20, width=5.0,
pressure_angle=20.0, bore_d=5.0,
missing_teeth=(0, 10))
wp = cq.Workplane('XY').gear(beaten_spur_gear)
show(wp)
```
## Helical Gear with spokes and extended hub
```
helical_gear = SpurGear(module=1.0, teeth_number=60, width=8.0,
pressure_angle=20.0, helix_angle=45.0,
bore_d=10.0, hub_d=16.0, hub_length=10.0,
recess_d=52.0, recess=3.0, n_spokes=5,
spoke_width=6.0, spoke_fillet=4.0,
spokes_id=23.0, spokes_od=48.0)
wp = cq.Workplane('XY').gear(helical_gear)
show(wp)
```
|
github_jupyter
|
import cadquery as cq
from jupyter_cadquery.cadquery import show
from jupyter_cadquery import set_defaults
set_defaults(display='cell', axes=False, axes0=True, grid=True,
default_color='lightgrey', tools=False)
from cq_gears import SpurGear, HerringboneGear
spur_gear = SpurGear(module=1.0, teeth_number=19, width=5.0, bore_d=5.0)
wp = cq.Workplane('XY').gear(spur_gear)
show(wp)
spur_gear = SpurGear(module=1.0, teeth_number=13, width=5.0, bore_d=5.0)
wp = (cq.Workplane('XY')
# Pushing an array of 4 points with spacing equal to the gear's pitch diameter
.rarray(xSpacing=spur_gear.r0 * 2.0,
ySpacing=1.0, xCount=4, yCount=1, center=False)
# Create 4 meshing gears
.gear(spur_gear)
.moveTo(spur_gear.r0 * 2 * 4, 0.0)
# Create an additional gear with the same profile but with different
# bore diameter and hub
.addGear(spur_gear, bore_d=3.0, hub_d=8.0, hub_length=4.0))
show(wp)
# Create two meshing helical gears with different tooth count and face width
helical_gear1 = SpurGear(module=1.0, teeth_number=29, width=16.0,
# the sign of helix_angle determines direction:
# positive=clockwise, negative=counterclockwise
helix_angle=40.0,
bore_d=20.0)
helical_gear2 = SpurGear(module=1.0, teeth_number=17, width=8.0,
helix_angle=-40.0,
bore_d=10.0)
wp = (cq.Workplane('XY')
.gear(helical_gear1)
.moveTo(helical_gear1.r0 + helical_gear2.r0, 0.0)
.addGear(helical_gear2))
show(wp)
# Herringbone helical gear variation
hb_gear = HerringboneGear(module=1.0, teeth_number=42, width=10.0,
helix_angle=-20.0, bore_d=28.0)
wp = cq.Workplane('XY').gear(hb_gear)
show(wp)
# Build a gear with some missing teeth
beaten_spur_gear = SpurGear(module=1.0, teeth_number=20, width=5.0,
pressure_angle=20.0, bore_d=5.0,
missing_teeth=(0, 10))
wp = cq.Workplane('XY').gear(beaten_spur_gear)
show(wp)
helical_gear = SpurGear(module=1.0, teeth_number=60, width=8.0,
pressure_angle=20.0, helix_angle=45.0,
bore_d=10.0, hub_d=16.0, hub_length=10.0,
recess_d=52.0, recess=3.0, n_spokes=5,
spoke_width=6.0, spoke_fillet=4.0,
spokes_id=23.0, spokes_od=48.0)
wp = cq.Workplane('XY').gear(helical_gear)
show(wp)
| 0.486819 | 0.845751 |
# Improving Data Quality
**Learning Objectives**
1. Resolve missing values
2. Convert the Date feature column to a datetime format
3. Rename a feature column, remove a value from a feature column
4. Create one-hot encoding features
5. Understand temporal feature conversions
## Introduction
Recall that machine learning models can only consume numeric data, and that numeric data should be "1"s or "0"s. Data is said to be "messy" or "untidy" if it is missing attribute values, contains noise or outliers, has duplicates, wrong data, upper/lower case column names, and is essentially not ready for ingestion by a machine learning algorithm.
This notebook presents and solves some of the most common issues of "untidy" data. Note that different problems will require different methods, and they are beyond the scope of this notebook.
Each learning objective will correspond to a _#TODO_ in this student lab notebook -- try to complete this notebook first and then review the [solution notebook](https://github.com/GoogleCloudPlatform/training-data-analyst/blob/master/courses/machine_learning/deepdive2/launching_into_ml/solutions/improve_data_quality.ipynb).
```
!sudo chown -R jupyter:jupyter /home/jupyter/training-data-analyst
```
Start by importing the necessary libraries for this lab.
### Import Libraries
```
# Importing necessary tensorflow library and printing the TF version.
import tensorflow as tf
print("TensorFlow version: ",tf.version.VERSION)
import os
import pandas as pd # First, we'll import Pandas, a data processing and CSV file I/O library
import numpy as np
from datetime import datetime
import matplotlib.pyplot as plt
import seaborn as sns
%matplotlib inline
```
### Load the Dataset
The dataset is based on California's [Vehicle Fuel Type Count by Zip Code](https://data.ca.gov/dataset/vehicle-fuel-type-count-by-zip-codeSynthetic) report. The dataset has been modified to make the data "untidy" and is thus a synthetic representation that can be used for learning purposes.
Let's download the raw .csv data by copying the data from a cloud storage bucket.
```
if not os.path.isdir("../data/transport"):
os.makedirs("../data/transport")
!gsutil cp gs://cloud-training-demos/feat_eng/transport/untidy_vehicle_data.csv ../data/transport
!ls -l ../data/transport
```
### Read Dataset into a Pandas DataFrame
Next, let's read in the dataset just copied from the cloud storage bucket and create a Pandas DataFrame. We also add a Pandas .head() function to show you the top 5 rows of data in the DataFrame. Head() and Tail() are "best-practice" functions used to investigate datasets.
```
df_transport = pd.read_csv('../data/transport/untidy_vehicle_data.csv')
df_transport.head() # Output the first five rows.
```
### DataFrame Column Data Types
DataFrames may have heterogenous or "mixed" data types, that is, some columns are numbers, some are strings, and some are dates etc. Because CSV files do not contain information on what data types are contained in each column, Pandas infers the data types when loading the data, e.g. if a column contains only numbers, Pandas will set that column’s data type to numeric: integer or float.
Run the next cell to see information on the DataFrame.
```
df_transport.info()
```
From what the .info() function shows us, we have six string objects and one float object. Let's print out the first and last five rows of each column. We can definitely see more of the "string" object values now!
```
print(df_transport)
```
### Summary Statistics
At this point, we have only one column which contains a numerical value (e.g. Vehicles). For features which contain numerical values, we are often interested in various statistical measures relating to those values. We can use .describe() to see some summary statistics for the numeric fields in our dataframe. Note, that because we only have one numeric feature, we see only one summary stastic - for now.
```
df_transport.describe()
```
Let's investigate a bit more of our data by using the .groupby() function.
```
df_transport.groupby('Fuel').first() # Get the first entry for each month.
```
#### Checking for Missing Values
Missing values adversely impact data quality, as they can lead the machine learning model to make inaccurate inferences about the data. Missing values can be the result of numerous factors, e.g. "bits" lost during streaming transmission, data entry, or perhaps a user forgot to fill in a field. Note that Pandas recognizes both empty cells and “NaN” types as missing values.
Let's show the null values for all features in the DataFrame.
```
df_transport.isnull().sum()
```
To see a sampling of which values are missing, enter the feature column name. You'll notice that "False" and "True" correpond to the presence or abscence of a value by index number.
```
print (df_transport['Date'])
print (df_transport['Date'].isnull())
print (df_transport['Make'])
print (df_transport['Make'].isnull())
print (df_transport['Model Year'])
print (df_transport['Model Year'].isnull())
```
### What can we deduce about the data at this point?
First, let's summarize our data by row, column, features, unique, and missing values,
```
print ("Rows : " ,df_transport.shape[0])
print ("Columns : " ,df_transport.shape[1])
print ("\nFeatures : \n" ,df_transport.columns.tolist())
print ("\nUnique values : \n",df_transport.nunique())
print ("\nMissing values : ", df_transport.isnull().sum().values.sum())
```
Let's see the data again -- this time the last five rows in the dataset.
```
df_transport.tail()
```
### What Are Our Data Quality Issues?
1. **Data Quality Issue #1**:
> **Missing Values**:
Each feature column has multiple missing values. In fact, we have a total of 18 missing values.
2. **Data Quality Issue #2**:
> **Date DataType**: Date is shown as an "object" datatype and should be a datetime. In addition, Date is in one column. Our business requirement is to see the Date parsed out to year, month, and day.
3. **Data Quality Issue #3**:
> **Model Year**: We are only interested in years greater than 2006, not "<2006".
4. **Data Quality Issue #4**:
> **Categorical Columns**: The feature column "Light_Duty" is categorical and has a "Yes/No" choice. We cannot feed values like this into a machine learning model. In addition, we need to "one-hot encode the remaining "string"/"object" columns.
5. **Data Quality Issue #5**:
> **Temporal Features**: How do we handle year, month, and day?
#### Data Quality Issue #1:
##### Resolving Missing Values
Most algorithms do not accept missing values. Yet, when we see missing values in our dataset, there is always a tendency to just "drop all the rows" with missing values. Although Pandas will fill in the blank space with “NaN", we should "handle" them in some way.
While all the methods to handle missing values is beyond the scope of this lab, there are a few methods you should consider. For numeric columns, use the "mean" values to fill in the missing numeric values. For categorical columns, use the "mode" (or most frequent values) to fill in missing categorical values.
In this lab, we use the .apply and Lambda functions to fill every column with its own most frequent value. You'll learn more about Lambda functions later in the lab.
Let's check again for missing values by showing how many rows contain NaN values for each feature column.
**Lab Task #1a:** Check for missing values by showing how many rows contain NaN values for each feature column.
```
# The isnull() method is used to check and manage NULL values in a data frame.
# TODO 1a
df_transport.isnull().sum()
```
**Lab Task #1b:** Apply the lambda function.
```
# Here we are using the apply function with lambda.
# We can use the apply() function to apply the lambda function to both rows and columns of a dataframe.
# TODO 1b
df_transport = df_transport.apply(lambda x:x.fillna(x.value_counts().index[0]))
df_transport.head()
```
**Lab Task #1c:** Check again for missing values.
```
# The isnull() method is used to check and manage NULL values in a data frame.
# TODO 1c
df_transport.isnull().sum()
```
#### Data Quality Issue #2:
##### Convert the Date Feature Column to a Datetime Format
The date column is indeed shown as a string object.
**Lab Task #2a:** Convert the datetime datatype with the to_datetime() function in Pandas.
```
# The date column is indeed shown as a string object. We can convert it to the datetime datatype with the to_datetime() function in Pandas.
# TODO 2a
df_transport['Date'] = pd.to_datetime(df_transport['Date'],
format='%m/%d/%Y')
```
**Lab Task #2b:** Show the converted Date.
```
# Date is now converted and will display the concise summary of an dataframe.
# TODO 2b
df_transport.info()
```
Let's parse Date into three columns, e.g. year, month, and day.
```
df_transport['year'] = df_transport['Date'].dt.year
df_transport['month'] = df_transport['Date'].dt.month
df_transport['day'] = df_transport['Date'].dt.day
#df['hour'] = df['date'].dt.hour - you could use this if your date format included hour.
#df['minute'] = df['date'].dt.minute - you could use this if your date format included minute.
df_transport.info()
```
Next, let's confirm the Date parsing. This will also give us a another visualization of the data.
```
# Here, we are creating a new dataframe called "grouped_data" and grouping by on the column "Make"
grouped_data = df_transport.groupby(['Make'])
# Get the first entry for each month.
df_transport.groupby('month').first()
```
Now that we have Dates as a integers, let's do some additional plotting.
```
plt.figure(figsize=(10,6))
sns.jointplot(x='month',y='Vehicles',data=df_transport)
#plt.title('Vehicles by Month')
```
#### Data Quality Issue #3:
##### Rename a Feature Column and Remove a Value.
Our feature columns have different "capitalizations" in their names, e.g. both upper and lower "case". In addition, there are "spaces" in some of the column names. In addition, we are only interested in years greater than 2006, not "<2006".
**Lab Task #3a:** Remove all the spaces for feature columns by renaming them.
```
# Let's remove all the spaces for feature columns by renaming them.
# TODO 3a
df_transport.rename(columns = { 'Date': 'date', 'Zip Code':'zipcode', 'Model Year': 'modelyear', 'Fuel': 'fuel', 'Make': 'make', 'Light_Duty': 'lightduty', 'Vehicles': 'vehicles'}, inplace = True)
# Output the first two rows.
df_transport.head(2)
```
**Note:** Next we create a copy of the dataframe to avoid the "SettingWithCopyWarning: A value is trying to be set on a copy of a slice from a DataFrame" warning. Run the cell to remove the value '<2006' from the modelyear feature column.
**Lab Task #3b:** Create a copy of the dataframe to avoid copy warning issues.
```
# Here, we create a copy of the dataframe to avoid copy warning issues.
# TODO 3b
df = df_transport.loc[df_transport.modelyear != '<2006'].copy()
```
Next, confirm that the modelyear value '<2006' has been removed by doing a value count.
```
df['modelyear'].value_counts(0)
```
#### Data Quality Issue #4:
##### Handling Categorical Columns
The feature column "lightduty" is categorical and has a "Yes/No" choice. We cannot feed values like this into a machine learning model. We need to convert the binary answers from strings of yes/no to integers of 1/0. There are various methods to achieve this. We will use the "apply" method with a lambda expression. Pandas. apply() takes a function and applies it to all values of a Pandas series.
##### What is a Lambda Function?
Typically, Python requires that you define a function using the def keyword. However, lambda functions are anonymous -- which means there is no need to name them. The most common use case for lambda functions is in code that requires a simple one-line function (e.g. lambdas only have a single expression).
As you progress through the Course Specialization, you will see many examples where lambda functions are being used. Now is a good time to become familiar with them.
First, lets count the number of "Yes" and"No's" in the 'lightduty' feature column.
```
df['lightduty'].value_counts(0)
```
Let's convert the Yes to 1 and No to 0. Pandas. apply() . apply takes a function and applies it to all values of a Pandas series (e.g. lightduty).
```
df.loc[:,'lightduty'] = df['lightduty'].apply(lambda x: 0 if x=='No' else 1)
df['lightduty'].value_counts(0)
# Confirm that "lightduty" has been converted.
df.head()
```
#### One-Hot Encoding Categorical Feature Columns
Machine learning algorithms expect input vectors and not categorical features. Specifically, they cannot handle text or string values. Thus, it is often useful to transform categorical features into vectors.
One transformation method is to create dummy variables for our categorical features. Dummy variables are a set of binary (0 or 1) variables that each represent a single class from a categorical feature. We simply encode the categorical variable as a one-hot vector, i.e. a vector where only one element is non-zero, or hot. With one-hot encoding, a categorical feature becomes an array whose size is the number of possible choices for that feature.
Panda provides a function called "get_dummies" to convert a categorical variable into dummy/indicator variables.
```
# Making dummy variables for categorical data with more inputs.
data_dummy = pd.get_dummies(df[['zipcode','modelyear', 'fuel', 'make']], drop_first=True)
data_dummy.head()
```
**Lab Task #4a:** Merge (concatenate) original data frame with 'dummy' dataframe.
```
# Merging (concatenate) original data frame with 'dummy' dataframe.
# TODO 4a
df = pd.concat([df,data_dummy], axis=1)
df.head()
```
**Lab Task #4b:** Drop attributes for which we made dummy variables.
```
# Dropping attributes for which we made dummy variables. Let's also drop the Date column.
# TODO 4b
df = df.drop(['date','zipcode','modelyear', 'fuel', 'make'], axis=1)
# Confirm that 'zipcode','modelyear', 'fuel', and 'make' have been dropped.
df.head()
```
#### Data Quality Issue #5:
##### Temporal Feature Columns
Our dataset now contains year, month, and day feature columns. Let's convert the month and day feature columns to meaningful representations as a way to get us thinking about changing temporal features -- as they are sometimes overlooked.
Note that the Feature Engineering course in this Specialization will provide more depth on methods to handle year, month, day, and hour feature columns.
First, let's print the unique values for "month" and "day" in our dataset.
```
print ('Unique values of month:',df.month.unique())
print ('Unique values of day:',df.day.unique())
print ('Unique values of year:',df.year.unique())
```
Next, we map each temporal variable onto a circle such that the lowest value for that variable appears right next to the largest value. We compute the x- and y- component of that point using sin and cos trigonometric functions. Don't worry, this is the last time we will use this code, as you can develop an input pipeline to address these temporal feature columns in TensorFlow and Keras - and it is much easier! But, sometimes you need to appreciate what you're not going to encounter as you move through the course!
Run the cell to view the output.
**Lab Task #5:** Drop month, and day
```
df['day_sin'] = np.sin(df.day*(2.*np.pi/31))
df['day_cos'] = np.cos(df.day*(2.*np.pi/31))
df['month_sin'] = np.sin((df.month-1)*(2.*np.pi/12))
df['month_cos'] = np.cos((df.month-1)*(2.*np.pi/12))
# TODO 5
df = df.drop(['month','day','year'], axis=1)
# scroll left to see the converted month and day coluumns.
df.tail(4)
```
### Conclusion
This notebook introduced a few concepts to improve data quality. We resolved missing values, converted the Date feature column to a datetime format, renamed feature columns, removed a value from a feature column, created one-hot encoding features, and converted temporal features to meaningful representations. By the end of our lab, we gained an understanding as to why data should be "cleaned" and "pre-processed" before input into a machine learning model.
Copyright 2020 Google Inc.
Licensed under the Apache License, Version 2.0 (the "License"); you may not use this file except in compliance with the License. You may obtain a copy of the License at
http://www.apache.org/licenses/LICENSE-2.0
Unless required by applicable law or agreed to in writing, software distributed under the License is distributed on an "AS IS" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. See the License for the specific language governing permissions and limitations under the License.
|
github_jupyter
|
!sudo chown -R jupyter:jupyter /home/jupyter/training-data-analyst
# Importing necessary tensorflow library and printing the TF version.
import tensorflow as tf
print("TensorFlow version: ",tf.version.VERSION)
import os
import pandas as pd # First, we'll import Pandas, a data processing and CSV file I/O library
import numpy as np
from datetime import datetime
import matplotlib.pyplot as plt
import seaborn as sns
%matplotlib inline
if not os.path.isdir("../data/transport"):
os.makedirs("../data/transport")
!gsutil cp gs://cloud-training-demos/feat_eng/transport/untidy_vehicle_data.csv ../data/transport
!ls -l ../data/transport
df_transport = pd.read_csv('../data/transport/untidy_vehicle_data.csv')
df_transport.head() # Output the first five rows.
df_transport.info()
print(df_transport)
df_transport.describe()
df_transport.groupby('Fuel').first() # Get the first entry for each month.
df_transport.isnull().sum()
print (df_transport['Date'])
print (df_transport['Date'].isnull())
print (df_transport['Make'])
print (df_transport['Make'].isnull())
print (df_transport['Model Year'])
print (df_transport['Model Year'].isnull())
print ("Rows : " ,df_transport.shape[0])
print ("Columns : " ,df_transport.shape[1])
print ("\nFeatures : \n" ,df_transport.columns.tolist())
print ("\nUnique values : \n",df_transport.nunique())
print ("\nMissing values : ", df_transport.isnull().sum().values.sum())
df_transport.tail()
# The isnull() method is used to check and manage NULL values in a data frame.
# TODO 1a
df_transport.isnull().sum()
# Here we are using the apply function with lambda.
# We can use the apply() function to apply the lambda function to both rows and columns of a dataframe.
# TODO 1b
df_transport = df_transport.apply(lambda x:x.fillna(x.value_counts().index[0]))
df_transport.head()
# The isnull() method is used to check and manage NULL values in a data frame.
# TODO 1c
df_transport.isnull().sum()
# The date column is indeed shown as a string object. We can convert it to the datetime datatype with the to_datetime() function in Pandas.
