code
stringlengths 2.5k
6.36M
| kind
stringclasses 2
values | parsed_code
stringlengths 0
404k
| quality_prob
float64 0
0.98
| learning_prob
float64 0.03
1
|
|---|---|---|---|---|
```
%reload_ext autoreload
%autoreload 2
from fastai.gen_doc.gen_notebooks import *
from pathlib import Path
```
#### To update this notebook
Run `tools/sgen_notebooks.py
Or run below:
You need to make sure to refresh right after
```
import glob
for f in Path().glob('*.ipynb'):
generate_missing_metadata(f)
```
# Metadata generated below
```
update_nb_metadata('tutorial.itemlist.ipynb',
summary='Advanced tutorial, explains how to create your custom `ItemBase` or `ItemList`',
title='Custom ItemList')
update_nb_metadata('tutorial.inference.ipynb',
summary='Intermediate tutorial, explains how to create a Learner for inference',
title='Inference Learner')
update_nb_metadata('tutorial.data.ipynb',
summary="Beginner's tutorial, explains how to quickly look at your data or model predictions",
title='Look at data')
update_nb_metadata('vision.gan.ipynb',
summary='All the modules and callbacks necessary to train a GAN',
title='vision.gan')
update_nb_metadata('callbacks.csv_logger.ipynb',
summary='Callbacks that saves the tracked metrics during training',
title='callbacks.csv_logger')
update_nb_metadata('callbacks.tracker.ipynb',
summary='Callbacks that take decisions depending on the evolution of metrics during training',
title='callbacks.tracker')
update_nb_metadata('torch_core.ipynb',
summary='Basic functions using pytorch',
title='torch_core')
update_nb_metadata('gen_doc.convert2html.ipynb',
summary='Converting the documentation notebooks to HTML pages',
title='gen_doc.convert2html')
update_nb_metadata('metrics.ipynb',
summary='Useful metrics for training',
title='metrics')
update_nb_metadata('callbacks.fp16.ipynb',
summary='Training in mixed precision implementation',
title='callbacks.fp16')
update_nb_metadata('callbacks.general_sched.ipynb',
summary='Implementation of a flexible training API',
title='callbacks.general_sched')
update_nb_metadata('text.ipynb',
keywords='fastai',
summary='Application to NLP, including ULMFiT fine-tuning',
title='text')
update_nb_metadata('callback.ipynb',
summary='Implementation of the callback system',
title='callback')
update_nb_metadata('tabular.models.ipynb',
keywords='fastai',
summary='Model for training tabular/structured data',
title='tabular.models')
update_nb_metadata('callbacks.mixup.ipynb',
summary='Implementation of mixup',
title='callbacks.mixup')
update_nb_metadata('applications.ipynb',
summary='Types of problems you can apply the fastai library to',
title='applications')
update_nb_metadata('vision.data.ipynb',
summary='Basic dataset for computer vision and helper function to get a DataBunch',
title='vision.data')
update_nb_metadata('overview.ipynb',
summary='Overview of the core modules',
title='overview')
update_nb_metadata('training.ipynb',
keywords='fastai',
summary='Overview of fastai training modules, including Learner, metrics, and callbacks',
title='training')
update_nb_metadata('text.transform.ipynb',
summary='NLP data processing; tokenizes text and creates vocab indexes',
title='text.transform')
# do not overwrite this notebook, or changes may get lost!
# update_nb_metadata('jekyll_metadata.ipynb')
update_nb_metadata('collab.ipynb',
summary='Application to collaborative filtering',
title='collab')
update_nb_metadata('text.learner.ipynb',
summary='Easy access of language models and ULMFiT',
title='text.learner')
update_nb_metadata('gen_doc.nbdoc.ipynb',
summary='Helper function to build the documentation',
title='gen_doc.nbdoc')
update_nb_metadata('vision.learner.ipynb',
summary='`Learner` support for computer vision',
title='vision.learner')
update_nb_metadata('core.ipynb',
summary='Basic helper functions for the fastai library',
title='core')
update_nb_metadata('fastai_typing.ipynb',
keywords='fastai',
summary='Type annotations names',
title='fastai_typing')
update_nb_metadata('gen_doc.gen_notebooks.ipynb',
summary='Generation of documentation notebook skeletons from python module',
title='gen_doc.gen_notebooks')
update_nb_metadata('basic_train.ipynb',
summary='Learner class and training loop',
title='basic_train')
update_nb_metadata('gen_doc.ipynb',
keywords='fastai',
summary='Documentation modules overview',
title='gen_doc')
update_nb_metadata('callbacks.rnn.ipynb',
summary='Implementation of a callback for RNN training',
title='callbacks.rnn')
update_nb_metadata('callbacks.one_cycle.ipynb',
summary='Implementation of the 1cycle policy',
title='callbacks.one_cycle')
update_nb_metadata('vision.ipynb',
summary='Application to Computer Vision',
title='vision')
update_nb_metadata('vision.transform.ipynb',
summary='List of transforms for data augmentation in CV',
title='vision.transform')
update_nb_metadata('callbacks.lr_finder.ipynb',
summary='Implementation of the LR Range test from Leslie Smith',
title='callbacks.lr_finder')
update_nb_metadata('text.data.ipynb',
summary='Basic dataset for NLP tasks and helper functions to create a DataBunch',
title='text.data')
update_nb_metadata('text.models.ipynb',
summary='Implementation of the AWD-LSTM and the RNN models',
title='text.models')
update_nb_metadata('tabular.data.ipynb',
summary='Base class to deal with tabular data and get a DataBunch',
title='tabular.data')
update_nb_metadata('callbacks.ipynb',
keywords='fastai',
summary='Callbacks implemented in the fastai library',
title='callbacks')
update_nb_metadata('train.ipynb',
summary='Extensions to Learner that easily implement Callback',
title='train')
update_nb_metadata('callbacks.hooks.ipynb',
summary='Implement callbacks using hooks',
title='callbacks.hooks')
update_nb_metadata('vision.image.ipynb',
summary='Image class, variants and internal data augmentation pipeline',
title='vision.image')
update_nb_metadata('vision.models.unet.ipynb',
summary='Dynamic Unet that can use any pretrained model as a backbone.',
title='vision.models.unet')
update_nb_metadata('vision.models.ipynb',
keywords='fastai',
summary='Overview of the models used for CV in fastai',
title='vision.models')
update_nb_metadata('tabular.transform.ipynb',
summary='Transforms to clean and preprocess tabular data',
title='tabular.transform')
update_nb_metadata('index.ipynb',
keywords='fastai',
toc='false',
title='Welcome to fastai')
update_nb_metadata('layers.ipynb',
summary='Provides essential functions to building and modifying `Model` architectures.',
title='layers')
update_nb_metadata('tabular.ipynb',
keywords='fastai',
summary='Application to tabular/structured data',
title='tabular')
update_nb_metadata('basic_data.ipynb',
summary='Basic classes to contain the data for model training.',
title='basic_data')
update_nb_metadata('datasets.ipynb')
update_nb_metadata('tmp.ipynb',
keywords='fastai')
update_nb_metadata('callbacks.tracking.ipynb')
update_nb_metadata('data_block.ipynb',
keywords='fastai',
summary='The data block API',
title='data_block')
update_nb_metadata('callbacks.tracker.ipynb',
keywords='fastai',
summary='Callbacks that take decisions depending on the evolution of metrics during training',
title='callbacks.tracking')
update_nb_metadata('widgets.ipynb')
update_nb_metadata('text_tmp.ipynb')
update_nb_metadata('tabular_tmp.ipynb')
update_nb_metadata('tutorial.data.ipynb')
update_nb_metadata('tutorial.itemlist.ipynb')
update_nb_metadata('tutorial.inference.ipynb')
update_nb_metadata('vision.gan.ipynb')
update_nb_metadata('utils.collect_env.ipynb')
update_nb_metadata('widgets.image_cleaner.ipynb')
update_nb_metadata('utils.mem.ipynb')
update_nb_metadata('callbacks.mem.ipynb')
update_nb_metadata('gen_doc.nbtest.ipynb',
summary='Helper functions to search for api tests',
title='gen_doc.nbtest')
update_nb_metadata('utils.ipython.ipynb')
update_nb_metadata('callbacks.misc.ipynb')
update_nb_metadata('utils.mod_display.ipynb')
update_nb_metadata('text.interpret.ipynb',
keywords='fastai',
summary='Easy access of language models and ULMFiT',
title='text.learner')
update_nb_metadata('vision.interpret.ipynb',
keywords='fastai',
summary='`Learner` support for computer vision',
title='vision.learner')
update_nb_metadata('widgets.class_confusion.ipynb')
```
|
github_jupyter
|
%reload_ext autoreload
%autoreload 2
from fastai.gen_doc.gen_notebooks import *
from pathlib import Path
import glob
for f in Path().glob('*.ipynb'):
generate_missing_metadata(f)
update_nb_metadata('tutorial.itemlist.ipynb',
summary='Advanced tutorial, explains how to create your custom `ItemBase` or `ItemList`',
title='Custom ItemList')
update_nb_metadata('tutorial.inference.ipynb',
summary='Intermediate tutorial, explains how to create a Learner for inference',
title='Inference Learner')
update_nb_metadata('tutorial.data.ipynb',
summary="Beginner's tutorial, explains how to quickly look at your data or model predictions",
title='Look at data')
update_nb_metadata('vision.gan.ipynb',
summary='All the modules and callbacks necessary to train a GAN',
title='vision.gan')
update_nb_metadata('callbacks.csv_logger.ipynb',
summary='Callbacks that saves the tracked metrics during training',
title='callbacks.csv_logger')
update_nb_metadata('callbacks.tracker.ipynb',
summary='Callbacks that take decisions depending on the evolution of metrics during training',
title='callbacks.tracker')
update_nb_metadata('torch_core.ipynb',
summary='Basic functions using pytorch',
title='torch_core')
update_nb_metadata('gen_doc.convert2html.ipynb',
summary='Converting the documentation notebooks to HTML pages',
title='gen_doc.convert2html')
update_nb_metadata('metrics.ipynb',
summary='Useful metrics for training',
title='metrics')
update_nb_metadata('callbacks.fp16.ipynb',
summary='Training in mixed precision implementation',
title='callbacks.fp16')
update_nb_metadata('callbacks.general_sched.ipynb',
summary='Implementation of a flexible training API',
title='callbacks.general_sched')
update_nb_metadata('text.ipynb',
keywords='fastai',
summary='Application to NLP, including ULMFiT fine-tuning',
title='text')
update_nb_metadata('callback.ipynb',
summary='Implementation of the callback system',
title='callback')
update_nb_metadata('tabular.models.ipynb',
keywords='fastai',
summary='Model for training tabular/structured data',
title='tabular.models')
update_nb_metadata('callbacks.mixup.ipynb',
summary='Implementation of mixup',
title='callbacks.mixup')
update_nb_metadata('applications.ipynb',
summary='Types of problems you can apply the fastai library to',
title='applications')
update_nb_metadata('vision.data.ipynb',
summary='Basic dataset for computer vision and helper function to get a DataBunch',
title='vision.data')
update_nb_metadata('overview.ipynb',
summary='Overview of the core modules',
title='overview')
update_nb_metadata('training.ipynb',
keywords='fastai',
summary='Overview of fastai training modules, including Learner, metrics, and callbacks',
title='training')
update_nb_metadata('text.transform.ipynb',
summary='NLP data processing; tokenizes text and creates vocab indexes',
title='text.transform')
# do not overwrite this notebook, or changes may get lost!
# update_nb_metadata('jekyll_metadata.ipynb')
update_nb_metadata('collab.ipynb',
summary='Application to collaborative filtering',
title='collab')
update_nb_metadata('text.learner.ipynb',
summary='Easy access of language models and ULMFiT',
title='text.learner')
update_nb_metadata('gen_doc.nbdoc.ipynb',
summary='Helper function to build the documentation',
title='gen_doc.nbdoc')
update_nb_metadata('vision.learner.ipynb',
summary='`Learner` support for computer vision',
title='vision.learner')
update_nb_metadata('core.ipynb',
summary='Basic helper functions for the fastai library',
title='core')
update_nb_metadata('fastai_typing.ipynb',
keywords='fastai',
summary='Type annotations names',
title='fastai_typing')
update_nb_metadata('gen_doc.gen_notebooks.ipynb',
summary='Generation of documentation notebook skeletons from python module',
title='gen_doc.gen_notebooks')
update_nb_metadata('basic_train.ipynb',
summary='Learner class and training loop',
title='basic_train')
update_nb_metadata('gen_doc.ipynb',
keywords='fastai',
summary='Documentation modules overview',
title='gen_doc')
update_nb_metadata('callbacks.rnn.ipynb',
summary='Implementation of a callback for RNN training',
title='callbacks.rnn')
update_nb_metadata('callbacks.one_cycle.ipynb',
summary='Implementation of the 1cycle policy',
title='callbacks.one_cycle')
update_nb_metadata('vision.ipynb',
summary='Application to Computer Vision',
title='vision')
update_nb_metadata('vision.transform.ipynb',
summary='List of transforms for data augmentation in CV',
title='vision.transform')
update_nb_metadata('callbacks.lr_finder.ipynb',
summary='Implementation of the LR Range test from Leslie Smith',
title='callbacks.lr_finder')
update_nb_metadata('text.data.ipynb',
summary='Basic dataset for NLP tasks and helper functions to create a DataBunch',
title='text.data')
update_nb_metadata('text.models.ipynb',
summary='Implementation of the AWD-LSTM and the RNN models',
title='text.models')
update_nb_metadata('tabular.data.ipynb',
summary='Base class to deal with tabular data and get a DataBunch',
title='tabular.data')
update_nb_metadata('callbacks.ipynb',
keywords='fastai',
summary='Callbacks implemented in the fastai library',
title='callbacks')
update_nb_metadata('train.ipynb',
summary='Extensions to Learner that easily implement Callback',
title='train')
update_nb_metadata('callbacks.hooks.ipynb',
summary='Implement callbacks using hooks',
title='callbacks.hooks')
update_nb_metadata('vision.image.ipynb',
summary='Image class, variants and internal data augmentation pipeline',
title='vision.image')
update_nb_metadata('vision.models.unet.ipynb',
summary='Dynamic Unet that can use any pretrained model as a backbone.',
title='vision.models.unet')
update_nb_metadata('vision.models.ipynb',
keywords='fastai',
summary='Overview of the models used for CV in fastai',
title='vision.models')
update_nb_metadata('tabular.transform.ipynb',
summary='Transforms to clean and preprocess tabular data',
title='tabular.transform')
update_nb_metadata('index.ipynb',
keywords='fastai',
toc='false',
title='Welcome to fastai')
update_nb_metadata('layers.ipynb',
summary='Provides essential functions to building and modifying `Model` architectures.',
title='layers')
update_nb_metadata('tabular.ipynb',
keywords='fastai',
summary='Application to tabular/structured data',
title='tabular')
update_nb_metadata('basic_data.ipynb',
summary='Basic classes to contain the data for model training.',
title='basic_data')
update_nb_metadata('datasets.ipynb')
update_nb_metadata('tmp.ipynb',
keywords='fastai')
update_nb_metadata('callbacks.tracking.ipynb')
update_nb_metadata('data_block.ipynb',
keywords='fastai',
summary='The data block API',
title='data_block')
update_nb_metadata('callbacks.tracker.ipynb',
keywords='fastai',
summary='Callbacks that take decisions depending on the evolution of metrics during training',
title='callbacks.tracking')
update_nb_metadata('widgets.ipynb')
update_nb_metadata('text_tmp.ipynb')
update_nb_metadata('tabular_tmp.ipynb')
update_nb_metadata('tutorial.data.ipynb')
update_nb_metadata('tutorial.itemlist.ipynb')
update_nb_metadata('tutorial.inference.ipynb')
update_nb_metadata('vision.gan.ipynb')
update_nb_metadata('utils.collect_env.ipynb')
update_nb_metadata('widgets.image_cleaner.ipynb')
update_nb_metadata('utils.mem.ipynb')
update_nb_metadata('callbacks.mem.ipynb')
update_nb_metadata('gen_doc.nbtest.ipynb',
summary='Helper functions to search for api tests',
title='gen_doc.nbtest')
update_nb_metadata('utils.ipython.ipynb')
update_nb_metadata('callbacks.misc.ipynb')
update_nb_metadata('utils.mod_display.ipynb')
update_nb_metadata('text.interpret.ipynb',
keywords='fastai',
summary='Easy access of language models and ULMFiT',
title='text.learner')
update_nb_metadata('vision.interpret.ipynb',
keywords='fastai',
summary='`Learner` support for computer vision',
title='vision.learner')
update_nb_metadata('widgets.class_confusion.ipynb')
| 0.80406 | 0.898366 |
# Subsetting the data
## About the Data
In this notebook, we will be working with earthquake data from August 18, 2021 - October 18, 2021 (obtained from the US Geological Survey (USGS) using the [USGS API](https://earthquake.usgs.gov/fdsnws/event/1/))
## Setup
We will be working with the `data/earthquakes.csv` file again, so we need to handle our imports and read it in.
```
import pandas as pd
df = pd.read_csv('earthquakes.csv')
```
## Selecting columns
Grab an entire column using attribute notation:
```
df.mag
```
Grab an entire column using dictionary syntax:
```
df['mag']
```
Selecting multiple columns:
```
df[['mag', 'title']]
```
Selecting columns using list comprehensions and string operations:
```
df[
['title', 'time']
+ [col for col in df.columns if col.startswith('mag')]
]
```
Breaking down this example:
1. the list comprehension
```
[col for col in df.columns if col.startswith('mag')]
```
2. assembling the list
```
['title', 'time'] \
+ [col for col in df.columns if col.startswith('mag')]
```
3. using this list as the list of columns
```
df[
['title', 'time']
+ [col for col in df.columns if col.startswith('mag')]
]
```
## Slicing
### Selecting rows
Using row numbers (inclusive of first index, exclusive of last):
```
df[100:103]
```
### Selecting rows and columns with chaining
```
df[['title', 'time']][100:103]
```
Order doesn't matter here:
```
df[100:103][['title', 'time']].equals(
df[['title', 'time']][100:103]
)
```
So we know how to select rows and columns, but can we update values? Well, if we try using what we have learned so far, we will see the following warning:
```
df[110:113]['title'] = df[110:113]['title'].str.lower()
```
Note that it worked here, but `pandas` says we were setting a value on a copy of a slice and that we should use `loc` instead (topic of the following section):
```
df[110:113]['title']
```
## Indexing
Now if we do this with `loc` as the warning suggests, everything goes smoothly. Note we have to lower the end index by one since `loc` is inclusive of endpoints:
```
df.loc[110:112, 'title'] = df.loc[110:112, 'title'].str.lower()
df.loc[110:112, 'title']
```
### Indexing with `loc`
Selection of the format `loc[row_indexer, column_indexer]` where `:` can be used to select all:
```
df.loc[:,'title']
```
We can use `loc` to select specific rows and columns without chaining. If we use row numbers with `loc`, they are now **inclusive** of the end index:
```
df.loc[10:15, ['title', 'mag']]
```
#### Indexing with `iloc`
Exclusive of the endpoint just as Python slicing:
```
df.iloc[10:15, [19, 8, 15]]
```
We can use slicing syntax with `iloc` for both rows and columns:
```
df.iloc[10:15, 6:10]
```
When using `loc`, we can slice on column names. This will be inclusive of the endpoint because you can't be expected to know what the next column name will be. As such, we have multiple ways to achieve the same end goal:
```
df.iloc[10:15, 6:10].equals(
df.loc[10:14, 'gap':'magType']
)
```
### Looking up scalar values
We used `loc` and `iloc` to grab subsets of the dataframe. However, if we are just interested in the specific value at a given `[row, column]`, then we can use `iat` and `at`. We use `at` with labels:
```
df.at[10, 'mag']
```
...and `iat` with integer indices:
```
df.iat[16, 24]
```
## Filtering
We can filter our dataframes using a **Boolean mask**, which can be made as follows:
```
df.mag > 2
```
To use a mask for selection, we simply place it inside the brackets:
```
df[df.mag >= 6.0]
```
We can use masks with `loc`:
```
df.loc[
df.mag >= 7.0,
['alert', 'mag', 'magType', 'title', 'tsunami', 'type']
]
```
Masks can be created using multiple criteria when combined with bitwise operators `&` for AND and `|` for OR. We must also surround each criterion with parentheses. We can't use `and`/`or` here because we need to evaluate row by row:
```
df.loc[
(df.tsunami == 1) & (df.alert == 'red'),
['alert', 'mag', 'magType', 'title', 'tsunami', 'type']
]
```
An example with an OR condition, which is less restrictive:
```
df.loc[
(df.tsunami == 1) | (df.alert == 'red'),
['alert', 'mag', 'magType', 'title', 'tsunami', 'type']
]
```
Masks can be created from any criteria that results in a Boolean. For example, we can select all earthquakes with the string `Alaska` in the `place` column with a non-null value for the `alert` column. To get non-nulls, we can use the `isnull()` method with the bitwise negation operator (`~`) or the `notnull()` method:
```
df.loc[
(df.place.str.contains('Alaska')) & (df.alert.notnull()),
['alert', 'mag', 'magType', 'title', 'tsunami', 'type']
]
```
We can even use regular expressions here:
```
df.loc[
(df.place.str.contains(r'CA|California$')) & (df.mag > 3.8),
['alert', 'mag', 'magType', 'title', 'tsunami', 'type']
]
```
We can use the `between()` method to turn 2 individual checks (is less than or equal to some maximum value and is greater than or equal to some minimum value) into a single one. Note this is inclusive of the endpoint by default:
```
df.loc[
df.mag.between(6.5, 7.5),
['alert', 'mag', 'magType', 'title', 'tsunami', 'type']
]
```
We can use the `isin()` method to check for membership in a list of values:
```
df.loc[
df.magType.isin(['mw', 'mwb']),
['alert', 'mag', 'magType', 'title', 'tsunami', 'type']
]
```
We can grab the index of the minimum and maximum values of a given column and use those to select the entire row where they occur:
```
[df.mag.idxmin(), df.mag.idxmax()]
df.loc[
[df.mag.idxmin(), df.mag.idxmax()],
['alert', 'mag', 'magType', 'title', 'tsunami', 'type']
]
```
Note that there is a `filter()` method, but it doesn't filter the data in the same sense as we discussed in this section. Here are a few things you can do with this method.
- grab columns of a dataframe by passing a list to `items`:
```
df.filter(items=['mag', 'magType']).head()
```
- grab all the columns that contain a string with the `like` parameter:
```
df.filter(like='mag').head()
```
- use regular expressions; here, we select any columns that start with `t`:
```
df.filter(regex=r'^t').head()
```
- use `filter()` along the rows, by passing in `axis=0`. Here, we will use the `place` column as the index (we will cover `set_index()` in chapter 3):
```
df.set_index('place').filter(like='Japan', axis=0).filter(items=['mag', 'magType', 'title']).head()
```
This also works on `Series` objects and will run on the index:
```
df.set_index('place').title.filter(like='Japan').head()
```
<hr>
<div>
<a href="./4-inspecting_dataframes.ipynb">
<button style="float: left;">← Previous Notebook</button>
</a>
<a href="./6-adding_and_removing_data.ipynb">
<button style="float: right;">Next Notebook →</button>
</a>
</div>
<br>
<hr>
|
github_jupyter
|
import pandas as pd
df = pd.read_csv('earthquakes.csv')
df.mag
df['mag']
df[['mag', 'title']]
df[
['title', 'time']
+ [col for col in df.columns if col.startswith('mag')]
]
[col for col in df.columns if col.startswith('mag')]
['title', 'time'] \
+ [col for col in df.columns if col.startswith('mag')]
df[
['title', 'time']
+ [col for col in df.columns if col.startswith('mag')]
]
df[100:103]
df[['title', 'time']][100:103]
df[100:103][['title', 'time']].equals(
df[['title', 'time']][100:103]
)
df[110:113]['title'] = df[110:113]['title'].str.lower()
df[110:113]['title']
df.loc[110:112, 'title'] = df.loc[110:112, 'title'].str.lower()
df.loc[110:112, 'title']
df.loc[:,'title']
df.loc[10:15, ['title', 'mag']]
df.iloc[10:15, [19, 8, 15]]
df.iloc[10:15, 6:10]
df.iloc[10:15, 6:10].equals(
df.loc[10:14, 'gap':'magType']
)
df.at[10, 'mag']
df.iat[16, 24]
df.mag > 2
df[df.mag >= 6.0]
df.loc[
df.mag >= 7.0,
['alert', 'mag', 'magType', 'title', 'tsunami', 'type']
]
df.loc[
(df.tsunami == 1) & (df.alert == 'red'),
['alert', 'mag', 'magType', 'title', 'tsunami', 'type']
]
df.loc[
(df.tsunami == 1) | (df.alert == 'red'),
['alert', 'mag', 'magType', 'title', 'tsunami', 'type']
]
df.loc[
(df.place.str.contains('Alaska')) & (df.alert.notnull()),
['alert', 'mag', 'magType', 'title', 'tsunami', 'type']
]
df.loc[
(df.place.str.contains(r'CA|California$')) & (df.mag > 3.8),
['alert', 'mag', 'magType', 'title', 'tsunami', 'type']
]
df.loc[
df.mag.between(6.5, 7.5),
['alert', 'mag', 'magType', 'title', 'tsunami', 'type']
]
df.loc[
df.magType.isin(['mw', 'mwb']),
['alert', 'mag', 'magType', 'title', 'tsunami', 'type']
]
[df.mag.idxmin(), df.mag.idxmax()]
df.loc[
[df.mag.idxmin(), df.mag.idxmax()],
['alert', 'mag', 'magType', 'title', 'tsunami', 'type']
]
df.filter(items=['mag', 'magType']).head()
df.filter(like='mag').head()
df.filter(regex=r'^t').head()
df.set_index('place').filter(like='Japan', axis=0).filter(items=['mag', 'magType', 'title']).head()
df.set_index('place').title.filter(like='Japan').head()
| 0.207616 | 0.987652 |
```
from pyscisci.datasource.MAG import MAG
from pyscisci.utils import groupby_count
import numpy as np
import matplotlib.pylab as plt
try:
import seaborn as sns
sns.set_style('white')
except:
pass
%matplotlib inline
# set this path to where the MAG database will be stored
path2mag = '/home/ajgates/MAG'
path2mag = '/Users/ajgates/Desktop/MAG'
mymag = MAG(path2mag, keep_in_memory=False)
# set keep_in_memory=False if you want to load the database each time its needed - good for when you
# cant keep more than one database in memory at a time
# otherwise keep_in_memory=True will keep each database in memory after its loaded
# before we can start running our analysis, we have to preprocess the raw data into
# DataFrames that are more convinent to work with
# we only need to run this for the first time, but it will take awhile
mymag.preprocess(verbose=True)
# MAG contains the following dataframes:
# pub_df - keeps all of the publication information
# columns : ['PublicationId', 'Year', 'JournalId', 'FamilyId', 'Doi', 'Title', 'Date', 'Volume', 'Issue', 'DocType']
# author_df - keeps all of the author information
# columns : ['AuthorId', 'FullName', 'LastName', 'FirstName', 'MiddleName']
# pub2ref_df - links publications to their references or citations
# columns : ['CitingPublicationId', 'CitedPublicationId']
# paa_df - links publications, authors, and affiliations
# columns : ['PublicationId', 'AuthorId', 'AffiliationId', 'AuthorSequence', 'OrigAuthorName', 'OrigAffiliationName']
# author2pub_df - links the authors to their publications
# columns : ['PublicationId', 'AuthorId', 'AuthorOrder']
# field_df - field information
# columns : ['FieldId', 'FieldLevel', 'NumberPublications', 'FieldName']
# pub2field_df - links publications to their fields
# columns : ['PublicationId', 'FieldId']
# affiliation_df - affiliation information
# columns : ['AffiliationId', 'NumberPublications', 'NumberCitations', 'FullName', 'GridId', 'OfficialPage', 'WikiPage', 'Latitude', 'Longitude']
# journal_df - journal information
# columns : ['JournalId', 'FullName', 'Issn', 'Publisher', 'Webpage']
# after additional processing, these DataFrames become available
# pub2refnoself_df - links publications to their references or citations with self-citations removed
# columns : ['CitingPublicationId', 'CitedPublicationId']
# impact_df - precomputed citation counts, columns will depend on which counts are computed
# columns : ['PublicationId', 'Year', ....]
# lets plot the number of publications each year
yearly_articles = groupby_count(df=mymag.pub_df, colgroupby='Year', colcountby='PublicationId', unique=True)
yearly_articles.sort_values(by='Year', inplace=True)
yearly_articles = yearly_articles.loc[yearly_articles['Year'] > 0]
fig, ax = plt.subplots(1,1,figsize=(8,5))
ax.plot(yearly_articles['Year'],yearly_articles['PublicationIdCount'])
ax.set_xlabel('Year')
ax.set_ylabel("# of publications")
ax.set_yscale('log')
plt.show()
# now we can see the distribution of author productivity
author_prod = mymag.author_productivity()
prodvalues, prodcounts = np.unique(author_prod['Productivity'].values, return_counts=True)
fig, ax = plt.subplots(1,1,figsize=(8,5))
ax.scatter(prodvalues, prodcounts)
ax.set_xlabel('Productivity')
ax.set_ylabel("# of authors")
ax.set_xscale('log')
ax.set_yscale('log')
plt.show()
```
|
github_jupyter
|
from pyscisci.datasource.MAG import MAG
from pyscisci.utils import groupby_count
import numpy as np
import matplotlib.pylab as plt
try:
import seaborn as sns
sns.set_style('white')
except:
pass
%matplotlib inline
# set this path to where the MAG database will be stored
path2mag = '/home/ajgates/MAG'
path2mag = '/Users/ajgates/Desktop/MAG'
mymag = MAG(path2mag, keep_in_memory=False)
# set keep_in_memory=False if you want to load the database each time its needed - good for when you
# cant keep more than one database in memory at a time
# otherwise keep_in_memory=True will keep each database in memory after its loaded
# before we can start running our analysis, we have to preprocess the raw data into
# DataFrames that are more convinent to work with
# we only need to run this for the first time, but it will take awhile
mymag.preprocess(verbose=True)
# MAG contains the following dataframes:
# pub_df - keeps all of the publication information
# columns : ['PublicationId', 'Year', 'JournalId', 'FamilyId', 'Doi', 'Title', 'Date', 'Volume', 'Issue', 'DocType']
# author_df - keeps all of the author information
# columns : ['AuthorId', 'FullName', 'LastName', 'FirstName', 'MiddleName']
# pub2ref_df - links publications to their references or citations
# columns : ['CitingPublicationId', 'CitedPublicationId']
# paa_df - links publications, authors, and affiliations
# columns : ['PublicationId', 'AuthorId', 'AffiliationId', 'AuthorSequence', 'OrigAuthorName', 'OrigAffiliationName']
# author2pub_df - links the authors to their publications
# columns : ['PublicationId', 'AuthorId', 'AuthorOrder']
# field_df - field information
# columns : ['FieldId', 'FieldLevel', 'NumberPublications', 'FieldName']
# pub2field_df - links publications to their fields
# columns : ['PublicationId', 'FieldId']
# affiliation_df - affiliation information
# columns : ['AffiliationId', 'NumberPublications', 'NumberCitations', 'FullName', 'GridId', 'OfficialPage', 'WikiPage', 'Latitude', 'Longitude']
# journal_df - journal information
# columns : ['JournalId', 'FullName', 'Issn', 'Publisher', 'Webpage']
# after additional processing, these DataFrames become available
# pub2refnoself_df - links publications to their references or citations with self-citations removed
# columns : ['CitingPublicationId', 'CitedPublicationId']
# impact_df - precomputed citation counts, columns will depend on which counts are computed
# columns : ['PublicationId', 'Year', ....]
# lets plot the number of publications each year
yearly_articles = groupby_count(df=mymag.pub_df, colgroupby='Year', colcountby='PublicationId', unique=True)
yearly_articles.sort_values(by='Year', inplace=True)
yearly_articles = yearly_articles.loc[yearly_articles['Year'] > 0]
fig, ax = plt.subplots(1,1,figsize=(8,5))
ax.plot(yearly_articles['Year'],yearly_articles['PublicationIdCount'])
ax.set_xlabel('Year')
ax.set_ylabel("# of publications")
ax.set_yscale('log')
plt.show()
# now we can see the distribution of author productivity
author_prod = mymag.author_productivity()
prodvalues, prodcounts = np.unique(author_prod['Productivity'].values, return_counts=True)
fig, ax = plt.subplots(1,1,figsize=(8,5))
ax.scatter(prodvalues, prodcounts)
ax.set_xlabel('Productivity')
ax.set_ylabel("# of authors")
ax.set_xscale('log')
ax.set_yscale('log')
plt.show()
| 0.415847 | 0.347911 |
# Raumluftqualität 3.0
In realen Gebäuden gibt es immer einen Luftaustausch mit der Umgebung. In modernen Gebäuden ist dieser aufgrund der hochwertigen Abdichtung nach einem Blowerdoortest sehr gering. Dann müssen Gebäude mechanisch belüftet werden, um Schadstoffe abzutransportieren und die Räume mit frischer Außenluft zu versorgen.
Wird ein Gebäude belüftet, so lässt sich bei bekannter Schadstoffproduktion und bei bekannten Konzentrationen der Raumluft und der Außenluft berechnen, welcher Volumenstrom zur Belüftung erforderlich ist.
Dazu diente die Gleichgewichtsbetrachtung, dass aus dem Raum genauso viel Schadstoff über die Abluft abtransportiert werden muss, wie über die Außenluft und durch die Schadstoffquellen zugeführt wird.
Der erforderliche Außenluftvolumenstrom ist:
$$
\dot V_{\rm au} = \dfrac{\dot V_{\rm sch}}{k_{\rm zul} - k_{\rm au}}.
$$
Im nächsten Schritt wird die Frage untersucht, wie sich die Schadstoffkonzentration in einem belüfteten Raum im Laufe der Zeit verändert. Dazu gehen wir zunächst von einem gut durchlüfteten Raum aus. Die CO_2-Konzentration in einem gut durchlüfteten Raum wird mit der CO_2-Konzentration der Außenluft übereinstimmen. Zu Beginn ist also
$$
k_0 = k_{\rm au}
$$
Zu einer beliebigen Zeit $t$ lässt sich die Schadstoffkonzentration $k(t)$ der Raumluft nach der folgenden Funktion berechnen:
$$
k(t)= k_\infty + (k_0-k_\infty)\cdot{\mathrm e}^{-\beta\,(t-t_0)}
$$
Dabei ist $k_\infty$ der Endwert, der sich nach (theoretisch unendlich) langer Zeit einstellt. Das Zeichen $\infty$ wird mathematisch als "Unendlich" gelesen. $k_0$ ist der Startwert zur Zeit $t=t_0$, wobei meistens $t_0=0$ gesetzt wird. Manchmal ist es aber bequem, nicht bei $t_0=0$ beginnen zu müssen, sondern zu einer beliebigen Zeit.
In einem gut durchlüfteten Raum ist $k_0 = k_\rm{au} \approx 400\,\rm{ppM}$.
Der Wert $k_\infty$ ist die Schadstoffkonzentration, die sich im Gleichgewicht zwischen Schadstoffzufuhr und -abtransport ergibt. Sie ist bekannt, wenn der Außenluftvolumenstrom $\dot V_{\rm au}$ und die Schadstoffproduktion $\dot V_\rm{sch}$ bekannt sind. Es ist nämlich
$$
\dot V_{\rm au} = \dfrac{\dot V_{\rm sch}}{k_\infty -k_{\rm au}}
$$
Nach $k_\infty$ umgestellt, ergibt sich
$$
k_\infty =
k_{\rm au} + \dfrac{\dot V_{\rm sch}}{\dot V_{\rm au}}
$$
### Beispiel
Ein Gebäude mit einer Grundfläche von $150\,{\rm m^2}$ und einer Geschosshöhe von $2.50\,{\rm m}$ wird mit
$180\,{\rm \dfrac{m^3}{h}}$ Außenluft belüftet. Zu Beginn ist die Schadstoffkonzentration $k_0=400\,\rm ppM$. Der Schadstoffvolumenstrom ist $540\,\rm\dfrac{\ell}{h}$ CO_2.
Berechnen Sie, welche Schadstoffkonzentration sich im Gebäude einstellt und stellen Sie den zeitlichenn Verlauf von $k(t)$ für einen 8-Stundentag dar.
#### Lösung
Zunächst wird das Raumvolumen berechnet. Es ergibt sich zu
$$
V_\rm{ra} = 150\cdot 2.5\,\rm{m}^2 = 375\,\rm{m}^2
$$
oder, mittels Jupyter:
```
# Raumvolumen
V_ra = 150*2.5 # m**3
V_ra
```
Im nächsten Schritt wird daraus die CO_2-Konzentration $k_\infty$ ermittelt:
$$
k_\infty = k_\rm{au} + \dfrac{\dot V_\rm{sch}}{\dot V_\rm{ra}}
= 400\,\rm{ppM} + \dfrac{540\cdot 10^{-3}\,\rm{m}^3}{375\,\rm{m}^3}
= 3400\,\rm{ppM}
$$
oder, mittels Jupyter:
```
# Schadstoffvolumenstrom (CO_2):
dV_sch = 540e-3 # 180 l/h
# CO_2-Konzentration der Außenluft
k_0 = 400e-6 # 400 ppM
# bekannter Außenluftvolumenstrom
dV_au = 180 # m**3/h
# Daraus berechneter Endwert der Konzentration
k_inf = k_0 + dV_sch/dV_au
# Kontrollausgabe in ppM
k_inf*1e6 # ppM
```
Die Luftwechselzahl ergibt sich durch
$$
\beta = \dfrac{\dot V_\rm{au}}{V_\rm{ra}} = 0.48\,\rm\dfrac{1}{h}
$$
oder, mittels Jupyter:
```
# Luftwechselzahl beta:
beta = dV_au/V_ra # 1/h
beta
```
Für das Plotten wird hier nur das Jupyter Notebook verwendet:
```
# Plotten vorbereiten
from matplotlib import pyplot as plt
%config InlineBackend.figure_format='retina'
import pandas as pd
import numpy as np
# Das Zeitintervall auf der x-Achse:
lt = np.linspace(0,4,17) # 0..4h
# Der Dataframe
df = pd.DataFrame(
{
't': lt,
'k': 1e6*(k_inf + (k_0-k_inf)*np.exp(-beta*lt)) # k in ppM
}
)
# Kontrollausdruck
df.head().T
# Zeitliche Entwicklung der CO_2-Konzentragion im Diagramm
ax = df.plot(x='t',y='k',label="$k = k(t)$")
ax.axhline(1e6*k_inf,c='r')
ax.grid()
ax.set(
ylim=(0,4000), ylabel=r'CO_2-Konzentration $k$ in $\rm ppM$',
xlim=(0,4), xlabel=r'Zeit $t$ in Stunden'
);
```
Man erkennt, dass bereits nach etwa 30 Minuten die Grenze von 1000 ppM erreicht wird. Nach etwas über 90 Minuten ist die Raumluftqualität nicht mehr akzeptabel.
## Die Schadstoffbilanz des Raumes nach einem Zeitschritt
Herleitung der Lösung.
Im nächsten Schritt wird untersucht, wie sich $k$ verändert, nachdem der Raum eine bestimmte Zeit $\Delta t$ belüftet worden ist.
- Von den Personen (oder sonstigen Schadstoffquellen) im Raum wird eine bestimmte Schadstoffmenge abgegeben,
nämlich
$$
\dot V{\rm sch}\cdot\Delta t
$$
- Mit der Außenluft wird eine bestimmte Schadstoffmenge in den Raum hineingetragen, nämlich
$$
k_{\rm au}\cdot\dot V_{\rm au}\cdot\Delta t
$$
- Mit der Abluft wird eine bestimmte Schadstoffmenge aus dem Raum herausgetragen, nämlich
$$
k_0\cdot\dot V_{\rm ab}\cdot\Delta t
$$
Damit ergibt sich:
$$
k_1 \cdot V_{\rm ra} =
k_0 \cdot V_{\rm ra}
+ \dot V{\rm sch}\cdot\Delta t
+ k_{\rm au}\cdot\dot V_{\rm au}\cdot\Delta t
- k_0\cdot\dot V_{\rm ab}\cdot\Delta t
$$
Oder, wenn durch das Raumvolumen dividiert wird:
$$
k_1 =
k_0
+ \dfrac{\dot V{\rm sch}}{V_{\rm ra}}\cdot\Delta t
+ k_{\rm au}\cdot\dfrac{\dot V_{\rm au}}{V_{\rm ra}}\cdot\Delta t
- k_0\cdot\dfrac{\dot V_{\rm ab}}{V_{\rm ra}}\cdot\Delta t
$$
Nun ist $\dfrac{\dot V_{\rm au}}{V_{\rm ra}} = \dfrac{\dot V_{\rm ab}}{V_{\rm ra}} = \beta$ gerade die Luftwechselzahl des Raumes. Deshalb ist
$$
k_1 =
k_0
+ \dfrac{\dot V{\rm sch}}{V_{\rm ra}}\cdot\Delta t
+ k_{\rm au}\cdot\beta\cdot\Delta t
- k_0\cdot\beta\cdot\Delta t
$$
oder, zusammengefasst:
$$
k_1 =
k_0\cdot\left(1 -\beta\cdot\Delta t \right)
+ \left(\dfrac{\dot V{\rm sch}}{V_{\rm ra}}
+ k_{\rm au}\cdot\beta\right)\cdot\Delta t
$$
Genau so, wie $k_1$ aus $k_0$ berechnet wurde, lässt sich $k_2$ aus $k_1$ berechnen, u.s.w. Das führt der Reihe nach auf
\begin{align}
k_1 &=
k_0\cdot\left(1 -\beta\cdot\Delta t \right)
+ \left(\dfrac{\dot V{\rm sch}}{V_{\rm ra}}
+ k_{\rm au}\cdot\beta\right)\cdot\Delta t
\\[2ex]
k_2 &=
k_1\cdot\left(1 -\beta\cdot\Delta t \right)
+ \left(\dfrac{\dot V{\rm sch}}{V_{\rm ra}}
+ k_{\rm au}\cdot\beta\right)\cdot\Delta t
\\[2ex]
k_3 &=
k_2\cdot\left(1 -\beta\cdot\Delta t \right)
+ \left(\dfrac{\dot V{\rm sch}}{V_{\rm ra}}
+ k_{\rm au}\cdot\beta\right)\cdot\Delta t
\\[2ex]
k_4 &=
k_3\cdot\left(1 -\beta\cdot\Delta t \right)
+ \left(\dfrac{\dot V{\rm sch}}{V_{\rm ra}}
+ k_{\rm au}\cdot\beta\right)\cdot\Delta t
\\[2ex]
&\,\,\vdots
\\
&=
\\
&\,\,\vdots
\\[2ex]
k_{n} &=
k_{n-1}\cdot\left(1 -\beta\cdot\Delta t \right)
+ \left(\dfrac{\dot V{\rm sch}}{V_{\rm ra}}
+ k_{\rm au}\cdot\beta\right)\cdot\Delta t
\end{align}
Da der Unterschied zwischen $k_{n}$ und $k_{n-1}$ immer kleiner wird, gilt schließlich
\begin{align}
k_{\infty} &=
k_{\infty}\cdot\left(1 -\beta\cdot\Delta t \right)
+ \left(\dfrac{\dot V{\rm sch}}{V_{\rm ra}}
+ k_{\rm au}\cdot\beta\right)\cdot\Delta t
\end{align}
wobei der Wert $k_\infty$ nach (theoretisch) unendlich langer Zeit erreicht wird.
Bildet man nun die Differenzen $k_1 - k_\infty$, $k_2-k_\infty$, $\ldots$ $k_{n+1} - k_\infty$, so ergeben sich die einfacheren Formeln
\begin{align}
k_1 - k_\infty &= (k_0 - k_\infty)\cdot\left(1 -\beta\cdot\Delta t \right) \\
k_2 - k_\infty &= (k_1 - k_\infty)\cdot\left(1 -\beta\cdot\Delta t \right)
= (k_0 - k_\infty)\cdot\left(1 -\beta\cdot\Delta t \right)^2\\
k_3 - k_\infty &= (k_2 - k_\infty)\cdot\left(1 -\beta\cdot\Delta t \right)
= (k_0 - k_\infty)\cdot\left(1 -\beta\cdot\Delta t \right)^3\\
k_4 - k_\infty &= (k_3 - k_\infty)\cdot\left(1 -\beta\cdot\Delta t \right)
= (k_0 - k_\infty)\cdot\left(1 -\beta\cdot\Delta t \right)^4\\
&\,\,\vdots\\
&= \\
&\,\,\vdots\\
k_n - k_\infty &= (k_{n-1} - k_\infty)\cdot\left(1 -\beta\cdot\Delta t \right)
= (k_0 - k_\infty)\cdot\left(1 -\beta\cdot\Delta t \right)^n\\
\end{align}
Lässt man nun das Zeitintervall $\Delta t$ immer kleiner werden, so werden immer mehr Zeitschritte benötigt, um den Zeitpunkt $t = n\cdot\Delta t$ zu erreichen. Es ergibt sich
$$
t = n\cdot\Delta t \implies \Delta t = \dfrac{t}{n}
$$
und, mit der Eulerschen Formel für die Funktion ${\rm e}^x$, nach der
$$
\lim_{n\to\infty}
\left(
1 + \dfrac{x}{n}
\right)^n
= {\rm e}^x
$$
ist:
$$
(1-\beta\cdot\Delta t)^n = \left(1-\dfrac{\beta\,t}{n}\right)^n \to {\mathrm e}^{-\beta\,t}
\quad\text{für}\quad n\to\infty
$$
Wegen $n\cdot\Delta t = t$ kann man $k_n$ durch $k(t)$ ersetzen. Damit ergibt sich schließlich die Formel
$$
k(t) - k_\infty = (k_0 - k_\infty)\cdot{\mathrm e}^{-\beta\,t}
$$
Dies Ergebnis schreibt man auch in der Form
$$
k(t) = k_\infty + (k_0 - k_\infty)\cdot{\mathrm e}^{-\beta\,t}
$$
|
github_jupyter
|
# Raumvolumen
V_ra = 150*2.5 # m**3
V_ra
# Schadstoffvolumenstrom (CO_2):
dV_sch = 540e-3 # 180 l/h
# CO_2-Konzentration der Außenluft
k_0 = 400e-6 # 400 ppM
# bekannter Außenluftvolumenstrom
dV_au = 180 # m**3/h
# Daraus berechneter Endwert der Konzentration
k_inf = k_0 + dV_sch/dV_au
# Kontrollausgabe in ppM
k_inf*1e6 # ppM
# Luftwechselzahl beta:
beta = dV_au/V_ra # 1/h
beta
# Plotten vorbereiten
from matplotlib import pyplot as plt
%config InlineBackend.figure_format='retina'
import pandas as pd
import numpy as np
# Das Zeitintervall auf der x-Achse:
lt = np.linspace(0,4,17) # 0..4h
# Der Dataframe
df = pd.DataFrame(
{
't': lt,
'k': 1e6*(k_inf + (k_0-k_inf)*np.exp(-beta*lt)) # k in ppM
}
)
# Kontrollausdruck
df.head().T
# Zeitliche Entwicklung der CO_2-Konzentragion im Diagramm
ax = df.plot(x='t',y='k',label="$k = k(t)$")
ax.axhline(1e6*k_inf,c='r')
ax.grid()
ax.set(
ylim=(0,4000), ylabel=r'CO_2-Konzentration $k$ in $\rm ppM$',
xlim=(0,4), xlabel=r'Zeit $t$ in Stunden'
);
| 0.219254 | 0.839668 |
```
# Importing Modules
import pandas as pd
from glob import glob
import numpy as np
from PIL import Image
import torch
data_file = glob('Dataset\\*')[0]
data_file
# Class Names
# 0=Angry, 1=Disgust, 2=Fear, 3=Happy, 4=Sad, 5=Surprise, 6=Neutral.
# Reading the data
df = pd.read_csv(data_file)
df.head()
mystring = df['pixels'].tolist()[0]
mylist = mystring.split(' ')
myarray = np.array([mylist], dtype = np.uint)
reshapedrray = myarray.reshape((48,48))
new_image = Image.fromarray(reshapedrray)
new_image.show()
def convert_str_pixels_to_array(str):
return np.array(mystring.split(' '), dtype = np.uint)
adf = df.head()
adf['img_arr'] = adf.apply(lambda x : convert_str_pixels_to_array(x['pixels']), axis = 1)
adf['img_arr'].dtypes
from torch.utils.data import Dataset
import numpy as np
# Borrowed from a medium article
class FER2013Dataset(Dataset):
"""Face Expression Recognition Dataset"""
def __init__(self, file_path):
"""
Args:
file_path (string): Path to the csv file with emotion, pixel & usage.
"""
self.file_path = file_path
self.classes = ('angry', 'disgust', 'fear', 'happy', 'sad', 'surprise', 'neutral') # Define the name of classes / expression
with open(self.file_path) as f: # load how many row / image the data contains
self.total_images = len(f.readlines()) - 1 #reduce 1 for row of column
def __len__(self): # to return total images when call `len(dataset)`
return self.total_images
def __getitem__(self, idx): # to return image and emotion when call `dataset[idx]`
if torch.is_tensor(idx):
idx = idx.tolist()
with open(fer_path) as f: # read all the csv using readlines
emotion, img, usage = f.readlines()[idx + 1].split(",") #plus 1 to skip first row (column name)
emotion = int(emotion) # just make sure it is int not str
img = img.split(" ") # because the pixels are seperated by space
img = np.array(img, 'int') # just make sure it is int not str
img = img.reshape(48,48) # change shape from 2304 to 48 * 48
sample = {'image': img, 'emotion': emotion}
return sample
train_file = 'Dataset/Training.csv'
val_file = 'Dataset/PrivateTest.csv'
test_file = 'Dataset/PublicTest.csv'
train_dataset = FER2013Dataset(file_path = train_file)
val_dataset = FER2013Dataset(file_path = val_file)
test_dataset = FER2013Dataset(file_path = test_file)
train_loader = torch.utils.data.DataLoader(train_dataset, batch_size = 32, shuffle = True)
val_loader = torch.utils.data.DataLoader(val_dataset, batch_size = 32, shuffle = True)
test_loader = torch.utils.data.DataLoader(test_dataset, batch_size = 32, shuffle = True)
train_loader.batch_size
df["Usage"].value_counts()
```
|
github_jupyter
|
# Importing Modules
import pandas as pd
from glob import glob
import numpy as np
from PIL import Image
import torch
data_file = glob('Dataset\\*')[0]
data_file
# Class Names
# 0=Angry, 1=Disgust, 2=Fear, 3=Happy, 4=Sad, 5=Surprise, 6=Neutral.
# Reading the data
df = pd.read_csv(data_file)
df.head()
mystring = df['pixels'].tolist()[0]
mylist = mystring.split(' ')
myarray = np.array([mylist], dtype = np.uint)
reshapedrray = myarray.reshape((48,48))
new_image = Image.fromarray(reshapedrray)
new_image.show()
def convert_str_pixels_to_array(str):
return np.array(mystring.split(' '), dtype = np.uint)
adf = df.head()
adf['img_arr'] = adf.apply(lambda x : convert_str_pixels_to_array(x['pixels']), axis = 1)
adf['img_arr'].dtypes
from torch.utils.data import Dataset
import numpy as np
# Borrowed from a medium article
class FER2013Dataset(Dataset):
"""Face Expression Recognition Dataset"""
def __init__(self, file_path):
"""
Args:
file_path (string): Path to the csv file with emotion, pixel & usage.
"""
self.file_path = file_path
self.classes = ('angry', 'disgust', 'fear', 'happy', 'sad', 'surprise', 'neutral') # Define the name of classes / expression
with open(self.file_path) as f: # load how many row / image the data contains
self.total_images = len(f.readlines()) - 1 #reduce 1 for row of column
def __len__(self): # to return total images when call `len(dataset)`
return self.total_images
def __getitem__(self, idx): # to return image and emotion when call `dataset[idx]`
if torch.is_tensor(idx):
idx = idx.tolist()
with open(fer_path) as f: # read all the csv using readlines
emotion, img, usage = f.readlines()[idx + 1].split(",") #plus 1 to skip first row (column name)
emotion = int(emotion) # just make sure it is int not str
img = img.split(" ") # because the pixels are seperated by space
img = np.array(img, 'int') # just make sure it is int not str
img = img.reshape(48,48) # change shape from 2304 to 48 * 48
sample = {'image': img, 'emotion': emotion}
return sample
train_file = 'Dataset/Training.csv'
val_file = 'Dataset/PrivateTest.csv'
test_file = 'Dataset/PublicTest.csv'
train_dataset = FER2013Dataset(file_path = train_file)
val_dataset = FER2013Dataset(file_path = val_file)
test_dataset = FER2013Dataset(file_path = test_file)
train_loader = torch.utils.data.DataLoader(train_dataset, batch_size = 32, shuffle = True)
val_loader = torch.utils.data.DataLoader(val_dataset, batch_size = 32, shuffle = True)
test_loader = torch.utils.data.DataLoader(test_dataset, batch_size = 32, shuffle = True)
train_loader.batch_size
df["Usage"].value_counts()
| 0.67662 | 0.432603 |
```
# reload packages
%load_ext autoreload
%autoreload 2
```
### Choose GPU
```
%env CUDA_DEVICE_ORDER=PCI_BUS_ID
%env CUDA_VISIBLE_DEVICES=1
import tensorflow as tf
gpu_devices = tf.config.experimental.list_physical_devices('GPU')
if len(gpu_devices)>0:
tf.config.experimental.set_memory_growth(gpu_devices[0], True)
print(gpu_devices)
tf.keras.backend.clear_session()
```
### dataset information
```
from datetime import datetime
dataset = "fmnist"
dims = (28, 28, 1)
num_classes = 10
labels_per_class = 64 # full
batch_size = 128
datestring = datetime.now().strftime("%Y_%m_%d_%H_%M_%S_%f")
datestring = (
str(dataset)
+ "_"
+ str(labels_per_class)
+ "____"
+ datestring
+ '_baseline_augmented'
)
print(datestring)
```
### Load packages
```
import tensorflow as tf
import numpy as np
import matplotlib.pyplot as plt
from tqdm.autonotebook import tqdm
from IPython import display
import pandas as pd
import umap
import copy
import os, tempfile
```
### Load dataset
```
from tfumap.load_datasets import load_FMNIST, mask_labels
X_train, X_test, X_valid, Y_train, Y_test, Y_valid = load_FMNIST(flatten=False)
X_train.shape
if labels_per_class == "full":
X_labeled = X_train
Y_masked = Y_labeled = Y_train
else:
X_labeled, Y_labeled, Y_masked = mask_labels(
X_train, Y_train, labels_per_class=labels_per_class
)
```
### Build network
```
from tensorflow.keras import datasets, layers, models
from tensorflow_addons.layers import WeightNormalization
def conv_block(filts, name, kernel_size = (3, 3), padding = "same", **kwargs):
return WeightNormalization(
layers.Conv2D(
filts, kernel_size, activation=None, padding=padding, **kwargs
),
name="conv"+name,
)
#CNN13
#See:
#https://github.com/vikasverma1077/ICT/blob/master/networks/lenet.py
#https://github.com/brain-research/realistic-ssl-evaluation
lr_alpha = 0.1
dropout_rate = 0.5
num_classes = 10
input_shape = dims
model = models.Sequential()
model.add(tf.keras.Input(shape=input_shape))
### conv1a
name = '1a'
model.add(conv_block(name = name, filts = 128, kernel_size = (3,3), padding="same"))
model.add(layers.BatchNormalization(name="bn"+name))
model.add(layers.LeakyReLU(alpha=lr_alpha, name = 'lrelu'+name))
### conv1b
name = '1b'
model.add(conv_block(name = name, filts = 128, kernel_size = (3,3), padding="same"))
model.add(layers.BatchNormalization(name="bn"+name))
model.add(layers.LeakyReLU(alpha=lr_alpha, name = 'lrelu'+name))
### conv1c
name = '1c'
model.add(conv_block(name = name, filts = 128, kernel_size = (3,3), padding="same"))
model.add(layers.BatchNormalization(name="bn"+name))
model.add(layers.LeakyReLU(alpha=lr_alpha, name = 'lrelu'+name))
# max pooling
model.add(layers.MaxPooling2D(pool_size=(2, 2), strides=2, padding='valid', name="mp1"))
# dropout
model.add(layers.Dropout(dropout_rate, name="drop1"))
### conv2a
name = '2a'
model.add(conv_block(name = name, filts = 256, kernel_size = (3,3), padding="same"))
model.add(layers.BatchNormalization(name="bn"+name))
model.add(layers.LeakyReLU(alpha=lr_alpha))
### conv2b
name = '2b'
model.add(conv_block(name = name, filts = 256, kernel_size = (3,3), padding="same"))
model.add(layers.BatchNormalization(name="bn"+name))
model.add(layers.LeakyReLU(alpha=lr_alpha, name = 'lrelu'+name))
### conv2c
name = '2c'
model.add(conv_block(name = name, filts = 256, kernel_size = (3,3), padding="same"))
model.add(layers.BatchNormalization(name="bn"+name))
model.add(layers.LeakyReLU(alpha=lr_alpha, name = 'lrelu'+name))
# max pooling
model.add(layers.MaxPooling2D(pool_size=(2, 2), strides=2, padding='valid', name="mp2"))
# dropout
model.add(layers.Dropout(dropout_rate, name="drop2"))
### conv3a
name = '3a'
model.add(conv_block(name = name, filts = 512, kernel_size = (3,3), padding="valid"))
model.add(layers.BatchNormalization(name="bn"+name))
model.add(layers.LeakyReLU(alpha=lr_alpha, name = 'lrelu'+name))
### conv3b
name = '3b'
model.add(conv_block(name = name, filts = 256, kernel_size = (1,1), padding="valid"))
model.add(layers.BatchNormalization(name="bn"+name))
model.add(layers.LeakyReLU(alpha=lr_alpha, name = 'lrelu'+name))
### conv3c
name = '3c'
model.add(conv_block(name = name, filts = 128, kernel_size = (1,1), padding="valid"))
model.add(layers.BatchNormalization(name="bn"+name))
model.add(layers.LeakyReLU(alpha=lr_alpha, name = 'lrelu'+name))
# max pooling
model.add(layers.AveragePooling2D(pool_size=(3, 3), strides=2, padding='valid'))
model.add(layers.Flatten())
model.add(layers.Dense(256, activation=None, name='z'))
model.add(WeightNormalization(layers.Dense(256, activation=None)))
model.add(layers.LeakyReLU(alpha=lr_alpha, name = 'lrelufc1'))
model.add(WeightNormalization(layers.Dense(256, activation=None)))
model.add(layers.LeakyReLU(alpha=lr_alpha, name = 'lrelufc2'))
model.add(WeightNormalization(layers.Dense(num_classes, activation=None)))
model.summary()
```
### Augmentation
```
import tensorflow_addons as tfa
def norm(x):
return( x - tf.reduce_min(x))#/(tf.reduce_max(x) - tf.reduce_min(x))
def augment(image, label):
if tf.random.uniform((1,), minval=0, maxval = 2, dtype=tf.int32)[0] == 0:
# stretch
randint_hor = tf.random.uniform((2,), minval=0, maxval = 8, dtype=tf.int32)[0]
randint_vert = tf.random.uniform((2,), minval=0, maxval = 8, dtype=tf.int32)[0]
image = tf.image.resize(image, (dims[0]+randint_vert*2, dims[1]+randint_hor*2))
#image = tf.image.crop_to_bounding_box(image, randint_vert,randint_hor,28,28)
image = tf.image.resize_with_pad(
image, dims[0], dims[1]
)
image = tf.image.resize_with_crop_or_pad(
image, dims[0] + 3, dims[1] + 3
) # crop 6 pixels
image = tf.image.random_crop(image, size=dims)
if tf.random.uniform((1,), minval=0, maxval = 2, dtype=tf.int32)[0] == 0:
image = tfa.image.rotate(
image,
tf.squeeze(tf.random.uniform(shape=(1, 1), minval=-0.25, maxval=0.25)),
interpolation="BILINEAR",
)
image = tf.image.random_flip_left_right(image)
image = tf.clip_by_value(image, 0, 1)
if tf.random.uniform((1,), minval=0, maxval = 2, dtype=tf.int32)[0] == 0:
image = tf.image.random_brightness(image, max_delta=0.5) # Random brightness
image = tf.image.random_contrast(image, lower=0.5, upper=1.75)
image = norm(image)
image = tf.clip_by_value(image, 0, 1)
if tf.random.uniform((1,), minval=0, maxval = 2, dtype=tf.int32)[0] == 0:
image = tfa.image.random_cutout(
tf.expand_dims(image, 0), (8, 8), constant_values=0.5
)[0]
image = tf.clip_by_value(image, 0, 1)
return image, label
nex = 10
for i in range(5):
fig, axs = plt.subplots(ncols=nex +1, figsize=((nex+1)*2, 2))
axs[0].imshow(np.squeeze(X_train[i]), cmap = plt.cm.Greys)
axs[0].axis('off')
for ax in axs.flatten()[1:]:
aug_img = np.squeeze(augment(X_train[i], Y_train[i])[0])
ax.matshow(aug_img, cmap = plt.cm.Greys, vmin=0, vmax=1)
ax.axis('off')
```
### train
```
early_stopping = tf.keras.callbacks.EarlyStopping(
monitor='val_accuracy', min_delta=0, patience=100, verbose=1, mode='auto',
baseline=None, restore_best_weights=True
)
import tensorflow_addons as tfa
opt = tf.keras.optimizers.Adam(1e-4)
opt = tfa.optimizers.MovingAverage(opt)
loss = tf.keras.losses.CategoricalCrossentropy(label_smoothing=0.2, from_logits=True)
model.compile(opt, loss = loss, metrics=['accuracy'])
Y_valid_one_hot = tf.keras.backend.one_hot(
Y_valid, num_classes
)
Y_labeled_one_hot = tf.keras.backend.one_hot(
Y_labeled, num_classes
)
from livelossplot import PlotLossesKerasTF
# plot losses callback
plotlosses = PlotLossesKerasTF()
train_ds = (
tf.data.Dataset.from_tensor_slices((X_labeled, Y_labeled_one_hot))
.repeat()
.shuffle(len(X_labeled))
.map(augment, num_parallel_calls=tf.data.experimental.AUTOTUNE)
.batch(batch_size)
.prefetch(tf.data.experimental.AUTOTUNE)
)
steps_per_epoch = int(len(X_train)/ batch_size)
history = model.fit(
train_ds,
epochs=500,
validation_data=(X_valid, Y_valid_one_hot),
callbacks = [early_stopping, plotlosses],
steps_per_epoch = steps_per_epoch,
)
plt.plot(history.history['loss'])
plt.plot(history.history['val_loss'])
plt.plot(history.history['accuracy'])
plt.plot(history.history['val_accuracy'])
submodel = tf.keras.models.Model(
[model.inputs[0]], [model.get_layer('z').output]
)
z = submodel.predict(X_train)
np.shape(z)
reducer = umap.UMAP(verbose=True)
embedding = reducer.fit_transform(z.reshape(len(z), np.product(np.shape(z)[1:])))
plt.scatter(embedding[:, 0], embedding[:, 1], c=Y_train.flatten(), s= 1, alpha = 0.1, cmap = plt.cm.tab10)
z_valid = submodel.predict(X_valid)
np.shape(z_valid)
reducer = umap.UMAP(verbose=True)
embedding = reducer.fit_transform(z_valid.reshape(len(z_valid), np.product(np.shape(z_valid)[1:])))
plt.scatter(embedding[:, 0], embedding[:, 1], c=Y_valid.flatten(), s= 1, alpha = 0.1, cmap = plt.cm.tab10)
fig, ax = plt.subplots(figsize=(10,10))
ax.scatter(embedding[:, 0], embedding[:, 1], c=Y_valid.flatten(), s= 1, alpha = 1, cmap = plt.cm.tab10)
predictions = model.predict(X_valid)
fig, ax = plt.subplots(figsize=(10,10))
ax.scatter(embedding[:, 0], embedding[:, 1], c=np.argmax(predictions, axis=1), s= 1, alpha = 1, cmap = plt.cm.tab10)
Y_test_one_hot = tf.keras.backend.one_hot(
Y_test, num_classes
)
result = model.evaluate(X_test, Y_test_one_hot)
```
### save results
```
# save score, valid embedding, weights, results
from tfumap.paths import MODEL_DIR, ensure_dir
save_folder = MODEL_DIR / 'semisupervised-keras' / dataset / str(labels_per_class) / datestring
ensure_dir(save_folder)
```
#### save weights
```
encoder = tf.keras.models.Model(
[model.inputs[0]], [model.get_layer('z').output]
)
encoder.save_weights((save_folder / "encoder").as_posix())
classifier = tf.keras.models.Model(
[tf.keras.Input(tensor=model.get_layer('weight_normalization').input)], [model.outputs[0]]
)
print([i.name for i in classifier.layers])
classifier.save_weights((save_folder / "classifier").as_posix())
```
#### save score
```
Y_test_one_hot = tf.keras.backend.one_hot(
Y_test, num_classes
)
result = model.evaluate(X_test, Y_test_one_hot)
np.save(save_folder / 'test_loss.npy', result)
```
#### save embedding
```
z = encoder.predict(X_train)
reducer = umap.UMAP(verbose=True)
embedding = reducer.fit_transform(z.reshape(len(z), np.product(np.shape(z)[1:])))
plt.scatter(embedding[:, 0], embedding[:, 1], c=Y_train.flatten(), s= 1, alpha = 0.1, cmap = plt.cm.tab10)
np.save(save_folder / 'train_embedding.npy', embedding)
```
#### save results
```
import pickle
with open(save_folder / 'history.pickle', 'wb') as file_pi:
pickle.dump(history.history, file_pi)
```
|
github_jupyter
|
# reload packages
%load_ext autoreload
%autoreload 2
%env CUDA_DEVICE_ORDER=PCI_BUS_ID
%env CUDA_VISIBLE_DEVICES=1
import tensorflow as tf
gpu_devices = tf.config.experimental.list_physical_devices('GPU')
if len(gpu_devices)>0:
tf.config.experimental.set_memory_growth(gpu_devices[0], True)
print(gpu_devices)
tf.keras.backend.clear_session()
from datetime import datetime
dataset = "fmnist"
dims = (28, 28, 1)
num_classes = 10
labels_per_class = 64 # full
batch_size = 128
datestring = datetime.now().strftime("%Y_%m_%d_%H_%M_%S_%f")
datestring = (
str(dataset)
+ "_"
+ str(labels_per_class)
+ "____"
+ datestring
+ '_baseline_augmented'
)
print(datestring)
import tensorflow as tf
import numpy as np
import matplotlib.pyplot as plt
from tqdm.autonotebook import tqdm
from IPython import display
import pandas as pd
import umap
import copy
import os, tempfile
from tfumap.load_datasets import load_FMNIST, mask_labels
X_train, X_test, X_valid, Y_train, Y_test, Y_valid = load_FMNIST(flatten=False)
X_train.shape
if labels_per_class == "full":
X_labeled = X_train
Y_masked = Y_labeled = Y_train
else:
X_labeled, Y_labeled, Y_masked = mask_labels(
X_train, Y_train, labels_per_class=labels_per_class
)
from tensorflow.keras import datasets, layers, models
from tensorflow_addons.layers import WeightNormalization
def conv_block(filts, name, kernel_size = (3, 3), padding = "same", **kwargs):
return WeightNormalization(
layers.Conv2D(
filts, kernel_size, activation=None, padding=padding, **kwargs
),
name="conv"+name,
)
#CNN13
#See:
#https://github.com/vikasverma1077/ICT/blob/master/networks/lenet.py
#https://github.com/brain-research/realistic-ssl-evaluation
lr_alpha = 0.1
dropout_rate = 0.5
num_classes = 10
input_shape = dims
model = models.Sequential()
model.add(tf.keras.Input(shape=input_shape))
### conv1a
name = '1a'
model.add(conv_block(name = name, filts = 128, kernel_size = (3,3), padding="same"))
model.add(layers.BatchNormalization(name="bn"+name))
model.add(layers.LeakyReLU(alpha=lr_alpha, name = 'lrelu'+name))
### conv1b
name = '1b'
model.add(conv_block(name = name, filts = 128, kernel_size = (3,3), padding="same"))
model.add(layers.BatchNormalization(name="bn"+name))
model.add(layers.LeakyReLU(alpha=lr_alpha, name = 'lrelu'+name))
### conv1c
name = '1c'
model.add(conv_block(name = name, filts = 128, kernel_size = (3,3), padding="same"))
model.add(layers.BatchNormalization(name="bn"+name))
model.add(layers.LeakyReLU(alpha=lr_alpha, name = 'lrelu'+name))
# max pooling
model.add(layers.MaxPooling2D(pool_size=(2, 2), strides=2, padding='valid', name="mp1"))
# dropout
model.add(layers.Dropout(dropout_rate, name="drop1"))
### conv2a
name = '2a'
model.add(conv_block(name = name, filts = 256, kernel_size = (3,3), padding="same"))
model.add(layers.BatchNormalization(name="bn"+name))
model.add(layers.LeakyReLU(alpha=lr_alpha))
### conv2b
name = '2b'
model.add(conv_block(name = name, filts = 256, kernel_size = (3,3), padding="same"))
model.add(layers.BatchNormalization(name="bn"+name))
model.add(layers.LeakyReLU(alpha=lr_alpha, name = 'lrelu'+name))
### conv2c
name = '2c'
model.add(conv_block(name = name, filts = 256, kernel_size = (3,3), padding="same"))
model.add(layers.BatchNormalization(name="bn"+name))
model.add(layers.LeakyReLU(alpha=lr_alpha, name = 'lrelu'+name))
# max pooling
model.add(layers.MaxPooling2D(pool_size=(2, 2), strides=2, padding='valid', name="mp2"))
# dropout
model.add(layers.Dropout(dropout_rate, name="drop2"))
### conv3a
name = '3a'
model.add(conv_block(name = name, filts = 512, kernel_size = (3,3), padding="valid"))
model.add(layers.BatchNormalization(name="bn"+name))
model.add(layers.LeakyReLU(alpha=lr_alpha, name = 'lrelu'+name))
### conv3b
name = '3b'
model.add(conv_block(name = name, filts = 256, kernel_size = (1,1), padding="valid"))
model.add(layers.BatchNormalization(name="bn"+name))
model.add(layers.LeakyReLU(alpha=lr_alpha, name = 'lrelu'+name))
### conv3c
name = '3c'
model.add(conv_block(name = name, filts = 128, kernel_size = (1,1), padding="valid"))
model.add(layers.BatchNormalization(name="bn"+name))
model.add(layers.LeakyReLU(alpha=lr_alpha, name = 'lrelu'+name))
# max pooling
model.add(layers.AveragePooling2D(pool_size=(3, 3), strides=2, padding='valid'))
model.add(layers.Flatten())
model.add(layers.Dense(256, activation=None, name='z'))
model.add(WeightNormalization(layers.Dense(256, activation=None)))
model.add(layers.LeakyReLU(alpha=lr_alpha, name = 'lrelufc1'))
model.add(WeightNormalization(layers.Dense(256, activation=None)))
model.add(layers.LeakyReLU(alpha=lr_alpha, name = 'lrelufc2'))
model.add(WeightNormalization(layers.Dense(num_classes, activation=None)))
model.summary()
import tensorflow_addons as tfa
def norm(x):
return( x - tf.reduce_min(x))#/(tf.reduce_max(x) - tf.reduce_min(x))
def augment(image, label):
if tf.random.uniform((1,), minval=0, maxval = 2, dtype=tf.int32)[0] == 0:
# stretch
randint_hor = tf.random.uniform((2,), minval=0, maxval = 8, dtype=tf.int32)[0]
randint_vert = tf.random.uniform((2,), minval=0, maxval = 8, dtype=tf.int32)[0]
image = tf.image.resize(image, (dims[0]+randint_vert*2, dims[1]+randint_hor*2))
#image = tf.image.crop_to_bounding_box(image, randint_vert,randint_hor,28,28)
image = tf.image.resize_with_pad(
image, dims[0], dims[1]
)
image = tf.image.resize_with_crop_or_pad(
image, dims[0] + 3, dims[1] + 3
) # crop 6 pixels
image = tf.image.random_crop(image, size=dims)
if tf.random.uniform((1,), minval=0, maxval = 2, dtype=tf.int32)[0] == 0:
image = tfa.image.rotate(
image,
tf.squeeze(tf.random.uniform(shape=(1, 1), minval=-0.25, maxval=0.25)),
interpolation="BILINEAR",
)
image = tf.image.random_flip_left_right(image)
image = tf.clip_by_value(image, 0, 1)
if tf.random.uniform((1,), minval=0, maxval = 2, dtype=tf.int32)[0] == 0:
image = tf.image.random_brightness(image, max_delta=0.5) # Random brightness
image = tf.image.random_contrast(image, lower=0.5, upper=1.75)
image = norm(image)
image = tf.clip_by_value(image, 0, 1)
if tf.random.uniform((1,), minval=0, maxval = 2, dtype=tf.int32)[0] == 0:
image = tfa.image.random_cutout(
tf.expand_dims(image, 0), (8, 8), constant_values=0.5
)[0]
image = tf.clip_by_value(image, 0, 1)
return image, label
nex = 10
for i in range(5):
fig, axs = plt.subplots(ncols=nex +1, figsize=((nex+1)*2, 2))
axs[0].imshow(np.squeeze(X_train[i]), cmap = plt.cm.Greys)
axs[0].axis('off')
for ax in axs.flatten()[1:]:
aug_img = np.squeeze(augment(X_train[i], Y_train[i])[0])
ax.matshow(aug_img, cmap = plt.cm.Greys, vmin=0, vmax=1)
ax.axis('off')
early_stopping = tf.keras.callbacks.EarlyStopping(
monitor='val_accuracy', min_delta=0, patience=100, verbose=1, mode='auto',
baseline=None, restore_best_weights=True
)
import tensorflow_addons as tfa
opt = tf.keras.optimizers.Adam(1e-4)
opt = tfa.optimizers.MovingAverage(opt)
loss = tf.keras.losses.CategoricalCrossentropy(label_smoothing=0.2, from_logits=True)
model.compile(opt, loss = loss, metrics=['accuracy'])
Y_valid_one_hot = tf.keras.backend.one_hot(
Y_valid, num_classes
)
Y_labeled_one_hot = tf.keras.backend.one_hot(
Y_labeled, num_classes
)
from livelossplot import PlotLossesKerasTF
# plot losses callback
plotlosses = PlotLossesKerasTF()
train_ds = (
tf.data.Dataset.from_tensor_slices((X_labeled, Y_labeled_one_hot))
.repeat()
.shuffle(len(X_labeled))
.map(augment, num_parallel_calls=tf.data.experimental.AUTOTUNE)
.batch(batch_size)
.prefetch(tf.data.experimental.AUTOTUNE)
)
steps_per_epoch = int(len(X_train)/ batch_size)
history = model.fit(
train_ds,
epochs=500,
validation_data=(X_valid, Y_valid_one_hot),
callbacks = [early_stopping, plotlosses],
steps_per_epoch = steps_per_epoch,
)
plt.plot(history.history['loss'])
plt.plot(history.history['val_loss'])
plt.plot(history.history['accuracy'])
plt.plot(history.history['val_accuracy'])
submodel = tf.keras.models.Model(
[model.inputs[0]], [model.get_layer('z').output]
)
z = submodel.predict(X_train)
np.shape(z)
reducer = umap.UMAP(verbose=True)
embedding = reducer.fit_transform(z.reshape(len(z), np.product(np.shape(z)[1:])))
plt.scatter(embedding[:, 0], embedding[:, 1], c=Y_train.flatten(), s= 1, alpha = 0.1, cmap = plt.cm.tab10)
z_valid = submodel.predict(X_valid)
np.shape(z_valid)
reducer = umap.UMAP(verbose=True)
embedding = reducer.fit_transform(z_valid.reshape(len(z_valid), np.product(np.shape(z_valid)[1:])))
plt.scatter(embedding[:, 0], embedding[:, 1], c=Y_valid.flatten(), s= 1, alpha = 0.1, cmap = plt.cm.tab10)
fig, ax = plt.subplots(figsize=(10,10))
ax.scatter(embedding[:, 0], embedding[:, 1], c=Y_valid.flatten(), s= 1, alpha = 1, cmap = plt.cm.tab10)
predictions = model.predict(X_valid)
fig, ax = plt.subplots(figsize=(10,10))
ax.scatter(embedding[:, 0], embedding[:, 1], c=np.argmax(predictions, axis=1), s= 1, alpha = 1, cmap = plt.cm.tab10)
Y_test_one_hot = tf.keras.backend.one_hot(
Y_test, num_classes
)
result = model.evaluate(X_test, Y_test_one_hot)
# save score, valid embedding, weights, results
from tfumap.paths import MODEL_DIR, ensure_dir
save_folder = MODEL_DIR / 'semisupervised-keras' / dataset / str(labels_per_class) / datestring
ensure_dir(save_folder)
encoder = tf.keras.models.Model(
[model.inputs[0]], [model.get_layer('z').output]
)
encoder.save_weights((save_folder / "encoder").as_posix())
classifier = tf.keras.models.Model(
[tf.keras.Input(tensor=model.get_layer('weight_normalization').input)], [model.outputs[0]]
)
print([i.name for i in classifier.layers])
classifier.save_weights((save_folder / "classifier").as_posix())
Y_test_one_hot = tf.keras.backend.one_hot(
Y_test, num_classes
)
result = model.evaluate(X_test, Y_test_one_hot)
np.save(save_folder / 'test_loss.npy', result)
z = encoder.predict(X_train)
reducer = umap.UMAP(verbose=True)
embedding = reducer.fit_transform(z.reshape(len(z), np.product(np.shape(z)[1:])))
plt.scatter(embedding[:, 0], embedding[:, 1], c=Y_train.flatten(), s= 1, alpha = 0.1, cmap = plt.cm.tab10)
np.save(save_folder / 'train_embedding.npy', embedding)
import pickle
with open(save_folder / 'history.pickle', 'wb') as file_pi:
pickle.dump(history.history, file_pi)
| 0.636692 | 0.742282 |

