code
stringlengths 2.5k
6.36M
| kind
stringclasses 2
values | parsed_code
stringlengths 0
404k
| quality_prob
float64 0
0.98
| learning_prob
float64 0.03
1
|
|---|---|---|---|---|
```
from __future__ import absolute_import, division, print_function
import codecs
import glob
import multiprocessing
import os
import pprint
import re
import nltk
import gensim.models.word2vec as w2v
import sklearn.manifold
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
import seaborn as sns
%pylab inline
nltk.download("punkt")
```
## Getting and cleaning data
```
hindi_filenames = sorted(glob.glob("../data/hin_corp_unicode/*txt"))
#hindi_filenames
corpus_raw = u""
for file_name in hindi_filenames:
print("Reading '{0}'...".format(file_name))
with codecs.open(file_name, "r", "utf-8") as f:
# Starting two lines are not useful in corpus
temp = f.readline()
temp = f.readline()
corpus_raw += f.read()
print("Corpus is now {0} characters long".format(len(corpus_raw)))
print()
tokenizer = nltk.data.load('tokenizers/punkt/english.pickle')
raw_sentences = tokenizer.tokenize(corpus_raw)
def sentence_to_wordlist(raw):
clean = re.sub("[.\r\n]"," ", raw)
words = clean.split()
return words
sentences = []
for raw_sentence in raw_sentences:
if len(raw_sentence) > 0:
sentences.append(sentence_to_wordlist(raw_sentence))
token_count = sum([len(sentence) for sentence in sentences])
print("The Hindi corpus contains {0:,} tokens".format(token_count))
sentences[0]
```
## Word Vectors
```
# Dimensionality of the resulting word vectors.
# More dimensions = more generalized
num_features = 50
# Minimum word count threshold.
min_word_count = 3
# Number of threads to run in parallel.
num_threads = multiprocessing.cpu_count()
# Context window length.
context_size = 8
# Downsample setting for frequent words.
#0 - 1e-5 is good for this
downsampling = 1e-3
# Seed for the RNG, to make the results reproducible.
# Random Number Generator
seed = 1
# Defining the model
model = w2v.Word2Vec(
sg=1,
seed=seed,
workers=num_threads,
size=num_features,
min_count=min_word_count,
window=context_size,
sample=downsampling
)
model.build_vocab(sentences)
model.train(sentences)
# Save our model
model.save(os.path.join("../data/", "hindi_word2Vec_small.w2v"))
```
## Explore the model
```
trained_model = w2v.Word2Vec.load(os.path.join("../data/", "hindi_word2Vec_small.w2v"))
# For reducing dimensiomns, to visualize vectors
tsne = sklearn.manifold.TSNE(n_components=2, random_state=0)
all_word_vectors_matrix = trained_model.syn1neg[:200] # Currently giving memory error for all words
# Reduced dimensions
all_word_vectors_matrix_2d = tsne.fit_transform(all_word_vectors_matrix)
points = pd.DataFrame(
[
(word, coords[0], coords[1])
for word, coords in [
(word, all_word_vectors_matrix_2d[trained_model.wv.vocab[word].index])
for word in trained_model.wv.vocab
if trained_model.wv.vocab[word].index < 200
]
],
columns=["word", "x", "y"]
)
s = trained_model.wv[u"आधार"]
```
|
github_jupyter
|
from __future__ import absolute_import, division, print_function
import codecs
import glob
import multiprocessing
import os
import pprint
import re
import nltk
import gensim.models.word2vec as w2v
import sklearn.manifold
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
import seaborn as sns
%pylab inline
nltk.download("punkt")
hindi_filenames = sorted(glob.glob("../data/hin_corp_unicode/*txt"))
#hindi_filenames
corpus_raw = u""
for file_name in hindi_filenames:
print("Reading '{0}'...".format(file_name))
with codecs.open(file_name, "r", "utf-8") as f:
# Starting two lines are not useful in corpus
temp = f.readline()
temp = f.readline()
corpus_raw += f.read()
print("Corpus is now {0} characters long".format(len(corpus_raw)))
print()
tokenizer = nltk.data.load('tokenizers/punkt/english.pickle')
raw_sentences = tokenizer.tokenize(corpus_raw)
def sentence_to_wordlist(raw):
clean = re.sub("[.\r\n]"," ", raw)
words = clean.split()
return words
sentences = []
for raw_sentence in raw_sentences:
if len(raw_sentence) > 0:
sentences.append(sentence_to_wordlist(raw_sentence))
token_count = sum([len(sentence) for sentence in sentences])
print("The Hindi corpus contains {0:,} tokens".format(token_count))
sentences[0]
# Dimensionality of the resulting word vectors.
# More dimensions = more generalized
num_features = 50
# Minimum word count threshold.
min_word_count = 3
# Number of threads to run in parallel.
num_threads = multiprocessing.cpu_count()
# Context window length.
context_size = 8
# Downsample setting for frequent words.
#0 - 1e-5 is good for this
downsampling = 1e-3
# Seed for the RNG, to make the results reproducible.
# Random Number Generator
seed = 1
# Defining the model
model = w2v.Word2Vec(
sg=1,
seed=seed,
workers=num_threads,
size=num_features,
min_count=min_word_count,
window=context_size,
sample=downsampling
)
model.build_vocab(sentences)
model.train(sentences)
# Save our model
model.save(os.path.join("../data/", "hindi_word2Vec_small.w2v"))
trained_model = w2v.Word2Vec.load(os.path.join("../data/", "hindi_word2Vec_small.w2v"))
# For reducing dimensiomns, to visualize vectors
tsne = sklearn.manifold.TSNE(n_components=2, random_state=0)
all_word_vectors_matrix = trained_model.syn1neg[:200] # Currently giving memory error for all words
# Reduced dimensions
all_word_vectors_matrix_2d = tsne.fit_transform(all_word_vectors_matrix)
points = pd.DataFrame(
[
(word, coords[0], coords[1])
for word, coords in [
(word, all_word_vectors_matrix_2d[trained_model.wv.vocab[word].index])
for word in trained_model.wv.vocab
if trained_model.wv.vocab[word].index < 200
]
],
columns=["word", "x", "y"]
)
s = trained_model.wv[u"आधार"]
| 0.373304 | 0.376394 |
<table class="ee-notebook-buttons" align="left">
<td><a target="_blank" href="https://github.com/giswqs/earthengine-py-notebooks/tree/master/AssetManagement/export_ImageCollection.ipynb"><img width=32px src="https://www.tensorflow.org/images/GitHub-Mark-32px.png" /> View source on GitHub</a></td>
<td><a target="_blank" href="https://nbviewer.jupyter.org/github/giswqs/earthengine-py-notebooks/blob/master/AssetManagement/export_ImageCollection.ipynb"><img width=26px src="https://upload.wikimedia.org/wikipedia/commons/thumb/3/38/Jupyter_logo.svg/883px-Jupyter_logo.svg.png" />Notebook Viewer</a></td>
<td><a target="_blank" href="https://mybinder.org/v2/gh/giswqs/earthengine-py-notebooks/master?filepath=AssetManagement/export_ImageCollection.ipynb"><img width=58px src="https://mybinder.org/static/images/logo_social.png" />Run in binder</a></td>
<td><a target="_blank" href="https://colab.research.google.com/github/giswqs/earthengine-py-notebooks/blob/master/AssetManagement/export_ImageCollection.ipynb"><img src="https://www.tensorflow.org/images/colab_logo_32px.png" /> Run in Google Colab</a></td>
</table>
## Install Earth Engine API and geemap
Install the [Earth Engine Python API](https://developers.google.com/earth-engine/python_install) and [geemap](https://github.com/giswqs/geemap). The **geemap** Python package is built upon the [ipyleaflet](https://github.com/jupyter-widgets/ipyleaflet) and [folium](https://github.com/python-visualization/folium) packages and implements several methods for interacting with Earth Engine data layers, such as `Map.addLayer()`, `Map.setCenter()`, and `Map.centerObject()`.
The following script checks if the geemap package has been installed. If not, it will install geemap, which automatically installs its [dependencies](https://github.com/giswqs/geemap#dependencies), including earthengine-api, folium, and ipyleaflet.
**Important note**: A key difference between folium and ipyleaflet is that ipyleaflet is built upon ipywidgets and allows bidirectional communication between the front-end and the backend enabling the use of the map to capture user input, while folium is meant for displaying static data only ([source](https://blog.jupyter.org/interactive-gis-in-jupyter-with-ipyleaflet-52f9657fa7a)). Note that [Google Colab](https://colab.research.google.com/) currently does not support ipyleaflet ([source](https://github.com/googlecolab/colabtools/issues/60#issuecomment-596225619)). Therefore, if you are using geemap with Google Colab, you should use [`import geemap.eefolium`](https://github.com/giswqs/geemap/blob/master/geemap/eefolium.py). If you are using geemap with [binder](https://mybinder.org/) or a local Jupyter notebook server, you can use [`import geemap`](https://github.com/giswqs/geemap/blob/master/geemap/geemap.py), which provides more functionalities for capturing user input (e.g., mouse-clicking and moving).
```
# Installs geemap package
import subprocess
try:
import geemap
except ImportError:
print('geemap package not installed. Installing ...')
subprocess.check_call(["python", '-m', 'pip', 'install', 'geemap'])
# Checks whether this notebook is running on Google Colab
try:
import google.colab
import geemap.eefolium as emap
except:
import geemap as emap
# Authenticates and initializes Earth Engine
import ee
try:
ee.Initialize()
except Exception as e:
ee.Authenticate()
ee.Initialize()
```
## Create an interactive map
The default basemap is `Google Satellite`. [Additional basemaps](https://github.com/giswqs/geemap/blob/master/geemap/geemap.py#L13) can be added using the `Map.add_basemap()` function.
```
Map = emap.Map(center=[40,-100], zoom=4)
Map.add_basemap('ROADMAP') # Add Google Map
Map
```
## Add Earth Engine Python script
```
# Add Earth Engine dataset
# USDA NAIP ImageCollection
collection = ee.ImageCollection('USDA/NAIP/DOQQ')
# create an roi
polys = ee.Geometry.Polygon(
[[[-99.29615020751953, 46.725459351792374],
[-99.2116928100586, 46.72404725733022],
[-99.21443939208984, 46.772037733479884],
[-99.30267333984375, 46.77321343419932]]])
# create a FeatureCollection based on the roi and center the map
centroid = polys.centroid()
lng, lat = centroid.getInfo()['coordinates']
print("lng = {}, lat = {}".format(lng, lat))
Map.setCenter(lng, lat, 12)
fc = ee.FeatureCollection(polys)
# filter the ImageCollection using the roi
naip = collection.filterBounds(polys)
naip_2015 = naip.filterDate('2015-01-01', '2015-12-31')
mosaic = naip_2015.mosaic()
# print out the number of images in the ImageCollection
count = naip_2015.size().getInfo()
print("Count: ", count)
# add the ImageCollection and the roi to the map
vis = {'bands': ['N', 'R', 'G']}
Map.addLayer(mosaic,vis)
Map.addLayer(fc)
# export the ImageCollection to Google Drive
downConfig = {'scale': 30, "maxPixels": 1.0E13, 'driveFolder': 'image'} # scale means resolution.
img_lst = naip_2015.toList(100)
for i in range(0, count):
image = ee.Image(img_lst.get(i))
name = image.get('system:index').getInfo()
# print(name)
task = ee.batch.Export.image(image, name, downConfig)
task.start()
```
## Display Earth Engine data layers
```
Map.addLayerControl() # This line is not needed for ipyleaflet-based Map.
Map
```
|
github_jupyter
|
# Installs geemap package
import subprocess
try:
import geemap
except ImportError:
print('geemap package not installed. Installing ...')
subprocess.check_call(["python", '-m', 'pip', 'install', 'geemap'])
# Checks whether this notebook is running on Google Colab
try:
import google.colab
import geemap.eefolium as emap
except:
import geemap as emap
# Authenticates and initializes Earth Engine
import ee
try:
ee.Initialize()
except Exception as e:
ee.Authenticate()
ee.Initialize()
Map = emap.Map(center=[40,-100], zoom=4)
Map.add_basemap('ROADMAP') # Add Google Map
Map
# Add Earth Engine dataset
# USDA NAIP ImageCollection
collection = ee.ImageCollection('USDA/NAIP/DOQQ')
# create an roi
polys = ee.Geometry.Polygon(
[[[-99.29615020751953, 46.725459351792374],
[-99.2116928100586, 46.72404725733022],
[-99.21443939208984, 46.772037733479884],
[-99.30267333984375, 46.77321343419932]]])
# create a FeatureCollection based on the roi and center the map
centroid = polys.centroid()
lng, lat = centroid.getInfo()['coordinates']
print("lng = {}, lat = {}".format(lng, lat))
Map.setCenter(lng, lat, 12)
fc = ee.FeatureCollection(polys)
# filter the ImageCollection using the roi
naip = collection.filterBounds(polys)
naip_2015 = naip.filterDate('2015-01-01', '2015-12-31')
mosaic = naip_2015.mosaic()
# print out the number of images in the ImageCollection
count = naip_2015.size().getInfo()
print("Count: ", count)
# add the ImageCollection and the roi to the map
vis = {'bands': ['N', 'R', 'G']}
Map.addLayer(mosaic,vis)
Map.addLayer(fc)
# export the ImageCollection to Google Drive
downConfig = {'scale': 30, "maxPixels": 1.0E13, 'driveFolder': 'image'} # scale means resolution.
img_lst = naip_2015.toList(100)
for i in range(0, count):
image = ee.Image(img_lst.get(i))
name = image.get('system:index').getInfo()
# print(name)
task = ee.batch.Export.image(image, name, downConfig)
task.start()
Map.addLayerControl() # This line is not needed for ipyleaflet-based Map.
Map
| 0.448909 | 0.947817 |
```
import pandas as pd
import numpy as np
df = pd.read_csv("C:\\ITM SPRING 2020\\ML\\sgemm_product_dataset\\processed_sgemm_product.csv")
y=df['MeanRun']
X=df.drop(columns ='MeanRun',axis=1)
from sklearn.model_selection import train_test_split
X_train,X_val, y_train,y_val = train_test_split(X,y,test_size=0.33,random_state=5)
X_train = X_train.T
y_train = np.array([y_train])
X_val=X_val.T
y_val=np.array([y_val])
print(y_val.shape)
print(X_val.shape)
print(y_train.shape)
print(X_train.shape)
```
# EXPERIMENT 1: Plotting training and validation cost against learning rates:
```
from LinearRegression_Threshold15 import *
Learning_rates= [.0001,.001,.01,.025,.05]
iterat =[]
for i in Learning_rates:
cost_t,cost_v,w,iteration = linear_regression_model(X_train,y_train,X_val,y_val,i,1000,.0000000000001)
iterat.append(len(cost_t))
plt.plot(cost_t,label='tc_lr = '+str(i))
plt.plot(cost_v,label='vc_lr='+str(i))
plt.xlabel('Iterations')
plt.ylabel('Cost')
plt.title('Training and Validation cost for varying learning rates')
plt.legend()
plt.show
iterat
cost_t,cost_v,w,iteration = linear_regression_model(X_train,y_train,X_val,y_val,.001,1000,.000000001)
print("Best paramaters based on Gradient Descent:")
w
print("coefficient of MWG_Ordinal : ", w[0,0])
print("coefficient of NWG_Ordinal : ", w[0,1])
print("coefficient of KWG_Ordinal : ", w[0,2])
print("coefficient of MDIMC_ordinal : ", w[0,3])
print("coefficient of NDIMC_ordinal : ", w[0,4])
print("coefficient of MDIMA_ordinal : ", w[0,5])
print("coefficient of NDIMB_ordinal : ", w[0,6])
print("coefficient of KWI_ordinal : ", w[0,7])
print("coefficient of VWM_ordinal : ", w[0,8])
print("coefficient of VWN_ordinal : ", w[0,9])
print("coefficient of STRM_1 : ", w[0,10])
print("coefficient of STRN_1 : ", w[0,11])
print("coefficient of SA_1 : ", w[0,12])
print("coefficient of SB_1 : ", w[0,13])
cost_v[-1]
Learning_rates= [.0001,.001,.01,.025,.05]
learning =[]
cost_t_min =[]
cost_v_min =[]
for i in Learning_rates:
cost_t,cost_v,w,iteration = linear_regression_model(X_train,y_train,X_val,y_val,i,2000,.00000001)
learning.append(i)
cost_t_min.append(cost_t[-1])
cost_v_min.append(cost_v[-1])
plt.plot(np.log10(learning),cost_t_min,label='Training Cost')
plt.plot(np.log10(learning),cost_v_min,label='Validation Cost')
plt.xlabel('Learning Rate (log scale)')
plt.ylabel('Cost')
plt.title('Cost(Validation and Training)as function of Learning rate')
plt.legend()
plt.show
cost_t_min
cost_v_min
```
# Experiment 2:
Plotting Cost of Training and Validation against varying Threshold
```
from LinearRegression_Threshold15 import *
threshold= [1,.1,.01]
thresh = []
cost_t_min =[]
cost_v_min =[]
iterate =[]
for i in threshold:
cost_t,cost_v,w,iteration = linear_regression_model(X_train,y_train,X_val,y_val,.025,20000,i)
thresh.append(i)
cost_t_min.append(cost_t[-1])
cost_v_min.append(cost_v[-1])
iterate.append(iteration)
plt.plot(np.log10(thresh),cost_t_min,label='Training Cost')
plt.plot(np.log10(thresh),cost_v_min,label='Validation Cost')
plt.xlabel('Threshold(log scale)')
plt.ylabel('Cost')
plt.legend()
plt.title('Cost(Training and Validation) as function of threshold')
plt.show
plt.plot(np.log10(thresh),iterate)
plt.xlabel('Threshold(log scale)')
plt.ylabel('Number of Iterations')
plt.title('Threshold Vs number of iterations')
plt.show
iterate
cost_t_min
cost_v_min
```
# EXPERIMENT 3 : Selecting 8 Random Features:
```
import pandas as pd
import numpy as np
df = pd.read_csv("C:\\ITM SPRING 2020\\ML\\sgemm_product_dataset\\processed_sgemm_product.csv")
y=df['MeanRun']
df.drop(columns ='MeanRun',axis=1,inplace = True)
X = df[df.columns.to_series().sample(8)]
X.columns
from sklearn.model_selection import train_test_split
X_train,X_val, y_train,y_val = train_test_split(X,y,test_size=0.33,random_state=5)
X_train = X_train.T
y_train = np.array([y_train])
X_val=X_val.T
y_val=np.array([y_val])
print(y_val.shape)
print(X_val.shape)
print(y_train.shape)
print(X_train.shape)
from LinearRegression_Threshold15 import *
Learning_rates= [.0001,.001,.01,.025,.05]
for i in Learning_rates:
cost_t,cost_v,w,iteration = linear_regression_model(X_train,y_train,X_val,y_val,i,1000,.0000000000001)
iterat.append(len(cost_t))
plt.plot(cost_t,label='tc_lr = '+str(i))
plt.plot(cost_v,label='vc_lr='+str(i))
plt.xlabel('Iterations')
plt.ylabel('Cost')
plt.title('Training and Validation cost for varying learning rates')
plt.legend()
plt.show
cost_t[-1]
cost_v[-1]
from LinearRegression_Threshold15 import *
threshold= [1,.1,.01]
thresh = []
cost_t_min =[]
cost_v_min =[]
iterate =[]
for i in threshold:
cost_t,cost_v,w,iteration = linear_regression_model(X_train,y_train,X_val,y_val,.025,20000,i)
thresh.append(i)
cost_t_min.append(cost_t[-1])
cost_v_min.append(cost_v[-1])
iterate.append(iteration)
plt.plot(np.log10(thresh),cost_t_min,label='Training Cost')
plt.plot(np.log10(thresh),cost_v_min,label='Validation Cost')
plt.xlabel('Threshold(log scale)')
plt.ylabel('Cost')
plt.legend()
plt.title('Cost(Training and Validation) as function of threshold')
plt.show
plt.plot(np.log10(thresh),iterate)
plt.xlabel('Threshold(log scale)')
plt.ylabel('Number of Iterations')
plt.title('Threshold Vs number of iterations')
plt.show
iterate
cost_t_min
cost_v_min
```
# Experiment - 4
```
Based on the exploratory data analysis it appears that variables like VWN, VWM,NDIMB,MDIMB, and binary variables, all other variables have some values which always result in low run time. Based on this observation, selected following 8 features:
MWG_Ordinal
NWG_Ordinal
KWG_Ordinal
MDIMC_Ordinal
NDIMC_Ordinal
MDIMA_Ordinal
NDIMA_Ordinal
VWM_Ordinal
import pandas as pd
import numpy as np
df = pd.read_csv("C:\\ITM SPRING 2020\\ML\\sgemm_product_dataset\\processed_sgemm_product.csv")
y=df['MeanRun']
df.drop(columns =['VWN_ordinal', 'STRM_1', 'STRN_1', 'SA_1', 'SB_1','MeanRun'],axis=1,inplace = True)
X.columns
from sklearn.model_selection import train_test_split
X_train,X_val, y_train,y_val = train_test_split(X,y,test_size=0.3,random_state=5)
X_train = X_train.T
y_train = np.array([y_train])
X_val=X_val.T
y_val=np.array([y_val])
from LinearRegression_Threshold15 import *
Learning_rates= [.0001,.001,.01,.025,.05]
for i in Learning_rates:
cost_t,cost_v,w,iteration = linear_regression_model(X_train,y_train,X_val,y_val,i,1000,.0000000000001)
iterat.append(len(cost_t))
plt.plot(cost_t,label='tc_lr = '+str(i))
plt.plot(cost_v,label='vc_lr='+str(i))
plt.xlabel('Iterations')
plt.ylabel('Cost')
plt.title('Training and Validation cost for varying learning rates')
plt.legend()
plt.show
Learning_rates= [.0001,.001,.01,.025,.05,.09]
learning =[]
cost_t_min =[]
cost_v_min =[]
for i in Learning_rates:
cost_t,cost_v,w,iteration = linear_regression_model(X_train,y_train,X_val,y_val,i,2000,.00000001)
learning.append(i)
cost_t_min.append(cost_t[-1])
cost_v_min.append(cost_v[-1])
plt.plot(np.log10(learning),cost_t_min,label='Training Cost')
plt.plot(np.log10(learning),cost_v_min,label='Validation Cost')
plt.xlabel('Learning Rate (log scale)')
plt.ylabel('Cost')
plt.title('Cost(Validation and Training)as function of Learning rate')
plt.legend()
plt.show
cost_t_min
cost_v_min
from LinearRegression_Threshold15 import *
threshold= [1,.1,.01]
thresh = []
cost_t_min =[]
cost_v_min =[]
iterate =[]
for i in threshold:
cost_t,cost_v,w,iteration = linear_regression_model(X_train,y_train,X_val,y_val,.025,20000,i)
thresh.append(i)
cost_t_min.append(cost_t[-1])
cost_v_min.append(cost_v[-1])
iterate.append(iteration)
plt.plot(np.log10(thresh),cost_t_min,label='Training Cost')
plt.plot(np.log10(thresh),cost_v_min,label='Validation Cost')
plt.xlabel('Threshold(log scale)')
plt.ylabel('Cost')
plt.legend()
plt.title('Cost(Training and Validation) as function of threshold')
plt.show
plt.plot(np.log10(thresh),iterate)
plt.xlabel('Threshold(log scale)')
plt.ylabel('Number of Iterations')
plt.title('Threshold Vs number of iterations')
plt.show
cost_t_min
cost_v_min
import pandas as pd
import numpy as np
df = pd.read_csv("C:\\ITM SPRING 2020\\ML\\sgemm_product_dataset\\processed_sgemm_product.csv")
```
|
github_jupyter
|
import pandas as pd
import numpy as np
df = pd.read_csv("C:\\ITM SPRING 2020\\ML\\sgemm_product_dataset\\processed_sgemm_product.csv")
y=df['MeanRun']
X=df.drop(columns ='MeanRun',axis=1)
from sklearn.model_selection import train_test_split
X_train,X_val, y_train,y_val = train_test_split(X,y,test_size=0.33,random_state=5)
X_train = X_train.T
y_train = np.array([y_train])
X_val=X_val.T
y_val=np.array([y_val])
print(y_val.shape)
print(X_val.shape)
print(y_train.shape)
print(X_train.shape)
from LinearRegression_Threshold15 import *
Learning_rates= [.0001,.001,.01,.025,.05]
iterat =[]
for i in Learning_rates:
cost_t,cost_v,w,iteration = linear_regression_model(X_train,y_train,X_val,y_val,i,1000,.0000000000001)
iterat.append(len(cost_t))
plt.plot(cost_t,label='tc_lr = '+str(i))
plt.plot(cost_v,label='vc_lr='+str(i))
plt.xlabel('Iterations')
plt.ylabel('Cost')
plt.title('Training and Validation cost for varying learning rates')
plt.legend()
plt.show
iterat
cost_t,cost_v,w,iteration = linear_regression_model(X_train,y_train,X_val,y_val,.001,1000,.000000001)
print("Best paramaters based on Gradient Descent:")
w
print("coefficient of MWG_Ordinal : ", w[0,0])
print("coefficient of NWG_Ordinal : ", w[0,1])
print("coefficient of KWG_Ordinal : ", w[0,2])
print("coefficient of MDIMC_ordinal : ", w[0,3])
print("coefficient of NDIMC_ordinal : ", w[0,4])
print("coefficient of MDIMA_ordinal : ", w[0,5])
print("coefficient of NDIMB_ordinal : ", w[0,6])
print("coefficient of KWI_ordinal : ", w[0,7])
print("coefficient of VWM_ordinal : ", w[0,8])
print("coefficient of VWN_ordinal : ", w[0,9])
print("coefficient of STRM_1 : ", w[0,10])
print("coefficient of STRN_1 : ", w[0,11])
print("coefficient of SA_1 : ", w[0,12])
print("coefficient of SB_1 : ", w[0,13])
cost_v[-1]
Learning_rates= [.0001,.001,.01,.025,.05]
learning =[]
cost_t_min =[]
cost_v_min =[]
for i in Learning_rates:
cost_t,cost_v,w,iteration = linear_regression_model(X_train,y_train,X_val,y_val,i,2000,.00000001)
learning.append(i)
cost_t_min.append(cost_t[-1])
cost_v_min.append(cost_v[-1])
plt.plot(np.log10(learning),cost_t_min,label='Training Cost')
plt.plot(np.log10(learning),cost_v_min,label='Validation Cost')
plt.xlabel('Learning Rate (log scale)')
plt.ylabel('Cost')
plt.title('Cost(Validation and Training)as function of Learning rate')
plt.legend()
plt.show
cost_t_min
cost_v_min
from LinearRegression_Threshold15 import *
threshold= [1,.1,.01]
thresh = []
cost_t_min =[]
cost_v_min =[]
iterate =[]
for i in threshold:
cost_t,cost_v,w,iteration = linear_regression_model(X_train,y_train,X_val,y_val,.025,20000,i)
thresh.append(i)
cost_t_min.append(cost_t[-1])
cost_v_min.append(cost_v[-1])
iterate.append(iteration)
plt.plot(np.log10(thresh),cost_t_min,label='Training Cost')
plt.plot(np.log10(thresh),cost_v_min,label='Validation Cost')
plt.xlabel('Threshold(log scale)')
plt.ylabel('Cost')
plt.legend()
plt.title('Cost(Training and Validation) as function of threshold')
plt.show
plt.plot(np.log10(thresh),iterate)
plt.xlabel('Threshold(log scale)')
plt.ylabel('Number of Iterations')
plt.title('Threshold Vs number of iterations')
plt.show
iterate
cost_t_min
cost_v_min
import pandas as pd
import numpy as np
df = pd.read_csv("C:\\ITM SPRING 2020\\ML\\sgemm_product_dataset\\processed_sgemm_product.csv")
y=df['MeanRun']
df.drop(columns ='MeanRun',axis=1,inplace = True)
X = df[df.columns.to_series().sample(8)]
X.columns
from sklearn.model_selection import train_test_split
X_train,X_val, y_train,y_val = train_test_split(X,y,test_size=0.33,random_state=5)
X_train = X_train.T
y_train = np.array([y_train])
X_val=X_val.T
y_val=np.array([y_val])
print(y_val.shape)
print(X_val.shape)
print(y_train.shape)
print(X_train.shape)
from LinearRegression_Threshold15 import *
Learning_rates= [.0001,.001,.01,.025,.05]
for i in Learning_rates:
cost_t,cost_v,w,iteration = linear_regression_model(X_train,y_train,X_val,y_val,i,1000,.0000000000001)
iterat.append(len(cost_t))
plt.plot(cost_t,label='tc_lr = '+str(i))
plt.plot(cost_v,label='vc_lr='+str(i))
plt.xlabel('Iterations')
plt.ylabel('Cost')
plt.title('Training and Validation cost for varying learning rates')
plt.legend()
plt.show
cost_t[-1]
cost_v[-1]
from LinearRegression_Threshold15 import *
threshold= [1,.1,.01]
thresh = []
cost_t_min =[]
cost_v_min =[]
iterate =[]
for i in threshold:
cost_t,cost_v,w,iteration = linear_regression_model(X_train,y_train,X_val,y_val,.025,20000,i)
thresh.append(i)
cost_t_min.append(cost_t[-1])
cost_v_min.append(cost_v[-1])
iterate.append(iteration)
plt.plot(np.log10(thresh),cost_t_min,label='Training Cost')
plt.plot(np.log10(thresh),cost_v_min,label='Validation Cost')
plt.xlabel('Threshold(log scale)')
plt.ylabel('Cost')
plt.legend()
plt.title('Cost(Training and Validation) as function of threshold')
plt.show
plt.plot(np.log10(thresh),iterate)
plt.xlabel('Threshold(log scale)')
plt.ylabel('Number of Iterations')
plt.title('Threshold Vs number of iterations')
plt.show
iterate
cost_t_min
cost_v_min
Based on the exploratory data analysis it appears that variables like VWN, VWM,NDIMB,MDIMB, and binary variables, all other variables have some values which always result in low run time. Based on this observation, selected following 8 features:
MWG_Ordinal
NWG_Ordinal
KWG_Ordinal
MDIMC_Ordinal
NDIMC_Ordinal
MDIMA_Ordinal
NDIMA_Ordinal
VWM_Ordinal
import pandas as pd
import numpy as np
df = pd.read_csv("C:\\ITM SPRING 2020\\ML\\sgemm_product_dataset\\processed_sgemm_product.csv")
y=df['MeanRun']
df.drop(columns =['VWN_ordinal', 'STRM_1', 'STRN_1', 'SA_1', 'SB_1','MeanRun'],axis=1,inplace = True)
X.columns
from sklearn.model_selection import train_test_split
X_train,X_val, y_train,y_val = train_test_split(X,y,test_size=0.3,random_state=5)
X_train = X_train.T
y_train = np.array([y_train])
X_val=X_val.T
y_val=np.array([y_val])
from LinearRegression_Threshold15 import *
Learning_rates= [.0001,.001,.01,.025,.05]
for i in Learning_rates:
cost_t,cost_v,w,iteration = linear_regression_model(X_train,y_train,X_val,y_val,i,1000,.0000000000001)
iterat.append(len(cost_t))
plt.plot(cost_t,label='tc_lr = '+str(i))
plt.plot(cost_v,label='vc_lr='+str(i))
plt.xlabel('Iterations')
plt.ylabel('Cost')
plt.title('Training and Validation cost for varying learning rates')
plt.legend()
plt.show
Learning_rates= [.0001,.001,.01,.025,.05,.09]
learning =[]
cost_t_min =[]
cost_v_min =[]
for i in Learning_rates:
cost_t,cost_v,w,iteration = linear_regression_model(X_train,y_train,X_val,y_val,i,2000,.00000001)
learning.append(i)
cost_t_min.append(cost_t[-1])
cost_v_min.append(cost_v[-1])
plt.plot(np.log10(learning),cost_t_min,label='Training Cost')
plt.plot(np.log10(learning),cost_v_min,label='Validation Cost')
plt.xlabel('Learning Rate (log scale)')
plt.ylabel('Cost')
plt.title('Cost(Validation and Training)as function of Learning rate')
plt.legend()
plt.show
cost_t_min
cost_v_min
from LinearRegression_Threshold15 import *
threshold= [1,.1,.01]
thresh = []
cost_t_min =[]
cost_v_min =[]
iterate =[]
for i in threshold:
cost_t,cost_v,w,iteration = linear_regression_model(X_train,y_train,X_val,y_val,.025,20000,i)
thresh.append(i)
cost_t_min.append(cost_t[-1])
cost_v_min.append(cost_v[-1])
iterate.append(iteration)
plt.plot(np.log10(thresh),cost_t_min,label='Training Cost')
plt.plot(np.log10(thresh),cost_v_min,label='Validation Cost')
plt.xlabel('Threshold(log scale)')
plt.ylabel('Cost')
plt.legend()
plt.title('Cost(Training and Validation) as function of threshold')
plt.show
plt.plot(np.log10(thresh),iterate)
plt.xlabel('Threshold(log scale)')
plt.ylabel('Number of Iterations')
plt.title('Threshold Vs number of iterations')
plt.show
cost_t_min
cost_v_min
import pandas as pd
import numpy as np
df = pd.read_csv("C:\\ITM SPRING 2020\\ML\\sgemm_product_dataset\\processed_sgemm_product.csv")
| 0.518302 | 0.655405 |
<a href="https://colab.research.google.com/github/aryamanak10/Natural-Language-Processing/blob/master/Stock%20Sentiment%20Analysis/Stock_Sentiment_Analysis_using_News_Headlines_NLP.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
### Importing the Dataset
```
import pandas as pd
df=pd.read_csv('Data.csv', encoding = "ISO-8859-1")
df.head()
df.shape
```
### Splitting the dataset into Training and Testing Set
```
train = df[df['Date'] < '20150101']
test = df[df['Date'] > '20141231']
```
### Cleaning the Texts
#### Removing punctuations
```
data=train.iloc[:,2:27]
data.replace("[^a-zA-Z]"," ",regex=True, inplace=True)
```
#### Renaming column names for ease of access
```
list1= [i for i in range(25)]
new_Index=[str(i) for i in list1]
data.columns= new_Index
data.head(5)
```
#### Convertng headlines to lower case
```
for index in new_Index:
data[index]=data[index].str.lower()
data.head(1)
' '.join(str(x) for x in data.iloc[0,0:25])
headlines = []
for row in range(0,len(data.index)):
headlines.append(' '.join(str(x) for x in data.iloc[row,0:25]))
headlines[1]
```
### Implementing BAGOFWORDS using CountVectorizer
```
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.ensemble import RandomForestClassifier
# ngram_range = 'bi-gram' words
countvector=CountVectorizer(ngram_range=(2,2))
traindataset=countvector.fit_transform(headlines)
```
### Implement RandomForest Classifier
```
randomclassifier=RandomForestClassifier(n_estimators=200,criterion='entropy')
randomclassifier.fit(traindataset, train['Label'])
```
### Predict for the Test Dataset
```
test_transform= []
for row in range(0,len(test.index)):
test_transform.append(' '.join(str(x) for x in test.iloc[row,2:27]))
test_dataset = countvector.transform(test_transform)
predictions = randomclassifier.predict(test_dataset)
```
### Evaluation Metrics
```
from sklearn.metrics import classification_report,confusion_matrix,accuracy_score
matrix=confusion_matrix(test['Label'],predictions)
print(matrix)
score=accuracy_score(test['Label'],predictions)
print(score)
report=classification_report(test['Label'],predictions)
print(report)
```
## Creating TF-IDF Model using TF-IDF Vectorizer
```
from sklearn.feature_extraction.text import TfidfVectorizer
tfidf_vector=CountVectorizer(ngram_range=(2,2))
traindataset=tfidf_vector.fit_transform(headlines)
```
### Implementing RandomForest Classifier
```
randomclassifier=RandomForestClassifier(n_estimators=200,criterion='entropy')
randomclassifier.fit(traindataset, train['Label'])
```
### Predict for the Test Dataset
```
test_transform= []
for row in range(0,len(test.index)):
test_transform.append(' '.join(str(x) for x in test.iloc[row,2:27]))
test_dataset = tfidf_vector.transform(test_transform)
predictions = randomclassifier.predict(test_dataset)
```
### Evaluation Metrics
```
matrix=confusion_matrix(test['Label'],predictions)
print(matrix)
score=accuracy_score(test['Label'],predictions)
print(score)
report=classification_report(test['Label'],predictions)
print(report)
```
### Implementing Naive-Bayes Classifier
```
from sklearn.naive_bayes import MultinomialNB
multi=MultinomialNB()
multi.fit(traindataset, train['Label'])
```
### Predict for the Test Dataset
```
test_transform= []
for row in range(0,len(test.index)):
test_transform.append(' '.join(str(x) for x in test.iloc[row,2:27]))
test_dataset = tfidf_vector.transform(test_transform)
predictions = multi.predict(test_dataset)
```
### Evaluation Metrics
```
matrix=confusion_matrix(test['Label'],predictions)
print(matrix)
score=accuracy_score(test['Label'],predictions)
print(score)
report=classification_report(test['Label'],predictions)
print(report)
```
|
github_jupyter
|
import pandas as pd
df=pd.read_csv('Data.csv', encoding = "ISO-8859-1")
df.head()
df.shape
train = df[df['Date'] < '20150101']
test = df[df['Date'] > '20141231']
data=train.iloc[:,2:27]
data.replace("[^a-zA-Z]"," ",regex=True, inplace=True)
list1= [i for i in range(25)]
new_Index=[str(i) for i in list1]
data.columns= new_Index
data.head(5)
for index in new_Index:
data[index]=data[index].str.lower()
data.head(1)
' '.join(str(x) for x in data.iloc[0,0:25])
headlines = []
for row in range(0,len(data.index)):
headlines.append(' '.join(str(x) for x in data.iloc[row,0:25]))
headlines[1]
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.ensemble import RandomForestClassifier
# ngram_range = 'bi-gram' words
countvector=CountVectorizer(ngram_range=(2,2))
traindataset=countvector.fit_transform(headlines)
randomclassifier=RandomForestClassifier(n_estimators=200,criterion='entropy')
randomclassifier.fit(traindataset, train['Label'])
test_transform= []
for row in range(0,len(test.index)):
test_transform.append(' '.join(str(x) for x in test.iloc[row,2:27]))
test_dataset = countvector.transform(test_transform)
predictions = randomclassifier.predict(test_dataset)
from sklearn.metrics import classification_report,confusion_matrix,accuracy_score
matrix=confusion_matrix(test['Label'],predictions)
print(matrix)
score=accuracy_score(test['Label'],predictions)
print(score)
report=classification_report(test['Label'],predictions)
print(report)
from sklearn.feature_extraction.text import TfidfVectorizer
tfidf_vector=CountVectorizer(ngram_range=(2,2))
traindataset=tfidf_vector.fit_transform(headlines)
randomclassifier=RandomForestClassifier(n_estimators=200,criterion='entropy')
randomclassifier.fit(traindataset, train['Label'])
test_transform= []
for row in range(0,len(test.index)):
test_transform.append(' '.join(str(x) for x in test.iloc[row,2:27]))
test_dataset = tfidf_vector.transform(test_transform)
predictions = randomclassifier.predict(test_dataset)
matrix=confusion_matrix(test['Label'],predictions)
print(matrix)
score=accuracy_score(test['Label'],predictions)
print(score)
report=classification_report(test['Label'],predictions)
print(report)
from sklearn.naive_bayes import MultinomialNB
multi=MultinomialNB()
multi.fit(traindataset, train['Label'])
test_transform= []
for row in range(0,len(test.index)):
test_transform.append(' '.join(str(x) for x in test.iloc[row,2:27]))
test_dataset = tfidf_vector.transform(test_transform)
predictions = multi.predict(test_dataset)
matrix=confusion_matrix(test['Label'],predictions)
print(matrix)
score=accuracy_score(test['Label'],predictions)
print(score)
report=classification_report(test['Label'],predictions)
print(report)
| 0.221267 | 0.969324 |
```
# This Python 3 environment comes with many helpful analytics libraries installed
# It is defined by the kaggle/python Docker image: https://github.com/kaggle/docker-python
# For example, here's several helpful packages to load
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import warnings
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk('/home/cristiandugacicu/projects/personal/zoomcamp/'):
for filename in filenames:
print(os.path.join(dirname, filename))
# !wget "https://raw.githubusercontent.com/alexeygrigorev/mlbookcamp-code/master/chapter-03-churn-prediction/WA_Fn-UseC_-Telco-Customer-Churn.csv" -O data-3.csv
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
df = pd.read_csv("/home/cristiandugacicu/projects/personal/zoomcamp/WA_Fn-UseC_-Telco-Customer-Churn.csv")
df.columns = df.columns.str.lower().str.replace(" ", "_")
categorical_columns = list(df.dtypes[df.dtypes == 'object'].index)
# fix categorical values
for c in categorical_columns:
df[c] = df[c].str.lower().str.replace(" ", "_")
# fix nan values if they are marked with another char
tc = pd.to_numeric(df.totalcharges, errors='coerce')
print("Missing totalcharges count:",tc.isnull().sum())
df.totalcharges = tc
df.totalcharges = df.totalcharges.fillna(0)
# fix yes/no with 0/1
df.churn = (df.churn == "yes").astype(int)
df.head().T
# df.isnull().sum()
# Setup validation framework
from sklearn.model_selection import train_test_split
df_full_train, df_test = train_test_split(df, test_size=0.2, random_state=1)
df_train, df_val = train_test_split(df_full_train, test_size=0.25, random_state=1)
len(df_train), len(df_val), len(df_test)
df_train = df_train.reset_index(drop="true")
df_val = df_val.reset_index(drop="true")
df_test = df_test.reset_index(drop="true")
y_full_train = df_full_train.churn.values
y_train = df_train.churn.values
y_val = df_val.churn.values
y_test = df_test.churn.values
del df_test["churn"]
del df_val["churn"]
del df_train["churn"]
df_train.columns
# EDA Exploritory Data Analysis
numerical_columns = ["monthlycharges", "totalcharges", "tenure"]
categorical_columns = ['gender', 'seniorcitizen', 'partner', 'dependents',
'phoneservice', 'multiplelines', 'internetservice',
'onlinesecurity', 'onlinebackup', 'deviceprotection', 'techsupport',
'streamingtv', 'streamingmovies', 'contract', 'paperlessbilling',
'paymentmethod']
df_full_train.churn.value_counts()
df_full_train.churn.value_counts(normalize=True)
df_full_train.churn.mean()
df_full_train.groupby("gender").churn.agg(["mean", "count"])
# Hot Encoding
from sklearn.feature_extraction import DictVectorizer
# train_dicts = df_train[["gender", "contract", "tenure"]].iloc[:100].to_dict(orient='records')
train_dicts = df_train[categorical_columns+numerical_columns].to_dict(orient='records')
dv = DictVectorizer(sparse = False) #sparse =True => enables sparse matrix by compressing 0 values
dv.fit(train_dicts)
dv.transform(train_dicts)
display("column names:",dv.get_feature_names()) # column names
X_train = dv.fit_transform(train_dicts)
val_dicts = df_val[categorical_columns+numerical_columns].to_dict(orient='records')
X_val = dv.transform(val_dicts)
#Logistic regretion
from sklearn import linear_model
model = linear_model.LogisticRegression()
model.fit(X_train, y_train)
weights = model.coef_[0].round(3) # weights
model.intercept_ # bias, w0
weights_with_featureNames = dict(zip(dv.get_feature_names(), weights))
display(weights_with_featureNames)
model.predict(X_val) #hard predictions: y= {0,1}
proba = model.predict_proba(X_val) #hard predictions: y= 0->1. Returns probablility as [% for 0,% for 1]
y_pred_val = proba[:,1] #take column 1
churn_decision = (y_pred_val >= 0.5)
customers_that_will_leave = df_val[churn_decision].customerid # the people who will churn
#check average accuracy on y_val
(churn_decision.astype(int) == y_val).mean()
df_pred = pd.DataFrame()
df_pred["probability"] = y_pred_val
df_pred["prediction"] = churn_decision.astype(int)
df_pred["actual"] = y_val
df_pred["prediction_correct"] = df_pred.prediction == df_pred.actual
print("Accuracy % on y_val:",df_pred.prediction_correct.mean())
# Small Model Interpretation
small = ["contract", "tenure", "monthlycharges"]
dicts_train_small = df_train[small].to_dict(orient="records")
dicts_val_small = df_val[small].to_dict(orient="records")
dv_small = DictVectorizer(sparse=False)
dv_small.fit(dicts_train_small)
dv_small.get_feature_names() # => ['contract=month-to-month', 'contract=one_year', 'contract=two_year', 'monthlycharges', 'tenure']
X_train_small = dv_small.transform(dicts_train_small)
model_small = linear_model.LogisticRegression()
model_small.fit(X_train_small, y_train)
w0 = model_small.intercept_[0] # bias, w0
w = model_small.coef_[0].round(3) # weights
weights_with_names = dict(zip(dv_small.get_feature_names(), w))
print("w0 =", w0, "weights_with_names: ", weights_with_names)
#Using the model
def train_model(dataFrame, y):
dicts = dataFrame[categorical_columns+numerical_columns].to_dict(orient="records")
dv = DictVectorizer(sparse=False)
dv.fit(dicts)
X = dv.transform(dicts)
model = linear_model.LogisticRegression()
model.fit(X, y)
return model
model_full_train = train_model(df_full_train,y_full_train)
w0 = model_full_train.intercept_[0] # bias, w0
w = model_full_train.coef_[0].round(3) # weigh
#test:
dicts_test = df_test[categorical_columns+numerical_columns].to_dict(orient="records")
X_test = dv.transform(dicts_test)
y_pred = model_full_train.predict_proba(X_test)[:,1]
churn_decision = (y_pred >= 0.5)
print("Model accuracy based on test data:", (churn_decision == y_test).mean())
print("Customers that will churn:", (churn_decision == y_test).sum(), "from", len(y_test))
#example: Prediction on 1 customer
customer = df_test.iloc[10]
X_customer = dv.transform([customer])
y_customer = model_full_train.predict_proba(X_customer)[0,1]
customer_churn = (y_customer>=0.5)
print("Probability of Customer #1 to churn:",customer_churn,",Actual churn:", y_test[10])
import matplotlib.pyplot as plt
from sklearn import metrics
thresholds = np.linspace(0, 1, 21)
scores = []
for t in thresholds:
# churn_decision = (y_pred >= t)
# score = (churn_decision == y_test).mean()
# OR using sklearn:
score = metrics.accuracy_score(y_test, y_pred >= t)
scores.append(score)
print('Model accuracy (thresholds=%.2f): %.3f' % (t, score))
print('Max Model accuracy is %.3f' % (max(scores)))
plt.plot(thresholds, scores)
# Confusion Table
# churn_decision:
t = 0.5
predict_positive = (y_pred >= t)
predict_negative = (y_pred < t)
actual_positive = (y_test >= t)
actual_negative = (y_test < t)
true_positive = (predict_positive & actual_positive).sum()
true_negative = (predict_negative & actual_negative).sum()
false_positive = (predict_positive & actual_negative).sum()
false_negative = (predict_negative & actual_positive).sum()
print('Customers that we predict will churn and they do churn (thresholds=%.2f): %1.0f' % (t, true_positive))
print('Customers that we predict will not churn and they do not churn (thresholds=%.2f): %1.0f' % (t, true_negative))
print('Customers that we predict will churn but they do not churn (thresholds=%.2f): %1.0f' % (t, false_positive))
print('Customers that we predict will not churn but they do churn (thresholds=%.2f): %1.0f' % (t, false_negative))
labels = ["true_positive", "false_positive", "true_negative", "false_negative"]
results = [true_positive, false_positive, true_negative, false_negative]
colors = ["lightgreen", "red", "green", "darkred"]
plt.pie(results, labels=labels, colors=colors, autopct='%1.0f%%')
confusion_matrix = np.array([
[true_negative, false_positive],
[false_negative, true_positive]
])
print("\nConfusion_matrix:\n",confusion_matrix)
print("\nConfusion_matrix %:\n",((confusion_matrix/confusion_matrix.sum()).round(2)))
#Precision #Recall
precission = true_positive / (true_positive + false_positive)
print("precission=",precission)
#Recall
recall = true_positive / (true_positive + false_negative)
print("recall=",recall)
#TP rate #FP rate
tpr = true_positive / (true_positive + false_negative)
print("tpr=",tpr)
fpr = false_positive / (false_positive + true_negative)
print("fpr=",fpr)
# thresholds / tpr # thresholds / tpr
def tpr_fpr_dataframe(y, y_pred1):
thresholds = np.linspace(0, 1, 101)
scores = []
for t in thresholds:
predict_positive = (y_pred1 >= t)
predict_negative = (y_pred1 < t)
actual_positive = (y >= t)
actual_negative = (y < t)
true_positive = (predict_positive & actual_positive).sum()
true_negative = (predict_negative & actual_negative).sum()
false_positive = (predict_positive & actual_negative).sum()
false_negative = (predict_negative & actual_positive).sum()
scores.append((t,true_positive,true_negative,false_positive,false_negative))
scores_columns = ["threshold","true_positive","true_negative","false_positive","false_negative"]
df_scores = pd.DataFrame(scores, columns=scores_columns)
df_scores["tpr"] = df_scores.true_positive / (df_scores.true_positive + df_scores.false_negative)
df_scores["fpr"] = df_scores.false_positive / (df_scores.false_positive + df_scores.true_negative)
return df_scores
df_total_scores = tpr_fpr_dataframe(y_test, y_pred)
display(df_total_scores[::10])
plt.plot(df_total_scores.threshold, df_total_scores.tpr, label="TPR")
plt.plot(df_total_scores.threshold, df_total_scores.fpr, label="FPR")
plt.xlabel("threshold")
plt.legend()
# Random prediction
np.random.seed(1)
y_rand = np.random.uniform(0,1,size=len(y_test))
((y_rand >= 0.5) == y_test).mean()
df_total_scores_random = tpr_fpr_dataframe(y_test, y_rand)
display(df_total_scores_random[::10])
plt.plot(df_total_scores_random.threshold, df_total_scores.tpr, label="TPR random")
plt.plot(df_total_scores_random.threshold, df_total_scores.fpr, label="FPR random")
plt.xlabel("threshold")
plt.legend()
# Ideal prediction for y_test data
# num_negative = (y_test == 0).sum()
# num_positive = (y_test == 1).sum()
num_negative = int(len(y_test) * 0.6) # 60% are not churn
num_positive = len(y_test)-num_negative # 40% are churn
display(num_negative, num_positive)
# % = 1-348/(1061+348)
y_ideal = np.repeat([0,1], [num_negative, num_positive])
y_ideal_pred = np.linspace(0, 1, len(y_test))
# ((y_ideal_pred >= 0.0.75301) == y_ideal).mean()
df_total_scores_ideal = tpr_fpr_dataframe(y_ideal, y_ideal_pred)
display(df_total_scores_ideal[::10])
plt.plot(df_total_scores_ideal.threshold, df_total_scores_ideal.tpr, label="TPR ideal")
plt.plot(df_total_scores_ideal.threshold, df_total_scores_ideal.fpr, label="FPR ideal")
plt.xlabel("threshold")
plt.legend()
# ROC Curve: FPT / TPR
plt.figure(figsize=(5,5))
plt.plot(df_total_scores.fpr, df_total_scores.tpr, label="ROC pred")
plt.plot(df_total_scores_random.fpr, df_total_scores_random.tpr, label="ROC random")
plt.plot(df_total_scores_ideal.fpr, df_total_scores_ideal.tpr, label="ROC ideal")
plt.xlabel("FPR")
plt.ylabel("TPR")
plt.legend()
import sklearn.metrics
df_total_scores_auc = metrics.auc(df_total_scores.fpr.fillna(1), df_total_scores.tpr)
df_total_scores_random_auc = metrics.auc(df_total_scores_random.fpr.fillna(1), df_total_scores_random.tpr)
df_total_scores_ideal_auc = metrics.auc(df_total_scores_ideal.fpr.fillna(1), df_total_scores_ideal.tpr)
print("df_total_scores_auc:", df_total_scores_auc)
print("df_total_scores_random_auc:", df_total_scores_random_auc)
print("df_total_scores_ideal_auc:", df_total_scores_ideal_auc)
# OR
fpr, tpr, thresholds = metrics.roc_curve(y_test, y_pred)
auc = metrics.auc(df_total_scores_random.fpr.fillna(1), df_total_scores_random.tpr)
# !pip install tqdm
import warnings
warnings.filterwarnings(action='once')
warnings.filterwarnings('ignore')
df_full_train, df_test = train_test_split(df, test_size=0.2, random_state=1)
#Cross Validation
from sklearn.model_selection import KFold
from tqdm.auto import tqdm
def train(dataFrame, y, C):
dicts = dataFrame.to_dict(orient="records")
dv = DictVectorizer(sparse=False)
X = dv.fit_transform(dicts)
model = linear_model.LogisticRegression(C=C)
model.fit(X, y)
return dv, model
def predict(dataFrame, dv, model):
dicts = dataFrame.to_dict(orient="records")
X = dv.transform(dicts)
y_pred = model.predict_proba(X)[:,1]
return y_pred
df_full_train_selected1 = df_full_train[categorical_columns+numerical_columns]
splits = 5
for C in tqdm([ 0.001, 0.01, 0.1, 0.5, 1, 5, 10], total=splits):
kf = KFold(n_splits=splits, shuffle=True, random_state=1)
auc_scores = []
for train_idx, val_idx in kf.split(df_full_train):
df_train_itter = df_full_train_selected1.iloc[train_idx]
df_val_itter = df_full_train_selected1.iloc[val_idx]
y_train_iter = df_full_train.iloc[train_idx].churn.values
y_val_iter = df_full_train.iloc[val_idx].churn.values
dv, model = train(df_train_itter, y_train_iter, C=C)
y_pred_iter = predict(df_val_itter, dv, model)
auc = metrics.roc_auc_score(y_val_iter, y_pred_iter)
auc_scores.append(auc)
print("C:%s, AUC mean: %.3f, AUC std: +-%.3f" % (C, np.mean(auc_scores), np.std(auc_scores)))
# train for 1 dataset:
dv, model = train(df_full_train_selected1, y_full_train, 1.0)
y_full_pred = predict(df_test, dv, model)
auc = metrics.roc_auc_score(y_test, y_full_pred)
print("C:%s, Full AUC mean: %.3f" % (1.0, auc))
```
|
github_jupyter
|
# This Python 3 environment comes with many helpful analytics libraries installed
# It is defined by the kaggle/python Docker image: https://github.com/kaggle/docker-python
# For example, here's several helpful packages to load
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import warnings
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk('/home/cristiandugacicu/projects/personal/zoomcamp/'):
for filename in filenames:
print(os.path.join(dirname, filename))
# !wget "https://raw.githubusercontent.com/alexeygrigorev/mlbookcamp-code/master/chapter-03-churn-prediction/WA_Fn-UseC_-Telco-Customer-Churn.csv" -O data-3.csv
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
df = pd.read_csv("/home/cristiandugacicu/projects/personal/zoomcamp/WA_Fn-UseC_-Telco-Customer-Churn.csv")
df.columns = df.columns.str.lower().str.replace(" ", "_")
categorical_columns = list(df.dtypes[df.dtypes == 'object'].index)
# fix categorical values
for c in categorical_columns:
df[c] = df[c].str.lower().str.replace(" ", "_")
# fix nan values if they are marked with another char
tc = pd.to_numeric(df.totalcharges, errors='coerce')
print("Missing totalcharges count:",tc.isnull().sum())
df.totalcharges = tc
df.totalcharges = df.totalcharges.fillna(0)
# fix yes/no with 0/1
df.churn = (df.churn == "yes").astype(int)
df.head().T
# df.isnull().sum()
# Setup validation framework
from sklearn.model_selection import train_test_split
df_full_train, df_test = train_test_split(df, test_size=0.2, random_state=1)
df_train, df_val = train_test_split(df_full_train, test_size=0.25, random_state=1)
len(df_train), len(df_val), len(df_test)
df_train = df_train.reset_index(drop="true")
df_val = df_val.reset_index(drop="true")
df_test = df_test.reset_index(drop="true")
y_full_train = df_full_train.churn.values
y_train = df_train.churn.values
y_val = df_val.churn.values
y_test = df_test.churn.values
del df_test["churn"]
del df_val["churn"]
del df_train["churn"]
df_train.columns
# EDA Exploritory Data Analysis
numerical_columns = ["monthlycharges", "totalcharges", "tenure"]
categorical_columns = ['gender', 'seniorcitizen', 'partner', 'dependents',
'phoneservice', 'multiplelines', 'internetservice',
'onlinesecurity', 'onlinebackup', 'deviceprotection', 'techsupport',
'streamingtv', 'streamingmovies', 'contract', 'paperlessbilling',
'paymentmethod']
df_full_train.churn.value_counts()
df_full_train.churn.value_counts(normalize=True)
df_full_train.churn.mean()
df_full_train.groupby("gender").churn.agg(["mean", "count"])
# Hot Encoding
from sklearn.feature_extraction import DictVectorizer
# train_dicts = df_train[["gender", "contract", "tenure"]].iloc[:100].to_dict(orient='records')
train_dicts = df_train[categorical_columns+numerical_columns].to_dict(orient='records')
dv = DictVectorizer(sparse = False) #sparse =True => enables sparse matrix by compressing 0 values
dv.fit(train_dicts)
dv.transform(train_dicts)
display("column names:",dv.get_feature_names()) # column names
X_train = dv.fit_transform(train_dicts)
val_dicts = df_val[categorical_columns+numerical_columns].to_dict(orient='records')
X_val = dv.transform(val_dicts)
#Logistic regretion
from sklearn import linear_model
model = linear_model.LogisticRegression()
model.fit(X_train, y_train)
weights = model.coef_[0].round(3) # weights
model.intercept_ # bias, w0
weights_with_featureNames = dict(zip(dv.get_feature_names(), weights))
display(weights_with_featureNames)
model.predict(X_val) #hard predictions: y= {0,1}
proba = model.predict_proba(X_val) #hard predictions: y= 0->1. Returns probablility as [% for 0,% for 1]
y_pred_val = proba[:,1] #take column 1
churn_decision = (y_pred_val >= 0.5)
customers_that_will_leave = df_val[churn_decision].customerid # the people who will churn
#check average accuracy on y_val
(churn_decision.astype(int) == y_val).mean()
df_pred = pd.DataFrame()
df_pred["probability"] = y_pred_val
df_pred["prediction"] = churn_decision.astype(int)
df_pred["actual"] = y_val
df_pred["prediction_correct"] = df_pred.prediction == df_pred.actual
print("Accuracy % on y_val:",df_pred.prediction_correct.mean())
# Small Model Interpretation
small = ["contract", "tenure", "monthlycharges"]
dicts_train_small = df_train[small].to_dict(orient="records")
dicts_val_small = df_val[small].to_dict(orient="records")
dv_small = DictVectorizer(sparse=False)
dv_small.fit(dicts_train_small)
dv_small.get_feature_names() # => ['contract=month-to-month', 'contract=one_year', 'contract=two_year', 'monthlycharges', 'tenure']
X_train_small = dv_small.transform(dicts_train_small)
model_small = linear_model.LogisticRegression()
model_small.fit(X_train_small, y_train)
w0 = model_small.intercept_[0] # bias, w0
w = model_small.coef_[0].round(3) # weights
weights_with_names = dict(zip(dv_small.get_feature_names(), w))
print("w0 =", w0, "weights_with_names: ", weights_with_names)
#Using the model
def train_model(dataFrame, y):
dicts = dataFrame[categorical_columns+numerical_columns].to_dict(orient="records")
dv = DictVectorizer(sparse=False)
dv.fit(dicts)
X = dv.transform(dicts)
model = linear_model.LogisticRegression()
model.fit(X, y)
return model
model_full_train = train_model(df_full_train,y_full_train)
w0 = model_full_train.intercept_[0] # bias, w0
w = model_full_train.coef_[0].round(3) # weigh
#test:
dicts_test = df_test[categorical_columns+numerical_columns].to_dict(orient="records")
X_test = dv.transform(dicts_test)
y_pred = model_full_train.predict_proba(X_test)[:,1]
churn_decision = (y_pred >= 0.5)
print("Model accuracy based on test data:", (churn_decision == y_test).mean())
print("Customers that will churn:", (churn_decision == y_test).sum(), "from", len(y_test))
#example: Prediction on 1 customer
customer = df_test.iloc[10]
X_customer = dv.transform([customer])
y_customer = model_full_train.predict_proba(X_customer)[0,1]
customer_churn = (y_customer>=0.5)
print("Probability of Customer #1 to churn:",customer_churn,",Actual churn:", y_test[10])
import matplotlib.pyplot as plt
from sklearn import metrics
thresholds = np.linspace(0, 1, 21)
scores = []
for t in thresholds:
# churn_decision = (y_pred >= t)
# score = (churn_decision == y_test).mean()
# OR using sklearn:
score = metrics.accuracy_score(y_test, y_pred >= t)
scores.append(score)
print('Model accuracy (thresholds=%.2f): %.3f' % (t, score))
print('Max Model accuracy is %.3f' % (max(scores)))
plt.plot(thresholds, scores)
# Confusion Table
# churn_decision:
t = 0.5
predict_positive = (y_pred >= t)
predict_negative = (y_pred < t)
actual_positive = (y_test >= t)
actual_negative = (y_test < t)
true_positive = (predict_positive & actual_positive).sum()
true_negative = (predict_negative & actual_negative).sum()
false_positive = (predict_positive & actual_negative).sum()
false_negative = (predict_negative & actual_positive).sum()
print('Customers that we predict will churn and they do churn (thresholds=%.2f): %1.0f' % (t, true_positive))
print('Customers that we predict will not churn and they do not churn (thresholds=%.2f): %1.0f' % (t, true_negative))
print('Customers that we predict will churn but they do not churn (thresholds=%.2f): %1.0f' % (t, false_positive))
print('Customers that we predict will not churn but they do churn (thresholds=%.2f): %1.0f' % (t, false_negative))
labels = ["true_positive", "false_positive", "true_negative", "false_negative"]
results = [true_positive, false_positive, true_negative, false_negative]
colors = ["lightgreen", "red", "green", "darkred"]
plt.pie(results, labels=labels, colors=colors, autopct='%1.0f%%')
confusion_matrix = np.array([
[true_negative, false_positive],
[false_negative, true_positive]
])
print("\nConfusion_matrix:\n",confusion_matrix)
print("\nConfusion_matrix %:\n",((confusion_matrix/confusion_matrix.sum()).round(2)))
#Precision #Recall
precission = true_positive / (true_positive + false_positive)
print("precission=",precission)
#Recall
recall = true_positive / (true_positive + false_negative)
print("recall=",recall)
#TP rate #FP rate
tpr = true_positive / (true_positive + false_negative)
print("tpr=",tpr)
fpr = false_positive / (false_positive + true_negative)
print("fpr=",fpr)
# thresholds / tpr # thresholds / tpr
def tpr_fpr_dataframe(y, y_pred1):
thresholds = np.linspace(0, 1, 101)
scores = []
for t in thresholds:
predict_positive = (y_pred1 >= t)
predict_negative = (y_pred1 < t)
actual_positive = (y >= t)
actual_negative = (y < t)
true_positive = (predict_positive & actual_positive).sum()
true_negative = (predict_negative & actual_negative).sum()
false_positive = (predict_positive & actual_negative).sum()
false_negative = (predict_negative & actual_positive).sum()
scores.append((t,true_positive,true_negative,false_positive,false_negative))
scores_columns = ["threshold","true_positive","true_negative","false_positive","false_negative"]
df_scores = pd.DataFrame(scores, columns=scores_columns)
df_scores["tpr"] = df_scores.true_positive / (df_scores.true_positive + df_scores.false_negative)
df_scores["fpr"] = df_scores.false_positive / (df_scores.false_positive + df_scores.true_negative)
return df_scores
df_total_scores = tpr_fpr_dataframe(y_test, y_pred)
display(df_total_scores[::10])
plt.plot(df_total_scores.threshold, df_total_scores.tpr, label="TPR")
plt.plot(df_total_scores.threshold, df_total_scores.fpr, label="FPR")
plt.xlabel("threshold")
plt.legend()
# Random prediction
np.random.seed(1)
y_rand = np.random.uniform(0,1,size=len(y_test))
((y_rand >= 0.5) == y_test).mean()
df_total_scores_random = tpr_fpr_dataframe(y_test, y_rand)
display(df_total_scores_random[::10])
plt.plot(df_total_scores_random.threshold, df_total_scores.tpr, label="TPR random")
plt.plot(df_total_scores_random.threshold, df_total_scores.fpr, label="FPR random")
plt.xlabel("threshold")
plt.legend()
# Ideal prediction for y_test data
# num_negative = (y_test == 0).sum()
# num_positive = (y_test == 1).sum()
num_negative = int(len(y_test) * 0.6) # 60% are not churn
num_positive = len(y_test)-num_negative # 40% are churn
display(num_negative, num_positive)
# % = 1-348/(1061+348)
y_ideal = np.repeat([0,1], [num_negative, num_positive])
y_ideal_pred = np.linspace(0, 1, len(y_test))
# ((y_ideal_pred >= 0.0.75301) == y_ideal).mean()
df_total_scores_ideal = tpr_fpr_dataframe(y_ideal, y_ideal_pred)
display(df_total_scores_ideal[::10])
plt.plot(df_total_scores_ideal.threshold, df_total_scores_ideal.tpr, label="TPR ideal")
plt.plot(df_total_scores_ideal.threshold, df_total_scores_ideal.fpr, label="FPR ideal")
plt.xlabel("threshold")
plt.legend()
# ROC Curve: FPT / TPR
plt.figure(figsize=(5,5))
plt.plot(df_total_scores.fpr, df_total_scores.tpr, label="ROC pred")
plt.plot(df_total_scores_random.fpr, df_total_scores_random.tpr, label="ROC random")
plt.plot(df_total_scores_ideal.fpr, df_total_scores_ideal.tpr, label="ROC ideal")
plt.xlabel("FPR")
plt.ylabel("TPR")
plt.legend()
import sklearn.metrics
df_total_scores_auc = metrics.auc(df_total_scores.fpr.fillna(1), df_total_scores.tpr)
df_total_scores_random_auc = metrics.auc(df_total_scores_random.fpr.fillna(1), df_total_scores_random.tpr)
df_total_scores_ideal_auc = metrics.auc(df_total_scores_ideal.fpr.fillna(1), df_total_scores_ideal.tpr)
print("df_total_scores_auc:", df_total_scores_auc)
print("df_total_scores_random_auc:", df_total_scores_random_auc)
print("df_total_scores_ideal_auc:", df_total_scores_ideal_auc)
# OR
fpr, tpr, thresholds = metrics.roc_curve(y_test, y_pred)
auc = metrics.auc(df_total_scores_random.fpr.fillna(1), df_total_scores_random.tpr)
# !pip install tqdm
import warnings
warnings.filterwarnings(action='once')
warnings.filterwarnings('ignore')
df_full_train, df_test = train_test_split(df, test_size=0.2, random_state=1)
#Cross Validation
from sklearn.model_selection import KFold
from tqdm.auto import tqdm
def train(dataFrame, y, C):
dicts = dataFrame.to_dict(orient="records")
dv = DictVectorizer(sparse=False)
X = dv.fit_transform(dicts)
model = linear_model.LogisticRegression(C=C)
model.fit(X, y)
return dv, model
def predict(dataFrame, dv, model):
dicts = dataFrame.to_dict(orient="records")
X = dv.transform(dicts)
y_pred = model.predict_proba(X)[:,1]
return y_pred
df_full_train_selected1 = df_full_train[categorical_columns+numerical_columns]
splits = 5
for C in tqdm([ 0.001, 0.01, 0.1, 0.5, 1, 5, 10], total=splits):
kf = KFold(n_splits=splits, shuffle=True, random_state=1)
auc_scores = []
for train_idx, val_idx in kf.split(df_full_train):
df_train_itter = df_full_train_selected1.iloc[train_idx]
df_val_itter = df_full_train_selected1.iloc[val_idx]
y_train_iter = df_full_train.iloc[train_idx].churn.values
y_val_iter = df_full_train.iloc[val_idx].churn.values
dv, model = train(df_train_itter, y_train_iter, C=C)
y_pred_iter = predict(df_val_itter, dv, model)
auc = metrics.roc_auc_score(y_val_iter, y_pred_iter)
auc_scores.append(auc)
print("C:%s, AUC mean: %.3f, AUC std: +-%.3f" % (C, np.mean(auc_scores), np.std(auc_scores)))
# train for 1 dataset:
dv, model = train(df_full_train_selected1, y_full_train, 1.0)
y_full_pred = predict(df_test, dv, model)
auc = metrics.roc_auc_score(y_test, y_full_pred)
print("C:%s, Full AUC mean: %.3f" % (1.0, auc))
| 0.608361 | 0.357371 |
```
import sys
sys.path.append('..')
import torch
import pandas as pd
import numpy as np
import pickle
import argparse
import networkx as nx
from collections import Counter
from torch_geometric.utils import dense_to_sparse, degree
import matplotlib.pyplot as plt
from src.gcn import GCNSynthetic
from src.utils.utils import normalize_adj, get_neighbourhood
```
### Syn1 dataset (BA houses) , best params so far: SGD + momentum=0.9, epochs=500, LR=0.1, beta=0.5
#### Uses correct version of symmetry constraint
#### For BA houses, class 0 = BA, class 1 = middle, class 2 = bottom, class 3 = top
```
header = ["node_idx", "new_idx", "cf_adj", "sub_adj", "y_pred_orig", "y_pred_new", "y_pred_new_actual",
"label", "num_nodes", "loss_total", "loss_pred", "loss_graph_dist"]
# For original model
dataset = "syn1"
hidden = 20
seed = 42
dropout = 0.0
# Load original dataset and model
with open("../data/gnn_explainer/{}.pickle".format(dataset), "rb") as f:
data = pickle.load(f)
adj = torch.Tensor(data["adj"]).squeeze() # Does not include self loops
features = torch.Tensor(data["feat"]).squeeze()
labels = torch.tensor(data["labels"]).squeeze()
idx_train = torch.tensor(data["train_idx"])
idx_test = torch.tensor(data["test_idx"])
edge_index = dense_to_sparse(adj)
norm_adj = normalize_adj(adj)
model = GCNSynthetic(nfeat=features.shape[1], nhid=hidden, nout=hidden,
nclass=len(labels.unique()), dropout=dropout)
model.load_state_dict(torch.load("../models/gcn_3layer_{}.pt".format(dataset)))
model.eval()
output = model(features, norm_adj)
y_pred_orig = torch.argmax(output, dim=1)
print("test set y_true counts: {}".format(np.unique(labels[idx_test].numpy(), return_counts=True)))
print("test set y_pred_orig counts: {}".format(np.unique(y_pred_orig[idx_test].numpy(), return_counts=True)))
print("Whole graph counts: {}".format(np.unique(labels.numpy(), return_counts=True)))
lr = 0.1
beta = 0.5
num_epochs = 500
mom = 0.9
# Load cf examples for test set
with open("../results/{}/correct_symm/SGD/{}_cf_examples_lr{}_beta{}_mom{}_epochs{}".format(dataset,
dataset, lr, beta, mom, num_epochs), "rb") as f:
cf_examples = pickle.load(f)
df_prep = []
for example in cf_examples:
if example != []:
df_prep.append(example[0])
df = pd.DataFrame(df_prep, columns=header)
print("Num cf examples found for best nesterov: {}/{}".format(len(df), len(idx_test)))
print("Average graph distance for best nesterov: {}".format(np.mean(df["loss_graph_dist"])))
# Add num edges to df
num_edges = []
for i in df.index:
num_edges.append(sum(sum(df["sub_adj"][i]))/2)
df["num_edges"] = num_edges
```
### FINAL NUMBERS
```
print("Num cf examples found: {}/{}".format(len(df), len(idx_test)))
print("Coverage: {}".format(len(df)/len(idx_test)))
print("Average graph distance: {}".format(np.mean(df["loss_graph_dist"])))
print("Average prop comp graph perturbed: {}".format(np.mean(df["loss_graph_dist"]/df["num_edges"])))
font = {'weight' : 'normal',
'size' : 18}
plt.rc('font', **font)
# Plot graph loss of cf examples
bins = [i+0.5 for i in range(11)]
# bins=[0, 20, 40, 60]
bins = [0, 10, 20, 30, 40, 50, 60, 70]
plt.hist(df["loss_graph_dist"], bins=bins, weights=np.ones(len(df))/len(df))
# plt.title("BA-SHAPES")
plt.xlabel("Explanation Size")
plt.xticks([0, 10, 20, 30, 40, 50, 60, 70])
plt.ylim(0, 1.1)
plt.ylabel("Prop CF examples")
# For accuracy, only look at motif nodes
df_motif = df[df["y_pred_orig"] != 0].reset_index(drop=True)
accuracy = []
# Get original predictions
dict_ypred_orig = dict(zip(sorted(np.concatenate((idx_train.numpy(), idx_test.numpy()))),
y_pred_orig.numpy()))
for i in range(len(df_motif)):
node_idx = df_motif["node_idx"][i]
new_idx = df_motif["new_idx"][i]
_, _, _, node_dict = get_neighbourhood(int(node_idx), edge_index, 4, features, labels)
# Confirm idx mapping is correct
if node_dict[node_idx] == df_motif["new_idx"][i]:
cf_adj = df_motif["cf_adj"][i]
sub_adj = df_motif["sub_adj"][i]
perturb = np.abs(cf_adj - sub_adj)
perturb_edges = np.nonzero(perturb) # Edge indices
nodes_involved = np.unique(np.concatenate((perturb_edges[0], perturb_edges[1]), axis=0))
perturb_nodes = nodes_involved[nodes_involved != new_idx] # Remove original node
# Retrieve original node idxs for original predictions
perturb_nodes_orig_idx = []
for j in perturb_nodes:
perturb_nodes_orig_idx.append([key for (key, value) in node_dict.items() if value == j])
perturb_nodes_orig_idx = np.array(perturb_nodes_orig_idx).flatten()
# Retrieve original predictions
perturb_nodes_orig_ypred = np.array([dict_ypred_orig[k] for k in perturb_nodes_orig_idx])
nodes_in_motif = perturb_nodes_orig_ypred[perturb_nodes_orig_ypred != 0]
prop_correct = len(nodes_in_motif)/len(perturb_nodes_orig_idx)
accuracy.append([node_idx, new_idx, perturb_nodes_orig_idx,
perturb_nodes_orig_ypred, nodes_in_motif, prop_correct])
df_accuracy = pd.DataFrame(accuracy, columns=["node_idx", "new_idx", "perturb_nodes_orig_idx",
"perturb_nodes_orig_ypred", "nodes_in_motif", "prop_correct"])
print("Accuracy", np.mean(df_accuracy["prop_correct"]))
```
### Dataset statistics
```
# Get full dataset statistics
full_dataset = pd.DataFrame()
idx_concat = torch.cat((idx_train, idx_test), 0)
full_dataset["node_idx"] = idx_concat
full_dataset["y_pred_orig"] = y_pred_orig[idx_concat]
full_dataset["label"] = labels[idx_concat]
full_dataset["node_degree"] = torch.sum(adj[idx_concat], dim=1).numpy()
num_nodes = []
sub_adj = []
sub_labels = []
for i in full_dataset["node_idx"]:
sub_adj0, _, sub_labels0, node_dict = get_neighbourhood(i,edge_index,4,
features,labels)
num_nodes.append(sub_adj0.shape[0])
# Need these for plotting later
sub_adj.append(sub_adj0.numpy())
# sub_labels.append(sub_labels0.numpy())
full_dataset["num_nodes"] = num_nodes
full_dataset["sub_adj"] = sub_adj
# Add num edges in computational graph
num_edges = []
for i in full_dataset.index:
num_edges.append(sum(sum(full_dataset["sub_adj"][i]))/2)
full_dataset["num_edges"] = num_edges
full_dataset.head()
print("Avg node degree: ", np.mean(full_dataset["node_degree"]))
print("Avg num nodes in computational graph: ", np.mean(full_dataset["num_nodes"]))
print("Avg num edges in computational graph: ", np.mean(full_dataset["num_edges"]))
```
|
github_jupyter
|
import sys
sys.path.append('..')
import torch
import pandas as pd
import numpy as np
import pickle
import argparse
import networkx as nx
from collections import Counter
from torch_geometric.utils import dense_to_sparse, degree
import matplotlib.pyplot as plt
from src.gcn import GCNSynthetic
from src.utils.utils import normalize_adj, get_neighbourhood
header = ["node_idx", "new_idx", "cf_adj", "sub_adj", "y_pred_orig", "y_pred_new", "y_pred_new_actual",
"label", "num_nodes", "loss_total", "loss_pred", "loss_graph_dist"]
# For original model
dataset = "syn1"
hidden = 20
seed = 42
dropout = 0.0
# Load original dataset and model
with open("../data/gnn_explainer/{}.pickle".format(dataset), "rb") as f:
data = pickle.load(f)
adj = torch.Tensor(data["adj"]).squeeze() # Does not include self loops
features = torch.Tensor(data["feat"]).squeeze()
labels = torch.tensor(data["labels"]).squeeze()
idx_train = torch.tensor(data["train_idx"])
idx_test = torch.tensor(data["test_idx"])
edge_index = dense_to_sparse(adj)
norm_adj = normalize_adj(adj)
model = GCNSynthetic(nfeat=features.shape[1], nhid=hidden, nout=hidden,
nclass=len(labels.unique()), dropout=dropout)
model.load_state_dict(torch.load("../models/gcn_3layer_{}.pt".format(dataset)))
model.eval()
output = model(features, norm_adj)
y_pred_orig = torch.argmax(output, dim=1)
print("test set y_true counts: {}".format(np.unique(labels[idx_test].numpy(), return_counts=True)))
print("test set y_pred_orig counts: {}".format(np.unique(y_pred_orig[idx_test].numpy(), return_counts=True)))
print("Whole graph counts: {}".format(np.unique(labels.numpy(), return_counts=True)))
lr = 0.1
beta = 0.5
num_epochs = 500
mom = 0.9
# Load cf examples for test set
with open("../results/{}/correct_symm/SGD/{}_cf_examples_lr{}_beta{}_mom{}_epochs{}".format(dataset,
dataset, lr, beta, mom, num_epochs), "rb") as f:
cf_examples = pickle.load(f)
df_prep = []
for example in cf_examples:
if example != []:
df_prep.append(example[0])
df = pd.DataFrame(df_prep, columns=header)
print("Num cf examples found for best nesterov: {}/{}".format(len(df), len(idx_test)))
print("Average graph distance for best nesterov: {}".format(np.mean(df["loss_graph_dist"])))
# Add num edges to df
num_edges = []
for i in df.index:
num_edges.append(sum(sum(df["sub_adj"][i]))/2)
df["num_edges"] = num_edges
print("Num cf examples found: {}/{}".format(len(df), len(idx_test)))
print("Coverage: {}".format(len(df)/len(idx_test)))
print("Average graph distance: {}".format(np.mean(df["loss_graph_dist"])))
print("Average prop comp graph perturbed: {}".format(np.mean(df["loss_graph_dist"]/df["num_edges"])))
font = {'weight' : 'normal',
'size' : 18}
plt.rc('font', **font)
# Plot graph loss of cf examples
bins = [i+0.5 for i in range(11)]
# bins=[0, 20, 40, 60]
bins = [0, 10, 20, 30, 40, 50, 60, 70]
plt.hist(df["loss_graph_dist"], bins=bins, weights=np.ones(len(df))/len(df))
# plt.title("BA-SHAPES")
plt.xlabel("Explanation Size")
plt.xticks([0, 10, 20, 30, 40, 50, 60, 70])
plt.ylim(0, 1.1)
plt.ylabel("Prop CF examples")
# For accuracy, only look at motif nodes
df_motif = df[df["y_pred_orig"] != 0].reset_index(drop=True)
accuracy = []
# Get original predictions
dict_ypred_orig = dict(zip(sorted(np.concatenate((idx_train.numpy(), idx_test.numpy()))),
y_pred_orig.numpy()))
for i in range(len(df_motif)):
node_idx = df_motif["node_idx"][i]
new_idx = df_motif["new_idx"][i]
_, _, _, node_dict = get_neighbourhood(int(node_idx), edge_index, 4, features, labels)
# Confirm idx mapping is correct
if node_dict[node_idx] == df_motif["new_idx"][i]:
cf_adj = df_motif["cf_adj"][i]
sub_adj = df_motif["sub_adj"][i]
perturb = np.abs(cf_adj - sub_adj)
perturb_edges = np.nonzero(perturb) # Edge indices
nodes_involved = np.unique(np.concatenate((perturb_edges[0], perturb_edges[1]), axis=0))
perturb_nodes = nodes_involved[nodes_involved != new_idx] # Remove original node
# Retrieve original node idxs for original predictions
perturb_nodes_orig_idx = []
for j in perturb_nodes:
perturb_nodes_orig_idx.append([key for (key, value) in node_dict.items() if value == j])
perturb_nodes_orig_idx = np.array(perturb_nodes_orig_idx).flatten()
# Retrieve original predictions
perturb_nodes_orig_ypred = np.array([dict_ypred_orig[k] for k in perturb_nodes_orig_idx])
nodes_in_motif = perturb_nodes_orig_ypred[perturb_nodes_orig_ypred != 0]
prop_correct = len(nodes_in_motif)/len(perturb_nodes_orig_idx)
accuracy.append([node_idx, new_idx, perturb_nodes_orig_idx,
perturb_nodes_orig_ypred, nodes_in_motif, prop_correct])
df_accuracy = pd.DataFrame(accuracy, columns=["node_idx", "new_idx", "perturb_nodes_orig_idx",
"perturb_nodes_orig_ypred", "nodes_in_motif", "prop_correct"])
print("Accuracy", np.mean(df_accuracy["prop_correct"]))
# Get full dataset statistics
full_dataset = pd.DataFrame()
idx_concat = torch.cat((idx_train, idx_test), 0)
full_dataset["node_idx"] = idx_concat
full_dataset["y_pred_orig"] = y_pred_orig[idx_concat]
full_dataset["label"] = labels[idx_concat]
full_dataset["node_degree"] = torch.sum(adj[idx_concat], dim=1).numpy()
num_nodes = []
sub_adj = []
sub_labels = []
for i in full_dataset["node_idx"]:
sub_adj0, _, sub_labels0, node_dict = get_neighbourhood(i,edge_index,4,
features,labels)
num_nodes.append(sub_adj0.shape[0])
# Need these for plotting later
sub_adj.append(sub_adj0.numpy())
# sub_labels.append(sub_labels0.numpy())
full_dataset["num_nodes"] = num_nodes
full_dataset["sub_adj"] = sub_adj
# Add num edges in computational graph
num_edges = []
for i in full_dataset.index:
num_edges.append(sum(sum(full_dataset["sub_adj"][i]))/2)
full_dataset["num_edges"] = num_edges
full_dataset.head()
print("Avg node degree: ", np.mean(full_dataset["node_degree"]))
print("Avg num nodes in computational graph: ", np.mean(full_dataset["num_nodes"]))
print("Avg num edges in computational graph: ", np.mean(full_dataset["num_edges"]))
| 0.566498 | 0.674329 |
<!--NOTEBOOK_HEADER-->
*This notebook contains course material from [CBE40455](https://jckantor.github.io/CBE40455) by
Jeffrey Kantor (jeff at nd.edu); the content is available [on Github](https://github.com/jckantor/CBE40455.git).
The text is released under the [CC-BY-NC-ND-4.0 license](https://creativecommons.org/licenses/by-nc-nd/4.0/legalcode),
and code is released under the [MIT license](https://opensource.org/licenses/MIT).*
<!--NAVIGATION-->
< [Log-Optimal Growth and the Kelly Criterion](http://nbviewer.jupyter.org/github/jckantor/CBE40455/blob/master/notebooks/07.08-Log-Optimal-Growth-and-the-Kelly-Criterion.ipynb) | [Contents](toc.ipynb) | [Optimization in Google Sheets](http://nbviewer.jupyter.org/github/jckantor/CBE40455/blob/master/notebooks/08.00-Optimization-in-Google-Sheets.ipynb) ><p><a href="https://colab.research.google.com/github/jckantor/CBE40455/blob/master/notebooks/07.09-Log-Optimal-Portfolios.ipynb"><img align="left" src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open in Colab" title="Open in Google Colaboratory"></a><p><a href="https://raw.githubusercontent.com/jckantor/CBE40455/master/notebooks/07.09-Log-Optimal-Portfolios.ipynb"><img align="left" src="https://img.shields.io/badge/Github-Download-blue.svg" alt="Download" title="Download Notebook"></a>
# Log-Optimal Portfolios
This notebook demonstrates the Kelly criterion and other phenomena associated with log-optimal growth.
## Initializations
```
%matplotlib notebook
import matplotlib.pyplot as plt
import numpy as np
import random
```
## Kelly's Criterion
In a nutshell, Kelly's criterion is to choose strategies that maximize expected log return.
$$\max E[\ln R]$$
where $R$ is total return. As we learned, Kelly's criterion has properties useful in the context of long-term investments.
## Example 1. Maximizing Return for a Game with Arbitrary Odds
Consider a game with two outcomes. For each \$1 wagered, a successful outcome with probability $p$ returns $b+1$ dollars. An unsuccessful outcome returns nothing. What fraction $w$ of our portfolio should we wager on each turn of the game?
There are two outcomes with returns
\begin{align*}
R_1 & = w(b+1) + 1 - w = 1+wb & \mbox{with probability }p\\
R_2 & = 1-w & \mbox{with probability }1-p
\end{align*}
The expected log return becomes
\begin{align*}
E[\ln R] & = p \ln R_1 + (1-p) \ln R_2 \\
& = p\ln(1+ wb) + (1-p)\ln(1-w)
\end{align*}
Applying Kelly's criterion, we seek a value for $w$ that maximizes $E[\ln R]$. Taking derivatives
\begin{align*}
\frac{\partial E[\ln R]}{\partial w} = \frac{pb}{1+w_{opt}b} - \frac{1-p}{1-w_{opt}} & = 0\\
\end{align*}
Solving for $w$
$$w_{opt} = \frac{p(b+1)-1}{b}$$
The growth rate is then the value of $E[\ln R]$ when $w = w_{opt}$, i.e.,
$$m = p\ln(1+ w_{opt}b) + (1-p)\ln(1-w_{opt})$$
You can test how well this works in the following cell. Fix $p$ and $b$, and let the code do a Monte Carlo simulation to show how well Kelly's criterion works.
```
%matplotlib inline
import numpy as np
import matplotlib.pyplot as plt
from numpy.random import uniform
p = 0.5075
b = 1
# Kelly criterion
w = (p*(b+1)-1)/b
# optimal growth rate
m = p*np.log(1+w*b) + (1-p)*np.log(1-w)
# number of plays to double wealth
K = int(np.log(2)/m)
# monte carlo simulation and plotting
for n in range(0,100):
W = [1]
for k in range(0,K):
if uniform() <= p:
W.append(W[-1]*(1+w*b))
else:
W.append(W[-1]*(1-w))
plt.semilogy(W,alpha=0.2)
plt.semilogy(np.linspace(0,K), np.exp(m*np.linspace(0,K)),'r',lw=3)
plt.title('Kelly Criterion w = ' + str(round(w,4)))
plt.xlabel('k')
plt.grid()
```
## Example 2. Betting Wheel
```
%matplotlib inline
import numpy as np
import matplotlib.pyplot as plt
w1 = np.linspace(0,1)
w2 = 0
w3 = 0
p1 = 1/2
p2 = 1/3
p3 = 1/6
R1 = 1 + 2*w1 - w2 - w3
R2 = 1 - w1 + w2 - w3
R3 = 1 - w1 - w2 + 5*w3
m = p1*np.log(R1) + p2*np.log(R2) + p3*np.log(R3)
plt.plot(w1,m)
plt.grid()
def wheel(w,N = 100):
w1,w2,w3 = w
```
## Example 3. Stock/Bond Portfolio in Continuous Time
```
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import datetime
from pandas_datareader import data, wb
from scipy.stats import norm
import requests
def get_symbol(symbol):
"""
get_symbol(symbol) uses Yahoo to look up a stock trading symbol and
return a description.
"""
url = "http://d.yimg.com/autoc.finance.yahoo.com/autoc?query={}®ion=1&lang=en".format(symbol)
result = requests.get(url).json()
for x in result['ResultSet']['Result']:
if x['symbol'] == symbol:
return x['name']
symbol = '^GSPC'
# end date is today
end = datetime.datetime.today().date()
# start date is three years prior
start = end-datetime.timedelta(1.5*365)
# get stock price data
S = data.DataReader(symbol,"yahoo",start,end)['Adj Close']
rlin = (S - S.shift(1))/S.shift(1)
rlog = np.log(S/S.shift(1))
rlin = rlin.dropna()
rlog = rlog.dropna()
# plot data
plt.figure(figsize=(10,6))
plt.subplot(3,1,1)
S.plot(title=get_symbol(symbol))
plt.ylabel('Adjusted Close')
plt.grid()
plt.subplot(3,1,2)
rlin.plot()
plt.title('Linear Returns (daily)')
plt.grid()
plt.tight_layout()
plt.subplot(3,1,3)
rlog.plot()
plt.title('Log Returns (daily)')
plt.grid()
plt.tight_layout()
print('Linear Returns')
mu,sigma = norm.fit(rlin)
print(' mu = {0:12.8f} (annualized = {1:.2f}%)'.format(mu,100*252*mu))
print('sigma = {0:12.8f} (annualized = {1:.2f}%)'.format(sigma,100*np.sqrt(252)*sigma))
print()
print('Log Returns')
nu,sigma = norm.fit(rlog)
print(' nu = {0:12.8f} (annualized = {1:.2f}%)'.format(nu,100*252*nu))
print('sigma = {0:12.8f} (annualized = {1:.2f}%)'.format(sigma,100*np.sqrt(252)*sigma))
mu = 252*mu
nu = 252*nu
sigma = np.sqrt(252)*sigma
rf = 0.04
mu = 0.08
sigma = 0.3
w = (mu-rf)/sigma**2
nu_opt = rf + (mu-rf)**2/2/sigma/sigma
sigma_opt = np.sqrt(mu-rf)/sigma
print(w,nu_opt,sigma_opt)
```
## Volatility Pumping
```
# payoffs for two states
u = 1.059
d = 1/u
p = 0.54
rf = 0.004
K = 100
ElnR = p*np.log(u) + (1-p)*np.log(d)
print("Expected return = {:0.5}".format(ElnR))
Z = np.array([float(random.random() <= p) for _ in range(0,K)])
R = d + (u-d)*Z
S = np.cumprod(np.concatenate(([1],R)))
ElnR = lambda alpha: p*np.log(alpha*u +(1-alpha)*np.exp(rf)) + \
(1-p)*np.log(alpha*d + (1-alpha)*np.exp(rf))
a = np.linspace(0,1)
plt.plot(a,map(ElnR,a))
from scipy.optimize import fminbound
alpha = fminbound(lambda(alpha): -ElnR(alpha),0,1)
print alpha
#plt.plot(alpha, ElnR(alpha),'r.',ms=10)
R = alpha*d + (1-alpha) + alpha*(u-d)*Z
S2 = np.cumprod(np.concatenate(([1],R)))
plt.figure(figsize=(10,4))
plt.plot(range(0,K+1),S,range(0,K+1),S2)
plt.legend(['Stock','Stock + Cash']);
```
<!--NAVIGATION-->
< [Log-Optimal Growth and the Kelly Criterion](http://nbviewer.jupyter.org/github/jckantor/CBE40455/blob/master/notebooks/07.08-Log-Optimal-Growth-and-the-Kelly-Criterion.ipynb) | [Contents](toc.ipynb) | [Optimization in Google Sheets](http://nbviewer.jupyter.org/github/jckantor/CBE40455/blob/master/notebooks/08.00-Optimization-in-Google-Sheets.ipynb) ><p><a href="https://colab.research.google.com/github/jckantor/CBE40455/blob/master/notebooks/07.09-Log-Optimal-Portfolios.ipynb"><img align="left" src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open in Colab" title="Open in Google Colaboratory"></a><p><a href="https://raw.githubusercontent.com/jckantor/CBE40455/master/notebooks/07.09-Log-Optimal-Portfolios.ipynb"><img align="left" src="https://img.shields.io/badge/Github-Download-blue.svg" alt="Download" title="Download Notebook"></a>
|
github_jupyter
|
%matplotlib notebook
import matplotlib.pyplot as plt
import numpy as np
import random
%matplotlib inline
import numpy as np
import matplotlib.pyplot as plt
from numpy.random import uniform
p = 0.5075
b = 1
# Kelly criterion
w = (p*(b+1)-1)/b
# optimal growth rate
m = p*np.log(1+w*b) + (1-p)*np.log(1-w)
# number of plays to double wealth
K = int(np.log(2)/m)
# monte carlo simulation and plotting
for n in range(0,100):
W = [1]
for k in range(0,K):
if uniform() <= p:
W.append(W[-1]*(1+w*b))
else:
W.append(W[-1]*(1-w))
plt.semilogy(W,alpha=0.2)
plt.semilogy(np.linspace(0,K), np.exp(m*np.linspace(0,K)),'r',lw=3)
plt.title('Kelly Criterion w = ' + str(round(w,4)))
plt.xlabel('k')
plt.grid()
%matplotlib inline
import numpy as np
import matplotlib.pyplot as plt
w1 = np.linspace(0,1)
w2 = 0
w3 = 0
p1 = 1/2
p2 = 1/3
p3 = 1/6
R1 = 1 + 2*w1 - w2 - w3
R2 = 1 - w1 + w2 - w3
R3 = 1 - w1 - w2 + 5*w3
m = p1*np.log(R1) + p2*np.log(R2) + p3*np.log(R3)
plt.plot(w1,m)
plt.grid()
def wheel(w,N = 100):
w1,w2,w3 = w
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import datetime
from pandas_datareader import data, wb
from scipy.stats import norm
import requests
def get_symbol(symbol):
"""
get_symbol(symbol) uses Yahoo to look up a stock trading symbol and
return a description.
"""
url = "http://d.yimg.com/autoc.finance.yahoo.com/autoc?query={}®ion=1&lang=en".format(symbol)
result = requests.get(url).json()
for x in result['ResultSet']['Result']:
if x['symbol'] == symbol:
return x['name']
symbol = '^GSPC'
# end date is today
end = datetime.datetime.today().date()
# start date is three years prior
start = end-datetime.timedelta(1.5*365)
# get stock price data
S = data.DataReader(symbol,"yahoo",start,end)['Adj Close']
rlin = (S - S.shift(1))/S.shift(1)
rlog = np.log(S/S.shift(1))
rlin = rlin.dropna()
rlog = rlog.dropna()
# plot data
plt.figure(figsize=(10,6))
plt.subplot(3,1,1)
S.plot(title=get_symbol(symbol))
plt.ylabel('Adjusted Close')
plt.grid()
plt.subplot(3,1,2)
rlin.plot()
plt.title('Linear Returns (daily)')
plt.grid()
plt.tight_layout()
plt.subplot(3,1,3)
rlog.plot()
plt.title('Log Returns (daily)')
plt.grid()
plt.tight_layout()
print('Linear Returns')
mu,sigma = norm.fit(rlin)
print(' mu = {0:12.8f} (annualized = {1:.2f}%)'.format(mu,100*252*mu))
print('sigma = {0:12.8f} (annualized = {1:.2f}%)'.format(sigma,100*np.sqrt(252)*sigma))
print()
print('Log Returns')
nu,sigma = norm.fit(rlog)
print(' nu = {0:12.8f} (annualized = {1:.2f}%)'.format(nu,100*252*nu))
print('sigma = {0:12.8f} (annualized = {1:.2f}%)'.format(sigma,100*np.sqrt(252)*sigma))
mu = 252*mu
nu = 252*nu
sigma = np.sqrt(252)*sigma
rf = 0.04
mu = 0.08
sigma = 0.3
w = (mu-rf)/sigma**2
nu_opt = rf + (mu-rf)**2/2/sigma/sigma
sigma_opt = np.sqrt(mu-rf)/sigma
print(w,nu_opt,sigma_opt)
# payoffs for two states
u = 1.059
d = 1/u
p = 0.54
rf = 0.004
K = 100
ElnR = p*np.log(u) + (1-p)*np.log(d)
print("Expected return = {:0.5}".format(ElnR))
Z = np.array([float(random.random() <= p) for _ in range(0,K)])
R = d + (u-d)*Z
S = np.cumprod(np.concatenate(([1],R)))
ElnR = lambda alpha: p*np.log(alpha*u +(1-alpha)*np.exp(rf)) + \
(1-p)*np.log(alpha*d + (1-alpha)*np.exp(rf))
a = np.linspace(0,1)
plt.plot(a,map(ElnR,a))
from scipy.optimize import fminbound
alpha = fminbound(lambda(alpha): -ElnR(alpha),0,1)
print alpha
#plt.plot(alpha, ElnR(alpha),'r.',ms=10)
R = alpha*d + (1-alpha) + alpha*(u-d)*Z
S2 = np.cumprod(np.concatenate(([1],R)))
plt.figure(figsize=(10,4))
plt.plot(range(0,K+1),S,range(0,K+1),S2)
plt.legend(['Stock','Stock + Cash']);
| 0.501953 | 0.938294 |
# Load Dependencies and Raw Data
```
import pandas as pd
import numpy as np
import dill, pickle
import copy
from collections import Counter
import itertools
from scipy import stats
import matplotlib.pyplot as plt
from sklearn.cluster import KMeans
from sklearn.cluster import AgglomerativeClustering
from sklearn.decomposition import FactorAnalysis
from skrebate import ReliefF, MultiSURF, MultiSURFstar
from sklearn.feature_selection import f_classif
from sklearn.decomposition import PCA
from sklearn.manifold import TSNE
from sklearn import metrics
from sklearn.metrics import adjusted_rand_score, rand_score
from sklearn.metrics.cluster import pair_confusion_matrix
from sklearn.preprocessing import StandardScaler, MinMaxScaler, RobustScaler
data = pd.read_excel('../data/GC-MS_data.xlsx')
# counts
ID_num = np.where(data.ID == 'Healthy', 0, 1)
data.insert(1, 'ID_num', ID_num)
```
# User Defined Functions
```
def do_factor_analysis(dataset):
fa = FactorAnalysis().fit(dataset)
return fa.mean_, fa.get_covariance()
def bhatt_dist(m1,cov1,m2,cov2):
cov = (1/2) * (cov1 + cov2)
Term1 = (1/8) * (m1 - m2).T @ np.linalg.inv(cov) @ (m1 - m2)
Term2 = (1 / 2) * np.log(np.linalg.det(cov) / np.sqrt(np.linalg.det(cov1) * np.linalg.det(cov2)))
return Term1+Term2, Term1, Term2
```
# Full Dataset
We need a metric that is better than randIndex. randIndex is label agnostic. in otherwords, if 2 instances for the negative class are clustered together it is a positive outcome for randIndex even if they are clustered in the same cluster as the positive instances.
Need to try out log transorfm of the data
```
data.head()
Counter(data.ID)
data.shape
```
# Scale the data
```
X_raw_all = data.values[:,2:]
X_scaled_all = StandardScaler().fit_transform(X_raw_all)
data_scaled_all = pd.DataFrame(X_scaled_all, columns = data.columns[2:])
data_scaled_all.insert(0, 'ID', data.ID.values)
data_scaled_all.insert(1, 'ID_num', data.ID_num.values)
data_scaled_all.head()
data_healthy_all_df = data_scaled_all.loc[data_scaled_all.ID == 'Healthy']
data_asthma_all_df = data_scaled_all.loc[data_scaled_all.ID == 'Asthmatic']
data_healthy_all = data_healthy_all_df.values[:,2:]
data_asthma_all = data_asthma_all_df.values[:, 2:]
healthy_mean, healthy_cov = do_factor_analysis(data_healthy_all)
asthma_mean, asthma_cov = do_factor_analysis(data_asthma_all)
dist, t1, t2 = bhatt_dist(healthy_mean, healthy_cov, asthma_mean, asthma_cov)
print(dist)
print(np.linalg.det(healthy_cov))
print(np.linalg.det(asthma_cov))
```
# Relief Methods
```
data_scaled_df = data_scaled_all
data_scaled_df.head()
```
## Relief-F
```
fs = ReliefF(discrete_threshold = 5, n_jobs=1)
fs.fit(data_scaled_df.values[:,2:].astype(float), data_scaled_df.ID_num.values)
feature_scores = fs.feature_importances_
feature_ids = np.where(feature_scores>=0)[0]
selected_features = np.array(data_scaled_df.columns[2:][feature_ids])
X_reliefF = data_scaled_df.values[:,2:][:,feature_ids]
X_reliefF.shape
X_reliefF_df = pd.DataFrame(X_reliefF, columns = selected_features)
X_reliefF_df.insert(0, 'ID', data.ID.values)
X_reliefF_df.head()
data_healthy_df = X_reliefF_df.loc[X_reliefF_df.ID == 'Healthy']
data_asthma_df = X_reliefF_df.loc[X_reliefF_df.ID == 'Asthmatic']
data_healthy = data_healthy_df.values[:,1:]
data_asthma = data_asthma_df.values[:, 1:]
healthy_mean, healthy_cov = do_factor_analysis(data_healthy)
asthma_mean, asthma_cov = do_factor_analysis(data_asthma)
dist, t1, t2 = bhatt_dist(healthy_mean, healthy_cov, asthma_mean, asthma_cov)
print(dist)
print(np.linalg.det(healthy_cov))
print(np.linalg.det(asthma_cov))
```
## MultiSURF
```
fs = MultiSURF(discrete_threshold = 5, n_jobs=1)
fs.fit(data_scaled_df.values[:,2:].astype(float), data_scaled_df.ID_num.values)
feature_scores = fs.feature_importances_
feature_ids = np.where(feature_scores>=0)[0]
selected_features = np.array(data_scaled_df.columns[2:][feature_ids])
X_MultiSURF = data_scaled_df.values[:,2:][:,feature_ids]
X_MultiSURF.shape
X_MultiSURF_df = pd.DataFrame(X_MultiSURF, columns = selected_features)
X_MultiSURF_df.insert(0, 'ID', data.ID.values)
X_MultiSURF_df.head()
data_healthy_df = X_MultiSURF_df.loc[X_reliefF_df.ID == 'Healthy']
data_asthma_df = X_MultiSURF_df.loc[X_reliefF_df.ID == 'Asthmatic']
data_healthy = data_healthy_df.values[:,1:]
data_asthma = data_asthma_df.values[:, 1:]
healthy_mean, healthy_cov = do_factor_analysis(data_healthy)
asthma_mean, asthma_cov = do_factor_analysis(data_asthma)
dist, t1, t2 = bhatt_dist(healthy_mean, healthy_cov, asthma_mean, asthma_cov)
print(dist)
print(np.linalg.det(healthy_cov))
print(np.linalg.det(asthma_cov))
```
## MultiSURFStar
```
fs = MultiSURFstar(discrete_threshold = 5, n_jobs=1)
fs.fit(data_scaled_df.values[:,2:].astype(float), data_scaled_df.ID_num.values)
feature_scores = fs.feature_importances_
feature_ids = np.where(feature_scores>=0)[0]
selected_features = np.array(data_scaled_df.columns[2:][feature_ids])
X_MultiSURFStar = data_scaled_df.values[:,2:][:,feature_ids]
X_MultiSURFStar.shape
X_MultiSURFStar_df = pd.DataFrame(X_MultiSURFStar, columns = selected_features)
X_MultiSURFStar_df.insert(0, 'ID', data.ID.values)
X_MultiSURFStar_df.head()
data_healthy_df = X_MultiSURFStar_df.loc[X_MultiSURFStar_df.ID == 'Healthy']
data_asthma_df = X_MultiSURFStar_df.loc[X_MultiSURFStar_df.ID == 'Asthmatic']
data_healthy = data_healthy_df.values[:,1:]
data_asthma = data_asthma_df.values[:, 1:]
healthy_mean, healthy_cov = do_factor_analysis(data_healthy)
asthma_mean, asthma_cov = do_factor_analysis(data_asthma)
dist, t1, t2 = bhatt_dist(healthy_mean, healthy_cov, asthma_mean, asthma_cov)
print(dist)
print(np.linalg.det(healthy_cov))
print(np.linalg.det(asthma_cov))
```
# Univariate Statistical Feature Selection
## Anova
```
f,p = f_classif(data_scaled_df.values[:,2:].astype(float), data.ID_num.values)
feature_ids = np.where(p<=0.05)[0]
selected_features = np.array(data_scaled_df.columns[2:][feature_ids])
X_anova = data_scaled_df.values[:,2:][:,feature_ids]
X_anova.shape
X_anova_df = pd.DataFrame(X_anova, columns = selected_features)
X_anova_df.insert(0, 'ID', data.ID.values)
X_anova_df.head()
data_healthy_df = X_anova_df.loc[X_anova_df.ID == 'Healthy']
data_asthma_df = X_anova_df.loc[X_anova_df.ID == 'Asthmatic']
data_healthy = data_healthy_df.values[:,1:]
data_asthma = data_asthma_df.values[:, 1:]
healthy_mean, healthy_cov = do_factor_analysis(data_healthy)
asthma_mean, asthma_cov = do_factor_analysis(data_asthma)
dist, t1, t2 = bhatt_dist(healthy_mean, healthy_cov, asthma_mean, asthma_cov)
print(dist)
print(np.linalg.det(healthy_cov))
print(np.linalg.det(asthma_cov))
```
# Combinations
## Anova + Relief-F
```
X_anova.shape
fs = ReliefF(discrete_threshold = 5, n_jobs=1)
fs.fit(X_anova.astype(float), data.ID_num.values)
feature_scores = fs.feature_importances_
feature_ids = np.where(feature_scores>=0)[0]
selected_features = selected_features[feature_ids]
X_ano_reliefF = X_anova[:,feature_ids]
X_ano_reliefF.shape
X_anova_relief_df = pd.DataFrame(X_ano_reliefF, columns = selected_features)
X_anova_relief_df.insert(0, 'ID', data.ID.values)
X_anova_relief_df.head()
data_healthy_df = X_anova_relief_df.loc[X_anova_relief_df.ID == 'Healthy']
data_asthma_df = X_anova_relief_df.loc[X_anova_relief_df.ID == 'Asthmatic']
data_healthy = data_healthy_df.values[:,1:]
data_asthma = data_asthma_df.values[:, 1:]
healthy_mean, healthy_cov = do_factor_analysis(data_healthy)
asthma_mean, asthma_cov = do_factor_analysis(data_asthma)
dist, t1, t2 = bhatt_dist(healthy_mean, healthy_cov, asthma_mean, asthma_cov)
print(dist)
print(np.linalg.det(healthy_cov))
print(np.linalg.det(asthma_cov))
```
# Dataset with linearly correlated features removed
```
with open('../data/independent_features.pik', "rb") as f:
independent_features = dill.load(f)
X_no_corr_df = independent_features['X_no_corr_df']
X_no_corr_df.shape
X_no_corr_df.head()
X_scaled = StandardScaler().fit_transform(X_no_corr_df.values)
data_scaled_df = pd.DataFrame(X_scaled, columns = X_no_corr_df.columns)
data_scaled_df.insert(0, 'ID', data.ID.values)
data_scaled_df.insert(1, 'ID_num', data.ID_num.values)
data_scaled_df.head()
data_healthy_df = data_scaled_df.loc[data_scaled_df.ID == 'Healthy']
data_asthma_df = data_scaled_df.loc[data_scaled_df.ID == 'Asthmatic']
data_healthy = data_healthy_df.values[:,2:]
data_asthma = data_asthma_df.values[:, 2:]
healthy_mean, healthy_cov = do_factor_analysis(data_healthy)
asthma_mean, asthma_cov = do_factor_analysis(data_asthma)
dist, t1, t2 = bhatt_dist(healthy_mean, healthy_cov, asthma_mean, asthma_cov)
dist
```
# Relief Methods
```
data_scaled_df.head()
```
## Relief-F
```
fs = ReliefF(discrete_threshold = 5, n_jobs=1)
fs.fit(data_scaled_df.values[:,2:].astype(float), data_scaled_df.ID_num.values)
feature_scores = fs.feature_importances_
feature_ids = np.where(feature_scores>=0)[0]
selected_features = np.array(X_no_corr_df.columns[feature_ids])
X_reliefF = data_scaled_df.values[:,2:][:,feature_ids]
X_reliefF.shape
X_reliefF_df = pd.DataFrame(X_reliefF, columns = selected_features)
X_reliefF_df.insert(0, 'ID', data.ID.values)
X_reliefF_df.head()
data_healthy_df = X_reliefF_df.loc[X_reliefF_df.ID == 'Healthy']
data_asthma_df = X_reliefF_df.loc[X_reliefF_df.ID == 'Asthmatic']
data_healthy = data_healthy_df.values[:,1:]
data_asthma = data_asthma_df.values[:, 1:]
healthy_mean, healthy_cov = do_factor_analysis(data_healthy)
asthma_mean, asthma_cov = do_factor_analysis(data_asthma)
dist, t1, t2 = bhatt_dist(healthy_mean, healthy_cov, asthma_mean, asthma_cov)
np.linalg.det(healthy_cov)
```
## MultiSURF
```
fs = MultiSURF(discrete_threshold = 5, n_jobs=1)
fs.fit(data_scaled_df.values[:,2:].astype(float), data_scaled_df.ID_num.values)
feature_scores = fs.feature_importances_
feature_ids = np.where(feature_scores>=0)[0]
selected_features = np.array(X_no_corr_df.columns[feature_ids])
X_MultiSURF = data_scaled_df.values[:,2:][:,feature_ids]
X_MultiSURF.shape
X_MultiSURF_df = pd.DataFrame(X_MultiSURF, columns = selected_features)
X_MultiSURF_df.insert(0, 'ID', data.ID.values)
X_MultiSURF_df.head()
data_healthy_df = X_MultiSURF_df.loc[X_reliefF_df.ID == 'Healthy']
data_asthma_df = X_MultiSURF_df.loc[X_reliefF_df.ID == 'Asthmatic']
data_healthy = data_healthy_df.values[:,1:]
data_asthma = data_asthma_df.values[:, 1:]
healthy_mean, healthy_cov = do_factor_analysis(data_healthy)
asthma_mean, asthma_cov = do_factor_analysis(data_asthma)
dist, t1, t2 = bhatt_dist(healthy_mean, healthy_cov, asthma_mean, asthma_cov)
dist
```
## MultiSURFStar
```
fs = MultiSURFstar(discrete_threshold = 5, n_jobs=1)
fs.fit(data_scaled_df.values[:,2:].astype(float), data_scaled_df.ID_num.values)
feature_scores = fs.feature_importances_
feature_ids = np.where(feature_scores>=0)[0]
selected_features = np.array(X_no_corr_df.columns[feature_ids])
X_MultiSURFStar = data_scaled_df.values[:,2:][:,feature_ids]
X_MultiSURFStar.shape
X_MultiSURFStar_df = pd.DataFrame(X_MultiSURFStar, columns = selected_features)
X_MultiSURFStar_df.insert(0, 'ID', data.ID.values)
X_MultiSURFStar_df.head()
data_healthy_df = X_MultiSURFStar_df.loc[X_MultiSURFStar_df.ID == 'Healthy']
data_asthma_df = X_MultiSURFStar_df.loc[X_MultiSURFStar_df.ID == 'Asthmatic']
data_healthy = data_healthy_df.values[:,1:]
data_asthma = data_asthma_df.values[:, 1:]
healthy_mean, healthy_cov = do_factor_analysis(data_healthy)
asthma_mean, asthma_cov = do_factor_analysis(data_asthma)
dist, t1, t2 = bhatt_dist(healthy_mean, healthy_cov, asthma_mean, asthma_cov)
dist
```
# Univariate Statistical Feature Selection
## Anova
```
f,p = f_classif(data_scaled_df.values[:,2:].astype(float), data.ID_num.values)
feature_ids = np.where(p<=0.05)[0]
selected_features = np.array(X_no_corr_df.columns[feature_ids])
X_anova = data_scaled_df.values[:,2:][:,feature_ids]
X_anova.shape
X_anova_df = pd.DataFrame(X_anova, columns = selected_features)
X_anova_df.insert(0, 'ID', data.ID.values)
X_anova_df.head()
data_healthy_df = X_anova_df.loc[X_anova_df.ID == 'Healthy']
data_asthma_df = X_anova_df.loc[X_anova_df.ID == 'Asthmatic']
data_healthy = data_healthy_df.values[:,1:]
data_asthma = data_asthma_df.values[:, 1:]
healthy_mean, healthy_cov = do_factor_analysis(data_healthy)
asthma_mean, asthma_cov = do_factor_analysis(data_asthma)
dist, t1, t2 = bhatt_dist(healthy_mean, healthy_cov, asthma_mean, asthma_cov)
dist
np.linalg.det(healthy_cov)
np.linalg.det(asthma_cov)
```
# Combinations
## Anova + Relief-F
```
X_anova.shape
fs = ReliefF(discrete_threshold = 5, n_jobs=1)
fs.fit(X_anova.astype(float), data.ID_num.values)
feature_scores = fs.feature_importances_
feature_ids = np.where(feature_scores>=0)[0]
selected_features = selected_features[feature_ids]
X_ano_reliefF = X_anova[:,feature_ids]
X_ano_reliefF.shape
X_anova_relief_df = pd.DataFrame(X_ano_reliefF, columns = selected_features)
X_anova_relief_df.insert(0, 'ID', data.ID.values)
X_anova_relief_df.head()
data_healthy_df = X_anova_relief_df.loc[X_anova_relief_df.ID == 'Healthy']
data_asthma_df = X_anova_relief_df.loc[X_anova_relief_df.ID == 'Asthmatic']
data_healthy = data_healthy_df.values[:,1:]
data_asthma = data_asthma_df.values[:, 1:]
data_scaled_df.head()
import random
random_feat_ids = list(range(681))
random.shuffle(random_feat_ids)
data_scaled_df.head()
data_healthy = data_scaled_df.loc[data_scaled_df.ID == 'Healthy'].values[:,2:]
data_asthma = data_scaled_df.loc[data_scaled_df.ID == 'Asthmatic'].values[:,2:]
healthy_mean, healthy_cov = do_factor_analysis(data_healthy[:,random_feat_ids[:30]])
asthma_mean, asthma_cov = do_factor_analysis(data_asthma[:,random_feat_ids[:30]])
dist, t1, t2 = bhatt_dist(healthy_mean, healthy_cov, asthma_mean, asthma_cov)
print(dist)
print(np.linalg.det(healthy_cov))
print(np.linalg.det(asthma_cov))
t2
np.linalg.det(asthma_cov)
```
|
github_jupyter
|
import pandas as pd
import numpy as np
import dill, pickle
import copy
from collections import Counter
import itertools
from scipy import stats
import matplotlib.pyplot as plt
from sklearn.cluster import KMeans
from sklearn.cluster import AgglomerativeClustering
from sklearn.decomposition import FactorAnalysis
from skrebate import ReliefF, MultiSURF, MultiSURFstar
from sklearn.feature_selection import f_classif
from sklearn.decomposition import PCA
from sklearn.manifold import TSNE
from sklearn import metrics
from sklearn.metrics import adjusted_rand_score, rand_score
from sklearn.metrics.cluster import pair_confusion_matrix
from sklearn.preprocessing import StandardScaler, MinMaxScaler, RobustScaler
data = pd.read_excel('../data/GC-MS_data.xlsx')
# counts
ID_num = np.where(data.ID == 'Healthy', 0, 1)
data.insert(1, 'ID_num', ID_num)
def do_factor_analysis(dataset):
fa = FactorAnalysis().fit(dataset)
return fa.mean_, fa.get_covariance()
def bhatt_dist(m1,cov1,m2,cov2):
cov = (1/2) * (cov1 + cov2)
Term1 = (1/8) * (m1 - m2).T @ np.linalg.inv(cov) @ (m1 - m2)
Term2 = (1 / 2) * np.log(np.linalg.det(cov) / np.sqrt(np.linalg.det(cov1) * np.linalg.det(cov2)))
return Term1+Term2, Term1, Term2
data.head()
Counter(data.ID)
data.shape
X_raw_all = data.values[:,2:]
X_scaled_all = StandardScaler().fit_transform(X_raw_all)
data_scaled_all = pd.DataFrame(X_scaled_all, columns = data.columns[2:])
data_scaled_all.insert(0, 'ID', data.ID.values)
data_scaled_all.insert(1, 'ID_num', data.ID_num.values)
data_scaled_all.head()
data_healthy_all_df = data_scaled_all.loc[data_scaled_all.ID == 'Healthy']
data_asthma_all_df = data_scaled_all.loc[data_scaled_all.ID == 'Asthmatic']
data_healthy_all = data_healthy_all_df.values[:,2:]
data_asthma_all = data_asthma_all_df.values[:, 2:]
healthy_mean, healthy_cov = do_factor_analysis(data_healthy_all)
asthma_mean, asthma_cov = do_factor_analysis(data_asthma_all)
dist, t1, t2 = bhatt_dist(healthy_mean, healthy_cov, asthma_mean, asthma_cov)
print(dist)
print(np.linalg.det(healthy_cov))
print(np.linalg.det(asthma_cov))
data_scaled_df = data_scaled_all
data_scaled_df.head()
fs = ReliefF(discrete_threshold = 5, n_jobs=1)
fs.fit(data_scaled_df.values[:,2:].astype(float), data_scaled_df.ID_num.values)
feature_scores = fs.feature_importances_
feature_ids = np.where(feature_scores>=0)[0]
selected_features = np.array(data_scaled_df.columns[2:][feature_ids])
X_reliefF = data_scaled_df.values[:,2:][:,feature_ids]
X_reliefF.shape
X_reliefF_df = pd.DataFrame(X_reliefF, columns = selected_features)
X_reliefF_df.insert(0, 'ID', data.ID.values)
X_reliefF_df.head()
data_healthy_df = X_reliefF_df.loc[X_reliefF_df.ID == 'Healthy']
data_asthma_df = X_reliefF_df.loc[X_reliefF_df.ID == 'Asthmatic']
data_healthy = data_healthy_df.values[:,1:]
data_asthma = data_asthma_df.values[:, 1:]
healthy_mean, healthy_cov = do_factor_analysis(data_healthy)
asthma_mean, asthma_cov = do_factor_analysis(data_asthma)
dist, t1, t2 = bhatt_dist(healthy_mean, healthy_cov, asthma_mean, asthma_cov)
print(dist)
print(np.linalg.det(healthy_cov))
print(np.linalg.det(asthma_cov))
fs = MultiSURF(discrete_threshold = 5, n_jobs=1)
fs.fit(data_scaled_df.values[:,2:].astype(float), data_scaled_df.ID_num.values)
feature_scores = fs.feature_importances_
feature_ids = np.where(feature_scores>=0)[0]
selected_features = np.array(data_scaled_df.columns[2:][feature_ids])
X_MultiSURF = data_scaled_df.values[:,2:][:,feature_ids]
X_MultiSURF.shape
X_MultiSURF_df = pd.DataFrame(X_MultiSURF, columns = selected_features)
X_MultiSURF_df.insert(0, 'ID', data.ID.values)
X_MultiSURF_df.head()
data_healthy_df = X_MultiSURF_df.loc[X_reliefF_df.ID == 'Healthy']
data_asthma_df = X_MultiSURF_df.loc[X_reliefF_df.ID == 'Asthmatic']
data_healthy = data_healthy_df.values[:,1:]
data_asthma = data_asthma_df.values[:, 1:]
healthy_mean, healthy_cov = do_factor_analysis(data_healthy)
asthma_mean, asthma_cov = do_factor_analysis(data_asthma)
dist, t1, t2 = bhatt_dist(healthy_mean, healthy_cov, asthma_mean, asthma_cov)
print(dist)
print(np.linalg.det(healthy_cov))
print(np.linalg.det(asthma_cov))
fs = MultiSURFstar(discrete_threshold = 5, n_jobs=1)
fs.fit(data_scaled_df.values[:,2:].astype(float), data_scaled_df.ID_num.values)
feature_scores = fs.feature_importances_
feature_ids = np.where(feature_scores>=0)[0]
selected_features = np.array(data_scaled_df.columns[2:][feature_ids])
X_MultiSURFStar = data_scaled_df.values[:,2:][:,feature_ids]
X_MultiSURFStar.shape
X_MultiSURFStar_df = pd.DataFrame(X_MultiSURFStar, columns = selected_features)
X_MultiSURFStar_df.insert(0, 'ID', data.ID.values)
X_MultiSURFStar_df.head()
data_healthy_df = X_MultiSURFStar_df.loc[X_MultiSURFStar_df.ID == 'Healthy']
data_asthma_df = X_MultiSURFStar_df.loc[X_MultiSURFStar_df.ID == 'Asthmatic']
data_healthy = data_healthy_df.values[:,1:]
data_asthma = data_asthma_df.values[:, 1:]
healthy_mean, healthy_cov = do_factor_analysis(data_healthy)
asthma_mean, asthma_cov = do_factor_analysis(data_asthma)
dist, t1, t2 = bhatt_dist(healthy_mean, healthy_cov, asthma_mean, asthma_cov)
print(dist)
print(np.linalg.det(healthy_cov))
print(np.linalg.det(asthma_cov))
f,p = f_classif(data_scaled_df.values[:,2:].astype(float), data.ID_num.values)
feature_ids = np.where(p<=0.05)[0]
selected_features = np.array(data_scaled_df.columns[2:][feature_ids])
X_anova = data_scaled_df.values[:,2:][:,feature_ids]
X_anova.shape
X_anova_df = pd.DataFrame(X_anova, columns = selected_features)
X_anova_df.insert(0, 'ID', data.ID.values)
X_anova_df.head()
data_healthy_df = X_anova_df.loc[X_anova_df.ID == 'Healthy']
data_asthma_df = X_anova_df.loc[X_anova_df.ID == 'Asthmatic']
data_healthy = data_healthy_df.values[:,1:]
data_asthma = data_asthma_df.values[:, 1:]
healthy_mean, healthy_cov = do_factor_analysis(data_healthy)
asthma_mean, asthma_cov = do_factor_analysis(data_asthma)
dist, t1, t2 = bhatt_dist(healthy_mean, healthy_cov, asthma_mean, asthma_cov)
print(dist)
print(np.linalg.det(healthy_cov))
print(np.linalg.det(asthma_cov))
X_anova.shape
fs = ReliefF(discrete_threshold = 5, n_jobs=1)
fs.fit(X_anova.astype(float), data.ID_num.values)
feature_scores = fs.feature_importances_
feature_ids = np.where(feature_scores>=0)[0]
selected_features = selected_features[feature_ids]
X_ano_reliefF = X_anova[:,feature_ids]
X_ano_reliefF.shape
X_anova_relief_df = pd.DataFrame(X_ano_reliefF, columns = selected_features)
X_anova_relief_df.insert(0, 'ID', data.ID.values)
X_anova_relief_df.head()
data_healthy_df = X_anova_relief_df.loc[X_anova_relief_df.ID == 'Healthy']
data_asthma_df = X_anova_relief_df.loc[X_anova_relief_df.ID == 'Asthmatic']
data_healthy = data_healthy_df.values[:,1:]
data_asthma = data_asthma_df.values[:, 1:]
healthy_mean, healthy_cov = do_factor_analysis(data_healthy)
asthma_mean, asthma_cov = do_factor_analysis(data_asthma)
dist, t1, t2 = bhatt_dist(healthy_mean, healthy_cov, asthma_mean, asthma_cov)
print(dist)
print(np.linalg.det(healthy_cov))
print(np.linalg.det(asthma_cov))
with open('../data/independent_features.pik', "rb") as f:
independent_features = dill.load(f)
X_no_corr_df = independent_features['X_no_corr_df']
X_no_corr_df.shape
X_no_corr_df.head()
X_scaled = StandardScaler().fit_transform(X_no_corr_df.values)
data_scaled_df = pd.DataFrame(X_scaled, columns = X_no_corr_df.columns)
data_scaled_df.insert(0, 'ID', data.ID.values)
data_scaled_df.insert(1, 'ID_num', data.ID_num.values)
data_scaled_df.head()
data_healthy_df = data_scaled_df.loc[data_scaled_df.ID == 'Healthy']
data_asthma_df = data_scaled_df.loc[data_scaled_df.ID == 'Asthmatic']
data_healthy = data_healthy_df.values[:,2:]
data_asthma = data_asthma_df.values[:, 2:]
healthy_mean, healthy_cov = do_factor_analysis(data_healthy)
asthma_mean, asthma_cov = do_factor_analysis(data_asthma)
dist, t1, t2 = bhatt_dist(healthy_mean, healthy_cov, asthma_mean, asthma_cov)
dist
data_scaled_df.head()
fs = ReliefF(discrete_threshold = 5, n_jobs=1)
fs.fit(data_scaled_df.values[:,2:].astype(float), data_scaled_df.ID_num.values)
feature_scores = fs.feature_importances_
feature_ids = np.where(feature_scores>=0)[0]
selected_features = np.array(X_no_corr_df.columns[feature_ids])
X_reliefF = data_scaled_df.values[:,2:][:,feature_ids]
X_reliefF.shape
X_reliefF_df = pd.DataFrame(X_reliefF, columns = selected_features)
X_reliefF_df.insert(0, 'ID', data.ID.values)
X_reliefF_df.head()
data_healthy_df = X_reliefF_df.loc[X_reliefF_df.ID == 'Healthy']
data_asthma_df = X_reliefF_df.loc[X_reliefF_df.ID == 'Asthmatic']
data_healthy = data_healthy_df.values[:,1:]
data_asthma = data_asthma_df.values[:, 1:]
healthy_mean, healthy_cov = do_factor_analysis(data_healthy)
asthma_mean, asthma_cov = do_factor_analysis(data_asthma)
dist, t1, t2 = bhatt_dist(healthy_mean, healthy_cov, asthma_mean, asthma_cov)
np.linalg.det(healthy_cov)
fs = MultiSURF(discrete_threshold = 5, n_jobs=1)
fs.fit(data_scaled_df.values[:,2:].astype(float), data_scaled_df.ID_num.values)
feature_scores = fs.feature_importances_
feature_ids = np.where(feature_scores>=0)[0]
selected_features = np.array(X_no_corr_df.columns[feature_ids])
X_MultiSURF = data_scaled_df.values[:,2:][:,feature_ids]
X_MultiSURF.shape
X_MultiSURF_df = pd.DataFrame(X_MultiSURF, columns = selected_features)
X_MultiSURF_df.insert(0, 'ID', data.ID.values)
X_MultiSURF_df.head()
data_healthy_df = X_MultiSURF_df.loc[X_reliefF_df.ID == 'Healthy']
data_asthma_df = X_MultiSURF_df.loc[X_reliefF_df.ID == 'Asthmatic']
data_healthy = data_healthy_df.values[:,1:]
data_asthma = data_asthma_df.values[:, 1:]
healthy_mean, healthy_cov = do_factor_analysis(data_healthy)
asthma_mean, asthma_cov = do_factor_analysis(data_asthma)
dist, t1, t2 = bhatt_dist(healthy_mean, healthy_cov, asthma_mean, asthma_cov)
dist
fs = MultiSURFstar(discrete_threshold = 5, n_jobs=1)
fs.fit(data_scaled_df.values[:,2:].astype(float), data_scaled_df.ID_num.values)
feature_scores = fs.feature_importances_
feature_ids = np.where(feature_scores>=0)[0]
selected_features = np.array(X_no_corr_df.columns[feature_ids])
X_MultiSURFStar = data_scaled_df.values[:,2:][:,feature_ids]
X_MultiSURFStar.shape
X_MultiSURFStar_df = pd.DataFrame(X_MultiSURFStar, columns = selected_features)
X_MultiSURFStar_df.insert(0, 'ID', data.ID.values)
X_MultiSURFStar_df.head()
data_healthy_df = X_MultiSURFStar_df.loc[X_MultiSURFStar_df.ID == 'Healthy']
data_asthma_df = X_MultiSURFStar_df.loc[X_MultiSURFStar_df.ID == 'Asthmatic']
data_healthy = data_healthy_df.values[:,1:]
data_asthma = data_asthma_df.values[:, 1:]
healthy_mean, healthy_cov = do_factor_analysis(data_healthy)
asthma_mean, asthma_cov = do_factor_analysis(data_asthma)
dist, t1, t2 = bhatt_dist(healthy_mean, healthy_cov, asthma_mean, asthma_cov)
dist
f,p = f_classif(data_scaled_df.values[:,2:].astype(float), data.ID_num.values)
feature_ids = np.where(p<=0.05)[0]
selected_features = np.array(X_no_corr_df.columns[feature_ids])
X_anova = data_scaled_df.values[:,2:][:,feature_ids]
X_anova.shape
X_anova_df = pd.DataFrame(X_anova, columns = selected_features)
X_anova_df.insert(0, 'ID', data.ID.values)
X_anova_df.head()
data_healthy_df = X_anova_df.loc[X_anova_df.ID == 'Healthy']
data_asthma_df = X_anova_df.loc[X_anova_df.ID == 'Asthmatic']
data_healthy = data_healthy_df.values[:,1:]
data_asthma = data_asthma_df.values[:, 1:]
healthy_mean, healthy_cov = do_factor_analysis(data_healthy)
asthma_mean, asthma_cov = do_factor_analysis(data_asthma)
dist, t1, t2 = bhatt_dist(healthy_mean, healthy_cov, asthma_mean, asthma_cov)
dist
np.linalg.det(healthy_cov)
np.linalg.det(asthma_cov)
X_anova.shape
fs = ReliefF(discrete_threshold = 5, n_jobs=1)
fs.fit(X_anova.astype(float), data.ID_num.values)
feature_scores = fs.feature_importances_
feature_ids = np.where(feature_scores>=0)[0]
selected_features = selected_features[feature_ids]
X_ano_reliefF = X_anova[:,feature_ids]
X_ano_reliefF.shape
X_anova_relief_df = pd.DataFrame(X_ano_reliefF, columns = selected_features)
X_anova_relief_df.insert(0, 'ID', data.ID.values)
X_anova_relief_df.head()
data_healthy_df = X_anova_relief_df.loc[X_anova_relief_df.ID == 'Healthy']
data_asthma_df = X_anova_relief_df.loc[X_anova_relief_df.ID == 'Asthmatic']
data_healthy = data_healthy_df.values[:,1:]
data_asthma = data_asthma_df.values[:, 1:]
data_scaled_df.head()
import random
random_feat_ids = list(range(681))
random.shuffle(random_feat_ids)
data_scaled_df.head()
data_healthy = data_scaled_df.loc[data_scaled_df.ID == 'Healthy'].values[:,2:]
data_asthma = data_scaled_df.loc[data_scaled_df.ID == 'Asthmatic'].values[:,2:]
healthy_mean, healthy_cov = do_factor_analysis(data_healthy[:,random_feat_ids[:30]])
asthma_mean, asthma_cov = do_factor_analysis(data_asthma[:,random_feat_ids[:30]])
dist, t1, t2 = bhatt_dist(healthy_mean, healthy_cov, asthma_mean, asthma_cov)
print(dist)
print(np.linalg.det(healthy_cov))
print(np.linalg.det(asthma_cov))
t2
np.linalg.det(asthma_cov)
| 0.455683 | 0.790085 |
```
#모듈 불러오기
import numpy as np
import pandas as pd
from sklearn.preprocessing import StandardScaler
from sklearn.preprocessing import MinMaxScaler
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LinearRegression
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import RepeatedStratifiedKFold
from sklearn.model_selection import cross_val_score
#데이터 불러오기
df_wine = pd.read_csv("../data/winequality-red.csv")
df_wine_test = pd.read_csv("../data/winequality-red_test.csv")
df_wine.head()
def evaluate_model(model, X, y):
# define the evaluation procedure
cv = RepeatedStratifiedKFold(n_splits=10, n_repeats=3, random_state=1)
# evaluate the model
scores = cross_val_score(model, X, y, scoring='accuracy', cv=cv, n_jobs=-1)
return scores
#공백 데이터 여부 체크
df_wine.isnull().sum()
#품질을 두개의 변수로 전환
bins = (0, 5, 10)
group_names = ['0', '1']
df_wine['quality'] = pd.cut(df_wine['quality'], bins = bins, labels=group_names)
df_wine
#예측하고자 하는 변수를 Y값으로, 학습 대상의 변수를 X값으로
X = df_wine.drop("quality", axis=1)
y = df_wine["quality"]
#표준화
scaler = MinMaxScaler()
X = scaler.fit_transform(X)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size = 0.1, random_state = 0, shuffle=False)
#모델링
linear = LinearRegression()
linear.fit(X,y)
logistic = LogisticRegression(C=1, penalty='l1', solver='liblinear', random_state=0, warm_start=False)
logistic.fit(X,y)
#모델평가
cv = RepeatedStratifiedKFold(n_splits=10, n_repeats=3, random_state=0)
n_scores = cross_val_score(logistic, X, y, scoring='accuracy', cv=cv, n_jobs=-1)
print('Mean Accuracy: %.3f (%.3f)' % (np.mean(n_scores), np.std(n_scores)))
#하이퍼 파라미터 튜닝
from sklearn.model_selection import GridSearchCV
parameters = [{'penalty':['l1','l2']},
{'C':[1, 10, 100, 1000]}]
grid_search = GridSearchCV(estimator = logistic,
param_grid = parameters,
scoring = 'accuracy',
cv = 5,
verbose=0)
grid_search.fit(X_train, y_train)
# 하이퍼 파라미터 결과 도출
# best score achieved during the GridSearchCV
print('GridSearch CV best score : {:.4f}\n\n'.format(grid_search.best_score_))
# print parameters that give the best results
print('Parameters that give the best results :','\n\n', (grid_search.best_params_))
# print estimator that was chosen by the GridSearch
print('\n\nEstimator that was chosen by the search :','\n\n', (grid_search.best_estimator_))
Y_pred = linear.predict(X_test)
acc_log = round(linear.score(X_train, y_train)* 100, 2)
acc_log
Y_pred = logistic.predict(X_test)
acc_log = round(logistic.score(X_train, y_train)* 100, 2)
acc_log
df_wine_test.head()
x_submission = df_wine_test.values
#Applying to the model
pred = logistic.predict(x_submission)
pred
pred.size
submission = pd.DataFrame(data=pred)
submission.index.name='ID'
submission.columns = ['quality']
submission.to_csv("submission.csv")
submission
```
|
github_jupyter
|
#모듈 불러오기
import numpy as np
import pandas as pd
from sklearn.preprocessing import StandardScaler
from sklearn.preprocessing import MinMaxScaler
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LinearRegression
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import RepeatedStratifiedKFold
from sklearn.model_selection import cross_val_score
#데이터 불러오기
df_wine = pd.read_csv("../data/winequality-red.csv")
df_wine_test = pd.read_csv("../data/winequality-red_test.csv")
df_wine.head()
def evaluate_model(model, X, y):
# define the evaluation procedure
cv = RepeatedStratifiedKFold(n_splits=10, n_repeats=3, random_state=1)
# evaluate the model
scores = cross_val_score(model, X, y, scoring='accuracy', cv=cv, n_jobs=-1)
return scores
#공백 데이터 여부 체크
df_wine.isnull().sum()
#품질을 두개의 변수로 전환
bins = (0, 5, 10)
group_names = ['0', '1']
df_wine['quality'] = pd.cut(df_wine['quality'], bins = bins, labels=group_names)
df_wine
#예측하고자 하는 변수를 Y값으로, 학습 대상의 변수를 X값으로
X = df_wine.drop("quality", axis=1)
y = df_wine["quality"]
#표준화
scaler = MinMaxScaler()
X = scaler.fit_transform(X)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size = 0.1, random_state = 0, shuffle=False)
#모델링
linear = LinearRegression()
linear.fit(X,y)
logistic = LogisticRegression(C=1, penalty='l1', solver='liblinear', random_state=0, warm_start=False)
logistic.fit(X,y)
#모델평가
cv = RepeatedStratifiedKFold(n_splits=10, n_repeats=3, random_state=0)
n_scores = cross_val_score(logistic, X, y, scoring='accuracy', cv=cv, n_jobs=-1)
print('Mean Accuracy: %.3f (%.3f)' % (np.mean(n_scores), np.std(n_scores)))
#하이퍼 파라미터 튜닝
from sklearn.model_selection import GridSearchCV
parameters = [{'penalty':['l1','l2']},
{'C':[1, 10, 100, 1000]}]
grid_search = GridSearchCV(estimator = logistic,
param_grid = parameters,
scoring = 'accuracy',
cv = 5,
verbose=0)
grid_search.fit(X_train, y_train)
# 하이퍼 파라미터 결과 도출
# best score achieved during the GridSearchCV
print('GridSearch CV best score : {:.4f}\n\n'.format(grid_search.best_score_))
# print parameters that give the best results
print('Parameters that give the best results :','\n\n', (grid_search.best_params_))
# print estimator that was chosen by the GridSearch
print('\n\nEstimator that was chosen by the search :','\n\n', (grid_search.best_estimator_))
Y_pred = linear.predict(X_test)
acc_log = round(linear.score(X_train, y_train)* 100, 2)
acc_log
Y_pred = logistic.predict(X_test)
acc_log = round(logistic.score(X_train, y_train)* 100, 2)
acc_log
df_wine_test.head()
x_submission = df_wine_test.values
#Applying to the model
pred = logistic.predict(x_submission)
pred
pred.size
submission = pd.DataFrame(data=pred)
submission.index.name='ID'
submission.columns = ['quality']
submission.to_csv("submission.csv")
submission
| 0.50708 | 0.626124 |