# TODO 2a
df_transport['Date'] = pd.to_datetime(df_transport['Date'],
format='%m/%d/%Y')
# Date is now converted and will display the concise summary of an dataframe.
# TODO 2b
df_transport.info()
df_transport['year'] = df_transport['Date'].dt.year
df_transport['month'] = df_transport['Date'].dt.month
df_transport['day'] = df_transport['Date'].dt.day
#df['hour'] = df['date'].dt.hour - you could use this if your date format included hour.
#df['minute'] = df['date'].dt.minute - you could use this if your date format included minute.
df_transport.info()
# Here, we are creating a new dataframe called "grouped_data" and grouping by on the column "Make"
grouped_data = df_transport.groupby(['Make'])
# Get the first entry for each month.
df_transport.groupby('month').first()
plt.figure(figsize=(10,6))
sns.jointplot(x='month',y='Vehicles',data=df_transport)
#plt.title('Vehicles by Month')
# Let's remove all the spaces for feature columns by renaming them.
# TODO 3a
df_transport.rename(columns = { 'Date': 'date', 'Zip Code':'zipcode', 'Model Year': 'modelyear', 'Fuel': 'fuel', 'Make': 'make', 'Light_Duty': 'lightduty', 'Vehicles': 'vehicles'}, inplace = True)
# Output the first two rows.
df_transport.head(2)
# Here, we create a copy of the dataframe to avoid copy warning issues.
# TODO 3b
df = df_transport.loc[df_transport.modelyear != '<2006'].copy()
df['modelyear'].value_counts(0)
df['lightduty'].value_counts(0)
df.loc[:,'lightduty'] = df['lightduty'].apply(lambda x: 0 if x=='No' else 1)
df['lightduty'].value_counts(0)
# Confirm that "lightduty" has been converted.
df.head()
# Making dummy variables for categorical data with more inputs.
data_dummy = pd.get_dummies(df[['zipcode','modelyear', 'fuel', 'make']], drop_first=True)
data_dummy.head()
# Merging (concatenate) original data frame with 'dummy' dataframe.
# TODO 4a
df = pd.concat([df,data_dummy], axis=1)
df.head()
# Dropping attributes for which we made dummy variables. Let's also drop the Date column.
# TODO 4b
df = df.drop(['date','zipcode','modelyear', 'fuel', 'make'], axis=1)
# Confirm that 'zipcode','modelyear', 'fuel', and 'make' have been dropped.
df.head()
print ('Unique values of month:',df.month.unique())
print ('Unique values of day:',df.day.unique())
print ('Unique values of year:',df.year.unique())
df['day_sin'] = np.sin(df.day*(2.*np.pi/31))
df['day_cos'] = np.cos(df.day*(2.*np.pi/31))
df['month_sin'] = np.sin((df.month-1)*(2.*np.pi/12))
df['month_cos'] = np.cos((df.month-1)*(2.*np.pi/12))
# TODO 5
df = df.drop(['month','day','year'], axis=1)
# scroll left to see the converted month and day coluumns.
df.tail(4)
| 0.233706 | 0.987412 |
# Lendo dados de geociência
## License
All content can be freely used and adapted under the terms of the
[Creative Commons Attribution 4.0 International License](http://creativecommons.org/licenses/by/4.0/).

## Imports
Coloque **todos** os `import` na célula abaixo. Não se esqueça do `%matplotlib inline` para que os gráficos apareçam no notebook.
```
import math
import matplotlib.pyplot as plt
%matplotlib inline
```
## IMPORTANTE
Agora que vocês sabem técnicas de programação defensiva, eu espero que todo o código que vocês fizerem abaixo utilizem essas técnicas. Crie docstrings para suas funções, cheque as entradas (quando for possível) e cheque as saídas. **Não esqueçam dos comentários**.
## Temperatura no Rio de Janeiro
O arquivo `data/23.31S-42.82W-TAVG-Trend.txt` contém dados de temperatura média mensal para a cidade do Rio de Janeiro. O arquivo também contém médias móveis anual, 5, 10 e 20 anos. Esses dados foram baixados do site Berkeley Earth (http://berkeleyearth.lbl.gov/locations/23.31S-42.82W).
### Tarefa
Faça duas funções, uma que lê os dados de temperatura mensal, outra que lê os dados da média móvel anual.
As duas funções devem:
* Receber como entrada **somente** o nome do arquivo de dados.
* Retornar duas listas: uma com as datas referentes aos dados e outra com os dados de temperatura.
* As datas retornadas devem ser em anos decimais. Ex: Janeiro de 1984 seria 1984.0833333333333 (1984 + 1/12).
* Datas sem valores de temperatura (NaN) devem ser ignoradas (não incluidas nas listas).
Utilize suas funções para carregar os dados e fazer um gráfico da temperatura média mensal e média movel anual pelo tempo.
```
arquivo = open("data/23.31S-42.82W-TAVG-Trend.txt")
numlin = 0
datas_mensais = []
Mtemperaturas = []
for linha in arquivo:
numlin = numlin + 1
blocos = linha.split()
if linha[0] != '%' and len(blocos) and blocos[2] != 'NaN':
Mtemperatura = 24.01 + float(blocos[2])
Mtemperaturas.append(Mtemperatura)
data = float(blocos[0]) + float(blocos[1])/12
datas_mensais.append(data)
arquivo.close()
N = 2096
for i in range(N):
print(datas_mensais[i],Mtemperaturas[i])
print(Mtemperaturas)
arquivo = open("data/23.31S-42.82W-TAVG-Trend.txt")
numlin = 0
datas_anuais = []
Atemperaturas = []
for linha in arquivo:
numlin = numlin + 1
blocos = linha.split()
if linha[0] != '%' and len(blocos) and blocos[4] != 'NaN':
Atemperatura = 24.01 + float(blocos[4])
Atemperaturas.append(Atemperatura)
data = float(blocos[0]) + float(blocos[1])/12
datas_anuais.append(data)
arquivo.close()
N = 2096
for i in range(N):
print(datas_mensais[i],Atemperaturas[i])
plt.figure()
plt.plot(datas_mensais, Mtemperaturas , ".k",label = "Média Mensal")
plt.plot(datas_anuais, Atemperaturas, "-r",label = "Média Móvel Anual")
plt.xlabel('Ano')
plt.ylabel('Temperatura Média (°C)')
plt.xlim(1820,2020)
plt.grid(axis = 'both')
plt.legend(loc='upper left', shadow=True, fontsize='large')
```
### Resultado esperado
O gráfico final deve ser parecido com o abaixo:

### Tarefa
Faça uma função que calcule a temperatura média anual a partir das temperaturas mensais. A sua função deve:
* Receber como entrada a lista das datas e a lista das temperaturas mensais.
* Retornar duas listas: uma com os anos e outra com as temperaturas médias correspondetes.
* Anos que não contem dados de todos os 12 meses devem ser ignorados (não incluídos nas listas retornadas).
Utilize sua função para calcular a média anual. Faça um gráfico da temperatura média anual por ano junto com a média móvel anual.
**Dica**: A função `math.floor` retorna o número inteiro que precede um número real. Ex: `math.floor(1984.23) == 1984`
```
resultado = []
for i in range(0,len(datas_mensais),1):
resultado.append(math.floor(datas_mensais[i]))
print(resultado)
Resposta = []
for i in range(0,len(datas_mensais),1):
if i + 11 < len(datas_mensais) and math.floor(datas_mensais[i]) == math.floor(datas_mensais[i+11]):
var = 0
for j in range(0,12,1):
var = (Mtemperaturas[i + j])/12 + var
lista = [var,math.floor(datas_mensais[i])]
Resposta.append(lista)
print(Resposta)
for i in range(0,len(datas_mensais),12):
print(i)
len(datas_mensais)
25%12
```
### Resultado esperado
O gráfico final deve ser parecido com o abaixo:

## Tarefa Bônus
Salve os dados da média anual em um arquivo CSV (comma separated values) chamado `temp-media-anual.csv`. Os valores devem ser separados por `,`. A primeira coluna deve conter os anos e a segunda as temperaturas. Esse arquivo deve estar presente em seu repositório (dê `git add` nele).
|
github_jupyter
|
import math
import matplotlib.pyplot as plt
%matplotlib inline
arquivo = open("data/23.31S-42.82W-TAVG-Trend.txt")
numlin = 0
datas_mensais = []
Mtemperaturas = []
for linha in arquivo:
numlin = numlin + 1
blocos = linha.split()
if linha[0] != '%' and len(blocos) and blocos[2] != 'NaN':
Mtemperatura = 24.01 + float(blocos[2])
Mtemperaturas.append(Mtemperatura)
data = float(blocos[0]) + float(blocos[1])/12
datas_mensais.append(data)
arquivo.close()
N = 2096
for i in range(N):
print(datas_mensais[i],Mtemperaturas[i])
print(Mtemperaturas)
arquivo = open("data/23.31S-42.82W-TAVG-Trend.txt")
numlin = 0
datas_anuais = []
Atemperaturas = []
for linha in arquivo:
numlin = numlin + 1
blocos = linha.split()
if linha[0] != '%' and len(blocos) and blocos[4] != 'NaN':
Atemperatura = 24.01 + float(blocos[4])
Atemperaturas.append(Atemperatura)
data = float(blocos[0]) + float(blocos[1])/12
datas_anuais.append(data)
arquivo.close()
N = 2096
for i in range(N):
print(datas_mensais[i],Atemperaturas[i])
plt.figure()
plt.plot(datas_mensais, Mtemperaturas , ".k",label = "Média Mensal")
plt.plot(datas_anuais, Atemperaturas, "-r",label = "Média Móvel Anual")
plt.xlabel('Ano')
plt.ylabel('Temperatura Média (°C)')
plt.xlim(1820,2020)
plt.grid(axis = 'both')
plt.legend(loc='upper left', shadow=True, fontsize='large')
resultado = []
for i in range(0,len(datas_mensais),1):
resultado.append(math.floor(datas_mensais[i]))
print(resultado)
Resposta = []
for i in range(0,len(datas_mensais),1):
if i + 11 < len(datas_mensais) and math.floor(datas_mensais[i]) == math.floor(datas_mensais[i+11]):
var = 0
for j in range(0,12,1):
var = (Mtemperaturas[i + j])/12 + var
lista = [var,math.floor(datas_mensais[i])]
Resposta.append(lista)
print(Resposta)
for i in range(0,len(datas_mensais),12):
print(i)
len(datas_mensais)
25%12
| 0.156395 | 0.909787 |
In this example we see how to robustly fit a linear model to faulty data using the RANSAC algorithm.
#### New to Plotly?
Plotly's Python library is free and open source! [Get started](https://plot.ly/python/getting-started/) by downloading the client and [reading the primer](https://plot.ly/python/getting-started/).
<br>You can set up Plotly to work in [online](https://plot.ly/python/getting-started/#initialization-for-online-plotting) or [offline](https://plot.ly/python/getting-started/#initialization-for-offline-plotting) mode, or in [jupyter notebooks](https://plot.ly/python/getting-started/#start-plotting-online).
<br>We also have a quick-reference [cheatsheet](https://images.plot.ly/plotly-documentation/images/python_cheat_sheet.pdf) (new!) to help you get started!
### Version
```
import sklearn
sklearn.__version__
```
### Imports
```
import plotly.plotly as py
import plotly.graph_objs as go
import numpy as np
from sklearn import linear_model, datasets
```
### Calculations
```
n_samples = 1000
n_outliers = 50
X, y, coef = datasets.make_regression(n_samples=n_samples, n_features=1,
n_informative=1, noise=10,
coef=True, random_state=0)
# Add outlier data
np.random.seed(0)
X[:n_outliers] = 3 + 0.5 * np.random.normal(size=(n_outliers, 1))
y[:n_outliers] = -3 + 10 * np.random.normal(size=n_outliers)
# Fit line using all data
model = linear_model.LinearRegression()
model.fit(X, y)
# Robustly fit linear model with RANSAC algorithm
model_ransac = linear_model.RANSACRegressor(linear_model.LinearRegression())
model_ransac.fit(X, y)
inlier_mask = model_ransac.inlier_mask_
outlier_mask = np.logical_not(inlier_mask)
# Predict data of estimated models
line_X = np.arange(-5, 5)
line_y = model.predict(line_X[:, np.newaxis])
line_y_ransac = model_ransac.predict(line_X[:, np.newaxis])
```
Compare estimated coefficients
```
print("Estimated coefficients (true, normal, RANSAC):")
print(coef, model.coef_, model_ransac.estimator_.coef_)
```
### Plot Results
```
def data_to_plotly(x):
k = []
for i in range(0, len(x)):
k.append(x[i][0])
return k
lw = 2
p1 = go.Scatter(x=data_to_plotly(X[inlier_mask]), y=y[inlier_mask],
mode='markers',
marker=dict(color='yellowgreen', size=6),
name='Inliers')
p2 = go.Scatter(x=data_to_plotly(X[outlier_mask]), y=y[outlier_mask],
mode='markers',
marker=dict(color='gold', size=6),
name='Outliers')
p3 = go.Scatter(x=line_X, y=line_y,
mode='lines',
line=dict(color='navy', width=lw,),
name='Linear regressor')
p4 = go.Scatter(x=line_X, y=line_y_ransac,
mode='lines',
line=dict(color='cornflowerblue', width=lw),
name='RANSAC regressor')
data = [p1, p2, p3, p4]
layout = go.Layout(xaxis=dict(zeroline=False, showgrid=False),
yaxis=dict(zeroline=False, showgrid=False)
)
fig = go.Figure(data=data, layout=layout)
py.iplot(fig)
from IPython.display import display, HTML
display(HTML('<link href="//fonts.googleapis.com/css?family=Open+Sans:600,400,300,200|Inconsolata|Ubuntu+Mono:400,700" rel="stylesheet" type="text/css" />'))
display(HTML('<link rel="stylesheet" type="text/css" href="http://help.plot.ly/documentation/all_static/css/ipython-notebook-custom.css">'))
! pip install git+https://github.com/plotly/publisher.git --upgrade
import publisher
publisher.publish(
'Robust Linear Model Estimation using RANSAC.ipynb', 'scikit-learn/plot-ransac/', 'Robust Linear Model Estimation using RANSAC | plotly',
'',
title = 'Robust Linear Model Estimation using RANSAC | plotly',
name = 'Robust Linear Model Estimation using RANSAC',
has_thumbnail='true', thumbnail='thumbnail/ransac.jpg',
language='scikit-learn', page_type='example_index',
display_as='linear_models', order=18,
ipynb= '~Diksha_Gabha/3223')
```
|
github_jupyter
|
import sklearn
sklearn.__version__
import plotly.plotly as py
import plotly.graph_objs as go
import numpy as np
from sklearn import linear_model, datasets
n_samples = 1000
n_outliers = 50
X, y, coef = datasets.make_regression(n_samples=n_samples, n_features=1,
n_informative=1, noise=10,
coef=True, random_state=0)
# Add outlier data
np.random.seed(0)
X[:n_outliers] = 3 + 0.5 * np.random.normal(size=(n_outliers, 1))
y[:n_outliers] = -3 + 10 * np.random.normal(size=n_outliers)
# Fit line using all data
model = linear_model.LinearRegression()
model.fit(X, y)
# Robustly fit linear model with RANSAC algorithm
model_ransac = linear_model.RANSACRegressor(linear_model.LinearRegression())
model_ransac.fit(X, y)
inlier_mask = model_ransac.inlier_mask_
outlier_mask = np.logical_not(inlier_mask)
# Predict data of estimated models
line_X = np.arange(-5, 5)
line_y = model.predict(line_X[:, np.newaxis])
line_y_ransac = model_ransac.predict(line_X[:, np.newaxis])
print("Estimated coefficients (true, normal, RANSAC):")
print(coef, model.coef_, model_ransac.estimator_.coef_)
def data_to_plotly(x):
k = []
for i in range(0, len(x)):
k.append(x[i][0])
return k
lw = 2
p1 = go.Scatter(x=data_to_plotly(X[inlier_mask]), y=y[inlier_mask],
mode='markers',
marker=dict(color='yellowgreen', size=6),
name='Inliers')
p2 = go.Scatter(x=data_to_plotly(X[outlier_mask]), y=y[outlier_mask],
mode='markers',
marker=dict(color='gold', size=6),
name='Outliers')
p3 = go.Scatter(x=line_X, y=line_y,
mode='lines',
line=dict(color='navy', width=lw,),
name='Linear regressor')
p4 = go.Scatter(x=line_X, y=line_y_ransac,
mode='lines',
line=dict(color='cornflowerblue', width=lw),
name='RANSAC regressor')
data = [p1, p2, p3, p4]
layout = go.Layout(xaxis=dict(zeroline=False, showgrid=False),
yaxis=dict(zeroline=False, showgrid=False)
)
fig = go.Figure(data=data, layout=layout)
py.iplot(fig)
from IPython.display import display, HTML
display(HTML('<link href="//fonts.googleapis.com/css?family=Open+Sans:600,400,300,200|Inconsolata|Ubuntu+Mono:400,700" rel="stylesheet" type="text/css" />'))
display(HTML('<link rel="stylesheet" type="text/css" href="http://help.plot.ly/documentation/all_static/css/ipython-notebook-custom.css">'))
! pip install git+https://github.com/plotly/publisher.git --upgrade
import publisher
publisher.publish(
'Robust Linear Model Estimation using RANSAC.ipynb', 'scikit-learn/plot-ransac/', 'Robust Linear Model Estimation using RANSAC | plotly',
'',
title = 'Robust Linear Model Estimation using RANSAC | plotly',
name = 'Robust Linear Model Estimation using RANSAC',
has_thumbnail='true', thumbnail='thumbnail/ransac.jpg',
language='scikit-learn', page_type='example_index',
display_as='linear_models', order=18,
ipynb= '~Diksha_Gabha/3223')
| 0.599837 | 0.966914 |
```
%load_ext autoreload
%autoreload 2
import matplotlib.pyplot as plt
import numpy as np
import sys
from ctapipe_io_lst import LSTEventSource
from lstchain.calib.camera.r0 import LSTR0Corrections
from traitlets.config.loader import Config
plt.rcParams['font.size'] = 18
```
# This is notebook to low level calibration R0->R1
Fill the R1 container ```event.r1.tel[0].waveform```.