# Qiskit Aer: Pulse simulation of a backend model
This notebook shows how to use the Aer pulse simulator using a model generated from a backend. In particular, we run a Rabi experiment on the Armonk one qubit backend, then simulate the same experiment on the pulse simulator, calibrating the model parameters to reproduce the results from the real backend.
## Table of contents
1) [Imports](#imports)
2) [Rabi oscillations on `ibmq_armonk`](#rabi)
3) [Reproducing Rabi oscillations on the simulator](#simulator)
## 1. Imports <a name='imports'></a>
Import general libraries:
```
import numpy as np
```
Import `IBMQ`, Rabi experiment generator and fitter from Ignis, and other functions for job submission:
```
from qiskit import IBMQ
from qiskit.ignis.characterization.calibrations import rabi_schedules, RabiFitter
from qiskit.pulse import DriveChannel
from qiskit.compiler import assemble
from qiskit.qobj.utils import MeasLevel, MeasReturnType
```
Import `PulseSimulator` and `PulseSystemModel` for pulse simulation:
```
# The pulse simulator
from qiskit.providers.aer import PulseSimulator
# object for representing physical models
from qiskit.providers.aer.pulse import PulseSystemModel
```
# 2. Rabi oscillations on `ibmq_armonk` backend <a name='rabi'></a>
First, we run a Rabi experiment on the `ibmq_armonk` backend using Ignis.
Get the `ibmq_armonk` backend:
```
provider = IBMQ.load_account()
armonk_backend = provider.get_backend('ibmq_armonk')
```
Construct Rabi experiment schedules.
```
# qubit list
qubits = [0]
# drive amplitudes to use
num_exps = 64
drive_amps = np.linspace(0, 1.0, num_exps)
# drive shape parameters
drive_duration = 2048
drive_sigma = 256
# list of drive channels
drive_channels = [DriveChannel(0)]
# construct the schedules
rabi_schedules, xdata = rabi_schedules(amp_list=drive_amps,
qubits=qubits,
pulse_width=drive_duration,
pulse_sigma=drive_sigma,
drives=drive_channels,
inst_map=armonk_backend.defaults().instruction_schedule_map,
meas_map=armonk_backend.configuration().meas_map)
```
Assemble the `qobj` for job submission.
```
rabi_qobj = assemble(rabi_schedules,
backend=armonk_backend,
meas_level=MeasLevel.KERNELED,
meas_return=MeasReturnType.AVERAGE,
shots=512)
```
Run the job on the backend.
```
job = armonk_backend.run(rabi_qobj)
job.status()
rabi_result = job.result(timeout=3600)
```
Fit and plot the results, getting the $\pi$-pulse amplitude.
```
rabi_backend_fit = RabiFitter(rabi_result, xdata, qubits, fit_p0 = [2.0e15, 2, 0, 0])
# get the pi amplitude
pi_amp = rabi_backend_fit.pi_amplitude(0)
# plot
rabi_backend_fit.plot(0)
print('Pi Amp: %f'%pi_amp)
```
# 3. Reproducing the Rabi oscillations with the simulator <a name='simulator'></a>
Next, we run the same experiments on the pulse simulator. This section demonstrates the use of the `PulseSystemModel.from_backend` function for generating `PulseSystemModel` objects from a backend.
**Note:** Currently not all system Hamiltonian information is available to the public, missing values have been replaced with $0$. As a result, in this notebook, we need to insert parameters into the backend object by hand. Specifically, we:
- Set the frequency of the qubit in the backend provided Hamiltonian to correspond with the backend provided estimate.
- Set the value of the drive strength to be consistent with the $\pi$-pulse amplitude found in the previous section. I.e. The drive strength $r$ is set so that: $r \times A = \pi/2$, where $A$ is the area under the $\pi$-pulse found above.
```
# A value to use if previous cells of notebook were not run
# pi_amp = 0.347467
# Infer the value of the drive strength from the pi pulse amplitude:
dt = getattr(armonk_backend.configuration(), 'dt')
from qiskit.pulse import pulse_lib
sample_array = pulse_lib.gaussian(duration=drive_duration, amp=1, sigma=drive_sigma).samples
A = pi_amp*sum(sample_array*dt) # area under curve
omegad0 = np.real(np.pi/(A * 2)) # inferred drive strength
# set drive strength omegad0 in backend object
getattr(armonk_backend.configuration(), 'hamiltonian')['vars']['omegad0'] = omegad0
# set the qubit frequency from the estimate in the defaults
freq_est = getattr(armonk_backend.defaults(), 'qubit_freq_est')[0]
getattr(armonk_backend.configuration(), 'hamiltonian')['vars']['wq0'] = 2*np.pi*freq_est
```
Construct a `PulseSystemModel` object from the backend, and instantiate the simulator.
```
armonk_model = PulseSystemModel.from_backend(armonk_backend)
backend_sim = PulseSimulator()
```
Assemble schedules as before, but now use `PulseSimulator` as the backend.
```
rabi_qobj_sim = assemble(rabi_schedules,
backend=backend_sim,
meas_level=1,
meas_return='avg',
shots=512)
```
Run the simulation and get the results.
```
sim_result = backend_sim.run(rabi_qobj_sim, armonk_model).result()
```
Generate the same plot.
```
rabi_sim_fit = RabiFitter(sim_result, xdata, qubits, fit_p0 = [1.5, 2, 0, 0])
rabi_sim_fit.plot(0)
```
Observe: the simulated results reproduce the oscillations of the device (the amplitude of the oscillation is arbitrary).
```
import qiskit.tools.jupyter
%qiskit_version_table
%qiskit_copyright
```
|
github_jupyter
|
import numpy as np
from qiskit import IBMQ
from qiskit.ignis.characterization.calibrations import rabi_schedules, RabiFitter
from qiskit.pulse import DriveChannel
from qiskit.compiler import assemble
from qiskit.qobj.utils import MeasLevel, MeasReturnType
# The pulse simulator
from qiskit.providers.aer import PulseSimulator
# object for representing physical models
from qiskit.providers.aer.pulse import PulseSystemModel
provider = IBMQ.load_account()
armonk_backend = provider.get_backend('ibmq_armonk')
# qubit list
qubits = [0]
# drive amplitudes to use
num_exps = 64
drive_amps = np.linspace(0, 1.0, num_exps)
# drive shape parameters
drive_duration = 2048
drive_sigma = 256
# list of drive channels
drive_channels = [DriveChannel(0)]
# construct the schedules
rabi_schedules, xdata = rabi_schedules(amp_list=drive_amps,
qubits=qubits,
pulse_width=drive_duration,
pulse_sigma=drive_sigma,
drives=drive_channels,
inst_map=armonk_backend.defaults().instruction_schedule_map,
meas_map=armonk_backend.configuration().meas_map)
rabi_qobj = assemble(rabi_schedules,
backend=armonk_backend,
meas_level=MeasLevel.KERNELED,
meas_return=MeasReturnType.AVERAGE,
shots=512)
job = armonk_backend.run(rabi_qobj)
job.status()
rabi_result = job.result(timeout=3600)
rabi_backend_fit = RabiFitter(rabi_result, xdata, qubits, fit_p0 = [2.0e15, 2, 0, 0])
# get the pi amplitude
pi_amp = rabi_backend_fit.pi_amplitude(0)
# plot
rabi_backend_fit.plot(0)
print('Pi Amp: %f'%pi_amp)
# A value to use if previous cells of notebook were not run
# pi_amp = 0.347467
# Infer the value of the drive strength from the pi pulse amplitude:
dt = getattr(armonk_backend.configuration(), 'dt')
from qiskit.pulse import pulse_lib
sample_array = pulse_lib.gaussian(duration=drive_duration, amp=1, sigma=drive_sigma).samples
A = pi_amp*sum(sample_array*dt) # area under curve
omegad0 = np.real(np.pi/(A * 2)) # inferred drive strength
# set drive strength omegad0 in backend object
getattr(armonk_backend.configuration(), 'hamiltonian')['vars']['omegad0'] = omegad0
# set the qubit frequency from the estimate in the defaults
freq_est = getattr(armonk_backend.defaults(), 'qubit_freq_est')[0]
getattr(armonk_backend.configuration(), 'hamiltonian')['vars']['wq0'] = 2*np.pi*freq_est
armonk_model = PulseSystemModel.from_backend(armonk_backend)
backend_sim = PulseSimulator()
rabi_qobj_sim = assemble(rabi_schedules,
backend=backend_sim,
meas_level=1,
meas_return='avg',
shots=512)
sim_result = backend_sim.run(rabi_qobj_sim, armonk_model).result()
rabi_sim_fit = RabiFitter(sim_result, xdata, qubits, fit_p0 = [1.5, 2, 0, 0])
rabi_sim_fit.plot(0)
import qiskit.tools.jupyter
%qiskit_version_table
%qiskit_copyright
| 0.704262 | 0.955131 |
```
import pandas as pd
import re
import numpy as np
import tensorflow as tf
import numpy as np
import operator
import os
import random
import matplotlib.pyplot as plt
from sklearn.cluster import KMeans, DBSCAN
from sklearn.manifold import TSNE
import pandas as pd
from sklearn.linear_model import LinearRegression
from scipy.spatial.distance import cdist
from tensorflow import keras
from tensorflow.keras import layers
from sklearn.model_selection import train_test_split
pip install --upgrade tensorflow==1.15.0
path = r'C:\Users\Yunseok Choi\Downloads\ESG Data'
os.listdir(path)
data1= pd.read_csv(path + '\RUSSELL 3000 DATA.csv')
data2= pd.read_csv(path + '\RUSSELL 3000 + @.csv')
data1.dropna(subset = ['PART'], inplace = True)
data2.dropna(subset = ['PART'], inplace = True)
rs100 = pd.read_csv(path + '\Industry.csv')
re100 = pd.read_csv(path + '\RE100.csv')
def preprocess(item):
global data1
global data2
global rs100
temp = pd.concat([data1[data1['Unnamed: 0'] == item].copy(),
data2[data2['Unnamed: 0'] == item].copy()])
temp.reset_index(inplace = True, drop = True)
temp = pd.DataFrame(np.insert(temp.values,0,values=[item, 'YEAR', 2016, 2017, 2018, 2019, 2020, 2021, 2022],axis=0))
temp = temp.transpose()
temp.reset_index(inplace = True, drop = True)
temp.drop([0], inplace = True)
temp.columns = list(temp.iloc[0].values)
temp.reset_index(inplace = True, drop = True)
temp.drop([0], inplace = True)
temp['INDUSTRY'] = rs100[rs100['COMPANY'] == item.replace('QUITY','quity')]['GICS Sector'].values[0]
temp['COMPANY'] = item
return temp
rus1000_list = [x.upper() for x in rs100['COMPANY'].values]
df = pd.DataFrame()
for item in set(data1['Unnamed: 0']):
if item.upper() in rus1000_list:
df = pd.concat([df, preprocess(item)])
df.shape
#df.drop(['IS_INT_EXPENSES'], axis = 1, inplace = True)
for column in df.columns:
print(column, ' / ', df.dropna(subset = [column]).shape)
year_list = list(set(df['YEAR'].values))
year_list
column_list = df.columns
column_list
for year in year_list:
print(year, '/', df[(df['TOTAL_GHG_EMISSIONS'].notna()) & (df['YEAR'] == year)].shape)
df.columns
df_reg = pd.DataFrame()
for column in column_list[1:-2]:
na_included = [np.nan if (len(re.findall(r'[0-9.]+', str(item))) != 1) else float(item) for item in df[column].values]
na_excluded = [x for x in na_included if np.isnan(float(x)) != True]
df_reg[column] = [x/np.mean(na_excluded) if (np.isnan(float(x)) != True) else np.nan for x in na_included]
for column in list(column_list[-2:]) + [column_list[0]]:
df_reg[column] = df[column].values
df_reg.reset_index(inplace = True, drop = True)
column_list = df_reg.columns
df_reg
for year in year_list:
print(year, ' ', df_reg[(df_reg['TOTAL_GHG_EMISSIONS'].notna()) & (df_reg['YEAR'] == year)].shape)
```
# t-SNE
```
df_all = df_reg[(df_reg['TOTAL_GHG_EMISSIONS'].notna())].fillna(0)
df_all.reset_index(inplace = True, drop = True)
x_all = df_all[column_list[:-3]].values
#차원 감축을 위한 t-sne 변수 결정. l_rate, iteration, perplexity을 다양하게 바꿔가며 output 관찰
perplexity = [10,20,30,40]
#l_rate = 1000
#iteration = 300
l_rate = 2000
iteration = 8000
data = x_all
plt.figure(figsize=(10,10))
for i in range(len(perplexity)):
plt.subplot(2,2,i+1)
if i == 0:
plt.subplots_adjust(hspace = 0.2, wspace = 0.2)
tsne = TSNE(n_components = 2, learning_rate = l_rate,
perplexity = perplexity[i], n_iter = iteration)
X = tsne.fit_transform(data)
plt.plot([x[0] for x in X], [x[1] for x in X], '.')
plt.title("Perplexity = {}".format(perplexity[i]))
df_all['TSNE'] = list(tsne.fit_transform(x_all))
```
# Clustering using Kmeans
### By Total Data
```
perplexity = 30
l_rate = 2000
iteration = 8000
tsne = TSNE(n_components = 2, learning_rate = l_rate,
perplexity = perplexity, n_iter = iteration)
#전체 데이터셋 클러스터 개수 결정을 위한 elbow method
k_t = 10
data = x_all
X = tsne.fit_transform(data)
df_all['TSNE'] = list(X)
distortions = []
K = range(1,51)
for k in K:
elbow = KMeans(n_clusters = k).fit(X)
distortions.append(sum(np.min(cdist(X, elbow.cluster_centers_, 'euclidean'), axis=1)) / X.shape[0])
plt.plot(K, distortions, 'go-')
plt.xlabel('k')
plt.ylabel('Distortion')
plt.title('The Elbow Method to determine k')
plt.show()
kmeans = KMeans(n_clusters = k_t).fit(X)
df_all['LABELS'] = kmeans.labels_
for i in range(k_t):
print('Cluster {} has {} companies.'.format(i,(kmeans.labels_ == i).sum()))
#전체 데이터셋 클러스터링 결과 시각화
data = df_all
for i in range(len(set(data['LABELS']))):
item = list(set(data['LABELS']))[i]
data[data['LABELS'] == item]['TSNE']
plt.plot([x[0] for x in data[data['LABELS'] == item]['TSNE'].values],[y[1] for y in data[data['LABELS'] == item]['TSNE'].values],
'.', color = 'C{}'.format(i))
plt.title('2016-2021 Results')
plt.show()
#년도별(16/17. 18/19, 20/21) 데이터 분리
df_c1 = df_all[(df_all['YEAR'] == 2016) | (df_all['YEAR'] == 2017)].reset_index(drop= True).copy()
df_c2 = df_all[(df_all['YEAR'] == 2018) | (df_all['YEAR'] == 2019)].reset_index(drop= True).copy()
df_c3 = df_all[(df_all['YEAR'] == 2020) | (df_all['YEAR'] == 2021)].reset_index(drop= True).copy()
#re100 선언 변수추가를 위한 processing
declare = []
for i in range(df_c1.shape[0]):
if df_c1.iloc[i]['COMPANY'].upper() in re100['COMPANY'].values:
if df_c1.iloc[i]['YEAR'] >= re100[re100['COMPANY'] == df_c1.iloc[i]['COMPANY']]['RE100 Declare Year']:
declare.append(1)
else:
declare.append(0)
else:
declare.append(0)
df_c1['RE100'] = declare
declare = []
for i in range(df_c2.shape[0]):
if df_c2.iloc[i]['COMPANY'].upper() in re100['COMPANY'].values:
if df_c2.iloc[i]['YEAR'] >= re100[re100['COMPANY'] == df_c2.iloc[i]['COMPANY']]['RE100 Declare Year']:
declare.append(1)
else:
declare.append(0)
else:
declare.append(0)
df_c2['RE100'] = declare
declare = []
for i in range(df_c3.shape[0]):
if df_c3.iloc[i]['COMPANY'].upper() in re100['COMPANY'].values:
if df_c3.iloc[i]['YEAR'] >= re100[re100['COMPANY'] == df_c3.iloc[i]['COMPANY']]['RE100 Declare Year']:
declare.append(1)
else:
declare.append(0)
else:
declare.append(0)
df_c3['RE100'] = declare
#16/17 클러스터링을 개수 결정을 위한 elbow method
X = np.array([(x[0], x[1]) for x in list(df_c1['TSNE'].values)])
distortions = []
K = range(1,51)
for k in K:
elbow = KMeans(n_clusters = k).fit(X)
distortions.append(sum(np.min(cdist(X, elbow.cluster_centers_, 'euclidean'), axis=1)) / X.shape[0])
plt.plot(K, distortions, 'go-')
plt.xlabel('k')
plt.ylabel('Distortion')
plt.title('The Elbow Method to determine k')
plt.show()
kmeans = KMeans(n_clusters = k_t).fit(X)
df_c1['CLUSTER LABELS'] = kmeans.labels_
for i in range(k_t):
print('Cluster {} has {} companies.'.format(i,(kmeans.labels_ == i).sum()))
#16/17 클러스터링 시각화
data = df_c1
for i in range(len(set(data['CLUSTER LABELS']))):
item = list(set(data['CLUSTER LABELS']))[i]
data[data['CLUSTER LABELS'] == item]['TSNE']
plt.plot([x[0] for x in data[data['CLUSTER LABELS'] == item]['TSNE'].values],[y[1] for y in data[data['CLUSTER LABELS'] == item]['TSNE'].values],
'.', color = 'C{}'.format(i))
plt.title('2016-2017 Results')
plt.show()
#20/21 클러스터링을 개수 결정을 위한 elbow method
X = np.array([(x[0], x[1]) for x in list(df_c2['TSNE'].values)])
distortions = []
K = range(1,51)
for k in K:
elbow = KMeans(n_clusters = k).fit(X)
distortions.append(sum(np.min(cdist(X, elbow.cluster_centers_, 'euclidean'), axis=1)) / X.shape[0])
plt.plot(K, distortions, 'go-')
plt.xlabel('k')
plt.ylabel('Distortion')
plt.title('The Elbow Method to determine k')
plt.show()
kmeans = KMeans(n_clusters = k_t).fit(X)
df_c2['CLUSTER LABELS'] = kmeans.labels_
for i in range(k_t):
print('Cluster {} has {} companies.'.format(i,(kmeans.labels_ == i).sum()))
#18/19 클러스터링 시각화
data = df_c2
for i in range(len(set(data['CLUSTER LABELS']))):
item = list(set(data['CLUSTER LABELS']))[i]
data[data['CLUSTER LABELS'] == item]['TSNE']
plt.plot([x[0] for x in data[data['CLUSTER LABELS'] == item]['TSNE'].values],[y[1] for y in data[data['CLUSTER LABELS'] == item]['TSNE'].values],
'.', color = 'C{}'.format(i))
plt.title('2018-2019 Results')
plt.show()
#20/21 클러스터링을 개수 결정을 위한 elbow method
X = np.array([(x[0], x[1]) for x in list(df_c3['TSNE'].values)])
distortions = []
K = range(1,51)
for k in K:
elbow = KMeans(n_clusters = k).fit(X)
distortions.append(sum(np.min(cdist(X, elbow.cluster_centers_, 'euclidean'), axis=1)) / X.shape[0])
plt.plot(K, distortions, 'go-')
plt.xlabel('k')
plt.ylabel('Distortion')
plt.title('The Elbow Method to determine k')
plt.show()
kmeans = KMeans(n_clusters = k_t).fit(X)
df_c3['CLUSTER LABELS'] = kmeans.labels_
for i in range(k_t):
print('Cluster {} has {} companies.'.format(i,(kmeans.labels_ == i).sum()))
#20/21 클러스터링 시각화
data = df_c3
for i in range(len(set(data['CLUSTER LABELS']))):
item = list(set(data['CLUSTER LABELS']))[i]
data[data['CLUSTER LABELS'] == item]['TSNE']
plt.plot([x[0] for x in data[data['CLUSTER LABELS'] == item]['TSNE'].values],[y[1] for y in data[data['CLUSTER LABELS'] == item]['TSNE'].values],
'.', color = 'C{}'.format(i))
plt.title('2020-2021 Results')
plt.show()
#현재 데이터 저장
df_c1.to_csv(path+('\df_c1.csv'))
df_c2.to_csv(path+('\df_c2.csv'))
df_c3.to_csv(path+('\df_c3.csv'))
df_all.to_csv(path+('\df_all.csv'))
```
2016~2017 data
```
#16/17 수익률 기준 top5 클러스터 각 클러스터별 회귀분석.
data = df_c1.copy()
company_list = []
for item in data['COMPANY'].values:
if item not in company_list:
company_list.append(item)
else:
pass
#company_list
avg_mkvalt = {}
n_mkvalt = {}
for company in company_list:
if data[data['COMPANY']==company].shape[0] == 2:
k = data[data['COMPANY']==company]['CLUSTER LABELS'].values[-1]
mkvalt0 = data[(data['COMPANY']==company) & (data['YEAR']==2016)]['TOT_MKT_VAL'].values[0]
mkvalt1 = data[(data['COMPANY']==company) & (data['YEAR']==2017)]['TOT_MKT_VAL'].values[0]
if mkvalt0 != 0:
if k not in avg_mkvalt.keys():
avg_mkvalt[k] = ((mkvalt1-mkvalt0)/mkvalt0)
n_mkvalt[k] = 1
else:
avg_mkvalt[k] += ((mkvalt1-mkvalt0)/mkvalt0)
n_mkvalt[k] += 1
rank = {}
for key in sorted(avg_mkvalt.keys()):
avg = (avg_mkvalt[key]/n_mkvalt[key])
rank[key] = avg
#print('Cluster {} has average mkvalt % change of {}'.format(key,avg))
print(sorted(rank.items(), key=operator.itemgetter(1), reverse = True))
top5 = [x[0] for x in sorted(rank.items(), key=operator.itemgetter(1), reverse = True)[:5]]
industry_list = list(set(data['INDUSTRY'].values))
for industry in industry_list:
data[str(industry).upper()] = list(((data['INDUSTRY'] == industry)*1).values)
data['CONSTANT'] = 1
columns = data.columns
iv = list(columns[1:15])+list(columns[16:20])+[columns[26]] + list(columns[-(len(industry_list)):])
dv = columns[15]
df_data = []
for k in top5:
temp = []
X = data[(data['CLUSTER LABELS']==k)&(data['YEAR']==2016)][iv].values
Y = data[(data['CLUSTER LABELS']==k)&(data['YEAR']==2016)][dv].values
lr = LinearRegression()
lr.fit(X, Y)
temp = list(round(x*100,2) for x in lr.coef_)
temp.append(round(lr.score(X,Y),2))
df_data.append(temp)
pd.DataFrame(np.array(df_data).transpose(), index = iv+['R^2'], columns = top5)
#연도별 '상' 그룸 회귀분석을 위한 변수선언
high_x = []
high_y = []
#16/17 클러스터 수익률 별 3:4:3 비율로 상 중 하 나눈 뒤 리그레션
data = df_c1.copy()
company_list = []
for item in data['COMPANY'].values:
if item not in company_list:
company_list.append(item)
else:
pass
#company_list
avg_mkvalt = {}
n_mkvalt = {}
for company in company_list:
if data[data['COMPANY']==company].shape[0] == 2:
k = data[data['COMPANY']==company]['CLUSTER LABELS'].values[-1]
mkvalt0 = data[(data['COMPANY']==company) & (data['YEAR']==2016)]['TOT_MKT_VAL'].values[0]
mkvalt1 = data[(data['COMPANY']==company) & (data['YEAR']==2017)]['TOT_MKT_VAL'].values[0]
if mkvalt0 != 0:
if k not in avg_mkvalt.keys():
avg_mkvalt[k] = ((mkvalt1-mkvalt0)/mkvalt0)
n_mkvalt[k] = 1
else:
avg_mkvalt[k] += ((mkvalt1-mkvalt0)/mkvalt0)
n_mkvalt[k] += 1
rank = {}
for key in sorted(avg_mkvalt.keys()):
avg = (avg_mkvalt[key]/n_mkvalt[key])
rank[key] = avg
#print('Cluster {} has average mkvalt % change of {}'.format(key,avg))
#print(sorted(rank.items(), key=operator.itemgetter(1), reverse = True))
top = [x[0] for x in sorted(rank.items(), key=operator.itemgetter(1), reverse = True)[0:3]]
mid = [x[0] for x in sorted(rank.items(), key=operator.itemgetter(1), reverse = True)[4:8]]
low = [x[0] for x in sorted(rank.items(), key=operator.itemgetter(1), reverse = True)[8:10]]
industry_list = list(set(data['INDUSTRY'].values)) + ['INFORMATION TECHNOLOGY']
for industry in industry_list:
data[str(industry).upper()] = list(((data['INDUSTRY'] == industry)*1).values)
data['CONSTANT'] = 1
columns = data.columns
iv = list(columns[1:15])+list(columns[16:20])+[columns[26]] + list(columns[-(len(industry_list)):])
dv = columns[15]
df_data = []
for item in [top, mid, low]:
X = []
Y = []
for k in item:
temp = []
X += [list(x) for x in data[(data['CLUSTER LABELS']==k)&(data['YEAR']==2016)][iv].values]
Y += list(data[(data['CLUSTER LABELS']==k)&(data['YEAR']==2016)][dv].values)
if item == top:
high_x += [list(x) for x in data[(data['CLUSTER LABELS']==k)&(data['YEAR']==2016)][iv].values]
high_y += list(data[(data['CLUSTER LABELS']==k)&(data['YEAR']==2016)][dv].values)
lr = LinearRegression()
lr.fit(X, Y)
temp = list(round(x*100,2) for x in lr.coef_)
temp.append(round(lr.score(X,Y),2))
df_data.append(temp)
pd.DataFrame(np.array(df_data).transpose(), index = iv+['R^2'], columns = ['Top','Mid','Low'])
```
2018~2019 data
```
#18/19 수익률 기준 top5 클러스터 각 클러스터별 회귀분석.
data = df_c2.copy()
company_list = []
for item in data['COMPANY'].values:
if item not in company_list:
company_list.append(item)
else:
pass
#company_list
avg_mkvalt = {}
n_mkvalt = {}
for company in company_list:
if data[data['COMPANY']==company].shape[0] == 2:
k = data[data['COMPANY']==company]['CLUSTER LABELS'].values[-1]
mkvalt0 = data[(data['COMPANY']==company) & (data['YEAR']==2018)]['TOT_MKT_VAL'].values[0]
mkvalt1 = data[(data['COMPANY']==company) & (data['YEAR']==2019)]['TOT_MKT_VAL'].values[0]
if mkvalt0 != 0:
if k not in avg_mkvalt.keys():
avg_mkvalt[k] = ((mkvalt1-mkvalt0)/mkvalt0)
n_mkvalt[k] = 1
else:
avg_mkvalt[k] += ((mkvalt1-mkvalt0)/mkvalt0)
n_mkvalt[k] += 1
rank = {}
for key in sorted(avg_mkvalt.keys()):
avg = (avg_mkvalt[key]/n_mkvalt[key])
rank[key] = avg
#print('Cluster {} has average mkvalt % change of {}'.format(key,avg))
print(sorted(rank.items(), key=operator.itemgetter(1), reverse = True))
top5 = [x[0] for x in sorted(rank.items(), key=operator.itemgetter(1), reverse = True)[:5]]
industry_list = list(set(data['INDUSTRY'].values))
for industry in industry_list:
data[str(industry).upper()] = list(((data['INDUSTRY'] == industry)*1).values)
data['CONSTANT'] = 1
columns = data.columns
iv = list(columns[1:15])+list(columns[16:20])+[columns[26]] + list(columns[-(len(industry_list)):])
dv = columns[15]
df_data = []
for k in top5:
temp = []
X = data[(data['CLUSTER LABELS']==k)&(data['YEAR']==2018)][iv].values
Y = data[(data['CLUSTER LABELS']==k)&(data['YEAR']==2018)][dv].values
lr = LinearRegression()
lr.fit(X, Y)
temp = list(round(x*100,2) for x in lr.coef_)
temp.append(round(lr.score(X,Y),2))
df_data.append(temp)
pd.DataFrame(np.array(df_data).transpose(), index = iv+['R^2'], columns = top5)
#18/19 클러스터 수익률 별 3:4:3 비율로 상 중 하 나눈 뒤 리그레션
data = df_c2.copy()
company_list = []
for item in data['COMPANY'].values:
if item not in company_list:
company_list.append(item)
else:
pass
#company_list
avg_mkvalt = {}
n_mkvalt = {}
for company in company_list:
if data[data['COMPANY']==company].shape[0] == 2:
k = data[data['COMPANY']==company]['CLUSTER LABELS'].values[-1]
mkvalt0 = data[(data['COMPANY']==company) & (data['YEAR']==2018)]['TOT_MKT_VAL'].values[0]
mkvalt1 = data[(data['COMPANY']==company) & (data['YEAR']==2019)]['TOT_MKT_VAL'].values[0]
if mkvalt0 != 0:
if k not in avg_mkvalt.keys():
avg_mkvalt[k] = ((mkvalt1-mkvalt0)/mkvalt0)
n_mkvalt[k] = 1
else:
avg_mkvalt[k] += ((mkvalt1-mkvalt0)/mkvalt0)
n_mkvalt[k] += 1
rank = {}
for key in sorted(avg_mkvalt.keys()):
avg = (avg_mkvalt[key]/n_mkvalt[key])
rank[key] = avg
#print('Cluster {} has average mkvalt % change of {}'.format(key,avg))
#print(sorted(rank.items(), key=operator.itemgetter(1), reverse = True))
top = [x[0] for x in sorted(rank.items(), key=operator.itemgetter(1), reverse = True)[0:3]]
mid = [x[0] for x in sorted(rank.items(), key=operator.itemgetter(1), reverse = True)[4:8]]
low = [x[0] for x in sorted(rank.items(), key=operator.itemgetter(1), reverse = True)[8:10]]
industry_list = list(set(data['INDUSTRY'].values))
for industry in industry_list:
data[str(industry).upper()] = list(((data['INDUSTRY'] == industry)*1).values)
data['CONSTANT'] = 1
columns = data.columns
iv = list(columns[1:15])+list(columns[16:20])+[columns[26]] + list(columns[-(len(industry_list)):])
dv = columns[15]
df_data = []
for item in [top, mid, low]:
X = []
Y = []
for k in item:
temp = []
X += [list(x) for x in data[(data['CLUSTER LABELS']==k)&(data['YEAR']==2018)][iv].values]
Y += list(data[(data['CLUSTER LABELS']==k)&(data['YEAR']==2018)][dv].values)
if item == top:
high_x += [list(x) for x in data[(data['CLUSTER LABELS']==k)&(data['YEAR']==2018)][iv].values]
high_y += list(data[(data['CLUSTER LABELS']==k)&(data['YEAR']==2018)][dv].values)
lr = LinearRegression()
lr.fit(X, Y)
temp = list(round(x*100,2) for x in lr.coef_)
temp.append(round(lr.score(X,Y),2))
df_data.append(temp)
pd.DataFrame(np.array(df_data).transpose(), index = iv+['R^2'], columns = ['Top','Mid','Low'])
```
2020~2021 data
```
#20/21 수익률 기준 top5 클러스터 각 클러스터별 회귀분석.
data = df_c3.copy()
company_list = []
for item in data['COMPANY'].values:
if item not in company_list:
company_list.append(item)
else:
pass
#company_list
avg_mkvalt = {}
n_mkvalt = {}
for company in company_list:
if data[data['COMPANY']==company].shape[0] == 2:
k = data[data['COMPANY']==company]['CLUSTER LABELS'].values[-1]
mkvalt0 = data[(data['COMPANY']==company) & (data['YEAR']==2020)]['TOT_MKT_VAL'].values[0]
mkvalt1 = data[(data['COMPANY']==company) & (data['YEAR']==2021)]['TOT_MKT_VAL'].values[0]
if mkvalt0 != 0:
if k not in avg_mkvalt.keys():
avg_mkvalt[k] = ((mkvalt1-mkvalt0)/mkvalt0)
n_mkvalt[k] = 1
else:
avg_mkvalt[k] += ((mkvalt1-mkvalt0)/mkvalt0)
n_mkvalt[k] += 1
rank = {}
for key in sorted(avg_mkvalt.keys()):
avg = (avg_mkvalt[key]/n_mkvalt[key])
rank[key] = avg
#print('Cluster {} has average mkvalt % change of {}'.format(key,avg))
print(sorted(rank.items(), key=operator.itemgetter(1), reverse = True))
top5 = [x[0] for x in sorted(rank.items(), key=operator.itemgetter(1), reverse = True)[:5]]
industry_list = list(set(data['INDUSTRY'].values))
for industry in industry_list:
data[str(industry).upper()] = list(((data['INDUSTRY'] == industry)*1).values)
data['CONSTANT'] = 1
columns = data.columns
iv = list(columns[1:15])+list(columns[16:20])+[columns[26]] + list(columns[-(len(industry_list)):])
dv = columns[15]
df_data = []
for k in top5:
temp = []
X = data[(data['CLUSTER LABELS']==k)&(data['YEAR']==2020)][iv].values
Y = data[(data['CLUSTER LABELS']==k)&(data['YEAR']==2020)][dv].values
lr = LinearRegression()
lr.fit(X, Y)
temp = list(round(x*100,2) for x in lr.coef_)
temp.append(round(lr.score(X,Y),2))
df_data.append(temp)
pd.DataFrame(np.array(df_data).transpose(), index = iv+['R^2'], columns = top5)
#20/21 클러스터 수익률 별 3:4:3 비율로 상 중 하 나눈 뒤 리그레션
data = df_c3.copy()
company_list = []
for item in data['COMPANY'].values:
if item not in company_list:
company_list.append(item)
else:
pass
#company_list
avg_mkvalt = {}
n_mkvalt = {}
for company in company_list:
if data[data['COMPANY']==company].shape[0] == 2:
k = data[data['COMPANY']==company]['CLUSTER LABELS'].values[-1]
mkvalt0 = data[(data['COMPANY']==company) & (data['YEAR']==2020)]['TOT_MKT_VAL'].values[0]
mkvalt1 = data[(data['COMPANY']==company) & (data['YEAR']==2021)]['TOT_MKT_VAL'].values[0]
if mkvalt0 != 0:
if k not in avg_mkvalt.keys():
avg_mkvalt[k] = ((mkvalt1-mkvalt0)/mkvalt0)
n_mkvalt[k] = 1
else:
avg_mkvalt[k] += ((mkvalt1-mkvalt0)/mkvalt0)
n_mkvalt[k] += 1
rank = {}
for key in sorted(avg_mkvalt.keys()):
avg = (avg_mkvalt[key]/n_mkvalt[key])
rank[key] = avg
#print('Cluster {} has average mkvalt % change of {}'.format(key,avg))
#print(sorted(rank.items(), key=operator.itemgetter(1), reverse = True))
top = [x[0] for x in sorted(rank.items(), key=operator.itemgetter(1), reverse = True)[0:3]]
mid = [x[0] for x in sorted(rank.items(), key=operator.itemgetter(1), reverse = True)[4:8]]
low = [x[0] for x in sorted(rank.items(), key=operator.itemgetter(1), reverse = True)[8:10]]
industry_list = list(set(data['INDUSTRY'].values))
for industry in industry_list:
data[str(industry).upper()] = list(((data['INDUSTRY'] == industry)*1).values)
data['CONSTANT'] = 1
columns = data.columns
iv = list(columns[1:15])+list(columns[16:20])+[columns[26]] + list(columns[-(len(industry_list)):])
dv = columns[15]
df_data = []
for item in [top, mid, low]:
X = []
Y = []
for k in item:
temp = []
X += [list(x) for x in data[(data['CLUSTER LABELS']==k)&(data['YEAR']==2020)][iv].values]
Y += list(data[(data['CLUSTER LABELS']==k)&(data['YEAR']==2020)][dv].values)
if item == top:
high_x += [list(x) for x in data[(data['CLUSTER LABELS']==k)&(data['YEAR']==2020)][iv].values]
high_y += list(data[(data['CLUSTER LABELS']==k)&(data['YEAR']==2020)][dv].values)
lr = LinearRegression()
lr.fit(X, Y)
temp = list(round(x*100,2) for x in lr.coef_)
temp.append(round(lr.score(X,Y),2))
df_data.append(temp)
pd.DataFrame(np.array(df_data).transpose(), index = iv+['R^2'], columns = ['Top','Mid','Low'])
# 16/17,18/19,20/21 수익률 기준 '상' 그룹 모아서 리그레션
temp = []
lr = LinearRegression()
lr.fit(high_x, high_y)
temp = list(round(x*100,2) for x in lr.coef_)
temp.append(round(lr.score(X,Y),2))
pd.DataFrame(temp,index = iv+['R^2'], columns = ['16-21 Top Clusters'])
```
Top MKVALT increase companies
```
data = df_c1
year0 = 2016
year1 = 2017
mkvalt = {}
for company in company_list:
if data[data['COMPANY']==company].shape[0] == 2:
mkvalt0 = data[(data['COMPANY']==company) & (data['YEAR']==2016)]['TOT_MKT_VAL'].values[0]
mkvalt1 = data[(data['COMPANY']==company) & (data['YEAR']==2017)]['TOT_MKT_VAL'].values[0]
if mkvalt0 != 0:
mkvalt[company] = ((mkvalt1-mkvalt0)/mkvalt0)
top10 = [x[0] for x in sorted(mkvalt.items(),key=operator.itemgetter(1),reverse = True)[:int(round(len(mkvalt.keys())/10,0))]]
industry_list = list(set(data['INDUSTRY'].values))
for industry in industry_list:
data[str(industry).upper()] = list(((data['INDUSTRY'] == industry)*1).values)
data['CONSTANT'] = 1
columns = data.columns
iv = list(columns[1:15])+list(columns[16:20])+[columns[26]] + list(columns[-(len(industry_list)):])
dv = columns[15]
df_data = []
X = []
Y = []
for company in top10:
temp = []
X+=[list(x) for x in (data[(data['COMPANY']==company) & (data['YEAR'] == year0)][iv].values)]
Y+=list(data[(data['COMPANY']==company) & (data['YEAR'] == year0)][dv].values)
lr = LinearRegression()
lr.fit(X, Y)
temp = list(round(x*100,2) for x in lr.coef_)
temp.append(round(lr.score(X,Y),2))
df_data.append(temp)
pd.DataFrame(np.array(df_data).transpose(), index = iv+['R^2'], columns = ['TOP10%'])
data = df_c2
year0 = 2018
year1 = 2019
mkvalt = {}
for company in company_list:
if data[data['COMPANY']==company].shape[0] == 2:
mkvalt0 = data[(data['COMPANY']==company) & (data['YEAR']==2018)]['TOT_MKT_VAL'].values[0]
mkvalt1 = data[(data['COMPANY']==company) & (data['YEAR']==2019)]['TOT_MKT_VAL'].values[0]
if mkvalt0 != 0:
mkvalt[company] = ((mkvalt1-mkvalt0)/mkvalt0)
top10 = [x[0] for x in sorted(mkvalt.items(),key=operator.itemgetter(1),reverse = True)[:int(round(len(mkvalt.keys())/10,0))]]
industry_list = list(set(data['INDUSTRY'].values))
for industry in industry_list:
data[str(industry).upper()] = list(((data['INDUSTRY'] == industry)*1).values)
data['CONSTANT'] = 1
columns = data.columns
iv = list(columns[1:15])+list(columns[16:20])+[columns[26]] + list(columns[-(len(industry_list)):])
dv = columns[15]
df_data = []
X = []
Y = []
for company in top10:
temp = []
X+=[list(x) for x in (data[(data['COMPANY']==company) & (data['YEAR'] == year0)][iv].values)]
Y+=list(data[(data['COMPANY']==company) & (data['YEAR'] == year0)][dv].values)
lr = LinearRegression()
lr.fit(X, Y)
temp = list(round(x*100,2) for x in lr.coef_)
temp.append(round(lr.score(X,Y),2))
df_data.append(temp)
pd.DataFrame(np.array(df_data).transpose(), index = iv+['R^2'], columns = ['TOP10%'])
data = df_c3
year0=2020
year1=2021
mkvalt = {}
for company in company_list:
if data[data['COMPANY']==company].shape[0] == 2:
mkvalt0 = data[(data['COMPANY']==company) & (data['YEAR']==year0)]['TOT_MKT_VAL'].values[0]
mkvalt1 = data[(data['COMPANY']==company) & (data['YEAR']==year1)]['TOT_MKT_VAL'].values[0]
if mkvalt0 != 0:
mkvalt[company] = ((mkvalt1-mkvalt0)/mkvalt0)
top10 = [x[0] for x in sorted(mkvalt.items(),key=operator.itemgetter(1),reverse = True)[:int(round(len(mkvalt.keys())/10,0))]]
industry_list = list(set(data['INDUSTRY'].values))
for industry in industry_list:
data[str(industry).upper()] = list(((data['INDUSTRY'] == industry)*1).values)
data['CONSTANT'] = 1
columns = data.columns
iv = list(columns[1:15])+list(columns[16:20])+[columns[26]] + list(columns[-(len(industry_list)):])
dv = columns[15]
df_data = []
X = []
Y = []
for company in top10:
temp = []
X+=[list(x) for x in (data[(data['COMPANY']==company) & (data['YEAR'] == year0)][iv].values)]
Y+=list(data[(data['COMPANY']==company) & (data['YEAR'] == year0)][dv].values)
lr = LinearRegression()
lr.fit(X, Y)
temp = list(round(x*100,2) for x in lr.coef_)
temp.append(round(lr.score(X,Y),2))
df_data.append(temp)
pd.DataFrame(np.array(df_data).transpose(), index = iv+['R^2'], columns = ['TOP10%'])
data = df_all
mkvalt = {}
year_list = [2016, 2017, 2018, 2019, 2020, 2021]
for company in company_list:
for i in range(len(year_list)-2):
year0 = year_list[i]
year1 = year_list[i+1]
if (data[(data['COMPANY']==company) & (data['YEAR']==year0)]['TOT_MKT_VAL'].shape[0] != 0) & (data[(data['COMPANY']==company) & (data['YEAR']==year1)]['TOT_MKT_VAL'].shape[0] != 0):
mkvalt0 = data[(data['COMPANY']==company) & (data['YEAR']==year0)]['TOT_MKT_VAL'].values[0]
mkvalt1 = data[(data['COMPANY']==company) & (data['YEAR']==year1)]['TOT_MKT_VAL'].values[0]
if mkvalt0 != 0:
mkvalt['{}/{}'.format(company,year0)] = ((mkvalt1-mkvalt0)/mkvalt0)
top10 = [x[0] for x in sorted(mkvalt.items(),key=operator.itemgetter(1),reverse = True)[:int(round(len(mkvalt.keys())/10,0))]]
industry_list = list(set(data['INDUSTRY'].values))
for industry in industry_list:
data[str(industry).upper()] = list(((data['INDUSTRY'] == industry)*1).values)
data['CONSTANT'] = 1
columns = data.columns
#iv = list(columns[0:15])+list(columns[16:20])+[columns[26]] + list(columns[-(len(industry_list)):])
iv = list(columns[1:15])+list(columns[16:20])+ list(columns[-(len(industry_list)):])
dv = columns[15]
df_data = []
X = []
Y = []
for i in range(len(top10)):
company, year = top10[i].split('/')[0],top10[i].split('/')[1]
temp = []
X+=[list(x) for x in (data[(data['COMPANY']==company) & (data['YEAR'] == float(year))][iv].values)]
Y+=list((data[(data['COMPANY']==company) & (data['YEAR'] == float(year))][dv].values))
lr = LinearRegression()
lr.fit(X, Y)
temp = list(round(x*100,2) for x in lr.coef_)
temp.append(round(lr.score(X,Y),2))
df_data.append(temp)
pd.DataFrame(np.array(df_data).transpose(), index = iv+['R^2'], columns = ['TOP10%'])
data = df_all
mkvalt = {}
year_list = [2016, 2017, 2018, 2019]
for company in company_list:
for i in range(len(year_list)-2):
year0 = year_list[i]
year1 = year_list[i+1]
if (data[(data['COMPANY']==company) & (data['YEAR']==year0)]['TOT_MKT_VAL'].shape[0] != 0) & (data[(data['COMPANY']==company) & (data['YEAR']==year1)]['TOT_MKT_VAL'].shape[0] != 0):
mkvalt0 = data[(data['COMPANY']==company) & (data['YEAR']==year0)]['TOT_MKT_VAL'].values[0]
mkvalt1 = data[(data['COMPANY']==company) & (data['YEAR']==year1)]['TOT_MKT_VAL'].values[0]
if mkvalt0 != 0:
mkvalt['{}/{}'.format(company,year0)] = ((mkvalt1-mkvalt0)/mkvalt0)
top10 = [x[0] for x in sorted(mkvalt.items(),key=operator.itemgetter(1),reverse = True)[:int(round(len(mkvalt.keys())/10,0))]]
industry_list = list(set(data['INDUSTRY'].values))
for industry in industry_list:
data[str(industry).upper()] = list(((data['INDUSTRY'] == industry)*1).values)
data['CONSTANT'] = 1
columns = data.columns
iv = list(columns[1:15])+list(columns[16:20])+[columns[26]] + list(columns[-(len(industry_list)):])
dv = columns[15]
df_data = []
X = []
Y = []
for i in range(len(top10)):
company, year = top10[i].split('/')[0],top10[i].split('/')[1]
temp = []
X+=[list(x) for x in (data[(data['COMPANY']==company) & (data['YEAR'] == float(year))][iv].values)]
Y+=list((data[(data['COMPANY']==company) & (data['YEAR'] == float(year))][dv].values))
lr = LinearRegression()
lr.fit(X, Y)
temp = list(round(x*100,2) for x in lr.coef_)
temp.append(round(lr.score(X,Y),2))
df_data.append(temp)
pd.DataFrame(np.array(df_data).transpose(), index = iv+['R^2'], columns = ['TOP10%'])
data = df_all
mkvalt = {}
year_list = [2018, 2019, 2020, 2021]
for company in company_list:
for i in range(len(year_list)-2):
year0 = year_list[i]
year1 = year_list[i+1]
if (data[(data['COMPANY']==company) & (data['YEAR']==year0)]['TOT_MKT_VAL'].shape[0] != 0) & (data[(data['COMPANY']==company) & (data['YEAR']==year1)]['TOT_MKT_VAL'].shape[0] != 0):
mkvalt0 = data[(data['COMPANY']==company) & (data['YEAR']==year0)]['TOT_MKT_VAL'].values[0]
mkvalt1 = data[(data['COMPANY']==company) & (data['YEAR']==year1)]['TOT_MKT_VAL'].values[0]
if mkvalt0 != 0:
mkvalt['{}/{}'.format(company,year0)] = ((mkvalt1-mkvalt0)/mkvalt0)
top10 = [x[0] for x in sorted(mkvalt.items(),key=operator.itemgetter(1),reverse = True)[:int(round(len(mkvalt.keys())/10,0))]]
industry_list = list(set(data['INDUSTRY'].values))
for industry in industry_list:
data[str(industry).upper()] = list(((data['INDUSTRY'] == industry)*1).values)
data['CONSTANT'] = 1
columns = data.columns
iv = list(columns[1:15])+list(columns[16:20])+[columns[26]] + list(columns[-(len(industry_list)):])
dv = columns[15]
df_data = []
X = []
Y = []
for i in range(len(top10)):
company, year = top10[i].split('/')[0],top10[i].split('/')[1]
temp = []
X+=[list(x) for x in (data[(data['COMPANY']==company) & (data['YEAR'] == float(year))][iv].values)]
Y+=list((data[(data['COMPANY']==company) & (data['YEAR'] == float(year))][dv].values))
lr = LinearRegression()
lr.fit(X, Y)
temp = list(round(x*100,2) for x in lr.coef_)
temp.append(round(lr.score(X,Y),2))
df_data.append(temp)
pd.DataFrame(np.array(df_data).transpose(), index = iv+['R^2'], columns = ['TOP10%'])
from tensorflow import keras
from tensorflow.keras import layers
from tensorflow.keras.models import Model
df_reg.shape
```
# 1. 재무데이터 및 esg데이터 분리하여 예측
# 2. 바이너리 말고, 수익구간 예측
```
df_all
#딥러닝을 위한 X, XX, Y 생성
data = df_all
mkvalt = {}
year_list = [2016, 2017, 2018, 2019, 2020, 2021]
industry_list = list(set(data['INDUSTRY'].values))
for industry in industry_list:
data[str(industry).upper()] = list(((data['INDUSTRY'] == industry)*1).values)
columns = data.columns
iv = list(columns[1:15])+list(columns[16:20])+ ['LABELS']
ivv = ['ROBECOSAM_ECON_DIMENSION_RANK','TOTAL_GHG_EMISSIONS'] + list(columns[3:15])+list(columns[16:20])
ivvv = iv[6:]+ ['LABELS']
X, XX, XXX = [], [], []
Y = []
for company in company_list:
for i in range(len(year_list)-2):
year0 = year_list[i]
year1 = year_list[i+1]
if data[(data['COMPANY']==company) & ((data['YEAR']==year0) | (data['YEAR']==year1))]['TOT_MKT_VAL'].shape[0] ==2:
mkvalt0 = data[(data['COMPANY']==company) & (data['YEAR']==year0)]['TOT_MKT_VAL'].values[0]
mkvalt1 = data[(data['COMPANY']==company) & (data['YEAR']==year1)]['TOT_MKT_VAL'].values[0]
if mkvalt0 != 0:
Y.append(float((mkvalt1-mkvalt0)/mkvalt0))
X.append(list(data[(data['COMPANY']==company)&(data['YEAR']==float(year0))][iv].values[0]))
XX.append(list(data[(data['COMPANY']==company)&(data['YEAR']==float(year0))][ivv].values[0]))
XXX.append(list(data[(data['COMPANY']==company)&(data['YEAR']==float(year0))][ivvv].values[0]))
```
수익률이 양수인 것을 1, 음수인것을 0으로 만들어 딥러닝으로 학습 및 예측
```
#Y를 바이너리로 제작, train/vaidation/test셋 구성
Y_bin = [1 if x > 0 else 0 for x in Y]
i = 169
x_train, x_tmp, y_train, y_tmp = train_test_split(X,Y_bin, test_size=0.2, shuffle=True, random_state=i)
x_val, x_test, y_val, y_test = train_test_split(x_tmp,y_tmp, test_size=0.5, shuffle=True, random_state=i)
# epochs, batch_size, learning rate를 정하기 위한 testing
keras.backend.clear_session()
inputs = Input(shape=(19,))
hidden1 = Dense(100, activation='swish')(inputs)
hidden2 = Dense(30, activation='swish')(hidden1)
hidden2 = Dense(30, activation='relu')(hidden1)
outputs = Dense(1, activation='sigmoid')(hidden2)
model1 = Model(inputs, outputs)
adam = keras.optimizers.Adam(learning_rate=0.00005)
model1.compile(loss='mse', optimizer= adam, metrics=['accuracy'])
hist = model1.fit(x_train, y_train, validation_data = (x_val, y_val),
epochs=10, batch_size=8, verbose=0)
fig, loss_ax = plt.subplots()
acc_ax = loss_ax.twinx()
loss_ax.plot(hist.history['loss'], 'y', label='train loss')
loss_ax.plot(hist.history['val_loss'], 'r', label='val loss')
loss_ax.set_xlabel('epoch')
loss_ax.set_ylabel('loss')
loss_ax.legend(loc='upper left')
acc_ax.plot(hist.history['accuracy'], 'b', label='train acc')
acc_ax.plot(hist.history['val_accuracy'], 'g', label='val acc')
acc_ax.set_ylabel('accuracy')
acc_ax.legend(loc='lower left')
plt.show()
prediction = model1.predict(x_test)
right = 0
for i in range(len([0 if x<0.5 else 1 for x in prediction])):
if [0 if x<0.5 else 1 for x in prediction][i] == y_test[i]:
right += 1
print('Accuracy: {}'.format(right/len(y_test)))
# Financial components만 가지고 각기다른 sample을 가지고 학습을 100번 반복하여 평균 정확도 및 std 계산
acc = []
for count in range(10):
i = random.randint(1,1000)
x_train, x_tmp, y_train, y_tmp = train_test_split(XXX,Y_bin, test_size=0.2, shuffle=True, random_state=i)
x_val, x_test, y_val, y_test = train_test_split(x_tmp,y_tmp, test_size=0.5, shuffle=True, random_state=i)
keras.backend.clear_session()
inputs = Input(shape=(14,))
hidden1 = Dense(100, activation='swish')(inputs)
hidden2 = Dense(30, activation='swish')(hidden1)
hidden2 = Dense(30, activation='relu')(hidden1)
outputs = Dense(1, activation='sigmoid')(hidden2)
model1 = Model(inputs, outputs)
adam = keras.optimizers.Adam(learning_rate=0.00005)
model1.compile(loss='mse', optimizer= adam, metrics=['accuracy'])
hist = model1.fit(x_train, y_train, validation_data = (x_val, y_val),
epochs=10, batch_size=8, verbose=0)
prediction = model1.predict(x_test)
right = 0
for i in range(len(y_test)):
if [0 if x<0.5 else 1 for x in prediction][i] == y_test[i]:
right += 1
acc.append((right/len(y_test)))
print("Financial Data's performance: {}".format(np.array(acc).mean()))
print("Financial Data's standard deviation: {}".format(np.array(acc).std()))
# Financial components + G Rank만 가지고 각기다른 sample을 가지고 학습을 100번 반복하여 평균 정확도 및 std 계산
acc = []
for count in range(10):
i = random.randint(1,1000)
x_train, x_tmp, y_train, y_tmp = train_test_split(XX,Y_bin, test_size=0.2, shuffle=True, random_state=i)
x_val, x_test, y_val, y_test = train_test_split(x_tmp,y_tmp, test_size=0.5, shuffle=True, random_state=i)
keras.backend.clear_session()
inputs = Input(shape=(18,))
hidden1 = Dense(100, activation='swish')(inputs)
hidden2 = Dense(30, activation='swish')(hidden1)
hidden2 = Dense(30, activation='relu')(hidden1)
outputs = Dense(1, activation='sigmoid')(hidden2)
model1 = Model(inputs, outputs)
adam = keras.optimizers.Adam(learning_rate=0.00005)
model1.compile(loss='mse', optimizer= adam, metrics=['accuracy'])
hist = model1.fit(x_train, y_train, validation_data = (x_val, y_val),
epochs=10, batch_size=8, verbose=0)
prediction = model1.predict(x_test)
right = 0
for i in range(len(y_test)):
if [0 if x<0.5 else 1 for x in prediction][i] == y_test[i]:
right += 1
acc.append((right/len(y_test)))
print("Financial+G Rank Data's performance: {}".format(np.array(acc).mean()))
print("Financial+G Rank Data's standard deviation: {}".format(np.array(acc).std()))
# Financial components + ESG Rank + etc 가지고 각기다른 sample을 가지고 학습을 100번 반복하여 평균 정확도 및 std 계산
acc = []
for count in range(10):
i = random.randint(1,1000)
x_train, x_tmp, y_train, y_tmp = train_test_split(X,Y_bin, test_size=0.2, shuffle=True, random_state=i)
x_val, x_test, y_val, y_test = train_test_split(x_tmp,y_tmp, test_size=0.5, shuffle=True, random_state=i)
keras.backend.clear_session()
inputs = Input(shape=(19,))
hidden1 = Dense(100, activation='swish')(inputs)
hidden2 = Dense(30, activation='swish')(hidden1)
hidden2 = Dense(30, activation='relu')(hidden1)
outputs = Dense(1, activation='sigmoid')(hidden2)
model1 = Model(inputs, outputs)
adam = keras.optimizers.Adam(learning_rate=0.00005)
model1.compile(loss='mse', optimizer= adam, metrics=['accuracy'])
hist = model1.fit(x_train, y_train, validation_data = (x_val, y_val),
epochs=10, batch_size=8, verbose=0)
prediction = model1.predict(x_test)
right = 0
for i in range(len(y_test)):
if [0 if x<0.5 else 1 for x in prediction][i] == y_test[i]:
right += 1
acc.append((right/len(y_test)))
print("Overall Data's performance: {}".format(np.array(acc).mean()))
print("Overall Data's standard deviation: {}".format(np.array(acc).std()))
#Y를 0(y<0%), 1(0%<y<10%), 2(10%<y<30%), 3(30%<y) 바이너리로 제작, train/vaidation/test셋 구성
y_class = []
for item in Y:
if item<=0 :
y_class.append(0)
elif 0<item<=0.3:
y_class.append(1)
elif 0.3<item<=0.6:
y_class.append(2)
elif 0.6<item:
y_class.append(3)
# epochs, batch_size, learning rate를 정하기 위한 testing
acc = []
i = 1
x_train, x_tmp, y_train, y_tmp = train_test_split(np.array(X),np.array(y_class), test_size=0.2, shuffle=True, random_state=i)
x_val, x_test, y_val, y_test = train_test_split(x_tmp,y_tmp, test_size=0.5, shuffle=True, random_state=i)
keras.backend.clear_session()
inputs = Input(shape=(19,))
hidden1 = Dense(100, activation='swish')(inputs)
hidden2 = Dense(30, activation='swish')(hidden1)
hidden3 = Dense(30, activation='swish')(hidden2)
outputs = Dense(4, activation='softmax')(hidden3)
model1 = Model(inputs, outputs)
adam = keras.optimizers.Adam(learning_rate=0.00001)
model1.compile(loss='sparse_categorical_crossentropy', optimizer= adam, metrics=['accuracy'])
hist = model1.fit(x_train, y_train, validation_data = (x_val, y_val),
epochs=100, batch_size=4, verbose=0)
fig, loss_ax = plt.subplots()
acc_ax = loss_ax.twinx()
loss_ax.plot(hist.history['loss'], 'y', label='train loss')
loss_ax.plot(hist.history['val_loss'], 'r', label='val loss')
loss_ax.set_xlabel('epoch')
loss_ax.set_ylabel('loss')
loss_ax.legend(loc='upper left')
acc_ax.plot(hist.history['accuracy'], 'b', label='train acc')
acc_ax.plot(hist.history['val_accuracy'], 'g', label='val acc')
acc_ax.set_ylabel('accuracy')
acc_ax.legend(loc='lower left')
plt.show()
# Financial components만 가지고 각기다른 sample을 가지고 학습을 100번 반복하여 평균 정확도 및 std 계산
acc = []
for count in range(100):
i = random.randint(1,1000)
x_train, x_tmp, y_train, y_tmp = train_test_split(XXX,y_class, test_size=0.2, shuffle=True, random_state=i)
x_val, x_test, y_val, y_test = train_test_split(x_tmp,y_tmp, test_size=0.5, shuffle=True, random_state=i)
keras.backend.clear_session()
inputs = Input(shape=(14,))
hidden1 = Dense(100, activation='swish')(inputs)
hidden2 = Dense(30, activation='relu')(hidden1)
outputs = Dense(4, activation='sigmoid')(hidden2)
model1 = Model(inputs, outputs)
adam = keras.optimizers.Adam(learning_rate=0.00005)
model1.compile(loss='mse', optimizer= adam, metrics=['accuracy'])
hist = model1.fit(x_train, y_train, validation_data = (x_val, y_val),
epochs=30, batch_size=8, verbose=0)
prediction = model1.predict(x_test)
right = 0
for i in range(len(y_test)):
if np.argmax(prediction[i]) == y_test[i]:
right += 1
acc.append((right/len(y_test)))
print("Financial Data's accuracy: {}".format(np.array(acc).mean()))
print("Financial Data's standard deviation: {}".format(np.array(acc).std()))
# Financial components+G Rank만 가지고 각기다른 sample을 가지고 학습을 100번 반복하여 평균 정확도 및 std 계산
acc = []
for count in range(100):
i = random.randint(1,1000)
x_train, x_tmp, y_train, y_tmp = train_test_split(XX,y_class, test_size=0.2, shuffle=True, random_state=i)
x_val, x_test, y_val, y_test = train_test_split(x_tmp,y_tmp, test_size=0.5, shuffle=True, random_state=i)
keras.backend.clear_session()
inputs = Input(shape=(18,))
hidden1 = Dense(100, activation='swish')(inputs)
hidden2 = Dense(30, activation='relu')(hidden1)
outputs = Dense(4, activation='sigmoid')(hidden2)
model1 = Model(inputs, outputs)
adam = keras.optimizers.Adam(learning_rate=0.00005)
model1.compile(loss='mse', optimizer= adam, metrics=['accuracy'])
hist = model1.fit(x_train, y_train, validation_data = (x_val, y_val),
epochs=30, batch_size=8, verbose=0)
prediction = model1.predict(x_test)
right = 0
for i in range(len(y_test)):
if np.argmax(prediction[i]) == y_test[i]:
right += 1
acc.append((right/len(y_test)))
print("Financial Data + G Rank's accuracy: {}".format(np.array(acc).mean()))
print("Financial Data + G Rank's standard deviation: {}".format(np.array(acc).std()))
# Financial components+G Rank만 가지고 각기다른 sample을 가지고 학습을 100번 반복하여 평균 정확도 및 std 계산
acc = []
for count in range(100):
i = random.randint(1,1000)
x_train, x_tmp, y_train, y_tmp = train_test_split(X,y_class, test_size=0.2, shuffle=True, random_state=i)
x_val, x_test, y_val, y_test = train_test_split(x_tmp,y_tmp, test_size=0.5, shuffle=True, random_state=i)
keras.backend.clear_session()
inputs = Input(shape=(19,))
hidden1 = Dense(100, activation='swish')(inputs)
hidden2 = Dense(30, activation='relu')(hidden1)
outputs = Dense(4, activation='sigmoid')(hidden2)
model1 = Model(inputs, outputs)
adam = keras.optimizers.Adam(learning_rate=0.00005)
model1.compile(loss='mse', optimizer= adam, metrics=['accuracy'])
hist = model1.fit(x_train, y_train, validation_data = (x_val, y_val),
epochs=30, batch_size=8, verbose=0)
prediction = model1.predict(x_test)
right = 0
for i in range(len(y_test)):
if np.argmax(prediction[i]) == y_test[i]:
right += 1
acc.append((right/len(y_test)))
print("Overall Data's accuracy: {}".format(np.array(acc).mean()))
print("Overall Data's's standard deviation: {}".format(np.array(acc).std()))
```
|
github_jupyter
|
import pandas as pd
import re
import numpy as np
import tensorflow as tf
import numpy as np
import operator
import os
import random
import matplotlib.pyplot as plt
from sklearn.cluster import KMeans, DBSCAN
from sklearn.manifold import TSNE
import pandas as pd
from sklearn.linear_model import LinearRegression
from scipy.spatial.distance import cdist
from tensorflow import keras
from tensorflow.keras import layers
from sklearn.model_selection import train_test_split
pip install --upgrade tensorflow==1.15.0
path = r'C:\Users\Yunseok Choi\Downloads\ESG Data'
os.listdir(path)
data1= pd.read_csv(path + '\RUSSELL 3000 DATA.csv')
data2= pd.read_csv(path + '\RUSSELL 3000 + @.csv')
data1.dropna(subset = ['PART'], inplace = True)
data2.dropna(subset = ['PART'], inplace = True)
rs100 = pd.read_csv(path + '\Industry.csv')
re100 = pd.read_csv(path + '\RE100.csv')
def preprocess(item):
global data1
global data2
global rs100
temp = pd.concat([data1[data1['Unnamed: 0'] == item].copy(),
data2[data2['Unnamed: 0'] == item].copy()])
temp.reset_index(inplace = True, drop = True)
temp = pd.DataFrame(np.insert(temp.values,0,values=[item, 'YEAR', 2016, 2017, 2018, 2019, 2020, 2021, 2022],axis=0))
temp = temp.transpose()
temp.reset_index(inplace = True, drop = True)
temp.drop([0], inplace = True)
temp.columns = list(temp.iloc[0].values)
temp.reset_index(inplace = True, drop = True)
temp.drop([0], inplace = True)
temp['INDUSTRY'] = rs100[rs100['COMPANY'] == item.replace('QUITY','quity')]['GICS Sector'].values[0]
temp['COMPANY'] = item
return temp
rus1000_list = [x.upper() for x in rs100['COMPANY'].values]
df = pd.DataFrame()
for item in set(data1['Unnamed: 0']):
if item.upper() in rus1000_list:
df = pd.concat([df, preprocess(item)])
df.shape
#df.drop(['IS_INT_EXPENSES'], axis = 1, inplace = True)
for column in df.columns:
print(column, ' / ', df.dropna(subset = [column]).shape)
year_list = list(set(df['YEAR'].values))
year_list
column_list = df.columns
column_list
for year in year_list:
print(year, '/', df[(df['TOTAL_GHG_EMISSIONS'].notna()) & (df['YEAR'] == year)].shape)
df.columns
df_reg = pd.DataFrame()
for column in column_list[1:-2]:
na_included = [np.nan if (len(re.findall(r'[0-9.]+', str(item))) != 1) else float(item) for item in df[column].values]
na_excluded = [x for x in na_included if np.isnan(float(x)) != True]
df_reg[column] = [x/np.mean(na_excluded) if (np.isnan(float(x)) != True) else np.nan for x in na_included]
for column in list(column_list[-2:]) + [column_list[0]]:
df_reg[column] = df[column].values
df_reg.reset_index(inplace = True, drop = True)
column_list = df_reg.columns
df_reg
for year in year_list:
print(year, ' ', df_reg[(df_reg['TOTAL_GHG_EMISSIONS'].notna()) & (df_reg['YEAR'] == year)].shape)
df_all = df_reg[(df_reg['TOTAL_GHG_EMISSIONS'].notna())].fillna(0)
df_all.reset_index(inplace = True, drop = True)
x_all = df_all[column_list[:-3]].values
#차원 감축을 위한 t-sne 변수 결정. l_rate, iteration, perplexity을 다양하게 바꿔가며 output 관찰
perplexity = [10,20,30,40]
#l_rate = 1000
#iteration = 300
l_rate = 2000
iteration = 8000
data = x_all
plt.figure(figsize=(10,10))
for i in range(len(perplexity)):
plt.subplot(2,2,i+1)
if i == 0:
plt.subplots_adjust(hspace = 0.2, wspace = 0.2)
tsne = TSNE(n_components = 2, learning_rate = l_rate,
perplexity = perplexity[i], n_iter = iteration)
X = tsne.fit_transform(data)
plt.plot([x[0] for x in X], [x[1] for x in X], '.')
plt.title("Perplexity = {}".format(perplexity[i]))
df_all['TSNE'] = list(tsne.fit_transform(x_all))
perplexity = 30
l_rate = 2000
iteration = 8000
tsne = TSNE(n_components = 2, learning_rate = l_rate,
perplexity = perplexity, n_iter = iteration)
#전체 데이터셋 클러스터 개수 결정을 위한 elbow method
k_t = 10
data = x_all
X = tsne.fit_transform(data)
df_all['TSNE'] = list(X)
distortions = []
K = range(1,51)
for k in K:
elbow = KMeans(n_clusters = k).fit(X)
distortions.append(sum(np.min(cdist(X, elbow.cluster_centers_, 'euclidean'), axis=1)) / X.shape[0])
plt.plot(K, distortions, 'go-')
plt.xlabel('k')
plt.ylabel('Distortion')
plt.title('The Elbow Method to determine k')
plt.show()
kmeans = KMeans(n_clusters = k_t).fit(X)
df_all['LABELS'] = kmeans.labels_
for i in range(k_t):
print('Cluster {} has {} companies.'.format(i,(kmeans.labels_ == i).sum()))
#전체 데이터셋 클러스터링 결과 시각화
data = df_all
for i in range(len(set(data['LABELS']))):
item = list(set(data['LABELS']))[i]
data[data['LABELS'] == item]['TSNE']
plt.plot([x[0] for x in data[data['LABELS'] == item]['TSNE'].values],[y[1] for y in data[data['LABELS'] == item]['TSNE'].values],
'.', color = 'C{}'.format(i))
plt.title('2016-2021 Results')
plt.show()
#년도별(16/17. 18/19, 20/21) 데이터 분리
df_c1 = df_all[(df_all['YEAR'] == 2016) | (df_all['YEAR'] == 2017)].reset_index(drop= True).copy()
df_c2 = df_all[(df_all['YEAR'] == 2018) | (df_all['YEAR'] == 2019)].reset_index(drop= True).copy()
df_c3 = df_all[(df_all['YEAR'] == 2020) | (df_all['YEAR'] == 2021)].reset_index(drop= True).copy()
#re100 선언 변수추가를 위한 processing
declare = []
for i in range(df_c1.shape[0]):
if df_c1.iloc[i]['COMPANY'].upper() in re100['COMPANY'].values:
if df_c1.iloc[i]['YEAR'] >= re100[re100['COMPANY'] == df_c1.iloc[i]['COMPANY']]['RE100 Declare Year']:
declare.append(1)
else:
declare.append(0)
else:
declare.append(0)
df_c1['RE100'] = declare
declare = []
for i in range(df_c2.shape[0]):
if df_c2.iloc[i]['COMPANY'].upper() in re100['COMPANY'].values:
if df_c2.iloc[i]['YEAR'] >= re100[re100['COMPANY'] == df_c2.iloc[i]['COMPANY']]['RE100 Declare Year']:
declare.append(1)
else:
declare.append(0)
else:
declare.append(0)
df_c2['RE100'] = declare
declare = []
for i in range(df_c3.shape[0]):
if df_c3.iloc[i]['COMPANY'].upper() in re100['COMPANY'].values:
if df_c3.iloc[i]['YEAR'] >= re100[re100['COMPANY'] == df_c3.iloc[i]['COMPANY']]['RE100 Declare Year']:
declare.append(1)
else:
declare.append(0)
else:
declare.append(0)
df_c3['RE100'] = declare
#16/17 클러스터링을 개수 결정을 위한 elbow method
X = np.array([(x[0], x[1]) for x in list(df_c1['TSNE'].values)])
distortions = []
K = range(1,51)
for k in K:
elbow = KMeans(n_clusters = k).fit(X)
distortions.append(sum(np.min(cdist(X, elbow.cluster_centers_, 'euclidean'), axis=1)) / X.shape[0])
plt.plot(K, distortions, 'go-')
plt.xlabel('k')
plt.ylabel('Distortion')
plt.title('The Elbow Method to determine k')
plt.show()
kmeans = KMeans(n_clusters = k_t).fit(X)
df_c1['CLUSTER LABELS'] = kmeans.labels_
for i in range(k_t):
print('Cluster {} has {} companies.'.format(i,(kmeans.labels_ == i).sum()))
#16/17 클러스터링 시각화
data = df_c1
for i in range(len(set(data['CLUSTER LABELS']))):
item = list(set(data['CLUSTER LABELS']))[i]
data[data['CLUSTER LABELS'] == item]['TSNE']
plt.plot([x[0] for x in data[data['CLUSTER LABELS'] == item]['TSNE'].values],[y[1] for y in data[data['CLUSTER LABELS'] == item]['TSNE'].values],
'.', color = 'C{}'.format(i))
plt.title('2016-2017 Results')
plt.show()
#20/21 클러스터링을 개수 결정을 위한 elbow method
X = np.array([(x[0], x[1]) for x in list(df_c2['TSNE'].values)])
distortions = []
K = range(1,51)
for k in K:
elbow = KMeans(n_clusters = k).fit(X)
distortions.append(sum(np.min(cdist(X, elbow.cluster_centers_, 'euclidean'), axis=1)) / X.shape[0])
plt.plot(K, distortions, 'go-')
plt.xlabel('k')
plt.ylabel('Distortion')
plt.title('The Elbow Method to determine k')
plt.show()
kmeans = KMeans(n_clusters = k_t).fit(X)
df_c2['CLUSTER LABELS'] = kmeans.labels_
for i in range(k_t):
print('Cluster {} has {} companies.'.format(i,(kmeans.labels_ == i).sum()))
#18/19 클러스터링 시각화
data = df_c2
for i in range(len(set(data['CLUSTER LABELS']))):
item = list(set(data['CLUSTER LABELS']))[i]
data[data['CLUSTER LABELS'] == item]['TSNE']
plt.plot([x[0] for x in data[data['CLUSTER LABELS'] == item]['TSNE'].values],[y[1] for y in data[data['CLUSTER LABELS'] == item]['TSNE'].values],
'.', color = 'C{}'.format(i))
plt.title('2018-2019 Results')
plt.show()
#20/21 클러스터링을 개수 결정을 위한 elbow method
X = np.array([(x[0], x[1]) for x in list(df_c3['TSNE'].values)])
distortions = []
K = range(1,51)
for k in K:
elbow = KMeans(n_clusters = k).fit(X)
distortions.append(sum(np.min(cdist(X, elbow.cluster_centers_, 'euclidean'), axis=1)) / X.shape[0])
plt.plot(K, distortions, 'go-')
plt.xlabel('k')
plt.ylabel('Distortion')
plt.title('The Elbow Method to determine k')
plt.show()
kmeans = KMeans(n_clusters = k_t).fit(X)
df_c3['CLUSTER LABELS'] = kmeans.labels_
for i in range(k_t):
print('Cluster {} has {} companies.'.format(i,(kmeans.labels_ == i).sum()))
#20/21 클러스터링 시각화
data = df_c3
for i in range(len(set(data['CLUSTER LABELS']))):
item = list(set(data['CLUSTER LABELS']))[i]
data[data['CLUSTER LABELS'] == item]['TSNE']
plt.plot([x[0] for x in data[data['CLUSTER LABELS'] == item]['TSNE'].values],[y[1] for y in data[data['CLUSTER LABELS'] == item]['TSNE'].values],
'.', color = 'C{}'.format(i))
plt.title('2020-2021 Results')
plt.show()
#현재 데이터 저장
df_c1.to_csv(path+('\df_c1.csv'))
df_c2.to_csv(path+('\df_c2.csv'))
df_c3.to_csv(path+('\df_c3.csv'))
df_all.to_csv(path+('\df_all.csv'))
#16/17 수익률 기준 top5 클러스터 각 클러스터별 회귀분석.
data = df_c1.copy()
company_list = []
for item in data['COMPANY'].values:
if item not in company_list:
company_list.append(item)
else:
pass
#company_list
avg_mkvalt = {}
n_mkvalt = {}
for company in company_list:
if data[data['COMPANY']==company].shape[0] == 2:
k = data[data['COMPANY']==company]['CLUSTER LABELS'].values[-1]
mkvalt0 = data[(data['COMPANY']==company) & (data['YEAR']==2016)]['TOT_MKT_VAL'].values[0]
mkvalt1 = data[(data['COMPANY']==company) & (data['YEAR']==2017)]['TOT_MKT_VAL'].values[0]
if mkvalt0 != 0:
if k not in avg_mkvalt.keys():
avg_mkvalt[k] = ((mkvalt1-mkvalt0)/mkvalt0)
n_mkvalt[k] = 1
else:
avg_mkvalt[k] += ((mkvalt1-mkvalt0)/mkvalt0)
n_mkvalt[k] += 1
rank = {}
for key in sorted(avg_mkvalt.keys()):
avg = (avg_mkvalt[key]/n_mkvalt[key])
rank[key] = avg
#print('Cluster {} has average mkvalt % change of {}'.format(key,avg))
print(sorted(rank.items(), key=operator.itemgetter(1), reverse = True))
top5 = [x[0] for x in sorted(rank.items(), key=operator.itemgetter(1), reverse = True)[:5]]
industry_list = list(set(data['INDUSTRY'].values))
for industry in industry_list:
data[str(industry).upper()] = list(((data['INDUSTRY'] == industry)*1).values)
data['CONSTANT'] = 1
columns = data.columns
iv = list(columns[1:15])+list(columns[16:20])+[columns[26]] + list(columns[-(len(industry_list)):])
dv = columns[15]
df_data = []
for k in top5:
temp = []
X = data[(data['CLUSTER LABELS']==k)&(data['YEAR']==2016)][iv].values
Y = data[(data['CLUSTER LABELS']==k)&(data['YEAR']==2016)][dv].values
lr = LinearRegression()
lr.fit(X, Y)
temp = list(round(x*100,2) for x in lr.coef_)
temp.append(round(lr.score(X,Y),2))
df_data.append(temp)
pd.DataFrame(np.array(df_data).transpose(), index = iv+['R^2'], columns = top5)
#연도별 '상' 그룸 회귀분석을 위한 변수선언
high_x = []
high_y = []
#16/17 클러스터 수익률 별 3:4:3 비율로 상 중 하 나눈 뒤 리그레션
data = df_c1.copy()
company_list = []
for item in data['COMPANY'].values:
if item not in company_list:
company_list.append(item)
else:
pass
#company_list
avg_mkvalt = {}
n_mkvalt = {}
for company in company_list:
if data[data['COMPANY']==company].shape[0] == 2:
k = data[data['COMPANY']==company]['CLUSTER LABELS'].values[-1]
mkvalt0 = data[(data['COMPANY']==company) & (data['YEAR']==2016)]['TOT_MKT_VAL'].values[0]
mkvalt1 = data[(data['COMPANY']==company) & (data['YEAR']==2017)]['TOT_MKT_VAL'].values[0]
if mkvalt0 != 0:
if k not in avg_mkvalt.keys():
avg_mkvalt[k] = ((mkvalt1-mkvalt0)/mkvalt0)
n_mkvalt[k] = 1
else:
avg_mkvalt[k] += ((mkvalt1-mkvalt0)/mkvalt0)
n_mkvalt[k] += 1
rank = {}
for key in sorted(avg_mkvalt.keys()):
avg = (avg_mkvalt[key]/n_mkvalt[key])
rank[key] = avg
#print('Cluster {} has average mkvalt % change of {}'.format(key,avg))
#print(sorted(rank.items(), key=operator.itemgetter(1), reverse = True))
top = [x[0] for x in sorted(rank.items(), key=operator.itemgetter(1), reverse = True)[0:3]]
mid = [x[0] for x in sorted(rank.items(), key=operator.itemgetter(1), reverse = True)[4:8]]
low = [x[0] for x in sorted(rank.items(), key=operator.itemgetter(1), reverse = True)[8:10]]
industry_list = list(set(data['INDUSTRY'].values)) + ['INFORMATION TECHNOLOGY']
for industry in industry_list:
data[str(industry).upper()] = list(((data['INDUSTRY'] == industry)*1).values)
data['CONSTANT'] = 1
columns = data.columns
iv = list(columns[1:15])+list(columns[16:20])+[columns[26]] + list(columns[-(len(industry_list)):])
dv = columns[15]
df_data = []
for item in [top, mid, low]:
X = []
Y = []
for k in item:
temp = []
X += [list(x) for x in data[(data['CLUSTER LABELS']==k)&(data['YEAR']==2016)][iv].values]
Y += list(data[(data['CLUSTER LABELS']==k)&(data['YEAR']==2016)][dv].values)
if item == top:
high_x += [list(x) for x in data[(data['CLUSTER LABELS']==k)&(data['YEAR']==2016)][iv].values]
high_y += list(data[(data['CLUSTER LABELS']==k)&(data['YEAR']==2016)][dv].values)
lr = LinearRegression()
lr.fit(X, Y)
temp = list(round(x*100,2) for x in lr.coef_)
temp.append(round(lr.score(X,Y),2))
df_data.append(temp)
pd.DataFrame(np.array(df_data).transpose(), index = iv+['R^2'], columns = ['Top','Mid','Low'])
#18/19 수익률 기준 top5 클러스터 각 클러스터별 회귀분석.
data = df_c2.copy()
company_list = []
for item in data['COMPANY'].values:
if item not in company_list:
company_list.append(item)
else:
pass
#company_list
avg_mkvalt = {}
n_mkvalt = {}
for company in company_list:
if data[data['COMPANY']==company].shape[0] == 2:
k = data[data['COMPANY']==company]['CLUSTER LABELS'].values[-1]
mkvalt0 = data[(data['COMPANY']==company) & (data['YEAR']==2018)]['TOT_MKT_VAL'].values[0]
mkvalt1 = data[(data['COMPANY']==company) & (data['YEAR']==2019)]['TOT_MKT_VAL'].values[0]
if mkvalt0 != 0:
if k not in avg_mkvalt.keys():
avg_mkvalt[k] = ((mkvalt1-mkvalt0)/mkvalt0)
n_mkvalt[k] = 1
else:
avg_mkvalt[k] += ((mkvalt1-mkvalt0)/mkvalt0)
n_mkvalt[k] += 1
rank = {}
for key in sorted(avg_mkvalt.keys()):
avg = (avg_mkvalt[key]/n_mkvalt[key])
rank[key] = avg
#print('Cluster {} has average mkvalt % change of {}'.format(key,avg))
print(sorted(rank.items(), key=operator.itemgetter(1), reverse = True))
top5 = [x[0] for x in sorted(rank.items(), key=operator.itemgetter(1), reverse = True)[:5]]
industry_list = list(set(data['INDUSTRY'].values))
for industry in industry_list:
data[str(industry).upper()] = list(((data['INDUSTRY'] == industry)*1).values)
data['CONSTANT'] = 1
columns = data.columns
iv = list(columns[1:15])+list(columns[16:20])+[columns[26]] + list(columns[-(len(industry_list)):])
dv = columns[15]
df_data = []
for k in top5:
temp = []
X = data[(data['CLUSTER LABELS']==k)&(data['YEAR']==2018)][iv].values
Y = data[(data['CLUSTER LABELS']==k)&(data['YEAR']==2018)][dv].values
lr = LinearRegression()
lr.fit(X, Y)
temp = list(round(x*100,2) for x in lr.coef_)
temp.append(round(lr.score(X,Y),2))
df_data.append(temp)
pd.DataFrame(np.array(df_data).transpose(), index = iv+['R^2'], columns = top5)
#18/19 클러스터 수익률 별 3:4:3 비율로 상 중 하 나눈 뒤 리그레션
data = df_c2.copy()
company_list = []
for item in data['COMPANY'].values:
if item not in company_list:
company_list.append(item)
else:
pass
#company_list
avg_mkvalt = {}
n_mkvalt = {}
for company in company_list:
if data[data['COMPANY']==company].shape[0] == 2:
k = data[data['COMPANY']==company]['CLUSTER LABELS'].values[-1]
mkvalt0 = data[(data['COMPANY']==company) & (data['YEAR']==2018)]['TOT_MKT_VAL'].values[0]
mkvalt1 = data[(data['COMPANY']==company) & (data['YEAR']==2019)]['TOT_MKT_VAL'].values[0]
if mkvalt0 != 0:
if k not in avg_mkvalt.keys():
avg_mkvalt[k] = ((mkvalt1-mkvalt0)/mkvalt0)
n_mkvalt[k] = 1
else:
avg_mkvalt[k] += ((mkvalt1-mkvalt0)/mkvalt0)
n_mkvalt[k] += 1
rank = {}
for key in sorted(avg_mkvalt.keys()):
avg = (avg_mkvalt[key]/n_mkvalt[key])
rank[key] = avg
#print('Cluster {} has average mkvalt % change of {}'.format(key,avg))
#print(sorted(rank.items(), key=operator.itemgetter(1), reverse = True))
top = [x[0] for x in sorted(rank.items(), key=operator.itemgetter(1), reverse = True)[0:3]]
mid = [x[0] for x in sorted(rank.items(), key=operator.itemgetter(1), reverse = True)[4:8]]
low = [x[0] for x in sorted(rank.items(), key=operator.itemgetter(1), reverse = True)[8:10]]
industry_list = list(set(data['INDUSTRY'].values))
for industry in industry_list:
data[str(industry).upper()] = list(((data['INDUSTRY'] == industry)*1).values)
data['CONSTANT'] = 1
columns = data.columns
iv = list(columns[1:15])+list(columns[16:20])+[columns[26]] + list(columns[-(len(industry_list)):])
dv = columns[15]
df_data = []
for item in [top, mid, low]:
X = []
Y = []
for k in item:
temp = []
X += [list(x) for x in data[(data['CLUSTER LABELS']==k)&(data['YEAR']==2018)][iv].values]
Y += list(data[(data['CLUSTER LABELS']==k)&(data['YEAR']==2018)][dv].values)
if item == top:
high_x += [list(x) for x in data[(data['CLUSTER LABELS']==k)&(data['YEAR']==2018)][iv].values]
high_y += list(data[(data['CLUSTER LABELS']==k)&(data['YEAR']==2018)][dv].values)
lr = LinearRegression()
lr.fit(X, Y)
temp = list(round(x*100,2) for x in lr.coef_)
temp.append(round(lr.score(X,Y),2))
df_data.append(temp)
pd.DataFrame(np.array(df_data).transpose(), index = iv+['R^2'], columns = ['Top','Mid','Low'])
#20/21 수익률 기준 top5 클러스터 각 클러스터별 회귀분석.
data = df_c3.copy()
company_list = []
for item in data['COMPANY'].values:
if item not in company_list:
company_list.append(item)
else:
pass
#company_list
avg_mkvalt = {}
n_mkvalt = {}
for company in company_list:
if data[data['COMPANY']==company].shape[0] == 2:
k = data[data['COMPANY']==company]['CLUSTER LABELS'].values[-1]
mkvalt0 = data[(data['COMPANY']==company) & (data['YEAR']==2020)]['TOT_MKT_VAL'].values[0]
mkvalt1 = data[(data['COMPANY']==company) & (data['YEAR']==2021)]['TOT_MKT_VAL'].values[0]
if mkvalt0 != 0:
if k not in avg_mkvalt.keys():
avg_mkvalt[k] = ((mkvalt1-mkvalt0)/mkvalt0)
n_mkvalt[k] = 1
else:
avg_mkvalt[k] += ((mkvalt1-mkvalt0)/mkvalt0)
n_mkvalt[k] += 1
rank = {}
for key in sorted(avg_mkvalt.keys()):
avg = (avg_mkvalt[key]/n_mkvalt[key])
rank[key] = avg
#print('Cluster {} has average mkvalt % change of {}'.format(key,avg))
print(sorted(rank.items(), key=operator.itemgetter(1), reverse = True))
top5 = [x[0] for x in sorted(rank.items(), key=operator.itemgetter(1), reverse = True)[:5]]
industry_list = list(set(data['INDUSTRY'].values))
for industry in industry_list:
data[str(industry).upper()] = list(((data['INDUSTRY'] == industry)*1).values)
data['CONSTANT'] = 1
columns = data.columns
iv = list(columns[1:15])+list(columns[16:20])+[columns[26]] + list(columns[-(len(industry_list)):])
dv = columns[15]
df_data = []
for k in top5:
temp = []
X = data[(data['CLUSTER LABELS']==k)&(data['YEAR']==2020)][iv].values
Y = data[(data['CLUSTER LABELS']==k)&(data['YEAR']==2020)][dv].values
lr = LinearRegression()
lr.fit(X, Y)
temp = list(round(x*100,2) for x in lr.coef_)
temp.append(round(lr.score(X,Y),2))
df_data.append(temp)
pd.DataFrame(np.array(df_data).transpose(), index = iv+['R^2'], columns = top5)
#20/21 클러스터 수익률 별 3:4:3 비율로 상 중 하 나눈 뒤 리그레션
data = df_c3.copy()
company_list = []
for item in data['COMPANY'].values:
if item not in company_list:
company_list.append(item)
else:
pass
#company_list
avg_mkvalt = {}
n_mkvalt = {}
for company in company_list:
if data[data['COMPANY']==company].shape[0] == 2:
k = data[data['COMPANY']==company]['CLUSTER LABELS'].values[-1]
mkvalt0 = data[(data['COMPANY']==company) & (data['YEAR']==2020)]['TOT_MKT_VAL'].values[0]
mkvalt1 = data[(data['COMPANY']==company) & (data['YEAR']==2021)]['TOT_MKT_VAL'].values[0]
if mkvalt0 != 0:
if k not in avg_mkvalt.keys():
avg_mkvalt[k] = ((mkvalt1-mkvalt0)/mkvalt0)
n_mkvalt[k] = 1
else:
avg_mkvalt[k] += ((mkvalt1-mkvalt0)/mkvalt0)
n_mkvalt[k] += 1
rank = {}
for key in sorted(avg_mkvalt.keys()):
avg = (avg_mkvalt[key]/n_mkvalt[key])
rank[key] = avg
#print('Cluster {} has average mkvalt % change of {}'.format(key,avg))
#print(sorted(rank.items(), key=operator.itemgetter(1), reverse = True))
top = [x[0] for x in sorted(rank.items(), key=operator.itemgetter(1), reverse = True)[0:3]]
mid = [x[0] for x in sorted(rank.items(), key=operator.itemgetter(1), reverse = True)[4:8]]
low = [x[0] for x in sorted(rank.items(), key=operator.itemgetter(1), reverse = True)[8:10]]
industry_list = list(set(data['INDUSTRY'].values))
for industry in industry_list:
data[str(industry).upper()] = list(((data['INDUSTRY'] == industry)*1).values)
data['CONSTANT'] = 1
columns = data.columns
iv = list(columns[1:15])+list(columns[16:20])+[columns[26]] + list(columns[-(len(industry_list)):])
dv = columns[15]
df_data = []
for item in [top, mid, low]:
X = []
Y = []
for k in item:
temp = []
X += [list(x) for x in data[(data['CLUSTER LABELS']==k)&(data['YEAR']==2020)][iv].values]
Y += list(data[(data['CLUSTER LABELS']==k)&(data['YEAR']==2020)][dv].values)
if item == top:
high_x += [list(x) for x in data[(data['CLUSTER LABELS']==k)&(data['YEAR']==2020)][iv].values]
high_y += list(data[(data['CLUSTER LABELS']==k)&(data['YEAR']==2020)][dv].values)
lr = LinearRegression()
lr.fit(X, Y)
temp = list(round(x*100,2) for x in lr.coef_)
temp.append(round(lr.score(X,Y),2))
df_data.append(temp)
pd.DataFrame(np.array(df_data).transpose(), index = iv+['R^2'], columns = ['Top','Mid','Low'])
# 16/17,18/19,20/21 수익률 기준 '상' 그룹 모아서 리그레션
temp = []
lr = LinearRegression()
lr.fit(high_x, high_y)
temp = list(round(x*100,2) for x in lr.coef_)
temp.append(round(lr.score(X,Y),2))
pd.DataFrame(temp,index = iv+['R^2'], columns = ['16-21 Top Clusters'])
data = df_c1
year0 = 2016
year1 = 2017
mkvalt = {}
for company in company_list:
if data[data['COMPANY']==company].shape[0] == 2:
mkvalt0 = data[(data['COMPANY']==company) & (data['YEAR']==2016)]['TOT_MKT_VAL'].values[0]
mkvalt1 = data[(data['COMPANY']==company) & (data['YEAR']==2017)]['TOT_MKT_VAL'].values[0]
if mkvalt0 != 0:
mkvalt[company] = ((mkvalt1-mkvalt0)/mkvalt0)
top10 = [x[0] for x in sorted(mkvalt.items(),key=operator.itemgetter(1),reverse = True)[:int(round(len(mkvalt.keys())/10,0))]]
industry_list = list(set(data['INDUSTRY'].values))
for industry in industry_list:
data[str(industry).upper()] = list(((data['INDUSTRY'] == industry)*1).values)
data['CONSTANT'] = 1
columns = data.columns
iv = list(columns[1:15])+list(columns[16:20])+[columns[26]] + list(columns[-(len(industry_list)):])
dv = columns[15]
df_data = []
X = []
Y = []
for company in top10:
temp = []
X+=[list(x) for x in (data[(data['COMPANY']==company) & (data['YEAR'] == year0)][iv].values)]
Y+=list(data[(data['COMPANY']==company) & (data['YEAR'] == year0)][dv].values)
lr = LinearRegression()
lr.fit(X, Y)
temp = list(round(x*100,2) for x in lr.coef_)
temp.append(round(lr.score(X,Y),2))
df_data.append(temp)
pd.DataFrame(np.array(df_data).transpose(), index = iv+['R^2'], columns = ['TOP10%'])
data = df_c2
year0 = 2018
year1 = 2019
mkvalt = {}
for company in company_list:
if data[data['COMPANY']==company].shape[0] == 2:
mkvalt0 = data[(data['COMPANY']==company) & (data['YEAR']==2018)]['TOT_MKT_VAL'].values[0]
mkvalt1 = data[(data['COMPANY']==company) & (data['YEAR']==2019)]['TOT_MKT_VAL'].values[0]
if mkvalt0 != 0:
mkvalt[company] = ((mkvalt1-mkvalt0)/mkvalt0)
top10 = [x[0] for x in sorted(mkvalt.items(),key=operator.itemgetter(1),reverse = True)[:int(round(len(mkvalt.keys())/10,0))]]
industry_list = list(set(data['INDUSTRY'].values))
for industry in industry_list:
data[str(industry).upper()] = list(((data['INDUSTRY'] == industry)*1).values)
data['CONSTANT'] = 1
columns = data.columns
iv = list(columns[1:15])+list(columns[16:20])+[columns[26]] + list(columns[-(len(industry_list)):])
dv = columns[15]
df_data = []
X = []
Y = []
for company in top10:
temp = []
X+=[list(x) for x in (data[(data['COMPANY']==company) & (data['YEAR'] == year0)][iv].values)]
Y+=list(data[(data['COMPANY']==company) & (data['YEAR'] == year0)][dv].values)
lr = LinearRegression()
lr.fit(X, Y)
temp = list(round(x*100,2) for x in lr.coef_)
temp.append(round(lr.score(X,Y),2))
df_data.append(temp)
pd.DataFrame(np.array(df_data).transpose(), index = iv+['R^2'], columns = ['TOP10%'])
data = df_c3
year0=2020
year1=2021
mkvalt = {}
for company in company_list:
if data[data['COMPANY']==company].shape[0] == 2:
mkvalt0 = data[(data['COMPANY']==company) & (data['YEAR']==year0)]['TOT_MKT_VAL'].values[0]
mkvalt1 = data[(data['COMPANY']==company) & (data['YEAR']==year1)]['TOT_MKT_VAL'].values[0]
if mkvalt0 != 0:
mkvalt[company] = ((mkvalt1-mkvalt0)/mkvalt0)
top10 = [x[0] for x in sorted(mkvalt.items(),key=operator.itemgetter(1),reverse = True)[:int(round(len(mkvalt.keys())/10,0))]]
industry_list = list(set(data['INDUSTRY'].values))
for industry in industry_list:
data[str(industry).upper()] = list(((data['INDUSTRY'] == industry)*1).values)
data['CONSTANT'] = 1
columns = data.columns
iv = list(columns[1:15])+list(columns[16:20])+[columns[26]] + list(columns[-(len(industry_list)):])
dv = columns[15]
df_data = []
X = []
Y = []
for company in top10:
temp = []
X+=[list(x) for x in (data[(data['COMPANY']==company) & (data['YEAR'] == year0)][iv].values)]
Y+=list(data[(data['COMPANY']==company) & (data['YEAR'] == year0)][dv].values)
lr = LinearRegression()
lr.fit(X, Y)
temp = list(round(x*100,2) for x in lr.coef_)
temp.append(round(lr.score(X,Y),2))
df_data.append(temp)
pd.DataFrame(np.array(df_data).transpose(), index = iv+['R^2'], columns = ['TOP10%'])
data = df_all
mkvalt = {}
year_list = [2016, 2017, 2018, 2019, 2020, 2021]
for company in company_list:
for i in range(len(year_list)-2):
year0 = year_list[i]
year1 = year_list[i+1]
if (data[(data['COMPANY']==company) & (data['YEAR']==year0)]['TOT_MKT_VAL'].shape[0] != 0) & (data[(data['COMPANY']==company) & (data['YEAR']==year1)]['TOT_MKT_VAL'].shape[0] != 0):
mkvalt0 = data[(data['COMPANY']==company) & (data['YEAR']==year0)]['TOT_MKT_VAL'].values[0]
mkvalt1 = data[(data['COMPANY']==company) & (data['YEAR']==year1)]['TOT_MKT_VAL'].values[0]
if mkvalt0 != 0:
mkvalt['{}/{}'.format(company,year0)] = ((mkvalt1-mkvalt0)/mkvalt0)
top10 = [x[0] for x in sorted(mkvalt.items(),key=operator.itemgetter(1),reverse = True)[:int(round(len(mkvalt.keys())/10,0))]]
industry_list = list(set(data['INDUSTRY'].values))
for industry in industry_list:
data[str(industry).upper()] = list(((data['INDUSTRY'] == industry)*1).values)
data['CONSTANT'] = 1
columns = data.columns
#iv = list(columns[0:15])+list(columns[16:20])+[columns[26]] + list(columns[-(len(industry_list)):])
iv = list(columns[1:15])+list(columns[16:20])+ list(columns[-(len(industry_list)):])
dv = columns[15]
df_data = []
X = []
Y = []
for i in range(len(top10)):
company, year = top10[i].split('/')[0],top10[i].split('/')[1]
temp = []
X+=[list(x) for x in (data[(data['COMPANY']==company) & (data['YEAR'] == float(year))][iv].values)]
Y+=list((data[(data['COMPANY']==company) & (data['YEAR'] == float(year))][dv].values))
lr = LinearRegression()
lr.fit(X, Y)
temp = list(round(x*100,2) for x in lr.coef_)
temp.append(round(lr.score(X,Y),2))
df_data.append(temp)
pd.DataFrame(np.array(df_data).transpose(), index = iv+['R^2'], columns = ['TOP10%'])
data = df_all
mkvalt = {}
year_list = [2016, 2017, 2018, 2019]
for company in company_list:
for i in range(len(year_list)-2):
year0 = year_list[i]
year1 = year_list[i+1]
if (data[(data['COMPANY']==company) & (data['YEAR']==year0)]['TOT_MKT_VAL'].shape[0] != 0) & (data[(data['COMPANY']==company) & (data['YEAR']==year1)]['TOT_MKT_VAL'].shape[0] != 0):
mkvalt0 = data[(data['COMPANY']==company) & (data['YEAR']==year0)]['TOT_MKT_VAL'].values[0]
mkvalt1 = data[(data['COMPANY']==company) & (data['YEAR']==year1)]['TOT_MKT_VAL'].values[0]
if mkvalt0 != 0:
mkvalt['{}/{}'.format(company,year0)] = ((mkvalt1-mkvalt0)/mkvalt0)
top10 = [x[0] for x in sorted(mkvalt.items(),key=operator.itemgetter(1),reverse = True)[:int(round(len(mkvalt.keys())/10,0))]]
industry_list = list(set(data['INDUSTRY'].values))
for industry in industry_list:
data[str(industry).upper()] = list(((data['INDUSTRY'] == industry)*1).values)
data['CONSTANT'] = 1
columns = data.columns
iv = list(columns[1:15])+list(columns[16:20])+[columns[26]] + list(columns[-(len(industry_list)):])
dv = columns[15]
df_data = []
X = []
Y = []
for i in range(len(top10)):
company, year = top10[i].split('/')[0],top10[i].split('/')[1]
temp = []
X+=[list(x) for x in (data[(data['COMPANY']==company) & (data['YEAR'] == float(year))][iv].values)]
Y+=list((data[(data['COMPANY']==company) & (data['YEAR'] == float(year))][dv].values))
lr = LinearRegression()
lr.fit(X, Y)
temp = list(round(x*100,2) for x in lr.coef_)
temp.append(round(lr.score(X,Y),2))
df_data.append(temp)
pd.DataFrame(np.array(df_data).transpose(), index = iv+['R^2'], columns = ['TOP10%'])
data = df_all
mkvalt = {}
year_list = [2018, 2019, 2020, 2021]
for company in company_list:
for i in range(len(year_list)-2):
year0 = year_list[i]
year1 = year_list[i+1]
if (data[(data['COMPANY']==company) & (data['YEAR']==year0)]['TOT_MKT_VAL'].shape[0] != 0) & (data[(data['COMPANY']==company) & (data['YEAR']==year1)]['TOT_MKT_VAL'].shape[0] != 0):
mkvalt0 = data[(data['COMPANY']==company) & (data['YEAR']==year0)]['TOT_MKT_VAL'].values[0]
mkvalt1 = data[(data['COMPANY']==company) & (data['YEAR']==year1)]['TOT_MKT_VAL'].values[0]
if mkvalt0 != 0:
mkvalt['{}/{}'.format(company,year0)] = ((mkvalt1-mkvalt0)/mkvalt0)
top10 = [x[0] for x in sorted(mkvalt.items(),key=operator.itemgetter(1),reverse = True)[:int(round(len(mkvalt.keys())/10,0))]]
industry_list = list(set(data['INDUSTRY'].values))
for industry in industry_list:
data[str(industry).upper()] = list(((data['INDUSTRY'] == industry)*1).values)
data['CONSTANT'] = 1
columns = data.columns
iv = list(columns[1:15])+list(columns[16:20])+[columns[26]] + list(columns[-(len(industry_list)):])
dv = columns[15]
df_data = []
X = []
Y = []
for i in range(len(top10)):
company, year = top10[i].split('/')[0],top10[i].split('/')[1]
temp = []
X+=[list(x) for x in (data[(data['COMPANY']==company) & (data['YEAR'] == float(year))][iv].values)]
Y+=list((data[(data['COMPANY']==company) & (data['YEAR'] == float(year))][dv].values))
lr = LinearRegression()
lr.fit(X, Y)
temp = list(round(x*100,2) for x in lr.coef_)
temp.append(round(lr.score(X,Y),2))
df_data.append(temp)
pd.DataFrame(np.array(df_data).transpose(), index = iv+['R^2'], columns = ['TOP10%'])
from tensorflow import keras
from tensorflow.keras import layers
from tensorflow.keras.models import Model
df_reg.shape
df_all
#딥러닝을 위한 X, XX, Y 생성
data = df_all
mkvalt = {}
year_list = [2016, 2017, 2018, 2019, 2020, 2021]
industry_list = list(set(data['INDUSTRY'].values))
for industry in industry_list:
data[str(industry).upper()] = list(((data['INDUSTRY'] == industry)*1).values)
columns = data.columns
iv = list(columns[1:15])+list(columns[16:20])+ ['LABELS']
ivv = ['ROBECOSAM_ECON_DIMENSION_RANK','TOTAL_GHG_EMISSIONS'] + list(columns[3:15])+list(columns[16:20])
ivvv = iv[6:]+ ['LABELS']
X, XX, XXX = [], [], []
Y = []
for company in company_list:
for i in range(len(year_list)-2):
year0 = year_list[i]
year1 = year_list[i+1]
if data[(data['COMPANY']==company) & ((data['YEAR']==year0) | (data['YEAR']==year1))]['TOT_MKT_VAL'].shape[0] ==2:
mkvalt0 = data[(data['COMPANY']==company) & (data['YEAR']==year0)]['TOT_MKT_VAL'].values[0]
mkvalt1 = data[(data['COMPANY']==company) & (data['YEAR']==year1)]['TOT_MKT_VAL'].values[0]
if mkvalt0 != 0:
Y.append(float((mkvalt1-mkvalt0)/mkvalt0))
X.append(list(data[(data['COMPANY']==company)&(data['YEAR']==float(year0))][iv].values[0]))
XX.append(list(data[(data['COMPANY']==company)&(data['YEAR']==float(year0))][ivv].values[0]))
XXX.append(list(data[(data['COMPANY']==company)&(data['YEAR']==float(year0))][ivvv].values[0]))
#Y를 바이너리로 제작, train/vaidation/test셋 구성
Y_bin = [1 if x > 0 else 0 for x in Y]
i = 169
x_train, x_tmp, y_train, y_tmp = train_test_split(X,Y_bin, test_size=0.2, shuffle=True, random_state=i)
x_val, x_test, y_val, y_test = train_test_split(x_tmp,y_tmp, test_size=0.5, shuffle=True, random_state=i)
# epochs, batch_size, learning rate를 정하기 위한 testing
keras.backend.clear_session()
inputs = Input(shape=(19,))
hidden1 = Dense(100, activation='swish')(inputs)
hidden2 = Dense(30, activation='swish')(hidden1)
hidden2 = Dense(30, activation='relu')(hidden1)
outputs = Dense(1, activation='sigmoid')(hidden2)
model1 = Model(inputs, outputs)
adam = keras.optimizers.Adam(learning_rate=0.00005)
model1.compile(loss='mse', optimizer= adam, metrics=['accuracy'])
hist = model1.fit(x_train, y_train, validation_data = (x_val, y_val),
epochs=10, batch_size=8, verbose=0)
fig, loss_ax = plt.subplots()
acc_ax = loss_ax.twinx()
loss_ax.plot(hist.history['loss'], 'y', label='train loss')
loss_ax.plot(hist.history['val_loss'], 'r', label='val loss')
loss_ax.set_xlabel('epoch')
loss_ax.set_ylabel('loss')
loss_ax.legend(loc='upper left')
acc_ax.plot(hist.history['accuracy'], 'b', label='train acc')
acc_ax.plot(hist.history['val_accuracy'], 'g', label='val acc')
acc_ax.set_ylabel('accuracy')
acc_ax.legend(loc='lower left')
plt.show()
prediction = model1.predict(x_test)
right = 0
for i in range(len([0 if x<0.5 else 1 for x in prediction])):
if [0 if x<0.5 else 1 for x in prediction][i] == y_test[i]:
right += 1
print('Accuracy: {}'.format(right/len(y_test)))
# Financial components만 가지고 각기다른 sample을 가지고 학습을 100번 반복하여 평균 정확도 및 std 계산
acc = []
for count in range(10):
i = random.randint(1,1000)
x_train, x_tmp, y_train, y_tmp = train_test_split(XXX,Y_bin, test_size=0.2, shuffle=True, random_state=i)
x_val, x_test, y_val, y_test = train_test_split(x_tmp,y_tmp, test_size=0.5, shuffle=True, random_state=i)
keras.backend.clear_session()
inputs = Input(shape=(14,))
hidden1 = Dense(100, activation='swish')(inputs)
hidden2 = Dense(30, activation='swish')(hidden1)
hidden2 = Dense(30, activation='relu')(hidden1)
outputs = Dense(1, activation='sigmoid')(hidden2)
model1 = Model(inputs, outputs)
adam = keras.optimizers.Adam(learning_rate=0.00005)
model1.compile(loss='mse', optimizer= adam, metrics=['accuracy'])
hist = model1.fit(x_train, y_train, validation_data = (x_val, y_val),
epochs=10, batch_size=8, verbose=0)
prediction = model1.predict(x_test)
right = 0
for i in range(len(y_test)):
if [0 if x<0.5 else 1 for x in prediction][i] == y_test[i]:
right += 1
acc.append((right/len(y_test)))
print("Financial Data's performance: {}".format(np.array(acc).mean()))
print("Financial Data's standard deviation: {}".format(np.array(acc).std()))
# Financial components + G Rank만 가지고 각기다른 sample을 가지고 학습을 100번 반복하여 평균 정확도 및 std 계산
acc = []
for count in range(10):
i = random.randint(1,1000)
x_train, x_tmp, y_train, y_tmp = train_test_split(XX,Y_bin, test_size=0.2, shuffle=True, random_state=i)
x_val, x_test, y_val, y_test = train_test_split(x_tmp,y_tmp, test_size=0.5, shuffle=True, random_state=i)
keras.backend.clear_session()
inputs = Input(shape=(18,))
hidden1 = Dense(100, activation='swish')(inputs)
hidden2 = Dense(30, activation='swish')(hidden1)
hidden2 = Dense(30, activation='relu')(hidden1)
outputs = Dense(1, activation='sigmoid')(hidden2)
model1 = Model(inputs, outputs)
adam = keras.optimizers.Adam(learning_rate=0.00005)
model1.compile(loss='mse', optimizer= adam, metrics=['accuracy'])
hist = model1.fit(x_train, y_train, validation_data = (x_val, y_val),
epochs=10, batch_size=8, verbose=0)
prediction = model1.predict(x_test)
right = 0
for i in range(len(y_test)):
if [0 if x<0.5 else 1 for x in prediction][i] == y_test[i]:
right += 1
acc.append((right/len(y_test)))
print("Financial+G Rank Data's performance: {}".format(np.array(acc).mean()))
print("Financial+G Rank Data's standard deviation: {}".format(np.array(acc).std()))
# Financial components + ESG Rank + etc 가지고 각기다른 sample을 가지고 학습을 100번 반복하여 평균 정확도 및 std 계산
acc = []
for count in range(10):
i = random.randint(1,1000)
x_train, x_tmp, y_train, y_tmp = train_test_split(X,Y_bin, test_size=0.2, shuffle=True, random_state=i)
x_val, x_test, y_val, y_test = train_test_split(x_tmp,y_tmp, test_size=0.5, shuffle=True, random_state=i)
keras.backend.clear_session()
inputs = Input(shape=(19,))
hidden1 = Dense(100, activation='swish')(inputs)
hidden2 = Dense(30, activation='swish')(hidden1)
hidden2 = Dense(30, activation='relu')(hidden1)
outputs = Dense(1, activation='sigmoid')(hidden2)
model1 = Model(inputs, outputs)
adam = keras.optimizers.Adam(learning_rate=0.00005)
model1.compile(loss='mse', optimizer= adam, metrics=['accuracy'])
hist = model1.fit(x_train, y_train, validation_data = (x_val, y_val),
epochs=10, batch_size=8, verbose=0)
prediction = model1.predict(x_test)
right = 0
for i in range(len(y_test)):
if [0 if x<0.5 else 1 for x in prediction][i] == y_test[i]:
right += 1
acc.append((right/len(y_test)))
print("Overall Data's performance: {}".format(np.array(acc).mean()))
print("Overall Data's standard deviation: {}".format(np.array(acc).std()))
#Y를 0(y<0%), 1(0%<y<10%), 2(10%<y<30%), 3(30%<y) 바이너리로 제작, train/vaidation/test셋 구성
y_class = []
for item in Y:
if item<=0 :
y_class.append(0)
elif 0<item<=0.3:
y_class.append(1)
elif 0.3<item<=0.6:
y_class.append(2)
elif 0.6<item:
y_class.append(3)
# epochs, batch_size, learning rate를 정하기 위한 testing
acc = []
i = 1
x_train, x_tmp, y_train, y_tmp = train_test_split(np.array(X),np.array(y_class), test_size=0.2, shuffle=True, random_state=i)
x_val, x_test, y_val, y_test = train_test_split(x_tmp,y_tmp, test_size=0.5, shuffle=True, random_state=i)
keras.backend.clear_session()
inputs = Input(shape=(19,))
hidden1 = Dense(100, activation='swish')(inputs)
hidden2 = Dense(30, activation='swish')(hidden1)
hidden3 = Dense(30, activation='swish')(hidden2)
outputs = Dense(4, activation='softmax')(hidden3)
model1 = Model(inputs, outputs)
adam = keras.optimizers.Adam(learning_rate=0.00001)
model1.compile(loss='sparse_categorical_crossentropy', optimizer= adam, metrics=['accuracy'])
hist = model1.fit(x_train, y_train, validation_data = (x_val, y_val),
epochs=100, batch_size=4, verbose=0)
fig, loss_ax = plt.subplots()
acc_ax = loss_ax.twinx()
loss_ax.plot(hist.history['loss'], 'y', label='train loss')
loss_ax.plot(hist.history['val_loss'], 'r', label='val loss')
loss_ax.set_xlabel('epoch')
loss_ax.set_ylabel('loss')
loss_ax.legend(loc='upper left')
acc_ax.plot(hist.history['accuracy'], 'b', label='train acc')
acc_ax.plot(hist.history['val_accuracy'], 'g', label='val acc')
acc_ax.set_ylabel('accuracy')
acc_ax.legend(loc='lower left')
plt.show()
# Financial components만 가지고 각기다른 sample을 가지고 학습을 100번 반복하여 평균 정확도 및 std 계산
acc = []
for count in range(100):
i = random.randint(1,1000)
x_train, x_tmp, y_train, y_tmp = train_test_split(XXX,y_class, test_size=0.2, shuffle=True, random_state=i)
x_val, x_test, y_val, y_test = train_test_split(x_tmp,y_tmp, test_size=0.5, shuffle=True, random_state=i)
keras.backend.clear_session()
inputs = Input(shape=(14,))
hidden1 = Dense(100, activation='swish')(inputs)
hidden2 = Dense(30, activation='relu')(hidden1)
outputs = Dense(4, activation='sigmoid')(hidden2)
model1 = Model(inputs, outputs)
adam = keras.optimizers.Adam(learning_rate=0.00005)
model1.compile(loss='mse', optimizer= adam, metrics=['accuracy'])
hist = model1.fit(x_train, y_train, validation_data = (x_val, y_val),
epochs=30, batch_size=8, verbose=0)
prediction = model1.predict(x_test)
right = 0
for i in range(len(y_test)):
if np.argmax(prediction[i]) == y_test[i]:
right += 1
acc.append((right/len(y_test)))
print("Financial Data's accuracy: {}".format(np.array(acc).mean()))
print("Financial Data's standard deviation: {}".format(np.array(acc).std()))
# Financial components+G Rank만 가지고 각기다른 sample을 가지고 학습을 100번 반복하여 평균 정확도 및 std 계산
acc = []
for count in range(100):
i = random.randint(1,1000)
x_train, x_tmp, y_train, y_tmp = train_test_split(XX,y_class, test_size=0.2, shuffle=True, random_state=i)
x_val, x_test, y_val, y_test = train_test_split(x_tmp,y_tmp, test_size=0.5, shuffle=True, random_state=i)
keras.backend.clear_session()
inputs = Input(shape=(18,))
hidden1 = Dense(100, activation='swish')(inputs)
hidden2 = Dense(30, activation='relu')(hidden1)
outputs = Dense(4, activation='sigmoid')(hidden2)
model1 = Model(inputs, outputs)
adam = keras.optimizers.Adam(learning_rate=0.00005)
model1.compile(loss='mse', optimizer= adam, metrics=['accuracy'])
hist = model1.fit(x_train, y_train, validation_data = (x_val, y_val),
epochs=30, batch_size=8, verbose=0)
prediction = model1.predict(x_test)
right = 0
for i in range(len(y_test)):
if np.argmax(prediction[i]) == y_test[i]:
right += 1
acc.append((right/len(y_test)))
print("Financial Data + G Rank's accuracy: {}".format(np.array(acc).mean()))
print("Financial Data + G Rank's standard deviation: {}".format(np.array(acc).std()))
# Financial components+G Rank만 가지고 각기다른 sample을 가지고 학습을 100번 반복하여 평균 정확도 및 std 계산
acc = []
for count in range(100):
i = random.randint(1,1000)
x_train, x_tmp, y_train, y_tmp = train_test_split(X,y_class, test_size=0.2, shuffle=True, random_state=i)
x_val, x_test, y_val, y_test = train_test_split(x_tmp,y_tmp, test_size=0.5, shuffle=True, random_state=i)
keras.backend.clear_session()
inputs = Input(shape=(19,))
hidden1 = Dense(100, activation='swish')(inputs)
hidden2 = Dense(30, activation='relu')(hidden1)
outputs = Dense(4, activation='sigmoid')(hidden2)
model1 = Model(inputs, outputs)
adam = keras.optimizers.Adam(learning_rate=0.00005)
model1.compile(loss='mse', optimizer= adam, metrics=['accuracy'])
hist = model1.fit(x_train, y_train, validation_data = (x_val, y_val),
epochs=30, batch_size=8, verbose=0)
prediction = model1.predict(x_test)
right = 0
for i in range(len(y_test)):
if np.argmax(prediction[i]) == y_test[i]:
right += 1
acc.append((right/len(y_test)))
print("Overall Data's accuracy: {}".format(np.array(acc).mean()))
print("Overall Data's's standard deviation: {}".format(np.array(acc).std()))
| 0.194597 | 0.470615 |
```
import nltk
from nltk import word_tokenize
from nltk.corpus import wordnet
from tqdm._tqdm_notebook import tqdm_notebook as tqdm
import pandas as pd
import numpy as np
import string
import random
import re
import gensim
import pickle
from nltk.stem import WordNetLemmatizer
word_lemm = WordNetLemmatizer()
from nltk.corpus import stopwords
en_stopwords = stopwords.words('english')
# nltk downloads
nltk.download('vader_lexicon')
nltk.download('stopwords')
nltk.download('averaged_perceptron_tagger')
nltk.download('wordnet')
#initialize tqdm
tqdm.pandas()
DATA_DIR = '../../data/reddit/comments/'
df = pd.read_pickle(DATA_DIR + 'reddit_2019_comments_clean1.pkl')
df['clean'] = df['clean'].progress_apply(lambda x: x.replace('\n\n',' ').replace('\n',' ').replace('\'s','s'))
df_dev = df.sample(1000)
def convert_to_valid_pos(x):
"""Converts the pos tag returned by the nltk.pos_tag function to a format accepted by wordNetLemmatizer"""
x = x[0].upper() # extract first character of the POS tag
# define mapping for the tag to correct tag.
tag_dict = {"J": wordnet.ADJ,
"N": wordnet.NOUN,
"R": wordnet.ADV,
"V": wordnet.VERB}
return tag_dict.get(x, wordnet.NOUN)
def get_lemma(sentence):
"""Given a sentence, derives the lemmatized version of the sentence"""
pos_tagged_text = nltk.pos_tag(word_tokenize(sentence))
lemm_list = []
for (word, tag) in pos_tagged_text:
lemm_list.append(word_lemm.lemmatize(word, pos = convert_to_valid_pos(tag)))
return lemm_list
def prepare_text_for_lda(text):
"""Lemmatizes text, removes stopwords and short words from given text."""
lemm_list = get_lemma(text)
tokens = [i for i in lemm_list if i not in en_stopwords]
tokens = [token for token in tokens if len(token) > 4]
return tokens
df['lemmas'] = df['clean'].progress_map(prepare_text_for_lda)
dictionary = gensim.corpora.Dictionary(df.lemmas)
corpus = list(df['lemmas'].progress_map(dictionary.doc2bow))
pickle.dump(corpus, open(DATA_DIR + 'reddit_2019_corpus.pkl', 'wb'))
dictionary.save(DATA_DIR + 'reddit_2019_dictionary.gensim')
#dictionary2 = gensim.corpora.Dictionary.load(DATA_DIR + 'reddit_2019_dictionary.gensim')
Topic_list = []
num_topics = 20
passes = 15
# https://radimrehurek.com/gensim/models/ldamulticore.html
ldamodel = gensim.models.ldamulticore.LdaMulticore(corpus,
num_topics = num_topics,
id2word = dictionary,
passes=passes,
workers = 3 ) #set this to cores - 1
ldamodel.save(DATA_DIR + 'models/{}_model_{}_{}.gensim'.format(num_topics, passes, 'reddit2019'))
topics = ldamodel.print_topics(num_words = 20)
for topic in topics:
Topic_list.append(topic[1])
import csv
# save the topics for later use.
topic_df = pd.DataFrame({'topics':Topic_list})
def clean_topic_words(x):
"""Clean topic words as output by the algorithm"""
clean_topic = re.findall("\".*?\"", x)
clean_topic = [s.replace('\"', '') for s in clean_topic]
return clean_topic
topic_df['topics'] = topic_df['topics'].map(clean_topic_words)
topic_df.to_csv(DATA_DIR + "topics/Topics_List_{}_model_{}_{}.csv".format(num_topics, passes, 'reddit2019'),
index=False, header=False,
quoting=csv.QUOTE_NONE, sep = '\n', escapechar='\\') # write out for later use
for topic in list(topic_df.topics):
print(topic)
```
|
github_jupyter
|
import nltk
from nltk import word_tokenize
from nltk.corpus import wordnet
from tqdm._tqdm_notebook import tqdm_notebook as tqdm
import pandas as pd
import numpy as np
import string
import random
import re
import gensim
import pickle
from nltk.stem import WordNetLemmatizer
word_lemm = WordNetLemmatizer()
from nltk.corpus import stopwords
en_stopwords = stopwords.words('english')
# nltk downloads
nltk.download('vader_lexicon')
nltk.download('stopwords')
nltk.download('averaged_perceptron_tagger')
nltk.download('wordnet')
#initialize tqdm
tqdm.pandas()
DATA_DIR = '../../data/reddit/comments/'
df = pd.read_pickle(DATA_DIR + 'reddit_2019_comments_clean1.pkl')
df['clean'] = df['clean'].progress_apply(lambda x: x.replace('\n\n',' ').replace('\n',' ').replace('\'s','s'))
df_dev = df.sample(1000)
def convert_to_valid_pos(x):
"""Converts the pos tag returned by the nltk.pos_tag function to a format accepted by wordNetLemmatizer"""
x = x[0].upper() # extract first character of the POS tag
# define mapping for the tag to correct tag.
tag_dict = {"J": wordnet.ADJ,
"N": wordnet.NOUN,
"R": wordnet.ADV,
"V": wordnet.VERB}
return tag_dict.get(x, wordnet.NOUN)
def get_lemma(sentence):
"""Given a sentence, derives the lemmatized version of the sentence"""
pos_tagged_text = nltk.pos_tag(word_tokenize(sentence))
lemm_list = []
for (word, tag) in pos_tagged_text:
lemm_list.append(word_lemm.lemmatize(word, pos = convert_to_valid_pos(tag)))
return lemm_list
def prepare_text_for_lda(text):
"""Lemmatizes text, removes stopwords and short words from given text."""
lemm_list = get_lemma(text)
tokens = [i for i in lemm_list if i not in en_stopwords]
tokens = [token for token in tokens if len(token) > 4]
return tokens
df['lemmas'] = df['clean'].progress_map(prepare_text_for_lda)
dictionary = gensim.corpora.Dictionary(df.lemmas)
corpus = list(df['lemmas'].progress_map(dictionary.doc2bow))
pickle.dump(corpus, open(DATA_DIR + 'reddit_2019_corpus.pkl', 'wb'))
dictionary.save(DATA_DIR + 'reddit_2019_dictionary.gensim')
#dictionary2 = gensim.corpora.Dictionary.load(DATA_DIR + 'reddit_2019_dictionary.gensim')
Topic_list = []
num_topics = 20
passes = 15
# https://radimrehurek.com/gensim/models/ldamulticore.html
ldamodel = gensim.models.ldamulticore.LdaMulticore(corpus,
num_topics = num_topics,
id2word = dictionary,
passes=passes,
workers = 3 ) #set this to cores - 1
ldamodel.save(DATA_DIR + 'models/{}_model_{}_{}.gensim'.format(num_topics, passes, 'reddit2019'))
topics = ldamodel.print_topics(num_words = 20)
for topic in topics:
Topic_list.append(topic[1])
import csv
# save the topics for later use.
topic_df = pd.DataFrame({'topics':Topic_list})
def clean_topic_words(x):
"""Clean topic words as output by the algorithm"""
clean_topic = re.findall("\".*?\"", x)
clean_topic = [s.replace('\"', '') for s in clean_topic]
return clean_topic
topic_df['topics'] = topic_df['topics'].map(clean_topic_words)
topic_df.to_csv(DATA_DIR + "topics/Topics_List_{}_model_{}_{}.csv".format(num_topics, passes, 'reddit2019'),
index=False, header=False,
quoting=csv.QUOTE_NONE, sep = '\n', escapechar='\\') # write out for later use
for topic in list(topic_df.topics):
print(topic)
| 0.421076 | 0.152442 |
```
input = '''x
x
ygzdvpxjk
bzdgq
aulrhfwi
tpasjur
jrutopas
rtjpaus
sraetpju
tpajsru
vus
uvs
vwups
r
r
r
d
j
qohtkgcpl
yukohqglp
dobtpghlkq
hglqpaosrnk
zathopqrnf
tfphoznqra
ornthfpqza
xymf
yfm
yaeflpmc
mxfy
xhjnyblvetrzcsdfgioqw
nyvjcwruaitzkbsfelxhqmo
vqlebsnycaijwxhftzor
pvnbhgqxokzfls
ktfiphrwzyu
snadfrzvj
huravjsfnz
wrfajsvznx
gzqpt
htzqp
pltxqz
dwf
fwvd
fwjxzd
wvkaetlonsmzyxp
flamyzwpnexosvk
ylsoxwnmkpegvazf
xnozlvwayepksmj
hegoanjwcuqr
unjghwraoecqm
vrmuincbeax
mrxcevanu
cmanxeuqvr
xmcrunveqa
rvxcunmea
edz
zgesn
ozqjtrewa
zdefv
czdenf
imnklhe
flsnkj
nkljfv
dtkhezmqarglsyujifncbpx
smyaqrdnjzutcpfkbxghl
nzupmhqkjfblrdasxgcty
ltbdpxsmhzkyjargfcunq
khbnlsgcwdepajfqry
fcglrsedyjkbnwpahz
rngyalswjpecfktbhd
eajkdwshrclpfgnby
eqvgcfpyxdjirlwhkabsn
nsxzlycfmhtkpr
wmxstkfoyrhlzp
jqxfycbl
fjztavkuxeh
mxdfipon
eyrvxswhgfa
yqutg
qytgu
ytugq
tlgquy
uqytg
lsnvemdftqgzk
ethkglvsqdf
edxfsltqgvk
zflnhgtsvkqde
zxqiowjp
ojziqxwp
pxwjizoq
jxfsvcu
scqufxjv
nmkbsdpth
ekqzlgwuyrcapt
bvxfjegzyawqtdpkrihonl
kgdxaywnmbthueqpfjvilo
wukhzt
ztqhipkexwo
zhkwltfu
dosntymrebgxip
vbakzmsqnxehtyg
besntwxymgju
pbsgyntoemx
ke
ke
ek
gkqrxeyhmpwoavszftnid
qablonwmkpridxgcztsehy
ejnuvyrgwkfphld
vyejdomzqcrtwuigs
sxjnructhkifz
umktfhirxnec
iunkcfhryxt
hftciukrnx
kuctixrfhn
m
c
ktwhurxsgza
huntakrzxg
bfiecpvksjzgxmtrhwndy
yewjdrpcfvmxub
kxv
bvkx
mpkdrilqnvgjszxhbceat
qplbdncjxgzvsimakehrt
abwpdvkcmjihleqsrxztgn
ejkotbiyranhvzpgmdsqlcx
upykgj
lkqbapvf
wpzmkr
upkngjw
henpvx
uhnepi
vhpn
unph
gacnoptsyh
va
v
v
v
mfgsejknphquwvtzic
nyfhzpkxqtmlwivsgec
l
l
lr
vneibmqwsuropl
qulpwvhbmirne
priwulqenbmv
uwbznlqrmpviea
nmvqulerpdbiw
zrcedpsjlgavqhkfwyxinobmut
wvzastimlhdyeobxckurfgpnjq
c
w
c
c
eavhgx
axvegh
xghvae
eagvhx
xaehvg
qhvryckj
cyqhvkj
vhyjmzcgqk
inyetbqzdcu
cezuhin
ejaurictfnym
hrngcezuia
xkliecvuwsn
qowvezrdukfbn
zewvyunqoambdr
zngwprusefvdbkqo
euwdbvqnzfro
xrqovndztwueb
klpubivcyfoemgdqzasn
stujebgynqmlxvak
nvmqsubergklwyh
pbsjdzutfgwmxieqcraklhv
bjcexgqskpvolmadnzryftwu
mvxrzwtqaujgpcebdklsf
balwcovj
jwobvalc
bwjaolvc
lbaojcvw
cvbwlajo
onvtx
xotnv
y
ydg
y
fy
ndluot
huotfw
gouqtl
usvy
qf
h
c
c
vjyolxpgqfk
qbugnhjvpx
xtzaprdcvjqigm
idryqgoevjwhpal
jwrledyhvgpaoqi
olgejvpdaqwiyrh
ajehiwrlodvypgq
oylrhqwajevgpid
x
x
p
r
rnqocxehf
yrjmlgdavuwihox
xrqpnkoht
yvackuxdrbespfoz
oyxcabeuszrfvpkd
utxobrfckapdvzsey
eoxsdayfpkucvbzr
jortnzkcqb
bznrjycktqo
zjqnkrtcob
qojktrnbcz
qktojzrncb
uokgp
krui
iqbhupfzg
fqklopwh
qstfhnp
nqykmsphf
jnzpybrvctmgx
pzjbyxmgkcvn
cvxjzgpnbmy
gvynmcxzbpj
zihyt
zihty
zhtyi
ohytiz
tyizh
hrtmfqdnbsjceuxgiakol
lxprsezfuyw
bnpesm
fqhcya
kdoy
hsfj
qecixr
dyzpo
nsviyq
isyqnv
cvqdsfnjyi
qvniys
qvysni
olsytfpiqbcgruavnmkdj
wobyanuximplfgtrc
fakvjrdemphinxzboygcq
rpxbevcfsodljzqmaigkh
osh
hoprni
efzcdv
jxih
p
qhamel
slehaz
mealyh
hela
belahtkn
ertwyfubagxcilm
gfytuxwmrbialec
cmxybwaptedlrufig
uciewrabfxymltg
geurytlicmbawxf
pqosuijf
qfsjiup
ifsrpujq
iv
vi
iv
gvi
vi
gcpsri
tsmvokr
usqagr
svtwqplur
thzwuporavyl
ltupsqrwv
utvwlsprq
apeidznxcr
imgtbyekslu
hseitolv
tfhpmels
haspqmfel
kplfrjymxeh
pthlvmqef
cehdmtfpsl
dj
caj
d
vohrlqx
kuzcdayghrqbtpe
kcpxwmaeruqzhvb
riwuxjg
puwhxdj
blknsvt
jtvyr
tjrvy
vjyrt
vytjr
jvrty
qngcme
qenc
knecpq
psetjy
xvikrzflgdm
yco
wconu
bucqa
kdnt
udnz
dnagt
xdkn
jsikbcef
ksefbi
yeikfbs
snevfqxiakb
wqtgfxruzyd
jrkfxwtzqdub
kemcnsaxjfiubg
mankwgqsuxibcj
sbgkvujnmcxai
fgyixmnkdacbujs
dmrkoujzbvtwsqyx
fqibxwkhmzcertd
jbw
wryg
w
w
y
g
bcfztdseowilyqapun
wpmsdbozlqvciku
xqsbloudprihcwz
intvfbkagzs
krweqxlajmtyu
r
p
zjcogl
pv
fljxyzkgt
ktzlcsigxyf
xygltfzki
lgyxtbkfzj
klmtrzdfegqxhny
ejfndtrmwlyzxqavhcbspoikg
ajnlbfrcswgmdkthpxiqvz
ysikgwrpx
rdmkqgsw
wgrchksjf
wykdasgr
emaizfwdc
adcwsozetmf
m
r
z
knvpqjbasl
jqnsvklbpa
dvksqnbljpa
pawskblnqjv
blakpsqjnfv
phioqsu
gunio
uoi
wlqhkndiuoxtgesrmpfv
wvgsmtolekfhaxnpqui
slu
uisal
fsepbqldiztcwanjohruky
lsbtpoenyvuazqghijfrkcwd
kqzrdphyoacenswumblxtij
zfbadvte
zteda
otazie
eturlyah
gt
tgd
gvta
tg
rjck
ymcv
lctoweb
cmqh
mruchy
iztpec
ipeztc
zceipt
icpzet
ecptzhi
mjubdrwxaitnvcekgslhqpz
ldhvzxbrqjtcmpuiasewk
dkjlrwqbpa
jqlzerbpa
svupyxrbk
grtnawqcybmdhfiz
qzjmpxinvwked
bxfuzhany
snxotzlgc
cfvejdxqrzlhobi
vrojlcdmzebuq
idrjshpbqtevogmy
ysdvpeuqlmhgbioj
pgwybvmosdjiehu
pvdhcseguyomjbi
kixjgompdbnvfhzseya
jgl
gjlez
glj
jgl
zrbxvcuwjidklfmnqotg
lmurpfvqzoxjgwbckndti
umlqjgbxdkrivcntwzfo
wsedxcrblhmotqjkavzfuing
tnqdxjmcrofvbzkyguwil
dmxcntqwauokif
tqmufiwnhk
inzeftquwmvpbjs
iwgfymqrutdn
ejsfxcgdtkhoqu
kfhitexsugdj
tnypmbxl
rxmbnltpy
xnbylmtp
c
ksc
thcpjf
gmucyaxdz
ck
xqizlfeksrjh
wgetzlkx
khnawcubgpx
knctwgupa
kpwngcau
apzwqckgnu
npkawcgu
itlec
lhectib
lctesi
mhanjrlytudgzovf
nzjgdapevrh
inhdrjeazgv
gznhdiajrv
jazvhdrng
itgemupjxbhvs
epsjthguxvimz
jmhxspugetvi
pjhiseuvtgmx
beln
le
xle
elhazjy
ecl
ev
v
ryzfswe
sylrkzc
vezqnasodyjxlfkgiup
vlkyixzeqdbajfruongp
td
tkqpl
tde
lcdkt
tcnkd
okciajvuxdqt
usoyw
owuys
ouswy
swyou
wsyou
xmfovdhgnlcja
qfympwaotzvchrlu
gcx
c
c
cxpanqwbzjeiruogsh
npqobaijrgwsecuh
vjfbmsagotwpyhckeriqu
biosrpqwgljexauch
kriedwmbz
ykhebdsjtz
inblqrgm
rqbimln
nbrqlim
wdlmasxozvngrkuy
zguaymvdolnkwsxr
gvxadrlowkmzunsy
frpv
vprasf
irapxf
unmfpe
icgmloskedvrhfq
xzitwrbjpny
qbvwlyext
qtblesvw
etlwbqv
tebvlw
hpbevwtl
bmnpvz
nbvmoz
nwczgybvlkd
abvztn
bnzve
b
c
b
tchlzb
gz
ndr
rdn
dnr
sctrbhpnmzev
qedsncvaruymg
dncrsemkvaox
yarlncvdwxsem
sfhunci
nsuhifc
siuhcfn
icufhns
gnheqlb
ukpd
lixtrswd
wntldgisx
spclikxftdhwzuejabo
stywildxv
stmlxdiwq
ehsqnvgzw
neszvwgqh
qwzhsevgn
etgcydsljko
xoyiqbumwanhr
hvbqfdkyzupwnctr
jnchwzpsobtkfeqruy
gvzipoldfjx
zodxivgpl
noibqzgxvldp
doqplvgzxi
olxpvdzig
sqri
qirs
nzhagdpqyrtuclsxojbfev
uqznepyjgfoavrthslbcdx
uxztlrfhgevopdbsancqjy
uqrtexnlzvjahcgbdpsfyo
jdhgotuxqvpzsrcfnaylbe
anytx
mgldfs
vn
jpxn
yujnw
zdgnamji
pmjnzgdwbcoi
jynmiguzd
jgrumdntiz
mganjdsiz
jikqaxmebd
uolhrpecyxvda
dafxhtrse
lbjmzepw
barycf
suxb
xuqb
xjkpdbsycqzfml
pszjxdkycmfq
hyqzxfpjctwkdsmn
fqsdkuyxcpjmez
tmxoylwjzas
gqbwlujd
pldwnjibrqe
oxtwegdyarcmufj
dcfuwrgxmytejo
mwutxyorcdgfej
dctrumjwxegyfo
twpyflemcjgourxzdk
ji
ij
hbjzg
czbtgj
bgjz
zjbg
hziwf
jqhgmlybvf
sgukx
jxqts
rbekhvqmjw
wjkmvbrghqye
qowekvthnjrb
jevhqkwbr
wreyhjvqkb
f
f
lfi
b
r
b
nim
ds
dms
nsd
reydi
clbyfjoqg
pzny
wekuiy
yhi
ujqkgvwhybdaxfoz
joaxzvkgbufqhw
zcyhdxriqtolasnwpmfvukg
tpolkuqmhzcyisvndargxfw
umphysgavwqixcdfrlkotzn
wgpruaotvcklnfidmxzyqhs
ubwgxvmkjz
gzkbmuxwvj
jyuxgbkvmwz
xvjkubzmgw
cjbalye
naero
aspe
ache
usfcwigx
wcistfguxz
ishfguxcw
vowtyizedpxjkuhgln
rtacogzuipjydhxlnew
gjmzenuwstlpxqbhyodfi
humbgtvizaqnrekx
urixmqaghktbnezv
geabrihuzvtqnsxdmk
oqdctfig
itmfgcqoyk
ijuoaqxcfthg
slbj
bslj
lbjs
jslb
dyhijmulgn
gcxrkbt
cogkr
l
hl
le
el
l
rd
dr
dr
sx
kzfyajw
cuplmx
yos
ys
jqyta
b
b
b
hwotauyxk
koufjyh
ouhyk
hklouy
sm
s
csig
sgi
esxy
rgukjdhytpmoewicz
dzgwtkejyacros
dwpuknfajbeztsvlcir
nbeilkzmyqswjfdapuvgt
zbvuifewstkadpnjl
lctyng
yncldht
hdusmgynzxba
auzvjdmhxybgns
hbumansdxogzyek
lnshkwetqz
enxhqolgkds
hxczvrwn
xzrwvnfoh
khrwsbeytzil
qr
o
o
hey
krtpbuzgy
buztrpkgy
tupybkrgzq
pyubztkogr
f
fx
m
x
uvbf
awy
odwf
fdwo
wodf
odwf
atnskdpybvlowehzmx
aopknhzybdwtsumlvx
lsbqxonmwgthdaykvzp
zcvgrlkiwdnjxuyeabftosqp
ijhuaexordwgtlqbpsynkcz
rb
b
e
e
p
rgfljswuqmnxdyzve
sdurkno
budprnsac
anruids
s
q
q
xq
x
mkcounerqzvywhb
rhmuwbylqcxzedovn
wcyrzehguqknbvmol
uiosmjcwyfqzvhptrben
cbafotnhw
ztbwievgscufo
kmrqfbwtcoy
ftwcosb
pqbdg
lvyknqjdt
okcdy
durhfwzamsi
edkt
rteknjgdziaxuvqpwms
euqatrgskzjdxpiwmnv
odpxerktavzujqiwsngm
xpajvmrwugsqzdyhitkne
zjpmskatnegrquxwifvd
ucp
puc
sbhrmvgkxlwcuizf
irughftcbsxvkzm
jgufnkhxcrwsqyav
rhjndfxayuqgsvc
wh
hw
hw
wh
hw
oskmgp
pskmorb
zbjxfa
afzujx
lrucpk
bosmyzdf
jxtmurs
kmou
uybvqmc
c
cha
h
t
yj
yj
ubhwxkosgq
giqwkhtuesbox
pwqjdvbzir
ijrzqdub
sjanrzbiq
jizqbry
m
m
um
m
m
hmgxpcyfzenjlavbrtd
yvtxmarbphjoefzgnl
dsmcohpqxzytbefwun
vkglfzxscniymhqurpdtbeoa
b
e
q
dr
rd
dr
dr
peqvdfnayt
qpjcsu
lfq
lqf
lfq
flq
qlf
thnkmpcl
fkiavtnux
tqpkn
zuxche
zcxuhr
xcznhua
xhuzcd
pxuhnczs
vdtesl
ktlmxsf
ekrfgonxblyuqwcmithazvdjp
awlpvtybicnhqdxeofzgjkmru
zdyfgtaoilqwuhkpbexcjvrnm
zutmxljqsbwevaog
jnouplrwkzq
bvkwprm
xiz
i
gdc
if
krm
rm
rmush
opjrnmeb
rmuki
iw
ysw
kl
gtjb
hrndzmf
z
sjcdb
d
cqs
hlot
ehs
yabvz
hjsgx
fgexm
xtjfe
ebfjxh
zsvfopulwnixc
kmqupeigvojczwbadlx
gxmawjupbvkedqlzco
exlzudcqbmvawjgokp
lgkconmhwxyvqtzuedbpaj
guofmwqdjxvezlapcbk
yzvldxhqpcregionbwmast
oxbnyzpseagchiqrtwvmld
ivmargzsdlcwhoxynpqbet
dfeczlatxrgybwpnmvisohq
sbdtompfakqzuw
odwukmshptfizq
pquszdktmifwno
uhgxdiptswayqref
waqdrxpltvufsciyge
lxkvhzeruigsboft
hobvxrieltumgyzfs
bovtrghfxeiulsz
mkxhjuygbvcno
ygoqnubvj
pyzdubtlnsjgvo
bnguvojy
pmuxezk
odpiyevns
iergxuojpqwskbdnyt
dtebiujnospkwyragq
roqbendjksgiwupty
nbhftycoqleusjdwpvgkri
qryzgjekwpubidtosn
hkosdxcnztelmp
ltxcshpodmkenz
aeokdzmhlxnctps
tcohzelnsxdpmk
rlmxzsdchoipenkt
klodupfxjg
dxouflgj
zjfugadelox
qe
eq
qey
nkemqr
kpyahrdgnv
vkrnygdpah
nygrkhdvpa
hngyqkrdcvpa
nhygkpravd
pojt
tjp
tpij
tpj
ptjx
ptudvwcsnbzlkmh
pnvuklmedhwcstzb
stpkbndzwcxulmhv
lshkvqbtnmauwpdzc
lvbkdstmhczunpw
cjki
i
dh
nayubo
zte
lrsyuzkehwmdi
uilhsekdmwr
ehlmdkiuwsr
mlwuiskedhr
remuhcdikwls
nevqwlas
tafkhjrndcsgm
aizsxn
anqlswvy
tdfxwikmqcygoja
pbmnevlsoxkiudhyctqf
uncjptla
snpj
mqnwhueayizvl
zwqlijnymve
pkloymxntbigqwzerdf
nvqyeilmzw
icjmlezwqyn
echgfxinmd
ftcmixeghn
xbikghlcpfnwem
jxcfgedimnh
jehnsmgxcdif
qorpli
ipohrq
pqirko
prioq
vcptybfelzaihduknmwgs
hsugpzyftmeiklbawnvcd
caukmbihneywsdvfztplg
tcziphfsbdkmavwulnyge
kcmfhlgbnaztdsyuvrpwei
yvzouqjilkgm
mgkpqyzotublsevi
iazkvluoygmq
wphvnftyxedckbzq
dbzhwqxnpvfyce
sqebwpydfvncxhz
jdtehlm
rz
rv
bw
wb
bw
btw
bw
esviqco
uceqvxj
qcvywrat
y
qf
y
wdopylavgxi
uomhenqjzbrcf
gyowtsk
hplycvznxtqw
xhlomdpznrcavebti
barocdnhpmgtlk
phrtmbdnacol
rgdomapnhbltc
sdqiauyzgvj
uvayizq
iqyuavz
qauzyvi
uvrs
usv
svu
suv
vsu
zg
mge
y
vcf
gunfyiqemws
esywnimfugq
mfwysnquieg
ag
gab
pga
agd
ga
npgitkbz
vxhzakcj
rkdst
onhzibjuyg
bha
la
iepquktyaxmfhnlobcrwjvds
kmfpuchdwbaxqviojynrstel
jwotpldbxqnkrichseyuvmfa
agunzk
izug
lawqrc
fdo
dp
ptmfuje
umfqjbedtlz
rmwxfjute
utsjmfer
fymhpzavr
lehormypnfwx
hpljurctygmf
upylfqtrdijhbm
ugscvjxfkh
sihzmngcjdv
pbwjhdvlntmxuykzcqf
nmisaefgqhcoz
orcfsxm
xcmfrs
syrxfcm
srfmxc
yjodxnav
smvkfxt
sutnflvk
rgb
b
b
uahjwtqbeklgfzixpocrvmn
kbjlqzxwuratocmfpinhegv
qrijtvkzbsclmefahpwugonx
cnvxwskaithmrpdjzegoufly
yojesiunmzclkvghxwtrdapf
fcazyohpuliwmgdnvsjterxk
padmkvwzgyxtjcselruhofni
thjrfauldyvpcisxngokmewz
jtr
tr
tr
rlt
o
fx
o
o
jviduxrqhpzykm
jrlhykixbv
hkyjilrvx
yhxivkrjl
xryhbkijv
jx
i
rekgmt
hdigybslxfozca
sbypgfvxzihcmlau
caliwbxzfgehy
xwbdprtsoqzuny
smkhxcjfaived
j
j
j
nvwbegrypaofxq
ofvqxeyrgp
klw
lkw
kwl
zyrfuqhnagk
ghfzeswqkc
ekqzohgf
rgumpkoehtndcb
gcdfkipuvbojlmn
uqlivpmac
vplcqimua
upmalcvqi
mcuvlpqai
woi
wio
zycvodbiatfr
boycztjaidfrv
kyimbvawqx
mvwkyjcqaiptr
vhiykmaqw
kqmvxyiwa
vakmqwiy
mb
axhbf
ldpczbr
vhfbym
zvibckofgyjqtw
fmqzswhpeklyjtbvuoi
fjixaznqkywtbvco
vxcnagsqdmhuotwe
envfkxdroqawcgsmty
aspitmqnxwedvcbl
tqsizydbaw
ohejgpk
xvrzbcpogdyfuhwknmiesqa
oigwafzqnmshkevcuxbypdrl
grhcbdxyuewisvazfqpkomn
mnyvbxcsaeoufpwrqzkdhgi
dxzyrsogubncfqtmvepihwka
n
n
n
n
n
bmpkgzfojxelrwtc
qairymcnsuxlokj
dewfscvkqbtzpx
cdpuewltsbzormq
wvtdngphbkeszcqj
disryhxvpbulzw
hbzlwdirsvpyxu
irzldphyubxswv
wihdrypxvuszlb
fxtqpck
xqkptcf
factlhdbwurgvxiekpszqjo
gjhxkdfpsltcvoibruqewaz
tcpfirvxkjwolzasbeguqhd
vjrfpclktdeshquigxbowza
pqabsimryhtd
uvlconjzxgwke
pmyjflwzckdvnergq
zjprwygclekfmqvdn
wykjldrvfqecngmzp
vjfmcynwegzprdklhq
cfqnmrldegyzpwkvj
xh
h
eh
nh
hldjq
kapbqmo
fbakptzq
okpcqbat
xjopbqka
ubpdqrkhga
wyaudlen
eawulynd
wueadlny
uaelndwy
aluewndy
byzxchgujesn
ygzxanucbejh
ehftw
lmvfa
hfsg
wvmfzghpsy
hfm
dhgmjzux
reablkontchmq
gzwqlcx
bgzlctwn
uzcwdpgaxlv
nukwtrszf
rfndkzcsomwu
rusfnwzk
ufzwlrnsk
bv
ai
b
v
v
uxcdomyb
qrwgzvfnai
lhntkrpjsegf
fqxugyni
ifynw
ayigfkn
qynafsgi
yfqcni
yecqrtdzbnhp
hcrzqodnv
qhnrzdcf
p
v
afoxchynvipsel
nsxlcohfpeiyav
sxalyfhpwvoneci
xislpcoyefvnha
najlhzu
zjyclua
tslumedaiq
k
k
k
k
k
odetwpfglrcjqmuihx
dyrqkfweansjvmhglzpu
igskahu
esihkglu
giushk
guhxkis
suknighx
oxemfvj
xomzsqv
mlnxvo
tlwfvk
tlvw
ztvlxcr
vwltf
vwlt
uwmpzlhenva
otzqhxmngylkup
whnufmlzpev
lfupmhnz
vmluszhpn
nqsireakdvpbtfcwgxy
bkqygapisnxfwrtecvodj
enbvtdisqwaxfpgrcyk
xvdbrwgimatefqcyspkn
ajumckognwqs
mcwbsjqtkgfdziro
acpzwt
zlcyaet
gvu
vg
gv
qgcjztkuo
ojbwtrxaq
tecrwjigkqpbnufmldavxh
fxvhgjetlrwdpziukbnc
xkvlhwrtbfydigpsocnje
kqyn
uw
caixsqhpgwvmo
uhxomepiqc
naud
numdat
vuzaednw
nagtdxu
guxdna
xdmkliqsw
kwlqxamti
vdqpmwlxik
mixyslpha
ayihosplx
xytbdakhiplrswz
povsiyhflxa
ynli
boyins
yin
iny
fgjmlirc
rjfilcgm
mohvfnrgzjkuc
ujroykmzfwa
xljiqtkpe
glymhtqpak
etpsklnq
dtpqkl
cefwpgnbamdzj
zyjeaxwumpdcfnbr
vxdhgi
hgdixv
dnxrhqvgim
ixvdghb
pgxihdv
ytbgfrm
mgoty
qjltepmgsy
tfymg
xlgpjbkwqeavsu
exjaugptqlsbkwv
jegvpwluibkaqysx
qwbexsvfprjulz
ugnwqbiyp
gnbqyipu
bygiqnpu
gybnpuqi
tmldrwhgivbnq
wtgdqhvrljb
cvlt
peoxb
rblnmixegdwafuoyq
qgoylwxbarumifedn
arfquebxydimnvclowg
fywzsnhitdcae
tadizcyfsnewh
sawtdziyfngpchx
vnuhpkriz
lrupivknj
hunripvk
vdkuqsrifnp
axdzckpvimueqjtgy
ctpymavjkxzreuqli
pedavgzqxthkcmjuiwy
kxperoiadjgsnfbm
jpikdgrexanmbfos
jrcedfukonpbgixsma
ugo
guos
guo
bmhnfupxotj
bjhftongxyupmz
uxbnmjocpfht
gdx
gk
gc
fvwc
wkqzcfvh
wcfv
vwecf
cvywf
w
w
w
c
eosuimrzpqvyk
kzmicpqvexours
ivrmseztophqkug
kipmquyorvezs
fpmkrd
tzupnklqxfa
ypfgek
jrfcpkbd
ofkgp
qrwoydm
bgtlcxpsh
gawqbjoyrcvizhpmsel
aveozrfgjpismdbcyhwl
rchmbojzqiweplygvsa
zbvclsxrmitehywjoapg
yjhimlberdvscgzpaowu
qawpmhydzn
rfbyw
wgokjfy
pnhbd
qfug
fdep
imowsvtxlzkry
ajg
nquegztm
srnj
wit
tusiv
qmcoftrljdyk
t
hxbsozmwegqjkicufl
likdywshmvxuzcfqb
vtaqrdcfoybz
cfozirq
frkqocz
qfhrzoc
t
t
xjqbomigwrfckhsynva
mqoniwygafbkcrsjvxh
aicnmeoqgvwjyfurxhztsbk
wfrmbcaivnskyhjxqgo
vzad
dvaqz
udazj
dqsvrj
jkcq
ezuqxjm
qdtavj
jtsq
qgawvsxo
cpmaxziubdewlfnt
rjsxaw
hkwxya
lmbopvufrxnw
mvdnioxjpaqeyfrlsgh
nzmtklorpxvf
eqiwxhrvudokt
xumrqhvnswiaky
vupkbiqrmwxnh
hixwqvzgck
ziedvfb
fvbezid
bidzefv
bfezvid
edizbfv
eglq
lgeq
qlge
pabzfmvsxdqoykhurtjl
zyleahvmbocxpkrtfs
g
g
g
gv
mgwxotefbiqjudn
uoniqfdtgxwbjeml
ryzmxkwosabecqvtj
bftceznowiyskpqvjxdm
ecmxqktvoswbzjy
czjhkyvsmwoetbaqx
zqrwuyesmvbcoktjx
ytanuz
byanf
jpoglevikmyhc
wsqdyx
uzwbmdsgafvyk
noqlhcjwx
qxrtpowh
vlsgakpqfjemz
kgvqauiesdjlfnz
fcegqzsjkvla
kglzqajpseyvf
hztmnjip
cfptrw
kaeubgqoxsfiyjtwmhcvzrnl
ikmhteubjczxyvoqsfln
vnxsukmbecqljfidozyth
ushnpjevlkbocxfmiqtyz
mye
ezmy
mye
meysnt
yme
xtvlmhrbdypeaf
bvxftlaephdmyr
dhuxmtryeplfvab
vxrmaybhplfdet
vxrfeyhplbdmta
pek
pe
dpo
gpdfjsaho
hnpo
dpamoy
wvueozqxp
wjdqzpftmhk
vmtod
tbdmg
rytuvcdbm
rtboyemdca
dxmsyfacqoigtuvjnwph
upojvbynselfdzhkwqrt
d
dmo
cymqgxwurzhdbteok
acydrugmobzlxek
xmugryzcobked
cmbyokexuldgzr
uebznq
ubqzen
qunbez
zuqebn
buzeqn
uewpvklzhoqifcsb
rlvdyhnkjptiwxgab
xgdazmoqwislvb
qxodszwlvbgai
azsdiwlgqoxvb
ldzvgwoqsxiba
ozbnvjquac
pgntbvw
aexsklbwjydt
akjbwedsxylt
keaxswdbtjyl
leykdbstjwxqam
xewlkdytasbj
ahzvbeqsctofui
ueabotfhvdzcsrqi
obzhuvteqapscfi
ovhbpuqcslzfieta
vbfhtqszaociue
ugilv
ivlgm
vxigrl
gvli
qjni
qongz
nyqj
sfdqwlvcxn
bcgdtuaxmlkrevof
ftorcabmwueykglxd
tahfxumdobrclzikneg
xakfmdtgocruleb
ihrobnmcpqtgwk
hwkrgmpoctuqni
nwiphcogrkqtm
whrqpktngmycboi
vjiwsothqgnkdrzecmp
fmvhjo
fomhvjq
pkvcijtamu
xijwkrbmluneta
evjomncktgpu
gumxn
mnuyqgfai
luqxgmwn
vgxsetchwpajodzmu
tubxpkqheniygjodwm
zugtaqdsofhpe
tuqpdhgar
ugnoadthq
yvkugjhdaxbcmqt
gthudaq
dzfmjsgaqubntpkvleo
zuwgvfbkarlyxpdihnme
z
z
zp
zb
z
khdj
dhj
hdkjrgz
eofuhjdta
gauozreitvy
oeidrzyv
zlvxcidkob
obdckzlvix
olckbzdxiv
lkxcboidvz
rpslxjnfh
bhvuytmosdzaqwgk
rbjloemzv
zlbeormj
rlmbjezo
djbyqfcoknxsmtzghrp
hksxmbyzgfnpdjqocr
cjodkxfrpgnmbqhyzs
ko
rok
kwo
kgio
ypocjw
hyw
pwy
iyfw
yqwgjv
hyto
hqtko
gw
g
ag
gj
sckdazw
fazodyc
wofxjgyqurha
fhxjboaerg
ztsgrohfjcal
hjgxarfou
rvqyegxakhbfoj
coat
coa
oca
coat
oack
tzcdyrhfuxisvlgm
txuplzrcsjhmyavi
ahcxfelbv
haxlbfevc
owfizxugtsemclrypjvahqdnb
ugncvldopbwqjzmxhirfaet
oupwmqnzilhdtcbavrkxejfg
fjyagkzt
gfyktjza
kayfgzqtj
iutxmzd
dmtuzix
tzmxidu
cndejklaphsquowi
pjocdlehukqai
hixujeqlrktapdco
xcapfuiljwgqsevot
tlfdxuwkjpnqvsmoeg
madurwbn
anwdbqmru
rdwubanm
gl
lanp
wlkijdruy
zlg
hlx
d
da
da
dmwp
e
ey
e
mcxndruabhvp
pnqudhcymb
qxldtifpvshowjbkemza
hvziwsglkjepobfmqaxd
hzxeslvdomagbrwkjifpq
hbxdsfkizqvljwopae
zucafhbkvelxoqipdsnjyw
hoeqg
fqghrt
gqh
v
r
r
iovxlnqsz
iosnqfxzl
ozxinsql
pqsvfxihdmzkrnacobetjluwy
wytcbhvxsrndpafleuiqozjkm
ngi
ogi
ticfga
qg
drl
mzbfwevsodakx
gdwvskin
vdskwli
jienvauxkgfdqh
qdhxjktavizcpnuef
fkbqueiladvjnxh
ukfwjndxhaieqv
gdlrowxahs
lusdcwhyg
dwalghs
hzdgitqeflws
dgkhlws
pmjkz
kzpmvj
pzkjmh
dshpxl
vbtmniafszewdkxup
opsqdxyj
etki
eikt
tiek
iket
ktei
yvfta
atypkqfv'''
groups = input.replace('\n', ' ').replace(' ', '\n').replace(' ', '').split('\n')
# part 1
groups = [set(group) for group in groups]
yes = sum([len(group) for group in groups])
print(yes)
# part 2
groups = input.split('\n\n')
count = 0
for group in groups:
people = group.split('\n')
people = [set(p) for p in people]
inter = people[0].intersection(*(people[1:]))
count += len(inter)
print(count)
```
|
github_jupyter
|
input = '''x
x
ygzdvpxjk
bzdgq
aulrhfwi
tpasjur
jrutopas
rtjpaus
sraetpju
tpajsru
vus
uvs
vwups
r
r
r
d
j
qohtkgcpl
yukohqglp
dobtpghlkq
hglqpaosrnk
zathopqrnf
tfphoznqra
ornthfpqza
xymf
yfm
yaeflpmc
mxfy
xhjnyblvetrzcsdfgioqw
nyvjcwruaitzkbsfelxhqmo
vqlebsnycaijwxhftzor
pvnbhgqxokzfls
ktfiphrwzyu
snadfrzvj
huravjsfnz
wrfajsvznx
gzqpt
htzqp
pltxqz
dwf
fwvd
fwjxzd
wvkaetlonsmzyxp
flamyzwpnexosvk
ylsoxwnmkpegvazf
xnozlvwayepksmj
hegoanjwcuqr
unjghwraoecqm
vrmuincbeax
mrxcevanu
cmanxeuqvr
xmcrunveqa
rvxcunmea
edz
zgesn
ozqjtrewa
zdefv
czdenf
imnklhe
flsnkj
nkljfv
dtkhezmqarglsyujifncbpx
smyaqrdnjzutcpfkbxghl
nzupmhqkjfblrdasxgcty
ltbdpxsmhzkyjargfcunq
khbnlsgcwdepajfqry
fcglrsedyjkbnwpahz
rngyalswjpecfktbhd
eajkdwshrclpfgnby
eqvgcfpyxdjirlwhkabsn
nsxzlycfmhtkpr
wmxstkfoyrhlzp
jqxfycbl
fjztavkuxeh
mxdfipon
eyrvxswhgfa
yqutg
qytgu
ytugq
tlgquy
uqytg
lsnvemdftqgzk
ethkglvsqdf
edxfsltqgvk
zflnhgtsvkqde
zxqiowjp
ojziqxwp
pxwjizoq
jxfsvcu
scqufxjv
nmkbsdpth
ekqzlgwuyrcapt
bvxfjegzyawqtdpkrihonl
kgdxaywnmbthueqpfjvilo
wukhzt
ztqhipkexwo
zhkwltfu
dosntymrebgxip
vbakzmsqnxehtyg
besntwxymgju
pbsgyntoemx
ke
ke
ek
gkqrxeyhmpwoavszftnid
qablonwmkpridxgcztsehy
ejnuvyrgwkfphld
vyejdomzqcrtwuigs
sxjnructhkifz
umktfhirxnec
iunkcfhryxt
hftciukrnx
kuctixrfhn
m
c
ktwhurxsgza
huntakrzxg
bfiecpvksjzgxmtrhwndy
yewjdrpcfvmxub
kxv
bvkx
mpkdrilqnvgjszxhbceat
qplbdncjxgzvsimakehrt
abwpdvkcmjihleqsrxztgn
ejkotbiyranhvzpgmdsqlcx
upykgj
lkqbapvf
wpzmkr
upkngjw
henpvx
uhnepi
vhpn
unph
gacnoptsyh
va
v
v
v
mfgsejknphquwvtzic
nyfhzpkxqtmlwivsgec
l
l
lr
vneibmqwsuropl
qulpwvhbmirne
priwulqenbmv
uwbznlqrmpviea
nmvqulerpdbiw
zrcedpsjlgavqhkfwyxinobmut
wvzastimlhdyeobxckurfgpnjq
c
w
c
c
eavhgx
axvegh
xghvae
eagvhx
xaehvg
qhvryckj
cyqhvkj
vhyjmzcgqk
inyetbqzdcu
cezuhin
ejaurictfnym
hrngcezuia
xkliecvuwsn
qowvezrdukfbn
zewvyunqoambdr
zngwprusefvdbkqo
euwdbvqnzfro
xrqovndztwueb
klpubivcyfoemgdqzasn
stujebgynqmlxvak
nvmqsubergklwyh
pbsjdzutfgwmxieqcraklhv
bjcexgqskpvolmadnzryftwu
mvxrzwtqaujgpcebdklsf
balwcovj
jwobvalc
bwjaolvc
lbaojcvw
cvbwlajo
onvtx
xotnv
y
ydg
y
fy
ndluot
huotfw
gouqtl
usvy
qf
h
c
c
vjyolxpgqfk
qbugnhjvpx
xtzaprdcvjqigm
idryqgoevjwhpal
jwrledyhvgpaoqi
olgejvpdaqwiyrh
ajehiwrlodvypgq
oylrhqwajevgpid
x
x
p
r
rnqocxehf
yrjmlgdavuwihox
xrqpnkoht
yvackuxdrbespfoz
oyxcabeuszrfvpkd
utxobrfckapdvzsey
eoxsdayfpkucvbzr
jortnzkcqb
bznrjycktqo
zjqnkrtcob
qojktrnbcz
qktojzrncb
uokgp
krui
iqbhupfzg
fqklopwh
qstfhnp
nqykmsphf
jnzpybrvctmgx
pzjbyxmgkcvn
cvxjzgpnbmy
gvynmcxzbpj
zihyt
zihty
zhtyi
ohytiz
tyizh
hrtmfqdnbsjceuxgiakol
lxprsezfuyw
bnpesm
fqhcya
kdoy
hsfj
qecixr
dyzpo
nsviyq
isyqnv
cvqdsfnjyi
qvniys
qvysni
olsytfpiqbcgruavnmkdj
wobyanuximplfgtrc
fakvjrdemphinxzboygcq
rpxbevcfsodljzqmaigkh
osh
hoprni
efzcdv
jxih
p
qhamel
slehaz
mealyh
hela
belahtkn
ertwyfubagxcilm
gfytuxwmrbialec
cmxybwaptedlrufig
uciewrabfxymltg
geurytlicmbawxf
pqosuijf
qfsjiup
ifsrpujq
iv
vi
iv
gvi
vi
gcpsri
tsmvokr
usqagr
svtwqplur
thzwuporavyl
ltupsqrwv
utvwlsprq
apeidznxcr
imgtbyekslu
hseitolv
tfhpmels
haspqmfel
kplfrjymxeh
pthlvmqef
cehdmtfpsl
dj
caj
d
vohrlqx
kuzcdayghrqbtpe
kcpxwmaeruqzhvb
riwuxjg
puwhxdj
blknsvt
jtvyr
tjrvy
vjyrt
vytjr
jvrty
qngcme
qenc
knecpq
psetjy
xvikrzflgdm
yco
wconu
bucqa
kdnt
udnz
dnagt
xdkn
jsikbcef
ksefbi
yeikfbs
snevfqxiakb
wqtgfxruzyd
jrkfxwtzqdub
kemcnsaxjfiubg
mankwgqsuxibcj
sbgkvujnmcxai
fgyixmnkdacbujs
dmrkoujzbvtwsqyx
fqibxwkhmzcertd
jbw
wryg
w
w
y
g
bcfztdseowilyqapun
wpmsdbozlqvciku
xqsbloudprihcwz
intvfbkagzs
krweqxlajmtyu
r
p
zjcogl
pv
fljxyzkgt
ktzlcsigxyf
xygltfzki
lgyxtbkfzj
klmtrzdfegqxhny
ejfndtrmwlyzxqavhcbspoikg
ajnlbfrcswgmdkthpxiqvz
ysikgwrpx
rdmkqgsw
wgrchksjf
wykdasgr
emaizfwdc
adcwsozetmf
m
r
z
knvpqjbasl
jqnsvklbpa
dvksqnbljpa
pawskblnqjv
blakpsqjnfv
phioqsu
gunio
uoi
wlqhkndiuoxtgesrmpfv
wvgsmtolekfhaxnpqui
slu
uisal
fsepbqldiztcwanjohruky
lsbtpoenyvuazqghijfrkcwd
kqzrdphyoacenswumblxtij
zfbadvte
zteda
otazie
eturlyah
gt
tgd
gvta
tg
rjck
ymcv
lctoweb
cmqh
mruchy
iztpec
ipeztc
zceipt
icpzet
ecptzhi
mjubdrwxaitnvcekgslhqpz
ldhvzxbrqjtcmpuiasewk
dkjlrwqbpa
jqlzerbpa
svupyxrbk
grtnawqcybmdhfiz
qzjmpxinvwked
bxfuzhany
snxotzlgc
cfvejdxqrzlhobi
vrojlcdmzebuq
idrjshpbqtevogmy
ysdvpeuqlmhgbioj
pgwybvmosdjiehu
pvdhcseguyomjbi
kixjgompdbnvfhzseya
jgl
gjlez
glj
jgl
zrbxvcuwjidklfmnqotg
lmurpfvqzoxjgwbckndti
umlqjgbxdkrivcntwzfo
wsedxcrblhmotqjkavzfuing
tnqdxjmcrofvbzkyguwil
dmxcntqwauokif
tqmufiwnhk
inzeftquwmvpbjs
iwgfymqrutdn
ejsfxcgdtkhoqu
kfhitexsugdj
tnypmbxl
rxmbnltpy
xnbylmtp
c
ksc
thcpjf
gmucyaxdz
ck
xqizlfeksrjh
wgetzlkx
khnawcubgpx
knctwgupa
kpwngcau
apzwqckgnu
npkawcgu
itlec
lhectib
lctesi
mhanjrlytudgzovf
nzjgdapevrh
inhdrjeazgv
gznhdiajrv
jazvhdrng
itgemupjxbhvs
epsjthguxvimz
jmhxspugetvi
pjhiseuvtgmx
beln
le
xle
elhazjy
ecl
ev
v
ryzfswe
sylrkzc
vezqnasodyjxlfkgiup
vlkyixzeqdbajfruongp
td
tkqpl
tde
lcdkt
tcnkd
okciajvuxdqt
usoyw
owuys
ouswy
swyou
wsyou
xmfovdhgnlcja
qfympwaotzvchrlu
gcx
c
c
cxpanqwbzjeiruogsh
npqobaijrgwsecuh
vjfbmsagotwpyhckeriqu
biosrpqwgljexauch
kriedwmbz
ykhebdsjtz
inblqrgm
rqbimln
nbrqlim
wdlmasxozvngrkuy
zguaymvdolnkwsxr
gvxadrlowkmzunsy
frpv
vprasf
irapxf
unmfpe
icgmloskedvrhfq
xzitwrbjpny
qbvwlyext
qtblesvw
etlwbqv
tebvlw
hpbevwtl
bmnpvz
nbvmoz
nwczgybvlkd
abvztn
bnzve
b
c
b
tchlzb
gz
ndr
rdn
dnr
sctrbhpnmzev
qedsncvaruymg
dncrsemkvaox
yarlncvdwxsem
sfhunci
nsuhifc
siuhcfn
icufhns
gnheqlb
ukpd
lixtrswd
wntldgisx
spclikxftdhwzuejabo
stywildxv
stmlxdiwq
ehsqnvgzw
neszvwgqh
qwzhsevgn
etgcydsljko
xoyiqbumwanhr
hvbqfdkyzupwnctr
jnchwzpsobtkfeqruy
gvzipoldfjx
zodxivgpl
noibqzgxvldp
doqplvgzxi
olxpvdzig
sqri
qirs
nzhagdpqyrtuclsxojbfev
uqznepyjgfoavrthslbcdx
uxztlrfhgevopdbsancqjy
uqrtexnlzvjahcgbdpsfyo
jdhgotuxqvpzsrcfnaylbe
anytx
mgldfs
vn
jpxn
yujnw
zdgnamji
pmjnzgdwbcoi
jynmiguzd
jgrumdntiz
mganjdsiz
jikqaxmebd
uolhrpecyxvda
dafxhtrse
lbjmzepw
barycf
suxb
xuqb
xjkpdbsycqzfml
pszjxdkycmfq
hyqzxfpjctwkdsmn
fqsdkuyxcpjmez
tmxoylwjzas
gqbwlujd
pldwnjibrqe
oxtwegdyarcmufj
dcfuwrgxmytejo
mwutxyorcdgfej
dctrumjwxegyfo
twpyflemcjgourxzdk
ji
ij
hbjzg
czbtgj
bgjz
zjbg
hziwf
jqhgmlybvf
sgukx
jxqts
rbekhvqmjw
wjkmvbrghqye
qowekvthnjrb
jevhqkwbr
wreyhjvqkb
f
f
lfi
b
r
b
nim
ds
dms
nsd
reydi
clbyfjoqg
pzny
wekuiy
yhi
ujqkgvwhybdaxfoz
joaxzvkgbufqhw
zcyhdxriqtolasnwpmfvukg
tpolkuqmhzcyisvndargxfw
umphysgavwqixcdfrlkotzn
wgpruaotvcklnfidmxzyqhs
ubwgxvmkjz
gzkbmuxwvj
jyuxgbkvmwz
xvjkubzmgw
cjbalye
naero
aspe
ache
usfcwigx
wcistfguxz
ishfguxcw
vowtyizedpxjkuhgln
rtacogzuipjydhxlnew
gjmzenuwstlpxqbhyodfi
humbgtvizaqnrekx
urixmqaghktbnezv
geabrihuzvtqnsxdmk
oqdctfig
itmfgcqoyk
ijuoaqxcfthg
slbj
bslj
lbjs
jslb
dyhijmulgn
gcxrkbt
cogkr
l
hl
le
el
l
rd
dr
dr
sx
kzfyajw
cuplmx
yos
ys
jqyta
b
b
b
hwotauyxk
koufjyh
ouhyk
hklouy
sm
s
csig
sgi
esxy
rgukjdhytpmoewicz
dzgwtkejyacros
dwpuknfajbeztsvlcir
nbeilkzmyqswjfdapuvgt
zbvuifewstkadpnjl
lctyng
yncldht
hdusmgynzxba
auzvjdmhxybgns
hbumansdxogzyek
lnshkwetqz
enxhqolgkds
hxczvrwn
xzrwvnfoh
khrwsbeytzil
qr
o
o
hey
krtpbuzgy
buztrpkgy
tupybkrgzq
pyubztkogr
f
fx
m
x
uvbf
awy
odwf
fdwo
wodf
odwf
atnskdpybvlowehzmx
aopknhzybdwtsumlvx
lsbqxonmwgthdaykvzp
zcvgrlkiwdnjxuyeabftosqp
ijhuaexordwgtlqbpsynkcz
rb
b
e
e
p
rgfljswuqmnxdyzve
sdurkno
budprnsac
anruids
s
q
q
xq
x
mkcounerqzvywhb
rhmuwbylqcxzedovn
wcyrzehguqknbvmol
uiosmjcwyfqzvhptrben
cbafotnhw
ztbwievgscufo
kmrqfbwtcoy
ftwcosb
pqbdg
lvyknqjdt
okcdy
durhfwzamsi
edkt
rteknjgdziaxuvqpwms
euqatrgskzjdxpiwmnv
odpxerktavzujqiwsngm
xpajvmrwugsqzdyhitkne
zjpmskatnegrquxwifvd
ucp
puc
sbhrmvgkxlwcuizf
irughftcbsxvkzm
jgufnkhxcrwsqyav
rhjndfxayuqgsvc
wh
hw
hw
wh
hw
oskmgp
pskmorb
zbjxfa
afzujx
lrucpk
bosmyzdf
jxtmurs
kmou
uybvqmc
c
cha
h
t
yj
yj
ubhwxkosgq
giqwkhtuesbox
pwqjdvbzir
ijrzqdub
sjanrzbiq
jizqbry
m
m
um
m
m
hmgxpcyfzenjlavbrtd
yvtxmarbphjoefzgnl
dsmcohpqxzytbefwun
vkglfzxscniymhqurpdtbeoa
b
e
q
dr
rd
dr
dr
peqvdfnayt
qpjcsu
lfq
lqf
lfq
flq
qlf
thnkmpcl
fkiavtnux
tqpkn
zuxche
zcxuhr
xcznhua
xhuzcd
pxuhnczs
vdtesl
ktlmxsf
ekrfgonxblyuqwcmithazvdjp
awlpvtybicnhqdxeofzgjkmru
zdyfgtaoilqwuhkpbexcjvrnm
zutmxljqsbwevaog
jnouplrwkzq
bvkwprm
xiz
i
gdc
if
krm
rm
rmush
opjrnmeb
rmuki
iw
ysw
kl
gtjb
hrndzmf
z
sjcdb
d
cqs
hlot
ehs
yabvz
hjsgx
fgexm
xtjfe
ebfjxh
zsvfopulwnixc
kmqupeigvojczwbadlx
gxmawjupbvkedqlzco
exlzudcqbmvawjgokp
lgkconmhwxyvqtzuedbpaj
guofmwqdjxvezlapcbk
yzvldxhqpcregionbwmast
oxbnyzpseagchiqrtwvmld
ivmargzsdlcwhoxynpqbet
dfeczlatxrgybwpnmvisohq
sbdtompfakqzuw
odwukmshptfizq
pquszdktmifwno
uhgxdiptswayqref
waqdrxpltvufsciyge
lxkvhzeruigsboft
hobvxrieltumgyzfs
bovtrghfxeiulsz
mkxhjuygbvcno
ygoqnubvj
pyzdubtlnsjgvo
bnguvojy
pmuxezk
odpiyevns
iergxuojpqwskbdnyt
dtebiujnospkwyragq
roqbendjksgiwupty
nbhftycoqleusjdwpvgkri
qryzgjekwpubidtosn
hkosdxcnztelmp
ltxcshpodmkenz
aeokdzmhlxnctps
tcohzelnsxdpmk
rlmxzsdchoipenkt
klodupfxjg
dxouflgj
zjfugadelox
qe
eq
qey
nkemqr
kpyahrdgnv
vkrnygdpah
nygrkhdvpa
hngyqkrdcvpa
nhygkpravd
pojt
tjp
tpij
tpj
ptjx
ptudvwcsnbzlkmh
pnvuklmedhwcstzb
stpkbndzwcxulmhv
lshkvqbtnmauwpdzc
lvbkdstmhczunpw
cjki
i
dh
nayubo
zte
lrsyuzkehwmdi
uilhsekdmwr
ehlmdkiuwsr
mlwuiskedhr
remuhcdikwls
nevqwlas
tafkhjrndcsgm
aizsxn
anqlswvy
tdfxwikmqcygoja
pbmnevlsoxkiudhyctqf
uncjptla
snpj
mqnwhueayizvl
zwqlijnymve
pkloymxntbigqwzerdf
nvqyeilmzw
icjmlezwqyn
echgfxinmd
ftcmixeghn
xbikghlcpfnwem
jxcfgedimnh
jehnsmgxcdif
qorpli
ipohrq
pqirko
prioq
vcptybfelzaihduknmwgs
hsugpzyftmeiklbawnvcd
caukmbihneywsdvfztplg
tcziphfsbdkmavwulnyge
kcmfhlgbnaztdsyuvrpwei
yvzouqjilkgm
mgkpqyzotublsevi
iazkvluoygmq
wphvnftyxedckbzq
dbzhwqxnpvfyce
sqebwpydfvncxhz
jdtehlm
rz
rv
bw
wb
bw
btw
bw
esviqco
uceqvxj
qcvywrat
y
qf
y
wdopylavgxi
uomhenqjzbrcf
gyowtsk
hplycvznxtqw
xhlomdpznrcavebti
barocdnhpmgtlk
phrtmbdnacol
rgdomapnhbltc
sdqiauyzgvj
uvayizq
iqyuavz
qauzyvi
uvrs
usv
svu
suv
vsu
zg
mge
y
vcf
gunfyiqemws
esywnimfugq
mfwysnquieg
ag
gab
pga
agd
ga
npgitkbz
vxhzakcj
rkdst
onhzibjuyg
bha
la
iepquktyaxmfhnlobcrwjvds
kmfpuchdwbaxqviojynrstel
jwotpldbxqnkrichseyuvmfa
agunzk
izug
lawqrc
fdo
dp
ptmfuje
umfqjbedtlz
rmwxfjute
utsjmfer
fymhpzavr
lehormypnfwx
hpljurctygmf
upylfqtrdijhbm
ugscvjxfkh
sihzmngcjdv
pbwjhdvlntmxuykzcqf
nmisaefgqhcoz
orcfsxm
xcmfrs
syrxfcm
srfmxc
yjodxnav
smvkfxt
sutnflvk
rgb
b
b
uahjwtqbeklgfzixpocrvmn
kbjlqzxwuratocmfpinhegv
qrijtvkzbsclmefahpwugonx
cnvxwskaithmrpdjzegoufly
yojesiunmzclkvghxwtrdapf
fcazyohpuliwmgdnvsjterxk
padmkvwzgyxtjcselruhofni
thjrfauldyvpcisxngokmewz
jtr
tr
tr
rlt
o
fx
o
o
jviduxrqhpzykm
jrlhykixbv
hkyjilrvx
yhxivkrjl
xryhbkijv
jx
i
rekgmt
hdigybslxfozca
sbypgfvxzihcmlau
caliwbxzfgehy
xwbdprtsoqzuny
smkhxcjfaived
j
j
j
nvwbegrypaofxq
ofvqxeyrgp
klw
lkw
kwl
zyrfuqhnagk
ghfzeswqkc
ekqzohgf
rgumpkoehtndcb
gcdfkipuvbojlmn
uqlivpmac
vplcqimua
upmalcvqi
mcuvlpqai
woi
wio
zycvodbiatfr
boycztjaidfrv
kyimbvawqx
mvwkyjcqaiptr
vhiykmaqw
kqmvxyiwa
vakmqwiy
mb
axhbf
ldpczbr
vhfbym
zvibckofgyjqtw
fmqzswhpeklyjtbvuoi
fjixaznqkywtbvco
vxcnagsqdmhuotwe
envfkxdroqawcgsmty
aspitmqnxwedvcbl
tqsizydbaw
ohejgpk
xvrzbcpogdyfuhwknmiesqa
oigwafzqnmshkevcuxbypdrl
grhcbdxyuewisvazfqpkomn
mnyvbxcsaeoufpwrqzkdhgi
dxzyrsogubncfqtmvepihwka
n
n
n
n
n
bmpkgzfojxelrwtc
qairymcnsuxlokj
dewfscvkqbtzpx
cdpuewltsbzormq
wvtdngphbkeszcqj
disryhxvpbulzw
hbzlwdirsvpyxu
irzldphyubxswv
wihdrypxvuszlb
fxtqpck
xqkptcf
factlhdbwurgvxiekpszqjo
gjhxkdfpsltcvoibruqewaz
tcpfirvxkjwolzasbeguqhd
vjrfpclktdeshquigxbowza
pqabsimryhtd
uvlconjzxgwke
pmyjflwzckdvnergq
zjprwygclekfmqvdn
wykjldrvfqecngmzp
vjfmcynwegzprdklhq
cfqnmrldegyzpwkvj
xh
h
eh
nh
hldjq
kapbqmo
fbakptzq
okpcqbat
xjopbqka
ubpdqrkhga
wyaudlen
eawulynd
wueadlny
uaelndwy
aluewndy
byzxchgujesn
ygzxanucbejh
ehftw
lmvfa
hfsg
wvmfzghpsy
hfm
dhgmjzux
reablkontchmq
gzwqlcx
bgzlctwn
uzcwdpgaxlv
nukwtrszf
rfndkzcsomwu
rusfnwzk
ufzwlrnsk
bv
ai
b
v
v
uxcdomyb
qrwgzvfnai
lhntkrpjsegf
fqxugyni
ifynw
ayigfkn
qynafsgi
yfqcni
yecqrtdzbnhp
hcrzqodnv
qhnrzdcf
p
v
afoxchynvipsel
nsxlcohfpeiyav
sxalyfhpwvoneci
xislpcoyefvnha
najlhzu
zjyclua
tslumedaiq
k
k
k
k
k
odetwpfglrcjqmuihx
dyrqkfweansjvmhglzpu
igskahu
esihkglu
giushk
guhxkis
suknighx
oxemfvj
xomzsqv
mlnxvo
tlwfvk
tlvw
ztvlxcr
vwltf
vwlt
uwmpzlhenva
otzqhxmngylkup
whnufmlzpev
lfupmhnz
vmluszhpn
nqsireakdvpbtfcwgxy
bkqygapisnxfwrtecvodj
enbvtdisqwaxfpgrcyk
xvdbrwgimatefqcyspkn
ajumckognwqs
mcwbsjqtkgfdziro
acpzwt
zlcyaet
gvu
vg
gv
qgcjztkuo
ojbwtrxaq
tecrwjigkqpbnufmldavxh
fxvhgjetlrwdpziukbnc
xkvlhwrtbfydigpsocnje
kqyn
uw
caixsqhpgwvmo
uhxomepiqc
naud
numdat
vuzaednw
nagtdxu
guxdna
xdmkliqsw
kwlqxamti
vdqpmwlxik
mixyslpha
ayihosplx
xytbdakhiplrswz
povsiyhflxa
ynli
boyins
yin
iny
fgjmlirc
rjfilcgm
mohvfnrgzjkuc
ujroykmzfwa
xljiqtkpe
glymhtqpak
etpsklnq
dtpqkl
cefwpgnbamdzj
zyjeaxwumpdcfnbr
vxdhgi
hgdixv
dnxrhqvgim
ixvdghb
pgxihdv
ytbgfrm
mgoty
qjltepmgsy
tfymg
xlgpjbkwqeavsu
exjaugptqlsbkwv
jegvpwluibkaqysx
qwbexsvfprjulz
ugnwqbiyp
gnbqyipu
bygiqnpu
gybnpuqi
tmldrwhgivbnq
wtgdqhvrljb
cvlt
peoxb
rblnmixegdwafuoyq
qgoylwxbarumifedn
arfquebxydimnvclowg
fywzsnhitdcae
tadizcyfsnewh
sawtdziyfngpchx
vnuhpkriz
lrupivknj
hunripvk
vdkuqsrifnp
axdzckpvimueqjtgy
ctpymavjkxzreuqli
pedavgzqxthkcmjuiwy
kxperoiadjgsnfbm
jpikdgrexanmbfos
jrcedfukonpbgixsma
ugo
guos
guo
bmhnfupxotj
bjhftongxyupmz
uxbnmjocpfht
gdx
gk
gc
fvwc
wkqzcfvh
wcfv
vwecf
cvywf
w
w
w
c
eosuimrzpqvyk
kzmicpqvexours
ivrmseztophqkug
kipmquyorvezs
fpmkrd
tzupnklqxfa
ypfgek
jrfcpkbd
ofkgp
qrwoydm
bgtlcxpsh
gawqbjoyrcvizhpmsel
aveozrfgjpismdbcyhwl
rchmbojzqiweplygvsa
zbvclsxrmitehywjoapg
yjhimlberdvscgzpaowu
qawpmhydzn
rfbyw
wgokjfy
pnhbd
qfug
fdep
imowsvtxlzkry
ajg
nquegztm
srnj
wit
tusiv
qmcoftrljdyk
t
hxbsozmwegqjkicufl
likdywshmvxuzcfqb
vtaqrdcfoybz
cfozirq
frkqocz
qfhrzoc
t
t
xjqbomigwrfckhsynva
mqoniwygafbkcrsjvxh
aicnmeoqgvwjyfurxhztsbk
wfrmbcaivnskyhjxqgo
vzad
dvaqz
udazj
dqsvrj
jkcq
ezuqxjm
qdtavj
jtsq
qgawvsxo
cpmaxziubdewlfnt
rjsxaw
hkwxya
lmbopvufrxnw
mvdnioxjpaqeyfrlsgh
nzmtklorpxvf
eqiwxhrvudokt
xumrqhvnswiaky
vupkbiqrmwxnh
hixwqvzgck
ziedvfb
fvbezid
bidzefv
bfezvid
edizbfv
eglq
lgeq
qlge
pabzfmvsxdqoykhurtjl
zyleahvmbocxpkrtfs
g
g
g
gv
mgwxotefbiqjudn
uoniqfdtgxwbjeml
ryzmxkwosabecqvtj
bftceznowiyskpqvjxdm
ecmxqktvoswbzjy
czjhkyvsmwoetbaqx
zqrwuyesmvbcoktjx
ytanuz
byanf
jpoglevikmyhc
wsqdyx
uzwbmdsgafvyk
noqlhcjwx
qxrtpowh
vlsgakpqfjemz
kgvqauiesdjlfnz
fcegqzsjkvla
kglzqajpseyvf
hztmnjip
cfptrw
kaeubgqoxsfiyjtwmhcvzrnl
ikmhteubjczxyvoqsfln
vnxsukmbecqljfidozyth
ushnpjevlkbocxfmiqtyz
mye
ezmy
mye
meysnt
yme
xtvlmhrbdypeaf
bvxftlaephdmyr
dhuxmtryeplfvab
vxrmaybhplfdet
vxrfeyhplbdmta
pek
pe
dpo
gpdfjsaho
hnpo
dpamoy
wvueozqxp
wjdqzpftmhk
vmtod
tbdmg
rytuvcdbm
rtboyemdca
dxmsyfacqoigtuvjnwph
upojvbynselfdzhkwqrt
d
dmo
cymqgxwurzhdbteok
acydrugmobzlxek
xmugryzcobked
cmbyokexuldgzr
uebznq
ubqzen
qunbez
zuqebn
buzeqn
uewpvklzhoqifcsb
rlvdyhnkjptiwxgab
xgdazmoqwislvb
qxodszwlvbgai
azsdiwlgqoxvb
ldzvgwoqsxiba
ozbnvjquac
pgntbvw
aexsklbwjydt
akjbwedsxylt
keaxswdbtjyl
leykdbstjwxqam
xewlkdytasbj
ahzvbeqsctofui
ueabotfhvdzcsrqi
obzhuvteqapscfi
ovhbpuqcslzfieta
vbfhtqszaociue
ugilv
ivlgm
vxigrl
gvli
qjni
qongz
nyqj
sfdqwlvcxn
bcgdtuaxmlkrevof
ftorcabmwueykglxd
tahfxumdobrclzikneg
xakfmdtgocruleb
ihrobnmcpqtgwk
hwkrgmpoctuqni
nwiphcogrkqtm
whrqpktngmycboi
vjiwsothqgnkdrzecmp
fmvhjo
fomhvjq
pkvcijtamu
xijwkrbmluneta
evjomncktgpu
gumxn
mnuyqgfai
luqxgmwn
vgxsetchwpajodzmu
tubxpkqheniygjodwm
zugtaqdsofhpe
tuqpdhgar
ugnoadthq
yvkugjhdaxbcmqt
gthudaq
dzfmjsgaqubntpkvleo
zuwgvfbkarlyxpdihnme
z
z
zp
zb
z
khdj
dhj
hdkjrgz
eofuhjdta
gauozreitvy
oeidrzyv
zlvxcidkob
obdckzlvix
olckbzdxiv
lkxcboidvz
rpslxjnfh
bhvuytmosdzaqwgk
rbjloemzv
zlbeormj
rlmbjezo
djbyqfcoknxsmtzghrp
hksxmbyzgfnpdjqocr
cjodkxfrpgnmbqhyzs
ko
rok
kwo
kgio
ypocjw
hyw
pwy
iyfw
yqwgjv
hyto
hqtko
gw
g
ag
gj
sckdazw
fazodyc
wofxjgyqurha
fhxjboaerg
ztsgrohfjcal
hjgxarfou
rvqyegxakhbfoj
coat
coa
oca
coat
oack
tzcdyrhfuxisvlgm
txuplzrcsjhmyavi
ahcxfelbv
haxlbfevc
owfizxugtsemclrypjvahqdnb
ugncvldopbwqjzmxhirfaet
oupwmqnzilhdtcbavrkxejfg
fjyagkzt
gfyktjza
kayfgzqtj
iutxmzd
dmtuzix
tzmxidu
cndejklaphsquowi
pjocdlehukqai
hixujeqlrktapdco
xcapfuiljwgqsevot
tlfdxuwkjpnqvsmoeg
madurwbn
anwdbqmru
rdwubanm
gl
lanp
wlkijdruy
zlg
hlx
d
da
da
dmwp
e
ey
e
mcxndruabhvp
pnqudhcymb
qxldtifpvshowjbkemza
hvziwsglkjepobfmqaxd
hzxeslvdomagbrwkjifpq
hbxdsfkizqvljwopae
zucafhbkvelxoqipdsnjyw
hoeqg
fqghrt
gqh
v
r
r
iovxlnqsz
iosnqfxzl
ozxinsql
pqsvfxihdmzkrnacobetjluwy
wytcbhvxsrndpafleuiqozjkm
ngi
ogi
ticfga
qg
drl
mzbfwevsodakx
gdwvskin
vdskwli
jienvauxkgfdqh
qdhxjktavizcpnuef
fkbqueiladvjnxh
ukfwjndxhaieqv
gdlrowxahs
lusdcwhyg
dwalghs
hzdgitqeflws
dgkhlws
pmjkz
kzpmvj
pzkjmh
dshpxl
vbtmniafszewdkxup
opsqdxyj
etki
eikt
tiek
iket
ktei
yvfta
atypkqfv'''
groups = input.replace('\n', ' ').replace(' ', '\n').replace(' ', '').split('\n')
# part 1
groups = [set(group) for group in groups]
yes = sum([len(group) for group in groups])
print(yes)
# part 2
groups = input.split('\n\n')
count = 0
for group in groups:
people = group.split('\n')
people = [set(p) for p in people]
inter = people[0].intersection(*(people[1:]))
count += len(inter)
print(count)
| 0.244273 | 0.157558 |
# SQL Data munging example
In this exercise, we will experiment with data in a sqlite database using pandas data queries.
## Slide example
Start by running the command from the slides. The following 2 cells setup the database.
```
!rm new.db
!sqlite3 new.db "create table customer \
(cid numeric, \
cust_name char(20), \
address varchar(256), \
primary key (cid));"
!sqlite3 new.db "create table product \
(pid numeric, \
prod_name char(20), \
price numeric, \
primary key (pid));"
!sqlite3 new.db "create table order_n \
(oid numeric, \
pid numeric references product, \
cid numeric references customer, \
quantity numeric, \
Primary key (oid));"
!sqlite3 new.db 'insert into customer values (1,"Joe Klein","USA");'
!sqlite3 new.db 'insert into customer values (2,"Rob Smith","CAN");'
!sqlite3 new.db 'insert into product values (1,"Pencil",1.23);'
!sqlite3 new.db 'insert into product values (2,"Pen",0.67);'
!sqlite3 new.db 'insert into product values (3,"Marker",1.03);'
!sqlite3 new.db 'insert into order_n values (1,2,1,13);'
!sqlite3 new.db 'insert into order_n values (2,3,2,45);'
```
## Experiments
In in the cell below, experiment with the commands from the slides.
```
# !sqlite3 new.db 'select...'
```
## Bank Data
Now we are going to experiment with more analytical data.
### Download data
```
wget http://www.fdic.gov/bank/individual/failed/banklist.csv
```
### Import into database
#### From command line
```
sqlite3 banks.db
```
#### Inside SQL terminal
```
.mode csv
.import banklist.csv bank
.schema bank
select * from bank limit 10;
```
```
!wget http://www.fdic.gov/bank/individual/failed/banklist.csv
!sqlite3 banks.db ".mode csv" ".import banklist.csv bank"
!sqlite3 banks.db ".schema bank"
# !sqlite3 banks.db "select ...
%matplotlib inline
import matplotlib.pyplot as plt
import pandas as pd
import sqlite3
```
## Db connection in python
Read through the API doc at https://docs.python.org/2/library/sqlite3.html and use that to:
1. List 10 banks
2. List all Chicago Banks
3. List all Wyoming Banks
```
# conn = sqlite3.connect...
```
## List 20 banks
```
# results = ...
print(results)
```
## List Chicago Banks
```
# results = ...
print(results)
```
## List Wyoming Banks
```
# results = ...
print(results)
```
## Use Pandas API to pull in Table data
Read API docs at https://pandas.pydata.org/pandas-docs/stable/generated/pandas.read_sql.html.
Use `pd.read_sql` to pull in Bank data as a DataFrame
```
# df = pd.read_sql_query(...
df.head()
```
## Using Closing Date column to make datetime index
Use https://pandas.pydata.org/pandas-docs/stable/generated/pandas.to_datetime.html and https://pandas.pydata.org/pandas-docs/version/0.22/generated/pandas.DatetimeIndex.html. You may have to "coerce" any errors.
```
# df.index = pd.DatetimeIndex(
# pd.to_datetime(...
df.head()
```
## Plot Monthly Bank Failures
See https://pandas.pydata.org/pandas-docs/stable/generated/pandas.Grouper.html for guidance.
```
# df.groupby(...
```
## Joining data
Let's say we want to analyze the bank failure with respect to the state population. For instance, we might want to understand the failure rate per person in a state. To do this we need to "join" the failure data with a table of state populations.
We can use the data from https://en.wikipedia.org/wiki/List_of_U.S._states_and_territories_by_population and https://www.infoplease.com/state-abbreviations-and-state-postal-codes to create a flat table with bank closures and population.
```
df_state_codes = pd.read_html(
'https://www.infoplease.com/state-abbreviations-and-state-postal-codes')[0]
df_state_codes
df_state_pop = pd.read_html(
'https://en.wikipedia.org/wiki/List_of_U.S._states_and_territories_by_population',
header=0)[0]
df_state_pop = df_state_pop[['Population estimate, July 1, 2017[4]', 'State or territory']].rename(columns={
'Population estimate, July 1, 2017[4]': 'population', 'State or territory':'State/District'
})
df_state_pop['population'] = pd.to_numeric(df_state_pop['population'],errors='coerce')
df_state_pop['population'].dtype
df_pop_codes = df_state_codes.merge(df_state_pop,on='State/District')
df_pop_codes.head()
```
## Use a similar process to join with bank df
```
# df_banks_pop = df_pop_codes.merge(...
# df_banks_pop.index = pd.DatetimeIndex(pd.to_datetime(...
df_banks_pop.head()
```
## Group by year and state and compute the failures per 1M people per year
```
# annual = df_banks_pop.groupby(...).agg({'population':'mean', 'Bank Name':len})
annual['fail_per_cap'] = (annual['Bank Name']/annual['population'])*1000000
annual.reset_index(inplace=True)
annual.head()
```
# Use seaborn to plot all states by year
See https://seaborn.pydata.org/generated/seaborn.FacetGrid.html
```
import seaborn as sns
# g = sns.FacetGrid(...
# g.map(...
```
## Bonus repeat join using sql
1. Create join tables as csv
2. Load to sqlite
3. Join using sqlite
4. Extract back to Pandas
|
github_jupyter
|
!rm new.db
!sqlite3 new.db "create table customer \
(cid numeric, \
cust_name char(20), \
address varchar(256), \
primary key (cid));"
!sqlite3 new.db "create table product \
(pid numeric, \
prod_name char(20), \
price numeric, \
primary key (pid));"
!sqlite3 new.db "create table order_n \
(oid numeric, \
pid numeric references product, \
cid numeric references customer, \
quantity numeric, \
Primary key (oid));"
!sqlite3 new.db 'insert into customer values (1,"Joe Klein","USA");'
!sqlite3 new.db 'insert into customer values (2,"Rob Smith","CAN");'
!sqlite3 new.db 'insert into product values (1,"Pencil",1.23);'
!sqlite3 new.db 'insert into product values (2,"Pen",0.67);'
!sqlite3 new.db 'insert into product values (3,"Marker",1.03);'
!sqlite3 new.db 'insert into order_n values (1,2,1,13);'
!sqlite3 new.db 'insert into order_n values (2,3,2,45);'
# !sqlite3 new.db 'select...'
wget http://www.fdic.gov/bank/individual/failed/banklist.csv
sqlite3 banks.db
.mode csv
.import banklist.csv bank
.schema bank
select * from bank limit 10;
!wget http://www.fdic.gov/bank/individual/failed/banklist.csv
!sqlite3 banks.db ".mode csv" ".import banklist.csv bank"
!sqlite3 banks.db ".schema bank"
# !sqlite3 banks.db "select ...
%matplotlib inline
import matplotlib.pyplot as plt
import pandas as pd
import sqlite3
# conn = sqlite3.connect...
# results = ...
print(results)
# results = ...
print(results)
# results = ...
print(results)
# df = pd.read_sql_query(...
df.head()
# df.index = pd.DatetimeIndex(
# pd.to_datetime(...
df.head()
# df.groupby(...
df_state_codes = pd.read_html(
'https://www.infoplease.com/state-abbreviations-and-state-postal-codes')[0]
df_state_codes
df_state_pop = pd.read_html(
'https://en.wikipedia.org/wiki/List_of_U.S._states_and_territories_by_population',
header=0)[0]
df_state_pop = df_state_pop[['Population estimate, July 1, 2017[4]', 'State or territory']].rename(columns={
'Population estimate, July 1, 2017[4]': 'population', 'State or territory':'State/District'
})
df_state_pop['population'] = pd.to_numeric(df_state_pop['population'],errors='coerce')
df_state_pop['population'].dtype
df_pop_codes = df_state_codes.merge(df_state_pop,on='State/District')
df_pop_codes.head()
# df_banks_pop = df_pop_codes.merge(...
# df_banks_pop.index = pd.DatetimeIndex(pd.to_datetime(...
df_banks_pop.head()
# annual = df_banks_pop.groupby(...).agg({'population':'mean', 'Bank Name':len})
annual['fail_per_cap'] = (annual['Bank Name']/annual['population'])*1000000
annual.reset_index(inplace=True)
annual.head()
import seaborn as sns
# g = sns.FacetGrid(...
# g.map(...
| 0.352425 | 0.894144 |
<img src='https://training.dwit.edu.np/frontend/images/computer-training-institute.png'>
<h1>Data Science and Machine learning in Python</h1>
<h3>Instructor: <a href='https://www.kaggle.com/atishadhikari'> Atish Adhikari</a></h3>
<hr>
```
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.datasets import load_iris
from sklearn.preprocessing import OneHotEncoder, StandardScaler
from sklearn.model_selection import train_test_split
from sklearn.metrics import classification_report, accuracy_score
X = load_iris().data
y = load_iris().target
y_cat = OneHotEncoder(sparse=False).fit_transform(y.reshape(-1, 1))
X_train, X_test, y_train, y_test = train_test_split(X, y_cat, test_size=0.2)
from tensorflow.keras.models import Sequential
from tensorflow.keras.optimizers import Nadam
from tensorflow.keras.layers import Dense, Dropout
model = Sequential()
model.add(Dense(input_dim=4, units=8, activation="tanh"))
model.add(Dropout(0.2))
model.add(Dense(units=8, activation="tanh"))
model.add(Dropout(0.2))
model.add(Dense(units=3, activation="softmax"))
model.compile(loss="categorical_crossentropy", optimizer=Nadam(), metrics=["accuracy"])
model.summary()
model.fit(X_train, y_train, epochs=500, validation_data=(X_test, y_test))
from tensorflow.keras.datasets import mnist, fashion_mnist
(X_train, y_train), (X_test, y_test) = mnist.load_data()
X_test.shape
plt.imshow(X_train[10], cmap=plt.cm.binary)
plt.show()
28*28
X_train = X_train.reshape(60000, 28*28)
X_test = X_test.reshape(10000, 28*28)
y_train_cat = OneHotEncoder(sparse=False).fit_transform(y_train.reshape(-1, 1))
y_test_cat = OneHotEncoder(sparse=False).fit_transform(y_test.reshape(-1, 1))
model = Sequential()
model.add( Dense(input_dim=784, units=32, activation="tanh"))
model.add(Dropout(0.2))
model.add( Dense(units=32, activation="tanh"))
model.add(Dropout(0.2))
model.add( Dense(units=16, activation="tanh"))
model.add(Dropout(0.2))
model.add( Dense(units=10, activation="softmax"))
model.compile(loss="categorical_crossentropy", optimizer="adam", metrics=["accuracy"])
model.summary()
model.fit(X_train, y_train_cat, validation_split=0.1, epochs=20)
y_pred = model.predict(X_test)
y_pred[0].round(2)
def plot_image(i, y_pred, y_test_class, img, class_names):
y_pred, y_test_class, img = y_pred[i], y_test_class[i], img[i]
plt.grid(False)
plt.xticks([])
plt.yticks([])
plt.imshow(img, cmap=plt.cm.binary)
predicted_label = np.argmax(y_pred)
if predicted_label == y_test_class:
color = 'blue'
else:
color = 'red'
plt.xlabel("{} {:2.0f}% ({})".format(class_names[predicted_label],
100*np.max(y_pred),
class_names[y_test_class]),
color=color)
class_names = [str(x) for x in range(10)]
num_rows = 5
num_cols = 8
num_images = num_rows*num_cols
plt.figure(figsize=(2*num_cols, 2*num_rows))
for i in range(num_images):
plt.subplot(num_rows, num_cols, i+1)
plot_image(i, y_pred, y_test, X_test.reshape(10000, 28, 28), class_names)
plt.show()
```
|
github_jupyter
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.datasets import load_iris
from sklearn.preprocessing import OneHotEncoder, StandardScaler
from sklearn.model_selection import train_test_split
from sklearn.metrics import classification_report, accuracy_score
X = load_iris().data
y = load_iris().target
y_cat = OneHotEncoder(sparse=False).fit_transform(y.reshape(-1, 1))
X_train, X_test, y_train, y_test = train_test_split(X, y_cat, test_size=0.2)
from tensorflow.keras.models import Sequential
from tensorflow.keras.optimizers import Nadam
from tensorflow.keras.layers import Dense, Dropout
model = Sequential()
model.add(Dense(input_dim=4, units=8, activation="tanh"))
model.add(Dropout(0.2))
model.add(Dense(units=8, activation="tanh"))
model.add(Dropout(0.2))
model.add(Dense(units=3, activation="softmax"))
model.compile(loss="categorical_crossentropy", optimizer=Nadam(), metrics=["accuracy"])
model.summary()
model.fit(X_train, y_train, epochs=500, validation_data=(X_test, y_test))
from tensorflow.keras.datasets import mnist, fashion_mnist
(X_train, y_train), (X_test, y_test) = mnist.load_data()
X_test.shape
plt.imshow(X_train[10], cmap=plt.cm.binary)
plt.show()
28*28
X_train = X_train.reshape(60000, 28*28)
X_test = X_test.reshape(10000, 28*28)
y_train_cat = OneHotEncoder(sparse=False).fit_transform(y_train.reshape(-1, 1))
y_test_cat = OneHotEncoder(sparse=False).fit_transform(y_test.reshape(-1, 1))
model = Sequential()
model.add( Dense(input_dim=784, units=32, activation="tanh"))
model.add(Dropout(0.2))
model.add( Dense(units=32, activation="tanh"))
model.add(Dropout(0.2))
model.add( Dense(units=16, activation="tanh"))
model.add(Dropout(0.2))
model.add( Dense(units=10, activation="softmax"))
model.compile(loss="categorical_crossentropy", optimizer="adam", metrics=["accuracy"])
model.summary()
model.fit(X_train, y_train_cat, validation_split=0.1, epochs=20)
y_pred = model.predict(X_test)
y_pred[0].round(2)
def plot_image(i, y_pred, y_test_class, img, class_names):
y_pred, y_test_class, img = y_pred[i], y_test_class[i], img[i]
plt.grid(False)
plt.xticks([])
plt.yticks([])
plt.imshow(img, cmap=plt.cm.binary)
predicted_label = np.argmax(y_pred)
if predicted_label == y_test_class:
color = 'blue'
else:
color = 'red'
plt.xlabel("{} {:2.0f}% ({})".format(class_names[predicted_label],
100*np.max(y_pred),
class_names[y_test_class]),
color=color)
class_names = [str(x) for x in range(10)]
num_rows = 5
num_cols = 8
num_images = num_rows*num_cols
plt.figure(figsize=(2*num_cols, 2*num_rows))
for i in range(num_images):
plt.subplot(num_rows, num_cols, i+1)
plot_image(i, y_pred, y_test, X_test.reshape(10000, 28, 28), class_names)
plt.show()
| 0.810666 | 0.97066 |
```
number_of_dimensions = 3
from coremltools.models.nearest_neighbors import KNearestNeighborsClassifierBuilder
builder = KNearestNeighborsClassifierBuilder(input_name='input',
output_name='output',
number_of_dimensions=number_of_dimensions,
default_class_label='00000',
number_of_neighbors=3,
weighting_scheme='inverse_distance',
index_type='linear')
builder.author = 'Willie Wu'
builder.license = 'MIT'
builder.description = 'Classifies {} dimension vector based on 3 nearest neighbors'.format(number_of_dimensions)
builder.spec.description.input[0].shortDescription = 'Input vector to classify'
builder.spec.description.output[0].shortDescription = 'Predicted label. Defaults to \'00000\''
builder.spec.description.output[1].shortDescription = 'Probabilities / score for each possible label.'
builder.spec.description.trainingInput[0].shortDescription = 'Example input vector'
builder.spec.description.trainingInput[1].shortDescription = 'Associated true label of each example vector'
import numpy as np
def give_random(high, target_percentile):
return int(np.random.normal(high*target_percentile, 15))
def give_random_all(target_percentile):
#weed, tree, grass
return [give_random(500, target_percentile),
give_random(1500, target_percentile),
give_random(200, target_percentile)]
# add_samples(data_points, labels)
# Add some samples to the KNearestNeighborsClassifier model
# :param data_points: List of input data points
# :param labels: List of corresponding labels
# :return: None
data = []
labels = []
user = {"Never": 0, "Rarely": 1, "Occasionally": 2, "Often": 3}
severity_options = [str(i) for i in list(range(2))]
for i in severity_options:
for j in severity_options:
for k in severity_options:
for h in severity_options:
for l in severity_options:
tot = int(i) + int(j) + int(k) + int(h)+ int(l)
percentile = (tot/5) /15
fuzzed = give_random_all(percentile)
labels.append(i+j+k+h+l)
data.append(fuzzed)
builder.add_samples(data, labels)
print(builder.is_updatable)
mlmodel_updatable_path = './UpdatableKNN.mlmodel'
# Save the updated spec
from coremltools.models import MLModel
mlmodel_updatable = MLModel(builder.spec)
mlmodel_updatable.save(mlmodel_updatable_path)
```
|
github_jupyter
|
number_of_dimensions = 3
from coremltools.models.nearest_neighbors import KNearestNeighborsClassifierBuilder
builder = KNearestNeighborsClassifierBuilder(input_name='input',
output_name='output',
number_of_dimensions=number_of_dimensions,
default_class_label='00000',
number_of_neighbors=3,
weighting_scheme='inverse_distance',
index_type='linear')
builder.author = 'Willie Wu'
builder.license = 'MIT'
builder.description = 'Classifies {} dimension vector based on 3 nearest neighbors'.format(number_of_dimensions)
builder.spec.description.input[0].shortDescription = 'Input vector to classify'
builder.spec.description.output[0].shortDescription = 'Predicted label. Defaults to \'00000\''
builder.spec.description.output[1].shortDescription = 'Probabilities / score for each possible label.'
builder.spec.description.trainingInput[0].shortDescription = 'Example input vector'
builder.spec.description.trainingInput[1].shortDescription = 'Associated true label of each example vector'
import numpy as np
def give_random(high, target_percentile):
return int(np.random.normal(high*target_percentile, 15))
def give_random_all(target_percentile):
#weed, tree, grass
return [give_random(500, target_percentile),
give_random(1500, target_percentile),
give_random(200, target_percentile)]
# add_samples(data_points, labels)
# Add some samples to the KNearestNeighborsClassifier model
# :param data_points: List of input data points
# :param labels: List of corresponding labels
# :return: None
data = []
labels = []
user = {"Never": 0, "Rarely": 1, "Occasionally": 2, "Often": 3}
severity_options = [str(i) for i in list(range(2))]
for i in severity_options:
for j in severity_options:
for k in severity_options:
for h in severity_options:
for l in severity_options:
tot = int(i) + int(j) + int(k) + int(h)+ int(l)
percentile = (tot/5) /15
fuzzed = give_random_all(percentile)
labels.append(i+j+k+h+l)
data.append(fuzzed)
builder.add_samples(data, labels)
print(builder.is_updatable)
mlmodel_updatable_path = './UpdatableKNN.mlmodel'
# Save the updated spec
from coremltools.models import MLModel
mlmodel_updatable = MLModel(builder.spec)
mlmodel_updatable.save(mlmodel_updatable_path)
| 0.715523 | 0.652363 |
```
import wandb
import nltk
from nltk.stem.porter import *
from torch.nn import *
from torch.optim import *
import numpy as np
import pandas as pd
import torch,torchvision
import random
from tqdm import *
from torch.utils.data import Dataset,DataLoader
stemmer = PorterStemmer()
PROJECT_NAME = 'Twitter-Sentiment-Analysis-V2'
device = 'cuda'
def tokenize(sentence):
return nltk.word_tokenize(sentence)
tokenize('$41000')
def stem(word):
return stemmer.stem(word.lower())
stem('organic')
def bag_of_words(tokenied_words,all_words):
tokenied_words = [stem(w) for w in tokenied_words]
bag = np.zeros(len(all_words))
for idx,w in enumerate(all_words):
if w in tokenied_words:
bag[idx] = 1.0
return bag
bag_of_words(['hi'],['how','hi','how'])
data = pd.read_csv('./data.csv').dropna()[:5000]
data
data['Positive'].value_counts()
X = data['im getting on borderlands and i will murder you all ,']
y = data['Positive']
words = []
data = []
idx = 0
labels = {}
labels_r = {}
for label in tqdm(y):
if label not in list(labels.keys()):
idx += 1
labels[label] = idx
labels_r[idx] = label
for X_batch,y_batch in tqdm(zip(X,y)):
X_batch = tokenize(X_batch)
new_X = []
for Xb in X_batch:
new_X.append(stem(Xb))
words.extend(new_X)
data.append([
new_X,
np.eye(labels[y_batch]+1,len(labels))[labels[y_batch]]
])
words = sorted(set(words))
np.random.shuffle(data)
X = []
y = []
for sentence,tag in tqdm(data):
X.append(bag_of_words(sentence,words))
y.append(tag)
from sklearn.model_selection import *
X_train,X_test,y_train,y_test = train_test_split(X,y,test_size=0.125,shuffle=False)
X_train = torch.from_numpy(np.array(X_train)).to(device).float()
y_train = torch.from_numpy(np.array(y_train)).to(device).float()
X_test = torch.from_numpy(np.array(X_test)).to(device).float()
y_test = torch.from_numpy(np.array(y_test)).to(device).float()
def get_loss(model,X,y,criterion):
preds = model(X)
loss = criterion(preds,y)
return loss.item()
def get_accuracy(model,X,y):
preds = model(X)
correct = 0
total = 0
for pred,yb in zip(preds,y):
pred = int(torch.argmax(pred))
yb = int(torch.argmax(yb))
if pred == yb:
correct += 1
total += 1
acc = round(correct/total,3)*100
return acc
labels
class Model(Module):
def __init__(self):
super().__init__()
self.activation = ReLU()
self.iters = 10
self.linear1 = Linear(len(words),512)
self.linear2 = Linear(512,512)
self.linear2bn = BatchNorm1d(512)
self.output = Linear(512,len(labels))
def forward(self,X):
preds = self.linear1(X)
for _ in range(self.iters):
preds = self.activation(self.linear2bn(self.linear2(preds)))
preds = self.output(preds)
return preds
model = Model().to(device)
criterion = MSELoss()
optimizer = Adam(model.parameters(),lr=0.001)
epochs = 100
batch_size = 32
wandb.init(project=PROJECT_NAME,name='baseline')
for _ in tqdm(range(epochs)):
for i in range(0,len(X_train),batch_size):
X_batch = X_train[i:i+batch_size]
y_batch = y_train[i:i+batch_size]
preds = model(X_batch)
loss = criterion(preds,y_batch)
optimizer.zero_grad()
loss.backward()
optimizer.step()
model.eval()
torch.cuda.empty_cache()
wandb.log({'Loss':(get_loss(model,X_train,y_train,criterion)+get_loss(model,X_batch,y_batch,criterion)/2)})
torch.cuda.empty_cache()
wandb.log({'Val Loss':get_loss(model,X_test,y_test,criterion)})
torch.cuda.empty_cache()
wandb.log({'Acc':(get_accuracy(model,X_train,y_train)+get_accuracy(model,X_batch,y_batch))/2})
torch.cuda.empty_cache()
wandb.log({'Val Acc':get_accuracy(model,X_test,y_test)})
torch.cuda.empty_cache()
model.train()
wandb.finish()
torch.cuda.empty_cache()
torch.save(model,'model.pt')
torch.save(model,'model.pth')
torch.save(model.state_dict(),'model-sd.pt')
torch.save(model.state_dict(),'model-sd.pth')
torch.save(words,'words.pt')
torch.save(words,'words.pth')
torch.save(data,'data.pt')
torch.save(data,'data.pth')
torch.save(labels,'labels.pt')
torch.save(labels,'labels.pth')
torch.save(idx,'idx.pt')
torch.save(idx,'idx.pth')
torch.save(y_train,'y_train.pt')
torch.save(y_test,'y_test.pth')
```
|
github_jupyter
|
import wandb
import nltk
from nltk.stem.porter import *
from torch.nn import *
from torch.optim import *
import numpy as np
import pandas as pd
import torch,torchvision
import random
from tqdm import *
from torch.utils.data import Dataset,DataLoader
stemmer = PorterStemmer()
PROJECT_NAME = 'Twitter-Sentiment-Analysis-V2'
device = 'cuda'
def tokenize(sentence):
return nltk.word_tokenize(sentence)
tokenize('$41000')
def stem(word):
return stemmer.stem(word.lower())
stem('organic')
def bag_of_words(tokenied_words,all_words):
tokenied_words = [stem(w) for w in tokenied_words]
bag = np.zeros(len(all_words))
for idx,w in enumerate(all_words):
if w in tokenied_words:
bag[idx] = 1.0
return bag
bag_of_words(['hi'],['how','hi','how'])
data = pd.read_csv('./data.csv').dropna()[:5000]
data
data['Positive'].value_counts()
X = data['im getting on borderlands and i will murder you all ,']
y = data['Positive']
words = []
data = []
idx = 0
labels = {}
labels_r = {}
for label in tqdm(y):
if label not in list(labels.keys()):
idx += 1
labels[label] = idx
labels_r[idx] = label
for X_batch,y_batch in tqdm(zip(X,y)):
X_batch = tokenize(X_batch)
new_X = []
for Xb in X_batch:
new_X.append(stem(Xb))
words.extend(new_X)
data.append([
new_X,
np.eye(labels[y_batch]+1,len(labels))[labels[y_batch]]
])
words = sorted(set(words))
np.random.shuffle(data)
X = []
y = []
for sentence,tag in tqdm(data):
X.append(bag_of_words(sentence,words))
y.append(tag)
from sklearn.model_selection import *
X_train,X_test,y_train,y_test = train_test_split(X,y,test_size=0.125,shuffle=False)
X_train = torch.from_numpy(np.array(X_train)).to(device).float()
y_train = torch.from_numpy(np.array(y_train)).to(device).float()
X_test = torch.from_numpy(np.array(X_test)).to(device).float()
y_test = torch.from_numpy(np.array(y_test)).to(device).float()
def get_loss(model,X,y,criterion):
preds = model(X)
loss = criterion(preds,y)
return loss.item()
def get_accuracy(model,X,y):
preds = model(X)
correct = 0
total = 0
for pred,yb in zip(preds,y):
pred = int(torch.argmax(pred))
yb = int(torch.argmax(yb))
if pred == yb:
correct += 1
total += 1
acc = round(correct/total,3)*100
return acc
labels
class Model(Module):
def __init__(self):
super().__init__()
self.activation = ReLU()
self.iters = 10
self.linear1 = Linear(len(words),512)
self.linear2 = Linear(512,512)
self.linear2bn = BatchNorm1d(512)
self.output = Linear(512,len(labels))
def forward(self,X):
preds = self.linear1(X)
for _ in range(self.iters):
preds = self.activation(self.linear2bn(self.linear2(preds)))
preds = self.output(preds)
return preds
model = Model().to(device)
criterion = MSELoss()
optimizer = Adam(model.parameters(),lr=0.001)
epochs = 100
batch_size = 32
wandb.init(project=PROJECT_NAME,name='baseline')
for _ in tqdm(range(epochs)):
for i in range(0,len(X_train),batch_size):
X_batch = X_train[i:i+batch_size]
y_batch = y_train[i:i+batch_size]
preds = model(X_batch)
loss = criterion(preds,y_batch)
optimizer.zero_grad()
loss.backward()
optimizer.step()
model.eval()
torch.cuda.empty_cache()
wandb.log({'Loss':(get_loss(model,X_train,y_train,criterion)+get_loss(model,X_batch,y_batch,criterion)/2)})
torch.cuda.empty_cache()
wandb.log({'Val Loss':get_loss(model,X_test,y_test,criterion)})
torch.cuda.empty_cache()
wandb.log({'Acc':(get_accuracy(model,X_train,y_train)+get_accuracy(model,X_batch,y_batch))/2})
torch.cuda.empty_cache()
wandb.log({'Val Acc':get_accuracy(model,X_test,y_test)})
torch.cuda.empty_cache()
model.train()
wandb.finish()
torch.cuda.empty_cache()
torch.save(model,'model.pt')
torch.save(model,'model.pth')
torch.save(model.state_dict(),'model-sd.pt')
torch.save(model.state_dict(),'model-sd.pth')
torch.save(words,'words.pt')
torch.save(words,'words.pth')
torch.save(data,'data.pt')
torch.save(data,'data.pth')
torch.save(labels,'labels.pt')
torch.save(labels,'labels.pth')
torch.save(idx,'idx.pt')
torch.save(idx,'idx.pth')
torch.save(y_train,'y_train.pt')
torch.save(y_test,'y_test.pth')
| 0.680348 | 0.246318 |
# Developing an AI application
Going forward, AI algorithms will be incorporated into more and more everyday applications. For example, you might want to include an image classifier in a smart phone app. To do this, you'd use a deep learning model trained on hundreds of thousands of images as part of the overall application architecture. A large part of software development in the future will be using these types of models as common parts of applications.
In this project, you'll train an image classifier to recognize different species of flowers. You can imagine using something like this in a phone app that tells you the name of the flower your camera is looking at. In practice you'd train this classifier, then export it for use in your application. We'll be using [this dataset](http://www.robots.ox.ac.uk/~vgg/data/flowers/102/index.html) of 102 flower categories, you can see a few examples below.
<img src='assets/Flowers.png' width=500px>
The project is broken down into multiple steps:
* Load and preprocess the image dataset
* Train the image classifier on your dataset
* Use the trained classifier to predict image content
We'll lead you through each part which you'll implement in Python.
When you've completed this project, you'll have an application that can be trained on any set of labeled images. Here your network will be learning about flowers and end up as a command line application. But, what you do with your new skills depends on your imagination and effort in building a dataset. For example, imagine an app where you take a picture of a car, it tells you what the make and model is, then looks up information about it. Go build your own dataset and make something new.
First up is importing the packages you'll need. It's good practice to keep all the imports at the beginning of your code. As you work through this notebook and find you need to import a package, make sure to add the import up here.
```
# Imports here
import torch
from torch import nn
from torch import optim
import torch.nn.functional as F
from torchvision import datasets, transforms, models
from collections import OrderedDict
import numpy as np
import pandas as pd
from PIL import Image
import matplotlib.pyplot as plt
import seaborn as sb
%matplotlib inline
```
## Load the data
Here you'll use `torchvision` to load the data ([documentation](http://pytorch.org/docs/0.3.0/torchvision/index.html)). The data should be included alongside this notebook, otherwise you can [download it here](https://s3.amazonaws.com/content.udacity-data.com/nd089/flower_data.tar.gz). The dataset is split into three parts, training, validation, and testing. For the training, you'll want to apply transformations such as random scaling, cropping, and flipping. This will help the network generalize leading to better performance. You'll also need to make sure the input data is resized to 224x224 pixels as required by the pre-trained networks.
The validation and testing sets are used to measure the model's performance on data it hasn't seen yet. For this you don't want any scaling or rotation transformations, but you'll need to resize then crop the images to the appropriate size.
The pre-trained networks you'll use were trained on the ImageNet dataset where each color channel was normalized separately. For all three sets you'll need to normalize the means and standard deviations of the images to what the network expects. For the means, it's `[0.485, 0.456, 0.406]` and for the standard deviations `[0.229, 0.224, 0.225]`, calculated from the ImageNet images. These values will shift each color channel to be centered at 0 and range from -1 to 1.
```
data_dir = 'flowers'
train_dir = data_dir + '/train'
valid_dir = data_dir + '/valid'
test_dir = data_dir + '/test'
# TODO: Define your transforms for the training, validation, and testing sets
data_transforms = {'train': transforms.Compose([transforms.RandomRotation(45),
transforms.RandomResizedCrop(224),
transforms.RandomHorizontalFlip(),transforms.ToTensor(),
transforms.Normalize([0.485, 0.456, 0.406], [0.229, 0.224, 0.225])]),
'valid_test': transforms.Compose([transforms.Resize(254),
transforms.CenterCrop(224),
transforms.ToTensor(),
transforms.Normalize([0.485, 0.456, 0.406],
[0.229, 0.224, 0.225])])
}
# TODO: Load the datasets with ImageFolder
image_datasets = {'train': datasets.ImageFolder(train_dir, transform=data_transforms['train']),
'valid': datasets.ImageFolder(valid_dir, transform=data_transforms['valid_test']),
'test': datasets.ImageFolder(test_dir, transform=data_transforms['valid_test'])
}
# TODO: Using the image datasets and the trainforms, define the dataloaders
dataloaders = {'train': torch.utils.data.DataLoader(image_datasets['train'], batch_size=32, shuffle=True),
'valid': torch.utils.data.DataLoader(image_datasets['valid'], batch_size=32),
'test': torch.utils.data.DataLoader(image_datasets['test'], batch_size=32)
}
trainloader, validloader, testloader= dataloaders['train'], dataloaders['valid'], dataloaders['test']
```
### Label mapping
You'll also need to load in a mapping from category label to category name. You can find this in the file `cat_to_name.json`. It's a JSON object which you can read in with the [`json` module](https://docs.python.org/2/library/json.html). This will give you a dictionary mapping the integer encoded categories to the actual names of the flowers.
```
import json
with open('cat_to_name.json', 'r') as f:
cat_to_name = json.load(f)
```
# Building and training the classifier
Now that the data is ready, it's time to build and train the classifier. As usual, you should use one of the pretrained models from `torchvision.models` to get the image features. Build and train a new feed-forward classifier using those features.
We're going to leave this part up to you. Refer to [the rubric](https://review.udacity.com/#!/rubrics/1663/view) for guidance on successfully completing this section. Things you'll need to do:
* Load a [pre-trained network](http://pytorch.org/docs/master/torchvision/models.html) (If you need a starting point, the VGG networks work great and are straightforward to use)
* Define a new, untrained feed-forward network as a classifier, using ReLU activations and dropout
* Train the classifier layers using backpropagation using the pre-trained network to get the features
* Track the loss and accuracy on the validation set to determine the best hyperparameters
We've left a cell open for you below, but use as many as you need. Our advice is to break the problem up into smaller parts you can run separately. Check that each part is doing what you expect, then move on to the next. You'll likely find that as you work through each part, you'll need to go back and modify your previous code. This is totally normal!
When training make sure you're updating only the weights of the feed-forward network. You should be able to get the validation accuracy above 70% if you build everything right. Make sure to try different hyperparameters (learning rate, units in the classifier, epochs, etc) to find the best model. Save those hyperparameters to use as default values in the next part of the project.
One last important tip if you're using the workspace to run your code: To avoid having your workspace disconnect during the long-running tasks in this notebook, please read in the earlier page in this lesson called Intro to
GPU Workspaces about Keeping Your Session Active. You'll want to include code from the workspace_utils.py module.
**Note for Workspace users:** If your network is over 1 GB when saved as a checkpoint, there might be issues with saving backups in your workspace. Typically this happens with wide dense layers after the convolutional layers. If your saved checkpoint is larger than 1 GB (you can open a terminal and check with `ls -lh`), you should reduce the size of your hidden layers and train again.
```
# TODO: Build and train your network
model = models.vgg16(pretrained=True)
# Frozen parameters
for parameters in model.parameters():
parameters.requires_grad = False
classifier = nn.Sequential(
nn.Linear(25088, 4096),
nn.ReLU(),
nn.Dropout(p=0.2),
nn.Linear(4096, 4096),
nn.ReLU(),
nn.Dropout(p=0.2),
nn.Linear(4096,102),
nn.LogSoftmax(dim=1)
)
model.classifier = classifier
model
criterion = nn.NLLLoss()
# Optimizing the classifier paramateres
optimizer = optim.Adam(model.classifier.parameters(), lr=0.001)
# Enable cuda if available
device = torch.device("cuda:0" if torch.cuda.is_available() else "cpu")
# change to cuda if available
model.to(device)
# Validation Function
def validation(model, validloader, criterion):
valid_loss = 0
accuracy = 0
for images, labels in validloader:
images, labels = images.to(device), labels.to(device)
output = model.forward(images)
valid_loss += criterion(output, labels).item()
ps = torch.exp(output)
equality = (labels.data == ps.max(dim=1)[1])
accuracy += equality.type(torch.FloatTensor).mean()
return valid_loss, accuracy
epochs = 15
print_every = 50
steps = 0
for e in range(epochs):
model.train()
running_loss = 0
for images, labels in iter(trainloader):
steps += 1
images, labels = images.to(device), labels.to(device)
# reset gradients to zeros
optimizer.zero_grad()
# feed-Forward and Backpropagation
outputs = model.forward(images)
loss = criterion(outputs, labels)
loss.backward()
# update the weights
optimizer.step()
running_loss += loss.item()
if steps % print_every == 0:
# evaluation mode
model.eval()
# Turning off the gradients for validation
with torch.no_grad():
valid_loss, accuracy = validation(model, validloader, criterion)
print("Epoch: {}/{}... ".format(e+1, epochs),
"Training Loss: {:.3f}".format(running_loss/print_every),
"Validation Loss: {:.3f}.. ".format(valid_loss/len(validloader)),
"Validation Accuracy: {:.3f}".format(accuracy/len(validloader)))
running_loss = 0
# turn back train mode
model.train()
```
## Testing your network
It's good practice to test your trained network on test data, images the network has never seen either in training or validation. This will give you a good estimate for the model's performance on completely new images. Run the test images through the network and measure the accuracy, the same way you did validation. You should be able to reach around 70% accuracy on the test set if the model has been trained well.
```
# TODO: Do validation on the test set
# evaluation mode
model.eval()
# Turning off the gradients for Testing the network
with torch.no_grad():
test_loss, test_accuracy = validation(model, testloader, criterion)
print("Test Loss: {:.3f}.. ".format(test_loss/len(testloader)),
"Test Accuracy: {:.3f}".format(test_accuracy/len(testloader)))
```
## Save the checkpoint
Now that your network is trained, save the model so you can load it later for making predictions. You probably want to save other things such as the mapping of classes to indices which you get from one of the image datasets: `image_datasets['train'].class_to_idx`. You can attach this to the model as an attribute which makes inference easier later on.
```model.class_to_idx = image_datasets['train'].class_to_idx```
Remember that you'll want to completely rebuild the model later so you can use it for inference. Make sure to include any information you need in the checkpoint. If you want to load the model and keep training, you'll want to save the number of epochs as well as the optimizer state, `optimizer.state_dict`. You'll likely want to use this trained model in the next part of the project, so best to save it now.
```
# TODO: Save the checkpoint
# Mapping classes to Indices
model.class_to_idx = image_datasets['train'].class_to_idx
checkpoint = {'cat_to_name': cat_to_name,
'class_to_idx': model.class_to_idx,
'model': model,
'classifier': model.classifier,
'state_dict': model.state_dict(),
'criterion': criterion,
'optimizer': optimizer,
'optimizer_state_dict': optimizer.state_dict(),
'epochs': epochs
}
torch.save(checkpoint, 'checkpoint.pth')
```
## Loading the checkpoint
At this point it's good to write a function that can load a checkpoint and rebuild the model. That way you can come back to this project and keep working on it without having to retrain the network.
```
# TODO: Write a function that loads a checkpoint and rebuilds the model
def load_checkpoint(filepath):
if torch.cuda.is_available():
map_location=lambda storage, loc: storage.cuda()
else:
map_location='cpu'
checkpoint = torch.load(filepath, map_location=map_location)
model = checkpoint['model']
model.load_state_dict(checkpoint['state_dict'])
model.cat_to_name = checkpoint['cat_to_name']
model.class_to_idx = checkpoint['class_to_idx']
# Frozen parameters for pre-trained feature network
for parameters in model.parameters():
parameters.requires_grad = False
model.classifier = checkpoint['classifier']
optimizer = checkpoint['optimizer']
optimizer.load_state_dict(checkpoint['optimizer_state_dict'])
model.epochs = checkpoint['epochs']
# Enable cuda if available
device = torch.device("cuda:0" if torch.cuda.is_available() else "cpu")
# change to cuda if available
model.to(device)
return model, optimizer
model, optimizer = load_checkpoint('checkpoint.pth')
```
# Inference for classification
Now you'll write a function to use a trained network for inference. That is, you'll pass an image into the network and predict the class of the flower in the image. Write a function called `predict` that takes an image and a model, then returns the top $K$ most likely classes along with the probabilities. It should look like
```python
probs, classes = predict(image_path, model)
print(probs)
print(classes)
> [ 0.01558163 0.01541934 0.01452626 0.01443549 0.01407339]
> ['70', '3', '45', '62', '55']
```
First you'll need to handle processing the input image such that it can be used in your network.
## Image Preprocessing
You'll want to use `PIL` to load the image ([documentation](https://pillow.readthedocs.io/en/latest/reference/Image.html)). It's best to write a function that preprocesses the image so it can be used as input for the model. This function should process the images in the same manner used for training.
First, resize the images where the shortest side is 256 pixels, keeping the aspect ratio. This can be done with the [`thumbnail`](http://pillow.readthedocs.io/en/3.1.x/reference/Image.html#PIL.Image.Image.thumbnail) or [`resize`](http://pillow.readthedocs.io/en/3.1.x/reference/Image.html#PIL.Image.Image.thumbnail) methods. Then you'll need to crop out the center 224x224 portion of the image.
Color channels of images are typically encoded as integers 0-255, but the model expected floats 0-1. You'll need to convert the values. It's easiest with a Numpy array, which you can get from a PIL image like so `np_image = np.array(pil_image)`.
As before, the network expects the images to be normalized in a specific way. For the means, it's `[0.485, 0.456, 0.406]` and for the standard deviations `[0.229, 0.224, 0.225]`. You'll want to subtract the means from each color channel, then divide by the standard deviation.
And finally, PyTorch expects the color channel to be the first dimension but it's the third dimension in the PIL image and Numpy array. You can reorder dimensions using [`ndarray.transpose`](https://docs.scipy.org/doc/numpy-1.13.0/reference/generated/numpy.ndarray.transpose.html). The color channel needs to be first and retain the order of the other two dimensions.
```
def process_image(image):
''' Scales, crops, and normalizes a PIL image for a PyTorch model,
returns an Numpy array
'''
# TODO: Process a PIL image for use in a PyTorch model
im = Image.open(image)
im.resize((256,256))
width, height = im.size # Get dimensions
new_width, new_height = 224, 224
left = (width - new_width)/2
top = (height - new_height)/2
right = (width + new_width)/2
bottom = (height + new_height)/2
# Crop the center of the image
im = im.crop((left, top, right, bottom))
# Convert PIL image to Numpy
np_image = np.array(im)
# Normalize the images
np_image = np_image / np_image.max()
mean = np.array([0.485, 0.456, 0.406])
std = np.array([0.229, 0.224, 0.225])
normalization = (np_image - mean) / std
pytorch_tensor = torch.Tensor(normalization.transpose((2,0,1)))
return pytorch_tensor
```
To check your work, the function below converts a PyTorch tensor and displays it in the notebook. If your `process_image` function works, running the output through this function should return the original image (except for the cropped out portions).
```
def imshow(image, ax=None, title=None):
"""Imshow for Tensor."""
if ax is None:
fig, ax = plt.subplots()
# PyTorch tensors assume the color channel is the first dimension
# but matplotlib assumes is the third dimension
image = image.numpy().transpose((1, 2, 0))
# Undo preprocessing
mean = np.array([0.485, 0.456, 0.406])
std = np.array([0.229, 0.224, 0.225])
image = std * image + mean
# Image needs to be clipped between 0 and 1 or it looks like noise when displayed
image = np.clip(image, 0, 1)
ax.imshow(image)
return ax
```
## Class Prediction
Once you can get images in the correct format, it's time to write a function for making predictions with your model. A common practice is to predict the top 5 or so (usually called top-$K$) most probable classes. You'll want to calculate the class probabilities then find the $K$ largest values.
To get the top $K$ largest values in a tensor use [`x.topk(k)`](http://pytorch.org/docs/master/torch.html#torch.topk). This method returns both the highest `k` probabilities and the indices of those probabilities corresponding to the classes. You need to convert from these indices to the actual class labels using `class_to_idx` which hopefully you added to the model or from an `ImageFolder` you used to load the data ([see here](#Save-the-checkpoint)). Make sure to invert the dictionary so you get a mapping from index to class as well.
Again, this method should take a path to an image and a model checkpoint, then return the probabilities and classes.
```python
probs, classes = predict(image_path, model)
print(probs)
print(classes)
> [ 0.01558163 0.01541934 0.01452626 0.01443549 0.01407339]
> ['70', '3', '45', '62', '55']
```
```
def predict(image_path, model, topk=5):
''' Predict the class (or classes) of an image using a trained deep learning model.
'''
# TODO: Implement the code to predict the class from an image file
device = torch.device("cuda:0" if torch.cuda.is_available() else "cpu")
# Loading Image and Model
im = process_image(image_path).to(device)
model, optimizer = load_checkpoint(model)
# evaluation mode
model.eval()
with torch.no_grad():
output = model.forward(im.unsqueeze_(0))
ps = torch.exp(output)
# top 𝐾 largest values
top_ps, topk = ps.topk(topk)
# reverse class_to_idx to idx_to_class
idx_to_class = {value : key for (key, value) in model.class_to_idx.items()}
# the highest k probabilities and classes
probs = top_ps.numpy()[0]
classes = [idx_to_class[_class] for _class in topk.numpy()[0]]
return probs, classes
```
## Sanity Checking
Now that you can use a trained model for predictions, check to make sure it makes sense. Even if the testing accuracy is high, it's always good to check that there aren't obvious bugs. Use `matplotlib` to plot the probabilities for the top 5 classes as a bar graph, along with the input image. It should look like this:
<img src='assets/inference_example.png' width=300px>
You can convert from the class integer encoding to actual flower names with the `cat_to_name.json` file (should have been loaded earlier in the notebook). To show a PyTorch tensor as an image, use the `imshow` function defined above.
```
# TODO: Display an image along with the top 5 classes
im = process_image('flowers/test/1/image_06743.jpg')
probs, classes = predict('flowers/test/1/image_06743.jpg', 'checkpoint.pth', topk=5 )
# Figure Object
fig = plt.figure(figsize = [4, 8]) # larger figure size for subplots
# Subplot 1
# Extract the name of the flower category with highest probability
# from cat_to_name.json file
cat_name = cat_to_name[classes[np.argmax(probs)]]
ax_1 = fig.add_subplot(2, 1, 1)
imshow(im, ax= ax_1)
ax_1.axis('off')
ax_1.set_title(cat_name)
# Subplot 2
ax_2 = fig.add_subplot(2, 1, 2)
y_pos = [i for i, _ in enumerate(probs)]
ax_2.barh(y_pos, probs)
ax_2.set_yticks(y_pos)
ax_2.set_yticklabels(cat_to_name[_class] for _class in classes)
ax_2.invert_yaxis();
```
|
github_jupyter
|
# Imports here
import torch
from torch import nn
from torch import optim
import torch.nn.functional as F
from torchvision import datasets, transforms, models
from collections import OrderedDict
import numpy as np
import pandas as pd
from PIL import Image
import matplotlib.pyplot as plt
import seaborn as sb
%matplotlib inline
data_dir = 'flowers'
train_dir = data_dir + '/train'
valid_dir = data_dir + '/valid'
test_dir = data_dir + '/test'
# TODO: Define your transforms for the training, validation, and testing sets
data_transforms = {'train': transforms.Compose([transforms.RandomRotation(45),
transforms.RandomResizedCrop(224),
transforms.RandomHorizontalFlip(),transforms.ToTensor(),
transforms.Normalize([0.485, 0.456, 0.406], [0.229, 0.224, 0.225])]),
'valid_test': transforms.Compose([transforms.Resize(254),
transforms.CenterCrop(224),
transforms.ToTensor(),
transforms.Normalize([0.485, 0.456, 0.406],
[0.229, 0.224, 0.225])])
}
# TODO: Load the datasets with ImageFolder
image_datasets = {'train': datasets.ImageFolder(train_dir, transform=data_transforms['train']),
'valid': datasets.ImageFolder(valid_dir, transform=data_transforms['valid_test']),
'test': datasets.ImageFolder(test_dir, transform=data_transforms['valid_test'])
}
# TODO: Using the image datasets and the trainforms, define the dataloaders
dataloaders = {'train': torch.utils.data.DataLoader(image_datasets['train'], batch_size=32, shuffle=True),
'valid': torch.utils.data.DataLoader(image_datasets['valid'], batch_size=32),
'test': torch.utils.data.DataLoader(image_datasets['test'], batch_size=32)
}
trainloader, validloader, testloader= dataloaders['train'], dataloaders['valid'], dataloaders['test']
import json
with open('cat_to_name.json', 'r') as f:
cat_to_name = json.load(f)
# TODO: Build and train your network
model = models.vgg16(pretrained=True)
# Frozen parameters
for parameters in model.parameters():
parameters.requires_grad = False
classifier = nn.Sequential(
nn.Linear(25088, 4096),
nn.ReLU(),
nn.Dropout(p=0.2),
nn.Linear(4096, 4096),
nn.ReLU(),
nn.Dropout(p=0.2),
nn.Linear(4096,102),
nn.LogSoftmax(dim=1)
)
model.classifier = classifier
model
criterion = nn.NLLLoss()
# Optimizing the classifier paramateres
optimizer = optim.Adam(model.classifier.parameters(), lr=0.001)
# Enable cuda if available
device = torch.device("cuda:0" if torch.cuda.is_available() else "cpu")
# change to cuda if available
model.to(device)
# Validation Function
def validation(model, validloader, criterion):
valid_loss = 0
accuracy = 0
for images, labels in validloader:
images, labels = images.to(device), labels.to(device)
output = model.forward(images)
valid_loss += criterion(output, labels).item()
ps = torch.exp(output)
equality = (labels.data == ps.max(dim=1)[1])
accuracy += equality.type(torch.FloatTensor).mean()
return valid_loss, accuracy
epochs = 15
print_every = 50
steps = 0
for e in range(epochs):
model.train()
running_loss = 0
for images, labels in iter(trainloader):
steps += 1
images, labels = images.to(device), labels.to(device)
# reset gradients to zeros
optimizer.zero_grad()
# feed-Forward and Backpropagation
outputs = model.forward(images)
loss = criterion(outputs, labels)
loss.backward()
# update the weights
optimizer.step()
running_loss += loss.item()
if steps % print_every == 0:
# evaluation mode
model.eval()
# Turning off the gradients for validation
with torch.no_grad():
valid_loss, accuracy = validation(model, validloader, criterion)
print("Epoch: {}/{}... ".format(e+1, epochs),
"Training Loss: {:.3f}".format(running_loss/print_every),
"Validation Loss: {:.3f}.. ".format(valid_loss/len(validloader)),
"Validation Accuracy: {:.3f}".format(accuracy/len(validloader)))
running_loss = 0
# turn back train mode
model.train()
# TODO: Do validation on the test set
# evaluation mode
model.eval()
# Turning off the gradients for Testing the network
with torch.no_grad():
test_loss, test_accuracy = validation(model, testloader, criterion)
print("Test Loss: {:.3f}.. ".format(test_loss/len(testloader)),
"Test Accuracy: {:.3f}".format(test_accuracy/len(testloader)))
Remember that you'll want to completely rebuild the model later so you can use it for inference. Make sure to include any information you need in the checkpoint. If you want to load the model and keep training, you'll want to save the number of epochs as well as the optimizer state, `optimizer.state_dict`. You'll likely want to use this trained model in the next part of the project, so best to save it now.
## Loading the checkpoint
At this point it's good to write a function that can load a checkpoint and rebuild the model. That way you can come back to this project and keep working on it without having to retrain the network.
# Inference for classification
Now you'll write a function to use a trained network for inference. That is, you'll pass an image into the network and predict the class of the flower in the image. Write a function called `predict` that takes an image and a model, then returns the top $K$ most likely classes along with the probabilities. It should look like
First you'll need to handle processing the input image such that it can be used in your network.
## Image Preprocessing
You'll want to use `PIL` to load the image ([documentation](https://pillow.readthedocs.io/en/latest/reference/Image.html)). It's best to write a function that preprocesses the image so it can be used as input for the model. This function should process the images in the same manner used for training.
First, resize the images where the shortest side is 256 pixels, keeping the aspect ratio. This can be done with the [`thumbnail`](http://pillow.readthedocs.io/en/3.1.x/reference/Image.html#PIL.Image.Image.thumbnail) or [`resize`](http://pillow.readthedocs.io/en/3.1.x/reference/Image.html#PIL.Image.Image.thumbnail) methods. Then you'll need to crop out the center 224x224 portion of the image.
Color channels of images are typically encoded as integers 0-255, but the model expected floats 0-1. You'll need to convert the values. It's easiest with a Numpy array, which you can get from a PIL image like so `np_image = np.array(pil_image)`.
As before, the network expects the images to be normalized in a specific way. For the means, it's `[0.485, 0.456, 0.406]` and for the standard deviations `[0.229, 0.224, 0.225]`. You'll want to subtract the means from each color channel, then divide by the standard deviation.
And finally, PyTorch expects the color channel to be the first dimension but it's the third dimension in the PIL image and Numpy array. You can reorder dimensions using [`ndarray.transpose`](https://docs.scipy.org/doc/numpy-1.13.0/reference/generated/numpy.ndarray.transpose.html). The color channel needs to be first and retain the order of the other two dimensions.
To check your work, the function below converts a PyTorch tensor and displays it in the notebook. If your `process_image` function works, running the output through this function should return the original image (except for the cropped out portions).
## Class Prediction
Once you can get images in the correct format, it's time to write a function for making predictions with your model. A common practice is to predict the top 5 or so (usually called top-$K$) most probable classes. You'll want to calculate the class probabilities then find the $K$ largest values.
To get the top $K$ largest values in a tensor use [`x.topk(k)`](http://pytorch.org/docs/master/torch.html#torch.topk). This method returns both the highest `k` probabilities and the indices of those probabilities corresponding to the classes. You need to convert from these indices to the actual class labels using `class_to_idx` which hopefully you added to the model or from an `ImageFolder` you used to load the data ([see here](#Save-the-checkpoint)). Make sure to invert the dictionary so you get a mapping from index to class as well.
Again, this method should take a path to an image and a model checkpoint, then return the probabilities and classes.
## Sanity Checking
Now that you can use a trained model for predictions, check to make sure it makes sense. Even if the testing accuracy is high, it's always good to check that there aren't obvious bugs. Use `matplotlib` to plot the probabilities for the top 5 classes as a bar graph, along with the input image. It should look like this:
<img src='assets/inference_example.png' width=300px>
You can convert from the class integer encoding to actual flower names with the `cat_to_name.json` file (should have been loaded earlier in the notebook). To show a PyTorch tensor as an image, use the `imshow` function defined above.
| 0.609524 | 0.992327 |
# Day 6: Custom Customs
As your flight approaches the regional airport where you'll switch to a much larger plane, [customs declaration forms](https://en.wikipedia.org/wiki/Customs_declaration) are distributed to the passengers.
The form asks a series of 26 yes-or-no questions marked `a` through `z`. All you need to do is identify the questions for which **anyone in your group** answers "yes". Since your group is just you, this doesn't take very long.
However, the person sitting next to you seems to be experiencing a language barrier and asks if you can help. For each of the people in their group, you write down the questions for which they answer "yes", one per line. For example:
abcx
abcy
abcz
In this group, there are **`6`** questions to which anyone answered "yes": `a`, `b`, `c`, `x`, `y`, and `z`. (Duplicate answers to the same question don't count extra; each question counts at most once.)
Another group asks for your help, then another, and eventually you've collected answers from every group on the plane (your puzzle input). Each group's answers are separated by a blank line, and within each group, each person's answers are on a single line. For example:
abc
a
b
c
ab
ac
a
a
a
a
b
This list represents answers from five groups:
* The first group contains one person who answered "yes" to **`3`** questions: `a`, `b`, and `c`.
* The second group contains three people; combined, they answered "yes" to **`3`** questions: `a`, `b`, and `c`.
* The third group contains two people; combined, they answered "yes" to **`3`** questions: `a`, `b`, and `c`.
* The fourth group contains four people; combined, they answered "yes" to only **`1`** question, `a`.
* The last group contains one person who answered "yes" to only **`1`** question, `b`.
In this example, the sum of these counts is `3 + 3 + 3 + 1 + 1` = **`11`**.
For each group, count the number of questions to which anyone answered "yes". What is the sum of those counts?
Your puzzle answer was `6291`.
```
input = """abc
a
b
c
ab
ac
a
a
a
a
b""".split('\n\n')
input = open('day.06.input.txt', 'r').read().split('\n\n')
from functools import reduce
import numpy as np
np.sum([len(reduce(set.union, [set(p)for p in g.split('\n')])) for g in input])
```
## Part Two
As you finish the last group's customs declaration, you notice that you misread one word in the instructions:
You don't need to identify the questions to which **anyone** answered "yes"; you need to identify the questions to which **everyone** answered "yes"!
Using the same example as above:
abc
a
b
c
ab
ac
a
a
a
a
b
This list represents answers from five groups:
* In the first group, everyone (all 1 person) answered "yes" to **`3`** questions: `a`, `b`, and `c`.
* In the second group, there is **no** question to which everyone answered "yes".
* In the third group, everyone answered yes to only **`1`** question, `a`. Since some people did not answer "yes" to `b` or `c`, they don't count.
* In the fourth group, everyone answered yes to only **`1`** question, `a`.
* In the fifth group, everyone (all 1 person) answered "yes" to **`1`** question, `b`.
In this example, the sum of these counts is 3 + 0 + 1 + 1 + 1 = 6.
For each group, count the number of questions to which everyone answered "yes". What is the sum of those counts?
Your puzzle answer was `3052`.
```
np.sum([len(reduce(set.intersection, [set(p)for p in g.split('\n')])) for g in input])
```
|
github_jupyter
|
input = """abc
a
b
c
ab
ac
a
a
a
a
b""".split('\n\n')
input = open('day.06.input.txt', 'r').read().split('\n\n')
from functools import reduce
import numpy as np
np.sum([len(reduce(set.union, [set(p)for p in g.split('\n')])) for g in input])
np.sum([len(reduce(set.intersection, [set(p)for p in g.split('\n')])) for g in input])
| 0.287968 | 0.962285 |
# Symbolic Computation
Symbolic computation deals with symbols, representing them exactly, instead of numerical approximations (floating point).
We will start with the following [borrowed](https://docs.sympy.org/latest/tutorial/intro.html) tutorial to introduce the concepts of SymPy. Devito uses SymPy heavily and builds upon it in its DSL.
```
import math
math.sqrt(3)
math.sqrt(8)
```
$\sqrt(8) = 2\sqrt(2)$, but it's hard to see that here
```
import sympy
sympy.sqrt(3)
```
SymPy can even simplify symbolic computations
```
sympy.sqrt(8)
from sympy import symbols
x, y = symbols('x y')
expr = x + 2*y
expr
```
Note that simply adding two symbols creates an expression. Now let's play around with it.
```
expr + 1
expr - x
```
Note that `expr - x` was not `x + 2y -x`
```
x*expr
from sympy import expand, factor
expanded_expr = expand(x*expr)
expanded_expr
factor(expanded_expr)
from sympy import diff, sin, exp
diff(sin(x)*exp(x), x)
from sympy import limit
limit(sin(x)/x, x, 0)
```
### Exercise
Solve $x^2 - 2 = 0$ using sympy.solve
```
# Type solution here
from sympy import solve
```
## Pretty printing
```
from sympy import init_printing, Integral, sqrt
init_printing(use_latex='mathjax')
Integral(sqrt(1/x), x)
from sympy import latex
latex(Integral(sqrt(1/x), x))
```
More symbols.
Exercise: fix the following piece of code
```
# NBVAL_SKIP
# The following piece of code is supposed to fail as it is
# The exercise is to fix the code
expr2 = x + 2*y +3*z
```
### Exercise
Solve $x + 2*y + 3*z$ for $x$
```
# Solution here
from sympy import solve
```
Difference between symbol name and python variable name
```
x, y = symbols("y z")
x
y
# NBVAL_SKIP
# The following code will error until the code in cell 16 above is
# fixed
z
```
Symbol names can be more than one character long
```
crazy = symbols('unrelated')
crazy + 1
x = symbols("x")
expr = x + 1
x = 2
```
What happens when I print expr now? Does it print 3?
```
print(expr)
```
How do we get 3?
```
x = symbols("x")
expr = x + 1
expr.subs(x, 2)
```
## Equalities
```
x + 1 == 4
from sympy import Eq
Eq(x + 1, 4)
```
Suppose we want to ask whether $(x + 1)^2 = x^2 + 2x + 1$
```
(x + 1)**2 == x**2 + 2*x + 1
from sympy import simplify
a = (x + 1)**2
b = x**2 + 2*x + 1
simplify(a-b)
```
### Exercise
Write a function that takes two expressions as input, and returns a tuple of two booleans. The first if they are equal symbolically, and the second if they are equal mathematically.
## More operations
```
z = symbols("z")
expr = x**3 + 4*x*y - z
expr.subs([(x, 2), (y, 4), (z, 0)])
from sympy import sympify
str_expr = "x**2 + 3*x - 1/2"
expr = sympify(str_expr)
expr
expr.subs(x, 2)
expr = sqrt(8)
expr
expr.evalf()
from sympy import pi
pi.evalf(100)
from sympy import cos
expr = cos(2*x)
expr.evalf(subs={x: 2.4})
```
### Exercise
```
from IPython.core.display import Image
Image(filename='figures/comic.png')
```
Write a function that takes a symbolic expression (like pi), and determines the first place where 789 appears.
Tip: Use the string representation of the number. Python starts counting at 0, but the decimal point offsets this
## Solving an ODE
```
from sympy import Function
f, g = symbols('f g', cls=Function)
f(x)
f(x).diff()
diffeq = Eq(f(x).diff(x, x) - 2*f(x).diff(x) + f(x), sin(x))
diffeq
from sympy import dsolve
dsolve(diffeq, f(x))
```
## Finite Differences
```
f = Function('f')
dfdx = f(x).diff(x)
dfdx.as_finite_difference()
from sympy import Symbol
d2fdx2 = f(x).diff(x, 2)
h = Symbol('h')
d2fdx2.as_finite_difference(h)
```
Now that we have seen some relevant features of vanilla SymPy, let's move on to Devito, which could be seen as SymPy finite differences on steroids!
|
github_jupyter
|
import math
math.sqrt(3)
math.sqrt(8)
import sympy
sympy.sqrt(3)
sympy.sqrt(8)
from sympy import symbols
x, y = symbols('x y')
expr = x + 2*y
expr
expr + 1
expr - x
x*expr
from sympy import expand, factor
expanded_expr = expand(x*expr)
expanded_expr
factor(expanded_expr)
from sympy import diff, sin, exp
diff(sin(x)*exp(x), x)
from sympy import limit
limit(sin(x)/x, x, 0)
# Type solution here
from sympy import solve
from sympy import init_printing, Integral, sqrt
init_printing(use_latex='mathjax')
Integral(sqrt(1/x), x)
from sympy import latex
latex(Integral(sqrt(1/x), x))
# NBVAL_SKIP
# The following piece of code is supposed to fail as it is
# The exercise is to fix the code
expr2 = x + 2*y +3*z
# Solution here
from sympy import solve
x, y = symbols("y z")
x
y
# NBVAL_SKIP
# The following code will error until the code in cell 16 above is
# fixed
z
crazy = symbols('unrelated')
crazy + 1
x = symbols("x")
expr = x + 1
x = 2
print(expr)
x = symbols("x")
expr = x + 1
expr.subs(x, 2)
x + 1 == 4
from sympy import Eq
Eq(x + 1, 4)
(x + 1)**2 == x**2 + 2*x + 1
from sympy import simplify
a = (x + 1)**2
b = x**2 + 2*x + 1
simplify(a-b)
z = symbols("z")
expr = x**3 + 4*x*y - z
expr.subs([(x, 2), (y, 4), (z, 0)])
from sympy import sympify
str_expr = "x**2 + 3*x - 1/2"
expr = sympify(str_expr)
expr
expr.subs(x, 2)
expr = sqrt(8)
expr
expr.evalf()
from sympy import pi
pi.evalf(100)
from sympy import cos
expr = cos(2*x)
expr.evalf(subs={x: 2.4})
from IPython.core.display import Image
Image(filename='figures/comic.png')
from sympy import Function
f, g = symbols('f g', cls=Function)
f(x)
f(x).diff()
diffeq = Eq(f(x).diff(x, x) - 2*f(x).diff(x) + f(x), sin(x))
diffeq
from sympy import dsolve
dsolve(diffeq, f(x))
f = Function('f')
dfdx = f(x).diff(x)
dfdx.as_finite_difference()
from sympy import Symbol
d2fdx2 = f(x).diff(x, 2)
h = Symbol('h')
d2fdx2.as_finite_difference(h)
| 0.487063 | 0.98222 |
# Scheduling Multipurpose Batch Processes using State-Task Networks
Keywords: cbc usage, state-task networks, gdp, disjunctive programming, batch processes
The State-Task Network (STN) is an approach to modeling multipurpose batch process for the purpose of short term scheduling. It was first developed by Kondili, et al., in 1993, and subsequently developed and extended by others.
## Imports
```
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
from IPython.display import display, HTML
import shutil
import sys
import os.path
if not shutil.which("pyomo"):
!pip install -q pyomo
assert(shutil.which("pyomo"))
if not (shutil.which("cbc") or os.path.isfile("cbc")):
if "google.colab" in sys.modules:
!apt-get install -y -qq coinor-cbc
else:
try:
!conda install -c conda-forge coincbc
except:
pass
assert(shutil.which("cbc") or os.path.isfile("cbc"))
from pyomo.environ import *
```
## References
Floudas, C. A., & Lin, X. (2005). Mixed integer linear programming in process scheduling: Modeling, algorithms, and applications. Annals of Operations Research, 139(1), 131-162.
Harjunkoski, I., Maravelias, C. T., Bongers, P., Castro, P. M., Engell, S., Grossmann, I. E., ... & Wassick, J. (2014). Scope for industrial applications of production scheduling models and solution methods. Computers & Chemical Engineering, 62, 161-193.
Kondili, E., Pantelides, C. C., & Sargent, R. W. H. (1993). A general algorithm for short-term scheduling of batch operations—I. MILP formulation. Computers & Chemical Engineering, 17(2), 211-227.
Méndez, C. A., Cerdá, J., Grossmann, I. E., Harjunkoski, I., & Fahl, M. (2006). State-of-the-art review of optimization methods for short-term scheduling of batch processes. Computers & Chemical Engineering, 30(6), 913-946.
Shah, N., Pantelides, C. C., & Sargent, R. W. H. (1993). A general algorithm for short-term scheduling of batch operations—II. Computational issues. Computers & Chemical Engineering, 17(2), 229-244.
Wassick, J. M., & Ferrio, J. (2011). Extending the resource task network for industrial applications. Computers & chemical engineering, 35(10), 2124-2140.
## Example (Kondili, et al., 1993)
A state-task network is a graphical representation of the activities in a multiproduct batch process. The representation includes the minimum details needed for short term scheduling of batch operations.
A well-studied example due to Kondili (1993) is shown below. Other examples are available in the references cited above.
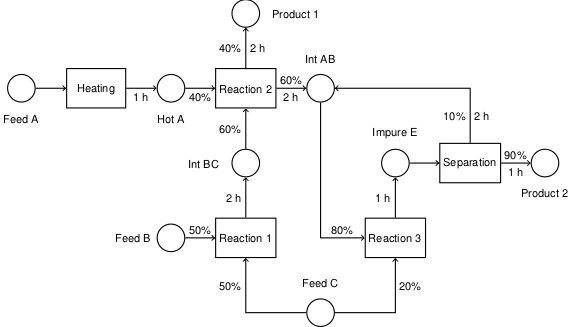
Each circular node in the diagram designates material in a particular state. The materials are generally held in suitable vessels with a known capacity. The relevant information for each state is the initial inventory, storage capacity, and the unit price of the material in each state. The price of materials in intermediate states may be assigned penalities in order to minimize the amount of work in progress.
The rectangular nodes denote process tasks. When scheduled for execution, each task is assigned an appropriate piece of equipment, and assigned a batch of material according to the incoming arcs. Each incoming arc begins at a state where the associated label indicates the mass fraction of the batch coming from that particular state. Outgoing arcs indicate the disposition of the batch to product states. The outgoing are labels indicate the fraction of the batch assigned to each product state, and the time necessary to produce that product.
Not shown in the diagram is the process equipment used to execute the tasks. A separate list of process units is available, each characterized by a capacity and list of tasks which can be performed in that unit.
### Exercise
Read this reciped for Hollandaise Sauce: http://www.foodnetwork.com/recipes/tyler-florence/hollandaise-sauce-recipe-1910043. Assume the available equipment consists of one sauce pan and a double-boiler on a stove. Draw a state-task network outlining the basic steps in the recipe.
## Encoding the STN data
The basic data structure specifies the states, tasks, and units comprising a state-task network. The intention is for all relevant problem data to be contained in a single JSON-like structure.
```
# planning horizon
H = 10
Kondili = {
# time grid
'TIME': range(0, H+1),
# states
'STATES': {
'Feed_A' : {'capacity': 500, 'initial': 500, 'price': 0},
'Feed_B' : {'capacity': 500, 'initial': 500, 'price': 0},
'Feed_C' : {'capacity': 500, 'initial': 500, 'price': 0},
'Hot_A' : {'capacity': 100, 'initial': 0, 'price': -100},
'Int_AB' : {'capacity': 200, 'initial': 0, 'price': -100},
'Int_BC' : {'capacity': 150, 'initial': 0, 'price': -100},
'Impure_E' : {'capacity': 100, 'initial': 0, 'price': -100},
'Product_1': {'capacity': 500, 'initial': 0, 'price': 10},
'Product_2': {'capacity': 500, 'initial': 0, 'price': 10},
},
# state-to-task arcs indexed by (state, task)
'ST_ARCS': {
('Feed_A', 'Heating') : {'rho': 1.0},
('Feed_B', 'Reaction_1'): {'rho': 0.5},
('Feed_C', 'Reaction_1'): {'rho': 0.5},
('Feed_C', 'Reaction_3'): {'rho': 0.2},
('Hot_A', 'Reaction_2'): {'rho': 0.4},
('Int_AB', 'Reaction_3'): {'rho': 0.8},
('Int_BC', 'Reaction_2'): {'rho': 0.6},
('Impure_E', 'Separation'): {'rho': 1.0},
},
# task-to-state arcs indexed by (task, state)
'TS_ARCS': {
('Heating', 'Hot_A') : {'dur': 1, 'rho': 1.0},
('Reaction_2', 'Product_1'): {'dur': 2, 'rho': 0.4},
('Reaction_2', 'Int_AB') : {'dur': 2, 'rho': 0.6},
('Reaction_1', 'Int_BC') : {'dur': 2, 'rho': 1.0},
('Reaction_3', 'Impure_E') : {'dur': 1, 'rho': 1.0},
('Separation', 'Int_AB') : {'dur': 2, 'rho': 0.1},
('Separation', 'Product_2'): {'dur': 1, 'rho': 0.9},
},
# unit data indexed by (unit, task)
'UNIT_TASKS': {
('Heater', 'Heating') : {'Bmin': 0, 'Bmax': 100, 'Cost': 1, 'vCost': 0, 'Tclean': 0},
('Reactor_1', 'Reaction_1'): {'Bmin': 0, 'Bmax': 80, 'Cost': 1, 'vCost': 0, 'Tclean': 0},
('Reactor_1', 'Reaction_2'): {'Bmin': 0, 'Bmax': 80, 'Cost': 1, 'vCost': 0, 'Tclean': 0},
('Reactor_1', 'Reaction_3'): {'Bmin': 0, 'Bmax': 80, 'Cost': 1, 'vCost': 0, 'Tclean': 0},
('Reactor_2', 'Reaction_1'): {'Bmin': 0, 'Bmax': 80, 'Cost': 1, 'vCost': 0, 'Tclean': 0},
('Reactor_2', 'Reaction_2'): {'Bmin': 0, 'Bmax': 80, 'Cost': 1, 'vCost': 0, 'Tclean': 0},
('Reactor_2', 'Reaction_3'): {'Bmin': 0, 'Bmax': 80, 'Cost': 1, 'vCost': 0, 'Tclean': 0},
('Still', 'Separation'): {'Bmin': 0, 'Bmax': 200, 'Cost': 1, 'vCost': 0, 'Tclean': 0},
},
}
STN = Kondili
H = 16
Hydrolubes = {
# time grid
'TIME': range(0, H+1),
# states
'STATES': {
'Feed_A' : {'capacity': 500, 'initial': 500, 'price': 0},
'Feed_B' : {'capacity': 500, 'initial': 500, 'price': 0},
'Feed_C' : {'capacity': 500, 'initial': 500, 'price': 0},
'Hot_A' : {'capacity': 100, 'initial': 0, 'price': -100},
'Int_AB' : {'capacity': 200, 'initial': 0, 'price': -100},
'Int_BC' : {'capacity': 150, 'initial': 0, 'price': -100},
'Impure_E' : {'capacity': 100, 'initial': 0, 'price': -100},
'Product_1': {'capacity': 500, 'initial': 0, 'price': 10},
'Product_2': {'capacity': 500, 'initial': 0, 'price': 10},
},
# state-to-task arcs indexed by (state, task)
'ST_ARCS': {
('Feed_A', 'Heating') : {'rho': 1.0},
('Feed_B', 'Reaction_1'): {'rho': 0.5},
('Feed_C', 'Reaction_1'): {'rho': 0.5},
('Feed_C', 'Reaction_3'): {'rho': 0.2},
('Hot_A', 'Reaction_2'): {'rho': 0.4},
('Int_AB', 'Reaction_3'): {'rho': 0.8},
('Int_BC', 'Reaction_2'): {'rho': 0.6},
('Impure_E', 'Separation'): {'rho': 1.0},
},
# task-to-state arcs indexed by (task, state)
'TS_ARCS': {
('Heating', 'Hot_A') : {'dur': 1, 'rho': 1.0},
('Reaction_2', 'Product_1'): {'dur': 2, 'rho': 0.4},
('Reaction_2', 'Int_AB') : {'dur': 2, 'rho': 0.6},
('Reaction_1', 'Int_BC') : {'dur': 2, 'rho': 1.0},
('Reaction_3', 'Impure_E') : {'dur': 1, 'rho': 1.0},
('Separation', 'Int_AB') : {'dur': 2, 'rho': 0.1},
('Separation', 'Product_2'): {'dur': 1, 'rho': 0.9},
},
# unit data indexed by (unit, task)
'UNIT_TASKS': {
('Heater', 'Heating') : {'Bmin': 0, 'Bmax': 100, 'Cost': 1, 'vCost': 0, 'Tclean': 0},
('Reactor_1', 'Reaction_1'): {'Bmin': 0, 'Bmax': 80, 'Cost': 1, 'vCost': 0, 'Tclean': 0},
('Reactor_1', 'Reaction_2'): {'Bmin': 0, 'Bmax': 80, 'Cost': 1, 'vCost': 0, 'Tclean': 0},
('Reactor_1', 'Reaction_3'): {'Bmin': 0, 'Bmax': 80, 'Cost': 1, 'vCost': 0, 'Tclean': 0},
('Reactor_2', 'Reaction_1'): {'Bmin': 0, 'Bmax': 80, 'Cost': 1, 'vCost': 0, 'Tclean': 0},
('Reactor_2', 'Reaction_2'): {'Bmin': 0, 'Bmax': 80, 'Cost': 1, 'vCost': 0, 'Tclean': 0},
('Reactor_2', 'Reaction_3'): {'Bmin': 0, 'Bmax': 80, 'Cost': 1, 'vCost': 0, 'Tclean': 0},
('Still', 'Separation'): {'Bmin': 0, 'Bmax': 200, 'Cost': 1, 'vCost': 0, 'Tclean': 0},
},
}
STN = Hydrolubes
```
### Setting a time grid
The following computations can be done on any time grid, including real-valued time points. TIME is a list of time points commencing at 0.
## Creating a Pyomo model
The following Pyomo model closely follows the development in Kondili, et al. (1993). In particular, the first step in the model is to process the STN data to create sets as given in Kondili.
One important difference from Kondili is the adoption of a more natural time scale that starts at $t = 0$ and extends to $t = H$ (rather than from 1 to H+1).
A second difference is the introduction of an additional decision variable denoted by $Q_{j,t}$ indicating the amount of material in unit $j$ at time $t$. A material balance then reads
\begin{align*}
Q_{jt} & = Q_{j(t-1)} + \sum_{i\in I_j}B_{ijt} - \sum_{i\in I_j}\sum_{\substack{s \in \bar{S}_i\\s\ni t-P_{is} \geq 0}}\bar{\rho}_{is}B_{ij(t-P_{is})} \qquad \forall j,t
\end{align*}
Following Kondili's notation, $I_j$ is the set of tasks that can be performed in unit $j$, and $\bar{S}_i$ is the set of states fed by task $j$. We assume the units are empty at the beginning and end of production period, i.e.,
\begin{align*}
Q_{j(-1)} & = 0 \qquad \forall j \\
Q_{j,H} & = 0 \qquad \forall j
\end{align*}
The unit allocation constraints are written the full backward aggregation method described by Shah (1993). The allocation constraint reads
\begin{align*}
\sum_{i \in I_j} \sum_{t'=t}^{t-p_i+1} W_{ijt'} & \leq 1 \qquad \forall j,t
\end{align*}
Each processing unit $j$ is tagged with a minimum and maximum capacity, $B_{ij}^{min}$ and $B_{ij}^{max}$, respectively, denoting the minimum and maximum batch sizes for each task $i$. A minimum capacity may be needed to cover heat exchange coils in a reactor or mixing blades in a blender, for example. The capacity may depend on the nature of the task being performed. These constraints are written
\begin{align*}
B_{ij}^{min}W_{ijt} & \leq B_{ijt} \leq B_{ij}^{max}W_{ijt} \qquad \forall j, \forall i\in I_j, \forall t
\end{align*}
### Characterization of tasks
```
STATES = STN['STATES']
ST_ARCS = STN['ST_ARCS']
TS_ARCS = STN['TS_ARCS']
UNIT_TASKS = STN['UNIT_TASKS']
TIME = STN['TIME']
H = max(TIME)
# set of tasks
TASKS = set([i for (j,i) in UNIT_TASKS])
# S[i] input set of states which feed task i
S = {i: set() for i in TASKS}
for (s,i) in ST_ARCS:
S[i].add(s)
# S_[i] output set of states fed by task i
S_ = {i: set() for i in TASKS}
for (i,s) in TS_ARCS:
S_[i].add(s)
# rho[(i,s)] input fraction of task i from state s
rho = {(i,s): ST_ARCS[(s,i)]['rho'] for (s,i) in ST_ARCS}
# rho_[(i,s)] output fraction of task i to state s
rho_ = {(i,s): TS_ARCS[(i,s)]['rho'] for (i,s) in TS_ARCS}
# P[(i,s)] time for task i output to state s
P = {(i,s): TS_ARCS[(i,s)]['dur'] for (i,s) in TS_ARCS}
# p[i] completion time for task i
p = {i: max([P[(i,s)] for s in S_[i]]) for i in TASKS}
# K[i] set of units capable of task i
K = {i: set() for i in TASKS}
for (j,i) in UNIT_TASKS:
K[i].add(j)
```
### Characterization of states
```
# T[s] set of tasks receiving material from state s
T = {s: set() for s in STATES}
for (s,i) in ST_ARCS:
T[s].add(i)
# set of tasks producing material for state s
T_ = {s: set() for s in STATES}
for (i,s) in TS_ARCS:
T_[s].add(i)
# C[s] storage capacity for state s
C = {s: STATES[s]['capacity'] for s in STATES}
```
### Characterization of units
```
UNITS = set([j for (j,i) in UNIT_TASKS])
# I[j] set of tasks performed with unit j
I = {j: set() for j in UNITS}
for (j,i) in UNIT_TASKS:
I[j].add(i)
# Bmax[(i,j)] maximum capacity of unit j for task i
Bmax = {(i,j):UNIT_TASKS[(j,i)]['Bmax'] for (j,i) in UNIT_TASKS}
# Bmin[(i,j)] minimum capacity of unit j for task i
Bmin = {(i,j):UNIT_TASKS[(j,i)]['Bmin'] for (j,i) in UNIT_TASKS}
```
### Pyomo model
```
TIME = np.array(TIME)
model = ConcreteModel()
# W[i,j,t] 1 if task i starts in unit j at time t
model.W = Var(TASKS, UNITS, TIME, domain=Boolean)
# B[i,j,t,] size of batch assigned to task i in unit j at time t
model.B = Var(TASKS, UNITS, TIME, domain=NonNegativeReals)
# S[s,t] inventory of state s at time t
model.S = Var(STATES.keys(), TIME, domain=NonNegativeReals)
# Q[j,t] inventory of unit j at time t
model.Q = Var(UNITS, TIME, domain=NonNegativeReals)
# Objective function
# project value
model.Value = Var(domain=NonNegativeReals)
model.valuec = Constraint(expr = model.Value == sum([STATES[s]['price']*model.S[s,H] for s in STATES]))
# project cost
model.Cost = Var(domain=NonNegativeReals)
model.costc = Constraint(expr = model.Cost == sum([UNIT_TASKS[(j,i)]['Cost']*model.W[i,j,t] +
UNIT_TASKS[(j,i)]['vCost']*model.B[i,j,t] for i in TASKS for j in K[i] for t in TIME]))
model.obj = Objective(expr = model.Value - model.Cost, sense = maximize)
# Constraints
model.cons = ConstraintList()
# a unit can only be allocated to one task
for j in UNITS:
for t in TIME:
lhs = 0
for i in I[j]:
for tprime in TIME:
if tprime >= (t-p[i]+1-UNIT_TASKS[(j,i)]['Tclean']) and tprime <= t:
lhs += model.W[i,j,tprime]
model.cons.add(lhs <= 1)
# state capacity constraint
model.sc = Constraint(STATES.keys(), TIME, rule = lambda model, s, t: model.S[s,t] <= C[s])
# state mass balances
for s in STATES.keys():
rhs = STATES[s]['initial']
for t in TIME:
for i in T_[s]:
for j in K[i]:
if t >= P[(i,s)]:
rhs += rho_[(i,s)]*model.B[i,j,max(TIME[TIME <= t-P[(i,s)]])]
for i in T[s]:
rhs -= rho[(i,s)]*sum([model.B[i,j,t] for j in K[i]])
model.cons.add(model.S[s,t] == rhs)
rhs = model.S[s,t]
# unit capacity constraints
for t in TIME:
for j in UNITS:
for i in I[j]:
model.cons.add(model.W[i,j,t]*Bmin[i,j] <= model.B[i,j,t])
model.cons.add(model.B[i,j,t] <= model.W[i,j,t]*Bmax[i,j])
# unit mass balances
for j in UNITS:
rhs = 0
for t in TIME:
rhs += sum([model.B[i,j,t] for i in I[j]])
for i in I[j]:
for s in S_[i]:
if t >= P[(i,s)]:
rhs -= rho_[(i,s)]*model.B[i,j,max(TIME[TIME <= t-P[(i,s)]])]
model.cons.add(model.Q[j,t] == rhs)
rhs = model.Q[j,t]
# unit terminal condition
model.tc = Constraint(UNITS, rule = lambda model, j: model.Q[j,H] == 0)
SolverFactory('cbc').solve(model).write()
```
## Analysis
### Profitability
```
print("Value of State Inventories = {0:12.2f}".format(model.Value()))
print(" Cost of Unit Assignments = {0:12.2f}".format(model.Cost()))
print(" Net Objective = {0:12.2f}".format(model.Value() - model.Cost()))
```
### Unit assignment
```
UnitAssignment = pd.DataFrame({j:[None for t in TIME] for j in UNITS}, index=TIME)
for t in TIME:
for j in UNITS:
for i in I[j]:
for s in S_[i]:
if t-p[i] >= 0:
if model.W[i,j,max(TIME[TIME <= t-p[i]])]() > 0:
UnitAssignment.loc[t,j] = None
for i in I[j]:
if model.W[i,j,t]() > 0:
UnitAssignment.loc[t,j] = (i,model.B[i,j,t]())
UnitAssignment
```
### State inventories
```
pd.DataFrame([[model.S[s,t]() for s in STATES.keys()] for t in TIME], columns = STATES.keys(), index = TIME)
plt.figure(figsize=(10,6))
for (s,idx) in zip(STATES.keys(),range(0,len(STATES.keys()))):
plt.subplot(ceil(len(STATES.keys())/3),3,idx+1)
tlast,ylast = 0,STATES[s]['initial']
for (t,y) in zip(list(TIME),[model.S[s,t]() for t in TIME]):
plt.plot([tlast,t,t],[ylast,ylast,y],'b')
#plt.plot([tlast,t],[ylast,y],'b.',ms=10)
tlast,ylast = t,y
plt.ylim(0,1.1*C[s])
plt.plot([0,H],[C[s],C[s]],'r--')
plt.title(s)
plt.tight_layout()
```
### Unit batch inventories
```
pd.DataFrame([[model.Q[j,t]() for j in UNITS] for t in TIME], columns = UNITS, index = TIME)
```
### Gannt chart
```
plt.figure(figsize=(12,6))
gap = H/500
idx = 1
lbls = []
ticks = []
for j in sorted(UNITS):
idx -= 1
for i in sorted(I[j]):
idx -= 1
ticks.append(idx)
lbls.append("{0:s} -> {1:s}".format(j,i))
plt.plot([0,H],[idx,idx],lw=20,alpha=.3,color='y')
for t in TIME:
if model.W[i,j,t]() > 0:
plt.plot([t+gap,t+p[i]-gap], [idx,idx],'b', lw=20, solid_capstyle='butt')
txt = "{0:.2f}".format(model.B[i,j,t]())
plt.text(t+p[i]/2, idx, txt, color='white', weight='bold', ha='center', va='center')
plt.xlim(0,H)
plt.gca().set_yticks(ticks)
plt.gca().set_yticklabels(lbls);
```
## Trace of events and states
```
sep = '\n--------------------------------------------------------------------------------------------\n'
print(sep)
print("Starting Conditions")
print(" Initial Inventories:")
for s in STATES.keys():
print(" {0:10s} {1:6.1f} kg".format(s,STATES[s]['initial']))
units = {j:{'assignment':'None', 't':0} for j in UNITS}
for t in TIME:
print(sep)
print("Time =",t,"hr")
print(" Instructions:")
for j in UNITS:
units[j]['t'] += 1
# transfer from unit to states
for i in I[j]:
for s in S_[i]:
if t-P[(i,s)] >= 0:
amt = rho_[(i,s)]*model.B[i,j,max(TIME[TIME <= t - P[(i,s)]])]()
if amt > 0:
print(" Transfer", amt, "kg from", j, "to", s)
for j in UNITS:
# release units from tasks
for i in I[j]:
if t-p[i] >= 0:
if model.W[i,j,max(TIME[TIME <= t-p[i]])]() > 0:
print(" Release", j, "from", i)
units[j]['assignment'] = 'None'
units[j]['t'] = 0
# assign units to tasks
for i in I[j]:
if model.W[i,j,t]() > 0:
print(" Assign", j, "with capacity", Bmax[(i,j)], "kg to task",i,"for",p[i],"hours")
units[j]['assignment'] = i
units[j]['t'] = 1
# transfer from states to starting tasks
for i in I[j]:
for s in S[i]:
amt = rho[(i,s)]*model.B[i,j,t]()
if amt > 0:
print(" Transfer", amt,"kg from", s, "to", j)
print("\n Inventories are now:")
for s in STATES.keys():
print(" {0:10s} {1:6.1f} kg".format(s,model.S[s,t]()))
print("\n Unit Assignments are now:")
for j in UNITS:
if units[j]['assignment'] != 'None':
fmt = " {0:s} performs the {1:s} task with a {2:.2f} kg batch for hour {3:f} of {4:f}"
i = units[j]['assignment']
print(fmt.format(j,i,model.Q[j,t](),units[j]['t'],p[i]))
print(sep)
print('Final Conditions')
print(" Final Inventories:")
for s in STATES.keys():
print(" {0:10s} {1:6.1f} kg".format(s,model.S[s,H]()))
```
|
github_jupyter
|
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
from IPython.display import display, HTML
import shutil
import sys
import os.path
if not shutil.which("pyomo"):
!pip install -q pyomo
assert(shutil.which("pyomo"))
if not (shutil.which("cbc") or os.path.isfile("cbc")):
if "google.colab" in sys.modules:
!apt-get install -y -qq coinor-cbc
else:
try:
!conda install -c conda-forge coincbc
except:
pass
assert(shutil.which("cbc") or os.path.isfile("cbc"))
from pyomo.environ import *
# planning horizon
H = 10
Kondili = {
# time grid
'TIME': range(0, H+1),
# states
'STATES': {
'Feed_A' : {'capacity': 500, 'initial': 500, 'price': 0},
'Feed_B' : {'capacity': 500, 'initial': 500, 'price': 0},
'Feed_C' : {'capacity': 500, 'initial': 500, 'price': 0},
'Hot_A' : {'capacity': 100, 'initial': 0, 'price': -100},
'Int_AB' : {'capacity': 200, 'initial': 0, 'price': -100},
'Int_BC' : {'capacity': 150, 'initial': 0, 'price': -100},
'Impure_E' : {'capacity': 100, 'initial': 0, 'price': -100},
'Product_1': {'capacity': 500, 'initial': 0, 'price': 10},
'Product_2': {'capacity': 500, 'initial': 0, 'price': 10},
},
# state-to-task arcs indexed by (state, task)
'ST_ARCS': {
('Feed_A', 'Heating') : {'rho': 1.0},
('Feed_B', 'Reaction_1'): {'rho': 0.5},
('Feed_C', 'Reaction_1'): {'rho': 0.5},
('Feed_C', 'Reaction_3'): {'rho': 0.2},
('Hot_A', 'Reaction_2'): {'rho': 0.4},
('Int_AB', 'Reaction_3'): {'rho': 0.8},
('Int_BC', 'Reaction_2'): {'rho': 0.6},
('Impure_E', 'Separation'): {'rho': 1.0},
},
# task-to-state arcs indexed by (task, state)
'TS_ARCS': {
('Heating', 'Hot_A') : {'dur': 1, 'rho': 1.0},
('Reaction_2', 'Product_1'): {'dur': 2, 'rho': 0.4},
('Reaction_2', 'Int_AB') : {'dur': 2, 'rho': 0.6},
('Reaction_1', 'Int_BC') : {'dur': 2, 'rho': 1.0},
('Reaction_3', 'Impure_E') : {'dur': 1, 'rho': 1.0},
('Separation', 'Int_AB') : {'dur': 2, 'rho': 0.1},
('Separation', 'Product_2'): {'dur': 1, 'rho': 0.9},
},
# unit data indexed by (unit, task)
'UNIT_TASKS': {
('Heater', 'Heating') : {'Bmin': 0, 'Bmax': 100, 'Cost': 1, 'vCost': 0, 'Tclean': 0},
('Reactor_1', 'Reaction_1'): {'Bmin': 0, 'Bmax': 80, 'Cost': 1, 'vCost': 0, 'Tclean': 0},
('Reactor_1', 'Reaction_2'): {'Bmin': 0, 'Bmax': 80, 'Cost': 1, 'vCost': 0, 'Tclean': 0},
('Reactor_1', 'Reaction_3'): {'Bmin': 0, 'Bmax': 80, 'Cost': 1, 'vCost': 0, 'Tclean': 0},
('Reactor_2', 'Reaction_1'): {'Bmin': 0, 'Bmax': 80, 'Cost': 1, 'vCost': 0, 'Tclean': 0},
('Reactor_2', 'Reaction_2'): {'Bmin': 0, 'Bmax': 80, 'Cost': 1, 'vCost': 0, 'Tclean': 0},
('Reactor_2', 'Reaction_3'): {'Bmin': 0, 'Bmax': 80, 'Cost': 1, 'vCost': 0, 'Tclean': 0},
('Still', 'Separation'): {'Bmin': 0, 'Bmax': 200, 'Cost': 1, 'vCost': 0, 'Tclean': 0},
},
}
STN = Kondili
H = 16
Hydrolubes = {
# time grid
'TIME': range(0, H+1),
# states
'STATES': {
'Feed_A' : {'capacity': 500, 'initial': 500, 'price': 0},
'Feed_B' : {'capacity': 500, 'initial': 500, 'price': 0},
'Feed_C' : {'capacity': 500, 'initial': 500, 'price': 0},
'Hot_A' : {'capacity': 100, 'initial': 0, 'price': -100},
'Int_AB' : {'capacity': 200, 'initial': 0, 'price': -100},
'Int_BC' : {'capacity': 150, 'initial': 0, 'price': -100},
'Impure_E' : {'capacity': 100, 'initial': 0, 'price': -100},
'Product_1': {'capacity': 500, 'initial': 0, 'price': 10},
'Product_2': {'capacity': 500, 'initial': 0, 'price': 10},
},
# state-to-task arcs indexed by (state, task)
'ST_ARCS': {
('Feed_A', 'Heating') : {'rho': 1.0},
('Feed_B', 'Reaction_1'): {'rho': 0.5},
('Feed_C', 'Reaction_1'): {'rho': 0.5},
('Feed_C', 'Reaction_3'): {'rho': 0.2},
('Hot_A', 'Reaction_2'): {'rho': 0.4},
('Int_AB', 'Reaction_3'): {'rho': 0.8},
('Int_BC', 'Reaction_2'): {'rho': 0.6},
('Impure_E', 'Separation'): {'rho': 1.0},
},
# task-to-state arcs indexed by (task, state)
'TS_ARCS': {
('Heating', 'Hot_A') : {'dur': 1, 'rho': 1.0},
('Reaction_2', 'Product_1'): {'dur': 2, 'rho': 0.4},
('Reaction_2', 'Int_AB') : {'dur': 2, 'rho': 0.6},
('Reaction_1', 'Int_BC') : {'dur': 2, 'rho': 1.0},
('Reaction_3', 'Impure_E') : {'dur': 1, 'rho': 1.0},
('Separation', 'Int_AB') : {'dur': 2, 'rho': 0.1},
('Separation', 'Product_2'): {'dur': 1, 'rho': 0.9},
},
# unit data indexed by (unit, task)
'UNIT_TASKS': {
('Heater', 'Heating') : {'Bmin': 0, 'Bmax': 100, 'Cost': 1, 'vCost': 0, 'Tclean': 0},
('Reactor_1', 'Reaction_1'): {'Bmin': 0, 'Bmax': 80, 'Cost': 1, 'vCost': 0, 'Tclean': 0},
('Reactor_1', 'Reaction_2'): {'Bmin': 0, 'Bmax': 80, 'Cost': 1, 'vCost': 0, 'Tclean': 0},
('Reactor_1', 'Reaction_3'): {'Bmin': 0, 'Bmax': 80, 'Cost': 1, 'vCost': 0, 'Tclean': 0},
('Reactor_2', 'Reaction_1'): {'Bmin': 0, 'Bmax': 80, 'Cost': 1, 'vCost': 0, 'Tclean': 0},
('Reactor_2', 'Reaction_2'): {'Bmin': 0, 'Bmax': 80, 'Cost': 1, 'vCost': 0, 'Tclean': 0},
('Reactor_2', 'Reaction_3'): {'Bmin': 0, 'Bmax': 80, 'Cost': 1, 'vCost': 0, 'Tclean': 0},
('Still', 'Separation'): {'Bmin': 0, 'Bmax': 200, 'Cost': 1, 'vCost': 0, 'Tclean': 0},
},
}
STN = Hydrolubes
STATES = STN['STATES']
ST_ARCS = STN['ST_ARCS']
TS_ARCS = STN['TS_ARCS']
UNIT_TASKS = STN['UNIT_TASKS']
TIME = STN['TIME']
H = max(TIME)
# set of tasks
TASKS = set([i for (j,i) in UNIT_TASKS])
# S[i] input set of states which feed task i
S = {i: set() for i in TASKS}
for (s,i) in ST_ARCS:
S[i].add(s)
# S_[i] output set of states fed by task i
S_ = {i: set() for i in TASKS}
for (i,s) in TS_ARCS:
S_[i].add(s)
# rho[(i,s)] input fraction of task i from state s
rho = {(i,s): ST_ARCS[(s,i)]['rho'] for (s,i) in ST_ARCS}
# rho_[(i,s)] output fraction of task i to state s
rho_ = {(i,s): TS_ARCS[(i,s)]['rho'] for (i,s) in TS_ARCS}
# P[(i,s)] time for task i output to state s
P = {(i,s): TS_ARCS[(i,s)]['dur'] for (i,s) in TS_ARCS}
# p[i] completion time for task i
p = {i: max([P[(i,s)] for s in S_[i]]) for i in TASKS}
# K[i] set of units capable of task i
K = {i: set() for i in TASKS}
for (j,i) in UNIT_TASKS:
K[i].add(j)
# T[s] set of tasks receiving material from state s
T = {s: set() for s in STATES}
for (s,i) in ST_ARCS:
T[s].add(i)
# set of tasks producing material for state s
T_ = {s: set() for s in STATES}
for (i,s) in TS_ARCS:
T_[s].add(i)
# C[s] storage capacity for state s
C = {s: STATES[s]['capacity'] for s in STATES}
UNITS = set([j for (j,i) in UNIT_TASKS])
# I[j] set of tasks performed with unit j
I = {j: set() for j in UNITS}
for (j,i) in UNIT_TASKS:
I[j].add(i)
# Bmax[(i,j)] maximum capacity of unit j for task i
Bmax = {(i,j):UNIT_TASKS[(j,i)]['Bmax'] for (j,i) in UNIT_TASKS}
# Bmin[(i,j)] minimum capacity of unit j for task i
Bmin = {(i,j):UNIT_TASKS[(j,i)]['Bmin'] for (j,i) in UNIT_TASKS}
TIME = np.array(TIME)
model = ConcreteModel()
# W[i,j,t] 1 if task i starts in unit j at time t
model.W = Var(TASKS, UNITS, TIME, domain=Boolean)
# B[i,j,t,] size of batch assigned to task i in unit j at time t
model.B = Var(TASKS, UNITS, TIME, domain=NonNegativeReals)
# S[s,t] inventory of state s at time t
model.S = Var(STATES.keys(), TIME, domain=NonNegativeReals)
# Q[j,t] inventory of unit j at time t
model.Q = Var(UNITS, TIME, domain=NonNegativeReals)
# Objective function
# project value
model.Value = Var(domain=NonNegativeReals)
model.valuec = Constraint(expr = model.Value == sum([STATES[s]['price']*model.S[s,H] for s in STATES]))
# project cost
model.Cost = Var(domain=NonNegativeReals)
model.costc = Constraint(expr = model.Cost == sum([UNIT_TASKS[(j,i)]['Cost']*model.W[i,j,t] +
UNIT_TASKS[(j,i)]['vCost']*model.B[i,j,t] for i in TASKS for j in K[i] for t in TIME]))
model.obj = Objective(expr = model.Value - model.Cost, sense = maximize)
# Constraints
model.cons = ConstraintList()
# a unit can only be allocated to one task
for j in UNITS:
for t in TIME:
lhs = 0
for i in I[j]:
for tprime in TIME:
if tprime >= (t-p[i]+1-UNIT_TASKS[(j,i)]['Tclean']) and tprime <= t:
lhs += model.W[i,j,tprime]
model.cons.add(lhs <= 1)
# state capacity constraint
model.sc = Constraint(STATES.keys(), TIME, rule = lambda model, s, t: model.S[s,t] <= C[s])
# state mass balances
for s in STATES.keys():
rhs = STATES[s]['initial']
for t in TIME:
for i in T_[s]:
for j in K[i]:
if t >= P[(i,s)]:
rhs += rho_[(i,s)]*model.B[i,j,max(TIME[TIME <= t-P[(i,s)]])]
for i in T[s]:
rhs -= rho[(i,s)]*sum([model.B[i,j,t] for j in K[i]])
model.cons.add(model.S[s,t] == rhs)
rhs = model.S[s,t]
# unit capacity constraints
for t in TIME:
for j in UNITS:
for i in I[j]:
model.cons.add(model.W[i,j,t]*Bmin[i,j] <= model.B[i,j,t])
model.cons.add(model.B[i,j,t] <= model.W[i,j,t]*Bmax[i,j])
# unit mass balances
for j in UNITS:
rhs = 0
for t in TIME:
rhs += sum([model.B[i,j,t] for i in I[j]])
for i in I[j]:
for s in S_[i]:
if t >= P[(i,s)]:
rhs -= rho_[(i,s)]*model.B[i,j,max(TIME[TIME <= t-P[(i,s)]])]
model.cons.add(model.Q[j,t] == rhs)
rhs = model.Q[j,t]
# unit terminal condition
model.tc = Constraint(UNITS, rule = lambda model, j: model.Q[j,H] == 0)
SolverFactory('cbc').solve(model).write()
print("Value of State Inventories = {0:12.2f}".format(model.Value()))
print(" Cost of Unit Assignments = {0:12.2f}".format(model.Cost()))
print(" Net Objective = {0:12.2f}".format(model.Value() - model.Cost()))
UnitAssignment = pd.DataFrame({j:[None for t in TIME] for j in UNITS}, index=TIME)
for t in TIME:
for j in UNITS:
for i in I[j]:
for s in S_[i]:
if t-p[i] >= 0:
if model.W[i,j,max(TIME[TIME <= t-p[i]])]() > 0:
UnitAssignment.loc[t,j] = None
for i in I[j]:
if model.W[i,j,t]() > 0:
UnitAssignment.loc[t,j] = (i,model.B[i,j,t]())
UnitAssignment
pd.DataFrame([[model.S[s,t]() for s in STATES.keys()] for t in TIME], columns = STATES.keys(), index = TIME)
plt.figure(figsize=(10,6))
for (s,idx) in zip(STATES.keys(),range(0,len(STATES.keys()))):
plt.subplot(ceil(len(STATES.keys())/3),3,idx+1)
tlast,ylast = 0,STATES[s]['initial']
for (t,y) in zip(list(TIME),[model.S[s,t]() for t in TIME]):
plt.plot([tlast,t,t],[ylast,ylast,y],'b')
#plt.plot([tlast,t],[ylast,y],'b.',ms=10)
tlast,ylast = t,y
plt.ylim(0,1.1*C[s])
plt.plot([0,H],[C[s],C[s]],'r--')
plt.title(s)
plt.tight_layout()
pd.DataFrame([[model.Q[j,t]() for j in UNITS] for t in TIME], columns = UNITS, index = TIME)
plt.figure(figsize=(12,6))
gap = H/500
idx = 1
lbls = []
ticks = []
for j in sorted(UNITS):
idx -= 1
for i in sorted(I[j]):
idx -= 1
ticks.append(idx)
lbls.append("{0:s} -> {1:s}".format(j,i))
plt.plot([0,H],[idx,idx],lw=20,alpha=.3,color='y')
for t in TIME:
if model.W[i,j,t]() > 0:
plt.plot([t+gap,t+p[i]-gap], [idx,idx],'b', lw=20, solid_capstyle='butt')
txt = "{0:.2f}".format(model.B[i,j,t]())
plt.text(t+p[i]/2, idx, txt, color='white', weight='bold', ha='center', va='center')
plt.xlim(0,H)
plt.gca().set_yticks(ticks)
plt.gca().set_yticklabels(lbls);
sep = '\n--------------------------------------------------------------------------------------------\n'
print(sep)
print("Starting Conditions")
print(" Initial Inventories:")
for s in STATES.keys():
print(" {0:10s} {1:6.1f} kg".format(s,STATES[s]['initial']))
units = {j:{'assignment':'None', 't':0} for j in UNITS}
for t in TIME:
print(sep)
print("Time =",t,"hr")
print(" Instructions:")
for j in UNITS:
units[j]['t'] += 1
# transfer from unit to states
for i in I[j]:
for s in S_[i]:
if t-P[(i,s)] >= 0:
amt = rho_[(i,s)]*model.B[i,j,max(TIME[TIME <= t - P[(i,s)]])]()
if amt > 0:
print(" Transfer", amt, "kg from", j, "to", s)
for j in UNITS:
# release units from tasks
for i in I[j]:
if t-p[i] >= 0:
if model.W[i,j,max(TIME[TIME <= t-p[i]])]() > 0:
print(" Release", j, "from", i)
units[j]['assignment'] = 'None'
units[j]['t'] = 0
# assign units to tasks
for i in I[j]:
if model.W[i,j,t]() > 0:
print(" Assign", j, "with capacity", Bmax[(i,j)], "kg to task",i,"for",p[i],"hours")
units[j]['assignment'] = i
units[j]['t'] = 1
# transfer from states to starting tasks
for i in I[j]:
for s in S[i]:
amt = rho[(i,s)]*model.B[i,j,t]()
if amt > 0:
print(" Transfer", amt,"kg from", s, "to", j)
print("\n Inventories are now:")
for s in STATES.keys():
print(" {0:10s} {1:6.1f} kg".format(s,model.S[s,t]()))
print("\n Unit Assignments are now:")
for j in UNITS:
if units[j]['assignment'] != 'None':
fmt = " {0:s} performs the {1:s} task with a {2:.2f} kg batch for hour {3:f} of {4:f}"
i = units[j]['assignment']
print(fmt.format(j,i,model.Q[j,t](),units[j]['t'],p[i]))
print(sep)
print('Final Conditions')
print(" Final Inventories:")
for s in STATES.keys():
print(" {0:10s} {1:6.1f} kg".format(s,model.S[s,H]()))
| 0.307878 | 0.897291 |
```
# TensorFlow and tf.keras
import tensorflow as tf
import random
import numpy as np
import numpy.matlib as matlib
import matplotlib.pyplot as plt
dataset = tf.keras.datasets.mnist
(train_images, train_labels), (test_images, test_labels) = dataset.load_data()
train_images = train_images / 256.0
test_images = test_images / 256.0
float_formatter = "{:.2f}".format
# plot functions
def plot_flat_images(images, dim, items=16, cmap=plt.cm.viridis):
plt.figure(figsize=(10, np.ceil(items/16)*10))
for i in range(items):
plt.subplot(np.ceil(items/4), 4,i+1)
plt.xticks([])
plt.yticks([])
plt.grid(False)
plt.imshow(images[i].reshape(dim), cmap)
plt.show()
def plot_vector(vector, cmap=plt.cm.viridis, colorBar=False):
plt.xticks([])
plt.yticks([])
plt.grid(False)
plt.imshow(vector.reshape(1, vector.shape[0]), cmap)
if colorBar:
plt.colorbar()
# neural network helper functions
def sigmoid(z):
"""The sigmoid function."""
return 1.0/(1.0+np.exp(-z))
def sigmoid_prime(z):
"""Derivative of the sigmoid function."""
return sigmoid(z)*(1-sigmoid(z))
def feedforward(a):
"""Return the output of the network if "a" is input."""
for b, w in zip(biases, weights):
a = sigmoid(np.dot(w, a)+b)
return a
def vectorized_result(j):
"""Return a 10-dimensional unit vector with a 1.0 in the jth
position and zeroes elsewhere. This is used to convert a digit
(0...9) into a corresponding desired output from the neural
network."""
e = np.zeros((10, 1))
e[j] = 1.0
return e
sizes = [28*28, 24, 10]
num_layers = len(sizes)
weights = [np.random.rand(y, x)*2-1 for x, y in zip(sizes[:-1], sizes[1:])]
biases = [np.random.rand(y, 1)*2-1 for y in sizes[1:]]
plot_flat_images(train_images, (train_images.shape[1], train_images.shape[2]), 4)
plot_flat_images(weights[0], (train_images.shape[1], train_images.shape[2]), 16, cmap=plt.cm.seismic)
def SGD(training_data, epochs, mini_batch_size, eta,
test_data=None):
"""Train the neural network using mini-batch stochastic
gradient descent. The ``training_data`` is a list of tuples
``(x, y)`` representing the training inputs and the desired
outputs. The other non-optional parameters are
self-explanatory. If ``test_data`` is provided then the
network will be evaluated against the test data after each
epoch, and partial progress printed out. This is useful for
tracking progress, but slows things down substantially."""
training_data = list(training_data)
n = len(training_data)
if test_data:
test_data = list(test_data)
n_test = len(test_data)
for j in range(epochs):
random.shuffle(training_data)
mini_batches = [
training_data[k:k+mini_batch_size]
for k in range(0, n, mini_batch_size)]
for mini_batch in mini_batches:
update_mini_batch(mini_batch, eta)
if test_data:
print("Epoch {} : {} / {}".format(j, evaluate(test_data),n_test))
else:
print("Epoch {} complete".format(j))
def update_mini_batch(mini_batch, eta):
"""Update the network's weights and biases by applying
gradient descent using backpropagation to a single mini batch.
The ``mini_batch`` is a list of tuples ``(x, y)``, and ``eta``
is the learning rate."""
global weights, biases
nabla_b = [np.zeros(b.shape) for b in biases]
nabla_w = [np.zeros(w.shape) for w in weights]
for x, y in mini_batch:
delta_nabla_b, delta_nabla_w = backprop(x, y)
nabla_b = [nb+dnb for nb, dnb in zip(nabla_b, delta_nabla_b)]
nabla_w = [nw+dnw for nw, dnw in zip(nabla_w, delta_nabla_w)]
weights = [w-(eta/len(mini_batch))*nw
for w, nw in zip(weights, nabla_w)]
biases = [b-(eta/len(mini_batch))*nb
for b, nb in zip(biases, nabla_b)]
def backprop(x, y):
"""Return a tuple ``(nabla_b, nabla_w)`` representing the
gradient for the cost function C_x. ``nabla_b`` and
``nabla_w`` are layer-by-layer lists of numpy arrays, similar
to ``self.biases`` and ``self.weights``."""
global weights, biases, num_layers
nabla_b = [np.zeros(b.shape) for b in biases]
nabla_w = [np.zeros(w.shape) for w in weights]
# feedforward
activation = x
activations = [x] # list to store all the activations, layer by layer
zs = [] # list to store all the z vectors, layer by layer
for b, w in zip(biases, weights):
z = np.dot(w, activation)+b
zs.append(z)
activation = sigmoid(z)
activations.append(activation)
# backward pass
delta = cost_derivative(activations[-1], y) * \
sigmoid_prime(zs[-1])
nabla_b[-1] = delta
nabla_w[-1] = np.dot(delta, activations[-2].transpose())
# Note that the variable l in the loop below is used a little
# differently to the notation in Chapter 2 of the book. Here,
# l = 1 means the last layer of neurons, l = 2 is the
# second-last layer, and so on. It's a renumbering of the
# scheme in the book, used here to take advantage of the fact
# that Python can use negative indices in lists.
for l in range(2, num_layers):
z = zs[-l]
sp = sigmoid_prime(z)
delta = np.dot(weights[-l+1].transpose(), delta) * sp
nabla_b[-l] = delta
nabla_w[-l] = np.dot(delta, activations[-l-1].transpose())
return (nabla_b, nabla_w)
def evaluate(test_data):
"""Return the number of test inputs for which the neural
network outputs the correct result. Note that the neural
network's output is assumed to be the index of whichever
neuron in the final layer has the highest activation."""
test_results = [(np.argmax(feedforward(x)), y)
for (x, y) in test_data]
return sum(int(x == y) for (x, y) in test_results)
def cost_derivative(output_activations, y):
"""Return the vector of partial derivatives \partial C_x /
\partial a for the output activations."""
return (output_activations-y)
training_inputs = [np.reshape(x, (784, 1)) for x in train_images]
training_results = [vectorized_result(y) for y in train_labels]
training_data = list(zip(training_inputs, training_results))
test_inputs = [np.reshape(x, (784, 1)) for x in test_images]
test_data = zip(test_inputs, test_labels)
SGD(training_data, epochs=10, mini_batch_size=10, eta=3.0, test_data=test_data)
plot_flat_images(weights[0], (train_images.shape[1], train_images.shape[2]), cmap=plt.cm.seismic)
plot_flat_images(weights[1], (3, 8), 10, cmap=plt.cm.bwr)
plot_flat_images(weights[2], (4, 4), 10, cmap=plt.cm.bwr)
np.argmax(feedforward(training_inputs[0]))
np.set_printoptions(formatter={'float_kind':float_formatter})
np.reshape(feedforward(training_inputs[0]), (10))
```
|
github_jupyter
|
# TensorFlow and tf.keras
import tensorflow as tf
import random
import numpy as np
import numpy.matlib as matlib
import matplotlib.pyplot as plt
dataset = tf.keras.datasets.mnist
(train_images, train_labels), (test_images, test_labels) = dataset.load_data()
train_images = train_images / 256.0
test_images = test_images / 256.0
float_formatter = "{:.2f}".format
# plot functions
def plot_flat_images(images, dim, items=16, cmap=plt.cm.viridis):
plt.figure(figsize=(10, np.ceil(items/16)*10))
for i in range(items):
plt.subplot(np.ceil(items/4), 4,i+1)
plt.xticks([])
plt.yticks([])
plt.grid(False)
plt.imshow(images[i].reshape(dim), cmap)
plt.show()
def plot_vector(vector, cmap=plt.cm.viridis, colorBar=False):
plt.xticks([])
plt.yticks([])
plt.grid(False)
plt.imshow(vector.reshape(1, vector.shape[0]), cmap)
if colorBar:
plt.colorbar()
# neural network helper functions
def sigmoid(z):
"""The sigmoid function."""
return 1.0/(1.0+np.exp(-z))
def sigmoid_prime(z):
"""Derivative of the sigmoid function."""
return sigmoid(z)*(1-sigmoid(z))
def feedforward(a):
"""Return the output of the network if "a" is input."""
for b, w in zip(biases, weights):
a = sigmoid(np.dot(w, a)+b)
return a
def vectorized_result(j):
"""Return a 10-dimensional unit vector with a 1.0 in the jth
position and zeroes elsewhere. This is used to convert a digit
(0...9) into a corresponding desired output from the neural
network."""
e = np.zeros((10, 1))
e[j] = 1.0
return e
sizes = [28*28, 24, 10]
num_layers = len(sizes)
weights = [np.random.rand(y, x)*2-1 for x, y in zip(sizes[:-1], sizes[1:])]
biases = [np.random.rand(y, 1)*2-1 for y in sizes[1:]]
plot_flat_images(train_images, (train_images.shape[1], train_images.shape[2]), 4)
plot_flat_images(weights[0], (train_images.shape[1], train_images.shape[2]), 16, cmap=plt.cm.seismic)
def SGD(training_data, epochs, mini_batch_size, eta,
test_data=None):
"""Train the neural network using mini-batch stochastic
gradient descent. The ``training_data`` is a list of tuples
``(x, y)`` representing the training inputs and the desired
outputs. The other non-optional parameters are
self-explanatory. If ``test_data`` is provided then the
network will be evaluated against the test data after each
epoch, and partial progress printed out. This is useful for
tracking progress, but slows things down substantially."""
training_data = list(training_data)
n = len(training_data)
if test_data:
test_data = list(test_data)
n_test = len(test_data)
for j in range(epochs):
random.shuffle(training_data)
mini_batches = [
training_data[k:k+mini_batch_size]
for k in range(0, n, mini_batch_size)]
for mini_batch in mini_batches:
update_mini_batch(mini_batch, eta)
if test_data:
print("Epoch {} : {} / {}".format(j, evaluate(test_data),n_test))
else:
print("Epoch {} complete".format(j))
def update_mini_batch(mini_batch, eta):
"""Update the network's weights and biases by applying
gradient descent using backpropagation to a single mini batch.
The ``mini_batch`` is a list of tuples ``(x, y)``, and ``eta``
is the learning rate."""
global weights, biases
nabla_b = [np.zeros(b.shape) for b in biases]
nabla_w = [np.zeros(w.shape) for w in weights]
for x, y in mini_batch:
delta_nabla_b, delta_nabla_w = backprop(x, y)
nabla_b = [nb+dnb for nb, dnb in zip(nabla_b, delta_nabla_b)]
nabla_w = [nw+dnw for nw, dnw in zip(nabla_w, delta_nabla_w)]
weights = [w-(eta/len(mini_batch))*nw
for w, nw in zip(weights, nabla_w)]
biases = [b-(eta/len(mini_batch))*nb
for b, nb in zip(biases, nabla_b)]
def backprop(x, y):
"""Return a tuple ``(nabla_b, nabla_w)`` representing the
gradient for the cost function C_x. ``nabla_b`` and
``nabla_w`` are layer-by-layer lists of numpy arrays, similar
to ``self.biases`` and ``self.weights``."""
global weights, biases, num_layers
nabla_b = [np.zeros(b.shape) for b in biases]
nabla_w = [np.zeros(w.shape) for w in weights]
# feedforward
activation = x
activations = [x] # list to store all the activations, layer by layer
zs = [] # list to store all the z vectors, layer by layer
for b, w in zip(biases, weights):
z = np.dot(w, activation)+b
zs.append(z)
activation = sigmoid(z)
activations.append(activation)
# backward pass
delta = cost_derivative(activations[-1], y) * \
sigmoid_prime(zs[-1])
nabla_b[-1] = delta
nabla_w[-1] = np.dot(delta, activations[-2].transpose())
# Note that the variable l in the loop below is used a little
# differently to the notation in Chapter 2 of the book. Here,
# l = 1 means the last layer of neurons, l = 2 is the
# second-last layer, and so on. It's a renumbering of the
# scheme in the book, used here to take advantage of the fact
# that Python can use negative indices in lists.
for l in range(2, num_layers):
z = zs[-l]
sp = sigmoid_prime(z)
delta = np.dot(weights[-l+1].transpose(), delta) * sp
nabla_b[-l] = delta
nabla_w[-l] = np.dot(delta, activations[-l-1].transpose())
return (nabla_b, nabla_w)
def evaluate(test_data):
"""Return the number of test inputs for which the neural
network outputs the correct result. Note that the neural
network's output is assumed to be the index of whichever
neuron in the final layer has the highest activation."""
test_results = [(np.argmax(feedforward(x)), y)
for (x, y) in test_data]
return sum(int(x == y) for (x, y) in test_results)
def cost_derivative(output_activations, y):
"""Return the vector of partial derivatives \partial C_x /
\partial a for the output activations."""
return (output_activations-y)
training_inputs = [np.reshape(x, (784, 1)) for x in train_images]
training_results = [vectorized_result(y) for y in train_labels]
training_data = list(zip(training_inputs, training_results))
test_inputs = [np.reshape(x, (784, 1)) for x in test_images]
test_data = zip(test_inputs, test_labels)
SGD(training_data, epochs=10, mini_batch_size=10, eta=3.0, test_data=test_data)
plot_flat_images(weights[0], (train_images.shape[1], train_images.shape[2]), cmap=plt.cm.seismic)
plot_flat_images(weights[1], (3, 8), 10, cmap=plt.cm.bwr)
plot_flat_images(weights[2], (4, 4), 10, cmap=plt.cm.bwr)
np.argmax(feedforward(training_inputs[0]))
np.set_printoptions(formatter={'float_kind':float_formatter})
np.reshape(feedforward(training_inputs[0]), (10))
| 0.86012 | 0.864825 |
<div style = "font-family:Georgia;
font-size:2.5vw;
color:lightblue;
font-style:bold;
text-align:center;
background:url('./Animations/Title Background.gif') no-repeat center;
background-size:cover)">
<br><br>
Histograms of Oriented Gradients (HOG)
<br><br><br>
</div>
<h1 style = "text-align:left">Introduction</h1>
As we saw with the ORB algorithm, we can use keypoints in images to do keypoint-based matching to detect objects in images. These type of algorithms work great when you want to detect objects that have a lot of consistent internal features that are not affected by the background. For example, these algorithms work well for facial detection because faces have a lot of consistent internal features that don’t get affected by the image background, such as the eyes, nose, and mouth. However, these type of algorithms don’t work so well when attempting to do more general object recognition, say for example, pedestrian detection in images. The reason is that people don’t have consistent internal features, like faces do, because the body shape and style of every person is different (see Fig. 1). This means that every person is going to have a different set of internal features, and so we need something that can more generally describe a person.
<br>
<figure>
<img src = "./Animations/pedestrians.jpeg" width = "100%" style = "border: thin silver solid; padding: 10px">
<figcaption style = "text-align:left; font-style:italic">Fig. 1. - Pedestrians.</figcaption>
</figure>
<br>
One option is to try to detect pedestrians by their contours instead. Detecting objects in images by their contours (boundaries) is very challenging because we have to deal with the difficulties brought about by the contrast between the background and the foreground. For example, suppose you wanted to detect a pedestrian in an image that is walking in front of a white building and she is wearing a white coat and black pants (see Fig. 2). We can see in Fig. 2, that since the background of the image is mostly white, the black pants are going to have a very high contrast, but the coat, since it is white as well, is going to have very low contrast. In this case, detecting the edges of pants is going to be easy but detecting the edges of the coat is going to be very difficult. This is where **HOG** comes in. HOG stands for **Histograms of Oriented Gradients** and it was first introduced by Navneet Dalal and Bill Triggs in 2005.
<br>
<figure>
<img src = "./Animations/woman.jpg" width = "100%" style = "border: thin silver solid; padding: 10px">
<figcaption style = "text-align:left; font-style:italic">Fig. 2. - High and Low Contrast.</figcaption>
</figure>
<br>
The HOG algorithm works by creating histograms of the distribution of gradient orientations in an image and then normalizing them in a very special way. This special normalization is what makes HOG so effective at detecting the edges of objects even in cases where the contrast is very low. These normalized histograms are put together into a feature vector, known as the HOG descriptor, that can be used to train a machine learning algorithm, such as a Support Vector Machine (SVM), to detect objects in images based on their boundaries (edges). Due to its great success and reliability, HOG has become one of the most widely used algorithms in computer vison for object detection.
In this notebook, you will learn:
* How the HOG algorithm works
* How to use OpenCV to create a HOG descriptor
* How to visualize the HOG descriptor.
# The HOG Algorithm
As its name suggests, the HOG algorithm, is based on creating histograms from the orientation of image gradients. The HOG algorithm is implemented in a series of steps:
1. Given the image of particular object, set a detection window (region of interest) that covers the entire object in the image (see Fig. 3).
2. Calculate the magnitude and direction of the gradient for each individual pixel in the detection window.
3. Divide the detection window into connected *cells* of pixels, with all cells being of the same size (see Fig. 3). The size of the cells is a free parameter and it is usually chosen so as to match the scale of the features that want to be detected. For example, in a 64 x 128 pixel detection window, square cells 6 to 8 pixels wide are suitable for detecting human limbs.
4. Create a Histogram for each cell, by first grouping the gradient directions of all pixels in each cell into a particular number of orientation (angular) bins; and then adding up the gradient magnitudes of the gradients in each angular bin (see Fig. 3). The number of bins in the histogram is a free parameter and it is usually set to 9 angular bins.
5. Group adjacent cells into *blocks* (see Fig. 3). The number of cells in each block is a free parameter and all blocks must be of the same size. The distance between each block (known as the stride) is a free parameter but it is usually set to half the block size, in which case you will get overlapping blocks (*see video below*). The HOG algorithm has been shown empirically to work better with overlapping blocks.
6. Use the cells contained within each block to normalize the cell histograms in that block (see Fig. 3). If you have overlapping blocks this means that most cells will be normalized with respect to different blocks (*see video below*). Therefore, the same cell may have several different normalizations.
7. Collect all the normalized histograms from all the blocks into a single feature vector called the HOG descriptor.
8. Use the resulting HOG descriptors from many images of the same type of object to train a machine learning algorithm, such as an SVM, to detect those type of objects in images. For example, you could use the HOG descriptors from many images of pedestrians to train an SVM to detect pedestrians in images. The training is done with both positive a negative examples of the object you want detect in the image.
9. Once the SVM has been trained, a sliding window approach is used to try to detect and locate objects in images. Detecting an object in the image entails finding the part of the image that looks similar to the HOG pattern learned by the SVM.
<br>
<figure>
<img src = "./Animations/HOG Diagram2.png" width = "100%" style = "border: thin silver solid; padding: 1px">
<figcaption style = "text-align:left; font-style:italic">Fig. 3. - HOG Diagram.</figcaption>
</figure>
<br>
<figure>
<video src = "./Animations/HOG Animation - Medium.mp4" width="100%" controls autoplay loop> </video>
<figcaption style = "text-align:left; font-style:italic">Vid. 1. - HOG Animation.</figcaption>
</figure>
# Why The HOG Algorithm Works
As we learned above, HOG creates histograms by adding the magnitude of the gradients in particular orientations in localized portions of the image called *cells*. By doing this we guarantee that stronger gradients will contribute more to the magnitude of their respective angular bin, while the effects of weak and randomly oriented gradients resulting from noise are minimized. In this manner the histograms tell us the dominant gradient orientation of each cell.
### Dealing with contrast
Now, the magnitude of the dominant orientation can vary widely due to variations in local illumination and the contrast between the background and the foreground.
To account for the background-foreground contrast differences, the HOG algorithm tries to detect edges locally. In order to do this, it defines groups of cells, called **blocks**, and normalizes the histograms using this local group of cells. By normalizing locally, the HOG algorithm can detect the edges in each block very reliably; this is called **block normalization**.
In addition to using block normalization, the HOG algorithm also uses overlapping blocks to increase its performance. By using overlapping blocks, each cell contributes several independent components to the final HOG descriptor, where each component corresponds to a cell being normalized with respect to a different block. This may seem redundant but, it has been shown empirically that by normalizing each cell several times with respect to different local blocks, the performance of the HOG algorithm increases dramatically.
### Loading Images and Importing Resources
The first step in building our HOG descriptor is to load the required packages into Python and to load our image.
We start by using OpenCV to load an image of a triangle tile. Since, the `cv2.imread()` function loads images as BGR we will convert our image to RGB so we can display it with the correct colors. As usual we will convert our BGR image to Gray Scale for analysis.
```
import cv2
import numpy as np
import matplotlib.pyplot as plt
# Set the default figure size
plt.rcParams['figure.figsize'] = [17.0, 7.0]
# Load the image
image = cv2.imread('./images/triangle_tile.jpeg')
# Convert the original image to RGB
original_image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
# Convert the original image to gray scale
gray_image = cv2.cvtColor(image, cv2.COLOR_BGR2GRAY)
# Print the shape of the original and gray scale images
print('The original image has shape: ', original_image.shape)
print('The gray scale image has shape: ', gray_image.shape)
# Display the images
plt.subplot(121)
plt.imshow(original_image)
plt.title('Original Image')
plt.subplot(122)
plt.imshow(gray_image, cmap='gray')
plt.title('Gray Scale Image')
plt.show()
```
# Creating The HOG Descriptor
We will be using OpenCV’s `HOGDescriptor` class to create the HOG descriptor. The parameters of the HOG descriptor are setup using the `HOGDescriptor()` function. The parameters of the `HOGDescriptor()` function and their default values are given below:
`cv2.HOGDescriptor(win_size = (64, 128),
block_size = (16, 16),
block_stride = (8, 8),
cell_size = (8, 8),
nbins = 9,
win_sigma = DEFAULT_WIN_SIGMA,
threshold_L2hys = 0.2,
gamma_correction = true,
nlevels = DEFAULT_NLEVELS)`
Parameters:
* **win_size** – *Size*
Size of detection window in pixels (*width, height*). Defines the region of interest. Must be an integer multiple of cell size.
* **block_size** – *Size*
Block size in pixels (*width, height*). Defines how many cells are in each block. Must be an integer multiple of cell size and it must be smaller than the detection window. The smaller the block the finer detail you will get.
* **block_stride** – *Size*
Block stride in pixels (*horizontal, vertical*). It must be an integer multiple of cell size. The `block_stride` defines the distance between adjecent blocks, for example, 8 pixels horizontally and 8 pixels vertically. Longer `block_strides` makes the algorithm run faster (because less blocks are evaluated) but the algorithm may not perform as well.
* **cell_size** – *Size*
Cell size in pixels (*width, height*). Determines the size fo your cell. The smaller the cell the finer detail you will get.
* **nbins** – *int*
Number of bins for the histograms. Determines the number of angular bins used to make the histograms. With more bins you capture more gradient directions. HOG uses unsigned gradients, so the angular bins will have values between 0 and 180 degrees.
* **win_sigma** – *double*
Gaussian smoothing window parameter. The performance of the HOG algorithm can be improved by smoothing the pixels near the edges of the blocks by applying a Gaussian spatial window to each pixel before computing the histograms.
* **threshold_L2hys** – *double*
L2-Hys (Lowe-style clipped L2 norm) normalization method shrinkage. The L2-Hys method is used to normalize the blocks and it consists of an L2-norm followed by clipping and a renormalization. The clipping limits the maximum value of the descriptor vector for each block to have the value of the given threshold (0.2 by default). After the clipping the descriptor vector is renormalized as described in *IJCV*, 60(2):91-110, 2004.
* **gamma_correction** – *bool*
Flag to specify whether the gamma correction preprocessing is required or not. Performing gamma correction slightly increases the performance of the HOG algorithm.
* **nlevels** – *int*
Maximum number of detection window increases.
As we can see, the `cv2.HOGDescriptor()`function supports a wide range of parameters. The first few arguments (`block_size, block_stride, cell_size`, and `nbins`) are probably the ones you are most likely to change. The other parameters can be safely left at their default values and you will get good results.
In the code below, we will use the `cv2.HOGDescriptor()`function to set the cell size, block size, block stride, and the number of bins for the histograms of the HOG descriptor. We will then use `.compute(image)`method to compute the HOG descriptor (feature vector) for the given `image`.
```
# Specify the parameters for our HOG descriptor
# Cell Size in pixels (width, height). Must be smaller than the size of the detection window
# and must be chosen so that the resulting Block Size is smaller than the detection window.
cell_size = (6, 6)
# Number of cells per block in each direction (x, y). Must be chosen so that the resulting
# Block Size is smaller than the detection window
num_cells_per_block = (2, 2)
# Block Size in pixels (width, height). Must be an integer multiple of Cell Size.
# The Block Size must be smaller than the detection window
block_size = (num_cells_per_block[0] * cell_size[0],
num_cells_per_block[1] * cell_size[1])
# Calculate the number of cells that fit in our image in the x and y directions
x_cells = gray_image.shape[1] // cell_size[0]
y_cells = gray_image.shape[0] // cell_size[1]
# Horizontal distance between blocks in units of Cell Size. Must be an integer and it must
# be set such that (x_cells - num_cells_per_block[0]) / h_stride = integer.
h_stride = 1
# Vertical distance between blocks in units of Cell Size. Must be an integer and it must
# be set such that (y_cells - num_cells_per_block[1]) / v_stride = integer.
v_stride = 1
# Block Stride in pixels (horizantal, vertical). Must be an integer multiple of Cell Size
block_stride = (cell_size[0] * h_stride, cell_size[1] * v_stride)
# Number of gradient orientation bins
num_bins = 9
# Specify the size of the detection window (Region of Interest) in pixels (width, height).
# It must be an integer multiple of Cell Size and it must cover the entire image. Because
# the detection window must be an integer multiple of cell size, depending on the size of
# your cells, the resulting detection window might be slightly smaller than the image.
# This is perfectly ok.
win_size = (x_cells * cell_size[0] , y_cells * cell_size[1])
# Print the shape of the gray scale image for reference
print('\nThe gray scale image has shape: ', gray_image.shape)
print()
# Print the parameters of our HOG descriptor
print('HOG Descriptor Parameters:\n')
print('Window Size:', win_size)
print('Cell Size:', cell_size)
print('Block Size:', block_size)
print('Block Stride:', block_stride)
print('Number of Bins:', num_bins)
print()
# Set the parameters of the HOG descriptor using the variables defined above
hog = cv2.HOGDescriptor(win_size, block_size, block_stride, cell_size, num_bins)
# Compute the HOG Descriptor for the gray scale image
hog_descriptor = hog.compute(gray_image)
```
# Number of Elements In The HOG Descriptor
The resulting HOG Descriptor (feature vector), contains the normalized histograms from all cells from all blocks in the detection window concatenated in one long vector. Therefore, the size of the HOG feature vector will be given by the total number of blocks in the detection window, multiplied by the number of cells per block, times the number of orientation bins:
<span class="mathquill">
\begin{equation}
\mbox{total_elements} = (\mbox{total_number_of_blocks})\mbox{ } \times \mbox{ } (\mbox{number_cells_per_block})\mbox{ } \times \mbox{ } (\mbox{number_of_bins})
\end{equation}
</span>
If we don’t have overlapping blocks (*i.e.* the `block_stride`equals the `block_size`), the total number of blocks can be easily calculated by dividing the size of the detection window by the block size. However, in the general case we have to take into account the fact that we have overlapping blocks. To find the total number of blocks in the general case (*i.e.* for any `block_stride` and `block_size`), we can use the formula given below:
<span class="mathquill">
\begin{equation}
\mbox{Total}_i = \left( \frac{\mbox{block_size}_i}{\mbox{block_stride}_i} \right)\left( \frac{\mbox{window_size}_i}{\mbox{block_size}_i} \right) - \left [\left( \frac{\mbox{block_size}_i}{\mbox{block_stride}_i} \right) - 1 \right]; \mbox{ for } i = x,y
\end{equation}
</span>
Where <span class="mathquill">Total$_x$</span>, is the total number of blocks along the width of the detection window, and <span class="mathquill">Total$_y$</span>, is the total number of blocks along the height of the detection window. This formula for <span class="mathquill">Total$_x$</span> and <span class="mathquill">Total$_y$</span>, takes into account the extra blocks that result from overlapping. After calculating <span class="mathquill">Total$_x$</span> and <span class="mathquill">Total$_y$</span>, we can get the total number of blocks in the detection window by multiplying <span class="mathquill">Total$_x$ $\times$ Total$_y$</span>. The above formula can be simplified considerably because the `block_size`, `block_stride`, and `window_size`are all defined in terms of the `cell_size`. By making all the appropriate substitutions and cancelations the above formula reduces to:
<span class="mathquill">
\begin{equation}
\mbox{Total}_i = \left(\frac{\mbox{cells}_i - \mbox{num_cells_per_block}_i}{N_i}\right) + 1\mbox{ }; \mbox{ for } i = x,y
\end{equation}
</span>
Where <span class="mathquill">cells$_x$</span> is the total number of cells along the width of the detection window, and <span class="mathquill">cells$_y$</span>, is the total number of cells along the height of the detection window. And <span class="mathquill">$N_x$</span> is the horizontal block stride in units of `cell_size` and <span class="mathquill">$N_y$</span> is the vertical block stride in units of `cell_size`.
Let's calculate what the number of elements for the HOG feature vector should be and check that it matches the shape of the HOG Descriptor calculated above.
```
# Calculate the total number of blocks along the width of the detection window
tot_bx = np.uint32(((x_cells - num_cells_per_block[0]) / h_stride) + 1)
# Calculate the total number of blocks along the height of the detection window
tot_by = np.uint32(((y_cells - num_cells_per_block[1]) / v_stride) + 1)
# Calculate the total number of elements in the feature vector
tot_els = (tot_bx) * (tot_by) * num_cells_per_block[0] * num_cells_per_block[1] * num_bins
# Print the total number of elements the HOG feature vector should have
print('\nThe total number of elements in the HOG Feature Vector should be: ',
tot_bx, 'x',
tot_by, 'x',
num_cells_per_block[0], 'x',
num_cells_per_block[1], 'x',
num_bins, '=',
tot_els)
# Print the shape of the HOG Descriptor to see that it matches the above
print('\nThe HOG Descriptor has shape:', hog_descriptor.shape)
print()
```
# Visualizing The HOG Descriptor
We can visualize the HOG Descriptor by plotting the histogram associated with each cell as a collection of vectors. To do this, we will plot each bin in the histogram as a single vector whose magnitude is given by the height of the bin and its orientation is given by the angular bin that its associated with. Since any given cell might have multiple histograms associated with it, due to the overlapping blocks, we will choose to average all the histograms for each cell to produce a single histogram for each cell.
OpenCV has no easy way to visualize the HOG Descriptor, so we have to do some manipulation first in order to visualize it. We will start by reshaping the HOG Descriptor in order to make our calculations easier. We will then compute the average histogram of each cell and finally we will convert the histogram bins into vectors. Once we have the vectors, we plot the corresponding vectors for each cell in an image.
The code below produces an interactive plot so that you can interact with the figure. The figure contains:
* the grayscale image,
* the HOG Descriptor (feature vector),
* a zoomed-in portion of the HOG Descriptor, and
* the histogram of the selected cell.
**You can click anywhere on the gray scale image or the HOG Descriptor image to select a particular cell**. Once you click on either image a *magenta* rectangle will appear showing the cell you selected. The Zoom Window will show you a zoomed in version of the HOG descriptor around the selected cell; and the histogram plot will show you the corresponding histogram for the selected cell. The interactive window also has buttons at the bottom that allow for other functionality, such as panning, and giving you the option to save the figure if desired. The home button returns the figure to its default value.
**NOTE**: If you are running this notebook in the Udacity workspace, there is around a 2 second lag in the interactive plot. This means that if you click in the image to zoom in, it will take about 2 seconds for the plot to refresh.
```
%matplotlib notebook
import copy
import matplotlib.patches as patches
# Set the default figure size
plt.rcParams['figure.figsize'] = [9.8, 9]
# Reshape the feature vector to [blocks_y, blocks_x, num_cells_per_block_x, num_cells_per_block_y, num_bins].
# The blocks_x and blocks_y will be transposed so that the first index (blocks_y) referes to the row number
# and the second index to the column number. This will be useful later when we plot the feature vector, so
# that the feature vector indexing matches the image indexing.
hog_descriptor_reshaped = hog_descriptor.reshape(tot_bx,
tot_by,
num_cells_per_block[0],
num_cells_per_block[1],
num_bins).transpose((1, 0, 2, 3, 4))
# Print the shape of the feature vector for reference
print('The feature vector has shape:', hog_descriptor.shape)
# Print the reshaped feature vector
print('The reshaped feature vector has shape:', hog_descriptor_reshaped.shape)
# Create an array that will hold the average gradients for each cell
ave_grad = np.zeros((y_cells, x_cells, num_bins))
# Print the shape of the ave_grad array for reference
print('The average gradient array has shape: ', ave_grad.shape)
# Create an array that will count the number of histograms per cell
hist_counter = np.zeros((y_cells, x_cells, 1))
# Add up all the histograms for each cell and count the number of histograms per cell
for i in range (num_cells_per_block[0]):
for j in range(num_cells_per_block[1]):
ave_grad[i:tot_by + i,
j:tot_bx + j] += hog_descriptor_reshaped[:, :, i, j, :]
hist_counter[i:tot_by + i,
j:tot_bx + j] += 1
# Calculate the average gradient for each cell
ave_grad /= hist_counter
# Calculate the total number of vectors we have in all the cells.
len_vecs = ave_grad.shape[0] * ave_grad.shape[1] * ave_grad.shape[2]
# Create an array that has num_bins equally spaced between 0 and 180 degress in radians.
deg = np.linspace(0, np.pi, num_bins, endpoint = False)
# Each cell will have a histogram with num_bins. For each cell, plot each bin as a vector (with its magnitude
# equal to the height of the bin in the histogram, and its angle corresponding to the bin in the histogram).
# To do this, create rank 1 arrays that will hold the (x,y)-coordinate of all the vectors in all the cells in the
# image. Also, create the rank 1 arrays that will hold all the (U,V)-components of all the vectors in all the
# cells in the image. Create the arrays that will hold all the vector positons and components.
U = np.zeros((len_vecs))
V = np.zeros((len_vecs))
X = np.zeros((len_vecs))
Y = np.zeros((len_vecs))
# Set the counter to zero
counter = 0
# Use the cosine and sine functions to calculate the vector components (U,V) from their maginitudes. Remember the
# cosine and sine functions take angles in radians. Calculate the vector positions and magnitudes from the
# average gradient array
for i in range(ave_grad.shape[0]):
for j in range(ave_grad.shape[1]):
for k in range(ave_grad.shape[2]):
U[counter] = ave_grad[i,j,k] * np.cos(deg[k])
V[counter] = ave_grad[i,j,k] * np.sin(deg[k])
X[counter] = (cell_size[0] / 2) + (cell_size[0] * i)
Y[counter] = (cell_size[1] / 2) + (cell_size[1] * j)
counter = counter + 1
# Create the bins in degress to plot our histogram.
angle_axis = np.linspace(0, 180, num_bins, endpoint = False)
angle_axis += ((angle_axis[1] - angle_axis[0]) / 2)
# Create a figure with 4 subplots arranged in 2 x 2
fig, ((a,b),(c,d)) = plt.subplots(2,2)
# Set the title of each subplot
a.set(title = 'Gray Scale Image\n(Click to Zoom)')
b.set(title = 'HOG Descriptor\n(Click to Zoom)')
c.set(title = 'Zoom Window', xlim = (0, 18), ylim = (0, 18), autoscale_on = False)
d.set(title = 'Histogram of Gradients')
# Plot the gray scale image
a.imshow(gray_image, cmap = 'gray')
a.set_aspect(aspect = 1)
# Plot the feature vector (HOG Descriptor)
b.quiver(Y, X, U, V, color = 'white', headwidth = 0, headlength = 0, scale_units = 'inches', scale = 5)
b.invert_yaxis()
b.set_aspect(aspect = 1)
b.set_facecolor('black')
# Define function for interactive zoom
def onpress(event):
#Unless the left mouse button is pressed do nothing
if event.button != 1:
return
# Only accept clicks for subplots a and b
if event.inaxes in [a, b]:
# Get mouse click coordinates
x, y = event.xdata, event.ydata
# Select the cell closest to the mouse click coordinates
cell_num_x = np.uint32(x / cell_size[0])
cell_num_y = np.uint32(y / cell_size[1])
# Set the edge coordinates of the rectangle patch
edgex = x - (x % cell_size[0])
edgey = y - (y % cell_size[1])
# Create a rectangle patch that matches the the cell selected above
rect = patches.Rectangle((edgex, edgey),
cell_size[0], cell_size[1],
linewidth = 1,
edgecolor = 'magenta',
facecolor='none')
# A single patch can only be used in a single plot. Create copies
# of the patch to use in the other subplots
rect2 = copy.copy(rect)
rect3 = copy.copy(rect)
# Update all subplots
a.clear()
a.set(title = 'Gray Scale Image\n(Click to Zoom)')
a.imshow(gray_image, cmap = 'gray')
a.set_aspect(aspect = 1)
a.add_patch(rect)
b.clear()
b.set(title = 'HOG Descriptor\n(Click to Zoom)')
b.quiver(Y, X, U, V, color = 'white', headwidth = 0, headlength = 0, scale_units = 'inches', scale = 5)
b.invert_yaxis()
b.set_aspect(aspect = 1)
b.set_facecolor('black')
b.add_patch(rect2)
c.clear()
c.set(title = 'Zoom Window')
c.quiver(Y, X, U, V, color = 'white', headwidth = 0, headlength = 0, scale_units = 'inches', scale = 1)
c.set_xlim(edgex - cell_size[0], edgex + (2 * cell_size[0]))
c.set_ylim(edgey - cell_size[1], edgey + (2 * cell_size[1]))
c.invert_yaxis()
c.set_aspect(aspect = 1)
c.set_facecolor('black')
c.add_patch(rect3)
d.clear()
d.set(title = 'Histogram of Gradients')
d.grid()
d.set_xlim(0, 180)
d.set_xticks(angle_axis)
d.set_xlabel('Angle')
d.bar(angle_axis,
ave_grad[cell_num_y, cell_num_x, :],
180 // num_bins,
align = 'center',
alpha = 0.5,
linewidth = 1.2,
edgecolor = 'k')
fig.canvas.draw()
# Create a connection between the figure and the mouse click
fig.canvas.mpl_connect('button_press_event', onpress)
plt.show()
```
# Understanding The Histograms
Let's take a look at a couple of snapshots of the above figure to see if the histograms for the selected cell make sense. Let's start looking at a cell that is inside a triangle and not near an edge:
<br>
<figure>
<img src = "./Animations/snapshot1.png" width = "70%" style = "border: thin silver solid; padding: 1px">
<figcaption style = "text-align:center; font-style:italic">Fig. 4. - Histograms Inside a Triangle.</figcaption>
</figure>
<br>
In this case, since the triangle is nearly all of the same color there shouldn't be any dominant gradient in the selected cell. As we can clearly see in the Zoom Window and the histogram, this is indeed the case. We have many gradients but none of them clearly dominates over the other.
Now let’s take a look at a cell that is near a horizontal edge:
<br>
<figure>
<img src = "./Animations/snapshot2.png" width = "70%" style = "border: thin silver solid; padding: 1px">
<figcaption style = "text-align:center; font-style:italic">Fig. 5. - Histograms Near a Horizontal Edge.</figcaption>
</figure>
<br>
Remember that edges are areas of an image where the intensity changes abruptly. In these cases, we will have a high intensity gradient in some particular direction. This is exactly what we see in the corresponding histogram and Zoom Window for the selected cell. In the Zoom Window, we can see that the dominant gradient is pointing up, almost at 90 degrees, since that’s the direction in which there is a sharp change in intensity. Therefore, we should expect to see the 90-degree bin in the histogram to dominate strongly over the others. This is in fact what we see.
Now let’s take a look at a cell that is near a vertical edge:
<br>
<figure>
<img src = "./Animations/snapshot3.png" width = "70%" style = "border: thin silver solid; padding: 1px">
<figcaption style = "text-align:center; font-style:italic">Fig. 6. - Histograms Near a Vertical Edge.</figcaption>
</figure>
<br>
In this case we expect the dominant gradient in the cell to be horizontal, close to 180 degrees, since that’s the direction in which there is a sharp change in intensity. Therefore, we should expect to see the 170-degree bin in the histogram to dominate strongly over the others. This is what we see in the histogram but we also see that there is another dominant gradient in the cell, namely the one in the 10-degree bin. The reason for this, is because the HOG algorithm is using unsigned gradients, which means 0 degrees and 180 degrees are considered the same. Therefore, when the histograms are being created, angles between 160 and 180 degrees, contribute proportionally to both the 10-degree bin and the 170-degree bin. This results in there being two dominant gradients in the cell near the vertical edge instead of just one.
To conclude let’s take a look at a cell that is near a diagonal edge.
<br>
<figure>
<img src = "./Animations/snapshot4.png" width = "70%" style = "border: thin silver solid; padding: 1px">
<figcaption style = "text-align:center; font-style:italic">Fig. 7. - Histograms Near a Diagonal Edge.</figcaption>
</figure>
<br>
To understand what we are seeing, let’s first remember that gradients have an *x*-component, and a *y*-component, just like vectors. Therefore, the resulting orientation of a gradient is going to be given by the vector sum of its components. For this reason, on vertical edges the gradients are horizontal, because they only have an x-component, as we saw in Figure 4. While on horizontal edges the gradients are vertical, because they only have a y-component, as we saw in Figure 3. Consequently, on diagonal edges, the gradients are also going to be diagonal because both the *x* and *y* components are non-zero. Since the diagonal edges in the image are close to 45 degrees, we should expect to see a dominant gradient orientation in the 50-degree bin. This is in fact what we see in the histogram but, just like in Figure 4., we see there are two dominant gradients instead of just one. The reason for this is that when the histograms are being created, angles that are near the boundaries of bins, contribute proportionally to the adjacent bins. For example, a gradient with an angle of 40 degrees, is right in the middle of the 30-degree and 50-degree bin. Therefore, the magnitude of the gradient is split evenly into the 30-degree and 50-degree bin. This results in there being two dominant gradients in the cell near the diagonal edge instead of just one.
Now that you know how HOG is implemented, in the workspace you will find a notebook named *Examples*. In there, you will be able set your own paramters for the HOG descriptor for various images. Have fun!
|
github_jupyter
|
import cv2
import numpy as np
import matplotlib.pyplot as plt
# Set the default figure size
plt.rcParams['figure.figsize'] = [17.0, 7.0]
# Load the image
image = cv2.imread('./images/triangle_tile.jpeg')
# Convert the original image to RGB
original_image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
# Convert the original image to gray scale
gray_image = cv2.cvtColor(image, cv2.COLOR_BGR2GRAY)
# Print the shape of the original and gray scale images
print('The original image has shape: ', original_image.shape)
print('The gray scale image has shape: ', gray_image.shape)
# Display the images
plt.subplot(121)
plt.imshow(original_image)
plt.title('Original Image')
plt.subplot(122)
plt.imshow(gray_image, cmap='gray')
plt.title('Gray Scale Image')
plt.show()
# Specify the parameters for our HOG descriptor
# Cell Size in pixels (width, height). Must be smaller than the size of the detection window
# and must be chosen so that the resulting Block Size is smaller than the detection window.
cell_size = (6, 6)
# Number of cells per block in each direction (x, y). Must be chosen so that the resulting
# Block Size is smaller than the detection window
num_cells_per_block = (2, 2)
# Block Size in pixels (width, height). Must be an integer multiple of Cell Size.
# The Block Size must be smaller than the detection window
block_size = (num_cells_per_block[0] * cell_size[0],
num_cells_per_block[1] * cell_size[1])
# Calculate the number of cells that fit in our image in the x and y directions
x_cells = gray_image.shape[1] // cell_size[0]
y_cells = gray_image.shape[0] // cell_size[1]
# Horizontal distance between blocks in units of Cell Size. Must be an integer and it must
# be set such that (x_cells - num_cells_per_block[0]) / h_stride = integer.
h_stride = 1
# Vertical distance between blocks in units of Cell Size. Must be an integer and it must
# be set such that (y_cells - num_cells_per_block[1]) / v_stride = integer.
v_stride = 1
# Block Stride in pixels (horizantal, vertical). Must be an integer multiple of Cell Size
block_stride = (cell_size[0] * h_stride, cell_size[1] * v_stride)
# Number of gradient orientation bins
num_bins = 9
# Specify the size of the detection window (Region of Interest) in pixels (width, height).
# It must be an integer multiple of Cell Size and it must cover the entire image. Because
# the detection window must be an integer multiple of cell size, depending on the size of
# your cells, the resulting detection window might be slightly smaller than the image.
# This is perfectly ok.
win_size = (x_cells * cell_size[0] , y_cells * cell_size[1])
# Print the shape of the gray scale image for reference
print('\nThe gray scale image has shape: ', gray_image.shape)
print()
# Print the parameters of our HOG descriptor
print('HOG Descriptor Parameters:\n')
print('Window Size:', win_size)
print('Cell Size:', cell_size)
print('Block Size:', block_size)
print('Block Stride:', block_stride)
print('Number of Bins:', num_bins)
print()
# Set the parameters of the HOG descriptor using the variables defined above
hog = cv2.HOGDescriptor(win_size, block_size, block_stride, cell_size, num_bins)
# Compute the HOG Descriptor for the gray scale image
hog_descriptor = hog.compute(gray_image)
# Calculate the total number of blocks along the width of the detection window
tot_bx = np.uint32(((x_cells - num_cells_per_block[0]) / h_stride) + 1)
# Calculate the total number of blocks along the height of the detection window
tot_by = np.uint32(((y_cells - num_cells_per_block[1]) / v_stride) + 1)
# Calculate the total number of elements in the feature vector
tot_els = (tot_bx) * (tot_by) * num_cells_per_block[0] * num_cells_per_block[1] * num_bins
# Print the total number of elements the HOG feature vector should have
print('\nThe total number of elements in the HOG Feature Vector should be: ',
tot_bx, 'x',
tot_by, 'x',
num_cells_per_block[0], 'x',
num_cells_per_block[1], 'x',
num_bins, '=',
tot_els)
# Print the shape of the HOG Descriptor to see that it matches the above
print('\nThe HOG Descriptor has shape:', hog_descriptor.shape)
print()
%matplotlib notebook
import copy
import matplotlib.patches as patches
# Set the default figure size
plt.rcParams['figure.figsize'] = [9.8, 9]
# Reshape the feature vector to [blocks_y, blocks_x, num_cells_per_block_x, num_cells_per_block_y, num_bins].
# The blocks_x and blocks_y will be transposed so that the first index (blocks_y) referes to the row number
# and the second index to the column number. This will be useful later when we plot the feature vector, so
# that the feature vector indexing matches the image indexing.
hog_descriptor_reshaped = hog_descriptor.reshape(tot_bx,
tot_by,
num_cells_per_block[0],
num_cells_per_block[1],
num_bins).transpose((1, 0, 2, 3, 4))
# Print the shape of the feature vector for reference
print('The feature vector has shape:', hog_descriptor.shape)
# Print the reshaped feature vector
print('The reshaped feature vector has shape:', hog_descriptor_reshaped.shape)
# Create an array that will hold the average gradients for each cell
ave_grad = np.zeros((y_cells, x_cells, num_bins))
# Print the shape of the ave_grad array for reference
print('The average gradient array has shape: ', ave_grad.shape)
# Create an array that will count the number of histograms per cell
hist_counter = np.zeros((y_cells, x_cells, 1))
# Add up all the histograms for each cell and count the number of histograms per cell
for i in range (num_cells_per_block[0]):
for j in range(num_cells_per_block[1]):
ave_grad[i:tot_by + i,
j:tot_bx + j] += hog_descriptor_reshaped[:, :, i, j, :]
hist_counter[i:tot_by + i,
j:tot_bx + j] += 1
# Calculate the average gradient for each cell
ave_grad /= hist_counter
# Calculate the total number of vectors we have in all the cells.
len_vecs = ave_grad.shape[0] * ave_grad.shape[1] * ave_grad.shape[2]
# Create an array that has num_bins equally spaced between 0 and 180 degress in radians.
deg = np.linspace(0, np.pi, num_bins, endpoint = False)
# Each cell will have a histogram with num_bins. For each cell, plot each bin as a vector (with its magnitude
# equal to the height of the bin in the histogram, and its angle corresponding to the bin in the histogram).
# To do this, create rank 1 arrays that will hold the (x,y)-coordinate of all the vectors in all the cells in the
# image. Also, create the rank 1 arrays that will hold all the (U,V)-components of all the vectors in all the
# cells in the image. Create the arrays that will hold all the vector positons and components.
U = np.zeros((len_vecs))
V = np.zeros((len_vecs))
X = np.zeros((len_vecs))
Y = np.zeros((len_vecs))
# Set the counter to zero
counter = 0
# Use the cosine and sine functions to calculate the vector components (U,V) from their maginitudes. Remember the
# cosine and sine functions take angles in radians. Calculate the vector positions and magnitudes from the
# average gradient array
for i in range(ave_grad.shape[0]):
for j in range(ave_grad.shape[1]):
for k in range(ave_grad.shape[2]):
U[counter] = ave_grad[i,j,k] * np.cos(deg[k])
V[counter] = ave_grad[i,j,k] * np.sin(deg[k])
X[counter] = (cell_size[0] / 2) + (cell_size[0] * i)
Y[counter] = (cell_size[1] / 2) + (cell_size[1] * j)
counter = counter + 1
# Create the bins in degress to plot our histogram.
angle_axis = np.linspace(0, 180, num_bins, endpoint = False)
angle_axis += ((angle_axis[1] - angle_axis[0]) / 2)
# Create a figure with 4 subplots arranged in 2 x 2
fig, ((a,b),(c,d)) = plt.subplots(2,2)
# Set the title of each subplot
a.set(title = 'Gray Scale Image\n(Click to Zoom)')
b.set(title = 'HOG Descriptor\n(Click to Zoom)')
c.set(title = 'Zoom Window', xlim = (0, 18), ylim = (0, 18), autoscale_on = False)
d.set(title = 'Histogram of Gradients')
# Plot the gray scale image
a.imshow(gray_image, cmap = 'gray')
a.set_aspect(aspect = 1)
# Plot the feature vector (HOG Descriptor)
b.quiver(Y, X, U, V, color = 'white', headwidth = 0, headlength = 0, scale_units = 'inches', scale = 5)
b.invert_yaxis()
b.set_aspect(aspect = 1)
b.set_facecolor('black')
# Define function for interactive zoom
def onpress(event):
#Unless the left mouse button is pressed do nothing
if event.button != 1:
return
# Only accept clicks for subplots a and b
if event.inaxes in [a, b]:
# Get mouse click coordinates
x, y = event.xdata, event.ydata
# Select the cell closest to the mouse click coordinates
cell_num_x = np.uint32(x / cell_size[0])
cell_num_y = np.uint32(y / cell_size[1])
# Set the edge coordinates of the rectangle patch
edgex = x - (x % cell_size[0])
edgey = y - (y % cell_size[1])
# Create a rectangle patch that matches the the cell selected above
rect = patches.Rectangle((edgex, edgey),
cell_size[0], cell_size[1],
linewidth = 1,
edgecolor = 'magenta',
facecolor='none')
# A single patch can only be used in a single plot. Create copies
# of the patch to use in the other subplots
rect2 = copy.copy(rect)
rect3 = copy.copy(rect)
# Update all subplots
a.clear()
a.set(title = 'Gray Scale Image\n(Click to Zoom)')
a.imshow(gray_image, cmap = 'gray')
a.set_aspect(aspect = 1)
a.add_patch(rect)
b.clear()
b.set(title = 'HOG Descriptor\n(Click to Zoom)')
b.quiver(Y, X, U, V, color = 'white', headwidth = 0, headlength = 0, scale_units = 'inches', scale = 5)
b.invert_yaxis()
b.set_aspect(aspect = 1)
b.set_facecolor('black')
b.add_patch(rect2)
c.clear()
c.set(title = 'Zoom Window')
c.quiver(Y, X, U, V, color = 'white', headwidth = 0, headlength = 0, scale_units = 'inches', scale = 1)
c.set_xlim(edgex - cell_size[0], edgex + (2 * cell_size[0]))
c.set_ylim(edgey - cell_size[1], edgey + (2 * cell_size[1]))
c.invert_yaxis()
c.set_aspect(aspect = 1)
c.set_facecolor('black')
c.add_patch(rect3)
d.clear()
d.set(title = 'Histogram of Gradients')
d.grid()
d.set_xlim(0, 180)
d.set_xticks(angle_axis)
d.set_xlabel('Angle')
d.bar(angle_axis,
ave_grad[cell_num_y, cell_num_x, :],
180 // num_bins,
align = 'center',
alpha = 0.5,
linewidth = 1.2,
edgecolor = 'k')
fig.canvas.draw()
# Create a connection between the figure and the mouse click
fig.canvas.mpl_connect('button_press_event', onpress)
plt.show()
| 0.719482 | 0.811153 |
```
import pandas as pd
from sklearn.feature_extraction.text import TfidfVectorizer as t
import numpy as np
import nltk
import pandas
import time
from nltk import tokenize,word_tokenize
from nltk.corpus import stopwords
import re
data = pd.read_csv('Final_dataset.csv')
data
data.columns
data.isna().sum()
data = data.fillna(' ')
def create_soup(x):
return ''.join(x['Description']) + '.' + ''.join(x['Storyline'])
data['soup'] = data.apply(create_soup, axis=1)
data
over = data['soup']
over_ = data['soup'].values
def process_sentence(text,stem = False,lem = True,remove_stop_words = True,stemmer = nltk.PorterStemmer(),wnl = nltk.WordNetLemmatizer(),stop_word = stopwords.words('english') ):
text = re.sub(r"[^A-Za-z0-9]"," ",text)
text = re.sub(r"\'s","",text)
text = re.sub(r"\'ve","have",text)
text = re.sub(r"can't", "cannot ", text)
text = re.sub(r"n't", " not ", text)
text = re.sub(r"I'm", "I am", text)
text = re.sub(r" m ", " am ", text)
text = re.sub(r"\'re", " are ", text)
text = re.sub(r"\'d", " would ", text)
text = re.sub(r"\'ll", " will ", text)
word = word_tokenize(text)
if remove_stop_words:
word = [wo for wo in word if not word in stop_word]
if lem:
word = [wnl.lemmatize(t) for t in word]
if stem:
word = [stemmer.stem(t) for t in word]
return ' '.join(word)
process_sentence(over_[0])
over_[0]
def clean_data():
for i in range(len(over_)):
over_[i] = process_sentence(over_[i])
len(over_)
clean_data()
tfidf_vec = t()
tfidf_vec.fit(over_)
tf_idf_mat = tfidf_vec.transform(over_)
tf_idf_mat.shape
indices = pd.Series(data.index, index=data['Title']).drop_duplicates()
indices['Before the Fall']
def suggest_movie(title):
ind = indices[title]
if type(ind) == pandas.core.series.Series:
ind = ind.values[0]
txt = process_sentence(over[ind])
#print(txt)
vec = tfidf_vec.transform([txt])
#print(vec.shape)
ar = np.dot(vec,tf_idf_mat.T)
ar =ar.toarray()
ar = np.reshape(ar,ar.shape[1])
ar = list(enumerate(ar))
sim_scores = sorted(ar, key=lambda x: x[1], reverse=True)
sim_scores = sim_scores[1:11]
movie_indices = [i[0] for i in sim_scores]
return data['Title'].iloc[movie_indices]
def pre_print(kk):
kk = kk.values
print()
for i in range(len(kk)):
print(i+1,kk[i])
print()
while True:
print('**--------------------**')
print('press 1 to continue')
print('press 2 to change module')
print('press 3 to exit')
print('**---------------------**')
ino = int(input())
take_screen_input = False
take_random_movie = True
if ino == 2:
print('Please enter correct name')
take_screen_input = True
take_random_movie = False
if ino == 1 or ino == 2:
if take_screen_input:
movie_ = str(input())
elif take_random_movie:
movie_ = data['Title'][np.random.randint(data.shape[0])]
else:
movie_ = 'The Godfather'
print('')
print('')
print('Movie Recommendation for \"{}\"'.format(movie_))
st = time.time()
lll = suggest_movie(movie_)
pre_print(lll)
print('Time taken to suggest an \"{}\": {}'.format(movie_,time.time() - st))
print('')
print('')
if ino ==3:
break
```
|
github_jupyter
|
import pandas as pd
from sklearn.feature_extraction.text import TfidfVectorizer as t
import numpy as np
import nltk
import pandas
import time
from nltk import tokenize,word_tokenize
from nltk.corpus import stopwords
import re
data = pd.read_csv('Final_dataset.csv')
data
data.columns
data.isna().sum()
data = data.fillna(' ')
def create_soup(x):
return ''.join(x['Description']) + '.' + ''.join(x['Storyline'])
data['soup'] = data.apply(create_soup, axis=1)
data
over = data['soup']
over_ = data['soup'].values
def process_sentence(text,stem = False,lem = True,remove_stop_words = True,stemmer = nltk.PorterStemmer(),wnl = nltk.WordNetLemmatizer(),stop_word = stopwords.words('english') ):
text = re.sub(r"[^A-Za-z0-9]"," ",text)
text = re.sub(r"\'s","",text)
text = re.sub(r"\'ve","have",text)
text = re.sub(r"can't", "cannot ", text)
text = re.sub(r"n't", " not ", text)
text = re.sub(r"I'm", "I am", text)
text = re.sub(r" m ", " am ", text)
text = re.sub(r"\'re", " are ", text)
text = re.sub(r"\'d", " would ", text)
text = re.sub(r"\'ll", " will ", text)
word = word_tokenize(text)
if remove_stop_words:
word = [wo for wo in word if not word in stop_word]
if lem:
word = [wnl.lemmatize(t) for t in word]
if stem:
word = [stemmer.stem(t) for t in word]
return ' '.join(word)
process_sentence(over_[0])
over_[0]
def clean_data():
for i in range(len(over_)):
over_[i] = process_sentence(over_[i])
len(over_)
clean_data()
tfidf_vec = t()
tfidf_vec.fit(over_)
tf_idf_mat = tfidf_vec.transform(over_)
tf_idf_mat.shape
indices = pd.Series(data.index, index=data['Title']).drop_duplicates()
indices['Before the Fall']
def suggest_movie(title):
ind = indices[title]
if type(ind) == pandas.core.series.Series:
ind = ind.values[0]
txt = process_sentence(over[ind])
#print(txt)
vec = tfidf_vec.transform([txt])
#print(vec.shape)
ar = np.dot(vec,tf_idf_mat.T)
ar =ar.toarray()
ar = np.reshape(ar,ar.shape[1])
ar = list(enumerate(ar))
sim_scores = sorted(ar, key=lambda x: x[1], reverse=True)
sim_scores = sim_scores[1:11]
movie_indices = [i[0] for i in sim_scores]
return data['Title'].iloc[movie_indices]
def pre_print(kk):
kk = kk.values
print()
for i in range(len(kk)):
print(i+1,kk[i])
print()
while True:
print('**--------------------**')
print('press 1 to continue')
print('press 2 to change module')
print('press 3 to exit')
print('**---------------------**')
ino = int(input())
take_screen_input = False
take_random_movie = True
if ino == 2:
print('Please enter correct name')
take_screen_input = True
take_random_movie = False
if ino == 1 or ino == 2:
if take_screen_input:
movie_ = str(input())
elif take_random_movie:
movie_ = data['Title'][np.random.randint(data.shape[0])]
else:
movie_ = 'The Godfather'
print('')
print('')
print('Movie Recommendation for \"{}\"'.format(movie_))
st = time.time()
lll = suggest_movie(movie_)
pre_print(lll)
print('Time taken to suggest an \"{}\": {}'.format(movie_,time.time() - st))
print('')
print('')
if ino ==3:
break
| 0.165694 | 0.235284 |
# Naive Bayes Classifier based on Term Frequencies
```
import pandas as pd
```
## 1. Load the Given data into a CSV File
To be done only once, later the data can be loaded on from the CSV file
```
# Create a Pandas dataframe and store the data
df = pd.DataFrame(columns=['TDP', 'Nifty', 'Sidhu', 'BJP', 'Sensex', 'Sixer', 'Congress', 'Century', 'Category'])
num_entries = 0
# The Given Data
data = [
[4,0,3,5,1,0,6,0,'Politics'],
[0,5,0,2,6,0,1,0,'Business'],
[0,0,6,1,0,4,1,2,'Sports'],
[4,1,0,1,1,0,6,0,'Politics'],
[0,0,0,0,0,5,0,6,'Sports'],
[0,4,0,2,6,0,0,1,'Business'],
[5,0,0,3,0,0,5,0,'Politics']
]
query_data = [0,3,0,2,6,0,2,1]
# Load the data into a Pandas dataframe and store it into a CSV File
for i in range(len(data)) :
df.loc[num_entries] = data[i]
num_entries += 1
df.to_csv('Data.csv', index=None)
```
## 2. Loading the dataset and getting the Queryset
```
df = pd.read_csv('Data.csv')
query_data = [0,3,0,2,6,0,2,1]
# Get the output labels
output_labels = df['Category'].unique()
# Get the list of all words taken into consideration from the documents
words = list(df.columns)[:-1]
# Get the number of documents in the whole dataset
num_train_documents = df.shape[0]
```
## 3. Calculating the Required Probabilities
```
# A dictionary to store the conditional probabilities
# Format : conditional_probability[(a, b)] = P(a/b) => Probability of occurance of event `a` given that the event `b` has occured
conditional_probability = {}
# A dictionary to store the probabilities
# Format : probablity[a] = P(a) => Probablity of occurance of event `a`
probability = {}
```
### 3.1 Calculate the Probability of occurance of the output labels (classes)
```
for output_class in output_labels :
temp_df = df.loc[df['Category'] == output_class]
probability[output_class] = (temp_df.shape[0] / num_train_documents)
# Display the Probability of each output class
probability
```
### 3.2 Calculate the Conditional Probabilities
```
# Set parameter for smoothing
# ALPHA = 0 for no smoothing
ALPHA = 0
# ALPHA = 1 for Laplace Smoothing
#ALPHA = 1
for output_class in output_labels :
temp_df = df.loc[df['Category'] == output_class]
# Find the total number of words in that category
total_word_count_in_category = 0
for i in range(temp_df.shape[0]) :
for word in words :
total_word_count_in_category += temp_df.iloc[i][word]
# For each word find the number of times it occurs in the current category output
for word in words :
current_word_count_in_category = 0
for i in range(temp_df.shape[0]) :
current_word_count_in_category += temp_df.iloc[i][word]
# Store the conditional probability
cur_prob = (current_word_count_in_category + ALPHA) / (total_word_count_in_category + (ALPHA * len(words)))
conditional_probability[(word, output_class)] = cur_prob
# Without Smoothing, Some values are zero
print("Conditional Probabilities without applying any smoothing : \n")
conditional_probability
# With Smoothing
print("Conditional Probabilities after applying smoothing : \n")
conditional_probability
```
## 4. Process the Query
```
# Convert the query array into a dictionary to index with the name of the word
query_dict = {}
for i, word in enumerate(words) :
query_dict[word] = query_data[i]
query_dict
```
## 5. Find the Probability of the result
```
categorical_result_probability = {}
for output_class in output_labels :
cur_prob = 1
for word in words :
cur_prob *= (conditional_probability[(word, output_class)] ** query_dict[word])
categorical_result_probability[output_class] = cur_prob
print("Categorical scores without applying any smoothing : \n", categorical_result_probability)
print("Categorical scores after applying Laplace smoothing : \n", categorical_result_probability)
# Find the maximum probability
result_category = max(categorical_result_probability, key=categorical_result_probability.get)
result_score = categorical_result_probability[result_category]
print(f"The query entered belongs to the category : {result_category}")
```
|
github_jupyter
|
import pandas as pd
# Create a Pandas dataframe and store the data
df = pd.DataFrame(columns=['TDP', 'Nifty', 'Sidhu', 'BJP', 'Sensex', 'Sixer', 'Congress', 'Century', 'Category'])
num_entries = 0
# The Given Data
data = [
[4,0,3,5,1,0,6,0,'Politics'],
[0,5,0,2,6,0,1,0,'Business'],
[0,0,6,1,0,4,1,2,'Sports'],
[4,1,0,1,1,0,6,0,'Politics'],
[0,0,0,0,0,5,0,6,'Sports'],
[0,4,0,2,6,0,0,1,'Business'],
[5,0,0,3,0,0,5,0,'Politics']
]
query_data = [0,3,0,2,6,0,2,1]
# Load the data into a Pandas dataframe and store it into a CSV File
for i in range(len(data)) :
df.loc[num_entries] = data[i]
num_entries += 1
df.to_csv('Data.csv', index=None)
df = pd.read_csv('Data.csv')
query_data = [0,3,0,2,6,0,2,1]
# Get the output labels
output_labels = df['Category'].unique()
# Get the list of all words taken into consideration from the documents
words = list(df.columns)[:-1]
# Get the number of documents in the whole dataset
num_train_documents = df.shape[0]
# A dictionary to store the conditional probabilities
# Format : conditional_probability[(a, b)] = P(a/b) => Probability of occurance of event `a` given that the event `b` has occured
conditional_probability = {}
# A dictionary to store the probabilities
# Format : probablity[a] = P(a) => Probablity of occurance of event `a`
probability = {}
for output_class in output_labels :
temp_df = df.loc[df['Category'] == output_class]
probability[output_class] = (temp_df.shape[0] / num_train_documents)
# Display the Probability of each output class
probability
# Set parameter for smoothing
# ALPHA = 0 for no smoothing
ALPHA = 0
# ALPHA = 1 for Laplace Smoothing
#ALPHA = 1
for output_class in output_labels :
temp_df = df.loc[df['Category'] == output_class]
# Find the total number of words in that category
total_word_count_in_category = 0
for i in range(temp_df.shape[0]) :
for word in words :
total_word_count_in_category += temp_df.iloc[i][word]
# For each word find the number of times it occurs in the current category output
for word in words :
current_word_count_in_category = 0
for i in range(temp_df.shape[0]) :
current_word_count_in_category += temp_df.iloc[i][word]
# Store the conditional probability
cur_prob = (current_word_count_in_category + ALPHA) / (total_word_count_in_category + (ALPHA * len(words)))
conditional_probability[(word, output_class)] = cur_prob
# Without Smoothing, Some values are zero
print("Conditional Probabilities without applying any smoothing : \n")
conditional_probability
# With Smoothing
print("Conditional Probabilities after applying smoothing : \n")
conditional_probability
# Convert the query array into a dictionary to index with the name of the word
query_dict = {}
for i, word in enumerate(words) :
query_dict[word] = query_data[i]
query_dict
categorical_result_probability = {}
for output_class in output_labels :
cur_prob = 1
for word in words :
cur_prob *= (conditional_probability[(word, output_class)] ** query_dict[word])
categorical_result_probability[output_class] = cur_prob
print("Categorical scores without applying any smoothing : \n", categorical_result_probability)
print("Categorical scores after applying Laplace smoothing : \n", categorical_result_probability)
# Find the maximum probability
result_category = max(categorical_result_probability, key=categorical_result_probability.get)
result_score = categorical_result_probability[result_category]
print(f"The query entered belongs to the category : {result_category}")
| 0.506347 | 0.94868 |
<a href="https://colab.research.google.com/github/TMUITLab/EAFR/blob/master/EA1.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
```
!git clone https://github.com/TMUITLab/EAFR
!wget -O 'data.zip' 'https://efsgpq-ch3301.files.1drv.com/y4muoyVficiYL6mAlmm7s9m65fhNRboLtxg7FmaufA9QzY2tVhsyi-nXNtgahgN8NhrumVKCHB-d_lfi_5OTy1e5NFe2walhCu2Z1zF3zcp_hammSHuJHk5BeG6YbT7STynmA3SDPP39sNzn9V2Iv2suqlHkIrDRvRuvvM_r6IKuiRmJ35YirCUrY_Rojf5d-oQrxyQTj86Wz70JyiwrAYxfA'
!unzip '/content/data.zip' -d '/content/EAFR'
!pip install torch-scatter -f https://data.pyg.org/whl/torch-1.10.0+cu111.html
!pip install torch-sparse -f https://data.pyg.org/whl/torch-1.10.0+cu111.html
!pip install torch-geometric
!pip install igraph
!git pull https://github.com/TMUITLab/EAFR
%cd '/content/EAFR'
!CUDA_VISIBLE_DEVICES=0 python3 run.py
!fuser -v /dev/nvidia*
!kill -9 1661
!nvcc --version https://github.com/MaoXinn/RREA
!python -c "import torch; print(torch.__version__)"
!python -c "import torch; print(torch.version.cuda)"
!git pull https://github.com/TMUITLab/EAFR
%cd '/content/EAFR'
!CUDA_VISIBLE_DEVICES=0 python3 run.py
!git clone 'https://github.com/vinhsuhi/EMGCN'
import requests
def download_file_from_google_drive(id, destination):
URL = "https://docs.google.com/uc?export=download"
session = requests.Session()
response = session.get(URL, params = { 'id' : id }, stream = True)
token = get_confirm_token(response)
if token:
params = { 'id' : id, 'confirm' : token }
response = session.get(URL, params = params, stream = True)
save_response_content(response, destination)
def get_confirm_token(response):
for key, value in response.cookies.items():
if key.startswith('download_warning'):
return value
return None
def save_response_content(response, destination):
CHUNK_SIZE = 32768
with open(destination, "wb") as f:
for chunk in response.iter_content(CHUNK_SIZE):
if chunk: # filter out keep-alive new chunks
f.write(chunk)
%cd /content/
download_file_from_google_drive('12XL08tB8zplCNhzLE-9qbsFFum7RoV6r','emgcn.rar')
!pip install patool
import patoolib
patoolib.extract_archive("/content/emgcn.rar", outdir="/content/EMGCN/")
%cd '/content/EMGCN'
!python -u network_alignment.py --dataset_name zh_en --source_dataset data/networkx/zh_enDI/zh/graphsage/ --target_dataset data/networkx/zh_enDI/en/graphsage --groundtruth data/networkx/zh_enDI/dictionaries/groundtruth EMGCN --sparse --log
!pip install torch-scatter -f https://data.pyg.org/whl/torch-1.10.0+cu111.html
!pip install torch-sparse -f https://data.pyg.org/whl/torch-1.10.0+cu111.html
!pip install torch-geometric
%cd '/content'
!git clone https://github.com/zhurboo/RAGA
download_file_from_google_drive('1uJ2omzIs0NCtJsGQsyFCBHCXUhoK1mkO','/content/RAGA/data.tar.gz')
%cd '/content/RAGA'
!tar -xf data.tar.gz
%%writefi!le setup.sh
git clone https://github.com/NVIDIA/apex
pip install -v --no-cache-dir --global-option="--cpp_ext" --global-option="--cuda_ext" ./apex
!sh setup.sh
%cd '/content/RAGA'
!python train.py
%cd '/content'
!git clone https://github.com/1049451037/GCN-Align
%cd '/content'
!git clone https://github.com/MaoXinn/RREA
import warnings
warnings.filterwarnings('ignore')
from importlib.machinery import SourceFileLoader
layer = SourceFileLoader("layer", "/content/RREA/CIKM/layer.py").load_module()
utils = SourceFileLoader("utils", "/content/RREA/CIKM/utils.py").load_module()
CSLS = SourceFileLoader("CSLS", "/content/RREA/CIKM/CSLS.py").load_module()
import tensorflow as tf
import os
import random
import keras
from tqdm import *
import numpy as np
from utils import *
from CSLS import *
import tensorflow as tf
import keras.backend as K
from keras.layers import *
from layer import NR_GraphAttention
os.environ["CUDA_VISIBLE_DEVICES"] = "0"
os.environ["TF_CPP_MIN_LOG_LEVEL"]="2"
tf.compat.v1.logging.set_verbosity(tf.compat.v1.logging.ERROR)
lang = 'zh'
train_pair,dev_pair,adj_matrix,r_index,r_val,adj_features,rel_features = load_data('/content/GCN-Align/data/%s_en/'%lang,train_ratio=0.30)
train_pair_main=train_pair
adj_matrix = np.stack(adj_matrix.nonzero(),axis = 1)
rel_matrix,rel_val = np.stack(rel_features.nonzero(),axis = 1),rel_features.data
ent_matrix,ent_val = np.stack(adj_features.nonzero(),axis = 1),adj_features.data
print(r_index1[:,0].max(),adj_matrix1.shape,r_val1.__len__())
print(r_index[:,0].max(),adj_matrix.shape,r_val.__len__())
entity1, rel1, triples1 = load_triples('/content/GCN-Align/data/%s_en/'%lang + 'triples_1')
num_entity_1 = len(entity1)
num_rel_1 = len(rel1)
layer = SourceFileLoader("layer", "/content/RREA/CIKM/layer.py").load_module()
from layer import NR_GraphAttention
tf.keras.backend.clear_session()
node_size = adj_features.shape[0]
rel_size = rel_features.shape[1]
triple_size = len(adj_matrix)
batch_size = node_size
class TokenEmbedding(keras.layers.Embedding):
"""Embedding layer with weights returned."""
def compute_output_shape(self, input_shape):
return self.input_dim, self.output_dim
def compute_mask(self, inputs, mask=None):
return None
def call(self, inputs):
return self.embeddings
def get_embedding():
inputs = [adj_matrix,r_index,r_val,rel_matrix,ent_matrix]
inputs = [np.expand_dims(item,axis=0) for item in inputs]
return get_emb.predict_on_batch(inputs)
def test(wrank = None):
vec = get_embedding()
return get_hits(vec,dev_pair,wrank=wrank)
def CSLS_test(thread_number = 16, csls=10,accurate = True):
vec = get_embedding()
Lvec = np.array([vec[e1] for e1, e2 in dev_pair])
Rvec = np.array([vec[e2] for e1, e2 in dev_pair])
Lvec = Lvec / np.linalg.norm(Lvec,axis=-1,keepdims=True)
Rvec = Rvec / np.linalg.norm(Rvec,axis=-1,keepdims=True)
eval_alignment_by_sim_mat(Lvec, Rvec, [1, 5, 10], thread_number, csls=csls, accurate=accurate)
return None
def get_train_set(batch_size = batch_size):