[](https://colab.research.google.com/github/JohnSnowLabs/spark-nlp-workshop/blob/master/jupyter/training/english/crf-ner/ner_dl_crf.ipynb)
## 0. Colab Setup
```
import os
# Install java
! apt-get install -y openjdk-8-jdk-headless -qq > /dev/null
os.environ["JAVA_HOME"] = "/usr/lib/jvm/java-8-openjdk-amd64"
os.environ["PATH"] = os.environ["JAVA_HOME"] + "/bin:" + os.environ["PATH"]
! java -version
# Install pyspark
! pip install --ignore-installed -q pyspark==2.4.4
# Install Spark NLP
! pip install --ignore-installed -q spark-nlp==2.5
```
## CRF Named Entity Recognition
In the following example, we walk-through a Conditional Random Fields NER model training and prediction.
This challenging annotator will require the user to provide either a labeled dataset during fit() stage, or use external CoNLL 2003 resources to train. It may optionally use an external word embeddings set and a list of additional entities.
The CRF Annotator will also require Part-of-speech tags so we add those in the same Pipeline. Also, we could use our special RecursivePipeline, which will tell SparkNLP's NER CRF approach to use the same pipeline for tagging external resources.
#### 1. Call necessary imports and set the resource path to read local data files
```
import os
import sys
from pyspark.sql import SparkSession
from pyspark.ml import Pipeline
from sparknlp.annotator import *
from sparknlp.common import *
from sparknlp.base import *
import time
import zipfile
```
#### 2. Download training dataset if not already there
```
# Download CoNLL 2003 Dataset
import os
from pathlib import Path
import urllib.request
if not Path("eng.train").is_file():
print("File Not found will downloading it!")
url = "https://github.com/patverga/torch-ner-nlp-from-scratch/raw/master/data/conll2003/eng.train"
urllib.request.urlretrieve(url, 'eng.train')
else:
print("File already present.")
```
#### 3. Load SparkSession if not already there
```
import sparknlp
spark = sparknlp.start()
print("Spark NLP version: ", sparknlp.version())
print("Apache Spark version: ", spark.version)
```
#### 4. Create annotator components in the right order, with their training Params. Finisher will output only NER. Put all in pipeline.
```
nerTagger = NerCrfApproach()\
.setInputCols(["sentence", "token", "pos", "embeddings"])\
.setLabelColumn("label")\
.setOutputCol("ner")\
.setMinEpochs(1)\
.setMaxEpochs(1)\
.setLossEps(1e-3)\
.setL2(1)\
.setC0(1250000)\
.setRandomSeed(0)\
.setVerbose(0)
```
#### 6. Load a dataset for prediction. Training is not relevant from this dataset.
```
from sparknlp.training import CoNLL
conll = CoNLL()
data = conll.readDataset(spark, path='eng.train')
embeddings = WordEmbeddingsModel.pretrained()\
.setOutputCol('embeddings')
ready_data = embeddings.transform(data)
ready_data.show(4)
```
#### 7. Training the model. Training doesn't really do anything from the dataset itself.
```
start = time.time()
print("Start fitting")
ner_model = nerTagger.fit(ready_data)
print("Fitting has ended")
print (time.time() - start)
```
#### 8. Save NerCrfModel into disk after training
```
ner_model.write().overwrite().save("./pip_wo_embedd/")
```
|
github_jupyter
|
import os
# Install java
! apt-get install -y openjdk-8-jdk-headless -qq > /dev/null
os.environ["JAVA_HOME"] = "/usr/lib/jvm/java-8-openjdk-amd64"
os.environ["PATH"] = os.environ["JAVA_HOME"] + "/bin:" + os.environ["PATH"]
! java -version
# Install pyspark
! pip install --ignore-installed -q pyspark==2.4.4
# Install Spark NLP
! pip install --ignore-installed -q spark-nlp==2.5
import os
import sys
from pyspark.sql import SparkSession
from pyspark.ml import Pipeline
from sparknlp.annotator import *
from sparknlp.common import *
from sparknlp.base import *
import time
import zipfile
# Download CoNLL 2003 Dataset
import os
from pathlib import Path
import urllib.request
if not Path("eng.train").is_file():
print("File Not found will downloading it!")
url = "https://github.com/patverga/torch-ner-nlp-from-scratch/raw/master/data/conll2003/eng.train"
urllib.request.urlretrieve(url, 'eng.train')
else:
print("File already present.")
import sparknlp
spark = sparknlp.start()
print("Spark NLP version: ", sparknlp.version())
print("Apache Spark version: ", spark.version)
nerTagger = NerCrfApproach()\
.setInputCols(["sentence", "token", "pos", "embeddings"])\
.setLabelColumn("label")\
.setOutputCol("ner")\
.setMinEpochs(1)\
.setMaxEpochs(1)\
.setLossEps(1e-3)\
.setL2(1)\
.setC0(1250000)\
.setRandomSeed(0)\
.setVerbose(0)
from sparknlp.training import CoNLL
conll = CoNLL()
data = conll.readDataset(spark, path='eng.train')
embeddings = WordEmbeddingsModel.pretrained()\
.setOutputCol('embeddings')
ready_data = embeddings.transform(data)
ready_data.show(4)
start = time.time()
print("Start fitting")
ner_model = nerTagger.fit(ready_data)
print("Fitting has ended")
print (time.time() - start)
ner_model.write().overwrite().save("./pip_wo_embedd/")
| 0.341912 | 0.878001 |
<img src=".\img\mioti.png">
# Proyecto Reconocimiento Facial: Almacenamiento de Caras
<img src="./img/emociones.png" style="width: 800px">
### Objetivos
En este notebook vamos a ir almacenando fotografías de las personas a las que queremos registrar para identificar más adelante.
Los pasos que se llevarán a cabo serán:
* Comprobar si esa persona está ya registrada en el sistema, en caso que no sea así se creará una carpeta con su nombre en el directorio de 'reconocimiento'
* A través de la video-cámara del equipo se irán tomando fotografías de la persona siempre que esta sea reconocida por el recuadro
* Estas fotografías se irán guardando en su carpeta correspondiente para el entrenamiento posterior de la red neuronal
### Importación de librerías
* Las librerías que vamos a utilizar son:
* cv2: para trabajar con imágenes y vídeos
* os: para trabajar con directorios
* numpy: para trabajar con arrays
* matplotlib: para visualizar las fotos tomadas
```
#Se importan las librerías necesarias
import cv2
import os
import numpy as np
import matplotlib.pyplot as plt
from keras.preprocessing.image import img_to_array
```
### Almacenado
* En primer lugar vamos a definir una ruta donde guardar las fotografías que se vayan generando. Estas se irán almacenando dentro de una carpeta con el nombre de la persona a la que se quiere registrar, en caso que esa carpeta no exista se creará
```
#Se define la ruta donde se va a crear la carpeta donde se alamcenarán los rostros del individuo
person_name='Ricardo' #Pon aqui tu nombre
data_path='./reconocimiento'
person_path=data_path+'/'+person_name
print(person_path)
#Creamos carpeta con nombre de la persona
if not os.path.exists(person_path):
print('carpeta creada: ',person_path)
os.makedirs(person_path)
```
* Para la captura de caras de las personas vamos a conectar la cámara del equipo y mantenerla activa mediante un bucle infinito, dentro de este bucle se van a invocar las siguientes funciones:
* face_detector: genera el cuadro cuando se reconoce un rostro, el tamaño será de 150x150
* draw_text_with_background: da formato al recuadro
* Mientras dure el bucle cada vez que sea identificado un rostro, este será capturado y añadido a la carpeta del nombre de la persona correspondiente. El ciclo termina en el momento en que se pulsa 'enter' o se llega a un número determinado de capturas, en nuestro caso hemos determinado 500 fotos como suficientes para que el modelo pueda hacer bien el reconocimiento
```
def draw_text_with_backgroud(img, text, x, y, font_scale, thickness=1, font=cv2.FONT_HERSHEY_SIMPLEX,
background=(175,50,200), foreground=(255,255,255), box_coords_1=(-5,5), box_coords_2=(5,-5)):
(text_width, text_height) = cv2.getTextSize(text, font, fontScale=font_scale, thickness=1)[0]
box_coords = ((x+box_coords_1[0], y+box_coords_1[1]), (x + text_width + box_coords_2[0], y - text_height + box_coords_2[1]))
cv2.rectangle(img, box_coords[0], box_coords[1], background, cv2.FILLED)
cv2.putText(img, text, (x, y), font, fontScale=font_scale, color=foreground, thickness=thickness)
face_classifier = cv2.CascadeClassifier(cv2.data.haarcascades + "haarcascade_frontalface_default.xml")
def face_detector(img):
# Convert image to grayscale
gray = cv2.cvtColor(img,cv2.COLOR_BGR2GRAY)
faces = face_classifier.detectMultiScale(gray, 1.5, 3)
if len(faces) == 0:
return (0,0,0,0), np.zeros((150,150), np.uint8), img
for idx,face in enumerate(faces):
x,y,w,h = face
cv2.rectangle(img,(x,y),(x+w,y+h),(255,0,0),2)
roi_gray = gray[y:y+h, x:x+w]
roi_gray = cv2.resize(roi_gray, (150, 150), interpolation = cv2.INTER_CUBIC)
if np.sum([roi_gray]) != 0.0:
roi = roi_gray.astype("float") / 255.0
roi = img_to_array(roi)
roi = np.expand_dims(roi, axis=0)
# make a prediction on the ROI, then lookup the class
label = f'Persona {idx} EMOCION'#class_labels[preds.argmax()]
draw_text_with_backgroud(img, label, x + 5, y, font_scale=0.4)
else:
cv2.putText(img, "No Face Found", (20, 60) , cv2.FONT_HERSHEY_SIMPLEX,2, (0,255,0), 3)
draw_text_with_backgroud(img, "No Face Found", x + 5, y, font_scale=0.4)
return (x,w,y,h), roi_gray, img
cap = cv2.VideoCapture(0)
count=0
while True:
ret, frame = cap.read()
rect, face, image = face_detector(frame)
if (face.sum()!=0):#Hay captura de cara
cv2.imwrite(person_path+'/rostro_{}.jpg'.format(count),face)
count+=1
cv2.imshow('Reconocedor de Emociones', image)
if cv2.waitKey(1) == 13 or count>=500: #13 is the Enter Key
break
cap.release()
cv2.destroyAllWindows()
```
### Visualización
* Una vez guardadas las fotografías de las personas que se quieren registrar vamos a pasar a realizar una visualización de una fracción de estas para comprobar que el proceso se está realizando correctamente. Para ello recorreremos todas las carpetas del directorio 'reconocimiento' y tomaremos las 5 primeras imágenes de cada una de ellas para después representarlas
```
lista_gente=os.listdir(data_path)
print(lista_gente)
photo=[]
names=[]
for name in lista_gente:
#Directorio de cada alumno
name_path='./reconocimiento/'+name
#Lista con los nombres de cada foto
photo_name=os.listdir(name_path)
#Bucle con las 5 primeras fotos de cada alumno
for image in photo_name[:5]:
#Ruta de cada foto
img=name_path+'/'+image
#Se añade foto a lista
photo.append(cv2.imread(img))
#Se añade nombre a lista
names.append(name)
row=len(lista_gente)
col=5
axes=[]
fig=plt.figure(figsize=(8,8))
#Recorremos la figura añadiendo en cada recuadro una imagen con su etiqueta
for i in range(row*col):
img=photo[i]
label=names[i]
axes.append(fig.add_subplot(row,col,i+1))
axes[-1].set_title(label)
plt.imshow(img)
plt.axis("off")
fig.tight_layout()
plt.show()
```
* Las caras se corresponden con el nombre de las carpetas donde fueron guardadas, el proceso se ha realizado correctamente
|
github_jupyter
|
#Se importan las librerías necesarias
import cv2
import os
import numpy as np
import matplotlib.pyplot as plt
from keras.preprocessing.image import img_to_array
#Se define la ruta donde se va a crear la carpeta donde se alamcenarán los rostros del individuo
person_name='Ricardo' #Pon aqui tu nombre
data_path='./reconocimiento'
person_path=data_path+'/'+person_name
print(person_path)
#Creamos carpeta con nombre de la persona
if not os.path.exists(person_path):
print('carpeta creada: ',person_path)
os.makedirs(person_path)
def draw_text_with_backgroud(img, text, x, y, font_scale, thickness=1, font=cv2.FONT_HERSHEY_SIMPLEX,
background=(175,50,200), foreground=(255,255,255), box_coords_1=(-5,5), box_coords_2=(5,-5)):
(text_width, text_height) = cv2.getTextSize(text, font, fontScale=font_scale, thickness=1)[0]
box_coords = ((x+box_coords_1[0], y+box_coords_1[1]), (x + text_width + box_coords_2[0], y - text_height + box_coords_2[1]))
cv2.rectangle(img, box_coords[0], box_coords[1], background, cv2.FILLED)
cv2.putText(img, text, (x, y), font, fontScale=font_scale, color=foreground, thickness=thickness)
face_classifier = cv2.CascadeClassifier(cv2.data.haarcascades + "haarcascade_frontalface_default.xml")
def face_detector(img):
# Convert image to grayscale
gray = cv2.cvtColor(img,cv2.COLOR_BGR2GRAY)
faces = face_classifier.detectMultiScale(gray, 1.5, 3)
if len(faces) == 0:
return (0,0,0,0), np.zeros((150,150), np.uint8), img
for idx,face in enumerate(faces):
x,y,w,h = face
cv2.rectangle(img,(x,y),(x+w,y+h),(255,0,0),2)
roi_gray = gray[y:y+h, x:x+w]
roi_gray = cv2.resize(roi_gray, (150, 150), interpolation = cv2.INTER_CUBIC)
if np.sum([roi_gray]) != 0.0:
roi = roi_gray.astype("float") / 255.0
roi = img_to_array(roi)
roi = np.expand_dims(roi, axis=0)
# make a prediction on the ROI, then lookup the class
label = f'Persona {idx} EMOCION'#class_labels[preds.argmax()]
draw_text_with_backgroud(img, label, x + 5, y, font_scale=0.4)
else:
cv2.putText(img, "No Face Found", (20, 60) , cv2.FONT_HERSHEY_SIMPLEX,2, (0,255,0), 3)
draw_text_with_backgroud(img, "No Face Found", x + 5, y, font_scale=0.4)
return (x,w,y,h), roi_gray, img
cap = cv2.VideoCapture(0)
count=0
while True:
ret, frame = cap.read()
rect, face, image = face_detector(frame)
if (face.sum()!=0):#Hay captura de cara
cv2.imwrite(person_path+'/rostro_{}.jpg'.format(count),face)
count+=1
cv2.imshow('Reconocedor de Emociones', image)
if cv2.waitKey(1) == 13 or count>=500: #13 is the Enter Key
break
cap.release()
cv2.destroyAllWindows()
lista_gente=os.listdir(data_path)
print(lista_gente)
photo=[]
names=[]
for name in lista_gente:
#Directorio de cada alumno
name_path='./reconocimiento/'+name
#Lista con los nombres de cada foto
photo_name=os.listdir(name_path)
#Bucle con las 5 primeras fotos de cada alumno
for image in photo_name[:5]:
#Ruta de cada foto
img=name_path+'/'+image
#Se añade foto a lista
photo.append(cv2.imread(img))
#Se añade nombre a lista
names.append(name)
row=len(lista_gente)
col=5
axes=[]
fig=plt.figure(figsize=(8,8))
#Recorremos la figura añadiendo en cada recuadro una imagen con su etiqueta
for i in range(row*col):
img=photo[i]
label=names[i]
axes.append(fig.add_subplot(row,col,i+1))
axes[-1].set_title(label)
plt.imshow(img)
plt.axis("off")
fig.tight_layout()
plt.show()
| 0.254787 | 0.907967 |
### <center>PANTELIDIS NIKOS AM2787
### <center>BOUTSIKARIS LEONIDAS AM2776
# <center>DATA MINING
# <center>ASSIGNMENT 3
### <center>EXERCISE 2
<br>
```
import numpy as np
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.naive_bayes import MultinomialNB
from sklearn import model_selection
from sklearn import preprocessing
```
# DATASET <br>
## TRAIN DATA
```
max_abs_scaler = preprocessing.MaxAbsScaler()
train_data = []
train_labels = []
with open("Data/training_data.txt", "r") as infile:
for i, line in enumerate(infile.readlines()):
temp = line.split('\t')
train_data.append(temp[1])
train_labels.append(temp[0])
train_vectorizer = CountVectorizer(ngram_range = (1,2),
analyzer = 'word',
stop_words = 'english')
X_train = train_vectorizer.fit_transform(train_data)
X_train = max_abs_scaler.fit_transform(X_train)
len(train_vectorizer.get_feature_names())
```
## TEST DATA
```
test_data = []
test_id = []
with open("Data/test_data.txt", "r") as infile:
for i, line in enumerate(infile.readlines()):
temp = line.split('\t')
test_data.append(temp[1])
test_id.append(temp[0])
test_vectorizer = CountVectorizer(vocabulary = train_vectorizer.get_feature_names(),
ngram_range = (1,2),
analyzer = 'word',
stop_words = 'english')
X_test = test_vectorizer.fit_transform(test_data)
X_test = max_abs_scaler.fit_transform(X_test)
```
# CLASSIFICATION
```
mNB_clf = MultinomialNB()
labels_tmp = [1 if i == "True" else 0 for i in train_labels]
scores = model_selection.cross_val_score(mNB_clf, X_train, labels_tmp, cv=5, scoring="f1")
mNB_clf.fit(X_train, train_labels)
y_pred = mNB_clf.predict(X_test)
scores.mean()
```
# OUTPUT
```
with open("submission.csv", "w+") as f:
f.write("id,label\n")
for i, l in enumerate(test_id):
f.write(str(l)+","+y_pred[i]+"\n")
```
## Previous Approaches
Firstly we tried vectorize the data with the binary vectorizer that we created for the previous assignment. The results were actually good(around 0.91 score in Kaggle). The problem was that it was hard to configure it. <br><br>
Then we decided to try vectorize the data with TFIDF- and Count-Vectorizer. We thought that TFIDF would do the work but we were wrong. We tried:<br>
1) With and without stopwords<br>
2) 1 and 2 grams of words <br><br>
But the results weren't good enough (0.89 - 0.95)
<br>
In the end we started trying different configurations with the CountVectorizer. The first configuration used a corpus without the stopwords (0.94). Then we tried n-grams. The (1,2) n-grams had the best score (0,96). We tried (1,3) but it gave lower score. Finally we tried the MaxAbsScaler on the vectorized data and we scored (0.97368)
## Best Approach Specs
1) CountVectorizer: Removed Stopwords, used (1,2) n-grams <br>
2) MaxAbsScaler <br>
3) Multinomial Naive Bayes
## Kaggle Info
- Team Name: Boutsikaris, Pantelidis
- Score: 0.97368
|
github_jupyter
|
import numpy as np
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.naive_bayes import MultinomialNB
from sklearn import model_selection
from sklearn import preprocessing
max_abs_scaler = preprocessing.MaxAbsScaler()
train_data = []
train_labels = []
with open("Data/training_data.txt", "r") as infile:
for i, line in enumerate(infile.readlines()):
temp = line.split('\t')
train_data.append(temp[1])
train_labels.append(temp[0])
train_vectorizer = CountVectorizer(ngram_range = (1,2),
analyzer = 'word',
stop_words = 'english')
X_train = train_vectorizer.fit_transform(train_data)
X_train = max_abs_scaler.fit_transform(X_train)
len(train_vectorizer.get_feature_names())
test_data = []
test_id = []
with open("Data/test_data.txt", "r") as infile:
for i, line in enumerate(infile.readlines()):
temp = line.split('\t')
test_data.append(temp[1])
test_id.append(temp[0])
test_vectorizer = CountVectorizer(vocabulary = train_vectorizer.get_feature_names(),
ngram_range = (1,2),
analyzer = 'word',
stop_words = 'english')
X_test = test_vectorizer.fit_transform(test_data)
X_test = max_abs_scaler.fit_transform(X_test)
mNB_clf = MultinomialNB()
labels_tmp = [1 if i == "True" else 0 for i in train_labels]
scores = model_selection.cross_val_score(mNB_clf, X_train, labels_tmp, cv=5, scoring="f1")
mNB_clf.fit(X_train, train_labels)
y_pred = mNB_clf.predict(X_test)
scores.mean()
with open("submission.csv", "w+") as f:
f.write("id,label\n")
for i, l in enumerate(test_id):
f.write(str(l)+","+y_pred[i]+"\n")
| 0.270191 | 0.785226 |
## 부록1. 파이썬 입문
#### 기본형
```
# 정수형
# 수치 표현이 정수인 경우, 이를 대입한 변수는 자동으로 정수형이 됨
a = 1
# 부동소수점 수형
# 수치 표현에 소수점이 포함되면, 이를 대입한 변수는 자동적으로 부동소수점 수형이 됨
b = 2.0
# 문자열형
# 문자열은 싱글 쿼트(')로 감싸서 표현함
# 또는 더블 쿼트(")로 감싸도 상관 없음
c = 'abc'
# 논리형
# True 또는 False 중 하나를 취하는 변수형
d = True
```
#### print 함수와 type 함수
```
# 정수형 변수 a의 값과 형
print(a)
print(type(a))
# 부동소수점 수형 변수 b의 값과 형
print(b)
print(type(b))
# 문자열형 변수 c의 값과 형
print(c)
print(type(c))
# 논리형 변수 d의 값과 형
print(d)
print(type(d))
```
#### 리스트
```
# 리스트의 정의
l = [1, 2, 3, 5, 8, 13]
# リストの値とtype
print(l)
print(type(l))
```
#### リストの要素数
```
# リストの要素数
print(len(l))
```
#### リストの要素参照
```
# リストの要素参照
# 最初の要素
print(l[0])
# 3番目の要素
print(l[2])
# 最後の要素 (こういう指定方法も可能)
print(l[-1])
```
#### 部分リスト参照1
```
# 部分リスト インデックス:2以上 インデックス: 5未満
print(l[2:5])
# 部分リスト インデックス:0以上 インデックス: 3未満
print(l[0:3])
# 開始インデックスが0の場合は省略可
print(l[:3])
```
#### 部分リスト参照2
```
# 部分リスト インデックス:4以上最後まで
# リストの長さを求める
n = len(l)
print(l[4:n])
# 最終インデックスが最終要素の場合は省略可
print(l[4:])
# 後ろから2つ
print(l[-2:])
#最初も最後も省略するとリスト全体になる
print(l[:])
```
#### タプル
```
# タプルの定義
t = (1, 2, 3, 5, 8, 13)
# タプルの値表示
print(t)
# タプルの型表示
print(type(t))
# タプルの要素数
print(len(t))
# タプルの要素参照
print(t[1])
t[1] = 1
x = 1
y = 2
z = (x, y)
print(type(z))
a, b = z
print(a)
print(b)
```
### 辞書
#### 辞書の定義
```
# 辞書の定義
my_dict = {'yes': 1, 'no': 0}
# print文の結果
print(my_dict)
# type関数の結果
print(type(my_dict))
```
#### 辞書の参照
```
# キーから値を参照
# key= 'yes'で検索
value1 = my_dict['yes']
print(value1)
# key='no'で検索
value2 = my_dict['no']
print(value2)
```
#### 辞書への項目追加
```
# 辞書への項目追加
my_dict['neutral'] = 2
# 結果確認
print(my_dict)
```
### 制御構造
#### ループ処理
```
# ループ処理
# リストの定義
list4 = ['One', 'Two', 'Three', 'Four']
# ループ処理
for item in list4:
print(item)
# range関数を使ったループ処理
for item in range(4):
print(item)
# 引数2つのrange関数
for item in range(1, 5):
print(item)
# 辞書とループ処理
# items関数
print(my_dict.items())
# items関数を使ったループ処理
for key, value in my_dict.items():
print(key, ':', value )
```
#### if文
```
# if文のサンプル
for i in range(1, 5):
if i % 2 == 0:
print(i, 'は偶数です')
else:
print(i, 'は奇数です')
```
#### 関数
```
# 関数の定義例1
def square(x):
p2 = x * x
return p2
# 関数の呼び出し例1
x1 = 13
r1 = square(x1)
print(x1, r1)
# 関数の定義例2
def squares(x):
p2 = x * x
p3= x * x * x
return (p2, p3)
# 関数の呼び出し例2
x1 = 13
p2, p3 = squares(x1)
print(x1, p2, p3)
```
#### ライプラリの導入
```
# 日本語化ライブラリ導入
!pip install japanize-matplotlib | tail -n 1
```
#### import文
```
# 必要ライブラリのimport
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
# matplotlib日本語化対応
import japanize_matplotlib
# データフレーム表示用関数
from IPython.display import display
```
#### ワーニング非表示
```
# 余分なワーニングを非表示にする
import warnings
warnings.filterwarnings('ignore')
```
#### 数値の整形表示
```
# f文字列の表示
a1 = 1.0/7.0
a2 = 123
str1 = f'a1 = {a1} a2 = {a2}'
print(str1)
# f文字列の詳細オプション
# .4f 小数点以下4桁の固定小数点表示
# 04 整数を0詰め4桁表示
str2 = f'a1 = {a1:.4f} a2 = {a2:04}'
print(str2)
# 04e 小数点以下4桁の浮動小数点表示
# #x 整数の16進数表示
str3 = f'a1 = {a1:.04e} a2 = {a2:#x}'
print(str3)
```
|
github_jupyter
|
# 정수형
# 수치 표현이 정수인 경우, 이를 대입한 변수는 자동으로 정수형이 됨
a = 1
# 부동소수점 수형
# 수치 표현에 소수점이 포함되면, 이를 대입한 변수는 자동적으로 부동소수점 수형이 됨
b = 2.0
# 문자열형
# 문자열은 싱글 쿼트(')로 감싸서 표현함
# 또는 더블 쿼트(")로 감싸도 상관 없음
c = 'abc'
# 논리형
# True 또는 False 중 하나를 취하는 변수형
d = True
# 정수형 변수 a의 값과 형
print(a)
print(type(a))
# 부동소수점 수형 변수 b의 값과 형
print(b)
print(type(b))
# 문자열형 변수 c의 값과 형
print(c)
print(type(c))
# 논리형 변수 d의 값과 형
print(d)
print(type(d))
# 리스트의 정의
l = [1, 2, 3, 5, 8, 13]
# リストの値とtype
print(l)
print(type(l))
# リストの要素数
print(len(l))
# リストの要素参照
# 最初の要素
print(l[0])
# 3番目の要素
print(l[2])
# 最後の要素 (こういう指定方法も可能)
print(l[-1])
# 部分リスト インデックス:2以上 インデックス: 5未満
print(l[2:5])
# 部分リスト インデックス:0以上 インデックス: 3未満
print(l[0:3])
# 開始インデックスが0の場合は省略可
print(l[:3])
# 部分リスト インデックス:4以上最後まで
# リストの長さを求める
n = len(l)
print(l[4:n])
# 最終インデックスが最終要素の場合は省略可
print(l[4:])
# 後ろから2つ
print(l[-2:])
#最初も最後も省略するとリスト全体になる
print(l[:])
# タプルの定義
t = (1, 2, 3, 5, 8, 13)
# タプルの値表示
print(t)
# タプルの型表示
print(type(t))
# タプルの要素数
print(len(t))
# タプルの要素参照
print(t[1])
t[1] = 1
x = 1
y = 2
z = (x, y)
print(type(z))
a, b = z
print(a)
print(b)
# 辞書の定義
my_dict = {'yes': 1, 'no': 0}
# print文の結果
print(my_dict)
# type関数の結果
print(type(my_dict))
# キーから値を参照
# key= 'yes'で検索
value1 = my_dict['yes']
print(value1)
# key='no'で検索
value2 = my_dict['no']
print(value2)
# 辞書への項目追加
my_dict['neutral'] = 2
# 結果確認
print(my_dict)
# ループ処理
# リストの定義
list4 = ['One', 'Two', 'Three', 'Four']
# ループ処理
for item in list4:
print(item)
# range関数を使ったループ処理
for item in range(4):
print(item)
# 引数2つのrange関数
for item in range(1, 5):
print(item)
# 辞書とループ処理
# items関数
print(my_dict.items())
# items関数を使ったループ処理
for key, value in my_dict.items():
print(key, ':', value )
# if文のサンプル
for i in range(1, 5):
if i % 2 == 0:
print(i, 'は偶数です')
else:
print(i, 'は奇数です')
# 関数の定義例1
def square(x):
p2 = x * x
return p2
# 関数の呼び出し例1
x1 = 13
r1 = square(x1)
print(x1, r1)
# 関数の定義例2
def squares(x):
p2 = x * x
p3= x * x * x
return (p2, p3)
# 関数の呼び出し例2
x1 = 13
p2, p3 = squares(x1)
print(x1, p2, p3)
# 日本語化ライブラリ導入
!pip install japanize-matplotlib | tail -n 1
# 必要ライブラリのimport
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
# matplotlib日本語化対応
import japanize_matplotlib
# データフレーム表示用関数
from IPython.display import display
# 余分なワーニングを非表示にする
import warnings
warnings.filterwarnings('ignore')
# f文字列の表示
a1 = 1.0/7.0
a2 = 123
str1 = f'a1 = {a1} a2 = {a2}'
print(str1)
# f文字列の詳細オプション
# .4f 小数点以下4桁の固定小数点表示
# 04 整数を0詰め4桁表示
str2 = f'a1 = {a1:.4f} a2 = {a2:04}'
print(str2)
# 04e 小数点以下4桁の浮動小数点表示
# #x 整数の16進数表示
str3 = f'a1 = {a1:.04e} a2 = {a2:#x}'
print(str3)
| 0.128552 | 0.925027 |
[](https://colab.research.google.com/github/ThomasAlbin/sandbox/blob/main/asteroid_taxonomy/5_2_ml_search.ipynb)
# Step 5.2: Machine Learning - Parameter Search / Optimization
In this step we are performing a GridSearch on the binary classification problem. Our considered metric: the F1 score!
```
# Import standard libraries
import os
# Import installed libraries
import numpy as np
import pandas as pd
import sklearn
from sklearn import preprocessing
from sklearn import svm
from sklearn.model_selection import GridSearchCV
# Let's mount the Google Drive, where we store files and models (if applicable, otherwise work
# locally)
try:
from google.colab import drive
drive.mount('/gdrive')
core_path = "/gdrive/MyDrive/Colab/asteroid_taxonomy/"
except ModuleNotFoundError:
core_path = ""
# Load the level 2 asteroid data
asteroids_df = pd.read_pickle(os.path.join(core_path, "data/lvl2/", "asteroids.pkl"))
# Now we add a binary classification schema, where we distinguish between e.g., X and non-X classes
asteroids_df.loc[:, "Class"] = asteroids_df["Main_Group"].apply(lambda x: 1 if x=="X" else 0)
# Allocate the spectra to one array and the classes to another one
asteroids_X = np.array([k["Reflectance_norm550nm"].tolist() for k in asteroids_df["SpectrumDF"]])
asteroids_y = np.array(asteroids_df["Class"].to_list())
# In this example we create a single test-training split with a ratio of 0.8 / 0.2
from sklearn.model_selection import StratifiedShuffleSplit
sss = StratifiedShuffleSplit(n_splits=1, test_size=0.2)
# Create a simple, single train / test split
for train_index, test_index in sss.split(asteroids_X, asteroids_y):
X_train, X_test = asteroids_X[train_index], asteroids_X[test_index]
y_train, y_test = asteroids_y[train_index], asteroids_y[test_index]
# Compute class weightning
positive_class_weight = int(1.0 / (sum(y_train) / len(X_train)))
# Perform now a GridSearch with the following parameter range and kernels
param_grid = [
{'C': [1, 10, 100, 1000], 'kernel': ['linear']},
{'C': [1, 10, 100, 1000], 'kernel': ['rbf']},
]
# Set the SVM classifier
svc = svm.SVC(class_weight={1: positive_class_weight})
# Instantiate the StandardScaler (mean 0, standard deviation 1) and use the training data to fit
# the scaler
scaler = preprocessing.StandardScaler().fit(X_train)
# Transform now the training data
X_train_scaled = scaler.transform(X_train)
# Set the GridSearch and ...
wclf = GridSearchCV(svc, param_grid, scoring='f1', verbose=3, cv=5)
# ... perform the training!
wclf.fit(X_train_scaled, y_train)
# Optional: get the best estimator
final_clf = wclf.best_estimator_
# Scale the testing data ...
X_test_scaled = scaler.transform(X_test)
# ... and perform a predicition
y_test_pred = final_clf.predict(X_test_scaled)
# Import the confusion matrix and perform the computation
from sklearn.metrics import confusion_matrix
conf_mat = confusion_matrix(y_test, y_test_pred)
print(conf_mat)
# The order of the confusion matrix is:
# - true negative (top left, tn)
# - false positive (top right, fp)
# - false negative (bottom left, fn)
# - true positive (bottom right, tp)
tn, fp, fn, tp = conf_mat.ravel()
# Recall: ratio of correctly classified X Class spectra, considering the false negatives
# (recall = tp / (tp + fn))
recall_score = round(sklearn.metrics.recall_score(y_test, y_test_pred), 3)
print(f"Recall Score: {recall_score}")
# Precision: ratio of correctly classified X Class spectra, considering the false positives
# (precision = tp / (tp + fp))
precision_score = round(sklearn.metrics.precision_score(y_test, y_test_pred), 3)
print(f"Precision Score: {precision_score}")
# A combined score
f1_score = round(sklearn.metrics.f1_score(y_test, y_test_pred), 3)
print(f"F1 Score: {f1_score}")
```
# Summary / Outlook:
To finalize: Apply all data (take care of scaling!) on the training and rerun it. Save it afterwards for further computations. Storing and using a model requires also to store the corresponding scaler (store them as pickle files)!
Further notes:<br>
- If you would like to improve the model later on, one needs to perform "partial fits" to improve / train the weight(s). A simple "fit" re-runs the training and computes the model from scratch. Only linear SVMs can be partially fitted using [SGDClassifiers](https://scikit-learn.org/stable/modules/generated/sklearn.linear_model.SGDClassifier.html)
- Using all data to train a final model requires a proper metric. Cross validation as shown above can be used, but one should consider "Nested Cross Validation" methods as shown [here](https://scikit-learn.org/stable/auto_examples/model_selection/plot_nested_cross_validation_iris.html)
- Also: a computational more extensive method called [HalvingGridSearchCV](https://scikit-learn.org/stable/modules/generated/sklearn.model_selection.HalvingGridSearchCV.html#sklearn.model_selection.HalvingGridSearchCV) may improve the model even further
|
github_jupyter
|
# Import standard libraries
import os
# Import installed libraries
import numpy as np
import pandas as pd
import sklearn
from sklearn import preprocessing
from sklearn import svm
from sklearn.model_selection import GridSearchCV
# Let's mount the Google Drive, where we store files and models (if applicable, otherwise work
# locally)
try:
from google.colab import drive
drive.mount('/gdrive')
core_path = "/gdrive/MyDrive/Colab/asteroid_taxonomy/"
except ModuleNotFoundError:
core_path = ""
# Load the level 2 asteroid data
asteroids_df = pd.read_pickle(os.path.join(core_path, "data/lvl2/", "asteroids.pkl"))
# Now we add a binary classification schema, where we distinguish between e.g., X and non-X classes
asteroids_df.loc[:, "Class"] = asteroids_df["Main_Group"].apply(lambda x: 1 if x=="X" else 0)
# Allocate the spectra to one array and the classes to another one
asteroids_X = np.array([k["Reflectance_norm550nm"].tolist() for k in asteroids_df["SpectrumDF"]])
asteroids_y = np.array(asteroids_df["Class"].to_list())
# In this example we create a single test-training split with a ratio of 0.8 / 0.2
from sklearn.model_selection import StratifiedShuffleSplit
sss = StratifiedShuffleSplit(n_splits=1, test_size=0.2)
# Create a simple, single train / test split
for train_index, test_index in sss.split(asteroids_X, asteroids_y):
X_train, X_test = asteroids_X[train_index], asteroids_X[test_index]
y_train, y_test = asteroids_y[train_index], asteroids_y[test_index]
# Compute class weightning
positive_class_weight = int(1.0 / (sum(y_train) / len(X_train)))
# Perform now a GridSearch with the following parameter range and kernels
param_grid = [
{'C': [1, 10, 100, 1000], 'kernel': ['linear']},
{'C': [1, 10, 100, 1000], 'kernel': ['rbf']},
]
# Set the SVM classifier
svc = svm.SVC(class_weight={1: positive_class_weight})
# Instantiate the StandardScaler (mean 0, standard deviation 1) and use the training data to fit
# the scaler
scaler = preprocessing.StandardScaler().fit(X_train)
# Transform now the training data
X_train_scaled = scaler.transform(X_train)
# Set the GridSearch and ...
wclf = GridSearchCV(svc, param_grid, scoring='f1', verbose=3, cv=5)
# ... perform the training!
wclf.fit(X_train_scaled, y_train)
# Optional: get the best estimator
final_clf = wclf.best_estimator_
# Scale the testing data ...
X_test_scaled = scaler.transform(X_test)
# ... and perform a predicition
y_test_pred = final_clf.predict(X_test_scaled)
# Import the confusion matrix and perform the computation
from sklearn.metrics import confusion_matrix
conf_mat = confusion_matrix(y_test, y_test_pred)
print(conf_mat)
# The order of the confusion matrix is:
# - true negative (top left, tn)
# - false positive (top right, fp)
# - false negative (bottom left, fn)
# - true positive (bottom right, tp)
tn, fp, fn, tp = conf_mat.ravel()
# Recall: ratio of correctly classified X Class spectra, considering the false negatives
# (recall = tp / (tp + fn))
recall_score = round(sklearn.metrics.recall_score(y_test, y_test_pred), 3)
print(f"Recall Score: {recall_score}")
# Precision: ratio of correctly classified X Class spectra, considering the false positives
# (precision = tp / (tp + fp))
precision_score = round(sklearn.metrics.precision_score(y_test, y_test_pred), 3)
print(f"Precision Score: {precision_score}")
# A combined score
f1_score = round(sklearn.metrics.f1_score(y_test, y_test_pred), 3)
print(f"F1 Score: {f1_score}")
| 0.772101 | 0.913638 |
```
import matplotlib.pyplot as plt
import numpy as np
import tensorflow as tf
from tensorflow.contrib.learn.python.learn.datasets.mnist import read_data_sets
sess = tf.Session()
data_dir = 'temp'
mnist = read_data_sets(data_dir)
train_xdata = np.array([np.reshape(x, (28,28)) for x in mnist.train.images])
test_xdata = np.array([np.reshape(x, (28,28)) for x in mnist.test.images])
train_labels = mnist.train.labels
test_labels = mnist.test.labels
batch_size = 100
learning_rate = 0.005
evaluation_size = 500
image_width = train_xdata[0].shape[0]
image_height = train_xdata[0].shape[1]
target_size = max(train_labels) + 1
num_channels = 1
generations = 500
eval_every = 5
conv1_features = 25
conv2_features = 50
max_pool_size1 = 2
max_pool_size2 = 2
fully_connected_size_1 = 100
x_input_shape = (batch_size, image_width, image_height, num_channels)
x_input = tf.placeholder(tf.float32, shape=x_input_shape)
y_target = tf.placeholder(tf.int32, shape=(batch_size))
eval_input_shape = (evaluation_size, image_width, image_height, num_channels)
eval_input = tf.placeholder(tf.float32, shape=eval_input_shape)
eval_target = tf.placeholder(tf.int32, shape=(evaluation_size))
conv1_weight = tf.Variable(tf.truncated_normal([4, 4, num_channels, conv1_features], stddev=0.1, dtype=tf.float32))
conv1_bias = tf.Variable(tf.zeros([conv1_features], dtype=tf.float32))
conv2_weight = tf.Variable(tf.truncated_normal([4, 4, conv1_features, conv2_features], stddev=0.1, dtype=tf.float32))
conv2_bias = tf.Variable(tf.zeros([conv2_features], dtype=tf.float32))
resulting_width = image_width // (max_pool_size1 * max_pool_size2)
resulting_height = image_height // (max_pool_size1 * max_pool_size2)
full1_input_size = resulting_width * resulting_height * conv2_features
full1_weight = tf.Variable(tf.truncated_normal([full1_input_size, fully_connected_size_1], stddev=0.1, dtype=tf.float32))
full1_bias = tf.Variable(tf.truncated_normal([fully_connected_size_1], stddev=0.1, dtype=tf.float32))
full2_weight = tf.Variable(tf.truncated_normal([fully_connected_size_1, target_size], stddev=0.1, dtype=tf.float32))
full2_bias = tf.Variable(tf.truncated_normal([target_size], stddev=0.1, dtype=tf.float32))
def my_conv_net(input_data):
conv1 = tf.nn.conv2d(
input_data,
conv1_weight,
strides=[1,1,1,1],
padding='SAME')
relu1 = tf.nn.relu(tf.nn.bias_add(conv1, conv1_bias))
max_pool1 = tf.nn.max_pool(
relu1,
ksize=[1, max_pool_size1, max_pool_size1, 1],
strides=[1, max_pool_size1, max_pool_size1, 1],
padding='SAME')
conv2 = tf.nn.conv2d(
max_pool1,
conv2_weight,
strides=[1,1,1,1],
padding='SAME')
relu2 = tf.nn.relu(tf.nn.bias_add(conv2, conv2_bias))
max_pool2 = tf.nn.max_pool(
relu2,
ksize=[1, max_pool_size2, max_pool_size2, 1],
strides=[1, max_pool_size2, max_pool_size2, 1],
padding='SAME')
final_conv_shape = max_pool2.get_shape().as_list()
final_shape = final_conv_shape[1] * final_conv_shape[2] * final_conv_shape[3]
flat_output = tf.reshape(max_pool2, [final_conv_shape[0], final_shape])
fully_connected1 = tf.nn.relu(tf.add(tf.matmul(flat_output, full1_weight), full1_bias))
final_model_output = tf.add(tf.matmul(fully_connected1, full2_weight), full2_bias)
return(final_model_output)
# Initialize Model Operations
def my_conv_net(input_data):
# First Conv-ReLU-MaxPool Layer
conv1 = tf.nn.conv2d(input_data, conv1_weight, strides=[1, 1, 1, 1], padding='SAME')
relu1 = tf.nn.relu(tf.nn.bias_add(conv1, conv1_bias))
max_pool1 = tf.nn.max_pool(relu1, ksize=[1, max_pool_size1, max_pool_size1, 1],
strides=[1, max_pool_size1, max_pool_size1, 1], padding='SAME')
# Second Conv-ReLU-MaxPool Layer
conv2 = tf.nn.conv2d(max_pool1, conv2_weight, strides=[1, 1, 1, 1], padding='SAME')
relu2 = tf.nn.relu(tf.nn.bias_add(conv2, conv2_bias))
max_pool2 = tf.nn.max_pool(relu2, ksize=[1, max_pool_size2, max_pool_size2, 1],
strides=[1, max_pool_size2, max_pool_size2, 1], padding='SAME')
# Transform Output into a 1xN layer for next fully connected layer
final_conv_shape = max_pool2.get_shape().as_list()
final_shape = final_conv_shape[1] * final_conv_shape[2] * final_conv_shape[3]
flat_output = tf.reshape(max_pool2, [final_conv_shape[0], final_shape])
# First Fully Connected Layer
fully_connected1 = tf.nn.relu(tf.add(tf.matmul(flat_output, full1_weight), full1_bias))
# Second Fully Connected Layer
final_model_output = tf.add(tf.matmul(fully_connected1, full2_weight), full2_bias)
return(final_model_output)
model_output = my_conv_net(x_input)
test_model_output = my_conv_net(eval_input)
model_output = my_conv_net(x_input)
test_model_output = my_conv_net(eval_input)
loss = tf.reduce_mean(tf.nn.sparse_softmax_cross_entropy_with_logits(logits=model_output, labels=y_target))
prediction = tf.nn.softmax(model_output)
test_prediction = tf.nn.softmax(test_model_output)
def get_accuracy(logits, targets):
batch_predictions = np.argmax(logits, axis=1)
num_correct = np.sum(np.equal(batch_predictions, targets))
return(100. * num_correct / batch_predictions.shape[0])
optimizer = tf.train.MomentumOptimizer(learning_rate, 0.9)
train_step = optimizer.minimize(loss)
init = tf.global_variables_initializer()
sess.run(init)
train_loss = []
train_acc = []
test_acc = []
for i in range(generations):
rand_index = np.random.choice(len(train_xdata), size=batch_size)
rand_x = train_xdata[rand_index]
rand_x = np.expand_dims(rand_x, 3)
rand_y = train_labels[rand_index]
train_dict = {x_input: rand_x, y_target: rand_y}
sess.run(train_step, feed_dict=train_dict)
temp_train_loss, temp_train_preds = sess.run([loss, prediction], feed_dict=train_dict)
temp_train_acc = get_accuracy(temp_train_preds, rand_y)
if (i+1) % eval_every == 0:
eval_index = np.random.choice(len(test_xdata), size=evaluation_size)
eval_x = test_xdata[eval_index]
eval_x = np.expand_dims(eval_x, 3)
eval_y = test_labels[eval_index]
test_dict = {eval_input: eval_x, eval_target: eval_y}
test_preds = sess.run(test_prediction, feed_dict=test_dict)
temp_test_acc = get_accuracy(test_preds, eval_y)
train_loss.append(temp_train_loss)
train_acc.append(temp_train_acc)
test_acc.append(temp_test_acc)
acc_and_loss = [(i+1), temp_train_loss, temp_train_acc, temp_test_acc]
acc_and_loss = [np.round(x, 2) for x in acc_and_loss]
print('Generation # {}. Train Loss: {:.2f}. Train Acc (Test Acc): {:.2f} ({:.2f})'.format(*acc_and_loss))
eval_indices = range(0, generations, eval_every)
plt.plot(eval_indices, train_loss, 'k-')
plt.title('Softmax Loss per Generation')
plt.xlabel('Generation')
plt.ylabel('Softmax Loss')
plt.show()
plt.plot(eval_indices, train_acc, 'k-', label='Train set Accuracy')
plt.plot(eval_indices, test_acc, 'r--', label='Test set Accuracy')
plt.title('Train and Test Accuracy')
plt.xlabel('Generation')
plt.ylabel('Accuracy')
plt.legend(loc='lower right')
plt.show()
actuals = rand_y[0:6]
predictions = np.argmax(temp_train_preds, axis=1)[0:6]
images = np.squeeze(rand_x[0:6])
Nrows = 2
Ncols = 3
for i in range(6):
plt.subplot(Nrows, Ncols, i+1)
plt.imshow(np.reshape(images[i], [28, 28]), cmap='Greys_r')
plt.title('Actual: {} Pred: {}'.format(actuals[i], predictions[i]), fontsize=10)
frame = plt.gca()
frame.axes.get_xaxis().set_visible(False)
frame.axes.get_yaxis().set_visible(False)
plt.show()
```
|
github_jupyter
|
import matplotlib.pyplot as plt
import numpy as np
import tensorflow as tf
from tensorflow.contrib.learn.python.learn.datasets.mnist import read_data_sets
sess = tf.Session()
data_dir = 'temp'
mnist = read_data_sets(data_dir)
train_xdata = np.array([np.reshape(x, (28,28)) for x in mnist.train.images])
test_xdata = np.array([np.reshape(x, (28,28)) for x in mnist.test.images])
train_labels = mnist.train.labels
test_labels = mnist.test.labels
batch_size = 100
learning_rate = 0.005
evaluation_size = 500
image_width = train_xdata[0].shape[0]
image_height = train_xdata[0].shape[1]
target_size = max(train_labels) + 1
num_channels = 1
generations = 500
eval_every = 5
conv1_features = 25
conv2_features = 50
max_pool_size1 = 2
max_pool_size2 = 2
fully_connected_size_1 = 100
x_input_shape = (batch_size, image_width, image_height, num_channels)
x_input = tf.placeholder(tf.float32, shape=x_input_shape)
y_target = tf.placeholder(tf.int32, shape=(batch_size))
eval_input_shape = (evaluation_size, image_width, image_height, num_channels)
eval_input = tf.placeholder(tf.float32, shape=eval_input_shape)
eval_target = tf.placeholder(tf.int32, shape=(evaluation_size))
conv1_weight = tf.Variable(tf.truncated_normal([4, 4, num_channels, conv1_features], stddev=0.1, dtype=tf.float32))
conv1_bias = tf.Variable(tf.zeros([conv1_features], dtype=tf.float32))
conv2_weight = tf.Variable(tf.truncated_normal([4, 4, conv1_features, conv2_features], stddev=0.1, dtype=tf.float32))
conv2_bias = tf.Variable(tf.zeros([conv2_features], dtype=tf.float32))
resulting_width = image_width // (max_pool_size1 * max_pool_size2)
resulting_height = image_height // (max_pool_size1 * max_pool_size2)
full1_input_size = resulting_width * resulting_height * conv2_features
full1_weight = tf.Variable(tf.truncated_normal([full1_input_size, fully_connected_size_1], stddev=0.1, dtype=tf.float32))
full1_bias = tf.Variable(tf.truncated_normal([fully_connected_size_1], stddev=0.1, dtype=tf.float32))
full2_weight = tf.Variable(tf.truncated_normal([fully_connected_size_1, target_size], stddev=0.1, dtype=tf.float32))
full2_bias = tf.Variable(tf.truncated_normal([target_size], stddev=0.1, dtype=tf.float32))
def my_conv_net(input_data):
conv1 = tf.nn.conv2d(
input_data,
conv1_weight,
strides=[1,1,1,1],
padding='SAME')
relu1 = tf.nn.relu(tf.nn.bias_add(conv1, conv1_bias))
max_pool1 = tf.nn.max_pool(
relu1,
ksize=[1, max_pool_size1, max_pool_size1, 1],
strides=[1, max_pool_size1, max_pool_size1, 1],
padding='SAME')
conv2 = tf.nn.conv2d(
max_pool1,
conv2_weight,
strides=[1,1,1,1],
padding='SAME')
relu2 = tf.nn.relu(tf.nn.bias_add(conv2, conv2_bias))
max_pool2 = tf.nn.max_pool(
relu2,
ksize=[1, max_pool_size2, max_pool_size2, 1],
strides=[1, max_pool_size2, max_pool_size2, 1],
padding='SAME')
final_conv_shape = max_pool2.get_shape().as_list()
final_shape = final_conv_shape[1] * final_conv_shape[2] * final_conv_shape[3]
flat_output = tf.reshape(max_pool2, [final_conv_shape[0], final_shape])
fully_connected1 = tf.nn.relu(tf.add(tf.matmul(flat_output, full1_weight), full1_bias))
final_model_output = tf.add(tf.matmul(fully_connected1, full2_weight), full2_bias)
return(final_model_output)
# Initialize Model Operations
def my_conv_net(input_data):
# First Conv-ReLU-MaxPool Layer
conv1 = tf.nn.conv2d(input_data, conv1_weight, strides=[1, 1, 1, 1], padding='SAME')
relu1 = tf.nn.relu(tf.nn.bias_add(conv1, conv1_bias))
max_pool1 = tf.nn.max_pool(relu1, ksize=[1, max_pool_size1, max_pool_size1, 1],
strides=[1, max_pool_size1, max_pool_size1, 1], padding='SAME')
# Second Conv-ReLU-MaxPool Layer
conv2 = tf.nn.conv2d(max_pool1, conv2_weight, strides=[1, 1, 1, 1], padding='SAME')
relu2 = tf.nn.relu(tf.nn.bias_add(conv2, conv2_bias))
max_pool2 = tf.nn.max_pool(relu2, ksize=[1, max_pool_size2, max_pool_size2, 1],
strides=[1, max_pool_size2, max_pool_size2, 1], padding='SAME')
# Transform Output into a 1xN layer for next fully connected layer
final_conv_shape = max_pool2.get_shape().as_list()
final_shape = final_conv_shape[1] * final_conv_shape[2] * final_conv_shape[3]
flat_output = tf.reshape(max_pool2, [final_conv_shape[0], final_shape])
# First Fully Connected Layer
fully_connected1 = tf.nn.relu(tf.add(tf.matmul(flat_output, full1_weight), full1_bias))
# Second Fully Connected Layer
final_model_output = tf.add(tf.matmul(fully_connected1, full2_weight), full2_bias)
return(final_model_output)
model_output = my_conv_net(x_input)
test_model_output = my_conv_net(eval_input)
model_output = my_conv_net(x_input)
test_model_output = my_conv_net(eval_input)
loss = tf.reduce_mean(tf.nn.sparse_softmax_cross_entropy_with_logits(logits=model_output, labels=y_target))
prediction = tf.nn.softmax(model_output)
test_prediction = tf.nn.softmax(test_model_output)
def get_accuracy(logits, targets):
batch_predictions = np.argmax(logits, axis=1)
num_correct = np.sum(np.equal(batch_predictions, targets))
return(100. * num_correct / batch_predictions.shape[0])
optimizer = tf.train.MomentumOptimizer(learning_rate, 0.9)
train_step = optimizer.minimize(loss)
init = tf.global_variables_initializer()
sess.run(init)
train_loss = []
train_acc = []
test_acc = []
for i in range(generations):
rand_index = np.random.choice(len(train_xdata), size=batch_size)
rand_x = train_xdata[rand_index]
rand_x = np.expand_dims(rand_x, 3)
rand_y = train_labels[rand_index]
train_dict = {x_input: rand_x, y_target: rand_y}
sess.run(train_step, feed_dict=train_dict)
temp_train_loss, temp_train_preds = sess.run([loss, prediction], feed_dict=train_dict)
temp_train_acc = get_accuracy(temp_train_preds, rand_y)
if (i+1) % eval_every == 0:
eval_index = np.random.choice(len(test_xdata), size=evaluation_size)
eval_x = test_xdata[eval_index]
eval_x = np.expand_dims(eval_x, 3)
eval_y = test_labels[eval_index]
test_dict = {eval_input: eval_x, eval_target: eval_y}
test_preds = sess.run(test_prediction, feed_dict=test_dict)
temp_test_acc = get_accuracy(test_preds, eval_y)
train_loss.append(temp_train_loss)
train_acc.append(temp_train_acc)
test_acc.append(temp_test_acc)
acc_and_loss = [(i+1), temp_train_loss, temp_train_acc, temp_test_acc]
acc_and_loss = [np.round(x, 2) for x in acc_and_loss]
print('Generation # {}. Train Loss: {:.2f}. Train Acc (Test Acc): {:.2f} ({:.2f})'.format(*acc_and_loss))
eval_indices = range(0, generations, eval_every)
plt.plot(eval_indices, train_loss, 'k-')
plt.title('Softmax Loss per Generation')
plt.xlabel('Generation')
plt.ylabel('Softmax Loss')
plt.show()
plt.plot(eval_indices, train_acc, 'k-', label='Train set Accuracy')
plt.plot(eval_indices, test_acc, 'r--', label='Test set Accuracy')
plt.title('Train and Test Accuracy')
plt.xlabel('Generation')
plt.ylabel('Accuracy')
plt.legend(loc='lower right')
plt.show()
actuals = rand_y[0:6]
predictions = np.argmax(temp_train_preds, axis=1)[0:6]
images = np.squeeze(rand_x[0:6])
Nrows = 2
Ncols = 3
for i in range(6):
plt.subplot(Nrows, Ncols, i+1)
plt.imshow(np.reshape(images[i], [28, 28]), cmap='Greys_r')
plt.title('Actual: {} Pred: {}'.format(actuals[i], predictions[i]), fontsize=10)
frame = plt.gca()
frame.axes.get_xaxis().set_visible(False)
frame.axes.get_yaxis().set_visible(False)
plt.show()
| 0.79649 | 0.644966 |
```
import matplotlib.pyplot as plt
from astropy.io import fits
from astropy.table import Table
import numpy as np
from marvin.tools.cube import Cube
with fits.open('./data2/galaxies_sorted.fits') as hdulist:
jelly = hdulist[1].data['plateifu']
with fits.open('./data2/manga_firefly-v2_4_3-STELLARPOP.fits') as fin:
plateifus = fin['GALAXY_INFO'].data['PLATEIFU']
spaxel_binid = fin['SPATIAL_BINID'].data
mstar_all = fin['SURFACE_MASS_DENSITY_VORONOI'].data
gal = jelly[0]
# Select galaxy and binids
ind1 = np.where(plateifus == gal)[0][0]
ind_binid = spaxel_binid[ind1, :, :, 0].astype(int)
# Create 2D stellar mass array
mstar = np.ones(ind_binid.shape) * np.nan
for row, inds in enumerate(ind_binid):
ind_nans = np.where(inds == -99)
mstar[row] = mstar_all[ind1, inds, 0]
mstar[row][ind_nans] = np.nan
# trim mstar to match size of DAP maps and write to csv
cube = Cube(gal)
len_x = int(cube.header['NAXIS1'])
mdens = mstar[:len_x, :len_x]
mdens_ma = np.ma.array(data=mdens, mask=mdens==-9999.)
plt.imshow(mdens_ma)
import mass_dens_mapext
gal = jelly[0]
massmap = mass_dens_mapext.get_massmap(gal)
plt.imshow(massmap)
plt.colorbar()
import mass_mapext
gal = jelly[0]
massmap = mass_mapext.get_massmap(gal)
plt.imshow(massmap)
plt.colorbar()
import jf
a = jf.Mymaps(plateifu=jelly[0],max_radii=1.5)
print(a.get_mass())
# a.nsa['z']
plt.imshow(a.spx_ellcoo_r_re.value>1.5)
plt.colorbar()
plt.imshow(a.get_mass_map_fromdens())
plt.colorbar()
massmap = a.get_mass_map()
plt.imshow(massmap)
plt.colorbar()
np.log10(np.ma.sum(np.power(massmap,10)))
jelly[0]
pipe3d_test = fits.open('manga-8944-6101.Pipe3D.cube.fits')
hdu0 = pipe3d_test[0]
import matplotlib.pyplot as plt
from astropy.io import fits
from astropy.table import Table
import numpy as np
from marvin.tools.cube import Cube
import pandas as pd
with fits.open('./data2/manga_firefly-v2_4_3-STELLARPOP.fits')as fin:
firefly_plateifus = fin['GALAXY_INFO'].data['PLATEIFU']
spaxel_binid = fin['SPATIAL_BINID'].data
mstar_all = fin['SURFACE_MASS_DENSITY_VORONOI'].data
spa_info = fin['SPATIAL_INFO'].data
def get_massmap(gal):
# Select galaxy and binids
ind1 = np.where(firefly_plateifus == gal)[0][0]
ind_binid = spaxel_binid[ind1, :, :].astype(int)
cells_binid = spa_info[ind1,:,0]
# Create 2D stellar mass array
mstar = np.ones(ind_binid.shape) * np.nan
for row, inds in enumerate(ind_binid):
inds[inds==-1]=-9999
ind_nans = np.where(np.logical_or(inds==-1,inds==-9999))
# print(inds)
cells=[]
for i in inds:
cells.append(np.where(cells_binid==(i+0.0))[0][0])
mstar[row] = mstar_all[ind1, cells, 0]
mstar[row][ind_nans] = 0.0
# trim mstar to match size of DAP maps
cube = Cube(gal)
len_x = int(cube.header['NAXIS1'])
mdens = mstar[:len_x, :len_x]
mdens_ma = np.ma.array(data=mdens, mask=mdens==0.0)
return mdens_ma
map_test = get_massmap(jelly[0])
plt.imshow(map_test)
plt.colorbar()
spaxel_binid_dis = spaxel_binid[np.where(firefly_plateifus == jelly[0])[0][0]].copy()
spaxel_binid_dis[spaxel_binid_dis==-1]=0
spaxel_binid_dis[spaxel_binid_dis==-9999]=0
cube = Cube(jelly[0])
len_x = int(cube.header['NAXIS1'])
spaxel_binid_dis = spaxel_binid_dis[:len_x, :len_x]
spaxel_binid_dis_ma = np.ma.array(data=spaxel_binid_dis, mask=spaxel_binid_dis==0.0)
plt.imshow(spaxel_binid_dis_ma)
mstar_all[0,:,0]
import mass_sum
mass_sum.get_mass(jelly[0])
import matplotlib.pyplot as plt
from astropy.io import fits
from astropy.table import Table
import numpy as np
from marvin.tools.cube import Cube
import pandas as pd
with fits.open('./data2/manga_firefly-v2_4_3-STELLARPOP.fits')as fin:
firefly_plateifus = fin['GALAXY_INFO'].data['PLATEIFU']
spaxel_binid = fin['SPATIAL_BINID'].data
mstar_all = fin['STELLAR_MASS_VORONOI'].data
spa_info = fin['SPATIAL_INFO'].data
ind1 = np.where(firefly_plateifus == jelly[0])[0][0]
mass_cell = mstar_all[ind1, :, 0]
mass_cell[mass_cell<0]=0.0
mass_list = np.ma.array(data=mass_cell, mask = mass_cell==0.0)
print(mass_list)
mass = np.log10(np.ma.sum(10**mass_list))
mass
import matplotlib.pyplot as plt
from astropy.io import fits
from astropy.table import Table
import numpy as np
from marvin.tools.cube import Cube
import pandas as pd
with fits.open('./data2/manga_firefly-v2_4_3-STELLARPOP.fits')as fin:
firefly_plateifus = fin['GALAXY_INFO'].data['PLATEIFU']
spaxel_binid = fin['SPATIAL_BINID'].data
mstar_all = fin['STELLAR_MASS_VORONOI'].data
spa_info = fin['SPATIAL_INFO'].data
ind1 = np.where(firefly_plateifus == jelly[0])[0][0]
mass_cell = mstar_all[ind1, :, 0]
mass_cell[mass_cell<0]=0.0
mass_list = np.ma.array(data=mass_cell, mask = mass_cell==0.0)
print(mass_list)
mass = np.log10(np.ma.sum(np.power(mass_list,10)))
mass
# bin number map
spin = spaxel_binid[1539]
np.savetxt('/Users/mlang/Desktop/jellydebug/spin.dat', spin)
spin_flat = spin.flatten()
bins = np.unique(spin_flat)
bins
np.unique(mstar_all[1539,:,0])
spa_info[1539][:,0]
hdu = fits.open('./data2/manga_firefly-v2_4_3-STELLARPOP.fits')
PLATEIFU_all = hdu[1].data['PLATEIFU']
my_galaxy = np.where(PLATEIFU_all=='0')[0]
# my_galaxy
hdu['SPATIAL_INFO'].data[my_galaxy]
from astropy.table import Table
b = np.arange(1,24)
dic={}
dic['s_num'] = b
dic['plate_ifu']=jelly
jjj = Table(dic)
from astropy.table import Table
t = Table()
t['s_num'] = b
t['plate_ifu'] = jelly
t.write('./jellyfish_ifu.fits')
import numpy as np
from astropy.io import fits
import matplotlib.pyplot as plt
from astropy.table import Table
def calc_mass(plateifu):
hdu = fits.open('./data2/manga_firefly-v2_4_3-STELLARPOP.fits')
PLATEIFU_all = hdu[1].data['PLATEIFU']
my_galaxy = np.where(PLATEIFU_all==plateifu)[0]
# print(hdu['SPATIAL_INFO'].data[my_galaxy][0,:,0])
remove_nan = ~np.isnan(hdu['SPATIAL_INFO'].data[my_galaxy][0,:,0])
vor_bin = hdu['SPATIAL_INFO'].data[my_galaxy][0,:,0][remove_nan]
logMstar = hdu['STELLAR_MASS_VORONOI'].data[my_galaxy][0,:,0][remove_nan]
vor_labels = np.unique(vor_bin)
print(vor_labels)
tot_Mstar = 0
for v in vor_labels:
ind = np.where(vor_bin==v)[0]
# print(ind)
tot_Mstar += np.average(10**logMstar[ind])
return np.log10(tot_Mstar)
t = Table.read('Jellyfish_GSWLC.fits')
plateifu_array = np.asarray(t['plateifu']).astype(str)
M = np.asarray(t['log_M'])
M_ff = np.zeros(M.size)
for i in range(M.size):
M_ff[i] = calc_mass(plateifu_array[i])
plt.scatter(M, M_ff)
plt.xlim(8.5,11)
plt.ylim(8.5,11)
plt.plot([8.5,11], [8.5,11], 'k--')
plt.xlabel('GSLWC')
plt.ylabel('Firefly')
plt.show()
import numpy as np
from astropy.io import fits
import matplotlib.pyplot as plt
from astropy.table import Table
t = Table.read('Jellyfish_GSWLC.fits')
plateifu_array = np.asarray(t['plateifu']).astype(str)
M = np.asarray(t['log_M'])
M_ff = np.zeros(M.size)
for i in range(M.size):
a = jf.Mymaps(plateifu=jelly[i], max_radii=5)
M_ff[i] = a.get_mass()
plt.scatter(M, M_ff)
plt.xlim(8.5,11)
plt.ylim(8.5,11)
plt.plot([8.5,11], [8.5,11], 'k--')
plt.xlabel('GSLWC')
plt.ylabel('Firefly')
plt.show()
import mass_sum
print(mass_sum.get_mass(jelly[1]))
np.power(10,2)
```
|
github_jupyter
|
import matplotlib.pyplot as plt
from astropy.io import fits
from astropy.table import Table
import numpy as np
from marvin.tools.cube import Cube
with fits.open('./data2/galaxies_sorted.fits') as hdulist:
jelly = hdulist[1].data['plateifu']
with fits.open('./data2/manga_firefly-v2_4_3-STELLARPOP.fits') as fin:
plateifus = fin['GALAXY_INFO'].data['PLATEIFU']
spaxel_binid = fin['SPATIAL_BINID'].data
mstar_all = fin['SURFACE_MASS_DENSITY_VORONOI'].data
gal = jelly[0]
# Select galaxy and binids
ind1 = np.where(plateifus == gal)[0][0]
ind_binid = spaxel_binid[ind1, :, :, 0].astype(int)
# Create 2D stellar mass array
mstar = np.ones(ind_binid.shape) * np.nan
for row, inds in enumerate(ind_binid):
ind_nans = np.where(inds == -99)
mstar[row] = mstar_all[ind1, inds, 0]
mstar[row][ind_nans] = np.nan
# trim mstar to match size of DAP maps and write to csv
cube = Cube(gal)
len_x = int(cube.header['NAXIS1'])
mdens = mstar[:len_x, :len_x]
mdens_ma = np.ma.array(data=mdens, mask=mdens==-9999.)
plt.imshow(mdens_ma)
import mass_dens_mapext
gal = jelly[0]
massmap = mass_dens_mapext.get_massmap(gal)
plt.imshow(massmap)
plt.colorbar()
import mass_mapext
gal = jelly[0]
massmap = mass_mapext.get_massmap(gal)
plt.imshow(massmap)
plt.colorbar()
import jf
a = jf.Mymaps(plateifu=jelly[0],max_radii=1.5)
print(a.get_mass())
# a.nsa['z']
plt.imshow(a.spx_ellcoo_r_re.value>1.5)
plt.colorbar()
plt.imshow(a.get_mass_map_fromdens())
plt.colorbar()
massmap = a.get_mass_map()
plt.imshow(massmap)
plt.colorbar()
np.log10(np.ma.sum(np.power(massmap,10)))
jelly[0]
pipe3d_test = fits.open('manga-8944-6101.Pipe3D.cube.fits')
hdu0 = pipe3d_test[0]
import matplotlib.pyplot as plt
from astropy.io import fits
from astropy.table import Table
import numpy as np
from marvin.tools.cube import Cube
import pandas as pd
with fits.open('./data2/manga_firefly-v2_4_3-STELLARPOP.fits')as fin:
firefly_plateifus = fin['GALAXY_INFO'].data['PLATEIFU']
spaxel_binid = fin['SPATIAL_BINID'].data
mstar_all = fin['SURFACE_MASS_DENSITY_VORONOI'].data
spa_info = fin['SPATIAL_INFO'].data
def get_massmap(gal):
# Select galaxy and binids
ind1 = np.where(firefly_plateifus == gal)[0][0]
ind_binid = spaxel_binid[ind1, :, :].astype(int)
cells_binid = spa_info[ind1,:,0]
# Create 2D stellar mass array
mstar = np.ones(ind_binid.shape) * np.nan
for row, inds in enumerate(ind_binid):
inds[inds==-1]=-9999
ind_nans = np.where(np.logical_or(inds==-1,inds==-9999))
# print(inds)
cells=[]
for i in inds:
cells.append(np.where(cells_binid==(i+0.0))[0][0])
mstar[row] = mstar_all[ind1, cells, 0]
mstar[row][ind_nans] = 0.0
# trim mstar to match size of DAP maps
cube = Cube(gal)
len_x = int(cube.header['NAXIS1'])
mdens = mstar[:len_x, :len_x]
mdens_ma = np.ma.array(data=mdens, mask=mdens==0.0)
return mdens_ma
map_test = get_massmap(jelly[0])
plt.imshow(map_test)
plt.colorbar()
spaxel_binid_dis = spaxel_binid[np.where(firefly_plateifus == jelly[0])[0][0]].copy()
spaxel_binid_dis[spaxel_binid_dis==-1]=0
spaxel_binid_dis[spaxel_binid_dis==-9999]=0
cube = Cube(jelly[0])
len_x = int(cube.header['NAXIS1'])
spaxel_binid_dis = spaxel_binid_dis[:len_x, :len_x]
spaxel_binid_dis_ma = np.ma.array(data=spaxel_binid_dis, mask=spaxel_binid_dis==0.0)
plt.imshow(spaxel_binid_dis_ma)
mstar_all[0,:,0]
import mass_sum
mass_sum.get_mass(jelly[0])
import matplotlib.pyplot as plt
from astropy.io import fits
from astropy.table import Table
import numpy as np
from marvin.tools.cube import Cube
import pandas as pd
with fits.open('./data2/manga_firefly-v2_4_3-STELLARPOP.fits')as fin:
firefly_plateifus = fin['GALAXY_INFO'].data['PLATEIFU']
spaxel_binid = fin['SPATIAL_BINID'].data
mstar_all = fin['STELLAR_MASS_VORONOI'].data
spa_info = fin['SPATIAL_INFO'].data
ind1 = np.where(firefly_plateifus == jelly[0])[0][0]
mass_cell = mstar_all[ind1, :, 0]
mass_cell[mass_cell<0]=0.0
mass_list = np.ma.array(data=mass_cell, mask = mass_cell==0.0)
print(mass_list)
mass = np.log10(np.ma.sum(10**mass_list))
mass
import matplotlib.pyplot as plt
from astropy.io import fits
from astropy.table import Table
import numpy as np
from marvin.tools.cube import Cube
import pandas as pd
with fits.open('./data2/manga_firefly-v2_4_3-STELLARPOP.fits')as fin:
firefly_plateifus = fin['GALAXY_INFO'].data['PLATEIFU']
spaxel_binid = fin['SPATIAL_BINID'].data
mstar_all = fin['STELLAR_MASS_VORONOI'].data
spa_info = fin['SPATIAL_INFO'].data
ind1 = np.where(firefly_plateifus == jelly[0])[0][0]
mass_cell = mstar_all[ind1, :, 0]
mass_cell[mass_cell<0]=0.0
mass_list = np.ma.array(data=mass_cell, mask = mass_cell==0.0)
print(mass_list)
mass = np.log10(np.ma.sum(np.power(mass_list,10)))
mass
# bin number map
spin = spaxel_binid[1539]
np.savetxt('/Users/mlang/Desktop/jellydebug/spin.dat', spin)
spin_flat = spin.flatten()
bins = np.unique(spin_flat)
bins
np.unique(mstar_all[1539,:,0])
spa_info[1539][:,0]
hdu = fits.open('./data2/manga_firefly-v2_4_3-STELLARPOP.fits')
PLATEIFU_all = hdu[1].data['PLATEIFU']
my_galaxy = np.where(PLATEIFU_all=='0')[0]
# my_galaxy
hdu['SPATIAL_INFO'].data[my_galaxy]
from astropy.table import Table
b = np.arange(1,24)
dic={}
dic['s_num'] = b
dic['plate_ifu']=jelly
jjj = Table(dic)
from astropy.table import Table
t = Table()
t['s_num'] = b
t['plate_ifu'] = jelly
t.write('./jellyfish_ifu.fits')
import numpy as np
from astropy.io import fits
import matplotlib.pyplot as plt
from astropy.table import Table
def calc_mass(plateifu):
hdu = fits.open('./data2/manga_firefly-v2_4_3-STELLARPOP.fits')
PLATEIFU_all = hdu[1].data['PLATEIFU']
my_galaxy = np.where(PLATEIFU_all==plateifu)[0]
# print(hdu['SPATIAL_INFO'].data[my_galaxy][0,:,0])
remove_nan = ~np.isnan(hdu['SPATIAL_INFO'].data[my_galaxy][0,:,0])
vor_bin = hdu['SPATIAL_INFO'].data[my_galaxy][0,:,0][remove_nan]
logMstar = hdu['STELLAR_MASS_VORONOI'].data[my_galaxy][0,:,0][remove_nan]
vor_labels = np.unique(vor_bin)
print(vor_labels)
tot_Mstar = 0
for v in vor_labels:
ind = np.where(vor_bin==v)[0]
# print(ind)
tot_Mstar += np.average(10**logMstar[ind])
return np.log10(tot_Mstar)
t = Table.read('Jellyfish_GSWLC.fits')
plateifu_array = np.asarray(t['plateifu']).astype(str)
M = np.asarray(t['log_M'])
M_ff = np.zeros(M.size)
for i in range(M.size):
M_ff[i] = calc_mass(plateifu_array[i])
plt.scatter(M, M_ff)
plt.xlim(8.5,11)
plt.ylim(8.5,11)
plt.plot([8.5,11], [8.5,11], 'k--')
plt.xlabel('GSLWC')
plt.ylabel('Firefly')
plt.show()
import numpy as np
from astropy.io import fits
import matplotlib.pyplot as plt
from astropy.table import Table
t = Table.read('Jellyfish_GSWLC.fits')
plateifu_array = np.asarray(t['plateifu']).astype(str)
M = np.asarray(t['log_M'])
M_ff = np.zeros(M.size)
for i in range(M.size):
a = jf.Mymaps(plateifu=jelly[i], max_radii=5)
M_ff[i] = a.get_mass()
plt.scatter(M, M_ff)
plt.xlim(8.5,11)
plt.ylim(8.5,11)
plt.plot([8.5,11], [8.5,11], 'k--')
plt.xlabel('GSLWC')
plt.ylabel('Firefly')
plt.show()
import mass_sum
print(mass_sum.get_mass(jelly[1]))
np.power(10,2)
| 0.243103 | 0.492249 |
# Random Forests
```
import numpy as np
import matplotlib.pyplot as plt
% matplotlib inline
plt.rcParams["figure.dpi"] = 200
np.set_printoptions(precision=3)
import pandas as pd
from sklearn.model_selection import train_test_split
from sklearn.pipeline import make_pipeline
from sklearn.preprocessing import scale, StandardScaler
from sklearn.datasets import load_breast_cancer
cancer = load_breast_cancer()
X_train, X_test, y_train, y_test = train_test_split(
cancer.data, cancer.target, stratify=cancer.target, random_state=0)
from sklearn.ensemble import RandomForestClassifier
from sklearn.datasets import load_digits
digits = load_digits()
X_train, X_test, y_train, y_test = train_test_split(
digits.data, digits.target, stratify=digits.target, random_state=0)
train_scores = []
test_scores = []
rf = RandomForestClassifier(warm_start=True)
estimator_range = range(1, 100, 5)
for n_estimators in estimator_range:
rf.n_estimators = n_estimators
rf.fit(X_train, y_train)
train_scores.append(rf.score(X_train, y_train))
test_scores.append(rf.score(X_test, y_test))
plt.plot(estimator_range, test_scores, label="test scores")
plt.plot(estimator_range, train_scores, label="train scores")
plt.ylabel("accuracy")
plt.xlabel("n_estimators")
plt.legend()
```
# out of bag predictions
```
train_scores = []
test_scores = []
oob_scores = []
feature_range = range(1, 64, 5)
for max_features in feature_range:
rf = RandomForestClassifier(max_features=max_features, oob_score=True, n_estimators=200, random_state=0)
rf.fit(X_train, y_train)
train_scores.append(rf.score(X_train, y_train))
test_scores.append(rf.score(X_test, y_test))
oob_scores.append(rf.oob_score_)
plt.plot(feature_range, test_scores, label="test scores")
plt.plot(feature_range, oob_scores, label="oob_scores scores")
plt.plot(feature_range, train_scores, label="train scores")
plt.legend()
plt.ylabel("accuracy")
plt.xlabel("max_features")
X_train, X_test, y_train, y_test = train_test_split(
cancer.data, cancer.target, stratify=cancer.target, random_state=1)
rf = RandomForestClassifier(n_estimators=100).fit(X_train, y_train)
rf.feature_importances_
pd.Series(rf.feature_importances_,
index=cancer.feature_names).plot(kind="barh")
```
# Exercise
Use a random forest classifier or random forest regressor on a dataset of your choice.
Try different values of n_estimators and max_depth and see how they impact performance and runtime.
Tune ``max_features`` with GridSearchCV.
```
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
import numpy as np
from sklearn.preprocessing import MinMaxScaler
dbf = pd.read_csv("data/adult.csv", index_col=0)
# identify the dependent variable
income = dbf.income
data_features = dbf.drop("income", axis=1)
# one hot encoding
data_one_hot = pd.get_dummies(data_features)
# Split data
X_train, X_test, y_train, y_test = train_test_split(data_one_hot, income
, stratify=income, random_state=0)
# Scale training data
scaler = MinMaxScaler().fit(X_train)
X_train_scaled = scaler.transform(X_train)
from sklearn.ensemble import RandomForestClassifier
train_scores = []
test_scores = []
rf = RandomForestClassifier(warm_start=True)
estimator_range = range(1, 20, 5)
for n_estimators in estimator_range:
rf.n_estimators = n_estimators
rf.fit(X_train, y_train)
train_scores.append(rf.score(X_train, y_train))
test_scores.append(rf.score(X_test, y_test))
plt.rcParams["figure.dpi"] = 300
rf = RandomForestClassifier(max_features=5, max_depth=3, min_samples_leaf=80).fit(X_train, y_train)
rf.feature_importances_[1:20]
pd.Series(rf.feature_importances_[1:20],
index=X_train.columns.values[1:20]).plot(kind="barh")
```
|
github_jupyter
|
import numpy as np
import matplotlib.pyplot as plt
% matplotlib inline
plt.rcParams["figure.dpi"] = 200
np.set_printoptions(precision=3)
import pandas as pd
from sklearn.model_selection import train_test_split
from sklearn.pipeline import make_pipeline
from sklearn.preprocessing import scale, StandardScaler
from sklearn.datasets import load_breast_cancer
cancer = load_breast_cancer()
X_train, X_test, y_train, y_test = train_test_split(
cancer.data, cancer.target, stratify=cancer.target, random_state=0)
from sklearn.ensemble import RandomForestClassifier
from sklearn.datasets import load_digits
digits = load_digits()
X_train, X_test, y_train, y_test = train_test_split(
digits.data, digits.target, stratify=digits.target, random_state=0)
train_scores = []
test_scores = []
rf = RandomForestClassifier(warm_start=True)
estimator_range = range(1, 100, 5)
for n_estimators in estimator_range:
rf.n_estimators = n_estimators
rf.fit(X_train, y_train)
train_scores.append(rf.score(X_train, y_train))
test_scores.append(rf.score(X_test, y_test))
plt.plot(estimator_range, test_scores, label="test scores")
plt.plot(estimator_range, train_scores, label="train scores")
plt.ylabel("accuracy")
plt.xlabel("n_estimators")
plt.legend()
train_scores = []
test_scores = []
oob_scores = []
feature_range = range(1, 64, 5)
for max_features in feature_range:
rf = RandomForestClassifier(max_features=max_features, oob_score=True, n_estimators=200, random_state=0)
rf.fit(X_train, y_train)
train_scores.append(rf.score(X_train, y_train))
test_scores.append(rf.score(X_test, y_test))
oob_scores.append(rf.oob_score_)
plt.plot(feature_range, test_scores, label="test scores")
plt.plot(feature_range, oob_scores, label="oob_scores scores")
plt.plot(feature_range, train_scores, label="train scores")
plt.legend()
plt.ylabel("accuracy")
plt.xlabel("max_features")
X_train, X_test, y_train, y_test = train_test_split(
cancer.data, cancer.target, stratify=cancer.target, random_state=1)
rf = RandomForestClassifier(n_estimators=100).fit(X_train, y_train)
rf.feature_importances_
pd.Series(rf.feature_importances_,
index=cancer.feature_names).plot(kind="barh")
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
import numpy as np
from sklearn.preprocessing import MinMaxScaler
dbf = pd.read_csv("data/adult.csv", index_col=0)
# identify the dependent variable
income = dbf.income
data_features = dbf.drop("income", axis=1)
# one hot encoding
data_one_hot = pd.get_dummies(data_features)
# Split data
X_train, X_test, y_train, y_test = train_test_split(data_one_hot, income
, stratify=income, random_state=0)
# Scale training data
scaler = MinMaxScaler().fit(X_train)
X_train_scaled = scaler.transform(X_train)
from sklearn.ensemble import RandomForestClassifier
train_scores = []
test_scores = []
rf = RandomForestClassifier(warm_start=True)
estimator_range = range(1, 20, 5)
for n_estimators in estimator_range:
rf.n_estimators = n_estimators
rf.fit(X_train, y_train)
train_scores.append(rf.score(X_train, y_train))
test_scores.append(rf.score(X_test, y_test))
plt.rcParams["figure.dpi"] = 300
rf = RandomForestClassifier(max_features=5, max_depth=3, min_samples_leaf=80).fit(X_train, y_train)
rf.feature_importances_[1:20]
pd.Series(rf.feature_importances_[1:20],
index=X_train.columns.values[1:20]).plot(kind="barh")
| 0.601477 | 0.918809 |
# Formulation of Maxwell's Equations in Cartesian Coordinates and Flat Spacetime - An Overview
## Author: Terrence Pierre Jacques
## This tutorial notebook outlines two formulations of Maxwell's equations.
### This module serves as a stepping stone towards solving Einstein's equations using NRPy+. As such, the [tutorial notebook on the scalar wave equation in curvilinear coordinates](Tutorial-ScalarWaveCurvilinear.ipynb) is *required* reading before beginning this module. That module, as well as its own prerequisite [module on reference metrics within NRPy+](Tutorial-Reference_Metric.ipynb) provides the needed overview of how NRPy+ handles reference metrics.
# Introduction
To numerically solve Einstein's equations, a system of coupled, non-linear equations, the spatial coordinates may be decoupled from the time coordinate. In doing so, numerical solutions may be marched forward in time, using the [Method of Lines](https://reference.wolfram.com/language/tutorial/NDSolveMethodOfLines.html) ([NRPy+ tutorial here](Tutorial-Method_of_Lines-C_Code_Generation.ipynb)). This is done using the 3 + 1 decomposition, originally done using the ADM formalism. However, it was found that using this formulation of Einstein's equations resulted in unstable simulations, due to its weakly hyperbolic system of equations. Hyperbolicity is the notion of the wave-like behavior of the numerical solution for a given system of PDE's. The ADM formulation of Einstein's equations involved mixed second derivatives, which give rise to a weakly hyperbolic system.
In contrast, the BSSN formalism removes these mixed second derivatives, resulting in a strongly hyperbolic system of equations. In this module, we follow the work of [Knapp, Walker & Baumgarte (2002)](https://arxiv.org/abs/gr-qc/0201051), showcasing the difference between using strongly versus weakly hyperbolic systems, when solving a physical problem with Maxwell's equations in vacuum.
### A Note on Notation:
As is standard in NRPy+,
* Greek indices refer to four-dimensional quantities where the zeroth component indicates temporal (time) component.
* Latin indices refer to three-dimensional quantities. This is somewhat counterintuitive since Python always indexes its lists starting from 0. As a result, the zeroth component of three-dimensional quantities will necessarily indicate the first *spatial* direction.
<a id='top'></a>
$$\label{top}$$
# Table of Contents:
1. [Step 1](#step1): Maxwell's Equations in Vacuum - System I (ADM-like)
1. [Step 2](#step2): Maxwell's Equations in Vacuum - System II (BSSN-like)
1. [Step 3](#latex_pdf_output): Output this notebook to $\LaTeX$-formatted PDF file
<a id='step1'></a>
# Step 1: Maxwell's Equations in Vacuum - System I (ADM-like) \[Back to [top](#top)\]
$$\label{step1}$$
We begin with Maxwell's equations in vacuum, i.e. no source terms, in flat space and in [Gaussian](https://en.wikipedia.org/wiki/Gaussian_units) and $c = 1$ units, given by
\begin{align}
&\vec{\nabla} \cdot \vec{E} = 0, \\
&\vec{\nabla} \cdot \vec{B} = 0, \\
&\frac{\partial \vec{B}}{\partial t} = -\vec{\nabla} \times \vec{E}, \\
&\frac{\partial \vec{E}}{\partial t} = \vec{\nabla} \times \vec{B},
\end{align}
and we have the associated potentials,
\begin{align}
\frac{\partial \vec{A}}{\partial t} &= -\vec{E} -\vec{\nabla}\varphi , \\
\vec{B} &= \vec{\nabla} \times \vec{A}.
\end{align}
Now, replacing $\vec{B}$ with $\vec{\nabla} \times \vec{A}$ in Ampere's law, and using the standard identity
$$
\vec{\nabla} \times \left( \vec{\nabla} \times \vec{A} \right) = \vec{\nabla} \left( \vec{\nabla} \cdot \vec{A} \right) - \nabla^2 \vec{A},
$$
we may write
$$
\vec{\nabla} \times \vec{B} = \vec{\nabla} \times \left( \vec{\nabla} \times \vec{A} \right) = \vec{\nabla} \left( \vec{\nabla} \cdot \vec{A} \right) - \nabla^2 \vec{A}.
$$
Thus, our time evolution equations become
\begin{align}
\frac{\partial \vec{A}}{\partial t} &= -\vec{E} -\vec{\nabla}\varphi , \\
\frac{\partial \vec{E}}{\partial t} &= \vec{\nabla} \left( \vec{\nabla} \cdot \vec{A} \right) - \nabla^2 \vec{A}.
\end{align}
Using index notation, in Cartesian coordinates we have
\begin{align}
\partial_t A^i &= -E^i - \partial^i \varphi, \\
\partial_t E^i &= \partial^i \partial_j A^j - \partial_j \partial^j A^i.
\end{align}
Note the presence of the mixed second derivative above, and that in our system of equations we have 6 equations (since $\vec{A}$ and $\vec{E}$ each have 3 components), but 7 unknowns (components of $\vec{A}$ and $\vec{E}$, and the scalar potential $\varphi$). Thus, we’ll add a time evolution equation to $\varphi$ as well, which amounts to choosing a gauge. In particular we’ll choose the [Lorenz gauge](https://en.wikipedia.org/wiki/Lorenz_gauge_condition):
$$
\partial_t \varphi = -\vec{\nabla} \cdot \vec{A},
$$
and using index notation, in Cartesian coordinates,
$$
\partial_t \varphi = -\partial_i A^i.
$$
Furthermore, note that because we are working in vacuum, in Cartesian coordinates we have the constraints
\begin{align}
\partial_i E^i &= 0, \\
\partial_i B^i &= 0,
\end{align}
But since $\vec{B} = \vec{\nabla} \times \vec{A}$, the divergence of $\vec{B}$ is automatically satisfied (excercise for the reader).
The right hand sides (RHSs) of our evolution equations are thus
\begin{align}
\partial_t A^i &= -E^i - \partial^i \varphi, \\
\partial_t E^i &= \partial^i \partial_j A^j - \partial_j \partial^j A^i, \\
\partial_t \varphi &= -\partial_i A^i,
\end{align}
subject to the constraint
$$
\mathcal{C} \equiv \partial_i E^i = 0.
$$
Tracking the departure of our numerical results from this constraint helps us keep track of the numerical error. We refer to this system of equations as System I.
In [this tutorial](Tutorial-VacuumMaxwell_Cartesian_RHSs.ipynb) the RHSs and constraint equations are implemented in NRPy+. The system is then evolved in time within a [start-to-finish notebook](Tutorial-Start_to_Finish-Solving_Maxwells_Equations_in_Vacuum-Cartesian.ipynb), using initial data defined in [this tutorial](Tutorial-VacuumMaxwell_InitialData.ipynb), showcasing the **instability** of the system as a result of the presence of the mixed second derivative for $E^i$.
<a id='step2'></a>
# Step 2: Maxwell's Equations in Vacuum - System II (BSSN - like) \[Back to [top](#top)\]
$$\label{step2}$$
As dicussed in the introduction and following the previous section, to maintain numerical stability and accuracy we remove the mixed 2nd derivative $\partial^i \partial_j A^j$ in the equation for $E^i$ by introducing
\begin{align}
\Gamma &\equiv \partial_i A^i, \\
\partial_t \Gamma &= \partial_i \partial_t A^i = -\partial_i E^i - \partial_i \partial^i \varphi \\
&= - \partial_i \partial^i \varphi.
\end{align}
Thus, our evolution equations are
\begin{align}
\partial_t A^i &= -E^i - \partial^i \varphi, \\
\partial_t E^i &= \partial^i \Gamma - \partial_j \partial^j A^i, \\
\partial_t \Gamma &= - \partial_i \partial^i \varphi, \\
\partial_t \varphi &= -\Gamma,
\end{align}
subject to the constraints
\begin{align}
\mathcal{G} &\equiv \Gamma - \partial_i A^i &= 0, \\
\mathcal{C} &\equiv \partial_i E^i &= 0.
\end{align}
We refer to this system of equations as System II.
In [this tutorial](Tutorial-VacuumMaxwell_Cartesian_RHSs.ipynb) the RHSs and constraints are implemented in NRPy+. The system is then evolved in time within a [start-to-finish notebook](Tutorial-Start_to_Finish-Solving_Maxwells_Equations_in_Vacuum-Cartesian.ipynb), using initial data defined in [this tutorial](Tutorial-VacuumMaxwell_InitialData.ipynb), showcasing the **stability** of the system as a result of the absence of the mixed second derivative for $E^i$.
<a id='latex_pdf_output'></a>
# Step 3: Output this notebook to $\LaTeX$-formatted PDF file \[Back to [top](#top)\]
$$\label{latex_pdf_output}$$
The following code cell converts this Jupyter notebook into a proper, clickable $\LaTeX$-formatted PDF file. After the cell is successfully run, the generated PDF may be found in the root NRPy+ tutorial directory, with filename
[Tutorial-VacuumMaxwell_formulation_Cartesian.pdf](Tutorial-VacuumMaxwell_formulation_Cartesian.pdf) (Note that clicking on this link may not work; you may need to open the PDF file through another means.)
```
import cmdline_helper as cmd # NRPy+: Multi-platform Python command-line interface
cmd.output_Jupyter_notebook_to_LaTeXed_PDF("Tutorial-VacuumMaxwell_formulation_Cartesian")
```
|
github_jupyter
|
import cmdline_helper as cmd # NRPy+: Multi-platform Python command-line interface
cmd.output_Jupyter_notebook_to_LaTeXed_PDF("Tutorial-VacuumMaxwell_formulation_Cartesian")
| 0.253953 | 0.993556 |
# Kalman FilterによるARMAモデル時系列の推定
## 関連
- `state_system.ipynb`
```
import numpy as np
import matplotlib.pyplot as plt
import sys
sys.path.append('../module')
from utils import cum_std
from kalman_filters import LinearKalmanFilter as LKF
from kalman_filters import ExtendedKalmanFilter as EnKF
np.random.seed(0)
def generate_state_data(F, G, q, x_0, size):
"""
モデルノイズは1次元を仮定
"""
data = np.zeros((size, len(x_0)))
x = x_0
data[0] = x
for i in range(1, size):
x = F@x + G@np.random.normal(loc=0, scale=q, size=(1,))
data[i] = x
return data
def generate_obs_data(H, r, series, noise=True):
"""
観測ノイズは1次元を仮定
"""
obs = (H@series.T).T
if noise:
obs += np.random.normal(loc=0, scale=r, size=(len(series),1))
return obs
```
## 1. ARMAモデルによるデータの生成
```
# ARモデル
p = 3
a = np.array([-0.9, -0.7, -0.5]).reshape(p, 1)
# MAモデル
q = 3
b = np.array([0.5, 0.5, 0.5])
N = max([p,q+1])
print(f'p = {p}, q = {q}, N = {N}')
# a, bを0拡張
if N > p:
a_N = np.vstack([a, np.zeros(N-p)])
else:
a_N = a
if N > q+1:
b_N = np.hstack([b, np.zeros(N-q-2)])
else:
b_N = b
# 状態遷移行列 (N, N)
F = np.block([a_N, np.vstack([np.eye(N-1), np.zeros((1, N-1))])])
print(f'F = \n{F}')
# ノイズ重み (N, 1), b_0=1を含む.
G = np.array([1, *b_N]).reshape(N, 1)
print(f'G = \n{G}')
# 観測モデル (1, N)
H = np.block([1, np.zeros((1, N-1))])
print(f'H_ma = \n{H}')
# モデルノイズstd
sigma_m = 1
q = sigma_m
Q = np.array([q])
# 観測ノイズstd
sigma_o = 0.5
r = sigma_o
R = np.array([r])
# データを生成
sample_size = 100
x_0 = np.ones(N)
state_data = generate_state_data(F, G, Q, x_0, sample_size)
obs_data = generate_obs_data(H, R, state_data)
true = generate_obs_data(H, R, state_data, noise=False)
plt.plot(true, label='true')
plt.plot(obs_data, label='obs')
_ = plt.title(f'$ARMA({p},{q}); \sigma_m={sigma_m}, \sigma_o={sigma_o}, a={a_N.T}, b={b_N.T}$')
plt.legend()
```
## 2. Kalman Filterによる状態推定
Linear Kalman Filterを使う
条件
- 1.でデータを生成したARMAモデルをそのまま使う.
- 初期値は
- $ x_0 $: state_dataからランダムに選ぶ.
- $ P_0 $: $ 10 I $とする.
```
%%time
y = obs_data
x_0 = state_data[np.random.randint(0, sample_size)]
P_0 = 10*np.eye(N)
lkf = LKF(F, H, G, Q, R, y, x_0, P_0, alpha=1)
lkf.forward_estimation()
estimate_data = generate_obs_data(H, R, np.array(lkf.x), noise=False)
```
### 結果のplot
```
fig = plt.figure(figsize=(15,12))
ax1 = fig.add_subplot(2,1,1)
ax1.plot(true, label='true', color='c')
# ax1.plot(obs_data, label='obs')
ax1.plot(estimate_data, label='est', color='g')
# ax1.plot(true - estimate_data, label='error', color='r')
plt.legend()
ax2 = fig.add_subplot(2,1,2)
# ax2.plot(np.abs(true - estimate_data)) 本来はこれ
ax2.plot(cum_std(true - estimate_data), label='est std')
# ax2.plot(np.abs(true - obs_data)) 本来はこれ
ax2.plot(cum_std(true - obs_data), label='obs std')
ax2.plot(lkf.trP, label='trP')
plt.legend()
```
### 発展
非線形モデルに拡張したExtended Kalman Filterでも確認.
今,モデルが線形なので同様の結果が得られた.
```
%%time
M = lambda x,t: F@x
y = obs_data
x_0 = state_data[np.random.randint(0, sample_size)]
P_0 = 10*np.eye(N)
ekf = EnKF(M, H, G, Q, R, y[:], x_0, P_0, alpha=1)
ekf.forward_estimation()
ekf_estimate_data = generate_obs_data(H, R, np.array(ekf.x), noise=False)
fig = plt.figure(figsize=(15,12))
ax1 = fig.add_subplot(2,1,1)
ax1.plot(true, label='true', color='c')
# ax1.plot(obs_data, label='obs')
ax1.plot(ekf_estimate_data, label='est', color='g')
# ax1.plot(true - ekf_estimate_data, label='error', color='r')
plt.legend()
ax2 = fig.add_subplot(2,1,2)
# ax2.plot(np.abs(true - ekf_estimate_data)) 本来はこれ
ax2.plot(cum_std(true - ekf_estimate_data), label='est std')
# ax2.plot(np.abs(true - obs_data)) 本来はこれ
ax2.plot(cum_std(true - obs_data), label='obs std')
ax2.plot(ekf.trP, label='trP')
plt.legend()
```
|
github_jupyter
|
import numpy as np
import matplotlib.pyplot as plt
import sys
sys.path.append('../module')
from utils import cum_std
from kalman_filters import LinearKalmanFilter as LKF
from kalman_filters import ExtendedKalmanFilter as EnKF
np.random.seed(0)
def generate_state_data(F, G, q, x_0, size):
"""
モデルノイズは1次元を仮定
"""
data = np.zeros((size, len(x_0)))
x = x_0
data[0] = x
for i in range(1, size):
x = F@x + G@np.random.normal(loc=0, scale=q, size=(1,))
data[i] = x
return data
def generate_obs_data(H, r, series, noise=True):
"""
観測ノイズは1次元を仮定
"""
obs = (H@series.T).T
if noise:
obs += np.random.normal(loc=0, scale=r, size=(len(series),1))
return obs
# ARモデル
p = 3
a = np.array([-0.9, -0.7, -0.5]).reshape(p, 1)
# MAモデル
q = 3
b = np.array([0.5, 0.5, 0.5])
N = max([p,q+1])
print(f'p = {p}, q = {q}, N = {N}')
# a, bを0拡張
if N > p:
a_N = np.vstack([a, np.zeros(N-p)])
else:
a_N = a
if N > q+1:
b_N = np.hstack([b, np.zeros(N-q-2)])
else:
b_N = b
# 状態遷移行列 (N, N)
F = np.block([a_N, np.vstack([np.eye(N-1), np.zeros((1, N-1))])])
print(f'F = \n{F}')
# ノイズ重み (N, 1), b_0=1を含む.
G = np.array([1, *b_N]).reshape(N, 1)
print(f'G = \n{G}')
# 観測モデル (1, N)
H = np.block([1, np.zeros((1, N-1))])
print(f'H_ma = \n{H}')
# モデルノイズstd
sigma_m = 1
q = sigma_m
Q = np.array([q])
# 観測ノイズstd
sigma_o = 0.5
r = sigma_o
R = np.array([r])
# データを生成
sample_size = 100
x_0 = np.ones(N)
state_data = generate_state_data(F, G, Q, x_0, sample_size)
obs_data = generate_obs_data(H, R, state_data)
true = generate_obs_data(H, R, state_data, noise=False)
plt.plot(true, label='true')
plt.plot(obs_data, label='obs')
_ = plt.title(f'$ARMA({p},{q}); \sigma_m={sigma_m}, \sigma_o={sigma_o}, a={a_N.T}, b={b_N.T}$')
plt.legend()
%%time
y = obs_data
x_0 = state_data[np.random.randint(0, sample_size)]
P_0 = 10*np.eye(N)
lkf = LKF(F, H, G, Q, R, y, x_0, P_0, alpha=1)
lkf.forward_estimation()
estimate_data = generate_obs_data(H, R, np.array(lkf.x), noise=False)
fig = plt.figure(figsize=(15,12))
ax1 = fig.add_subplot(2,1,1)
ax1.plot(true, label='true', color='c')
# ax1.plot(obs_data, label='obs')
ax1.plot(estimate_data, label='est', color='g')
# ax1.plot(true - estimate_data, label='error', color='r')
plt.legend()
ax2 = fig.add_subplot(2,1,2)
# ax2.plot(np.abs(true - estimate_data)) 本来はこれ
ax2.plot(cum_std(true - estimate_data), label='est std')
# ax2.plot(np.abs(true - obs_data)) 本来はこれ
ax2.plot(cum_std(true - obs_data), label='obs std')
ax2.plot(lkf.trP, label='trP')
plt.legend()
%%time
M = lambda x,t: F@x
y = obs_data
x_0 = state_data[np.random.randint(0, sample_size)]
P_0 = 10*np.eye(N)
ekf = EnKF(M, H, G, Q, R, y[:], x_0, P_0, alpha=1)
ekf.forward_estimation()
ekf_estimate_data = generate_obs_data(H, R, np.array(ekf.x), noise=False)
fig = plt.figure(figsize=(15,12))
ax1 = fig.add_subplot(2,1,1)
ax1.plot(true, label='true', color='c')
# ax1.plot(obs_data, label='obs')
ax1.plot(ekf_estimate_data, label='est', color='g')
# ax1.plot(true - ekf_estimate_data, label='error', color='r')
plt.legend()
ax2 = fig.add_subplot(2,1,2)
# ax2.plot(np.abs(true - ekf_estimate_data)) 本来はこれ
ax2.plot(cum_std(true - ekf_estimate_data), label='est std')
# ax2.plot(np.abs(true - obs_data)) 本来はこれ
ax2.plot(cum_std(true - obs_data), label='obs std')
ax2.plot(ekf.trP, label='trP')
plt.legend()
| 0.311532 | 0.9255 |
```
from pysurvival.datasets import Dataset
%pylab inline
raw_dataset = Dataset('churn').load()
print('The raw dataset has the following shape: {}.'.format(raw_dataset.shape))
raw_dataset.head(2)
raw_dataset.product_travel_expense.value_counts()
raw_dataset.product_payroll.value_counts()
raw_dataset.product_accounting.value_counts()
raw_dataset.company_size.value_counts()
raw_dataset.us_region.value_counts()
# Creating one-hot vectors
categories = ['product_travel_expense', 'product_payroll', 'product_accounting', 'company_size', 'us_region']
dataset = pd.get_dummies(raw_dataset, columns=categories, drop_first=True)
# Creating time and event columns
time_column = 'months_active'
event_column = 'churned'
# Extracting the features
features = np.setdiff1d(dataset.columns, [time_column, event_column]).tolist()
# Checking for null values
n_null = sum(dataset[features].isnull().sum())
print('The dataset contains {} null values'.format(n_null))
# Removing duplicates if there exist
n_dupli = sum(dataset.duplicated(keep='first'))
dataset = dataset.drop_duplicates(keep='first').reset_index(drop=True)
print('The dataset contains {} duplicates'.format(n_dupli))
n = dataset.shape[0]
from pysurvival.utils.display import correlation_matrix
correlation_matrix(dataset[features], figure_size=(30, 15), text_fontsize=10)
from sklearn.model_selection import train_test_split
# Building training and testing sets
index_train, index_test = train_test_split(range(n), test_size=0.35)
data_train = dataset.loc[index_train].reset_index(drop=True)
data_test = dataset.loc[index_test].reset_index(drop=True)
# Creating the X, T and E inputs
X_train, X_test = data_train[features], data_test[features]
T_train, T_test = data_train[time_column], data_test[time_column]
E_train, E_test = data_train[event_column], data_test[event_column]
from pysurvival.models.survival_forest import ConditionalSurvivalForestModel
# Fitting the model
csf = ConditionalSurvivalForestModel(num_trees=200)
csf.fit(X_train, T_train, E_train, max_features='sqrt',
max_depth=5, min_node_size=20, alpha=0.05, minprop=0.1)
# Computing variables importance
csf.variable_importance_table.head()
from pysurvival.utils.metrics import concordance_index
c_index = concordance_index(csf, X_test, T_test, E_test)
print('C-index: {:.2f}'.format(c_index))
from pysurvival.utils.display import integrated_brier_score
ibs = integrated_brier_score(csf, X_test, T_test, E_test, t_max=12, figure_size=(12, 4))
print('IBS: {:.2f}'.format(ibs))
from pysurvival.utils.display import compare_to_actual
results = compare_to_actual(csf, X_test, T_test, E_test,
is_at_risk = False, figure_size=(12, 5),
metrics=['rmse', 'mean', 'median'])
from pysurvival.utils.display import create_risk_groups
risk_groups = create_risk_groups(model=csf, X=X_test, use_log=False, num_bins=30,
figure_size=(20, 4),
low={'lower_bound':0,'upper_bound':8.5, 'color':'red'},
medium={'lower_bound':8.5, 'upper_bound':12., 'color':'green'},
high={'lower_bound':12., 'upper_bound':25, 'color':'blue'})
# Initializing the figure
fig, ax = plt.subplots(figsize=(13, 5))
# Selecting a random individual that experienced an event from each group
groups=[]
for i, (label, (color, indexes)) in enumerate(risk_groups.items()):
if len(indexes) == 0:
continue
X=X_test.values[indexes, :]
T=T_test.values[indexes]
E=E_test.values[indexes]
# Randomly extracting an individual that experienced an event
choices = np.argwhere((E==1.)).flatten()
if len(choices) == 0:
continue
k = np.random.choice(choices, 1)[0]
t = T[k]
# Computing the Survival function for all times t
survival = csf.predict_survival(X[k, :]).flatten()
# Displaying the functions
label_='{} risk'.format(label)
plt.plot(csf.times, survival, color=color, label=label_, lw=2)
groups.append(label)
# Actual time
plt.axvline(x=t, color=color, ls='--')
ax.annotate('T={:.1f}'.format(t), xy=(t, 0.5*(1.+0.2*i)),
xytext=(t, 0.5*(1.+0.2*i)), fontsize=12)
# Show everything
groups_str = ', '.join(groups)
title='Comparing Survival functions between {} risk grades'.format(groups_str)
plt.legend(fontsize=12)
plt.title(title, fontsize=15)
plt.ylim(0, 1.05)
plt.show();
```
|
github_jupyter
|
from pysurvival.datasets import Dataset
%pylab inline
raw_dataset = Dataset('churn').load()
print('The raw dataset has the following shape: {}.'.format(raw_dataset.shape))
raw_dataset.head(2)
raw_dataset.product_travel_expense.value_counts()
raw_dataset.product_payroll.value_counts()
raw_dataset.product_accounting.value_counts()
raw_dataset.company_size.value_counts()
raw_dataset.us_region.value_counts()
# Creating one-hot vectors
categories = ['product_travel_expense', 'product_payroll', 'product_accounting', 'company_size', 'us_region']
dataset = pd.get_dummies(raw_dataset, columns=categories, drop_first=True)
# Creating time and event columns
time_column = 'months_active'
event_column = 'churned'
# Extracting the features
features = np.setdiff1d(dataset.columns, [time_column, event_column]).tolist()
# Checking for null values
n_null = sum(dataset[features].isnull().sum())
print('The dataset contains {} null values'.format(n_null))
# Removing duplicates if there exist
n_dupli = sum(dataset.duplicated(keep='first'))
dataset = dataset.drop_duplicates(keep='first').reset_index(drop=True)
print('The dataset contains {} duplicates'.format(n_dupli))
n = dataset.shape[0]
from pysurvival.utils.display import correlation_matrix
correlation_matrix(dataset[features], figure_size=(30, 15), text_fontsize=10)
from sklearn.model_selection import train_test_split
# Building training and testing sets
index_train, index_test = train_test_split(range(n), test_size=0.35)
data_train = dataset.loc[index_train].reset_index(drop=True)
data_test = dataset.loc[index_test].reset_index(drop=True)
# Creating the X, T and E inputs
X_train, X_test = data_train[features], data_test[features]
T_train, T_test = data_train[time_column], data_test[time_column]
E_train, E_test = data_train[event_column], data_test[event_column]
from pysurvival.models.survival_forest import ConditionalSurvivalForestModel
# Fitting the model
csf = ConditionalSurvivalForestModel(num_trees=200)
csf.fit(X_train, T_train, E_train, max_features='sqrt',
max_depth=5, min_node_size=20, alpha=0.05, minprop=0.1)
# Computing variables importance
csf.variable_importance_table.head()
from pysurvival.utils.metrics import concordance_index
c_index = concordance_index(csf, X_test, T_test, E_test)
print('C-index: {:.2f}'.format(c_index))
from pysurvival.utils.display import integrated_brier_score
ibs = integrated_brier_score(csf, X_test, T_test, E_test, t_max=12, figure_size=(12, 4))
print('IBS: {:.2f}'.format(ibs))
from pysurvival.utils.display import compare_to_actual
results = compare_to_actual(csf, X_test, T_test, E_test,
is_at_risk = False, figure_size=(12, 5),
metrics=['rmse', 'mean', 'median'])
from pysurvival.utils.display import create_risk_groups
risk_groups = create_risk_groups(model=csf, X=X_test, use_log=False, num_bins=30,
figure_size=(20, 4),
low={'lower_bound':0,'upper_bound':8.5, 'color':'red'},
medium={'lower_bound':8.5, 'upper_bound':12., 'color':'green'},
high={'lower_bound':12., 'upper_bound':25, 'color':'blue'})
# Initializing the figure
fig, ax = plt.subplots(figsize=(13, 5))
# Selecting a random individual that experienced an event from each group
groups=[]
for i, (label, (color, indexes)) in enumerate(risk_groups.items()):
if len(indexes) == 0:
continue
X=X_test.values[indexes, :]
T=T_test.values[indexes]
E=E_test.values[indexes]
# Randomly extracting an individual that experienced an event
choices = np.argwhere((E==1.)).flatten()
if len(choices) == 0:
continue
k = np.random.choice(choices, 1)[0]
t = T[k]
# Computing the Survival function for all times t
survival = csf.predict_survival(X[k, :]).flatten()
# Displaying the functions
label_='{} risk'.format(label)
plt.plot(csf.times, survival, color=color, label=label_, lw=2)
groups.append(label)
# Actual time
plt.axvline(x=t, color=color, ls='--')
ax.annotate('T={:.1f}'.format(t), xy=(t, 0.5*(1.+0.2*i)),
xytext=(t, 0.5*(1.+0.2*i)), fontsize=12)
# Show everything
groups_str = ', '.join(groups)
title='Comparing Survival functions between {} risk grades'.format(groups_str)
plt.legend(fontsize=12)
plt.title(title, fontsize=15)
plt.ylim(0, 1.05)
plt.show();
| 0.718496 | 0.693771 |
```
import tensorflow as tf
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import pandas as pd
from sklearn.preprocessing import MinMaxScaler
from datetime import datetime
from datetime import timedelta
sns.set()
df = pd.read_csv('../dataset/GOOG-year.csv')
date_ori = pd.to_datetime(df.iloc[:, 0]).tolist()
df.head()
minmax = MinMaxScaler().fit(df.iloc[:, 1:].astype('float32'))
df_log = minmax.transform(df.iloc[:, 1:].astype('float32'))
df_log = pd.DataFrame(df_log)
df_log.head()
timestamp = 5
epoch = 500
future_day = 50
def sinusoidal_positional_encoding(
inputs, num_units, zero_pad = False, scale = False
):
T = inputs.get_shape().as_list()[1]
position_idx = tf.tile(
tf.expand_dims(tf.range(T), 0), [tf.shape(inputs)[0], 1]
)
position_enc = np.array(
[
[
pos / np.power(10000, 2.0 * i / num_units)
for i in range(num_units)
]
for pos in range(T)
]
)
position_enc[:, 0::2] = np.sin(position_enc[:, 0::2])
position_enc[:, 1::2] = np.cos(position_enc[:, 1::2])
lookup_table = tf.convert_to_tensor(position_enc, tf.float32)
if zero_pad:
lookup_table = tf.concat(
[tf.zeros([1, num_units]), lookup_table[1:, :]], axis = 0
)
outputs = tf.nn.embedding_lookup(lookup_table, position_idx)
if scale:
outputs = outputs * num_units ** 0.5
return outputs
class Model:
def __init__(
self, seq_len, learning_rate, dimension_input, dimension_output
):
self.X = tf.placeholder(tf.float32, [None, seq_len, dimension_input])
self.Y = tf.placeholder(tf.float32, [None, dimension_output])
x = self.X
x += sinusoidal_positional_encoding(x, dimension_input)
masks = tf.sign(self.X[:, :, 0])
align = tf.squeeze(tf.layers.dense(x, 1, tf.tanh), -1)
paddings = tf.fill(tf.shape(align), float('-inf'))
align = tf.where(tf.equal(masks, 0), paddings, align)
align = tf.expand_dims(tf.nn.softmax(align), -1)
x = tf.squeeze(tf.matmul(tf.transpose(x, [0, 2, 1]), align), -1)
self.logits = tf.layers.dense(x, dimension_output)
self.cost = tf.reduce_mean(tf.square(self.Y - self.logits))
self.optimizer = tf.train.AdamOptimizer(
learning_rate = learning_rate
).minimize(self.cost)
tf.reset_default_graph()
modelnn = Model(timestamp, 0.01, df_log.shape[1], df_log.shape[1])
sess = tf.InteractiveSession()
sess.run(tf.global_variables_initializer())
for i in range(epoch):
total_loss = 0
for k in range(0, (df_log.shape[0] // timestamp) * timestamp, timestamp):
batch_x = np.expand_dims(
df_log.iloc[k : k + timestamp].values, axis = 0
)
batch_y = df_log.iloc[k + 1 : k + timestamp + 1].values
_, loss = sess.run(
[modelnn.optimizer, modelnn.cost],
feed_dict = {modelnn.X: batch_x, modelnn.Y: batch_y},
)
loss = np.mean(loss)
total_loss += loss
total_loss /= df_log.shape[0] // timestamp
if (i + 1) % 100 == 0:
print('epoch:', i + 1, 'avg loss:', total_loss)
output_predict = np.zeros((df_log.shape[0] + future_day, df_log.shape[1]))
output_predict[0] = df_log.iloc[0]
upper_b = (df_log.shape[0] // timestamp) * timestamp
for k in range(0, (df_log.shape[0] // timestamp) * timestamp, timestamp):
out_logits = sess.run(
modelnn.logits,
feed_dict = {
modelnn.X: np.expand_dims(df_log.iloc[k : k + timestamp], axis = 0)
},
)
output_predict[k + 1 : k + timestamp + 1] = out_logits
df_log.loc[df_log.shape[0]] = out_logits[-1]
date_ori.append(date_ori[-1] + timedelta(days = 1))
for i in range(future_day - 1):
out_logits = sess.run(
modelnn.logits,
feed_dict = {
modelnn.X: np.expand_dims(df_log.iloc[-timestamp:], axis = 0)
},
)
output_predict[df_log.shape[0]] = out_logits[-1]
df_log.loc[df_log.shape[0]] = out_logits[-1]
date_ori.append(date_ori[-1] + timedelta(days = 1))
df_log = minmax.inverse_transform(df_log.values)
date_ori = pd.Series(date_ori).dt.strftime(date_format = '%Y-%m-%d').tolist()
def anchor(signal, weight):
buffer = []
last = signal[0]
for i in signal:
smoothed_val = last * weight + (1 - weight) * i
buffer.append(smoothed_val)
last = smoothed_val
return buffer
current_palette = sns.color_palette('Paired', 12)
fig = plt.figure(figsize = (15, 10))
ax = plt.subplot(111)
x_range_original = np.arange(df.shape[0])
x_range_future = np.arange(df_log.shape[0])
ax.plot(
x_range_original,
df.iloc[:, 1],
label = 'true Open',
color = current_palette[0],
)
ax.plot(
x_range_future,
anchor(df_log[:, 0], 0.5),
label = 'predict Open',
color = current_palette[1],
)
ax.plot(
x_range_original,
df.iloc[:, 2],
label = 'true High',
color = current_palette[2],
)
ax.plot(
x_range_future,
anchor(df_log[:, 1], 0.5),
label = 'predict High',
color = current_palette[3],
)
ax.plot(
x_range_original,
df.iloc[:, 3],
label = 'true Low',
color = current_palette[4],
)
ax.plot(
x_range_future,
anchor(df_log[:, 2], 0.5),
label = 'predict Low',
color = current_palette[5],
)
ax.plot(
x_range_original,
df.iloc[:, 4],
label = 'true Close',
color = current_palette[6],
)
ax.plot(
x_range_future,
anchor(df_log[:, 3], 0.5),
label = 'predict Close',
color = current_palette[7],
)
ax.plot(
x_range_original,
df.iloc[:, 5],
label = 'true Adj Close',
color = current_palette[8],
)
ax.plot(
x_range_future,
anchor(df_log[:, 4], 0.5),
label = 'predict Adj Close',
color = current_palette[9],
)
box = ax.get_position()
ax.set_position(
[box.x0, box.y0 + box.height * 0.1, box.width, box.height * 0.9]
)
ax.legend(
loc = 'upper center',
bbox_to_anchor = (0.5, -0.05),
fancybox = True,
shadow = True,
ncol = 5,
)
plt.title('overlap stock market')
plt.xticks(x_range_future[::30], date_ori[::30])
plt.show()
fig = plt.figure(figsize = (20, 8))
plt.subplot(1, 2, 1)
plt.plot(
x_range_original,
df.iloc[:, 1],
label = 'true Open',
color = current_palette[0],
)
plt.plot(
x_range_original,
df.iloc[:, 2],
label = 'true High',
color = current_palette[2],
)
plt.plot(
x_range_original,
df.iloc[:, 3],
label = 'true Low',
color = current_palette[4],
)
plt.plot(
x_range_original,
df.iloc[:, 4],
label = 'true Close',
color = current_palette[6],
)
plt.plot(
x_range_original,
df.iloc[:, 5],
label = 'true Adj Close',
color = current_palette[8],
)
plt.xticks(x_range_original[::60], df.iloc[:, 0].tolist()[::60])
plt.legend()
plt.title('true market')
plt.subplot(1, 2, 2)
plt.plot(
x_range_future,
anchor(df_log[:, 0], 0.5),
label = 'predict Open',
color = current_palette[1],
)
plt.plot(
x_range_future,
anchor(df_log[:, 1], 0.5),
label = 'predict High',
color = current_palette[3],
)
plt.plot(
x_range_future,
anchor(df_log[:, 2], 0.5),
label = 'predict Low',
color = current_palette[5],
)
plt.plot(
x_range_future,
anchor(df_log[:, 3], 0.5),
label = 'predict Close',
color = current_palette[7],
)
plt.plot(
x_range_future,
anchor(df_log[:, 4], 0.5),
label = 'predict Adj Close',
color = current_palette[9],
)
plt.xticks(x_range_future[::60], date_ori[::60])
plt.legend()
plt.title('predict market')
plt.show()
fig = plt.figure(figsize = (15, 10))
ax = plt.subplot(111)
ax.plot(x_range_original, df.iloc[:, -1], label = 'true Volume')
ax.plot(x_range_future, anchor(df_log[:, -1], 0.5), label = 'predict Volume')
box = ax.get_position()
ax.set_position(
[box.x0, box.y0 + box.height * 0.1, box.width, box.height * 0.9]
)
ax.legend(
loc = 'upper center',
bbox_to_anchor = (0.5, -0.05),
fancybox = True,
shadow = True,
ncol = 5,
)
plt.xticks(x_range_future[::30], date_ori[::30])
plt.title('overlap market volume')
plt.show()
fig = plt.figure(figsize = (20, 8))
plt.subplot(1, 2, 1)
plt.plot(x_range_original, df.iloc[:, -1], label = 'true Volume')
plt.xticks(x_range_original[::60], df.iloc[:, 0].tolist()[::60])
plt.legend()
plt.title('true market volume')
plt.subplot(1, 2, 2)
plt.plot(x_range_future, anchor(df_log[:, -1], 0.5), label = 'predict Volume')
plt.xticks(x_range_future[::60], date_ori[::60])
plt.legend()
plt.title('predict market volume')
plt.show()
```
|
github_jupyter
|
import tensorflow as tf
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import pandas as pd
from sklearn.preprocessing import MinMaxScaler
from datetime import datetime
from datetime import timedelta
sns.set()
df = pd.read_csv('../dataset/GOOG-year.csv')
date_ori = pd.to_datetime(df.iloc[:, 0]).tolist()
df.head()
minmax = MinMaxScaler().fit(df.iloc[:, 1:].astype('float32'))
df_log = minmax.transform(df.iloc[:, 1:].astype('float32'))
df_log = pd.DataFrame(df_log)
df_log.head()
timestamp = 5
epoch = 500
future_day = 50
def sinusoidal_positional_encoding(
inputs, num_units, zero_pad = False, scale = False
):
T = inputs.get_shape().as_list()[1]
position_idx = tf.tile(
tf.expand_dims(tf.range(T), 0), [tf.shape(inputs)[0], 1]
)
position_enc = np.array(
[
[
pos / np.power(10000, 2.0 * i / num_units)
for i in range(num_units)
]
for pos in range(T)
]
)
position_enc[:, 0::2] = np.sin(position_enc[:, 0::2])
position_enc[:, 1::2] = np.cos(position_enc[:, 1::2])
lookup_table = tf.convert_to_tensor(position_enc, tf.float32)
if zero_pad:
lookup_table = tf.concat(
[tf.zeros([1, num_units]), lookup_table[1:, :]], axis = 0
)
outputs = tf.nn.embedding_lookup(lookup_table, position_idx)
if scale:
outputs = outputs * num_units ** 0.5
return outputs
class Model:
def __init__(
self, seq_len, learning_rate, dimension_input, dimension_output
):
self.X = tf.placeholder(tf.float32, [None, seq_len, dimension_input])
self.Y = tf.placeholder(tf.float32, [None, dimension_output])
x = self.X
x += sinusoidal_positional_encoding(x, dimension_input)
masks = tf.sign(self.X[:, :, 0])
align = tf.squeeze(tf.layers.dense(x, 1, tf.tanh), -1)
paddings = tf.fill(tf.shape(align), float('-inf'))
align = tf.where(tf.equal(masks, 0), paddings, align)
align = tf.expand_dims(tf.nn.softmax(align), -1)
x = tf.squeeze(tf.matmul(tf.transpose(x, [0, 2, 1]), align), -1)
self.logits = tf.layers.dense(x, dimension_output)
self.cost = tf.reduce_mean(tf.square(self.Y - self.logits))
self.optimizer = tf.train.AdamOptimizer(
learning_rate = learning_rate
).minimize(self.cost)
tf.reset_default_graph()
modelnn = Model(timestamp, 0.01, df_log.shape[1], df_log.shape[1])
sess = tf.InteractiveSession()
sess.run(tf.global_variables_initializer())
for i in range(epoch):
total_loss = 0
for k in range(0, (df_log.shape[0] // timestamp) * timestamp, timestamp):
batch_x = np.expand_dims(
df_log.iloc[k : k + timestamp].values, axis = 0
)
batch_y = df_log.iloc[k + 1 : k + timestamp + 1].values
_, loss = sess.run(
[modelnn.optimizer, modelnn.cost],
feed_dict = {modelnn.X: batch_x, modelnn.Y: batch_y},
)
loss = np.mean(loss)
total_loss += loss
total_loss /= df_log.shape[0] // timestamp
if (i + 1) % 100 == 0:
print('epoch:', i + 1, 'avg loss:', total_loss)
output_predict = np.zeros((df_log.shape[0] + future_day, df_log.shape[1]))
output_predict[0] = df_log.iloc[0]
upper_b = (df_log.shape[0] // timestamp) * timestamp
for k in range(0, (df_log.shape[0] // timestamp) * timestamp, timestamp):
out_logits = sess.run(
modelnn.logits,
feed_dict = {
modelnn.X: np.expand_dims(df_log.iloc[k : k + timestamp], axis = 0)
},
)
output_predict[k + 1 : k + timestamp + 1] = out_logits
df_log.loc[df_log.shape[0]] = out_logits[-1]
date_ori.append(date_ori[-1] + timedelta(days = 1))
for i in range(future_day - 1):
out_logits = sess.run(
modelnn.logits,
feed_dict = {
modelnn.X: np.expand_dims(df_log.iloc[-timestamp:], axis = 0)
},
)
output_predict[df_log.shape[0]] = out_logits[-1]
df_log.loc[df_log.shape[0]] = out_logits[-1]
date_ori.append(date_ori[-1] + timedelta(days = 1))
df_log = minmax.inverse_transform(df_log.values)
date_ori = pd.Series(date_ori).dt.strftime(date_format = '%Y-%m-%d').tolist()
def anchor(signal, weight):
buffer = []
last = signal[0]
for i in signal:
smoothed_val = last * weight + (1 - weight) * i
buffer.append(smoothed_val)
last = smoothed_val
return buffer
current_palette = sns.color_palette('Paired', 12)
fig = plt.figure(figsize = (15, 10))
ax = plt.subplot(111)
x_range_original = np.arange(df.shape[0])
x_range_future = np.arange(df_log.shape[0])
ax.plot(
x_range_original,
df.iloc[:, 1],
label = 'true Open',
color = current_palette[0],
)
ax.plot(
x_range_future,
anchor(df_log[:, 0], 0.5),
label = 'predict Open',
color = current_palette[1],
)
ax.plot(
x_range_original,
df.iloc[:, 2],
label = 'true High',
color = current_palette[2],
)
ax.plot(
x_range_future,
anchor(df_log[:, 1], 0.5),
label = 'predict High',
color = current_palette[3],
)
ax.plot(
x_range_original,
df.iloc[:, 3],
label = 'true Low',
color = current_palette[4],
)
ax.plot(
x_range_future,
anchor(df_log[:, 2], 0.5),
label = 'predict Low',
color = current_palette[5],
)
ax.plot(
x_range_original,
df.iloc[:, 4],
label = 'true Close',
color = current_palette[6],
)
ax.plot(
x_range_future,
anchor(df_log[:, 3], 0.5),
label = 'predict Close',
color = current_palette[7],
)
ax.plot(
x_range_original,
df.iloc[:, 5],
label = 'true Adj Close',
color = current_palette[8],
)
ax.plot(
x_range_future,
anchor(df_log[:, 4], 0.5),
label = 'predict Adj Close',
color = current_palette[9],
)
box = ax.get_position()
ax.set_position(
[box.x0, box.y0 + box.height * 0.1, box.width, box.height * 0.9]
)
ax.legend(
loc = 'upper center',
bbox_to_anchor = (0.5, -0.05),
fancybox = True,
shadow = True,
ncol = 5,
)
plt.title('overlap stock market')
plt.xticks(x_range_future[::30], date_ori[::30])
plt.show()
fig = plt.figure(figsize = (20, 8))
plt.subplot(1, 2, 1)
plt.plot(
x_range_original,
df.iloc[:, 1],
label = 'true Open',
color = current_palette[0],
)
plt.plot(
x_range_original,
df.iloc[:, 2],
label = 'true High',
color = current_palette[2],
)
plt.plot(
x_range_original,
df.iloc[:, 3],
label = 'true Low',
color = current_palette[4],
)
plt.plot(
x_range_original,
df.iloc[:, 4],
label = 'true Close',
color = current_palette[6],
)
plt.plot(
x_range_original,
df.iloc[:, 5],
label = 'true Adj Close',
color = current_palette[8],
)
plt.xticks(x_range_original[::60], df.iloc[:, 0].tolist()[::60])
plt.legend()
plt.title('true market')
plt.subplot(1, 2, 2)
plt.plot(
x_range_future,
anchor(df_log[:, 0], 0.5),
label = 'predict Open',
color = current_palette[1],
)
plt.plot(
x_range_future,
anchor(df_log[:, 1], 0.5),
label = 'predict High',
color = current_palette[3],
)
plt.plot(
x_range_future,
anchor(df_log[:, 2], 0.5),
label = 'predict Low',
color = current_palette[5],
)
plt.plot(
x_range_future,
anchor(df_log[:, 3], 0.5),
label = 'predict Close',
color = current_palette[7],
)
plt.plot(
x_range_future,
anchor(df_log[:, 4], 0.5),
label = 'predict Adj Close',
color = current_palette[9],
)
plt.xticks(x_range_future[::60], date_ori[::60])
plt.legend()
plt.title('predict market')
plt.show()
fig = plt.figure(figsize = (15, 10))
ax = plt.subplot(111)
ax.plot(x_range_original, df.iloc[:, -1], label = 'true Volume')
ax.plot(x_range_future, anchor(df_log[:, -1], 0.5), label = 'predict Volume')
box = ax.get_position()
ax.set_position(
[box.x0, box.y0 + box.height * 0.1, box.width, box.height * 0.9]
)
ax.legend(
loc = 'upper center',
bbox_to_anchor = (0.5, -0.05),
fancybox = True,
shadow = True,
ncol = 5,
)
plt.xticks(x_range_future[::30], date_ori[::30])
plt.title('overlap market volume')
plt.show()
fig = plt.figure(figsize = (20, 8))
plt.subplot(1, 2, 1)
plt.plot(x_range_original, df.iloc[:, -1], label = 'true Volume')
plt.xticks(x_range_original[::60], df.iloc[:, 0].tolist()[::60])
plt.legend()
plt.title('true market volume')
plt.subplot(1, 2, 2)
plt.plot(x_range_future, anchor(df_log[:, -1], 0.5), label = 'predict Volume')
plt.xticks(x_range_future[::60], date_ori[::60])
plt.legend()
plt.title('predict market volume')
plt.show()
| 0.742702 | 0.490907 |
```
import os
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib as mpl
import matplotlib.pyplot as plt
%matplotlib inline
sns.set_context("talk")
sns.set_style("white")
dpi=100
fig_width = 10
fig_height = 6
model_names = ["STSNet", "ECGNet"]
model_names_lookup = ["deep-sts-preop-v13-swish", "v30"]
path_to_predictions_prefix = os.path.expanduser("~/dropbox/sts-ecg/predictions")
path_to_figures_prefix = os.path.expanduser("~/dropbox/sts-ecg/figures-and-tables")
csv_name = "predictions_test.csv"
```
## Parse predictions for each bootstrap into one df containing `y`, `y_hat`, `brier`, and `y_hat_delta`
```
dfs = []
for bootstrap in range(10):
dfs_bootstrap = {}
for model_name, lookup_name in zip(model_names, model_names_lookup):
path_to_predictions = os.path.join(path_to_predictions_prefix, lookup_name, str(bootstrap), csv_name)
# Get CSV into df
dfs_bootstrap[model_name] = pd.read_csv(path_to_predictions)
# Rename columns
dfs_bootstrap[model_name].columns = ["mrn", f"y_{model_name}", f"y_hat_{model_name}"]
# Merge model results into one df
df_both_models = dfs_bootstrap[model_names[0]].merge(right=dfs_bootstrap[model_names[1]], on="mrn")
# Append df to list of dfs
dfs.append(df_both_models)
print(f"Parsing predictions from bootstrap {bootstrap}")
df = pd.concat(dfs)
```
## Scale predictions (min-max) and calculate error
```
y_hat_min = 0.01
y_hat_max = 0.2
for model_name in model_names:
df[df[f'y_hat_{model_name}'] > 0.2] = 0.2
df[f'y_hat_{model_name}_scaled'] = (df[f'y_hat_{model_name}'] - y_hat_min) / (y_hat_max - y_hat_min)
# Calculate delta between y_hat values of each model
df[f'squared_error_{model_name}'] = (df[f"y_{model_name}"] - df[f"y_hat_{model_name}_scaled"])**2
print(f'{model_name} pre-scaling range: [{y_hat_min:0.3f} {y_hat_max:0.3f}]')
print(f'{model_name} pre-scaling range: [{y_hat_min_new:0.3f} {y_hat_max_new:0.3f}]')
print('\n')
df[f'squared_error_between_models'] = (df[f"y_hat_{model_names[0]}_scaled"] - df[f"y_hat_{model_names[1]}_scaled"])**2
```
## Plot of y_hat
```
for model_name in model_names:
fig, ax = plt.subplots(figsize=(fig_width, fig_height))
sns.distplot(df[f'y_hat_{model_name}'], ax=ax)
plt.xlim([-0.05, 1.05])
plt.title(f"{model_name}")
plt.xlabel("y_hat")
plt.ylabel("Counts")
plt.tight_layout()
fpath = os.path.join(path_to_figures_prefix, f"y_hat_{model_name}.png").lower()
plt.savefig(fpath, dpi=dpi, transparent=False)
for model_name in model_names:
fig, ax = plt.subplots(figsize=(fig_width, fig_height))
sns.distplot(df[f'y_hat_{model_name}_scaled'], ax=ax)
plt.xlim([-0.05, 1.05])
plt.title(f"{model_name}")
plt.xlabel("y_hat")
plt.ylabel("Counts")
plt.tight_layout()
fpath = os.path.join(path_to_figures_prefix, f"y_hat_scaled_{model_name}.png").lower()
plt.savefig(fpath, dpi=dpi, transparent=False)
df
```
## Scatterplot of model squared error vs (STSNet - ECGNet)^2
```
from scipy import stats
def calc_r2(x, y):
return stats.pearsonr(x, y)[0] ** 2
r2 = calc_r2(
x=df[f"squared_error_{model_names[0]}"],
y=df[f"squared_error_between_models"],
)
fig, ax = plt.subplots(figsize=(fig_width, fig_height))
sns.scatterplot(
ax=ax,
x=df[f"squared_error_{model_names[0]}"],
y=df[f"squared_error_between_models"],
cmap="Blues",
alpha=0.75,
)
ax.set_title(f"STSNet error vs difference between STSNet and ECGNet")
ax.set_xlabel(f"{model_names[0]}: (y - y_hat)^2")
ax.set_ylabel(f"(STSNet - ECGNet)^2")
ax.set_xlim([-0.025, 1.025])
ax.set_ylim([-0.025, 1.025])
fpath = os.path.join(path_to_figures_prefix, f"
.png")
plt.tight_layout()
plt.savefig(fname=fpath, dpi=dpi, transparent=False)
print(f"Saved {fpath}")
```
|
github_jupyter
|
import os
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib as mpl
import matplotlib.pyplot as plt
%matplotlib inline
sns.set_context("talk")
sns.set_style("white")
dpi=100
fig_width = 10
fig_height = 6
model_names = ["STSNet", "ECGNet"]
model_names_lookup = ["deep-sts-preop-v13-swish", "v30"]
path_to_predictions_prefix = os.path.expanduser("~/dropbox/sts-ecg/predictions")
path_to_figures_prefix = os.path.expanduser("~/dropbox/sts-ecg/figures-and-tables")
csv_name = "predictions_test.csv"
dfs = []
for bootstrap in range(10):
dfs_bootstrap = {}
for model_name, lookup_name in zip(model_names, model_names_lookup):
path_to_predictions = os.path.join(path_to_predictions_prefix, lookup_name, str(bootstrap), csv_name)
# Get CSV into df
dfs_bootstrap[model_name] = pd.read_csv(path_to_predictions)
# Rename columns
dfs_bootstrap[model_name].columns = ["mrn", f"y_{model_name}", f"y_hat_{model_name}"]
# Merge model results into one df
df_both_models = dfs_bootstrap[model_names[0]].merge(right=dfs_bootstrap[model_names[1]], on="mrn")
# Append df to list of dfs
dfs.append(df_both_models)
print(f"Parsing predictions from bootstrap {bootstrap}")
df = pd.concat(dfs)
y_hat_min = 0.01
y_hat_max = 0.2
for model_name in model_names:
df[df[f'y_hat_{model_name}'] > 0.2] = 0.2
df[f'y_hat_{model_name}_scaled'] = (df[f'y_hat_{model_name}'] - y_hat_min) / (y_hat_max - y_hat_min)
# Calculate delta between y_hat values of each model
df[f'squared_error_{model_name}'] = (df[f"y_{model_name}"] - df[f"y_hat_{model_name}_scaled"])**2
print(f'{model_name} pre-scaling range: [{y_hat_min:0.3f} {y_hat_max:0.3f}]')
print(f'{model_name} pre-scaling range: [{y_hat_min_new:0.3f} {y_hat_max_new:0.3f}]')
print('\n')
df[f'squared_error_between_models'] = (df[f"y_hat_{model_names[0]}_scaled"] - df[f"y_hat_{model_names[1]}_scaled"])**2
for model_name in model_names:
fig, ax = plt.subplots(figsize=(fig_width, fig_height))
sns.distplot(df[f'y_hat_{model_name}'], ax=ax)
plt.xlim([-0.05, 1.05])
plt.title(f"{model_name}")
plt.xlabel("y_hat")
plt.ylabel("Counts")
plt.tight_layout()
fpath = os.path.join(path_to_figures_prefix, f"y_hat_{model_name}.png").lower()
plt.savefig(fpath, dpi=dpi, transparent=False)
for model_name in model_names:
fig, ax = plt.subplots(figsize=(fig_width, fig_height))
sns.distplot(df[f'y_hat_{model_name}_scaled'], ax=ax)
plt.xlim([-0.05, 1.05])
plt.title(f"{model_name}")
plt.xlabel("y_hat")
plt.ylabel("Counts")
plt.tight_layout()
fpath = os.path.join(path_to_figures_prefix, f"y_hat_scaled_{model_name}.png").lower()
plt.savefig(fpath, dpi=dpi, transparent=False)
df
from scipy import stats
def calc_r2(x, y):
return stats.pearsonr(x, y)[0] ** 2
r2 = calc_r2(
x=df[f"squared_error_{model_names[0]}"],
y=df[f"squared_error_between_models"],
)
fig, ax = plt.subplots(figsize=(fig_width, fig_height))
sns.scatterplot(
ax=ax,
x=df[f"squared_error_{model_names[0]}"],
y=df[f"squared_error_between_models"],
cmap="Blues",
alpha=0.75,
)
ax.set_title(f"STSNet error vs difference between STSNet and ECGNet")
ax.set_xlabel(f"{model_names[0]}: (y - y_hat)^2")
ax.set_ylabel(f"(STSNet - ECGNet)^2")
ax.set_xlim([-0.025, 1.025])
ax.set_ylim([-0.025, 1.025])
fpath = os.path.join(path_to_figures_prefix, f"
.png")
plt.tight_layout()
plt.savefig(fname=fpath, dpi=dpi, transparent=False)
print(f"Saved {fpath}")
| 0.495117 | 0.679941 |
# Predictive Models 101
## Machine Learning in the Industry
The focus of this chapter is to talk about how we normally use **machine learning** in the industry. If you are not familiar with machine learning, you can see this chapter as a machine learning crash course. And if you've never worked with ML before, I strongly recommend you learn at least the basics to get the most out of what is to come. The rest of this book is aimed at Data Scientists who want to solve causal problems in their workplace. For this reason, I'll tend to assume you already have basic data science knowledge, including Machine Learning.
But this doesn't mean you should skip this chapter if you are already versed in ML. I still think you will benefit from reading it through. Differently from other machine learning material, this one will **not** discuss the ins and outs of algorithms like decision trees or neural networks. Instead, it will be laser focused on **how machine learning is applied in the real world**.