Following function exist:
1. to subtraction pedestal ```subtract_pedestal(event)```
2. to dt correction ```time_lapse_corr(event)```
3. to interpolate spikes ```interpolate_spikes(event)```
or you can call method ```calibrate(event)```, then baseline value will be around 0
To subtraction pedestal you need pedestal file from cta-lstchain-extra or you can create pedestal file yourself using scirpt ```create_pedestal_file.py```
In ```config``` you can set how many samples in waveform keep to R1 container (first two and last two samples in waveform are noisy)
```
# Give path to real LST data
# We have to use LSTEventSource: LST-1.1.Run00xxx.xxx.fits.fz
# because for dt correction and interpolate spikes events must be in turn (event_id = 0,1,2, ...)
reader = LSTEventSource(input_url="/media/pawel1/ADATA HD330/20190215/LST-1.1.Run00097.0001.fits.fz",
max_events=5)
# Otherwise, you can give your own input file
#reader = LSTEventSource(input_url= )#Any .fits.fz you might have
offset_value = 400
# Give path to pedestal file and set offset to baseline
# Assuming that you are running the Notebook from ~/cta-lstchain/notebooks
config = Config({
"LSTR0Corrections": {
"pedestal_path": "../../cta-lstchain-extra/calib/camera/pedestal_run97_new.fits",
"offset": offset_value,
"tel_id": 0, # for EVB3 will be 1
"r1_sample_start": None,
"r1_sample_end": None
}
})
# Give path to pedestal file
lst_r0 = LSTR0Corrections(config=config)
t = np.linspace(2, 37, 36)
for ev in reader:
plt.figure(figsize=(12, 7))
plt.step(t, ev.r0.tel[0].waveform[0, 7, 2:38], color="blue", label="raw waveform")
lst_r0.subtract_pedestal(ev, tel_id=0)
plt.step(t, ev.r1.tel[0].waveform[0, 7, 2:38], color="red", label="after pedestal substraction ")
lst_r0.time_lapse_corr(ev, tel_id=0)
lst_r0.interpolate_spikes(ev, tel_id=0)
plt.step(t, ev.r1.tel[0].waveform[0, 7, 2:38], color="green", label="after pedestal substraction + dt corr + interpolate spikes")
plt.plot([0, 40], [offset_value, offset_value], 'k--', label="offset")
plt.xlabel("time sample [ns]")
plt.ylabel("counts [ADC]")
plt.legend()
plt.ylim([-50, 500])
plt.show()
```
# Call ```calibrate``` method
```
# Give path to real LST data
reader = LSTEventSource(
input_url="/media/pawel1/ADATA HD330/20190215/LST-1.1.Run00097.0001.fits.fz",
max_events=5)
# Again, you can give your own input file
#reader = LSTEventSource(input_url= )#Any .fits.fz you might have
# Give path to pedestal file and set offset to baseline
config = Config({
"LSTR0Corrections": {
"pedestal_path": "../../cta-lstchain-extra/calib/camera/pedestal_run97_new.fits",
"offset": 300,
"tel_id": 0, # for EVB3 will be 1
"r1_sample_start": 2,
"r1_sample_end": 38
}
})
lst_r0 = LSTR0Corrections(config=config)
t = np.linspace(2, 37, 36)
for ev in reader:
print(ev.r0.event_id)
plt.figure(figsize=(12, 7))
plt.step(t, ev.r0.tel[0].waveform[0, 7, 2:38], color="blue", label="raw waveform")
lst_r0.calibrate(ev)
plt.step(t, ev.r1.tel[0].waveform[0, 7, :], color="red", label="after calib")
plt.xlabel("time sample [ns]")
plt.ylabel("counts [ADC]")
plt.legend()
plt.ylim([-50, 500])
plt.show()
```
|
github_jupyter
|
%load_ext autoreload
%autoreload 2
import matplotlib.pyplot as plt
import numpy as np
import sys
from ctapipe_io_lst import LSTEventSource
from lstchain.calib.camera.r0 import LSTR0Corrections
from traitlets.config.loader import Config
plt.rcParams['font.size'] = 18
2. to dt correction ```time_lapse_corr(event)```
3. to interpolate spikes ```interpolate_spikes(event)```
or you can call method ```calibrate(event)```, then baseline value will be around 0
To subtraction pedestal you need pedestal file from cta-lstchain-extra or you can create pedestal file yourself using scirpt ```create_pedestal_file.py```
In ```config``` you can set how many samples in waveform keep to R1 container (first two and last two samples in waveform are noisy)
# Call ```calibrate``` method
| 0.378229 | 0.82226 |
# Detection of changes using the Cumulative Sum (CUSUM)
> Marcos Duarte
> Laboratory of Biomechanics and Motor Control ([http://demotu.org/](http://demotu.org/))
> Federal University of ABC, Brazil
[Change detection](http://en.wikipedia.org/wiki/Change_detection) refers to procedures to identify abrupt changes in a phenomenon (Basseville and Nikiforov 1993, Gustafsson 2000). By abrupt change it is meant any difference in relation to previous known data faster than expected of some characteristic of the data such as amplitude, mean, variance, frequency, etc.
The [Cumulative sum (CUSUM)](http://en.wikipedia.org/wiki/CUSUM) algorithm is a classical technique for monitoring change detection. One form of implementing the CUSUM algorithm involves the calculation of the cumulative sum of positive and negative changes ($g_t^+$ and $g_t^-$) in the data ($x$) and comparison to a $threshold$. When this threshold is exceeded a change is detected ($t_{talarm}$) and the cumulative sum restarts from zero. To avoid the detection of a change in absence of an actual change or a slow drift, this algorithm also depends on a parameter $drift$ for drift correction. This form of the CUSUM algorithm is given by:
$$ \begin{array}{l l}
\left\{ \begin{array}{l l}
s[t] = x[t] - x[t-1] \\
g^+[t] = max\left(g^+[t-1] + s[t]-drift,\; 0\right) \\
g^-[t] = max\left(g^-[t-1] - s[t]-drift,\; 0\right)
\end{array} \right. \\
\\
\; if \;\;\; g^+[t] > threshold \;\;\; or \;\;\; g^-[t] > threshold: \\
\\
\left\{ \begin{array}{l l}
t_{talarm}=t \\
g^+[t] = 0 \\
g^-[t] = 0
\end{array} \right.
\end{array} $$
<!-- TEASER_END -->
There are different implementations of the CUSUM algorithm; for example, the term for the sum of the last elements ($s[t]$ above) can have a longer history (with filtering), it can be normalized by removing the data mean and then divided by the data variance), or this sum term can be squared for detecting both variance and parameter changes, etc.
For the CUSUM algorithm to work properly, it depends on tuning the parameters $h$ and $v$ to what is meant by a change in the data. According to Gustafsson (2000), this tuning can be performed following these steps:
- Start with a very large $threshold$.
- Choose $drift$ to one half of the expected change, or adjust $drift$ such that $g$ = 0 more than 50% of the time.
- Then set the $threshold$ so the required number of false alarms (this can be done automatically) or delay for detection is obtained.
- If faster detection is sought, try to decrease $drift$.
- If fewer false alarms are wanted, try to increase $drift$.
- If there is a subset of the change times that does not make sense, try to increase $drift$.
The function `detect_cusum.py` (code at the end of this text) implements the CUSUM algorithm and a procedure to calculate the ending of the detected change. The function signature is:
```python
ta, tai, taf, amp = detect_cusum(x, threshold=1, drift=0, ending=False, show=True, ax=None)
```
Let's see how to use `detect_cusum.py`; first let's import the necessary Python libraries and configure the environment:
```
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
import sys
sys.path.insert(1, r'./../functions') # add to pythonpath
from detect_cusum import detect_cusum
```
Running the function examples:
```
>>> x = np.random.randn(300)/5
>>> x[100:200] += np.arange(0, 4, 4/100)
>>> ta, tai, taf, amp = detect_cusum(x, 2, .02, True, True)
>>> x = np.random.randn(300)
>>> x[100:200] += 6
>>> detect_cusum(x, 4, 1.5, True, True)
>>> x = 2*np.sin(2*np.pi*np.arange(0, 3, .01))
>>> ta, tai, taf, amp = detect_cusum(x, 1, .05, True, True)
```
## Function performance
Here is a poor test of the `detect_cusum.py` performance:
```
x = np.random.randn(10000)
x[400:600] += 6
print('Detection of onset (data size = %d):' %x.size)
%timeit detect_cusum(x, 4, 1.5, True, False)
```
## References
- Michèle Basseville and Igor V. Nikiforov (1993). [Detection of Abrupt Changes: Theory and Application](http://books.google.com.br/books/about/Detection_of_abrupt_changes.html?id=Vu5SAAAAMAAJ). Prentice-Hall.
- Fredrik Gustafsson (2000) [Adaptive Filtering and Change Detection](http://books.google.com.br/books?id=cyNTAAAAMAAJ). Wiley.
## Function `detect_cusum.py`
```
# %load ./../functions/detect_cusum.py
"""Cumulative sum algorithm (CUSUM) to detect abrupt changes in data."""
from __future__ import division, print_function
import numpy as np
__author__ = 'Marcos Duarte, https://github.com/demotu/BMC'
__version__ = "1.0.4"
__license__ = "MIT"
def detect_cusum(x, threshold=1, drift=0, ending=False, show=True, ax=None):
"""Cumulative sum algorithm (CUSUM) to detect abrupt changes in data.
Parameters
----------
x : 1D array_like
data.
threshold : positive number, optional (default = 1)
amplitude threshold for the change in the data.
drift : positive number, optional (default = 0)
drift term that prevents any change in the absence of change.
ending : bool, optional (default = False)
True (1) to estimate when the change ends; False (0) otherwise.
show : bool, optional (default = True)
True (1) plots data in matplotlib figure, False (0) don't plot.
ax : a matplotlib.axes.Axes instance, optional (default = None).
Returns
-------
ta : 1D array_like [indi, indf], int
alarm time (index of when the change was detected).
tai : 1D array_like, int
index of when the change started.
taf : 1D array_like, int
index of when the change ended (if `ending` is True).
amp : 1D array_like, float
amplitude of changes (if `ending` is True).
Notes
-----
Tuning of the CUSUM algorithm according to Gustafsson (2000)[1]_:
Start with a very large `threshold`.
Choose `drift` to one half of the expected change, or adjust `drift` such
that `g` = 0 more than 50% of the time.
Then set the `threshold` so the required number of false alarms (this can
be done automatically) or delay for detection is obtained.
If faster detection is sought, try to decrease `drift`.
If fewer false alarms are wanted, try to increase `drift`.
If there is a subset of the change times that does not make sense,
try to increase `drift`.
Note that by default repeated sequential changes, i.e., changes that have
the same beginning (`tai`) are not deleted because the changes were
detected by the alarm (`ta`) at different instants. This is how the
classical CUSUM algorithm operates.
If you want to delete the repeated sequential changes and keep only the
beginning of the first sequential change, set the parameter `ending` to
True. In this case, the index of the ending of the change (`taf`) and the
amplitude of the change (or of the total amplitude for a repeated
sequential change) are calculated and only the first change of the repeated
sequential changes is kept. In this case, it is likely that `ta`, `tai`,
and `taf` will have less values than when `ending` was set to False.
See this IPython Notebook [2]_.
References
----------
.. [1] Gustafsson (2000) Adaptive Filtering and Change Detection.
.. [2] hhttp://nbviewer.ipython.org/github/demotu/BMC/blob/master/notebooks/DetectCUSUM.ipynb
Examples
--------
>>> from detect_cusum import detect_cusum
>>> x = np.random.randn(300)/5
>>> x[100:200] += np.arange(0, 4, 4/100)
>>> ta, tai, taf, amp = detect_cusum(x, 2, .02, True, True)
>>> x = np.random.randn(300)
>>> x[100:200] += 6
>>> detect_cusum(x, 4, 1.5, True, True)
>>> x = 2*np.sin(2*np.pi*np.arange(0, 3, .01))
>>> ta, tai, taf, amp = detect_cusum(x, 1, .05, True, True)
"""
x = np.atleast_1d(x).astype('float64')
gp, gn = np.zeros(x.size), np.zeros(x.size)
ta, tai, taf = np.array([[], [], []], dtype=int)
tap, tan = 0, 0
amp = np.array([])
# Find changes (online form)
for i in range(1, x.size):
s = x[i] - x[i-1]
gp[i] = gp[i-1] + s - drift # cumulative sum for + change
gn[i] = gn[i-1] - s - drift # cumulative sum for - change
if gp[i] < 0:
gp[i], tap = 0, i
if gn[i] < 0:
gn[i], tan = 0, i
if gp[i] > threshold or gn[i] > threshold: # change detected!
ta = np.append(ta, i) # alarm index
tai = np.append(tai, tap if gp[i] > threshold else tan) # start
gp[i], gn[i] = 0, 0 # reset alarm
# THE CLASSICAL CUSUM ALGORITHM ENDS HERE
# Estimation of when the change ends (offline form)
if tai.size and ending:
_, tai2, _, _ = detect_cusum(x[::-1], threshold, drift, show=False)
taf = x.size - tai2[::-1] - 1
# Eliminate repeated changes, changes that have the same beginning
tai, ind = np.unique(tai, return_index=True)
ta = ta[ind]
# taf = np.unique(taf, return_index=False) # corect later
if tai.size != taf.size:
if tai.size < taf.size:
taf = taf[[np.argmax(taf >= i) for i in ta]]
else:
ind = [np.argmax(i >= ta[::-1])-1 for i in taf]
ta = ta[ind]
tai = tai[ind]
# Delete intercalated changes (the ending of the change is after
# the beginning of the next change)
ind = taf[:-1] - tai[1:] > 0
if ind.any():
ta = ta[~np.append(False, ind)]
tai = tai[~np.append(False, ind)]
taf = taf[~np.append(ind, False)]
# Amplitude of changes
amp = x[taf] - x[tai]
if show:
_plot(x, threshold, drift, ending, ax, ta, tai, taf, gp, gn)
return ta, tai, taf, amp
def _plot(x, threshold, drift, ending, ax, ta, tai, taf, gp, gn):
"""Plot results of the detect_cusum function, see its help."""
try:
import matplotlib.pyplot as plt
except ImportError:
print('matplotlib is not available.')
else:
if ax is None:
_, (ax1, ax2) = plt.subplots(2, 1, figsize=(8, 6))
t = range(x.size)
ax1.plot(t, x, 'b-', lw=2)
if len(ta):
ax1.plot(tai, x[tai], '>', mfc='g', mec='g', ms=10,
label='Start')
if ending:
ax1.plot(taf, x[taf], '<', mfc='g', mec='g', ms=10,
label='Ending')
ax1.plot(ta, x[ta], 'o', mfc='r', mec='r', mew=1, ms=5,
label='Alarm')
ax1.legend(loc='best', framealpha=.5, numpoints=1)
ax1.set_xlim(-.01*x.size, x.size*1.01-1)
ax1.set_xlabel('Data #', fontsize=14)
ax1.set_ylabel('Amplitude', fontsize=14)
ymin, ymax = x[np.isfinite(x)].min(), x[np.isfinite(x)].max()
yrange = ymax - ymin if ymax > ymin else 1
ax1.set_ylim(ymin - 0.1*yrange, ymax + 0.1*yrange)
ax1.set_title('Time series and detected changes ' +
'(threshold= %.3g, drift= %.3g): N changes = %d'
% (threshold, drift, len(tai)))
ax2.plot(t, gp, 'y-', label='+')
ax2.plot(t, gn, 'm-', label='-')
ax2.set_xlim(-.01*x.size, x.size*1.01-1)
ax2.set_xlabel('Data #', fontsize=14)
ax2.set_ylim(-0.01*threshold, 1.1*threshold)
ax2.axhline(threshold, color='r')
ax1.set_ylabel('Amplitude', fontsize=14)
ax2.set_title('Time series of the cumulative sums of ' +
'positive and negative changes')
ax2.legend(loc='best', framealpha=.5, numpoints=1)
plt.tight_layout()
plt.show()
```
|
github_jupyter
|
ta, tai, taf, amp = detect_cusum(x, threshold=1, drift=0, ending=False, show=True, ax=None)
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
import sys
sys.path.insert(1, r'./../functions') # add to pythonpath
from detect_cusum import detect_cusum
>>> x = np.random.randn(300)/5
>>> x[100:200] += np.arange(0, 4, 4/100)
>>> ta, tai, taf, amp = detect_cusum(x, 2, .02, True, True)
>>> x = np.random.randn(300)
>>> x[100:200] += 6
>>> detect_cusum(x, 4, 1.5, True, True)
>>> x = 2*np.sin(2*np.pi*np.arange(0, 3, .01))
>>> ta, tai, taf, amp = detect_cusum(x, 1, .05, True, True)
x = np.random.randn(10000)
x[400:600] += 6
print('Detection of onset (data size = %d):' %x.size)
%timeit detect_cusum(x, 4, 1.5, True, False)
# %load ./../functions/detect_cusum.py
"""Cumulative sum algorithm (CUSUM) to detect abrupt changes in data."""
from __future__ import division, print_function
import numpy as np
__author__ = 'Marcos Duarte, https://github.com/demotu/BMC'
__version__ = "1.0.4"
__license__ = "MIT"
def detect_cusum(x, threshold=1, drift=0, ending=False, show=True, ax=None):
"""Cumulative sum algorithm (CUSUM) to detect abrupt changes in data.
Parameters
----------
x : 1D array_like
data.
threshold : positive number, optional (default = 1)
amplitude threshold for the change in the data.
drift : positive number, optional (default = 0)
drift term that prevents any change in the absence of change.
ending : bool, optional (default = False)
True (1) to estimate when the change ends; False (0) otherwise.
show : bool, optional (default = True)
True (1) plots data in matplotlib figure, False (0) don't plot.
ax : a matplotlib.axes.Axes instance, optional (default = None).
Returns
-------
ta : 1D array_like [indi, indf], int
alarm time (index of when the change was detected).
tai : 1D array_like, int
index of when the change started.
taf : 1D array_like, int
index of when the change ended (if `ending` is True).
amp : 1D array_like, float
amplitude of changes (if `ending` is True).
Notes
-----
Tuning of the CUSUM algorithm according to Gustafsson (2000)[1]_:
Start with a very large `threshold`.
Choose `drift` to one half of the expected change, or adjust `drift` such
that `g` = 0 more than 50% of the time.
Then set the `threshold` so the required number of false alarms (this can
be done automatically) or delay for detection is obtained.
If faster detection is sought, try to decrease `drift`.
If fewer false alarms are wanted, try to increase `drift`.
If there is a subset of the change times that does not make sense,
try to increase `drift`.
Note that by default repeated sequential changes, i.e., changes that have
the same beginning (`tai`) are not deleted because the changes were
detected by the alarm (`ta`) at different instants. This is how the
classical CUSUM algorithm operates.
If you want to delete the repeated sequential changes and keep only the
beginning of the first sequential change, set the parameter `ending` to
True. In this case, the index of the ending of the change (`taf`) and the
amplitude of the change (or of the total amplitude for a repeated
sequential change) are calculated and only the first change of the repeated
sequential changes is kept. In this case, it is likely that `ta`, `tai`,
and `taf` will have less values than when `ending` was set to False.
See this IPython Notebook [2]_.
References
----------
.. [1] Gustafsson (2000) Adaptive Filtering and Change Detection.