negative_ratio = batch_size // len(train_pair) + 1
train_set = np.reshape(np.repeat(np.expand_dims(train_pair,axis=0),axis=0,repeats=negative_ratio),newshape=(-1,2))
np.random.shuffle(train_set); train_set = train_set[:batch_size]
train_set = np.concatenate([train_set,np.random.randint(0,node_size,train_set.shape)],axis = -1)
return train_set
def get_trgat(node_size,rel_size,node_hidden,rel_hidden,triple_size,n_attn_heads = 2,dropout_rate = 0,gamma = 3,lr = 0.005,depth = 2):
adj_input = Input(shape=(None,2))
index_input = Input(shape=(None,2),dtype='int64')
val_input = Input(shape = (None,))
rel_adj = Input(shape=(None,2))
ent_adj = Input(shape=(None,2))
ent_emb = TokenEmbedding(node_size,node_hidden,trainable = True)(val_input)
rel_emb = TokenEmbedding(rel_size,node_hidden,trainable = True)(val_input)
E = TokenEmbedding(node_hidden,node_hidden,trainable = True)(val_input)
R = TokenEmbedding(node_hidden,node_hidden,trainable = True)(val_input)
E = tf.linalg.expm(E - tf.transpose(E))
R = tf.linalg.expm(R - tf.transpose(R))
ent_emb = tf.concat([tf.matmul(ent_emb[:num_entity_1,:] ,E),ent_emb[num_entity_1:,:]],axis=0)
rel_emb = tf.concat([tf.matmul(rel_emb[:num_rel_1,:] ,R),rel_emb[num_rel_1:,:]],axis=0)
def avg(tensor,size):
adj = K.cast(K.squeeze(tensor[0],axis = 0),dtype = "int64")
adj = tf.SparseTensor(indices=adj, values=tf.ones_like(adj[:,0],dtype = 'float32'), dense_shape=(node_size,size))
adj = tf.compat.v1.sparse_softmax(adj)
return tf.compat.v1.sparse_tensor_dense_matmul(adj,tensor[1])
opt = [rel_emb,adj_input,index_input,val_input,ent_emb]
ent_feature = Lambda(avg,arguments={'size':node_size})([ent_adj,ent_emb])
rel_feature = Lambda(avg,arguments={'size':rel_size})([rel_adj,rel_emb])
tot_feature = tf.concat([rel_feature,ent_feature],axis=-1)
encoder = NR_GraphAttention(node_size,activation="relu",
rel_size = rel_size,
depth = depth,
attn_heads=n_attn_heads,
triple_size = triple_size,
attn_heads_reduction='average',
dropout_rate=dropout_rate)
encoder1 = NR_GraphAttention(node_size,activation="relu",
rel_size = rel_size,
depth = depth,
attn_heads=n_attn_heads,
triple_size = triple_size,
attn_heads_reduction='average',
dropout_rate=dropout_rate)
elemetns = [encoder([ent_emb]+opt),encoder([rel_feature]+opt),encoder([ent_feature]+opt)]
#x = [tf.keras.utils.normalize(x) for el in elemetns]
out_feature = Concatenate(-1)(elemetns)
out_feature = Dropout(dropout_rate)(out_feature)
alignment_input = Input(shape=(None,4))
find = Lambda(lambda x:K.gather(reference=x[0],indices=K.cast(K.squeeze(x[1],axis=0), 'int32')))([out_feature,alignment_input])
def align_loss(tensor):
def _cosine(x):
dot1 = K.batch_dot(x[0], x[1], axes=1)
dot2 = K.batch_dot(x[0], x[0], axes=1)
dot3 = K.batch_dot(x[1], x[1], axes=1)
max_ = K.maximum(K.sqrt(dot2 * dot3), K.epsilon())
return dot1 / max_
def l1(ll,rr):
return K.sum(K.abs(ll-rr),axis=-1,keepdims=True)
def l2(ll,rr):
return K.sum(K.square(ll-rr),axis=-1,keepdims=True)
l,r,fl,fr = [tensor[:,0,:],tensor[:,1,:],tensor[:,2 ,:],tensor[:,3,:]]
loss = K.relu(gamma + l1(l,r) - l1(l,fr)) + K.relu(gamma + l1(l,r) - l1(fl,r))
return tf.compat.v1.reduce_sum(loss,keep_dims=True) / (batch_size)
ent_mean_1 = tf.reduce_mean(ent_emb[:num_entity_1,:],axis=0)
ent_mean_2 = tf.reduce_mean(ent_emb[num_entity_1:,:],axis=0)
rel_mean_1 = tf.reduce_mean(rel_emb[:num_rel_1,:],axis=0)
rel_mean_2 = tf.reduce_mean(rel_emb[num_rel_1:,:],axis=0)
entf_mean_1 = tf.reduce_mean(ent_feature[:num_entity_1,:],axis=0)
entf_mean_2 = tf.reduce_mean(ent_feature[num_entity_1:,:],axis=0)
relf_mean_1 = tf.reduce_mean(rel_feature[:num_rel_1,:],axis=0)
relf_mean_2 = tf.reduce_mean(rel_feature[num_rel_1:,:],axis=0)
reg_term = K.sum(tf.square(rel_mean_1-rel_mean_2))
#reg_term += K.mean(K.sum(tf.square(ent_feature-ent_emb),axis=-1))
#reg_term += K.mean(K.sum(tf.square(rel_feature-ent_emb),axis=-1))
reg_term += K.sum(tf.square(ent_mean_1-ent_mean_2))
reg_term += K.sum(tf.square(entf_mean_1-entf_mean_2)) + K.sum(tf.square(relf_mean_1-relf_mean_2));
loss = Lambda(align_loss)(find) + 0.1* reg_term;
inputs = [adj_input,index_input,val_input,rel_adj,ent_adj]
train_model = keras.Model(inputs = inputs + [alignment_input],outputs = loss)
train_model.compile(loss=lambda y_true,y_pred: y_pred,optimizer=tf.keras.optimizers.RMSprop(lr=lr))
feature_model = keras.Model(inputs = inputs,outputs = out_feature)
return train_model,feature_model
model,get_emb = get_trgat(dropout_rate=0.30,node_size=node_size,rel_size=rel_size,n_attn_heads = 1,depth=2,gamma =3,node_hidden=100,rel_hidden = 100,triple_size = triple_size)
model.summary(); initial_weights = model.get_weights()
train_pair = train_pair_main
config = tf.compat.v1.ConfigProto()
config.gpu_options.allow_growth=True
sess = tf.compat.v1.Session(config=config)
rest_set_1 = [e1 for e1, e2 in dev_pair]
rest_set_2 = [e2 for e1, e2 in dev_pair]
np.random.shuffle(rest_set_1)
np.random.shuffle(rest_set_2)
epoch = 1200
for turn in range(5):
print("iteration %d start."%turn)
for i in trange(epoch):
train_set = get_train_set()
inputs = [adj_matrix,r_index,r_val,rel_matrix,ent_matrix,train_set]
inputs = [np.expand_dims(item,axis=0) for item in inputs]
model.train_on_batch(inputs,np.zeros((1,1)))
if i%100 == 99:
CSLS_test()
new_pair = []
vec = get_embedding()
Lvec = np.array([vec[e] for e in rest_set_1])
Rvec = np.array([vec[e] for e in rest_set_2])
Lvec = Lvec / np.linalg.norm(Lvec,axis=-1,keepdims=True)
Rvec = Rvec / np.linalg.norm(Rvec,axis=-1,keepdims=True)
A,_ = eval_alignment_by_sim_mat(Lvec, Rvec, [1, 5, 10], 16,10,True,False)
B,_ = eval_alignment_by_sim_mat(Rvec, Lvec,[1, 5, 10], 16,10,True,False)
A = sorted(list(A)); B = sorted(list(B))
for a,b in A:
if B[b][1] == a:
new_pair.append([rest_set_1[a],rest_set_2[b]])
print("generate new semi-pairs: %d." % len(new_pair))
train_pair = np.concatenate([train_pair,np.array(new_pair)],axis = 0)
for e1,e2 in new_pair:
if e1 in rest_set_1:
rest_set_1.remove(e1)
for e1,e2 in new_pair:
if e2 in rest_set_2:
rest_set_2.remove(e2)
!git clone https://github.com/MaoXinn/MRAEA
import os
import tqdm
import numpy as np
import tensorflow as tf
import keras
from importlib.machinery import SourceFileLoader
utils = SourceFileLoader("utils", "/content/MRAEA/utils.py").load_module()
model = SourceFileLoader("model", "/content/MRAEA/model.py").load_module()
from utils import *
from model import *
os.environ["CUDA_VISIBLE_DEVICES"] = "0"
os.environ["TF_CPP_MIN_LOG_LEVEL"]="2"
tf.compat.v1.logging.set_verbosity(tf.compat.v1.logging.ERROR)
config = tf.compat.v1.ConfigProto()
config.gpu_options.allow_growth=True
sess = tf.compat.v1.Session(config=config)
lang = 'zh'
train_pair,test_pair,adj_matrix,r_index,r_val,adj_features,rel_features = load_data('/content/GCN-Align/data/%s_en/'%lang,train_ratio=0.3)
adj_matrix = np.stack(adj_matrix.nonzero(),axis = 1)
rel_matrix,rel_val = np.stack(rel_features.nonzero(),axis = 1),rel_features.data
ent_matrix,ent_val = np.stack(adj_features.nonzero(),axis = 1),adj_features.data
model = SourceFileLoader("model", "/content/MRAEA/model.py").load_module()
node_size = adj_features.shape[1]
rel_size = rel_features.shape[1]
triple_size = len(adj_matrix)
batch_size = node_size
model,get_emb = get_model(lr=0.001,dropout_rate=0.30,node_size=node_size,rel_size=rel_size,n_attn_heads = 2,
depth=2,gamma = 3,node_hidden=100,rel_hidden = 100,triple_size = triple_size,batch_size = batch_size)
model.summary();
def get_train_set(batch_size,train_pair):
negative_ratio = batch_size // len(train_pair) + 1
train_set = np.reshape(np.repeat(np.expand_dims(train_pair,axis=0),axis=0,repeats=negative_ratio),newshape=(-1,2))
np.random.shuffle(train_set); train_set = train_set[:batch_size]
train_set = np.concatenate([train_set,np.random.randint(0,node_size,train_set.shape)],axis = -1)
return train_set
def test():
inputs = [adj_matrix,r_index,r_val,rel_matrix,ent_matrix]
inputs = [np.expand_dims(item,axis=0) for item in inputs]
se_vec = get_emb.predict_on_batch(inputs)
get_hits(se_vec,test_pair)
print()
return se_vec
for epoch in tqdm.tnrange(5000):
train_set = get_train_set(batch_size,train_pair)
inputs = [adj_matrix,r_index,r_val,rel_matrix,ent_matrix,train_set]
inputs = [np.expand_dims(item,axis=0) for item in inputs]
model.train_on_batch(inputs,np.zeros((1,1)))
if (epoch%1000 == 999):
test()
!git clone https://github.com/MaoXinn/Dual-AMN
import warnings
warnings.filterwarnings('ignore')
from importlib.machinery import SourceFileLoader
utils = SourceFileLoader("utils", "/content/Dual-AMN/utils.py").load_module()
evaluate = SourceFileLoader("evaluate", "/content/Dual-AMN/evaluate.py").load_module()
layer = SourceFileLoader("layer", "/content/Dual-AMN/layer.py").load_module()
import os
import keras
import numpy as np
import numba as nb
from utils import *
from tqdm import *
from evaluate import evaluate
import tensorflow as tf
import keras.backend as K
from keras.layers import *
from layer import NR_GraphAttention
os.environ["CUDA_VISIBLE_DEVICES"] = "0"
os.environ["TF_CPP_MIN_LOG_LEVEL"]="2"
tf.compat.v1.logging.set_verbosity(tf.compat.v1.logging.ERROR)
config = tf.compat.v1.ConfigProto()
config.gpu_options.allow_growth=True
sess = tf.compat.v1.Session(config=config)
seed = 12306
np.random.seed(seed)
tf.compat.v1.set_random_seed(seed)
train_pair,dev_pair,adj_matrix,r_index,r_val,adj_features,rel_features = load_data("/content/GCN-Align/data/zh_en/",train_ratio=0.30)
adj_matrix = np.stack(adj_matrix.nonzero(),axis = 1)
rel_matrix,rel_val = np.stack(rel_features.nonzero(),axis = 1),rel_features.data
ent_matrix,ent_val = np.stack(adj_features.nonzero(),axis = 1),adj_features.data
node_size = adj_features.shape[0]
rel_size = rel_features.shape[1]
triple_size = len(adj_matrix)
node_hidden = 100
rel_hidden = 100
batch_size = 1024
dropout_rate = 0.3
lr = 0.005
gamma = 1
depth = 2
layer = SourceFileLoader("layer", "/content/Dual-AMN/layer.py").load_module()
from layer import NR_GraphAttention
def get_embedding(index_a,index_b,vec = None):
if vec is None:
inputs = [adj_matrix,r_index,r_val,rel_matrix,ent_matrix]
inputs = [np.expand_dims(item,axis=0) for item in inputs]
vec = get_emb.predict_on_batch(inputs)
Lvec = np.array([vec[e] for e in index_a])
Rvec = np.array([vec[e] for e in index_b])
Lvec = Lvec / (np.linalg.norm(Lvec,axis=-1,keepdims=True)+1e-5)
Rvec = Rvec / (np.linalg.norm(Rvec,axis=-1,keepdims=True)+1e-5)
return Lvec,Rvec
class TokenEmbedding(keras.layers.Embedding):
"""Embedding layer with weights returned."""
def compute_output_shape(self, input_shape):
return self.input_dim, self.output_dim
def compute_mask(self, inputs, mask=None):
return None
def call(self, inputs):
return self.embeddings
def get_trgat(node_hidden,rel_hidden,triple_size=triple_size,node_size=node_size,rel_size=rel_size,dropout_rate = 0,gamma = 3,lr = 0.005,depth = 2):
adj_input = Input(shape=(None,2))
index_input = Input(shape=(None,2),dtype='int64')
val_input = Input(shape = (None,))
rel_adj = Input(shape=(None,2))
ent_adj = Input(shape=(None,2))
ent_emb = TokenEmbedding(node_size,node_hidden,trainable = True)(val_input)
rel_emb = TokenEmbedding(rel_size,node_hidden,trainable = True)(val_input)
def avg(tensor,size):
adj = K.cast(K.squeeze(tensor[0],axis = 0),dtype = "int64")
adj = tf.SparseTensor(indices=adj, values=tf.ones_like(adj[:,0],dtype = 'float32'), dense_shape=(node_size,size))
adj = tf.compat.v1.sparse_softmax(adj)
return tf.compat.v1.sparse_tensor_dense_matmul(adj,tensor[1])
opt = [rel_emb,adj_input,index_input,val_input]
ent_feature = Lambda(avg,arguments={'size':node_size})([ent_adj,ent_emb])
rel_feature = Lambda(avg,arguments={'size':rel_size})([rel_adj,rel_emb])
e_encoder = NR_GraphAttention(node_size,activation="tanh",
rel_size = rel_size,
use_bias = True,
depth = depth,
triple_size = triple_size)
r_encoder = NR_GraphAttention(node_size,activation="tanh",
rel_size = rel_size,
use_bias = True,
depth = depth,
triple_size = triple_size)
out_feature = Concatenate(-1)([e_encoder([ent_feature]+opt),r_encoder([rel_feature]+opt)])
out_feature = Dropout(dropout_rate)(out_feature)
alignment_input = Input(shape=(None,2))
def align_loss(tensor):
def squared_dist(x):
A,B = x
row_norms_A = tf.reduce_sum(tf.square(A), axis=1)
row_norms_A = tf.reshape(row_norms_A, [-1, 1]) # Column vector.
row_norms_B = tf.reduce_sum(tf.square(B), axis=1)
row_norms_B = tf.reshape(row_norms_B, [1, -1]) # Row vector.
return row_norms_A + row_norms_B - 2 * tf.matmul(A, B,transpose_b=True)
emb = tensor[1]
l,r = K.cast(tensor[0][0,:,0],'int32'),K.cast(tensor[0][0,:,1],'int32')
l_emb,r_emb = K.gather(reference=emb,indices=l),K.gather(reference=emb,indices=r)
pos_dis = K.sum(K.square(l_emb-r_emb),axis=-1,keepdims=True)
r_neg_dis = squared_dist([r_emb,emb])
l_neg_dis = squared_dist([l_emb,emb])
l_loss = pos_dis - l_neg_dis + gamma
l_loss = l_loss *(1 - K.one_hot(indices=l,num_classes=node_size) - K.one_hot(indices=r,num_classes=node_size))
r_loss = pos_dis - r_neg_dis + gamma
r_loss = r_loss *(1 - K.one_hot(indices=l,num_classes=node_size) - K.one_hot(indices=r,num_classes=node_size))
r_loss = (r_loss - K.stop_gradient(K.mean(r_loss,axis=-1,keepdims=True))) / K.stop_gradient(K.std(r_loss,axis=-1,keepdims=True))
l_loss = (l_loss - K.stop_gradient(K.mean(l_loss,axis=-1,keepdims=True))) / K.stop_gradient(K.std(l_loss,axis=-1,keepdims=True))
lamb,tau = 30, 10
l_loss = K.logsumexp(lamb*l_loss+tau,axis=-1)
r_loss = K.logsumexp(lamb*r_loss+tau,axis=-1)
return K.mean(l_loss + r_loss)
loss = Lambda(align_loss)([alignment_input,out_feature])
inputs = [adj_input,index_input,val_input,rel_adj,ent_adj]
train_model = keras.Model(inputs = inputs + [alignment_input],outputs = loss)
train_model.compile(loss=lambda y_true,y_pred: y_pred,optimizer=tf.keras.optimizers.RMSprop(lr))
feature_model = keras.Model(inputs = inputs,outputs = out_feature)
return train_model,feature_model
model,get_emb = get_trgat(dropout_rate=dropout_rate,
node_size=node_size,
rel_size=rel_size,
depth=depth,
gamma =gamma,
node_hidden=node_hidden,
rel_hidden=rel_hidden,
lr=lr)
evaluater = evaluate(dev_pair)
model.summary()
```
```
rest_set_1 = [e1 for e1, e2 in dev_pair]
rest_set_2 = [e2 for e1, e2 in dev_pair]
np.random.shuffle(rest_set_1)
np.random.shuffle(rest_set_2)
epoch = 20
for turn in range(10):
for i in trange(epoch):
np.random.shuffle(train_pair)
for pairs in [train_pair[i*batch_size:(i+1)*batch_size] for i in range(len(train_pair)//batch_size + 1)]:
if len(pairs) == 0:
continue
inputs = [adj_matrix,r_index,r_val,rel_matrix,ent_matrix,pairs]
inputs = [np.expand_dims(item,axis=0) for item in inputs]
model.train_on_batch(inputs,np.zeros((1,1)))
if i==epoch-1:
Lvec,Rvec = get_embedding(dev_pair[:,0],dev_pair[:,1])
evaluater.test(Lvec,Rvec)
new_pair = []
Lvec,Rvec = get_embedding(rest_set_1,rest_set_2)
A,B = evaluater.CSLS_cal(Lvec,Rvec,False)
for i,j in enumerate(A):
if B[j] == i:
new_pair.append([rest_set_1[j],rest_set_2[i]])
train_pair = np.concatenate([train_pair,np.array(new_pair)],axis = 0)
for e1,e2 in new_pair:
if e1 in rest_set_1:
rest_set_1.remove(e1)
for e1,e2 in new_pair:
if e2 in rest_set_2:
rest_set_2.remove(e2)
epoch = 5
```
|
github_jupyter
|
!git clone https://github.com/TMUITLab/EAFR
!wget -O 'data.zip' 'https://efsgpq-ch3301.files.1drv.com/y4muoyVficiYL6mAlmm7s9m65fhNRboLtxg7FmaufA9QzY2tVhsyi-nXNtgahgN8NhrumVKCHB-d_lfi_5OTy1e5NFe2walhCu2Z1zF3zcp_hammSHuJHk5BeG6YbT7STynmA3SDPP39sNzn9V2Iv2suqlHkIrDRvRuvvM_r6IKuiRmJ35YirCUrY_Rojf5d-oQrxyQTj86Wz70JyiwrAYxfA'
!unzip '/content/data.zip' -d '/content/EAFR'
!pip install torch-scatter -f https://data.pyg.org/whl/torch-1.10.0+cu111.html
!pip install torch-sparse -f https://data.pyg.org/whl/torch-1.10.0+cu111.html
!pip install torch-geometric
!pip install igraph
!git pull https://github.com/TMUITLab/EAFR
%cd '/content/EAFR'
!CUDA_VISIBLE_DEVICES=0 python3 run.py
!fuser -v /dev/nvidia*
!kill -9 1661
!nvcc --version https://github.com/MaoXinn/RREA
!python -c "import torch; print(torch.__version__)"
!python -c "import torch; print(torch.version.cuda)"
!git pull https://github.com/TMUITLab/EAFR
%cd '/content/EAFR'
!CUDA_VISIBLE_DEVICES=0 python3 run.py
!git clone 'https://github.com/vinhsuhi/EMGCN'
import requests
def download_file_from_google_drive(id, destination):
URL = "https://docs.google.com/uc?export=download"
session = requests.Session()
response = session.get(URL, params = { 'id' : id }, stream = True)
token = get_confirm_token(response)
if token:
params = { 'id' : id, 'confirm' : token }
response = session.get(URL, params = params, stream = True)
save_response_content(response, destination)
def get_confirm_token(response):
for key, value in response.cookies.items():
if key.startswith('download_warning'):
return value
return None
def save_response_content(response, destination):
CHUNK_SIZE = 32768
with open(destination, "wb") as f:
for chunk in response.iter_content(CHUNK_SIZE):
if chunk: # filter out keep-alive new chunks
f.write(chunk)
%cd /content/
download_file_from_google_drive('12XL08tB8zplCNhzLE-9qbsFFum7RoV6r','emgcn.rar')
!pip install patool
import patoolib
patoolib.extract_archive("/content/emgcn.rar", outdir="/content/EMGCN/")
%cd '/content/EMGCN'
!python -u network_alignment.py --dataset_name zh_en --source_dataset data/networkx/zh_enDI/zh/graphsage/ --target_dataset data/networkx/zh_enDI/en/graphsage --groundtruth data/networkx/zh_enDI/dictionaries/groundtruth EMGCN --sparse --log
!pip install torch-scatter -f https://data.pyg.org/whl/torch-1.10.0+cu111.html
!pip install torch-sparse -f https://data.pyg.org/whl/torch-1.10.0+cu111.html
!pip install torch-geometric
%cd '/content'
!git clone https://github.com/zhurboo/RAGA
download_file_from_google_drive('1uJ2omzIs0NCtJsGQsyFCBHCXUhoK1mkO','/content/RAGA/data.tar.gz')
%cd '/content/RAGA'
!tar -xf data.tar.gz
%%writefi!le setup.sh
git clone https://github.com/NVIDIA/apex
pip install -v --no-cache-dir --global-option="--cpp_ext" --global-option="--cuda_ext" ./apex
!sh setup.sh
%cd '/content/RAGA'
!python train.py
%cd '/content'
!git clone https://github.com/1049451037/GCN-Align
%cd '/content'
!git clone https://github.com/MaoXinn/RREA
import warnings
warnings.filterwarnings('ignore')
from importlib.machinery import SourceFileLoader
layer = SourceFileLoader("layer", "/content/RREA/CIKM/layer.py").load_module()
utils = SourceFileLoader("utils", "/content/RREA/CIKM/utils.py").load_module()
CSLS = SourceFileLoader("CSLS", "/content/RREA/CIKM/CSLS.py").load_module()
import tensorflow as tf
import os
import random
import keras
from tqdm import *
import numpy as np
from utils import *
from CSLS import *
import tensorflow as tf
import keras.backend as K
from keras.layers import *
from layer import NR_GraphAttention
os.environ["CUDA_VISIBLE_DEVICES"] = "0"
os.environ["TF_CPP_MIN_LOG_LEVEL"]="2"
tf.compat.v1.logging.set_verbosity(tf.compat.v1.logging.ERROR)
lang = 'zh'
train_pair,dev_pair,adj_matrix,r_index,r_val,adj_features,rel_features = load_data('/content/GCN-Align/data/%s_en/'%lang,train_ratio=0.30)
train_pair_main=train_pair
adj_matrix = np.stack(adj_matrix.nonzero(),axis = 1)
rel_matrix,rel_val = np.stack(rel_features.nonzero(),axis = 1),rel_features.data
ent_matrix,ent_val = np.stack(adj_features.nonzero(),axis = 1),adj_features.data
print(r_index1[:,0].max(),adj_matrix1.shape,r_val1.__len__())
print(r_index[:,0].max(),adj_matrix.shape,r_val.__len__())
entity1, rel1, triples1 = load_triples('/content/GCN-Align/data/%s_en/'%lang + 'triples_1')
num_entity_1 = len(entity1)
num_rel_1 = len(rel1)
layer = SourceFileLoader("layer", "/content/RREA/CIKM/layer.py").load_module()
from layer import NR_GraphAttention
tf.keras.backend.clear_session()
node_size = adj_features.shape[0]
rel_size = rel_features.shape[1]
triple_size = len(adj_matrix)
batch_size = node_size
class TokenEmbedding(keras.layers.Embedding):
"""Embedding layer with weights returned."""
def compute_output_shape(self, input_shape):
return self.input_dim, self.output_dim
def compute_mask(self, inputs, mask=None):
return None
def call(self, inputs):
return self.embeddings
def get_embedding():
inputs = [adj_matrix,r_index,r_val,rel_matrix,ent_matrix]
inputs = [np.expand_dims(item,axis=0) for item in inputs]
return get_emb.predict_on_batch(inputs)
def test(wrank = None):
vec = get_embedding()
return get_hits(vec,dev_pair,wrank=wrank)
def CSLS_test(thread_number = 16, csls=10,accurate = True):
vec = get_embedding()
Lvec = np.array([vec[e1] for e1, e2 in dev_pair])
Rvec = np.array([vec[e2] for e1, e2 in dev_pair])
Lvec = Lvec / np.linalg.norm(Lvec,axis=-1,keepdims=True)
Rvec = Rvec / np.linalg.norm(Rvec,axis=-1,keepdims=True)
eval_alignment_by_sim_mat(Lvec, Rvec, [1, 5, 10], thread_number, csls=csls, accurate=accurate)
return None
def get_train_set(batch_size = batch_size):
negative_ratio = batch_size // len(train_pair) + 1
train_set = np.reshape(np.repeat(np.expand_dims(train_pair,axis=0),axis=0,repeats=negative_ratio),newshape=(-1,2))
np.random.shuffle(train_set); train_set = train_set[:batch_size]
train_set = np.concatenate([train_set,np.random.randint(0,node_size,train_set.shape)],axis = -1)
return train_set
def get_trgat(node_size,rel_size,node_hidden,rel_hidden,triple_size,n_attn_heads = 2,dropout_rate = 0,gamma = 3,lr = 0.005,depth = 2):
adj_input = Input(shape=(None,2))
index_input = Input(shape=(None,2),dtype='int64')
val_input = Input(shape = (None,))
rel_adj = Input(shape=(None,2))
ent_adj = Input(shape=(None,2))
ent_emb = TokenEmbedding(node_size,node_hidden,trainable = True)(val_input)
rel_emb = TokenEmbedding(rel_size,node_hidden,trainable = True)(val_input)
E = TokenEmbedding(node_hidden,node_hidden,trainable = True)(val_input)
R = TokenEmbedding(node_hidden,node_hidden,trainable = True)(val_input)
E = tf.linalg.expm(E - tf.transpose(E))
R = tf.linalg.expm(R - tf.transpose(R))
ent_emb = tf.concat([tf.matmul(ent_emb[:num_entity_1,:] ,E),ent_emb[num_entity_1:,:]],axis=0)
rel_emb = tf.concat([tf.matmul(rel_emb[:num_rel_1,:] ,R),rel_emb[num_rel_1:,:]],axis=0)
def avg(tensor,size):
adj = K.cast(K.squeeze(tensor[0],axis = 0),dtype = "int64")
adj = tf.SparseTensor(indices=adj, values=tf.ones_like(adj[:,0],dtype = 'float32'), dense_shape=(node_size,size))
adj = tf.compat.v1.sparse_softmax(adj)
return tf.compat.v1.sparse_tensor_dense_matmul(adj,tensor[1])
opt = [rel_emb,adj_input,index_input,val_input,ent_emb]
ent_feature = Lambda(avg,arguments={'size':node_size})([ent_adj,ent_emb])
rel_feature = Lambda(avg,arguments={'size':rel_size})([rel_adj,rel_emb])
tot_feature = tf.concat([rel_feature,ent_feature],axis=-1)
encoder = NR_GraphAttention(node_size,activation="relu",
rel_size = rel_size,
depth = depth,
attn_heads=n_attn_heads,
triple_size = triple_size,
attn_heads_reduction='average',
dropout_rate=dropout_rate)
encoder1 = NR_GraphAttention(node_size,activation="relu",
rel_size = rel_size,
depth = depth,
attn_heads=n_attn_heads,
triple_size = triple_size,
attn_heads_reduction='average',
dropout_rate=dropout_rate)
elemetns = [encoder([ent_emb]+opt),encoder([rel_feature]+opt),encoder([ent_feature]+opt)]
#x = [tf.keras.utils.normalize(x) for el in elemetns]
out_feature = Concatenate(-1)(elemetns)
out_feature = Dropout(dropout_rate)(out_feature)
alignment_input = Input(shape=(None,4))
find = Lambda(lambda x:K.gather(reference=x[0],indices=K.cast(K.squeeze(x[1],axis=0), 'int32')))([out_feature,alignment_input])
def align_loss(tensor):
def _cosine(x):
dot1 = K.batch_dot(x[0], x[1], axes=1)
dot2 = K.batch_dot(x[0], x[0], axes=1)
dot3 = K.batch_dot(x[1], x[1], axes=1)
max_ = K.maximum(K.sqrt(dot2 * dot3), K.epsilon())
return dot1 / max_
def l1(ll,rr):
return K.sum(K.abs(ll-rr),axis=-1,keepdims=True)
def l2(ll,rr):
return K.sum(K.square(ll-rr),axis=-1,keepdims=True)
l,r,fl,fr = [tensor[:,0,:],tensor[:,1,:],tensor[:,2 ,:],tensor[:,3,:]]
loss = K.relu(gamma + l1(l,r) - l1(l,fr)) + K.relu(gamma + l1(l,r) - l1(fl,r))
return tf.compat.v1.reduce_sum(loss,keep_dims=True) / (batch_size)
ent_mean_1 = tf.reduce_mean(ent_emb[:num_entity_1,:],axis=0)
ent_mean_2 = tf.reduce_mean(ent_emb[num_entity_1:,:],axis=0)
rel_mean_1 = tf.reduce_mean(rel_emb[:num_rel_1,:],axis=0)
rel_mean_2 = tf.reduce_mean(rel_emb[num_rel_1:,:],axis=0)
entf_mean_1 = tf.reduce_mean(ent_feature[:num_entity_1,:],axis=0)
entf_mean_2 = tf.reduce_mean(ent_feature[num_entity_1:,:],axis=0)
relf_mean_1 = tf.reduce_mean(rel_feature[:num_rel_1,:],axis=0)
relf_mean_2 = tf.reduce_mean(rel_feature[num_rel_1:,:],axis=0)
reg_term = K.sum(tf.square(rel_mean_1-rel_mean_2))
#reg_term += K.mean(K.sum(tf.square(ent_feature-ent_emb),axis=-1))
#reg_term += K.mean(K.sum(tf.square(rel_feature-ent_emb),axis=-1))
reg_term += K.sum(tf.square(ent_mean_1-ent_mean_2))
reg_term += K.sum(tf.square(entf_mean_1-entf_mean_2)) + K.sum(tf.square(relf_mean_1-relf_mean_2));
loss = Lambda(align_loss)(find) + 0.1* reg_term;
inputs = [adj_input,index_input,val_input,rel_adj,ent_adj]
train_model = keras.Model(inputs = inputs + [alignment_input],outputs = loss)
train_model.compile(loss=lambda y_true,y_pred: y_pred,optimizer=tf.keras.optimizers.RMSprop(lr=lr))
feature_model = keras.Model(inputs = inputs,outputs = out_feature)
return train_model,feature_model
model,get_emb = get_trgat(dropout_rate=0.30,node_size=node_size,rel_size=rel_size,n_attn_heads = 1,depth=2,gamma =3,node_hidden=100,rel_hidden = 100,triple_size = triple_size)
model.summary(); initial_weights = model.get_weights()
train_pair = train_pair_main
config = tf.compat.v1.ConfigProto()
config.gpu_options.allow_growth=True
sess = tf.compat.v1.Session(config=config)
rest_set_1 = [e1 for e1, e2 in dev_pair]
rest_set_2 = [e2 for e1, e2 in dev_pair]
np.random.shuffle(rest_set_1)
np.random.shuffle(rest_set_2)
epoch = 1200
for turn in range(5):
print("iteration %d start."%turn)
for i in trange(epoch):
train_set = get_train_set()
inputs = [adj_matrix,r_index,r_val,rel_matrix,ent_matrix,train_set]
inputs = [np.expand_dims(item,axis=0) for item in inputs]
model.train_on_batch(inputs,np.zeros((1,1)))
if i%100 == 99:
CSLS_test()
new_pair = []
vec = get_embedding()
Lvec = np.array([vec[e] for e in rest_set_1])
Rvec = np.array([vec[e] for e in rest_set_2])
Lvec = Lvec / np.linalg.norm(Lvec,axis=-1,keepdims=True)
Rvec = Rvec / np.linalg.norm(Rvec,axis=-1,keepdims=True)
A,_ = eval_alignment_by_sim_mat(Lvec, Rvec, [1, 5, 10], 16,10,True,False)
B,_ = eval_alignment_by_sim_mat(Rvec, Lvec,[1, 5, 10], 16,10,True,False)
A = sorted(list(A)); B = sorted(list(B))
for a,b in A:
if B[b][1] == a:
new_pair.append([rest_set_1[a],rest_set_2[b]])
print("generate new semi-pairs: %d." % len(new_pair))
train_pair = np.concatenate([train_pair,np.array(new_pair)],axis = 0)
for e1,e2 in new_pair:
if e1 in rest_set_1:
rest_set_1.remove(e1)
for e1,e2 in new_pair:
if e2 in rest_set_2:
rest_set_2.remove(e2)
!git clone https://github.com/MaoXinn/MRAEA
import os
import tqdm
import numpy as np
import tensorflow as tf
import keras
from importlib.machinery import SourceFileLoader
utils = SourceFileLoader("utils", "/content/MRAEA/utils.py").load_module()
model = SourceFileLoader("model", "/content/MRAEA/model.py").load_module()
from utils import *
from model import *
os.environ["CUDA_VISIBLE_DEVICES"] = "0"
os.environ["TF_CPP_MIN_LOG_LEVEL"]="2"
tf.compat.v1.logging.set_verbosity(tf.compat.v1.logging.ERROR)
config = tf.compat.v1.ConfigProto()
config.gpu_options.allow_growth=True
sess = tf.compat.v1.Session(config=config)
lang = 'zh'
train_pair,test_pair,adj_matrix,r_index,r_val,adj_features,rel_features = load_data('/content/GCN-Align/data/%s_en/'%lang,train_ratio=0.3)
adj_matrix = np.stack(adj_matrix.nonzero(),axis = 1)
rel_matrix,rel_val = np.stack(rel_features.nonzero(),axis = 1),rel_features.data
ent_matrix,ent_val = np.stack(adj_features.nonzero(),axis = 1),adj_features.data
model = SourceFileLoader("model", "/content/MRAEA/model.py").load_module()
node_size = adj_features.shape[1]
rel_size = rel_features.shape[1]
triple_size = len(adj_matrix)
batch_size = node_size
model,get_emb = get_model(lr=0.001,dropout_rate=0.30,node_size=node_size,rel_size=rel_size,n_attn_heads = 2,
depth=2,gamma = 3,node_hidden=100,rel_hidden = 100,triple_size = triple_size,batch_size = batch_size)
model.summary();
def get_train_set(batch_size,train_pair):
negative_ratio = batch_size // len(train_pair) + 1
train_set = np.reshape(np.repeat(np.expand_dims(train_pair,axis=0),axis=0,repeats=negative_ratio),newshape=(-1,2))
np.random.shuffle(train_set); train_set = train_set[:batch_size]
train_set = np.concatenate([train_set,np.random.randint(0,node_size,train_set.shape)],axis = -1)
return train_set
def test():
inputs = [adj_matrix,r_index,r_val,rel_matrix,ent_matrix]
inputs = [np.expand_dims(item,axis=0) for item in inputs]
se_vec = get_emb.predict_on_batch(inputs)
get_hits(se_vec,test_pair)
print()
return se_vec
for epoch in tqdm.tnrange(5000):
train_set = get_train_set(batch_size,train_pair)
inputs = [adj_matrix,r_index,r_val,rel_matrix,ent_matrix,train_set]
inputs = [np.expand_dims(item,axis=0) for item in inputs]
model.train_on_batch(inputs,np.zeros((1,1)))
if (epoch%1000 == 999):
test()
!git clone https://github.com/MaoXinn/Dual-AMN
import warnings
warnings.filterwarnings('ignore')
from importlib.machinery import SourceFileLoader
utils = SourceFileLoader("utils", "/content/Dual-AMN/utils.py").load_module()
evaluate = SourceFileLoader("evaluate", "/content/Dual-AMN/evaluate.py").load_module()
layer = SourceFileLoader("layer", "/content/Dual-AMN/layer.py").load_module()
import os
import keras
import numpy as np
import numba as nb
from utils import *
from tqdm import *
from evaluate import evaluate
import tensorflow as tf
import keras.backend as K
from keras.layers import *
from layer import NR_GraphAttention
os.environ["CUDA_VISIBLE_DEVICES"] = "0"
os.environ["TF_CPP_MIN_LOG_LEVEL"]="2"
tf.compat.v1.logging.set_verbosity(tf.compat.v1.logging.ERROR)
config = tf.compat.v1.ConfigProto()
config.gpu_options.allow_growth=True
sess = tf.compat.v1.Session(config=config)
seed = 12306
np.random.seed(seed)
tf.compat.v1.set_random_seed(seed)
train_pair,dev_pair,adj_matrix,r_index,r_val,adj_features,rel_features = load_data("/content/GCN-Align/data/zh_en/",train_ratio=0.30)
adj_matrix = np.stack(adj_matrix.nonzero(),axis = 1)
rel_matrix,rel_val = np.stack(rel_features.nonzero(),axis = 1),rel_features.data
ent_matrix,ent_val = np.stack(adj_features.nonzero(),axis = 1),adj_features.data
node_size = adj_features.shape[0]
rel_size = rel_features.shape[1]
triple_size = len(adj_matrix)
node_hidden = 100
rel_hidden = 100
batch_size = 1024
dropout_rate = 0.3
lr = 0.005
gamma = 1
depth = 2
layer = SourceFileLoader("layer", "/content/Dual-AMN/layer.py").load_module()
from layer import NR_GraphAttention
def get_embedding(index_a,index_b,vec = None):
if vec is None:
inputs = [adj_matrix,r_index,r_val,rel_matrix,ent_matrix]
inputs = [np.expand_dims(item,axis=0) for item in inputs]
vec = get_emb.predict_on_batch(inputs)
Lvec = np.array([vec[e] for e in index_a])
Rvec = np.array([vec[e] for e in index_b])
Lvec = Lvec / (np.linalg.norm(Lvec,axis=-1,keepdims=True)+1e-5)
Rvec = Rvec / (np.linalg.norm(Rvec,axis=-1,keepdims=True)+1e-5)
return Lvec,Rvec
class TokenEmbedding(keras.layers.Embedding):
"""Embedding layer with weights returned."""
def compute_output_shape(self, input_shape):
return self.input_dim, self.output_dim
def compute_mask(self, inputs, mask=None):
return None
def call(self, inputs):
return self.embeddings
def get_trgat(node_hidden,rel_hidden,triple_size=triple_size,node_size=node_size,rel_size=rel_size,dropout_rate = 0,gamma = 3,lr = 0.005,depth = 2):
adj_input = Input(shape=(None,2))
index_input = Input(shape=(None,2),dtype='int64')
val_input = Input(shape = (None,))
rel_adj = Input(shape=(None,2))
ent_adj = Input(shape=(None,2))
ent_emb = TokenEmbedding(node_size,node_hidden,trainable = True)(val_input)
rel_emb = TokenEmbedding(rel_size,node_hidden,trainable = True)(val_input)
def avg(tensor,size):
adj = K.cast(K.squeeze(tensor[0],axis = 0),dtype = "int64")
adj = tf.SparseTensor(indices=adj, values=tf.ones_like(adj[:,0],dtype = 'float32'), dense_shape=(node_size,size))
adj = tf.compat.v1.sparse_softmax(adj)
return tf.compat.v1.sparse_tensor_dense_matmul(adj,tensor[1])
opt = [rel_emb,adj_input,index_input,val_input]
ent_feature = Lambda(avg,arguments={'size':node_size})([ent_adj,ent_emb])
rel_feature = Lambda(avg,arguments={'size':rel_size})([rel_adj,rel_emb])
e_encoder = NR_GraphAttention(node_size,activation="tanh",
rel_size = rel_size,
use_bias = True,
depth = depth,
triple_size = triple_size)
r_encoder = NR_GraphAttention(node_size,activation="tanh",
rel_size = rel_size,
use_bias = True,
depth = depth,
triple_size = triple_size)
out_feature = Concatenate(-1)([e_encoder([ent_feature]+opt),r_encoder([rel_feature]+opt)])
out_feature = Dropout(dropout_rate)(out_feature)
alignment_input = Input(shape=(None,2))
def align_loss(tensor):
def squared_dist(x):
A,B = x
row_norms_A = tf.reduce_sum(tf.square(A), axis=1)
row_norms_A = tf.reshape(row_norms_A, [-1, 1]) # Column vector.
row_norms_B = tf.reduce_sum(tf.square(B), axis=1)
row_norms_B = tf.reshape(row_norms_B, [1, -1]) # Row vector.
return row_norms_A + row_norms_B - 2 * tf.matmul(A, B,transpose_b=True)
emb = tensor[1]
l,r = K.cast(tensor[0][0,:,0],'int32'),K.cast(tensor[0][0,:,1],'int32')
l_emb,r_emb = K.gather(reference=emb,indices=l),K.gather(reference=emb,indices=r)
pos_dis = K.sum(K.square(l_emb-r_emb),axis=-1,keepdims=True)
r_neg_dis = squared_dist([r_emb,emb])
l_neg_dis = squared_dist([l_emb,emb])
l_loss = pos_dis - l_neg_dis + gamma
l_loss = l_loss *(1 - K.one_hot(indices=l,num_classes=node_size) - K.one_hot(indices=r,num_classes=node_size))
r_loss = pos_dis - r_neg_dis + gamma
r_loss = r_loss *(1 - K.one_hot(indices=l,num_classes=node_size) - K.one_hot(indices=r,num_classes=node_size))
r_loss = (r_loss - K.stop_gradient(K.mean(r_loss,axis=-1,keepdims=True))) / K.stop_gradient(K.std(r_loss,axis=-1,keepdims=True))
l_loss = (l_loss - K.stop_gradient(K.mean(l_loss,axis=-1,keepdims=True))) / K.stop_gradient(K.std(l_loss,axis=-1,keepdims=True))
lamb,tau = 30, 10
l_loss = K.logsumexp(lamb*l_loss+tau,axis=-1)
r_loss = K.logsumexp(lamb*r_loss+tau,axis=-1)
return K.mean(l_loss + r_loss)
loss = Lambda(align_loss)([alignment_input,out_feature])
inputs = [adj_input,index_input,val_input,rel_adj,ent_adj]
train_model = keras.Model(inputs = inputs + [alignment_input],outputs = loss)
train_model.compile(loss=lambda y_true,y_pred: y_pred,optimizer=tf.keras.optimizers.RMSprop(lr))
feature_model = keras.Model(inputs = inputs,outputs = out_feature)
return train_model,feature_model
model,get_emb = get_trgat(dropout_rate=dropout_rate,
node_size=node_size,
rel_size=rel_size,
depth=depth,
gamma =gamma,
node_hidden=node_hidden,
rel_hidden=rel_hidden,
lr=lr)
evaluater = evaluate(dev_pair)
model.summary()
rest_set_1 = [e1 for e1, e2 in dev_pair]
rest_set_2 = [e2 for e1, e2 in dev_pair]
np.random.shuffle(rest_set_1)
np.random.shuffle(rest_set_2)
epoch = 20
for turn in range(10):
for i in trange(epoch):
np.random.shuffle(train_pair)
for pairs in [train_pair[i*batch_size:(i+1)*batch_size] for i in range(len(train_pair)//batch_size + 1)]:
if len(pairs) == 0:
continue
inputs = [adj_matrix,r_index,r_val,rel_matrix,ent_matrix,pairs]
inputs = [np.expand_dims(item,axis=0) for item in inputs]
model.train_on_batch(inputs,np.zeros((1,1)))
if i==epoch-1:
Lvec,Rvec = get_embedding(dev_pair[:,0],dev_pair[:,1])
evaluater.test(Lvec,Rvec)
new_pair = []
Lvec,Rvec = get_embedding(rest_set_1,rest_set_2)
A,B = evaluater.CSLS_cal(Lvec,Rvec,False)
for i,j in enumerate(A):
if B[j] == i:
new_pair.append([rest_set_1[j],rest_set_2[i]])
train_pair = np.concatenate([train_pair,np.array(new_pair)],axis = 0)
for e1,e2 in new_pair:
if e1 in rest_set_1:
rest_set_1.remove(e1)
for e1,e2 in new_pair:
if e2 in rest_set_2:
rest_set_2.remove(e2)
epoch = 5
| 0.557604 | 0.561335 |
# Stack Semantics in Trax: Ungraded Lab
In this ungraded lab, we will explain the stack semantics in Trax. This will help in understanding how to use layers like `Select` and `Residual` which operates on elements in the stack. If you've taken a computer science class before, you will recall that a stack is a data structure that follows the Last In, First Out (LIFO) principle. That is, whatever is the latest element that is pushed into the stack will also be the first one to be popped out. If you're not yet familiar with stacks, then you may find this [short tutorial](https://www.tutorialspoint.com/python_data_structure/python_stack.htm) useful. In a nutshell, all you really need to remember is it puts elements one on top of the other. You should be aware of what is on top of the stack to know which element you will be popping first. You will see this in the discussions below. Let's get started!
## Imports
```
import numpy as np # regular ol' numpy
from trax import layers as tl # core building block
from trax import shapes # data signatures: dimensionality and type
from trax import fastmath # uses jax, offers numpy on steroids
```
## 1. The tl.Serial Combinator is Stack Oriented.
To understand how stack-orientation works in [Trax](https://trax-ml.readthedocs.io/en/latest/) most times one will be using the `Serial` layer. In order to explain the way stack orientation works we will define two simple `Function` layers: 1) Addition and 2) Multiplication.
Suppose we want to make the simple calculation (3 + 4) * 15 + 3. `Serial` will perform the calculations in the following manner `3` `4` `add` `15` `mul` `3` `add`. The steps of the calculation are shown in the table below. The first column shows the operations made on the stack and the second column the output of those operations. **Moreover, the rightmost element in the second column represents the top of the stack** (e.g. in the second row, `Push(3)` pushes `3 ` on top of the stack and `4` is now under it). In the case of operations such as `add` or `mul`, we will need to `pop` the elements to operate before making the operation. That is the reason why inside `add` you will find two `pop` operations, meaning that we will pop the two elements at the top of the stack and sum them. Then, the result is pushed back to the top of the stack.
<div style="text-align:center" width="50px"><img src="images/Stack1.png" /></div>
After processing all, the stack contains 108 which is the answer to our simple computation.
From this, the following can be concluded: a stack-based layer has only one way to handle data, by taking one piece of data from atop the stack, termed popping, and putting data back atop the stack, termed pushing. Any expression that can be written conventionally, can be written in this form and thus be amenable to being interpreted by a stack-oriented layer like `Serial`.
### Coding the example in the table:
**Defining addition**
```
def Addition():
layer_name = "Addition" # don't forget to give your custom layer a name to identify
# Custom function for the custom layer
def func(x, y):
return x + y
return tl.Fn(layer_name, func)
# Test it
add = Addition()
# Inspect properties
print("-- Properties --")
print("name :", add.name)
print("expected inputs :", add.n_in)
print("promised outputs :", add.n_out, "\n")
# Inputs
x = np.array([3])
y = np.array([4])
print("-- Inputs --")
print("x :", x, "\n")
print("y :", y, "\n")
# Outputs
z = add((x, y))
print("-- Outputs --")
print("z :", z)
```
**Defining multiplication**
```
def Multiplication():
layer_name = (
"Multiplication" # don't forget to give your custom layer a name to identify
)
# Custom function for the custom layer
def func(x, y):
return x * y
return tl.Fn(layer_name, func)
# Test it
mul = Multiplication()
# Inspect properties
print("-- Properties --")
print("name :", mul.name)
print("expected inputs :", mul.n_in)
print("promised outputs :", mul.n_out, "\n")
# Inputs
x = np.array([7])
y = np.array([15])
print("-- Inputs --")
print("x :", x, "\n")
print("y :", y, "\n")
# Outputs
z = mul((x, y))
print("-- Outputs --")
print("z :", z)
```
**Implementing the computations using Serial combinator.**
```
# Serial combinator
serial = tl.Serial(
Addition(), Multiplication(), Addition() # add 3 + 4 # multiply result by 15 and then add 3
)
# Initialization
x = (np.array([3]), np.array([4]), np.array([15]), np.array([3])) # input
serial.init(shapes.signature(x)) # initializing serial instance
print("-- Serial Model --")
print(serial, "\n")
print("-- Properties --")
print("name :", serial.name)
print("sublayers :", serial.sublayers)
print("expected inputs :", serial.n_in)
print("promised outputs :", serial.n_out, "\n")
# Inputs
print("-- Inputs --")
print("x :", x, "\n")
# Outputs
y = serial(x)
print("-- Outputs --")
print("y :", y)
```
The example with the two simple adition and multiplication functions that where coded together with the serial combinator show how stack semantics work in `Trax`.
## 2. The tl.Select combinator in the context of the serial combinator
Having understood how stack semantics work in `Trax`, we will demonstrate how the [tl.Select](https://trax-ml.readthedocs.io/en/latest/trax.layers.html?highlight=select#trax.layers.combinators.Select) combinator works.
### First example of tl.Select
Suppose we want to make the simple calculation (3 + 4) * 3 + 4. We can use `Select` to perform the calculations in the following manner:
1. `4`
2. `3`
3. `tl.Select([0,1,0,1])`
4. `add`
5. `mul`
6. `add`.
The `tl.Select` requires a list or tuple of 0-based indices to select elements relative to the top of the stack. For our example, the top of the stack is `3` (which is at index 0) then `4` (index 1); remember that the rightmost element in the second column corresponds to the top of the stack. Then, we Select to add in an ordered manner to the top of the stack which after the command is `3` `4` `3` `4`. The steps of the calculation for our example are shown in the table below. As in the previous table each column shows the contents of the stack and the outputs after the operations are carried out. Remember that for `add` or `mul`, we will need to `pop` the elements to operate before making the operation. So the two `pop` operations inside the `add`/`mul` will mean that the two elements at the top of the stack will be popped and them operated; the other elements will keep at their positions in the stack. Finally, the result of the operation is pushed back to the top of the stack.
<div style="text-align:center" width="20px"><img src="images/Stack2.png" /></div>
After processing all the inputs, the stack contains 25 which is the answer we get above.
```
serial = tl.Serial(tl.Select([0, 1, 0, 1]), Addition(), Multiplication(), Addition())
# Initialization
x = (np.array([3]), np.array([4])) # input
serial.init(shapes.signature(x)) # initializing serial instance
print("-- Serial Model --")
print(serial, "\n")
print("-- Properties --")
print("name :", serial.name)
print("sublayers :", serial.sublayers)
print("expected inputs :", serial.n_in)
print("promised outputs :", serial.n_out, "\n")
# Inputs
print("-- Inputs --")
print("x :", x, "\n")
# Outputs
y = serial(x)
print("-- Outputs --")
print("y :", y)
```
### Second example of tl.Select
Suppose we want to make the simple calculation (3 + 4) * 4. We can use `Select` to perform the calculations in the following manner:
1. `4`
2. `3`
3. `tl.Select([0,1,0,1])`
4. `add`
5. `tl.Select([0], n_in=2)`
6. `mul`
From the [documentation](https://trax-ml.readthedocs.io/en/latest/trax.layers.html#trax.layers.combinators.Select), you will see that `n_in` refers to the number of input elements to pop from the stack, and replace with those specified by the indices.
The following example is a bit contrived but it demonstrates the flexibility of the command. The second `tl.Select` pops two elements (specified in `n_in`) from the stack starting from index `[0]` (i.e. top of the stack) and replaces them with the element in index `[0]`. This means that `7` (index `[0]`) and `3` (index `[1]`) will be popped out (because `n_in = 2`) but only `7` is placed back on top of the stack because it only selects element at index `[0]` to replace the popped elements. As in the previous table each column shows the contents of the stack and the outputs after the operations are carried out.
<div style="text-align:center" width="20px"><img src="images/Stack3.png" /></div>
After processing all the inputs, the stack contains 28 which is the answer we get above.
```
serial = tl.Serial(
tl.Select([0, 1, 0, 1]), Addition(), tl.Select([0], n_in=2), Multiplication()
)
# Initialization
x = (np.array([3]), np.array([4])) # input
serial.init(shapes.signature(x)) # initializing serial instance
print("-- Serial Model --")
print(serial, "\n")
print("-- Properties --")
print("name :", serial.name)
print("sublayers :", serial.sublayers)
print("expected inputs :", serial.n_in)
print("promised outputs :", serial.n_out, "\n")
# Inputs
print("-- Inputs --")
print("x :", x, "\n")
# Outputs
y = serial(x)
print("-- Outputs --")
print("y :", y)
```
**In summary what select does in this example is a copy of the inputs in order to be used further along in the stack of operations.**
## 3. The tl.Residual combinator in the context of the serial combinator
### tl.Residual
[Residual networks](https://arxiv.org/pdf/1512.03385.pdf) are frequently used to make deep models easier to train by utilizing *skip connections*, or *shortcuts* to jump over some layers and you will be using it in the assignment as well. Trax already has a built in layer for this (`tl.Residual`). The [Residual layer](https://trax-ml.readthedocs.io/en/latest/trax.layers.html?highlight=residual#trax.layers.combinators.Residual) allows to create a *skip connection* so we can compute the element-wise *sum* of the *stack-top* input with the output of the layer series. Let's first see how it is used in the code below:
```
# Let's define a Serial network
serial = tl.Serial(
# Practice using Select again by duplicating the first two inputs
tl.Select([0, 1, 0, 1]),
# Place a Residual layer that skips over the Fn: Addition() layer
tl.Residual(Addition())
)
print("-- Serial Model --")
print(serial, "\n")
print("-- Properties --")
print("name :", serial.name)
print("expected inputs :", serial.n_in)
print("promised outputs :", serial.n_out, "\n")
```
Here, we use the Serial combinator to define our model. The inputs first goes through a `Select` layer, followed by a `Residual` layer which passes the `Fn: Addition()` layer as an argument. What this means is the `Residual` layer will take the stack top input at that point and add it to the output of the `Fn: Addition()` layer. You can picture it like the diagram the below, where `x1` and `x2` are the inputs to the model:
<img src="images/residual_example_add.png" width="400"/></div>
Now, let's try running our model with some sample inputs and see the result:
```
# Inputs
x1 = np.array([3])
x2 = np.array([4])
print("-- Inputs --")
print("(x1, x2) :", (x1, x2), "\n")
# Outputs
y = serial((x1, x2))
print("-- Outputs --")
print("y :", y)
```
As you can see, the `Residual` layer remembers the stack top input (i.e. `3`) and adds it to the result of the `Fn: Addition()` layer (i.e. `3 + 4 = 7`). The output of `Residual(Addition()` is then `3 + 7 = 10` and is pushed onto the stack.
On a different note, you'll notice that the `Select` layer has 4 outputs but the `Fn: Addition()` layer only pops 2 inputs from the stack. This means the duplicate inputs (i.e. the 2 rightmost arrows of the `Select` outputs in the figure above) remain in the stack. This is why you still see it in the output of our simple serial network (i.e. `array([3]), array([4])`). This is useful if you want to use these duplicate inputs in another layer further down the network.
### Modifying the network
To strengthen your understanding, you can modify the network above and examine the outputs you get. For example, you can pass the `Fn: Multiplication()` layer instead in the `Residual` block:
```
# model definition
serial = tl.Serial(
tl.Select([0, 1, 0, 1]),
tl.Residual(Multiplication())
)
print("-- Serial Model --")
print(serial, "\n")
print("-- Properties --")
print("name :", serial.name)
print("expected inputs :", serial.n_in)
print("promised outputs :", serial.n_out, "\n")
```
This means you'll have a different output that will be added to the stack top input saved by the Residual block. The diagram becomes like this:
<img src="images/residual_example_multiply.png" width="400"/></div>
```
# Inputs
x1 = np.array([3])
x2 = np.array([4])
print("-- Inputs --")
print("(x1, x2) :", (x1, x2), "\n")
# Outputs
y = serial((x1, x2))
print("-- Outputs --")
print("y :", y)
```
#### Congratulations! In this lab, we described how stack semantics work with Trax layers such as Select and Residual. You will be using these in the assignment and you can go back to this lab in case you want to review its usage.
|
github_jupyter
|
import numpy as np # regular ol' numpy
from trax import layers as tl # core building block
from trax import shapes # data signatures: dimensionality and type
from trax import fastmath # uses jax, offers numpy on steroids
def Addition():
layer_name = "Addition" # don't forget to give your custom layer a name to identify
# Custom function for the custom layer
def func(x, y):
return x + y
return tl.Fn(layer_name, func)
# Test it
add = Addition()
# Inspect properties
print("-- Properties --")
print("name :", add.name)
print("expected inputs :", add.n_in)
print("promised outputs :", add.n_out, "\n")
# Inputs
x = np.array([3])
y = np.array([4])
print("-- Inputs --")
print("x :", x, "\n")
print("y :", y, "\n")
# Outputs
z = add((x, y))
print("-- Outputs --")
print("z :", z)
def Multiplication():
layer_name = (
"Multiplication" # don't forget to give your custom layer a name to identify
)
# Custom function for the custom layer
def func(x, y):
return x * y
return tl.Fn(layer_name, func)
# Test it
mul = Multiplication()
# Inspect properties
print("-- Properties --")
print("name :", mul.name)
print("expected inputs :", mul.n_in)
print("promised outputs :", mul.n_out, "\n")
# Inputs
x = np.array([7])
y = np.array([15])
print("-- Inputs --")
print("x :", x, "\n")
print("y :", y, "\n")
# Outputs
z = mul((x, y))
print("-- Outputs --")
print("z :", z)
# Serial combinator
serial = tl.Serial(
Addition(), Multiplication(), Addition() # add 3 + 4 # multiply result by 15 and then add 3
)
# Initialization
x = (np.array([3]), np.array([4]), np.array([15]), np.array([3])) # input
serial.init(shapes.signature(x)) # initializing serial instance
print("-- Serial Model --")
print(serial, "\n")
print("-- Properties --")
print("name :", serial.name)
print("sublayers :", serial.sublayers)
print("expected inputs :", serial.n_in)
print("promised outputs :", serial.n_out, "\n")
# Inputs
print("-- Inputs --")
print("x :", x, "\n")
# Outputs
y = serial(x)
print("-- Outputs --")
print("y :", y)
serial = tl.Serial(tl.Select([0, 1, 0, 1]), Addition(), Multiplication(), Addition())
# Initialization
x = (np.array([3]), np.array([4])) # input
serial.init(shapes.signature(x)) # initializing serial instance
print("-- Serial Model --")
print(serial, "\n")
print("-- Properties --")
print("name :", serial.name)
print("sublayers :", serial.sublayers)
print("expected inputs :", serial.n_in)
print("promised outputs :", serial.n_out, "\n")
# Inputs
print("-- Inputs --")
print("x :", x, "\n")
# Outputs
y = serial(x)
print("-- Outputs --")
print("y :", y)
serial = tl.Serial(
tl.Select([0, 1, 0, 1]), Addition(), tl.Select([0], n_in=2), Multiplication()
)
# Initialization
x = (np.array([3]), np.array([4])) # input
serial.init(shapes.signature(x)) # initializing serial instance
print("-- Serial Model --")
print(serial, "\n")
print("-- Properties --")
print("name :", serial.name)
print("sublayers :", serial.sublayers)
print("expected inputs :", serial.n_in)
print("promised outputs :", serial.n_out, "\n")
# Inputs
print("-- Inputs --")
print("x :", x, "\n")
# Outputs
y = serial(x)
print("-- Outputs --")
print("y :", y)
# Let's define a Serial network
serial = tl.Serial(
# Practice using Select again by duplicating the first two inputs
tl.Select([0, 1, 0, 1]),
# Place a Residual layer that skips over the Fn: Addition() layer
tl.Residual(Addition())
)
print("-- Serial Model --")
print(serial, "\n")
print("-- Properties --")
print("name :", serial.name)
print("expected inputs :", serial.n_in)
print("promised outputs :", serial.n_out, "\n")
# Inputs
x1 = np.array([3])
x2 = np.array([4])
print("-- Inputs --")
print("(x1, x2) :", (x1, x2), "\n")
# Outputs
y = serial((x1, x2))
print("-- Outputs --")
print("y :", y)
# model definition
serial = tl.Serial(
tl.Select([0, 1, 0, 1]),
tl.Residual(Multiplication())
)
print("-- Serial Model --")
print(serial, "\n")
print("-- Properties --")
print("name :", serial.name)
print("expected inputs :", serial.n_in)
print("promised outputs :", serial.n_out, "\n")
# Inputs
x1 = np.array([3])
x2 = np.array([4])
print("-- Inputs --")
print("(x1, x2) :", (x1, x2), "\n")
# Outputs
y = serial((x1, x2))
print("-- Outputs --")
print("y :", y)
| 0.644673 | 0.982691 |
# Live Inference and Benchmark CT-scan Data with OpenVINO
## Kidney Segmentation with PyTorch Lightning and OpenVINO™ - Part 4
This tutorial is part of a series on how to train, optimize, quantize and show live inference on a medical segmentation model. The goal is to accelerate inference on a kidney segmentation model. The [UNet](https://arxiv.org/abs/1505.04597) model is trained from scratch; the data is from [Kits19](https://github.com/neheller/kits19).
This tutorial shows how to
- Visually compare inference results of an FP16 and INT8 OpenVINO IR model
- Benchmark performance of the original model and the quantized model
- Show live inference with OpenVINO's async API and MULTI plugin
To learn how this model was quantized, please see the [Convert and Quantize a UNet Model and Show Live Inference](../110-ct-segmentation-quantize/110-ct-segmentation-quantize.ipynb) tutorial. The content of the current tutorial partly overlaps with that. It demonstrates how to visualize the results and show benchmark information when you already have a quantized model.
All notebooks in this series:
- [Data Preparation for 2D Segmentation of 3D Medical Data](../110-ct-segmentation-quantize/data-preparation-ct-scan.ipynb)
- Train a 2D-UNet Medical Imaging Model with PyTorch Lightning (will be published soon)
- [Convert and Quantize a UNet Model and Show Live Inference](../110-ct-segmentation-quantize/110-ct-segmentation-quantize.ipynb)
- Live Inference and Benchmark CT-scan data (this notebook)
## Instructions
This notebook needs a quantized OpenVINO IR model. We provide a pretrained model trained for 20 epochs with the full [Kits-19](https://github.com/neheller/kits19) frames dataset, which has an F1 score on the validation set of 0.9. The training code will be made available soon. It also needs images from the Kits19 dataset, converted to 2D images. For demonstration purposes, this tutorial will download one converted CT scan to use for inference.
To install the requirements for running this notebook, please follow the instructions in the README.
## Imports
```
import glob
import os
import random
import sys
import time
import zipfile
from pathlib import Path
from typing import List
import cv2
import matplotlib.pyplot as plt
import numpy as np
from async_inference import CTAsyncPipeline, SegModel
from IPython.display import Image, display
from omz_python.models import model as omz_model
from openvino.inference_engine import IECore
sys.path.append("../utils")
from notebook_utils import benchmark_model, download_file
```
## Settings
To use the pretrained models, set `IR_PATH` to `"pretrained_model/unet44.xml"` and `COMPRESSED_MODEL_PATH` to `"pretrained_model/quantized_unet44.xml"`. To use a model that you trained or optimized yourself, adjust the model paths.
```
# Directory that contains the CT scan data. This directory should contain subdirectories
# case_00XXX where XXX is between 000 and 299
BASEDIR = "kits19_frames_1"
# The directory that contains the IR model files. Should contain unet44.xml and bin
# and quantized_unet44.xml and bin.
IR_PATH = "pretrained_model/unet44.xml"
COMPRESSED_MODEL_PATH = "pretrained_model/quantized_unet44.xml"
```
## Download and Prepare Data
Download one validation video for live inference. We reuse the KitsDataset class that was also used in the training and quantization notebook that will be released later.
Data is expected in `BASEDIR` defined in the cell above. `BASEDIR` should contain directories named `case_00000` to `case_00299`. If data for the case specified above does not exist yet, it will be downloaded and extracted in the next cell.
```
# The CT scan case number. For example: 16 for data from the case_00016 directory
# Currently only 16 is supported
case = 16
if not Path(f"{BASEDIR}/case_{case:05d}").exists():
filename = download_file(
f"https://s3.us-west-1.amazonaws.com/openvino.notebooks/case_{case:05d}.zip"
)
with zipfile.ZipFile(filename, "r") as zip_ref:
zip_ref.extractall(path=BASEDIR)
os.remove(filename) # remove zipfile
print(f"Downloaded and extracted data for case_{case:05d}")
else:
print(f"Data for case_{case:05d} exists")
class KitsDataset(object):
def __init__(self, basedir: str, dataset_type: str, transforms=None):
"""
Dataset class for prepared Kits19 data, for binary segmentation (background/kidney)
:param basedir: Directory that contains the prepared CT scans, in subdirectories
case_00000 until case_00210
:param dataset_type: either "train" or "val"
:param transforms: Compose object with augmentations
"""
allmasks = sorted(glob.glob(f"{basedir}/case_*/segmentation_frames/*png"))
if len(allmasks) == 0:
raise ValueError(
f"basedir: '{basedir}' does not contain data for type '{dataset_type}'"
)
self.valpatients = [11, 15, 16, 49, 50, 79, 81, 89, 106, 108, 112, 126, 129, 133,
141, 166, 169, 170, 192, 202, 204] # fmt: skip
valcases = [f"case_{i:05d}" for i in self.valpatients]
if dataset_type == "train":
masks = [mask for mask in allmasks if Path(mask).parents[1].name not in valcases]
elif dataset_type == "val":
masks = [mask for mask in allmasks if Path(mask).parents[1].name in valcases]
else:
raise ValueError("Please choose train or val dataset split")
if dataset_type == "train":
random.shuffle(masks)
self.basedir = basedir
self.dataset_type = dataset_type
self.dataset = masks
self.transforms = transforms
print(
f"Created {dataset_type} dataset with {len(self.dataset)} items. Base directory for data: {basedir}"
)
def __getitem__(self, index):
"""
Get an item from the dataset at the specified index.
:return: (annotation, input_image, metadata) where annotation is (index, segmentation_mask)
and metadata a dictionary with case and slice number
"""
mask_path = self.dataset[index]
# Open the image with OpenCV with `cv2.IMREAD_UNCHANGED` to prevent automatic
# conversion of 1-channel black and white images to 3-channel BGR images.
mask = cv2.imread(mask_path, cv2.IMREAD_UNCHANGED)
image_path = str(Path(mask_path.replace("segmentation", "imaging")).with_suffix(".jpg"))
img = cv2.imread(image_path, cv2.IMREAD_UNCHANGED)
if img.shape[:2] != (512, 512):
img = cv2.resize(img, (512, 512))
mask = cv2.resize(mask, (512, 512))
annotation = (index, mask.astype(np.uint8))
input_image = np.expand_dims(img, axis=0).astype(np.float32)
return (
annotation,
input_image,
{"case": Path(mask_path).parents[1].name, "slice": Path(mask_path).stem},
)
def __len__(self):
return len(self.dataset)
# The sigmoid function is used to transform the result of the network
# to binary segmentation masks
def sigmoid(x):
return np.exp(-np.logaddexp(0, -x))
# Create an instance of the KitsDataset class
# If you set dataset_type to train, make sure that `basedir` contains training data
dataset = KitsDataset(basedir=BASEDIR, dataset_type="val", transforms=None)
```
## Load Model
```
num_images = 4
colormap = "gray"
ie = IECore()
net_ir = ie.read_network(IR_PATH)
net_pot = ie.read_network(COMPRESSED_MODEL_PATH)
exec_net_ir = ie.load_network(network=net_ir, device_name="CPU")
exec_net_pot = ie.load_network(network=net_pot, device_name="CPU")
input_layer = next(iter(net_ir.input_info))
output_layer_ir = next(iter(net_ir.outputs))
output_layer_pot = next(iter(net_pot.outputs))
```
## Show Results
Visualize the results of the model on four slices of the validation set. Compare the results of the FP16 IR model with the results of the quantized INT8 model and the reference segmentation annotation.
Medical imaging datasets tend to be very imbalanced: most of the slices in a CT scan do not contain kidney data. The segmentation model should be good at finding kidneys where they exist (in medical terms: have good sensitivity) but also not find spurious kidneys that do not exist (have good specificity). In the next cell, we show four slices: two slices that have no kidney data, and two slices that contain kidney data. For this example, a slice has kidney data if at least 50 pixels in the slices are annotated as kidney.
Run this cell again to show results on a different subset. The random seed is displayed to allow reproducing specific runs of this cell.
> Note: the images are shown after optional augmenting and resizing. In the Kits19 dataset all but one of the cases has input shape `(512, 512)`.
```
# Create a dataset, and make a subset of the dataset for visualization
# The dataset items are (annotation, image) where annotation is (index, mask)
background_slices = (item for item in dataset if np.count_nonzero(item[0][1]) == 0)
kidney_slices = (item for item in dataset if np.count_nonzero(item[0][1]) > 50)
# Set seed to current time. To reproduce specific results, copy the printed seed
# and manually set `seed` to that value.
seed = int(time.time())
random.seed(seed)
print(f"Visualizing results with seed {seed}")
data_subset = random.sample(list(background_slices), 2) + random.sample(list(kidney_slices), 2)
fig, ax = plt.subplots(nrows=num_images, ncols=4, figsize=(24, num_images * 4))
for i, (annotation, image, meta) in enumerate(data_subset):
mask = annotation[1]
res_ir = exec_net_ir.infer(inputs={input_layer: image})
res_pot = exec_net_pot.infer(inputs={input_layer: image})
target_mask = mask.astype(np.uint8)
result_mask_ir = sigmoid(res_ir[output_layer_ir]).round().astype(np.uint8)[0, 0, ::]
result_mask_pot = sigmoid(res_pot[output_layer_pot]).round().astype(np.uint8)[0, 0, ::]
ax[i, 0].imshow(image[0, ::], cmap=colormap)
ax[i, 1].imshow(target_mask, cmap=colormap)
ax[i, 2].imshow(result_mask_ir, cmap=colormap)
ax[i, 3].imshow(result_mask_pot, cmap=colormap)
ax[i, 0].set_title(f"{meta['slice']}")
ax[i, 1].set_title("Annotation")
ax[i, 2].set_title("Prediction on FP16 model")
ax[i, 3].set_title("Prediction on INT8 model")
```
### Compare Performance of the Original and Quantized Models
To measure the inference performance of the FP16 and INT8 models, we use [Benchmark Tool](https://docs.openvinotoolkit.org/latest/openvino_inference_engine_tools_benchmark_tool_README.html), OpenVINO's inference performance measurement tool. Benchmark tool is a command line application that can be run in the notebook with `! benchmark_app` or `%sx benchmark_app`.
In this tutorial, we use a wrapper function from [Notebook Utils](https://github.com/openvinotoolkit/openvino_notebooks/blob/main/notebooks/utils/notebook_utils.ipynb). It prints the `benchmark_app` command with the chosen parameters.
> NOTE: For the most accurate performance estimation, we recommended running `benchmark_app` in a terminal/command prompt after closing other applications. Run `benchmark_app --help` to see all command line options.
```
# By default, benchmark on MULTI:CPU,GPU if a GPU is available, otherwise on CPU.
device = "MULTI:CPU,GPU" if "GPU" in ie.available_devices else "CPU"
# Uncomment one of the options below to benchmark on other devices
# device = "GPU"
# device = "CPU"
# device = "AUTO"
# Benchmark FP16 model
benchmark_model(model_path=IR_PATH, device=device, seconds=15)
# Benchmark INT8 model
benchmark_model(model_path=COMPRESSED_MODEL_PATH, device=device, seconds=15)
```
## Show Live Inference
To show live inference on the model in the notebook, we use the asynchronous processing feature of OpenVINO Inference Engine.
If you use a GPU device, with `device="GPU"` or `device="MULTI:CPU,GPU"` to do inference on an integrated graphics card, model loading will be slow the first time you run this code. The model will be cached, so after the first time model loading will be fast. See the [OpenVINO API tutorial](../002-openvino-api/002-openvino-api.ipynb) for more information on Inference Engine, including Model Caching.
#### Visualization Functions
We define a helper function `show_array` to efficiently show images in the notebook. The `do_inference` function uses [Open Model Zoo](https://github.com/openvinotoolkit/open_model_zoo/)'s AsyncPipeline to perform asynchronous inference. After inference on the specified CT scan has completed, the total time and throughput (fps), including preprocessing and displaying, will be printed.
```
def showarray(frame: np.ndarray, display_handle=None):
"""
Display array `frame`. Replace information at `display_handle` with `frame`
encoded as jpeg image
Create a display_handle with: `display_handle = display(display_id=True)`
"""
_, frame = cv2.imencode(ext=".jpeg", img=frame)
if display_handle is None:
display_handle = display(Image(data=frame.tobytes()), display_id=True)
else:
display_handle.update(Image(data=frame.tobytes()))
return display_handle
def do_inference(imagelist: List, model: omz_model.Model, device: str):
"""
Do inference of images in `imagelist` on `model` on the given `device` and show
the results in real time in a Jupyter Notebook
:param imagelist: list of images/frames to do inference on
:param model: Model instance for inference
:param device: Name of device to perform inference on. For example: "CPU"
"""
display_handle = None
next_frame_id = 0
next_frame_id_to_show = 0
input_layer = next(iter(model.net.input_info))
# Create asynchronous pipeline and print time it takes to load the model
load_start_time = time.perf_counter()
pipeline = CTAsyncPipeline(
ie=ie, model=model, plugin_config={}, device=device, max_num_requests=0
)
load_end_time = time.perf_counter()
# Perform asynchronous inference
start_time = time.perf_counter()
while next_frame_id < len(imagelist) - 1:
results = pipeline.get_result(next_frame_id_to_show)
if results:
# Show next result from async pipeline
result, meta = results
display_handle = showarray(result, display_handle)
next_frame_id_to_show += 1
if pipeline.is_ready():
# Submit new image to async pipeline
image = imagelist[next_frame_id]
pipeline.submit_data(
inputs={input_layer: image}, id=next_frame_id, meta={"frame": image}
)
next_frame_id += 1
else:
# If the pipeline is not ready yet and there are no results: wait
pipeline.await_any()
pipeline.await_all()
# Show all frames that are in the pipeline after all images have been submitted
while pipeline.has_completed_request():
results = pipeline.get_result(next_frame_id_to_show)
if results:
result, meta = results
display_handle = showarray(result, display_handle)
next_frame_id_to_show += 1
end_time = time.perf_counter()
duration = end_time - start_time
fps = len(imagelist) / duration
print(f"Loaded model to {device} in {load_end_time-load_start_time:.2f} seconds.")
print(f"Total time for {next_frame_id+1} frames: {duration:.2f} seconds, fps:{fps:.2f}")
```
#### Load Model and Images
Load the segmentation model with `SegModel`, based on the [Open Model Zoo](https://github.com/openvinotoolkit/open_model_zoo/) Model API. Load a CT scan from the `BASEDIR` directory (by default: _kits19_frames_) to a list.
```
ie = IECore()
segmentation_model = SegModel(ie=ie, model_path=Path(COMPRESSED_MODEL_PATH))
case = 16
demopattern = f"{BASEDIR}/case_{case:05d}/imaging_frames/*jpg"
imlist = sorted(glob.glob(demopattern))
images = [cv2.imread(im, cv2.IMREAD_UNCHANGED) for im in imlist]
```
#### Show Inference
In the next cell, we run the `do inference` function, which loads the model to the specified device (using caching for faster model loading on GPU devices), performs inference, and displays the results in real-time.
```
# Possible options for device include "CPU", "GPU", "AUTO", "MULTI"
device = "MULTI:CPU,GPU" if "GPU" in ie.available_devices else "CPU"
do_inference(imagelist=images, model=segmentation_model, device=device)
```
|
github_jupyter
|
import glob
import os
import random
import sys
import time
import zipfile
from pathlib import Path
from typing import List
import cv2
import matplotlib.pyplot as plt
import numpy as np
from async_inference import CTAsyncPipeline, SegModel
from IPython.display import Image, display
from omz_python.models import model as omz_model
from openvino.inference_engine import IECore
sys.path.append("../utils")
from notebook_utils import benchmark_model, download_file
# Directory that contains the CT scan data. This directory should contain subdirectories
# case_00XXX where XXX is between 000 and 299
BASEDIR = "kits19_frames_1"
# The directory that contains the IR model files. Should contain unet44.xml and bin
# and quantized_unet44.xml and bin.
IR_PATH = "pretrained_model/unet44.xml"
COMPRESSED_MODEL_PATH = "pretrained_model/quantized_unet44.xml"
# The CT scan case number. For example: 16 for data from the case_00016 directory
# Currently only 16 is supported
case = 16
if not Path(f"{BASEDIR}/case_{case:05d}").exists():
filename = download_file(
f"https://s3.us-west-1.amazonaws.com/openvino.notebooks/case_{case:05d}.zip"
)
with zipfile.ZipFile(filename, "r") as zip_ref:
zip_ref.extractall(path=BASEDIR)
os.remove(filename) # remove zipfile
print(f"Downloaded and extracted data for case_{case:05d}")
else:
print(f"Data for case_{case:05d} exists")
class KitsDataset(object):
def __init__(self, basedir: str, dataset_type: str, transforms=None):
"""
Dataset class for prepared Kits19 data, for binary segmentation (background/kidney)
:param basedir: Directory that contains the prepared CT scans, in subdirectories
case_00000 until case_00210
:param dataset_type: either "train" or "val"
:param transforms: Compose object with augmentations
"""
allmasks = sorted(glob.glob(f"{basedir}/case_*/segmentation_frames/*png"))
if len(allmasks) == 0:
raise ValueError(
f"basedir: '{basedir}' does not contain data for type '{dataset_type}'"
)
self.valpatients = [11, 15, 16, 49, 50, 79, 81, 89, 106, 108, 112, 126, 129, 133,
141, 166, 169, 170, 192, 202, 204] # fmt: skip
valcases = [f"case_{i:05d}" for i in self.valpatients]
if dataset_type == "train":
masks = [mask for mask in allmasks if Path(mask).parents[1].name not in valcases]
elif dataset_type == "val":
masks = [mask for mask in allmasks if Path(mask).parents[1].name in valcases]
else:
raise ValueError("Please choose train or val dataset split")
if dataset_type == "train":
random.shuffle(masks)
self.basedir = basedir
self.dataset_type = dataset_type
self.dataset = masks
self.transforms = transforms
print(
f"Created {dataset_type} dataset with {len(self.dataset)} items. Base directory for data: {basedir}"
)
def __getitem__(self, index):
"""
Get an item from the dataset at the specified index.
:return: (annotation, input_image, metadata) where annotation is (index, segmentation_mask)
and metadata a dictionary with case and slice number
"""
mask_path = self.dataset[index]
# Open the image with OpenCV with `cv2.IMREAD_UNCHANGED` to prevent automatic
# conversion of 1-channel black and white images to 3-channel BGR images.
mask = cv2.imread(mask_path, cv2.IMREAD_UNCHANGED)
image_path = str(Path(mask_path.replace("segmentation", "imaging")).with_suffix(".jpg"))
img = cv2.imread(image_path, cv2.IMREAD_UNCHANGED)
if img.shape[:2] != (512, 512):
img = cv2.resize(img, (512, 512))
mask = cv2.resize(mask, (512, 512))
annotation = (index, mask.astype(np.uint8))
input_image = np.expand_dims(img, axis=0).astype(np.float32)
return (
annotation,
input_image,
{"case": Path(mask_path).parents[1].name, "slice": Path(mask_path).stem},
)
def __len__(self):
return len(self.dataset)
# The sigmoid function is used to transform the result of the network
# to binary segmentation masks
def sigmoid(x):
return np.exp(-np.logaddexp(0, -x))
# Create an instance of the KitsDataset class
# If you set dataset_type to train, make sure that `basedir` contains training data
dataset = KitsDataset(basedir=BASEDIR, dataset_type="val", transforms=None)
num_images = 4
colormap = "gray"
ie = IECore()
net_ir = ie.read_network(IR_PATH)
net_pot = ie.read_network(COMPRESSED_MODEL_PATH)
exec_net_ir = ie.load_network(network=net_ir, device_name="CPU")
exec_net_pot = ie.load_network(network=net_pot, device_name="CPU")
input_layer = next(iter(net_ir.input_info))
output_layer_ir = next(iter(net_ir.outputs))
output_layer_pot = next(iter(net_pot.outputs))
# Create a dataset, and make a subset of the dataset for visualization
# The dataset items are (annotation, image) where annotation is (index, mask)
background_slices = (item for item in dataset if np.count_nonzero(item[0][1]) == 0)
kidney_slices = (item for item in dataset if np.count_nonzero(item[0][1]) > 50)
# Set seed to current time. To reproduce specific results, copy the printed seed
# and manually set `seed` to that value.
seed = int(time.time())
random.seed(seed)
print(f"Visualizing results with seed {seed}")
data_subset = random.sample(list(background_slices), 2) + random.sample(list(kidney_slices), 2)
fig, ax = plt.subplots(nrows=num_images, ncols=4, figsize=(24, num_images * 4))
for i, (annotation, image, meta) in enumerate(data_subset):
mask = annotation[1]
res_ir = exec_net_ir.infer(inputs={input_layer: image})
res_pot = exec_net_pot.infer(inputs={input_layer: image})
target_mask = mask.astype(np.uint8)
result_mask_ir = sigmoid(res_ir[output_layer_ir]).round().astype(np.uint8)[0, 0, ::]
result_mask_pot = sigmoid(res_pot[output_layer_pot]).round().astype(np.uint8)[0, 0, ::]
ax[i, 0].imshow(image[0, ::], cmap=colormap)
ax[i, 1].imshow(target_mask, cmap=colormap)
ax[i, 2].imshow(result_mask_ir, cmap=colormap)
ax[i, 3].imshow(result_mask_pot, cmap=colormap)
ax[i, 0].set_title(f"{meta['slice']}")
ax[i, 1].set_title("Annotation")
ax[i, 2].set_title("Prediction on FP16 model")
ax[i, 3].set_title("Prediction on INT8 model")
# By default, benchmark on MULTI:CPU,GPU if a GPU is available, otherwise on CPU.
device = "MULTI:CPU,GPU" if "GPU" in ie.available_devices else "CPU"
# Uncomment one of the options below to benchmark on other devices
# device = "GPU"
# device = "CPU"
# device = "AUTO"
# Benchmark FP16 model
benchmark_model(model_path=IR_PATH, device=device, seconds=15)
# Benchmark INT8 model
benchmark_model(model_path=COMPRESSED_MODEL_PATH, device=device, seconds=15)
def showarray(frame: np.ndarray, display_handle=None):
"""
Display array `frame`. Replace information at `display_handle` with `frame`
encoded as jpeg image
Create a display_handle with: `display_handle = display(display_id=True)`
"""
_, frame = cv2.imencode(ext=".jpeg", img=frame)
if display_handle is None:
display_handle = display(Image(data=frame.tobytes()), display_id=True)
else:
display_handle.update(Image(data=frame.tobytes()))
return display_handle
def do_inference(imagelist: List, model: omz_model.Model, device: str):
"""
Do inference of images in `imagelist` on `model` on the given `device` and show
the results in real time in a Jupyter Notebook
:param imagelist: list of images/frames to do inference on
:param model: Model instance for inference
:param device: Name of device to perform inference on. For example: "CPU"
"""
display_handle = None
next_frame_id = 0
next_frame_id_to_show = 0
input_layer = next(iter(model.net.input_info))
# Create asynchronous pipeline and print time it takes to load the model
load_start_time = time.perf_counter()
pipeline = CTAsyncPipeline(
ie=ie, model=model, plugin_config={}, device=device, max_num_requests=0
)
load_end_time = time.perf_counter()
# Perform asynchronous inference
start_time = time.perf_counter()
while next_frame_id < len(imagelist) - 1:
results = pipeline.get_result(next_frame_id_to_show)
if results:
# Show next result from async pipeline
result, meta = results
display_handle = showarray(result, display_handle)
next_frame_id_to_show += 1
if pipeline.is_ready():
# Submit new image to async pipeline
image = imagelist[next_frame_id]
pipeline.submit_data(
inputs={input_layer: image}, id=next_frame_id, meta={"frame": image}
)
next_frame_id += 1
else:
# If the pipeline is not ready yet and there are no results: wait
pipeline.await_any()
pipeline.await_all()
# Show all frames that are in the pipeline after all images have been submitted
while pipeline.has_completed_request():
results = pipeline.get_result(next_frame_id_to_show)
if results:
result, meta = results
display_handle = showarray(result, display_handle)
next_frame_id_to_show += 1
end_time = time.perf_counter()
duration = end_time - start_time
fps = len(imagelist) / duration
print(f"Loaded model to {device} in {load_end_time-load_start_time:.2f} seconds.")
print(f"Total time for {next_frame_id+1} frames: {duration:.2f} seconds, fps:{fps:.2f}")
ie = IECore()
segmentation_model = SegModel(ie=ie, model_path=Path(COMPRESSED_MODEL_PATH))
case = 16
demopattern = f"{BASEDIR}/case_{case:05d}/imaging_frames/*jpg"
imlist = sorted(glob.glob(demopattern))
images = [cv2.imread(im, cv2.IMREAD_UNCHANGED) for im in imlist]
# Possible options for device include "CPU", "GPU", "AUTO", "MULTI"
device = "MULTI:CPU,GPU" if "GPU" in ie.available_devices else "CPU"
do_inference(imagelist=images, model=segmentation_model, device=device)
| 0.588298 | 0.983738 |
# Beginning Programming in Python
### Dictionaries/Sets
#### CSE20 - Spring 2021
Interactive Slides: [https://tinyurl.com/cse20-spr21-dictionaries-sets](https://tinyurl.com/cse20-spr21-dictionaries-sets)
# Dictionaries
- A mapping type that defines **key:value** relationships between values
- A **key** is a unique value that is associated with its respective **value**
- All of the **key**s in a dictionary have to be unique and can be any hashable type. **Value**s do not have to be unique.
- **key**s can be any type and a single dictionary can use multiple types as keys
- Dictionaries are a mutable type
- No particular naming convention for variables, just choose informative variable names
# Dictionaries: Instantiation (Creation)
- There are a few ways to instantiate a dictionary. The way we'll go over today is using curly braces `{key:value}`
```
nums_to_strings = {1:"one", 2:"two", 3:"three"}
strings_to_nums = {
"one":1,
"two":2,
"three":3
}
```
# Dictionaries: Instantiation (Creation)
```
mixed_types_dict = {
1: "one",
"two": 2.0,
"another_dict": {"three": 3},
"a list": [1, 2, 3]
}
print(mixed_types_dict)
```
# Dictionaries: Access
- To access/retrieve the **value**s stored in a dictionary we use square brackets with the **key** `[key]`
- We can only access a single value at a time using a single key
- Dictionaries are considered **unordered** so the keys are not guaranteed to be in the order that they were added in.
# Dictionaries: Access
```
nums_to_strings = {1:"one", 2:"two", 3:"three"}
strings_to_nums = {
"one":1,
"two":2,
"three":3
}
print(nums_to_strings[2])
print(strings_to_nums["two"])
```
# Dictionaries: Access
- If you try to use a key doesn't exists in the dictionary it will raise a `KeyError`
- If your not sure if a key is in a dictionary you can use the membership operators `in` and `not in` to check for the key
```
nums_to_strings = {1:"one"}
print(2 in nums_to_strings)
print(nums_to_strings[1])
```
# Dictionaries: Updates
- To update/add **value**s stored in a dictionary we use square brackets with the **key** `[key]`
```
nums_to_strings = {1:"one"}
print(nums_to_strings)
nums_to_strings[1] = "ONE"
nums_to_strings["2"] = "TWO"
print(nums_to_strings)
```
# `dict`: Methods
- Dictionaries are considered `object`s, we'll go over `object`s in more detail when we go over Object Oriented Programming (OOP).
- For now you need to know that objects can have functions called `methods`, which can be "called" by using the `dict_variable.method_name()` notation.
# `dict` Methods: `clear()`
- `clear()`removes all items from the dictionary
```
nums_to_strings = {1:"one"}
print(nums_to_strings)
nums_to_strings.clear()
print(nums_to_strings)
```
# `dict` Methods: `get()`
- `get()` works just like using square brackets `[]`
```
nums_to_strings = {1:"one", 2:"two"}
print(nums_to_strings.get(1))
print(nums_to_strings[1])
```
# `dict` Methods: `items()`
- Returns a "view" of the key:value pairs in the dictionary.
- Note these views are dynamic and change as the dictionary changes
```
nums_to_strings = {1:"one", 2:"two"}
items = nums_to_strings.items()
print(nums_to_strings, items)
nums_to_strings[3] = "three"
print(nums_to_strings, items)
```
# `dict` Methods: `keys()`
- Returns a "view" of the keys in the dictionary.
- Note these views are dynamic and change as the dictionary changes
```
nums_to_strings = {1:"one", 2:"two"}
keys = nums_to_strings.keys()
print(nums_to_strings, keys)
nums_to_strings[3] = "three"
print(nums_to_strings, keys)
```
# `dict` Methods: `values()`
- Returns a "view" of the keys in the dictionary.
- Note these views are dynamic and change as the dictionary changes
```
nums_to_strings = {1:"one", 2:"two"}
vals = nums_to_strings.values()
print(nums_to_strings, vals)
nums_to_strings[3] = "three"
print(nums_to_strings, vals)
```
# `dict` Methods: `pop()`
- Returns the value at the given key while removing the key
```
nums_to_strings = {1:"one", 2:"two"}
val_at_1 = nums_to_strings.pop(1)
print(nums_to_strings, val_at_1)
```
# `dict` Methods: `popitem()`
- Returns the key/value:
- Last In First Out (LIFO) order. (Python 3.7+)
- Arbitrary order (Python < 3.7)
```
nums_to_strings = {1:"one", 2:"two", 3:"three"}
val_at_1 = nums_to_strings.popitem()
print(nums_to_strings)
print(val_at_1)
```
# `dict` Methods: `setdefault()`
- If the key argument that is passed to the method exists in the dictionary, then it is returned. If it not in the dictionary, then an optional default argument can be used. If no default argument is provided, the it is set to `None`
# Quick aside: `None` and `pass`
- `None` is a special type in python that is intended to represent the absence information.
- It usually means, "Not Anything" or "null"
- `pass` is a placeholder statement in Python that doesn't perform any actions but can be used to ensure no syntax errors, i.e:
```python
if [boolean]:
pass
else:
pass
```
# `dict` Methods: `setdefault()`
- If the key argument that is passed to the method exists in the dictionary, then it is returned. If it not in the dictionary, then an optional default argument can be used. If no default argument is provided, the it is set to `None`
```
nums_to_strings = {1:"one", 2:"two"}
val_at_2 = nums_to_strings.setdefault(2, "three")
print(nums_to_strings, val_at_2)
val_at_3 = nums_to_strings.setdefault(3, "three")
print(nums_to_strings, val_at_3)
```
# `dict` Methods: `update()`
- Merges the dictionary that the method is being called on with the one that is passed as an argument in the method
```
nums_to_strings = {1:"one", 2:"two"}
print(nums_to_strings)
nums_to_strings_three = {3:"three"}
nums_to_strings.update(nums_to_strings_three)
print(nums_to_strings)
```
# Built-in Functions That are Compatible With `dict`s
- `len()` returns the number of items in a `dict`
- `list()` returns the keys in the dictionary as a list
```
nums_to_strings = {1:"one", 2:"two"}
print(nums_to_strings)
print("There are", len(nums_to_strings), "items in the dictionary")
print("The following keys are in the dictionary:", list(nums_to_strings))
```
# Sets
- `set`s are an **unordered** collection containing **no** duplicates
- They are functionally similar to mathematical sets. We can perform common set operations like:
- membership
- subsets
- supersets
- intersections
- etc ...
# `set`: instantiation (creation)
- Sets can be created a few ways. Today we'll go over two ways: curly braces `{}` and the type constructor `set()`
```
a_set = {1, 2, 5}
another_set = set([1, 2, 5])
print(a_set, another_set)
```
# Common `set` operations: Membership
- You can use the membership operators `in` and `not in` to see if a value is a member of the set
```
a_set = {1, 2, 5}
val = 2
print(val in a_set)
```
# Common `set` operations: isdisjoint()
- The `isdisjoint()` methods returns `True` if the two sets don't have any elements in common
```
a_set = {1, 2, 3}
another_set = {4, 5, 6}
a_set.isdisjoint(another_set)
```
# Common `set` operations: subset
- `issubset()` and `<=` will test if all of the elements in the set on the left side are in the set on the right side.
- `<` will test if the set on the left is a proper subset.
```
a_set = {1, 2, 3}
another_set = {1, 2, 3}
print(a_set.issubset(another_set))
print(a_set <= another_set)
print(a_set < another_set)
```
# Common `set` operations: superset
- `issuperset()` and `>=` will test if all of the elements in the set on the right side are in the set on the left side.
- `>` will test if the set on the left is a proper superset.
```
a_set = {1, 2, 3, 4}
another_set = {1, 2, 3}
print(a_set.issuperset(another_set))
print(a_set >= another_set)
print(a_set > another_set)
```
# Common `set` operations: union and intersection
- `union()` and `|` will return the union of the set on the left and the set on the right.
- `intersection()` and `&` will return the intersection of the set on the left and on the right
```
a_set = {1, 2, 3}
another_set = {2, 3, 4}
print(a_set | another_set)
print(a_set & another_set)
```
# Common `set` operations: difference and symmetric difference
- `difference()` and `-` will return a new set with the elements in the set on the left that are not in the set on the right
- `symmetric_difference()` and `^` will return a new set containing the elements that both sets don't have in common
```
a_set = {1, 2, 3}
another_set = {2, 3, 4}
print(a_set - another_set)
print(a_set ^ another_set)
```
# `set` Methods: `add()`
- `add()` adds the given element to the set
```
a_set = {1, 2, 3}
a_set.add("4")
a_set.add(5.0)
a_set.add(2)
print(a_set)
```
# `set` Methods: `remove()` & `discard()`
- `remove()` removes the given element from the set and raises an error if the element isn't in the set
- `discard()` removes the given element from the set and **doesn't** raise an error if the element isn't in the set
```
a_set = {1, 2, 3, 5}
a_set.remove(2)
a_set.discard(4)
print(a_set)
```
# `set` Methods: `pop()` & `clear()`
- `pop()` removes and returns an arbitrary element from the set
- `clear()` removes all elements from the set
```
a_set = {1, 2, 3}
print(a_set.pop())
print(a_set)
a_set.clear()
print(a_set)
```
# Built-in Functions That are Compatible With `set`s
- `len()` returns the number of items in a `set`
```
a_set = {1, 2, 3}
print(a_set)
print("There are", len(a_set), "elements in the set")
```
# What's Due Next?
- zybooks Chapter 3 due April 18th 11:59 PM
- Assignment 2 due April 25th 11:59 PM
|
github_jupyter
|
nums_to_strings = {1:"one", 2:"two", 3:"three"}
strings_to_nums = {
"one":1,
"two":2,
"three":3
}
mixed_types_dict = {
1: "one",
"two": 2.0,
"another_dict": {"three": 3},
"a list": [1, 2, 3]
}
print(mixed_types_dict)
nums_to_strings = {1:"one", 2:"two", 3:"three"}
strings_to_nums = {
"one":1,
"two":2,
"three":3
}
print(nums_to_strings[2])
print(strings_to_nums["two"])
nums_to_strings = {1:"one"}
print(2 in nums_to_strings)
print(nums_to_strings[1])
nums_to_strings = {1:"one"}
print(nums_to_strings)
nums_to_strings[1] = "ONE"
nums_to_strings["2"] = "TWO"
print(nums_to_strings)
nums_to_strings = {1:"one"}
print(nums_to_strings)
nums_to_strings.clear()
print(nums_to_strings)
nums_to_strings = {1:"one", 2:"two"}
print(nums_to_strings.get(1))
print(nums_to_strings[1])
nums_to_strings = {1:"one", 2:"two"}
items = nums_to_strings.items()
print(nums_to_strings, items)
nums_to_strings[3] = "three"
print(nums_to_strings, items)
nums_to_strings = {1:"one", 2:"two"}
keys = nums_to_strings.keys()
print(nums_to_strings, keys)
nums_to_strings[3] = "three"
print(nums_to_strings, keys)
nums_to_strings = {1:"one", 2:"two"}
vals = nums_to_strings.values()
print(nums_to_strings, vals)
nums_to_strings[3] = "three"
print(nums_to_strings, vals)
nums_to_strings = {1:"one", 2:"two"}
val_at_1 = nums_to_strings.pop(1)
print(nums_to_strings, val_at_1)
nums_to_strings = {1:"one", 2:"two", 3:"three"}
val_at_1 = nums_to_strings.popitem()
print(nums_to_strings)
print(val_at_1)
if [boolean]:
pass
else:
pass
nums_to_strings = {1:"one", 2:"two"}
val_at_2 = nums_to_strings.setdefault(2, "three")
print(nums_to_strings, val_at_2)
val_at_3 = nums_to_strings.setdefault(3, "three")
print(nums_to_strings, val_at_3)
nums_to_strings = {1:"one", 2:"two"}
print(nums_to_strings)
nums_to_strings_three = {3:"three"}
nums_to_strings.update(nums_to_strings_three)
print(nums_to_strings)
nums_to_strings = {1:"one", 2:"two"}
print(nums_to_strings)
print("There are", len(nums_to_strings), "items in the dictionary")
print("The following keys are in the dictionary:", list(nums_to_strings))
a_set = {1, 2, 5}
another_set = set([1, 2, 5])
print(a_set, another_set)
a_set = {1, 2, 5}
val = 2
print(val in a_set)
a_set = {1, 2, 3}
another_set = {4, 5, 6}
a_set.isdisjoint(another_set)
a_set = {1, 2, 3}
another_set = {1, 2, 3}
print(a_set.issubset(another_set))
print(a_set <= another_set)
print(a_set < another_set)
a_set = {1, 2, 3, 4}
another_set = {1, 2, 3}
print(a_set.issuperset(another_set))
print(a_set >= another_set)
print(a_set > another_set)
a_set = {1, 2, 3}
another_set = {2, 3, 4}
print(a_set | another_set)
print(a_set & another_set)
a_set = {1, 2, 3}
another_set = {2, 3, 4}
print(a_set - another_set)
print(a_set ^ another_set)
a_set = {1, 2, 3}
a_set.add("4")
a_set.add(5.0)
a_set.add(2)
print(a_set)
a_set = {1, 2, 3, 5}
a_set.remove(2)
a_set.discard(4)
print(a_set)
a_set = {1, 2, 3}
print(a_set.pop())
print(a_set)
a_set.clear()
print(a_set)
a_set = {1, 2, 3}
print(a_set)
print("There are", len(a_set), "elements in the set")
| 0.192957 | 0.987711 |
# Developing an AI application
Going forward, AI algorithms will be incorporated into more and more everyday applications. For example, you might want to include an image classifier in a smart phone app. To do this, you'd use a deep learning model trained on hundreds of thousands of images as part of the overall application architecture. A large part of software development in the future will be using these types of models as common parts of applications.
In this project, you'll train an image classifier to recognize different species of flowers. You can imagine using something like this in a phone app that tells you the name of the flower your camera is looking at. In practice you'd train this classifier, then export it for use in your application. We'll be using [this dataset](http://www.robots.ox.ac.uk/~vgg/data/flowers/102/index.html) of 102 flower categories, you can see a few examples below.
<img src='assets/Flowers.png' width=500px>
The project is broken down into multiple steps:
* Load and preprocess the image dataset
* Train the image classifier on your dataset
* Use the trained classifier to predict image content
We'll lead you through each part which you'll implement in Python.
When you've completed this project, you'll have an application that can be trained on any set of labeled images. Here your network will be learning about flowers and end up as a command line application. But, what you do with your new skills depends on your imagination and effort in building a dataset. For example, imagine an app where you take a picture of a car, it tells you what the make and model is, then looks up information about it. Go build your own dataset and make something new.
First up is importing the packages you'll need. It's good practice to keep all the imports at the beginning of your code. As you work through this notebook and find you need to import a package, make sure to add the import up here.
```
# Imports here
%matplotlib inline
%config InlineBackend.figure_format = 'retina'
import numpy as np
import matplotlib.pyplot as plt
import torch
from torch import nn, optim
import torch.nn.functional as F
from collections import OrderedDict
from torchvision import transforms, models, datasets
from PIL import Image
from os import listdir
import random
import json
```
## Load the data
Here you'll use `torchvision` to load the data ([documentation](http://pytorch.org/docs/0.3.0/torchvision/index.html)). The data should be included alongside this notebook, otherwise you can [download it here](https://s3.amazonaws.com/content.udacity-data.com/nd089/flower_data.tar.gz). The dataset is split into three parts, training, validation, and testing. For the training, you'll want to apply transformations such as random scaling, cropping, and flipping. This will help the network generalize leading to better performance. You'll also need to make sure the input data is resized to 224x224 pixels as required by the pre-trained networks.
The validation and testing sets are used to measure the model's performance on data it hasn't seen yet. For this you don't want any scaling or rotation transformations, but you'll need to resize then crop the images to the appropriate size.
The pre-trained networks you'll use were trained on the ImageNet dataset where each color channel was normalized separately. For all three sets you'll need to normalize the means and standard deviations of the images to what the network expects. For the means, it's `[0.485, 0.456, 0.406]` and for the standard deviations `[0.229, 0.224, 0.225]`, calculated from the ImageNet images. These values will shift each color channel to be centered at 0 and range from -1 to 1.
```
data_dir = 'flowers'
train_dir = data_dir + '/train'
valid_dir = data_dir + '/valid'
test_dir = data_dir + '/test'
# TODO: Define your transforms for the training, validation, and testing sets
data_transforms = transforms.Compose([transforms.ToTensor(),
transforms.Normalize([0.485, 0.456, 0.406], [0.229, 0.224, 0.225])
])
train_transforms = transforms.Compose([transforms.Resize(225),
transforms.RandomCrop(224),
transforms.RandomHorizontalFlip(),
transforms.ToTensor(),
transforms.Normalize([0.485, 0.456, 0.406], [0.229, 0.224, 0.225])
])
valid_transforms = transforms.Compose([transforms.Resize(225),
transforms.RandomCrop(224),
transforms.RandomHorizontalFlip(),
transforms.ToTensor(),
transforms.Normalize([0.485, 0.456, 0.406], [0.229, 0.224, 0.225])
])
test_transforms = transforms.Compose([transforms.Resize(225),
transforms.RandomCrop(224),
transforms.RandomHorizontalFlip(),
transforms.ToTensor(),
transforms.Normalize([0.485, 0.456, 0.406], [0.229, 0.224, 0.225])
])
# TODO: Load the datasets with ImageFolder
data = datasets.ImageFolder(data_dir, transform = data_transforms)
train_data = datasets.ImageFolder(train_dir, transform = train_transforms)
valid_data = datasets.ImageFolder(valid_dir, transform = valid_transforms)
test_data = datasets.ImageFolder(test_dir, transform = test_transforms)
# TODO: Using the image datasets and the trainforms, define the dataloaders
dataloader = torch.utils.data.DataLoader(data, batch_size = 32, shuffle = True)
trainloader = torch.utils.data.DataLoader(train_data, batch_size = 32, shuffle = True)
validloader = torch.utils.data.DataLoader(valid_data, batch_size = 32, shuffle = True)
testloader = torch.utils.data.DataLoader(test_data, batch_size = 32, shuffle = True)
```
### Label mapping
You'll also need to load in a mapping from category label to category name. You can find this in the file `cat_to_name.json`. It's a JSON object which you can read in with the [`json` module](https://docs.python.org/2/library/json.html). This will give you a dictionary mapping the integer encoded categories to the actual names of the flowers.
```
with open('cat_to_name.json', 'r') as f:
cat_to_name = json.load(f)
```
# Building and training the classifier
Now that the data is ready, it's time to build and train the classifier. As usual, you should use one of the pretrained models from `torchvision.models` to get the image features. Build and train a new feed-forward classifier using those features.
We're going to leave this part up to you. Refer to [the rubric](https://review.udacity.com/#!/rubrics/1663/view) for guidance on successfully completing this section. Things you'll need to do:
* Load a [pre-trained network](http://pytorch.org/docs/master/torchvision/models.html) (If you need a starting point, the VGG networks work great and are straightforward to use)
* Define a new, untrained feed-forward network as a classifier, using ReLU activations and dropout
* Train the classifier layers using backpropagation using the pre-trained network to get the features
* Track the loss and accuracy on the validation set to determine the best hyperparameters
We've left a cell open for you below, but use as many as you need. Our advice is to break the problem up into smaller parts you can run separately. Check that each part is doing what you expect, then move on to the next. You'll likely find that as you work through each part, you'll need to go back and modify your previous code. This is totally normal!
When training make sure you're updating only the weights of the feed-forward network. You should be able to get the validation accuracy above 70% if you build everything right. Make sure to try different hyperparameters (learning rate, units in the classifier, epochs, etc) to find the best model. Save those hyperparameters to use as default values in the next part of the project.
One last important tip if you're using the workspace to run your code: To avoid having your workspace disconnect during the long-running tasks in this notebook, please read in the earlier page in this lesson called Intro to
GPU Workspaces about Keeping Your Session Active. You'll want to include code from the workspace_utils.py module.
**Note for Workspace users:** If your network is over 1 GB when saved as a checkpoint, there might be issues with saving backups in your workspace. Typically this happens with wide dense layers after the convolutional layers. If your saved checkpoint is larger than 1 GB (you can open a terminal and check with `ls -lh`), you should reduce the size of your hidden layers and train again.
```
# TODO: Build and train your network
model = models.vgg19(pretrained = True)
print(model)
for param in model.parameters():
param.requires_grad = False
model.classifier = nn.Sequential(OrderedDict
([('fc1', nn.Linear(25088, 4096)),
('relu', nn.ReLU()),
('fc2', nn.Linear(4096, 102)),
('out', nn.LogSoftmax(dim = 1))
]))
criterion = nn.NLLLoss()
optimizer = optim.Adam(model.classifier.parameters(), lr = 0.001)
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
model.to(device);
epochs = 3
steps = 0
running_loss = 0
print_every = 40
for i in range(epochs):
for inputs, labels in trainloader:
steps += 1
inputs, labels = inputs.to(device), labels.to(device)
optimizer.zero_grad()
logprobs = model.forward(inputs)
loss = criterion(logprobs, labels)
loss.backward()
optimizer.step()
running_loss += loss.item()
if steps % print_every == 0:
valid_loss = 0
accuracy = 0
model.eval()
with torch.no_grad():
for inputs, labels in validloader:
inputs, labels = inputs.to(device), labels.to(device)
logprobs = model.forward(inputs)
batch_loss = criterion(logprobs, labels)
valid_loss += batch_loss.item()
prob = torch.exp(logprobs)
top_prob, top_class = prob.topk(1, dim=1)
equals = top_class == labels.view(*top_class.shape)
accuracy += torch.mean(equals.type(torch.FloatTensor)).item()
print(f"Epoch {i+1}/{epochs}.. "
f"Train loss: {running_loss/print_every:.3f}.. "
f"Validation loss: {valid_loss/len(validloader):.3f}.. "
f"Validation accuracy: {accuracy/len(validloader):.3f}")
running_loss = 0
model.train()
print("\nTraining Complete!")
```
## Testing your network
It's good practice to test your trained network on test data, images the network has never seen either in training or validation. This will give you a good estimate for the model's performance on completely new images. Run the test images through the network and measure the accuracy, the same way you did validation. You should be able to reach around 70% accuracy on the test set if the model has been trained well.
```
# TODO: Do validation on the test set
def testing(testloader, device='cpu'):
correct = 0
total = 0
with torch.no_grad():
for data in testloader:
images, labels = data[0].to(device), data[1].to(device)
outputs = model(images)
_, predicted = torch.max(outputs.data, 1)
total += labels.size(0)
correct += (predicted == labels).sum().item()
print('Accuracy of the network: %d %%' % (100 * correct / total))
return correct / total
testing(testloader,'cuda')
```
## Save the checkpoint
Now that your network is trained, save the model so you can load it later for making predictions. You probably want to save other things such as the mapping of classes to indices which you get from one of the image datasets: `image_datasets['train'].class_to_idx`. You can attach this to the model as an attribute which makes inference easier later on.
```model.class_to_idx = image_datasets['train'].class_to_idx```
Remember that you'll want to completely rebuild the model later so you can use it for inference. Make sure to include any information you need in the checkpoint. If you want to load the model and keep training, you'll want to save the number of epochs as well as the optimizer state, `optimizer.state_dict`. You'll likely want to use this trained model in the next part of the project, so best to save it now.
```
# TODO: Save the checkpoint
model.class_to_idx = train_data.class_to_idx
checkpoint = {'transfer_model' : 'vgg19',
'input_size' : 25088,
'output_size' : 102,
'features' : model.features,
'classifier' : model.classifier,
'optimizer' : optimizer.state_dict(),
'state_dict' : model.state_dict(),
'idx_to_class' : {v: k for k, v in train_data.class_to_idx.items()}
}
torch.save(checkpoint, 'CheckPoint.pth')
```
## Loading the checkpoint
At this point it's good to write a function that can load a checkpoint and rebuild the model. That way you can come back to this project and keep working on it without having to retrain the network.
```
# TODO: Write a function that loads a checkpoint and rebuilds the model
def load_checkpoint(path):
model_info = torch.load(path)
model = models.vgg19(pretrained = True)
model.classifier = nn.Sequential(OrderedDict([
('fc1', nn.Linear(25088, 4096)),
('relu', nn.ReLU()),
('fc2', nn.Linear(4096, 102)),
('out', nn.LogSoftmax(dim = 1))
]))
model.load_state_dict(model_info['state_dict'])
return model, model_info
model, model_info = load_checkpoint('CheckPoint.pth')
print(model)
```
# Inference for classification
Now you'll write a function to use a trained network for inference. That is, you'll pass an image into the network and predict the class of the flower in the image. Write a function called `predict` that takes an image and a model, then returns the top $K$ most likely classes along with the probabilities. It should look like
```python
probs, classes = predict(image_path, model)
print(probs)
print(classes)
> [ 0.01558163 0.01541934 0.01452626 0.01443549 0.01407339]
> ['70', '3', '45', '62', '55']
```
First you'll need to handle processing the input image such that it can be used in your network.
## Image Preprocessing
You'll want to use `PIL` to load the image ([documentation](https://pillow.readthedocs.io/en/latest/reference/Image.html)). It's best to write a function that preprocesses the image so it can be used as input for the model. This function should process the images in the same manner used for training.
First, resize the images where the shortest side is 256 pixels, keeping the aspect ratio. This can be done with the [`thumbnail`](http://pillow.readthedocs.io/en/3.1.x/reference/Image.html#PIL.Image.Image.thumbnail) or [`resize`](http://pillow.readthedocs.io/en/3.1.x/reference/Image.html#PIL.Image.Image.thumbnail) methods. Then you'll need to crop out the center 224x224 portion of the image.
Color channels of images are typically encoded as integers 0-255, but the model expected floats 0-1. You'll need to convert the values. It's easiest with a Numpy array, which you can get from a PIL image like so `np_image = np.array(pil_image)`.
As before, the network expects the images to be normalized in a specific way. For the means, it's `[0.485, 0.456, 0.406]` and for the standard deviations `[0.229, 0.224, 0.225]`. You'll want to subtract the means from each color channel, then divide by the standard deviation.
And finally, PyTorch expects the color channel to be the first dimension but it's the third dimension in the PIL image and Numpy array. You can reorder dimensions using [`ndarray.transpose`](https://docs.scipy.org/doc/numpy-1.13.0/reference/generated/numpy.ndarray.transpose.html). The color channel needs to be first and retain the order of the other two dimensions.
```
def process_image(image):
''' Scales, crops, and normalizes a PIL image for a PyTorch model,
returns an Numpy array
'''
# TODO: Process a PIL image for use in a PyTorch model
im = Image.open(image)
width, height = im.size
picture_coords = [width, height]
max_span = max(picture_coords)
max_element = picture_coords.index(max_span)
if max_element == 0:
min_element = 1
else:
min_element = 0
aspect_ratio=picture_coords[max_element]/picture_coords[min_element]
new_picture_coords = [0,0]
new_picture_coords[min_element] = 256
new_picture_coords[max_element] = int(256 * aspect_ratio)
im = im.resize(new_picture_coords)
width, height = new_picture_coords
left = (width - 244)/2
top = (height - 244)/2
right = (width + 244)/2
bottom = (height + 244)/2
im = im.crop((left, top, right, bottom))
np_image = np.array(im)
np_image = np_image.astype('float64')
np_image = np_image / [255,255,255]
np_image = (np_image - [0.485, 0.456, 0.406])/ [0.229, 0.224, 0.225]
np_image = np_image.transpose((2, 0, 1))
return np_image
new_image = process_image('flowers/train/1/image_06734.jpg')
```
To check your work, the function below converts a PyTorch tensor and displays it in the notebook. If your `process_image` function works, running the output through this function should return the original image (except for the cropped out portions).
```
def imshow(image, ax=None, title=None):
"""Imshow for Tensor."""
if ax is None:
fig, ax = plt.subplots()
# PyTorch tensors assume the color channel is the first dimension
# but matplotlib assumes is the third dimension
image = image.transpose((1, 2, 0))
# Undo preprocessing
mean = np.array([0.485, 0.456, 0.406])
std = np.array([0.229, 0.224, 0.225])
image = std * image + mean
# Image needs to be clipped between 0 and 1 or it looks like noise when displayed
image = np.clip(image, 0, 1)
ax.imshow(image)
return ax
imshow(new_image)
```
## Class Prediction
Once you can get images in the correct format, it's time to write a function for making predictions with your model. A common practice is to predict the top 5 or so (usually called top-$K$) most probable classes. You'll want to calculate the class probabilities then find the $K$ largest values.
To get the top $K$ largest values in a tensor use [`x.topk(k)`](http://pytorch.org/docs/master/torch.html#torch.topk). This method returns both the highest `k` probabilities and the indices of those probabilities corresponding to the classes. You need to convert from these indices to the actual class labels using `class_to_idx` which hopefully you added to the model or from an `ImageFolder` you used to load the data ([see here](#Save-the-checkpoint)). Make sure to invert the dictionary so you get a mapping from index to class as well.
Again, this method should take a path to an image and a model checkpoint, then return the probabilities and classes.
```python
probs, classes = predict(image_path, model)
print(probs)
print(classes)
> [ 0.01558163 0.01541934 0.01452626 0.01443549 0.01407339]
> ['70', '3', '45', '62', '55']
```
```
def predict(image_path, model, topk=5):
''' Predict the class (or classes) of an image using a trained deep learning model.
'''
# TODO: Implement the code to predict the class from an image file
with torch.no_grad():
image = process_image(image_path)
image = torch.from_numpy(image)
image.unsqueeze_(0)
image = image.float()
model, _ = load_checkpoint(model)
outputs = model(image)
probs, classes = torch.exp(outputs).topk(topk)
return probs[0].tolist(), classes[0].add(1).tolist()
predict('flowers/train/1/image_06734.jpg','CheckPoint.pth')
```
## Sanity Checking
Now that you can use a trained model for predictions, check to make sure it makes sense. Even if the testing accuracy is high, it's always good to check that there aren't obvious bugs. Use `matplotlib` to plot the probabilities for the top 5 classes as a bar graph, along with the input image. It should look like this:
<img src='assets/inference_example.png' width=300px>
You can convert from the class integer encoding to actual flower names with the `cat_to_name.json` file (should have been loaded earlier in the notebook). To show a PyTorch tensor as an image, use the `imshow` function defined above.
```
# TODO: Display an image along with the top 5 classes
def display_pred(image_path,model):
probs, classes = predict(image_path,'CheckPoint.pth')
plant_classes = [cat_to_name[str(cls)] + "({})".format(str(cls)) for cls in classes]
im = Image.open(image_path)
fig, ax = plt.subplots(2,1)
ax[0].imshow(im);
y_positions = np.arange(len(plant_classes))
ax[1].barh(y_positions,probs,color='blue')
ax[1].set_yticks(y_positions)
ax[1].set_yticklabels(plant_classes)
ax[1].invert_yaxis()
ax[1].set_xlabel('Accuracy %')
ax[0].set_title('Top 5 Flower Predictions')
return None
display_pred('flowers/train/1/image_06772.jpg','CheckPoint.pth')
```
|
github_jupyter
|
# Imports here
%matplotlib inline
%config InlineBackend.figure_format = 'retina'
import numpy as np
import matplotlib.pyplot as plt
import torch
from torch import nn, optim
import torch.nn.functional as F
from collections import OrderedDict
from torchvision import transforms, models, datasets
from PIL import Image
from os import listdir
import random
import json
data_dir = 'flowers'
train_dir = data_dir + '/train'
valid_dir = data_dir + '/valid'
test_dir = data_dir + '/test'
# TODO: Define your transforms for the training, validation, and testing sets
data_transforms = transforms.Compose([transforms.ToTensor(),
transforms.Normalize([0.485, 0.456, 0.406], [0.229, 0.224, 0.225])
])
train_transforms = transforms.Compose([transforms.Resize(225),
transforms.RandomCrop(224),
transforms.RandomHorizontalFlip(),
transforms.ToTensor(),
transforms.Normalize([0.485, 0.456, 0.406], [0.229, 0.224, 0.225])
])
valid_transforms = transforms.Compose([transforms.Resize(225),
transforms.RandomCrop(224),
transforms.RandomHorizontalFlip(),
transforms.ToTensor(),
transforms.Normalize([0.485, 0.456, 0.406], [0.229, 0.224, 0.225])
])
test_transforms = transforms.Compose([transforms.Resize(225),
transforms.RandomCrop(224),
transforms.RandomHorizontalFlip(),
transforms.ToTensor(),
transforms.Normalize([0.485, 0.456, 0.406], [0.229, 0.224, 0.225])
])
# TODO: Load the datasets with ImageFolder
data = datasets.ImageFolder(data_dir, transform = data_transforms)
train_data = datasets.ImageFolder(train_dir, transform = train_transforms)
valid_data = datasets.ImageFolder(valid_dir, transform = valid_transforms)
test_data = datasets.ImageFolder(test_dir, transform = test_transforms)
# TODO: Using the image datasets and the trainforms, define the dataloaders
dataloader = torch.utils.data.DataLoader(data, batch_size = 32, shuffle = True)
trainloader = torch.utils.data.DataLoader(train_data, batch_size = 32, shuffle = True)
validloader = torch.utils.data.DataLoader(valid_data, batch_size = 32, shuffle = True)
testloader = torch.utils.data.DataLoader(test_data, batch_size = 32, shuffle = True)
with open('cat_to_name.json', 'r') as f:
cat_to_name = json.load(f)
# TODO: Build and train your network
model = models.vgg19(pretrained = True)
print(model)
for param in model.parameters():
param.requires_grad = False
model.classifier = nn.Sequential(OrderedDict
([('fc1', nn.Linear(25088, 4096)),
('relu', nn.ReLU()),
('fc2', nn.Linear(4096, 102)),
('out', nn.LogSoftmax(dim = 1))
]))
criterion = nn.NLLLoss()
optimizer = optim.Adam(model.classifier.parameters(), lr = 0.001)
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
model.to(device);
epochs = 3
steps = 0
running_loss = 0
print_every = 40
for i in range(epochs):
for inputs, labels in trainloader:
steps += 1
inputs, labels = inputs.to(device), labels.to(device)
optimizer.zero_grad()
logprobs = model.forward(inputs)
loss = criterion(logprobs, labels)
loss.backward()
optimizer.step()
running_loss += loss.item()
if steps % print_every == 0:
valid_loss = 0
accuracy = 0
model.eval()
with torch.no_grad():
for inputs, labels in validloader:
inputs, labels = inputs.to(device), labels.to(device)
logprobs = model.forward(inputs)
batch_loss = criterion(logprobs, labels)
valid_loss += batch_loss.item()
prob = torch.exp(logprobs)
top_prob, top_class = prob.topk(1, dim=1)
equals = top_class == labels.view(*top_class.shape)
accuracy += torch.mean(equals.type(torch.FloatTensor)).item()
print(f"Epoch {i+1}/{epochs}.. "
f"Train loss: {running_loss/print_every:.3f}.. "
f"Validation loss: {valid_loss/len(validloader):.3f}.. "
f"Validation accuracy: {accuracy/len(validloader):.3f}")
running_loss = 0
model.train()
print("\nTraining Complete!")
# TODO: Do validation on the test set
def testing(testloader, device='cpu'):
correct = 0
total = 0
with torch.no_grad():
for data in testloader:
images, labels = data[0].to(device), data[1].to(device)
outputs = model(images)
_, predicted = torch.max(outputs.data, 1)
total += labels.size(0)
correct += (predicted == labels).sum().item()
print('Accuracy of the network: %d %%' % (100 * correct / total))
return correct / total
testing(testloader,'cuda')
Remember that you'll want to completely rebuild the model later so you can use it for inference. Make sure to include any information you need in the checkpoint. If you want to load the model and keep training, you'll want to save the number of epochs as well as the optimizer state, `optimizer.state_dict`. You'll likely want to use this trained model in the next part of the project, so best to save it now.
## Loading the checkpoint
At this point it's good to write a function that can load a checkpoint and rebuild the model. That way you can come back to this project and keep working on it without having to retrain the network.
# Inference for classification
Now you'll write a function to use a trained network for inference. That is, you'll pass an image into the network and predict the class of the flower in the image. Write a function called `predict` that takes an image and a model, then returns the top $K$ most likely classes along with the probabilities. It should look like
First you'll need to handle processing the input image such that it can be used in your network.
## Image Preprocessing
You'll want to use `PIL` to load the image ([documentation](https://pillow.readthedocs.io/en/latest/reference/Image.html)). It's best to write a function that preprocesses the image so it can be used as input for the model. This function should process the images in the same manner used for training.
First, resize the images where the shortest side is 256 pixels, keeping the aspect ratio. This can be done with the [`thumbnail`](http://pillow.readthedocs.io/en/3.1.x/reference/Image.html#PIL.Image.Image.thumbnail) or [`resize`](http://pillow.readthedocs.io/en/3.1.x/reference/Image.html#PIL.Image.Image.thumbnail) methods. Then you'll need to crop out the center 224x224 portion of the image.
Color channels of images are typically encoded as integers 0-255, but the model expected floats 0-1. You'll need to convert the values. It's easiest with a Numpy array, which you can get from a PIL image like so `np_image = np.array(pil_image)`.
As before, the network expects the images to be normalized in a specific way. For the means, it's `[0.485, 0.456, 0.406]` and for the standard deviations `[0.229, 0.224, 0.225]`. You'll want to subtract the means from each color channel, then divide by the standard deviation.
And finally, PyTorch expects the color channel to be the first dimension but it's the third dimension in the PIL image and Numpy array. You can reorder dimensions using [`ndarray.transpose`](https://docs.scipy.org/doc/numpy-1.13.0/reference/generated/numpy.ndarray.transpose.html). The color channel needs to be first and retain the order of the other two dimensions.
To check your work, the function below converts a PyTorch tensor and displays it in the notebook. If your `process_image` function works, running the output through this function should return the original image (except for the cropped out portions).
## Class Prediction
Once you can get images in the correct format, it's time to write a function for making predictions with your model. A common practice is to predict the top 5 or so (usually called top-$K$) most probable classes. You'll want to calculate the class probabilities then find the $K$ largest values.
To get the top $K$ largest values in a tensor use [`x.topk(k)`](http://pytorch.org/docs/master/torch.html#torch.topk). This method returns both the highest `k` probabilities and the indices of those probabilities corresponding to the classes. You need to convert from these indices to the actual class labels using `class_to_idx` which hopefully you added to the model or from an `ImageFolder` you used to load the data ([see here](#Save-the-checkpoint)). Make sure to invert the dictionary so you get a mapping from index to class as well.
Again, this method should take a path to an image and a model checkpoint, then return the probabilities and classes.
## Sanity Checking
Now that you can use a trained model for predictions, check to make sure it makes sense. Even if the testing accuracy is high, it's always good to check that there aren't obvious bugs. Use `matplotlib` to plot the probabilities for the top 5 classes as a bar graph, along with the input image. It should look like this:
<img src='assets/inference_example.png' width=300px>
You can convert from the class integer encoding to actual flower names with the `cat_to_name.json` file (should have been loaded earlier in the notebook). To show a PyTorch tensor as an image, use the `imshow` function defined above.
| 0.628407 | 0.981666 |
```
# Importing SparkContext, SparkConf
from pyspark import SparkContext, SparkConf
# To stop any existing spark Context. Only single Spark context can run per JVM.
sc.stop()
```
# Understanding Configuration of Spark for local mode
#### In local mode, we have one JVM machine, which has as Executor/ Driver( so we have one executor)
#### Task slots = Executor cores = Available threads != CPU cores
#### On each slot/core, you can allocate multiple tasks
# Understanding your own machine
#### In Windows Power Shell
wmic
wmic:root\cli> CPU Get NumberOfCores,NumberOfLogicalProcessors /Format:List
#### It will show
NumberOfCores=4
NumberOfLogicalProcessors=8
#### Hyper-Threading is enabled. With Hyper-Threading, a microprocessor's "core" processor can execute two (rather than one) concurrent streams (or threads) of instructions sent by the operating system
## Setting up Spark Local Cluster configuration:
#### One way to set my local mode : keep 2 processors for OS, 6 processors for Spark JVM, set 6 task slots.
#### if we need to utilize all the processors efficently(CPU utilization)
#### Set 12 or 18 task slots with memory allocation per CPU processor
```
conf_spark = SparkConf().set("spark.driver.host", "127.0.0.1").setMaster("local[12]").setAppName("myapp")
conf_spark.getAll()
sc = SparkContext(conf=conf_spark)
```
#### If we look in Spark Web UI, we can spot 12 slots, with some memory allocation
<img src="Data/Executors_SparkWebUI.PNG">
#### We have 2 other options for local let us understand other 2 options
#### local[*] or local
```
sc.stop()
conf_spark = SparkConf().set("spark.driver.host", "127.0.0.1").setMaster("local[*]").setAppName("myapp")
sc = SparkContext(conf=conf_spark)
```
<img src="Data/Executors_SparkWebUI2.PNG">
```
## Shows ALL 8 AVALIABLE coreS, with 366.3MB Memory
sc.stop()
conf_spark = SparkConf().set("spark.driver.host", "127.0.0.1").setMaster("local").setAppName("myapp")
sc = SparkContext(conf=conf_spark)
## Shows 1 core, with 366.3MB Memory
```
#### Standard: Number of cores = Concurrent tasks a executor can run (Source:Stackoverflow)
### Let us understand the memory of spark slots
#### There will be no storage memory. Spark does n't have storage system as HDFS, but there can be disk spillage.So, we are talking about only Cache Memory and overall memory of executor used during execution of tasks.We say overall memory is allocated to Executor/Driver.We have one JVM, which has one executor/driver.We need to find driver.memory/executor.memory.Driver collects the results and Executor stores the partitions of data in memory for computations.
#### On-Heap memory management: Objects are allocated on the JVM heap and bound by GC.
#### Off-Heap memory management: Objects are allocated in memory outside the JVM by serialization,managed by the application, and are not bound by GC. This memory management method can avoid frequent GC, but the disadvantage is that you have to write the logic of memory allocation and memory release.
#### speed On-Heap>Off-Heap>Disk
#### Unified Memory Manager mechanism:
#### The Storage memory and Execution memory share a memory area, and both can occupy each other's free area.
```
# Setting memory for application
Maximum heap size settings can be set with spark.driver.memory in the cluster mode and through
the --driver-memory command line option in the client mode.
Note: In client mode, this config must not be set through the SparkConf directly in your application,
because the driver JVM has already started at that point.
Instead, please set this through the --driver-java-options command line option or in your default properties file.
## Testing above point?
# Yes we need to change the memory in conf file.
sc.stop()
conf_spark = SparkConf().set("spark.driver.host", "127.0.0.1").setMaster("local[5]").setAppName("myapp")
sc = SparkContext(conf=conf_spark)
conf_spark.getAll()
```
<img src="Data/Executors_SparkWebUI3.PNG">
```
## How to choose 5G memory. It depends on tasks and size of dataset.
## Will vary for different data sizes and number of operations such as shuffle, cache and computations
## More can be found on https://spark.apache.org/docs/latest/tuning.html#memory-management-overview
## http://spark.apache.org/docs/latest/configuration.html#memory-management
```
##### References: Databricks, StackOverflow, Apache Spark Documentation, www.tutorialdocs.com
|
github_jupyter
|
# Importing SparkContext, SparkConf
from pyspark import SparkContext, SparkConf
# To stop any existing spark Context. Only single Spark context can run per JVM.
sc.stop()
conf_spark = SparkConf().set("spark.driver.host", "127.0.0.1").setMaster("local[12]").setAppName("myapp")
conf_spark.getAll()
sc = SparkContext(conf=conf_spark)
sc.stop()
conf_spark = SparkConf().set("spark.driver.host", "127.0.0.1").setMaster("local[*]").setAppName("myapp")
sc = SparkContext(conf=conf_spark)
## Shows ALL 8 AVALIABLE coreS, with 366.3MB Memory
sc.stop()
conf_spark = SparkConf().set("spark.driver.host", "127.0.0.1").setMaster("local").setAppName("myapp")
sc = SparkContext(conf=conf_spark)
## Shows 1 core, with 366.3MB Memory
# Setting memory for application
Maximum heap size settings can be set with spark.driver.memory in the cluster mode and through
the --driver-memory command line option in the client mode.
Note: In client mode, this config must not be set through the SparkConf directly in your application,
because the driver JVM has already started at that point.
Instead, please set this through the --driver-java-options command line option or in your default properties file.
## Testing above point?
# Yes we need to change the memory in conf file.
sc.stop()
conf_spark = SparkConf().set("spark.driver.host", "127.0.0.1").setMaster("local[5]").setAppName("myapp")
sc = SparkContext(conf=conf_spark)
conf_spark.getAll()
## How to choose 5G memory. It depends on tasks and size of dataset.
## Will vary for different data sizes and number of operations such as shuffle, cache and computations
## More can be found on https://spark.apache.org/docs/latest/tuning.html#memory-management-overview
## http://spark.apache.org/docs/latest/configuration.html#memory-management
| 0.663342 | 0.886764 |
```
from scipy.optimize import minimize
import numpy as np
import matplotlib.pyplot as plt
from matplotlib.collections import LineCollection
from matplotlib.colors import ListedColormap, BoundaryNorm
import random
class GPR:
def __init__(self, optimize=True):
self.is_fit = False
self.train_X, self.train_y = None, None
self.params = {"l": 0.5, "sigma_f": 0.2}
self.optimize = optimize
def fit(self, X, y):
# store train data
self.train_X = np.asarray(X)
self.train_y = np.asarray(y)
self.is_fit = True
def predict(self, X):
if not self.is_fit:
print("GPR Model not fit yet.")
return
X = np.asarray(X)
Kff = self.kernel(X, X) # (N,N)
Kyy = self.kernel(self.train_X, self.train_X) # (k,k)
Kfy = self.kernel(X, self.train_X) # (N,k)
Kyy_inv = np.linalg.inv(Kyy + 1e-8 * np.eye(len(self.train_X))) # (k,k)
mu = Kfy.dot(Kyy_inv).dot(self.train_y)
cov = self.kernel(X, X) - Kfy.dot(Kyy_inv).dot(Kfy.T)
return mu, cov
def kernel(self, x1, x2):
dist_matrix = np.sum(x1**2, 1).reshape(-1, 1) + np.sum(x2**2, 1) - 2 * np.dot(x1, x2.T)
return self.params["sigma_f"] ** 2 * np.exp(-0.5 / self.params["l"] ** 2 * dist_matrix)
def y(x, noise_sigma=0.0):
x = np.asarray(x)
y = np.cos(x) + np.random.normal(0, noise_sigma, size=x.shape)
return y.tolist()
# 生成主要曲线
# point:随机点数,点数越多曲线越曲折
# length:曲线总长度
def generate_main(point=5,length=20):
trx0 = []
try0 = []
for i in range(point):
trx0.append(random.random()*length)
gsran = random.gauss(0,1)
if gsran > 10 or gsran<-10:
gsran = random.random()*10
try0.append(gsran)
train_X = np.array(trx0).reshape(-1,1)
train_y = np.array(try0)
test_X = np.arange(0, length, 0.1).reshape(-1, 1)
# print('max,',np.max(train_y))
# print('min,',np.min(train_y))
gpr = GPR()
gpr.fit(train_X, train_y)
mu, cov = gpr.predict(test_X)
test_y = mu.ravel()
return test_X[:,0],test_y
def scale_wave(x):
a = -x**2+2*x
return np.sqrt(a)
def ex0(wave):
num0 = 0
for i in range(len(wave)):
if wave[i] < 0.05:
num0 += 1
return num0 / len(wave)
# 生成随机波动强度
# wave_point,拨动点数,点数越多波动越曲折
# length,曲线长度
def generate_wave(wave_point=12,length=20):
# 曲线的幅度
trx1 = []
for i in range(wave_point):
trx1.append(int(random.random()*length))
trx1 = np.array(trx1).reshape(-1, 1)
try1 = [0]*wave_point
for i in range(len(try1)):
try1[i] = random.random()*0.5+0.5
gpr1 = GPR()
testx1 = np.arange(0, length, 0.1).reshape(-1, 1)
gpr1.fit(trx1,try1)
mu1,cov1 = gpr1.predict(testx1)
testy1 = mu1.ravel()
return testx1[:,0],testy1
# 曲线的颜色
# color_point,颜色的波动幅度,越多颜色波动越剧烈
# length,总长度,三个函数的总长度要相同
def generate_color(color_point=5,length=20):
trx2 = []
for i in range(color_point):
trx2.append(int(random.random()*length))
trx2 = np.array(trx2).reshape(-1, 1)
try2 = []
for i in range(color_point):
try2.append(random.random())
gpr2 = GPR()
testx2 = np.arange(0, length, 0.1).reshape(-1, 1)
gpr2.fit(trx2,try2)
mu2,cov2 = gpr2.predict(testx2)
testy2 = mu2.ravel()
return testx2[:,0],np.abs(testy2)
np.abs([-5,3])
mys = []
wys = []
cys = []
count = 0
while len(wys) < 40:
count += 1
# print(count)
mx,my = generate_main()
wx,wy = generate_wave()
cx,cy = generate_color()
if np.max(my) > 3 or np.min(my) < -3:
continue
if ex0(wy) > 0.2:
continue
# print(np.max(wy))
print(np.min(wy))
mys.append(my)
cys.append(cy)
wys.append(wy)
print('count,',count)
# 横坐标总长,变长
def get_tri(x,edge):
gen3 = 1.71828
# 上,左下,右下
return [[x/2,edge/gen3-0.6],[x/2-edge/2,-edge / 2 /gen3-0.6],[x/2+edge/2,-edge/2/gen3-0.6]]
# para:上,左下,右下的节点坐标,h是缩小的幅度
def tri_shrink(pos0,pos1,pos2,h):
gen3 = 1.71828
return [[pos0[0],pos0[1]-h],[pos1[0]+gen3 * h / 2,pos1[1]+h/2],[pos2[0]-gen3 * h / 2,pos2[1]+h/2]]
def norm(ys):
m1 = np.max(ys)
m2 = abs(np.min(ys))
m = max(m1,m2)
ys = ys / m
return ys
%matplotlib inline
# plt.figure()
fig, ax = plt.subplots(figsize=(16,16))
# plt.fill_between(test_X.ravel(), test_y + uncertainty, test_y - uncertainty, alpha=0.1)
# plt.plot(test_X,test_y+uncertainty)
# u = wave_y*6
# Use a boundary norm instead
# 主线的循环
for l in range(len(mys)):
main_x = mx
main_y = mys[l]
wave_y = wys[l]*2
# print(wave_y)
# wave_y = wys[l]*4
# 每条主线,不同波动的循环
for i in range(10):
if i > 6:
continue
plt.plot(main_x,main_y+wave_y*i/10,color='black',alpha=0.1)
plt.plot(main_x,main_y-wave_y*i/10,color='black',alpha=0.1)
# 画三角形
pos = get_tri(20,12.56)
h = 0.2
for i in range(5):
thish = h*i
posnew = tri_shrink(pos[0],pos[1],pos[2],thish)
trixs = [posnew[0][0],posnew[1][0],posnew[2][0],posnew[0][0]]
triys = [posnew[0][1],posnew[1][1],posnew[2][1],posnew[0][1]]
plt.plot(trixs,triys,color='gray',alpha=0.3)
plt.xticks(())
plt.yticks(())
# 取消边框
for key, spine in ax.spines.items():
# 'left', 'right', 'bottom', 'top'
if key == 'right' or key == 'top' or key == 'bottom' or key == 'left':
spine.set_visible(False)
ax.set_xlim(0, 20)
ax.set_ylim(-10,10)
plt.show()
import numpy as np
import matplotlib.pyplot as plt
from matplotlib.collections import LineCollection
from matplotlib.colors import ListedColormap, BoundaryNorm
# dydx = color_y # first derivative ,used for colormap
# dydx_used = dydx*0.5+np.mean(dydx)
fig, axs = plt.subplots(figsize=(16,16))
line_num = 8
for l in range(len(mys)):
main_x = mx
main_y = mys[l]
wave_y = wys[l]*3
dydx = cys[l]
for i in range(line_num):
x = main_x
if i < line_num/2:
y = main_y+wave_y*i/10
else:
y = main_y-wave_y*(i-line_num/2)/10
points = np.array([x, y]).T.reshape(-1, 1, 2)
segments = np.concatenate([points[:-1], points[1:]], axis=1)
# Create a continuous norm to map from data points to colors
norm = plt.Normalize(dydx.min(), dydx.max())
lc = LineCollection(segments, cmap='summer', norm=norm,alpha=0.15)
# Set the values used for colormapping
lc.set_array(dydx)
line = axs.add_collection(lc)
plt.xticks(())
plt.yticks(())
# 画三角形
pos = get_tri(20,12.56)
h = 0.2
for i in range(5):
thish = h*i
posnew = tri_shrink(pos[0],pos[1],pos[2],thish)
trixs = [posnew[0][0],posnew[1][0],posnew[2][0],posnew[0][0]]
triys = [posnew[0][1],posnew[1][1],posnew[2][1],posnew[0][1]]
plt.plot(trixs,triys,color='gray',alpha=0.3)
# 取消边框
for key, spine in axs.spines.items():
# 'left', 'right', 'bottom', 'top'
if key == 'right' or key == 'top' or key == 'bottom' or key == 'left':
spine.set_visible(False)
axs.set_xlim(0, 20)
axs.set_ylim(-10,10)
plt.savefig('lines.png',bbox_inches='tight',dpi=300)
plt.show()
for i in range(1):
plt.plot(scale_wave(wys[i]),color='red')
plt.plot(wys[i],color='green')
# plt.plot(mys[i],color='green')
# plt.plot(mys[i]*wys[i]*2,color='blue')
plt.show()
for wave in wys:
print(ex0(wave))
scale_wave(np.array([0.1,0.5]))
```
|
github_jupyter
|
from scipy.optimize import minimize
import numpy as np
import matplotlib.pyplot as plt
from matplotlib.collections import LineCollection
from matplotlib.colors import ListedColormap, BoundaryNorm
import random
class GPR:
def __init__(self, optimize=True):
self.is_fit = False
self.train_X, self.train_y = None, None
self.params = {"l": 0.5, "sigma_f": 0.2}
self.optimize = optimize
def fit(self, X, y):
# store train data
self.train_X = np.asarray(X)
self.train_y = np.asarray(y)
self.is_fit = True
def predict(self, X):
if not self.is_fit:
print("GPR Model not fit yet.")
return
X = np.asarray(X)
Kff = self.kernel(X, X) # (N,N)
Kyy = self.kernel(self.train_X, self.train_X) # (k,k)
Kfy = self.kernel(X, self.train_X) # (N,k)
Kyy_inv = np.linalg.inv(Kyy + 1e-8 * np.eye(len(self.train_X))) # (k,k)
mu = Kfy.dot(Kyy_inv).dot(self.train_y)
cov = self.kernel(X, X) - Kfy.dot(Kyy_inv).dot(Kfy.T)
return mu, cov
def kernel(self, x1, x2):
dist_matrix = np.sum(x1**2, 1).reshape(-1, 1) + np.sum(x2**2, 1) - 2 * np.dot(x1, x2.T)
return self.params["sigma_f"] ** 2 * np.exp(-0.5 / self.params["l"] ** 2 * dist_matrix)
def y(x, noise_sigma=0.0):
x = np.asarray(x)
y = np.cos(x) + np.random.normal(0, noise_sigma, size=x.shape)
return y.tolist()
# 生成主要曲线
# point:随机点数,点数越多曲线越曲折
# length:曲线总长度
def generate_main(point=5,length=20):
trx0 = []
try0 = []
for i in range(point):
trx0.append(random.random()*length)
gsran = random.gauss(0,1)
if gsran > 10 or gsran<-10:
gsran = random.random()*10
try0.append(gsran)
train_X = np.array(trx0).reshape(-1,1)
train_y = np.array(try0)
test_X = np.arange(0, length, 0.1).reshape(-1, 1)
# print('max,',np.max(train_y))
# print('min,',np.min(train_y))
gpr = GPR()
gpr.fit(train_X, train_y)
mu, cov = gpr.predict(test_X)
test_y = mu.ravel()
return test_X[:,0],test_y
def scale_wave(x):
a = -x**2+2*x
return np.sqrt(a)
def ex0(wave):
num0 = 0
for i in range(len(wave)):
if wave[i] < 0.05:
num0 += 1
return num0 / len(wave)
# 生成随机波动强度
# wave_point,拨动点数,点数越多波动越曲折
# length,曲线长度
def generate_wave(wave_point=12,length=20):
# 曲线的幅度
trx1 = []
for i in range(wave_point):
trx1.append(int(random.random()*length))
trx1 = np.array(trx1).reshape(-1, 1)
try1 = [0]*wave_point
for i in range(len(try1)):
try1[i] = random.random()*0.5+0.5
gpr1 = GPR()
testx1 = np.arange(0, length, 0.1).reshape(-1, 1)
gpr1.fit(trx1,try1)
mu1,cov1 = gpr1.predict(testx1)
testy1 = mu1.ravel()
return testx1[:,0],testy1
# 曲线的颜色
# color_point,颜色的波动幅度,越多颜色波动越剧烈
# length,总长度,三个函数的总长度要相同
def generate_color(color_point=5,length=20):
trx2 = []
for i in range(color_point):
trx2.append(int(random.random()*length))
trx2 = np.array(trx2).reshape(-1, 1)
try2 = []
for i in range(color_point):
try2.append(random.random())
gpr2 = GPR()
testx2 = np.arange(0, length, 0.1).reshape(-1, 1)
gpr2.fit(trx2,try2)
mu2,cov2 = gpr2.predict(testx2)
testy2 = mu2.ravel()
return testx2[:,0],np.abs(testy2)
np.abs([-5,3])
mys = []
wys = []
cys = []
count = 0
while len(wys) < 40:
count += 1
# print(count)
mx,my = generate_main()
wx,wy = generate_wave()
cx,cy = generate_color()
if np.max(my) > 3 or np.min(my) < -3:
continue
if ex0(wy) > 0.2:
continue
# print(np.max(wy))
print(np.min(wy))
mys.append(my)
cys.append(cy)
wys.append(wy)
print('count,',count)
# 横坐标总长,变长
def get_tri(x,edge):
gen3 = 1.71828
# 上,左下,右下
return [[x/2,edge/gen3-0.6],[x/2-edge/2,-edge / 2 /gen3-0.6],[x/2+edge/2,-edge/2/gen3-0.6]]
# para:上,左下,右下的节点坐标,h是缩小的幅度
def tri_shrink(pos0,pos1,pos2,h):
gen3 = 1.71828
return [[pos0[0],pos0[1]-h],[pos1[0]+gen3 * h / 2,pos1[1]+h/2],[pos2[0]-gen3 * h / 2,pos2[1]+h/2]]
def norm(ys):
m1 = np.max(ys)
m2 = abs(np.min(ys))
m = max(m1,m2)
ys = ys / m
return ys
%matplotlib inline
# plt.figure()
fig, ax = plt.subplots(figsize=(16,16))
# plt.fill_between(test_X.ravel(), test_y + uncertainty, test_y - uncertainty, alpha=0.1)
# plt.plot(test_X,test_y+uncertainty)
# u = wave_y*6
# Use a boundary norm instead
# 主线的循环
for l in range(len(mys)):
main_x = mx
main_y = mys[l]
wave_y = wys[l]*2
# print(wave_y)
# wave_y = wys[l]*4
# 每条主线,不同波动的循环
for i in range(10):
if i > 6:
continue
plt.plot(main_x,main_y+wave_y*i/10,color='black',alpha=0.1)
plt.plot(main_x,main_y-wave_y*i/10,color='black',alpha=0.1)
# 画三角形
pos = get_tri(20,12.56)
h = 0.2
for i in range(5):
thish = h*i
posnew = tri_shrink(pos[0],pos[1],pos[2],thish)
trixs = [posnew[0][0],posnew[1][0],posnew[2][0],posnew[0][0]]
triys = [posnew[0][1],posnew[1][1],posnew[2][1],posnew[0][1]]
plt.plot(trixs,triys,color='gray',alpha=0.3)
plt.xticks(())
plt.yticks(())
# 取消边框
for key, spine in ax.spines.items():
# 'left', 'right', 'bottom', 'top'
if key == 'right' or key == 'top' or key == 'bottom' or key == 'left':
spine.set_visible(False)
ax.set_xlim(0, 20)
ax.set_ylim(-10,10)
plt.show()
import numpy as np
import matplotlib.pyplot as plt
from matplotlib.collections import LineCollection
from matplotlib.colors import ListedColormap, BoundaryNorm
# dydx = color_y # first derivative ,used for colormap
# dydx_used = dydx*0.5+np.mean(dydx)
fig, axs = plt.subplots(figsize=(16,16))
line_num = 8
for l in range(len(mys)):
main_x = mx
main_y = mys[l]
wave_y = wys[l]*3
dydx = cys[l]
for i in range(line_num):
x = main_x
if i < line_num/2:
y = main_y+wave_y*i/10
else:
y = main_y-wave_y*(i-line_num/2)/10
points = np.array([x, y]).T.reshape(-1, 1, 2)
segments = np.concatenate([points[:-1], points[1:]], axis=1)
# Create a continuous norm to map from data points to colors
norm = plt.Normalize(dydx.min(), dydx.max())
lc = LineCollection(segments, cmap='summer', norm=norm,alpha=0.15)
# Set the values used for colormapping
lc.set_array(dydx)
line = axs.add_collection(lc)
plt.xticks(())
plt.yticks(())
# 画三角形
pos = get_tri(20,12.56)
h = 0.2
for i in range(5):
thish = h*i
posnew = tri_shrink(pos[0],pos[1],pos[2],thish)
trixs = [posnew[0][0],posnew[1][0],posnew[2][0],posnew[0][0]]
triys = [posnew[0][1],posnew[1][1],posnew[2][1],posnew[0][1]]
plt.plot(trixs,triys,color='gray',alpha=0.3)
# 取消边框
for key, spine in axs.spines.items():
# 'left', 'right', 'bottom', 'top'
if key == 'right' or key == 'top' or key == 'bottom' or key == 'left':
spine.set_visible(False)
axs.set_xlim(0, 20)
axs.set_ylim(-10,10)
plt.savefig('lines.png',bbox_inches='tight',dpi=300)
plt.show()
for i in range(1):
plt.plot(scale_wave(wys[i]),color='red')
plt.plot(wys[i],color='green')
# plt.plot(mys[i],color='green')
# plt.plot(mys[i]*wys[i]*2,color='blue')
plt.show()
for wave in wys:
print(ex0(wave))
scale_wave(np.array([0.1,0.5]))
| 0.327131 | 0.519826 |
<a href="https://colab.research.google.com/github/Praxis-QR/RDWH/blob/main/MySQL_Tutorial_Getting_Started.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
<br>
<hr>
[Prithwis Mukerjee](http://www.linkedin.com/in/prithwis)<br>
#MySQL Tutorials <br>
1. https://www.mysqltutorial.org/
2. https://www.mysqltutorial.org/basic-mysql-tutorial.aspx
```
#!cat /proc/cpuinfo
#!cat /proc/meminfo
#!ls /proc/*info
```
#Section 1 : Getting started with MySQL
##Install MySQL in local Colab VM
```
# this will take some time and show an error.
# dont worry, just carry on
!apt install -y mysql-server > /dev/null
#!ls /etc/init.d
!/etc/init.d/mysql restart
```
## Download Sample Database - Classicmodels <br>
Create Tables <br>
https://www.mysqltutorial.org/mysql-sample-database.aspx
```
#!gdown https://drive.google.com/uc?id=1Ik3QuYTd52M5qI9D4aOCfGcQ7v2AB9wx
#!wget https://sp.mysqltutorial.org/wp-content/uploads/2018/03/mysqlsampledatabase.zip
#!wget https://github.com/Praxis-QR/RDWH/raw/main/mysqlsampledatabase.zip
!wget -q https://github.com/Praxis-QR/RDWH/raw/main/data/mysqlsampledatabase.zip
!unzip mysqlsampledatabase.zip
#!cat /content/mysqlsampledatabase.sql
!head /content/mysqlsampledatabase.sql
```
## Loading Sample Database into MySQL server
```
#https://www.mysqltutorial.org/how-to-load-sample-database-into-mysql-database-server.aspx
!mysql --table < mysqlsampledatabase.sql
```
###Check contents of sample database - Classicmodels
```
#New database classicmodels has been created
!mysql -e 'show databases'
!mysql classicmodels -e 'show tables'
```
This is what the database looks like : <br><br>
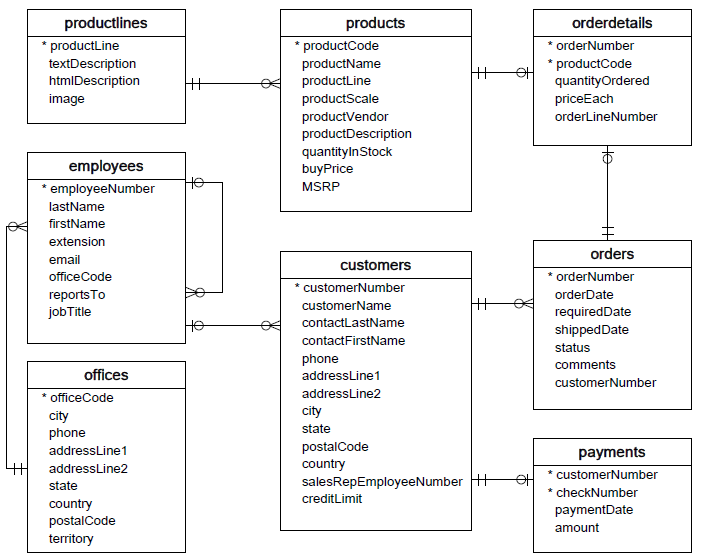<br>
```
!mysql classicmodels -e 'select * from employees'
```
##Install Python Client <br>
```
!apt install libmysqlclient-dev > /dev/null
!pip install mysqlclient
import pandas as pd
import MySQLdb
#To run any non-SELECT SQL command
def runCMD (DDL):
#DBConn= MySQLdb.connect(hostName,userName,passWord,dbName)
DBConn= MySQLdb.connect(db='classicmodels') # since local MySQL server, userid, password not needed
myCursor = DBConn.cursor()
retcode = myCursor.execute(DDL)
print (retcode)
DBConn.commit()
DBConn.close()
#To run any SELECT SQL command
def runSELECT (CMD):
#DBConn= MySQLdb.connect(hostName,userName,passWord,dbName)
DBConn= MySQLdb.connect(db='classicmodels') # since local MySQL server, userid, password not needed
df_mysql = pd.read_sql(CMD, con=DBConn)
DBConn.close()
return df_mysql
```
###Check out all tables
```
!mysql classicmodels -e 'show tables'
#runSELECT('select * from customers limit 5')
#runSELECT('select * from employees limit 5')
#runSELECT('select * from offices limit 5')
#runSELECT('select * from orderdetails limit 5')
#runSELECT('select * from orders limit 5')
#runSELECT('select * from payments limit 5')
#runSELECT('select * from productlines limit 5')
runSELECT('select * from products limit 5')
```
#Section 2 : Querying Data
#Section 3 - Sorting Data
#Chronobooks <br>
<hr>
Chronotantra and Chronoyantra are two science fiction novels that explore the collapse of human civilisation on Earth and then its rebirth and reincarnation both on Earth as well as on the distant worlds of Mars, Titan and Enceladus. But is it the human civilisation that is being reborn? Or is it some other sentience that is revealing itself.
If you have an interest in AI and found this material useful, you may consider buying these novel, in paperback or kindle, from [http://bit.ly/chronobooks](http://bit.ly/chronobooks)
|
github_jupyter
|
#!cat /proc/cpuinfo
#!cat /proc/meminfo
#!ls /proc/*info
# this will take some time and show an error.
# dont worry, just carry on
!apt install -y mysql-server > /dev/null
#!ls /etc/init.d
!/etc/init.d/mysql restart
#!gdown https://drive.google.com/uc?id=1Ik3QuYTd52M5qI9D4aOCfGcQ7v2AB9wx
#!wget https://sp.mysqltutorial.org/wp-content/uploads/2018/03/mysqlsampledatabase.zip
#!wget https://github.com/Praxis-QR/RDWH/raw/main/mysqlsampledatabase.zip
!wget -q https://github.com/Praxis-QR/RDWH/raw/main/data/mysqlsampledatabase.zip
!unzip mysqlsampledatabase.zip
#!cat /content/mysqlsampledatabase.sql
!head /content/mysqlsampledatabase.sql
#https://www.mysqltutorial.org/how-to-load-sample-database-into-mysql-database-server.aspx
!mysql --table < mysqlsampledatabase.sql
#New database classicmodels has been created
!mysql -e 'show databases'
!mysql classicmodels -e 'show tables'
!mysql classicmodels -e 'select * from employees'
!apt install libmysqlclient-dev > /dev/null
!pip install mysqlclient
import pandas as pd
import MySQLdb
#To run any non-SELECT SQL command
def runCMD (DDL):
#DBConn= MySQLdb.connect(hostName,userName,passWord,dbName)
DBConn= MySQLdb.connect(db='classicmodels') # since local MySQL server, userid, password not needed
myCursor = DBConn.cursor()
retcode = myCursor.execute(DDL)
print (retcode)
DBConn.commit()
DBConn.close()
#To run any SELECT SQL command
def runSELECT (CMD):
#DBConn= MySQLdb.connect(hostName,userName,passWord,dbName)
DBConn= MySQLdb.connect(db='classicmodels') # since local MySQL server, userid, password not needed
df_mysql = pd.read_sql(CMD, con=DBConn)
DBConn.close()
return df_mysql
!mysql classicmodels -e 'show tables'
#runSELECT('select * from customers limit 5')
#runSELECT('select * from employees limit 5')
#runSELECT('select * from offices limit 5')
#runSELECT('select * from orderdetails limit 5')
#runSELECT('select * from orders limit 5')
#runSELECT('select * from payments limit 5')
#runSELECT('select * from productlines limit 5')
runSELECT('select * from products limit 5')
| 0.162879 | 0.845241 |
This code snippet will test an already trained DQ-DTC agent on the validation profile:
<img src="Figures/Validation_Profile.png" width="600">
```
import numpy as np
from pathlib import Path
import tensorflow as tf
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Dense, Flatten, LeakyReLU, ELU
from tensorflow.keras.optimizers import Adam
from rl.agents.dqn import DQNAgent
from rl.policy import LinearAnnealedPolicy, EpsGreedyQPolicy
from rl.memory import SequentialMemory, EpisodeParameterMemory
from CustomKerasRL2Callbacks_torqueCtrl import StoreEpisodeLogger
from gym.wrappers import FlattenObservation
from gym.core import Wrapper
from gym.spaces import Box, Tuple
import sys, os
import h5py
sys.path.append(os.path.abspath(os.path.join('..')))
import gym_electric_motor as gem
from gym_electric_motor.reward_functions import WeightedSumOfErrors
from gym_electric_motor.physical_systems import ConstantSpeedLoad, ExternalSpeedLoad
from gym_electric_motor.reference_generators import WienerProcessReferenceGenerator, ConstReferenceGenerator, \
MultipleReferenceGenerator, StepReferenceGenerator
def test_profile_speed(t):
"""
This function defines the speed profile of the validation episode.
"""
lim = 12000 * 2 * np.pi / 60
niveau0 = 00
niveau1 = 0.15 * lim
niveau2 = 0.5 * lim
if t <= 0.05:
omega = niveau0
elif t <= 0.20:
omega = (t - 0.05) * (niveau1 - niveau0) / 0.15 + niveau0
elif t <= 1.3:
omega = niveau1
elif t <= 1.45:
omega = (t - 1.3) * -2 * niveau1 / 0.15 + niveau1
elif t <= 2.55:
omega = - niveau1
elif t <= 2.7:
omega = (t - 2.55) * (niveau1 + niveau2) / 0.15 - niveau1
elif t <= 3.8:
omega = niveau2
elif t <= 3.95:
omega = (t - 3.8) * -2 * niveau2 / 0.15 + niveau2
elif t <= 5.05:
omega = - niveau2
elif t <= 5.2:
omega = (t - 5.05) * (niveau0 + niveau2) / 0.15 - niveau2
else:
omega = niveau0
return omega
class TransformObservationWrapper(Wrapper):
"""
The following environment considers the dead time in the real-world motor control systems.
The real-world system changes its state, while the agent calculates the next action based on a previously measured
observation. Therefore, for the agent it seems as if the applied action effects the state one step delayed.
(with a dead time of one time-step)
For complete observability of the system at each time-step we append the last played action of the agent to the
observation, because this action will be the one that is active in the next step.
"""
def __init__(self, environment):
super().__init__(environment)
# reduced observation space [w_me, i_d, i_q, u_d, u_q, cos(eps), sin(eps), T_ref] (all normalized)
self.observation_space = Tuple((Box(
np.concatenate(([environment.observation_space[0].low[0]],
environment.observation_space[0].low[5:7],
environment.observation_space[0].low[10:12],
[-1, -1],
[-1])),
np.concatenate(([environment.observation_space[0].high[0]],
environment.observation_space[0].high[5:7],
environment.observation_space[0].high[10:12],
[+1, +1],
[+1])),
), environment.observation_space[1]))
self.subactions = -np.power(-1, self.env.physical_system._converter._subactions)
# gamma = 0 is assumed for calculating the return G in the test case
self.gamma = 0
self.test = True
def step(self, action):
(state, ref), rew, term, info = self.env.step(action)
self._obs_logger = np.concatenate((state, ref))
eps = state[12] * np.pi
angle_scale = 0.1
angles = [angle_scale * np.cos(eps), angle_scale * np.sin(eps)]
u_abc = self.subactions[action]
u_dq = self.env.physical_system.abc_to_dq_space(u_abc, epsilon_el=eps)
now_requested_voltage = u_dq
i_d = state[5]
i_q = state[6]
T = state[1]
T_ref = ref[0]
current_total = np.sqrt(i_d ** 2 + i_q ** 2)
# building the custom observation vector
observable_state = np.concatenate(([state[0]],
state[5:7],
now_requested_voltage,
angles,
[2 * current_total - 1]))
# as this script is only for testing there is no benefit in defining the reward
reward = None
return (observable_state, ref), rew, term, info
def reset(self, **kwargs):
state, ref = self.env.reset()
self._obs_logger = np.concatenate((state, ref))
eps = state[12] * np.pi
angle_scale = 0.1
angles = [angle_scale * np.cos(eps), angle_scale * np.sin(eps)]
torque_error = [(ref[0] - state[1]) / 2]
u_abc = self.subactions[0]
u_dq = self.env.physical_system.abc_to_dq_space(u_abc, epsilon_el=eps)
now_requested_voltage = u_dq # reduced observation
i_d = state[5]
i_q = state[6]
current_total = np.sqrt(i_d ** 2 + i_q ** 2)
observable_state = np.concatenate(([state[0]],
state[5:7],
now_requested_voltage,
angles,
[2 * current_total - 1])) # reduced observation
return (observable_state, ref)
torque_ref_generator = ConstReferenceGenerator(reference_state='torque', reference_value=np.random.uniform(-1, 1))
motor_parameter = dict(p=3, # [p] = 1, nb of pole pairs
r_s=17.932e-3, # [r_s] = Ohm, stator resistance
l_d=0.37e-3, # [l_d] = H, d-axis inductance
l_q=1.2e-3, # [l_q] = H, q-axis inductance
psi_p=65.65e-3, # [psi_p] = Vs, magnetic flux of the permanent magnet
) # BRUSA
u_sup = 350
nominal_values=dict(omega=12000 * 2 * np.pi / 60,
i=240,
u=u_sup
)
limit_values=nominal_values.copy()
limit_values["i"] = 270
limit_values["torque"] = 200
def test_agent(param_dict):
"""
this function is used to perform one validation episode with predefined network weights
although these weights were saved in beforehand one still needs to initialize a network of the
corresponding form to load the weights into
"""
# unpack the parameters
subfolder_name = param_dict["subfolder_name"]
layers = param_dict["layers"]
neurons = param_dict["neurons"]
activation_fcn = param_dict["activation_fcn"]
activation_fcn_parameter = param_dict["activation_fcn_parameter"]
tf.config.set_visible_devices([], 'GPU')
Path(subfolder_name).mkdir(parents=True, exist_ok=True)
# build the environment for the validation profile
env = gem.make("emotor-pmsm-disc-v1",
motor_parameter=motor_parameter,
nominal_values=nominal_values,
limit_values=limit_values,
u_sup=u_sup,
load=ExternalSpeedLoad(speed_profile=test_profile_speed, # here, the speed profile is implemented
tau=50e-6),
tau=50e-6,
reward_function=WeightedSumOfErrors(observed_states=None,
reward_weights={'torque': 1},
gamma=0),
reference_generator=torque_ref_generator,
ode_solver='scipy.solve_ivp',
dead_time=True
)
(x, r) = env.reset()
tau=env._physical_system.tau
limits = env.physical_system.limits
# wrap the environment for proper
env = FlattenObservation(TransformObservationWrapper(env))
# define the network, has to be the same as in the training profile
# select special procedure for parameterized activations
if activation_fcn == "leaky_relu" or activation_fcn == "elu":
dense_activation_fcn = 'linear'
else:
dense_activation_fcn = activation_fcn
nb_actions = env.action_space.n
window_length = 1
model = Sequential()
model.add(Flatten(input_shape=(window_length,) + env.observation_space.shape))
for i in range(layers):
model.add(Dense(neurons, activation=dense_activation_fcn))
if activation_fcn == 'leaky_relu':
model.add(LeakyReLU(alpha=activation_fcn_parameter))
elif activation_fcn == 'elu':
model.add(ELU(alpha=activation_fcn_parameter))
model.add(Dense(nb_actions,
activation='linear'
))
# memory will not be used in testing episodes, probably one could avoid initializing it
memory = SequentialMemory(limit=0, window_length=window_length)
policy = LinearAnnealedPolicy(EpsGreedyQPolicy(eps=0),
attr='eps',
value_max=0,
value_min=0,
value_test=0, # this is the epsilon used for testing episodes, 0 means deterministic operation
nb_steps=0)
# define the agent
agent = DQNAgent(model=model,
nb_actions=nb_actions,
gamma=0,
batch_size=4,
memory=memory,
memory_interval=1,
policy=policy,
train_interval=1,
target_model_update=0,
enable_double_dqn=False)
# compile the agent and load the weights that were learned during training
agent.compile(Adam(lr=0), metrics=['mse'])
agent.load_weights(filepath=subfolder_name + "/" + "weights.hdf5")
# define the callback for the testing routine
logger = StoreEpisodeLogger(folder_name=subfolder_name,
file_name="DQ_DTC_validation_episode",
tau=tau, limits=limits, training=True,
lr_max=0, lr_min=0,
nb_steps_start=0,
nb_steps_reduction=0,
speed_generator=None,
create_eps_logs=True,
test=True)
callbacks = [logger]
# perform one testing episode, the length of the episode "nb_max_episode_steps" was adjusted to the speed / torque profile
history = agent.test(env,
nb_episodes=1,
action_repetition=1,
verbose=0,
visualize=False,
nb_max_episode_steps=130000,
callbacks=callbacks)
# parameterize the agent
# the "subfolder_name" is the directory where the network weights "weights.hdf5" will be taken from
subfolder_name = "Exemplary_Weights"
# this short block will read out the weights dimensions to set the correct network geometry
# only activation fcn and activation fcn parameter will need to be set manually
with h5py.File(subfolder_name + "/weights.hdf5", "r") as f:
dense = np.copy(f["dense"]["dense"]["kernel:0"])
nb_neurons = np.shape(dense)[1]
keys = list(f.keys())
nb_layers = -1
for key in keys:
if "dense" in key:
nb_layers += 1
param_dict = {"subfolder_name": subfolder_name,
"layers": nb_layers,
"neurons": nb_neurons,
"activation_fcn": "leaky_relu",
"activation_fcn_parameter": 0.3425,
}
# this will run the test episode
# run time depends on the CPU speed, might take more than 20 minutes
# please stay patient although no progress bar is displayed
test_agent(param_dict)
from Plot_TimeDomain_torqueCtrl import plot_episode
# this function will save a pdf of the validation episode to the "Plots" folder
# a "Plots" folder will be created if there is none
plot_episode(training_folder = "Exemplary_Weights",
episode_number = 0,
episode_type = "DQ_DTC_validation_episode")
```
|
github_jupyter
|
import numpy as np
from pathlib import Path
import tensorflow as tf
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Dense, Flatten, LeakyReLU, ELU
from tensorflow.keras.optimizers import Adam
from rl.agents.dqn import DQNAgent
from rl.policy import LinearAnnealedPolicy, EpsGreedyQPolicy
from rl.memory import SequentialMemory, EpisodeParameterMemory
from CustomKerasRL2Callbacks_torqueCtrl import StoreEpisodeLogger
from gym.wrappers import FlattenObservation
from gym.core import Wrapper
from gym.spaces import Box, Tuple
import sys, os
import h5py
sys.path.append(os.path.abspath(os.path.join('..')))
import gym_electric_motor as gem
from gym_electric_motor.reward_functions import WeightedSumOfErrors
from gym_electric_motor.physical_systems import ConstantSpeedLoad, ExternalSpeedLoad
from gym_electric_motor.reference_generators import WienerProcessReferenceGenerator, ConstReferenceGenerator, \
MultipleReferenceGenerator, StepReferenceGenerator
def test_profile_speed(t):
"""
This function defines the speed profile of the validation episode.
"""
lim = 12000 * 2 * np.pi / 60
niveau0 = 00
niveau1 = 0.15 * lim
niveau2 = 0.5 * lim
if t <= 0.05:
omega = niveau0
elif t <= 0.20:
omega = (t - 0.05) * (niveau1 - niveau0) / 0.15 + niveau0
elif t <= 1.3:
omega = niveau1
elif t <= 1.45:
omega = (t - 1.3) * -2 * niveau1 / 0.15 + niveau1
elif t <= 2.55:
omega = - niveau1
elif t <= 2.7:
omega = (t - 2.55) * (niveau1 + niveau2) / 0.15 - niveau1
elif t <= 3.8:
omega = niveau2
elif t <= 3.95:
omega = (t - 3.8) * -2 * niveau2 / 0.15 + niveau2
elif t <= 5.05:
omega = - niveau2
elif t <= 5.2:
omega = (t - 5.05) * (niveau0 + niveau2) / 0.15 - niveau2
else:
omega = niveau0
return omega
class TransformObservationWrapper(Wrapper):
"""
The following environment considers the dead time in the real-world motor control systems.
The real-world system changes its state, while the agent calculates the next action based on a previously measured
observation. Therefore, for the agent it seems as if the applied action effects the state one step delayed.
(with a dead time of one time-step)
For complete observability of the system at each time-step we append the last played action of the agent to the
observation, because this action will be the one that is active in the next step.
"""
def __init__(self, environment):
super().__init__(environment)
# reduced observation space [w_me, i_d, i_q, u_d, u_q, cos(eps), sin(eps), T_ref] (all normalized)
self.observation_space = Tuple((Box(
np.concatenate(([environment.observation_space[0].low[0]],
environment.observation_space[0].low[5:7],
environment.observation_space[0].low[10:12],
[-1, -1],
[-1])),
np.concatenate(([environment.observation_space[0].high[0]],
environment.observation_space[0].high[5:7],
environment.observation_space[0].high[10:12],
[+1, +1],
[+1])),
), environment.observation_space[1]))
self.subactions = -np.power(-1, self.env.physical_system._converter._subactions)
# gamma = 0 is assumed for calculating the return G in the test case
self.gamma = 0
self.test = True
def step(self, action):
(state, ref), rew, term, info = self.env.step(action)
self._obs_logger = np.concatenate((state, ref))
eps = state[12] * np.pi
angle_scale = 0.1
angles = [angle_scale * np.cos(eps), angle_scale * np.sin(eps)]
u_abc = self.subactions[action]
u_dq = self.env.physical_system.abc_to_dq_space(u_abc, epsilon_el=eps)
now_requested_voltage = u_dq
i_d = state[5]
i_q = state[6]
T = state[1]
T_ref = ref[0]
current_total = np.sqrt(i_d ** 2 + i_q ** 2)
# building the custom observation vector
observable_state = np.concatenate(([state[0]],
state[5:7],
now_requested_voltage,
angles,
[2 * current_total - 1]))
# as this script is only for testing there is no benefit in defining the reward
reward = None
return (observable_state, ref), rew, term, info
def reset(self, **kwargs):
state, ref = self.env.reset()
self._obs_logger = np.concatenate((state, ref))
eps = state[12] * np.pi
angle_scale = 0.1
angles = [angle_scale * np.cos(eps), angle_scale * np.sin(eps)]
torque_error = [(ref[0] - state[1]) / 2]
u_abc = self.subactions[0]
u_dq = self.env.physical_system.abc_to_dq_space(u_abc, epsilon_el=eps)
now_requested_voltage = u_dq # reduced observation
i_d = state[5]
i_q = state[6]
current_total = np.sqrt(i_d ** 2 + i_q ** 2)
observable_state = np.concatenate(([state[0]],
state[5:7],
now_requested_voltage,
angles,
[2 * current_total - 1])) # reduced observation
return (observable_state, ref)
torque_ref_generator = ConstReferenceGenerator(reference_state='torque', reference_value=np.random.uniform(-1, 1))
motor_parameter = dict(p=3, # [p] = 1, nb of pole pairs
r_s=17.932e-3, # [r_s] = Ohm, stator resistance
l_d=0.37e-3, # [l_d] = H, d-axis inductance
l_q=1.2e-3, # [l_q] = H, q-axis inductance
psi_p=65.65e-3, # [psi_p] = Vs, magnetic flux of the permanent magnet
) # BRUSA
u_sup = 350
nominal_values=dict(omega=12000 * 2 * np.pi / 60,
i=240,
u=u_sup
)
limit_values=nominal_values.copy()
limit_values["i"] = 270
limit_values["torque"] = 200
def test_agent(param_dict):
"""
this function is used to perform one validation episode with predefined network weights
although these weights were saved in beforehand one still needs to initialize a network of the
corresponding form to load the weights into
"""
# unpack the parameters
subfolder_name = param_dict["subfolder_name"]
layers = param_dict["layers"]
neurons = param_dict["neurons"]
activation_fcn = param_dict["activation_fcn"]
activation_fcn_parameter = param_dict["activation_fcn_parameter"]
tf.config.set_visible_devices([], 'GPU')
Path(subfolder_name).mkdir(parents=True, exist_ok=True)
# build the environment for the validation profile
env = gem.make("emotor-pmsm-disc-v1",
motor_parameter=motor_parameter,
nominal_values=nominal_values,
limit_values=limit_values,
u_sup=u_sup,
load=ExternalSpeedLoad(speed_profile=test_profile_speed, # here, the speed profile is implemented
tau=50e-6),
tau=50e-6,
reward_function=WeightedSumOfErrors(observed_states=None,
reward_weights={'torque': 1},
gamma=0),
reference_generator=torque_ref_generator,
ode_solver='scipy.solve_ivp',
dead_time=True
)
(x, r) = env.reset()
tau=env._physical_system.tau
limits = env.physical_system.limits
# wrap the environment for proper
env = FlattenObservation(TransformObservationWrapper(env))
# define the network, has to be the same as in the training profile
# select special procedure for parameterized activations
if activation_fcn == "leaky_relu" or activation_fcn == "elu":
dense_activation_fcn = 'linear'
else:
dense_activation_fcn = activation_fcn
nb_actions = env.action_space.n
window_length = 1
model = Sequential()
model.add(Flatten(input_shape=(window_length,) + env.observation_space.shape))
for i in range(layers):
model.add(Dense(neurons, activation=dense_activation_fcn))
if activation_fcn == 'leaky_relu':
model.add(LeakyReLU(alpha=activation_fcn_parameter))
elif activation_fcn == 'elu':
model.add(ELU(alpha=activation_fcn_parameter))
model.add(Dense(nb_actions,
activation='linear'
))
# memory will not be used in testing episodes, probably one could avoid initializing it
memory = SequentialMemory(limit=0, window_length=window_length)
policy = LinearAnnealedPolicy(EpsGreedyQPolicy(eps=0),
attr='eps',
value_max=0,
value_min=0,
value_test=0, # this is the epsilon used for testing episodes, 0 means deterministic operation
nb_steps=0)
# define the agent
agent = DQNAgent(model=model,
nb_actions=nb_actions,
gamma=0,
batch_size=4,
memory=memory,
memory_interval=1,
policy=policy,
train_interval=1,
target_model_update=0,
enable_double_dqn=False)
# compile the agent and load the weights that were learned during training
agent.compile(Adam(lr=0), metrics=['mse'])
agent.load_weights(filepath=subfolder_name + "/" + "weights.hdf5")
# define the callback for the testing routine
logger = StoreEpisodeLogger(folder_name=subfolder_name,
file_name="DQ_DTC_validation_episode",
tau=tau, limits=limits, training=True,
lr_max=0, lr_min=0,
nb_steps_start=0,
nb_steps_reduction=0,
speed_generator=None,
create_eps_logs=True,
test=True)
callbacks = [logger]
# perform one testing episode, the length of the episode "nb_max_episode_steps" was adjusted to the speed / torque profile
history = agent.test(env,
nb_episodes=1,
action_repetition=1,
verbose=0,
visualize=False,
nb_max_episode_steps=130000,
callbacks=callbacks)
# parameterize the agent
# the "subfolder_name" is the directory where the network weights "weights.hdf5" will be taken from
subfolder_name = "Exemplary_Weights"
# this short block will read out the weights dimensions to set the correct network geometry
# only activation fcn and activation fcn parameter will need to be set manually
with h5py.File(subfolder_name + "/weights.hdf5", "r") as f:
dense = np.copy(f["dense"]["dense"]["kernel:0"])
nb_neurons = np.shape(dense)[1]
keys = list(f.keys())
nb_layers = -1
for key in keys:
if "dense" in key:
nb_layers += 1
param_dict = {"subfolder_name": subfolder_name,
"layers": nb_layers,
"neurons": nb_neurons,
"activation_fcn": "leaky_relu",
"activation_fcn_parameter": 0.3425,
}
# this will run the test episode
# run time depends on the CPU speed, might take more than 20 minutes
# please stay patient although no progress bar is displayed
test_agent(param_dict)
from Plot_TimeDomain_torqueCtrl import plot_episode
# this function will save a pdf of the validation episode to the "Plots" folder
# a "Plots" folder will be created if there is none
plot_episode(training_folder = "Exemplary_Weights",
episode_number = 0,
episode_type = "DQ_DTC_validation_episode")
| 0.649467 | 0.878366 |
# A Brief Introduction to Jupyter Notebooks
Jupyter Notebooks combine an execution environment for R/Python/Julia/Haskell with written instructions/documentation/descriptions as markdown. They are organized in _cells_, and each cell has a type. For use, two types of cells are relevant, Markdown and Code.
## Markdown Cells
The Markdown cells contain textual information that is formatted using Markdown. An explanation for how Markdown works can be found as part of the [Juypter Documentation](https://jupyter-notebook.readthedocs.io/en/stable/examples/Notebook/Working%20With%20Markdown%20Cells.html). For this exercise, we use the Markdown cells to provide explanations, define tasks, and ask questions.
## Code Cells
The code cells contain exectuable code. Each cell is executed on its own and there is no fixed order for the execution. To execute the code in a cell, you simply have to click on the _Run_ button at the top of the page or hit Ctrl+Enter. Code has to be written in R, Python, Julia, or Haskell. All cells within the same notebook must use the same language, which is defined by the kernel of the notebook. We provide the notebooks to you with the Python kernel enabled. You may switch to a different kernel using the menu bar by clicking on Kernel-->Change Kernel and then selecting the language of your choice.
## The internal state
While the cells are executed on their own, they all share the same state. For example, if you load a library in one cell, it will be available in all cells. Similarly, if you store something in a variable, this variable will be globally accessible from all other cells. The state can be reset by restarting the kernel. This usually happens automatically, when you re-open the notebook. You can also trigger this manually using the menu bar by clicking on Kernel-->Restart.
## The Output
The output of a code cell appears below the cell. By default, the result of the last executed line is printed. Other textual outputs can be generated by using the print-commands of the programming languange. The generation of plots and similar elements will be covered in this exercise.
## Hello World
Below this cell, you find a code cell that contains the code for printing "Hello World" in Python. Execute the cell and see what happens.
```
print("Hello World")
```
You actually do not require the print to achieve the same result, because the return value of the final line is printed automatically. The cell below accomplishes (almost) the same thing.
```
"Hello World"
```
# Programming in this Exercise
You may solve the problems in whichever programming language you desire. You may or may not use Jupyter Notebooks. Within this exercise, we are interested in the interpretation of results, not the programming to achieve this. We give general guidance on how to solve problems with Python as part the the descriptions of the exercises. However, other languages, especially R, are also suited for solving the exerices. However, in case you have programming problems, we will only help you (within reasonable limits) if you use Python.
Please be reminded that these exercises are primarily designed for Computer Science M.Sc. students. Thus, we assume that all students poses basic programming skills and are able to learn new programming languages on their own. We will also usually not comment on you code quality. In case you have no experience in programming (at all) you may find this exercise difficult. If you only have experience with other languages, but not Python, you should be able to solve all tasks using help from the internet without problems. Google and StackOverflow are your friends.
|
github_jupyter
|
print("Hello World")
"Hello World"
| 0.19433 | 0.941007 |
# K-means with Silhouette analysis
***
- 我們試著以輪廓分析 (Silhouette analysis) 來觀察 K-mean 分群時不同 K 值的比較
```
# 載入套件
import numpy as np
import matplotlib
import matplotlib.pyplot as plt
import matplotlib.cm as cm
from sklearn.cluster import KMeans
from sklearn import datasets
from sklearn.metrics import silhouette_samples, silhouette_score
np.random.seed(5)
%matplotlib inline
# 載入 iris 資料集
iris = datasets.load_iris()
X = iris.data
y = iris.target
# 設定需要計算的 K 值集合
range_n_clusters = [2, 3, 4, 5, 6, 7, 8]
# 計算並繪製輪廓分析的結果
for n_clusters in range_n_clusters:
# Create a subplot with 1 row and 2 columns
fig, (ax1, ax2) = plt.subplots(1, 2)
fig.set_size_inches(18, 7)
# The 1st subplot is the silhouette plot
# The silhouette coefficient can range from -1, 1 but in this example all
# lie within [-0.1, 1]
ax1.set_xlim([-0.1, 1])
# The (n_clusters+1)*10 is for inserting blank space between silhouette
# plots of individual clusters, to demarcate them clearly.
ax1.set_ylim([0, len(X) + (n_clusters + 1) * 10])
# Initialize the clusterer with n_clusters value and a random generator
# seed of 10 for reproducibility.
clusterer = KMeans(n_clusters=n_clusters, random_state=10)
cluster_labels = clusterer.fit_predict(X)
# The silhouette_score gives the average value for all the samples.
# This gives a perspective into the density and separation of the formed
# clusters
silhouette_avg = silhouette_score(X, cluster_labels)
print("For n_clusters =", n_clusters,
"The average silhouette_score is :", silhouette_avg)
# Compute the silhouette scores for each sample
sample_silhouette_values = silhouette_samples(X, cluster_labels)
y_lower = 10
for i in range(n_clusters):
# Aggregate the silhouette scores for samples belonging to
# cluster i, and sort them
ith_cluster_silhouette_values = \
sample_silhouette_values[cluster_labels == i]
ith_cluster_silhouette_values.sort()
size_cluster_i = ith_cluster_silhouette_values.shape[0]
y_upper = y_lower + size_cluster_i
color = cm.nipy_spectral(float(i) / n_clusters)
ax1.fill_betweenx(np.arange(y_lower, y_upper),
0, ith_cluster_silhouette_values,
facecolor=color, edgecolor=color, alpha=0.7)
# Label the silhouette plots with their cluster numbers at the middle
ax1.text(-0.05, y_lower + 0.5 * size_cluster_i, str(i))
# Compute the new y_lower for next plot
y_lower = y_upper + 10 # 10 for the 0 samples
ax1.set_title("The silhouette plot for the various clusters.")
ax1.set_xlabel("The silhouette coefficient values")
ax1.set_ylabel("Cluster label")
# The vertical line for average silhouette score of all the values
ax1.axvline(x=silhouette_avg, color="red", linestyle="--")
ax1.set_yticks([]) # Clear the yaxis labels / ticks
ax1.set_xticks([-0.1, 0, 0.2, 0.4, 0.6, 0.8, 1])
# 2nd Plot showing the actual clusters formed
colors = cm.nipy_spectral(cluster_labels.astype(float) / n_clusters)
ax2.scatter(X[:, 0], X[:, 1], marker='.', s=30, lw=0, alpha=0.7,
c=colors, edgecolor='k')
# Labeling the clusters
centers = clusterer.cluster_centers_
# Draw white circles at cluster centers
ax2.scatter(centers[:, 0], centers[:, 1], marker='o',
c="white", alpha=1, s=200, edgecolor='k')
for i, c in enumerate(centers):
ax2.scatter(c[0], c[1], marker='$%d$' % i, alpha=1,
s=50, edgecolor='k')
ax2.set_title("The visualization of the clustered data.")
ax2.set_xlabel("Feature space for the 1st feature")
ax2.set_ylabel("Feature space for the 2nd feature")
plt.suptitle(("Silhouette analysis for KMeans clustering on sample data "
"with n_clusters = %d" % n_clusters),
fontsize=14, fontweight='bold')
plt.show()
```
# 分析結果解說
* 觀察輸出值:silhouette_score, 如果是一個適合的分群值, 應該要比下一個分群值的分數大很多
* 由結果可以看出 : 2, 3, 5 都是不錯的分群值(因為比 3, 4, 6的分數都高很多), 相形之下, 4, 6, 7 作為分群的效果就不明顯
# 作業
* 試著模擬出 5 群高斯分布的資料, 並以此觀察 K-mean 與輪廓分析的結果
|
github_jupyter
|
# 載入套件
import numpy as np
import matplotlib
import matplotlib.pyplot as plt
import matplotlib.cm as cm
from sklearn.cluster import KMeans
from sklearn import datasets
from sklearn.metrics import silhouette_samples, silhouette_score
np.random.seed(5)
%matplotlib inline
# 載入 iris 資料集
iris = datasets.load_iris()
X = iris.data
y = iris.target
# 設定需要計算的 K 值集合
range_n_clusters = [2, 3, 4, 5, 6, 7, 8]
# 計算並繪製輪廓分析的結果
for n_clusters in range_n_clusters:
# Create a subplot with 1 row and 2 columns
fig, (ax1, ax2) = plt.subplots(1, 2)
fig.set_size_inches(18, 7)
# The 1st subplot is the silhouette plot
# The silhouette coefficient can range from -1, 1 but in this example all
# lie within [-0.1, 1]
ax1.set_xlim([-0.1, 1])
# The (n_clusters+1)*10 is for inserting blank space between silhouette
# plots of individual clusters, to demarcate them clearly.
ax1.set_ylim([0, len(X) + (n_clusters + 1) * 10])
# Initialize the clusterer with n_clusters value and a random generator
# seed of 10 for reproducibility.
clusterer = KMeans(n_clusters=n_clusters, random_state=10)
cluster_labels = clusterer.fit_predict(X)
# The silhouette_score gives the average value for all the samples.
# This gives a perspective into the density and separation of the formed
# clusters
silhouette_avg = silhouette_score(X, cluster_labels)
print("For n_clusters =", n_clusters,
"The average silhouette_score is :", silhouette_avg)
# Compute the silhouette scores for each sample
sample_silhouette_values = silhouette_samples(X, cluster_labels)
y_lower = 10
for i in range(n_clusters):
# Aggregate the silhouette scores for samples belonging to
# cluster i, and sort them
ith_cluster_silhouette_values = \
sample_silhouette_values[cluster_labels == i]
ith_cluster_silhouette_values.sort()
size_cluster_i = ith_cluster_silhouette_values.shape[0]
y_upper = y_lower + size_cluster_i
color = cm.nipy_spectral(float(i) / n_clusters)
ax1.fill_betweenx(np.arange(y_lower, y_upper),
0, ith_cluster_silhouette_values,
facecolor=color, edgecolor=color, alpha=0.7)
# Label the silhouette plots with their cluster numbers at the middle
ax1.text(-0.05, y_lower + 0.5 * size_cluster_i, str(i))
# Compute the new y_lower for next plot
y_lower = y_upper + 10 # 10 for the 0 samples
ax1.set_title("The silhouette plot for the various clusters.")
ax1.set_xlabel("The silhouette coefficient values")
ax1.set_ylabel("Cluster label")
# The vertical line for average silhouette score of all the values
ax1.axvline(x=silhouette_avg, color="red", linestyle="--")
ax1.set_yticks([]) # Clear the yaxis labels / ticks
ax1.set_xticks([-0.1, 0, 0.2, 0.4, 0.6, 0.8, 1])
# 2nd Plot showing the actual clusters formed
colors = cm.nipy_spectral(cluster_labels.astype(float) / n_clusters)
ax2.scatter(X[:, 0], X[:, 1], marker='.', s=30, lw=0, alpha=0.7,
c=colors, edgecolor='k')
# Labeling the clusters
centers = clusterer.cluster_centers_
# Draw white circles at cluster centers
ax2.scatter(centers[:, 0], centers[:, 1], marker='o',
c="white", alpha=1, s=200, edgecolor='k')
for i, c in enumerate(centers):
ax2.scatter(c[0], c[1], marker='$%d$' % i, alpha=1,
s=50, edgecolor='k')
ax2.set_title("The visualization of the clustered data.")
ax2.set_xlabel("Feature space for the 1st feature")
ax2.set_ylabel("Feature space for the 2nd feature")
plt.suptitle(("Silhouette analysis for KMeans clustering on sample data "
"with n_clusters = %d" % n_clusters),
fontsize=14, fontweight='bold')
plt.show()
| 0.718002 | 0.873862 |
```
import csv
import datetime
import h5py
import mir_eval
import numpy as np
import os
import pandas as pd
import sys
import time
sys.path.append('../src')
import localmodule
# Define constants.
data_dir = localmodule.get_data_dir()
dataset_name = localmodule.get_dataset_name()
models_dir = localmodule.get_models_dir()
negative_labels = localmodule.get_negative_labels()
tolerances = localmodule.get_tolerances()
n_thresholds = 100
# Read command-line arguments. ENABLE
#args = sys.argv[1:]
#unit_str = args[0]
#odf_str = args[1]
#suppressor_str = args[2]
unit_str = "unit01"
odf_str = "tseep"
clip_suppressor_str = "clip-suppressor"
# Print header.
start_time = int(time.time())
print(str(datetime.datetime.now()) + " Start.")
print("Evaluating Old Bird on " + dataset_name + ", " + unit_str + ".")
print('h5py version: {:s}'.format(h5py.__version__))
print('mir_eval version: {:s}'.format(mir_eval.__version__))
print('numpy version: {:s}'.format(np.__version__))
print('pandas version: {:s}'.format(pd.__version__))
print("")
# Define directory for predictions.
oldbird_models_dir = os.path.join(models_dir, "oldbird")
unit_dir = os.path.join(oldbird_models_dir, unit_str)
predictions_name = "_".join(["predictions", clip_suppressor_str])
predictions_dir = os.path.join(unit_dir, predictions_name)
# Open annotation as Pandas DataFrame.
annotations_name = "_".join([dataset_name, "annotations"])
annotations_dir = os.path.join(data_dir, annotations_name)
annotation_name = unit_str + ".txt"
annotation_path = os.path.join(annotations_dir, annotation_name)
ann_df = pd.read_csv(annotation_path, delimiter="\t")
# Restrict rows to negative labels.
if "Calls" in ann_df.columns:
ann_df = ann_df.loc[~ann_df["Calls"].isin(negative_labels)]
# Restrict rows to frequency range of interest.
if odf_str in ["thrush", "tseep"]:
oldbird_data_name = "_".join([dataset_name, "oldbird"])
oldbird_data_dir = os.path.join(data_dir, oldbird_data_name)
oldbird_data_path = os.path.join(oldbird_data_dir, unit_str + ".hdf5")
oldbird_hdf5 = h5py.File(oldbird_data_path, "r")
settings_key = "_".join([odf_str, "settings"])
settings = oldbird_hdf5[settings_key]
filter_f0 = settings["filter_f0"].value
filter_f1 = settings["filter_f1"].value
ann_df = ann_df[
((0.5*(ann_df["Low Freq (Hz)"]+ann_df["High Freq (Hz)"])) > filter_f0) &
((0.5*(ann_df["Low Freq (Hz)"]+ann_df["High Freq (Hz)"])) < filter_f1)]
filter_f1
# Load middle times of true events.
begin_times = np.array(ann_df["Begin Time (s)"])
end_times = np.array(ann_df["End Time (s)"])
relevant = 0.5 * (begin_times+end_times)
n_relevant = len(relevant)
# Prepare header for metrics.
csv_header = [
"Dataset", "Unit", "ODF", "Clip suppressor", "Tolerance",
"Threshold ID", "Relevant", "Selected", "True positives",
"False positives", "False negatives", "Precision", "Recall", "F1 Score"]
# Loop over tolerances.
for tolerance in tolerances:
# Create a CSV file for metrics.
tolerance_str = "tol-" + str(int(np.round(1000*tolerance)))
csv_file_name = "_".join([dataset_name, "oldbird", odf_str,
clip_suppressor_str, unit_str, tolerance_str, "metrics.csv"])
csv_file_path = os.path.join(unit_dir, csv_file_name)
csv_file = open(csv_file_path, 'w')
csv_writer = csv.writer(csv_file, delimiter=',')
csv_writer.writerow(csv_header)
# Loop over thresholds.
for threshold_id in range(n_thresholds):
# Load middle times of prediction.
threshold_str = "th-" + str(threshold_id).zfill(2)
prediction_name_components = [dataset_name, "oldbird", odf_str,
unit_str, threshold_str, "predictions"]
if clip_suppressor_str == "clip-suppressor":
prediction_name_components.append(clip_suppressor_str)
prediction_name = "_".join(prediction_name_components) + ".csv"
prediction_path = os.path.join(predictions_dir, prediction_name)
prediction_df = pd.read_csv(prediction_path)
selected = prediction_df["Time (s)"]
# Match selected events with relevant events using the mir_eval toolbox.
selected_relevant = mir_eval.util.match_events(
relevant, selected, tolerance)
# Define metrics.
true_positives = len(selected_relevant)
n_selected = len(selected)
false_positives = n_selected - true_positives
false_negatives = n_relevant - true_positives
if n_selected == 0 or true_positives == 0:
precision = 0.0
recall = 0.0
f1_score = 0.0
else:
precision = 100 * true_positives / n_selected
recall = 100 * true_positives / n_relevant
f1_score = 2*precision*recall / (precision+recall)
# Write row.
row = [
dataset_name,
unit_str,
clip_suppressor_str,
str(int(np.round(1000*tolerance))).rjust(4),
threshold_str,
str(n_relevant).rjust(5),
str(n_selected).rjust(6),
str(true_positives).rjust(5),
str(false_positives).rjust(5),
str(false_negatives).rjust(5),
format(precision, ".6f"),
format(recall, ".6f"),
format(f1_score, ".6f")
]
csv_writer.writerow(row)
# Close CSV file.
csv_file.close()
# Print elapsed time.
print(str(datetime.datetime.now()) + " Finish.")
elapsed_time = time.time() - int(start_time)
elapsed_hours = int(elapsed_time / (60 * 60))
elapsed_minutes = int((elapsed_time % (60 * 60)) / 60)
elapsed_seconds = elapsed_time % 60.
elapsed_str = "{:>02}:{:>02}:{:>05.2f}".format(elapsed_hours,
elapsed_minutes,
elapsed_seconds)
print("Total elapsed time: " + elapsed_str + ".")
csv_file_path
```
|
github_jupyter
|
import csv
import datetime
import h5py
import mir_eval
import numpy as np
import os
import pandas as pd
import sys
import time
sys.path.append('../src')
import localmodule
# Define constants.
data_dir = localmodule.get_data_dir()
dataset_name = localmodule.get_dataset_name()
models_dir = localmodule.get_models_dir()
negative_labels = localmodule.get_negative_labels()
tolerances = localmodule.get_tolerances()
n_thresholds = 100
# Read command-line arguments. ENABLE
#args = sys.argv[1:]
#unit_str = args[0]
#odf_str = args[1]
#suppressor_str = args[2]
unit_str = "unit01"
odf_str = "tseep"
clip_suppressor_str = "clip-suppressor"
# Print header.
start_time = int(time.time())
print(str(datetime.datetime.now()) + " Start.")
print("Evaluating Old Bird on " + dataset_name + ", " + unit_str + ".")
print('h5py version: {:s}'.format(h5py.__version__))
print('mir_eval version: {:s}'.format(mir_eval.__version__))
print('numpy version: {:s}'.format(np.__version__))
print('pandas version: {:s}'.format(pd.__version__))
print("")
# Define directory for predictions.
oldbird_models_dir = os.path.join(models_dir, "oldbird")
unit_dir = os.path.join(oldbird_models_dir, unit_str)
predictions_name = "_".join(["predictions", clip_suppressor_str])
predictions_dir = os.path.join(unit_dir, predictions_name)
# Open annotation as Pandas DataFrame.
annotations_name = "_".join([dataset_name, "annotations"])
annotations_dir = os.path.join(data_dir, annotations_name)
annotation_name = unit_str + ".txt"
annotation_path = os.path.join(annotations_dir, annotation_name)
ann_df = pd.read_csv(annotation_path, delimiter="\t")
# Restrict rows to negative labels.
if "Calls" in ann_df.columns:
ann_df = ann_df.loc[~ann_df["Calls"].isin(negative_labels)]
# Restrict rows to frequency range of interest.
if odf_str in ["thrush", "tseep"]:
oldbird_data_name = "_".join([dataset_name, "oldbird"])
oldbird_data_dir = os.path.join(data_dir, oldbird_data_name)
oldbird_data_path = os.path.join(oldbird_data_dir, unit_str + ".hdf5")
oldbird_hdf5 = h5py.File(oldbird_data_path, "r")
settings_key = "_".join([odf_str, "settings"])
settings = oldbird_hdf5[settings_key]
filter_f0 = settings["filter_f0"].value
filter_f1 = settings["filter_f1"].value
ann_df = ann_df[
((0.5*(ann_df["Low Freq (Hz)"]+ann_df["High Freq (Hz)"])) > filter_f0) &
((0.5*(ann_df["Low Freq (Hz)"]+ann_df["High Freq (Hz)"])) < filter_f1)]
filter_f1
# Load middle times of true events.
begin_times = np.array(ann_df["Begin Time (s)"])
end_times = np.array(ann_df["End Time (s)"])
relevant = 0.5 * (begin_times+end_times)
n_relevant = len(relevant)
# Prepare header for metrics.
csv_header = [
"Dataset", "Unit", "ODF", "Clip suppressor", "Tolerance",
"Threshold ID", "Relevant", "Selected", "True positives",
"False positives", "False negatives", "Precision", "Recall", "F1 Score"]
# Loop over tolerances.
for tolerance in tolerances:
# Create a CSV file for metrics.
tolerance_str = "tol-" + str(int(np.round(1000*tolerance)))
csv_file_name = "_".join([dataset_name, "oldbird", odf_str,
clip_suppressor_str, unit_str, tolerance_str, "metrics.csv"])
csv_file_path = os.path.join(unit_dir, csv_file_name)
csv_file = open(csv_file_path, 'w')
csv_writer = csv.writer(csv_file, delimiter=',')
csv_writer.writerow(csv_header)
# Loop over thresholds.
for threshold_id in range(n_thresholds):
# Load middle times of prediction.
threshold_str = "th-" + str(threshold_id).zfill(2)
prediction_name_components = [dataset_name, "oldbird", odf_str,
unit_str, threshold_str, "predictions"]
if clip_suppressor_str == "clip-suppressor":
prediction_name_components.append(clip_suppressor_str)
prediction_name = "_".join(prediction_name_components) + ".csv"
prediction_path = os.path.join(predictions_dir, prediction_name)
prediction_df = pd.read_csv(prediction_path)
selected = prediction_df["Time (s)"]
# Match selected events with relevant events using the mir_eval toolbox.
selected_relevant = mir_eval.util.match_events(
relevant, selected, tolerance)
# Define metrics.
true_positives = len(selected_relevant)
n_selected = len(selected)
false_positives = n_selected - true_positives
false_negatives = n_relevant - true_positives
if n_selected == 0 or true_positives == 0:
precision = 0.0
recall = 0.0
f1_score = 0.0
else:
precision = 100 * true_positives / n_selected
recall = 100 * true_positives / n_relevant
f1_score = 2*precision*recall / (precision+recall)
# Write row.
row = [
dataset_name,
unit_str,
clip_suppressor_str,
str(int(np.round(1000*tolerance))).rjust(4),
threshold_str,
str(n_relevant).rjust(5),
str(n_selected).rjust(6),
str(true_positives).rjust(5),
str(false_positives).rjust(5),
str(false_negatives).rjust(5),
format(precision, ".6f"),
format(recall, ".6f"),
format(f1_score, ".6f")
]
csv_writer.writerow(row)
# Close CSV file.
csv_file.close()
# Print elapsed time.
print(str(datetime.datetime.now()) + " Finish.")
elapsed_time = time.time() - int(start_time)
elapsed_hours = int(elapsed_time / (60 * 60))
elapsed_minutes = int((elapsed_time % (60 * 60)) / 60)
elapsed_seconds = elapsed_time % 60.
elapsed_str = "{:>02}:{:>02}:{:>05.2f}".format(elapsed_hours,
elapsed_minutes,
elapsed_seconds)
print("Total elapsed time: " + elapsed_str + ".")
csv_file_path
| 0.418459 | 0.154249 |
## Coding Exercise #0205
### 1. Pandas DataFrame basics:
```
import pandas as pd
import numpy as np
import os
```
#### 1.1. Creating a new DataFrame:
From a dictionary:
```
data = { 'NAME' : ['Jake', 'Jennifer', 'Paul', 'Andrew'], 'AGE': [24,21,25,19], 'GENDER':['M','F','M','M']}
df = pd.DataFrame(data)
df
```
From a NumPy array:
```
df = pd.DataFrame(np.random.rand(10,5), columns=['A','B','C','D','E'])
df.head(3)
```
#### 1.2. Reading data into a DataFrame:
```
!wget --no-clobber https://raw.githubusercontent.com/stefannae/SIC-Artificial-Intelligence/main/SIC_AI_Coding_Exercises/SIC_AI_Chapter_03_Coding_Exercises/data_studentlist.csv
df = pd.read_csv('data_studentlist.csv', header='infer')
```
Check for some of the DataFrame attributes:
```
df.shape
df.size
df.ndim
df.columns
df.index
type(df)
```
Summarize the DataFrame:
```
df.info()
df.describe()
```
Show the head and tail parts:
```
df.head(3)
df.tail(3)
```
Replacing the 'columns' (header):
```
header = df.columns
header
df.columns = ['NAME', 'GENDER' , 'AGE', 'GRADE', 'ABSENCE', 'BLOODTYPE', 'HEIGHT', 'WEIGHT']
df.head(3)
```
#### 1.3. Indexing and slicing DataFrames:
```
# A column.
df.NAME
# This is in fact a Series:
type(df.NAME)
# This is also a Series:
df['NAME']
# However, this is a DataFrame with one column, not a Series:
df[['NAME']]
# Another way of getting a columns as Series:
df.loc[:,'NAME']
df.loc[:,['NAME','GENDER']]
df.iloc[:,[0,1]]
df.loc[:,(header =='NAME') | (header == 'GENDER')]
# This is a row.
df.loc[2]
df.loc[2:4]
df.iloc[2:4]
df.drop(columns=['NAME','GENDER']) # This is just a view.
#df.drop(columns=['NAME','GENDER'],inplace=True) # => To remove permanently.
df.loc[:, (header!='NAME') & (header!='GENDER')]
```
Conditional slicing:
```
# Males only.
df[df.GENDER == 'M']
# Only non-males.
df[-(df.GENDER == 'M')]
df[df.GENDER == 'F']
df[df.HEIGHT > 170]
df[ (df.WEIGHT > 70) & (df.WEIGHT < 80)]
df[ (df.GENDER == 'M') & (df.HEIGHT > 175)]
# Opposite criteria to the previous one.
df[ -((df.GENDER == 'M') & (df.HEIGHT > 175))]
```
#### 1.4. File reading and writing:
Read and write in the CSV format:
```
df2 = df.drop(columns=['GRADE','ABSENCE'])
df2.to_csv('data_mine.csv',index=False)
df3 = pd.read_csv('data_mine.csv',encoding='latin1',header='infer')
df3.head(3)
```
Read and write as a Excel document:
```
!wget --no-clobber https://github.com/stefannae/SIC-Artificial-Intelligence/blob/main/SIC_AI_Coding_Exercises/SIC_AI_Chapter_03_Coding_Exercises/data_studentlist.xlsx
dfx = pd.read_excel('data_studentlist.xlsx')
dfx.head(5)
dfx.to_excel('data_studentlist2.xlsx',sheet_name='NewSheet', index=False)
```
|
github_jupyter
|
import pandas as pd
import numpy as np
import os
data = { 'NAME' : ['Jake', 'Jennifer', 'Paul', 'Andrew'], 'AGE': [24,21,25,19], 'GENDER':['M','F','M','M']}
df = pd.DataFrame(data)
df
df = pd.DataFrame(np.random.rand(10,5), columns=['A','B','C','D','E'])
df.head(3)
!wget --no-clobber https://raw.githubusercontent.com/stefannae/SIC-Artificial-Intelligence/main/SIC_AI_Coding_Exercises/SIC_AI_Chapter_03_Coding_Exercises/data_studentlist.csv
df = pd.read_csv('data_studentlist.csv', header='infer')
df.shape
df.size
df.ndim
df.columns
df.index
type(df)
df.info()
df.describe()
df.head(3)
df.tail(3)
header = df.columns
header
df.columns = ['NAME', 'GENDER' , 'AGE', 'GRADE', 'ABSENCE', 'BLOODTYPE', 'HEIGHT', 'WEIGHT']
df.head(3)
# A column.
df.NAME
# This is in fact a Series:
type(df.NAME)
# This is also a Series:
df['NAME']
# However, this is a DataFrame with one column, not a Series:
df[['NAME']]
# Another way of getting a columns as Series:
df.loc[:,'NAME']
df.loc[:,['NAME','GENDER']]
df.iloc[:,[0,1]]
df.loc[:,(header =='NAME') | (header == 'GENDER')]
# This is a row.
df.loc[2]
df.loc[2:4]
df.iloc[2:4]
df.drop(columns=['NAME','GENDER']) # This is just a view.
#df.drop(columns=['NAME','GENDER'],inplace=True) # => To remove permanently.
df.loc[:, (header!='NAME') & (header!='GENDER')]
# Males only.
df[df.GENDER == 'M']
# Only non-males.
df[-(df.GENDER == 'M')]
df[df.GENDER == 'F']
df[df.HEIGHT > 170]
df[ (df.WEIGHT > 70) & (df.WEIGHT < 80)]
df[ (df.GENDER == 'M') & (df.HEIGHT > 175)]
# Opposite criteria to the previous one.
df[ -((df.GENDER == 'M') & (df.HEIGHT > 175))]
df2 = df.drop(columns=['GRADE','ABSENCE'])
df2.to_csv('data_mine.csv',index=False)
df3 = pd.read_csv('data_mine.csv',encoding='latin1',header='infer')
df3.head(3)
!wget --no-clobber https://github.com/stefannae/SIC-Artificial-Intelligence/blob/main/SIC_AI_Coding_Exercises/SIC_AI_Chapter_03_Coding_Exercises/data_studentlist.xlsx
dfx = pd.read_excel('data_studentlist.xlsx')
dfx.head(5)
dfx.to_excel('data_studentlist2.xlsx',sheet_name='NewSheet', index=False)
| 0.466359 | 0.876423 |
```
import gc, os
from tqdm import tqdm
import pandas as pd
import numpy as np
import sys
sys.path.append(f'/home/{os.environ.get("USER")}/PythonLibrary')
import lgbextension as ex
import lightgbm as lgb
from matplotlib import pyplot as plt
from multiprocessing import cpu_count, Pool
from sklearn.preprocessing import LabelEncoder
from sklearn.model_selection import GroupKFold
from glob import glob
NFOLD = 5
train = pd.read_csv('../input/application_train.csv.zip')
prev = pd.read_csv('../input/previous_application.csv.zip')
X_train, X_test = prev.align(train, join='inner', axis=1)
X_train.drop('SK_ID_CURR', axis=1, inplace=True)
X_train.head()
prev[X_test.columns.tolist()+['NAME_SELLER_INDUSTRY']].head()
col_cat = X_train.head().select_dtypes('O').columns.tolist()
col_cat
le = LabelEncoder()
for c in col_cat:
X_train[c].fillna('na dayo', inplace=True)
X_test[c].fillna('na dayo', inplace=True)
le.fit( X_train[c].append(X_test[c]) )
X_train[c] = le.transform(X_train[c])
X_test[c] = le.transform(X_test[c])
y_names = prev.columns.difference(X_train.columns).tolist()
y_names
prev[y_names].dtypes
prev[y_names].isnull().sum()
prev[y_names].head()
SEED = 71
param_bin = {
'objective': 'binary',
'metric': 'auc',
'learning_rate': 0.01,
'max_depth': 6,
'num_leaves': 63,
'max_bin': 255,
'min_child_weight': 10,
'min_data_in_leaf': 150,
'reg_lambda': 0.5, # L2 regularization term on weights.
'reg_alpha': 0.5, # L1 regularization term on weights.
'colsample_bytree': 0.9,
'subsample': 0.9,
# 'nthread': 32,
'nthread': cpu_count(),
'bagging_freq': 1,
'verbose':-1,
'seed': SEED
}
param_reg = {
'objective': 'regression',
'metric': 'rmse',
'learning_rate': 0.01,
'max_depth': 6,
'num_leaves': 63,
'max_bin': 255,
'min_child_weight': 10,
'min_data_in_leaf': 150,
'reg_lambda': 0.5, # L2 regularization term on weights.
'reg_alpha': 0.5, # L1 regularization term on weights.
'colsample_bytree': 0.9,
'subsample': 0.9,
# 'nthread': 32,
'nthread': cpu_count(),
'bagging_freq': 1,
'verbose':-1,
'seed': SEED
}
group_kfold = GroupKFold(n_splits=NFOLD)
sub_train = prev[['SK_ID_CURR']]
sub_train['g'] = sub_train.SK_ID_CURR % NFOLD
y_name = y_names[0]
y = prev[y_name]
dtrain = lgb.Dataset(X_train, y.map(np.log1p), categorical_feature=col_cat )
gc.collect()
ret, models = lgb.cv(param_reg, dtrain, 99999, stratified=False,
folds=group_kfold.split(X_train, y,
sub_train['g']),
early_stopping_rounds=100, verbose_eval=50,
seed=111)
X_train.head()
y
```
|
github_jupyter
|
import gc, os
from tqdm import tqdm
import pandas as pd
import numpy as np
import sys
sys.path.append(f'/home/{os.environ.get("USER")}/PythonLibrary')
import lgbextension as ex
import lightgbm as lgb
from matplotlib import pyplot as plt
from multiprocessing import cpu_count, Pool
from sklearn.preprocessing import LabelEncoder
from sklearn.model_selection import GroupKFold
from glob import glob
NFOLD = 5
train = pd.read_csv('../input/application_train.csv.zip')
prev = pd.read_csv('../input/previous_application.csv.zip')
X_train, X_test = prev.align(train, join='inner', axis=1)
X_train.drop('SK_ID_CURR', axis=1, inplace=True)
X_train.head()
prev[X_test.columns.tolist()+['NAME_SELLER_INDUSTRY']].head()
col_cat = X_train.head().select_dtypes('O').columns.tolist()
col_cat
le = LabelEncoder()
for c in col_cat:
X_train[c].fillna('na dayo', inplace=True)
X_test[c].fillna('na dayo', inplace=True)
le.fit( X_train[c].append(X_test[c]) )
X_train[c] = le.transform(X_train[c])
X_test[c] = le.transform(X_test[c])
y_names = prev.columns.difference(X_train.columns).tolist()
y_names
prev[y_names].dtypes
prev[y_names].isnull().sum()
prev[y_names].head()
SEED = 71
param_bin = {
'objective': 'binary',
'metric': 'auc',
'learning_rate': 0.01,
'max_depth': 6,
'num_leaves': 63,
'max_bin': 255,
'min_child_weight': 10,
'min_data_in_leaf': 150,
'reg_lambda': 0.5, # L2 regularization term on weights.
'reg_alpha': 0.5, # L1 regularization term on weights.
'colsample_bytree': 0.9,
'subsample': 0.9,
# 'nthread': 32,
'nthread': cpu_count(),
'bagging_freq': 1,
'verbose':-1,
'seed': SEED
}
param_reg = {
'objective': 'regression',
'metric': 'rmse',
'learning_rate': 0.01,
'max_depth': 6,
'num_leaves': 63,
'max_bin': 255,
'min_child_weight': 10,
'min_data_in_leaf': 150,
'reg_lambda': 0.5, # L2 regularization term on weights.
'reg_alpha': 0.5, # L1 regularization term on weights.
'colsample_bytree': 0.9,
'subsample': 0.9,
# 'nthread': 32,
'nthread': cpu_count(),
'bagging_freq': 1,
'verbose':-1,
'seed': SEED
}
group_kfold = GroupKFold(n_splits=NFOLD)
sub_train = prev[['SK_ID_CURR']]
sub_train['g'] = sub_train.SK_ID_CURR % NFOLD
y_name = y_names[0]
y = prev[y_name]
dtrain = lgb.Dataset(X_train, y.map(np.log1p), categorical_feature=col_cat )
gc.collect()
ret, models = lgb.cv(param_reg, dtrain, 99999, stratified=False,
folds=group_kfold.split(X_train, y,
sub_train['g']),
early_stopping_rounds=100, verbose_eval=50,
seed=111)
X_train.head()
y
| 0.311741 | 0.130535 |
# Implement a Neural Network
This notebook contains useful information and testing code to help you to develop a neural network by implementing the forward pass and backpropagation algorithm in the `models/neural_net.py` file.
```
import matplotlib.pyplot as plt
import numpy as np
from models.neural_net import NeuralNetwork
%matplotlib inline
plt.rcParams['figure.figsize'] = (10.0, 8.0) # set default size of plots
# For auto-reloading external modules
# See http://stackoverflow.com/questions/1907993/autoreload-of-modules-in-ipython
%load_ext autoreload
%autoreload 2
def rel_error(x, y):
"""Returns relative error"""
return np.max(np.abs(x - y) / (np.maximum(1e-8, np.abs(x) + np.abs(y))))
```
You will implement your network in the class `NeuralNetwork` inside the file `models/neural_net.py` to represent instances of the network. The network parameters are stored in the instance variable `self.params` where keys are string parameter names and values are numpy arrays.
The cell below initializes a toy dataset and corresponding model which will allow you to check your forward and backward pass by using a numeric gradient check.
```
# Create a small net and some toy data to check your implementations.
# Note that we set the random seed for repeatable experiments.
input_size = 4
hidden_size = 10
num_classes = 3
num_inputs = 5
def init_toy_model(num_layers):
np.random.seed(0)
hidden_sizes = [hidden_size] * (num_layers - 1)
return NeuralNetwork(input_size, hidden_sizes, num_classes, num_layers)
def init_toy_data():
np.random.seed(0)
X = 10 * np.random.randn(num_inputs, input_size)
y = np.random.randint(num_classes, size=num_inputs)
return X, y
```
# Implement forward and backward pass
The first thing you will do is implement the forward pass of your neural network along with the loss calculation. The forward pass should be implemented in the `forward` function. You can use helper functions like `linear`, `relu`, and `softmax` to help organize your code.
Next, you will implement the backward pass using the backpropagation algorithm. Backpropagation will compute the gradient of the loss with respect to the model parameters `W1`, `b1`, ... etc. Use a softmax fuction with cross entropy loss for loss calcuation. Fill in the code blocks in `NeuralNetwork.backward`.
# Train the network
To train the network we will use stochastic gradient descent (SGD), similar to the SVM and Softmax classifiers you trained. This should be similar to the training procedure you used for the SVM and Softmax classifiers.
Once you have implemented SGD, run the code below to train a two-layer network on toy data. You should achieve a training loss less than 0.2 using a two-layer network with relu activation.
```
# Hyperparameters
epochs = 100
batch_size = 1
learning_rate = 5e-3
learning_rate_decay = 0.95
regularization = 5e-6
# Initialize a new neural network model
net = init_toy_model(3)
X, y = init_toy_data()
# Variables to store performance for each epoch
train_loss = np.zeros(epochs)
train_accuracy = np.zeros(epochs)
def softmax(X: np.ndarray) -> np.ndarray:
return np.divide(np.exp(X), np.sum(np.exp(X), axis=1, keepdims=True))
# For each epoch...
for epoch in range(epochs):
# Training
# Run the forward pass of the model to get a prediction and compute the accuracy
# Run the backward pass of the model to update the weights and compute the loss
train_accuracy[epoch] = np.mean(np.argmax(net.forward(X), axis=1) == y)
train_loss[epoch] = net.backward(X, y, learning_rate, regularization)
for key in net.params:
net.params[key] -= learning_rate * net.gradients[key]
#learning_rate *= learning_rate_decay
# Plot the loss function and train / validation accuracies
plt.subplot(2, 1, 1)
plt.plot(train_loss)
plt.title('Loss history')
plt.xlabel('Iteration')
plt.ylabel('Loss')
plt.subplot(2, 1, 2)
plt.plot(train_accuracy)
plt.title('Classification accuracy history')
plt.xlabel('Epoch')
plt.ylabel('Classification accuracy')
plt.show()
```
|
github_jupyter
|
import matplotlib.pyplot as plt
import numpy as np
from models.neural_net import NeuralNetwork
%matplotlib inline
plt.rcParams['figure.figsize'] = (10.0, 8.0) # set default size of plots
# For auto-reloading external modules
# See http://stackoverflow.com/questions/1907993/autoreload-of-modules-in-ipython
%load_ext autoreload
%autoreload 2
def rel_error(x, y):
"""Returns relative error"""
return np.max(np.abs(x - y) / (np.maximum(1e-8, np.abs(x) + np.abs(y))))
# Create a small net and some toy data to check your implementations.
# Note that we set the random seed for repeatable experiments.
input_size = 4
hidden_size = 10
num_classes = 3
num_inputs = 5
def init_toy_model(num_layers):
np.random.seed(0)
hidden_sizes = [hidden_size] * (num_layers - 1)
return NeuralNetwork(input_size, hidden_sizes, num_classes, num_layers)
def init_toy_data():
np.random.seed(0)
X = 10 * np.random.randn(num_inputs, input_size)
y = np.random.randint(num_classes, size=num_inputs)
return X, y
# Hyperparameters
epochs = 100
batch_size = 1
learning_rate = 5e-3
learning_rate_decay = 0.95
regularization = 5e-6
# Initialize a new neural network model
net = init_toy_model(3)
X, y = init_toy_data()
# Variables to store performance for each epoch
train_loss = np.zeros(epochs)
train_accuracy = np.zeros(epochs)
def softmax(X: np.ndarray) -> np.ndarray:
return np.divide(np.exp(X), np.sum(np.exp(X), axis=1, keepdims=True))
# For each epoch...
for epoch in range(epochs):
# Training
# Run the forward pass of the model to get a prediction and compute the accuracy
# Run the backward pass of the model to update the weights and compute the loss
train_accuracy[epoch] = np.mean(np.argmax(net.forward(X), axis=1) == y)
train_loss[epoch] = net.backward(X, y, learning_rate, regularization)
for key in net.params:
net.params[key] -= learning_rate * net.gradients[key]
#learning_rate *= learning_rate_decay
# Plot the loss function and train / validation accuracies
plt.subplot(2, 1, 1)
plt.plot(train_loss)
plt.title('Loss history')
plt.xlabel('Iteration')
plt.ylabel('Loss')
plt.subplot(2, 1, 2)
plt.plot(train_accuracy)
plt.title('Classification accuracy history')
plt.xlabel('Epoch')
plt.ylabel('Classification accuracy')
plt.show()
| 0.903035 | 0.991915 |
# online nets
Deep learning is powerful but computationally expensive, frequently requiring massive compute budgets. In persuit of cost-effective-yet-powerful AI, this work explores and evaluates a heuristic which should lend to more-efficient use of data through online learning.
Goal: evaluate a deep learning alternative capable of true online learning. Solution requirements:
1. catastrophic forgetting should be impossible;
2. all data is integrated into sufficient statistics of fixed dimension;
3. and our solution should have predictive power comparable to deep learning.
## modeling strategy
We will not attempt to derive sufficient statistics for an entire deep net, but instead leverage well-known sufficient statistics for least squares models,
so will have sufficient statistics per deep net layer. If this can be empirically shown effective, we'll build-out the theory afterwards.
Recognizing a deep net as a series of compositions, as follows.
$ Y + \varepsilon \approx \mathbb{E}Y = \sigma_3 \circ \beta_3^T \circ \sigma_2 \circ \beta_2^T \circ \sigma_1 \circ \beta_1^T X $
So, we can isolate invidivdual $\beta_j$ matrices using (psuedo-)inverses $\beta_j^{-1}$ like so.
$ \sigma_2^{-1} \circ \beta_3^{-1} \circ \sigma_3^{-1} (Y) \approx \beta_2^T \circ \sigma_1 \circ \beta_1^T X $
In this example, if we freeze all $\beta_j$'s except $\beta_2$, we are free to update $\hat \beta_2$ using $\tilde Y = \sigma_2^{-1} \circ \beta_3^{-1} \circ \sigma_3^{-1} (Y) $
and $\tilde X = \sigma_1 \circ \beta_1^T X $.
Using a least squares formulation for fitting to $\left( \tilde X, \tilde Y \right)$, we get sufficient statistics per layer.
# model code definitions
```
import torch
TORCH_TENSOR_TYPE = type(torch.tensor(1))
def padded_diagonal(diag_value, n_rows, n_cols):
## construct diagonal matrix
n_diag = min(n_rows, n_cols)
diag = torch.diag(torch.tensor([diag_value]*n_diag))
if n_rows > n_cols:
## pad rows
pad = n_rows - n_cols
return torch.cat([diag, torch.zeros((pad, n_cols))], 0)
if n_cols > n_rows:
## pad cols
pad = n_cols - n_rows
return torch.cat([diag, torch.zeros((n_rows, pad))], 1)
## no padding
return diag
class OnlineDenseLayer:
'''
A single dense net, formulated as a least squares model.
'''
def __init__(self, p, q, activation=lambda x:x, activation_inverse=lambda x:x, lam=1.):
'''
inputs:
- p: input dimension
- q: output dimension
- activation: non-linear function, from R^p to R^q. Default is identity.
- activation_inverse: inverse of the activation function. Default is identity.
- lam: regularization term
'''
self.__validate_inputs(p=p, q=q, lam=lam)
self.p = p
self.q = q
self.activation = activation
self.activation_inverse = activation_inverse
self.lam = lam
self.xTy = padded_diagonal(lam, p+1,q) # +1 for intercept
self.yTx = padded_diagonal(lam, q+1,p)
self.xTx_inv = torch.diag(torch.tensor([1./lam]*(p+1)))
self.yTy_inv = torch.diag(torch.tensor([1./lam]*(q+1)))
self.betaT_forward = torch.matmul(self.xTx_inv, self.xTy)
self.betaT_forward = torch.transpose(self.betaT_forward, 0, 1)
self.betaT_backward = torch.matmul(self.yTy_inv, self.yTx)
self.betaT_backward = torch.transpose(self.betaT_backward, 0, 1)
self.x_forward = None
self.y_forward = None
self.x_backward = None
self.y_backward = None
pass
def forward(self, x):
'creates and stores x_forward and y_forward, then returns activation(y_forward)'
self.__validate_inputs(x=x, p=self.p)
self.x_forward = x
x = torch.cat((torch.tensor([[1.]]), x), dim=0) # intercept
self.y_forward = torch.matmul(self.betaT_forward, x) # predict
return self.activation(self.y_forward)
def backward(self, y):
'creates and stores x_backward and y_backward, then returns y_backward'
y = self.activation_inverse(y)
self.__validate_inputs(y=y, q=self.q)
self.y_backward = y
y = torch.cat((torch.tensor([[1.]]), y), dim=0)
self.x_backward = torch.matmul(self.betaT_backward, y)
return self.x_backward
def forward_fit(self):
'uses x_forward and y_backward to update forward model, then returns Sherman Morrison denominator'
self.__validate_inputs(x=self.x_forward, y=self.y_backward, p=self.p, q=self.q)
x = torch.cat((torch.tensor([[1.]]), self.x_forward), dim=0)
self.xTx_inv, sm_denom = self.sherman_morrison(self.xTx_inv, x, x)
self.xTy += torch.matmul(x, torch.transpose(self.y_backward, 0, 1))
self.betaT_forward = torch.matmul(self.xTx_inv, self.xTy)
self.betaT_forward = torch.transpose(self.betaT_forward, 0, 1)
return sm_denom
def backward_fit(self):
'uses x_backward and y_forward to update backward model, then returns Sherman Morrison denominator'
self.__validate_inputs(x=self.x_forward, y=self.y_backward, p=self.p, q=self.q)
y = torch.cat((torch.tensor([[1.]]), self.y_backward), dim=0)
self.yTy_inv, sm_denom = self.sherman_morrison(self.yTy_inv, y, y)
self.yTx += torch.matmul(y, torch.transpose(self.x_backward, 0, 1))
self.betaT_backward = torch.matmul(self.yTy_inv, self.yTx)
self.betaT_backward = torch.transpose(self.betaT_backward, 0, 1)
return sm_denom
@staticmethod
def sherman_morrison(inv_mat, vec1, vec2):
'''
applies Sherman Morrison updates, (mat + vec1 vec2^T)^{-1}
inputs:
- inv_mat: an inverted matrix
- vec1: a column vector
- vec2: a column vector
returns:
- updated matrix
- the Sherman Morrison denominator, for tracking numerical stability
'''
v2t = torch.transpose(vec2, 0, 1)
denominator = 1. + torch.matmul(torch.matmul(v2t, inv_mat), vec1)
numerator = torch.matmul(torch.matmul(inv_mat, vec1), torch.matmul(v2t, inv_mat))
updated_inv_mat = inv_mat - numerator / denominator
return updated_inv_mat, float(denominator)
def __validate_inputs(self, p=None, q=None, lam=None, x=None, y=None):
'raises value exceptions if provided parameters are invalid'
if q is not None:
if not isinstance(q, int):
raise ValueError('`q` must be int!')
if q <= 0:
raise ValueError('`q` must be greater than zero!')
if p is not None:
if not isinstance(p, int):
raise ValueError('`p` must be int!')
if p <= 0:
raise ValueError('`p` must be greater than zero!')
if lam is not None:
if not (isinstance(lam, float) or isinstance(lam, int)):
raise ValueError('`lam` must be float or int!')
if lam < 0:
raise ValueError('`lam` must be non-negative!')
if x is not None and p is not None:
if type(x) != TORCH_TENSOR_TYPE:
raise ValueError('`x` must be of type `torch.tensor`!')
if list(x.shape) != [p,1]:
raise ValueError('`x.shape` must be `[p,1]`!')
if torch.isnan(x).any():
raise ValueError('`x` contains `nan`!')
pass
if y is not None and q is not None:
if type(y) != TORCH_TENSOR_TYPE:
raise ValueError('`y` must be of type `torch.tensor`!')
if list(y.shape) != [q,1]:
raise ValueError('`y.shape` must be `[q,1]`')
if torch.isnan(y).any():
raise ValueError('`y` contains `nan`!')
pass
pass
pass
class OnlineNet:
'online, sequential dense net'
def __init__(self, layer_list):
## validate inputs
if type(layer_list) != list:
raise ValueError('`layer_list` must be of type list!')
for layer in layer_list:
if not issubclass(type(layer), OnlineDenseLayer):
raise ValueError('each item in `layer_list` must be an instance of a subclass of `OnlineDenseLayer`!')
## assign
self.layer_list = layer_list
pass
def forward(self, x):
'predict forward'
for layer in self.layer_list:
x = layer.forward(x)
return x
def backward(self, y):
'predict backward'
for layer in reversed(self.layer_list):
y = layer.backward(y)
return y
def fit(self):
'assumes layers x & y targets have already been set. Returns Sherman Morrison denominators per layer in (forward, backward) pairs in a list'
sherman_morrison_denominator_list = []
for layer in self.layer_list:
forward_smd = layer.forward_fit()
backward_smd = layer.backward_fit()
sherman_morrison_denominator_list.append((forward_smd, backward_smd))
return sherman_morrison_denominator_list
def __reduce_sherman_morrison_denominator_list(self, smd_pair_list):
'returns the value closest to zero'
if type(smd_pair_list) != list:
raise ValueError('`smd_pair_list` must be of type `list`!')
if len(smd_pair_list) == 0:
return None
smallest_smd = None
for smd_pair in smd_pair_list:
if type(smd_pair) != tuple:
raise ValueError('`smd_pair_list` must be list of tuples!')
if smallest_smd is None:
smallest_smd = smd_pair[0]
if abs(smallest_smd) > abs(smd_pair[0]):
smallest_smd = smd_pair[0]
if abs(smallest_smd) > abs(smd_pair[1]):
smallest_smd = smd_pair[1]
return float(smallest_smd)
def __call__(self, x, y=None):
'''
If only x is given, a prediction is made and returned.
If x and y are given, then the model is updated, and returns
- the prediction
- the sherman morrison denominator closest to zero, for tracking numerical stability
'''
y_hat = self.forward(x)
if y is None:
return y_hat
self.backward(y)
self.layer_list[0].x_forward = x
self.layer_list[0].x_backward = x
self.layer_list[-1].y_forward = y
self.layer_list[-1].y_backward = y
smd_pair_list = self.fit()
smallest_smd = self.__reduce_sherman_morrison_denominator_list(smd_pair_list)
return y_hat, smallest_smd
## tests
## test 1: sherman morrison
a = torch.tensor([[2., 1.], [1., 2.]])
b = torch.tensor([[.1],[.2]])
sm_inv, _ = OnlineDenseLayer.sherman_morrison(torch.inverse(a),b,b)
num_inv = torch.inverse(a+torch.matmul(b, torch.transpose(b,0,1)))
err = float(torch.abs(sm_inv - num_inv).sum())
assert(err < 1e-5)
```
# first experiment: mnist classification
```
from tqdm import tqdm
from torchvision import datasets, transforms
transform=transforms.Compose([
transforms.ToTensor(),
transforms.Normalize((0.1307,), (0.3081,))
])
dataset1 = datasets.MNIST('../../data', train=True, download=True, transform=transform)
dataset2 = datasets.MNIST('../../data', train=False, transform=transform)
train_loader = torch.utils.data.DataLoader(dataset1)
test_loader = torch.utils.data.DataLoader(dataset2)
n_labels = 10
## activation functions
## torch.sigmoid
inv_sigmoid = lambda x: -torch.log((1/(x+1e-8))-1)
leaky_relu_alpha = .1
leaky_relu = lambda x: (x > 0)*x + (x <= 0)*x*leaky_relu_alpha
inv_leaky_relu = lambda x: (x > 0)*x + (x <= 0)*x/leaky_relu_alpha
model = OnlineNet(
[
OnlineDenseLayer(p=1*1*28*28, q=1000, activation=torch.sigmoid, activation_inverse=inv_sigmoid),
OnlineDenseLayer(p=1000, q=5000, activation=torch.sigmoid, activation_inverse=inv_sigmoid),
OnlineDenseLayer(p=5000, q=100, activation=torch.sigmoid, activation_inverse=inv_sigmoid),
OnlineDenseLayer(p=100, q=n_labels, activation=torch.sigmoid, activation_inverse=inv_sigmoid) ## TODO invert correctly, or else get NANs
#OnlineDenseLayer(p=3, q=2, activation=leaky_relu, activation_inverse=inv_leaky_relu),
#OnlineDenseLayer(p=2, q=1)
]
)
def build_data(image, label):
'format data from iterator for model'
y = torch.tensor([1. if int(label[0]) == idx else 0. for idx in range(n_labels)]) ## one-hot representation
x = image.reshape([-1]) ## flatten
## shrink so sigmoid inverse is well-defined
y = y*.90 + .05
## reshape to column vectors
x = x.reshape([-1,1])
y = y.reshape([-1,1])
return x, y
def build_test_data():
x = torch.normal(mean=torch.zeros([3,1]))
y = torch.sigmoid(3. + 5.*x[0] - 10.*x[1])
y = y + 3*x[2]
y = y.reshape([-1,1])
return x, y
errs = []
stab = []
pbar = tqdm(train_loader)
for [image, label] in pbar:
x, y = build_data(image, label)
#x, y = build_test_data()
## fit
y_hat, stability = model(x, y)
err = float((y - y_hat).abs().sum())
errs.append(err)
stab.append(stability)
pbar.set_description(f'err: {err}, stab: {stability}')
## train error
## TODO
import matplotlib.pyplot as plt
cs = torch.cumsum(torch.tensor(errs), dim=0)
ma = (cs[100:] - cs[:-100])/100.
#list(ma)
plt.plot(errs)
#plt.plot(stab)
plt.show()
```
## scratch space
$x_1, X_2, X_3, \ldots, X_{p-1}, x_p = y$
$\beta_1^F, \beta_1^B, \beta_2^F, \beta_2^B, \ldots, \beta_{p-1}^F, \beta_{p-1}^B$
$\hat x_{j+1} = \sigma \left( \beta_j^{FT} x_j \right)$
forward series: $x_1, \hat x_2, \hat x_3, \ldots, \hat x_{p-1}, x_p = y$
$ \hat \beta_{p-1}^{FT} = \text{argmin}_\beta \| x_p - \beta^{T} \hat x_{p-1} \|^2 $
$ \hat \beta_{p-2}^{FT} = \text{argmin}_\beta \| \hat x_{p-1} - \beta^{T} \hat x_{p-2} \|^2 $ We won't do this.
$ \tilde x_{j-1} = \sigma^{-1}\left( \beta_j^{BT} x_j \right)$
backward series: $x_1, \tilde x_2, \tilde x_3, \ldots, \tilde x_{p-1}, x_p = y$
$ \hat \beta_{p-2}^{F} = \text{argmin}_\beta \| \tilde x_{p-1} - \beta^{T} \hat x_{p-2} \|^2 $
$ \hat \beta_{p-2}^{B} = \text{argmin}_\beta \| \hat x_{p-2} - \beta^{T} \tilde x_{p-1} \|^2 $
$ \hat x_3 = \sigma \left( \beta_2^{FT} \hat x_2 \right) $
$ \hat \beta_2^F = \text{argmin}_\beta \| \sigma^{-1}\left( \hat x_3 \right) - \beta^T \hat x_2 \|^2 $ Useless without $\tilde x$
$ \tilde x_2 = \sigma^{-1}\left( \beta_3^{BT} \tilde x_3 \right) $
$ \hat \beta_2^B = \text{argmin}_\beta \| \sigma\left( \hat x_2 \right) - \beta^T \hat x_3 \|^2 $ Useless without $\hat x$
So, use these estimates instead.
$ \hat \beta_2^F = \text{argmin}_\beta \| \sigma^{-1}\left( \tilde x_3 \right) - \beta^T \hat x_2 \|^2 $
$ \hat \beta_2^B = \text{argmin}_\beta \| \sigma\left( \hat x_2 \right) - \beta^T \tilde x_3 \|^2 $
$\mathbb{E} l(X;\theta) \approx l(X;\theta_0) + \left( \theta - \theta_0 \right)^T \mathbb{E} \nabla_\theta l(X;\theta_0) + \left( \theta - \theta_0 \right)^T \mathbb{E} \nabla^2_\theta l(X;\theta_0) \left( \theta - \theta_0 \right)/2 $
$ = \mathbb{E}l(X;\theta_0) + 0 - \left( \theta - \theta_0 \right)^T \mathcal{I}_{\theta_0} \left( \theta - \theta_0 \right)/2 $
$ X_{j+1} = \beta^T X_j + \sigma \varepsilon, \; \varepsilon \sim N(0, \mathcal{I}) $
$ X_{j+k} = \beta^{kT} X_j \Rightarrow \mathbb{E}[X_j|X_0] = \mathbb{E}[\beta^{jT} X_0|X_0] = \beta^{jT} X_0 $
$ \mathbb{E}\left[ f(X_0) \; | \; X_0 \right] = f(X_0) $
|
github_jupyter
|
import torch
TORCH_TENSOR_TYPE = type(torch.tensor(1))
def padded_diagonal(diag_value, n_rows, n_cols):
## construct diagonal matrix
n_diag = min(n_rows, n_cols)
diag = torch.diag(torch.tensor([diag_value]*n_diag))
if n_rows > n_cols:
## pad rows
pad = n_rows - n_cols
return torch.cat([diag, torch.zeros((pad, n_cols))], 0)
if n_cols > n_rows:
## pad cols
pad = n_cols - n_rows
return torch.cat([diag, torch.zeros((n_rows, pad))], 1)
## no padding
return diag
class OnlineDenseLayer:
'''
A single dense net, formulated as a least squares model.
'''
def __init__(self, p, q, activation=lambda x:x, activation_inverse=lambda x:x, lam=1.):
'''
inputs:
- p: input dimension
- q: output dimension
- activation: non-linear function, from R^p to R^q. Default is identity.
- activation_inverse: inverse of the activation function. Default is identity.
- lam: regularization term
'''
self.__validate_inputs(p=p, q=q, lam=lam)
self.p = p
self.q = q
self.activation = activation
self.activation_inverse = activation_inverse
self.lam = lam
self.xTy = padded_diagonal(lam, p+1,q) # +1 for intercept
self.yTx = padded_diagonal(lam, q+1,p)
self.xTx_inv = torch.diag(torch.tensor([1./lam]*(p+1)))
self.yTy_inv = torch.diag(torch.tensor([1./lam]*(q+1)))
self.betaT_forward = torch.matmul(self.xTx_inv, self.xTy)
self.betaT_forward = torch.transpose(self.betaT_forward, 0, 1)
self.betaT_backward = torch.matmul(self.yTy_inv, self.yTx)
self.betaT_backward = torch.transpose(self.betaT_backward, 0, 1)
self.x_forward = None
self.y_forward = None
self.x_backward = None
self.y_backward = None
pass
def forward(self, x):
'creates and stores x_forward and y_forward, then returns activation(y_forward)'
self.__validate_inputs(x=x, p=self.p)
self.x_forward = x
x = torch.cat((torch.tensor([[1.]]), x), dim=0) # intercept
self.y_forward = torch.matmul(self.betaT_forward, x) # predict
return self.activation(self.y_forward)
def backward(self, y):
'creates and stores x_backward and y_backward, then returns y_backward'
y = self.activation_inverse(y)
self.__validate_inputs(y=y, q=self.q)
self.y_backward = y
y = torch.cat((torch.tensor([[1.]]), y), dim=0)
self.x_backward = torch.matmul(self.betaT_backward, y)
return self.x_backward
def forward_fit(self):
'uses x_forward and y_backward to update forward model, then returns Sherman Morrison denominator'
self.__validate_inputs(x=self.x_forward, y=self.y_backward, p=self.p, q=self.q)
x = torch.cat((torch.tensor([[1.]]), self.x_forward), dim=0)
self.xTx_inv, sm_denom = self.sherman_morrison(self.xTx_inv, x, x)
self.xTy += torch.matmul(x, torch.transpose(self.y_backward, 0, 1))
self.betaT_forward = torch.matmul(self.xTx_inv, self.xTy)
self.betaT_forward = torch.transpose(self.betaT_forward, 0, 1)
return sm_denom
def backward_fit(self):
'uses x_backward and y_forward to update backward model, then returns Sherman Morrison denominator'
self.__validate_inputs(x=self.x_forward, y=self.y_backward, p=self.p, q=self.q)
y = torch.cat((torch.tensor([[1.]]), self.y_backward), dim=0)
self.yTy_inv, sm_denom = self.sherman_morrison(self.yTy_inv, y, y)
self.yTx += torch.matmul(y, torch.transpose(self.x_backward, 0, 1))
self.betaT_backward = torch.matmul(self.yTy_inv, self.yTx)
self.betaT_backward = torch.transpose(self.betaT_backward, 0, 1)
return sm_denom
@staticmethod
def sherman_morrison(inv_mat, vec1, vec2):
'''
applies Sherman Morrison updates, (mat + vec1 vec2^T)^{-1}
inputs:
- inv_mat: an inverted matrix
- vec1: a column vector
- vec2: a column vector
returns:
- updated matrix
- the Sherman Morrison denominator, for tracking numerical stability
'''
v2t = torch.transpose(vec2, 0, 1)
denominator = 1. + torch.matmul(torch.matmul(v2t, inv_mat), vec1)
numerator = torch.matmul(torch.matmul(inv_mat, vec1), torch.matmul(v2t, inv_mat))
updated_inv_mat = inv_mat - numerator / denominator
return updated_inv_mat, float(denominator)
def __validate_inputs(self, p=None, q=None, lam=None, x=None, y=None):
'raises value exceptions if provided parameters are invalid'
if q is not None:
if not isinstance(q, int):
raise ValueError('`q` must be int!')
if q <= 0:
raise ValueError('`q` must be greater than zero!')
if p is not None:
if not isinstance(p, int):
raise ValueError('`p` must be int!')
if p <= 0:
raise ValueError('`p` must be greater than zero!')
if lam is not None:
if not (isinstance(lam, float) or isinstance(lam, int)):
raise ValueError('`lam` must be float or int!')
if lam < 0:
raise ValueError('`lam` must be non-negative!')
if x is not None and p is not None:
if type(x) != TORCH_TENSOR_TYPE:
raise ValueError('`x` must be of type `torch.tensor`!')
if list(x.shape) != [p,1]:
raise ValueError('`x.shape` must be `[p,1]`!')
if torch.isnan(x).any():
raise ValueError('`x` contains `nan`!')
pass
if y is not None and q is not None:
if type(y) != TORCH_TENSOR_TYPE:
raise ValueError('`y` must be of type `torch.tensor`!')
if list(y.shape) != [q,1]:
raise ValueError('`y.shape` must be `[q,1]`')
if torch.isnan(y).any():
raise ValueError('`y` contains `nan`!')
pass
pass
pass
class OnlineNet:
'online, sequential dense net'
def __init__(self, layer_list):
## validate inputs
if type(layer_list) != list:
raise ValueError('`layer_list` must be of type list!')
for layer in layer_list:
if not issubclass(type(layer), OnlineDenseLayer):
raise ValueError('each item in `layer_list` must be an instance of a subclass of `OnlineDenseLayer`!')
## assign
self.layer_list = layer_list
pass
def forward(self, x):
'predict forward'
for layer in self.layer_list:
x = layer.forward(x)
return x
def backward(self, y):
'predict backward'
for layer in reversed(self.layer_list):
y = layer.backward(y)
return y
def fit(self):
'assumes layers x & y targets have already been set. Returns Sherman Morrison denominators per layer in (forward, backward) pairs in a list'
sherman_morrison_denominator_list = []
for layer in self.layer_list:
forward_smd = layer.forward_fit()
backward_smd = layer.backward_fit()
sherman_morrison_denominator_list.append((forward_smd, backward_smd))
return sherman_morrison_denominator_list
def __reduce_sherman_morrison_denominator_list(self, smd_pair_list):
'returns the value closest to zero'
if type(smd_pair_list) != list:
raise ValueError('`smd_pair_list` must be of type `list`!')
if len(smd_pair_list) == 0:
return None
smallest_smd = None
for smd_pair in smd_pair_list:
if type(smd_pair) != tuple:
raise ValueError('`smd_pair_list` must be list of tuples!')
if smallest_smd is None:
smallest_smd = smd_pair[0]
if abs(smallest_smd) > abs(smd_pair[0]):
smallest_smd = smd_pair[0]
if abs(smallest_smd) > abs(smd_pair[1]):
smallest_smd = smd_pair[1]
return float(smallest_smd)
def __call__(self, x, y=None):
'''
If only x is given, a prediction is made and returned.
If x and y are given, then the model is updated, and returns
- the prediction
- the sherman morrison denominator closest to zero, for tracking numerical stability
'''
y_hat = self.forward(x)
if y is None:
return y_hat
self.backward(y)
self.layer_list[0].x_forward = x
self.layer_list[0].x_backward = x
self.layer_list[-1].y_forward = y
self.layer_list[-1].y_backward = y
smd_pair_list = self.fit()
smallest_smd = self.__reduce_sherman_morrison_denominator_list(smd_pair_list)
return y_hat, smallest_smd
## tests
## test 1: sherman morrison
a = torch.tensor([[2., 1.], [1., 2.]])
b = torch.tensor([[.1],[.2]])
sm_inv, _ = OnlineDenseLayer.sherman_morrison(torch.inverse(a),b,b)
num_inv = torch.inverse(a+torch.matmul(b, torch.transpose(b,0,1)))
err = float(torch.abs(sm_inv - num_inv).sum())
assert(err < 1e-5)
from tqdm import tqdm
from torchvision import datasets, transforms
transform=transforms.Compose([
transforms.ToTensor(),
transforms.Normalize((0.1307,), (0.3081,))
])
dataset1 = datasets.MNIST('../../data', train=True, download=True, transform=transform)
dataset2 = datasets.MNIST('../../data', train=False, transform=transform)
train_loader = torch.utils.data.DataLoader(dataset1)
test_loader = torch.utils.data.DataLoader(dataset2)
n_labels = 10
## activation functions
## torch.sigmoid
inv_sigmoid = lambda x: -torch.log((1/(x+1e-8))-1)
leaky_relu_alpha = .1
leaky_relu = lambda x: (x > 0)*x + (x <= 0)*x*leaky_relu_alpha
inv_leaky_relu = lambda x: (x > 0)*x + (x <= 0)*x/leaky_relu_alpha
model = OnlineNet(
[
OnlineDenseLayer(p=1*1*28*28, q=1000, activation=torch.sigmoid, activation_inverse=inv_sigmoid),
OnlineDenseLayer(p=1000, q=5000, activation=torch.sigmoid, activation_inverse=inv_sigmoid),
OnlineDenseLayer(p=5000, q=100, activation=torch.sigmoid, activation_inverse=inv_sigmoid),
OnlineDenseLayer(p=100, q=n_labels, activation=torch.sigmoid, activation_inverse=inv_sigmoid) ## TODO invert correctly, or else get NANs
#OnlineDenseLayer(p=3, q=2, activation=leaky_relu, activation_inverse=inv_leaky_relu),
#OnlineDenseLayer(p=2, q=1)
]
)
def build_data(image, label):
'format data from iterator for model'
y = torch.tensor([1. if int(label[0]) == idx else 0. for idx in range(n_labels)]) ## one-hot representation
x = image.reshape([-1]) ## flatten
## shrink so sigmoid inverse is well-defined
y = y*.90 + .05
## reshape to column vectors
x = x.reshape([-1,1])
y = y.reshape([-1,1])
return x, y
def build_test_data():
x = torch.normal(mean=torch.zeros([3,1]))
y = torch.sigmoid(3. + 5.*x[0] - 10.*x[1])
y = y + 3*x[2]
y = y.reshape([-1,1])
return x, y
errs = []
stab = []
pbar = tqdm(train_loader)
for [image, label] in pbar:
x, y = build_data(image, label)
#x, y = build_test_data()
## fit
y_hat, stability = model(x, y)
err = float((y - y_hat).abs().sum())
errs.append(err)
stab.append(stability)
pbar.set_description(f'err: {err}, stab: {stability}')
## train error
## TODO
import matplotlib.pyplot as plt
cs = torch.cumsum(torch.tensor(errs), dim=0)
ma = (cs[100:] - cs[:-100])/100.
#list(ma)
plt.plot(errs)
#plt.plot(stab)
plt.show()
| 0.831143 | 0.973494 |
# Tutorial 4. Immediate mode
In this tutorial we will talk about a cute feature about Caffe2: immediate mode.
From the previous tutorials you have seen that Caffe2 *declares* a network, and during this declaration phase, nothing gets actually executed - it's like writing the source of a program, and "compilation/execution" only happens later.
This sometimes gets a bit tricky if we are in a researchy mind, and want to inspect typical intermediate outputs as we go. This is when the immediate mode come to help. At a high level, what the immediate mode does is to run the corresponding operators as you write them. The results live under a special workspace that can then be accessed via `FetchImmediate()` and `FeedImmediate()` runs.
Let's show some examples.
```
%matplotlib inline
from caffe2.python import cnn, core, visualize, workspace, model_helper, brew
import numpy as np
import os
core.GlobalInit(['caffe2', '--caffe2_log_level=-1'])
```
Now, as we have known before, in the normal mode, when you create an operator, we are *declaring* it only, and nothing gets actually executed. Let's re-confirm that.
```
workspace.ResetWorkspace()
# declaration
op = core.CreateOperator("GaussianFill", [], "X", shape=[3, 5])
print('Before execution, workspace contains X: {}'
.format(workspace.HasBlob("X")))
# execution
workspace.RunOperatorOnce(op)
print('After execution, workspace contains X: {}'
.format(workspace.HasBlob("X")))
```
## Entering and exiting immediate mode.
Entering immediate mode is easy: you basically invoke `workspace.StartImmediate()`. Since immediate mode has quite a lot of side effects, it would be good to read through the warning message to make sure you understand the implications.
(If you don't want to see the messages, pass `i_know=True` to `StartImmediate` to suppress that.)
```
workspace.StartImmediate()
```
Now that you have enabled immediate mode, any operators you run will simultaneously be executed in a separate immediate workspace. Note - the main workspace that you are working on is not affected. We designed the immediate workspace to be separate from the main workspace, so that nothing in the main workspace gets polluted.
```
# declaration, and since we are in immediate mode, run it in the immediate workspace.
op = core.CreateOperator("GaussianFill", [], "X", shape=[3, 5])
print('Before execution, does workspace contain X? {}'
.format(workspace.HasBlob("X")))
print('But we can access it using the Immediate related functions.'
'Here is a list of immediate blobs:')
print(workspace.ImmediateBlobs())
print('The content is like this:')
print(workspace.FetchImmediate('X'))
# After the immediate execution, you can invoke StopImmediate() to clean up.
workspace.StopImmediate()
```
## Manually feeding blobs
But wait, you say - what if I want to create an operator that uses an input that is "declared" but not present yet? Since the immediate workspace does not have the input, we will encounter an exception:
```
workspace.StartImmediate(i_know=True)
op = core.CreateOperator("Relu", "X", "Y")
```
This is because immediate mode, being completely imperative, requires any input to be used to already exist in the immediate workspace. To make the immediate mode aware of such external inputs, we can manually feed blobs to the immediate workspace.
```
X = np.random.randn(2, 3).astype(np.float32)
workspace.FeedImmediate("X", X)
# Now, we can safely run CreateOperator since immediate mode knows what X looks like
op = core.CreateOperator("Relu", "X", "Y")
print("Example input is:\n{}".format(workspace.FetchImmediate("X")))
print("Example output is:\n{}".format(workspace.FetchImmediate("Y")))
workspace.StopImmediate()
```
## When is immediate mode useful?
You might want to use immediate mode when you are not very sure about the shape of the intermediate results, such as in a CNN where there are multiple convolution and pooling layers. Let's say that you are creating an MNIST convnet model but don't want to calculate the number of dimensions for the final FC layer. Here is what you might want to do.
```
model = model_helper.ModelHelper(name="mnist")
# Start the immediate mode.
workspace.StartImmediate(i_know=True)
data_folder = os.path.join(os.path.expanduser('~'), 'caffe2_notebooks', 'tutorial_data')
data_uint8, label = model.TensorProtosDBInput(
[], ["data_uint8", "label"], batch_size=64,
db=os.path.join(data_folder, 'mnist/mnist-train-nchw-leveldb'),
db_type='leveldb')
data = model.net.Cast(data_uint8, "data", to=core.DataType.FLOAT)
data = model.net.Scale(data, data, scale=float(1./256))
data = model.net.StopGradient(data, data)
conv1 = brew.conv(model, data, 'conv1', 1, 20, 5)
pool1 = brew.max_pool(model, conv1, 'pool1', kernel=2, stride=2)
conv2 = brew.conv(model, pool1, 'conv2', 20, 50, 5)
pool2 = brew.max_pool(model, conv2, 'pool2', kernel=2, stride=2)
# What is the shape of pool2 again...?
feature_dimensions = workspace.FetchImmediate("pool2").shape[1:]
print("Feature dimensions before FC layer: {}".format(feature_dimensions))
fc3 = brew.fc(model, pool2, 'fc3', int(np.prod(feature_dimensions)), 500)
fc3 = brew.relu(model, fc3, fc3)
pred = brew.fc(model, fc3, 'pred', 500, 10)
softmax = brew.softmax(model, pred, 'softmax')
# Let's see if the dimensions are all correct:
for blob in ["data", "conv1", "pool1", "conv2", "pool2", "fc3", "pred"]:
print("Blob {} has shape: {}".format(
blob, workspace.FetchImmediate(blob).shape))
# Let's also visualize a sample input.
print("Sample input:")
visualize.NCHW.ShowMultiple(workspace.FetchImmediate("data"))
workspace.StopImmediate()
```
Remember, immediate mode is only intended to be used in debugging mode, and are only intended for you to verify things interactively. For example, in the use case above, what you want to do eventually is to remove the feature_dimensions argument and replace it with code that do not depend on immediate mode, such as hard-coding it.
## Departing words
Immediate mode could be a useful tool for quick iterations. But it could also easily go wrong. Make sure that you understand its purpose, and never abuse it in real product environments. The philosophy of Caffe2 is to make things very flexible and this is one example of it, but it also makes you easy to shoot yourself in the foot. Take care :)
|
github_jupyter
|
%matplotlib inline
from caffe2.python import cnn, core, visualize, workspace, model_helper, brew
import numpy as np
import os
core.GlobalInit(['caffe2', '--caffe2_log_level=-1'])
workspace.ResetWorkspace()
# declaration
op = core.CreateOperator("GaussianFill", [], "X", shape=[3, 5])
print('Before execution, workspace contains X: {}'
.format(workspace.HasBlob("X")))
# execution
workspace.RunOperatorOnce(op)
print('After execution, workspace contains X: {}'
.format(workspace.HasBlob("X")))
workspace.StartImmediate()
# declaration, and since we are in immediate mode, run it in the immediate workspace.
op = core.CreateOperator("GaussianFill", [], "X", shape=[3, 5])
print('Before execution, does workspace contain X? {}'
.format(workspace.HasBlob("X")))
print('But we can access it using the Immediate related functions.'
'Here is a list of immediate blobs:')
print(workspace.ImmediateBlobs())
print('The content is like this:')
print(workspace.FetchImmediate('X'))
# After the immediate execution, you can invoke StopImmediate() to clean up.
workspace.StopImmediate()
workspace.StartImmediate(i_know=True)
op = core.CreateOperator("Relu", "X", "Y")
X = np.random.randn(2, 3).astype(np.float32)
workspace.FeedImmediate("X", X)
# Now, we can safely run CreateOperator since immediate mode knows what X looks like
op = core.CreateOperator("Relu", "X", "Y")
print("Example input is:\n{}".format(workspace.FetchImmediate("X")))
print("Example output is:\n{}".format(workspace.FetchImmediate("Y")))
workspace.StopImmediate()
model = model_helper.ModelHelper(name="mnist")
# Start the immediate mode.
workspace.StartImmediate(i_know=True)
data_folder = os.path.join(os.path.expanduser('~'), 'caffe2_notebooks', 'tutorial_data')
data_uint8, label = model.TensorProtosDBInput(
[], ["data_uint8", "label"], batch_size=64,
db=os.path.join(data_folder, 'mnist/mnist-train-nchw-leveldb'),
db_type='leveldb')
data = model.net.Cast(data_uint8, "data", to=core.DataType.FLOAT)
data = model.net.Scale(data, data, scale=float(1./256))
data = model.net.StopGradient(data, data)
conv1 = brew.conv(model, data, 'conv1', 1, 20, 5)
pool1 = brew.max_pool(model, conv1, 'pool1', kernel=2, stride=2)
conv2 = brew.conv(model, pool1, 'conv2', 20, 50, 5)
pool2 = brew.max_pool(model, conv2, 'pool2', kernel=2, stride=2)
# What is the shape of pool2 again...?
feature_dimensions = workspace.FetchImmediate("pool2").shape[1:]
print("Feature dimensions before FC layer: {}".format(feature_dimensions))
fc3 = brew.fc(model, pool2, 'fc3', int(np.prod(feature_dimensions)), 500)
fc3 = brew.relu(model, fc3, fc3)
pred = brew.fc(model, fc3, 'pred', 500, 10)
softmax = brew.softmax(model, pred, 'softmax')
# Let's see if the dimensions are all correct:
for blob in ["data", "conv1", "pool1", "conv2", "pool2", "fc3", "pred"]:
print("Blob {} has shape: {}".format(
blob, workspace.FetchImmediate(blob).shape))
# Let's also visualize a sample input.
print("Sample input:")
visualize.NCHW.ShowMultiple(workspace.FetchImmediate("data"))
workspace.StopImmediate()
| 0.471953 | 0.978219 |
```
flex_title = "Altair plots"
flex_author = "built using jupyter-flex"
flex_source_link = "https://github.com/danielfrg/jupyter-flex/blob/master/examples/plots/altair.ipynb"
flex_include_source = True
```
# Simple charts
### Simple Scatter Plot with Tooltips
```
import numpy as np
import pandas as pd
import altair as alt
from vega_datasets import data
alt.renderers.set_embed_options(actions=False)
np.random.seed(42)
source = data.cars()
plot = alt.Chart(source).mark_circle(size=60).encode(
x='Horsepower',
y='Miles_per_Gallon',
color='Origin',
tooltip=['Name', 'Origin', 'Horsepower', 'Miles_per_Gallon']
)
plot
plot.properties(
width='container',
height='container'
).interactive()
```
## Col 2
### Simple bar chart
```
source = pd.DataFrame({
'a': ['A', 'B', 'C', 'D', 'E', 'F', 'G', 'H', 'I'],
'b': [28, 55, 43, 91, 81, 53, 19, 87, 52]
})
plot = alt.Chart(source).mark_bar().encode(
x='a',
y='b'
)
plot
plot.properties(
width='container',
height='container'
)
```
### Simple Heatmap
```
# Compute x^2 + y^2 across a 2D grid
x, y = np.meshgrid(range(-5, 5), range(-5, 5))
z = x ** 2 + y ** 2
# Convert this grid to columnar data expected by Altair
source = pd.DataFrame({'x': x.ravel(),
'y': y.ravel(),
'z': z.ravel()})
plot = alt.Chart(source).mark_rect().encode(
x='x:O',
y='y:O',
color='z:Q'
)
plot
plot.properties(
width='container',
height='container'
)
```
# Bar Charts
### Bar Chart with Negative Values
```
source = data.us_employment()
alt.Chart(source).mark_bar().encode(
x="month:T",
y="nonfarm_change:Q",
color=alt.condition(
alt.datum.nonfarm_change > 0,
alt.value("steelblue"), # The positive color
alt.value("orange") # The negative color
)
).properties(
width="container",
height="container"
)
```
### Horizontal Bar Chart
```
source = data.wheat()
alt.Chart(source).mark_bar().encode(
x='wheat:Q',
y="year:O"
).properties(
width="container",
height="container"
)
```
## Col 2
### Stacked Bar Chart
```
source = data.barley()
alt.Chart(source).mark_bar().encode(
x='variety',
y='sum(yield)',
color='site'
).properties(
width="container",
height="container"
)
```
# Line and Area Charts
### Filled Step Chart
```
source = data.stocks()
alt.Chart(source).mark_area(
color="lightblue",
interpolate='step-after',
line=True
).encode(
x='date',
y='price'
).transform_filter(alt.datum.symbol == 'GOOG').properties(
width="container",
height="container"
)
```
### Multi Series Line Chart
```
source = data.stocks()
alt.Chart(source).mark_line().encode(
x='date',
y='price',
color='symbol'
).properties(
width="container",
height="container"
)
```
## Col 2
### Cumulative Count Chart
```
source = data.movies.url
alt.Chart(source).transform_window(
cumulative_count="count()",
sort=[{"field": "IMDB_Rating"}],
).mark_area().encode(
x="IMDB_Rating:Q",
y="cumulative_count:Q"
).properties(
width="container",
height="container"
)
```
### Stacked Density Estimates
```
source = data.iris()
alt.Chart(source).transform_fold(
['petalWidth',
'petalLength',
'sepalWidth',
'sepalLength'],
as_ = ['Measurement_type', 'value']
).transform_density(
density='value',
bandwidth=0.3,
groupby=['Measurement_type'],
extent= [0, 8],
counts = True,
steps=200
).mark_area().encode(
alt.X('value:Q'),
alt.Y('density:Q', stack='zero'),
alt.Color('Measurement_type:N')
).properties(
width="container",
height="container"
)
```
# Scatter and Maps
### Binned Scatterplot
```
source = data.movies.url
alt.Chart(source).mark_circle().encode(
alt.X('IMDB_Rating:Q', bin=True),
alt.Y('Rotten_Tomatoes_Rating:Q', bin=True),
size='count()'
).properties(
width="container",
height="container"
)
```
### Multifeature Scatter Plot
```
source = data.iris()
alt.Chart(source).mark_circle().encode(
alt.X('sepalLength', scale=alt.Scale(zero=False)),
alt.Y('sepalWidth', scale=alt.Scale(zero=False, padding=1)),
color='species',
size='petalWidth'
).properties(
width="container",
height="container"
)
```
## Col 2
### Choropleth Map
```
from vega_datasets import data
counties = alt.topo_feature(data.us_10m.url, 'counties')
source = data.unemployment.url
alt.Chart(counties).mark_geoshape().encode(
color='rate:Q'
).transform_lookup(
lookup='id',
from_=alt.LookupData(source, 'id', ['rate'])
).project(
type='albersUsa'
).properties(
width="container",
height="container"
)
```
### Layered Histogram
```
# Generating Data
source = pd.DataFrame({
'Trial A': np.random.normal(0, 0.8, 1000),
'Trial B': np.random.normal(-2, 1, 1000),
'Trial C': np.random.normal(3, 2, 1000)
})
alt.Chart(source).transform_fold(
['Trial A', 'Trial B', 'Trial C'],
as_=['Experiment', 'Measurement']
).mark_area(
opacity=0.3,
interpolate='step'
).encode(
alt.X('Measurement:Q', bin=alt.Bin(maxbins=100)),
alt.Y('count()', stack=None),
alt.Color('Experiment:N')
).properties(
width="container",
height="container"
)
```
# Scatter Matrix
```
source = data.cars()
alt.Chart(source).mark_circle().encode(
alt.X(alt.repeat("column"), type='quantitative'),
alt.Y(alt.repeat("row"), type='quantitative'),
color='Origin:N'
).repeat(
row=['Horsepower', 'Acceleration', 'Miles_per_Gallon'],
column=['Miles_per_Gallon', 'Acceleration', 'Horsepower']
).interactive()
```
# Faceted Density Estimates
```
source = data.iris()
alt.Chart(source).transform_fold(
['petalWidth',
'petalLength',
'sepalWidth',
'sepalLength'],
as_ = ['Measurement_type', 'value']
).transform_density(
density='value',
bandwidth=0.3,
groupby=['Measurement_type'],
extent= [0, 8]
).mark_area().encode(
alt.X('value:Q'),
alt.Y('density:Q'),
alt.Row('Measurement_type:N')
).properties(width=600, height=180)
```
# Interactive
### Interactive Crossfilter
```
source = alt.UrlData(
data.flights_2k.url,
format={'parse': {'date': 'date'}}
)
brush = alt.selection(type='interval', encodings=['x'])
# Define the base chart, with the common parts of the
# background and highlights
base = alt.Chart().mark_bar().encode(
x=alt.X(alt.repeat('column'), type='quantitative', bin=alt.Bin(maxbins=20)),
y='count()'
).properties(
width=200,
height=300
)
# gray background with selection
background = base.encode(
color=alt.value('#ddd')
).add_selection(brush)
# blue highlights on the transformed data
highlight = base.transform_filter(brush)
# layer the two charts & repeat
alt.layer(
background,
highlight,
data=source
).transform_calculate(
"time",
"hours(datum.date)"
).repeat(column=["distance", "delay", "time"])
```
### Scatter Plot and Histogram with Interval Selection
```
x = np.random.normal(size=100)
y = np.random.normal(size=100)
m = np.random.normal(15, 1, size=100)
source = pd.DataFrame({"x": x, "y":y, "m":m})
# interval selection in the scatter plot
pts = alt.selection(type="interval", encodings=["x"])
# left panel: scatter plot
points = alt.Chart().mark_point(filled=True, color="black").encode(
x='x',
y='y'
).transform_filter(
pts
).properties(
width=300,
height=300
)
# right panel: histogram
mag = alt.Chart().mark_bar().encode(
x='mbin:N',
y="count()",
color=alt.condition(pts, alt.value("black"), alt.value("lightgray"))
).properties(
width=300,
height=300
).add_selection(pts)
# build the chart:
alt.hconcat(
points,
mag,
data=source
).transform_bin(
"mbin",
field="m",
bin=alt.Bin(maxbins=20)
)
```
## Col 2
### Interactive average
```
source = data.seattle_weather()
brush = alt.selection(type='interval', encodings=['x'])
bars = alt.Chart().mark_bar().encode(
x='month(date):O',
y='mean(precipitation):Q',
opacity=alt.condition(brush, alt.OpacityValue(1), alt.OpacityValue(0.7)),
).add_selection(
brush
).properties(width=700, height=300)
line = alt.Chart().mark_rule(color='firebrick').encode(
y='mean(precipitation):Q',
size=alt.SizeValue(3)
).transform_filter(
brush
).properties(width=700, height=300)
alt.layer(bars, line, data=source)
```
### Interactive Legend
```
source = data.unemployment_across_industries.url
selection = alt.selection_multi(fields=['series'], bind='legend')
alt.Chart(source).mark_area().encode(
alt.X('yearmonth(date):T', axis=alt.Axis(domain=False, format='%Y', tickSize=0)),
alt.Y('sum(count):Q', stack='center', axis=None),
alt.Color('series:N', scale=alt.Scale(scheme='category20b')),
opacity=alt.condition(selection, alt.value(1), alt.value(0.2))
).properties(
width="container",
height="container"
).add_selection(
selection
)
```
|
github_jupyter
|
flex_title = "Altair plots"
flex_author = "built using jupyter-flex"
flex_source_link = "https://github.com/danielfrg/jupyter-flex/blob/master/examples/plots/altair.ipynb"
flex_include_source = True
import numpy as np
import pandas as pd
import altair as alt
from vega_datasets import data
alt.renderers.set_embed_options(actions=False)
np.random.seed(42)
source = data.cars()
plot = alt.Chart(source).mark_circle(size=60).encode(
x='Horsepower',
y='Miles_per_Gallon',
color='Origin',
tooltip=['Name', 'Origin', 'Horsepower', 'Miles_per_Gallon']
)
plot
plot.properties(
width='container',
height='container'
).interactive()
source = pd.DataFrame({
'a': ['A', 'B', 'C', 'D', 'E', 'F', 'G', 'H', 'I'],
'b': [28, 55, 43, 91, 81, 53, 19, 87, 52]
})
plot = alt.Chart(source).mark_bar().encode(
x='a',
y='b'
)
plot
plot.properties(
width='container',
height='container'
)
# Compute x^2 + y^2 across a 2D grid
x, y = np.meshgrid(range(-5, 5), range(-5, 5))
z = x ** 2 + y ** 2
# Convert this grid to columnar data expected by Altair
source = pd.DataFrame({'x': x.ravel(),
'y': y.ravel(),
'z': z.ravel()})
plot = alt.Chart(source).mark_rect().encode(
x='x:O',
y='y:O',
color='z:Q'
)
plot
plot.properties(
width='container',
height='container'
)
source = data.us_employment()
alt.Chart(source).mark_bar().encode(
x="month:T",
y="nonfarm_change:Q",
color=alt.condition(
alt.datum.nonfarm_change > 0,
alt.value("steelblue"), # The positive color
alt.value("orange") # The negative color
)
).properties(
width="container",
height="container"
)
source = data.wheat()
alt.Chart(source).mark_bar().encode(
x='wheat:Q',
y="year:O"
).properties(
width="container",
height="container"
)
source = data.barley()
alt.Chart(source).mark_bar().encode(
x='variety',
y='sum(yield)',
color='site'
).properties(
width="container",
height="container"
)
source = data.stocks()
alt.Chart(source).mark_area(
color="lightblue",
interpolate='step-after',
line=True
).encode(
x='date',
y='price'
).transform_filter(alt.datum.symbol == 'GOOG').properties(
width="container",
height="container"
)
source = data.stocks()
alt.Chart(source).mark_line().encode(
x='date',
y='price',
color='symbol'
).properties(
width="container",
height="container"
)
source = data.movies.url
alt.Chart(source).transform_window(
cumulative_count="count()",
sort=[{"field": "IMDB_Rating"}],
).mark_area().encode(
x="IMDB_Rating:Q",
y="cumulative_count:Q"
).properties(
width="container",
height="container"
)
source = data.iris()
alt.Chart(source).transform_fold(
['petalWidth',
'petalLength',
'sepalWidth',
'sepalLength'],
as_ = ['Measurement_type', 'value']
).transform_density(
density='value',
bandwidth=0.3,
groupby=['Measurement_type'],
extent= [0, 8],
counts = True,
steps=200
).mark_area().encode(
alt.X('value:Q'),
alt.Y('density:Q', stack='zero'),
alt.Color('Measurement_type:N')
).properties(
width="container",
height="container"
)
source = data.movies.url
alt.Chart(source).mark_circle().encode(
alt.X('IMDB_Rating:Q', bin=True),
alt.Y('Rotten_Tomatoes_Rating:Q', bin=True),
size='count()'
).properties(
width="container",
height="container"
)
source = data.iris()
alt.Chart(source).mark_circle().encode(
alt.X('sepalLength', scale=alt.Scale(zero=False)),
alt.Y('sepalWidth', scale=alt.Scale(zero=False, padding=1)),
color='species',
size='petalWidth'
).properties(
width="container",
height="container"
)
from vega_datasets import data
counties = alt.topo_feature(data.us_10m.url, 'counties')
source = data.unemployment.url
alt.Chart(counties).mark_geoshape().encode(
color='rate:Q'
).transform_lookup(
lookup='id',
from_=alt.LookupData(source, 'id', ['rate'])
).project(
type='albersUsa'
).properties(
width="container",
height="container"
)
# Generating Data
source = pd.DataFrame({
'Trial A': np.random.normal(0, 0.8, 1000),
'Trial B': np.random.normal(-2, 1, 1000),
'Trial C': np.random.normal(3, 2, 1000)
})
alt.Chart(source).transform_fold(
['Trial A', 'Trial B', 'Trial C'],
as_=['Experiment', 'Measurement']
).mark_area(
opacity=0.3,
interpolate='step'
).encode(
alt.X('Measurement:Q', bin=alt.Bin(maxbins=100)),
alt.Y('count()', stack=None),
alt.Color('Experiment:N')
).properties(
width="container",
height="container"
)
source = data.cars()
alt.Chart(source).mark_circle().encode(
alt.X(alt.repeat("column"), type='quantitative'),
alt.Y(alt.repeat("row"), type='quantitative'),
color='Origin:N'
).repeat(
row=['Horsepower', 'Acceleration', 'Miles_per_Gallon'],
column=['Miles_per_Gallon', 'Acceleration', 'Horsepower']
).interactive()
source = data.iris()
alt.Chart(source).transform_fold(
['petalWidth',
'petalLength',
'sepalWidth',
'sepalLength'],
as_ = ['Measurement_type', 'value']
).transform_density(
density='value',
bandwidth=0.3,
groupby=['Measurement_type'],
extent= [0, 8]
).mark_area().encode(
alt.X('value:Q'),
alt.Y('density:Q'),
alt.Row('Measurement_type:N')
).properties(width=600, height=180)
source = alt.UrlData(
data.flights_2k.url,
format={'parse': {'date': 'date'}}
)
brush = alt.selection(type='interval', encodings=['x'])
# Define the base chart, with the common parts of the
# background and highlights
base = alt.Chart().mark_bar().encode(
x=alt.X(alt.repeat('column'), type='quantitative', bin=alt.Bin(maxbins=20)),
y='count()'
).properties(
width=200,
height=300
)
# gray background with selection
background = base.encode(
color=alt.value('#ddd')
).add_selection(brush)
# blue highlights on the transformed data
highlight = base.transform_filter(brush)
# layer the two charts & repeat
alt.layer(
background,
highlight,
data=source
).transform_calculate(
"time",
"hours(datum.date)"
).repeat(column=["distance", "delay", "time"])
x = np.random.normal(size=100)
y = np.random.normal(size=100)
m = np.random.normal(15, 1, size=100)
source = pd.DataFrame({"x": x, "y":y, "m":m})
# interval selection in the scatter plot
pts = alt.selection(type="interval", encodings=["x"])
# left panel: scatter plot
points = alt.Chart().mark_point(filled=True, color="black").encode(
x='x',
y='y'
).transform_filter(
pts
).properties(
width=300,
height=300
)
# right panel: histogram
mag = alt.Chart().mark_bar().encode(
x='mbin:N',
y="count()",
color=alt.condition(pts, alt.value("black"), alt.value("lightgray"))
).properties(
width=300,
height=300
).add_selection(pts)
# build the chart:
alt.hconcat(
points,
mag,
data=source
).transform_bin(
"mbin",
field="m",
bin=alt.Bin(maxbins=20)
)
source = data.seattle_weather()
brush = alt.selection(type='interval', encodings=['x'])
bars = alt.Chart().mark_bar().encode(
x='month(date):O',
y='mean(precipitation):Q',
opacity=alt.condition(brush, alt.OpacityValue(1), alt.OpacityValue(0.7)),
).add_selection(
brush
).properties(width=700, height=300)
line = alt.Chart().mark_rule(color='firebrick').encode(
y='mean(precipitation):Q',
size=alt.SizeValue(3)
).transform_filter(
brush
).properties(width=700, height=300)
alt.layer(bars, line, data=source)
source = data.unemployment_across_industries.url
selection = alt.selection_multi(fields=['series'], bind='legend')
alt.Chart(source).mark_area().encode(
alt.X('yearmonth(date):T', axis=alt.Axis(domain=False, format='%Y', tickSize=0)),
alt.Y('sum(count):Q', stack='center', axis=None),
alt.Color('series:N', scale=alt.Scale(scheme='category20b')),
opacity=alt.condition(selection, alt.value(1), alt.value(0.2))
).properties(
width="container",
height="container"
).add_selection(
selection
)
| 0.887424 | 0.88263 |
# We illustrate the use of SlackMinimizer in two ways.
## The script slack_minimizer.py can be run directly, showing the results on a random forest (see source comments around main() for details).
```
%run slack_minimizer.py data/a1a_all.csv 1000 20000 -f 0.01 -k -1 -d 40 --tree_node_specialists
```
## We also give an example in which many heterogeneous non-tree classifiers are combined with SlackMinimizer.
```
import composite_feature
import numpy as np
import scipy as sp
import sklearn.linear_model, sklearn.ensemble
import muffled_utils
import time
import sklearn.metrics
labeled_file = 'data/a1a_all.csv'
labeled_set_size = 1000
unlabeled_set_size = 10000
holdout_set_size = 500
validation_set_size = 1000
inittime = time.time()
(x_train, y_train, x_unl, y_unl, x_out, y_out, x_validate, y_validate) = muffled_utils.read_random_data_from_csv(
labeled_file, labeled_set_size, unlabeled_set_size, holdout_set_size, validation_set_size)
print('Data loaded. \tTime = ' + str(time.time() - inittime))
# Now train a few different base classifiers
inittime = time.time()
skcl = []
clrf = sklearn.ensemble.RandomForestClassifier(n_estimators=50, n_jobs=-1)
skcl.append(('Plain RF', clrf))
cldt = sklearn.tree.DecisionTreeClassifier()
skcl.append(('DT', cldt))
cletf = sklearn.ensemble.AdaBoostClassifier(n_estimators=50, algorithm='SAMME')
skcl.append(('AdaBoost', cletf))
clgb = sklearn.ensemble.GradientBoostingClassifier(n_estimators=50)#, loss='deviance')
skcl.append(('LogitBoost', clgb))
cllogistic = sklearn.linear_model.LogisticRegression()#(loss='log')
skcl.append(('Logistic regression', cllogistic))
#clgp = sklearn.gaussian_process.GaussianProcessClassifier()
#skcl.append(('Gaussian process', clgp))
# Now x_train is a (LABELED_SET_SIZE x d) matrix, and y_train a vector of size LABELED_SET_SIZE.
for i in range(len(skcl)):
skcl[i][1].fit(x_train, y_train)
print(skcl[i][0] + ' trained', time.time() - inittime)
classifier_list = list(zip(*skcl)[1])
print [sklearn.metrics.roc_auc_score(y_validate, c.predict(x_validate)) for c in classifier_list]
k = 0
failure_prob = 0.0005
inittime = time.time()
(b_vector, allfeats_out, allfeats_unl, allfeats_val) = composite_feature.predict_multiple(
classifier_list, x_out, x_unl, x_validate, y_out=y_out, k=k,
failure_prob=failure_prob, from_sklearn_rf=False, use_tree_partition=False)
print ('Featurizing done. \tTime = ' + str(time.time() - inittime))
import slack_minimizer
gradh = slack_minimizer.SlackMinimizer(
b_vector, allfeats_unl, allfeats_out, y_out, unlabeled_labels=y_unl,
validation_set=allfeats_val, validation_labels=y_validate)
statauc = gradh.sgd(50, unl_stride_size=100, linesearch=True, logging_interval=5)
```
|
github_jupyter
|
%run slack_minimizer.py data/a1a_all.csv 1000 20000 -f 0.01 -k -1 -d 40 --tree_node_specialists
import composite_feature
import numpy as np
import scipy as sp
import sklearn.linear_model, sklearn.ensemble
import muffled_utils
import time
import sklearn.metrics
labeled_file = 'data/a1a_all.csv'
labeled_set_size = 1000
unlabeled_set_size = 10000
holdout_set_size = 500
validation_set_size = 1000
inittime = time.time()
(x_train, y_train, x_unl, y_unl, x_out, y_out, x_validate, y_validate) = muffled_utils.read_random_data_from_csv(
labeled_file, labeled_set_size, unlabeled_set_size, holdout_set_size, validation_set_size)
print('Data loaded. \tTime = ' + str(time.time() - inittime))
# Now train a few different base classifiers
inittime = time.time()
skcl = []
clrf = sklearn.ensemble.RandomForestClassifier(n_estimators=50, n_jobs=-1)
skcl.append(('Plain RF', clrf))
cldt = sklearn.tree.DecisionTreeClassifier()
skcl.append(('DT', cldt))
cletf = sklearn.ensemble.AdaBoostClassifier(n_estimators=50, algorithm='SAMME')
skcl.append(('AdaBoost', cletf))
clgb = sklearn.ensemble.GradientBoostingClassifier(n_estimators=50)#, loss='deviance')
skcl.append(('LogitBoost', clgb))
cllogistic = sklearn.linear_model.LogisticRegression()#(loss='log')
skcl.append(('Logistic regression', cllogistic))
#clgp = sklearn.gaussian_process.GaussianProcessClassifier()
#skcl.append(('Gaussian process', clgp))
# Now x_train is a (LABELED_SET_SIZE x d) matrix, and y_train a vector of size LABELED_SET_SIZE.
for i in range(len(skcl)):
skcl[i][1].fit(x_train, y_train)
print(skcl[i][0] + ' trained', time.time() - inittime)
classifier_list = list(zip(*skcl)[1])
print [sklearn.metrics.roc_auc_score(y_validate, c.predict(x_validate)) for c in classifier_list]
k = 0
failure_prob = 0.0005
inittime = time.time()
(b_vector, allfeats_out, allfeats_unl, allfeats_val) = composite_feature.predict_multiple(
classifier_list, x_out, x_unl, x_validate, y_out=y_out, k=k,
failure_prob=failure_prob, from_sklearn_rf=False, use_tree_partition=False)
print ('Featurizing done. \tTime = ' + str(time.time() - inittime))
import slack_minimizer
gradh = slack_minimizer.SlackMinimizer(
b_vector, allfeats_unl, allfeats_out, y_out, unlabeled_labels=y_unl,
validation_set=allfeats_val, validation_labels=y_validate)
statauc = gradh.sgd(50, unl_stride_size=100, linesearch=True, logging_interval=5)
| 0.30632 | 0.842863 |
```
# The code starting from here is for setting up the environment of Google colab
# We need to connect google colab to google drive so that we can save the files and call scripts in the google drive
# We also need to install the related package: for example allennlp
# There will be an indicator to show where this code part ends
from google.colab import drive
drive.mount('/content/gdrive')
# Install a Drive FUSE wrapper.
# https://github.com/astrada/google-drive-ocamlfuse
!apt-get install -y -qq software-properties-common python-software-properties module-init-tools
!add-apt-repository -y ppa:alessandro-strada/ppa 2>&1 > /dev/null
!apt-get update -qq 2>&1 > /dev/null
!apt-get -y install -qq google-drive-ocamlfuse fuse
# Generate auth tokens for Colab
from google.colab import auth
auth.authenticate_user()
# Generate creds for the Drive FUSE library.
from oauth2client.client import GoogleCredentials
creds = GoogleCredentials.get_application_default()
import getpass
!google-drive-ocamlfuse -headless -id={creds.client_id} -secret={creds.client_secret} < /dev/null 2>&1 | grep URL
vcode = getpass.getpass()
!echo {vcode} | google-drive-ocamlfuse -headless -id={creds.client_id} -secret={creds.client_secret}
# Create a directory and mount Google Drive using that directory.
!mkdir -p drive
!google-drive-ocamlfuse drive
# http://pytorch.org/
from os import path
from wheel.pep425tags import get_abbr_impl, get_impl_ver, get_abi_tag
platform = '{}{}-{}'.format(get_abbr_impl(), get_impl_ver(), get_abi_tag())
accelerator = 'cu80' if path.exists('/opt/bin/nvidia-smi') else 'cpu'
!pip3 install -q http://download.pytorch.org/whl/{accelerator}/torch-0.4.0-{platform}-linux_x86_64.whl torchvision
!pip install overrides
!pip install allennlp
import os
!ls
os.chdir('./drive/269')
# Here we end the code for environment setting and begin the main part of the code
# In this code, we use ELMO as word embedding and LEAM as our model
# The tutorial of AllenNLP is http://mlexplained.com/2019/01/30/an-in-depth-tutorial-to-allennlp-from-basics-to-elmo-and-bert/
# In this tutorial there is also instructions about how to change ELMO to BERT
# to change the model to BERT, code need to be changed includes following codes in this notebook, model.py maybe also trainer.py
import os
from main import main
from src.model import *
from src.utils import *
from src.trainer import *
# we first deal with the data
# to satisfy the requirements of AllenNLP api, we first save our data to pickle file
if not os.path.exists('train.pickle') or not os.path.exists('test.pickle') or not os.path.exists('class_name.pickle'):
data_dir = 'data/reuters'
vocab_url = os.path.join(data_dir, 'vocab.pkl')
train_url = os.path.join(data_dir, 'train')
test_url = os.path.join(data_dir, 'test')
vocab = pickle.load(open(vocab_url, 'rb'))
vocab = list(zip(*sorted(vocab.items(), key=lambda x: x[1])))[0]
vocab_size = len(vocab)
train_set, train_labels, class_names = dataset(train_url)
test_set, test_labels, _ = dataset(test_url)
train_text = []
for idx_list in train_set:
s = ""
for idx in idx_list:
s += vocab[idx] + " "
train_text.append(s)
test_text = []
for idx_list in test_set:
s = ""
for idx in idx_list:
s += vocab[idx] + " "
test_text.append(s)
train = {'text': train_text,
'id': np.arange(len(train_text)),
'labels': train_labels}
with open('train.pickle', 'wb') as handle:
pickle.dump(train, handle, protocol=pickle.HIGHEST_PROTOCOL)
test = {'text': test_text,
'id': np.arange(len(test_text)),
'labels': test_labels}
with open('test.pickle', 'wb') as handle:
pickle.dump(test, handle, protocol=pickle.HIGHEST_PROTOCOL)
class_name = {'name': class_names}
with open('class_name.pickle', 'wb') as handle:
pickle.dump(class_name, handle, protocol=pickle.HIGHEST_PROTOCOL)
from pathlib import Path
from typing import *
import torch
import torch.optim as optim
import numpy as np
import pandas as pd
import pickle
from functools import partial
from overrides import overrides
from allennlp.data import Instance
from allennlp.data.token_indexers import TokenIndexer
from allennlp.data.tokenizers import Token
from allennlp.nn import util as nn_util
from allennlp.data.vocabulary import Vocabulary
from allennlp.data.dataset_readers import DatasetReader
# allennlp API for data preprocessing
from allennlp.data.fields import TextField, MetadataField, ArrayField
class JigsawDatasetReader(DatasetReader):
def __init__(self, tokenizer: Callable[[str], List[str]]=lambda x: x.split(),
token_indexers: Dict[str, TokenIndexer] = None,
max_seq_len: Optional[int]=80) -> None:
super().__init__(lazy=False)
self.tokenizer = tokenizer
self.token_indexers = token_indexers or {"tokens": SingleIdTokenIndexer()}
self.max_seq_len = max_seq_len
@overrides
def text_to_instance(self, tokens: List[Token], id: str,
labels: np.ndarray) -> Instance:
sentence_field = TextField(tokens, self.token_indexers)
fields = {"tokens": sentence_field}
id_field = MetadataField(id)
fields["id"] = id_field
label_field = ArrayField(array=labels)
fields["label"] = label_field
return Instance(fields)
@overrides
def _read(self, file_path: str) -> Iterator[Instance]:
with open(file_path, 'rb') as handle:
b = pickle.load(handle)
text = b['text']
idx = b['id']
labels = b['labels']
assert len(text) == len(idx) and len(text) == len(labels)
for i in range(len(text)):
yield self.text_to_instance(
[Token(x) for x in self.tokenizer(text[i])],
idx[i], labels[i],
)
from allennlp.data.tokenizers.word_splitter import SpacyWordSplitter
from allennlp.data.token_indexers.elmo_indexer import ELMoCharacterMapper, ELMoTokenCharactersIndexer
# the token indexer is responsible for mapping tokens to integers
token_indexer = ELMoTokenCharactersIndexer()
reader = JigsawDatasetReader(
token_indexers={"tokens": token_indexer}
)
with open('class_name.pickle', 'rb') as handle:
tmp = pickle.load(handle)
class_names = tmp['name']
print(class_names)
train_ds, test_ds = (reader.read(fname) for fname in ["train.pickle", "test.pickle"])
val_ds = None
vocab = Vocabulary()
from allennlp.data.iterators import BucketIterator
iterator = BucketIterator(batch_size=30, sorting_keys=[("tokens", "num_tokens")])
iterator.index_with(vocab)
model = Leam_Classifier(len(class_names), 256, 256, 10,
n_layer=1, dropout_rate=0.5, embpath=None, label_att=True, multilabel=True)
model.load_state_dict(torch.load('model1.pth'))
trainer = Trainer(iterator, batch_size=30, num_epoches=50, learning_rate=3e-4, valid_freq=10, model_type='embed')
trainer.set_validation(test_ds)
trainer.init_model(model)
# train the model and save the result
#trainer.model.load_state_dict(torch.load('model1.pth'))
train_prob, train_beta = trainer.fit(train_ds, class_names)
torch.save(trainer.model.state_dict(), 'model.pth')
# here we load the pretrained model intorch.save(trainer.model.state_dict(), 'model.pth')
# trainer.model.load_state_dict(torch.load('model.pth'))
# first test it on the validation set to see wheter we have loaded in the right model
prob, _ = trainer.predict()
tmp = trainer.val_y.detach().cpu().numpy()
pred = prob > 0.5
val_match = np.mean([(pred[i][tmp[i]==1]==1).any() for i in range(len(pred))])
val_hs = (((pred==1)*(tmp==1)).sum(1)/(((pred==1)+(tmp==1))>0).sum(1)).mean()
print("Validation result: ",
"val_match : %0.4f" % val_match,
"val_hs : %0.4f" % val_hs
)
```
```
# load in the testset
# original res testing result: test_match : 0.9156 test_hs : 0.8540
if not os.path.exists('task.pickle'):
task_url = os.path.join(data_dir, 'task')
seq_task, task_labels, _ = dataset(task_url, monitor=False)
vocab = pickle.load(open(vocab_url, 'rb'))
vocab = list(zip(*sorted(vocab.items(), key=lambda x: x[1])))[0]
task_text = []
for idx_list in seq_task:
s = ""
for idx in idx_list:
s += vocab[idx] + " "
task_text.append(s)
task = {'text': task_text,
'id': np.arange(len(task_text)),
'labels': task_labels}
with open('task.pickle', 'wb') as handle:
pickle.dump(task, handle, protocol=pickle.HIGHEST_PROTOCOL)
task_ds = reader.read('task.pickle')
'''
trainer.set_validation(task_ds)
prob, _ = trainer.predict()
tmp = trainer.val_y.detach().cpu().numpy()
pred = prob > 0.5
test_match = np.mean([(pred[i][tmp[i]==1]==1).any() for i in range(len(pred))])
test_hs = (((pred==1)*(tmp==1)).sum(1)/(((pred==1)+(tmp==1))>0).sum(1)).mean()
print("testing result: ",
"test_match : %0.4f" % test_match,
"test_hs : %0.4f" % test_hs
)
'''
# seems the original code (code used when doing this experiments) doesn't save the final model to trainer.model
# so here we manually load the model in trainer and do evaluation here
# best testing result: test_match : 0.9162 test_hs : 0.8686
from allennlp.data.iterators import BucketIterator
iterator = BucketIterator(batch_size=30, sorting_keys=[("tokens", "num_tokens")])
iterator.index_with(vocab)
model = Leam_Classifier(len(class_names), 256, 256, 10,
n_layer=1, dropout_rate=0.5, embpath=None, label_att=True, multilabel=True)
model.load_state_dict(torch.load('model.pth'))
trainer = Trainer(iterator, batch_size=30, num_epoches=50, learning_rate=3e-4, valid_freq=10, model_type='embed')
trainer.init_model(model)
trainer.set_validation(task_ds)
prob, _ = trainer.predict()
tmp = trainer.val_y.detach().cpu().numpy()
pred = prob > 0.5
test_match = np.mean([(pred[i][tmp[i]==1]==1).any() for i in range(len(pred))])
test_hs = (((pred==1)*(tmp==1)).sum(1)/(((pred==1)+(tmp==1))>0).sum(1)).mean()
precision_list = []
recall_list = []
for i in range(101):
cur_thresh = i * 0.01
pred = prob > cur_thresh
precision = (((pred==1)*(tmp==1)).sum(1) / ((pred==1)).sum(1)).mean()
recall = (((pred==1)*(tmp==1)).sum(1) / ((tmp==1)).sum(1)).mean()
precision_list.append(precision)
recall_list.append(recall)
print("testing result: ",
"test_match : %0.4f" % test_match,
"test_hs : %0.4f" % test_hs
)
precision_list = []
recall_list = []
fp_list = []
for i in range(0, 10001):
cur_thresh = i * 0.0001
pred = (prob > cur_thresh).astype(np.int)
precision = ((pred * tmp).sum(1) / (pred.sum(1) + 1e-5)).mean()
recall = ((pred * tmp).sum(1) / (tmp.sum(1) + 1e-5)).mean()
fp = 1 - ((((pred == 0) * (tmp ==0)).sum(1)) / ((tmp==0).sum(1) + 1e-5)).mean()
precision_list.append(precision)
recall_list.append(recall)
fp_list.append(fp)
#print(precision_list)
#print(recall_list)
from sklearn.metrics import auc
from matplotlib import pyplot as plt
myauc = auc(recall_list, precision_list)
print(myauc)
plt.figure()
plt.plot(recall_list, precision_list)
plt.ylabel('precision')
plt.xlabel('recall')
plt.title('precision recall curve')
myauc= auc(fp_list, recall_list)
print(myauc)
plt.figure()
plt.plot(fp_list, recall_list)
plt.ylabel('recall')
plt.xlabel('false positive rate')
plt.title('ROC curve')
torch.save(model.state_dict(), 'model.pth')
# here we try to use ELMO without LEAM
from allennlp.data.iterators import BucketIterator
iterator = BucketIterator(batch_size=30, sorting_keys=[("tokens", "num_tokens")])
iterator.index_with(vocab)
model = Leam_Classifier(len(class_names), 256, 256, 10,
n_layer=1, dropout_rate=0.5, embpath=None, label_att=False, multilabel=True)
#model.load_state_dict(torch.load('model1.pth'))
trainer = Trainer(iterator, batch_size=30, num_epoches=120, learning_rate=1e-3, valid_freq=10, model_type='noembed')
trainer.set_validation(test_ds)
trainer.init_model(model)
train_prob = trainer.fit(train_ds, class_names)
torch.save(trainer.model.state_dict(), 'model_no_LEAM.pth')
prob, _ = trainer.predict()
tmp = trainer.val_y.detach().cpu().numpy()
pred = prob > 0.5
val_match = np.mean([(pred[i][tmp[i]==1]==1).any() for i in range(len(pred))])
val_hs = (((pred==1)*(tmp==1)).sum(1)/(((pred==1)+(tmp==1))>0).sum(1)).mean()
print("Validation result: ",
"val_match : %0.4f" % val_match,
"val_hs : %0.4f" % val_hs
)
task_ds = reader.read('task.pickle')
trainer.set_validation(task_ds)
prob, _ = trainer.predict()
tmp = trainer.val_y.detach().cpu().numpy()
pred = prob > 0.5
test_match = np.mean([(pred[i][tmp[i]==1]==1).any() for i in range(len(pred))])
test_hs = (((pred==1)*(tmp==1)).sum(1)/(((pred==1)+(tmp==1))>0).sum(1)).mean()
print("testing result: ",
"test_match : %0.4f" % test_match,
"test_hs : %0.4f" % test_hs
)
```
|
github_jupyter
|
# The code starting from here is for setting up the environment of Google colab
# We need to connect google colab to google drive so that we can save the files and call scripts in the google drive
# We also need to install the related package: for example allennlp
# There will be an indicator to show where this code part ends
from google.colab import drive
drive.mount('/content/gdrive')
# Install a Drive FUSE wrapper.
# https://github.com/astrada/google-drive-ocamlfuse
!apt-get install -y -qq software-properties-common python-software-properties module-init-tools
!add-apt-repository -y ppa:alessandro-strada/ppa 2>&1 > /dev/null
!apt-get update -qq 2>&1 > /dev/null
!apt-get -y install -qq google-drive-ocamlfuse fuse
# Generate auth tokens for Colab
from google.colab import auth
auth.authenticate_user()
# Generate creds for the Drive FUSE library.
from oauth2client.client import GoogleCredentials
creds = GoogleCredentials.get_application_default()
import getpass
!google-drive-ocamlfuse -headless -id={creds.client_id} -secret={creds.client_secret} < /dev/null 2>&1 | grep URL
vcode = getpass.getpass()
!echo {vcode} | google-drive-ocamlfuse -headless -id={creds.client_id} -secret={creds.client_secret}
# Create a directory and mount Google Drive using that directory.
!mkdir -p drive
!google-drive-ocamlfuse drive
# http://pytorch.org/
from os import path
from wheel.pep425tags import get_abbr_impl, get_impl_ver, get_abi_tag
platform = '{}{}-{}'.format(get_abbr_impl(), get_impl_ver(), get_abi_tag())
accelerator = 'cu80' if path.exists('/opt/bin/nvidia-smi') else 'cpu'
!pip3 install -q http://download.pytorch.org/whl/{accelerator}/torch-0.4.0-{platform}-linux_x86_64.whl torchvision
!pip install overrides
!pip install allennlp
import os
!ls
os.chdir('./drive/269')
# Here we end the code for environment setting and begin the main part of the code
# In this code, we use ELMO as word embedding and LEAM as our model
# The tutorial of AllenNLP is http://mlexplained.com/2019/01/30/an-in-depth-tutorial-to-allennlp-from-basics-to-elmo-and-bert/
# In this tutorial there is also instructions about how to change ELMO to BERT
# to change the model to BERT, code need to be changed includes following codes in this notebook, model.py maybe also trainer.py
import os
from main import main
from src.model import *
from src.utils import *
from src.trainer import *
# we first deal with the data
# to satisfy the requirements of AllenNLP api, we first save our data to pickle file
if not os.path.exists('train.pickle') or not os.path.exists('test.pickle') or not os.path.exists('class_name.pickle'):
data_dir = 'data/reuters'
vocab_url = os.path.join(data_dir, 'vocab.pkl')
train_url = os.path.join(data_dir, 'train')
test_url = os.path.join(data_dir, 'test')
vocab = pickle.load(open(vocab_url, 'rb'))
vocab = list(zip(*sorted(vocab.items(), key=lambda x: x[1])))[0]
vocab_size = len(vocab)
train_set, train_labels, class_names = dataset(train_url)
test_set, test_labels, _ = dataset(test_url)
train_text = []
for idx_list in train_set:
s = ""
for idx in idx_list:
s += vocab[idx] + " "
train_text.append(s)
test_text = []
for idx_list in test_set:
s = ""
for idx in idx_list:
s += vocab[idx] + " "
test_text.append(s)
train = {'text': train_text,
'id': np.arange(len(train_text)),
'labels': train_labels}
with open('train.pickle', 'wb') as handle:
pickle.dump(train, handle, protocol=pickle.HIGHEST_PROTOCOL)
test = {'text': test_text,
'id': np.arange(len(test_text)),
'labels': test_labels}
with open('test.pickle', 'wb') as handle:
pickle.dump(test, handle, protocol=pickle.HIGHEST_PROTOCOL)
class_name = {'name': class_names}
with open('class_name.pickle', 'wb') as handle:
pickle.dump(class_name, handle, protocol=pickle.HIGHEST_PROTOCOL)
from pathlib import Path
from typing import *
import torch
import torch.optim as optim
import numpy as np
import pandas as pd
import pickle
from functools import partial
from overrides import overrides
from allennlp.data import Instance
from allennlp.data.token_indexers import TokenIndexer
from allennlp.data.tokenizers import Token
from allennlp.nn import util as nn_util
from allennlp.data.vocabulary import Vocabulary
from allennlp.data.dataset_readers import DatasetReader
# allennlp API for data preprocessing
from allennlp.data.fields import TextField, MetadataField, ArrayField
class JigsawDatasetReader(DatasetReader):
def __init__(self, tokenizer: Callable[[str], List[str]]=lambda x: x.split(),
token_indexers: Dict[str, TokenIndexer] = None,
max_seq_len: Optional[int]=80) -> None:
super().__init__(lazy=False)
self.tokenizer = tokenizer
self.token_indexers = token_indexers or {"tokens": SingleIdTokenIndexer()}
self.max_seq_len = max_seq_len
@overrides
def text_to_instance(self, tokens: List[Token], id: str,
labels: np.ndarray) -> Instance:
sentence_field = TextField(tokens, self.token_indexers)
fields = {"tokens": sentence_field}
id_field = MetadataField(id)
fields["id"] = id_field
label_field = ArrayField(array=labels)
fields["label"] = label_field
return Instance(fields)
@overrides
def _read(self, file_path: str) -> Iterator[Instance]:
with open(file_path, 'rb') as handle:
b = pickle.load(handle)
text = b['text']
idx = b['id']
labels = b['labels']
assert len(text) == len(idx) and len(text) == len(labels)
for i in range(len(text)):
yield self.text_to_instance(
[Token(x) for x in self.tokenizer(text[i])],
idx[i], labels[i],
)
from allennlp.data.tokenizers.word_splitter import SpacyWordSplitter
from allennlp.data.token_indexers.elmo_indexer import ELMoCharacterMapper, ELMoTokenCharactersIndexer
# the token indexer is responsible for mapping tokens to integers
token_indexer = ELMoTokenCharactersIndexer()
reader = JigsawDatasetReader(
token_indexers={"tokens": token_indexer}
)
with open('class_name.pickle', 'rb') as handle:
tmp = pickle.load(handle)
class_names = tmp['name']
print(class_names)
train_ds, test_ds = (reader.read(fname) for fname in ["train.pickle", "test.pickle"])
val_ds = None
vocab = Vocabulary()
from allennlp.data.iterators import BucketIterator
iterator = BucketIterator(batch_size=30, sorting_keys=[("tokens", "num_tokens")])
iterator.index_with(vocab)
model = Leam_Classifier(len(class_names), 256, 256, 10,
n_layer=1, dropout_rate=0.5, embpath=None, label_att=True, multilabel=True)
model.load_state_dict(torch.load('model1.pth'))
trainer = Trainer(iterator, batch_size=30, num_epoches=50, learning_rate=3e-4, valid_freq=10, model_type='embed')
trainer.set_validation(test_ds)
trainer.init_model(model)
# train the model and save the result
#trainer.model.load_state_dict(torch.load('model1.pth'))
train_prob, train_beta = trainer.fit(train_ds, class_names)
torch.save(trainer.model.state_dict(), 'model.pth')
# here we load the pretrained model intorch.save(trainer.model.state_dict(), 'model.pth')
# trainer.model.load_state_dict(torch.load('model.pth'))
# first test it on the validation set to see wheter we have loaded in the right model
prob, _ = trainer.predict()
tmp = trainer.val_y.detach().cpu().numpy()
pred = prob > 0.5
val_match = np.mean([(pred[i][tmp[i]==1]==1).any() for i in range(len(pred))])
val_hs = (((pred==1)*(tmp==1)).sum(1)/(((pred==1)+(tmp==1))>0).sum(1)).mean()
print("Validation result: ",
"val_match : %0.4f" % val_match,
"val_hs : %0.4f" % val_hs
)
# load in the testset
# original res testing result: test_match : 0.9156 test_hs : 0.8540
if not os.path.exists('task.pickle'):
task_url = os.path.join(data_dir, 'task')
seq_task, task_labels, _ = dataset(task_url, monitor=False)
vocab = pickle.load(open(vocab_url, 'rb'))
vocab = list(zip(*sorted(vocab.items(), key=lambda x: x[1])))[0]
task_text = []
for idx_list in seq_task:
s = ""
for idx in idx_list:
s += vocab[idx] + " "
task_text.append(s)
task = {'text': task_text,
'id': np.arange(len(task_text)),
'labels': task_labels}
with open('task.pickle', 'wb') as handle:
pickle.dump(task, handle, protocol=pickle.HIGHEST_PROTOCOL)
task_ds = reader.read('task.pickle')
'''
trainer.set_validation(task_ds)
prob, _ = trainer.predict()
tmp = trainer.val_y.detach().cpu().numpy()
pred = prob > 0.5
test_match = np.mean([(pred[i][tmp[i]==1]==1).any() for i in range(len(pred))])
test_hs = (((pred==1)*(tmp==1)).sum(1)/(((pred==1)+(tmp==1))>0).sum(1)).mean()
print("testing result: ",
"test_match : %0.4f" % test_match,
"test_hs : %0.4f" % test_hs
)
'''
# seems the original code (code used when doing this experiments) doesn't save the final model to trainer.model
# so here we manually load the model in trainer and do evaluation here
# best testing result: test_match : 0.9162 test_hs : 0.8686
from allennlp.data.iterators import BucketIterator
iterator = BucketIterator(batch_size=30, sorting_keys=[("tokens", "num_tokens")])
iterator.index_with(vocab)
model = Leam_Classifier(len(class_names), 256, 256, 10,
n_layer=1, dropout_rate=0.5, embpath=None, label_att=True, multilabel=True)
model.load_state_dict(torch.load('model.pth'))
trainer = Trainer(iterator, batch_size=30, num_epoches=50, learning_rate=3e-4, valid_freq=10, model_type='embed')
trainer.init_model(model)
trainer.set_validation(task_ds)
prob, _ = trainer.predict()
tmp = trainer.val_y.detach().cpu().numpy()
pred = prob > 0.5
test_match = np.mean([(pred[i][tmp[i]==1]==1).any() for i in range(len(pred))])
test_hs = (((pred==1)*(tmp==1)).sum(1)/(((pred==1)+(tmp==1))>0).sum(1)).mean()
precision_list = []
recall_list = []
for i in range(101):
cur_thresh = i * 0.01
pred = prob > cur_thresh
precision = (((pred==1)*(tmp==1)).sum(1) / ((pred==1)).sum(1)).mean()
recall = (((pred==1)*(tmp==1)).sum(1) / ((tmp==1)).sum(1)).mean()
precision_list.append(precision)
recall_list.append(recall)
print("testing result: ",
"test_match : %0.4f" % test_match,
"test_hs : %0.4f" % test_hs
)
precision_list = []
recall_list = []
fp_list = []
for i in range(0, 10001):
cur_thresh = i * 0.0001
pred = (prob > cur_thresh).astype(np.int)
precision = ((pred * tmp).sum(1) / (pred.sum(1) + 1e-5)).mean()
recall = ((pred * tmp).sum(1) / (tmp.sum(1) + 1e-5)).mean()
fp = 1 - ((((pred == 0) * (tmp ==0)).sum(1)) / ((tmp==0).sum(1) + 1e-5)).mean()
precision_list.append(precision)
recall_list.append(recall)
fp_list.append(fp)
#print(precision_list)
#print(recall_list)
from sklearn.metrics import auc
from matplotlib import pyplot as plt
myauc = auc(recall_list, precision_list)
print(myauc)
plt.figure()
plt.plot(recall_list, precision_list)
plt.ylabel('precision')
plt.xlabel('recall')
plt.title('precision recall curve')
myauc= auc(fp_list, recall_list)
print(myauc)
plt.figure()
plt.plot(fp_list, recall_list)
plt.ylabel('recall')
plt.xlabel('false positive rate')
plt.title('ROC curve')
torch.save(model.state_dict(), 'model.pth')
# here we try to use ELMO without LEAM
from allennlp.data.iterators import BucketIterator
iterator = BucketIterator(batch_size=30, sorting_keys=[("tokens", "num_tokens")])
iterator.index_with(vocab)
model = Leam_Classifier(len(class_names), 256, 256, 10,
n_layer=1, dropout_rate=0.5, embpath=None, label_att=False, multilabel=True)
#model.load_state_dict(torch.load('model1.pth'))
trainer = Trainer(iterator, batch_size=30, num_epoches=120, learning_rate=1e-3, valid_freq=10, model_type='noembed')
trainer.set_validation(test_ds)
trainer.init_model(model)
train_prob = trainer.fit(train_ds, class_names)
torch.save(trainer.model.state_dict(), 'model_no_LEAM.pth')
prob, _ = trainer.predict()
tmp = trainer.val_y.detach().cpu().numpy()
pred = prob > 0.5
val_match = np.mean([(pred[i][tmp[i]==1]==1).any() for i in range(len(pred))])
val_hs = (((pred==1)*(tmp==1)).sum(1)/(((pred==1)+(tmp==1))>0).sum(1)).mean()
print("Validation result: ",
"val_match : %0.4f" % val_match,
"val_hs : %0.4f" % val_hs
)
task_ds = reader.read('task.pickle')
trainer.set_validation(task_ds)
prob, _ = trainer.predict()
tmp = trainer.val_y.detach().cpu().numpy()
pred = prob > 0.5
test_match = np.mean([(pred[i][tmp[i]==1]==1).any() for i in range(len(pred))])
test_hs = (((pred==1)*(tmp==1)).sum(1)/(((pred==1)+(tmp==1))>0).sum(1)).mean()
print("testing result: ",
"test_match : %0.4f" % test_match,
"test_hs : %0.4f" % test_hs
)
| 0.480235 | 0.224544 |
# Step 1:
Install `jupyter_contrib_nbextensions` di **base** dengan command ini
```bash
conda install -c conda-forge jupyter_contrib_nbextensions
```
# Step 2:
Centang:
- Include custom menu content parsed from JSON string below
- Insert the new menu(s) before their sibling (the default value of false means they are inserted after the sibling)
- Snippets menus are often quite big, and positioned at the right side of the menu bar, so by default they open to the left of the menu. Set this to false to get them to open to the right as normal.
Sisanya di un-check
# Step 3:
Isi code ini ke dalam `JSON string parsed to define custom menus (only used if the option above is checked)`
```json
{
"name": "J.COp Snippets",
"sub-menu": [
{
"name": "Import common packages",
"snippet": [
"import numpy as np",
"import pandas as pd",
"",
"from sklearn.model_selection import train_test_split",
"from sklearn.pipeline import Pipeline",
"from sklearn.compose import ColumnTransformer",
"",
"from jcopml.pipeline import num_pipe, cat_pipe",
"from jcopml.utils import save_model, load_model",
"from jcopml.plot import plot_missing_value",
"from jcopml.feature_importance import mean_score_decrease"
]
},
{
"name": "Import csv data",
"snippet": [
"df = pd.read_csv(\"____________\", index_col=\"___________\", parse_dates=[\"____________\"])",
"df.head()"
]
},
{
"name": "Dataset Splitting",
"sub-menu": [
{
"name": "Shuffle Split",
"snippet": [
"X = df.drop(columns=\"___________\")",
"y = \"_____________\"",
"",
"X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42)",
"X_train.shape, X_test.shape, y_train.shape, y_test.shape"
]
},
{
"name": "Stratified Shuffle Split",
"snippet": [
"X = df.drop(columns=\"___________\")",
"y = \"_____________\"",
"",
"X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, stratify=y, random_state=42)",
"X_train.shape, X_test.shape, y_train.shape, y_test.shape"
]
}
]
},
{
"name": "Preprocessor",
"sub-menu": [
{
"name": "Common",
"snippet": [
"preprocessor = ColumnTransformer([",
" ('numeric', num_pipe(), [\"______________\"]),",
" ('categoric', cat_pipe(encoder='onehot'), [\"_____________\"]),",
"])"
]
},
{
"name": "Advance example",
"snippet": [
"# Note: You could not use gsp, rsp, and bsp recommendation in advance mode",
"# You should specify your own parameter grid / interval when tuning",
"preprocessor = ColumnTransformer([",
" ('numeric1', num_pipe(impute='mean', poly=2, scaling='standard', transform='yeo-johnson'), [\"______________\"]),",
" ('numeric2', num_pipe(impute='median', poly=2, scaling='robust'), [\"______________\"]),",
" ('categoric1', cat_pipe(encoder='ordinal'), [\"_____________\"]),",
" ('categoric2', cat_pipe(encoder='onehot'), [\"_____________\"]) ",
"])"
]
}
]
},
{
"name": "Supervised Learning Pipeline",
"sub-menu": [
{
"name": "Regression",
"sub-menu": [
{
"name": "K-Nearest Neighbor (KNN)",
"snippet": [
"from sklearn.neighbors import KNeighborsRegressor",
"pipeline = Pipeline([",
" ('prep', preprocessor),",
" ('algo', KNeighborsRegressor())",
"])"
]
},
{
"name": "Support Vector Machine (SVM)",
"snippet": [
"from sklearn.svm import SVR",
"pipeline = Pipeline([",
" ('prep', preprocessor),",
" ('algo', SVR(max_iter=500))",
"])"
]
},
{
"name": "Random Forest (RF)",
"snippet": [
"from sklearn.ensemble import RandomForestRegressor",
"pipeline = Pipeline([",
" ('prep', preprocessor),",
" ('algo', RandomForestRegressor(n_jobs=-1, random_state=42))",
"])"
]
},
{
"name": "Extreme Gradient Boosting (XGBoost)",
"snippet": [
"from xgboost import XGBRegressor",
"pipeline = Pipeline([",
" ('prep', preprocessor),",
" ('algo', XGBRegressor(n_jobs=-1, random_state=42))",
"])"
]
},
{
"name": "Linear Regression",
"snippet": [
"from sklearn.linear_model import LinearRegression",
"pipeline = Pipeline([",
" ('prep', preprocessor),",
" ('algo', LinearRegression())",
"])"
]
},
{
"name": "ElasticNet Regression",
"snippet": [
"from sklearn.linear_model import ElasticNet",
"pipeline = Pipeline([",
" ('prep', preprocessor),",
" ('algo', ElasticNet())",
"])"
]
}
]
},
{
"name": "Classification",
"sub-menu": [
{
"name": "K-Nearest Neighbor (KNN)",
"snippet": [
"from sklearn.neighbors import KNeighborsClassifier",
"pipeline = Pipeline([",
" ('prep', preprocessor),",
" ('algo', KNeighborsClassifier())",
"])"
]
},
{
"name": "Support Vector Machine (SVM)",
"snippet": [
"from sklearn.svm import SVC",
"pipeline = Pipeline([",
" ('prep', preprocessor),",
" ('algo', SVC(max_iter=500))",
"])"
]
},
{
"name": "Random Forest (RF)",
"snippet": [
"from sklearn.ensemble import RandomForestClassifier",
"pipeline = Pipeline([",
" ('prep', preprocessor),",
" ('algo', RandomForestClassifier(n_jobs=-1, random_state=42))",
"])"
]
},
{
"name": "Extreme Gradient Boosting (XGBoost)",
"snippet": [
"from xgboost import XGBClassifier",
"pipeline = Pipeline([",
" ('prep', preprocessor),",
" ('algo', XGBClassifier(n_jobs=-1, random_state=42))",
"])"
]
},
{
"name": "Logistic Regression",
"snippet": [
"from sklearn.linear_model import LogisticRegression",
"pipeline = Pipeline([",
" ('prep', preprocessor),",
" ('algo', LogisticRegression(solver='lbfgs', n_jobs=-1, random_state=42))",
"])"
]
}
]
}
]
},
{
"name": "Hyperparameter Tuning",
"sub-menu": [
{
"name": "Grid Search",
"snippet": [
"from sklearn.model_selection import GridSearchCV",
"from jcopml.tuning import grid_search_params as gsp",
"",
"model = GridSearchCV(pipeline, gsp.\"_______________\", cv=\"___\", scoring='___', n_jobs=-1, verbose=1)",
"model.fit(X_train, y_train)",
"",
"print(model.best_params_)",
"print(model.score(X_train, y_train), model.best_score_, model.score(X_test, y_test))"
]
},
{
"name": "Randomized Search",
"snippet": [
"from sklearn.model_selection import RandomizedSearchCV",
"from jcopml.tuning import random_search_params as rsp",
"",
"model = RandomizedSearchCV(pipeline, rsp.\"_______________\", cv=\"___\", scoring='___', n_iter=\"___\", n_jobs=-1, verbose=1, random_state=42)",
"model.fit(X_train, y_train)",
"",
"print(model.best_params_)",
"print(model.score(X_train, y_train), model.best_score_, model.score(X_test, y_test))"
]
},
{
"name": "Bayesian Search",
"snippet": [
"from jcopml.tuning.skopt import BayesSearchCV",
"from jcopml.tuning import bayes_search_params as bsp",
"",
"model = BayesSearchCV(pipeline, bsp.\"_______________\", cv=\"___\", scoring=\"__\", n_iter=\"___\", n_jobs=-1, verbose=1, random_state=42)",
"model.fit(X_train, y_train)",
"",
"print(model.best_params_)",
"print(model.score(X_train, y_train), model.best_score_, model.score(X_test, y_test))"
]
}
]
},
{
"name": "Save model",
"sub-menu": [
{
"name": "Save the whole search object",
"snippet": ["save_model(model, \"__________.pkl\")"]
},
{
"name": "Save best estimator only",
"snippet": ["save_model(model.best_estimator_, \"__________.pkl\")"]
}
]
}
]
}
```
```
import numpy as np
import pandas as pd
from sklearn.model_selection import train_test_split
from sklearn.pipeline import Pipeline
from sklearn.compose import ColumnTransformer
from jcopml.pipeline import num_pipe, cat_pipe
from jcopml.utils import save_model, load_model
from jcopml.plot import plot_missing_value
from jcopml.feature_importance import mean_score_decrease
```
# Import Data
```
df = pd.read_csv("____________", index_col="___________", parse_dates=["____________"])
df.head()
plot_missing_value(df)
```
# Dataset Splitting
```
X = df.drop(columns="___________")
y = "_____________"
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, stratify=y, random_state=42)
X_train.shape, X_test.shape, y_train.shape, y_test.shape
```
# Preprocessor
```
preprocessor = ColumnTransformer([
('numeric', num_pipe(), ["______________"]),
('categoric', cat_pipe(encoder='onehot'), ["_____________"]),
])
```
# Training
```
from sklearn.neighbors import KNeighborsClassifier
from sklearn.model_selection import GridSearchCV
from jcopml.tuning import grid_search_params as gsp
pipeline = Pipeline([
('prep', preprocessor),
('algo', KNeighborsClassifier())
])
model = GridSearchCV(pipeline, gsp."_______________", cv="___", scoring='___', n_jobs=-1, verbose=1)
model.fit(X_train, y_train)
print(model.best_params_)
print(model.score(X_train, y_train), model.best_score_, model.score(X_test, y_test))
```
# Save Model
```
save_model(model.best_estimator_, "__________.pkl")
```
|
github_jupyter
|
conda install -c conda-forge jupyter_contrib_nbextensions
{
"name": "J.COp Snippets",
"sub-menu": [
{
"name": "Import common packages",
"snippet": [
"import numpy as np",
"import pandas as pd",
"",
"from sklearn.model_selection import train_test_split",
"from sklearn.pipeline import Pipeline",
"from sklearn.compose import ColumnTransformer",
"",
"from jcopml.pipeline import num_pipe, cat_pipe",
"from jcopml.utils import save_model, load_model",
"from jcopml.plot import plot_missing_value",
"from jcopml.feature_importance import mean_score_decrease"
]
},
{
"name": "Import csv data",
"snippet": [
"df = pd.read_csv(\"____________\", index_col=\"___________\", parse_dates=[\"____________\"])",
"df.head()"
]
},
{
"name": "Dataset Splitting",
"sub-menu": [
{
"name": "Shuffle Split",
"snippet": [
"X = df.drop(columns=\"___________\")",
"y = \"_____________\"",
"",
"X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42)",
"X_train.shape, X_test.shape, y_train.shape, y_test.shape"
]
},
{
"name": "Stratified Shuffle Split",
"snippet": [
"X = df.drop(columns=\"___________\")",
"y = \"_____________\"",
"",
"X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, stratify=y, random_state=42)",
"X_train.shape, X_test.shape, y_train.shape, y_test.shape"
]
}
]
},
{
"name": "Preprocessor",
"sub-menu": [
{
"name": "Common",
"snippet": [
"preprocessor = ColumnTransformer([",
" ('numeric', num_pipe(), [\"______________\"]),",
" ('categoric', cat_pipe(encoder='onehot'), [\"_____________\"]),",
"])"
]
},
{
"name": "Advance example",
"snippet": [
"# Note: You could not use gsp, rsp, and bsp recommendation in advance mode",
"# You should specify your own parameter grid / interval when tuning",
"preprocessor = ColumnTransformer([",
" ('numeric1', num_pipe(impute='mean', poly=2, scaling='standard', transform='yeo-johnson'), [\"______________\"]),",
" ('numeric2', num_pipe(impute='median', poly=2, scaling='robust'), [\"______________\"]),",
" ('categoric1', cat_pipe(encoder='ordinal'), [\"_____________\"]),",
" ('categoric2', cat_pipe(encoder='onehot'), [\"_____________\"]) ",
"])"
]
}
]
},
{
"name": "Supervised Learning Pipeline",
"sub-menu": [
{
"name": "Regression",
"sub-menu": [
{
"name": "K-Nearest Neighbor (KNN)",
"snippet": [
"from sklearn.neighbors import KNeighborsRegressor",
"pipeline = Pipeline([",
" ('prep', preprocessor),",
" ('algo', KNeighborsRegressor())",
"])"
]
},
{
"name": "Support Vector Machine (SVM)",
"snippet": [
"from sklearn.svm import SVR",
"pipeline = Pipeline([",
" ('prep', preprocessor),",
" ('algo', SVR(max_iter=500))",
"])"
]
},
{
"name": "Random Forest (RF)",
"snippet": [
"from sklearn.ensemble import RandomForestRegressor",
"pipeline = Pipeline([",
" ('prep', preprocessor),",
" ('algo', RandomForestRegressor(n_jobs=-1, random_state=42))",
"])"
]
},
{
"name": "Extreme Gradient Boosting (XGBoost)",
"snippet": [
"from xgboost import XGBRegressor",
"pipeline = Pipeline([",
" ('prep', preprocessor),",
" ('algo', XGBRegressor(n_jobs=-1, random_state=42))",
"])"
]
},
{
"name": "Linear Regression",
"snippet": [
"from sklearn.linear_model import LinearRegression",
"pipeline = Pipeline([",
" ('prep', preprocessor),",
" ('algo', LinearRegression())",
"])"
]
},
{
"name": "ElasticNet Regression",
"snippet": [
"from sklearn.linear_model import ElasticNet",
"pipeline = Pipeline([",
" ('prep', preprocessor),",
" ('algo', ElasticNet())",
"])"
]
}
]
},
{
"name": "Classification",
"sub-menu": [
{
"name": "K-Nearest Neighbor (KNN)",
"snippet": [
"from sklearn.neighbors import KNeighborsClassifier",
"pipeline = Pipeline([",
" ('prep', preprocessor),",
" ('algo', KNeighborsClassifier())",
"])"
]
},
{
"name": "Support Vector Machine (SVM)",
"snippet": [
"from sklearn.svm import SVC",
"pipeline = Pipeline([",
" ('prep', preprocessor),",
" ('algo', SVC(max_iter=500))",
"])"
]
},
{
"name": "Random Forest (RF)",
"snippet": [
"from sklearn.ensemble import RandomForestClassifier",
"pipeline = Pipeline([",
" ('prep', preprocessor),",
" ('algo', RandomForestClassifier(n_jobs=-1, random_state=42))",
"])"
]
},
{
"name": "Extreme Gradient Boosting (XGBoost)",
"snippet": [
"from xgboost import XGBClassifier",
"pipeline = Pipeline([",
" ('prep', preprocessor),",
" ('algo', XGBClassifier(n_jobs=-1, random_state=42))",
"])"
]
},
{
"name": "Logistic Regression",
"snippet": [
"from sklearn.linear_model import LogisticRegression",
"pipeline = Pipeline([",
" ('prep', preprocessor),",
" ('algo', LogisticRegression(solver='lbfgs', n_jobs=-1, random_state=42))",
"])"
]
}
]
}
]
},
{
"name": "Hyperparameter Tuning",
"sub-menu": [
{
"name": "Grid Search",
"snippet": [
"from sklearn.model_selection import GridSearchCV",
"from jcopml.tuning import grid_search_params as gsp",
"",
"model = GridSearchCV(pipeline, gsp.\"_______________\", cv=\"___\", scoring='___', n_jobs=-1, verbose=1)",
"model.fit(X_train, y_train)",
"",
"print(model.best_params_)",
"print(model.score(X_train, y_train), model.best_score_, model.score(X_test, y_test))"
]
},
{
"name": "Randomized Search",
"snippet": [
"from sklearn.model_selection import RandomizedSearchCV",
"from jcopml.tuning import random_search_params as rsp",
"",
"model = RandomizedSearchCV(pipeline, rsp.\"_______________\", cv=\"___\", scoring='___', n_iter=\"___\", n_jobs=-1, verbose=1, random_state=42)",
"model.fit(X_train, y_train)",
"",
"print(model.best_params_)",
"print(model.score(X_train, y_train), model.best_score_, model.score(X_test, y_test))"
]
},
{
"name": "Bayesian Search",
"snippet": [
"from jcopml.tuning.skopt import BayesSearchCV",
"from jcopml.tuning import bayes_search_params as bsp",
"",
"model = BayesSearchCV(pipeline, bsp.\"_______________\", cv=\"___\", scoring=\"__\", n_iter=\"___\", n_jobs=-1, verbose=1, random_state=42)",
"model.fit(X_train, y_train)",
"",
"print(model.best_params_)",
"print(model.score(X_train, y_train), model.best_score_, model.score(X_test, y_test))"
]
}
]
},
{
"name": "Save model",
"sub-menu": [
{
"name": "Save the whole search object",
"snippet": ["save_model(model, \"__________.pkl\")"]
},
{
"name": "Save best estimator only",
"snippet": ["save_model(model.best_estimator_, \"__________.pkl\")"]
}
]
}
]
}
import numpy as np
import pandas as pd
from sklearn.model_selection import train_test_split
from sklearn.pipeline import Pipeline
from sklearn.compose import ColumnTransformer
from jcopml.pipeline import num_pipe, cat_pipe
from jcopml.utils import save_model, load_model
from jcopml.plot import plot_missing_value
from jcopml.feature_importance import mean_score_decrease
df = pd.read_csv("____________", index_col="___________", parse_dates=["____________"])
df.head()
plot_missing_value(df)
X = df.drop(columns="___________")
y = "_____________"
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, stratify=y, random_state=42)
X_train.shape, X_test.shape, y_train.shape, y_test.shape
preprocessor = ColumnTransformer([
('numeric', num_pipe(), ["______________"]),
('categoric', cat_pipe(encoder='onehot'), ["_____________"]),
])
from sklearn.neighbors import KNeighborsClassifier
from sklearn.model_selection import GridSearchCV
from jcopml.tuning import grid_search_params as gsp
pipeline = Pipeline([
('prep', preprocessor),
('algo', KNeighborsClassifier())
])
model = GridSearchCV(pipeline, gsp."_______________", cv="___", scoring='___', n_jobs=-1, verbose=1)
model.fit(X_train, y_train)
print(model.best_params_)
print(model.score(X_train, y_train), model.best_score_, model.score(X_test, y_test))
save_model(model.best_estimator_, "__________.pkl")
| 0.581778 | 0.832679 |
[](https://mybinder.org/v2/gh/oddrationale/AdventOfCode2020CSharp/main?urlpath=lab%2Ftree%2FDay04.ipynb)
# --- Day 4: Passport Processing ---
```
using System.IO;
using System.Text.RegularExpressions;
```
Read the input file and split into separate passport entries.
```
var batchFile = File.ReadAllText(@"input/04.txt").Split("\n\n");
```
Fields for the passport:
- byr (Birth Year)
- iyr (Issue Year)
- eyr (Expiration Year)
- hgt (Height)
- hcl (Hair Color)
- ecl (Eye Color)
- pid (Passport ID)
- cid (Country ID)
For Part 1, just check if the string contains each of the fields above, except for `cid`.
```
bool IsPassportValid(string passport)
{
return passport.Contains("byr") &&
passport.Contains("iyr") &&
passport.Contains("eyr") &&
passport.Contains("hgt") &&
passport.Contains("hcl") &&
passport.Contains("ecl") &&
passport.Contains("pid");
}
batchFile.Where(IsPassportValid).Count()
```
# --- Part Two ---
For Part 2, create a proper `Passport` object. Here, decided to use C# 9's new `record` feature.
```
record Passport
{
public int? BirthYear { get; init; }
public int? IssueYear { get; init; }
public int? ExpirationYear { get; init; }
public string Height { get; init; }
public string HairColor { get; init; }
public string EyeColor { get; init; }
public string PassportId { get; init; }
public string CountryId { get; init; }
public bool IsValid
{
get
{
return IsBirthYearValid(BirthYear) &&
IsIssueYearValid(IssueYear) &&
IsExpirationYearValid(ExpirationYear) &&
IsHeightValid(Height) &&
IsHairColorValid(HairColor) &&
IsEyeColorValid(EyeColor) &&
IsPassportIdValid(PassportId);
}
}
public Passport(string passport)
{
Dictionary<string,string> tempDict = Regex
.Split(passport, @"[ \r\n]")
.Select(f => f.Split(":"))
.ToDictionary(kv => kv.First(), kv => kv.Last());
// Maybe possible to use reflection here?
foreach (KeyValuePair<string, string> kv in tempDict)
{
switch (kv.Key)
{
case "byr":
BirthYear = int.Parse(kv.Value);
break;
case "iyr":
IssueYear = int.Parse(kv.Value);
break;
case "eyr":
ExpirationYear = int.Parse(kv.Value);
break;
case "hgt":
Height = kv.Value;
break;
case "hcl":
HairColor = kv.Value;
break;
case "ecl":
EyeColor = kv.Value;
break;
case "pid":
PassportId = kv.Value;
break;
case "cid":
CountryId = kv.Value;
break;
}
}
}
private bool IsBirthYearValid(int? birthYear) => 1920 <= birthYear && birthYear <= 2002;
private bool IsIssueYearValid(int? issueYear) => 2010 <= issueYear && issueYear <= 2020;
private bool IsExpirationYearValid(int? expirationYear) => 2020 <= expirationYear && expirationYear <= 2030;
private bool IsHeightValid(string height)
{
if (height is null)
{
return false;
}
else if (height.EndsWith("cm"))
{
var cm = Convert.ToInt32(height[0..^2]);
return 150 <= cm && cm <= 193;
}
else if (height.EndsWith("in"))
{
var inches = Convert.ToInt32(height[0..^2]);
return 59 <= inches && inches <= 76;
}
else
{
return false;
}
}
private bool IsHairColorValid(string hairColor) => Regex.IsMatch(hairColor ?? string.Empty, @"^#[0-9a-f]{6}$");
private bool IsEyeColorValid(string eyeColor) => Regex.IsMatch(eyeColor ?? string.Empty, @"^(amb|blu|brn|gry|grn|hzl|oth)$");
private bool IsPassportIdValid(string passportId) => Regex.IsMatch(passportId ?? string.Empty, @"^\d{9}$");
}
batchFile.Select(line => new Passport(line)).Where(passport => passport.IsValid).Count()
```
|
github_jupyter
|
using System.IO;
using System.Text.RegularExpressions;
var batchFile = File.ReadAllText(@"input/04.txt").Split("\n\n");
bool IsPassportValid(string passport)
{
return passport.Contains("byr") &&
passport.Contains("iyr") &&
passport.Contains("eyr") &&
passport.Contains("hgt") &&
passport.Contains("hcl") &&
passport.Contains("ecl") &&
passport.Contains("pid");
}
batchFile.Where(IsPassportValid).Count()
record Passport
{
public int? BirthYear { get; init; }
public int? IssueYear { get; init; }
public int? ExpirationYear { get; init; }
public string Height { get; init; }
public string HairColor { get; init; }
public string EyeColor { get; init; }
public string PassportId { get; init; }
public string CountryId { get; init; }
public bool IsValid
{
get
{
return IsBirthYearValid(BirthYear) &&
IsIssueYearValid(IssueYear) &&
IsExpirationYearValid(ExpirationYear) &&
IsHeightValid(Height) &&
IsHairColorValid(HairColor) &&
IsEyeColorValid(EyeColor) &&
IsPassportIdValid(PassportId);
}
}
public Passport(string passport)
{
Dictionary<string,string> tempDict = Regex
.Split(passport, @"[ \r\n]")
.Select(f => f.Split(":"))
.ToDictionary(kv => kv.First(), kv => kv.Last());
// Maybe possible to use reflection here?
foreach (KeyValuePair<string, string> kv in tempDict)
{
switch (kv.Key)
{
case "byr":
BirthYear = int.Parse(kv.Value);
break;
case "iyr":
IssueYear = int.Parse(kv.Value);
break;
case "eyr":
ExpirationYear = int.Parse(kv.Value);
break;
case "hgt":
Height = kv.Value;
break;
case "hcl":
HairColor = kv.Value;
break;
case "ecl":
EyeColor = kv.Value;
break;
case "pid":
PassportId = kv.Value;
break;
case "cid":
CountryId = kv.Value;
break;
}
}
}
private bool IsBirthYearValid(int? birthYear) => 1920 <= birthYear && birthYear <= 2002;
private bool IsIssueYearValid(int? issueYear) => 2010 <= issueYear && issueYear <= 2020;
private bool IsExpirationYearValid(int? expirationYear) => 2020 <= expirationYear && expirationYear <= 2030;
private bool IsHeightValid(string height)
{
if (height is null)
{
return false;
}
else if (height.EndsWith("cm"))
{
var cm = Convert.ToInt32(height[0..^2]);
return 150 <= cm && cm <= 193;
}
else if (height.EndsWith("in"))
{
var inches = Convert.ToInt32(height[0..^2]);
return 59 <= inches && inches <= 76;
}
else
{
return false;
}
}
private bool IsHairColorValid(string hairColor) => Regex.IsMatch(hairColor ?? string.Empty, @"^#[0-9a-f]{6}$");
private bool IsEyeColorValid(string eyeColor) => Regex.IsMatch(eyeColor ?? string.Empty, @"^(amb|blu|brn|gry|grn|hzl|oth)$");
private bool IsPassportIdValid(string passportId) => Regex.IsMatch(passportId ?? string.Empty, @"^\d{9}$");
}
batchFile.Select(line => new Passport(line)).Where(passport => passport.IsValid).Count()
| 0.362292 | 0.854642 |