The first thing I want to adress is why are we talking about machine learning in a causal inference book? The short answer is because I think one of the best ways to understand causality is to put it in contrast with the predictive models approach brought by machine learning. The long answer is twofold. First, if you've got to this point in this book, there is a high chance you are already familiar with machine learning. Second, even if you aren't, given the current popularity of these topics, you probably already have some idea on what they are. The only problem is that, with all the hype around machine learning, I might have to bring you back to earth and explain what it really does in very practical terms. Finally, more recent developments causal inference make heavy use of machine learning algorithms, so there is that too.
As I've said in the beginning of the book, **machine learning is a way to make fast, automatic and good predictions**. That's not the entire picture, but we could say that it covers 90% of it. It's in the field of supervised machine learning where most of the cool advancements, like computer vision, self-driving cars, language translation and diagnostics, have been made. Notice how at first these might not seem like prediction tasks. How is language translation a prediction? And that's the beauty of machine learning. We can solve more problems with prediction than what is initially apparent. In the case of language translation, you can frame it as a prediction problem where you present a machine with one sentence and it has to predict the same sentence in another language. Notice that I'm **not** using the word prediction in a forecasting or anticipating the future sense. Prediction is simply mapping from one defined input to an initially unknown but equally well defined output.

What machine learning really does is it learns this mapping function, even if it is a very complicated mapping function. The bottom line is that if you can frame a problem as this mapping from an input to an output, then machine learning might be a good candidate to solve it. As for self driving cars, you can think of it as not one, but multiple complex prediction problems: predicting the correct angle of the wheel from sensors in the front of the car, predicting the pressure in the brakes from cameras around the car, predicting the pressure in the accelerator from gps data. Solving those (and a tone more) of prediction problems is what makes a self driving car.
OK… You now understand how prediction can be more powerful than we first though. Self-driving cars and language translation are cool and all, but they are quite distant, unless you work at a major tech company like Google or Uber. So, to make this more relatable, let's talk in terms of problems almost every company has: customer acquisition (that is getting new customers).
From the customer acquisition perspective, what you often have to do is figure out who are the profitable customers. In this problem, each customer has a cost of acquisition (maybe marketing costs, onboarding costs, shipping costs...) and will hopefully generate a positive cashflow for the company. For example, let's say you are an internet provider or a gas company. Your typical customer might have a cash flow that looks something like this.

Each bar represents a monetary event in the life of your relationship with the customer. For example, to get a customer, right off the bet, you need to invest in marketing. Then, after someone decides to do business with you, you might incur in some sort of onboarding cost (where you have to explain to your customer how to use your product) or installation costs. Only then, the customer starts to generate monthly revenues. At some point, the customer might need some assistance and you will have maintenance costs. Finally, if the customer decides to end the contract, you might have some final costs for that too.
To see if this is a profitable customer, we can rearrange the bar in what is called a cascade plot. Hopefully, the sum of the cash events end way up above the zero line.

In contrast, it could very well be that the customer will generate much more costs than revenues. If he or she uses very little of your product and has high maintenance demands, when we pile up the cash events, they could end up below the zero line.

Of course, this cash flow could be simpler or much more complicated, depending on the type of business. You can do stuff like time discounts with an interest rate and get all crazy about it, but I think the point here is made.
But what can you do about this? Well, if you have many examples of profitable and non profitable customers, you can train a machine learning model to identify them. That way, you can focus your marketing strategies that engage only on the profitable customers. Or, if your contract permits, you can end relations with a customer before he or she generates more costs. Essentially, what you are doing here is framing the business problem as a prediction problem so that you can solve it with machine learning: you want to predict or identify profitable and unprofitable customers so that you only engage with the profitable ones.
```
import pandas as pd
import numpy as np
from sklearn import ensemble
from sklearn.model_selection import train_test_split
from sklearn.pipeline import Pipeline
from sklearn.metrics import r2_score
import seaborn as sns
from matplotlib import pyplot as plt
from matplotlib import style
style.use("ggplot")
```
For instance, suppose you have 30 days of transactional data on 10000 customers. You also have the cost of acquisition `cacq`. This could be the bid you place for them if you are doing online marketing, it could be the cost of shipping or any training you have to do with your customer so they can use your product. Also, for the sake of simplicity (this is a crash course, not a semester on customer valuation), let's pretend you have total control of the customer that you do business with. In other words, you have the power to deny a customer even if he or she wants to do business with you. If that's the case, your task now becomes identifying who will be profitable beforehand, so you can choose to engage only with them.
```
transactions = pd.read_csv("data/customer_transactions.csv")
print(transactions.shape)
transactions.head()
```
What we need to do now is distinguish the good from the bad customers according to this transactional data. For the sake of simplicity, I'll just sum up all transactions and the CACQ. Keep in mind that this throws under the rug a lot of nuances, like distinguishing customers that churned from those that are in a break between one purchase and the next.
I'll then join this sum, which I call `net_value`, with customer specific features. Since my goals is to figure out which customer will be profitable **before** deciding to engage with them, you can only use data prior to the acquisition period. In our case, these features are age, region and income, which are all available at another `csv` file.
```
profitable = (transactions[["customer_id"]]
.assign(net_value = transactions
.drop(columns="customer_id")
.sum(axis=1)))
customer_features = (pd.read_csv("data/customer_features.csv")
.merge(profitable, on="customer_id"))
customer_features.head()
```
Good! Our task is becoming less abstract. We wish to identify the profitable customers (`net_value > 0`) from the non profitable ones. Let's try different things and see which one works better. But before that, we need to take a quick look into Machine Learning (feel free skip if you know how ML works)
## Machine Learning Crash Course
For our intent and purpose, we can think of ML as an overpowered way of making predictions. For it to work, you need some data with labels or the ground truth of what you are predicting. Then, you can train a ML model on that data and use it to make predictions where the ground truth is not yet known. The image below exemplifies the typical machine learning flow.

First, you need data where the ground truth, `net_value` here is known. Then, you train a ML model that will use features - region, income and age in our case - to predict `net_value`. This training or estimating step will produce a machine learning model that can be used to make predictions about `net_value` when you don't yet have the true `net_value`. This is shown in the left part of the image. You have some new data where you have the features (region, income and age) but you don't know the `net_value` yet. So you pass this data through your model and it provides you with `net_value` predictions.
One tricky thing with ML models is that they can approximate almost any function. Another way of saying this is that they can be made so powerful as to perfectly fit the data in the training set. Machine learning models often have what we call complexity hyperparameters. These things adjust how powerful or complex the model can be. In the image below, you can see examples of a simple model, an intermediate model and a complex and powerful model. Notice how the complex model has a perfect fit of the training data.

This raises some problems. Namely, how can we know if our model is any good before using it to make predictions in the real world? One way we have is to compare the predictions with the actual values on the dataset where we have the ground truth. But remember that the model can be made so powerful as to perfectly fit the data. If this happens, the predictions will perfectly match the ground truth. This is problematic, because it means this validation is misleading, since I can nail it just by making my model more powerful and complex.
Besides, it is generally **not** a good thing to have a very complex model. And you already have some intuition into why that is the case. In the image above, for instance, which model do you prefer? The more complex one that gets all the predictions right? Probably not. You probably prefer the middle one. It's smoother and simpler and yet, it still makes some good predictions, even if it doesn't perfectly fit the data.

Your intuition is in the right place. What happens if you give too much power to your model, is that it will not only learn the patterns in your data, but it also learns the random noise. Since the noise will be different when you use the model to make predictions in the real world (it's random after all), your "perfect" model will make mistakes. In ML terms, we say that models that are too complex are overfitting and don't generalize well. So, what can we do?
The idea is to split the dataset for which we have the ground truth into two. Then, we can give one part for the model to train on and the other part we can use to validate the model predictions. This is called cross validation as we will discuss it more later. For now, you can see what happens when we do that.

In the dataset above, which the model didn't see during training, the complex model doesn't do a very good job. The model in the middle, on the other hand, seems to perform better. To choose the right model complexity, we can train different models, each one with a different complexity, and see how they perform on some data that we have the ground truth, but that was not used for training the model.
## Cross Validation
Cross validation is essential for selecting the model complexity but it's more generally useful. We can use it whenever we want to try many different things and estimate how they would play out in the real world. The idea being that we pretend not to have access to some of the data when estimating our methods. Then, we can use this holdout data for evaluation. We can apply this to the whole problem of figuring out which customers are profitable or not. Here is an outline of what we should do
1. We have data on existing customers. On this data, we know which ones are profitables and which ones are not (we know the ground truth). Let's call our internal data the training set.
2. We will use the internal data to *learn* a rule that tells us which customer is profitable (hence training).
3. We will apply the rule to the holdout data that was **not** used for learning the rule. This should simulate the process of learning a rule in one dataset and applying it to another, a process that will be inevitable when we go to production and score truly unseen data.
Here is a picture of what cross validation looks like. There is the truly unseen data at the leftmost part of the image and then there is data that we only pretend not to have at learning time.