.. [2] hhttp://nbviewer.ipython.org/github/demotu/BMC/blob/master/notebooks/DetectCUSUM.ipynb
Examples
--------
>>> from detect_cusum import detect_cusum
>>> x = np.random.randn(300)/5
>>> x[100:200] += np.arange(0, 4, 4/100)
>>> ta, tai, taf, amp = detect_cusum(x, 2, .02, True, True)
>>> x = np.random.randn(300)
>>> x[100:200] += 6
>>> detect_cusum(x, 4, 1.5, True, True)
>>> x = 2*np.sin(2*np.pi*np.arange(0, 3, .01))
>>> ta, tai, taf, amp = detect_cusum(x, 1, .05, True, True)
"""
x = np.atleast_1d(x).astype('float64')
gp, gn = np.zeros(x.size), np.zeros(x.size)
ta, tai, taf = np.array([[], [], []], dtype=int)
tap, tan = 0, 0
amp = np.array([])
# Find changes (online form)
for i in range(1, x.size):
s = x[i] - x[i-1]
gp[i] = gp[i-1] + s - drift # cumulative sum for + change
gn[i] = gn[i-1] - s - drift # cumulative sum for - change
if gp[i] < 0:
gp[i], tap = 0, i
if gn[i] < 0:
gn[i], tan = 0, i
if gp[i] > threshold or gn[i] > threshold: # change detected!
ta = np.append(ta, i) # alarm index
tai = np.append(tai, tap if gp[i] > threshold else tan) # start
gp[i], gn[i] = 0, 0 # reset alarm
# THE CLASSICAL CUSUM ALGORITHM ENDS HERE
# Estimation of when the change ends (offline form)
if tai.size and ending:
_, tai2, _, _ = detect_cusum(x[::-1], threshold, drift, show=False)
taf = x.size - tai2[::-1] - 1
# Eliminate repeated changes, changes that have the same beginning
tai, ind = np.unique(tai, return_index=True)
ta = ta[ind]
# taf = np.unique(taf, return_index=False) # corect later
if tai.size != taf.size:
if tai.size < taf.size:
taf = taf[[np.argmax(taf >= i) for i in ta]]
else:
ind = [np.argmax(i >= ta[::-1])-1 for i in taf]
ta = ta[ind]
tai = tai[ind]
# Delete intercalated changes (the ending of the change is after
# the beginning of the next change)
ind = taf[:-1] - tai[1:] > 0
if ind.any():
ta = ta[~np.append(False, ind)]
tai = tai[~np.append(False, ind)]
taf = taf[~np.append(ind, False)]
# Amplitude of changes
amp = x[taf] - x[tai]
if show:
_plot(x, threshold, drift, ending, ax, ta, tai, taf, gp, gn)
return ta, tai, taf, amp
def _plot(x, threshold, drift, ending, ax, ta, tai, taf, gp, gn):
"""Plot results of the detect_cusum function, see its help."""
try:
import matplotlib.pyplot as plt
except ImportError:
print('matplotlib is not available.')
else:
if ax is None:
_, (ax1, ax2) = plt.subplots(2, 1, figsize=(8, 6))
t = range(x.size)
ax1.plot(t, x, 'b-', lw=2)
if len(ta):
ax1.plot(tai, x[tai], '>', mfc='g', mec='g', ms=10,
label='Start')
if ending:
ax1.plot(taf, x[taf], '<', mfc='g', mec='g', ms=10,
label='Ending')
ax1.plot(ta, x[ta], 'o', mfc='r', mec='r', mew=1, ms=5,
label='Alarm')
ax1.legend(loc='best', framealpha=.5, numpoints=1)
ax1.set_xlim(-.01*x.size, x.size*1.01-1)
ax1.set_xlabel('Data #', fontsize=14)
ax1.set_ylabel('Amplitude', fontsize=14)
ymin, ymax = x[np.isfinite(x)].min(), x[np.isfinite(x)].max()
yrange = ymax - ymin if ymax > ymin else 1
ax1.set_ylim(ymin - 0.1*yrange, ymax + 0.1*yrange)
ax1.set_title('Time series and detected changes ' +
'(threshold= %.3g, drift= %.3g): N changes = %d'
% (threshold, drift, len(tai)))
ax2.plot(t, gp, 'y-', label='+')
ax2.plot(t, gn, 'm-', label='-')
ax2.set_xlim(-.01*x.size, x.size*1.01-1)
ax2.set_xlabel('Data #', fontsize=14)
ax2.set_ylim(-0.01*threshold, 1.1*threshold)
ax2.axhline(threshold, color='r')
ax1.set_ylabel('Amplitude', fontsize=14)
ax2.set_title('Time series of the cumulative sums of ' +
'positive and negative changes')
ax2.legend(loc='best', framealpha=.5, numpoints=1)
plt.tight_layout()
plt.show()
| 0.685529 | 0.983629 |
# 3 The design engineer as detective
If you have never before experienced a disappointment when building systems, then welcome to the world of engineering design!
If you struggled to complete any of the previous challenges, then take heart. Solving practical engineering problems often requires several attempts as you come to understand the problem better. In some cases, you may even come to realise that the problem you first thought you were trying to solve turns out not to be the actual problem at all.
Even if you have identified the problem correctly, designing the solution may not be straightforward. In many cases, design is characterised by defining a set of *requirements*, translating these into a *specification*, generating one or more possible *solutions* to the specified problem, identifying which one to implement, and then *evaluating* the outcome.
If the ‘solution’ is not satisfactory, then it is necessary to go back and try to formulate another possible solution, or maybe even check that you have correctly identified the requirements based on what you may have learned about the problem by trying to meet the requirements of the previously specified design.
This is called the *design cycle*.

During the design cycle, candidate solutions are generated and evaluated. If the design fails to meet expectations, then a new or updated design must be generated. If the evaluation of the design is satisfactory, then the design can be accepted and the process may end.
In a continuous improvement design cycle, the original design may be used but it may also be reconsidered. If an improvement is found, then the new design may be both adopted and passed back round the cycle for further reconsideration.
### 3.1 Following the design cycle
In the lollipop line-following challenge, the system was specified according to the requirement that the robot would go all the way round the track and stop at the red bar. I generated a solution based on my ‘model’ of the system.
When I first ran the program, my evaluation was that the system did not work: for some reason, the robot kept stopping! (Even if it had worked first time, it’s possible that the solution was not very good either in terms of the way the robot behaved, or in terms of how the program was written.)
Under these circumstances, the design engineer looks for reasons why things went wrong, using all the available information. Sometimes this involves devising experiments. It’s rather like being a detective, trying to piece together the solution.
```
from nbev3devsim.load_nbev3devwidget import roboSim, eds
%load_ext nbev3devsim
%load_ext nbtutor
```
Here’s the program I used for the lollipop challenge at first:
```
%%sim_magic_preloaded --background Lollipop
colorLeft = ColorSensor(INPUT_2)
colorRight = ColorSensor(INPUT_3)
print(colorLeft.reflected_light_intensity)
while ((colorLeft.full_reflected_light_intensity < 30)
or (colorLeft.full_reflected_light_intensity > 40)):
intensity_left = colorLeft.full_reflected_light_intensity
#intensity_right = colorRight.reflected_light_intensity
print(intensity_left)
if intensity_left < 50:
left_motor_speed = SpeedPercent(0)
right_motor_speed = SpeedPercent(40)
else:
left_motor_speed = SpeedPercent(40)
right_motor_speed = SpeedPercent(0)
tank_drive.on(left_motor_speed, right_motor_speed)
```
One of the main assumptions underlying my first program was that the colours could be separated by thresholds and that I would use a while loop to loop through some edge-follower code if I didn’t see red.
For the stopping decision (‘Have I seen red?’) I checked whether or not the full reflected light percentage was between 30% and 40%. This was based on the fact that the red line gives a full reflected light percentage of 33.3%, the solid grey background has a reading of 82.75% and the black background has a reading of 0%.
But when I downloaded and ran the program (which you can try too), the robot kept stopping on the line. So what was going on?
Let’s try a simple test program to get a better idea of what the robot is actually perceiving.
We’ll explore the data at leisure in the notebook, so run the following code cell to clear the notebook datalog:
```
# Clear the datalog
%sim_data --clear
```
For our test case, we’ll get the robot to turn on the spot so that the sensor crosses over the black line and the grey background a couple of times.
Download and run the following program in the simulator to collect sensor data as the robot spins slowly on the spot for three seconds, grabbing data into the datalog by printing it to the robot display terminal:
```
%%sim_magic_preloaded --background Lollipop -R
import time
tank_drive.on(SpeedPercent(10), SpeedPercent(-10))
for i in range(150):
print('Colour: ' + str(colorLeft.full_reflected_light_intensity ))
time.sleep(0.02)
say("All done")
```
We can now use some magic to grab the data from the datalog and put it into a *pandas* dataframe. Let’s preview the first few rows of the dataframe:
```
data_df = %sim_data
data_df.head()
```
The `cufflinks` package adds support for plotting charts using the `plotly` package directly from *pandas* dataframes. Whilst recent updates to the *plotly* package add native support for `plotly` charts to *pandas* plotting, `cufflinks` still has some additional nice features that are useful for our purposes, such as the ability to overplot individual data point markers on a plotly chart as well as highlight different value ranges.
```
import cufflinks as cf
# By default, cufflinks will try to use
# an online plotly mode
cf.go_offline()
```
We can now create a `plotly` chart using *markers* to identify each sensor reading sample point as well as a *line* that connects them.
A horizontal band, identified by the `hband=[(MIN_Y, MAX_Y)]` parameter, adds a horizontal band to the chart corresponding to the thresholded range I was using to identify whether the robot had encountered the red line:
```
data_df.iplot( x = 'time', y = 'value',
mode='markers+lines', hspan=[(30,40)])
```
Here’s a screenshot of part of the trace of light readings I got from an earlier experiment I carried out as the simulated robot went from the grey background, over the black line and back to the grey background:

The vertical scale shows sensor values on a vertical *y*-axis scale ranging from 0 to 85 or so (the actual readings in principle range form 0...100 per cent). The horizontal scale is a time base showing a time of 0 to 2 seconds, with a plot every 0.02 s (that is, about every fiftieth of a second). The sensor readings are also joined to form a line. The line starts with high values, starting at a *y*-value of just over 80. There is then an abrupt fall in the values until they reach a low point of 0 for 3 samples, followed by a return to the high values around 80. On the diagram, I have marked a horizontal band highlighting values between 30% and 35%.
This chart helps to explain why my original program did not work as intended. On the right of the black line, in the transition from black to grey, the simulated robot has recorded a value of about 34, within the thresholded band value I was using to identify the red line.
The sensor value is calculated as some function of the value of several pixels in view of the sensor at any one time, and it just so happens that the calculation may return a reading I had associated with the red line.
If a sampled data point falls between the values I used for my threshold settings that were intended to identify the red line, then the robot would have a ‘false positive’ match and stop before it was supposed to.
In fact, the `.reflected_light` mode in either raw or percentage form or full percentage form is *not* a good indicator of colour at all.
Based on this investigation, I ended up using a different approach to identify the stopping condition (that is, the presence of the red band); originally, I just tested that the red RGB component was set to 255 but then I realised that this could give a false positive if a solid yellow colour was in view of the sensor (the RGB value for yellow is `(255, 255, 0)`). So I then iterated my design again and tested that the red value was 255 and the green component was 0. (I really should extend it again to check that the blue value is also zero.)
However, following the discussion at the end of the last notebook regarding noise, I think I should probably change the exact equality test on the red value to a threshold test, such as `color_sensor.rgb[0]>250` and that the other values are less than some other threshold value (for example, `color_sensor.rgb[1]<15`) to allow for noise in the background or the sensor.
As you can see, designing a program often requires a continual process of iterative improvement.
## 3.2 Optional activity
Create, download and run a program in the simulator that drives the robot over the *Rainbow_bands* background, logging the reflected light sensor data as it does so.
Then either chart the data in the simulator to review it, or grab the datalog into the notebook and view it here at your leisure.
Does the reflected light sensor data allow you to reliably identify each colour band?
## 3.3 Summary
In this notebook, you have seen how an investigative process may be required to help us better understand why any assumptions made about how our program might work need checking against how it actually works in practice. This is particularly true in cases where the robot appears to behave counter to the way we intended it to behave when we created our program.
We have also seen how the process of design is often an iterative one in which we repeatedly refine and improve our program as we better understand the problem and what we are actually trying to achieve.
<!-- JD: add something about what's in the next notebook? -->
|
github_jupyter
|
from nbev3devsim.load_nbev3devwidget import roboSim, eds
%load_ext nbev3devsim
%load_ext nbtutor
%%sim_magic_preloaded --background Lollipop
colorLeft = ColorSensor(INPUT_2)
colorRight = ColorSensor(INPUT_3)
print(colorLeft.reflected_light_intensity)
while ((colorLeft.full_reflected_light_intensity < 30)
or (colorLeft.full_reflected_light_intensity > 40)):
intensity_left = colorLeft.full_reflected_light_intensity
#intensity_right = colorRight.reflected_light_intensity
print(intensity_left)
if intensity_left < 50:
left_motor_speed = SpeedPercent(0)
right_motor_speed = SpeedPercent(40)
else:
left_motor_speed = SpeedPercent(40)
right_motor_speed = SpeedPercent(0)
tank_drive.on(left_motor_speed, right_motor_speed)
# Clear the datalog
%sim_data --clear
%%sim_magic_preloaded --background Lollipop -R
import time
tank_drive.on(SpeedPercent(10), SpeedPercent(-10))
for i in range(150):
print('Colour: ' + str(colorLeft.full_reflected_light_intensity ))
time.sleep(0.02)
say("All done")
data_df = %sim_data
data_df.head()
import cufflinks as cf
# By default, cufflinks will try to use
# an online plotly mode
cf.go_offline()
data_df.iplot( x = 'time', y = 'value',
mode='markers+lines', hspan=[(30,40)])
| 0.382603 | 0.971699 |
# fairlearn Trial
fairnessを実現するためのmethodを実行。
- CorrelationRemover
- GridSearch
- ThresholdOptimizer
```
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.base import clone
from lightgbm import LGBMClassifier
%matplotlib inline
# fairlearnで用いるmodel関連methodのimport
from fairlearn.preprocessing import CorrelationRemover
from fairlearn.postprocessing import ThresholdOptimizer
from fairlearn.reductions import DemographicParity, EqualizedOdds, TruePositiveRateParity
from fairlearn.reductions import GridSearch
def logistic(x, a=1):
return 1/(1 + np.exp(-a*x))
# サンプルデータ生成
def make_sample_data(N=10000, p_s=0.1):
s = np.r_[np.zeros(int(N*p_s)) , np.ones(int(N*(1-p_s)))] # sensitive
np.random.seed(0)
x = np.random.normal(0, 1, size=N) - 0.2*s # correlated with s
np.random.seed(1)
y = 0.3*x + 0.5*s + np.random.normal(0,1,size=N) # outcome y is correlated with x and s
y -= np.mean(y)
y = np.array(logistic(y) > 0.5, dtype=int) # flg
np.random.seed(2)
train_idx = np.random.choice(N, size=N//2, replace=False) # random split
test_idx = np.array(list(set(np.arange(N)) - set(train_idx)))
return x[train_idx], y[train_idx], s[train_idx], x[test_idx], y[test_idx], s[test_idx]
# 公平性の各種metricsをprint
def print_result_summary(y_true, y_pred, s):
# Demographic Parityで用いる差
dp_ave = y_pred.mean()
dp_s1 = y_pred[s==1].mean()
dp_s0 = y_pred[s==0].mean()
dp_diff = np.abs(dp_s1 - dp_s0)
# TruePositiveRate Parityで用いる差 (Equalized Opportunity)
tpr_ave = y_pred[y_true==1].mean()
tpr_s1 = y_pred[np.all([y_true==1, s==1],axis=0)].mean()
tpr_s0 = y_pred[np.all([y_true==1, s==0],axis=0)].mean()
tpr_diff = np.abs(tpr_s1 - tpr_s0)
# FalsePositiveRate Parityで用いる差
fpr_ave = y_pred[y_true==0].mean()
fpr_s1 = y_pred[np.all([y_true==0, s==1],axis=0)].mean()
fpr_s0 = y_pred[np.all([y_true==0, s==0],axis=0)].mean()
fpr_diff = np.abs(fpr_s1 - fpr_s0)
# print result
dp_text = f"Demographic Parity:\t\t[mean] {dp_ave:.3f},\t[s=1] {dp_s1:.3f},\t[s=0] {dp_s0:.3f},\t[abs_diff] {dp_diff:.3f}"
tpr_text = f"TruePositiveRate Parity:\t[mean] {tpr_ave:.3f},\t[s=1] {tpr_s1:.3f},\t[s=0] {tpr_s0:.3f},\t[abs_diff] {tpr_diff:.3f}"
fpr_text = f"FalsePositiveRate Parity:\t[mean] {fpr_ave:.3f},\t[s=1] {fpr_s1:.3f},\t[s=0] {fpr_s0:.3f},\t[abs_diff] {fpr_diff:.3f}"
print(dp_text)
print(tpr_text)
print(fpr_text)
```
# Metricsの確認
```
# Metrics
from fairlearn.metrics import (
demographic_parity_difference,
true_positive_rate_difference,
false_positive_rate_difference,
equalized_odds_difference)
# テストデータ
y_true = np.array([0,0,1,1,1,0,0])
y_pred = np.array([0,0,0,1,1,1,1])
s = np.array([1,0,1,0,1,0,0])
# 自作
print_result_summary(y_true, y_pred, s)
# fairlearnでのmetrics実装, 自作funcとの[abs_diff]との一致が確認できる
print("Demographic Parity Diff :\t\t %.3f"% demographic_parity_difference(y_true=y_true, y_pred=y_pred, sensitive_features=s))