## 一. Normal Equation
### 1. 正规方程
正规方程法相对梯度下降法,它可以一步找到最小值。而且它也不需要进行特征值的缩放。
样本集是 $ m * n $ 的矩阵,每行样本表示为 $ \vec{x^{(i)}} $ ,第 i 行第 n 列分别表示为 $ x^{(i)}_{0} , x^{(i)}_{1} , x^{(i)}_{2} , x^{(i)}_{3} \cdots x^{(i)}_{n} $, m 行向量分别表示为 $ \vec{x^{(1)}} , \vec{x^{(2)}} , \vec{x^{(3)}} , \cdots \vec{x^{(m)}} $
令
$$ \vec{x^{(i)}} = \begin{bmatrix} x^{(i)}_{0}\\ x^{(i)}_{1}\\ \vdots \\ x^{(i)}_{n}\\ \end{bmatrix} $$
$ \vec{x^{(i)}} $ 是这样一个 $(n+1)*1$ 维向量。每行都对应着 i 行 0-n 个变量。
再构造几个矩阵:
$$ X = \begin{bmatrix} (\vec{x^{(1)}})^{T}\\ \vdots \\ (\vec{x^{(m)}})^{T} \end{bmatrix} \;\;\;\;
\Theta = \begin{bmatrix} \theta_{0}\\ \theta_{1}\\ \vdots \\ \theta_{n}\\ \end{bmatrix} \;\;\;\;
Y = \begin{bmatrix} y^{(1)}\\ y^{(2)}\\ \vdots \\ y^{(m)}\\ \end{bmatrix}
$$
X 是一个 $ m * (n+1)$ 的矩阵,$ \Theta $ 是一个 $ (n+1) * 1$ 的向量,Y 是一个 $ m * 1$的矩阵。
对比之前代价函数中,$$ \rm{CostFunction} = \rm{F}({\theta_{0}},{\theta_{1}}) = \frac{1}{2m}\sum_{i = 1}^{m} (h_{\theta}(x^{(i)})-y^{(i)})^2 $$
$$
\begin{align*}
X \cdot \Theta - Y =
\begin{bmatrix}
(\vec{x^{(1)}})^{T}\\
\vdots \\
(\vec{x^{(m)}})^{T}
\end{bmatrix} \cdot
\begin{bmatrix}
\theta_{0}\\
\theta_{1}\\
\vdots \\
\theta_{n}\\
\end{bmatrix} -
\begin{bmatrix}
y^{(1)}\\
y^{(2)}\\
\vdots \\
y^{(m)}\\
\end{bmatrix} =
\begin{bmatrix}
h_{\theta}(x^{(1)})-y^{(1)}\\
h_{\theta}(x^{(2)})-y^{(2)}\\
\vdots \\
h_{\theta}(x^{(m)})-y^{(m)}\\
\end{bmatrix}
\end{align*}$$
代入到之前代价函数中,
$$
\begin{align*}
\rm{CostFunction} = \rm{F}({\theta_{0}},{\theta_{1}}) &= \frac{1}{2m}\sum_{i = 1}^{m} (h_{\theta}(x^{(i)})-y^{(i)})^2\\
& = \frac{1}{2m} (X \cdot \Theta - Y)^{T}(X \cdot \Theta - Y)\\
\end{align*}
$$
----------------------------------------------------------------------------------------------------------------
### 2. 矩阵的微分和矩阵的迹
接下来在进行推导之前,需要引入矩阵迹的概念,因为迹是求解一阶矩阵微分的工具。
矩阵迹的定义是
$$ \rm{tr} A = \sum_{i=1}^{n}A_{ii}$$
简单的说就是左上角到右下角对角线上元素的和。
接下来有几个性质在下面推导过程中需要用到:
1. $ \rm{tr}\;a = a $ , a 是标量 ( $ a \in \mathbb{R} $)
2. $ \rm{tr}\;AB = \rm{tr}\;BA $ 更近一步 $ \rm{tr}\;ABC = \rm{tr}\;CAB = \rm{tr}\;BCA $
证明:假设 A 是 $n * m$ 矩阵, B 是 $m * n$ 矩阵,则有
$$ \rm{tr}\;AB = \sum_{i=1}^{n}\sum_{j=1}^{m}A_{ij}B_{ji} = \sum_{j=1}^{n} \sum_{i=1}^{m}B_{ji}A_{ij}= \rm{tr}\;BA $$
同理:$$ \rm{tr}\;ABC = \rm{tr}\;(AB)C = \rm{tr}\;C(AB) = \rm{tr}\;CAB$$
$$ \rm{tr}\;ABC = \rm{tr}\;A(BC) = \rm{tr}\;(BC)A = \rm{tr}\;BCA$$
连起来,即 $$ \rm{tr}\;ABC = \rm{tr}\;CAB = \rm{tr}\;BCA $$
3. $ \triangledown_{A}\rm{tr}\;AB = \triangledown_{A}\rm{tr}\;BA = B^{T}$
证明:按照矩阵梯度的定义:
$$\triangledown_{X}f(X) = \begin{bmatrix}
\frac{\partial f(X) }{\partial x_{11}} & \cdots & \frac{\partial f(X) }{\partial x_{1n}}\\
\vdots & \ddots & \vdots \\
\frac{\partial f(X) }{\partial x_{m1}} & \cdots & \frac{\partial f(X) }{\partial x_{mn}}
\end{bmatrix} = \frac{\partial f(X) }{\partial X}$$
假设 A 是 $n * m$ 矩阵, B 是 $m * n$ 矩阵,则有
$$\begin{align*}\triangledown_{A}\rm{tr}\;AB &= \triangledown_{A} \sum_{i=1}^{n}\sum_{j=1}^{m}A_{ij}B_{ji} = \frac{\partial}{\partial A}(\sum_{i=1}^{n}\sum_{j=1}^{m}A_{ij}B_{ji})\\ & = \begin{bmatrix}
\frac{\partial}{\partial A_{11}}(\sum_{i=1}^{n}\sum_{j=1}^{m}A_{ij}B_{ji}) & \cdots & \frac{\partial}{\partial A_{1m}}(\sum_{i=1}^{n}\sum_{j=1}^{m}A_{ij}B_{ji})\\
\vdots & \ddots & \vdots \\
\frac{\partial}{\partial A_{n1}}(\sum_{i=1}^{n}\sum_{j=1}^{m}A_{ij}B_{ji}) & \cdots & \frac{\partial}{\partial A_{nm}}(\sum_{i=1}^{n}\sum_{j=1}^{m}A_{ij}B_{ji})
\end{bmatrix} \\ & = \begin{bmatrix}
B_{11} & \cdots & B_{m1} \\
\vdots & \ddots & \vdots \\
B_{1n} & \cdots & B_{mn}
\end{bmatrix} = B^{T}\\ \end{align*}$$
$$\begin{align*}\triangledown_{A}\rm{tr}\;BA &= \triangledown_{A} \sum_{i=1}^{m}\sum_{j=1}^{n}B_{ij}A_{ji} = \frac{\partial}{\partial A}(\sum_{i=1}^{m}\sum_{j=1}^{n}B_{ij}A_{ji})\\ & = \begin{bmatrix}
\frac{\partial}{\partial A_{11}}(\sum_{i=1}^{m}\sum_{j=1}^{n}B_{ij}A_{ji}) & \cdots & \frac{\partial}{\partial A_{1m}}(\sum_{i=1}^{m}\sum_{j=1}^{n}B_{ij}A_{ji})\\
\vdots & \ddots & \vdots \\
\frac{\partial}{\partial A_{n1}}(\sum_{i=1}^{m}\sum_{j=1}^{n}B_{ij}A_{ji}) & \cdots & \frac{\partial}{\partial A_{nm}}(\sum_{i=1}^{m}\sum_{j=1}^{n}B_{ij}A_{ji})
\end{bmatrix} \\ & = \begin{bmatrix}
B_{11} & \cdots & B_{m1} \\
\vdots & \ddots & \vdots \\
B_{1n} & \cdots & B_{mn}
\end{bmatrix} = B^{T}\\ \end{align*}$$
所以有 $ \triangledown_{A}\rm{tr}\;AB = \triangledown_{A}\rm{tr}\;BA = B^{T}$
4. $\triangledown_{A^{T}}a = (\triangledown_{A}a)^{T}\;\;\;\; (a \in \mathbb{R})$
证明:假设 A 是 $n * m$ 矩阵
$$\begin{align*}\triangledown_{A^{T}}a & = \begin{bmatrix}
\frac{\partial}{\partial A_{11}}a & \cdots & \frac{\partial}{\partial A_{1n}}a\\
\vdots & \ddots & \vdots \\
\frac{\partial}{\partial A_{m1}}a & \cdots & \frac{\partial}{\partial A_{mn}}a
\end{bmatrix} = (\begin{bmatrix}
\frac{\partial}{\partial A_{11}}a & \cdots & \frac{\partial}{\partial A_{1m}}a\\
\vdots & \ddots & \vdots \\
\frac{\partial}{\partial A_{n1}}a & \cdots & \frac{\partial}{\partial A_{nm}}a
\end{bmatrix})^{T} \\ & = (\triangledown_{A}a)^{T}\\ \end{align*}$$
5. $\mathrm{d}(\rm{tr}\;A) = \rm{tr}(\mathrm{d}A)$
证明:
$$\mathrm{d}(\rm{tr}\;A) = \mathrm{d}(\sum_{i=1}^{n}a_{ii}) = \sum_{i=1}^{n}\mathrm{d}a_{ii} = \rm{tr}(\mathrm{d}A)$$
矩阵的迹的微分等于矩阵的微分的迹。
6. $\triangledown_{A}\rm{tr}\;ABA^{T}C = CAB + C^{T}AB^{T}$
证明:
根据实标量函数梯度的乘法法则:
若 f(A)、g(A)、h(A) 分别是矩阵 A 的实标量函数,则有
$$\begin{align*}\frac{\partial f(A)g(A)}{\partial A} &= g(A)\frac{\partial f(A)}{\partial A} + f(A)\frac{\partial g(A)}{\partial A}\\ \frac{\partial f(A)g(A)h(A)}{\partial A} &= g(A)h(A)\frac{\partial f(A)}{\partial A} + f(A)h(A)\frac{\partial g(A)}{\partial A}+ f(A)g(A)\frac{\partial h(A)}{\partial A}\\ \end{align*}$$
令 $f(A) = AB,g(A) = A^{T}C$,由性质5,矩阵的迹的微分等于矩阵的微分的迹,那么则有:
$$\begin{align*} \triangledown_{A}\rm{tr}\;ABA^{T}C & = \rm{tr}(\triangledown_{A}ABA^{T}C) = \rm{tr}(\triangledown_{A}f(A)g(A)) = \rm{tr}\triangledown_{A_{1}}(A_{1}BA^{T}C) + \rm{tr}\triangledown_{A_{2}}(ABA_{2}^{T}C) \\ & = (BA^{T}C)^{T} + \rm{tr}\triangledown_{A_{2}}(ABA_{2}^{T}C) = C^{T}AB^{T} + \triangledown_{A_{2}}\rm{tr}(ABA_{2}^{T}C)\\ & = C^{T}AB^{T} + \triangledown_{A_{2}}\rm{tr}(A_{2}^{T}CAB) = C^{T}AB^{T} + (\triangledown_{{A_{2}}^{T}}\;\rm{tr}\;A_{2}^{T}CAB)^{T} \\ & = C^{T}AB^{T} + ((CAB)^{T})^{T} \\ & = C^{T}AB^{T} + CAB \\ \end{align*}$$
------------------------------------------------------------------------------------------------------------
### 3. 推导
回到之前的代价函数中:
$$
\rm{CostFunction} = \rm{F}({\theta_{0}},{\theta_{1}}) = \frac{1}{2m} (X \cdot \Theta - Y)^{T}(X \cdot \Theta - Y)
$$
求导:
$$
\begin{align*}
\triangledown_{\theta}\rm{F}(\theta) & = \frac{1}{2m} \triangledown_{\theta}(X \cdot \Theta - Y)^{T}(X \cdot \Theta - Y) = \frac{1}{2m}\triangledown_{\theta}(\Theta^{T}X^{T}-Y^{T})(X\Theta-Y)\\
& = \frac{1}{2m}\triangledown_{\theta}(\Theta^{T}X^{T}X\Theta-Y^{T}X\Theta-\Theta^{T}X^{T}Y+Y^{T}Y) \\ \end{align*}
$$
上式中,对 $\Theta $矩阵求导,$ Y^{T}Y $ 与 $\Theta $ 无关,所以这一项为 0 。 $Y^{T}X\Theta$ 是标量,由性质4可以知道,$Y^{T}X\Theta = (Y^{T}X\Theta)^{T} = \Theta^{T}X^{T}Y$,因为 $\Theta^{T}X^{T}X\Theta , Y^{T}X\Theta $都是标量,所以它们的也等于它们的迹,(处理矩阵微分的问题常常引入矩阵的迹),于是有
$$
\begin{align*}
\triangledown_{\theta}\rm{F}(\theta) & = \frac{1}{2m}\triangledown_{\theta}(\Theta^{T}X^{T}X\Theta-2Y^{T}X\Theta) \\
& = \frac{1}{2m}\triangledown_{\theta}\rm{tr}\;(\Theta^{T}X^{T}X\Theta-2Y^{T}X\Theta) \\ & = \frac{1}{2m}\triangledown_{\theta}\rm{tr}\;(\Theta\Theta^{T}X^{T}X-2Y^{T}X\Theta) \\ & = \frac{1}{m}(\frac{1}{2}\triangledown_{\theta}\rm{tr}\;\Theta\Theta^{T}X^{T}X -\triangledown_{\theta}\rm{tr}\;Y^{T}X\Theta) \\ & = \frac{1}{m}(\frac{1}{2}\triangledown_{\theta}\rm{tr}\;\Theta\Theta^{T}X^{T}X -(Y^{T}X)^{T}) = \frac{1}{m}(\frac{1}{2}\triangledown_{\theta}\rm{tr}\;\Theta\Theta^{T}X^{T}X -X^{T}Y)\\ \end{align*}
$$
上面第三步用的性质2矩阵迹的交换律,第五步用的性质3。
为了能进一步化简矩阵的微分,我们在矩阵的迹上面乘以一个单位矩阵,不影响结果。于是:
$$
\begin{align*}
\triangledown_{\theta}\rm{F}(\theta) & = \frac{1}{m}(\frac{1}{2}\triangledown_{\theta}\rm{tr}\;\Theta\Theta^{T}X^{T}X -X^{T}Y) \\ &= \frac{1}{m}(\frac{1}{2}\triangledown_{\theta}\rm{tr}\;\Theta I \Theta^{T}X^{T}X -X^{T}Y) \end{align*}
$$
利用性质6 展开上面的式子,令 $ A = \Theta , B = I , C = X^{T}X $。
$$
\begin{align*}
\triangledown_{\theta}\rm{F}(\theta) &= \frac{1}{m}(\frac{1}{2}\triangledown_{\theta}\rm{tr}\;\Theta I \Theta^{T}X^{T}X -X^{T}Y) \\ & = \frac{1}{m}(\frac{1}{2}(X^{T}X\Theta I + (X^{T}X)^{T}\Theta I^{T}) -X^{T}Y) \\ & = \frac{1}{m}(\frac{1}{2}(X^{T}X\Theta I + (X^{T}X)^{T}\Theta I^{T}) -X^{T}Y) \\ & = \frac{1}{m}(\frac{1}{2}(X^{T}X\Theta + X^{T}X\Theta) -X^{T}Y) = \frac{1}{m}(X^{T}X\Theta -X^{T}Y) \\ \end{align*}
$$
令 $\triangledown_{\theta}\rm{F}(\theta) = 0$,即 $X^{T}X\Theta -X^{T}Y = 0$, 于是 $ X^{T}X\Theta = X^{T}Y $ ,这里假设 $ X^{T}X$ 这个矩阵是可逆的,等号两边同时左乘$ X^{T}X$的逆矩阵,得到 $\Theta = (X^{T}X)^{-1}X^{T}Y$
最终结果也就推导出来了,$$\Theta = (X^{T}X)^{-1}X^{T}Y$$
但是这里有一个**前提条件是 $X^{T}X$ 是非奇异(非退化)矩阵, 即 $ \left | X^{T}X \right | \neq 0 $**
------------------------------------------------------------------------------------------------------------
### 4. 梯度下降和正规方程法比较:
优点:
梯度下降在超大数据集面前也能运行的很良好。
正规方程在超大数据集合面前性能会变得很差,因为需要计算 $(x^{T}x)^{-1}$,时间复杂度在 $O(n^{3})$ 这个级别。
缺点:
梯度下降需要合理的选择学习速率 $\alpha$ , 需要很多次迭代的操作去选择合理的 $\alpha$,寻找最小值的时候也需要迭代很多次才能收敛。
正规方程的优势相比而言,不需要选择学习速率 $\alpha$,也不需要多次的迭代或者画图检测是否收敛。
------------------------------------------------------------------------------------------------------------
## 二. Normal Equation Noninvertibility
上一章谈到了如何利用正规方程法求解 $\Theta $,但是在线性代数中存在这样一个问题,如果是奇异(退化)矩阵,是不存在逆矩阵的。也就是说用上面正规方程的公式是不一定能求解出正确结果的。
在 Octave 软件中,存在2个求解逆矩阵的函数,一个是 pinv 和 inv。pinv (pseudo-inverse)求解的是**伪逆矩阵**,inv 求解的是逆矩阵,所以用 pinv 求解问题,就算是 $ X^{T}X $ 不存在逆矩阵,也一样可以得到最后的结果。
导致$ X^{T}X $ 不存在逆矩阵有2种情况:
1. 多余的特征。特征之间呈倍数关系,线性依赖。
2. 过多的特征。当 $ m \leqslant n $ 的时候,会导致过多的特征。解决办法是删除一些特征,或者进行正则化。
所以解决$ X^{T}X $ 不存在逆矩阵的办法也就是对应上面2种情况:
1. 删掉多余的特征,线性相关的,倍数关系的。直到没有多余的特征
2. 再删除一些不影响结果的特征,或者进行正则化。
------------------------------------------------------------------------------------------------------------
## 三. Linear Regression with Multiple Variables 测试
### 1. Question 1
Suppose m=4 students have taken some class, and the class had a midterm exam and a final exam. You have collected a dataset of their scores on the two exams, which is as follows:
midterm exam (midterm exam)2 final exam
89 7921 96
72 5184 74
94 8836 87
69 4761 78
You'd like to use polynomial regression to predict a student's final exam score from their midterm exam score. Concretely, suppose you want to fit a model of the form hθ(x)=θ0+θ1x1+θ2x2, where x1 is the midterm score and x2 is (midterm score)2. Further, you plan to use both feature scaling (dividing by the "max-min", or range, of a feature) and mean normalization.
What is the normalized feature x(2)2? (Hint: midterm = 72, final = 74 is training example 2.) Please round off your answer to two decimal places and enter in the text box below.
解答:
标准化 $$x = \frac{x_{2}^{2}-\frac{(7921+5184+8836+4761)}{4}}{\max - \min } = \frac{5184 - 6675.5}{8836-4761} = -0.37$$
### 2. Question 2
You run gradient descent for 15 iterations
with α=0.3 and compute J(θ) after each
iteration. You find that the value of J(θ) increases over
time. Based on this, which of the following conclusions seems
most plausible?
A. Rather than use the current value of α, it'd be more promising to try a smaller value of α (say α=0.1).
B. α=0.3 is an effective choice of learning rate.
C. Rather than use the current value of α, it'd be more promising to try a larger value of α (say α=1.0).
解答: A
下降太快所以a下降速率过大,a越大下降越快,a小下降慢,在本题中,代价函数快速收敛到最小值,代表此时a最合适。
### 3. Question 3
Suppose you have m=28 training examples with n=4 features (excluding the additional all-ones feature for the intercept term, which you should add). The normal equation is θ=(XTX)−1XTy. For the given values of m and n, what are the dimensions of θ, X, and y in this equation?
A. X is 28×4, y is 28×1, θ is 4×4
B. X is 28×5, y is 28×5, θ is 5×5
C. X is 28×5, y is 28×1, θ is 5×1
D. X is 28×4, y is 28×1, θ is4×1
解答: C
这里需要注意的是,题目中说了额外添加一列全部为1的,所以列数是5 。
### 4. Question 4
Suppose you have a dataset with m=50 examples and n=15 features for each example. You want to use multivariate linear regression to fit the parameters θ to our data. Should you prefer gradient descent or the normal equation?
A. Gradient descent, since it will always converge to the optimal θ.
B. Gradient descent, since (XTX)−1 will be very slow to compute in the normal equation.
C. The normal equation, since it provides an efficient way to directly find the solution.
D. The normal equation, since gradient descent might be unable to find the optimal θ.
解答: C
数据量少,选择正规方程法更加高效
### 5. Question 5
Which of the following are reasons for using feature scaling?
A. It prevents the matrix XTX (used in the normal equation) from being non-invertable (singular/degenerate).
B. It is necessary to prevent the normal equation from getting stuck in local optima.
C. It speeds up gradient descent by making it require fewer iterations to get to a good solution.
D. It speeds up gradient descent by making each iteration of gradient descent less expensive to compute.
解答: C
normal equation 不需要 Feature Scaling,排除AB, 特征缩放减少迭代数量,加快梯度下降,然而不能防止梯度下降陷入局部最优。
------------------------------------------------------
> GitHub Repo:[Halfrost-Field](https://github.com/halfrost/Halfrost-Field)
>
> Follow: [halfrost · GitHub](https://github.com/halfrost)
>
> Source: [https://github.com/halfrost/Halfrost-Field/blob/master/contents/Machine\_Learning/Computing\_Parameters\_Analytically.ipynb](https://github.com/halfrost/Halfrost-Field/blob/master/contents/Machine_Learning/Computing_Parameters_Analytically.ipynb)
|
github_jupyter
|