To summarize, we will partition our internal data into a training and a test set. We can use the training set to come up with models or rules that predict if a customer is profitable or not, but we will validate those rules in another partition of the dataset: the test set. This test set will be hidden from our learning procedure. The hope is that this will mimic the situation we will encounter once we go to production.
Just as a side note here, there are tons of ways to make cross validation better other than this simple train test split (k-fold cross-validation or temporal cross validation, for instance), but for the sake of what we will do here, this is enough. Remember that the spirit of cross validation is to simulate what would happen once we go to a production environment. By doing that we hope to get more realistic estimates.
For our case, I won't do anything fancy. I'll just divide the dataset into two. 70% will be used to build a method that allows us to identify profitable customers and 30% will be used to evaluate how good that method is.
```
train, test = train_test_split(customer_features, test_size=0.3, random_state=13)
train.shape, test.shape
```
## Predictions and Policies
We've been talking about methods and approaches to identify profitable customers but it is time we get more precise with our concepts. Let's introduce two. A **prediction** is a number that estimates or predicts something. For example, we can try to predict the profitability of a customer and the prediction would be something like 16 reais, meaning that we predict this customer will generate 16 reais in revenue. The point here is that prediction is a simple number. The second concept is that of a **policy**. A policy is an automatic decision rule. While a prediction is a number, a policy is a decision. For example, we can have a policy that engages with customers with income greater than 1000 and doesn't engage otherwise. We usually build policies on top of predictions: engage with all customers that have profitability predictions above 10 and don't engage otherwise. Machine learning will usually take care of the first, that is, of prediction. But notice that predictions alone are useless. We need to attach some decision, or policy to it.
We can do very simple policies and models or very complicated ones. For both policies and predictions, we need to use cross validation, that is, estimate the policy or prediction in one partition of the data and validate its usefulness in another. Since we've already partitioned our data into two, we are good to go.
## One Feature Policies
Before we go machine learning crazy on this profitability problem, let's try the simple stuff first. The 80% gain with 20% effort stuff. They often work wonders and, surprising, most data scientists forget about them. So, what is the simplest thing we can do? Naturally, **just engage with all the customers!** Instead of figuring out which ones are profitable, let's just do business with everyone and hope the profitable customers more than compensate for the non profitable ones.
To check if this is a good idea, we can see the average net value of the customers. If that turns out to be positive, it means that, on average, we will make money on our customers. Sure, there will be profitable and non profitable ones but, on average, if we have enough customers, we will make money. On the other hand, if this value is negative, it means that we will lose money if we engage with all the customers.
```
train["net_value"].mean()
```
That's a bummer... If we engage with everyone, we would lose about 30 reais for customers we do business with. Our first, very simple thing didn't work and we better find something more promising if we don't want to go out of business. Just a quick side note here, keep in mind that this is a pedagogical example. Although the very simple, "treat everyone the same" kind of policy didn't work here, they often do in real life. It is usually the case that sending a marketing email to everyone is better than not sending it, or giving discounts coupons to everyone is often better than not giving them.
Moving forward, what is the next simplest thing we can think of? One idea is taking our features and seeing if they alone distinguish the good from the bad customers. Take `income`, for instance. It's intuitive that richer customers should be more profitable, right? What if we do business only with the top richest customers? Would that be a good idea?
To figure this out we can partition our data into income quantiles (a quantile has the propriety of dividing the data into partitions of equal size, that's why I like them). Then, for each income quantile, let's compute the average net value. The hope here is that, although the average net value in negative, \\(E[NetValue]<0\\), there might be some subpopulation defined by income where the net value is positive, \\(E[NetValue|Income=x]<0\\), probably, higher income levels.
```
plt.figure(figsize=(12,6))
np.random.seed(123) ## seed because the CIs from seaborn uses boostrap
# pd.qcut create quantiles of a column
sns.barplot(data=train.assign(income_quantile=pd.qcut(train["income"], q=20)),
x="income_quantile", y="net_value")
plt.title("Profitability by Income")
plt.xticks(rotation=70);
```
And, sadly, nope. Yet again, all levels of income have negative average net value. Although it is true that richer customers are "less bad" than non rich customers, they still generate, on average, negative net value. So income didn't help us much here, but what about the other variables, like region? If most of our costs come, say, from having to serve customers in far away places, we should expect that the region distinguishes the profitable from the unprofitable customers.
Since region is already a categorical variable, we don't need to use quantiles here. Let's just see the average net value per region.
```
plt.figure(figsize=(12,6))
np.random.seed(123)
region_plot = sns.barplot(data=train, x="region", y="net_value")
plt.title("Profitability by Region");
```
Bingo! We can clearly see that some regions are profitable, like regions 2, 17, 39, and some are not profitable, like region 0, 9, 29 and the specially bad region 26. This is looking super promising! We can take this and transform into a policy: only do business with the regions that showed to be profitable *according to the data that we have here*.
To construct this policy, we will be conservative and take only the regions where the lower end of the confidence interval is above zero. A region like 44 and 27, although slightly positive, will be left out according to this policy.
The following code extracts the lowest y value for each point in the plot above. The `enumerate` thing just zips each point with an index (0 to 49, since there are 50 points). We can do this only because the points are ordered from region 0 to 49. Then, the second dictionary generator filters only those regions where the lower end of the confidence interval is above zero. The result is the regions we will do business with according to our policy.
```
# extract the lower bound of the 95% CI from the plot above
regions_to_net = {region: line.get_ydata().min()
for region, line in enumerate(region_plot.lines)}
# filters regions where the net value is > 0.
regions_to_invest = {region: net
for region, net in regions_to_net.items()
if net > 0}
regions_to_invest
```
`regions_to_invest` has all the regions we will engage with. Lets now see how this policy would have performed in our test set, the one we pretend not to have. This is a key step in evaluating our policy, because it could very well be that, simply by chance, a region in our training set is appearing to be profitable. If that is only due to randomness, it will be unlikely that we will find that same pattern in the test set.
To do so, we will filter the test set to contain only the customers in the regions defined as profitable (according to our training set). Then, we will plot the distribution of net income for those customers and also show the average net income of our policy.
```
region_policy = (test[test["region"]
# filter regions in regions_to_invest
.isin(regions_to_invest.keys())])
sns.histplot(data=region_policy, x="net_value")
# average has to be over all customers, not just the one we've filtered with the policy
plt.title("Average Net Income: %.2f" % (region_policy["net_value"].sum() / test.shape[0]));
```
Not bad! If we use this very simple rule of doing business with only those regions, we can expect to gain about 15 reais per customer. Sure, out of the 3000 customers in the test set, we chose not to engage with some of them (gain of zero) and there will be some customers that we choose to engage with but made us lose money (the ones below zero in our histogram). However, in aggregate, the profitable ones will more than compensate for that.
Now that you've figured this out, pat yourself in the back! It's already a super useful policy. Your boss will be super happy because your company can make money instead of losing it on customers. What an achievement! Let this be a lesson to never underestimate the value of simple policies.
## Machine Learning Models as Policy Inputs
If you are willing to do even better, we can now use the power of machine learning. Keep in mind that this will add tones of complexity to the whole thing and usually only marginal gains. But, depending on the circumstances, marginal gains can be translated into huge piles of money and that's why machine learning is so valuable these days.
Here, I'll use a Gradient Boosting model. It's a fairly complicated model to explain, but one that is very simple to use. For our purpose here, we don't need to get into the details of how it works. Instead, just remember what we've seen in our ML Crash course: a ML model is a super powerful predictive machine that has some complexity parameters. The more complex, the more powerful the model becomes. However, if the complexity is too high, the model will learn noise and not generalize well to unseen data. Hence, we need to use cross validation here to see if the model has the right complexity.
Now, we need to ask, how can good predictions be used to improve upon our simple region policy to identify and engage with profitable customers? I think there are two main improvements that we can make here. First, you will have to agree that going through all the features looking for one that distinguishes good from bad customers is a cumbersome process. Here, we had only 3 of them (age, income and region), so it wasn't that bad, but imagine if we had more than 100. Also, you have to be careful with issues of [multiple testing](https://en.wikipedia.org/wiki/Multiple_comparisons_problem) and false positive rates. The second reason is that it is probably the case that you need more than one feature to distinguish between customers. In our example, we believe that features other than region also have some information on customer profitability. Sure, when we looked at income alone it didn't give us much, but what about income on those regions that are just barely unprofitable? Maybe, on those regions, if we focus only on richer customers, we could still get some profit. Coming up with these more complicated policies that involve interacting more than one feature can be super complex. The combinations we have to look at grows exponentially with the number of features and it is simply not a practical thing to do. Istead, what we can do is throw all those features into a machine learning model and have it learn those interactions for us. This is precisely what we will do next.
The goal of this model will be to predict `net_value` using `region`, `income`, `age`. To help it, we will take the region feature, which is categorical, and encode it with the lower end of the confidence interval of net income for that region. Remember that we have those stored in the `regions_to_net` dictionary? With this, all we have to do is call the method `.replace()` and pass this dictionary as the argument. I'll create a function for this, because we will do this replacement multiple times. This process of transforming features to facilitate learning is generally called feature engineering.
```
def encode(df):
return df.replace({"region": regions_to_net})
```
Next, our model will be imported from [Sklearn](https://scikit-learn.org/stable/). All their models have a pretty standard usage. First, you instantiate the model passing in the complexity parameters. For this model, we will set the number of estimators to 400, the max depth to 4 and so on. The deeper the model and the greater the number of estimators, the more powerful the model will be. Of course, we can't let it be too powerful, otherwise it will learn the noise in the training data or overfit to it. Again, you don't need to know the details of what these parameters do. Just keep in mind that this is a very good prediction model. Then, to train our model, we will call the `.fit()` method, passing the features `X` and the variable we want to predict - or target variable - `net_value`.
```
model_params = {'n_estimators': 400,
'max_depth': 4,
'min_samples_split': 10,
'learning_rate': 0.01,
'loss': 'ls'}
features = ["region", "income", "age"]
target = "net_value"
np.random.seed(123)
reg = ensemble.GradientBoostingRegressor(**model_params)
# fit model on the training set
encoded_train = train[features].pipe(encode)
reg.fit(encoded_train, train[target]);
```
The model is trained. Now, we need to check if it is any good. To do this, we can look at the predictive performance of this model **on the test set**. There are tons of metrics to evaluate the predictive performance of a machine learning model. Here, I'll use one which is called \\(R^2\\). We don't need to get into much detail here. It suffices to say that the \\(R^2\\) is used to evaluate models that predict a continuous variable (like `net_income`). Also, \\(R^2\\) can go from minus infinity (it will be negative if the prediction is worse than the average) to 1.0. The \\(R^2\\) tells us how much of the variance in `net_income` is explained by our model.
```
train_pred = (encoded_train
.assign(predictions=reg.predict(encoded_train[features])))
print("Train R2: ", r2_score(y_true=train[target], y_pred=train_pred["predictions"]))
print("Test R2: ", r2_score(y_true=test[target], y_pred=reg.predict(test[features].pipe(encode))))
```
In this case, the model explains about 71% of the `net_income` variance in the training set but only about 69% of the `net_income` variance in the test set. This is expected. Since the model had access to the training set, the performance there will often be overestimated. Just for fun (and to learn more about overfitting), try setting the 'max_depth' of the model to 14 and see what happens. You will likely see that the train \\(R^2\\) skyrockets but the test set \\(R^2\\) gets lower. This is what overfitting looks like.
Next, in order to make our policy, we will store the test set predictions in a `prediction` column.
```
model_policy = test.assign(prediction=reg.predict(test[features].pipe(encode)))
model_policy.head()
```
Just like we did with the `regions` feature, we can show the average net value by predictions of our model. Since the model is continuous and not categorical, we need to make it discrete first. One way of doing so is using pandas `pd.qcut` (by golly! I love this function!), which partitions the data into quantiles using the model prediction. Let's use 50 quantiles because 50 is the number of regions that we had. And just as a convention, I tend to call these model quantiles **model bands**, because it gives the intuition that this group has model predictions within a band, say, from -10 to 200.
```
plt.figure(figsize=(12,6))
n_bands = 50
bands = [f"band_{b}" for b in range(1,n_bands+1)]
np.random.seed(123)
model_plot = sns.barplot(data=model_policy
.assign(model_band = pd.qcut(model_policy["prediction"], q=n_bands)),
x="model_band", y="net_value")
plt.title("Profitability by Model Prediction Quantiles")
plt.xticks(rotation=70);
```
Here, notice how there are model bands where the net value is super negative, while there are also bands where it is very positive. Also, there are bands where we don't know exactly if the net value is negative or positive. Finally, notice how they have an upward trend, from left to right. Since we are predicting net income, it is expected that the prediction will be proportional to what it predicts.
Now, to compare this policy using a machine learning model with the one using only the regions we can also show the histogram of net gains, along with the total net value in the test set.
```
plt.figure(figsize=(10,6))
model_plot_df = (model_policy[model_policy["prediction"]>0])
sns.histplot(data=model_plot_df, x="net_value", color="C2", label="model_policy")
region_plot_df = (model_policy[model_policy["region"].isin(regions_to_invest.keys())])
sns.histplot(data=region_plot_df, x="net_value", label="region_policy")
plt.title("Model Net Income: %.2f; Region Policy Net Income %.2f." %
(model_plot_df["net_value"].sum() / test.shape[0],
region_plot_df["net_value"].sum() / test.shape[0]))
plt.legend();
```
As we can see, the model generates a better policy than just using the regions feature, but not by much. While the model policy would have made us about 16.6 reais / customer on the test set, the region policy would have made us only 15.5 / customer. It's just slightly better, but if you have tons and tons of customers, this might already justify using a model instead of a simple one feature policy.
## Fine Grain Policy
As a recap, so far, we tested the most simple of all policies, which is just engaging with all the customers. Since that didn't work (the average net income per customer was negative), we developed a single feature policy that was based on regions: we would do business in some regions, but not in others. This already gave us very good results. Next, we went full machine learning, with a predictive model. Then, we used that model as an input to a policy and chose to do business with all the customers whose net income predictions were above zero.
Here, the decision which the policy handles is very simple: engage with a customer or don't engage. The policies we had so far dealt with the binary case. They were in the form of
```
if prediction > 0 then do business else don't do business.
```
This is something we call **thresholding**. If the prediction exceeds a certain threshold (zero in our case, but could be something else), we take one decision, if it doesn't, we take another. One other example of where this could be applied in real life is transactional fraud detection: if the prediction score of a model that detects fraud is above some threshold `X`, we deny the transaction, otherwise we approve it.
Thresholding works in lots of real case scenarios and it is particularly useful when the nature of the decision is binary. However, we can think of cases where things tend to be more nuanced. For example, you might be willing to spend more in marketing to get the attention of very profitable customers. Or you might want to add them to some prime customers list, where you give special treatment to them, but it also costs you more to do so. Notice that if we include these possibilities, your decision goes from binary (engage vs don't engage) to continuous: how much should you invest in a customer.
Here, for the next example, suppose your decision is not just who to do business with, but how much marketing costs you should invest in each customer. And for the sake of the example, assume that you are competing with other firms and whoever spends more on marketing in a particular customer wins that customer (much like a bidding mechanism). In that case, it makes sense to invest more in highly profitable customers, less in marginally profitable customers and not at all in non profitable customers.
One way to do that is to descritize your predictions into bands. We've done this previously for the purpose of model comparison, but here we'll do it for decision making. Let's create 20 bands. We can think of those as quantiles or equal size groups. The first band will contain the 5% less profitable customers *according to our predictions*, the second band will contain from the 5% to the 10% less profitable and so on. The last band, 20, will contain the most profitable customers.
Notice that the binning too has to be estimated on the training set and applied on the test set! For this reason, we will compute the bins using `pd.qcut` on the training set. To actually do the binning, we will use `np.digitize`, passing the bins that where precomputed on the training set.
```
def model_binner(prediction_column, bins):
# find the bins according to the training set
bands = pd.qcut(prediction_column, q=bins, retbins=True)[1]
def binner_function(prediction_column):
return np.digitize(prediction_column, bands)
return binner_function
# train the bining function
binner_fn = model_binner(train_pred["predictions"], 20)
# apply the binning
model_band = model_policy.assign(bands = binner_fn(model_policy["prediction"]))
model_band.head()
plt.figure(figsize=(10,6))
sns.barplot(data=model_band, x="bands", y="net_value")
plt.title("Model Bands");
```
With these bands, we can allocate the bulk of our marketing investments to band 20 and 19. Notice how we went from a binary decision (engage vs not engage), to a continuous one: how much to invest on marketing for each customer. Of course you can fine tune this even more, adding more bands. In the limit, you are not binning at all. Instead, you are using the raw prediction of the model and you can create decision rules like
```
mkt_investments_i = model_prediction_i * 0.3
```
where for each customer \\(i\\), you invest 30% of the net_value predicted by the model (30% was an arbitrary number, but you get the point).
## Key Ideas
We've covered A LOT of ground here in a very short time, so I think this recap is extremely relevant for us to see what we accomplished here. First, we learned how the majority of machine learning applications involves nothing more than making good predictions, where prediction is understood as mapping from a known input to an initially unknown, but well defined output. But when I say "nothing more", I'm not being entirely fair. We also saw how good predictions can solve more problems than we might think at first, like language translation and self driving cars.
Then, we got back down to earth and looked at how good predictions can help us with more common tasks, like figuring out which customer we should bring in and which to avoid. Specifically, we looked at how we could predict customer profit. With that prediction, we've built a policy that decides who we should do business with. Notice that this is just an example of where prediction models can be applied. There are sure tones of other ones, like credit card underwriting, fraud detection, cancer diagnostics and anything else where good predictions might be useful.
The key takeaway here is that **if you can frame your business problem as a prediction problem, then machine learning is probably the right tool for the job**. I really can't emphasize this enough. With all the hype around machine learning, I feel that people forgot about this very important point and often end up making models that are very good in predicting something totally useless. Instead of thinking about how to frame a business problem as a prediction problem and *then* solving it with machine learning, they often build a prediction model and try to see what business problem could benefit from that prediction. This might work, but, more often than not, is a shot in the dark that only generates solutions in search of a problem.
## References
The things I've written here are mostly stuff from my head. I've learned them through experience. This means there isn't a direct reference I can point you to. It also means that the things I wrote here have **not** passed the academic scrutiny that good science often goes through. Instead, notice how I'm talking about things that work in practice, but I don't spend too much time explaining why that is the case. It's a sort of science from the streets, if you will. However, I am putting this up for public scrutiny, so, by all means, if you find something preposterous, open an issue and I'll address it to the best of my efforts.
Finally, I believe I might have been too quick for those who were hoping for a comprehensive and detailed introduction of machine learning. To be honest, I believe that where I can truly generate value is teaching about causal inference, not machine learning. For the latter, there are tons of amazing online resources, much better than I could ever dream of creating. The classical one is [Andrew Ng's course on Machine Learning](https://www.coursera.org/learn/machine-learning) and I definitely recommend you take a look into it if you are new to machine learning.
## Contribute
Causal Inference for the Brave and True is an open-source material on causal inference, the statistics of science. It uses only free software, based in Python. Its goal is to be accessible monetarily and intellectually.
If you found this book valuable and you want to support it, please go to [Patreon](https://www.patreon.com/causal_inference_for_the_brave_and_true). If you are not ready to contribute financially, you can also help by fixing typos, suggesting edits or giving feedback on passages you didn't understand. Just go to the book's repository and [open an issue](https://github.com/matheusfacure/python-causality-handbook/issues). Finally, if you liked this content, please share it with others who might find it useful and give it a [star on GitHub](https://github.com/matheusfacure/python-causality-handbook/stargazers).
|
github_jupyter
|
import pandas as pd
import numpy as np
from sklearn import ensemble
from sklearn.model_selection import train_test_split
from sklearn.pipeline import Pipeline
from sklearn.metrics import r2_score
import seaborn as sns
from matplotlib import pyplot as plt
from matplotlib import style
style.use("ggplot")
transactions = pd.read_csv("data/customer_transactions.csv")
print(transactions.shape)
transactions.head()
profitable = (transactions[["customer_id"]]
.assign(net_value = transactions
.drop(columns="customer_id")
.sum(axis=1)))
customer_features = (pd.read_csv("data/customer_features.csv")
.merge(profitable, on="customer_id"))
customer_features.head()
train, test = train_test_split(customer_features, test_size=0.3, random_state=13)
train.shape, test.shape
train["net_value"].mean()
plt.figure(figsize=(12,6))
np.random.seed(123) ## seed because the CIs from seaborn uses boostrap
# pd.qcut create quantiles of a column
sns.barplot(data=train.assign(income_quantile=pd.qcut(train["income"], q=20)),
x="income_quantile", y="net_value")
plt.title("Profitability by Income")
plt.xticks(rotation=70);
plt.figure(figsize=(12,6))
np.random.seed(123)
region_plot = sns.barplot(data=train, x="region", y="net_value")
plt.title("Profitability by Region");
# extract the lower bound of the 95% CI from the plot above
regions_to_net = {region: line.get_ydata().min()
for region, line in enumerate(region_plot.lines)}
# filters regions where the net value is > 0.
regions_to_invest = {region: net
for region, net in regions_to_net.items()
if net > 0}
regions_to_invest
region_policy = (test[test["region"]
# filter regions in regions_to_invest
.isin(regions_to_invest.keys())])
sns.histplot(data=region_policy, x="net_value")
# average has to be over all customers, not just the one we've filtered with the policy
plt.title("Average Net Income: %.2f" % (region_policy["net_value"].sum() / test.shape[0]));
def encode(df):
return df.replace({"region": regions_to_net})
model_params = {'n_estimators': 400,
'max_depth': 4,
'min_samples_split': 10,
'learning_rate': 0.01,
'loss': 'ls'}
features = ["region", "income", "age"]
target = "net_value"
np.random.seed(123)
reg = ensemble.GradientBoostingRegressor(**model_params)
# fit model on the training set
encoded_train = train[features].pipe(encode)
reg.fit(encoded_train, train[target]);
train_pred = (encoded_train
.assign(predictions=reg.predict(encoded_train[features])))
print("Train R2: ", r2_score(y_true=train[target], y_pred=train_pred["predictions"]))
print("Test R2: ", r2_score(y_true=test[target], y_pred=reg.predict(test[features].pipe(encode))))
model_policy = test.assign(prediction=reg.predict(test[features].pipe(encode)))
model_policy.head()
plt.figure(figsize=(12,6))
n_bands = 50
bands = [f"band_{b}" for b in range(1,n_bands+1)]
np.random.seed(123)
model_plot = sns.barplot(data=model_policy
.assign(model_band = pd.qcut(model_policy["prediction"], q=n_bands)),
x="model_band", y="net_value")
plt.title("Profitability by Model Prediction Quantiles")
plt.xticks(rotation=70);
plt.figure(figsize=(10,6))
model_plot_df = (model_policy[model_policy["prediction"]>0])
sns.histplot(data=model_plot_df, x="net_value", color="C2", label="model_policy")
region_plot_df = (model_policy[model_policy["region"].isin(regions_to_invest.keys())])
sns.histplot(data=region_plot_df, x="net_value", label="region_policy")
plt.title("Model Net Income: %.2f; Region Policy Net Income %.2f." %
(model_plot_df["net_value"].sum() / test.shape[0],
region_plot_df["net_value"].sum() / test.shape[0]))
plt.legend();
if prediction > 0 then do business else don't do business.
def model_binner(prediction_column, bins):
# find the bins according to the training set
bands = pd.qcut(prediction_column, q=bins, retbins=True)[1]
def binner_function(prediction_column):
return np.digitize(prediction_column, bands)
return binner_function
# train the bining function
binner_fn = model_binner(train_pred["predictions"], 20)
# apply the binning
model_band = model_policy.assign(bands = binner_fn(model_policy["prediction"]))
model_band.head()
plt.figure(figsize=(10,6))
sns.barplot(data=model_band, x="bands", y="net_value")
plt.title("Model Bands");
mkt_investments_i = model_prediction_i * 0.3
| 0.74055 | 0.987005 |
<a href="https://colab.research.google.com/github/jonkrohn/DLTFpT/blob/master/notebooks/object_detection.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# Object Detection
Based on Renu Khandelwal's YOLOv3 demo provided [here](https://medium.com/datadriveninvestor/object-detection-using-yolov3-using-keras-80bf35e61ce1).
#### Load dependencies
```
import os
import scipy.io
import scipy.misc
import numpy as np
from numpy import expand_dims
import pandas as pd
import PIL
import struct
import cv2
from numpy import expand_dims
import tensorflow as tf
from tensorflow.keras import backend as K
from tensorflow.keras.layers import Input, Lambda, Conv2D, BatchNormalization, LeakyReLU, ZeroPadding2D, UpSampling2D
from tensorflow.keras.models import load_model, Model
from tensorflow.keras.layers import add, concatenate
from tensorflow.keras.preprocessing.image import load_img
from tensorflow.keras.preprocessing.image import img_to_array
import matplotlib.pyplot as plt
from matplotlib.pyplot import imshow
from matplotlib.patches import Rectangle
from skimage.transform import resize
%matplotlib inline
```
#### Set hyperparameters
```
net_h, net_w = 416, 416
obj_thresh, nms_thresh = 0.5, 0.45
# there are 80 class labels in the MS COCO dataset:
labels = ["person", "bicycle", "car", "motorbike", "aeroplane", "bus", "train", "truck", \
"boat", "traffic light", "fire hydrant", "stop sign", "parking meter", "bench", \
"bird", "cat", "dog", "horse", "sheep", "cow", "elephant", "bear", "zebra", "giraffe", \
"backpack", "umbrella", "handbag", "tie", "suitcase", "frisbee", "skis", "snowboard", \
"sports ball", "kite", "baseball bat", "baseball glove", "skateboard", "surfboard", \
"tennis racket", "bottle", "wine glass", "cup", "fork", "knife", "spoon", "bowl", "banana", \
"apple", "sandwich", "orange", "broccoli", "carrot", "hot dog", "pizza", "donut", "cake", \
"chair", "sofa", "pottedplant", "bed", "diningtable", "toilet", "tvmonitor", "laptop", "mouse", \
"remote", "keyboard", "cell phone", "microwave", "oven", "toaster", "sink", "refrigerator", \
"book", "clock", "vase", "scissors", "teddy bear", "hair drier", "toothbrush"]
```
#### Design model architecture
```
# define block of conv layers:
def _conv_block(inp, convs, skip=True):
x = inp
count = 0
for conv in convs:
if count == (len(convs) - 2) and skip:
skip_connection = x
count += 1
if conv['stride'] > 1: x = ZeroPadding2D(((1,0),(1,0)))(x) # peculiar padding as darknet prefer left and top
x = Conv2D(conv['filter'],
conv['kernel'],
strides=conv['stride'],
padding='valid' if conv['stride'] > 1 else 'same', # peculiar padding as darknet prefer left and top
name='conv_' + str(conv['layer_idx']),
use_bias=False if conv['bnorm'] else True)(x)
if conv['bnorm']: x = BatchNormalization(epsilon=0.001, name='bnorm_' + str(conv['layer_idx']))(x)
if conv['leaky']: x = LeakyReLU(alpha=0.1, name='leaky_' + str(conv['layer_idx']))(x)
return add([skip_connection, x]) if skip else x
# use _conv_block() to define model architecture:
def make_yolov3_model():
input_image = Input(shape=(None, None, 3))
# Layer 0 => 4
x = _conv_block(input_image, [{'filter': 32, 'kernel': 3, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 0},
{'filter': 64, 'kernel': 3, 'stride': 2, 'bnorm': True, 'leaky': True, 'layer_idx': 1},
{'filter': 32, 'kernel': 1, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 2},
{'filter': 64, 'kernel': 3, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 3}])
# Layer 5 => 8
x = _conv_block(x, [{'filter': 128, 'kernel': 3, 'stride': 2, 'bnorm': True, 'leaky': True, 'layer_idx': 5},
{'filter': 64, 'kernel': 1, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 6},
{'filter': 128, 'kernel': 3, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 7}])
# Layer 9 => 11
x = _conv_block(x, [{'filter': 64, 'kernel': 1, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 9},
{'filter': 128, 'kernel': 3, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 10}])
# Layer 12 => 15
x = _conv_block(x, [{'filter': 256, 'kernel': 3, 'stride': 2, 'bnorm': True, 'leaky': True, 'layer_idx': 12},
{'filter': 128, 'kernel': 1, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 13},
{'filter': 256, 'kernel': 3, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 14}])
# Layer 16 => 36
for i in range(7):
x = _conv_block(x, [{'filter': 128, 'kernel': 1, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 16+i*3},
{'filter': 256, 'kernel': 3, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 17+i*3}])
skip_36 = x
# Layer 37 => 40
x = _conv_block(x, [{'filter': 512, 'kernel': 3, 'stride': 2, 'bnorm': True, 'leaky': True, 'layer_idx': 37},
{'filter': 256, 'kernel': 1, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 38},
{'filter': 512, 'kernel': 3, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 39}])
# Layer 41 => 61
for i in range(7):
x = _conv_block(x, [{'filter': 256, 'kernel': 1, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 41+i*3},
{'filter': 512, 'kernel': 3, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 42+i*3}])
skip_61 = x
# Layer 62 => 65
x = _conv_block(x, [{'filter': 1024, 'kernel': 3, 'stride': 2, 'bnorm': True, 'leaky': True, 'layer_idx': 62},
{'filter': 512, 'kernel': 1, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 63},
{'filter': 1024, 'kernel': 3, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 64}])
# Layer 66 => 74
for i in range(3):
x = _conv_block(x, [{'filter': 512, 'kernel': 1, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 66+i*3},
{'filter': 1024, 'kernel': 3, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 67+i*3}])
# Layer 75 => 79
x = _conv_block(x, [{'filter': 512, 'kernel': 1, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 75},
{'filter': 1024, 'kernel': 3, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 76},
{'filter': 512, 'kernel': 1, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 77},
{'filter': 1024, 'kernel': 3, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 78},
{'filter': 512, 'kernel': 1, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 79}], skip=False)
# Layer 80 => 82
yolo_82 = _conv_block(x, [{'filter': 1024, 'kernel': 3, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 80},
{'filter': 255, 'kernel': 1, 'stride': 1, 'bnorm': False, 'leaky': False, 'layer_idx': 81}], skip=False)
# Layer 83 => 86
x = _conv_block(x, [{'filter': 256, 'kernel': 1, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 84}], skip=False)
x = UpSampling2D(2)(x)
x = concatenate([x, skip_61])
# Layer 87 => 91
x = _conv_block(x, [{'filter': 256, 'kernel': 1, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 87},
{'filter': 512, 'kernel': 3, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 88},
{'filter': 256, 'kernel': 1, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 89},
{'filter': 512, 'kernel': 3, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 90},
{'filter': 256, 'kernel': 1, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 91}], skip=False)
# Layer 92 => 94
yolo_94 = _conv_block(x, [{'filter': 512, 'kernel': 3, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 92},
{'filter': 255, 'kernel': 1, 'stride': 1, 'bnorm': False, 'leaky': False, 'layer_idx': 93}], skip=False)
# Layer 95 => 98
x = _conv_block(x, [{'filter': 128, 'kernel': 1, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 96}], skip=False)
x = UpSampling2D(2)(x)
x = concatenate([x, skip_36])
# Layer 99 => 106
yolo_106 = _conv_block(x, [{'filter': 128, 'kernel': 1, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 99},
{'filter': 256, 'kernel': 3, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 100},
{'filter': 128, 'kernel': 1, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 101},
{'filter': 256, 'kernel': 3, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 102},
{'filter': 128, 'kernel': 1, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 103},
{'filter': 256, 'kernel': 3, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 104},
{'filter': 255, 'kernel': 1, 'stride': 1, 'bnorm': False, 'leaky': False, 'layer_idx': 105}], skip=False)
model = Model(input_image, [yolo_82, yolo_94, yolo_106])
return model
yolov3 = make_yolov3_model()
# N.B.: uncomment the following line of code to download yolov3 model weights:
! wget -c https://www.dropbox.com/s/88xnszqf7xkf70j/yolov3.h5
yolov3.load_weights('yolov3.h5')
```
#### Define object detection-specific functions
```
def load_image_pixels(filename, shape):
# load the image to get its shape
image = load_img(filename)
width, height = image.size
# load the image with the required size
image = load_img(filename, target_size=shape)
# convert to numpy array
image = img_to_array(image)
# scale pixel values to [0, 1]
image = image.astype('float32')
image /= 255.0
# add a dimension so that we have one sample
image = expand_dims(image, 0)
return image, width, height
class BoundBox:
def __init__(self, xmin, ymin, xmax, ymax, objness = None, classes = None):
self.xmin = xmin
self.ymin = ymin
self.xmax = xmax
self.ymax = ymax
self.objness = objness
self.classes = classes
self.label = -1
self.score = -1
def get_label(self):
if self.label == -1:
self.label = np.argmax(self.classes)
return self.label
def get_score(self):
if self.score == -1:
self.score = self.classes[self.get_label()]
return self.score
def _sigmoid(x):
return 1. / (1. + np.exp(-x))
def _interval_overlap(interval_a, interval_b):
x1, x2 = interval_a
x3, x4 = interval_b
if x3 < x1:
if x4 < x1:
return 0
else:
return min(x2,x4) - x1
else:
if x2 < x3:
return 0
else:
return min(x2,x4) - x3
def bbox_iou(box1, box2):
intersect_w = _interval_overlap([box1.xmin, box1.xmax], [box2.xmin, box2.xmax])
intersect_h = _interval_overlap([box1.ymin, box1.ymax], [box2.ymin, box2.ymax])
intersect = intersect_w * intersect_h
w1, h1 = box1.xmax-box1.xmin, box1.ymax-box1.ymin
w2, h2 = box2.xmax-box2.xmin, box2.ymax-box2.ymin
union = w1*h1 + w2*h2 - intersect
return float(intersect) / union
def do_nms(boxes, nms_thresh):
if len(boxes) > 0:
nb_class = len(boxes[0].classes)
else:
return
for c in range(nb_class):
sorted_indices = np.argsort([-box.classes[c] for box in boxes])
for i in range(len(sorted_indices)):
index_i = sorted_indices[i]
if boxes[index_i].classes[c] == 0: continue
for j in range(i+1, len(sorted_indices)):
index_j = sorted_indices[j]
if bbox_iou(boxes[index_i], boxes[index_j]) >= nms_thresh:
boxes[index_j].classes[c] = 0
# decode_netout() takes each one of the NumPy arrays, one at a time,
# and decodes the candidate bounding boxes and class predictions
def decode_netout(netout, anchors, obj_thresh, net_h, net_w):
grid_h, grid_w = netout.shape[:2]
nb_box = 3
netout = netout.reshape((grid_h, grid_w, nb_box, -1))
nb_class = netout.shape[-1] - 5
boxes = []
netout[..., :2] = _sigmoid(netout[..., :2])
netout[..., 4:] = _sigmoid(netout[..., 4:])
netout[..., 5:] = netout[..., 4][..., np.newaxis] * netout[..., 5:]
netout[..., 5:] *= netout[..., 5:] > obj_thresh
for i in range(grid_h*grid_w):
row = i / grid_w
col = i % grid_w
for b in range(nb_box):
# 4th element is objectness score
objectness = netout[int(row)][int(col)][b][4]
#objectness = netout[..., :4]
if(objectness.all() <= obj_thresh): continue
# first 4 elements are x, y, w, and h
x, y, w, h = netout[int(row)][int(col)][b][:4]
x = (col + x) / grid_w # center position, unit: image width
y = (row + y) / grid_h # center position, unit: image height
w = anchors[2 * b + 0] * np.exp(w) / net_w # unit: image width
h = anchors[2 * b + 1] * np.exp(h) / net_h # unit: image height
# last elements are class probabilities
classes = netout[int(row)][col][b][5:]
box = BoundBox(x-w/2, y-h/2, x+w/2, y+h/2, objectness, classes)
#box = BoundBox(x-w/2, y-h/2, x+w/2, y+h/2, None, classes)
boxes.append(box)
return boxes
# to stretch bounding boxes back into the shape of the original image,
# enabling plotting of the original image and with bounding boxes overlain
def correct_yolo_boxes(boxes, image_h, image_w, net_h, net_w):
if (float(net_w)/image_w) < (float(net_h)/image_h):
new_w = net_w
new_h = (image_h*net_w)/image_w
else:
new_h = net_w
new_w = (image_w*net_h)/image_h
for i in range(len(boxes)):
x_offset, x_scale = (net_w - new_w)/2./net_w, float(new_w)/net_w
y_offset, y_scale = (net_h - new_h)/2./net_h, float(new_h)/net_h
boxes[i].xmin = int((boxes[i].xmin - x_offset) / x_scale * image_w)
boxes[i].xmax = int((boxes[i].xmax - x_offset) / x_scale * image_w)
boxes[i].ymin = int((boxes[i].ymin - y_offset) / y_scale * image_h)
boxes[i].ymax = int((boxes[i].ymax - y_offset) / y_scale * image_h)
def draw_boxes(filename, v_boxes, v_labels, v_scores):
# load the image
data = plt.imread(filename)
# plot the image
plt.imshow(data)
# get the context for drawing boxes
ax = plt.gca()
# plot each box
for i in range(len(v_boxes)):
box = v_boxes[i]
# get coordinates
y1, x1, y2, x2 = box.ymin, box.xmin, box.ymax, box.xmax
# calculate width and height of the box
width, height = x2 - x1, y2 - y1
# create the shape
rect = Rectangle((x1, y1), width, height, fill=False, color='red')
# draw the box
ax.add_patch(rect)
# draw text and score in top left corner
label = "%s (%.3f)" % (v_labels[i], v_scores[i])
plt.text(x1, y1, label, color='red')
# show the plot
plt.show()
# get all of the results above a threshold
# takes the list of boxes, known labels,
# and our classification threshold as arguments
# and returns parallel lists of boxes, labels, and scores.
def get_boxes(boxes, labels, thresh):
v_boxes, v_labels, v_scores = list(), list(), list()
# enumerate all boxes
for box in boxes:
# enumerate all possible labels
for i in range(len(labels)):
# check if the threshold for this label is high enough
if box.classes[i] > thresh:
v_boxes.append(box)
v_labels.append(labels[i])
v_scores.append(box.classes[i]*100)
# don't break, many labels may trigger for one box
return v_boxes, v_labels, v_scores
```
#### Load sample image
```
! wget -c https://raw.githubusercontent.com/jonkrohn/DLTFpT/master/notebooks/oboe-with-book.jpg
# define the expected input shape for the model
input_w, input_h = 416, 416
# define our new photo
photo_filename = 'oboe-with-book.jpg'
# load and prepare image
image, image_w, image_h = load_image_pixels(photo_filename, (net_w, net_w))
plt.imshow(plt.imread(photo_filename))
```
#### Perform inference
```
# make prediction
yolos = yolov3.predict(image)
# define the anchors
anchors = [[116,90, 156,198, 373,326], [30,61, 62,45, 59,119], [10,13, 16,30, 33,23]]
# define the probability threshold for detected objects
class_threshold = 0.6
boxes = list()
for i in range(len(yolos)):
# decode the output of the network
boxes += decode_netout(yolos[i][0], anchors[i], obj_thresh, net_h, net_w)
# correct the sizes of the bounding boxes
correct_yolo_boxes(boxes, image_h, image_w, net_h, net_w)
# suppress non-maximal boxes
do_nms(boxes, nms_thresh)
# extract the details of the detected objects
v_boxes, v_labels, v_scores = get_boxes(boxes, labels, class_threshold)
# summarize what model found
for i in range(len(v_boxes)):
print(v_labels[i], v_scores[i])
# draw what model found
draw_boxes(photo_filename, v_boxes, v_labels, v_scores)
```
|
github_jupyter
|
import os
import scipy.io
import scipy.misc
import numpy as np
from numpy import expand_dims
import pandas as pd
import PIL
import struct
import cv2
from numpy import expand_dims
import tensorflow as tf
from tensorflow.keras import backend as K
from tensorflow.keras.layers import Input, Lambda, Conv2D, BatchNormalization, LeakyReLU, ZeroPadding2D, UpSampling2D
from tensorflow.keras.models import load_model, Model
from tensorflow.keras.layers import add, concatenate
from tensorflow.keras.preprocessing.image import load_img
from tensorflow.keras.preprocessing.image import img_to_array
import matplotlib.pyplot as plt
from matplotlib.pyplot import imshow
from matplotlib.patches import Rectangle
from skimage.transform import resize
%matplotlib inline
net_h, net_w = 416, 416
obj_thresh, nms_thresh = 0.5, 0.45
# there are 80 class labels in the MS COCO dataset:
labels = ["person", "bicycle", "car", "motorbike", "aeroplane", "bus", "train", "truck", \
"boat", "traffic light", "fire hydrant", "stop sign", "parking meter", "bench", \
"bird", "cat", "dog", "horse", "sheep", "cow", "elephant", "bear", "zebra", "giraffe", \
"backpack", "umbrella", "handbag", "tie", "suitcase", "frisbee", "skis", "snowboard", \
"sports ball", "kite", "baseball bat", "baseball glove", "skateboard", "surfboard", \
"tennis racket", "bottle", "wine glass", "cup", "fork", "knife", "spoon", "bowl", "banana", \
"apple", "sandwich", "orange", "broccoli", "carrot", "hot dog", "pizza", "donut", "cake", \
"chair", "sofa", "pottedplant", "bed", "diningtable", "toilet", "tvmonitor", "laptop", "mouse", \
"remote", "keyboard", "cell phone", "microwave", "oven", "toaster", "sink", "refrigerator", \
"book", "clock", "vase", "scissors", "teddy bear", "hair drier", "toothbrush"]
# define block of conv layers:
def _conv_block(inp, convs, skip=True):
x = inp
count = 0
for conv in convs:
if count == (len(convs) - 2) and skip:
skip_connection = x
count += 1
if conv['stride'] > 1: x = ZeroPadding2D(((1,0),(1,0)))(x) # peculiar padding as darknet prefer left and top
x = Conv2D(conv['filter'],
conv['kernel'],
strides=conv['stride'],
padding='valid' if conv['stride'] > 1 else 'same', # peculiar padding as darknet prefer left and top
name='conv_' + str(conv['layer_idx']),
use_bias=False if conv['bnorm'] else True)(x)
if conv['bnorm']: x = BatchNormalization(epsilon=0.001, name='bnorm_' + str(conv['layer_idx']))(x)
if conv['leaky']: x = LeakyReLU(alpha=0.1, name='leaky_' + str(conv['layer_idx']))(x)
return add([skip_connection, x]) if skip else x
# use _conv_block() to define model architecture:
def make_yolov3_model():
input_image = Input(shape=(None, None, 3))
# Layer 0 => 4
x = _conv_block(input_image, [{'filter': 32, 'kernel': 3, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 0},
{'filter': 64, 'kernel': 3, 'stride': 2, 'bnorm': True, 'leaky': True, 'layer_idx': 1},
{'filter': 32, 'kernel': 1, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 2},
{'filter': 64, 'kernel': 3, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 3}])
# Layer 5 => 8
x = _conv_block(x, [{'filter': 128, 'kernel': 3, 'stride': 2, 'bnorm': True, 'leaky': True, 'layer_idx': 5},
{'filter': 64, 'kernel': 1, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 6},
{'filter': 128, 'kernel': 3, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 7}])
# Layer 9 => 11
x = _conv_block(x, [{'filter': 64, 'kernel': 1, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 9},
{'filter': 128, 'kernel': 3, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 10}])
# Layer 12 => 15
x = _conv_block(x, [{'filter': 256, 'kernel': 3, 'stride': 2, 'bnorm': True, 'leaky': True, 'layer_idx': 12},
{'filter': 128, 'kernel': 1, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 13},
{'filter': 256, 'kernel': 3, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 14}])
# Layer 16 => 36
for i in range(7):
x = _conv_block(x, [{'filter': 128, 'kernel': 1, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 16+i*3},
{'filter': 256, 'kernel': 3, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 17+i*3}])
skip_36 = x
# Layer 37 => 40
x = _conv_block(x, [{'filter': 512, 'kernel': 3, 'stride': 2, 'bnorm': True, 'leaky': True, 'layer_idx': 37},
{'filter': 256, 'kernel': 1, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 38},
{'filter': 512, 'kernel': 3, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 39}])
# Layer 41 => 61
for i in range(7):
x = _conv_block(x, [{'filter': 256, 'kernel': 1, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 41+i*3},
{'filter': 512, 'kernel': 3, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 42+i*3}])
skip_61 = x
# Layer 62 => 65
x = _conv_block(x, [{'filter': 1024, 'kernel': 3, 'stride': 2, 'bnorm': True, 'leaky': True, 'layer_idx': 62},
{'filter': 512, 'kernel': 1, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 63},
{'filter': 1024, 'kernel': 3, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 64}])
# Layer 66 => 74
for i in range(3):
x = _conv_block(x, [{'filter': 512, 'kernel': 1, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 66+i*3},
{'filter': 1024, 'kernel': 3, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 67+i*3}])
# Layer 75 => 79
x = _conv_block(x, [{'filter': 512, 'kernel': 1, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 75},
{'filter': 1024, 'kernel': 3, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 76},
{'filter': 512, 'kernel': 1, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 77},
{'filter': 1024, 'kernel': 3, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 78},
{'filter': 512, 'kernel': 1, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 79}], skip=False)
# Layer 80 => 82
yolo_82 = _conv_block(x, [{'filter': 1024, 'kernel': 3, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 80},
{'filter': 255, 'kernel': 1, 'stride': 1, 'bnorm': False, 'leaky': False, 'layer_idx': 81}], skip=False)
# Layer 83 => 86
x = _conv_block(x, [{'filter': 256, 'kernel': 1, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 84}], skip=False)
x = UpSampling2D(2)(x)
x = concatenate([x, skip_61])
# Layer 87 => 91
x = _conv_block(x, [{'filter': 256, 'kernel': 1, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 87},
{'filter': 512, 'kernel': 3, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 88},
{'filter': 256, 'kernel': 1, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 89},
{'filter': 512, 'kernel': 3, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 90},
{'filter': 256, 'kernel': 1, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 91}], skip=False)
# Layer 92 => 94
yolo_94 = _conv_block(x, [{'filter': 512, 'kernel': 3, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 92},
{'filter': 255, 'kernel': 1, 'stride': 1, 'bnorm': False, 'leaky': False, 'layer_idx': 93}], skip=False)
# Layer 95 => 98
x = _conv_block(x, [{'filter': 128, 'kernel': 1, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 96}], skip=False)
x = UpSampling2D(2)(x)
x = concatenate([x, skip_36])
# Layer 99 => 106
yolo_106 = _conv_block(x, [{'filter': 128, 'kernel': 1, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 99},
{'filter': 256, 'kernel': 3, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 100},
{'filter': 128, 'kernel': 1, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 101},
{'filter': 256, 'kernel': 3, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 102},
{'filter': 128, 'kernel': 1, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 103},
{'filter': 256, 'kernel': 3, 'stride': 1, 'bnorm': True, 'leaky': True, 'layer_idx': 104},
{'filter': 255, 'kernel': 1, 'stride': 1, 'bnorm': False, 'leaky': False, 'layer_idx': 105}], skip=False)
model = Model(input_image, [yolo_82, yolo_94, yolo_106])
return model
yolov3 = make_yolov3_model()
# N.B.: uncomment the following line of code to download yolov3 model weights:
! wget -c https://www.dropbox.com/s/88xnszqf7xkf70j/yolov3.h5
yolov3.load_weights('yolov3.h5')
def load_image_pixels(filename, shape):
# load the image to get its shape
image = load_img(filename)
width, height = image.size
# load the image with the required size
image = load_img(filename, target_size=shape)
# convert to numpy array
image = img_to_array(image)
# scale pixel values to [0, 1]
image = image.astype('float32')
image /= 255.0
# add a dimension so that we have one sample
image = expand_dims(image, 0)
return image, width, height
class BoundBox:
def __init__(self, xmin, ymin, xmax, ymax, objness = None, classes = None):
self.xmin = xmin
self.ymin = ymin
self.xmax = xmax
self.ymax = ymax
self.objness = objness
self.classes = classes
self.label = -1
self.score = -1
def get_label(self):
if self.label == -1:
self.label = np.argmax(self.classes)
return self.label
def get_score(self):
if self.score == -1:
self.score = self.classes[self.get_label()]
return self.score
def _sigmoid(x):
return 1. / (1. + np.exp(-x))
def _interval_overlap(interval_a, interval_b):
x1, x2 = interval_a
x3, x4 = interval_b
if x3 < x1:
if x4 < x1:
return 0
else:
return min(x2,x4) - x1
else:
if x2 < x3:
return 0
else:
return min(x2,x4) - x3
def bbox_iou(box1, box2):
intersect_w = _interval_overlap([box1.xmin, box1.xmax], [box2.xmin, box2.xmax])
intersect_h = _interval_overlap([box1.ymin, box1.ymax], [box2.ymin, box2.ymax])
intersect = intersect_w * intersect_h
w1, h1 = box1.xmax-box1.xmin, box1.ymax-box1.ymin
w2, h2 = box2.xmax-box2.xmin, box2.ymax-box2.ymin
union = w1*h1 + w2*h2 - intersect
return float(intersect) / union
def do_nms(boxes, nms_thresh):
if len(boxes) > 0:
nb_class = len(boxes[0].classes)
else:
return
for c in range(nb_class):
sorted_indices = np.argsort([-box.classes[c] for box in boxes])
for i in range(len(sorted_indices)):
index_i = sorted_indices[i]
if boxes[index_i].classes[c] == 0: continue
for j in range(i+1, len(sorted_indices)):
index_j = sorted_indices[j]
if bbox_iou(boxes[index_i], boxes[index_j]) >= nms_thresh:
boxes[index_j].classes[c] = 0
# decode_netout() takes each one of the NumPy arrays, one at a time,
# and decodes the candidate bounding boxes and class predictions
def decode_netout(netout, anchors, obj_thresh, net_h, net_w):
grid_h, grid_w = netout.shape[:2]
nb_box = 3
netout = netout.reshape((grid_h, grid_w, nb_box, -1))
nb_class = netout.shape[-1] - 5
boxes = []
netout[..., :2] = _sigmoid(netout[..., :2])
netout[..., 4:] = _sigmoid(netout[..., 4:])
netout[..., 5:] = netout[..., 4][..., np.newaxis] * netout[..., 5:]
netout[..., 5:] *= netout[..., 5:] > obj_thresh
for i in range(grid_h*grid_w):
row = i / grid_w
col = i % grid_w
for b in range(nb_box):
# 4th element is objectness score
objectness = netout[int(row)][int(col)][b][4]
#objectness = netout[..., :4]
if(objectness.all() <= obj_thresh): continue
# first 4 elements are x, y, w, and h
x, y, w, h = netout[int(row)][int(col)][b][:4]
x = (col + x) / grid_w # center position, unit: image width
y = (row + y) / grid_h # center position, unit: image height
w = anchors[2 * b + 0] * np.exp(w) / net_w # unit: image width
h = anchors[2 * b + 1] * np.exp(h) / net_h # unit: image height
# last elements are class probabilities
classes = netout[int(row)][col][b][5:]
box = BoundBox(x-w/2, y-h/2, x+w/2, y+h/2, objectness, classes)
#box = BoundBox(x-w/2, y-h/2, x+w/2, y+h/2, None, classes)
boxes.append(box)
return boxes
# to stretch bounding boxes back into the shape of the original image,
# enabling plotting of the original image and with bounding boxes overlain
def correct_yolo_boxes(boxes, image_h, image_w, net_h, net_w):
if (float(net_w)/image_w) < (float(net_h)/image_h):
new_w = net_w
new_h = (image_h*net_w)/image_w
else:
new_h = net_w
new_w = (image_w*net_h)/image_h
for i in range(len(boxes)):
x_offset, x_scale = (net_w - new_w)/2./net_w, float(new_w)/net_w
y_offset, y_scale = (net_h - new_h)/2./net_h, float(new_h)/net_h
boxes[i].xmin = int((boxes[i].xmin - x_offset) / x_scale * image_w)
boxes[i].xmax = int((boxes[i].xmax - x_offset) / x_scale * image_w)
boxes[i].ymin = int((boxes[i].ymin - y_offset) / y_scale * image_h)
boxes[i].ymax = int((boxes[i].ymax - y_offset) / y_scale * image_h)
def draw_boxes(filename, v_boxes, v_labels, v_scores):
# load the image
data = plt.imread(filename)
# plot the image
plt.imshow(data)
# get the context for drawing boxes
ax = plt.gca()
# plot each box
for i in range(len(v_boxes)):
box = v_boxes[i]
# get coordinates
y1, x1, y2, x2 = box.ymin, box.xmin, box.ymax, box.xmax
# calculate width and height of the box
width, height = x2 - x1, y2 - y1
# create the shape
rect = Rectangle((x1, y1), width, height, fill=False, color='red')
# draw the box
ax.add_patch(rect)
# draw text and score in top left corner
label = "%s (%.3f)" % (v_labels[i], v_scores[i])
plt.text(x1, y1, label, color='red')
# show the plot
plt.show()
# get all of the results above a threshold
# takes the list of boxes, known labels,
# and our classification threshold as arguments
# and returns parallel lists of boxes, labels, and scores.
def get_boxes(boxes, labels, thresh):
v_boxes, v_labels, v_scores = list(), list(), list()
# enumerate all boxes
for box in boxes:
# enumerate all possible labels
for i in range(len(labels)):
# check if the threshold for this label is high enough
if box.classes[i] > thresh:
v_boxes.append(box)
v_labels.append(labels[i])
v_scores.append(box.classes[i]*100)
# don't break, many labels may trigger for one box
return v_boxes, v_labels, v_scores
! wget -c https://raw.githubusercontent.com/jonkrohn/DLTFpT/master/notebooks/oboe-with-book.jpg
# define the expected input shape for the model
input_w, input_h = 416, 416
# define our new photo
photo_filename = 'oboe-with-book.jpg'
# load and prepare image
image, image_w, image_h = load_image_pixels(photo_filename, (net_w, net_w))
plt.imshow(plt.imread(photo_filename))
# make prediction
yolos = yolov3.predict(image)
# define the anchors
anchors = [[116,90, 156,198, 373,326], [30,61, 62,45, 59,119], [10,13, 16,30, 33,23]]
# define the probability threshold for detected objects
class_threshold = 0.6
boxes = list()
for i in range(len(yolos)):
# decode the output of the network
boxes += decode_netout(yolos[i][0], anchors[i], obj_thresh, net_h, net_w)
# correct the sizes of the bounding boxes
correct_yolo_boxes(boxes, image_h, image_w, net_h, net_w)
# suppress non-maximal boxes
do_nms(boxes, nms_thresh)
# extract the details of the detected objects
v_boxes, v_labels, v_scores = get_boxes(boxes, labels, class_threshold)
# summarize what model found
for i in range(len(v_boxes)):
print(v_labels[i], v_scores[i])
# draw what model found
draw_boxes(photo_filename, v_boxes, v_labels, v_scores)
| 0.566019 | 0.888178 |
```
import pandas as pd
import numpy as np
from sklearn.svm import SVC
from sklearn.model_selection import train_test_split
from sklearn import metrics
from sklearn.metrics import confusion_matrix
from sklearn.model_selection import KFold
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import GridSearchCV
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.preprocessing import scale
```
#### Loading Data
```
df=pd.read_csv("train.csv")
df.head()
df.shape
# Getting the 20 % of data
n=20
df=df.head(int(len(df.index)*(n/100)))
df.shape
# Checking for null values
df.isnull().sum().sort_values(ascending=False)
```
### Model Building
```
# splitting into X and y
X = df.drop("label", axis = 1)
y = df.label.values.astype(int)
# scaling the features
X_scaled = scale(X)
# train test split
X_train, X_test, y_train, y_test = train_test_split(X_scaled, y, test_size = 0.3, random_state = 4)
# using rbf kernel, C=1, default value of gamma
model = SVC(C = 1, kernel='rbf')
model.fit(X_train, y_train)
y_pred = model.predict(X_test)
# confusion matrix
confusion_matrix(y_true=y_test, y_pred=y_pred)
```
### Model Evaluation
```
#Visualising the confusion matrix on the heatmap
plt.figure(figsize=(12,10))
corr=confusion_matrix(y_true=y_test, y_pred=y_pred)
sns.heatmap(corr,annot=True)
# accuracy
print("accuracy", metrics.accuracy_score(y_test, y_pred))
# Calculating precision
TP=0 #Initializing True positives
FP=0 #Initializing False positives
for i in range (corr.shape[0]):
TP=corr[i][i]
FP=corr.sum(axis=1)[i]-TP
print ("Precision for {}, is {ratio} ".format(i,ratio=TP/(TP+FP)))
# Calculating recall
TP=0 #Initializing True positives
FN=0 #Initializing False negatives
for i in range (corr.shape[0]):
TP=corr[i][i]
FN=corr.sum(axis=0)[i]-TP
print ("Recall for {}, is {ratio} ".format(i,ratio=TP/(TP+FN)))
```
### Hyperparameter Tuning
#### Grid Search to find optimal hyperparameters
```
# creating a KFold object with 5 splits
folds = KFold(n_splits = 5, shuffle = True, random_state = 4)
# specify range of hyperparameters
# Set the parameters by cross-validation
hyper_params = [ {'gamma': [1e-2, 1e-3, 1e-4],
'C': [1, 10, 100, 1000]}]
# specify model
model = SVC(kernel="rbf")
# set up GridSearchCV()
model_cv = GridSearchCV(estimator = model,
param_grid = hyper_params,
scoring= 'accuracy',
cv = folds,
verbose = 1,
return_train_score=True)
# fit the model
model_cv.fit(X_train, y_train)
# cv results
cv_results = pd.DataFrame(model_cv.cv_results_)
cv_results
# converting C to numeric type for plotting on x-axis
cv_results['param_C'] = cv_results['param_C'].astype('int')
# # plotting
plt.figure(figsize=(16,6))
# subplot 1/3
plt.subplot(131)
gamma_01 = cv_results[cv_results['param_gamma']==0.01]
plt.plot(gamma_01["param_C"], gamma_01["mean_test_score"])
plt.plot(gamma_01["param_C"], gamma_01["mean_train_score"])
plt.xlabel('C')
plt.ylabel('Accuracy')
plt.title("Gamma=0.01")
plt.ylim([0.80, 1])
plt.legend(['test accuracy', 'train accuracy'], loc='upper left')
plt.xscale('log')
# subplot 2/3
plt.subplot(132)
gamma_001 = cv_results[cv_results['param_gamma']==0.001]
plt.plot(gamma_001["param_C"], gamma_001["mean_test_score"])
plt.plot(gamma_001["param_C"], gamma_001["mean_train_score"])
plt.xlabel('C')
plt.ylabel('Accuracy')
plt.title("Gamma=0.001")
plt.ylim([0.80, 1])
plt.legend(['test accuracy', 'train accuracy'], loc='upper left')
plt.xscale('log')
# subplot 3/3
plt.subplot(133)
gamma_0001 = cv_results[cv_results['param_gamma']==0.0001]
plt.plot(gamma_0001["param_C"], gamma_0001["mean_test_score"])
plt.plot(gamma_0001["param_C"], gamma_0001["mean_train_score"])
plt.xlabel('C')
plt.ylabel('Accuracy')
plt.title("Gamma=0.0001")
plt.ylim([0.80, 1])
plt.legend(['test accuracy', 'train accuracy'], loc='upper left')
plt.xscale('log')
# printing the optimal accuracy score and hyperparameters
best_score = model_cv.best_score_
best_hyperparams = model_cv.best_params_
print("The best test score is {0} corresponding to hyperparameters {1}".format(best_score, best_hyperparams))
```
### Building the model using the best parameters
```
model = SVC(C = 100,gamma=.001, kernel='rbf')
model.fit(X_train, y_train)
y_pred = model.predict(X_test)
# metrics
print("accuracy", metrics.accuracy_score(y_test, y_pred), "\n")
print(metrics.confusion_matrix(y_test, y_pred), "\n")
```
#### The accuracy has improved by 1%
```
corr=confusion_matrix(y_test, y_pred)
# Calculating precision
TP=0 #Initializing True positives
FP=0 #Initializing False positives
for i in range (corr.shape[0]):
TP=corr[i][i]
FP=corr.sum(axis=1)[i]-TP
print ("Precision for {}, is {ratio} ".format(i,ratio=TP/(TP+FP)))
# Calculating recall
TP=0 #Initializing True positives
FN=0 #Initializing False negatives
for i in range (corr.shape[0]):
TP=corr[i][i]
FN=corr.sum(axis=0)[i]-TP
print ("Recall for {}, is {ratio} ".format(i,ratio=TP/(TP+FN)))
```
### Conclusion : The Accuracy,Precision and Recall have increased after hyperparameter tuning
### Using the actual train data,test data as provided separately, and predicting the labels of test data
```
# Preparing the test data
test=pd.read_csv("test.csv")
print(test.shape)
X_test=test
X_test=scale(X_test)#Scaling
model = SVC(C = 100,gamma=.001, kernel='rbf')
model.fit(X_train, y_train)
y_pred = model.predict(X_test)
# adding the predicted labels as an additional column to the test data
test['label']=pd.Series(y_pred)
test.head()
submission=pd.DataFrame(test.label)
submission.insert(0, 'ImageID', range(1, 1 + len(submission)))
submission.head()
submission.to_csv('submission.csv',sep=',')
```
## Kaggle Accuracy Score : 93.8%
|
github_jupyter
|
import pandas as pd
import numpy as np
from sklearn.svm import SVC
from sklearn.model_selection import train_test_split
from sklearn import metrics
from sklearn.metrics import confusion_matrix
from sklearn.model_selection import KFold
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import GridSearchCV
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.preprocessing import scale
df=pd.read_csv("train.csv")
df.head()
df.shape
# Getting the 20 % of data
n=20
df=df.head(int(len(df.index)*(n/100)))
df.shape
# Checking for null values
df.isnull().sum().sort_values(ascending=False)
# splitting into X and y
X = df.drop("label", axis = 1)
y = df.label.values.astype(int)
# scaling the features
X_scaled = scale(X)
# train test split
X_train, X_test, y_train, y_test = train_test_split(X_scaled, y, test_size = 0.3, random_state = 4)
# using rbf kernel, C=1, default value of gamma
model = SVC(C = 1, kernel='rbf')
model.fit(X_train, y_train)
y_pred = model.predict(X_test)
# confusion matrix
confusion_matrix(y_true=y_test, y_pred=y_pred)
#Visualising the confusion matrix on the heatmap
plt.figure(figsize=(12,10))
corr=confusion_matrix(y_true=y_test, y_pred=y_pred)
sns.heatmap(corr,annot=True)
# accuracy
print("accuracy", metrics.accuracy_score(y_test, y_pred))
# Calculating precision
TP=0 #Initializing True positives
FP=0 #Initializing False positives
for i in range (corr.shape[0]):
TP=corr[i][i]
FP=corr.sum(axis=1)[i]-TP
print ("Precision for {}, is {ratio} ".format(i,ratio=TP/(TP+FP)))
# Calculating recall
TP=0 #Initializing True positives
FN=0 #Initializing False negatives
for i in range (corr.shape[0]):
TP=corr[i][i]
FN=corr.sum(axis=0)[i]-TP
print ("Recall for {}, is {ratio} ".format(i,ratio=TP/(TP+FN)))
# creating a KFold object with 5 splits
folds = KFold(n_splits = 5, shuffle = True, random_state = 4)
# specify range of hyperparameters
# Set the parameters by cross-validation
hyper_params = [ {'gamma': [1e-2, 1e-3, 1e-4],
'C': [1, 10, 100, 1000]}]
# specify model
model = SVC(kernel="rbf")
# set up GridSearchCV()
model_cv = GridSearchCV(estimator = model,
param_grid = hyper_params,
scoring= 'accuracy',
cv = folds,
verbose = 1,
return_train_score=True)
# fit the model
model_cv.fit(X_train, y_train)
# cv results
cv_results = pd.DataFrame(model_cv.cv_results_)
cv_results
# converting C to numeric type for plotting on x-axis
cv_results['param_C'] = cv_results['param_C'].astype('int')
# # plotting
plt.figure(figsize=(16,6))
# subplot 1/3
plt.subplot(131)
gamma_01 = cv_results[cv_results['param_gamma']==0.01]
plt.plot(gamma_01["param_C"], gamma_01["mean_test_score"])
plt.plot(gamma_01["param_C"], gamma_01["mean_train_score"])
plt.xlabel('C')
plt.ylabel('Accuracy')
plt.title("Gamma=0.01")
plt.ylim([0.80, 1])
plt.legend(['test accuracy', 'train accuracy'], loc='upper left')
plt.xscale('log')
# subplot 2/3
plt.subplot(132)
gamma_001 = cv_results[cv_results['param_gamma']==0.001]
plt.plot(gamma_001["param_C"], gamma_001["mean_test_score"])
plt.plot(gamma_001["param_C"], gamma_001["mean_train_score"])
plt.xlabel('C')
plt.ylabel('Accuracy')
plt.title("Gamma=0.001")
plt.ylim([0.80, 1])
plt.legend(['test accuracy', 'train accuracy'], loc='upper left')
plt.xscale('log')
# subplot 3/3
plt.subplot(133)
gamma_0001 = cv_results[cv_results['param_gamma']==0.0001]
plt.plot(gamma_0001["param_C"], gamma_0001["mean_test_score"])
plt.plot(gamma_0001["param_C"], gamma_0001["mean_train_score"])
plt.xlabel('C')
plt.ylabel('Accuracy')
plt.title("Gamma=0.0001")
plt.ylim([0.80, 1])
plt.legend(['test accuracy', 'train accuracy'], loc='upper left')
plt.xscale('log')
# printing the optimal accuracy score and hyperparameters
best_score = model_cv.best_score_
best_hyperparams = model_cv.best_params_
print("The best test score is {0} corresponding to hyperparameters {1}".format(best_score, best_hyperparams))
model = SVC(C = 100,gamma=.001, kernel='rbf')
model.fit(X_train, y_train)
y_pred = model.predict(X_test)
# metrics
print("accuracy", metrics.accuracy_score(y_test, y_pred), "\n")
print(metrics.confusion_matrix(y_test, y_pred), "\n")
corr=confusion_matrix(y_test, y_pred)
# Calculating precision
TP=0 #Initializing True positives
FP=0 #Initializing False positives
for i in range (corr.shape[0]):
TP=corr[i][i]
FP=corr.sum(axis=1)[i]-TP
print ("Precision for {}, is {ratio} ".format(i,ratio=TP/(TP+FP)))
# Calculating recall
TP=0 #Initializing True positives
FN=0 #Initializing False negatives
for i in range (corr.shape[0]):
TP=corr[i][i]
FN=corr.sum(axis=0)[i]-TP
print ("Recall for {}, is {ratio} ".format(i,ratio=TP/(TP+FN)))
# Preparing the test data
test=pd.read_csv("test.csv")
print(test.shape)
X_test=test
X_test=scale(X_test)#Scaling
model = SVC(C = 100,gamma=.001, kernel='rbf')
model.fit(X_train, y_train)
y_pred = model.predict(X_test)
# adding the predicted labels as an additional column to the test data
test['label']=pd.Series(y_pred)
test.head()
submission=pd.DataFrame(test.label)
submission.insert(0, 'ImageID', range(1, 1 + len(submission)))
submission.head()
submission.to_csv('submission.csv',sep=',')
| 0.667798 | 0.759292 |
<a href="https://colab.research.google.com/github/saptarshidatta96/Sentiment-Analysis/blob/main/Sentiment_Analysis_with_LSTM.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
```
import os
import random
import numpy as np
import pandas as pd
from scipy.sparse import csr_matrix
from tensorflow.python.keras.preprocessing import sequence
from tensorflow.python.keras.preprocessing import text
import tensorflow as tf
from tensorflow import keras
from keras import models
from keras import initializers
from keras import regularizers
from keras.layers import Dense, Embedding, LSTM, SpatialDropout1D
from keras.layers import CuDNNLSTM
from keras.layers import Dropout
from tensorflow.keras.optimizers import Adam
from tensorflow.keras.callbacks import EarlyStopping
from sklearn.metrics import accuracy_score, confusion_matrix, classification_report
random.seed(42)
from google.colab import drive
drive.mount('/content/gdrive')
!tar -xvf "/content/gdrive/MyDrive/aclImdb_v1.tar.gz" -C "/content/"
def load_dataset(dataset):
data = []
label = []
for item in os.listdir('/content/aclImdb/{}/'.format(dataset)):
if item == 'pos':
tweet_txt = os.path.join('/content/aclImdb/{}/'.format(dataset), item)
for tweets in os.listdir(tweet_txt):
if tweets.endswith('.txt'):
with open(os.path.join(tweet_txt, tweets)) as f:
data.append(f.read())
label.append(1)
elif item == 'neg':
tweet_txt = os.path.join('/content/aclImdb/{}/'.format(dataset), item)
for tweets in os.listdir(tweet_txt):
if tweets.endswith('.txt'):
with open(os.path.join(tweet_txt, tweets)) as f:
data.append(f.read())
label.append(0)
return data, label
train_data, train_label = load_dataset('train')
test_data, test_label = load_dataset('test')
def split_training_and_validation_sets(data, label, validation_split):
num_training_samples = int((1 - validation_split) * len(data))
return ((data[:num_training_samples], label[:num_training_samples]),
(data[num_training_samples:], label[num_training_samples:]))
(train_data, train_label), (valid_data, valid_label) = split_training_and_validation_sets(train_data, train_label, 0.1)
random.seed(42)
random.shuffle(train_data)
random.seed(42)
random.shuffle(train_label)
train_label = tf.convert_to_tensor(train_label, dtype=tf.float32)
valid_label = tf.convert_to_tensor(valid_label, dtype=tf.float32)
def sequence_vectorizer(train_data, valid_data):
# Create vocabulary with training texts.
tokenizer = text.Tokenizer(num_words=20000)
tokenizer.fit_on_texts(train_data)
# Vectorize training and validation texts.
x_train = tokenizer.texts_to_sequences(train_data)
x_val = tokenizer.texts_to_sequences(valid_data)
# Get max sequence length.
max_length = len(max(x_train, key=len))
if max_length > 500:
max_length = 500
# Fix sequence length to max value. Sequences shorter than the length are
# padded in the beginning and sequences longer are truncated
# at the beginning.
x_train = sequence.pad_sequences(x_train, maxlen=max_length)
x_val = sequence.pad_sequences(x_val, maxlen=max_length)
x_train = tf.convert_to_tensor(x_train, dtype=tf.float32)
x_val = tf.convert_to_tensor(x_val, dtype=tf.float32)
return x_train, x_val, tokenizer.word_index
x_train, x_val, word_index = sequence_vectorizer(train_data, valid_data)
def LSTM_Model():
model = models.Sequential()
model.add(Embedding(20000, 120, input_length=500))
model.add(SpatialDropout1D(0.4))
model.add(CuDNNLSTM(176, return_sequences=True))
model.add(Dropout(0.8))
model.add(CuDNNLSTM(32))
model.add(Dropout(0.8))
model.add(Dense(1,activation='sigmoid'))
return model
model = LSTM_Model()
model.summary()
model.compile(loss='binary_crossentropy', optimizer='adam', metrics = ['accuracy'])
callbacks = [EarlyStopping(monitor='val_loss', patience=2)]
model.fit(x_train, train_label,
epochs=20,
callbacks=callbacks,
validation_data=(x_val, valid_label),
verbose=2,
batch_size=512)
model.save('/content/gdrive/MyDrive/models/sentiment_analysis_LSTM_trained_model.h5',save_format= 'tf')
```
Load Model
```
loaded_model = keras.models.load_model('/content/gdrive/MyDrive/models/sentiment_analysis_LSTM_trained_model.h5')
x_test, _, _ = sequence_vectorizer(test_data, valid_data)
predictions = loaded_model.predict(x_test)
pred = [1 if a>0.5 else 0 for a in predictions]
print(pred)
print(test_label)
accuracy_score(pred, test_label)
print(classification_report(pred, test_label))
confusion_matrix(pred, test_label)
```
|
github_jupyter
|
import os
import random
import numpy as np
import pandas as pd
from scipy.sparse import csr_matrix
from tensorflow.python.keras.preprocessing import sequence
from tensorflow.python.keras.preprocessing import text
import tensorflow as tf
from tensorflow import keras
from keras import models
from keras import initializers
from keras import regularizers
from keras.layers import Dense, Embedding, LSTM, SpatialDropout1D
from keras.layers import CuDNNLSTM
from keras.layers import Dropout
from tensorflow.keras.optimizers import Adam
from tensorflow.keras.callbacks import EarlyStopping
from sklearn.metrics import accuracy_score, confusion_matrix, classification_report
random.seed(42)
from google.colab import drive
drive.mount('/content/gdrive')
!tar -xvf "/content/gdrive/MyDrive/aclImdb_v1.tar.gz" -C "/content/"
def load_dataset(dataset):
data = []
label = []
for item in os.listdir('/content/aclImdb/{}/'.format(dataset)):
if item == 'pos':
tweet_txt = os.path.join('/content/aclImdb/{}/'.format(dataset), item)
for tweets in os.listdir(tweet_txt):
if tweets.endswith('.txt'):
with open(os.path.join(tweet_txt, tweets)) as f:
data.append(f.read())
label.append(1)
elif item == 'neg':
tweet_txt = os.path.join('/content/aclImdb/{}/'.format(dataset), item)
for tweets in os.listdir(tweet_txt):
if tweets.endswith('.txt'):
with open(os.path.join(tweet_txt, tweets)) as f:
data.append(f.read())
label.append(0)
return data, label
train_data, train_label = load_dataset('train')
test_data, test_label = load_dataset('test')
def split_training_and_validation_sets(data, label, validation_split):
num_training_samples = int((1 - validation_split) * len(data))
return ((data[:num_training_samples], label[:num_training_samples]),
(data[num_training_samples:], label[num_training_samples:]))
(train_data, train_label), (valid_data, valid_label) = split_training_and_validation_sets(train_data, train_label, 0.1)
random.seed(42)
random.shuffle(train_data)
random.seed(42)
random.shuffle(train_label)
train_label = tf.convert_to_tensor(train_label, dtype=tf.float32)
valid_label = tf.convert_to_tensor(valid_label, dtype=tf.float32)
def sequence_vectorizer(train_data, valid_data):
# Create vocabulary with training texts.
tokenizer = text.Tokenizer(num_words=20000)
tokenizer.fit_on_texts(train_data)
# Vectorize training and validation texts.
x_train = tokenizer.texts_to_sequences(train_data)
x_val = tokenizer.texts_to_sequences(valid_data)
# Get max sequence length.
max_length = len(max(x_train, key=len))
if max_length > 500:
max_length = 500
# Fix sequence length to max value. Sequences shorter than the length are
# padded in the beginning and sequences longer are truncated
# at the beginning.
x_train = sequence.pad_sequences(x_train, maxlen=max_length)
x_val = sequence.pad_sequences(x_val, maxlen=max_length)
x_train = tf.convert_to_tensor(x_train, dtype=tf.float32)
x_val = tf.convert_to_tensor(x_val, dtype=tf.float32)
return x_train, x_val, tokenizer.word_index
x_train, x_val, word_index = sequence_vectorizer(train_data, valid_data)
def LSTM_Model():
model = models.Sequential()
model.add(Embedding(20000, 120, input_length=500))
model.add(SpatialDropout1D(0.4))
model.add(CuDNNLSTM(176, return_sequences=True))
model.add(Dropout(0.8))
model.add(CuDNNLSTM(32))
model.add(Dropout(0.8))
model.add(Dense(1,activation='sigmoid'))
return model
model = LSTM_Model()
model.summary()
model.compile(loss='binary_crossentropy', optimizer='adam', metrics = ['accuracy'])
callbacks = [EarlyStopping(monitor='val_loss', patience=2)]
model.fit(x_train, train_label,
epochs=20,
callbacks=callbacks,
validation_data=(x_val, valid_label),
verbose=2,
batch_size=512)
model.save('/content/gdrive/MyDrive/models/sentiment_analysis_LSTM_trained_model.h5',save_format= 'tf')
loaded_model = keras.models.load_model('/content/gdrive/MyDrive/models/sentiment_analysis_LSTM_trained_model.h5')
x_test, _, _ = sequence_vectorizer(test_data, valid_data)
predictions = loaded_model.predict(x_test)
pred = [1 if a>0.5 else 0 for a in predictions]
print(pred)
print(test_label)
accuracy_score(pred, test_label)
print(classification_report(pred, test_label))
confusion_matrix(pred, test_label)
| 0.629547 | 0.749683 |
# Introduction to Probability and Statistics
|
In this notebook, we will play around with some of the concepts we have previously discussed. Many concepts from probability and statistics are well-represented in major libraries for data processing in Python, such as `numpy` and `pandas`.
```
import numpy as np
import pandas as pd
import random
import matplotlib.pyplot as plt
```
## Random Variables and Distributions
Let's start with drawing a sample of 30 variables from a uniform disribution from 0 to 9. We will also compute mean and variance.
```
sample = [ random.randint(0,10) for _ in range(30) ]
print(f"Sample: {sample}")
print(f"Mean = {np.mean(sample)}")
print(f"Variance = {np.var(sample)}")
```
To visually estimate how many different values are there in the sample, we can plot the **histogram**:
```
plt.hist(sample)
plt.show()
```
## Analyzing Real Data
Mean and variance are very important when analyzing real-world data. Let's load the data about baseball players from [SOCR MLB Height/Weight Data](http://wiki.stat.ucla.edu/socr/index.php/SOCR_Data_MLB_HeightsWeights)
```
df = pd.read_csv("../../data/SOCR_MLB.tsv",sep='\t',header=None,names=['Name','Team','Role','Height','Weight','Age'])
df
```
> We are using a package called **Pandas** here for data analysis. We will talk more about Pandas and working with data in Python later in this course.
Let's compute average values for age, height and weight:
```
df[['Age','Height','Weight']].mean()
```
Now let's focus on height, and compute standard deviation and variance:
```
print(list(df['Height'])[:20])
mean = df['Height'].mean()
var = df['Height'].var()
std = df['Height'].std()
print(f"Mean = {mean}\nVariance = {var}\nStandard Deviation = {std}")
```
In addition to mean, it makes sense to look at median value and quartiles. They can be visualized using **box plot**:
```
plt.figure(figsize=(10,2))
plt.boxplot(df['Height'],vert=False,showmeans=True)
plt.grid(color='gray',linestyle='dotted')
plt.show()
```
We can also make box plots of subsets of our dataset, for example, grouped by player role.
```
df.boxplot(column='Height',by='Role')
plt.xticks(rotation='vertical')
plt.show()
```
> **Note**: This diagram suggests, that on average, height of first basemen is higher that height of second basemen. Later we will learn how we can test this hypothesis more formally, and how to demonstrate that our data is statistically significant to show that.
Age, height and weight are all continuous random variables. What do you think their distribution is? A good way to find out is to plot the histogram of values:
```
df['Weight'].hist(bins=15)
plt.suptitle('Weight distribution of MLB Players')
plt.xlabel('Weight')
plt.ylabel('Count')
plt.show()
```
## Normal Distribution
Let's create an artificial sample of weights that follows normal distribution with the same mean and variance as real data:
```
generated = np.random.normal(mean,std,1000)
generated[:20]
plt.hist(generated,bins=15)
plt.show()
plt.hist(np.random.normal(0,1,50000),bins=300)
plt.show()
```
Since most values in real life are normally distributed, it means we should not use uniform random number generator to generate sample data. Here is what happens if we try to generate weights with uniform distribution (generated by `np.random.rand`):
```
wrong_sample = np.random.rand(1000)*2*std+mean-std
plt.hist(wrong_sample)
plt.show()
```
## Confidence Intervals
Let's now calculate confidence intervals for the weights and heights of baseball players. We will use the code [from this stackoverflow discussion](https://stackoverflow.com/questions/15033511/compute-a-confidence-interval-from-sample-data):
```
import scipy.stats
def mean_confidence_interval(data, confidence=0.95):
a = 1.0 * np.array(data)
n = len(a)
m, se = np.mean(a), scipy.stats.sem(a)
h = se * scipy.stats.t.ppf((1 + confidence) / 2., n-1)
return m, h
for p in [0.85, 0.9, 0.95]:
m, h = mean_confidence_interval(df['Weight'].fillna(method='pad'),p)
print(f"p={p:.2f}, mean = {m:.2f}±{h:.2f}")
```
## Hypothesis Testing
Let's explore different roles in our baseball players dataset:
```
df.groupby('Role').agg({ 'Height' : 'mean', 'Weight' : 'mean', 'Age' : 'count'}).rename(columns={ 'Age' : 'Count'})
```
Let's test the hypothesis that First Basemen are higher then Second Basemen. The simplest way to do it is to test the confidence intervals:
```
for p in [0.85,0.9,0.95]:
m1, h1 = mean_confidence_interval(df.loc[df['Role']=='First_Baseman',['Height']],p)
m2, h2 = mean_confidence_interval(df.loc[df['Role']=='Second_Baseman',['Height']],p)
print(f'Conf={p:.2f}, 1st basemen height: {m1-h1[0]:.2f}..{m1+h1[0]:.2f}, 2nd basemen height: {m2-h2[0]:.2f}..{m2+h2[0]:.2f}')
```
We can see that intervals do not overlap.
More statistically correct way to prove the hypothesis is to use **Student t-test**:
```
from scipy.stats import ttest_ind
tval, pval = ttest_ind(df.loc[df['Role']=='First_Baseman',['Height']], df.loc[df['Role']=='Second_Baseman',['Height']],equal_var=False)
print(f"T-value = {tval[0]:.2f}\nP-value: {pval[0]}")
```
Two values returned by the `ttest_ind` functions are:
* p-value can be considered as the probability of two distributions having the same mean. In our case, it is very low, meaning that there is strong evidence supporting that first basemen are taller
* t-value is the intermediate value of normalized mean difference that is used in t-test, and it is compared against threshold value for a given confidence value
## Simulating Normal Distribution with Central Limit Theorem
Pseudo-random generator in Python is designed to give us uniform distribution. If we want to create a generator for normal distribution, we can use central limit theorem. To get a normally distributed value we will just compute a mean of a uniform-generated sample.
```
def normal_random(sample_size=100):
sample = [random.uniform(0,1) for _ in range(sample_size) ]
return sum(sample)/sample_size
sample = [normal_random() for _ in range(100)]
plt.hist(sample)
plt.show()
```
## Correlation and Evil Baseball Corp
Correlation allows us to find inner connection between data sequences. In our toy example, let's pretend there is an evil baseball corporation that pays it's players according to their height - the taller the player is, the more money he/she gets. Suppose there is a base salary of $1000, and an additional bonus from $0 to $100, depending on height. We will take the real players from MLB, and compute their imaginary salaries:
```
heights = df['Height']
salaries = 1000+(heights-heights.min())/(heights.max()-heights.mean())*100
print(list(zip(heights,salaries))[:10])
```
Let's now compute covariance and correlation of those sequences. `np.cov` will give us so-called **covariance matrix**, which is an extension of covariance to multiple variables. The element $M_{ij}$ of the covariance matrix $M$ is a correlation between input variables $X_i$ and $X_j$, and diagonal values $M_{ii}$ is the variance of $X_{i}$. Similarly, `np.corrcoef` will give us **correlation matrix**.
```
print(f"Covariance matrix:\n{np.cov(heights,salaries)}")
print(f"Covariance = {np.cov(heights,salaries)[0,1]}")
print(f"Correlation = {np.corrcoef(heights,salaries)[0,1]}")
```
Correlation equal to 1 means that there is a strong **linear relation** between two variables. We can visually see the linear relation by plotting one value against the other:
```
plt.scatter(heights,salaries)
plt.show()
```
Let's see what happens if the relation is not linear. Suppose that our corporation decided to hide the obvious linear dependency between heights and salaries, and introduced some non-linearity into the formula, such as `sin`:
```
salaries = 1000+np.sin((heights-heights.min())/(heights.max()-heights.mean()))*100
print(f"Correlation = {np.corrcoef(heights,salaries)[0,1]}")
```
In this case, the correlation is slightly smaller, but it is still quite high. Now, to make the relation even less obvious, we might want to add some extra randomness by adding some random variable to the salary. Let's see what happens:
```
salaries = 1000+np.sin((heights-heights.min())/(heights.max()-heights.mean()))*100+np.random.random(size=len(heights))*20-10
print(f"Correlation = {np.corrcoef(heights,salaries)[0,1]}")
plt.scatter(heights, salaries)
plt.show()
```
> Can you guess why the dots line up into vertical lines like this?
We have observed the correlation between artificially engineered concept like salary and the observed variable *height*. Let's also see if the two observed variables, such as height and weight, also correlate:
```
np.corrcoef(df['Height'],df['Weight'])
```
Unfortunately, we did not get any results - only some strange `nan` values. This is due to the fact that some of the values in our series are undefined, represented as `nan`, which causes the result of the operation to be undefined as well. By looking at the matrix we can see that `Weight` is problematic column, because self-correlation between `Height` values has been computed.
> This example shows the importance of **data preparation** and **cleaning**. Without proper data we cannot compute anything.
Let's use `fillna` method to fill the missing values, and compute the correlation:
```
np.corrcoef(df['Height'],df['Weight'].fillna(method='pad'))
```
The is indeed a correlation, but not such a strong one as in our artificial example. Indeed, if we look at the scatter plot of one value against the other, the relation would be much less obvious:
```
plt.scatter(df['Height'],df['Weight'])
plt.xlabel('Height')
plt.ylabel('Weight')
plt.show()
```
## Conclusion
In this notebook, we have learnt how to perform basic operations on data to compute statistical functions. We now know how to use sound apparatus of math and statistics in order to prove some hypotheses, and how to compute confidence intervals for random variable given data sample.
|
github_jupyter
|
import numpy as np
import pandas as pd
import random
import matplotlib.pyplot as plt
sample = [ random.randint(0,10) for _ in range(30) ]
print(f"Sample: {sample}")
print(f"Mean = {np.mean(sample)}")
print(f"Variance = {np.var(sample)}")
plt.hist(sample)
plt.show()
df = pd.read_csv("../../data/SOCR_MLB.tsv",sep='\t',header=None,names=['Name','Team','Role','Height','Weight','Age'])
df
df[['Age','Height','Weight']].mean()
print(list(df['Height'])[:20])
mean = df['Height'].mean()
var = df['Height'].var()
std = df['Height'].std()
print(f"Mean = {mean}\nVariance = {var}\nStandard Deviation = {std}")
plt.figure(figsize=(10,2))
plt.boxplot(df['Height'],vert=False,showmeans=True)
plt.grid(color='gray',linestyle='dotted')
plt.show()
df.boxplot(column='Height',by='Role')
plt.xticks(rotation='vertical')
plt.show()
df['Weight'].hist(bins=15)
plt.suptitle('Weight distribution of MLB Players')
plt.xlabel('Weight')
plt.ylabel('Count')
plt.show()
generated = np.random.normal(mean,std,1000)
generated[:20]
plt.hist(generated,bins=15)
plt.show()
plt.hist(np.random.normal(0,1,50000),bins=300)
plt.show()
wrong_sample = np.random.rand(1000)*2*std+mean-std
plt.hist(wrong_sample)
plt.show()
import scipy.stats
def mean_confidence_interval(data, confidence=0.95):
a = 1.0 * np.array(data)
n = len(a)
m, se = np.mean(a), scipy.stats.sem(a)
h = se * scipy.stats.t.ppf((1 + confidence) / 2., n-1)
return m, h
for p in [0.85, 0.9, 0.95]:
m, h = mean_confidence_interval(df['Weight'].fillna(method='pad'),p)
print(f"p={p:.2f}, mean = {m:.2f}±{h:.2f}")
df.groupby('Role').agg({ 'Height' : 'mean', 'Weight' : 'mean', 'Age' : 'count'}).rename(columns={ 'Age' : 'Count'})
for p in [0.85,0.9,0.95]:
m1, h1 = mean_confidence_interval(df.loc[df['Role']=='First_Baseman',['Height']],p)
m2, h2 = mean_confidence_interval(df.loc[df['Role']=='Second_Baseman',['Height']],p)
print(f'Conf={p:.2f}, 1st basemen height: {m1-h1[0]:.2f}..{m1+h1[0]:.2f}, 2nd basemen height: {m2-h2[0]:.2f}..{m2+h2[0]:.2f}')
from scipy.stats import ttest_ind
tval, pval = ttest_ind(df.loc[df['Role']=='First_Baseman',['Height']], df.loc[df['Role']=='Second_Baseman',['Height']],equal_var=False)
print(f"T-value = {tval[0]:.2f}\nP-value: {pval[0]}")
def normal_random(sample_size=100):
sample = [random.uniform(0,1) for _ in range(sample_size) ]
return sum(sample)/sample_size
sample = [normal_random() for _ in range(100)]
plt.hist(sample)
plt.show()
heights = df['Height']
salaries = 1000+(heights-heights.min())/(heights.max()-heights.mean())*100
print(list(zip(heights,salaries))[:10])
print(f"Covariance matrix:\n{np.cov(heights,salaries)}")
print(f"Covariance = {np.cov(heights,salaries)[0,1]}")
print(f"Correlation = {np.corrcoef(heights,salaries)[0,1]}")
plt.scatter(heights,salaries)
plt.show()
salaries = 1000+np.sin((heights-heights.min())/(heights.max()-heights.mean()))*100
print(f"Correlation = {np.corrcoef(heights,salaries)[0,1]}")
salaries = 1000+np.sin((heights-heights.min())/(heights.max()-heights.mean()))*100+np.random.random(size=len(heights))*20-10
print(f"Correlation = {np.corrcoef(heights,salaries)[0,1]}")
plt.scatter(heights, salaries)
plt.show()
np.corrcoef(df['Height'],df['Weight'])
np.corrcoef(df['Height'],df['Weight'].fillna(method='pad'))
plt.scatter(df['Height'],df['Weight'])
plt.xlabel('Height')
plt.ylabel('Weight')
plt.show()
| 0.460289 | 0.989024 |
# Credit
Implemente un programa que determine si un número de tarjeta de crédito proporcionado es válido según el algoritmo de Luhn.
<code>$ python credit.py
Number: 378282246310005
AMEX</code>
Una tarjeta de crédito (o débito), por supuesto, es una tarjeta de plástico con la que puede pagar bienes y servicios. Impreso en esa tarjeta hay un número que también está almacenado en una base de datos en algún lugar, de modo que cuando su tarjeta se usa para comprar algo, el acreedor sabe a quién facturar. Hay muchas personas con tarjetas de crédito en este mundo, por lo que esos números son bastante largos: American Express usa números de 15 dígitos, MasterCard usa números de 16 dígitos y Visa usa números de 13 y 16 dígitos. Y esos dígitos son números decimales (0 a 9), no binarios, lo que significa, por ejemplo, que American Express podría imprimir hasta 10 ^ 15 = 1,000,000,000,000,000 tarjetas únicas. (Eso es, um, un billón).
En realidad, eso es un poco exagerado, porque los números de las tarjetas de crédito en realidad tienen cierta estructura. Todos los números de American Express comienzan con 34 o 37; la mayoría de los números de MasterCard comienzan con 51, 52, 53, 54 o 55; y todos los números de Visa comienzan con 4. Pero los números de tarjetas de crédito también tienen una "suma de verificación" incorporada, una relación matemática entre al menos un número y otros. Esa suma de comprobación permite a las computadoras (o humanos a los que les gustan las matemáticas) detectar errores tipográficos (por ejemplo, transposiciones), si no números fraudulentos, sin tener que consultar una base de datos, lo que puede ser lento. Por supuesto, un matemático deshonesto podría sin duda crear un número falso que, sin embargo, respete la restricción matemática, por lo que una búsqueda en la base de datos aún es necesaria para verificaciones más rigurosas.
## Especificaciones
- En <code>credit.py</code> escribir un programa que solicita al usuario un número de tarjeta de crédito y luego informes (a través de <code>print</code>) si se trata de una válida American Express, MasterCard, o número de tarjeta Visa.
- Para que podamos automatizar algunas pruebas de su código, le pedimos que la última línea de salida de su programa sea <code>AMEX\n</code>o <code>MASTERCARD\n</code> o <code>VISA\n</code> o <code>INVALID\n</code> , nada más, nada menos.
- Para simplificar, puede suponer que la entrada del usuario será completamente numérica (es decir, sin guiones, como podría estar impreso en una tarjeta real).
## Uso
Su programa debería comportarse según el ejemplo siguiente.
<code>$ python credit.py
Number: 378282246310005
AMEX</code>
## Algoritmo de Luhn
Entonces, ¿cuál es la fórmula secreta? Bueno, la mayoría de las tarjetas utilizan un algoritmo inventado por Hans Peter Luhn de IBM. De acuerdo con el algoritmo de Luhn, puede determinar si un número de tarjeta de crédito es (sintácticamente) válido de la siguiente manera:
1. Multiplica cada dos dígitos por 2, comenzando con el penúltimo dígito del número y luego suma los dígitos de esos productos.
2. Suma la suma a la suma de los dígitos que no se multiplicaron por 2.
3. Si el último dígito del total es 0 (o, dicho de manera más formal, si el módulo total 10 es congruente con 0), ¡el número es válido!
Eso es un poco confuso, así que probemos un ejemplo con la visa de David: 4003600000000014.
1. Por el bien de la discusión, primero subrayemos cada dos dígitos, comenzando con el penúltimo dígito del número:
**4** 0 **0** 3 **6** 0 **0** 0 **0** 0 **0** 0 **0** 0 **1** 4
Bien, multipliquemos cada uno de los dígitos subrayados por 2:
1 • 2 + 0 • 2 + 0 • 2 + 0 • 2 + 0 • 2 + 6 • 2 + 0 • 2 + 4 • 2
Eso nos da:
2 + 0 + 0 + 0 + 0 + 12 + 0 + 8
2. Ahora agreguemos los dígitos de esos productos (es decir, no los productos en sí) juntos:
2 + 0 + 0 + 0 + 0 + 1 + 2 + 0 + 8 = 13
Ahora agreguemos esa suma (13) a la suma de los dígitos que no fueron multiplicados por 2 (comenzando desde el final):
13 + 4 + 0 + 0 + 0 + 0 + 0 + 3 + 0 = 20
3. Sí, el último dígito de esa suma (20) es un 0, ¡así que la tarjeta de David es legítima!
Por lo tanto, validar los números de tarjetas de crédito no es difícil, pero se vuelve un poco tedioso a mano. Escribamos un programa.
## Pruebas
- Ejecute su programa como python <code>credit.py</code>y espere a que se le solicite la entrada. Escribe <code>378282246310005</code> y presiona enter. Su programa debería generar <code>AMEX</code>.
- Ejecute su programa como python <code>credit.py</code> y espere a que se le solicite la entrada. Escribe <code>371449635398431</code> y presiona enter. Su programa debería generar <code>AMEX</code>.
- Ejecute su programa como python <code>credit.py</code> y espere a que se le solicite la entrada. Escribe <code>5555555555554444</code> y presiona enter. Su programa debería generar <code>MASTERCARD</code>.
- Ejecute su programa como python <code>credit.py</code> y espere a que se le solicite la entrada. Escribe <code>5105105105105100</code> y presiona enter. Su programa debería generar <code>MASTERCARD</code>.
- Ejecute su programa como python <code>credit.py</code> y espere a que se le solicite la entrada. Escribe <code>4111111111111111</code> y presiona enter. Su programa debería generar <code>VISA</code>.
- Ejecute su programa como python <code>credit.py</code> y espere a que se le solicite la entrada. Escribe <code>4012888888881881</code> y presiona enter. Su programa debería generar <code>VISA</code>.
- Ejecute su programa como python <code>credit.py</code> y espere a que se le solicite la entrada. Escribe <code>1234567890</code> y presiona enter. Su programa debería generar <code>INVALID</code> .
Aqui más códigos de tarjetas de paypal para validar <a href='https://developer.paypal.com/docs/payflow/payflow-pro/payflow-pro-testing/#credit-card-numbers-for-testing'>link</a>
```
def s_digitos(num):
if (num==(num%10)):
return num
else:
return (num%10)+ s_digitos(int(num/10))
Ntarjeta= ""
def v_tarjeta():
Reverse_Num_Tarjeta = Ntarjeta[::-1]
suma=0
for i in range(1,len(Reverse_Num_Tarjeta),2):
suma+=s_digitos(int(Reverse_Num_Tarjeta[i])*2)
for i in range(0,len(Reverse_Num_Tarjeta),2):
suma+=int(Reverse_Num_Tarjeta[i])
if (suma%10)==0:
return True
else:
return False
def t_tarjeta():
if Ntarjeta.startswith("34") or Ntarjeta.startswith("37"):
return "AMEX"
elif Ntarjeta.startswith("51") or Ntarjeta.startswith("52") or Ntarjeta.startswith("53") or Ntarjeta.startswith("54") or Ntarjeta.startswith("55"):
return "MASTERCARD"
elif Ntarjeta.startswith("4"):
return "VISA"
else:
return "Numero invalido"
if __name__ == "__main__":
Ntarjeta=input("Ingrese número de tarjeta: ")
if v_tarjeta():
print(t_tarjeta())
else:
print("Numero invalido")
def s_digitos(num):
if (num==(num%10)):
return num
else:
return (num%10)+ s_digitos(int(num/10))
Ntarjeta= ""
def v_tarjeta():
Reverse_Num_Tarjeta = Ntarjeta[::-1]
suma=0
for i in range(1,len(Reverse_Num_Tarjeta),2):
suma+=s_digitos(int(Reverse_Num_Tarjeta[i])*2)
for i in range(0,len(Reverse_Num_Tarjeta),2):
suma+=int(Reverse_Num_Tarjeta[i])
if (suma%10)==0:
return True
else:
return False
def t_tarjeta():
if Ntarjeta.startswith("34") or Ntarjeta.startswith("37"):
return "AMEX"
elif Ntarjeta.startswith("51") or Ntarjeta.startswith("52") or Ntarjeta.startswith("53") or Ntarjeta.startswith("54") or Ntarjeta.startswith("55"):
return "MASTERCARD"
elif Ntarjeta.startswith("4"):
return "VISA"
else:
return "Numero invalido"
if __name__ == "__main__":
Ntarjeta=input("Ingrese número de tarjeta: ")
if v_tarjeta():
print(t_tarjeta())
else:
print("Numero invalido")
```
|
github_jupyter
|
def s_digitos(num):
if (num==(num%10)):
return num
else:
return (num%10)+ s_digitos(int(num/10))
Ntarjeta= ""
def v_tarjeta():
Reverse_Num_Tarjeta = Ntarjeta[::-1]
suma=0
for i in range(1,len(Reverse_Num_Tarjeta),2):
suma+=s_digitos(int(Reverse_Num_Tarjeta[i])*2)
for i in range(0,len(Reverse_Num_Tarjeta),2):
suma+=int(Reverse_Num_Tarjeta[i])
if (suma%10)==0:
return True
else:
return False
def t_tarjeta():
if Ntarjeta.startswith("34") or Ntarjeta.startswith("37"):
return "AMEX"
elif Ntarjeta.startswith("51") or Ntarjeta.startswith("52") or Ntarjeta.startswith("53") or Ntarjeta.startswith("54") or Ntarjeta.startswith("55"):
return "MASTERCARD"
elif Ntarjeta.startswith("4"):
return "VISA"
else:
return "Numero invalido"
if __name__ == "__main__":
Ntarjeta=input("Ingrese número de tarjeta: ")
if v_tarjeta():
print(t_tarjeta())
else:
print("Numero invalido")
def s_digitos(num):
if (num==(num%10)):
return num
else:
return (num%10)+ s_digitos(int(num/10))
Ntarjeta= ""
def v_tarjeta():
Reverse_Num_Tarjeta = Ntarjeta[::-1]
suma=0
for i in range(1,len(Reverse_Num_Tarjeta),2):
suma+=s_digitos(int(Reverse_Num_Tarjeta[i])*2)
for i in range(0,len(Reverse_Num_Tarjeta),2):
suma+=int(Reverse_Num_Tarjeta[i])
if (suma%10)==0:
return True
else:
return False
def t_tarjeta():
if Ntarjeta.startswith("34") or Ntarjeta.startswith("37"):
return "AMEX"
elif Ntarjeta.startswith("51") or Ntarjeta.startswith("52") or Ntarjeta.startswith("53") or Ntarjeta.startswith("54") or Ntarjeta.startswith("55"):
return "MASTERCARD"
elif Ntarjeta.startswith("4"):
return "VISA"
else:
return "Numero invalido"
if __name__ == "__main__":
Ntarjeta=input("Ingrese número de tarjeta: ")
if v_tarjeta():
print(t_tarjeta())
else:
print("Numero invalido")
| 0.074787 | 0.795817 |
# Instrumental Noise in _Kepler_ and _K2_ #4: Electronic Noise
## Learning Goals
By the end of this tutorial, you will be able to:
- Explain electronic crosstalk and how it manifests in _Kepler_ data.
- Identify rolling bands in _Kepler_ data and compare their effect to other sources of noise.
- Understand the short- and long-term consequences of cosmic rays on the _Kepler_ detector.
## Introduction
This tutorial is the fourth part of a series on identifying instrumental and systematic sources of noise in _Kepler_ and _K2_ data. The first three tutorials in this series are suggested (but not necessary) reading for working through this one. Assumed knowledge for this tutorial is a working familiarity with _Kepler_ light curve files, target pixel files, and their associated metadata.
## Imports
We'll use **[Lightkurve](https://docs.lightkurve.org/)** for downloading and handling _Kepler_ data throughout this tutorial. We'll also use **[NumPy](https://numpy.org/)** to handle arrays for aperture masks, and **[Matplotlib](https://matplotlib.org/)** to help with some plotting.
```
import lightkurve as lk
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
```
---
## 1. Background
The _Kepler_ space telescope operated as a _photometer_: using charge-coupled devices (CCDs), _Kepler_ collected photons and processed these as flux measurements. The detector array consisted of 25 modules, and 94.6 million pixels. With an instrument of this size, electronic noise issues are inevitable. In this tutorial, we'll look at three major sources of electronic noise that occurred throughout both the _Kepler_ and _K2_ missions.
## 2. Crosstalk
Crosstalk refers to the phenomenon where an electronic signal from one channel of a device is transferred to another channel and causes noise. In _Kepler_ and _K2_ data, this often manifests as signal from one (usually bright) target appearing elsewhere on the detector. This is known as "video crosstalk," and only has a significant effect when it occurs on the same module, due to the shielding between modules ([_Kepler_ Instrument Handbook](https://archive.stsci.edu/files/live/sites/mast/files/home/missions-and-data/kepler/_documents/KSCI-19033-002-instrument-hb.pdf) Section 6.3). _Kepler_ crosstalk can also occur between the CCD modules and the Fine Guidance Sensors (FGS), and between either of these and the onboard clock. However, these kinds of crosstalk are well understood and corrected by the pipeline before data delivery. If you are interested in reading more about these types of crosstalk, they're covered in Sections 6.2 and 6.4 of the [_Kepler_ Instrument Handbook](https://archive.stsci.edu/files/live/sites/mast/files/home/missions-and-data/kepler/_documents/KSCI-19033-002-instrument-hb.pdf).
To understand what video crosstalk looks like, let's take an example from the _K2_ mission, as documented [here](https://github.com/KeplerGO/lightkurve/issues/160). UGC 7394 (EPIC 200084891) is a galaxy that was observed during _K2_ Campaign 10. Campaign 10 was delivered in two parts, as observation began with a pointing error of 3.5 pixels ([_K2_ Data Release Notes 15](https://archive.stsci.edu/missions/k2/doc/drn/KSCI-19131-001_K2-DRN15_C10.pdf), Section 2.1), so when we download our data using Lightkurve we have to specify which file we want. Let's begin with Campaign 10a:
```
tpf_ct = lk.search_targetpixelfile('EPIC 200084891', campaign=10)[0].download()
tpf_ct.plot();
```
UGC 7394 is very clear in this image, but there's also a column of what looks like saturation bleed down the middle. In fact, this is video crosstalk from a nearby bright star. The separation between these two sources becomes even more clear when we check Campaign 10b, which used a slightly different pointing:
```
tpf_ct = lk.search_targetpixelfile('EPIC 200084891', campaign=10)[1].download()
tpf_ct.plot();
```
You can also check for yourself that the crosstalk source is unrelated to the galaxy by using Lightkurve's [`interact()`](https://docs.lightkurve.org/api/lightkurve.targetpixelfile.KeplerTargetPixelFile.html#lightkurve.targetpixelfile.KeplerTargetPixelFile.interact) function offline and noting that the spreads of the two sources on the detector differ independently.
## 3. Rolling Bands
The _Kepler_ detector electronics have a high-frequency noise feature due to circuit self-resonance in the GHz range, which is aliased to lower frequencies by the pixel sampling rate for detector readout. These aliases lead to changing temperatures and pixel sensitivities on the focal plane, which appear as horizontal stripes in motion across on the detector. For a more technical exploration of this phenomenon, see Section 6.7 of the [_Kepler_ Instrument Handbook](https://archive.stsci.edu/files/live/sites/mast/files/home/missions-and-data/kepler/_documents/KSCI-19033-002-instrument-hb.pdf).
Let's have a look at an example of rolling bands in _K2_ data (as documented [here](https://github.com/KeplerGO/lightkurve/issues/160)). This star, EPIC 211741417, has a rolling band passing across its aperture, beginning around 3306 BKJD (Barycentric _Kepler_ Julian Date) days. First we'll download some data, and see if the rolling bands are visible in a cadence taken from before the rolling band passes (left), and after it passes (right). We'll also make sure the images have the same color scale, so we can make a fair comparison.
```
tpf_rbs = lk.search_targetpixelfile('EPIC 211741417', campaign=16).download()
fig, ax = plt.subplots(1,2, figsize=(15,6))
tpf_rbs.plot(ax=ax[0], cadenceno=156405, vmin=0, vmax=600)
tpf_rbs.plot(ax=ax[1], cadenceno=156556, vmin=0, vmax=600);
```
There's nothing obvious here; the background is a little darker during the rolling band event, but not noticeably so on this scale. To get a better look at this, you can use Lightkurve's [`interact()`](https://docs.lightkurve.org/tutorials/04-interact-with-lightcurves-and-tpf.html) function in an offline notebook. However, it's also possible to observe the rolling band by looking at some light curves.
We'll extract two single-pixel light curves for two pixels in this target pixel file (TPF): the brightest pixel where the target lies, and the bottom left pixel as representative of the background flux. First, we'll make an aperture mask for each; you can read more about this process in the custom aperture photometry tutorial.
```
target_mask = np.zeros((tpf_rbs[0].shape[1:]), dtype='bool')
target_mask[2,3] = True
background_mask = np.zeros((tpf_rbs[0].shape[1:]), dtype='bool')
background_mask[0,0] = True
```
Now let's look at the bright pixel and highlight the region where we expect to see the effects of the rolling band:
```
ax = tpf_rbs.to_lightcurve(aperture_mask=target_mask).plot()
ax.set_ylim(360, 960)
ax.fill_betweenx(ax.get_ylim(), 3306, 3311, facecolor='r', alpha=0.3)
```
This light curve shows the _K2_ six-hour pointing drift, and an overall trend due to temperature changes on the detector. There is no noticeable change during the rolling band crossing. This tells us that this star is bright enough that the effect of the rolling band is negligible in comparison.
Now let's look at the background pixel light curve:
```
ax = tpf_rbs.to_lightcurve(aperture_mask=background_mask).remove_outliers().plot()
ax.set_ylim(-20, 40)
ax.fill_betweenx(ax.get_ylim(), 3306, 3311, facecolor='r', alpha=0.3);
```
Sure enough, we can see a dip in background flux as the rolling band passes over the TPF. Note the scale of this dip; this low amplitude confirms what we saw above, that rolling bands are negligible in amplitude compared to target flux, six-hour drift, and temperature-induced flux variations. Nevertheless, it's important to be aware of rolling bands in your data when searching for noise sources and working on background modeling.
Rolling bands are identified by quality flags 18 and 19 in light curve and TPF metadata ([MAST Kepler Archive Manual](https://archive.stsci.edu/files/live/sites/mast/files/home/missions-and-data/k2/_documents/MAST_Kepler_Archive_Manual_2020.pdf), Table 2-3), but the automated flags are incomplete and do not detect all rolling bands. For further reading, there is a [Lightkurve tutorial](https://docs.lightkurve.org/tutorials/04-identify-rolling-band.html) on checking for rolling bands in _Kepler_ data.
## 4. Cosmic Rays and Sudden Pixel Sensitivity Dropout
In the first tutorial in this series, we covered cosmic rays as single-cadence quality events. We saw that when a cosmic ray hits a pixel on the detector, it can momentarily increase that pixel's flux reading by injecting noise in between readouts. However, there are multiple other consequences of cosmic rays on the detector that occurred over the course of the _Kepler_ and _K2_ missions.
Sudden Pixel Sensitivity Dropout (SPSD) occurs when a cosmic ray _reduces_ the sensitivity of a group of pixels where it lands. SPSD events are categorized as medium-term and long-term (or permanent) damage. In medium-term events, the effected pixels will gradually return to their pre-SPSD sensitivity. Long-term damage would require [annealing](https://en.wikipedia.org/wiki/Annealing_(metallurgy)) to fix, which could not be done on the _Kepler_ telescope, and as such is materially equivalent to permanent damage. Pixels affected by long-term or permanent damage retained their decreased sensitivity throughout the mission. For further reading, see the [_Kepler_ Data Characteristics Handbook](https://archive.stsci.edu/files/live/sites/mast/files/home/missions-and-data/kepler/_documents/Data_Characteristics.pdf), Section 5.9.
To observe an SPSD event, let's download some data first:
```
cr = lk.search_lightcurve('KIC 7461601', quarter=6).download(quality_bitmask=0)
```
Recalling from the first tutorial in this series that cosmic ray events are flagged in the `QUALITY` column of light curve and TPF metadata, let's look at the particular quality flag we're interested in:
```
lk.KeplerQualityFlags.decode(1152)
```
Now let's plot the simple aperture photometry (SAP) flux of the star we downloaded, with a dashed line where we expect to see a SPSD event:
```
ax = cr.plot(column='sap_flux')
ymin, ymax = ax.get_ylim()
ax.axvline(cr.time.value[cr.quality.value==1152][0], c='r', ls='--');
```
In the above image, we can see that the flux level drops abruptly around 547 BKJD days. This is due to a cosmic ray hitting the optimal aperture containing pixels used for photometry, and causing a sudden decrease in pixel sensitivity.
This effect is corrected by the presearch data conditioning (PDC) pipeline:
```
cr.plot();
```
## About this Notebook
**Author:** [Isabel Colman](http://orcid.org/0000-0001-8196-516X) (`isabel.colman@sydney.edu.au`)
**Updated on:** 2020-09-29
## Citing Lightkurve and Astropy
If you use `lightkurve` or `astropy` for published research, please cite the authors. Click the buttons below to copy BibTeX entries to your clipboard.
```
lk.show_citation_instructions()
```
<img style="float: right;" src="https://raw.githubusercontent.com/spacetelescope/notebooks/master/assets/stsci_pri_combo_mark_horizonal_white_bkgd.png" alt="Space Telescope Logo" width="200px"/>
|
github_jupyter
|
import lightkurve as lk
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
tpf_ct = lk.search_targetpixelfile('EPIC 200084891', campaign=10)[0].download()
tpf_ct.plot();
tpf_ct = lk.search_targetpixelfile('EPIC 200084891', campaign=10)[1].download()
tpf_ct.plot();
tpf_rbs = lk.search_targetpixelfile('EPIC 211741417', campaign=16).download()
fig, ax = plt.subplots(1,2, figsize=(15,6))
tpf_rbs.plot(ax=ax[0], cadenceno=156405, vmin=0, vmax=600)
tpf_rbs.plot(ax=ax[1], cadenceno=156556, vmin=0, vmax=600);
target_mask = np.zeros((tpf_rbs[0].shape[1:]), dtype='bool')
target_mask[2,3] = True
background_mask = np.zeros((tpf_rbs[0].shape[1:]), dtype='bool')
background_mask[0,0] = True
ax = tpf_rbs.to_lightcurve(aperture_mask=target_mask).plot()
ax.set_ylim(360, 960)
ax.fill_betweenx(ax.get_ylim(), 3306, 3311, facecolor='r', alpha=0.3)
ax = tpf_rbs.to_lightcurve(aperture_mask=background_mask).remove_outliers().plot()
ax.set_ylim(-20, 40)
ax.fill_betweenx(ax.get_ylim(), 3306, 3311, facecolor='r', alpha=0.3);
cr = lk.search_lightcurve('KIC 7461601', quarter=6).download(quality_bitmask=0)
lk.KeplerQualityFlags.decode(1152)
ax = cr.plot(column='sap_flux')
ymin, ymax = ax.get_ylim()
ax.axvline(cr.time.value[cr.quality.value==1152][0], c='r', ls='--');
cr.plot();
lk.show_citation_instructions()
| 0.369088 | 0.990441 |
# Creating interactive dashboards
```
import pandas as pd
import holoviews as hv
from bokeh.sampledata import stocks
from holoviews.operation.timeseries import rolling, rolling_outlier_std
hv.extension('bokeh')
```
In the [Data Processing Pipelines section](./14-Data_Pipelines.ipynb) we discovered how to declare a ``DynamicMap`` and control multiple processing steps with the use of custom streams as described in the [Responding to Events](./12-Responding_to_Events.ipynb) guide. Here we will use the same example exploring a dataset of stock timeseries and build a small dashboard using the [Panel](https://panel.pyviz.org) library, which allows us to declare easily declare custom widgets and link them to our streams. We will begin by once again declaring our function that loads the stock data:
```
def load_symbol(symbol, variable='adj_close', **kwargs):
df = pd.DataFrame(getattr(stocks, symbol))
df['date'] = df.date.astype('datetime64[ns]')
return hv.Curve(df, ('date', 'Date'), variable)
stock_symbols = ['AAPL', 'IBM', 'FB', 'GOOG', 'MSFT']
dmap = hv.DynamicMap(load_symbol, kdims='Symbol').redim.values(Symbol=stock_symbols)
dmap.opts(framewise=True)
```
## Building dashboards
Controlling stream events manually from the Python prompt can be a bit cumbersome. However since you can now trigger events from Python we can easily bind any Python based widget framework to the stream. HoloViews itself is based on param and param has various UI toolkits that accompany it and allow you to quickly generate a set of widgets. Here we will use ``panel``, which is based on bokeh to control our stream values.
To do so we will declare a ``StockExplorer`` class subclassing ``Parameterized`` and defines two parameters, the ``rolling_window`` as an integer and the ``symbol`` as an ObjectSelector. Additionally we define a view method, which defines the DynamicMap and applies the two operations we have already played with, returning an overlay of the smoothed ``Curve`` and outlier ``Scatter``.
```
import param
import panel as pn
variables = ['open', 'high', 'low', 'close', 'volume', 'adj_close']
class StockExplorer(param.Parameterized):
rolling_window = param.Integer(default=10, bounds=(1, 365))
symbol = param.ObjectSelector(default='AAPL', objects=stock_symbols)
variable = param.ObjectSelector(default='adj_close', objects=variables)
@param.depends('symbol', 'variable')
def load_symbol(self):
df = pd.DataFrame(getattr(stocks, self.symbol))
df['date'] = df.date.astype('datetime64[ns]')
return hv.Curve(df, ('date', 'Date'), self.variable).opts(framewise=True)
```
You will have noticed the ``param.depends`` decorator on the ``load_symbol`` method above, this declares that the method depends on these two parameters. When we pass the method to a ``DynamicMap`` it will now automatically listen to changes to the 'symbol', and 'variable' parameters. To generate a set of widgets to control these parameters we can simply supply the ``explorer.param`` accessor to a panel layout, and combining the two we can quickly build a little GUI:
```
explorer = StockExplorer()
stock_dmap = hv.DynamicMap(explorer.load_symbol)
pn.Row(explorer.param, stock_dmap)
```
The ``rolling_window`` parameter is not yet connected to anything however, so just like in the [Data Processing Pipelines section](./14-Data_Pipelines.ipynb) we will see how we can get the widget to control the parameters of an operation. Both the ``rolling`` and ``rolling_outlier_std`` operations accept a ``rolling_window`` parameter, so we simply pass that parameter into the operation. Finally we compose everything into a panel ``Row``:
```
# Apply rolling mean
smoothed = rolling(stock_dmap, rolling_window=explorer.param.rolling_window)
# Find outliers
outliers = rolling_outlier_std(stock_dmap, rolling_window=explorer.param.rolling_window).opts(
color='red', marker='triangle')
pn.Row(explorer.param, (smoothed * outliers).opts(width=600, padding=0.1))
```
## A function based approach
Instead of defining a whole Parameterized class we can also use the ``depends`` decorator to directly link the widgets to a DynamicMap callback function. This approach makes the link between the widgets and the computation very explicit at the cost of tying the widget and display code very closely together.
Instead of declaring the dependencies as strings we map the parameter instance to a particular keyword argument in the ``depends`` call. In this way we can link the symbol to the ``RadioButtonGroup`` value and the ``variable`` to the ``Select`` widget value:
```
symbol = pn.widgets.RadioButtonGroup(options=stock_symbols)
variable = pn.widgets.Select(options=variables)
rolling_window = pn.widgets.IntSlider(name='Rolling Window', value=10, start=1, end=365)
@pn.depends(symbol=symbol.param.value, variable=variable.param.value)
def load_symbol_cb(symbol, variable):
return load_symbol(symbol, variable)
dmap = hv.DynamicMap(load_symbol_cb)
smoothed = rolling(stock_dmap, rolling_window=rolling_window.param.value)
pn.Row(pn.WidgetBox('## Stock Explorer', symbol, variable, rolling_window), smoothed.opts(width=500))
```
## Replacing the output
Updating plots using a ``DynamicMap`` is a very efficient means of updating a plot since it will only update the data that has changed. In some cases it is either necessary or more convenient to redraw a plot entirely. ``Panel`` makes this easy by annotating a method with any dependencies that should trigger the plot to be redrawn. In the example below we extend the ``StockExplorer`` by adding a ``datashade`` boolean and a view method which will flip between a datashaded and regular view of the plot:
```
from holoviews.operation.datashader import datashade, dynspread
class AdvancedStockExplorer(StockExplorer):
datashade = param.Boolean(default=False)
@param.depends('datashade')
def view(self):
stocks = hv.DynamicMap(self.load_symbol)
# Apply rolling mean
smoothed = rolling(stocks, rolling_window=self.param.rolling_window)
if self.datashade:
smoothed = dynspread(datashade(smoothed, aggregator='any')).opts(framewise=True)
# Find outliers
outliers = rolling_outlier_std(stocks, rolling_window=self.param.rolling_window).opts(
width=600, color='red', marker='triangle', framewise=True)
return (smoothed * outliers)
```
In the previous example we explicitly called the ``view`` method, but to allow ``panel`` to update the plot when the datashade parameter is toggled we instead pass it the actual view method. Whenever the datashade parameter is toggled ``panel`` will call the method and update the plot with whatever is returned:
```
explorer = AdvancedStockExplorer()
pn.Row(explorer.param, explorer.view)
```
As you can see using streams we have bound the widgets to the streams letting us easily control the stream values and making it trivial to define complex dashboards. For more information on how to deploy bokeh apps from HoloViews and build dashboards see the [Deploying Bokeh Apps](./Deploying_Bokeh_Apps.ipynb).
|
github_jupyter
|
import pandas as pd
import holoviews as hv
from bokeh.sampledata import stocks
from holoviews.operation.timeseries import rolling, rolling_outlier_std
hv.extension('bokeh')
def load_symbol(symbol, variable='adj_close', **kwargs):
df = pd.DataFrame(getattr(stocks, symbol))
df['date'] = df.date.astype('datetime64[ns]')
return hv.Curve(df, ('date', 'Date'), variable)
stock_symbols = ['AAPL', 'IBM', 'FB', 'GOOG', 'MSFT']
dmap = hv.DynamicMap(load_symbol, kdims='Symbol').redim.values(Symbol=stock_symbols)
dmap.opts(framewise=True)
import param
import panel as pn
variables = ['open', 'high', 'low', 'close', 'volume', 'adj_close']
class StockExplorer(param.Parameterized):
rolling_window = param.Integer(default=10, bounds=(1, 365))
symbol = param.ObjectSelector(default='AAPL', objects=stock_symbols)
variable = param.ObjectSelector(default='adj_close', objects=variables)
@param.depends('symbol', 'variable')
def load_symbol(self):
df = pd.DataFrame(getattr(stocks, self.symbol))
df['date'] = df.date.astype('datetime64[ns]')
return hv.Curve(df, ('date', 'Date'), self.variable).opts(framewise=True)
explorer = StockExplorer()
stock_dmap = hv.DynamicMap(explorer.load_symbol)
pn.Row(explorer.param, stock_dmap)
# Apply rolling mean
smoothed = rolling(stock_dmap, rolling_window=explorer.param.rolling_window)
# Find outliers
outliers = rolling_outlier_std(stock_dmap, rolling_window=explorer.param.rolling_window).opts(
color='red', marker='triangle')
pn.Row(explorer.param, (smoothed * outliers).opts(width=600, padding=0.1))
symbol = pn.widgets.RadioButtonGroup(options=stock_symbols)
variable = pn.widgets.Select(options=variables)
rolling_window = pn.widgets.IntSlider(name='Rolling Window', value=10, start=1, end=365)
@pn.depends(symbol=symbol.param.value, variable=variable.param.value)
def load_symbol_cb(symbol, variable):
return load_symbol(symbol, variable)
dmap = hv.DynamicMap(load_symbol_cb)
smoothed = rolling(stock_dmap, rolling_window=rolling_window.param.value)
pn.Row(pn.WidgetBox('## Stock Explorer', symbol, variable, rolling_window), smoothed.opts(width=500))
from holoviews.operation.datashader import datashade, dynspread
class AdvancedStockExplorer(StockExplorer):
datashade = param.Boolean(default=False)
@param.depends('datashade')
def view(self):
stocks = hv.DynamicMap(self.load_symbol)
# Apply rolling mean
smoothed = rolling(stocks, rolling_window=self.param.rolling_window)
if self.datashade:
smoothed = dynspread(datashade(smoothed, aggregator='any')).opts(framewise=True)
# Find outliers
outliers = rolling_outlier_std(stocks, rolling_window=self.param.rolling_window).opts(
width=600, color='red', marker='triangle', framewise=True)
return (smoothed * outliers)
explorer = AdvancedStockExplorer()
pn.Row(explorer.param, explorer.view)
| 0.801625 | 0.951953 |
```
# coding: utf-8
import pandas as pd
import numpy as np
from keras.preprocessing import sequence
from keras.models import Sequential
from keras.layers import Dense, Dropout, Activation, Lambda, Flatten
from keras.layers import Embedding
from keras.layers import Convolution1D, MaxPooling1D
from keras.datasets import imdb
from keras import backend as K
import re
from keras.utils import np_utils
from keras.preprocessing import text
from keras.callbacks import ModelCheckpoint
from keras.regularizers import l2
# 生成的 word vector 的 dimension
maxlen = 1000
alphabet = 'abcdefghijklmnopqrstuvwxyz0123456789,.!? '
datatrain = pd.read_csv("new_CSV_Data/Tone2_train.csv", header=0)
datatest = pd.read_csv("new_CSV_Data/Tone2_test.csv", header=0)
chars = set(alphabet)
print('total chars:', len(chars))
char_indices = dict((c, i) for i, c in enumerate(chars))
indices_char = dict((i, c) for i, c in enumerate(chars))
# 创建 len(docs)个, 1 * maxlen 的矩阵
X_train = np.ones((datatrain.shape[0], maxlen), dtype = np.int64) * 0
docs = []
labels = []
print('zipping the data:')
epoch = 0
for label, content in zip(datatrain.classes, datatrain.content):
content = re.sub("[^a-z0-9\,\.\!\?]", " ", content)
docs.append(content)
labels.append(label)
epoch = epoch + 1
if (epoch % 20000 == 0):
print('zipping the training data:', epoch)
print('Success!')
print('There are training set:', datatrain.shape[0])
print('Doing one hot encoding:')
# One-Hot encoding 另外应该是反过来进行 encode 的,,稀疏部分用0代替
for i, doc in enumerate(docs):
# 倒着数后面的maxlen个数字,但是输出顺序不变
for t, char in enumerate(doc[-maxlen:]):
X_train[i, (maxlen-1-t)] = char_indices[char]
print('Success!')
Y_train = np.array(labels)
print('Convert class vector to binary class matrix (for use with categorical_crossentropy)')
nb_classes = 5
print(nb_classes, 'classes in the dataset')
Y_train = np_utils.to_categorical(Y_train, nb_classes)
print('Success!')
X_test = np.ones((datatest.shape[0], maxlen), dtype = np.int64) * 0
docs = []
labels = []
print('zipping the test data:')
epoch = 0
for label, content in zip(datatest.classes, datatest.content):
content = re.sub("[^a-z0-9\,\.\!\?]", " ", content)
docs.append(content)
labels.append(label)
epoch = epoch + 1
if (epoch % 20000 == 0):
print('zipping the test data:', epoch)
print('Success!')
print('There are test set:', datatest.shape[0])
print('Doing one hot encoding:')
# One-Hot encoding 另外应该是反过来进行 encode 的,,稀疏部分用-1代替
for i, doc in enumerate(docs):
# 倒着数后面的maxlen个数字,但是输出顺序不变
for t, char in enumerate(doc[-maxlen:]):
X_test[i, (maxlen-1-t)] = char_indices[char]
print('Success!')
Y_test = np.array(labels)
print('Convert class vector to binary class matrix (for use with categorical_crossentropy)')
nb_classes = 5
print(nb_classes, 'classes in the dataset')
Y_test = np_utils.to_categorical(Y_test, nb_classes)
print('Success!')
print("All of the pre-processde work is done.")
model = Sequential()
# we start off with an efficient embedding layer which maps
# our vocab indices into embedding_dims dimensions
model.add(Embedding(input_dim = 41, output_dim = 50, input_length = maxlen, init = 'he_normal', W_regularizer=l2(0.01)) )
# we add a Convolution1D, which will learn nb_filter
# word group filters of size filter_length:
model.add(Convolution1D(nb_filter = 128, filter_length = 3, W_regularizer=l2(0.01), init = 'he_normal', border_mode='same', activation='relu', subsample_length=1))
model.add(Convolution1D(nb_filter = 128, filter_length = 3, W_regularizer=l2(0.01), init = 'he_normal', border_mode='same', activation='relu', subsample_length=1))
model.add(Convolution1D(nb_filter = 128, filter_length = 3, W_regularizer=l2(0.01), init = 'he_normal', border_mode='same', activation='relu', subsample_length=1))
# we use max pooling:
model.add(MaxPooling1D(pool_length = model.output_shape[1]))
#model.add(MaxPooling1D(pool_length = 2))
#print(model.output_shape[1], "pooling shape")
# We flatten the output of the conv layer,
# so that we can add a vanilla dense layer:
model.add(Flatten())
# We add a vanilla hidden layer:
model.add(Dense(100))
model.add(Dropout(0.1))
model.add(Activation('relu'))
# We add a vanilla hidden layer:
model.add(Dense(100))
model.add(Dropout(0.1))
model.add(Activation('relu'))
# We project onto a single unit output layer, and squash it with a sigmoid:
model.add(Dense(5))
model.add(Activation('softmax'))
checkpointers = ModelCheckpoint("parameters/weights.{epoch:02d}-{val_acc:.4f}.hdf5", monitor='val_acc', verbose=0, save_best_only=False, mode='auto')
#model.load_weights("parameters/weights.39-0.32.hdf5")
model.summary()
model.compile(loss='categorical_crossentropy', optimizer='adam', metrics=['accuracy'])
model.fit(X_train, Y_train, batch_size = 128, nb_epoch = 20, validation_data=(X_test, Y_test), callbacks = [checkpointers])
```
|
github_jupyter
|
# coding: utf-8
import pandas as pd
import numpy as np
from keras.preprocessing import sequence
from keras.models import Sequential
from keras.layers import Dense, Dropout, Activation, Lambda, Flatten
from keras.layers import Embedding
from keras.layers import Convolution1D, MaxPooling1D
from keras.datasets import imdb
from keras import backend as K
import re
from keras.utils import np_utils
from keras.preprocessing import text
from keras.callbacks import ModelCheckpoint
from keras.regularizers import l2
# 生成的 word vector 的 dimension
maxlen = 1000
alphabet = 'abcdefghijklmnopqrstuvwxyz0123456789,.!? '
datatrain = pd.read_csv("new_CSV_Data/Tone2_train.csv", header=0)
datatest = pd.read_csv("new_CSV_Data/Tone2_test.csv", header=0)
chars = set(alphabet)
print('total chars:', len(chars))
char_indices = dict((c, i) for i, c in enumerate(chars))
indices_char = dict((i, c) for i, c in enumerate(chars))
# 创建 len(docs)个, 1 * maxlen 的矩阵
X_train = np.ones((datatrain.shape[0], maxlen), dtype = np.int64) * 0
docs = []
labels = []
print('zipping the data:')
epoch = 0
for label, content in zip(datatrain.classes, datatrain.content):
content = re.sub("[^a-z0-9\,\.\!\?]", " ", content)
docs.append(content)
labels.append(label)
epoch = epoch + 1
if (epoch % 20000 == 0):
print('zipping the training data:', epoch)
print('Success!')
print('There are training set:', datatrain.shape[0])
print('Doing one hot encoding:')
# One-Hot encoding 另外应该是反过来进行 encode 的,,稀疏部分用0代替
for i, doc in enumerate(docs):
# 倒着数后面的maxlen个数字,但是输出顺序不变
for t, char in enumerate(doc[-maxlen:]):
X_train[i, (maxlen-1-t)] = char_indices[char]
print('Success!')
Y_train = np.array(labels)
print('Convert class vector to binary class matrix (for use with categorical_crossentropy)')
nb_classes = 5
print(nb_classes, 'classes in the dataset')
Y_train = np_utils.to_categorical(Y_train, nb_classes)
print('Success!')
X_test = np.ones((datatest.shape[0], maxlen), dtype = np.int64) * 0
docs = []
labels = []
print('zipping the test data:')
epoch = 0
for label, content in zip(datatest.classes, datatest.content):
content = re.sub("[^a-z0-9\,\.\!\?]", " ", content)
docs.append(content)
labels.append(label)
epoch = epoch + 1
if (epoch % 20000 == 0):
print('zipping the test data:', epoch)
print('Success!')
print('There are test set:', datatest.shape[0])
print('Doing one hot encoding:')
# One-Hot encoding 另外应该是反过来进行 encode 的,,稀疏部分用-1代替
for i, doc in enumerate(docs):
# 倒着数后面的maxlen个数字,但是输出顺序不变
for t, char in enumerate(doc[-maxlen:]):
X_test[i, (maxlen-1-t)] = char_indices[char]
print('Success!')
Y_test = np.array(labels)
print('Convert class vector to binary class matrix (for use with categorical_crossentropy)')
nb_classes = 5
print(nb_classes, 'classes in the dataset')
Y_test = np_utils.to_categorical(Y_test, nb_classes)
print('Success!')
print("All of the pre-processde work is done.")
model = Sequential()
# we start off with an efficient embedding layer which maps
# our vocab indices into embedding_dims dimensions
model.add(Embedding(input_dim = 41, output_dim = 50, input_length = maxlen, init = 'he_normal', W_regularizer=l2(0.01)) )
# we add a Convolution1D, which will learn nb_filter
# word group filters of size filter_length:
model.add(Convolution1D(nb_filter = 128, filter_length = 3, W_regularizer=l2(0.01), init = 'he_normal', border_mode='same', activation='relu', subsample_length=1))
model.add(Convolution1D(nb_filter = 128, filter_length = 3, W_regularizer=l2(0.01), init = 'he_normal', border_mode='same', activation='relu', subsample_length=1))
model.add(Convolution1D(nb_filter = 128, filter_length = 3, W_regularizer=l2(0.01), init = 'he_normal', border_mode='same', activation='relu', subsample_length=1))
# we use max pooling:
model.add(MaxPooling1D(pool_length = model.output_shape[1]))
#model.add(MaxPooling1D(pool_length = 2))
#print(model.output_shape[1], "pooling shape")
# We flatten the output of the conv layer,
# so that we can add a vanilla dense layer:
model.add(Flatten())
# We add a vanilla hidden layer:
model.add(Dense(100))
model.add(Dropout(0.1))
model.add(Activation('relu'))
# We add a vanilla hidden layer:
model.add(Dense(100))
model.add(Dropout(0.1))
model.add(Activation('relu'))
# We project onto a single unit output layer, and squash it with a sigmoid:
model.add(Dense(5))
model.add(Activation('softmax'))
checkpointers = ModelCheckpoint("parameters/weights.{epoch:02d}-{val_acc:.4f}.hdf5", monitor='val_acc', verbose=0, save_best_only=False, mode='auto')
#model.load_weights("parameters/weights.39-0.32.hdf5")
model.summary()
model.compile(loss='categorical_crossentropy', optimizer='adam', metrics=['accuracy'])
model.fit(X_train, Y_train, batch_size = 128, nb_epoch = 20, validation_data=(X_test, Y_test), callbacks = [checkpointers])
| 0.513425 | 0.334644 |
### Recommender Systems
Personalization of the user experience has been at high priority and has become the new mantra in consumer focused industry. You might have observed e-commerce companies casting personalized ads for you suggesting what to buy, which news to read, which video to watch, where/what to eat and who you might be interested in networking (friends/professionals) on social media sites. Recommender systems are the core information filtering system designed to predict the user preference and help to recommend right items to create user specific personalization experience. There are two types of recommendation systems i.e., 1) content based filtering 2) collaborative filtering
```
from IPython.display import Image
Image(filename='../Chapter 5 Figures/Recommender_System.png', width=600)
```
Let's consider a movie rating dataset for 6 movies and 7 users as shown in below table.
```
from IPython.display import Image
Image(filename='../Chapter 5 Figures/Movie_Rating.png', width=900)
import numpy as np
import pandas as pd
df = pd.read_csv('Data/movie_rating.csv')
n_users = df.userID.unique().shape[0]
n_items = df.itemID.unique().shape[0]
print '\nNumber of users = ' + str(n_users) + ' | Number of movies = ' + str(n_items)
# Create user-item matrices
df_matrix = np.zeros((n_users, n_items))
for line in df.itertuples():
df_matrix[line[1]-1, line[2]-1] = line[3]
from sklearn.metrics.pairwise import pairwise_distances
user_similarity = pairwise_distances(df_matrix, metric='euclidean')
item_similarity = pairwise_distances(df_matrix.T, metric='euclidean')
# Top 3 similar users for user id 7
print "Similar users for user id 7: \n", pd.DataFrame(user_similarity).loc[6,pd.DataFrame(user_similarity).loc[6,:] > 0].sort_values(ascending=False)[0:3]
# Top 3 similar items for item id 6
print "Similar items for item id 6: \n", pd.DataFrame(item_similarity).loc[5,pd.DataFrame(item_similarity).loc[5,:] > 0].sort_values(ascending=False)[0:3]
```
### Item based similarity
```
from IPython.display import Image
Image(filename='../Chapter 5 Figures/Item_Similarity_Formula.png', width=300)
# Function for item based rating prediction
def item_based_prediction(rating_matrix, similarity_matrix):
return rating_matrix.dot(similarity_matrix) / np.array([np.abs(similarity_matrix).sum(axis=1)])
item_based_prediction = item_based_prediction(df_matrix, item_similarity)
```
### Memory based collaborative filtering (user based similarity)
```
from IPython.display import Image
Image(filename='../Chapter 5 Figures/User_Similarity_Formula.png', width=300)
# Function for user based rating prediction
def user_based_prediction(rating_matrix, similarity_matrix):
mean_user_rating = rating_matrix.mean(axis=1)
ratings_diff = (rating_matrix - mean_user_rating[:, np.newaxis])
return mean_user_rating[:, np.newaxis] + similarity_matrix.dot(ratings_diff) / np.array([np.abs(similarity_matrix).sum(axis=1)]).T
user_based_prediction = user_based_prediction(df_matrix, user_similarity)
# Calculate the RMSE
from sklearn.metrics import mean_squared_error
from math import sqrt
def rmse(prediction, actual):
prediction = prediction[actual.nonzero()].flatten()
actual = actual[actual.nonzero()].flatten()
return sqrt(mean_squared_error(prediction, actual))
print 'User-based CF RMSE: ' + str(rmse(user_based_prediction, df_matrix))
print 'Item-based CF RMSE: ' + str(rmse(item_based_prediction, df_matrix))
y_user_based = pd.DataFrame(user_based_prediction)
# Predictions for movies that the user 6 hasn't rated yet
predictions = y_user_based.loc[6,pd.DataFrame(df_matrix).loc[6,:] == 0]
top = predictions.sort_values(ascending=False).head(n=1)
recommendations = pd.DataFrame(data=top)
recommendations.columns = ['Predicted Rating']
print recommendations
y_item_based = pd.DataFrame(item_based_prediction)
# Predictions for movies that the user 6 hasn't rated yet
predictions = y_item_based.loc[6,pd.DataFrame(df_matrix).loc[6,:] == 0]
top = predictions.sort_values(ascending=False).head(n=1)
recommendations = pd.DataFrame(data=top)
recommendations.columns = ['Predicted Rating']
print recommendations
```
### Model based collaborative filtering (user based similarity)
```
# calculate sparsity level
sparsity=round(1.0-len(df)/float(n_users*n_items),3)
print 'The sparsity level of is ' + str(sparsity*100) + '%'
import scipy.sparse as sp
from scipy.sparse.linalg import svds
# Get SVD components from train matrix. Choose k.
u, s, vt = svds(df_matrix, k = 5)
s_diag_matrix=np.diag(s)
X_pred = np.dot(np.dot(u, s_diag_matrix), vt)
print 'User-based CF MSE: ' + str(rmse(X_pred, df_matrix))
```
In our case since the data set is small sparsity level is 0%. I recommend you to try this method on the MovieLens 100k dataset which you can download from https://grouplens.org/datasets/movielens/100k/
|
github_jupyter
|
from IPython.display import Image
Image(filename='../Chapter 5 Figures/Recommender_System.png', width=600)
from IPython.display import Image
Image(filename='../Chapter 5 Figures/Movie_Rating.png', width=900)
import numpy as np
import pandas as pd
df = pd.read_csv('Data/movie_rating.csv')
n_users = df.userID.unique().shape[0]
n_items = df.itemID.unique().shape[0]
print '\nNumber of users = ' + str(n_users) + ' | Number of movies = ' + str(n_items)
# Create user-item matrices
df_matrix = np.zeros((n_users, n_items))
for line in df.itertuples():
df_matrix[line[1]-1, line[2]-1] = line[3]
from sklearn.metrics.pairwise import pairwise_distances
user_similarity = pairwise_distances(df_matrix, metric='euclidean')
item_similarity = pairwise_distances(df_matrix.T, metric='euclidean')
# Top 3 similar users for user id 7
print "Similar users for user id 7: \n", pd.DataFrame(user_similarity).loc[6,pd.DataFrame(user_similarity).loc[6,:] > 0].sort_values(ascending=False)[0:3]
# Top 3 similar items for item id 6
print "Similar items for item id 6: \n", pd.DataFrame(item_similarity).loc[5,pd.DataFrame(item_similarity).loc[5,:] > 0].sort_values(ascending=False)[0:3]
from IPython.display import Image
Image(filename='../Chapter 5 Figures/Item_Similarity_Formula.png', width=300)
# Function for item based rating prediction
def item_based_prediction(rating_matrix, similarity_matrix):
return rating_matrix.dot(similarity_matrix) / np.array([np.abs(similarity_matrix).sum(axis=1)])
item_based_prediction = item_based_prediction(df_matrix, item_similarity)
from IPython.display import Image
Image(filename='../Chapter 5 Figures/User_Similarity_Formula.png', width=300)
# Function for user based rating prediction
def user_based_prediction(rating_matrix, similarity_matrix):
mean_user_rating = rating_matrix.mean(axis=1)
ratings_diff = (rating_matrix - mean_user_rating[:, np.newaxis])
return mean_user_rating[:, np.newaxis] + similarity_matrix.dot(ratings_diff) / np.array([np.abs(similarity_matrix).sum(axis=1)]).T
user_based_prediction = user_based_prediction(df_matrix, user_similarity)
# Calculate the RMSE
from sklearn.metrics import mean_squared_error
from math import sqrt
def rmse(prediction, actual):
prediction = prediction[actual.nonzero()].flatten()
actual = actual[actual.nonzero()].flatten()
return sqrt(mean_squared_error(prediction, actual))
print 'User-based CF RMSE: ' + str(rmse(user_based_prediction, df_matrix))
print 'Item-based CF RMSE: ' + str(rmse(item_based_prediction, df_matrix))
y_user_based = pd.DataFrame(user_based_prediction)
# Predictions for movies that the user 6 hasn't rated yet
predictions = y_user_based.loc[6,pd.DataFrame(df_matrix).loc[6,:] == 0]
top = predictions.sort_values(ascending=False).head(n=1)
recommendations = pd.DataFrame(data=top)
recommendations.columns = ['Predicted Rating']
print recommendations
y_item_based = pd.DataFrame(item_based_prediction)
# Predictions for movies that the user 6 hasn't rated yet
predictions = y_item_based.loc[6,pd.DataFrame(df_matrix).loc[6,:] == 0]
top = predictions.sort_values(ascending=False).head(n=1)
recommendations = pd.DataFrame(data=top)
recommendations.columns = ['Predicted Rating']
print recommendations
# calculate sparsity level
sparsity=round(1.0-len(df)/float(n_users*n_items),3)
print 'The sparsity level of is ' + str(sparsity*100) + '%'
import scipy.sparse as sp
from scipy.sparse.linalg import svds
# Get SVD components from train matrix. Choose k.
u, s, vt = svds(df_matrix, k = 5)
s_diag_matrix=np.diag(s)
X_pred = np.dot(np.dot(u, s_diag_matrix), vt)
print 'User-based CF MSE: ' + str(rmse(X_pred, df_matrix))
| 0.679498 | 0.91302 |
# Outlier Detection
```
import pandas as pd
import numpy as np
from sklearn import preprocessing
import sys
import math
import matplotlib.pyplot as plt
import datetime
inputFile = "data/two-hour-sample.csv"
df = pd.read_csv(inputFile, sep=",")
print(df.shape)
print(df.head())
# Rename the columns because when I did this work I liked my names better
colnames = ["StartTime", "Dur", "Proto", "SrcAddr", "Sport", "Dir", "DstAddr",
"Dport", "TotPkts", "TotBytes", "SrcBytes"]
df = df[colnames]
df.columns = ['timestamp', 'duration', 'proto', 'src_ip', 'src_port', 'direction', 'dest_ip',
'dest_port', 'tot_pkts', 'tot_bytes', 'bytes_toclient']
df['row_id'] = df.index
# Clean up missing ports
df.src_port.fillna(0)
df.src_port.fillna(0)
df.replace(to_replace={'src_port': {float('NaN'): 0},
'dest_port': {float('NaN'): 0}}, inplace=True)
# Set a place holder for the example, normally this would be extracted from the timestamp
df['day'] = 1
```
## Feature Creation
```
#### Add Total Counts (How much overall traffic to this IP?)
totalCount = df.shape[0]
srcDf = df[['src_ip', 'proto']].groupby(
'src_ip', as_index=False).count().rename({"proto": "src_count"}, axis=1)
print(srcDf.head())
destDf = df[['dest_ip', 'proto']].groupby(
'dest_ip', as_index=False).count().rename({"proto": "dest_count"}, axis=1)
print(destDf.head())
src_joined = pd.merge(df, srcDf, how='left',
on='src_ip', suffixes=('', '_count'))
df2 = pd.merge(src_joined, destDf, how='left', on=[
'dest_ip'], suffixes=('', '_count'))
##### Compute IP percentages
srcCol = df2.columns.get_loc('src_count')
destCol = df2.columns.get_loc('dest_count')
print(str(srcCol) + " " + str(destCol))
dfa = df2.assign(src_pct=df2.src_count / totalCount)
dfb = dfa.assign(dest_pct=dfa.dest_count / totalCount)
#### Compute Protocol Percentages
srcDf = dfb[['src_ip', 'proto', "day"]].groupby(
['src_ip', 'proto'], as_index=False).count().rename({"day": "src_proto_count"}, axis=1)
# print(srcDf.head())
destDf = dfb[['dest_ip', 'proto', 'day']].groupby(
['dest_ip', 'proto'], as_index=False).count().rename({"day": "dest_proto_count"}, axis=1)
# print(destDf.head())
src_joined = pd.merge(dfb, srcDf, how='left', on=[
'src_ip', 'proto'], suffixes=('', '_count'))
df3 = pd.merge(src_joined, destDf, how='left', on=[
'dest_ip', 'proto'], suffixes=('', '_count'))
df4 = df3.assign(src_proto_pct=df3.src_proto_count / df3.src_count)
df5 = df4.assign(dest_proto_pct=df3.dest_proto_count / df3.dest_count)
#### Compute Protocol Port Percentages
### First compute total protocol counts overall
protoDf = df5[['proto', 'src_port']].groupby(
'proto', as_index=False).count().rename({"src_port": "proto_count"}, axis=1)
df6 = pd.merge(df5, protoDf, how='left',
on='proto', suffixes=('', '_count'))
protoSPortDf = df6[['proto', 'src_port', 'day']].groupby(
['proto', 'src_port'], as_index=False).count().rename({"day": "proto_src_port_count"}, axis=1)
df7 = pd.merge(df6, protoSPortDf, how='left', on=[
'proto', 'src_port'], suffixes=('', '_count'))
df8 = df7.assign(
proto_src_port_pct=df7.proto_src_port_count/df7.proto_count)
print(df8.head())
protoDPortDf = df8[['proto', 'dest_port', 'day']].groupby(
['proto', 'dest_port'], as_index=False).count().rename({"day": "proto_dest_port_count"}, axis=1)
df9 = pd.merge(df8, protoDPortDf, how='left', on=[
'proto', 'dest_port'], suffixes=('', '_count'))
df10 = df9.assign(
proto_dest_port_pct=df9.proto_dest_port_count/df9.proto_count)
# Compute standardized counts for number based features
scaler = preprocessing.StandardScaler()
df10['pkts_scaled'] = scaler.fit_transform(df10[['tot_pkts']])
df10['bytes_scaled'] = scaler.fit_transform(df10[['tot_bytes']])
df10['duration_scaled'] = scaler.fit_transform(df10[['duration']])
df = df10.assign(abs_pkts=abs(df10.pkts_scaled))
df = df.assign(abs_bytes=abs(df.bytes_scaled))
df = df.assign(abs_dur=abs(df.duration_scaled))
featureList = ['src_pct', 'dest_pct', 'src_proto_pct', 'dest_proto_pct',
'proto_src_port_pct', 'proto_dest_port_pct', 'abs_pkts']
# Check the shape of the full data
print(df.shape)
# Create a subset of the variables for training
trainDf = df[featureList]
print(trainDf.shape)
print(trainDf.head())
# Import Outlier Math
from scipy import stats
from sklearn import svm
from sklearn.covariance import EllipticEnvelope
from sklearn.ensemble import IsolationForest
from sklearn.neighbors import LocalOutlierFactor
rng = np.random.RandomState(42)
# Example settings
n_samples = 100000
outliers_fraction = 0.01 # TODO: Tweak this parameter
clusters_separation = [0, 1, 2]
# Set up the possibility to run multiple outlier detectors
# For the purposes of time we will only run Local Outlier Factor
# Isolation Forest is another quick and easy one to try
classifiers = {
# "svm": svm.OneClassSVM(nu=0.95 * outliers_fraction + 0.05,
# kernel="rbf", gamma=0.1),
# "rc": EllipticEnvelope(contamination=outliers_fraction),
# "iso": IsolationForest(max_samples=n_samples,
# contamination=outliers_fraction,
# random_state=rng),
"lof": LocalOutlierFactor(
n_neighbors=25,
contamination=outliers_fraction)
}
## Run the Model
for i, (clf_name, clf) in enumerate(classifiers.items()):
now = datetime.datetime.now()
print("Starting " + clf_name + " " + str(now))
# fit the data and tag outliers
if clf_name == "lof":
y_pred = clf.fit_predict(trainDf)
scores_pred = clf.negative_outlier_factor_
else:
clf.fit(trainDf)
scores_pred = clf.decision_function(trainDf)
y_pred = clf.predict(trainDf)
threshold = stats.scoreatpercentile(scores_pred,
100 * outliers_fraction)
print(clf_name)
print(threshold)
print(scores_pred)
df[clf_name] = scores_pred
df[clf_name + "_pred"] = y_pred
print(df.head())
print(df.shape)
print(df.size)
df.head()
now = datetime.datetime.now()
print("Complete " + str(now))
df.groupby("lof_pred").size()
plt.hist(df["lof_pred"])
```
|
github_jupyter
|
import pandas as pd
import numpy as np
from sklearn import preprocessing
import sys
import math
import matplotlib.pyplot as plt
import datetime
inputFile = "data/two-hour-sample.csv"
df = pd.read_csv(inputFile, sep=",")
print(df.shape)
print(df.head())
# Rename the columns because when I did this work I liked my names better
colnames = ["StartTime", "Dur", "Proto", "SrcAddr", "Sport", "Dir", "DstAddr",
"Dport", "TotPkts", "TotBytes", "SrcBytes"]
df = df[colnames]
df.columns = ['timestamp', 'duration', 'proto', 'src_ip', 'src_port', 'direction', 'dest_ip',
'dest_port', 'tot_pkts', 'tot_bytes', 'bytes_toclient']
df['row_id'] = df.index
# Clean up missing ports
df.src_port.fillna(0)
df.src_port.fillna(0)
df.replace(to_replace={'src_port': {float('NaN'): 0},
'dest_port': {float('NaN'): 0}}, inplace=True)
# Set a place holder for the example, normally this would be extracted from the timestamp
df['day'] = 1
#### Add Total Counts (How much overall traffic to this IP?)
totalCount = df.shape[0]
srcDf = df[['src_ip', 'proto']].groupby(
'src_ip', as_index=False).count().rename({"proto": "src_count"}, axis=1)
print(srcDf.head())
destDf = df[['dest_ip', 'proto']].groupby(
'dest_ip', as_index=False).count().rename({"proto": "dest_count"}, axis=1)
print(destDf.head())
src_joined = pd.merge(df, srcDf, how='left',
on='src_ip', suffixes=('', '_count'))
df2 = pd.merge(src_joined, destDf, how='left', on=[
'dest_ip'], suffixes=('', '_count'))
##### Compute IP percentages
srcCol = df2.columns.get_loc('src_count')
destCol = df2.columns.get_loc('dest_count')
print(str(srcCol) + " " + str(destCol))
dfa = df2.assign(src_pct=df2.src_count / totalCount)
dfb = dfa.assign(dest_pct=dfa.dest_count / totalCount)
#### Compute Protocol Percentages
srcDf = dfb[['src_ip', 'proto', "day"]].groupby(
['src_ip', 'proto'], as_index=False).count().rename({"day": "src_proto_count"}, axis=1)
# print(srcDf.head())
destDf = dfb[['dest_ip', 'proto', 'day']].groupby(
['dest_ip', 'proto'], as_index=False).count().rename({"day": "dest_proto_count"}, axis=1)
# print(destDf.head())
src_joined = pd.merge(dfb, srcDf, how='left', on=[
'src_ip', 'proto'], suffixes=('', '_count'))
df3 = pd.merge(src_joined, destDf, how='left', on=[
'dest_ip', 'proto'], suffixes=('', '_count'))
df4 = df3.assign(src_proto_pct=df3.src_proto_count / df3.src_count)
df5 = df4.assign(dest_proto_pct=df3.dest_proto_count / df3.dest_count)
#### Compute Protocol Port Percentages
### First compute total protocol counts overall
protoDf = df5[['proto', 'src_port']].groupby(
'proto', as_index=False).count().rename({"src_port": "proto_count"}, axis=1)
df6 = pd.merge(df5, protoDf, how='left',
on='proto', suffixes=('', '_count'))
protoSPortDf = df6[['proto', 'src_port', 'day']].groupby(
['proto', 'src_port'], as_index=False).count().rename({"day": "proto_src_port_count"}, axis=1)
df7 = pd.merge(df6, protoSPortDf, how='left', on=[
'proto', 'src_port'], suffixes=('', '_count'))
df8 = df7.assign(
proto_src_port_pct=df7.proto_src_port_count/df7.proto_count)
print(df8.head())
protoDPortDf = df8[['proto', 'dest_port', 'day']].groupby(
['proto', 'dest_port'], as_index=False).count().rename({"day": "proto_dest_port_count"}, axis=1)
df9 = pd.merge(df8, protoDPortDf, how='left', on=[
'proto', 'dest_port'], suffixes=('', '_count'))
df10 = df9.assign(
proto_dest_port_pct=df9.proto_dest_port_count/df9.proto_count)
# Compute standardized counts for number based features
scaler = preprocessing.StandardScaler()
df10['pkts_scaled'] = scaler.fit_transform(df10[['tot_pkts']])
df10['bytes_scaled'] = scaler.fit_transform(df10[['tot_bytes']])
df10['duration_scaled'] = scaler.fit_transform(df10[['duration']])
df = df10.assign(abs_pkts=abs(df10.pkts_scaled))
df = df.assign(abs_bytes=abs(df.bytes_scaled))
df = df.assign(abs_dur=abs(df.duration_scaled))
featureList = ['src_pct', 'dest_pct', 'src_proto_pct', 'dest_proto_pct',
'proto_src_port_pct', 'proto_dest_port_pct', 'abs_pkts']
# Check the shape of the full data
print(df.shape)
# Create a subset of the variables for training
trainDf = df[featureList]
print(trainDf.shape)
print(trainDf.head())
# Import Outlier Math
from scipy import stats
from sklearn import svm
from sklearn.covariance import EllipticEnvelope
from sklearn.ensemble import IsolationForest
from sklearn.neighbors import LocalOutlierFactor
rng = np.random.RandomState(42)
# Example settings
n_samples = 100000
outliers_fraction = 0.01 # TODO: Tweak this parameter
clusters_separation = [0, 1, 2]
# Set up the possibility to run multiple outlier detectors
# For the purposes of time we will only run Local Outlier Factor
# Isolation Forest is another quick and easy one to try
classifiers = {
# "svm": svm.OneClassSVM(nu=0.95 * outliers_fraction + 0.05,
# kernel="rbf", gamma=0.1),
# "rc": EllipticEnvelope(contamination=outliers_fraction),
# "iso": IsolationForest(max_samples=n_samples,
# contamination=outliers_fraction,
# random_state=rng),
"lof": LocalOutlierFactor(
n_neighbors=25,
contamination=outliers_fraction)
}
## Run the Model
for i, (clf_name, clf) in enumerate(classifiers.items()):
now = datetime.datetime.now()
print("Starting " + clf_name + " " + str(now))
# fit the data and tag outliers
if clf_name == "lof":
y_pred = clf.fit_predict(trainDf)
scores_pred = clf.negative_outlier_factor_
else:
clf.fit(trainDf)
scores_pred = clf.decision_function(trainDf)
y_pred = clf.predict(trainDf)
threshold = stats.scoreatpercentile(scores_pred,
100 * outliers_fraction)
print(clf_name)
print(threshold)
print(scores_pred)
df[clf_name] = scores_pred
df[clf_name + "_pred"] = y_pred
print(df.head())
print(df.shape)
print(df.size)
df.head()
now = datetime.datetime.now()
print("Complete " + str(now))
df.groupby("lof_pred").size()
plt.hist(df["lof_pred"])
| 0.372391 | 0.748099 |
This notebook contains scripts which evaluate the performance of a classification model on the test set.
```
from train_classifier import *
import matplotlib.pyplot as plt
import seaborn as sns
import pandas as pd
from sklearn.metrics import roc_auc_score, roc_curve
from scipy.stats import gaussian_kde
import numpy as np
```
Start by loading the fine-tuned model and the test dataset.
```
model_dir = '../data/overflow/BERT'
model, collate_fn = get_bert_model(model_dir)
test_dataset = mongo_dataset.MongoDataset().get_partition('classification_test', projection)
```
Use Huggingface's Trainer interface to make predictions on the test set.
```
train_args = TrainingArguments(**default_training_args)
trainer = Trainer(model=model, args=train_args, data_collator=collate_fn)
predictions = trainer.predict(test_dataset)
```
Convert this output into a pandas dataframe so it's easier to work with.
```
probs = softmax(predictions.predictions, axis=1)[:,1]
labels = predictions.label_ids.astype(bool)
df = pd.DataFrame(zip(probs, labels), columns=['AnswerProbability', 'Answered'])
df['Title'] = pd.DataFrame(test_dataset).Title
```
Plot the distributions of predicted probabilities for the two classes (answered and unanswered questions).
```
sns.displot(df,x='AnswerProbability', bins=50, hue='Answered', kde=True);
```
Because we'd like to interpret the model's output as a probability of getting an answer, it is important to verify that the probabilities are calibrated. This means, for example, that if we choose 100 titles to which the model assigned a probability of ~90%, approximately 90/100 of those should actually be answered. Poorly calibrated probabilities can be a sign of overfitting.
```
width = 0.1
class_ratio = 1/df.Answered.mean() - 1
kde0 = gaussian_kde(df.AnswerProbability[~df.Answered],width)
kde1 = gaussian_kde(df.AnswerProbability[df.Answered],width)
p_up = np.linspace(0.60,1,1000)
plt.plot(p_up,1/(class_ratio*kde0(p_up)/kde1(p_up)+1))
plt.plot(p_up,p_up,linestyle='dashed')
plt.xlabel('Predicted probability');
plt.ylabel('Actual probability');
plt.title('Calibration of probabilities');
plt.legend(['Calibration curve', 'Ideal']);
```
Due to the class imbalance, accuracy is not an effective metric (the model would maximize accuracy by predicting that every question is answered). Instead, the ROC curve and ROC-AUC score are a better metric for model performance.
```
score = roc_auc_score(labels, probs)
fpr, tpr, thresholds = roc_curve(labels, probs)
plt.plot(fpr,tpr);
plt.plot(fpr,fpr, color='red', linestyle='dashed');
plt.xlabel('False positive rate');
plt.ylabel('True positive rate');
plt.legend(['ROC curve', 'Baseline']);
plt.title(f'ROC-AUC = {score:.4f}');
```
To better understand what makes a title good or bad, it's helpful to examine questions which the model considers most and least likely to be answered.
```
num_examples = 10
# Sample from the bottom 5% of titles
bottom_quantile = df.AnswerProbability.quantile(0.05)
bad_titles = df.Title[df.AnswerProbability<=bottom_quantile].sample(num_examples)
# Sample from the top 5% of titles
top_quantile = df.AnswerProbability.quantile(0.95)
good_titles = df.Title[df.AnswerProbability>=top_quantile].sample(num_examples)
# Print the examples
print(f'Most likely to be answered (>{top_quantile:.0%} chance):')
for title in good_titles:
print(' '+title)
print()
print(f'Least likely to be answered (<{bottom_quantile:.0%} chance):')
for title in bad_titles:
print(' '+title)
```
|
github_jupyter
|
from train_classifier import *
import matplotlib.pyplot as plt
import seaborn as sns
import pandas as pd
from sklearn.metrics import roc_auc_score, roc_curve
from scipy.stats import gaussian_kde
import numpy as np
model_dir = '../data/overflow/BERT'
model, collate_fn = get_bert_model(model_dir)
test_dataset = mongo_dataset.MongoDataset().get_partition('classification_test', projection)
train_args = TrainingArguments(**default_training_args)
trainer = Trainer(model=model, args=train_args, data_collator=collate_fn)
predictions = trainer.predict(test_dataset)
probs = softmax(predictions.predictions, axis=1)[:,1]
labels = predictions.label_ids.astype(bool)
df = pd.DataFrame(zip(probs, labels), columns=['AnswerProbability', 'Answered'])
df['Title'] = pd.DataFrame(test_dataset).Title
sns.displot(df,x='AnswerProbability', bins=50, hue='Answered', kde=True);
width = 0.1
class_ratio = 1/df.Answered.mean() - 1
kde0 = gaussian_kde(df.AnswerProbability[~df.Answered],width)
kde1 = gaussian_kde(df.AnswerProbability[df.Answered],width)
p_up = np.linspace(0.60,1,1000)
plt.plot(p_up,1/(class_ratio*kde0(p_up)/kde1(p_up)+1))
plt.plot(p_up,p_up,linestyle='dashed')
plt.xlabel('Predicted probability');
plt.ylabel('Actual probability');
plt.title('Calibration of probabilities');
plt.legend(['Calibration curve', 'Ideal']);
score = roc_auc_score(labels, probs)
fpr, tpr, thresholds = roc_curve(labels, probs)
plt.plot(fpr,tpr);
plt.plot(fpr,fpr, color='red', linestyle='dashed');
plt.xlabel('False positive rate');
plt.ylabel('True positive rate');
plt.legend(['ROC curve', 'Baseline']);
plt.title(f'ROC-AUC = {score:.4f}');
num_examples = 10
# Sample from the bottom 5% of titles
bottom_quantile = df.AnswerProbability.quantile(0.05)
bad_titles = df.Title[df.AnswerProbability<=bottom_quantile].sample(num_examples)
# Sample from the top 5% of titles
top_quantile = df.AnswerProbability.quantile(0.95)
good_titles = df.Title[df.AnswerProbability>=top_quantile].sample(num_examples)
# Print the examples
print(f'Most likely to be answered (>{top_quantile:.0%} chance):')
for title in good_titles:
print(' '+title)
print()
print(f'Least likely to be answered (<{bottom_quantile:.0%} chance):')
for title in bad_titles:
print(' '+title)
| 0.715026 | 0.962708 |
# Introduction to Deep Learning with PyTorch
In this notebook, you'll get introduced to [PyTorch](http://pytorch.org/), a framework for building and training neural networks. PyTorch in a lot of ways behaves like the arrays you love from Numpy. These Numpy arrays, after all, are just tensors. PyTorch takes these tensors and makes it simple to move them to GPUs for the faster processing needed when training neural networks. It also provides a module that automatically calculates gradients (for backpropagation!) and another module specifically for building neural networks. All together, PyTorch ends up being more coherent with Python and the Numpy/Scipy stack compared to TensorFlow and other frameworks.
## Neural Networks
Deep Learning is based on artificial neural networks which have been around in some form since the late 1950s. The networks are built from individual parts approximating neurons, typically called units or simply "neurons." Each unit has some number of weighted inputs. These weighted inputs are summed together (a linear combination) then passed through an activation function to get the unit's output.
<img src="assets/simple_neuron.png" width=400px>
Mathematically this looks like:
$$
\begin{align}
y &= f(w_1 x_1 + w_2 x_2 + b) \\
y &= f\left(\sum_i w_i x_i +b \right)
\end{align}
$$
With vectors this is the dot/inner product of two vectors:
$$
h = \begin{bmatrix}
x_1 \, x_2 \cdots x_n
\end{bmatrix}
\cdot
\begin{bmatrix}
w_1 \\
w_2 \\
\vdots \\
w_n
\end{bmatrix}
$$
## Tensors
It turns out neural network computations are just a bunch of linear algebra operations on *tensors*, a generalization of matrices. A vector is a 1-dimensional tensor, a matrix is a 2-dimensional tensor, an array with three indices is a 3-dimensional tensor (RGB color images for example). The fundamental data structure for neural networks are tensors and PyTorch (as well as pretty much every other deep learning framework) is built around tensors.
<img src="assets/tensor_examples.svg" width=600px>
With the basics covered, it's time to explore how we can use PyTorch to build a simple neural network.
```
# First, import PyTorch
import torch
def activation(x):
""" Sigmoid activation function
Arguments
---------
x: torch.Tensor
"""
return 1/(1+torch.exp(-x))
### Generate some data
torch.manual_seed(7) # Set the random seed so things are predictable
# Features are 5 random normal variables
features = torch.randn((1, 5))
# True weights for our data, random normal variables again
weights = torch.randn_like(features)
# and a true bias term
bias = torch.randn((1, 1))
```
Above I generated data we can use to get the output of our simple network. This is all just random for now, going forward we'll start using normal data. Going through each relevant line:
`features = torch.randn((1, 5))` creates a tensor with shape `(1, 5)`, one row and five columns, that contains values randomly distributed according to the normal distribution with a mean of zero and standard deviation of one.
`weights = torch.randn_like(features)` creates another tensor with the same shape as `features`, again containing values from a normal distribution.
Finally, `bias = torch.randn((1, 1))` creates a single value from a normal distribution.
PyTorch tensors can be added, multiplied, subtracted, etc, just like Numpy arrays. In general, you'll use PyTorch tensors pretty much the same way you'd use Numpy arrays. They come with some nice benefits though such as GPU acceleration which we'll get to later. For now, use the generated data to calculate the output of this simple single layer network.
> **Exercise**: Calculate the output of the network with input features `features`, weights `weights`, and bias `bias`. Similar to Numpy, PyTorch has a [`torch.sum()`](https://pytorch.org/docs/stable/torch.html#torch.sum) function, as well as a `.sum()` method on tensors, for taking sums. Use the function `activation` defined above as the activation function.
```
## Calculate the output of this network using the weights and bias tensors
output = activation(torch.sum( features * weights ) + bias)
print(output)
```
You can do the multiplication and sum in the same operation using a matrix multiplication. In general, you'll want to use matrix multiplications since they are more efficient and accelerated using modern libraries and high-performance computing on GPUs.
Here, we want to do a matrix multiplication of the features and the weights. For this we can use [`torch.mm()`](https://pytorch.org/docs/stable/torch.html#torch.mm) or [`torch.matmul()`](https://pytorch.org/docs/stable/torch.html#torch.matmul) which is somewhat more complicated and supports broadcasting. If we try to do it with `features` and `weights` as they are, we'll get an error
```python
>> torch.mm(features, weights)
---------------------------------------------------------------------------
RuntimeError Traceback (most recent call last)
<ipython-input-13-15d592eb5279> in <module>()
----> 1 torch.mm(features, weights)
RuntimeError: size mismatch, m1: [1 x 5], m2: [1 x 5] at /Users/soumith/minicondabuild3/conda-bld/pytorch_1524590658547/work/aten/src/TH/generic/THTensorMath.c:2033
```
As you're building neural networks in any framework, you'll see this often. Really often. What's happening here is our tensors aren't the correct shapes to perform a matrix multiplication. Remember that for matrix multiplications, the number of columns in the first tensor must equal to the number of rows in the second column. Both `features` and `weights` have the same shape, `(1, 5)`. This means we need to change the shape of `weights` to get the matrix multiplication to work.
**Note:** To see the shape of a tensor called `tensor`, use `tensor.shape`. If you're building neural networks, you'll be using this method often.
There are a few options here: [`weights.reshape()`](https://pytorch.org/docs/stable/tensors.html#torch.Tensor.reshape), [`weights.resize_()`](https://pytorch.org/docs/stable/tensors.html#torch.Tensor.resize_), and [`weights.view()`](https://pytorch.org/docs/stable/tensors.html#torch.Tensor.view).
* `weights.reshape(a, b)` will return a new tensor with the same data as `weights` with size `(a, b)` sometimes, and sometimes a clone, as in it copies the data to another part of memory.
* `weights.resize_(a, b)` returns the same tensor with a different shape. However, if the new shape results in fewer elements than the original tensor, some elements will be removed from the tensor (but not from memory). If the new shape results in more elements than the original tensor, new elements will be uninitialized in memory. Here I should note that the underscore at the end of the method denotes that this method is performed **in-place**. Here is a great forum thread to [read more about in-place operations](https://discuss.pytorch.org/t/what-is-in-place-operation/16244) in PyTorch.
* `weights.view(a, b)` will return a new tensor with the same data as `weights` with size `(a, b)`.
I usually use `.view()`, but any of the three methods will work for this. So, now we can reshape `weights` to have five rows and one column with something like `weights.view(5, 1)`.
> **Exercise**: Calculate the output of our little network using matrix multiplication.
```
## Calculate the output of this network using matrix multiplication
output = activation(torch.mm(features, weights.view(5, 1)) + bias)
print(output)
```
### Stack them up!
That's how you can calculate the output for a single neuron. The real power of this algorithm happens when you start stacking these individual units into layers and stacks of layers, into a network of neurons. The output of one layer of neurons becomes the input for the next layer. With multiple input units and output units, we now need to express the weights as a matrix.
<img src='assets/multilayer_diagram_weights.png' width=450px>
The first layer shown on the bottom here are the inputs, understandably called the **input layer**. The middle layer is called the **hidden layer**, and the final layer (on the right) is the **output layer**. We can express this network mathematically with matrices again and use matrix multiplication to get linear combinations for each unit in one operation. For example, the hidden layer ($h_1$ and $h_2$ here) can be calculated
$$
\vec{h} = [h_1 \, h_2] =
\begin{bmatrix}
x_1 \, x_2 \cdots \, x_n
\end{bmatrix}
\cdot
\begin{bmatrix}
w_{11} & w_{12} \\
w_{21} &w_{22} \\
\vdots &\vdots \\
w_{n1} &w_{n2}
\end{bmatrix}
$$
The output for this small network is found by treating the hidden layer as inputs for the output unit. The network output is expressed simply
$$
y = f_2 \! \left(\, f_1 \! \left(\vec{x} \, \mathbf{W_1}\right) \mathbf{W_2} \right)
$$
```
### Generate some data
torch.manual_seed(7) # Set the random seed so things are predictable
# Features are 3 random normal variables
features = torch.randn((1, 3))
# Define the size of each layer in our network
n_input = features.shape[1] # Number of input units, must match number of input features
n_hidden = 2 # Number of hidden units
n_output = 1 # Number of output units
# Weights for inputs to hidden layer
W1 = torch.randn(n_input, n_hidden)
# Weights for hidden layer to output layer
W2 = torch.randn(n_hidden, n_output)
# and bias terms for hidden and output layers
B1 = torch.randn((1, n_hidden))
B2 = torch.randn((1, n_output))
```
> **Exercise:** Calculate the output for this multi-layer network using the weights `W1` & `W2`, and the biases, `B1` & `B2`.
```
## Your solution here
layer_1 = activation(torch.mm(features, W1) + B1) # 1x2
layer_2 = activation(torch.mm(layer_1, W2) + B2) # 1x1
print(layer_2)
```
If you did this correctly, you should see the output `tensor([[ 0.3171]])`.
The number of hidden units a parameter of the network, often called a **hyperparameter** to differentiate it from the weights and biases parameters. As you'll see later when we discuss training a neural network, the more hidden units a network has, and the more layers, the better able it is to learn from data and make accurate predictions.
## Numpy to Torch and back
Special bonus section! PyTorch has a great feature for converting between Numpy arrays and Torch tensors. To create a tensor from a Numpy array, use `torch.from_numpy()`. To convert a tensor to a Numpy array, use the `.numpy()` method.
```
import numpy as np
a = np.random.rand(4,3)
a
b = torch.from_numpy(a)
b
b.numpy()
```
The memory is shared between the Numpy array and Torch tensor, so if you change the values in-place of one object, the other will change as well.
```
# Multiply PyTorch Tensor by 2, in place
b.mul_(2)
# Numpy array matches new values from Tensor
a
```
|
github_jupyter
|
# First, import PyTorch
import torch
def activation(x):
""" Sigmoid activation function
Arguments
---------
x: torch.Tensor
"""
return 1/(1+torch.exp(-x))
### Generate some data
torch.manual_seed(7) # Set the random seed so things are predictable
# Features are 5 random normal variables
features = torch.randn((1, 5))
# True weights for our data, random normal variables again
weights = torch.randn_like(features)
# and a true bias term
bias = torch.randn((1, 1))
## Calculate the output of this network using the weights and bias tensors
output = activation(torch.sum( features * weights ) + bias)
print(output)
>> torch.mm(features, weights)
---------------------------------------------------------------------------
RuntimeError Traceback (most recent call last)
<ipython-input-13-15d592eb5279> in <module>()
----> 1 torch.mm(features, weights)
RuntimeError: size mismatch, m1: [1 x 5], m2: [1 x 5] at /Users/soumith/minicondabuild3/conda-bld/pytorch_1524590658547/work/aten/src/TH/generic/THTensorMath.c:2033
## Calculate the output of this network using matrix multiplication
output = activation(torch.mm(features, weights.view(5, 1)) + bias)
print(output)
### Generate some data
torch.manual_seed(7) # Set the random seed so things are predictable
# Features are 3 random normal variables
features = torch.randn((1, 3))
# Define the size of each layer in our network
n_input = features.shape[1] # Number of input units, must match number of input features
n_hidden = 2 # Number of hidden units
n_output = 1 # Number of output units
# Weights for inputs to hidden layer
W1 = torch.randn(n_input, n_hidden)
# Weights for hidden layer to output layer
W2 = torch.randn(n_hidden, n_output)
# and bias terms for hidden and output layers
B1 = torch.randn((1, n_hidden))
B2 = torch.randn((1, n_output))
## Your solution here
layer_1 = activation(torch.mm(features, W1) + B1) # 1x2
layer_2 = activation(torch.mm(layer_1, W2) + B2) # 1x1
print(layer_2)
import numpy as np
a = np.random.rand(4,3)
a
b = torch.from_numpy(a)
b
b.numpy()
# Multiply PyTorch Tensor by 2, in place
b.mul_(2)
# Numpy array matches new values from Tensor
a
| 0.792825 | 0.994964 |
# 2D jet data
In this tutorial we will explore a small dataset provided with this package that contains pressure data of the flow exiting a nozzle (also referred to as a jet). In particular, we want to identify whether the data contains spatio-temporal coherent structures.
## Loading and configuring data
The dataset is part of the data used for the regression tests that come with this library and is stored into `tests/data/fluidmechanic_data.mat`. The first step to anlyze this dataset is to import the required libraries, including the custom libraries
- `from pyspod.spod_low_storage import SPOD_low_storage`
- `from pyspod.spod_low_ram import SPOD_low_ram`
- `from pyspod.spod_streaming import SPOD_streaming`
that contain three different implementations of the SPOD algorithm, the first requiring low storage memory (intended for large RAM machines or small amount of data), the second requiring low RAM (intended for large dataset or small RAM machines), and the third being a streaming algorithm, that required little amount of memory (both storage and RAM) but runs typically slower than the other two.
```
import os
import sys
import time
import h5py
import warnings
import xarray as xr
import numpy as np
from pathlib import Path
# Paths
CWD = os.getcwd()
sys.path.insert(0, os.path.join(CWD, "../../../"))
# Import library specific modules
from pyspod.spod_low_storage import SPOD_low_storage
from pyspod.spod_low_ram import SPOD_low_ram
from pyspod.spod_streaming import SPOD_streaming
```
We then need to load the data from the `.mat` file and inspect it:
```
# Inspect and load data
file = os.path.join(CWD,'../../../tests/data/fluidmechanics_data.mat')
variables = ['p']
with h5py.File(file, 'r') as f:
data_arrays = dict()
for k, v in f.items():
data_arrays[k] = np.array(v)
dt = data_arrays['dt'][0,0]
block_dimension = 64 * dt
x1 = data_arrays['r'].T; x1 = x1[:,0]
x2 = data_arrays['x'].T; x2 = x2[0,:]
X = data_arrays[variables[0]].T
t = dt * np.arange(0,X.shape[0]); t = t.T
nt = t.shape[0]
print('t.shape = ', t.shape)
print('x1.shape = ', x1.shape)
print('x2.shape = ', x2.shape)
print('X.shape = ', X.shape)
```
the `mat` file contains **3 coordinates**:
- r, (radial coordinate)
- x, (axial coordinate)
- time,
along with **1 variable**:
- p (pressure).
In order for the data matrix `X` to be suitable to the `PySPOD` library the
- first dimension must correspond to the number of time snapshots (1000 in our case)
- last dimension should corresponds to the number of variables (1 in our case)
- the remaining dimensions corresponds to the spatial dimensions (20, and 88 in our case, that correspond to radial and axial spatial coordinates).
We note that the data matrix `X` used is already in a shape that is suitable to `PySPOD`, as its dimension is:
$$\text{$X$ dimensions} = 1000 \times 20 \times 88 $$
It is important to note at this point that we loaded all the required data into RAM, and stored it into a `numpy.ndarray`. We will later pass this array to the constructor of the `PySPOD` class for running our analysis. However, we could have used a different approach to load the data. In fact, the constructor to the `PySPOD` class accepts an argument called `data_handler`, that points to a function whose objective is to read the data at run time. This is particularly useful for large datasets, where it might be not possible to load all the data in RAM upfront. Therefore, in this case, we could simply define a data reader function as the following:
```
def read_data(data, t_0, t_end, variables):
... implement here your method
data: path to the data file
t_0: start time slicing
t_end: end time slicing
variables: list with names of the variables
return X
```
and pass it to the `PySPOD` constructor under the argument `data_handler`. The path to the data file, will then be specified in place of the data, under the argument `X`. See below, when we setup the analysis and call the constructor for a more detailed explantion of the parameters `X` and `data_handler`. In summary, if `X` is a numpy.ndarray containing your data, `data_handler` is set to `False`, if `X` is a `str` containing the path to your data file, `data_handler` is a function that reads your data, and whose arguments must be: (1.) `str` containing the path to the data file, (2) `int` containing the start time snapshot for slicing the data sequentially at run time, (3) `int` containing the end time snapshot for slicing the data sequentially at run time, and (4) a `list` containing the name of the variables in your data file.
## Setting required and optional parameters
Once our data is in a shape suitable to the `PySPOD` library, we define the **required** and **optional parameters**. In particular, we define a dictionary of parameters, that will be passed to the constructor of `PySPOD`.
The required parameters are as follows:
- `time_step`: time-sampling of the data (for now this must be constant)
- `n_space_dims`: number of spatial dimensions
- `n_variables`: number of variables
- `n_DFT`: length of FFT blocks
The optional parameters are as follows:
- `overlap`: dimension of the overlap region between adjacent blocks in percentage (0 to 100)
- `mean_type`: type of mean to be subtracted from the data (`longtime`, `blockwise` or `zero`)
- `normalize_weights`: weights normalization by data variance
- `normalize_data`: normalize data by variance
- `n_modes_save`: number of modes to be saved
- `conf_level`: calculate confidence level of modes
- `reuse_blocks`: whether to attempt reusing FFT blocks previously computed (if found)
- `savefft`: save FFT blocks to reuse them in the future (to save time)
- `savedir`: where to save the data
**Note that we do not set any parameter for the Weights adopted to compute th einner product in the SPOD calculation. In this case, the algorithm will use automatically uniform weighting (weighting equal 1), and it will prompt a warning stating the use of default uniform weighting.**
```
# define required and optional parameters
params = dict()
# -- required parameters
params['time_step' ] = dt # data time-sampling
params['n_snapshots' ] = t.shape[0] # number of time snapshots (we consider all data)
params['n_space_dims'] = 2 # number of spatial dimensions (longitude and latitude)
params['n_variables' ] = len(variables) # number of variables
params['n_DFT' ] = np.ceil(block_dimension / dt) # length of FFT blocks (100 time-snapshots)
# -- optional parameters
params['overlap' ] = 50 # dimension block overlap region
params['mean_type' ] = 'blockwise' # type of mean to subtract to the data
params['normalize_weights'] = False # normalization of weights by data variance
params['normalize_data' ] = False # normalize data by data variance
params['n_modes_save' ] = 3 # modes to be saved
params['conf_level' ] = 0.95 # calculate confidence level
params['reuse_blocks' ] = False # whether to reuse blocks if present
params['savefft' ] = False # save FFT blocks to reuse them in the future (saves time)
params['savedir' ] = os.path.join(CWD, 'results', Path(file).stem) # folder where to save results
```
## Running the SPOD analysis
Once we have loaded the data and defined the required and optional parameters, we can perform the analysis. This step is accomplished by calling the `PySPOD` constructor, `SPOD_streaming(params=params, data_handler=False, variables=variables)` and the `fit` method, `SPOD_analysis.fit(data=X, nt=nt)`.
The `PySPOD` constructor takes the parameters `params`, a parameter called `data_handler` that can be either `False` or a function to read the data, and `variables` that is the list containing the names of our variables. If, as `data_handler`, we pass `False`, then we need to load the entire matrix of data into RAM, and that must comply with the **PySPOD** input data requirements (i.e. the dimension of the data matrix must correspond to (time $\times$ spatial dimension shape $\times$ number of variables).
The method `fit` takes as inputs `data`, that can either be a `numpy.ndarray` containing the data or the path to the data file (if `data_handler` is not set to `False`), and the number of time snapshots `nt`.
In more detail, the arguments to the constructor are defined as follows:
- `params`: must be a dictionary and contains the parameters that we have just defined.
- `data_handler`: can be either `False` or a function handler. If it is a function handler, it must hold the function to read the data. The template for the function to read the data must have as first argument the data file, as second and third the time indices through which we will slice the data in time, and as fourth argument a list containing the name of the variables. See hour data reader as an example and modify it according to your needs.
- `variables`: is a list containing our variables.
The arguments to the `fit` method are:
- `data`: it can either be a `numpy.ndarray` and contain all data required for the analysis or a `str` containing the path to the data file. If we pass a `numpy.ndarray`, its dimensions must be equal to (time $\times$ spatial dimension shape $\times$ number of variables), and the argument `file_handler` must be set to `False`. If we pass a `str` containing the path to the data file, we need also to provide a data reader through the argument `data_handler`. The data reader must conform to reading the file and storing the data in memory according to the shape of data just described: **(number of time snapshots $\times$ shape of spatial dimensions $\times$ number of variables)**. Note that the template for the data reader must have as first argument the path to the data file, as second and third the time indices through which we will slice the data in time, and as fourth argument a list containing the name of the variables. An example of data reader was provided above. You can readily modify it according to your needs. See the sections above for a template example of the data reader function.
- `nt`: the number of time snapshots to consider.
The `fit` method returns a `PySPOD` object containg the results.
```
# Perform SPOD analysis using low storage module
SPOD_analysis = SPOD_streaming(params=params, data_handler=False, variables=variables)
spod = SPOD_analysis.fit(data=X, nt=nt)
```
## Postprocessing and visualizing results
The results are stored in a `PySPOD` objcet that is composed by:
- a set of eigenvalues per each frequency computed, and
- a set of modes, per each frequency computed.
In order to visualize them, we can use the built-in plotting functionalities of `PySPOD`.
We first select the frequency (equivalently period T_approx), that we want to investigate, and identify the nearest frequency in the results by using the built-in functions `find_nearest_freq`, and `get_modes_at_freq`, that are part of the `postprocessing` module, and can be directly called from the `PySPOD` object returned once the `fit` method has completed.
```
# Show results
T_approx = 12.5 # approximate period
freq_found, freq_idx = spod.find_nearest_freq(freq_required=1/T_approx, freq=spod.freq)
modes_at_freq = spod.get_modes_at_freq(freq_idx=freq_idx)
```
We can then plot the **eigenvalues in the complex plane**, using the built-in function `plot_eigs`, that is part of the `postprocessing` module. We note that the eigenvalues are all real.
```
spod.plot_eigs()
```
We can then plot the **eigenvalues as a function of frequency and period**. Again, we can see how thorough the `PySPOD` object returned after the computation we can access the frequency array (`spod.freq`) along with the plotting methods `spod.plot_eigs_vs_frequency` and `spod.plot_eigs_vs_period`.
```
freq = spod.freq
spod.plot_eigs_vs_frequency(freq=freq)
spod.plot_eigs_vs_period (freq=freq, xticks=[1, 0.5, 0.2, 0.1, 0.05, 0.02])
```
We can then plot the **modes** that were computed by the SPOD algorithm via the built-in `plot_2D_modes_at_frequency` method, that can again be accessed via the `PySPOD` object returned after the computation. To this method, we pass the frequency of the modes we are interested in. This corresponds to the frequency associated to the T_approx of 12.5 time units that we requested, and stored in the variable `freq_found` that we calculated above. Note that we also pass the `vars_idx` corresponding to the variable we are interested in, modes_idx corresponding to the modes we are interested in, as well as `x1`, and `x2`, that correspond to radial and axial coordinates.
```
spod.plot_2D_modes_at_frequency(
freq_required=freq_found,
freq=freq,
x1=x1,
x2=x2,
modes_idx=[0,1],
vars_idx=[0])
```
Note that we can also plot the **original data** by
```
spod.plot_2D_data(x1=x1, x2=x2, vars_idx=[0], time_idx=[0,100,200])
```
Along with a video of the original data
```
# spod.generate_2D_data_video(x1=x1, x2=x2, vars_idx=[0])
```
## Using modes for reduced order modeling
It is possible to use the modes generated by SPOD to construct reduced order models. In particular, we can assume that the original data is composed by a temporal mean and a fluctuating component:
$$\mathbf{q}(t) = \bar{\mathbf{q}} + \mathbf{q}'(t)$$
We can use a reduced number of modes $\boldsymbol{\Phi}_r$, with $r < n$, where $n$ is the original dimension of the data $\mathbf{q}(t)$ to approximate $\mathbf{q}'(t)$, hence the dynamics of the system. Following [Chu and Schmidt, 2020](https://arxiv.org/pdf/2012.02902.pdf):
- we construct a vector $\tilde{\mathbf{q}}$ that approximates the original data $\mathbf{q}'$ as follows
$$ \tilde{\mathbf{q}} = \boldsymbol{\Phi}_r(\boldsymbol{\Phi}_r^{*} \mathbf{W}\boldsymbol{\Phi}_r)^{-1}\boldsymbol{\Phi}_r^{*}\mathbf{W}\mathbf{q}' = \mathbf{P}\mathbf{q}'$$
- we couple the approximated vector $\tilde{q}$ with the underlying dynamics (eventually approximated) of the problem being studied
$$ \frac{\text{d}}{\text{d}t}\mathbf{q}' = f(\bar{\mathbf{q}}, \mathbf{q}', t) \longrightarrow \frac{\text{d}}{\text{d}t}\tilde{\mathbf{q}} = \mathbf{P}f(\bar{\mathbf{q}}, \tilde{\mathbf{q}}, t)$$
Obviously, the second step depends on the knowledge we have of the system and how well our knowledge approximates the original dynamics.
## Final notes
The results are stored in the results folder defined in the parameter `params[savedir]` you specified. We can load the results for both modes and eigenvalues, and use any other postprocessing tool that is more suitable to your application. The files are stored in `numpy` binary format `.npy`. There exists several tools to convert them in `netCDF`, `MATLAB` and several other formats that can be better suited to you specific post-processing pipeline.
This tutorial was intended to help you setup your own 2D case. You can play with the parameters we explored above to gain more insights into the capabilities of the library.
|
github_jupyter
|
import os
import sys
import time
import h5py
import warnings
import xarray as xr
import numpy as np
from pathlib import Path
# Paths
CWD = os.getcwd()
sys.path.insert(0, os.path.join(CWD, "../../../"))
# Import library specific modules
from pyspod.spod_low_storage import SPOD_low_storage
from pyspod.spod_low_ram import SPOD_low_ram
from pyspod.spod_streaming import SPOD_streaming
# Inspect and load data
file = os.path.join(CWD,'../../../tests/data/fluidmechanics_data.mat')
variables = ['p']
with h5py.File(file, 'r') as f:
data_arrays = dict()
for k, v in f.items():
data_arrays[k] = np.array(v)
dt = data_arrays['dt'][0,0]
block_dimension = 64 * dt
x1 = data_arrays['r'].T; x1 = x1[:,0]
x2 = data_arrays['x'].T; x2 = x2[0,:]
X = data_arrays[variables[0]].T
t = dt * np.arange(0,X.shape[0]); t = t.T
nt = t.shape[0]
print('t.shape = ', t.shape)
print('x1.shape = ', x1.shape)
print('x2.shape = ', x2.shape)
print('X.shape = ', X.shape)
def read_data(data, t_0, t_end, variables):
... implement here your method
data: path to the data file
t_0: start time slicing
t_end: end time slicing
variables: list with names of the variables
return X
# define required and optional parameters
params = dict()
# -- required parameters
params['time_step' ] = dt # data time-sampling
params['n_snapshots' ] = t.shape[0] # number of time snapshots (we consider all data)
params['n_space_dims'] = 2 # number of spatial dimensions (longitude and latitude)
params['n_variables' ] = len(variables) # number of variables
params['n_DFT' ] = np.ceil(block_dimension / dt) # length of FFT blocks (100 time-snapshots)
# -- optional parameters
params['overlap' ] = 50 # dimension block overlap region
params['mean_type' ] = 'blockwise' # type of mean to subtract to the data
params['normalize_weights'] = False # normalization of weights by data variance
params['normalize_data' ] = False # normalize data by data variance
params['n_modes_save' ] = 3 # modes to be saved
params['conf_level' ] = 0.95 # calculate confidence level
params['reuse_blocks' ] = False # whether to reuse blocks if present
params['savefft' ] = False # save FFT blocks to reuse them in the future (saves time)
params['savedir' ] = os.path.join(CWD, 'results', Path(file).stem) # folder where to save results
# Perform SPOD analysis using low storage module
SPOD_analysis = SPOD_streaming(params=params, data_handler=False, variables=variables)
spod = SPOD_analysis.fit(data=X, nt=nt)
# Show results
T_approx = 12.5 # approximate period
freq_found, freq_idx = spod.find_nearest_freq(freq_required=1/T_approx, freq=spod.freq)
modes_at_freq = spod.get_modes_at_freq(freq_idx=freq_idx)
spod.plot_eigs()
freq = spod.freq
spod.plot_eigs_vs_frequency(freq=freq)
spod.plot_eigs_vs_period (freq=freq, xticks=[1, 0.5, 0.2, 0.1, 0.05, 0.02])
spod.plot_2D_modes_at_frequency(
freq_required=freq_found,
freq=freq,
x1=x1,
x2=x2,
modes_idx=[0,1],
vars_idx=[0])
spod.plot_2D_data(x1=x1, x2=x2, vars_idx=[0], time_idx=[0,100,200])
# spod.generate_2D_data_video(x1=x1, x2=x2, vars_idx=[0])
| 0.352982 | 0.990102 |
# Introduction
You've built a model to identify clothing types in the **MNIST for Fashion** dataset. Now you will make your model bigger, specify larger stride lengths and apply dropout. These changes will make your model faster and more accurate.
This is a last step in the **[Deep Learning Track](https://www.kaggle.com/learn/deep-learning)**.
## Data Preparation
**Run this cell of code.**
```
import numpy as np
from sklearn.model_selection import train_test_split
from tensorflow import keras
# Set up code checking
from learntools.core import binder
binder.bind(globals())
from learntools.deep_learning.exercise_8 import *
print("Setup Complete")
img_rows, img_cols = 28, 28
num_classes = 10
def prep_data(raw):
y = raw[:, 0]
out_y = keras.utils.to_categorical(y, num_classes)
x = raw[:,1:]
num_images = raw.shape[0]
out_x = x.reshape(num_images, img_rows, img_cols, 1)
out_x = out_x / 255
return out_x, out_y
fashion_file = "../input/fashionmnist/fashion-mnist_train.csv"
fashion_data = np.loadtxt(fashion_file, skiprows=1, delimiter=',')
x, y = prep_data(fashion_data)
```
# 1) Increasing Stride Size in A Layer
Below is a model without strides (or more accurately, with a stride length of 1)
Run it. Notice it's accuracy and how long it takes per epoch. Then you will change the stride length in one of the layers.
```
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Dense, Flatten, Conv2D, Dropout
batch_size = 16
fashion_model = Sequential()
fashion_model.add(Conv2D(16, kernel_size=(3, 3),
activation='relu',
input_shape=(img_rows, img_cols, 1)))
fashion_model.add(Conv2D(16, (3, 3), activation='relu'))
fashion_model.add(Flatten())
fashion_model.add(Dense(128, activation='relu'))
fashion_model.add(Dense(num_classes, activation='softmax'))
fashion_model.compile(loss=keras.losses.categorical_crossentropy,
optimizer='adam',
metrics=['accuracy'])
fashion_model.fit(x, y,
batch_size=batch_size,
epochs=3,
validation_split = 0.2)
```
You have the same code in the cell below, but the model is now called `fashion_model_1`. Change the specification of `fashion_model_1` so the second convolutional layer has a stride length of 2.
Run the cell after you have done that. How does the speed and accuracy change compared to the first model you ran above?
```
fashion_model_1 = Sequential()
fashion_model_1.add(Conv2D(16, kernel_size=(3, 3),
activation='relu',
input_shape=(img_rows, img_cols, 1)))
fashion_model_1.add(Conv2D(16, (3, 3), activation='relu', strides=2))
fashion_model_1.add(Flatten())
fashion_model_1.add(Dense(128, activation='relu'))
fashion_model_1.add(Dense(num_classes, activation='softmax'))
fashion_model_1.compile(loss=keras.losses.categorical_crossentropy,
optimizer='adam',
metrics=['accuracy'])
fashion_model_1.fit(x, y,
batch_size=batch_size,
epochs=3,
validation_split = 0.2)
# Check your answer
q_1.check()
```
For the solution, uncomment and run the cell below:
```
#_COMMENT_IF(PROD)_
q_1.solution()
```
You should notice that your model training ran about twice as fast, but the accuracy change was trivial.
In addition to being faster to train, this model is also faster at making predictions. This is very important in many scenarios. In practice, you'll need to decide whether that type of speed is important in the applications where you eventually apply deep learning models.
You could experiment with more layers or more convolutions in each layer. With some fine-tuning, you can build a model that is both faster and more accurate than the original model.
# Congrats
You've finished the Deep Learning course. You have the tools to create and tune computer vision models.
If you feel like playing more with this dataset, you can open up a new code cell to experiment with different models (adding dropout, adding layers, etc.) Or pick a new project and try out your skills.