print("TruePositiveRate Parity Diff :\t %.3f"% true_positive_rate_difference(y_true=y_true, y_pred=y_pred, sensitive_features=s))
print("FalsePositiveRate Parity Diff :\t %.3f"% false_positive_rate_difference(y_true=y_true, y_pred=y_pred, sensitive_features=s))
# TruePositiveRate DiffとFalsePositiveRate Diffのうち大きい方の値が入る
print("EqualizedOdds Parity Diff :\t\t %.3f"% equalized_odds_difference(y_true=y_true, y_pred=y_pred, sensitive_features=s))
```
# データ生成
```
# sample data
x_train, y_train, s_train, x_test, y_test, s_test = make_sample_data(N=10000, p_s=0.1)
X_train = np.c_[x_train, s_train]
X_test = np.c_[x_test, s_test]
```
# ベースラインモデル, そのままモデルfit
### センシティブ変数も入力
```
# model fit
baseline_model = LGBMClassifier(random_state=1)
baseline_model.fit(X_train, y_train)
# predict
y_train_pred_baseline = baseline_model.predict(X_train)
y_test_pred_baseline = baseline_model.predict(X_test)
print("Train")
print_result_summary(y_true=y_train, y_pred=y_train_pred_baseline, s=s_train)
print("\nTest")
print_result_summary(y_true=y_test, y_pred=y_test_pred_baseline, s=s_test)
```
### センシティブ変数をDrop
```
# model fit
baseline_model_wos = LGBMClassifier(random_state=1)
baseline_model_wos.fit(X_train[:,0].reshape(-1,1), y_train)
# predict
y_train_pred_baseline_wos = baseline_model_wos.predict(X_train[:,0].reshape(-1,1))
y_test_pred_baseline_wos = baseline_model_wos.predict(X_test[:,0].reshape(-1,1))
print("Train")
print_result_summary(y_true=y_train, y_pred=y_train_pred_baseline_wos, s=s_train)
print("\nTest")
print_result_summary(y_true=y_test, y_pred=y_test_pred_baseline_wos, s=s_test)
```
# 公平性を加味したモデルfit
## [Pre-process] CorrelationRemover
```
# 相関を除去
corr_remover = CorrelationRemover(sensitive_feature_ids=[1]) # X_trainのうち2列目がsに該当
X_train_rmcorr = corr_remover.fit_transform(X_train)
X_test_rmcorr = corr_remover.transform(X_test)
# model fit
clf = LGBMClassifier()
clf.fit(X_train_rmcorr, y_train)
# predict
y_train_pred_rmcorr = clf.predict(X_train_rmcorr)
y_test_pred_rmcorr = clf.predict(X_test_rmcorr)
print("Train")
print_result_summary(y_true=y_train, y_pred=y_train_pred_rmcorr, s=s_train)
print("\nTest")
print_result_summary(y_true=y_test, y_pred=y_test_pred_rmcorr, s=s_test)
```
## [In-process] Reduction Method
### Demographic Parity
```
# model fit
sweep = GridSearch(
estimator=LGBMClassifier(random_state=1), # estimator needs `sample_weight` at fit
constraints=DemographicParity(), # EqualizedOdds, TruePositiveRateParityなども使用可
grid_size=50,
grid_limit=1
)
sweep.fit(X_train, y_train, sensitive_features=s_train)
# predict
y_train_pred_indp = sweep.predict(X_train) # 最もDemographicParityが小さいものが選ばれる
y_test_pred_indp = sweep.predict(X_test)
print("Train")
print_result_summary(y_true=y_train, y_pred=y_train_pred_indp, s=s_train)
print("\nTest")
print_result_summary(y_true=y_test, y_pred=y_test_pred_indp, s=s_test)
# 各モデルの予測結果metricsを取得
sweep_preds = [predictor.predict(X_train) for predictor in sweep.predictors_] # 各predictorの予測値を取得
dp_diff_list = [
demographic_parity_difference(y_train, preds, sensitive_features=s_train)
for preds in sweep_preds
]
np.sort(dp_diff_list)[:5] #top-5のdemographic_parity_diffを確認, sweep.predictによる結果と同一であることを確認できる
```
### Equalized Odds
```
# model fit
sweep = GridSearch(
estimator=LGBMClassifier(random_state=1),
constraints=EqualizedOdds(), # EqualizedOdds, TruePositiveRateParityなども使用可
grid_size=50,
grid_limit=1
)
sweep.fit(X_train, y_train, sensitive_features=s_train)
# predict
y_train_pred_ineo = sweep.predict(X_train) # 最もDemographicParityが小さいものが選ばれる
y_test_pred_ineo = sweep.predict(X_test)
print("Train")
print_result_summary(y_true=y_train, y_pred=y_train_pred_ineo, s=s_train)
print("\nTest")
print_result_summary(y_true=y_test, y_pred=y_test_pred_ineo, s=s_test)
```
## [Post-process]
### Demographic Parity
```
# set
optimizer = ThresholdOptimizer(
estimator=baseline_model,
constraints="demographic_parity", # 他に’{false,true}_{positive,negative}_rate_parity’’equalized_odds’ が使用可能
prefit=True # 学習済みモデルを渡している場合 True, prefit=Falseでestimator=LGBMClassifier(random_state=1)でも同じ結果
)
# fit optimizer
optimizer.fit(X=X_train,y=y_train, sensitive_features=s_train)
y_test_pred_postdp = optimizer.predict(X_test, sensitive_features=s_test, random_state=20) #乱数固定
y_train_pred_postdp = optimizer.predict(X_train, sensitive_features=s_train, random_state=100) # 乱数固定
print("Train")
print_result_summary(y_true=y_train, y_pred=y_train_pred_postdp, s=s_train)
print("\nTest")
print_result_summary(y_true=y_test, y_pred=y_test_pred_postdp, s=s_test)
```
### Equalized Odds
```
optimizer = ThresholdOptimizer(
estimator=clone(baseline_model),
constraints="equalized_odds",
prefit=False
)
optimizer.fit(X=X_train,y=y_train.reshape(-1,1), sensitive_features=s_train)
y_test_pred_posteo = optimizer.predict(X_test, sensitive_features=s_test, random_state=20)
y_train_pred_posteo = optimizer.predict(X_train, sensitive_features=s_train, random_state=20)
print("Train")
print_result_summary(y_true=y_train, y_pred=y_train_pred_posteo, s=s_train)
print("\nTest")
print_result_summary(y_true=y_test, y_pred=y_test_pred_posteo, s=s_test)
```
|
github_jupyter
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.base import clone
from lightgbm import LGBMClassifier
%matplotlib inline
# fairlearnで用いるmodel関連methodのimport
from fairlearn.preprocessing import CorrelationRemover
from fairlearn.postprocessing import ThresholdOptimizer
from fairlearn.reductions import DemographicParity, EqualizedOdds, TruePositiveRateParity
from fairlearn.reductions import GridSearch
def logistic(x, a=1):
return 1/(1 + np.exp(-a*x))
# サンプルデータ生成
def make_sample_data(N=10000, p_s=0.1):
s = np.r_[np.zeros(int(N*p_s)) , np.ones(int(N*(1-p_s)))] # sensitive
np.random.seed(0)
x = np.random.normal(0, 1, size=N) - 0.2*s # correlated with s
np.random.seed(1)
y = 0.3*x + 0.5*s + np.random.normal(0,1,size=N) # outcome y is correlated with x and s
y -= np.mean(y)
y = np.array(logistic(y) > 0.5, dtype=int) # flg
np.random.seed(2)
train_idx = np.random.choice(N, size=N//2, replace=False) # random split
test_idx = np.array(list(set(np.arange(N)) - set(train_idx)))
return x[train_idx], y[train_idx], s[train_idx], x[test_idx], y[test_idx], s[test_idx]
# 公平性の各種metricsをprint
def print_result_summary(y_true, y_pred, s):
# Demographic Parityで用いる差
dp_ave = y_pred.mean()
dp_s1 = y_pred[s==1].mean()
dp_s0 = y_pred[s==0].mean()
dp_diff = np.abs(dp_s1 - dp_s0)
# TruePositiveRate Parityで用いる差 (Equalized Opportunity)
tpr_ave = y_pred[y_true==1].mean()
tpr_s1 = y_pred[np.all([y_true==1, s==1],axis=0)].mean()
tpr_s0 = y_pred[np.all([y_true==1, s==0],axis=0)].mean()
tpr_diff = np.abs(tpr_s1 - tpr_s0)
# FalsePositiveRate Parityで用いる差
fpr_ave = y_pred[y_true==0].mean()
fpr_s1 = y_pred[np.all([y_true==0, s==1],axis=0)].mean()
fpr_s0 = y_pred[np.all([y_true==0, s==0],axis=0)].mean()
fpr_diff = np.abs(fpr_s1 - fpr_s0)
# print result
dp_text = f"Demographic Parity:\t\t[mean] {dp_ave:.3f},\t[s=1] {dp_s1:.3f},\t[s=0] {dp_s0:.3f},\t[abs_diff] {dp_diff:.3f}"
tpr_text = f"TruePositiveRate Parity:\t[mean] {tpr_ave:.3f},\t[s=1] {tpr_s1:.3f},\t[s=0] {tpr_s0:.3f},\t[abs_diff] {tpr_diff:.3f}"
fpr_text = f"FalsePositiveRate Parity:\t[mean] {fpr_ave:.3f},\t[s=1] {fpr_s1:.3f},\t[s=0] {fpr_s0:.3f},\t[abs_diff] {fpr_diff:.3f}"
print(dp_text)
print(tpr_text)
print(fpr_text)
# Metrics
from fairlearn.metrics import (
demographic_parity_difference,
true_positive_rate_difference,
false_positive_rate_difference,
equalized_odds_difference)
# テストデータ
y_true = np.array([0,0,1,1,1,0,0])
y_pred = np.array([0,0,0,1,1,1,1])
s = np.array([1,0,1,0,1,0,0])
# 自作
print_result_summary(y_true, y_pred, s)
# fairlearnでのmetrics実装, 自作funcとの[abs_diff]との一致が確認できる
print("Demographic Parity Diff :\t\t %.3f"% demographic_parity_difference(y_true=y_true, y_pred=y_pred, sensitive_features=s))
print("TruePositiveRate Parity Diff :\t %.3f"% true_positive_rate_difference(y_true=y_true, y_pred=y_pred, sensitive_features=s))
print("FalsePositiveRate Parity Diff :\t %.3f"% false_positive_rate_difference(y_true=y_true, y_pred=y_pred, sensitive_features=s))
# TruePositiveRate DiffとFalsePositiveRate Diffのうち大きい方の値が入る
print("EqualizedOdds Parity Diff :\t\t %.3f"% equalized_odds_difference(y_true=y_true, y_pred=y_pred, sensitive_features=s))
# sample data
x_train, y_train, s_train, x_test, y_test, s_test = make_sample_data(N=10000, p_s=0.1)
X_train = np.c_[x_train, s_train]
X_test = np.c_[x_test, s_test]
# model fit
baseline_model = LGBMClassifier(random_state=1)
baseline_model.fit(X_train, y_train)
# predict
y_train_pred_baseline = baseline_model.predict(X_train)
y_test_pred_baseline = baseline_model.predict(X_test)
print("Train")
print_result_summary(y_true=y_train, y_pred=y_train_pred_baseline, s=s_train)
print("\nTest")
print_result_summary(y_true=y_test, y_pred=y_test_pred_baseline, s=s_test)
# model fit
baseline_model_wos = LGBMClassifier(random_state=1)
baseline_model_wos.fit(X_train[:,0].reshape(-1,1), y_train)
# predict
y_train_pred_baseline_wos = baseline_model_wos.predict(X_train[:,0].reshape(-1,1))
y_test_pred_baseline_wos = baseline_model_wos.predict(X_test[:,0].reshape(-1,1))
print("Train")
print_result_summary(y_true=y_train, y_pred=y_train_pred_baseline_wos, s=s_train)
print("\nTest")
print_result_summary(y_true=y_test, y_pred=y_test_pred_baseline_wos, s=s_test)
# 相関を除去
corr_remover = CorrelationRemover(sensitive_feature_ids=[1]) # X_trainのうち2列目がsに該当
X_train_rmcorr = corr_remover.fit_transform(X_train)
X_test_rmcorr = corr_remover.transform(X_test)
# model fit
clf = LGBMClassifier()
clf.fit(X_train_rmcorr, y_train)
# predict
y_train_pred_rmcorr = clf.predict(X_train_rmcorr)
y_test_pred_rmcorr = clf.predict(X_test_rmcorr)
print("Train")
print_result_summary(y_true=y_train, y_pred=y_train_pred_rmcorr, s=s_train)
print("\nTest")
print_result_summary(y_true=y_test, y_pred=y_test_pred_rmcorr, s=s_test)
# model fit
sweep = GridSearch(
estimator=LGBMClassifier(random_state=1), # estimator needs `sample_weight` at fit
constraints=DemographicParity(), # EqualizedOdds, TruePositiveRateParityなども使用可
grid_size=50,
grid_limit=1
)
sweep.fit(X_train, y_train, sensitive_features=s_train)
# predict
y_train_pred_indp = sweep.predict(X_train) # 最もDemographicParityが小さいものが選ばれる
y_test_pred_indp = sweep.predict(X_test)
print("Train")
print_result_summary(y_true=y_train, y_pred=y_train_pred_indp, s=s_train)
print("\nTest")
print_result_summary(y_true=y_test, y_pred=y_test_pred_indp, s=s_test)
# 各モデルの予測結果metricsを取得
sweep_preds = [predictor.predict(X_train) for predictor in sweep.predictors_] # 各predictorの予測値を取得
dp_diff_list = [
demographic_parity_difference(y_train, preds, sensitive_features=s_train)
for preds in sweep_preds
]
np.sort(dp_diff_list)[:5] #top-5のdemographic_parity_diffを確認, sweep.predictによる結果と同一であることを確認できる
# model fit
sweep = GridSearch(
estimator=LGBMClassifier(random_state=1),
constraints=EqualizedOdds(), # EqualizedOdds, TruePositiveRateParityなども使用可
grid_size=50,
grid_limit=1
)
sweep.fit(X_train, y_train, sensitive_features=s_train)
# predict
y_train_pred_ineo = sweep.predict(X_train) # 最もDemographicParityが小さいものが選ばれる
y_test_pred_ineo = sweep.predict(X_test)
print("Train")
print_result_summary(y_true=y_train, y_pred=y_train_pred_ineo, s=s_train)
print("\nTest")
print_result_summary(y_true=y_test, y_pred=y_test_pred_ineo, s=s_test)
# set
optimizer = ThresholdOptimizer(
estimator=baseline_model,
constraints="demographic_parity", # 他に’{false,true}_{positive,negative}_rate_parity’’equalized_odds’ が使用可能
prefit=True # 学習済みモデルを渡している場合 True, prefit=Falseでestimator=LGBMClassifier(random_state=1)でも同じ結果
)
# fit optimizer
optimizer.fit(X=X_train,y=y_train, sensitive_features=s_train)
y_test_pred_postdp = optimizer.predict(X_test, sensitive_features=s_test, random_state=20) #乱数固定
y_train_pred_postdp = optimizer.predict(X_train, sensitive_features=s_train, random_state=100) # 乱数固定
print("Train")
print_result_summary(y_true=y_train, y_pred=y_train_pred_postdp, s=s_train)
print("\nTest")
print_result_summary(y_true=y_test, y_pred=y_test_pred_postdp, s=s_test)
optimizer = ThresholdOptimizer(
estimator=clone(baseline_model),
constraints="equalized_odds",
prefit=False
)
optimizer.fit(X=X_train,y=y_train.reshape(-1,1), sensitive_features=s_train)
y_test_pred_posteo = optimizer.predict(X_test, sensitive_features=s_test, random_state=20)
y_train_pred_posteo = optimizer.predict(X_train, sensitive_features=s_train, random_state=20)
print("Train")
print_result_summary(y_true=y_train, y_pred=y_train_pred_posteo, s=s_train)
print("\nTest")
print_result_summary(y_true=y_test, y_pred=y_test_pred_posteo, s=s_test)
| 0.399343 | 0.765769 |
```
lines = '''start-A
start-b
A-c
A-b
b-d
A-end
b-end'''.splitlines()
lines='''dc-end
HN-start
start-kj
dc-start
dc-HN
LN-dc
HN-end
kj-sa
kj-HN
kj-dc'''.splitlines()
lines = '''fs-end
he-DX
fs-he
start-DX
pj-DX
end-zg
zg-sl
zg-pj
pj-he
RW-he
fs-DX
pj-RW
zg-RW
start-pj
he-WI
zg-he
pj-fs
start-RW'''.splitlines()
lines = '''start-A
start-b
A-c
A-b
b-d
A-end
b-end'''.splitlines()
#read from day12.txt then split lines
with open('day12.txt', 'r') as f:
lines = f.read().splitlines()
# Fortunately, the sensors are still mostly working, and so you build a rough map of the remaining caves (your puzzle input). For example:
# start-A
# start-b
# A-c
# A-b
# b-d
# A-end
# b-end
# This is a list of how all of the caves are connected. You start in the cave named start, and your destination is the cave named end. An entry like b-d means that cave b is connected to cave d - that is, you can move between them.
#split by -
connectes = [line.split('-') for line in lines]
# So, the above cave system looks roughly like this:
# start
# / \
# c--A-----b--d
# \ /
# end
# Your goal is to find the number of distinct paths that start at start, end at end, and don't visit small caves more than once. There are two types of caves: big caves (written in uppercase, like A) and small caves (written in lowercase, like b). It would be a waste of time to visit any small cave more than once, but big caves are large enough that it might be worth visiting them multiple times. So, all paths you find should visit small caves at most once, and can visit big caves any number of times.
def is_small(cave):
if cave == 'start':return False
return cave[0].islower()
#build a graph of caves
def build_graph(connectes):
graph = {}
caves= set()
for connect in connectes:
caves.add(connect[0])
caves.add(connect[1])
if connect[0] not in graph:
graph[connect[0]] = []
if connect[1] not in graph:
graph[connect[1]] = []
graph[connect[0]].append(connect[1])
graph[connect[1]].append(connect[0])
return caves, graph
caves, cave_system = build_graph(connectes)
#remove a small cave from cave_system
def remove_cave(small_cave, cave_system):
for cave in cave_system:
if small_cave in cave_system[cave]:
cave_system[cave].remove(small_cave)
return cave_system
#remove start
cave_system = remove_cave('start', cave_system)
#get all paths from start to end
end='end'
def get_paths(caves, cave_system, start='start', path=[], small_caves=[]):
# print(small_caves, path)
path = path + [start]
if start == end:
return [path]
small_caves = small_caves + [start] if is_small(start) else small_caves
paths = []
for node in cave_system[start]:
if node not in small_caves:
newpaths = get_paths(caves, cave_system, node, path, small_caves)
for newpath in newpaths:
paths.append(newpath)
return paths
paths = get_paths(caves, cave_system)
print(len(paths))
# Given these rules, there are 10 paths through this example cave system:
# start,A,b,A,c,A,end
# start,A,b,A,end
# start,A,b,end
# start,A,c,A,b,A,end
# start,A,c,A,b,end
# start,A,c,A,end
# start,A,end
# start,b,A,c,A,end
# start,b,A,end
# start,b,end
# (Each line in the above list corresponds to a single path; the caves visited by that path are listed in the order they are visited and separated by commas.)
# Note that in this cave system, cave d is never visited by any path: to do so, cave b would need to be visited twice (once on the way to cave d and a second time when returning from cave d), and since cave b is small, this is not allowed.
# Here is a slightly larger example:
# dc-end
# HN-start
# start-kj
# dc-start
# dc-HN
# LN-dc
# HN-end
# kj-sa
# kj-HN
# kj-dc
# The 19 paths through it are as follows:
# start,HN,dc,HN,end
# start,HN,dc,HN,kj,HN,end
# start,HN,dc,end
# start,HN,dc,kj,HN,end
# start,HN,end
# start,HN,kj,HN,dc,HN,end
# start,HN,kj,HN,dc,end
# start,HN,kj,HN,end
# start,HN,kj,dc,HN,end
# start,HN,kj,dc,end
# start,dc,HN,end
# start,dc,HN,kj,HN,end
# start,dc,end
# start,dc,kj,HN,end
# start,kj,HN,dc,HN,end
# start,kj,HN,dc,end
# start,kj,HN,end
# start,kj,dc,HN,end
# start,kj,dc,end
# Finally, this even larger example has 226 paths through it:
# fs-end
# he-DX
# fs-he
# start-DX
# pj-DX
# end-zg
# zg-sl
# zg-pj
# pj-he
# RW-he
# fs-DX
# pj-RW
# zg-RW
# start-pj
# he-WI
# zg-he
# pj-fs
# start-RW
# How many paths through this cave system are there that visit small caves at most once?
# --- Part Two ---
# After reviewing the available paths, you realize you might have time to visit a single small cave twice. Specifically, big caves can be visited any number of times, a single small cave can be visited at most twice, and the remaining small caves can be visited at most once. However, the caves named start and end can only be visited exactly once each: once you leave the start cave, you may not return to it, and once you reach the end cave, the path must end immediately.