## 一. Normal Equation
### 1. 正规方程
正规方程法相对梯度下降法,它可以一步找到最小值。而且它也不需要进行特征值的缩放。
样本集是 $ m * n $ 的矩阵,每行样本表示为 $ \vec{x^{(i)}} $ ,第 i 行第 n 列分别表示为 $ x^{(i)}_{0} , x^{(i)}_{1} , x^{(i)}_{2} , x^{(i)}_{3} \cdots x^{(i)}_{n} $, m 行向量分别表示为 $ \vec{x^{(1)}} , \vec{x^{(2)}} , \vec{x^{(3)}} , \cdots \vec{x^{(m)}} $
令
$$ \vec{x^{(i)}} = \begin{bmatrix} x^{(i)}_{0}\\ x^{(i)}_{1}\\ \vdots \\ x^{(i)}_{n}\\ \end{bmatrix} $$
$ \vec{x^{(i)}} $ 是这样一个 $(n+1)*1$ 维向量。每行都对应着 i 行 0-n 个变量。
再构造几个矩阵:
$$ X = \begin{bmatrix} (\vec{x^{(1)}})^{T}\\ \vdots \\ (\vec{x^{(m)}})^{T} \end{bmatrix} \;\;\;\;
\Theta = \begin{bmatrix} \theta_{0}\\ \theta_{1}\\ \vdots \\ \theta_{n}\\ \end{bmatrix} \;\;\;\;
Y = \begin{bmatrix} y^{(1)}\\ y^{(2)}\\ \vdots \\ y^{(m)}\\ \end{bmatrix}
$$
X 是一个 $ m * (n+1)$ 的矩阵,$ \Theta $ 是一个 $ (n+1) * 1$ 的向量,Y 是一个 $ m * 1$的矩阵。
对比之前代价函数中,$$ \rm{CostFunction} = \rm{F}({\theta_{0}},{\theta_{1}}) = \frac{1}{2m}\sum_{i = 1}^{m} (h_{\theta}(x^{(i)})-y^{(i)})^2 $$
$$
\begin{align*}
X \cdot \Theta - Y =
\begin{bmatrix}
(\vec{x^{(1)}})^{T}\\
\vdots \\
(\vec{x^{(m)}})^{T}
\end{bmatrix} \cdot
\begin{bmatrix}
\theta_{0}\\
\theta_{1}\\
\vdots \\
\theta_{n}\\
\end{bmatrix} -
\begin{bmatrix}
y^{(1)}\\
y^{(2)}\\
\vdots \\
y^{(m)}\\
\end{bmatrix} =
\begin{bmatrix}
h_{\theta}(x^{(1)})-y^{(1)}\\
h_{\theta}(x^{(2)})-y^{(2)}\\
\vdots \\
h_{\theta}(x^{(m)})-y^{(m)}\\
\end{bmatrix}
\end{align*}$$
代入到之前代价函数中,
$$
\begin{align*}
\rm{CostFunction} = \rm{F}({\theta_{0}},{\theta_{1}}) &= \frac{1}{2m}\sum_{i = 1}^{m} (h_{\theta}(x^{(i)})-y^{(i)})^2\\
& = \frac{1}{2m} (X \cdot \Theta - Y)^{T}(X \cdot \Theta - Y)\\
\end{align*}
$$
----------------------------------------------------------------------------------------------------------------
### 2. 矩阵的微分和矩阵的迹
接下来在进行推导之前,需要引入矩阵迹的概念,因为迹是求解一阶矩阵微分的工具。
矩阵迹的定义是
$$ \rm{tr} A = \sum_{i=1}^{n}A_{ii}$$
简单的说就是左上角到右下角对角线上元素的和。
接下来有几个性质在下面推导过程中需要用到:
1. $ \rm{tr}\;a = a $ , a 是标量 ( $ a \in \mathbb{R} $)
2. $ \rm{tr}\;AB = \rm{tr}\;BA $ 更近一步 $ \rm{tr}\;ABC = \rm{tr}\;CAB = \rm{tr}\;BCA $
证明:假设 A 是 $n * m$ 矩阵, B 是 $m * n$ 矩阵,则有
$$ \rm{tr}\;AB = \sum_{i=1}^{n}\sum_{j=1}^{m}A_{ij}B_{ji} = \sum_{j=1}^{n} \sum_{i=1}^{m}B_{ji}A_{ij}= \rm{tr}\;BA $$
同理:$$ \rm{tr}\;ABC = \rm{tr}\;(AB)C = \rm{tr}\;C(AB) = \rm{tr}\;CAB$$
$$ \rm{tr}\;ABC = \rm{tr}\;A(BC) = \rm{tr}\;(BC)A = \rm{tr}\;BCA$$
连起来,即 $$ \rm{tr}\;ABC = \rm{tr}\;CAB = \rm{tr}\;BCA $$
3. $ \triangledown_{A}\rm{tr}\;AB = \triangledown_{A}\rm{tr}\;BA = B^{T}$
证明:按照矩阵梯度的定义:
$$\triangledown_{X}f(X) = \begin{bmatrix}
\frac{\partial f(X) }{\partial x_{11}} & \cdots & \frac{\partial f(X) }{\partial x_{1n}}\\
\vdots & \ddots & \vdots \\
\frac{\partial f(X) }{\partial x_{m1}} & \cdots & \frac{\partial f(X) }{\partial x_{mn}}
\end{bmatrix} = \frac{\partial f(X) }{\partial X}$$
假设 A 是 $n * m$ 矩阵, B 是 $m * n$ 矩阵,则有
$$\begin{align*}\triangledown_{A}\rm{tr}\;AB &= \triangledown_{A} \sum_{i=1}^{n}\sum_{j=1}^{m}A_{ij}B_{ji} = \frac{\partial}{\partial A}(\sum_{i=1}^{n}\sum_{j=1}^{m}A_{ij}B_{ji})\\ & = \begin{bmatrix}
\frac{\partial}{\partial A_{11}}(\sum_{i=1}^{n}\sum_{j=1}^{m}A_{ij}B_{ji}) & \cdots & \frac{\partial}{\partial A_{1m}}(\sum_{i=1}^{n}\sum_{j=1}^{m}A_{ij}B_{ji})\\
\vdots & \ddots & \vdots \\
\frac{\partial}{\partial A_{n1}}(\sum_{i=1}^{n}\sum_{j=1}^{m}A_{ij}B_{ji}) & \cdots & \frac{\partial}{\partial A_{nm}}(\sum_{i=1}^{n}\sum_{j=1}^{m}A_{ij}B_{ji})
\end{bmatrix} \\ & = \begin{bmatrix}
B_{11} & \cdots & B_{m1} \\
\vdots & \ddots & \vdots \\
B_{1n} & \cdots & B_{mn}
\end{bmatrix} = B^{T}\\ \end{align*}$$
$$\begin{align*}\triangledown_{A}\rm{tr}\;BA &= \triangledown_{A} \sum_{i=1}^{m}\sum_{j=1}^{n}B_{ij}A_{ji} = \frac{\partial}{\partial A}(\sum_{i=1}^{m}\sum_{j=1}^{n}B_{ij}A_{ji})\\ & = \begin{bmatrix}
\frac{\partial}{\partial A_{11}}(\sum_{i=1}^{m}\sum_{j=1}^{n}B_{ij}A_{ji}) & \cdots & \frac{\partial}{\partial A_{1m}}(\sum_{i=1}^{m}\sum_{j=1}^{n}B_{ij}A_{ji})\\
\vdots & \ddots & \vdots \\
\frac{\partial}{\partial A_{n1}}(\sum_{i=1}^{m}\sum_{j=1}^{n}B_{ij}A_{ji}) & \cdots & \frac{\partial}{\partial A_{nm}}(\sum_{i=1}^{m}\sum_{j=1}^{n}B_{ij}A_{ji})
\end{bmatrix} \\ & = \begin{bmatrix}
B_{11} & \cdots & B_{m1} \\
\vdots & \ddots & \vdots \\
B_{1n} & \cdots & B_{mn}
\end{bmatrix} = B^{T}\\ \end{align*}$$
所以有 $ \triangledown_{A}\rm{tr}\;AB = \triangledown_{A}\rm{tr}\;BA = B^{T}$
4. $\triangledown_{A^{T}}a = (\triangledown_{A}a)^{T}\;\;\;\; (a \in \mathbb{R})$
证明:假设 A 是 $n * m$ 矩阵
$$\begin{align*}\triangledown_{A^{T}}a & = \begin{bmatrix}
\frac{\partial}{\partial A_{11}}a & \cdots & \frac{\partial}{\partial A_{1n}}a\\
\vdots & \ddots & \vdots \\
\frac{\partial}{\partial A_{m1}}a & \cdots & \frac{\partial}{\partial A_{mn}}a
\end{bmatrix} = (\begin{bmatrix}
\frac{\partial}{\partial A_{11}}a & \cdots & \frac{\partial}{\partial A_{1m}}a\\
\vdots & \ddots & \vdots \\
\frac{\partial}{\partial A_{n1}}a & \cdots & \frac{\partial}{\partial A_{nm}}a
\end{bmatrix})^{T} \\ & = (\triangledown_{A}a)^{T}\\ \end{align*}$$
5. $\mathrm{d}(\rm{tr}\;A) = \rm{tr}(\mathrm{d}A)$
证明:
$$\mathrm{d}(\rm{tr}\;A) = \mathrm{d}(\sum_{i=1}^{n}a_{ii}) = \sum_{i=1}^{n}\mathrm{d}a_{ii} = \rm{tr}(\mathrm{d}A)$$
矩阵的迹的微分等于矩阵的微分的迹。
6. $\triangledown_{A}\rm{tr}\;ABA^{T}C = CAB + C^{T}AB^{T}$
证明:
根据实标量函数梯度的乘法法则:
若 f(A)、g(A)、h(A) 分别是矩阵 A 的实标量函数,则有
$$\begin{align*}\frac{\partial f(A)g(A)}{\partial A} &= g(A)\frac{\partial f(A)}{\partial A} + f(A)\frac{\partial g(A)}{\partial A}\\ \frac{\partial f(A)g(A)h(A)}{\partial A} &= g(A)h(A)\frac{\partial f(A)}{\partial A} + f(A)h(A)\frac{\partial g(A)}{\partial A}+ f(A)g(A)\frac{\partial h(A)}{\partial A}\\ \end{align*}$$
令 $f(A) = AB,g(A) = A^{T}C$,由性质5,矩阵的迹的微分等于矩阵的微分的迹,那么则有:
$$\begin{align*} \triangledown_{A}\rm{tr}\;ABA^{T}C & = \rm{tr}(\triangledown_{A}ABA^{T}C) = \rm{tr}(\triangledown_{A}f(A)g(A)) = \rm{tr}\triangledown_{A_{1}}(A_{1}BA^{T}C) + \rm{tr}\triangledown_{A_{2}}(ABA_{2}^{T}C) \\ & = (BA^{T}C)^{T} + \rm{tr}\triangledown_{A_{2}}(ABA_{2}^{T}C) = C^{T}AB^{T} + \triangledown_{A_{2}}\rm{tr}(ABA_{2}^{T}C)\\ & = C^{T}AB^{T} + \triangledown_{A_{2}}\rm{tr}(A_{2}^{T}CAB) = C^{T}AB^{T} + (\triangledown_{{A_{2}}^{T}}\;\rm{tr}\;A_{2}^{T}CAB)^{T} \\ & = C^{T}AB^{T} + ((CAB)^{T})^{T} \\ & = C^{T}AB^{T} + CAB \\ \end{align*}$$
------------------------------------------------------------------------------------------------------------
### 3. 推导
回到之前的代价函数中:
$$
\rm{CostFunction} = \rm{F}({\theta_{0}},{\theta_{1}}) = \frac{1}{2m} (X \cdot \Theta - Y)^{T}(X \cdot \Theta - Y)
$$
求导:
$$
\begin{align*}
\triangledown_{\theta}\rm{F}(\theta) & = \frac{1}{2m} \triangledown_{\theta}(X \cdot \Theta - Y)^{T}(X \cdot \Theta - Y) = \frac{1}{2m}\triangledown_{\theta}(\Theta^{T}X^{T}-Y^{T})(X\Theta-Y)\\
& = \frac{1}{2m}\triangledown_{\theta}(\Theta^{T}X^{T}X\Theta-Y^{T}X\Theta-\Theta^{T}X^{T}Y+Y^{T}Y) \\ \end{align*}
$$
上式中,对 $\Theta $矩阵求导,$ Y^{T}Y $ 与 $\Theta $ 无关,所以这一项为 0 。 $Y^{T}X\Theta$ 是标量,由性质4可以知道,$Y^{T}X\Theta = (Y^{T}X\Theta)^{T} = \Theta^{T}X^{T}Y$,因为 $\Theta^{T}X^{T}X\Theta , Y^{T}X\Theta $都是标量,所以它们的也等于它们的迹,(处理矩阵微分的问题常常引入矩阵的迹),于是有
$$
\begin{align*}
\triangledown_{\theta}\rm{F}(\theta) & = \frac{1}{2m}\triangledown_{\theta}(\Theta^{T}X^{T}X\Theta-2Y^{T}X\Theta) \\
& = \frac{1}{2m}\triangledown_{\theta}\rm{tr}\;(\Theta^{T}X^{T}X\Theta-2Y^{T}X\Theta) \\ & = \frac{1}{2m}\triangledown_{\theta}\rm{tr}\;(\Theta\Theta^{T}X^{T}X-2Y^{T}X\Theta) \\ & = \frac{1}{m}(\frac{1}{2}\triangledown_{\theta}\rm{tr}\;\Theta\Theta^{T}X^{T}X -\triangledown_{\theta}\rm{tr}\;Y^{T}X\Theta) \\ & = \frac{1}{m}(\frac{1}{2}\triangledown_{\theta}\rm{tr}\;\Theta\Theta^{T}X^{T}X -(Y^{T}X)^{T}) = \frac{1}{m}(\frac{1}{2}\triangledown_{\theta}\rm{tr}\;\Theta\Theta^{T}X^{T}X -X^{T}Y)\\ \end{align*}
$$
上面第三步用的性质2矩阵迹的交换律,第五步用的性质3。
为了能进一步化简矩阵的微分,我们在矩阵的迹上面乘以一个单位矩阵,不影响结果。于是:
$$
\begin{align*}
\triangledown_{\theta}\rm{F}(\theta) & = \frac{1}{m}(\frac{1}{2}\triangledown_{\theta}\rm{tr}\;\Theta\Theta^{T}X^{T}X -X^{T}Y) \\ &= \frac{1}{m}(\frac{1}{2}\triangledown_{\theta}\rm{tr}\;\Theta I \Theta^{T}X^{T}X -X^{T}Y) \end{align*}
$$
利用性质6 展开上面的式子,令 $ A = \Theta , B = I , C = X^{T}X $。
$$
\begin{align*}
\triangledown_{\theta}\rm{F}(\theta) &= \frac{1}{m}(\frac{1}{2}\triangledown_{\theta}\rm{tr}\;\Theta I \Theta^{T}X^{T}X -X^{T}Y) \\ & = \frac{1}{m}(\frac{1}{2}(X^{T}X\Theta I + (X^{T}X)^{T}\Theta I^{T}) -X^{T}Y) \\ & = \frac{1}{m}(\frac{1}{2}(X^{T}X\Theta I + (X^{T}X)^{T}\Theta I^{T}) -X^{T}Y) \\ & = \frac{1}{m}(\frac{1}{2}(X^{T}X\Theta + X^{T}X\Theta) -X^{T}Y) = \frac{1}{m}(X^{T}X\Theta -X^{T}Y) \\ \end{align*}
$$
令 $\triangledown_{\theta}\rm{F}(\theta) = 0$,即 $X^{T}X\Theta -X^{T}Y = 0$, 于是 $ X^{T}X\Theta = X^{T}Y $ ,这里假设 $ X^{T}X$ 这个矩阵是可逆的,等号两边同时左乘$ X^{T}X$的逆矩阵,得到 $\Theta = (X^{T}X)^{-1}X^{T}Y$
最终结果也就推导出来了,$$\Theta = (X^{T}X)^{-1}X^{T}Y$$
但是这里有一个**前提条件是 $X^{T}X$ 是非奇异(非退化)矩阵, 即 $ \left | X^{T}X \right | \neq 0 $**
------------------------------------------------------------------------------------------------------------
### 4. 梯度下降和正规方程法比较:
优点:
梯度下降在超大数据集面前也能运行的很良好。
正规方程在超大数据集合面前性能会变得很差,因为需要计算 $(x^{T}x)^{-1}$,时间复杂度在 $O(n^{3})$ 这个级别。
缺点:
梯度下降需要合理的选择学习速率 $\alpha$ , 需要很多次迭代的操作去选择合理的 $\alpha$,寻找最小值的时候也需要迭代很多次才能收敛。
正规方程的优势相比而言,不需要选择学习速率 $\alpha$,也不需要多次的迭代或者画图检测是否收敛。
------------------------------------------------------------------------------------------------------------
## 二. Normal Equation Noninvertibility
上一章谈到了如何利用正规方程法求解 $\Theta $,但是在线性代数中存在这样一个问题,如果是奇异(退化)矩阵,是不存在逆矩阵的。也就是说用上面正规方程的公式是不一定能求解出正确结果的。
在 Octave 软件中,存在2个求解逆矩阵的函数,一个是 pinv 和 inv。pinv (pseudo-inverse)求解的是**伪逆矩阵**,inv 求解的是逆矩阵,所以用 pinv 求解问题,就算是 $ X^{T}X $ 不存在逆矩阵,也一样可以得到最后的结果。
导致$ X^{T}X $ 不存在逆矩阵有2种情况:
1. 多余的特征。特征之间呈倍数关系,线性依赖。
2. 过多的特征。当 $ m \leqslant n $ 的时候,会导致过多的特征。解决办法是删除一些特征,或者进行正则化。
所以解决$ X^{T}X $ 不存在逆矩阵的办法也就是对应上面2种情况:
1. 删掉多余的特征,线性相关的,倍数关系的。直到没有多余的特征
2. 再删除一些不影响结果的特征,或者进行正则化。
------------------------------------------------------------------------------------------------------------
## 三. Linear Regression with Multiple Variables 测试
### 1. Question 1
Suppose m=4 students have taken some class, and the class had a midterm exam and a final exam. You have collected a dataset of their scores on the two exams, which is as follows:
midterm exam (midterm exam)2 final exam
89 7921 96
72 5184 74
94 8836 87
69 4761 78
You'd like to use polynomial regression to predict a student's final exam score from their midterm exam score. Concretely, suppose you want to fit a model of the form hθ(x)=θ0+θ1x1+θ2x2, where x1 is the midterm score and x2 is (midterm score)2. Further, you plan to use both feature scaling (dividing by the "max-min", or range, of a feature) and mean normalization.
What is the normalized feature x(2)2? (Hint: midterm = 72, final = 74 is training example 2.) Please round off your answer to two decimal places and enter in the text box below.
解答:
标准化 $$x = \frac{x_{2}^{2}-\frac{(7921+5184+8836+4761)}{4}}{\max - \min } = \frac{5184 - 6675.5}{8836-4761} = -0.37$$
### 2. Question 2
You run gradient descent for 15 iterations
with α=0.3 and compute J(θ) after each
iteration. You find that the value of J(θ) increases over
time. Based on this, which of the following conclusions seems
most plausible?
A. Rather than use the current value of α, it'd be more promising to try a smaller value of α (say α=0.1).
B. α=0.3 is an effective choice of learning rate.
C. Rather than use the current value of α, it'd be more promising to try a larger value of α (say α=1.0).
解答: A
下降太快所以a下降速率过大,a越大下降越快,a小下降慢,在本题中,代价函数快速收敛到最小值,代表此时a最合适。
### 3. Question 3
Suppose you have m=28 training examples with n=4 features (excluding the additional all-ones feature for the intercept term, which you should add). The normal equation is θ=(XTX)−1XTy. For the given values of m and n, what are the dimensions of θ, X, and y in this equation?
A. X is 28×4, y is 28×1, θ is 4×4
B. X is 28×5, y is 28×5, θ is 5×5
C. X is 28×5, y is 28×1, θ is 5×1
D. X is 28×4, y is 28×1, θ is4×1
解答: C
这里需要注意的是,题目中说了额外添加一列全部为1的,所以列数是5 。
### 4. Question 4
Suppose you have a dataset with m=50 examples and n=15 features for each example. You want to use multivariate linear regression to fit the parameters θ to our data. Should you prefer gradient descent or the normal equation?
A. Gradient descent, since it will always converge to the optimal θ.
B. Gradient descent, since (XTX)−1 will be very slow to compute in the normal equation.
C. The normal equation, since it provides an efficient way to directly find the solution.
D. The normal equation, since gradient descent might be unable to find the optimal θ.
解答: C
数据量少,选择正规方程法更加高效
### 5. Question 5
Which of the following are reasons for using feature scaling?
A. It prevents the matrix XTX (used in the normal equation) from being non-invertable (singular/degenerate).
B. It is necessary to prevent the normal equation from getting stuck in local optima.
C. It speeds up gradient descent by making it require fewer iterations to get to a good solution.
D. It speeds up gradient descent by making each iteration of gradient descent less expensive to compute.
解答: C
normal equation 不需要 Feature Scaling,排除AB, 特征缩放减少迭代数量,加快梯度下降,然而不能防止梯度下降陷入局部最优。
------------------------------------------------------
> GitHub Repo:[Halfrost-Field](https://github.com/halfrost/Halfrost-Field)
>
> Follow: [halfrost · GitHub](https://github.com/halfrost)
>
> Source: [https://github.com/halfrost/Halfrost-Field/blob/master/contents/Machine\_Learning/Computing\_Parameters\_Analytically.ipynb](https://github.com/halfrost/Halfrost-Field/blob/master/contents/Machine_Learning/Computing_Parameters_Analytically.ipynb)
| 0.478529 | 0.876476 |
# Calculate Race Track from logs and SVG
Given the svg file from the track selection in the DeepRacer console, this
code will generate a passable `.npy` file with waypoints to use in log
analysis.
```
import svgpathtools as svg
from shapely.geometry import Point, Polygon
from shapely.geometry.polygon import LinearRing, LineString
from shapely import affinity
from shapely import ops
import numpy as np
import math
```
## Inputs
```
# Set this to the track svg file
source_track_name_svg = "canada_race"
target_track_name_npy = "Canada_Training"
# These should come from (a) circuit track logs for # of waypoints and track length,
# and (b) the companion training track for track width
DESIRED_LENGTH = 18.45
DESIRED_NUMBER_WAYPOINTS = 202
```
## Load SVG Source Track
```
# Load and parse the svg
fname = "./tracks/%s.svg" % source_track_name_svg
paths,attributes = svg.svg2paths(fname)
# Discover which path has a 'stroke-dasharray', indicating the center line
center_line_path = None
for i in range(len(attributes)):
if 'stroke-dasharray' in attributes[i]:
center_line_path = paths[i]
type(center_line_path)
```
## Convert SVG Path to Shapely
```
#Interpolate the SVG Path into waypoints
points = list()
svg_length = center_line_path.length()
for i in range(DESIRED_NUMBER_WAYPOINTS):
center_point = center_line_path.point(i/DESIRED_NUMBER_WAYPOINTS)
center_x, center_y = center_point.real, center_point.imag
points.append((center_x, center_y))
# svg.path doesn't quite have final point the same as first point so we
# be sure to close the path by repeating the zero coordinate
point = center_line_path.point(0)
points.append((point.real, point.imag))
# Make shapely geometry out of waypoints
ls_source_center_line = LineString(points)
# Scale to desired size
scale_factor = DESIRED_LENGTH / ls_source_center_line.length
ls_source_center_line = affinity.scale(ls_source_center_line, xfact=scale_factor, yfact=scale_factor)
print("Center length: ", ls_source_center_line.length)
# SVG has (always?) mirrored image
ls_source_center_line = affinity.scale(ls_source_center_line, xfact=-1.0)
ls_source_center_line
```
# Fit the track to another similar track
Usually applicable for race tracks vs training tracks for virtual summit
```
# Load the target shape
target_track_waypoints = np.load("tracks/%s.npy" % target_track_name_npy)
ls_target_center_line = LineString(target_track_waypoints[:,0:2])
target_track_width = Point(target_track_waypoints[0,2:4]).distance(Point(target_track_waypoints[0,4:6]))
ls_target_center_line
from bayes_opt import BayesianOptimization
# Bayes tuning
N_ITER=30
INIT_POINTS=800
KAPPA=1
# Convert source and target to polygons for optimization function
p_source = Polygon(ls_source_center_line)
p_target = Polygon(ls_target_center_line)
# Initialize our optimization by forcing at least one overlapping point
target_center = p_target.centroid
source_center = p_source.centroid
p_source = affinity.translate(p_source,
xoff=(target_center.x - source_center.x),
yoff=(target_center.y - source_center.y))
minx, miny, maxx, maxy = p_target.bounds
mintx = minx - target_center.x
maxtx = maxx - target_center.x
minty = miny - target_center.y
maxty = maxy - target_center.y
pbounds = {
'r': (-180,180), # degree of rotation
'x': (mintx, maxtx), # centroid definitely won't move outside of target bounds
'y': (minty, maxty)
}
print("Searching pbounds: ", pbounds)
print("Target bounds: ", p_target.bounds)
def coincident_area(r,x,y):
global p_source
global p_target
p_source_t = affinity.translate(p_source, xoff=x, yoff=y)
p_source_r = affinity.rotate(p_source_t, r, origin='centroid')
intersection = p_source_r.intersection(p_target)
difference = p_source_r.difference(p_target)
return intersection.area - 20*difference.area # penalize overhang so optimization doesn't find some odd orientation that has largest overlap
# Start the optimization
optimizer = BayesianOptimization(f=coincident_area, pbounds=pbounds, random_state=1)
optimizer.maximize(n_iter=N_ITER, init_points=INIT_POINTS, kappa=KAPPA)
# Apply results of optimization
values = optimizer.max['params']
print(optimizer.max)
values['r'] = 90
p_applied = affinity.translate(p_source, xoff=values['x'], yoff=values['y'])
p_applied = affinity.rotate(p_applied, values['r'], origin='centroid')
# Visually inspect the translated, rotated, and scaled track
import matplotlib.pyplot as plt
tx,ty = p_target.exterior.xy
plt.plot(tx,ty)
sx,sy = p_applied.exterior.xy
plt.plot(sx,sy)
plt.show()
# Make a poly out of our center line, expanding its dimensions to have outer and inner boundaries
ls_center = p_applied.exterior
poly = ls_center.buffer(target_track_width/2.0)
print("Center Closed: ", ls_center.is_ring)
print("Center Length: ", ls_center.length)
print("Center Bounds: ", ls_center.bounds)
ls_outer = poly.exterior
ls_inner = poly.interiors[0]
print("Outer Length: ", ls_outer.length)
print("Innter Length: ", ls_inner.length)
# Generate final waypoint list as [center_x, center_y, inner_x, inner_y, outer_x, outer_y]
all_waypoints = list()
for i in range(len(ls_center.coords)):
distance = i / len(ls_center.coords)
center_point = ls_center.coords[i]
outer_point = ls_outer.interpolate(distance, normalized=True)
inner_point = ls_inner.interpolate(distance, normalized=True)
all_waypoints.append(list(sum((center_point, inner_point.coords[0], outer_point.coords[0]), ())))
poly
```
## Write out the resulting geometry
```
# Save resulting waypoints to file
fname = "./tracks/%s.npy"% source_track_name_svg
np.save(fname, all_waypoints)
```
|
github_jupyter
|
import svgpathtools as svg
from shapely.geometry import Point, Polygon
from shapely.geometry.polygon import LinearRing, LineString
from shapely import affinity
from shapely import ops
import numpy as np
import math
# Set this to the track svg file
source_track_name_svg = "canada_race"
target_track_name_npy = "Canada_Training"
# These should come from (a) circuit track logs for # of waypoints and track length,
# and (b) the companion training track for track width
DESIRED_LENGTH = 18.45
DESIRED_NUMBER_WAYPOINTS = 202
# Load and parse the svg
fname = "./tracks/%s.svg" % source_track_name_svg
paths,attributes = svg.svg2paths(fname)
# Discover which path has a 'stroke-dasharray', indicating the center line
center_line_path = None
for i in range(len(attributes)):
if 'stroke-dasharray' in attributes[i]:
center_line_path = paths[i]
type(center_line_path)
#Interpolate the SVG Path into waypoints
points = list()
svg_length = center_line_path.length()
for i in range(DESIRED_NUMBER_WAYPOINTS):
center_point = center_line_path.point(i/DESIRED_NUMBER_WAYPOINTS)
center_x, center_y = center_point.real, center_point.imag
points.append((center_x, center_y))
# svg.path doesn't quite have final point the same as first point so we
# be sure to close the path by repeating the zero coordinate
point = center_line_path.point(0)
points.append((point.real, point.imag))
# Make shapely geometry out of waypoints
ls_source_center_line = LineString(points)
# Scale to desired size
scale_factor = DESIRED_LENGTH / ls_source_center_line.length
ls_source_center_line = affinity.scale(ls_source_center_line, xfact=scale_factor, yfact=scale_factor)
print("Center length: ", ls_source_center_line.length)
# SVG has (always?) mirrored image
ls_source_center_line = affinity.scale(ls_source_center_line, xfact=-1.0)
ls_source_center_line
# Load the target shape
target_track_waypoints = np.load("tracks/%s.npy" % target_track_name_npy)
ls_target_center_line = LineString(target_track_waypoints[:,0:2])
target_track_width = Point(target_track_waypoints[0,2:4]).distance(Point(target_track_waypoints[0,4:6]))
ls_target_center_line
from bayes_opt import BayesianOptimization
# Bayes tuning
N_ITER=30
INIT_POINTS=800
KAPPA=1
# Convert source and target to polygons for optimization function
p_source = Polygon(ls_source_center_line)
p_target = Polygon(ls_target_center_line)
# Initialize our optimization by forcing at least one overlapping point
target_center = p_target.centroid
source_center = p_source.centroid
p_source = affinity.translate(p_source,
xoff=(target_center.x - source_center.x),
yoff=(target_center.y - source_center.y))
minx, miny, maxx, maxy = p_target.bounds
mintx = minx - target_center.x
maxtx = maxx - target_center.x
minty = miny - target_center.y
maxty = maxy - target_center.y
pbounds = {
'r': (-180,180), # degree of rotation
'x': (mintx, maxtx), # centroid definitely won't move outside of target bounds
'y': (minty, maxty)
}
print("Searching pbounds: ", pbounds)
print("Target bounds: ", p_target.bounds)
def coincident_area(r,x,y):
global p_source
global p_target
p_source_t = affinity.translate(p_source, xoff=x, yoff=y)
p_source_r = affinity.rotate(p_source_t, r, origin='centroid')
intersection = p_source_r.intersection(p_target)
difference = p_source_r.difference(p_target)
return intersection.area - 20*difference.area # penalize overhang so optimization doesn't find some odd orientation that has largest overlap
# Start the optimization
optimizer = BayesianOptimization(f=coincident_area, pbounds=pbounds, random_state=1)
optimizer.maximize(n_iter=N_ITER, init_points=INIT_POINTS, kappa=KAPPA)
# Apply results of optimization
values = optimizer.max['params']
print(optimizer.max)
values['r'] = 90
p_applied = affinity.translate(p_source, xoff=values['x'], yoff=values['y'])
p_applied = affinity.rotate(p_applied, values['r'], origin='centroid')
# Visually inspect the translated, rotated, and scaled track
import matplotlib.pyplot as plt
tx,ty = p_target.exterior.xy
plt.plot(tx,ty)
sx,sy = p_applied.exterior.xy
plt.plot(sx,sy)
plt.show()
# Make a poly out of our center line, expanding its dimensions to have outer and inner boundaries
ls_center = p_applied.exterior
poly = ls_center.buffer(target_track_width/2.0)
print("Center Closed: ", ls_center.is_ring)
print("Center Length: ", ls_center.length)
print("Center Bounds: ", ls_center.bounds)
ls_outer = poly.exterior
ls_inner = poly.interiors[0]
print("Outer Length: ", ls_outer.length)
print("Innter Length: ", ls_inner.length)
# Generate final waypoint list as [center_x, center_y, inner_x, inner_y, outer_x, outer_y]
all_waypoints = list()
for i in range(len(ls_center.coords)):
distance = i / len(ls_center.coords)
center_point = ls_center.coords[i]
outer_point = ls_outer.interpolate(distance, normalized=True)
inner_point = ls_inner.interpolate(distance, normalized=True)
all_waypoints.append(list(sum((center_point, inner_point.coords[0], outer_point.coords[0]), ())))
poly
# Save resulting waypoints to file
fname = "./tracks/%s.npy"% source_track_name_svg
np.save(fname, all_waypoints)
| 0.67405 | 0.875255 |
# Import Modules
```
import tensorflow as tf
from tensorflow.keras.layers import Conv2D, Conv2DTranspose, BatchNormalization, Activation, MaxPooling2D, concatenate, Dropout, Input
from tensorflow.keras.models import Model
```
# Model
```
class Blocks:
def __init__(self, n_filters):
self.n_filters = n_filters
self.n = 1
self.convLayers = {}
def pool_block(self, input_x, pool_size=(2, 2), dropout=0.5):
x = MaxPooling2D(pool_size=pool_size)(input_x)
x = Dropout(dropout)(x)
return x
def conv2d_block(self, input_x, kernel_size=(3,3), pad='same', count=True):
if count:
name = f'conv_{(self.n)}'
else:
name = f'conv_ePath_{(self.n // 2)}'
x = Conv2D(filters=self.n_filters * self.n, kernel_size=kernel_size, padding=pad)(input_x)
x = Activation('relu')(x)
x = BatchNormalization()(x)
x = Conv2D(filters=self.n_filters * self.n, kernel_size=kernel_size, padding=pad, name=name)(x)
x = Activation('relu')(x)
x = BatchNormalization()(x)
if count:
self.convLayers[name] = x
self.n *=2
return x
def convTrans_block(self, input_x, kernel_size=(3,3), strides=(2, 2), pad='same', dropout=0.5):
assert self.n >= 2, f'n = {self.n}'
self.n //=2
conv_name = f'conv_{self.n // 2}'
x = Conv2DTranspose(filters=self.n_filters * self.n, kernel_size=kernel_size, strides = strides, padding=pad)(input_x)
x = concatenate([x, self.convLayers[conv_name]])
x = Dropout(dropout)(x)
return x
SHAPE = (256, 256, 3)
block = Blocks(n_filters=16)
inputs = Input(shape=SHAPE)
# contracting path
x = block.conv2d_block(inputs)
x = block.pool_block(x)
x = block.conv2d_block(x)
x = block.pool_block(x)
x = block.conv2d_block(x)
x = block.pool_block(x)
x = block.conv2d_block(x)
x = block.pool_block(x)
x = block.conv2d_block(x)
# expansive path
x = block.convTrans_block(x)
x = block.conv2d_block(x, count=False)
x = block.convTrans_block(x)
x = block.conv2d_block(x, count=False)
x = block.convTrans_block(x)
x = block.conv2d_block(x, count=False)
x = block.convTrans_block(x)
x = block.conv2d_block(x, count=False)
outputs = Conv2D(1, (1, 1), activation='sigmoid')(x)
model = Model(inputs=[inputs], outputs=[outputs])
model.summary()
```
|
github_jupyter
|
import tensorflow as tf
from tensorflow.keras.layers import Conv2D, Conv2DTranspose, BatchNormalization, Activation, MaxPooling2D, concatenate, Dropout, Input
from tensorflow.keras.models import Model
class Blocks:
def __init__(self, n_filters):
self.n_filters = n_filters
self.n = 1
self.convLayers = {}
def pool_block(self, input_x, pool_size=(2, 2), dropout=0.5):
x = MaxPooling2D(pool_size=pool_size)(input_x)
x = Dropout(dropout)(x)
return x
def conv2d_block(self, input_x, kernel_size=(3,3), pad='same', count=True):
if count:
name = f'conv_{(self.n)}'
else:
name = f'conv_ePath_{(self.n // 2)}'
x = Conv2D(filters=self.n_filters * self.n, kernel_size=kernel_size, padding=pad)(input_x)
x = Activation('relu')(x)
x = BatchNormalization()(x)
x = Conv2D(filters=self.n_filters * self.n, kernel_size=kernel_size, padding=pad, name=name)(x)
x = Activation('relu')(x)
x = BatchNormalization()(x)
if count:
self.convLayers[name] = x
self.n *=2
return x
def convTrans_block(self, input_x, kernel_size=(3,3), strides=(2, 2), pad='same', dropout=0.5):
assert self.n >= 2, f'n = {self.n}'
self.n //=2
conv_name = f'conv_{self.n // 2}'
x = Conv2DTranspose(filters=self.n_filters * self.n, kernel_size=kernel_size, strides = strides, padding=pad)(input_x)
x = concatenate([x, self.convLayers[conv_name]])
x = Dropout(dropout)(x)
return x
SHAPE = (256, 256, 3)
block = Blocks(n_filters=16)
inputs = Input(shape=SHAPE)
# contracting path
x = block.conv2d_block(inputs)
x = block.pool_block(x)
x = block.conv2d_block(x)
x = block.pool_block(x)
x = block.conv2d_block(x)
x = block.pool_block(x)
x = block.conv2d_block(x)
x = block.pool_block(x)
x = block.conv2d_block(x)
# expansive path
x = block.convTrans_block(x)
x = block.conv2d_block(x, count=False)
x = block.convTrans_block(x)
x = block.conv2d_block(x, count=False)
x = block.convTrans_block(x)
x = block.conv2d_block(x, count=False)
x = block.convTrans_block(x)
x = block.conv2d_block(x, count=False)
outputs = Conv2D(1, (1, 1), activation='sigmoid')(x)
model = Model(inputs=[inputs], outputs=[outputs])
model.summary()
| 0.932844 | 0.908334 |
# Construir, compilar y entrenar modelos de Machine Learning con TensorFlow
# Preprocesar información para tenerla lista y utilizarla en un modelo.
```
# Import constant from TensorFlow
from tensorflow import constant
# Convert the credit_numpy array into a tensorflow constant
credit_constant = constant(credit_numpy)
# Print constant datatype
print('\n The datatype is:', credit_constant.dtype)
# Print constant shape
print('\n The shape is:', credit_constant.shape)
# Define the 1-dimensional variable A1
A1 = Variable([1, 2, 3, 4])
# Print the variable A1
print('\n A1: ', A1)
# Convert A1 to a numpy array and assign it to B1
B1 = A1.numpy()
# Print B1
print('\n B1: ', B1)
# Define tensors A1 and A23 as constants
A1 = constant([1, 2, 3, 4])
A23 = constant([[1, 2, 3], [1, 6, 4]])
# Define B1 and B23 to have the correct shape
B1 = ones_like(A1)
B23 = ones_like(A23)
# Perform element-wise multiplication
C1 = multiply(A1,B1)
C23 = multiply(A23, B23)
# Print the tensors C1 and C23
print('\n C1: {}'.format(C1.numpy()))
print('\n C23: {}'.format(C23.numpy()))
```
## Making predictions with matrix multiplication
```
# Define features, params, and bill as constants
features = constant([[2, 24], [2, 26], [2, 57], [1, 37]])
params = constant([[1000], [150]])
bill = constant([[3913], [2682], [8617], [64400]])
# Compute billpred using features and params
billpred = matmul(features, params)
# Compute and print the error
error = bill - billpred
print(error.numpy())
```
## Transformar datos de entrada
```
# Reshape the grayscale image tensor into a vector
gray_vector = reshape(gray_tensor, (784, 1))
# Reshape the color image tensor into a vector
color_vector = reshape(color_tensor, (2352, 1))
```
## Optimizando con gradientes
```
def compute_gradient(x0):
# Define x as a variable with an initial value of x0
x = Variable(x0)
with GradientTape() as tape:
tape.watch(x)
# Define y using the multiply operation
y = multiply(x, x)
# Return the gradient of y with respect to x
return tape.gradient(y, x).numpy()
# Compute and print gradients at x = -1, 1, and 0
print(compute_gradient(-1.0))
print(compute_gradient(1.0))
print(compute_gradient(0.0))
```
## Trabajando con datos de imágenes
```
# Reshape model from a 1x3 to a 3x1 tensor
model = reshape(model, (3, 1))
# Multiply letter by model
output = matmul(letter, model)
# Sum over output and print prediction using the numpy method
prediction = reduce_sum(output)
print(prediction.numpy())
```
|
github_jupyter
|
# Import constant from TensorFlow
from tensorflow import constant
# Convert the credit_numpy array into a tensorflow constant
credit_constant = constant(credit_numpy)
# Print constant datatype
print('\n The datatype is:', credit_constant.dtype)
# Print constant shape
print('\n The shape is:', credit_constant.shape)
# Define the 1-dimensional variable A1
A1 = Variable([1, 2, 3, 4])
# Print the variable A1
print('\n A1: ', A1)
# Convert A1 to a numpy array and assign it to B1
B1 = A1.numpy()
# Print B1
print('\n B1: ', B1)
# Define tensors A1 and A23 as constants
A1 = constant([1, 2, 3, 4])
A23 = constant([[1, 2, 3], [1, 6, 4]])
# Define B1 and B23 to have the correct shape
B1 = ones_like(A1)
B23 = ones_like(A23)
# Perform element-wise multiplication
C1 = multiply(A1,B1)
C23 = multiply(A23, B23)
# Print the tensors C1 and C23
print('\n C1: {}'.format(C1.numpy()))
print('\n C23: {}'.format(C23.numpy()))
# Define features, params, and bill as constants
features = constant([[2, 24], [2, 26], [2, 57], [1, 37]])
params = constant([[1000], [150]])
bill = constant([[3913], [2682], [8617], [64400]])
# Compute billpred using features and params
billpred = matmul(features, params)
# Compute and print the error
error = bill - billpred
print(error.numpy())
# Reshape the grayscale image tensor into a vector
gray_vector = reshape(gray_tensor, (784, 1))
# Reshape the color image tensor into a vector
color_vector = reshape(color_tensor, (2352, 1))
def compute_gradient(x0):
# Define x as a variable with an initial value of x0
x = Variable(x0)
with GradientTape() as tape:
tape.watch(x)
# Define y using the multiply operation
y = multiply(x, x)
# Return the gradient of y with respect to x
return tape.gradient(y, x).numpy()
# Compute and print gradients at x = -1, 1, and 0
print(compute_gradient(-1.0))
print(compute_gradient(1.0))
print(compute_gradient(0.0))
# Reshape model from a 1x3 to a 3x1 tensor
model = reshape(model, (3, 1))
# Multiply letter by model
output = matmul(letter, model)
# Sum over output and print prediction using the numpy method
prediction = reduce_sum(output)
print(prediction.numpy())
| 0.743354 | 0.938294 |
```
import tensorflow as tf
import pandas as pd
import matplotlib.pyplot as plt
from sklearn import preprocessing
import numpy as np
import pandas as pd
data = pd.read_csv('data_stocks.csv')
data.info()
data.head()
%matplotlib inline
plt.grid()
plt.plot(data['SP500'])
#每次仅能运行一次
data.drop('DATE', axis=1, inplace=True)
data_train = data.iloc[:int(data.shape[0] * 0.8), :]
data_test = data.iloc[int(data.shape[0] * 0.8):, :]
data_train
data_train.shape
print(data_test)
data_test.shape
scaler = preprocessing.MinMaxScaler(feature_range=(-1, 1))
scaler.fit(data_train)
data_train = scaler.transform(data_train)
data_test = scaler.transform(data_test)
# print(data_train)
# print(data_test)
X_train = data_train[:, 1:]
y_train = data_train[:, 0]
X_test = data_test[:, 1:]
y_test = data_test[:, 0]
input_dim = X_train.shape[1]
output_dim = 1
hidden_1 = 1024
hidden_2 = 512
hidden_3 = 256
hidden_4 = 128
batch_size = 256
epochs = 10
X = tf.placeholder(shape=[None, input_dim], dtype=tf.float32)
Y = tf.placeholder(shape=[None], dtype=tf.float32)
# 第一层
W1 = tf.get_variable('W1', [input_dim, hidden_1], initializer=tf.contrib.layers.xavier_initializer(seed=1))
b1 = tf.get_variable('b1', [hidden_1], initializer=tf.zeros_initializer())
# 第二层
W2 = tf.get_variable('W2', [hidden_1, hidden_2], initializer=tf.contrib.layers.xavier_initializer(seed=1))
b2 = tf.get_variable('b2', [hidden_2], initializer=tf.zeros_initializer())
# 第三层
W3 = tf.get_variable('W3', [hidden_2, hidden_3], initializer=tf.contrib.layers.xavier_initializer(seed=1))
b3 = tf.get_variable('b3', [hidden_3], initializer=tf.zeros_initializer())
# 第四层
W4 = tf.get_variable('W4', [hidden_3, hidden_4], initializer=tf.contrib.layers.xavier_initializer(seed=1))
b4 = tf.get_variable('b4', [hidden_4], initializer=tf.zeros_initializer())
# 输出层
W5 = tf.get_variable('W5', [hidden_4, output_dim], initializer=tf.contrib.layers.xavier_initializer(seed=1))
b5 = tf.get_variable('b5', [output_dim], initializer=tf.zeros_initializer())
h1 = tf.nn.relu(tf.add(tf.matmul(X, W1), b1))
h2 = tf.nn.relu(tf.add(tf.matmul(h1, W2), b2))
h3 = tf.nn.relu(tf.add(tf.matmul(h2, W3), b3))
h4 = tf.nn.relu(tf.add(tf.matmul(h3, W4), b4))
out = tf.transpose(tf.add(tf.matmul(h4, W5), b5))
loss = tf.reduce_mean(tf.squared_difference(out, Y))
optimizer = tf.train.AdamOptimizer().minimize(loss)
with tf.Session() as sess:
# 初始化所有变量
sess.run(tf.global_variables_initializer())
for e in range(epochs):
# 将数据打乱
shuffle_indices = np.random.permutation(np.arange(y_train.shape[0]))
X_train = X_train[shuffle_indices]
y_train = y_train[shuffle_indices]
for i in range(y_train.shape[0] // batch_size):
start = i * batch_size
batch_x = X_train[start : start + batch_size]
batch_y = y_train[start : start + batch_size]
sess.run(optimizer, feed_dict={X: batch_x, Y: batch_y})
if i % 50 == 0:
print('MSE Train:', sess.run(loss, feed_dict={X: X_train, Y: y_train}))
print('MSE Test:', sess.run(loss, feed_dict={X: X_test, Y: y_test}))
y_pred = sess.run(out, feed_dict={X: X_test})
y_pred = np.squeeze(y_pred)
plt.plot(y_test, label='test')
plt.plot(y_pred, label='pred')
plt.title('Epoch ' + str(e) + ', Batch ' + str(i))
plt.legend()
plt.grid()
plt.show()
```
|
github_jupyter
|
import tensorflow as tf
import pandas as pd
import matplotlib.pyplot as plt
from sklearn import preprocessing
import numpy as np
import pandas as pd
data = pd.read_csv('data_stocks.csv')
data.info()
data.head()
%matplotlib inline
plt.grid()
plt.plot(data['SP500'])
#每次仅能运行一次
data.drop('DATE', axis=1, inplace=True)
data_train = data.iloc[:int(data.shape[0] * 0.8), :]
data_test = data.iloc[int(data.shape[0] * 0.8):, :]
data_train
data_train.shape
print(data_test)
data_test.shape
scaler = preprocessing.MinMaxScaler(feature_range=(-1, 1))
scaler.fit(data_train)
data_train = scaler.transform(data_train)
data_test = scaler.transform(data_test)
# print(data_train)
# print(data_test)
X_train = data_train[:, 1:]
y_train = data_train[:, 0]
X_test = data_test[:, 1:]
y_test = data_test[:, 0]
input_dim = X_train.shape[1]
output_dim = 1
hidden_1 = 1024
hidden_2 = 512
hidden_3 = 256
hidden_4 = 128
batch_size = 256
epochs = 10
X = tf.placeholder(shape=[None, input_dim], dtype=tf.float32)
Y = tf.placeholder(shape=[None], dtype=tf.float32)
# 第一层
W1 = tf.get_variable('W1', [input_dim, hidden_1], initializer=tf.contrib.layers.xavier_initializer(seed=1))
b1 = tf.get_variable('b1', [hidden_1], initializer=tf.zeros_initializer())
# 第二层
W2 = tf.get_variable('W2', [hidden_1, hidden_2], initializer=tf.contrib.layers.xavier_initializer(seed=1))
b2 = tf.get_variable('b2', [hidden_2], initializer=tf.zeros_initializer())
# 第三层
W3 = tf.get_variable('W3', [hidden_2, hidden_3], initializer=tf.contrib.layers.xavier_initializer(seed=1))
b3 = tf.get_variable('b3', [hidden_3], initializer=tf.zeros_initializer())
# 第四层
W4 = tf.get_variable('W4', [hidden_3, hidden_4], initializer=tf.contrib.layers.xavier_initializer(seed=1))
b4 = tf.get_variable('b4', [hidden_4], initializer=tf.zeros_initializer())
# 输出层
W5 = tf.get_variable('W5', [hidden_4, output_dim], initializer=tf.contrib.layers.xavier_initializer(seed=1))
b5 = tf.get_variable('b5', [output_dim], initializer=tf.zeros_initializer())
h1 = tf.nn.relu(tf.add(tf.matmul(X, W1), b1))
h2 = tf.nn.relu(tf.add(tf.matmul(h1, W2), b2))
h3 = tf.nn.relu(tf.add(tf.matmul(h2, W3), b3))
h4 = tf.nn.relu(tf.add(tf.matmul(h3, W4), b4))
out = tf.transpose(tf.add(tf.matmul(h4, W5), b5))
loss = tf.reduce_mean(tf.squared_difference(out, Y))
optimizer = tf.train.AdamOptimizer().minimize(loss)
with tf.Session() as sess:
# 初始化所有变量
sess.run(tf.global_variables_initializer())
for e in range(epochs):
# 将数据打乱
shuffle_indices = np.random.permutation(np.arange(y_train.shape[0]))
X_train = X_train[shuffle_indices]
y_train = y_train[shuffle_indices]
for i in range(y_train.shape[0] // batch_size):
start = i * batch_size
batch_x = X_train[start : start + batch_size]
batch_y = y_train[start : start + batch_size]
sess.run(optimizer, feed_dict={X: batch_x, Y: batch_y})
if i % 50 == 0:
print('MSE Train:', sess.run(loss, feed_dict={X: X_train, Y: y_train}))
print('MSE Test:', sess.run(loss, feed_dict={X: X_test, Y: y_test}))
y_pred = sess.run(out, feed_dict={X: X_test})
y_pred = np.squeeze(y_pred)
plt.plot(y_test, label='test')
plt.plot(y_pred, label='pred')
plt.title('Epoch ' + str(e) + ', Batch ' + str(i))
plt.legend()
plt.grid()
plt.show()
| 0.47098 | 0.590632 |
# Base enem 2016
## Predição se o aluno é treineiro.
## Segundo teste:
### * Limpeza dos dados
### * Uso do balanceamento
### * Algoritmos:
* Regressão Logística (Score obtido: 96.083151)
* Decision tree (Score obtido: 96.105033)
* Random Forest (Score obtido: 96.630197)
* Nearest Neighbors (Score obtido: 64.135667)
```
import pandas as pd
import numpy as np
import warnings
from sklearn.preprocessing import OneHotEncoder
from sklearn.linear_model import LogisticRegression
from imblearn.over_sampling import SMOTE
from sklearn.ensemble import BaggingClassifier, RandomForestClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.tree import DecisionTreeClassifier, plot_tree
warnings.filterwarnings('ignore')
pd.set_option('display.max_columns', 300)
pd.set_option('display.max_rows', 300)
file_train = "train.csv"
file_test = "test.csv"
df_raw_train = pd.read_csv(file_train, index_col=False)
df_raw_test = pd.read_csv(file_test, index_col=False)
df_raw_train.shape, df_raw_test.shape
columns_used=['NU_INSCRICAO','CO_UF_RESIDENCIA', 'SG_UF_RESIDENCIA', 'NU_IDADE', 'TP_SEXO', 'TP_COR_RACA',
'TP_NACIONALIDADE','TP_ST_CONCLUSAO','TP_ANO_CONCLUIU', 'TP_ESCOLA', 'TP_ENSINO',
'TP_PRESENCA_CN', 'TP_PRESENCA_CH', 'TP_PRESENCA_LC','NU_NOTA_CN', 'NU_NOTA_CH',
'NU_NOTA_LC','TP_LINGUA','TP_STATUS_REDACAO', 'NU_NOTA_COMP1', 'NU_NOTA_COMP2',
'NU_NOTA_COMP3','NU_NOTA_COMP4', 'NU_NOTA_COMP5', 'NU_NOTA_REDACAO','Q001', 'Q002',
'Q006', 'Q024', 'Q025', 'Q026', 'Q027', 'Q047', 'IN_TREINEIRO']
numerical_vars = ['NU_NOTA_CN', 'NU_NOTA_CH', 'NU_NOTA_LC','NU_NOTA_COMP1', 'NU_NOTA_COMP2', 'NU_NOTA_COMP3',
'NU_NOTA_COMP4','NU_NOTA_COMP5', 'NU_NOTA_REDACAO']
target = ['IN_TREINEIRO']
df_train=df_raw_train[columns_used]
df_train.drop(['SG_UF_RESIDENCIA','TP_ENSINO'], inplace=True, axis=1)
df_test=df_raw_test[columns_used[:-1]]
df_test.drop(['SG_UF_RESIDENCIA','TP_ENSINO'], inplace=True, axis=1)
df_train[numerical_vars] = df_train[numerical_vars].fillna(0)
df_test[numerical_vars] = df_test[numerical_vars].fillna(0)
df_train_clean = pd.DataFrame(index=df_train.index)
df_test_clean = pd.DataFrame(index=df_test.index)
df_train_clean['NU_INSCRICAO'] = df_raw_train['NU_INSCRICAO']
df_test_clean['NU_INSCRICAO'] = df_raw_test['NU_INSCRICAO']
def create_encoder(column, prefix):
train_column_df = pd.get_dummies(df_train[column])
test_column_df = pd.get_dummies(df_test[column])
train_name_columns = df_train[column].sort_values().unique()
train_name_columns_co = [str(prefix) + str(train_name_column) for train_name_column in train_name_columns]
test_name_columns = df_test[column].sort_values().unique()
test_name_columns_co = [str(prefix) + str(test_name_column) for test_name_column in test_name_columns]
train_column_df.columns=train_name_columns_co
test_column_df.columns=test_name_columns_co
global df_train_clean
global df_test_clean
df_train_clean = pd.concat([df_train_clean, train_column_df ], axis=1)
df_test_clean = pd.concat([df_test_clean, test_column_df ], axis=1)
categorical_vars = {'CO_UF_RESIDENCIA' : 'co_uf_', 'TP_SEXO' : 'sexo_', 'TP_COR_RACA': 'raca_', 'TP_ST_CONCLUSAO': 'tp_st_con_',
'TP_ANO_CONCLUIU': 'tp_ano_con_', 'TP_ESCOLA': 'tp_esc_','TP_PRESENCA_CN': 'tp_pres_cn',
'TP_PRESENCA_CH': 'tp_pres_ch', 'TP_PRESENCA_LC': 'tp_pres_lc', 'TP_LINGUA': 'tp_ling_',
'Q001': 'q001_', 'Q002': 'q002_', 'Q006': 'q006_', 'Q024': 'q024_',
'Q025': 'q025_', 'Q026': 'q026_', 'Q047': 'q047_'}
for column, prefix in categorical_vars.items():
create_encoder(column, prefix)
numerical_vars = ['NU_NOTA_CN', 'NU_NOTA_CH', 'NU_NOTA_LC','NU_NOTA_COMP1', 'NU_NOTA_COMP2', 'NU_NOTA_COMP3',
'NU_NOTA_COMP4','NU_NOTA_COMP5', 'NU_NOTA_REDACAO']
df_train_clean = pd.concat([df_train_clean, df_train[numerical_vars]], axis=1)
df_test_clean = pd.concat([df_test_clean, df_test[numerical_vars]], axis=1)
X_train = df_train_clean.loc[:,'co_uf_11':]
y_train = df_train['IN_TREINEIRO']
X_test = df_test_clean.loc[:,'co_uf_11':]
X_train.shape, y_train.shape, X_test.shape
X_train_comp_X_test = X_train[X_test.columns]
X_train_comp_X_test.shape, y_train.shape, X_test.shape
df_train['IN_TREINEIRO'].value_counts()
df_result_insc = pd.DataFrame(df_test_clean['NU_INSCRICAO'])
```
## Aplicando SMOTE
```
#smote = SMOTE(ratio="minority")
smote = SMOTE(sampling_strategy="minority")
X_smote, y_smote = smote.fit_resample(X_train_comp_X_test, y_train)
```
## Logistic Regression
```
regressor = LogisticRegression()
regressor.fit(X_smote, y_smote)
y_pred = regressor.predict(X_test)
resultado = pd.concat([df_result_insc, pd.DataFrame(np.round(y_pred,3))], axis=1)
resultado.reset_index(inplace=True, drop=True)
resultado.columns=['NU_INSCRICAO', 'IN_TREINEIRO']
resultado.to_csv("answer.csv", index=False) # Score obtido: 96.083151
```
## Decision Tree
```
decision_tree = DecisionTreeClassifier(max_depth=2)
decision_tree_fitted = decision_tree.fit(X_smote, y_smote)
y_pred = decision_tree.predict(X_test)
resultado = pd.concat([df_result_insc, pd.DataFrame(np.round(y_pred,3))], axis=1)
resultado.reset_index(inplace=True, drop=True)
resultado.columns=['NU_INSCRICAO', 'IN_TREINEIRO']
resultado.to_csv("answer.csv", index=False) # Score 96.105033
```
## Random Forest
```
random_forest = RandomForestClassifier(n_estimators=500)
random_forest.fit(X_smote, y_smote)
y_pred = random_forest.predict(X_test)
resultado = pd.concat([df_result_insc, pd.DataFrame(np.round(y_pred,3))], axis=1)
resultado.reset_index(inplace=True, drop=True)
resultado.columns=['NU_INSCRICAO', 'IN_TREINEIRO']
resultado.to_csv("answer.csv", index=False) # Score obtido: 96.630197
```
## Random Forest - parametrização
```
random_forest = RandomForestClassifier(n_estimators=1000, min_samples_leaf=1 ,random_state=0, class_weight="balanced", n_jobs=6)
random_forest.fit(X_smote, y_smote)
y_pred = random_forest.predict(X_test)
resultado = pd.concat([df_result_insc, pd.DataFrame(np.round(y_pred,3))], axis=1)
resultado.reset_index(inplace=True, drop=True)
resultado.columns=['NU_INSCRICAO', 'IN_TREINEIRO']
resultado.to_csv("answer.csv", index=False) # Score Obtido 96.586433
```
## Nearest Neighbors
```
knn = KNeighborsClassifier(n_neighbors=5)
knn.fit(X_smote, y_smote)
y_pred = knn.predict(X_test)
resultado = pd.concat([df_result_insc, pd.DataFrame(np.round(y_pred,3))], axis=1)
resultado.reset_index(inplace=True, drop=True)
resultado.columns=['NU_INSCRICAO', 'IN_TREINEIRO']
resultado.to_csv("answer.csv", index=False) # Score Obtido 64.135667
```
|
github_jupyter
|
import pandas as pd
import numpy as np
import warnings
from sklearn.preprocessing import OneHotEncoder
from sklearn.linear_model import LogisticRegression
from imblearn.over_sampling import SMOTE
from sklearn.ensemble import BaggingClassifier, RandomForestClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.tree import DecisionTreeClassifier, plot_tree
warnings.filterwarnings('ignore')
pd.set_option('display.max_columns', 300)
pd.set_option('display.max_rows', 300)
file_train = "train.csv"
file_test = "test.csv"
df_raw_train = pd.read_csv(file_train, index_col=False)
df_raw_test = pd.read_csv(file_test, index_col=False)
df_raw_train.shape, df_raw_test.shape
columns_used=['NU_INSCRICAO','CO_UF_RESIDENCIA', 'SG_UF_RESIDENCIA', 'NU_IDADE', 'TP_SEXO', 'TP_COR_RACA',
'TP_NACIONALIDADE','TP_ST_CONCLUSAO','TP_ANO_CONCLUIU', 'TP_ESCOLA', 'TP_ENSINO',
'TP_PRESENCA_CN', 'TP_PRESENCA_CH', 'TP_PRESENCA_LC','NU_NOTA_CN', 'NU_NOTA_CH',
'NU_NOTA_LC','TP_LINGUA','TP_STATUS_REDACAO', 'NU_NOTA_COMP1', 'NU_NOTA_COMP2',
'NU_NOTA_COMP3','NU_NOTA_COMP4', 'NU_NOTA_COMP5', 'NU_NOTA_REDACAO','Q001', 'Q002',
'Q006', 'Q024', 'Q025', 'Q026', 'Q027', 'Q047', 'IN_TREINEIRO']
numerical_vars = ['NU_NOTA_CN', 'NU_NOTA_CH', 'NU_NOTA_LC','NU_NOTA_COMP1', 'NU_NOTA_COMP2', 'NU_NOTA_COMP3',
'NU_NOTA_COMP4','NU_NOTA_COMP5', 'NU_NOTA_REDACAO']
target = ['IN_TREINEIRO']
df_train=df_raw_train[columns_used]
df_train.drop(['SG_UF_RESIDENCIA','TP_ENSINO'], inplace=True, axis=1)
df_test=df_raw_test[columns_used[:-1]]
df_test.drop(['SG_UF_RESIDENCIA','TP_ENSINO'], inplace=True, axis=1)
df_train[numerical_vars] = df_train[numerical_vars].fillna(0)
df_test[numerical_vars] = df_test[numerical_vars].fillna(0)
df_train_clean = pd.DataFrame(index=df_train.index)
df_test_clean = pd.DataFrame(index=df_test.index)
df_train_clean['NU_INSCRICAO'] = df_raw_train['NU_INSCRICAO']
df_test_clean['NU_INSCRICAO'] = df_raw_test['NU_INSCRICAO']
def create_encoder(column, prefix):
train_column_df = pd.get_dummies(df_train[column])
test_column_df = pd.get_dummies(df_test[column])
train_name_columns = df_train[column].sort_values().unique()
train_name_columns_co = [str(prefix) + str(train_name_column) for train_name_column in train_name_columns]
test_name_columns = df_test[column].sort_values().unique()
test_name_columns_co = [str(prefix) + str(test_name_column) for test_name_column in test_name_columns]
train_column_df.columns=train_name_columns_co
test_column_df.columns=test_name_columns_co
global df_train_clean
global df_test_clean
df_train_clean = pd.concat([df_train_clean, train_column_df ], axis=1)
df_test_clean = pd.concat([df_test_clean, test_column_df ], axis=1)
categorical_vars = {'CO_UF_RESIDENCIA' : 'co_uf_', 'TP_SEXO' : 'sexo_', 'TP_COR_RACA': 'raca_', 'TP_ST_CONCLUSAO': 'tp_st_con_',
'TP_ANO_CONCLUIU': 'tp_ano_con_', 'TP_ESCOLA': 'tp_esc_','TP_PRESENCA_CN': 'tp_pres_cn',
'TP_PRESENCA_CH': 'tp_pres_ch', 'TP_PRESENCA_LC': 'tp_pres_lc', 'TP_LINGUA': 'tp_ling_',
'Q001': 'q001_', 'Q002': 'q002_', 'Q006': 'q006_', 'Q024': 'q024_',
'Q025': 'q025_', 'Q026': 'q026_', 'Q047': 'q047_'}
for column, prefix in categorical_vars.items():
create_encoder(column, prefix)
numerical_vars = ['NU_NOTA_CN', 'NU_NOTA_CH', 'NU_NOTA_LC','NU_NOTA_COMP1', 'NU_NOTA_COMP2', 'NU_NOTA_COMP3',
'NU_NOTA_COMP4','NU_NOTA_COMP5', 'NU_NOTA_REDACAO']
df_train_clean = pd.concat([df_train_clean, df_train[numerical_vars]], axis=1)
df_test_clean = pd.concat([df_test_clean, df_test[numerical_vars]], axis=1)
X_train = df_train_clean.loc[:,'co_uf_11':]
y_train = df_train['IN_TREINEIRO']
X_test = df_test_clean.loc[:,'co_uf_11':]
X_train.shape, y_train.shape, X_test.shape
X_train_comp_X_test = X_train[X_test.columns]
X_train_comp_X_test.shape, y_train.shape, X_test.shape
df_train['IN_TREINEIRO'].value_counts()
df_result_insc = pd.DataFrame(df_test_clean['NU_INSCRICAO'])
#smote = SMOTE(ratio="minority")
smote = SMOTE(sampling_strategy="minority")
X_smote, y_smote = smote.fit_resample(X_train_comp_X_test, y_train)
regressor = LogisticRegression()
regressor.fit(X_smote, y_smote)
y_pred = regressor.predict(X_test)
resultado = pd.concat([df_result_insc, pd.DataFrame(np.round(y_pred,3))], axis=1)
resultado.reset_index(inplace=True, drop=True)
resultado.columns=['NU_INSCRICAO', 'IN_TREINEIRO']
resultado.to_csv("answer.csv", index=False) # Score obtido: 96.083151
decision_tree = DecisionTreeClassifier(max_depth=2)
decision_tree_fitted = decision_tree.fit(X_smote, y_smote)
y_pred = decision_tree.predict(X_test)
resultado = pd.concat([df_result_insc, pd.DataFrame(np.round(y_pred,3))], axis=1)
resultado.reset_index(inplace=True, drop=True)
resultado.columns=['NU_INSCRICAO', 'IN_TREINEIRO']
resultado.to_csv("answer.csv", index=False) # Score 96.105033
random_forest = RandomForestClassifier(n_estimators=500)
random_forest.fit(X_smote, y_smote)
y_pred = random_forest.predict(X_test)
resultado = pd.concat([df_result_insc, pd.DataFrame(np.round(y_pred,3))], axis=1)
resultado.reset_index(inplace=True, drop=True)
resultado.columns=['NU_INSCRICAO', 'IN_TREINEIRO']
resultado.to_csv("answer.csv", index=False) # Score obtido: 96.630197
random_forest = RandomForestClassifier(n_estimators=1000, min_samples_leaf=1 ,random_state=0, class_weight="balanced", n_jobs=6)
random_forest.fit(X_smote, y_smote)
y_pred = random_forest.predict(X_test)
resultado = pd.concat([df_result_insc, pd.DataFrame(np.round(y_pred,3))], axis=1)
resultado.reset_index(inplace=True, drop=True)
resultado.columns=['NU_INSCRICAO', 'IN_TREINEIRO']
resultado.to_csv("answer.csv", index=False) # Score Obtido 96.586433
knn = KNeighborsClassifier(n_neighbors=5)
knn.fit(X_smote, y_smote)
y_pred = knn.predict(X_test)
resultado = pd.concat([df_result_insc, pd.DataFrame(np.round(y_pred,3))], axis=1)
resultado.reset_index(inplace=True, drop=True)
resultado.columns=['NU_INSCRICAO', 'IN_TREINEIRO']
resultado.to_csv("answer.csv", index=False) # Score Obtido 64.135667
| 0.344113 | 0.696062 |
```
from pathlib import PurePath
from palettable.colorbrewer.diverging import RdBu_5
from palettable.colorbrewer.sequential import Reds_5
from raster_compare.base import PixelClassifier, RasterFile
from raster_compare.plots import PlotBase
from plot_helpers import *
RED_BLUE = RdBu_5.mpl_colormap
REDS = Reds_5.mpl_colormap
ortho_image = plt.imread(
# str(HOME_DIR / 'ERW_20180524_Agisoft_rgb_5m_ortho_2kx2k.tif')
str(HOME_DIR / 'ERW_20180524_Agisoft_rgb_5m_ortho_2kx2k_tree.tif')
)
hillshade = RasterFile(HILLSHADE, band_number=1).band_values()
def style_area_axes(axes):
set_axes_style(axes)
axes.set_xticks([])
axes.set_yticks([])
casi_data = PixelClassifier(
# HOME_DIR / 'ERW_20180524/CASI/CASI_2018_05_24_weekly_mosaic_classified_lidargrid_2kx2k_1m.tif',
HOME_DIR / 'ERW_20180524/CASI/CASI_2018_05_24_weekly_mosaic_classified_lidargrid_2kx2k_1m_trees.tif',
HOME_DIR / 'cloud_mask/cloud_mask_lidargrid_2kx2k_1m.vrt',
# HOME_DIR / 'ERW_20180524/Lidar/ERW_20180524_Lidar_fr__lr_diff_2kx2k_1m.tif'
HOME_DIR / 'ERW_20180524/Lidar/ERW_20180524_Lidar_fr__lr_diff_2kx2k_1m_tree.tif'
)
lidar_data = RasterFile(
# HOME_DIR / 'ERW_snow_depth_lidar/ERW_snow_depth_lidar_lr_2kx2k_1m.tif',
HOME_DIR / 'ERW_snow_depth_lidar/ERW_snow_depth_lidar_lr_2kx2k_1m_tree.vrt',
band_number=1
)
band_values_lidar = lidar_data.band_values()
band_values_lidar_mask = band_values_lidar.mask.copy()
sfm_data = RasterFile(
# HOME_DIR / 'ERW_snow_depth_sfm/ERW_snow_depth_sfm_lr_2kx2k_1m.tif',
HOME_DIR / 'ERW_snow_depth_sfm/ERW_snow_depth_sfm_lr_2kx2k_1m_tree.vrt',
band_number=1
)
band_values_sfm = sfm_data.band_values()
band_values_sfm_mask = band_values_sfm.mask.copy()
sd_diff_data = RasterFile(
# HOME_DIR / 'ERW_snow_depth_diff/ERW_snow_depth_diff_2kx2k_1m.tif',
HOME_DIR / 'ERW_snow_depth_diff/ERW_snow_depth_diff_2kx2k_1m_tree.vrt',
band_number=1
)
band_values_sd_diff = sd_diff_data.band_values()
band_values_sd_diff_mask = band_values_sd_diff.mask.copy()
band_values_lidar.mask = casi_data.snow_surfaces(band_values_lidar_mask)
band_values_sfm.mask = casi_data.snow_surfaces(band_values_sfm_mask)
band_values_sd_diff.mask = casi_data.snow_surfaces(band_values_sd_diff_mask)
ticks = [-1.5, -1, -0.5, 0, 0.5, 1, 1.5]
tick_labels = ['< -1.5', '-1', '-0.5', '0', '0.5', '1', '> 1.5']
bins = np.arange(-1.50, 1.50 + 0.05, step=0.05)
bounds = dict(
norm=colors.BoundaryNorm(
boundaries=bins, ncolors=RED_BLUE.N,
)
)
def add_hillshade(ax):
ax.imshow(hillshade, cmap='gray', clim=(1, 255), alpha=0.5)
figure = plt.figure(
figsize=(12, 8),
dpi=150,
constrained_layout=False,
)
grid_spec = figure.add_gridspec(
nrows=2,
ncols=3,
wspace=0.0
)
ax1 = figure.add_subplot(grid_spec[0, 0])
add_hillshade(ax1)
ax1.imshow(
band_values_lidar,
cmap=RED_BLUE,
**bounds
)
ax1.set_title('Lidar Snow Depth')
ax2 = figure.add_subplot(grid_spec[0, 1])
add_hillshade(ax2)
ax2.imshow(
band_values_sfm,
cmap=RED_BLUE,
**bounds
)
ax2.set_title('SfM Snow Depth')
ax3 = figure.add_subplot(grid_spec[0, -1])
ax3.imshow(ortho_image)
ax3.add_artist(
ScaleBar(5.0, location='lower left')
)
ax4 = figure.add_subplot(grid_spec[1, :-1])
data = prepare_data([
band_values_lidar, band_values_sfm
])
hist_bins = np.arange(
np.nanmin(data[0]),
np.nanmax(data[0]),
0.01
)
hist_lidar = ax4.hist(data[0], bins=hist_bins, color='dodgerblue', label='Lidar')
hist_sfm = ax4.hist(data[1], bins=hist_bins, color='brown', alpha=0.6, label='SfM')
vline_opts = dict(color='darkorange', linewidth=2)
label_y = max(hist_lidar[0].max(), hist_sfm[0].max())
# ax4.axvline(x=data[0].mean(), **vline_opts, ymax=0.82, label='Mean')
# ax4.annotate('Lidar', (data[0].mean() + .05, label_y * 0.78))
# ax4.axvline(x=data[1].mean(), **vline_opts, ymax=0.92)
# ax4.annotate('SfM', (data[1].mean() + .05, label_y * 0.9))
ax4.axvline(x=data[0].mean(), **vline_opts, ymax=0.95, label='Mean')
ax4.annotate('Lidar', (data[0].mean() + .05, label_y * 0.92))
ax4.axvline(x=data[1].mean(), **vline_opts, ymax=0.7)
ax4.annotate('SfM', (data[1].mean() - .25, label_y * 0.67))
ax4.axvline(x=0, color='black', linewidth=1)
ax4.ticklabel_format(style='sci', axis='y', scilimits=(4, 4))
ax4.yaxis.get_offset_text().set_visible(False)
ax4.set_xlabel("Snow Depth (m)")
ax4.set_ylabel("Count $(10^4)$")
ax4.set_xlim(-1.5, 3.5)
ax4.set_xticks([-1, -.5, 0, .5, 1, 2, 3])
ax4.legend()
ax4.set_title("Vegetated Area Distribution")
# ax4.set_title("High Elevation Distribution")
ax5 = figure.add_subplot(grid_spec[1, -1])
add_hillshade(ax5)
diff_plot = ax5.imshow(
band_values_sd_diff,
cmap=RED_BLUE,
**bounds
)
ax5.set_title('Snow Depth Difference')
cbar = PlotBase.insert_colorbar(
ax5, diff_plot,
'Depth Difference (m)',
ticks=ticks,
)
cbar.ax.set_yticklabels(tick_labels)
[style_area_axes(ax) for ax in [ax1, ax2, ax3, ax5]];
```
## Show positive snow depth values
```
band_values_lidar.mask = (
casi_data.snow_surfaces(np.ma.masked_less(band_values_lidar, 0).mask)
)
band_values_sfm.mask = (
casi_data.snow_surfaces(np.ma.masked_less(band_values_sfm, 0).mask)
)
band_values_sd_diff.mask = (
casi_data.snow_surfaces(np.ma.masked_less(band_values_sd_diff, 0).mask)
)
figure = plt.figure(
figsize=(12, 8),
dpi=150,
constrained_layout=False,
)
grid_spec = figure.add_gridspec(
nrows=2,
ncols=3,
wspace=0.0
)
ax1 = figure.add_subplot(grid_spec[0, 0])
add_hillshade(ax1)
ax1.imshow(
band_values_lidar,
cmap=RED_BLUE,
**bounds
)
ax1.set_title('Lidar Snow Depth')
ax2 = figure.add_subplot(grid_spec[0, 1])
add_hillshade(ax2)
ax2.imshow(
band_values_sfm,
cmap=RED_BLUE,
**bounds,
)
ax2.set_title('SfM Snow Depth')
ax3 = figure.add_subplot(grid_spec[0, -1])
ax3.imshow(ortho_image)
ax3.add_artist(
ScaleBar(5.0, location='upper left')
)
ax4 = figure.add_subplot(grid_spec[1, :-1])
hist_bins = np.arange(
np.nanmin(data[0]),
np.nanmax(data[0]),
0.01
)
ax4.hist(data[0], bins=hist_bins, color='dodgerblue', label='Lidar')
ax4.hist(data[1], bins=hist_bins, color='brown', alpha=0.6, label='SfM')
ax4.axvline(x=0, color='black', linewidth=1)
ax4.axvspan(-1.5, 0, alpha=0.5, color='grey')
ax4.ticklabel_format(style='sci', axis='y', scilimits=(4, 4))
ax4.yaxis.get_offset_text().set_visible(False)
ax4.set_xlabel("Snow Depth (m)")
ax4.set_ylabel("Count $(10^4)$")
ax4.set_xlim(-1.5, 3.5)
ax4.set_xticks([-1, -.5, 0, .5, 1, 2, 3])
ax4.legend()
ax4.set_title("High Elevation Distribution")
ax5 = figure.add_subplot(grid_spec[1, -1])
add_hillshade(ax5)
diff_plot = ax5.imshow(
band_values_sd_diff,
cmap=RED_BLUE,
**bounds
)
ax5.set_title('Snow Depth Difference')
cbar = PlotBase.insert_colorbar(
ax5, diff_plot,
'Depth Difference (m)',
ticks=ticks,
)
cbar.ax.set_yticklabels(tick_labels)
[style_area_axes(ax) for ax in [ax1, ax2, ax3, ax5]];
```
## Show areas of negative snow depth
```
color_map = LinearSegmentedColormap.from_list(
'snow_pixels',
['none', 'firebrick'],
N=2
)
figure = plt.figure(
figsize=(12, 8),
dpi=150,
constrained_layout=False,
)
grid_spec = figure.add_gridspec(
nrows=2,
ncols=3,
wspace=0.0
)
ax1 = figure.add_subplot(grid_spec[0, 0])
add_hillshade(ax1)
ax1.imshow(
band_values_lidar < 0,
cmap=color_map,
)
ax1.set_title('Lidar')
ax2 = figure.add_subplot(grid_spec[0, 1])
add_hillshade(ax2)
ax2.imshow(
band_values_sfm < 0,
cmap=color_map,
)
ax2.set_title('SfM')
ax3 = figure.add_subplot(grid_spec[0, -1])
ax3.imshow(ortho_image)
ax3.add_artist(
ScaleBar(5.0, location='upper left')
)
ax4 = figure.add_subplot(grid_spec[1, :-1])
ax4.hist(data[0], bins=hist_bins, color='dodgerblue', label='Lidar')
ax4.hist(data[1], bins=hist_bins, color='brown', alpha=0.6, label='SfM')
ax4.axvline(x=0, color='black', linewidth=1)
ax4.axvspan(0, 3.5, alpha=0.5, color='grey')
ax4.ticklabel_format(style='sci', axis='y', scilimits=(4, 4))
ax4.yaxis.get_offset_text().set_visible(False)
ax4.set_xlabel("Snow Depth (m)")
ax4.set_ylabel("Count $(10^4)$")
ax4.set_xlim(-1.5, 3.5)
ax4.set_xticks([-1, -.5, 0, .5, 1, 2, 3])
ax4.legend()
ax4.set_title("High Elevation Distribution")
[style_area_axes(ax) for ax in [ax1, ax2, ax3]];
color_map = LinearSegmentedColormap.from_list(
'snow_pixels',
['none', 'mediumblue'],
N=2
)
figure = plt.figure(
figsize=(12, 8),
dpi=150,
constrained_layout=False,
)
grid_spec = figure.add_gridspec(
nrows=2,
ncols=3,
wspace=0.0
)
ax1 = figure.add_subplot(grid_spec[0, 0])
add_hillshade(ax1)
ax1.imshow(
band_values_lidar > .75,
cmap=color_map,
alpha=0.9,
)
ax1.set_title('Lidar Snow Depth')
ax2 = figure.add_subplot(grid_spec[0, 1])
add_hillshade(ax2)
ax2.imshow(
band_values_sfm > 0.75,
cmap=color_map,
)
ax2.set_title('SfM Snow Depth')
ax3 = figure.add_subplot(grid_spec[0, -1])
ax3.imshow(ortho_image)
ax3.add_artist(
ScaleBar(5.0, location='upper left')
)
ax4 = figure.add_subplot(grid_spec[1, :-1])
hist_bins = np.arange(
np.nanmin(data[0]),
np.nanmax(data[0]),
0.01
)
ax4.hist(data[0], bins=hist_bins, color='dodgerblue', label='Lidar')
ax4.hist(data[1], bins=hist_bins, color='brown', alpha=0.6, label='SfM')
ax4.axvline(x=0.75, color='black', linewidth=1)
ax4.ticklabel_format(style='sci', axis='y', scilimits=(4, 4))
ax4.yaxis.get_offset_text().set_visible(False)
ax4.set_xlabel("Snow Depth (m)")
ax4.set_ylabel("Count $(10^4)$")
ax4.set_xlim(-1.5, 3.5)
ax4.set_xticks([-1, -.5, 0, .5, 1, 2, 3])
ax4.legend()
[style_area_axes(ax) for ax in [ax1, ax2, ax3]];
```
|
github_jupyter
|
from pathlib import PurePath
from palettable.colorbrewer.diverging import RdBu_5
from palettable.colorbrewer.sequential import Reds_5
from raster_compare.base import PixelClassifier, RasterFile
from raster_compare.plots import PlotBase
from plot_helpers import *
RED_BLUE = RdBu_5.mpl_colormap
REDS = Reds_5.mpl_colormap
ortho_image = plt.imread(
# str(HOME_DIR / 'ERW_20180524_Agisoft_rgb_5m_ortho_2kx2k.tif')
str(HOME_DIR / 'ERW_20180524_Agisoft_rgb_5m_ortho_2kx2k_tree.tif')
)
hillshade = RasterFile(HILLSHADE, band_number=1).band_values()
def style_area_axes(axes):
set_axes_style(axes)
axes.set_xticks([])
axes.set_yticks([])
casi_data = PixelClassifier(
# HOME_DIR / 'ERW_20180524/CASI/CASI_2018_05_24_weekly_mosaic_classified_lidargrid_2kx2k_1m.tif',
HOME_DIR / 'ERW_20180524/CASI/CASI_2018_05_24_weekly_mosaic_classified_lidargrid_2kx2k_1m_trees.tif',
HOME_DIR / 'cloud_mask/cloud_mask_lidargrid_2kx2k_1m.vrt',
# HOME_DIR / 'ERW_20180524/Lidar/ERW_20180524_Lidar_fr__lr_diff_2kx2k_1m.tif'
HOME_DIR / 'ERW_20180524/Lidar/ERW_20180524_Lidar_fr__lr_diff_2kx2k_1m_tree.tif'
)
lidar_data = RasterFile(
# HOME_DIR / 'ERW_snow_depth_lidar/ERW_snow_depth_lidar_lr_2kx2k_1m.tif',
HOME_DIR / 'ERW_snow_depth_lidar/ERW_snow_depth_lidar_lr_2kx2k_1m_tree.vrt',
band_number=1
)
band_values_lidar = lidar_data.band_values()
band_values_lidar_mask = band_values_lidar.mask.copy()
sfm_data = RasterFile(
# HOME_DIR / 'ERW_snow_depth_sfm/ERW_snow_depth_sfm_lr_2kx2k_1m.tif',
HOME_DIR / 'ERW_snow_depth_sfm/ERW_snow_depth_sfm_lr_2kx2k_1m_tree.vrt',
band_number=1
)
band_values_sfm = sfm_data.band_values()
band_values_sfm_mask = band_values_sfm.mask.copy()
sd_diff_data = RasterFile(
# HOME_DIR / 'ERW_snow_depth_diff/ERW_snow_depth_diff_2kx2k_1m.tif',
HOME_DIR / 'ERW_snow_depth_diff/ERW_snow_depth_diff_2kx2k_1m_tree.vrt',
band_number=1
)
band_values_sd_diff = sd_diff_data.band_values()
band_values_sd_diff_mask = band_values_sd_diff.mask.copy()
band_values_lidar.mask = casi_data.snow_surfaces(band_values_lidar_mask)
band_values_sfm.mask = casi_data.snow_surfaces(band_values_sfm_mask)
band_values_sd_diff.mask = casi_data.snow_surfaces(band_values_sd_diff_mask)
ticks = [-1.5, -1, -0.5, 0, 0.5, 1, 1.5]
tick_labels = ['< -1.5', '-1', '-0.5', '0', '0.5', '1', '> 1.5']
bins = np.arange(-1.50, 1.50 + 0.05, step=0.05)
bounds = dict(
norm=colors.BoundaryNorm(
boundaries=bins, ncolors=RED_BLUE.N,
)
)
def add_hillshade(ax):
ax.imshow(hillshade, cmap='gray', clim=(1, 255), alpha=0.5)
figure = plt.figure(
figsize=(12, 8),
dpi=150,
constrained_layout=False,
)
grid_spec = figure.add_gridspec(
nrows=2,
ncols=3,
wspace=0.0
)
ax1 = figure.add_subplot(grid_spec[0, 0])
add_hillshade(ax1)
ax1.imshow(
band_values_lidar,
cmap=RED_BLUE,
**bounds
)
ax1.set_title('Lidar Snow Depth')
ax2 = figure.add_subplot(grid_spec[0, 1])
add_hillshade(ax2)
ax2.imshow(
band_values_sfm,
cmap=RED_BLUE,
**bounds
)
ax2.set_title('SfM Snow Depth')
ax3 = figure.add_subplot(grid_spec[0, -1])
ax3.imshow(ortho_image)
ax3.add_artist(
ScaleBar(5.0, location='lower left')
)
ax4 = figure.add_subplot(grid_spec[1, :-1])
data = prepare_data([
band_values_lidar, band_values_sfm
])
hist_bins = np.arange(
np.nanmin(data[0]),
np.nanmax(data[0]),
0.01
)
hist_lidar = ax4.hist(data[0], bins=hist_bins, color='dodgerblue', label='Lidar')
hist_sfm = ax4.hist(data[1], bins=hist_bins, color='brown', alpha=0.6, label='SfM')
vline_opts = dict(color='darkorange', linewidth=2)
label_y = max(hist_lidar[0].max(), hist_sfm[0].max())
# ax4.axvline(x=data[0].mean(), **vline_opts, ymax=0.82, label='Mean')
# ax4.annotate('Lidar', (data[0].mean() + .05, label_y * 0.78))
# ax4.axvline(x=data[1].mean(), **vline_opts, ymax=0.92)
# ax4.annotate('SfM', (data[1].mean() + .05, label_y * 0.9))
ax4.axvline(x=data[0].mean(), **vline_opts, ymax=0.95, label='Mean')
ax4.annotate('Lidar', (data[0].mean() + .05, label_y * 0.92))
ax4.axvline(x=data[1].mean(), **vline_opts, ymax=0.7)
ax4.annotate('SfM', (data[1].mean() - .25, label_y * 0.67))
ax4.axvline(x=0, color='black', linewidth=1)
ax4.ticklabel_format(style='sci', axis='y', scilimits=(4, 4))
ax4.yaxis.get_offset_text().set_visible(False)
ax4.set_xlabel("Snow Depth (m)")
ax4.set_ylabel("Count $(10^4)$")
ax4.set_xlim(-1.5, 3.5)
ax4.set_xticks([-1, -.5, 0, .5, 1, 2, 3])
ax4.legend()
ax4.set_title("Vegetated Area Distribution")
# ax4.set_title("High Elevation Distribution")
ax5 = figure.add_subplot(grid_spec[1, -1])
add_hillshade(ax5)
diff_plot = ax5.imshow(
band_values_sd_diff,
cmap=RED_BLUE,
**bounds
)
ax5.set_title('Snow Depth Difference')
cbar = PlotBase.insert_colorbar(
ax5, diff_plot,
'Depth Difference (m)',
ticks=ticks,
)
cbar.ax.set_yticklabels(tick_labels)
[style_area_axes(ax) for ax in [ax1, ax2, ax3, ax5]];
band_values_lidar.mask = (
casi_data.snow_surfaces(np.ma.masked_less(band_values_lidar, 0).mask)
)
band_values_sfm.mask = (
casi_data.snow_surfaces(np.ma.masked_less(band_values_sfm, 0).mask)
)
band_values_sd_diff.mask = (
casi_data.snow_surfaces(np.ma.masked_less(band_values_sd_diff, 0).mask)
)
figure = plt.figure(
figsize=(12, 8),
dpi=150,
constrained_layout=False,
)
grid_spec = figure.add_gridspec(
nrows=2,
ncols=3,
wspace=0.0
)
ax1 = figure.add_subplot(grid_spec[0, 0])
add_hillshade(ax1)
ax1.imshow(
band_values_lidar,
cmap=RED_BLUE,
**bounds
)
ax1.set_title('Lidar Snow Depth')
ax2 = figure.add_subplot(grid_spec[0, 1])
add_hillshade(ax2)
ax2.imshow(
band_values_sfm,
cmap=RED_BLUE,
**bounds,
)
ax2.set_title('SfM Snow Depth')
ax3 = figure.add_subplot(grid_spec[0, -1])
ax3.imshow(ortho_image)
ax3.add_artist(
ScaleBar(5.0, location='upper left')
)
ax4 = figure.add_subplot(grid_spec[1, :-1])
hist_bins = np.arange(
np.nanmin(data[0]),
np.nanmax(data[0]),
0.01
)
ax4.hist(data[0], bins=hist_bins, color='dodgerblue', label='Lidar')
ax4.hist(data[1], bins=hist_bins, color='brown', alpha=0.6, label='SfM')
ax4.axvline(x=0, color='black', linewidth=1)
ax4.axvspan(-1.5, 0, alpha=0.5, color='grey')
ax4.ticklabel_format(style='sci', axis='y', scilimits=(4, 4))
ax4.yaxis.get_offset_text().set_visible(False)
ax4.set_xlabel("Snow Depth (m)")
ax4.set_ylabel("Count $(10^4)$")
ax4.set_xlim(-1.5, 3.5)
ax4.set_xticks([-1, -.5, 0, .5, 1, 2, 3])
ax4.legend()
ax4.set_title("High Elevation Distribution")
ax5 = figure.add_subplot(grid_spec[1, -1])
add_hillshade(ax5)
diff_plot = ax5.imshow(
band_values_sd_diff,
cmap=RED_BLUE,
**bounds
)
ax5.set_title('Snow Depth Difference')
cbar = PlotBase.insert_colorbar(
ax5, diff_plot,
'Depth Difference (m)',
ticks=ticks,
)
cbar.ax.set_yticklabels(tick_labels)
[style_area_axes(ax) for ax in [ax1, ax2, ax3, ax5]];
color_map = LinearSegmentedColormap.from_list(
'snow_pixels',
['none', 'firebrick'],
N=2
)
figure = plt.figure(
figsize=(12, 8),
dpi=150,
constrained_layout=False,
)
grid_spec = figure.add_gridspec(
nrows=2,
ncols=3,
wspace=0.0
)
ax1 = figure.add_subplot(grid_spec[0, 0])
add_hillshade(ax1)
ax1.imshow(
band_values_lidar < 0,
cmap=color_map,
)
ax1.set_title('Lidar')
ax2 = figure.add_subplot(grid_spec[0, 1])
add_hillshade(ax2)
ax2.imshow(
band_values_sfm < 0,
cmap=color_map,
)
ax2.set_title('SfM')
ax3 = figure.add_subplot(grid_spec[0, -1])
ax3.imshow(ortho_image)
ax3.add_artist(
ScaleBar(5.0, location='upper left')
)
ax4 = figure.add_subplot(grid_spec[1, :-1])
ax4.hist(data[0], bins=hist_bins, color='dodgerblue', label='Lidar')
ax4.hist(data[1], bins=hist_bins, color='brown', alpha=0.6, label='SfM')
ax4.axvline(x=0, color='black', linewidth=1)
ax4.axvspan(0, 3.5, alpha=0.5, color='grey')
ax4.ticklabel_format(style='sci', axis='y', scilimits=(4, 4))
ax4.yaxis.get_offset_text().set_visible(False)
ax4.set_xlabel("Snow Depth (m)")
ax4.set_ylabel("Count $(10^4)$")
ax4.set_xlim(-1.5, 3.5)
ax4.set_xticks([-1, -.5, 0, .5, 1, 2, 3])
ax4.legend()
ax4.set_title("High Elevation Distribution")
[style_area_axes(ax) for ax in [ax1, ax2, ax3]];
color_map = LinearSegmentedColormap.from_list(
'snow_pixels',
['none', 'mediumblue'],
N=2
)
figure = plt.figure(
figsize=(12, 8),
dpi=150,
constrained_layout=False,
)
grid_spec = figure.add_gridspec(
nrows=2,
ncols=3,
wspace=0.0
)
ax1 = figure.add_subplot(grid_spec[0, 0])
add_hillshade(ax1)
ax1.imshow(
band_values_lidar > .75,
cmap=color_map,
alpha=0.9,
)
ax1.set_title('Lidar Snow Depth')
ax2 = figure.add_subplot(grid_spec[0, 1])
add_hillshade(ax2)
ax2.imshow(
band_values_sfm > 0.75,
cmap=color_map,
)
ax2.set_title('SfM Snow Depth')
ax3 = figure.add_subplot(grid_spec[0, -1])
ax3.imshow(ortho_image)
ax3.add_artist(
ScaleBar(5.0, location='upper left')
)
ax4 = figure.add_subplot(grid_spec[1, :-1])
hist_bins = np.arange(
np.nanmin(data[0]),
np.nanmax(data[0]),
0.01
)
ax4.hist(data[0], bins=hist_bins, color='dodgerblue', label='Lidar')
ax4.hist(data[1], bins=hist_bins, color='brown', alpha=0.6, label='SfM')
ax4.axvline(x=0.75, color='black', linewidth=1)
ax4.ticklabel_format(style='sci', axis='y', scilimits=(4, 4))
ax4.yaxis.get_offset_text().set_visible(False)
ax4.set_xlabel("Snow Depth (m)")
ax4.set_ylabel("Count $(10^4)$")
ax4.set_xlim(-1.5, 3.5)
ax4.set_xticks([-1, -.5, 0, .5, 1, 2, 3])
ax4.legend()
[style_area_axes(ax) for ax in [ax1, ax2, ax3]];
| 0.458834 | 0.279837 |
# Automatic Model Tuning : Automatic training job early stopping
_**Using automatic training job early stopping to speed up the tuning of an end-to-end multiclass image classification task**_
---
## Important notes:
* Two hyperparameter tuning jobs will be created in this sample notebook. With current setting, each tuning job takes around 2 hours to complete and may cost you up to 16 USD depending on which region you are in.
* Due to cost consideration, the goal of this example is to show you how to use the new feature, not necessarily to achieve the best result.
* The built-in image classification algorithm on GPU instance will be used in this example.
* Different runs of this notebook may lead to different results, due to the non-deterministic nature of Automatic Model Tuning. But it is fair to assume some training jobs will be stopped by automatic early stopping.
---
## Contents
1. [Background](#Background)
1. [Set_up](#Set-up)
1. [Data_preparation](#Data-preparation)
1. [Set_up_hyperparameter_tuning_job](#Set-up-hyperparameter-tuning-job)
1. [Launch_hyperparameter_tuning_job](#Launch-hyperparameter-tuning-job)
1. [Launch_hyperparameter_tuning_job_with_automatic_early_stopping](#Launch-hyperparameter-tuning-job-with-automatic-early-stopping)
1. [Wrap_up](#Wrap-up)
---
## Background
Selecting the right hyperparameter values for machine learning model can be difficult. The right answer dependes on the algorithm and the data; Some algorithms have many tuneable hyperparameters; Some are very sensitive to the hyperparameter values selected; and yet most have a non-linear relationship between model fit and hyperparameter values. Amazon SageMaker Automatic Model Tuning helps by automating the hyperparameter tuning process.
Experienced data scientist often stop a training when it is not promising based on the first few validation metrics emitted during the training. This notebook will demonstrate how to use the automatic training job early stopping of Amazon SageMaker Automatic Model Tuning to speed up the tuning process with a simple switch.
---
## Set up
Let us start by specifying:
- The role that is used to give learning and hosting the access to the data. This will automatically be obtained from the role used to start the notebook.
- The S3 bucket that will be used for loading training data and saving model data.
- The Amazon SageMaker image classification docker image which need not to be changed.
```
import boto3
import sagemaker
from sagemaker import get_execution_role
from sagemaker.image_uris import retrieve
sess = sagemaker.Session()
role = get_execution_role()
bucket = sess.default_bucket()
training_image = retrieve("image-classification", boto3.Session().region_name, "1")
print(training_image)
```
## Data preparation
In this example, [caltech-256 dataset](http://www.vision.caltech.edu/Image_Datasets/Caltech256/) dataset will be used, which contains 30608 images of 256 objects.
```
import os
import urllib.request
import boto3
def download(url):
filename = url.split("/")[-1]
if not os.path.exists(filename):
urllib.request.urlretrieve(url, filename)
def upload_to_s3(channel, file):
s3 = boto3.resource("s3")
data = open(file, "rb")
key = channel + "/" + file
s3.Bucket(bucket).put_object(Key=key, Body=data)
s3_train_key = "image-classification-full-training/train"
s3_validation_key = "image-classification-full-training/validation"
download("http://data.mxnet.io/data/caltech-256/caltech-256-60-train.rec")
upload_to_s3(s3_train_key, "caltech-256-60-train.rec")
download("http://data.mxnet.io/data/caltech-256/caltech-256-60-val.rec")
upload_to_s3(s3_validation_key, "caltech-256-60-val.rec")
```
## Set up hyperparameter tuning job
For this example, three hyperparameters will be tuned: learning_rate, mini_batch_size and optimizer, which has the greatest impact on the objective metric. See [here](https://docs.aws.amazon.com/sagemaker/latest/dg/IC-tuning.html) for more detail and the full list of hyperparameters that can be tuned.
Before launching the tuning job, training jobs that the hyperparameter tuning job will launch need to be configured by defining an estimator that specifies the following information:
* The container image for the algorithm (image-classification).
* The s3 location for training and validation data.
* The type and number of instances to use for the training jobs.
* The output specification where the output can be stored after training.
The values of any hyperparameters that are not tuned in the tuning job (StaticHyperparameters):
* **num_layers**: The number of layers (depth) for the network. We use 18 in this samples but other values such as 50, 152 can be used.
* **image_shape**: The input image dimensions,'num_channels, height, width', for the network. It should be no larger than the actual image size. The number of channels should be same as in the actual image.
* **num_classes**: This is the number of output classes for the new dataset. For caltech, we use 257 because it has 256 object categories + 1 clutter class.
* **num_training_samples**: This is the total number of training samples. It is set to 15240 for caltech dataset with the current split.
* **epochs**: Number of training epochs. In this example we set it to only 10 to save the cost. If you would like to get higher accuracy the number of epochs can be increased.
* **top_k**: Report the top-k accuracy during training.
* **precision_dtype**: Training data type precision (default: float32). If set to 'float16', the training will be done in mixed_precision mode and will be faster than float32 mode.
* **augmentation_type**: 'crop'. Randomly crop the image and flip the image horizontally.
```
s3_train_data = "s3://{}/{}/".format(bucket, s3_train_key)
s3_validation_data = "s3://{}/{}/".format(bucket, s3_validation_key)
s3_output_key = "image-classification-full-training/output"
s3_output = "s3://{}/{}/".format(bucket, s3_output_key)
s3_input_train = sagemaker.TrainingInput(
s3_data=s3_train_data, content_type="application/x-recordio"
)
s3_input_validation = sagemaker.TrainingInput(
s3_data=s3_validation_data, content_type="application/x-recordio"
)
sess = sagemaker.Session()
imageclassification = sagemaker.estimator.Estimator(
training_image,
role,
instance_count=1,
instance_type="ml.p3.2xlarge",
output_path=s3_output,
sagemaker_session=sess,
)
imageclassification.set_hyperparameters(
num_layers=18,
image_shape="3,224,224",
num_classes=257,
epochs=10,
top_k="2",
num_training_samples=15420,
precision_dtype="float32",
augmentation_type="crop",
)
```
Next, the tuning job with the following configurations need to be specified:
* the hyperparameters that SageMaker Automatic Model Tuning will tune: learning_rate, mini_batch_size and optimizer
* the maximum number of training jobs it will run to optimize the objective metric: 20
* the number of parallel training jobs that will run in the tuning job: 2
* the objective metric that Automatic Model Tuning will use: validation:accuracy
```
from time import gmtime, strftime
from sagemaker.tuner import (
IntegerParameter,
CategoricalParameter,
ContinuousParameter,
HyperparameterTuner,
)
tuning_job_name = "imageclassif-job-{}".format(strftime("%d-%H-%M-%S", gmtime()))
hyperparameter_ranges = {
"learning_rate": ContinuousParameter(0.00001, 1.0),
"mini_batch_size": IntegerParameter(16, 64),
"optimizer": CategoricalParameter(["sgd", "adam", "rmsprop", "nag"]),
}
objective_metric_name = "validation:accuracy"
tuner = HyperparameterTuner(
imageclassification,
objective_metric_name,
hyperparameter_ranges,
objective_type="Maximize",
max_jobs=20,
max_parallel_jobs=2,
)
```
## Launch hyperparameter tuning job
Now we can launch a hyperparameter tuning job by calling fit in tuner. We will wait until the tuning finished, which may take around 2 hours.
```
tuner.fit(
{"train": s3_input_train, "validation": s3_input_validation},
job_name=tuning_job_name,
include_cls_metadata=False,
)
tuner.wait()
```
After the tuning finished, the top 5 performing hyperparameters can be listed below. One can analyse the results deeper by using [HPO_Analyze_TuningJob_Results.ipynb notebook](https://github.com/awslabs/amazon-sagemaker-examples/blob/master/hyperparameter_tuning/analyze_results/HPO_Analyze_TuningJob_Results.ipynb).
```
tuner_metrics = sagemaker.HyperparameterTuningJobAnalytics(tuning_job_name)
tuner_metrics.dataframe().sort_values(["FinalObjectiveValue"], ascending=False).head(5)
```
The total training time and training jobs status can be checked with the following script. Because automatic early stopping is by default off, all the training jobs should be completed normally.
```
total_time = tuner_metrics.dataframe()["TrainingElapsedTimeSeconds"].sum() / 3600
print("The total training time is {:.2f} hours".format(total_time))
tuner_metrics.dataframe()["TrainingJobStatus"].value_counts()
```
## Launch hyperparameter tuning job with automatic early stopping
Now we lunch the same tuning job with only one difference: setting **early_stopping_type**=**'Auto'** to enable automatic training job early stopping.
```
tuning_job_name_es = "imageclassif-job-{}-es".format(strftime("%d-%H-%M-%S", gmtime()))
tuner_es = HyperparameterTuner(
imageclassification,
objective_metric_name,
hyperparameter_ranges,
objective_type="Maximize",
max_jobs=20,
max_parallel_jobs=2,
early_stopping_type="Auto",
)
tuner_es.fit(
{"train": s3_input_train, "validation": s3_input_validation},
job_name=tuning_job_name_es,
include_cls_metadata=False,
)
tuner_es.wait()
```
After the tuning job finished, we again list the top 5 performing training jobs.
```
tuner_metrics_es = sagemaker.HyperparameterTuningJobAnalytics(tuning_job_name_es)
tuner_metrics_es.dataframe().sort_values(["FinalObjectiveValue"], ascending=False).head(5)
```
The total training time and training jobs status can be checked with the following script.
```
df = tuner_metrics_es.dataframe()
total_time_es = df["TrainingElapsedTimeSeconds"].sum() / 3600
print("The total training time with early stopping is {:.2f} hours".format(total_time_es))
df["TrainingJobStatus"].value_counts()
```
The stopped training jobs can be listed using the following scripts.
```
df[df.TrainingJobStatus == "Stopped"]
```
## Wrap up
In this notebook, we demonstrated how to use automatic early stopping to speed up model tuning. One thing to keep in
mind is as the training time for each training job gets longer, the benefit of training job early stopping becomes more significant. On the other hand, smaller training jobs won’t benefit as much due to infrastructure overhead. For example, our experiments show that the effect of training job early stopping typically becomes noticeable when the training jobs last longer than **4 minutes**. To enable automatic early stopping, one can simply set **early_stopping_type** to **'Auto'**.
For more information on using SageMaker's Automatic Model Tuning, see our other [example notebooks](https://github.com/awslabs/amazon-sagemaker-examples/tree/master/hyperparameter_tuning) and [documentation](https://docs.aws.amazon.com/sagemaker/latest/dg/automatic-model-tuning.html).
|
github_jupyter
|
import boto3
import sagemaker
from sagemaker import get_execution_role
from sagemaker.image_uris import retrieve
sess = sagemaker.Session()
role = get_execution_role()
bucket = sess.default_bucket()
training_image = retrieve("image-classification", boto3.Session().region_name, "1")
print(training_image)
import os
import urllib.request
import boto3
def download(url):
filename = url.split("/")[-1]
if not os.path.exists(filename):
urllib.request.urlretrieve(url, filename)
def upload_to_s3(channel, file):
s3 = boto3.resource("s3")
data = open(file, "rb")
key = channel + "/" + file
s3.Bucket(bucket).put_object(Key=key, Body=data)
s3_train_key = "image-classification-full-training/train"
s3_validation_key = "image-classification-full-training/validation"
download("http://data.mxnet.io/data/caltech-256/caltech-256-60-train.rec")
upload_to_s3(s3_train_key, "caltech-256-60-train.rec")
download("http://data.mxnet.io/data/caltech-256/caltech-256-60-val.rec")
upload_to_s3(s3_validation_key, "caltech-256-60-val.rec")
s3_train_data = "s3://{}/{}/".format(bucket, s3_train_key)
s3_validation_data = "s3://{}/{}/".format(bucket, s3_validation_key)
s3_output_key = "image-classification-full-training/output"
s3_output = "s3://{}/{}/".format(bucket, s3_output_key)
s3_input_train = sagemaker.TrainingInput(
s3_data=s3_train_data, content_type="application/x-recordio"
)
s3_input_validation = sagemaker.TrainingInput(
s3_data=s3_validation_data, content_type="application/x-recordio"
)
sess = sagemaker.Session()
imageclassification = sagemaker.estimator.Estimator(
training_image,
role,
instance_count=1,
instance_type="ml.p3.2xlarge",
output_path=s3_output,
sagemaker_session=sess,
)
imageclassification.set_hyperparameters(
num_layers=18,
image_shape="3,224,224",
num_classes=257,
epochs=10,
top_k="2",
num_training_samples=15420,
precision_dtype="float32",
augmentation_type="crop",
)
from time import gmtime, strftime
from sagemaker.tuner import (
IntegerParameter,
CategoricalParameter,
ContinuousParameter,
HyperparameterTuner,
)
tuning_job_name = "imageclassif-job-{}".format(strftime("%d-%H-%M-%S", gmtime()))
hyperparameter_ranges = {
"learning_rate": ContinuousParameter(0.00001, 1.0),
"mini_batch_size": IntegerParameter(16, 64),
"optimizer": CategoricalParameter(["sgd", "adam", "rmsprop", "nag"]),
}
objective_metric_name = "validation:accuracy"
tuner = HyperparameterTuner(
imageclassification,
objective_metric_name,
hyperparameter_ranges,
objective_type="Maximize",
max_jobs=20,
max_parallel_jobs=2,
)
tuner.fit(
{"train": s3_input_train, "validation": s3_input_validation},
job_name=tuning_job_name,
include_cls_metadata=False,
)
tuner.wait()
tuner_metrics = sagemaker.HyperparameterTuningJobAnalytics(tuning_job_name)
tuner_metrics.dataframe().sort_values(["FinalObjectiveValue"], ascending=False).head(5)
total_time = tuner_metrics.dataframe()["TrainingElapsedTimeSeconds"].sum() / 3600
print("The total training time is {:.2f} hours".format(total_time))
tuner_metrics.dataframe()["TrainingJobStatus"].value_counts()
tuning_job_name_es = "imageclassif-job-{}-es".format(strftime("%d-%H-%M-%S", gmtime()))
tuner_es = HyperparameterTuner(
imageclassification,
objective_metric_name,
hyperparameter_ranges,
objective_type="Maximize",
max_jobs=20,
max_parallel_jobs=2,
early_stopping_type="Auto",
)
tuner_es.fit(
{"train": s3_input_train, "validation": s3_input_validation},
job_name=tuning_job_name_es,
include_cls_metadata=False,
)
tuner_es.wait()
tuner_metrics_es = sagemaker.HyperparameterTuningJobAnalytics(tuning_job_name_es)
tuner_metrics_es.dataframe().sort_values(["FinalObjectiveValue"], ascending=False).head(5)
df = tuner_metrics_es.dataframe()
total_time_es = df["TrainingElapsedTimeSeconds"].sum() / 3600
print("The total training time with early stopping is {:.2f} hours".format(total_time_es))
df["TrainingJobStatus"].value_counts()
df[df.TrainingJobStatus == "Stopped"]
| 0.314471 | 0.978975 |
# MaestroSDK
## In this notebook you will:
- Connect with Maestro SDK
- Create and Finish a Task
- Create a new Log and Log Entries
- Fetch entries from a Log
- Delete a Log
- Emit Alerts
- Send Messages
- List, Download and Upload Artifacts / Result Files
## Prerequisites:
- **Python 3.7+**
- **BotCity Maestro SDK Python Package - botcity-maestro-sdk**
If you don't have the package installed yet, follow the instructions on the [project documentation website](https://botcity-dev.github.io/botcity-maestro-sdk-python/index.html).
- **Bot Maestro Account**
In order to follow this tutorial you must have a BotCity account.
If you don't have one yet, sign up now for a FREE Community Edition account by [clicking here](https://developers.botcity.dev/app/signup).
```
# For simplicity let's import everything from the Maestro SDK
from botcity.maestro import *
```
## BotMaestroSDK
The `BotMaestroSDK` is the main class to be used when interacting with BotMaestro.
```
maestro = BotMaestroSDK()
```
## Login
The login information is available when you access BotMaestro and click on the `Dev. Environment` menu.

```
maestro.login("YOUR_SERVER_HERE", "YOUR_USER_HERE", "YOUR_KEY_HERE")
```
If everything went well with the login, your maestro object now should have an `access_token`. Check it out by printing the current token assigned to your connection:
```
print(maestro.access_token)
```
## Activity
For the subsequent parts of this tutorial I will use an existing Activity available on my BotMaestro instance.
Looking at the image below you can notice the **Label** column. This is the value which we will refer to during the next steps as `activity label`. So in my case it will be `DemoActivity`. Make sure to adjust as needed for your case.