A few fun datasets you might try include:
- [Written letter recognition](https://www.kaggle.com/olgabelitskaya/classification-of-handwritten-letters)
- [Flower Identification](https://www.kaggle.com/alxmamaev/flowers-recognition)
- [Cats vs Dogs](https://www.kaggle.com/c/dogs-vs-cats-redux-kernels-edition)
- [10 Monkeys](https://www.kaggle.com/slothkong/10-monkey-species)
- [Predict Bone Age from X-Rays](https://www.kaggle.com/kmader/rsna-bone-age)
You have learned a lot, and you'll learn it as you practice. Have fun with it!
|
github_jupyter
|
import numpy as np
from sklearn.model_selection import train_test_split
from tensorflow import keras
# Set up code checking
from learntools.core import binder
binder.bind(globals())
from learntools.deep_learning.exercise_8 import *
print("Setup Complete")
img_rows, img_cols = 28, 28
num_classes = 10
def prep_data(raw):
y = raw[:, 0]
out_y = keras.utils.to_categorical(y, num_classes)
x = raw[:,1:]
num_images = raw.shape[0]
out_x = x.reshape(num_images, img_rows, img_cols, 1)
out_x = out_x / 255
return out_x, out_y
fashion_file = "../input/fashionmnist/fashion-mnist_train.csv"
fashion_data = np.loadtxt(fashion_file, skiprows=1, delimiter=',')
x, y = prep_data(fashion_data)
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Dense, Flatten, Conv2D, Dropout
batch_size = 16
fashion_model = Sequential()
fashion_model.add(Conv2D(16, kernel_size=(3, 3),
activation='relu',
input_shape=(img_rows, img_cols, 1)))
fashion_model.add(Conv2D(16, (3, 3), activation='relu'))
fashion_model.add(Flatten())
fashion_model.add(Dense(128, activation='relu'))
fashion_model.add(Dense(num_classes, activation='softmax'))
fashion_model.compile(loss=keras.losses.categorical_crossentropy,
optimizer='adam',
metrics=['accuracy'])
fashion_model.fit(x, y,
batch_size=batch_size,
epochs=3,
validation_split = 0.2)
fashion_model_1 = Sequential()
fashion_model_1.add(Conv2D(16, kernel_size=(3, 3),
activation='relu',
input_shape=(img_rows, img_cols, 1)))
fashion_model_1.add(Conv2D(16, (3, 3), activation='relu', strides=2))
fashion_model_1.add(Flatten())
fashion_model_1.add(Dense(128, activation='relu'))
fashion_model_1.add(Dense(num_classes, activation='softmax'))
fashion_model_1.compile(loss=keras.losses.categorical_crossentropy,
optimizer='adam',
metrics=['accuracy'])
fashion_model_1.fit(x, y,
batch_size=batch_size,
epochs=3,
validation_split = 0.2)
# Check your answer
q_1.check()
#_COMMENT_IF(PROD)_
q_1.solution()
| 0.8119 | 0.975923 |
```
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
from skbio.stats.composition import clr, centralize, ilr
from skbio import OrdinationResults
%matplotlib inline
def cart2polar(x, y):
theta = np.arctan2(y, x)
rho = np.sqrt(x**2 + y**2)
return rho, theta
species = 'pelag'
category = 'latitude'
N = 50
legend_loc = 'lower right'
species = 'pelag'
category = 'temperature'
N = 50
legend_loc = 'lower right'
species = 'proch'
category = 'latitude'
N = 50
legend_loc = 'upper left'
species = 'proch'
category = 'temperature'
N = 50
legend_loc = 'upper left'
counts = pd.read_csv('/Users/luke/singlecell/notebooks/tara_%s_nonzero_SRF.csv' % species)
tara_metadata = pd.read_csv('/Users/luke/singlecell/notebooks/tara_metadata_SRF.tsv', sep='\t')
tara_metadata = tara_metadata.sort_values(by=r'Sample label [TARA_station#_environmental-feature_size-fraction]')
og_metadata = pd.read_csv('/Users/luke/singlecell/notebooks/og_metadata.tsv', sep='\t', index_col=0)
f = lambda x: x.split('_')[1]
tara_metadata['SampleNumber'] = tara_metadata[r'Sample label [TARA_station#_environmental-feature_size-fraction]'].apply(f)
counts['SampleNumber'] = counts[r'Unnamed: 0'].apply(f)
tara_metadata = tara_metadata.sort_values(by=r'SampleNumber')
counts = counts.sort_values(by=r'SampleNumber')
mat = counts.values[:, 1:-1]
mat = mat.astype(dtype=np.float)
u1, k1, v1 = np.linalg.svd(clr(centralize(mat+1)))
n = len(u1)
G = np.sqrt(n - 1) * u1[:, :2]
H = np.vstack(((np.sqrt(k1[0]) * v1[0, :]) / np.sqrt(n - 1), (np.sqrt(k1[1]) * v1[1, :]) / np.sqrt(n - 1)))
# Get top N hits
rho, theta = cart2polar(H[0, :], H[1, :])
z = rho.argsort()[-N:]
# Calculate radii and degrees to easier identification on the biplot
feats = pd.DataFrame({'rho':rho[z], 'theta':theta[z]*180/np.pi}, index=counts.columns[1:-1][z])
feats['Description'] = og_metadata['Description'][feats.index]
feats.sort_values(by='rho', ascending=False, inplace=True)
feats.to_csv('/Users/luke/singlecell/notebooks/ordination_of_tara_biplot_ogs_%s_%s.tsv' % (species, category), sep='\t')
# cat lookup
if category == 'temperature':
dict_cat = {
'polar': u"<10 \N{DEGREE SIGN}C, 'polar' samples",
'temperate': u"10-20 \N{DEGREE SIGN}C, 'temperate' samples",
'tropical': u">20 \N{DEGREE SIGN}C, 'tropical' samples"
}
colors = ['light orange', 'light blue', 'blue']
elif category == 'latitude':
dict_cat = {
'temperate': u'temperate samples',
'subtropical': u'subtropical samples',
'tropical': u'tropical samples'
}
colors = ['blue', 'light blue', 'light orange']
# plot
plt.figure(figsize=(10, 10))
marker_rs = dict(color=sns.xkcd_rgb['light orange'], marker='o', markeredgewidth=1, markeredgecolor='black')
idx = tara_metadata['category_redsea'] == True
p1 = plt.plot(u1[idx.values, 0], u1[idx.values, 1], '.', label='Red Sea samples', markersize=10, zorder=10, **marker_rs)
i = 0
for cat in set(tara_metadata['category_%s' % category]):
idx = tara_metadata['category_%s' % category] == cat
plt.plot(u1[idx.values, 0], u1[idx.values, 1], 'o', label=dict_cat[cat], markersize=10, color=sns.xkcd_rgb[colors[i]])
i += 1
for j in z:
_x, _y = H[0, j], H[1, j]
plt.arrow(0, 0, _x, _y, head_width=0.01)
perc_explained = k1**2 / (k1**2).sum()
plt.xlabel('PC 1 ({:.2%})'.format(perc_explained[0]), fontsize=18)
plt.ylabel('PC 2 ({:.2%})'.format(perc_explained[1]), fontsize=18)
plt.xticks(fontsize=14)
plt.yticks(fontsize=14)
plt.legend(fontsize=16, loc=legend_loc)
plt.savefig('/Users/luke/singlecell/notebooks/ordination_of_tara_samples_%s_%s.pdf' % (species, category))
# ordination results
u = pd.DataFrame(u1, index=counts.index)
proportion_explained=pd.Series(perc_explained, index=counts.index)
eigvals=pd.Series(k1, index=counts.index)
res = OrdinationResults('','',samples=u, proportion_explained=proportion_explained, eigvals=eigvals)
res.write('/Users/luke/singlecell/notebooks/ordination_of_tara_results_%s_%s.pc' % (species, category))
```
|
github_jupyter
|
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
from skbio.stats.composition import clr, centralize, ilr
from skbio import OrdinationResults
%matplotlib inline
def cart2polar(x, y):
theta = np.arctan2(y, x)
rho = np.sqrt(x**2 + y**2)
return rho, theta
species = 'pelag'
category = 'latitude'
N = 50
legend_loc = 'lower right'
species = 'pelag'
category = 'temperature'
N = 50
legend_loc = 'lower right'
species = 'proch'
category = 'latitude'
N = 50
legend_loc = 'upper left'
species = 'proch'
category = 'temperature'
N = 50
legend_loc = 'upper left'
counts = pd.read_csv('/Users/luke/singlecell/notebooks/tara_%s_nonzero_SRF.csv' % species)
tara_metadata = pd.read_csv('/Users/luke/singlecell/notebooks/tara_metadata_SRF.tsv', sep='\t')
tara_metadata = tara_metadata.sort_values(by=r'Sample label [TARA_station#_environmental-feature_size-fraction]')
og_metadata = pd.read_csv('/Users/luke/singlecell/notebooks/og_metadata.tsv', sep='\t', index_col=0)
f = lambda x: x.split('_')[1]
tara_metadata['SampleNumber'] = tara_metadata[r'Sample label [TARA_station#_environmental-feature_size-fraction]'].apply(f)
counts['SampleNumber'] = counts[r'Unnamed: 0'].apply(f)
tara_metadata = tara_metadata.sort_values(by=r'SampleNumber')
counts = counts.sort_values(by=r'SampleNumber')
mat = counts.values[:, 1:-1]
mat = mat.astype(dtype=np.float)
u1, k1, v1 = np.linalg.svd(clr(centralize(mat+1)))
n = len(u1)
G = np.sqrt(n - 1) * u1[:, :2]
H = np.vstack(((np.sqrt(k1[0]) * v1[0, :]) / np.sqrt(n - 1), (np.sqrt(k1[1]) * v1[1, :]) / np.sqrt(n - 1)))
# Get top N hits
rho, theta = cart2polar(H[0, :], H[1, :])
z = rho.argsort()[-N:]
# Calculate radii and degrees to easier identification on the biplot
feats = pd.DataFrame({'rho':rho[z], 'theta':theta[z]*180/np.pi}, index=counts.columns[1:-1][z])
feats['Description'] = og_metadata['Description'][feats.index]
feats.sort_values(by='rho', ascending=False, inplace=True)
feats.to_csv('/Users/luke/singlecell/notebooks/ordination_of_tara_biplot_ogs_%s_%s.tsv' % (species, category), sep='\t')
# cat lookup
if category == 'temperature':
dict_cat = {
'polar': u"<10 \N{DEGREE SIGN}C, 'polar' samples",
'temperate': u"10-20 \N{DEGREE SIGN}C, 'temperate' samples",
'tropical': u">20 \N{DEGREE SIGN}C, 'tropical' samples"
}
colors = ['light orange', 'light blue', 'blue']
elif category == 'latitude':
dict_cat = {
'temperate': u'temperate samples',
'subtropical': u'subtropical samples',
'tropical': u'tropical samples'
}
colors = ['blue', 'light blue', 'light orange']
# plot
plt.figure(figsize=(10, 10))
marker_rs = dict(color=sns.xkcd_rgb['light orange'], marker='o', markeredgewidth=1, markeredgecolor='black')
idx = tara_metadata['category_redsea'] == True
p1 = plt.plot(u1[idx.values, 0], u1[idx.values, 1], '.', label='Red Sea samples', markersize=10, zorder=10, **marker_rs)
i = 0
for cat in set(tara_metadata['category_%s' % category]):
idx = tara_metadata['category_%s' % category] == cat
plt.plot(u1[idx.values, 0], u1[idx.values, 1], 'o', label=dict_cat[cat], markersize=10, color=sns.xkcd_rgb[colors[i]])
i += 1
for j in z:
_x, _y = H[0, j], H[1, j]
plt.arrow(0, 0, _x, _y, head_width=0.01)
perc_explained = k1**2 / (k1**2).sum()
plt.xlabel('PC 1 ({:.2%})'.format(perc_explained[0]), fontsize=18)
plt.ylabel('PC 2 ({:.2%})'.format(perc_explained[1]), fontsize=18)
plt.xticks(fontsize=14)
plt.yticks(fontsize=14)
plt.legend(fontsize=16, loc=legend_loc)
plt.savefig('/Users/luke/singlecell/notebooks/ordination_of_tara_samples_%s_%s.pdf' % (species, category))
# ordination results
u = pd.DataFrame(u1, index=counts.index)
proportion_explained=pd.Series(perc_explained, index=counts.index)
eigvals=pd.Series(k1, index=counts.index)
res = OrdinationResults('','',samples=u, proportion_explained=proportion_explained, eigvals=eigvals)
res.write('/Users/luke/singlecell/notebooks/ordination_of_tara_results_%s_%s.pc' % (species, category))
| 0.523177 | 0.514522 |
# Intel® Extension for Scikit-learn KNN for MNIST dataset
```
from timeit import default_timer as timer
from IPython.display import HTML
from sklearn import metrics
from sklearn.datasets import fetch_openml
from sklearn.model_selection import train_test_split
```
### Download the data
```
x, y = fetch_openml(name='mnist_784', return_X_y=True)
```
Split the data into train and test sets
```
x_train, x_test, y_train, y_test = train_test_split(x, y, test_size=0.2, random_state=72)
x_train.shape, x_test.shape, y_train.shape, y_test.shape
```
### Patch original Scikit-learn with Intel® Extension for Scikit-learn
Intel® Extension for Scikit-learn (previously known as daal4py) contains drop-in replacement functionality for the stock Scikit-learn package. You can take advantage of the performance optimizations of Intel® Extension for Scikit-learn by adding just two lines of code before the usual Scikit-learn imports:
```
from sklearnex import patch_sklearn
patch_sklearn()
```
Intel® Extension for Scikit-learn patching affects performance of specific Scikit-learn functionality. Refer to the [list of supported algorithms and parameters](https://intel.github.io/scikit-learn-intelex/algorithms.html) for details. In cases when unsupported parameters are used, the package fallbacks into original Scikit-learn. If the patching does not cover your scenarios, [submit an issue on GitHub](https://github.com/intel/scikit-learn-intelex/issues).
Training and predict KNN algorithm with Intel® Extension for Scikit-learn for MNIST dataset
```
from sklearn.neighbors import KNeighborsClassifier
params = {
'n_neighbors': 40,
'weights': 'distance',
'n_jobs': -1
}
start = timer()
knn = KNeighborsClassifier(**params).fit(x_train, y_train)
predicted = knn.predict(x_test)
time_opt = timer() - start
f"Intel® extension for Scikit-learn time: {time_opt:.2f} s"
report = metrics.classification_report(y_test, predicted)
print(f"Classification report for Intel® extension for Scikit-learn KNN:\n{report}\n")
```
*The first column of the classification report above is the class labels.*
### Train the same algorithm with original Scikit-learn
In order to cancel optimizations, we use *unpatch_sklearn* and reimport the class KNeighborsClassifier.
```
from sklearnex import unpatch_sklearn
unpatch_sklearn()
```
Training and predict KNN algorithm with original Scikit-learn library for MNSIT dataset
```
from sklearn.neighbors import KNeighborsClassifier
start = timer()
knn = KNeighborsClassifier(**params).fit(x_train, y_train)
predicted = knn.predict(x_test)
time_original = timer() - start
f"Original Scikit-learn time: {time_original:.2f} s"
report = metrics.classification_report(y_test, predicted)
print(f"Classification report for original Scikit-learn KNN:\n{report}\n")
HTML(f'<h2>With scikit-learn-intelex patching you can:</h2>'
f"<ul>"
f"<li>Use your Scikit-learn code for training and prediction with minimal changes (a couple of lines of code);</li>"
f"<li>Fast execution training and prediction of Scikit-learn models;</li>"
f"<li>Get the similar quality</li>"
f"<li>Get speedup in <strong>{(time_original/time_opt):.1f}</strong> times.</li>"
f"</ul>")
```
|
github_jupyter
|
from timeit import default_timer as timer
from IPython.display import HTML
from sklearn import metrics
from sklearn.datasets import fetch_openml
from sklearn.model_selection import train_test_split
x, y = fetch_openml(name='mnist_784', return_X_y=True)
x_train, x_test, y_train, y_test = train_test_split(x, y, test_size=0.2, random_state=72)
x_train.shape, x_test.shape, y_train.shape, y_test.shape
from sklearnex import patch_sklearn
patch_sklearn()
from sklearn.neighbors import KNeighborsClassifier
params = {
'n_neighbors': 40,
'weights': 'distance',
'n_jobs': -1
}
start = timer()
knn = KNeighborsClassifier(**params).fit(x_train, y_train)
predicted = knn.predict(x_test)
time_opt = timer() - start
f"Intel® extension for Scikit-learn time: {time_opt:.2f} s"
report = metrics.classification_report(y_test, predicted)
print(f"Classification report for Intel® extension for Scikit-learn KNN:\n{report}\n")
from sklearnex import unpatch_sklearn
unpatch_sklearn()
from sklearn.neighbors import KNeighborsClassifier
start = timer()
knn = KNeighborsClassifier(**params).fit(x_train, y_train)
predicted = knn.predict(x_test)
time_original = timer() - start
f"Original Scikit-learn time: {time_original:.2f} s"
report = metrics.classification_report(y_test, predicted)
print(f"Classification report for original Scikit-learn KNN:\n{report}\n")
HTML(f'<h2>With scikit-learn-intelex patching you can:</h2>'
f"<ul>"
f"<li>Use your Scikit-learn code for training and prediction with minimal changes (a couple of lines of code);</li>"
f"<li>Fast execution training and prediction of Scikit-learn models;</li>"
f"<li>Get the similar quality</li>"
f"<li>Get speedup in <strong>{(time_original/time_opt):.1f}</strong> times.</li>"
f"</ul>")
| 0.651022 | 0.965835 |
# Linked List
```
class Node:
def __init__(self, value):
self.value = value
self.nextNode = None
def cycleCheck(node):
p1 = node
p2 = node
while p2 != None and p2.nextNode != None:
p1 = p1.nextNode
p2 = p2.nextNode.nextNode
return p1 == p2
def reverseLinkedList(node):
curr = head
prev = None
while curr:
temp = curr.nextNode
curr.nextNode = prev
prev = curr
curr = temp
return prev
def nth_to_last(n):
lp = head
rp = head
for i in range(n-1):
if not rp.nextNode:
raise LookupError('Error n: is greater than linked list')
rp = rp.nextNode
while rp.nextNode:
lp = lp.nextNode
rp = rp.nextNode
return lp
```
<br>
# Pairwise Swap Elements
```
class Node:
def __init__(self, data):
self.data = data
self.next = None
class LinkedList:
def __init__(self):
self.head = None
def printList(self):
temp = self.head
while(temp):
print(temp.data)
temp = temp.next
def push(self, new_data):
new_node = Node(new_data)
new_node.next = self.head
self.head = new_node
def pairwiseSwap(self):
temp = self.head
if self.head is None:
return
while temp and temp.next:
if temp.data != temp.next.data:
temp.data, temp.next.data = temp.next.data, temp.data
temp = temp.next.next
llist = LinkedList()
llist.push(5)
llist.push(4)
llist.push(3)
llist.push(2)
llist.push(1)
llist.pairwiseSwap()
llist.printList()
```
# Merge two sorted linked list
```
class Node:
def __init__(self, data):
self.data = data
self.next = None
class LinkedList:
def __init__(self):
self.head = None
def printList(self):
temp = self.head
while temp:
print(temp.data, end=" ")
temp = temp.next
def addToList(self, data):
newNode = Node(data)
if self.head is None:
self.head = newNode
return
last = self.head
while last.next:
last = last.next
last.next = newNode
def mergeLists(headA, headB):
dummyNode = Node(0)
tail = dummyNode
while True:
if headA is None:
tail.next = headB
break
if headB is None:
tail.next = headA
break
if headA.data < headB.data:
tail.next = headA
headA = headA.next
else:
tail.next = headB
headB = headB.next
tail = tail.next
return dummyNode.next
def mergeLists_Rec(head1, head2):
temp = None
if head1 is None:
return head2
if head2 is None:
return head1
if head1.data <= head2.data:
temp = head1
temp.next = mergeLists_Rec(head1.next, head2)
else:
temp = head2
temp.next = mergeLists_Rec(head1, head2.next)
return temp
listA = LinkedList()
listB = LinkedList()
listC = LinkedList()
listA.addToList(5)
listA.addToList(10)
listA.addToList(15)
listB.addToList(2)
listB.addToList(3)
listB.addToList(20)
listA.head = mergeLists(listA.head, listB.head)
listA.printList()
#print("\n\n")
#listC.head = mergeLists_Rec(listA.head, listB.head)
#listC.printList()
```
|
github_jupyter
|
class Node:
def __init__(self, value):
self.value = value
self.nextNode = None
def cycleCheck(node):
p1 = node
p2 = node
while p2 != None and p2.nextNode != None:
p1 = p1.nextNode
p2 = p2.nextNode.nextNode
return p1 == p2
def reverseLinkedList(node):
curr = head
prev = None
while curr:
temp = curr.nextNode
curr.nextNode = prev
prev = curr
curr = temp
return prev
def nth_to_last(n):
lp = head
rp = head
for i in range(n-1):
if not rp.nextNode:
raise LookupError('Error n: is greater than linked list')
rp = rp.nextNode
while rp.nextNode:
lp = lp.nextNode
rp = rp.nextNode
return lp
class Node:
def __init__(self, data):
self.data = data
self.next = None
class LinkedList:
def __init__(self):
self.head = None
def printList(self):
temp = self.head
while(temp):
print(temp.data)
temp = temp.next
def push(self, new_data):
new_node = Node(new_data)
new_node.next = self.head
self.head = new_node
def pairwiseSwap(self):
temp = self.head
if self.head is None:
return
while temp and temp.next:
if temp.data != temp.next.data:
temp.data, temp.next.data = temp.next.data, temp.data
temp = temp.next.next
llist = LinkedList()
llist.push(5)
llist.push(4)
llist.push(3)
llist.push(2)
llist.push(1)
llist.pairwiseSwap()
llist.printList()
class Node:
def __init__(self, data):
self.data = data
self.next = None
class LinkedList:
def __init__(self):
self.head = None
def printList(self):
temp = self.head
while temp:
print(temp.data, end=" ")
temp = temp.next
def addToList(self, data):
newNode = Node(data)
if self.head is None:
self.head = newNode
return
last = self.head
while last.next:
last = last.next
last.next = newNode
def mergeLists(headA, headB):
dummyNode = Node(0)
tail = dummyNode
while True:
if headA is None:
tail.next = headB
break
if headB is None:
tail.next = headA
break
if headA.data < headB.data:
tail.next = headA
headA = headA.next
else:
tail.next = headB
headB = headB.next
tail = tail.next
return dummyNode.next
def mergeLists_Rec(head1, head2):
temp = None
if head1 is None:
return head2
if head2 is None:
return head1
if head1.data <= head2.data:
temp = head1
temp.next = mergeLists_Rec(head1.next, head2)
else:
temp = head2
temp.next = mergeLists_Rec(head1, head2.next)
return temp
listA = LinkedList()
listB = LinkedList()
listC = LinkedList()
listA.addToList(5)
listA.addToList(10)
listA.addToList(15)
listB.addToList(2)
listB.addToList(3)
listB.addToList(20)
listA.head = mergeLists(listA.head, listB.head)
listA.printList()
#print("\n\n")
#listC.head = mergeLists_Rec(listA.head, listB.head)
#listC.printList()
| 0.421076 | 0.78695 |
# Overview
This week we'll get started on modern network science. We'll focus on two key results that kick-started a revolution in our understanding of networks.
* Problems with random networks as models for real networks and the Watts-Strogatz model
* Scale-free networks and the Barabasi-Albert model
But before we can get started, there's the bookkeeping stuff, the admin things. Do watch it - the material below tells you about key elements of how to do well in class, assignments and stuff.
And the good news is that after today, we can take it easy with admin stuff for a while. We'll restart this aspect when we get closer to the project assignments that we finish the class with.
# Part 0: The admin stuff
### (Why we use Peer Evaluations Edition)
We use the system [Peergrade.io](http://peergrade.io/) to get you better feedback and make you smarter. In the video below, I explain why that is the case. There are a lot of good reasons that peer evaluations are great, so watch the video :)
```
from IPython.display import YouTubeVideo
YouTubeVideo("-TC18KgpiIQ",width=800, height=450)
```
# Part 1: Small world networks
Once again, we'll start with some lecturing. So it's time to watch a little video to get you started.
> **_Video Lecture_**: Some properties of real world networks. Clustering and small paths. The Watts-Strogatz model.
>
```
YouTubeVideo("tMLJ2NYI1FU",width=800, height=450)
```
Next up is fun with reading the textbook. There's lots of goodies left in Chapter 3 that covers the stuff I've just covered in the video from a slightly different angle.
>
> *Reading*: For this part, we'll read the remaining part of *Network Science* Chapter 3, Section 3.5 - 3.10, with ***emphasis*** on 3.8 and 3.9\.
>
> *Exercises*: Did you really read the text? Answer the following questions (no calculations needed) in your IPython notebook.
>
> * What's the problem with random networks as a model for real-world networks according to the argument in section 3.5 (near the end)?
> * List the four regimes that characterize random networks as a function of $\langle k \rangle$.
> * According to the book, why is it a problem for random networks (in terms of being a model for real-world networks) that the degree-dependent clustering $C(k)$ decreases as a function of $k$ in real-world networks?
>
The next set of exercises departs from the book by presenting a little study of the Watts-Strogatz (WS) model. We're going to see just how few random links the WS model needs to dramatically shorten the path-lengths in the network. And while doing that, we'll practice using `networkx`, writing loops, and plotting nice figures.
> *Exercises*: WS edition.
>
> * First, let's use `networkx` to play around with WS graphs. Use `nx.watts_strogatz_graph` to generate 3 graphs with 500 nodes each, average degree = 4, and rewiring probablity $p = 0, 0.1,$ and $1$. Calculate the average shortest path length $\langle d \rangle$ for each one. Describe what happens to the network when $p = 1$.
> * Generate a lot of networks with different values of $p$. You will notice that paths are short when $p$ is close to one and they are long when $p = 0$. What's the value of $p$ for which the average shortest path length gets close to the short paths we find in a fully randomized network.
> * Let's investigate this behavior in detail. Generate 50 networks with $N = 500$, $\langle k \rangle = 4$, for each of $p = \{0, 0.01, 0.03, 0.05, 0.1, 0.2\}$. Calculate the average of $\langle d \rangle$ as well as the standard deviation over the 50 networks, to create a plot that shows how the path length decreases very quickly with only a little fraction of re-wiring. Use the standard deviation to add [errorbars](https://matplotlib.org/3.1.1/api/_as_gen/matplotlib.pyplot.errorbar.html) to the plot. My version of the plot is below (since a picture's worth 1000 words).
>
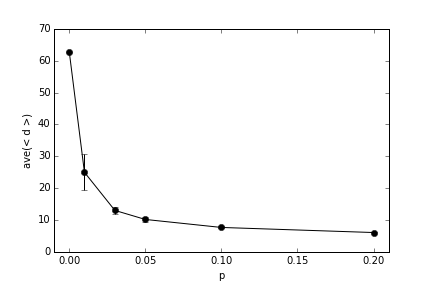
```
import networkx as nx
import matplotlib.pyplot as plt
nodes = 500
avgdegree = 4
p = [0, 0.1, 1]
fig, ax = plt.subplots(2,2)
axs = [ax[0,0], ax[0,1], ax[1,0]]
ax[1,1].set_visible(False)
for prob, axis in zip(p,axs):
ws = nx.watts_strogatz_graph(nodes, avgdegree, prob)
print("For graph with p= {}, the average shortest path is {}".format(prob, nx.average_shortest_path_length(ws)))
nx.draw(ws, node_size=10, ax = axis)
import statistics
nodes = 500
avgdegree = 4
p = [0,0.01,0.03,0.05,0.1,0.2]
d = {}
for prob in p:
d[prob] = []
for i in range(50):
ws = nx.watts_strogatz_graph(nodes, avgdegree, prob)
d[prob].append(nx.average_shortest_path_length(ws))
Davg = []
Dstd = []
for L in d.values():
Davg.append(sum(L) / len(L))
Dstd.append(statistics.stdev(L))
plt.errorbar(p, Davg, yerr=Dstd)
plt.xlabel("p")
plt.ylabel("average(< d >)")
```
## Part 2: Scale-free networks
The text book uses two whole chapters on the scale free property. We'll try and power through during the remainder of this lecture. As always, let's start by getting my take on the whole thing.
> **_Video Lecture_**: The scale free property and the Barabasi-Albert Model.
>
```
YouTubeVideo("myLgzbXxhOQ",width=800, height=450)
```
And now it's time for you guys to read. Recall that Barabasi (who wrote the textbook) discovered power-laws.
> *Reading*: Now we dig into the extended history and and theory behind Scale-Free networks and the Barabasi-Albert Model.
>
> * Chapter 4, Section 4.1 - 4.7\.
> * Chapter 5, section 5.1 - 5.5\.
>
> *Exercises*: BA edition.
>
> First a couple of questions to make sure that you've actually read the text.
>
> * What are the three slope dependent regimes of complex networks with power-law degree distributions? Briefly describe each one. (You will have to skim chp 4.7 to answer this one).
> * What are the three regimes we find in non-linear preferential attachement? (chapter 5) Briefly describe each one.
>
> We're going to create our own Barabasi-Albert model (a special case) in right in a `notebook`. Follow the recipe below for success:
>
> * First create a graph consisting of a single link. (You can call the nodes anything, but I would simply use integers as names).
> * Now add another node, connecting one of the existing nodes in proportion to their degree.
> * Keep going until you have a 100 node network.
> * *Hint*: The difficult part here is connecting to each node according to their degree. The way I do it is: generate a list of all edges (e.g. pairs of nodes), then flatten it (e.g. remove connection information). That list contains each node in proportion to its connections, thus drawing a random node from that list (e.g. using `random.choice`) corresponds to selecting a node with probability proportional to it's degree.
> * Plot the network.
> * Add more nodes until you have a 5000 node network.
> * What's the maximum and minimum degree?
> * Now, bin the degree distribution using `numpy.histogram`.
> * Plot the distribution. Plot it with both linear and log-log axes.
>
> 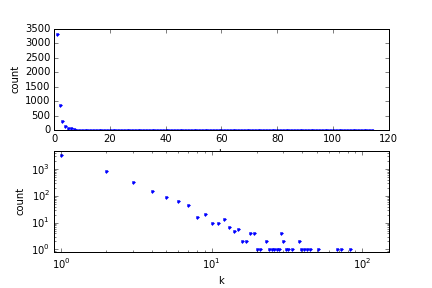
>
> Next step is to explore the [Friendship paradox](https://en.wikipedia.org/wiki/Friendship_paradox). This paradox states that _almost everyone_ has fewer friends than their friends have, on average\*. This sounds crazy, but is actually an almost trivial consequence of living in a social network with a power-law degree distribution. The explanation is that almost everyone is friends with a hub, that drives up the average degree of the friends. Let's explore that in the 5000 node BA network we've just generated. Do the following:
>
> * Pick a node $i$ at random (e.g. use `random.choice`). [Find its degree](https://networkx.github.io/documentation/stable/reference/classes/generated/networkx.Graph.degree.html).
> * Find $i$'s [neighbors](https://networkx.github.io/documentation/networkx-1.10/reference/generated/networkx.Graph.neighbors.html?highlight=neighbors#networkx.Graph.neighbors). And calculate their average degree.
> * Compare the two numbers to check if it's true that $i$'s friends (on average) have more friends than $i$.
> * Do this 1000 times. How many out of those 1000 times is the friendship paradox true?
>
> Finally, we'll build a network of same size and degree, using the growth mechanism without the preferential attachment.
>
> * Compare to the ER network of same size and same $p$. What are the differences? Explain in your own words. *Hint*: To get started, take a look at the degree distribution, and study the number of connected components.
>
```
import random
G = nx.Graph()
G.add_nodes_from([1,2])
G.add_edge(1,2)
for i in range (3,101):
edgelist = nx.utils.misc.flatten(G.edges)
randChoice = random.choice(edgelist)
G.add_node(i)
G.add_edge(i, randChoice)
nx.draw(G, node_size=20)
for i in range (101,5001):
edgelist = nx.utils.misc.flatten(G.edges)
randChoice = random.choice(edgelist)
G.add_node(i)
G.add_edge(i, randChoice)
NodeDegree = G.degree
print(f"Max degree is {max(NodeDegree)}, min degree is {min(NodeDegree)} (Node, Degree)")
degreeList = [val for (node, val) in NodeDegree]
import numpy as np
NodeHist, bin_edges = np.histogram(degreeList, bins='auto')
print(NodeHist)
print(bin_edges)
plt.hist(degreeList, bins="auto")
plt.hist(degreeList, bins="auto")
plt.xscale('log')
plt.yscale('log')
from networkx.algorithms.assortativity import average_neighbor_degree
nodes = nx.utils.misc.flatten(G.nodes)
tries = 1000
count = 0
for i in range(tries):
randNode = random.choice(nodes)
mainDegree = G.degree(randNode)
neighborhoodDegree = average_neighbor_degree(G, nodes=[randNode])
if mainDegree < neighborhoodDegree[randNode]:
count += 1
print(f"Out of {tries} tries the average neighborhood degree \nwas higher than the original node {count} times. ({count/tries * 100}%)")
```
|
github_jupyter
|
from IPython.display import YouTubeVideo
YouTubeVideo("-TC18KgpiIQ",width=800, height=450)
YouTubeVideo("tMLJ2NYI1FU",width=800, height=450)
import networkx as nx
import matplotlib.pyplot as plt
nodes = 500
avgdegree = 4
p = [0, 0.1, 1]
fig, ax = plt.subplots(2,2)
axs = [ax[0,0], ax[0,1], ax[1,0]]
ax[1,1].set_visible(False)
for prob, axis in zip(p,axs):
ws = nx.watts_strogatz_graph(nodes, avgdegree, prob)
print("For graph with p= {}, the average shortest path is {}".format(prob, nx.average_shortest_path_length(ws)))
nx.draw(ws, node_size=10, ax = axis)
import statistics
nodes = 500
avgdegree = 4
p = [0,0.01,0.03,0.05,0.1,0.2]
d = {}
for prob in p:
d[prob] = []
for i in range(50):
ws = nx.watts_strogatz_graph(nodes, avgdegree, prob)
d[prob].append(nx.average_shortest_path_length(ws))
Davg = []
Dstd = []
for L in d.values():
Davg.append(sum(L) / len(L))
Dstd.append(statistics.stdev(L))
plt.errorbar(p, Davg, yerr=Dstd)
plt.xlabel("p")
plt.ylabel("average(< d >)")
YouTubeVideo("myLgzbXxhOQ",width=800, height=450)
import random
G = nx.Graph()
G.add_nodes_from([1,2])
G.add_edge(1,2)
for i in range (3,101):
edgelist = nx.utils.misc.flatten(G.edges)
randChoice = random.choice(edgelist)
G.add_node(i)
G.add_edge(i, randChoice)
nx.draw(G, node_size=20)
for i in range (101,5001):
edgelist = nx.utils.misc.flatten(G.edges)
randChoice = random.choice(edgelist)
G.add_node(i)
G.add_edge(i, randChoice)
NodeDegree = G.degree
print(f"Max degree is {max(NodeDegree)}, min degree is {min(NodeDegree)} (Node, Degree)")
degreeList = [val for (node, val) in NodeDegree]
import numpy as np
NodeHist, bin_edges = np.histogram(degreeList, bins='auto')
print(NodeHist)
print(bin_edges)
plt.hist(degreeList, bins="auto")
plt.hist(degreeList, bins="auto")
plt.xscale('log')
plt.yscale('log')
from networkx.algorithms.assortativity import average_neighbor_degree
nodes = nx.utils.misc.flatten(G.nodes)
tries = 1000
count = 0
for i in range(tries):
randNode = random.choice(nodes)
mainDegree = G.degree(randNode)
neighborhoodDegree = average_neighbor_degree(G, nodes=[randNode])
if mainDegree < neighborhoodDegree[randNode]:
count += 1
print(f"Out of {tries} tries the average neighborhood degree \nwas higher than the original node {count} times. ({count/tries * 100}%)")
| 0.323915 | 0.987946 |
# Teaching machine learning with weather predicition dataset
- Task: predict BBQ weather (True/False) for the next day based on single-day weather observations.
- Technique: Random forest classifier
- Tool: Python + Scikit-learn
```
import pandas as pd
import os
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import MinMaxScaler
from sklearn.pipeline import Pipeline
from sklearn.metrics import plot_confusion_matrix
from sklearn.ensemble import RandomForestClassifier
```
## Pipeline for using machine learning on weather prediction dataset
### Load data
```
# Read data
path_data = os.path.join('..', "dataset")
filename_data = os.path.join(path_data, "weather_prediction_dataset.csv")
data = pd.read_csv(filename_data)
data.head()
```
### Load extra BBQ labels
```
filename_data = os.path.join(path_data, "weather_prediction_bbq_labels.csv")
labels_bbq = pd.read_csv(filename_data)
labels_bbq.head()
```
### Split the data
We only select the first three years of data. We split into train and test, and then into X and y.
```
data_3years = data[:365*3].drop(columns=['DATE', 'MONTH'])
labels_3years = labels_bbq[1:(365 * 3 + 1)].drop(columns=['DATE'])
X, X_test, y, y_test = train_test_split(data_3years, labels_3years["DUSSELDORF_BBQ_weather"],
test_size=0.3, random_state=0)
#y = y.astype(int)
#y_test = y_test.astype(int)
X.shape, X_test.shape, y.shape, y_test.shape
y
```
## Define a pipeline
We define an sklearn pipeline with two steps:
- normalize the features to a 0-1 range
- A [Random Forest classifier](https://scikit-learn.org/stable/modules/generated/sklearn.ensemble.RandomForestClassifier.html) with 10 trees with a maximum depth of 5
```
# We give the pipeline tuples of step names, and step objects
pipe = Pipeline([
('scale', MinMaxScaler()),
('model', RandomForestClassifier(n_estimators=50, max_depth=6, random_state=0))
])
pipe.fit(X, y)
```
We can use this pipe directly to predict the class label, or plot a confusion matrix. We first plot the confusion matrix for the train set:
```
predictions = pipe.predict(X)
predictions[:10]
from matplotlib import pyplot as plt
labels = ["True month", "Predicted month"]
conf_mat = plot_confusion_matrix(pipe, X, y)
plt.xlabel("Predicted BBQ weather")
plt.ylabel("True BBQ weather")
plt.savefig("random_forest_month_classification_bbq.png", dpi=300)
```
To now test in on our test set:
```
pred_test = pipe.predict(X_test)
```
We plot a confusion matrix for the predictions on the test set. To make sure the classes are nicely ordered, we define the labels explitely.
```
labels = y.unique()
labels = labels.sort()
conf_mat = plot_confusion_matrix(pipe, X_test, y_test,
labels=labels,
cmap="Blues", colorbar=False)
plt.xlabel("Predicted BBQ weather")
plt.ylabel("True BBQ weather")
plt.tight_layout()
plt.savefig("random_forest_month_classification_bbq_testset.png", dpi=300)
plt.savefig("random_forest_month_classification_bbq_testset.pdf")
import numpy as np
recall = conf_mat.confusion_matrix[1,1] / np.sum(conf_mat.confusion_matrix[1,:])
print(f"recall: {recall:.4f}")
precision = conf_mat.confusion_matrix[1,1] / np.sum(conf_mat.confusion_matrix[:, 1])
print(f"precision: {precision:.4f}")
```
It seems that the confusion matrix is worse than the one on the train set, but it is difficult to draw this conclusion from the confusion matrix alone. It would be nice to have one score that expresses how well our model does.
We calculate *accuracy*: this is the number of correctly labeled instances, divided the total number of instances. Note that this score works well when we have a balanced dataset (similar number of instances per class) but if we have imbalanced class, the majority class can dominate the score.
```
from sklearn.metrics import accuracy_score
pred_test = pipe.predict(X_test)
print('Train accuracy:', accuracy_score(y, predictions))
print('Test accuracy:', accuracy_score(y_test, pred_test))
```
This results looks quiet OK-ish, but
- there is a clear sign of overfitting (performance on training data much better than on test data)
- there is a bias in the data (most days are not BBQ weather).
|
github_jupyter
|
import pandas as pd
import os
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import MinMaxScaler
from sklearn.pipeline import Pipeline
from sklearn.metrics import plot_confusion_matrix
from sklearn.ensemble import RandomForestClassifier
# Read data
path_data = os.path.join('..', "dataset")
filename_data = os.path.join(path_data, "weather_prediction_dataset.csv")
data = pd.read_csv(filename_data)
data.head()
filename_data = os.path.join(path_data, "weather_prediction_bbq_labels.csv")
labels_bbq = pd.read_csv(filename_data)
labels_bbq.head()
data_3years = data[:365*3].drop(columns=['DATE', 'MONTH'])
labels_3years = labels_bbq[1:(365 * 3 + 1)].drop(columns=['DATE'])
X, X_test, y, y_test = train_test_split(data_3years, labels_3years["DUSSELDORF_BBQ_weather"],
test_size=0.3, random_state=0)
#y = y.astype(int)
#y_test = y_test.astype(int)
X.shape, X_test.shape, y.shape, y_test.shape
y
# We give the pipeline tuples of step names, and step objects
pipe = Pipeline([
('scale', MinMaxScaler()),
('model', RandomForestClassifier(n_estimators=50, max_depth=6, random_state=0))
])
pipe.fit(X, y)
predictions = pipe.predict(X)
predictions[:10]
from matplotlib import pyplot as plt
labels = ["True month", "Predicted month"]
conf_mat = plot_confusion_matrix(pipe, X, y)
plt.xlabel("Predicted BBQ weather")
plt.ylabel("True BBQ weather")
plt.savefig("random_forest_month_classification_bbq.png", dpi=300)
pred_test = pipe.predict(X_test)
labels = y.unique()
labels = labels.sort()
conf_mat = plot_confusion_matrix(pipe, X_test, y_test,
labels=labels,
cmap="Blues", colorbar=False)
plt.xlabel("Predicted BBQ weather")
plt.ylabel("True BBQ weather")
plt.tight_layout()
plt.savefig("random_forest_month_classification_bbq_testset.png", dpi=300)
plt.savefig("random_forest_month_classification_bbq_testset.pdf")
import numpy as np
recall = conf_mat.confusion_matrix[1,1] / np.sum(conf_mat.confusion_matrix[1,:])
print(f"recall: {recall:.4f}")
precision = conf_mat.confusion_matrix[1,1] / np.sum(conf_mat.confusion_matrix[:, 1])
print(f"precision: {precision:.4f}")
from sklearn.metrics import accuracy_score
pred_test = pipe.predict(X_test)
print('Train accuracy:', accuracy_score(y, predictions))
print('Test accuracy:', accuracy_score(y_test, pred_test))
| 0.698329 | 0.984246 |
# Mask R-CNN - Inspect Nucleus Trained Model
Code and visualizations to test, debug, and evaluate the Mask R-CNN model.
```
import os
import sys
import random
import math
import re
import time
import numpy as np
import tensorflow as tf
import matplotlib
import matplotlib.pyplot as plt
import matplotlib.patches as patches
# Root directory of the project
ROOT_DIR = os.path.abspath("../../")
# Import Mask RCNN
sys.path.append(ROOT_DIR) # To find local version of the library
from mrcnn import utils
from mrcnn import visualize
from mrcnn.visualize import display_images
import mrcnn.model as modellib
from mrcnn.model import log
import nucleus
%matplotlib inline
# Directory to save logs and trained model
LOGS_DIR = os.path.join(ROOT_DIR, "logs")
# Comment out to reload imported modules if they change
# %load_ext autoreload
# %autoreload 2
```
## Configurations
```
# Dataset directory
DATASET_DIR = os.path.join(ROOT_DIR, "datasets/nucleus")
# Inference Configuration
config = nucleus.NucleusInferenceConfig()
config.display()
```
## Notebook Preferences
```
# Device to load the neural network on.
# Useful if you're training a model on the same
# machine, in which case use CPU and leave the
# GPU for training.
DEVICE = "/gpu:0" # /cpu:0 or /gpu:0
# Inspect the model in training or inference modes
# values: 'inference' or 'training'
# Only inference mode is supported right now
TEST_MODE = "inference"
def get_ax(rows=1, cols=1, size=16):
"""Return a Matplotlib Axes array to be used in
all visualizations in the notebook. Provide a
central point to control graph sizes.
Adjust the size attribute to control how big to render images
"""
fig, ax = plt.subplots(rows, cols, figsize=(size*cols, size*rows))
fig.tight_layout()
return ax
```
## Load Validation Dataset
```
# Load validation dataset
dataset = nucleus.NucleusDataset()
dataset.load_nucleus(DATASET_DIR, "val")
dataset.prepare()
print("Images: {}\nClasses: {}".format(len(dataset.image_ids), dataset.class_names))
```
## Load Model
```
# Create model in inference mode
with tf.device(DEVICE):
model = modellib.MaskRCNN(mode="inference",
model_dir=LOGS_DIR,
config=config)
# Path to a specific weights file
# weights_path = "/path/to/mask_rcnn_nucleus.h5"
# Or, load the last model you trained
weights_path = model.find_last()
# Load weights
print("Loading weights ", weights_path)
model.load_weights(weights_path, by_name=True)
```
## Run Detection
```
image_id = random.choice(dataset.image_ids)
image, image_meta, gt_class_id, gt_bbox, gt_mask =\
modellib.load_image_gt(dataset, config, image_id, use_mini_mask=False)
info = dataset.image_info[image_id]
print("image ID: {}.{} ({}) {}".format(info["source"], info["id"], image_id,
dataset.image_reference(image_id)))
print("Original image shape: ", modellib.parse_image_meta(image_meta[np.newaxis,...])["original_image_shape"][0])
# Run object detection
results = model.detect_molded(np.expand_dims(image, 0), np.expand_dims(image_meta, 0), verbose=1)
# Display results
r = results[0]
log("gt_class_id", gt_class_id)
log("gt_bbox", gt_bbox)
log("gt_mask", gt_mask)
# Compute AP over range 0.5 to 0.95 and print it
utils.compute_ap_range(gt_bbox, gt_class_id, gt_mask,
r['rois'], r['class_ids'], r['scores'], r['masks'],
verbose=1)
visualize.display_differences(
image,
gt_bbox, gt_class_id, gt_mask,
r['rois'], r['class_ids'], r['scores'], r['masks'],
dataset.class_names, ax=get_ax(),
show_box=False, show_mask=False,
iou_threshold=0.5, score_threshold=0.5)
# Display predictions only
# visualize.display_instances(image, r['rois'], r['masks'], r['class_ids'],
# dataset.class_names, r['scores'], ax=get_ax(1),
# show_bbox=False, show_mask=False,
# title="Predictions")
# Display Ground Truth only
# visualize.display_instances(image, gt_bbox, gt_mask, gt_class_id,
# dataset.class_names, ax=get_ax(1),
# show_bbox=False, show_mask=False,
# title="Ground Truth")
```
### Compute AP on Batch of Images
```
def compute_batch_ap(dataset, image_ids, verbose=1):
APs = []
for image_id in image_ids:
# Load image
image, image_meta, gt_class_id, gt_bbox, gt_mask =\
modellib.load_image_gt(dataset, config,
image_id, use_mini_mask=False)
# Run object detection
results = model.detect_molded(image[np.newaxis], image_meta[np.newaxis], verbose=0)
# Compute AP over range 0.5 to 0.95
r = results[0]
ap = utils.compute_ap_range(
gt_bbox, gt_class_id, gt_mask,
r['rois'], r['class_ids'], r['scores'], r['masks'],
verbose=0)
APs.append(ap)
if verbose:
info = dataset.image_info[image_id]
meta = modellib.parse_image_meta(image_meta[np.newaxis,...])
print("{:3} {} AP: {:.2f}".format(
meta["image_id"][0], meta["original_image_shape"][0], ap))
return APs
# Run on validation set
limit = 5
APs = compute_batch_ap(dataset, dataset.image_ids[:limit])
print("Mean AP overa {} images: {:.4f}".format(len(APs), np.mean(APs)))
```
## Step by Step Prediction
## Stage 1: Region Proposal Network
The Region Proposal Network (RPN) runs a lightweight binary classifier on a lot of boxes (anchors) over the image and returns object/no-object scores. Anchors with high *objectness* score (positive anchors) are passed to the stage two to be classified.
Often, even positive anchors don't cover objects fully. So the RPN also regresses a refinement (a delta in location and size) to be applied to the anchors to shift it and resize it a bit to the correct boundaries of the object.
### 1.a RPN Targets
The RPN targets are the training values for the RPN. To generate the targets, we start with a grid of anchors that cover the full image at different scales, and then we compute the IoU of the anchors with ground truth object. Positive anchors are those that have an IoU >= 0.7 with any ground truth object, and negative anchors are those that don't cover any object by more than 0.3 IoU. Anchors in between (i.e. cover an object by IoU >= 0.3 but < 0.7) are considered neutral and excluded from training.
To train the RPN regressor, we also compute the shift and resizing needed to make the anchor cover the ground truth object completely.
```
# Get anchors and convert to pixel coordinates
anchors = model.get_anchors(image.shape)
anchors = utils.denorm_boxes(anchors, image.shape[:2])
log("anchors", anchors)
# Generate RPN trainig targets
# target_rpn_match is 1 for positive anchors, -1 for negative anchors
# and 0 for neutral anchors.
target_rpn_match, target_rpn_bbox = modellib.build_rpn_targets(
image.shape, anchors, gt_class_id, gt_bbox, model.config)
log("target_rpn_match", target_rpn_match)
log("target_rpn_bbox", target_rpn_bbox)
positive_anchor_ix = np.where(target_rpn_match[:] == 1)[0]
negative_anchor_ix = np.where(target_rpn_match[:] == -1)[0]
neutral_anchor_ix = np.where(target_rpn_match[:] == 0)[0]
positive_anchors = anchors[positive_anchor_ix]
negative_anchors = anchors[negative_anchor_ix]
neutral_anchors = anchors[neutral_anchor_ix]
log("positive_anchors", positive_anchors)
log("negative_anchors", negative_anchors)
log("neutral anchors", neutral_anchors)
# Apply refinement deltas to positive anchors
refined_anchors = utils.apply_box_deltas(
positive_anchors,
target_rpn_bbox[:positive_anchors.shape[0]] * model.config.RPN_BBOX_STD_DEV)
log("refined_anchors", refined_anchors, )
# Display positive anchors before refinement (dotted) and
# after refinement (solid).
visualize.draw_boxes(
image, ax=get_ax(),
boxes=positive_anchors,
refined_boxes=refined_anchors)
```
### 1.b RPN Predictions
Here we run the RPN graph and display its predictions.
```
# Run RPN sub-graph
pillar = model.keras_model.get_layer("ROI").output # node to start searching from
# TF 1.4 introduces a new version of NMS. Search for both names to support TF 1.3 and 1.4
nms_node = model.ancestor(pillar, "ROI/rpn_non_max_suppression:0")
if nms_node is None:
nms_node = model.ancestor(pillar, "ROI/rpn_non_max_suppression/NonMaxSuppressionV2:0")
rpn = model.run_graph(image[np.newaxis], [
("rpn_class", model.keras_model.get_layer("rpn_class").output),
("pre_nms_anchors", model.ancestor(pillar, "ROI/pre_nms_anchors:0")),
("refined_anchors", model.ancestor(pillar, "ROI/refined_anchors:0")),
("refined_anchors_clipped", model.ancestor(pillar, "ROI/refined_anchors_clipped:0")),
("post_nms_anchor_ix", nms_node),
("proposals", model.keras_model.get_layer("ROI").output),
], image_metas=image_meta[np.newaxis])
# Show top anchors by score (before refinement)
limit = 100
sorted_anchor_ids = np.argsort(rpn['rpn_class'][:,:,1].flatten())[::-1]
visualize.draw_boxes(image, boxes=anchors[sorted_anchor_ids[:limit]], ax=get_ax())
# Show top anchors with refinement. Then with clipping to image boundaries
limit = 50
ax = get_ax(1, 2)
visualize.draw_boxes(
image, ax=ax[0],
boxes=utils.denorm_boxes(rpn["pre_nms_anchors"][0, :limit], image.shape[:2]),
refined_boxes=utils.denorm_boxes(rpn["refined_anchors"][0, :limit], image.shape[:2]))
visualize.draw_boxes(
image, ax=ax[1],
refined_boxes=utils.denorm_boxes(rpn["refined_anchors_clipped"][0, :limit], image.shape[:2]))
# Show refined anchors after non-max suppression
limit = 50
ixs = rpn["post_nms_anchor_ix"][:limit]
visualize.draw_boxes(
image, ax=get_ax(),
refined_boxes=utils.denorm_boxes(rpn["refined_anchors_clipped"][0, ixs], image.shape[:2]))
# Show final proposals
# These are the same as the previous step (refined anchors
# after NMS) but with coordinates normalized to [0, 1] range.
limit = 50
# Convert back to image coordinates for display
# h, w = config.IMAGE_SHAPE[:2]
# proposals = rpn['proposals'][0, :limit] * np.array([h, w, h, w])
visualize.draw_boxes(
image, ax=get_ax(),
refined_boxes=utils.denorm_boxes(rpn['proposals'][0, :limit], image.shape[:2]))
```
## Stage 2: Proposal Classification
This stage takes the region proposals from the RPN and classifies them.
### 2.a Proposal Classification
Run the classifier heads on proposals to generate class propbabilities and bounding box regressions.
```
# Get input and output to classifier and mask heads.
mrcnn = model.run_graph([image], [
("proposals", model.keras_model.get_layer("ROI").output),
("probs", model.keras_model.get_layer("mrcnn_class").output),
("deltas", model.keras_model.get_layer("mrcnn_bbox").output),
("masks", model.keras_model.get_layer("mrcnn_mask").output),
("detections", model.keras_model.get_layer("mrcnn_detection").output),
])
# Get detection class IDs. Trim zero padding.
det_class_ids = mrcnn['detections'][0, :, 4].astype(np.int32)
det_count = np.where(det_class_ids == 0)[0][0]
det_class_ids = det_class_ids[:det_count]
detections = mrcnn['detections'][0, :det_count]
print("{} detections: {}".format(
det_count, np.array(dataset.class_names)[det_class_ids]))
captions = ["{} {:.3f}".format(dataset.class_names[int(c)], s) if c > 0 else ""
for c, s in zip(detections[:, 4], detections[:, 5])]
visualize.draw_boxes(
image,
refined_boxes=utils.denorm_boxes(detections[:, :4], image.shape[:2]),
visibilities=[2] * len(detections),
captions=captions, title="Detections",
ax=get_ax())
```
### 2.c Step by Step Detection
Here we dive deeper into the process of processing the detections.
```
# Proposals are in normalized coordinates
proposals = mrcnn["proposals"][0]
# Class ID, score, and mask per proposal
roi_class_ids = np.argmax(mrcnn["probs"][0], axis=1)
roi_scores = mrcnn["probs"][0, np.arange(roi_class_ids.shape[0]), roi_class_ids]
roi_class_names = np.array(dataset.class_names)[roi_class_ids]
roi_positive_ixs = np.where(roi_class_ids > 0)[0]
# How many ROIs vs empty rows?
print("{} Valid proposals out of {}".format(np.sum(np.any(proposals, axis=1)), proposals.shape[0]))
print("{} Positive ROIs".format(len(roi_positive_ixs)))
# Class counts
print(list(zip(*np.unique(roi_class_names, return_counts=True))))
# Display a random sample of proposals.
# Proposals classified as background are dotted, and
# the rest show their class and confidence score.
limit = 200
ixs = np.random.randint(0, proposals.shape[0], limit)
captions = ["{} {:.3f}".format(dataset.class_names[c], s) if c > 0 else ""
for c, s in zip(roi_class_ids[ixs], roi_scores[ixs])]
visualize.draw_boxes(
image,
boxes=utils.denorm_boxes(proposals[ixs], image.shape[:2]),
visibilities=np.where(roi_class_ids[ixs] > 0, 2, 1),
captions=captions, title="ROIs Before Refinement",
ax=get_ax())
```
#### Apply Bounding Box Refinement
```
# Class-specific bounding box shifts.
roi_bbox_specific = mrcnn["deltas"][0, np.arange(proposals.shape[0]), roi_class_ids]
log("roi_bbox_specific", roi_bbox_specific)
# Apply bounding box transformations
# Shape: [N, (y1, x1, y2, x2)]
refined_proposals = utils.apply_box_deltas(
proposals, roi_bbox_specific * config.BBOX_STD_DEV)
log("refined_proposals", refined_proposals)
# Show positive proposals
# ids = np.arange(roi_boxes.shape[0]) # Display all
limit = 5
ids = np.random.randint(0, len(roi_positive_ixs), limit) # Display random sample
captions = ["{} {:.3f}".format(dataset.class_names[c], s) if c > 0 else ""
for c, s in zip(roi_class_ids[roi_positive_ixs][ids], roi_scores[roi_positive_ixs][ids])]
visualize.draw_boxes(
image, ax=get_ax(),
boxes=utils.denorm_boxes(proposals[roi_positive_ixs][ids], image.shape[:2]),
refined_boxes=utils.denorm_boxes(refined_proposals[roi_positive_ixs][ids], image.shape[:2]),
visibilities=np.where(roi_class_ids[roi_positive_ixs][ids] > 0, 1, 0),
captions=captions, title="ROIs After Refinement")
```
#### Filter Low Confidence Detections
```
# Remove boxes classified as background
keep = np.where(roi_class_ids > 0)[0]
print("Keep {} detections:\n{}".format(keep.shape[0], keep))
# Remove low confidence detections
keep = np.intersect1d(keep, np.where(roi_scores >= config.DETECTION_MIN_CONFIDENCE)[0])
print("Remove boxes below {} confidence. Keep {}:\n{}".format(
config.DETECTION_MIN_CONFIDENCE, keep.shape[0], keep))
```
#### Per-Class Non-Max Suppression
```
# Apply per-class non-max suppression
pre_nms_boxes = refined_proposals[keep]
pre_nms_scores = roi_scores[keep]
pre_nms_class_ids = roi_class_ids[keep]
nms_keep = []
for class_id in np.unique(pre_nms_class_ids):
# Pick detections of this class
ixs = np.where(pre_nms_class_ids == class_id)[0]
# Apply NMS
class_keep = utils.non_max_suppression(pre_nms_boxes[ixs],
pre_nms_scores[ixs],
config.DETECTION_NMS_THRESHOLD)
# Map indicies
class_keep = keep[ixs[class_keep]]
nms_keep = np.union1d(nms_keep, class_keep)
print("{:22}: {} -> {}".format(dataset.class_names[class_id][:20],
keep[ixs], class_keep))
keep = np.intersect1d(keep, nms_keep).astype(np.int32)
print("\nKept after per-class NMS: {}\n{}".format(keep.shape[0], keep))
# Show final detections
ixs = np.arange(len(keep)) # Display all
# ixs = np.random.randint(0, len(keep), 10) # Display random sample
captions = ["{} {:.3f}".format(dataset.class_names[c], s) if c > 0 else ""
for c, s in zip(roi_class_ids[keep][ixs], roi_scores[keep][ixs])]
visualize.draw_boxes(
image,
boxes=utils.denorm_boxes(proposals[keep][ixs], image.shape[:2]),
refined_boxes=utils.denorm_boxes(refined_proposals[keep][ixs], image.shape[:2]),
visibilities=np.where(roi_class_ids[keep][ixs] > 0, 1, 0),
captions=captions, title="Detections after NMS",
ax=get_ax())
```
## Stage 3: Generating Masks
This stage takes the detections (refined bounding boxes and class IDs) from the previous layer and runs the mask head to generate segmentation masks for every instance.
### 3.a Mask Targets
These are the training targets for the mask branch
```
limit = 8
display_images(np.transpose(gt_mask[..., :limit], [2, 0, 1]), cmap="Blues")
```
### 3.b Predicted Masks
```
# Get predictions of mask head
mrcnn = model.run_graph([image], [
("detections", model.keras_model.get_layer("mrcnn_detection").output),
("masks", model.keras_model.get_layer("mrcnn_mask").output),
])
# Get detection class IDs. Trim zero padding.
det_class_ids = mrcnn['detections'][0, :, 4].astype(np.int32)
det_count = np.where(det_class_ids == 0)[0][0]
det_class_ids = det_class_ids[:det_count]
print("{} detections: {}".format(
det_count, np.array(dataset.class_names)[det_class_ids]))
# Masks
det_boxes = utils.denorm_boxes(mrcnn["detections"][0, :, :4], image.shape[:2])
det_mask_specific = np.array([mrcnn["masks"][0, i, :, :, c]
for i, c in enumerate(det_class_ids)])
det_masks = np.array([utils.unmold_mask(m, det_boxes[i], image.shape)
for i, m in enumerate(det_mask_specific)])
log("det_mask_specific", det_mask_specific)
log("det_masks", det_masks)
display_images(det_mask_specific[:4] * 255, cmap="Blues", interpolation="none")
display_images(det_masks[:4] * 255, cmap="Blues", interpolation="none")
```
## Visualize Activations
In some cases it helps to look at the output from different layers and visualize them to catch issues and odd patterns.
```
# Get activations of a few sample layers
activations = model.run_graph([image], [
("input_image", model.keras_model.get_layer("input_image").output),
("res2c_out", model.keras_model.get_layer("res2c_out").output),
("res3c_out", model.keras_model.get_layer("res3c_out").output),
("rpn_bbox", model.keras_model.get_layer("rpn_bbox").output),
("roi", model.keras_model.get_layer("ROI").output),
])
# Backbone feature map
display_images(np.transpose(activations["res2c_out"][0,:,:,:4], [2, 0, 1]), cols=4)
```
|
github_jupyter
|
import os
import sys
import random
import math
import re
import time
import numpy as np
import tensorflow as tf
import matplotlib
import matplotlib.pyplot as plt
import matplotlib.patches as patches
# Root directory of the project
ROOT_DIR = os.path.abspath("../../")
# Import Mask RCNN
sys.path.append(ROOT_DIR) # To find local version of the library
from mrcnn import utils
from mrcnn import visualize
from mrcnn.visualize import display_images
import mrcnn.model as modellib
from mrcnn.model import log
import nucleus
%matplotlib inline
# Directory to save logs and trained model
LOGS_DIR = os.path.join(ROOT_DIR, "logs")
# Comment out to reload imported modules if they change
# %load_ext autoreload
# %autoreload 2
# Dataset directory
DATASET_DIR = os.path.join(ROOT_DIR, "datasets/nucleus")
# Inference Configuration
config = nucleus.NucleusInferenceConfig()
config.display()
# Device to load the neural network on.
# Useful if you're training a model on the same
# machine, in which case use CPU and leave the
# GPU for training.
DEVICE = "/gpu:0" # /cpu:0 or /gpu:0
# Inspect the model in training or inference modes
# values: 'inference' or 'training'
# Only inference mode is supported right now
TEST_MODE = "inference"
def get_ax(rows=1, cols=1, size=16):
"""Return a Matplotlib Axes array to be used in
all visualizations in the notebook. Provide a
central point to control graph sizes.
Adjust the size attribute to control how big to render images
"""
fig, ax = plt.subplots(rows, cols, figsize=(size*cols, size*rows))
fig.tight_layout()
return ax
# Load validation dataset
dataset = nucleus.NucleusDataset()
dataset.load_nucleus(DATASET_DIR, "val")
dataset.prepare()
print("Images: {}\nClasses: {}".format(len(dataset.image_ids), dataset.class_names))
# Create model in inference mode
with tf.device(DEVICE):
model = modellib.MaskRCNN(mode="inference",
model_dir=LOGS_DIR,
config=config)
# Path to a specific weights file
# weights_path = "/path/to/mask_rcnn_nucleus.h5"
# Or, load the last model you trained
weights_path = model.find_last()
# Load weights
print("Loading weights ", weights_path)
model.load_weights(weights_path, by_name=True)
image_id = random.choice(dataset.image_ids)
image, image_meta, gt_class_id, gt_bbox, gt_mask =\
modellib.load_image_gt(dataset, config, image_id, use_mini_mask=False)
info = dataset.image_info[image_id]
print("image ID: {}.{} ({}) {}".format(info["source"], info["id"], image_id,
dataset.image_reference(image_id)))
print("Original image shape: ", modellib.parse_image_meta(image_meta[np.newaxis,...])["original_image_shape"][0])
# Run object detection
results = model.detect_molded(np.expand_dims(image, 0), np.expand_dims(image_meta, 0), verbose=1)
# Display results
r = results[0]
log("gt_class_id", gt_class_id)
log("gt_bbox", gt_bbox)
log("gt_mask", gt_mask)
# Compute AP over range 0.5 to 0.95 and print it
utils.compute_ap_range(gt_bbox, gt_class_id, gt_mask,
r['rois'], r['class_ids'], r['scores'], r['masks'],
verbose=1)
visualize.display_differences(
image,
gt_bbox, gt_class_id, gt_mask,
r['rois'], r['class_ids'], r['scores'], r['masks'],
dataset.class_names, ax=get_ax(),
show_box=False, show_mask=False,
iou_threshold=0.5, score_threshold=0.5)
# Display predictions only
# visualize.display_instances(image, r['rois'], r['masks'], r['class_ids'],
# dataset.class_names, r['scores'], ax=get_ax(1),
# show_bbox=False, show_mask=False,
# title="Predictions")
# Display Ground Truth only
# visualize.display_instances(image, gt_bbox, gt_mask, gt_class_id,
# dataset.class_names, ax=get_ax(1),
# show_bbox=False, show_mask=False,
# title="Ground Truth")
def compute_batch_ap(dataset, image_ids, verbose=1):
APs = []
for image_id in image_ids:
# Load image
image, image_meta, gt_class_id, gt_bbox, gt_mask =\
modellib.load_image_gt(dataset, config,
image_id, use_mini_mask=False)
# Run object detection
results = model.detect_molded(image[np.newaxis], image_meta[np.newaxis], verbose=0)
# Compute AP over range 0.5 to 0.95
r = results[0]
ap = utils.compute_ap_range(
gt_bbox, gt_class_id, gt_mask,
r['rois'], r['class_ids'], r['scores'], r['masks'],
verbose=0)
APs.append(ap)
if verbose:
info = dataset.image_info[image_id]
meta = modellib.parse_image_meta(image_meta[np.newaxis,...])
print("{:3} {} AP: {:.2f}".format(
meta["image_id"][0], meta["original_image_shape"][0], ap))
return APs
# Run on validation set
limit = 5
APs = compute_batch_ap(dataset, dataset.image_ids[:limit])
print("Mean AP overa {} images: {:.4f}".format(len(APs), np.mean(APs)))
# Get anchors and convert to pixel coordinates
anchors = model.get_anchors(image.shape)
anchors = utils.denorm_boxes(anchors, image.shape[:2])
log("anchors", anchors)
# Generate RPN trainig targets
# target_rpn_match is 1 for positive anchors, -1 for negative anchors
# and 0 for neutral anchors.
target_rpn_match, target_rpn_bbox = modellib.build_rpn_targets(
image.shape, anchors, gt_class_id, gt_bbox, model.config)
log("target_rpn_match", target_rpn_match)
log("target_rpn_bbox", target_rpn_bbox)
positive_anchor_ix = np.where(target_rpn_match[:] == 1)[0]
negative_anchor_ix = np.where(target_rpn_match[:] == -1)[0]
neutral_anchor_ix = np.where(target_rpn_match[:] == 0)[0]
positive_anchors = anchors[positive_anchor_ix]
negative_anchors = anchors[negative_anchor_ix]
neutral_anchors = anchors[neutral_anchor_ix]
log("positive_anchors", positive_anchors)
log("negative_anchors", negative_anchors)
log("neutral anchors", neutral_anchors)
# Apply refinement deltas to positive anchors
refined_anchors = utils.apply_box_deltas(
positive_anchors,
target_rpn_bbox[:positive_anchors.shape[0]] * model.config.RPN_BBOX_STD_DEV)
log("refined_anchors", refined_anchors, )
# Display positive anchors before refinement (dotted) and
# after refinement (solid).
visualize.draw_boxes(
image, ax=get_ax(),
boxes=positive_anchors,
refined_boxes=refined_anchors)
# Run RPN sub-graph
pillar = model.keras_model.get_layer("ROI").output # node to start searching from
# TF 1.4 introduces a new version of NMS. Search for both names to support TF 1.3 and 1.4
nms_node = model.ancestor(pillar, "ROI/rpn_non_max_suppression:0")
if nms_node is None:
nms_node = model.ancestor(pillar, "ROI/rpn_non_max_suppression/NonMaxSuppressionV2:0")
rpn = model.run_graph(image[np.newaxis], [
("rpn_class", model.keras_model.get_layer("rpn_class").output),
("pre_nms_anchors", model.ancestor(pillar, "ROI/pre_nms_anchors:0")),
("refined_anchors", model.ancestor(pillar, "ROI/refined_anchors:0")),
("refined_anchors_clipped", model.ancestor(pillar, "ROI/refined_anchors_clipped:0")),
("post_nms_anchor_ix", nms_node),
("proposals", model.keras_model.get_layer("ROI").output),
], image_metas=image_meta[np.newaxis])
# Show top anchors by score (before refinement)
limit = 100
sorted_anchor_ids = np.argsort(rpn['rpn_class'][:,:,1].flatten())[::-1]
visualize.draw_boxes(image, boxes=anchors[sorted_anchor_ids[:limit]], ax=get_ax())
# Show top anchors with refinement. Then with clipping to image boundaries
limit = 50
ax = get_ax(1, 2)
visualize.draw_boxes(
image, ax=ax[0],
boxes=utils.denorm_boxes(rpn["pre_nms_anchors"][0, :limit], image.shape[:2]),
refined_boxes=utils.denorm_boxes(rpn["refined_anchors"][0, :limit], image.shape[:2]))
visualize.draw_boxes(
image, ax=ax[1],
refined_boxes=utils.denorm_boxes(rpn["refined_anchors_clipped"][0, :limit], image.shape[:2]))
# Show refined anchors after non-max suppression
limit = 50
ixs = rpn["post_nms_anchor_ix"][:limit]
visualize.draw_boxes(
image, ax=get_ax(),
refined_boxes=utils.denorm_boxes(rpn["refined_anchors_clipped"][0, ixs], image.shape[:2]))
# Show final proposals
# These are the same as the previous step (refined anchors
# after NMS) but with coordinates normalized to [0, 1] range.
limit = 50
# Convert back to image coordinates for display
# h, w = config.IMAGE_SHAPE[:2]
# proposals = rpn['proposals'][0, :limit] * np.array([h, w, h, w])
visualize.draw_boxes(
image, ax=get_ax(),
refined_boxes=utils.denorm_boxes(rpn['proposals'][0, :limit], image.shape[:2]))
# Get input and output to classifier and mask heads.
mrcnn = model.run_graph([image], [
("proposals", model.keras_model.get_layer("ROI").output),
("probs", model.keras_model.get_layer("mrcnn_class").output),
("deltas", model.keras_model.get_layer("mrcnn_bbox").output),
("masks", model.keras_model.get_layer("mrcnn_mask").output),
("detections", model.keras_model.get_layer("mrcnn_detection").output),
])
# Get detection class IDs. Trim zero padding.
det_class_ids = mrcnn['detections'][0, :, 4].astype(np.int32)
det_count = np.where(det_class_ids == 0)[0][0]
det_class_ids = det_class_ids[:det_count]
detections = mrcnn['detections'][0, :det_count]
print("{} detections: {}".format(
det_count, np.array(dataset.class_names)[det_class_ids]))
captions = ["{} {:.3f}".format(dataset.class_names[int(c)], s) if c > 0 else ""
for c, s in zip(detections[:, 4], detections[:, 5])]
visualize.draw_boxes(
image,
refined_boxes=utils.denorm_boxes(detections[:, :4], image.shape[:2]),
visibilities=[2] * len(detections),
captions=captions, title="Detections",
ax=get_ax())
# Proposals are in normalized coordinates
proposals = mrcnn["proposals"][0]
# Class ID, score, and mask per proposal
roi_class_ids = np.argmax(mrcnn["probs"][0], axis=1)
roi_scores = mrcnn["probs"][0, np.arange(roi_class_ids.shape[0]), roi_class_ids]
roi_class_names = np.array(dataset.class_names)[roi_class_ids]
roi_positive_ixs = np.where(roi_class_ids > 0)[0]
# How many ROIs vs empty rows?
print("{} Valid proposals out of {}".format(np.sum(np.any(proposals, axis=1)), proposals.shape[0]))
print("{} Positive ROIs".format(len(roi_positive_ixs)))
# Class counts
print(list(zip(*np.unique(roi_class_names, return_counts=True))))
# Display a random sample of proposals.
# Proposals classified as background are dotted, and
# the rest show their class and confidence score.
limit = 200
ixs = np.random.randint(0, proposals.shape[0], limit)
captions = ["{} {:.3f}".format(dataset.class_names[c], s) if c > 0 else ""
for c, s in zip(roi_class_ids[ixs], roi_scores[ixs])]
visualize.draw_boxes(
image,
boxes=utils.denorm_boxes(proposals[ixs], image.shape[:2]),
visibilities=np.where(roi_class_ids[ixs] > 0, 2, 1),
captions=captions, title="ROIs Before Refinement",
ax=get_ax())
# Class-specific bounding box shifts.
roi_bbox_specific = mrcnn["deltas"][0, np.arange(proposals.shape[0]), roi_class_ids]
log("roi_bbox_specific", roi_bbox_specific)
# Apply bounding box transformations
# Shape: [N, (y1, x1, y2, x2)]
refined_proposals = utils.apply_box_deltas(
proposals, roi_bbox_specific * config.BBOX_STD_DEV)
log("refined_proposals", refined_proposals)
# Show positive proposals
# ids = np.arange(roi_boxes.shape[0]) # Display all
limit = 5
ids = np.random.randint(0, len(roi_positive_ixs), limit) # Display random sample
captions = ["{} {:.3f}".format(dataset.class_names[c], s) if c > 0 else ""
for c, s in zip(roi_class_ids[roi_positive_ixs][ids], roi_scores[roi_positive_ixs][ids])]
visualize.draw_boxes(
image, ax=get_ax(),
boxes=utils.denorm_boxes(proposals[roi_positive_ixs][ids], image.shape[:2]),
refined_boxes=utils.denorm_boxes(refined_proposals[roi_positive_ixs][ids], image.shape[:2]),
visibilities=np.where(roi_class_ids[roi_positive_ixs][ids] > 0, 1, 0),
captions=captions, title="ROIs After Refinement")
# Remove boxes classified as background
keep = np.where(roi_class_ids > 0)[0]
print("Keep {} detections:\n{}".format(keep.shape[0], keep))
# Remove low confidence detections
keep = np.intersect1d(keep, np.where(roi_scores >= config.DETECTION_MIN_CONFIDENCE)[0])
print("Remove boxes below {} confidence. Keep {}:\n{}".format(
config.DETECTION_MIN_CONFIDENCE, keep.shape[0], keep))
# Apply per-class non-max suppression
pre_nms_boxes = refined_proposals[keep]
pre_nms_scores = roi_scores[keep]
pre_nms_class_ids = roi_class_ids[keep]
nms_keep = []
for class_id in np.unique(pre_nms_class_ids):
# Pick detections of this class
ixs = np.where(pre_nms_class_ids == class_id)[0]
# Apply NMS
class_keep = utils.non_max_suppression(pre_nms_boxes[ixs],
pre_nms_scores[ixs],
config.DETECTION_NMS_THRESHOLD)
# Map indicies
class_keep = keep[ixs[class_keep]]
nms_keep = np.union1d(nms_keep, class_keep)
print("{:22}: {} -> {}".format(dataset.class_names[class_id][:20],
keep[ixs], class_keep))
keep = np.intersect1d(keep, nms_keep).astype(np.int32)
print("\nKept after per-class NMS: {}\n{}".format(keep.shape[0], keep))
# Show final detections
ixs = np.arange(len(keep)) # Display all
# ixs = np.random.randint(0, len(keep), 10) # Display random sample
captions = ["{} {:.3f}".format(dataset.class_names[c], s) if c > 0 else ""
for c, s in zip(roi_class_ids[keep][ixs], roi_scores[keep][ixs])]
visualize.draw_boxes(
image,
boxes=utils.denorm_boxes(proposals[keep][ixs], image.shape[:2]),
refined_boxes=utils.denorm_boxes(refined_proposals[keep][ixs], image.shape[:2]),
visibilities=np.where(roi_class_ids[keep][ixs] > 0, 1, 0),
captions=captions, title="Detections after NMS",
ax=get_ax())
limit = 8
display_images(np.transpose(gt_mask[..., :limit], [2, 0, 1]), cmap="Blues")
# Get predictions of mask head
mrcnn = model.run_graph([image], [
("detections", model.keras_model.get_layer("mrcnn_detection").output),
("masks", model.keras_model.get_layer("mrcnn_mask").output),
])
# Get detection class IDs. Trim zero padding.
det_class_ids = mrcnn['detections'][0, :, 4].astype(np.int32)
det_count = np.where(det_class_ids == 0)[0][0]
det_class_ids = det_class_ids[:det_count]
print("{} detections: {}".format(
det_count, np.array(dataset.class_names)[det_class_ids]))
# Masks
det_boxes = utils.denorm_boxes(mrcnn["detections"][0, :, :4], image.shape[:2])
det_mask_specific = np.array([mrcnn["masks"][0, i, :, :, c]
for i, c in enumerate(det_class_ids)])
det_masks = np.array([utils.unmold_mask(m, det_boxes[i], image.shape)
for i, m in enumerate(det_mask_specific)])
log("det_mask_specific", det_mask_specific)
log("det_masks", det_masks)
display_images(det_mask_specific[:4] * 255, cmap="Blues", interpolation="none")
display_images(det_masks[:4] * 255, cmap="Blues", interpolation="none")
# Get activations of a few sample layers
activations = model.run_graph([image], [
("input_image", model.keras_model.get_layer("input_image").output),
("res2c_out", model.keras_model.get_layer("res2c_out").output),
("res3c_out", model.keras_model.get_layer("res3c_out").output),
("rpn_bbox", model.keras_model.get_layer("rpn_bbox").output),
("roi", model.keras_model.get_layer("ROI").output),
])
# Backbone feature map
display_images(np.transpose(activations["res2c_out"][0,:,:,:4], [2, 0, 1]), cols=4)
| 0.61115 | 0.88263 |
# Using PyTorch-based models in GluonTS
This notebook illustrates how one can implement a time series model using PyTorch, and use it together with the rest of the GluonTS ecosystem for data loading, feature processing, and model evaluation.
```
from typing import List, Optional, Callable
from itertools import islice
import torch
import torch.nn as nn
import numpy as np
import pandas as pd
from matplotlib import pyplot as plt
import matplotlib.dates as mdates
from gluonts.dataset.field_names import FieldName
from gluonts.dataset.loader import TrainDataLoader
from gluonts.dataset.repository.datasets import get_dataset
from gluonts.evaluation import Evaluator
from gluonts.evaluation.backtest import make_evaluation_predictions
from gluonts.torch.batchify import batchify
from gluonts.torch.support.util import copy_parameters
from gluonts.torch.model.predictor import PyTorchPredictor
from gluonts.transform import Chain, AddObservedValuesIndicator, InstanceSplitter, ExpectedNumInstanceSampler
```
For this example we will use the "electricity" dataset, which can be loaded as follows.
```
dataset = get_dataset("electricity")
```
This is what the first time series from the training portion of the dataset look like:
```
date_formater = mdates.DateFormatter('%Y')
fig = plt.figure(figsize=(12,8))
for idx, entry in enumerate(islice(dataset.train, 9)):
ax = plt.subplot(3, 3, idx+1)
t = pd.date_range(start=entry["start"], periods=len(entry["target"]), freq=entry["start"].freq)
plt.plot(t, entry["target"])
plt.xticks(pd.date_range(start=pd.to_datetime("2011-12-31"), periods=3, freq="AS"))
ax.xaxis.set_major_formatter(date_formater)
```
## Probabilistic feed-forward network using PyTorch
We will use a pretty simple model, based on a feed-forward network whose output layer produces the parameters of a Student's t-distribution at each time step in the prediction range. We will define two networks based on this idea:
* The `TrainingFeedForwardNetwork` computes the loss associated with given observations, i.e. the negative log-likelihood of the observations according to the output distribution; this will be used during training.
* The `SamplingFeedForwardNetwork` will be used at inference time: this uses the output distribution to draw a sample of a given size, as a way to encode the predicted distribution.
```
def mean_abs_scaling(context, min_scale=1e-5):
return context.abs().mean(1).clamp(min_scale, None).unsqueeze(1)
def no_scaling(context):
return torch.ones(context.shape[0], 1)
class TrainingFeedForwardNetwork(nn.Module):
distr_type = torch.distributions.StudentT
def __init__(
self,
prediction_length: int,
context_length: int,
hidden_dimensions: List[int],
batch_norm: bool=False,
scaling: Callable=mean_abs_scaling,
) -> None:
super().__init__()
assert prediction_length > 0
assert context_length > 0
assert len(hidden_dimensions) > 0
self.prediction_length = prediction_length
self.context_length = context_length
self.hidden_dimensions = hidden_dimensions
self.batch_norm = batch_norm
self.scaling = scaling
dimensions = [context_length] + hidden_dimensions[:-1]
modules = []
for in_size, out_size in zip(dimensions[:-1], dimensions[1:]):
modules += [self.__make_lin(in_size, out_size), nn.ReLU()]
if batch_norm:
modules.append(nn.BatchNorm1d(units))
modules.append(self.__make_lin(dimensions[-1], prediction_length * hidden_dimensions[-1]))
self.nn = nn.Sequential(*modules)
self.df_proj = nn.Sequential(self.__make_lin(hidden_dimensions[-1], 1), nn.Softplus())
self.loc_proj = self.__make_lin(hidden_dimensions[-1], 1)
self.scale_proj = nn.Sequential(self.__make_lin(hidden_dimensions[-1], 1), nn.Softplus())
@staticmethod
def __make_lin(dim_in, dim_out):
lin = nn.Linear(dim_in, dim_out)
torch.nn.init.uniform_(lin.weight, -0.07, 0.07)
torch.nn.init.zeros_(lin.bias)
return lin
def distr_and_scale(self, context):
scale = self.scaling(context)
scaled_context = context / scale
nn_out = self.nn(scaled_context)
nn_out_reshaped = nn_out.reshape(-1, self.prediction_length, self.hidden_dimensions[-1])
distr_args = (
2.0 + self.df_proj(nn_out_reshaped).squeeze(dim=-1),
self.loc_proj(nn_out_reshaped).squeeze(dim=-1),
self.scale_proj(nn_out_reshaped).squeeze(dim=-1),
)
distr = net.distr_type(*distr_args)
return distr, scale
def forward(self, context, target):
assert context.shape[-1] == self.context_length
assert target.shape[-1] == self.prediction_length
distr, scale = self.distr_and_scale(context)
loss = (-distr.log_prob(target / scale) + torch.log(scale)).mean(dim=1)
return loss
class SamplingFeedForwardNetwork(TrainingFeedForwardNetwork):
def __init__(self, *args, num_samples: int = 1000, **kwargs):
super().__init__(*args, **kwargs)
self.num_samples = num_samples
def forward(self, context):
assert context.shape[-1] == self.context_length
distr, scale = self.distr_and_scale(context)
sample = distr.sample((self.num_samples, )) * scale
return sample.permute(1, 0, 2)
```
We can now instantiate the training network, and explore its set of parameters.
```
context_length = 2 * 7 * 24
prediction_length = dataset.metadata.prediction_length
hidden_dimensions = [96, 48]
batch_size = 32
num_batches_per_epoch = 100
net = TrainingFeedForwardNetwork(
prediction_length=prediction_length,
context_length=context_length,
hidden_dimensions=hidden_dimensions,
batch_norm=False,
scaling=mean_abs_scaling,
)
sum(np.prod(p.shape) for p in net.parameters())
for p in net.parameters():
print(p.shape)
```
## Defining the training data loader
We now set up the data loader which will yield batches of data to train on. Starting from the original dataset, the data loader is configured to apply the following transformation, which does essentially two things:
* Replaces `nan`s in the target field with a dummy value (zero), and adds a field indicating which values were actually observed vs imputed this way.
* Slices out training instances of a fixed length randomly from the given dataset; these will be stacked into batches by the data loader itself.
```
transformation = Chain([
AddObservedValuesIndicator(
target_field=FieldName.TARGET,
output_field=FieldName.OBSERVED_VALUES,
),
InstanceSplitter(
target_field=FieldName.TARGET,
is_pad_field=FieldName.IS_PAD,
start_field=FieldName.START,
forecast_start_field=FieldName.FORECAST_START,
train_sampler=ExpectedNumInstanceSampler(num_instances=1),
past_length=context_length,
future_length=prediction_length,
time_series_fields=[FieldName.OBSERVED_VALUES],
),
])
data_loader = TrainDataLoader(
dataset.train,
batch_size=batch_size,
stack_fn=batchify,
transform=transformation,
num_batches_per_epoch=num_batches_per_epoch
)
```
## Train the model
We can now train the model using any of the available optimizers from PyTorch:
```
optimizer = torch.optim.Adam(net.parameters())
for epoch_no in range(10):
sum_epoch_loss = 0.0
for batch_no, batch in enumerate(data_loader, start=1):
optimizer.zero_grad()
context = batch["past_target"]
target = batch["future_target"]
loss_vec = net(context, target)
loss = loss_vec.mean()
loss.backward()
optimizer.step()
sum_epoch_loss += loss.detach().numpy().item()
print(f"{epoch_no}: {sum_epoch_loss / num_batches_per_epoch}")
```
## Create predictor out of the trained model, and test it
We now have a trained model, whose parameters can be copied over to a `SamplingFeedForwardNetwork` object: we will wrap this into a `PyTorchPredictor` that can be used for inference tasks.
```
pred_net = SamplingFeedForwardNetwork(
prediction_length=net.prediction_length,
context_length=net.context_length,
hidden_dimensions=net.hidden_dimensions,
batch_norm=net.batch_norm,
)
copy_parameters(net, pred_net)
predictor_pytorch = PyTorchPredictor(
prediction_length=prediction_length, freq = dataset.metadata.freq,
input_names = ["past_target"], prediction_net=pred_net, batch_size=32, input_transform=transformation,
device=None
)
```
For example, we can do backtesting on the test dataset: in what follows, `make_evaluation_predictions` will slice out the trailing `prediction_length` observations from the test time series, and use the given predictor to obtain forecasts for the same time range.
```
forecast_it, ts_it = make_evaluation_predictions(
dataset=dataset.test,
predictor=predictor_pytorch,
num_samples=1000,
)
forecasts_pytorch = list(forecast_it)
tss_pytorch = list(ts_it)
```
Once we have the forecasts, we can plot them:
```
plt.figure(figsize=(20, 15))
date_formater = mdates.DateFormatter('%b, %d')
plt.rcParams.update({'font.size': 15})
for idx, (forecast, ts) in islice(enumerate(zip(forecasts_pytorch, tss_pytorch)), 9):
ax =plt.subplot(3, 3, idx+1)
plt.plot(ts[-5 * prediction_length:], label="target")
forecast.plot()
plt.xticks(rotation=60)
ax.xaxis.set_major_formatter(date_formater)
plt.gcf().tight_layout()
plt.legend()
plt.show()
```
And we can compute evaluation metrics, that summarize the performance of the model on our test data.
```
evaluator = Evaluator(quantiles=[0.1, 0.5, 0.9])
metrics_pytorch, _ = evaluator(iter(tss_pytorch), iter(forecasts_pytorch), num_series=len(dataset.test))
pd.DataFrame.from_records(metrics_pytorch, index=["FeedForward"]).transpose()
```
|
github_jupyter
|
from typing import List, Optional, Callable
from itertools import islice
import torch
import torch.nn as nn
import numpy as np
import pandas as pd
from matplotlib import pyplot as plt
import matplotlib.dates as mdates
from gluonts.dataset.field_names import FieldName
from gluonts.dataset.loader import TrainDataLoader
from gluonts.dataset.repository.datasets import get_dataset
from gluonts.evaluation import Evaluator
from gluonts.evaluation.backtest import make_evaluation_predictions
from gluonts.torch.batchify import batchify
from gluonts.torch.support.util import copy_parameters
from gluonts.torch.model.predictor import PyTorchPredictor
from gluonts.transform import Chain, AddObservedValuesIndicator, InstanceSplitter, ExpectedNumInstanceSampler
dataset = get_dataset("electricity")
date_formater = mdates.DateFormatter('%Y')
fig = plt.figure(figsize=(12,8))
for idx, entry in enumerate(islice(dataset.train, 9)):
ax = plt.subplot(3, 3, idx+1)
t = pd.date_range(start=entry["start"], periods=len(entry["target"]), freq=entry["start"].freq)
plt.plot(t, entry["target"])
plt.xticks(pd.date_range(start=pd.to_datetime("2011-12-31"), periods=3, freq="AS"))
ax.xaxis.set_major_formatter(date_formater)
def mean_abs_scaling(context, min_scale=1e-5):
return context.abs().mean(1).clamp(min_scale, None).unsqueeze(1)
def no_scaling(context):
return torch.ones(context.shape[0], 1)
class TrainingFeedForwardNetwork(nn.Module):
distr_type = torch.distributions.StudentT
def __init__(
self,
prediction_length: int,
context_length: int,
hidden_dimensions: List[int],
batch_norm: bool=False,
scaling: Callable=mean_abs_scaling,
) -> None:
super().__init__()
assert prediction_length > 0
assert context_length > 0
assert len(hidden_dimensions) > 0
self.prediction_length = prediction_length
self.context_length = context_length
self.hidden_dimensions = hidden_dimensions
self.batch_norm = batch_norm
self.scaling = scaling
dimensions = [context_length] + hidden_dimensions[:-1]
modules = []
for in_size, out_size in zip(dimensions[:-1], dimensions[1:]):
modules += [self.__make_lin(in_size, out_size), nn.ReLU()]
if batch_norm:
modules.append(nn.BatchNorm1d(units))
modules.append(self.__make_lin(dimensions[-1], prediction_length * hidden_dimensions[-1]))
self.nn = nn.Sequential(*modules)
self.df_proj = nn.Sequential(self.__make_lin(hidden_dimensions[-1], 1), nn.Softplus())
self.loc_proj = self.__make_lin(hidden_dimensions[-1], 1)
self.scale_proj = nn.Sequential(self.__make_lin(hidden_dimensions[-1], 1), nn.Softplus())
@staticmethod
def __make_lin(dim_in, dim_out):
lin = nn.Linear(dim_in, dim_out)
torch.nn.init.uniform_(lin.weight, -0.07, 0.07)
torch.nn.init.zeros_(lin.bias)
return lin
def distr_and_scale(self, context):
scale = self.scaling(context)
scaled_context = context / scale
nn_out = self.nn(scaled_context)
nn_out_reshaped = nn_out.reshape(-1, self.prediction_length, self.hidden_dimensions[-1])
distr_args = (
2.0 + self.df_proj(nn_out_reshaped).squeeze(dim=-1),
self.loc_proj(nn_out_reshaped).squeeze(dim=-1),
self.scale_proj(nn_out_reshaped).squeeze(dim=-1),
)
distr = net.distr_type(*distr_args)
return distr, scale
def forward(self, context, target):
assert context.shape[-1] == self.context_length
assert target.shape[-1] == self.prediction_length
distr, scale = self.distr_and_scale(context)
loss = (-distr.log_prob(target / scale) + torch.log(scale)).mean(dim=1)
return loss
class SamplingFeedForwardNetwork(TrainingFeedForwardNetwork):
def __init__(self, *args, num_samples: int = 1000, **kwargs):
super().__init__(*args, **kwargs)
self.num_samples = num_samples
def forward(self, context):
assert context.shape[-1] == self.context_length
distr, scale = self.distr_and_scale(context)
sample = distr.sample((self.num_samples, )) * scale
return sample.permute(1, 0, 2)
context_length = 2 * 7 * 24
prediction_length = dataset.metadata.prediction_length
hidden_dimensions = [96, 48]
batch_size = 32
num_batches_per_epoch = 100
net = TrainingFeedForwardNetwork(
prediction_length=prediction_length,
context_length=context_length,
hidden_dimensions=hidden_dimensions,
batch_norm=False,
scaling=mean_abs_scaling,
)
sum(np.prod(p.shape) for p in net.parameters())
for p in net.parameters():
print(p.shape)
transformation = Chain([
AddObservedValuesIndicator(
target_field=FieldName.TARGET,
output_field=FieldName.OBSERVED_VALUES,
),
InstanceSplitter(
target_field=FieldName.TARGET,
is_pad_field=FieldName.IS_PAD,
start_field=FieldName.START,
forecast_start_field=FieldName.FORECAST_START,
train_sampler=ExpectedNumInstanceSampler(num_instances=1),
past_length=context_length,
future_length=prediction_length,
time_series_fields=[FieldName.OBSERVED_VALUES],
),
])
data_loader = TrainDataLoader(
dataset.train,
batch_size=batch_size,
stack_fn=batchify,
transform=transformation,
num_batches_per_epoch=num_batches_per_epoch
)
optimizer = torch.optim.Adam(net.parameters())
for epoch_no in range(10):
sum_epoch_loss = 0.0
for batch_no, batch in enumerate(data_loader, start=1):
optimizer.zero_grad()
context = batch["past_target"]
target = batch["future_target"]
loss_vec = net(context, target)
loss = loss_vec.mean()
loss.backward()
optimizer.step()
sum_epoch_loss += loss.detach().numpy().item()
print(f"{epoch_no}: {sum_epoch_loss / num_batches_per_epoch}")
pred_net = SamplingFeedForwardNetwork(
prediction_length=net.prediction_length,
context_length=net.context_length,
hidden_dimensions=net.hidden_dimensions,
batch_norm=net.batch_norm,
)
copy_parameters(net, pred_net)
predictor_pytorch = PyTorchPredictor(
prediction_length=prediction_length, freq = dataset.metadata.freq,
input_names = ["past_target"], prediction_net=pred_net, batch_size=32, input_transform=transformation,
device=None
)
forecast_it, ts_it = make_evaluation_predictions(
dataset=dataset.test,
predictor=predictor_pytorch,
num_samples=1000,
)
forecasts_pytorch = list(forecast_it)
tss_pytorch = list(ts_it)
plt.figure(figsize=(20, 15))
date_formater = mdates.DateFormatter('%b, %d')
plt.rcParams.update({'font.size': 15})
for idx, (forecast, ts) in islice(enumerate(zip(forecasts_pytorch, tss_pytorch)), 9):
ax =plt.subplot(3, 3, idx+1)
plt.plot(ts[-5 * prediction_length:], label="target")
forecast.plot()
plt.xticks(rotation=60)
ax.xaxis.set_major_formatter(date_formater)
plt.gcf().tight_layout()
plt.legend()
plt.show()
evaluator = Evaluator(quantiles=[0.1, 0.5, 0.9])
metrics_pytorch, _ = evaluator(iter(tss_pytorch), iter(forecasts_pytorch), num_series=len(dataset.test))
pd.DataFrame.from_records(metrics_pytorch, index=["FeedForward"]).transpose()
| 0.938209 | 0.953751 |
```
!apt install libasound2-dev portaudio19-dev libportaudio2 libportaudiocpp0 ffmpeg
!pip install pyaudio
import pyaudio
import wave
import numpy as np
import torch
import torch.nn as nn
import torch.optim as optim
import torch.nn.functional as F
from torchvision import datasets,transforms
class Net(nn.Module):
def __init__(self):
super(Net,self).__init__()
self.conv1=nn.Conv2d(1,20,5,1)
self.conv2=nn.Conv2d(20,50,5,1)
self.fc1=nn.Linear(4*4*50,500)
self.fc2=nn.Linear(500,10)
self.ordered_layers=[self.conv1,self.conv2,self.fc1,self.fc2]
def forward(self,x):
x=F.relu(self.conv1(x))
x=F.max_pool2D(x,2,2)
x=F.relu(self.conv2(x))
x=F.max_pool2D(x,2,2)
x=x.view(-1,4*4*50)
x=F.relu(self.fc1(x))
x=self.fc2(x)
return F.log_softmax(x,dim=1)
def open_stream(fs):
p=pyaudio.PyAudio()
stream=p.open(format=pyaudio.paFloat32,channels=1,rate=fs,output=True,output_device_index=0)
return p,stream
def gen(fs,freq,duration):
sinwave=np.sin(2*np.pi*np.arrange(fs*duration)*freq/fs)
sample=sinwave.astype(np.float32)
return 0.1*sample
def train(model,device,train_loader,optimizer,epoch):
model.train()
fs=44100
duration=0.01
f=200.0
p,stream=open_stream(fs)
frames=[]
for batch_idx, (data, target) in enumerate(train_loader):
data,target=data.to(device),target.to(device)
optimizer.zero_grad()
output=model(data)
loss=F.nll_loss(output,target)
loss.backward()
norms=[]
for layer in model.ordered_layers:
norm_grad=layer.weight.grad.norm()
norms.append(norm_grad)
tone=f+(norms.numpy()*100.0)
tone=tone.astype(np.float32)
samples=gen(fs,tone,duration)
frames.append(samples)
silence=np.zeros(samples.shape[0]*2,dtyple=np.float32)
frames.append(silence)
optimizer.step()
if batch_idx==150:
break
wf=wave.open("sgdlr 0.01.wav","wb")
wf.setnchannels(1)
wf.setsampwidth(p.get_sample_size(pyaudio.paFloat32))
wf.setframerate(fs)
wf.writeframes(b''.join(frames))
wf.close()
stream.stop_stream()
stream.close()
p.terminate()
train_loader = torch.utils.data.DataLoader(
datasets.MNIST('../data', train=True, download=True,
transform=transforms.Compose([
transforms.ToTensor(),
transforms.Normalize((0.1307,), (0.3081,))
])),
batch_size=256, shuffle=True)
device=torch.device('cuda')
model=Net().to(device)
optimizer = optim.SGD(model.parameters(), lr=0.01, momentum=0.5)
for epoch in range(1, 10):
train(model, device, train_loader, optimizer, epoch)
```
|
github_jupyter
|
!apt install libasound2-dev portaudio19-dev libportaudio2 libportaudiocpp0 ffmpeg
!pip install pyaudio
import pyaudio
import wave
import numpy as np
import torch
import torch.nn as nn
import torch.optim as optim
import torch.nn.functional as F
from torchvision import datasets,transforms
class Net(nn.Module):
def __init__(self):
super(Net,self).__init__()
self.conv1=nn.Conv2d(1,20,5,1)
self.conv2=nn.Conv2d(20,50,5,1)
self.fc1=nn.Linear(4*4*50,500)
self.fc2=nn.Linear(500,10)
self.ordered_layers=[self.conv1,self.conv2,self.fc1,self.fc2]
def forward(self,x):
x=F.relu(self.conv1(x))
x=F.max_pool2D(x,2,2)
x=F.relu(self.conv2(x))
x=F.max_pool2D(x,2,2)
x=x.view(-1,4*4*50)
x=F.relu(self.fc1(x))
x=self.fc2(x)
return F.log_softmax(x,dim=1)
def open_stream(fs):
p=pyaudio.PyAudio()
stream=p.open(format=pyaudio.paFloat32,channels=1,rate=fs,output=True,output_device_index=0)
return p,stream
def gen(fs,freq,duration):
sinwave=np.sin(2*np.pi*np.arrange(fs*duration)*freq/fs)
sample=sinwave.astype(np.float32)
return 0.1*sample
def train(model,device,train_loader,optimizer,epoch):
model.train()
fs=44100
duration=0.01
f=200.0
p,stream=open_stream(fs)
frames=[]
for batch_idx, (data, target) in enumerate(train_loader):
data,target=data.to(device),target.to(device)
optimizer.zero_grad()
output=model(data)
loss=F.nll_loss(output,target)
loss.backward()
norms=[]
for layer in model.ordered_layers:
norm_grad=layer.weight.grad.norm()
norms.append(norm_grad)
tone=f+(norms.numpy()*100.0)
tone=tone.astype(np.float32)
samples=gen(fs,tone,duration)
frames.append(samples)
silence=np.zeros(samples.shape[0]*2,dtyple=np.float32)
frames.append(silence)
optimizer.step()
if batch_idx==150:
break
wf=wave.open("sgdlr 0.01.wav","wb")
wf.setnchannels(1)
wf.setsampwidth(p.get_sample_size(pyaudio.paFloat32))
wf.setframerate(fs)
wf.writeframes(b''.join(frames))
wf.close()
stream.stop_stream()
stream.close()
p.terminate()
train_loader = torch.utils.data.DataLoader(
datasets.MNIST('../data', train=True, download=True,
transform=transforms.Compose([
transforms.ToTensor(),
transforms.Normalize((0.1307,), (0.3081,))
])),
batch_size=256, shuffle=True)
device=torch.device('cuda')
model=Net().to(device)
optimizer = optim.SGD(model.parameters(), lr=0.01, momentum=0.5)
for epoch in range(1, 10):
train(model, device, train_loader, optimizer, epoch)
| 0.913678 | 0.319546 |
# Metodos sobre cadenas
En Python los objetos tipo cadena de texto (str) poseen diversos metodos que permiten manipularlas para llevar a cabo diferentes propositos. A continuación exploraremos los que consideramos mas importantes en esta introducción.
<img src="./string.jpg" width="500">
## count()
El metodo count nos indica cuantas veces se repite un caracter o una palabra en una oración.
```
Mensaje = "hola mundo"
print(Mensaje.count("o"))
print(Mensaje.count("hola"))
```
## find()
El metodo find nos indicara la posición en la que se encuentra el elemento en la cadena indicada. Si el elemento se encuentra repetido en la cadena de texto, el metodo regresara el primer elemento encontrado.
```
# Con elementos unicos
print(Mensaje.find("a"))
print(Mensaje.find("mundo"))
# Con un elemento repetido
print(Mensaje.find("o"))
# Por su parte rfind() regresa la ultima ocurrencia
print(Mensaje.rfind("o"))
```
## isnumeric()
Es un metodo que nos indica si los caracteres de la cadena corresponden a valores numericos.
```
Num = "1234"
NotNum = "abc123"
print(Num.isnumeric())
print(NotNum.isnumeric())
```
## startswith() y endswith()
Son metodos que nos indicaran si una cadena inicia o termina con el caracter o palabra indicado.
```
# Para el inicio
print(Mensaje.startswith("h"))
print(Mensaje.startswith("hola"))
print(Mensaje.startswith("mundo"))
# Para el final
print(Mensaje.endswith("o"))
print(Mensaje.endswith("mundo"))
print(Mensaje.endswith("hola"))
```
## capitalize(), lower(), upper() y swapcase()
**capitalize()** es metodo se encarga de cambiar la primer letra de la cadena de caracteres por una mayuscula.
**lower()** es un metodo que se encarga de cambiar todas las letras de la cadena de texto por minusculas.
**upper()** es un metodo que se encarga de cambiar todas las letras de la cadena de texto por mayusculas.
**swapcase()** es un metodo que se encarga por cambiar las minusculas por mayusculas y mayusculas por minusculas en una cadena.
```
# Capitalize
print(Mensaje)
print(Mensaje.capitalize())
# Lower
print("HOLA".lower())
# Upper
print("hola".upper())
# Swapcase
print("HoLa")
print("HoLa".swapcase())
```
## center(), ljust() y rjust()
**center()** Este metodo se encarga de ajustar la posición de un caracter o palabra en el centro de acuerdo a la cantidad de espacios indicados.
**ljust()** Este metodo se encarga de ajustar la posición de un caracter o palabra en la izquierda de acuerdo a la cantidad de espacios indicados.
**rjust()** Este metodo se encarga de ajustar la posición de un caracter o palabra en la derecha de acuerdo a la cantidad de espacios indicados.
```
# Ljust
print("Hola".ljust(10))
# Center
print("Hola".center(10))
# Rjust
print("Hola".rjust(10))
```
## strip(), lstrip() y rstrip()
**strip()** es un metodo que se encarga de eliminar el primer y ultimo caracter de la cadena de texto.
**lstrip()** es un metodo que se encarga de eliminar el primer caracter de la cadena de texto.
**rstrip()** es un metodo que se encarga de eliminar el ultimo caracter de la cadena de texto.
```
Texto = " Mundo "
# strip()
print(Texto.strip() + "Hola")
# lstrip()
print(Texto.lstrip() + "Hola")
# rstrip()
print(Texto.rstrip() + "Hola")
```
## replace()
Este metodo se encarga de reemplazar el caracter o palabra indicada por otro elemento proporcionado como parametro.
```
Enunciado = "Hxla mundx"
# Reemplazar caracter x por o
print(Enunciado.replace("x","o"))
# Eliminar caracter x
print(Enunciado.replace("x",""))
# Cambiar una palabra
print(Enunciado.replace("mundx","amigo"))
# Reemplazo doble
print(Enunciado.replace("x","o").replace("mundo","amigo"))
```
## split()
Es un metodo que se encarga de generar una lista de elementos a partir de una cadena se texto, lo cual se logra indicando el separador que delimitara cada elemento.
```
Variable = "Me gusta comer chocolates"
# Si queremos generar una lista de cada palabra indicamos como separador un espacio simple
print(Variable.split(" "))
# Si queremos una separación que genere una lista de dos elementos separamos con una palabra
print(Variable.split("comer"))
# Si queremos mas elementos tambien podemos usar como separador un caracter
print(Variable.split("c"))
# Podemos indicar cuantos separadores utilizaremos por medio de un segundo parametro
print(Variable.split("c",1))
Lineas = "Hola\nComo\nEstas?"
# Separar por saltos de linea
print(Lineas)
print(Lineas.splitlines())
```
## join()
Este metodo podria considerarse lo opuesto al metodo split, ya que parte de una lista de elementos regresando una cadena de texto por medio de la union de los elementos con el separador indicado.
```
Values = ["Los", "dulces", "son", "de", "sabor", "a", "moras"]
" ".join(Values)
```
|
github_jupyter
|
Mensaje = "hola mundo"
print(Mensaje.count("o"))
print(Mensaje.count("hola"))
# Con elementos unicos
print(Mensaje.find("a"))
print(Mensaje.find("mundo"))
# Con un elemento repetido
print(Mensaje.find("o"))
# Por su parte rfind() regresa la ultima ocurrencia
print(Mensaje.rfind("o"))
Num = "1234"
NotNum = "abc123"
print(Num.isnumeric())
print(NotNum.isnumeric())
# Para el inicio
print(Mensaje.startswith("h"))
print(Mensaje.startswith("hola"))
print(Mensaje.startswith("mundo"))
# Para el final
print(Mensaje.endswith("o"))
print(Mensaje.endswith("mundo"))
print(Mensaje.endswith("hola"))
# Capitalize
print(Mensaje)
print(Mensaje.capitalize())
# Lower
print("HOLA".lower())
# Upper
print("hola".upper())
# Swapcase
print("HoLa")
print("HoLa".swapcase())
# Ljust
print("Hola".ljust(10))
# Center
print("Hola".center(10))
# Rjust
print("Hola".rjust(10))
Texto = " Mundo "
# strip()
print(Texto.strip() + "Hola")
# lstrip()
print(Texto.lstrip() + "Hola")
# rstrip()
print(Texto.rstrip() + "Hola")
Enunciado = "Hxla mundx"
# Reemplazar caracter x por o
print(Enunciado.replace("x","o"))
# Eliminar caracter x
print(Enunciado.replace("x",""))
# Cambiar una palabra
print(Enunciado.replace("mundx","amigo"))
# Reemplazo doble
print(Enunciado.replace("x","o").replace("mundo","amigo"))
Variable = "Me gusta comer chocolates"
# Si queremos generar una lista de cada palabra indicamos como separador un espacio simple
print(Variable.split(" "))
# Si queremos una separación que genere una lista de dos elementos separamos con una palabra
print(Variable.split("comer"))
# Si queremos mas elementos tambien podemos usar como separador un caracter
print(Variable.split("c"))
# Podemos indicar cuantos separadores utilizaremos por medio de un segundo parametro
print(Variable.split("c",1))
Lineas = "Hola\nComo\nEstas?"
# Separar por saltos de linea
print(Lineas)
print(Lineas.splitlines())
Values = ["Los", "dulces", "son", "de", "sabor", "a", "moras"]
" ".join(Values)
| 0.123036 | 0.837487 |
#### Generate some data
```
import pandas as pd
import numpy as np
from pandas import Timestamp
mycols = ['store', 'type', 'department', 'date', 'weekly_sales', 'is_holiday']
content = np.array([[1, 'A', 1, Timestamp('2010-02-05 00:00:00'), 24924.5, False],[1, 'B', 1, Timestamp('2010-03-05 00:00:00'), 21827.9, True],[1, 'A', 1, Timestamp('2010-04-02 00:00:00'), 57258.43, False],[1, 'C', 1, Timestamp('2010-05-07 00:00:00'), 17413.94, False],[1, 'C', 1, Timestamp('2010-06-04 00:00:00'), 17558.09, False],[1, 'C', 1, Timestamp('2010-07-02 00:00:00'), 16333.14, False],[1, 'C', 1, Timestamp('2010-08-06 00:00:00'), 17508.41, False],[1, 'B', 1, Timestamp('2010-09-03 00:00:00'), 16241.78, False],[1, 'A', 1, Timestamp('2010-10-01 00:00:00'), 20094.19, True],[1, 'B', 1, Timestamp('2010-11-05 00:00:00'), 34238.88, False],[1, 'C', 1, Timestamp('2010-12-03 00:00:00'), 22517.56, False],[1, 'A', 1, Timestamp('2011-01-07 00:00:00'), 15984.24, False],[1, 'C', 2, Timestamp('2010-02-05 00:00:00'), 50605.27, True],[1, 'A', 2, Timestamp('2010-03-05 00:00:00'), 48397.98, False],[1, 'A', 2, Timestamp('2010-04-02 00:00:00'), 47450.5, False],[1, 'A', 2, Timestamp('2010-05-07 00:00:00'), 47903.01, False],[1, 'A', 2, Timestamp('2010-06-04 00:00:00'), 48754.47, False],[1, 'A', 2, Timestamp('2010-07-02 00:00:00'), 47077.72, False],[1, 'A', 2, Timestamp('2010-08-06 00:00:00'), 50031.73, False],[1, 'A', 2, Timestamp('2010-09-03 00:00:00'), 49015.05, False],[1, 'A', 2, Timestamp('2010-10-01 00:00:00'), 45829.02, False],[1, 'A', 2, Timestamp('2010-11-05 00:00:00'), 46381.43, True],[1, 'A', 2, Timestamp('2010-12-03 00:00:00'), 44405.02, False],[1, 'A', 2, Timestamp('2011-01-07 00:00:00'), 43202.29, False],[1, 'A', 3, Timestamp('2010-02-05 00:00:00'), 13740.12, False],[1, 'A', 3, Timestamp('2010-03-05 00:00:00'), 12275.58, False],[1, 'A', 3, Timestamp('2010-04-02 00:00:00'), 11157.08, False],[1, 'A', 3, Timestamp('2010-05-07 00:00:00'), 9372.8, True],[1, 'A', 3, Timestamp('2010-06-04 00:00:00'), 8001.41, False],[1, 'C', 3, Timestamp('2010-07-02 00:00:00'), 7857.88, True],[1, 'A', 3, Timestamp('2010-08-06 00:00:00'), 26719.02, False],[1, 'A', 3, Timestamp('2010-09-03 00:00:00'), 19081.8, False],[1, 'B', 3, Timestamp('2010-10-01 00:00:00'), 9775.17, False],[1, 'A', 3, Timestamp('2010-11-05 00:00:00'), 9825.22, False],[1, 'A', 3, Timestamp('2010-12-03 00:00:00'), 10856.85, False],[1, 'A', 3, Timestamp('2011-01-07 00:00:00'), 15808.15, False],[1, 'A', 4, Timestamp('2010-02-05 00:00:00'), 39954.04, False],[1, 'A', 4, Timestamp('2010-03-05 00:00:00'), 38086.19, False],[1, 'A', 4, Timestamp('2010-04-02 00:00:00'), 37809.49, False],[1, 'A', 4, Timestamp('2010-05-07 00:00:00'), 37168.34, False],[1, 'A', 4, Timestamp('2010-06-04 00:00:00'), 40548.19, False],[1, 'B', 4, Timestamp('2010-07-02 00:00:00'), 39773.71, False],[1, 'A', 4, Timestamp('2010-08-06 00:00:00'), 40973.88, False],[1, 'A', 4, Timestamp('2010-09-03 00:00:00'), 38321.88, False],[1, 'A', 4, Timestamp('2010-10-01 00:00:00'), 34912.45, False],[1, 'A', 4, Timestamp('2010-11-05 00:00:00'), 37980.55, False],[1, 'A', 4, Timestamp('2010-12-03 00:00:00'), 37110.55, False],[1, 'A', 4, Timestamp('2011-01-07 00:00:00'), 37947.8, False],[1, 'A', 5, Timestamp('2010-02-05 00:00:00'), 32229.38, False],[1, 'A', 5, Timestamp('2010-03-05 00:00:00'), 23082.14, False],[1, 'B', 5, Timestamp('2010-04-02 00:00:00'), 29967.92, False],[1, 'A', 5, Timestamp('2010-05-07 00:00:00'), 19260.44, False],[1, 'A', 5, Timestamp('2010-06-04 00:00:00'), 22932.26, False],[1, 'A', 5, Timestamp('2010-07-02 00:00:00'), 18887.71, False],[1, 'A', 5, Timestamp('2010-08-06 00:00:00'), 16926.17, False],[1, 'A', 5, Timestamp('2010-09-03 00:00:00'), 15390.52, False],[1, 'A', 5, Timestamp('2010-10-01 00:00:00'), 23381.38, False],[1, 'A', 5, Timestamp('2010-11-05 00:00:00'), 23903.81, False],[1, 'A', 5, Timestamp('2010-12-03 00:00:00'), 36472.02, False],[1, 'A', 5, Timestamp('2011-01-07 00:00:00'), 22699.69, False],[1, 'A', 6, Timestamp('2010-02-05 00:00:00'), 5749.03, False],[1, 'A', 6, Timestamp('2010-03-05 00:00:00'), 4221.25, False],[1, 'A', 6, Timestamp('2010-04-02 00:00:00'), 4132.61, False],[1, 'A', 6, Timestamp('2010-05-07 00:00:00'), 7477.7, False],[1, 'A', 6, Timestamp('2010-06-04 00:00:00'), 5484.9, False],[1, 'A', 6, Timestamp('2010-07-02 00:00:00'), 4541.91, False],[1, 'A', 6, Timestamp('2010-08-06 00:00:00'), 4700.38, False],[1, 'A', 6, Timestamp('2010-09-03 00:00:00'), 3553.75, False],[1, 'B', 6, Timestamp('2010-10-01 00:00:00'), 2876.19, False],[1, 'A', 6, Timestamp('2010-11-05 00:00:00'), 5036.99, False],[1, 'A', 6, Timestamp('2010-12-03 00:00:00'), 6356.96, False],[1, 'A', 6, Timestamp('2011-01-07 00:00:00'), 1376.15, False],[1, 'A', 7, Timestamp('2010-02-05 00:00:00'), 21084.08, False],[1, 'A', 7, Timestamp('2010-03-05 00:00:00'), 19659.7, False],[1, 'A', 7, Timestamp('2010-04-02 00:00:00'), 22427.62, False],[1, 'A', 7, Timestamp('2010-05-07 00:00:00'), 20457.62, False],[1, 'A', 7, Timestamp('2010-06-04 00:00:00'), 44563.68, False],[1, 'A', 7, Timestamp('2010-07-02 00:00:00'), 22589.0, False],[1, 'A', 7, Timestamp('2010-08-06 00:00:00'), 21842.57, False],[1, 'A', 7, Timestamp('2010-09-03 00:00:00'), 18005.65, False],[1, 'A', 7, Timestamp('2010-10-01 00:00:00'), 16481.79, False],[1, 'A', 7, Timestamp('2010-11-05 00:00:00'), 19136.58, False],[1, 'A', 7, Timestamp('2010-12-03 00:00:00'), 47406.83, False],[1, 'A', 7, Timestamp('2011-01-07 00:00:00'), 17516.16, False],[1, 'A', 8, Timestamp('2010-02-05 00:00:00'), 40129.01, False],[1, 'A', 8, Timestamp('2010-03-05 00:00:00'), 38776.09, False],[1, 'A', 8, Timestamp('2010-04-02 00:00:00'), 38151.58, False],[1, 'A', 8, Timestamp('2010-05-07 00:00:00'), 35393.78, False],[1, 'A', 8, Timestamp('2010-06-04 00:00:00'), 35181.47, False],[1, 'A', 8, Timestamp('2010-07-02 00:00:00'), 35580.01, False],[1, 'A', 8, Timestamp('2010-08-06 00:00:00'), 34833.35, False],[1, 'A', 8, Timestamp('2010-09-03 00:00:00'), 35562.68, False],[1, 'A', 8, Timestamp('2010-10-01 00:00:00'), 34658.25, False],[1, 'A', 8, Timestamp('2010-11-05 00:00:00'), 36182.58, False],[1, 'A', 8, Timestamp('2010-12-03 00:00:00'), 36222.74, False],[1, 'A', 8, Timestamp('2011-01-07 00:00:00'), 36599.46, False],[1, 'A', 9, Timestamp('2010-02-05 00:00:00'), 16930.99, False],[1, 'A', 9, Timestamp('2010-03-05 00:00:00'), 24064.7, False],[1, 'A', 9, Timestamp('2010-04-02 00:00:00'), 25435.02, False],[1, 'A', 9, Timestamp('2010-05-07 00:00:00'), 27588.34, False]])
df = pd.DataFrame(content, columns=mycols)
df['frac_sales'] = df['weekly_sales']*np.random.rand()
df.head()
```
### `loc[]`
DataFrames can be sliced by index values using `loc[]`
- You can only slice an index if the index is sorted (using .sort_index())
- To slice at the outer level, first and last can be strings
- To slice at inner levels, first and last should be tuples
- If you pass a single slice to .loc[], it will slice the rows
```
df_srt = df.set_index(['type','department']).sort_index()
df_srt.loc['A':'B']
df_srt.loc[('A',2):('B',1)]
df_srt.loc[('A',2):('B',1),['weekly_sales','frac_sales']]
df_srt.loc[('A',2):('B',1),'weekly_sales':'frac_sales']
```
### `iloc[]`
```
df.iloc[:5,2:3]
```
|
github_jupyter
|
import pandas as pd
import numpy as np
from pandas import Timestamp
mycols = ['store', 'type', 'department', 'date', 'weekly_sales', 'is_holiday']
content = np.array([[1, 'A', 1, Timestamp('2010-02-05 00:00:00'), 24924.5, False],[1, 'B', 1, Timestamp('2010-03-05 00:00:00'), 21827.9, True],[1, 'A', 1, Timestamp('2010-04-02 00:00:00'), 57258.43, False],[1, 'C', 1, Timestamp('2010-05-07 00:00:00'), 17413.94, False],[1, 'C', 1, Timestamp('2010-06-04 00:00:00'), 17558.09, False],[1, 'C', 1, Timestamp('2010-07-02 00:00:00'), 16333.14, False],[1, 'C', 1, Timestamp('2010-08-06 00:00:00'), 17508.41, False],[1, 'B', 1, Timestamp('2010-09-03 00:00:00'), 16241.78, False],[1, 'A', 1, Timestamp('2010-10-01 00:00:00'), 20094.19, True],[1, 'B', 1, Timestamp('2010-11-05 00:00:00'), 34238.88, False],[1, 'C', 1, Timestamp('2010-12-03 00:00:00'), 22517.56, False],[1, 'A', 1, Timestamp('2011-01-07 00:00:00'), 15984.24, False],[1, 'C', 2, Timestamp('2010-02-05 00:00:00'), 50605.27, True],[1, 'A', 2, Timestamp('2010-03-05 00:00:00'), 48397.98, False],[1, 'A', 2, Timestamp('2010-04-02 00:00:00'), 47450.5, False],[1, 'A', 2, Timestamp('2010-05-07 00:00:00'), 47903.01, False],[1, 'A', 2, Timestamp('2010-06-04 00:00:00'), 48754.47, False],[1, 'A', 2, Timestamp('2010-07-02 00:00:00'), 47077.72, False],[1, 'A', 2, Timestamp('2010-08-06 00:00:00'), 50031.73, False],[1, 'A', 2, Timestamp('2010-09-03 00:00:00'), 49015.05, False],[1, 'A', 2, Timestamp('2010-10-01 00:00:00'), 45829.02, False],[1, 'A', 2, Timestamp('2010-11-05 00:00:00'), 46381.43, True],[1, 'A', 2, Timestamp('2010-12-03 00:00:00'), 44405.02, False],[1, 'A', 2, Timestamp('2011-01-07 00:00:00'), 43202.29, False],[1, 'A', 3, Timestamp('2010-02-05 00:00:00'), 13740.12, False],[1, 'A', 3, Timestamp('2010-03-05 00:00:00'), 12275.58, False],[1, 'A', 3, Timestamp('2010-04-02 00:00:00'), 11157.08, False],[1, 'A', 3, Timestamp('2010-05-07 00:00:00'), 9372.8, True],[1, 'A', 3, Timestamp('2010-06-04 00:00:00'), 8001.41, False],[1, 'C', 3, Timestamp('2010-07-02 00:00:00'), 7857.88, True],[1, 'A', 3, Timestamp('2010-08-06 00:00:00'), 26719.02, False],[1, 'A', 3, Timestamp('2010-09-03 00:00:00'), 19081.8, False],[1, 'B', 3, Timestamp('2010-10-01 00:00:00'), 9775.17, False],[1, 'A', 3, Timestamp('2010-11-05 00:00:00'), 9825.22, False],[1, 'A', 3, Timestamp('2010-12-03 00:00:00'), 10856.85, False],[1, 'A', 3, Timestamp('2011-01-07 00:00:00'), 15808.15, False],[1, 'A', 4, Timestamp('2010-02-05 00:00:00'), 39954.04, False],[1, 'A', 4, Timestamp('2010-03-05 00:00:00'), 38086.19, False],[1, 'A', 4, Timestamp('2010-04-02 00:00:00'), 37809.49, False],[1, 'A', 4, Timestamp('2010-05-07 00:00:00'), 37168.34, False],[1, 'A', 4, Timestamp('2010-06-04 00:00:00'), 40548.19, False],[1, 'B', 4, Timestamp('2010-07-02 00:00:00'), 39773.71, False],[1, 'A', 4, Timestamp('2010-08-06 00:00:00'), 40973.88, False],[1, 'A', 4, Timestamp('2010-09-03 00:00:00'), 38321.88, False],[1, 'A', 4, Timestamp('2010-10-01 00:00:00'), 34912.45, False],[1, 'A', 4, Timestamp('2010-11-05 00:00:00'), 37980.55, False],[1, 'A', 4, Timestamp('2010-12-03 00:00:00'), 37110.55, False],[1, 'A', 4, Timestamp('2011-01-07 00:00:00'), 37947.8, False],[1, 'A', 5, Timestamp('2010-02-05 00:00:00'), 32229.38, False],[1, 'A', 5, Timestamp('2010-03-05 00:00:00'), 23082.14, False],[1, 'B', 5, Timestamp('2010-04-02 00:00:00'), 29967.92, False],[1, 'A', 5, Timestamp('2010-05-07 00:00:00'), 19260.44, False],[1, 'A', 5, Timestamp('2010-06-04 00:00:00'), 22932.26, False],[1, 'A', 5, Timestamp('2010-07-02 00:00:00'), 18887.71, False],[1, 'A', 5, Timestamp('2010-08-06 00:00:00'), 16926.17, False],[1, 'A', 5, Timestamp('2010-09-03 00:00:00'), 15390.52, False],[1, 'A', 5, Timestamp('2010-10-01 00:00:00'), 23381.38, False],[1, 'A', 5, Timestamp('2010-11-05 00:00:00'), 23903.81, False],[1, 'A', 5, Timestamp('2010-12-03 00:00:00'), 36472.02, False],[1, 'A', 5, Timestamp('2011-01-07 00:00:00'), 22699.69, False],[1, 'A', 6, Timestamp('2010-02-05 00:00:00'), 5749.03, False],[1, 'A', 6, Timestamp('2010-03-05 00:00:00'), 4221.25, False],[1, 'A', 6, Timestamp('2010-04-02 00:00:00'), 4132.61, False],[1, 'A', 6, Timestamp('2010-05-07 00:00:00'), 7477.7, False],[1, 'A', 6, Timestamp('2010-06-04 00:00:00'), 5484.9, False],[1, 'A', 6, Timestamp('2010-07-02 00:00:00'), 4541.91, False],[1, 'A', 6, Timestamp('2010-08-06 00:00:00'), 4700.38, False],[1, 'A', 6, Timestamp('2010-09-03 00:00:00'), 3553.75, False],[1, 'B', 6, Timestamp('2010-10-01 00:00:00'), 2876.19, False],[1, 'A', 6, Timestamp('2010-11-05 00:00:00'), 5036.99, False],[1, 'A', 6, Timestamp('2010-12-03 00:00:00'), 6356.96, False],[1, 'A', 6, Timestamp('2011-01-07 00:00:00'), 1376.15, False],[1, 'A', 7, Timestamp('2010-02-05 00:00:00'), 21084.08, False],[1, 'A', 7, Timestamp('2010-03-05 00:00:00'), 19659.7, False],[1, 'A', 7, Timestamp('2010-04-02 00:00:00'), 22427.62, False],[1, 'A', 7, Timestamp('2010-05-07 00:00:00'), 20457.62, False],[1, 'A', 7, Timestamp('2010-06-04 00:00:00'), 44563.68, False],[1, 'A', 7, Timestamp('2010-07-02 00:00:00'), 22589.0, False],[1, 'A', 7, Timestamp('2010-08-06 00:00:00'), 21842.57, False],[1, 'A', 7, Timestamp('2010-09-03 00:00:00'), 18005.65, False],[1, 'A', 7, Timestamp('2010-10-01 00:00:00'), 16481.79, False],[1, 'A', 7, Timestamp('2010-11-05 00:00:00'), 19136.58, False],[1, 'A', 7, Timestamp('2010-12-03 00:00:00'), 47406.83, False],[1, 'A', 7, Timestamp('2011-01-07 00:00:00'), 17516.16, False],[1, 'A', 8, Timestamp('2010-02-05 00:00:00'), 40129.01, False],[1, 'A', 8, Timestamp('2010-03-05 00:00:00'), 38776.09, False],[1, 'A', 8, Timestamp('2010-04-02 00:00:00'), 38151.58, False],[1, 'A', 8, Timestamp('2010-05-07 00:00:00'), 35393.78, False],[1, 'A', 8, Timestamp('2010-06-04 00:00:00'), 35181.47, False],[1, 'A', 8, Timestamp('2010-07-02 00:00:00'), 35580.01, False],[1, 'A', 8, Timestamp('2010-08-06 00:00:00'), 34833.35, False],[1, 'A', 8, Timestamp('2010-09-03 00:00:00'), 35562.68, False],[1, 'A', 8, Timestamp('2010-10-01 00:00:00'), 34658.25, False],[1, 'A', 8, Timestamp('2010-11-05 00:00:00'), 36182.58, False],[1, 'A', 8, Timestamp('2010-12-03 00:00:00'), 36222.74, False],[1, 'A', 8, Timestamp('2011-01-07 00:00:00'), 36599.46, False],[1, 'A', 9, Timestamp('2010-02-05 00:00:00'), 16930.99, False],[1, 'A', 9, Timestamp('2010-03-05 00:00:00'), 24064.7, False],[1, 'A', 9, Timestamp('2010-04-02 00:00:00'), 25435.02, False],[1, 'A', 9, Timestamp('2010-05-07 00:00:00'), 27588.34, False]])
df = pd.DataFrame(content, columns=mycols)
df['frac_sales'] = df['weekly_sales']*np.random.rand()
df.head()
df_srt = df.set_index(['type','department']).sort_index()
df_srt.loc['A':'B']
df_srt.loc[('A',2):('B',1)]
df_srt.loc[('A',2):('B',1),['weekly_sales','frac_sales']]
df_srt.loc[('A',2):('B',1),'weekly_sales':'frac_sales']
df.iloc[:5,2:3]
| 0.374104 | 0.725016 |
## Calculate terminated results Exiobase v.3.3.11b1 exc. iLUC, electricity markets and social extensions
Investments are not integrated in the MR_HIOT table. They are accounted for in the Final Demand activities.
This tutorial is divided in 4 sections.
1. Extract numbers from Excel files
2. Replace 0s with 1s in norm0 file
3. Read all the csv files as matrices
4. Run operations
## 1. Extract numbers from Excel files
#### Give the name and location of the excel file containing the HIOT and the FD tables.
```
HIOT_FD = "/Users/marie/Desktop/MR_HIOT_2011_v3.3.11.xlsx"
import pandas as pd
import csv
### MR-HIOT.csv is created because the excel is too heavy
data_xls = pd.read_excel(HIOT_FD, 'HIOT', index_col=None)
data_xls.to_csv('MR_HIOT.csv', encoding='utf-8')
### FD.csv is created because the excel is too heavy
data_xls = pd.read_excel(HIOT_FD, 'FD', index_col=None, header = None)
data_xls.to_csv('FD.csv', encoding='utf-8')
```
From MR_HIOT_2011_v3.3.11.xlsx (MR_HIOT.csv & FD.csv), we create:
- Zn_tonorm.csv (7872 columns & rows)
- norm.csv (7872 columns)
- FD.csv (288 columns & 7872 rows)
```
outfile1 ="/Users/marie/Desktop/Zn_tonorm.csv"
source = pd.read_csv('MR_HIOT.csv', index_col = None, header = None, low_memory = False)
Zn_tonorm = source.iloc[7:7879, 5:7877]
Zn_tonorm.to_csv(outfile1, header = None, index = None)
outfile2 ="/Users/marie/Desktop/norm.csv"
norm = source.iloc[1:2, 5:7877]
norm.to_csv(outfile2, header = None, index = None)
outfile3 ="/Users/marie/Desktop/FD.csv"
source1 = pd.read_csv('FD.csv', index_col = None, header = None, low_memory = False)
FD = source1.iloc[8:7880, 6:294]
FD.to_csv(outfile3, header = None, index = None)
```
#### Give the name of the excel file from which the extensions should be extracted
The file from Stefano cannot be used as it is.
Water extensions should be corrected, biogenic carbon relocated, biogenic methane recalculated and land occupation flows summed up.
```
extensions = "/Users/marie/Desktop/MR_HIOT_2011_v3.3.11_extensions_MS.xlsx"
```
We create Bn_tonorm.csv including the extensions for the 7872 producing activities:
- 30 resource flows (green water was excluded)
- 240 land occupation flows
- 62 direct emissions to Air, Water and Soil
```
import pandas as pd
data_xls = pd.read_excel(extensions, 'resource_act', index_col=None, header = None, encoding='utf-8')
##outfile4 ="/Users/marie/Desktop/Bn_tonorm_resource.csv"
Bn_tonorm_resource = data_xls.iloc[7:37, 5:7877]
##Bn_tonorm_resource.to_csv(outfile4, header = None, index = None)
data_xls = pd.read_excel(extensions, 'Land_act', index_col=None, header = None, encoding='utf-8')
##outfile5 ="/Users/marie/Desktop/Bn_tonorm_land.csv"
Bn_tonorm_land = data_xls.iloc[247:251, 5:7877]
##Bn_tonorm_land.to_csv(outfile5, header = None, Index = None)
data_xls = pd.read_excel(extensions, 'Emiss_act', index_col=None, header = None, encoding='utf-8')
##outfile6 ="/Users/marie/Desktop/Bn_tonorm_emiss.csv"
Bn_tonorm_emiss = data_xls.iloc[7:70, 5:7877]
##Bn_tonorm_emiss.to_csv(outfile6, header = None, index = None)
outfile ="/Users/marie/Desktop/Bn_tonorm.csv"
frame = [Bn_tonorm_resource, Bn_tonorm_land, Bn_tonorm_emiss]
Bn_tonorm = pd.concat(frame)
Bn_tonorm.to_csv(outfile, header = None, index = None)
```
We create FD_ext.csv including the extensions for the 288 Final Demand activities.
```
data_xls = pd.read_excel(extensions, 'resource_FD', index_col=None, header = None, encoding='utf-8')
##outfile8 ="/Users/marie/Desktop/FD_resource.csv"
FD_resource = data_xls.iloc[7:37, 5:293]
##FD_resource.to_csv(outfile8, header = None, index = None)
data_xls = pd.read_excel(extensions, 'Land_FD', index_col=None, header = None, encoding='utf-8')
##outfile9 ="/Users/marie/Desktop/FD_land.csv"
FD_land = data_xls.iloc[247:251, 5:293]
##FD_land.to_csv(outfile9, header = None, index = None)
data_xls = pd.read_excel(extensions, 'Emiss_FD', index_col=None, header = None,encoding='utf-8')
##outfile10 ="/Users/marie/Desktop/FD_emiss.csv"
FD_emiss = data_xls.iloc[7:70, 5:293]
##FD_emiss.to_csv(outfile10, header = None, Index = None)
outfile ="/Users/marie/Desktop/FD_ext.csv"
frame = [FD_resource, FD_land, FD_emiss]
FD_ext = pd.concat(frame)
FD_ext.to_csv(outfile, header = None, index = None)
```
## 2. Replace 0s with 1s in norm0 file
Replace 0s with 1s in norm.csv (matrices can't be divided by 0)
```
def replace_0with1(source, result):
with open(source,"r") as source:
rdr = csv.reader(source)
with open (result, "w") as result:
wtr = csv.writer(result)
for row in rdr:
row = [x.replace('0', '1') if x == '0' else x for x in row]
wtr.writerow(row)
replace_0with1("norm0.csv", "norm1.csv")
```
## 3. Read CSV files as matrices
To make operations with the numpy package, read the following files extracted previously:
- Zn_tonorm.csv as a matrice
- norm1.csv as a vector
- Bn_tonorm.csv as a matrice
- FD.csv as a matrice
- FD_ext.csv as a matrice
```
import csv
import numpy as np
with open('norm1.csv','r') as dest_f:
data_iter = csv.reader(dest_f,
delimiter = ',',
quotechar = '"')
data = [data for data in data_iter]
nor = np.asarray(data, dtype='float')
with open("Zn_tonorm.csv",'r') as dest_f:
data_iter = csv.reader(dest_f,
delimiter = ',',
quotechar = '"')
data = [data for data in data_iter]
Zn_tonorm = np.array(list(data)).astype('float')
with open("Bn_tonorm.csv",'r') as dest_f:
data_iter = csv.reader(dest_f,
delimiter = ',',
quotechar = '"')
data = [data for data in data_iter]
Bn_tonorm = np.array(list(data)).astype('float')
with open("FD.csv",'r') as dest_f:
data_iter = csv.reader(dest_f,
delimiter = ',',
quotechar = '"')
data = [data for data in data_iter]
f_cons = np.array(list(data)).astype('float')
with open("FD_ext.csv",'r') as dest_f:
data_iter = csv.reader(dest_f,
delimiter = ',',
quotechar = '"')
data = [data for data in data_iter]
f_em = np.array(list(data)).astype('float')
```
## 4. Run operations
To obtain Zn and Bn, Zn_tonorm and Bn_tonorm needs to be didvided by the norm vector.
```
Zn = Zn_tonorm/nor
Bn = Bn_tonorm/nor
```
We create the identity matrice
```
identity = np.matrix(np.identity(7872), copy=False)
An = identity-Zn
S = np.linalg.inv(An)
BLCI = Bn*S
from io import StringIO
import numpy as np
s=StringIO()
np.savetxt('BLCI.csv', BLCI, fmt='%.10f', delimiter=',', newline="\n")
F = BLCI*f_cons
F2 = F+f_em
from io import StringIO
import numpy as np
s=StringIO()
np.savetxt('F2.csv', F2, fmt='%.10f', delimiter=',', newline="\n")
```
|
github_jupyter
|
HIOT_FD = "/Users/marie/Desktop/MR_HIOT_2011_v3.3.11.xlsx"
import pandas as pd
import csv
### MR-HIOT.csv is created because the excel is too heavy
data_xls = pd.read_excel(HIOT_FD, 'HIOT', index_col=None)
data_xls.to_csv('MR_HIOT.csv', encoding='utf-8')
### FD.csv is created because the excel is too heavy
data_xls = pd.read_excel(HIOT_FD, 'FD', index_col=None, header = None)
data_xls.to_csv('FD.csv', encoding='utf-8')
outfile1 ="/Users/marie/Desktop/Zn_tonorm.csv"
source = pd.read_csv('MR_HIOT.csv', index_col = None, header = None, low_memory = False)
Zn_tonorm = source.iloc[7:7879, 5:7877]
Zn_tonorm.to_csv(outfile1, header = None, index = None)
outfile2 ="/Users/marie/Desktop/norm.csv"
norm = source.iloc[1:2, 5:7877]
norm.to_csv(outfile2, header = None, index = None)
outfile3 ="/Users/marie/Desktop/FD.csv"
source1 = pd.read_csv('FD.csv', index_col = None, header = None, low_memory = False)
FD = source1.iloc[8:7880, 6:294]
FD.to_csv(outfile3, header = None, index = None)
extensions = "/Users/marie/Desktop/MR_HIOT_2011_v3.3.11_extensions_MS.xlsx"
import pandas as pd
data_xls = pd.read_excel(extensions, 'resource_act', index_col=None, header = None, encoding='utf-8')
##outfile4 ="/Users/marie/Desktop/Bn_tonorm_resource.csv"
Bn_tonorm_resource = data_xls.iloc[7:37, 5:7877]
##Bn_tonorm_resource.to_csv(outfile4, header = None, index = None)
data_xls = pd.read_excel(extensions, 'Land_act', index_col=None, header = None, encoding='utf-8')
##outfile5 ="/Users/marie/Desktop/Bn_tonorm_land.csv"
Bn_tonorm_land = data_xls.iloc[247:251, 5:7877]
##Bn_tonorm_land.to_csv(outfile5, header = None, Index = None)
data_xls = pd.read_excel(extensions, 'Emiss_act', index_col=None, header = None, encoding='utf-8')
##outfile6 ="/Users/marie/Desktop/Bn_tonorm_emiss.csv"
Bn_tonorm_emiss = data_xls.iloc[7:70, 5:7877]
##Bn_tonorm_emiss.to_csv(outfile6, header = None, index = None)
outfile ="/Users/marie/Desktop/Bn_tonorm.csv"
frame = [Bn_tonorm_resource, Bn_tonorm_land, Bn_tonorm_emiss]
Bn_tonorm = pd.concat(frame)
Bn_tonorm.to_csv(outfile, header = None, index = None)
data_xls = pd.read_excel(extensions, 'resource_FD', index_col=None, header = None, encoding='utf-8')
##outfile8 ="/Users/marie/Desktop/FD_resource.csv"
FD_resource = data_xls.iloc[7:37, 5:293]
##FD_resource.to_csv(outfile8, header = None, index = None)
data_xls = pd.read_excel(extensions, 'Land_FD', index_col=None, header = None, encoding='utf-8')
##outfile9 ="/Users/marie/Desktop/FD_land.csv"
FD_land = data_xls.iloc[247:251, 5:293]
##FD_land.to_csv(outfile9, header = None, index = None)
data_xls = pd.read_excel(extensions, 'Emiss_FD', index_col=None, header = None,encoding='utf-8')
##outfile10 ="/Users/marie/Desktop/FD_emiss.csv"
FD_emiss = data_xls.iloc[7:70, 5:293]
##FD_emiss.to_csv(outfile10, header = None, Index = None)
outfile ="/Users/marie/Desktop/FD_ext.csv"
frame = [FD_resource, FD_land, FD_emiss]
FD_ext = pd.concat(frame)
FD_ext.to_csv(outfile, header = None, index = None)
def replace_0with1(source, result):
with open(source,"r") as source:
rdr = csv.reader(source)
with open (result, "w") as result:
wtr = csv.writer(result)
for row in rdr:
row = [x.replace('0', '1') if x == '0' else x for x in row]
wtr.writerow(row)
replace_0with1("norm0.csv", "norm1.csv")
import csv
import numpy as np
with open('norm1.csv','r') as dest_f:
data_iter = csv.reader(dest_f,
delimiter = ',',
quotechar = '"')
data = [data for data in data_iter]
nor = np.asarray(data, dtype='float')
with open("Zn_tonorm.csv",'r') as dest_f:
data_iter = csv.reader(dest_f,
delimiter = ',',
quotechar = '"')
data = [data for data in data_iter]
Zn_tonorm = np.array(list(data)).astype('float')
with open("Bn_tonorm.csv",'r') as dest_f:
data_iter = csv.reader(dest_f,
delimiter = ',',
quotechar = '"')
data = [data for data in data_iter]
Bn_tonorm = np.array(list(data)).astype('float')
with open("FD.csv",'r') as dest_f:
data_iter = csv.reader(dest_f,
delimiter = ',',
quotechar = '"')
data = [data for data in data_iter]
f_cons = np.array(list(data)).astype('float')
with open("FD_ext.csv",'r') as dest_f:
data_iter = csv.reader(dest_f,
delimiter = ',',
quotechar = '"')
data = [data for data in data_iter]
f_em = np.array(list(data)).astype('float')
Zn = Zn_tonorm/nor
Bn = Bn_tonorm/nor
identity = np.matrix(np.identity(7872), copy=False)
An = identity-Zn
S = np.linalg.inv(An)
BLCI = Bn*S
from io import StringIO
import numpy as np
s=StringIO()
np.savetxt('BLCI.csv', BLCI, fmt='%.10f', delimiter=',', newline="\n")
F = BLCI*f_cons
F2 = F+f_em
from io import StringIO
import numpy as np
s=StringIO()
np.savetxt('F2.csv', F2, fmt='%.10f', delimiter=',', newline="\n")
| 0.220175 | 0.789984 |
```
### ADLFRAMEWORK
import adlframework
from adlframework.retrievals.BlobLocalCache import BlobLocalCache
from adlframework.datasource import DataSource
from adlframework.dataentity.audio_de import AudioRecordingDataEntity
from adlframework.experiment import SimpleExperiment
from adlframework.processors.general_processors import reshape, to_np_arr
from adlframework.filters.general_filters import min_array_shape
### KERAS
from keras.losses import KLD, MAE
from keras.optimizers import Adadelta, Adam
import keras.backend as K
from keras.models import Sequential
from keras.layers import *
from keras.callbacks import ModelCheckpoint, TensorBoard
from keras.utils.training_utils import multi_gpu_model
### UTILS
import pdb
import numpy as np
from functools import partial
import tensorflow as tf
from tqdm import tqdm_notebook as tqdm
config = tf.ConfigProto()
config.gpu_options.allow_growth=True
tfsession = tf.Session(config=config)
K.set_session(tfsession)
### Hyperparameters
input_window = 3 # seconds
input_time = 3
drop_out_prob = .2
stride = 8
padding = 'same'
activation = 'relu'
optimizer = 'adam'
loss = 'MAE'
epochs = 100
steps_per_epoch = 100
val_steps = 2
fs = 44100
input_shape = (fs*input_window, 1)
```
## DATA!
```
controllers = [to_np_arr, partial(reshape, shape=(-1, 1)), partial(min_array_shape, min_shape=input_shape)]
cache_path = 'local_cache/AVEC/'
train_retrieval = BlobLocalCache(cache_path+'wav/train', cache_path+'labels/train')
val_retrieval = BlobLocalCache(cache_path+'wav/dev', cache_path+'labels/dev')
# test_retrieval = BlobLocalCache(cache_path+'wav/test', cache_path+'labels/test')
epochs = 100
max_mem = .5
universal_args = {
'window_size': input_window,
'timestamp_column': 'Timestamps',
'sampling_method': 'linear_interpolation',
'ignore_cache': True,
'verbosity': 3,
'max_mem_percent': max_mem,
'controllers': controllers,
'batch_size': 100,
'workers': 16
}
## Creating and splitting datasets
train_ds = DataSource(train_retrieval, AudioRecordingDataEntity,
**universal_args)
val_ds = DataSource(val_retrieval, AudioRecordingDataEntity,
**universal_args)
# test_ds = DataSource(test_retrieval, AudioRecordingDataEntity,
# ignore_cache=True,
# batch_size=30,
# verbosity=3,
# max_mem_percent=max_mem,
# workers=workers,
# controllers=controllers,
# **universal_args)
### Callbacks
callbacks = [#ModelCheckpoint('weights/weights.{epoch:02d}-{val_loss:.2f}.hdf5'),
TensorBoard(log_dir='./Graph', histogram_freq=0, write_graph=True, write_images=True)]
```
## Define Network
```
with tf.device("/cpu:0"):
model = Sequential()
model.add(Conv1D(240, 128, strides=stride, padding=padding, input_shape=input_shape))
model.add(BatchNormalization())
model.add(Activation(activation))
model.add(MaxPooling1D(pool_size=8))
model.add(Conv1D(360, 64, strides=stride, padding=padding))
model.add(BatchNormalization())
model.add(Activation(activation))
model.add(MaxPooling1D(pool_size=8))
model.add(Dropout(rate=drop_out_prob))
model.add(Conv1D(512, 32, strides=stride, padding=padding))
model.add(BatchNormalization())
model.add(Activation(activation))
model.add(Dropout(rate=drop_out_prob))
model.add(Conv1D(1024, 16, strides=stride, padding=padding))
model.add(BatchNormalization())
model.add(Activation(activation))
model.add(Dropout(rate=drop_out_prob))
model.add(Conv1D(512, 8, strides=stride, padding=padding))
model.add(BatchNormalization())
model.add(Activation(activation))
model.add(Flatten())
model.add(Dense(256, activation=activation))
model.add(Dropout(rate=drop_out_prob))
model.add(Dense(2, activation=activation, name='emotion_cnn_output'))
# make the model parallel
model = multi_gpu_model(model, gpus=8)
# we'll store a copy of the model on *every* GPU and then combine
# the results from the gradient updates on the CPU
# initialize the model
model.compile(optimizer, loss)
```
## Train!
```
model.fit_generator(train_ds,
steps_per_epoch=steps_per_epoch,
epochs=epochs,
callbacks=callbacks,
validation_data=val_ds,
validation_steps=val_steps)
```
|
github_jupyter
|
### ADLFRAMEWORK
import adlframework
from adlframework.retrievals.BlobLocalCache import BlobLocalCache
from adlframework.datasource import DataSource
from adlframework.dataentity.audio_de import AudioRecordingDataEntity
from adlframework.experiment import SimpleExperiment
from adlframework.processors.general_processors import reshape, to_np_arr
from adlframework.filters.general_filters import min_array_shape
### KERAS
from keras.losses import KLD, MAE
from keras.optimizers import Adadelta, Adam
import keras.backend as K
from keras.models import Sequential
from keras.layers import *
from keras.callbacks import ModelCheckpoint, TensorBoard
from keras.utils.training_utils import multi_gpu_model
### UTILS
import pdb
import numpy as np
from functools import partial
import tensorflow as tf
from tqdm import tqdm_notebook as tqdm
config = tf.ConfigProto()
config.gpu_options.allow_growth=True
tfsession = tf.Session(config=config)
K.set_session(tfsession)
### Hyperparameters
input_window = 3 # seconds
input_time = 3
drop_out_prob = .2
stride = 8
padding = 'same'
activation = 'relu'
optimizer = 'adam'
loss = 'MAE'
epochs = 100
steps_per_epoch = 100
val_steps = 2
fs = 44100
input_shape = (fs*input_window, 1)
controllers = [to_np_arr, partial(reshape, shape=(-1, 1)), partial(min_array_shape, min_shape=input_shape)]
cache_path = 'local_cache/AVEC/'
train_retrieval = BlobLocalCache(cache_path+'wav/train', cache_path+'labels/train')
val_retrieval = BlobLocalCache(cache_path+'wav/dev', cache_path+'labels/dev')
# test_retrieval = BlobLocalCache(cache_path+'wav/test', cache_path+'labels/test')
epochs = 100
max_mem = .5
universal_args = {
'window_size': input_window,
'timestamp_column': 'Timestamps',
'sampling_method': 'linear_interpolation',
'ignore_cache': True,
'verbosity': 3,
'max_mem_percent': max_mem,
'controllers': controllers,
'batch_size': 100,
'workers': 16
}
## Creating and splitting datasets
train_ds = DataSource(train_retrieval, AudioRecordingDataEntity,
**universal_args)
val_ds = DataSource(val_retrieval, AudioRecordingDataEntity,
**universal_args)
# test_ds = DataSource(test_retrieval, AudioRecordingDataEntity,
# ignore_cache=True,
# batch_size=30,
# verbosity=3,
# max_mem_percent=max_mem,
# workers=workers,
# controllers=controllers,
# **universal_args)
### Callbacks
callbacks = [#ModelCheckpoint('weights/weights.{epoch:02d}-{val_loss:.2f}.hdf5'),
TensorBoard(log_dir='./Graph', histogram_freq=0, write_graph=True, write_images=True)]
with tf.device("/cpu:0"):
model = Sequential()
model.add(Conv1D(240, 128, strides=stride, padding=padding, input_shape=input_shape))
model.add(BatchNormalization())
model.add(Activation(activation))
model.add(MaxPooling1D(pool_size=8))
model.add(Conv1D(360, 64, strides=stride, padding=padding))
model.add(BatchNormalization())
model.add(Activation(activation))
model.add(MaxPooling1D(pool_size=8))
model.add(Dropout(rate=drop_out_prob))
model.add(Conv1D(512, 32, strides=stride, padding=padding))
model.add(BatchNormalization())
model.add(Activation(activation))
model.add(Dropout(rate=drop_out_prob))
model.add(Conv1D(1024, 16, strides=stride, padding=padding))
model.add(BatchNormalization())
model.add(Activation(activation))
model.add(Dropout(rate=drop_out_prob))
model.add(Conv1D(512, 8, strides=stride, padding=padding))
model.add(BatchNormalization())
model.add(Activation(activation))
model.add(Flatten())
model.add(Dense(256, activation=activation))
model.add(Dropout(rate=drop_out_prob))
model.add(Dense(2, activation=activation, name='emotion_cnn_output'))
# make the model parallel
model = multi_gpu_model(model, gpus=8)
# we'll store a copy of the model on *every* GPU and then combine
# the results from the gradient updates on the CPU
# initialize the model
model.compile(optimizer, loss)
model.fit_generator(train_ds,
steps_per_epoch=steps_per_epoch,
epochs=epochs,
callbacks=callbacks,
validation_data=val_ds,
validation_steps=val_steps)
| 0.764452 | 0.420124 |
# 1次元のデータの整理
## データの中心の指標
```
import numpy as np
import pandas as pd
# Jupyter Notebookの出力を小数点以下3桁に抑える
%precision 3
# Dataframeの出力を小数点以下3桁に抑える
pd.set_option('precision', 3)
df = pd.read_csv('../data/ch2_scores_em.csv',
index_col='生徒番号')
# dfの最初の5行を表示
df.head()
scores = np.array(df['英語'])[:10]
scores
scores_df = pd.DataFrame({'点数':scores},
index=pd.Index(['A', 'B', 'C', 'D', 'E',
'F', 'G', 'H', 'I', 'J'],
name='生徒'))
scores_df
```
### 平均値
```
sum(scores) / len(scores)
np.mean(scores)
scores_df.mean()
```
### 中央値
```
sorted_scores = np.sort(scores)
sorted_scores
n = len(sorted_scores)
if n % 2 == 0:
m0 = sorted_scores[n//2 - 1]
m1 = sorted_scores[n//2]
median = (m0 + m1) / 2
else:
median = sorted_scores[(n+1)//2 - 1]
median
np.median(scores)
scores_df.median()
```
### 最頻値
```
pd.Series([1, 1, 1, 2, 2, 3]).mode()
pd.Series([1, 2, 3, 4, 5]).mode()
```
## データのばらつきの指標
### 分散と標準偏差
#### 偏差
```
mean = np.mean(scores)
deviation = scores - mean
deviation
another_scores = [50, 60, 58, 54, 51, 56, 57, 53, 52, 59]
another_mean = np.mean(another_scores)
another_deviation = another_scores - another_mean
another_deviation
np.mean(deviation)
np.mean(another_deviation)
summary_df = scores_df.copy()
summary_df['偏差'] = deviation
summary_df
summary_df.mean()
```
#### 分散
```
np.mean(deviation ** 2)
np.var(scores)
scores_df.var()
summary_df['偏差二乗'] = np.square(deviation)
summary_df
summary_df.mean()
```
#### 標準偏差
```
np.sqrt(np.var(scores, ddof=0))
np.std(scores, ddof=0)
```
### 範囲と四分位範囲
#### 範囲
```
np.max(scores) - np.min(scores)
```
#### 四分位範囲
```
scores_Q1 = np.percentile(scores, 25)
scores_Q3 = np.percentile(scores, 75)
scores_IQR = scores_Q3 - scores_Q1
scores_IQR
```
### データの指標のまとめ
```
pd.Series(scores).describe()
```
## データの正規化
### 標準化
```
z = (scores - np.mean(scores)) / np.std(scores)
z
np.mean(z), np.std(z, ddof=0)
```
### 偏差値
```
z = 50 + 10 * (scores - np.mean(scores)) / np.std(scores)
scores_df['偏差値'] = z
scores_df
```
## データの視覚化
```
# 50人分の英語の点数のarray
english_scores = np.array(df['英語'])
# Seriesに変換してdescribeを表示
pd.Series(english_scores).describe()
```
### 度数分布表
```
freq, _ = np.histogram(english_scores, bins=10, range=(0, 100))
freq
# 0~10, 10~20, ... といった文字列のリストを作成
freq_class = [f'{i}~{i+10}' for i in range(0, 100, 10)]
# freq_classをインデックスにしてfreqでDataFrameを作成
freq_dist_df = pd.DataFrame({'度数':freq},
index=pd.Index(freq_class,
name='階級'))
freq_dist_df
class_value = [(i+(i+10))//2 for i in range(0, 100, 10)]
class_value
rel_freq = freq / freq.sum()
rel_freq
cum_rel_freq = np.cumsum(rel_freq)
cum_rel_freq
freq_dist_df['階級値'] = class_value
freq_dist_df['相対度数'] = rel_freq
freq_dist_df['累積相対度数'] = cum_rel_freq
freq_dist_df = freq_dist_df[['階級値', '度数',
'相対度数', '累積相対度数']]
freq_dist_df
```
#### 最頻値ふたたび
```
freq_dist_df.loc[freq_dist_df['度数'].idxmax(), '階級値']
```
### ヒストグラム
```
# Matplotlibのpyplotモジュールをpltという名前でインポート
import matplotlib.pyplot as plt
# グラフがnotebook上に表示されるようにする
%matplotlib inline
# キャンバスを作る
# figsizeで横・縦の大きさを指定
fig = plt.figure(figsize=(10, 6))
# キャンバス上にグラフを描画するための領域を作る
# 引数は領域を1×1個作り、1つめの領域に描画することを意味する
ax = fig.add_subplot(111)
# 階級数を10にしてヒストグラムを描画
freq, _, _ = ax.hist(english_scores, bins=10, range=(0, 100))
# X軸にラベルをつける
ax.set_xlabel('点数')
# Y軸にラベルをつける
ax.set_ylabel('人数')
# X軸に0, 10, 20, ..., 100の目盛りをふる
ax.set_xticks(np.linspace(0, 100, 10+1))
# Y軸に0, 1, 2, ...の目盛りをふる
ax.set_yticks(np.arange(0, freq.max()+1))
# グラフの表示
plt.show()
fig = plt.figure(figsize=(10, 6))
ax = fig.add_subplot(111)
freq, _ , _ = ax.hist(english_scores, bins=25, range=(0, 100))
ax.set_xlabel('点数')
ax.set_ylabel('人数')
ax.set_xticks(np.linspace(0, 100, 25+1))
ax.set_yticks(np.arange(0, freq.max()+1))
plt.show()
fig = plt.figure(figsize=(10, 6))
ax1 = fig.add_subplot(111)
# Y軸のスケールが違うグラフをax1と同じ領域上に書けるようにする
ax2 = ax1.twinx()
# 相対度数のヒストグラムにするためには、度数をデータの数で割る必要がある
# これはhistの引数weightを指定することで実現できる
weights = np.ones_like(english_scores) / len(english_scores)
rel_freq, _, _ = ax1.hist(english_scores, bins=25,
range=(0, 100), weights=weights)
cum_rel_freq = np.cumsum(rel_freq)
class_value = [(i+(i+4))//2 for i in range(0, 100, 4)]
# 折れ線グラフの描画
# 引数lsを'--'にすることで線が点線に
# 引数markerを'o'にすることでデータ点を丸に
# 引数colorを'gray'にすることで灰色に
ax2.plot(class_value, cum_rel_freq,
ls='--', marker='o', color='gray')
# 折れ線グラフの罫線を消去
ax2.grid(visible=False)
ax1.set_xlabel('点数')
ax1.set_ylabel('相対度数')
ax2.set_ylabel('累積相対度数')
ax1.set_xticks(np.linspace(0, 100, 25+1))
plt.show()
```
### 箱ひげ図
```
fig = plt.figure(figsize=(5, 6))
ax = fig.add_subplot(111)
ax.boxplot(english_scores, labels=['英語'])
plt.show()
```
|
github_jupyter
|
import numpy as np
import pandas as pd
# Jupyter Notebookの出力を小数点以下3桁に抑える
%precision 3
# Dataframeの出力を小数点以下3桁に抑える
pd.set_option('precision', 3)
df = pd.read_csv('../data/ch2_scores_em.csv',
index_col='生徒番号')
# dfの最初の5行を表示
df.head()
scores = np.array(df['英語'])[:10]
scores
scores_df = pd.DataFrame({'点数':scores},
index=pd.Index(['A', 'B', 'C', 'D', 'E',
'F', 'G', 'H', 'I', 'J'],
name='生徒'))
scores_df
sum(scores) / len(scores)
np.mean(scores)
scores_df.mean()
sorted_scores = np.sort(scores)
sorted_scores
n = len(sorted_scores)
if n % 2 == 0:
m0 = sorted_scores[n//2 - 1]
m1 = sorted_scores[n//2]
median = (m0 + m1) / 2
else:
median = sorted_scores[(n+1)//2 - 1]
median
np.median(scores)
scores_df.median()
pd.Series([1, 1, 1, 2, 2, 3]).mode()
pd.Series([1, 2, 3, 4, 5]).mode()
mean = np.mean(scores)
deviation = scores - mean
deviation
another_scores = [50, 60, 58, 54, 51, 56, 57, 53, 52, 59]
another_mean = np.mean(another_scores)
another_deviation = another_scores - another_mean
another_deviation
np.mean(deviation)
np.mean(another_deviation)
summary_df = scores_df.copy()
summary_df['偏差'] = deviation
summary_df
summary_df.mean()
np.mean(deviation ** 2)
np.var(scores)
scores_df.var()
summary_df['偏差二乗'] = np.square(deviation)
summary_df
summary_df.mean()
np.sqrt(np.var(scores, ddof=0))
np.std(scores, ddof=0)
np.max(scores) - np.min(scores)
scores_Q1 = np.percentile(scores, 25)
scores_Q3 = np.percentile(scores, 75)
scores_IQR = scores_Q3 - scores_Q1
scores_IQR
pd.Series(scores).describe()
z = (scores - np.mean(scores)) / np.std(scores)
z
np.mean(z), np.std(z, ddof=0)
z = 50 + 10 * (scores - np.mean(scores)) / np.std(scores)
scores_df['偏差値'] = z
scores_df
# 50人分の英語の点数のarray
english_scores = np.array(df['英語'])
# Seriesに変換してdescribeを表示
pd.Series(english_scores).describe()
freq, _ = np.histogram(english_scores, bins=10, range=(0, 100))
freq
# 0~10, 10~20, ... といった文字列のリストを作成
freq_class = [f'{i}~{i+10}' for i in range(0, 100, 10)]
# freq_classをインデックスにしてfreqでDataFrameを作成
freq_dist_df = pd.DataFrame({'度数':freq},
index=pd.Index(freq_class,
name='階級'))
freq_dist_df
class_value = [(i+(i+10))//2 for i in range(0, 100, 10)]
class_value
rel_freq = freq / freq.sum()
rel_freq
cum_rel_freq = np.cumsum(rel_freq)
cum_rel_freq
freq_dist_df['階級値'] = class_value
freq_dist_df['相対度数'] = rel_freq
freq_dist_df['累積相対度数'] = cum_rel_freq
freq_dist_df = freq_dist_df[['階級値', '度数',
'相対度数', '累積相対度数']]
freq_dist_df
freq_dist_df.loc[freq_dist_df['度数'].idxmax(), '階級値']
# Matplotlibのpyplotモジュールをpltという名前でインポート
import matplotlib.pyplot as plt
# グラフがnotebook上に表示されるようにする
%matplotlib inline
# キャンバスを作る
# figsizeで横・縦の大きさを指定
fig = plt.figure(figsize=(10, 6))
# キャンバス上にグラフを描画するための領域を作る
# 引数は領域を1×1個作り、1つめの領域に描画することを意味する
ax = fig.add_subplot(111)
# 階級数を10にしてヒストグラムを描画
freq, _, _ = ax.hist(english_scores, bins=10, range=(0, 100))
# X軸にラベルをつける
ax.set_xlabel('点数')
# Y軸にラベルをつける
ax.set_ylabel('人数')
# X軸に0, 10, 20, ..., 100の目盛りをふる
ax.set_xticks(np.linspace(0, 100, 10+1))
# Y軸に0, 1, 2, ...の目盛りをふる
ax.set_yticks(np.arange(0, freq.max()+1))
# グラフの表示
plt.show()
fig = plt.figure(figsize=(10, 6))
ax = fig.add_subplot(111)
freq, _ , _ = ax.hist(english_scores, bins=25, range=(0, 100))
ax.set_xlabel('点数')
ax.set_ylabel('人数')
ax.set_xticks(np.linspace(0, 100, 25+1))
ax.set_yticks(np.arange(0, freq.max()+1))
plt.show()
fig = plt.figure(figsize=(10, 6))
ax1 = fig.add_subplot(111)
# Y軸のスケールが違うグラフをax1と同じ領域上に書けるようにする
ax2 = ax1.twinx()
# 相対度数のヒストグラムにするためには、度数をデータの数で割る必要がある
# これはhistの引数weightを指定することで実現できる
weights = np.ones_like(english_scores) / len(english_scores)
rel_freq, _, _ = ax1.hist(english_scores, bins=25,
range=(0, 100), weights=weights)
cum_rel_freq = np.cumsum(rel_freq)
class_value = [(i+(i+4))//2 for i in range(0, 100, 4)]
# 折れ線グラフの描画
# 引数lsを'--'にすることで線が点線に
# 引数markerを'o'にすることでデータ点を丸に
# 引数colorを'gray'にすることで灰色に
ax2.plot(class_value, cum_rel_freq,
ls='--', marker='o', color='gray')
# 折れ線グラフの罫線を消去
ax2.grid(visible=False)
ax1.set_xlabel('点数')
ax1.set_ylabel('相対度数')
ax2.set_ylabel('累積相対度数')
ax1.set_xticks(np.linspace(0, 100, 25+1))
plt.show()
fig = plt.figure(figsize=(5, 6))
ax = fig.add_subplot(111)
ax.boxplot(english_scores, labels=['英語'])
plt.show()
| 0.339609 | 0.928862 |
```
import numpy as np
from matplotlib import pyplot as plt
import copy
import random
def initialise_state(N): #N is the grid dimension (in the above example, N=4)
'''
Author: Siddharth Bachoti
~Function Description~
'''
grid = np.ones((N,N,2),dtype=int)
return np.array(grid)
def plot_vector(p1,p2):
'''
Author: Siddharth Chaini
'''
p1 = np.array(p1)
p2 = np.array(p2)
dp = p2-p1
plt.quiver(p1[0], p1[1], dp[0], dp[1],angles='xy', scale_units='xy', scale=1, headwidth = 5, headlength = 7)
def get_coord_list(arr):
'''
Author: Siddharth Chaini
'''
coord_list=[]
num = len(arr)
for i in range(num):
temp_coord = []
for j in range(num):
current_elems = arr[i][j]
xpt = (num-1)-i
ypt = j
temp_coord.append((xpt,ypt))
coord_list.append(temp_coord)
return coord_list
def visualise_2d_model(arr):
'''
Author: Siddharth Chaini
'''
num = len(arr)
plt.axes().set_aspect('equal')
coord_list = get_coord_list(arr)
for i in range(num):
for j in range(num):
current_up_state = arr[i][j][0]
current_right_state = arr[i][j][1]
x_current = coord_list[i][j][1]
y_current = coord_list[i][j][0]
lower_neighbour_up_state = arr[(i+1)%num][j][0]
x_up = coord_list[(i+1)%num][j][1]
y_up = coord_list[(i+1)%num][j][0]
left_neighbour_right_state = arr[i][j-1][1]
x_left = coord_list[i][j-1][1]
y_left = coord_list[i][j-1][0]
current_down_state = -(lower_neighbour_up_state)
current_left_state = -(left_neighbour_right_state)
# plt.plot(x_current,y_current,'ob')
plt.plot(x_current,y_current,
marker="o", markersize=9, markeredgecolor="k",
markerfacecolor="red",
zorder=1)
if current_up_state == 1:
plot_vector([x_current,y_current],[x_current,y_current+1])
elif current_up_state == -1:
plot_vector([x_current,y_current+1],[x_current,y_current])
if current_right_state == 1:
plot_vector([x_current,y_current],[x_current+1,y_current])
elif current_right_state == -1:
plot_vector([x_current+1,y_current],[x_current,y_current])
if current_down_state == 1:
plot_vector([x_current,y_current],[x_current,y_current-1])
elif current_down_state == -1:
plot_vector([x_current,y_current-1],[x_current,y_current])
if current_left_state == 1:
plot_vector([x_current,y_current],[x_current-1,y_current])
elif current_left_state == -1:
plot_vector([x_current-1,y_current],[x_current,y_current])
plt.xlim(-1,num+1)
plt.ylim(-1,num+1)
plt.axis('off')
plt.show()
plt.close()
def check_config(arr):
'''
Author: Tanmay Bhore
'''
flag=True
N=len(arr)
for i in range(len(arr)):
for j in range(len(arr)):
current_up_state = arr[i][j][0]
current_right_state = arr[i][j][1]
lower_neighbour_up_state = arr[(i+1)%N][j][0]
left_neighbour_right_state = arr[i][j-1][1]
current_left_state = -(left_neighbour_right_state)
current_down_state = -(lower_neighbour_up_state)
if (current_up_state + current_right_state + current_left_state + current_down_state) != 0:
flag=False
break
return flag
def long_loop(arr2, verbose=True):
'''
Author: Team ℏ
'''
arr = copy.deepcopy(arr2)
N=len(arr)
iters=0
n1 = np.random.randint(low=0, high=N)
n2 = np.random.randint(low=0, high=N)
inital_pt =(n1,n2)
prev_choice=None
while True:
iters+=1
if n1==inital_pt[0] and n2==inital_pt[1] and iters!=1:
if verbose:
print(f"Completed in {iters} iterations.")
# assert(check_config(arr))
break
current_up_state = arr[n1][n2][0]
current_right_state = arr[n1][n2][1]
lower_neighbour_up_state = arr[(n1+1)%N][n2][0]
left_neighbour_right_state = arr[n1][n2-1][1]
current_down_state = -(lower_neighbour_up_state)
current_left_state = -(left_neighbour_right_state)
current_states_dict = {"up":current_up_state,"right":current_right_state,"down":current_down_state,"left":current_left_state}
outgoing_state_dict={}
incoming_state_dict={}
for key in current_states_dict.keys():
if current_states_dict[key]==1: #current state is outgoing
outgoing_state_dict[key]=current_states_dict[key]
else:
incoming_state_dict[key]=current_states_dict[key]
if prev_choice =="right":
forbidden_choice="left"
if prev_choice =="up":
forbidden_choice="down"
if prev_choice =="left":
forbidden_choice="right"
if prev_choice =="down":
forbidden_choice="up"
else:
forbidden_choice=None
while True:
out_choice = np.random.choice(list(outgoing_state_dict.keys()))
if out_choice !=forbidden_choice:
break
prev_choice=out_choice
if out_choice == "up":
arr[n1][n2][0]= - (arr[n1][n2][0])
n1=(n1-1)%N
n2=n2
continue
if out_choice == "right":
arr[n1][n2][1]= - (arr[n1][n2][1])
n1=n1
n2=(n2+1)%N
continue
if out_choice == "down":
arr[(n1+1)%N][n2][0]= - (arr[(n1+1)%N][n2][0])
n1=(n1+1)%N
n2=n2
continue
if out_choice == "left":
arr[n1][(n2-1)%N][1]= - (arr[n1][(n2-1)%N][1])
n1=n1
n2=(n2-1)%N
continue
return arr
def count_states(num,total_counts,return_dict = False,verbose=False): #Change total_counts parameter to an error percentage later to make it independent of num
'''
Author: Team ℏ
'''
state_dict={}
i=0
oldarr = long_loop(initialise_state(num), verbose=False)
while True:
if verbose:
print(f"Iteration Number = {i} and Total states found = {len(state_dict)}", end="\r")
newarr = long_loop(oldarr,verbose=False)
name =arr_to_string(newarr)
if name not in state_dict:
count_repetitions=0
state_dict[name]=1
else:
count_repetitions+=1
state_dict[name]+=1
if count_repetitions==total_counts:
break
i+=1
oldarr=newarr
if return_dict:
return len(state_dict),state_dict
else:
return len(state_dict)
def print_acche_se(arr):
for elem1 in arr:
for elem2 in elem1:
print(f"[{elem2[0]},{elem2[1]}]",end=",\t")
print()
print()
def state2to4(arr):
'''
Author: Siddharth Chaini
Examine once.
'''
fourstatearr=np.zeros((arr.shape[0],arr.shape[1],4))
N=len(arr)
for i in range(len(arr)):
for j in range(len(arr)):
current_up_state = arr[i][j][0]
current_right_state = arr[i][j][1]
lower_neighbour_up_state = arr[(i+1)%N][j][0]
left_neighbour_right_state = arr[i][j-1][1]
current_left_state = -(left_neighbour_right_state)
current_down_state = -(lower_neighbour_up_state)
fourstatearr[i][j][0] = current_up_state
fourstatearr[i][j][1] = current_right_state
fourstatearr[i][j][2] = current_down_state
fourstatearr[i][j][3] = current_left_state
return fourstatearr
#Rot 90 anticlock
#Up becomes left, left becomes down, down becomes right, right becomes up
def rot90_anticlock(arr2):
'''
Author: Siddharth Chaini
'''
fourstatearr = state2to4(arr2)
fourstatearr = np.rot90(fourstatearr,1)
arr=np.zeros((fourstatearr.shape[0],fourstatearr.shape[1],2))
N=len(arr)
for i in range(len(arr)):
for j in range(len(arr)):
current_up_state = fourstatearr[i][j][0]
current_right_state = fourstatearr[i][j][1]
current_down_state = fourstatearr[i][j][2]
current_left_state = fourstatearr[i][j][3]
new_up_state = current_right_state
new_right_state = current_down_state
arr[i][j][0]=new_up_state
arr[i][j][1]=new_right_state
return arr.astype(int)
#Rot 180 anticlock
#Up becomes down, left becomes right, down becomes up, right becomes left
def rot180_anticlock(arr2):
'''
Author: Siddharth Chaini
'''
fourstatearr = state2to4(arr2)
fourstatearr = np.rot90(fourstatearr,2)
arr=np.zeros((fourstatearr.shape[0],fourstatearr.shape[1],2))
N=len(arr)
for i in range(len(arr)):
for j in range(len(arr)):
current_up_state = fourstatearr[i][j][0]
current_right_state = fourstatearr[i][j][1]
current_down_state = fourstatearr[i][j][2]
current_left_state = fourstatearr[i][j][3]
new_up_state = current_down_state
new_right_state = current_left_state
arr[i][j][0]=new_up_state
arr[i][j][1]=new_right_state
return arr.astype(int)
#Rot 270 anticlock
#Up becomes right, left becomes up, down becomes left, right becomes down
def rot270_anticlock(arr2):
'''
Author: Siddharth Chaini
'''
fourstatearr = state2to4(arr2)
fourstatearr = np.rot90(fourstatearr,3)
arr=np.zeros((fourstatearr.shape[0],fourstatearr.shape[1],2))
N=len(arr)
for i in range(len(arr)):
for j in range(len(arr)):
current_up_state = fourstatearr[i][j][0]
current_right_state = fourstatearr[i][j][1]
current_down_state = fourstatearr[i][j][2]
current_left_state = fourstatearr[i][j][3]
new_up_state = current_left_state
new_right_state = current_up_state
arr[i][j][0]=new_up_state
arr[i][j][1]=new_right_state
return arr.astype(int)
#Flip horizontally
#Up becomes right, left becomes up, down becomes left, right becomes down
def hor_flip(arr2):
'''
Author: Siddharth Chaini
'''
arr = np.flip(arr2,1)
proper_arr=np.zeros_like(arr2)
num = len(arr)
for i in range(num):
for j in range(num):
current_up_state = arr[i][j][0]
current_left_state = arr[i][j][1]
right_neighbour_left_state = arr[i][(j+1)%num][1]
current_right_state = - (right_neighbour_left_state)
proper_arr[i][j][0]=current_up_state
proper_arr[i][j][1]=current_right_state
return proper_arr.astype(int)
#Flip vertically
#Up becomes right, left becomes up, down becomes left, right becomes down
def ver_flip(arr2):
'''
Author: Siddharth Chaini
'''
arr = np.flip(arr2,0)
proper_arr=np.zeros_like(arr2)
num = len(arr)
for i in range(num):
for j in range(num):
current_down_state = arr[i][j][0]
current_right_state = arr[i][j][1]
upper_neighbour_down_state = arr[i-1][j][0]
current_up_state = - (upper_neighbour_down_state)
proper_arr[i][j][0]=current_up_state
proper_arr[i][j][1]=current_right_state
return proper_arr.astype(int)
def flip_secondary_diag(arr2):
'''
Author: Siddharth Bachoti
'''
arr = copy.deepcopy(arr2)
N = len(arr)
for i in range(N):
for j in range(N):
if (i+j)<=N-1:
dist = N-(i+j+1)
arr[i][j][0], arr[i+dist][j+dist][0], arr[i][j][1], arr[i+dist][j+dist][1] = arr[i+dist][j+dist][1], arr[i][j][1], arr[i+dist][j+dist][0], arr[i][j][0]
return arr.astype(int)
def flip_primary_diag(arr2):
'''
Author: Siddharth Bachoti
'''
arr = copy.deepcopy(arr2)
N = len(arr)
arr = rot90_anticlock(flip_secondary_diag(rot270_anticlock(arr)))
return arr.astype(int)
def get_all_column_translations(arr):
result_arr_list=[]
N=len(arr)
for i in range(1,N):
a1 = arr[:,0:i].reshape(N,-1,2)
a2 = arr[:,i:].reshape(N,-1,2)
res = np.hstack([a2,a1])
result_arr_list.append(res)
return result_arr_list
def get_all_row_translations(arr):
result_arr_list=[]
N=len(arr)
for i in range(1,N):
a1 = arr[0:i,:].reshape(-1,N,2)
a2 = arr[i:,:].reshape(-1,N,2)
res = np.vstack([a2,a1])
result_arr_list.append(res)
return result_arr_list
def arr_to_string(arr2):
arr = copy.deepcopy(arr2)
name = ' '.join(map(str, arr.flatten())).replace(' ','')
return name
def string_to_arr(s):
'''
Author: Siddharth Chaini
'''
replaced_str = s.replace("-1","0")
arr=[]
for i in replaced_str:
if i=='1':
arr.append(1)
elif i=="0":
arr.append(-1)
else:
print("ERROR")
assert(1==0)
arr = np.array(arr)
arr = arr.reshape(int(np.sqrt(len(arr)/2)),int(np.sqrt(len(arr)/2)),2)
return arr
def remove_symmetries(all_names):
'''
Author: Team ℏ
'''
assert type(all_names)==list
for i,given_name in enumerate(all_names):
# print("*******************************")
# print(f"Original Name = {given_name}")
arr = string_to_arr(given_name)
#Column Translation symmetries
templist=get_all_column_translations(arr)
for newarr in templist:
name = arr_to_string(newarr)
# print(f"Col Trans Name = {name}")
if name in all_names[i+1:]:
idx = all_names[i+1:].index(name) + i+1
del all_names[idx]
i = all_names.index(given_name)
#Row Translation symmetries
templist=get_all_row_translations(arr)
for newarr in templist:
name = arr_to_string(newarr)
# print(f"Row Trans Name = {name}")
if name in all_names[i+1:]:
idx = all_names[i+1:].index(name) + i+1
del all_names[idx]
i = all_names.index(given_name)
#Check 90 degree rotation symmetry
name = arr_to_string(rot90_anticlock(arr))
# print(f"Rot 90 Name = {name}")
if name in all_names[i+1:]:
idx = all_names[i+1:].index(name) + i+1
del all_names[idx]
i = all_names.index(given_name)
#Check 180 degree rotation symmetry
name = arr_to_string(rot180_anticlock(arr))
# print(f"Rot 180 Name = {name}")
if name in all_names[i+1:]:
idx = all_names[i+1:].index(name) + i+1
del all_names[idx]
i = all_names.index(given_name)
#Check 270 degree rotation symmetry
name = arr_to_string(rot270_anticlock(arr))
# print(f"Rot 270 Name = {name}")
if name in all_names[i+1:]:
idx = all_names[i+1:].index(name) + i+1
del all_names[idx]
i = all_names.index(given_name)
#Check horizontal flip symmetry
name = arr_to_string(hor_flip(arr))
# print(f"Flip Hor Name = {name}")
if name in all_names[i+1:]:
idx = all_names[i+1:].index(name) + i+1
del all_names[idx]
i = all_names.index(given_name)
#Check vertical flip symmetry
name = arr_to_string(ver_flip(arr))
# print(f"Flip Ver Name = {name}")
if name in all_names[i+1:]:
idx = all_names[i+1:].index(name) + i+1
del all_names[idx]
i = all_names.index(given_name)
#Check secondary diagonal flip symmetry
name = arr_to_string(flip_secondary_diag(arr))
# print(f"Sec Diag Name = {name}")
if name in all_names[i+1:]:
idx = all_names[i+1:].index(name) + i+1
del all_names[idx]
i = all_names.index(given_name)
#Check primary diagonal flip symmetry
name = arr_to_string(flip_primary_diag(arr))
# print(f"Prim Diag Name = {name}")
if name in all_names[i+1:]:
idx = all_names[i+1:].index(name) + i+1
del all_names[idx]
i = all_names.index(given_name)
return all_names
tot_states,state_dict = count_states(3,10000,return_dict=True)
tot_states
all_n = list(state_dict.keys())
random.shuffle(all_n)
len(all_n)
short_arrays = remove_symmetries(all_n)
len(short_arrays)
for name in short_arrays:
arr=string_to_arr(name)
visualise_2d_model(arr)
```
|
github_jupyter
|
import numpy as np
from matplotlib import pyplot as plt
import copy
import random
def initialise_state(N): #N is the grid dimension (in the above example, N=4)
'''
Author: Siddharth Bachoti
~Function Description~
'''
grid = np.ones((N,N,2),dtype=int)
return np.array(grid)
def plot_vector(p1,p2):
'''
Author: Siddharth Chaini
'''
p1 = np.array(p1)
p2 = np.array(p2)
dp = p2-p1
plt.quiver(p1[0], p1[1], dp[0], dp[1],angles='xy', scale_units='xy', scale=1, headwidth = 5, headlength = 7)
def get_coord_list(arr):
'''
Author: Siddharth Chaini
'''
coord_list=[]
num = len(arr)
for i in range(num):
temp_coord = []
for j in range(num):
current_elems = arr[i][j]
xpt = (num-1)-i
ypt = j
temp_coord.append((xpt,ypt))
coord_list.append(temp_coord)
return coord_list
def visualise_2d_model(arr):
'''
Author: Siddharth Chaini
'''
num = len(arr)
plt.axes().set_aspect('equal')
coord_list = get_coord_list(arr)
for i in range(num):
for j in range(num):
current_up_state = arr[i][j][0]
current_right_state = arr[i][j][1]
x_current = coord_list[i][j][1]
y_current = coord_list[i][j][0]
lower_neighbour_up_state = arr[(i+1)%num][j][0]
x_up = coord_list[(i+1)%num][j][1]
y_up = coord_list[(i+1)%num][j][0]
left_neighbour_right_state = arr[i][j-1][1]
x_left = coord_list[i][j-1][1]
y_left = coord_list[i][j-1][0]
current_down_state = -(lower_neighbour_up_state)
current_left_state = -(left_neighbour_right_state)
# plt.plot(x_current,y_current,'ob')
plt.plot(x_current,y_current,
marker="o", markersize=9, markeredgecolor="k",
markerfacecolor="red",
zorder=1)
if current_up_state == 1:
plot_vector([x_current,y_current],[x_current,y_current+1])
elif current_up_state == -1:
plot_vector([x_current,y_current+1],[x_current,y_current])
if current_right_state == 1:
plot_vector([x_current,y_current],[x_current+1,y_current])
elif current_right_state == -1:
plot_vector([x_current+1,y_current],[x_current,y_current])
if current_down_state == 1:
plot_vector([x_current,y_current],[x_current,y_current-1])
elif current_down_state == -1:
plot_vector([x_current,y_current-1],[x_current,y_current])
if current_left_state == 1:
plot_vector([x_current,y_current],[x_current-1,y_current])
elif current_left_state == -1:
plot_vector([x_current-1,y_current],[x_current,y_current])
plt.xlim(-1,num+1)
plt.ylim(-1,num+1)
plt.axis('off')
plt.show()
plt.close()
def check_config(arr):
'''
Author: Tanmay Bhore
'''
flag=True
N=len(arr)
for i in range(len(arr)):
for j in range(len(arr)):
current_up_state = arr[i][j][0]
current_right_state = arr[i][j][1]
lower_neighbour_up_state = arr[(i+1)%N][j][0]
left_neighbour_right_state = arr[i][j-1][1]
current_left_state = -(left_neighbour_right_state)
current_down_state = -(lower_neighbour_up_state)
if (current_up_state + current_right_state + current_left_state + current_down_state) != 0:
flag=False
break
return flag
def long_loop(arr2, verbose=True):
'''
Author: Team ℏ
'''
arr = copy.deepcopy(arr2)
N=len(arr)
iters=0
n1 = np.random.randint(low=0, high=N)
n2 = np.random.randint(low=0, high=N)
inital_pt =(n1,n2)
prev_choice=None
while True:
iters+=1
if n1==inital_pt[0] and n2==inital_pt[1] and iters!=1:
if verbose:
print(f"Completed in {iters} iterations.")
# assert(check_config(arr))
break
current_up_state = arr[n1][n2][0]
current_right_state = arr[n1][n2][1]
lower_neighbour_up_state = arr[(n1+1)%N][n2][0]
left_neighbour_right_state = arr[n1][n2-1][1]
current_down_state = -(lower_neighbour_up_state)
current_left_state = -(left_neighbour_right_state)
current_states_dict = {"up":current_up_state,"right":current_right_state,"down":current_down_state,"left":current_left_state}
outgoing_state_dict={}
incoming_state_dict={}
for key in current_states_dict.keys():
if current_states_dict[key]==1: #current state is outgoing
outgoing_state_dict[key]=current_states_dict[key]
else:
incoming_state_dict[key]=current_states_dict[key]
if prev_choice =="right":
forbidden_choice="left"
if prev_choice =="up":
forbidden_choice="down"
if prev_choice =="left":
forbidden_choice="right"
if prev_choice =="down":
forbidden_choice="up"
else:
forbidden_choice=None
while True:
out_choice = np.random.choice(list(outgoing_state_dict.keys()))
if out_choice !=forbidden_choice:
break
prev_choice=out_choice
if out_choice == "up":
arr[n1][n2][0]= - (arr[n1][n2][0])
n1=(n1-1)%N
n2=n2
continue
if out_choice == "right":
arr[n1][n2][1]= - (arr[n1][n2][1])
n1=n1
n2=(n2+1)%N
continue
if out_choice == "down":
arr[(n1+1)%N][n2][0]= - (arr[(n1+1)%N][n2][0])
n1=(n1+1)%N
n2=n2
continue
if out_choice == "left":
arr[n1][(n2-1)%N][1]= - (arr[n1][(n2-1)%N][1])
n1=n1
n2=(n2-1)%N
continue
return arr
def count_states(num,total_counts,return_dict = False,verbose=False): #Change total_counts parameter to an error percentage later to make it independent of num
'''
Author: Team ℏ
'''
state_dict={}
i=0
oldarr = long_loop(initialise_state(num), verbose=False)
while True:
if verbose:
print(f"Iteration Number = {i} and Total states found = {len(state_dict)}", end="\r")
newarr = long_loop(oldarr,verbose=False)
name =arr_to_string(newarr)
if name not in state_dict:
count_repetitions=0
state_dict[name]=1
else:
count_repetitions+=1
state_dict[name]+=1
if count_repetitions==total_counts:
break
i+=1
oldarr=newarr
if return_dict:
return len(state_dict),state_dict
else:
return len(state_dict)
def print_acche_se(arr):
for elem1 in arr:
for elem2 in elem1:
print(f"[{elem2[0]},{elem2[1]}]",end=",\t")
print()
print()
def state2to4(arr):
'''
Author: Siddharth Chaini
Examine once.
'''
fourstatearr=np.zeros((arr.shape[0],arr.shape[1],4))
N=len(arr)
for i in range(len(arr)):
for j in range(len(arr)):
current_up_state = arr[i][j][0]
current_right_state = arr[i][j][1]
lower_neighbour_up_state = arr[(i+1)%N][j][0]
left_neighbour_right_state = arr[i][j-1][1]
current_left_state = -(left_neighbour_right_state)
current_down_state = -(lower_neighbour_up_state)
fourstatearr[i][j][0] = current_up_state
fourstatearr[i][j][1] = current_right_state
fourstatearr[i][j][2] = current_down_state
fourstatearr[i][j][3] = current_left_state
return fourstatearr
#Rot 90 anticlock
#Up becomes left, left becomes down, down becomes right, right becomes up
def rot90_anticlock(arr2):
'''
Author: Siddharth Chaini
'''
fourstatearr = state2to4(arr2)
fourstatearr = np.rot90(fourstatearr,1)
arr=np.zeros((fourstatearr.shape[0],fourstatearr.shape[1],2))
N=len(arr)
for i in range(len(arr)):
for j in range(len(arr)):
current_up_state = fourstatearr[i][j][0]
current_right_state = fourstatearr[i][j][1]
current_down_state = fourstatearr[i][j][2]
current_left_state = fourstatearr[i][j][3]
new_up_state = current_right_state
new_right_state = current_down_state
arr[i][j][0]=new_up_state
arr[i][j][1]=new_right_state
return arr.astype(int)
#Rot 180 anticlock
#Up becomes down, left becomes right, down becomes up, right becomes left
def rot180_anticlock(arr2):
'''
Author: Siddharth Chaini
'''
fourstatearr = state2to4(arr2)
fourstatearr = np.rot90(fourstatearr,2)
arr=np.zeros((fourstatearr.shape[0],fourstatearr.shape[1],2))
N=len(arr)
for i in range(len(arr)):
for j in range(len(arr)):
current_up_state = fourstatearr[i][j][0]
current_right_state = fourstatearr[i][j][1]
current_down_state = fourstatearr[i][j][2]
current_left_state = fourstatearr[i][j][3]
new_up_state = current_down_state
new_right_state = current_left_state
arr[i][j][0]=new_up_state
arr[i][j][1]=new_right_state
return arr.astype(int)
#Rot 270 anticlock
#Up becomes right, left becomes up, down becomes left, right becomes down
def rot270_anticlock(arr2):
'''
Author: Siddharth Chaini
'''
fourstatearr = state2to4(arr2)
fourstatearr = np.rot90(fourstatearr,3)
arr=np.zeros((fourstatearr.shape[0],fourstatearr.shape[1],2))
N=len(arr)
for i in range(len(arr)):
for j in range(len(arr)):
current_up_state = fourstatearr[i][j][0]
current_right_state = fourstatearr[i][j][1]
current_down_state = fourstatearr[i][j][2]
current_left_state = fourstatearr[i][j][3]
new_up_state = current_left_state
new_right_state = current_up_state
arr[i][j][0]=new_up_state
arr[i][j][1]=new_right_state
return arr.astype(int)
#Flip horizontally
#Up becomes right, left becomes up, down becomes left, right becomes down
def hor_flip(arr2):
'''
Author: Siddharth Chaini
'''
arr = np.flip(arr2,1)
proper_arr=np.zeros_like(arr2)
num = len(arr)
for i in range(num):
for j in range(num):
current_up_state = arr[i][j][0]
current_left_state = arr[i][j][1]
right_neighbour_left_state = arr[i][(j+1)%num][1]
current_right_state = - (right_neighbour_left_state)
proper_arr[i][j][0]=current_up_state
proper_arr[i][j][1]=current_right_state
return proper_arr.astype(int)
#Flip vertically
#Up becomes right, left becomes up, down becomes left, right becomes down
def ver_flip(arr2):
'''
Author: Siddharth Chaini
'''
arr = np.flip(arr2,0)
proper_arr=np.zeros_like(arr2)
num = len(arr)
for i in range(num):
for j in range(num):
current_down_state = arr[i][j][0]
current_right_state = arr[i][j][1]
upper_neighbour_down_state = arr[i-1][j][0]
current_up_state = - (upper_neighbour_down_state)
proper_arr[i][j][0]=current_up_state
proper_arr[i][j][1]=current_right_state
return proper_arr.astype(int)
def flip_secondary_diag(arr2):
'''
Author: Siddharth Bachoti
'''
arr = copy.deepcopy(arr2)
N = len(arr)
for i in range(N):
for j in range(N):
if (i+j)<=N-1:
dist = N-(i+j+1)
arr[i][j][0], arr[i+dist][j+dist][0], arr[i][j][1], arr[i+dist][j+dist][1] = arr[i+dist][j+dist][1], arr[i][j][1], arr[i+dist][j+dist][0], arr[i][j][0]
return arr.astype(int)
def flip_primary_diag(arr2):
'''
Author: Siddharth Bachoti
'''
arr = copy.deepcopy(arr2)
N = len(arr)
arr = rot90_anticlock(flip_secondary_diag(rot270_anticlock(arr)))
return arr.astype(int)
def get_all_column_translations(arr):
result_arr_list=[]
N=len(arr)
for i in range(1,N):
a1 = arr[:,0:i].reshape(N,-1,2)
a2 = arr[:,i:].reshape(N,-1,2)
res = np.hstack([a2,a1])
result_arr_list.append(res)
return result_arr_list
def get_all_row_translations(arr):
result_arr_list=[]
N=len(arr)
for i in range(1,N):
a1 = arr[0:i,:].reshape(-1,N,2)
a2 = arr[i:,:].reshape(-1,N,2)
res = np.vstack([a2,a1])
result_arr_list.append(res)
return result_arr_list
def arr_to_string(arr2):
arr = copy.deepcopy(arr2)
name = ' '.join(map(str, arr.flatten())).replace(' ','')
return name
def string_to_arr(s):
'''
Author: Siddharth Chaini
'''
replaced_str = s.replace("-1","0")
arr=[]
for i in replaced_str:
if i=='1':
arr.append(1)
elif i=="0":
arr.append(-1)
else:
print("ERROR")
assert(1==0)
arr = np.array(arr)
arr = arr.reshape(int(np.sqrt(len(arr)/2)),int(np.sqrt(len(arr)/2)),2)
return arr
def remove_symmetries(all_names):
'''
Author: Team ℏ
'''
assert type(all_names)==list
for i,given_name in enumerate(all_names):
# print("*******************************")
# print(f"Original Name = {given_name}")
arr = string_to_arr(given_name)
#Column Translation symmetries
templist=get_all_column_translations(arr)
for newarr in templist:
name = arr_to_string(newarr)
# print(f"Col Trans Name = {name}")
if name in all_names[i+1:]:
idx = all_names[i+1:].index(name) + i+1
del all_names[idx]
i = all_names.index(given_name)
#Row Translation symmetries
templist=get_all_row_translations(arr)
for newarr in templist:
name = arr_to_string(newarr)
# print(f"Row Trans Name = {name}")
if name in all_names[i+1:]:
idx = all_names[i+1:].index(name) + i+1
del all_names[idx]
i = all_names.index(given_name)
#Check 90 degree rotation symmetry
name = arr_to_string(rot90_anticlock(arr))
# print(f"Rot 90 Name = {name}")
if name in all_names[i+1:]:
idx = all_names[i+1:].index(name) + i+1
del all_names[idx]
i = all_names.index(given_name)
#Check 180 degree rotation symmetry
name = arr_to_string(rot180_anticlock(arr))
# print(f"Rot 180 Name = {name}")
if name in all_names[i+1:]:
idx = all_names[i+1:].index(name) + i+1
del all_names[idx]
i = all_names.index(given_name)
#Check 270 degree rotation symmetry
name = arr_to_string(rot270_anticlock(arr))
# print(f"Rot 270 Name = {name}")
if name in all_names[i+1:]:
idx = all_names[i+1:].index(name) + i+1
del all_names[idx]
i = all_names.index(given_name)
#Check horizontal flip symmetry
name = arr_to_string(hor_flip(arr))
# print(f"Flip Hor Name = {name}")
if name in all_names[i+1:]:
idx = all_names[i+1:].index(name) + i+1
del all_names[idx]
i = all_names.index(given_name)
#Check vertical flip symmetry
name = arr_to_string(ver_flip(arr))
# print(f"Flip Ver Name = {name}")
if name in all_names[i+1:]:
idx = all_names[i+1:].index(name) + i+1
del all_names[idx]
i = all_names.index(given_name)
#Check secondary diagonal flip symmetry
name = arr_to_string(flip_secondary_diag(arr))
# print(f"Sec Diag Name = {name}")
if name in all_names[i+1:]:
idx = all_names[i+1:].index(name) + i+1
del all_names[idx]
i = all_names.index(given_name)
#Check primary diagonal flip symmetry
name = arr_to_string(flip_primary_diag(arr))
# print(f"Prim Diag Name = {name}")
if name in all_names[i+1:]:
idx = all_names[i+1:].index(name) + i+1
del all_names[idx]
i = all_names.index(given_name)
return all_names
tot_states,state_dict = count_states(3,10000,return_dict=True)
tot_states
all_n = list(state_dict.keys())
random.shuffle(all_n)
len(all_n)
short_arrays = remove_symmetries(all_n)
len(short_arrays)
for name in short_arrays:
arr=string_to_arr(name)
visualise_2d_model(arr)
| 0.11235 | 0.560373 |
<a href="https://colab.research.google.com/github/donalrinho/Bc2JpsiMuNu/blob/main/Bc2JpsiMuNu_RapidSim_LHCb_templates.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# Making histograms for use in a binned fit
In the last notebook, we learned how to calculate weights which help us project out template shapes for each term in our angular decay rate. In this notebook, we use some cutting-edge histogramming tools in Python (the `hist` package) to create histograms which can be used in a `zfit` binned fit.
```
!pip install -q uproot
!pip install -q tensorflow==2.6.2 #specific versions for compatability with zfit
!pip install -q hist
!pip install -q mplhep
!pip install -q zfit
!pip install -q uncertainties
import uproot
import numpy as np
import tensorflow as tf
import zfit
import hist
from hist import Hist
import mplhep
import pandas as pd
import pickle
import json
#Load ROOT file
drive_dir = "/content/drive/MyDrive/Bc2JpsiMuNu_ROOT_files"
file_path = f"{drive_dir}/Bc2JpsiMuNu_RapidSim_LHCb_Vars_Weights"
print(f"Loading ROOT file {file_path}.root")
tree_name = "DecayTree"
events = uproot.open(f"{file_path}.root:{tree_name}")
events
#Make pandas DataFrame
df = events.arrays(library="pd")
```
Let's make a 1D histogram of the reconstructed $\cos(\theta_{J/\psi})$, with 100 bins and a range from -1 to +1:
```
#Define a histogram object
h = Hist(hist.axis.Regular(bins=100, start=-1., stop=1., name="costheta_Jpsi"))
#Fill our hist with the variable of interest, taking values from our DataFrame
h.fill(df["costheta_Jpsi_reco"])
```
We can use the `mplhep` (matplotlib for high energy physics) package to quickly plot our hist:
```
mplhep.histplot(h)
```
We can also make histograms with weights applied, which will be important for the purposes of our fit. To do this, we supply a weight column of our DataFrame to the `fill()` funcion.
```
#A histogram of costheta_Jpsi_reco with weight applied for term 1 in the angular fit
h_1 = (
Hist.new
.Regular(100, -1., 1., name="costheta_Jpsi_reco_1")
.Weight()
)
h_1.fill(df["costheta_Jpsi_reco"], weight=df["weight_1"])
mplhep.histplot(h_1)
```
We aren't limited to 1D histograms either! We can make a histogram with any number of dimensions using `Hist`. In this analysis, we'll want to fit the 3D angular distribution. So we can try to make a 3D histogram.
```
#3D histogram in all three decay angles with weights applied for term 0
#10 bins in each angle = 10^3 = 1000 bins in total
hist3D_0 = (
Hist.new
.Regular(10, -1., 1., name="costheta_Jpsi")
.Regular(10, -1., 1., name="costheta_W")
.Regular(10, -np.pi, np.pi, name="chi")
.Weight()
)
hist3D_0.fill(df["costheta_Jpsi_reco"], df["costheta_W_reco"], df["chi_reco"], weight=df["weight_0"])
```
It can be useful to keep sets of useful objects, like histograms, in a dictionary. Then we can access them via their `key` (the name we give them inside square brackets) in the dictionary at a later stage.
Here we loop over the 6 angular terms we have, making histograms for each angular term. Notice that we are using our `weight_{0..5}` branches here, to apply the corresponding weights for each angular term.
```
#Make histograms for each angular term
all_h = {}
for i in range(0,6):
all_h[i] = (
Hist.new
.Regular(10, -1., 1., name="costheta_Jpsi")
.Regular(10, -1., 1., name="costheta_W")
.Regular(10, -np.pi, np.pi, name="chi")
.Weight()
)
all_h[i].fill(df["costheta_Jpsi_reco"], df["costheta_W_reco"], df["chi_reco"], weight=df[f"weight_{i}"])
all_h
```
We can also be a bit smarter and define a general dictionary of variables. Inside this, we can put names for our x, y, and z variables, number of bins, min and max values, and a LaTeX name for plotting.
```
vars = {}
vars["x_var"] = {"name": "costheta_Jpsi_reco", "min": -1., "max": 1., "bins": 10, "latex": "$\\cos(\\theta_{J/\\psi})$"}
vars["y_var"] = {"name": "costheta_W_reco", "min": -1., "max": 1., "bins": 10, "latex": "$\\cos(\\theta_{W})$"}
vars["z_var"] = {"name": "chi_reco", "min": -np.pi, "max": np.pi, "bins": 10, "latex": "$\\chi$ [rad]"}
vars
```
Now we can use values from this `vars` dictionary when making our set of histograms:
```
all_h = {}
#Loop over each of our angular decay rate terms
for i in range(0,6):
all_h[i] = (
Hist.new
.Regular(vars["x_var"]["bins"], vars["x_var"]["min"], vars["x_var"]["max"], name=vars["x_var"]["name"])
.Regular(vars["y_var"]["bins"], vars["y_var"]["min"], vars["y_var"]["max"], name=vars["y_var"]["name"])
.Regular(vars["z_var"]["bins"], vars["z_var"]["min"], vars["z_var"]["max"], name=vars["z_var"]["name"])
.Weight()
)
all_h[i].fill(df[vars["x_var"]["name"]],
df[vars["y_var"]["name"]],
df[vars["z_var"]["name"]],
weight=df[f"weight_{i}"])
```
So far we have made histograms that have equal width bins. It is also possible to make histograms where you pass in a list of the bin edges. One useful binning scheme is to place equal numbers of events into each bin. We can make use of the `qcut` functionality in `pandas` to help us do this.
```
#Above binning schemes have equal width bins - we can also make binnings that have equal events per bin
def get_binning(df):
qc_bin_vals = {}
for v in vars:
qc = pd.qcut(df[vars[v]["name"]], q=vars[v]["bins"], precision=5)
qc_bins = qc.unique()
qc_bin_vals[v] = []
for j in range(0,vars[v]["bins"]):
qc_bin_vals[v].append(qc_bins[j].left)
qc_bin_vals[v].append(qc_bins[j].right)
#Retain unique values then sort
qc_bin_vals[v] = list(set(qc_bin_vals[v]))
qc_bin_vals[v].sort()
qc_bin_vals[v][0] = vars[v]["min"]
qc_bin_vals[v][-1] = vars[v]["max"]
print(f"Binning for {vars[v]['name']}: {qc_bin_vals[v]}")
return qc_bin_vals
binnings = get_binning(df)
#Save to JSON for use in future analyses
json_path = "/content/drive/MyDrive/Bc2JpsiMuNu_Analysis/json"
with open(f"{json_path}/binnings.json", "w") as fp:
json.dump(binnings, fp)
binnings
```
Now we can use the values from our `binnings` dictionary to make a set of histograms with our new non-uniform binning scheme.
```
all_h = {}
#Loop over each of our angular decay rate terms
for i in range(0,6):
all_h[i] = (
Hist.new
.Variable(binnings["x_var"], name=vars["x_var"]["name"])
.Variable(binnings["y_var"], name=vars["y_var"]["name"])
.Variable(binnings["z_var"], name=vars["z_var"]["name"])
.Weight()
)
all_h[i].fill(df[vars["x_var"]["name"]],
df[vars["y_var"]["name"]],
df[vars["z_var"]["name"]],
weight=df[f"weight_{i}"])
```
One final step to prepare our histograms for use as fit templates is to normalise them all. We do this by calculating the sum of all bins in the first histogram we make, and then dividing all of the histograms by that same sum.
```
norm = None
all_h_norm = {}
for i in range(0,6):
if not norm:
norm = np.sum(all_h[i].values())
print(f"Normalisation: {norm}")
all_h_norm[i] = all_h[i] / norm
```
Let's save our histograms for use in future analysis. We can store them in a pickle file:
```
hist_path = "/content/drive/MyDrive/Bc2JpsiMuNu_Analysis/pickle"
for h in all_h_norm:
with open(f"{hist_path}/hist_{h}.pkl", "wb") as f:
pickle.dump(all_h_norm[h], f)
```
We have now created 3D histogram templates for each term in our angular decay rate. The histograms are defined in our reconstructed angular variable space, which is the space that a real data analysis would operate in (we don't have access to the true angles in real data!). We will want to use these templates to try and perform a fit to the 3D reconstructed angular distribution, in order to measure the helicity amplitudes. This fit will be the binned equivalent of the fit we did to the truth-level angles in our earlier zfit notebook.
|
github_jupyter
|
!pip install -q uproot
!pip install -q tensorflow==2.6.2 #specific versions for compatability with zfit
!pip install -q hist
!pip install -q mplhep
!pip install -q zfit
!pip install -q uncertainties
import uproot
import numpy as np
import tensorflow as tf
import zfit
import hist
from hist import Hist
import mplhep
import pandas as pd
import pickle
import json
#Load ROOT file
drive_dir = "/content/drive/MyDrive/Bc2JpsiMuNu_ROOT_files"
file_path = f"{drive_dir}/Bc2JpsiMuNu_RapidSim_LHCb_Vars_Weights"
print(f"Loading ROOT file {file_path}.root")
tree_name = "DecayTree"
events = uproot.open(f"{file_path}.root:{tree_name}")
events
#Make pandas DataFrame
df = events.arrays(library="pd")
#Define a histogram object
h = Hist(hist.axis.Regular(bins=100, start=-1., stop=1., name="costheta_Jpsi"))
#Fill our hist with the variable of interest, taking values from our DataFrame
h.fill(df["costheta_Jpsi_reco"])
mplhep.histplot(h)
#A histogram of costheta_Jpsi_reco with weight applied for term 1 in the angular fit
h_1 = (
Hist.new
.Regular(100, -1., 1., name="costheta_Jpsi_reco_1")
.Weight()
)
h_1.fill(df["costheta_Jpsi_reco"], weight=df["weight_1"])
mplhep.histplot(h_1)
#3D histogram in all three decay angles with weights applied for term 0
#10 bins in each angle = 10^3 = 1000 bins in total
hist3D_0 = (
Hist.new
.Regular(10, -1., 1., name="costheta_Jpsi")
.Regular(10, -1., 1., name="costheta_W")
.Regular(10, -np.pi, np.pi, name="chi")
.Weight()
)
hist3D_0.fill(df["costheta_Jpsi_reco"], df["costheta_W_reco"], df["chi_reco"], weight=df["weight_0"])
#Make histograms for each angular term
all_h = {}
for i in range(0,6):
all_h[i] = (
Hist.new
.Regular(10, -1., 1., name="costheta_Jpsi")
.Regular(10, -1., 1., name="costheta_W")
.Regular(10, -np.pi, np.pi, name="chi")
.Weight()
)
all_h[i].fill(df["costheta_Jpsi_reco"], df["costheta_W_reco"], df["chi_reco"], weight=df[f"weight_{i}"])
all_h
vars = {}
vars["x_var"] = {"name": "costheta_Jpsi_reco", "min": -1., "max": 1., "bins": 10, "latex": "$\\cos(\\theta_{J/\\psi})$"}
vars["y_var"] = {"name": "costheta_W_reco", "min": -1., "max": 1., "bins": 10, "latex": "$\\cos(\\theta_{W})$"}
vars["z_var"] = {"name": "chi_reco", "min": -np.pi, "max": np.pi, "bins": 10, "latex": "$\\chi$ [rad]"}
vars
all_h = {}
#Loop over each of our angular decay rate terms
for i in range(0,6):
all_h[i] = (
Hist.new
.Regular(vars["x_var"]["bins"], vars["x_var"]["min"], vars["x_var"]["max"], name=vars["x_var"]["name"])
.Regular(vars["y_var"]["bins"], vars["y_var"]["min"], vars["y_var"]["max"], name=vars["y_var"]["name"])
.Regular(vars["z_var"]["bins"], vars["z_var"]["min"], vars["z_var"]["max"], name=vars["z_var"]["name"])
.Weight()
)
all_h[i].fill(df[vars["x_var"]["name"]],
df[vars["y_var"]["name"]],
df[vars["z_var"]["name"]],
weight=df[f"weight_{i}"])
#Above binning schemes have equal width bins - we can also make binnings that have equal events per bin
def get_binning(df):
qc_bin_vals = {}
for v in vars:
qc = pd.qcut(df[vars[v]["name"]], q=vars[v]["bins"], precision=5)
qc_bins = qc.unique()
qc_bin_vals[v] = []
for j in range(0,vars[v]["bins"]):
qc_bin_vals[v].append(qc_bins[j].left)
qc_bin_vals[v].append(qc_bins[j].right)
#Retain unique values then sort
qc_bin_vals[v] = list(set(qc_bin_vals[v]))
qc_bin_vals[v].sort()
qc_bin_vals[v][0] = vars[v]["min"]
qc_bin_vals[v][-1] = vars[v]["max"]
print(f"Binning for {vars[v]['name']}: {qc_bin_vals[v]}")
return qc_bin_vals
binnings = get_binning(df)
#Save to JSON for use in future analyses
json_path = "/content/drive/MyDrive/Bc2JpsiMuNu_Analysis/json"
with open(f"{json_path}/binnings.json", "w") as fp:
json.dump(binnings, fp)
binnings
all_h = {}
#Loop over each of our angular decay rate terms
for i in range(0,6):
all_h[i] = (
Hist.new
.Variable(binnings["x_var"], name=vars["x_var"]["name"])
.Variable(binnings["y_var"], name=vars["y_var"]["name"])
.Variable(binnings["z_var"], name=vars["z_var"]["name"])
.Weight()
)
all_h[i].fill(df[vars["x_var"]["name"]],
df[vars["y_var"]["name"]],
df[vars["z_var"]["name"]],
weight=df[f"weight_{i}"])
norm = None
all_h_norm = {}
for i in range(0,6):
if not norm:
norm = np.sum(all_h[i].values())
print(f"Normalisation: {norm}")
all_h_norm[i] = all_h[i] / norm
hist_path = "/content/drive/MyDrive/Bc2JpsiMuNu_Analysis/pickle"
for h in all_h_norm:
with open(f"{hist_path}/hist_{h}.pkl", "wb") as f:
pickle.dump(all_h_norm[h], f)
| 0.317955 | 0.942401 |
# Plot CPU utilization of experiment 2.2 version 3
```
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import requests
import io
import glob
def readCSV2pd_CPU(directoryPath,tf_load,edge_name):
"""
This function is to read a CSV file and return the average value and varience
input: directoryPath : path of file names
tf_load : list of traffic load
"""
avg_cpu = []
var_cpu = []
for tf in tf_load:
cpu_data = pd.DataFrame()
for file_name in glob.glob(directoryPath+edge_name+str(tf)+'.csv'):
x = pd.read_csv(file_name, low_memory=False)
cpu_data = pd.concat([cpu_data,x],axis=0)
Row, Col = cpu_data.shape
df = cpu_data.drop(range(22,Row))
df = df.drop(range(0,8))
#print(df['percentage_cpu_utilisation'])
cpu_mean = df.mean(axis=0)
cpu_var = df.var(axis=0)
avg_cpu.append(cpu_mean['percentage_cpu_utilisation'])
var_cpu.append(cpu_var['percentage_cpu_utilisation'])
return avg_cpu, var_cpu
```
## Read file CVS
```
directoryPath = '/Users/kalika/PycharmProjects/Privacy_SDN_Edge_IoT/PlanB/CPU_utilization_Experiment/version3_Experiment_style/Experiment2_2/Edge1_CPU/'
tf_load = [i*2 for i in range(1,20)]
edge_name = 'edge1CPU_M'
avg_cpu, var_cpu = readCSV2pd_CPU(directoryPath,tf_load,edge_name)
directoryPath = '/Users/kalika/PycharmProjects/Privacy_SDN_Edge_IoT/PlanB/CPU_utilization_Experiment/version3_Experiment_style/Experiment2_2/Edge4_CPU/'
tf_load = [i*2 for i in range(1,20)]
edge_name = 'edge4CPU_M'
avg_cpu4, var_cpu4 = readCSV2pd_CPU(directoryPath,tf_load,edge_name)
```
## Plot CPU utilization
```
fig = plt.figure()
ax = fig.add_subplot(1, 1, 1)
ax.plot(tf_load, avg_cpu, color='green', linestyle='dashed', linewidth = 2,
marker='o', markerfacecolor='green', markersize=10,label="Edge 1")
ax.plot(tf_load, avg_cpu4, color='red', linestyle='dashed', linewidth = 2,
marker='x', markerfacecolor='red', markersize=10,label="Edge 4")
plt.ylim(0,30)
plt.xlim(0,40)
plt.xlabel('Traffic load $\lambda_{4,1}$ (Mbps)')
# naming the y axis
plt.ylabel('Average of CPU utilization (%)')
plt.legend()
plt.show()
fig = plt.figure()
ax = fig.add_subplot(1, 1, 1)
ax.plot(tf_load, var_cpu, color='green', linestyle='dashed', linewidth = 2,
marker='o', markerfacecolor='green', markersize=10,label="Edge 1")
ax.plot(tf_load, var_cpu4, color='red', linestyle='dashed', linewidth = 2,
marker='x', markerfacecolor='red', markersize=10,label="Edge 4")
#plt.ylim(0,20)
plt.xlim(0,40)
plt.xlabel('Traffic load $\lambda_{1,2}$ (Mbps)')
# naming the y axis
plt.ylabel('Variance of CPU utilization (%)')
plt.legend()
plt.show()
```
|
github_jupyter
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import requests
import io
import glob
def readCSV2pd_CPU(directoryPath,tf_load,edge_name):
"""
This function is to read a CSV file and return the average value and varience
input: directoryPath : path of file names
tf_load : list of traffic load
"""
avg_cpu = []
var_cpu = []
for tf in tf_load:
cpu_data = pd.DataFrame()
for file_name in glob.glob(directoryPath+edge_name+str(tf)+'.csv'):
x = pd.read_csv(file_name, low_memory=False)
cpu_data = pd.concat([cpu_data,x],axis=0)
Row, Col = cpu_data.shape
df = cpu_data.drop(range(22,Row))
df = df.drop(range(0,8))
#print(df['percentage_cpu_utilisation'])
cpu_mean = df.mean(axis=0)
cpu_var = df.var(axis=0)
avg_cpu.append(cpu_mean['percentage_cpu_utilisation'])
var_cpu.append(cpu_var['percentage_cpu_utilisation'])
return avg_cpu, var_cpu
directoryPath = '/Users/kalika/PycharmProjects/Privacy_SDN_Edge_IoT/PlanB/CPU_utilization_Experiment/version3_Experiment_style/Experiment2_2/Edge1_CPU/'
tf_load = [i*2 for i in range(1,20)]
edge_name = 'edge1CPU_M'
avg_cpu, var_cpu = readCSV2pd_CPU(directoryPath,tf_load,edge_name)
directoryPath = '/Users/kalika/PycharmProjects/Privacy_SDN_Edge_IoT/PlanB/CPU_utilization_Experiment/version3_Experiment_style/Experiment2_2/Edge4_CPU/'
tf_load = [i*2 for i in range(1,20)]
edge_name = 'edge4CPU_M'
avg_cpu4, var_cpu4 = readCSV2pd_CPU(directoryPath,tf_load,edge_name)
fig = plt.figure()
ax = fig.add_subplot(1, 1, 1)
ax.plot(tf_load, avg_cpu, color='green', linestyle='dashed', linewidth = 2,
marker='o', markerfacecolor='green', markersize=10,label="Edge 1")
ax.plot(tf_load, avg_cpu4, color='red', linestyle='dashed', linewidth = 2,
marker='x', markerfacecolor='red', markersize=10,label="Edge 4")
plt.ylim(0,30)
plt.xlim(0,40)
plt.xlabel('Traffic load $\lambda_{4,1}$ (Mbps)')
# naming the y axis
plt.ylabel('Average of CPU utilization (%)')
plt.legend()
plt.show()
fig = plt.figure()
ax = fig.add_subplot(1, 1, 1)
ax.plot(tf_load, var_cpu, color='green', linestyle='dashed', linewidth = 2,
marker='o', markerfacecolor='green', markersize=10,label="Edge 1")
ax.plot(tf_load, var_cpu4, color='red', linestyle='dashed', linewidth = 2,
marker='x', markerfacecolor='red', markersize=10,label="Edge 4")
#plt.ylim(0,20)
plt.xlim(0,40)
plt.xlabel('Traffic load $\lambda_{1,2}$ (Mbps)')
# naming the y axis
plt.ylabel('Variance of CPU utilization (%)')
plt.legend()
plt.show()
| 0.394784 | 0.756852 |
# Search in the INSEE database
The code here-in-below shows how to search in the csv file from INSEE after they have been formatted in Insee_formatting.ipynb.
Various methods to look for data are timed and compared
Dr. M. Fortin, Sept. 2020
### Importing libraries
```
import pandas as pd
import numpy as np
```
### List of formatted csv in the zip file obtained as explained in Insee_formatting.ipynb
```
from zipfile import ZipFile
zip_file = ZipFile('Insee.zip')
file_name=[text_file.filename for text_file in zip_file.infolist()]
print(file_name)
```
# 1. French presidents
## 1.1 Georges Pompidou
```
nom_recherche="Pompidou"
prenom_recherche="Georges"
genre="1" # only male presidents so far :(
decade='70s'
insee = pd.DataFrame()
insee=pd.read_csv(zip_file.open(file_name[file_name.index('Insee_'+decade+'.csv')]),dtype=str, index_col=0)
insee
```
#### Columns
1. nom: family name
2. prenom(s): first name(s)
3. sexe: gender 1 for male and 2 for female
4. journaiss: day of birth
5. monthnaiss: month of birth
6. yearnaiss: year of birth
7. lieunaiss: postcode of the place of birth
8. commnaiss: place of birth
9. paysnaiss: country of birth
10. jourdeces: day of death
11. monthdeces: month of death
12. yeardeces: year of death
14. lieudeces: postcode of the place of death
15. actedeces: reference of the death record
### 1.1.1. Time the search for gender
#### Using where()
```
%%time
insee[insee.sexe.where(insee.sexe==genre).notnull()].head()
```
#### Using a list comprehension
```
%%time
insee[insee.sexe==genre].head()
```
#### Using Vectorization
```
%%time
f = np.vectorize(lambda haystack, needle: needle in haystack)
insee[f(insee.sexe, genre)].head()
```
#### Conclusion: The list comprehension appears to be the fastest
### 1.1.2 Time the search for the family name (nom)
#### Using a list comprehension
```
%%time
insee[(insee['nom'] == nom_recherche.upper())].head()
```
#### Using find()
```
%%time
insee[ insee['nom'].str.find(nom_recherche.upper())==0].head()
```
#### Using match()
```
%%time
insee[insee.nom.str.match(nom_recherche.upper())==True].head()
```
#### Conclusion: again the list comprehension is the fastest
### 1.1.3. Check if adding the constraint on gender slows down the search
#### Case A:
1. gender
2. family name
3. first name
```
%%time
insee = pd.DataFrame()
insee=pd.read_csv(zip_file.open(file_name[file_name.index('Insee_'+decade+'.csv')]),dtype=str, index_col=0)
insee=insee[(insee['nom'] == nom_recherche.upper())]
insee=insee[insee.prenoms.str.contains(prenom_recherche.upper())]
insee=insee[insee.sexe==genre]
insee
```
#### Case B:
1. family name
2. first name
```
%%time
insee = pd.DataFrame()
insee=pd.read_csv(zip_file.open(file_name[file_name.index('Insee_'+decade+'.csv')]),dtype=str, index_col=0)
insee=insee[(insee['nom'] == nom_recherche.upper())]
insee=insee[insee.prenoms.str.contains(prenom_recherche.upper())]
insee
```
**Conclusion** comparing cases A and B: adding the gender seems to slow down the search a little
### 1.1.4. Compare the search order
#### Case C:
1. first name
2. family name
```
%%time
insee = pd.DataFrame()
insee=pd.read_csv(zip_file.open(file_name[file_name.index('Insee_'+decade+'.csv')]),dtype=str, index_col=0)
insee=insee[insee.prenoms.str.contains(prenom_recherche.upper(), na=False)]
insee=insee[(insee['nom'] == nom_recherche.upper())]
insee
```
**Conclusion** comparing cases B and C: family name then first name is faster (the search of the family name is more restrictive than the one of the first name obviously)
### 1.1.5. Check if one should combine the searches or do it one by one
#### Case D: first name & family name simultaneously
```
%%time
insee = pd.DataFrame()
insee=pd.read_csv(zip_file.open(file_name[file_name.index('Insee_'+decade+'.csv')]),dtype=str, index_col=0)
insee[(insee['nom'] == nom_recherche.upper()) &(insee.prenoms.str.contains(prenom_recherche.upper()))]
```
#### Conclusion: comparing cases D and B: non-simultaneous searches are faster
### 1.1.6. Conclusions:
All in all it is faster to:
* Use list comprehension when possible
* Do searches one by one, starting with the most restrictive one here the family name
## 1.2. Francois Mitterand
```
nom_recherche="Mitterrand"
prenom_recherche="Francois"
genre="1"
decade='90s'
%%time
insee = pd.DataFrame()
insee=pd.read_csv(zip_file.open(file_name[file_name.index('Insee_'+decade+'.csv')]),dtype=str, index_col=0)
insee=insee[(insee['nom'] == nom_recherche.upper())]
insee[insee.prenoms.str.contains(prenom_recherche.upper())]
```
## 1.3. Jacques Chirac
```
nom_recherche="Chirac"
prenom_recherche="Jacques"
genre="1"
decade='10s'
%%time
insee = pd.DataFrame()
insee=pd.read_csv(zip_file.open(file_name[file_name.index('Insee_'+decade+'.csv')]),dtype=str, index_col=0)
insee=insee[(insee['nom'] == nom_recherche.upper())]
insee[insee.prenoms.str.contains(prenom_recherche.upper())]
```
# 2. Other personalities
## 2.1. Agnes Varda
I know that her first name is not necessarily the real one. Hence I use her name and gender for the search
```
nom_recherche="Varda"
genre="2"
decade='10s'
%%time
insee = pd.DataFrame()
insee=pd.read_csv(zip_file.open(file_name[file_name.index('Insee_'+decade+'.csv')]),dtype=str, index_col=0)
insee=insee[insee.sexe==genre]
insee[(insee['nom'] == nom_recherche.upper())]
```
I know that Agnès Varda was born in Belgium so her civil name was Arlette Varda and she was on May 30, 1928 and passed on March 29, 2019.
## 2.2. Francois Truffaut
I do not know when he passed hence I am checking all files.
Also I want to find people whose first names start with Francois, hence the use of startwith().
Finally in French the female name Francoise starts similarly to male name Francois, hence I restrict to male, checking for gender.
```
nom_recherche="Truffaut"
prenom_recherche="Francois"
genre="1"
%%time
results = pd.DataFrame()
for filename in file_name:
insee = pd.DataFrame()
insee = pd.read_csv(zip_file.open(filename),dtype=str, index_col=0)
insee=insee[insee['nom'] == nom_recherche.upper()]
insee=insee[insee.prenoms.str.startswith(prenom_recherche.upper(),na=False)]
results = pd.concat([results,insee[insee.sexe==genre]])
results
```
Since I have no recollection of Truffaut passing in 2008 when I lived in Paris, I conclude that the screen director passed in 1984.
# 3. My paternal grandfather
I know his gender obviously and his family name and that he passed in the 70s.
```
genre="1"
nom_recherche="FORTIN"
decade='70s'
insee = pd.DataFrame()
insee=pd.read_csv(zip_file.open(file_name[file_name.index('Insee_'+decade+'.csv')]),dtype=str, index_col=0)
insee=insee[(insee.nom == nom_recherche.upper())]
fortin=insee[(insee.sexe ==genre)]
fortin
```
I also know he was born and died in the same departement (territorial subdivision) in Normandy so the citycode (lieunaiss/lieudeces) where he was born starts with 14, 61, 50, 76, 27.
```
results = pd.DataFrame()
departement=["14","61","50","76","27"]
for dep in departement:
results=pd.concat([results,fortin[(fortin.lieunaiss.str.startswith(dep)) & (fortin.lieudeces.str.startswith(dep))]])
results
```
I also know his first names (prenoms) and the name of his birthplace (commnaiss) but I will keep that secret... All I can say is that my grandfather is indeed in the list just above :)
|
github_jupyter
|
import pandas as pd
import numpy as np
from zipfile import ZipFile
zip_file = ZipFile('Insee.zip')
file_name=[text_file.filename for text_file in zip_file.infolist()]
print(file_name)
nom_recherche="Pompidou"
prenom_recherche="Georges"
genre="1" # only male presidents so far :(
decade='70s'
insee = pd.DataFrame()
insee=pd.read_csv(zip_file.open(file_name[file_name.index('Insee_'+decade+'.csv')]),dtype=str, index_col=0)
insee
%%time
insee[insee.sexe.where(insee.sexe==genre).notnull()].head()
%%time
insee[insee.sexe==genre].head()
%%time
f = np.vectorize(lambda haystack, needle: needle in haystack)
insee[f(insee.sexe, genre)].head()
%%time
insee[(insee['nom'] == nom_recherche.upper())].head()
%%time
insee[ insee['nom'].str.find(nom_recherche.upper())==0].head()
%%time
insee[insee.nom.str.match(nom_recherche.upper())==True].head()
%%time
insee = pd.DataFrame()
insee=pd.read_csv(zip_file.open(file_name[file_name.index('Insee_'+decade+'.csv')]),dtype=str, index_col=0)
insee=insee[(insee['nom'] == nom_recherche.upper())]
insee=insee[insee.prenoms.str.contains(prenom_recherche.upper())]
insee=insee[insee.sexe==genre]
insee
%%time
insee = pd.DataFrame()
insee=pd.read_csv(zip_file.open(file_name[file_name.index('Insee_'+decade+'.csv')]),dtype=str, index_col=0)
insee=insee[(insee['nom'] == nom_recherche.upper())]
insee=insee[insee.prenoms.str.contains(prenom_recherche.upper())]
insee
%%time
insee = pd.DataFrame()
insee=pd.read_csv(zip_file.open(file_name[file_name.index('Insee_'+decade+'.csv')]),dtype=str, index_col=0)
insee=insee[insee.prenoms.str.contains(prenom_recherche.upper(), na=False)]
insee=insee[(insee['nom'] == nom_recherche.upper())]
insee
%%time
insee = pd.DataFrame()
insee=pd.read_csv(zip_file.open(file_name[file_name.index('Insee_'+decade+'.csv')]),dtype=str, index_col=0)
insee[(insee['nom'] == nom_recherche.upper()) &(insee.prenoms.str.contains(prenom_recherche.upper()))]
nom_recherche="Mitterrand"
prenom_recherche="Francois"
genre="1"
decade='90s'
%%time
insee = pd.DataFrame()
insee=pd.read_csv(zip_file.open(file_name[file_name.index('Insee_'+decade+'.csv')]),dtype=str, index_col=0)
insee=insee[(insee['nom'] == nom_recherche.upper())]
insee[insee.prenoms.str.contains(prenom_recherche.upper())]
nom_recherche="Chirac"
prenom_recherche="Jacques"
genre="1"
decade='10s'
%%time
insee = pd.DataFrame()
insee=pd.read_csv(zip_file.open(file_name[file_name.index('Insee_'+decade+'.csv')]),dtype=str, index_col=0)
insee=insee[(insee['nom'] == nom_recherche.upper())]
insee[insee.prenoms.str.contains(prenom_recherche.upper())]
nom_recherche="Varda"
genre="2"
decade='10s'
%%time
insee = pd.DataFrame()
insee=pd.read_csv(zip_file.open(file_name[file_name.index('Insee_'+decade+'.csv')]),dtype=str, index_col=0)
insee=insee[insee.sexe==genre]
insee[(insee['nom'] == nom_recherche.upper())]
nom_recherche="Truffaut"
prenom_recherche="Francois"
genre="1"
%%time
results = pd.DataFrame()
for filename in file_name:
insee = pd.DataFrame()
insee = pd.read_csv(zip_file.open(filename),dtype=str, index_col=0)
insee=insee[insee['nom'] == nom_recherche.upper()]
insee=insee[insee.prenoms.str.startswith(prenom_recherche.upper(),na=False)]
results = pd.concat([results,insee[insee.sexe==genre]])
results
genre="1"
nom_recherche="FORTIN"
decade='70s'
insee = pd.DataFrame()
insee=pd.read_csv(zip_file.open(file_name[file_name.index('Insee_'+decade+'.csv')]),dtype=str, index_col=0)
insee=insee[(insee.nom == nom_recherche.upper())]
fortin=insee[(insee.sexe ==genre)]
fortin
results = pd.DataFrame()
departement=["14","61","50","76","27"]
for dep in departement:
results=pd.concat([results,fortin[(fortin.lieunaiss.str.startswith(dep)) & (fortin.lieudeces.str.startswith(dep))]])
results
| 0.09067 | 0.942188 |
<center>