#get all paths from start to end with small caves visited at most twice
end='end'
def get_paths2(caves, cave_system, start='start', path=[], small_caves=[]):
path = path + [start]
if start == end:
return 1
if is_small(start): small_caves = small_caves + [start]
small_caves.sort()
hasTwice = False
for i in range(len(small_caves)-1):
if small_caves[i] == small_caves[i+1]:
hasTwice = True
break
paths = 0
for node in cave_system[start]:
if (not is_small(node)) or (not hasTwice) or (node not in small_caves):
newpaths = get_paths2(caves, cave_system, node, path, small_caves)
paths += newpaths
return paths
paths = get_paths2(caves, cave_system)
print(paths)
# Now, the 36 possible paths through the first example above are:
# start,A,b,A,b,A,c,A,end
# start,A,b,A,b,A,end
# start,A,b,A,b,end
# start,A,b,A,c,A,b,A,end
# start,A,b,A,c,A,b,end
# start,A,b,A,c,A,c,A,end
# start,A,b,A,c,A,end
# start,A,b,A,end
# start,A,b,d,b,A,c,A,end
# start,A,b,d,b,A,end
# start,A,b,d,b,end
# start,A,b,end
# start,A,c,A,b,A,b,A,end
# start,A,c,A,b,A,b,end
# start,A,c,A,b,A,c,A,end
# start,A,c,A,b,A,end
# start,A,c,A,b,d,b,A,end
# start,A,c,A,b,d,b,end
# start,A,c,A,b,end
# start,A,c,A,c,A,b,A,end
# start,A,c,A,c,A,b,end
# start,A,c,A,c,A,end
# start,A,c,A,end
# start,A,end
# start,b,A,b,A,c,A,end
# start,b,A,b,A,end
# start,b,A,b,end
# start,b,A,c,A,b,A,end
# start,b,A,c,A,b,end
# start,b,A,c,A,c,A,end
# start,b,A,c,A,end
# start,b,A,end
# start,b,d,b,A,c,A,end
# start,b,d,b,A,end
# start,b,d,b,end
# start,b,end
# The slightly larger example above now has 103 paths through it, and the even larger example now has 3509 paths through it.
# Given these new rules, how many paths through this cave system are there?
```
|
github_jupyter
|
lines = '''start-A
start-b
A-c
A-b
b-d
A-end
b-end'''.splitlines()
lines='''dc-end
HN-start
start-kj
dc-start
dc-HN
LN-dc
HN-end
kj-sa
kj-HN
kj-dc'''.splitlines()
lines = '''fs-end
he-DX
fs-he
start-DX
pj-DX
end-zg
zg-sl
zg-pj
pj-he
RW-he
fs-DX
pj-RW
zg-RW
start-pj
he-WI
zg-he
pj-fs
start-RW'''.splitlines()
lines = '''start-A
start-b
A-c
A-b
b-d
A-end
b-end'''.splitlines()
#read from day12.txt then split lines
with open('day12.txt', 'r') as f:
lines = f.read().splitlines()
# Fortunately, the sensors are still mostly working, and so you build a rough map of the remaining caves (your puzzle input). For example:
# start-A
# start-b
# A-c
# A-b
# b-d
# A-end
# b-end
# This is a list of how all of the caves are connected. You start in the cave named start, and your destination is the cave named end. An entry like b-d means that cave b is connected to cave d - that is, you can move between them.
#split by -
connectes = [line.split('-') for line in lines]
# So, the above cave system looks roughly like this:
# start
# / \
# c--A-----b--d
# \ /
# end
# Your goal is to find the number of distinct paths that start at start, end at end, and don't visit small caves more than once. There are two types of caves: big caves (written in uppercase, like A) and small caves (written in lowercase, like b). It would be a waste of time to visit any small cave more than once, but big caves are large enough that it might be worth visiting them multiple times. So, all paths you find should visit small caves at most once, and can visit big caves any number of times.
def is_small(cave):
if cave == 'start':return False
return cave[0].islower()
#build a graph of caves
def build_graph(connectes):
graph = {}
caves= set()
for connect in connectes:
caves.add(connect[0])
caves.add(connect[1])
if connect[0] not in graph:
graph[connect[0]] = []
if connect[1] not in graph:
graph[connect[1]] = []
graph[connect[0]].append(connect[1])
graph[connect[1]].append(connect[0])
return caves, graph
caves, cave_system = build_graph(connectes)
#remove a small cave from cave_system
def remove_cave(small_cave, cave_system):
for cave in cave_system:
if small_cave in cave_system[cave]:
cave_system[cave].remove(small_cave)
return cave_system
#remove start
cave_system = remove_cave('start', cave_system)
#get all paths from start to end
end='end'
def get_paths(caves, cave_system, start='start', path=[], small_caves=[]):
# print(small_caves, path)
path = path + [start]
if start == end:
return [path]
small_caves = small_caves + [start] if is_small(start) else small_caves
paths = []
for node in cave_system[start]:
if node not in small_caves:
newpaths = get_paths(caves, cave_system, node, path, small_caves)
for newpath in newpaths:
paths.append(newpath)
return paths
paths = get_paths(caves, cave_system)
print(len(paths))
# Given these rules, there are 10 paths through this example cave system:
# start,A,b,A,c,A,end
# start,A,b,A,end
# start,A,b,end
# start,A,c,A,b,A,end
# start,A,c,A,b,end
# start,A,c,A,end
# start,A,end
# start,b,A,c,A,end
# start,b,A,end
# start,b,end
# (Each line in the above list corresponds to a single path; the caves visited by that path are listed in the order they are visited and separated by commas.)
# Note that in this cave system, cave d is never visited by any path: to do so, cave b would need to be visited twice (once on the way to cave d and a second time when returning from cave d), and since cave b is small, this is not allowed.
# Here is a slightly larger example:
# dc-end
# HN-start
# start-kj
# dc-start
# dc-HN
# LN-dc
# HN-end
# kj-sa
# kj-HN
# kj-dc
# The 19 paths through it are as follows:
# start,HN,dc,HN,end
# start,HN,dc,HN,kj,HN,end
# start,HN,dc,end
# start,HN,dc,kj,HN,end
# start,HN,end
# start,HN,kj,HN,dc,HN,end
# start,HN,kj,HN,dc,end
# start,HN,kj,HN,end
# start,HN,kj,dc,HN,end
# start,HN,kj,dc,end
# start,dc,HN,end
# start,dc,HN,kj,HN,end
# start,dc,end
# start,dc,kj,HN,end
# start,kj,HN,dc,HN,end
# start,kj,HN,dc,end
# start,kj,HN,end
# start,kj,dc,HN,end
# start,kj,dc,end
# Finally, this even larger example has 226 paths through it:
# fs-end
# he-DX
# fs-he
# start-DX
# pj-DX
# end-zg
# zg-sl
# zg-pj
# pj-he
# RW-he
# fs-DX
# pj-RW
# zg-RW
# start-pj
# he-WI
# zg-he
# pj-fs
# start-RW
# How many paths through this cave system are there that visit small caves at most once?
# --- Part Two ---
# After reviewing the available paths, you realize you might have time to visit a single small cave twice. Specifically, big caves can be visited any number of times, a single small cave can be visited at most twice, and the remaining small caves can be visited at most once. However, the caves named start and end can only be visited exactly once each: once you leave the start cave, you may not return to it, and once you reach the end cave, the path must end immediately.
#get all paths from start to end with small caves visited at most twice
end='end'
def get_paths2(caves, cave_system, start='start', path=[], small_caves=[]):
path = path + [start]
if start == end:
return 1
if is_small(start): small_caves = small_caves + [start]
small_caves.sort()
hasTwice = False
for i in range(len(small_caves)-1):
if small_caves[i] == small_caves[i+1]:
hasTwice = True
break
paths = 0
for node in cave_system[start]:
if (not is_small(node)) or (not hasTwice) or (node not in small_caves):
newpaths = get_paths2(caves, cave_system, node, path, small_caves)
paths += newpaths
return paths
paths = get_paths2(caves, cave_system)
print(paths)
# Now, the 36 possible paths through the first example above are:
# start,A,b,A,b,A,c,A,end
# start,A,b,A,b,A,end
# start,A,b,A,b,end
# start,A,b,A,c,A,b,A,end
# start,A,b,A,c,A,b,end
# start,A,b,A,c,A,c,A,end
# start,A,b,A,c,A,end
# start,A,b,A,end
# start,A,b,d,b,A,c,A,end
# start,A,b,d,b,A,end
# start,A,b,d,b,end
# start,A,b,end
# start,A,c,A,b,A,b,A,end
# start,A,c,A,b,A,b,end
# start,A,c,A,b,A,c,A,end
# start,A,c,A,b,A,end
# start,A,c,A,b,d,b,A,end
# start,A,c,A,b,d,b,end
# start,A,c,A,b,end
# start,A,c,A,c,A,b,A,end
# start,A,c,A,c,A,b,end
# start,A,c,A,c,A,end
# start,A,c,A,end
# start,A,end
# start,b,A,b,A,c,A,end
# start,b,A,b,A,end
# start,b,A,b,end
# start,b,A,c,A,b,A,end
# start,b,A,c,A,b,end
# start,b,A,c,A,c,A,end
# start,b,A,c,A,end
# start,b,A,end
# start,b,d,b,A,c,A,end
# start,b,d,b,A,end
# start,b,d,b,end
# start,b,end
# The slightly larger example above now has 103 paths through it, and the even larger example now has 3509 paths through it.
# Given these new rules, how many paths through this cave system are there?
| 0.370795 | 0.334372 |
# Dave's Data Science Project, Spring 2019
Obisety is a huge problem in America and leads to diseases that kill Americans. Many people estimate that the percentage of people obese is over 30% in America. America is above the world average and is first of the major world powers. Many factors have been linked to obesity including portion sizes, wealth, laziness, more inactive jobs, cost of healthy food and many more. For my project I wanted to put some of these factors to the test against obesity. Instead of doing the whole country I wanted to focus on one part or region to see if any specific factors effected obesity more since that seems to be less common in obesity research. I grew up in New Mexico and saw that many of the states around it had a wide range of obesity from being well above to well below the national average. Those two factors made me decide to focus on the south west region. Specifically the counties in Oklahoma, Kansas, Nevada, Wyoming, Colorado and Utah. I decided to exclude California do to its massive size and used Texas as a boarder to the more south east cities.
I collected my data using policy map through Tufts University. Policy map is a website that provides different sets of public data mostly from censuses in America to provide data for scientific and educational purposes. Policy map is a business it makes data interactive and people subscribe to it like universities and research labs.
The three factors I will be comparing to percent of the population that is obese are percent of adults that engage in binge drinking, percent of adults reporting to be physically inactive and rate of fast food restaurants. Drinking, physical inactivity and fast food are all well known as being healthy. (ADD MORE LATER)
## Files Included
The included packages that help with creating and visualizing my analysis.
```
from io_hw import io_hw
import pandas as pd
import matplotlib.pyplot as pyplot
import matplotlib.cm as cm
import numpy as np
import math
```
## Initial Data Parsing
Here, the io_hw.p file is used, and the desired fields are then extracted from the dataframe.
```
def read_data():
df, head_df = io_hw('data.csv')
obese = df.iloc[:, 3].tolist()
drink = df.iloc[:, 4].tolist()
inactive = df.iloc[:, 5].tolist()
fastfood = df.iloc[:, 6].tolist()
return obese, drink, inactive, fastfood
tempdf, temphead = io_hw('data.csv')
tempdf
```
## Data Cleaning (removing NaN values):
NaN values can hurt the accuracy of my analysis, as well as cause problems in code, so I remove them. However, I want to use as much data as possible during each peice of analysis, and so I rerun this clean code for each y vector.
```
def clean(x_init, y_init):
ziplist = zip(x_init, y_init)
x_clean = [x for (x, y) in ziplist if not math.isnan(y)]
ziplist = zip(x_init, y_init)
y_clean = [y for (x, y) in ziplist if not math.isnan(y)]
return x_clean, y_clean
```
## Creating Correlation Graphs:
These graphs display the correlation between obesity and the desired y variable.
```
def plot(x_init, y_init, name):
x, y = clean(x_init, y_init)
pyplot.title(name + " vs Obesity by County in the Midwest")
pyplot.xlabel("Obesity (%)")
pyplot.ylabel(name + "(%)")
curr_plot = pyplot.scatter(x, y)
```
## Finding Variable Correlation:
This function assists in figuring out the correlation coefficients between the two inputted variables. For this use, x_init will always be the obesity vector.
```
def correlation(x_init, y_init):
x, y = clean(x_init, y_init)
x_np = np.array(x)
y_np = np.array(y)
return np.corrcoef(x_np, y_np)[0][1]
```
## Plotting Figures:
```
obese, drink, inactive, fastfood = read_data()
plot(obese, drink, "Binge Drinking")
plot(obese, inactive, "Inactivity")
plot(obese, fastfood, "Fast Food Proximity")
```
## Correlation
This piece of code finds each of the test factors' correlation with obesity, and then prints the factor with the highest magnitude of correlation.
```
drink_corr = correlation(obese, drink)
inactive_corr = correlation(obese, inactive)
fastfood_corr = correlation(obese, fastfood)
highest_corr = max([drink_corr, inactive_corr, fastfood_corr], key=abs)
zip_corr = [("Drink Correlation", drink_corr),
("Inactivity Correlation", inactive_corr),
("Fast Food Correlation", fastfood_corr)]
highest_corr_name = [x for (x, y) in zip_corr if highest_corr==y]
print(highest_corr_name[0] + ": " + str(highest_corr))
```
As we can see from the (talk about correlation)
```
drink_corr
inactive_corr
fastfood_corr
```
## Plotting All Correlation using Heat Map
The pyplot package is used again here to map my four initial variables against eachother in a heat map. This is useful for visualizing the correlation of each of the test variables with eachother, as well as with obesity.
```
def plot_all(obese, drink, inactive, fastfood):
ziplist = zip(obese, drink, inactive, fastfood)
o_clean = [x for (x, y, z, w) in ziplist if not math.isnan(w)]
ziplist = zip(obese, drink, inactive, fastfood)
d_clean = [y for (x, y, z, w) in ziplist if not math.isnan(w)]
ziplist = zip(obese, drink, inactive, fastfood)
i_clean = [z for (x, y, z, w) in ziplist if not math.isnan(w)]
ziplist = zip(obese, drink, inactive, fastfood)
f_clean = [w for (x, y, z, w) in ziplist if not math.isnan(w)]
df_dict = {"Obesity": obese, "Binge Drinking": drink, "Inactivity": inactive, "Fast Food Proximity": fastfood}
df = pd.DataFrame(df_dict)
fig = pyplot.figure()
ax = fig.add_subplot(111)
cmap = cm.get_cmap('jet', 30)
cax = ax.imshow(df.corr(), interpolation = "nearest", cmap = cmap)
ax.grid(True)
pyplot.title('Correlation Between Test Variables and Obesity')
labels = ["", "Obesity", "", "Binge Drinking", "", "Inactivity", "", "Fast Food", ""]
ax.set_xticklabels(labels)
ax.set_yticklabels(labels)
fig.colorbar(cax)
pyplot.show()
plot_all(obese, drink, inactive, fastfood)
```
## Adding in New Data
Below, new data is added in. The location stays as the same as desired, but new factors are added to see if they contribute to obesity.
```
new_df = pd.read_csv('merge dataset.csv')
new_df
```
## Plotting New Data
Similarly to above, the new data is inputted into the plot function and the correlation between each new variable and obesity is shown.