If you need help creating an Activity please refer to the BotMaestro documentation available [here](https://botcity.atlassian.net/wiki/spaces/CBG/overview).
## Tasks
Tasks are instances of an Activity.
### Creating a Task
We can create a new task via the BotMaestro SDK using the following code:
(More information and details on the parameters used can be found [here](https://botcity-dev.github.io/botcity-maestro-sdk-python/api/sdk.html#botcity.maestro.sdk.BotMaestroSDK.create_task).)
```
task = maestro.create_task(activity_label="DemoActivity", parameters={}, test=False)
```
Inspecting the returned object we can find interesting information that will be used on the following steps when we interact with the task.
```
print(task)
```
### Finishing a Task
Once tasks are created, they are queued on the BotMaestro and collected for execution by the **BotRunner**.
The tasks that are collected for execution move forward to the `Running` state.
It is the bot developer responsibility to inform the BotMaestro via the SDK of the proper final status of a task.
This allow for better control and also to add details about the task completion that can later be inspected by users via the BotMaestro portal.
A task can be finished with one of the following Status:
- `SUCCESS`: The task finished successfully.
- `FAILED`: The task failed to finish.
- `PARTIALLY_COMPLETED`: The task completed part of the expected steps.
The possible finishing statuses are available via code using the `AutomationTaskFinishStatus` Enum.
Here is how we finish the task created on the previous step:
(More information and details on the parameters used can be found [here](https://botcity-dev.github.io/botcity-maestro-sdk-python/api/sdk.html#botcity.maestro.sdk.BotMaestroSDK.finish_task).)
> **⚠ PRO TIP**
> When implementing your bot, the `task_id` is available via the `execution` object
> and you can access it like this:
> `execution.task_id`.
```
maestro.finish_task(task_id=task.id, status=AutomationTaskFinishStatus.SUCCESS, message="Task Finished OK.")
```
### Retrieving a Task
You can fetch more information about a task with the following code:
```
maestro.get_task(task.id)
```
## Logs
Logs are an excellent way to keep track of a bot execution and provide insightiful information for
operation and monitoring.
BotMaestro offers a very flexible log implementation that is very easy to use and create the log
which best fit your use case.
As of now, a log is associated with an activity. That means that the following commands will refer to the `activity label` mentioned on the first steps of this tutorial.
Over the following sections we will show you how to create a log, insert log entries and delete a log.
### Creating a Log
To create a new Log for an Activity we need to provide the following information:
- Activity Label
- List of Columns
The SDK provides the [Columns](https://botcity-dev.github.io/botcity-maestro-sdk-python/api/model.html#botcity.maestro.model.Column) class which helps to create new entries.
A `Column` instance holds the following information:
- `name`: Text to be displayed on the BotMaestro Web Portal
- `label`: Unique Identifier for this column on this log
- `width`: Suggested width in pixels.
Here is how we can create a new Log:
(More information and details on the parameters used can be found [here](https://botcity-dev.github.io/botcity-maestro-sdk-python/api/sdk.html#botcity.maestro.sdk.BotMaestroSDK.new_log).)
```
# Create a list of columns
columns = [
Column(name="Date/Time", label="timestamp", width=300),
Column(name="# Records", label="records", width=200),
Column(name="Status", label="status", width=100),
]
# Create a new log
maestro.new_log(
"DemoActivity",
columns
)
```
#### Your new Log is Ready
If no errors happened during the processing of the request, you should now be able to see a new log when
accessing the **Execution Log** menu.

### Creating new Log Entries
With your shiny new log ready, it is time to create some log entries.
Here is how you can insert new log entries:
(More information and details on the parameters used can be found [here](https://botcity-dev.github.io/botcity-maestro-sdk-python/api/sdk.html#botcity.maestro.sdk.BotMaestroSDK.new_log_entry).)
```
import datetime
maestro.new_log_entry(
activity_label="DemoActivity",
values = {
"timestamp": datetime.datetime.now().strftime("%Y-%m-%d_%H-%M"),
"records": "10",
"status": "SUCCESS"
}
)
```
#### Your new Log Entry is ready
The new entry is now displayed on the `Demo Activity` logbook.
> **⚠ PRO TIP**
> Using strings like `SUCCESS`, `FAILED`, `YES`, `NO`, `true` or `false` will render icons
> such as the green check mark displayed below.

### Fetch Log Data
Retrieving log data is as easy as creating log entries.
Optionally we can also pass a `date` argument which acts as filters the initial date for log to be retrieved.
The `date` parameter must use the `DD/MM/YYYY` format. If `date` is not informed, all data from the log is retrieved.
(More information and details on the parameters used can be found [here](https://botcity-dev.github.io/botcity-maestro-sdk-python/api/sdk.html#botcity.maestro.sdk.BotMaestroSDK.get_log).)
```
maestro.get_log(activity_label='DemoActivity')
# Get today's date in the DD/MM/YYYY format.
today = datetime.datetime.now().strftime("%d/%m/%Y")
maestro.get_log(activity_label='DemoActivity', date=today)
```
### Deleting an entire Log
If by any reason you need to completely remove the log along with all its entries,
you can do so using the command below.
(More information and details on the parameters used can be found [here](https://botcity-dev.github.io/botcity-maestro-sdk-python/api/sdk.html#botcity.maestro.sdk.BotMaestroSDK.delete_log).)
```
maestro.delete_log(activity_label="DemoActivity")
```
## Alerts
Alerts are customized messages for a task with an `alert level`, `title` and `message`.
The alerts are displayed on the `Alerts` menu of the BotMaestro portal and they can provide clear and fast information about the status of a task and activity.

### Emitting Alerts
An alert can be emitted with one of the following types:
- `INFO`: Information alert.
- `WARN`: Warning alert.
- `ERROR`: Error alert.
The possible alert types are available via code using the `AlertType` Enum.
Here is how we emit alerts for the task created on the previous step:
(More information and details on the parameters used can be found [here](https://botcity-dev.github.io/botcity-maestro-sdk-python/api/sdk.html#botcity.maestro.sdk.BotMaestroSDK.finish_task).)
> **⚠ PRO TIP**
> When implementing your bot, the `task_id` is available via the `execution` object
> and you can access it like this:
> `execution.task_id`.
#### Info Alert
```
maestro.alert(task_id=task.id, title="Info Alert", message="This is an info alert", alert_type=AlertType.INFO)
```
#### Warn Alert
```
maestro.alert(task_id=task.id, title="Warn Alert", message="This is a warn alert", alert_type=AlertType.WARN)
```
#### Error Alert
```
maestro.alert(task_id=task.id, title="Error Alert", message="This is an error alert", alert_type=AlertType.ERROR)
```
#### Your alerts are ready
When you navigate into the BotMaestro portal and click over Alerts, the recently generated alerts will be available.

## Messages
By using the BotMaestro SDK, users can send messages to each other or even external email addresses in a very simple way.
The Maestro SDK can send two types of messages:
- `TEXT`: Plain text message body.
- `HTML`: HTML message body.
The possible message types are available via code using the `MessageType` Enum.
Here is how we send messages via the Maestro SDK:
(More information and details on the parameters used can be found [here](https://botcity-dev.github.io/botcity-maestro-sdk-python/api/sdk.html#botcity.maestro.sdk.BotMaestroSDK.message).)
```
emails = ["your_email@your_provider.com"] # List of emails, if not using, pass an empty list
users = ["maestro_user1", "maestro_user2"] # List of usernames, if not using, pass an empty list
subject = "Test Message"
body = "This is the message content."
maestro.message(emails, users, subject, body, MessageType.TEXT)
```
#### You've got mail
If everything went well with the parameters informed, you probably received an email like this:

## Artifacts / Result Files
During execution of your bot, it can upload or download any type of files to/from the BotMaestro portal.
As a general term, we refer to those files are `artifacts` or `result files`.
They can be accessed via the menu `Result Files` on the BotMaestro portal.

You can download the files directly from the web interface or using the Maestro SDK.
Over the next steps we will show you how to upload and download artifacts using the SDK API.
### Uploading an Artifact
In this example we will generate a temporary text file and upload this artifact to BotMaestro.
(More information and details on the parameters used can be found [here](https://botcity-dev.github.io/botcity-maestro-sdk-python/api/sdk.html#botcity.maestro.sdk.BotMaestroSDK.post_artifact).)
#### Creating a Temporary File
```
with open("my_artifact.txt", "w") as f:
f.writelines(["My First Artifact", " with BotMaestro SDK Python!"])
```
#### Uploading the Artifact
> **⚠ PRO TIP**
> When implementing your bot, the `task_id` is available via the `execution` object
> and you can access it like this:
> `execution.task_id`.
```
maestro.post_artifact(task_id=task.id, artifact_name="My Artifact", filepath="my_artifact.txt")
```
#### Viewing the new Artifact
If we now go into the BotMaestro web portal, the new artifact will be displayed.

### Listing all Artifacts
The `list_artifacts` method returns all artifacts available to your organization.
Here is how we list all artifacts via the BotMaestro SDK:
(More info and details on the parameters used can be found [here](https://botcity-dev.github.io/botcity-maestro-sdk-python/api/sdk.html#botcity.maestro.sdk.BotMaestroSDK.list_artifacts).)
```
artifacts = maestro.list_artifacts()
print(artifacts)
```
### Downloading an Artifact
In order to download an artifact you will need the `artifact id`.
The `get_artifact` method returns the artifact name and the file content as an array of bytes.
Here is how we download an artifact via the BotMaestro SDK:
(More information and details on the parameters used can be found [here](https://botcity-dev.github.io/botcity-maestro-sdk-python/api/sdk.html#botcity.maestro.sdk.BotMaestroSDK.get_artifact).)
```
name, content = maestro.get_artifact(artifact_id=artifacts[0].id)
```
#### Saving to Disk
```
with open(f"{name}.txt", "wb") as f:
f.write(content)
```
## Next Steps
This concludes this tutorial. Thank you for following along.
As next step we recommend that you try to integrate the BotMaestro SDK into your Bot.
In case you have questions, suggestions or bump into an issue, feel free to [reach out in our community](http://community.botcity.dev/).
Have fun Automating!
|
github_jupyter
|
# For simplicity let's import everything from the Maestro SDK
from botcity.maestro import *
maestro = BotMaestroSDK()
maestro.login("YOUR_SERVER_HERE", "YOUR_USER_HERE", "YOUR_KEY_HERE")
print(maestro.access_token)
task = maestro.create_task(activity_label="DemoActivity", parameters={}, test=False)
print(task)
maestro.finish_task(task_id=task.id, status=AutomationTaskFinishStatus.SUCCESS, message="Task Finished OK.")
maestro.get_task(task.id)
# Create a list of columns
columns = [
Column(name="Date/Time", label="timestamp", width=300),
Column(name="# Records", label="records", width=200),
Column(name="Status", label="status", width=100),
]
# Create a new log
maestro.new_log(
"DemoActivity",
columns
)
import datetime
maestro.new_log_entry(
activity_label="DemoActivity",
values = {
"timestamp": datetime.datetime.now().strftime("%Y-%m-%d_%H-%M"),
"records": "10",
"status": "SUCCESS"
}
)
maestro.get_log(activity_label='DemoActivity')
# Get today's date in the DD/MM/YYYY format.
today = datetime.datetime.now().strftime("%d/%m/%Y")
maestro.get_log(activity_label='DemoActivity', date=today)
maestro.delete_log(activity_label="DemoActivity")
maestro.alert(task_id=task.id, title="Info Alert", message="This is an info alert", alert_type=AlertType.INFO)
maestro.alert(task_id=task.id, title="Warn Alert", message="This is a warn alert", alert_type=AlertType.WARN)
maestro.alert(task_id=task.id, title="Error Alert", message="This is an error alert", alert_type=AlertType.ERROR)
emails = ["your_email@your_provider.com"] # List of emails, if not using, pass an empty list
users = ["maestro_user1", "maestro_user2"] # List of usernames, if not using, pass an empty list
subject = "Test Message"
body = "This is the message content."
maestro.message(emails, users, subject, body, MessageType.TEXT)
with open("my_artifact.txt", "w") as f:
f.writelines(["My First Artifact", " with BotMaestro SDK Python!"])
maestro.post_artifact(task_id=task.id, artifact_name="My Artifact", filepath="my_artifact.txt")
artifacts = maestro.list_artifacts()
print(artifacts)
name, content = maestro.get_artifact(artifact_id=artifacts[0].id)
with open(f"{name}.txt", "wb") as f:
f.write(content)
| 0.35031 | 0.918114 |
# Исключения
Исключения это ситуации, которые могут возникнуть во время выполнения программы и приводят к невозможности дальнейшей корректной работы. Обычно их используют для описания крайних ситуаций, например деление на ноль, отсутствия файла и т.д.
Все исключения в Python наследуются от базового класса ```BaseException```, от которого в свою очередь наследуется класс ```Exception```, объединяющий большинство исключений.
В Python существует большое количество возможных исключений, например:
- ошибки типов ```TypeError```;
- ошибки значений ```ValueError```;
- синтаксические ошибки ```SyntaxError```;
- предупреждения ```Warning```;
- и т.д.
Подробнее об их иерархии и назначении можно прочитать в [документации](https://docs.python.org/3.9/library/exceptions.html#exception-hierarchy).
Вот примеры некоторых исключительных ситуаций:
```
'1' + 1 # TypeError
1.e # SyntaxError
```
Для самостоятельного возбуждения исключений используется инструкция ```raise```, после которой указывается тип исключения. Например:
```
raise ValueError
```
# Обработка исключений
Для обработки исключений используется конструкцию ```try ... except ... else ... finally```. Ветки ```try``` и ```except``` являются обязательными, в то время как остальные опциональны. Веток ```except``` может быть несколько, остальные ветки могут присутствовать в единственном экземпляре.
В ветке ```try``` располагается код, который может вызывать исключение. Хорошим тоном является расположение минимального количества кода в ```try```.
Ветка ```except``` предназначена для отслеживания исключений. После ключевого слова ```except``` желательно указывать тип исключения, которое нужно обработать. Если после ```except``` не указать какое исключение ожидается, то он примет любое исключение. Если указано несколько веток ```except``` с разными типами исключений, то они будут проверяться последовательно пока тип исклбчения не совпадет, что аналогично выполнению условного оператора.
Последовательность выполнения конструкции ```try ... except ... else ... finally``` следующая. Выполняется код внутри ```try```, если он вызывает исключение происходит последовательная проверка на совпадения типа исключения и типа указанного после ```except```. Если типы совпадают, код внутри соответствующего ```except``` выполняется. Если исключения не произошло выполняется код внутри ```else```. В заключении выполняется код внутри ```finaly```. Ветка ```finaly``` выполняется в любом случае, в независимости от того было исключение или нет.
```
x = input()
print(f'Ввод пользователя: {repr(x)}')
try:
x = int(x)
except ValueError:
print('Введено не целое число')
x = input()
print(f'Ввод пользователя: {repr(x)}')
try:
x = int(x)
except ValueError:
print('Введено не целое число')
else:
print(f'Введено целое число: {x}')
finally:
print('Выполняется в любом случае')
```
Использовать ```except``` без указания исключения считается плохой практикой, также как и указывать в качестве исключения ```Exception```, так как он является предком для большого семейства исключений. Также не рекомендуется помещать в блок ```try``` большие блоки кода. Это ухудшит читаемость и затруднит отладку. Нужно выделять только те участки, где нужно обработать исключение. Стоит помнить, что исключения нужны для исключительных ситуаций, т.е. не нужно использовать их повсеместно (особенно там, где можно обойтись условным оператором).
С исключением можно работать, для этого в ```except``` пишут выражение ```Exception as e```. На место ```Exception``` записывается тип нужного исключения. Переменная ```e``` может иметь и другое имя, в большинстве случаем принято называть так.
```
x = input()
print(f'Ввод пользователя: {repr(x)}')
try:
x = int(x)
except ValueError as e:
print(f'Что-то пошло не так: {e}')
```
Время жизни это переменной ограничено веткой ```except```. В этом можно убедиться на следующем примере.
```
x = input()
print(f'Ввод пользователя: {repr(x)}')
e = 0
try:
x = int(x)
except ValueError as e:
print(f'Что-то пошло не так: {e}')
print(f'{e = }') # переменная e удалена
```
В случае если возникла необходимость работать с исключением вне блока ```except``` нужно его сохранить в дополнительной переменной.
```
exception = None
try:
1/0
except ZeroDivisionError as e:
exception = e
print(f'{exception = }')
print(f'{type(exception) = }')
```
Иногда бывает необходимость возбудить исключение в связи с другим исключением, т.е. связать их, для этого используется оператор ```raise ... from ...```. Это бывает полезно для отслеживания причины появления ошибок.
Могут возникнуть случаи, когда исключения возникают в неожиданных местах, например в ветках except или finally. Тогда интерпретатор использует связывание исключений и выводит осмысленное сообщение об ошибке без лишних затруднений.
```
try:
{}['a']
except KeyError as e:
raise RuntimeError('Что-то пошло не так') from e
```
Обратите внимание, что переменные, созданные внутри блока ```try```, остаются доступны после его выполнения только в том случае, если операция связывания была выполнена до возбуждения исключения.
```
try:
a = 'до'
b = 1/0
c = 'после'
except ZeroDivisionError:
print('Деление на ноль!')
print(f'{a = }')
print(f'{b = }')
print(f'{c = }')
```
Поэтому хорошим тоном будет создать переменные заранее.
```
a = None
try:
a = 1/0
except ZeroDivisionError:
print('Деление на ноль!')
print(f'{a = }')
```
# Принцип **EAFP**
Принцип [**EAFP**](https://docs.python.org/3.9/glossary.html#term-eafp) (easier to ask for forgiveness than permission) или "проще просить прощения, чем разрешения". Этот принцип основан на предположении, что определенная операция всегда выполняется корректно. В случае не корректного выполнения просто обрабатывается ошибка. Таким образом этот принцип отдает предпочтение конструкциям ```try ... except ...```, а не ```if .. else ...```. Код, следующий этому принципу, считается немного более читаемым (субъективно), а также следования ему значит следование питоническому пути (хотя Гвидо [не считает](https://mail.python.org/pipermail/python-dev/2014-March/133118.html), что **EAFP** необходимо строго следовать).
```
d = {'a': 1, 'b': 2}
try:
d['c']
except KeyError:
print('Ключ не найден')
```
# Принцип **LBYL**
Противоположностью принципу **EAFP** выступает принцип [**LBYL**](https://docs.python.org/3/glossary.html#term-lbyl) (look before you leap) или "смотри прежде чем прыгать" (~~смотри куда прешь~~). Следуя этому принципу, необходимо сначала выполнить проверку, а затем действие.
```
d = {'a': 1, 'b': 2}
if 'c' not in d:
print('Ключ не найден')
```
# Полезные ссылки
- [Гвидо о EAFP](https://mail.python.org/pipermail/python-dev/2014-March/133118.html)
- [What is the EAFP principle in Python?](https://stackoverflow.com/questions/11360858/what-is-the-eafp-principle-in-python)
- [Как быстро работают исключения? (сравнение ```if ... else ...``` и ```try ... except ...```)](https://stackoverflow.com/questions/8107695/python-faq-how-fast-are-exceptions)
|
github_jupyter
|
'1' + 1 # TypeError
1.e # SyntaxError
raise ValueError
x = input()
print(f'Ввод пользователя: {repr(x)}')
try:
x = int(x)
except ValueError:
print('Введено не целое число')
x = input()
print(f'Ввод пользователя: {repr(x)}')
try:
x = int(x)
except ValueError:
print('Введено не целое число')
else:
print(f'Введено целое число: {x}')
finally:
print('Выполняется в любом случае')
x = input()
print(f'Ввод пользователя: {repr(x)}')
try:
x = int(x)
except ValueError as e:
print(f'Что-то пошло не так: {e}')
x = input()
print(f'Ввод пользователя: {repr(x)}')
e = 0
try:
x = int(x)
except ValueError as e:
print(f'Что-то пошло не так: {e}')
print(f'{e = }') # переменная e удалена
exception = None
try:
1/0
except ZeroDivisionError as e:
exception = e
print(f'{exception = }')
print(f'{type(exception) = }')
try:
{}['a']
except KeyError as e:
raise RuntimeError('Что-то пошло не так') from e
try:
a = 'до'
b = 1/0
c = 'после'
except ZeroDivisionError:
print('Деление на ноль!')
print(f'{a = }')
print(f'{b = }')
print(f'{c = }')
a = None
try:
a = 1/0
except ZeroDivisionError:
print('Деление на ноль!')
print(f'{a = }')
d = {'a': 1, 'b': 2}
try:
d['c']
except KeyError:
print('Ключ не найден')
d = {'a': 1, 'b': 2}
if 'c' not in d:
print('Ключ не найден')
| 0.174797 | 0.981364 |
# Introduction to linear regression
See Géron, A. (2017). *Hands-on machine learning with Scikit-Learn and TensorFlow: concepts, tools, and techniques to build intelligent systems.* " O'Reilly Media, Inc.".
Linear regression is a simple way of predicting the values of a variable by computing a weighted sum of the features, as follows:
$$
\hat{y} = \theta_0 + \theta_1 x_1 + \dots + \theta_n x_n
$$
or
$$
\hat{y} = \theta^{T} \cdot \vec{x}
$$
In order to train a model and learn $\theta$, we need a measure of quality of the prediction, such as the Mean Square Error (MSE). The values of $\theta$ that **minimize** MSE is the final prediction model.
$$
MSE(\theta) = \frac{1}{m} \sum\limits_{i = 1}^{m} \left(\theta^T \cdot x^i - y^i \right)^{2}
$$
```
def mse(theta, x, y):
s = np.array([np.power(theta.T.dot(p) - y[i], 2) for i, p in enumerate(x)]).sum()
return s / len(x)
```
## Example
```
import numpy as np
import matplotlib.pyplot as plt
x = np.random.rand(100, 1)
y = x + np.random.rand(100, 1)
X = np.array([x.ravel(), y.ravel()]).T
plt.scatter(X[:,0], X[:,1])
plt.show()
X[:10,:]
```
## Learning $\theta$
There are several solutions for learning $\theta$. One is to calculate the partial derivatives of the cost function for each parameter and then change the parameters in the direction that minimizes the MSE function.
The partial derivative with respect to one of the parameters $\theta_j$ is calculated as:
$$
\frac{\partial}{\partial \theta_j} MSE(\theta) = \frac{2}{m}\sum\limits_{i=1}^{m}\left(\theta^T \cdot \vec{x}^i -
y^i \right)x_{j}^{i}
$$
which becomes for all the features
$$
\nabla_{\theta} MSE(\theta) = \left(
\begin{array}{c}
\frac{\partial}{\partial \theta_0}MSE(\theta)\\
\frac{\partial}{\partial \theta_1}MSE(\theta)\\
\dots\\
\frac{\partial}{\partial \theta_n}MSE(\theta)\\
\end{array}
\right) = \frac{2}{m} X^T \cdot \left( X \cdot \theta - y \right)
$$
Then, given the gradient vector, we change $\theta$ in the opposite direction by a given factor $\eta$ (called learning rate).
$$
\theta_{\textrm i+1 step} = \theta - \eta \nabla_{\theta} MSE(\theta)
$$
## Example
We need to add x0 = 1 to each point
```
eta, iterations, size = 0.05, 100, 100
theta = np.random.randn(2, 1)
x_b = np.c_[np.ones((100, 1)), x]
mse(theta, x_b, y)
theta
mse_values = []
for i in range(iterations):
gradients = 2/size * x_b.T.dot(x_b.dot(theta) - y)
theta = theta - eta * gradients
mse_values.append(mse(theta, x_b, y))
plt.plot(mse_values)
plt.show()
theta
```
### Prediction
We need to add x0 = 1 to each point
```
X_new = np.array([[0], [2]])
X_new_b = np.c_[np.ones((2, 1)), X_new]
X_new_b
y_pred = X_new_b.dot(theta)
y_pred
plt.plot(X_new, y_pred, "r-")
plt.plot(x, y, "b.")
plt.show()
theta
```
# Sklearn implementation
```
from sklearn.linear_model import LinearRegression
lr = LinearRegression()
lr.fit(x, y)
print(lr.intercept_, lr.coef_)
y_pred = lr.predict(X_new)
plt.plot(X_new, y_pred, "r-")
plt.plot(x, y, "b.")
plt.show()
```
# Application to real data
```
import pandas as pd
d = '/Users/alfio/Dati/kaggle/world-happiness-report/2017.csv'
F = pd.read_csv(d, low_memory=False)
cols = ['Happiness.Score', 'Economy..GDP.per.Capita.', 'Freedom']
S = F[cols]
S.head()
fig, axes = plt.subplots(nrows=1, ncols=2, figsize=(14, 4))
axes[0].scatter(S.iloc[:,1], S.iloc[:,0], alpha=0.4)
axes[0].set_xlabel(cols[1])
axes[0].set_ylabel(cols[0])
axes[1].scatter(S.iloc[:,2], S.iloc[:,0], alpha=0.4)
axes[1].set_xlabel(cols[2])
axes[1].set_ylabel(cols[0])
plt.show()
def predict(data, features, target):
lr = LinearRegression()
if S.iloc[:100,features].shape[1] == 1:
train_x, train_y = np.array(S.iloc[:100,features]).reshape(-1, 1), S.iloc[:100,target]
test_x, test_y = np.array(S.iloc[100:,features]).reshape(-1, 1), S.iloc[100:,target]
else:
train_x, train_y = np.array(S.iloc[:100,features]), S.iloc[:100,target]
test_x, test_y = np.array(S.iloc[100:,features]), S.iloc[100:,target]
lr.fit(train_x, train_y)
y_pred = lr.predict(test_x)
y_pred_t = lr.predict(train_x)
return y_pred, train_x, train_y, test_x, test_y, y_pred_t
```
## GDP, Freedom & both
```
gdp = predict(S, [1], 0)
free = predict(S, [2], 0)
both = predict(S, [1,2], 0)
def visualize(ax, test_x, test_y, pred_y):
ax.scatter(test_x, test_y, alpha=0.2)
ax.scatter(test_x, pred_y, alpha=0.4)
fig, axes = plt.subplots(nrows=2, ncols=3, figsize=(12, 8))
visualize(axes[0,0], gdp[1], gdp[2], gdp[5])
axes[0,0].set_title('Train GDP')
visualize(axes[0,1], free[1], free[2], free[5])
axes[0,1].set_title('Train Freedom')
visualize(axes[0,2], both[1].mean(axis=1), both[2], both[5])
axes[0,2].set_title('Train Both')
visualize(axes[1,0], gdp[3], gdp[4], gdp[0])
axes[1,0].set_title('Test GDP')
visualize(axes[1,1], free[3], free[4], free[0])
axes[1,1].set_title('Test Freedom')
visualize(axes[1,2], both[3].mean(axis=1), both[4], both[0])
axes[1,2].set_title('Test Both')
plt.tight_layout()
plt.show()
from sklearn.metrics import mean_squared_error
print(mean_squared_error(gdp[4], gdp[0]))
print(mean_squared_error(free[4], free[0]))
print(mean_squared_error(both[4], both[0]))
```
|
github_jupyter
|
def mse(theta, x, y):
s = np.array([np.power(theta.T.dot(p) - y[i], 2) for i, p in enumerate(x)]).sum()
return s / len(x)
import numpy as np
import matplotlib.pyplot as plt
x = np.random.rand(100, 1)
y = x + np.random.rand(100, 1)
X = np.array([x.ravel(), y.ravel()]).T
plt.scatter(X[:,0], X[:,1])
plt.show()
X[:10,:]
eta, iterations, size = 0.05, 100, 100
theta = np.random.randn(2, 1)
x_b = np.c_[np.ones((100, 1)), x]
mse(theta, x_b, y)
theta
mse_values = []
for i in range(iterations):
gradients = 2/size * x_b.T.dot(x_b.dot(theta) - y)
theta = theta - eta * gradients
mse_values.append(mse(theta, x_b, y))
plt.plot(mse_values)
plt.show()
theta
X_new = np.array([[0], [2]])
X_new_b = np.c_[np.ones((2, 1)), X_new]
X_new_b
y_pred = X_new_b.dot(theta)
y_pred
plt.plot(X_new, y_pred, "r-")
plt.plot(x, y, "b.")
plt.show()
theta
from sklearn.linear_model import LinearRegression
lr = LinearRegression()
lr.fit(x, y)
print(lr.intercept_, lr.coef_)
y_pred = lr.predict(X_new)
plt.plot(X_new, y_pred, "r-")
plt.plot(x, y, "b.")
plt.show()
import pandas as pd
d = '/Users/alfio/Dati/kaggle/world-happiness-report/2017.csv'
F = pd.read_csv(d, low_memory=False)
cols = ['Happiness.Score', 'Economy..GDP.per.Capita.', 'Freedom']
S = F[cols]
S.head()
fig, axes = plt.subplots(nrows=1, ncols=2, figsize=(14, 4))
axes[0].scatter(S.iloc[:,1], S.iloc[:,0], alpha=0.4)
axes[0].set_xlabel(cols[1])
axes[0].set_ylabel(cols[0])
axes[1].scatter(S.iloc[:,2], S.iloc[:,0], alpha=0.4)
axes[1].set_xlabel(cols[2])
axes[1].set_ylabel(cols[0])
plt.show()
def predict(data, features, target):
lr = LinearRegression()
if S.iloc[:100,features].shape[1] == 1:
train_x, train_y = np.array(S.iloc[:100,features]).reshape(-1, 1), S.iloc[:100,target]
test_x, test_y = np.array(S.iloc[100:,features]).reshape(-1, 1), S.iloc[100:,target]
else:
train_x, train_y = np.array(S.iloc[:100,features]), S.iloc[:100,target]
test_x, test_y = np.array(S.iloc[100:,features]), S.iloc[100:,target]
lr.fit(train_x, train_y)
y_pred = lr.predict(test_x)
y_pred_t = lr.predict(train_x)
return y_pred, train_x, train_y, test_x, test_y, y_pred_t
gdp = predict(S, [1], 0)
free = predict(S, [2], 0)
both = predict(S, [1,2], 0)
def visualize(ax, test_x, test_y, pred_y):
ax.scatter(test_x, test_y, alpha=0.2)
ax.scatter(test_x, pred_y, alpha=0.4)
fig, axes = plt.subplots(nrows=2, ncols=3, figsize=(12, 8))
visualize(axes[0,0], gdp[1], gdp[2], gdp[5])
axes[0,0].set_title('Train GDP')
visualize(axes[0,1], free[1], free[2], free[5])
axes[0,1].set_title('Train Freedom')
visualize(axes[0,2], both[1].mean(axis=1), both[2], both[5])
axes[0,2].set_title('Train Both')
visualize(axes[1,0], gdp[3], gdp[4], gdp[0])
axes[1,0].set_title('Test GDP')
visualize(axes[1,1], free[3], free[4], free[0])
axes[1,1].set_title('Test Freedom')
visualize(axes[1,2], both[3].mean(axis=1), both[4], both[0])
axes[1,2].set_title('Test Both')
plt.tight_layout()
plt.show()
from sklearn.metrics import mean_squared_error
print(mean_squared_error(gdp[4], gdp[0]))
print(mean_squared_error(free[4], free[0]))
print(mean_squared_error(both[4], both[0]))
| 0.595257 | 0.992788 |
Do this to get wikipedia's latest dump of english language data:
``` wget http://download.wikimedia.org/enwiki/latest/enwiki-latest-pages-articles.xml.bz2 ```
It's going to be a very large file, several gigabytes.
Replace "en" by the appropriate [language code](https://en.wikipedia.org/wiki/List_of_ISO_639-1_codes) to grab it for a different language.
Run
```WikiExtractor.py -cb 250K -o extracted itwiki-latest-pages-articles.xml.bz2```
to get a cleaned up version.
Alternatively, there is some pre-cleaned text available for download at the [polyglot project](https://sites.google.com/site/rmyeid/projects/polyglot).
```
import re
def extract_words(line):
line = line.lower()
words = line.split(' ')
words = [word.strip(' .()!;\n') for word in words]
words = list(filter(lambda w:w.isalpha(),words))
return words
words=[]
with open("peuptext.txt") as f:
for line in f.readlines():
words = words + extract_words(line)
glyphs = set(c for word in words for c in word)
glyphs.add('WORD_START')
glyphs.add('WORD_END')
num_glyphs = len(glyphs)
int_to_glyph = dict(enumerate(glyphs))
glyph_to_int = {v:k for k,v in int_to_glyph.items()}
import numpy as np
counts = np.zeros((num_glyphs,num_glyphs,num_glyphs,num_glyphs),dtype=np.dtype('u8'))
for word in words:
for i in range(len(word)+1):
c1 = glyph_to_int['WORD_START'] if i-3<0 else glyph_to_int[word[i-3]]
c2 = glyph_to_int['WORD_START'] if i-2<0 else glyph_to_int[word[i-2]]
c3 = glyph_to_int['WORD_START'] if i-1<0 else glyph_to_int[word[i-1]]
c4 = glyph_to_int['WORD_END'] if i>=len(word) else glyph_to_int[word[i]]
counts[c1,c2,c3,c4] += 1
totals = counts.sum(axis=3)
distribution = counts / (np.vectorize(lambda x : x if x!=0 else 1)(totals[:,:,:,np.newaxis]))
def generate_word(dist):
c1 = c2 = c3 = glyph_to_int['WORD_START']
word = []
while c3!=glyph_to_int['WORD_END']:
if distribution[c1,c2,c3].sum()==0:
next_char = np.random.choice(range(num_glyphs))
else:
next_char = np.random.choice(range(num_glyphs),p=distribution[c1,c2,c3])
c1=c2
c2=c3
c3=next_char
word.append(next_char)
return ''.join(int_to_glyph[c] for c in word[:-1])
generate_word(distribution)
import numpy as np
import pickle
filename = 'indonesian' #filename without txt extension
# First build alphabet. Right now this is done here by hand to suit english; e.g. no unicode.
glyphs = set(map(chr,range(ord('a'),ord('z')+1)))
glyphs.add('WORD_START')
glyphs.add('WORD_END')
num_glyphs = len(glyphs)
int_to_glyph = dict(enumerate(glyphs))
glyph_to_int = {v:k for k,v in int_to_glyph.items()}
def extract_words(line):
line = line.lower()
words = line.split(' ')
words = [word.strip(' .()!;\n') for word in words]
words = list(filter(lambda w:w and all(c in glyphs for c in w),words))
# TODO: make that filter smart enough to handle unicode
return words
# Intitalize counts
counts = np.zeros((num_glyphs,num_glyphs,num_glyphs,num_glyphs),dtype=np.dtype('u8'))
# Now go through file and build up distributon
with open(filename+".txt") as f:
for line in f.readlines():
for word in extract_words(line):
for i in range(len(word)+1):
c1 = glyph_to_int['WORD_START'] if i-3<0 else glyph_to_int[word[i-3]]
c2 = glyph_to_int['WORD_START'] if i-2<0 else glyph_to_int[word[i-2]]
c3 = glyph_to_int['WORD_START'] if i-1<0 else glyph_to_int[word[i-1]]
c4 = glyph_to_int['WORD_END'] if i>=len(word) else glyph_to_int[word[i]]
counts[c1,c2,c3,c4] += 1
totals = counts.sum(axis=3)
distribution = counts / (np.vectorize(lambda x : x if x!=0 else 1)(totals[:,:,:,np.newaxis]))
with open(filename+".pkl",'wb') as pickle_file:
pickle.dump(distribution,pickle_file)
for i in range(100):
word = generate_word(distribution)
if len(word)>4:
print(word)
import os
import sys
module_path = os.path.abspath(os.path.join('epitran'))
if module_path not in sys.path:
sys.path.append(module_path)
import epitran
epi = epitran.Epitran("eng-Latn",ligatures=False)
epi.trans_list("The cutest thing is really cute!! Potassium. George.")
simple = epitran.simple.SimpleEpitran("ind-Latn")
ipa_chars=set(p for l in simple._load_g2p_map("ind-Latn",False).values() for p in l)
ipa_chars=ipa_chars.union(map(epitran.ligaturize.ligaturize,ipa_chars))
for p in list(ipa_chars): ipa_chars.update(set(p))
ipa_chars
'eng-Latn' in epitran.Epitran.special
import panphon
ft = panphon.featuretable.FeatureTable()
def load_ipa_chars(lang_code):
"""Return set of characters that epitran will use for phonemes for the given language code"""
if lang_code in epitran.Epitran.special:
if lang_code == "eng-Latn":
flite = epitran.flite.Flite()
ipa_chars = set(flite._read_arpabet("epitran/epitran/data/arpabet.csv").values())
else:
raise NotImplementedError
else:
simple = epitran.simple.SimpleEpitran(lang_code)
ipa_chars=set(p for l in simple._load_g2p_map(lang_code,False).values() for p in l)
# ipa_chars=set(map(epitran.ligaturize.ligaturize,ipa_chars))
# ipa_chars_single = set()
# for p in ipa_chars: ipa_chars_single.update(set(p))
if '' in ipa_chars : ipa_chars.remove('')
ipa_chars_segmented = set()
for p in ipa_chars: ipa_chars_segmented.update(ft.segs_safe(p))
return ipa_chars_segmented
# Hmmm now I am seeing that setting up IPA as glyphs will lose some things.
# And not doing so is not so bad for any language that isn't english.
# Maybe I'll do IPA for english only?
# Then for other languages they just have their own graphemes,
# however when we do merging we identify them via a blurry version of IPA
# idea: generate a phonology at the start.
# this contains the "blur" i.e. the projection from ipa to a smaller set of phonemes
# to help generate this projection, use the phonetic features like place of articulation, etc.
import panphon
ft = panphon.featuretable.FeatureTable()
print(load_ipa_chars('eng-Latn'))
print(ft.segs_safe("alsˤbaːħ"))
epi = epitran.Epitran("ara-Arab")
epi.transliterate("وكيل") # The README for epitran warns against using arabic and some other languages.
import numpy as np
import pickle
import os
import sys
module_path = os.path.abspath(os.path.join('epitran'))
if module_path not in sys.path:
sys.path.append(module_path)
import epitran
filename = 'peuptext' #filename without txt extension
lang_code = "eng-Latn" #language code for epitran
epi = epitran.Epitran(lang_code)
# TODO: Now I want a more automated way to build the alphabet of IPA glyphs...
# Hmm you should be able to check if something is an IPA character by collecting all IPA characters
# (including ligatures? if you use this)
# Some IPA characters can be gathered from epitran's maps that are used for simple phonetic things
# English is exceptional. To get those, use Flite._read_arpabet
glyphs = load_ipa_chars(lang_code)
glyphs.add('WORD_START')
glyphs.add('WORD_END')
num_glyphs = len(glyphs)
int_to_glyph = dict(enumerate(glyphs))
glyph_to_int = {v:k for k,v in int_to_glyph.items()}
def extract_words(line):
# line = line.lower()
words = []
for word in line.split(' '):
orig_word = word # testing line
word = word.strip(' .()!:;,\n')
word = epi.trans_list(word)
if word and all(c in glyphs for c in word): words.append(word)
else : print(orig_word) # testing line. do report *some* of these in full thing.
return words
extract_words("this is a line And IT SERVES, AS SOM;ekindof EXA33mple. example. father.")
window_size = 3 # How many adjacent characters in each group considered for the distribution.
# Intitalize counts
counts = np.zeros((num_glyphs,)*window_size,dtype=np.dtype('u8')) # TODO use scipy sparse array instead
# Now go through file and build up distributon
with open(filename+".txt") as f:
for line in f.readlines():
for word in extract_words(line):
for i in range(len(word)+1):
group = []
for lookback in range(window_size-1,0,-1):
group.append(glyph_to_int['WORD_START'] if (i-lookback)<0 else glyph_to_int[word[i-lookback]])
group.append(glyph_to_int['WORD_END'] if i>=len(word) else glyph_to_int[word[i]])
counts[tuple(group)] += 1
totals = counts.sum(axis=window_size-1)
distribution = counts / (np.vectorize(lambda x : x if x!=0 else 1)(totals.reshape(totals.shape+(1,))))
with open(filename+".pkl",'wb') as pickle_file:
pickle.dump(distribution,pickle_file)
def generate_word(dist):
previous = [glyph_to_int['WORD_START']]*(window_size-1)
word = []
while previous[-1]!=glyph_to_int['WORD_END']:
if distribution[tuple(previous)].sum()==0:
next_char = np.random.choice(range(num_glyphs))
print("Uh oh! This shouldn't happen, right?")
else:
next_char = np.random.choice(range(num_glyphs),p=distribution[tuple(previous)])
previous = previous[1:]+[next_char]
word.append(next_char)
return ''.join(int_to_glyph[c] for c in word[:-1])
for i in range(100):
word = generate_word(distribution)
if len(word)>4:
print(word)
```
Next: Look into what words are getting dropped by extract_words, just in case you're still dropping things you shouldn't. Then generate dist (probably with window size of 3 not 4) for english by using wikipedia data and pickle that for later. Have a progress bar for this, Do the same with at least one other language, avoiding the ones that the epitran docs suggest to avoid. Then look into generating phonology and orthography, by using panphon somehow, and try the distribution merging idea.
```
np.count_nonzero(distribution)
```
|
github_jupyter
|
It's going to be a very large file, several gigabytes.
Replace "en" by the appropriate [language code](https://en.wikipedia.org/wiki/List_of_ISO_639-1_codes) to grab it for a different language.
Run
to get a cleaned up version.
Alternatively, there is some pre-cleaned text available for download at the [polyglot project](https://sites.google.com/site/rmyeid/projects/polyglot).
Next: Look into what words are getting dropped by extract_words, just in case you're still dropping things you shouldn't. Then generate dist (probably with window size of 3 not 4) for english by using wikipedia data and pickle that for later. Have a progress bar for this, Do the same with at least one other language, avoiding the ones that the epitran docs suggest to avoid. Then look into generating phonology and orthography, by using panphon somehow, and try the distribution merging idea.
| 0.41834 | 0.734072 |
## Basics of Calculus
Calculus is the study of how things change. It provides a framework to construct relatively simple quantitative models of change, and to deduce their consequences. If you graph a quadratic you will notice that you do not get a straight line. Think of this like you moving on earth. Though the earth is round, as we walk down the street it looks pretty flat to us.
If you look at a quadratic function $f$ at some particular argument, call it $z$, and very close to $z$, then $f$ will look like a straight line. This slope is often written as
$f'\left( x \right) = y' = \frac{{df}}{{dx}} = \frac{{dy}}{{dx}} = \frac{d}{{dx}}\left( {f\left( x \right)} \right) = \frac{d}{{dx}}\left( y \right)$
### What is the rate of change?
On if the important applications of derivatives is finding the rate of change. That is the fact that $f'\left( x \right)$ represents the rate of change of $f\left( x \right)$.
We can think of it by using an example. Think of a car, in finding the velocity of the car, we are given the position of the car at time $t$. To compute the velocity of the car we just need find the rate at which the position is changing. This can be cacluated using the function below.
$\begin{align*}Velocity & = \frac{{{\mbox{change in position}}}}{{{\mbox{time traveled}}}}\end{align*}$
$\begin{align*}Rate of Change = \frac{{f\left( t \right) - f\left( a \right)}}{{t - a}}\end{align*}$
```
from IPython.display import YouTubeVideo
YouTubeVideo('K2jQ0AGbYaA', width=860, height=460)
import numpy as np
import matplotlib.pyplot as plt
from sympy import *
x = symbols('x')
expr = cos(x)
print("Expression : {}".format(expr))
limit_expr = limit(expr, x, 0)
print("Limit of expression tends to 0 : {}".format(limit_expr))
```
### Introduction to differentiation
So far we've learned how to evaluate limits for points on a line. Now we're going to build on that knowledge and look at a calculus technique called differentiation. In differentiation, we use our knowledge of limits to calculate the derivative of a function in order to determine the rate of change at an individual point on its line. The word differentiation is derived from the word difference (subtraction). The known difference of function $f(x)$ at a point $x$ given a chnage of $\Delta x$ is simply the difference between the values of the function at those points:
The derivative of $f\left( x \right)$ with respect to x is the function $f'\left( x \right)$ and is defined as, $\begin{equation}f'\left( x \right) = \mathop {\lim }\limits_{h \to 0} \frac{{f\left( {x + h} \right) - f\left( x \right)}}{h} \label{eq:eq2}\end{equation}$
```
from IPython.display import YouTubeVideo
YouTubeVideo('a5WVw9vmGHU', width=860, height=460)
import numpy as np
import matplotlib.pyplot as plt
from sympy import *
x = symbols('x')
expr = diff(x**2 +x , x)
print("Expression : {}".format(expr))
print(expr.subs(x,3))
expr = diff(cos(x) +x , x)
print("Expression : {}".format(expr))
print(expr.subs(x,0))
```
### Introduction to Integration
Lets say we want to find the area under a function between the function line and the X axis. This is called the integral of the function and it's expressed like this. $\int{{f\left( x \right)\,dx}}$ . To calculate the integral of $f(x)$ with respect to $x$, we need to find the anti-derivative of $f(x)$ , which means we needs a function whose derivative is $f(x)$ . By calculating the parallel in reverse, we can figure out that this is one half X squared.
To do so there are many techniques and methods. Lets discuss them in the video in detail.
```
from IPython.display import YouTubeVideo
YouTubeVideo('5pwZchmmgF0', width=860, height=460)
x_range = np.linspace(0,2,1000)
f = lambda x:np.exp(x)
plt.plot(x_range, f(x_range))
plt.fill_between(x_range, 0, f(x_range), alpha = 0.25)
plt.title("Area below the curve")
import numpy as np
import matplotlib.pyplot as plt
from sympy import *
x = symbols('x')
expr = integrate(x**2 +x , x)
print("Expression : {}".format(expr))
print(expr.subs(x,3))
expr = integrate(cos(x) +x , x)
print("Expression : {}".format(expr))
print(expr.subs(x,3))
```
### Exercises
1. Determine where the following function is increasing and decreasing : $ A\left( t \right) = 17{t^3} - 45{t^2} - 130{t} + 150$. Integration can be used to find areas, volumes, central points and many useful things.
2. Sketch the graph of $f\left( x \right) = 3 - {\frac{1}{2}}x $ and identify all the minima and maxima of the function on each of the following intervals.
a). $\left( { - \infty ,\infty } \right)$
b). $\left[ { - 1,1} \right]$
c). $\left[ {1,4} \right]$
3. Evaluate the following indefinite integrals: $\displaystyle \int{{7{x^6} - 12{x^3} - 5\,dx}}$
**Note** : The following tutorial is a brief introduction to Calculus. You Can dive more deeper using the various resources and books availabe [online](https://www.sydney.edu.au/stuserv/documents/maths_learning_centre/)
|
github_jupyter
|
from IPython.display import YouTubeVideo
YouTubeVideo('K2jQ0AGbYaA', width=860, height=460)
import numpy as np
import matplotlib.pyplot as plt
from sympy import *
x = symbols('x')
expr = cos(x)
print("Expression : {}".format(expr))
limit_expr = limit(expr, x, 0)
print("Limit of expression tends to 0 : {}".format(limit_expr))
from IPython.display import YouTubeVideo
YouTubeVideo('a5WVw9vmGHU', width=860, height=460)
import numpy as np
import matplotlib.pyplot as plt
from sympy import *
x = symbols('x')
expr = diff(x**2 +x , x)
print("Expression : {}".format(expr))
print(expr.subs(x,3))
expr = diff(cos(x) +x , x)
print("Expression : {}".format(expr))
print(expr.subs(x,0))
from IPython.display import YouTubeVideo
YouTubeVideo('5pwZchmmgF0', width=860, height=460)
x_range = np.linspace(0,2,1000)
f = lambda x:np.exp(x)
plt.plot(x_range, f(x_range))
plt.fill_between(x_range, 0, f(x_range), alpha = 0.25)
plt.title("Area below the curve")
import numpy as np
import matplotlib.pyplot as plt
from sympy import *
x = symbols('x')
expr = integrate(x**2 +x , x)
print("Expression : {}".format(expr))
print(expr.subs(x,3))
expr = integrate(cos(x) +x , x)
print("Expression : {}".format(expr))
print(expr.subs(x,3))
| 0.377541 | 0.993765 |
<h1 style='color:brown;'>---------------Language translation using Deep learning LSTM's----------------</h1>
<h1>Table of contents</h1>
<a href='#intro' style="text-decoration:none;color:purple;">1) Intro to Seq2Seq models </a>
<a href='#trad' style="text-decoration:none;color:purple;">2) Traditional phrase based statistical translation model</a>
<a href='#arch' style="text-decoration:none;color:purple;">3) Architecture of Sequence to Sequence model </a>
<a href='#keras' style="text-decoration:none;color:purple;">4) Keras and tensorflow support</a>
<a href='#other' style="text-decoration:none;color:purple;">5) Other methods for machine translation</a>
<a href='#data' style="text-decoration:none;color:purple;">6) Datasets available for language translation</a>
<a href='#code' style="text-decoration:none;color:purple;">7) Code snippet in keras </a>
<a href='#transfer' style="text-decoration:none;color:purple;">8) Transfer learning for machine translation</a>
<a href='#metric' style="text-decoration:none;color:purple;">9) Evaluation metrics and loss function</a>
<a id='intro'></a>
## 1) Intro to Seq2Seq models for language translation
### Sequence-to-sequence (seq2seq) models have enjoyed great success in a variety of tasks such as machine translation, speech recognition, and text summarization.
Use of Seq2Seq models in language translation: Converting english text to french language:
### "the cat sat on the mat" -> [Seq2Seq model] -> "le chat etait assis sur le tapis"

Encoder-decoder architecture – example of a general approach for NMT. An encoder converts a source sentence into a "meaning" vector which is passed through a decoder to produce a translation.
<a id='trad'></a>
## 2) Traditional method Phrase based translation A.K.A Statistical machine translation
For more check out:
https://en.wikipedia.org/wiki/Statistical_machine_translation

<h3>Back in the old days, traditional phrase-based translation systems performed their task by breaking up source sentences into multiple chunks and then translated them phrase-by-phrase. This led to disfluency in the translation outputs and was not quite like how we, humans, translate. We read the entire source sentence, understand its meaning, and then produce a translation. Neural Machine Translation (NMT) mimics that!</h3>
<a id='arch'></a>
## 3) Architecture of Seq2Seq model:


Above is the basic architecture of Seq2Seq model which has an:
1) Encoder which converts input(Source language sentence) into meaning vector(Context).
2) Note that here output of encoder is discarded, but returns the internal state as a context to decoder.
3) Then decoder translates the Source language to target language by predicting the next output in a sequential manner.
<a id='keras'></a>
## 4) Keras and tensorflow support for seq2seq models
Seq2Seq models can be created both in tensorflow as well as in keras.
Steps in creating a Seq2Seq model:
1) Prepare encoder input data, decoder input data, decoder target data.
2) Train a basic LSTM-based Seq2Seq model to predict decoder_target_data given encoder_input_data and decoder_input_data.
3) Decode some sentences to check that the model is working
For more information checkout:
https://www.tensorflow.org/tutorials/seq2seq
https://blog.keras.io/a-ten-minute-introduction-to-sequence-to-sequence-learning-in-keras.html
<a id='other'></a>
## 5) Other translation methods available:
<ul>
<li><a href="https://en.wikipedia.org/wiki/Machine_translation" title="Machine translation">Machine translation</a></li>
<li><a href="https://en.wikipedia.org/wiki/Rule-based_machine_translation" title="Rule-based machine translation">Rule-based machine translation</a></li>
<li><a href="https://en.wikipedia.org/wiki/Transfer-based_machine_translation" title="Transfer-based machine translation">Transfer-based machine translation</a></li>
<li><a href="https://en.wikipedia.org/wiki/Interlingual_machine_translation" title="Interlingual machine translation">Interlingual machine translation</a></li>
<li><a href="https://en.wikipedia.org/wiki/Statistical_machine_translation" title="Statistical machine translation">Statistical machine translation</a></li>
<li><a href="https://en.wikipedia.org/wiki/Example-based_machine_translation" title="Example-based machine translation">Example-based machine translation</a></li>
</ul>
<a id='data'></a>
## 6) Datasets available for Neural machine translation:
1) Small-scale: English-Vietnamese parallel corpus of TED talks (133K sentence pairs) provided by the IWSLT Evaluation Campaign.
2) Large-scale: German-English parallel corpus (4.5M sentence pairs) provided by the WMT Evaluation Campaign.
3) Stanford NLP group: https://nlp.stanford.edu/projects/nmt/
4) Data sets for various languages for creating base models: http://www.manythings.org/anki/
<a id='code'></a>
## 7) Code snippet for Seq2Seq model in keras:
<code style='color:purple'>
from keras.models import Model
from keras.layers import Input, LSTM, Dense
#Define an input sequence and process it.
encoder_inputs = Input(shape=(None, num_encoder_tokens))
encoder = LSTM(latent_dim, return_state=True)
encoder_outputs, state_h, state_c = encoder(encoder_inputs)
#We discard `encoder_outputs` and only keep the states.
encoder_states = [state_h, state_c]
#Set up the decoder, using `encoder_states` as initial state.
decoder_inputs = Input(shape=(None, num_decoder_tokens))
#We set up our decoder to return full output sequences,
and to return internal states as well. We don't use the
return states in the training model, but we will use them in inference.
decoder_lstm = LSTM(latent_dim, return_sequences=True, return_state=True)
decoder_outputs, _, _ = decoder_lstm(decoder_inputs,
initial_state=encoder_states)
decoder_dense = Dense(num_decoder_tokens, activation='softmax')
decoder_outputs = decoder_dense(decoder_outputs)
#Define the model that will turn
#`encoder_input_data` & `decoder_input_data` into `decoder_target_data`
model = Model([encoder_inputs, decoder_inputs], decoder_outputs)
</code>
<a id='transfer'></a>
## 8) Transfer learning in Machine translation:
The encoder-decoder framework for neural
machine translation (NMT) has been shown
effective in large data scenarios, but is much
less effective for low-resource languages. We
present a transfer learning method that signifi-
cantly improves BLEU scores across a range
of low-resource languages. Our key idea is
to first train a high-resource language pair
(the parent model), then transfer some of the
learned parameters to the low-resource pair
(the child model) to initialize and constrain
training.
http://www.aclweb.org/anthology/D16-1163
https://research.googleblog.com/2016/11/zero-shot-translation-with-googles.html
#### A spectacular example of Transfer Learning would be <h3>Google’s Multilingual Neural Machine Translation (GNMT) system.</h3>
In GNMT, a single model is trained to translate between language pairs such as English⇄Korean and English⇄Japanese. That is, samples consisting of translation pairs of English⇄Korean and English⇄Japanese are used to train a unified model.

Zero-shot translation
The GNMT system is said to represent an improvement over the former Google Translate in that it can handle "zero-shot translation", that is it directly translates one language into another (for example, Japanese to Korean).[2] Google Translate previously first translated the source language into English and then translated the English into the target language rather than translating directly from one language to another.[4]
## Model consists of a deep LSTM network with 8 encoder and 8 decoder layers using residual connections as well as attention connections from the decoder network to the encoder.
By using a single model to translate between any two languages, the model is forced to learn universal features common to all languages. This enables the system to do “Zero-Shot Translation”: the model is able to translate between a language pair for which it hasn’t explicitly seen any training data. In the case of English, Japanese and Korean, a model trained to translate between English⇄Korean and English⇄Japanese is also able to translate between Korean⇄Japanese without any explicit supervised training.

https://research.googleblog.com/2016/11/zero-shot-translation-with-googles.html
https://arxiv.org/pdf/1611.04558.pdf
<a id='metric'></a>
## 9) Evaluation metics and loss function for Language translation model:
Automatic evaluation
1 BLEU
2 NIST
3 Word error rate
4 METEOR
5 LEPOR
Most commonly used metric is the BLEU score, it stands for (bilingual evaluation understudy).
BLEU was one of the first metrics to report high correlation with human judgments of quality. The metric is currently one of the most popular in the field. The central idea behind the metric is that "the closer a machine translation is to a professional human translation, the better it is". The metric calculates scores for individual segments, generally sentences—then averages these scores over the whole corpus for a final score. It has been shown to correlate highly with human judgments of quality at the corpus level.
BLEU uses a modified form of precision to compare a candidate translation against multiple reference translations. The metric modifies simple precision since machine translation systems have been known to generate more words than appear in a reference text. No other machine translation metric is yet to significantly outperform BLEU with respect to correlation with human judgment across language pairs.
<h2>BLEU’s evaluation system requires two inputs:
<li>
a numerical translation closeness metric, which is then assigned and measured against
</li>
<li>
a corpus of human reference translations.
</li>
</h2>
Loss is the categorical cross entropy loss.
## Time taken to train On WMT En→Fr, the training set contains 36M sentence pairs
### ---On WMT En→Fr, it takes around 6 days to train a basic model using 96 NVIDIA K80 GPUs.
Attention is all you need paper on using attention mechanism on seq2seq models
https://arxiv.org/pdf/1706.03762.pdf

## Google neural machine translator architecture:

|
github_jupyter
|
<h1 style='color:brown;'>---------------Language translation using Deep learning LSTM's----------------</h1>
<h1>Table of contents</h1>
<a href='#intro' style="text-decoration:none;color:purple;">1) Intro to Seq2Seq models </a>
<a href='#trad' style="text-decoration:none;color:purple;">2) Traditional phrase based statistical translation model</a>
<a href='#arch' style="text-decoration:none;color:purple;">3) Architecture of Sequence to Sequence model </a>
<a href='#keras' style="text-decoration:none;color:purple;">4) Keras and tensorflow support</a>
<a href='#other' style="text-decoration:none;color:purple;">5) Other methods for machine translation</a>
<a href='#data' style="text-decoration:none;color:purple;">6) Datasets available for language translation</a>
<a href='#code' style="text-decoration:none;color:purple;">7) Code snippet in keras </a>
<a href='#transfer' style="text-decoration:none;color:purple;">8) Transfer learning for machine translation</a>
<a href='#metric' style="text-decoration:none;color:purple;">9) Evaluation metrics and loss function</a>
<a id='intro'></a>
## 1) Intro to Seq2Seq models for language translation
### Sequence-to-sequence (seq2seq) models have enjoyed great success in a variety of tasks such as machine translation, speech recognition, and text summarization.
Use of Seq2Seq models in language translation: Converting english text to french language:
### "the cat sat on the mat" -> [Seq2Seq model] -> "le chat etait assis sur le tapis"

Encoder-decoder architecture – example of a general approach for NMT. An encoder converts a source sentence into a "meaning" vector which is passed through a decoder to produce a translation.
<a id='trad'></a>
## 2) Traditional method Phrase based translation A.K.A Statistical machine translation
For more check out:
https://en.wikipedia.org/wiki/Statistical_machine_translation

<h3>Back in the old days, traditional phrase-based translation systems performed their task by breaking up source sentences into multiple chunks and then translated them phrase-by-phrase. This led to disfluency in the translation outputs and was not quite like how we, humans, translate. We read the entire source sentence, understand its meaning, and then produce a translation. Neural Machine Translation (NMT) mimics that!</h3>
<a id='arch'></a>
## 3) Architecture of Seq2Seq model:


Above is the basic architecture of Seq2Seq model which has an:
1) Encoder which converts input(Source language sentence) into meaning vector(Context).
2) Note that here output of encoder is discarded, but returns the internal state as a context to decoder.
3) Then decoder translates the Source language to target language by predicting the next output in a sequential manner.
<a id='keras'></a>
## 4) Keras and tensorflow support for seq2seq models
Seq2Seq models can be created both in tensorflow as well as in keras.
Steps in creating a Seq2Seq model:
1) Prepare encoder input data, decoder input data, decoder target data.
2) Train a basic LSTM-based Seq2Seq model to predict decoder_target_data given encoder_input_data and decoder_input_data.
3) Decode some sentences to check that the model is working
For more information checkout:
https://www.tensorflow.org/tutorials/seq2seq
https://blog.keras.io/a-ten-minute-introduction-to-sequence-to-sequence-learning-in-keras.html
<a id='other'></a>
## 5) Other translation methods available:
<ul>
<li><a href="https://en.wikipedia.org/wiki/Machine_translation" title="Machine translation">Machine translation</a></li>
<li><a href="https://en.wikipedia.org/wiki/Rule-based_machine_translation" title="Rule-based machine translation">Rule-based machine translation</a></li>
<li><a href="https://en.wikipedia.org/wiki/Transfer-based_machine_translation" title="Transfer-based machine translation">Transfer-based machine translation</a></li>
<li><a href="https://en.wikipedia.org/wiki/Interlingual_machine_translation" title="Interlingual machine translation">Interlingual machine translation</a></li>
<li><a href="https://en.wikipedia.org/wiki/Statistical_machine_translation" title="Statistical machine translation">Statistical machine translation</a></li>
<li><a href="https://en.wikipedia.org/wiki/Example-based_machine_translation" title="Example-based machine translation">Example-based machine translation</a></li>
</ul>
<a id='data'></a>
## 6) Datasets available for Neural machine translation:
1) Small-scale: English-Vietnamese parallel corpus of TED talks (133K sentence pairs) provided by the IWSLT Evaluation Campaign.
2) Large-scale: German-English parallel corpus (4.5M sentence pairs) provided by the WMT Evaluation Campaign.
3) Stanford NLP group: https://nlp.stanford.edu/projects/nmt/
4) Data sets for various languages for creating base models: http://www.manythings.org/anki/
<a id='code'></a>
## 7) Code snippet for Seq2Seq model in keras:
<code style='color:purple'>
from keras.models import Model
from keras.layers import Input, LSTM, Dense
#Define an input sequence and process it.
encoder_inputs = Input(shape=(None, num_encoder_tokens))
encoder = LSTM(latent_dim, return_state=True)
encoder_outputs, state_h, state_c = encoder(encoder_inputs)
#We discard `encoder_outputs` and only keep the states.
encoder_states = [state_h, state_c]
#Set up the decoder, using `encoder_states` as initial state.
decoder_inputs = Input(shape=(None, num_decoder_tokens))
#We set up our decoder to return full output sequences,
and to return internal states as well. We don't use the
return states in the training model, but we will use them in inference.
decoder_lstm = LSTM(latent_dim, return_sequences=True, return_state=True)
decoder_outputs, _, _ = decoder_lstm(decoder_inputs,
initial_state=encoder_states)
decoder_dense = Dense(num_decoder_tokens, activation='softmax')
decoder_outputs = decoder_dense(decoder_outputs)
#Define the model that will turn
#`encoder_input_data` & `decoder_input_data` into `decoder_target_data`
model = Model([encoder_inputs, decoder_inputs], decoder_outputs)
</code>
<a id='transfer'></a>
## 8) Transfer learning in Machine translation:
The encoder-decoder framework for neural
machine translation (NMT) has been shown
effective in large data scenarios, but is much
less effective for low-resource languages. We
present a transfer learning method that signifi-
cantly improves BLEU scores across a range
of low-resource languages. Our key idea is
to first train a high-resource language pair
(the parent model), then transfer some of the
learned parameters to the low-resource pair
(the child model) to initialize and constrain
training.
http://www.aclweb.org/anthology/D16-1163
https://research.googleblog.com/2016/11/zero-shot-translation-with-googles.html
#### A spectacular example of Transfer Learning would be <h3>Google’s Multilingual Neural Machine Translation (GNMT) system.</h3>
In GNMT, a single model is trained to translate between language pairs such as English⇄Korean and English⇄Japanese. That is, samples consisting of translation pairs of English⇄Korean and English⇄Japanese are used to train a unified model.

Zero-shot translation
The GNMT system is said to represent an improvement over the former Google Translate in that it can handle "zero-shot translation", that is it directly translates one language into another (for example, Japanese to Korean).[2] Google Translate previously first translated the source language into English and then translated the English into the target language rather than translating directly from one language to another.[4]
## Model consists of a deep LSTM network with 8 encoder and 8 decoder layers using residual connections as well as attention connections from the decoder network to the encoder.
By using a single model to translate between any two languages, the model is forced to learn universal features common to all languages. This enables the system to do “Zero-Shot Translation”: the model is able to translate between a language pair for which it hasn’t explicitly seen any training data. In the case of English, Japanese and Korean, a model trained to translate between English⇄Korean and English⇄Japanese is also able to translate between Korean⇄Japanese without any explicit supervised training.

https://research.googleblog.com/2016/11/zero-shot-translation-with-googles.html
https://arxiv.org/pdf/1611.04558.pdf
<a id='metric'></a>
## 9) Evaluation metics and loss function for Language translation model:
Automatic evaluation
1 BLEU
2 NIST
3 Word error rate
4 METEOR
5 LEPOR
Most commonly used metric is the BLEU score, it stands for (bilingual evaluation understudy).
BLEU was one of the first metrics to report high correlation with human judgments of quality. The metric is currently one of the most popular in the field. The central idea behind the metric is that "the closer a machine translation is to a professional human translation, the better it is". The metric calculates scores for individual segments, generally sentences—then averages these scores over the whole corpus for a final score. It has been shown to correlate highly with human judgments of quality at the corpus level.
BLEU uses a modified form of precision to compare a candidate translation against multiple reference translations. The metric modifies simple precision since machine translation systems have been known to generate more words than appear in a reference text. No other machine translation metric is yet to significantly outperform BLEU with respect to correlation with human judgment across language pairs.
<h2>BLEU’s evaluation system requires two inputs:
<li>
a numerical translation closeness metric, which is then assigned and measured against
</li>
<li>
a corpus of human reference translations.
</li>
</h2>
Loss is the categorical cross entropy loss.
## Time taken to train On WMT En→Fr, the training set contains 36M sentence pairs
### ---On WMT En→Fr, it takes around 6 days to train a basic model using 96 NVIDIA K80 GPUs.
Attention is all you need paper on using attention mechanism on seq2seq models
https://arxiv.org/pdf/1706.03762.pdf