</center>
# Book Organization
PART 1CORE PYTORCH
1. Intro to deep learning and PYTORCH
2. Pretrained models
3. Tensors
4. Real word data representation using Tensors
5. The mechanics of learning
6. Using neural networks to fit data
7. Learning from Images
8. Using convolutional to generalize
Part 2 Learning from images in the real world: Early detection of lung
1. Using Pytorch to fight Cancer
2.
Part 3 DEPLOYMENT
# Chapter 01 : Introduction to deep learning and PYTORCH
This chapter objectives
> How deep learning changes our approach to machine learning
> Understanding why PyTorch is a good fit for deep learning
> Examining a typical deep learning project
> The hardware you’ll need to follow along with the examples
>> This book focuses on practical PyTorch, with the aim of covering enough ground to allow you to solve real-world machine learning problems, such as in vision, with deep learning or explore new models as they pop up in research literature
## Pre-requisites:
- Experience with some Python programming: datatypes,classes,functions, loops, lists, dictionaries, etc.
- A willingness to learn
## Deep learning and machine learning: deep learning revolution
- Machine learning relied heavily on feature engineering. Features are transformations on input data that facilitate a downstream algorithm, like a classifier, to produce correct outcomes on new data.
- Deep learning, on the other hand, deals with finding such representations automatically, from raw data, in order to successfully perform a task.
- is is not to say that feature engineering has no place with deep learning; we often need to inject some form of prior knowledge in a learning system.
> Often, these automatically created features are better than those that are handcrafted! As with many disruptive technologies, this fact has led to a change in perspective.
<center>