```
education = new_df.iloc[:, 3].tolist()
income = new_df.iloc[:, 5].tolist()
plot(obese, education, "College Education")
plot(obese, income, "Low Income")
education_corr = correlation(obese, education)
education_corr
income_corr = correlation(obese, income)
income_corr
```
|
github_jupyter
|
from io_hw import io_hw
import pandas as pd
import matplotlib.pyplot as pyplot
import matplotlib.cm as cm
import numpy as np
import math
def read_data():
df, head_df = io_hw('data.csv')
obese = df.iloc[:, 3].tolist()
drink = df.iloc[:, 4].tolist()
inactive = df.iloc[:, 5].tolist()
fastfood = df.iloc[:, 6].tolist()
return obese, drink, inactive, fastfood
tempdf, temphead = io_hw('data.csv')
tempdf
def clean(x_init, y_init):
ziplist = zip(x_init, y_init)
x_clean = [x for (x, y) in ziplist if not math.isnan(y)]
ziplist = zip(x_init, y_init)
y_clean = [y for (x, y) in ziplist if not math.isnan(y)]
return x_clean, y_clean
def plot(x_init, y_init, name):
x, y = clean(x_init, y_init)
pyplot.title(name + " vs Obesity by County in the Midwest")
pyplot.xlabel("Obesity (%)")
pyplot.ylabel(name + "(%)")
curr_plot = pyplot.scatter(x, y)
def correlation(x_init, y_init):
x, y = clean(x_init, y_init)
x_np = np.array(x)
y_np = np.array(y)
return np.corrcoef(x_np, y_np)[0][1]
obese, drink, inactive, fastfood = read_data()
plot(obese, drink, "Binge Drinking")
plot(obese, inactive, "Inactivity")
plot(obese, fastfood, "Fast Food Proximity")
drink_corr = correlation(obese, drink)
inactive_corr = correlation(obese, inactive)
fastfood_corr = correlation(obese, fastfood)
highest_corr = max([drink_corr, inactive_corr, fastfood_corr], key=abs)
zip_corr = [("Drink Correlation", drink_corr),
("Inactivity Correlation", inactive_corr),
("Fast Food Correlation", fastfood_corr)]
highest_corr_name = [x for (x, y) in zip_corr if highest_corr==y]
print(highest_corr_name[0] + ": " + str(highest_corr))
drink_corr
inactive_corr
fastfood_corr
def plot_all(obese, drink, inactive, fastfood):
ziplist = zip(obese, drink, inactive, fastfood)
o_clean = [x for (x, y, z, w) in ziplist if not math.isnan(w)]
ziplist = zip(obese, drink, inactive, fastfood)
d_clean = [y for (x, y, z, w) in ziplist if not math.isnan(w)]
ziplist = zip(obese, drink, inactive, fastfood)
i_clean = [z for (x, y, z, w) in ziplist if not math.isnan(w)]
ziplist = zip(obese, drink, inactive, fastfood)
f_clean = [w for (x, y, z, w) in ziplist if not math.isnan(w)]
df_dict = {"Obesity": obese, "Binge Drinking": drink, "Inactivity": inactive, "Fast Food Proximity": fastfood}
df = pd.DataFrame(df_dict)
fig = pyplot.figure()
ax = fig.add_subplot(111)
cmap = cm.get_cmap('jet', 30)
cax = ax.imshow(df.corr(), interpolation = "nearest", cmap = cmap)
ax.grid(True)
pyplot.title('Correlation Between Test Variables and Obesity')
labels = ["", "Obesity", "", "Binge Drinking", "", "Inactivity", "", "Fast Food", ""]
ax.set_xticklabels(labels)
ax.set_yticklabels(labels)
fig.colorbar(cax)
pyplot.show()
plot_all(obese, drink, inactive, fastfood)
new_df = pd.read_csv('merge dataset.csv')
new_df
education = new_df.iloc[:, 3].tolist()
income = new_df.iloc[:, 5].tolist()
plot(obese, education, "College Education")
plot(obese, income, "Low Income")
education_corr = correlation(obese, education)
education_corr
income_corr = correlation(obese, income)
income_corr
| 0.411347 | 0.981203 |
# Candlestick Breakaway
https://patternswizard.com/breakaway-candlestick-pattern/
```
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import talib
import warnings
warnings.filterwarnings("ignore")
# yahoo finance is used to fetch data
import yfinance as yf
yf.pdr_override()
# input
symbol = 'MPHASIS.NS'
start = '2020-01-01'
end = '2021-10-11'
# Read data
df = yf.download(symbol,start,end)
# View Columns
df.head()
```
## Candlestick with Breakaway
```
from matplotlib import dates as mdates
import datetime as dt
dfc = df.copy()
dfc['VolumePositive'] = dfc['Open'] < dfc['Adj Close']
#dfc = dfc.dropna()
dfc = dfc.reset_index()
dfc['Date'] = pd.to_datetime(dfc['Date'])
dfc['Date'] = dfc['Date'].apply(mdates.date2num)
dfc.head()
from mplfinance.original_flavor import candlestick_ohlc
fig = plt.figure(figsize=(14,10))
ax = plt.subplot(2, 1, 1)
candlestick_ohlc(ax,dfc.values, width=0.5, colorup='g', colordown='r', alpha=1.0)
ax.xaxis_date()
ax.xaxis.set_major_formatter(mdates.DateFormatter('%d-%m-%Y'))
ax.grid(True, which='both')
ax.minorticks_on()
axv = ax.twinx()
colors = dfc.VolumePositive.map({True: 'g', False: 'r'})
axv.bar(dfc.Date, dfc['Volume'], color=colors, alpha=0.4)
axv.axes.yaxis.set_ticklabels([])
axv.set_ylim(0, 3*df.Volume.max())
ax.set_title('Stock '+ symbol +' Closing Price')
ax.set_ylabel('Price')
breakaway = talib.CDLBREAKAWAY(df['Open'], df['High'], df['Low'], df['Close'])
breakaway = breakaway[breakaway != 0]
df['breakaway'] = talib.CDLBREAKAWAY(df['Open'], df['High'], df['Low'], df['Close'])
df.loc[df['breakaway'] !=0]
df['Adj Close'].loc[df['breakaway'] !=0]
df['Adj Close'].loc[df['breakaway'] !=0].index
breakaway
breakaway.index
df
fig = plt.figure(figsize=(20,16))
ax = plt.subplot(2, 1, 1)
candlestick_ohlc(ax,dfc.values, width=0.5, colorup='g', colordown='r', alpha=1.0)
ax.xaxis_date()
ax.xaxis.set_major_formatter(mdates.DateFormatter('%d-%m-%Y'))
ax.grid(True, which='both')
ax.minorticks_on()
axv = ax.twinx()
ax.plot_date(df['Adj Close'].loc[df['breakaway'] !=0].index, df['Adj Close'].loc[df['breakaway'] !=0],
'or', # marker style 'o', color 'g'
fillstyle='none', # circle is not filled (with color)
ms=10.0)
colors = dfc.VolumePositive.map({True: 'g', False: 'r'})
axv.bar(dfc.Date, dfc['Volume'], color=colors, alpha=0.4)
axv.axes.yaxis.set_ticklabels([])
axv.set_ylim(0, 3*df.Volume.max())
ax.set_title('Stock '+ symbol +' Closing Price')
ax.set_ylabel('Price')
```
## Plot Certain dates
```
df = df['2020-05-01':'2020-06-01']
dfc = df.copy()
dfc['VolumePositive'] = dfc['Open'] < dfc['Adj Close']
#dfc = dfc.dropna()
dfc = dfc.reset_index()
dfc['Date'] = pd.to_datetime(dfc['Date'])
dfc['Date'] = dfc['Date'].apply(mdates.date2num)
dfc.head()
fig = plt.figure(figsize=(20,16))
ax = plt.subplot(2, 1, 1)
ax.set_facecolor('pink')
candlestick_ohlc(ax,dfc.values, width=0.5, colorup='g', colordown='r', alpha=1.0)
ax.xaxis_date()
ax.xaxis.set_major_formatter(mdates.DateFormatter('%d-%m-%Y'))
#ax.grid(True, which='both')
#ax.minorticks_on()
axv = ax.twinx()
ax.plot_date(df['Adj Close'].loc[df['breakaway'] !=0].index, df['Adj Close'].loc[df['breakaway'] !=0],
'or', # marker style 'o', color 'g'
fillstyle='none', # circle is not filled (with color)
ms=30.0)
colors = dfc.VolumePositive.map({True: 'g', False: 'r'})
axv.bar(dfc.Date, dfc['Volume'], color=colors, alpha=0.4)
axv.axes.yaxis.set_ticklabels([])
axv.set_ylim(0, 3*df.Volume.max())
ax.set_title('Stock '+ symbol +' Closing Price')
ax.set_ylabel('Price')
```
# Highlight Candlestick
```
from matplotlib.dates import date2num
from datetime import datetime
fig = plt.figure(figsize=(20,16))
ax = plt.subplot(2, 1, 1)
candlestick_ohlc(ax,dfc.values, width=0.5, colorup='g', colordown='r', alpha=1.0)
ax.xaxis_date()
ax.xaxis.set_major_formatter(mdates.DateFormatter('%d-%m-%Y'))
#ax.grid(True, which='both')
#ax.minorticks_on()
axv = ax.twinx()
ax.axvspan(date2num(datetime(2020,5,4)), date2num(datetime(2020,5,5)),
label="breakaway",color="red", alpha=0.3)
ax.legend(loc='upper center')
colors = dfc.VolumePositive.map({True: 'g', False: 'r'})
axv.bar(dfc.Date, dfc['Volume'], color=colors, alpha=0.4)
axv.axes.yaxis.set_ticklabels([])
axv.set_ylim(0, 3*df.Volume.max())
ax.set_title('Stock '+ symbol +' Closing Price')
ax.set_ylabel('Price')
```
|
github_jupyter
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import talib
import warnings
warnings.filterwarnings("ignore")
# yahoo finance is used to fetch data
import yfinance as yf
yf.pdr_override()
# input
symbol = 'MPHASIS.NS'
start = '2020-01-01'
end = '2021-10-11'
# Read data
df = yf.download(symbol,start,end)
# View Columns
df.head()
from matplotlib import dates as mdates
import datetime as dt
dfc = df.copy()
dfc['VolumePositive'] = dfc['Open'] < dfc['Adj Close']
#dfc = dfc.dropna()
dfc = dfc.reset_index()
dfc['Date'] = pd.to_datetime(dfc['Date'])
dfc['Date'] = dfc['Date'].apply(mdates.date2num)
dfc.head()
from mplfinance.original_flavor import candlestick_ohlc
fig = plt.figure(figsize=(14,10))
ax = plt.subplot(2, 1, 1)
candlestick_ohlc(ax,dfc.values, width=0.5, colorup='g', colordown='r', alpha=1.0)
ax.xaxis_date()
ax.xaxis.set_major_formatter(mdates.DateFormatter('%d-%m-%Y'))
ax.grid(True, which='both')
ax.minorticks_on()
axv = ax.twinx()
colors = dfc.VolumePositive.map({True: 'g', False: 'r'})
axv.bar(dfc.Date, dfc['Volume'], color=colors, alpha=0.4)
axv.axes.yaxis.set_ticklabels([])
axv.set_ylim(0, 3*df.Volume.max())
ax.set_title('Stock '+ symbol +' Closing Price')
ax.set_ylabel('Price')
breakaway = talib.CDLBREAKAWAY(df['Open'], df['High'], df['Low'], df['Close'])
breakaway = breakaway[breakaway != 0]
df['breakaway'] = talib.CDLBREAKAWAY(df['Open'], df['High'], df['Low'], df['Close'])
df.loc[df['breakaway'] !=0]
df['Adj Close'].loc[df['breakaway'] !=0]
df['Adj Close'].loc[df['breakaway'] !=0].index
breakaway
breakaway.index
df
fig = plt.figure(figsize=(20,16))
ax = plt.subplot(2, 1, 1)
candlestick_ohlc(ax,dfc.values, width=0.5, colorup='g', colordown='r', alpha=1.0)
ax.xaxis_date()
ax.xaxis.set_major_formatter(mdates.DateFormatter('%d-%m-%Y'))
ax.grid(True, which='both')
ax.minorticks_on()
axv = ax.twinx()
ax.plot_date(df['Adj Close'].loc[df['breakaway'] !=0].index, df['Adj Close'].loc[df['breakaway'] !=0],
'or', # marker style 'o', color 'g'
fillstyle='none', # circle is not filled (with color)
ms=10.0)
colors = dfc.VolumePositive.map({True: 'g', False: 'r'})
axv.bar(dfc.Date, dfc['Volume'], color=colors, alpha=0.4)
axv.axes.yaxis.set_ticklabels([])
axv.set_ylim(0, 3*df.Volume.max())
ax.set_title('Stock '+ symbol +' Closing Price')
ax.set_ylabel('Price')
df = df['2020-05-01':'2020-06-01']
dfc = df.copy()
dfc['VolumePositive'] = dfc['Open'] < dfc['Adj Close']
#dfc = dfc.dropna()
dfc = dfc.reset_index()
dfc['Date'] = pd.to_datetime(dfc['Date'])
dfc['Date'] = dfc['Date'].apply(mdates.date2num)
dfc.head()
fig = plt.figure(figsize=(20,16))
ax = plt.subplot(2, 1, 1)
ax.set_facecolor('pink')
candlestick_ohlc(ax,dfc.values, width=0.5, colorup='g', colordown='r', alpha=1.0)
ax.xaxis_date()
ax.xaxis.set_major_formatter(mdates.DateFormatter('%d-%m-%Y'))
#ax.grid(True, which='both')
#ax.minorticks_on()
axv = ax.twinx()
ax.plot_date(df['Adj Close'].loc[df['breakaway'] !=0].index, df['Adj Close'].loc[df['breakaway'] !=0],
'or', # marker style 'o', color 'g'
fillstyle='none', # circle is not filled (with color)
ms=30.0)
colors = dfc.VolumePositive.map({True: 'g', False: 'r'})
axv.bar(dfc.Date, dfc['Volume'], color=colors, alpha=0.4)
axv.axes.yaxis.set_ticklabels([])
axv.set_ylim(0, 3*df.Volume.max())
ax.set_title('Stock '+ symbol +' Closing Price')
ax.set_ylabel('Price')
from matplotlib.dates import date2num
from datetime import datetime
fig = plt.figure(figsize=(20,16))
ax = plt.subplot(2, 1, 1)
candlestick_ohlc(ax,dfc.values, width=0.5, colorup='g', colordown='r', alpha=1.0)
ax.xaxis_date()
ax.xaxis.set_major_formatter(mdates.DateFormatter('%d-%m-%Y'))
#ax.grid(True, which='both')
#ax.minorticks_on()
axv = ax.twinx()
ax.axvspan(date2num(datetime(2020,5,4)), date2num(datetime(2020,5,5)),
label="breakaway",color="red", alpha=0.3)
ax.legend(loc='upper center')
colors = dfc.VolumePositive.map({True: 'g', False: 'r'})
axv.bar(dfc.Date, dfc['Volume'], color=colors, alpha=0.4)
axv.axes.yaxis.set_ticklabels([])
axv.set_ylim(0, 3*df.Volume.max())
ax.set_title('Stock '+ symbol +' Closing Price')
ax.set_ylabel('Price')
| 0.361841 | 0.712332 |
# Auto-encoding of Julia Source Code
We investigate the ability of deep neural network architectures to capture key syntactic and semantic information in Julia source code using relatively low-dimensional representations. These representations can be potentially very powerful tools for automated program analysis and curation, especially across heterogeneous research fields and repositories.
There is in fact a wealth of [research](https://github.com/src-d/awesome-machine-learning-on-source-code) in this area, though to date there has been no application of these ideas or approaches to the Julia language itself. In this experiment, we study the makeup and patterns of the base Julia language source code in the hopes of identifying futher avenues of research.
We use the Keras deep learning API to construct our autoencoding model, and Numpy and Pandas libraries to manage the data inputs and outputs.
```
from keras import regularizers
from keras.callbacks import EarlyStopping
from keras.layers import Input, GRU, RepeatVector, Activation, CuDNNGRU
from keras.layers import Dense, BatchNormalization, Embedding
from keras.models import Model
from keras.optimizers import Adam
from keras.preprocessing.text import Tokenizer
from keras.preprocessing.sequence import pad_sequences
import numpy as np
import pandas as pd
```
The shape of the lower-dimensional representation we wish to create is a somewhat arbitrary choice; we chose 64 dimensions as a reasonably rich but still manageable number. Identifying optimal dimensionality is an area for further research.
We read our saved code snippets and source filepath labels in to a Pandas DataFrame. We also truncate any path information above the Julia repo itself as irrelevant (and necessarily uniformly repeated). Given large number of unique sources filepaths (546) and the low number of snippets in the median source file (19), we compute two aggregated groups of source files: top-level folders in the Julia repo and second-level divisions, which include both folders (with multiple files and folders within them) as well as single source code files in the top level folders themselves.
There are 7 top level folders with a wide range of amount of code within each, and 273 second-level divisions:
```
test 9391
stdlib 7846
base 7486
doc 30
contrib 25
src 7
etc 2
base_gcutils 5
base_array 170
test_meta 70
test_spawn 85
test_project 8
test_stacktraces 12
base_sysinfo 34
[...]```
```
latent_dim = 64
with open("all_funcs.csv", "r") as f:
funcs = f.read()
funcs = funcs.split(".jl\n")
funcs = funcs[:-1] # remove trailing empty item
funcs = pd.DataFrame([x.rsplit("\t",1) for x in funcs])
funcs.columns = ['code','source']
funcs = funcs[funcs.code.apply(lambda x: len(x)<=500)]
funcs.reset_index(drop=True, inplace=True)
funcs.source = funcs.source.apply(lambda x: x[x.index("julia/")+6:])
funcs["top_folder"] = funcs.source.apply(lambda x: x[:x.index("/")])
funcs['top2'] = funcs.source.apply(lambda x: '_'.join(x.split("/")[:2]))
```
We begin with a common sequence-to-sequence modeling approach as represented by [recent work](https://towardsdatascience.com/how-to-create-data-products-that-are-magical-using-sequence-to-sequence-models-703f86a231f8) at GitHub itself. This system was originally built to summarize Github issues, using the issue text as input sequences and issue titles as the target sequences. We adapt this to use as an auto-encoding system by using the same source code snippets as both inputs and targets.
<img src="../img/autoenc_diagram.png" />
_(Please note, if you have Keras v. 2.2.4 installed that there is an issue with the accuracy calculation (https://github.com/keras-team/keras/issues/11749). This is resolvable by upgrading to the latest version available via git or by downgrading to an earlier version of Keras. Given that the problem is already fixed in the latest unreleased code, we expect this issue to be resolved in the next pip-able release as well (i.e., 2.2.5).)_
We define two utility functions to aid in our encoding: `chars_to_indices()` which translates Julia source code into integers representing each character, and `ae_models` which build our autoencoder architecture. This second function returns two models - the full autoencoder, as well as the encoder sub-component. We use this second model to encode our Julia source code sequences after training is complete.
```
def chars_to_indices(data, tok=None, max_len=None):
if max_len is None:
max_len = max(data.apply(lambda x: len(x)))
if tok is None:
tok = Tokenizer(num_words=None,
filters="",
lower=False,
split='',
char_level=True)
data = data.values
tok.fit_on_texts(data)
sequences = tok.texts_to_sequences(data)
sequences = pad_sequences(sequences,
maxlen=max_len,
padding='post')
sequences = np.array(sequences, dtype='int16')
return sequences, tok
def ae_models(maxlen, latent_dim, N, use_gpu=False):
inputs = Input((maxlen,), name='Encoder_Inputs')
encoded = Embedding(N,
latent_dim,
name='Char_Embedding',
mask_zero=False)(inputs)
encoded = BatchNormalization(name='BatchNorm_Encoder')(encoded)
if use_gpu:
_, state_h = CuDNNGRU(latent_dim, return_state=True)(encoded)
else:
_, state_h = GRU(latent_dim, return_state=True)(encoded)
enc = Model(inputs=inputs, outputs=state_h, name='Encoder_Model')
enc_out = enc(inputs)
dec_inputs = Input(shape=(None,), name='Decoder_Inputs')
decoded = Embedding(N,
latent_dim,
name='Decoder_Embedding',
mask_zero=False)(dec_inputs)
decoded = BatchNormalization(name='BatchNorm_Decoder_1')(decoded)
if use_gpu:
dec_out, _ = CuDNNGRU(latent_dim,
return_state=True,
return_sequences=True)(decoded, initial_state=enc_out)
else:
dec_out, _ = GRU(latent_dim,
return_state=True,
return_sequences=True)(decoded, initial_state=enc_out)
dec_out = BatchNormalization(name='BatchNorm_Decoder_2')(dec_out)
dec_out = Dense(N, activation='softmax', name='Final_Out')(dec_out)
sequence_autoencoder = Model(inputs=[inputs, dec_inputs], outputs=dec_out)
return sequence_autoencoder, enc
```
We convert our Julia code snippets to vectors of integers, tokenized at the character level. E.g.,
```Julia
a::BitSet ⊊ b::BitSet = begin
#= none:414 =#
a <= b && a != b
end
```
becomes:
```Python
array([ 11, 8, 8, 52, 9, 4, 48, 2, 4, 1, 135, 1, 28,
8, 8, 52, 9, 4, 48, 2, 4, 1, 5, 1, 28, 2,
24, 9, 3, 13, 1, 1, 1, 1, 1, 1, 1, 1, 10,
5, 1, 3, 6, 3, 2, 8, 37, 21, 37, 1, 5, 10,
13, 1, 1, 1, 1, 1, 1, 1, 1, 11, 1, 65, 5,
1, 28, 1, 76, 76, 1, 11, 1, 67, 5, 1, 28, 13,
1, 1, 1, 1, 2, 3, 17])```
There are 235 unique characters in our Julia source code data (343 in the entire source code corpus, though over 100 of these are contained solely in a handful of excessively long expressions that were excluded for this experiment). To leverage teacher forcing in our seq2seq autoencoder training, we include the original input sequence as an input to the decoder portion of the autoencoder, and our target output is the the same input sequence one character offset.
```
seqs, tok = chars_to_indices(funcs.iloc[:,0])
N = len(np.unique(seqs))
decoder_inputs = seqs[:, :-1]
Y = seqs[:, 1: ]
```
We then build our model. The system can be set to run on a GPU or a CPU, depending on the hardware available. On an Nvidia Quadro P4000 training this model takes roughly 50 seconds per epoch.
```
# autoencoder, enc = ae_models(max_len, 64, N, use_gpu=True)
autoencoder, enc = ae_models(500, 64, 235, use_gpu=False)
autoencoder.summary()
```
We found that Adam optimization with amsgrad gave the best results for training efficiency and accuracy. We use sparse categorical crossentropy as our loss function, with the aim to reproduce input code snippets as exactly as possible in our outputs, character by character, over the 235-character "vocabulary."
We train for up to 100 epochs, using a batch size of 32 and validating on 12% of the total data. We also use early stopping to settle on an optimal set of model weights should learning level out before the 100th epoch. In various tests this was always the case, with our final model achieving a 93% level of per-character accuracy on the 44th epoch. In some cases it is useful to reduce the learning rate to 0.0001 after the first training session has stopped, but in our experiment this did not improve the model's performance.
```
opt = Adam(lr=0.001, amsgrad=True)
autoencoder.compile(loss='sparse_categorical_crossentropy',
optimizer=opt,
metrics=['accuracy'])
early_stop = EarlyStopping(monitor='val_acc',
min_delta=0.0001,
patience=10,
verbose=1,
mode='auto',
restore_best_weights=True)
autoencoder.fit([seqs, decoder_inputs],
np.expand_dims(Y, -1),
epochs = 100,
batch_size = 32,
validation_split=0.12,
callbacks=[early_stop],
shuffle=True)
autoencoder.save("autoencoder.h5")
enc.save("encoder.h5")
```
We encode our Julia source code snippet sequences with our newly trained encoder model. This gives us a two-dimensional Numpy array of shape (24787, 64) - each snippet being one row, represented as a vector of 64 float values.