## Google neural machine translator architecture:

| 0.872714 | 0.802168 |
```
#hide
!pip install -Uqq fastbook
import fastbook
fastbook.setup_book()
#hide
from fastbook import *
```
# Your Deep Learning Journey
Hello, and thank you for letting us join you on your deep learning journey, however far along that you may be! In this chapter, we will tell you a little bit more about what to expect in this book, introduce the key concepts behind deep learning, and train our first models on different tasks. It doesn't matter if you don't come from a technical or a mathematical background (though it's okay if you do too!); we wrote this book to make deep learning accessible to as many people as possible.
## Deep Learning Is for Everyone
A lot of people assume that you need all kinds of hard-to-find stuff to get great results with deep learning, but as you'll see in this book, those people are wrong. <<myths>> is a list of a few thing you *absolutely don't need* to do world-class deep learning.
```asciidoc
[[myths]]
.What you don't need to do deep learning
[options="header"]
|======
| Myth (don't need) | Truth
| Lots of math | Just high school math is sufficient
| Lots of data | We've seen record-breaking results with <50 items of data
| Lots of expensive computers | You can get what you need for state of the art work for free
|======
```
Deep learning is a computer technique to extract and transform data–-with use cases ranging from human speech recognition to animal imagery classification–-by using multiple layers of neural networks. Each of these layers takes its inputs from previous layers and progressively refines them. The layers are trained by algorithms that minimize their errors and improve their accuracy. In this way, the network learns to perform a specified task. We will discuss training algorithms in detail in the next section.
Deep learning has power, flexibility, and simplicity. That's why we believe it should be applied across many disciplines. These include the social and physical sciences, the arts, medicine, finance, scientific research, and many more. To give a personal example, despite having no background in medicine, Jeremy started Enlitic, a company that uses deep learning algorithms to diagnose illness and disease. Within months of starting the company, it was announced that its algorithm could identify malignant tumors [more accurately than radiologists](https://www.nytimes.com/2016/02/29/technology/the-promise-of-artificial-intelligence-unfolds-in-small-steps.html).
Here's a list of some of the thousands of tasks in different areas at which deep learning, or methods heavily using deep learning, is now the best in the world:
- Natural language processing (NLP):: Answering questions; speech recognition; summarizing documents; classifying documents; finding names, dates, etc. in documents; searching for articles mentioning a concept
- Computer vision:: Satellite and drone imagery interpretation (e.g., for disaster resilience); face recognition; image captioning; reading traffic signs; locating pedestrians and vehicles in autonomous vehicles
- Medicine:: Finding anomalies in radiology images, including CT, MRI, and X-ray images; counting features in pathology slides; measuring features in ultrasounds; diagnosing diabetic retinopathy
- Biology:: Folding proteins; classifying proteins; many genomics tasks, such as tumor-normal sequencing and classifying clinically actionable genetic mutations; cell classification; analyzing protein/protein interactions
- Image generation:: Colorizing images; increasing image resolution; removing noise from images; converting images to art in the style of famous artists
- Recommendation systems:: Web search; product recommendations; home page layout
- Playing games:: Chess, Go, most Atari video games, and many real-time strategy games
- Robotics:: Handling objects that are challenging to locate (e.g., transparent, shiny, lacking texture) or hard to pick up
- Other applications:: Financial and logistical forecasting, text to speech, and much more...
What is remarkable is that deep learning has such varied application yet nearly all of deep learning is based on a single type of model, the neural network.
But neural networks are not in fact completely new. In order to have a wider perspective on the field, it is worth it to start with a bit of history.
## Neural Networks: A Brief History
In 1943 Warren McCulloch, a neurophysiologist, and Walter Pitts, a logician, teamed up to develop a mathematical model of an artificial neuron. In their [paper](https://link.springer.com/article/10.1007/BF02478259) "A Logical Calculus of the Ideas Immanent in Nervous Activity" they declared that:
> : Because of the “all-or-none” character of nervous activity, neural events and the relations among them can be treated by means of propositional logic. It is found that the behavior of every net can be described in these terms.
McCulloch and Pitts realized that a simplified model of a real neuron could be represented using simple addition and thresholding, as shown in <<neuron>>. Pitts was self-taught, and by age 12, had received an offer to study at Cambridge University with the great Bertrand Russell. He did not take up this invitation, and indeed throughout his life did not accept any offers of advanced degrees or positions of authority. Most of his famous work was done while he was homeless. Despite his lack of an officially recognized position and increasing social isolation, his work with McCulloch was influential, and was taken up by a psychologist named Frank Rosenblatt.
<img alt="Natural and artificial neurons" width="500" caption="Natural and artificial neurons" src="images/chapter7_neuron.png" id="neuron"/>
Rosenblatt further developed the artificial neuron to give it the ability to learn. Even more importantly, he worked on building the first device that actually used these principles, the Mark I Perceptron. In "The Design of an Intelligent Automaton" Rosenblatt wrote about this work: "We are now about to witness the birth of such a machine–-a machine capable of perceiving, recognizing and identifying its surroundings without any human training or control." The perceptron was built, and was able to successfully recognize simple shapes.
An MIT professor named Marvin Minsky (who was a grade behind Rosenblatt at the same high school!), along with Seymour Papert, wrote a book called _Perceptrons_ (MIT Press), about Rosenblatt's invention. They showed that a single layer of these devices was unable to learn some simple but critical mathematical functions (such as XOR). In the same book, they also showed that using multiple layers of the devices would allow these limitations to be addressed. Unfortunately, only the first of these insights was widely recognized. As a result, the global academic community nearly entirely gave up on neural networks for the next two decades.
Perhaps the most pivotal work in neural networks in the last 50 years was the multi-volume *Parallel Distributed Processing* (PDP) by David Rumelhart, James McClellan, and the PDP Research Group, released in 1986 by MIT Press. Chapter 1 lays out a similar hope to that shown by Rosenblatt:
> : People are smarter than today's computers because the brain employs a basic computational architecture that is more suited to deal with a central aspect of the natural information processing tasks that people are so good at. ...We will introduce a computational framework for modeling cognitive processes that seems… closer than other frameworks to the style of computation as it might be done by the brain.
The premise that PDP is using here is that traditional computer programs work very differently to brains, and that might be why computer programs had been (at that point) so bad at doing things that brains find easy (such as recognizing objects in pictures). The authors claimed that the PDP approach was "closer
than other frameworks" to how the brain works, and therefore it might be better able to handle these kinds of tasks.
In fact, the approach laid out in PDP is very similar to the approach used in today's neural networks. The book defined parallel distributed processing as requiring:
1. A set of *processing units*
1. A *state of activation*
1. An *output function* for each unit
1. A *pattern of connectivity* among units
1. A *propagation rule* for propagating patterns of activities through the network of connectivities
1. An *activation rule* for combining the inputs impinging on a unit with the current state of that unit to produce an output for the unit
1. A *learning rule* whereby patterns of connectivity are modified by experience
1. An *environment* within which the system must operate
We will see in this book that modern neural networks handle each of these requirements.
In the 1980's most models were built with a second layer of neurons, thus avoiding the problem that had been identified by Minsky and Papert (this was their "pattern of connectivity among units," to use the framework above). And indeed, neural networks were widely used during the '80s and '90s for real, practical projects. However, again a misunderstanding of the theoretical issues held back the field. In theory, adding just one extra layer of neurons was enough to allow any mathematical function to be approximated with these neural networks, but in practice such networks were often too big and too slow to be useful.
Although researchers showed 30 years ago that to get practical good performance you need to use even more layers of neurons, it is only in the last decade that this principle has been more widely appreciated and applied. Neural networks are now finally living up to their potential, thanks to the use of more layers, coupled with the capacity to do so due to improvements in computer hardware, increases in data availability, and algorithmic tweaks that allow neural networks to be trained faster and more easily. We now have what Rosenblatt promised: "a machine capable of perceiving, recognizing, and identifying its surroundings without any human training or control."
This is what you will learn how to build in this book. But first, since we are going to be spending a lot of time together, let's get to know each other a bit…
## Who We Are
We are Sylvain and Jeremy, your guides on this journey. We hope that you will find us well suited for this position.
Jeremy has been using and teaching machine learning for around 30 years. He started using neural networks 25 years ago. During this time, he has led many companies and projects that have machine learning at their core, including founding the first company to focus on deep learning and medicine, Enlitic, and taking on the role of President and Chief Scientist of the world's largest machine learning community, Kaggle. He is the co-founder, along with Dr. Rachel Thomas, of fast.ai, the organization that built the course this book is based on.
From time to time you will hear directly from us, in sidebars like this one from Jeremy:
> J: Hi everybody, I'm Jeremy! You might be interested to know that I do not have any formal technical education. I completed a BA, with a major in philosophy, and didn't have great grades. I was much more interested in doing real projects, rather than theoretical studies, so I worked full time at a management consulting firm called McKinsey and Company throughout my university years. If you're somebody who would rather get their hands dirty building stuff than spend years learning abstract concepts, then you will understand where I am coming from! Look out for sidebars from me to find information most suited to people with a less mathematical or formal technical background—that is, people like me…
Sylvain, on the other hand, knows a lot about formal technical education. In fact, he has written 10 math textbooks, covering the entire advanced French maths curriculum!
> S: Unlike Jeremy, I have not spent many years coding and applying machine learning algorithms. Rather, I recently came to the machine learning world, by watching Jeremy's fast.ai course videos. So, if you are somebody who has not opened a terminal and written commands at the command line, then you will understand where I am coming from! Look out for sidebars from me to find information most suited to people with a more mathematical or formal technical background, but less real-world coding experience—that is, people like me…
The fast.ai course has been studied by hundreds of thousands of students, from all walks of life, from all parts of the world. Sylvain stood out as the most impressive student of the course that Jeremy had ever seen, which led to him joining fast.ai, and then becoming the coauthor, along with Jeremy, of the fastai software library.
All this means that between us you have the best of both worlds: the people who know more about the software than anybody else, because they wrote it; an expert on math, and an expert on coding and machine learning; and also people who understand both what it feels like to be a relative outsider in math, and a relative outsider in coding and machine learning.
Anybody who has watched sports knows that if you have a two-person commentary team then you also need a third person to do "special comments." Our special commentator is Alexis Gallagher. Alexis has a very diverse background: he has been a researcher in mathematical biology, a screenplay writer, an improv performer, a McKinsey consultant (like Jeremy!), a Swift coder, and a CTO.
> A: I've decided it's time for me to learn about this AI stuff! After all, I've tried pretty much everything else… But I don't really have a background in building machine learning models. Still… how hard can it be? I'm going to be learning throughout this book, just like you are. Look out for my sidebars for learning tips that I found helpful on my journey, and hopefully you will find helpful too.
## How to Learn Deep Learning
Harvard professor David Perkins, who wrote _Making Learning Whole_ (Jossey-Bass), has much to say about teaching. The basic idea is to teach the *whole game*. That means that if you're teaching baseball, you first take people to a baseball game or get them to play it. You don't teach them how to wind twine to make a baseball from scratch, the physics of a parabola, or the coefficient of friction of a ball on a bat.
Paul Lockhart, a Columbia math PhD, former Brown professor, and K-12 math teacher, imagines in the influential [essay](https://www.maa.org/external_archive/devlin/LockhartsLament.pdf) "A Mathematician's Lament" a nightmare world where music and art are taught the way math is taught. Children are not allowed to listen to or play music until they have spent over a decade mastering music notation and theory, spending classes transposing sheet music into a different key. In art class, students study colors and applicators, but aren't allowed to actually paint until college. Sound absurd? This is how math is taught–-we require students to spend years doing rote memorization and learning dry, disconnected *fundamentals* that we claim will pay off later, long after most of them quit the subject.
Unfortunately, this is where many teaching resources on deep learning begin–-asking learners to follow along with the definition of the Hessian and theorems for the Taylor approximation of your loss functions, without ever giving examples of actual working code. We're not knocking calculus. We love calculus, and Sylvain has even taught it at the college level, but we don't think it's the best place to start when learning deep learning!
In deep learning, it really helps if you have the motivation to fix your model to get it to do better. That's when you start learning the relevant theory. But you need to have the model in the first place. We teach almost everything through real examples. As we build out those examples, we go deeper and deeper, and we'll show you how to make your projects better and better. This means that you'll be gradually learning all the theoretical foundations you need, in context, in such a way that you'll see why it matters and how it works.
So, here's our commitment to you. Throughout this book, we will follow these principles:
- Teaching the *whole game*. We'll start by showing how to use a complete, working, very usable, state-of-the-art deep learning network to solve real-world problems, using simple, expressive tools. And then we'll gradually dig deeper and deeper into understanding how those tools are made, and how the tools that make those tools are made, and so on…
- Always teaching through examples. We'll ensure that there is a context and a purpose that you can understand intuitively, rather than starting with algebraic symbol manipulation.
- Simplifying as much as possible. We've spent years building tools and teaching methods that make previously complex topics very simple.
- Removing barriers. Deep learning has, until now, been a very exclusive game. We're breaking it open, and ensuring that everyone can play.
The hardest part of deep learning is artisanal: how do you know if you've got enough data, whether it is in the right format, if your model is training properly, and, if it's not, what you should do about it? That is why we believe in learning by doing. As with basic data science skills, with deep learning you only get better through practical experience. Trying to spend too much time on the theory can be counterproductive. The key is to just code and try to solve problems: the theory can come later, when you have context and motivation.
There will be times when the journey will feel hard. Times where you feel stuck. Don't give up! Rewind through the book to find the last bit where you definitely weren't stuck, and then read slowly through from there to find the first thing that isn't clear. Then try some code experiments yourself, and Google around for more tutorials on whatever the issue you're stuck with is—often you'll find some different angle on the material might help it to click. Also, it's expected and normal to not understand everything (especially the code) on first reading. Trying to understand the material serially before proceeding can sometimes be hard. Sometimes things click into place after you get more context from parts down the road, from having a bigger picture. So if you do get stuck on a section, try moving on anyway and make a note to come back to it later.
Remember, you don't need any particular academic background to succeed at deep learning. Many important breakthroughs are made in research and industry by folks without a PhD, such as ["Unsupervised Representation Learning with Deep Convolutional Generative Adversarial Networks"](https://arxiv.org/abs/1511.06434)—one of the most influential papers of the last decade—with over 5,000 citations, which was written by Alec Radford when he was an undergraduate. Even at Tesla, where they're trying to solve the extremely tough challenge of making a self-driving car, CEO [Elon Musk says](https://twitter.com/elonmusk/status/1224089444963311616):
> : A PhD is definitely not required. All that matters is a deep understanding of AI & ability to implement NNs in a way that is actually useful (latter point is what’s truly hard). Don’t care if you even graduated high school.
What you will need to do to succeed however is to apply what you learn in this book to a personal project, and always persevere.
### Your Projects and Your Mindset
Whether you're excited to identify if plants are diseased from pictures of their leaves, auto-generate knitting patterns, diagnose TB from X-rays, or determine when a raccoon is using your cat door, we will get you using deep learning on your own problems (via pre-trained models from others) as quickly as possible, and then will progressively drill into more details. You'll learn how to use deep learning to solve your own problems at state-of-the-art accuracy within the first 30 minutes of the next chapter! (And feel free to skip straight there now if you're dying to get coding right away.) There is a pernicious myth out there that you need to have computing resources and datasets the size of those at Google to be able to do deep learning, but it's not true.
So, what sorts of tasks make for good test cases? You could train your model to distinguish between Picasso and Monet paintings or to pick out pictures of your daughter instead of pictures of your son. It helps to focus on your hobbies and passions–-setting yourself four or five little projects rather than striving to solve a big, grand problem tends to work better when you're getting started. Since it is easy to get stuck, trying to be too ambitious too early can often backfire. Then, once you've got the basics mastered, aim to complete something you're really proud of!
> J: Deep learning can be set to work on almost any problem. For instance, my first startup was a company called FastMail, which provided enhanced email services when it launched in 1999 (and still does to this day). In 2002 I set it up to use a primitive form of deep learning, single-layer neural networks, to help categorize emails and stop customers from receiving spam.
Common character traits in the people that do well at deep learning include playfulness and curiosity. The late physicist Richard Feynman is an example of someone who we'd expect to be great at deep learning: his development of an understanding of the movement of subatomic particles came from his amusement at how plates wobble when they spin in the air.
Let's now focus on what you will learn, starting with the software.
## The Software: PyTorch, fastai, and Jupyter
(And Why It Doesn't Matter)
We've completed hundreds of machine learning projects using dozens of different packages, and many different programming languages. At fast.ai, we have written courses using most of the main deep learning and machine learning packages used today. After PyTorch came out in 2017 we spent over a thousand hours testing it before deciding that we would use it for future courses, software development, and research. Since that time PyTorch has become the world's fastest-growing deep learning library and is already used for most research papers at top conferences. This is generally a leading indicator of usage in industry, because these are the papers that end up getting used in products and services commercially. We have found that PyTorch is the most flexible and expressive library for deep learning. It does not trade off speed for simplicity, but provides both.
PyTorch works best as a low-level foundation library, providing the basic operations for higher-level functionality. The fastai library is the most popular library for adding this higher-level functionality on top of PyTorch. It's also particularly well suited to the purposes of this book, because it is unique in providing a deeply layered software architecture (there's even a [peer-reviewed academic paper](https://arxiv.org/abs/2002.04688) about this layered API). In this book, as we go deeper and deeper into the foundations of deep learning, we will also go deeper and deeper into the layers of fastai. This book covers version 2 of the fastai library, which is a from-scratch rewrite providing many unique features.
However, it doesn't really matter what software you learn, because it takes only a few days to learn to switch from one library to another. What really matters is learning the deep learning foundations and techniques properly. Our focus will be on using code that clearly expresses the concepts that you need to learn. Where we are teaching high-level concepts, we will use high-level fastai code. Where we are teaching low-level concepts, we will use low-level PyTorch, or even pure Python code.
If it feels like new deep learning libraries are appearing at a rapid pace nowadays, then you need to be prepared for a much faster rate of change in the coming months and years. As more people enter the field, they will bring more skills and ideas, and try more things. You should assume that whatever specific libraries and software you learn today will be obsolete in a year or two. Just think about the number of changes in libraries and technology stacks that occur all the time in the world of web programming—a much more mature and slow-growing area than deep learning. We strongly believe that the focus in learning needs to be on understanding the underlying techniques and how to apply them in practice, and how to quickly build expertise in new tools and techniques as they are released.
By the end of the book, you'll understand nearly all the code that's inside fastai (and much of PyTorch too), because in each chapter we'll be digging a level deeper to show you exactly what's going on as we build and train our models. This means that you'll have learned the most important best practices used in modern deep learning—not just how to use them, but how they really work and are implemented. If you want to use those approaches in another framework, you'll have the knowledge you need to do so if needed.
Since the most important thing for learning deep learning is writing code and experimenting, it's important that you have a great platform for experimenting with code. The most popular programming experimentation platform is called Jupyter. This is what we will be using throughout this book. We will show you how you can use Jupyter to train and experiment with models and introspect every stage of the data pre-processing and model development pipeline. [Jupyter Notebook](https://jupyter.org/) is the most popular tool for doing data science in Python, for good reason. It is powerful, flexible, and easy to use. We think you will love it!
Let's see it in practice and train our first model.
## Your First Model
As we said before, we will teach you how to do things before we explain why they work. Following this top-down approach, we will begin by actually training an image classifier to recognize dogs and cats with almost 100% accuracy. To train this model and run our experiments, you will need to do some initial setup. Don't worry, it's not as hard as it looks.
> s: Do not skip the setup part even if it looks intimidating at first, especially if you have little or no experience using things like a terminal or the command line. Most of that is actually not necessary and you will find that the easiest servers can be set up with just your usual web browser. It is crucial that you run your own experiments in parallel with this book in order to learn.
### Getting a GPU Deep Learning Server
To do nearly everything in this book, you'll need access to a computer with an NVIDIA GPU (unfortunately other brands of GPU are not fully supported by the main deep learning libraries). However, we don't recommend you buy one; in fact, even if you already have one, we don't suggest you use it just yet! Setting up a computer takes time and energy, and you want all your energy to focus on deep learning right now. Therefore, we instead suggest you rent access to a computer that already has everything you need preinstalled and ready to go. Costs can be as little as US$0.25 per hour while you're using it, and some options are even free.
> jargon: Graphics Processing Unit (GPU): Also known as a _graphics card_. A special kind of processor in your computer that can handle thousands of single tasks at the same time, especially designed for displaying 3D environments on a computer for playing games. These same basic tasks are very similar to what neural networks do, such that GPUs can run neural networks hundreds of times faster than regular CPUs. All modern computers contain a GPU, but few contain the right kind of GPU necessary for deep learning.
The best choice of GPU servers to use with this book will change over time, as companies come and go and prices change. We maintain a list of our recommended options on the [book's website](https://book.fast.ai/), so go there now and follow the instructions to get connected to a GPU deep learning server. Don't worry, it only takes about two minutes to get set up on most platforms, and many don't even require any payment, or even a credit card, to get started.
> A: My two cents: heed this advice! If you like computers you will be tempted to set up your own box. Beware! It is feasible but surprisingly involved and distracting. There is a good reason this book is not titled, _Everything You Ever Wanted to Know About Ubuntu System Administration, NVIDIA Driver Installation, apt-get, conda, pip, and Jupyter Notebook Configuration_. That would be a book of its own. Having designed and deployed our production machine learning infrastructure at work, I can testify it has its satisfactions, but it is as unrelated to modeling as maintaining an airplane is to flying one.
Each option shown on the website includes a tutorial; after completing the tutorial, you will end up with a screen looking like <<notebook_init>>.
<img alt="Initial view of Jupyter Notebook" width="658" caption="Initial view of Jupyter Notebook" id="notebook_init" src="images/att_00057.png">
You are now ready to run your first Jupyter notebook!
> jargon: Jupyter Notebook: A piece of software that allows you to include formatted text, code, images, videos, and much more, all within a single interactive document. Jupyter received the highest honor for software, the ACM Software System Award, thanks to its wide use and enormous impact in many academic fields and in industry. Jupyter Notebook is the software most widely used by data scientists for developing and interacting with deep learning models.
### Running Your First Notebook
The notebooks are labeled by chapter and then by notebook number, so that they are in the same order as they are presented in this book. So, the very first notebook you will see listed is the notebook that you need to use now. You will be using this notebook to train a model that can recognize dog and cat photos. To do this, you'll be downloading a _dataset_ of dog and cat photos, and using that to _train a model_. A dataset is simply a bunch of data—it could be images, emails, financial indicators, sounds, or anything else. There are many datasets made freely available that are suitable for training models. Many of these datasets are created by academics to help advance research, many are made available for competitions (there are competitions where data scientists can compete to see who has the most accurate model!), and some are by-products of other processes (such as financial filings).
> note: Full and Stripped Notebooks: There are two folders containing different versions of the notebooks. The _full_ folder contains the exact notebooks used to create the book you're reading now, with all the prose and outputs. The _stripped_ version has the same headings and code cells, but all outputs and prose have been removed. After reading a section of the book, we recommend working through the stripped notebooks, with the book closed, and seeing if you can figure out what each cell will show before you execute it. Also try to recall what the code is demonstrating.
To open a notebook, just click on it. The notebook will open, and it will look something like <<jupyter>> (note that there may be slight differences in details across different platforms; you can ignore those differences).
<img alt="An example of notebook" width="700" caption="A Jupyter notebook" src="images/0_jupyter.png" id="jupyter"/>
A notebook consists of _cells_. There are two main types of cell:
- Cells containing formatted text, images, and so forth. These use a format called *markdown*, which you will learn about soon.
- Cells containing code that can be executed, and outputs will appear immediately underneath (which could be plain text, tables, images, animations, sounds, or even interactive applications).
Jupyter notebooks can be in one of two modes: edit mode or command mode. In edit mode typing on your keyboard enters the letters into the cell in the usual way. However, in command mode, you will not see any flashing cursor, and the keys on your keyboard will each have a special function.
Before continuing, press the Escape key on your keyboard to switch to command mode (if you are already in command mode, this does nothing, so press it now just in case). To see a complete list of all of the functions available, press H; press Escape to remove this help screen. Notice that in command mode, unlike most programs, commands do not require you to hold down Control, Alt, or similar—you simply press the required letter key.
You can make a copy of a cell by pressing C (the cell needs to be selected first, indicated with an outline around it; if it is not already selected, click on it once). Then press V to paste a copy of it.
Click on the cell that begins with the line "# CLICK ME" to select it. The first character in that line indicates that what follows is a comment in Python, so it is ignored when executing the cell. The rest of the cell is, believe it or not, a complete system for creating and training a state-of-the-art model for recognizing cats versus dogs. So, let's train it now! To do so, just press Shift-Enter on your keyboard, or press the Play button on the toolbar. Then wait a few minutes while the following things happen:
1. A dataset called the [Oxford-IIIT Pet Dataset](http://www.robots.ox.ac.uk/~vgg/data/pets/) that contains 7,349 images of cats and dogs from 37 different breeds will be downloaded from the fast.ai datasets collection to the GPU server you are using, and will then be extracted.
2. A *pretrained model* that has already been trained on 1.3 million images, using a competition-winning model will be downloaded from the internet.
3. The pretrained model will be *fine-tuned* using the latest advances in transfer learning, to create a model that is specially customized for recognizing dogs and cats.
The first two steps only need to be run once on your GPU server. If you run the cell again, it will use the dataset and model that have already been downloaded, rather than downloading them again. Let's take a look at the contents of the cell, and the results (<<first_training>>):
```
#id first_training
#caption Results from the first training
# CLICK ME
from fastai.vision.all import *
path = untar_data(URLs.PETS)/'images'
def is_cat(x): return x[0].isupper()
dls = ImageDataLoaders.from_name_func(
path, get_image_files(path), valid_pct=0.2, seed=42,
label_func=is_cat, item_tfms=Resize(224))
learn = cnn_learner(dls, resnet34, metrics=error_rate)
learn.fine_tune(1)
```
You will probably not see exactly the same results that are in the book. There are a lot of sources of small random variation involved in training models. We generally see an error rate of well less than 0.02 in this example, however.
> important: Training Time: Depending on your network speed, it might take a few minutes to download the pretrained model and dataset. Running `fine_tune` might take a minute or so. Often models in this book take a few minutes to train, as will your own models, so it's a good idea to come up with good techniques to make the most of this time. For instance, keep reading the next section while your model trains, or open up another notebook and use it for some coding experiments.
### Sidebar: This Book Was Written in Jupyter Notebooks
We wrote this book using Jupyter notebooks, so for nearly every chart, table, and calculation in this book, we'll be showing you the exact code required to replicate it yourself. That's why very often in this book, you will see some code immediately followed by a table, a picture or just some text. If you go on the [book's website](https://book.fast.ai) you will find all the code, and you can try running and modifying every example yourself.
You just saw how a cell that outputs a table looks inside the book. Here is an example of a cell that outputs text:
```
1+1
```
Jupyter will always print or show the result of the last line (if there is one). For instance, here is an example of a cell that outputs an image:
```
img = PILImage.create(image_cat())
img.to_thumb(192)
```
### End sidebar
So, how do we know if this model is any good? In the last column of the table you can see the error rate, which is the proportion of images that were incorrectly identified. The error rate serves as our metric—our measure of model quality, chosen to be intuitive and comprehensible. As you can see, the model is nearly perfect, even though the training time was only a few seconds (not including the one-time downloading of the dataset and the pretrained model). In fact, the accuracy you've achieved already is far better than anybody had ever achieved just 10 years ago!
Finally, let's check that this model actually works. Go and get a photo of a dog, or a cat; if you don't have one handy, just search Google Images and download an image that you find there. Now execute the cell with `uploader` defined. It will output a button you can click, so you can select the image you want to classify:
```
#hide_output
uploader = widgets.FileUpload()
uploader
```
<img alt="An upload button" width="159" id="upload" src="images/att_00008.png">
Now you can pass the uploaded file to the model. Make sure that it is a clear photo of a single dog or a cat, and not a line drawing, cartoon, or similar. The notebook will tell you whether it thinks it is a dog or a cat, and how confident it is. Hopefully, you'll find that your model did a great job:
```
#hide
# For the book, we can't actually click an upload button, so we fake it
uploader = SimpleNamespace(data = ['images/chapter1_cat_example.jpg'])
img = PILImage.create(uploader.data[0])
is_cat,_,probs = learn.predict(img)
print(f"Is this a cat?: {is_cat}.")
print(f"Probability it's a cat: {probs[1].item():.6f}")
```
Congratulations on your first classifier!
But what does this mean? What did you actually do? In order to explain this, let's zoom out again to take in the big picture.
### What Is Machine Learning?
Your classifier is a deep learning model. As was already mentioned, deep learning models use neural networks, which originally date from the 1950s and have become powerful very recently thanks to recent advancements.
Another key piece of context is that deep learning is just a modern area in the more general discipline of *machine learning*. To understand the essence of what you did when you trained your own classification model, you don't need to understand deep learning. It is enough to see how your model and your training process are examples of the concepts that apply to machine learning in general.
So in this section, we will describe what machine learning is. We will look at the key concepts, and show how they can be traced back to the original essay that introduced them.
*Machine learning* is, like regular programming, a way to get computers to complete a specific task. But how would we use regular programming to do what we just did in the last section: recognize dogs versus cats in photos? We would have to write down for the computer the exact steps necessary to complete the task.
Normally, it's easy enough for us to write down the steps to complete a task when we're writing a program. We just think about the steps we'd take if we had to do the task by hand, and then we translate them into code. For instance, we can write a function that sorts a list. In general, we'd write a function that looks something like <<basic_program>> (where *inputs* might be an unsorted list, and *results* a sorted list).
```
#hide_input
#caption A traditional program
#id basic_program
#alt Pipeline inputs, program, results
gv('''program[shape=box3d width=1 height=0.7]
inputs->program->results''')
```
But for recognizing objects in a photo that's a bit tricky; what *are* the steps we take when we recognize an object in a picture? We really don't know, since it all happens in our brain without us being consciously aware of it!
Right back at the dawn of computing, in 1949, an IBM researcher named Arthur Samuel started working on a different way to get computers to complete tasks, which he called *machine learning*. In his classic 1962 essay "Artificial Intelligence: A Frontier of Automation", he wrote:
> : Programming a computer for such computations is, at best, a difficult task, not primarily because of any inherent complexity in the computer itself but, rather, because of the need to spell out every minute step of the process in the most exasperating detail. Computers, as any programmer will tell you, are giant morons, not giant brains.
His basic idea was this: instead of telling the computer the exact steps required to solve a problem, show it examples of the problem to solve, and let it figure out how to solve it itself. This turned out to be very effective: by 1961 his checkers-playing program had learned so much that it beat the Connecticut state champion! Here's how he described his idea (from the same essay as above):
> : Suppose we arrange for some automatic means of testing the effectiveness of any current weight assignment in terms of actual performance and provide a mechanism for altering the weight assignment so as to maximize the performance. We need not go into the details of such a procedure to see that it could be made entirely automatic and to see that a machine so programmed would "learn" from its experience.
There are a number of powerful concepts embedded in this short statement:
- The idea of a "weight assignment"
- The fact that every weight assignment has some "actual performance"
- The requirement that there be an "automatic means" of testing that performance,
- The need for a "mechanism" (i.e., another automatic process) for improving the performance by changing the weight assignments
Let us take these concepts one by one, in order to understand how they fit together in practice. First, we need to understand what Samuel means by a *weight assignment*.
Weights are just variables, and a weight assignment is a particular choice of values for those variables. The program's inputs are values that it processes in order to produce its results—for instance, taking image pixels as inputs, and returning the classification "dog" as a result. The program's weight assignments are other values that define how the program will operate.
Since they will affect the program they are in a sense another kind of input, so we will update our basic picture in <<basic_program>> and replace it with <<weight_assignment>> in order to take this into account.
```
#hide_input
#caption A program using weight assignment
#id weight_assignment
gv('''model[shape=box3d width=1 height=0.7]
inputs->model->results; weights->model''')
```
We've changed the name of our box from *program* to *model*. This is to follow modern terminology and to reflect that the *model* is a special kind of program: it's one that can do *many different things*, depending on the *weights*. It can be implemented in many different ways. For instance, in Samuel's checkers program, different values of the weights would result in different checkers-playing strategies.
(By the way, what Samuel called "weights" are most generally referred to as model *parameters* these days, in case you have encountered that term. The term *weights* is reserved for a particular type of model parameter.)
Next, Samuel said we need an *automatic means of testing the effectiveness of any current weight assignment in terms of actual performance*. In the case of his checkers program, the "actual performance" of a model would be how well it plays. And you could automatically test the performance of two models by setting them to play against each other, and seeing which one usually wins.
Finally, he says we need *a mechanism for altering the weight assignment so as to maximize the performance*. For instance, we could look at the difference in weights between the winning model and the losing model, and adjust the weights a little further in the winning direction.
We can now see why he said that such a procedure *could be made entirely automatic and... a machine so programmed would "learn" from its experience*. Learning would become entirely automatic when the adjustment of the weights was also automatic—when instead of us improving a model by adjusting its weights manually, we relied on an automated mechanism that produced adjustments based on performance.
<<training_loop>> shows the full picture of Samuel's idea of training a machine learning model.
```
#hide_input
#caption Training a machine learning model
#id training_loop
#alt The basic training loop
gv('''ordering=in
model[shape=box3d width=1 height=0.7]
inputs->model->results; weights->model; results->performance
performance->weights[constraint=false label=update]''')
```
Notice the distinction between the model's *results* (e.g., the moves in a checkers game) and its *performance* (e.g., whether it wins the game, or how quickly it wins).
Also note that once the model is trained—that is, once we've chosen our final, best, favorite weight assignment—then we can think of the weights as being *part of the model*, since we're not varying them any more.
Therefore, actually *using* a model after it's trained looks like <<using_model>>.
```
#hide_input
#caption Using a trained model as a program
#id using_model
gv('''model[shape=box3d width=1 height=0.7]
inputs->model->results''')
```
This looks identical to our original diagram in <<basic_program>>, just with the word *program* replaced with *model*. This is an important insight: *a trained model can be treated just like a regular computer program*.
> jargon: Machine Learning: The training of programs developed by allowing a computer to learn from its experience, rather than through manually coding the individual steps.
### What Is a Neural Network?
It's not too hard to imagine what the model might look like for a checkers program. There might be a range of checkers strategies encoded, and some kind of search mechanism, and then the weights could vary how strategies are selected, what parts of the board are focused on during a search, and so forth. But it's not at all obvious what the model might look like for an image recognition program, or for understanding text, or for many other interesting problems we might imagine.
What we would like is some kind of function that is so flexible that it could be used to solve any given problem, just by varying its weights. Amazingly enough, this function actually exists! It's the neural network, which we already discussed. That is, if you regard a neural network as a mathematical function, it turns out to be a function which is extremely flexible depending on its weights. A mathematical proof called the *universal approximation theorem* shows that this function can solve any problem to any level of accuracy, in theory. The fact that neural networks are so flexible means that, in practice, they are often a suitable kind of model, and you can focus your effort on the process of training them—that is, of finding good weight assignments.
But what about that process? One could imagine that you might need to find a new "mechanism" for automatically updating weights for every problem. This would be laborious. What we'd like here as well is a completely general way to update the weights of a neural network, to make it improve at any given task. Conveniently, this also exists!
This is called *stochastic gradient descent* (SGD). We'll see how neural networks and SGD work in detail in <<chapter_mnist_basics>>, as well as explaining the universal approximation theorem. For now, however, we will instead use Samuel's own words: *We need not go into the details of such a procedure to see that it could be made entirely automatic and to see that a machine so programmed would "learn" from its experience.*
> J: Don't worry, neither SGD nor neural nets are mathematically complex. Both nearly entirely rely on addition and multiplication to do their work (but they do a _lot_ of addition and multiplication!). The main reaction we hear from students when they see the details is: "Is that all it is?"
In other words, to recap, a neural network is a particular kind of machine learning model, which fits right in to Samuel's original conception. Neural networks are special because they are highly flexible, which means they can solve an unusually wide range of problems just by finding the right weights. This is powerful, because stochastic gradient descent provides us a way to find those weight values automatically.
Having zoomed out, let's now zoom back in and revisit our image classification problem using Samuel's framework.
Our inputs are the images. Our weights are the weights in the neural net. Our model is a neural net. Our results are the values that are calculated by the neural net, like "dog" or "cat."
What about the next piece, an *automatic means of testing the effectiveness of any current weight assignment in terms of actual performance*? Determining "actual performance" is easy enough: we can simply define our model's performance as its accuracy at predicting the correct answers.
Putting this all together, and assuming that SGD is our mechanism for updating the weight assignments, we can see how our image classifier is a machine learning model, much like Samuel envisioned.
### A Bit of Deep Learning Jargon
Samuel was working in the 1960s, and since then terminology has changed. Here is the modern deep learning terminology for all the pieces we have discussed:
- The functional form of the *model* is called its *architecture* (but be careful—sometimes people use *model* as a synonym of *architecture*, so this can get confusing).
- The *weights* are called *parameters*.
- The *predictions* are calculated from the *independent variable*, which is the *data* not including the *labels*.
- The *results* of the model are called *predictions*.
- The measure of *performance* is called the *loss*.
- The loss depends not only on the predictions, but also the correct *labels* (also known as *targets* or the *dependent variable*); e.g., "dog" or "cat."
After making these changes, our diagram in <<training_loop>> looks like <<detailed_loop>>.
```
#hide_input
#caption Detailed training loop
#id detailed_loop
gv('''ordering=in
model[shape=box3d width=1 height=0.7 label=architecture]
inputs->model->predictions; parameters->model; labels->loss; predictions->loss
loss->parameters[constraint=false label=update]''')
```
### Limitations Inherent To Machine Learning
From this picture we can now see some fundamental things about training a deep learning model:
- A model cannot be created without data.
- A model can only learn to operate on the patterns seen in the input data used to train it.
- This learning approach only creates *predictions*, not recommended *actions*.
- It's not enough to just have examples of input data; we need *labels* for that data too (e.g., pictures of dogs and cats aren't enough to train a model; we need a label for each one, saying which ones are dogs, and which are cats).
Generally speaking, we've seen that most organizations that say they don't have enough data, actually mean they don't have enough *labeled* data. If any organization is interested in doing something in practice with a model, then presumably they have some inputs they plan to run their model against. And presumably they've been doing that some other way for a while (e.g., manually, or with some heuristic program), so they have data from those processes! For instance, a radiology practice will almost certainly have an archive of medical scans (since they need to be able to check how their patients are progressing over time), but those scans may not have structured labels containing a list of diagnoses or interventions (since radiologists generally create free-text natural language reports, not structured data). We'll be discussing labeling approaches a lot in this book, because it's such an important issue in practice.
Since these kinds of machine learning models can only make *predictions* (i.e., attempt to replicate labels), this can result in a significant gap between organizational goals and model capabilities. For instance, in this book you'll learn how to create a *recommendation system* that can predict what products a user might purchase. This is often used in e-commerce, such as to customize products shown on a home page by showing the highest-ranked items. But such a model is generally created by looking at a user and their buying history (*inputs*) and what they went on to buy or look at (*labels*), which means that the model is likely to tell you about products the user already has or already knows about, rather than new products that they are most likely to be interested in hearing about. That's very different to what, say, an expert at your local bookseller might do, where they ask questions to figure out your taste, and then tell you about authors or series that you've never heard of before.
Another critical insight comes from considering how a model interacts with its environment. This can create *feedback loops*, as described here:
- A *predictive policing* model is created based on where arrests have been made in the past. In practice, this is not actually predicting crime, but rather predicting arrests, and is therefore partially simply reflecting biases in existing policing processes.
- Law enforcement officers then might use that model to decide where to focus their police activity, resulting in increased arrests in those areas.
- Data on these additional arrests would then be fed back in to retrain future versions of the model.
This is a *positive feedback loop*, where the more the model is used, the more biased the data becomes, making the model even more biased, and so forth.
Feedback loops can also create problems in commercial settings. For instance, a video recommendation system might be biased toward recommending content consumed by the biggest watchers of video (e.g., conspiracy theorists and extremists tend to watch more online video content than the average), resulting in those users increasing their video consumption, resulting in more of those kinds of videos being recommended. We'll consider this topic more in detail in <<chapter_ethics>>.
Now that you have seen the base of the theory, let's go back to our code example and see in detail how the code corresponds to the process we just described.
### How Our Image Recognizer Works
Let's see just how our image recognizer code maps to these ideas. We'll put each line into a separate cell, and look at what each one is doing (we won't explain every detail of every parameter yet, but will give a description of the important bits; full details will come later in the book).
The first line imports all of the fastai.vision library.
```python
from fastai.vision.all import *
```
This gives us all of the functions and classes we will need to create a wide variety of computer vision models.
> J: A lot of Python coders recommend avoiding importing a whole library like this (using the `import *` syntax), because in large software projects it can cause problems. However, for interactive work such as in a Jupyter notebook, it works great. The fastai library is specially designed to support this kind of interactive use, and it will only import the necessary pieces into your environment.
The second line downloads a standard dataset from the [fast.ai datasets collection](https://course.fast.ai/datasets) (if not previously downloaded) to your server, extracts it (if not previously extracted), and returns a `Path` object with the extracted location:
```python
path = untar_data(URLs.PETS)/'images'
```
> S: Throughout my time studying at fast.ai, and even still today, I've learned a lot about productive coding practices. The fastai library and fast.ai notebooks are full of great little tips that have helped make me a better programmer. For instance, notice that the fastai library doesn't just return a string containing the path to the dataset, but a `Path` object. This is a really useful class from the Python 3 standard library that makes accessing files and directories much easier. If you haven't come across it before, be sure to check out its documentation or a tutorial and try it out. Note that the https://book.fast.ai[website] contains links to recommended tutorials for each chapter. I'll keep letting you know about little coding tips I've found useful as we come across them.
In the third line we define a function, `is_cat`, labels cats based on a filename rule provided by the dataset creators:
```python
def is_cat(x): return x[0].isupper()
```
We use that function in the fourth line, which tells fastai what kind of dataset we have, and how it is structured:
```python
dls = ImageDataLoaders.from_name_func(
path, get_image_files(path), valid_pct=0.2, seed=42,
label_func=is_cat, item_tfms=Resize(224))
```
There are various different classes for different kinds of deep learning datasets and problems—here we're using `ImageDataLoaders`. The first part of the class name will generally be the type of data you have, such as image, or text.
The other important piece of information that we have to tell fastai is how to get the labels from the dataset. Computer vision datasets are normally structured in such a way that the label for an image is part of the filename, or path—most commonly the parent folder name. fastai comes with a number of standardized labeling methods, and ways to write your own. Here we're telling fastai to use the `is_cat` function we just defined.
Finally, we define the `Transform`s that we need. A `Transform` contains code that is applied automatically during training; fastai includes many predefined `Transform`s, and adding new ones is as simple as creating a Python function. There are two kinds: `item_tfms` are applied to each item (in this case, each item is resized to a 224-pixel square), while `batch_tfms` are applied to a *batch* of items at a time using the GPU, so they're particularly fast (we'll see many examples of these throughout this book).
Why 224 pixels? This is the standard size for historical reasons (old pretrained models require this size exactly), but you can pass pretty much anything. If you increase the size, you'll often get a model with better results (since it will be able to focus on more details), but at the price of speed and memory consumption; the opposite is true if you decrease the size.
> Note: Classification and Regression: _classification_ and _regression_ have very specific meanings in machine learning. These are the two main types of model that we will be investigating in this book. A classification model is one which attempts to predict a class, or category. That is, it's predicting from a number of discrete possibilities, such as "dog" or "cat." A regression model is one which attempts to predict one or more numeric quantities, such as a temperature or a location. Sometimes people use the word _regression_ to refer to a particular kind of model called a _linear regression model_; this is a bad practice, and we won't be using that terminology in this book!
The Pet dataset contains 7,390 pictures of dogs and cats, consisting of 37 different breeds. Each image is labeled using its filename: for instance the file *great\_pyrenees\_173.jpg* is the 173rd example of an image of a Great Pyrenees breed dog in the dataset. The filenames start with an uppercase letter if the image is a cat, and a lowercase letter otherwise. We have to tell fastai how to get labels from the filenames, which we do by calling `from_name_func` (which means that labels can be extracted using a function applied to the filename), and passing `x[0].isupper()`, which evaluates to `True` if the first letter is uppercase (i.e., it's a cat).
The most important parameter to mention here is `valid_pct=0.2`. This tells fastai to hold out 20% of the data and *not use it for training the model at all*. This 20% of the data is called the *validation set*; the remaining 80% is called the *training set*. The validation set is used to measure the accuracy of the model. By default, the 20% that is held out is selected randomly. The parameter `seed=42` sets the *random seed* to the same value every time we run this code, which means we get the same validation set every time we run it—this way, if we change our model and retrain it, we know that any differences are due to the changes to the model, not due to having a different random validation set.
fastai will *always* show you your model's accuracy using *only* the validation set, *never* the training set. This is absolutely critical, because if you train a large enough model for a long enough time, it will eventually memorize the label of every item in your dataset! The result will not actually be a useful model, because what we care about is how well our model works on *previously unseen images*. That is always our goal when creating a model: for it to be useful on data that the model only sees in the future, after it has been trained.
Even when your model has not fully memorized all your data, earlier on in training it may have memorized certain parts of it. As a result, the longer you train for, the better your accuracy will get on the training set; the validation set accuracy will also improve for a while, but eventually it will start getting worse as the model starts to memorize the training set, rather than finding generalizable underlying patterns in the data. When this happens, we say that the model is *overfitting*.
<<img_overfit>> shows what happens when you overfit, using a simplified example where we have just one parameter, and some randomly generated data based on the function `x**2`. As you can see, although the predictions in the overfit model are accurate for data near the observed data points, they are way off when outside of that range.
<img src="images/att_00000.png" alt="Example of overfitting" caption="Example of overfitting" id="img_overfit" width="700">
**Overfitting is the single most important and challenging issue** when training for all machine learning practitioners, and all algorithms. As you will see, it is very easy to create a model that does a great job at making predictions on the exact data it has been trained on, but it is much harder to make accurate predictions on data the model has never seen before. And of course, this is the data that will actually matter in practice. For instance, if you create a handwritten digit classifier (as we will very soon!) and use it to recognize numbers written on checks, then you are never going to see any of the numbers that the model was trained on—check will have slightly different variations of writing to deal with. You will learn many methods to avoid overfitting in this book. However, you should only use those methods after you have confirmed that overfitting is actually occurring (i.e., you have actually observed the validation accuracy getting worse during training). We often see practitioners using over-fitting avoidance techniques even when they have enough data that they didn't need to do so, ending up with a model that may be less accurate than what they could have achieved.
> important: Validation Set: When you train a model, you must _always_ have both a training set and a validation set, and must measure the accuracy of your model only on the validation set. If you train for too long, with not enough data, you will see the accuracy of your model start to get worse; this is called _overfitting_. fastai defaults `valid_pct` to `0.2`, so even if you forget, fastai will create a validation set for you!
The fifth line of the code training our image recognizer tells fastai to create a *convolutional neural network* (CNN) and specifies what *architecture* to use (i.e. what kind of model to create), what data we want to train it on, and what *metric* to use:
```python
learn = cnn_learner(dls, resnet34, metrics=error_rate)
```
Why a CNN? It's the current state-of-the-art approach to creating computer vision models. We'll be learning all about how CNNs work in this book. Their structure is inspired by how the human vision system works.
There are many different architectures in fastai, which we will introduce in this book (as well as discussing how to create your own). Most of the time, however, picking an architecture isn't a very important part of the deep learning process. It's something that academics love to talk about, but in practice it is unlikely to be something you need to spend much time on. There are some standard architectures that work most of the time, and in this case we're using one called _ResNet_ that we'll be talking a lot about during the book; it is both fast and accurate for many datasets and problems. The `34` in `resnet34` refers to the number of layers in this variant of the architecture (other options are `18`, `50`, `101`, and `152`). Models using architectures with more layers take longer to train, and are more prone to overfitting (i.e. you can't train them for as many epochs before the accuracy on the validation set starts getting worse). On the other hand, when using more data, they can be quite a bit more accurate.
What is a metric? A *metric* is a function that measures the quality of the model's predictions using the validation set, and will be printed at the end of each *epoch*. In this case, we're using `error_rate`, which is a function provided by fastai that does just what it says: tells you what percentage of images in the validation set are being classified incorrectly. Another common metric for classification is `accuracy` (which is just `1.0 - error_rate`). fastai provides many more, which will be discussed throughout this book.
The concept of a metric may remind you of *loss*, but there is an important distinction. The entire purpose of loss is to define a "measure of performance" that the training system can use to update weights automatically. In other words, a good choice for loss is a choice that is easy for stochastic gradient descent to use. But a metric is defined for human consumption, so a good metric is one that is easy for you to understand, and that hews as closely as possible to what you want the model to do. At times, you might decide that the loss function is a suitable metric, but that is not necessarily the case.
`cnn_learner` also has a parameter `pretrained`, which defaults to `True` (so it's used in this case, even though we haven't specified it), which sets the weights in your model to values that have already been trained by experts to recognize a thousand different categories across 1.3 million photos (using the famous [*ImageNet* dataset](http://www.image-net.org/)). A model that has weights that have already been trained on some other dataset is called a *pretrained model*. You should nearly always use a pretrained model, because it means that your model, before you've even shown it any of your data, is already very capable. And, as you'll see, in a deep learning model many of these capabilities are things you'll need, almost regardless of the details of your project. For instance, parts of pretrained models will handle edge, gradient, and color detection, which are needed for many tasks.
When using a pretrained model, `cnn_learner` will remove the last layer, since that is always specifically customized to the original training task (i.e. ImageNet dataset classification), and replace it with one or more new layers with randomized weights, of an appropriate size for the dataset you are working with. This last part of the model is known as the *head*.
Using pretrained models is the *most* important method we have to allow us to train more accurate models, more quickly, with less data, and less time and money. You might think that would mean that using pretrained models would be the most studied area in academic deep learning... but you'd be very, very wrong! The importance of pretrained models is generally not recognized or discussed in most courses, books, or software library features, and is rarely considered in academic papers. As we write this at the start of 2020, things are just starting to change, but it's likely to take a while. So be careful: most people you speak to will probably greatly underestimate what you can do in deep learning with few resources, because they probably won't deeply understand how to use pretrained models.
Using a pretrained model for a task different to what it was originally trained for is known as *transfer learning*. Unfortunately, because transfer learning is so under-studied, few domains have pretrained models available. For instance, there are currently few pretrained models available in medicine, making transfer learning challenging to use in that domain. In addition, it is not yet well understood how to use transfer learning for tasks such as time series analysis.
> jargon: Transfer learning: Using a pretrained model for a task different to what it was originally trained for.
The sixth line of our code tells fastai how to *fit* the model:
```python
learn.fine_tune(1)
```
As we've discussed, the architecture only describes a *template* for a mathematical function; it doesn't actually do anything until we provide values for the millions of parameters it contains.
This is the key to deep learning—determining how to fit the parameters of a model to get it to solve your problem. In order to fit a model, we have to provide at least one piece of information: how many times to look at each image (known as number of *epochs*). The number of epochs you select will largely depend on how much time you have available, and how long you find it takes in practice to fit your model. If you select a number that is too small, you can always train for more epochs later.
But why is the method called `fine_tune`, and not `fit`? fastai actually *does* have a method called `fit`, which does indeed fit a model (i.e. look at images in the training set multiple times, each time updating the parameters to make the predictions closer and closer to the target labels). But in this case, we've started with a pretrained model, and we don't want to throw away all those capabilities that it already has. As you'll learn in this book, there are some important tricks to adapt a pretrained model for a new dataset—a process called *fine-tuning*.
> jargon: Fine-tuning: A transfer learning technique where the parameters of a pretrained model are updated by training for additional epochs using a different task to that used for pretraining.
When you use the `fine_tune` method, fastai will use these tricks for you. There are a few parameters you can set (which we'll discuss later), but in the default form shown here, it does two steps:
1. Use one epoch to fit just those parts of the model necessary to get the new random head to work correctly with your dataset.
1. Use the number of epochs requested when calling the method to fit the entire model, updating the weights of the later layers (especially the head) faster than the earlier layers (which, as we'll see, generally don't require many changes from the pretrained weights).
The *head* of a model is the part that is newly added to be specific to the new dataset. An *epoch* is one complete pass through the dataset. After calling `fit`, the results after each epoch are printed, showing the epoch number, the training and validation set losses (the "measure of performance" used for training the model), and any *metrics* you've requested (error rate, in this case).
So, with all this code our model learned to recognize cats and dogs just from labeled examples. But how did it do it?
### What Our Image Recognizer Learned
At this stage we have an image recognizer that is working very well, but we have no idea what it is actually doing! Although many people complain that deep learning results in impenetrable "black box" models (that is, something that gives predictions but that no one can understand), this really couldn't be further from the truth. There is a vast body of research showing how to deeply inspect deep learning models, and get rich insights from them. Having said that, all kinds of machine learning models (including deep learning, and traditional statistical models) can be challenging to fully understand, especially when considering how they will behave when coming across data that is very different to the data used to train them. We'll be discussing this issue throughout this book.
In 2013 a PhD student, Matt Zeiler, and his supervisor, Rob Fergus, published the paper ["Visualizing and Understanding Convolutional Networks"](https://arxiv.org/pdf/1311.2901.pdf), which showed how to visualize the neural network weights learned in each layer of a model. They carefully analyzed the model that won the 2012 ImageNet competition, and used this analysis to greatly improve the model, such that they were able to go on to win the 2013 competition! <<img_layer1>> is the picture that they published of the first layer's weights.
<img src="images/layer1.png" alt="Activations of the first layer of a CNN" width="300" caption="Activations of the first layer of a CNN (courtesy of Matthew D. Zeiler and Rob Fergus)" id="img_layer1">
This picture requires some explanation. For each layer, the image part with the light gray background shows the reconstructed weights pictures, and the larger section at the bottom shows the parts of the training images that most strongly matched each set of weights. For layer 1, what we can see is that the model has discovered weights that represent diagonal, horizontal, and vertical edges, as well as various different gradients. (Note that for each layer only a subset of the features are shown; in practice there are thousands across all of the layers.) These are the basic building blocks that the model has learned for computer vision. They have been widely analyzed by neuroscientists and computer vision researchers, and it turns out that these learned building blocks are very similar to the basic visual machinery in the human eye, as well as the handcrafted computer vision features that were developed prior to the days of deep learning. The next layer is represented in <<img_layer2>>.
<img src="images/layer2.png" alt="Activations of the second layer of a CNN" width="800" caption="Activations of the second layer of a CNN (courtesy of Matthew D. Zeiler and Rob Fergus)" id="img_layer2">
For layer 2, there are nine examples of weight reconstructions for each of the features found by the model. We can see that the model has learned to create feature detectors that look for corners, repeating lines, circles, and other simple patterns. These are built from the basic building blocks developed in the first layer. For each of these, the right-hand side of the picture shows small patches from actual images which these features most closely match. For instance, the particular pattern in row 2, column 1 matches the gradients and textures associated with sunsets.
<<img_layer3>> shows the image from the paper showing the results of reconstructing the features of layer 3.
<img src="images/chapter2_layer3.PNG" alt="Activations of the third layer of a CNN" width="800" caption="Activations of the third layer of a CNN (courtesy of Matthew D. Zeiler and Rob Fergus)" id="img_layer3">
As you can see by looking at the righthand side of this picture, the features are now able to identify and match with higher-level semantic components, such as car wheels, text, and flower petals. Using these components, layers four and five can identify even higher-level concepts, as shown in <<img_layer4>>.
<img src="images/chapter2_layer4and5.PNG" alt="Activations of layers 4 and 5 of a CNN" width="800" caption="Activations of layers 4 and 5 of a CNN (courtesy of Matthew D. Zeiler and Rob Fergus)" id="img_layer4">
This article was studying an older model called *AlexNet* that only contained five layers. Networks developed since then can have hundreds of layers—so you can imagine how rich the features developed by these models can be!
When we fine-tuned our pretrained model earlier, we adapted what those last layers focus on (flowers, humans, animals) to specialize on the cats versus dogs problem. More generally, we could specialize such a pretrained model on many different tasks. Let's have a look at some examples.
### Image Recognizers Can Tackle Non-Image Tasks
An image recognizer can, as its name suggests, only recognize images. But a lot of things can be represented as images, which means that an image recogniser can learn to complete many tasks.
For instance, a sound can be converted to a spectrogram, which is a chart that shows the amount of each frequency at each time in an audio file. Fast.ai student Ethan Sutin used this approach to easily beat the published accuracy of a state-of-the-art [environmental sound detection model](https://medium.com/@etown/great-results-on-audio-classification-with-fastai-library-ccaf906c5f52) using a dataset of 8,732 urban sounds. fastai's `show_batch` clearly shows how each different sound has a quite distinctive spectrogram, as you can see in <<img_spect>>.
<img alt="show_batch with spectrograms of sounds" width="400" caption="show_batch with spectrograms of sounds" id="img_spect" src="images/att_00012.png">
A time series can easily be converted into an image by simply plotting the time series on a graph. However, it is often a good idea to try to represent your data in a way that makes it as easy as possible to pull out the most important components. In a time series, things like seasonality and anomalies are most likely to be of interest. There are various transformations available for time series data. For instance, fast.ai student Ignacio Oguiza created images from a time series dataset for olive oil classification, using a technique called Gramian Angular Difference Field (GADF); you can see the result in <<ts_image>>. He then fed those images to an image classification model just like the one you see in this chapter. His results, despite having only 30 training set images, were well over 90% accurate, and close to the state of the art.
<img alt="Converting a time series into an image" width="700" caption="Converting a time series into an image" id="ts_image" src="images/att_00013.png">
Another interesting fast.ai student project example comes from Gleb Esman. He was working on fraud detection at Splunk, using a dataset of users' mouse movements and mouse clicks. He turned these into pictures by drawing an image where the position, speed, and acceleration of the mouse pointer was displayed using coloured lines, and the clicks were displayed using [small colored circles](https://www.splunk.com/en_us/blog/security/deep-learning-with-splunk-and-tensorflow-for-security-catching-the-fraudster-in-neural-networks-with-behavioral-biometrics.html), as shown in <<splunk>>. He then fed this into an image recognition model just like the one we've used in this chapter, and it worked so well that it led to a patent for this approach to fraud analytics!
<img alt="Converting computer mouse behavior to an image" width="450" caption="Converting computer mouse behavior to an image" id="splunk" src="images/att_00014.png">
Another example comes from the paper ["Malware Classification with Deep Convolutional Neural Networks"](https://ieeexplore.ieee.org/abstract/document/8328749) by Mahmoud Kalash et al., which explains that "the malware binary file is divided into 8-bit sequences which are then converted to equivalent decimal values. This decimal vector is reshaped and a gray-scale image is generated that represents the malware sample," like in <<malware_proc>>.
<img alt="Malware classification process" width="623" caption="Malware classification process" id="malware_proc" src="images/att_00055.png">
The authors then show "pictures" generated through this process of malware in different categories, as shown in <<malware_eg>>.
<img alt="Malware examples" width="650" caption="Malware examples" id="malware_eg" src="images/att_00056.png">
As you can see, the different types of malware look very distinctive to the human eye. The model the researchers trained based on this image representation was more accurate at malware classification than any previous approach shown in the academic literature. This suggests a good rule of thumb for converting a dataset into an image representation: if the human eye can recognize categories from the images, then a deep learning model should be able to do so too.
In general, you'll find that a small number of general approaches in deep learning can go a long way, if you're a bit creative in how you represent your data! You shouldn't think of approaches like the ones described here as "hacky workarounds," because actually they often (as here) beat previously state-of-the-art results. These really are the right ways to think about these problem domains.
### Jargon Recap
We just covered a lot of information so let's recap briefly, <<dljargon>> provides a handy vocabulary.
```asciidoc
[[dljargon]]
.Deep learning vocabulary
[options="header"]
|=====
| Term | Meaning
|Label | The data that we're trying to predict, such as "dog" or "cat"
|Architecture | The _template_ of the model that we're trying to fit; the actual mathematical function that we're passing the input data and parameters to
|Model | The combination of the architecture with a particular set of parameters
|Parameters | The values in the model that change what task it can do, and are updated through model training
|Fit | Update the parameters of the model such that the predictions of the model using the input data match the target labels
|Train | A synonym for _fit_
|Pretrained model | A model that has already been trained, generally using a large dataset, and will be fine-tuned
|Fine-tune | Update a pretrained model for a different task
|Epoch | One complete pass through the input data
|Loss | A measure of how good the model is, chosen to drive training via SGD
|Metric | A measurement of how good the model is, using the validation set, chosen for human consumption
|Validation set | A set of data held out from training, used only for measuring how good the model is
|Training set | The data used for fitting the model; does not include any data from the validation set
|Overfitting | Training a model in such a way that it _remembers_ specific features of the input data, rather than generalizing well to data not seen during training
|CNN | Convolutional neural network; a type of neural network that works particularly well for computer vision tasks
|=====
```
With this vocabulary in hand, we are now in a position to bring together all the key concepts introduced so far. Take a moment to review those definitions and read the following summary. If you can follow the explanation, then you're well equipped to understand the discussions to come.
*Machine learning* is a discipline where we define a program not by writing it entirely ourselves, but by learning from data. *Deep learning* is a specialty within machine learning that uses *neural networks* with multiple *layers*. *Image classification* is a representative example (also known as *image recognition*). We start with *labeled data*; that is, a set of images where we have assigned a *label* to each image indicating what it represents. Our goal is to produce a program, called a *model*, which, given a new image, will make an accurate *prediction* regarding what that new image represents.
Every model starts with a choice of *architecture*, a general template for how that kind of model works internally. The process of *training* (or *fitting*) the model is the process of finding a set of *parameter values* (or *weights*) that specialize that general architecture into a model that works well for our particular kind of data. In order to define how well a model does on a single prediction, we need to define a *loss function*, which determines how we score a prediction as good or bad.
To make the training process go faster, we might start with a *pretrained model*—a model that has already been trained on someone else's data. We can then adapt it to our data by training it a bit more on our data, a process called *fine-tuning*.
When we train a model, a key concern is to ensure that our model *generalizes*—that is, that it learns general lessons from our data which also apply to new items it will encounter, so that it can make good predictions on those items. The risk is that if we train our model badly, instead of learning general lessons it effectively memorizes what it has already seen, and then it will make poor predictions about new images. Such a failure is called *overfitting*. In order to avoid this, we always divide our data into two parts, the *training set* and the *validation set*. We train the model by showing it only the training set and then we evaluate how well the model is doing by seeing how well it performs on items from the validation set. In this way, we check if the lessons the model learns from the training set are lessons that generalize to the validation set. In order for a person to assess how well the model is doing on the validation set overall, we define a *metric*. During the training process, when the model has seen every item in the training set, we call that an *epoch*.
All these concepts apply to machine learning in general. That is, they apply to all sorts of schemes for defining a model by training it with data. What makes deep learning distinctive is a particular class of architectures: the architectures based on *neural networks*. In particular, tasks like image classification rely heavily on *convolutional neural networks*, which we will discuss shortly.
## Deep Learning Is Not Just for Image Classification
Deep learning's effectiveness for classifying images has been widely discussed in recent years, even showing _superhuman_ results on complex tasks like recognizing malignant tumors in CT scans. But it can do a lot more than this, as we will show here.
For instance, let's talk about something that is critically important for autonomous vehicles: localizing objects in a picture. If a self-driving car doesn't know where a pedestrian is, then it doesn't know how to avoid one! Creating a model that can recognize the content of every individual pixel in an image is called *segmentation*. Here is how we can train a segmentation model with fastai, using a subset of the [*Camvid* dataset](http://www0.cs.ucl.ac.uk/staff/G.Brostow/papers/Brostow_2009-PRL.pdf) from the paper "Semantic Object Classes in Video: A High-Definition Ground Truth Database" by Gabruel J. Brostow, Julien Fauqueur, and Roberto Cipolla:
```
path = untar_data(URLs.CAMVID_TINY)
dls = SegmentationDataLoaders.from_label_func(
path, bs=8, fnames = get_image_files(path/"images"),
label_func = lambda o: path/'labels'/f'{o.stem}_P{o.suffix}',
codes = np.loadtxt(path/'codes.txt', dtype=str)
)
learn = unet_learner(dls, resnet34)
learn.fine_tune(8)
```
We are not even going to walk through this code line by line, because it is nearly identical to our previous example! (Although we will be doing a deep dive into segmentation models in <<chapter_arch_details>>, along with all of the other models that we are briefly introducing in this chapter, and many, many more.)
We can visualize how well it achieved its task, by asking the model to color-code each pixel of an image. As you can see, it nearly perfectly classifies every pixel in every object. For instance, notice that all of the cars are overlaid with the same color and all of the trees are overlaid with the same color (in each pair of images, the lefthand image is the ground truth label and the right is the prediction from the model):
```
learn.show_results(max_n=6, figsize=(7,8))
```
One other area where deep learning has dramatically improved in the last couple of years is natural language processing (NLP). Computers can now generate text, translate automatically from one language to another, analyze comments, label words in sentences, and much more. Here is all of the code necessary to train a model that can classify the sentiment of a movie review better than anything that existed in the world just five years ago:
```
from fastai.text.all import *
dls = TextDataLoaders.from_folder(untar_data(URLs.IMDB), valid='test')
learn = text_classifier_learner(dls, AWD_LSTM, drop_mult=0.5, metrics=accuracy)
learn.fine_tune(4, 1e-2)
```
#clean
If you hit a "CUDA out of memory error" after running this cell, click on the menu Kernel, then restart. Instead of executing the cell above, copy and paste the following code in it:
```
from fastai.text.all import *
dls = TextDataLoaders.from_folder(untar_data(URLs.IMDB), valid='test', bs=32)
learn = text_classifier_learner(dls, AWD_LSTM, drop_mult=0.5, metrics=accuracy)
learn.fine_tune(4, 1e-2)
```
This reduces the batch size to 32 (we will explain this later). If you keep hitting the same error, change 32 to 16.
This model is using the ["IMDb Large Movie Review dataset"](https://ai.stanford.edu/~ang/papers/acl11-WordVectorsSentimentAnalysis.pdf) from the paper "Learning Word Vectors for Sentiment Analysis" by Andrew Maas et al. It works well with movie reviews of many thousands of words, but let's test it out on a very short one to see how it does its thing:
```
learn.predict("I really liked that movie!")
```
Here we can see the model has considered the review to be positive. The second part of the result is the index of "pos" in our data vocabulary and the last part is the probabilities attributed to each class (99.6% for "pos" and 0.4% for "neg").
Now it's your turn! Write your own mini movie review, or copy one from the internet, and you can see what this model thinks about it.
### Sidebar: The Order Matters
In a Jupyter notebook, the order in which you execute each cell is very important. It's not like Excel, where everything gets updated as soon as you type something anywhere—it has an inner state that gets updated each time you execute a cell. For instance, when you run the first cell of the notebook (with the "CLICK ME" comment), you create an object called `learn` that contains a model and data for an image classification problem. If we were to run the cell just shown in the text (the one that predicts if a review is good or not) straight after, we would get an error as this `learn` object does not contain a text classification model. This cell needs to be run after the one containing:
```python
from fastai.text.all import *
dls = TextDataLoaders.from_folder(untar_data(URLs.IMDB), valid='test')
learn = text_classifier_learner(dls, AWD_LSTM, drop_mult=0.5,
metrics=accuracy)
learn.fine_tune(4, 1e-2)
```
The outputs themselves can be deceiving, because they include the results of the last time the cell was executed; if you change the code inside a cell without executing it, the old (misleading) results will remain.
Except when we mention it explicitly, the notebooks provided on the [book website](https://book.fast.ai/) are meant to be run in order, from top to bottom. In general, when experimenting, you will find yourself executing cells in any order to go fast (which is a super neat feature of Jupyter Notebook), but once you have explored and arrived at the final version of your code, make sure you can run the cells of your notebooks in order (your future self won't necessarily remember the convoluted path you took otherwise!).
In command mode, pressing `0` twice will restart the *kernel* (which is the engine powering your notebook). This will wipe your state clean and make it as if you had just started in the notebook. Choose Run All Above from the Cell menu to run all cells above the point where you are. We have found this to be very useful when developing the fastai library.
### End sidebar
If you ever have any questions about a fastai method, you should use the function `doc`, passing it the method name:
```python
doc(learn.predict)
```
This will make a small window pop up with content like this:
<img src="images/doc_ex.png" width="600">
A brief one-line explanation is provided by `doc`. The "Show in docs" link takes you to the full documentation, where you'll find all the details and lots of examples. Also, most of fastai's methods are just a handful of lines, so you can click the "source" link to see exactly what's going on behind the scenes.
Let's move on to something much less sexy, but perhaps significantly more widely commercially useful: building models from plain *tabular* data.
> jargon: Tabular: Data that is in the form of a table, such as from a spreadsheet, database, or CSV file. A tabular model is a model that tries to predict one column of a table based on information in other columns of the table.
It turns out that looks very similar too. Here is the code necessary to train a model that will predict whether a person is a high-income earner, based on their socioeconomic background:
```
from fastai.tabular.all import *
path = untar_data(URLs.ADULT_SAMPLE)
dls = TabularDataLoaders.from_csv(path/'adult.csv', path=path, y_names="salary",
cat_names = ['workclass', 'education', 'marital-status', 'occupation',
'relationship', 'race'],
cont_names = ['age', 'fnlwgt', 'education-num'],
procs = [Categorify, FillMissing, Normalize])
learn = tabular_learner(dls, metrics=accuracy)
```
As you see, we had to tell fastai which columns are *categorical* (that is, contain values that are one of a discrete set of choices, such as `occupation`) and which are *continuous* (that is, contain a number that represents a quantity, such as `age`).
There is no pretrained model available for this task (in general, pretrained models are not widely available for any tabular modeling tasks, although some organizations have created them for internal use), so we don't use `fine_tune` in this case. Instead we use `fit_one_cycle`, the most commonly used method for training fastai models *from scratch* (i.e. without transfer learning):
```
learn.fit_one_cycle(3)
```
This model is using the [*Adult* dataset](http://robotics.stanford.edu/~ronnyk/nbtree.pdf), from the paper "Scaling Up the Accuracy of Naive-Bayes Classifiers: a Decision-Tree Hybrid" by Rob Kohavi, which contains some demographic data about individuals (like their education, marital status, race, sex, and whether or not they have an annual income greater than \$50k). The model is over 80\% accurate, and took around 30 seconds to train.
Let's look at one more. Recommendation systems are very important, particularly in e-commerce. Companies like Amazon and Netflix try hard to recommend products or movies that users might like. Here's how to train a model that will predict movies people might like, based on their previous viewing habits, using the [MovieLens dataset](https://doi.org/10.1145/2827872):
```
from fastai.collab import *
path = untar_data(URLs.ML_SAMPLE)
dls = CollabDataLoaders.from_csv(path/'ratings.csv')
learn = collab_learner(dls, y_range=(0.5,5.5))
learn.fine_tune(10)
```
This model is predicting movie ratings on a scale of 0.5 to 5.0 to within around 0.6 average error. Since we're predicting a continuous number, rather than a category, we have to tell fastai what range our target has, using the `y_range` parameter.
Although we're not actually using a pretrained model (for the same reason that we didn't for the tabular model), this example shows that fastai lets us use `fine_tune` anyway in this case (you'll learn how and why this works in <<chapter_pet_breeds>>). Sometimes it's best to experiment with `fine_tune` versus `fit_one_cycle` to see which works best for your dataset.
We can use the same `show_results` call we saw earlier to view a few examples of user and movie IDs, actual ratings, and predictions:
```
learn.show_results()
```
### Sidebar: Datasets: Food for Models
You’ve already seen quite a few models in this section, each one trained using a different dataset to do a different task. In machine learning and deep learning, we can’t do anything without data. So, the people that create datasets for us to train our models on are the (often underappreciated) heroes. Some of the most useful and important datasets are those that become important *academic baselines*; that is, datasets that are widely studied by researchers and used to compare algorithmic changes. Some of these become household names (at least, among households that train models!), such as MNIST, CIFAR-10, and ImageNet.
The datasets used in this book have been selected because they provide great examples of the kinds of data that you are likely to encounter, and the academic literature has many examples of model results using these datasets to which you can compare your work.
Most datasets used in this book took the creators a lot of work to build. For instance, later in the book we’ll be showing you how to create a model that can translate between French and English. The key input to this is a French/English parallel text corpus prepared back in 2009 by Professor Chris Callison-Burch of the University of Pennsylvania. This dataset contains over 20 million sentence pairs in French and English. He built the dataset in a really clever way: by crawling millions of Canadian web pages (which are often multilingual) and then using a set of simple heuristics to transform URLs of French content onto URLs pointing to the same content in English.
As you look at datasets throughout this book, think about where they might have come from, and how they might have been curated. Then think about what kinds of interesting datasets you could create for your own projects. (We’ll even take you step by step through the process of creating your own image dataset soon.)
fast.ai has spent a lot of time creating cut-down versions of popular datasets that are specially designed to support rapid prototyping and experimentation, and to be easier to learn with. In this book we will often start by using one of the cut-down versions and later scale up to the full-size version (just as we're doing in this chapter!). In fact, this is how the world’s top practitioners do their modeling in practice; they do most of their experimentation and prototyping with subsets of their data, and only use the full dataset when they have a good understanding of what they have to do.
### End sidebar
Each of the models we trained showed a training and validation loss. A good validation set is one of the most important pieces of the training process. Let's see why and learn how to create one.
## Validation Sets and Test Sets
As we've discussed, the goal of a model is to make predictions about data. But the model training process is fundamentally dumb. If we trained a model with all our data, and then evaluated the model using that same data, we would not be able to tell how well our model can perform on data it hasn’t seen. Without this very valuable piece of information to guide us in training our model, there is a very good chance it would become good at making predictions about that data but would perform poorly on new data.
To avoid this, our first step was to split our dataset into two sets: the *training set* (which our model sees in training) and the *validation set*, also known as the *development set* (which is used only for evaluation). This lets us test that the model learns lessons from the training data that generalize to new data, the validation data.
One way to understand this situation is that, in a sense, we don't want our model to get good results by "cheating." If it makes an accurate prediction for a data item, that should be because it has learned characteristics of that kind of item, and not because the model has been shaped by *actually having seen that particular item*.
Splitting off our validation data means our model never sees it in training and so is completely untainted by it, and is not cheating in any way. Right?
In fact, not necessarily. The situation is more subtle. This is because in realistic scenarios we rarely build a model just by training its weight parameters once. Instead, we are likely to explore many versions of a model through various modeling choices regarding network architecture, learning rates, data augmentation strategies, and other factors we will discuss in upcoming chapters. Many of these choices can be described as choices of *hyperparameters*. The word reflects that they are parameters about parameters, since they are the higher-level choices that govern the meaning of the weight parameters.
The problem is that even though the ordinary training process is only looking at predictions on the training data when it learns values for the weight parameters, the same is not true of us. We, as modelers, are evaluating the model by looking at predictions on the validation data when we decide to explore new hyperparameter values! So subsequent versions of the model are, indirectly, shaped by us having seen the validation data. Just as the automatic training process is in danger of overfitting the training data, we are in danger of overfitting the validation data through human trial and error and exploration.
The solution to this conundrum is to introduce another level of even more highly reserved data, the *test set*. Just as we hold back the validation data from the training process, we must hold back the test set data even from ourselves. It cannot be used to improve the model; it can only be used to evaluate the model at the very end of our efforts. In effect, we define a hierarchy of cuts of our data, based on how fully we want to hide it from training and modeling processes: training data is fully exposed, the validation data is less exposed, and test data is totally hidden. This hierarchy parallels the different kinds of modeling and evaluation processes themselves—the automatic training process with back propagation, the more manual process of trying different hyper-parameters between training sessions, and the assessment of our final result.
The test and validation sets should have enough data to ensure that you get a good estimate of your accuracy. If you're creating a cat detector, for instance, you generally want at least 30 cats in your validation set. That means that if you have a dataset with thousands of items, using the default 20% validation set size may be more than you need. On the other hand, if you have lots of data, using some of it for validation probably doesn't have any downsides.
Having two levels of "reserved data"—a validation set and a test set, with one level representing data that you are virtually hiding from yourself—may seem a bit extreme. But the reason it is often necessary is because models tend to gravitate toward the simplest way to do good predictions (memorization), and we as fallible humans tend to gravitate toward fooling ourselves about how well our models are performing. The discipline of the test set helps us keep ourselves intellectually honest. That doesn't mean we *always* need a separate test set—if you have very little data, you may need to just have a validation set—but generally it's best to use one if at all possible.
This same discipline can be critical if you intend to hire a third party to perform modeling work on your behalf. A third party might not understand your requirements accurately, or their incentives might even encourage them to misunderstand them. A good test set can greatly mitigate these risks and let you evaluate whether their work solves your actual problem.
To put it bluntly, if you're a senior decision maker in your organization (or you're advising senior decision makers), the most important takeaway is this: if you ensure that you really understand what test and validation sets are and why they're important, then you'll avoid the single biggest source of failures we've seen when organizations decide to use AI. For instance, if you're considering bringing in an external vendor or service, make sure that you hold out some test data that the vendor *never gets to see*. Then *you* check their model on your test data, using a metric that *you* choose based on what actually matters to you in practice, and *you* decide what level of performance is adequate. (It's also a good idea for you to try out some simple baseline yourself, so you know what a really simple model can achieve. Often it'll turn out that your simple model performs just as well as one produced by an external "expert"!)
### Use Judgment in Defining Test Sets
To do a good job of defining a validation set (and possibly a test set), you will sometimes want to do more than just randomly grab a fraction of your original dataset. Remember: a key property of the validation and test sets is that they must be representative of the new data you will see in the future. This may sound like an impossible order! By definition, you haven’t seen this data yet. But you usually still do know some things.
It's instructive to look at a few example cases. Many of these examples come from predictive modeling competitions on the [Kaggle](https://www.kaggle.com/) platform, which is a good representation of problems and methods you might see in practice.
One case might be if you are looking at time series data. For a time series, choosing a random subset of the data will be both too easy (you can look at the data both before and after the dates your are trying to predict) and not representative of most business use cases (where you are using historical data to build a model for use in the future). If your data includes the date and you are building a model to use in the future, you will want to choose a continuous section with the latest dates as your validation set (for instance, the last two weeks or last month of available data).
Suppose you want to split the time series data in <<timeseries1>> into training and validation sets.
<img src="images/timeseries1.png" width="400" id="timeseries1" caption="A time series" alt="A serie of values">
A random subset is a poor choice (too easy to fill in the gaps, and not indicative of what you'll need in production), as we can see in <<timeseries2>>.
<img src="images/timeseries2.png" width="400" id="timeseries2" caption="A poor training subset" alt="Random training subset">
Instead, use the earlier data as your training set (and the later data for the validation set), as shown in <<timeseries3>>.
<img src="images/timeseries3.png" width="400" id="timeseries3" caption="A good training subset" alt="Training subset using the data up to a certain timestamp">
For example, Kaggle had a competition to [predict the sales in a chain of Ecuadorian grocery stores](https://www.kaggle.com/c/favorita-grocery-sales-forecasting). Kaggle's training data ran from Jan 1 2013 to Aug 15 2017, and the test data spanned Aug 16 2017 to Aug 31 2017. That way, the competition organizer ensured that entrants were making predictions for a time period that was *in the future*, from the perspective of their model. This is similar to the way quant hedge fund traders do *back-testing* to check whether their models are predictive of future periods, based on past data.
A second common case is when you can easily anticipate ways the data you will be making predictions for in production may be *qualitatively different* from the data you have to train your model with.
In the Kaggle [distracted driver competition](https://www.kaggle.com/c/state-farm-distracted-driver-detection), the independent variables are pictures of drivers at the wheel of a car, and the dependent variables are categories such as texting, eating, or safely looking ahead. Lots of pictures are of the same drivers in different positions, as we can see in <<img_driver>>. If you were an insurance company building a model from this data, note that you would be most interested in how the model performs on drivers it hasn't seen before (since you would likely have training data only for a small group of people). In recognition of this, the test data for the competition consists of images of people that don't appear in the training set.
<img src="images/driver.PNG" width="600" id="img_driver" caption="Two pictures from the training data" alt="Two pictures from the training data, showing the same driver">
If you put one of the images in <<img_driver>> in your training set and one in the validation set, your model will have an easy time making a prediction for the one in the validation set, so it will seem to be performing better than it would on new people. Another perspective is that if you used all the people in training your model, your model might be overfitting to particularities of those specific people, and not just learning the states (texting, eating, etc.).
A similar dynamic was at work in the [Kaggle fisheries competition](https://www.kaggle.com/c/the-nature-conservancy-fisheries-monitoring) to identify the species of fish caught by fishing boats in order to reduce illegal fishing of endangered populations. The test set consisted of boats that didn't appear in the training data. This means that you'd want your validation set to include boats that are not in the training set.
Sometimes it may not be clear how your validation data will differ. For instance, for a problem using satellite imagery, you'd need to gather more information on whether the training set just contained certain geographic locations, or if it came from geographically scattered data.
Now that you have gotten a taste of how to build a model, you can decide what you want to dig into next.
## A _Choose Your Own Adventure_ moment
If you would like to learn more about how to use deep learning models in practice, including how to identify and fix errors, create a real working web application, and avoid your model causing unexpected harm to your organization or society more generally, then keep reading the next two chapters. If you would like to start learning the foundations of how deep learning works under the hood, skip to <<chapter_mnist_basics>>. (Did you ever read _Choose Your Own Adventure_ books as a kid? Well, this is kind of like that… except with more deep learning than that book series contained.)
You will need to read all these chapters to progress further in the book, but it is totally up to you which order you read them in. They don't depend on each other. If you skip ahead to <<chapter_mnist_basics>>, we will remind you at the end to come back and read the chapters you skipped over before you go any further.
## Questionnaire
It can be hard to know in pages and pages of prose what the key things are that you really need to focus on and remember. So, we've prepared a list of questions and suggested steps to complete at the end of each chapter. All the answers are in the text of the chapter, so if you're not sure about anything here, reread that part of the text and make sure you understand it. Answers to all these questions are also available on the [book's website](https://book.fast.ai). You can also visit [the forums](https://forums.fast.ai) if you get stuck to get help from other folks studying this material.
For more questions, including detailed answers and links to the video timeline, have a look at Radek Osmulski's [aiquizzes](http://aiquizzes.com/howto).
1. Do you need these for deep learning?
- Lots of math T / F
- Lots of data T / F
- Lots of expensive computers T / F
- A PhD T / F
1. Name five areas where deep learning is now the best in the world.
1. What was the name of the first device that was based on the principle of the artificial neuron?
1. Based on the book of the same name, what are the requirements for parallel distributed processing (PDP)?
1. What were the two theoretical misunderstandings that held back the field of neural networks?
1. What is a GPU?
1. Open a notebook and execute a cell containing: `1+1`. What happens?
1. Follow through each cell of the stripped version of the notebook for this chapter. Before executing each cell, guess what will happen.
1. Complete the Jupyter Notebook online appendix.
1. Why is it hard to use a traditional computer program to recognize images in a photo?
1. What did Samuel mean by "weight assignment"?
1. What term do we normally use in deep learning for what Samuel called "weights"?
1. Draw a picture that summarizes Samuel's view of a machine learning model.
1. Why is it hard to understand why a deep learning model makes a particular prediction?
1. What is the name of the theorem that shows that a neural network can solve any mathematical problem to any level of accuracy?
1. What do you need in order to train a model?
1. How could a feedback loop impact the rollout of a predictive policing model?
1. Do we always have to use 224×224-pixel images with the cat recognition model?
1. What is the difference between classification and regression?
1. What is a validation set? What is a test set? Why do we need them?
1. What will fastai do if you don't provide a validation set?
1. Can we always use a random sample for a validation set? Why or why not?
1. What is overfitting? Provide an example.
1. What is a metric? How does it differ from "loss"?
1. How can pretrained models help?
1. What is the "head" of a model?
1. What kinds of features do the early layers of a CNN find? How about the later layers?
1. Are image models only useful for photos?
1. What is an "architecture"?
1. What is segmentation?
1. What is `y_range` used for? When do we need it?
1. What are "hyperparameters"?
1. What's the best way to avoid failures when using AI in an organization?
### Further Research
Each chapter also has a "Further Research" section that poses questions that aren't fully answered in the text, or gives more advanced assignments. Answers to these questions aren't on the book's website; you'll need to do your own research!
1. Why is a GPU useful for deep learning? How is a CPU different, and why is it less effective for deep learning?
1. Try to think of three areas where feedback loops might impact the use of machine learning. See if you can find documented examples of that happening in practice.
|
github_jupyter
|
#hide
!pip install -Uqq fastbook
import fastbook
fastbook.setup_book()
#hide
from fastbook import *
[[myths]]
.What you don't need to do deep learning
[options="header"]
|======
| Myth (don't need) | Truth
| Lots of math | Just high school math is sufficient
| Lots of data | We've seen record-breaking results with <50 items of data
| Lots of expensive computers | You can get what you need for state of the art work for free
|======
#id first_training
#caption Results from the first training
# CLICK ME
from fastai.vision.all import *
path = untar_data(URLs.PETS)/'images'
def is_cat(x): return x[0].isupper()
dls = ImageDataLoaders.from_name_func(
path, get_image_files(path), valid_pct=0.2, seed=42,
label_func=is_cat, item_tfms=Resize(224))
learn = cnn_learner(dls, resnet34, metrics=error_rate)
learn.fine_tune(1)
1+1
img = PILImage.create(image_cat())
img.to_thumb(192)
#hide_output
uploader = widgets.FileUpload()
uploader
#hide
# For the book, we can't actually click an upload button, so we fake it
uploader = SimpleNamespace(data = ['images/chapter1_cat_example.jpg'])
img = PILImage.create(uploader.data[0])
is_cat,_,probs = learn.predict(img)
print(f"Is this a cat?: {is_cat}.")
print(f"Probability it's a cat: {probs[1].item():.6f}")
#hide_input
#caption A traditional program
#id basic_program
#alt Pipeline inputs, program, results
gv('''program[shape=box3d width=1 height=0.7]
inputs->program->results''')
#hide_input
#caption A program using weight assignment
#id weight_assignment
gv('''model[shape=box3d width=1 height=0.7]
inputs->model->results; weights->model''')
#hide_input
#caption Training a machine learning model
#id training_loop
#alt The basic training loop
gv('''ordering=in
model[shape=box3d width=1 height=0.7]
inputs->model->results; weights->model; results->performance
performance->weights[constraint=false label=update]''')
#hide_input
#caption Using a trained model as a program
#id using_model
gv('''model[shape=box3d width=1 height=0.7]
inputs->model->results''')
#hide_input
#caption Detailed training loop
#id detailed_loop
gv('''ordering=in
model[shape=box3d width=1 height=0.7 label=architecture]
inputs->model->predictions; parameters->model; labels->loss; predictions->loss
loss->parameters[constraint=false label=update]''')
from fastai.vision.all import *
path = untar_data(URLs.PETS)/'images'
def is_cat(x): return x[0].isupper()
dls = ImageDataLoaders.from_name_func(
path, get_image_files(path), valid_pct=0.2, seed=42,
label_func=is_cat, item_tfms=Resize(224))
learn = cnn_learner(dls, resnet34, metrics=error_rate)
learn.fine_tune(1)
[[dljargon]]
.Deep learning vocabulary
[options="header"]
|=====
| Term | Meaning
|Label | The data that we're trying to predict, such as "dog" or "cat"
|Architecture | The _template_ of the model that we're trying to fit; the actual mathematical function that we're passing the input data and parameters to
|Model | The combination of the architecture with a particular set of parameters
|Parameters | The values in the model that change what task it can do, and are updated through model training
|Fit | Update the parameters of the model such that the predictions of the model using the input data match the target labels
|Train | A synonym for _fit_
|Pretrained model | A model that has already been trained, generally using a large dataset, and will be fine-tuned
|Fine-tune | Update a pretrained model for a different task
|Epoch | One complete pass through the input data
|Loss | A measure of how good the model is, chosen to drive training via SGD
|Metric | A measurement of how good the model is, using the validation set, chosen for human consumption
|Validation set | A set of data held out from training, used only for measuring how good the model is
|Training set | The data used for fitting the model; does not include any data from the validation set
|Overfitting | Training a model in such a way that it _remembers_ specific features of the input data, rather than generalizing well to data not seen during training
|CNN | Convolutional neural network; a type of neural network that works particularly well for computer vision tasks
|=====
path = untar_data(URLs.CAMVID_TINY)
dls = SegmentationDataLoaders.from_label_func(
path, bs=8, fnames = get_image_files(path/"images"),
label_func = lambda o: path/'labels'/f'{o.stem}_P{o.suffix}',
codes = np.loadtxt(path/'codes.txt', dtype=str)
)
learn = unet_learner(dls, resnet34)
learn.fine_tune(8)
learn.show_results(max_n=6, figsize=(7,8))
from fastai.text.all import *
dls = TextDataLoaders.from_folder(untar_data(URLs.IMDB), valid='test')
learn = text_classifier_learner(dls, AWD_LSTM, drop_mult=0.5, metrics=accuracy)
learn.fine_tune(4, 1e-2)
from fastai.text.all import *
dls = TextDataLoaders.from_folder(untar_data(URLs.IMDB), valid='test', bs=32)
learn = text_classifier_learner(dls, AWD_LSTM, drop_mult=0.5, metrics=accuracy)
learn.fine_tune(4, 1e-2)
learn.predict("I really liked that movie!")
from fastai.text.all import *
dls = TextDataLoaders.from_folder(untar_data(URLs.IMDB), valid='test')
learn = text_classifier_learner(dls, AWD_LSTM, drop_mult=0.5,
metrics=accuracy)
learn.fine_tune(4, 1e-2)
doc(learn.predict)
from fastai.tabular.all import *
path = untar_data(URLs.ADULT_SAMPLE)
dls = TabularDataLoaders.from_csv(path/'adult.csv', path=path, y_names="salary",
cat_names = ['workclass', 'education', 'marital-status', 'occupation',
'relationship', 'race'],
cont_names = ['age', 'fnlwgt', 'education-num'],
procs = [Categorify, FillMissing, Normalize])
learn = tabular_learner(dls, metrics=accuracy)
learn.fit_one_cycle(3)
from fastai.collab import *
path = untar_data(URLs.ML_SAMPLE)
dls = CollabDataLoaders.from_csv(path/'ratings.csv')
learn = collab_learner(dls, y_range=(0.5,5.5))
learn.fine_tune(10)
learn.show_results()
| 0.777807 | 0.960952 |
```
# default_exp update
```
# Calculating Carbon Intensity
<br>
### Imports
```
#exports
import pandas as pd
import numpy as np
import json
import dask
import dask.dataframe as dd
import matplotlib.pyplot as plt
import seaborn as sns
import os
from ipypb import track
from warnings import warn
import FEAutils as hlp
from IPython.display import JSON
# Loading
df_emissions = pd.read_csv('../data/raw/pi/pi_emissions.csv').set_index('authorisation id / permit id')
# Checking
assert (df_emissions<0).sum().sum() == 0, 'Negative emissions are present'
df_emissions.head()
#exports
def get_B1610_columns(data_dir):
B1610_files = [f for f in os.listdir(data_dir) if '.csv' in f]
columns = []
for B1610_file in track(B1610_files, label='columns'):
df_B1610_week = pd.read_csv(f'{data_dir}/{B1610_file}')
columns += list(df_B1610_week.columns)
columns = ['datetime'] + sorted(list(set(columns)-set(['datetime'])))
return columns
B1610_dir = '../data/raw/elexon'
B1610_columns = get_B1610_columns(B1610_dir)
B1610_columns[:5]
#exports
@dask.delayed
def read_B1610_file(filename, columns):
df_B1610_week = pd.read_csv(filename)
cols_to_add = list(set(columns) - set(df_B1610_week.columns))
df_B1610_week[cols_to_add] = np.NaN
df_B1610_week = df_B1610_week[columns]
return df_B1610_week
def load_B1610_dask_stream_df(data_dir, dt_col='datetime', columns=None):
# Identifying columns
if columns is None:
columns = get_B1610_columns(data_dir)
# Loading data
B1610_files = [f for f in os.listdir(data_dir) if '.csv' in f]
df_B1610 = dd.from_delayed([read_B1610_file(f'{data_dir}/{B1610_file}', columns) for B1610_file in B1610_files])
# Formatting date index
if dt_col is not None:
df_B1610[dt_col] = df_B1610[dt_col].map(lambda dt: pd.to_datetime(dt, format='%Y-%m-%d %H:%M:%S', errors='coerce', utc=True))
df_B1610 = df_B1610.set_index(dt_col)
return df_B1610
%%time
ddf_B1610 = load_B1610_dask_stream_df(B1610_dir, columns=B1610_columns)
df_B1610 = ddf_B1610.compute()
df_B1610.head()
with open('../data/mappings/ngc_bmu_id_to_pi_permits.json', 'r') as fp:
ngc_bmu_id_to_pi_permits = json.load(fp)
JSON([ngc_bmu_id_to_pi_permits])
with open('../data/mappings/ngc_bmu_id_to_fuel.json', 'r') as fp:
ngc_bmu_id_to_fuel = json.load(fp)
JSON([ngc_bmu_id_to_fuel])
s_ngc_bmu_id_to_fuel = pd.Series(ngc_bmu_id_to_fuel)
potential_emitting_ngc_bmu_ids = s_ngc_bmu_id_to_fuel[~s_ngc_bmu_id_to_fuel.isin(['wind', 'npshyd', 'nuclear', 'ps', 'hydro'])].index
print(f'There are {len(potential_emitting_ngc_bmu_ids)} NGC BMU ids that are potentially carbon emitting')
print(f'There are {len(ngc_bmu_id_to_pi_permits)} NGC BMU ids that have been mapped to a PI permit')
ngc_bmu_ids_in_B1610_without_fuel_mapping = sorted(list(set(df_B1610.columns) - set(ngc_bmu_id_to_fuel.keys())))
assert len(ngc_bmu_ids_in_B1610_without_fuel_mapping) == 0, f'The following NGC BMU ids are associated with power plants in the B1610 dataframe but do not have an associated fuel type:\n{", ".join(ngc_bmu_ids_in_B1610_without_fuel_mapping)}'
flatten_list = lambda t: [item for sublist in t for item in sublist]
account_ids_not_in_emissions_df = sorted(list(set(flatten_list([[elem] if isinstance(elem, str) else elem for elem in ngc_bmu_id_to_pi_permits.values()])) - set(df_emissions.index)))
assert len(account_ids_not_in_emissions_df) == 0, f'The following account ids are associated with power plants but do not have an entry in the emissions dataframe:\n{", ".join(account_ids_not_in_emissions_df)}'
ngc_bmu_ids_not_in_B1610_df = sorted(list(set(ngc_bmu_id_to_pi_permits.keys()) - set(potential_emitting_ngc_bmu_ids)))
if len(ngc_bmu_ids_not_in_B1610_df) > 0:
warn(f'The following account ids are associated with carbon emitting power plants but do not have an entry in the emissions dataframe:\n{", ".join(ngc_bmu_ids_not_in_B1610_df)}')
relevant_ngc_bmu_ids = sorted(list(set(potential_emitting_ngc_bmu_ids) & set(B1610_columns) & set(ngc_bmu_id_to_pi_permits.keys())))
s_ngc_bmu_id_to_pi_permits = pd.Series(ngc_bmu_id_to_pi_permits).loc[relevant_ngc_bmu_ids]
s_ngc_bmu_id_to_pi_permit_groups = s_ngc_bmu_id_to_pi_permits.apply(lambda x: '__'.join(x) if isinstance(x, list) else x)
unique_accounts_ids = sorted(list(set(flatten_list([[elem] if isinstance(elem, str) else elem for elem in s_ngc_bmu_id_to_pi_permits.values]))))
df_power = pd.DataFrame()
for accounts_id in track(unique_accounts_ids):
ngc_bmu_ids_associated_with_account = list(s_ngc_bmu_id_to_pi_permits[s_ngc_bmu_id_to_pi_permit_groups.str.contains(accounts_id)].index)
df_power[accounts_id] = df_B1610[ngc_bmu_ids_associated_with_account].sum(axis=1)
df_power.head()
df_annual_power = df_power['2016':'2020'].resample('Y').sum()/2 # /2 to convert MW to MWh
df_annual_power.index = df_annual_power.index.year
df_annual_power
grouped_pi_permits = list(s_ngc_bmu_id_to_pi_permit_groups[s_ngc_bmu_id_to_pi_permit_groups.str.contains('__')].drop_duplicates().str.split('__').values)
print(f'There are {len(grouped_pi_permits)} installations which are associated with multiple PI permits')
df_grouped_emissions = pd.DataFrame()
df_grouped_power = pd.DataFrame()
non_grouped_permits = list(s_ngc_bmu_id_to_pi_permit_groups[~s_ngc_bmu_id_to_pi_permit_groups.str.contains('__')].unique())
df_grouped_emissions[non_grouped_permits] = df_emissions.T[non_grouped_permits]
df_grouped_power[non_grouped_permits] = df_annual_power[non_grouped_permits]
for pi_permits_group in grouped_pi_permits:
group_name = '__'.join(pi_permits_group)
s_group_emissions = df_emissions.T[pi_permits_group].sum(axis=1)
df_grouped_emissions[group_name] = s_group_emissions
df_grouped_power[group_name] = df_annual_power[pi_permits_group].sum(axis=1)
df_grouped_emissions.index = df_grouped_emissions.index.astype(int)
df_grouped_emissions.head(3)
s_emissions
df_annual_carbon_intensity = pd.DataFrame()
s_carbon_intensity = pd.Series(index=df_grouped_emissions.columns, dtype='float64')
for account_id in df_grouped_emissions.columns:
s_emissions = df_grouped_emissions.loc[2016:2020, account_id]
s_power = df_grouped_power.loc[2016:2020, account_id]
df_annual_carbon_intensity[account_id] = s_emissions/s_power
weights = s_power.loc[df_annual_carbon_intensity[account_id].dropna().index]
if len(weights) > 0:
s_carbon_intensity[account_id] = np.average(df_annual_carbon_intensity[account_id].dropna(), weights=weights)
df_annual_carbon_intensity = df_annual_carbon_intensity.replace(np.inf, np.nan)
df_annual_carbon_intensity.head()
df_ngc_bmu_emissions = pd.DataFrame({
'fuel': s_ngc_bmu_id_to_fuel.loc[s_ngc_bmu_id_to_pi_permit_groups.index],
'gco2_per_kWh': s_ngc_bmu_id_to_pi_permit_groups.map(s_carbon_intensity),
'pi_permits': s_ngc_bmu_id_to_pi_permit_groups
})
df_ngc_bmu_emissions.index.name = 'ngc_bmu_id'
df_ngc_bmu_emissions.head()
# want to find eutl_account groups that have only a single fuel type
# then report the value_counts of the pure types
s_pi_permits_fuel_types = df_ngc_bmu_emissions.groupby('pi_permits')['fuel'].unique()
s_pi_permits_pure_fuel_types = s_pi_permits_fuel_types[s_pi_permits_fuel_types.apply(len)==1].apply(lambda x: x[0])
s_pi_permits_pure_fuel_types.value_counts()
# need to focus on the permits rather than ngc when plotting
ocgt_permits = s_pi_permits_pure_fuel_types[s_pi_permits_pure_fuel_types=='coal'].index
df_filtered_emissions = df_ngc_bmu_emissions[df_ngc_bmu_emissions['pi_permits'].isin(ocgt_permits)]
df_filtered_emissions
missing_relevant_bmus_from_B1610 = sorted(list((set(potential_emitting_ngc_bmu_ids) & set(B1610_columns)) - set(df_ngc_bmu_emissions.index)))
pct_missing = len(missing_relevant_bmus_from_B1610)/len(set(potential_emitting_ngc_bmu_ids) & set(B1610_columns))
df_ngc_bmu_emissions.to_csv('../data/intermediate/pi_first_estimate.csv')
```
|
github_jupyter
|
# default_exp update
#exports
import pandas as pd
import numpy as np
import json
import dask
import dask.dataframe as dd
import matplotlib.pyplot as plt
import seaborn as sns
import os
from ipypb import track
from warnings import warn
import FEAutils as hlp
from IPython.display import JSON
# Loading
df_emissions = pd.read_csv('../data/raw/pi/pi_emissions.csv').set_index('authorisation id / permit id')
# Checking
assert (df_emissions<0).sum().sum() == 0, 'Negative emissions are present'
df_emissions.head()
#exports
def get_B1610_columns(data_dir):
B1610_files = [f for f in os.listdir(data_dir) if '.csv' in f]
columns = []
for B1610_file in track(B1610_files, label='columns'):
df_B1610_week = pd.read_csv(f'{data_dir}/{B1610_file}')
columns += list(df_B1610_week.columns)
columns = ['datetime'] + sorted(list(set(columns)-set(['datetime'])))
return columns
B1610_dir = '../data/raw/elexon'
B1610_columns = get_B1610_columns(B1610_dir)
B1610_columns[:5]
#exports
@dask.delayed
def read_B1610_file(filename, columns):
df_B1610_week = pd.read_csv(filename)
cols_to_add = list(set(columns) - set(df_B1610_week.columns))
df_B1610_week[cols_to_add] = np.NaN
df_B1610_week = df_B1610_week[columns]
return df_B1610_week
def load_B1610_dask_stream_df(data_dir, dt_col='datetime', columns=None):
# Identifying columns
if columns is None:
columns = get_B1610_columns(data_dir)
# Loading data
B1610_files = [f for f in os.listdir(data_dir) if '.csv' in f]
df_B1610 = dd.from_delayed([read_B1610_file(f'{data_dir}/{B1610_file}', columns) for B1610_file in B1610_files])
# Formatting date index
if dt_col is not None:
df_B1610[dt_col] = df_B1610[dt_col].map(lambda dt: pd.to_datetime(dt, format='%Y-%m-%d %H:%M:%S', errors='coerce', utc=True))
df_B1610 = df_B1610.set_index(dt_col)
return df_B1610
%%time
ddf_B1610 = load_B1610_dask_stream_df(B1610_dir, columns=B1610_columns)
df_B1610 = ddf_B1610.compute()
df_B1610.head()
with open('../data/mappings/ngc_bmu_id_to_pi_permits.json', 'r') as fp:
ngc_bmu_id_to_pi_permits = json.load(fp)
JSON([ngc_bmu_id_to_pi_permits])
with open('../data/mappings/ngc_bmu_id_to_fuel.json', 'r') as fp:
ngc_bmu_id_to_fuel = json.load(fp)
JSON([ngc_bmu_id_to_fuel])
s_ngc_bmu_id_to_fuel = pd.Series(ngc_bmu_id_to_fuel)
potential_emitting_ngc_bmu_ids = s_ngc_bmu_id_to_fuel[~s_ngc_bmu_id_to_fuel.isin(['wind', 'npshyd', 'nuclear', 'ps', 'hydro'])].index
print(f'There are {len(potential_emitting_ngc_bmu_ids)} NGC BMU ids that are potentially carbon emitting')
print(f'There are {len(ngc_bmu_id_to_pi_permits)} NGC BMU ids that have been mapped to a PI permit')
ngc_bmu_ids_in_B1610_without_fuel_mapping = sorted(list(set(df_B1610.columns) - set(ngc_bmu_id_to_fuel.keys())))
assert len(ngc_bmu_ids_in_B1610_without_fuel_mapping) == 0, f'The following NGC BMU ids are associated with power plants in the B1610 dataframe but do not have an associated fuel type:\n{", ".join(ngc_bmu_ids_in_B1610_without_fuel_mapping)}'
flatten_list = lambda t: [item for sublist in t for item in sublist]
account_ids_not_in_emissions_df = sorted(list(set(flatten_list([[elem] if isinstance(elem, str) else elem for elem in ngc_bmu_id_to_pi_permits.values()])) - set(df_emissions.index)))
assert len(account_ids_not_in_emissions_df) == 0, f'The following account ids are associated with power plants but do not have an entry in the emissions dataframe:\n{", ".join(account_ids_not_in_emissions_df)}'
ngc_bmu_ids_not_in_B1610_df = sorted(list(set(ngc_bmu_id_to_pi_permits.keys()) - set(potential_emitting_ngc_bmu_ids)))
if len(ngc_bmu_ids_not_in_B1610_df) > 0:
warn(f'The following account ids are associated with carbon emitting power plants but do not have an entry in the emissions dataframe:\n{", ".join(ngc_bmu_ids_not_in_B1610_df)}')
relevant_ngc_bmu_ids = sorted(list(set(potential_emitting_ngc_bmu_ids) & set(B1610_columns) & set(ngc_bmu_id_to_pi_permits.keys())))
s_ngc_bmu_id_to_pi_permits = pd.Series(ngc_bmu_id_to_pi_permits).loc[relevant_ngc_bmu_ids]
s_ngc_bmu_id_to_pi_permit_groups = s_ngc_bmu_id_to_pi_permits.apply(lambda x: '__'.join(x) if isinstance(x, list) else x)
unique_accounts_ids = sorted(list(set(flatten_list([[elem] if isinstance(elem, str) else elem for elem in s_ngc_bmu_id_to_pi_permits.values]))))
df_power = pd.DataFrame()
for accounts_id in track(unique_accounts_ids):
ngc_bmu_ids_associated_with_account = list(s_ngc_bmu_id_to_pi_permits[s_ngc_bmu_id_to_pi_permit_groups.str.contains(accounts_id)].index)
df_power[accounts_id] = df_B1610[ngc_bmu_ids_associated_with_account].sum(axis=1)
df_power.head()
df_annual_power = df_power['2016':'2020'].resample('Y').sum()/2 # /2 to convert MW to MWh
df_annual_power.index = df_annual_power.index.year
df_annual_power
grouped_pi_permits = list(s_ngc_bmu_id_to_pi_permit_groups[s_ngc_bmu_id_to_pi_permit_groups.str.contains('__')].drop_duplicates().str.split('__').values)
print(f'There are {len(grouped_pi_permits)} installations which are associated with multiple PI permits')
df_grouped_emissions = pd.DataFrame()
df_grouped_power = pd.DataFrame()
non_grouped_permits = list(s_ngc_bmu_id_to_pi_permit_groups[~s_ngc_bmu_id_to_pi_permit_groups.str.contains('__')].unique())
df_grouped_emissions[non_grouped_permits] = df_emissions.T[non_grouped_permits]
df_grouped_power[non_grouped_permits] = df_annual_power[non_grouped_permits]
for pi_permits_group in grouped_pi_permits:
group_name = '__'.join(pi_permits_group)
s_group_emissions = df_emissions.T[pi_permits_group].sum(axis=1)
df_grouped_emissions[group_name] = s_group_emissions
df_grouped_power[group_name] = df_annual_power[pi_permits_group].sum(axis=1)
df_grouped_emissions.index = df_grouped_emissions.index.astype(int)
df_grouped_emissions.head(3)
s_emissions
df_annual_carbon_intensity = pd.DataFrame()
s_carbon_intensity = pd.Series(index=df_grouped_emissions.columns, dtype='float64')
for account_id in df_grouped_emissions.columns:
s_emissions = df_grouped_emissions.loc[2016:2020, account_id]
s_power = df_grouped_power.loc[2016:2020, account_id]
df_annual_carbon_intensity[account_id] = s_emissions/s_power
weights = s_power.loc[df_annual_carbon_intensity[account_id].dropna().index]
if len(weights) > 0:
s_carbon_intensity[account_id] = np.average(df_annual_carbon_intensity[account_id].dropna(), weights=weights)
df_annual_carbon_intensity = df_annual_carbon_intensity.replace(np.inf, np.nan)
df_annual_carbon_intensity.head()
df_ngc_bmu_emissions = pd.DataFrame({
'fuel': s_ngc_bmu_id_to_fuel.loc[s_ngc_bmu_id_to_pi_permit_groups.index],
'gco2_per_kWh': s_ngc_bmu_id_to_pi_permit_groups.map(s_carbon_intensity),
'pi_permits': s_ngc_bmu_id_to_pi_permit_groups
})
df_ngc_bmu_emissions.index.name = 'ngc_bmu_id'
df_ngc_bmu_emissions.head()
# want to find eutl_account groups that have only a single fuel type
# then report the value_counts of the pure types
s_pi_permits_fuel_types = df_ngc_bmu_emissions.groupby('pi_permits')['fuel'].unique()
s_pi_permits_pure_fuel_types = s_pi_permits_fuel_types[s_pi_permits_fuel_types.apply(len)==1].apply(lambda x: x[0])
s_pi_permits_pure_fuel_types.value_counts()
# need to focus on the permits rather than ngc when plotting
ocgt_permits = s_pi_permits_pure_fuel_types[s_pi_permits_pure_fuel_types=='coal'].index
df_filtered_emissions = df_ngc_bmu_emissions[df_ngc_bmu_emissions['pi_permits'].isin(ocgt_permits)]
df_filtered_emissions
missing_relevant_bmus_from_B1610 = sorted(list((set(potential_emitting_ngc_bmu_ids) & set(B1610_columns)) - set(df_ngc_bmu_emissions.index)))
pct_missing = len(missing_relevant_bmus_from_B1610)/len(set(potential_emitting_ngc_bmu_ids) & set(B1610_columns))
df_ngc_bmu_emissions.to_csv('../data/intermediate/pi_first_estimate.csv')
| 0.288369 | 0.613468 |
# Transfer Learning
In this notebook, you'll learn how to use pre-trained networks to solved challenging problems in computer vision. Specifically, you'll use networks trained on [ImageNet](http://www.image-net.org/) [available from torchvision](http://pytorch.org/docs/0.3.0/torchvision/models.html).
ImageNet is a massive dataset with over 1 million labeled images in 1000 categories. It's used to train deep neural networks using an architecture called convolutional layers. I'm not going to get into the details of convolutional networks here, but if you want to learn more about them, please [watch this](https://www.youtube.com/watch?v=2-Ol7ZB0MmU).
Once trained, these models work astonishingly well as feature detectors for images they weren't trained on. Using a pre-trained network on images not in the training set is called transfer learning. Here we'll use transfer learning to train a network that can classify our cat and dog photos with near perfect accuracy.
With `torchvision.models` you can download these pre-trained networks and use them in your applications. We'll include `models` in our imports now.
```
%matplotlib inline
%config InlineBackend.figure_format = 'retina'
import matplotlib.pyplot as plt
import torch
from torch import nn
from torch import optim
import torch.nn.functional as F
from torchvision import datasets, transforms, models
```
Most of the pretrained models require the input to be 224x224 images. Also, we'll need to match the normalization used when the models were trained. Each color channel was normalized separately, the means are `[0.485, 0.456, 0.406]` and the standard deviations are `[0.229, 0.224, 0.225]`.
```
BATCH_SIZE = 64
N_CLASSES = 2
data_dir = 'Cat_Dog_data'
norm_means = (0.485, 0.456, 0.406)
norm_sigmas = (0.229, 0.224, 0.225)
# TODO: Define transforms for the training data and testing data
train_transforms = transforms.Compose(
[
transforms.Resize(255),
transforms.CenterCrop(224),
transforms.RandomRotation(30),
transforms.RandomHorizontalFlip(),
transforms.ToTensor(),
transforms.Normalize(norm_means, norm_sigmas)
]
)
test_transforms = transforms.Compose(
[
transforms.Resize(255),
transforms.CenterCrop(224),
transforms.ToTensor(),
transforms.Normalize(norm_means, norm_sigmas)
]
)
# Pass transforms in here, then run the next cell to see how the transforms look
train_data = datasets.ImageFolder(data_dir + '/train', transform=train_transforms)
test_data = datasets.ImageFolder(data_dir + '/test', transform=test_transforms)
trainloader = torch.utils.data.DataLoader(train_data, batch_size=BATCH_SIZE, shuffle=True)
testloader = torch.utils.data.DataLoader(test_data, batch_size=int(BATCH_SIZE/2))
Xs, ys = next(iter(trainloader))
Xs.shape # ensure that input is 224x224
```
We can load in a model such as [DenseNet](http://pytorch.org/docs/0.3.0/torchvision/models.html#id5). Let's print out the model architecture so we can see what's going on.
```
model = models.densenet121(pretrained=True)
model
```
This model is built out of two main parts, the features and the classifier. The features part is a stack of convolutional layers and overall works as a feature detector that can be fed into a classifier. The classifier part is a single fully-connected layer `(classifier): Linear(in_features=1024, out_features=1000)`. This layer was trained on the ImageNet dataset, so it won't work for our specific problem. That means we need to replace the classifier, but the features will work perfectly on their own. In general, I think about pre-trained networks as amazingly good feature detectors that can be used as the input for simple feed-forward classifiers.
```
# Freeze parameters so we don't backprop through them
for param in model.parameters():
param.requires_grad = False
# Replace classifier
from collections import OrderedDict
classifier = nn.Sequential(OrderedDict([
('fc1', nn.Linear(1024, 500)),
('relu', nn.ReLU()),
('fc2', nn.Linear(500, N_CLASSES)),
('output', nn.LogSoftmax(dim=1))
]))
model.classifier = classifier
```
With our model built, we need to train the classifier. However, now we're using a **really deep** neural network. If you try to train this on a CPU like normal, it will take a long, long time. Instead, we're going to use the GPU to do the calculations. The linear algebra computations are done in parallel on the GPU leading to 100x increased training speeds. It's also possible to train on multiple GPUs, further decreasing training time.
PyTorch, along with pretty much every other deep learning framework, uses [CUDA](https://developer.nvidia.com/cuda-zone) to efficiently compute the forward and backwards passes on the GPU. In PyTorch, you move your model parameters and other tensors to the GPU memory using `model.to('cuda')`. You can move them back from the GPU with `model.to('cpu')` which you'll commonly do when you need to operate on the network output outside of PyTorch. As a demonstration of the increased speed, I'll compare how long it takes to perform a forward and backward pass with and without a GPU.
```
import time
for device in ['cpu', 'cuda']:
criterion = nn.NLLLoss()
# Only train the classifier parameters, feature parameters are frozen
optimizer = optim.Adam(model.classifier.parameters(), lr=0.001)
model.to(device)
for ii, (inputs, labels) in enumerate(trainloader):
# Move input and label tensors to the GPU
inputs, labels = inputs.to(device), labels.to(device)
start = time.time()
outputs = model.forward(inputs)
loss = criterion(outputs, labels)
loss.backward()
optimizer.step()
if ii==3:
break
print(f"Device = {device}; Time per batch: {(time.time() - start)/3:.3f} seconds")
```
You can write device agnostic code which will automatically use CUDA if it's enabled like so:
```python
# at beginning of the script
device = torch.device("cuda:0" if torch.cuda.is_available() else "cpu")
...
# then whenever you get a new Tensor or Module
# this won't copy if they are already on the desired device
input = data.to(device)
model = MyModule(...).to(device)
```
From here, I'll let you finish training the model. The process is the same as before except now your model is much more powerful. You should get better than 95% accuracy easily.
>**Exercise:** Train a pretrained models to classify the cat and dog images. Continue with the DenseNet model, or try ResNet, it's also a good model to try out first. Make sure you are only training the classifier and the parameters for the features part are frozen.
```
# TODO: Train a model with a pre-trained network
```
|
github_jupyter
|
%matplotlib inline
%config InlineBackend.figure_format = 'retina'
import matplotlib.pyplot as plt
import torch
from torch import nn
from torch import optim
import torch.nn.functional as F
from torchvision import datasets, transforms, models
BATCH_SIZE = 64
N_CLASSES = 2
data_dir = 'Cat_Dog_data'
norm_means = (0.485, 0.456, 0.406)
norm_sigmas = (0.229, 0.224, 0.225)
# TODO: Define transforms for the training data and testing data
train_transforms = transforms.Compose(
[
transforms.Resize(255),
transforms.CenterCrop(224),
transforms.RandomRotation(30),
transforms.RandomHorizontalFlip(),
transforms.ToTensor(),
transforms.Normalize(norm_means, norm_sigmas)
]
)
test_transforms = transforms.Compose(
[
transforms.Resize(255),
transforms.CenterCrop(224),
transforms.ToTensor(),
transforms.Normalize(norm_means, norm_sigmas)
]
)
# Pass transforms in here, then run the next cell to see how the transforms look
train_data = datasets.ImageFolder(data_dir + '/train', transform=train_transforms)
test_data = datasets.ImageFolder(data_dir + '/test', transform=test_transforms)
trainloader = torch.utils.data.DataLoader(train_data, batch_size=BATCH_SIZE, shuffle=True)
testloader = torch.utils.data.DataLoader(test_data, batch_size=int(BATCH_SIZE/2))
Xs, ys = next(iter(trainloader))
Xs.shape # ensure that input is 224x224
model = models.densenet121(pretrained=True)
model
# Freeze parameters so we don't backprop through them
for param in model.parameters():
param.requires_grad = False
# Replace classifier
from collections import OrderedDict
classifier = nn.Sequential(OrderedDict([
('fc1', nn.Linear(1024, 500)),
('relu', nn.ReLU()),
('fc2', nn.Linear(500, N_CLASSES)),
('output', nn.LogSoftmax(dim=1))
]))
model.classifier = classifier
import time
for device in ['cpu', 'cuda']:
criterion = nn.NLLLoss()
# Only train the classifier parameters, feature parameters are frozen
optimizer = optim.Adam(model.classifier.parameters(), lr=0.001)
model.to(device)
for ii, (inputs, labels) in enumerate(trainloader):
# Move input and label tensors to the GPU
inputs, labels = inputs.to(device), labels.to(device)
start = time.time()
outputs = model.forward(inputs)
loss = criterion(outputs, labels)
loss.backward()
optimizer.step()
if ii==3:
break
print(f"Device = {device}; Time per batch: {(time.time() - start)/3:.3f} seconds")
# at beginning of the script
device = torch.device("cuda:0" if torch.cuda.is_available() else "cpu")
...
# then whenever you get a new Tensor or Module
# this won't copy if they are already on the desired device
input = data.to(device)
model = MyModule(...).to(device)
# TODO: Train a model with a pre-trained network
| 0.540924 | 0.987676 |
# Useful Scripts for Research
_Summary:_
This notebook is updated regularly based on the necessity and anytime I find anything useful for my learning and research using Python, git-bash and github.
> ## Table of Contents
> - Markdown Shorcuts
> - Data Preparation
> - Advance Scripts
> - Git & Github
## Markdown Shortcuts
- Run = `shift` + 'enter'/'return'
- insert tab above = `esc` + a
- insert tab below = `esc` + b
- convert cell into `Markdown` = `esc` + m
- convert cell into 'Code' = `esc` + y
- undo = `esc` + z
- redo = `esc` + shift + z
- copy = `esc` + c
- inserting equation stats and end with 'dollar sign', such as $f(x) = x^2$
- inserting picture:
- inserting web/links:
## Data Preparation (Python)
getting the column name
This following code is from this [source](https://github.com/gedeck/practical-statistics-for-data-scientists/blob/master/python/notebooks/Chapter%201%20-%20Exploratory%20Data%20Analysis.ipynb). It is used to create a relative directory, to access data contained in the folder called `./data`
```
try:
import common
DATA = common.dataDirectory()
except ImportError:
DATA = Path().resolve() / 'data'
```
Parsing multiple name
Cek on: https://www.youtube.com/watch?v=ve2pmm5JqmI
# Advance Scripts
### Processing a single script for multiple similar files
## Matrix Operation
The following codes are for conducting matrix operation using Python.
1. Using `numpy`
2. Using `pytorch`
3. Using `tensorflow`
```
import numpy as np
A = np.array([[3, 4],[5, 6],[7, 8]]) # A matrix with 3x2 dimension
B = np.array([[1, 9], [2, 0]]) # B matrix with 2x2 dimension
AB = np.dot(A,B)
AB
Ainv = np.linalg.inv(B)
Ainv
```
2. Using `torch`
```
import torch
B_pt = torch.from_numpy(B) # much cleaner than TF conversion
B_pt
!pip install tensorflow
import tensorflow as tf
```
## Machine Learning
### ML using `keras`
```
import keras
from keras.datasets import mnist
from keras.models import Sequential
from keras.layers import Dense
from keras.optimizers import SGD
from matplotlib import pyplot as plt
(X_train, y_train), (X_valid, y_valid) = mnist.load_data()
```
Let’s examine these data. As mentioned in Chapter 4, the mathematical notation x is used to represent the data we’re feeding into a model as input, while y is used for the labeled output that we’re training the model to predict. With this in mind, `X_train` stores the MNIST digits we’ll be training our model on.9 Executing `X_train.shape` yields the output `(60000, 28, 28)`. This shows us that, as expected, we have 60,000 images in our training dataset, each of which is a 28×28 matrix of values. Running y_train.shape, we unsurprisingly discover we have 60,000 labels indicating what digit is contained in each of the 60,000 training images. y_train[0:12] outputs an array of 12 integers representing the first dozen labels, so we can see that the first handwritten digit in the training set `(X_train[0])` is the number five, the second is a zero, the third is a four, and so on.
9. The convention is to use an uppercase letter like $X$ when the variable being represented is a __two-dimensional matrix__ or a __data structure with even higher dimensionality__. In contrast, a lowercase letter like $x$ is used to represent a __single value (a scalar)__ or a __one-dimensional array__.
```
plt.figure(figsize=(5,5))
for k in range(12):
plt.subplot(3,4, k+1)
plt.imshow(X_train[k], cmap='Greys')
plt.axis('off')
plt.tight_layout()
plt.show()
X_train = X_train.reshape(60000, 784).astype('float32')
X_valid = X_valid.reshape(10000, 784).astype('float32')
```
# Git and Github
## Git Command
* `mkdir` --> to make directory/folder
* `cd {dirName}` --> to go to {dirName}
* `cd ..` --> to go to upper folder
* `ls` --> to get list of folder & files
* `ls -a` --> to get all lists includes git
* `touch {fileName.ext}` --> to create a file with specific extension
* `mv {oldFileName.ext} {newFileName.ext}` --> Change the name of file
* `touch new-folder/style.css` --> create file in specific folder
* `ls new-folder/` --> check list of files in specific folder
* `cp style.css` .. --> copy file in the upper folder
* `rm style.css` --> to remove a file
* `rm -rf {folderName}` --> to remove folder
|
github_jupyter
|
try:
import common
DATA = common.dataDirectory()
except ImportError:
DATA = Path().resolve() / 'data'
import numpy as np
A = np.array([[3, 4],[5, 6],[7, 8]]) # A matrix with 3x2 dimension
B = np.array([[1, 9], [2, 0]]) # B matrix with 2x2 dimension
AB = np.dot(A,B)
AB
Ainv = np.linalg.inv(B)
Ainv
import torch
B_pt = torch.from_numpy(B) # much cleaner than TF conversion
B_pt
!pip install tensorflow
import tensorflow as tf
import keras
from keras.datasets import mnist
from keras.models import Sequential
from keras.layers import Dense
from keras.optimizers import SGD
from matplotlib import pyplot as plt
(X_train, y_train), (X_valid, y_valid) = mnist.load_data()
plt.figure(figsize=(5,5))
for k in range(12):
plt.subplot(3,4, k+1)
plt.imshow(X_train[k], cmap='Greys')
plt.axis('off')
plt.tight_layout()
plt.show()
X_train = X_train.reshape(60000, 784).astype('float32')
X_valid = X_valid.reshape(10000, 784).astype('float32')
| 0.596081 | 0.958847 |
## Hierarchical Topic Models and the Nested Chinese Restaurant Process
#### Tun-Chieh Hsu, Xialingzi Jin, Yen-Hua Chen
### I. Background
Recently, complex probabilistic models are increasingly prevalent in various of domains. However, there are several challenges that should be dealt with due to their open-ended nature. That is, the data sets often grow over time, as they growing, they bring new entities and new structures to the fore. Take the problem of learning a topic hierarchy from data for example. Given a collection of __*documents*__, each of which contains a set of __*words*__ and the goal is to discover common usage patterns or __*topics*__ in the documents, and to organize these topics into a hierarchy.
This paper proposes a new method that specified a generative probabilistic model for hierarchical structures and adopt Bayesian perspective to learn such structures from data. The hierarchies in this case could be considered as random variables and specified procedurally. In addition, the underlying approach of constructing the probabilistic object is __Chinese restaurant process (CRP)__, a distribution on partitions of integers. In this paper, they extend CRP to a hierarchy of partitions, known as __nested Chinese restaruant process (nCRP)__, and apply it as a representation of prior and posterior distributions for topic hierarchies. To be more specific, each node in the hierarchy is associated with a topic, where a topic is a distribution across words. A document is generated by choosing a path from the root to a leaf, repeatedly sampling topics along that path, and sampling the words from the selected topics. Thus the orga- nization of topics into a hierarchy aims to capture the breadth of usage of topics across the corpus, reflecting underlying syntactic and semantic notions of generality and specificity.
### II. Algorithm Description
#### A. Chinese Restaurant Process
CRP is an analogous to seating customers at tables in a Chinese restaurant. Imagine there is a Chinese restaurant with an infinite number of circular tables, each with infinite capacity. Customer 1 sits at the first table. The next customer either sits at the same table as customer 1, or the next table. The $m$th subsequent customer sits at a table drawn from the following distribution:
\begin{align*}
p(\text{occupied table}\hspace{0.5ex}i\hspace{0.5ex}\text{ | previous customers}) = \frac{m_i}{\gamma+m-1}\\
p(\text{next unoccupied table | previous customers}) = \frac{\gamma}{\gamma + m -1}
\end{align*}
where $m_i$ is the number of previous customers at table $i$, and $\gamma$ is a parameter. After $M$
customers sit down, the seating plan gives a partition of $M$ items. This distribution gives
the same partition structure as draws from a Dirichlet process.
#### B. Nested Chinese Restaurant Process
A nested Chinese restaurant process (nCRP) is an extended version of CRP. Suppose that there are an infinite number of infinite-table Chinese restaurants in a city. A restaurant is determined to be the root restaurant and on each of its infinite tables is a card with the name of another restaurant. On each of the tables in those restaurants are cards that refer to other restaurants, and this structure repeats infinitely. Each restaurant is referred to exactly once. As a result, the whole process could be imagined as an infinitely-branched tree.
Now, consider a tourist arrives in the city for a culinary vacation. On the first first day, he select a root Chinese restaurant and selects a table from the equation above. On the second day, he enters to the restaurant refered by previous restaurant , again from the above equation. This process was repeated for $L$ days, and at the end, the tourist has sat at L restaurants which constitute a path from the root to a restaurant at the $L$th level in the infinite tree. After M tourists take L-day vacations, the collection of paths describe a particular L-level subtree of the infinite tree.
#### C. Hierarchical Topic Model (hLDA)
The hierarchical latent Dirichlet allocation model (hLDA) together with nested Chinese restaruant process (nCRP) illustrate the pattern of words from the collection of documents. There are 3 procedures in hLDA: (1) Draw a path from root-node to a leaf; (2) Select a specific path, draw a vector of topic along the path; (3) Draw the words from the topic. In addition, all documents share the topic associated with the root restaurant.
1. Let $c_1$ be the root restaurant.
+ For each level $\ell\in\{2,...,L\}$:
1. Draw a table from restaurant $c_{\ell-1}$ using CRP. Set $c_{\ell}$ to be the restaurant reffered to by that table.
+ Draw an L-dimensional topic proportion vector $\theta$ from Dir($\alpha$).
+ For each word $n\in\{1,...,N\}$:
1. Draw $z\in\{1,...,L\}$ from Mult($\theta$).
+ Draw $w_n$ from the topic associated with restaurant $c_z$.
<img src="hLDA.png" style="width:400px">
* Notation:
* $T$ : L-level infinite-tree - drawn from CRP($\gamma$)
* $\theta$ : L-dimensional topic propotional distribution - drawn from Dir($\alpha$)
* $\beta$ : probability of words for each topic - drawn from $\eta$
* $c_{\ell}$ : L-level paths, given $T$
* $z$ : actual number of topics for each level - drawn from Mult($\theta$)
* $w$ : word distribution for each topic at each level
* $N$ : number of words - $n\in\{1,...,N\}$
* $M$ : number of documents - $m\in\{1,...,M\}$
### III. Approximate Inference by Gibbs Sampling
Gibbs sampling will sample from the posterior nCRP and corresponding topics in the hLDA model. The sampler are divided into 2 parts -- $z_{m,n}$ and $ c_{m,\ell}$. In addition, variables $\theta$ and $\beta$ are integrated out.
#### A. Notation
* $w_{m,n}$ : the $n$th word in the $m$th documnt
* $c_{m,\ell}$ : the restaurant corresponding to the $\ell$th topic in document $m$
* $z_{m,n}$ : the assignment of the $n$th word in the $m$th document to one of the $L$ available topics
#### B. Topic distribution : $z_{m,n}$
\begin{align*}
p(z_{i}=j\hspace{0.5ex}|\hspace{0.5ex}{\bf z}_{-i},{\bf w})\propto\frac{n_{-i,j}^{(w_{i})}+\beta}{n_{-i,j}^{(\cdot)}+W\beta}\frac{n_{-i,j}^{(d_{i})}+\alpha}{n_{-i,\cdot}^{(d_{i})}+T\alpha}
\end{align*}
* $z_{i}$ : assignments of words to topics
* $n_{-i,j}^{(w_{i})}$ : number of words assigned to topic $j$ that are the same as $w_i$
* $n_{-i,j}^{(\cdot)}$ : total number of words assigned to topic $j$
* $n_{-i,j}^{(d_{i})}$ : number of words from document $d_i$ assigned to topic $j$
* $n_{-i,\cdot}^{(d_{i})}$ : total number of words in document $d_i$
* $W$ : number of words have been assigned
#### C. Path : ${\bf c}_{m}$
$$p({\bf c}_{m}\hspace{0.5ex}|\hspace{0.5ex}{\bf w}, {\bf c}_{-m}, {\bf z})\propto p({\bf w}_{m}\hspace{0.5ex}|\hspace{0.5ex}{\bf c}, {\bf w}_{-m}, {\bf z})\cdot p({\bf c}_{m}\hspace{0.5ex}|\hspace{0.5ex}{\bf c}_{-m})$$
* $p({\bf c}_{m}\hspace{0.5ex}|\hspace{0.5ex}{\bf w}, {\bf c}_{-m}, {\bf z})$ : posterior of the set of probabilities of possible novel paths
* $p({\bf w}_{m}\hspace{0.5ex}|\hspace{0.5ex}{\bf c}, {\bf w}_{-m}, {\bf z})$ : likelihood of the data given a particular choice of ${\bf c}_{m}$
* $p({\bf c}_{m}\hspace{0.5ex}|\hspace{0.5ex}{\bf c}_{-m})$ : prior on ${\bf c}_{m}$ which implies by the nCRP
$$p({\bf w}_{m}\hspace{0.5ex}|\hspace{0.5ex}{\bf c}, {\bf w}_{-m}, {\bf z})=\prod_{\ell=1}^{L}\left(\frac{\Gamma(n_{c_{m,\ell},-m}^{(\cdot)}+W\eta)}{\prod_{w}\Gamma(n_{c_{m,\ell},-m}^{(w)}+\eta)}\frac{\prod_{w}\Gamma(n_{c_{m,\ell},-m}^{(w)}+n_{c_{m,\ell},m}^{(w)}+\eta)}{\Gamma(n_{c_{m,\ell},-m}^{(\cdot)}+n_{c_{m,\ell},m}^{(\cdot)}+W\eta)}\right)$$
* $p({\bf w}_{m}\hspace{0.5ex}|\hspace{0.5ex}{\bf c}, {\bf w}_{-m}, {\bf z})$ : joint distribution of likelihood
* $n_{c_{m,\ell},-m}^{(w)}$ : number of instances of word $w$ that have been assigned to the topic indexed by $c_{m,\ell}$, not in the document $m$
* $W$ : total vocabulary size
### IV. Implementation
#### A. Package import
```
import numpy as np
from scipy.special import gammaln
import random
from collections import Counter
import string
import graphviz
import pygraphviz
import pydot
```
#### B. Function construction
#### B.1 Chinese Restaurant Process (CRP)
```
def CRP(topic, phi):
'''
CRP gives the probability of topic assignment for specific vocabulary
Return a 1 * j array, where j is the number of topic
Parameter
---------
topic: a list of lists, contains assigned words in each sublist (topic)
phi: double, parameter for CRP
Return
------
p_crp: the probability of topic assignments for new word
'''
p_crp = np.empty(len(topic)+1)
m = sum([len(x) for x in topic])
p_crp[0] = phi / (phi + m)
for i, word in enumerate(topic):
p_crp[i+1] = len(word) / (phi + m)
return p_crp
```
#### B.2 Node Sampling
```
def node_sampling(corpus_s, phi):
'''
Node sampling samples the number of topics, L
return a j-layer list of lists, where j is the number of topics
Parameter
---------
corpus_s: a list of lists, contains words in each sublist (document)
phi: double, parameter for CRP
Return
------
topic: a list of lists, contains assigned words in each sublist (topic)
'''
topic = []
for corpus in corpus_s:
for word in corpus:
cm = CRP(topic, phi)
theta = np.random.multinomial(1, (cm/sum(cm))).argmax()
if theta == 0:
topic.append([word])
else:
topic[theta-1].append(word)
return topic
```
#### B.3 Gibbs sampling -- $z_{m,n}$
```
def Z(corpus_s, topic, alpha, beta):
'''
Z samples from LDA model
Return two j-layer list of lists, where j is the number of topics
Parameter
---------
corpus_s: a list of lists, contains words in each sublist (document)
topic: a L-dimensional list of lists, sample from node_sampling
alpha: double, parameter
beta: double, parameter
Return
------
z_topic: a j-dimensional list of lists, drawn from L-dimensioanl topic, j<L
z_doc: a j-dimensioanl list of lists, report from which document the word is assigned to each topic
'''
n_vocab = sum([len(x) for x in corpus_s])
t_zm = np.zeros(n_vocab).astype('int')
z_topic = [[] for _ in topic]
z_doc = [[] for _ in topic]
z_tmp = np.zeros((n_vocab, len(topic)))
assigned = np.zeros((len(corpus_s), len(topic)))
n = 0
for i in range(len(corpus_s)):
for d in range(len(corpus_s[i])):
wi = corpus_s[i][d]
for j in range(len(topic)):
lik = (z_topic[j].count(wi) + beta) / (assigned[i, j] + n_vocab * beta)
pri = (len(z_topic[j]) + alpha) / ((len(corpus_s[i]) - 1) + len(topic) * alpha)
z_tmp[n, j] = lik * pri
t_zm[n] = np.random.multinomial(1, (z_tmp[n,:]/sum(z_tmp[n,:]))).argmax()
z_topic[t_zm[n]].append(wi)
z_doc[t_zm[n]].append(i)
assigned[i, t_zm[n]] += 1
n += 1
z_topic = [x for x in z_topic if x != []]
z_doc = [x for x in z_doc if x != []]
return z_topic, z_doc
```
#### B.4 Gibbs sampling -- ${\bf c}_{m}$, CRP prior
```
def CRP_prior(corpus_s, doc, phi):
'''
CRP_prior implies by nCRP
Return a m*j array, whre m is the number of documents and j is the number of topics
Parameter
---------
corpus_s: a list of lists, contains words in each sublist (document)
doc: a j-dimensioanl list of lists, drawn from Z function (z_doc)
phi: double, parameter for CRP
Return
------
c_p: a m*j array, for each document the probability of the topics
'''
c_p = np.empty((len(corpus_s), len(doc)))
for i, corpus in enumerate(corpus_s):
p_topic = [[x for x in doc[j] if x != i] for j in range(len(doc))]
tmp = CRP(p_topic, phi)
c_p[i,:] = tmp[1:]
return c_p
```
#### B.5 Gibbs sampling -- ${\bf c}_{m}$, likelihood
```
def likelihood(corpus_s, topic, eta):
'''
likelihood gives the propability of data given a particular choice of c
Return a m*j array, whre m is the number of documents and j is the number of topics
Parameter
---------
corpus_s: a list of lists, contains words in each sublist (document)
topic: a j-dimensional list of lists, drawn from Z function (z_assigned)
eta: double, parameter
Return
------
w_m: a m*j array
'''
w_m = np.empty((len(corpus_s), len(topic)))
allword_topic = [word for t in topic for word in t]
n_vocab = sum([len(x) for x in corpus_s])
for i, corpus in enumerate(corpus_s):
prob_result = []
for j in range(len(topic)):
current_topic = topic[j]
n_word_topic = len(current_topic)
prev_dominator = 1
later_numerator = 1
prob_word = 1
overlap = [val for val in set(corpus) if val in current_topic]
prev_numerator = gammaln(len(current_topic) - len(overlap) + n_vocab * eta)
later_dominator = gammaln(len(current_topic) + n_vocab * eta)
for word in corpus:
corpus_list = corpus
if current_topic.count(word) - corpus_list.count(word) < 0 :
a = 0
else:
a = current_topic.count(word) - corpus_list.count(word)
prev_dominator += gammaln(a + eta)
later_numerator += gammaln(current_topic.count(word) + eta)
prev = prev_numerator - prev_dominator
later = later_numerator - later_dominator
like = prev + later
w_m[i, j] = like
w_m[i, :] = w_m[i, :] + abs(min(w_m[i, :]) + 0.1)
w_m = w_m/w_m.sum(axis = 1)[:, np.newaxis]
return w_m
```
#### B.6 Gibbs sampling -- ${\bf c}_{m}$, posterior
```
def post(w_m, c_p):
'''
Parameter
---------
w_m: likelihood, drawn from likelihood function
c_p: prior, drawn from CRP_prior function
Return
------
c_m, a m*j list of lists
'''
c_m = (w_m * c_p) / (w_m * c_p).sum(axis = 1)[:, np.newaxis]
return np.array(c_m)
```
#### B.7 Gibbs sampling -- $w_{n}$
```
def wn(c_m, corpus_s):
'''
wn return the assignment of words for topics, drawn from multinomial distribution
Return a n*1 array, where n is the total number of word
Parameter
---------
c_m: a m*j list of lists, drawn from post function
corpus_s: a list of lists, contains words in each sublist (document)
Return
------
wn_ass: a n*1 array, report the topic assignment for each word
'''
wn_ass = []
for i, corpus in enumerate(corpus_s):
for word in corpus:
theta = np.random.multinomial(1, c_m[i]).argmax()
wn_ass.append(theta)
return np.array(wn_ass)
```
#### C. Gibbs sampling
#### C.1 Find most common value
```
most_common = lambda x: Counter(x).most_common(1)[0][0]
```
#### C.2 Gibbs sampling
```
def gibbs(corpus_s, topic, alpha, beta, phi, eta, ite):
'''
gibbs will return the distribution of words for topics
Return a j-dimensional list of lists, where j is the number of topics
Parameter
---------
corpus_s: a list of lists, contains words in each sublist (document)
topic: a j-dimensional list of lists, drawn from Z function (z_assigned)
alpha: double, parameter for Z function
beta: double, parameter for Z function
phi: double, parameter fro CRP_prior function
eta: double, parameter for w_n function
ite: int, number of iteration
Return
------
wn_topic: a j-dimensional list of lists, the distribution of words for topics
'''
n_vocab = sum([len(x) for x in corpus_s])
gibbs = np.empty((n_vocab, ite)).astype('int')
for i in range(ite):
z_topic, z_doc = Z(corpus_s, topic, alpha, beta)
c_p = CRP_prior(corpus_s, z_doc, phi)
w_m = likelihood(corpus_s, z_topic, eta)
c_m = post(w_m, c_p)
gibbs[:, i] = wn(c_m, corpus_s)
# drop first 1/10 data
gibbs = gibbs[:, int(ite/10):]
theta = [most_common(gibbs[x]) for x in range(n_vocab)]
n_topic = max(theta)+1
wn_topic = [[] for _ in range(n_topic)]
wn_doc_topic = [[] for _ in range(n_topic)]
doc = 0
n = 0
for i, corpus_s in enumerate(corpus_s):
if doc == i:
for word in corpus_s:
wn_doc_topic[theta[n]].append(word)
n += 1
for j in range(n_topic):
if wn_doc_topic[j] != []:
wn_topic[j].append(wn_doc_topic[j])
wn_doc_topic = [[] for _ in range(n_topic)]
doc += 1
wn_topic = [x for x in wn_topic if x != []]
return wn_topic
```
### V. Topic Model with hLDA
Gibbs sampling in section __`IV`__ distributes the input __*vocabularies*__ from __*documents*__ in __*corpus*__ to available __*topics*__, which sampled from $L$-dimensional topics. In section __`V`__, an $n$-level tree will be presented by tree plot, which the root-node will be more general and the leaves will be more specific. In addition, tree plot will return the words sorted by their frequencies for each node.
#### A. hLDA model
```
def hLDA(corpus_s, alpha, beta, phi, eta, ite, level):
'''
hLDA generates an n*1 list of lists, where n is the number of level
Parameter
---------
corpus_s: a list of lists, contains words in each sublist (document)
alpha: double, parameter for Z function
beta: double, parameter for Z function
phi: double, parameter fro CRP_prior function
eta: double, parameter for w_n function
ite: int, number of iteration
level: int, number of level
Return
hLDA_tree: an n*1 list of lists, each sublist represents a level, the sublist in each level represents a topic
node: an n*1 list of lists, returns how many nodes there are in each level
'''
topic = node_sampling(corpus_s, phi)
print(len(topic))
hLDA_tree = [[] for _ in range(level)]
tmp_tree = []
node = [[] for _ in range(level+1)]
node[0].append(1)
for i in range(level):
if i == 0:
wn_topic = gibbs(corpus_s, topic, alpha, beta, phi, eta, ite)
node_topic = [x for word in wn_topic[0] for x in word]
hLDA_tree[0].append(node_topic)
tmp_tree.append(wn_topic[1:])
tmp_tree = tmp_tree[0]
node[1].append(len(wn_topic[1:]))
else:
for j in range(sum(node[i])):
if tmp_tree == []:
break
wn_topic = gibbs(tmp_tree[0], topic, alpha, beta, phi, eta, ite)
node_topic = [x for word in wn_topic[0] for x in word]
hLDA_tree[i].append(node_topic)
tmp_tree.remove(tmp_tree[0])
if wn_topic[1:] != []:
tmp_tree.extend(wn_topic[1:])
node[i+1].append(len(wn_topic[1:]))
return hLDA_tree, node[:level]
```
#### B. hLDA plot
```
def HLDA_plot(hLDA_object, Len = 8, save = False):
from IPython.display import Image, display
def viewPydot(pdot):
plt = Image(pdot.create_png())
display(plt)
words = hLDA_object[0]
struc = hLDA_object[1]
graph = pydot.Dot(graph_type='graph')
end_index = [np.insert(np.cumsum(i),0,0) for i in struc]
for level in range(len(struc)-1):
leaf_level = level + 1
leaf_word = words[leaf_level]
leaf_struc = struc[leaf_level]
word = words[level]
end_leaf_index = end_index[leaf_level]
for len_root in range(len(word)):
root_word = '\n'.join([x[0] for x in Counter(word[len_root]).most_common(Len)])
leaf_index = leaf_struc[len_root]
start = end_leaf_index[len_root]
end = end_leaf_index[len_root+1]
lf = leaf_word[start:end]
for l in lf:
leaf_w = '\n'.join([x[0] for x in Counter(list(l)).most_common(Len)])
edge = pydot.Edge(root_word, leaf_w)
graph.add_edge(edge)
if save == True:
graph.write_png('graph.png')
viewPydot(graph)
```
### VI. Empirical Example
#### A. Simulated data
For simulated data example, each document, $d$, in corpus is generated by normal distribution with different size of words, $w_{d,n}$, where $n\in\{10,...,200\}$ and ${\bf w}_{d}\sim N(0, 1)$. In this example, by generating 35 documents in the corpus, we are able to see the simulated tree with the number near mean, $0$, such as `{w0, w1, w-1}` in the root node and the number far from mean such as `{w10, w-10, w15}` in the leaves.
```
def sim_corpus(n):
n_rows = n
corpus = [[] for _ in range(n_rows)]
for i in range(n_rows):
n_cols = np.random.randint(10, 200, 1, dtype = 'int')[0]
for j in range(n_cols):
num = np.random.normal(0, 1, n_cols)
word = 'w%s' % int(round(num[j], 1)*10)
corpus[i].append(word)
return corpus
corpus_0 = sim_corpus(35)
tree_0 = hLDA(corpus_0, 0.1, 0.01, 2, 0.01, 100, 3)
HLDA_plot(tree_0, 5, False)
```
#### B. Real data
For real data example, the corpus of documents is generated from [Blei's sample data](https://github.com/blei-lab/lda-c). The documents are splitted by paragraph; that is, each paragraph reprents one document. We take first 11 documents to form the sample corpus used in the hLDA model. To form the corpus, we read the corpus as a large list of lists. The sublists in the nested list represent the documents; the elements in each sublist represent the words in specific document. Note that the punctuations are removed from the corpus.
```
def read_corpus(corpus_path):
punc = ['`', ',', "'", '.', '!', '?']
corpus = []
with open(corpus_path, 'r') as f:
for line in f:
for x in punc:
line = line.replace(x, '')
line = line.strip('\n')
word = line.split(' ')
corpus.append(word)
return(corpus)
corpus_1 = read_corpus('sample.txt')
tree_1 = hLDA(corpus_1, 0.1, 0.01, 1, 0.01, 100, 3)
HLDA_plot(tree_1, 5, False)
```
### VII. Download and Install from Github
The hLDA code of the paper __Hierarchical Topic Models and the Nested Chinese Restaurant Process__ is released on github with the package named [hLDA](https://github.com/Yen-HuaChen/STA663-Final-Project.git) (click to clone). One can easily [download](https://github.com/Yen-HuaChen/STA663-Final-Project/archive/v0.0.1.tar.gz) (click to download) and install by running `python setup.py install`. The package provides 4 functions:
1. `hLDA.sim_corpus(n)`: return a simulated corpus with $n$ number of documents
1. inputs:
1. `n`: `int`, number of documents in the corpus
2. `hLDA.read_corpus(corpus_path)`: return a list of lists of corpus with length $n$, where $n$ is the number of documents.
1. inputs:
1. corpus_path: the path of txt file, note that each paragraph represents a document
3. `hLDA.hLDA(corpus, alpha, beta, phi, eta, iteration, level)`: return a $n$-level tree, where $n$ is the input level
1. inputs:
1. corpus: corpus read from `hLDA.read_corpus` or simulated from `sim_corpus`
2. alpha: `double`, parameter for `Z` function
3. beta: `double`, parameter for `Z` function
4. phi: `double`, parameter fro `CRP_prior` function
5. eta: `double`, parameter for `w_n` function
6. iteration: `int`, number of iteration for gibbs sampling
7. level: `int`, number of level
4. `hLDA.HLDA_plot(hLDA_result, n_words, save)`: return a tree plot from hLDA topic model
1. inputs:
1. hLDA_result: the hLDA result generated from `hLDA.hLDA`
2. n_words: `int`, how many words to show in each node (sorted by frequency), default with 5
3. save: `boolean`, save the plot or not, default with `False`
Note that the requirement packages for hLDA are: (1) `numpy`; (2) `scipy`; (3) `collections`; (4) `string`; (5) `pygraphviz`; (6) `pydot`.
```
import hLDA
sim = hLDA.sim_corpus(5)
print(sim[0])
corpus = hLDA.read_corpus('sample.txt')
print(corpus[0])
tree = hLDA.hLDA(corpus, 0.1, 0.01, 1, 0.01, 10, 3)
hLDA.HLDA_plot(tree)
```
### VIII. Optimization
To optimize the hLDA model, we choose cython to speed the functions up, since the only matrix calculation function, __`c_m`__, was already vectorized. However, after applying cython, the code is not able to speed up efficiently. The possible reasons are shown as follows.
First, if we simply speed up single function, cython does it well. Take the first function, __`node_sampling`__, for example, the run time decreased from 52.2 ms to 47.2ms, which menas cython is about 10% faster than python code. On the other hand, if we try to speed up all the functions used in gibbs sampling function, __`gibbs`__, the run time is similar or even slower, since it has to import external cython function to complete the work.
Second, most of the variables used in hLDA are lists. When coding cython in python, we fail to initialize the data type for the list variables efficiently.
```
%load_ext Cython
%%cython -a
cimport cython
cimport numpy as np
import numpy as np
@cython.cdivision
@cython.boundscheck(False)
@cython.wraparound(False)
def CRP_c(list topic, double phi):
cdef double[:] cm = np.empty(len(topic)+1)
cdef int m = sum([len(x) for x in topic])
cm[0] = phi / (phi + m)
cdef int i
cdef list word
for i, word in enumerate(topic):
cm[i+1] = len(word) / (phi + m)
return np.array(cm)
def node_sampling_c(list corpus_s, double phi):
cdef list topic = []
cdef int theta
cdef list corpus
cdef str word
for corpus in corpus_s:
for word in corpus:
cm = CRP_c(topic, phi)
theta = np.random.multinomial(1, (cm/sum(cm))).argmax()
if theta == 0:
topic.append([word])
else:
topic[theta-1].append(word)
return topic
%timeit node_sampling_c(corpus_1, 1)
%timeit node_sampling(corpus_1, 1)
```
### IX. Code Comparison
This section will introduce LDA model as the comparison with hLDA model. The __LDA model__ needs user to *specify the number of topics* and returns the *probability of the words* in each topic, which are the most different parts compares to hLDA model. The __hLDA model__ applies nonparametric prior which allows arbitrary factors and readily accommodates growing data collections. That is , the hLDA model will *sample the number of topics by nCRP* and return a *topic hierarchy tree*.
The `lda_topic` function returns a single-layer word distributions for topics, which number is specified as parameter in the function. In each topic, the LDA model gives the probability distribution of possible words. In LDA model, it treats corpus as a big document, instead of consider each document by it own. Furthermore, the model is not able to illustrate the relationship between topics and words which are provided in hLDA model.
```
import matplotlib.pyplot as plt
from nltk.tokenize import RegexpTokenizer
from stop_words import get_stop_words
from nltk.stem.porter import PorterStemmer
from gensim import corpora, models
import gensim
def lda_topic(corpus_s, dic, n_topics, ite):
lda = gensim.models.ldamodel.LdaModel(corpus = corpus_s,
id2word = dic,
num_topics = n_topics,
update_every = 1,
chunksize = 1,
passes = 1,
iterations = ite)
return lda.print_topics()
corpus = read_corpus('sample.txt')
def lda_corpus(corpus_s):
texts = []
tokenizer = RegexpTokenizer(r'\w+')
for doc in corpus_s:
for word in doc:
raw = word.lower()
tokens = tokenizer.tokenize(raw)
texts.append(tokens)
dictionary = corpora.Dictionary(texts)
n_corpus = [dictionary.doc2bow(text) for text in texts]
corpora.MmCorpus.serialize('sample.mm', n_corpus)
sample = gensim.corpora.MmCorpus('sample.mm')
return sample, dictionary
sample, dic = lda_corpus(corpus)
lda_topic(sample, dic, 3, 5000)
```
### X. Conclusion
By introducing nCRP as the nonparametric prior for hierarchical extension to the LDA, here forms the hLDA. First, in the hLDA topic model, the words are allocated by Gibbs sampling of two critical variable -- ${\bf z}$ and ${\bf c}_{m}$. The formor variable, ${\bf z}$, illustrates how words are allocated to each topic, thus finding the number of topics for each parent node. The later variable, ${\bf c}_{m}$, the posterior of likelihood (${\bf w}_{m}$) and nCRP prior (${\bf c}_{m}$), is a set of possible values correspondings to the topics simulated from ${\bf z}$ for each document $m$. After setting up ${\bf z}$ and ${\bf c}_{m}$, the hLDA then runs the Gibbs sampling to draw $w_{n}$, the distribution of the words to the topics drawn from ${\bf z}$ and ${\bf c}_{m}$. Last, we write the `hLDA` function and `HLDA_plot` function to print the result in list and plot it as a topic tree.
### References
[1] Griffiths, Thomas L., and Mark Steyvers. "A probabilistic approach to semantic representation." Proceedings of the 24th annual conference of the cognitive science society. 2002.
[2] Griffiths, D. M. B. T. L., and M. I. J. J. B. Tenenbaum. "Hierarchical topic models and the nested chinese restaurant process." Advances in neural information processing systems 16 (2004): 17.
[3] Blei, David M., Thomas L. Griffiths, and Michael I. Jordan. "The nested chinese restaurant process and bayesian nonparametric inference of topic hierarchies." Journal of the ACM (JACM) 57.2 (2010): 7.
|
github_jupyter
|
import numpy as np
from scipy.special import gammaln
import random
from collections import Counter
import string
import graphviz
import pygraphviz
import pydot
def CRP(topic, phi):
'''
CRP gives the probability of topic assignment for specific vocabulary
Return a 1 * j array, where j is the number of topic
Parameter
---------
topic: a list of lists, contains assigned words in each sublist (topic)
phi: double, parameter for CRP
Return
------
p_crp: the probability of topic assignments for new word
'''
p_crp = np.empty(len(topic)+1)
m = sum([len(x) for x in topic])
p_crp[0] = phi / (phi + m)
for i, word in enumerate(topic):
p_crp[i+1] = len(word) / (phi + m)
return p_crp
def node_sampling(corpus_s, phi):
'''
Node sampling samples the number of topics, L
return a j-layer list of lists, where j is the number of topics
Parameter
---------
corpus_s: a list of lists, contains words in each sublist (document)
phi: double, parameter for CRP
Return
------
topic: a list of lists, contains assigned words in each sublist (topic)
'''
topic = []
for corpus in corpus_s:
for word in corpus:
cm = CRP(topic, phi)
theta = np.random.multinomial(1, (cm/sum(cm))).argmax()
if theta == 0:
topic.append([word])
else:
topic[theta-1].append(word)
return topic
def Z(corpus_s, topic, alpha, beta):
'''
Z samples from LDA model
Return two j-layer list of lists, where j is the number of topics
Parameter
---------
corpus_s: a list of lists, contains words in each sublist (document)
topic: a L-dimensional list of lists, sample from node_sampling
alpha: double, parameter
beta: double, parameter
Return
------
z_topic: a j-dimensional list of lists, drawn from L-dimensioanl topic, j<L
z_doc: a j-dimensioanl list of lists, report from which document the word is assigned to each topic
'''
n_vocab = sum([len(x) for x in corpus_s])
t_zm = np.zeros(n_vocab).astype('int')
z_topic = [[] for _ in topic]
z_doc = [[] for _ in topic]
z_tmp = np.zeros((n_vocab, len(topic)))
assigned = np.zeros((len(corpus_s), len(topic)))
n = 0
for i in range(len(corpus_s)):
for d in range(len(corpus_s[i])):
wi = corpus_s[i][d]
for j in range(len(topic)):
lik = (z_topic[j].count(wi) + beta) / (assigned[i, j] + n_vocab * beta)
pri = (len(z_topic[j]) + alpha) / ((len(corpus_s[i]) - 1) + len(topic) * alpha)
z_tmp[n, j] = lik * pri
t_zm[n] = np.random.multinomial(1, (z_tmp[n,:]/sum(z_tmp[n,:]))).argmax()
z_topic[t_zm[n]].append(wi)
z_doc[t_zm[n]].append(i)
assigned[i, t_zm[n]] += 1
n += 1
z_topic = [x for x in z_topic if x != []]
z_doc = [x for x in z_doc if x != []]
return z_topic, z_doc
def CRP_prior(corpus_s, doc, phi):
'''
CRP_prior implies by nCRP
Return a m*j array, whre m is the number of documents and j is the number of topics
Parameter
---------
corpus_s: a list of lists, contains words in each sublist (document)
doc: a j-dimensioanl list of lists, drawn from Z function (z_doc)
phi: double, parameter for CRP
Return
------
c_p: a m*j array, for each document the probability of the topics
'''
c_p = np.empty((len(corpus_s), len(doc)))
for i, corpus in enumerate(corpus_s):
p_topic = [[x for x in doc[j] if x != i] for j in range(len(doc))]
tmp = CRP(p_topic, phi)
c_p[i,:] = tmp[1:]
return c_p
def likelihood(corpus_s, topic, eta):
'''
likelihood gives the propability of data given a particular choice of c
Return a m*j array, whre m is the number of documents and j is the number of topics
Parameter
---------
corpus_s: a list of lists, contains words in each sublist (document)
topic: a j-dimensional list of lists, drawn from Z function (z_assigned)
eta: double, parameter
Return
------
w_m: a m*j array
'''
w_m = np.empty((len(corpus_s), len(topic)))
allword_topic = [word for t in topic for word in t]
n_vocab = sum([len(x) for x in corpus_s])
for i, corpus in enumerate(corpus_s):
prob_result = []
for j in range(len(topic)):
current_topic = topic[j]
n_word_topic = len(current_topic)
prev_dominator = 1
later_numerator = 1
prob_word = 1
overlap = [val for val in set(corpus) if val in current_topic]
prev_numerator = gammaln(len(current_topic) - len(overlap) + n_vocab * eta)
later_dominator = gammaln(len(current_topic) + n_vocab * eta)
for word in corpus:
corpus_list = corpus
if current_topic.count(word) - corpus_list.count(word) < 0 :
a = 0
else:
a = current_topic.count(word) - corpus_list.count(word)
prev_dominator += gammaln(a + eta)
later_numerator += gammaln(current_topic.count(word) + eta)
prev = prev_numerator - prev_dominator
later = later_numerator - later_dominator
like = prev + later
w_m[i, j] = like
w_m[i, :] = w_m[i, :] + abs(min(w_m[i, :]) + 0.1)
w_m = w_m/w_m.sum(axis = 1)[:, np.newaxis]
return w_m
def post(w_m, c_p):
'''
Parameter
---------
w_m: likelihood, drawn from likelihood function
c_p: prior, drawn from CRP_prior function
Return
------
c_m, a m*j list of lists
'''
c_m = (w_m * c_p) / (w_m * c_p).sum(axis = 1)[:, np.newaxis]
return np.array(c_m)
def wn(c_m, corpus_s):
'''
wn return the assignment of words for topics, drawn from multinomial distribution
Return a n*1 array, where n is the total number of word
Parameter
---------
c_m: a m*j list of lists, drawn from post function
corpus_s: a list of lists, contains words in each sublist (document)
Return
------
wn_ass: a n*1 array, report the topic assignment for each word
'''
wn_ass = []
for i, corpus in enumerate(corpus_s):
for word in corpus:
theta = np.random.multinomial(1, c_m[i]).argmax()
wn_ass.append(theta)
return np.array(wn_ass)
most_common = lambda x: Counter(x).most_common(1)[0][0]
def gibbs(corpus_s, topic, alpha, beta, phi, eta, ite):
'''
gibbs will return the distribution of words for topics
Return a j-dimensional list of lists, where j is the number of topics
Parameter
---------
corpus_s: a list of lists, contains words in each sublist (document)
topic: a j-dimensional list of lists, drawn from Z function (z_assigned)
alpha: double, parameter for Z function
beta: double, parameter for Z function
phi: double, parameter fro CRP_prior function
eta: double, parameter for w_n function
ite: int, number of iteration
Return
------
wn_topic: a j-dimensional list of lists, the distribution of words for topics
'''
n_vocab = sum([len(x) for x in corpus_s])
gibbs = np.empty((n_vocab, ite)).astype('int')
for i in range(ite):
z_topic, z_doc = Z(corpus_s, topic, alpha, beta)
c_p = CRP_prior(corpus_s, z_doc, phi)
w_m = likelihood(corpus_s, z_topic, eta)
c_m = post(w_m, c_p)
gibbs[:, i] = wn(c_m, corpus_s)
# drop first 1/10 data
gibbs = gibbs[:, int(ite/10):]
theta = [most_common(gibbs[x]) for x in range(n_vocab)]
n_topic = max(theta)+1
wn_topic = [[] for _ in range(n_topic)]
wn_doc_topic = [[] for _ in range(n_topic)]
doc = 0
n = 0
for i, corpus_s in enumerate(corpus_s):
if doc == i:
for word in corpus_s:
wn_doc_topic[theta[n]].append(word)
n += 1
for j in range(n_topic):
if wn_doc_topic[j] != []:
wn_topic[j].append(wn_doc_topic[j])
wn_doc_topic = [[] for _ in range(n_topic)]
doc += 1
wn_topic = [x for x in wn_topic if x != []]
return wn_topic
def hLDA(corpus_s, alpha, beta, phi, eta, ite, level):
'''
hLDA generates an n*1 list of lists, where n is the number of level
Parameter
---------
corpus_s: a list of lists, contains words in each sublist (document)
alpha: double, parameter for Z function
beta: double, parameter for Z function
phi: double, parameter fro CRP_prior function
eta: double, parameter for w_n function
ite: int, number of iteration
level: int, number of level
Return
hLDA_tree: an n*1 list of lists, each sublist represents a level, the sublist in each level represents a topic
node: an n*1 list of lists, returns how many nodes there are in each level
'''
topic = node_sampling(corpus_s, phi)
print(len(topic))
hLDA_tree = [[] for _ in range(level)]
tmp_tree = []
node = [[] for _ in range(level+1)]
node[0].append(1)
for i in range(level):
if i == 0:
wn_topic = gibbs(corpus_s, topic, alpha, beta, phi, eta, ite)
node_topic = [x for word in wn_topic[0] for x in word]
hLDA_tree[0].append(node_topic)
tmp_tree.append(wn_topic[1:])
tmp_tree = tmp_tree[0]
node[1].append(len(wn_topic[1:]))
else:
for j in range(sum(node[i])):
if tmp_tree == []:
break
wn_topic = gibbs(tmp_tree[0], topic, alpha, beta, phi, eta, ite)
node_topic = [x for word in wn_topic[0] for x in word]
hLDA_tree[i].append(node_topic)
tmp_tree.remove(tmp_tree[0])
if wn_topic[1:] != []:
tmp_tree.extend(wn_topic[1:])
node[i+1].append(len(wn_topic[1:]))
return hLDA_tree, node[:level]
def HLDA_plot(hLDA_object, Len = 8, save = False):
from IPython.display import Image, display
def viewPydot(pdot):
plt = Image(pdot.create_png())
display(plt)
words = hLDA_object[0]
struc = hLDA_object[1]
graph = pydot.Dot(graph_type='graph')
end_index = [np.insert(np.cumsum(i),0,0) for i in struc]
for level in range(len(struc)-1):
leaf_level = level + 1
leaf_word = words[leaf_level]
leaf_struc = struc[leaf_level]
word = words[level]
end_leaf_index = end_index[leaf_level]
for len_root in range(len(word)):
root_word = '\n'.join([x[0] for x in Counter(word[len_root]).most_common(Len)])
leaf_index = leaf_struc[len_root]
start = end_leaf_index[len_root]
end = end_leaf_index[len_root+1]
lf = leaf_word[start:end]
for l in lf:
leaf_w = '\n'.join([x[0] for x in Counter(list(l)).most_common(Len)])
edge = pydot.Edge(root_word, leaf_w)
graph.add_edge(edge)
if save == True:
graph.write_png('graph.png')
viewPydot(graph)
def sim_corpus(n):
n_rows = n
corpus = [[] for _ in range(n_rows)]
for i in range(n_rows):
n_cols = np.random.randint(10, 200, 1, dtype = 'int')[0]
for j in range(n_cols):
num = np.random.normal(0, 1, n_cols)
word = 'w%s' % int(round(num[j], 1)*10)
corpus[i].append(word)
return corpus
corpus_0 = sim_corpus(35)
tree_0 = hLDA(corpus_0, 0.1, 0.01, 2, 0.01, 100, 3)
HLDA_plot(tree_0, 5, False)
def read_corpus(corpus_path):
punc = ['`', ',', "'", '.', '!', '?']
corpus = []
with open(corpus_path, 'r') as f:
for line in f:
for x in punc:
line = line.replace(x, '')
line = line.strip('\n')
word = line.split(' ')
corpus.append(word)
return(corpus)
corpus_1 = read_corpus('sample.txt')
tree_1 = hLDA(corpus_1, 0.1, 0.01, 1, 0.01, 100, 3)
HLDA_plot(tree_1, 5, False)
import hLDA
sim = hLDA.sim_corpus(5)
print(sim[0])
corpus = hLDA.read_corpus('sample.txt')
print(corpus[0])
tree = hLDA.hLDA(corpus, 0.1, 0.01, 1, 0.01, 10, 3)
hLDA.HLDA_plot(tree)
%load_ext Cython
%%cython -a
cimport cython
cimport numpy as np
import numpy as np
@cython.cdivision
@cython.boundscheck(False)
@cython.wraparound(False)
def CRP_c(list topic, double phi):
cdef double[:] cm = np.empty(len(topic)+1)
cdef int m = sum([len(x) for x in topic])
cm[0] = phi / (phi + m)
cdef int i
cdef list word
for i, word in enumerate(topic):
cm[i+1] = len(word) / (phi + m)
return np.array(cm)
def node_sampling_c(list corpus_s, double phi):
cdef list topic = []
cdef int theta
cdef list corpus
cdef str word
for corpus in corpus_s:
for word in corpus:
cm = CRP_c(topic, phi)
theta = np.random.multinomial(1, (cm/sum(cm))).argmax()
if theta == 0:
topic.append([word])
else:
topic[theta-1].append(word)
return topic
%timeit node_sampling_c(corpus_1, 1)
%timeit node_sampling(corpus_1, 1)
import matplotlib.pyplot as plt
from nltk.tokenize import RegexpTokenizer
from stop_words import get_stop_words
from nltk.stem.porter import PorterStemmer
from gensim import corpora, models
import gensim
def lda_topic(corpus_s, dic, n_topics, ite):
lda = gensim.models.ldamodel.LdaModel(corpus = corpus_s,
id2word = dic,
num_topics = n_topics,
update_every = 1,
chunksize = 1,
passes = 1,
iterations = ite)
return lda.print_topics()
corpus = read_corpus('sample.txt')
def lda_corpus(corpus_s):
texts = []
tokenizer = RegexpTokenizer(r'\w+')
for doc in corpus_s:
for word in doc:
raw = word.lower()
tokens = tokenizer.tokenize(raw)
texts.append(tokens)
dictionary = corpora.Dictionary(texts)
n_corpus = [dictionary.doc2bow(text) for text in texts]
corpora.MmCorpus.serialize('sample.mm', n_corpus)
sample = gensim.corpora.MmCorpus('sample.mm')
return sample, dictionary
sample, dic = lda_corpus(corpus)
lda_topic(sample, dic, 3, 5000)
| 0.518302 | 0.98985 |