</center>
### How feautures are learned?
<center>

</center>
### Deep Learning steps
- We need a way to ingest whatever data we have at hand.
- We somehow need to define the deep learning machine.
- We must have an automated way, training, to obtain useful representations and make the machine produce desired outputs” (
<center>

</center>
> Training consists of driving the criterion toward lower and lower scores by incrementally modifying our deep learning machine until it achieves low scores, even on data not seen during training
## PyTorch for deep learning
- PyTorch is a library for Python programs that facilitates building deep learning projects.
- As Python does for programming, PyTorch provides an excellent introduction to deep learning.
- At its core, the deep learning machine in figure above is a rather complex mathematical function mapping inputs to an output. To facilitate expressing this function, PyTorch provides a core data structure, the tensor, which is a multidimensional array that shares many similarities with NumPy arrays.
- Then, PyTorch comes with features to perform accelerated mathematical operations on dedicated hardware, which makes it convenient to design neural network architectures and train them on individual machines or parallel computing resources.
## Why PyTorch?
- It’s Pythonic and easy to learn, and using the library generally feels familiar to developers who have used Python previously.
- PyTorch gives us a data type, the **Tensor**, to hold numbers, vectors, matrices, or arrays in general. In addition, it provides functions for operating on them
- But PyTorch offers two things that make it particularly relevant for deep learning:
- it provides accelerated computation using graphical processing units (GPUs), often yielding speedups in the range of 50x over doing the same calculation on a CPU.
- PyTorch provides facilities that support numerical optimization on generic mathematical expressions, which deep learning uses for training (we can safely characterize PyTorch as a high-performance library with optimization (e.g RMSProp and Adam) support for scientific computing in Python.)
- While it was initially focused on research workflows, PyTorch has been equipped with a high-performance C++ runtime that can be used to deploy models for inference without relying on Python, and can be used for designing and training models in C++.
- It also has bindings to other languages and an interface for deploying to mobile devices.
### The deep learning Learnscapes
<center>

</center>
- Theano and TensorFlow were the premiere low-level libraries, working with a model that had the user define a computational graph and then execute it.
- Lasagne and Keras were high-level wrappers around Theano, with Keras wrapping TensorFlow and CNTK as well.
- Caffe, Chainer, DyNet, Torch (the Lua-based precursor to PyTorch), MXNet, CNTK, DL4J, and others filled various niches in the ecosystem.
> The community largely consolidated behind either **PyTorch** or **TensorFlow**, with the adoption of other libraries dwindling, except for those filling specific niches.
- Theano, one of the first deep learning frameworks, has ceased active development.
- TensorFlow:
- Consumed Keras entirely, promoting it to a first-class API (means you can operate on them in the usual manner)
- Released TF 2.0 with [eager mode](https://towardsdatascience.com/eager-execution-vs-graph-execution-which-is-better-38162ea4dbf6) by default (even though slower than graph mode/execution)
- JAX,
- a library by Google that was developed independently from TensorFlow, has started gaining traction as a NumPy equivalent with GPU, autograd and JIT capabilities.
- PyTorch:
- Consumed Caffe2 for its backend
- Replaced most of the low-level code reused from the Lua-based Torch project
- Added support for ONNX, a vendor-neutral model description and exchange format
- Added a delayed-execution “graph mode” runtime called TorchScript
- Released version 1.0
- Replaced CNTK and Chainer as the framework of choice by their respective corporate sponsors
> TensorFlow has a robust pipeline to production, an extensive industry-wide community, and massive mindshare. PyTorch has made huge inroads with the research and teaching communities, thanks to its ease of use, and has picked up momentum since, as researchers and graduates train students and move to industry. It has also built up steam in terms of production solutions.
> Interestingly, with the advent of TorchScript and eager mode, both PyTorch and TensorFlow have seen their feature sets start to converge with the other’s, though the presentation of these features and the overall experience is still quite different between the two
<center>
)
</center>
## 1.4 An overview of how PyTorch supports deep learning
- Pytorch is written in Python, but there’s a lot of non-Python code in it.
- For performance reasons, most of PyTorch is written in C++ and [CUDA](www.geforce.com/hardware/technology/cuda), a C++-like language from NVIDIA that can be compiled to run with massive parallelism on GPUs.
- However, the Python API is where PyTorch shines in term of usability and integration with the wider Python ecosystem
### PyTorch’s support for deep learning : Tensors, autograd, and distributed computing
- PyTorch is a library that provides multidimensional arrays, or tensors and an extensive library of operations on them, provided by the torch module.
> Tensors is just an n-dimensional array in PyTorch. Tensors support some additional enhancements which make them unique: Apart from CPU, they can be loaded on the GPU for faster computations

> A tensor is a generalization of vectors and matrices and is easily understood as a multidimensional array. Tensors are the building blocks of deep learning, and they are the most fundamental data structures in PyTorch (Tensorflow named after Tensor).
<center>

</center>
### PyTorch’s support for deep learning : Autograd, and distributed computing
- The second core thing that PyTorch provides is the ability of tensors to keep track of the operations performed on them and to analytically compute derivatives of an output of a computation with respect to any of its inputs.
- This is used for numerical optimization, and it is provided natively by tensors by virtue of dispatching through [PyTorch’s autograd](https://towardsdatascience.com/pytorch-autograd-understanding-the-heart-of-pytorchs-magic-2686cd94ec95) engine under the hood.
> [PyTorch’s autograd](https://towardsdatascience.com/pytorch-autograd-understanding-the-heart-of-pytorchs-magic-2686cd94ec95) abstracts the complicated mathematics and helps us “magically” calculate gradients of high dimensional curves with only a few lines of code.
<center>

</center>
On setting **.requires_grad = True** they start forming a backward graph that tracks every operation applied on them to calculate the gradients using something called a dynamic computation graph (DCG).
#### 1 Torch.nn
> The core PyTorch modules for building neural networks are located in torch.nn, which provides common neural network layers and other architectural components. Fully connected layers, convolutional layers, activation functions, and loss functions can all be found here
<center>

</center>
#### 2. Fetching data
> We need to convert each sample from our data into a something PyTorch can actually handle: tensors.
> This bridge between our **custom data** (in whatever format it might be) and a** standardized PyTorch tensor** is the **Dataset** class PyTorch provides in torch.utils.data. (Discuss in chapter 4). For example, text data can (), image data (), vedio data ()
As this process is wildly different from one problem to the next, we will have to implement this data sourcing ourselves
For exaple, Vector data—Rank-2 tensors of shape (samples, features), where each sample is a vector of numerical attributes (“features”)
Timeseries data or sequence data—Rank-3 tensors of shape (samples, timesteps, features), where each sample is a sequence (of length timesteps) of feature vectors
Images—Rank-4 tensors of shape (samples, height, width, channels), where each sample is a 2D grid of pixels, and each pixel is represented by a vector of values (“channels”)
Video—Rank-5 tensors of shape (samples, frames, height, width, channels), where each sample is a sequence (of length frames) of images
#### 2. Data Batches
> As data storage is often slow, we want to parallelize data loading. However, Python does not provide easy, efficient, parallel processing, we will need multiple processes to load our data. In order to assemble them into batches (tensors that encompass several samples). PyTorch provides all that magic in the **DataLoader** class (Chapt 7).
<center>

</center>
#### Training (several steps)
> At each step in the training loop, we evaluate our model on the samples we got from the data loader. We then compare the outputs of our model to the desired output (the targets) using some criterion or loss function. PyTorch also has a variety of loss functions at our disposal (Torch.nn).
<center>

</center>
> After we have compared our actual outputs to the ideal with the loss functions, we need to push the model a little to move its outputs to better resemble the target. This is where the PyTorch autograd engine comes in; but we also need an optimizer doing the updates, and that is what PyTorch offers us in torch.optim. (Chapt 5,6,8)
How you should change your weights or learning rates of your neural network to reduce the losses is defined by the **optimizers** you use !!! The right optimization algorithm can reduce training time exponentially. Optimization algorithms or strategies are responsible for reducing the losses and to provide the most accurate results possible.
>> [Stochastic Gradient Descent, Mini-Batch Gradient Descent, Momentum, Adagrad , AdaDelta , Adam](https://towardsdatascience.com/optimizers-for-training-neural-network-59450d71caf6)
> Gradient Descent is the most basic but most used optimization algorithm. It’s used heavily in linear regression and classification algorithms. Backpropagation in neural networks also uses a gradient descent algorithm
<center>

</center>
##### Parallel training
> Sometimes, we need to train on multiple GPU : **torch.nn.parallel.DistributedDataParallel** and the **torch.distributed** submodule can be employed to use the additional hardware.
#### 4. Trained Model
> The training loop is the most time-consuming part of a deep learning project. At the end of it, we are rewarded with a model whose parameters have been **optimized on our task**. This is the **Trained Model**.
#### 5. Deploying the model
> Thi may involve putting the model on a server or exporting it to load it to a cloud engine, Or we might integrate it with a larger application, or run it on a phone.
PyTorch defaults to an [immediate execution model (eager mode)](https://towardsdatascience.com/eager-execution-vs-graph-execution-which-is-better-38162ea4dbf6). Whenever an instruction involving PyTorch is executed by the Python interpreter, the corresponding operation is immediately carried out by the underlying C++ or CUDA implementation. As more instructions operate on tensors, more operations are executed by the backend implementation.
## 1.5 Hardware and software requirements
- First part can be use on CPU
- Second part you may need CUDA-enabled GPU with atleast 8GB of memory or better
- To be clear: GPU is not mandatory if you’re willing to wait, but running on a GPU cuts training time by at least an order of magnitude (and usually it’s 40–50x faster).
- Colab is a Google-owned cloud environment that runs on Google Cloud Platform. It is a free and open-source platform for creating and sharing interactive and reproducible workflows.
- Again, training reduced by using multiple GPUs on the same machine, and even further on clusters of machines equipped with multiple GPUs.
- Part 2 has some nontrivial download bandwidth and disk space requirements as well. The raw data needed for the cancer-detection project in part 2 is about 60 GB to download, and when uncompressed it requires about 120 GB of space.
- While it is possible to use network storage for this, there might be training speed penalties if the network access is slower than local disk. Preferably you will have space on a local SSD to store the data for fast retrieval
## Exercises
```
import torch
x = torch.rand(5, 3)
print(x)
```
|
github_jupyter
|
import torch
x = torch.rand(5, 3)
print(x)
| 0.276886 | 0.949949 |
# Introduction
In this notebook, we will do a comprehensive analysis of the Android app market by comparing thousands of apps in the Google Play store.
# About the Dataset of Google Play Store Apps & Reviews
**Data Source:** <br>
App and review data was scraped from the Google Play Store by Lavanya Gupta in 2018. Original files listed [here](
https://www.kaggle.com/lava18/google-play-store-apps).
# Import Statements
```
import pandas as pd
import plotly.express as px
```
# Notebook Presentation
```
# Show numeric output in decimal format e.g., 2.15
pd.options.display.float_format = '{:,.2f}'.format
```
# Read the Dataset
```
df_apps = pd.read_csv('data/apps.csv')
```
# Data Cleaning
**Challenge**: How many rows and columns does `df_apps` have? What are the column names? Look at a random sample of 5 different rows with [.sample()](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.DataFrame.sample.html).
```
print(df_apps.shape)
df_apps.head()
df_apps.sample(5)
```
### Drop Unused Columns
**Challenge**: Remove the columns called `Last_Updated` and `Android_Version` from the DataFrame. We will not use these columns.
```
df_apps.drop(['Last_Updated', 'Android_Ver'], axis=1, inplace=True)
df_apps.head()
```
### Find and Remove NaN values in Ratings
**Challenge**: How may rows have a NaN value (not-a-number) in the Ratings column? Create DataFrame called `df_apps_clean` that does not include these rows.
```
nan_rows = df_apps[df_apps.Rating.isna()]
print(nan_rows.shape)
nan_rows.head()
df_apps_clean = df_apps.dropna()
print(df_apps_clean.shape)
```
### Find and Remove Duplicates
**Challenge**: Are there any duplicates in data? Check for duplicates using the [.duplicated()](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.DataFrame.duplicated.html) function. How many entries can you find for the "Instagram" app? Use [.drop_duplicates()](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.DataFrame.drop_duplicates.html) to remove any duplicates from `df_apps_clean`.
```
duplicated_rows = df_apps_clean[df_apps_clean.duplicated()]
print(duplicated_rows.shape)
duplicated_rows.head()
df_apps_clean[df_apps_clean.App == 'Instagram']
df_apps_clean = df_apps_clean.drop_duplicates() # Not enough
df_apps_clean[df_apps_clean.App == 'Instagram']
# We need to specify the subset got identifying duplicates
df_apps_clean = df_apps_clean.drop_duplicates(subset=['App', 'Type', 'Price'])
df_apps_clean[df_apps_clean.App == 'Instagram']
print(df_apps_clean.shape)
```
# Find Highest Rated Apps
**Challenge**: Identify which apps are the highest rated. What problem might you encounter if you rely exclusively on ratings alone to determine the quality of an app?
```
df_apps_clean.sort_values('Rating', ascending=False).head()
```
# Find 5 Largest Apps in terms of Size (MBs)
**Challenge**: What's the size in megabytes (MB) of the largest Android apps in the Google Play Store. Based on the data, do you think there could be limit in place or can developers make apps as large as they please?
```
df_apps_clean.sort_values('Size_MBs', ascending=False).head()
```
# Find the 5 App with Most Reviews
**Challenge**: Which apps have the highest number of reviews? Are there any paid apps among the top 50?
```
df_apps_clean.sort_values('Reviews', ascending=False).head(50)
```
# Plotly Pie and Donut Charts - Visualise Categorical Data: Content Ratings
```
ratings = df_apps_clean.Content_Rating.value_counts()
print(ratings)
fig = px.pie(labels=ratings.index, values=ratings.values)
fig.show()
fig = px.pie(labels=ratings.index, values=ratings.values, title="Content Rating", names=ratings.index)
fig.update_traces(textposition='outside', textinfo='percent+label')
fig.show()
fig = px.pie(labels=ratings.index, values=ratings.values, title="Content Rating", names=ratings.index, hole=0.6)
fig.update_traces(textposition='inside', textfont_size=15, textinfo='percent')
fig.show()
```
# Numeric Type Conversion: Examine the Number of Installs
**Challenge**: How many apps had over 1 billion (that's right - BILLION) installations? How many apps just had a single install?
Check the datatype of the Installs column.
Count the number of apps at each level of installations.
Convert the number of installations (the Installs column) to a numeric data type. Hint: this is a 2-step process. You'll have make sure you remove non-numeric characters first.
```
print(df_apps_clean.Installs.describe())
print(df_apps_clean.info())
df_apps_clean[['App', 'Installs']].groupby('Installs').count()
df_apps_clean.Installs = df_apps_clean.Installs.astype(str).str.replace(',', '')
df_apps_clean.Installs = pd.to_numeric(df_apps_clean.Installs)
df_apps_clean[['App', 'Installs']].groupby('Installs').count()
```
# Find the Most Expensive Apps, Filter out the Junk, and Calculate a (ballpark) Sales Revenue Estimate
Let's examine the Price column more closely.
**Challenge**: Convert the price column to numeric data. Then investigate the top 20 most expensive apps in the dataset.
Remove all apps that cost more than $250 from the `df_apps_clean` DataFrame.
Add a column called 'Revenue_Estimate' to the DataFrame. This column should hold the price of the app times the number of installs. What are the top 10 highest grossing paid apps according to this estimate? Out of the top 10 highest grossing paid apps, how many are games?
```
print(df_apps_clean.Price.describe())
df_apps_clean.Price = df_apps_clean.Price.astype(str).str.replace('$', '')
df_apps_clean.Price = pd.to_numeric(df_apps_clean.Price)
df_apps_clean.sort_values('Price', ascending=False).head(20)
```
### The most expensive apps sub $250
```
df_apps_clean = df_apps_clean[df_apps_clean['Price'] < 250]
df_apps_clean.sort_values('Price', ascending=False).head(50)
```
### Highest Grossing Paid Apps (ballpark estimate)
```
df_apps_clean['Revenue_Estimate'] = df_apps_clean.Installs.mul(df_apps_clean.Price)
df_apps_clean.sort_values('Revenue_Estimate', ascending=False)[:10]
```
# Plotly Bar Charts & Scatter Plots: Analysing App Categories
```
print(df_apps_clean.Category.nunique())
top10_category = df_apps_clean.Category.value_counts()[:10]
print(top10_category)
```
### Vertical Bar Chart - Highest Competition (Number of Apps)
```
bar = px.bar(x = top10_category.index, y = top10_category.values)
bar.show()
```
### Horizontal Bar Chart - Most Popular Categories (Highest Downloads)
```
category_installs = df_apps_clean.groupby('Category').agg({'Installs': pd.Series.sum})
category_installs.sort_values('Installs', ascending=True, inplace=True)
h_bar = px.bar(x = category_installs.Installs, y = category_installs.index, orientation = "h")
h_bar.show()
h_bar = px.bar(x = category_installs.Installs, y = category_installs.index, orientation = "h", title='Category Popularity')
h_bar.update_layout(xaxis_title="Number of Downloads", yaxis_title="Category")
h_bar.show()
```
### Category Concentration - Downloads vs. Competition
**Challenge**:
* First, create a DataFrame that has the number of apps in one column and the number of installs in another:
<img src=https://imgur.com/uQRSlXi.png width="350">
* Then use the [plotly express examples from the documentation](https://plotly.com/python/line-and-scatter/) alongside the [.scatter() API reference](https://plotly.com/python-api-reference/generated/plotly.express.scatter.html)to create scatter plot that looks like this.
<img src=https://imgur.com/cHsqh6a.png>
*Hint*: Use the size, hover_name and color parameters in .scatter(). To scale the yaxis, call .update_layout() and specify that the yaxis should be on a log-scale like so: yaxis=dict(type='log')
```
cat_number = df_apps_clean.groupby('Category').agg({'App': pd.Series.count})
cat_number
cat_merged_df = pd.merge(cat_number, category_installs, on='Category', how='inner')
print(f'The dimensions of the DataFrame are: {cat_merged_df.shape}')
cat_merged_df.sort_values('Installs', ascending=False)
scatter = px.scatter(cat_merged_df, # data
x='App', # column name
y='Installs',
title='Category Concentration',
size='App',
hover_name=cat_merged_df.index,
color='Installs')
scatter.update_layout(xaxis_title="Number of Apps (Lower=More Concentrated)",
yaxis_title="Installs",
yaxis=dict(type='log'))
scatter.show()
```
# Extracting Nested Data from a Column
**Challenge**: How many different types of genres are there? Can an app belong to more than one genre? Check what happens when you use .value_counts() on a column with nested values? See if you can work around this problem by using the .split() function and the DataFrame's [.stack() method](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.DataFrame.stack.html).
```
print(len(df_apps_clean.Genres.unique()))
df_apps_clean.Genres.value_counts().sort_values(ascending=True)[:5]
# Split the strings on the semi-colon and then .stack them.
stack = df_apps_clean.Genres.str.split(';', expand=True).stack()
print(f'We now have a single column with shape: {stack.shape}')
num_genres = stack.value_counts()
print(f'Number of genres: {len(num_genres)}')
```
# Colour Scales in Plotly Charts - Competition in Genres
**Challenge**: Can you create this chart with the Series containing the genre data?
<img src=https://imgur.com/DbcoQli.png width=400>
Try experimenting with the built in colour scales in Plotly. You can find a full list [here](https://plotly.com/python/builtin-colorscales/).
* Find a way to set the colour scale using the color_continuous_scale parameter.
* Find a way to make the color axis disappear by using coloraxis_showscale.
```
bar = px.bar(x = num_genres.index[:15], # index = category name
y = num_genres.values[:15], # count
title='Top Genres',
hover_name=num_genres.index[:15],
color=num_genres.values[:15],
color_continuous_scale='Agsunset')
bar.update_layout(xaxis_title='Genre',
yaxis_title='Number of Apps',
coloraxis_showscale=False)
bar.show()
```
# Grouped Bar Charts: Free vs. Paid Apps per Category
```
print(df_apps_clean.Type.value_counts())
df_free_vs_paid = df_apps_clean.groupby(["Category", "Type"], as_index=False).agg({'App': pd.Series.count})
df_free_vs_paid.head()
```
**Challenge**: Use the plotly express bar [chart examples](https://plotly.com/python/bar-charts/#bar-chart-with-sorted-or-ordered-categories) and the [.bar() API reference](https://plotly.com/python-api-reference/generated/plotly.express.bar.html#plotly.express.bar) to create this bar chart:
<img src=https://imgur.com/LE0XCxA.png>
You'll want to use the `df_free_vs_paid` DataFrame that you created above that has the total number of free and paid apps per category.
See if you can figure out how to get the look above by changing the `categoryorder` to 'total descending' as outlined in the documentation here [here](https://plotly.com/python/categorical-axes/#automatically-sorting-categories-by-name-or-total-value).
```
g_bar = px.bar(df_free_vs_paid,
x='Category',
y='App',
title='Free vs Paid Apps by Category',
color='Type',
barmode='group')
g_bar.update_layout(xaxis_title='Category',
yaxis_title='Number of Apps',
xaxis={'categoryorder':'total descending'},
yaxis=dict(type='log'))
g_bar.show()
```
# Plotly Box Plots: Lost Downloads for Paid Apps
**Challenge**: Create a box plot that shows the number of Installs for free versus paid apps. How does the median number of installations compare? Is the difference large or small?
Use the [Box Plots Guide](https://plotly.com/python/box-plots/) and the [.box API reference](https://plotly.com/python-api-reference/generated/plotly.express.box.html) to create the following chart.
<img src=https://imgur.com/uVsECT3.png>
```
box = px.box(df_apps_clean,
y='Installs',
x='Type',
color='Type',
notched=True,
points='all',
title='How Many Downloads are Paid Apps Giving Up?')
box.update_layout(yaxis=dict(type='log'))
box.show()
```
# Plotly Box Plots: Revenue by App Category
**Challenge**: See if you can generate the chart below:
<img src=https://imgur.com/v4CiNqX.png>
Looking at the hover text, how much does the median app earn in the Tools category? If developing an Android app costs $30,000 or thereabouts, does the average photography app recoup its development costs?
Hint: I've used 'min ascending' to sort the categories.
```
df_paid_apps = df_apps_clean[df_apps_clean['Type'] == 'Paid']
box = px.box(df_paid_apps,
x='Category',
y='Revenue_Estimate',
title='How Much Can Paid Apps Earn?')
box.update_layout(xaxis_title='Category',
yaxis_title='Paid App Ballpark Revenue',
xaxis={'categoryorder':'min ascending'},
yaxis=dict(type='log'))
box.show()
```
# How Much Can You Charge? Examine Paid App Pricing Strategies by Category
**Challenge**: What is the median price price for a paid app? Then compare pricing by category by creating another box plot. But this time examine the prices (instead of the revenue estimates) of the paid apps. I recommend using `{categoryorder':'max descending'}` to sort the categories.
```
print(df_paid_apps.Price.median())
box = px.box(df_paid_apps,
x='Category',
y="Price",
title='Price per Category')
box.update_layout(xaxis_title='Category',
yaxis_title='Paid App Price',
xaxis={'categoryorder':'max descending'},
yaxis=dict(type='log'))
box.show()
```
|
github_jupyter
|
import pandas as pd
import plotly.express as px
# Show numeric output in decimal format e.g., 2.15
pd.options.display.float_format = '{:,.2f}'.format
df_apps = pd.read_csv('data/apps.csv')
print(df_apps.shape)
df_apps.head()
df_apps.sample(5)
df_apps.drop(['Last_Updated', 'Android_Ver'], axis=1, inplace=True)
df_apps.head()
nan_rows = df_apps[df_apps.Rating.isna()]
print(nan_rows.shape)
nan_rows.head()
df_apps_clean = df_apps.dropna()
print(df_apps_clean.shape)
duplicated_rows = df_apps_clean[df_apps_clean.duplicated()]
print(duplicated_rows.shape)
duplicated_rows.head()
df_apps_clean[df_apps_clean.App == 'Instagram']
df_apps_clean = df_apps_clean.drop_duplicates() # Not enough
df_apps_clean[df_apps_clean.App == 'Instagram']
# We need to specify the subset got identifying duplicates
df_apps_clean = df_apps_clean.drop_duplicates(subset=['App', 'Type', 'Price'])
df_apps_clean[df_apps_clean.App == 'Instagram']
print(df_apps_clean.shape)
df_apps_clean.sort_values('Rating', ascending=False).head()
df_apps_clean.sort_values('Size_MBs', ascending=False).head()
df_apps_clean.sort_values('Reviews', ascending=False).head(50)
ratings = df_apps_clean.Content_Rating.value_counts()
print(ratings)
fig = px.pie(labels=ratings.index, values=ratings.values)
fig.show()
fig = px.pie(labels=ratings.index, values=ratings.values, title="Content Rating", names=ratings.index)
fig.update_traces(textposition='outside', textinfo='percent+label')
fig.show()
fig = px.pie(labels=ratings.index, values=ratings.values, title="Content Rating", names=ratings.index, hole=0.6)
fig.update_traces(textposition='inside', textfont_size=15, textinfo='percent')
fig.show()
print(df_apps_clean.Installs.describe())
print(df_apps_clean.info())
df_apps_clean[['App', 'Installs']].groupby('Installs').count()
df_apps_clean.Installs = df_apps_clean.Installs.astype(str).str.replace(',', '')
df_apps_clean.Installs = pd.to_numeric(df_apps_clean.Installs)
df_apps_clean[['App', 'Installs']].groupby('Installs').count()
print(df_apps_clean.Price.describe())
df_apps_clean.Price = df_apps_clean.Price.astype(str).str.replace('$', '')
df_apps_clean.Price = pd.to_numeric(df_apps_clean.Price)
df_apps_clean.sort_values('Price', ascending=False).head(20)
df_apps_clean = df_apps_clean[df_apps_clean['Price'] < 250]
df_apps_clean.sort_values('Price', ascending=False).head(50)
df_apps_clean['Revenue_Estimate'] = df_apps_clean.Installs.mul(df_apps_clean.Price)
df_apps_clean.sort_values('Revenue_Estimate', ascending=False)[:10]
print(df_apps_clean.Category.nunique())
top10_category = df_apps_clean.Category.value_counts()[:10]
print(top10_category)
bar = px.bar(x = top10_category.index, y = top10_category.values)
bar.show()
category_installs = df_apps_clean.groupby('Category').agg({'Installs': pd.Series.sum})
category_installs.sort_values('Installs', ascending=True, inplace=True)
h_bar = px.bar(x = category_installs.Installs, y = category_installs.index, orientation = "h")
h_bar.show()
h_bar = px.bar(x = category_installs.Installs, y = category_installs.index, orientation = "h", title='Category Popularity')
h_bar.update_layout(xaxis_title="Number of Downloads", yaxis_title="Category")
h_bar.show()
cat_number = df_apps_clean.groupby('Category').agg({'App': pd.Series.count})
cat_number
cat_merged_df = pd.merge(cat_number, category_installs, on='Category', how='inner')
print(f'The dimensions of the DataFrame are: {cat_merged_df.shape}')
cat_merged_df.sort_values('Installs', ascending=False)
scatter = px.scatter(cat_merged_df, # data
x='App', # column name
y='Installs',
title='Category Concentration',
size='App',
hover_name=cat_merged_df.index,
color='Installs')
scatter.update_layout(xaxis_title="Number of Apps (Lower=More Concentrated)",
yaxis_title="Installs",
yaxis=dict(type='log'))
scatter.show()
print(len(df_apps_clean.Genres.unique()))
df_apps_clean.Genres.value_counts().sort_values(ascending=True)[:5]
# Split the strings on the semi-colon and then .stack them.
stack = df_apps_clean.Genres.str.split(';', expand=True).stack()
print(f'We now have a single column with shape: {stack.shape}')
num_genres = stack.value_counts()
print(f'Number of genres: {len(num_genres)}')
bar = px.bar(x = num_genres.index[:15], # index = category name
y = num_genres.values[:15], # count
title='Top Genres',
hover_name=num_genres.index[:15],
color=num_genres.values[:15],
color_continuous_scale='Agsunset')
bar.update_layout(xaxis_title='Genre',
yaxis_title='Number of Apps',
coloraxis_showscale=False)
bar.show()
print(df_apps_clean.Type.value_counts())
df_free_vs_paid = df_apps_clean.groupby(["Category", "Type"], as_index=False).agg({'App': pd.Series.count})
df_free_vs_paid.head()
g_bar = px.bar(df_free_vs_paid,
x='Category',
y='App',
title='Free vs Paid Apps by Category',
color='Type',
barmode='group')
g_bar.update_layout(xaxis_title='Category',
yaxis_title='Number of Apps',
xaxis={'categoryorder':'total descending'},
yaxis=dict(type='log'))
g_bar.show()
box = px.box(df_apps_clean,
y='Installs',
x='Type',
color='Type',
notched=True,
points='all',
title='How Many Downloads are Paid Apps Giving Up?')
box.update_layout(yaxis=dict(type='log'))
box.show()
df_paid_apps = df_apps_clean[df_apps_clean['Type'] == 'Paid']
box = px.box(df_paid_apps,
x='Category',
y='Revenue_Estimate',
title='How Much Can Paid Apps Earn?')
box.update_layout(xaxis_title='Category',
yaxis_title='Paid App Ballpark Revenue',
xaxis={'categoryorder':'min ascending'},
yaxis=dict(type='log'))
box.show()
print(df_paid_apps.Price.median())
box = px.box(df_paid_apps,
x='Category',
y="Price",
title='Price per Category')
box.update_layout(xaxis_title='Category',
yaxis_title='Paid App Price',
xaxis={'categoryorder':'max descending'},
yaxis=dict(type='log'))
box.show()
| 0.436862 | 0.98222 |
# DaCe with Explicit Dataflow in Python
In this tutorial, we will use the explicit dataflow specification in Python to construct DaCe programs.
```
import dace
```
Explicit dataflow is a Python-based syntax that is close to defining SDFGs. In explicit ` @dace.program `s, the code (Tasklets) and memory movement (Memlets) are specified separately, as we show below.
## Matrix Transposition
We begin with a simple example, transposing a matrix (out-of-place).
First, since we do not know what the matrix sizes will be, we define symbolic sizes:
```
M = dace.symbol('M')
N = dace.symbol('N')
```
We now proceed to define the data-centric part of the application (i.e., the part that can be optimized by DaCe). It is a simple function which, when called, invokes the compilation and optimization procedure. It can also be compiled explicitly, as we show in the next example.
DaCe programs use explicit types, so that they can be compiled. We provide a numpy-compatible set of types that can define N-dimensional tensors. For example, `dace.int64` defines a 64-bit signed integer scalar, and `dace.float32[133,8]` defines a 133-row and 8-column 2D array.
```
@dace.program
def transpose(A: dace.float32[M, N], B: dace.float32[N, M]):
# Inside the function we will define a tasklet in a map, which is shortened
# to dace.map. We define the map range in the arguments:
@dace.map
def mytasklet(i: _[0:M], j: _[0:N]):
# Pre-declaring the memlets is required in explicit dataflow, tasklets
# cannot use any external memory apart from data flowing to/from it.
a << A[i,j] # Input memlet (<<)
b >> B[j,i] # Output memlet (>>)
# The code
b = a
```
And that's it! We will now define some regression test using numpy:
```
import numpy as np
A = np.random.rand(37, 11).astype(np.float32)
expected = A.transpose()
# Define an array for the output of the dace program
B = np.random.rand(11, 37).astype(np.float32)
```
Before we call `transpose`, we can inspect the SDFG:
```
sdfg = transpose.to_sdfg()
sdfg
```
We can now call `transpose` directly, or using the SDFG we created. When calling `transpose`, we need to feed the symbols as well as the arguments (since the arrays are `numpy` rather than symbolic `dace` arrays, see below tutorials). When prompted for transformations, we will now just press the "Enter" key to skip them.
```
sdfg(A=A, B=B, M=A.shape[0], N=A.shape[1])
print('Difference:', np.linalg.norm(expected - B))
```
## Query (using Streams)
In this example, we will use the Stream construct and symbolic dace ND arrays to create a simple parallel filter. We first define a symbolic size and a symbolically-sized array:
```
N = dace.symbol('N')
N.set(255)
storage = dace.ndarray(shape=[N], dtype=dace.int32)
# The size of "output" will actually be lesser or equal to N, but we need to
# statically allocate the memory.
output = dace.ndarray(shape=[N], dtype=dace.int32)
# The size is a scalar
output_size = dace.scalar(dtype=dace.uint32)
```
As with `transpose`, the DaCe program also consists of a tasklet nested in a Map, but also includes a Stream (to which we push outputs as necessary) that is directly connected to the output array, as well as a conflict-resolution output (because all tasklets in the map write to the same address:
```
@dace.program
def query(data: dace.int32[N], output: dace.int32[N], outsz: dace.int32[1],
threshold: dace.int32):
# Define a local, unbounded (buffer_size=0) stream
S = dace.define_stream(dace.int32, 0)
# Filtering tasklet
@dace.map
def filter(i: _[0:N]):
a << data[i]
# Writing to S (no location necessary) a dynamic number of times (-1)
out >> S(-1)
# Writing to outsz dynamically (-1), if there is a conflict, we will sum the results
osz >> outsz(-1, lambda a,b: a+b)
if a > threshold:
# Pushing to a stream or writing with a conflict use the assignment operator
out = a
osz = 1
# Define a memlet from S to the output
S >> output
```
We can compile `query` without defining anything further. However, before we call `query`, we will need to set the symbol sizes.
```
qfunc = query.compile()
thres = 50
# Define some random integers and zero outputs
import numpy as np
storage[:] = np.random.randint(0, 100, size=N.get())
output_size[0] = 0
output[:] = np.zeros(N.get()).astype(np.int32)
# Compute expected output using numpy
expected = storage[np.where(storage > thres)]
```
Here we will just use the Python function prototype to call the code, since we do not invoke it through the SDFG:
```
qfunc(data=storage, output=output, outsz=output_size, threshold=thres, N=N)
output_size
filtered_output = output[:output_size[0]]
# Sorting outputs to avoid concurrency-based reordering
print('Difference:', np.linalg.norm(np.sort(expected) - np.sort(filtered_output)))
```
|
github_jupyter
|
import dace
M = dace.symbol('M')
N = dace.symbol('N')
@dace.program
def transpose(A: dace.float32[M, N], B: dace.float32[N, M]):
# Inside the function we will define a tasklet in a map, which is shortened
# to dace.map. We define the map range in the arguments:
@dace.map
def mytasklet(i: _[0:M], j: _[0:N]):
# Pre-declaring the memlets is required in explicit dataflow, tasklets
# cannot use any external memory apart from data flowing to/from it.
a << A[i,j] # Input memlet (<<)
b >> B[j,i] # Output memlet (>>)
# The code
b = a
import numpy as np
A = np.random.rand(37, 11).astype(np.float32)
expected = A.transpose()
# Define an array for the output of the dace program
B = np.random.rand(11, 37).astype(np.float32)
sdfg = transpose.to_sdfg()
sdfg
sdfg(A=A, B=B, M=A.shape[0], N=A.shape[1])
print('Difference:', np.linalg.norm(expected - B))
N = dace.symbol('N')
N.set(255)
storage = dace.ndarray(shape=[N], dtype=dace.int32)
# The size of "output" will actually be lesser or equal to N, but we need to
# statically allocate the memory.
output = dace.ndarray(shape=[N], dtype=dace.int32)
# The size is a scalar
output_size = dace.scalar(dtype=dace.uint32)
@dace.program
def query(data: dace.int32[N], output: dace.int32[N], outsz: dace.int32[1],
threshold: dace.int32):
# Define a local, unbounded (buffer_size=0) stream
S = dace.define_stream(dace.int32, 0)
# Filtering tasklet
@dace.map
def filter(i: _[0:N]):
a << data[i]
# Writing to S (no location necessary) a dynamic number of times (-1)
out >> S(-1)
# Writing to outsz dynamically (-1), if there is a conflict, we will sum the results
osz >> outsz(-1, lambda a,b: a+b)
if a > threshold:
# Pushing to a stream or writing with a conflict use the assignment operator
out = a
osz = 1
# Define a memlet from S to the output
S >> output
qfunc = query.compile()
thres = 50
# Define some random integers and zero outputs
import numpy as np
storage[:] = np.random.randint(0, 100, size=N.get())
output_size[0] = 0
output[:] = np.zeros(N.get()).astype(np.int32)
# Compute expected output using numpy
expected = storage[np.where(storage > thres)]
qfunc(data=storage, output=output, outsz=output_size, threshold=thres, N=N)
output_size
filtered_output = output[:output_size[0]]
# Sorting outputs to avoid concurrency-based reordering
print('Difference:', np.linalg.norm(np.sort(expected) - np.sort(filtered_output)))
| 0.596433 | 0.987387 |
# Author: Sanjay Kazi
## Task_6: Prediction using Decision Tree Algorithm
### Problem Statement:
#### 1. Create the Decision Tree classifier and visualize it graphically.
#### 2. The purpose is if we feed any new data to this classifier, it would be able to predict the right class accordingly.
# ***GRIP*** @ The Spark Foundation
# Importing Libraries
```
# Importing the required Libraries
from sklearn.datasets import load_iris
from sklearn.tree import DecisionTreeClassifier, export_graphviz
from sklearn.model_selection import train_test_split
import sklearn.metrics as sm
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
import pydot
from IPython.display import Image
```
# Loading Data
```
data=pd.DataFrame(iris['data'],columns=["Petal length","Petal Width","Sepal Length","Sepal Width"])
data['Species']=iris['target']
data['Species']=data['Species'].apply(lambda x: iris['target_names'][x])
data.head()
y=iris.target
print(y)
data.shape
data.info()
data.describe()
data.isna().sum()
```
- there is no NaN value present
- Now lets visualize the the data above using seaborn library
```
sns.pairplot(data)
```
- there is a high correlation between sepal length and sepal width which is easily teraceble.
## Let's plot sepal length vs sepal width data and see if there is any relevent clusters we obtain
```
iris["target"]
iris.feature_names
# Scatter plot of data based on Sepal Length and Width features
sns.FacetGrid(data,hue='Species').map(plt.scatter,'Sepal Length','Sepal Width').add_legend()
plt.show()
```
# Model Building
```
# Model Training
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.1, random_state=1)
tree_classifier = DecisionTreeClassifier()
tree_classifier.fit(X_train,y_train)
y_pred = tree_classifier.predict(X_test)
df = pd.DataFrame({'Actual': y_test, 'Predicted': y_pred})
df
```
## Let's visualize the model
### Text Representation of the tree
```
from sklearn import tree
text_representation = tree.export_text(tree_classifier)
print(text_representation)
```
### Visual Representation
```
fig = plt.figure(figsize=(25,20))
_ = tree.plot_tree(tree_classifier,
feature_names=iris.feature_names,
class_names=iris.target_names,
filled=True)
fig.savefig("decistion_tree.png")
```
- The plot_tree method was added to sklearn in version 0.21. It requires matplotlib to be installed. It allows us to easily produce figure of the tree
## Let's see our model's prediction
```
# Estimating class probabilities
print(tree_classifier.predict([[4.7, 3.2, 1.3, 0.2]]))
print("Our model predicts the class as 0, that is, setosa.")
```
- The data fed are petals length width are 4.7cm and 3.2cm and sepal length and width are 1.3cm and 0.2cm.
## Let's also apply performance metrics to check how good our model is
```
# Model Accuracy
print("Accuracy:",sm.accuracy_score(y_test, y_pred))
```
- this might be a case of over fitting but as we know this is a little dataset and it fits the model very well hence this might be case that we are getting 100% sccuracy.
# Summary
- Thish model is successfylly developed
|
github_jupyter
|
# Importing the required Libraries
from sklearn.datasets import load_iris
from sklearn.tree import DecisionTreeClassifier, export_graphviz
from sklearn.model_selection import train_test_split
import sklearn.metrics as sm
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
import pydot
from IPython.display import Image
data=pd.DataFrame(iris['data'],columns=["Petal length","Petal Width","Sepal Length","Sepal Width"])
data['Species']=iris['target']
data['Species']=data['Species'].apply(lambda x: iris['target_names'][x])
data.head()
y=iris.target
print(y)
data.shape
data.info()
data.describe()
data.isna().sum()
sns.pairplot(data)
iris["target"]
iris.feature_names
# Scatter plot of data based on Sepal Length and Width features
sns.FacetGrid(data,hue='Species').map(plt.scatter,'Sepal Length','Sepal Width').add_legend()
plt.show()
# Model Training
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.1, random_state=1)
tree_classifier = DecisionTreeClassifier()
tree_classifier.fit(X_train,y_train)
y_pred = tree_classifier.predict(X_test)
df = pd.DataFrame({'Actual': y_test, 'Predicted': y_pred})
df
from sklearn import tree
text_representation = tree.export_text(tree_classifier)
print(text_representation)
fig = plt.figure(figsize=(25,20))
_ = tree.plot_tree(tree_classifier,
feature_names=iris.feature_names,
class_names=iris.target_names,
filled=True)
fig.savefig("decistion_tree.png")
# Estimating class probabilities
print(tree_classifier.predict([[4.7, 3.2, 1.3, 0.2]]))
print("Our model predicts the class as 0, that is, setosa.")
# Model Accuracy
print("Accuracy:",sm.accuracy_score(y_test, y_pred))
| 0.652795 | 0.977883 |
# Think Python, Week 2: Functions
<img src='../meta/images/python-logo.png' style="float:right">
## Objectives
---
* Understand functions
* how they're defined and called
* parameters and arguments
* local values
* Return values
* Understand the `import` statement and *dot notation*
### Contents
---
* [Chapter 3: Functions](#Chapter-3%3A-Functions)
* [Concept: Composition](#Concept%3A-Composition)
* [Homework](#Homework)
## Questions from Last Week
---
> Confusion charges compound interest.
### `git pull`
What if both the repo (cloud version) and your local file have changed?
Symptom: `pull` raises an error like this.
```
> git pull
error: cannot pull with rebase: You have unstaged changes.
error: please commit or stash them.
```
One resolution (we'll see others later): in a terminal in your `ThinkPython-2019` directory, ...
* Use `git status` to see which files have changed.
* Make a local copy if you want to keep yours (suggestion: prefix `my-` to the filename)
* Discard your local file: `git checkout -- 01-Types-and-Variables/01-Index.ipynb`
* This overwrites your local version of `01-Index.ipynb` with the repo version
## Chapter 3: Functions
---
> “a function is a named sequence of statements that performs a computation. When you define a function, you specify the name and the sequence of statements. Later, you can “call” the function by name.”
### Exercise: Cost of books and shipping at quantity
* Bulk book purchases cost 40% of their retail price.
* Shipping is \\$3 for the first copy, and \\$0.75 for each additional copy.
* Write a function to compute the total cost of an order, given two parameters, `price` and `quantity`.

```
def totalcost(price, quantity):
cost = (price * 0.4) * quantity
shippingcost = 3.0 + ((quantity - 1) * 0.75)
total = cost + shippingcost
print('Cost:', cost)
print('Shipping cost:', shippingcost)
print('Total:', total)
totalcost(27.69, 20)
```
How'd you do? If you couldn't define the function or made mistakes, do you understand what went wrong?
### Anatomy of a Function Definition

* Required syntax: `def`, parenthesis around the parameter list, and a colon.
* Function definitions are stored as the value of a variable, so variable naming rules apply.
### Calling Functions (Simplified)
```
>>> totalcost(27.69, 4*5)
```
1. Evaluate the argument list
2. Look up the function object for the name
4. Assign values from the argument list to the parameters of the function
* `price = 27.69`
* `quantity = 20`
5. Execute the statements in the body
6. Return a value (if any)
### Testing Your Understanding
```
# what happens?
totalcost(27.69)
# what happens?
totalcost(27.69, 0)
# what's the type of totalcost?
type(totalcost)
# what happens?
totalcost = 27.69
totalcost
```
### Function Variables and Parameters are *Local*
* Parameters are only defined within the body of the function
* After the function has been executed, the local variables are no longer defined
```
def totalcost(price, quantity):
cost = (price * 0.4) * quantity
shippingcost = 3.0 + ((quantity - 1) * 0.75)
total = cost + shippingcost
print('Cost:', cost)
print('Shipping cost:', shippingcost)
print('Total:', total)
totalcost(27.69, 20)
price
cost
```
### Variables and Parameters are Distinct
> "The name of the variable we pass as an argument ... has nothing to do with the name of the parameter"
```
thinkpythoncost = 27.69
def doubler(cost):
cost = cost * 2
print("Inside the function, cost is: ", cost)
doubler(thinkpythoncost)
thinkpythoncost
```
## *All* Functions Return a Value
> "Other functions, like print_twice, perform an action but don’t return a value. They are called void functions."
* Misleading: even if we don't specify a value to return, they *do* return a value! Better:
> Other functions, like print_twice, perform an action but **return the special value None.**
The intepreter doesn't display the None value returned by void functions.
## `import`
---
> “A module is a file that contains a collection of related functions.”
The `import` statement imports a Python *module*. Python is a "batteries included" language, and comes with many dozens of modules. A significant part of learning to program in Python is learning about these modules and what they do.
```
# The `operator` module exports a set of efficient functions corresponding to the intrinsic operators of Python.
import operator
operator.add(2, 2)
operator
```
## Debugging: `type`
```
x = totalcost(27.69, 20)
print(x)
type(x)
```
## Best Practices: Functions
---
* Provide a simple name for a complex sequence of statements
* Write once and re-use many times
* Compose different functions
* Assemble larger programs
* Debug once
## Homework
---
* Rewrite `totalcost` and add parameters for the cost of the first book and the cost of additional books.
* See if you can identify some smallish functions in your area of work (even if you don't yet know how to express them in Python). Try writing out in words what parameters you'd want, and what the sequence of steps would be.
* Example: use the Faithlife platform to send you the message "Hello world!"
* Go to https://faithlife.com in a browser
* Click on the word bubble near the upper right
* Click on "New Message"
* In the "To:" line, type ...
* etc
* Bonus points: give someone else your function description and ask them to execute it while you watch (without providing extra input!). How did they do? Did you get ideas for how to improve your function?
* Read Chapter 4. He uses some math concepts that may be intimidating, but don't let them scare you! Even if you don't get all the math bits, make sure you understand the principles.
* When you're done with the `turtle` examples, use `turtle.bye()` to close the Turtle graphics window.
## Additional Resources
---
* <img src="../meta/images/bd.png" style="display: inline;" /><img src="../meta/images/bd.png" style="display: inline;" /> [The Python Standard Library](https://docs.python.org/3/library/index.html) has a long list of modules. Even if you're not yet ready to understand everything they do, it may be useful to know they're there for later.