```
encoded_reps = enc.predict(seqs)
encoded_reps = pd.DataFrame(encoded_reps)
encoded_reps.to_csv("encoded_reps.csv", index=False)
```
## Visualization and Analysis
We use Python's `matplotlib` library for plotting our visualizations, and [`UMAP`](https://arxiv.org/abs/1802.03426) (Uniform Manifold Approximation and Projection for Dimension Reduction) to represent our findings in a two-dimensional space. We also include code to produce three-dimensional interactive visualizations which can be even more informative, but the interactivity is beyond the scope of this static notebook documentation.
UMAP is a novel manifold learning technique for dimension reduction constructed from a theoretical framework based in Riemannian geometry and algebraic topology. The UMAP algorithm is competitive with the popular t-SNE method for visualization quality, while seeking to preserve more of the global structure with superior run time performance.
For visualizing our findings here, we limit our test to the "base" top-level folder (7486 code snippets), and label our cases by their second-level folder/file. We further limit our test cases to those labels with at least 100 examples for better visualization and comparison (21 categories).
```
from matplotlib import colors as mcolors
from matplotlib import pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
from sklearn.metrics import silhouette_score, silhouette_samples
from sklearn.metrics import adjusted_rand_score, adjusted_mutual_info_score
import sklearn.cluster as cluster
from umap import UMAP
X_test = encoded_reps[funcs.top_folder=="base"]
Y_test = funcs.top2[funcs.top_folder=="base"]
code_test = funcs.code[funcs.top_folder=="base"]
top_cats = list(Y_test.value_counts()[Y_test.value_counts()>=100].index)
X_test = X_test[Y_test.apply(lambda x: x in top_cats)]
code_test = code_test[Y_test.apply(lambda x: x in top_cats)]
Y_test = Y_test[Y_test.apply(lambda x: x in top_cats)]
```
We use cosine distance to compare the 64-dimensional vectors for our code snippets, and compute UMAP projections in two- and three-dimensional representations. Cosine distance is a measure of distnace between two vectors of an inner product space that measures the cosine of the angle between them, and is one of the most common distance metrics for comparing sequences in machine learning.
```
reducer = UMAP(random_state=42,
metric='cosine',
n_neighbors=30,
n_components=2)
embedding = reducer.fit_transform(X_test)
reducer_3d = UMAP(random_state=42,
metric='cosine',
n_neighbors=30,
n_components=3)
embedding_3d = reducer_3d.fit_transform(X_test)
```
We compute silhouette scores, which measure cohesion vs. overlap in multi-dimensional clusters on a scale from -1 (no separation and perfect overlap) to 1 (perfectly separated and compact clusters), both for our latent 64-dimensional space and the two-dimensional UMAP representation for our 21 test categories. We then visualize the UMAP representation (optionally in three dimensions).
```
sils = silhouette_samples(X_test, Y_test, metric='cosine')
clusts = pd.concat([X_test.reset_index(drop=True),
Y_test.reset_index(drop=True),
pd.Series(sils)], axis=1, ignore_index=True)
centroids = clusts.groupby(64).agg('mean').sort_values(65)
sils2 = silhouette_samples(embedding, Y_test, metric='cosine')
clusts2 = pd.concat([X_test.reset_index(drop=True),
Y_test.reset_index(drop=True),
pd.Series(sils2)], axis=1, ignore_index=True)
centroids2 = clusts2.groupby(64).agg('mean').sort_values(65)
src = list(centroids.index)
colors = dict(mcolors.BASE_COLORS, **mcolors.CSS4_COLORS)
by_hsv = sorted((tuple(mcolors.rgb_to_hsv(mcolors.to_rgba(color)[:3])), name)
for name, color in colors.items())
sorted_names = [name for hsv, name in by_hsv]
NUM_COLORS = len(src)
step = int(np.floor(len(sorted_names)/NUM_COLORS))
my_cols = [sorted_names[i]
for i in range(0, len(sorted_names), step)]
fig, ax = plt.subplots(figsize=(12, 10))
for i, s in enumerate(src):
ax.scatter(embedding[Y_test==s, 0],
embedding[Y_test==s, 1],
c=my_cols[i],
linewidths=0.1,
edgecolors='k',
label=s)
plt.setp(ax, xticks=[], yticks=[])
plt.title("Julia source code data embedded into two dimensions by UMAP",
fontsize=18)
plt.legend(loc="upper left", bbox_to_anchor=(1,1))
plt.subplots_adjust(right=0.75)
plt.show()
# #
# # 3d Plot
# #
# fig, ax = plt.subplots(figsize=(12, 10))
# ax2 = fig.add_subplot(111, projection="3d")
# for i, s in enumerate(src):
# ax2.scatter(embedding_3d[Y_test==s, 0],
# embedding_3d[Y_test==s, 1],
# embedding_3d[Y_test==s, 2],
# c=my_cols[i],
# linewidths=0.1,
# edgecolors='k',
# label=s)
# plt.setp(ax2, xticks=[], yticks=[])
# plt.title("Julia source code data embedded into two dimensions by UMAP",
# fontsize=18)
# plt.legend(loc="upper left", bbox_to_anchor=(1,1))
# plt.subplots_adjust(right=0.75)
# plt.show()
```
## Findings and Analysis
Encoded code snippets sort into several distinct groups based on syntactic and semantic content, but not necessarily in line with their source in the Julia source code directory tree. There is in fact a large degree of overlap across our directory-structure-defined categories.
<img src="../img/julia_code_map100.png" />
Those in base/Base.jl exhibited a great degree of intra-group similarity and cohesion (this group had a silhouette score of 0.74 in the UMAP embedding space, and 0.39 in the auto encoded 64-dimensional representation space). The only other source code group with somewhat similar cohesion was base/docs/ (silhouette of 0.07 in UMAP space, and 0.28 in latent space), while the rest of the top level code groups were far more intermixed with each other.
<img src="../img/silhouettes.png" />
Using some quick k-means clustering on the UMAP embeddings allows us to zero in on some of these groups and inspect the code snippets for what common content and structures.
<img src="../img/k_clusts.png" />
Clusters 3, 15, 18, and 37 overlap most significantly with the bulk of code groups base_compiler, base_Base, base_boot, and base_docs, respectively. Looking at example snippets from these clusters shows some obvious consistencies in form and content.
Cluster 3 is largely assignments, often of constants, and these would naturally feature prominently in the base_compiler code group. Referenceing our original UMAP projection graphic, it's also unsurprising to see many other code groups featured in cluster 3, as this is a very common type of expression across the Julia language.
```Julia
primitive type UInt128 <: Unsigned 128 end
const UTF8PROC_COMPOSE = 1 << 3
const _tvarnames = Symbol[:_A, :_B, :_C, :_D, :_E, :_F, :_G, :_H, :_I, :_J, :_K, :_L, :_M, :_N, :_O, :_P, :_Q, :_R, :_S, :_T, :_U, :_V, :_W, :_X, :_Y, :_Z]
const LOG2_E = 1.4426950408889634
const DATATYPE_PARAMETERS_FIELDINDEX = fieldindex(DataType, :parameters)```
Cluster 15 is largely `include` statements, and again this makes intuitive sense that these would comprise much of the base_Base code snippets. This is very tightly grouped cluster, as these statements are syntacticly simple and very uniform.
```Julia
include("strings/basic.jl")
include("process.jl")
include("gmp.jl")
include("arraymath.jl")
include("asyncmap.jl")```
Cluster 18, heavily identified with the base_boot code group, appears to consiste primarily of short type conversion functions.
```Julia
toUInt16(x::UInt16) = begin
x
end
toUInt32(x::UInt8) = begin
zext_int(UInt32, x)
end
toInt16(x::Bool) = begin
and_int(zext_int(Int16, x), Int16(1))
end
toInt16(x::Int32) = begin
checked_trunc_sint(Int16, x)
end
toInt16(x::Int16) = begin
x
end```
Finally, Cluster 37, where we find most of our base_docs snippets, consists primarily of doc-string prefaced code snippets. The base_docs code group is almost all doc-strings itself, and thus the tight grouping of its code snippets in this cluster.
```Julia
"""
Signed <: Integer
Abstract supertype for all signed integers.
"""
Signed
"""
copy(x)
Create a shallow copy of `x`: the outer structure is copied, but not all internal values.
For example, copying an array produces a new array with identically-same elements as the
original.
"""
copy
"""
cis(z)
Return ``\\exp(iz)``.
# Examples
``jldoctest
julia> cis(π) ≈ -1
true
``
"""
function cis(z::Complex)
v = exp(-imag(z))
s, c = sincos(real(z))
Complex(v * c, v * s)
end
"""
InitError(mod::Symbol, error)
An error occurred when running a module's `__init__` function. The actual error thrown is
available in the `.error` field.
"""
InitError
"""
StridedVecOrMat{T}
Union type of [`StridedVector`](@ref) and [`StridedMatrix`](@ref) with elements of type `T`.
"""
StridedVecOrMat
```
Finally, we look at different code snippets from the large agglomeration at the top of our UMAP visualization, represented in our cluster graphic as clusters 29, 13, 33, and 0 (from left to right). These clusters are all moderately short functions of similar complexity and layout, and these are found across all of our various code groups irrespective of purpose.
Cluster 29:
```Julia
function userefs(@nospecialize(x))
relevant = x isa Expr && is_relevant_expr(x) || (x isa GotoIfNot || (x isa ReturnNode || (x isa PiNode || (x isa PhiNode || (x isa PhiCNode || x isa UpsilonNode)))))
return UseRefIterator(x, relevant)
end
function adce_erase!(phi_uses, extra_worklist, compact, idx)
if compact.result[idx] isa PhiNode
maybe_erase_unused!(extra_worklist, compact, idx, (val->begin
phi_uses[val.id] -= 1
end))
else
maybe_erase_unused!(extra_worklist, compact, idx)
end
end```
Cluster 13:
```Julia
function gettypeinfos(io::IO, p::Pair)
typeinfo = get(io, :typeinfo, Any)
if p isa typeinfo <: Pair
fieldtype(typeinfo, 1) => fieldtype(typeinfo, 2)
else
Any => Any
end
end
function _maxlength(t::Tuple, t2::Tuple, t3::Tuple...)
@_inline_meta
max(length(t), _maxlength(t2, t3...))
end
function Base.deepcopy_internal(x::BigInt, stackdict::IdDict)
if haskey(stackdict, x)
return stackdict[x]
end
y = MPZ.set(x)
stackdict[x] = y
return y
end```
Cluster 33:
```Julia
function adduint64!(x::Bignum, operand::UInt64)
operand == 0 && return
other = Bignum()
assignuint64!(other, operand)
addbignum!(x, other)
end
function show(io::IO, r::LinRange)
print(io, ""range("")
show(io, first(r))
print(io, "", stop="")
show(io, last(r))
print(io, "", length="")
show(io, length(r))
print(io, ')')
end```
Cluster 0:
```Julia
function length(s::AbstractString, i::Int, j::Int)
@boundscheck begin
0 < i ≤ ncodeunits(s) + 1 || throw(BoundsError(s, i))
0 ≤ j < ncodeunits(s) + 1 || throw(BoundsError(s, j))
end
n = 0
for k = i:j
@inbounds n += isvalid(s, k)
end
return n
end
function fma(x::BigFloat, y::BigFloat, z::BigFloat)
r = BigFloat()
ccall(("mpfr_fma", :libmpfr), Int32, (Ref{BigFloat}, Ref{BigFloat}, Ref{BigFloat}, Ref{BigFloat}, MPFRRoundingMode), r, x, y, z, ROUNDING_MODE[])
return r
end```
## Discussion and Future Avenues for Research
From this brief initial analysis, we can conclude a few things. First, as with other major programming languages previously tested, deep neural networks can in fact learn to recognize Julia code structures and content in potentially useful ways. Second, we confirm that UMAP clusters in our latent space successfully capture important characteristics of the code. And third - contrary to some of our prior expectations - while the latent representations of Julia source code capture important information and patterns in Julia expressions, these do not consistently point back to particular source code packages or groups in the Julia language ontology. Instead, these representations capture more low-level differencies and similarities, finding similar code structures that more often than not repeat across different regions of the code base. As with its human creators, there is much more difference within Julia source code groups than between them.
Given these findings, it would be interesting to compare similar efforts with other languages to analyze how true this is across languages, or whether it is significantly more true in some languages than others. For instance, it would be interesting to explore if more parsimonious languages like Julia (or high level languages generally) exhibit the kind of wide disparity in syntactic patterns even within semantically similar code groups that we have seen here, while lower languages would see more syntactic similarity and semantic diversity. With respect to Julia code in particular and meta-modeling generally, we expect that further effort will enable us to leverage these embeddings for higher level tasks like recommending modifications, synthesizing models, identifying similar models, and validating the correctness of synthesized models.
|
github_jupyter
|
from keras import regularizers
from keras.callbacks import EarlyStopping
from keras.layers import Input, GRU, RepeatVector, Activation, CuDNNGRU
from keras.layers import Dense, BatchNormalization, Embedding
from keras.models import Model
from keras.optimizers import Adam
from keras.preprocessing.text import Tokenizer
from keras.preprocessing.sequence import pad_sequences
import numpy as np
import pandas as pd
test 9391
stdlib 7846
base 7486
doc 30
contrib 25
src 7
etc 2
base_gcutils 5
base_array 170
test_meta 70
test_spawn 85
test_project 8
test_stacktraces 12
base_sysinfo 34
[...]```
We begin with a common sequence-to-sequence modeling approach as represented by [recent work](https://towardsdatascience.com/how-to-create-data-products-that-are-magical-using-sequence-to-sequence-models-703f86a231f8) at GitHub itself. This system was originally built to summarize Github issues, using the issue text as input sequences and issue titles as the target sequences. We adapt this to use as an auto-encoding system by using the same source code snippets as both inputs and targets.
<img src="../img/autoenc_diagram.png" />
_(Please note, if you have Keras v. 2.2.4 installed that there is an issue with the accuracy calculation (https://github.com/keras-team/keras/issues/11749). This is resolvable by upgrading to the latest version available via git or by downgrading to an earlier version of Keras. Given that the problem is already fixed in the latest unreleased code, we expect this issue to be resolved in the next pip-able release as well (i.e., 2.2.5).)_
We define two utility functions to aid in our encoding: `chars_to_indices()` which translates Julia source code into integers representing each character, and `ae_models` which build our autoencoder architecture. This second function returns two models - the full autoencoder, as well as the encoder sub-component. We use this second model to encode our Julia source code sequences after training is complete.
We convert our Julia code snippets to vectors of integers, tokenized at the character level. E.g.,
becomes:
There are 235 unique characters in our Julia source code data (343 in the entire source code corpus, though over 100 of these are contained solely in a handful of excessively long expressions that were excluded for this experiment). To leverage teacher forcing in our seq2seq autoencoder training, we include the original input sequence as an input to the decoder portion of the autoencoder, and our target output is the the same input sequence one character offset.
We then build our model. The system can be set to run on a GPU or a CPU, depending on the hardware available. On an Nvidia Quadro P4000 training this model takes roughly 50 seconds per epoch.
We found that Adam optimization with amsgrad gave the best results for training efficiency and accuracy. We use sparse categorical crossentropy as our loss function, with the aim to reproduce input code snippets as exactly as possible in our outputs, character by character, over the 235-character "vocabulary."
We train for up to 100 epochs, using a batch size of 32 and validating on 12% of the total data. We also use early stopping to settle on an optimal set of model weights should learning level out before the 100th epoch. In various tests this was always the case, with our final model achieving a 93% level of per-character accuracy on the 44th epoch. In some cases it is useful to reduce the learning rate to 0.0001 after the first training session has stopped, but in our experiment this did not improve the model's performance.
We encode our Julia source code snippet sequences with our newly trained encoder model. This gives us a two-dimensional Numpy array of shape (24787, 64) - each snippet being one row, represented as a vector of 64 float values.
## Visualization and Analysis
We use Python's `matplotlib` library for plotting our visualizations, and [`UMAP`](https://arxiv.org/abs/1802.03426) (Uniform Manifold Approximation and Projection for Dimension Reduction) to represent our findings in a two-dimensional space. We also include code to produce three-dimensional interactive visualizations which can be even more informative, but the interactivity is beyond the scope of this static notebook documentation.
UMAP is a novel manifold learning technique for dimension reduction constructed from a theoretical framework based in Riemannian geometry and algebraic topology. The UMAP algorithm is competitive with the popular t-SNE method for visualization quality, while seeking to preserve more of the global structure with superior run time performance.
For visualizing our findings here, we limit our test to the "base" top-level folder (7486 code snippets), and label our cases by their second-level folder/file. We further limit our test cases to those labels with at least 100 examples for better visualization and comparison (21 categories).
We use cosine distance to compare the 64-dimensional vectors for our code snippets, and compute UMAP projections in two- and three-dimensional representations. Cosine distance is a measure of distnace between two vectors of an inner product space that measures the cosine of the angle between them, and is one of the most common distance metrics for comparing sequences in machine learning.
We compute silhouette scores, which measure cohesion vs. overlap in multi-dimensional clusters on a scale from -1 (no separation and perfect overlap) to 1 (perfectly separated and compact clusters), both for our latent 64-dimensional space and the two-dimensional UMAP representation for our 21 test categories. We then visualize the UMAP representation (optionally in three dimensions).
## Findings and Analysis
Encoded code snippets sort into several distinct groups based on syntactic and semantic content, but not necessarily in line with their source in the Julia source code directory tree. There is in fact a large degree of overlap across our directory-structure-defined categories.
<img src="../img/julia_code_map100.png" />
Those in base/Base.jl exhibited a great degree of intra-group similarity and cohesion (this group had a silhouette score of 0.74 in the UMAP embedding space, and 0.39 in the auto encoded 64-dimensional representation space). The only other source code group with somewhat similar cohesion was base/docs/ (silhouette of 0.07 in UMAP space, and 0.28 in latent space), while the rest of the top level code groups were far more intermixed with each other.
<img src="../img/silhouettes.png" />
Using some quick k-means clustering on the UMAP embeddings allows us to zero in on some of these groups and inspect the code snippets for what common content and structures.
<img src="../img/k_clusts.png" />
Clusters 3, 15, 18, and 37 overlap most significantly with the bulk of code groups base_compiler, base_Base, base_boot, and base_docs, respectively. Looking at example snippets from these clusters shows some obvious consistencies in form and content.
Cluster 3 is largely assignments, often of constants, and these would naturally feature prominently in the base_compiler code group. Referenceing our original UMAP projection graphic, it's also unsurprising to see many other code groups featured in cluster 3, as this is a very common type of expression across the Julia language.
Cluster 15 is largely `include` statements, and again this makes intuitive sense that these would comprise much of the base_Base code snippets. This is very tightly grouped cluster, as these statements are syntacticly simple and very uniform.
Cluster 18, heavily identified with the base_boot code group, appears to consiste primarily of short type conversion functions.
Finally, Cluster 37, where we find most of our base_docs snippets, consists primarily of doc-string prefaced code snippets. The base_docs code group is almost all doc-strings itself, and thus the tight grouping of its code snippets in this cluster.
Finally, we look at different code snippets from the large agglomeration at the top of our UMAP visualization, represented in our cluster graphic as clusters 29, 13, 33, and 0 (from left to right). These clusters are all moderately short functions of similar complexity and layout, and these are found across all of our various code groups irrespective of purpose.
Cluster 29:
Cluster 13:
Cluster 33:
Cluster 0:
| 0.901529 | 0.96378 |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.