## Init
```
import logging
from importlib import reload
import sonosco
reload(logging)
logging.basicConfig(level=logging.INFO)
import IPython.display as ipd
import os
from sonosco.common.constants import SONOSCO
from sonosco.common.utils import setup_logging
from sonosco.datasets.download_datasets.create_manifest import create_manifest
import pandas as pd
import torch
from sonosco.training.experiment import Experiment
from sonosco.training.trainer import ModelTrainer
from sonosco.model.deserializer import Deserializer
from sonosco.decoders import GreedyDecoder
from sonosco.models.seq2seq_las import Seq2Seq
from sonosco.common.path_utils import parse_yaml
from sonosco.datasets import create_data_loaders
from sonosco.training.word_error_rate import word_error_rate
from sonosco.training.character_error_rate import character_error_rate
from sonosco.training.losses import cross_entropy_loss
file_id = "1995-1826-0007"
logging.info("Manifest:")
audio_file = os.path.expanduser(f"~/temp/data/libri_speech/test_clean/wav/{file_id}.wav")
transcript_file = os.path.expanduser(f"~/temp/data/libri_speech/test_clean/txt/{file_id}.txt")
# path_to_data = '/Users/yuriy/Documents/University/ss2019/sonosco/demo/data/'
# output_file = '/Users/yuriy/Documents/University/ss2019/sonosco/demo/data/manifest.csv'
path_to_data = '/Users/w.jurasz/temp/data/demo'
output_file = '/Users/w.jurasz/temp/data/demo/manifest.csv'
```
## Data
```
with open(transcript_file, 'r') as f:
print(f'Transcription: {f.read()}')
ipd.Audio(audio_file)
create_manifest(path_to_data, output_file)
pd.read_csv(output_file, names=["Audio", "Text", "Duration"])
```
# Training
```
config = parse_yaml('./train_seq2seq_demo.yaml')['train']
device = torch.device("cpu")
train_loader, val_loader, test_loader = create_data_loaders(**config)
training_args = {
'loss': cross_entropy_loss,
'epochs': config["max_epochs"],
'train_data_loader': train_loader,
'val_data_loader': val_loader,
'test_data_loader': test_loader,
'lr': config["learning_rate"],
'custom_model_eval': True,
'metrics': [word_error_rate, character_error_rate],
'decoder': GreedyDecoder(config['labels']),
'device': device,
'test_step': config["test_step"]}
experiment = Experiment.create(config, logging.getLogger())
CONTINUE = True
if CONTINUE:
loader = Deserializer()
trainer, config = loader.deserialize(ModelTrainer, config["checkpoint_path"], {
'train_data_loader': train_loader,'val_data_loader': val_loader, 'test_data_loader': test_loader,
}, with_config=True)
else:
model = Seq2Seq(config["encoder"], config["decoder"])
trainer = ModelTrainer(model, **training_args)
experiment.setup_model_trainer(trainer, checkpoints=True, tensorboard=True)
try:
experiment.start()
except KeyboardInterrupt:
experiment.stop()
```
|
github_jupyter
|
import logging
from importlib import reload
import sonosco
reload(logging)
logging.basicConfig(level=logging.INFO)
import IPython.display as ipd
import os
from sonosco.common.constants import SONOSCO
from sonosco.common.utils import setup_logging
from sonosco.datasets.download_datasets.create_manifest import create_manifest
import pandas as pd
import torch
from sonosco.training.experiment import Experiment
from sonosco.training.trainer import ModelTrainer
from sonosco.model.deserializer import Deserializer
from sonosco.decoders import GreedyDecoder
from sonosco.models.seq2seq_las import Seq2Seq
from sonosco.common.path_utils import parse_yaml
from sonosco.datasets import create_data_loaders
from sonosco.training.word_error_rate import word_error_rate
from sonosco.training.character_error_rate import character_error_rate
from sonosco.training.losses import cross_entropy_loss
file_id = "1995-1826-0007"
logging.info("Manifest:")
audio_file = os.path.expanduser(f"~/temp/data/libri_speech/test_clean/wav/{file_id}.wav")
transcript_file = os.path.expanduser(f"~/temp/data/libri_speech/test_clean/txt/{file_id}.txt")
# path_to_data = '/Users/yuriy/Documents/University/ss2019/sonosco/demo/data/'
# output_file = '/Users/yuriy/Documents/University/ss2019/sonosco/demo/data/manifest.csv'
path_to_data = '/Users/w.jurasz/temp/data/demo'
output_file = '/Users/w.jurasz/temp/data/demo/manifest.csv'
with open(transcript_file, 'r') as f:
print(f'Transcription: {f.read()}')
ipd.Audio(audio_file)
create_manifest(path_to_data, output_file)
pd.read_csv(output_file, names=["Audio", "Text", "Duration"])
config = parse_yaml('./train_seq2seq_demo.yaml')['train']
device = torch.device("cpu")
train_loader, val_loader, test_loader = create_data_loaders(**config)
training_args = {
'loss': cross_entropy_loss,
'epochs': config["max_epochs"],
'train_data_loader': train_loader,
'val_data_loader': val_loader,
'test_data_loader': test_loader,
'lr': config["learning_rate"],
'custom_model_eval': True,
'metrics': [word_error_rate, character_error_rate],
'decoder': GreedyDecoder(config['labels']),
'device': device,
'test_step': config["test_step"]}
experiment = Experiment.create(config, logging.getLogger())
CONTINUE = True
if CONTINUE:
loader = Deserializer()
trainer, config = loader.deserialize(ModelTrainer, config["checkpoint_path"], {
'train_data_loader': train_loader,'val_data_loader': val_loader, 'test_data_loader': test_loader,
}, with_config=True)
else:
model = Seq2Seq(config["encoder"], config["decoder"])
trainer = ModelTrainer(model, **training_args)
experiment.setup_model_trainer(trainer, checkpoints=True, tensorboard=True)
try:
experiment.start()
except KeyboardInterrupt:
experiment.stop()
| 0.340595 | 0.372705 |
```
%%javascript
//Imports the javascript code - not required if the extensions are already installed
require(['/tree/py3dmol/py3dmol/molviz3d.js','/tree/py3dmol/py3dmol/molviz2d.js',
'/tree/py3dmol/py3dmol/3Dmol-min.js','/tree/py3dmol/py3dmol/d3.v3.min.js'],
function(){console.log('JS extensions loaded.')})
%pylab inline
import cStringIO as cio
import numpy as np
import pandas
import numba
import buckyball as bb
import buckyball.units as u
"""Set up our unit system"""
u.ureg.define('earth_mass = 5.97219e24 * kg')
u.em = u.ureg.earth_mass
u.ureg.define('earth_radius = 7917.5/2.0 * miles')
u.au = u.ureg.au
u.yj = u.ureg.yottajoule
u.G = u.ureg.newtonian_constant_of_gravitation
u.default.length = u.au
u.default.mass = u.em
u.default.energy = u.yj
u.default.time = u.ureg.day
"""Set up astronomical data"""
data = pandas.read_csv('data/solar.csv')
planets = {}
for name,mass,orbit,velocity in zip(data.Name,
data['Mass (Earth=1)'],
data['Mean Distance from Sun (AU)'],
data['Mean Orbital Velocity (km/sec)']):
planets[name] = {'mass':mass*u.em,
'orbit':orbit*u.au,
'speed':velocity*u.ureg.km/u.ureg.seconds}
def make_planet(name):
planet = planets[name]
atom = bb.Atom(name=name, mass=planet['mass'], atnum=0)
atom.x = planet['orbit']
atom.momentum[1] = planet['speed'] * atom.mass
return atom
"""Define the potential"""
class Gravity(bb.basemethods.EnergyModelBase):
def prep(self):
n = self.mol.num_atoms
self.indices = np.triu_indices(n,k=1)
self.forcebuffer = np.zeros((n,3)) * u.default.force
self._prepped = True
mg = u.to_units_array([u.G*self.mol.atoms[i].mass*self.mol.atoms[j].mass
for i,j in zip(*self.indices)])
self.mass_grav = mg
def calculate(self,requests=None):
if not self._prepped: self.prep()
energy,forces = self.fast_gravity()
return {'potential_energy':u.default.convert(energy),
'forces':u.default.convert(forces)}
def fast_gravity(self):
dists = self.mol.calc_distance_array(self.mol)[self.indices]
disps = self.mol.calc_displacements()[self.indices]
base_array = self.mass_grav/dists
potential = -np.sum(base_array)
f1 = base_array/(dists**2)
f2 = np.multiply(f1,disps.T).T
self.forcebuffer *= 0.0
for ind,(i,j) in enumerate(zip(*self.indices)):
self.forcebuffer[3*i:3*i+3] += f2[ind]
self.forcebuffer[3*j:3*j+3] -= f2[ind]
return potential,self.forcebuffer
"""Timing..."""
twobody = bb.Molecule([make_planet('Earth'),make_planet('Sun')],
name='EarthAndSun')
twobody.set_energy_model(Gravity())
#twobody.energy_model.calculate()
#%timeit twobody.energy_model.calculate()
"""Run a simulation of the earth and sun"""
twobody = bb.Molecule([make_planet('Earth'),make_planet('Sun')],
name='EarthAndSun')
twobody.set_energy_model(Gravity())
print 'Starting potential energy: ',twobody.calc_potential_energy()
vv = bb.integrators.VelocityVerlet(frame_interval=20, timestep=1.0*u.ureg.day)
twobody.set_integrator(vv)
duration = 5.0*u.ureg.year
traj = twobody.run(duration)
print 'Created trajectory with %d frames over %s'%(len(traj),duration)
twobody.energy_model.mass_grav
"""Plot the results"""
print 'There are %d frames.'%len(traj)
plot(traj.time,traj.positions[:,0],label='x')
plot(traj.time,traj.positions[:,1],label='y')
xlabel('time / days'); ylabel('coordinate / Ast. Unit');grid(); legend(loc='lower left')
title("Earth's location relative to the sun")
"""A bigger simulation"""
energy_model=Gravity()
integrator=bb.integrators.VelocityVerlet(timestep=1.0*u.ureg.day,
frame_interval=10)
solar_system = bb.Molecule([make_planet(n) for n in data.Name],
energy_model=energy_model,
integrator=integrator)
traj = solar_system.run(0.5*u.ureg.year)
```
|
github_jupyter
|
%%javascript
//Imports the javascript code - not required if the extensions are already installed
require(['/tree/py3dmol/py3dmol/molviz3d.js','/tree/py3dmol/py3dmol/molviz2d.js',
'/tree/py3dmol/py3dmol/3Dmol-min.js','/tree/py3dmol/py3dmol/d3.v3.min.js'],
function(){console.log('JS extensions loaded.')})
%pylab inline
import cStringIO as cio
import numpy as np
import pandas
import numba
import buckyball as bb
import buckyball.units as u
"""Set up our unit system"""
u.ureg.define('earth_mass = 5.97219e24 * kg')
u.em = u.ureg.earth_mass
u.ureg.define('earth_radius = 7917.5/2.0 * miles')
u.au = u.ureg.au
u.yj = u.ureg.yottajoule
u.G = u.ureg.newtonian_constant_of_gravitation
u.default.length = u.au
u.default.mass = u.em
u.default.energy = u.yj
u.default.time = u.ureg.day
"""Set up astronomical data"""
data = pandas.read_csv('data/solar.csv')
planets = {}
for name,mass,orbit,velocity in zip(data.Name,
data['Mass (Earth=1)'],
data['Mean Distance from Sun (AU)'],
data['Mean Orbital Velocity (km/sec)']):
planets[name] = {'mass':mass*u.em,
'orbit':orbit*u.au,
'speed':velocity*u.ureg.km/u.ureg.seconds}
def make_planet(name):
planet = planets[name]
atom = bb.Atom(name=name, mass=planet['mass'], atnum=0)
atom.x = planet['orbit']
atom.momentum[1] = planet['speed'] * atom.mass
return atom
"""Define the potential"""
class Gravity(bb.basemethods.EnergyModelBase):
def prep(self):
n = self.mol.num_atoms
self.indices = np.triu_indices(n,k=1)
self.forcebuffer = np.zeros((n,3)) * u.default.force
self._prepped = True
mg = u.to_units_array([u.G*self.mol.atoms[i].mass*self.mol.atoms[j].mass
for i,j in zip(*self.indices)])
self.mass_grav = mg
def calculate(self,requests=None):
if not self._prepped: self.prep()
energy,forces = self.fast_gravity()
return {'potential_energy':u.default.convert(energy),
'forces':u.default.convert(forces)}
def fast_gravity(self):
dists = self.mol.calc_distance_array(self.mol)[self.indices]
disps = self.mol.calc_displacements()[self.indices]
base_array = self.mass_grav/dists
potential = -np.sum(base_array)
f1 = base_array/(dists**2)
f2 = np.multiply(f1,disps.T).T
self.forcebuffer *= 0.0
for ind,(i,j) in enumerate(zip(*self.indices)):
self.forcebuffer[3*i:3*i+3] += f2[ind]
self.forcebuffer[3*j:3*j+3] -= f2[ind]
return potential,self.forcebuffer
"""Timing..."""
twobody = bb.Molecule([make_planet('Earth'),make_planet('Sun')],
name='EarthAndSun')
twobody.set_energy_model(Gravity())
#twobody.energy_model.calculate()
#%timeit twobody.energy_model.calculate()
"""Run a simulation of the earth and sun"""
twobody = bb.Molecule([make_planet('Earth'),make_planet('Sun')],
name='EarthAndSun')
twobody.set_energy_model(Gravity())
print 'Starting potential energy: ',twobody.calc_potential_energy()
vv = bb.integrators.VelocityVerlet(frame_interval=20, timestep=1.0*u.ureg.day)
twobody.set_integrator(vv)
duration = 5.0*u.ureg.year
traj = twobody.run(duration)
print 'Created trajectory with %d frames over %s'%(len(traj),duration)
twobody.energy_model.mass_grav
"""Plot the results"""
print 'There are %d frames.'%len(traj)
plot(traj.time,traj.positions[:,0],label='x')
plot(traj.time,traj.positions[:,1],label='y')
xlabel('time / days'); ylabel('coordinate / Ast. Unit');grid(); legend(loc='lower left')
title("Earth's location relative to the sun")
"""A bigger simulation"""
energy_model=Gravity()
integrator=bb.integrators.VelocityVerlet(timestep=1.0*u.ureg.day,
frame_interval=10)
solar_system = bb.Molecule([make_planet(n) for n in data.Name],
energy_model=energy_model,
integrator=integrator)
traj = solar_system.run(0.5*u.ureg.year)
| 0.497559 | 0.434701 |
# Network Traffic Forecasting (using time series data)
In telco, accurate forecast of KPIs (e.g. network traffic, utilizations, user experience, etc.) for communication networks ( 2G/3G/4G/5G/wired) can help predict network failures, allocate resource, or save energy.
In this notebook, we demonstrate a reference use case where we use the network traffic KPI(s) in the past to predict traffic KPI(s) in the future. We demostrate how to do **univariate forecasting** (predict only 1 series), and **multivariate forecasting** (predicts more than 1 series at the same time) using Project Chronos.
For demonstration, we use the publicly available network traffic data repository maintained by the [WIDE project](http://mawi.wide.ad.jp/mawi/) and in particular, the network traffic traces aggregated every 2 hours (i.e. AverageRate in Mbps/Gbps and Total Bytes) in year 2018 and 2019 at the transit link of WIDE to the upstream ISP ([dataset link](http://mawi.wide.ad.jp/~agurim/dataset/)).
## Helper functions
This section defines some helper functions to be used in the following procedures. You can refer to it later when they're used.
```
def plot_predict_actual_values(date, y_pred, y_test, ylabel):
"""
plot the predicted values and actual values (for the test data)
"""
fig, axs = plt.subplots(figsize=(16,6))
axs.plot(date, y_pred, color='red', label='predicted values')
axs.plot(date, y_test, color='blue', label='actual values')
axs.set_title('the predicted values and actual values (for the test data)')
plt.xlabel('test datetime')
plt.ylabel(ylabel)
plt.legend(loc='upper left')
plt.show()
```
## Download raw dataset and load into dataframe
Now we download the dataset and load it into a pandas dataframe. Steps are as below.
* First, run the script `get_data.sh` to download the raw data. It will download the monthly aggregated traffic data in year 2018 and 2019 into `data` folder. The raw data contains aggregated network traffic (average MBPs and total bytes) as well as other metrics.
* Second, run `extract_data.sh` to extract relavant traffic KPI's from raw data, i.e. `AvgRate` for average use rate, and `total` for total bytes. The script will extract the KPI's with timestamps into `data/data.csv`.
* Finally, use pandas to load `data/data.csv` into a dataframe as shown below
```
import os
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
%matplotlib inline
raw_df = pd.read_csv("data/data.csv")
```
Below are some example records of the data
```
raw_df.head()
```
## Data pre-processing
Now we need to do data cleaning and preprocessing on the raw data. Note that this part could vary for different dataset.
For the network traffic data we're using, the processing contains 2 parts:
1. Convert string datetime to TimeStamp
2. Unify the measurement scale for `AvgRate` value - some uses Mbps, some uses Gbps
```
df = pd.DataFrame(pd.to_datetime(raw_df.StartTime))
# we can find 'AvgRate' is of two scales: 'Mbps' and 'Gbps'
raw_df.AvgRate.str[-4:].unique()
# Unify AvgRate value
df['AvgRate'] = raw_df.AvgRate.apply(lambda x:float(x[:-4]) if x.endswith("Mbps") else float(x[:-4])*1000)
df["total"] = raw_df["total"]
df.head()
```
Plot the data to see how the KPI's look like
```
ax = df.plot(y='AvgRate',figsize=(16,6), title="AvgRate of network traffic data")
ax = df.plot(y='total',figsize=(16,6), title="total bytes of network traffic data")
```
## Feature Engineering & Data Preperation
For feature engineering, we use year, month, week, day of week and hour as features in addition to the target KPI values.
For data preperation, we impute the data to handle missing data and scale the data.
We generate a built-in TSDataset to complete the whole processing.
```
from zoo.chronos.data import TSDataset
from sklearn.preprocessing import StandardScaler
# we look back one week data which is of the frequency of 2h.
look_back = 84
# specify the number of steps to be predicted,one day is selected by default.
horizon = 1
tsdata_train, _, tsdata_test = TSDataset.from_pandas(df, dt_col="StartTime", target_col=["AvgRate","total"], with_split=True, test_ratio=0.1)
standard_scaler = StandardScaler()
for tsdata in [tsdata_train, tsdata_test]:
tsdata.gen_dt_feature()\
.impute(mode="last")\
.scale(standard_scaler, fit=(tsdata is tsdata_train))
```
# Time series forecasting
### Univariate forecasting
For _univariate_ forecasting, we forecast ``AvgRate`` only. We need to roll the data on corresponding target column with tsdataset.
```
for tsdata in [tsdata_train, tsdata_test]:
tsdata.roll(lookback=look_back, horizon=horizon, target_col="AvgRate")
x_train, y_train = tsdata_train.to_numpy()
x_test, y_test = tsdata_test.to_numpy()
y_train, y_test = y_train[:, 0, :], y_test[:, 0, :]
x_train.shape, y_train.shape, x_test.shape, y_test.shape
```
For univariate forcasting, we use LSTMForecaster for forecasting.
```
from zoo.chronos.model.forecast.lstm_forecaster import LSTMForecaster
```
First we initiate a LSTMForecaster.
* `feature_dim` should match the training data input feature, so we just use the last dimension of train data shape.
* `target_dim` equals the variate num we want to predict. We set target_dim=1 for univariate forecasting.
```
# build model
lstm_config = {"lstm_units": [32] * 2, "lr":0.001}
forecaster = LSTMForecaster(target_dim=1, feature_dim=x_train.shape[-1], **lstm_config)
```
Then we use fit to train the model. Wait sometime for it to finish.
```
%%time
forecaster.fit(x=x_train, y=y_train, batch_size=1024, epochs=50, distributed=False)
```
After training is finished. You can use the forecaster to do prediction and evaluation.
```
# make prediction
y_pred = forecaster.predict(x_test)
```
Since we have used standard scaler to scale the input data (including the target values), we need to inverse the scaling on the predicted values too.
```
y_pred_unscale = tsdata_test.unscale_numpy(np.expand_dims(y_pred, axis=1))[:, 0, :]
y_test_unscale = tsdata_test.unscale_numpy(np.expand_dims(y_test, axis=1))[:, 0, :]
```
calculate the symetric mean absolute percentage error.
```
from zoo.automl.common.metrics import Evaluator
# evaluate with sMAPE
print("sMAPE is", Evaluator.evaluate("smape", y_test_unscale, y_pred_unscale))
# evaluate with mean_squared_error
print("mean_squared error is", Evaluator.evaluate("mse", y_test_unscale, y_pred_unscale))
```
### multivariate forecasting
For _multivariate_ forecasting, we forecast `AvgRate` and `total` at the same time. We need to roll the data on corresponding target column with tsdataset.
```
for tsdata in [tsdata_train, tsdata_test]:
tsdata.roll(lookback=look_back, horizon=horizon, target_col=["AvgRate","total"])
x_train_m, y_train_m = tsdata_train.to_numpy()
x_test_m, y_test_m = tsdata_test.to_numpy()
y_train_m, y_test_m = y_train_m[:, 0, :], y_test_m[:, 0, :]
x_train_m.shape, y_train_m.shape, x_test_m.shape, y_test_m.shape
```
For multivariate forecasting, we use MTNetForecaster for forecasting.
```
from zoo.chronos.model.forecast.mtnet_forecaster import MTNetForecaster
```
First, we initialize a mtnet_forecaster according to input data shape. The lookback length is equal to `(long_series_num+1)*series_length` Details refer to [chronos docs](https://github.com/intel-analytics/analytics-zoo/tree/master/pyzoo/zoo/chronos).
```
mtnet_forecaster = MTNetForecaster(target_dim=y_train_m.shape[-1],
feature_dim=x_train_m.shape[-1],
long_series_num=6,
series_length=12,
ar_window_size=6,
cnn_height=4
)
```
MTNet needs to preprocess the X into another format, so we call `MTNetForecaster.preprocess_input` on train_x and test_x.
```
# mtnet requires reshape of input x before feeding into model.
x_train_mtnet = mtnet_forecaster.preprocess_input(x_train_m)
x_test_mtnet = mtnet_forecaster.preprocess_input(x_test_m)
```
Now we train the model and wait till it finished.
```
%%time
hist = mtnet_forecaster.fit(x = x_train_mtnet, y = y_train_m, batch_size=1024, epochs=20)
```
Use the model for prediction and inverse the scaling of the prediction results
```
y_pred_m = mtnet_forecaster.predict(x_test_mtnet)
y_pred_m_unscale = tsdata_test.unscale_numpy(np.expand_dims(y_pred_m, axis=1))[:, 0, :]
y_test_m_unscale = tsdata_test.unscale_numpy(np.expand_dims(y_test_m, axis=1))[:, 0, :]
from zoo.automl.common.metrics import Evaluator
# evaluate with sMAPE
print("sMAPE is", Evaluator.evaluate("smape", y_test_m_unscale, y_pred_m_unscale, multioutput="raw_values"))
# evaluate with mean_squared_error
print("mean_squared error is", Evaluator.evaluate("mse", y_test_m_unscale, y_pred_m_unscale, multioutput="raw_values"))
```
plot actual and prediction values for `AvgRate` KPI
```
multi_target_value = ["AvgRate","total"]
test_date=df[-y_test_m_unscale.shape[0]:].index
plot_predict_actual_values(test_date, y_pred_m_unscale[:,0], y_test_m_unscale[:,0], ylabel=multi_target_value[0])
```
plot actual and prediction values for `total bytes` KPI
```
plot_predict_actual_values(test_date, y_pred_m_unscale[:,1], y_test_m_unscale[:,1], ylabel=multi_target_value[1])
```
|
github_jupyter
|
def plot_predict_actual_values(date, y_pred, y_test, ylabel):
"""
plot the predicted values and actual values (for the test data)
"""
fig, axs = plt.subplots(figsize=(16,6))
axs.plot(date, y_pred, color='red', label='predicted values')
axs.plot(date, y_test, color='blue', label='actual values')
axs.set_title('the predicted values and actual values (for the test data)')
plt.xlabel('test datetime')
plt.ylabel(ylabel)
plt.legend(loc='upper left')
plt.show()
import os
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
%matplotlib inline
raw_df = pd.read_csv("data/data.csv")
raw_df.head()
df = pd.DataFrame(pd.to_datetime(raw_df.StartTime))
# we can find 'AvgRate' is of two scales: 'Mbps' and 'Gbps'
raw_df.AvgRate.str[-4:].unique()
# Unify AvgRate value
df['AvgRate'] = raw_df.AvgRate.apply(lambda x:float(x[:-4]) if x.endswith("Mbps") else float(x[:-4])*1000)
df["total"] = raw_df["total"]
df.head()
ax = df.plot(y='AvgRate',figsize=(16,6), title="AvgRate of network traffic data")
ax = df.plot(y='total',figsize=(16,6), title="total bytes of network traffic data")
from zoo.chronos.data import TSDataset
from sklearn.preprocessing import StandardScaler
# we look back one week data which is of the frequency of 2h.
look_back = 84
# specify the number of steps to be predicted,one day is selected by default.
horizon = 1
tsdata_train, _, tsdata_test = TSDataset.from_pandas(df, dt_col="StartTime", target_col=["AvgRate","total"], with_split=True, test_ratio=0.1)
standard_scaler = StandardScaler()
for tsdata in [tsdata_train, tsdata_test]:
tsdata.gen_dt_feature()\
.impute(mode="last")\
.scale(standard_scaler, fit=(tsdata is tsdata_train))
for tsdata in [tsdata_train, tsdata_test]:
tsdata.roll(lookback=look_back, horizon=horizon, target_col="AvgRate")
x_train, y_train = tsdata_train.to_numpy()
x_test, y_test = tsdata_test.to_numpy()
y_train, y_test = y_train[:, 0, :], y_test[:, 0, :]
x_train.shape, y_train.shape, x_test.shape, y_test.shape
from zoo.chronos.model.forecast.lstm_forecaster import LSTMForecaster
# build model
lstm_config = {"lstm_units": [32] * 2, "lr":0.001}
forecaster = LSTMForecaster(target_dim=1, feature_dim=x_train.shape[-1], **lstm_config)
%%time
forecaster.fit(x=x_train, y=y_train, batch_size=1024, epochs=50, distributed=False)
# make prediction
y_pred = forecaster.predict(x_test)
y_pred_unscale = tsdata_test.unscale_numpy(np.expand_dims(y_pred, axis=1))[:, 0, :]
y_test_unscale = tsdata_test.unscale_numpy(np.expand_dims(y_test, axis=1))[:, 0, :]
from zoo.automl.common.metrics import Evaluator
# evaluate with sMAPE
print("sMAPE is", Evaluator.evaluate("smape", y_test_unscale, y_pred_unscale))
# evaluate with mean_squared_error
print("mean_squared error is", Evaluator.evaluate("mse", y_test_unscale, y_pred_unscale))
for tsdata in [tsdata_train, tsdata_test]:
tsdata.roll(lookback=look_back, horizon=horizon, target_col=["AvgRate","total"])
x_train_m, y_train_m = tsdata_train.to_numpy()
x_test_m, y_test_m = tsdata_test.to_numpy()
y_train_m, y_test_m = y_train_m[:, 0, :], y_test_m[:, 0, :]
x_train_m.shape, y_train_m.shape, x_test_m.shape, y_test_m.shape
from zoo.chronos.model.forecast.mtnet_forecaster import MTNetForecaster
mtnet_forecaster = MTNetForecaster(target_dim=y_train_m.shape[-1],
feature_dim=x_train_m.shape[-1],
long_series_num=6,
series_length=12,
ar_window_size=6,
cnn_height=4
)
# mtnet requires reshape of input x before feeding into model.
x_train_mtnet = mtnet_forecaster.preprocess_input(x_train_m)
x_test_mtnet = mtnet_forecaster.preprocess_input(x_test_m)
%%time
hist = mtnet_forecaster.fit(x = x_train_mtnet, y = y_train_m, batch_size=1024, epochs=20)
y_pred_m = mtnet_forecaster.predict(x_test_mtnet)
y_pred_m_unscale = tsdata_test.unscale_numpy(np.expand_dims(y_pred_m, axis=1))[:, 0, :]
y_test_m_unscale = tsdata_test.unscale_numpy(np.expand_dims(y_test_m, axis=1))[:, 0, :]
from zoo.automl.common.metrics import Evaluator
# evaluate with sMAPE
print("sMAPE is", Evaluator.evaluate("smape", y_test_m_unscale, y_pred_m_unscale, multioutput="raw_values"))
# evaluate with mean_squared_error
print("mean_squared error is", Evaluator.evaluate("mse", y_test_m_unscale, y_pred_m_unscale, multioutput="raw_values"))
multi_target_value = ["AvgRate","total"]
test_date=df[-y_test_m_unscale.shape[0]:].index
plot_predict_actual_values(test_date, y_pred_m_unscale[:,0], y_test_m_unscale[:,0], ylabel=multi_target_value[0])
plot_predict_actual_values(test_date, y_pred_m_unscale[:,1], y_test_m_unscale[:,1], ylabel=multi_target_value[1])
| 0.722037 | 0.98945 |
## Data Source
Dataset is derived from Fannie Mae’s [Single-Family Loan Performance Data](http://www.fanniemae.com/portal/funding-the-market/data/loan-performance-data.html) with all rights reserved by Fannie Mae. This processed dataset is redistributed with permission and consent from Fannie Mae. For the full raw dataset visit [Fannie Mae]() to register for an account and to download
Instruction is available at NVIDIA [RAPIDS demo site](https://docs.rapids.ai/datasets/mortgage-data).
## Prerequisite
This notebook runs in a AWS EMR cluster with GPU nodes, with [Spark RAPIDS](https://https://docs.aws.amazon.com/emr/index.html) set up.
### Define Spark conf and Create Spark Session
For details explanation for spark conf, please go to Spark RAPIDS config guide.
Please customeize your Spark configurations based on your GPU cluster.
```
%%configure -f
{
"driverMemory": "4000M",
"driverCores": 2,
"executorMemory": "4000M",
"conf": {"spark.sql.adaptive.enabled": false, "spark.dynamicAllocation.enabled": false, "spark.executor.instances":2, "spark.executor.cores":2, "spark.rapids.sql.explain":"ALL", "spark.task.cpus":"1", "spark.rapids.sql.concurrentGpuTasks":"2", "spark.rapids.memory.pinnedPool.size":"2G", "spark.executor.memoryOverhead":"2G", "spark.executor.extraJavaOptions":"-Dai.rapids.cudf.prefer-pinned=true", "spark.locality.wait":"0s", "spark.sql.files.maxPartitionBytes":"512m", "spark.executor.resource.gpu.amount":"1", "spark.task.resource.gpu.amount":"0.5", "spark.plugins":"com.nvidia.spark.SQLPlugin", "spark.rapids.sql.hasNans":"false", "spark.rapids.sql.batchSizeBytes":"512M", "spark.rapids.sql.reader.batchSizeBytes":"768M", "spark.rapids.sql.variableFloatAgg.enabled":"true"}
}
%%info
import time
from pyspark import broadcast
from pyspark.sql import SparkSession
from pyspark.sql.functions import *
from pyspark.sql.types import *
from pyspark.sql.window import Window
```
### Define ETL Process
Define data schema and steps to do the ETL process:
```
def _get_quarter_from_csv_file_name():
return substring_index(substring_index(input_file_name(), '.', 1), '_', -1)
_csv_perf_schema = StructType([
StructField('loan_id', LongType()),
StructField('monthly_reporting_period', StringType()),
StructField('servicer', StringType()),
StructField('interest_rate', DoubleType()),
StructField('current_actual_upb', DoubleType()),
StructField('loan_age', DoubleType()),
StructField('remaining_months_to_legal_maturity', DoubleType()),
StructField('adj_remaining_months_to_maturity', DoubleType()),
StructField('maturity_date', StringType()),
StructField('msa', DoubleType()),
StructField('current_loan_delinquency_status', IntegerType()),
StructField('mod_flag', StringType()),
StructField('zero_balance_code', StringType()),
StructField('zero_balance_effective_date', StringType()),
StructField('last_paid_installment_date', StringType()),
StructField('foreclosed_after', StringType()),
StructField('disposition_date', StringType()),
StructField('foreclosure_costs', DoubleType()),
StructField('prop_preservation_and_repair_costs', DoubleType()),
StructField('asset_recovery_costs', DoubleType()),
StructField('misc_holding_expenses', DoubleType()),
StructField('holding_taxes', DoubleType()),
StructField('net_sale_proceeds', DoubleType()),
StructField('credit_enhancement_proceeds', DoubleType()),
StructField('repurchase_make_whole_proceeds', StringType()),
StructField('other_foreclosure_proceeds', DoubleType()),
StructField('non_interest_bearing_upb', DoubleType()),
StructField('principal_forgiveness_upb', StringType()),
StructField('repurchase_make_whole_proceeds_flag', StringType()),
StructField('foreclosure_principal_write_off_amount', StringType()),
StructField('servicing_activity_indicator', StringType())])
_csv_acq_schema = StructType([
StructField('loan_id', LongType()),
StructField('orig_channel', StringType()),
StructField('seller_name', StringType()),
StructField('orig_interest_rate', DoubleType()),
StructField('orig_upb', IntegerType()),
StructField('orig_loan_term', IntegerType()),
StructField('orig_date', StringType()),
StructField('first_pay_date', StringType()),
StructField('orig_ltv', DoubleType()),
StructField('orig_cltv', DoubleType()),
StructField('num_borrowers', DoubleType()),
StructField('dti', DoubleType()),
StructField('borrower_credit_score', DoubleType()),
StructField('first_home_buyer', StringType()),
StructField('loan_purpose', StringType()),
StructField('property_type', StringType()),
StructField('num_units', IntegerType()),
StructField('occupancy_status', StringType()),
StructField('property_state', StringType()),
StructField('zip', IntegerType()),
StructField('mortgage_insurance_percent', DoubleType()),
StructField('product_type', StringType()),
StructField('coborrow_credit_score', DoubleType()),
StructField('mortgage_insurance_type', DoubleType()),
StructField('relocation_mortgage_indicator', StringType())])
_name_mapping = [
("WITMER FUNDING, LLC", "Witmer"),
("WELLS FARGO CREDIT RISK TRANSFER SECURITIES TRUST 2015", "Wells Fargo"),
("WELLS FARGO BANK, NA" , "Wells Fargo"),
("WELLS FARGO BANK, N.A." , "Wells Fargo"),
("WELLS FARGO BANK, NA" , "Wells Fargo"),
("USAA FEDERAL SAVINGS BANK" , "USAA"),
("UNITED SHORE FINANCIAL SERVICES, LLC D\\/B\\/A UNITED WHOLESALE MORTGAGE" , "United Seq(e"),
("U.S. BANK N.A." , "US Bank"),
("SUNTRUST MORTGAGE INC." , "Suntrust"),
("STONEGATE MORTGAGE CORPORATION" , "Stonegate Mortgage"),
("STEARNS LENDING, LLC" , "Stearns Lending"),
("STEARNS LENDING, INC." , "Stearns Lending"),
("SIERRA PACIFIC MORTGAGE COMPANY, INC." , "Sierra Pacific Mortgage"),
("REGIONS BANK" , "Regions"),
("RBC MORTGAGE COMPANY" , "RBC"),
("QUICKEN LOANS INC." , "Quicken Loans"),
("PULTE MORTGAGE, L.L.C." , "Pulte Mortgage"),
("PROVIDENT FUNDING ASSOCIATES, L.P." , "Provident Funding"),
("PROSPECT MORTGAGE, LLC" , "Prospect Mortgage"),
("PRINCIPAL RESIDENTIAL MORTGAGE CAPITAL RESOURCES, LLC" , "Principal Residential"),
("PNC BANK, N.A." , "PNC"),
("PMT CREDIT RISK TRANSFER TRUST 2015-2" , "PennyMac"),
("PHH MORTGAGE CORPORATION" , "PHH Mortgage"),
("PENNYMAC CORP." , "PennyMac"),
("PACIFIC UNION FINANCIAL, LLC" , "Other"),
("OTHER" , "Other"),
("NYCB MORTGAGE COMPANY, LLC" , "NYCB"),
("NEW YORK COMMUNITY BANK" , "NYCB"),
("NETBANK FUNDING SERVICES" , "Netbank"),
("NATIONSTAR MORTGAGE, LLC" , "Nationstar Mortgage"),
("METLIFE BANK, NA" , "Metlife"),
("LOANDEPOT.COM, LLC" , "LoanDepot.com"),
("J.P. MORGAN MADISON AVENUE SECURITIES TRUST, SERIES 2015-1" , "JP Morgan Chase"),
("J.P. MORGAN MADISON AVENUE SECURITIES TRUST, SERIES 2014-1" , "JP Morgan Chase"),
("JPMORGAN CHASE BANK, NATIONAL ASSOCIATION" , "JP Morgan Chase"),
("JPMORGAN CHASE BANK, NA" , "JP Morgan Chase"),
("JP MORGAN CHASE BANK, NA" , "JP Morgan Chase"),
("IRWIN MORTGAGE, CORPORATION" , "Irwin Mortgage"),
("IMPAC MORTGAGE CORP." , "Impac Mortgage"),
("HSBC BANK USA, NATIONAL ASSOCIATION" , "HSBC"),
("HOMEWARD RESIDENTIAL, INC." , "Homeward Mortgage"),
("HOMESTREET BANK" , "Other"),
("HOMEBRIDGE FINANCIAL SERVICES, INC." , "HomeBridge"),
("HARWOOD STREET FUNDING I, LLC" , "Harwood Mortgage"),
("GUILD MORTGAGE COMPANY" , "Guild Mortgage"),
("GMAC MORTGAGE, LLC (USAA FEDERAL SAVINGS BANK)" , "GMAC"),
("GMAC MORTGAGE, LLC" , "GMAC"),
("GMAC (USAA)" , "GMAC"),
("FREMONT BANK" , "Fremont Bank"),
("FREEDOM MORTGAGE CORP." , "Freedom Mortgage"),
("FRANKLIN AMERICAN MORTGAGE COMPANY" , "Franklin America"),
("FLEET NATIONAL BANK" , "Fleet National"),
("FLAGSTAR CAPITAL MARKETS CORPORATION" , "Flagstar Bank"),
("FLAGSTAR BANK, FSB" , "Flagstar Bank"),
("FIRST TENNESSEE BANK NATIONAL ASSOCIATION" , "Other"),
("FIFTH THIRD BANK" , "Fifth Third Bank"),
("FEDERAL HOME LOAN BANK OF CHICAGO" , "Fedral Home of Chicago"),
("FDIC, RECEIVER, INDYMAC FEDERAL BANK FSB" , "FDIC"),
("DOWNEY SAVINGS AND LOAN ASSOCIATION, F.A." , "Downey Mortgage"),
("DITECH FINANCIAL LLC" , "Ditech"),
("CITIMORTGAGE, INC." , "Citi"),
("CHICAGO MORTGAGE SOLUTIONS DBA INTERFIRST MORTGAGE COMPANY" , "Chicago Mortgage"),
("CHICAGO MORTGAGE SOLUTIONS DBA INTERBANK MORTGAGE COMPANY" , "Chicago Mortgage"),
("CHASE HOME FINANCE, LLC" , "JP Morgan Chase"),
("CHASE HOME FINANCE FRANKLIN AMERICAN MORTGAGE COMPANY" , "JP Morgan Chase"),
("CHASE HOME FINANCE (CIE 1)" , "JP Morgan Chase"),
("CHASE HOME FINANCE" , "JP Morgan Chase"),
("CASHCALL, INC." , "CashCall"),
("CAPITAL ONE, NATIONAL ASSOCIATION" , "Capital One"),
("CALIBER HOME LOANS, INC." , "Caliber Funding"),
("BISHOPS GATE RESIDENTIAL MORTGAGE TRUST" , "Bishops Gate Mortgage"),
("BANK OF AMERICA, N.A." , "Bank of America"),
("AMTRUST BANK" , "AmTrust"),
("AMERISAVE MORTGAGE CORPORATION" , "Amerisave"),
("AMERIHOME MORTGAGE COMPANY, LLC" , "AmeriHome Mortgage"),
("ALLY BANK" , "Ally Bank"),
("ACADEMY MORTGAGE CORPORATION" , "Academy Mortgage"),
("NO CASH-OUT REFINANCE" , "OTHER REFINANCE"),
("REFINANCE - NOT SPECIFIED" , "OTHER REFINANCE"),
("Other REFINANCE" , "OTHER REFINANCE")]
cate_col_names = [
"orig_channel",
"first_home_buyer",
"loan_purpose",
"property_type",
"occupancy_status",
"property_state",
"relocation_mortgage_indicator",
"seller_name",
"mod_flag"
]
# Numberic columns
label_col_name = "delinquency_12"
numeric_col_names = [
"orig_interest_rate",
"orig_upb",
"orig_loan_term",
"orig_ltv",
"orig_cltv",
"num_borrowers",
"dti",
"borrower_credit_score",
"num_units",
"zip",
"mortgage_insurance_percent",
"current_loan_delinquency_status",
"current_actual_upb",
"interest_rate",
"loan_age",
"msa",
"non_interest_bearing_upb",
label_col_name
]
all_col_names = cate_col_names + numeric_col_names
def read_perf_csv(spark, path):
return spark.read.format('csv') \
.option('nullValue', '') \
.option('header', 'false') \
.option('delimiter', '|') \
.schema(_csv_perf_schema) \
.load(path) \
.withColumn('quarter', _get_quarter_from_csv_file_name())
def read_acq_csv(spark, path):
return spark.read.format('csv') \
.option('nullValue', '') \
.option('header', 'false') \
.option('delimiter', '|') \
.schema(_csv_acq_schema) \
.load(path) \
.withColumn('quarter', _get_quarter_from_csv_file_name())
def _parse_dates(perf):
return perf \
.withColumn('monthly_reporting_period', to_date(col('monthly_reporting_period'), 'MM/dd/yyyy')) \
.withColumn('monthly_reporting_period_month', month(col('monthly_reporting_period'))) \
.withColumn('monthly_reporting_period_year', year(col('monthly_reporting_period'))) \
.withColumn('monthly_reporting_period_day', dayofmonth(col('monthly_reporting_period'))) \
.withColumn('last_paid_installment_date', to_date(col('last_paid_installment_date'), 'MM/dd/yyyy')) \
.withColumn('foreclosed_after', to_date(col('foreclosed_after'), 'MM/dd/yyyy')) \
.withColumn('disposition_date', to_date(col('disposition_date'), 'MM/dd/yyyy')) \
.withColumn('maturity_date', to_date(col('maturity_date'), 'MM/yyyy')) \
.withColumn('zero_balance_effective_date', to_date(col('zero_balance_effective_date'), 'MM/yyyy'))
def _create_perf_deliquency(spark, perf):
aggDF = perf.select(
col("quarter"),
col("loan_id"),
col("current_loan_delinquency_status"),
when(col("current_loan_delinquency_status") >= 1, col("monthly_reporting_period")).alias("delinquency_30"),
when(col("current_loan_delinquency_status") >= 3, col("monthly_reporting_period")).alias("delinquency_90"),
when(col("current_loan_delinquency_status") >= 6, col("monthly_reporting_period")).alias("delinquency_180")) \
.groupBy("quarter", "loan_id") \
.agg(
max("current_loan_delinquency_status").alias("delinquency_12"),
min("delinquency_30").alias("delinquency_30"),
min("delinquency_90").alias("delinquency_90"),
min("delinquency_180").alias("delinquency_180")) \
.select(
col("quarter"),
col("loan_id"),
(col("delinquency_12") >= 1).alias("ever_30"),
(col("delinquency_12") >= 3).alias("ever_90"),
(col("delinquency_12") >= 6).alias("ever_180"),
col("delinquency_30"),
col("delinquency_90"),
col("delinquency_180"))
joinedDf = perf \
.withColumnRenamed("monthly_reporting_period", "timestamp") \
.withColumnRenamed("monthly_reporting_period_month", "timestamp_month") \
.withColumnRenamed("monthly_reporting_period_year", "timestamp_year") \
.withColumnRenamed("current_loan_delinquency_status", "delinquency_12") \
.withColumnRenamed("current_actual_upb", "upb_12") \
.select("quarter", "loan_id", "timestamp", "delinquency_12", "upb_12", "timestamp_month", "timestamp_year") \
.join(aggDF, ["loan_id", "quarter"], "left_outer")
# calculate the 12 month delinquency and upb values
months = 12
monthArray = [lit(x) for x in range(0, 12)]
# explode on a small amount of data is actually slightly more efficient than a cross join
testDf = joinedDf \
.withColumn("month_y", explode(array(monthArray))) \
.select(
col("quarter"),
floor(((col("timestamp_year") * 12 + col("timestamp_month")) - 24000) / months).alias("josh_mody"),
floor(((col("timestamp_year") * 12 + col("timestamp_month")) - 24000 - col("month_y")) / months).alias("josh_mody_n"),
col("ever_30"),
col("ever_90"),
col("ever_180"),
col("delinquency_30"),
col("delinquency_90"),
col("delinquency_180"),
col("loan_id"),
col("month_y"),
col("delinquency_12"),
col("upb_12")) \
.groupBy("quarter", "loan_id", "josh_mody_n", "ever_30", "ever_90", "ever_180", "delinquency_30", "delinquency_90", "delinquency_180", "month_y") \
.agg(max("delinquency_12").alias("delinquency_12"), min("upb_12").alias("upb_12")) \
.withColumn("timestamp_year", floor((lit(24000) + (col("josh_mody_n") * lit(months)) + (col("month_y") - 1)) / lit(12))) \
.selectExpr('*', 'pmod(24000 + (josh_mody_n * {}) + month_y, 12) as timestamp_month_tmp'.format(months)) \
.withColumn("timestamp_month", when(col("timestamp_month_tmp") == lit(0), lit(12)).otherwise(col("timestamp_month_tmp"))) \
.withColumn("delinquency_12", ((col("delinquency_12") > 3).cast("int") + (col("upb_12") == 0).cast("int")).alias("delinquency_12")) \
.drop("timestamp_month_tmp", "josh_mody_n", "month_y")
return perf.withColumnRenamed("monthly_reporting_period_month", "timestamp_month") \
.withColumnRenamed("monthly_reporting_period_year", "timestamp_year") \
.join(testDf, ["quarter", "loan_id", "timestamp_year", "timestamp_month"], "left") \
.drop("timestamp_year", "timestamp_month")
def _create_acquisition(spark, acq):
nameMapping = spark.createDataFrame(_name_mapping, ["from_seller_name", "to_seller_name"])
return acq.join(nameMapping, col("seller_name") == col("from_seller_name"), "left") \
.drop("from_seller_name") \
.withColumn("old_name", col("seller_name")) \
.withColumn("seller_name", coalesce(col("to_seller_name"), col("seller_name"))) \
.drop("to_seller_name") \
.withColumn("orig_date", to_date(col("orig_date"), "MM/yyyy")) \
.withColumn("first_pay_date", to_date(col("first_pay_date"), "MM/yyyy"))
def _gen_dictionary(etl_df, col_names):
cnt_table = etl_df.select(posexplode(array([col(i) for i in col_names])))\
.withColumnRenamed("pos", "column_id")\
.withColumnRenamed("col", "data")\
.filter("data is not null")\
.groupBy("column_id", "data")\
.count()
windowed = Window.partitionBy("column_id").orderBy(desc("count"))
return cnt_table.withColumn("id", row_number().over(windowed)).drop("count")
def _cast_string_columns_to_numeric(spark, input_df):
cached_dict_df = _gen_dictionary(input_df, cate_col_names).cache()
output_df = input_df
# Generate the final table with all columns being numeric.
for col_pos, col_name in enumerate(cate_col_names):
col_dict_df = cached_dict_df.filter(col("column_id") == col_pos)\
.drop("column_id")\
.withColumnRenamed("data", col_name)
output_df = output_df.join(broadcast(col_dict_df), col_name, "left")\
.drop(col_name)\
.withColumnRenamed("id", col_name)
return output_df
def run_mortgage(spark, perf, acq):
parsed_perf = _parse_dates(perf)
perf_deliqency = _create_perf_deliquency(spark, parsed_perf)
cleaned_acq = _create_acquisition(spark, acq)
df = perf_deliqency.join(cleaned_acq, ["loan_id", "quarter"], "inner")
# change to this for 20 year mortgage data - test_quarters = ['2016Q1','2016Q2','2016Q3','2016Q4']
test_quarters = ['2000Q4']
train_df = df.filter(df.quarter.isin(test_quarters)).drop("quarter")
test_df = df.filter(df.quarter.isin(test_quarters)).drop("quarter")
casted_train_df = _cast_string_columns_to_numeric(spark, train_df)\
.select(all_col_names)\
.withColumn(label_col_name, when(col(label_col_name) > 0, 1).otherwise(0))\
.fillna(float(0))
casted_test_df = _cast_string_columns_to_numeric(spark, test_df)\
.select(all_col_names)\
.withColumn(label_col_name, when(col(label_col_name) > 0, 1).otherwise(0))\
.fillna(float(0))
return casted_train_df, casted_test_df
```
### Define Data Input/Output location
This example is using one year mortgage data (year 2000) for GPU Spark cluster (2x g4dn.2xlarge). Please use large GPU cluster to process the full mortgage data.
```
orig_perf_path = 's3://spark-xgboost-mortgage-dataset-east1/mortgage-etl-demo/perf/*'
orig_acq_path = 's3://spark-xgboost-mortgage-dataset-east1/mortgage-etl-demo/acq/*'
train_path = 's3://spark-xgboost-mortgage-dataset-east1/mortgage-xgboost-demo/train/'
test_path = 's3://spark-xgboost-mortgage-dataset-east1/mortgage-xgboost-demo/test/'
tmp_perf_path = 's3://spark-xgboost-mortgage-dataset-east1/mortgage_parquet_gpu/perf/'
tmp_acq_path = 's3://spark-xgboost-mortgage-dataset-east1/mortgage_parquet_gpu/acq/'
```
### Read CSV data and Transcode to Parquet
```
# Lets transcode the data first
start = time.time()
# we want a few big files instead of lots of small files
spark.conf.set('spark.sql.files.maxPartitionBytes', '200G')
acq = read_acq_csv(spark, orig_acq_path)
acq.repartition(20).write.parquet(tmp_acq_path, mode='overwrite')
perf = read_perf_csv(spark, orig_perf_path)
perf.coalesce(80).write.parquet(tmp_perf_path, mode='overwrite')
end = time.time()
print(end - start)
```
### Execute ETL Code Defined in 1st Cell
```
# Now lets actually process the data\n",
start = time.time()
spark.conf.set('spark.sql.files.maxPartitionBytes', '1G')
spark.conf.set('spark.sql.shuffle.partitions', '160')
perf = spark.read.parquet(tmp_perf_path)
acq = spark.read.parquet(tmp_acq_path)
train_out, test_out = run_mortgage(spark, perf, acq)
train_out.write.parquet(train_path, mode='overwrite')
end = time.time()
print(end - start)
test_out.write.parquet(test_path, mode='overwrite')
end = time.time()
print(end - start)
```
### Print Physical Plan
```
#train_out.explain()
print(spark._jvm.org.apache.spark.sql.api.python.PythonSQLUtils.explainString(train_out._jdf.queryExecution(), 'simple'))
```
|
github_jupyter
|
%%configure -f
{
"driverMemory": "4000M",
"driverCores": 2,
"executorMemory": "4000M",
"conf": {"spark.sql.adaptive.enabled": false, "spark.dynamicAllocation.enabled": false, "spark.executor.instances":2, "spark.executor.cores":2, "spark.rapids.sql.explain":"ALL", "spark.task.cpus":"1", "spark.rapids.sql.concurrentGpuTasks":"2", "spark.rapids.memory.pinnedPool.size":"2G", "spark.executor.memoryOverhead":"2G", "spark.executor.extraJavaOptions":"-Dai.rapids.cudf.prefer-pinned=true", "spark.locality.wait":"0s", "spark.sql.files.maxPartitionBytes":"512m", "spark.executor.resource.gpu.amount":"1", "spark.task.resource.gpu.amount":"0.5", "spark.plugins":"com.nvidia.spark.SQLPlugin", "spark.rapids.sql.hasNans":"false", "spark.rapids.sql.batchSizeBytes":"512M", "spark.rapids.sql.reader.batchSizeBytes":"768M", "spark.rapids.sql.variableFloatAgg.enabled":"true"}
}
%%info
import time
from pyspark import broadcast
from pyspark.sql import SparkSession
from pyspark.sql.functions import *
from pyspark.sql.types import *
from pyspark.sql.window import Window
def _get_quarter_from_csv_file_name():
return substring_index(substring_index(input_file_name(), '.', 1), '_', -1)
_csv_perf_schema = StructType([
StructField('loan_id', LongType()),
StructField('monthly_reporting_period', StringType()),
StructField('servicer', StringType()),
StructField('interest_rate', DoubleType()),
StructField('current_actual_upb', DoubleType()),
StructField('loan_age', DoubleType()),
StructField('remaining_months_to_legal_maturity', DoubleType()),
StructField('adj_remaining_months_to_maturity', DoubleType()),
StructField('maturity_date', StringType()),
StructField('msa', DoubleType()),
StructField('current_loan_delinquency_status', IntegerType()),
StructField('mod_flag', StringType()),
StructField('zero_balance_code', StringType()),
StructField('zero_balance_effective_date', StringType()),
StructField('last_paid_installment_date', StringType()),
StructField('foreclosed_after', StringType()),
StructField('disposition_date', StringType()),
StructField('foreclosure_costs', DoubleType()),
StructField('prop_preservation_and_repair_costs', DoubleType()),
StructField('asset_recovery_costs', DoubleType()),
StructField('misc_holding_expenses', DoubleType()),
StructField('holding_taxes', DoubleType()),
StructField('net_sale_proceeds', DoubleType()),
StructField('credit_enhancement_proceeds', DoubleType()),
StructField('repurchase_make_whole_proceeds', StringType()),
StructField('other_foreclosure_proceeds', DoubleType()),
StructField('non_interest_bearing_upb', DoubleType()),
StructField('principal_forgiveness_upb', StringType()),
StructField('repurchase_make_whole_proceeds_flag', StringType()),
StructField('foreclosure_principal_write_off_amount', StringType()),
StructField('servicing_activity_indicator', StringType())])
_csv_acq_schema = StructType([
StructField('loan_id', LongType()),
StructField('orig_channel', StringType()),
StructField('seller_name', StringType()),
StructField('orig_interest_rate', DoubleType()),
StructField('orig_upb', IntegerType()),
StructField('orig_loan_term', IntegerType()),
StructField('orig_date', StringType()),
StructField('first_pay_date', StringType()),
StructField('orig_ltv', DoubleType()),
StructField('orig_cltv', DoubleType()),
StructField('num_borrowers', DoubleType()),
StructField('dti', DoubleType()),
StructField('borrower_credit_score', DoubleType()),
StructField('first_home_buyer', StringType()),
StructField('loan_purpose', StringType()),
StructField('property_type', StringType()),
StructField('num_units', IntegerType()),
StructField('occupancy_status', StringType()),
StructField('property_state', StringType()),
StructField('zip', IntegerType()),
StructField('mortgage_insurance_percent', DoubleType()),
StructField('product_type', StringType()),
StructField('coborrow_credit_score', DoubleType()),
StructField('mortgage_insurance_type', DoubleType()),
StructField('relocation_mortgage_indicator', StringType())])
_name_mapping = [
("WITMER FUNDING, LLC", "Witmer"),
("WELLS FARGO CREDIT RISK TRANSFER SECURITIES TRUST 2015", "Wells Fargo"),
("WELLS FARGO BANK, NA" , "Wells Fargo"),
("WELLS FARGO BANK, N.A." , "Wells Fargo"),
("WELLS FARGO BANK, NA" , "Wells Fargo"),
("USAA FEDERAL SAVINGS BANK" , "USAA"),
("UNITED SHORE FINANCIAL SERVICES, LLC D\\/B\\/A UNITED WHOLESALE MORTGAGE" , "United Seq(e"),
("U.S. BANK N.A." , "US Bank"),
("SUNTRUST MORTGAGE INC." , "Suntrust"),
("STONEGATE MORTGAGE CORPORATION" , "Stonegate Mortgage"),
("STEARNS LENDING, LLC" , "Stearns Lending"),
("STEARNS LENDING, INC." , "Stearns Lending"),
("SIERRA PACIFIC MORTGAGE COMPANY, INC." , "Sierra Pacific Mortgage"),
("REGIONS BANK" , "Regions"),
("RBC MORTGAGE COMPANY" , "RBC"),
("QUICKEN LOANS INC." , "Quicken Loans"),
("PULTE MORTGAGE, L.L.C." , "Pulte Mortgage"),
("PROVIDENT FUNDING ASSOCIATES, L.P." , "Provident Funding"),
("PROSPECT MORTGAGE, LLC" , "Prospect Mortgage"),
("PRINCIPAL RESIDENTIAL MORTGAGE CAPITAL RESOURCES, LLC" , "Principal Residential"),
("PNC BANK, N.A." , "PNC"),
("PMT CREDIT RISK TRANSFER TRUST 2015-2" , "PennyMac"),
("PHH MORTGAGE CORPORATION" , "PHH Mortgage"),
("PENNYMAC CORP." , "PennyMac"),
("PACIFIC UNION FINANCIAL, LLC" , "Other"),
("OTHER" , "Other"),
("NYCB MORTGAGE COMPANY, LLC" , "NYCB"),
("NEW YORK COMMUNITY BANK" , "NYCB"),
("NETBANK FUNDING SERVICES" , "Netbank"),
("NATIONSTAR MORTGAGE, LLC" , "Nationstar Mortgage"),
("METLIFE BANK, NA" , "Metlife"),
("LOANDEPOT.COM, LLC" , "LoanDepot.com"),
("J.P. MORGAN MADISON AVENUE SECURITIES TRUST, SERIES 2015-1" , "JP Morgan Chase"),
("J.P. MORGAN MADISON AVENUE SECURITIES TRUST, SERIES 2014-1" , "JP Morgan Chase"),
("JPMORGAN CHASE BANK, NATIONAL ASSOCIATION" , "JP Morgan Chase"),
("JPMORGAN CHASE BANK, NA" , "JP Morgan Chase"),
("JP MORGAN CHASE BANK, NA" , "JP Morgan Chase"),
("IRWIN MORTGAGE, CORPORATION" , "Irwin Mortgage"),
("IMPAC MORTGAGE CORP." , "Impac Mortgage"),
("HSBC BANK USA, NATIONAL ASSOCIATION" , "HSBC"),
("HOMEWARD RESIDENTIAL, INC." , "Homeward Mortgage"),
("HOMESTREET BANK" , "Other"),
("HOMEBRIDGE FINANCIAL SERVICES, INC." , "HomeBridge"),
("HARWOOD STREET FUNDING I, LLC" , "Harwood Mortgage"),
("GUILD MORTGAGE COMPANY" , "Guild Mortgage"),
("GMAC MORTGAGE, LLC (USAA FEDERAL SAVINGS BANK)" , "GMAC"),
("GMAC MORTGAGE, LLC" , "GMAC"),
("GMAC (USAA)" , "GMAC"),
("FREMONT BANK" , "Fremont Bank"),
("FREEDOM MORTGAGE CORP." , "Freedom Mortgage"),
("FRANKLIN AMERICAN MORTGAGE COMPANY" , "Franklin America"),
("FLEET NATIONAL BANK" , "Fleet National"),
("FLAGSTAR CAPITAL MARKETS CORPORATION" , "Flagstar Bank"),
("FLAGSTAR BANK, FSB" , "Flagstar Bank"),
("FIRST TENNESSEE BANK NATIONAL ASSOCIATION" , "Other"),
("FIFTH THIRD BANK" , "Fifth Third Bank"),
("FEDERAL HOME LOAN BANK OF CHICAGO" , "Fedral Home of Chicago"),
("FDIC, RECEIVER, INDYMAC FEDERAL BANK FSB" , "FDIC"),
("DOWNEY SAVINGS AND LOAN ASSOCIATION, F.A." , "Downey Mortgage"),
("DITECH FINANCIAL LLC" , "Ditech"),
("CITIMORTGAGE, INC." , "Citi"),
("CHICAGO MORTGAGE SOLUTIONS DBA INTERFIRST MORTGAGE COMPANY" , "Chicago Mortgage"),
("CHICAGO MORTGAGE SOLUTIONS DBA INTERBANK MORTGAGE COMPANY" , "Chicago Mortgage"),
("CHASE HOME FINANCE, LLC" , "JP Morgan Chase"),
("CHASE HOME FINANCE FRANKLIN AMERICAN MORTGAGE COMPANY" , "JP Morgan Chase"),
("CHASE HOME FINANCE (CIE 1)" , "JP Morgan Chase"),
("CHASE HOME FINANCE" , "JP Morgan Chase"),
("CASHCALL, INC." , "CashCall"),
("CAPITAL ONE, NATIONAL ASSOCIATION" , "Capital One"),
("CALIBER HOME LOANS, INC." , "Caliber Funding"),
("BISHOPS GATE RESIDENTIAL MORTGAGE TRUST" , "Bishops Gate Mortgage"),
("BANK OF AMERICA, N.A." , "Bank of America"),
("AMTRUST BANK" , "AmTrust"),
("AMERISAVE MORTGAGE CORPORATION" , "Amerisave"),
("AMERIHOME MORTGAGE COMPANY, LLC" , "AmeriHome Mortgage"),
("ALLY BANK" , "Ally Bank"),
("ACADEMY MORTGAGE CORPORATION" , "Academy Mortgage"),
("NO CASH-OUT REFINANCE" , "OTHER REFINANCE"),
("REFINANCE - NOT SPECIFIED" , "OTHER REFINANCE"),
("Other REFINANCE" , "OTHER REFINANCE")]
cate_col_names = [
"orig_channel",
"first_home_buyer",
"loan_purpose",
"property_type",
"occupancy_status",
"property_state",
"relocation_mortgage_indicator",
"seller_name",
"mod_flag"
]
# Numberic columns
label_col_name = "delinquency_12"
numeric_col_names = [
"orig_interest_rate",
"orig_upb",
"orig_loan_term",
"orig_ltv",
"orig_cltv",
"num_borrowers",
"dti",
"borrower_credit_score",
"num_units",
"zip",
"mortgage_insurance_percent",
"current_loan_delinquency_status",
"current_actual_upb",
"interest_rate",
"loan_age",
"msa",
"non_interest_bearing_upb",
label_col_name
]
all_col_names = cate_col_names + numeric_col_names
def read_perf_csv(spark, path):
return spark.read.format('csv') \
.option('nullValue', '') \
.option('header', 'false') \
.option('delimiter', '|') \
.schema(_csv_perf_schema) \
.load(path) \
.withColumn('quarter', _get_quarter_from_csv_file_name())
def read_acq_csv(spark, path):
return spark.read.format('csv') \
.option('nullValue', '') \
.option('header', 'false') \
.option('delimiter', '|') \
.schema(_csv_acq_schema) \
.load(path) \
.withColumn('quarter', _get_quarter_from_csv_file_name())
def _parse_dates(perf):
return perf \
.withColumn('monthly_reporting_period', to_date(col('monthly_reporting_period'), 'MM/dd/yyyy')) \
.withColumn('monthly_reporting_period_month', month(col('monthly_reporting_period'))) \
.withColumn('monthly_reporting_period_year', year(col('monthly_reporting_period'))) \
.withColumn('monthly_reporting_period_day', dayofmonth(col('monthly_reporting_period'))) \
.withColumn('last_paid_installment_date', to_date(col('last_paid_installment_date'), 'MM/dd/yyyy')) \
.withColumn('foreclosed_after', to_date(col('foreclosed_after'), 'MM/dd/yyyy')) \
.withColumn('disposition_date', to_date(col('disposition_date'), 'MM/dd/yyyy')) \
.withColumn('maturity_date', to_date(col('maturity_date'), 'MM/yyyy')) \
.withColumn('zero_balance_effective_date', to_date(col('zero_balance_effective_date'), 'MM/yyyy'))
def _create_perf_deliquency(spark, perf):
aggDF = perf.select(
col("quarter"),
col("loan_id"),
col("current_loan_delinquency_status"),
when(col("current_loan_delinquency_status") >= 1, col("monthly_reporting_period")).alias("delinquency_30"),
when(col("current_loan_delinquency_status") >= 3, col("monthly_reporting_period")).alias("delinquency_90"),
when(col("current_loan_delinquency_status") >= 6, col("monthly_reporting_period")).alias("delinquency_180")) \
.groupBy("quarter", "loan_id") \
.agg(
max("current_loan_delinquency_status").alias("delinquency_12"),
min("delinquency_30").alias("delinquency_30"),
min("delinquency_90").alias("delinquency_90"),
min("delinquency_180").alias("delinquency_180")) \
.select(
col("quarter"),
col("loan_id"),
(col("delinquency_12") >= 1).alias("ever_30"),
(col("delinquency_12") >= 3).alias("ever_90"),
(col("delinquency_12") >= 6).alias("ever_180"),
col("delinquency_30"),
col("delinquency_90"),
col("delinquency_180"))
joinedDf = perf \
.withColumnRenamed("monthly_reporting_period", "timestamp") \
.withColumnRenamed("monthly_reporting_period_month", "timestamp_month") \
.withColumnRenamed("monthly_reporting_period_year", "timestamp_year") \
.withColumnRenamed("current_loan_delinquency_status", "delinquency_12") \
.withColumnRenamed("current_actual_upb", "upb_12") \
.select("quarter", "loan_id", "timestamp", "delinquency_12", "upb_12", "timestamp_month", "timestamp_year") \
.join(aggDF, ["loan_id", "quarter"], "left_outer")
# calculate the 12 month delinquency and upb values
months = 12
monthArray = [lit(x) for x in range(0, 12)]
# explode on a small amount of data is actually slightly more efficient than a cross join
testDf = joinedDf \
.withColumn("month_y", explode(array(monthArray))) \
.select(
col("quarter"),
floor(((col("timestamp_year") * 12 + col("timestamp_month")) - 24000) / months).alias("josh_mody"),
floor(((col("timestamp_year") * 12 + col("timestamp_month")) - 24000 - col("month_y")) / months).alias("josh_mody_n"),
col("ever_30"),
col("ever_90"),
col("ever_180"),
col("delinquency_30"),
col("delinquency_90"),
col("delinquency_180"),
col("loan_id"),
col("month_y"),
col("delinquency_12"),
col("upb_12")) \
.groupBy("quarter", "loan_id", "josh_mody_n", "ever_30", "ever_90", "ever_180", "delinquency_30", "delinquency_90", "delinquency_180", "month_y") \
.agg(max("delinquency_12").alias("delinquency_12"), min("upb_12").alias("upb_12")) \
.withColumn("timestamp_year", floor((lit(24000) + (col("josh_mody_n") * lit(months)) + (col("month_y") - 1)) / lit(12))) \
.selectExpr('*', 'pmod(24000 + (josh_mody_n * {}) + month_y, 12) as timestamp_month_tmp'.format(months)) \
.withColumn("timestamp_month", when(col("timestamp_month_tmp") == lit(0), lit(12)).otherwise(col("timestamp_month_tmp"))) \
.withColumn("delinquency_12", ((col("delinquency_12") > 3).cast("int") + (col("upb_12") == 0).cast("int")).alias("delinquency_12")) \
.drop("timestamp_month_tmp", "josh_mody_n", "month_y")
return perf.withColumnRenamed("monthly_reporting_period_month", "timestamp_month") \
.withColumnRenamed("monthly_reporting_period_year", "timestamp_year") \
.join(testDf, ["quarter", "loan_id", "timestamp_year", "timestamp_month"], "left") \
.drop("timestamp_year", "timestamp_month")
def _create_acquisition(spark, acq):
nameMapping = spark.createDataFrame(_name_mapping, ["from_seller_name", "to_seller_name"])
return acq.join(nameMapping, col("seller_name") == col("from_seller_name"), "left") \
.drop("from_seller_name") \
.withColumn("old_name", col("seller_name")) \
.withColumn("seller_name", coalesce(col("to_seller_name"), col("seller_name"))) \
.drop("to_seller_name") \
.withColumn("orig_date", to_date(col("orig_date"), "MM/yyyy")) \
.withColumn("first_pay_date", to_date(col("first_pay_date"), "MM/yyyy"))
def _gen_dictionary(etl_df, col_names):
cnt_table = etl_df.select(posexplode(array([col(i) for i in col_names])))\
.withColumnRenamed("pos", "column_id")\
.withColumnRenamed("col", "data")\
.filter("data is not null")\
.groupBy("column_id", "data")\
.count()
windowed = Window.partitionBy("column_id").orderBy(desc("count"))
return cnt_table.withColumn("id", row_number().over(windowed)).drop("count")
def _cast_string_columns_to_numeric(spark, input_df):
cached_dict_df = _gen_dictionary(input_df, cate_col_names).cache()
output_df = input_df
# Generate the final table with all columns being numeric.
for col_pos, col_name in enumerate(cate_col_names):
col_dict_df = cached_dict_df.filter(col("column_id") == col_pos)\
.drop("column_id")\
.withColumnRenamed("data", col_name)
output_df = output_df.join(broadcast(col_dict_df), col_name, "left")\
.drop(col_name)\
.withColumnRenamed("id", col_name)
return output_df
def run_mortgage(spark, perf, acq):
parsed_perf = _parse_dates(perf)
perf_deliqency = _create_perf_deliquency(spark, parsed_perf)
cleaned_acq = _create_acquisition(spark, acq)
df = perf_deliqency.join(cleaned_acq, ["loan_id", "quarter"], "inner")
# change to this for 20 year mortgage data - test_quarters = ['2016Q1','2016Q2','2016Q3','2016Q4']
test_quarters = ['2000Q4']
train_df = df.filter(df.quarter.isin(test_quarters)).drop("quarter")
test_df = df.filter(df.quarter.isin(test_quarters)).drop("quarter")
casted_train_df = _cast_string_columns_to_numeric(spark, train_df)\
.select(all_col_names)\
.withColumn(label_col_name, when(col(label_col_name) > 0, 1).otherwise(0))\
.fillna(float(0))
casted_test_df = _cast_string_columns_to_numeric(spark, test_df)\
.select(all_col_names)\
.withColumn(label_col_name, when(col(label_col_name) > 0, 1).otherwise(0))\
.fillna(float(0))
return casted_train_df, casted_test_df
orig_perf_path = 's3://spark-xgboost-mortgage-dataset-east1/mortgage-etl-demo/perf/*'
orig_acq_path = 's3://spark-xgboost-mortgage-dataset-east1/mortgage-etl-demo/acq/*'
train_path = 's3://spark-xgboost-mortgage-dataset-east1/mortgage-xgboost-demo/train/'
test_path = 's3://spark-xgboost-mortgage-dataset-east1/mortgage-xgboost-demo/test/'
tmp_perf_path = 's3://spark-xgboost-mortgage-dataset-east1/mortgage_parquet_gpu/perf/'
tmp_acq_path = 's3://spark-xgboost-mortgage-dataset-east1/mortgage_parquet_gpu/acq/'
# Lets transcode the data first
start = time.time()
# we want a few big files instead of lots of small files
spark.conf.set('spark.sql.files.maxPartitionBytes', '200G')
acq = read_acq_csv(spark, orig_acq_path)
acq.repartition(20).write.parquet(tmp_acq_path, mode='overwrite')
perf = read_perf_csv(spark, orig_perf_path)
perf.coalesce(80).write.parquet(tmp_perf_path, mode='overwrite')
end = time.time()
print(end - start)
# Now lets actually process the data\n",
start = time.time()
spark.conf.set('spark.sql.files.maxPartitionBytes', '1G')
spark.conf.set('spark.sql.shuffle.partitions', '160')
perf = spark.read.parquet(tmp_perf_path)
acq = spark.read.parquet(tmp_acq_path)
train_out, test_out = run_mortgage(spark, perf, acq)
train_out.write.parquet(train_path, mode='overwrite')
end = time.time()
print(end - start)
test_out.write.parquet(test_path, mode='overwrite')
end = time.time()
print(end - start)
#train_out.explain()
print(spark._jvm.org.apache.spark.sql.api.python.PythonSQLUtils.explainString(train_out._jdf.queryExecution(), 'simple'))
| 0.484868 | 0.870432 |
# Rational Expectations Agricultural Market Model
**Randall Romero Aguilar, PhD**
This demo is based on the original Matlab demo accompanying the <a href="https://mitpress.mit.edu/books/applied-computational-economics-and-finance">Computational Economics and Finance</a> 2001 textbook by Mario Miranda and Paul Fackler.
Original (Matlab) CompEcon file: **demintro01.m**
Running this file requires the Python version of CompEcon. This can be installed with pip by running
!pip install compecon --upgrade
<i>Last updated: 2021-Oct-01</i>
<hr>
```
import numpy as np
import matplotlib.pyplot as plt
from compecon import demo, qnwlogn, discmoments
%matplotlib inline
plt.style.use('seaborn')
```
Generate yield distribution
```
sigma2 = 0.2 ** 2
y, w = qnwlogn(25, -0.5 * sigma2, sigma2)
```
Compute rational expectations equilibrium using function iteration, iterating on acreage planted
```
A = lambda aa, pp: 0.5 + 0.5 * np.dot(w, np.maximum(1.5 - 0.5 * aa * y, pp))
ptarg = 1
a = 1
for it in range(50):
aold = a
a = A(a, ptarg)
print('{:3d} {:8.4f} {:8.1e}'.format(it, a, np.linalg.norm(a - aold)))
if np.linalg.norm(a - aold) < 1.e-8:
break
```
Intermediate outputs
```
q = a * y # quantity produced in each state
p = 1.5 - 0.5 * a * y # market price in each state
f = np.maximum(p, ptarg) # farm price in each state
r = f * q # farm revenue in each state
g = (f - p) * q #government expenditures
xavg, xstd = discmoments(w, np.vstack((p, f, r, g)))
varnames = ['Market Price', 'Farm Price', 'Farm Revenue', 'Government Expenditures']
```
Print results
```
print('\n{:24s} {:8s} {:8s}'.format('Variable', 'Expect', 'Std Dev'))
for varname, av, sd in zip(varnames, xavg, xstd):
print(f'{varname:24s} {av:8.4f} {sd:8.4f}')
```
Generate fixed-point mapping
```
aeq = a
a = np.linspace(0, 2, 100)
g = np.array([A(k, ptarg) for k in a])
```
### Graph rational expectations equilibrium
```
fig1 = plt.figure(figsize=[6, 6])
ax = fig1.add_subplot(111, title='Rational expectations equilibrium', aspect=1,
xlabel='Acreage Planted', xticks=[0, aeq, 2], xticklabels=['0', '$a^{*}$', '2'],
ylabel='Rational Acreage Planted', yticks=[0, aeq, 2],yticklabels=['0', '$a^{*}$', '2'])
ax.plot(a, g, 'b', linewidth=4)
ax.plot(a, a, ':', color='grey', linewidth=2)
ax.plot([0, aeq, aeq], [aeq, aeq, 0], 'r--', linewidth=3)
ax.plot([aeq], [aeq], 'ro', markersize=12)
ax.text(0.05, 0, '45${}^o$', color='grey')
ax.text(1.85, aeq - 0.15,'$g(a)$', color='blue')
fig1.show()
```
### Compute rational expectations equilibrium as a function of the target price
```
nplot = 50
ptarg = np.linspace(0, 2, nplot)
a = 1
Ep, Ef, Er, Eg, Sp, Sf, Sr, Sg = (np.empty(nplot) for k in range(8))
for ip in range(nplot):
for it in range(50):
aold = a
a = A(a, ptarg[ip])
if np.linalg.norm((a - aold) < 1.e-10):
break
q = a * y # quantity produced
p = 1.5 - 0.5 * a * y # market price
f = np.maximum(p, ptarg[ip]) # farm price
r = f * q # farm revenue
g = (f - p) * q # government expenditures
xavg, xstd = discmoments(w, np.vstack((p, f, r, g)))
Ep[ip], Ef[ip], Er[ip], Eg[ip] = tuple(xavg)
Sp[ip], Sf[ip], Sr[ip], Sg[ip] = tuple(xstd)
zeroline = lambda y: plt.axhline(y[0], linestyle=':', color='gray')
```
### Graph expected prices vs target price
```
fig2 = plt.figure(figsize=[8, 6])
ax1 = fig2.add_subplot(121, title='Expected price',
xlabel='Target price', xticks=[0, 1, 2],
ylabel='Expectation', yticks=[0.5, 1, 1.5, 2], ylim=[0.5, 2.0])
zeroline(Ep)
ax1.plot(ptarg, Ep, linewidth=4, label='Market Price')
ax1.plot(ptarg, Ef, linewidth=4, label='Farm Price')
ax1.legend(loc='upper left')
# Graph expected prices vs target price
ax2 = fig2.add_subplot(122, title='Price variabilities',
xlabel='Target price', xticks=[0, 1, 2],
ylabel='Standard deviation', yticks=[0, 0.1, 0.2]) #plt.ylim(0.5, 2.0)
zeroline(Sf)
ax2.plot(ptarg, Sp, linewidth=4, label='Market Price')
ax2.plot(ptarg, Sf, linewidth=4, label='Farm Price')
ax2.legend(loc='upper left')
fig2.show()
# Graph expected farm revenue vs target price
fig3 = plt.figure(figsize=[12, 6])
ax1 = fig3.add_subplot(131, title='Expected revenue',
xlabel='Target price', xticks=[0, 1, 2],
ylabel='Expectation', yticks=[1, 2, 3], ylim=[0.8, 3.0])
zeroline(Er)
ax1.plot(ptarg, Er, linewidth=4)
# Graph standard deviation of farm revenue vs target price
ax2 = fig3.add_subplot(132, title='Farm Revenue Variability',
xlabel='Target price', xticks=[0, 1, 2],
ylabel='Standard deviation', yticks=[0, 0.2, 0.4])
zeroline(Sr)
ax2.plot(ptarg, Sr, linewidth=4)
# Graph expected government expenditures vs target price
ax3 = fig3.add_subplot(133, title='Expected Government Expenditures',
xlabel='Target price', xticks=[0, 1, 2],
ylabel='Expectation', yticks=[0, 1, 2], ylim=[-0.05, 2.0])
zeroline(Eg)
ax3.plot(ptarg, Eg, linewidth=4)
plt.show()
#fig1.savefig('demintro02--01.png')
#fig2.savefig('demintro02--02.png')
#fig3.savefig('demintro02--03.png')
```
|
github_jupyter
|
import numpy as np
import matplotlib.pyplot as plt
from compecon import demo, qnwlogn, discmoments
%matplotlib inline
plt.style.use('seaborn')
sigma2 = 0.2 ** 2
y, w = qnwlogn(25, -0.5 * sigma2, sigma2)
A = lambda aa, pp: 0.5 + 0.5 * np.dot(w, np.maximum(1.5 - 0.5 * aa * y, pp))
ptarg = 1
a = 1
for it in range(50):
aold = a
a = A(a, ptarg)
print('{:3d} {:8.4f} {:8.1e}'.format(it, a, np.linalg.norm(a - aold)))
if np.linalg.norm(a - aold) < 1.e-8:
break
q = a * y # quantity produced in each state
p = 1.5 - 0.5 * a * y # market price in each state
f = np.maximum(p, ptarg) # farm price in each state
r = f * q # farm revenue in each state
g = (f - p) * q #government expenditures
xavg, xstd = discmoments(w, np.vstack((p, f, r, g)))
varnames = ['Market Price', 'Farm Price', 'Farm Revenue', 'Government Expenditures']
print('\n{:24s} {:8s} {:8s}'.format('Variable', 'Expect', 'Std Dev'))
for varname, av, sd in zip(varnames, xavg, xstd):
print(f'{varname:24s} {av:8.4f} {sd:8.4f}')
aeq = a
a = np.linspace(0, 2, 100)
g = np.array([A(k, ptarg) for k in a])
fig1 = plt.figure(figsize=[6, 6])
ax = fig1.add_subplot(111, title='Rational expectations equilibrium', aspect=1,
xlabel='Acreage Planted', xticks=[0, aeq, 2], xticklabels=['0', '$a^{*}$', '2'],
ylabel='Rational Acreage Planted', yticks=[0, aeq, 2],yticklabels=['0', '$a^{*}$', '2'])
ax.plot(a, g, 'b', linewidth=4)
ax.plot(a, a, ':', color='grey', linewidth=2)
ax.plot([0, aeq, aeq], [aeq, aeq, 0], 'r--', linewidth=3)
ax.plot([aeq], [aeq], 'ro', markersize=12)
ax.text(0.05, 0, '45${}^o$', color='grey')
ax.text(1.85, aeq - 0.15,'$g(a)$', color='blue')
fig1.show()
nplot = 50
ptarg = np.linspace(0, 2, nplot)
a = 1
Ep, Ef, Er, Eg, Sp, Sf, Sr, Sg = (np.empty(nplot) for k in range(8))
for ip in range(nplot):
for it in range(50):
aold = a
a = A(a, ptarg[ip])
if np.linalg.norm((a - aold) < 1.e-10):
break
q = a * y # quantity produced
p = 1.5 - 0.5 * a * y # market price
f = np.maximum(p, ptarg[ip]) # farm price
r = f * q # farm revenue
g = (f - p) * q # government expenditures
xavg, xstd = discmoments(w, np.vstack((p, f, r, g)))
Ep[ip], Ef[ip], Er[ip], Eg[ip] = tuple(xavg)
Sp[ip], Sf[ip], Sr[ip], Sg[ip] = tuple(xstd)
zeroline = lambda y: plt.axhline(y[0], linestyle=':', color='gray')
fig2 = plt.figure(figsize=[8, 6])
ax1 = fig2.add_subplot(121, title='Expected price',
xlabel='Target price', xticks=[0, 1, 2],
ylabel='Expectation', yticks=[0.5, 1, 1.5, 2], ylim=[0.5, 2.0])
zeroline(Ep)
ax1.plot(ptarg, Ep, linewidth=4, label='Market Price')
ax1.plot(ptarg, Ef, linewidth=4, label='Farm Price')
ax1.legend(loc='upper left')
# Graph expected prices vs target price
ax2 = fig2.add_subplot(122, title='Price variabilities',
xlabel='Target price', xticks=[0, 1, 2],
ylabel='Standard deviation', yticks=[0, 0.1, 0.2]) #plt.ylim(0.5, 2.0)
zeroline(Sf)
ax2.plot(ptarg, Sp, linewidth=4, label='Market Price')
ax2.plot(ptarg, Sf, linewidth=4, label='Farm Price')
ax2.legend(loc='upper left')
fig2.show()
# Graph expected farm revenue vs target price
fig3 = plt.figure(figsize=[12, 6])
ax1 = fig3.add_subplot(131, title='Expected revenue',
xlabel='Target price', xticks=[0, 1, 2],
ylabel='Expectation', yticks=[1, 2, 3], ylim=[0.8, 3.0])
zeroline(Er)
ax1.plot(ptarg, Er, linewidth=4)
# Graph standard deviation of farm revenue vs target price
ax2 = fig3.add_subplot(132, title='Farm Revenue Variability',
xlabel='Target price', xticks=[0, 1, 2],
ylabel='Standard deviation', yticks=[0, 0.2, 0.4])
zeroline(Sr)
ax2.plot(ptarg, Sr, linewidth=4)
# Graph expected government expenditures vs target price
ax3 = fig3.add_subplot(133, title='Expected Government Expenditures',
xlabel='Target price', xticks=[0, 1, 2],
ylabel='Expectation', yticks=[0, 1, 2], ylim=[-0.05, 2.0])
zeroline(Eg)
ax3.plot(ptarg, Eg, linewidth=4)
plt.show()
#fig1.savefig('demintro02--01.png')
#fig2.savefig('demintro02--02.png')
#fig3.savefig('demintro02--03.png')
| 0.632049 | 0.93337 |
## Exploratory Data Analysis
Use this notebook to get familiar with the datasets we have. There is 10 questions we need to answer during the EDA.
We shouldn't limit our EDA to these 10 questions. Let's be creative :).
#### **Task 1**: Test the hypothesis that the delay is from Normal distribution. and that **mean** of the delay is 0. Be careful about the outliers.
#### **Task 2**: Is average/median monthly delay different during the year? If yes, which are months with the biggest delays and what could be the reason?
#### **Task 3**: Does the weather affect the delay?
Use the API to pull the weather information for flights. There is no need to get weather for ALL flights. We can choose the right representative sample. Let's focus on four weather types:
- sunny
- cloudy
- rainy
- snow.
Test the hypothesis that these 4 delays are from the same distribution. If they are not, which ones are significantly different?
#### **Task 4**: How taxi times changing during the day? Does higher traffic lead to bigger taxi times?
#### **Task 5**: What is the average percentage of delays that is already created before departure? (aka are arrival delays caused by departure delays?) Are airlines able to lower the delay during the flights?
```
import pandas as pd
import matplotlib.pyplot as plt
df = pd.read_csv('data/flights_random_50k_cleaned.csv').drop(columns='Unnamed: 0')
a, b = len(df), len(df[df.arr_delay > df.dep_delay])
print(f'Arrival delay is greater than departure delay in {b} of {a} flights ({b/a*100}%).')
print(f'In other words, in {100 - b/a*100}% of flights, the delay happens before the plane has left the gate. Origin airport factors are more significant than destination airport factors.')
```
75% of delays happen before the plane has left the gate.
#### **Task 6**: How many states cover 50% of US air traffic?
#### **Task 7**: Test the hypothesis whether planes fly faster when there is the departure delay?
#### **Task 8**: When (which hour) do most 'LONG', 'SHORT', 'MEDIUM' haul flights take off?
#### **Task 9**: Find the top 10 the bussiest airports. Does the biggest number of flights mean that the biggest number of passengers went through the particular airport? How much traffic do these 10 airports cover?
#### **Task 10**: Do bigger delays lead to bigger fuel comsumption per passenger?
We need to do four things to answer this as accurate as possible:
- Find out average monthly delay per air carrier (monthly delay is sum of all delays in 1 month)
- Find out distance covered monthly by different air carriers
- Find out number of passengers that were carried by different air carriers
- Find out total fuel comsumption per air carrier.
Use this information to get the average fuel comsumption per passenger per km. Is this higher for the airlines with bigger average delays?
|
github_jupyter
|
import pandas as pd
import matplotlib.pyplot as plt
df = pd.read_csv('data/flights_random_50k_cleaned.csv').drop(columns='Unnamed: 0')
a, b = len(df), len(df[df.arr_delay > df.dep_delay])
print(f'Arrival delay is greater than departure delay in {b} of {a} flights ({b/a*100}%).')
print(f'In other words, in {100 - b/a*100}% of flights, the delay happens before the plane has left the gate. Origin airport factors are more significant than destination airport factors.')
| 0.345878 | 0.991389 |
Load up a test dataframe:
```
import pandas as pd
df = pd.read_csv('https://gist.githubusercontent.com/netj/8836201/raw/6f9306ad21398ea43cba4f7d537619d0e07d5ae3/iris.csv')
df = df.iloc[:10]
```
Set some sizes. Ideally these can be set automatically (along with the fontsize) and later use the textwrap library to handle long strings
```
figwidth = 400
figheight = 300
ncols = len(df.columns)
colwidth = figwidth/ncols
rowheight = figheight/ (len(df)+1) #plus one for the header row.
```
These handle the `<text>`, `<line>` and `<rect>` elements. Together these can make any dataframe!
```
def gettxt(st, x, y, length, height, va='central', cl='normal'):
"""Adds a text element. x and y refer to the bottom left of the cell.
The actual x,y position of the text is inferred from the cell
type (i.e. is it a column header or not, and is it a string or number) """
_y = y - height/2 #y alignment is always the same
if cl == 'heavy': #it's a header cell
_x = x + length/2
ha = 'middle'
else: #it's a value cell.
if isinstance(st, str): #strings go in the middle of the cell
_x = x + length/2
ha = 'middle'
else: #its a float.
_x = x+length - length/10
ha = 'end'
ln = f" <text x=\"{_x}\" y=\"{_y}\" text-anchor=\"{ha}\" class=\"{cl}\">{st}</text> \n"
return ln
def hline(x1,y1, x2, y2):
vln = f" <line x1=\"{x1}\" y1=\"{y1}\" x2=\"{x2}\" y2=\"{y2}\" style=\"stroke:rgb(0,0,0);stroke-width:0.5\" />"
return vln
def hbox(nrow, rowheight, figwidth, ):
hbox = f""" <rect x="0" y="{(nrow+1)*rowheight}" width="{figwidth}" height="{rowheight}" style="fill:#eee;fill-opacity:0.8;stroke-width:0;stroke:rgb(0,0,0)" />\n"""
return hbox
```
Open up an svg file and add the SVG header:
```
f = open('svg_directly.svg', 'w')
f.write(f"""<svg version="1.1"
baseProfile="full"
width="{figwidth}" height="{figheight}"
xmlns="http://www.w3.org/2000/svg">
<style>
.normal {{ font: normal 12px sans-serif; fill: black; dominant-baseline: central; }}
.heavy {{ font: bold 12px sans-serif; fill: black; dominant-baseline: central; }}
</style>
""")
```
Write the dataframe column headers with `<text>` in bold and the horizontal line under the header:
```
for count, col in enumerate(df.columns):
f.write( gettxt(col, count*colwidth, rowheight,colwidth, rowheight, cl='heavy') )
f.write( hline(0, rowheight, figwidth, rowheight) )
f.write('\n')
```
Write the table values and horizontal shaded boxes:
```
shaded = True
for rownum in range(df.shape[0]):
row = df.iloc[rownum]
if shaded:
f.write(hbox(rownum, rowheight, figwidth))
shaded = not shaded
for count, value in enumerate(row):
f.write( gettxt(value, (count)*colwidth, (rownum+2)*rowheight, colwidth, rowheight, cl='normal') )
```
Done. close file
```
f.write(""" </svg>""")
f.close()
```
# Now compare the pair:
```
from IPython.display import SVG
s = open('svg_directly.svg').read()
SVG(s)
df
```
|
github_jupyter
|
import pandas as pd
df = pd.read_csv('https://gist.githubusercontent.com/netj/8836201/raw/6f9306ad21398ea43cba4f7d537619d0e07d5ae3/iris.csv')
df = df.iloc[:10]
figwidth = 400
figheight = 300
ncols = len(df.columns)
colwidth = figwidth/ncols
rowheight = figheight/ (len(df)+1) #plus one for the header row.
def gettxt(st, x, y, length, height, va='central', cl='normal'):
"""Adds a text element. x and y refer to the bottom left of the cell.
The actual x,y position of the text is inferred from the cell
type (i.e. is it a column header or not, and is it a string or number) """
_y = y - height/2 #y alignment is always the same
if cl == 'heavy': #it's a header cell
_x = x + length/2
ha = 'middle'
else: #it's a value cell.
if isinstance(st, str): #strings go in the middle of the cell
_x = x + length/2
ha = 'middle'
else: #its a float.
_x = x+length - length/10
ha = 'end'
ln = f" <text x=\"{_x}\" y=\"{_y}\" text-anchor=\"{ha}\" class=\"{cl}\">{st}</text> \n"
return ln
def hline(x1,y1, x2, y2):
vln = f" <line x1=\"{x1}\" y1=\"{y1}\" x2=\"{x2}\" y2=\"{y2}\" style=\"stroke:rgb(0,0,0);stroke-width:0.5\" />"
return vln
def hbox(nrow, rowheight, figwidth, ):
hbox = f""" <rect x="0" y="{(nrow+1)*rowheight}" width="{figwidth}" height="{rowheight}" style="fill:#eee;fill-opacity:0.8;stroke-width:0;stroke:rgb(0,0,0)" />\n"""
return hbox
f = open('svg_directly.svg', 'w')
f.write(f"""<svg version="1.1"
baseProfile="full"
width="{figwidth}" height="{figheight}"
xmlns="http://www.w3.org/2000/svg">
<style>
.normal {{ font: normal 12px sans-serif; fill: black; dominant-baseline: central; }}
.heavy {{ font: bold 12px sans-serif; fill: black; dominant-baseline: central; }}
</style>
""")
for count, col in enumerate(df.columns):
f.write( gettxt(col, count*colwidth, rowheight,colwidth, rowheight, cl='heavy') )
f.write( hline(0, rowheight, figwidth, rowheight) )
f.write('\n')
shaded = True
for rownum in range(df.shape[0]):
row = df.iloc[rownum]
if shaded:
f.write(hbox(rownum, rowheight, figwidth))
shaded = not shaded
for count, value in enumerate(row):
f.write( gettxt(value, (count)*colwidth, (rownum+2)*rowheight, colwidth, rowheight, cl='normal') )
f.write(""" </svg>""")
f.close()
from IPython.display import SVG
s = open('svg_directly.svg').read()
SVG(s)
df
| 0.488527 | 0.814311 |
# ⚖️ 6. Visualize communities on cooperation-competition plot
You are now familiar with how the SMETANA global algorithm works, next we will inspect the generated output. This notebook loads the global simulation results for the different communities directly from the `/data/cooccurrence/simulation/mip_mro` subfolders. You may follow along passively on GitHub or launch the jupyter notebook from your terminal to interactively run through each code chunk. To do so, launch jupyter from your terminal:
```bash
$ cd $ROOT
$ jupyter notebook --browser firefox
```
This should launch a browser window where you can navigate to the `SymbNET/scripts/` folder and click on the appropriate file to launch this notebook interactively.
Note: this script is forked from Daniel's [cooccurrence repo](https://github.com/cdanielmachado/cooccurrence/blob/master/notebooks/Figure%201.ipynb).
### Load libraries
```
%matplotlib inline
import pandas as pd
from glob import glob
import seaborn as sns
import matplotlib.pyplot as plt
import numpy as np
import matplotlib.lines as mlines
from random import sample
sns.set_palette('deep')
```
## 🍯 Simulation results
```
types = [ "gut_normal" ,"gut_impaired","gut_t2d","gut_refseq","kefir","soil"]
dfs = []
for commtype in types:
dfi = pd.concat(pd.read_csv(filename, sep='\t', dtype={'mip': float, 'mro': float})
for filename in glob(f"../data/cooccurrence/simulation/mip_mro/{commtype}/*_global.tsv"))
dfi['type'] = commtype
dfs.append(dfi)
df = pd.concat(dfs)
df.reset_index(inplace=True)
df.dropna(inplace=True)
df["community size"] = df["size"]
df
```
## 🤓 Plot communities on cooperation-competition plot
```
sizes = [5]
subdf = df[df["size"].isin(sizes)].copy()
subdf["Competition"] = subdf["mro"]
subdf["Cooperation"] = subdf["mip"]
palette = {"gut_normal": '#1ba055', "gut_impaired": '#ff7f00', "gut_t2d": '#e41a1c', "gut_refseq": '#cccccc',"kefir":'#984ea3',"soil":'#a65628'}
g = sns.FacetGrid(subdf, col="community size", hue="type", height=3,hue_order=types, palette=palette)
g.map(plt.scatter, "Competition", "Cooperation", alpha=0.5, s=25, linewidths=0.7, edgecolors='w');
g.add_legend()
g.savefig("../plots/global_polarization/mip_mro.png", dpi=300)
```
## 🍷 Discussion questions
- How did your predicted competitiveness/cooperativeness rankings compare with the results from the plot above? Are you surprised by the results?
- Why is there variation within communities, given that simulations were carried out using the same models?
## 🍾 You have completed the core of this tutorial, move on to [the general discussion](https://github.com/franciscozorrilla/SymbNET/blob/main/scripts/9.wrap_up_discussion.md) or continue below for optional bonus exercises 👇
In this tutorial we only considered communities of size 5, lets see how communities polarize at larger sizes. We can also plot our simulation results against the plot generated in the [original plublication](https://www.nature.com/articles/s41559-020-01353-4/figures/1).
```
types = ["random", "bin_rnd_01", "bin_rnd_001", "gut_normal" ,"gut_impaired","gut_t2d","gut_refseq","kefir","soil"]
dfs = []
for commtype in types:
dfi = pd.concat(pd.read_csv(filename, sep='\t', dtype={'mip': float, 'mro': float})
for filename in glob(f"../data/cooccurrence/simulation/mip_mro/{commtype}/*_global.tsv"))
dfi['type'] = commtype
dfs.append(dfi)
df = pd.concat(dfs)
df.reset_index(inplace=True)
df.dropna(inplace=True)
df["community size"] = df["size"]
df
sizes = [2,5,10,20,30,40]
subdf = df[df["size"].isin(sizes)].copy()
subdf["Competition"] = subdf["mro"]
subdf["Cooperation"] = subdf["mip"]
palette = {"random": '#cccccc', "bin_rnd_01": '#ed7e17', "bin_rnd_001": '#1ba055', "gut_normal": '#000',"gut_impaired":'#377eb8',"gut_t2d":'#e41a1c',"gut_refseq":'#f781bf',"kefir": '#984ea3',"soil": '#a65628'}
g = sns.FacetGrid(subdf, col="community size", hue="type", height=5,hue_order=types, palette=palette, col_wrap=3, sharex=False, sharey=False)
g.map(plt.scatter, "Competition", "Cooperation", alpha=0.5, s=25, linewidths=0.7, edgecolors='w');
g.fig.subplots_adjust(top=0.9)
g.add_legend()
g.savefig("../plots/global_polarization/mip_mro_background.png", dpi=300)
```
Additonally, we can recreate the remaining two panels in this figure from the publication showing how cooperation and competition scores vary with community size.
```
f, axs = plt.subplots(2,1, figsize=(4,6))
sns.lineplot(data=df, x="community size", y="mip", hue="type", ci="sd", palette=palette, legend=False, ax=axs[0])
sns.lineplot(data=df, x="community size", y="mro", hue="type", ci="sd", palette=palette, legend=False, ax=axs[1])
axs[0].set_xlim(2,40)
axs[0].set_ylim(0,45)
axs[0].set_ylabel("Cooperation")
axs[0].set_yticks([0, 10, 20, 30, 40])
axs[1].set_xlim(2,40)
axs[1].set_ylabel("Competition")
axs[1].set_yticks([0.3, 0.4, 0.5, 0.6])
plt.tight_layout()
plt.savefig("../plots/global_polarization/cooccurrence_plots.png", dpi=300)
```
## 🍷 Discussion questions (optional)
- Why might the simulations generated by some of our communities fall outside of the original publication's cooperation-coompetition plot (community size N=5) ?
## Move on to [optional exercise 7](https://github.com/franciscozorrilla/SymbNET/blob/main/scripts/7.generate_ensemble_models.md)
|
github_jupyter
|
$ cd $ROOT
$ jupyter notebook --browser firefox
%matplotlib inline
import pandas as pd
from glob import glob
import seaborn as sns
import matplotlib.pyplot as plt
import numpy as np
import matplotlib.lines as mlines
from random import sample
sns.set_palette('deep')
types = [ "gut_normal" ,"gut_impaired","gut_t2d","gut_refseq","kefir","soil"]
dfs = []
for commtype in types:
dfi = pd.concat(pd.read_csv(filename, sep='\t', dtype={'mip': float, 'mro': float})
for filename in glob(f"../data/cooccurrence/simulation/mip_mro/{commtype}/*_global.tsv"))
dfi['type'] = commtype
dfs.append(dfi)
df = pd.concat(dfs)
df.reset_index(inplace=True)
df.dropna(inplace=True)
df["community size"] = df["size"]
df
sizes = [5]
subdf = df[df["size"].isin(sizes)].copy()
subdf["Competition"] = subdf["mro"]
subdf["Cooperation"] = subdf["mip"]
palette = {"gut_normal": '#1ba055', "gut_impaired": '#ff7f00', "gut_t2d": '#e41a1c', "gut_refseq": '#cccccc',"kefir":'#984ea3',"soil":'#a65628'}
g = sns.FacetGrid(subdf, col="community size", hue="type", height=3,hue_order=types, palette=palette)
g.map(plt.scatter, "Competition", "Cooperation", alpha=0.5, s=25, linewidths=0.7, edgecolors='w');
g.add_legend()
g.savefig("../plots/global_polarization/mip_mro.png", dpi=300)
types = ["random", "bin_rnd_01", "bin_rnd_001", "gut_normal" ,"gut_impaired","gut_t2d","gut_refseq","kefir","soil"]
dfs = []
for commtype in types:
dfi = pd.concat(pd.read_csv(filename, sep='\t', dtype={'mip': float, 'mro': float})
for filename in glob(f"../data/cooccurrence/simulation/mip_mro/{commtype}/*_global.tsv"))
dfi['type'] = commtype
dfs.append(dfi)
df = pd.concat(dfs)
df.reset_index(inplace=True)
df.dropna(inplace=True)
df["community size"] = df["size"]
df
sizes = [2,5,10,20,30,40]
subdf = df[df["size"].isin(sizes)].copy()
subdf["Competition"] = subdf["mro"]
subdf["Cooperation"] = subdf["mip"]
palette = {"random": '#cccccc', "bin_rnd_01": '#ed7e17', "bin_rnd_001": '#1ba055', "gut_normal": '#000',"gut_impaired":'#377eb8',"gut_t2d":'#e41a1c',"gut_refseq":'#f781bf',"kefir": '#984ea3',"soil": '#a65628'}
g = sns.FacetGrid(subdf, col="community size", hue="type", height=5,hue_order=types, palette=palette, col_wrap=3, sharex=False, sharey=False)
g.map(plt.scatter, "Competition", "Cooperation", alpha=0.5, s=25, linewidths=0.7, edgecolors='w');
g.fig.subplots_adjust(top=0.9)
g.add_legend()
g.savefig("../plots/global_polarization/mip_mro_background.png", dpi=300)
f, axs = plt.subplots(2,1, figsize=(4,6))
sns.lineplot(data=df, x="community size", y="mip", hue="type", ci="sd", palette=palette, legend=False, ax=axs[0])
sns.lineplot(data=df, x="community size", y="mro", hue="type", ci="sd", palette=palette, legend=False, ax=axs[1])
axs[0].set_xlim(2,40)
axs[0].set_ylim(0,45)
axs[0].set_ylabel("Cooperation")
axs[0].set_yticks([0, 10, 20, 30, 40])
axs[1].set_xlim(2,40)
axs[1].set_ylabel("Competition")
axs[1].set_yticks([0.3, 0.4, 0.5, 0.6])
plt.tight_layout()
plt.savefig("../plots/global_polarization/cooccurrence_plots.png", dpi=300)
| 0.350866 | 0.859133 |
```
# For changes in .py
%reload_ext autoreload
%autoreload 2
import numpy as np
import pandas as pd
import os
pd.options.display.max_columns = 999
pd.options.display.max_rows = 1999
pd.options.display.max_colwidth = 200
import warnings
warnings.filterwarnings('ignore')
import sys
sys.path.insert(0, "../")
from scripts import manipulation, alesp_parser, alesp_tamitacao_parser
import basedosdados as bd
```
## Deputados
Possíveis valores para o campo Situacao
- 'REL' = Renunciou para assumir outro cargo eletivo
- 'OUT' = Outros'FAL' = Falecido
- 'REN' = Renunciou'LIC' = Licenciado
- 'EXE' = No exercíciodo mandato
- 'CAS' = Cassado
- ' ' = Não categorizad
```
%%time
deputados = alesp_parser.parse_deputados(True)
tb = bd.Table('deputados','br_sp_alesp')
tb.create(
path='../data/servidores/deputados_alesp.csv',
if_table_exists='replace',
if_storage_data_exists='replace',
if_table_config_exists='pass'
)
tb.publish('replace')
```
## Servidores
```
%%time
alesp_parser.parse_servidores()
tb = bd.Table('assessores_parlamentares','br_sp_alesp')
tb.create(
path='../data/servidores/assessores_parlamentares.csv',
if_table_exists='replace',
if_storage_data_exists='replace',
if_table_config_exists='pass'
)
tb.publish('replace')
tb = bd.Table('assessores_lideranca','br_sp_alesp')
tb.create(
path='../data/servidores/assessores_lideranca.csv',
if_table_exists='replace',
if_storage_data_exists='repalce',
if_table_config_exists='pass'
)
tb.publish('replace')
```
## Despesas ALL
https://www.al.sp.gov.br/dados-abertos/recurso/21
```
%%time
alesp_parser.parse_despesas(False)
tb = bd.Table('despesas_gabinete_atual','br_sp_alesp')
tb.create(
path='../data/gastos/despesas_gabinetes_mandato.csv',
if_table_exists='replace',
if_storage_data_exists='pass',
if_table_config_exists='pass'
)
tb.publish('replace')
tb = bd.Table('despesas_gabinete','br_sp_alesp')
tb.create(
path='../data/gastos/despesas_gabinete/',
partitioned=True,
if_table_exists='replace',
if_storage_data_exists='pass',
if_table_config_exists='pass'
)
tb.publish('replace')
```
# Tramitacao
https://www.al.sp.gov.br/dados-abertos/grupo/1
## AUTORES
Lista de deputados autores e apoiadores das proposituras.
https://www.al.sp.gov.br/dados-abertos/recurso/81
```
%%time
alesp_tamitacao_parser.parse_autores(False)
tb = bd.Table('tramitacao_documento_autor','br_sp_alesp')
tb.create(
path='../data/tramitacoes/documento_autor.csv',
if_table_exists='replace',
if_storage_data_exists='replace',
if_table_config_exists='replace'
)
tb.publish('replace')
```
## Comissoes
https://www.al.sp.gov.br/dados-abertos/recurso/43
```
%%time
alesp_tamitacao_parser.parse_comissoes(False)
tb = bd.Table('tramitacao_comissoes','br_sp_alesp')
tb.create(
path='../data/tramitacoes/comissoes.csv',
if_table_exists='replace',
if_storage_data_exists='replace',
if_table_config_exists='replace'
)
tb.publish('replace')
```
## Deliberações nas Comissões
Lista das deliberações sobre as matérias que tramitam nas Comissões Permanentes da Alesp.
https://www.al.sp.gov.br/dados-abertos/recurso/52
```
%%time
alesp_tamitacao_parser.parse_deliberacoes_comissoes(False)
tb = bd.Table('tramitacao_comissoes_permanentes_deliberacoes','br_sp_alesp')
tb.create(
path='../data/tramitacoes/comissoes_permanentes_deliberacoes.csv',
if_table_exists='replace',
if_storage_data_exists='replace',
if_table_config_exists='replace'
)
tb.publish('replace')
```
## Membros de Comissões Permanentes
Lista de membros das Comissões da Alesp.
https://www.al.sp.gov.br/dados-abertos/recurso/45
```
%%time
alesp_tamitacao_parser.parse_comissoes_membros(False)
tb = bd.Table('tramitacao_comissoes_membros','br_sp_alesp')
tb.create(
path='../data/tramitacoes/comissoes_membros.csv',
if_table_exists='replace',
if_storage_data_exists='replace',
if_table_config_exists='replace'
)
tb.publish('replace')
```
## Natureza do Documento
Lista das naturezas (tipos) dos documentos que fazem parte do Processo Legislativo
https://www.al.sp.gov.br/dados-abertos/recurso/44
```
%%time
alesp_tamitacao_parser.parse_naturezasSpl(download=False)
tb = bd.Table('tramitacao_natureza','br_sp_alesp')
tb.create(
path='../data/tramitacoes/naturezasSpl.csv',
if_table_exists='replace',
if_storage_data_exists='replace',
if_table_config_exists='replace'
)
tb.publish('replace')
```
## Documentos Palavras Chave
Lista de palavras-chave associadas aos documentos e proposituras que tramitam no processo legislativo. Essa indexação é realizada pela Divisão de Biblioteca e Documentação.
https://www.al.sp.gov.br/dados-abertos/recurso/42
```
%%time
alesp_tamitacao_parser.parse_documento_palavras(False)
tb = bd.Table('tramitacao_documento_palavras_chave','br_sp_alesp')
tb.create(
path='../data/tramitacoes/documento_palavras.csv',
if_table_exists='replace',
if_storage_data_exists='replace',
if_table_config_exists='replace'
)
tb.publish('replace')
```
## Index Palavras Chave
Lista de palavras-chave que podem ser associadas aos documentos que tramitam no Processo Legislativo no sistema SPL. A lista é definida e a indexação realizada pela equipe do DDI (Departamento de Documentação e Informação) e da DBD (Divisão de Biblioteca e Documentação) Alesp.
https://www.al.sp.gov.br/dados-abertos/recurso/41
```
%%time
alesp_tamitacao_parser.parse_documento_index_palavras(False)
tb = bd.Table('tramitacao_index_palavras_chave','br_sp_alesp')
tb.create(
path='../data/tramitacoes/index_palavras_chave.csv',
if_table_exists='replace',
if_storage_data_exists='replace',
if_table_config_exists='replace'
)
tb.publish('replace')
```
## Pareceres
Recurso com a lista dos pareceres elaborados nas Comissões das matérias que tramitam ou tramitaram na Alesp.
https://www.al.sp.gov.br/dados-abertos/recurso/103
http://www.al.sp.gov.br/repositorioDados/processo_legislativo/propositura_parecer.zip
```
%%time
alesp_tamitacao_parser.parse_propositura_parecer(False)
tb = bd.Table('tramitacao_propositura_parecer','br_sp_alesp')
tb.create(
path='../data/tramitacoes/propositura_parecer.csv',
if_table_exists='replace',
if_storage_data_exists='replace',
if_table_config_exists='replace'
)
tb.publish('replace')
```
## Presença nas Comissões
Presença dos Deputados Estaduais nas reuniões das Comissões Permanentes da Alesp.
https://www.al.sp.gov.br/dados-abertos/recurso/53
```
%%time
alesp_tamitacao_parser.parse_comissoes_permanentes_presencas(False)
tb = bd.Table('tramitacao_comissoes_permanentes_presencas','br_sp_alesp')
tb.create(
path='../data/tramitacoes/comissoes_permanentes_presencas.csv',
if_table_exists='replace',
if_storage_data_exists='replace',
if_table_config_exists='replace'
)
tb.publish('replace')
```
## Proposituras
Lista de proposituras apresentadas pelo Deputados Estaduais e que tramitam ou tramitaram no Processo Legislativo.
https://www.al.sp.gov.br/dados-abertos/recurso/56
```
%%time
df = alesp_tamitacao_parser.parse_proposituras(False)
tb = bd.Table('tramitacao_proposituras','br_sp_alesp')
tb.create(
path='../data/tramitacoes/proposituras.csv',
if_table_exists='replace',
if_storage_data_exists='replace',
if_table_config_exists='replace'
)
tb.publish('replace')
```
## Regimes de Tramitação das Proposituras
Dados dos regimes de tramitação das proposituras.
https://www.al.sp.gov.br/dados-abertos/recurso/56
```
%%time
alesp_tamitacao_parser.parse_documento_regime(False)
tb = bd.Table('tramitacao_documento_regime','br_sp_alesp')
tb.create(
path='../data/tramitacoes/documento_regime.csv',
if_table_exists='replace',
if_storage_data_exists='replace',
if_table_config_exists='replace'
)
tb.publish('replace')
```
## Reuniões de Comissão
Lista das reuniões realizadas nas Comissões Permanentes da Alesp.
https://www.al.sp.gov.br/dados-abertos/recurso/56
```
%%time
alesp_tamitacao_parser.parse_comissoes_permanentes_reunioes(False)
tb = bd.Table('tramitacao_comissoes_permanentes_reunioes','br_sp_alesp')
tb.create(
path='../data/tramitacoes/comissoes_permanentes_reunioes.csv',
if_table_exists='replace',
if_storage_data_exists='replace',
if_table_config_exists='replace'
)
tb.publish('replace')
```
## Votações nas Comissões
Lista das votações nas deliberações das matérias que tramitam nas Comissões Permanentes da Alesp.
https://www.al.sp.gov.br/dados-abertos/recurso/55
```
%%time
alesp_tamitacao_parser.parse_comissoes_permanentes_votacoes(False)
tb = bd.Table('tramitacao_comissoes_permanentes_votacoes','br_sp_alesp')
tb.create(
path='../data/tramitacoes/comissoes_permanentes_votacoes.csv',
if_table_exists='replace',
if_storage_data_exists='replace',
if_table_config_exists='replace'
)
tb.publish('replace')
```
## --->>> Tramitações atuais
https://www.al.sp.gov.br/dados-abertos/recurso/221
```
%%time
alesp_tamitacao_parser.parse_documento_andamento_atual(False)
tb = bd.Table('tramitacao_documento_andamento_atual','br_sp_alesp')
tb.create(
path='../data/tramitacoes/documento_andamento_atual.csv',
if_table_exists='replace',
if_storage_data_exists='replace',
if_table_config_exists='replace'
)
tb.publish('replace')
```
## Tramitações
Recurso com os andamentos das matérias que tramitam ou tramitaram na Alesp.
https://www.al.sp.gov.br/dados-abertos/recurso/101
```
# %%time
# alesp_tamitacao_parser.parse_documento_andamento(False)
# tb = bd.Table('tramitacao_documento_andamento','br_sp_alesp')
# tb.create(
# path='../data/tramitacoes/documento_andamento.csv',
# if_table_exists='replace',
# if_storage_data_exists='replace',
# if_table_config_exists='raise'
# )
# tb.publish('replace')
```
|
github_jupyter
|
# For changes in .py
%reload_ext autoreload
%autoreload 2
import numpy as np
import pandas as pd
import os
pd.options.display.max_columns = 999
pd.options.display.max_rows = 1999
pd.options.display.max_colwidth = 200
import warnings
warnings.filterwarnings('ignore')
import sys
sys.path.insert(0, "../")
from scripts import manipulation, alesp_parser, alesp_tamitacao_parser
import basedosdados as bd
%%time
deputados = alesp_parser.parse_deputados(True)
tb = bd.Table('deputados','br_sp_alesp')
tb.create(
path='../data/servidores/deputados_alesp.csv',
if_table_exists='replace',
if_storage_data_exists='replace',
if_table_config_exists='pass'
)
tb.publish('replace')
%%time
alesp_parser.parse_servidores()
tb = bd.Table('assessores_parlamentares','br_sp_alesp')
tb.create(
path='../data/servidores/assessores_parlamentares.csv',
if_table_exists='replace',
if_storage_data_exists='replace',
if_table_config_exists='pass'
)
tb.publish('replace')
tb = bd.Table('assessores_lideranca','br_sp_alesp')
tb.create(
path='../data/servidores/assessores_lideranca.csv',
if_table_exists='replace',
if_storage_data_exists='repalce',
if_table_config_exists='pass'
)
tb.publish('replace')
%%time
alesp_parser.parse_despesas(False)
tb = bd.Table('despesas_gabinete_atual','br_sp_alesp')
tb.create(
path='../data/gastos/despesas_gabinetes_mandato.csv',
if_table_exists='replace',
if_storage_data_exists='pass',
if_table_config_exists='pass'
)
tb.publish('replace')
tb = bd.Table('despesas_gabinete','br_sp_alesp')
tb.create(
path='../data/gastos/despesas_gabinete/',
partitioned=True,
if_table_exists='replace',
if_storage_data_exists='pass',
if_table_config_exists='pass'
)
tb.publish('replace')
%%time
alesp_tamitacao_parser.parse_autores(False)
tb = bd.Table('tramitacao_documento_autor','br_sp_alesp')
tb.create(
path='../data/tramitacoes/documento_autor.csv',
if_table_exists='replace',
if_storage_data_exists='replace',
if_table_config_exists='replace'
)
tb.publish('replace')
%%time
alesp_tamitacao_parser.parse_comissoes(False)
tb = bd.Table('tramitacao_comissoes','br_sp_alesp')
tb.create(
path='../data/tramitacoes/comissoes.csv',
if_table_exists='replace',
if_storage_data_exists='replace',
if_table_config_exists='replace'
)
tb.publish('replace')
%%time
alesp_tamitacao_parser.parse_deliberacoes_comissoes(False)
tb = bd.Table('tramitacao_comissoes_permanentes_deliberacoes','br_sp_alesp')
tb.create(
path='../data/tramitacoes/comissoes_permanentes_deliberacoes.csv',
if_table_exists='replace',
if_storage_data_exists='replace',
if_table_config_exists='replace'
)
tb.publish('replace')
%%time
alesp_tamitacao_parser.parse_comissoes_membros(False)
tb = bd.Table('tramitacao_comissoes_membros','br_sp_alesp')
tb.create(
path='../data/tramitacoes/comissoes_membros.csv',
if_table_exists='replace',
if_storage_data_exists='replace',
if_table_config_exists='replace'
)
tb.publish('replace')
%%time
alesp_tamitacao_parser.parse_naturezasSpl(download=False)
tb = bd.Table('tramitacao_natureza','br_sp_alesp')
tb.create(
path='../data/tramitacoes/naturezasSpl.csv',
if_table_exists='replace',
if_storage_data_exists='replace',
if_table_config_exists='replace'
)
tb.publish('replace')
%%time
alesp_tamitacao_parser.parse_documento_palavras(False)
tb = bd.Table('tramitacao_documento_palavras_chave','br_sp_alesp')
tb.create(
path='../data/tramitacoes/documento_palavras.csv',
if_table_exists='replace',
if_storage_data_exists='replace',
if_table_config_exists='replace'
)
tb.publish('replace')
%%time
alesp_tamitacao_parser.parse_documento_index_palavras(False)
tb = bd.Table('tramitacao_index_palavras_chave','br_sp_alesp')
tb.create(
path='../data/tramitacoes/index_palavras_chave.csv',
if_table_exists='replace',
if_storage_data_exists='replace',
if_table_config_exists='replace'
)
tb.publish('replace')
%%time
alesp_tamitacao_parser.parse_propositura_parecer(False)
tb = bd.Table('tramitacao_propositura_parecer','br_sp_alesp')
tb.create(
path='../data/tramitacoes/propositura_parecer.csv',
if_table_exists='replace',
if_storage_data_exists='replace',
if_table_config_exists='replace'
)
tb.publish('replace')
%%time
alesp_tamitacao_parser.parse_comissoes_permanentes_presencas(False)
tb = bd.Table('tramitacao_comissoes_permanentes_presencas','br_sp_alesp')
tb.create(
path='../data/tramitacoes/comissoes_permanentes_presencas.csv',
if_table_exists='replace',
if_storage_data_exists='replace',
if_table_config_exists='replace'
)
tb.publish('replace')
%%time
df = alesp_tamitacao_parser.parse_proposituras(False)
tb = bd.Table('tramitacao_proposituras','br_sp_alesp')
tb.create(
path='../data/tramitacoes/proposituras.csv',
if_table_exists='replace',
if_storage_data_exists='replace',
if_table_config_exists='replace'
)
tb.publish('replace')
%%time
alesp_tamitacao_parser.parse_documento_regime(False)
tb = bd.Table('tramitacao_documento_regime','br_sp_alesp')
tb.create(
path='../data/tramitacoes/documento_regime.csv',
if_table_exists='replace',
if_storage_data_exists='replace',
if_table_config_exists='replace'
)
tb.publish('replace')
%%time
alesp_tamitacao_parser.parse_comissoes_permanentes_reunioes(False)
tb = bd.Table('tramitacao_comissoes_permanentes_reunioes','br_sp_alesp')
tb.create(
path='../data/tramitacoes/comissoes_permanentes_reunioes.csv',
if_table_exists='replace',
if_storage_data_exists='replace',
if_table_config_exists='replace'
)
tb.publish('replace')
%%time
alesp_tamitacao_parser.parse_comissoes_permanentes_votacoes(False)
tb = bd.Table('tramitacao_comissoes_permanentes_votacoes','br_sp_alesp')
tb.create(
path='../data/tramitacoes/comissoes_permanentes_votacoes.csv',
if_table_exists='replace',
if_storage_data_exists='replace',
if_table_config_exists='replace'
)
tb.publish('replace')
%%time
alesp_tamitacao_parser.parse_documento_andamento_atual(False)
tb = bd.Table('tramitacao_documento_andamento_atual','br_sp_alesp')
tb.create(
path='../data/tramitacoes/documento_andamento_atual.csv',
if_table_exists='replace',
if_storage_data_exists='replace',
if_table_config_exists='replace'
)
tb.publish('replace')
# %%time
# alesp_tamitacao_parser.parse_documento_andamento(False)
# tb = bd.Table('tramitacao_documento_andamento','br_sp_alesp')
# tb.create(
# path='../data/tramitacoes/documento_andamento.csv',
# if_table_exists='replace',
# if_storage_data_exists='replace',
# if_table_config_exists='raise'
# )
# tb.publish('replace')
| 0.092301 | 0.403156 |
# Recursion
[Click here to run this chapter on Colab](https://colab.research.google.com/github/AllenDowney/DSIRP/blob/main/notebooks/recursion.ipynb)
## Example 1
Here's an example of recursion from [this section of Think Python](https://greenteapress.com/thinkpython2/html/thinkpython2006.html#sec62).
```
def countdown(n):
if n == 0:
print('Blastoff!')
else:
print(n)
countdown(n-1)
countdown(3)
```
To understand recursion, it's important to have a good mental model of what happens when you run a function:
1. Python interprets the arguments.
2. It creates a stack frame, which will contain the parameters and local variables.
3. Next it assigns the values of the arguments to the parameters.
4. Python runs the body of the function.
5. Then it recycles the stack frame.
The runtime stack contains the stack frames of currently-running functions.
Here's a stack diagram that shows what happens when this `countdown` runs.
<img src="https://greenteapress.com/thinkpython2/html/thinkpython2005.png">
**Exercise:** What happens if you run countdown with a negative number? [See here for more info]()
## Example 2
Here's an example of recursion with a function that returns a value, from [this section of Think Python](https://greenteapress.com/thinkpython2/html/thinkpython2007.html#sec74).
```
def factorial(n):
if n == 0:
print(n, 1)
return 1
else:
recurse = factorial(n-1)
result = n * recurse
print(n, recurse, result)
return result
factorial(3)
```
Here's the stack frame.
<img src="https://greenteapress.com/thinkpython2/html/thinkpython2007.png">
**Exercise:** Suppose you want to raise a number, `x`, to an integer power, `k`. An efficient way to do that is:
* If `k` is even, raise `n` to `k/2` and square it.
* If `k` is odd, raise `n` to `(k-1)/2`, square it, and multiply by `x` one more time.
Write a recursive function that implements this algorithm.
What is the order of growth of this algorithm?
To keep it simple, suppose `k` is a power of two.
How many times do we have to divide `k` by two before we get to 1?
Thinking about it in reverse, starting with 1, how many times do we have to double 1 before we get to `k`? In math notation, the question is
$$2^y = k$$
where `y` is the unknown number of steps. Taking the log of both sides, base 2:
$$y = log_2 k$$
In terms of order of growth, this algorithm is in `O(log k)`. We don't have to specify the base of the logarithm, because a log in one base is a constant multiple of a log in any other base.
## Example 3
Here's another example of recursion from [this section of Think Python](https://greenteapress.com/thinkpython2/html/thinkpython2007.html#sec76).
```
def fibonacci(n):
print(n)
if n == 0:
return 0
elif n == 1:
return 1
else:
return fibonacci(n-1) + fibonacci(n-2)
fibonacci(4)
```
Here's a stack graph that shows all stack frames created during this function call.
Note that these frames are not all on the stack at the same time.
<img src="https://greenteapress.com/thinkpython2/html/thinkpython2017.png">
Here's the [section from Think Python](https://greenteapress.com/thinkpython2/html/thinkpython2012.html#sec135) that shows how we can make fibonacci faster by "memoizing" it. That's not a typo; the word is really [memoize](https://en.wikipedia.org/wiki/Memoization).
```
known = {0:0, 1:1}
def fibonacci_memo(n):
if n in known:
return known[n]
print(n)
res = fibonacci_memo(n-1) + fibonacci_memo(n-2)
known[n] = res
return res
fibonacci_memo(4)
```
**Exercise:** The [Ackermann function](http://en.wikipedia.org/wiki/Ackermann_function), $A(m, n)$, is defined:
$$
A(m, n) = \begin{cases}
n+1 & \mbox{if } m = 0 \\
A(m-1, 1) & \mbox{if } m > 0 \mbox{ and } n = 0 \\
A(m-1, A(m, n-1)) & \mbox{if } m > 0 \mbox{ and } n > 0.
\end{cases}
$$
Write a function named `ackermann` that evaluates the Ackermann function.
Use your function to evaluate `ackermann(3, 4)`, which should be 125.
What happens for larger values of `m` and `n`?
If you memoize it, can you evaluate the function with bigger values?
## String functions
Many things we do iteratively can be expressed recursively as well.
```
def reverse(s):
if len(s) < 2:
return s
first, rest = s[0], s[1:]
return reverse(rest) + first
reverse('reverse')
```
For sequences and mapping types, there's usually no advantage of the recursive version. But for trees and graphs, a recursive implementation can be clearer, more concise, and more demonstrably correct.
**Exercise:** Here's an exercise from, of all places, [StackOverflow](https://stackoverflow.com/questions/28977737/writing-a-recursive-string-function):
> Write a recursive, string-valued function, `replace`, that accepts a string and returns a new string consisting of the original string with each blank replaced with an asterisk (*)
>
> Replacing the blanks in a string involves:
>
> 1. Nothing if the string is empty
>
> 2. Otherwise: If the first character is not a blank, simply concatenate it with the result of replacing the rest of the string
>
> 3. If the first character IS a blank, concatenate an * with the result of replacing the rest of the string
## Exercises
This one is from [Structure and Interpretation of Computer Programs](https://mitpress.mit.edu/sites/default/files/sicp/index.html):
> The greatest common divisor (GCD) of two integers `a` and `b` is defined to be the largest integer that divides both `a` and `b` with no remainder. For example, the GCD of 16 and 28 is 4. [...] One way to find the GCD of two integers is to factor them and search for common factors, but there is a [famous algorithm](https://en.wikipedia.org/wiki/Euclidean_algorithm) that is much more efficient.
>
> The idea of the algorithm is based on the observation that, if `r` is the remainder when `a` is divided by `b`, then the common divisors of `a` and `b` are precisely the same as the common divisors of `b` and `r`.
>
> Thus, we can use the equation
>
> $$GCD(a, b) = GCD(b, r)$$
>
>to successively reduce the problem of computing a GCD to the problem of computing the GCD of smaller and smaller pairs of integers.
>
> It is possible to show that starting with any two positive integers and performing repeated reductions will always eventually produce a pair where the second number is 0. Then the GCD is the other number in the pair.
Write a function called `gcd` that takes two integers and uses this algorithm to compute their greatest common divisor.
This one is from [Structure and Interpretation of Computer Programs](https://mitpress.mit.edu/sites/default/files/sicp/index.html):
> How many different ways can we make change of \$1.00, given half-dollars, quarters, dimes, nickels, and pennies? [...]
>
>[...] Suppose we think of the types of coins available as arranged in some order. [..] observe that the ways to make change can be divided into two groups: those that do not use any of the first kind of coin, and those that do. Therefore, the total number of ways to make change for some amount is equal to the number of ways to make change for the amount without using any of the first kind of coin, plus the number of ways to make change assuming that we do use the first kind of coin.
Write a function that takes as parameters an amount of money in cents and a sequence of coin denominations. It should return the number of combinations of coins that add up to the given amount.
The result for one dollar (`100` cents) with coins of denominations `(50, 25, 10, 5, 1)` should be `292`.
You might have to give some thought to the base cases.
**Exercise:** Here's one of my favorite Car Talk Puzzlers (http://www.cartalk.com/content/puzzlers):
>What is the longest English word, that remains a valid English word, as you remove its letters one at a time?
>
>Now, letters can be removed from either end, or the middle, but you can’t rearrange any of the letters. Every time you drop a letter, you wind up with another English word. If you do that, you’re eventually going to wind up with one letter and that too is going to be an English word—one that’s found in the dictionary. I want to know what’s the longest word and how many letters does it have?
>
>I’m going to give you a little modest example: Sprite. Ok? You start off with sprite, you take a letter off, one from the interior of the word, take the r away, and we’re left with the word spite, then we take the e off the end, we’re left with spit, we take the s off, we’re left with pit, it, and I.
Write a program to find all words that can be reduced in this way, and then find the longest one.
This exercise is a little more challenging than most, so here are some suggestions:
* You might want to write a function that takes a word and computes a list of all the words that can be formed by removing one letter. These are the “children” of the word.
* Recursively, a word is reducible if any of its children are reducible. As base cases, you can consider the single letter words “I”, “a” to be reducible.
* To improve the performance of your program, you might want to memoize the words that are known to be reducible.
```
from os.path import basename, exists
def download(url):
filename = basename(url)
if not exists(filename):
from urllib.request import urlretrieve
local, _ = urlretrieve(url, filename)
print('Downloaded ' + local)
download('https://github.com/AllenDowney/DSIRP/raw/main/american-english')
def read_words(filename):
"""Read lines from a file and split them into words."""
res = set()
for line in open(filename):
for word in line.split():
res.add(word.strip().lower())
return res
word_set = read_words('american-english')
len(word_set)
```
*Data Structures and Information Retrieval in Python*
Copyright 2021 Allen Downey
License: [Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International](https://creativecommons.org/licenses/by-nc-sa/4.0/)
|
github_jupyter
|
def countdown(n):
if n == 0:
print('Blastoff!')
else:
print(n)
countdown(n-1)
countdown(3)
def factorial(n):
if n == 0:
print(n, 1)
return 1
else:
recurse = factorial(n-1)
result = n * recurse
print(n, recurse, result)
return result
factorial(3)
def fibonacci(n):
print(n)
if n == 0:
return 0
elif n == 1:
return 1
else:
return fibonacci(n-1) + fibonacci(n-2)
fibonacci(4)
known = {0:0, 1:1}
def fibonacci_memo(n):
if n in known:
return known[n]
print(n)
res = fibonacci_memo(n-1) + fibonacci_memo(n-2)
known[n] = res
return res
fibonacci_memo(4)
def reverse(s):
if len(s) < 2:
return s
first, rest = s[0], s[1:]
return reverse(rest) + first
reverse('reverse')
from os.path import basename, exists
def download(url):
filename = basename(url)
if not exists(filename):
from urllib.request import urlretrieve
local, _ = urlretrieve(url, filename)
print('Downloaded ' + local)
download('https://github.com/AllenDowney/DSIRP/raw/main/american-english')
def read_words(filename):
"""Read lines from a file and split them into words."""
res = set()
for line in open(filename):
for word in line.split():
res.add(word.strip().lower())
return res
word_set = read_words('american-english')
len(word_set)
| 0.227727 | 0.99402 |
# "You think that's funny?" Topic modelling and text generation using Amazon and Netflix stand-up comedy scripts
# 1) Construction of the dataset
In this section, I have built the dataset used in this project. To do that, I combined the list of the stand-up comedy specials and their release date extracted from Wikipedia (`List_stand_up_comedy_full.csv` file) together with the text of the shows extracted by a series of .srt files (e.g., subtitles scripts) retrieved from the croudsourced community Subscene.com.
The final dataframe is then saved in the file `Stand_up_comedy_dataset.csv`.
The final dataset for the analysis contains the following features:
| Column name | Datatype | Definition |
| :- | :- | :- |
| `Title` | object | Title of the stand-up comedy special. |
| `Producer` | object | Platform that produced and released the show. |
| `Comedian` | object | Name and surname of the comedian. |
| `Gender` | object | Gender of the comedian. |
| `Release date` | object | Release date of the show. |
| `Original language` | object | Original language of the audio of the show.|
| `Text` | object | Full text/transcript of the show. |
| `Len_Hours` | int64 | Number of runtime hours. |
| `Len_Minutes` | int64 | Number of runtime minutes. |
| `Len_Minutes` | int64 | Number of runtime seconds. |
| `File_name` | object | Name of the .srt file (subtitles/transcript) in the working folder.|
The sources used to construct this dataset are Wikipedia (i.e., [Amazon stand-up comedy specials list](https://en.wikipedia.org/wiki/List_of_Amazon_original_programming#Stand-up_comedy_specials) and [Netflix stand up comedy list](https://en.wikipedia.org/wiki/List_of_Netflix_original_stand-up_comedy_specials)) for the first six features listed above. The remaining five features have been instead extracted from `.srt` files (subtitles files) downloaded from the website [Subscene.com](https://subscene.com). Subscene is a crowdsourced community of translators that hosts thousands of transcripts produced by its members. These `.srt` files contain the audio transcript in English language of the show and information on the lenght/timing of each sentence present in the show.
Notice that the dataset includes all the stand-up comedy shows for which an English transcript (.srt file) was available on Subscene. The stand-up comedy specials that do not meet this criterion were not included in the dataset.
```
import pysrt # Necessary to read .srt files
from tqdm import tqdm # Progress bar for loop
import os
import re
import string
import pandas as pd
import numpy as np
import warnings
warnings.filterwarnings('ignore')
# Import list of stand up shows from .csv file (source: Wikipedia + manual integrations/adjustments)
df = pd.read_csv('List_stand_up_comedy_full.csv')
print('Total number of stand-up comedy shows:', df.shape[0])
df.head()
# Example .srt file
example = pysrt.open('Stand_up_specials_subs/Ali.Wong.Baby.Cobra.2016.720p.WEBRip.x264-JAWN.srt')
print('Starting time of sentence in position 0:', example[0].start)
print('Ending time of sentence in position 0:', example[0].end)
print('\nSentence example up to position 0:\n\n', example[0].text)
```
## 1.1) Extract text data from .srt files and create dataset for analysis
```
# Define cleaning function for .srt text files
def clean_srt(text):
text = text.encode("ascii", "ignore") # Remove non-ASCII characters
text = text.decode()
text = text.replace('\n', ' ') # Remove new row escape
text = re.sub(r'<[^>]+>', '', text) # Eliminate text within < > characters (often contaning info on subs font/color)
text = re.sub(r'\[[^]]+\]', '', text) # Eliminate text within squared brackets (often contaning audio description for hearing-impaired individuals)
text = re.sub(r'\([^)]+\)', '', text) # Eliminate text within parentheses (often contaning audio description for hearing-impaired individuals)
text = text.replace('-', '') # Remove dialogue dashes
text = ' '.join(text.split()) # Reduce all double/triple whitespacing to single
return text
# Clean_srt function test on example file
clean_srt(example[0].text)
# Extract text data from .srt files and add to the dataset
main_dir = 'Stand_up_specials_subs'
# Get file names from folders
file_path = [os.path.join(root,f) for root,dirs,files in os.walk(main_dir) for f in files]
text_file = []
file_names = []
show_lenght = []
for file in tqdm(file_path):
# Get file name to use as common column to merge with df
file_name = os.path.basename(file)
file_names.append(file_name)
# Open .srt file
subs = pysrt.open(file, encoding='iso-8859-1')
# Extract text content and append to list
text = subs.text
text = clean_srt(text)
text_file.append(text)
# Extract lenght
lenght = subs[len(subs)-1].end
show_lenght.append(lenght)
# Transform list in a dataframe and rename columns
df_fn = pd.DataFrame(file_names)
df_len = pd.DataFrame(show_lenght)
df_txt = pd.DataFrame(text_file)
extract_df = pd.concat([df_fn, df_len, df_txt], ignore_index=True, axis=1).rename(columns={0 :'File_name', 1 : 'Len_Hours', 2 : 'Len_Minutes', 3 : 'Len_Seconds', 4 :'Len_Milliseconds', 5 : 'Text'})
print('Number of stand-up comedy shows with text:', extract_df.shape[0])
extract_df.head()
```
## 1.2) Merge dataframe with extracted text data and stand-up shows dataset
```
# Remove file extension from file name for merging
extract_df['File_name'] = extract_df['File_name'].str.replace('.srt', '', regex=False)
# Concatenate the two dataframes to obtain final dataset
df_final = pd.merge(df, extract_df, how='left', on='File_name')
# Generate comedian's name column
df_final['Comedian'] = df_final['Title'].str.split(':', expand=True)[0]
# Reorder columns, drop milliseconds column and print basic info
df_final = df_final[['Title', 'Producer', 'Comedian', 'Gender', 'Release date', 'Original language',
'Text', 'Len_Hours', 'Len_Minutes', 'Len_Seconds', 'File_name']]
print('Final dataset shape:\n', df_final.shape)
df_final.info()
df_final.head()
# Save to .csv file
df_final.to_csv('Stand_up_comedy_dataset.csv', index=False)
```
|
github_jupyter
|
import pysrt # Necessary to read .srt files
from tqdm import tqdm # Progress bar for loop
import os
import re
import string
import pandas as pd
import numpy as np
import warnings
warnings.filterwarnings('ignore')
# Import list of stand up shows from .csv file (source: Wikipedia + manual integrations/adjustments)
df = pd.read_csv('List_stand_up_comedy_full.csv')
print('Total number of stand-up comedy shows:', df.shape[0])
df.head()
# Example .srt file
example = pysrt.open('Stand_up_specials_subs/Ali.Wong.Baby.Cobra.2016.720p.WEBRip.x264-JAWN.srt')
print('Starting time of sentence in position 0:', example[0].start)
print('Ending time of sentence in position 0:', example[0].end)
print('\nSentence example up to position 0:\n\n', example[0].text)
# Define cleaning function for .srt text files
def clean_srt(text):
text = text.encode("ascii", "ignore") # Remove non-ASCII characters
text = text.decode()
text = text.replace('\n', ' ') # Remove new row escape
text = re.sub(r'<[^>]+>', '', text) # Eliminate text within < > characters (often contaning info on subs font/color)
text = re.sub(r'\[[^]]+\]', '', text) # Eliminate text within squared brackets (often contaning audio description for hearing-impaired individuals)
text = re.sub(r'\([^)]+\)', '', text) # Eliminate text within parentheses (often contaning audio description for hearing-impaired individuals)
text = text.replace('-', '') # Remove dialogue dashes
text = ' '.join(text.split()) # Reduce all double/triple whitespacing to single
return text
# Clean_srt function test on example file
clean_srt(example[0].text)
# Extract text data from .srt files and add to the dataset
main_dir = 'Stand_up_specials_subs'
# Get file names from folders
file_path = [os.path.join(root,f) for root,dirs,files in os.walk(main_dir) for f in files]
text_file = []
file_names = []
show_lenght = []
for file in tqdm(file_path):
# Get file name to use as common column to merge with df
file_name = os.path.basename(file)
file_names.append(file_name)
# Open .srt file
subs = pysrt.open(file, encoding='iso-8859-1')
# Extract text content and append to list
text = subs.text
text = clean_srt(text)
text_file.append(text)
# Extract lenght
lenght = subs[len(subs)-1].end
show_lenght.append(lenght)
# Transform list in a dataframe and rename columns
df_fn = pd.DataFrame(file_names)
df_len = pd.DataFrame(show_lenght)
df_txt = pd.DataFrame(text_file)
extract_df = pd.concat([df_fn, df_len, df_txt], ignore_index=True, axis=1).rename(columns={0 :'File_name', 1 : 'Len_Hours', 2 : 'Len_Minutes', 3 : 'Len_Seconds', 4 :'Len_Milliseconds', 5 : 'Text'})
print('Number of stand-up comedy shows with text:', extract_df.shape[0])
extract_df.head()
# Remove file extension from file name for merging
extract_df['File_name'] = extract_df['File_name'].str.replace('.srt', '', regex=False)
# Concatenate the two dataframes to obtain final dataset
df_final = pd.merge(df, extract_df, how='left', on='File_name')
# Generate comedian's name column
df_final['Comedian'] = df_final['Title'].str.split(':', expand=True)[0]
# Reorder columns, drop milliseconds column and print basic info
df_final = df_final[['Title', 'Producer', 'Comedian', 'Gender', 'Release date', 'Original language',
'Text', 'Len_Hours', 'Len_Minutes', 'Len_Seconds', 'File_name']]
print('Final dataset shape:\n', df_final.shape)
df_final.info()
df_final.head()
# Save to .csv file
df_final.to_csv('Stand_up_comedy_dataset.csv', index=False)
| 0.446253 | 0.888566 |
```
# hide ssl warnings for this example.
import requests
requests.packages.urllib3.disable_warnings()
```
# Turning python-fmrest foundset into DataFrame
This is a short example on how to easily build a Pandas DataFrame from a Foundset.
Here you can find information about [Pandas](http://pandas.pydata.org) and [DataFrames](http://pandas.pydata.org/pandas-docs/stable/dsintro.html) and how you can work with these data structures when analysing data.
For this example, make sure to first install Pandas (`pip install pandas`) and optionally Matplotlib (`pip install matplotlib`).
```
import fmrest
from pandas.plotting import parallel_coordinates
from matplotlib import pyplot as plt
%matplotlib inline
```
## Create server instance and login
First, we create our server instance just like in the examples before.
```
fms = fmrest.Server('https://10.211.55.15',
user='admin',
password='admin',
database='Contacts',
layout='Iris',
verify_ssl=False,
#type_conversion=True #Danger!
)
fms.login()
```
If you have numbers in text fields or want to work with dates, you can try out `type_conversion=True`.
**Note**: As this "type-guessing" can, at times, lead to unexpected results, I strongly recommended you to do the type conversion yourself when working with your own "known" data fields.
## Fetch records from server
Let's fetch all data from our Iris layout as specified above.
(When you have more than 100 records to fetch, make sure to specify the `limit` parameter as the Data API defaults to 100 records.)
```
foundset = fms.get_records(limit=150)
foundset
```
## Turn foundset into DataFrame
Now that we have our Foundset instance, the only thing we need to do is to call the `to_df` method. This will give us a Pandas DataFrame.
Let's look at the first 5 rows.
```
df = foundset.to_df()
df.head()
```
Since we are not interested in `recordId` or `modId`, we just drop them.
```
df.drop(['recordId', 'modId'], axis=1, inplace=True)
df.head()
```
Before working with your data, check that the types are OK (previous versions of the Data API always returned strings and ignored the data type set in your DB schema).
```
df.dtypes
```
## Going from here
With your DataFrame ready, you can go on analyzing and plotting your data as you wish...
```
plt.figure(figsize=(12, 12))
parallel_coordinates(df, 'class')
```
Learn more here: https://pandas.pydata.org/pandas-docs/stable/index.html
|
github_jupyter
|
# hide ssl warnings for this example.
import requests
requests.packages.urllib3.disable_warnings()
import fmrest
from pandas.plotting import parallel_coordinates
from matplotlib import pyplot as plt
%matplotlib inline
fms = fmrest.Server('https://10.211.55.15',
user='admin',
password='admin',
database='Contacts',
layout='Iris',
verify_ssl=False,
#type_conversion=True #Danger!
)
fms.login()
foundset = fms.get_records(limit=150)
foundset
df = foundset.to_df()
df.head()
df.drop(['recordId', 'modId'], axis=1, inplace=True)
df.head()
df.dtypes
plt.figure(figsize=(12, 12))
parallel_coordinates(df, 'class')
| 0.36886 | 0.90878 |
# 1. Import libraries
```
#----------------------------Reproducible----------------------------------------------------------------------------------------
import numpy as np
import tensorflow as tf
import random as rn
import os
seed=0
os.environ['PYTHONHASHSEED'] = str(seed)
np.random.seed(seed)
rn.seed(seed)
#session_conf = tf.ConfigProto(intra_op_parallelism_threads=1, inter_op_parallelism_threads=1)
session_conf =tf.compat.v1.ConfigProto(intra_op_parallelism_threads=1, inter_op_parallelism_threads=1)
from keras import backend as K
#tf.set_random_seed(seed)
tf.compat.v1.set_random_seed(seed)
#sess = tf.Session(graph=tf.get_default_graph(), config=session_conf)
sess = tf.compat.v1.Session(graph=tf.compat.v1.get_default_graph(), config=session_conf)
K.set_session(sess)
#----------------------------Reproducible----------------------------------------------------------------------------------------
os.environ['TF_CPP_MIN_LOG_LEVEL'] = '3'
#--------------------------------------------------------------------------------------------------------------------------------
from keras.datasets import mnist
from keras.models import Model
from keras.layers import Dense, Input, Flatten, Activation, Dropout, Layer
from keras.layers.normalization import BatchNormalization
from keras.utils import to_categorical
from keras import optimizers,initializers,constraints,regularizers
from keras import backend as K
from keras.callbacks import LambdaCallback,ModelCheckpoint
from keras.utils import plot_model
from sklearn.model_selection import StratifiedKFold
from sklearn.ensemble import ExtraTreesClassifier
from sklearn import svm
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import ShuffleSplit
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
from sklearn.svm import SVC
import h5py
import math
import matplotlib
import matplotlib.pyplot as plt
import matplotlib.cm as cm
%matplotlib inline
matplotlib.style.use('ggplot')
import pandas as pd
from sklearn.impute import SimpleImputer
from sklearn.preprocessing import MinMaxScaler
from sklearn.model_selection import train_test_split
import scipy.sparse as sparse
#--------------------------------------------------------------------------------------------------------------------------------
#Import ourslef defined methods
import sys
sys.path.append(r"./Defined")
import Functions as F
# The following code should be added before the keras model
#np.random.seed(seed)
```
# 2. Loading data
```
data_frame=pd.read_excel('./Dataset/Data_Cortex_Nuclear.xls',sheet_name='Hoja1')
data_arr=(np.array(data_frame)[:,1:78]).copy()
label_arr=(np.array(data_frame)[:,81]).copy()
for index_i in np.arange(len(label_arr)):
if label_arr[index_i]=='c-CS-s':
label_arr[index_i]='0'
if label_arr[index_i]=='c-CS-m':
label_arr[index_i]='1'
if label_arr[index_i]=='c-SC-s':
label_arr[index_i]='2'
if label_arr[index_i]=='c-SC-m':
label_arr[index_i]='3'
if label_arr[index_i]=='t-CS-s':
label_arr[index_i]='4'
if label_arr[index_i]=='t-CS-m':
label_arr[index_i]='5'
if label_arr[index_i]=='t-SC-s':
label_arr[index_i]='6'
if label_arr[index_i]=='t-SC-m':
label_arr[index_i]='7'
label_arr_onehot=label_arr#to_categorical(label_arr)
# Show before Imputer
#print(data_arr[558])
imp_mean = SimpleImputer(missing_values=np.nan, strategy='mean')
imp_mean.fit(data_arr)
data_arr=imp_mean.transform(data_arr)
# Show after Imputer
#print(data_arr[558])
data_arr=MinMaxScaler(feature_range=(0,1)).fit_transform(data_arr)
C_train_x,C_test_x,C_train_y,C_test_y= train_test_split(data_arr,label_arr_onehot,test_size=0.2,random_state=seed)
x_train,x_validate,y_train_onehot,y_validate_onehot= train_test_split(C_train_x,C_train_y,test_size=0.1,random_state=seed)
x_test=C_test_x
y_test_onehot=C_test_y
print('Shape of x_train: ' + str(x_train.shape))
print('Shape of x_validate: ' + str(x_validate.shape))
print('Shape of x_test: ' + str(x_test.shape))
print('Shape of y_train: ' + str(y_train_onehot.shape))
print('Shape of y_validate: ' + str(y_validate_onehot.shape))
print('Shape of y_test: ' + str(y_test_onehot.shape))
print('Shape of C_train_x: ' + str(C_train_x.shape))
print('Shape of C_train_y: ' + str(C_train_y.shape))
print('Shape of C_test_x: ' + str(C_test_x.shape))
print('Shape of C_test_y: ' + str(C_test_y.shape))
key_feture_number=10
```
# 3.Model
```
np.random.seed(seed)
#--------------------------------------------------------------------------------------------------------------------------------
class Feature_Select_Layer(Layer):
def __init__(self, output_dim, **kwargs):
super(Feature_Select_Layer, self).__init__(**kwargs)
self.output_dim = output_dim
def build(self, input_shape):
self.kernel = self.add_weight(name='kernel',
shape=(input_shape[1],),
initializer=initializers.RandomUniform(minval=0.999999, maxval=0.9999999, seed=seed),
trainable=True)
super(Feature_Select_Layer, self).build(input_shape)
def call(self, x, selection=False,k=key_feture_number):
kernel=K.abs(self.kernel)
if selection:
kernel_=K.transpose(kernel)
kth_largest = tf.math.top_k(kernel_, k=k)[0][-1]
kernel = tf.where(condition=K.less(kernel,kth_largest),x=K.zeros_like(kernel),y=kernel)
return K.dot(x, tf.linalg.tensor_diag(kernel))
def compute_output_shape(self, input_shape):
return (input_shape[0], self.output_dim)
#--------------------------------------------------------------------------------------------------------------------------------
def Autoencoder(p_data_feature=x_train.shape[1],\
p_encoding_dim=key_feture_number,\
p_learning_rate= 1E-3):
input_img = Input(shape=(p_data_feature,), name='input_img')
encoded = Dense(p_encoding_dim, activation='linear',kernel_initializer=initializers.glorot_uniform(seed))(input_img)
bottleneck=encoded
decoded = Dense(p_data_feature, activation='linear',kernel_initializer=initializers.glorot_uniform(seed))(encoded)
latent_encoder = Model(input_img, bottleneck)
autoencoder = Model(input_img, decoded)
autoencoder.compile(loss='mean_squared_error', optimizer=optimizers.Adam(lr=p_learning_rate))
print('Autoencoder Structure-------------------------------------')
autoencoder.summary()
#print('Latent Encoder Structure-------------------------------------')
#latent_encoder.summary()
return autoencoder,latent_encoder
#--------------------------------------------------------------------------------------------------------------------------------
def Identity_Autoencoder(p_data_feature=x_train.shape[1],\
p_encoding_dim=key_feture_number,\
p_learning_rate= 1E-3):
input_img = Input(shape=(p_data_feature,), name='autoencoder_input')
feature_selection = Feature_Select_Layer(output_dim=p_data_feature,\
input_shape=(p_data_feature,),\
name='feature_selection')
feature_selection_score=feature_selection(input_img)
encoded = Dense(p_encoding_dim,\
activation='linear',\
kernel_initializer=initializers.glorot_uniform(seed),\
name='autoencoder_hidden_layer')
encoded_score=encoded(feature_selection_score)
bottleneck_score=encoded_score
decoded = Dense(p_data_feature,\
activation='linear',\
kernel_initializer=initializers.glorot_uniform(seed),\
name='autoencoder_output')
decoded_score =decoded(bottleneck_score)
latent_encoder_score = Model(input_img, bottleneck_score)
autoencoder = Model(input_img, decoded_score)
autoencoder.compile(loss='mean_squared_error',\
optimizer=optimizers.Adam(lr=p_learning_rate))
print('Autoencoder Structure-------------------------------------')
autoencoder.summary()
return autoencoder,latent_encoder_score
#--------------------------------------------------------------------------------------------------------------------------------
def Fractal_Autoencoder(p_data_feature=x_train.shape[1],\
p_feture_number=key_feture_number,\
p_encoding_dim=key_feture_number,\
p_learning_rate=1E-3,\
p_loss_weight_1=1,\
p_loss_weight_2=2):
input_img = Input(shape=(p_data_feature,), name='autoencoder_input')
feature_selection = Feature_Select_Layer(output_dim=p_data_feature,\
input_shape=(p_data_feature,),\
name='feature_selection')
feature_selection_score=feature_selection(input_img)
feature_selection_choose=feature_selection(input_img,selection=True,k=p_feture_number)
encoded = Dense(p_encoding_dim,\
activation='linear',\
kernel_initializer=initializers.glorot_uniform(seed),\
name='autoencoder_hidden_layer')
encoded_score=encoded(feature_selection_score)
encoded_choose=encoded(feature_selection_choose)
bottleneck_score=encoded_score
bottleneck_choose=encoded_choose
decoded = Dense(p_data_feature,\
activation='linear',\
kernel_initializer=initializers.glorot_uniform(seed),\
name='autoencoder_output')
decoded_score =decoded(bottleneck_score)
decoded_choose =decoded(bottleneck_choose)
latent_encoder_score = Model(input_img, bottleneck_score)
latent_encoder_choose = Model(input_img, bottleneck_choose)
feature_selection_output=Model(input_img,feature_selection_choose)
autoencoder = Model(input_img, [decoded_score,decoded_choose])
autoencoder.compile(loss=['mean_squared_error','mean_squared_error'],\
loss_weights=[p_loss_weight_1, p_loss_weight_2],\
optimizer=optimizers.Adam(lr=p_learning_rate))
print('Autoencoder Structure-------------------------------------')
autoencoder.summary()
return autoencoder,feature_selection_output,latent_encoder_score,latent_encoder_choose
```
## 3.1 Structure and paramter testing
```
epochs_number=200
batch_size_value=128
```
---
### 3.1.1 Fractal Autoencoder
---
```
loss_weight_1=0.0078125
F_AE,\
feature_selection_output,\
latent_encoder_score_F_AE,\
latent_encoder_choose_F_AE=Fractal_Autoencoder(p_data_feature=x_train.shape[1],\
p_feture_number=key_feture_number,\
p_encoding_dim=key_feture_number,\
p_learning_rate= 1E-3,\
p_loss_weight_1=loss_weight_1,\
p_loss_weight_2=1)
file_name="./log/F_AE_"+str(key_feture_number)+".png"
plot_model(F_AE, to_file=file_name,show_shapes=True)
model_checkpoint=ModelCheckpoint('./log_weights/F_AE_'+str(key_feture_number)+'_weights_'+str(loss_weight_1)+'.{epoch:04d}.hdf5',period=100,save_weights_only=True,verbose=1)
#print_weights = LambdaCallback(on_epoch_end=lambda batch, logs: print(F_AE.layers[1].get_weights()))
F_AE_history = F_AE.fit(x_train, [x_train,x_train],\
epochs=epochs_number,\
batch_size=batch_size_value,\
shuffle=True,\
validation_data=(x_validate, [x_validate,x_validate]),\
callbacks=[model_checkpoint])
loss = F_AE_history.history['loss']
val_loss = F_AE_history.history['val_loss']
epochs = range(epochs_number)
plt.plot(epochs, loss, 'bo', label='Training Loss')
plt.plot(epochs, val_loss, 'r', label='Validation Loss')
plt.xlabel('Epochs')
plt.ylabel('Loss')
plt.legend()
plt.show()
plt.plot(epochs[250:], loss[250:], 'bo', label='Training Loss')
plt.plot(epochs[250:], val_loss[250:], 'r', label='Validation Loss')
plt.xlabel('Epochs')
plt.ylabel('Loss')
plt.legend()
plt.show()
p_data=F_AE.predict(x_test)
numbers=x_test.shape[0]*x_test.shape[1]
print("MSE for one-to-one map layer",np.sum(np.power(np.array(p_data)[0]-x_test,2))/numbers)
print("MSE for feature selection layer",np.sum(np.power(np.array(p_data)[1]-x_test,2))/numbers)
```
---
### 3.1.2 Feature selection layer output
---
```
FS_layer_output=feature_selection_output.predict(x_test)
print(np.sum(FS_layer_output[0]>0))
```
---
### 3.1.3 Key features show
---
```
key_features=F.top_k_keepWeights_1(F_AE.get_layer(index=1).get_weights()[0],key_feture_number)
print(np.sum(F_AE.get_layer(index=1).get_weights()[0]>0))
```
# 4 Classifying
### 4.1 Extra Trees
```
train_feature=C_train_x
train_label=C_train_y
test_feature=C_test_x
test_label=C_test_y
p_seed=seed
F.ETree(train_feature,train_label,test_feature,test_label,p_seed)
selected_position_list=np.where(key_features>0)[0]
```
---
#### 4.1.1. On Identity Selection layer
---
a) with zeros
```
train_feature=feature_selection_output.predict(C_train_x)
print("train_feature>0: ",np.sum(train_feature[0]>0))
print(train_feature.shape)
train_label=C_train_y
test_feature=feature_selection_output.predict(C_test_x)
print("test_feature>0: ",np.sum(test_feature[0]>0))
print(test_feature.shape)
test_label=C_test_y
p_seed=seed
F.ETree(train_feature,train_label,test_feature,test_label,p_seed)
```
---
b) Sparse matrix
```
train_feature=feature_selection_output.predict(C_train_x)
print(train_feature.shape)
train_label=C_train_y
test_feature=feature_selection_output.predict(C_test_x)
print(test_feature.shape)
test_label=C_test_y
train_feature_sparse=sparse.coo_matrix(train_feature)
test_feature_sparse=sparse.coo_matrix(test_feature)
p_seed=seed
F.ETree(train_feature_sparse,train_label,test_feature_sparse,test_label,p_seed)
```
---
c) Compression
```
train_feature_=feature_selection_output.predict(C_train_x)
train_feature=F.compress_zero(train_feature_,key_feture_number)
print(train_feature.shape)
train_label=C_train_y
test_feature_=feature_selection_output.predict(C_test_x)
test_feature=F.compress_zero(test_feature_,key_feture_number)
print(test_feature.shape)
test_label=C_test_y
p_seed=seed
F.ETree(train_feature,train_label,test_feature,test_label,p_seed)
```
---
d) Compression with structure
```
train_feature_=feature_selection_output.predict(C_train_x)
train_feature=F.compress_zero_withkeystructure(train_feature_,selected_position_list)
print(train_feature.shape)
train_label=C_train_y
test_feature_=feature_selection_output.predict(C_test_x)
test_feature=F.compress_zero_withkeystructure(test_feature_,selected_position_list)
print(test_feature.shape)
test_label=C_test_y
p_seed=seed
F.ETree(train_feature,train_label,test_feature,test_label,p_seed)
```
---
#### 4.1.2. On Original Selection
---
a) with zeros
```
train_feature=np.multiply(C_train_x, key_features)
print("train_feature>0: ",np.sum(train_feature[0]>0))
print(train_feature.shape)
train_label=C_train_y
test_feature=np.multiply(C_test_x, key_features)
print("test_feature>0: ",np.sum(test_feature[0]>0))
print(test_feature.shape)
test_label=C_test_y
p_seed=seed
F.ETree(train_feature,train_label,test_feature,test_label,p_seed)
```
---
b) Sparse matrix
```
train_feature=np.multiply(C_train_x, key_features)
print(train_feature.shape)
train_label=C_train_y
test_feature=np.multiply(C_test_x, key_features)
print(test_feature.shape)
test_label=C_test_y
train_feature_sparse=sparse.coo_matrix(train_feature)
test_feature_sparse=sparse.coo_matrix(test_feature)
p_seed=seed
F.ETree(train_feature_sparse,train_label,test_feature_sparse,test_label,p_seed)
```
---
c) Compression
```
train_feature_=np.multiply(C_train_x, key_features)
train_feature=F.compress_zero(train_feature_,key_feture_number)
print(train_feature.shape)
train_label=C_train_y
test_feature_=np.multiply(C_test_x, key_features)
test_feature=F.compress_zero(test_feature_,key_feture_number)
print(test_feature.shape)
test_label=C_test_y
p_seed=seed
F.ETree(train_feature,train_label,test_feature,test_label,p_seed)
```
---
d) Compression with structure
```
train_feature_=np.multiply(C_train_x, key_features)
train_feature=F.compress_zero_withkeystructure(train_feature_,selected_position_list)
print(train_feature.shape)
train_label=C_train_y
test_feature_=np.multiply(C_test_x, key_features)
test_feature=F.compress_zero_withkeystructure(test_feature_,selected_position_list)
print(test_feature.shape)
test_label=C_test_y
p_seed=seed
F.ETree(train_feature,train_label,test_feature,test_label,p_seed)
```
---
#### 4.1.3. Latent space
---
```
train_feature=latent_encoder_score_F_AE.predict(C_train_x)
print(train_feature.shape)
train_label=C_train_y
test_feature=latent_encoder_score_F_AE.predict(C_test_x)
print(test_feature.shape)
test_label=C_test_y
p_seed=seed
F.ETree(train_feature,train_label,test_feature,test_label,p_seed)
train_feature=latent_encoder_choose_F_AE.predict(C_train_x)
print(train_feature.shape)
train_label=C_train_y
test_feature=latent_encoder_choose_F_AE.predict(C_test_x)
print(test_feature.shape)
test_label=C_test_y
p_seed=seed
F.ETree(train_feature,train_label,test_feature,test_label,p_seed)
```
---
# 6 Feature group compare
---
```
Selected_Weights=F.top_k_keep(F_AE.get_layer(index=1).get_weights()[0],key_feture_number)
selected_position_group=F.k_index_argsort_1d(Selected_Weights,key_feture_number)
train_feature_=np.multiply(C_train_x, key_features)
train_feature=F.compress_zero_withkeystructure(train_feature_,selected_position_group)
print(train_feature.shape)
train_label=C_train_y
test_feature_=np.multiply(C_test_x, key_features)
test_feature=F.compress_zero_withkeystructure(test_feature_,selected_position_group)
print(test_feature.shape)
test_label=C_test_y
p_seed=seed
F.ETree(train_feature[:,0:5],train_label,test_feature[:,0:5],test_label,p_seed)
p_seed=seed
F.ETree(train_feature[:,5:],train_label,test_feature[:,5:],test_label,p_seed)
p_seed=seed
F.ETree(train_feature[:,0:6],train_label,test_feature[:,0:6],test_label,p_seed)
p_seed=seed
F.ETree(train_feature[:,6:],train_label,test_feature[:,6:],test_label,p_seed)
```
# 6. Reconstruction loss
```
from sklearn.linear_model import LinearRegression
def mse_check(train, test):
LR = LinearRegression(n_jobs = -1)
LR.fit(train[0], train[1])
MSELR = ((LR.predict(test[0]) - test[1]) ** 2).mean()
return MSELR
train_feature_=np.multiply(C_train_x, key_features)
C_train_selected_x=F.compress_zero_withkeystructure(train_feature_,selected_position_list)
print(C_train_selected_x.shape)
test_feature_=np.multiply(C_test_x, key_features)
C_test_selected_x=F.compress_zero_withkeystructure(test_feature_,selected_position_list)
print(C_test_selected_x.shape)
train_feature_tuple=(C_train_selected_x,C_train_x)
test_feature_tuple=(C_test_selected_x,C_test_x)
reconstruction_loss=mse_check(train_feature_tuple, test_feature_tuple)
print(reconstruction_loss)
```
|
github_jupyter
|
#----------------------------Reproducible----------------------------------------------------------------------------------------
import numpy as np
import tensorflow as tf
import random as rn
import os
seed=0
os.environ['PYTHONHASHSEED'] = str(seed)
np.random.seed(seed)
rn.seed(seed)
#session_conf = tf.ConfigProto(intra_op_parallelism_threads=1, inter_op_parallelism_threads=1)
session_conf =tf.compat.v1.ConfigProto(intra_op_parallelism_threads=1, inter_op_parallelism_threads=1)
from keras import backend as K
#tf.set_random_seed(seed)
tf.compat.v1.set_random_seed(seed)
#sess = tf.Session(graph=tf.get_default_graph(), config=session_conf)
sess = tf.compat.v1.Session(graph=tf.compat.v1.get_default_graph(), config=session_conf)
K.set_session(sess)
#----------------------------Reproducible----------------------------------------------------------------------------------------
os.environ['TF_CPP_MIN_LOG_LEVEL'] = '3'
#--------------------------------------------------------------------------------------------------------------------------------
from keras.datasets import mnist
from keras.models import Model
from keras.layers import Dense, Input, Flatten, Activation, Dropout, Layer
from keras.layers.normalization import BatchNormalization
from keras.utils import to_categorical
from keras import optimizers,initializers,constraints,regularizers
from keras import backend as K
from keras.callbacks import LambdaCallback,ModelCheckpoint
from keras.utils import plot_model
from sklearn.model_selection import StratifiedKFold
from sklearn.ensemble import ExtraTreesClassifier
from sklearn import svm
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import ShuffleSplit
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
from sklearn.svm import SVC
import h5py
import math
import matplotlib
import matplotlib.pyplot as plt
import matplotlib.cm as cm
%matplotlib inline
matplotlib.style.use('ggplot')
import pandas as pd
from sklearn.impute import SimpleImputer
from sklearn.preprocessing import MinMaxScaler
from sklearn.model_selection import train_test_split
import scipy.sparse as sparse
#--------------------------------------------------------------------------------------------------------------------------------
#Import ourslef defined methods
import sys
sys.path.append(r"./Defined")
import Functions as F
# The following code should be added before the keras model
#np.random.seed(seed)
data_frame=pd.read_excel('./Dataset/Data_Cortex_Nuclear.xls',sheet_name='Hoja1')
data_arr=(np.array(data_frame)[:,1:78]).copy()
label_arr=(np.array(data_frame)[:,81]).copy()
for index_i in np.arange(len(label_arr)):
if label_arr[index_i]=='c-CS-s':
label_arr[index_i]='0'
if label_arr[index_i]=='c-CS-m':
label_arr[index_i]='1'
if label_arr[index_i]=='c-SC-s':
label_arr[index_i]='2'
if label_arr[index_i]=='c-SC-m':
label_arr[index_i]='3'
if label_arr[index_i]=='t-CS-s':
label_arr[index_i]='4'
if label_arr[index_i]=='t-CS-m':
label_arr[index_i]='5'
if label_arr[index_i]=='t-SC-s':
label_arr[index_i]='6'
if label_arr[index_i]=='t-SC-m':
label_arr[index_i]='7'
label_arr_onehot=label_arr#to_categorical(label_arr)
# Show before Imputer
#print(data_arr[558])
imp_mean = SimpleImputer(missing_values=np.nan, strategy='mean')
imp_mean.fit(data_arr)
data_arr=imp_mean.transform(data_arr)
# Show after Imputer
#print(data_arr[558])
data_arr=MinMaxScaler(feature_range=(0,1)).fit_transform(data_arr)
C_train_x,C_test_x,C_train_y,C_test_y= train_test_split(data_arr,label_arr_onehot,test_size=0.2,random_state=seed)
x_train,x_validate,y_train_onehot,y_validate_onehot= train_test_split(C_train_x,C_train_y,test_size=0.1,random_state=seed)
x_test=C_test_x
y_test_onehot=C_test_y
print('Shape of x_train: ' + str(x_train.shape))
print('Shape of x_validate: ' + str(x_validate.shape))
print('Shape of x_test: ' + str(x_test.shape))
print('Shape of y_train: ' + str(y_train_onehot.shape))
print('Shape of y_validate: ' + str(y_validate_onehot.shape))
print('Shape of y_test: ' + str(y_test_onehot.shape))
print('Shape of C_train_x: ' + str(C_train_x.shape))
print('Shape of C_train_y: ' + str(C_train_y.shape))
print('Shape of C_test_x: ' + str(C_test_x.shape))
print('Shape of C_test_y: ' + str(C_test_y.shape))
key_feture_number=10
np.random.seed(seed)
#--------------------------------------------------------------------------------------------------------------------------------
class Feature_Select_Layer(Layer):
def __init__(self, output_dim, **kwargs):
super(Feature_Select_Layer, self).__init__(**kwargs)
self.output_dim = output_dim
def build(self, input_shape):
self.kernel = self.add_weight(name='kernel',
shape=(input_shape[1],),
initializer=initializers.RandomUniform(minval=0.999999, maxval=0.9999999, seed=seed),
trainable=True)
super(Feature_Select_Layer, self).build(input_shape)
def call(self, x, selection=False,k=key_feture_number):
kernel=K.abs(self.kernel)
if selection:
kernel_=K.transpose(kernel)
kth_largest = tf.math.top_k(kernel_, k=k)[0][-1]
kernel = tf.where(condition=K.less(kernel,kth_largest),x=K.zeros_like(kernel),y=kernel)
return K.dot(x, tf.linalg.tensor_diag(kernel))
def compute_output_shape(self, input_shape):
return (input_shape[0], self.output_dim)
#--------------------------------------------------------------------------------------------------------------------------------
def Autoencoder(p_data_feature=x_train.shape[1],\
p_encoding_dim=key_feture_number,\
p_learning_rate= 1E-3):
input_img = Input(shape=(p_data_feature,), name='input_img')
encoded = Dense(p_encoding_dim, activation='linear',kernel_initializer=initializers.glorot_uniform(seed))(input_img)
bottleneck=encoded
decoded = Dense(p_data_feature, activation='linear',kernel_initializer=initializers.glorot_uniform(seed))(encoded)
latent_encoder = Model(input_img, bottleneck)
autoencoder = Model(input_img, decoded)
autoencoder.compile(loss='mean_squared_error', optimizer=optimizers.Adam(lr=p_learning_rate))
print('Autoencoder Structure-------------------------------------')
autoencoder.summary()
#print('Latent Encoder Structure-------------------------------------')
#latent_encoder.summary()
return autoencoder,latent_encoder
#--------------------------------------------------------------------------------------------------------------------------------
def Identity_Autoencoder(p_data_feature=x_train.shape[1],\
p_encoding_dim=key_feture_number,\
p_learning_rate= 1E-3):
input_img = Input(shape=(p_data_feature,), name='autoencoder_input')
feature_selection = Feature_Select_Layer(output_dim=p_data_feature,\
input_shape=(p_data_feature,),\
name='feature_selection')
feature_selection_score=feature_selection(input_img)
encoded = Dense(p_encoding_dim,\
activation='linear',\
kernel_initializer=initializers.glorot_uniform(seed),\
name='autoencoder_hidden_layer')
encoded_score=encoded(feature_selection_score)
bottleneck_score=encoded_score
decoded = Dense(p_data_feature,\
activation='linear',\
kernel_initializer=initializers.glorot_uniform(seed),\
name='autoencoder_output')
decoded_score =decoded(bottleneck_score)
latent_encoder_score = Model(input_img, bottleneck_score)
autoencoder = Model(input_img, decoded_score)
autoencoder.compile(loss='mean_squared_error',\
optimizer=optimizers.Adam(lr=p_learning_rate))
print('Autoencoder Structure-------------------------------------')
autoencoder.summary()
return autoencoder,latent_encoder_score
#--------------------------------------------------------------------------------------------------------------------------------
def Fractal_Autoencoder(p_data_feature=x_train.shape[1],\
p_feture_number=key_feture_number,\
p_encoding_dim=key_feture_number,\
p_learning_rate=1E-3,\
p_loss_weight_1=1,\
p_loss_weight_2=2):
input_img = Input(shape=(p_data_feature,), name='autoencoder_input')
feature_selection = Feature_Select_Layer(output_dim=p_data_feature,\
input_shape=(p_data_feature,),\
name='feature_selection')
feature_selection_score=feature_selection(input_img)
feature_selection_choose=feature_selection(input_img,selection=True,k=p_feture_number)
encoded = Dense(p_encoding_dim,\
activation='linear',\
kernel_initializer=initializers.glorot_uniform(seed),\
name='autoencoder_hidden_layer')
encoded_score=encoded(feature_selection_score)
encoded_choose=encoded(feature_selection_choose)
bottleneck_score=encoded_score
bottleneck_choose=encoded_choose
decoded = Dense(p_data_feature,\
activation='linear',\
kernel_initializer=initializers.glorot_uniform(seed),\
name='autoencoder_output')
decoded_score =decoded(bottleneck_score)
decoded_choose =decoded(bottleneck_choose)
latent_encoder_score = Model(input_img, bottleneck_score)
latent_encoder_choose = Model(input_img, bottleneck_choose)
feature_selection_output=Model(input_img,feature_selection_choose)
autoencoder = Model(input_img, [decoded_score,decoded_choose])
autoencoder.compile(loss=['mean_squared_error','mean_squared_error'],\
loss_weights=[p_loss_weight_1, p_loss_weight_2],\
optimizer=optimizers.Adam(lr=p_learning_rate))
print('Autoencoder Structure-------------------------------------')
autoencoder.summary()
return autoencoder,feature_selection_output,latent_encoder_score,latent_encoder_choose
epochs_number=200
batch_size_value=128
loss_weight_1=0.0078125
F_AE,\
feature_selection_output,\
latent_encoder_score_F_AE,\
latent_encoder_choose_F_AE=Fractal_Autoencoder(p_data_feature=x_train.shape[1],\
p_feture_number=key_feture_number,\
p_encoding_dim=key_feture_number,\
p_learning_rate= 1E-3,\
p_loss_weight_1=loss_weight_1,\
p_loss_weight_2=1)
file_name="./log/F_AE_"+str(key_feture_number)+".png"
plot_model(F_AE, to_file=file_name,show_shapes=True)
model_checkpoint=ModelCheckpoint('./log_weights/F_AE_'+str(key_feture_number)+'_weights_'+str(loss_weight_1)+'.{epoch:04d}.hdf5',period=100,save_weights_only=True,verbose=1)
#print_weights = LambdaCallback(on_epoch_end=lambda batch, logs: print(F_AE.layers[1].get_weights()))
F_AE_history = F_AE.fit(x_train, [x_train,x_train],\
epochs=epochs_number,\
batch_size=batch_size_value,\
shuffle=True,\
validation_data=(x_validate, [x_validate,x_validate]),\
callbacks=[model_checkpoint])
loss = F_AE_history.history['loss']
val_loss = F_AE_history.history['val_loss']
epochs = range(epochs_number)
plt.plot(epochs, loss, 'bo', label='Training Loss')
plt.plot(epochs, val_loss, 'r', label='Validation Loss')
plt.xlabel('Epochs')
plt.ylabel('Loss')
plt.legend()
plt.show()
plt.plot(epochs[250:], loss[250:], 'bo', label='Training Loss')
plt.plot(epochs[250:], val_loss[250:], 'r', label='Validation Loss')
plt.xlabel('Epochs')
plt.ylabel('Loss')
plt.legend()
plt.show()
p_data=F_AE.predict(x_test)
numbers=x_test.shape[0]*x_test.shape[1]
print("MSE for one-to-one map layer",np.sum(np.power(np.array(p_data)[0]-x_test,2))/numbers)
print("MSE for feature selection layer",np.sum(np.power(np.array(p_data)[1]-x_test,2))/numbers)
FS_layer_output=feature_selection_output.predict(x_test)
print(np.sum(FS_layer_output[0]>0))
key_features=F.top_k_keepWeights_1(F_AE.get_layer(index=1).get_weights()[0],key_feture_number)
print(np.sum(F_AE.get_layer(index=1).get_weights()[0]>0))
train_feature=C_train_x
train_label=C_train_y
test_feature=C_test_x
test_label=C_test_y
p_seed=seed
F.ETree(train_feature,train_label,test_feature,test_label,p_seed)
selected_position_list=np.where(key_features>0)[0]
train_feature=feature_selection_output.predict(C_train_x)
print("train_feature>0: ",np.sum(train_feature[0]>0))
print(train_feature.shape)
train_label=C_train_y
test_feature=feature_selection_output.predict(C_test_x)
print("test_feature>0: ",np.sum(test_feature[0]>0))
print(test_feature.shape)
test_label=C_test_y
p_seed=seed
F.ETree(train_feature,train_label,test_feature,test_label,p_seed)
train_feature=feature_selection_output.predict(C_train_x)
print(train_feature.shape)
train_label=C_train_y
test_feature=feature_selection_output.predict(C_test_x)
print(test_feature.shape)
test_label=C_test_y
train_feature_sparse=sparse.coo_matrix(train_feature)
test_feature_sparse=sparse.coo_matrix(test_feature)
p_seed=seed
F.ETree(train_feature_sparse,train_label,test_feature_sparse,test_label,p_seed)
train_feature_=feature_selection_output.predict(C_train_x)
train_feature=F.compress_zero(train_feature_,key_feture_number)
print(train_feature.shape)
train_label=C_train_y
test_feature_=feature_selection_output.predict(C_test_x)
test_feature=F.compress_zero(test_feature_,key_feture_number)
print(test_feature.shape)
test_label=C_test_y
p_seed=seed
F.ETree(train_feature,train_label,test_feature,test_label,p_seed)
train_feature_=feature_selection_output.predict(C_train_x)
train_feature=F.compress_zero_withkeystructure(train_feature_,selected_position_list)
print(train_feature.shape)
train_label=C_train_y
test_feature_=feature_selection_output.predict(C_test_x)
test_feature=F.compress_zero_withkeystructure(test_feature_,selected_position_list)
print(test_feature.shape)
test_label=C_test_y
p_seed=seed
F.ETree(train_feature,train_label,test_feature,test_label,p_seed)
train_feature=np.multiply(C_train_x, key_features)
print("train_feature>0: ",np.sum(train_feature[0]>0))
print(train_feature.shape)
train_label=C_train_y
test_feature=np.multiply(C_test_x, key_features)
print("test_feature>0: ",np.sum(test_feature[0]>0))
print(test_feature.shape)
test_label=C_test_y
p_seed=seed
F.ETree(train_feature,train_label,test_feature,test_label,p_seed)
train_feature=np.multiply(C_train_x, key_features)
print(train_feature.shape)
train_label=C_train_y
test_feature=np.multiply(C_test_x, key_features)
print(test_feature.shape)
test_label=C_test_y
train_feature_sparse=sparse.coo_matrix(train_feature)
test_feature_sparse=sparse.coo_matrix(test_feature)
p_seed=seed
F.ETree(train_feature_sparse,train_label,test_feature_sparse,test_label,p_seed)
train_feature_=np.multiply(C_train_x, key_features)
train_feature=F.compress_zero(train_feature_,key_feture_number)
print(train_feature.shape)
train_label=C_train_y
test_feature_=np.multiply(C_test_x, key_features)
test_feature=F.compress_zero(test_feature_,key_feture_number)
print(test_feature.shape)
test_label=C_test_y
p_seed=seed
F.ETree(train_feature,train_label,test_feature,test_label,p_seed)
train_feature_=np.multiply(C_train_x, key_features)
train_feature=F.compress_zero_withkeystructure(train_feature_,selected_position_list)
print(train_feature.shape)
train_label=C_train_y
test_feature_=np.multiply(C_test_x, key_features)
test_feature=F.compress_zero_withkeystructure(test_feature_,selected_position_list)
print(test_feature.shape)
test_label=C_test_y
p_seed=seed
F.ETree(train_feature,train_label,test_feature,test_label,p_seed)
train_feature=latent_encoder_score_F_AE.predict(C_train_x)
print(train_feature.shape)
train_label=C_train_y
test_feature=latent_encoder_score_F_AE.predict(C_test_x)
print(test_feature.shape)
test_label=C_test_y
p_seed=seed
F.ETree(train_feature,train_label,test_feature,test_label,p_seed)
train_feature=latent_encoder_choose_F_AE.predict(C_train_x)
print(train_feature.shape)
train_label=C_train_y
test_feature=latent_encoder_choose_F_AE.predict(C_test_x)
print(test_feature.shape)
test_label=C_test_y
p_seed=seed
F.ETree(train_feature,train_label,test_feature,test_label,p_seed)
Selected_Weights=F.top_k_keep(F_AE.get_layer(index=1).get_weights()[0],key_feture_number)
selected_position_group=F.k_index_argsort_1d(Selected_Weights,key_feture_number)
train_feature_=np.multiply(C_train_x, key_features)
train_feature=F.compress_zero_withkeystructure(train_feature_,selected_position_group)
print(train_feature.shape)
train_label=C_train_y
test_feature_=np.multiply(C_test_x, key_features)
test_feature=F.compress_zero_withkeystructure(test_feature_,selected_position_group)
print(test_feature.shape)
test_label=C_test_y
p_seed=seed
F.ETree(train_feature[:,0:5],train_label,test_feature[:,0:5],test_label,p_seed)
p_seed=seed
F.ETree(train_feature[:,5:],train_label,test_feature[:,5:],test_label,p_seed)
p_seed=seed
F.ETree(train_feature[:,0:6],train_label,test_feature[:,0:6],test_label,p_seed)
p_seed=seed
F.ETree(train_feature[:,6:],train_label,test_feature[:,6:],test_label,p_seed)
from sklearn.linear_model import LinearRegression
def mse_check(train, test):
LR = LinearRegression(n_jobs = -1)
LR.fit(train[0], train[1])
MSELR = ((LR.predict(test[0]) - test[1]) ** 2).mean()
return MSELR
train_feature_=np.multiply(C_train_x, key_features)
C_train_selected_x=F.compress_zero_withkeystructure(train_feature_,selected_position_list)
print(C_train_selected_x.shape)
test_feature_=np.multiply(C_test_x, key_features)
C_test_selected_x=F.compress_zero_withkeystructure(test_feature_,selected_position_list)
print(C_test_selected_x.shape)
train_feature_tuple=(C_train_selected_x,C_train_x)
test_feature_tuple=(C_test_selected_x,C_test_x)
reconstruction_loss=mse_check(train_feature_tuple, test_feature_tuple)
print(reconstruction_loss)
| 0.434461 | 0.576631 |
```
import pandas as pd, numpy as np
import os, time, random,gc
from tqdm import tqdm
import matplotlib.pyplot as plt
import seaborn as sns
pd.options.mode.chained_assignment = None
os.listdir('input')
def seed_everything(seed):
random.seed(seed)
os.environ['PYTHONHASHSEED'] = str(seed)
np.random.seed(seed)
SEED = 42
seed_everything(SEED)
%%time
tr = pd.read_feather('../dacon_sc/input/tr.feather')
te = pd.read_feather('../dacon_sc/input/te.feather')
tr.head()
tr_y = tr.groupby(['game_id'])['winner'].max()
tr_y = tr_y.reset_index()
train_Y = tr_y['winner']
tr_y.head()
tr.shape
tr.head()
def make_timespan(df):
df['time_span'] = 0
df['time_span'] = np.where(df['time']<1, 0,
np.where(df['time']<2, 1,
np.where(df['time']<3, 2,
np.where(df['time']<4, 3,
np.where(df['time']<5, 4,
np.where(df['time']<6, 5,
np.where(df['time']<7, 6,
np.where(df['time']<8, 7,
np.where(df['time']<9, 8,
np.where(df['time']<10, 9, 10))))))))))
return df
def tsp_cnt(df):
df = make_timespan(df)
new_p0 = pd.DataFrame({"game_id":df['game_id'].unique()})
p0 = df[df['player']==0]
for tsp in tqdm(range(11)):
tmp = p0[p0['time_span']==tsp].groupby(['game_id'])['time'].count().reset_index().rename(columns={"time":"p0_tsp_{}_cnt".format(tsp)})
# tmp.drop(['time_span'],axis=1, inplace=True)
new_p0 = new_p0.merge(tmp, on='game_id', how='left')
new_p1 = pd.DataFrame({"game_id":df['game_id'].unique()})
p1 = df[df['player']==1]
for tsp in tqdm(range(11)):
tmp = p1[p1['time_span']==tsp].groupby(['game_id'])['time'].count().reset_index().rename(columns={"time":"p1_tsp_{}_cnt".format(tsp)})
# tmp.drop(['time_span'],axis=1, inplace=True)
new_p1 = new_p1.merge(tmp, on='game_id', how='left')
new_df = new_p0.merge(new_p1, on='game_id', how='left')
return new_df
tr_tsp = tsp_cnt(tr)
te_tsp = tsp_cnt(te)
tr_tsp.head()
te_tsp.head()
def get_sp(df):
df_tmp = pd.DataFrame(df.game_id.unique(), columns=['game_id'])
df_tmp.index = df_tmp.game_id
df_tmp = df_tmp.drop(['game_id'], axis = 1)
p0 = df[(df.event=='Camera')&(df.player==0)]
p0 = p0[p0.shift(1).game_id!=p0.game_id] # 쉬프트를 이용하여 각 게임의 첫번째 데이터 찾기
p0 = p0.loc[:, ['game_id','event_contents']].rename({'event_contents':'player0_starting'}, axis = 1)
p0.index = p0['game_id']
p0 = p0.drop(['game_id'], axis=1)
df_tmp = pd.merge(df_tmp, p0, on='game_id', how='left')
del p0
p1 = df[(df.event=='Camera')&(df.player==1)]
p1 = p1[p1.shift(1).game_id!=p1.game_id]
p1 = p1.loc[:, ['game_id','event_contents']].rename({'event_contents':'player1_starting'}, axis = 1)
p1.index = p1['game_id']
p1 = p1.drop(['game_id'], axis=1)
df_tmp = pd.merge(df_tmp, p1, on='game_id', how='left')
del p1
df_tmp['player0_starting'] = df_tmp.player0_starting.str.split('(').str[1]
df_tmp['player0_starting'] = df_tmp.player0_starting.str.split(')').str[0]
split_xy = df_tmp.player0_starting.str.split(',')
df_tmp['player0_x'] = split_xy.str[0].astype('float')
df_tmp['player0_y'] = split_xy.str[1].astype('float')
del split_xy
df_tmp['player1_starting'] = df_tmp.player1_starting.str.split('(').str[1]
df_tmp['player1_starting'] = df_tmp.player1_starting.str.split(')').str[0]
split_xy = df_tmp.player1_starting.str.split(',')
df_tmp['player1_x'] = split_xy.str[0].astype('float')
df_tmp['player1_y'] = split_xy.str[1].astype('float')
del split_xy
# location_p0 = df_tmp.loc[:, ['player0_x', 'player0_y']]
# location_p0 = location_p0.rename({'player0_x':'location_x', 'player0_y':'location_y'}, axis=1)
# location_p1 = df_tmp.loc[:, ['player1_x', 'player1_y']]
# location_p1 = location_p1.rename({'player1_x':'location_x', 'player1_y':'location_y'}, axis=1)
# location_p1.index += location_p0.index[-1]+1
# location = pd.concat([location_p0, location_p1])
# location = location.dropna()
# del location_p0, location_p1, df_tmp
df_tmp.fillna(-999, inplace=True)
df_tmp['p0_sp'] = df_tmp['player0_x'].astype(int).astype(str) + df_tmp['player0_y'].astype(int).astype(str)
df_tmp['p1_sp'] = df_tmp['player1_x'].astype(int).astype(str) + df_tmp['player1_y'].astype(int).astype(str)
df_tmp['p0_sp2'] = np.round(df_tmp['player0_x'],2).astype(str) + np.round(df_tmp['player0_y'],2).astype(str)
df_tmp['p1_sp2'] = np.round(df_tmp['player1_x'],2).astype(str) + np.round(df_tmp['player1_y'],2).astype(str)
df_tmp['all_sp'] = df_tmp['p0_sp']+df_tmp['p1_sp']
df_tmp['all_sp2'] = df_tmp['p0_sp2']+df_tmp['p1_sp2']
return df_tmp
%%time
tr_sp = get_sp(tr)
te_sp = get_sp(te)
tr_sp.head()
te_sp.head()
tr_sp.nunique()
te_sp.nunique()
tra = tr.merge(tr_sp.iloc[:,-6:], on='game_id', how='left')
tra.head()
trp0 = tr[tr['player']==0].groupby(['game_id','time_span'])['time'].count().reset_index().rename(columns={"time":"p0_cnt"})
trp1 = tr[tr['player']==1].groupby(['game_id','time_span'])['time'].count().reset_index().rename(columns={"time":"p1_cnt"})
trcnt = trp0.merge(trp1, on=['game_id','time_span'], how='left')
trcnt = trcnt.merge(tr_sp.iloc[:,-6:], on='game_id', how='left')
tep0 = te[te['player']==0].groupby(['game_id','time_span'])['time'].count().reset_index().rename(columns={"time":"p0_cnt"})
tep1 = te[te['player']==1].groupby(['game_id','time_span'])['time'].count().reset_index().rename(columns={"time":"p1_cnt"})
tecnt = tep0.merge(tep1, on=['game_id','time_span'], how='left')
tecnt = tecnt.merge(te_sp.iloc[:,-6:], on='game_id', how='left')
tecnt.head(20)
def get_spc(df):
p0_spc = df[df['player']==0].groupby(['game_id'])['species'].max().reset_index().rename(columns={"species":'p0_spc'})
p1_spc = df[df['player']==1].groupby(['game_id'])['species'].max().reset_index().rename(columns={"species":'p1_spc'})
p0_spc = p0_spc.merge(p1_spc, on='game_id', how='left')
return p0_spc
%%time
tr_spc = get_spc(tr)
te_spc = get_spc(te)
tr_spc.head()
trcnt = trcnt.merge(tr_spc, on='game_id', how='left')
tecnt = tecnt.merge(te_spc, on='game_id', how='left')
trcnt['game_spc'] = trcnt['p0_spc']+trcnt['p1_spc']
tecnt['game_spc'] = tecnt['p0_spc']+tecnt['p1_spc']
trcnt['game_allsp_spc'] = trcnt['all_sp']+trcnt['p0_spc']+trcnt['p1_spc']
trcnt['game_allsp2_spc'] = trcnt['all_sp2']+trcnt['p0_spc']+trcnt['p1_spc']
tecnt['game_allsp_spc'] = tecnt['all_sp']+tecnt['p0_spc']+tecnt['p1_spc']
tecnt['game_allsp2_spc'] = tecnt['all_sp2']+tecnt['p0_spc']+tecnt['p1_spc']
trcnt['game_spc_sort'] = trcnt['game_spc'].apply(lambda x: sorted(x))
tecnt['game_spc_sort'] = tecnt['game_spc'].apply(lambda x: sorted(x))
trcnt.head()
tecnt.tail()
trcnt.shape,tecnt.shape
def df_copy(tr_df, te_df):
tr = tr_df.copy();te = te_df.copy()
return tr, te
def sub_day_agg(tr_df, te_df, merge_columns, date_columns, columns, aggs=['mean']):
tr, te = df_copy(tr_df, te_df)
for merge_column in merge_columns:
for date in date_columns:
tr['mc_date'] = tr[merge_column].astype(str) + '_' +tr[date].astype(str)
te['mc_date'] = te[merge_column].astype(str) + '_' +te[date].astype(str)
for col in columns:
for agg in aggs:
valid = pd.concat([tr[['mc_date', col]], te[['mc_date', col]]])
new_cn = merge_column + '_' + date + '_' + col + '_' + agg
if agg=='quantile':
valid = valid.groupby('mc_date')[col].quantile(0.8).reset_index().rename(columns={col:new_cn})
else:
valid = valid.groupby('mc_date')[col].agg([agg]).reset_index().rename(columns={agg:new_cn})
valid.index = valid['mc_date'].tolist()
valid = valid[new_cn].to_dict()
tr[new_cn] = tr['mc_date'].map(valid)
te[new_cn] = te['mc_date'].map(valid)
tr = tr.drop(columns=['mc_date'])
te = te.drop(columns=['mc_date'])
return tr, te
traa, teaa = sub_day_agg(trcnt, tecnt, merge_columns=['all_sp','all_sp2','p0_spc','p1_spc','game_allsp_spc','game_spc_sort','game_allsp2_spc'],
date_columns=['time_span'], columns=['p0_cnt', 'p1_cnt'], aggs=['mean'])
traa.head()
def sub_day_agg_flat(df):
new_df = pd.DataFrame({"game_id":df['game_id'].unique()})
for tsp in tqdm(range(11)):
tmp = df[df['time_span']==tsp][['game_id','all_sp_time_span_p0_cnt_mean'
,'all_sp_time_span_p1_cnt_mean'
,'all_sp2_time_span_p0_cnt_mean'
,'all_sp2_time_span_p1_cnt_mean'
,'p0_spc_time_span_p0_cnt_mean'
,'p0_spc_time_span_p1_cnt_mean'
,'p1_spc_time_span_p0_cnt_mean'
,'p1_spc_time_span_p1_cnt_mean'
,'game_allsp_spc_time_span_p0_cnt_mean'
,'game_allsp_spc_time_span_p1_cnt_mean'
,'game_spc_sort_time_span_p0_cnt_mean'
,'game_spc_sort_time_span_p1_cnt_mean'
,'game_allsp2_spc_time_span_p0_cnt_mean'
,'game_allsp2_spc_time_span_p1_cnt_mean'
]]
tmp.rename(columns={"all_sp_time_span_p0_cnt_mean":"all_sp_time_span_p0_cnt_mean_t{}".format(tsp)
,"all_sp_time_span_p1_cnt_mean":"all_sp_time_span_p1_cnt_mean_t{}".format(tsp)
,"all_sp2_time_span_p0_cnt_mean":"all_sp2_time_span_p0_cnt_mean_t{}".format(tsp)
,"all_sp2_time_span_p1_cnt_mean":"all_sp2_time_span_p1_cnt_mean_t{}".format(tsp)
,"p0_spc_time_span_p0_cnt_mean":"p0_spc_time_span_p0_cnt_mean_t{}".format(tsp)
,"p0_spc_time_span_p1_cnt_mean":"p0_spc_time_span_p1_cnt_mean_t{}".format(tsp)
,"p1_spc_time_span_p0_cnt_mean":"p1_spc_time_span_p0_cnt_mean_t{}".format(tsp)
,"p1_spc_time_span_p1_cnt_mean":"p1_spc_time_span_p1_cnt_mean_t{}".format(tsp)
,"game_allsp_spc_time_span_p0_cnt_mean":"game_allsp_spc_time_span_p0_cnt_mean_t{}".format(tsp)
,"game_allsp_spc_time_span_p1_cnt_mean":"game_allsp_spc_time_span_p1_cnt_mean_t{}".format(tsp)
,"game_spc_sort_time_span_p0_cnt_mean":"game_spc_sort_time_span_p0_cnt_mean_t{}".format(tsp)
,"game_spc_sort_time_span_p1_cnt_mean":"game_spc_sort_time_span_p1_cnt_mean_t{}".format(tsp)
,"game_allsp2_spc_time_span_p0_cnt_mean":"game_allsp2_spc_time_span_p0_cnt_mean_t{}".format(tsp)
,"game_allsp2_spc_time_span_p1_cnt_mean":"game_allsp2_spc_time_span_p1_cnt_mean_t{}".format(tsp)
},inplace=True)
new_df = new_df.merge(tmp, on='game_id', how='left')
return new_df
tr_sub = sub_day_agg_flat(traa)
te_sub = sub_day_agg_flat(teaa)
tr_sub.head()
len(list(set(tr_sub.columns.tolist())))
te_sub.head()
tr_tsp.head()
tr_df = tr_tsp.merge(tr_sub, on='game_id', how='left')
te_df = te_tsp.merge(te_sub, on='game_id', how='left')
tr_df.head()
tr_df = tr_df.merge(tr_spc, on='game_id', how='left')
te_df = te_df.merge(te_spc, on='game_id', how='left')
tr_df.head()
tr_sp.dtypes
tr_sp.select_dtypes(include=[object]).columns
tr_df = tr_df.merge(tr_sp.iloc[:,-6:], on='game_id', how='left')
te_df = te_df.merge(te_sp.iloc[:,-6:], on='game_id', how='left')
for df in tr_df, te_df:
for c in df.select_dtypes(include=[object]).columns.tolist():
df[c] = df[c].astype('category')
def get_spc_sort(df):
df['game_spc'] = df['p0_spc'].astype(str)+df['p1_spc'].astype(str)
df['game_allsp_spc'] = df['all_sp'].astype(str)+df['p0_spc'].astype(str)+df['p1_spc'].astype(str)
df['game_allsp2_spc'] = df['all_sp2'].astype(str)+df['p0_spc'].astype(str)+df['p1_spc'].astype(str)
df['game_spc_sort'] = df['game_spc'].astype(str).apply(lambda x: sorted(x))
df['game_spc_sort'] = df['game_spc_sort'].astype(str)
return df
tr_df = get_spc_sort(tr_df)
te_df = get_spc_sort(te_df)
for df in tr_df, te_df:
for c in df.select_dtypes(include=[object]).columns.tolist():
df[c] = df[c].astype('category')
tr_df.dtypes.value_counts()
tr_df.head()
tr_df.columns
tr_df.to_feather('../dacon_sc/input/tr_df.feather')
te_df.to_feather('../dacon_sc/input/te_df.feather')
tr_time_eve = pd.read_feather("../dacon_sc/input/tr_time_eve.feather")
te_time_eve = pd.read_feather("../dacon_sc/input/te_time_eve.feather")
tr_df = tr_df.merge(tr_time_eve, on='game_id', how='left')
te_df = te_df.merge(te_time_eve, on='game_id', how='left')
tr_df.head()
tr_df.head()
%%time
tr_v7 = pd.read_feather('../dacon_sc/input/tr_v7.feather')
te_v7 = pd.read_feather('../dacon_sc/input/te_v7.feather')
def diff_all(df):
p0_cols = df[df.filter(regex='p0').columns.tolist()].select_dtypes(exclude=[object,'category']).columns.tolist()
p1_cols = df[df.filter(regex='p1').columns.tolist()].select_dtypes(exclude=[object,'category']).columns.tolist()
for a, b in tqdm(zip(p0_cols, p1_cols), total=len(p0_cols)):
df["{}_{}_diff".format(a,b)] = df[a] - df[b]
return df
tr_v7 = diff_all(tr_v7)
te_v7 = diff_all(te_v7)
best_feat = pd.read_csv('../dacon_sc/feats/feat2k.csv')
best_feat.head()
tr_v7 = tr_v7.merge(tr_df, on='game_id', how='left')
te_v7 = te_v7.merge(te_df, on='game_id', how='left')
tr_v7.head()
len(best_feat['Feature'].tolist())
base_cols = tr_df.columns[1:].tolist()
sel_cols = base_cols + best_feat['Feature'].tolist()
print(len(sel_cols))
import time
from sklearn import metrics
from operator import itemgetter
import lightgbm as lgb
from sklearn.model_selection import KFold
def LGB_KFOLD_BINA(n_fold, train_X, test_X, metric, lr, num_leaves, max_depth):
folds = KFold(n_splits=n_fold, shuffle=True, random_state=42)
oof_lgb = np.zeros(len(train_X))
predictions = np.zeros(len(test_X))
feature_importance_df = pd.DataFrame()
cv_score_df = []
# Model parameters
lgb_params = {'num_leaves': num_leaves,
'min_data_in_leaf': 20,
'objective':'binary',
'max_depth': max_depth,
'learning_rate': lr,
"boosting": "gbdt",
"feature_fraction": 0.3,
"bagging_freq": 1,
"bagging_fraction": 0.7,
"bagging_seed": 42,
"metric": metric,
"lambda_l1": 0.0,
"verbosity": 300,
"nthread": -1,
"random_state": 42}
model_start = time.time()
for fold_, (trn_idx, val_idx) in enumerate(folds.split(train_X)):
strLog = "fold {}".format(fold_)
print(strLog+ "-" * 50)
X_tr, X_val = train_X.iloc[trn_idx], train_X.iloc[val_idx]
y_tr, y_val = train_Y.iloc[trn_idx], train_Y.iloc[val_idx]
model = lgb.LGBMClassifier(**lgb_params, n_estimators = 200000, n_jobs = -1)
model.fit(X_tr,
y_tr,
eval_set=[(X_tr, y_tr), (X_val, y_val)],
eval_metric=metric,
verbose=300,
early_stopping_rounds=200)
oof_lgb[val_idx] = model.predict_proba(X_val, num_iteration=model.best_iteration_)[:,1]
cv_score_df.append(model.best_score_)
#feature importance
fold_importance_df = pd.DataFrame()
fold_importance_df["Feature"] = train_X.columns
fold_importance_df["importance"] = model.feature_importances_[:len(train_X.columns)]
fold_importance_df["fold"] = fold_ + 1
feature_importance_df = pd.concat([feature_importance_df, fold_importance_df], axis=0)
#predictions
predictions += model.predict_proba(test_X, num_iteration=model.best_iteration_)[:,1] / folds.n_splits
cv_score_df = pd.DataFrame.from_dict(cv_score_df)
cv_score_df = cv_score_df.valid_1.tolist()
cv_score_df = list(map(itemgetter(metric),cv_score_df))
print("-" * 50)
#print("SF RMSE = {}".format(oof_score))
print("Mean "+ metric + " = {}".format(np.mean(cv_score_df)))
print("Std "+ metric + " = {}".format(np.std(cv_score_df)))
# lgb.plot_metric(model, metric=metric, title='auc plot', xlabel='Iterations', ylabel='auto', figsize=(10,8), grid=False)
model_end = time.time()
model_elapsed = model_end - model_start
print('Model elapsed {0:0.2f}'.format(model_elapsed/60), "minutes.")
cols = (feature_importance_df[["Feature", "importance"]]
.groupby("Feature")
.mean()
.sort_values(by="importance", ascending=False)[:].index)
best_features = feature_importance_df.loc[feature_importance_df.Feature.isin(cols)]
pd.set_option('display.max_rows', 500)
best_features = best_features.drop(['fold'],axis=1)
best_features = best_features.groupby(['Feature'], as_index = False).mean()
best_features['Feature Rank'] = best_features['importance'].rank(ascending=0)
best_features = best_features.sort_values('Feature Rank', ascending = True)
print(best_features.loc[best_features['importance']!=0].head(100))
return predictions, oof_lgb, np.mean(cv_score_df), np.std(cv_score_df), best_features
tr_v7[sel_cols].dtypes.value_counts()
tr_df.dtypes.value_counts()
# tr_tsp.fillna(0, inplace=True)
# te_tsp.fillna(0, inplace=True)
#early stopping 200, lr=0.01
pred, oof, cv, cv_std, best_feat = LGB_KFOLD_BINA(5, tr_v7[sel_cols], te_v7[sel_cols], "auc", lr=0.03, num_leaves=700, max_depth=-1)
submission = pd.read_csv('../dacon_sc/input/sample_submission.csv')
submission['winner'] = pred
submission.describe()
sns.distplot(submission['winner'])
cv
submission.to_csv("../dacon_sc/sub/final_{}.csv".format(cv), index=False)
```
|
github_jupyter
|
import pandas as pd, numpy as np
import os, time, random,gc
from tqdm import tqdm
import matplotlib.pyplot as plt
import seaborn as sns
pd.options.mode.chained_assignment = None
os.listdir('input')
def seed_everything(seed):
random.seed(seed)
os.environ['PYTHONHASHSEED'] = str(seed)
np.random.seed(seed)
SEED = 42
seed_everything(SEED)
%%time
tr = pd.read_feather('../dacon_sc/input/tr.feather')
te = pd.read_feather('../dacon_sc/input/te.feather')
tr.head()
tr_y = tr.groupby(['game_id'])['winner'].max()
tr_y = tr_y.reset_index()
train_Y = tr_y['winner']
tr_y.head()
tr.shape
tr.head()
def make_timespan(df):
df['time_span'] = 0
df['time_span'] = np.where(df['time']<1, 0,
np.where(df['time']<2, 1,
np.where(df['time']<3, 2,
np.where(df['time']<4, 3,
np.where(df['time']<5, 4,
np.where(df['time']<6, 5,
np.where(df['time']<7, 6,
np.where(df['time']<8, 7,
np.where(df['time']<9, 8,
np.where(df['time']<10, 9, 10))))))))))
return df
def tsp_cnt(df):
df = make_timespan(df)
new_p0 = pd.DataFrame({"game_id":df['game_id'].unique()})
p0 = df[df['player']==0]
for tsp in tqdm(range(11)):
tmp = p0[p0['time_span']==tsp].groupby(['game_id'])['time'].count().reset_index().rename(columns={"time":"p0_tsp_{}_cnt".format(tsp)})
# tmp.drop(['time_span'],axis=1, inplace=True)
new_p0 = new_p0.merge(tmp, on='game_id', how='left')
new_p1 = pd.DataFrame({"game_id":df['game_id'].unique()})
p1 = df[df['player']==1]
for tsp in tqdm(range(11)):
tmp = p1[p1['time_span']==tsp].groupby(['game_id'])['time'].count().reset_index().rename(columns={"time":"p1_tsp_{}_cnt".format(tsp)})
# tmp.drop(['time_span'],axis=1, inplace=True)
new_p1 = new_p1.merge(tmp, on='game_id', how='left')
new_df = new_p0.merge(new_p1, on='game_id', how='left')
return new_df
tr_tsp = tsp_cnt(tr)
te_tsp = tsp_cnt(te)
tr_tsp.head()
te_tsp.head()
def get_sp(df):
df_tmp = pd.DataFrame(df.game_id.unique(), columns=['game_id'])
df_tmp.index = df_tmp.game_id
df_tmp = df_tmp.drop(['game_id'], axis = 1)
p0 = df[(df.event=='Camera')&(df.player==0)]
p0 = p0[p0.shift(1).game_id!=p0.game_id] # 쉬프트를 이용하여 각 게임의 첫번째 데이터 찾기
p0 = p0.loc[:, ['game_id','event_contents']].rename({'event_contents':'player0_starting'}, axis = 1)
p0.index = p0['game_id']
p0 = p0.drop(['game_id'], axis=1)
df_tmp = pd.merge(df_tmp, p0, on='game_id', how='left')
del p0
p1 = df[(df.event=='Camera')&(df.player==1)]
p1 = p1[p1.shift(1).game_id!=p1.game_id]
p1 = p1.loc[:, ['game_id','event_contents']].rename({'event_contents':'player1_starting'}, axis = 1)
p1.index = p1['game_id']
p1 = p1.drop(['game_id'], axis=1)
df_tmp = pd.merge(df_tmp, p1, on='game_id', how='left')
del p1
df_tmp['player0_starting'] = df_tmp.player0_starting.str.split('(').str[1]
df_tmp['player0_starting'] = df_tmp.player0_starting.str.split(')').str[0]
split_xy = df_tmp.player0_starting.str.split(',')
df_tmp['player0_x'] = split_xy.str[0].astype('float')
df_tmp['player0_y'] = split_xy.str[1].astype('float')
del split_xy
df_tmp['player1_starting'] = df_tmp.player1_starting.str.split('(').str[1]
df_tmp['player1_starting'] = df_tmp.player1_starting.str.split(')').str[0]
split_xy = df_tmp.player1_starting.str.split(',')
df_tmp['player1_x'] = split_xy.str[0].astype('float')
df_tmp['player1_y'] = split_xy.str[1].astype('float')
del split_xy
# location_p0 = df_tmp.loc[:, ['player0_x', 'player0_y']]
# location_p0 = location_p0.rename({'player0_x':'location_x', 'player0_y':'location_y'}, axis=1)
# location_p1 = df_tmp.loc[:, ['player1_x', 'player1_y']]
# location_p1 = location_p1.rename({'player1_x':'location_x', 'player1_y':'location_y'}, axis=1)
# location_p1.index += location_p0.index[-1]+1
# location = pd.concat([location_p0, location_p1])
# location = location.dropna()
# del location_p0, location_p1, df_tmp
df_tmp.fillna(-999, inplace=True)
df_tmp['p0_sp'] = df_tmp['player0_x'].astype(int).astype(str) + df_tmp['player0_y'].astype(int).astype(str)
df_tmp['p1_sp'] = df_tmp['player1_x'].astype(int).astype(str) + df_tmp['player1_y'].astype(int).astype(str)
df_tmp['p0_sp2'] = np.round(df_tmp['player0_x'],2).astype(str) + np.round(df_tmp['player0_y'],2).astype(str)
df_tmp['p1_sp2'] = np.round(df_tmp['player1_x'],2).astype(str) + np.round(df_tmp['player1_y'],2).astype(str)
df_tmp['all_sp'] = df_tmp['p0_sp']+df_tmp['p1_sp']
df_tmp['all_sp2'] = df_tmp['p0_sp2']+df_tmp['p1_sp2']
return df_tmp
%%time
tr_sp = get_sp(tr)
te_sp = get_sp(te)
tr_sp.head()
te_sp.head()
tr_sp.nunique()
te_sp.nunique()
tra = tr.merge(tr_sp.iloc[:,-6:], on='game_id', how='left')
tra.head()
trp0 = tr[tr['player']==0].groupby(['game_id','time_span'])['time'].count().reset_index().rename(columns={"time":"p0_cnt"})
trp1 = tr[tr['player']==1].groupby(['game_id','time_span'])['time'].count().reset_index().rename(columns={"time":"p1_cnt"})
trcnt = trp0.merge(trp1, on=['game_id','time_span'], how='left')
trcnt = trcnt.merge(tr_sp.iloc[:,-6:], on='game_id', how='left')
tep0 = te[te['player']==0].groupby(['game_id','time_span'])['time'].count().reset_index().rename(columns={"time":"p0_cnt"})
tep1 = te[te['player']==1].groupby(['game_id','time_span'])['time'].count().reset_index().rename(columns={"time":"p1_cnt"})
tecnt = tep0.merge(tep1, on=['game_id','time_span'], how='left')
tecnt = tecnt.merge(te_sp.iloc[:,-6:], on='game_id', how='left')
tecnt.head(20)
def get_spc(df):
p0_spc = df[df['player']==0].groupby(['game_id'])['species'].max().reset_index().rename(columns={"species":'p0_spc'})
p1_spc = df[df['player']==1].groupby(['game_id'])['species'].max().reset_index().rename(columns={"species":'p1_spc'})
p0_spc = p0_spc.merge(p1_spc, on='game_id', how='left')
return p0_spc
%%time
tr_spc = get_spc(tr)
te_spc = get_spc(te)
tr_spc.head()
trcnt = trcnt.merge(tr_spc, on='game_id', how='left')
tecnt = tecnt.merge(te_spc, on='game_id', how='left')
trcnt['game_spc'] = trcnt['p0_spc']+trcnt['p1_spc']
tecnt['game_spc'] = tecnt['p0_spc']+tecnt['p1_spc']
trcnt['game_allsp_spc'] = trcnt['all_sp']+trcnt['p0_spc']+trcnt['p1_spc']
trcnt['game_allsp2_spc'] = trcnt['all_sp2']+trcnt['p0_spc']+trcnt['p1_spc']
tecnt['game_allsp_spc'] = tecnt['all_sp']+tecnt['p0_spc']+tecnt['p1_spc']
tecnt['game_allsp2_spc'] = tecnt['all_sp2']+tecnt['p0_spc']+tecnt['p1_spc']
trcnt['game_spc_sort'] = trcnt['game_spc'].apply(lambda x: sorted(x))
tecnt['game_spc_sort'] = tecnt['game_spc'].apply(lambda x: sorted(x))
trcnt.head()
tecnt.tail()
trcnt.shape,tecnt.shape
def df_copy(tr_df, te_df):
tr = tr_df.copy();te = te_df.copy()
return tr, te
def sub_day_agg(tr_df, te_df, merge_columns, date_columns, columns, aggs=['mean']):
tr, te = df_copy(tr_df, te_df)
for merge_column in merge_columns:
for date in date_columns:
tr['mc_date'] = tr[merge_column].astype(str) + '_' +tr[date].astype(str)
te['mc_date'] = te[merge_column].astype(str) + '_' +te[date].astype(str)
for col in columns:
for agg in aggs:
valid = pd.concat([tr[['mc_date', col]], te[['mc_date', col]]])
new_cn = merge_column + '_' + date + '_' + col + '_' + agg
if agg=='quantile':
valid = valid.groupby('mc_date')[col].quantile(0.8).reset_index().rename(columns={col:new_cn})
else:
valid = valid.groupby('mc_date')[col].agg([agg]).reset_index().rename(columns={agg:new_cn})
valid.index = valid['mc_date'].tolist()
valid = valid[new_cn].to_dict()
tr[new_cn] = tr['mc_date'].map(valid)
te[new_cn] = te['mc_date'].map(valid)
tr = tr.drop(columns=['mc_date'])
te = te.drop(columns=['mc_date'])
return tr, te
traa, teaa = sub_day_agg(trcnt, tecnt, merge_columns=['all_sp','all_sp2','p0_spc','p1_spc','game_allsp_spc','game_spc_sort','game_allsp2_spc'],
date_columns=['time_span'], columns=['p0_cnt', 'p1_cnt'], aggs=['mean'])
traa.head()
def sub_day_agg_flat(df):
new_df = pd.DataFrame({"game_id":df['game_id'].unique()})
for tsp in tqdm(range(11)):
tmp = df[df['time_span']==tsp][['game_id','all_sp_time_span_p0_cnt_mean'
,'all_sp_time_span_p1_cnt_mean'
,'all_sp2_time_span_p0_cnt_mean'
,'all_sp2_time_span_p1_cnt_mean'
,'p0_spc_time_span_p0_cnt_mean'
,'p0_spc_time_span_p1_cnt_mean'
,'p1_spc_time_span_p0_cnt_mean'
,'p1_spc_time_span_p1_cnt_mean'
,'game_allsp_spc_time_span_p0_cnt_mean'
,'game_allsp_spc_time_span_p1_cnt_mean'
,'game_spc_sort_time_span_p0_cnt_mean'
,'game_spc_sort_time_span_p1_cnt_mean'
,'game_allsp2_spc_time_span_p0_cnt_mean'
,'game_allsp2_spc_time_span_p1_cnt_mean'
]]
tmp.rename(columns={"all_sp_time_span_p0_cnt_mean":"all_sp_time_span_p0_cnt_mean_t{}".format(tsp)
,"all_sp_time_span_p1_cnt_mean":"all_sp_time_span_p1_cnt_mean_t{}".format(tsp)
,"all_sp2_time_span_p0_cnt_mean":"all_sp2_time_span_p0_cnt_mean_t{}".format(tsp)
,"all_sp2_time_span_p1_cnt_mean":"all_sp2_time_span_p1_cnt_mean_t{}".format(tsp)
,"p0_spc_time_span_p0_cnt_mean":"p0_spc_time_span_p0_cnt_mean_t{}".format(tsp)
,"p0_spc_time_span_p1_cnt_mean":"p0_spc_time_span_p1_cnt_mean_t{}".format(tsp)
,"p1_spc_time_span_p0_cnt_mean":"p1_spc_time_span_p0_cnt_mean_t{}".format(tsp)
,"p1_spc_time_span_p1_cnt_mean":"p1_spc_time_span_p1_cnt_mean_t{}".format(tsp)
,"game_allsp_spc_time_span_p0_cnt_mean":"game_allsp_spc_time_span_p0_cnt_mean_t{}".format(tsp)
,"game_allsp_spc_time_span_p1_cnt_mean":"game_allsp_spc_time_span_p1_cnt_mean_t{}".format(tsp)
,"game_spc_sort_time_span_p0_cnt_mean":"game_spc_sort_time_span_p0_cnt_mean_t{}".format(tsp)
,"game_spc_sort_time_span_p1_cnt_mean":"game_spc_sort_time_span_p1_cnt_mean_t{}".format(tsp)
,"game_allsp2_spc_time_span_p0_cnt_mean":"game_allsp2_spc_time_span_p0_cnt_mean_t{}".format(tsp)
,"game_allsp2_spc_time_span_p1_cnt_mean":"game_allsp2_spc_time_span_p1_cnt_mean_t{}".format(tsp)
},inplace=True)
new_df = new_df.merge(tmp, on='game_id', how='left')
return new_df
tr_sub = sub_day_agg_flat(traa)
te_sub = sub_day_agg_flat(teaa)
tr_sub.head()
len(list(set(tr_sub.columns.tolist())))
te_sub.head()
tr_tsp.head()
tr_df = tr_tsp.merge(tr_sub, on='game_id', how='left')
te_df = te_tsp.merge(te_sub, on='game_id', how='left')
tr_df.head()
tr_df = tr_df.merge(tr_spc, on='game_id', how='left')
te_df = te_df.merge(te_spc, on='game_id', how='left')
tr_df.head()
tr_sp.dtypes
tr_sp.select_dtypes(include=[object]).columns
tr_df = tr_df.merge(tr_sp.iloc[:,-6:], on='game_id', how='left')
te_df = te_df.merge(te_sp.iloc[:,-6:], on='game_id', how='left')
for df in tr_df, te_df:
for c in df.select_dtypes(include=[object]).columns.tolist():
df[c] = df[c].astype('category')
def get_spc_sort(df):
df['game_spc'] = df['p0_spc'].astype(str)+df['p1_spc'].astype(str)
df['game_allsp_spc'] = df['all_sp'].astype(str)+df['p0_spc'].astype(str)+df['p1_spc'].astype(str)
df['game_allsp2_spc'] = df['all_sp2'].astype(str)+df['p0_spc'].astype(str)+df['p1_spc'].astype(str)
df['game_spc_sort'] = df['game_spc'].astype(str).apply(lambda x: sorted(x))
df['game_spc_sort'] = df['game_spc_sort'].astype(str)
return df
tr_df = get_spc_sort(tr_df)
te_df = get_spc_sort(te_df)
for df in tr_df, te_df:
for c in df.select_dtypes(include=[object]).columns.tolist():
df[c] = df[c].astype('category')
tr_df.dtypes.value_counts()
tr_df.head()
tr_df.columns
tr_df.to_feather('../dacon_sc/input/tr_df.feather')
te_df.to_feather('../dacon_sc/input/te_df.feather')
tr_time_eve = pd.read_feather("../dacon_sc/input/tr_time_eve.feather")
te_time_eve = pd.read_feather("../dacon_sc/input/te_time_eve.feather")
tr_df = tr_df.merge(tr_time_eve, on='game_id', how='left')
te_df = te_df.merge(te_time_eve, on='game_id', how='left')
tr_df.head()
tr_df.head()
%%time
tr_v7 = pd.read_feather('../dacon_sc/input/tr_v7.feather')
te_v7 = pd.read_feather('../dacon_sc/input/te_v7.feather')
def diff_all(df):
p0_cols = df[df.filter(regex='p0').columns.tolist()].select_dtypes(exclude=[object,'category']).columns.tolist()
p1_cols = df[df.filter(regex='p1').columns.tolist()].select_dtypes(exclude=[object,'category']).columns.tolist()
for a, b in tqdm(zip(p0_cols, p1_cols), total=len(p0_cols)):
df["{}_{}_diff".format(a,b)] = df[a] - df[b]
return df
tr_v7 = diff_all(tr_v7)
te_v7 = diff_all(te_v7)
best_feat = pd.read_csv('../dacon_sc/feats/feat2k.csv')
best_feat.head()
tr_v7 = tr_v7.merge(tr_df, on='game_id', how='left')
te_v7 = te_v7.merge(te_df, on='game_id', how='left')
tr_v7.head()
len(best_feat['Feature'].tolist())
base_cols = tr_df.columns[1:].tolist()
sel_cols = base_cols + best_feat['Feature'].tolist()
print(len(sel_cols))
import time
from sklearn import metrics
from operator import itemgetter
import lightgbm as lgb
from sklearn.model_selection import KFold
def LGB_KFOLD_BINA(n_fold, train_X, test_X, metric, lr, num_leaves, max_depth):
folds = KFold(n_splits=n_fold, shuffle=True, random_state=42)
oof_lgb = np.zeros(len(train_X))
predictions = np.zeros(len(test_X))
feature_importance_df = pd.DataFrame()
cv_score_df = []
# Model parameters
lgb_params = {'num_leaves': num_leaves,
'min_data_in_leaf': 20,
'objective':'binary',
'max_depth': max_depth,
'learning_rate': lr,
"boosting": "gbdt",
"feature_fraction": 0.3,
"bagging_freq": 1,
"bagging_fraction": 0.7,
"bagging_seed": 42,
"metric": metric,
"lambda_l1": 0.0,
"verbosity": 300,
"nthread": -1,
"random_state": 42}
model_start = time.time()
for fold_, (trn_idx, val_idx) in enumerate(folds.split(train_X)):
strLog = "fold {}".format(fold_)
print(strLog+ "-" * 50)
X_tr, X_val = train_X.iloc[trn_idx], train_X.iloc[val_idx]
y_tr, y_val = train_Y.iloc[trn_idx], train_Y.iloc[val_idx]
model = lgb.LGBMClassifier(**lgb_params, n_estimators = 200000, n_jobs = -1)
model.fit(X_tr,
y_tr,
eval_set=[(X_tr, y_tr), (X_val, y_val)],
eval_metric=metric,
verbose=300,
early_stopping_rounds=200)
oof_lgb[val_idx] = model.predict_proba(X_val, num_iteration=model.best_iteration_)[:,1]
cv_score_df.append(model.best_score_)
#feature importance
fold_importance_df = pd.DataFrame()
fold_importance_df["Feature"] = train_X.columns
fold_importance_df["importance"] = model.feature_importances_[:len(train_X.columns)]
fold_importance_df["fold"] = fold_ + 1
feature_importance_df = pd.concat([feature_importance_df, fold_importance_df], axis=0)
#predictions
predictions += model.predict_proba(test_X, num_iteration=model.best_iteration_)[:,1] / folds.n_splits
cv_score_df = pd.DataFrame.from_dict(cv_score_df)
cv_score_df = cv_score_df.valid_1.tolist()
cv_score_df = list(map(itemgetter(metric),cv_score_df))
print("-" * 50)
#print("SF RMSE = {}".format(oof_score))
print("Mean "+ metric + " = {}".format(np.mean(cv_score_df)))
print("Std "+ metric + " = {}".format(np.std(cv_score_df)))
# lgb.plot_metric(model, metric=metric, title='auc plot', xlabel='Iterations', ylabel='auto', figsize=(10,8), grid=False)
model_end = time.time()
model_elapsed = model_end - model_start
print('Model elapsed {0:0.2f}'.format(model_elapsed/60), "minutes.")
cols = (feature_importance_df[["Feature", "importance"]]
.groupby("Feature")
.mean()
.sort_values(by="importance", ascending=False)[:].index)
best_features = feature_importance_df.loc[feature_importance_df.Feature.isin(cols)]
pd.set_option('display.max_rows', 500)
best_features = best_features.drop(['fold'],axis=1)
best_features = best_features.groupby(['Feature'], as_index = False).mean()
best_features['Feature Rank'] = best_features['importance'].rank(ascending=0)
best_features = best_features.sort_values('Feature Rank', ascending = True)
print(best_features.loc[best_features['importance']!=0].head(100))
return predictions, oof_lgb, np.mean(cv_score_df), np.std(cv_score_df), best_features
tr_v7[sel_cols].dtypes.value_counts()
tr_df.dtypes.value_counts()
# tr_tsp.fillna(0, inplace=True)
# te_tsp.fillna(0, inplace=True)
#early stopping 200, lr=0.01
pred, oof, cv, cv_std, best_feat = LGB_KFOLD_BINA(5, tr_v7[sel_cols], te_v7[sel_cols], "auc", lr=0.03, num_leaves=700, max_depth=-1)
submission = pd.read_csv('../dacon_sc/input/sample_submission.csv')
submission['winner'] = pred
submission.describe()
sns.distplot(submission['winner'])
cv
submission.to_csv("../dacon_sc/sub/final_{}.csv".format(cv), index=False)
| 0.099623 | 0.284571 |
```
from numpy import *
import matplotlib.cm as cm
import matplotlib.pyplot as plt
import AxionFuncs
import Like
# Generate Tabulated spectra
nm = 10000
m_vals = logspace(-4.0,2e0,nm)
E_max = 20.0
E0= 50.0e-3
nE_bins = 100
E_bins,R1_tab,R0 = AxionFuncs.BinnedPhotonNumberTable(m_vals,E0,E_max,nE_bins,nfine=500,res_on=False)
m_DL_vals = [3e-3,8e-3,2e-2,1.3e-1]
DL = Like.MassDiscoveryLimit_Simple(m_vals,R1_tab,R0,m_DL_vals)
err_m = [1,1,1,1]
err_g = [0.01,0.3,0.6,0.99]
Lmin = [-1000,-1000,-1000,-1000]
Lmax = [100,100,100,100]
fig = plt.figure(figsize=(5,5))
ax = fig.add_subplot(111)
isel = 3
m0 = m_DL_vals[isel]
g0 = DL[isel]
nL = 500
N_obs = Like.InterpExpectedEvents(g0,m0,m_vals,R1_tab)
mi = linspace(m_vals[1],(m0*(1+err_m[isel])),nL)
gi = linspace(g0*(1-err_g[isel]),g0*(1+err_g[isel]),nL)
LL = zeros(shape=(nL,nL))
for ii in range(0,nL):
for jj in range(0,nL):
m = mi[jj]
g = gi[ii]
LL[ii,jj] = Like.llhood2([log10(g),m],N_obs,m_vals,R1_tab)
LL = -2*(LL-LL.min())
Lprof_m = zeros(shape=nL)
Lprof_g = zeros(shape=nL)
for i in range(0,nL):
imin = argmax(LL[i,:])
jmin = argmax(LL[:,i])
Lprof_g[i] = LL[i,jmin]
Lprof_m[i] = Like.llhood2_marg(mi[i],N_obs,m_vals,R1_tab)
Lprof_m = -2*(Lprof_m-Lprof_m.min())
plt.contourf(mi,gi,LL,50)
plt.contour(mi,gi,LL,levels = [-6.2,-2.3],colors="Red")
plt.show()
from matplotlib.ticker import NullFormatter, NullLocator, MultipleLocator
import matplotlib.colors as colors
markerz = ['o','s','^','d']
y = 100*(gi-g0)/g0
# plot the result
fig = plt.figure(figsize=(14.3, 13))
plt.rcParams['axes.linewidth'] = 2
plt.rcParams['axes.labelsize'] = 30
plt.rc('text', usetex=True)
plt.rc('font', family='serif')
# define axes
width_main = 0.52
height_main = 0.52
width_cb = 0.02
width_side = 0.2
ax_Pxy = plt.axes((width_side, width_side, width_main, height_main))
ax_Px = plt.axes((width_side,0, width_main, width_side))
ax_Py = plt.axes((0,width_side, width_side, height_main))
ax_cb = plt.axes((width_side+width_main, width_side-0.025, width_cb, height_main+0.025))
# draw the joint probability
plt.axes(ax_Pxy)
cmap = plt.get_cmap('Greens')
new_cmap = colors.LinearSegmentedColormap.from_list(
'trunc({n},{a:.2f},{b:.2f})'.format(n=cmap.name, a=0, b=0.9),
cmap(linspace(0, 0.9, 100)))
plt.contourf(mi,y,(LL),linspace(Lmin[isel],0,30),cmap=new_cmap,extend='min')
plt.contourf(mi,y,(LL),linspace(Lmin[isel],0,30),cmap=new_cmap,extend='min')
plt.contourf(mi,y,(LL),linspace(Lmin[isel],0,30),cmap=new_cmap,extend='min')
plt.plot([m0,m0],[y[0],0],'k-')
plt.plot([mi[0],m0],[0,0],'k-')
plt.plot(m0,0,markerz[isel],color='Crimson',markersize=25,markeredgecolor='k',markeredgewidth=2)
plt.text(mi[0]+0.05*(mi[-1]-mi[0]),y[0]+0.9*(y[-1]-y[0]),r'$m_a = $ '+str(m_DL_vals[isel])+' eV',fontsize=35)
cb = plt.colorbar(cax=ax_cb)
cb = plt.colorbar(cax=ax_cb)
cb = plt.colorbar(cax=ax_cb,ticks=arange(-1000,0,100))
cb.set_label(r'$2 \Delta \ln \mathcal{L}(m_a,g_{a\gamma})$',labelpad=35,rotation=-90)
# Draw extra contour lines
plt.axes(ax_Pxy)
plt.contourf(mi,y,(LL),levels = [-6.1801,0],colors="Crimson",linestyles = 'none',alpha=0.6)
plt.contourf(mi,y,(LL),levels = [-2.2958,0],colors="Crimson",linestyles = 'none',alpha=0.7)
plt.contour(mi,y,(LL),levels = [-2.2958],colors="k",linestyles = 'solid')
plt.contour(mi,y,(LL),levels = [-6.1801],colors="k",linestyles = 'solid')
# draw p(x) distribution
ax_Px.plot(mi, Lprof_m, '-',color='Crimson',linewidth=3)
# draw p(y) distribution
ax_Py.plot(Lprof_g, y, '-',color='Crimson',linewidth=3)
# define axis limits
ax_Pxy.set_xlim(mi[0], mi[-1])
ax_Pxy.set_ylim(y[0], y[-1])
ax_Px.set_xlim(mi[0], mi[-1])
ax_Px.set_ylim(-10,0)
ax_Py.set_ylim(y[0], y[-1])
ax_Py.set_xlim(-10,0)
# label axes
ax_Px.set_xlabel(r'$m_a$ [eV]')
ax_Px.yaxis.set_label_position('left')
ax_Py.set_ylabel(r'$(g_{a\gamma}-\hat{g}_{a\gamma})/\hat{g}_{a\gamma}$ [\%]')
ax_Py.xaxis.set_label_position('top')
ax_Py.set_xlabel('$2\Delta \ln \mathcal{L}(g_{a\gamma})$',labelpad=10,fontsize=27)
ax_Px.set_ylabel('$2 \Delta \ln \mathcal{L}(m_a)$',fontsize=27)
ax_Py.set_xticks([-9,-4,-1])
ax_Py.set_xticklabels(['-9','-4',''])
ax_Px.set_yticks([-9,-4,-1])
ax_Px.yaxis.grid()
ax_Py.xaxis.grid()
ax_Pxy.xaxis.set_major_formatter(NullFormatter())
ax_Pxy.yaxis.set_major_formatter(NullFormatter())
ax_Py.tick_params(which='major',direction='in',width=2,length=10,labelsize=25)
ax_Px.tick_params(which='major',direction='in',width=2,length=10,labelsize=25)
ax_cb.tick_params(which='major',direction='out',width=2,length=10,labelsize=20)
plt.show()
fig.savefig('../plots/Like'+str(isel)+'.pdf',bbox_inches='tight')
```
|
github_jupyter
|
from numpy import *
import matplotlib.cm as cm
import matplotlib.pyplot as plt
import AxionFuncs
import Like
# Generate Tabulated spectra
nm = 10000
m_vals = logspace(-4.0,2e0,nm)
E_max = 20.0
E0= 50.0e-3
nE_bins = 100
E_bins,R1_tab,R0 = AxionFuncs.BinnedPhotonNumberTable(m_vals,E0,E_max,nE_bins,nfine=500,res_on=False)
m_DL_vals = [3e-3,8e-3,2e-2,1.3e-1]
DL = Like.MassDiscoveryLimit_Simple(m_vals,R1_tab,R0,m_DL_vals)
err_m = [1,1,1,1]
err_g = [0.01,0.3,0.6,0.99]
Lmin = [-1000,-1000,-1000,-1000]
Lmax = [100,100,100,100]
fig = plt.figure(figsize=(5,5))
ax = fig.add_subplot(111)
isel = 3
m0 = m_DL_vals[isel]
g0 = DL[isel]
nL = 500
N_obs = Like.InterpExpectedEvents(g0,m0,m_vals,R1_tab)
mi = linspace(m_vals[1],(m0*(1+err_m[isel])),nL)
gi = linspace(g0*(1-err_g[isel]),g0*(1+err_g[isel]),nL)
LL = zeros(shape=(nL,nL))
for ii in range(0,nL):
for jj in range(0,nL):
m = mi[jj]
g = gi[ii]
LL[ii,jj] = Like.llhood2([log10(g),m],N_obs,m_vals,R1_tab)
LL = -2*(LL-LL.min())
Lprof_m = zeros(shape=nL)
Lprof_g = zeros(shape=nL)
for i in range(0,nL):
imin = argmax(LL[i,:])
jmin = argmax(LL[:,i])
Lprof_g[i] = LL[i,jmin]
Lprof_m[i] = Like.llhood2_marg(mi[i],N_obs,m_vals,R1_tab)
Lprof_m = -2*(Lprof_m-Lprof_m.min())
plt.contourf(mi,gi,LL,50)
plt.contour(mi,gi,LL,levels = [-6.2,-2.3],colors="Red")
plt.show()
from matplotlib.ticker import NullFormatter, NullLocator, MultipleLocator
import matplotlib.colors as colors
markerz = ['o','s','^','d']
y = 100*(gi-g0)/g0
# plot the result
fig = plt.figure(figsize=(14.3, 13))
plt.rcParams['axes.linewidth'] = 2
plt.rcParams['axes.labelsize'] = 30
plt.rc('text', usetex=True)
plt.rc('font', family='serif')
# define axes
width_main = 0.52
height_main = 0.52
width_cb = 0.02
width_side = 0.2
ax_Pxy = plt.axes((width_side, width_side, width_main, height_main))
ax_Px = plt.axes((width_side,0, width_main, width_side))
ax_Py = plt.axes((0,width_side, width_side, height_main))
ax_cb = plt.axes((width_side+width_main, width_side-0.025, width_cb, height_main+0.025))
# draw the joint probability
plt.axes(ax_Pxy)
cmap = plt.get_cmap('Greens')
new_cmap = colors.LinearSegmentedColormap.from_list(
'trunc({n},{a:.2f},{b:.2f})'.format(n=cmap.name, a=0, b=0.9),
cmap(linspace(0, 0.9, 100)))
plt.contourf(mi,y,(LL),linspace(Lmin[isel],0,30),cmap=new_cmap,extend='min')
plt.contourf(mi,y,(LL),linspace(Lmin[isel],0,30),cmap=new_cmap,extend='min')
plt.contourf(mi,y,(LL),linspace(Lmin[isel],0,30),cmap=new_cmap,extend='min')
plt.plot([m0,m0],[y[0],0],'k-')
plt.plot([mi[0],m0],[0,0],'k-')
plt.plot(m0,0,markerz[isel],color='Crimson',markersize=25,markeredgecolor='k',markeredgewidth=2)
plt.text(mi[0]+0.05*(mi[-1]-mi[0]),y[0]+0.9*(y[-1]-y[0]),r'$m_a = $ '+str(m_DL_vals[isel])+' eV',fontsize=35)
cb = plt.colorbar(cax=ax_cb)
cb = plt.colorbar(cax=ax_cb)
cb = plt.colorbar(cax=ax_cb,ticks=arange(-1000,0,100))
cb.set_label(r'$2 \Delta \ln \mathcal{L}(m_a,g_{a\gamma})$',labelpad=35,rotation=-90)
# Draw extra contour lines
plt.axes(ax_Pxy)
plt.contourf(mi,y,(LL),levels = [-6.1801,0],colors="Crimson",linestyles = 'none',alpha=0.6)
plt.contourf(mi,y,(LL),levels = [-2.2958,0],colors="Crimson",linestyles = 'none',alpha=0.7)
plt.contour(mi,y,(LL),levels = [-2.2958],colors="k",linestyles = 'solid')
plt.contour(mi,y,(LL),levels = [-6.1801],colors="k",linestyles = 'solid')
# draw p(x) distribution
ax_Px.plot(mi, Lprof_m, '-',color='Crimson',linewidth=3)
# draw p(y) distribution
ax_Py.plot(Lprof_g, y, '-',color='Crimson',linewidth=3)
# define axis limits
ax_Pxy.set_xlim(mi[0], mi[-1])
ax_Pxy.set_ylim(y[0], y[-1])
ax_Px.set_xlim(mi[0], mi[-1])
ax_Px.set_ylim(-10,0)
ax_Py.set_ylim(y[0], y[-1])
ax_Py.set_xlim(-10,0)
# label axes
ax_Px.set_xlabel(r'$m_a$ [eV]')
ax_Px.yaxis.set_label_position('left')
ax_Py.set_ylabel(r'$(g_{a\gamma}-\hat{g}_{a\gamma})/\hat{g}_{a\gamma}$ [\%]')
ax_Py.xaxis.set_label_position('top')
ax_Py.set_xlabel('$2\Delta \ln \mathcal{L}(g_{a\gamma})$',labelpad=10,fontsize=27)
ax_Px.set_ylabel('$2 \Delta \ln \mathcal{L}(m_a)$',fontsize=27)
ax_Py.set_xticks([-9,-4,-1])
ax_Py.set_xticklabels(['-9','-4',''])
ax_Px.set_yticks([-9,-4,-1])
ax_Px.yaxis.grid()
ax_Py.xaxis.grid()
ax_Pxy.xaxis.set_major_formatter(NullFormatter())
ax_Pxy.yaxis.set_major_formatter(NullFormatter())
ax_Py.tick_params(which='major',direction='in',width=2,length=10,labelsize=25)
ax_Px.tick_params(which='major',direction='in',width=2,length=10,labelsize=25)
ax_cb.tick_params(which='major',direction='out',width=2,length=10,labelsize=20)
plt.show()
fig.savefig('../plots/Like'+str(isel)+'.pdf',bbox_inches='tight')
| 0.504883 | 0.432483 |
```
import math
import random
import gym
import numpy as np
import torch
import torch.nn as nn
import torch.optim as optim
import torch.nn.functional as F
from torch.distributions import Normal
from IPython.display import clear_output
import matplotlib.pyplot as plt
%matplotlib inline
```
<h2>Use CUDA</h2>
```
use_cuda = torch.cuda.is_available()
device = torch.device("cuda" if use_cuda else "cpu")
```
<h2>Create Environments</h2>
```
from common.multiprocessing_env import SubprocVecEnv
num_envs = 16
env_name = "Pendulum-v0"
def make_env():
def _thunk():
env = gym.make(env_name)
return env
return _thunk
envs = [make_env() for i in range(num_envs)]
envs = SubprocVecEnv(envs)
env = gym.make(env_name)
```
<h2>Neural Network</h2>
```
def init_weights(m):
if isinstance(m, nn.Linear):
nn.init.normal_(m.weight, mean=0., std=0.1)
nn.init.constant_(m.bias, 0.1)
class ActorCritic(nn.Module):
def __init__(self, num_inputs, num_outputs, hidden_size, std=0.0):
super(ActorCritic, self).__init__()
self.critic = nn.Sequential(
nn.Linear(num_inputs, hidden_size),
nn.ReLU(),
nn.Linear(hidden_size, 1)
)
self.actor = nn.Sequential(
nn.Linear(num_inputs, hidden_size),
nn.ReLU(),
nn.Linear(hidden_size, num_outputs),
)
self.log_std = nn.Parameter(torch.ones(1, num_outputs) * std)
self.apply(init_weights)
def forward(self, x):
value = self.critic(x)
mu = self.actor(x)
std = self.log_std.exp().expand_as(mu)
dist = Normal(mu, std)
return dist, value
def plot(frame_idx, rewards):
clear_output(True)
plt.figure(figsize=(20,5))
plt.subplot(131)
plt.title('frame %s. reward: %s' % (frame_idx, rewards[-1]))
plt.plot(rewards)
plt.show()
def test_env(vis=False):
state = env.reset()
if vis: env.render()
done = False
total_reward = 0
while not done:
state = torch.FloatTensor(state).unsqueeze(0).to(device)
dist, _ = model(state)
next_state, reward, done, _ = env.step(dist.sample().cpu().numpy()[0])
state = next_state
if vis: env.render()
total_reward += reward
return total_reward
```
<h1>High-Dimensional Continuous Control Using Generalized Advantage Estimation</h1>
<h3><a href="https://arxiv.org/abs/1506.02438">Arxiv</a></h3>
```
def compute_gae(next_value, rewards, masks, values, gamma=0.99, tau=0.95):
values = values + [next_value]
gae = 0
returns = []
for step in reversed(range(len(rewards))):
delta = rewards[step] + gamma * values[step + 1] * masks[step] - values[step]
gae = delta + gamma * tau * masks[step] * gae
returns.insert(0, gae + values[step])
return returns
num_inputs = envs.observation_space.shape[0]
num_outputs = envs.action_space.shape[0]
#Hyper params:
hidden_size = 256
lr = 3e-2
num_steps = 20
model = ActorCritic(num_inputs, num_outputs, hidden_size).to(device)
optimizer = optim.Adam(model.parameters())
max_frames = 100000
frame_idx = 0
test_rewards = []
state = envs.reset()
while frame_idx < max_frames:
log_probs = []
values = []
rewards = []
masks = []
entropy = 0
for _ in range(num_steps):
state = torch.FloatTensor(state).to(device)
dist, value = model(state)
action = dist.sample()
next_state, reward, done, _ = envs.step(action.cpu().numpy())
log_prob = dist.log_prob(action)
entropy += dist.entropy().mean()
log_probs.append(log_prob)
values.append(value)
rewards.append(torch.FloatTensor(reward).unsqueeze(1).to(device))
masks.append(torch.FloatTensor(1 - done).unsqueeze(1).to(device))
state = next_state
frame_idx += 1
if frame_idx % 1000 == 0:
test_rewards.append(np.mean([test_env() for _ in range(10)]))
plot(frame_idx, test_rewards)
next_state = torch.FloatTensor(next_state).to(device)
_, next_value = model(next_state)
returns = compute_gae(next_value, rewards, masks, values)
log_probs = torch.cat(log_probs)
returns = torch.cat(returns).detach()
values = torch.cat(values)
advantage = returns - values
actor_loss = -(log_probs * advantage.detach()).mean()
critic_loss = advantage.pow(2).mean()
loss = actor_loss + 0.5 * critic_loss - 0.001 * entropy
optimizer.zero_grad()
loss.backward()
optimizer.step()
test_env(True)
```
|
github_jupyter
|
import math
import random
import gym
import numpy as np
import torch
import torch.nn as nn
import torch.optim as optim
import torch.nn.functional as F
from torch.distributions import Normal
from IPython.display import clear_output
import matplotlib.pyplot as plt
%matplotlib inline
use_cuda = torch.cuda.is_available()
device = torch.device("cuda" if use_cuda else "cpu")
from common.multiprocessing_env import SubprocVecEnv
num_envs = 16
env_name = "Pendulum-v0"
def make_env():
def _thunk():
env = gym.make(env_name)
return env
return _thunk
envs = [make_env() for i in range(num_envs)]
envs = SubprocVecEnv(envs)
env = gym.make(env_name)
def init_weights(m):
if isinstance(m, nn.Linear):
nn.init.normal_(m.weight, mean=0., std=0.1)
nn.init.constant_(m.bias, 0.1)
class ActorCritic(nn.Module):
def __init__(self, num_inputs, num_outputs, hidden_size, std=0.0):
super(ActorCritic, self).__init__()
self.critic = nn.Sequential(
nn.Linear(num_inputs, hidden_size),
nn.ReLU(),
nn.Linear(hidden_size, 1)
)
self.actor = nn.Sequential(
nn.Linear(num_inputs, hidden_size),
nn.ReLU(),
nn.Linear(hidden_size, num_outputs),
)
self.log_std = nn.Parameter(torch.ones(1, num_outputs) * std)
self.apply(init_weights)
def forward(self, x):
value = self.critic(x)
mu = self.actor(x)
std = self.log_std.exp().expand_as(mu)
dist = Normal(mu, std)
return dist, value
def plot(frame_idx, rewards):
clear_output(True)
plt.figure(figsize=(20,5))
plt.subplot(131)
plt.title('frame %s. reward: %s' % (frame_idx, rewards[-1]))
plt.plot(rewards)
plt.show()
def test_env(vis=False):
state = env.reset()
if vis: env.render()
done = False
total_reward = 0
while not done:
state = torch.FloatTensor(state).unsqueeze(0).to(device)
dist, _ = model(state)
next_state, reward, done, _ = env.step(dist.sample().cpu().numpy()[0])
state = next_state
if vis: env.render()
total_reward += reward
return total_reward
def compute_gae(next_value, rewards, masks, values, gamma=0.99, tau=0.95):
values = values + [next_value]
gae = 0
returns = []
for step in reversed(range(len(rewards))):
delta = rewards[step] + gamma * values[step + 1] * masks[step] - values[step]
gae = delta + gamma * tau * masks[step] * gae
returns.insert(0, gae + values[step])
return returns
num_inputs = envs.observation_space.shape[0]
num_outputs = envs.action_space.shape[0]
#Hyper params:
hidden_size = 256
lr = 3e-2
num_steps = 20
model = ActorCritic(num_inputs, num_outputs, hidden_size).to(device)
optimizer = optim.Adam(model.parameters())
max_frames = 100000
frame_idx = 0
test_rewards = []
state = envs.reset()
while frame_idx < max_frames:
log_probs = []
values = []
rewards = []
masks = []
entropy = 0
for _ in range(num_steps):
state = torch.FloatTensor(state).to(device)
dist, value = model(state)
action = dist.sample()
next_state, reward, done, _ = envs.step(action.cpu().numpy())
log_prob = dist.log_prob(action)
entropy += dist.entropy().mean()
log_probs.append(log_prob)
values.append(value)
rewards.append(torch.FloatTensor(reward).unsqueeze(1).to(device))
masks.append(torch.FloatTensor(1 - done).unsqueeze(1).to(device))
state = next_state
frame_idx += 1
if frame_idx % 1000 == 0:
test_rewards.append(np.mean([test_env() for _ in range(10)]))
plot(frame_idx, test_rewards)
next_state = torch.FloatTensor(next_state).to(device)
_, next_value = model(next_state)
returns = compute_gae(next_value, rewards, masks, values)
log_probs = torch.cat(log_probs)
returns = torch.cat(returns).detach()
values = torch.cat(values)
advantage = returns - values
actor_loss = -(log_probs * advantage.detach()).mean()
critic_loss = advantage.pow(2).mean()
loss = actor_loss + 0.5 * critic_loss - 0.001 * entropy
optimizer.zero_grad()
loss.backward()
optimizer.step()
test_env(True)
| 0.895283 | 0.837055 |
# Matplotlib
matplotlib is a python 2D plotting library which produces publication quality figures in a variety of hardcopy formats and interactive environments across platforms. matplotlib can be used in python scripts, the python and ipython shell, web application servers, and many graphical user interface toolkits.
matplotlib tries to make easy things easy and hard things possible. You can generate plots, histograms, power spectra, bar charts, errorcharts, scatterplots, etc, with just a few lines of code. For a sampling, see the screenshots, thumbnail gallery, and examples directory
```
import numpy as np
%pylab inline
```
Let's create some graphics using a function that increase in its randomness in "linear space":
```
plot(np.linspace(0, 10, 100) * np.random.random(100))
```
Notice that many "defaults" such as color, line style, markings, etc. have been made for you -- and all of them look pretty good.
Let's try to modify these defaults.
```
plot(np.linspace(0, 10, 100) * np.random.random(100),
color='red', linewidth=2, linestyle="--")
```
Let's plot more than 1 line graph at a time where noise variance does NOT increase over the horizontal axis:
```
plot(np.linspace(0, 10, 100) + np.random.random(100),
color='red', linewidth=2, linestyle="--")
plot(np.linspace(0, 10, 100) - np.random.random(100),
color='blue', linewidth=2, linestyle="-")
```
Let's look at another type of visualization: histogram:
```
hist(np.linspace(0, 10, 100) + np.random.random(100) * 3,
color='green')
```
matplotlib has a lot of options, particularly with the recent release of matplotlib 2.0.
# Bokeh
Bokeh is a Python interactive visualization library that targets modern web browsers for presentation. Its goal is to provide elegant, concise construction of novel graphics in the style of D3.js, and to extend this capability with high-performance interactivity over very large or streaming datasets. Bokeh can help anyone who would like to quickly and easily create interactive plots, dashboards, and data applications.
```
import numpy as np
from bokeh.plotting import figure, output_file, show
```
Plotting data in basic Python lists as a line chart including zoom, pan, resize, save, and other tools is simple and straightforward:
```
a = np.arange(10)
b = np.random.randn(10)
```
Let's create a HTML page with interactive graphics.
```
output_file("lines.html")
```
create a new plot with a title and axis labels
```
p = figure(title="Bokeh Line",
x_axis_label='x',
y_axis_label='y')
```
add a line renderer with legend and line thickness
```
p.line(a, b, legend="Random Walk", line_width=2)
# let's finally see the results
show(p)
```
When you execute this script, you will see that a new output file "lines.html" is created, and that a browser automatically opens a new tab to display it.
That's pretty cool with almost no web development!
The basic steps to creating plots with the bokeh.plotting interface are:
- Prepare some data (in this case plain python lists).
- Tell Bokeh where to generate output (in this case using output_file(), with the filename "lines.html").
- Call figure() to create a plot with some overall options like title, tools and axes labels.
- Add renderers (in this case, Figure.line) for our data, with visual customizations like colors, legends and widths to the plot.
- Ask Bokeh to show() or save() the results.
- Steps three and four can be repeated to create more than one plot, as shown in some of the examples below.
The bokeh.plotting interface is also quite handy if we need to customize the output a bit more by adding more data series, glyphs, logarithmic axis, and so on. It’s also possible to easily combine multiple glyphs together on one plot as shown below:
```
x = np.arange(10)
y0 = [i**2 for i in x]
y1 = [10**i for i in x]
y2 = [10**(i**2) for i in x]
```
Output to a static HTML file
```
output_file("loglines.html")
```
Let's be specific about the "tools" we are enabling in this graphics: pan and zoom, box zoom, reset, and save.
```
p = figure(
tools="pan,box_zoom,reset,save",
y_axis_type="log", y_range=[0.001, 10**11], title="Bokeh with log axis",
x_axis_label='sections', y_axis_label='particles'
)
p.line(x, x, legend="y=x")
p.circle(x, x, legend="y=x", fill_color="white", size=8)
p.line(x, y0, legend="y=x^2", line_width=3)
p.line(x, y1, legend="y=10^x", line_color="red")
p.circle(x, y1, legend="y=10^x", fill_color="red", line_color="red", size=6)
p.line(x, y2, legend="y=10^x^2", line_color="orange", line_dash="4 4")
```
Remember to call show on the figure:
```
show(p)
```
## Bokeh Glyphs
Glyphs are the basic visual marks that Bokeh can display. At the lowest level, there are glyph objects, such as Line. If you are using the low-level bokeh.models interface, it is your responsibility to create and coordinate all the various Bokeh objects, including glyph objects and their data sources. To make life easier, the bokeh.plotting interface exposes higher level glyph methods such as the Figure.line method used in the first example. The second example also adds in calls to Figure.circle to display circle and line glyphs together on the same plot. Besides lines and circles, Bokeh makes many additional glyphs and markers available.
The visual appearance of a glyph is tied directly to the data values that are associated with the glyph’s various attributes. In the example above we see that positional attributes like x and y can be set to vectors of data. But glyphs also have some combination of Line Properties, Fill Properties, and Text Properties to control their appearance. All of these attributes can be set with “vectorized” values as well. We will show examples of this below.
#### Guides and Annotations
Bokeh plots can also have other visual components that aid presentation or help the user make comparisons. These fall into two categories.
1. Guides are visual aids that help users judge distances, angles, etc. These include grid lines or bands, axes (such as linear, log, or datetime) that may have ticks and tick labels as well.
2. Annotations are visual aids that label or name parts of the plot. These include titles, legends, etc.
#### Ranges
Ranges describe the data-space bounds of a plot. By default, plots generated with the bokeh.plotting interface come configured with DataRange1d objects that try to automatically set the plot bounds to encompass all the available data. But it is possible to supply explicit Range1d objects for fixed bounds. As a convenience these can also typically be spelled as 2-tuples or lists:
> ``` p = figure(x_range=[0,10], y_range=(10, 20)) ```
#### Resources
To generate plots, the client library BokehJS JavaScript and CSS code must be loaded into the browser. By default, the output_file() function will load BokehJS from http://cdn.pydata.org . However, you can also configure Bokeh to generate static HTML files with BokehJS resources embedded directly inside, by passing the argument mode="inline" to the output_file() function.
#### Vectorized colors and sizes
Let's see how to provide sequences of data values for glyph
attributes like fill_color and radius.
```
# let's create and x and y random array with length 1000
N = 4000
x = np.random.random(size=N) * 100
y = np.random.random(size=N) * 100
# let's also randomly determine the radii for the bubbles
radii = np.random.random(size=N) * 1.5
# create some random RGB hex codes to create color variation
colors = [
"#%02x%02x%02x" % (int(r), int(g), 150) for r, g in zip(50+2*x, 30+2*y)
]
```
Next: let's define the specific user controls that we want to reify in this graphics:
```
TOOLS='resize,pan,wheel_zoom,box_zoom,reset,hover'
```
Now is time to actually create the graphics:
```
p = figure(tools=TOOLS,
x_range=(0,100),
y_range=(0,100),
title="Interactive Bubble Chart",
x_axis_type="linear",
y_axis_type="linear"
)
```
Finally, let's add bubbles as represented by circles to the chart:
```
p.circle(x, y, radius=radii, fill_color=colors,
fill_alpha=0.5, line_color=None)
output_file('bubbles.html', title="Bokeh Bubble Chart", mode='cdn')
show(p)
```
#### Now, that's pretty cool!
For more information on how to use Bokeh to impress your colleagues and bosses, check out resources here: http://bokeh.pydata.org/
|
github_jupyter
|
import numpy as np
%pylab inline
plot(np.linspace(0, 10, 100) * np.random.random(100))
plot(np.linspace(0, 10, 100) * np.random.random(100),
color='red', linewidth=2, linestyle="--")
plot(np.linspace(0, 10, 100) + np.random.random(100),
color='red', linewidth=2, linestyle="--")
plot(np.linspace(0, 10, 100) - np.random.random(100),
color='blue', linewidth=2, linestyle="-")
hist(np.linspace(0, 10, 100) + np.random.random(100) * 3,
color='green')
import numpy as np
from bokeh.plotting import figure, output_file, show
a = np.arange(10)
b = np.random.randn(10)
output_file("lines.html")
p = figure(title="Bokeh Line",
x_axis_label='x',
y_axis_label='y')
p.line(a, b, legend="Random Walk", line_width=2)
# let's finally see the results
show(p)
x = np.arange(10)
y0 = [i**2 for i in x]
y1 = [10**i for i in x]
y2 = [10**(i**2) for i in x]
output_file("loglines.html")
p = figure(
tools="pan,box_zoom,reset,save",
y_axis_type="log", y_range=[0.001, 10**11], title="Bokeh with log axis",
x_axis_label='sections', y_axis_label='particles'
)
p.line(x, x, legend="y=x")
p.circle(x, x, legend="y=x", fill_color="white", size=8)
p.line(x, y0, legend="y=x^2", line_width=3)
p.line(x, y1, legend="y=10^x", line_color="red")
p.circle(x, y1, legend="y=10^x", fill_color="red", line_color="red", size=6)
p.line(x, y2, legend="y=10^x^2", line_color="orange", line_dash="4 4")
show(p)
#### Resources
To generate plots, the client library BokehJS JavaScript and CSS code must be loaded into the browser. By default, the output_file() function will load BokehJS from http://cdn.pydata.org . However, you can also configure Bokeh to generate static HTML files with BokehJS resources embedded directly inside, by passing the argument mode="inline" to the output_file() function.
#### Vectorized colors and sizes
Let's see how to provide sequences of data values for glyph
attributes like fill_color and radius.
Next: let's define the specific user controls that we want to reify in this graphics:
Now is time to actually create the graphics:
Finally, let's add bubbles as represented by circles to the chart:
| 0.536556 | 0.987676 |
This study guide should reinforce and provide practice for all of the concepts you have seen in Unit 1 Sprint 1. There are a mix of written questions and coding exercises, both are equally important to prepare you for the sprint challenge as well as to be able to speak on these topics comfortably in interviews and on the job.
If you get stuck or are unsure of something remember the 20 minute rule. If that doesn't help, then research a solution with google and stackoverflow. Only once you have exausted these methods should you turn to your track team and mentor - they won't be there on your SC or during an interview. That being said, don't hesitate to ask for help if you truly are stuck.
Have fun studying!
## Questions
```
```
When completing this section, try to limit your answers to 2-3 sentences max and use plain english as much as possible. It's very easy to hide incomplete knowledge and undertanding behind fancy or technical words, so imagine you are explaining these things to a non-technical interviewer.
1. What is a Data Frame?
```
your answer
```
2. What is Pandas?
```
your answer
```
3. How do you check for missing values?
```
your answer
```
4. What is numpy?
```
your answer
```
5. Explain the difference between tidy and wide (summary) data.
```
your answer
```
6. Explain the difference between categorical and quantitative data.
```
your answer
```
7. For categorical variables, explain the difference between an ordinal, nominal or identifier variable.
```
your answer
```
8. For quantitative variables, explain the difference between a discrete and a continuous variable.
```
your answer
```
9. Explain the differnece between an inner, outer, left and right merge.
```
your answer
```
10. Explain the differnece between merging and concatenating data.
```
your answer
```
11. Explain the purpose of a function.
```
your answer
```
12. Explain what .apply() does.
```
your answer
```
13. Explain what .strip() does.
```
your answer
```
14. Explain what .strip('%') does.
```
your answer
```
15. Explain what .split('-') does.
```
your answer
```
16. Give an example of a misleading figure and how you would fix it.
```
your answer
```
17. Describe the important fetures of the distribution of a quantitative variable.
```
your answer
```
## Coding problems
Import pandas, numpy, matplotlib, etc.
```
```
Import a dataset from a link
```
```
Import a dataset from a .csv file saved on your personal computer.
```
```
Import matplotlib
```
```
Loading and viewing a Dataframe
```
```
Using the loaded DataFrame to create and display a plot or graph.
```
```
Print the first five rows of a dataset
```
```
Print the last five rows of a dataset
```
```
Print a single variable in a dataset
```
```
Drop rows from a dataset
```
```
Find the dimensions of a dataframe
```
```
Identify the data types for each column in a dataframe
```
```
Display summary statstics for a dataset.
```
```
Create a new variable that is a linear combination of other variables.
```
```
Create a new variable using the .apply() function.
```
```
Create a new variable using if-then statments with .loc
```
```
Add and and or statements to your if-then statement with .loc
```
```
Convert a date to a datetime format.
```
```
Make a histogram
```
```
Make a box plot
```
```
Make a bar plot
```
```
Make a line plot
```
```
Print axis and figure legends
```
```
Identify missing data in a dataframe.
|
github_jupyter
|
```
When completing this section, try to limit your answers to 2-3 sentences max and use plain english as much as possible. It's very easy to hide incomplete knowledge and undertanding behind fancy or technical words, so imagine you are explaining these things to a non-technical interviewer.
1. What is a Data Frame?
2. What is Pandas?
3. How do you check for missing values?
4. What is numpy?
5. Explain the difference between tidy and wide (summary) data.
6. Explain the difference between categorical and quantitative data.
7. For categorical variables, explain the difference between an ordinal, nominal or identifier variable.
8. For quantitative variables, explain the difference between a discrete and a continuous variable.
9. Explain the differnece between an inner, outer, left and right merge.
10. Explain the differnece between merging and concatenating data.
11. Explain the purpose of a function.
12. Explain what .apply() does.
13. Explain what .strip() does.
14. Explain what .strip('%') does.
15. Explain what .split('-') does.
16. Give an example of a misleading figure and how you would fix it.
17. Describe the important fetures of the distribution of a quantitative variable.
## Coding problems
Import pandas, numpy, matplotlib, etc.
Import a dataset from a link
Import a dataset from a .csv file saved on your personal computer.
Import matplotlib
Loading and viewing a Dataframe
Using the loaded DataFrame to create and display a plot or graph.
Print the first five rows of a dataset
Print the last five rows of a dataset
Print a single variable in a dataset
Drop rows from a dataset
Find the dimensions of a dataframe
Identify the data types for each column in a dataframe
Display summary statstics for a dataset.
Create a new variable that is a linear combination of other variables.
Create a new variable using the .apply() function.
Create a new variable using if-then statments with .loc
Add and and or statements to your if-then statement with .loc
Convert a date to a datetime format.
Make a histogram
Make a box plot
Make a bar plot
Make a line plot
Print axis and figure legends
| 0.69987 | 0.990263 |
#Implementacja YOLOV4
na bazie notatnika udostępnionego przez firmę Roboflow
#Konfiguracja cuDNN dla YOLOv4
```
!/usr/local/cuda/bin/nvcc --version
!nvidia-smi
# This cell ensures you have the correct architecture for your respective GPU
# If you command is not found, look through these GPUs, find the respective
# GPU and add them to the archTypes dictionary
# Tesla V100
# ARCH= -gencode arch=compute_70,code=[sm_70,compute_70]
# Tesla K80
# ARCH= -gencode arch=compute_37,code=sm_37
# GeForce RTX 2080 Ti, RTX 2080, RTX 2070, Quadro RTX 8000, Quadro RTX 6000, Quadro RTX 5000, Tesla T4, XNOR Tensor Cores
# ARCH= -gencode arch=compute_75,code=[sm_75,compute_75]
# Jetson XAVIER
# ARCH= -gencode arch=compute_72,code=[sm_72,compute_72]
# GTX 1080, GTX 1070, GTX 1060, GTX 1050, GTX 1030, Titan Xp, Tesla P40, Tesla P4
# ARCH= -gencode arch=compute_61,code=sm_61
# GP100/Tesla P100 - DGX-1
# ARCH= -gencode arch=compute_60,code=sm_60
# For Jetson TX1, Tegra X1, DRIVE CX, DRIVE PX - uncomment:
# ARCH= -gencode arch=compute_53,code=[sm_53,compute_53]
# For Jetson Tx2 or Drive-PX2 uncomment:
# ARCH= -gencode arch=compute_62,code=[sm_62,compute_62]
import os
os.environ['GPU_TYPE'] = str(os.popen('nvidia-smi --query-gpu=name --format=csv,noheader').read())
def getGPUArch(argument):
try:
argument = argument.strip()
# All Colab GPUs
archTypes = {
"Tesla V100-SXM2-16GB": "-gencode arch=compute_70,code=[sm_70,compute_70]",
"Tesla K80": "-gencode arch=compute_37,code=sm_37",
"Tesla T4": "-gencode arch=compute_75,code=[sm_75,compute_75]",
"Tesla P40": "-gencode arch=compute_61,code=sm_61",
"Tesla P4": "-gencode arch=compute_61,code=sm_61",
"Tesla P100-PCIE-16GB": "-gencode arch=compute_60,code=sm_60"
}
return archTypes[argument]
except KeyError:
return "GPU must be added to GPU Commands"
os.environ['ARCH_VALUE'] = getGPUArch(os.environ['GPU_TYPE'])
print("GPU Type: " + os.environ['GPU_TYPE'])
print("ARCH Value: " + os.environ['ARCH_VALUE'])
```
# Instalacja DarkNet do YOLOV4
```
%cd /content/
%rm -rf darknet
!git clone https://github.com/roboflow-ai/darknet.git
%cd /content/darknet/
%rm Makefile
%%writefile Makefile
GPU=1
CUDNN=1
CUDNN_HALF=0
OPENCV=1
AVX=0
OPENMP=0
LIBSO=1
ZED_CAMERA=0
ZED_CAMERA_v2_8=0
USE_CPP=0
DEBUG=0
ARCH= -gencode arch=compute_35,code=sm_35 \
-gencode arch=compute_50,code=[sm_50,compute_50] \
-gencode arch=compute_52,code=[sm_52,compute_52] \
-gencode arch=compute_61,code=[sm_61,compute_61] \
-gencode arch=compute_37,code=sm_37
ARCH= -gencode arch=compute_60,code=sm_60
OS := $(shell uname)
VPATH=./src/
EXEC=darknet
OBJDIR=./obj/
ifeq ($(LIBSO), 1)
LIBNAMESO=libdarknet.so
APPNAMESO=uselib
endif
ifeq ($(USE_CPP), 1)
CC=g++
else
CC=gcc
endif
CPP=g++ -std=c++11
NVCC=nvcc
OPTS=-Ofast
LDFLAGS= -lm -pthread
COMMON= -Iinclude/ -I3rdparty/stb/include
CFLAGS=-Wall -Wfatal-errors -Wno-unused-result -Wno-unknown-pragmas -fPIC
ifeq ($(DEBUG), 1)
#OPTS= -O0 -g
#OPTS= -Og -g
COMMON+= -DDEBUG
CFLAGS+= -DDEBUG
else
ifeq ($(AVX), 1)
CFLAGS+= -ffp-contract=fast -mavx -mavx2 -msse3 -msse4.1 -msse4.2 -msse4a
endif
endif
CFLAGS+=$(OPTS)
ifneq (,$(findstring MSYS_NT,$(OS)))
LDFLAGS+=-lws2_32
endif
ifeq ($(OPENCV), 1)
COMMON+= -DOPENCV
CFLAGS+= -DOPENCV
LDFLAGS+= `pkg-config --libs opencv4 2> /dev/null || pkg-config --libs opencv`
COMMON+= `pkg-config --cflags opencv4 2> /dev/null || pkg-config --cflags opencv`
endif
ifeq ($(OPENMP), 1)
CFLAGS+= -fopenmp
LDFLAGS+= -lgomp
endif
ifeq ($(GPU), 1)
COMMON+= -DGPU -I/usr/local/cuda/include/
CFLAGS+= -DGPU
ifeq ($(OS),Darwin) #MAC
LDFLAGS+= -L/usr/local/cuda/lib -lcuda -lcudart -lcublas -lcurand
else
LDFLAGS+= -L/usr/local/cuda/lib64 -lcuda -lcudart -lcublas -lcurand
endif
endif
ifeq ($(CUDNN), 1)
COMMON+= -DCUDNN
ifeq ($(OS),Darwin) #MAC
CFLAGS+= -DCUDNN -I/usr/local/cuda/include
LDFLAGS+= -L/usr/local/cuda/lib -lcudnn
else
CFLAGS+= -DCUDNN -I/usr/local/cudnn/include
LDFLAGS+= -L/usr/local/cudnn/lib64 -lcudnn
endif
endif
ifeq ($(CUDNN_HALF), 1)
COMMON+= -DCUDNN_HALF
CFLAGS+= -DCUDNN_HALF
ARCH+= -gencode arch=compute_70,code=[sm_70,compute_70]
endif
ifeq ($(ZED_CAMERA), 1)
CFLAGS+= -DZED_STEREO -I/usr/local/zed/include
ifeq ($(ZED_CAMERA_v2_8), 1)
LDFLAGS+= -L/usr/local/zed/lib -lsl_core -lsl_input -lsl_zed
#-lstdc++ -D_GLIBCXX_USE_CXX11_ABI=0
else
LDFLAGS+= -L/usr/local/zed/lib -lsl_zed
#-lstdc++ -D_GLIBCXX_USE_CXX11_ABI=0
endif
endif
OBJ=image_opencv.o http_stream.o gemm.o utils.o dark_cuda.o convolutional_layer.o list.o image.o activations.o im2col.o col2im.o blas.o crop_layer.o dropout_layer.o maxpool_layer.o softmax_layer.o data.o matrix.o network.o connected_layer.o cost_layer.o parser.o option_list.o darknet.o detection_layer.o captcha.o route_layer.o writing.o box.o nightmare.o normalization_layer.o avgpool_layer.o coco.o dice.o yolo.o detector.o layer.o compare.o classifier.o local_layer.o swag.o shortcut_layer.o activation_layer.o rnn_layer.o gru_layer.o rnn.o rnn_vid.o crnn_layer.o demo.o tag.o cifar.o go.o batchnorm_layer.o art.o region_layer.o reorg_layer.o reorg_old_layer.o super.o voxel.o tree.o yolo_layer.o gaussian_yolo_layer.o upsample_layer.o lstm_layer.o conv_lstm_layer.o scale_channels_layer.o sam_layer.o
ifeq ($(GPU), 1)
LDFLAGS+= -lstdc++
OBJ+=convolutional_kernels.o activation_kernels.o im2col_kernels.o col2im_kernels.o blas_kernels.o crop_layer_kernels.o dropout_layer_kernels.o maxpool_layer_kernels.o network_kernels.o avgpool_layer_kernels.o
endif
OBJS = $(addprefix $(OBJDIR), $(OBJ))
DEPS = $(wildcard src/*.h) Makefile include/darknet.h
all: $(OBJDIR) backup results setchmod $(EXEC) $(LIBNAMESO) $(APPNAMESO)
ifeq ($(LIBSO), 1)
CFLAGS+= -fPIC
$(LIBNAMESO): $(OBJDIR) $(OBJS) include/yolo_v2_class.hpp src/yolo_v2_class.cpp
$(CPP) -shared -std=c++11 -fvisibility=hidden -DLIB_EXPORTS $(COMMON) $(CFLAGS) $(OBJS) src/yolo_v2_class.cpp -o $@ $(LDFLAGS)
$(APPNAMESO): $(LIBNAMESO) include/yolo_v2_class.hpp src/yolo_console_dll.cpp
$(CPP) -std=c++11 $(COMMON) $(CFLAGS) -o $@ src/yolo_console_dll.cpp $(LDFLAGS) -L ./ -l:$(LIBNAMESO)
endif
$(EXEC): $(OBJS)
$(CPP) -std=c++11 $(COMMON) $(CFLAGS) $^ -o $@ $(LDFLAGS)
$(OBJDIR)%.o: %.c $(DEPS)
$(CC) $(COMMON) $(CFLAGS) -c $< -o $@
$(OBJDIR)%.o: %.cpp $(DEPS)
$(CPP) -std=c++11 $(COMMON) $(CFLAGS) -c $< -o $@
$(OBJDIR)%.o: %.cu $(DEPS)
$(NVCC) $(ARCH) $(COMMON) --compiler-options "$(CFLAGS)" -c $< -o $@
$(OBJDIR):
mkdir -p $(OBJDIR)
backup:
mkdir -p backup
results:
mkdir -p results
setchmod:
chmod +x *.sh
.PHONY: clean
clean:
rm -rf $(OBJS) $(EXEC) $(LIBNAMESO) $(APPNAMESO)
%cd /content/darknet/
!sed -i 's/OPENCV=0/OPENCV=1/g' Makefile
!sed -i 's/GPU=0/GPU=1/g' Makefile
!sed -i 's/CUDNN=0/CUDNN=1/g' Makefile
!sed -i "s/ARCH= -gencode arch=compute_60,code=sm_60/ARCH= ${ARCH_VALUE}/g" Makefile
!make
#Pobranie wag do YOLOV4
%cd /content/darknet
!wget https://github.com/AlexeyAB/darknet/releases/download/darknet_yolo_v3_optimal/yolov4.conv.137
```
# Ustawienie zbioru danych
```
#Zaladowanie przy pomocy Roboflow
%cd /content/darknet
!curl -L "https://app.roboflow.com/ds/nmN6nlXic6?key=MfDugCvuC5" > roboflow.zip; unzip roboflow.zip; rm roboflow.zip
%cd /content/darknet/
%cp train/_darknet.labels data/obj.names
%mkdir data/obj
%cp train/*.jpg data/obj/
%cp valid/*.jpg data/obj/
%cp train/*.txt data/obj/
%cp valid/*.txt data/obj/
with open('data/obj.data', 'w') as out:
out.write('classes = 3\n')
out.write('train = data/train.txt\n')
out.write('valid = data/valid.txt\n')
out.write('names = data/obj.names\n')
out.write('backup = backup/')
import os
with open('data/train.txt', 'w') as out:
for img in [f for f in os.listdir('train') if f.endswith('jpg')]:
out.write('data/obj/' + img + '\n')
import os
with open('data/valid.txt', 'w') as out:
for img in [f for f in os.listdir('valid') if f.endswith('jpg')]:
out.write('data/obj/' + img + '\n')
```
# Konfiguracja treningowa
```
def file_len(fname):
with open(fname) as f:
for i, l in enumerate(f):
pass
return i + 1
num_classes = file_len('train/_darknet.labels')
print("writing config for a custom YOLOv4 detector detecting number of classes: " + str(num_classes))
if os.path.exists('./cfg/custom-yolov4-detector.cfg'): os.remove('./cfg/custom-yolov4-detector.cfg')
with open('./cfg/custom-yolov4-detector.cfg', 'a') as f:
f.write('[net]' + '\n')
f.write('batch=64' + '\n')
f.write('subdivisions=24' + '\n')
f.write('width=416' + '\n')
f.write('height=416' + '\n')
f.write('channels=3' + '\n')
f.write('momentum=0.949' + '\n')
f.write('decay=0.0005' + '\n')
f.write('angle=0' + '\n')
f.write('saturation = 1.5' + '\n')
f.write('exposure = 1.5' + '\n')
f.write('hue = .1' + '\n')
f.write('\n')
f.write('learning_rate=0.001' + '\n')
f.write('burn_in=1000' + '\n')
max_batches = 2000
f.write('max_batches=' + str(max_batches) + '\n')
f.write('policy=steps' + '\n')
steps1 = .8 * max_batches
steps2 = .9 * max_batches
f.write('steps='+str(steps1)+','+str(steps2) + '\n')
with open('cfg/yolov4-custom2.cfg', 'r') as f2:
content = f2.readlines()
for line in content:
f.write(line)
num_filters = (num_classes + 5) * 3
f.write('filters='+str(num_filters) + '\n')
f.write('activation=linear')
f.write('\n')
f.write('\n')
f.write('[yolo]' + '\n')
f.write('mask = 0,1,2' + '\n')
f.write('anchors = 12, 16, 19, 36, 40, 28, 36, 75, 76, 55, 72, 146, 142, 110, 192, 243, 459, 401' + '\n')
f.write('classes=' + str(num_classes) + '\n')
with open('cfg/yolov4-custom3.cfg', 'r') as f3:
content = f3.readlines()
for line in content:
f.write(line)
num_filters = (num_classes + 5) * 3
f.write('filters='+str(num_filters) + '\n')
f.write('activation=linear')
f.write('\n')
f.write('\n')
f.write('[yolo]' + '\n')
f.write('mask = 3,4,5' + '\n')
f.write('anchors = 12, 16, 19, 36, 40, 28, 36, 75, 76, 55, 72, 146, 142, 110, 192, 243, 459, 401' + '\n')
f.write('classes=' + str(num_classes) + '\n')
with open('cfg/yolov4-custom4.cfg', 'r') as f4:
content = f4.readlines()
for line in content:
f.write(line)
num_filters = (num_classes + 5) * 3
f.write('filters='+str(num_filters) + '\n')
f.write('activation=linear')
f.write('\n')
f.write('\n')
f.write('[yolo]' + '\n')
f.write('mask = 6,7,8' + '\n')
f.write('anchors = 12, 16, 19, 36, 40, 28, 36, 75, 76, 55, 72, 146, 142, 110, 192, 243, 459, 401' + '\n')
f.write('classes=' + str(num_classes) + '\n')
with open('cfg/yolov4-custom5.cfg', 'r') as f5:
content = f5.readlines()
for line in content:
f.write(line)
print("file is written!")
#Mozliwosc edycji pliku konfiguracyjnego
%cat cfg/custom-yolov4-detector.cfg
```
# Pętla treningowa
```
!./darknet detector train data/obj.data cfg/custom-yolov4-detector.cfg yolov4.conv.137 -dont_show -map
def imShow(path):
import cv2
import matplotlib.pyplot as plt
%matplotlib inline
image = cv2.imread(path)
height, width = image.shape[:2]
resized_image = cv2.resize(image,(3*width, 3*height), interpolation = cv2.INTER_CUBIC)
fig = plt.gcf()
fig.set_size_inches(18, 10)
plt.axis("off")
#plt.rcParams['figure.figsize'] = [10, 5]
plt.imshow(cv2.cvtColor(resized_image, cv2.COLOR_BGR2RGB))
plt.show()
!ls backup
%cp data/obj.names data/coco.names
```
#Test na danych testowych
```
test_images = [f for f in os.listdir('test') if f.endswith('.jpg')]
import random
img_path = "test/" + random.choice(test_images);
!./darknet detect cfg/custom-yolov4-detector.cfg backup/custom-yolov4-detector_final.weights {img_path} -dont-show
imShow('/content/darknet/predictions.jpg')
```
# Zapis wag
```
from google.colab import files
files.download('./backup/custom-yolov4-detector_best.weights')
```
|
github_jupyter
|
!/usr/local/cuda/bin/nvcc --version
!nvidia-smi
# This cell ensures you have the correct architecture for your respective GPU
# If you command is not found, look through these GPUs, find the respective
# GPU and add them to the archTypes dictionary
# Tesla V100
# ARCH= -gencode arch=compute_70,code=[sm_70,compute_70]
# Tesla K80
# ARCH= -gencode arch=compute_37,code=sm_37
# GeForce RTX 2080 Ti, RTX 2080, RTX 2070, Quadro RTX 8000, Quadro RTX 6000, Quadro RTX 5000, Tesla T4, XNOR Tensor Cores
# ARCH= -gencode arch=compute_75,code=[sm_75,compute_75]
# Jetson XAVIER
# ARCH= -gencode arch=compute_72,code=[sm_72,compute_72]
# GTX 1080, GTX 1070, GTX 1060, GTX 1050, GTX 1030, Titan Xp, Tesla P40, Tesla P4
# ARCH= -gencode arch=compute_61,code=sm_61
# GP100/Tesla P100 - DGX-1
# ARCH= -gencode arch=compute_60,code=sm_60
# For Jetson TX1, Tegra X1, DRIVE CX, DRIVE PX - uncomment:
# ARCH= -gencode arch=compute_53,code=[sm_53,compute_53]
# For Jetson Tx2 or Drive-PX2 uncomment:
# ARCH= -gencode arch=compute_62,code=[sm_62,compute_62]
import os
os.environ['GPU_TYPE'] = str(os.popen('nvidia-smi --query-gpu=name --format=csv,noheader').read())
def getGPUArch(argument):
try:
argument = argument.strip()
# All Colab GPUs
archTypes = {
"Tesla V100-SXM2-16GB": "-gencode arch=compute_70,code=[sm_70,compute_70]",
"Tesla K80": "-gencode arch=compute_37,code=sm_37",
"Tesla T4": "-gencode arch=compute_75,code=[sm_75,compute_75]",
"Tesla P40": "-gencode arch=compute_61,code=sm_61",
"Tesla P4": "-gencode arch=compute_61,code=sm_61",
"Tesla P100-PCIE-16GB": "-gencode arch=compute_60,code=sm_60"
}
return archTypes[argument]
except KeyError:
return "GPU must be added to GPU Commands"
os.environ['ARCH_VALUE'] = getGPUArch(os.environ['GPU_TYPE'])
print("GPU Type: " + os.environ['GPU_TYPE'])
print("ARCH Value: " + os.environ['ARCH_VALUE'])
%cd /content/
%rm -rf darknet
!git clone https://github.com/roboflow-ai/darknet.git
%cd /content/darknet/
%rm Makefile
%%writefile Makefile
GPU=1
CUDNN=1
CUDNN_HALF=0
OPENCV=1
AVX=0
OPENMP=0
LIBSO=1
ZED_CAMERA=0
ZED_CAMERA_v2_8=0
USE_CPP=0
DEBUG=0
ARCH= -gencode arch=compute_35,code=sm_35 \
-gencode arch=compute_50,code=[sm_50,compute_50] \
-gencode arch=compute_52,code=[sm_52,compute_52] \
-gencode arch=compute_61,code=[sm_61,compute_61] \
-gencode arch=compute_37,code=sm_37
ARCH= -gencode arch=compute_60,code=sm_60
OS := $(shell uname)
VPATH=./src/
EXEC=darknet
OBJDIR=./obj/
ifeq ($(LIBSO), 1)
LIBNAMESO=libdarknet.so
APPNAMESO=uselib
endif
ifeq ($(USE_CPP), 1)
CC=g++
else
CC=gcc
endif
CPP=g++ -std=c++11
NVCC=nvcc
OPTS=-Ofast
LDFLAGS= -lm -pthread
COMMON= -Iinclude/ -I3rdparty/stb/include
CFLAGS=-Wall -Wfatal-errors -Wno-unused-result -Wno-unknown-pragmas -fPIC
ifeq ($(DEBUG), 1)
#OPTS= -O0 -g
#OPTS= -Og -g
COMMON+= -DDEBUG
CFLAGS+= -DDEBUG
else
ifeq ($(AVX), 1)
CFLAGS+= -ffp-contract=fast -mavx -mavx2 -msse3 -msse4.1 -msse4.2 -msse4a
endif
endif
CFLAGS+=$(OPTS)
ifneq (,$(findstring MSYS_NT,$(OS)))
LDFLAGS+=-lws2_32
endif
ifeq ($(OPENCV), 1)
COMMON+= -DOPENCV
CFLAGS+= -DOPENCV
LDFLAGS+= `pkg-config --libs opencv4 2> /dev/null || pkg-config --libs opencv`
COMMON+= `pkg-config --cflags opencv4 2> /dev/null || pkg-config --cflags opencv`
endif
ifeq ($(OPENMP), 1)
CFLAGS+= -fopenmp
LDFLAGS+= -lgomp
endif
ifeq ($(GPU), 1)
COMMON+= -DGPU -I/usr/local/cuda/include/
CFLAGS+= -DGPU
ifeq ($(OS),Darwin) #MAC
LDFLAGS+= -L/usr/local/cuda/lib -lcuda -lcudart -lcublas -lcurand
else
LDFLAGS+= -L/usr/local/cuda/lib64 -lcuda -lcudart -lcublas -lcurand
endif
endif
ifeq ($(CUDNN), 1)
COMMON+= -DCUDNN
ifeq ($(OS),Darwin) #MAC
CFLAGS+= -DCUDNN -I/usr/local/cuda/include
LDFLAGS+= -L/usr/local/cuda/lib -lcudnn
else
CFLAGS+= -DCUDNN -I/usr/local/cudnn/include
LDFLAGS+= -L/usr/local/cudnn/lib64 -lcudnn
endif
endif
ifeq ($(CUDNN_HALF), 1)
COMMON+= -DCUDNN_HALF
CFLAGS+= -DCUDNN_HALF
ARCH+= -gencode arch=compute_70,code=[sm_70,compute_70]
endif
ifeq ($(ZED_CAMERA), 1)
CFLAGS+= -DZED_STEREO -I/usr/local/zed/include
ifeq ($(ZED_CAMERA_v2_8), 1)
LDFLAGS+= -L/usr/local/zed/lib -lsl_core -lsl_input -lsl_zed
#-lstdc++ -D_GLIBCXX_USE_CXX11_ABI=0
else
LDFLAGS+= -L/usr/local/zed/lib -lsl_zed
#-lstdc++ -D_GLIBCXX_USE_CXX11_ABI=0
endif
endif
OBJ=image_opencv.o http_stream.o gemm.o utils.o dark_cuda.o convolutional_layer.o list.o image.o activations.o im2col.o col2im.o blas.o crop_layer.o dropout_layer.o maxpool_layer.o softmax_layer.o data.o matrix.o network.o connected_layer.o cost_layer.o parser.o option_list.o darknet.o detection_layer.o captcha.o route_layer.o writing.o box.o nightmare.o normalization_layer.o avgpool_layer.o coco.o dice.o yolo.o detector.o layer.o compare.o classifier.o local_layer.o swag.o shortcut_layer.o activation_layer.o rnn_layer.o gru_layer.o rnn.o rnn_vid.o crnn_layer.o demo.o tag.o cifar.o go.o batchnorm_layer.o art.o region_layer.o reorg_layer.o reorg_old_layer.o super.o voxel.o tree.o yolo_layer.o gaussian_yolo_layer.o upsample_layer.o lstm_layer.o conv_lstm_layer.o scale_channels_layer.o sam_layer.o
ifeq ($(GPU), 1)
LDFLAGS+= -lstdc++
OBJ+=convolutional_kernels.o activation_kernels.o im2col_kernels.o col2im_kernels.o blas_kernels.o crop_layer_kernels.o dropout_layer_kernels.o maxpool_layer_kernels.o network_kernels.o avgpool_layer_kernels.o
endif
OBJS = $(addprefix $(OBJDIR), $(OBJ))
DEPS = $(wildcard src/*.h) Makefile include/darknet.h
all: $(OBJDIR) backup results setchmod $(EXEC) $(LIBNAMESO) $(APPNAMESO)
ifeq ($(LIBSO), 1)
CFLAGS+= -fPIC
$(LIBNAMESO): $(OBJDIR) $(OBJS) include/yolo_v2_class.hpp src/yolo_v2_class.cpp
$(CPP) -shared -std=c++11 -fvisibility=hidden -DLIB_EXPORTS $(COMMON) $(CFLAGS) $(OBJS) src/yolo_v2_class.cpp -o $@ $(LDFLAGS)
$(APPNAMESO): $(LIBNAMESO) include/yolo_v2_class.hpp src/yolo_console_dll.cpp
$(CPP) -std=c++11 $(COMMON) $(CFLAGS) -o $@ src/yolo_console_dll.cpp $(LDFLAGS) -L ./ -l:$(LIBNAMESO)
endif
$(EXEC): $(OBJS)
$(CPP) -std=c++11 $(COMMON) $(CFLAGS) $^ -o $@ $(LDFLAGS)
$(OBJDIR)%.o: %.c $(DEPS)
$(CC) $(COMMON) $(CFLAGS) -c $< -o $@
$(OBJDIR)%.o: %.cpp $(DEPS)
$(CPP) -std=c++11 $(COMMON) $(CFLAGS) -c $< -o $@
$(OBJDIR)%.o: %.cu $(DEPS)
$(NVCC) $(ARCH) $(COMMON) --compiler-options "$(CFLAGS)" -c $< -o $@
$(OBJDIR):
mkdir -p $(OBJDIR)
backup:
mkdir -p backup
results:
mkdir -p results
setchmod:
chmod +x *.sh
.PHONY: clean
clean:
rm -rf $(OBJS) $(EXEC) $(LIBNAMESO) $(APPNAMESO)
%cd /content/darknet/
!sed -i 's/OPENCV=0/OPENCV=1/g' Makefile
!sed -i 's/GPU=0/GPU=1/g' Makefile
!sed -i 's/CUDNN=0/CUDNN=1/g' Makefile
!sed -i "s/ARCH= -gencode arch=compute_60,code=sm_60/ARCH= ${ARCH_VALUE}/g" Makefile
!make
#Pobranie wag do YOLOV4
%cd /content/darknet
!wget https://github.com/AlexeyAB/darknet/releases/download/darknet_yolo_v3_optimal/yolov4.conv.137
#Zaladowanie przy pomocy Roboflow
%cd /content/darknet
!curl -L "https://app.roboflow.com/ds/nmN6nlXic6?key=MfDugCvuC5" > roboflow.zip; unzip roboflow.zip; rm roboflow.zip
%cd /content/darknet/
%cp train/_darknet.labels data/obj.names
%mkdir data/obj
%cp train/*.jpg data/obj/
%cp valid/*.jpg data/obj/
%cp train/*.txt data/obj/
%cp valid/*.txt data/obj/
with open('data/obj.data', 'w') as out:
out.write('classes = 3\n')
out.write('train = data/train.txt\n')
out.write('valid = data/valid.txt\n')
out.write('names = data/obj.names\n')
out.write('backup = backup/')
import os
with open('data/train.txt', 'w') as out:
for img in [f for f in os.listdir('train') if f.endswith('jpg')]:
out.write('data/obj/' + img + '\n')
import os
with open('data/valid.txt', 'w') as out:
for img in [f for f in os.listdir('valid') if f.endswith('jpg')]:
out.write('data/obj/' + img + '\n')
def file_len(fname):
with open(fname) as f:
for i, l in enumerate(f):
pass
return i + 1
num_classes = file_len('train/_darknet.labels')
print("writing config for a custom YOLOv4 detector detecting number of classes: " + str(num_classes))
if os.path.exists('./cfg/custom-yolov4-detector.cfg'): os.remove('./cfg/custom-yolov4-detector.cfg')
with open('./cfg/custom-yolov4-detector.cfg', 'a') as f:
f.write('[net]' + '\n')
f.write('batch=64' + '\n')
f.write('subdivisions=24' + '\n')
f.write('width=416' + '\n')
f.write('height=416' + '\n')
f.write('channels=3' + '\n')
f.write('momentum=0.949' + '\n')
f.write('decay=0.0005' + '\n')
f.write('angle=0' + '\n')
f.write('saturation = 1.5' + '\n')
f.write('exposure = 1.5' + '\n')
f.write('hue = .1' + '\n')
f.write('\n')
f.write('learning_rate=0.001' + '\n')
f.write('burn_in=1000' + '\n')
max_batches = 2000
f.write('max_batches=' + str(max_batches) + '\n')
f.write('policy=steps' + '\n')
steps1 = .8 * max_batches
steps2 = .9 * max_batches
f.write('steps='+str(steps1)+','+str(steps2) + '\n')
with open('cfg/yolov4-custom2.cfg', 'r') as f2:
content = f2.readlines()
for line in content:
f.write(line)
num_filters = (num_classes + 5) * 3
f.write('filters='+str(num_filters) + '\n')
f.write('activation=linear')
f.write('\n')
f.write('\n')
f.write('[yolo]' + '\n')
f.write('mask = 0,1,2' + '\n')
f.write('anchors = 12, 16, 19, 36, 40, 28, 36, 75, 76, 55, 72, 146, 142, 110, 192, 243, 459, 401' + '\n')
f.write('classes=' + str(num_classes) + '\n')
with open('cfg/yolov4-custom3.cfg', 'r') as f3:
content = f3.readlines()
for line in content:
f.write(line)
num_filters = (num_classes + 5) * 3
f.write('filters='+str(num_filters) + '\n')
f.write('activation=linear')
f.write('\n')
f.write('\n')
f.write('[yolo]' + '\n')
f.write('mask = 3,4,5' + '\n')
f.write('anchors = 12, 16, 19, 36, 40, 28, 36, 75, 76, 55, 72, 146, 142, 110, 192, 243, 459, 401' + '\n')
f.write('classes=' + str(num_classes) + '\n')
with open('cfg/yolov4-custom4.cfg', 'r') as f4:
content = f4.readlines()
for line in content:
f.write(line)
num_filters = (num_classes + 5) * 3
f.write('filters='+str(num_filters) + '\n')
f.write('activation=linear')
f.write('\n')
f.write('\n')
f.write('[yolo]' + '\n')
f.write('mask = 6,7,8' + '\n')
f.write('anchors = 12, 16, 19, 36, 40, 28, 36, 75, 76, 55, 72, 146, 142, 110, 192, 243, 459, 401' + '\n')
f.write('classes=' + str(num_classes) + '\n')
with open('cfg/yolov4-custom5.cfg', 'r') as f5:
content = f5.readlines()
for line in content:
f.write(line)
print("file is written!")
#Mozliwosc edycji pliku konfiguracyjnego
%cat cfg/custom-yolov4-detector.cfg
!./darknet detector train data/obj.data cfg/custom-yolov4-detector.cfg yolov4.conv.137 -dont_show -map
def imShow(path):
import cv2
import matplotlib.pyplot as plt
%matplotlib inline
image = cv2.imread(path)
height, width = image.shape[:2]
resized_image = cv2.resize(image,(3*width, 3*height), interpolation = cv2.INTER_CUBIC)
fig = plt.gcf()
fig.set_size_inches(18, 10)
plt.axis("off")
#plt.rcParams['figure.figsize'] = [10, 5]
plt.imshow(cv2.cvtColor(resized_image, cv2.COLOR_BGR2RGB))
plt.show()
!ls backup
%cp data/obj.names data/coco.names
test_images = [f for f in os.listdir('test') if f.endswith('.jpg')]
import random
img_path = "test/" + random.choice(test_images);
!./darknet detect cfg/custom-yolov4-detector.cfg backup/custom-yolov4-detector_final.weights {img_path} -dont-show
imShow('/content/darknet/predictions.jpg')
from google.colab import files
files.download('./backup/custom-yolov4-detector_best.weights')
| 0.274449 | 0.279952 |
```
import torch
from torch import nn
from torch.utils.data import DataLoader
from torchvision import datasets
from torchvision.transforms import ToTensor
from torchvision.utils import save_image
cuda = torch.cuda.is_available()
device = torch.device("cuda" if cuda else "cpu")
print(device)
training_data = datasets.FashionMNIST(root="data", train=True, download=True, transform=ToTensor())
test_data = datasets.FashionMNIST(root="data", train=False, download=True, transform=ToTensor())
class Model(nn.Module):
def __init__(self, n_features: int):
super().__init__()
self.n_features = n_features
self.encoder = nn.Sequential(nn.Linear(n_features, 128),
nn.ReLU(),
nn.Linear(128, 64),
nn.ReLU(),
nn.Linear(64, 32),
nn.ReLU(),
nn.Linear(32, 16),
nn.ReLU(),
nn.Linear(16, 8)
)
self.decoder = nn.Sequential(nn.Linear(8, 16),
nn.ReLU(),
nn.Linear(16, 32),
nn.ReLU(),
nn.Linear(32, 64),
nn.ReLU(),
nn.Linear(64, 128),
nn.ReLU(),
nn.Linear(128, self.n_features),
nn.Sigmoid()
)
def forward(self, x):
x = self.encoder(x)
x = self.decoder(x)
return x
model = Model(n_features=28 * 28).to(device)
print(model)
def train(dataloader, model, loss_fn, optimizer):
size = len(dataloader.dataset)
model.train()
for batch, (X, y) in enumerate(dataloader):
X = X.reshape(-1, 28 * 28)
X = X.to(device)
pred = model(X)
loss = loss_fn(pred, X)
# Backpropagation
optimizer.zero_grad()
loss.backward()
optimizer.step()
if batch % 100 == 0:
loss, current = loss.item(), batch * len(X)
print(f"loss: {loss:>7f} [{current:>5d}/{size:>5d}]")
def test(dataloader, model, loss_fn):
num_batches = len(dataloader)
model.eval()
test_loss = 0
with torch.no_grad():
for X, y in dataloader:
X = X.reshape(-1, 28 * 28)
X = X.to(device)
pred = model(X)
test_loss += loss_fn(pred, X).item()
test_loss /= num_batches
print(f"Test Error: \n Avg loss: {test_loss:>8f} \n")
def test_with_testimage(model, test_image, epoch: int, filename: str):
model.eval()
with torch.no_grad():
test_image = test_image.reshape(-1, 28 * 28)
test_image = test_image.to(device)
predicted_test_image = model(test_image)
predicted_test_image = predicted_test_image.reshape(-1, 28, 28)
save_image(predicted_test_image[0], f"epoch_{epoch}_{filename}.png")
def save_weights(model: nn.Module, filename: str) -> None:
torch.save(model.state_dict(), filename)
loss_fn = nn.MSELoss()
optimizer = torch.optim.Adam(model.parameters(), lr=1e-3) # step, update von params = params - lr * grad
batch_size = 64
train_dataloader = DataLoader(training_data, batch_size=batch_size)
test_dataloader = DataLoader(test_data, batch_size=1, shuffle=True)
test_image = test_data.data[0, :, :].float()
save_image(test_image, "testimage.png")
noise = torch.randn(test_image.shape) * 0.1
noisy_test_image = torch.add(test_image, noise)
save_image(noisy_test_image, "noisy_testimage.png")
for epoch in range(100):
print(f"Epoch {epoch}\n-------------------------------")
train(dataloader=train_dataloader, model=model, loss_fn=loss_fn, optimizer=optimizer)
# test(dataloader=test_dataloader, model=model, loss_fn=loss_fn)
test_with_testimage(model=model, test_image=test_image, epoch=epoch, filename="no_noise")
test_with_testimage(model=model, test_image=noisy_test_image, epoch=epoch, filename="noise")
save_weights(model, filename=f"model_epoch_{epoch}.pth")
```
|
github_jupyter
|
import torch
from torch import nn
from torch.utils.data import DataLoader
from torchvision import datasets
from torchvision.transforms import ToTensor
from torchvision.utils import save_image
cuda = torch.cuda.is_available()
device = torch.device("cuda" if cuda else "cpu")
print(device)
training_data = datasets.FashionMNIST(root="data", train=True, download=True, transform=ToTensor())
test_data = datasets.FashionMNIST(root="data", train=False, download=True, transform=ToTensor())
class Model(nn.Module):
def __init__(self, n_features: int):
super().__init__()
self.n_features = n_features
self.encoder = nn.Sequential(nn.Linear(n_features, 128),
nn.ReLU(),
nn.Linear(128, 64),
nn.ReLU(),
nn.Linear(64, 32),
nn.ReLU(),
nn.Linear(32, 16),
nn.ReLU(),
nn.Linear(16, 8)
)
self.decoder = nn.Sequential(nn.Linear(8, 16),
nn.ReLU(),
nn.Linear(16, 32),
nn.ReLU(),
nn.Linear(32, 64),
nn.ReLU(),
nn.Linear(64, 128),
nn.ReLU(),
nn.Linear(128, self.n_features),
nn.Sigmoid()
)
def forward(self, x):
x = self.encoder(x)
x = self.decoder(x)
return x
model = Model(n_features=28 * 28).to(device)
print(model)
def train(dataloader, model, loss_fn, optimizer):
size = len(dataloader.dataset)
model.train()
for batch, (X, y) in enumerate(dataloader):
X = X.reshape(-1, 28 * 28)
X = X.to(device)
pred = model(X)
loss = loss_fn(pred, X)
# Backpropagation
optimizer.zero_grad()
loss.backward()
optimizer.step()
if batch % 100 == 0:
loss, current = loss.item(), batch * len(X)
print(f"loss: {loss:>7f} [{current:>5d}/{size:>5d}]")
def test(dataloader, model, loss_fn):
num_batches = len(dataloader)
model.eval()
test_loss = 0
with torch.no_grad():
for X, y in dataloader:
X = X.reshape(-1, 28 * 28)
X = X.to(device)
pred = model(X)
test_loss += loss_fn(pred, X).item()
test_loss /= num_batches
print(f"Test Error: \n Avg loss: {test_loss:>8f} \n")
def test_with_testimage(model, test_image, epoch: int, filename: str):
model.eval()
with torch.no_grad():
test_image = test_image.reshape(-1, 28 * 28)
test_image = test_image.to(device)
predicted_test_image = model(test_image)
predicted_test_image = predicted_test_image.reshape(-1, 28, 28)
save_image(predicted_test_image[0], f"epoch_{epoch}_{filename}.png")
def save_weights(model: nn.Module, filename: str) -> None:
torch.save(model.state_dict(), filename)
loss_fn = nn.MSELoss()
optimizer = torch.optim.Adam(model.parameters(), lr=1e-3) # step, update von params = params - lr * grad
batch_size = 64
train_dataloader = DataLoader(training_data, batch_size=batch_size)
test_dataloader = DataLoader(test_data, batch_size=1, shuffle=True)
test_image = test_data.data[0, :, :].float()
save_image(test_image, "testimage.png")
noise = torch.randn(test_image.shape) * 0.1
noisy_test_image = torch.add(test_image, noise)
save_image(noisy_test_image, "noisy_testimage.png")
for epoch in range(100):
print(f"Epoch {epoch}\n-------------------------------")
train(dataloader=train_dataloader, model=model, loss_fn=loss_fn, optimizer=optimizer)
# test(dataloader=test_dataloader, model=model, loss_fn=loss_fn)
test_with_testimage(model=model, test_image=test_image, epoch=epoch, filename="no_noise")
test_with_testimage(model=model, test_image=noisy_test_image, epoch=epoch, filename="noise")
save_weights(model, filename=f"model_epoch_{epoch}.pth")
| 0.942981 | 0.784113 |
<a href="https://githubtocolab.com/giswqs/geemap/blob/master/examples/notebooks/37_pydeck_3d.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open in Colab"/></a>
Uncomment the following line to install [geemap](https://geemap.org) if needed.
```
# !pip install geemap
```
# How to use Earth Engine with pydeck for 3D terrain visualization
Pydeck + Earth Engine: Terrain Visualization
**Requirements**
- [earthengine-api](https://github.com/google/earthengine-api): a Python client library for calling the Google Earth Engine API.
- [pydeck](https://pydeck.gl/index.html): a WebGL-powered framework for visual exploratory data analysis of large datasets.
- [pydeck-earthengine-layers](https://github.com/UnfoldedInc/earthengine-layers/tree/master/py): a pydekc wrapper for Google Earth Engine. For documentation please visit this [website](https://earthengine-layers.com/).
- [Mapbox API key](https://pydeck.gl/installation.html#getting-a-mapbox-api-key): you will need this add basemap tiles to pydeck.
**Installation**
- conda create -n deck python
- conda activate deck
- conda install mamba -c conda-forge
- mamba install earthengine-api pydeck pydeck-earthengine-layers -c conda-forge
- jupyter nbextension install --sys-prefix --symlink --overwrite --py pydeck
- jupyter nbextension enable --sys-prefix --py pydeck
This example is adopted from [here](https://github.com/UnfoldedInc/earthengine-layers/blob/master/py/examples/terrain.ipynb). Credits to the developers of the [pydeck-earthengine-layers](https://github.com/UnfoldedInc/earthengine-layers) package.
## 2D Visualization
```
import ee
import geemap
Map = geemap.Map()
Map
image = ee.Image('USGS/NED').select('elevation')
vis_params = {
"min": 0,
"max": 4000,
"palette": ['006633', 'E5FFCC', '662A00', 'D8D8D8', 'F5F5F5'],
}
Map.addLayer(image, vis_params, 'NED')
```
## 3D Visualization
### Import libraries
First, import required packages. Note that here we import the `EarthEngineTerrainLayer` instead of the `EarthEngineLayer`.
```
import ee
import pydeck as pdk
from pydeck_earthengine_layers import EarthEngineTerrainLayer
```
### Authenticate with Earth Engine
Using Earth Engine requires authentication. If you don't have a Google account approved for use with Earth Engine, you'll need to request access. For more information and to sign up, go to https://signup.earthengine.google.com/.
```
try:
ee.Initialize()
except Exception as e:
ee.Authenticate()
ee.Initialize()
```
### Terrain Example
In contrast to the `EarthEngineLayer`, where you need to supply _one_ Earth Engine object to render, with the `EarthEngineTerrainLayer` you must supply **two** Earth Engine objects. The first is used to render the image in the same way as the `EarthEngineLayer`; the second supplies elevation values used to extrude terrain in 3D. Hence the former can be any `Image` object; the latter must be an `Image` object whose values represents terrain heights.
It's important for the terrain source to have relatively high spatial resolution. In previous examples, we used [SRTM90][srtm90] as an elevation source, but that only has a resolution of 90 meters. When used as an elevation source, it looks very blocky/pixelated at high zoom levels. In this example we'll use [SRTM30][srtm30] (30-meter resolution) as the `Image` source and the [USGS's National Elevation Dataset][ned] (10-meter resolution, U.S. only) as the terrain source. SRTM30 is generally the best-resolution worldwide data source available.
[srtm90]: https://developers.google.com/earth-engine/datasets/catalog/CGIAR_SRTM90_V4
[srtm30]: https://developers.google.com/earth-engine/datasets/catalog/USGS_SRTMGL1_003
[ned]: https://developers.google.com/earth-engine/datasets/catalog/USGS_NED
```
# image = ee.Image('USGS/SRTMGL1_003')
image = ee.Image('USGS/NED').select('elevation')
terrain = ee.Image('USGS/NED').select('elevation')
```
Here `vis_params` consists of parameters that will be passed to the Earth Engine [`visParams` argument][visparams]. Any parameters that you could pass directly to Earth Engine in the code editor, you can also pass here to the `EarthEngineLayer`.
[visparams]: https://developers.google.com/earth-engine/image_visualization
```
vis_params = {
"min": 0,
"max": 4000,
"palette": ['006633', 'E5FFCC', '662A00', 'D8D8D8', 'F5F5F5'],
}
```
Now we're ready to create the Pydeck layer. The `EarthEngineLayer` makes this simple. Just pass the Earth Engine object to the class.
Including the `id` argument isn't necessary when you only have one pydeck layer, but it is necessary to distinguish multiple layers, so it's good to get into the habit of including an `id` parameter.
```
ee_layer = EarthEngineTerrainLayer(image, terrain, vis_params, id="EETerrainLayer")
```
Then just pass this layer to a `pydeck.Deck` instance, and call `.show()` to create a map:
```
view_state = pdk.ViewState(
latitude=36.15, longitude=-111.96, zoom=10.5, bearing=-66.16, pitch=60
)
r = pdk.Deck(layers=[ee_layer], initial_view_state=view_state)
r.show()
```
|
github_jupyter
|
# !pip install geemap
import ee
import geemap
Map = geemap.Map()
Map
image = ee.Image('USGS/NED').select('elevation')
vis_params = {
"min": 0,
"max": 4000,
"palette": ['006633', 'E5FFCC', '662A00', 'D8D8D8', 'F5F5F5'],
}
Map.addLayer(image, vis_params, 'NED')
import ee
import pydeck as pdk
from pydeck_earthengine_layers import EarthEngineTerrainLayer
try:
ee.Initialize()
except Exception as e:
ee.Authenticate()
ee.Initialize()
# image = ee.Image('USGS/SRTMGL1_003')
image = ee.Image('USGS/NED').select('elevation')
terrain = ee.Image('USGS/NED').select('elevation')
vis_params = {
"min": 0,
"max": 4000,
"palette": ['006633', 'E5FFCC', '662A00', 'D8D8D8', 'F5F5F5'],
}
ee_layer = EarthEngineTerrainLayer(image, terrain, vis_params, id="EETerrainLayer")
view_state = pdk.ViewState(
latitude=36.15, longitude=-111.96, zoom=10.5, bearing=-66.16, pitch=60
)
r = pdk.Deck(layers=[ee_layer], initial_view_state=view_state)
r.show()
| 0.414662 | 0.977479 |
# Mapboxgl Python Library for location data visualization
https://github.com/mapbox/mapboxgl-jupyter
# Add a scale annotation to the map
| Property | Description | Example |
|:--------- |:-------------|:--------|
| scale | controls visibility of map scale annotation | True |
| scale_position | controls the position of the map scale in the map controls pane | 'bottom-right' |
| scale_background_color | string background color for scale | 'white' |
| scale_border_color | string border color for scale | 'white' |
| scale_text_color | string color for legend text | '#6e6e6e' |
scale=True,
scale_border_color='#eee',
scale_background_color='red',
## Create a visualization from example data file
```
import pandas as pd
import os
from mapboxgl.utils import *
from mapboxgl.viz import *
# Set Mapbox Acces Token; Must be a public token, starting with `pk`
token = os.getenv('MAPBOX_ACCESS_TOKEN')
# Load data from sample csv
data_url = 'https://raw.githubusercontent.com/mapbox/mapboxgl-jupyter/master/examples/data/points.csv'
df = pd.read_csv(data_url).round(3)
# Generate data breaks using numpy quantiles and color stops from colorBrewer
measure = 'Avg Medicare Payments'
color_breaks = [round(df[measure].quantile(q=x*0.1), 2) for x in range(1,9)]
color_stops = create_color_stops(color_breaks, colors='YlOrRd')
# Create a geojson Feature Collection from the current dataframe
geodata = df_to_geojson(df,
properties=['Avg Medicare Payments', 'Avg Covered Charges', 'date'],
lat='lat',
lon='lon',
precision=3)
# Create the viz from the dataframe
viz = CircleViz(geodata,
access_token=token,
color_property='Avg Medicare Payments',
color_stops=color_stops,
radius=2.5,
stroke_width=0.2,
center=(-95, 40),
zoom=2.5,
scale=True,
scale_unit_system='imperial',
below_layer='waterway-label',
height='300px')
# Show the viz
viz.show()
```
## Update scale to match Mapbox Dark-v9 style
```
# Map settings
viz.style='mapbox://styles/mapbox/dark-v9?optimize=true'
viz.label_color = 'hsl(0, 0%, 70%)'
viz.label_halo_color = 'hsla(0, 0%, 10%, 0.75)'
viz.height = '400px'
# Legend settings
viz.legend_gradient = False
viz.legend_fill = '#343332'
viz.legend_header_fill = '#343332'
viz.legend_text_color = 'hsl(0, 0%, 70%)'
viz.legend_key_borders_on = False
viz.legend_title_halo_color = 'hsla(0, 0%, 10%, 0.75)'
# Scale settings
viz.scale_border_color = 'hsla(0, 0%, 10%, 0.75)'
viz.scale_position = 'top-left'
viz.scale_background_color = '#343332'
viz.scale_text_color = 'hsl(0, 0%, 70%)'
# Render map
viz.show()
```
|
github_jupyter
|
import pandas as pd
import os
from mapboxgl.utils import *
from mapboxgl.viz import *
# Set Mapbox Acces Token; Must be a public token, starting with `pk`
token = os.getenv('MAPBOX_ACCESS_TOKEN')
# Load data from sample csv
data_url = 'https://raw.githubusercontent.com/mapbox/mapboxgl-jupyter/master/examples/data/points.csv'
df = pd.read_csv(data_url).round(3)
# Generate data breaks using numpy quantiles and color stops from colorBrewer
measure = 'Avg Medicare Payments'
color_breaks = [round(df[measure].quantile(q=x*0.1), 2) for x in range(1,9)]
color_stops = create_color_stops(color_breaks, colors='YlOrRd')
# Create a geojson Feature Collection from the current dataframe
geodata = df_to_geojson(df,
properties=['Avg Medicare Payments', 'Avg Covered Charges', 'date'],
lat='lat',
lon='lon',
precision=3)
# Create the viz from the dataframe
viz = CircleViz(geodata,
access_token=token,
color_property='Avg Medicare Payments',
color_stops=color_stops,
radius=2.5,
stroke_width=0.2,
center=(-95, 40),
zoom=2.5,
scale=True,
scale_unit_system='imperial',
below_layer='waterway-label',
height='300px')
# Show the viz
viz.show()
# Map settings
viz.style='mapbox://styles/mapbox/dark-v9?optimize=true'
viz.label_color = 'hsl(0, 0%, 70%)'
viz.label_halo_color = 'hsla(0, 0%, 10%, 0.75)'
viz.height = '400px'
# Legend settings
viz.legend_gradient = False
viz.legend_fill = '#343332'
viz.legend_header_fill = '#343332'
viz.legend_text_color = 'hsl(0, 0%, 70%)'
viz.legend_key_borders_on = False
viz.legend_title_halo_color = 'hsla(0, 0%, 10%, 0.75)'
# Scale settings
viz.scale_border_color = 'hsla(0, 0%, 10%, 0.75)'
viz.scale_position = 'top-left'
viz.scale_background_color = '#343332'
viz.scale_text_color = 'hsl(0, 0%, 70%)'
# Render map
viz.show()
| 0.710327 | 0.845241 |
<a href="https://colab.research.google.com/github/nguyenvuong1122000/deep-text-recognition-benchmark/blob/master/deep_text_recognition_benchmarkNTV.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
This can be executed in https://colab.research.google.com "Python 3 / GPU" runtime.
```
!git clone https://github.com/roatienza/deep-text-recognition-benchmark
%cd deep-text-recognition-benchmark
```
Next, download large model files from Google Drive, using hack: https://stackoverflow.com/questions/20665881/direct-download-from-google-drive-using-google-drive-api/32742700#32742700
```
!wget https://github.com/roatienza/deep-text-recognition-benchmark/releases/download/v0.1.0/vitstr_small_patch16_224_aug.pth
models = {
'data_training': 'https://drive.google.com/u/0/uc?id=1MVBWIxsMyAGckPZdkMxdT4KO89na1Js0&export=download',
}
for k, v in models.items():
doc_id = v[v.find('=')+1:]
!curl -c /tmp/cookies "https://drive.google.com/uc?export=download&id=$doc_id" > /tmp/intermezzo.html
!curl -L -b /tmp/cookies "https://drive.google.com$(cat /tmp/intermezzo.html | grep -Po 'uc-download-link" [^>]* href="\K[^"]*' | sed 's/\&/\&/g')" > $k
!ls -al *.pth
pip install -r /content/deep-text-recognition-benchmark/requirements.txt
```
```
!apt-get update
!sudo apt-get install imagemagick imagemagick-doc
# !wget https://www.dropbox.com/sh/i39abvnefllx2si/AADCgxB8t7jk37_1qBC5E9FEa/evaluation.zip?dl=0
# !wget https://thor.robots.ox.ac.uk/~vgg/data/text/mjsynth.tar.gz
# !gunzip mjsynth.tar.gz
# !unzip evaluation.zip?dl=0
!CUDA_VISIBLE_DEVICES=0 python3 test.py --eval_data evaluation \
--benchmark_all_eval --Transformation None --FeatureExtraction None \
--SequenceModeling None --Prediction None --Transformer \
--sensitive --data_filtering_off --imgH 224 --imgW 224 \
--TransformerModel=vitstr_small_patch16_224 --saved_model vitstr_small_patch16_224_aug.pth
output = !CUDA_VISIBLE_DEVICES=0 python3 demo.py --image_folder demo_image/ \
-- None --FeatureExtraction None \
--SequenceModeling None --Prediction None \
--sensitive --imgH 224 --imgW 224 \
--saved_model vitstr_small_patch16_224_aug.pth
```
```
```
```
output
from IPython.core.display import display, HTML
from PIL import Image
import base64
import io
import pandas as pd
data = pd.DataFrame()
for ind, row in enumerate(output[output.index('image_path \tpredicted_labels \tconfidence score')+2:]):
row = row.split('\t')
filename = row[0].strip()
label = row[1].strip()
conf = row[2].strip()
img = Image.open(filename)
img_buffer = io.BytesIO()
img.save(img_buffer, format="PNG")
imgStr = base64.b64encode(img_buffer.getvalue()).decode("utf-8")
data.loc[ind, 'img'] = '<img src="data:image/png;base64,{0:s}">'.format(imgStr)
data.loc[ind, 'id'] = filename
data.loc[ind, 'label'] = label
data.loc[ind, 'conf'] = conf
html_all = data.to_html(escape=False)
display(HTML(html_all))
```
|
github_jupyter
|
!git clone https://github.com/roatienza/deep-text-recognition-benchmark
%cd deep-text-recognition-benchmark
!wget https://github.com/roatienza/deep-text-recognition-benchmark/releases/download/v0.1.0/vitstr_small_patch16_224_aug.pth
models = {
'data_training': 'https://drive.google.com/u/0/uc?id=1MVBWIxsMyAGckPZdkMxdT4KO89na1Js0&export=download',
}
for k, v in models.items():
doc_id = v[v.find('=')+1:]
!curl -c /tmp/cookies "https://drive.google.com/uc?export=download&id=$doc_id" > /tmp/intermezzo.html
!curl -L -b /tmp/cookies "https://drive.google.com$(cat /tmp/intermezzo.html | grep -Po 'uc-download-link" [^>]* href="\K[^"]*' | sed 's/\&/\&/g')" > $k
!ls -al *.pth
pip install -r /content/deep-text-recognition-benchmark/requirements.txt
!apt-get update
!sudo apt-get install imagemagick imagemagick-doc
# !wget https://www.dropbox.com/sh/i39abvnefllx2si/AADCgxB8t7jk37_1qBC5E9FEa/evaluation.zip?dl=0
# !wget https://thor.robots.ox.ac.uk/~vgg/data/text/mjsynth.tar.gz
# !gunzip mjsynth.tar.gz
# !unzip evaluation.zip?dl=0
!CUDA_VISIBLE_DEVICES=0 python3 test.py --eval_data evaluation \
--benchmark_all_eval --Transformation None --FeatureExtraction None \
--SequenceModeling None --Prediction None --Transformer \
--sensitive --data_filtering_off --imgH 224 --imgW 224 \
--TransformerModel=vitstr_small_patch16_224 --saved_model vitstr_small_patch16_224_aug.pth
output = !CUDA_VISIBLE_DEVICES=0 python3 demo.py --image_folder demo_image/ \
-- None --FeatureExtraction None \
--SequenceModeling None --Prediction None \
--sensitive --imgH 224 --imgW 224 \
--saved_model vitstr_small_patch16_224_aug.pth
```
| 0.513181 | 0.81772 |
# Pengenalan Pemrosesan Bahasa Alami
[](https://colab.research.google.com/github/aliakbars/talks/blob/master/Intro%20to%20NLP.ipynb)
Oleh: Ali Akbar Septiandri
```
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import seaborn as sns
%matplotlib inline
```
## Memuat Dokumen
```
from sklearn.datasets import fetch_20newsgroups
categories = [
'alt.atheism',
'soc.religion.christian',
'comp.sys.ibm.pc.hardware',
'comp.windows.x',
'rec.sport.baseball',
'rec.sport.hockey',
]
newsgroups_train = fetch_20newsgroups(subset='train', remove=('headers', 'footers', 'quotes'), categories=categories)
newsgroups_test = fetch_20newsgroups(subset='test', remove=('headers', 'footers', 'quotes'), categories=categories)
X_train, y_train = newsgroups_train['data'], newsgroups_train['target']
X_test, y_test = newsgroups_test['data'], newsgroups_test['target']
newsgroups_train['target_names']
```
## Tokenizing, Stemming, Lemmatizing
Umumnya, kata "token" merujuk pada satuan kata. Namun, tidak jarang kita perlu memecah paragraf menjadi kalimat sehingga kita memerlukan *sentence tokenizer*. Contoh penggunaannya dengan NLTK adalah sebagai berikut.
```
import nltk
nltk.sent_tokenize(X_train[3])
sentence = nltk.sent_tokenize(X_train[3])[1]
```
### spaCy Tokenizer
Untuk berbagai bahasa (termasuk bahasa Indonesia), salah satu tokenizer terbaik yang dapat digunakan adalah dari spaCy. Dengan satu kali masukan, Anda dapat menghasilkan token yang langsung mempunyai atribut seperti dijelaskan di [dokumentasinya](https://spacy.io/api/token#attributes).
```
from spacy.lang.en import English
nlp = English()
[token for token in nlp(sentence) if not token.is_space]
```
Perhatikan bahwa tanda baca juga dihitung sebagai token!
### Lemmatizer
Dengan menggunakan kode yang sama, Anda hanya perlu mengganti bagian `print(token)` menjadi `print(token.lemma_)` untuk melihat bentuk kamus dari tiap token.
```
[token.lemma_ for token in nlp(sentence) if not token.is_space]
```
### Stemmer
Stemmer berfungsi untuk memotong imbuhan. Di pengolahan teks modern, prapemrosesan ini jarang dilakukan.
```
stemmer = nltk.stem.porter.PorterStemmer()
tokens = ['regarding', 'programming', 'denied', 'flew']
[stemmer.stem(token) for token in tokens]
```
## POS Tagging
```
import spacy
nlp = spacy.load('en_core_web_sm')
doc = nlp(u'Apple is looking at buying U.K. startup for $1 billion')
rows = []
for token in doc:
rows.append([token.text, token.lemma_, token.pos_, token.tag_, token.dep_, token.shape_, token.is_alpha, token.is_stop])
pd.DataFrame(rows, columns=['text','lemma','pos','tag','dep','shape','alpha','stopword'])
```
## Klasifikasi Dokumen
Pertama, kita sebaiknya melihat sebaran dari kategori dokumen pada data latih.
```
sns.countplot(y=y_train)
plt.yticks(range(6), newsgroups_train['target_names']);
```
Datanya terlihat tersebar cukup merata, kecuali untuk topik `alt.atheism`. Namun, kita tidak perlu menyeimbangkan kelasnya untuk saat ini dan kita dapat menggunakan akurasi sebagai metrik evaluasi.
```
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import cross_val_score
from sklearn.pipeline import make_pipeline
vec = CountVectorizer()
vec.fit_transform(X_train)
```
Hasil transformasi bag-of-words dari teks menghasilkan *sparse matrix*. Informasi di tiap sel dalam matriks tersebut adalah jumlah kemunculan suatu kata dalam suatu dokumen. Sekarang, kita akan mencoba melakukan klasifikasi datanya.
```
clf = make_pipeline(
CountVectorizer(),
LogisticRegression(solver='lbfgs', max_iter=300, multi_class='auto', random_state=42)
)
acc = cross_val_score(clf, X_train, y_train, cv=3, n_jobs=2)
print('Akurasi: {:.2%} ± {:.2%}'.format(acc.mean(), acc.std()))
```
Apa kelemahan dari model ini? Beberapa hal yang dapat dicoba:
1. Bag-of-words $\rightarrow$ n-gram
2. Bag-of-words $\rightarrow$ TF-IDF
3. Buang stopwords
4. Tokenizer $\rightarrow$ Lemmatizer
```
nlp = English()
def lemmatizer(text):
"""Mengembalikan list of lemmas"""
pass
# Kode Anda di sini
```
## Reduksi Dimensi
Salah satu cara mudah untuk mengevaluasi data sebelum melakukan pemodelan adalah dengan visualisasi. Namun, dengan dimensi yang begitu besar, bagaimana cara memvisualisasikannya sementara manusia hanya baik mengolah gambar dalam dua dimensi?
Untuk kebutuhan tersebut, Anda dapat menggunakan metode reduksi dimensi seperti *Principal Component Analysis* (PCA). Dalam dunia pengolahan teks, menerapkan PCA pada matriks hasil n-gram dikenal juga dengan nama *Latent Semantic Analysis* (LSA). Berhubung matriks yang digunakan jarang, alternatif PCA yang digunakan adalah [Truncated SVD](https://scikit-learn.org/stable/modules/generated/sklearn.decomposition.TruncatedSVD.html).
```
from sklearn.decomposition import TruncatedSVD
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.manifold import TSNE
pipe = make_pipeline(
TfidfVectorizer(stop_words='english'),
TruncatedSVD(2, random_state=42)
)
X_map = pipe.fit_transform(X_train)
X_map.shape
```
Sekarang, kita sudah dapat memvisualisasikan datanya.
```
fig, ax = plt.subplots(figsize=(10,10))
for i, label in enumerate(newsgroups_train['target_names']):
ax.scatter(*X_map[y_train == i].T, marker='.', label=label)
plt.legend();
```
Perhatikan bahwa dokumen-dokumen `rec.sport` berdekatan, begitu juga dokumen-dokumen di bawah kategori `comp`. Sudah sesuai dengan intuisi kita bukan? Pertanyaannya, apakah modelnya juga akan lebih baik dengan LSA?
```
from sklearn.neighbors import KNeighborsClassifier
clf = make_pipeline(
TfidfVectorizer(stop_words='english'),
TruncatedSVD(100, random_state=42),
LogisticRegression(solver='lbfgs', max_iter=300, multi_class='auto', random_state=42)
)
acc = cross_val_score(clf, X_train, y_train, cv=3, n_jobs=2)
print('Akurasi: {:.2%} ± {:.2%}'.format(acc.mean(), acc.std()))
```
## Word2Vec
```
sentences = []
for doc in X_train:
sentences.append([token for token in nlp(doc) if not token.is_space])
!python -m spacy download en_core_web_md
import spacy
nlp = spacy.load('en_core_web_md')
for token in nlp('cat dog banana'):
print(token.vector.shape)
```
|
github_jupyter
|
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import seaborn as sns
%matplotlib inline
from sklearn.datasets import fetch_20newsgroups
categories = [
'alt.atheism',
'soc.religion.christian',
'comp.sys.ibm.pc.hardware',
'comp.windows.x',
'rec.sport.baseball',
'rec.sport.hockey',
]
newsgroups_train = fetch_20newsgroups(subset='train', remove=('headers', 'footers', 'quotes'), categories=categories)
newsgroups_test = fetch_20newsgroups(subset='test', remove=('headers', 'footers', 'quotes'), categories=categories)
X_train, y_train = newsgroups_train['data'], newsgroups_train['target']
X_test, y_test = newsgroups_test['data'], newsgroups_test['target']
newsgroups_train['target_names']
import nltk
nltk.sent_tokenize(X_train[3])
sentence = nltk.sent_tokenize(X_train[3])[1]
from spacy.lang.en import English
nlp = English()
[token for token in nlp(sentence) if not token.is_space]
[token.lemma_ for token in nlp(sentence) if not token.is_space]
stemmer = nltk.stem.porter.PorterStemmer()
tokens = ['regarding', 'programming', 'denied', 'flew']
[stemmer.stem(token) for token in tokens]
import spacy
nlp = spacy.load('en_core_web_sm')
doc = nlp(u'Apple is looking at buying U.K. startup for $1 billion')
rows = []
for token in doc:
rows.append([token.text, token.lemma_, token.pos_, token.tag_, token.dep_, token.shape_, token.is_alpha, token.is_stop])
pd.DataFrame(rows, columns=['text','lemma','pos','tag','dep','shape','alpha','stopword'])
sns.countplot(y=y_train)
plt.yticks(range(6), newsgroups_train['target_names']);
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import cross_val_score
from sklearn.pipeline import make_pipeline
vec = CountVectorizer()
vec.fit_transform(X_train)
clf = make_pipeline(
CountVectorizer(),
LogisticRegression(solver='lbfgs', max_iter=300, multi_class='auto', random_state=42)
)
acc = cross_val_score(clf, X_train, y_train, cv=3, n_jobs=2)
print('Akurasi: {:.2%} ± {:.2%}'.format(acc.mean(), acc.std()))
nlp = English()
def lemmatizer(text):
"""Mengembalikan list of lemmas"""
pass
# Kode Anda di sini
from sklearn.decomposition import TruncatedSVD
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.manifold import TSNE
pipe = make_pipeline(
TfidfVectorizer(stop_words='english'),
TruncatedSVD(2, random_state=42)
)
X_map = pipe.fit_transform(X_train)
X_map.shape
fig, ax = plt.subplots(figsize=(10,10))
for i, label in enumerate(newsgroups_train['target_names']):
ax.scatter(*X_map[y_train == i].T, marker='.', label=label)
plt.legend();
from sklearn.neighbors import KNeighborsClassifier
clf = make_pipeline(
TfidfVectorizer(stop_words='english'),
TruncatedSVD(100, random_state=42),
LogisticRegression(solver='lbfgs', max_iter=300, multi_class='auto', random_state=42)
)
acc = cross_val_score(clf, X_train, y_train, cv=3, n_jobs=2)
print('Akurasi: {:.2%} ± {:.2%}'.format(acc.mean(), acc.std()))
sentences = []
for doc in X_train:
sentences.append([token for token in nlp(doc) if not token.is_space])
!python -m spacy download en_core_web_md
import spacy
nlp = spacy.load('en_core_web_md')
for token in nlp('cat dog banana'):
print(token.vector.shape)
| 0.509276 | 0.919823 |
```
import sys
sys.path.append("../../../")
sys.path.append("../../hyperLAI/")
sys.path.append("../../../libraries/")
from utils.model_utils import *
from utils.sim_funcs import sim_func_dict
from models.fc_model import fc_model
from features.hyperLAIdataset import HyperLoader
from sklearn.feature_selection import VarianceThreshold
import torch
from torch.utils import data
from features.hyperLAIdataset import HyperLoader
import numpy as np
import json
dataset = HyperLoader("/scratch/users/patelas/hyperLAI/snp_data/", chromosome=22)
train_indices, valid_indices, test_indices = train_valid_test(len(dataset), 0.8, 0.1)
variance_filter(dataset, train_indices, 5000)
snps_train = dataset.snps[train_indices,:]
vt_var = np.var(dataset.snps, axis=0)
var_sorted = np.argsort(vt_var)[::-1]
dataset.snps.shape
vt_var[77852]
import matplotlib.pyplot as plt
plt.scatter(range(len(vt_var)), sorted(vt_var))
plt.show()
train_size = int(0.8 * len(dataset))
valid_size = int(0.1 * len(dataset))
test_size = len(dataset) - train_size - valid_size
np.random.seed(0)
indices = np.array(list(range(len(dataset))))
np.random.shuffle(indices)
train_indices = indices[0 : train_size]
valid_indices = indices[train_size : train_size + valid_size]
test_indices = indices[train_size + valid_size : ]
# sfunc = lambda x,y: (torch.sum(x == y)).float() / len(x)
sfunc = lambda x,y: torch.true_divide(torch.sum(x==y), len(x))
snp_data = torch.tensor(np.random.randint(0,2,size=(4,4)))
snp_data
smat = make_pairwise_similarities(snp_data, sfunc)
trips, sims = trips_and_sims(snp_data, sfunc)
trips
sims
sims[[2,3],:]
input_size=20
num_int_layers=1
int_layer_sizes=[5]
embedding_size=2
dropout_vals=[0.8]
fctest = fc_model(input_size, num_int_layers, int_layer_sizes, embedding_size, dropout_vals)
list(fctest.parameters())
def run_epoch(model, dloader, device, sim_func, optimizer=None):
batch_hyp_losses = []
for i, (snp_data, suppop_labels, pop_labels) in enumerate(dloader):
if model.training:
assert optimizer is not None
optimizer.zero_grad()
else:
assert optimizer is None
triple_ids, similarities = trips_and_sims(snp_data, sim_func)
snp_data = snp_data.to(device)
triple_ids = triple_ids.to(device)
similarities = similarities.to(device)
embeddings_pred = model(snp_data)
hyp_loss = model.loss(embeddings_pred, triple_ids, similarities)
batch_hyp_losses.append(hyp_loss.item())
if model.training:
hyp_loss.backward()
optimizer.step()
return batch_hyp_losses
def train_model(model, train_loader, valid_loader, num_epochs, learning_rate, sim_func,
patience, txt_writer, output_dir, early_stopping,
patience, early_stop_min_delta, optimizer=None):
device = torch.device("cuda") if torch.cuda.is_available() else torch.device("cpu")
if device == torch.device("cuda"):
print("Training occurring on GPU")
model = model.to(device)
if optimizer is None:
optimizer = RAdam(model.parameters(), lr=learning_rate)
if early_stopping:
valid_loss_history = []
best_valid_epoch_loss, best_model = float("inf"), None
for epoch in range(num_epochs):
if torch.cuda.is_available:
torch.cuda.empty_cache()
model.train()
train_batch_losses = run_epoch(model, train_loader, device, sim_func, optimizer)
train_epoch_loss = np.nanmean(train_batch_losses)
print_and_log("Train: epoch %d: average loss = %6.10f" % (epoch + 1, train_epoch_loss), txt_writer)
with torch.no_grad():
model.eval()
valid_batch_losses = run_epoch(model, valid_loader, device, sim_func, optimizer=None)
valid_epoch_loss = np.nanmean(valid_batch_losses)
print_and_log("Valid: epoch %d: average loss = %6.10f" % (epoch + 1, valid_epoch_loss), txt_writer)
if valid_epoch_loss < best_valid_epoch_loss:
best_valid_epoch_loss = valid_epoch_loss
best_model = model
save_model(model, optimizer, valid_epoch_loss, epoch + 1, output_dir+"model.pt")
if early_stopping:
if len(valid_loss_history) < patience + 1:
# Not enough history yet; tack on the loss
valid_loss_history = [valid_epoch_loss] + valid_loss_history
else:
# Tack on the new validation loss, kicking off the old one
valid_loss_history = \
[valid_epoch_loss] + valid_loss_history[:-1]
if len(valid_loss_history) == patience + 1:
# There is sufficient history to check for improvement
best_delta = np.max(np.diff(valid_loss_history))
if best_delta < early_stop_min_delta:
break # Not improving enough
txt_writer.flush()
json_file = open("../../hyperLAI/models/fc_config.json")
vars_json = json.load(json_file)
json_file.close()
eval(vars_json["sim_func"])(np.array([2,3]),np.array([3,3]))
vars_json["restrict_labels"]
import sys
sys.path.append("../../../")
sys.path.append("../../hyperLAI")
%run "../../hyperLAI/models/train_fc_model.py"
dataset = HyperLoader("/scratch/users/patelas/hyperLAI/snp_data/whole_genome/variance_filtered_500000/",
[0,1,2,3,4,5,6], "all")
train_inds = np.load("/scratch/users/patelas/hyperLAI/ancestry_training_splits/80_10_10/train_indices.npy")
test_inds = np.load("/scratch/users/patelas/hyperLAI/ancestry_training_splits/80_10_10/test_indices.npy")
hl2 = HyperLoader("/scratch/users/patelas/hyperLAI/snp_data/whole_genome/variance_filtered_500000_updated/",
train_inds,[0,1,2,3,4,5,6], "all")
hl2.snps.shape
hl2_top = hl2.snps[:,:5]
hl2_bottom = hl2.snps[:,-5:]
np.sum(hl2_top, axis=0)
np.var(top5_snps, axis=0)
np.var(bottom5_snps, axis=0)
hl = HyperLoader("/scratch/users/patelas/hyperLAI/snp_data/whole_genome/variance_filtered_500000/",
[0,1,2], "all")
len(hl)
data_dir = "/scratch/users/patelas/hyperLAI/snp_data/whole_genome/variance_filtered_500000_updated/"
train_inds = np.load("/scratch/users/patelas/hyperLAI/ancestry_training_splits/80_10_10/train_indices.npy")
valid_inds = np.load("/scratch/users/patelas/hyperLAI/ancestry_training_splits/80_10_10/valid_indices.npy")
test_inds = np.load("/scratch/users/patelas/hyperLAI/ancestry_training_splits/80_10_10/test_indices.npy")
all_inds = np.sort(np.concatenate([train_inds, valid_inds, test_inds]))
print(all_inds[0], all_inds[-1])
#Create the dataset
# dataset = HyperLoader(data_dir, all_inds, [0,1,2,3,4,5,6], "all")
# dataset = HyperLoader(data_dir, test_inds, [0], "all")
top5_snps = dataset.snps[:,:5]
bottom5_snps = dataset.snps[:,-5:]
np.random.seed(0)
train_shortened = np.random.choice(train_inds, size=2000, replace=False)
print(np.var(top5_snps, axis=0))
print(np.var(bottom5_snps, axis=0))
np.var(top5_snps[train_shortened,:], axis=0)
np.var(bottom5_snps[train_shortened], axis=0)
print(np.sum(top5_snps, axis=0))
print(np.sum(top5_snps[train_shortened], axis=0))
print(np.sum(bottom5_snps, axis=0))
print(np.sum(bottom5_snps[train_shortened], axis=0))
print(np.sum(top5_snps[train_inds], axis=0))
v0 = testk2[:,0]
v0_mean = np.mean(v0)
print(np.sum([(int(x) - v0_mean)**2 for x in v0]) / len(v0))
v0
v0_mean
len(np.unique(train_shortened))
snps_shortened = dataset.snps[train_shortened]
np.sort(np.sum(snps_shortened, axis=0))
hl_eur = HyperLoader("/scratch/users/patelas/hyperLAI/snp_data/whole_genome/variance_filtered_500000_subpops/european/",
train_inds,[0], "all")
eur_top5 = hl_eur.snps[:,:5]
eur_bottom5 = hl_eur.snps[:,-5:]
print(np.var(eur_top5, axis=0))
print(np.var(eur_bottom5, axis=0))
short_sums = np.sum(dataset.snps[train_shortened], axis=0)
import matplotlib.pyplot as plt
plt.scatter(np.arange(len(short_sums)), np.sort(short_sums))
plt.show()
np.sort(short_sums)[::-1][:2000]
np.sum(short_sums < 1050)
np.sum(short_sums > 1000)
378041+234941-500000
hl_sub = HyperLoader("/scratch/users/patelas/hyperLAI/snp_data/whole_genome/variance_filtered_500000_subpops/european/",
train_inds,[6], "all")
len(hl_sub), hl_sub.pop_labels[2]
loader22 = HyperLoader("/scratch/users/patelas/hyperLAI/snp_data/",
train_inds,[0,1,2,3,4,5,6], 22)
import matplotlib.pyplot as plt
plt.plot(np.sort(np.var(loader22.snps, axis=0)))
plt.show()
import numpy as np
np.var([1] * 4743 + [0])
ftest = sim_func_dict["ancestry_label"]
ftest == sim_func_dict["ancestry_label"]
ones = torch.ones([64,1])
zeros = torch.zeros([64,1])
torch.cat([ones, zeros], axis=1).shape
tloader = data.DataLoader(dataset, batch_size = 64)
for i, (snp_data, suppop_labels, pop_labels) in enumerate(tloader):
tcat = torch.stack([suppop_labels, pop_labels], dim=1)
print(tcat.shape)
break
tcat
```
Try European Population
```
euro_train_data = HyperLoader("/scratch/users/patelas/hyperLAI/snp_data/whole_genome/variance_filtered_500000_subpops/european/",
train_inds,[0], "all")
euro_test_data = HyperLoader("/scratch/users/patelas/hyperLAI/snp_data/whole_genome/variance_filtered_500000_subpops/european/",
test_inds,[0], "all")
from sklearn.decomposition import PCA
euro_pca = PCA(n_components=2)
euro_pca.fit(euro_train_data.snps)
euro_test_trans = euro_pca.transform(euro_test_data.snps)
def plot_weights_pca(embeddings, labels, annotations=None):
'''
Plot embeddings. Uncomment/comment as necessary depending on if you want raw embeddings or PCA
'''
# weights_pca = PCA().fit_transform(embeddings)
# scplot = sns.scatterplot(x=weights_pca[:,0], y=weights_pca[:,1], hue=labels)
scplot = sns.scatterplot(x=embeddings[:,0], y=embeddings[:,1], hue=labels)
# plt.xlabel("PC1")
# plt.ylabel("PC2")
# plt.title("PCA of Embedding Weights")
plt.xlabel("Embedding 1")
plt.ylabel("Embedding 2")
plt.title("Embedding Weights")
if annotations is not None:
for line in range(len(labels)):
# if weights_pca[line,1] > -0.3:
# if annotations[line] != "Hazara":
# continue
scplot.text(embeddings[line,0]+0.001, embeddings[line,1],
annotations[line], horizontalalignment='left', color='black', size=8)
import matplotlib.pyplot as plt
import seaborn as sns
plt.figure(figsize=[15, 15], dpi=300)
sp_full = [euro_test_data.suppop_label_index[x] for x in euro_test_data.suppop_labels]
p_full = [euro_test_data.pop_label_index[x] for x in euro_test_data.pop_labels]
plot_weights_pca(euro_test_trans, p_full, annotations=p_full)
plt.show()
euro_test_data = HyperLoader("/scratch/users/patelas/hyperLAI/snp_data/whole_genome/variance_filtered_500000_subpops/european/",
test_inds,[0], "all")
len(euro_data)
```
Dasgupta's Cost
```
hl_sub = HyperLoader("/scratch/users/patelas/hyperLAI/snp_data/whole_genome/variance_filtered_500000_updated/",
test_inds,[0,1,2,3,4,5,6], "all")
from sklearn.decomposition import PCA
all_pca = PCA(n_components=2)
fit_subset = np.random.choice(len(hl2), 2000, replace=False)
all_pca.fit_transform(hl2.snps[fit_subset])
all_test_trans = all_pca.transform(hl_sub.snps)
import matplotlib.pyplot as plt
import seaborn as sns
plt.figure(figsize=[15, 15], dpi=300)
sp_full = [hl_sub.suppop_label_index[x] for x in hl_sub.suppop_labels]
p_full = [hl_sub.pop_label_index[x] for x in hl_sub.pop_labels]
plot_weights_pca(all_test_trans, sp_full, annotations=p_full)
plt.show()
```
|
github_jupyter
|
import sys
sys.path.append("../../../")
sys.path.append("../../hyperLAI/")
sys.path.append("../../../libraries/")
from utils.model_utils import *
from utils.sim_funcs import sim_func_dict
from models.fc_model import fc_model
from features.hyperLAIdataset import HyperLoader
from sklearn.feature_selection import VarianceThreshold
import torch
from torch.utils import data
from features.hyperLAIdataset import HyperLoader
import numpy as np
import json
dataset = HyperLoader("/scratch/users/patelas/hyperLAI/snp_data/", chromosome=22)
train_indices, valid_indices, test_indices = train_valid_test(len(dataset), 0.8, 0.1)
variance_filter(dataset, train_indices, 5000)
snps_train = dataset.snps[train_indices,:]
vt_var = np.var(dataset.snps, axis=0)
var_sorted = np.argsort(vt_var)[::-1]
dataset.snps.shape
vt_var[77852]
import matplotlib.pyplot as plt
plt.scatter(range(len(vt_var)), sorted(vt_var))
plt.show()
train_size = int(0.8 * len(dataset))
valid_size = int(0.1 * len(dataset))
test_size = len(dataset) - train_size - valid_size
np.random.seed(0)
indices = np.array(list(range(len(dataset))))
np.random.shuffle(indices)
train_indices = indices[0 : train_size]
valid_indices = indices[train_size : train_size + valid_size]
test_indices = indices[train_size + valid_size : ]
# sfunc = lambda x,y: (torch.sum(x == y)).float() / len(x)
sfunc = lambda x,y: torch.true_divide(torch.sum(x==y), len(x))
snp_data = torch.tensor(np.random.randint(0,2,size=(4,4)))
snp_data
smat = make_pairwise_similarities(snp_data, sfunc)
trips, sims = trips_and_sims(snp_data, sfunc)
trips
sims
sims[[2,3],:]
input_size=20
num_int_layers=1
int_layer_sizes=[5]
embedding_size=2
dropout_vals=[0.8]
fctest = fc_model(input_size, num_int_layers, int_layer_sizes, embedding_size, dropout_vals)
list(fctest.parameters())
def run_epoch(model, dloader, device, sim_func, optimizer=None):
batch_hyp_losses = []
for i, (snp_data, suppop_labels, pop_labels) in enumerate(dloader):
if model.training:
assert optimizer is not None
optimizer.zero_grad()
else:
assert optimizer is None
triple_ids, similarities = trips_and_sims(snp_data, sim_func)
snp_data = snp_data.to(device)
triple_ids = triple_ids.to(device)
similarities = similarities.to(device)
embeddings_pred = model(snp_data)
hyp_loss = model.loss(embeddings_pred, triple_ids, similarities)
batch_hyp_losses.append(hyp_loss.item())
if model.training:
hyp_loss.backward()
optimizer.step()
return batch_hyp_losses
def train_model(model, train_loader, valid_loader, num_epochs, learning_rate, sim_func,
patience, txt_writer, output_dir, early_stopping,
patience, early_stop_min_delta, optimizer=None):
device = torch.device("cuda") if torch.cuda.is_available() else torch.device("cpu")
if device == torch.device("cuda"):
print("Training occurring on GPU")
model = model.to(device)
if optimizer is None:
optimizer = RAdam(model.parameters(), lr=learning_rate)
if early_stopping:
valid_loss_history = []
best_valid_epoch_loss, best_model = float("inf"), None
for epoch in range(num_epochs):
if torch.cuda.is_available:
torch.cuda.empty_cache()
model.train()
train_batch_losses = run_epoch(model, train_loader, device, sim_func, optimizer)
train_epoch_loss = np.nanmean(train_batch_losses)
print_and_log("Train: epoch %d: average loss = %6.10f" % (epoch + 1, train_epoch_loss), txt_writer)
with torch.no_grad():
model.eval()
valid_batch_losses = run_epoch(model, valid_loader, device, sim_func, optimizer=None)
valid_epoch_loss = np.nanmean(valid_batch_losses)
print_and_log("Valid: epoch %d: average loss = %6.10f" % (epoch + 1, valid_epoch_loss), txt_writer)
if valid_epoch_loss < best_valid_epoch_loss:
best_valid_epoch_loss = valid_epoch_loss
best_model = model
save_model(model, optimizer, valid_epoch_loss, epoch + 1, output_dir+"model.pt")
if early_stopping:
if len(valid_loss_history) < patience + 1:
# Not enough history yet; tack on the loss
valid_loss_history = [valid_epoch_loss] + valid_loss_history
else:
# Tack on the new validation loss, kicking off the old one
valid_loss_history = \
[valid_epoch_loss] + valid_loss_history[:-1]
if len(valid_loss_history) == patience + 1:
# There is sufficient history to check for improvement
best_delta = np.max(np.diff(valid_loss_history))
if best_delta < early_stop_min_delta:
break # Not improving enough
txt_writer.flush()
json_file = open("../../hyperLAI/models/fc_config.json")
vars_json = json.load(json_file)
json_file.close()
eval(vars_json["sim_func"])(np.array([2,3]),np.array([3,3]))
vars_json["restrict_labels"]
import sys
sys.path.append("../../../")
sys.path.append("../../hyperLAI")
%run "../../hyperLAI/models/train_fc_model.py"
dataset = HyperLoader("/scratch/users/patelas/hyperLAI/snp_data/whole_genome/variance_filtered_500000/",
[0,1,2,3,4,5,6], "all")
train_inds = np.load("/scratch/users/patelas/hyperLAI/ancestry_training_splits/80_10_10/train_indices.npy")
test_inds = np.load("/scratch/users/patelas/hyperLAI/ancestry_training_splits/80_10_10/test_indices.npy")
hl2 = HyperLoader("/scratch/users/patelas/hyperLAI/snp_data/whole_genome/variance_filtered_500000_updated/",
train_inds,[0,1,2,3,4,5,6], "all")
hl2.snps.shape
hl2_top = hl2.snps[:,:5]
hl2_bottom = hl2.snps[:,-5:]
np.sum(hl2_top, axis=0)
np.var(top5_snps, axis=0)
np.var(bottom5_snps, axis=0)
hl = HyperLoader("/scratch/users/patelas/hyperLAI/snp_data/whole_genome/variance_filtered_500000/",
[0,1,2], "all")
len(hl)
data_dir = "/scratch/users/patelas/hyperLAI/snp_data/whole_genome/variance_filtered_500000_updated/"
train_inds = np.load("/scratch/users/patelas/hyperLAI/ancestry_training_splits/80_10_10/train_indices.npy")
valid_inds = np.load("/scratch/users/patelas/hyperLAI/ancestry_training_splits/80_10_10/valid_indices.npy")
test_inds = np.load("/scratch/users/patelas/hyperLAI/ancestry_training_splits/80_10_10/test_indices.npy")
all_inds = np.sort(np.concatenate([train_inds, valid_inds, test_inds]))
print(all_inds[0], all_inds[-1])
#Create the dataset
# dataset = HyperLoader(data_dir, all_inds, [0,1,2,3,4,5,6], "all")
# dataset = HyperLoader(data_dir, test_inds, [0], "all")
top5_snps = dataset.snps[:,:5]
bottom5_snps = dataset.snps[:,-5:]
np.random.seed(0)
train_shortened = np.random.choice(train_inds, size=2000, replace=False)
print(np.var(top5_snps, axis=0))
print(np.var(bottom5_snps, axis=0))
np.var(top5_snps[train_shortened,:], axis=0)
np.var(bottom5_snps[train_shortened], axis=0)
print(np.sum(top5_snps, axis=0))
print(np.sum(top5_snps[train_shortened], axis=0))
print(np.sum(bottom5_snps, axis=0))
print(np.sum(bottom5_snps[train_shortened], axis=0))
print(np.sum(top5_snps[train_inds], axis=0))
v0 = testk2[:,0]
v0_mean = np.mean(v0)
print(np.sum([(int(x) - v0_mean)**2 for x in v0]) / len(v0))
v0
v0_mean
len(np.unique(train_shortened))
snps_shortened = dataset.snps[train_shortened]
np.sort(np.sum(snps_shortened, axis=0))
hl_eur = HyperLoader("/scratch/users/patelas/hyperLAI/snp_data/whole_genome/variance_filtered_500000_subpops/european/",
train_inds,[0], "all")
eur_top5 = hl_eur.snps[:,:5]
eur_bottom5 = hl_eur.snps[:,-5:]
print(np.var(eur_top5, axis=0))
print(np.var(eur_bottom5, axis=0))
short_sums = np.sum(dataset.snps[train_shortened], axis=0)
import matplotlib.pyplot as plt
plt.scatter(np.arange(len(short_sums)), np.sort(short_sums))
plt.show()
np.sort(short_sums)[::-1][:2000]
np.sum(short_sums < 1050)
np.sum(short_sums > 1000)
378041+234941-500000
hl_sub = HyperLoader("/scratch/users/patelas/hyperLAI/snp_data/whole_genome/variance_filtered_500000_subpops/european/",
train_inds,[6], "all")
len(hl_sub), hl_sub.pop_labels[2]
loader22 = HyperLoader("/scratch/users/patelas/hyperLAI/snp_data/",
train_inds,[0,1,2,3,4,5,6], 22)
import matplotlib.pyplot as plt
plt.plot(np.sort(np.var(loader22.snps, axis=0)))
plt.show()
import numpy as np
np.var([1] * 4743 + [0])
ftest = sim_func_dict["ancestry_label"]
ftest == sim_func_dict["ancestry_label"]
ones = torch.ones([64,1])
zeros = torch.zeros([64,1])
torch.cat([ones, zeros], axis=1).shape
tloader = data.DataLoader(dataset, batch_size = 64)
for i, (snp_data, suppop_labels, pop_labels) in enumerate(tloader):
tcat = torch.stack([suppop_labels, pop_labels], dim=1)
print(tcat.shape)
break
tcat
euro_train_data = HyperLoader("/scratch/users/patelas/hyperLAI/snp_data/whole_genome/variance_filtered_500000_subpops/european/",
train_inds,[0], "all")
euro_test_data = HyperLoader("/scratch/users/patelas/hyperLAI/snp_data/whole_genome/variance_filtered_500000_subpops/european/",
test_inds,[0], "all")
from sklearn.decomposition import PCA
euro_pca = PCA(n_components=2)
euro_pca.fit(euro_train_data.snps)
euro_test_trans = euro_pca.transform(euro_test_data.snps)
def plot_weights_pca(embeddings, labels, annotations=None):
'''
Plot embeddings. Uncomment/comment as necessary depending on if you want raw embeddings or PCA
'''
# weights_pca = PCA().fit_transform(embeddings)
# scplot = sns.scatterplot(x=weights_pca[:,0], y=weights_pca[:,1], hue=labels)
scplot = sns.scatterplot(x=embeddings[:,0], y=embeddings[:,1], hue=labels)
# plt.xlabel("PC1")
# plt.ylabel("PC2")
# plt.title("PCA of Embedding Weights")
plt.xlabel("Embedding 1")
plt.ylabel("Embedding 2")
plt.title("Embedding Weights")
if annotations is not None:
for line in range(len(labels)):
# if weights_pca[line,1] > -0.3:
# if annotations[line] != "Hazara":
# continue
scplot.text(embeddings[line,0]+0.001, embeddings[line,1],
annotations[line], horizontalalignment='left', color='black', size=8)
import matplotlib.pyplot as plt
import seaborn as sns
plt.figure(figsize=[15, 15], dpi=300)
sp_full = [euro_test_data.suppop_label_index[x] for x in euro_test_data.suppop_labels]
p_full = [euro_test_data.pop_label_index[x] for x in euro_test_data.pop_labels]
plot_weights_pca(euro_test_trans, p_full, annotations=p_full)
plt.show()
euro_test_data = HyperLoader("/scratch/users/patelas/hyperLAI/snp_data/whole_genome/variance_filtered_500000_subpops/european/",
test_inds,[0], "all")
len(euro_data)
hl_sub = HyperLoader("/scratch/users/patelas/hyperLAI/snp_data/whole_genome/variance_filtered_500000_updated/",
test_inds,[0,1,2,3,4,5,6], "all")
from sklearn.decomposition import PCA
all_pca = PCA(n_components=2)
fit_subset = np.random.choice(len(hl2), 2000, replace=False)
all_pca.fit_transform(hl2.snps[fit_subset])
all_test_trans = all_pca.transform(hl_sub.snps)
import matplotlib.pyplot as plt
import seaborn as sns
plt.figure(figsize=[15, 15], dpi=300)
sp_full = [hl_sub.suppop_label_index[x] for x in hl_sub.suppop_labels]
p_full = [hl_sub.pop_label_index[x] for x in hl_sub.pop_labels]
plot_weights_pca(all_test_trans, sp_full, annotations=p_full)
plt.show()
| 0.437824 | 0.352954 |
# Cross-subject decoding Motor responses
### (LINDEX v. LMID and RINDEX v. RMID)
#### Takuya Ito
#### 2/28/2018
```
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
import multiprocessing as mp
import scipy.stats as stats
import os
os.environ['OMP_NUM_THREADS'] = str(1)
import statsmodels.sandbox.stats.multicomp as mc
import seaborn as sns
import h5py
import tools_group_rsa_python3 as tools_group
from importlib import reload
import nibabel as nib
sns.set_style("whitegrid")
plt.rcParams["font.family"] = "FreeSans"
```
## 0.1 Load data
```
# Excluding 084
subjNums = ['013','014','016','017','018','021','023','024','026','027','028','030','031','032','033',
'034','035','037','038','039','040','041','042','043','045','046','047','048','049','050',
'053','055','056','057','058','062','063','066','067','068','069','070','072','074','075',
'076','077','081','085','086','087','088','090','092','093','094','095','097','098','099',
'101','102','103','104','105','106','108','109','110','111','112','114','115','117','119',
'120','121','122','123','124','125','126','127','128','129','130','131','132','134','135',
'136','137','138','139','140','141']
basedir = '/projects3/SRActFlow/'
# Using final partition
networkdef = np.loadtxt('/projects3/NetworkDiversity/data/network_partition.txt')
networkorder = np.asarray(sorted(range(len(networkdef)), key=lambda k: networkdef[k]))
networkorder.shape = (len(networkorder),1)
# network mappings for final partition set
networkmappings = {'fpn':7, 'vis1':1, 'vis2':2, 'smn':3, 'aud':8, 'lan':6, 'dan':5, 'con':4, 'dmn':9,
'pmulti':10, 'none1':11, 'none2':12}
networks = networkmappings.keys()
xticks = {}
reorderednetworkaffil = networkdef[networkorder]
for net in networks:
netNum = networkmappings[net]
netind = np.where(reorderednetworkaffil==netNum)[0]
tick = np.max(netind)
xticks[tick] = net
## General parameters/variables
nParcels = 360
nSubjs = len(subjNums)
glasserfile2 = '/projects/AnalysisTools/ParcelsGlasser2016/Q1-Q6_RelatedParcellation210.LR.CorticalAreas_dil_Colors.32k_fs_RL.dlabel.nii'
glasser2 = nib.load(glasserfile2).get_data()
glasser2 = np.squeeze(glasser2)
sortednets = np.sort(list(xticks.keys()))
orderednetworks = []
for net in sortednets: orderednetworks.append(xticks[net])
networkpalette = ['royalblue','slateblue','paleturquoise','darkorchid','limegreen',
'lightseagreen','yellow','orchid','r','peru','orange','olivedrab']
networkpalette = np.asarray(networkpalette)
OrderedNetworks = ['VIS1','VIS2','SMN','CON','DAN','LAN','FPN','AUD','DMN','PMM','VMM','ORA']
# gsr = True
nResponses = 2
data_task_rh = np.zeros((len(glasser2),nResponses,len(subjNums)))
data_task_lh = np.zeros((len(glasser2),nResponses,len(subjNums)))
scount = 0
for subj in subjNums:
data_task_rh[:,:,scount] = tools_group.loadMotorResponses(subj,hand='Right')
data_task_lh[:,:,scount] = tools_group.loadMotorResponses(subj,hand='Left')
scount += 1
```
## 1.1 Run across subject decoding on hand-specific motor responses
```
nproc = 30
# ncvs = 50
tools_group = reload(tools_group)
ncvs = 1
rois = np.where(networkdef==networkmappings['smn'])[0]
print('Running decoding for Right hand responses')
distances_baseline_rh, rmatches_rh, rmismatches_rh = tools_group.conditionDecodings(data_task_rh, rois, ncvs=ncvs, effects=True, motorOutput=True, nproc=nproc)
print('Running decoding for Left hand responses')
distances_baseline_lh, rmatches_lh, rmismatches_lh = tools_group.conditionDecodings(data_task_lh, rois, ncvs=ncvs, effects=True, motorOutput=True, nproc=nproc)
```
## 1.2 Compute statistics
#### Right-hand decoding statistics
```
smnROIs = np.where(networkdef==networkmappings['smn'])[0]
ntrials = distances_baseline_rh.shape[1]
statistics_rh = np.zeros((len(smnROIs),3)) # acc, q, acc_thresh
for roicount in range(len(smnROIs)):
p = stats.binom_test(np.mean(distances_baseline_rh[roicount,:])*ntrials,n=ntrials,p=1/float(data_task_rh.shape[1]))
if np.mean(distances_baseline_rh[roicount,:])>1/float(data_task_rh.shape[1]):
p = p/2.0
else:
p = 1.0-p/2.0
statistics_rh[roicount,0] = np.mean(distances_baseline_rh[roicount,:])
statistics_rh[roicount,1] = p
h0, qs = mc.fdrcorrection0(statistics_rh[:,1])
for roicount in range(len(smnROIs)):
statistics_rh[roicount,1] = qs[roicount]
statistics_rh[roicount,2] = h0[roicount]*statistics_rh[roicount,0]
# Count number of significant ROIs for RH decoding
sig_ind = np.where(statistics_rh[:,1]<0.05)[0]
sig_ind = np.asarray(sig_ind,dtype=int)
print('Number of ROIs significant for right hand responses:', sig_ind.shape[0])
if sig_ind.shape[0]>0:
print('Significant ROIs:', smnROIs[sig_ind]+1)
# print('R_matched effect-size:', rmatches_rh[sig_ind])
# print('R_mismatched effect-size:', rmismatches_rh[sig_ind])
print('Accuracies:', statistics_rh[sig_ind,0])
```
#### Left-hand decoding statistics
```
smnROIs = np.where(networkdef==networkmappings['smn'])[0]
ntrials = distances_baseline_lh.shape[1]
statistics_lh = np.zeros((len(smnROIs),3)) # acc, q, acc_thresh
for roicount in range(len(smnROIs)):
p = stats.binom_test(np.mean(distances_baseline_lh[roicount,:])*ntrials,n=ntrials,p=1/float(data_task_lh.shape[1]))
if np.mean(distances_baseline_lh[roicount,:])>1/float(data_task_lh.shape[1]):
p = p/2.0
else:
p = 1.0-p/2.0
statistics_lh[roicount,0] = np.mean(distances_baseline_lh[roicount,:])
statistics_lh[roicount,1] = p
h0, qs = mc.fdrcorrection0(statistics_lh[:,1])
for roicount in range(len(smnROIs)):
statistics_lh[roicount,1] = qs[roicount]
statistics_lh[roicount,2] = h0[roicount]*statistics_lh[roicount,0]
# Count number of significant ROIs for LH decoding
sig_ind = np.where(statistics_lh[:,1]<0.05)[0]
print('Number of ROIs significant for right hand responses:', sig_ind.shape[0])
if sig_ind.shape[0]>0:
print('Significant ROIs:', smnROIs[sig_ind] + 1)
# print 'R_matched effect-size:', rmatches_lh[sig_ind]
# print 'R_mismatched effect-size:', rmismatches_lh[sig_ind]
print('Accuracies:', statistics_lh[sig_ind,0])
```
## 1.3 Map accuracies back to cortical surface
```
# Put all data into a single matrix (since we only run a single classification)
lefthand = np.zeros((glasser2.shape[0],3))
righthand = np.zeros((glasser2.shape[0],3))
roicount = 0
for roi in smnROIs:
# Print significant parcel number
vertex_ind = np.where(glasser2==roi+1)[0]
lefthand[vertex_ind,0] = statistics_lh[roicount,0]
lefthand[vertex_ind,1] = statistics_lh[roicount,1]
lefthand[vertex_ind,2] = statistics_lh[roicount,2]
righthand[vertex_ind,0] = statistics_rh[roicount,0]
righthand[vertex_ind,1] = statistics_rh[roicount,1]
righthand[vertex_ind,2] = statistics_rh[roicount,2]
roicount += 1
np.savetxt('/projects3/SRActFlow/data/results/GroupfMRI/MotorResponseDecoding/MotorResponseRegions_LH.csv', smnROIs[np.where(statistics_lh[:,1]<0.05)[0]], delimiter=',')
np.savetxt('/projects3/SRActFlow/data/results/GroupfMRI/MotorResponseDecoding/MotorResponseRegions_RH.csv', smnROIs[np.where(statistics_rh[:,1]<0.05)[0]], delimiter=',')
####
# Write file to csv and run wb_command
outdir = '/projects3/SRActFlow/data/results/GroupfMRI/MotorResponseDecoding/'
filename = 'MotorResponseDecodingsLH'
np.savetxt(outdir + filename + '.csv', lefthand,fmt='%s')
wb_file = filename + '.dscalar.nii'
wb_command = 'wb_command -cifti-convert -from-text ' + outdir + filename + '.csv ' + glasserfile2 + ' ' + outdir + wb_file + ' -reset-scalars'
os.system(wb_command)
filename = 'MotorResponseDecodingsRH'
np.savetxt(outdir + filename + '.csv', righthand,fmt='%s')
wb_file = filename + '.dscalar.nii'
wb_command = 'wb_command -cifti-convert -from-text ' + outdir + filename + '.csv ' + glasserfile2 + ' ' + outdir + wb_file + ' -reset-scalars'
os.system(wb_command)
```
|
github_jupyter
|
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
import multiprocessing as mp
import scipy.stats as stats
import os
os.environ['OMP_NUM_THREADS'] = str(1)
import statsmodels.sandbox.stats.multicomp as mc
import seaborn as sns
import h5py
import tools_group_rsa_python3 as tools_group
from importlib import reload
import nibabel as nib
sns.set_style("whitegrid")
plt.rcParams["font.family"] = "FreeSans"
# Excluding 084
subjNums = ['013','014','016','017','018','021','023','024','026','027','028','030','031','032','033',
'034','035','037','038','039','040','041','042','043','045','046','047','048','049','050',
'053','055','056','057','058','062','063','066','067','068','069','070','072','074','075',
'076','077','081','085','086','087','088','090','092','093','094','095','097','098','099',
'101','102','103','104','105','106','108','109','110','111','112','114','115','117','119',
'120','121','122','123','124','125','126','127','128','129','130','131','132','134','135',
'136','137','138','139','140','141']
basedir = '/projects3/SRActFlow/'
# Using final partition
networkdef = np.loadtxt('/projects3/NetworkDiversity/data/network_partition.txt')
networkorder = np.asarray(sorted(range(len(networkdef)), key=lambda k: networkdef[k]))
networkorder.shape = (len(networkorder),1)
# network mappings for final partition set
networkmappings = {'fpn':7, 'vis1':1, 'vis2':2, 'smn':3, 'aud':8, 'lan':6, 'dan':5, 'con':4, 'dmn':9,
'pmulti':10, 'none1':11, 'none2':12}
networks = networkmappings.keys()
xticks = {}
reorderednetworkaffil = networkdef[networkorder]
for net in networks:
netNum = networkmappings[net]
netind = np.where(reorderednetworkaffil==netNum)[0]
tick = np.max(netind)
xticks[tick] = net
## General parameters/variables
nParcels = 360
nSubjs = len(subjNums)
glasserfile2 = '/projects/AnalysisTools/ParcelsGlasser2016/Q1-Q6_RelatedParcellation210.LR.CorticalAreas_dil_Colors.32k_fs_RL.dlabel.nii'
glasser2 = nib.load(glasserfile2).get_data()
glasser2 = np.squeeze(glasser2)
sortednets = np.sort(list(xticks.keys()))
orderednetworks = []
for net in sortednets: orderednetworks.append(xticks[net])
networkpalette = ['royalblue','slateblue','paleturquoise','darkorchid','limegreen',
'lightseagreen','yellow','orchid','r','peru','orange','olivedrab']
networkpalette = np.asarray(networkpalette)
OrderedNetworks = ['VIS1','VIS2','SMN','CON','DAN','LAN','FPN','AUD','DMN','PMM','VMM','ORA']
# gsr = True
nResponses = 2
data_task_rh = np.zeros((len(glasser2),nResponses,len(subjNums)))
data_task_lh = np.zeros((len(glasser2),nResponses,len(subjNums)))
scount = 0
for subj in subjNums:
data_task_rh[:,:,scount] = tools_group.loadMotorResponses(subj,hand='Right')
data_task_lh[:,:,scount] = tools_group.loadMotorResponses(subj,hand='Left')
scount += 1
nproc = 30
# ncvs = 50
tools_group = reload(tools_group)
ncvs = 1
rois = np.where(networkdef==networkmappings['smn'])[0]
print('Running decoding for Right hand responses')
distances_baseline_rh, rmatches_rh, rmismatches_rh = tools_group.conditionDecodings(data_task_rh, rois, ncvs=ncvs, effects=True, motorOutput=True, nproc=nproc)
print('Running decoding for Left hand responses')
distances_baseline_lh, rmatches_lh, rmismatches_lh = tools_group.conditionDecodings(data_task_lh, rois, ncvs=ncvs, effects=True, motorOutput=True, nproc=nproc)
smnROIs = np.where(networkdef==networkmappings['smn'])[0]
ntrials = distances_baseline_rh.shape[1]
statistics_rh = np.zeros((len(smnROIs),3)) # acc, q, acc_thresh
for roicount in range(len(smnROIs)):
p = stats.binom_test(np.mean(distances_baseline_rh[roicount,:])*ntrials,n=ntrials,p=1/float(data_task_rh.shape[1]))
if np.mean(distances_baseline_rh[roicount,:])>1/float(data_task_rh.shape[1]):
p = p/2.0
else:
p = 1.0-p/2.0
statistics_rh[roicount,0] = np.mean(distances_baseline_rh[roicount,:])
statistics_rh[roicount,1] = p
h0, qs = mc.fdrcorrection0(statistics_rh[:,1])
for roicount in range(len(smnROIs)):
statistics_rh[roicount,1] = qs[roicount]
statistics_rh[roicount,2] = h0[roicount]*statistics_rh[roicount,0]
# Count number of significant ROIs for RH decoding
sig_ind = np.where(statistics_rh[:,1]<0.05)[0]
sig_ind = np.asarray(sig_ind,dtype=int)
print('Number of ROIs significant for right hand responses:', sig_ind.shape[0])
if sig_ind.shape[0]>0:
print('Significant ROIs:', smnROIs[sig_ind]+1)
# print('R_matched effect-size:', rmatches_rh[sig_ind])
# print('R_mismatched effect-size:', rmismatches_rh[sig_ind])
print('Accuracies:', statistics_rh[sig_ind,0])
smnROIs = np.where(networkdef==networkmappings['smn'])[0]
ntrials = distances_baseline_lh.shape[1]
statistics_lh = np.zeros((len(smnROIs),3)) # acc, q, acc_thresh
for roicount in range(len(smnROIs)):
p = stats.binom_test(np.mean(distances_baseline_lh[roicount,:])*ntrials,n=ntrials,p=1/float(data_task_lh.shape[1]))
if np.mean(distances_baseline_lh[roicount,:])>1/float(data_task_lh.shape[1]):
p = p/2.0
else:
p = 1.0-p/2.0
statistics_lh[roicount,0] = np.mean(distances_baseline_lh[roicount,:])
statistics_lh[roicount,1] = p
h0, qs = mc.fdrcorrection0(statistics_lh[:,1])
for roicount in range(len(smnROIs)):
statistics_lh[roicount,1] = qs[roicount]
statistics_lh[roicount,2] = h0[roicount]*statistics_lh[roicount,0]
# Count number of significant ROIs for LH decoding
sig_ind = np.where(statistics_lh[:,1]<0.05)[0]
print('Number of ROIs significant for right hand responses:', sig_ind.shape[0])
if sig_ind.shape[0]>0:
print('Significant ROIs:', smnROIs[sig_ind] + 1)
# print 'R_matched effect-size:', rmatches_lh[sig_ind]
# print 'R_mismatched effect-size:', rmismatches_lh[sig_ind]
print('Accuracies:', statistics_lh[sig_ind,0])
# Put all data into a single matrix (since we only run a single classification)
lefthand = np.zeros((glasser2.shape[0],3))
righthand = np.zeros((glasser2.shape[0],3))
roicount = 0
for roi in smnROIs:
# Print significant parcel number
vertex_ind = np.where(glasser2==roi+1)[0]
lefthand[vertex_ind,0] = statistics_lh[roicount,0]
lefthand[vertex_ind,1] = statistics_lh[roicount,1]
lefthand[vertex_ind,2] = statistics_lh[roicount,2]
righthand[vertex_ind,0] = statistics_rh[roicount,0]
righthand[vertex_ind,1] = statistics_rh[roicount,1]
righthand[vertex_ind,2] = statistics_rh[roicount,2]
roicount += 1
np.savetxt('/projects3/SRActFlow/data/results/GroupfMRI/MotorResponseDecoding/MotorResponseRegions_LH.csv', smnROIs[np.where(statistics_lh[:,1]<0.05)[0]], delimiter=',')
np.savetxt('/projects3/SRActFlow/data/results/GroupfMRI/MotorResponseDecoding/MotorResponseRegions_RH.csv', smnROIs[np.where(statistics_rh[:,1]<0.05)[0]], delimiter=',')
####
# Write file to csv and run wb_command
outdir = '/projects3/SRActFlow/data/results/GroupfMRI/MotorResponseDecoding/'
filename = 'MotorResponseDecodingsLH'
np.savetxt(outdir + filename + '.csv', lefthand,fmt='%s')
wb_file = filename + '.dscalar.nii'
wb_command = 'wb_command -cifti-convert -from-text ' + outdir + filename + '.csv ' + glasserfile2 + ' ' + outdir + wb_file + ' -reset-scalars'
os.system(wb_command)
filename = 'MotorResponseDecodingsRH'
np.savetxt(outdir + filename + '.csv', righthand,fmt='%s')
wb_file = filename + '.dscalar.nii'
wb_command = 'wb_command -cifti-convert -from-text ' + outdir + filename + '.csv ' + glasserfile2 + ' ' + outdir + wb_file + ' -reset-scalars'
os.system(wb_command)
| 0.161585 | 0.664186 |
#### - Sobhan Moradian Daghigh
#### - 12/4/2021
#### - ML - EX02 - Q3
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from sklearn import datasets
from sklearn.metrics import mean_absolute_error, mean_squared_error, accuracy_score
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import Normalizer
import random
import operator
```
#### Reading data
```
iris = datasets.load_iris()
dataset = pd.DataFrame(data= np.c_[iris.data, iris.target], columns= iris.feature_names + ['target'])
dataset.head()
dataset.info()
```
<!-- #### Normalizing -->
```
# scaler = Normalizer().fit(dataset.iloc[:, :-1])
# normalized_dataset = scaler.transform(dataset.iloc[:, :-1])
# normalized_dataset = pd.DataFrame(normalized_dataset, columns=dataset.columns[:-1])
# normalized_dataset['target'] = dataset.iloc[:, -1]
# normalized_dataset
```
#### Implementation of cross validation spliter
```
def cross_validation(dataset, k_folds=10):
# Shuffling the dataset
shuffled = dataset.sample(frac=1)
remind = shuffled.shape[0] % k_folds
shuffled = shuffled.iloc[:-remind, :] if remind != 0 else shuffled
return np.array_split(shuffled, k_folds)
```
#### As a test:
```
cross_vals = cross_validation(normalized_dataset)
cross_vals[0].head()
```
#### Implementation of euclidean distance
```
def euclideanDistance(vectorA, vectorB):
distance = 0
for i in range(len(vectorA) - 1):
distance += np.power(np.subtract(vectorA[i], vectorB[i]), 2)
return np.sqrt(distance)
```
#### Implementation of KNNClassifier
```
class kNNClassifier:
def __init__(self, k_neighbors):
self.k = k_neighbors
def fit(self, train):
self.train = train
def getNeighbors(self, test):
distances = []
for i, trainRow in self.train.iterrows():
distances.append([trainRow, euclideanDistance(test, trainRow), self.train.iloc[:, -1][i]])
distances.sort(key=operator.itemgetter(1))
neighbors = []
for index in range(self.k):
neighbors.append(distances[index])
return neighbors
def predict(self, test):
self.test = test
predictions = []
for i, testRow in self.test.iterrows():
neighbors = self.getNeighbors(testRow)
output= [row[-1] for row in neighbors]
prediction = max(set(output), key=output.count)
predictions.append(prediction)
return predictions
```
#### Find the optimal k_neighbors using cross validation
```
cross_vals = cross_validation(normalized_dataset)
k_range = 31
acc = []
acc_index = []
for i in range(1,k_range, 2):
accus = []
for fold in cross_vals:
train = normalized_dataset[~normalized_dataset.index.isin(fold.index)]
test = fold
knn = kNNClassifier(k_neighbors=i)
knn.fit(train)
predictions = knn.predict(test)
accuracy = accuracy_score(test.iloc[:, -1], predictions)
accus.append(accuracy)
max_accu = np.mean(accus)
acc.append(max_accu)
acc_index.append(i)
plt.figure(figsize=(10,6))
plt.plot(acc_index, acc, color = 'blue', linestyle='dashed',
marker='o', markerfacecolor='red', markersize=10)
plt.title('accuracy vs. K Value')
plt.xlabel('K')
plt.ylabel('Accuracy')
print("Maximum accuracy: {:.3f} at K = {}".format(max(acc), np.multiply(acc.index(max(acc)), 2) + 1))
```
#### So the winner k is 3.
#### Part B:
#### Split data into 80% of train and 20% of test.
```
x_train, x_test, y_train, y_test = train_test_split(normalized_dataset.iloc[:, :-1], normalized_dataset.iloc[:, -1], train_size=0.8, shuffle=True)
train = pd.DataFrame(x_train)
train['target'] = y_train
test = pd.DataFrame(x_test)
test['target'] = y_test
knn = kNNClassifier(k_neighbors=3)
knn.fit(train)
ts_predictions = knn.predict(test)
accuracy = accuracy_score(test.iloc[:, -1], ts_predictions)
ts_MSE = mean_squared_error(y_test, ts_predictions)
ts_RMSE = mean_squared_error(y_test, ts_predictions, squared=False)
ts_MAE = mean_absolute_error(y_test, ts_predictions)
tr_predictions = knn.predict(train)
tr_MSE = mean_squared_error(y_train, tr_predictions)
tr_RMSE = mean_squared_error(y_train, tr_predictions, squared=False)
tr_MAE = mean_absolute_error(y_train, tr_predictions)
print('Train:')
print('MSE: {:.3f}'.format(tr_MSE))
print('RMSE: {:.3f}'.format(tr_RMSE))
print('MAE: {:.3f}'.format(tr_MAE))
print('\nTest:')
print('Accuracy: {:.3f}'.format(accuracy))
print('MSE: {:.3f}'.format(ts_MSE))
print('RMSE: {:.3f}'.format(ts_RMSE))
print('MAE: {:.3f}'.format(ts_MAE))
```
|
github_jupyter
|
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from sklearn import datasets
from sklearn.metrics import mean_absolute_error, mean_squared_error, accuracy_score
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import Normalizer
import random
import operator
iris = datasets.load_iris()
dataset = pd.DataFrame(data= np.c_[iris.data, iris.target], columns= iris.feature_names + ['target'])
dataset.head()
dataset.info()
# scaler = Normalizer().fit(dataset.iloc[:, :-1])
# normalized_dataset = scaler.transform(dataset.iloc[:, :-1])
# normalized_dataset = pd.DataFrame(normalized_dataset, columns=dataset.columns[:-1])
# normalized_dataset['target'] = dataset.iloc[:, -1]
# normalized_dataset
def cross_validation(dataset, k_folds=10):
# Shuffling the dataset
shuffled = dataset.sample(frac=1)
remind = shuffled.shape[0] % k_folds
shuffled = shuffled.iloc[:-remind, :] if remind != 0 else shuffled
return np.array_split(shuffled, k_folds)
cross_vals = cross_validation(normalized_dataset)
cross_vals[0].head()
def euclideanDistance(vectorA, vectorB):
distance = 0
for i in range(len(vectorA) - 1):
distance += np.power(np.subtract(vectorA[i], vectorB[i]), 2)
return np.sqrt(distance)
class kNNClassifier:
def __init__(self, k_neighbors):
self.k = k_neighbors
def fit(self, train):
self.train = train
def getNeighbors(self, test):
distances = []
for i, trainRow in self.train.iterrows():
distances.append([trainRow, euclideanDistance(test, trainRow), self.train.iloc[:, -1][i]])
distances.sort(key=operator.itemgetter(1))
neighbors = []
for index in range(self.k):
neighbors.append(distances[index])
return neighbors
def predict(self, test):
self.test = test
predictions = []
for i, testRow in self.test.iterrows():
neighbors = self.getNeighbors(testRow)
output= [row[-1] for row in neighbors]
prediction = max(set(output), key=output.count)
predictions.append(prediction)
return predictions
cross_vals = cross_validation(normalized_dataset)
k_range = 31
acc = []
acc_index = []
for i in range(1,k_range, 2):
accus = []
for fold in cross_vals:
train = normalized_dataset[~normalized_dataset.index.isin(fold.index)]
test = fold
knn = kNNClassifier(k_neighbors=i)
knn.fit(train)
predictions = knn.predict(test)
accuracy = accuracy_score(test.iloc[:, -1], predictions)
accus.append(accuracy)
max_accu = np.mean(accus)
acc.append(max_accu)
acc_index.append(i)
plt.figure(figsize=(10,6))
plt.plot(acc_index, acc, color = 'blue', linestyle='dashed',
marker='o', markerfacecolor='red', markersize=10)
plt.title('accuracy vs. K Value')
plt.xlabel('K')
plt.ylabel('Accuracy')
print("Maximum accuracy: {:.3f} at K = {}".format(max(acc), np.multiply(acc.index(max(acc)), 2) + 1))
x_train, x_test, y_train, y_test = train_test_split(normalized_dataset.iloc[:, :-1], normalized_dataset.iloc[:, -1], train_size=0.8, shuffle=True)
train = pd.DataFrame(x_train)
train['target'] = y_train
test = pd.DataFrame(x_test)
test['target'] = y_test
knn = kNNClassifier(k_neighbors=3)
knn.fit(train)
ts_predictions = knn.predict(test)
accuracy = accuracy_score(test.iloc[:, -1], ts_predictions)
ts_MSE = mean_squared_error(y_test, ts_predictions)
ts_RMSE = mean_squared_error(y_test, ts_predictions, squared=False)
ts_MAE = mean_absolute_error(y_test, ts_predictions)
tr_predictions = knn.predict(train)
tr_MSE = mean_squared_error(y_train, tr_predictions)
tr_RMSE = mean_squared_error(y_train, tr_predictions, squared=False)
tr_MAE = mean_absolute_error(y_train, tr_predictions)
print('Train:')
print('MSE: {:.3f}'.format(tr_MSE))
print('RMSE: {:.3f}'.format(tr_RMSE))
print('MAE: {:.3f}'.format(tr_MAE))
print('\nTest:')
print('Accuracy: {:.3f}'.format(accuracy))
print('MSE: {:.3f}'.format(ts_MSE))
print('RMSE: {:.3f}'.format(ts_RMSE))
print('MAE: {:.3f}'.format(ts_MAE))
| 0.78316 | 0.915545 |
```
# To push the split for training and developing data faster
# we need to skip the index when the timeline is not continuous. (This is covered by dropping all na values)
# The problem arise when there is not enough data after skipping.
# When that happen, the program skip again, leaving some valuable data behind.
# The best thing that can happen is when we skip the second time, if the timeline is not continuous, it will skip
# to the earliest not continous timeline. If we doesn't have any marker, we must find it manually by looping each index.
# So we need to have a marker. It will be put in the raw data files.
# Put timeskip marker in the raw datafiles. (New columns name Timeskip, value is the first time data appear again after the index)
# If after the skip, the timeline is not continous, find the time skip marker and put the position there.
# The refined file doesn't have data in PM (Missing data -> np.nan), findout why *** Not Done 2018 have some problem
# The split for training and developing data has index out of bound, fix it.
# The split for target have some difficulty:
# we can't shift the data so we have to merge it with splitting training data
# When splitting the data
# We don't need to remove the data, we can just calculate like normal.
# Then in the finished file, we drop all data which deemed Missing.
# When putting data to the model, we need to figure out a way to know which data is dropped
# !pip install --upgrade sklearn
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from datetime import datetime
import seaborn as sns
import plotly.express as px
from itertools import product
import warnings
import statsmodels.api as sm
from sklearn.preprocessing import MinMaxScaler
from sklearn.model_selection import GridSearchCV
import holidays
plt.style.use('seaborn-darkgrid')
import joblib
import pandas as pd
import numpy as np
import glob
import xarray as xr
idx = pd.IndexSlice
root_path = r'/mnt/4ba37af6-51fd-47bc-8321-8c500c229114/study/School/KHOA LUAN TOT NGHIEP/'
_thudohanoi_data_path = r'/mnt/4ba37af6-51fd-47bc-8321-8c500c229114/study/School/KHOA LUAN TOT NGHIEP/Data/thudohanoi/refined_data'
_thudohanoi_files = glob.glob(_thudohanoi_data_path + '/*.csv')
thudohanoi_df = pd.DataFrame()
for file in _thudohanoi_files:
print('Currently processing file \n{}'.format(file))
thudohanoi_df = thudohanoi_df.append(pd.read_csv(file, parse_dates=True, index_col=['site_id', 'time'],
dtype={'CO': np.float64, 'NO2': np.float64,'PM25': np.float64,
'AQI_h': np.float64, 'AQI_h_I': np.int, 'site_id': np.int}))
# Remove site 16 because of some inconsistency in data
hanoi_df = thudohanoi_df[(thudohanoi_df.index.get_level_values(0) != 49)].copy()
hcm_df = thudohanoi_df[(thudohanoi_df.index.get_level_values(0) == 49)].copy()
def extract_time_features(df):
# Job: Expand time in data
time_index = df.index.get_level_values(1)
df_time_features = pd.DataFrame()
df_time_features['Hour'] = time_index.hour.astype(float)
df_time_features['Month'] = time_index.month.astype(float)
df_time_features['Day of Week'] = time_index.dayofweek.astype(float)
df_time_features['Day of Month'] = time_index.day.astype(float)
df_time_features['Days in Month'] = time_index.daysinmonth.astype(float)
df_time_features['Year'] = time_index.year.astype(float)
# Job: Encode time cyclical data
hour_in_day = 23
df_time_features['sin_hour'] = np.sin(2*np.pi*df_time_features['Hour']/hour_in_day)
df_time_features['cos_hour'] = np.cos(2*np.pi*df_time_features['Hour']/hour_in_day)
month_in_year = 12
df_time_features['sin_month'] = np.sin(2*np.pi*df_time_features['Month']/month_in_year)
df_time_features['cos_month'] = np.cos(2*np.pi*df_time_features['Month']/month_in_year)
day_in_week = 6
df_time_features['sin_dayweek'] = np.sin(2*np.pi*df_time_features['Day of Week']/day_in_week)
df_time_features['cos_dayweek'] = np.cos(2*np.pi*df_time_features['Day of Week']/day_in_week)
df_time_features['sin_daymonth'] = np.sin(2*np.pi*df_time_features['Day of Month']/df_time_features['Days in Month'])
df_time_features['cos_daymonth'] = np.cos(2*np.pi*df_time_features['Day of Month']/df_time_features['Days in Month'])
# One hot encode year data
one_hot_df = pd.get_dummies(df_time_features['Year'], drop_first=True, prefix='year')
df_time_features = df_time_features.join(one_hot_df)
# Input weekday/weekend/holiday data
vn_holidays = np.array(list(holidays.VN(years=[2015,2016,2017,2018,2019,2020,2021]).keys()))
holiday_mask = np.isin(time_index.date, vn_holidays)
masks = (holiday_mask) | (df_time_features['Day of Week'].values == 5) | (df_time_features['Day of Week'].values == 6)
df_time_features['day_off'] = np.where(masks == True, 1, 0)
df_time_features = df_time_features.drop(columns=['Day of Month', 'Month', 'Day of Week', 'Days in Month', 'Year', 'Hour'])
# Input lagged data
windows = list(range(1,13))
windows.append(24)
for window in windows:
feature = 'AQI_h'
series_rolled = df['AQI_h'].rolling(window=window, min_periods=0)
series_mean = series_rolled.mean().shift(1).reset_index()
series_std = series_rolled.std().shift(1).reset_index()
df_time_features[f"{feature}_mean_lag{window}"] = series_mean['AQI_h'].values
# df_time_features[f"{feature}_std_lag{window}"] = series_std['AQI_h'].values
df_time_features.fillna(df_time_features.mean(), inplace=True)
df_time_features.fillna(df['AQI_h'].mean(), inplace=True)
return df_time_features.values, df_time_features.columns
def add_features(df):
# Change all data to numpy, then concatenate those numpy.
# Then construct the dataframe to old frame. This can work
data_df = df[['AQI_h']].copy()
# Job: Normalize train data
scaler = MinMaxScaler(feature_range=(-1,1))
for col in ['AQI_h']:
data_df[[col]] = scaler.fit_transform(data_df[[col]])
columns = ['site_id', 'time', 'AQI_h']
df_numpy = data_df.reset_index().to_numpy()
# Add onehot site label
one_hot_site = pd.get_dummies(data_df.index.get_level_values(0), prefix='site', drop_first=True).astype(int)
columns.extend(one_hot_site.columns)
# Add onehot air category
one_hot_cat = pd.get_dummies(df['AQI_h_I'], drop_first=True, prefix='cat').astype(int)
columns.extend(one_hot_cat.columns)
# Add time features
time_features, time_columns = extract_time_features(data_df)
columns.extend(time_columns)
df_numpy = np.concatenate([df_numpy, one_hot_site.values, one_hot_cat.values, time_features], axis=1)
final_df = pd.DataFrame(df_numpy, columns=columns).set_index(['site_id', 'time'])
for float_col in final_df.loc[:, final_df.dtypes == float].columns:
final_df.loc[:, float_col] = final_df.loc[:, float_col].values.round(6)
return final_df
def generate_train_test_set_by_time(df, site='hcm' ,ratio = 0.1):
# Generate test set by taking the lastest 10% data from each site
if site=='hanoi':
train_df = df[df.index.get_level_values(0) != 48]
latest_time = train_df.index.get_level_values(1).max()
oldest_time = train_df.index.get_level_values(1).min()
cutoff_hour = (latest_time - oldest_time).total_seconds()
cutoff_hour = cutoff_hour // 3600
cutoff_hour = cutoff_hour * ratio
test_df = train_df[train_df.index.get_level_values(1) >= (latest_time - pd.Timedelta(hours=cutoff_hour))]
train_df = train_df[train_df.index.get_level_values(1) < (latest_time - pd.Timedelta(hours=cutoff_hour))]
# Generate train_test set for site 48
train_df_48 = df[df.index.get_level_values(0) == 48]
latest_time = train_df_48.index.get_level_values(1).max()
oldest_time = train_df_48.index.get_level_values(1).min()
cutoff_hour = (latest_time - oldest_time).total_seconds()
cutoff_hour = cutoff_hour // 3600
cutoff_hour = cutoff_hour * ratio
test_df = test_df.append(train_df_48[train_df_48.index.get_level_values(1) >= (latest_time - pd.Timedelta(hours=cutoff_hour))])
train_df = train_df.append(train_df_48[train_df_48.index.get_level_values(1) < (latest_time - pd.Timedelta(hours=cutoff_hour))])
elif site=='hcm':
train_df = df.copy()
latest_time = train_df.index.get_level_values(1).max()
oldest_time = train_df.index.get_level_values(1).min()
cutoff_hour = (latest_time - oldest_time).total_seconds()
cutoff_hour = cutoff_hour // 3600
cutoff_hour = cutoff_hour * ratio
test_df = train_df[train_df.index.get_level_values(1) >= (latest_time - pd.Timedelta(hours=cutoff_hour))]
train_df = train_df[train_df.index.get_level_values(1) < (latest_time - pd.Timedelta(hours=cutoff_hour))]
return train_df, test_df
def generate_data_label(df):
data = df.copy()
label = df.AQI_h.copy()
site_ids = data.index.get_level_values(0).unique()
columns_data = data.columns
for site in site_ids:
data.loc[data.index.get_level_values(0) == site] = data[data.index.get_level_values(0) == site][:-1]
label.loc[label.index.get_level_values(0) == site] = label[label.index.get_level_values(0) == site].shift(-1)
data = data.dropna()
label = label.dropna()
data['label'] = label
return data
def convert_types_hcm(df):
converted_df = df.copy()
int_columns = ['cat_2.0', 'cat_3.0', 'cat_4.0', 'cat_5.0', 'cat_6.0', 'year_2017.0',
'year_2018.0', 'year_2019.0', 'year_2020.0', 'year_2021.0', 'day_off']
float_columns = ['AQI_h', 'sin_hour', 'cos_hour',
'sin_month', 'cos_month', 'sin_dayweek', 'cos_dayweek', 'sin_daymonth',
'cos_daymonth', 'AQI_h_mean_lag1', 'AQI_h_mean_lag2', 'AQI_h_mean_lag3',
'AQI_h_mean_lag4', 'AQI_h_mean_lag5', 'AQI_h_mean_lag6',
'AQI_h_mean_lag7', 'AQI_h_mean_lag8', 'AQI_h_mean_lag9',
'AQI_h_mean_lag10', 'AQI_h_mean_lag11', 'AQI_h_mean_lag12',
'AQI_h_mean_lag24', 'label']
converted_df[int_columns] = converted_df.loc[:,int_columns].astype(int)
converted_df[float_columns] = converted_df.loc[:,float_columns].astype(float)
return converted_df
def convert_types_hanoi(df):
converted_df = df.copy()
int_columns = ['site_7', 'site_8', 'site_9', 'site_10', 'site_11', 'site_12',
'site_13', 'site_14', 'site_15', 'site_16', 'site_24', 'site_25',
'site_26', 'site_27', 'site_28', 'site_29', 'site_30', 'site_31',
'site_32', 'site_33', 'site_34', 'site_35', 'site_36', 'site_37',
'site_38', 'site_39', 'site_40', 'site_41', 'site_42', 'site_43',
'site_44', 'site_45', 'site_46', 'site_47', 'site_48', 'cat_2.0',
'cat_3.0', 'cat_4.0', 'cat_5.0', 'cat_6.0', 'year_2016.0', 'year_2017.0',
'year_2018.0', 'year_2019.0', 'year_2020.0', 'year_2021.0', 'day_off']
float_columns = ['AQI_h', 'sin_hour', 'cos_hour',
'sin_month', 'cos_month', 'sin_dayweek', 'cos_dayweek', 'sin_daymonth',
'cos_daymonth', 'AQI_h_mean_lag1', 'AQI_h_mean_lag2', 'AQI_h_mean_lag3',
'AQI_h_mean_lag4', 'AQI_h_mean_lag5', 'AQI_h_mean_lag6',
'AQI_h_mean_lag7', 'AQI_h_mean_lag8', 'AQI_h_mean_lag9',
'AQI_h_mean_lag10', 'AQI_h_mean_lag11', 'AQI_h_mean_lag12',
'AQI_h_mean_lag24', 'label']
converted_df[int_columns] = converted_df.loc[:,int_columns].astype(int)
converted_df[float_columns] = converted_df.loc[:,float_columns].astype(float)
return converted_df
def generate_data_to_model(df, site='hanoi'):
df_train, df_test = generate_train_test_set_by_time(df, site=site)
df_train, df_valid = generate_train_test_set_by_time(df_train, site=site)
features = df_train.columns
features = features[:-1]
X_train, y_train = df_train[features], df_train.label
X_valid, y_valid = df_valid[features], df_valid.label
X_test, y_test = df_test[features], df_test.label
return X_train, y_train, X_valid, y_valid, X_test, y_test, features
def write_and_plot_result(model, X_test, y_test, scaler, model_type):
if model_type == 'auto_arima':
transformed_predict = model.predict(n_periods=len(X_test), exogenous=X_test)
else:
transformed_predict = model.predict(X_test)
transformed_predict = hanoi_scaler.inverse_transform(transformed_predict.reshape(-1,1))
transformed_test = scaler.inverse_transform(y_test.values.reshape(-1,1))
# Write result
from sklearn.metrics import mean_squared_error, mean_absolute_error, r2_score
from math import sqrt
rmse_xgboost = mean_squared_error(transformed_predict,transformed_test, squared=False)
mae_xgboost = mean_absolute_error(transformed_predict,transformed_test)
r2_xgboost = r2_score(transformed_predict,transformed_test)
print("R2: {} / RMSE: {} / MAE: {}".format(r2_xgboost, rmse_xgboost, mae_xgboost))
# Plot model
fig = plt.figure(figsize=(16, 9))
ax = fig.add_subplot()
ax.plot(transformed_test.reshape(-1,1), label='label')
ax.plot(transformed_predict.reshape(-1,1), label='predict')
fig.legend()
plt.show()
return None
hanoi_scaler = MinMaxScaler(feature_range=(-1,1))
hanoi_scaler.fit(hanoi_df[['AQI_h']])
hcm_scaler = MinMaxScaler(feature_range=(-1,1))
hcm_scaler.fit(hcm_df[['AQI_h']])
series_hanoi = add_features(hanoi_df).copy()
series_hanoi = generate_data_label(series_hanoi)
series_hanoi = convert_types_hanoi(series_hanoi)
series_hcm = add_features(hcm_df).copy()
series_hcm = generate_data_label(series_hcm)
series_hcm = convert_types_hcm(series_hcm)
X_train, y_train, X_valid, y_valid, X_test, y_test, features = generate_data_to_model(series_hanoi, site='hanoi')
result_df = pd.DataFrame()
```
<a id="subsection-eight"></a>
# AUTO ARIMA
```
! pip install pmdarima
import pmdarima as pm
model_pmdarima = pm.auto_arima(y=y_train.values, exogenous=X_train, trace=2, n_jobs=-1, stepwise=True)
model_pmdarima.fit(y_train, exogenous=X_train)
forecast = model_pmdarima.predict(n_periods=len(X_valid), exogenous=X_valid)
result_df["ARIMAX_AQI"] = forecast
joblib.dump(model_pmdarima, 'auto_arima.pkl')
write_and_plot_result(model_pmdarima, X_test, y_test, hanoi_scaler, model_type='auto_arima')
result_df["Forecast_ARIMAX"].plot(figsize=(16, 9))
result_df[["Forecast_ARIMAX", "AQI_h"]].plot(figsize=(16, 9))
df_train = series_hanoi_train
df_valid = series_hanoi_valid
df_test = series_hanoi_test
```
<a id="subsection-eight"></a>
# RANDOM FOREST REGRESSOR
```
from sklearn.ensemble import RandomForestRegressor
rnd_reg = RandomForestRegressor()
params={
"max_depth" : [1, 3, 4, 5, 6, 7],
"min_samples_split": [1, 3, 4, 5, 6, 7],
"min_samples_leaf" : [1, 3, 4, 5, 6, 7]
}
model_rnd = GridSearchCV(
rnd_reg,
param_grid=params,
n_jobs=-1,
cv=5,
verbose=3,
)
model_rnd.fit(X_train, y_train)
model_rnd = joblib.load('./hanoi_reg_models/rnd_model.pkl')
write_and_plot_result(model_rnd, X_test, y_test, hanoi_scaler, model_type="none")
write_and_plot_result(model_rnd, X_test, y_test, hcm_scaler, model_type="none")
joblib.dump(model_rnd, 'rnd_model.pkl')
```
<a id="subsection-eight"></a>
# CART
```
from sklearn import tree
from sklearn.model_selection import GridSearchCV
from sklearn.metrics import mean_squared_error, mean_absolute_error
plt.style.use('fivethirtyeight')
from datetime import datetime
tree_reg = tree.DecisionTreeRegressor()
## Hyper Parameter Optimization Grid
params={
"max_depth" : [1, 3, 4, 5, 6, 7],
"min_samples_split": [1, 3, 4, 5, 6, 7],
"min_samples_leaf" : [1, 3, 4, 5, 6, 7]
}
best_params_hanoi={'ccp_alpha': 0.0, 'criterion': 'mse', 'max_depth': 7, 'max_features': None, 'max_leaf_nodes': None, 'min_impurity_decrease': 0.0, 'min_impurity_split': None, 'min_samples_leaf': 5, 'min_samples_split': 3, 'min_weight_fraction_leaf': 0.0, 'presort': 'deprecated', 'random_state': None, 'splitter': 'best'}
tree_reg = tree.DecisionTreeRegressor(**best_params)
model_CART = GridSearchCV(
tree_reg,
param_grid=params,
n_jobs=-1,
cv=5,
verbose=3,
)
model_CART.fit(X_train, y_train)
tree_reg.fit(X_train, y_train)
tree_reg = joblib.load('./hanoi_reg_models/cart_model.pkl')
write_and_plot_result(tree_reg, X_test, y_test, hanoi_scaler, model_type="none")
joblib.dump(model_CART, 'cart_model.pkl')
write_and_plot_result(model_CART, X_test, y_test, hcm_scaler, model_type='none')
print(f"Model Best Score : {model_CART.best_score_}")
print(f"Model Best Parameters : {model_CART.best_estimator_.get_params()}")
```
<a id="subsection-eight"></a>
# XG Boost
```
from sklearn import ensemble
from sklearn import metrics
from sklearn.model_selection import RandomizedSearchCV
import xgboost as xgb
from xgboost import plot_importance, plot_tree
from sklearn.metrics import mean_squared_error, mean_absolute_error, r2_score
plt.style.use('fivethirtyeight')
from datetime import datetime
reg = xgb.XGBRegressor()
## Hyper Parameter Optimization Grid
params={
"learning_rate" : [0.05, 0.10, 0.15, 0.20, 0.25, 0.30],
"max_depth" : [1, 3, 4, 5, 6, 7],
"n_estimators" : [int(x) for x in np.linspace(start=100, stop=2000, num=10)],
"min_child_weight" : [int(x) for x in np.arange(3, 15, 1)],
"gamma" : [ 0.0, 0.1, 0.2 , 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9, 1],
"subsample" : [0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9, 1],
"colsample_bytree" : [0.5, 0.6, 0.7, 0.8, 0.9, 1],
"colsample_bylevel": [0.5, 0.6, 0.7, 0.8, 0.9, 1],
}
model_xgboost = RandomizedSearchCV(
reg,
param_distributions=params,
n_iter=10,
n_jobs=-1,
cv=5,
verbose=3,
)
model_xgboost.fit(X_train, y_train)
import joblib
joblib.dump(model_xgboost, 'xgboost_model.pkl')
loaded_model = joblib.load("xgboost_model.pkl")
model_xgboost = joblib.load('./hanoi_reg_models/xgboost_model.pkl')
write_and_plot_result(model_xgboost, X_test, y_test, hanoi_scaler, model_type='none')
f = open('xg_models.txt'.format(root_path, str(np.datetime64('today', 'D'))), 'w')
f.write("Model Best Score : {}".format(model_xgboost.best_score_))
f.write("Model best param: {}".format(model_xgboost.best_estimator_.get_params()))
f.close()
print(f"Model Best Score : {model_xgboost.best_score_}")
print(f"Model Best Parameters : {model_xgboost.best_estimator_.get_params()}")
```
|
github_jupyter
|
# To push the split for training and developing data faster
# we need to skip the index when the timeline is not continuous. (This is covered by dropping all na values)
# The problem arise when there is not enough data after skipping.
# When that happen, the program skip again, leaving some valuable data behind.
# The best thing that can happen is when we skip the second time, if the timeline is not continuous, it will skip
# to the earliest not continous timeline. If we doesn't have any marker, we must find it manually by looping each index.
# So we need to have a marker. It will be put in the raw data files.
# Put timeskip marker in the raw datafiles. (New columns name Timeskip, value is the first time data appear again after the index)
# If after the skip, the timeline is not continous, find the time skip marker and put the position there.
# The refined file doesn't have data in PM (Missing data -> np.nan), findout why *** Not Done 2018 have some problem
# The split for training and developing data has index out of bound, fix it.
# The split for target have some difficulty:
# we can't shift the data so we have to merge it with splitting training data
# When splitting the data
# We don't need to remove the data, we can just calculate like normal.
# Then in the finished file, we drop all data which deemed Missing.
# When putting data to the model, we need to figure out a way to know which data is dropped
# !pip install --upgrade sklearn
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from datetime import datetime
import seaborn as sns
import plotly.express as px
from itertools import product
import warnings
import statsmodels.api as sm
from sklearn.preprocessing import MinMaxScaler
from sklearn.model_selection import GridSearchCV
import holidays
plt.style.use('seaborn-darkgrid')
import joblib
import pandas as pd
import numpy as np
import glob
import xarray as xr
idx = pd.IndexSlice
root_path = r'/mnt/4ba37af6-51fd-47bc-8321-8c500c229114/study/School/KHOA LUAN TOT NGHIEP/'
_thudohanoi_data_path = r'/mnt/4ba37af6-51fd-47bc-8321-8c500c229114/study/School/KHOA LUAN TOT NGHIEP/Data/thudohanoi/refined_data'
_thudohanoi_files = glob.glob(_thudohanoi_data_path + '/*.csv')
thudohanoi_df = pd.DataFrame()
for file in _thudohanoi_files:
print('Currently processing file \n{}'.format(file))
thudohanoi_df = thudohanoi_df.append(pd.read_csv(file, parse_dates=True, index_col=['site_id', 'time'],
dtype={'CO': np.float64, 'NO2': np.float64,'PM25': np.float64,
'AQI_h': np.float64, 'AQI_h_I': np.int, 'site_id': np.int}))
# Remove site 16 because of some inconsistency in data
hanoi_df = thudohanoi_df[(thudohanoi_df.index.get_level_values(0) != 49)].copy()
hcm_df = thudohanoi_df[(thudohanoi_df.index.get_level_values(0) == 49)].copy()
def extract_time_features(df):
# Job: Expand time in data
time_index = df.index.get_level_values(1)
df_time_features = pd.DataFrame()
df_time_features['Hour'] = time_index.hour.astype(float)
df_time_features['Month'] = time_index.month.astype(float)
df_time_features['Day of Week'] = time_index.dayofweek.astype(float)
df_time_features['Day of Month'] = time_index.day.astype(float)
df_time_features['Days in Month'] = time_index.daysinmonth.astype(float)
df_time_features['Year'] = time_index.year.astype(float)
# Job: Encode time cyclical data
hour_in_day = 23
df_time_features['sin_hour'] = np.sin(2*np.pi*df_time_features['Hour']/hour_in_day)
df_time_features['cos_hour'] = np.cos(2*np.pi*df_time_features['Hour']/hour_in_day)
month_in_year = 12
df_time_features['sin_month'] = np.sin(2*np.pi*df_time_features['Month']/month_in_year)
df_time_features['cos_month'] = np.cos(2*np.pi*df_time_features['Month']/month_in_year)
day_in_week = 6
df_time_features['sin_dayweek'] = np.sin(2*np.pi*df_time_features['Day of Week']/day_in_week)
df_time_features['cos_dayweek'] = np.cos(2*np.pi*df_time_features['Day of Week']/day_in_week)
df_time_features['sin_daymonth'] = np.sin(2*np.pi*df_time_features['Day of Month']/df_time_features['Days in Month'])
df_time_features['cos_daymonth'] = np.cos(2*np.pi*df_time_features['Day of Month']/df_time_features['Days in Month'])
# One hot encode year data
one_hot_df = pd.get_dummies(df_time_features['Year'], drop_first=True, prefix='year')
df_time_features = df_time_features.join(one_hot_df)
# Input weekday/weekend/holiday data
vn_holidays = np.array(list(holidays.VN(years=[2015,2016,2017,2018,2019,2020,2021]).keys()))
holiday_mask = np.isin(time_index.date, vn_holidays)
masks = (holiday_mask) | (df_time_features['Day of Week'].values == 5) | (df_time_features['Day of Week'].values == 6)
df_time_features['day_off'] = np.where(masks == True, 1, 0)
df_time_features = df_time_features.drop(columns=['Day of Month', 'Month', 'Day of Week', 'Days in Month', 'Year', 'Hour'])
# Input lagged data
windows = list(range(1,13))
windows.append(24)
for window in windows:
feature = 'AQI_h'
series_rolled = df['AQI_h'].rolling(window=window, min_periods=0)
series_mean = series_rolled.mean().shift(1).reset_index()
series_std = series_rolled.std().shift(1).reset_index()
df_time_features[f"{feature}_mean_lag{window}"] = series_mean['AQI_h'].values
# df_time_features[f"{feature}_std_lag{window}"] = series_std['AQI_h'].values
df_time_features.fillna(df_time_features.mean(), inplace=True)
df_time_features.fillna(df['AQI_h'].mean(), inplace=True)
return df_time_features.values, df_time_features.columns
def add_features(df):
# Change all data to numpy, then concatenate those numpy.
# Then construct the dataframe to old frame. This can work
data_df = df[['AQI_h']].copy()
# Job: Normalize train data
scaler = MinMaxScaler(feature_range=(-1,1))
for col in ['AQI_h']:
data_df[[col]] = scaler.fit_transform(data_df[[col]])
columns = ['site_id', 'time', 'AQI_h']
df_numpy = data_df.reset_index().to_numpy()
# Add onehot site label
one_hot_site = pd.get_dummies(data_df.index.get_level_values(0), prefix='site', drop_first=True).astype(int)
columns.extend(one_hot_site.columns)
# Add onehot air category
one_hot_cat = pd.get_dummies(df['AQI_h_I'], drop_first=True, prefix='cat').astype(int)
columns.extend(one_hot_cat.columns)
# Add time features
time_features, time_columns = extract_time_features(data_df)
columns.extend(time_columns)
df_numpy = np.concatenate([df_numpy, one_hot_site.values, one_hot_cat.values, time_features], axis=1)
final_df = pd.DataFrame(df_numpy, columns=columns).set_index(['site_id', 'time'])
for float_col in final_df.loc[:, final_df.dtypes == float].columns:
final_df.loc[:, float_col] = final_df.loc[:, float_col].values.round(6)
return final_df
def generate_train_test_set_by_time(df, site='hcm' ,ratio = 0.1):
# Generate test set by taking the lastest 10% data from each site
if site=='hanoi':
train_df = df[df.index.get_level_values(0) != 48]
latest_time = train_df.index.get_level_values(1).max()
oldest_time = train_df.index.get_level_values(1).min()
cutoff_hour = (latest_time - oldest_time).total_seconds()
cutoff_hour = cutoff_hour // 3600
cutoff_hour = cutoff_hour * ratio
test_df = train_df[train_df.index.get_level_values(1) >= (latest_time - pd.Timedelta(hours=cutoff_hour))]
train_df = train_df[train_df.index.get_level_values(1) < (latest_time - pd.Timedelta(hours=cutoff_hour))]
# Generate train_test set for site 48
train_df_48 = df[df.index.get_level_values(0) == 48]
latest_time = train_df_48.index.get_level_values(1).max()
oldest_time = train_df_48.index.get_level_values(1).min()
cutoff_hour = (latest_time - oldest_time).total_seconds()
cutoff_hour = cutoff_hour // 3600
cutoff_hour = cutoff_hour * ratio
test_df = test_df.append(train_df_48[train_df_48.index.get_level_values(1) >= (latest_time - pd.Timedelta(hours=cutoff_hour))])
train_df = train_df.append(train_df_48[train_df_48.index.get_level_values(1) < (latest_time - pd.Timedelta(hours=cutoff_hour))])
elif site=='hcm':
train_df = df.copy()
latest_time = train_df.index.get_level_values(1).max()
oldest_time = train_df.index.get_level_values(1).min()
cutoff_hour = (latest_time - oldest_time).total_seconds()
cutoff_hour = cutoff_hour // 3600
cutoff_hour = cutoff_hour * ratio
test_df = train_df[train_df.index.get_level_values(1) >= (latest_time - pd.Timedelta(hours=cutoff_hour))]
train_df = train_df[train_df.index.get_level_values(1) < (latest_time - pd.Timedelta(hours=cutoff_hour))]
return train_df, test_df
def generate_data_label(df):
data = df.copy()
label = df.AQI_h.copy()
site_ids = data.index.get_level_values(0).unique()
columns_data = data.columns
for site in site_ids:
data.loc[data.index.get_level_values(0) == site] = data[data.index.get_level_values(0) == site][:-1]
label.loc[label.index.get_level_values(0) == site] = label[label.index.get_level_values(0) == site].shift(-1)
data = data.dropna()
label = label.dropna()
data['label'] = label
return data
def convert_types_hcm(df):
converted_df = df.copy()
int_columns = ['cat_2.0', 'cat_3.0', 'cat_4.0', 'cat_5.0', 'cat_6.0', 'year_2017.0',
'year_2018.0', 'year_2019.0', 'year_2020.0', 'year_2021.0', 'day_off']
float_columns = ['AQI_h', 'sin_hour', 'cos_hour',
'sin_month', 'cos_month', 'sin_dayweek', 'cos_dayweek', 'sin_daymonth',
'cos_daymonth', 'AQI_h_mean_lag1', 'AQI_h_mean_lag2', 'AQI_h_mean_lag3',
'AQI_h_mean_lag4', 'AQI_h_mean_lag5', 'AQI_h_mean_lag6',
'AQI_h_mean_lag7', 'AQI_h_mean_lag8', 'AQI_h_mean_lag9',
'AQI_h_mean_lag10', 'AQI_h_mean_lag11', 'AQI_h_mean_lag12',
'AQI_h_mean_lag24', 'label']
converted_df[int_columns] = converted_df.loc[:,int_columns].astype(int)
converted_df[float_columns] = converted_df.loc[:,float_columns].astype(float)
return converted_df
def convert_types_hanoi(df):
converted_df = df.copy()
int_columns = ['site_7', 'site_8', 'site_9', 'site_10', 'site_11', 'site_12',
'site_13', 'site_14', 'site_15', 'site_16', 'site_24', 'site_25',
'site_26', 'site_27', 'site_28', 'site_29', 'site_30', 'site_31',
'site_32', 'site_33', 'site_34', 'site_35', 'site_36', 'site_37',
'site_38', 'site_39', 'site_40', 'site_41', 'site_42', 'site_43',
'site_44', 'site_45', 'site_46', 'site_47', 'site_48', 'cat_2.0',
'cat_3.0', 'cat_4.0', 'cat_5.0', 'cat_6.0', 'year_2016.0', 'year_2017.0',
'year_2018.0', 'year_2019.0', 'year_2020.0', 'year_2021.0', 'day_off']
float_columns = ['AQI_h', 'sin_hour', 'cos_hour',
'sin_month', 'cos_month', 'sin_dayweek', 'cos_dayweek', 'sin_daymonth',
'cos_daymonth', 'AQI_h_mean_lag1', 'AQI_h_mean_lag2', 'AQI_h_mean_lag3',
'AQI_h_mean_lag4', 'AQI_h_mean_lag5', 'AQI_h_mean_lag6',
'AQI_h_mean_lag7', 'AQI_h_mean_lag8', 'AQI_h_mean_lag9',
'AQI_h_mean_lag10', 'AQI_h_mean_lag11', 'AQI_h_mean_lag12',
'AQI_h_mean_lag24', 'label']
converted_df[int_columns] = converted_df.loc[:,int_columns].astype(int)
converted_df[float_columns] = converted_df.loc[:,float_columns].astype(float)
return converted_df
def generate_data_to_model(df, site='hanoi'):
df_train, df_test = generate_train_test_set_by_time(df, site=site)
df_train, df_valid = generate_train_test_set_by_time(df_train, site=site)
features = df_train.columns
features = features[:-1]
X_train, y_train = df_train[features], df_train.label
X_valid, y_valid = df_valid[features], df_valid.label
X_test, y_test = df_test[features], df_test.label
return X_train, y_train, X_valid, y_valid, X_test, y_test, features
def write_and_plot_result(model, X_test, y_test, scaler, model_type):
if model_type == 'auto_arima':
transformed_predict = model.predict(n_periods=len(X_test), exogenous=X_test)
else:
transformed_predict = model.predict(X_test)
transformed_predict = hanoi_scaler.inverse_transform(transformed_predict.reshape(-1,1))
transformed_test = scaler.inverse_transform(y_test.values.reshape(-1,1))
# Write result
from sklearn.metrics import mean_squared_error, mean_absolute_error, r2_score
from math import sqrt
rmse_xgboost = mean_squared_error(transformed_predict,transformed_test, squared=False)
mae_xgboost = mean_absolute_error(transformed_predict,transformed_test)
r2_xgboost = r2_score(transformed_predict,transformed_test)
print("R2: {} / RMSE: {} / MAE: {}".format(r2_xgboost, rmse_xgboost, mae_xgboost))
# Plot model
fig = plt.figure(figsize=(16, 9))
ax = fig.add_subplot()
ax.plot(transformed_test.reshape(-1,1), label='label')
ax.plot(transformed_predict.reshape(-1,1), label='predict')
fig.legend()
plt.show()
return None
hanoi_scaler = MinMaxScaler(feature_range=(-1,1))
hanoi_scaler.fit(hanoi_df[['AQI_h']])
hcm_scaler = MinMaxScaler(feature_range=(-1,1))
hcm_scaler.fit(hcm_df[['AQI_h']])
series_hanoi = add_features(hanoi_df).copy()
series_hanoi = generate_data_label(series_hanoi)
series_hanoi = convert_types_hanoi(series_hanoi)
series_hcm = add_features(hcm_df).copy()
series_hcm = generate_data_label(series_hcm)
series_hcm = convert_types_hcm(series_hcm)
X_train, y_train, X_valid, y_valid, X_test, y_test, features = generate_data_to_model(series_hanoi, site='hanoi')
result_df = pd.DataFrame()
! pip install pmdarima
import pmdarima as pm
model_pmdarima = pm.auto_arima(y=y_train.values, exogenous=X_train, trace=2, n_jobs=-1, stepwise=True)
model_pmdarima.fit(y_train, exogenous=X_train)
forecast = model_pmdarima.predict(n_periods=len(X_valid), exogenous=X_valid)
result_df["ARIMAX_AQI"] = forecast
joblib.dump(model_pmdarima, 'auto_arima.pkl')
write_and_plot_result(model_pmdarima, X_test, y_test, hanoi_scaler, model_type='auto_arima')
result_df["Forecast_ARIMAX"].plot(figsize=(16, 9))
result_df[["Forecast_ARIMAX", "AQI_h"]].plot(figsize=(16, 9))
df_train = series_hanoi_train
df_valid = series_hanoi_valid
df_test = series_hanoi_test
from sklearn.ensemble import RandomForestRegressor
rnd_reg = RandomForestRegressor()
params={
"max_depth" : [1, 3, 4, 5, 6, 7],
"min_samples_split": [1, 3, 4, 5, 6, 7],
"min_samples_leaf" : [1, 3, 4, 5, 6, 7]
}
model_rnd = GridSearchCV(
rnd_reg,
param_grid=params,
n_jobs=-1,
cv=5,
verbose=3,
)
model_rnd.fit(X_train, y_train)
model_rnd = joblib.load('./hanoi_reg_models/rnd_model.pkl')
write_and_plot_result(model_rnd, X_test, y_test, hanoi_scaler, model_type="none")
write_and_plot_result(model_rnd, X_test, y_test, hcm_scaler, model_type="none")
joblib.dump(model_rnd, 'rnd_model.pkl')
from sklearn import tree
from sklearn.model_selection import GridSearchCV
from sklearn.metrics import mean_squared_error, mean_absolute_error
plt.style.use('fivethirtyeight')
from datetime import datetime
tree_reg = tree.DecisionTreeRegressor()
## Hyper Parameter Optimization Grid
params={
"max_depth" : [1, 3, 4, 5, 6, 7],
"min_samples_split": [1, 3, 4, 5, 6, 7],
"min_samples_leaf" : [1, 3, 4, 5, 6, 7]
}
best_params_hanoi={'ccp_alpha': 0.0, 'criterion': 'mse', 'max_depth': 7, 'max_features': None, 'max_leaf_nodes': None, 'min_impurity_decrease': 0.0, 'min_impurity_split': None, 'min_samples_leaf': 5, 'min_samples_split': 3, 'min_weight_fraction_leaf': 0.0, 'presort': 'deprecated', 'random_state': None, 'splitter': 'best'}
tree_reg = tree.DecisionTreeRegressor(**best_params)
model_CART = GridSearchCV(
tree_reg,
param_grid=params,
n_jobs=-1,
cv=5,
verbose=3,
)
model_CART.fit(X_train, y_train)
tree_reg.fit(X_train, y_train)
tree_reg = joblib.load('./hanoi_reg_models/cart_model.pkl')
write_and_plot_result(tree_reg, X_test, y_test, hanoi_scaler, model_type="none")
joblib.dump(model_CART, 'cart_model.pkl')
write_and_plot_result(model_CART, X_test, y_test, hcm_scaler, model_type='none')
print(f"Model Best Score : {model_CART.best_score_}")
print(f"Model Best Parameters : {model_CART.best_estimator_.get_params()}")
from sklearn import ensemble
from sklearn import metrics
from sklearn.model_selection import RandomizedSearchCV
import xgboost as xgb
from xgboost import plot_importance, plot_tree
from sklearn.metrics import mean_squared_error, mean_absolute_error, r2_score
plt.style.use('fivethirtyeight')
from datetime import datetime
reg = xgb.XGBRegressor()
## Hyper Parameter Optimization Grid
params={
"learning_rate" : [0.05, 0.10, 0.15, 0.20, 0.25, 0.30],
"max_depth" : [1, 3, 4, 5, 6, 7],
"n_estimators" : [int(x) for x in np.linspace(start=100, stop=2000, num=10)],
"min_child_weight" : [int(x) for x in np.arange(3, 15, 1)],
"gamma" : [ 0.0, 0.1, 0.2 , 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9, 1],
"subsample" : [0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9, 1],
"colsample_bytree" : [0.5, 0.6, 0.7, 0.8, 0.9, 1],
"colsample_bylevel": [0.5, 0.6, 0.7, 0.8, 0.9, 1],
}
model_xgboost = RandomizedSearchCV(
reg,
param_distributions=params,
n_iter=10,
n_jobs=-1,
cv=5,
verbose=3,
)
model_xgboost.fit(X_train, y_train)
import joblib
joblib.dump(model_xgboost, 'xgboost_model.pkl')
loaded_model = joblib.load("xgboost_model.pkl")
model_xgboost = joblib.load('./hanoi_reg_models/xgboost_model.pkl')
write_and_plot_result(model_xgboost, X_test, y_test, hanoi_scaler, model_type='none')
f = open('xg_models.txt'.format(root_path, str(np.datetime64('today', 'D'))), 'w')
f.write("Model Best Score : {}".format(model_xgboost.best_score_))
f.write("Model best param: {}".format(model_xgboost.best_estimator_.get_params()))
f.close()
print(f"Model Best Score : {model_xgboost.best_score_}")
print(f"Model Best Parameters : {model_xgboost.best_estimator_.get_params()}")
| 0.489015 | 0.723358 |
## Building a machine learning text classifier
```
!pip install pandas seaborn nltk scikit-learn==1.0.2 transformers tensorflow emoji torch
import numpy as np
import pandas as pd
import nltk
import nltk.corpus
import sklearn
import sklearn.pipeline
import sklearn.feature_extraction.text
import sklearn.naive_bayes
import sklearn.model_selection
import sklearn.metrics
import tensorflow
import transformers
```
## Loading and exploring data
```
data = pd.read_excel("comm106e_happysad.xlsx")
data.head()
data['label'].value_counts()
data['label'] = data['label'].str.lower()
data['label'].value_counts()
```
## Training the model pipeline
```
pipeline = sklearn.pipeline.Pipeline([
('vect', sklearn.feature_extraction.text.CountVectorizer()),
('tfidf', sklearn.feature_extraction.text.TfidfTransformer()),
('clf', sklearn.naive_bayes.MultinomialNB()),
])
pipeline.fit(data['text'], data['label'])
```
## Scoring some text
```
emails_to_score = ['I need this report by 9am or else I\'ll be mad!',
'I love it, I\'m so happy!',
'our next conference should be in France',
'our next conference should be in Germany'
]
prediction = pipeline.predict_proba(emails_to_score)
prediction
```
# Re-training with only 80% of the texts, leaving the other 20% for testing
### Split the data into training and testing sets
```
X_train, X_test, y_train, y_test = sklearn.model_selection.train_test_split(data['text'],
data['label'],
test_size=0.2,
random_state=42)
```
### Train the model by fitting the pipeline to the 80% training data
```
pipeline.fit(X_train, y_train)
```
### Use the pipeline to predict the labels of the 20% testing data that was not used to train it
```
y_pred = pipeline.predict(X_test)
y_pred
```
### Score the accuracy of the model with the results of the predictions made from the 20% testing data
```
pipeline.score(X_test, y_test)
```
### Print a classification report for how the 20% test data did, showing various statistics
```
print(sklearn.metrics.classification_report(y_test, y_pred))
```
# Some definitions
But please look at the chart at https://en.wikipedia.org/wiki/Precision_and_recall#Definition_(classification_context)
## Accuracy:
$\frac{\mbox{number of correct predictions}}{\mbox{total number of items predicted}}$
## Precision or positive predictive value (PPV):
$\frac{\mbox{number of true positives for a label}}{\mbox{number of true + false positives for that label}}$
## True positive rate (TPR) or recall, sensitivity, hit rate:
$\frac{\mbox{number of true positives for a label}}{\mbox{number of actual positives for that label}}$
## F1 score (harmonic mean between precision/PPV and recall/TPR):
$2*\frac{\mbox{precision * recall}}{\mbox{precision + recall}}$
## Support: Number of cases in each label or all labels
# Running our classification report
```
print(sklearn.metrics.classification_report(y_test, y_pred))
sklearn.metrics.precision_score(y_test, y_pred, pos_label='happy')
```
# Auditing the model for variations in locations
```
countries = pd.read_csv("countries.csv")
countries
results_list = []
for country_name in countries['country']:
sample_text = "It is so nice in " + country_name + ", I love it there!"
probability = pipeline.predict_proba([sample_text])[0][0]
result = {'country':country_name,
'happy_prediction':probability}
results_list.append(result)
country_audit_data = pd.DataFrame(results_list)
country_audit_data
country_audit_data.sort_values('happy_prediction')
```
|
github_jupyter
|
!pip install pandas seaborn nltk scikit-learn==1.0.2 transformers tensorflow emoji torch
import numpy as np
import pandas as pd
import nltk
import nltk.corpus
import sklearn
import sklearn.pipeline
import sklearn.feature_extraction.text
import sklearn.naive_bayes
import sklearn.model_selection
import sklearn.metrics
import tensorflow
import transformers
data = pd.read_excel("comm106e_happysad.xlsx")
data.head()
data['label'].value_counts()
data['label'] = data['label'].str.lower()
data['label'].value_counts()
pipeline = sklearn.pipeline.Pipeline([
('vect', sklearn.feature_extraction.text.CountVectorizer()),
('tfidf', sklearn.feature_extraction.text.TfidfTransformer()),
('clf', sklearn.naive_bayes.MultinomialNB()),
])
pipeline.fit(data['text'], data['label'])
emails_to_score = ['I need this report by 9am or else I\'ll be mad!',
'I love it, I\'m so happy!',
'our next conference should be in France',
'our next conference should be in Germany'
]
prediction = pipeline.predict_proba(emails_to_score)
prediction
X_train, X_test, y_train, y_test = sklearn.model_selection.train_test_split(data['text'],
data['label'],
test_size=0.2,
random_state=42)
pipeline.fit(X_train, y_train)
y_pred = pipeline.predict(X_test)
y_pred
pipeline.score(X_test, y_test)
print(sklearn.metrics.classification_report(y_test, y_pred))
print(sklearn.metrics.classification_report(y_test, y_pred))
sklearn.metrics.precision_score(y_test, y_pred, pos_label='happy')
countries = pd.read_csv("countries.csv")
countries
results_list = []
for country_name in countries['country']:
sample_text = "It is so nice in " + country_name + ", I love it there!"
probability = pipeline.predict_proba([sample_text])[0][0]
result = {'country':country_name,
'happy_prediction':probability}
results_list.append(result)
country_audit_data = pd.DataFrame(results_list)
country_audit_data
country_audit_data.sort_values('happy_prediction')
| 0.390127 | 0.855066 |
# Download biorxiv preprint table from the PrePubMed repository
```
import os
import json
import logging
import pandas
import requests
import utilities
# Configure logging to write to file
logging.basicConfig(level=logging.INFO, filename=os.path.join('logs/donwload.log'), filemode='w')
```
## Get `OmnesRes/prepub` version
```
url = 'https://api.github.com/repos/OmnesRes/prepub/git/refs/heads/master'
response = requests.get(url)
response = response.json()
response['object']
```
## Load bioRxiv data
```
url = 'https://github.com/OmnesRes/prepub/raw/master/biorxiv/biorxiv_licenses.tsv'
biorxiv_df = pandas.read_table(url)
# Limit to preprints through November 2016
biorxiv_df = biorxiv_df.query("Date <= '2017-06-07'")
biorxiv_df.head(2)
```
## Processing
```
# Remove URL from DOIs
biorxiv_df.DOI = biorxiv_df.DOI.str.extract(r'(10\.[0-9]+/[0-9]+)', expand=False)
biorxiv_df.License = biorxiv_df.License.str.replace('CC-BY', 'CC BY')
biorxiv_df.License = biorxiv_df.License.fillna('None')
```
## Authors
```
author_df = (biorxiv_df
.pipe(utilities.tidy_split, column='Authors')
.rename(columns={'Authors': 'Author'})
[['DOI', 'Author']]
.sort_values(['DOI', 'Author'])
.drop_duplicates()
.reset_index(drop=True)
)
# Standardize author names
author_df['Standard_Author'] = author_df.Author.map(utilities.get_standard_author)
author_df.tail(2)
# Authors with the most preprints
author_df.Standard_Author.value_counts().head(2)
```
## Subjects
```
# Create a TSV with a row per preprint-subject pair
subject_df = (biorxiv_df
.pipe(utilities.tidy_split, column='Subjects')
.rename(columns={'Subjects': 'Subject'})
[['DOI', 'Subject']]
.sort_values(['DOI', 'Subject'])
.reset_index(drop=True)
)
subject_df.tail(2)
# Number of subjects per preprint
subject_df.DOI.value_counts().value_counts()
# Number of preprints by subject
subject_df.Subject.value_counts()
```
## Preprints
```
preprint_df = (biorxiv_df
[['DOI', 'Date', 'License']]
.sort_values('DOI')
.reset_index(drop=True)
)
preprint_df.tail(4)
len(preprint_df)
# Preprints by license
preprint_df.License.value_counts(normalize=True).reset_index()
# Preprints by year
preprint_df.Date.map(lambda date: date.split('-')[0]).value_counts()
```
## Save as a TSVs
```
path = os.path.join('data', 'preprints.tsv')
preprint_df.to_csv(path, sep='\t', index=False)
path = os.path.join('data', 'subjects.tsv')
subject_df.to_csv(path, sep='\t', index=False)
path = os.path.join('data', 'authors.tsv')
author_df.to_csv(path, sep='\t', index=False)
```
|
github_jupyter
|
import os
import json
import logging
import pandas
import requests
import utilities
# Configure logging to write to file
logging.basicConfig(level=logging.INFO, filename=os.path.join('logs/donwload.log'), filemode='w')
url = 'https://api.github.com/repos/OmnesRes/prepub/git/refs/heads/master'
response = requests.get(url)
response = response.json()
response['object']
url = 'https://github.com/OmnesRes/prepub/raw/master/biorxiv/biorxiv_licenses.tsv'
biorxiv_df = pandas.read_table(url)
# Limit to preprints through November 2016
biorxiv_df = biorxiv_df.query("Date <= '2017-06-07'")
biorxiv_df.head(2)
# Remove URL from DOIs
biorxiv_df.DOI = biorxiv_df.DOI.str.extract(r'(10\.[0-9]+/[0-9]+)', expand=False)
biorxiv_df.License = biorxiv_df.License.str.replace('CC-BY', 'CC BY')
biorxiv_df.License = biorxiv_df.License.fillna('None')
author_df = (biorxiv_df
.pipe(utilities.tidy_split, column='Authors')
.rename(columns={'Authors': 'Author'})
[['DOI', 'Author']]
.sort_values(['DOI', 'Author'])
.drop_duplicates()
.reset_index(drop=True)
)
# Standardize author names
author_df['Standard_Author'] = author_df.Author.map(utilities.get_standard_author)
author_df.tail(2)
# Authors with the most preprints
author_df.Standard_Author.value_counts().head(2)
# Create a TSV with a row per preprint-subject pair
subject_df = (biorxiv_df
.pipe(utilities.tidy_split, column='Subjects')
.rename(columns={'Subjects': 'Subject'})
[['DOI', 'Subject']]
.sort_values(['DOI', 'Subject'])
.reset_index(drop=True)
)
subject_df.tail(2)
# Number of subjects per preprint
subject_df.DOI.value_counts().value_counts()
# Number of preprints by subject
subject_df.Subject.value_counts()
preprint_df = (biorxiv_df
[['DOI', 'Date', 'License']]
.sort_values('DOI')
.reset_index(drop=True)
)
preprint_df.tail(4)
len(preprint_df)
# Preprints by license
preprint_df.License.value_counts(normalize=True).reset_index()
# Preprints by year
preprint_df.Date.map(lambda date: date.split('-')[0]).value_counts()
path = os.path.join('data', 'preprints.tsv')
preprint_df.to_csv(path, sep='\t', index=False)
path = os.path.join('data', 'subjects.tsv')
subject_df.to_csv(path, sep='\t', index=False)
path = os.path.join('data', 'authors.tsv')
author_df.to_csv(path, sep='\t', index=False)
| 0.29584 | 0.663062 |
# Double pendulum
Consider the pendulum suspended on the pendulum. We weill use dAlembert principle to derive eqaution of motion in generalized coordinaes. Naturally, we choose two angles as coordinates which comply with contraints.
```
var('t')
var('l1 l2 m1 m2 g')
xy_names = [('x1','x1'),('y1','y1'),('x2','x2'),('y2','y2')]
uv_names = [('phi1','\\varphi_1'),('phi2','\\varphi_2')]
load('cas_utils.sage')
to_fun, to_var = make_symbols(xy_names,uv_names)
x2u = {x1:l1*sin(phi1),\
y1:-l1*cos(phi1),\
x2:l1*sin(phi1)+l2*sin(phi2),\
y2:-l1*cos(phi1)-l2*cos(phi2)}
transform_virtual_displacements(xy_names,uv_names,verbose=True)
dAlemb = (m1*x1.subs(x2u).subs(to_fun).diff(t,2))*dx1_polar + \
(m1*y1.subs(x2u).subs(to_fun).diff(t,2)+m1*g)*dy1_polar+\
(m2*x2.subs(x2u).subs(to_fun).diff(t,2))*dx2_polar + \
(m2*y2.subs(x2u).subs(to_fun).diff(t,2)+m2*g)*dy2_polar
dAlemb = dAlemb.subs(to_var)
showmath(dAlemb)
eq1 = dAlemb.expand().coefficient(dphi1).trig_simplify()
eq2 = dAlemb.expand().coefficient(dphi2).trig_simplify()
showmath(eq1)
showmath(eq2)
sol = solve([eq1,eq2],[phi1dd,phi2dd])[0]
showmath(sol)
showmath( sol[0].rhs().denominator() )
(l1*(2*m1+m2-m2*cos(2*phi1-2*phi2))).expand_trig().expand_trig().expand().show()
bool ( -2*sol[0].rhs().denominator()==(l1*(2*m1+m2-m2*cos(2*phi1-2*phi2))).expand_trig().expand_trig().expand() )
```
Since the "textbook" solution contains a slightly different form, let's check if we have these formulas:
$$T(\varphi_1,\varphi_2,\dot{\varphi}_1,\dot{\varphi}_2) = \frac{m_1}{2} l_1^2 \dot{\varphi}_1^2 + \frac{m_2}{2} \left( l_1^2 \dot{\varphi}_1^2 + l_2^2 \dot{\varphi}_2^2 + 2 l_1 l_2 \dot{\varphi}_1 \dot{\varphi}_2 \cos(\varphi_1-\varphi_2) \right)$$ $$V(\varphi_1,\varphi_2) = -(m_1+m_2) g l_1 \cos\varphi_1 - m_2 g l_2 \cos\varphi_2$$
$$m_{2}l_{2}\ddot{\varphi}_{2}\cos\left(\varphi_{1}-\varphi_{2}\right)+\left(m_{1}+m_{2}\right)l_{1}\ddot{\varphi}_{1}+m_{2}l_{2}\dot{\varphi}_{2}^{2}\sin\left(\varphi_{1}-\varphi_{2}\right)+\left(m_{1}+m_{2}\right)g\sin\varphi_{1}=0$$
$$l_{2}\ddot{\varphi}_{2}+l_{1}\ddot{\varphi}_{1}\cos\left(\varphi_{1}-\varphi_{2}\right)-l_{1}\dot{\varphi}_{1}^{2}\sin\left(\varphi_{1}-\varphi_{2}\right)+g\sin\varphi_{2}=0$$
```
rown_wiki = [m2*l2*cos(phi1-phi2)*phi2dd+(m1+m2)*l1*phi1dd+m2*l2*phi2d^2 * sin(phi1-phi2)+ (m1+m2)*g*sin(phi1),\
l2*phi2dd+l1*cos(phi1-phi2)*phi1dd-l1*phi1d^2*sin(phi1-phi2)+g*sin(phi2)]
showmath(rown_wiki[0])
showmath(rown_wiki[1])
rown_wiki[0].show()
(eq1/l1).reduce_trig().show()
rown_wiki[0].show()
bool((eq1/l1) == rown_wiki[0] )
(eq2/l2/m2).reduce_trig().show()
rown_wiki[1].show()
bool((eq2/l2/m2) == rown_wiki[1] )
```
## Euler -Langrange
```
Ekin = 1/2*(m1*x1.subs(x2u).subs(to_fun).diff(t).subs(to_var)^2+\
m1*y1.subs(x2u).subs(to_fun).diff(t).subs(to_var)^2+\
m2*x2.subs(x2u).subs(to_fun).diff(t).subs(to_var)^2+\
m2*y2.subs(x2u).subs(to_fun).diff(t).subs(to_var)^2 )
Epot = m1*g*y1.subs(x2u)+m2*g*y2.subs(x2u)
showmath( Epot.collect(cos(phi1)) )
showmath( Epot )
showmath( Ekin.trig_simplify() )
L = Ekin - Epot
len(L.expand().operands())
EL1 = L.diff(phi1d).subs(to_fun).diff(t).subs(to_var) - L.diff(phi1)
EL2 = L.diff(phi2d).subs(to_fun).diff(t).subs(to_var) - L.diff(phi2)
EL1.expand().operands()
EL1 = (EL1/l1).trig_reduce()
EL2 = (EL2/l2).trig_reduce()
showmath(EL1)
sol = solve([EL1,EL2],[phi1dd,phi2dd])[0]
show(sol)
expr = sol[0].rhs()
for ex_ in expr.factor().numerator().operands():
show(ex_)
```
## Numerical analysis
```
import numpy as np
ode = [phi1d,phi2d]+[sol[0].rhs(),sol[1].rhs()]
ode = map(lambda x:x.subs({l1:1,l2:1,m1:1,m2:1,g:9.81}),ode)
times = srange(0,5,.01)
numsol = desolve_odeint(ode,[2.1,0,0,0],times,[phi1,phi2,phi1d,phi2d])
p = line ( zip(np.sin(numsol[:,0])+np.sin(numsol[:,1]),\
-np.cos(numsol[:,0])-np.cos(numsol[:,1])), color='gray' )
p.show(figsize=4)
def plot_dp(f1,f2,pars):
mass1 = vector([x1,y1]).subs(x2u).subs(pars).subs({phi1:f1,phi2:f2})
mass2 = vector([x2,y2]).subs(x2u).subs(pars).subs({phi1:f1,phi2:f2})
plt = point([(0,0),mass1],aspect_ratio=1,size=40)
plt += point(mass2,xmin=-2,xmax=2,ymin=-2,ymax=2,size=40)
plt += line([(0,0),mass1,mass2],color='gray')
return plt
plot_dp(numsol[213,0],numsol[213,1],{l1:1,l2:1})+p
@interact
def _(ith=slider(0,numsol.shape[0]-1)):
f1,f2 = numsol[ith,:2]
plot_dp(f1,f2,{l1:1,l2:1}).show(axes=False)
```
\newpage
|
github_jupyter
|
var('t')
var('l1 l2 m1 m2 g')
xy_names = [('x1','x1'),('y1','y1'),('x2','x2'),('y2','y2')]
uv_names = [('phi1','\\varphi_1'),('phi2','\\varphi_2')]
load('cas_utils.sage')
to_fun, to_var = make_symbols(xy_names,uv_names)
x2u = {x1:l1*sin(phi1),\
y1:-l1*cos(phi1),\
x2:l1*sin(phi1)+l2*sin(phi2),\
y2:-l1*cos(phi1)-l2*cos(phi2)}
transform_virtual_displacements(xy_names,uv_names,verbose=True)
dAlemb = (m1*x1.subs(x2u).subs(to_fun).diff(t,2))*dx1_polar + \
(m1*y1.subs(x2u).subs(to_fun).diff(t,2)+m1*g)*dy1_polar+\
(m2*x2.subs(x2u).subs(to_fun).diff(t,2))*dx2_polar + \
(m2*y2.subs(x2u).subs(to_fun).diff(t,2)+m2*g)*dy2_polar
dAlemb = dAlemb.subs(to_var)
showmath(dAlemb)
eq1 = dAlemb.expand().coefficient(dphi1).trig_simplify()
eq2 = dAlemb.expand().coefficient(dphi2).trig_simplify()
showmath(eq1)
showmath(eq2)
sol = solve([eq1,eq2],[phi1dd,phi2dd])[0]
showmath(sol)
showmath( sol[0].rhs().denominator() )
(l1*(2*m1+m2-m2*cos(2*phi1-2*phi2))).expand_trig().expand_trig().expand().show()
bool ( -2*sol[0].rhs().denominator()==(l1*(2*m1+m2-m2*cos(2*phi1-2*phi2))).expand_trig().expand_trig().expand() )
rown_wiki = [m2*l2*cos(phi1-phi2)*phi2dd+(m1+m2)*l1*phi1dd+m2*l2*phi2d^2 * sin(phi1-phi2)+ (m1+m2)*g*sin(phi1),\
l2*phi2dd+l1*cos(phi1-phi2)*phi1dd-l1*phi1d^2*sin(phi1-phi2)+g*sin(phi2)]
showmath(rown_wiki[0])
showmath(rown_wiki[1])
rown_wiki[0].show()
(eq1/l1).reduce_trig().show()
rown_wiki[0].show()
bool((eq1/l1) == rown_wiki[0] )
(eq2/l2/m2).reduce_trig().show()
rown_wiki[1].show()
bool((eq2/l2/m2) == rown_wiki[1] )
Ekin = 1/2*(m1*x1.subs(x2u).subs(to_fun).diff(t).subs(to_var)^2+\
m1*y1.subs(x2u).subs(to_fun).diff(t).subs(to_var)^2+\
m2*x2.subs(x2u).subs(to_fun).diff(t).subs(to_var)^2+\
m2*y2.subs(x2u).subs(to_fun).diff(t).subs(to_var)^2 )
Epot = m1*g*y1.subs(x2u)+m2*g*y2.subs(x2u)
showmath( Epot.collect(cos(phi1)) )
showmath( Epot )
showmath( Ekin.trig_simplify() )
L = Ekin - Epot
len(L.expand().operands())
EL1 = L.diff(phi1d).subs(to_fun).diff(t).subs(to_var) - L.diff(phi1)
EL2 = L.diff(phi2d).subs(to_fun).diff(t).subs(to_var) - L.diff(phi2)
EL1.expand().operands()
EL1 = (EL1/l1).trig_reduce()
EL2 = (EL2/l2).trig_reduce()
showmath(EL1)
sol = solve([EL1,EL2],[phi1dd,phi2dd])[0]
show(sol)
expr = sol[0].rhs()
for ex_ in expr.factor().numerator().operands():
show(ex_)
import numpy as np
ode = [phi1d,phi2d]+[sol[0].rhs(),sol[1].rhs()]
ode = map(lambda x:x.subs({l1:1,l2:1,m1:1,m2:1,g:9.81}),ode)
times = srange(0,5,.01)
numsol = desolve_odeint(ode,[2.1,0,0,0],times,[phi1,phi2,phi1d,phi2d])
p = line ( zip(np.sin(numsol[:,0])+np.sin(numsol[:,1]),\
-np.cos(numsol[:,0])-np.cos(numsol[:,1])), color='gray' )
p.show(figsize=4)
def plot_dp(f1,f2,pars):
mass1 = vector([x1,y1]).subs(x2u).subs(pars).subs({phi1:f1,phi2:f2})
mass2 = vector([x2,y2]).subs(x2u).subs(pars).subs({phi1:f1,phi2:f2})
plt = point([(0,0),mass1],aspect_ratio=1,size=40)
plt += point(mass2,xmin=-2,xmax=2,ymin=-2,ymax=2,size=40)
plt += line([(0,0),mass1,mass2],color='gray')
return plt
plot_dp(numsol[213,0],numsol[213,1],{l1:1,l2:1})+p
@interact
def _(ith=slider(0,numsol.shape[0]-1)):
f1,f2 = numsol[ith,:2]
plot_dp(f1,f2,{l1:1,l2:1}).show(axes=False)
| 0.250913 | 0.961025 |
```
import scipy.io as sio
import numpy as np
import matplotlib.pyplot as plt
from numpy import matmul as mm
import math
from scipy.linalg import cho_factor,cho_solve
train6 = sio.loadmat('training6.mat')
sol6 = sio.loadmat('solution6.mat')
ball = train6['ball']; rgb = train6['rgb']; t = train6['t']
predictions = sol6['predictions']
def rdivide(A,B):
c,low = cho_factor(B.T)
C = cho_solve((c,low),A.T).T
return C
def ldivide(A,B):
c,low = cho_factor(A)
C = cho_solve((c,low),B)
return C
```
### Path of the ball
```
plt.figure(figsize=(20,10))
plt.plot(ball[0,:],ball[1,:],'bo-')
plt.plot(ball[0,-1],ball[1,-1],'s',
markersize=10,markeredgecolor=[0,.5,0],
markerfacecolor='r')
plt.plot(ball[0,0],ball[1,0],'s',
markersize=10,markeredgecolor=[0,.5,0],
markerfacecolor='g')
plt.title('Ball Position tracks')
plt.xlabel('X (meters)')
plt.ylabel('Y (meters)')
plt.axis('equal')
```
### Run Algorithm -- Kalman Filter
```
def kalmanFilter(t,x,y,state,param,previous_t):
dt = t-previous_t
C = np.array([[1,0,0,0],[0,1,0,0]])
A = np.array([[1,0,dt,0],[0,1,0,dt],[0,0,1,0],[0,0,0,1]])
Af = np.array([[1,0,.33,0],[0,1,0,.33],[0,0,1,0],[0,0,0,1]])
Sm = np.diag([.02,.001,.09,.01])
R = np.diag([.002,.002])
if previous_t < 0 :
state = np.array([x,y,0,0])
param['P'] = .1*np.eye(4)
predictx = x
predicty = y
return predictx,predicty,state,param
P = param['P']
P = mm(mm(A,P),A.T)+Sm
K = rdivide(mm(P,C.T),R+mm(mm(C,P),C.T))
xt = state.T
z = np.array([[x],[y]])
x_hat = mm(A,xt).reshape(-1,1) + mm(K,z-mm(mm(C,A),xt).reshape(-1,1))
x_f = mm(Af,xt).reshape(-1,1) + mm(K,z-mm(mm(C,Af),xt).reshape(-1,1))
state = x_hat.T
predictx,predicty = x_f[0],x_f[1]
P -= mm(mm(K,C),P)
param['P'] = P
return predictx,predicty,state,param
state = np.array([0,0,0,0])
last_t,N = -1,91
myPredictions = np.zeros((2,N))
param = {}
for i in range(N):
px,py,state,param = kalmanFilter(t[0,i],ball[0,i],
ball[1,i],state,param,last_t)
last_t = t[0,i]
myPredictions[0,i] = px
myPredictions[1,i] = py
plt.figure(figsize=(20,10))
plt.plot(ball[0,:],ball[1,:],'bo-')
plt.plot(ball[0,-1],ball[1,-1],'s',
markersize=10,markeredgecolor=[0,.5,0],
markerfacecolor='r')
plt.plot(ball[0,0],ball[1,0],'s',
markersize=10,markeredgecolor=[0,.5,0],
markerfacecolor='g')
plt.plot(myPredictions[0,:],myPredictions[1,:],'k+-')
plt.title('Ball Position tracks')
plt.xlabel('X (meters)')
plt.ylabel('Y (meters)')
plt.axis('equal')
```
### Show the error
```
nSkip = 10
myError = myPredictions[:,:-11]-ball[:,nSkip:-1]
myError_dist = np.sqrt(myError[0,:]**2+myError[1,:]**2)
myError_mean = np.mean(myError_dist)
plt.plot(myError_dist)
plt.title('Prediction Error over Time')
plt.xlabel('Frame')
plt.ylabel('Error (meters)')
print('Your Prediction: '+str(myError_mean))
```
### Solution comparison
```
error = predictions[:,:-11]-ball[:,nSkip:-1]
error_dist = np.sqrt(error[0,:]**2+error[1,:]**2)
error_mean = np.mean(error_dist)
print('Another Kalman Prediction: '+str(error_mean))
plt.plot(myError_dist)
plt.plot(error_dist)
plt.title('Prediction Error over Time')
plt.xlabel('Frame')
plt.ylabel('Error (meters)')
plt.figure(figsize=(15,10))
plt.plot(ball[0,:],ball[1,:],'bo-')
plt.plot(predictions[0,:],predictions[1,:],'mo-')
plt.plot(ball[0,-1],ball[1,-1],'s',
markersize=10,markeredgecolor=[0,.5,0],
markerfacecolor='r')
plt.plot(ball[0,0],ball[1,0],'s',
markersize=10,markeredgecolor=[0,.5,0],
markerfacecolor='g')
plt.plot(myPredictions[0,:],myPredictions[1,:],'k+-')
plt.title('Ball Position tracks')
plt.legend(['Observed','End','Start',
'Your Prediction','Another Kalman Prediction'])
plt.xlabel('X (meters)')
plt.ylabel('Y (meters)')
plt.axis('equal')
```
|
github_jupyter
|
import scipy.io as sio
import numpy as np
import matplotlib.pyplot as plt
from numpy import matmul as mm
import math
from scipy.linalg import cho_factor,cho_solve
train6 = sio.loadmat('training6.mat')
sol6 = sio.loadmat('solution6.mat')
ball = train6['ball']; rgb = train6['rgb']; t = train6['t']
predictions = sol6['predictions']
def rdivide(A,B):
c,low = cho_factor(B.T)
C = cho_solve((c,low),A.T).T
return C
def ldivide(A,B):
c,low = cho_factor(A)
C = cho_solve((c,low),B)
return C
plt.figure(figsize=(20,10))
plt.plot(ball[0,:],ball[1,:],'bo-')
plt.plot(ball[0,-1],ball[1,-1],'s',
markersize=10,markeredgecolor=[0,.5,0],
markerfacecolor='r')
plt.plot(ball[0,0],ball[1,0],'s',
markersize=10,markeredgecolor=[0,.5,0],
markerfacecolor='g')
plt.title('Ball Position tracks')
plt.xlabel('X (meters)')
plt.ylabel('Y (meters)')
plt.axis('equal')
def kalmanFilter(t,x,y,state,param,previous_t):
dt = t-previous_t
C = np.array([[1,0,0,0],[0,1,0,0]])
A = np.array([[1,0,dt,0],[0,1,0,dt],[0,0,1,0],[0,0,0,1]])
Af = np.array([[1,0,.33,0],[0,1,0,.33],[0,0,1,0],[0,0,0,1]])
Sm = np.diag([.02,.001,.09,.01])
R = np.diag([.002,.002])
if previous_t < 0 :
state = np.array([x,y,0,0])
param['P'] = .1*np.eye(4)
predictx = x
predicty = y
return predictx,predicty,state,param
P = param['P']
P = mm(mm(A,P),A.T)+Sm
K = rdivide(mm(P,C.T),R+mm(mm(C,P),C.T))
xt = state.T
z = np.array([[x],[y]])
x_hat = mm(A,xt).reshape(-1,1) + mm(K,z-mm(mm(C,A),xt).reshape(-1,1))
x_f = mm(Af,xt).reshape(-1,1) + mm(K,z-mm(mm(C,Af),xt).reshape(-1,1))
state = x_hat.T
predictx,predicty = x_f[0],x_f[1]
P -= mm(mm(K,C),P)
param['P'] = P
return predictx,predicty,state,param
state = np.array([0,0,0,0])
last_t,N = -1,91
myPredictions = np.zeros((2,N))
param = {}
for i in range(N):
px,py,state,param = kalmanFilter(t[0,i],ball[0,i],
ball[1,i],state,param,last_t)
last_t = t[0,i]
myPredictions[0,i] = px
myPredictions[1,i] = py
plt.figure(figsize=(20,10))
plt.plot(ball[0,:],ball[1,:],'bo-')
plt.plot(ball[0,-1],ball[1,-1],'s',
markersize=10,markeredgecolor=[0,.5,0],
markerfacecolor='r')
plt.plot(ball[0,0],ball[1,0],'s',
markersize=10,markeredgecolor=[0,.5,0],
markerfacecolor='g')
plt.plot(myPredictions[0,:],myPredictions[1,:],'k+-')
plt.title('Ball Position tracks')
plt.xlabel('X (meters)')
plt.ylabel('Y (meters)')
plt.axis('equal')
nSkip = 10
myError = myPredictions[:,:-11]-ball[:,nSkip:-1]
myError_dist = np.sqrt(myError[0,:]**2+myError[1,:]**2)
myError_mean = np.mean(myError_dist)
plt.plot(myError_dist)
plt.title('Prediction Error over Time')
plt.xlabel('Frame')
plt.ylabel('Error (meters)')
print('Your Prediction: '+str(myError_mean))
error = predictions[:,:-11]-ball[:,nSkip:-1]
error_dist = np.sqrt(error[0,:]**2+error[1,:]**2)
error_mean = np.mean(error_dist)
print('Another Kalman Prediction: '+str(error_mean))
plt.plot(myError_dist)
plt.plot(error_dist)
plt.title('Prediction Error over Time')
plt.xlabel('Frame')
plt.ylabel('Error (meters)')
plt.figure(figsize=(15,10))
plt.plot(ball[0,:],ball[1,:],'bo-')
plt.plot(predictions[0,:],predictions[1,:],'mo-')
plt.plot(ball[0,-1],ball[1,-1],'s',
markersize=10,markeredgecolor=[0,.5,0],
markerfacecolor='r')
plt.plot(ball[0,0],ball[1,0],'s',
markersize=10,markeredgecolor=[0,.5,0],
markerfacecolor='g')
plt.plot(myPredictions[0,:],myPredictions[1,:],'k+-')
plt.title('Ball Position tracks')
plt.legend(['Observed','End','Start',
'Your Prediction','Another Kalman Prediction'])
plt.xlabel('X (meters)')
plt.ylabel('Y (meters)')
plt.axis('equal')
| 0.467089 | 0.835148 |
# Continuous Control
---
In this notebook, you will learn how to use the Unity ML-Agents environment for the second project of the [Deep Reinforcement Learning Nanodegree](https://www.udacity.com/course/deep-reinforcement-learning-nanodegree--nd893) program.
### 1. Start the Environment
We begin by importing the necessary packages. If the code cell below returns an error, please revisit the project instructions to double-check that you have installed [Unity ML-Agents](https://github.com/Unity-Technologies/ml-agents/blob/master/docs/Installation.md) and [NumPy](http://www.numpy.org/).
```
from unityagents import UnityEnvironment
import numpy as np
```
Next, we will start the environment! **_Before running the code cell below_**, change the `file_name` parameter to match the location of the Unity environment that you downloaded.
- **Mac**: `"path/to/Reacher.app"`
- **Windows** (x86): `"path/to/Reacher_Windows_x86/Reacher.exe"`
- **Windows** (x86_64): `"path/to/Reacher_Windows_x86_64/Reacher.exe"`
- **Linux** (x86): `"path/to/Reacher_Linux/Reacher.x86"`
- **Linux** (x86_64): `"path/to/Reacher_Linux/Reacher.x86_64"`
- **Linux** (x86, headless): `"path/to/Reacher_Linux_NoVis/Reacher.x86"`
- **Linux** (x86_64, headless): `"path/to/Reacher_Linux_NoVis/Reacher.x86_64"`
For instance, if you are using a Mac, then you downloaded `Reacher.app`. If this file is in the same folder as the notebook, then the line below should appear as follows:
```
env = UnityEnvironment(file_name="Reacher.app")
```
```
env = UnityEnvironment(file_name='Reacher_Linux/Reacher.x86_64')
```
Environments contain **_brains_** which are responsible for deciding the actions of their associated agents. Here we check for the first brain available, and set it as the default brain we will be controlling from Python.
```
# get the default brain
brain_name = env.brain_names[0]
brain = env.brains[brain_name]
```
### 2. Examine the State and Action Spaces
In this environment, a double-jointed arm can move to target locations. A reward of `+0.1` is provided for each step that the agent's hand is in the goal location. Thus, the goal of your agent is to maintain its position at the target location for as many time steps as possible.
The observation space consists of `33` variables corresponding to position, rotation, velocity, and angular velocities of the arm. Each action is a vector with four numbers, corresponding to torque applicable to two joints. Every entry in the action vector must be a number between `-1` and `1`.
Run the code cell below to print some information about the environment.
```
# reset the environment
env_info = env.reset(train_mode=True)[brain_name]
# number of agents
num_agents = len(env_info.agents)
print('Number of agents:', num_agents)
# size of each action
action_size = brain.vector_action_space_size
print('Size of each action:', action_size)
# examine the state space
states = env_info.vector_observations
state_size = states.shape[1]
print('There are {} agents. Each observes a state with length: {}'.format(states.shape[0], state_size))
print('The state for the first agent looks like:', states[0])
```
### 3. Take Random Actions in the Environment
In the next code cell, you will learn how to use the Python API to control the agent and receive feedback from the environment.
Once this cell is executed, you will watch the agent's performance, if it selects an action at random with each time step. A window should pop up that allows you to observe the agent, as it moves through the environment.
Of course, as part of the project, you'll have to change the code so that the agent is able to use its experience to gradually choose better actions when interacting with the environment!
```
'''
env_info = env.reset(train_mode=False)[brain_name] # reset the environment
states = env_info.vector_observations # get the current state (for each agent)
scores = np.zeros(num_agents) # initialize the score (for each agent)
while True:
actions = np.random.randn(num_agents, action_size) # select an action (for each agent)
actions = np.clip(actions, -1, 1) # all actions between -1 and 1
env_info = env.step(actions)[brain_name] # send all actions to tne environment
next_states = env_info.vector_observations # get next state (for each agent)
rewards = env_info.rewards # get reward (for each agent)
dones = env_info.local_done # see if episode finished
scores += env_info.rewards # update the score (for each agent)
states = next_states # roll over states to next time step
if np.any(dones): # exit loop if episode finished
break
print('Total score (averaged over agents) this episode: {}'.format(np.mean(scores)))
'''
```
When finished, you can close the environment.
```
#env.close()
```
### 4. It's Your Turn!
Now it's your turn to train your own agent to solve the environment! When training the environment, set `train_mode=True`, so that the line for resetting the environment looks like the following:
```python
env_info = env.reset(train_mode=True)[brain_name]
```
```
import torch
from ddpg_agent import Agent
from collections import deque
import matplotlib.pyplot as plt
%matplotlib inline
agent = Agent(state_size=state_size, action_size=action_size, random_seed=10)
def ddpg(n_episodes=10000, print_every=100):
scores_deque = deque(maxlen=print_every)
scores = []
for i_episode in range(1, n_episodes+1):
env_info = env.reset(train_mode=True)[brain_name]
state = env_info.vector_observations[0]
agent.reset()
score = 0
while True:
# get agent action
action = agent.act(state)
# get environment info
env_info = env.step(action)[brain_name] # send the action to the environment
next_state = env_info.vector_observations[0] # get the next state
reward = env_info.rewards[0] # get the reward
done = env_info.local_done[0] # see if episode has finished
#set agent
agent.step(state, action, reward, next_state, done)
state = next_state
score += reward
if done:
break
scores_deque.append(score)
scores.append(score)
mean_score = np.mean(scores_deque)
print('\rEpisode {}\tAverage Score: {:.2f} Epsisode Score: {:.2f}'.format(i_episode, mean_score, score), end="")
if i_episode % print_every == 0:
print('\rEpisode {}\tAverage Score: {:.2f} Epsisode Score: {:.2f}'.format(i_episode, mean_score, score))
if(mean_score > 30.0):
torch.save(agent.actor_local.state_dict(), 'checkpoint_actor.pth')
torch.save(agent.critic_local.state_dict(), 'checkpoint_critic.pth')
print('Continuous Control Enviroment solved in {} episodes with a mean score of {:.2f} over 100 epsiodes'.format(i_episode, mean_score))
return scores
return scores
scores = ddpg()
fig = plt.figure()
ax = fig.add_subplot(111)
plt.plot(np.arange(1, len(scores)+1), scores)
plt.ylabel('Score')
plt.xlabel('Episode #')
plt.show()
agent.actor_local.load_state_dict(torch.load('checkpoint_actor.pth'))
agent.critic_local.load_state_dict(torch.load('checkpoint_critic.pth'))
env_info = env.reset(train_mode=False)[brain_name] # reset the environment
state = env_info.vector_observations[0] # get the current state
episode_score = 0
while True:
action = agent.act(state, add_noise=False) # get agent's action
env_info = env.step(action)[brain_name] # send the action to the environment
next_state = env_info.vector_observations[0] # get the next state
reward = env_info.rewards[0] # get the reward
done = env_info.local_done[0] # see if episode has finished
episode_score += reward # update the score
state = next_state # roll over the state to next time step
if done: # exit loop if episode finished
break
```
|
github_jupyter
|
from unityagents import UnityEnvironment
import numpy as np
env = UnityEnvironment(file_name="Reacher.app")
env = UnityEnvironment(file_name='Reacher_Linux/Reacher.x86_64')
# get the default brain
brain_name = env.brain_names[0]
brain = env.brains[brain_name]
# reset the environment
env_info = env.reset(train_mode=True)[brain_name]
# number of agents
num_agents = len(env_info.agents)
print('Number of agents:', num_agents)
# size of each action
action_size = brain.vector_action_space_size
print('Size of each action:', action_size)
# examine the state space
states = env_info.vector_observations
state_size = states.shape[1]
print('There are {} agents. Each observes a state with length: {}'.format(states.shape[0], state_size))
print('The state for the first agent looks like:', states[0])
'''
env_info = env.reset(train_mode=False)[brain_name] # reset the environment
states = env_info.vector_observations # get the current state (for each agent)
scores = np.zeros(num_agents) # initialize the score (for each agent)
while True:
actions = np.random.randn(num_agents, action_size) # select an action (for each agent)
actions = np.clip(actions, -1, 1) # all actions between -1 and 1
env_info = env.step(actions)[brain_name] # send all actions to tne environment
next_states = env_info.vector_observations # get next state (for each agent)
rewards = env_info.rewards # get reward (for each agent)
dones = env_info.local_done # see if episode finished
scores += env_info.rewards # update the score (for each agent)
states = next_states # roll over states to next time step
if np.any(dones): # exit loop if episode finished
break
print('Total score (averaged over agents) this episode: {}'.format(np.mean(scores)))
'''
#env.close()
env_info = env.reset(train_mode=True)[brain_name]
import torch
from ddpg_agent import Agent
from collections import deque
import matplotlib.pyplot as plt
%matplotlib inline
agent = Agent(state_size=state_size, action_size=action_size, random_seed=10)
def ddpg(n_episodes=10000, print_every=100):
scores_deque = deque(maxlen=print_every)
scores = []
for i_episode in range(1, n_episodes+1):
env_info = env.reset(train_mode=True)[brain_name]
state = env_info.vector_observations[0]
agent.reset()
score = 0
while True:
# get agent action
action = agent.act(state)
# get environment info
env_info = env.step(action)[brain_name] # send the action to the environment
next_state = env_info.vector_observations[0] # get the next state
reward = env_info.rewards[0] # get the reward
done = env_info.local_done[0] # see if episode has finished
#set agent
agent.step(state, action, reward, next_state, done)
state = next_state
score += reward
if done:
break
scores_deque.append(score)
scores.append(score)
mean_score = np.mean(scores_deque)
print('\rEpisode {}\tAverage Score: {:.2f} Epsisode Score: {:.2f}'.format(i_episode, mean_score, score), end="")
if i_episode % print_every == 0:
print('\rEpisode {}\tAverage Score: {:.2f} Epsisode Score: {:.2f}'.format(i_episode, mean_score, score))
if(mean_score > 30.0):
torch.save(agent.actor_local.state_dict(), 'checkpoint_actor.pth')
torch.save(agent.critic_local.state_dict(), 'checkpoint_critic.pth')
print('Continuous Control Enviroment solved in {} episodes with a mean score of {:.2f} over 100 epsiodes'.format(i_episode, mean_score))
return scores
return scores
scores = ddpg()
fig = plt.figure()
ax = fig.add_subplot(111)
plt.plot(np.arange(1, len(scores)+1), scores)
plt.ylabel('Score')
plt.xlabel('Episode #')
plt.show()
agent.actor_local.load_state_dict(torch.load('checkpoint_actor.pth'))
agent.critic_local.load_state_dict(torch.load('checkpoint_critic.pth'))
env_info = env.reset(train_mode=False)[brain_name] # reset the environment
state = env_info.vector_observations[0] # get the current state
episode_score = 0
while True:
action = agent.act(state, add_noise=False) # get agent's action
env_info = env.step(action)[brain_name] # send the action to the environment
next_state = env_info.vector_observations[0] # get the next state
reward = env_info.rewards[0] # get the reward
done = env_info.local_done[0] # see if episode has finished
episode_score += reward # update the score
state = next_state # roll over the state to next time step
if done: # exit loop if episode finished
break
| 0.431105 | 0.976129 |
# TensorFlow script mode training and serving
Script mode is a training script format for TensorFlow that lets you execute any TensorFlow training script in SageMaker with minimal modification. The [SageMaker Python SDK](https://github.com/aws/sagemaker-python-sdk) handles transferring your script to a SageMaker training instance. On the training instance, SageMaker's native TensorFlow support sets up training-related environment variables and executes your training script. In this tutorial, we use the SageMaker Python SDK to launch a training job and deploy the trained model.
Script mode supports training with a Python script, a Python module, or a shell script. In this example, we use a Python script to train a classification model on the [MNIST dataset](http://yann.lecun.com/exdb/mnist/). In this example, we will show how easily you can train a SageMaker using TensorFlow 1.x and TensorFlow 2.0 scripts with SageMaker Python SDK. In addition, this notebook demonstrates how to perform real time inference with the [SageMaker TensorFlow Serving container](https://github.com/aws/sagemaker-tensorflow-serving-container). The TensorFlow Serving container is the default inference method for script mode. For full documentation on the TensorFlow Serving container, please visit [here](https://github.com/aws/sagemaker-python-sdk/blob/master/src/sagemaker/tensorflow/deploying_tensorflow_serving.rst).
# Set up the environment
Let's start by setting up the environment:
```
import os
import sagemaker
from sagemaker import get_execution_role
sagemaker_session = sagemaker.Session()
role = get_execution_role()
region = sagemaker_session.boto_session.region_name
```
## Training Data
The MNIST dataset has been loaded to the public S3 buckets ``sagemaker-sample-data-<REGION>`` under the prefix ``tensorflow/mnist``. There are four ``.npy`` file under this prefix:
* ``train_data.npy``
* ``eval_data.npy``
* ``train_labels.npy``
* ``eval_labels.npy``
```
training_data_uri = 's3://sagemaker-sample-data-{}/tensorflow/mnist'.format(region)
```
# Construct a script for distributed training
This tutorial's training script was adapted from TensorFlow's official [CNN MNIST example](https://github.com/tensorflow/tensorflow/blob/master/tensorflow/examples/tutorials/layers/cnn_mnist.py). We have modified it to handle the ``model_dir`` parameter passed in by SageMaker. This is an S3 path which can be used for data sharing during distributed training and checkpointing and/or model persistence. We have also added an argument-parsing function to handle processing training-related variables.
At the end of the training job we have added a step to export the trained model to the path stored in the environment variable ``SM_MODEL_DIR``, which always points to ``/opt/ml/model``. This is critical because SageMaker uploads all the model artifacts in this folder to S3 at end of training.
Here is the entire script:
```
!pygmentize 'mnist.py'
# TensorFlow 2.1 script
!pygmentize 'mnist-2.py'
```
# Create a training job using the `TensorFlow` estimator
The `sagemaker.tensorflow.TensorFlow` estimator handles locating the script mode container, uploading your script to a S3 location and creating a SageMaker training job. Let's call out a couple important parameters here:
* `py_version` is set to `'py3'` to indicate that we are using script mode since legacy mode supports only Python 2. Though Python 2 will be deprecated soon, you can use script mode with Python 2 by setting `py_version` to `'py2'` and `script_mode` to `True`.
* `distributions` is used to configure the distributed training setup. It's required only if you are doing distributed training either across a cluster of instances or across multiple GPUs. Here we are using parameter servers as the distributed training schema. SageMaker training jobs run on homogeneous clusters. To make parameter server more performant in the SageMaker setup, we run a parameter server on every instance in the cluster, so there is no need to specify the number of parameter servers to launch. Script mode also supports distributed training with [Horovod](https://github.com/horovod/horovod). You can find the full documentation on how to configure `distributions` [here](https://github.com/aws/sagemaker-python-sdk/tree/master/src/sagemaker/tensorflow#distributed-training).
```
from sagemaker.tensorflow import TensorFlow
mnist_estimator = TensorFlow(entry_point='mnist.py',
role=role,
train_instance_count=2,
train_instance_type='ml.p3.2xlarge',
framework_version='1.15.2',
py_version='py3',
distributions={'parameter_server': {'enabled': True}})
```
You can also initiate an estimator to train with TensorFlow 2.1 script. The only things that you will need to change are the script name and ``framewotk_version``
```
mnist_estimator2 = TensorFlow(entry_point='mnist-2.py',
role=role,
train_instance_count=2,
train_instance_type='ml.p3.2xlarge',
framework_version='2.1.0',
py_version='py3',
distributions={'parameter_server': {'enabled': True}})
```
## Calling ``fit``
To start a training job, we call `estimator.fit(training_data_uri)`.
An S3 location is used here as the input. `fit` creates a default channel named `'training'`, which points to this S3 location. In the training script we can then access the training data from the location stored in `SM_CHANNEL_TRAINING`. `fit` accepts a couple other types of input as well. See the API doc [here](https://sagemaker.readthedocs.io/en/stable/estimators.html#sagemaker.estimator.EstimatorBase.fit) for details.
When training starts, the TensorFlow container executes mnist.py, passing `hyperparameters` and `model_dir` from the estimator as script arguments. Because we didn't define either in this example, no hyperparameters are passed, and `model_dir` defaults to `s3://<DEFAULT_BUCKET>/<TRAINING_JOB_NAME>`, so the script execution is as follows:
```bash
python mnist.py --model_dir s3://<DEFAULT_BUCKET>/<TRAINING_JOB_NAME>
```
When training is complete, the training job will upload the saved model for TensorFlow serving.
```
mnist_estimator.fit(training_data_uri)
```
Calling fit to train a model with TensorFlow 2.1 scroipt.
```
mnist_estimator2.fit(training_data_uri)
```
# Deploy the trained model to an endpoint
The `deploy()` method creates a SageMaker model, which is then deployed to an endpoint to serve prediction requests in real time. We will use the TensorFlow Serving container for the endpoint, because we trained with script mode. This serving container runs an implementation of a web server that is compatible with SageMaker hosting protocol. The [Using your own inference code]() document explains how SageMaker runs inference containers.
```
predictor = mnist_estimator.deploy(initial_instance_count=1, instance_type='ml.p2.xlarge')
```
Deployed the trained TensorFlow 2.1 model to an endpoint.
```
predictor2 = mnist_estimator2.deploy(initial_instance_count=1, instance_type='ml.p2.xlarge')
```
# Invoke the endpoint
Let's download the training data and use that as input for inference.
```
import numpy as np
!aws --region {region} s3 cp s3://sagemaker-sample-data-{region}/tensorflow/mnist/train_data.npy train_data.npy
!aws --region {region} s3 cp s3://sagemaker-sample-data-{region}/tensorflow/mnist/train_labels.npy train_labels.npy
train_data = np.load('train_data.npy')
train_labels = np.load('train_labels.npy')
```
The formats of the input and the output data correspond directly to the request and response formats of the `Predict` method in the [TensorFlow Serving REST API](https://www.tensorflow.org/serving/api_rest). SageMaker's TensforFlow Serving endpoints can also accept additional input formats that are not part of the TensorFlow REST API, including the simplified JSON format, line-delimited JSON objects ("jsons" or "jsonlines"), and CSV data.
In this example we are using a `numpy` array as input, which will be serialized into the simplified JSON format. In addtion, TensorFlow serving can also process multiple items at once as you can see in the following code. You can find the complete documentation on how to make predictions against a TensorFlow serving SageMaker endpoint [here](https://github.com/aws/sagemaker-python-sdk/blob/master/src/sagemaker/tensorflow/deploying_tensorflow_serving.rst#making-predictions-against-a-sagemaker-endpoint).
```
predictions = predictor.predict(train_data[:50])
for i in range(0, 50):
prediction = predictions['predictions'][i]['classes']
label = train_labels[i]
print('prediction is {}, label is {}, matched: {}'.format(prediction, label, prediction == label))
```
Examine the prediction result from the TensorFlow 2.1 model.
```
predictions2 = predictor2.predict(train_data[:50])
for i in range(0, 50):
prediction = predictions2['predictions'][i]
label = train_labels[i]
print('prediction is {}, label is {}, matched: {}'.format(prediction, label, prediction == label))
```
# Delete the endpoint
Let's delete the endpoint we just created to prevent incurring any extra costs.
```
sagemaker.Session().delete_endpoint(predictor.endpoint)
```
Delete the TensorFlow 2.1 endpoint as well.
```
sagemaker.Session().delete_endpoint(predictor2.endpoint)
```
|
github_jupyter
|
import os
import sagemaker
from sagemaker import get_execution_role
sagemaker_session = sagemaker.Session()
role = get_execution_role()
region = sagemaker_session.boto_session.region_name
training_data_uri = 's3://sagemaker-sample-data-{}/tensorflow/mnist'.format(region)
!pygmentize 'mnist.py'
# TensorFlow 2.1 script
!pygmentize 'mnist-2.py'
from sagemaker.tensorflow import TensorFlow
mnist_estimator = TensorFlow(entry_point='mnist.py',
role=role,
train_instance_count=2,
train_instance_type='ml.p3.2xlarge',
framework_version='1.15.2',
py_version='py3',
distributions={'parameter_server': {'enabled': True}})
mnist_estimator2 = TensorFlow(entry_point='mnist-2.py',
role=role,
train_instance_count=2,
train_instance_type='ml.p3.2xlarge',
framework_version='2.1.0',
py_version='py3',
distributions={'parameter_server': {'enabled': True}})
python mnist.py --model_dir s3://<DEFAULT_BUCKET>/<TRAINING_JOB_NAME>
mnist_estimator.fit(training_data_uri)
mnist_estimator2.fit(training_data_uri)
predictor = mnist_estimator.deploy(initial_instance_count=1, instance_type='ml.p2.xlarge')
predictor2 = mnist_estimator2.deploy(initial_instance_count=1, instance_type='ml.p2.xlarge')
import numpy as np
!aws --region {region} s3 cp s3://sagemaker-sample-data-{region}/tensorflow/mnist/train_data.npy train_data.npy
!aws --region {region} s3 cp s3://sagemaker-sample-data-{region}/tensorflow/mnist/train_labels.npy train_labels.npy
train_data = np.load('train_data.npy')
train_labels = np.load('train_labels.npy')
predictions = predictor.predict(train_data[:50])
for i in range(0, 50):
prediction = predictions['predictions'][i]['classes']
label = train_labels[i]
print('prediction is {}, label is {}, matched: {}'.format(prediction, label, prediction == label))
predictions2 = predictor2.predict(train_data[:50])
for i in range(0, 50):
prediction = predictions2['predictions'][i]
label = train_labels[i]
print('prediction is {}, label is {}, matched: {}'.format(prediction, label, prediction == label))
sagemaker.Session().delete_endpoint(predictor.endpoint)
sagemaker.Session().delete_endpoint(predictor2.endpoint)
| 0.382372 | 0.993103 |
<a href="https://colab.research.google.com/github/gtbook/gtsam-examples/blob/main/GaussianMRFExample_lbp.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# Clustered Belief Propagation
Instead of "vanilla" loopy belief propagation, we now introduce more a structured approximation. In particular, we could treat a number of variables $X_c$ as a cluster and use a Gaussian density for the entire cluster. This could easily be implemented using vanilla LBP, by introducing new clustered variables and proceeding as before. However, here we instead adopt a different approximate density as a collection of Bayes trees.
If we group several variables together, and build a variational mean field approximation on those clusters, we can get much more accurate marginals, much faster. This is because we are making less of an approximation, and because we use the power of GTSAM to optimize over a bunch of variables at the same time.
```
%pip -q install gtbook # also installs latest gtsam pre-release
from collections import defaultdict
from dataclasses import dataclass
from typing import FrozenSet
import matplotlib.pyplot as plt
import gtsam
import numpy as np
from gtbook.driving import planar_example, marginals_figure
from gtbook.display import show
import gtsam.utils.plot as gtsam_plot
GaussianPrior = gtsam.GaussianDensity.FromMeanAndStddev
ConstrainedOrdering = gtsam.Ordering.ColamdConstrainedLastGaussianFactorGraph
Keys = FrozenSet[int]
Edge = FrozenSet[int]
```
## A Linear SLAM example
We use a planar SLAM example that we linearize at the ground truth, in order to illustrate structured LBP.
```
nonlinear_graph, truth, graph_keys = planar_example()
x1, x2, x3, l1, l2 = graph_keys
graph = nonlinear_graph.linearize(truth)
show(nonlinear_graph, truth, binary_edges=True)
```
The Gaussian marginals around the non-linear solution are shown below:
```
marginals = gtsam.Marginals(graph, truth)
marginals_figure(truth, marginals, graph_keys)
```
## Cluster Nodes
In the example below we create three clusters: the Markov chain x1-x2-x3 in one cluster, and the two landmarks l1 and l2 each in their own separate cluster. For each we create an initial belief *in the form of a Gaussian Bayes net*:
```
belief0 = gtsam.GaussianBayesNet()
zero = np.zeros((3,), float)
belief0.push_back(GaussianPrior(x1, zero, 1.5))
belief0.push_back(GaussianPrior(x2, zero, 1.5))
belief0.push_back(GaussianPrior(x3, zero, 1.5))
belief1 = gtsam.GaussianBayesNet()
belief1.push_back(GaussianPrior(l1, zero[:2], 1.5))
belief2 = gtsam.GaussianBayesNet()
belief2.push_back(GaussianPrior(l2, zero[:2], 1.5))
key_sets = [{x1, x2, x3}, {l1}, {l2}]
beliefs = [belief0, belief1, belief2]
node_specs = list(zip(key_sets, beliefs))
```
Let's then create a set of `Clusters` that each approximate the density of a cluster:
```
@dataclass(frozen=True)
class Cluster:
keys: Keys
belief: gtsam.GaussianBayesNet
variable_nodes = [Cluster(keys, initial_belief)
for keys, initial_belief in node_specs]
```
We can't currently update a Bayes tree in python yet, but in C++ we would use a `gtsam.BayesTree` so we can easily get covariances and incrementally update the custer. Soon!
## Visualizing the Covariances
For now, we can visualize the covariances by converting to a Bayes tree. Note that we add the linear solution to the non-linear values for visualization purposes:
```
def plot_covariances(variable_nodes):
"""Plot all covariances."""
graph = gtsam.GaussianFactorGraph()
for q_j in variable_nodes:
graph.push_back(q_j.belief)
bt = graph.eliminateMultifrontal()
mean = bt.optimize()
cov = {key: bt.marginalCovariance(key)
for q_j in variable_nodes for key in q_j.keys}
plt.figure(0, figsize=(12, 7), dpi=80)
for key in [x1, x2, x3]:
gtsam_plot.plot_point2(0, truth.atPose2(key).translation() + mean.at(
key)[:2], 0.5, bt.marginalCovariance(key))
for key in [l1, l2]:
gtsam_plot.plot_point2(0, truth.atPoint2(key) + mean.at(key), 0.5,
bt.marginalCovariance(key))
plt.axis('equal')
plt.xlim([-0.8, 6])
plt.ylim([-0.8, 3])
plot_covariances(variable_nodes)
```
Note that above all beliefs are just uniform Gaussians with a large standard deviation.
## (Super) Factor Nodes
We also parse the factor graph in "super-factors" that bridge different cluster nodes:
```
# First figure out which nodes every node is connected to.
factor_indices = defaultdict(set)
for i in range(graph.size()):
factor = graph.at(i)
for j, (keys, _) in enumerate(node_specs):
if keys.intersection(factor.keys()):
factor_indices[i].add(j)
# Then store based on those index tuples.
factor_nodes = defaultdict(gtsam.GaussianFactorGraph)
for i, edge in factor_indices.items():
assert len(edge) <= 2, "Invalid interaction between nodes."
factor_nodes[frozenset(edge)].push_back(graph.at(i))
```
Every super-factor is really a factor graph. For example, one of the super-factors is intra-cluster:
```
edge0, edge01, edge02 = frozenset({0}), frozenset({0, 1}), frozenset({0, 2})
show(factor_nodes[edge0], hints={'x':0}, binary_edges=True)
```
And another one is between cluster 0 and 1:
```
show(factor_nodes[edge01], hints={'x':0, 'l':1}, binary_edges=True)
```
## Messages
Finally, as in our vanilla LBP implementation, we only have one type of message, from factors (indexed by `frozenset` edges) to clusters (indexed by int). Again, messages are entire factor *graphs* now:
```
Message = gtsam.GaussianFactorGraph
messages = defaultdict(dict)
for i, factors in factor_nodes.items():
if len(i) == 1:
j, *others = i
messages[j][i] = factors
continue
for j in i:
messages[j][i] = Message()
```
We initialized the messages $m_{i->j}$ above to empty, except the ones that correspond to unary "factors". There is only one here, which is exactly the intra-cluster factor graph from above:
```
for j, messages_j in messages.items():
for i, message_ij in messages_j.items():
print(f"F_{list(i)} -> X_{j}: size = {message_ij.size()}")
```
## Cluster Belief Propagation
With these three data structures in hand, the code to update the belief of one cluster is easy: just add all "message" factor graphs to a big graph, and then eliminate:
```
def update_node(j: int):
"""Update cluster j."""
graph = gtsam.GaussianFactorGraph()
for message in messages[j].values():
for i in range(message.size()):
graph.push_back(message.at(i))
# Eliminate into Bayes net.
new_belief = graph.eliminateSequential(gtsam.Ordering.OrderingType.NATURAL)
variable_nodes[j] = Cluster(variable_nodes[j].keys, new_belief)
```
For example, updating cluster 0 we get an updated Bayes net on the Markov chain x1-x2-x3, which we show below as a Bayes net (it was eliminated in the x1,x2,x3 order, which is why the past is conditioned on the present), and below that we show the updated covariances:
```
update_node(0)
show(variable_nodes[0].belief, hints={'x':0})
plot_covariances(variable_nodes)
```
Above we see that the Markov chain has been updated in one fell swoop, and has the correct covariance structure. However, we did not use all information: we should have updated the messages using the other cluster's beliefs first. Let's correct that now.
Calculating a new messages $m_{i\rightarrow j}$ takes the super-factor and adds the belief of the cluster "on the other side" to it, *minus* the messages that was sent to that other cluster:
```
def calculate_new_message(factors: gtsam.GaussianFactorGraph,
keys: Keys, downdated_belief: gtsam.GaussianFactorGraph):
"""Calculate message from factors and belief for sending node."""
graph = gtsam.GaussianFactorGraph()
graph.push_back(downdated_belief)
graph.push_back(factors)
ordering = gtsam.Ordering()
for key in keys:
ordering.push_back(key)
_, remaining = graph.eliminatePartialSequential(ordering)
return remaining
```
With that, we can write our final iteration code:
```
def update_messages_and_node(j: int):
"""Update messages into cluster j and calculate belief q(X_j)."""
for edge in messages[j].keys():
if len(edge) == 2: # only update binary messages
j1, j2 = edge
k = j2 if j1 == j else j1
other_cluster = variable_nodes[k]
downdated_belief = gtsam.GaussianFactorGraph()
downdated_belief.push_back(other_cluster.belief)
message_to_subtract = messages[k][edge]
for i in range(message_to_subtract.size()):
downdated_belief.push_back(message_to_subtract.at(i).negate())
messages[j][edge] = calculate_new_message(factor_nodes[edge],
other_cluster.keys,
downdated_belief)
update_node(j)
```
Note that above we down-date the belief for the "other node" before eliminating the variables associated with it. Otherwise we are double-counting information.
## Running Cluster-BP
Below we run this to convergence, which is very fast.
```
update_messages_and_node(0)
plot_covariances(variable_nodes)
```
We update the two other clusters as well:
```
for j in [1,2]:
update_messages_and_node(j)
plot_covariances(variable_nodes)
```
As you can see, these are remarkably close, after *one* round-robin iteration. Doing a few more rounds does not make a difference, and when overlaid on the *true* covariances you can hardly see the difference:
```
for round in range(5):
for j in range(3):
update_messages_and_node(j)
marginals_figure(truth, marginals, graph_keys)
plot_covariances(variable_nodes)
```
## Different Initial Beliefs
Of course, we started from beliefs that already had the mean correct. But, We get a more interesting animation when we start different points and pretend to be rather confident:
```
rng = np.random.default_rng(42)
new_belief0 = gtsam.GaussianBayesNet()
minmax = -3.0, 3.0
new_belief0.push_back(GaussianPrior(x1, rng.uniform(*minmax, size=(3,)), 0.1))
new_belief0.push_back(GaussianPrior(x2, rng.uniform(*minmax, size=(3,)), 0.1))
new_belief0.push_back(GaussianPrior(x3, rng.uniform(*minmax, size=(3,)), 0.1))
new_belief1 = gtsam.GaussianBayesNet()
new_belief1.push_back(GaussianPrior(l1, rng.uniform(*minmax, size=(2,)), 0.1))
new_belief2 = gtsam.GaussianBayesNet()
new_belief2.push_back(GaussianPrior(l2, rng.uniform(*minmax, size=(2,)), 0.1))
key_sets = [{x1, x2, x3}, {l1}, {l2}]
new_beliefs = [new_belief0, new_belief1, new_belief2]
new_node_specs = list(zip(key_sets, new_beliefs))
variable_nodes = [Cluster(keys, initial_belief)
for keys, initial_belief in new_node_specs]
messages = defaultdict(dict)
for i, factors in factor_nodes.items():
if len(i) == 1:
j, *others = i
messages[j][i] = factors
continue
for j in i:
messages[j][i] = Message()
# marginals_figure(truth, marginals, graph_keys)
plot_covariances(variable_nodes)
for round in range(5):
for j in range(3):
update_messages_and_node(j)
plot_covariances(variable_nodes)
```
Still, for this small graph we converge rather fast.
## Summary
A cluster-based belief propagation approach is still a "mean-field" approximation, but on clusters rather than individual variables. We recover "vanilla" BP if make the clusters contain only one variable, but note that even then these variables are multivariate. Finally, if we use *one* cluster, because we call sparse factorization (`eliminateSequential`!), we converge to the *true* posterior density.
We'll do a larger example in a non-linear SLAM notebook.
|
github_jupyter
|
%pip -q install gtbook # also installs latest gtsam pre-release
from collections import defaultdict
from dataclasses import dataclass
from typing import FrozenSet
import matplotlib.pyplot as plt
import gtsam
import numpy as np
from gtbook.driving import planar_example, marginals_figure
from gtbook.display import show
import gtsam.utils.plot as gtsam_plot
GaussianPrior = gtsam.GaussianDensity.FromMeanAndStddev
ConstrainedOrdering = gtsam.Ordering.ColamdConstrainedLastGaussianFactorGraph
Keys = FrozenSet[int]
Edge = FrozenSet[int]
nonlinear_graph, truth, graph_keys = planar_example()
x1, x2, x3, l1, l2 = graph_keys
graph = nonlinear_graph.linearize(truth)
show(nonlinear_graph, truth, binary_edges=True)
marginals = gtsam.Marginals(graph, truth)
marginals_figure(truth, marginals, graph_keys)
belief0 = gtsam.GaussianBayesNet()
zero = np.zeros((3,), float)
belief0.push_back(GaussianPrior(x1, zero, 1.5))
belief0.push_back(GaussianPrior(x2, zero, 1.5))
belief0.push_back(GaussianPrior(x3, zero, 1.5))
belief1 = gtsam.GaussianBayesNet()
belief1.push_back(GaussianPrior(l1, zero[:2], 1.5))
belief2 = gtsam.GaussianBayesNet()
belief2.push_back(GaussianPrior(l2, zero[:2], 1.5))
key_sets = [{x1, x2, x3}, {l1}, {l2}]
beliefs = [belief0, belief1, belief2]
node_specs = list(zip(key_sets, beliefs))
@dataclass(frozen=True)
class Cluster:
keys: Keys
belief: gtsam.GaussianBayesNet
variable_nodes = [Cluster(keys, initial_belief)
for keys, initial_belief in node_specs]
def plot_covariances(variable_nodes):
"""Plot all covariances."""
graph = gtsam.GaussianFactorGraph()
for q_j in variable_nodes:
graph.push_back(q_j.belief)
bt = graph.eliminateMultifrontal()
mean = bt.optimize()
cov = {key: bt.marginalCovariance(key)
for q_j in variable_nodes for key in q_j.keys}
plt.figure(0, figsize=(12, 7), dpi=80)
for key in [x1, x2, x3]:
gtsam_plot.plot_point2(0, truth.atPose2(key).translation() + mean.at(
key)[:2], 0.5, bt.marginalCovariance(key))
for key in [l1, l2]:
gtsam_plot.plot_point2(0, truth.atPoint2(key) + mean.at(key), 0.5,
bt.marginalCovariance(key))
plt.axis('equal')
plt.xlim([-0.8, 6])
plt.ylim([-0.8, 3])
plot_covariances(variable_nodes)
# First figure out which nodes every node is connected to.
factor_indices = defaultdict(set)
for i in range(graph.size()):
factor = graph.at(i)
for j, (keys, _) in enumerate(node_specs):
if keys.intersection(factor.keys()):
factor_indices[i].add(j)
# Then store based on those index tuples.
factor_nodes = defaultdict(gtsam.GaussianFactorGraph)
for i, edge in factor_indices.items():
assert len(edge) <= 2, "Invalid interaction between nodes."
factor_nodes[frozenset(edge)].push_back(graph.at(i))
edge0, edge01, edge02 = frozenset({0}), frozenset({0, 1}), frozenset({0, 2})
show(factor_nodes[edge0], hints={'x':0}, binary_edges=True)
show(factor_nodes[edge01], hints={'x':0, 'l':1}, binary_edges=True)
Message = gtsam.GaussianFactorGraph
messages = defaultdict(dict)
for i, factors in factor_nodes.items():
if len(i) == 1:
j, *others = i
messages[j][i] = factors
continue
for j in i:
messages[j][i] = Message()
for j, messages_j in messages.items():
for i, message_ij in messages_j.items():
print(f"F_{list(i)} -> X_{j}: size = {message_ij.size()}")
def update_node(j: int):
"""Update cluster j."""
graph = gtsam.GaussianFactorGraph()
for message in messages[j].values():
for i in range(message.size()):
graph.push_back(message.at(i))
# Eliminate into Bayes net.
new_belief = graph.eliminateSequential(gtsam.Ordering.OrderingType.NATURAL)
variable_nodes[j] = Cluster(variable_nodes[j].keys, new_belief)
update_node(0)
show(variable_nodes[0].belief, hints={'x':0})
plot_covariances(variable_nodes)
def calculate_new_message(factors: gtsam.GaussianFactorGraph,
keys: Keys, downdated_belief: gtsam.GaussianFactorGraph):
"""Calculate message from factors and belief for sending node."""
graph = gtsam.GaussianFactorGraph()
graph.push_back(downdated_belief)
graph.push_back(factors)
ordering = gtsam.Ordering()
for key in keys:
ordering.push_back(key)
_, remaining = graph.eliminatePartialSequential(ordering)
return remaining
def update_messages_and_node(j: int):
"""Update messages into cluster j and calculate belief q(X_j)."""
for edge in messages[j].keys():
if len(edge) == 2: # only update binary messages
j1, j2 = edge
k = j2 if j1 == j else j1
other_cluster = variable_nodes[k]
downdated_belief = gtsam.GaussianFactorGraph()
downdated_belief.push_back(other_cluster.belief)
message_to_subtract = messages[k][edge]
for i in range(message_to_subtract.size()):
downdated_belief.push_back(message_to_subtract.at(i).negate())
messages[j][edge] = calculate_new_message(factor_nodes[edge],
other_cluster.keys,
downdated_belief)
update_node(j)
update_messages_and_node(0)
plot_covariances(variable_nodes)
for j in [1,2]:
update_messages_and_node(j)
plot_covariances(variable_nodes)
for round in range(5):
for j in range(3):
update_messages_and_node(j)
marginals_figure(truth, marginals, graph_keys)
plot_covariances(variable_nodes)
rng = np.random.default_rng(42)
new_belief0 = gtsam.GaussianBayesNet()
minmax = -3.0, 3.0
new_belief0.push_back(GaussianPrior(x1, rng.uniform(*minmax, size=(3,)), 0.1))
new_belief0.push_back(GaussianPrior(x2, rng.uniform(*minmax, size=(3,)), 0.1))
new_belief0.push_back(GaussianPrior(x3, rng.uniform(*minmax, size=(3,)), 0.1))
new_belief1 = gtsam.GaussianBayesNet()
new_belief1.push_back(GaussianPrior(l1, rng.uniform(*minmax, size=(2,)), 0.1))
new_belief2 = gtsam.GaussianBayesNet()
new_belief2.push_back(GaussianPrior(l2, rng.uniform(*minmax, size=(2,)), 0.1))
key_sets = [{x1, x2, x3}, {l1}, {l2}]
new_beliefs = [new_belief0, new_belief1, new_belief2]
new_node_specs = list(zip(key_sets, new_beliefs))
variable_nodes = [Cluster(keys, initial_belief)
for keys, initial_belief in new_node_specs]
messages = defaultdict(dict)
for i, factors in factor_nodes.items():
if len(i) == 1:
j, *others = i
messages[j][i] = factors
continue
for j in i:
messages[j][i] = Message()
# marginals_figure(truth, marginals, graph_keys)
plot_covariances(variable_nodes)
for round in range(5):
for j in range(3):
update_messages_and_node(j)
plot_covariances(variable_nodes)
| 0.766992 | 0.981648 |
# Notation
___
## Reference for tricky symbols
[Tricky symbols](http://web.ift.uib.no/Teori/KURS/WRK/TeX/symALL.html)
[Table Generator](http://www.tablesgenerator.com/)
[Fonts in Latex](https://www.sharelatex.com/learn/Mathematical_fonts)
[Integrals, sums and limits](https://www.sharelatex.com/learn/Integrals,_sums_and_limits)
$\mathbb{R}$ = `$\mathbb{R}$`
## Model structure
### Some general rules
We use square brackets for layers, and round brackets for training examples.
Training examples are spaced out horizontally as column vectors - i.e. the second value in the matrix shape $(n_x, m)$ generally corresponds to the number of training examples.
### The general matrices
We have 6 key matrices that we'll play with, which we can divide into two groups.
The first four are representations of our features:
* $X$, representing the input to our model - the training data. This has the shape $(n_x, m)$ where $n_x$ is the number of features, and $m$ is the number of training samples.
* $Y$, representing the output to our model - the model outputs. This may have a number of shapes, depending on on the model structure we're chasing, but we can generally describe it as $(n_{x\ final}, m)$ where $n_{x\ final}$ is the number of neurons in our final (output) layer and $m$ is the number of training samples.
* $Z$, representing the inputs to a layer that has undergone a linear transformation, shape $(n_x, m)$, where $n_x$ is the number of neurons on a given layer.
* $A$, representing the activated linearised inputs, shape $(n_x, m)$, where $n_x$ is the number of neurons on a given layer and $m$ is the number of samples. This is essentially $Z$ but transformed.
The second group of two are the parameters of our model:
* $W$, representing the weights of the model. This is of shape $(n_x, n_{x-1})$, where $n_x$ and $n_{x-1}$ are the number of neurons in the current and previous layers, respectively.
* $B$, representing the bias for each layer of the model. This is of shape $(n_x, 1)$, where $n_x$ is the number of neurons on the relevant layer. The second dimension is 1 because this is a bias relevant to each neuron.
So given this:
$X$ = $\begin{bmatrix}
\vdots & \vdots & \vdots & & \vdots \\
x^{(1)} & x^{(2)} & x^{(3)} & \dots & x^{(m)} \\
\vdots & \vdots & \vdots & & \vdots \\
\end{bmatrix}$ = sum of feature vectors
We would have
$Z^{[1]} = W^{[1]}X^{[0]} + B^{[1]}$
and
$A^{[1]} = {\sigma}(Z^{[1]})$
### Some special cases
Some simple cases:
$A^{[0]} = X$, i.e. the 'activated output' of the zeroth layer is the input to the model.
$A^{[n]} = \hat{Y}$, i.e. the output of the last activation layer in the prediction.
### Backward propagation
$dz^{[l]} = d^{[l]}\times g^{[l]\prime}(z^{[l]})$, or
$dz^{[l]} = W^{[l+1]T}dz^{[l+1]}\times g^{[l]\prime}(z^{[l]})$
$dW^{[l]} = dz^{[l]}.a^{[l-1]}$
$db^{[l]} = dz^{[l]}$
$da^{[l-1]} = W^{[l]T}.dz^{[l]}$
## Binary classification
$(x,y)$ = training set, where $x\in\mathbb{R}^{n_x}, y\in{0,1}$
$(x^1, y^1)$ = single training example
$(x^{\{1\}}, y^{\{1\}})$ = single mini-batch extracted from a full training set
$x$ = feature vector
$X$ = $\begin{bmatrix}
\vdots & \vdots & \vdots & & \vdots \\
x^{1} & x^{2} & x^{3} & \dots & x^{m} \\
\vdots & \vdots & \vdots & & \vdots \\
\end{bmatrix}$ = sum of feature vectors
Note that we have $m$ samples as columns and $n_x$ features as rows. In Python, this comes out as `X.shape=(n_x, m)`.
$Y$ = $\begin{bmatrix} y^1 & y^2 & y^3 & \dots & y^m \end{bmatrix}$ = sum of labels.
In Python, this comes out as `Y.shape=(1,m)`.
$m$ = number of sample pairs
$m_{train}$ = number of sample pairs in train set
|
github_jupyter
|
# Notation
___
## Reference for tricky symbols
[Tricky symbols](http://web.ift.uib.no/Teori/KURS/WRK/TeX/symALL.html)
[Table Generator](http://www.tablesgenerator.com/)
[Fonts in Latex](https://www.sharelatex.com/learn/Mathematical_fonts)
[Integrals, sums and limits](https://www.sharelatex.com/learn/Integrals,_sums_and_limits)
$\mathbb{R}$ = `$\mathbb{R}$`
## Model structure
### Some general rules
We use square brackets for layers, and round brackets for training examples.
Training examples are spaced out horizontally as column vectors - i.e. the second value in the matrix shape $(n_x, m)$ generally corresponds to the number of training examples.
### The general matrices
We have 6 key matrices that we'll play with, which we can divide into two groups.
The first four are representations of our features:
* $X$, representing the input to our model - the training data. This has the shape $(n_x, m)$ where $n_x$ is the number of features, and $m$ is the number of training samples.
* $Y$, representing the output to our model - the model outputs. This may have a number of shapes, depending on on the model structure we're chasing, but we can generally describe it as $(n_{x\ final}, m)$ where $n_{x\ final}$ is the number of neurons in our final (output) layer and $m$ is the number of training samples.
* $Z$, representing the inputs to a layer that has undergone a linear transformation, shape $(n_x, m)$, where $n_x$ is the number of neurons on a given layer.
* $A$, representing the activated linearised inputs, shape $(n_x, m)$, where $n_x$ is the number of neurons on a given layer and $m$ is the number of samples. This is essentially $Z$ but transformed.
The second group of two are the parameters of our model:
* $W$, representing the weights of the model. This is of shape $(n_x, n_{x-1})$, where $n_x$ and $n_{x-1}$ are the number of neurons in the current and previous layers, respectively.
* $B$, representing the bias for each layer of the model. This is of shape $(n_x, 1)$, where $n_x$ is the number of neurons on the relevant layer. The second dimension is 1 because this is a bias relevant to each neuron.
So given this:
$X$ = $\begin{bmatrix}
\vdots & \vdots & \vdots & & \vdots \\
x^{(1)} & x^{(2)} & x^{(3)} & \dots & x^{(m)} \\
\vdots & \vdots & \vdots & & \vdots \\
\end{bmatrix}$ = sum of feature vectors
We would have
$Z^{[1]} = W^{[1]}X^{[0]} + B^{[1]}$
and
$A^{[1]} = {\sigma}(Z^{[1]})$
### Some special cases
Some simple cases:
$A^{[0]} = X$, i.e. the 'activated output' of the zeroth layer is the input to the model.
$A^{[n]} = \hat{Y}$, i.e. the output of the last activation layer in the prediction.
### Backward propagation
$dz^{[l]} = d^{[l]}\times g^{[l]\prime}(z^{[l]})$, or
$dz^{[l]} = W^{[l+1]T}dz^{[l+1]}\times g^{[l]\prime}(z^{[l]})$
$dW^{[l]} = dz^{[l]}.a^{[l-1]}$
$db^{[l]} = dz^{[l]}$
$da^{[l-1]} = W^{[l]T}.dz^{[l]}$
## Binary classification
$(x,y)$ = training set, where $x\in\mathbb{R}^{n_x}, y\in{0,1}$
$(x^1, y^1)$ = single training example
$(x^{\{1\}}, y^{\{1\}})$ = single mini-batch extracted from a full training set
$x$ = feature vector
$X$ = $\begin{bmatrix}
\vdots & \vdots & \vdots & & \vdots \\
x^{1} & x^{2} & x^{3} & \dots & x^{m} \\
\vdots & \vdots & \vdots & & \vdots \\
\end{bmatrix}$ = sum of feature vectors
Note that we have $m$ samples as columns and $n_x$ features as rows. In Python, this comes out as `X.shape=(n_x, m)`.
$Y$ = $\begin{bmatrix} y^1 & y^2 & y^3 & \dots & y^m \end{bmatrix}$ = sum of labels.
In Python, this comes out as `Y.shape=(1,m)`.
$m$ = number of sample pairs
$m_{train}$ = number of sample pairs in train set
| 0.875202 | 0.962391 |
This Notebook is accompanied by the following blog-post:
http://ataspinar.com/2018/04/04/machine-learning-with-signal-processing-techniques/
```
from siml.sk_utils import *
import numpy as np
import matplotlib.pyplot as plt
from scipy.signal import welch
from scipy.fftpack import fft
from scipy import signal
from collections import defaultdict, Counter
from detect_peaks import *
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import classification_report
```
# 0. Loading the signals from file
```
activities_description = {
1: 'walking',
2: 'walking upstairs',
3: 'walking downstairs',
4: 'sitting',
5: 'standing',
6: 'laying'
}
def read_signals(filename):
with open(filename, 'r') as fp:
data = fp.read().splitlines()
data = map(lambda x: x.rstrip().lstrip().split(), data)
data = [list(map(float, line)) for line in data]
return data
def read_labels(filename):
with open(filename, 'r') as fp:
activities = fp.read().splitlines()
activities = list(map(int, activities))
return activities
def randomize(dataset, labels):
permutation = np.random.permutation(labels.shape[0])
shuffled_dataset = dataset[permutation, :, :]
shuffled_labels = labels[permutation]
return shuffled_dataset, shuffled_labels
INPUT_FOLDER_TRAIN = '../datasets/UCI_HAR/train/InertialSignals/'
INPUT_FOLDER_TEST = '../datasets/UCI_HAR/test/InertialSignals/'
INPUT_FILES_TRAIN = ['body_acc_x_train.txt', 'body_acc_y_train.txt', 'body_acc_z_train.txt',
'body_gyro_x_train.txt', 'body_gyro_y_train.txt', 'body_gyro_z_train.txt',
'total_acc_x_train.txt', 'total_acc_y_train.txt', 'total_acc_z_train.txt']
INPUT_FILES_TEST = ['body_acc_x_test.txt', 'body_acc_y_test.txt', 'body_acc_z_test.txt',
'body_gyro_x_test.txt', 'body_gyro_y_test.txt', 'body_gyro_z_test.txt',
'total_acc_x_test.txt', 'total_acc_y_test.txt', 'total_acc_z_test.txt']
LABELFILE_TRAIN = '../datasets/UCI_HAR/train/y_train.txt'
LABELFILE_TEST = '../datasets/UCI_HAR/test/y_test.txt'
train_signals, test_signals = [], []
for input_file in INPUT_FILES_TRAIN:
signal = read_signals(INPUT_FOLDER_TRAIN + input_file)
train_signals.append(signal)
train_signals = np.transpose(np.array(train_signals), (1, 2, 0))
for input_file in INPUT_FILES_TEST:
signal = read_signals(INPUT_FOLDER_TEST + input_file)
test_signals.append(signal)
test_signals = np.transpose(np.array(test_signals), (1, 2, 0))
train_labels = read_labels(LABELFILE_TRAIN)
test_labels = read_labels(LABELFILE_TEST)
[no_signals_train, no_steps_train, no_components_train] = np.shape(train_signals)
[no_signals_test, no_steps_test, no_components_test] = np.shape(train_signals)
no_labels = len(np.unique(train_labels[:]))
print("The train dataset contains {} signals, each one of length {} and {} components ".format(no_signals_train, no_steps_train, no_components_train))
print("The test dataset contains {} signals, each one of length {} and {} components ".format(no_signals_test, no_steps_test, no_components_test))
print("The train dataset contains {} labels, with the following distribution:\n {}".format(np.shape(train_labels)[0], Counter(train_labels[:])))
print("The test dataset contains {} labels, with the following distribution:\n {}".format(np.shape(test_labels)[0], Counter(test_labels[:])))
train_signals, train_labels = randomize(train_signals, np.array(train_labels))
test_signals, test_labels = randomize(test_signals, np.array(test_labels))
```
# 1. Visualizations
```
N = 128
f_s = 50
t_n = 2.56
T = t_n / N
sample_rate = T
denominator = 10
signal_no = 15
signals = train_signals[signal_no, :, :]
signal = signals[:, 3]
label = train_labels[signal_no]
activity_name = activities_description[label]
def get_values(y_values, T, N, f_s):
y_values = y_values
x_values = [sample_rate * kk for kk in range(0,len(y_values))]
return x_values, y_values
def get_fft_values(y_values, T, N, f_s):
f_values = np.linspace(0.0, 1.0/(2.0*T), N//2)
fft_values_ = fft(y_values)
fft_values = 2.0/N * np.abs(fft_values_[0:N//2])
return f_values, fft_values
def get_psd_values(y_values, T, N, f_s):
f_values, psd_values = welch(y_values, fs=f_s)
return f_values, psd_values
def autocorr(x):
result = np.correlate(x, x, mode='full')
return result[len(result)//2:]
def get_autocorr_values(y_values, T, N, f_s):
autocorr_values = autocorr(y_values)
x_values = np.array([T * jj for jj in range(0, N)])
return x_values, autocorr_values
```
## 1a. Visualization of the FFT
```
f_values, fft_values = get_fft_values(signal, T, N, f_s)
plt.plot(f_values, fft_values, linestyle='-', color='blue')
plt.xlabel('Frequency [Hz]', fontsize=16)
plt.ylabel('Amplitude', fontsize=16)
plt.title("Frequency domain of the signal", fontsize=16)
plt.show()
```
## 1b. Visualization of the PSD
```
f_values, psd_values = get_psd_values(signal, T, N, f_s)
plt.plot(f_values, psd_values, linestyle='-', color='blue')
plt.xlabel('Frequency [Hz]')
plt.ylabel('PSD [V**2 / Hz]')
plt.show()
```
## 1c. Visualization of the Autocorrelation
```
t_values, autocorr_values = get_autocorr_values(signal, T, N, f_s)
plt.plot(t_values, autocorr_values, linestyle='-', color='blue')
plt.xlabel('time delay [s]')
plt.ylabel('Autocorrelation amplitude')
plt.show()
```
## 1d. Visualization of all transformations on all components
```
labels = ['x-component', 'y-component', 'z-component']
colors = ['r', 'g', 'b']
suptitle = "Different signals for the activity: {}"
xlabels = ['Time [sec]', 'Freq [Hz]', 'Freq [Hz]', 'Time lag [s]']
ylabel = 'Amplitude'
axtitles = [['Acceleration', 'Gyro', 'Total acceleration'],
['FFT acc', 'FFT gyro', 'FFT total acc'],
['PSD acc', 'PSD gyro', 'PSD total acc'],
['Autocorr acc', 'Autocorr gyro', 'Autocorr total acc']
]
list_functions = [get_values, get_fft_values, get_psd_values, get_autocorr_values]
f, axarr = plt.subplots(nrows=4, ncols=3, figsize=(12,12))
f.suptitle(suptitle.format(activity_name), fontsize=16)
for row_no in range(0,4):
for comp_no in range(0,9):
col_no = comp_no // 3
plot_no = comp_no % 3
color = colors[plot_no]
label = labels[plot_no]
axtitle = axtitles[row_no][col_no]
xlabel = xlabels[row_no]
value_retriever = list_functions[row_no]
ax = axarr[row_no, col_no]
ax.set_title(axtitle, fontsize=16)
ax.set_xlabel(xlabel, fontsize=16)
if col_no == 0:
ax.set_ylabel(ylabel, fontsize=16)
signal_component = signals[:, comp_no]
x_values, y_values = value_retriever(signal_component, T, N, f_s)
ax.plot(x_values, y_values, linestyle='-', color=color, label=label)
if row_no > 0:
max_peak_height = 0.1 * np.nanmax(y_values)
indices_peaks = detect_peaks(y_values, mph=max_peak_height)
ax.scatter(x_values[indices_peaks], y_values[indices_peaks], c=color, marker='*', s=60)
if col_no == 2:
ax.legend(loc='center left', bbox_to_anchor=(1, 0.5))
plt.tight_layout()
plt.subplots_adjust(top=0.90, hspace=0.6)
plt.show()
```
# 2. Extract Features from the signals
```
def get_first_n_peaks(x,y,no_peaks=5):
x_, y_ = list(x), list(y)
if len(x_) >= no_peaks:
return x_[:no_peaks], y_[:no_peaks]
else:
missing_no_peaks = no_peaks-len(x_)
return x_ + [0]*missing_no_peaks, y_ + [0]*missing_no_peaks
def get_features(x_values, y_values, mph):
indices_peaks = detect_peaks(y_values, mph=mph)
peaks_x, peaks_y = get_first_n_peaks(x_values[indices_peaks], y_values[indices_peaks])
return peaks_x + peaks_y
def extract_features_labels(dataset, labels, T, N, f_s, denominator):
percentile = 5
list_of_features = []
list_of_labels = []
for signal_no in range(0, len(dataset)):
features = []
list_of_labels.append(labels[signal_no])
for signal_comp in range(0,dataset.shape[2]):
signal = dataset[signal_no, :, signal_comp]
signal_min = np.nanpercentile(signal, percentile)
signal_max = np.nanpercentile(signal, 100-percentile)
#ijk = (100 - 2*percentile)/10
mph = signal_min + (signal_max - signal_min)/denominator
features += get_features(*get_psd_values(signal, T, N, f_s), mph)
features += get_features(*get_fft_values(signal, T, N, f_s), mph)
features += get_features(*get_autocorr_values(signal, T, N, f_s), mph)
list_of_features.append(features)
return np.array(list_of_features), np.array(list_of_labels)
X_train, Y_train = extract_features_labels(train_signals, train_labels, T, N, f_s, denominator)
X_test, Y_test = extract_features_labels(test_signals, test_labels, T, N, f_s, denominator)
```
# 3. Classification of the signals
## 3.1 Try classification with Random Forest
```
clf = RandomForestClassifier(n_estimators=1000)
clf.fit(X_train, Y_train)
print("Accuracy on training set is : {}".format(clf.score(X_train, Y_train)))
print("Accuracy on test set is : {}".format(clf.score(X_test, Y_test)))
Y_test_pred = clf.predict(X_test)
print(classification_report(Y_test, Y_test_pred))
```
## 3.2 Try out several classifiers (to see which one initially scores best)
```
#See https://github.com/taspinar/siml
dict_results = batch_classify(X_train, Y_train, X_test, Y_test)
display_dict_models(dict_results)
```
## 3.3 Hyperparameter optimization of best classifier
```
GDB_params = {
'n_estimators': [100, 500, 1000],
'learning_rate': [0.5, 0.1, 0.01, 0.001],
'criterion': ['friedman_mse', 'mse', 'mae']
}
for n_est in GDB_params['n_estimators']:
for lr in GDB_params['learning_rate']:
for crit in GDB_params['criterion']:
clf = GradientBoostingClassifier(n_estimators=n_est,
learning_rate = lr,
criterion = crit)
clf.fit(X_train, Y_train)
train_score = clf.score(X_train, Y_train)
test_score = clf.score(X_test, Y_test)
print("For ({}, {}, {}) - train, test score: \t {:.5f} \t-\t {:.5f}".format(
n_est, lr, crit[:4], train_score, test_score)
)
```
|
github_jupyter
|
from siml.sk_utils import *
import numpy as np
import matplotlib.pyplot as plt
from scipy.signal import welch
from scipy.fftpack import fft
from scipy import signal
from collections import defaultdict, Counter
from detect_peaks import *
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import classification_report
activities_description = {
1: 'walking',
2: 'walking upstairs',
3: 'walking downstairs',
4: 'sitting',
5: 'standing',
6: 'laying'
}
def read_signals(filename):
with open(filename, 'r') as fp:
data = fp.read().splitlines()
data = map(lambda x: x.rstrip().lstrip().split(), data)
data = [list(map(float, line)) for line in data]
return data
def read_labels(filename):
with open(filename, 'r') as fp:
activities = fp.read().splitlines()
activities = list(map(int, activities))
return activities
def randomize(dataset, labels):
permutation = np.random.permutation(labels.shape[0])
shuffled_dataset = dataset[permutation, :, :]
shuffled_labels = labels[permutation]
return shuffled_dataset, shuffled_labels
INPUT_FOLDER_TRAIN = '../datasets/UCI_HAR/train/InertialSignals/'
INPUT_FOLDER_TEST = '../datasets/UCI_HAR/test/InertialSignals/'
INPUT_FILES_TRAIN = ['body_acc_x_train.txt', 'body_acc_y_train.txt', 'body_acc_z_train.txt',
'body_gyro_x_train.txt', 'body_gyro_y_train.txt', 'body_gyro_z_train.txt',
'total_acc_x_train.txt', 'total_acc_y_train.txt', 'total_acc_z_train.txt']
INPUT_FILES_TEST = ['body_acc_x_test.txt', 'body_acc_y_test.txt', 'body_acc_z_test.txt',
'body_gyro_x_test.txt', 'body_gyro_y_test.txt', 'body_gyro_z_test.txt',
'total_acc_x_test.txt', 'total_acc_y_test.txt', 'total_acc_z_test.txt']
LABELFILE_TRAIN = '../datasets/UCI_HAR/train/y_train.txt'
LABELFILE_TEST = '../datasets/UCI_HAR/test/y_test.txt'
train_signals, test_signals = [], []
for input_file in INPUT_FILES_TRAIN:
signal = read_signals(INPUT_FOLDER_TRAIN + input_file)
train_signals.append(signal)
train_signals = np.transpose(np.array(train_signals), (1, 2, 0))
for input_file in INPUT_FILES_TEST:
signal = read_signals(INPUT_FOLDER_TEST + input_file)
test_signals.append(signal)
test_signals = np.transpose(np.array(test_signals), (1, 2, 0))
train_labels = read_labels(LABELFILE_TRAIN)
test_labels = read_labels(LABELFILE_TEST)
[no_signals_train, no_steps_train, no_components_train] = np.shape(train_signals)
[no_signals_test, no_steps_test, no_components_test] = np.shape(train_signals)
no_labels = len(np.unique(train_labels[:]))
print("The train dataset contains {} signals, each one of length {} and {} components ".format(no_signals_train, no_steps_train, no_components_train))
print("The test dataset contains {} signals, each one of length {} and {} components ".format(no_signals_test, no_steps_test, no_components_test))
print("The train dataset contains {} labels, with the following distribution:\n {}".format(np.shape(train_labels)[0], Counter(train_labels[:])))
print("The test dataset contains {} labels, with the following distribution:\n {}".format(np.shape(test_labels)[0], Counter(test_labels[:])))
train_signals, train_labels = randomize(train_signals, np.array(train_labels))
test_signals, test_labels = randomize(test_signals, np.array(test_labels))
N = 128
f_s = 50
t_n = 2.56
T = t_n / N
sample_rate = T
denominator = 10
signal_no = 15
signals = train_signals[signal_no, :, :]
signal = signals[:, 3]
label = train_labels[signal_no]
activity_name = activities_description[label]
def get_values(y_values, T, N, f_s):
y_values = y_values
x_values = [sample_rate * kk for kk in range(0,len(y_values))]
return x_values, y_values
def get_fft_values(y_values, T, N, f_s):
f_values = np.linspace(0.0, 1.0/(2.0*T), N//2)
fft_values_ = fft(y_values)
fft_values = 2.0/N * np.abs(fft_values_[0:N//2])
return f_values, fft_values
def get_psd_values(y_values, T, N, f_s):
f_values, psd_values = welch(y_values, fs=f_s)
return f_values, psd_values
def autocorr(x):
result = np.correlate(x, x, mode='full')
return result[len(result)//2:]
def get_autocorr_values(y_values, T, N, f_s):
autocorr_values = autocorr(y_values)
x_values = np.array([T * jj for jj in range(0, N)])
return x_values, autocorr_values
f_values, fft_values = get_fft_values(signal, T, N, f_s)
plt.plot(f_values, fft_values, linestyle='-', color='blue')
plt.xlabel('Frequency [Hz]', fontsize=16)
plt.ylabel('Amplitude', fontsize=16)
plt.title("Frequency domain of the signal", fontsize=16)
plt.show()
f_values, psd_values = get_psd_values(signal, T, N, f_s)
plt.plot(f_values, psd_values, linestyle='-', color='blue')
plt.xlabel('Frequency [Hz]')
plt.ylabel('PSD [V**2 / Hz]')
plt.show()
t_values, autocorr_values = get_autocorr_values(signal, T, N, f_s)
plt.plot(t_values, autocorr_values, linestyle='-', color='blue')
plt.xlabel('time delay [s]')
plt.ylabel('Autocorrelation amplitude')
plt.show()
labels = ['x-component', 'y-component', 'z-component']
colors = ['r', 'g', 'b']
suptitle = "Different signals for the activity: {}"
xlabels = ['Time [sec]', 'Freq [Hz]', 'Freq [Hz]', 'Time lag [s]']
ylabel = 'Amplitude'
axtitles = [['Acceleration', 'Gyro', 'Total acceleration'],
['FFT acc', 'FFT gyro', 'FFT total acc'],
['PSD acc', 'PSD gyro', 'PSD total acc'],
['Autocorr acc', 'Autocorr gyro', 'Autocorr total acc']
]
list_functions = [get_values, get_fft_values, get_psd_values, get_autocorr_values]
f, axarr = plt.subplots(nrows=4, ncols=3, figsize=(12,12))
f.suptitle(suptitle.format(activity_name), fontsize=16)
for row_no in range(0,4):
for comp_no in range(0,9):
col_no = comp_no // 3
plot_no = comp_no % 3
color = colors[plot_no]
label = labels[plot_no]
axtitle = axtitles[row_no][col_no]
xlabel = xlabels[row_no]
value_retriever = list_functions[row_no]
ax = axarr[row_no, col_no]
ax.set_title(axtitle, fontsize=16)
ax.set_xlabel(xlabel, fontsize=16)
if col_no == 0:
ax.set_ylabel(ylabel, fontsize=16)
signal_component = signals[:, comp_no]
x_values, y_values = value_retriever(signal_component, T, N, f_s)
ax.plot(x_values, y_values, linestyle='-', color=color, label=label)
if row_no > 0:
max_peak_height = 0.1 * np.nanmax(y_values)
indices_peaks = detect_peaks(y_values, mph=max_peak_height)
ax.scatter(x_values[indices_peaks], y_values[indices_peaks], c=color, marker='*', s=60)
if col_no == 2:
ax.legend(loc='center left', bbox_to_anchor=(1, 0.5))
plt.tight_layout()
plt.subplots_adjust(top=0.90, hspace=0.6)
plt.show()
def get_first_n_peaks(x,y,no_peaks=5):
x_, y_ = list(x), list(y)
if len(x_) >= no_peaks:
return x_[:no_peaks], y_[:no_peaks]
else:
missing_no_peaks = no_peaks-len(x_)
return x_ + [0]*missing_no_peaks, y_ + [0]*missing_no_peaks
def get_features(x_values, y_values, mph):
indices_peaks = detect_peaks(y_values, mph=mph)
peaks_x, peaks_y = get_first_n_peaks(x_values[indices_peaks], y_values[indices_peaks])
return peaks_x + peaks_y
def extract_features_labels(dataset, labels, T, N, f_s, denominator):
percentile = 5
list_of_features = []
list_of_labels = []
for signal_no in range(0, len(dataset)):
features = []
list_of_labels.append(labels[signal_no])
for signal_comp in range(0,dataset.shape[2]):
signal = dataset[signal_no, :, signal_comp]
signal_min = np.nanpercentile(signal, percentile)
signal_max = np.nanpercentile(signal, 100-percentile)
#ijk = (100 - 2*percentile)/10
mph = signal_min + (signal_max - signal_min)/denominator
features += get_features(*get_psd_values(signal, T, N, f_s), mph)
features += get_features(*get_fft_values(signal, T, N, f_s), mph)
features += get_features(*get_autocorr_values(signal, T, N, f_s), mph)
list_of_features.append(features)
return np.array(list_of_features), np.array(list_of_labels)
X_train, Y_train = extract_features_labels(train_signals, train_labels, T, N, f_s, denominator)
X_test, Y_test = extract_features_labels(test_signals, test_labels, T, N, f_s, denominator)
clf = RandomForestClassifier(n_estimators=1000)
clf.fit(X_train, Y_train)
print("Accuracy on training set is : {}".format(clf.score(X_train, Y_train)))
print("Accuracy on test set is : {}".format(clf.score(X_test, Y_test)))
Y_test_pred = clf.predict(X_test)
print(classification_report(Y_test, Y_test_pred))
#See https://github.com/taspinar/siml
dict_results = batch_classify(X_train, Y_train, X_test, Y_test)
display_dict_models(dict_results)
GDB_params = {
'n_estimators': [100, 500, 1000],
'learning_rate': [0.5, 0.1, 0.01, 0.001],
'criterion': ['friedman_mse', 'mse', 'mae']
}
for n_est in GDB_params['n_estimators']:
for lr in GDB_params['learning_rate']:
for crit in GDB_params['criterion']:
clf = GradientBoostingClassifier(n_estimators=n_est,
learning_rate = lr,
criterion = crit)
clf.fit(X_train, Y_train)
train_score = clf.score(X_train, Y_train)
test_score = clf.score(X_test, Y_test)
print("For ({}, {}, {}) - train, test score: \t {:.5f} \t-\t {:.5f}".format(
n_est, lr, crit[:4], train_score, test_score)
)
| 0.504394 | 0.958226 |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.