|
github_jupyter
|
> git pull
error: cannot pull with rebase: You have unstaged changes.
error: please commit or stash them.
def totalcost(price, quantity):
cost = (price * 0.4) * quantity
shippingcost = 3.0 + ((quantity - 1) * 0.75)
total = cost + shippingcost
print('Cost:', cost)
print('Shipping cost:', shippingcost)
print('Total:', total)
totalcost(27.69, 20)
>>> totalcost(27.69, 4*5)
# what happens?
totalcost(27.69)
# what happens?
totalcost(27.69, 0)
# what's the type of totalcost?
type(totalcost)
# what happens?
totalcost = 27.69
totalcost
def totalcost(price, quantity):
cost = (price * 0.4) * quantity
shippingcost = 3.0 + ((quantity - 1) * 0.75)
total = cost + shippingcost
print('Cost:', cost)
print('Shipping cost:', shippingcost)
print('Total:', total)
totalcost(27.69, 20)
price
cost
thinkpythoncost = 27.69
def doubler(cost):
cost = cost * 2
print("Inside the function, cost is: ", cost)
doubler(thinkpythoncost)
thinkpythoncost
# The `operator` module exports a set of efficient functions corresponding to the intrinsic operators of Python.
import operator
operator.add(2, 2)
operator
x = totalcost(27.69, 20)
print(x)
type(x)
| 0.347537 | 0.969814 |
```
import numpy as np
import random
import pandas as pd
import matplotlib.pyplot as plt
from plotly.subplots import make_subplots
import plotly.graph_objects as go
def replay_plot(negatives, positives, survivors, deaths, df, grid_max, quar_grid):
fig = make_subplots(rows=2, cols=1)
fig.add_trace(
go.Scatter(x=[i for i in range(len(negatives))], y=negatives,
mode="lines",
line=dict(width=2, color="blue"), name='Susceptible',),
row=1, col=1
)
fig.add_trace(
go.Scatter(x=[i for i in range(len(negatives))], y=positives,
mode="lines",
line=dict(width=2, color="green"), name='Infected',),
row=1, col=1
)
fig.add_trace(
go.Scatter(x=[i for i in range(len(negatives))], y=survivors,
mode="lines",
line=dict(width=2, color="orange"), name='Recovered',),
row=1, col=1
)
fig.add_trace(
go.Scatter(x=[i for i in range(len(negatives))], y=deaths,
mode="lines",
line=dict(width=2, color="black"), name='Died',),
row=1, col=1
)
fig.add_trace(
go.Scatter(
x=df[(df['index']==len(df['index'].unique())-1) & (df['state']=='Susceptible')]['x_pos'],
y=df[(df['index']==len(df['index'].unique())-1) & (df['state']=='Susceptible')]['y_pos'],
name='Susceptible',
mode='markers',
marker=dict(
color="blue"),
showlegend=False
),
row=2, col=1)
fig.add_trace(go.Scatter(
x=df[(df['index']==len(df['index'].unique())-1) & (df['state']=='Infected')]['x_pos'],
y=df[(df['index']==len(df['index'].unique())-1) & (df['state']=='Infected')]['y_pos'],
name='Infected',
mode='markers',
marker=dict(
color="green"),
showlegend=False
),
row=2, col=1)
fig.add_trace(go.Scatter(
x=df[(df['index']==len(df['index'].unique())-1) & (df['state']=='Recovered')]['x_pos'],
y=df[(df['index']==len(df['index'].unique())-1) & (df['state']=='Recovered')]['y_pos'],
name='Recovered',
mode='markers',
marker=dict(
color="orange"),
showlegend=False
),
row=2, col=1)
fig.add_trace(go.Scatter(
x=quar_grid[0],
y=quar_grid[1],
name='Quarantene Area',
mode='markers',
marker=dict(size=1,
color="red"),
showlegend=True
),
row=2, col=1)
frames =[go.Frame(
data=[go.Scatter(
x=[i for i in range(k)],
y=negatives,
mode="lines",
line=dict(width=2, color="blue")),
go.Scatter(
x=[i for i in range(k)],
y=positives,
mode="lines",
line=dict(width=2, color="green")),
go.Scatter(
x=[i for i in range(k)],
y=survivors,
mode="lines",
line=dict(width=2, color="orange")),
go.Scatter(
x=[i for i in range(k)],
y=deaths,
mode="lines",
line=dict(width=2, color="black")),
go.Scatter(
x=df[(df['index']==k) & (df['state']=='Susceptible')]['x_pos'],
y=df[(df['index']==k) & (df['state']=='Susceptible')]['y_pos'],
mode='markers',
marker=dict(
color="blue")
),
go.Scatter(
x=df[(df['index']==k) & (df['state']=='Infected')]['x_pos'],
y=df[(df['index']==k) & (df['state']=='Infected')]['y_pos'],
mode='markers',
marker=dict(
color="green")
),
go.Scatter(
x=df[(df['index']==k) & (df['state']=='Recovered')]['x_pos'],
y=df[(df['index']==k) & (df['state']=='Recovered')]['y_pos'],
mode='markers',
marker=dict(
color="orange")
),
],
traces=[0,1,2,3,4,5,6])
for k in range(len(negatives))]
fig.frames=frames
fig.update_layout(
shapes=[
# unfilled circle
dict(
type="rect",
xref="x2",
yref="y2",
x0=quar_grid[0][0],
y0=quar_grid[1][0],
x1=quar_grid[0][1],
y1=quar_grid[1][1],
line_color="red",
line=dict(
width=2,
),
),
],
updatemenus= [
{
"buttons": [
{
"args": [None, {"frame": {"duration": 100, "redraw": False},
"fromcurrent": True,
"transition": {"duration": 10,
"easing": "quadratic-in-out"}}],
"label": "Play",
"method": "animate"
},
{
"args": [[None], {"frame": {"duration": 0, "redraw": False},
"mode": "immediate",
"transition": {"duration": 0}}],
"label": "Pause",
"method": "animate"
}
],
"direction": "left",
"pad": {"r": 10, "t": 87},
"showactive": False,
"type": "buttons",
"x": 0.14,
"xanchor": "right",
"y": 1.4,
"yanchor": "top"
}
],)
fig.update_xaxes(title_text="Simulation Steps", row=1, col=1)
fig.update_yaxes(title_text="Number of Cases", row=1, col=1)
fig.update_xaxes(title_text="X", range=[0,grid_max[0][1]], row=2, col=1)
fig.update_yaxes(title_text="Y", range=[0,grid_max[1][1]], row=2, col=1)
fig.update_layout(height=600, width=800, title_text="Track and Trace Modelling")
fig.show()
class Person:
def __init__(self, situation, position, speed, extremes, age):
self.situation, self.position = situation, position
self.speed, self.extremes = speed, extremes
angle = np.random.uniform(0, 2*np.pi)
self.x_dir, self.y_dir = np.cos(angle), np.sin(angle)
self.age = age
self.reabilitation = 0
def step_ahead(self, community, proximity, contagiousness, no_move, deaths_p,
quarantene_grid, prob_to_be_untracked):
self = contacts(self, community, proximity, contagiousness, deaths_p)
self = new_pos(self, no_move, quarantene_grid, prob_to_be_untracked)
def contacts(ind, community, proximity, unlikelyness_of_contact, p_died):
if (ind.situation == 2) or (ind.situation == 3):
pass
elif ind.situation == 1:
if np.random.choice(2, 1, p=[1-p_died*ind.age, p_died*ind.age])[0] == 0:
ind.reabilitation += 1
if ind.reabilitation >= 14:
ind.situation = 2
else:
ind.situation = 3
else:
close_people = 0
for friend in community:
xx = (ind.position[0]-friend.position[0])
yy = (ind.position[1]-friend.position[1])
if (friend.situation == 1) and ((np.sqrt(xx**2 + yy**2) < proximity)):
close_people += 1
if sum([1 for x in np.random.random(close_people) if x >unlikelyness_of_contact]) > 0:
ind.situation = 1
return ind
def angle_between2(p1, p2):
#ang = np.arctan2(p1,p2)
ang = np.arctan2(p2,p1)
return ang
def check_bounds(ind, x_or_y, travelled_dist, i, no_move, quarantene_grid, prob_to_be_untracked):
if (ind.situation == 1) or (ind.situation == 2):
if np.random.choice(2, 1, p=[prob_to_be_untracked, 1-prob_to_be_untracked])[0] ==1:
if ind.position[i] == ind.position[0]:
des_pos = random.uniform(quarantene_grid[0][0], quarantene_grid[0][1])
if ind.position[i] < des_pos:
updated_dir = angle_between2(ind.position[i], des_pos)
else:
updated_dir = angle_between2(des_pos, ind.position[i])
#updated_dir = angle_between2(des_pos, ind.position[i])
if ind.position[i] < quarantene_grid[0][0] or ind.position[i] > quarantene_grid[0][1]:
whitin_bounds = False
else:
ind.extremes = quarantene_grid
whitin_bounds = True
else:
des_pos = random.uniform(quarantene_grid[1][0], quarantene_grid[1][1])
#updated_dir = angle_between2(ind.position[i], des_pos)
if ind.position[i] < des_pos:
updated_dir = angle_between2(ind.position[i], des_pos)
else:
updated_dir = angle_between2(des_pos, ind.position[i])
if ind.position[i] < quarantene_grid[1][0] or ind.position[i] > quarantene_grid[1][1]:
whitin_bounds = False
else:
ind.extremes = quarantene_grid
whitin_bounds = True
if whitin_bounds:
if ind.position[i] + updated_dir*travelled_dist < ind.extremes[i][0]:
updated_pos = -ind.position[i] + (-updated_dir*travelled_dist) + 2*ind.extremes[i][0]
updated_dir = -updated_dir
elif ind.position[i] + updated_dir*travelled_dist > ind.extremes[i][1]:
updated_pos = -ind.position[i] + (-updated_dir*travelled_dist) + 2*ind.extremes[i][1]
updated_dir = -updated_dir
else:
if ind.position[i] > des_pos:
updated_pos = ind.position[i] - updated_dir*travelled_dist
else:
updated_pos = ind.position[i] + updated_dir*travelled_dist
else:
if ind.position[i] > des_pos:
updated_pos = ind.position[i] - updated_dir*travelled_dist
else:
updated_pos = ind.position[i] + updated_dir*travelled_dist
else:
if ind.position[i] + x_or_y*travelled_dist < ind.extremes[i][0]:
updated_pos = -ind.position[i] + (-x_or_y*travelled_dist) + 2*ind.extremes[i][0]
updated_dir = -x_or_y
elif ind.position[i] + x_or_y*travelled_dist > ind.extremes[i][1]:
updated_pos = -ind.position[i] + (-x_or_y*travelled_dist) + 2*ind.extremes[i][1]
updated_dir = -x_or_y
else:
updated_pos = ind.position[i] + x_or_y*travelled_dist
if no_move:
updated_dir = 0
else:
updated_dir = x_or_y
elif ind.position[i] + x_or_y*travelled_dist < ind.extremes[i][0]:
updated_pos = -ind.position[i] + (-x_or_y*travelled_dist) + 2*ind.extremes[i][0]
updated_dir = -x_or_y
elif ind.position[i] + x_or_y*travelled_dist > ind.extremes[i][1]:
updated_pos = -ind.position[i] + (-x_or_y*travelled_dist) + 2*ind.extremes[i][1]
updated_dir = -x_or_y
else:
updated_pos = ind.position[i] + x_or_y*travelled_dist
if no_move:
updated_dir = 0
else:
updated_dir = x_or_y
return updated_pos, updated_dir
def new_pos(ind, no_move, quarantene_grid, prob_to_be_untracked):
travelled_dist = ind.speed*np.random.random()
ind.position[0], ind.x_dir = check_bounds(ind, ind.x_dir, travelled_dist, 0, no_move,
quarantene_grid, prob_to_be_untracked)
ind.position[1], ind.y_dir = check_bounds(ind, ind.y_dir, travelled_dist, 1, no_move,
quarantene_grid, prob_to_be_untracked)
return ind
def pop_simulation(size, iterations, probs_positives,
grid_lists, min_contact_radious,
unlikelyness_of_spread, static, d_p, avg_age, quarantene_grid,
prob_to_be_untracked):
population = []
for grid_l in grid_lists:
for i in range(0, size//len(grid_lists)):
population.append(Person(np.random.choice(2, 1, p=[1-probs_positives, probs_positives])[0],
[random.uniform(grid_l[0][0], grid_l[0][1]),
random.uniform(grid_l[1][0], grid_l[1][1])],
random.uniform(0, 1), grid_l,
min(2, max(1, np.random.normal(1+avg_age, 0.12, 1)[0]))))
negatives, positives, survivors = [], [], []
x_res, y_res, state = [], [], []
index, deaths = [], []
for it in range(iterations):
it_negative, it_positives, it_survivors, it_dead = 0, 0, 0, 0
for i, single in enumerate(population):
x_res.append(single.position[0])
y_res.append(single.position[1])
index.append(it)
if single.situation == 0:
it_negative += 1
state.append('Susceptible')
elif single.situation == 1:
it_positives += 1
state.append('Infected')
elif single.situation == 3:
it_dead += 1
state.append('Died')
else:
it_survivors += 1
state.append('Recovered')
single.step_ahead(population[:i]+population[i+1:], min_contact_radious,
unlikelyness_of_spread, no_move=static, deaths_p=d_p,
quarantene_grid=quarantene_grid, prob_to_be_untracked=prob_to_be_untracked)
negatives.append(it_negative)
positives.append(it_positives)
survivors.append(it_survivors)
deaths.append(it_dead)
return negatives, positives, survivors, deaths, x_res, y_res, state, index
probs_positives = 0.04
# The first pair represents the bounding X coordinates, while the second pair represents the Y coordinates
grid_max = [[0, 5], [0, 5]]
grid_limits = [[[0, 1], [0,1]], [[4, 5], [4, 5]], [[0, 1], [4, 5]]]
min_contact_radious = 0.3
unlikelyness_of_spread = 0.9
iterations = 50
size = 50
static = False
d_p = 0.02
avg_age = 0.9
quarantene_grid = [[0,1], [0,1]] #[[4, 5], [0, 1]] # [[2,3],[2,3]] # [[1,2], [0,1]]
prob_to_be_untracked = 0#0.5
negatives, positives, survivors, deaths, x_res, y_res, state, index = pop_simulation(size, iterations,
probs_positives,
grid_limits,
min_contact_radious,
unlikelyness_of_spread,
static, d_p, avg_age,
quarantene_grid,
prob_to_be_untracked)
print(deaths[len(deaths)-1])
plt.plot(negatives, label='Susceptible')
plt.plot(positives, label='Infected')
plt.plot(survivors, label='Recovered')
plt.plot(deaths, label='Died')
plt.legend()
plt.show()
len(grid_limits)
d = {'x_pos': x_res, 'y_pos': y_res, 'state': state, 'index':index}
df = pd.DataFrame(data=d)
#df.head()
for i in list(df['index'].unique()):
for j in list(df['state'].unique()):
if len(df[(df['index']==i) & (df['state'] == j)]) == 0:
df = df.append(pd.DataFrame([[grid_max[0][1]+5, grid_max[1][1]+5, j, i]],
columns=df.columns))
replay_plot(negatives, positives, survivors, deaths, df, grid_max=grid_max, quar_grid=quarantene_grid)
grids_types = {
'Single Community': [grid_max],
'2 Isolated Communities': [[[0, 1], [0, 1]], [[4, 5], [4, 5]]],
'2 Communities with shared central point': [grid_max, [[0, 1], [0, 1]], [[4, 5], [4, 5]]],
'4 Isolated Communities': [[[0, 1], [0, 1]], [[4, 5], [4, 5]], [[0, 1], [4, 5]], [[4, 5], [0, 1]]],
'4 Communities with shared central point': [grid_max, [[0, 1], [0, 1]], [[4, 5], [4, 5]], [[0, 1], [4, 5]], [[4, 5], [0, 1]]]
}
list(grids_types.keys())
quarantine_locations = {
'Single Community': [[4, 5], [0, 1]],
'2 Isolated Communities': [[4, 5], [0, 1]],
'2 Communities with shared central point': [[4, 5], [0, 1]],
'4 Isolated Communities': [[2, 3], [2, 3]],
'4 Communities with shared central point': [[2, 3], [2, 3]]
}
```
|
github_jupyter
|
import numpy as np
import random
import pandas as pd
import matplotlib.pyplot as plt
from plotly.subplots import make_subplots
import plotly.graph_objects as go
def replay_plot(negatives, positives, survivors, deaths, df, grid_max, quar_grid):
fig = make_subplots(rows=2, cols=1)
fig.add_trace(
go.Scatter(x=[i for i in range(len(negatives))], y=negatives,
mode="lines",
line=dict(width=2, color="blue"), name='Susceptible',),
row=1, col=1
)
fig.add_trace(
go.Scatter(x=[i for i in range(len(negatives))], y=positives,
mode="lines",
line=dict(width=2, color="green"), name='Infected',),
row=1, col=1
)
fig.add_trace(
go.Scatter(x=[i for i in range(len(negatives))], y=survivors,
mode="lines",
line=dict(width=2, color="orange"), name='Recovered',),
row=1, col=1
)
fig.add_trace(
go.Scatter(x=[i for i in range(len(negatives))], y=deaths,
mode="lines",
line=dict(width=2, color="black"), name='Died',),
row=1, col=1
)
fig.add_trace(
go.Scatter(
x=df[(df['index']==len(df['index'].unique())-1) & (df['state']=='Susceptible')]['x_pos'],
y=df[(df['index']==len(df['index'].unique())-1) & (df['state']=='Susceptible')]['y_pos'],
name='Susceptible',
mode='markers',
marker=dict(
color="blue"),
showlegend=False
),
row=2, col=1)
fig.add_trace(go.Scatter(
x=df[(df['index']==len(df['index'].unique())-1) & (df['state']=='Infected')]['x_pos'],
y=df[(df['index']==len(df['index'].unique())-1) & (df['state']=='Infected')]['y_pos'],
name='Infected',
mode='markers',
marker=dict(
color="green"),
showlegend=False
),
row=2, col=1)
fig.add_trace(go.Scatter(
x=df[(df['index']==len(df['index'].unique())-1) & (df['state']=='Recovered')]['x_pos'],
y=df[(df['index']==len(df['index'].unique())-1) & (df['state']=='Recovered')]['y_pos'],
name='Recovered',
mode='markers',
marker=dict(
color="orange"),
showlegend=False
),
row=2, col=1)
fig.add_trace(go.Scatter(
x=quar_grid[0],
y=quar_grid[1],
name='Quarantene Area',
mode='markers',
marker=dict(size=1,
color="red"),
showlegend=True
),
row=2, col=1)
frames =[go.Frame(
data=[go.Scatter(
x=[i for i in range(k)],
y=negatives,
mode="lines",
line=dict(width=2, color="blue")),
go.Scatter(
x=[i for i in range(k)],
y=positives,
mode="lines",
line=dict(width=2, color="green")),
go.Scatter(
x=[i for i in range(k)],
y=survivors,
mode="lines",
line=dict(width=2, color="orange")),
go.Scatter(
x=[i for i in range(k)],
y=deaths,
mode="lines",
line=dict(width=2, color="black")),
go.Scatter(
x=df[(df['index']==k) & (df['state']=='Susceptible')]['x_pos'],
y=df[(df['index']==k) & (df['state']=='Susceptible')]['y_pos'],
mode='markers',
marker=dict(
color="blue")
),
go.Scatter(
x=df[(df['index']==k) & (df['state']=='Infected')]['x_pos'],
y=df[(df['index']==k) & (df['state']=='Infected')]['y_pos'],
mode='markers',
marker=dict(
color="green")
),
go.Scatter(
x=df[(df['index']==k) & (df['state']=='Recovered')]['x_pos'],
y=df[(df['index']==k) & (df['state']=='Recovered')]['y_pos'],
mode='markers',
marker=dict(
color="orange")
),
],
traces=[0,1,2,3,4,5,6])
for k in range(len(negatives))]
fig.frames=frames
fig.update_layout(
shapes=[
# unfilled circle
dict(
type="rect",
xref="x2",
yref="y2",
x0=quar_grid[0][0],
y0=quar_grid[1][0],
x1=quar_grid[0][1],
y1=quar_grid[1][1],
line_color="red",
line=dict(
width=2,
),
),
],
updatemenus= [
{
"buttons": [
{
"args": [None, {"frame": {"duration": 100, "redraw": False},
"fromcurrent": True,
"transition": {"duration": 10,
"easing": "quadratic-in-out"}}],
"label": "Play",
"method": "animate"
},
{
"args": [[None], {"frame": {"duration": 0, "redraw": False},
"mode": "immediate",
"transition": {"duration": 0}}],
"label": "Pause",
"method": "animate"
}
],
"direction": "left",
"pad": {"r": 10, "t": 87},
"showactive": False,
"type": "buttons",
"x": 0.14,
"xanchor": "right",
"y": 1.4,
"yanchor": "top"
}
],)
fig.update_xaxes(title_text="Simulation Steps", row=1, col=1)
fig.update_yaxes(title_text="Number of Cases", row=1, col=1)
fig.update_xaxes(title_text="X", range=[0,grid_max[0][1]], row=2, col=1)
fig.update_yaxes(title_text="Y", range=[0,grid_max[1][1]], row=2, col=1)
fig.update_layout(height=600, width=800, title_text="Track and Trace Modelling")
fig.show()
class Person:
def __init__(self, situation, position, speed, extremes, age):
self.situation, self.position = situation, position
self.speed, self.extremes = speed, extremes
angle = np.random.uniform(0, 2*np.pi)
self.x_dir, self.y_dir = np.cos(angle), np.sin(angle)
self.age = age
self.reabilitation = 0
def step_ahead(self, community, proximity, contagiousness, no_move, deaths_p,
quarantene_grid, prob_to_be_untracked):
self = contacts(self, community, proximity, contagiousness, deaths_p)
self = new_pos(self, no_move, quarantene_grid, prob_to_be_untracked)
def contacts(ind, community, proximity, unlikelyness_of_contact, p_died):
if (ind.situation == 2) or (ind.situation == 3):
pass
elif ind.situation == 1:
if np.random.choice(2, 1, p=[1-p_died*ind.age, p_died*ind.age])[0] == 0:
ind.reabilitation += 1
if ind.reabilitation >= 14:
ind.situation = 2
else:
ind.situation = 3
else:
close_people = 0
for friend in community:
xx = (ind.position[0]-friend.position[0])
yy = (ind.position[1]-friend.position[1])
if (friend.situation == 1) and ((np.sqrt(xx**2 + yy**2) < proximity)):
close_people += 1
if sum([1 for x in np.random.random(close_people) if x >unlikelyness_of_contact]) > 0:
ind.situation = 1
return ind
def angle_between2(p1, p2):
#ang = np.arctan2(p1,p2)
ang = np.arctan2(p2,p1)
return ang
def check_bounds(ind, x_or_y, travelled_dist, i, no_move, quarantene_grid, prob_to_be_untracked):
if (ind.situation == 1) or (ind.situation == 2):
if np.random.choice(2, 1, p=[prob_to_be_untracked, 1-prob_to_be_untracked])[0] ==1:
if ind.position[i] == ind.position[0]:
des_pos = random.uniform(quarantene_grid[0][0], quarantene_grid[0][1])
if ind.position[i] < des_pos:
updated_dir = angle_between2(ind.position[i], des_pos)
else:
updated_dir = angle_between2(des_pos, ind.position[i])
#updated_dir = angle_between2(des_pos, ind.position[i])
if ind.position[i] < quarantene_grid[0][0] or ind.position[i] > quarantene_grid[0][1]:
whitin_bounds = False
else:
ind.extremes = quarantene_grid
whitin_bounds = True
else:
des_pos = random.uniform(quarantene_grid[1][0], quarantene_grid[1][1])
#updated_dir = angle_between2(ind.position[i], des_pos)
if ind.position[i] < des_pos:
updated_dir = angle_between2(ind.position[i], des_pos)
else:
updated_dir = angle_between2(des_pos, ind.position[i])
if ind.position[i] < quarantene_grid[1][0] or ind.position[i] > quarantene_grid[1][1]:
whitin_bounds = False
else:
ind.extremes = quarantene_grid
whitin_bounds = True
if whitin_bounds:
if ind.position[i] + updated_dir*travelled_dist < ind.extremes[i][0]:
updated_pos = -ind.position[i] + (-updated_dir*travelled_dist) + 2*ind.extremes[i][0]
updated_dir = -updated_dir
elif ind.position[i] + updated_dir*travelled_dist > ind.extremes[i][1]:
updated_pos = -ind.position[i] + (-updated_dir*travelled_dist) + 2*ind.extremes[i][1]
updated_dir = -updated_dir
else:
if ind.position[i] > des_pos:
updated_pos = ind.position[i] - updated_dir*travelled_dist
else:
updated_pos = ind.position[i] + updated_dir*travelled_dist
else:
if ind.position[i] > des_pos:
updated_pos = ind.position[i] - updated_dir*travelled_dist
else:
updated_pos = ind.position[i] + updated_dir*travelled_dist
else:
if ind.position[i] + x_or_y*travelled_dist < ind.extremes[i][0]:
updated_pos = -ind.position[i] + (-x_or_y*travelled_dist) + 2*ind.extremes[i][0]
updated_dir = -x_or_y
elif ind.position[i] + x_or_y*travelled_dist > ind.extremes[i][1]:
updated_pos = -ind.position[i] + (-x_or_y*travelled_dist) + 2*ind.extremes[i][1]
updated_dir = -x_or_y
else:
updated_pos = ind.position[i] + x_or_y*travelled_dist
if no_move:
updated_dir = 0
else:
updated_dir = x_or_y
elif ind.position[i] + x_or_y*travelled_dist < ind.extremes[i][0]:
updated_pos = -ind.position[i] + (-x_or_y*travelled_dist) + 2*ind.extremes[i][0]
updated_dir = -x_or_y
elif ind.position[i] + x_or_y*travelled_dist > ind.extremes[i][1]:
updated_pos = -ind.position[i] + (-x_or_y*travelled_dist) + 2*ind.extremes[i][1]
updated_dir = -x_or_y
else:
updated_pos = ind.position[i] + x_or_y*travelled_dist
if no_move:
updated_dir = 0
else:
updated_dir = x_or_y
return updated_pos, updated_dir
def new_pos(ind, no_move, quarantene_grid, prob_to_be_untracked):
travelled_dist = ind.speed*np.random.random()
ind.position[0], ind.x_dir = check_bounds(ind, ind.x_dir, travelled_dist, 0, no_move,
quarantene_grid, prob_to_be_untracked)
ind.position[1], ind.y_dir = check_bounds(ind, ind.y_dir, travelled_dist, 1, no_move,
quarantene_grid, prob_to_be_untracked)
return ind
def pop_simulation(size, iterations, probs_positives,
grid_lists, min_contact_radious,
unlikelyness_of_spread, static, d_p, avg_age, quarantene_grid,
prob_to_be_untracked):
population = []
for grid_l in grid_lists:
for i in range(0, size//len(grid_lists)):
population.append(Person(np.random.choice(2, 1, p=[1-probs_positives, probs_positives])[0],
[random.uniform(grid_l[0][0], grid_l[0][1]),
random.uniform(grid_l[1][0], grid_l[1][1])],
random.uniform(0, 1), grid_l,
min(2, max(1, np.random.normal(1+avg_age, 0.12, 1)[0]))))
negatives, positives, survivors = [], [], []
x_res, y_res, state = [], [], []
index, deaths = [], []
for it in range(iterations):
it_negative, it_positives, it_survivors, it_dead = 0, 0, 0, 0
for i, single in enumerate(population):
x_res.append(single.position[0])
y_res.append(single.position[1])
index.append(it)
if single.situation == 0:
it_negative += 1
state.append('Susceptible')
elif single.situation == 1:
it_positives += 1
state.append('Infected')
elif single.situation == 3:
it_dead += 1
state.append('Died')
else:
it_survivors += 1
state.append('Recovered')
single.step_ahead(population[:i]+population[i+1:], min_contact_radious,
unlikelyness_of_spread, no_move=static, deaths_p=d_p,
quarantene_grid=quarantene_grid, prob_to_be_untracked=prob_to_be_untracked)
negatives.append(it_negative)
positives.append(it_positives)
survivors.append(it_survivors)
deaths.append(it_dead)
return negatives, positives, survivors, deaths, x_res, y_res, state, index
probs_positives = 0.04
# The first pair represents the bounding X coordinates, while the second pair represents the Y coordinates
grid_max = [[0, 5], [0, 5]]
grid_limits = [[[0, 1], [0,1]], [[4, 5], [4, 5]], [[0, 1], [4, 5]]]
min_contact_radious = 0.3
unlikelyness_of_spread = 0.9
iterations = 50
size = 50
static = False
d_p = 0.02
avg_age = 0.9
quarantene_grid = [[0,1], [0,1]] #[[4, 5], [0, 1]] # [[2,3],[2,3]] # [[1,2], [0,1]]
prob_to_be_untracked = 0#0.5
negatives, positives, survivors, deaths, x_res, y_res, state, index = pop_simulation(size, iterations,
probs_positives,
grid_limits,
min_contact_radious,
unlikelyness_of_spread,
static, d_p, avg_age,
quarantene_grid,
prob_to_be_untracked)
print(deaths[len(deaths)-1])
plt.plot(negatives, label='Susceptible')
plt.plot(positives, label='Infected')
plt.plot(survivors, label='Recovered')
plt.plot(deaths, label='Died')
plt.legend()
plt.show()
len(grid_limits)
d = {'x_pos': x_res, 'y_pos': y_res, 'state': state, 'index':index}
df = pd.DataFrame(data=d)
#df.head()
for i in list(df['index'].unique()):
for j in list(df['state'].unique()):
if len(df[(df['index']==i) & (df['state'] == j)]) == 0:
df = df.append(pd.DataFrame([[grid_max[0][1]+5, grid_max[1][1]+5, j, i]],
columns=df.columns))
replay_plot(negatives, positives, survivors, deaths, df, grid_max=grid_max, quar_grid=quarantene_grid)
grids_types = {
'Single Community': [grid_max],
'2 Isolated Communities': [[[0, 1], [0, 1]], [[4, 5], [4, 5]]],
'2 Communities with shared central point': [grid_max, [[0, 1], [0, 1]], [[4, 5], [4, 5]]],
'4 Isolated Communities': [[[0, 1], [0, 1]], [[4, 5], [4, 5]], [[0, 1], [4, 5]], [[4, 5], [0, 1]]],
'4 Communities with shared central point': [grid_max, [[0, 1], [0, 1]], [[4, 5], [4, 5]], [[0, 1], [4, 5]], [[4, 5], [0, 1]]]
}
list(grids_types.keys())
quarantine_locations = {
'Single Community': [[4, 5], [0, 1]],
'2 Isolated Communities': [[4, 5], [0, 1]],
'2 Communities with shared central point': [[4, 5], [0, 1]],
'4 Isolated Communities': [[2, 3], [2, 3]],
'4 Communities with shared central point': [[2, 3], [2, 3]]
}
| 0.330255 | 0.36815 |
# Explore difference between PHONEIX-ACES two spectra of incremental temperature
#### Aim:
In this Notebook I explore the differences in the combined models when there are incremental changes of one component.
For example. The difference between a model with I_host([5200, 4.50, 0.0]) and companions I_companion([2500, 4.5, 0.0]) and I_companion([2600, 4.5, 0.0]). Where the temperature changes by 100K (one model grid step)
####Method:
Create combined spectral models, that differ in grid parameters incremented by 1 step (all else fixed). Take the difference between the two and analsyse the differences. In this case we take the mean difference and the standard deviation and see how these change for different host and companion parameters.
At this stage just for a single radial velocity point of both components.
#### Results:
For now this is with a fixed radial velocity of each component=0. Possibly this will be bigger if offset.
The Trends:
Host differences:
- Teff -
- logg -
- feh -
Companion differences:
- Teff -
- logg -
- feh -
The small difference in companion parametes make it harder to distinguish the correct parameters. The shape of $\chi**2$ will be shallower. We see this for companion radial velocity were there is not a clear minimum and large error bars.
```
from simulators.iam_module import prepare_iam_model_spectra, continuum_alpha, continuum, inherent_alpha_model
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
class Spectrum():
pass
fudge=None
obs_spec = Spectrum()
obs_spec.xaxis = np.linspace(2110, 2130, 1000)
chip = 1
def combine_spectrum(params1, params2, rvs, gammas, plot=True):
# Load phoenix models and scale by area and wavelength limit
mod1_spec, mod2_spec = prepare_iam_model_spectra(params1, params2, limits=[2110, 2130],
area_scale=True, wav_scale=True)
inherent_alpha = continuum_alpha(mod1_spec, mod2_spec, chip)
# Combine model spectra with iam model
if plot:
plt_spec = mod1_spec.copy()
plt_spec2 = mod2_spec.copy()
max_host = np.max(plt_spec.flux)
ax = plt.plot()
plt_spec /= max_host
plt_spec2 /= max_host
plt_spec.plot(label=params1)
plt_spec2.plot(label=params2)
SNR = 500
std_noise = np.random.normal(0, np.ones_like(plt_spec.xaxis)/SNR)
plt.plot(plt_spec.xaxis, std_noise, label="Noise with std=1/{}".format(SNR))
plt.title("Spectra before combination\n (Scaled by area already)")
plt.ylabel("Normalzied flux (peak host)")
plt.legend()
plt.show()
if fudge or (fudge is not None):
fudge_factor = float(fudge)
mod2_spec.flux *= fudge_factor # fudge factor multiplication
mod2_spec.plot(label="fudged {0}".format(params2))
plt.title("fudges models")
plt.legend()
warnings.warn("Using a fudge factor = {0}".format(fudge_factor))
iam_grid_func = inherent_alpha_model(mod1_spec.xaxis, mod1_spec.flux, mod2_spec.flux,
rvs=rvs, gammas=gammas)
iam_grid_models = iam_grid_func(obs_spec.xaxis)
iam_grid_models = iam_grid_func(obs_spec.xaxis)
# Continuum normalize all iam_gird_models
def axis_continuum(flux):
"""Continuum to apply along axis with predefined variables parameters."""
return continuum(obs_spec.xaxis, flux, splits=20, method="exponential", top=5)
iam_grid_continuum = np.apply_along_axis(axis_continuum, 0, iam_grid_models)
iam_grid_models = iam_grid_models / iam_grid_continuum
return iam_grid_models
host1 = [5200, 4.5, 0.0]
host2 = [5100, 4.5, 0.0]
host3 = [5000, 4.5, 0.0]
comp1 = [2500, 4.5, 0.0]
comp2 = [2400, 4.5, 0.0]
comp3 = [2300, 4.5, 0.0]
rvs = [0]
gammas = [0]
model11 = combine_spectrum(host1, comp1, rvs, gammas).squeeze()
model12 = combine_spectrum(host1, comp2, rvs, gammas).squeeze()
model13 = combine_spectrum(host1, comp3, rvs, gammas).squeeze()
model21 = combine_spectrum(host2, comp1, rvs, gammas).squeeze()
model22 = combine_spectrum(host2, comp2, rvs, gammas).squeeze()
model23 = combine_spectrum(host2, comp3, rvs, gammas).squeeze()
model31 = combine_spectrum(host3, comp1, rvs, gammas).squeeze()
model32 = combine_spectrum(host3, comp2, rvs, gammas).squeeze()
model33 = combine_spectrum(host3, comp3, rvs, gammas).squeeze()
# print(model22.shape)
plt.legend()
def plot_components(params1, params2, rvs, gammas, plot=True, snr=500):
# Load phoenix models and scale by area and wavelength limit
mod1_spec, mod2_spec = prepare_iam_model_spectra(params1, params2, limits=[2110, 2130],
area_scale=True, wav_scale=True)
inherent_alpha = continuum_alpha(mod1_spec, mod2_spec, chip)
# Combine model spectra with iam model
if plot:
plt_spec = mod1_spec.copy()
plt_spec2 = mod2_spec.copy()
max_host = np.median(plt_spec.flux)
ax = plt.plot()
plt_spec /= max_host
plt_spec2 /= max_host
plt_spec.plot(label=params1)
plt_spec2.plot(label=params2)
std_noise = np.random.normal(0.5, np.ones_like(plt_spec.xaxis)/snr)
plt.plot(plt_spec.xaxis, std_noise, label="Noise with std=1/{} (offset)".format(snr))
plt.title("Spectra before combination\n (Scaled by emited area)")
plt.ylabel("Normalzied flux (peak host)")
plt.xlim([2110, 2120])
plt.legend(loc=5)
#plt.ylim([0,1.3])
plt.show()
plot_components(host3, comp3, rvs, gammas, snr=500)
plot_components(comp3, comp3, rvs, gammas, snr=500)
# Standard devition of median normalized spectrum of companion
mod1_spec, mod2_spec = prepare_iam_model_spectra(host3, comp3, limits=[2110, 2130],
area_scale=True, wav_scale=True)
print("std of companion spectrum", np.std(mod2_spec.flux/np.median(mod2_spec.flux)-1))
print("std of companion spectrum", (np.max(mod2_spec.flux)- np.min(mod2_spec.flux))/np.median(mod2_spec.flux))
print("Very little difference")
plt.plot(model11, label="11")
plt.plot(model12,":", label="12")
plt.plot(model21, label="21")
plt.plot(model22, "--", label="22")
plt.legend()
plt.ylim([.9, 1.01])
plt.xlim(0,200)
plt.show()
plt.plot(model11-model12, label="11-12")
plt.plot(model21-model22 + 0.005 , label="21-22")
plt.plot(model31-model33 + 0.010, label="31-32")
plt.plot(model11-model13 + 0.015, label="11-13")
plt.plot(model21-model23 + 0.02 , label="21-23")
plt.plot(model31-model33 + 0.025, label="31-33")
plt.legend()
plt.show()
stat = lambda x: (np.nanmean(x), np.nanstd(x))
print("Same host teff, companion spectra changes teff 100K")
print("comp = 5200")
print("diff mean {0:5.05f}, std {1:5.05f}".format(*stat(model11-model12)))
print("comp = 5100")
print("diff mean {0:5.05f}, std {1:5.05f}".format(*stat(model21-model22)))
# Same comp temperature, companion changes by teff 100K
print("comp = 2500")
print("diff mean {0:5.05f}, std {1:5.05f}".format(*stat(model11-model21)))
print("comp = 2400")
print("diff mean {0:5.05f}, std {1:5.05f}".format(*stat(model12-model22)))
```
## Plots
```
linestyle = ["-", "--", "-."]
```
# Tempetature Changes
```
offset = [-20, 0, 20]
def diff_comp100(params1, params2):
"""Differnece in parameters when param2teff changes bu +100"""
rvs = [0]
gammas = [0]
params_new = [params2[0]+100, *params2[1:]]
s1 = combine_spectrum(params1, params2, rvs, gammas, plot=False).squeeze()
s2 = combine_spectrum(params1, params_new, rvs, gammas, plot=False).squeeze()
diff = s1-s2
return diff
# Make plots of t
host = [5100, 6000, 7000]
comp = np.arange(2300, 5000, 100)
for i, hteff in enumerate(host):
cmp_mean = []
cmp_std = []
for comp_teff in comp:
diff = diff_comp100([hteff, 4.5, 0.0], [comp_teff, 4.5, 0.0])
mean, std = stat(diff)
cmp_mean.append(mean)
cmp_std.append(std)
# print(len(comp), len(cmp_mean), len(cmp_std))
plt.errorbar(comp+offset[i], cmp_mean, cmp_std , label=hteff, linestyle=linestyle[i])
plt.legend(title="Host temperature")
plt.ylabel("Flux Diff and std")
plt.xlabel("Companion Spectrum")
plt.title("Companion change by 100K")
plt.show()
for i, hteff in enumerate(host):
cmp_mean = []
cmp_std = []
for comp_teff in comp:
diff = diff_comp100([hteff, 4.5, 0.0], [comp_teff, 4.5, 0.0])
mean, std = stat(diff)
cmp_mean.append(mean)
cmp_std.append(std)
# print(len(comp), len(cmp_mean), len(cmp_std))
plt.plot(comp, cmp_std , label=hteff, linestyle=linestyle[i])
plt.legend(title="Host temperature")
plt.ylabel("Std")
plt.xlabel("Companion Spectrum")
plt.title("Companion change by 100K")
plt.show()
def diff_host100(params1, params2):
"""Differnece in parameters when param2teff changes bu +100"""
rvs = [0]
gammas = [0]
params_new = [params1[0]+100, *params1[1:]]
s1 = combine_spectrum(params1, params2, rvs, gammas, plot=False).squeeze()
s2 = combine_spectrum(params_new, params2, rvs, gammas, plot=False).squeeze()
diff = s1-s2
return diff
# Make plots of t
comp = [2500, 3400, 4000]
host = np.arange(5100, 7000, 100)
for i, cteff in enumerate(comp):
host_mean = []
host_std = []
for hteff in host:
diff = diff_comp100([hteff, 4.5, 0.0], [cteff, 4.5, 0.0])
mean, std = stat(diff)
host_mean.append(mean)
host_std.append(std)
# print(len(comp), len(cmp_mean), len(cmp_std))
plt.errorbar(host+offset[i], host_mean, host_std , label=cteff, ls=linestyle[i], marker=".")
plt.legend(title="Comp temperature")
plt.ylabel("Flux Diff and std")
plt.xlabel("Host Spectrum Teff")
plt.title("Host change by 100K")
plt.show()
for i, cteff in enumerate(comp):
host_mean = []
host_std = []
for hteff in host:
diff = diff_host100([hteff, 4.5, 0.0], [cteff, 4.5, 0.0])
mean, std = stat(diff)
host_mean.append(mean)
host_std.append(std)
plt.plot(host, host_std , label=cteff, ls=linestyle[i], ms=3)
plt.legend(title="Host Teff")
plt.ylabel("Std")
plt.xlabel("Host Teff")
plt.title("Host change by 100K")
plt.show()
```
I do not know why there is a large spike at 5000 for the companion.
# Logg change
```
offset = [-0.05, 0, 0.05]
def diff_complogg(params1, params2):
"""Differnece in parameters when param2teff changes bu +100"""
rvs = [0]
gammas = [0]
params_new = [params2[0], params2[1]+0.5, params2[2]]
s1 = combine_spectrum(params1, params2, rvs, gammas, plot=False).squeeze()
s2 = combine_spectrum(params1, params_new, rvs, gammas, plot=False).squeeze()
diff = s2-s1
return diff
# Make plots of t
host = [2.5, 3, 5]
comp = np.arange(2.5, 6, 0.5)
for i, hlogg in enumerate(host):
cmp_mean = []
cmp_std = []
for clogg in comp:
try:
diff = diff_complogg([6000, hlogg, 0.0], [2500, clogg, 0.0])
mean, std = stat(diff)
cmp_mean.append(mean)
cmp_std.append(std)
except:
cmp_mean.append(np.nan)
cmp_std.append(np.nan)
# print(len(comp), len(cmp_mean), len(cmp_std))
plt.errorbar(comp+offset[i], cmp_mean, cmp_std , label=hlogg, linestyle=linestyle[i])
plt.legend(title="Host logg")
plt.ylabel("Flux Diff and std")
plt.xlabel("Companion Logg")
plt.title("Companion increase by logg=0.5")
plt.show()
for i, hlogg in enumerate(host):
cmp_mean = []
cmp_std = []
for clogg in comp:
try:
diff = diff_complogg([6000, hlogg, 0.0], [2500, clogg, 0.0])
mean, std = stat(diff)
cmp_mean.append(mean)
cmp_std.append(std)
except:
cmp_mean.append(np.nan)
cmp_std.append(np.nan)
# print(len(comp), len(cmp_mean), len(cmp_std))
plt.plot(comp, cmp_std , label=hlogg, linestyle=linestyle[i])
plt.legend(title="Host logg")
plt.ylabel("Std")
plt.xlabel("Companion Logg")
plt.title("Companion increase by logg=0.5")
plt.show()
def diff_hostlogg(params1, params2):
"""Differnece in parameters when param2teff changes bu +100"""
rvs = [0]
gammas = [0]
s1 = combine_spectrum(params1, params2, rvs, gammas, plot=False).squeeze()
params_new = [params1[0], params1[1]+0.5, params1[2]]
s2 = combine_spectrum(params_new, params2, rvs, gammas, plot=False).squeeze()
diff = s2-s1
return diff
# Make plots of t
comp = [ 3, 5]
host = np.arange(3, 6, 0.5)
for i, clogg in enumerate(comp):
host_mean = []
host_std = []
for hlogg in host:
diff = diff_complogg([6000, hlogg, 0.0], [2500, clogg, 0.0])
mean, std = stat(diff)
host_mean.append(mean)
host_std.append(std)
# print(len(host), len(host_mean), len(host_std))
plt.errorbar(host+offset[i], host_mean, host_std , label=clogg, linestyle=linestyle[i])
plt.legend(title="Companion logg")
plt.ylabel("Flux Diff and std")
plt.xlabel("Host Logg")
plt.title("Companion increase by logg=0.5")
plt.show()
for i, clogg in enumerate(comp):
host_mean = []
host_std = []
for hlogg in host:
diff = diff_complogg([6000, hlogg, 0.0], [2500, clogg, 0.0])
mean, std = stat(diff)
host_mean.append(mean)
host_std.append(std)
plt.plot(host, host_std , label=clogg, linestyle=linestyle[i])
plt.legend(title="Companion logg")
plt.ylabel("Std")
plt.xlabel("Host Logg")
plt.title("Companion increase by logg=0.5")
plt.show()
```
# Feh changes
```
def diff_compfeh(params1, params2):
"""Differnece in parameters when param2teff changes bu +100"""
rvs = [0]
gammas = [0]
s1 = combine_spectrum(params1, params2, rvs, gammas, plot=False).squeeze()
params_new = [params2[0], params2[1], params2[2]+0.5]
s2 = combine_spectrum(params1, params_new, rvs, gammas, plot=False).squeeze()
diff = s2 - s1
return diff
# Make plots of t
host = [-2, -0.5, 1]
comp = np.arange(-2.5, 1.5, 0.5)
for i, hfeh in enumerate(host):
comp_mean = []
comp_std = []
for cfeh in comp:
try:
diff = diff_compfeh([6000, 4.5, hfeh], [2500, 4.5, cfeh])
mean, std = stat(diff)
comp_mean.append(mean)
comp_std.append(std)
except:
comp_mean.append(np.nan)
comp_std.append(np.nan)
print(len(comp), len(comp_mean), len(comp_std))
plt.errorbar(comp+offset[i], comp_mean, comp_std , label=hfeh, linestyle=linestyle[i])
plt.legend(title="Companion logg")
plt.ylabel("Flux Diff and std")
plt.xlabel("Host feh")
plt.title("Companion increase by feh=0.5")
plt.show()
for i, hfeh in enumerate(host):
comp_mean = []
comp_std = []
for cfeh in comp:
try:
diff = diff_compfeh([6000, 4.5, hfeh], [2500, 4.5, cfeh])
mean, std = stat(diff)
comp_mean.append(mean)
comp_std.append(std)
except:
comp_mean.append(np.nan)
comp_std.append(np.nan)
print(len(comp), len(comp_mean), len(comp_std))
plt.errorbar(comp, comp_std , label=hfeh, linestyle=linestyle[i])
plt.legend(title="Companion logg")
plt.ylabel("Std")
plt.xlabel("Host feh")
plt.title("Companion increase by feh=0.5")
plt.show()
def diff_hostfeh(params1, params2):
"""Differnece in parameters when param2 teff changes by +100"""
rvs = [0]
gammas = [0]
s1 = combine_spectrum(params1, params2, rvs, gammas, plot=False).squeeze()
params_new = [params1[0], params1[1], params1[2]+0.5]
s2 = combine_spectrum(params_new, params2, rvs, gammas, plot=False).squeeze()
diff = s2 - s1
return diff
# Make plots of t
comp = [-2, -0.5, 1]
host = np.arange(-2.5, 1.5, 0.5)
for i, cfeh in enumerate(comp):
host_mean = []
host_std = []
for hfeh in host:
try:
diff = diff_hostfeh([6000, 4.5, hfeh], [2500, 4.5, cfeh])
mean, std = stat(diff)
host_mean.append(mean)
host_std.append(std)
except:
host_mean.append(np.nan)
host_std.append(np.nan)
print(len(host), len(host_mean), len(host_std))
plt.errorbar(host+offset[i], host_mean, host_std, label=cfeh, linestyle=linestyle[i])
plt.legend(title="Companion logg")
plt.ylabel("Flux Diff and std")
plt.xlabel("Host logg")
plt.title("Host increase by logg=0.5")
plt.show()
for i, cfeh in enumerate(comp):
host_mean = []
host_std = []
for hfeh in host:
try:
diff = diff_hostfeh([6000, 4.5, hfeh], [2500, 4.5, cfeh])
mean, std = stat(diff)
host_mean.append(mean)
host_std.append(std)
except:
host_mean.append(np.nan)
host_std.append(np.nan)
print(len(host), len(host_mean), len(host_std))
plt.errorbar(host, host_std, label=cfeh, linestyle=linestyle[i])
plt.legend(title="Companion feh")
plt.ylabel("Std")
plt.xlabel("Host feh")
plt.title("Host increase by feh=0.5")
plt.show()
mod1_spec, mod2_spec = prepare_iam_model_spectra([5000, 4.5, 0.0], [5000, 5., 0.0], limits=[2010, 2430],
area_scale=True, wav_scale=True)
mod1_spec.plot(label="1")
mod2_spec.plot(label="2")
plt.legend()
plt.show()
```
|
github_jupyter
|
from simulators.iam_module import prepare_iam_model_spectra, continuum_alpha, continuum, inherent_alpha_model
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
class Spectrum():
pass
fudge=None
obs_spec = Spectrum()
obs_spec.xaxis = np.linspace(2110, 2130, 1000)
chip = 1
def combine_spectrum(params1, params2, rvs, gammas, plot=True):
# Load phoenix models and scale by area and wavelength limit
mod1_spec, mod2_spec = prepare_iam_model_spectra(params1, params2, limits=[2110, 2130],
area_scale=True, wav_scale=True)
inherent_alpha = continuum_alpha(mod1_spec, mod2_spec, chip)
# Combine model spectra with iam model
if plot:
plt_spec = mod1_spec.copy()
plt_spec2 = mod2_spec.copy()
max_host = np.max(plt_spec.flux)
ax = plt.plot()
plt_spec /= max_host
plt_spec2 /= max_host
plt_spec.plot(label=params1)
plt_spec2.plot(label=params2)
SNR = 500
std_noise = np.random.normal(0, np.ones_like(plt_spec.xaxis)/SNR)
plt.plot(plt_spec.xaxis, std_noise, label="Noise with std=1/{}".format(SNR))
plt.title("Spectra before combination\n (Scaled by area already)")
plt.ylabel("Normalzied flux (peak host)")
plt.legend()
plt.show()
if fudge or (fudge is not None):
fudge_factor = float(fudge)
mod2_spec.flux *= fudge_factor # fudge factor multiplication
mod2_spec.plot(label="fudged {0}".format(params2))
plt.title("fudges models")
plt.legend()
warnings.warn("Using a fudge factor = {0}".format(fudge_factor))
iam_grid_func = inherent_alpha_model(mod1_spec.xaxis, mod1_spec.flux, mod2_spec.flux,
rvs=rvs, gammas=gammas)
iam_grid_models = iam_grid_func(obs_spec.xaxis)
iam_grid_models = iam_grid_func(obs_spec.xaxis)
# Continuum normalize all iam_gird_models
def axis_continuum(flux):
"""Continuum to apply along axis with predefined variables parameters."""
return continuum(obs_spec.xaxis, flux, splits=20, method="exponential", top=5)
iam_grid_continuum = np.apply_along_axis(axis_continuum, 0, iam_grid_models)
iam_grid_models = iam_grid_models / iam_grid_continuum
return iam_grid_models
host1 = [5200, 4.5, 0.0]
host2 = [5100, 4.5, 0.0]
host3 = [5000, 4.5, 0.0]
comp1 = [2500, 4.5, 0.0]
comp2 = [2400, 4.5, 0.0]
comp3 = [2300, 4.5, 0.0]
rvs = [0]
gammas = [0]
model11 = combine_spectrum(host1, comp1, rvs, gammas).squeeze()
model12 = combine_spectrum(host1, comp2, rvs, gammas).squeeze()
model13 = combine_spectrum(host1, comp3, rvs, gammas).squeeze()
model21 = combine_spectrum(host2, comp1, rvs, gammas).squeeze()
model22 = combine_spectrum(host2, comp2, rvs, gammas).squeeze()
model23 = combine_spectrum(host2, comp3, rvs, gammas).squeeze()
model31 = combine_spectrum(host3, comp1, rvs, gammas).squeeze()
model32 = combine_spectrum(host3, comp2, rvs, gammas).squeeze()
model33 = combine_spectrum(host3, comp3, rvs, gammas).squeeze()
# print(model22.shape)
plt.legend()
def plot_components(params1, params2, rvs, gammas, plot=True, snr=500):
# Load phoenix models and scale by area and wavelength limit
mod1_spec, mod2_spec = prepare_iam_model_spectra(params1, params2, limits=[2110, 2130],
area_scale=True, wav_scale=True)
inherent_alpha = continuum_alpha(mod1_spec, mod2_spec, chip)
# Combine model spectra with iam model
if plot:
plt_spec = mod1_spec.copy()
plt_spec2 = mod2_spec.copy()
max_host = np.median(plt_spec.flux)
ax = plt.plot()
plt_spec /= max_host
plt_spec2 /= max_host
plt_spec.plot(label=params1)
plt_spec2.plot(label=params2)
std_noise = np.random.normal(0.5, np.ones_like(plt_spec.xaxis)/snr)
plt.plot(plt_spec.xaxis, std_noise, label="Noise with std=1/{} (offset)".format(snr))
plt.title("Spectra before combination\n (Scaled by emited area)")
plt.ylabel("Normalzied flux (peak host)")
plt.xlim([2110, 2120])
plt.legend(loc=5)
#plt.ylim([0,1.3])
plt.show()
plot_components(host3, comp3, rvs, gammas, snr=500)
plot_components(comp3, comp3, rvs, gammas, snr=500)
# Standard devition of median normalized spectrum of companion
mod1_spec, mod2_spec = prepare_iam_model_spectra(host3, comp3, limits=[2110, 2130],
area_scale=True, wav_scale=True)
print("std of companion spectrum", np.std(mod2_spec.flux/np.median(mod2_spec.flux)-1))
print("std of companion spectrum", (np.max(mod2_spec.flux)- np.min(mod2_spec.flux))/np.median(mod2_spec.flux))
print("Very little difference")
plt.plot(model11, label="11")
plt.plot(model12,":", label="12")
plt.plot(model21, label="21")
plt.plot(model22, "--", label="22")
plt.legend()
plt.ylim([.9, 1.01])
plt.xlim(0,200)
plt.show()
plt.plot(model11-model12, label="11-12")
plt.plot(model21-model22 + 0.005 , label="21-22")
plt.plot(model31-model33 + 0.010, label="31-32")
plt.plot(model11-model13 + 0.015, label="11-13")
plt.plot(model21-model23 + 0.02 , label="21-23")
plt.plot(model31-model33 + 0.025, label="31-33")
plt.legend()
plt.show()
stat = lambda x: (np.nanmean(x), np.nanstd(x))
print("Same host teff, companion spectra changes teff 100K")
print("comp = 5200")
print("diff mean {0:5.05f}, std {1:5.05f}".format(*stat(model11-model12)))
print("comp = 5100")
print("diff mean {0:5.05f}, std {1:5.05f}".format(*stat(model21-model22)))
# Same comp temperature, companion changes by teff 100K
print("comp = 2500")
print("diff mean {0:5.05f}, std {1:5.05f}".format(*stat(model11-model21)))
print("comp = 2400")
print("diff mean {0:5.05f}, std {1:5.05f}".format(*stat(model12-model22)))
linestyle = ["-", "--", "-."]
offset = [-20, 0, 20]
def diff_comp100(params1, params2):
"""Differnece in parameters when param2teff changes bu +100"""
rvs = [0]
gammas = [0]
params_new = [params2[0]+100, *params2[1:]]
s1 = combine_spectrum(params1, params2, rvs, gammas, plot=False).squeeze()
s2 = combine_spectrum(params1, params_new, rvs, gammas, plot=False).squeeze()
diff = s1-s2
return diff
# Make plots of t
host = [5100, 6000, 7000]
comp = np.arange(2300, 5000, 100)
for i, hteff in enumerate(host):
cmp_mean = []
cmp_std = []
for comp_teff in comp:
diff = diff_comp100([hteff, 4.5, 0.0], [comp_teff, 4.5, 0.0])
mean, std = stat(diff)
cmp_mean.append(mean)
cmp_std.append(std)
# print(len(comp), len(cmp_mean), len(cmp_std))
plt.errorbar(comp+offset[i], cmp_mean, cmp_std , label=hteff, linestyle=linestyle[i])
plt.legend(title="Host temperature")
plt.ylabel("Flux Diff and std")
plt.xlabel("Companion Spectrum")
plt.title("Companion change by 100K")
plt.show()
for i, hteff in enumerate(host):
cmp_mean = []
cmp_std = []
for comp_teff in comp:
diff = diff_comp100([hteff, 4.5, 0.0], [comp_teff, 4.5, 0.0])
mean, std = stat(diff)
cmp_mean.append(mean)
cmp_std.append(std)
# print(len(comp), len(cmp_mean), len(cmp_std))
plt.plot(comp, cmp_std , label=hteff, linestyle=linestyle[i])
plt.legend(title="Host temperature")
plt.ylabel("Std")
plt.xlabel("Companion Spectrum")
plt.title("Companion change by 100K")
plt.show()
def diff_host100(params1, params2):
"""Differnece in parameters when param2teff changes bu +100"""
rvs = [0]
gammas = [0]
params_new = [params1[0]+100, *params1[1:]]
s1 = combine_spectrum(params1, params2, rvs, gammas, plot=False).squeeze()
s2 = combine_spectrum(params_new, params2, rvs, gammas, plot=False).squeeze()
diff = s1-s2
return diff
# Make plots of t
comp = [2500, 3400, 4000]
host = np.arange(5100, 7000, 100)
for i, cteff in enumerate(comp):
host_mean = []
host_std = []
for hteff in host:
diff = diff_comp100([hteff, 4.5, 0.0], [cteff, 4.5, 0.0])
mean, std = stat(diff)
host_mean.append(mean)
host_std.append(std)
# print(len(comp), len(cmp_mean), len(cmp_std))
plt.errorbar(host+offset[i], host_mean, host_std , label=cteff, ls=linestyle[i], marker=".")
plt.legend(title="Comp temperature")
plt.ylabel("Flux Diff and std")
plt.xlabel("Host Spectrum Teff")
plt.title("Host change by 100K")
plt.show()
for i, cteff in enumerate(comp):
host_mean = []
host_std = []
for hteff in host:
diff = diff_host100([hteff, 4.5, 0.0], [cteff, 4.5, 0.0])
mean, std = stat(diff)
host_mean.append(mean)
host_std.append(std)
plt.plot(host, host_std , label=cteff, ls=linestyle[i], ms=3)
plt.legend(title="Host Teff")
plt.ylabel("Std")
plt.xlabel("Host Teff")
plt.title("Host change by 100K")
plt.show()
offset = [-0.05, 0, 0.05]
def diff_complogg(params1, params2):
"""Differnece in parameters when param2teff changes bu +100"""
rvs = [0]
gammas = [0]
params_new = [params2[0], params2[1]+0.5, params2[2]]
s1 = combine_spectrum(params1, params2, rvs, gammas, plot=False).squeeze()
s2 = combine_spectrum(params1, params_new, rvs, gammas, plot=False).squeeze()
diff = s2-s1
return diff
# Make plots of t
host = [2.5, 3, 5]
comp = np.arange(2.5, 6, 0.5)
for i, hlogg in enumerate(host):
cmp_mean = []
cmp_std = []
for clogg in comp:
try:
diff = diff_complogg([6000, hlogg, 0.0], [2500, clogg, 0.0])
mean, std = stat(diff)
cmp_mean.append(mean)
cmp_std.append(std)
except:
cmp_mean.append(np.nan)
cmp_std.append(np.nan)
# print(len(comp), len(cmp_mean), len(cmp_std))
plt.errorbar(comp+offset[i], cmp_mean, cmp_std , label=hlogg, linestyle=linestyle[i])
plt.legend(title="Host logg")
plt.ylabel("Flux Diff and std")
plt.xlabel("Companion Logg")
plt.title("Companion increase by logg=0.5")
plt.show()
for i, hlogg in enumerate(host):
cmp_mean = []
cmp_std = []
for clogg in comp:
try:
diff = diff_complogg([6000, hlogg, 0.0], [2500, clogg, 0.0])
mean, std = stat(diff)
cmp_mean.append(mean)
cmp_std.append(std)
except:
cmp_mean.append(np.nan)
cmp_std.append(np.nan)
# print(len(comp), len(cmp_mean), len(cmp_std))
plt.plot(comp, cmp_std , label=hlogg, linestyle=linestyle[i])
plt.legend(title="Host logg")
plt.ylabel("Std")
plt.xlabel("Companion Logg")
plt.title("Companion increase by logg=0.5")
plt.show()
def diff_hostlogg(params1, params2):
"""Differnece in parameters when param2teff changes bu +100"""
rvs = [0]
gammas = [0]
s1 = combine_spectrum(params1, params2, rvs, gammas, plot=False).squeeze()
params_new = [params1[0], params1[1]+0.5, params1[2]]
s2 = combine_spectrum(params_new, params2, rvs, gammas, plot=False).squeeze()
diff = s2-s1
return diff
# Make plots of t
comp = [ 3, 5]
host = np.arange(3, 6, 0.5)
for i, clogg in enumerate(comp):
host_mean = []
host_std = []
for hlogg in host:
diff = diff_complogg([6000, hlogg, 0.0], [2500, clogg, 0.0])
mean, std = stat(diff)
host_mean.append(mean)
host_std.append(std)
# print(len(host), len(host_mean), len(host_std))
plt.errorbar(host+offset[i], host_mean, host_std , label=clogg, linestyle=linestyle[i])
plt.legend(title="Companion logg")
plt.ylabel("Flux Diff and std")
plt.xlabel("Host Logg")
plt.title("Companion increase by logg=0.5")
plt.show()
for i, clogg in enumerate(comp):
host_mean = []
host_std = []
for hlogg in host:
diff = diff_complogg([6000, hlogg, 0.0], [2500, clogg, 0.0])
mean, std = stat(diff)
host_mean.append(mean)
host_std.append(std)
plt.plot(host, host_std , label=clogg, linestyle=linestyle[i])
plt.legend(title="Companion logg")
plt.ylabel("Std")
plt.xlabel("Host Logg")
plt.title("Companion increase by logg=0.5")
plt.show()
def diff_compfeh(params1, params2):
"""Differnece in parameters when param2teff changes bu +100"""
rvs = [0]
gammas = [0]
s1 = combine_spectrum(params1, params2, rvs, gammas, plot=False).squeeze()
params_new = [params2[0], params2[1], params2[2]+0.5]
s2 = combine_spectrum(params1, params_new, rvs, gammas, plot=False).squeeze()
diff = s2 - s1
return diff
# Make plots of t
host = [-2, -0.5, 1]
comp = np.arange(-2.5, 1.5, 0.5)
for i, hfeh in enumerate(host):
comp_mean = []
comp_std = []
for cfeh in comp:
try:
diff = diff_compfeh([6000, 4.5, hfeh], [2500, 4.5, cfeh])
mean, std = stat(diff)
comp_mean.append(mean)
comp_std.append(std)
except:
comp_mean.append(np.nan)
comp_std.append(np.nan)
print(len(comp), len(comp_mean), len(comp_std))
plt.errorbar(comp+offset[i], comp_mean, comp_std , label=hfeh, linestyle=linestyle[i])
plt.legend(title="Companion logg")
plt.ylabel("Flux Diff and std")
plt.xlabel("Host feh")
plt.title("Companion increase by feh=0.5")
plt.show()
for i, hfeh in enumerate(host):
comp_mean = []
comp_std = []
for cfeh in comp:
try:
diff = diff_compfeh([6000, 4.5, hfeh], [2500, 4.5, cfeh])
mean, std = stat(diff)
comp_mean.append(mean)
comp_std.append(std)
except:
comp_mean.append(np.nan)
comp_std.append(np.nan)
print(len(comp), len(comp_mean), len(comp_std))
plt.errorbar(comp, comp_std , label=hfeh, linestyle=linestyle[i])
plt.legend(title="Companion logg")
plt.ylabel("Std")
plt.xlabel("Host feh")
plt.title("Companion increase by feh=0.5")
plt.show()
def diff_hostfeh(params1, params2):
"""Differnece in parameters when param2 teff changes by +100"""
rvs = [0]
gammas = [0]
s1 = combine_spectrum(params1, params2, rvs, gammas, plot=False).squeeze()
params_new = [params1[0], params1[1], params1[2]+0.5]
s2 = combine_spectrum(params_new, params2, rvs, gammas, plot=False).squeeze()
diff = s2 - s1
return diff
# Make plots of t
comp = [-2, -0.5, 1]
host = np.arange(-2.5, 1.5, 0.5)
for i, cfeh in enumerate(comp):
host_mean = []
host_std = []
for hfeh in host:
try:
diff = diff_hostfeh([6000, 4.5, hfeh], [2500, 4.5, cfeh])
mean, std = stat(diff)
host_mean.append(mean)
host_std.append(std)
except:
host_mean.append(np.nan)
host_std.append(np.nan)
print(len(host), len(host_mean), len(host_std))
plt.errorbar(host+offset[i], host_mean, host_std, label=cfeh, linestyle=linestyle[i])
plt.legend(title="Companion logg")
plt.ylabel("Flux Diff and std")
plt.xlabel("Host logg")
plt.title("Host increase by logg=0.5")
plt.show()
for i, cfeh in enumerate(comp):
host_mean = []
host_std = []
for hfeh in host:
try:
diff = diff_hostfeh([6000, 4.5, hfeh], [2500, 4.5, cfeh])
mean, std = stat(diff)
host_mean.append(mean)
host_std.append(std)
except:
host_mean.append(np.nan)
host_std.append(np.nan)
print(len(host), len(host_mean), len(host_std))
plt.errorbar(host, host_std, label=cfeh, linestyle=linestyle[i])
plt.legend(title="Companion feh")
plt.ylabel("Std")
plt.xlabel("Host feh")
plt.title("Host increase by feh=0.5")
plt.show()
mod1_spec, mod2_spec = prepare_iam_model_spectra([5000, 4.5, 0.0], [5000, 5., 0.0], limits=[2010, 2430],
area_scale=True, wav_scale=True)
mod1_spec.plot(label="1")
mod2_spec.plot(label="2")
plt.legend()
plt.show()
| 0.787441 | 0.971402 |
```
pip install facebook-scraper
```
FABRICATIONS ABOUT THE PAP
```
import requests
import json
import os
import pandas as pd
import re
from facebook_scraper import get_posts
FAP_comments=[]
FAP_image=[]
FAP_likes=[]
FAP_postid=[]
FAP_posttext=[]
FAP_posturl=[]
FAP_sharedtext=[]
FAP_shares=[]
FAP_text=[]
FAP_time=[]
for post in get_posts('FabricationsAboutThePAP', pages=50000):
FAP_comments.append(post['comments'])
FAP_image.append(post['image'])
FAP_likes.append(post['likes'])
FAP_postid.append(post['post_id'])
FAP_posttext.append(post['post_text'])
FAP_posturl.append(post['post_url'])
FAP_sharedtext.append(post['shared_text'])
FAP_shares.append(post['shares'])
FAP_text.append(post['text'])
FAP_time.append(post['time'])
print(post['text'][:50])
frames=[pd.DataFrame(FAP_comments),pd.DataFrame(FAP_image),pd.DataFrame(FAP_likes),pd.DataFrame(FAP_postid),pd.DataFrame(FAP_posttext),pd.DataFrame(FAP_posturl),
pd.DataFrame(FAP_sharedtext),pd.DataFrame(FAP_shares),pd.DataFrame(FAP_text),pd.DataFrame(FAP_time)]
FAP_db_all=pd.concat(frames,axis=1)
# Import Drive API and authenticate.
from google.colab import drive
# Mount your Drive to the Colab VM.
drive.mount('/gdrive')
# Write the DataFrame to CSV file.
with open('/gdrive/My Drive/FAP_DB.csv', 'w') as f:
FAP_db_all.to_csv(f)
```
FACTUALLY SINGAPORE
```
from facebook_scraper import get_posts
FA_comments=[]
FA_image=[]
FA_likes=[]
FA_postid=[]
FA_posttext=[]
FA_posturl=[]
FA_sharedtext=[]
FA_shares=[]
FA_text=[]
FA_time=[]
for post in get_posts('factually.sg', pages=50000):
FA_comments.append(post['comments'])
FA_image.append(post['image'])
FA_likes.append(post['likes'])
FA_postid.append(post['post_id'])
FA_posttext.append(post['post_text'])
FA_posturl.append(post['post_url'])
FA_sharedtext.append(post['shared_text'])
FA_shares.append(post['shares'])
FA_text.append(post['text'])
FA_time.append(post['time'])
print(post['text'][:50])
frames=[pd.DataFrame(FA_comments),pd.DataFrame(FA_image),pd.DataFrame(FA_likes),pd.DataFrame(FA_postid),pd.DataFrame(FA_posttext),pd.DataFrame(FA_posturl),
pd.DataFrame(FA_sharedtext),pd.DataFrame(FA_shares),pd.DataFrame(FA_text),pd.DataFrame(FA_time)]
FA_db_all=pd.concat(frames,axis=1)
# Write the DataFrame to CSV file.
with open('/gdrive/My Drive/FA_DB.csv', 'w') as f:
FA_db_all.to_csv(f)
```
Global Times Singapore
```
from facebook_scraper import get_posts
GT_comments=[]
GT_image=[]
GT_likes=[]
GT_postid=[]
GT_posttext=[]
GT_posturl=[]
GT_sharedtext=[]
GT_shares=[]
GT_text=[]
GT_time=[]
for post in get_posts('GlobalTimesSingapore', pages=50000):
GT_comments.append(post['comments'])
GT_image.append(post['image'])
GT_likes.append(post['likes'])
GT_postid.append(post['post_id'])
GT_posttext.append(post['post_text'])
GT_posturl.append(post['post_url'])
GT_sharedtext.append(post['shared_text'])
GT_shares.append(post['shares'])
GT_text.append(post['text'])
GT_time.append(post['time'])
print(post['text'][:50])
frames=[pd.DataFrame(GT_comments),pd.DataFrame(GT_image),pd.DataFrame(GT_likes),pd.DataFrame(GT_postid),pd.DataFrame(GT_posttext),pd.DataFrame(GT_posturl),
pd.DataFrame(GT_sharedtext),pd.DataFrame(GT_shares),pd.DataFrame(GT_text),pd.DataFrame(GT_time)]
GT_db_all=pd.concat(frames,axis=1)
# Write the DataFrame to CSV file.
with open('/gdrive/My Drive/GTS_DB.csv', 'w') as f:
GT_db_all.to_csv(f)
```
FactCheckerSG
```
from facebook_scraper import get_posts
FCheck_comments=[]
FCheck_image=[]
FCheck_likes=[]
FCheck_postid=[]
FCheck_posttext=[]
FCheck_posturl=[]
FCheck_sharedtext=[]
FCheck_shares=[]
FCheck_text=[]
FCheck_time=[]
for post in get_posts('factchecker.sg', pages=50000):
FCheck_comments.append(post['comments'])
FCheck_image.append(post['image'])
FCheck_likes.append(post['likes'])
FCheck_postid.append(post['post_id'])
FCheck_posttext.append(post['post_text'])
FCheck_posturl.append(post['post_url'])
FCheck_sharedtext.append(post['shared_text'])
FCheck_shares.append(post['shares'])
FCheck_text.append(post['text'])
FCheck_time.append(post['time'])
print(post['text'][:50])
frames=[pd.DataFrame(FCheck_comments),pd.DataFrame(FCheck_image),pd.DataFrame(FCheck_likes),pd.DataFrame(FCheck_postid),pd.DataFrame(FCheck_posttext),pd.DataFrame(FCheck_posturl),
pd.DataFrame(FCheck_sharedtext),pd.DataFrame(FCheck_shares),pd.DataFrame(FCheck_text),pd.DataFrame(FCheck_time)]
FCheck_db_all=pd.concat(frames,axis=1)
# Write the DataFrame to CSV file.
with open('/gdrive/My Drive/FChecker_DB.csv', 'w') as f:
FCheck_db_all.to_csv(f)
```
EverydaySG
```
from facebook_scraper import get_posts
everysg_comments=[]
everysg_image=[]
everysg_likes=[]
everysg_postid=[]
everysg_posttext=[]
everysg_posturl=[]
everysg_sharedtext=[]
everysg_shares=[]
everysg_text=[]
everysg_time=[]
for post in get_posts('everydaysg', pages=50000):
everysg_comments.append(post['comments'])
everysg_image.append(post['image'])
everysg_likes.append(post['likes'])
everysg_postid.append(post['post_id'])
everysg_posttext.append(post['post_text'])
everysg_posturl.append(post['post_url'])
everysg_sharedtext.append(post['shared_text'])
everysg_shares.append(post['shares'])
everysg_text.append(post['text'])
everysg_time.append(post['time'])
print(post['text'][:50])
frames=[pd.DataFrame(everysg_comments),pd.DataFrame(everysg_image),pd.DataFrame(everysg_likes),pd.DataFrame(everysg_postid),pd.DataFrame(everysg_posttext),pd.DataFrame(everysg_posturl),
pd.DataFrame(everysg_sharedtext),pd.DataFrame(everysg_shares),pd.DataFrame(everysg_text),pd.DataFrame(everysg_time)]
everysg_db_all=pd.concat(frames,axis=1)
# Write the DataFrame to CSV file.
with open('/gdrive/My Drive/everydaysg_DB.csv', 'w') as f:
everysg_db_all.to_csv(f)
```
SGMatters
```
from facebook_scraper import get_posts
sgmatters_comments=[]
sgmatters_image=[]
sgmatters_likes=[]
sgmatters_postid=[]
sgmatters_posttext=[]
sgmatters_posturl=[]
sgmatters_sharedtext=[]
sgmatters_shares=[]
sgmatters_text=[]
sgmatters_time=[]
for post in get_posts('SingaporeMatters', pages=50000):
sgmatters_comments.append(post['comments'])
sgmatters_image.append(post['image'])
sgmatters_likes.append(post['likes'])
sgmatters_postid.append(post['post_id'])
sgmatters_posttext.append(post['post_text'])
sgmatters_posturl.append(post['post_url'])
sgmatters_sharedtext.append(post['shared_text'])
sgmatters_shares.append(post['shares'])
sgmatters_text.append(post['text'])
sgmatters_time.append(post['time'])
print(post['text'][:50])
frames=[pd.DataFrame(sgmatters_comments),pd.DataFrame(sgmatters_image),pd.DataFrame(sgmatters_likes),pd.DataFrame(sgmatters_postid),pd.DataFrame(sgmatters_posttext),pd.DataFrame(sgmatters_posturl),
pd.DataFrame(sgmatters_sharedtext),pd.DataFrame(sgmatters_shares),pd.DataFrame(sgmatters_text),pd.DataFrame(sgmatters_time)]
sgmatters_db_all=pd.concat(frames,axis=1)
# Write the DataFrame to CSV file.
with open('/gdrive/My Drive/sgmatters_DB.csv', 'w') as f:
sgmatters_db_all.to_csv(f)
from facebook_scraper import get_posts
TOC_comments=[]
TOC_image=[]
TOC_likes=[]
TOC_postid=[]
TOC_posttext=[]
TOC_posturl=[]
TOC_sharedtext=[]
TOC_shares=[]
TOC_text=[]
TOC_time=[]
for post in get_posts('theonlinecitizen', pages=50000):
TOC_comments.append(post['comments'])
TOC_image.append(post['image'])
TOC_likes.append(post['likes'])
TOC_postid.append(post['post_id'])
TOC_posttext.append(post['post_text'])
TOC_posturl.append(post['post_url'])
TOC_sharedtext.append(post['shared_text'])
TOC_shares.append(post['shares'])
TOC_text.append(post['text'])
TOC_time.append(post['time'])
print(post['text'][:50])
frames=[pd.DataFrame(TOC_comments),pd.DataFrame(TOC_image),pd.DataFrame(TOC_likes),pd.DataFrame(TOC_postid),pd.DataFrame(TOC_posttext),pd.DataFrame(TOC_posturl),
pd.DataFrame(TOC_sharedtext),pd.DataFrame(TOC_shares),pd.DataFrame(TOC_text),pd.DataFrame(TOC_time)]
TOC_db_all=pd.concat(frames,axis=1)
# Write the DataFrame to CSV file.
with open('/gdrive/My Drive/TOC_DB.csv', 'w') as f:
TOC_db_all.to_csv(f)
```
WAKE UP SG
```
from facebook_scraper import get_posts
WUSG_comments=[]
WUSG_image=[]
WUSG_likes=[]
WUSG_postid=[]
WUSG_posttext=[]
WUSG_posturl=[]
WUSG_sharedtext=[]
WUSG_shares=[]
WUSG_text=[]
WUSG_time=[]
for post in get_posts('wakeupSG', pages=50000):
WUSG_comments.append(post['comments'])
WUSG_image.append(post['image'])
WUSG_likes.append(post['likes'])
WUSG_postid.append(post['post_id'])
WUSG_posttext.append(post['post_text'])
WUSG_posturl.append(post['post_url'])
WUSG_sharedtext.append(post['shared_text'])
WUSG_shares.append(post['shares'])
WUSG_text.append(post['text'])
WUSG_time.append(post['time'])
print(post['text'][:50])
frames=[pd.DataFrame(WUSG_comments),pd.DataFrame(WUSG_image),pd.DataFrame(WUSG_likes),pd.DataFrame(WUSG_postid),pd.DataFrame(WUSG_posttext),pd.DataFrame(WUSG_posturl),
pd.DataFrame(WUSG_sharedtext),pd.DataFrame(WUSG_shares),pd.DataFrame(WUSG_text),pd.DataFrame(WUSG_time)]
WUSG_db_all=pd.concat(frames,axis=1)
# Write the DataFrame to CSV file.
with open('/gdrive/My Drive/WUSG_DB.csv', 'w') as f:
WUSG_db_all.to_csv(f)
from facebook_scraper import get_posts
TISG_comments=[]
TISG_image=[]
TISG_likes=[]
TISG_postid=[]
TISG_posttext=[]
TISG_posturl=[]
TISG_sharedtext=[]
TISG_shares=[]
TISG_text=[]
TISG_time=[]
for post in get_posts("TheIndependentSG", pages=50000):
TISG_comments.append(post['comments'])
TISG_image.append(post['image'])
TISG_likes.append(post['likes'])
TISG_postid.append(post['post_id'])
TISG_posttext.append(post['post_text'])
TISG_posturl.append(post['post_url'])
TISG_sharedtext.append(post['shared_text'])
TISG_shares.append(post['shares'])
TISG_text.append(post['text'])
TISG_time.append(post['time'])
print(post['text'][:50])
frames=[pd.DataFrame(TISG_comments),pd.DataFrame(TISG_image),pd.DataFrame(TISG_likes),pd.DataFrame(TISG_postid),pd.DataFrame(TISG_posttext),pd.DataFrame(TISG_posturl),
pd.DataFrame(TISG_sharedtext),pd.DataFrame(TISG_shares),pd.DataFrame(TISG_text),pd.DataFrame(TISG_time)]
TISG_db_all=pd.concat(frames,axis=1)
# Write the DataFrame to CSV file.
with open('/gdrive/My Drive/TISG_DB.csv', 'w') as f:
TISG_db_all.to_csv(f)
from facebook_scraper import get_posts
STRU_comments=[]
STRU_image=[]
STRU_likes=[]
STRU_postid=[]
STRU_posttext=[]
STRU_posturl=[]
STRU_sharedtext=[]
STRU_shares=[]
STRU_text=[]
STRU_time=[]
for post in get_posts("STReviewUncensored", pages=50000):
STRU_comments.append(post['comments'])
STRU_image.append(post['image'])
STRU_likes.append(post['likes'])
STRU_postid.append(post['post_id'])
STRU_posttext.append(post['post_text'])
STRU_posturl.append(post['post_url'])
STRU_sharedtext.append(post['shared_text'])
STRU_shares.append(post['shares'])
STRU_text.append(post['text'])
STRU_time.append(post['time'])
print(post['text'][:50])
frames=[pd.DataFrame(STRU_comments),pd.DataFrame(STRU_image),pd.DataFrame(STRU_likes),pd.DataFrame(STRU_postid),pd.DataFrame(STRU_posttext),pd.DataFrame(STRU_posturl),
pd.DataFrame(STRU_sharedtext),pd.DataFrame(STRU_shares),pd.DataFrame(STRU_text),pd.DataFrame(STRU_time)]
STRU_db_all=pd.concat(frames,axis=1)
# Write the DataFrame to CSV file.
with open('/gdrive/My Drive/STRU_DB.csv', 'w') as f:
STRU_db_all.to_csv(f)
from facebook_scraper import get_posts
DHRS_comments=[]
DHRS_image=[]
DHRS_likes=[]
DHRS_postid=[]
DHRS_posttext=[]
DHRS_posturl=[]
DHRS_sharedtext=[]
DHRS_shares=[]
DHRS_text=[]
DHRS_time=[]
for post in get_posts("Democracy-and-Human-Rights-for-Singapore-2529543017080086", pages=50000):
DHRS_comments.append(post['comments'])
DHRS_image.append(post['image'])
DHRS_likes.append(post['likes'])
DHRS_postid.append(post['post_id'])
DHRS_posttext.append(post['post_text'])
DHRS_posturl.append(post['post_url'])
DHRS_sharedtext.append(post['shared_text'])
DHRS_shares.append(post['shares'])
DHRS_text.append(post['text'])
DHRS_time.append(post['time'])
print(post['text'][:50])
frames=[pd.DataFrame(DHRS_comments),pd.DataFrame(DHRS_image),pd.DataFrame(DHRS_likes),pd.DataFrame(DHRS_postid),pd.DataFrame(DHRS_posttext),pd.DataFrame(DHRS_posturl),
pd.DataFrame(DHRS_sharedtext),pd.DataFrame(DHRS_shares),pd.DataFrame(DHRS_text),pd.DataFrame(DHRS_time)]
DHRS_db_all=pd.concat(frames,axis=1)
# Write the DataFrame to CSV file.
with open('/gdrive/My Drive/DHRS_DB.csv', 'w') as f:
DHRS_db_all.to_csv(f)
from facebook_scraper import get_posts
SNSG_comments=[]
SNSG_image=[]
SNSG_likes=[]
SNSG_postid=[]
SNSG_posttext=[]
SNSG_posturl=[]
SNSG_sharedtext=[]
SNSG_shares=[]
SNSG_text=[]
SNSG_time=[]
for post in get_posts("State-News-Singapore-108794737524065", pages=50000):
SNSG_comments.append(post['comments'])
SNSG_image.append(post['image'])
SNSG_likes.append(post['likes'])
SNSG_postid.append(post['post_id'])
SNSG_posttext.append(post['post_text'])
SNSG_posturl.append(post['post_url'])
SNSG_sharedtext.append(post['shared_text'])
SNSG_shares.append(post['shares'])
SNSG_text.append(post['text'])
SNSG_time.append(post['time'])
print(post['text'][:50])
frames=[pd.DataFrame(SNSG_comments),pd.DataFrame(SNSG_image),pd.DataFrame(SNSG_likes),pd.DataFrame(SNSG_postid),pd.DataFrame(SNSG_posttext),pd.DataFrame(SNSG_posturl),
pd.DataFrame(SNSG_sharedtext),pd.DataFrame(SNSG_shares),pd.DataFrame(SNSG_text),pd.DataFrame(SNSG_time)]
SNSG_db_all=pd.concat(frames,axis=1)
# Write the DataFrame to CSV file.
with open('/gdrive/My Drive/SNSG_DB.csv', 'w') as f:
SNSG_db_all.to_csv(f)
from facebook_scraper import get_posts
FBP_comments=[]
FBP_image=[]
FBP_likes=[]
FBP_postid=[]
FBP_posttext=[]
FBP_posturl=[]
FBP_sharedtext=[]
FBP_shares=[]
FBP_text=[]
FBP_time=[]
for post in get_posts("FabricationsByThePAP", pages=50000):
FBP_comments.append(post['comments'])
FBP_image.append(post['image'])
FBP_likes.append(post['likes'])
FBP_postid.append(post['post_id'])
FBP_posttext.append(post['post_text'])
FBP_posturl.append(post['post_url'])
FBP_sharedtext.append(post['shared_text'])
FBP_shares.append(post['shares'])
FBP_text.append(post['text'])
FBP_time.append(post['time'])
print(post['text'][:50])
frames=[pd.DataFrame(FBP_comments),pd.DataFrame(FBP_image),pd.DataFrame(FBP_likes),pd.DataFrame(FBP_postid),pd.DataFrame(FBP_posttext),pd.DataFrame(FBP_posturl),
pd.DataFrame(FBP_sharedtext),pd.DataFrame(FBP_shares),pd.DataFrame(FBP_text),pd.DataFrame(FBP_time)]
FBP_db_all=pd.concat(frames,axis=1)
# Write the DataFrame to CSV file.
with open('/gdrive/My Drive/FBP_DB.csv', 'w') as f:
FBP_db_all.to_csv(f)
```
|
github_jupyter
|
pip install facebook-scraper
import requests
import json
import os
import pandas as pd
import re
from facebook_scraper import get_posts
FAP_comments=[]
FAP_image=[]
FAP_likes=[]
FAP_postid=[]
FAP_posttext=[]
FAP_posturl=[]
FAP_sharedtext=[]
FAP_shares=[]
FAP_text=[]
FAP_time=[]
for post in get_posts('FabricationsAboutThePAP', pages=50000):
FAP_comments.append(post['comments'])
FAP_image.append(post['image'])
FAP_likes.append(post['likes'])
FAP_postid.append(post['post_id'])
FAP_posttext.append(post['post_text'])
FAP_posturl.append(post['post_url'])
FAP_sharedtext.append(post['shared_text'])
FAP_shares.append(post['shares'])
FAP_text.append(post['text'])
FAP_time.append(post['time'])
print(post['text'][:50])
frames=[pd.DataFrame(FAP_comments),pd.DataFrame(FAP_image),pd.DataFrame(FAP_likes),pd.DataFrame(FAP_postid),pd.DataFrame(FAP_posttext),pd.DataFrame(FAP_posturl),
pd.DataFrame(FAP_sharedtext),pd.DataFrame(FAP_shares),pd.DataFrame(FAP_text),pd.DataFrame(FAP_time)]
FAP_db_all=pd.concat(frames,axis=1)
# Import Drive API and authenticate.
from google.colab import drive
# Mount your Drive to the Colab VM.
drive.mount('/gdrive')
# Write the DataFrame to CSV file.
with open('/gdrive/My Drive/FAP_DB.csv', 'w') as f:
FAP_db_all.to_csv(f)
from facebook_scraper import get_posts
FA_comments=[]
FA_image=[]
FA_likes=[]
FA_postid=[]
FA_posttext=[]
FA_posturl=[]
FA_sharedtext=[]
FA_shares=[]
FA_text=[]
FA_time=[]
for post in get_posts('factually.sg', pages=50000):
FA_comments.append(post['comments'])
FA_image.append(post['image'])
FA_likes.append(post['likes'])
FA_postid.append(post['post_id'])
FA_posttext.append(post['post_text'])
FA_posturl.append(post['post_url'])
FA_sharedtext.append(post['shared_text'])
FA_shares.append(post['shares'])
FA_text.append(post['text'])
FA_time.append(post['time'])
print(post['text'][:50])
frames=[pd.DataFrame(FA_comments),pd.DataFrame(FA_image),pd.DataFrame(FA_likes),pd.DataFrame(FA_postid),pd.DataFrame(FA_posttext),pd.DataFrame(FA_posturl),
pd.DataFrame(FA_sharedtext),pd.DataFrame(FA_shares),pd.DataFrame(FA_text),pd.DataFrame(FA_time)]
FA_db_all=pd.concat(frames,axis=1)
# Write the DataFrame to CSV file.
with open('/gdrive/My Drive/FA_DB.csv', 'w') as f:
FA_db_all.to_csv(f)
from facebook_scraper import get_posts
GT_comments=[]
GT_image=[]
GT_likes=[]
GT_postid=[]
GT_posttext=[]
GT_posturl=[]
GT_sharedtext=[]
GT_shares=[]
GT_text=[]
GT_time=[]
for post in get_posts('GlobalTimesSingapore', pages=50000):
GT_comments.append(post['comments'])
GT_image.append(post['image'])
GT_likes.append(post['likes'])
GT_postid.append(post['post_id'])
GT_posttext.append(post['post_text'])
GT_posturl.append(post['post_url'])
GT_sharedtext.append(post['shared_text'])
GT_shares.append(post['shares'])
GT_text.append(post['text'])
GT_time.append(post['time'])
print(post['text'][:50])
frames=[pd.DataFrame(GT_comments),pd.DataFrame(GT_image),pd.DataFrame(GT_likes),pd.DataFrame(GT_postid),pd.DataFrame(GT_posttext),pd.DataFrame(GT_posturl),
pd.DataFrame(GT_sharedtext),pd.DataFrame(GT_shares),pd.DataFrame(GT_text),pd.DataFrame(GT_time)]
GT_db_all=pd.concat(frames,axis=1)
# Write the DataFrame to CSV file.
with open('/gdrive/My Drive/GTS_DB.csv', 'w') as f:
GT_db_all.to_csv(f)
from facebook_scraper import get_posts
FCheck_comments=[]
FCheck_image=[]
FCheck_likes=[]
FCheck_postid=[]
FCheck_posttext=[]
FCheck_posturl=[]
FCheck_sharedtext=[]
FCheck_shares=[]
FCheck_text=[]
FCheck_time=[]
for post in get_posts('factchecker.sg', pages=50000):
FCheck_comments.append(post['comments'])
FCheck_image.append(post['image'])
FCheck_likes.append(post['likes'])
FCheck_postid.append(post['post_id'])
FCheck_posttext.append(post['post_text'])
FCheck_posturl.append(post['post_url'])
FCheck_sharedtext.append(post['shared_text'])
FCheck_shares.append(post['shares'])
FCheck_text.append(post['text'])
FCheck_time.append(post['time'])
print(post['text'][:50])
frames=[pd.DataFrame(FCheck_comments),pd.DataFrame(FCheck_image),pd.DataFrame(FCheck_likes),pd.DataFrame(FCheck_postid),pd.DataFrame(FCheck_posttext),pd.DataFrame(FCheck_posturl),
pd.DataFrame(FCheck_sharedtext),pd.DataFrame(FCheck_shares),pd.DataFrame(FCheck_text),pd.DataFrame(FCheck_time)]
FCheck_db_all=pd.concat(frames,axis=1)
# Write the DataFrame to CSV file.
with open('/gdrive/My Drive/FChecker_DB.csv', 'w') as f:
FCheck_db_all.to_csv(f)
from facebook_scraper import get_posts
everysg_comments=[]
everysg_image=[]
everysg_likes=[]
everysg_postid=[]
everysg_posttext=[]
everysg_posturl=[]
everysg_sharedtext=[]
everysg_shares=[]
everysg_text=[]
everysg_time=[]
for post in get_posts('everydaysg', pages=50000):
everysg_comments.append(post['comments'])
everysg_image.append(post['image'])
everysg_likes.append(post['likes'])
everysg_postid.append(post['post_id'])
everysg_posttext.append(post['post_text'])
everysg_posturl.append(post['post_url'])
everysg_sharedtext.append(post['shared_text'])
everysg_shares.append(post['shares'])
everysg_text.append(post['text'])
everysg_time.append(post['time'])
print(post['text'][:50])
frames=[pd.DataFrame(everysg_comments),pd.DataFrame(everysg_image),pd.DataFrame(everysg_likes),pd.DataFrame(everysg_postid),pd.DataFrame(everysg_posttext),pd.DataFrame(everysg_posturl),
pd.DataFrame(everysg_sharedtext),pd.DataFrame(everysg_shares),pd.DataFrame(everysg_text),pd.DataFrame(everysg_time)]
everysg_db_all=pd.concat(frames,axis=1)
# Write the DataFrame to CSV file.
with open('/gdrive/My Drive/everydaysg_DB.csv', 'w') as f:
everysg_db_all.to_csv(f)
from facebook_scraper import get_posts
sgmatters_comments=[]
sgmatters_image=[]
sgmatters_likes=[]
sgmatters_postid=[]
sgmatters_posttext=[]
sgmatters_posturl=[]
sgmatters_sharedtext=[]
sgmatters_shares=[]
sgmatters_text=[]
sgmatters_time=[]
for post in get_posts('SingaporeMatters', pages=50000):
sgmatters_comments.append(post['comments'])
sgmatters_image.append(post['image'])
sgmatters_likes.append(post['likes'])
sgmatters_postid.append(post['post_id'])
sgmatters_posttext.append(post['post_text'])
sgmatters_posturl.append(post['post_url'])
sgmatters_sharedtext.append(post['shared_text'])
sgmatters_shares.append(post['shares'])
sgmatters_text.append(post['text'])
sgmatters_time.append(post['time'])
print(post['text'][:50])
frames=[pd.DataFrame(sgmatters_comments),pd.DataFrame(sgmatters_image),pd.DataFrame(sgmatters_likes),pd.DataFrame(sgmatters_postid),pd.DataFrame(sgmatters_posttext),pd.DataFrame(sgmatters_posturl),
pd.DataFrame(sgmatters_sharedtext),pd.DataFrame(sgmatters_shares),pd.DataFrame(sgmatters_text),pd.DataFrame(sgmatters_time)]
sgmatters_db_all=pd.concat(frames,axis=1)
# Write the DataFrame to CSV file.
with open('/gdrive/My Drive/sgmatters_DB.csv', 'w') as f:
sgmatters_db_all.to_csv(f)
from facebook_scraper import get_posts
TOC_comments=[]
TOC_image=[]
TOC_likes=[]
TOC_postid=[]
TOC_posttext=[]
TOC_posturl=[]
TOC_sharedtext=[]
TOC_shares=[]
TOC_text=[]
TOC_time=[]
for post in get_posts('theonlinecitizen', pages=50000):
TOC_comments.append(post['comments'])
TOC_image.append(post['image'])
TOC_likes.append(post['likes'])
TOC_postid.append(post['post_id'])
TOC_posttext.append(post['post_text'])
TOC_posturl.append(post['post_url'])
TOC_sharedtext.append(post['shared_text'])
TOC_shares.append(post['shares'])
TOC_text.append(post['text'])
TOC_time.append(post['time'])
print(post['text'][:50])
frames=[pd.DataFrame(TOC_comments),pd.DataFrame(TOC_image),pd.DataFrame(TOC_likes),pd.DataFrame(TOC_postid),pd.DataFrame(TOC_posttext),pd.DataFrame(TOC_posturl),
pd.DataFrame(TOC_sharedtext),pd.DataFrame(TOC_shares),pd.DataFrame(TOC_text),pd.DataFrame(TOC_time)]
TOC_db_all=pd.concat(frames,axis=1)
# Write the DataFrame to CSV file.
with open('/gdrive/My Drive/TOC_DB.csv', 'w') as f:
TOC_db_all.to_csv(f)
from facebook_scraper import get_posts
WUSG_comments=[]
WUSG_image=[]
WUSG_likes=[]
WUSG_postid=[]
WUSG_posttext=[]
WUSG_posturl=[]
WUSG_sharedtext=[]
WUSG_shares=[]
WUSG_text=[]
WUSG_time=[]
for post in get_posts('wakeupSG', pages=50000):
WUSG_comments.append(post['comments'])
WUSG_image.append(post['image'])
WUSG_likes.append(post['likes'])
WUSG_postid.append(post['post_id'])
WUSG_posttext.append(post['post_text'])
WUSG_posturl.append(post['post_url'])
WUSG_sharedtext.append(post['shared_text'])
WUSG_shares.append(post['shares'])
WUSG_text.append(post['text'])
WUSG_time.append(post['time'])
print(post['text'][:50])
frames=[pd.DataFrame(WUSG_comments),pd.DataFrame(WUSG_image),pd.DataFrame(WUSG_likes),pd.DataFrame(WUSG_postid),pd.DataFrame(WUSG_posttext),pd.DataFrame(WUSG_posturl),
pd.DataFrame(WUSG_sharedtext),pd.DataFrame(WUSG_shares),pd.DataFrame(WUSG_text),pd.DataFrame(WUSG_time)]
WUSG_db_all=pd.concat(frames,axis=1)
# Write the DataFrame to CSV file.
with open('/gdrive/My Drive/WUSG_DB.csv', 'w') as f:
WUSG_db_all.to_csv(f)
from facebook_scraper import get_posts
TISG_comments=[]
TISG_image=[]
TISG_likes=[]
TISG_postid=[]
TISG_posttext=[]
TISG_posturl=[]
TISG_sharedtext=[]
TISG_shares=[]
TISG_text=[]
TISG_time=[]
for post in get_posts("TheIndependentSG", pages=50000):
TISG_comments.append(post['comments'])
TISG_image.append(post['image'])
TISG_likes.append(post['likes'])
TISG_postid.append(post['post_id'])
TISG_posttext.append(post['post_text'])
TISG_posturl.append(post['post_url'])
TISG_sharedtext.append(post['shared_text'])
TISG_shares.append(post['shares'])
TISG_text.append(post['text'])
TISG_time.append(post['time'])
print(post['text'][:50])
frames=[pd.DataFrame(TISG_comments),pd.DataFrame(TISG_image),pd.DataFrame(TISG_likes),pd.DataFrame(TISG_postid),pd.DataFrame(TISG_posttext),pd.DataFrame(TISG_posturl),
pd.DataFrame(TISG_sharedtext),pd.DataFrame(TISG_shares),pd.DataFrame(TISG_text),pd.DataFrame(TISG_time)]
TISG_db_all=pd.concat(frames,axis=1)
# Write the DataFrame to CSV file.
with open('/gdrive/My Drive/TISG_DB.csv', 'w') as f:
TISG_db_all.to_csv(f)
from facebook_scraper import get_posts
STRU_comments=[]
STRU_image=[]
STRU_likes=[]
STRU_postid=[]
STRU_posttext=[]
STRU_posturl=[]
STRU_sharedtext=[]
STRU_shares=[]
STRU_text=[]
STRU_time=[]
for post in get_posts("STReviewUncensored", pages=50000):
STRU_comments.append(post['comments'])
STRU_image.append(post['image'])
STRU_likes.append(post['likes'])
STRU_postid.append(post['post_id'])
STRU_posttext.append(post['post_text'])
STRU_posturl.append(post['post_url'])
STRU_sharedtext.append(post['shared_text'])
STRU_shares.append(post['shares'])
STRU_text.append(post['text'])
STRU_time.append(post['time'])
print(post['text'][:50])
frames=[pd.DataFrame(STRU_comments),pd.DataFrame(STRU_image),pd.DataFrame(STRU_likes),pd.DataFrame(STRU_postid),pd.DataFrame(STRU_posttext),pd.DataFrame(STRU_posturl),
pd.DataFrame(STRU_sharedtext),pd.DataFrame(STRU_shares),pd.DataFrame(STRU_text),pd.DataFrame(STRU_time)]
STRU_db_all=pd.concat(frames,axis=1)
# Write the DataFrame to CSV file.
with open('/gdrive/My Drive/STRU_DB.csv', 'w') as f:
STRU_db_all.to_csv(f)
from facebook_scraper import get_posts
DHRS_comments=[]
DHRS_image=[]
DHRS_likes=[]
DHRS_postid=[]
DHRS_posttext=[]
DHRS_posturl=[]
DHRS_sharedtext=[]
DHRS_shares=[]
DHRS_text=[]
DHRS_time=[]
for post in get_posts("Democracy-and-Human-Rights-for-Singapore-2529543017080086", pages=50000):
DHRS_comments.append(post['comments'])
DHRS_image.append(post['image'])
DHRS_likes.append(post['likes'])
DHRS_postid.append(post['post_id'])
DHRS_posttext.append(post['post_text'])
DHRS_posturl.append(post['post_url'])
DHRS_sharedtext.append(post['shared_text'])
DHRS_shares.append(post['shares'])
DHRS_text.append(post['text'])
DHRS_time.append(post['time'])
print(post['text'][:50])
frames=[pd.DataFrame(DHRS_comments),pd.DataFrame(DHRS_image),pd.DataFrame(DHRS_likes),pd.DataFrame(DHRS_postid),pd.DataFrame(DHRS_posttext),pd.DataFrame(DHRS_posturl),
pd.DataFrame(DHRS_sharedtext),pd.DataFrame(DHRS_shares),pd.DataFrame(DHRS_text),pd.DataFrame(DHRS_time)]
DHRS_db_all=pd.concat(frames,axis=1)
# Write the DataFrame to CSV file.
with open('/gdrive/My Drive/DHRS_DB.csv', 'w') as f:
DHRS_db_all.to_csv(f)
from facebook_scraper import get_posts
SNSG_comments=[]
SNSG_image=[]
SNSG_likes=[]
SNSG_postid=[]
SNSG_posttext=[]
SNSG_posturl=[]
SNSG_sharedtext=[]
SNSG_shares=[]
SNSG_text=[]
SNSG_time=[]
for post in get_posts("State-News-Singapore-108794737524065", pages=50000):
SNSG_comments.append(post['comments'])
SNSG_image.append(post['image'])
SNSG_likes.append(post['likes'])
SNSG_postid.append(post['post_id'])
SNSG_posttext.append(post['post_text'])
SNSG_posturl.append(post['post_url'])
SNSG_sharedtext.append(post['shared_text'])
SNSG_shares.append(post['shares'])
SNSG_text.append(post['text'])
SNSG_time.append(post['time'])
print(post['text'][:50])
frames=[pd.DataFrame(SNSG_comments),pd.DataFrame(SNSG_image),pd.DataFrame(SNSG_likes),pd.DataFrame(SNSG_postid),pd.DataFrame(SNSG_posttext),pd.DataFrame(SNSG_posturl),
pd.DataFrame(SNSG_sharedtext),pd.DataFrame(SNSG_shares),pd.DataFrame(SNSG_text),pd.DataFrame(SNSG_time)]
SNSG_db_all=pd.concat(frames,axis=1)
# Write the DataFrame to CSV file.
with open('/gdrive/My Drive/SNSG_DB.csv', 'w') as f:
SNSG_db_all.to_csv(f)
from facebook_scraper import get_posts
FBP_comments=[]
FBP_image=[]
FBP_likes=[]
FBP_postid=[]
FBP_posttext=[]
FBP_posturl=[]
FBP_sharedtext=[]
FBP_shares=[]
FBP_text=[]
FBP_time=[]
for post in get_posts("FabricationsByThePAP", pages=50000):
FBP_comments.append(post['comments'])
FBP_image.append(post['image'])
FBP_likes.append(post['likes'])
FBP_postid.append(post['post_id'])
FBP_posttext.append(post['post_text'])
FBP_posturl.append(post['post_url'])
FBP_sharedtext.append(post['shared_text'])
FBP_shares.append(post['shares'])
FBP_text.append(post['text'])
FBP_time.append(post['time'])
print(post['text'][:50])
frames=[pd.DataFrame(FBP_comments),pd.DataFrame(FBP_image),pd.DataFrame(FBP_likes),pd.DataFrame(FBP_postid),pd.DataFrame(FBP_posttext),pd.DataFrame(FBP_posturl),
pd.DataFrame(FBP_sharedtext),pd.DataFrame(FBP_shares),pd.DataFrame(FBP_text),pd.DataFrame(FBP_time)]
FBP_db_all=pd.concat(frames,axis=1)
# Write the DataFrame to CSV file.
with open('/gdrive/My Drive/FBP_DB.csv', 'w') as f:
FBP_db_all.to_csv(f)
| 0.168686 | 0.33089 |
In [The Mean as Predictor](mean_meaning), we found that the mean had some good properties as a single best predictor for a whole distribution.
* The mean gives a total prediction error of zero. Put otherwise, on
average, your prediction error is zero.
* The mean gives the lowest squared error. Put otherwise, the mean
gives the lowest average squared difference from the observed value.
Now we can consider what predictor we should use when predicting one set of values, from a different set of values.
We load our usual libraries.
```
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
# Make plots look a little bit more fancy
plt.style.use('fivethirtyeight')
# Print to 2 decimal places, show tiny values as 0
np.set_printoptions(precision=2, suppress=True)
import pandas as pd
```
Again, we use the gender data that we first saw in the [data frame introduction](../04/data_frame_intro).
```
# Load the data file
gender_data = pd.read_csv('gender_stats.csv')
```
We have already seen the data for the Maternal Mortality Ratio
`mat_mort_ratio`. Now we will look to see whether we can predict the Maternal Mortality Ratio (MMR) with the Fertility Rate. The Fertility Rate (FR) is the total number of births per woman. We predict that countries with many births per woman will also tend to have high Maternal Mortality Ratios. Fertility Rate is `fert_rate` in the `gender_data` data frame.
```
# Show the column names
gender_data.columns
```
To start, we make a new data frame that contains only the columns corresponding to MMR and FR. To get two columns out of a data frame, we pass a list of column names inside square brackets:
```
# Make new data frame with only MMR and FR
just_mmr_fr = gender_data[['mat_mort_ratio', 'fert_rate']]
```
To save space, we only show the first five rows of the data frame, using the `head` method of the data frame.
```
# Show the first five rows
just_mmr_fr.head()
```
Looking at these values, we see that there are missing values: `NaN`. These will make our life harder. For now, we drop all rows that have any missing values, using the `dropna` method of the data frame.
```
# Drop all rows with any NaN values
clean_mmr_fr = just_mmr_fr.dropna()
clean_mmr_fr.head()
```
We fetch out the columns of MMR and FR data. These are each Pandas Series:
```
mmr_series = clean_mmr_fr['mat_mort_ratio']
fert_series = clean_mmr_fr['fert_rate']
type(fert_series)
```
The last step in our data munging is to convert the columns of MMR and FR data into arrays, to make them simpler to work with. We do this with the Numpy `array` function, that makes arrays from many other types of object.
```
mmr = np.array(mmr_series)
fert = np.array(fert_series)
```
## Looking for straight lines
Now we are ready to look for a relationship between MMR and FR.
As usual, we start with a plot. This time, for fun, we add a label to the X and Y axes with `xlabel` and `ylabel`.
```
# Plot FR on the x axis, MMR on the y axis
plt.plot(fert, mmr, 'o')
plt.xlabel('Fertility rate')
plt.ylabel('Maternal mortality ratio')
```
The `'o'` argument to the plot function above is a "plot marker". It tells Matplotlib to plot the points as points, rather than joining them with lines. The markers for the points will be filled circles, with `'o'`, but we can also ask for other symbols such as plus marks (with `'+'`) and crosses (with `'x'`).
## Putting points on plots
Before we go on, we will need some machinery to plot arbitrary points on plots.
In fact this works in exactly the same way as the points you have already seen on plots. We use the `plot` function, with a suitable plot marker. The x coordinates of the points go in the first argument, and the y coordinates go in the second.
To plot a single point, pass a single x and y coordinate value:
```
plt.plot(fert, mmr, 'o')
# A green point at x=2, y=1000
plt.plot(2, 1000, 'o', color='green')
```
To plot more than one point, pass multiple x and y coordinate values:
```
plt.plot(fert, mmr, 'o')
# Two green points, one at [2, 1000], the other at [3, 1200]
plt.plot([2, 3], [1000, 1200], 'o', color='green')
```
## The mean as applied to plots
We want a straight line that fits these points.
The straight line should do the best job it can in *predicting* the MMR values from the FP values.
We found that the mean was a good predictor. We could try and find a line or something similar that went through the mean of the MMR values, at any given FR value.
Let's split the FR values up into bins centered on 1.5, 2.5 and so on. Then we take the mean of all the MMR values corresponding to FR values between 1 and 2, 2 and 3 and so on.
```
# The centers for our FR bins
fert_bin_centers = np.arange(7) + 1.5
fert_bin_centers
# The number of bins
n_bins = len(fert_bin_centers)
n_bins
```
Show the center of the bins on the x axis of the plot.
```
plt.plot(fert, mmr, 'o')
plt.plot(fert_bin_centers, np.zeros(n_bins), 'o', color='green')
```
Take the mean of the MMR values for each bin.
```
mmr_means = np.zeros(n_bins)
for i in np.arange(n_bins):
mid = fert_bin_centers[i]
# Boolean identifing indices withing the FR bin
fr_within_bin = (fert >= mid - 0.5) & (fert < mid + 0.5)
# Take the mean of the corresponding MMR values
mmr_means[i] = np.mean(mmr[fr_within_bin])
mmr_means
```
These means should be good predictors for MMR values, given an FR value. We check the bin of the FR value and take the corresponding MMR mean as the prediction.
Here is a plot of the means of MMR for every bin:
```
plt.plot(fert, mmr, 'o')
plt.plot(fert_bin_centers, mmr_means, 'o', color='green')
```
## Finding a predicting line
The means per bin give some prediction of the MMR values from the FR. Can we do better? Can we find a line that predicts the MMR data from the FR data?
Remember, any line can be fully described by an *intercept* and a *slope*. A line predicts the $y$ values from the $x$ values, using the slope $s$ and the intercept $I$:
$$
y = I + x * s
$$
The *intercept* is the value of the line when x is equal to 0. It is therefore where the line crosses the y axis.
In our case, let us assume the intercept is 0. We will assume MMR of 0 if there are no births. We will see that things are more complicated than that, but we will start there.
Now we want to find a good *slope*. The *slope* is the amount that the y values increase for a one unit increase in the x values. In our case, it is the increase in the MMR for a 1 child increase in the FR.
Let's guess the slope is 100.
```
slope = 100
```
Remember our line prediction for y (MMR) is:
$$
y = I + x * s
$$
where x is the FR. In our case we assume the intercept is 0, so:
```
mmr_predicted = fert * slope
```
Plot the predictions in red on the original data in blue.
```
plt.plot(fert, mmr, 'o')
plt.plot(fert, mmr_predicted, 'o', color='red')
```
The red are the predictions, the blue are the original data. At each MMR value we have a prediction, and therefore, an error in our prediction; the difference between the predicted value and the actual values.
```
error = mmr - mmr_predicted
error[:10]
```
In this plot, for each point, we draw a thin dotted line between the prediction of MMR for each point, and its actual value.
```
plt.plot(fert, mmr, 'o')
plt.plot(fert, mmr_predicted, 'o', color='red')
# Draw a line between predicted and actual
for i in np.arange(len(fert)):
x = fert[i]
y_0 = mmr_predicted[i]
y_1 = mmr[i]
plt.plot([x, x], [y_0, y_1], ':', color='black', linewidth=1)
```
## What is a good line?
We have guessed a slope, and so defined a line. We calculated the errors from our guessed line.
How would we decide whether our slope was a good one? Put otherwise, how would we decide when we have a good line?
A good line should have small prediction errors. That is, the line should give a good prediction of the points. That is, the line should result in small *errors*.
We would like a slope that gives us the smallest error.
## One metric for the line
[The Mean as Predictor](mean_meaning) section showed that the mean is the value with the smallest squared distance from the other values in the distribution. The mean is the predictor value that minimizes the sum of squared distances from the other values.
We can use the same metric for our line. Instead of using a single vector as a predictor, now we are using the values on the line as predictors. We want the FR slope, in our case, that gives the best predictors of the MMR values. Specifically, we want the slope that gives the smallest sum of squares difference between the line prediction and the actual values.
We have already calculated the prediction and error for our slope of 100, but let's do it again, and then calculate the *sum of squares* of the error:
```
slope = 100
mmr_predicted = fert * slope
error = mmr - mmr_predicted
# The sum of squared error
sum(error ** 2)
```
We are about to do this calculation many times, for many different slopes. We need a *function*.
In the function below, we are using [function world](../07/functions)
to get the values of `fert` and `mmr` defined here at the top level,
outside *function world*. The function can see these values, from
function world.
```
def sos_error(slope):
fitted = fert * slope # 'fert' comes from the top level
error = mmr - fitted # 'mmr' comes from the top level
return np.sum(error ** 2)
```
First check we get the same answer as the calculation above:
```
sos_error(100)
```
Does 200 give a higher or lower sum of squared error?
```
sos_error(200)
```
Now we can use the same strategy as we used in the [mean meaning](mean_meaning) page, to try lots of slopes, and find the one that gives the smallest sum of squared error.
```
# Slopes to try
some_slopes = np.arange(50, 110, 0.1)
n_slopes = len(some_slopes)
# Try all these slopes, calculate and record sum of squared error
sos_errors = np.zeros(n_slopes)
for i in np.arange(n_slopes):
slope = some_slopes[i]
sos_errors[i] = sos_error(slope)
# Show the first 10 values
sos_errors[:10]
```
We plot the slopes we have tried, on the x axis, against the sum of squared error, on the y-axis.
```
plt.plot(some_slopes, sos_errors)
plt.xlabel('Candidate slopes')
plt.ylabel('Sum of squared error')
```
The minimum of the sum of squared error is:
```
np.min(sos_errors)
```
We want to find the slope that corresponds to this minimum. We can use [argmin](where_and_argmin).
```
# Index of minumum value
i_of_min = np.argmin(sos_errors)
i_of_min
```
This is the index position of the minimum. We will therefore get the minimum (again) if we index into the original array with the index we just found:
```
# Check we do in fact get the minimum at this index
sos_errors[i_of_min]
```
Now we can get and show the slope value that corresponds the minimum sum of squared error:
```
best_slope = some_slopes[i_of_min]
best_slope
```
Plot the data, predictions and errors for the line that minimizes the sum of squared error:
```
best_predicted = fert * best_slope
plt.plot(fert, mmr, 'o')
plt.plot(fert, best_predicted, 'o', color='red')
for i in np.arange(len(fert)):
x = fert[i]
y_0 = best_predicted[i]
y_1 = mmr[i]
plt.plot([x, x], [y_0, y_1], ':', color='black', linewidth=1)
plt.title('The best-fit line using least-squared error')
```
The algorithm we have used so far, is rather slow and clunky, because we had to make an array with lots of slopes to try, and then go through each one to find the slope that minimizes the squared error.
In fact, we will soon see, we can use some tricks to get Python to do all this work for us, much more quickly.
Finding techniques for doing this automatically is a whole
mathematical field, called
[optimization](https://en.wikipedia.org/wiki/Mathematical_optimization).
For now, let's leap to using these techniques on our problem, of finding the best slope:
```
from scipy.optimize import minimize
# 100 below is the slope value to start the search.
minimize(sos_error, 100)
```
|
github_jupyter
|
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
# Make plots look a little bit more fancy
plt.style.use('fivethirtyeight')
# Print to 2 decimal places, show tiny values as 0
np.set_printoptions(precision=2, suppress=True)
import pandas as pd
# Load the data file
gender_data = pd.read_csv('gender_stats.csv')
# Show the column names
gender_data.columns
# Make new data frame with only MMR and FR
just_mmr_fr = gender_data[['mat_mort_ratio', 'fert_rate']]
# Show the first five rows
just_mmr_fr.head()
# Drop all rows with any NaN values
clean_mmr_fr = just_mmr_fr.dropna()
clean_mmr_fr.head()
mmr_series = clean_mmr_fr['mat_mort_ratio']
fert_series = clean_mmr_fr['fert_rate']
type(fert_series)
mmr = np.array(mmr_series)
fert = np.array(fert_series)
# Plot FR on the x axis, MMR on the y axis
plt.plot(fert, mmr, 'o')
plt.xlabel('Fertility rate')
plt.ylabel('Maternal mortality ratio')
plt.plot(fert, mmr, 'o')
# A green point at x=2, y=1000
plt.plot(2, 1000, 'o', color='green')
plt.plot(fert, mmr, 'o')
# Two green points, one at [2, 1000], the other at [3, 1200]
plt.plot([2, 3], [1000, 1200], 'o', color='green')
# The centers for our FR bins
fert_bin_centers = np.arange(7) + 1.5
fert_bin_centers
# The number of bins
n_bins = len(fert_bin_centers)
n_bins
plt.plot(fert, mmr, 'o')
plt.plot(fert_bin_centers, np.zeros(n_bins), 'o', color='green')
mmr_means = np.zeros(n_bins)
for i in np.arange(n_bins):
mid = fert_bin_centers[i]
# Boolean identifing indices withing the FR bin
fr_within_bin = (fert >= mid - 0.5) & (fert < mid + 0.5)
# Take the mean of the corresponding MMR values
mmr_means[i] = np.mean(mmr[fr_within_bin])
mmr_means
plt.plot(fert, mmr, 'o')
plt.plot(fert_bin_centers, mmr_means, 'o', color='green')
slope = 100
mmr_predicted = fert * slope
plt.plot(fert, mmr, 'o')
plt.plot(fert, mmr_predicted, 'o', color='red')
error = mmr - mmr_predicted
error[:10]
plt.plot(fert, mmr, 'o')
plt.plot(fert, mmr_predicted, 'o', color='red')
# Draw a line between predicted and actual
for i in np.arange(len(fert)):
x = fert[i]
y_0 = mmr_predicted[i]
y_1 = mmr[i]
plt.plot([x, x], [y_0, y_1], ':', color='black', linewidth=1)
slope = 100
mmr_predicted = fert * slope
error = mmr - mmr_predicted
# The sum of squared error
sum(error ** 2)
def sos_error(slope):
fitted = fert * slope # 'fert' comes from the top level
error = mmr - fitted # 'mmr' comes from the top level
return np.sum(error ** 2)
sos_error(100)
sos_error(200)
# Slopes to try
some_slopes = np.arange(50, 110, 0.1)
n_slopes = len(some_slopes)
# Try all these slopes, calculate and record sum of squared error
sos_errors = np.zeros(n_slopes)
for i in np.arange(n_slopes):
slope = some_slopes[i]
sos_errors[i] = sos_error(slope)
# Show the first 10 values
sos_errors[:10]
plt.plot(some_slopes, sos_errors)
plt.xlabel('Candidate slopes')
plt.ylabel('Sum of squared error')
np.min(sos_errors)
# Index of minumum value
i_of_min = np.argmin(sos_errors)
i_of_min
# Check we do in fact get the minimum at this index
sos_errors[i_of_min]
best_slope = some_slopes[i_of_min]
best_slope
best_predicted = fert * best_slope
plt.plot(fert, mmr, 'o')
plt.plot(fert, best_predicted, 'o', color='red')
for i in np.arange(len(fert)):
x = fert[i]
y_0 = best_predicted[i]
y_1 = mmr[i]
plt.plot([x, x], [y_0, y_1], ':', color='black', linewidth=1)
plt.title('The best-fit line using least-squared error')
from scipy.optimize import minimize
# 100 below is the slope value to start the search.
minimize(sos_error, 100)
| 0.762513 | 0.995532 |
# Kalman Filter
Kalman filters are linear models for state estimation of dynamic systems [1]. They have been the <i>de facto</i> standard in many robotics and tracking/prediction applications because they are well suited for systems with uncertainty about an observable dynamic process. They use a "observe, predict, correct" paradigm to extract information from an otherwise noisy signal. In Pyro, we can build differentiable Kalman filters with learnable parameters using the `pyro.contrib.tracking` [library](http://docs.pyro.ai/en/dev/contrib.tracking.html#module-pyro.contrib.tracking.extended_kalman_filter)
## Dynamic process
To start, consider this simple motion model:
$$ X_{k+1} = FX_k + \mathbf{W}_k $$
$$ \mathbf{Z}_k = HX_k + \mathbf{V}_k $$
where $k$ is the state, $X$ is the signal estimate, $Z_k$ is the observed value at timestep $k$, $\mathbf{W}_k$ and $\mathbf{V}_k$ are independent noise processes (ie $\mathbb{E}[w_k v_j^T] = 0$ for all $j, k$) which we'll approximate as Gaussians. Note that the state transitions are linear.
## Kalman Update
At each time step, we perform a prediction for the mean and covariance:
$$ \hat{X}_k = F\hat{X}_{k-1}$$
$$\hat{P}_k = FP_{k-1}F^T + Q$$
and a correction for the measurement:
$$ K_k = \hat{P}_k H^T(H\hat{P}_k H^T + R)^{-1}$$
$$ X_k = \hat{X}_k + K_k(z_k - H\hat{X}_k)$$
$$ P_k = (I-K_k H)\hat{P}_k$$
where $X$ is the position estimate, $P$ is the covariance matrix, $K$ is the Kalman Gain, and $Q$ and $R$ are covariance matrices.
For an in-depth derivation, see \[2\]
## Nonlinear Estimation: Extended Kalman Filter
What if our system is non-linear, eg in GPS navigation? Consider the following non-linear system:
$$ X_{k+1} = \mathbf{f}(X_k) + \mathbf{W}_k $$
$$ \mathbf{Z}_k = \mathbf{h}(X_k) + \mathbf{V}_k $$
Notice that $\mathbf{f}$ and $\mathbf{h}$ are now (smooth) non-linear functions.
The Extended Kalman Filter (EKF) attacks this problem by using a local linearization of the Kalman filter via a [Taylors Series expansion](https://en.wikipedia.org/wiki/Taylor_series).
$$ f(X_k, k) \approx f(x_k^R, k) + \mathbf{H}_k(X_k - x_k^R) + \cdots$$
where $\mathbf{H}_k$ is the Jacobian matrix at time $k$, $x_k^R$ is the previous optimal estimate, and we ignore the higher order terms. At each time step, we compute a Jacobian conditioned the previous predictions (this computation is handled by Pyro under the hood), and use the result to perform a prediction and update.
Omitting the derivations, the modification to the above predictions are now:
$$ \hat{X}_k \approx \mathbf{f}(X_{k-1}^R)$$
$$ \hat{P}_k = \mathbf{H}_\mathbf{f}(X_{k-1})P_{k-1}\mathbf{H}_\mathbf{f}^T(X_{k-1}) + Q$$
and the updates are now:
$$ X_k \approx \hat{X}_k + K_k\big(z_k - \mathbf{h}(\hat{X}_k)\big)$$
$$ K_k = \hat{P}_k \mathbf{H}_\mathbf{h}(\hat{X}_k) \Big(\mathbf{H}_\mathbf{h}(\hat{X}_k)\hat{P}_k \mathbf{H}_\mathbf{h}(\hat{X}_k) + R_k\Big)^{-1} $$
$$ P_k = \big(I - K_k \mathbf{H}_\mathbf{h}(\hat{X}_k)\big)\hat{P}_K$$
In Pyro, all we need to do is create an `EKFState` object and use its `predict` and `update` methods. Pyro will do exact inference to compute the innovations and we will use SVI to learn a MAP estimate of the position and measurement covariances.
As an example, let's look at an object moving at near-constant velocity in 2-D in a discrete time space over 100 time steps.
```
import os
import math
import torch
import pyro
import pyro.distributions as dist
from pyro.infer.autoguide import AutoDelta
from pyro.optim import Adam
from pyro.infer import SVI, Trace_ELBO, config_enumerate
from pyro.contrib.tracking.extended_kalman_filter import EKFState
from pyro.contrib.tracking.distributions import EKFDistribution
from pyro.contrib.tracking.dynamic_models import NcvContinuous
from pyro.contrib.tracking.measurements import PositionMeasurement
smoke_test = ('CI' in os.environ)
assert pyro.__version__.startswith('1.8.1')
dt = 1e-2
num_frames = 10
dim = 4
# Continuous model
ncv = NcvContinuous(dim, 2.0)
# Truth trajectory
xs_truth = torch.zeros(num_frames, dim)
# initial direction
theta0_truth = 0.0
# initial state
with torch.no_grad():
xs_truth[0, :] = torch.tensor([0.0, 0.0, math.cos(theta0_truth), math.sin(theta0_truth)])
for frame_num in range(1, num_frames):
# sample independent process noise
dx = pyro.sample('process_noise_{}'.format(frame_num), ncv.process_noise_dist(dt))
xs_truth[frame_num, :] = ncv(xs_truth[frame_num-1, :], dt=dt) + dx
```
Next, let's specify the measurements. Notice that we only measure the positions of the particle.
```
# Measurements
measurements = []
mean = torch.zeros(2)
# no correlations
cov = 1e-5 * torch.eye(2)
with torch.no_grad():
# sample independent measurement noise
dzs = pyro.sample('dzs', dist.MultivariateNormal(mean, cov).expand((num_frames,)))
# compute measurement means
zs = xs_truth[:, :2] + dzs
```
We'll use a [Delta autoguide](http://docs.pyro.ai/en/dev/infer.autoguide.html#autodelta) to learn MAP estimates of the position and measurement covariances. The `EKFDistribution` computes the joint log density of all of the EKF states given a tensor of sequential measurements.
```
def model(data):
# a HalfNormal can be used here as well
R = pyro.sample('pv_cov', dist.HalfCauchy(2e-6)) * torch.eye(4)
Q = pyro.sample('measurement_cov', dist.HalfCauchy(1e-6)) * torch.eye(2)
# observe the measurements
pyro.sample('track_{}'.format(i), EKFDistribution(xs_truth[0], R, ncv,
Q, time_steps=num_frames),
obs=data)
guide = AutoDelta(model) # MAP estimation
optim = pyro.optim.Adam({'lr': 2e-2})
svi = SVI(model, guide, optim, loss=Trace_ELBO(retain_graph=True))
pyro.set_rng_seed(0)
pyro.clear_param_store()
for i in range(250 if not smoke_test else 2):
loss = svi.step(zs)
if not i % 10:
print('loss: ', loss)
# retrieve states for visualization
R = guide()['pv_cov'] * torch.eye(4)
Q = guide()['measurement_cov'] * torch.eye(2)
ekf_dist = EKFDistribution(xs_truth[0], R, ncv, Q, time_steps=num_frames)
states= ekf_dist.filter_states(zs)
```
## References
\[1\] Kalman, R. E. *A New Approach to Linear Filtering and Prediction Problems.* 1960
\[2\] Welch, Greg, and Bishop, Gary. *An Introduction to the Kalman Filter.* 2006.
|
github_jupyter
|
import os
import math
import torch
import pyro
import pyro.distributions as dist
from pyro.infer.autoguide import AutoDelta
from pyro.optim import Adam
from pyro.infer import SVI, Trace_ELBO, config_enumerate
from pyro.contrib.tracking.extended_kalman_filter import EKFState
from pyro.contrib.tracking.distributions import EKFDistribution
from pyro.contrib.tracking.dynamic_models import NcvContinuous
from pyro.contrib.tracking.measurements import PositionMeasurement
smoke_test = ('CI' in os.environ)
assert pyro.__version__.startswith('1.8.1')
dt = 1e-2
num_frames = 10
dim = 4
# Continuous model
ncv = NcvContinuous(dim, 2.0)
# Truth trajectory
xs_truth = torch.zeros(num_frames, dim)
# initial direction
theta0_truth = 0.0
# initial state
with torch.no_grad():
xs_truth[0, :] = torch.tensor([0.0, 0.0, math.cos(theta0_truth), math.sin(theta0_truth)])
for frame_num in range(1, num_frames):
# sample independent process noise
dx = pyro.sample('process_noise_{}'.format(frame_num), ncv.process_noise_dist(dt))
xs_truth[frame_num, :] = ncv(xs_truth[frame_num-1, :], dt=dt) + dx
# Measurements
measurements = []
mean = torch.zeros(2)
# no correlations
cov = 1e-5 * torch.eye(2)
with torch.no_grad():
# sample independent measurement noise
dzs = pyro.sample('dzs', dist.MultivariateNormal(mean, cov).expand((num_frames,)))
# compute measurement means
zs = xs_truth[:, :2] + dzs
def model(data):
# a HalfNormal can be used here as well
R = pyro.sample('pv_cov', dist.HalfCauchy(2e-6)) * torch.eye(4)
Q = pyro.sample('measurement_cov', dist.HalfCauchy(1e-6)) * torch.eye(2)
# observe the measurements
pyro.sample('track_{}'.format(i), EKFDistribution(xs_truth[0], R, ncv,
Q, time_steps=num_frames),
obs=data)
guide = AutoDelta(model) # MAP estimation
optim = pyro.optim.Adam({'lr': 2e-2})
svi = SVI(model, guide, optim, loss=Trace_ELBO(retain_graph=True))
pyro.set_rng_seed(0)
pyro.clear_param_store()
for i in range(250 if not smoke_test else 2):
loss = svi.step(zs)
if not i % 10:
print('loss: ', loss)
# retrieve states for visualization
R = guide()['pv_cov'] * torch.eye(4)
Q = guide()['measurement_cov'] * torch.eye(2)
ekf_dist = EKFDistribution(xs_truth[0], R, ncv, Q, time_steps=num_frames)
states= ekf_dist.filter_states(zs)
| 0.639849 | 0.991891 |
```
%load_ext autoreload
%autoreload 2
%matplotlib inline
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from tqdm import tqdm_notebook as tqdm
# get the metadata
md = pd.read_excel('/home/jvdzwaan/ownCloud/Shared/OCR/Selection_OCR_GT_metadata.xlsx')
md
def convert_identifier(identifier):
parts = identifier.split(':')
return '_'.join([parts[0].upper(), parts[1]])
convert_identifier('ddd:010010081:mpeg21')
%%time
import os
import glob
in_dir = '/home/jvdzwaan/data/kb-ocr/text_aligned_blocks-match_gs/aligned/'
not_found = 0
for ident in md['Identifier']:
#print(ident)
#print(convert_identifier(ident))
c = convert_identifier(ident)
g = '{}{}*'.format(in_dir, c)
files = glob.glob(g)
if len(files) == 0:
not_found += 1
elif len(files) == 1:
pass
else:
print('Trouble with', identifier)
not_found
md['file_prefix'] = md.apply(lambda row: convert_identifier(row['Identifier']), axis=1)
md
def file_available(file_prefix, indir):
g = '{}{}*'.format(in_dir, file_prefix)
files = glob.glob(g)
if len(files) == 0:
return False
elif len(files) == 1:
return True
else:
print('Trouble with', file_prefix)
return 'ERROR'
in_dir = '/home/jvdzwaan/data/kb-ocr/text_aligned_blocks-match_gs/aligned/'
md['file_available'] = md.apply(lambda row: file_available(row['file_prefix'], in_dir), axis=1)
md
import os
def file_name(file_prefix, indir):
g = '{}{}*'.format(in_dir, file_prefix)
files = glob.glob(g)
if len(files) == 0:
return 'UNKNOWN'
elif len(files) == 1:
return os.path.basename(files[0])
else:
print('Trouble with', file_prefix)
return 'ERROR'
in_dir = '/home/jvdzwaan/data/kb-ocr/text_aligned_blocks-match_gs/aligned/'
md['file_name'] = md.apply(lambda row: file_name(row['file_prefix'], in_dir), axis=1)
md
import datetime
def get_period(year):
#print(type(year))
if isinstance(year, datetime.datetime):
year = int(year.strftime("%Y"))
elif isinstance(year, str):
year = int(year)
if year < 1883:
return 1
elif year >= 1883 and year < 1947:
return 2
else:
return 3
md['period'] = md.apply(lambda row: get_period(row['Date of publication']), axis=1)
md
def get_year(year):
#print(type(year))
if isinstance(year, datetime.datetime):
year = int(year.strftime("%Y"))
elif isinstance(year, str):
year = int(year)
return year
md['year'] = md.apply(lambda row: get_year(row['Date of publication']), axis=1)
md
md = md.query('file_available == True').copy()
md
md.columns
md.to_csv('/home/jvdzwaan/data/kb-ocr/metadata.csv')
grouped = md[['Date of publication', 'file_prefix', 'file_name', 'ABBYY Version', 'period']].groupby(['ABBYY Version', 'period'])
grouped.count()
def create_datadivision(in_files, SEED=4, TEST_PERCENTAGE=10, VAL_PERCENTAGE=10):
np.random.seed(SEED)
np.random.shuffle(in_files)
n_test = int(len(in_files)/100.0 * TEST_PERCENTAGE)
n_val = int(len(in_files)/100.0 * VAL_PERCENTAGE)
validation_texts = in_files[0:n_val]
test_texts = in_files[n_val:n_val+n_test]
train_texts = in_files[n_val+n_test:]
division = {
'train': [os.path.basename(t) for t in train_texts],
'val': [os.path.basename(t) for t in validation_texts],
'test': [os.path.basename(t) for t in test_texts]
}
return division
in_files = list(grouped.get_group((8.1, 1))['file_name'])
print(len(in_files))
division = create_datadivision(in_files)
print('# train', len(division['train']))
print('# val', len(division['val']))
print('# test', len(division['test']))
# make a datadivision for all periods and Abby 8
import json
for p in (1, 2, 3):
print('Period', p)
in_files = list(grouped.get_group((8.1, p))['file_name'])
print(len(in_files))
division = create_datadivision(in_files)
print('# train', len(division['train']))
print('# val', len(division['val']))
print('# test', len(division['test']))
out_file = f'/home/jvdzwaan/data/kb-ocr/text_aligned_blocks-match_gs/datadivision-A8P{p}.json'
print(out_file)
with open(out_file, 'w') as f:
json.dump(division, f, indent=4)
print()
```
|
github_jupyter
|
%load_ext autoreload
%autoreload 2
%matplotlib inline
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from tqdm import tqdm_notebook as tqdm
# get the metadata
md = pd.read_excel('/home/jvdzwaan/ownCloud/Shared/OCR/Selection_OCR_GT_metadata.xlsx')
md
def convert_identifier(identifier):
parts = identifier.split(':')
return '_'.join([parts[0].upper(), parts[1]])
convert_identifier('ddd:010010081:mpeg21')
%%time
import os
import glob
in_dir = '/home/jvdzwaan/data/kb-ocr/text_aligned_blocks-match_gs/aligned/'
not_found = 0
for ident in md['Identifier']:
#print(ident)
#print(convert_identifier(ident))
c = convert_identifier(ident)
g = '{}{}*'.format(in_dir, c)
files = glob.glob(g)
if len(files) == 0:
not_found += 1
elif len(files) == 1:
pass
else:
print('Trouble with', identifier)
not_found
md['file_prefix'] = md.apply(lambda row: convert_identifier(row['Identifier']), axis=1)
md
def file_available(file_prefix, indir):
g = '{}{}*'.format(in_dir, file_prefix)
files = glob.glob(g)
if len(files) == 0:
return False
elif len(files) == 1:
return True
else:
print('Trouble with', file_prefix)
return 'ERROR'
in_dir = '/home/jvdzwaan/data/kb-ocr/text_aligned_blocks-match_gs/aligned/'
md['file_available'] = md.apply(lambda row: file_available(row['file_prefix'], in_dir), axis=1)
md
import os
def file_name(file_prefix, indir):
g = '{}{}*'.format(in_dir, file_prefix)
files = glob.glob(g)
if len(files) == 0:
return 'UNKNOWN'
elif len(files) == 1:
return os.path.basename(files[0])
else:
print('Trouble with', file_prefix)
return 'ERROR'
in_dir = '/home/jvdzwaan/data/kb-ocr/text_aligned_blocks-match_gs/aligned/'
md['file_name'] = md.apply(lambda row: file_name(row['file_prefix'], in_dir), axis=1)
md
import datetime
def get_period(year):
#print(type(year))
if isinstance(year, datetime.datetime):
year = int(year.strftime("%Y"))
elif isinstance(year, str):
year = int(year)
if year < 1883:
return 1
elif year >= 1883 and year < 1947:
return 2
else:
return 3
md['period'] = md.apply(lambda row: get_period(row['Date of publication']), axis=1)
md
def get_year(year):
#print(type(year))
if isinstance(year, datetime.datetime):
year = int(year.strftime("%Y"))
elif isinstance(year, str):
year = int(year)
return year
md['year'] = md.apply(lambda row: get_year(row['Date of publication']), axis=1)
md
md = md.query('file_available == True').copy()
md
md.columns
md.to_csv('/home/jvdzwaan/data/kb-ocr/metadata.csv')
grouped = md[['Date of publication', 'file_prefix', 'file_name', 'ABBYY Version', 'period']].groupby(['ABBYY Version', 'period'])
grouped.count()
def create_datadivision(in_files, SEED=4, TEST_PERCENTAGE=10, VAL_PERCENTAGE=10):
np.random.seed(SEED)
np.random.shuffle(in_files)
n_test = int(len(in_files)/100.0 * TEST_PERCENTAGE)
n_val = int(len(in_files)/100.0 * VAL_PERCENTAGE)
validation_texts = in_files[0:n_val]
test_texts = in_files[n_val:n_val+n_test]
train_texts = in_files[n_val+n_test:]
division = {
'train': [os.path.basename(t) for t in train_texts],
'val': [os.path.basename(t) for t in validation_texts],
'test': [os.path.basename(t) for t in test_texts]
}
return division
in_files = list(grouped.get_group((8.1, 1))['file_name'])
print(len(in_files))
division = create_datadivision(in_files)
print('# train', len(division['train']))
print('# val', len(division['val']))
print('# test', len(division['test']))
# make a datadivision for all periods and Abby 8
import json
for p in (1, 2, 3):
print('Period', p)
in_files = list(grouped.get_group((8.1, p))['file_name'])
print(len(in_files))
division = create_datadivision(in_files)
print('# train', len(division['train']))
print('# val', len(division['val']))
print('# test', len(division['test']))
out_file = f'/home/jvdzwaan/data/kb-ocr/text_aligned_blocks-match_gs/datadivision-A8P{p}.json'
print(out_file)
with open(out_file, 'w') as f:
json.dump(division, f, indent=4)
print()
| 0.193223 | 0.212722 |
# Handling Missing Data
The difference between data found in many tutorials and data in the real world is that real-world data is rarely clean and homogeneous.
In particular, **many interesting datasets will have some amount of data missing**.
To make matters even more complicated, different data sources may indicate missing data in different ways.
In this section, we will discuss some general considerations for missing data, discuss how Pandas chooses to represent it, and demonstrate some built-in Pandas tools for handling missing data in Python.
**Here and throughout the book, we'll refer to missing data in general as *null*, *NaN*, or *NA* values.**
## Trade-Offs in Missing Data Conventions
There are a number of schemes that have been developed to indicate the presence of missing data in a table or DataFrame.
Generally, they revolve around one of two strategies: **using a *mask* that globally indicates missing values, or choosing a *sentinel value* that indicates a missing entry**.
In the masking approach, **the mask** might be an entirely separate Boolean array, or it may involve appropriation of one bit in the data representation to locally indicate the null status of a value.
In the sentinel approach, the sentinel value **could be some data-specific convention, such as indicating a missing integer value with -9999** or some rare bit pattern, or it could be a more global convention, such as indicating a missing floating-point value with **NaN (Not a Number)**, a special value which is part of the IEEE floating-point specification.
None of these approaches is without trade-offs: use of a separate mask array requires allocation of an additional Boolean array, which adds overhead in both storage and computation. A sentinel value reduces the range of valid values that can be represented, and may require extra (often non-optimized) logic in CPU and GPU arithmetic. Common special values like NaN are not available for all data types.
As in most cases where no universally optimal choice exists, different languages and systems use different conventions.
**For example, the R language uses reserved bit patterns within each data type as sentinel values indicating missing data**, while the SciDB system uses an extra byte attached to every cell which indicates a NA state.
## Missing Data in Pandas
The way in which Pandas **handles missing values is constrained by its reliance on the NumPy package**, which does not have a built-in notion of NA values for non-floating-point data types.
Pandas could have followed **R's lead in specifying bit patterns for each individual data type** to indicate nullness, but this approach turns out to be rather unwieldy.
While R contains four basic data types, **NumPy supports *far* more than this**: for example, while R has a single integer type, NumPy supports *fourteen* basic integer types once you account for available precisions, signedness, and endianness of the encoding.
**Reserving a specific bit pattern in all available NumPy types would lead to an unwieldy amount of overhead** in special-casing various operations for various types, likely even requiring a new fork of the NumPy package. Further, for the smaller data types (such as 8-bit integers), sacrificing a bit to use as a mask will significantly reduce the range of values it can represent.
**NumPy does have support for masked arrays** – that is, arrays that have a separate Boolean mask array attached for marking data as "good" or "bad."
Pandas could have derived from this, but the overhead in both storage, computation, and code maintenance makes that an unattractive choice.
With these constraints in mind, **Pandas chose to use sentinels for missing data**, and further chose to use two already-existing Python null values: the special floating-point **``NaN`` value, and the Python ``None`` object**.
This choice has some side effects, as we will see, but in practice ends up being a good compromise in most cases of interest.
### ``None``: Pythonic missing data
The first sentinel value used by Pandas is **``None``**, a Python singleton object that is often used for missing data in Python code.
Because it is a Python object, ``None`` cannot be used in any arbitrary NumPy/Pandas array, but only in arrays with data type ``'object'`` (i.e., arrays of Python objects):
```
import numpy as np
import pandas as pd
vals1 = np.array([1, None, 3, 4])
vals1
```
This ``dtype=object`` means that the best common type representation NumPy could infer for the contents of the array is that they are Python objects.
While this kind of object array is useful for some purposes, any operations on the data will be done at the Python level, with much more overhead than the typically fast operations seen for arrays with native types:
```
for dtype in ['object', 'int']:
print("dtype =", dtype)
%timeit np.arange(1E6, dtype=dtype).sum()
print()
```
The use of Python objects in an array also means that if you perform aggregations like ``sum()`` or ``min()`` across an array with a ``None`` value, you will generally get an error:
```
vals1.sum()
```
This reflects the fact that addition between an integer and ``None`` is undefined.
### ``NaN``: Missing numerical data
The other missing data representation, ``NaN`` (acronym for *Not a Number*), is different; it is a special floating-point value recognized by all systems that use the standard IEEE floating-point representation:
```
vals2 = np.array([1, np.nan, 2, 3])
vals2.dtype
```
Notice that NumPy chose a native floating-point type for this array: this means that **unlike the object array from before, this array supports fast operations pushed into compiled code.**
You should be aware that ``NaN`` is a bit like a data virus–it infects any other object it touches.
Regardless of the operation, the result of arithmetic with ``NaN`` will be another ``NaN``:
```
1 + np.nan
0 * np.nan
```
Note that this means that aggregates over the values are well defined (i.e., they don't result in an error) but not always useful:
```
vals2.sum(), vals2.min(), vals2.max()
```
NumPy does provide some special aggregations that will ignore these missing values:
```
np.nansum(vals2), np.nanmin(vals2), np.nanmax(vals2)
```
Keep in mind that ``NaN`` is specifically a floating-point value; there is no equivalent NaN value for integers, strings, or other types.
### NaN and None in Pandas
``NaN`` and ``None`` both have their place, and Pandas is built to handle the two of them nearly interchangeably, converting between them where appropriate:
```
pd.Series([1, np.nan, 2, None])
my_series = pd.Series([1, np.nan, 2, None])
print(type(my_series[1]))
print(type(my_series[3]))
```
For types that don't have an available sentinel value, Pandas automatically type-casts when NA values are present.
For example, **if we set a value in an integer array to ``np.nan``, it will automatically be upcast to a floating-point type to accommodate the NA**:
```
x = pd.Series(range(2), dtype=int)
x[0] = None
x
a = pd.Series(['1', np.nan, '2', None])
print(a)
print(type(a.iloc[0]))
print(type(a.iloc[1]))
print(type(a.iloc[3]))
```
Notice that **in addition to casting the integer array to floating point, Pandas automatically converts the ``None`` to a ``NaN`` value.**
(Be aware that there is a proposal to add a native integer NA to Pandas in the future; as of this writing, it has not been included).
While this type of magic may feel a bit hackish compared to the more unified approach to NA values in domain-specific languages like R, the Pandas sentinel/casting approach works quite well in practice and in my experience only rarely causes issues.
The following table lists the upcasting conventions in Pandas when NA values are introduced:
|Typeclass | Conversion When Storing NAs | NA Sentinel Value |
|--------------|-----------------------------|------------------------|
| ``floating`` | No change | ``np.nan`` |
| ``object`` | No change | ``None`` or ``np.nan`` |
| ``integer`` | Cast to ``float64`` | ``np.nan`` |
| ``boolean`` | Cast to ``float64`` or false| ``np.nan`` or ``None`` respectively|
Keep in mind that in Pandas, string data is always stored with an ``object`` dtype.
## Operating on Null Values
As we have seen, **Pandas treats ``None`` and ``NaN`` as essentially interchangeable for indicating missing or null values.**
To facilitate this convention, there are several useful methods for detecting, removing, and replacing null values in Pandas data structures.
They are:
- ``isnull()``: Generate a boolean mask indicating missing values
- ``notnull()``: Opposite of ``isnull()``
- ``dropna()``: Return a filtered version of the data
- ``fillna()``: Return a copy of the data with missing values filled or imputed
We will conclude this section with a brief exploration and demonstration of these routines.
### Detecting null values
Pandas data structures have two useful methods for detecting null data: ``isnull()`` and ``notnull()``.
Either one will return a Boolean mask over the data. For example:
```
data = pd.Series([1, np.nan, 'hello', None])
data
data.isnull()
```
As mentioned in [Data Indexing and Selection](03.02-Data-Indexing-and-Selection.ipynb), Boolean masks can be used directly as a ``Series`` or ``DataFrame`` index:
```
data[data.isnull()]
data.isnull()
data
data[data.notnull()]
```
The ``isnull()`` and ``notnull()`` methods produce similar Boolean results for ``DataFrame``s.
### Dropping null values
In addition to the masking used before, there are the convenience methods, ``dropna()``
(which removes NA values) and ``fillna()`` (which fills in NA values). For a ``Series``,
the result is straightforward:
```
data.dropna()
data
```
For a ``DataFrame``, there are more options.
Consider the following ``DataFrame``:
```
df = pd.DataFrame([[1, np.nan, 2],
[2, 3, 5],
[np.nan, 4, 6]])
df
```
We cannot drop single values from a ``DataFrame``; we can only drop full rows or full columns.
Depending on the application, you might want one or the other, so ``dropna()`` gives a number of options for a ``DataFrame``.
By default, ``dropna()`` will drop all rows in which *any* null value is present:
```
df.dropna()
```
Alternatively, you can drop NA values along a different axis; ``axis=1`` drops all columns containing a null value:
```
df.dropna(axis = 1)
df.dropna(axis = 'columns')
```
But this drops some good data as well; you might rather be interested in dropping rows or columns with *all* NA values, or a majority of NA values.
This can be specified through the ``how`` or ``thresh`` parameters, which allow fine control of the number of nulls to allow through.
The default is ``how='any'``, such that any row or column (depending on the ``axis`` keyword) containing a null value will be dropped.
You can also specify ``how='all'``, which will only drop rows/columns that are *all* null values:
```
df[3] = np.nan
df
df.dropna(axis = 'columns', how = 'all')
```
For finer-grained control, the ``thresh`` parameter lets you specify a minimum number of non-null values for the row/column to be kept:
```
df
df.dropna(axis = 'rows', thresh=3)
```
Here the first and last row have been dropped, because they contain only two non-null values.
### Filling null values
Sometimes rather than dropping NA values, you'd rather replace them with a valid value.
This value might be a single number like zero, or it might be some sort of imputation or interpolation from the good values.
You could do this in-place using the ``isnull()`` method as a mask, but because it is such a common operation Pandas provides the ``fillna()`` method, which returns a copy of the array with the null values replaced.
Consider the following ``Series``:
```
data = pd.Series([1, np.nan, 2, None, 3], index=list('abcde'))
data
data = pd.Series([1, np.nan, 2, None, 3], index=list('abcde'))
data
```
We can fill NA entries with a single value, such as zero:
```
data.fillna(0)
```
We can specify a forward-fill to propagate the previous value forward:
```
data
# forward-fill
data.fillna(method = 'ffill')
```
Or we can specify a back-fill to propagate the next values backward:
```
# back-fill
data.fillna(method = 'bfill')
```
For ``DataFrame``s, the options are similar, but we can also specify an ``axis`` along which the fills take place:
```
df
df.fillna(method = 'ffill', axis=1)
```
Notice that if a previous value is not available during a forward fill, the NA value remains.
|
github_jupyter
|
import numpy as np
import pandas as pd
vals1 = np.array([1, None, 3, 4])
vals1
for dtype in ['object', 'int']:
print("dtype =", dtype)
%timeit np.arange(1E6, dtype=dtype).sum()
print()
vals1.sum()
vals2 = np.array([1, np.nan, 2, 3])
vals2.dtype
1 + np.nan
0 * np.nan
vals2.sum(), vals2.min(), vals2.max()
np.nansum(vals2), np.nanmin(vals2), np.nanmax(vals2)
pd.Series([1, np.nan, 2, None])
my_series = pd.Series([1, np.nan, 2, None])
print(type(my_series[1]))
print(type(my_series[3]))
x = pd.Series(range(2), dtype=int)
x[0] = None
x
a = pd.Series(['1', np.nan, '2', None])
print(a)
print(type(a.iloc[0]))
print(type(a.iloc[1]))
print(type(a.iloc[3]))
data = pd.Series([1, np.nan, 'hello', None])
data
data.isnull()
data[data.isnull()]
data.isnull()
data
data[data.notnull()]
data.dropna()
data
df = pd.DataFrame([[1, np.nan, 2],
[2, 3, 5],
[np.nan, 4, 6]])
df
df.dropna()
df.dropna(axis = 1)
df.dropna(axis = 'columns')
df[3] = np.nan
df
df.dropna(axis = 'columns', how = 'all')
df
df.dropna(axis = 'rows', thresh=3)
data = pd.Series([1, np.nan, 2, None, 3], index=list('abcde'))
data
data = pd.Series([1, np.nan, 2, None, 3], index=list('abcde'))
data
data.fillna(0)
data
# forward-fill
data.fillna(method = 'ffill')
# back-fill
data.fillna(method = 'bfill')
df
df.fillna(method = 'ffill', axis=1)
| 0.261519 | 0.994129 |
```
import numpy as np
my_list=[1,2,3]
```
my_array=np.array(my_list)
```
my_array
type(my_array)
type(my_list)
```
# np.arange()
```
# np.arange(4) 默认返回0-4 左闭右开
np.arange(4)
np.arrange
# np.arange(4.0) 返回浮点数字
np.arange(4.0)
# np.arange(3,7) 从3开始,返回到7 左闭右开
np.arange(3,7)
np.arange(3.0,7.0)
# np.arange(3,10,2) 从3开始返回到7 间隔为2
np.arange(3,10,2)
np.arange(3,11,2)
```
# np.zeros() & np.ones()
#### np.zeros(shape,dtype=float,ordert='C')
order = 'C', 'F; default is 'C'
```
# np.zeros(5) 返回5个float 0 默认是浮点数字
np.zeros(5)
# np.zeros(5,dtype=int)
np.zeros(5,dtype=int)
np.zeros((5,))
#注意观察 (5,)和(5,1) 不一样,(5,1)是一位数组的意思
np.zeros((5,1))
np.zeros((2,5))
np.zeros((2,5),dtype=int)
# np.ones 是一样的用法
np.ones(3)
np.ones(3,dtype=int)
np.ones((2,2),dtype=int)
```
# np.linspace()
```
np.linspace(3,9,3)
np.linspace(3,11,3)
np.linspace(3,12,3)
np.arange(3,12,3)
np.arange(3,12,4)
```
### arange()类似于内置函数range(),通过指定开始值、终值和步长创建表示等差数列的一维数组,注意得到的结果数组不包含终值。
### linspace()通过指定开始值、终值和元素个数创建表示等差数列的一维数组,可以通过endpoint参数指定是否包含终值,默认值为True,即包含终值。
## np.eye()
### Create an identity matrix; Returns a 2D array with ones on the diagonal and zeros elsewhere;
```
# 创建一个 以1为对角线的 二维矩阵;其他位置 用0代替;k是缩进值
np.eye(2)
np.eye(4,dtype=int)
# K=1 缩进 1
np.eye(4,k=1,dtype=int)
np.eye(4,dtype=int)
```
# Random
## np.random.rand()
```
# 再「0,1」 范围内随机生成值
np.random.rand(3)
np.random.rand(3,3)
```
## np.random.randn()
```
# 在正态分布 sigma=1 内返回值
np.random.randn(4)
np.random.randn(3,3)
import numpy as np
np.random.rand(2,4)
```
## np.random.randint()
```
# 左开右闭返回 int 函数, 默认从0开始;
np.random.randint(5)
np.random.randint(5)
np.random.randint(5,10)
# 加上size 能返回特定数量的值
np.random.randint(5,10,size=(3,2))
np.random.randint(5,100,size=(2,5))
```
## np.random.seed()
```
# can be used to set the random state, so that tne same 'random'
#results canbe reproduced
np.random.seed(1)
np.random.randint(0,100,size=2)
np.random.randint(0,100,size=2)
# 相当于编号一样 很好理解
np.random.seed(1)
np.random.randint(0,100,size=2)
```
# Array Attributes and Methods
## Reshape
```
arr=np.arange(25)
arr
ranarr=np.random.randint(0,50,10)
ranarr
arr
# reshape
arr.reshape(5,5)
arr.reshape((5,5))
arr.reshape(5,4)
# 会报错事因为 reshape 包含20个 但是arr有25个。
ranarr
ranarr.reshape(2,5)
ranarr
```
## max() min() argmax() argmin()
```
ranarr.max()
ranarr.min()
ranarr.argmax()
ranarr.argmin()
```
# Shape
```
arr
arr.shape
arr.reshape(25,1)
arr.reshape(1,25)
arr.reshape(25,1).shape
```
# dtype
```
arr.dtype
arr2=np.array([1.0,2,3,4])
arr2
arr2.dtype
```
## Exercise
```
myarray=np.linspace(0,10,101)
myarray
```
|
github_jupyter
|
import numpy as np
my_list=[1,2,3]
my_array
type(my_array)
type(my_list)
# np.arange(4) 默认返回0-4 左闭右开
np.arange(4)
np.arrange
# np.arange(4.0) 返回浮点数字
np.arange(4.0)
# np.arange(3,7) 从3开始,返回到7 左闭右开
np.arange(3,7)
np.arange(3.0,7.0)
# np.arange(3,10,2) 从3开始返回到7 间隔为2
np.arange(3,10,2)
np.arange(3,11,2)
# np.zeros(5) 返回5个float 0 默认是浮点数字
np.zeros(5)
# np.zeros(5,dtype=int)
np.zeros(5,dtype=int)
np.zeros((5,))
#注意观察 (5,)和(5,1) 不一样,(5,1)是一位数组的意思
np.zeros((5,1))
np.zeros((2,5))
np.zeros((2,5),dtype=int)
# np.ones 是一样的用法
np.ones(3)
np.ones(3,dtype=int)
np.ones((2,2),dtype=int)
np.linspace(3,9,3)
np.linspace(3,11,3)
np.linspace(3,12,3)
np.arange(3,12,3)
np.arange(3,12,4)
# 创建一个 以1为对角线的 二维矩阵;其他位置 用0代替;k是缩进值
np.eye(2)
np.eye(4,dtype=int)
# K=1 缩进 1
np.eye(4,k=1,dtype=int)
np.eye(4,dtype=int)
# 再「0,1」 范围内随机生成值
np.random.rand(3)
np.random.rand(3,3)
# 在正态分布 sigma=1 内返回值
np.random.randn(4)
np.random.randn(3,3)
import numpy as np
np.random.rand(2,4)
# 左开右闭返回 int 函数, 默认从0开始;
np.random.randint(5)
np.random.randint(5)
np.random.randint(5,10)
# 加上size 能返回特定数量的值
np.random.randint(5,10,size=(3,2))
np.random.randint(5,100,size=(2,5))
# can be used to set the random state, so that tne same 'random'
#results canbe reproduced
np.random.seed(1)
np.random.randint(0,100,size=2)
np.random.randint(0,100,size=2)
# 相当于编号一样 很好理解
np.random.seed(1)
np.random.randint(0,100,size=2)
arr=np.arange(25)
arr
ranarr=np.random.randint(0,50,10)
ranarr
arr
# reshape
arr.reshape(5,5)
arr.reshape((5,5))
arr.reshape(5,4)
# 会报错事因为 reshape 包含20个 但是arr有25个。
ranarr
ranarr.reshape(2,5)
ranarr
ranarr.max()
ranarr.min()
ranarr.argmax()
ranarr.argmin()
arr
arr.shape
arr.reshape(25,1)
arr.reshape(1,25)
arr.reshape(25,1).shape
arr.dtype
arr2=np.array([1.0,2,3,4])
arr2
arr2.dtype
myarray=np.linspace(0,10,101)
myarray
| 0.203312 | 0.801509 |
Sascha Spors,
Professorship Signal Theory and Digital Signal Processing,
Institute of Communications Engineering (INT),
Faculty of Computer Science and Electrical Engineering (IEF),
University of Rostock,
Germany
# Tutorial Selected Topics in Audio Signal Processing
Winter Semester 2021/22 (Master Course)
- lecture: https://github.com/spatialaudio/selected-topics-in-audio-signal-processing-lecture
- tutorial: https://github.com/spatialaudio/selected-topics-in-audio-signal-processing-exercises
WIP...
The project is currently under heavy development while adding new material for the winter term 2021/22
Feel free to contact lecturer frank.schultz@uni-rostock.de
For usage of `%matplotlib widget` for convenient rotation of the 3D plot below we should `conda install -c conda-forge ipympl`, cf. [https://github.com/matplotlib/ipympl](https://github.com/matplotlib/ipympl)
# Exercise 7: Create Orthonormal Column Space Vectors
- Gram-Schmidt QR vs.
- SVD
```
import numpy as np
import matplotlib.pyplot as plt
from scipy.linalg import inv, norm, qr, svd, svdvals
from numpy.linalg import matrix_rank
# %matplotlib widget
# create matrix with full rank and very congruent-like columns
A = np.array([[0.95, 0.85, 1.05], [1.125, 0.8, 0.9], [0.925, 1.1, 0.8]])
svd_equal_qr_flag = False
if svd_equal_qr_flag:
# a made up example where SVD's U approx QR's Q (besides polarity!!!)
# note that condition number of this A is very large and
# ortho Q suffers from numerical precision
A[:, 0] = [-0.597426625235553, -0.534589417708599, -0.59774206973714]
print('A\n', A)
print('rank of A =', matrix_rank(A))
[u, s, vh] = svd(A)
[q, r] = qr(A)
print('Q\n', q)
print('R\n', r)
print('sing vals of A =', s, '==\nsing vals of R =', svdvals(r))
```
### Gram-Schmidt procedure
hard coded (we should program a non-hard coded 4D example to double check the projection/subtraction routine)
```
Q, R = np.zeros(A.shape), np.zeros(A.shape)
# 1st q
# polarity is free to choose, so make it consistent with qr(A)
R[0, 0] = - norm(A[:, 0], 2)
Q[:, 0] = A[:, 0] / R[0, 0]
# 2nd q
R[0, 1] = np.inner(Q[:, 0], A[:, 1]) # 2nd A col onto q1
tmp = A[:, 1] - R[0, 1]*Q[:, 0] # subtract projection
# polarity is free to choose, so make it consistent with qr(A)
R[1, 1] = + norm(tmp, 2)
Q[:, 1] = tmp / R[1, 1]
# 3rd q
R[0, 2] = np.inner(Q[:, 0], A[:, 2]) # 3rd A col onto q1
R[1, 2] = np.inner(Q[:, 1], A[:, 2]) # 3rd A col onto q2
tmp = A[:, 2] - R[0, 2]*Q[:, 0] - R[1, 2]*Q[:, 1] # % subtract projections
# polarity is free to choose, so make it consistent with qr(A)
R[2, 2] = - norm(tmp, 2)
Q[:, 2] = tmp / R[2, 2]
print('check if our QR == qr():',
np.allclose(r, R),
np.allclose(q, Q))
print('check if Q is orthonormal:',
np.allclose(np.eye(3), Q.T @ Q),
np.allclose(np.eye(3), Q @ Q.T),
np.allclose(inv(Q), Q.T))
# check rank1 matrix superposition:
# A1 has all entries, first col of A2 is zero...
A1 = np.outer(Q[:,0], R[0,:])
A2 = np.outer(Q[:,1], R[1,:])
A3 = np.outer(Q[:,2], R[2,:])
print(A1, '\n\n', A2, '\n\n', A3)
np.allclose(A1+A2+A3, A)
```
### Plot the 3 columns for the differently spanned column spaces
```
fig = plt.figure()
ax = plt.axes(projection='3d')
ax.view_init(elev=25, azim=-160)
for n in range(2): # plot vecs dim 1&2
ax.plot([0, A[0, n]], [0, A[1, n]], [0, A[2, n]], 'C0', lw=1)
ax.plot([0, u[0, n]], [0, u[1, n]], [0, u[2, n]], 'C1', lw=2)
ax.plot([0, q[0, n]], [0, q[1, n]], [0, q[2, n]], 'C3', lw=3)
# plot vecs dim 3, add label
ax.plot([0, A[0, 2]], [0, A[1, 2]], [0, A[2, 2]], 'C0', lw=1, label='A')
ax.plot([0, u[0, 2]], [0, u[1, 2]], [0, u[2, 2]], 'C1', lw=2, label='SVD U')
ax.plot([0, q[0, 2]], [0, q[1, 2]], [0, q[2, 2]],
'C3', lw=3, label='Gram-Schmidt Q')
ax.set_xlim(-1.2, 1.2)
ax.set_ylim(-1.2, 1.2)
ax.set_zlim(-1.2, 1.2)
ax.set_xlabel('x')
ax.set_ylabel('y')
ax.set_zlabel('z')
ax.legend()
plt.close(fig)
```
## **Copyright**
The notebooks are provided as [Open Educational Resources](https://en.wikipedia.org/wiki/Open_educational_resources). Feel free to use the notebooks for your own purposes. The text is licensed under [Creative Commons Attribution 4.0](https://creativecommons.org/licenses/by/4.0/), the code of the IPython examples under the [MIT license](https://opensource.org/licenses/MIT). Please attribute the work as follows: *Frank Schultz, Audio Signal Processing - A Tutorial Featuring Computational Examples* with the URL https://github.com/spatialaudio/selected-topics-in-audio-signal-processing-exercises
|
github_jupyter
|
import numpy as np
import matplotlib.pyplot as plt
from scipy.linalg import inv, norm, qr, svd, svdvals
from numpy.linalg import matrix_rank
# %matplotlib widget
# create matrix with full rank and very congruent-like columns
A = np.array([[0.95, 0.85, 1.05], [1.125, 0.8, 0.9], [0.925, 1.1, 0.8]])
svd_equal_qr_flag = False
if svd_equal_qr_flag:
# a made up example where SVD's U approx QR's Q (besides polarity!!!)
# note that condition number of this A is very large and
# ortho Q suffers from numerical precision
A[:, 0] = [-0.597426625235553, -0.534589417708599, -0.59774206973714]
print('A\n', A)
print('rank of A =', matrix_rank(A))
[u, s, vh] = svd(A)
[q, r] = qr(A)
print('Q\n', q)
print('R\n', r)
print('sing vals of A =', s, '==\nsing vals of R =', svdvals(r))
Q, R = np.zeros(A.shape), np.zeros(A.shape)
# 1st q
# polarity is free to choose, so make it consistent with qr(A)
R[0, 0] = - norm(A[:, 0], 2)
Q[:, 0] = A[:, 0] / R[0, 0]
# 2nd q
R[0, 1] = np.inner(Q[:, 0], A[:, 1]) # 2nd A col onto q1
tmp = A[:, 1] - R[0, 1]*Q[:, 0] # subtract projection
# polarity is free to choose, so make it consistent with qr(A)
R[1, 1] = + norm(tmp, 2)
Q[:, 1] = tmp / R[1, 1]
# 3rd q
R[0, 2] = np.inner(Q[:, 0], A[:, 2]) # 3rd A col onto q1
R[1, 2] = np.inner(Q[:, 1], A[:, 2]) # 3rd A col onto q2
tmp = A[:, 2] - R[0, 2]*Q[:, 0] - R[1, 2]*Q[:, 1] # % subtract projections
# polarity is free to choose, so make it consistent with qr(A)
R[2, 2] = - norm(tmp, 2)
Q[:, 2] = tmp / R[2, 2]
print('check if our QR == qr():',
np.allclose(r, R),
np.allclose(q, Q))
print('check if Q is orthonormal:',
np.allclose(np.eye(3), Q.T @ Q),
np.allclose(np.eye(3), Q @ Q.T),
np.allclose(inv(Q), Q.T))
# check rank1 matrix superposition:
# A1 has all entries, first col of A2 is zero...
A1 = np.outer(Q[:,0], R[0,:])
A2 = np.outer(Q[:,1], R[1,:])
A3 = np.outer(Q[:,2], R[2,:])
print(A1, '\n\n', A2, '\n\n', A3)
np.allclose(A1+A2+A3, A)
fig = plt.figure()
ax = plt.axes(projection='3d')
ax.view_init(elev=25, azim=-160)
for n in range(2): # plot vecs dim 1&2
ax.plot([0, A[0, n]], [0, A[1, n]], [0, A[2, n]], 'C0', lw=1)
ax.plot([0, u[0, n]], [0, u[1, n]], [0, u[2, n]], 'C1', lw=2)
ax.plot([0, q[0, n]], [0, q[1, n]], [0, q[2, n]], 'C3', lw=3)
# plot vecs dim 3, add label
ax.plot([0, A[0, 2]], [0, A[1, 2]], [0, A[2, 2]], 'C0', lw=1, label='A')
ax.plot([0, u[0, 2]], [0, u[1, 2]], [0, u[2, 2]], 'C1', lw=2, label='SVD U')
ax.plot([0, q[0, 2]], [0, q[1, 2]], [0, q[2, 2]],
'C3', lw=3, label='Gram-Schmidt Q')
ax.set_xlim(-1.2, 1.2)
ax.set_ylim(-1.2, 1.2)
ax.set_zlim(-1.2, 1.2)
ax.set_xlabel('x')
ax.set_ylabel('y')
ax.set_zlabel('z')
ax.legend()
plt.close(fig)
| 0.473414 | 0.952574 |
<a href="https://colab.research.google.com/github/African-Quant/FOREX_RelativeStrengthOscillator/blob/main/Oanda_RelativeStrength_EN.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
```
#@title Installation
!pip install git+https://github.com/yhilpisch/tpqoa.git --upgrade --quiet
!pip install pykalman --quiet
!pip install --upgrade mplfinance --quiet
#@title Imports
import tpqoa
import numpy as np
import pandas as pd
from pykalman import KalmanFilter
%matplotlib inline
from pylab import mpl, plt
plt.style.use('seaborn')
mpl.rcParams['savefig.dpi'] = 300
mpl.rcParams['font.family'] = 'serif'
from datetime import date, timedelta
import warnings
warnings.filterwarnings("ignore")
#@title Oanda API
path = '/content/drive/MyDrive/Oanda_Algo/pyalgo.cfg'
api = tpqoa.tpqoa(path)
#@title Symbols/Currency Pairs
def symbolsList():
symbols = []
syms = api.get_instruments()
for x in syms:
symbols.append(x[1])
return symbols
symbols = symbolsList()
pairs = ['AUD_CAD', 'AUD_CHF', 'AUD_JPY', 'AUD_NZD', 'AUD_USD', 'CAD_CHF',
'CAD_JPY', 'CHF_JPY', 'EUR_AUD', 'EUR_CAD', 'EUR_CHF', 'EUR_GBP',
'EUR_JPY', 'EUR_NZD', 'EUR_USD', 'GBP_AUD', 'GBP_CAD', 'GBP_CHF',
'GBP_JPY', 'GBP_NZD', 'GBP_USD', 'NZD_CAD', 'NZD_CHF', 'NZD_JPY',
'NZD_USD', 'USD_CAD', 'USD_CHF', 'USD_JPY',]
#@title getData(instr, gran = 'D', td=1000)
def getData(instr, gran = 'D', td=1000):
start = f"{date.today() - timedelta(td)}"
end = f"{date.today() - timedelta(1)}"
granularity = gran
price = 'M' # price: string one of 'A' (ask), 'B' (bid) or 'M' (middle)
data = api.get_history(instr, start, end, granularity, price)
data.drop(['complete'], axis=1, inplace=True)
data.reset_index(inplace=True)
data.rename(columns = {'time':'Date','o':'Open','c': 'Close', 'h':'High', 'l': 'Low'}, inplace = True)
data.set_index('Date', inplace=True)
return data
#@title Indexes
def USD_Index():
'''Creating a USD Index from a basket of instruments
denominated in dollars
'''
USD = ['EUR_USD', 'GBP_USD', 'USD_CAD', 'USD_CHF',
'USD_JPY', 'AUD_USD', 'NZD_USD']
df = pd.DataFrame()
for i in USD:
data = getData(i).ffill(axis='rows')
# setting the Dollar as the base
if '_USD' == i[-4:]:
data[f'{i}'] = (data['Close'])**(-1)
else:
data[f'{i}'] = data['Close']
df = pd.concat([df, data.loc[:,f'{i}']], axis=1)
df['US_index'] = 1
for i in range(len(USD)):
df['US_index'] *= df[USD[i]]
return ((df['US_index'])**(1/(len(USD)))).to_frame()
def EURO_Index():
'''Creating a EUR Index from a basket of instruments
denominated in EUROs
'''
EUR = ['EUR_USD', 'EUR_GBP', 'EUR_JPY', 'EUR_CHF', 'EUR_CAD',
'EUR_AUD', 'EUR_NZD']
df = pd.DataFrame()
for i in EUR:
data = getData(i).ffill(axis='rows')
data[f'{i}'] = data['Close']
df = pd.concat([df, data.loc[:,f'{i}']], axis=1)
df['EUR_index'] = 1
for i in range(len(EUR)):
df['EUR_index'] *= df[EUR[i]]
return ((df['EUR_index'])**(1/(len(EUR)))).to_frame()
def GBP_Index():
'''Creating a GBP Index from a basket of instruments
denominated in Pound Sterling
'''
GBP = ['GBP_USD', 'EUR_GBP', 'GBP_JPY', 'GBP_CHF', 'GBP_CAD',
'GBP_AUD', 'GBP_NZD']
df = pd.DataFrame()
for i in GBP:
data = getData(i).ffill(axis='rows')
# setting the Dollar as the base
if '_GBP' == i[-4:]:
data[f'{i}'] = (data['Close'])**(-1)
else:
data[f'{i}'] = data['Close']
df = pd.concat([df, data.loc[:,f'{i}']], axis=1)
df['GBP_index'] = 1
for i in range(len(GBP)):
df['GBP_index'] *= df[GBP[i]]
return ((df['GBP_index'])**(1/(len(GBP)))).to_frame()
def CHF_Index():
'''Creating a CHF Index from a basket of instruments
denominated in Swiss Francs
'''
CHF = ['CHF_JPY', 'EUR_CHF', 'GBP_CHF', 'USD_CHF', 'CAD_CHF',
'AUD_CHF', 'NZD_CHF']
df = pd.DataFrame()
for i in CHF:
data = getData(i).ffill(axis='rows')
# setting the Dollar as the base
if '_CHF' == i[-4:]:
data[f'{i}'] = (data['Close'])**(-1)
else:
data[f'{i}'] = data['Close']
df = pd.concat([df, data.loc[:,f'{i}']], axis=1)
df['CHF_index'] = 1
for i in range(len(CHF)):
df['CHF_index'] *= df[CHF[i]]
return ((df['CHF_index'])**(1/(len(CHF)))).to_frame()
def CAD_Index():
'''Creating a CAD Index from a basket of instruments
denominated in Canadian Dollars
'''
CAD = ['CAD_JPY', 'EUR_CAD', 'GBP_CAD', 'USD_CAD', 'CAD_CHF',
'AUD_CAD', 'NZD_CAD']
df = pd.DataFrame()
for i in CAD:
data = getData(i).ffill(axis='rows')
# setting the Dollar as the base
if '_CAD' == i[-4:]:
data[f'{i}'] = (data['Close'])**(-1)
else:
data[f'{i}'] = data['Close']
df = pd.concat([df, data.loc[:,f'{i}']], axis=1)
df['CAD_index'] = 1
for i in range(len(CAD)):
df['CAD_index'] *= df[CAD[i]]
return ((df['CAD_index'])**(1/(len(CAD)))).to_frame()
def JPY_Index():
'''Creating a JPY Index from a basket of instruments
denominated in Swiss Francs
'''
JPY = ['CAD_JPY', 'EUR_JPY', 'GBP_JPY', 'USD_JPY', 'CHF_JPY',
'AUD_JPY', 'NZD_JPY']
df = pd.DataFrame()
for i in JPY:
data = getData(i).ffill(axis='rows')
# setting the Japanese Yen as the base
data[f'{i}'] = (data['Close'])**(-1)
df = pd.concat([df, data.loc[:,f'{i}']], axis=1)
df['JPY_index'] = 1
for i in range(len(JPY)):
df['JPY_index'] *= df[JPY[i]]
return ((df['JPY_index'])**(1/(len(JPY)))).to_frame()
def AUD_Index():
'''Creating a AUD Index from a basket of instruments
denominated in Australian Dollar
'''
AUD = ['AUD_JPY', 'EUR_AUD', 'GBP_AUD', 'AUD_USD', 'AUD_CAD',
'AUD_CHF', 'AUD_NZD']
df = pd.DataFrame()
for i in AUD:
data = getData(i).ffill(axis='rows')
# setting the Aussie Dollar as the base
if '_AUD' == i[-4:]:
data[f'{i}'] = (data['Close'])**(-1)
else:
data[f'{i}'] = data['Close']
df = pd.concat([df, data.loc[:,f'{i}']], axis=1)
df['AUD_index'] = 1
for i in range(len(AUD)):
df['AUD_index'] *= df[AUD[i]]
return ((df['AUD_index'])**(1/(len(AUD)))).to_frame()
def NZD_Index():
'''Creating a NZD Index from a basket of instruments
denominated in New Zealand Dollar
'''
NZD = ['NZD_JPY', 'EUR_NZD', 'GBP_NZD', 'NZD_USD', 'NZD_CAD',
'NZD_CHF', 'AUD_NZD']
df = pd.DataFrame()
for i in NZD:
data = getData(i).ffill(axis='rows')
# setting the Dollar as the base
if '_NZD' == i[-4:]:
data[f'{i}'] = (data['Close'])**(-1)
else:
data[f'{i}'] = data['Close']
df = pd.concat([df, data.loc[:,f'{i}']], axis=1)
df['NZD_index'] = 1
for i in range(len(NZD)):
df['NZD_index'] *= df[ NZD[i]]
return ((df['NZD_index'])**(1/(len(NZD)))).to_frame()
def eSuperRCS(df):
"""
This code computes the super smoother introduced by John Ehlers
"""
spr = df.to_frame().copy()
# HighPass filter cyclic components whose periods are shorter than 48 bars
alpha1 = (np.cos(0.707*2*np.pi/48) + np.sin(0.707*2*np.pi/48) - 1)/np.cos(0.707*2*np.pi/48)
hiPass = pd.DataFrame(None, index=spr.index, columns=['filtered'])
for i in range(len(spr)):
if i < 3:
hiPass.iloc[i, 0] = spr.iat[i, 0]
else:
hiPass.iloc[i, 0] = ((1 - alpha1/2)*(1 - alpha1/2)*(spr.iat[i, 0]
- 2*spr.iat[i-1, 0] + spr.iat[i-2, 0] + 2*(1 - alpha1)*hiPass.iat[i-1, 0]
- (1 - alpha1)**2 *hiPass.iat[i-2, 0]))
# SuperSmoother
a1 = np.exp(-1.414*(np.pi) / 10)
b1 = 2*a1*np.cos(1.414*(np.pi) / 10)
c2 = b1
c3 = -a1*a1
c1 = 1 - c2 - c3
Filt = pd.DataFrame(None, index=spr.index, columns=['filtered'])
for i in range(len(spr)):
if i < 3:
Filt.iloc[i, 0] = hiPass.iat[i, 0]
else:
Filt.iloc[i, 0] = c1*(hiPass.iat[i, 0] + hiPass.iat[i - 1, 0]/ 2 + c2*Filt.iat[i-1, 0] + c3*Filt.iat[i-2, 0])
Filt['eSuperRCS'] = RSI(Filt['filtered'])
return Filt['eSuperRCS']
def RSI(series, period=25):
delta = series.diff()
up = delta.clip(lower=0)
dn = -1*delta.clip(upper=0)
ema_up = up.ewm(com=period-1, adjust=False).mean()
ewm_dn = dn.ewm(com=period-1, adjust=False).mean()
rs = (ema_up/ewm_dn)
return 100 - 100 / (1 + rs)
def will_pr(data, lb=14):
df = data[['High', 'Low', 'Close']].copy()
df['max_hi'] = data['High'].rolling(window=lb).max()
df['min_lo'] = data['Low'].rolling(window=lb).min()
df['will_pr'] = 0
for i in range(len(df)):
try:
df.iloc[i, 5] = ((df.iat[i, 3] - df.iat[i, 2])/(df.iat[i, 3] - df.iat[i, 4])) * (-100)
except ValueError:
pass
return df['will_pr']
en = getData('EUR_NZD')
eur = EURO_Index()
nzd = NZD_Index()
df = pd.concat((en, eur, nzd), axis=1).ffill(axis='rows')
df
tickers = ['EUR_index', 'NZD_index']
cumm_rtn = (1 + df[tickers].pct_change()).cumprod()
cumm_rtn.plot();
plt.ylabel('Cumulative Return');
plt.xlabel('Time');
plt.title('Cummulative Plot of EUR_index & NZD_index');
import statsmodels.api as sm
obs_mat = sm.add_constant(df[tickers[0]].values, prepend=False)[:, np.newaxis]
# y is 1-dimensional, (alpha, beta) is 2-dimensional
kf = KalmanFilter(n_dim_obs=1, n_dim_state=2,
initial_state_mean=np.ones(2),
initial_state_covariance=np.ones((2, 2)),
transition_matrices=np.eye(2),
observation_matrices=obs_mat,
observation_covariance=10**2,
transition_covariance=0.01**2 * np.eye(2))
state_means, state_covs = kf.filter(df[tickers[1]])
beta_kf = pd.DataFrame({'Slope': state_means[:, 0], 'Intercept': state_means[:, 1]},
index=df.index)
spread_kf = df[tickers[0]] - df[tickers[1]] * beta_kf['Slope'] - beta_kf['Intercept']
spread_kf = spread_kf
spread_kf.plot();
len(df)
df['spread'] = spread_kf
df['EUR/NZD'] = df['EUR_index']/df['NZD_index']
df['eSuperRCS'] = eSuperRCS(df['spread'])
df = df.iloc[-700:]
fig = plt.figure(figsize=(10, 7))
ax1, ax2 = fig.subplots(nrows=2, ncols=1)
ax1.plot(df.index, df['Close'],color='cyan' )
ax2.plot(df.index, df['EUR/NZD'].values, color='maroon')
ax1.set_title('EUR_NZD')
ax2.set_title('EUR/NZD')
plt.show()
def viewPlot(data, win = 150):
fig = plt.figure(figsize=(17, 10))
ax1, ax2 = fig.subplots(nrows=2, ncols=1)
df1 = data.iloc[-win:, ]
# High and Low prices are plotted
for i in range(len(df1)):
ax1.vlines(x = df1.index[i], ymin = df1.iat[i, 2], ymax = df1.iat[i, 1], color = 'magenta', linewidth = 2)
ax2.plot(df1.index, df1['eSuperRCS'].values, color='maroon')
ax2.axhline(55, color='green')
ax2.axhline(45, color='green')
ax2.axhline(50, color='orange')
ax1.set_title('EUR_NZD')
ax2.set_title('spread oscillator')
return plt.show()
viewPlot(df, win = 150)
```
|
github_jupyter
|
#@title Installation
!pip install git+https://github.com/yhilpisch/tpqoa.git --upgrade --quiet
!pip install pykalman --quiet
!pip install --upgrade mplfinance --quiet
#@title Imports
import tpqoa
import numpy as np
import pandas as pd
from pykalman import KalmanFilter
%matplotlib inline
from pylab import mpl, plt
plt.style.use('seaborn')
mpl.rcParams['savefig.dpi'] = 300
mpl.rcParams['font.family'] = 'serif'
from datetime import date, timedelta
import warnings
warnings.filterwarnings("ignore")
#@title Oanda API
path = '/content/drive/MyDrive/Oanda_Algo/pyalgo.cfg'
api = tpqoa.tpqoa(path)
#@title Symbols/Currency Pairs
def symbolsList():
symbols = []
syms = api.get_instruments()
for x in syms:
symbols.append(x[1])
return symbols
symbols = symbolsList()
pairs = ['AUD_CAD', 'AUD_CHF', 'AUD_JPY', 'AUD_NZD', 'AUD_USD', 'CAD_CHF',
'CAD_JPY', 'CHF_JPY', 'EUR_AUD', 'EUR_CAD', 'EUR_CHF', 'EUR_GBP',
'EUR_JPY', 'EUR_NZD', 'EUR_USD', 'GBP_AUD', 'GBP_CAD', 'GBP_CHF',
'GBP_JPY', 'GBP_NZD', 'GBP_USD', 'NZD_CAD', 'NZD_CHF', 'NZD_JPY',
'NZD_USD', 'USD_CAD', 'USD_CHF', 'USD_JPY',]
#@title getData(instr, gran = 'D', td=1000)
def getData(instr, gran = 'D', td=1000):
start = f"{date.today() - timedelta(td)}"
end = f"{date.today() - timedelta(1)}"
granularity = gran
price = 'M' # price: string one of 'A' (ask), 'B' (bid) or 'M' (middle)
data = api.get_history(instr, start, end, granularity, price)
data.drop(['complete'], axis=1, inplace=True)
data.reset_index(inplace=True)
data.rename(columns = {'time':'Date','o':'Open','c': 'Close', 'h':'High', 'l': 'Low'}, inplace = True)
data.set_index('Date', inplace=True)
return data
#@title Indexes
def USD_Index():
'''Creating a USD Index from a basket of instruments
denominated in dollars
'''
USD = ['EUR_USD', 'GBP_USD', 'USD_CAD', 'USD_CHF',
'USD_JPY', 'AUD_USD', 'NZD_USD']
df = pd.DataFrame()
for i in USD:
data = getData(i).ffill(axis='rows')
# setting the Dollar as the base
if '_USD' == i[-4:]:
data[f'{i}'] = (data['Close'])**(-1)
else:
data[f'{i}'] = data['Close']
df = pd.concat([df, data.loc[:,f'{i}']], axis=1)
df['US_index'] = 1
for i in range(len(USD)):
df['US_index'] *= df[USD[i]]
return ((df['US_index'])**(1/(len(USD)))).to_frame()
def EURO_Index():
'''Creating a EUR Index from a basket of instruments
denominated in EUROs
'''
EUR = ['EUR_USD', 'EUR_GBP', 'EUR_JPY', 'EUR_CHF', 'EUR_CAD',
'EUR_AUD', 'EUR_NZD']
df = pd.DataFrame()
for i in EUR:
data = getData(i).ffill(axis='rows')
data[f'{i}'] = data['Close']
df = pd.concat([df, data.loc[:,f'{i}']], axis=1)
df['EUR_index'] = 1
for i in range(len(EUR)):
df['EUR_index'] *= df[EUR[i]]
return ((df['EUR_index'])**(1/(len(EUR)))).to_frame()
def GBP_Index():
'''Creating a GBP Index from a basket of instruments
denominated in Pound Sterling
'''
GBP = ['GBP_USD', 'EUR_GBP', 'GBP_JPY', 'GBP_CHF', 'GBP_CAD',
'GBP_AUD', 'GBP_NZD']
df = pd.DataFrame()
for i in GBP:
data = getData(i).ffill(axis='rows')
# setting the Dollar as the base
if '_GBP' == i[-4:]:
data[f'{i}'] = (data['Close'])**(-1)
else:
data[f'{i}'] = data['Close']
df = pd.concat([df, data.loc[:,f'{i}']], axis=1)
df['GBP_index'] = 1
for i in range(len(GBP)):
df['GBP_index'] *= df[GBP[i]]
return ((df['GBP_index'])**(1/(len(GBP)))).to_frame()
def CHF_Index():
'''Creating a CHF Index from a basket of instruments
denominated in Swiss Francs
'''
CHF = ['CHF_JPY', 'EUR_CHF', 'GBP_CHF', 'USD_CHF', 'CAD_CHF',
'AUD_CHF', 'NZD_CHF']
df = pd.DataFrame()
for i in CHF:
data = getData(i).ffill(axis='rows')
# setting the Dollar as the base
if '_CHF' == i[-4:]:
data[f'{i}'] = (data['Close'])**(-1)
else:
data[f'{i}'] = data['Close']
df = pd.concat([df, data.loc[:,f'{i}']], axis=1)
df['CHF_index'] = 1
for i in range(len(CHF)):
df['CHF_index'] *= df[CHF[i]]
return ((df['CHF_index'])**(1/(len(CHF)))).to_frame()
def CAD_Index():
'''Creating a CAD Index from a basket of instruments
denominated in Canadian Dollars
'''
CAD = ['CAD_JPY', 'EUR_CAD', 'GBP_CAD', 'USD_CAD', 'CAD_CHF',
'AUD_CAD', 'NZD_CAD']
df = pd.DataFrame()
for i in CAD:
data = getData(i).ffill(axis='rows')
# setting the Dollar as the base
if '_CAD' == i[-4:]:
data[f'{i}'] = (data['Close'])**(-1)
else:
data[f'{i}'] = data['Close']
df = pd.concat([df, data.loc[:,f'{i}']], axis=1)
df['CAD_index'] = 1
for i in range(len(CAD)):
df['CAD_index'] *= df[CAD[i]]
return ((df['CAD_index'])**(1/(len(CAD)))).to_frame()
def JPY_Index():
'''Creating a JPY Index from a basket of instruments
denominated in Swiss Francs
'''
JPY = ['CAD_JPY', 'EUR_JPY', 'GBP_JPY', 'USD_JPY', 'CHF_JPY',
'AUD_JPY', 'NZD_JPY']
df = pd.DataFrame()
for i in JPY:
data = getData(i).ffill(axis='rows')
# setting the Japanese Yen as the base
data[f'{i}'] = (data['Close'])**(-1)
df = pd.concat([df, data.loc[:,f'{i}']], axis=1)
df['JPY_index'] = 1
for i in range(len(JPY)):
df['JPY_index'] *= df[JPY[i]]
return ((df['JPY_index'])**(1/(len(JPY)))).to_frame()
def AUD_Index():
'''Creating a AUD Index from a basket of instruments
denominated in Australian Dollar
'''
AUD = ['AUD_JPY', 'EUR_AUD', 'GBP_AUD', 'AUD_USD', 'AUD_CAD',
'AUD_CHF', 'AUD_NZD']
df = pd.DataFrame()
for i in AUD:
data = getData(i).ffill(axis='rows')
# setting the Aussie Dollar as the base
if '_AUD' == i[-4:]:
data[f'{i}'] = (data['Close'])**(-1)
else:
data[f'{i}'] = data['Close']
df = pd.concat([df, data.loc[:,f'{i}']], axis=1)
df['AUD_index'] = 1
for i in range(len(AUD)):
df['AUD_index'] *= df[AUD[i]]
return ((df['AUD_index'])**(1/(len(AUD)))).to_frame()
def NZD_Index():
'''Creating a NZD Index from a basket of instruments
denominated in New Zealand Dollar
'''
NZD = ['NZD_JPY', 'EUR_NZD', 'GBP_NZD', 'NZD_USD', 'NZD_CAD',
'NZD_CHF', 'AUD_NZD']
df = pd.DataFrame()
for i in NZD:
data = getData(i).ffill(axis='rows')
# setting the Dollar as the base
if '_NZD' == i[-4:]:
data[f'{i}'] = (data['Close'])**(-1)
else:
data[f'{i}'] = data['Close']
df = pd.concat([df, data.loc[:,f'{i}']], axis=1)
df['NZD_index'] = 1
for i in range(len(NZD)):
df['NZD_index'] *= df[ NZD[i]]
return ((df['NZD_index'])**(1/(len(NZD)))).to_frame()
def eSuperRCS(df):
"""
This code computes the super smoother introduced by John Ehlers
"""
spr = df.to_frame().copy()
# HighPass filter cyclic components whose periods are shorter than 48 bars
alpha1 = (np.cos(0.707*2*np.pi/48) + np.sin(0.707*2*np.pi/48) - 1)/np.cos(0.707*2*np.pi/48)
hiPass = pd.DataFrame(None, index=spr.index, columns=['filtered'])
for i in range(len(spr)):
if i < 3:
hiPass.iloc[i, 0] = spr.iat[i, 0]
else:
hiPass.iloc[i, 0] = ((1 - alpha1/2)*(1 - alpha1/2)*(spr.iat[i, 0]
- 2*spr.iat[i-1, 0] + spr.iat[i-2, 0] + 2*(1 - alpha1)*hiPass.iat[i-1, 0]
- (1 - alpha1)**2 *hiPass.iat[i-2, 0]))
# SuperSmoother
a1 = np.exp(-1.414*(np.pi) / 10)
b1 = 2*a1*np.cos(1.414*(np.pi) / 10)
c2 = b1
c3 = -a1*a1
c1 = 1 - c2 - c3
Filt = pd.DataFrame(None, index=spr.index, columns=['filtered'])
for i in range(len(spr)):
if i < 3:
Filt.iloc[i, 0] = hiPass.iat[i, 0]
else:
Filt.iloc[i, 0] = c1*(hiPass.iat[i, 0] + hiPass.iat[i - 1, 0]/ 2 + c2*Filt.iat[i-1, 0] + c3*Filt.iat[i-2, 0])
Filt['eSuperRCS'] = RSI(Filt['filtered'])
return Filt['eSuperRCS']
def RSI(series, period=25):
delta = series.diff()
up = delta.clip(lower=0)
dn = -1*delta.clip(upper=0)
ema_up = up.ewm(com=period-1, adjust=False).mean()
ewm_dn = dn.ewm(com=period-1, adjust=False).mean()
rs = (ema_up/ewm_dn)
return 100 - 100 / (1 + rs)
def will_pr(data, lb=14):
df = data[['High', 'Low', 'Close']].copy()
df['max_hi'] = data['High'].rolling(window=lb).max()
df['min_lo'] = data['Low'].rolling(window=lb).min()
df['will_pr'] = 0
for i in range(len(df)):
try:
df.iloc[i, 5] = ((df.iat[i, 3] - df.iat[i, 2])/(df.iat[i, 3] - df.iat[i, 4])) * (-100)
except ValueError:
pass
return df['will_pr']
en = getData('EUR_NZD')
eur = EURO_Index()
nzd = NZD_Index()
df = pd.concat((en, eur, nzd), axis=1).ffill(axis='rows')
df
tickers = ['EUR_index', 'NZD_index']
cumm_rtn = (1 + df[tickers].pct_change()).cumprod()
cumm_rtn.plot();
plt.ylabel('Cumulative Return');
plt.xlabel('Time');
plt.title('Cummulative Plot of EUR_index & NZD_index');
import statsmodels.api as sm
obs_mat = sm.add_constant(df[tickers[0]].values, prepend=False)[:, np.newaxis]
# y is 1-dimensional, (alpha, beta) is 2-dimensional
kf = KalmanFilter(n_dim_obs=1, n_dim_state=2,
initial_state_mean=np.ones(2),
initial_state_covariance=np.ones((2, 2)),
transition_matrices=np.eye(2),
observation_matrices=obs_mat,
observation_covariance=10**2,
transition_covariance=0.01**2 * np.eye(2))
state_means, state_covs = kf.filter(df[tickers[1]])
beta_kf = pd.DataFrame({'Slope': state_means[:, 0], 'Intercept': state_means[:, 1]},
index=df.index)
spread_kf = df[tickers[0]] - df[tickers[1]] * beta_kf['Slope'] - beta_kf['Intercept']
spread_kf = spread_kf
spread_kf.plot();
len(df)
df['spread'] = spread_kf
df['EUR/NZD'] = df['EUR_index']/df['NZD_index']
df['eSuperRCS'] = eSuperRCS(df['spread'])
df = df.iloc[-700:]
fig = plt.figure(figsize=(10, 7))
ax1, ax2 = fig.subplots(nrows=2, ncols=1)
ax1.plot(df.index, df['Close'],color='cyan' )
ax2.plot(df.index, df['EUR/NZD'].values, color='maroon')
ax1.set_title('EUR_NZD')
ax2.set_title('EUR/NZD')
plt.show()
def viewPlot(data, win = 150):
fig = plt.figure(figsize=(17, 10))
ax1, ax2 = fig.subplots(nrows=2, ncols=1)
df1 = data.iloc[-win:, ]
# High and Low prices are plotted
for i in range(len(df1)):
ax1.vlines(x = df1.index[i], ymin = df1.iat[i, 2], ymax = df1.iat[i, 1], color = 'magenta', linewidth = 2)
ax2.plot(df1.index, df1['eSuperRCS'].values, color='maroon')
ax2.axhline(55, color='green')
ax2.axhline(45, color='green')
ax2.axhline(50, color='orange')
ax1.set_title('EUR_NZD')
ax2.set_title('spread oscillator')
return plt.show()
viewPlot(df, win = 150)
| 0.293911 | 0.728145 |
```
import sys
import os
module_path = os.path.abspath(os.path.join('..'))
if module_path not in sys.path:
sys.path.append(module_path + "/src/simulations_v2")
from stochastic_simulation import StochasticSimulation
import numpy as np
from scipy.stats import poisson
def E_dist(max_time, mean_time):
pmf = list()
for i in range(1, max_time):
pmf.append(poisson.pmf(i, mean_time))
pmf.append(1-np.sum(pmf))
return np.array(pmf)
dist = E_dist(1, 1)
params = {
'max_time_exposed': 1,
'exposed_time_function': (lambda n: np.random.multinomial(n, dist)),
'max_time_pre_ID': 2,
'pre_ID_time_function': (lambda n: np.random.multinomial(n, np.array([0.8, 0.2]))),
'max_time_ID': 10,
'ID_time_function': (lambda n: np.random.multinomial(n, [0]*3+[1/7]*7)),
'sample_QI_exit_function': (lambda n: np.random.binomial(n, 0.05)),
'sample_QS_exit_function': (lambda n: np.random.binomial(n, 0.3)),
'exposed_infection_p': 0.1,
'expected_contacts_per_day': 3,
'days_between_tests': 1,
'test_population_fraction': 1/6,
'test_protocol_QFNR': 0.1,
'test_protocol_QFPR': 0.005,
'perform_contact_tracing': False,
'contact_tracing_constant': None,
'pre_ID_state': 'detectable',
'population_size': 34000,
'initial_E_count': 0,
'initial_pre_ID_count': 10,
'initial_ID_count': 0
}
sim = StochasticSimulation(params)
dfs = []
for _ in range(100):
dfs.append(sim.run_new_trajectory(250))
new_params = params.copy()
new_params['days_between_tests'] = 6
new_params['test_population_fraction'] = 1
new_sim = StochasticSimulation(new_params)
new_dfs = []
for _ in range(100):
new_dfs.append(new_sim.run_new_trajectory(250))
params_trace = params.copy()
params_trace['perform_contact_tracing'] = True
params_trace['contact_tracing_constant'] = 10
sim_trace = StochasticSimulation(params_trace)
dfs_trace = []
for _ in range(100):
dfs_trace.append(sim_trace.run_new_trajectory(250))
new_params_trace = params_trace.copy()
new_params_trace['days_between_tests'] = 6
new_params_trace['test_population_fraction'] = 1
new_sim_trace = StochasticSimulation(new_params_trace)
new_dfs_trace = []
for _ in range(100):
new_dfs_trace.append(new_sim_trace.run_new_trajectory(250))
params_more_test = params.copy()
params_more_test['test_population_fraction'] = 1/3
sim_more_test = StochasticSimulation(params_more_test)
dfs_more_test = []
for _ in range(100):
dfs_more_test.append(sim_more_test.run_new_trajectory(250))
new_params_more_test = params_more_test.copy()
new_params_more_test['days_between_tests'] = 3
new_params_more_test['test_population_fraction'] = 1
new_sim_more_test = StochasticSimulation(new_params_more_test)
new_dfs_more_test = []
for _ in range(100):
new_dfs_more_test.append(new_sim_more_test.run_new_trajectory(250))
import matplotlib.pyplot as plt
%matplotlib inline
plt.rcParams['font.size'] = 12
def add_plot(df, params, color='blue'):
cols = ['ID_{}'.format(x) for x in range(params['max_time_ID'])] + \
['pre_ID_{}'.format(x) for x in range(params['max_time_pre_ID'])]
plt.plot(df[cols].sum(axis=1), linewidth=10.0, alpha=0.1, color=color)
plt.figure(figsize=(20,12))
plt.subplot(2,1,1)
for df in dfs:
add_plot(df, params)
plt.subplot(2,1,2)
for df in new_dfs:
add_plot(df, params, color='green')
plt.show()
import matplotlib.pyplot as plt
%matplotlib inline
plt.rcParams['font.size'] = 12
def add_plot(df, params, color='blue'):
cols = ['ID_{}'.format(x) for x in range(params['max_time_ID'])] + \
['pre_ID_{}'.format(x) for x in range(params['max_time_pre_ID'])]
plt.plot(df[cols].sum(axis=1), linewidth=10.0, alpha=0.1, color=color)
plt.figure(figsize=(20,12))
plt.subplot(2,1,1)
for df in dfs_trace:
add_plot(df, params)
plt.subplot(2,1,2)
for df in new_dfs_trace:
add_plot(df, params, color='green')
plt.show()
import matplotlib.pyplot as plt
%matplotlib inline
plt.rcParams['font.size'] = 12
def add_plot(df, params, color='blue'):
cols = ['ID_{}'.format(x) for x in range(params['max_time_ID'])] + \
['pre_ID_{}'.format(x) for x in range(params['max_time_pre_ID'])]
plt.plot(df[cols].sum(axis=1), linewidth=10.0, alpha=0.1, color=color)
plt.figure(figsize=(20,12))
plt.subplot(2,1,1)
for df in dfs_more_test:
add_plot(df, params)
plt.subplot(2,1,2)
for df in new_dfs_more_test:
add_plot(df, params, color='green')
plt.show()
new_dfs[0].head(n=50)
df = dfs[0]
import matplotlib.pyplot as plt
%matplotlib inline
plt.plot(df[['ID_0','ID_1']].sum(axis=1))
df = sim.run_new_trajectory(50)
df
sim.sim_df
sim.step()
sim.sim_df
data = sim.get_current_state_vector()
labels = sim.get_state_vector_labels()
sim.E
labels
labels
import pandas as pd
df = pd.DataFrame(columns=labels)
newdf = pd.DataFrame([data], columns=labels)
df
newdf
df.append(newdf)
df
new_df = pd.DataFrame([data], columns=labels)
df.append(new_df)
df
df.concat(pd.DataFrame(data))
import numpy as np
a = np.array([1,2,4])
b = [2]
c = np.array([1,3,3])
d = np.concatenate([a,b,c])
d
np.random.binomial(10,0.3)
np.random.binomial(np.array([0,6,10]), 0.3)
a = np.array([1,2,3])
b = np.array([2,2,2])
c = np.array([4,4,4])
d = c - a
d - b
b = np.minimum(d,b)
b
min(b)
np.random.choice([0,3,4], size=6, replace=False)
```
|
github_jupyter
|
import sys
import os
module_path = os.path.abspath(os.path.join('..'))
if module_path not in sys.path:
sys.path.append(module_path + "/src/simulations_v2")
from stochastic_simulation import StochasticSimulation
import numpy as np
from scipy.stats import poisson
def E_dist(max_time, mean_time):
pmf = list()
for i in range(1, max_time):
pmf.append(poisson.pmf(i, mean_time))
pmf.append(1-np.sum(pmf))
return np.array(pmf)
dist = E_dist(1, 1)
params = {
'max_time_exposed': 1,
'exposed_time_function': (lambda n: np.random.multinomial(n, dist)),
'max_time_pre_ID': 2,
'pre_ID_time_function': (lambda n: np.random.multinomial(n, np.array([0.8, 0.2]))),
'max_time_ID': 10,
'ID_time_function': (lambda n: np.random.multinomial(n, [0]*3+[1/7]*7)),
'sample_QI_exit_function': (lambda n: np.random.binomial(n, 0.05)),
'sample_QS_exit_function': (lambda n: np.random.binomial(n, 0.3)),
'exposed_infection_p': 0.1,
'expected_contacts_per_day': 3,
'days_between_tests': 1,
'test_population_fraction': 1/6,
'test_protocol_QFNR': 0.1,
'test_protocol_QFPR': 0.005,
'perform_contact_tracing': False,
'contact_tracing_constant': None,
'pre_ID_state': 'detectable',
'population_size': 34000,
'initial_E_count': 0,
'initial_pre_ID_count': 10,
'initial_ID_count': 0
}
sim = StochasticSimulation(params)
dfs = []
for _ in range(100):
dfs.append(sim.run_new_trajectory(250))
new_params = params.copy()
new_params['days_between_tests'] = 6
new_params['test_population_fraction'] = 1
new_sim = StochasticSimulation(new_params)
new_dfs = []
for _ in range(100):
new_dfs.append(new_sim.run_new_trajectory(250))
params_trace = params.copy()
params_trace['perform_contact_tracing'] = True
params_trace['contact_tracing_constant'] = 10
sim_trace = StochasticSimulation(params_trace)
dfs_trace = []
for _ in range(100):
dfs_trace.append(sim_trace.run_new_trajectory(250))
new_params_trace = params_trace.copy()
new_params_trace['days_between_tests'] = 6
new_params_trace['test_population_fraction'] = 1
new_sim_trace = StochasticSimulation(new_params_trace)
new_dfs_trace = []
for _ in range(100):
new_dfs_trace.append(new_sim_trace.run_new_trajectory(250))
params_more_test = params.copy()
params_more_test['test_population_fraction'] = 1/3
sim_more_test = StochasticSimulation(params_more_test)
dfs_more_test = []
for _ in range(100):
dfs_more_test.append(sim_more_test.run_new_trajectory(250))
new_params_more_test = params_more_test.copy()
new_params_more_test['days_between_tests'] = 3
new_params_more_test['test_population_fraction'] = 1
new_sim_more_test = StochasticSimulation(new_params_more_test)
new_dfs_more_test = []
for _ in range(100):
new_dfs_more_test.append(new_sim_more_test.run_new_trajectory(250))
import matplotlib.pyplot as plt
%matplotlib inline
plt.rcParams['font.size'] = 12
def add_plot(df, params, color='blue'):
cols = ['ID_{}'.format(x) for x in range(params['max_time_ID'])] + \
['pre_ID_{}'.format(x) for x in range(params['max_time_pre_ID'])]
plt.plot(df[cols].sum(axis=1), linewidth=10.0, alpha=0.1, color=color)
plt.figure(figsize=(20,12))
plt.subplot(2,1,1)
for df in dfs:
add_plot(df, params)
plt.subplot(2,1,2)
for df in new_dfs:
add_plot(df, params, color='green')
plt.show()
import matplotlib.pyplot as plt
%matplotlib inline
plt.rcParams['font.size'] = 12
def add_plot(df, params, color='blue'):
cols = ['ID_{}'.format(x) for x in range(params['max_time_ID'])] + \
['pre_ID_{}'.format(x) for x in range(params['max_time_pre_ID'])]
plt.plot(df[cols].sum(axis=1), linewidth=10.0, alpha=0.1, color=color)
plt.figure(figsize=(20,12))
plt.subplot(2,1,1)
for df in dfs_trace:
add_plot(df, params)
plt.subplot(2,1,2)
for df in new_dfs_trace:
add_plot(df, params, color='green')
plt.show()
import matplotlib.pyplot as plt
%matplotlib inline
plt.rcParams['font.size'] = 12
def add_plot(df, params, color='blue'):
cols = ['ID_{}'.format(x) for x in range(params['max_time_ID'])] + \
['pre_ID_{}'.format(x) for x in range(params['max_time_pre_ID'])]
plt.plot(df[cols].sum(axis=1), linewidth=10.0, alpha=0.1, color=color)
plt.figure(figsize=(20,12))
plt.subplot(2,1,1)
for df in dfs_more_test:
add_plot(df, params)
plt.subplot(2,1,2)
for df in new_dfs_more_test:
add_plot(df, params, color='green')
plt.show()
new_dfs[0].head(n=50)
df = dfs[0]
import matplotlib.pyplot as plt
%matplotlib inline
plt.plot(df[['ID_0','ID_1']].sum(axis=1))
df = sim.run_new_trajectory(50)
df
sim.sim_df
sim.step()
sim.sim_df
data = sim.get_current_state_vector()
labels = sim.get_state_vector_labels()
sim.E
labels
labels
import pandas as pd
df = pd.DataFrame(columns=labels)
newdf = pd.DataFrame([data], columns=labels)
df
newdf
df.append(newdf)
df
new_df = pd.DataFrame([data], columns=labels)
df.append(new_df)
df
df.concat(pd.DataFrame(data))
import numpy as np
a = np.array([1,2,4])
b = [2]
c = np.array([1,3,3])
d = np.concatenate([a,b,c])
d
np.random.binomial(10,0.3)
np.random.binomial(np.array([0,6,10]), 0.3)
a = np.array([1,2,3])
b = np.array([2,2,2])
c = np.array([4,4,4])
d = c - a
d - b
b = np.minimum(d,b)
b
min(b)
np.random.choice([0,3,4], size=6, replace=False)
| 0.178168 | 0.461138 |
# Logistic Regression Example
A logistic regression learning algorithm example using TensorFlow library.
This example is using the MNIST database of handwritten digits (http://yann.lecun.com/exdb/mnist/)
- Author: Aymeric Damien
- Project: https://github.com/aymericdamien/TensorFlow-Examples/
```
import tensorflow as tf
import matplotlib.pyplot as plt
plt.rcParams["figure.figsize"] = (15,9)
# Import MINST data
from tensorflow.examples.tutorials.mnist import input_data
mnist = input_data.read_data_sets("/tmp/data/", one_hot=True)
# Parameters
learning_rate = 0.01
training_epochs = 25
batch_size = 100
display_step = 1
# tf Graph Input
x = tf.placeholder(tf.float32, [None, 784]) # mnist data image of shape 28*28=784
y = tf.placeholder(tf.float32, [None, 10]) # 0-9 digits recognition => 10 classes
# Set model weights
W = tf.Variable(tf.zeros([784, 10]))
b = tf.Variable(tf.zeros([10]))
# Construct model
pred = tf.nn.softmax(tf.matmul(x, W) + b) # Softmax
# Minimize error using cross entropy
cost = tf.reduce_mean(-tf.reduce_sum(y*tf.log(pred), axis=1))
# Gradient Descent
optimizer = tf.train.RMSPropOptimizer(learning_rate).minimize(cost)
# Initialize the variables (i.e. assign their default value)
init = tf.global_variables_initializer()
# Start training
with tf.Session() as sess:
sess.run(init)
# Training cycle
for epoch in range(training_epochs):
avg_cost = 0.
total_batch = int(mnist.train.num_examples/batch_size)
# Loop over all batches
for i in range(total_batch):
batch_xs, batch_ys = mnist.train.next_batch(batch_size)
# Fit training using batch data
_, c = sess.run([optimizer, cost], feed_dict={x: batch_xs, y: batch_ys})
# Compute average loss
avg_cost += c / total_batch
# Display logs per epoch step
if (epoch+1) % display_step == 0:
print ("Epoch:", '%04d' % (epoch+1), "cost =", "{:.9f}".format(avg_cost))
print ("Optimization Finished!")
# Test model
correct_prediction = tf.equal(tf.argmax(pred, 1), tf.argmax(y, 1))
# Calculate accuracy for 3000 examples
accuracy = tf.reduce_mean(tf.cast(correct_prediction, tf.float32))
print ("Accuracy:", accuracy.eval({x: mnist.test.images[:3000], y: mnist.test.labels[:3000]}))
```
AdamOptimizer
```python
Epoch: 0001 cost = 0.361675045
Epoch: 0002 cost = 0.298970559
Epoch: 0003 cost = 0.288769366
Epoch: 0004 cost = 0.285408240
Epoch: 0005 cost = 0.283692228
Epoch: 0006 cost = 0.276906072
Epoch: 0007 cost = 0.276427931
Epoch: 0008 cost = nan
Epoch: 0009 cost = nan
Epoch: 0010 cost = nan
Epoch: 0011 cost = nan
Epoch: 0012 cost = nan
Epoch: 0013 cost = nan
Epoch: 0014 cost = nan
Epoch: 0015 cost = nan
Epoch: 0016 cost = nan
Epoch: 0017 cost = nan
Epoch: 0018 cost = nan
Epoch: 0019 cost = nan
Epoch: 0020 cost = nan
Epoch: 0021 cost = nan
Epoch: 0022 cost = nan
Epoch: 0023 cost = nan
Epoch: 0024 cost = nan
Epoch: 0025 cost = nan
Optimization Finished!
Accuracy: 0.0903333```
GradientDescentOptimizer
```python
Epoch: 0001 cost = 1.183696304
Epoch: 0002 cost = 0.665356710
Epoch: 0003 cost = 0.552879873
Epoch: 0004 cost = 0.498750605
Epoch: 0005 cost = 0.465504965
Epoch: 0006 cost = 0.442631221
Epoch: 0007 cost = 0.425511703
Epoch: 0008 cost = 0.412157585
Epoch: 0009 cost = 0.401433004
Epoch: 0010 cost = 0.392410344
Epoch: 0011 cost = 0.384753543
Epoch: 0012 cost = 0.378148372
Epoch: 0013 cost = 0.372378035
Epoch: 0014 cost = 0.367284291
Epoch: 0015 cost = 0.362740744
Epoch: 0016 cost = 0.358625480
Epoch: 0017 cost = 0.354877754
Epoch: 0018 cost = 0.351465592
Epoch: 0019 cost = 0.348312076
Epoch: 0020 cost = 0.345416591
Epoch: 0021 cost = 0.342766170
Epoch: 0022 cost = 0.340245704
Epoch: 0023 cost = 0.337928486
Epoch: 0024 cost = 0.335732542
Epoch: 0025 cost = 0.333686892
Optimization Finished!
Accuracy: 0.889
```
RMSPropOptimizer
|
github_jupyter
|
import tensorflow as tf
import matplotlib.pyplot as plt
plt.rcParams["figure.figsize"] = (15,9)
# Import MINST data
from tensorflow.examples.tutorials.mnist import input_data
mnist = input_data.read_data_sets("/tmp/data/", one_hot=True)
# Parameters
learning_rate = 0.01
training_epochs = 25
batch_size = 100
display_step = 1
# tf Graph Input
x = tf.placeholder(tf.float32, [None, 784]) # mnist data image of shape 28*28=784
y = tf.placeholder(tf.float32, [None, 10]) # 0-9 digits recognition => 10 classes
# Set model weights
W = tf.Variable(tf.zeros([784, 10]))
b = tf.Variable(tf.zeros([10]))
# Construct model
pred = tf.nn.softmax(tf.matmul(x, W) + b) # Softmax
# Minimize error using cross entropy
cost = tf.reduce_mean(-tf.reduce_sum(y*tf.log(pred), axis=1))
# Gradient Descent
optimizer = tf.train.RMSPropOptimizer(learning_rate).minimize(cost)
# Initialize the variables (i.e. assign their default value)
init = tf.global_variables_initializer()
# Start training
with tf.Session() as sess:
sess.run(init)
# Training cycle
for epoch in range(training_epochs):
avg_cost = 0.
total_batch = int(mnist.train.num_examples/batch_size)
# Loop over all batches
for i in range(total_batch):
batch_xs, batch_ys = mnist.train.next_batch(batch_size)
# Fit training using batch data
_, c = sess.run([optimizer, cost], feed_dict={x: batch_xs, y: batch_ys})
# Compute average loss
avg_cost += c / total_batch
# Display logs per epoch step
if (epoch+1) % display_step == 0:
print ("Epoch:", '%04d' % (epoch+1), "cost =", "{:.9f}".format(avg_cost))
print ("Optimization Finished!")
# Test model
correct_prediction = tf.equal(tf.argmax(pred, 1), tf.argmax(y, 1))
# Calculate accuracy for 3000 examples
accuracy = tf.reduce_mean(tf.cast(correct_prediction, tf.float32))
print ("Accuracy:", accuracy.eval({x: mnist.test.images[:3000], y: mnist.test.labels[:3000]}))
Epoch: 0001 cost = 0.361675045
Epoch: 0002 cost = 0.298970559
Epoch: 0003 cost = 0.288769366
Epoch: 0004 cost = 0.285408240
Epoch: 0005 cost = 0.283692228
Epoch: 0006 cost = 0.276906072
Epoch: 0007 cost = 0.276427931
Epoch: 0008 cost = nan
Epoch: 0009 cost = nan
Epoch: 0010 cost = nan
Epoch: 0011 cost = nan
Epoch: 0012 cost = nan
Epoch: 0013 cost = nan
Epoch: 0014 cost = nan
Epoch: 0015 cost = nan
Epoch: 0016 cost = nan
Epoch: 0017 cost = nan
Epoch: 0018 cost = nan
Epoch: 0019 cost = nan
Epoch: 0020 cost = nan
Epoch: 0021 cost = nan
Epoch: 0022 cost = nan
Epoch: 0023 cost = nan
Epoch: 0024 cost = nan
Epoch: 0025 cost = nan
Optimization Finished!
Accuracy: 0.0903333```
GradientDescentOptimizer
| 0.79534 | 0.981076 |
```
from IPython.display import SVG, display
svg_code = '''
<svg height="2024" width="2024">
<polygon points="1030,1045
1017,1060
1014,1079
1015,1098
1015,1117
1007,1136
994,1155
983,1174
975,1193
975,1212
980,1231
984,1250
986,1269
993,1288
995,1307
1001,1326
1005,1345
1007,1364
1014,1383
1015,1402
1016,1421
1017,1440
1023,1459
1027,1478
1033,1497
1037,1516
1040,1535
1046,1554
1049,1573
1055,1592
1058,1611
1065,1630
1069,1649
1075,1668
1087,1687
1106,1693
1125,1693
1144,1692
1163,1687
1181,1672
1200,1670
1219,1669
1238,1662
1254,1648
1263,1630
1269,1611
1269,1592
1266,1573
1262,1554
1261,1535
1260,1516
1257,1497
1251,1478
1249,1459
1243,1440
1238,1421
1234,1402
1228,1383
1220,1364
1214,1345
1207,1326
1204,1307
1196,1288
1190,1269
1185,1250
1178,1231
1174,1212
1167,1193
1164,1174
1154,1155
1143,1139
1124,1136
1106,1126
1089,1112
1079,1095
1072,1076
1060,1057
1045,1045" style="fill:lime;stroke:purple;stroke-width:1" />
</svg>
'''
display(SVG(svg_code))
%cd /content/drive/MyDrive/datasets/Data
!ls
```
# [7줄로 객채 영역분할 따라하기](https://towardsdatascience.com/custom-instance-segmentation-training-with-7-lines-of-code-ff340851e99b)
```
%cd /content/drive/MyDrive/Segmentation
'''의존 라이브러리 설치'''
# !pip install tensorflow>=2.0
# !pip install imgaug
!pip install pixellib
```
### 🔺 위의 [RESTART RUNTIME] 을 눌러줘야 아래에서 pixellib 사용 가능.
```
'''데이터 준비'''
%cd /content/drive/MyDrive/Segmentation
!wget https://github.com/ayoolaolafenwa/PixelLib/releases/download/1.0.0/Nature.zip
!unzip Nature.zip
import pixellib
from pixellib.custom_train import instance_custom_training
vis_img = instance_custom_training()
vis_img.load_dataset("Nature")
vis_img.visualize_sample()
'''모델 다운로드'''
%cd /content/drive/MyDrive/Segmentation
!wget https://github.com/ayoolaolafenwa/PixelLib/releases/download/1.2/mask_rcnn_coco.h5
'''모델을 데이터셋으로 학습'''
%cd /content/drive/MyDrive/Segmentation
import pixellib
from pixellib.custom_train import instance_custom_training
train_maskrcnn = instance_custom_training()
train_maskrcnn.modelConfig(network_backbone = "resnet101", num_classes= 2, batch_size = 2)
train_maskrcnn.load_pretrained_model("mask_rcnn_coco.h5")
train_maskrcnn.load_dataset("Nature")
train_maskrcnn.train_model(num_epochs = 300, augmentation=True, path_trained_models = "mask_rcnn_models")
!nvidia-smi
```
### 😱 5에폭에 30분이상 300에폭 30시간이상?
```
'''학습이 오래걸리니 학습된 모델 다운로드'''
%cd /content/drive/MyDrive/Segmentation
!wget -P mask_rcnn_models https://github.com/ayoolaolafenwa/PixelLib/releases/download/1.0.0/Nature_model_resnet101.h5
'''모델평가'''
%cd /content/drive/MyDrive/Segmentation
import pixellib
from pixellib.custom_train import instance_custom_training
train_maskrcnn = instance_custom_training()
train_maskrcnn.modelConfig(network_backbone = "resnet101", num_classes= 2)
train_maskrcnn.load_dataset("Nature")
train_maskrcnn.evaluate_model("mask_rcnn_models/Nature_model_resnet101.h5")
'''추론 샘플 다운로드'''
%cd /content/drive/MyDrive/Segmentation
!wget -O sample1.jpg https://github.com/ayoolaolafenwa/PixelLib/raw/master/Tutorials/Images/squirrel.jpg
!wget -O sample2.jpg https://upload.wikimedia.org/wikipedia/commons/f/f6/Mimicry_in_Nature_%283172759556%29.jpg
'''다운받은 모델로 추론하기'''
%cd /content/drive/MyDrive/Segmentation
import pixellib
from pixellib.instance import custom_segmentation
segment_image = custom_segmentation()
segment_image.inferConfig(num_classes= 2, class_names= ["BG", "butterfly", "squirrel"])
segment_image.load_model("mask_rcnn_models/Nature_model_resnet101.h5")
# segment_image.segmentImage("sample1.jpg", show_bboxes=True, output_image_name="sample_out.jpg")
segment_image.segmentImage("sample2.jpg", show_bboxes=True, output_image_name="sample2_out.jpg")
%cd /content/drive/MyDrive/Segmentation
from IPython.display import Image
# Image('/content/sample_out.jpg')
Image('/content/sample2_out.jpg')
'''학습한 모델로 추론하기'''
%cd /content/drive/MyDrive/Segmentation
import pixellib
from pixellib.instance import custom_segmentation
segment_image = custom_segmentation()
segment_image.inferConfig(num_classes= 2, class_names= ["BG", "butterfly", "squirrel"])
segment_image.load_model("./mask_rcnn_models/mask_rcnn_model.004-2.073580.h5")
segment_image.segmentImage("sample1.jpg", show_bboxes=False, output_image_name="sample_out.jpg")
%cd /content/drive/MyDrive/Segmentation
from IPython.display import Image
Image('./sample_out.jpg')
```
* 예제가 잘 나오면, 문제 데이터를 이용해 학습하려 했으나<br> 위와 같이 사전 학습모델이 이상한 출력을 보였기에 다시 학습 시켜봅니다.
* 그 다음 Mask R-CNN보다 좋아보이는 PointRend를 학습시켜보고 싶고 https://github.com/facebookresearch/detectron2/tree/main/projects/PointRend 이것도 학습에 gpu 8개 쓰네요.
* 그 다음 https://github.com/hustvl/QueryInst 이것도 해보고 싶었어요.
|
github_jupyter
|
from IPython.display import SVG, display
svg_code = '''
<svg height="2024" width="2024">
<polygon points="1030,1045
1017,1060
1014,1079
1015,1098
1015,1117
1007,1136
994,1155
983,1174
975,1193
975,1212
980,1231
984,1250
986,1269
993,1288
995,1307
1001,1326
1005,1345
1007,1364
1014,1383
1015,1402
1016,1421
1017,1440
1023,1459
1027,1478
1033,1497
1037,1516
1040,1535
1046,1554
1049,1573
1055,1592
1058,1611
1065,1630
1069,1649
1075,1668
1087,1687
1106,1693
1125,1693
1144,1692
1163,1687
1181,1672
1200,1670
1219,1669
1238,1662
1254,1648
1263,1630
1269,1611
1269,1592
1266,1573
1262,1554
1261,1535
1260,1516
1257,1497
1251,1478
1249,1459
1243,1440
1238,1421
1234,1402
1228,1383
1220,1364
1214,1345
1207,1326
1204,1307
1196,1288
1190,1269
1185,1250
1178,1231
1174,1212
1167,1193
1164,1174
1154,1155
1143,1139
1124,1136
1106,1126
1089,1112
1079,1095
1072,1076
1060,1057
1045,1045" style="fill:lime;stroke:purple;stroke-width:1" />
</svg>
'''
display(SVG(svg_code))
%cd /content/drive/MyDrive/datasets/Data
!ls
%cd /content/drive/MyDrive/Segmentation
'''의존 라이브러리 설치'''
# !pip install tensorflow>=2.0
# !pip install imgaug
!pip install pixellib
'''데이터 준비'''
%cd /content/drive/MyDrive/Segmentation
!wget https://github.com/ayoolaolafenwa/PixelLib/releases/download/1.0.0/Nature.zip
!unzip Nature.zip
import pixellib
from pixellib.custom_train import instance_custom_training
vis_img = instance_custom_training()
vis_img.load_dataset("Nature")
vis_img.visualize_sample()
'''모델 다운로드'''
%cd /content/drive/MyDrive/Segmentation
!wget https://github.com/ayoolaolafenwa/PixelLib/releases/download/1.2/mask_rcnn_coco.h5
'''모델을 데이터셋으로 학습'''
%cd /content/drive/MyDrive/Segmentation
import pixellib
from pixellib.custom_train import instance_custom_training
train_maskrcnn = instance_custom_training()
train_maskrcnn.modelConfig(network_backbone = "resnet101", num_classes= 2, batch_size = 2)
train_maskrcnn.load_pretrained_model("mask_rcnn_coco.h5")
train_maskrcnn.load_dataset("Nature")
train_maskrcnn.train_model(num_epochs = 300, augmentation=True, path_trained_models = "mask_rcnn_models")
!nvidia-smi
'''학습이 오래걸리니 학습된 모델 다운로드'''
%cd /content/drive/MyDrive/Segmentation
!wget -P mask_rcnn_models https://github.com/ayoolaolafenwa/PixelLib/releases/download/1.0.0/Nature_model_resnet101.h5
'''모델평가'''
%cd /content/drive/MyDrive/Segmentation
import pixellib
from pixellib.custom_train import instance_custom_training
train_maskrcnn = instance_custom_training()
train_maskrcnn.modelConfig(network_backbone = "resnet101", num_classes= 2)
train_maskrcnn.load_dataset("Nature")
train_maskrcnn.evaluate_model("mask_rcnn_models/Nature_model_resnet101.h5")
'''추론 샘플 다운로드'''
%cd /content/drive/MyDrive/Segmentation
!wget -O sample1.jpg https://github.com/ayoolaolafenwa/PixelLib/raw/master/Tutorials/Images/squirrel.jpg
!wget -O sample2.jpg https://upload.wikimedia.org/wikipedia/commons/f/f6/Mimicry_in_Nature_%283172759556%29.jpg
'''다운받은 모델로 추론하기'''
%cd /content/drive/MyDrive/Segmentation
import pixellib
from pixellib.instance import custom_segmentation
segment_image = custom_segmentation()
segment_image.inferConfig(num_classes= 2, class_names= ["BG", "butterfly", "squirrel"])
segment_image.load_model("mask_rcnn_models/Nature_model_resnet101.h5")
# segment_image.segmentImage("sample1.jpg", show_bboxes=True, output_image_name="sample_out.jpg")
segment_image.segmentImage("sample2.jpg", show_bboxes=True, output_image_name="sample2_out.jpg")
%cd /content/drive/MyDrive/Segmentation
from IPython.display import Image
# Image('/content/sample_out.jpg')
Image('/content/sample2_out.jpg')
'''학습한 모델로 추론하기'''
%cd /content/drive/MyDrive/Segmentation
import pixellib
from pixellib.instance import custom_segmentation
segment_image = custom_segmentation()
segment_image.inferConfig(num_classes= 2, class_names= ["BG", "butterfly", "squirrel"])
segment_image.load_model("./mask_rcnn_models/mask_rcnn_model.004-2.073580.h5")
segment_image.segmentImage("sample1.jpg", show_bboxes=False, output_image_name="sample_out.jpg")
%cd /content/drive/MyDrive/Segmentation
from IPython.display import Image
Image('./sample_out.jpg')
| 0.385722 | 0.246874 |
# ALENN - Replication Notebook
## Brock and Hommes (1998) Model, KDE
Donovan Platt
<br>
Mathematical Institute, University of Oxford
<br>
Institute for New Economic Thinking at the Oxford Martin School
<br>
<br>
Copyright (c) 2020, University of Oxford. All rights reserved.
<br>
Distributed under a BSD 3-Clause licence. See the accompanying LICENCE file for further details.
# 1. Modules and Packages
Load all required modules and packages.
```
# Import the ALENN ABM Estimation Package
import alenn
# Import Numerical Computation Libraries
import numpy as np
import pandas as pd
# Import General Mathematical Libraries
from scipy import stats
```
# 2. Estimation Experiments
Replication of the KDE experiments. Note that here we generate only a single Markov Chain as opposed to the 5 considered in the original paper.
## 2.1. Free Parameter Set 1
### Model Specification
```
# Specify the Simulated Data Characteristics
T_emp = 1000 # Pseudo-empirical series length
T_sim = 1000 # Length of each Monte Carlo replication
n = 100 # Number of Monte Carlo replications
# Specify the Pseudo-Empirical Data
empirical = alenn.models.brock_hommes(0, 0, -0.7, -0.4, 0.5, 0.3, 1.01, 0, 0.01, 10, 0.04, 10, T_emp, n, 1)[:, 0]
# Define the Candidate Model Function
def model(theta):
return alenn.models.brock_hommes(0, 0, theta[0], theta[1], theta[2], theta[3], 1.01, 0, 0.01, 10, 0.04, 10, T_sim, n, 7)
# Define Parameter Priors
priors = [stats.uniform(loc = -2.5, scale = 2.5).pdf,
stats.uniform(loc = -1.5, scale = 1.5).pdf,
stats.uniform(loc = 0, scale = 2.5).pdf,
stats.uniform(loc = 0, scale = 1.5).pdf]
# Define the Parameter Bounds
theta_lower = np.array([-2.5, -1.5, 0, 0])
theta_upper = np.array([0, 0, 2.5, 1.5])
```
### Posterior Specification
```
# Create a KDE Posterior Approximator Object (Uses Default Settings from the Paper)
posterior = alenn.kde.KDEPosterior()
# Add the Model, Priors, and Empirical Data to the Newly-created Object
posterior.set_model(model)
posterior.set_prior(priors)
posterior.load_data(empirical)
```
### Sampler Specification
```
# Create an Adaptive MCMC Sampler Object
sampler = alenn.mcmc.AdaptiveMCMC(K = 70, S = 10000)
# Add the Posterior Approximator and Parameter Ranges to the Newly-created Object
sampler.set_posterior(posterior)
sampler.set_initialisation_ranges(theta_lower, theta_upper)
# Initiate the Sampling Process
sampler.sample_posterior()
```
### Result Processing
```
# Process the Sampler Output
samples = sampler.process_samples(burn_in = 5000)
# Calculate the Posterior Mean
pos_mean = samples[:, :posterior.num_param].mean(axis = 0)
# Calculate the Posterior Standard Deviation
pos_std = samples[:, :posterior.num_param].std(axis = 0)
# Construct a Result Table
result_table = pd.DataFrame(np.array([pos_mean, pos_std]).transpose(), columns = ['Posterior Mean', 'Posterior Std. Dev.'])
result_table.index.name = 'Parameter'
result_table.index += 1
# Display the Result Table
print('Final Estimation Results:')
print('')
print(result_table)
```
## 2.2. Free Parameter Set 2
### Model Specification
```
# Specify the Simulated Data Characteristics
T_emp = 1000 # Pseudo-empirical series length
T_sim = 1000 # Length of each Monte Carlo replication
n = 100 # Number of Monte Carlo replications
# Specify the Pseudo-Empirical Data
empirical = alenn.models.brock_hommes(0, 0, 0.6, 0.65, 0.7, -0.55, 1.01, 0, 0.01, 10, 0.04, 10, T_emp, n, 1)[:, 0]
# Define the Candidate Model Function
def model(theta):
return alenn.models.brock_hommes(0, 0, theta[0], theta[1], theta[2], theta[3], 1.01, 0, 0.01, 10, 0.04, 10, T_sim, n, 7)
# Define Parameter Priors
priors = [stats.uniform(loc = 0, scale = 2.5).pdf,
stats.uniform(loc = 0, scale = 1.5).pdf,
stats.uniform(loc = 0, scale = 2.5).pdf,
stats.uniform(loc = -1.5, scale = 1.5).pdf]
# Define the Parameter Bounds
theta_lower = np.array([0, 0, 0, -1.5])
theta_upper = np.array([2.5, 1.5, 2.5, 0])
```
### Posterior Specification
```
# Create a KDE Posterior Approximator Object (Uses Default Settings from the Paper)
posterior = alenn.kde.KDEPosterior()
# Add the Model, Priors, and Empirical Data to the Newly-created Object
posterior.set_model(model)
posterior.set_prior(priors)
posterior.load_data(empirical)
```
### Sampler Specification
```
# Create an Adaptive MCMC Sampler Object
sampler = alenn.mcmc.AdaptiveMCMC(K = 70, S = 10000)
# Add the Posterior Approximator and Parameter Ranges to the Newly-created Object
sampler.set_posterior(posterior)
sampler.set_initialisation_ranges(theta_lower, theta_upper)
# Initiate the Sampling Process
sampler.sample_posterior()
```
### Result Processing
```
# Process the Sampler Output
samples = sampler.process_samples(burn_in = 5000)
# Calculate the Posterior Mean
pos_mean = samples[:, :posterior.num_param].mean(axis = 0)
# Calculate the Posterior Standard Deviation
pos_std = samples[:, :posterior.num_param].std(axis = 0)
# Construct a Result Table
result_table = pd.DataFrame(np.array([pos_mean, pos_std]).transpose(), columns = ['Posterior Mean', 'Posterior Std. Dev.'])
result_table.index.name = 'Parameter'
result_table.index += 1
# Display the Result Table
print('Final Estimation Results:')
print('')
print(result_table)
```
|
github_jupyter
|
# Import the ALENN ABM Estimation Package
import alenn
# Import Numerical Computation Libraries
import numpy as np
import pandas as pd
# Import General Mathematical Libraries
from scipy import stats
# Specify the Simulated Data Characteristics
T_emp = 1000 # Pseudo-empirical series length
T_sim = 1000 # Length of each Monte Carlo replication
n = 100 # Number of Monte Carlo replications
# Specify the Pseudo-Empirical Data
empirical = alenn.models.brock_hommes(0, 0, -0.7, -0.4, 0.5, 0.3, 1.01, 0, 0.01, 10, 0.04, 10, T_emp, n, 1)[:, 0]
# Define the Candidate Model Function
def model(theta):
return alenn.models.brock_hommes(0, 0, theta[0], theta[1], theta[2], theta[3], 1.01, 0, 0.01, 10, 0.04, 10, T_sim, n, 7)
# Define Parameter Priors
priors = [stats.uniform(loc = -2.5, scale = 2.5).pdf,
stats.uniform(loc = -1.5, scale = 1.5).pdf,
stats.uniform(loc = 0, scale = 2.5).pdf,
stats.uniform(loc = 0, scale = 1.5).pdf]
# Define the Parameter Bounds
theta_lower = np.array([-2.5, -1.5, 0, 0])
theta_upper = np.array([0, 0, 2.5, 1.5])
# Create a KDE Posterior Approximator Object (Uses Default Settings from the Paper)
posterior = alenn.kde.KDEPosterior()
# Add the Model, Priors, and Empirical Data to the Newly-created Object
posterior.set_model(model)
posterior.set_prior(priors)
posterior.load_data(empirical)
# Create an Adaptive MCMC Sampler Object
sampler = alenn.mcmc.AdaptiveMCMC(K = 70, S = 10000)
# Add the Posterior Approximator and Parameter Ranges to the Newly-created Object
sampler.set_posterior(posterior)
sampler.set_initialisation_ranges(theta_lower, theta_upper)
# Initiate the Sampling Process
sampler.sample_posterior()
# Process the Sampler Output
samples = sampler.process_samples(burn_in = 5000)
# Calculate the Posterior Mean
pos_mean = samples[:, :posterior.num_param].mean(axis = 0)
# Calculate the Posterior Standard Deviation
pos_std = samples[:, :posterior.num_param].std(axis = 0)
# Construct a Result Table
result_table = pd.DataFrame(np.array([pos_mean, pos_std]).transpose(), columns = ['Posterior Mean', 'Posterior Std. Dev.'])
result_table.index.name = 'Parameter'
result_table.index += 1
# Display the Result Table
print('Final Estimation Results:')
print('')
print(result_table)
# Specify the Simulated Data Characteristics
T_emp = 1000 # Pseudo-empirical series length
T_sim = 1000 # Length of each Monte Carlo replication
n = 100 # Number of Monte Carlo replications
# Specify the Pseudo-Empirical Data
empirical = alenn.models.brock_hommes(0, 0, 0.6, 0.65, 0.7, -0.55, 1.01, 0, 0.01, 10, 0.04, 10, T_emp, n, 1)[:, 0]
# Define the Candidate Model Function
def model(theta):
return alenn.models.brock_hommes(0, 0, theta[0], theta[1], theta[2], theta[3], 1.01, 0, 0.01, 10, 0.04, 10, T_sim, n, 7)
# Define Parameter Priors
priors = [stats.uniform(loc = 0, scale = 2.5).pdf,
stats.uniform(loc = 0, scale = 1.5).pdf,
stats.uniform(loc = 0, scale = 2.5).pdf,
stats.uniform(loc = -1.5, scale = 1.5).pdf]
# Define the Parameter Bounds
theta_lower = np.array([0, 0, 0, -1.5])
theta_upper = np.array([2.5, 1.5, 2.5, 0])
# Create a KDE Posterior Approximator Object (Uses Default Settings from the Paper)
posterior = alenn.kde.KDEPosterior()
# Add the Model, Priors, and Empirical Data to the Newly-created Object
posterior.set_model(model)
posterior.set_prior(priors)
posterior.load_data(empirical)
# Create an Adaptive MCMC Sampler Object
sampler = alenn.mcmc.AdaptiveMCMC(K = 70, S = 10000)
# Add the Posterior Approximator and Parameter Ranges to the Newly-created Object
sampler.set_posterior(posterior)
sampler.set_initialisation_ranges(theta_lower, theta_upper)
# Initiate the Sampling Process
sampler.sample_posterior()
# Process the Sampler Output
samples = sampler.process_samples(burn_in = 5000)
# Calculate the Posterior Mean
pos_mean = samples[:, :posterior.num_param].mean(axis = 0)
# Calculate the Posterior Standard Deviation
pos_std = samples[:, :posterior.num_param].std(axis = 0)
# Construct a Result Table
result_table = pd.DataFrame(np.array([pos_mean, pos_std]).transpose(), columns = ['Posterior Mean', 'Posterior Std. Dev.'])
result_table.index.name = 'Parameter'
result_table.index += 1
# Display the Result Table
print('Final Estimation Results:')
print('')
print(result_table)
| 0.716516 | 0.955319 |
# Predicting Heart Disease with ML
This notebook will use various python machine learning and datascience libraries and tools to build a algorithm that will attempt to predict if a person has heart disease based on their medical attributes
```
def what():
'''Info Function to list out process steps"what()" '''
print(f" 1. Problem definition: \n",
"2. Data retrival and exploration: \n",
"3. Evaluation Goal: \n",
"4. Feature Engineering: \n",
"5. Modeling: \n",
"6. Expirementation: \n"
)
what()
```
## Problem definition:
>Given clinical parameters about a patient attempt to predict if they have heart disease or not
### Data Description & Data Dictionary
>The Data used in this notebook comes from the Clevland Heart disease dataset availble from the UCI Machine Learning repository. This dataset contains 76 attributes, However the author the the dataset states all published experiments refer to using a subset of 14 of these attributes. This dataset too uses only these 14 attributes. The "goal" field refers to the presence of heart disease in the patient.
* age
* sex - (1 = male; 0 = female)
* cp - chest pain type (4 values)
* trestbps - resting blood pressure
* chol - serum cholestoral in mg/dl
* fbs - fasting blood sugar > 120 mg/dl
* restecg - resting electrocardiographic results (values 0,1,2)
* thalach - maximum heart rate achieved
* exang - exercise induced angina
* oldpeak - oldpeak = ST depression induced by exercise relative to rest
* slope - the slope of the peak exercise ST segment
* ca - number of major vessels (0-3) colored by flourosopy
* thal - thal: 3 = normal; 6 = fixed defect; 7 = reversable defect
* num/target
>https://archive.ics.uci.edu/ml/datasets/heart+disease
### Evaluation Goals
>Attempt to get 90% accuracy on data before moving to QA
### Feature Engineering
```
# Exploritoray and data analyasis and plotting libraries
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
%matplotlib inline
#models from scikit-learn
from sklearn.linear_model import LogisticRegression
from sklearn.neighbors import KNeighborsClassifier
from sklearn.ensemble import RandomForestClassifier
#Model Evaluations
from sklearn.model_selection import train_test_split, cross_val_score
from sklearn.metrics import confusion_matrix, classification_report
from sklearn.metrics import precision_score, recall_score, f1_score
from sklearn.metrics import plot_roc_curve
# Get Data
df = pd.read_csv("data/heart-disease.csv")
```
## Data Exploration
>Goal: Understand the Data
1. What questions are you trying to answer?
2. What kind of data do we have and how do we treat different types?
3. What's missing from the data and how do we deal with it?
4. What are the outliers in the data and why should we care?
5. How can we add change, or remove features to get more accuracy?
6. How many different classes does our data have?
```
df.shape
df
df.head()
df.tail()
df["target"].value_counts()
df["target"].value_counts().plot( kind="bar", color=["salmon", "lightblue"]);
df.info()
df.isna().sum()
df.describe()
```
### Heart Disease Frequency based on sex
```
df.sex.value_counts()
pd.crosstab(df.target, df.sex)
pd.crosstab(df.target, df.sex).plot(kind="bar", figsize=(10,6), color=["salmon", "lightblue"]);
plt.title("Heart Disease Frequency based on Sex")
plt.legend(["Male", "Female"])
plt.xlabel("0 = Heart Disease AND 1 = No Heart Disease")
plt.ylabel("Number of entries in Dataset")
plt.xticks(rotation= 0);
```
### SWAG or intuition based on the current data..
>_Total Female entries = 96
> * 76 have heart disease
> * 24 Do not have heart disease
> * This shows about 75% have heart disese in this dataset
>_Total Male entries = 207
> * 114 have heart disease
> * 96 Do not have heart disease
> * This shows about 50% have heart disease within this dataset
>_Total entries = 303
> * 165 have heart disease
> * 138 Do not have heart disease
> * This shows about 54% total have heart disease within this dataset
> These intuitions will be helpfuls as baselines later when we evaluate our models accuracy
### Age vs Max Heart Rate Achieved (thalach) Heart Disease correlation
```
plt.figure(figsize=(10,6));
# Create plot with positive entries
plt.scatter(df.age[df.target == 1],
df.thalach[df.target == 1],
color="salmon");
# Scatter with negative entries
plt.scatter(df.age[df.target == 0],
df.thalach[df.target == 0],
c='lightblue');
plt.title("Age vs Max Heart Rate For Heart Disease");
plt.xlabel("AGE");
plt.ylabel("Maximum heart rate achieved");
plt.legend(["Heart Disease","No Heart Disease"]);
#distribution of age
df.age.plot.hist();
```
### Heart Disease frequency per chest pain type breakdown
###### cp: chest pain type.
>1: typical angina.
>2: atypical angina.
>3: non-anginal pain.
>4: asymptomatic.
```
pd.crosstab(df.cp, df.target)
pd.crosstab(df.cp, df.target).plot(kind="bar", figsize=(10,6), color=["salmon","lightblue"]);
plt.title("Heart Disease frequency per chest pain type");
plt.xlabel("Chest Pain Types");
plt.ylabel("Amount");
plt.xticks(rotation="0");
plt.legend(["No Heart Disease", "Heart Disease"]);
```
### Check the correlation of all the features
```
df.corr()
## Plot the correlations
corr_matrix = df.corr()
fig, ax = plt.subplots(figsize=(15,10))
ax = sns.heatmap(corr_matrix,
annot = True,
linewidths = 0.5,
fmt = ".2f",
cmap = "YlGnBu");
bottom, top = ax.get_ylim();
ax.set_ylim(bottom + 0.5, top - 0.5);
```
|
github_jupyter
|
def what():
'''Info Function to list out process steps"what()" '''
print(f" 1. Problem definition: \n",
"2. Data retrival and exploration: \n",
"3. Evaluation Goal: \n",
"4. Feature Engineering: \n",
"5. Modeling: \n",
"6. Expirementation: \n"
)
what()
# Exploritoray and data analyasis and plotting libraries
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
%matplotlib inline
#models from scikit-learn
from sklearn.linear_model import LogisticRegression
from sklearn.neighbors import KNeighborsClassifier
from sklearn.ensemble import RandomForestClassifier
#Model Evaluations
from sklearn.model_selection import train_test_split, cross_val_score
from sklearn.metrics import confusion_matrix, classification_report
from sklearn.metrics import precision_score, recall_score, f1_score
from sklearn.metrics import plot_roc_curve
# Get Data
df = pd.read_csv("data/heart-disease.csv")
df.shape
df
df.head()
df.tail()
df["target"].value_counts()
df["target"].value_counts().plot( kind="bar", color=["salmon", "lightblue"]);
df.info()
df.isna().sum()
df.describe()
df.sex.value_counts()
pd.crosstab(df.target, df.sex)
pd.crosstab(df.target, df.sex).plot(kind="bar", figsize=(10,6), color=["salmon", "lightblue"]);
plt.title("Heart Disease Frequency based on Sex")
plt.legend(["Male", "Female"])
plt.xlabel("0 = Heart Disease AND 1 = No Heart Disease")
plt.ylabel("Number of entries in Dataset")
plt.xticks(rotation= 0);
plt.figure(figsize=(10,6));
# Create plot with positive entries
plt.scatter(df.age[df.target == 1],
df.thalach[df.target == 1],
color="salmon");
# Scatter with negative entries
plt.scatter(df.age[df.target == 0],
df.thalach[df.target == 0],
c='lightblue');
plt.title("Age vs Max Heart Rate For Heart Disease");
plt.xlabel("AGE");
plt.ylabel("Maximum heart rate achieved");
plt.legend(["Heart Disease","No Heart Disease"]);
#distribution of age
df.age.plot.hist();
pd.crosstab(df.cp, df.target)
pd.crosstab(df.cp, df.target).plot(kind="bar", figsize=(10,6), color=["salmon","lightblue"]);
plt.title("Heart Disease frequency per chest pain type");
plt.xlabel("Chest Pain Types");
plt.ylabel("Amount");
plt.xticks(rotation="0");
plt.legend(["No Heart Disease", "Heart Disease"]);
df.corr()
## Plot the correlations
corr_matrix = df.corr()
fig, ax = plt.subplots(figsize=(15,10))
ax = sns.heatmap(corr_matrix,
annot = True,
linewidths = 0.5,
fmt = ".2f",
cmap = "YlGnBu");
bottom, top = ax.get_ylim();
ax.set_ylim(bottom + 0.5, top - 0.5);
| 0.757166 | 0.97959 |
## Tutorial 3
In this tutorial we will learn how to
* Use dropout in fully connected networks
* Use custom datasets in PyTorch
* Implement KL divergence
So far, we used datasets stored in PyTorch datasets. What happens if we want to use different data?
```
import torch
from torch.utils.data import Dataset, DataLoader
from torch.utils.data.dataset import Dataset
class SampleDataset(Dataset):
def __init__(self):
pass
def __getitem__(self, index):
return (img, label)
def __len__(self):
return count
```
Let's take a look at a sample dataset
```
class SampleDataset(Dataset):
def __init__(self, n_features: int = 1024, n_samples: int = 1000):
self.n_features = n_features
self.n_samples = n_samples
self.entries = self._create_entries()
def _create_entries(self):
entries = []
for i in range(self.n_samples):
entries.append({'x': torch.randn(self.n_features), 'y': 1})
return entries
def __getitem__(self, index):
entry = self.entries[index]
return entry['x'], entry['y']
def __len__(self):
return len(self.entries)
```
Now, let's check that it works
```
sample_dataset = SampleDataset(n_features=5, n_samples=100)
sample_loader = DataLoader(sample_dataset, batch_size=4, shuffle=True, num_workers=0)
for x, y in sample_loader:
print(f'Input batch: {x}')
print(f'Label batch {y}')
break
```
## Dropout
Let's add dropout to the model you saw a week ago
```
import torchvision
from torch import nn
from torchvision import datasets, transforms
input_size = 784
num_classes = 10
num_epochs = 5
batch_size = 100
learning_rate = 0.001
# MNIST Dataset (Images and Labels)
transform = transforms.Compose([transforms.ToTensor(), transforms.Normalize(mean=[0.3081,],std=[0.1306,])])
train_dataset = datasets.MNIST(root='./data', train=True, transform=transform, download=True)
test_dataset = datasets.MNIST(root='./data', train=False, transform=transform, download=True)
# Dataset Loader (Input Pipline)
train_loader = torch.utils.data.DataLoader(dataset=train_dataset, batch_size=batch_size, shuffle=True)
test_loader = torch.utils.data.DataLoader(dataset=test_dataset, batch_size=batch_size, shuffle=False)
class TwoLayers(nn.Module):
def __init__(self, input_size, output_size):
super(TwoLayers, self).__init__()
self.linear1 = nn.Linear(input_size, 100)
self.linear2 = nn.Linear(100, output_size)
# The only difference from the previous TA
self.dropout = nn.Dropout(0.5)
def forward(self, x):
out = self.linear1(x)
out = self.dropout(out)
return self.linear2(torch.tanh(out))
model = TwoLayers(input_size, num_classes)
# Loss and Optimizer
# Softmax is internally computed.
ce_loss = nn.CrossEntropyLoss()
optimizer = torch.optim.SGD(model.parameters(), lr=learning_rate)
param= [i.nelement() for i in model.parameters()]
print ("number of parameters: ", sum(param))
for epoch in range(num_epochs):
for i, (images, labels) in enumerate(train_loader):
images = images.view(-1, 28*28)
# Forward + Backward + Optimize
optimizer.zero_grad()
outputs = model(images)
loss = ce_loss(outputs, labels)
loss.backward()
optimizer.step()
if i % 100 == 0:
print ('Epoch: [{}/{}], Step: [{}/{}], Loss: {:.4}'.format(epoch+1, num_epochs,
i+1, len(train_dataset)//batch_size,
loss.item()))
model.eval()
correct = 0
total = 0
for images, labels in test_loader:
images = images.view(-1, 28*28)
outputs = model(images)
predicted = torch.argmax(outputs, 1)
total += labels.size(0)
correct += (predicted == labels).sum()
print('Accuracy of the model on the 10000 test images: ', float(correct) / total)
torch.save(model.state_dict(), 'model.pkl')
```
## KL divergence
```
P = torch.Tensor([0.36, 0.48, 0.16])
Q = torch.Tensor([0.333, 0.333, 0.333])
(P * (P / Q).log()).sum()
import torch.nn.functional as F
F.kl_div(Q.log(), P, None, None, 'sum')
```
|
github_jupyter
|
import torch
from torch.utils.data import Dataset, DataLoader
from torch.utils.data.dataset import Dataset
class SampleDataset(Dataset):
def __init__(self):
pass
def __getitem__(self, index):
return (img, label)
def __len__(self):
return count
class SampleDataset(Dataset):
def __init__(self, n_features: int = 1024, n_samples: int = 1000):
self.n_features = n_features
self.n_samples = n_samples
self.entries = self._create_entries()
def _create_entries(self):
entries = []
for i in range(self.n_samples):
entries.append({'x': torch.randn(self.n_features), 'y': 1})
return entries
def __getitem__(self, index):
entry = self.entries[index]
return entry['x'], entry['y']
def __len__(self):
return len(self.entries)
sample_dataset = SampleDataset(n_features=5, n_samples=100)
sample_loader = DataLoader(sample_dataset, batch_size=4, shuffle=True, num_workers=0)
for x, y in sample_loader:
print(f'Input batch: {x}')
print(f'Label batch {y}')
break
import torchvision
from torch import nn
from torchvision import datasets, transforms
input_size = 784
num_classes = 10
num_epochs = 5
batch_size = 100
learning_rate = 0.001
# MNIST Dataset (Images and Labels)
transform = transforms.Compose([transforms.ToTensor(), transforms.Normalize(mean=[0.3081,],std=[0.1306,])])
train_dataset = datasets.MNIST(root='./data', train=True, transform=transform, download=True)
test_dataset = datasets.MNIST(root='./data', train=False, transform=transform, download=True)
# Dataset Loader (Input Pipline)
train_loader = torch.utils.data.DataLoader(dataset=train_dataset, batch_size=batch_size, shuffle=True)
test_loader = torch.utils.data.DataLoader(dataset=test_dataset, batch_size=batch_size, shuffle=False)
class TwoLayers(nn.Module):
def __init__(self, input_size, output_size):
super(TwoLayers, self).__init__()
self.linear1 = nn.Linear(input_size, 100)
self.linear2 = nn.Linear(100, output_size)
# The only difference from the previous TA
self.dropout = nn.Dropout(0.5)
def forward(self, x):
out = self.linear1(x)
out = self.dropout(out)
return self.linear2(torch.tanh(out))
model = TwoLayers(input_size, num_classes)
# Loss and Optimizer
# Softmax is internally computed.
ce_loss = nn.CrossEntropyLoss()
optimizer = torch.optim.SGD(model.parameters(), lr=learning_rate)
param= [i.nelement() for i in model.parameters()]
print ("number of parameters: ", sum(param))
for epoch in range(num_epochs):
for i, (images, labels) in enumerate(train_loader):
images = images.view(-1, 28*28)
# Forward + Backward + Optimize
optimizer.zero_grad()
outputs = model(images)
loss = ce_loss(outputs, labels)
loss.backward()
optimizer.step()
if i % 100 == 0:
print ('Epoch: [{}/{}], Step: [{}/{}], Loss: {:.4}'.format(epoch+1, num_epochs,
i+1, len(train_dataset)//batch_size,
loss.item()))
model.eval()
correct = 0
total = 0
for images, labels in test_loader:
images = images.view(-1, 28*28)
outputs = model(images)
predicted = torch.argmax(outputs, 1)
total += labels.size(0)
correct += (predicted == labels).sum()
print('Accuracy of the model on the 10000 test images: ', float(correct) / total)
torch.save(model.state_dict(), 'model.pkl')
P = torch.Tensor([0.36, 0.48, 0.16])
Q = torch.Tensor([0.333, 0.333, 0.333])
(P * (P / Q).log()).sum()
import torch.nn.functional as F
F.kl_div(Q.log(), P, None, None, 'sum')
| 0.935649 | 0.972934 |
```
%%capture
# Comment this out if you don't want to install pennylane from this notebook
!pip install pennylane
# Comment this out if you don't want to install matplotlib from this notebook
!pip install matplotlib
```
# Differentiable quantum computing with PennyLane
In this tutorial we will:
* learn step-by-step how quantum computations are implemented in PennyLane,
* understand parameter-dependent quantum computations ("variational circuits"),
* build our first quantum machine learning model, and
* compute its gradient.
We need the following imports:
```
import pennylane as qml
from pennylane import numpy as np
```
## 1. Quantum nodes
In PennyLane, a *quantum node* is a computational unit that involves the construction, evaluation, pre- and postprocessing of quantum computations.
A quantum node consists of a *quantum function* that defines a circuit, as well as a *device* on which it is run.
There is a growing [device ecosystem](https://pennylane.ai/plugins.html) which allows you to change only one line of code to dispatch your quantum computation to local simulators, remote simulators and remote hardware from different vendors.
Here we will use the built-in `default.qubit` device.
```
dev = qml.device('default.qubit', wires=2)
```
To combine the device with a quantum function to a quantum node we can use the `qml.qnode` decorator. The function can then be evaluated as if it was any other python function. Internally, it will construct a circuit and run it on the device.
```
@qml.qnode(dev)
def circuit():
qml.Hadamard(wires=0)
return qml.probs(wires=[0, 1])
circuit()
```
## 2. Building quantum circuits
### The initial state
<br />
<img src="figures/1.png" width="500" height="100">
<br />
The initial state has 100% probability to be measured in the "0..0" configuration. Let's see how we can verify this with PennyLane.
```
@qml.qnode(dev)
def circuit():
return qml.probs(wires=[0, 1])
circuit()
```
The internal state vector that we use to mathematically keep track of probabilities is complex-valued. Since `default.qubit` is a simulator we can have a look at the state, for example by checking the device's `state` attribute.
```
dev.state
```
### Unitary evolutions
<br />
<img src="figures/2.png" width="500">
<br />
Quantum circuits are represented by unitary matrices. We can evolve the initial state by an arbitrary unitrary matrix as follows:
```
s = 1/np.sqrt(2)
U = np.array([[0., -s, 0., s],
[ s, 0., -s, 0.],
[ s, 0., s, 0.],
[0., -s, 0., -s]])
@qml.qnode(dev)
def circuit():
qml.QubitUnitary(U, wires=[0, 1])
return qml.probs(wires=[0, 1])
circuit()
```
The internal quantum state changed.
```
dev.state
```
### Measurements sample outcomes from the distribution
<br />
<img src="figures/3.png" width="500">
<br />
The most common measurement takes samples $-1, 1$ from the "Pauli-Z" observable. The samples indicate if the qubit was measured in state $| 0 \rangle$ or $| 1 \rangle$.
```
@qml.qnode(dev)
def circuit():
qml.QubitUnitary(U, wires=[0, 1])
return qml.sample(qml.PauliZ(wires=0)), qml.sample(qml.PauliZ(wires=1))
circuit()
```
The quantum state should be still the same as above.
```
dev.state
```
### Computing expectation values
<br />
<img src="figures/4.png" width="500">
<br />
When we want outputs of computations to be deterministic, we often interpret the expected measurement outcome as the result. This value is estimated by taking lots of samples and averaging over them.
```
@qml.qnode(dev)
def circuit():
qml.QubitUnitary(U, wires=[0, 1])
return qml.expval(qml.PauliZ(wires=0)), qml.expval(qml.PauliZ(wires=1))
circuit()
```
Again, the quantum state should be the same as above.
```
dev.state
```
### Quantum circuits are decomposed into gates
<br />
<img src="figures/5.png" width="500">
<br />
Quantum circuits rarely consist of one large unitary (which quickly becomes intractably large as the number of qubits grow). Instead, they are composed of *quantum gates*.
```
@qml.qnode(dev)
def circuit():
qml.PauliX(wires=0)
qml.CNOT(wires=[0,1])
qml.Hadamard(wires=0)
qml.PauliZ(wires=1)
return qml.expval(qml.PauliZ(wires=0)), qml.expval(qml.PauliZ(wires=1))
circuit()
```
### Some gates depend on "control" parameters
<br />
<img src="figures/6.png" width="500">
<br />
To train circuits, there is a special subset of gates which is of particular interest: the Pauli rotation gates. These "rotate" a special representation of the quantum state around a specific axis. The gates depend on a scalar parameter which is the angle of the rotation.
```
@qml.qnode(dev)
def circuit(w1, w2):
qml.RX(w1, wires=0)
qml.CNOT(wires=[0,1])
qml.RY(w2, wires=1)
return qml.expval(qml.PauliZ(wires=0)), qml.expval(qml.PauliZ(wires=1))
circuit(0.2, 1.3)
```
The names `w1`, `w2` are already suggestive that these can be used like the trainable parameters of a classical machine learning model. But we could also call the control parameters `x1`, `x2` and encode data features into quantum states.
## 3. A full quantum machine learning model and its gradient
Finally, we can use pre-coded routines or [templates](https://pennylane.readthedocs.io/en/stable/introduction/templates.html) to conveniently build full quantum machine learning model that include a data encoding part, and a trainable part.
<br />
<img src="figures/7.png" width="500">
<br />
Here, we will use the `AngleEmbedding` template to load the data, and the `BasicEntanglingLayers` as the trainable part of the circuit.
```
@qml.qnode(dev)
def quantum_model(x, w):
qml.templates.AngleEmbedding(x, wires=[0, 1])
qml.templates.BasicEntanglerLayers(w, wires=[0, 1])
return qml.expval(qml.PauliZ(wires=0))
x = np.array([0.1, 0.2], requires_grad=False)
w = np.array([[-2.1, 1.2], [-1.4, -3.9], [0.5, 0.2]])
quantum_model(x, w)
```
We can draw the circuit.
```
print(quantum_model.draw())
```
The best thing is that by using PennyLane, we can easily compute its gradient!
```
gradient_fn = qml.grad(quantum_model)
gradient_fn(x, w)
```
This allows us to slot the quantum circuit into the machine learning example from the previous notebook.
# TASKS
1. Copy and paste the code from the previous notebook to here and replace the classical model by
the `quantum_model` function. This will allow you to train the model!
2. Add a bias term to the quantum model.
3. Replace the hand-coded optimisation step by a native [PennyLane optimiser](https://pennylane.readthedocs.io/en/stable/introduction/optimizers.html).
4. Rewrite the entire example in PyTorch.
Tipp: You must set the qnode to the correct interface via `@qml.qnode(dev, interface='tf')`.
```
import matplotlib.pyplot as plt
%matplotlib inline
# Create sample data
np.random.seed(42)
# Wield samples
n_samples = 100
X0 = np.array([[np.random.normal(loc=-1, scale=1),
np.random.normal(loc=1, scale=1)] for i in range(n_samples//2)])
X1 = np.array([[np.random.normal(loc=1, scale=1),
np.random.normal(loc=-1, scale=1)] for i in range(n_samples//2)])
X = np.concatenate([X0, X1], axis=0)
Y = np.concatenate([-np.ones(50), np.ones(50)], axis=0)
data = list(zip(X, Y))
plt.scatter(X0[:,0], X0[:,1])
plt.scatter(X1[:,0], X1[:,1])
plt.show()
def loss(a, b):
return (a - b)**2
def average_loss(w, data):
c = 0
for x, y in data:
prediction = quantum_model(x, w)
c += loss(prediction, y)
return c/len(data)
average_loss(w, data)
gradient_fn = qml.grad(average_loss, argnum=0)
gradient_fn(w, data)
w_new = w - 0.05*gradient_fn(w, data)
history = []
for i in range(15):
w_new = w - 0.05*gradient_fn(w, data)
print(average_loss(w_new, data))
history.append(w_new)
w = w_new
```
|
github_jupyter
|
%%capture
# Comment this out if you don't want to install pennylane from this notebook
!pip install pennylane
# Comment this out if you don't want to install matplotlib from this notebook
!pip install matplotlib
import pennylane as qml
from pennylane import numpy as np
dev = qml.device('default.qubit', wires=2)
@qml.qnode(dev)
def circuit():
qml.Hadamard(wires=0)
return qml.probs(wires=[0, 1])
circuit()
@qml.qnode(dev)
def circuit():
return qml.probs(wires=[0, 1])
circuit()
dev.state
s = 1/np.sqrt(2)
U = np.array([[0., -s, 0., s],
[ s, 0., -s, 0.],
[ s, 0., s, 0.],
[0., -s, 0., -s]])
@qml.qnode(dev)
def circuit():
qml.QubitUnitary(U, wires=[0, 1])
return qml.probs(wires=[0, 1])
circuit()
dev.state
@qml.qnode(dev)
def circuit():
qml.QubitUnitary(U, wires=[0, 1])
return qml.sample(qml.PauliZ(wires=0)), qml.sample(qml.PauliZ(wires=1))
circuit()
dev.state
@qml.qnode(dev)
def circuit():
qml.QubitUnitary(U, wires=[0, 1])
return qml.expval(qml.PauliZ(wires=0)), qml.expval(qml.PauliZ(wires=1))
circuit()
dev.state
@qml.qnode(dev)
def circuit():
qml.PauliX(wires=0)
qml.CNOT(wires=[0,1])
qml.Hadamard(wires=0)
qml.PauliZ(wires=1)
return qml.expval(qml.PauliZ(wires=0)), qml.expval(qml.PauliZ(wires=1))
circuit()
@qml.qnode(dev)
def circuit(w1, w2):
qml.RX(w1, wires=0)
qml.CNOT(wires=[0,1])
qml.RY(w2, wires=1)
return qml.expval(qml.PauliZ(wires=0)), qml.expval(qml.PauliZ(wires=1))
circuit(0.2, 1.3)
@qml.qnode(dev)
def quantum_model(x, w):
qml.templates.AngleEmbedding(x, wires=[0, 1])
qml.templates.BasicEntanglerLayers(w, wires=[0, 1])
return qml.expval(qml.PauliZ(wires=0))
x = np.array([0.1, 0.2], requires_grad=False)
w = np.array([[-2.1, 1.2], [-1.4, -3.9], [0.5, 0.2]])
quantum_model(x, w)
print(quantum_model.draw())
gradient_fn = qml.grad(quantum_model)
gradient_fn(x, w)
import matplotlib.pyplot as plt
%matplotlib inline
# Create sample data
np.random.seed(42)
# Wield samples
n_samples = 100
X0 = np.array([[np.random.normal(loc=-1, scale=1),
np.random.normal(loc=1, scale=1)] for i in range(n_samples//2)])
X1 = np.array([[np.random.normal(loc=1, scale=1),
np.random.normal(loc=-1, scale=1)] for i in range(n_samples//2)])
X = np.concatenate([X0, X1], axis=0)
Y = np.concatenate([-np.ones(50), np.ones(50)], axis=0)
data = list(zip(X, Y))
plt.scatter(X0[:,0], X0[:,1])
plt.scatter(X1[:,0], X1[:,1])
plt.show()
def loss(a, b):
return (a - b)**2
def average_loss(w, data):
c = 0
for x, y in data:
prediction = quantum_model(x, w)
c += loss(prediction, y)
return c/len(data)
average_loss(w, data)
gradient_fn = qml.grad(average_loss, argnum=0)
gradient_fn(w, data)
w_new = w - 0.05*gradient_fn(w, data)
history = []
for i in range(15):
w_new = w - 0.05*gradient_fn(w, data)
print(average_loss(w_new, data))
history.append(w_new)
w = w_new
| 0.551332 | 0.954942 |
<img src="Images/GMIT-logo.png" width="500" align="center" />
# Programming for Data Analysis
## Project
<br/>
#### Lecturer: Brian McGinley
#### Student: Damien Connolly
#### Student number: G00340321
<br/>
**************************************************************************************************************************
### Introduction
The aim of this project is to create a dataset by simulating a real-world phenomenon. It was important when selecting a database to ensure the dataset created can be measured and created using the numpy random package.
****************************************************************************************************************************
#### Task:
Choose a real-world phenomenon that can be measured and for which you could collect at least one-hundred data points across at least four different variables.
Investigate the types of variables involved, their likely distributions, and their relationships with each other.
Synthesise/simulate a data set as closely matching their properties as possible.
Detail your research and implement the simulation in a Jupyter notebook – the data set itself can simply be displayed in an output cell within the notebook.
****************************************************************************************************************************
#### Data to be simulated
I have chosen to simulate data about homeless persons in Ireland between the ages 15 - 64. I have chosen the following variables:
* Gender
* Age
* Location
* Accomodation Type
The statistics used in this project can be found here: [CSO.ie](https://www.cso.ie/en/releasesandpublications/ep/p-cp5hpi/cp5hpi/hpi/)
*****************************************************************************************************************************
#### Background
The Central Statistics Office (CSO) is Ireland's national statistical office and its purpose is to impartially collect, analyse and make available statistics about Ireland’s people, society and economy. At national level, CSO official statistics inform decision making across a range of areas including construction, health, welfare, the environment and the economy. At European level they provide an accurate picture of Ireland’s economic and social performance and enable comparisons between Ireland and other countries <sup>1</sup>.
The homeless persons in Ireland stats used for this project are taken from the 2016 census. Of the 4,761,865 persons enumerated in the State on Census Night, April 24 2016, 6,906 were either counted in accommodation providing shelter for homeless persons or were identified sleeping rough <sup>1</sup>.
******************************************************************************************************************************
```
# Import libraries used in this notebook
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from scipy import stats
from matplotlib import rcParams
# Magic command to ensure that plots render inline [2].
%matplotlib inline
# Set seaborn style
# Use darkplot plot style for contrast
sns.set_style("darkgrid")
# Set the default colour palette
sns.set_palette("colorblind")
# Set figure size
rcParams['figure.figsize'] = 11.7,8.27
```
### Variables
***************************************************************************************************************************
#### Gender
According to the figures, 6,906 people in total were counted to be homeless on census night in 2016. Males accounted for 4,018 of those people while the total number of females was 2,888 <sup>1</sup>.
For the purpose of this project I will be looking at people between the ages of 15 and 64. The age profile statistics provided allow us to remove the people outside of the specified age range easily. The total number of people between the ages 15 - 64 to be simulated will be 4,969. This includes 2,980 males and 1,989 females, meaning males account for approximately 60% and females 40%.
In order to simulate an accurate number of both genders I have decided to use the numpy.random.choice function. This allows us to distribute the data to match the real world figures and it can also return a string which is useful when there are no numerical values involved.
```
# Set the random seed so the numbers will stay the same each time the cell is executed
np.random.seed(1234)
# Define the gender variables
choice = ["Male", "Female"]
# Simulate the gender with correct probability
gender = np.random.choice(choice, 4969, p=[0.599, 0.401])
# Plot results
sns.countplot(x=gender)
# Set title
plt.title('Distribution of Gender')
# Show plot
plt.show
# Visualise the data on a pie-chart [3].
# Data to plot
labels = 'Male', 'Female'
slices_gender = [0.5997, 0.4003]
colors = ['lightskyblue', 'lightcoral']
explode = (0.1, 0) # explode 1st slice
# Plot
plt.pie(slices_gender, explode=explode, labels=labels, colors=colors,
autopct='%1.1f%%', shadow=True, startangle=140, textprops={'fontsize': 30})
plt.title("\nGender of people homeless\n", fontweight="bold", fontsize="30", color="g")
plt.axis('equal')
plt.show()
```
This distribution looks good. We can see from the distribution plot that the number of males (2980) is just below 3000 and the number of females (1989) just below 2000. The pie-chart also shows that the number of males and females has been distributed correctly.
```
# Begin to create the dataset by adding Gender
df = pd.DataFrame(data={"Gender": gender})
df
```
*****************************************************************************************************************************
#### Age Group
According to the CSO, the homeless population tended to be younger than the general population. The average age of a homeless person was 30.5 years compared to the higher 37.4 average age for the general population. There were 4,464 homeless persons in the age group 20-59, accounting for 64.6 per cent of the total homeless count. The corresponding proportion for the overall population of the State was 54.1 per cent. The average age for females was 26.8 years compared with the higher 33.2 years for males. This contrasted with the general population where the average female age was 38.0 years compared with the lower average age of 36.7 years for males <sup>1</sup>.
```
# View age data
agefile = pd.read_excel("ageStatsTotal.xlsx")
agefile
# Remove final total row for plotting purposes
af = agefile.drop([16])
# Plot the total column to visualise the age distribution
sns.relplot(x="Age-group", y="Total", kind="line", data=af, height=5, aspect=2)
# Add labels and title
plt.xlabel('Age-group')
plt.ylabel('Total')
plt.title('Distribution of Ages', fontsize=14)
```
As stated before we are looking only at people within the age range 15 - 64. By looking at the distribution starting from the age of 15 up to 64, it seems likely that the distribution could be replicated using a triangular distribution. The triangular distribution is a continuous probability distribution with lower limit left, peak at mode, and upper limit right <sup>4</sup>. It should be possible to simulate the distribution using these parameters.
* Lower limit: The Starting point of the triangle. We will be starting from 15.
* Mode: The highest frequency i.e. the age with the greatest proportion of the population. From the graph above we can see that the age-range 25 - 29 has the highest number of males and females combined accounting for 730. So for the purpose of this project I will set the mode to 27.
* Upper limit: The Ending point of the triangle. 64 in this case.
```
# Simulate ages using the triangular distribution [5].
# Set the seed
np.random.seed(1)
# Set the parameters and generate ages using the triangular distribution
age = np.random.triangular(15, 27, 64, 4969)
# Print the min, max and median age generated from the array
print("The minimum age is :", min(age),"\nThe maximum age is :", max(age),"\nThe median age is :", np.median(age))
# Plot distribution
sns.displot(kde="line", data=age, aspect = 2)
# Add labels and title
plt.xlabel('Age-group')
plt.ylabel('Count')
plt.title('Distribution of Ages', fontsize=14)
```
The distribution created above looks fairly similar to the original distribution. This should work. The array currently only contains floats, I can easily round them off to their nearest integer using np.rint.
```
# Round to nearest integer
fAge = np.rint(age)
# Print the min, max and median age generated from the array
print("The minimum age is :", min(fAge),"\nThe maximum age is :", max(fAge),"\nThe median age is :", np.median(fAge))
# Add Age to the dataframe
df['Age'] = fAge
# View dataframe
df
```
****************************************************************************************************************************
#### Location
The table below shows the distribution of homeless persons by region. The results show that 72.5 percent of the people who were homeless were based in Dublin. This accounted for 5,009 people, of that amount 55.9 percent or 2,802 were male. The next largest region was the South West. Of the 471 homeless persons enumerated in this region, 285 were male <sup>1</sup>.
```
# View location data
ls = pd.read_csv("location.stats.csv")
ls
# Drop row 9 (Total) for plotting purposes
locStat = ls.drop([9])
# Create plot
locStat.plot(x='Region', y='Total', kind = 'line')
```
From the distribution it is clear to see that Dublin contains the majority of the homeless people at 72.5%. This distribution could be difficult to replicate due to such a big fall off to the rest of the regions. In order to simulate this data I will use the numpy.random.choice function and select items from a list based on the correct probability. Numpy.random.choice takes an array as a parameter and randomly returns one of its values, it can also be used to return multiple values. This is uselful when you want to choose multiple items from a particular list <sup>6</sup>.
```
# Generate the location data [6].
# Set the location variables
location = ['Dublin', 'South West', 'Mid West', 'South East', 'Mid East', 'West', 'North East', 'Midlands', 'North West']
# Set the parameters and generate array with locations
loc = np.random.choice(location, 4969, p=[0.7251, 0.0683, 0.0574, 0.0467, 0.0334, 0.0274, 0.0222, 0.0137, 0.0058])
# Add location to dataframe
df['Location'] = loc
# View dataframe
df
```
**************************************************************************************************************************
#### Accomodation Type
Homeless persons were identified based on where they were on Census Night rather than by self-identification. This means that people in Long Term Accommodation (LTA) are excluded from the main results in this report, an approach agreed in advance with all the major stakeholders working in the area of homelessness <sup>1</sup>. In this report people are divided into two categories, Sleeping in accomodation or Sleeping rough.
```
# View accomodation data
accType = pd.read_excel("AccomodationType.xlsx")
accType
```
The table above shows that males account for the majority of the rough sleepers. Of the 123 that were included in the report 104 were male while only 19 were female. In order to replicate this data I have decided to use the numpy.random.choice function and try simulate the data using the correct probability for each gender.
```
# Set Accomodation Type variables
type = ['In Accomodation', 'Sleeping Rough']
# Loop through the data set and simulate each gender with correct probability [7].
# Add AccomodationType to the dataset
for idx, val in enumerate(df.itertuples()):
if df.Gender[idx] == "Male":
df.loc[idx, 'AccomodationType'] = np.random.choice(type, p=[0.9740, 0.0260])
elif df.Gender[idx] == "Female":
df.loc[idx, 'AccomodationType'] = np.random.choice(type, p=[0.9934, 0.0066])
# Disply dataframe
df
```
*****************************************************************************************************************************
### Assesment of simulated data
```
# Check count for males and females [8].
df['Gender'].value_counts()
```
The count above is correct for males and females.
*********************************************************************************************************
```
# Count each unique value within Age [8].
df['Age'].value_counts()
```
27 has the highest number of people at that age as it was set as the mode when the data was been simulated. The highest number overall comes in the 26 - 30 age bracket matching that of the original age data.
*******************************************************************************************
```
# Mean Age
df['Age'].mean()
```
The mean age of the simulated dataset is 35.3 and according to the CSO the average age across the whole age range was 30.5. Since I simulated only the ages ranging from 15 to 64, while losing a higher number of people from the under 15 age-group, an increase in the mean is to be expected.
*******************************************************************************************
```
# Count the Location figures [8].
df['Location'].value_counts()
```
According the the stats Dublin accounted for 72.5 percent of the total number of homeless people. The simulated data returns approximately the same percentage of the total number simulated. The rest of the list is also distributed matching the original figures.
****************************************************************************************************************
```
# View the AccomodationType data [8].
df['AccomodationType'].value_counts()
```
In the original data people sleeping rough accounted for 1.8 percent of homeless people, the simiulated data is very similiar again.
********************************************************************************************************************
```
# Group by Gender value [9].
dfGen = df.groupby('Gender')
# Check Gender distributions by location [9].
dfGen['Location'].value_counts()
```
This table is very alike to the original dataset with a similar distribtion across all of the locations.
***********************************************************************************************************
```
# Check Gender distributions by AccomodationType [9].
dfGen['AccomodationType'].value_counts()
```
The values are distributed relatively similiar to the original data set.
************************************************************************************************************************
### Conclusion
```
# View dataframe
df
```
The aim of this project was to create a dataset by simulating a real-world phenomenon. The final result is shown above - a table with 4969 rows, each containing 4 variables.
Overall, it was a very interesting project and it allowed me to combine the knowledge and skills I have learned over the course of the semester and apply it to real world data. This project has shown me that it is possible to generate simulated data using numpy.random in order to represent a real-world phenomenon.
******************************************************************************************************************************
### References
[1]. cso.ie https://www.cso.ie/en/releasesandpublications/ep/p-cp5hpi/cp5hpi/hpi/
[2]. stackoverflow.com https://stackoverflow.com/questions/43027980/purpose-of-matplotlib-inline
[3]. pythonspot.com https://pythonspot.com/matplotlib-pie-chart/
[4]. docs.scipy.org https://docs.scipy.org/doc/numpy-1.14.1/reference/generated/numpy.random.triangular.html
[5]. geekforgeeks.com https://www.geeksforgeeks.org/numpy-random-triangular-in-python/
[6]. docs.scipy.org https://docs.scipy.org/doc//numpy-1.10.4/reference/generated/numpy.random.choice.html
[7]. stackoverflow.com https://stackoverflow.com/questions/16476924/how-to-iterate-over-rows-in-a-dataframe-in-pandas
[8]. pandas.pydata.org https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.DataFrame.count.html
[9]. pandas.pydata.org https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.DataFrame.groupby.html
|
github_jupyter
|
# Import libraries used in this notebook
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from scipy import stats
from matplotlib import rcParams
# Magic command to ensure that plots render inline [2].
%matplotlib inline
# Set seaborn style
# Use darkplot plot style for contrast
sns.set_style("darkgrid")
# Set the default colour palette
sns.set_palette("colorblind")
# Set figure size
rcParams['figure.figsize'] = 11.7,8.27
# Set the random seed so the numbers will stay the same each time the cell is executed
np.random.seed(1234)
# Define the gender variables
choice = ["Male", "Female"]
# Simulate the gender with correct probability
gender = np.random.choice(choice, 4969, p=[0.599, 0.401])
# Plot results
sns.countplot(x=gender)
# Set title
plt.title('Distribution of Gender')
# Show plot
plt.show
# Visualise the data on a pie-chart [3].
# Data to plot
labels = 'Male', 'Female'
slices_gender = [0.5997, 0.4003]
colors = ['lightskyblue', 'lightcoral']
explode = (0.1, 0) # explode 1st slice
# Plot
plt.pie(slices_gender, explode=explode, labels=labels, colors=colors,
autopct='%1.1f%%', shadow=True, startangle=140, textprops={'fontsize': 30})
plt.title("\nGender of people homeless\n", fontweight="bold", fontsize="30", color="g")
plt.axis('equal')
plt.show()
# Begin to create the dataset by adding Gender
df = pd.DataFrame(data={"Gender": gender})
df
# View age data
agefile = pd.read_excel("ageStatsTotal.xlsx")
agefile
# Remove final total row for plotting purposes
af = agefile.drop([16])
# Plot the total column to visualise the age distribution
sns.relplot(x="Age-group", y="Total", kind="line", data=af, height=5, aspect=2)
# Add labels and title
plt.xlabel('Age-group')
plt.ylabel('Total')
plt.title('Distribution of Ages', fontsize=14)
# Simulate ages using the triangular distribution [5].
# Set the seed
np.random.seed(1)
# Set the parameters and generate ages using the triangular distribution
age = np.random.triangular(15, 27, 64, 4969)
# Print the min, max and median age generated from the array
print("The minimum age is :", min(age),"\nThe maximum age is :", max(age),"\nThe median age is :", np.median(age))
# Plot distribution
sns.displot(kde="line", data=age, aspect = 2)
# Add labels and title
plt.xlabel('Age-group')
plt.ylabel('Count')
plt.title('Distribution of Ages', fontsize=14)
# Round to nearest integer
fAge = np.rint(age)
# Print the min, max and median age generated from the array
print("The minimum age is :", min(fAge),"\nThe maximum age is :", max(fAge),"\nThe median age is :", np.median(fAge))
# Add Age to the dataframe
df['Age'] = fAge
# View dataframe
df
# View location data
ls = pd.read_csv("location.stats.csv")
ls
# Drop row 9 (Total) for plotting purposes
locStat = ls.drop([9])
# Create plot
locStat.plot(x='Region', y='Total', kind = 'line')
# Generate the location data [6].
# Set the location variables
location = ['Dublin', 'South West', 'Mid West', 'South East', 'Mid East', 'West', 'North East', 'Midlands', 'North West']
# Set the parameters and generate array with locations
loc = np.random.choice(location, 4969, p=[0.7251, 0.0683, 0.0574, 0.0467, 0.0334, 0.0274, 0.0222, 0.0137, 0.0058])
# Add location to dataframe
df['Location'] = loc
# View dataframe
df
# View accomodation data
accType = pd.read_excel("AccomodationType.xlsx")
accType
# Set Accomodation Type variables
type = ['In Accomodation', 'Sleeping Rough']
# Loop through the data set and simulate each gender with correct probability [7].
# Add AccomodationType to the dataset
for idx, val in enumerate(df.itertuples()):
if df.Gender[idx] == "Male":
df.loc[idx, 'AccomodationType'] = np.random.choice(type, p=[0.9740, 0.0260])
elif df.Gender[idx] == "Female":
df.loc[idx, 'AccomodationType'] = np.random.choice(type, p=[0.9934, 0.0066])
# Disply dataframe
df
# Check count for males and females [8].
df['Gender'].value_counts()
# Count each unique value within Age [8].
df['Age'].value_counts()
# Mean Age
df['Age'].mean()
# Count the Location figures [8].
df['Location'].value_counts()
# View the AccomodationType data [8].
df['AccomodationType'].value_counts()
# Group by Gender value [9].
dfGen = df.groupby('Gender')
# Check Gender distributions by location [9].
dfGen['Location'].value_counts()
# Check Gender distributions by AccomodationType [9].
dfGen['AccomodationType'].value_counts()
# View dataframe
df
| 0.83752 | 0.989192 |
# 第二十三讲:微分方程和$e^{At}$
## 微分方程$\frac{\mathrm{d}u}{\mathrm{d}t}=Au$
本讲主要讲解解一阶方程(first-order system)一阶倒数(first derivative)常系数(constant coefficient)线性方程,上一讲介绍了如何计算矩阵的幂,本讲将进一步涉及矩阵的指数形式。我们通过解一个例子来详细介绍计算方法。
有方程组$\begin{cases}\frac{\mathrm{d}u_1}{\mathrm{d}t}&=-u_1+2u_2\\\frac{\mathrm{d}u_2}{\mathrm{d}t}&=u_1-2u_2\end{cases}$,则系数矩阵是$A=\begin{bmatrix}-1&2\\1&-2\end{bmatrix}$,设初始条件为在$0$时刻$u(0)=\begin{bmatrix}u_1\\u_2\end{bmatrix}=\begin{bmatrix}1\\0\end{bmatrix}$。
* 这个初始条件的意义可以看做在开始时一切都在$u_1$中,但随着时间的推移,将有$\frac{\mathrm{d}u_2}{\mathrm{d}t}>0$,因为$u_1$项初始为正,$u_1$中的事物会流向$u_2$。随着时间的发展我们可以追踪流动的变化。
* 根据上一讲所学的知识,我们知道第一步需要找到特征值与特征向量。$A=\begin{bmatrix}-1&2\\1&-2\end{bmatrix}$,很明显这是一个奇异矩阵,所以第一个特征值是$\lambda_1=0$,另一个特征向量可以从迹得到$tr(A)=-3$。当然我们也可以用一般方法计算$\left|A-\lambda I\right|=\begin{vmatrix}-1-\lambda&2\\1&-2-\lambda\end{vmatrix}=\lambda^2+3\lambda=0$。
(教授提前剧透,特征值$\lambda_2=-3$将会逐渐消失,因为答案中将会有一项为$e^{-3t}$,该项会随着时间的推移趋近于$0$。答案的另一部分将有一项为$e^{0t}$,该项是一个常数,其值为$1$,并不随时间而改变。通常含有$0$特征值的矩阵会随着时间的推移达到稳态。)
* 求特征向量,$\lambda_1=0$时,即求$A$的零空间,很明显$x_1=\begin{bmatrix}2\\1\end{bmatrix}$;$\lambda_2=-3$时,求$A+3I$的零空间,$\begin{bmatrix}2&2\\1&1\end{bmatrix}$的零空间为$x_2=\begin{bmatrix}1\\-1\end{bmatrix}$。
* 则方程组的通解为:$u(t)=c_1e^{\lambda_1t}x_1+c_2e^{\lambda_2t}x_2$,通解的前后两部分都是该方程组的纯解,即方程组的通解就是两个与特征值、特征向量相关的纯解的线性组合。我们来验证一下,比如取$u=e^{\lambda_1t}x_1$带入$\frac{\mathrm{d}u}{\mathrm{d}t}=Au$,对时间求导得到$\lambda_1e^{\lambda_1t}x_1=Ae^{\lambda_1t}x_1$,化简得$\lambda_1x_1=Ax_1$。
对比上一讲,解$u_{k+1}=Au_k$时得到$u_k=c_1\lambda^kx_1+c_2\lambda^kx_2$,而解$\frac{\mathrm{d}u}{\mathrm{d}t}=Au$我们得到$u(t)=c_1e^{\lambda_1t}x_1+c_2e^{\lambda_2t}x_2$。
* 继续求$c_1,c_2$,$u(t)=c_1\cdot 1\cdot\begin{bmatrix}2\\1\end{bmatrix}+c_2\cdot e^{-3t}\cdot\begin{bmatrix}1\\-1\end{bmatrix}$,已知$t=0$时,$\begin{bmatrix}1\\0\end{bmatrix}=c_1\begin{bmatrix}2\\1\end{bmatrix}+c_2\begin{bmatrix}1\\-1\end{bmatrix}$($Sc=u(0)$),所以$c_1=\frac{1}{3}, c_2=\frac{1}{3}$。
* 于是我们写出最终结果,$u(t)=\frac{1}{3}\begin{bmatrix}2\\1\end{bmatrix}+\frac{1}{3}e^{-3t}\begin{bmatrix}1\\-1\end{bmatrix}$。
稳定性:这个流动过程从$u(0)=\begin{bmatrix}1\\0\end{bmatrix}$开始,初始值$1$的一部分流入初始值$0$中,经过无限的时间最终达到稳态$u(\infty)=\begin{bmatrix}\frac{2}{3}\\\frac{1}{3}\end{bmatrix}$。所以,要使得$u(t)\to 0$,则需要负的特征值。但如果特征值为复数呢?如$\lambda=-3+6i$,我们来计算$\left|e^{(-3+6i)t}\right|$,其中的$\left|e^{6it}\right|$部分为$\left|\cos 6t+i\sin 6t\right|=1$,因为这部分的模为$\cos^2\alpha+\sin^2\alpha=1$,这个虚部就在单位圆上转悠。所以只有实数部分才是重要的。所以我们可以把前面的结论改为**需要实部为负数的特征值**。实部会决定最终结果趋近于$0$或$\infty$,虚部不过是一些小杂音。
收敛态:需要其中一个特征值实部为$0$,而其他特征值的实部皆小于$0$。
发散态:如果某个特征值实部大于$0$。上面的例子中,如果将$A$变为$-A$,特征值也会变号,结果发散。
再进一步,我们想知道如何从直接判断任意二阶矩阵的特征值是否均小于零。对于二阶矩阵$A=\begin{bmatrix}a&b\\c&d\end{bmatrix}$,矩阵的迹为$a+d=\lambda_1+\lambda_2$,如果矩阵稳定,则迹应为负数。但是这个条件还不够,有反例迹小于$0$依然发散:$\begin{bmatrix}-2&0\\0&1\end{bmatrix}$,迹为$-1$但是仍然发散。还需要加上一个条件,因为$\det A=\lambda_1\cdot\lambda_2$,所以还需要行列式为正数。
总结:原方程组有两个相互耦合的未知函数,$u_1, u_2$相互耦合,而特征值和特征向量的作则就是解耦,也就是对角化(diagonalize)。回到原方程组$\frac{\mathrm{d}u}{\mathrm{d}t}=Au$,将$u$表示为特征向量的线性组合$u=Sv$,代入原方程有$S\frac{\mathrm{d}v}{\mathrm{d}t}=ASv$,两边同乘以$S^{-1}$得$\frac{\mathrm{d}v}{\mathrm{d}t}=S^{-1}ASv=\Lambda v$。以特征向量为基,将$u$表示为$Sv$,得到关于$v$的对角化方程组,新方程组不存在耦合,此时$\begin{cases}\frac{\mathrm{d}v_1}{\mathrm{d}t}&=\lambda_1v_1\\\frac{\mathrm{d}v_2}{\mathrm{d}t}&=\lambda_2v_2\\\vdots&\vdots\\\frac{\mathrm{d}v_n}{\mathrm{d}t}&=\lambda_nv_n\end{cases}$,这是一个各未知函数间没有联系的方程组,它们的解的一般形式为$v(t)=e^{\Lambda t}v(0)$,则原方程组的解的一般形式为$u(t)=e^{At}u(0)=Se^{\Lambda t}S^{-1}u(0)$。这里引入了指数部分为矩阵的形式。
## 指数矩阵$e^{At}$
在上面的结论中,我们见到了$e^{At}$。这种指数部分带有矩阵的情况称为指数矩阵(exponential matrix)。
理解指数矩阵的关键在于,将指数形式展开称为幂基数形式,就像$e^x=1+x+\frac{x^2}{2}+\frac{x^3}{6}+\cdots$一样,将$e^{At}$展开成幂级数的形式为:
$$e^{At}=I+At+\frac{(At)^2}{2}+\frac{(At)^3}{6}+\cdots+\frac{(At)^n}{n!}+\cdots$$
再说些题外话,有两个极具美感的泰勒级数:$e^x=\sum \frac{x^n}{n!}$与$\frac{1}{1-x}=\sum x^n$,如果把第二个泰勒级数写成指数矩阵形式,有$(I-At)^{-1}=I+At+(At)^2+(At)^3+\cdots$,这个式子在$t$非常小的时候,后面的高次项近似等于零,所以可以用来近似$I-At$的逆矩阵,通常近似为$I+At$,当然也可以再加几项。第一个级数对我们而言比第二个级数好,因为第一个级数总会收敛于某个值,所以$e^x$总会有意义,而第二个级数需要$A$特征值的绝对值小于$1$(因为涉及矩阵的幂运算)。我们看到这些泰勒级数的公式对矩阵同样适用。
回到正题,我们需要证明$Se^{\Lambda t}S^{-1}=e^{At}$,继续使用泰勒级数:
$$
e^{At}=I+At+\frac{(At)^2}{2}+\frac{(At)^3}{6}+\cdots+\frac{(At)^n}{n!}+\cdots\\
e^{At}=SS^{-1}+S\Lambda S^{-1}t+\frac{S\Lambda^2S^{-1}}{2}t^2+\frac{S\Lambda^3S^{-1}}{6}t^3+\cdots+\frac{S\Lambda^nS^{-1}}{n!}t^n+\cdots\\
e^{At}=S\left(I+\Lambda t+\frac{\Lambda^2t^2}{2}+\frac{\Lambda^3t^3}{3}+\cdots+\frac{\Lambda^nt^n}{n}+\cdots\right)S^{-1}\\
e^{At}=Se^{\Lambda t}S^{-1}
$$
需要注意的是,$e^{At}$的泰勒级数展开是恒成立的,但我们推出的版本却需要**矩阵可对角化**这个前提条件。
最后,我们来看看什么是$e^{\Lambda t}$,我们将$e^{At}$变为对角矩阵就是因为对角矩阵简单、没有耦合,$e^{\Lambda t}=\begin{bmatrix}e^{\lambda_1t}&0&\cdots&0\\0&e^{\lambda_2t}&\cdots&0\\\vdots&\vdots&\ddots&\vdots\\0&0&\cdots&e^{\lambda_nt}\end{bmatrix}$。
有了$u(t)=Se^{\Lambda t}S^{-1}u(0)$,再来看矩阵的稳定性可知,所有特征值的实部均为负数时矩阵收敛,此时对角线上的指数收敛为$0$。如果我们画出复平面,则要使微分方程存在稳定解,则特征值存在于复平面的左侧(即实部为负);要使矩阵的幂收敛于$0$,则特征值存在于单位圆内部(即模小于$1$),这是幂稳定区域。(上一讲的差分方程需要计算矩阵的幂。)
同差分方程一样,我们来看二阶情况如何计算,有$y''+by'+k=0$。我们也模仿差分方程的情形,构造方程组$\begin{cases}y''&=-by'-ky\\y'&=y'\end{cases}$,写成矩阵形式有$\begin{bmatrix}y''\\y'\end{bmatrix}=\begin{bmatrix}-b&-k\\1&0\end{bmatrix}\begin{bmatrix}y'\\y\end{bmatrix}$,令$u'=\begin{bmatrix}y''\\y'\end{bmatrix}, \ u=\begin{bmatrix}y'\\y\end{bmatrix}$。
继续推广,对于$5$阶微分方程$y'''''+by''''+cy'''+dy''+ey'+f=0$,则可以写作$\begin{bmatrix}y'''''\\y''''\\y'''\\y''\\y'\end{bmatrix}=\begin{bmatrix}-b&-c&-d&-e&-f\\1&0&0&0&0\\0&1&0&0&0\\0&0&1&0&0\\0&0&0&1&0\end{bmatrix}\begin{bmatrix}y''''\\y'''\\y''\\y'\\y\end{bmatrix}$,这样我们就把一个五阶微分方程化为$5\times 5$一阶方程组了,然后就是求特征值、特征向量了步骤了。
|
github_jupyter
|
# 第二十三讲:微分方程和$e^{At}$
## 微分方程$\frac{\mathrm{d}u}{\mathrm{d}t}=Au$
本讲主要讲解解一阶方程(first-order system)一阶倒数(first derivative)常系数(constant coefficient)线性方程,上一讲介绍了如何计算矩阵的幂,本讲将进一步涉及矩阵的指数形式。我们通过解一个例子来详细介绍计算方法。
有方程组$\begin{cases}\frac{\mathrm{d}u_1}{\mathrm{d}t}&=-u_1+2u_2\\\frac{\mathrm{d}u_2}{\mathrm{d}t}&=u_1-2u_2\end{cases}$,则系数矩阵是$A=\begin{bmatrix}-1&2\\1&-2\end{bmatrix}$,设初始条件为在$0$时刻$u(0)=\begin{bmatrix}u_1\\u_2\end{bmatrix}=\begin{bmatrix}1\\0\end{bmatrix}$。
* 这个初始条件的意义可以看做在开始时一切都在$u_1$中,但随着时间的推移,将有$\frac{\mathrm{d}u_2}{\mathrm{d}t}>0$,因为$u_1$项初始为正,$u_1$中的事物会流向$u_2$。随着时间的发展我们可以追踪流动的变化。
* 根据上一讲所学的知识,我们知道第一步需要找到特征值与特征向量。$A=\begin{bmatrix}-1&2\\1&-2\end{bmatrix}$,很明显这是一个奇异矩阵,所以第一个特征值是$\lambda_1=0$,另一个特征向量可以从迹得到$tr(A)=-3$。当然我们也可以用一般方法计算$\left|A-\lambda I\right|=\begin{vmatrix}-1-\lambda&2\\1&-2-\lambda\end{vmatrix}=\lambda^2+3\lambda=0$。
(教授提前剧透,特征值$\lambda_2=-3$将会逐渐消失,因为答案中将会有一项为$e^{-3t}$,该项会随着时间的推移趋近于$0$。答案的另一部分将有一项为$e^{0t}$,该项是一个常数,其值为$1$,并不随时间而改变。通常含有$0$特征值的矩阵会随着时间的推移达到稳态。)
* 求特征向量,$\lambda_1=0$时,即求$A$的零空间,很明显$x_1=\begin{bmatrix}2\\1\end{bmatrix}$;$\lambda_2=-3$时,求$A+3I$的零空间,$\begin{bmatrix}2&2\\1&1\end{bmatrix}$的零空间为$x_2=\begin{bmatrix}1\\-1\end{bmatrix}$。
* 则方程组的通解为:$u(t)=c_1e^{\lambda_1t}x_1+c_2e^{\lambda_2t}x_2$,通解的前后两部分都是该方程组的纯解,即方程组的通解就是两个与特征值、特征向量相关的纯解的线性组合。我们来验证一下,比如取$u=e^{\lambda_1t}x_1$带入$\frac{\mathrm{d}u}{\mathrm{d}t}=Au$,对时间求导得到$\lambda_1e^{\lambda_1t}x_1=Ae^{\lambda_1t}x_1$,化简得$\lambda_1x_1=Ax_1$。
对比上一讲,解$u_{k+1}=Au_k$时得到$u_k=c_1\lambda^kx_1+c_2\lambda^kx_2$,而解$\frac{\mathrm{d}u}{\mathrm{d}t}=Au$我们得到$u(t)=c_1e^{\lambda_1t}x_1+c_2e^{\lambda_2t}x_2$。
* 继续求$c_1,c_2$,$u(t)=c_1\cdot 1\cdot\begin{bmatrix}2\\1\end{bmatrix}+c_2\cdot e^{-3t}\cdot\begin{bmatrix}1\\-1\end{bmatrix}$,已知$t=0$时,$\begin{bmatrix}1\\0\end{bmatrix}=c_1\begin{bmatrix}2\\1\end{bmatrix}+c_2\begin{bmatrix}1\\-1\end{bmatrix}$($Sc=u(0)$),所以$c_1=\frac{1}{3}, c_2=\frac{1}{3}$。
* 于是我们写出最终结果,$u(t)=\frac{1}{3}\begin{bmatrix}2\\1\end{bmatrix}+\frac{1}{3}e^{-3t}\begin{bmatrix}1\\-1\end{bmatrix}$。
稳定性:这个流动过程从$u(0)=\begin{bmatrix}1\\0\end{bmatrix}$开始,初始值$1$的一部分流入初始值$0$中,经过无限的时间最终达到稳态$u(\infty)=\begin{bmatrix}\frac{2}{3}\\\frac{1}{3}\end{bmatrix}$。所以,要使得$u(t)\to 0$,则需要负的特征值。但如果特征值为复数呢?如$\lambda=-3+6i$,我们来计算$\left|e^{(-3+6i)t}\right|$,其中的$\left|e^{6it}\right|$部分为$\left|\cos 6t+i\sin 6t\right|=1$,因为这部分的模为$\cos^2\alpha+\sin^2\alpha=1$,这个虚部就在单位圆上转悠。所以只有实数部分才是重要的。所以我们可以把前面的结论改为**需要实部为负数的特征值**。实部会决定最终结果趋近于$0$或$\infty$,虚部不过是一些小杂音。
收敛态:需要其中一个特征值实部为$0$,而其他特征值的实部皆小于$0$。
发散态:如果某个特征值实部大于$0$。上面的例子中,如果将$A$变为$-A$,特征值也会变号,结果发散。
再进一步,我们想知道如何从直接判断任意二阶矩阵的特征值是否均小于零。对于二阶矩阵$A=\begin{bmatrix}a&b\\c&d\end{bmatrix}$,矩阵的迹为$a+d=\lambda_1+\lambda_2$,如果矩阵稳定,则迹应为负数。但是这个条件还不够,有反例迹小于$0$依然发散:$\begin{bmatrix}-2&0\\0&1\end{bmatrix}$,迹为$-1$但是仍然发散。还需要加上一个条件,因为$\det A=\lambda_1\cdot\lambda_2$,所以还需要行列式为正数。
总结:原方程组有两个相互耦合的未知函数,$u_1, u_2$相互耦合,而特征值和特征向量的作则就是解耦,也就是对角化(diagonalize)。回到原方程组$\frac{\mathrm{d}u}{\mathrm{d}t}=Au$,将$u$表示为特征向量的线性组合$u=Sv$,代入原方程有$S\frac{\mathrm{d}v}{\mathrm{d}t}=ASv$,两边同乘以$S^{-1}$得$\frac{\mathrm{d}v}{\mathrm{d}t}=S^{-1}ASv=\Lambda v$。以特征向量为基,将$u$表示为$Sv$,得到关于$v$的对角化方程组,新方程组不存在耦合,此时$\begin{cases}\frac{\mathrm{d}v_1}{\mathrm{d}t}&=\lambda_1v_1\\\frac{\mathrm{d}v_2}{\mathrm{d}t}&=\lambda_2v_2\\\vdots&\vdots\\\frac{\mathrm{d}v_n}{\mathrm{d}t}&=\lambda_nv_n\end{cases}$,这是一个各未知函数间没有联系的方程组,它们的解的一般形式为$v(t)=e^{\Lambda t}v(0)$,则原方程组的解的一般形式为$u(t)=e^{At}u(0)=Se^{\Lambda t}S^{-1}u(0)$。这里引入了指数部分为矩阵的形式。
## 指数矩阵$e^{At}$
在上面的结论中,我们见到了$e^{At}$。这种指数部分带有矩阵的情况称为指数矩阵(exponential matrix)。
理解指数矩阵的关键在于,将指数形式展开称为幂基数形式,就像$e^x=1+x+\frac{x^2}{2}+\frac{x^3}{6}+\cdots$一样,将$e^{At}$展开成幂级数的形式为:
$$e^{At}=I+At+\frac{(At)^2}{2}+\frac{(At)^3}{6}+\cdots+\frac{(At)^n}{n!}+\cdots$$
再说些题外话,有两个极具美感的泰勒级数:$e^x=\sum \frac{x^n}{n!}$与$\frac{1}{1-x}=\sum x^n$,如果把第二个泰勒级数写成指数矩阵形式,有$(I-At)^{-1}=I+At+(At)^2+(At)^3+\cdots$,这个式子在$t$非常小的时候,后面的高次项近似等于零,所以可以用来近似$I-At$的逆矩阵,通常近似为$I+At$,当然也可以再加几项。第一个级数对我们而言比第二个级数好,因为第一个级数总会收敛于某个值,所以$e^x$总会有意义,而第二个级数需要$A$特征值的绝对值小于$1$(因为涉及矩阵的幂运算)。我们看到这些泰勒级数的公式对矩阵同样适用。
回到正题,我们需要证明$Se^{\Lambda t}S^{-1}=e^{At}$,继续使用泰勒级数:
$$
e^{At}=I+At+\frac{(At)^2}{2}+\frac{(At)^3}{6}+\cdots+\frac{(At)^n}{n!}+\cdots\\
e^{At}=SS^{-1}+S\Lambda S^{-1}t+\frac{S\Lambda^2S^{-1}}{2}t^2+\frac{S\Lambda^3S^{-1}}{6}t^3+\cdots+\frac{S\Lambda^nS^{-1}}{n!}t^n+\cdots\\
e^{At}=S\left(I+\Lambda t+\frac{\Lambda^2t^2}{2}+\frac{\Lambda^3t^3}{3}+\cdots+\frac{\Lambda^nt^n}{n}+\cdots\right)S^{-1}\\
e^{At}=Se^{\Lambda t}S^{-1}
$$
需要注意的是,$e^{At}$的泰勒级数展开是恒成立的,但我们推出的版本却需要**矩阵可对角化**这个前提条件。
最后,我们来看看什么是$e^{\Lambda t}$,我们将$e^{At}$变为对角矩阵就是因为对角矩阵简单、没有耦合,$e^{\Lambda t}=\begin{bmatrix}e^{\lambda_1t}&0&\cdots&0\\0&e^{\lambda_2t}&\cdots&0\\\vdots&\vdots&\ddots&\vdots\\0&0&\cdots&e^{\lambda_nt}\end{bmatrix}$。
有了$u(t)=Se^{\Lambda t}S^{-1}u(0)$,再来看矩阵的稳定性可知,所有特征值的实部均为负数时矩阵收敛,此时对角线上的指数收敛为$0$。如果我们画出复平面,则要使微分方程存在稳定解,则特征值存在于复平面的左侧(即实部为负);要使矩阵的幂收敛于$0$,则特征值存在于单位圆内部(即模小于$1$),这是幂稳定区域。(上一讲的差分方程需要计算矩阵的幂。)
同差分方程一样,我们来看二阶情况如何计算,有$y''+by'+k=0$。我们也模仿差分方程的情形,构造方程组$\begin{cases}y''&=-by'-ky\\y'&=y'\end{cases}$,写成矩阵形式有$\begin{bmatrix}y''\\y'\end{bmatrix}=\begin{bmatrix}-b&-k\\1&0\end{bmatrix}\begin{bmatrix}y'\\y\end{bmatrix}$,令$u'=\begin{bmatrix}y''\\y'\end{bmatrix}, \ u=\begin{bmatrix}y'\\y\end{bmatrix}$。
继续推广,对于$5$阶微分方程$y'''''+by''''+cy'''+dy''+ey'+f=0$,则可以写作$\begin{bmatrix}y'''''\\y''''\\y'''\\y''\\y'\end{bmatrix}=\begin{bmatrix}-b&-c&-d&-e&-f\\1&0&0&0&0\\0&1&0&0&0\\0&0&1&0&0\\0&0&0&1&0\end{bmatrix}\begin{bmatrix}y''''\\y'''\\y''\\y'\\y\end{bmatrix}$,这样我们就把一个五阶微分方程化为$5\times 5$一阶方程组了,然后就是求特征值、特征向量了步骤了。
| 0.318591 | 0.908537 |
```
import tensorflow as tf
import tensorflow.keras as keras
import tensorflow.keras.layers as layers
class Attention(layers.Layer):
def __init__(self, unit):
super(Attention, self).__init__()
self.unit = unit
def build(self, input_shape):
assert len(input_shape) == 3
self.weight = self.add_weight(shape=(input_shape[-1], self.unit),
initializer=keras.initializers.RandomNormal(),
trainable=True)
self.features_dim = input_shape[-1]
self.bias = self.add_weight(shape=(self.unit,),
initializer=keras.initializers.Zeros(),
trainable=True)
self.u = self.add_weight(shape=(self.unit,),
initializer=keras.initializers.RandomNormal(),
trainable=True)
def call(self, inputs, mask=None):
v = tf.tanh(tf.tensordot(inputs, self.weight, axes=1) + self.bias)
vu = tf.tensordot(v, self.u, axes=1)
alphas = tf.nn.softmax(vu)
output = tf.reduce_sum(inputs * tf.expand_dims(alphas, axis=-1), axis=1)
return output
def compute_output_shape(self, input_shape):
return input_shape[0], self.features_dim
from tensorflow.keras import Input, Model
from tensorflow.keras.layers import Embedding, Dense, Dropout, Bidirectional, LSTM
class TextAttBiRNN(object):
def __init__(self, maxlen, max_features, embedding_dims,
class_num=1,
last_activation='sigmoid'):
self.maxlen = maxlen
self.max_features = max_features
self.embedding_dims = embedding_dims
self.class_num = class_num
self.last_activation = last_activation
def get_model(self):
input = Input((self.maxlen,))
embedding = Embedding(self.max_features, self.embedding_dims, input_length=self.maxlen)(input)
x = Bidirectional(LSTM(128, return_sequences=True))(embedding) # LSTM or GRU
x = Attention(self.maxlen)(x)
output = Dense(self.class_num, activation=self.last_activation)(x)
model = Model(inputs=input, outputs=output)
return model
from tensorflow.keras.callbacks import EarlyStopping
from tensorflow.keras.datasets import imdb
from tensorflow.keras.preprocessing import sequence
max_features = 5000
maxlen = 400
batch_size = 32
embedding_dims = 50
epochs = 10
print('Loading data...')
(x_train, y_train), (x_test, y_test) = imdb.load_data(num_words=max_features)
print(len(x_train), 'train sequences')
print(len(x_test), 'test sequences')
print('Pad sequences (samples x time)...')
x_train = sequence.pad_sequences(x_train, maxlen=maxlen)
x_test = sequence.pad_sequences(x_test, maxlen=maxlen)
print('x_train shape:', x_train.shape)
print('x_test shape:', x_test.shape)
print('Build model...')
model = TextAttBiRNN(maxlen, max_features, embedding_dims).get_model()
model.summary()
model.compile('adam', 'binary_crossentropy', metrics=['accuracy'])
print('Train...')
early_stopping = EarlyStopping(monitor='val_acc', patience=3, mode='max')
model.fit(x_train, y_train,
batch_size=batch_size,
epochs=epochs,
callbacks=[early_stopping],
validation_data=(x_test, y_test))
print('Test...')
result = model.predict(x_test)
```
|
github_jupyter
|
import tensorflow as tf
import tensorflow.keras as keras
import tensorflow.keras.layers as layers
class Attention(layers.Layer):
def __init__(self, unit):
super(Attention, self).__init__()
self.unit = unit
def build(self, input_shape):
assert len(input_shape) == 3
self.weight = self.add_weight(shape=(input_shape[-1], self.unit),
initializer=keras.initializers.RandomNormal(),
trainable=True)
self.features_dim = input_shape[-1]
self.bias = self.add_weight(shape=(self.unit,),
initializer=keras.initializers.Zeros(),
trainable=True)
self.u = self.add_weight(shape=(self.unit,),
initializer=keras.initializers.RandomNormal(),
trainable=True)
def call(self, inputs, mask=None):
v = tf.tanh(tf.tensordot(inputs, self.weight, axes=1) + self.bias)
vu = tf.tensordot(v, self.u, axes=1)
alphas = tf.nn.softmax(vu)
output = tf.reduce_sum(inputs * tf.expand_dims(alphas, axis=-1), axis=1)
return output
def compute_output_shape(self, input_shape):
return input_shape[0], self.features_dim
from tensorflow.keras import Input, Model
from tensorflow.keras.layers import Embedding, Dense, Dropout, Bidirectional, LSTM
class TextAttBiRNN(object):
def __init__(self, maxlen, max_features, embedding_dims,
class_num=1,
last_activation='sigmoid'):
self.maxlen = maxlen
self.max_features = max_features
self.embedding_dims = embedding_dims
self.class_num = class_num
self.last_activation = last_activation
def get_model(self):
input = Input((self.maxlen,))
embedding = Embedding(self.max_features, self.embedding_dims, input_length=self.maxlen)(input)
x = Bidirectional(LSTM(128, return_sequences=True))(embedding) # LSTM or GRU
x = Attention(self.maxlen)(x)
output = Dense(self.class_num, activation=self.last_activation)(x)
model = Model(inputs=input, outputs=output)
return model
from tensorflow.keras.callbacks import EarlyStopping
from tensorflow.keras.datasets import imdb
from tensorflow.keras.preprocessing import sequence
max_features = 5000
maxlen = 400
batch_size = 32
embedding_dims = 50
epochs = 10
print('Loading data...')
(x_train, y_train), (x_test, y_test) = imdb.load_data(num_words=max_features)
print(len(x_train), 'train sequences')
print(len(x_test), 'test sequences')
print('Pad sequences (samples x time)...')
x_train = sequence.pad_sequences(x_train, maxlen=maxlen)
x_test = sequence.pad_sequences(x_test, maxlen=maxlen)
print('x_train shape:', x_train.shape)
print('x_test shape:', x_test.shape)
print('Build model...')
model = TextAttBiRNN(maxlen, max_features, embedding_dims).get_model()
model.summary()
model.compile('adam', 'binary_crossentropy', metrics=['accuracy'])
print('Train...')
early_stopping = EarlyStopping(monitor='val_acc', patience=3, mode='max')
model.fit(x_train, y_train,
batch_size=batch_size,
epochs=epochs,
callbacks=[early_stopping],
validation_data=(x_test, y_test))
print('Test...')
result = model.predict(x_test)
| 0.848298 | 0.65546 |
# Generative Model Benchmarking
The goal here is to use the [data programing paradigm](https://arxiv.org/abs/1605.07723) to probabilistically label our training dataset for the disease associates gene realtionship. The label functions have already been generated and now it is time to train the generative model. This model captures important features such as agreements and disagreements between label functions; furthermore, this model can capture the dependency structure between label functions (i.e. correlations between label functions). More information can be found in this [blog post](https://hazyresearch.github.io/snorkel/blog/structure_learning.html) or in this [paper](https://arxiv.org/abs/1703.00854). The underlying hypothesis here is: **Modeling dependency structure between label functions has better performance compared to the conditionally independent model.**
## Set up The Environment
The few blocks below sets up our python environment to perform the experiment.
```
%load_ext autoreload
%autoreload 2
%matplotlib inline
from itertools import product
import os
import sys
sys.path.append(os.path.abspath('../../../modules'))
import pandas as pd
from tqdm import tqdm_notebook
#Set up the environment
username = "danich1"
password = "snorkel"
dbname = "pubmeddb"
#Path subject to change for different os
database_str = "postgresql+psycopg2://{}:{}@/{}?host=/var/run/postgresql".format(username, password, dbname)
os.environ['SNORKELDB'] = database_str
from snorkel import SnorkelSession
session = SnorkelSession()
from snorkel.annotations import LabelAnnotator
from snorkel.learning.structure import DependencySelector
from snorkel.models import candidate_subclass
from utils.label_functions import DG_LFS
from utils.notebook_utils.dataframe_helper import load_candidate_dataframes
from utils.notebook_utils.label_matrix_helper import (
get_auc_significant_stats,
get_overlap_matrix,
get_conflict_matrix,
make_cids_query,
label_candidates
)
from utils.notebook_utils.train_model_helper import train_generative_model
from utils.notebook_utils.plot_helper import (
plot_label_matrix_heatmap,
plot_roc_curve,
plot_generative_model_weights,
plot_pr_curve
)
DiseaseGene = candidate_subclass('DiseaseGene', ['Disease', 'Gene'])
quick_load = True
```
## Load the data for Generative Model Experiments
```
spreadsheet_names = {
'train': '../../sentence_labels_train.xlsx',
'dev': '../../sentence_labels_train_dev.xlsx',
'test': '../../sentence_labels_dev.xlsx'
}
candidate_dfs = {
key:load_candidate_dataframes(spreadsheet_names[key])
for key in spreadsheet_names
}
for key in candidate_dfs:
print("Size of {} set: {}".format(key, candidate_dfs[key].shape[0]))
label_functions = (
list(DG_LFS["DaG_DB"].values()) +
list(DG_LFS["DaG_TEXT"].values())
)
if quick_load:
labeler = LabelAnnotator(lfs=[])
label_matricies = {
key:labeler.load_matrix(session, cids_query=make_cids_query(session, candidate_dfs[key]))
for key in candidate_dfs
}
else:
labeler = LabelAnnotator(lfs=label_functions)
label_matricies = {
key:label_candidates(
labeler,
cids_query=make_cids_query(session, candidate_dfs[key]),
label_functions=label_functions,
apply_existing=(key!='train')
)
for key in candidate_dfs
}
lf_names = [
label_matricies['test'].get_key(session, index).name
for index in range(label_matricies['test'].shape[1])
]
```
## Visualize Label Functions
Before training the generative model, here are some visualizations for the given label functions. These visualizations are helpful in determining the efficacy of each label functions as well as observing the overlaps and conflicts between each function.
```
plot_label_matrix_heatmap(label_matricies['train'].T,
yaxis_tick_labels=lf_names,
figsize=(10,8))
```
Looking at the heatmap above, this is a decent distribution of labels. Some of the label functions are outputting a lot of labels (distant supervision ones) and some are very sparse in their output. Nevertheless, nothing shocking scream out here in terms of label function performance.
```
plot_label_matrix_heatmap(get_overlap_matrix(label_matricies['train'], normalize=True),
yaxis_tick_labels=lf_names, xaxis_tick_labels=lf_names,
figsize=(10,8), colorbar=False, plot_title="Overlap Matrix")
```
The overlap matrix above shows how two label functions overlap with each other. The brighter the color the more overlaps a label function has with another label function. Ignoring the diagonals, there isn't much overlap between functions as expected.
```
plot_label_matrix_heatmap(get_conflict_matrix(label_matricies['train'], normalize=True),
yaxis_tick_labels=lf_names, xaxis_tick_labels=lf_names,
figsize=(10,8), colorbar=False, plot_title="Conflict Matrix")
```
The conflict matrix above shows how often label functions conflict with each other. The brighter the color the more conflict a label function has with another function. Ignoring the diagonals, there isn't many conflicts between functions except for the LF_DG_NO_CONCLUSION and LF_DG_ALLOWED_DISTANCE. Possible reasons for lack of conflicts could be lack of coverage a few functions have, which is shown in the cell below.
```
label_matricies['train'].lf_stats(session)
```
# Train the Generative Model
After visualizing the label functions and their associated properties, now it is time to work on the generative model. AS with common machine learning pipelines, the first step is to find the best hyperparameters for this model. Using the grid search algorithm, the follow parameters were optimized: amount of burnin, strength of regularization, number of epochs to run the model.
## Set the hyperparameter grid search
```
regularization_grid = pd.np.round(pd.np.linspace(0.001, 0.8, num=25), 3)
```
## What are the best hyperparameters for the conditionally independent model?
```
gen_ci_models = {
"{}".format(str(parameter)):train_generative_model(
label_matricies['train'],
burn_in=100,
epochs=100,
reg_param=parameter,
step_size=1/label_matricies['train'].shape[0]
)
for parameter in tqdm_notebook(regularization_grid)
}
ci_marginal_df = pd.DataFrame(pd.np.array([
gen_ci_models[model_name].marginals(label_matricies['dev'])
for model_name in sorted(gen_ci_models.keys())
]).T, columns=sorted(gen_ci_models.keys()))
ci_marginal_df['candidate_id'] = candidate_dfs['dev'].candidate_id.values
ci_marginal_df.head(2)
ci_aucs = plot_roc_curve(
ci_marginal_df.drop("candidate_id", axis=1),
candidate_dfs['dev'].curated_dsh,
model_type='scatterplot', xlim=[0,0.7], figsize=(14,8),
plot_title="Disease Associates Gene CI AUROC"
)
ci_auc_stats_df = get_auc_significant_stats(candidate_dfs['dev'], ci_aucs).sort_values('auroc', ascending=False)
ci_auc_stats_df
```
From this data frame, the best performing model had the following parameters: 50-burnin, 50-epochs, 0.2-regularization. By looking at the top five models, the regularization parameter stays at 0.2. The amount of epochs and burnin varies, but the regularization parameter is important to note.
```
plot_pr_curve(
ci_marginal_df.drop("candidate_id", axis=1),
candidate_dfs['dev'].curated_dsh,
model_type='scatterplot', xlim=[0, 1], figsize=(14,8),
plot_title="Disease Associates Gene CI AUPRC"
)
```
## Does modeling dependencies aid in performance?
```
from snorkel.learning.structure import DependencySelector
gen_da_models = {
"{}".format(parameter):train_generative_model(
label_matricies['train'],
burn_in=100,
epochs=100,
reg_param=parameter,
step_size=1/label_matricies['train'].shape[0],
deps=DependencySelector().select(label_matricies['train']),
lf_propensity=True
)
for parameter in tqdm_notebook(regularization_grid)
}
da_marginal_df = pd.DataFrame(pd.np.array([
gen_da_models[model_name].marginals(label_matricies['dev'])
for model_name in sorted(gen_da_models.keys())
]).T, columns=sorted(gen_da_models.keys()))
da_marginal_df['candidate_id'] = candidate_dfs['dev'].candidate_id.values
da_marginal_df.head(2)
da_aucs = plot_roc_curve(
da_marginal_df.drop("candidate_id", axis=1),
candidate_dfs['dev'].curated_dsh,
model_type='scatterplot', xlim=[0,1], figsize=(14,8),
plot_title="Disease Associates Gene DA AUROC"
)
da_auc_stats_df = get_auc_significant_stats(candidate_dfs['dev'], da_aucs).sort_values('auroc', ascending=False)
da_auc_stats_df
```
From this data frame, the best performing model had the following parameters: 100-burnin, 100-epochs, 0.2-regularization. By looking at the top nine models, the regularization parameter stays at 0.2. The pattern of regularization is the same with the conditionally independent model. This means using 0.2 is a good choice for regularization. The amount of burnin and epochs can vary.
```
plot_pr_curve(
da_marginal_df.drop("candidate_id", axis=1),
candidate_dfs['dev'].curated_dsh,
model_type='scatterplot', xlim=[0, 1], figsize=(14,8),
plot_title="Disease Associates Gene DA AUPRC"
)
best_model_ci = '0.334'
best_model_da = '0.401'
test_marginals_df = pd.DataFrame(pd.np.array([
gen_ci_models[best_model_ci].marginals(label_matricies['test']),
gen_da_models[best_model_da].marginals(label_matricies['test'])
]).T, columns=['CI', 'DA'])
test_marginals_df['candidate_id'] = candidate_dfs['test'].candidate_id.values
test_marginals_df.head(2)
_ = plot_roc_curve(
test_marginals_df.drop('candidate_id',axis=1),
candidate_dfs['test'].curated_dsh,
model_type='curve', xlim=[0,0.7], figsize=(10,8),
plot_title="Disease Associates Gene Test AUROC"
)
```
Printed above are the best performing models from the conditinally independent model and the dependency aware model. These reults support the hypothesis that modeling depenency structure improves performance compared to the conditionally indepent assumption. Now that the best parameters are found the next step is to begin training the discriminator model to make the actual classification of sentneces.
|
github_jupyter
|
%load_ext autoreload
%autoreload 2
%matplotlib inline
from itertools import product
import os
import sys
sys.path.append(os.path.abspath('../../../modules'))
import pandas as pd
from tqdm import tqdm_notebook
#Set up the environment
username = "danich1"
password = "snorkel"
dbname = "pubmeddb"
#Path subject to change for different os
database_str = "postgresql+psycopg2://{}:{}@/{}?host=/var/run/postgresql".format(username, password, dbname)
os.environ['SNORKELDB'] = database_str
from snorkel import SnorkelSession
session = SnorkelSession()
from snorkel.annotations import LabelAnnotator
from snorkel.learning.structure import DependencySelector
from snorkel.models import candidate_subclass
from utils.label_functions import DG_LFS
from utils.notebook_utils.dataframe_helper import load_candidate_dataframes
from utils.notebook_utils.label_matrix_helper import (
get_auc_significant_stats,
get_overlap_matrix,
get_conflict_matrix,
make_cids_query,
label_candidates
)
from utils.notebook_utils.train_model_helper import train_generative_model
from utils.notebook_utils.plot_helper import (
plot_label_matrix_heatmap,
plot_roc_curve,
plot_generative_model_weights,
plot_pr_curve
)
DiseaseGene = candidate_subclass('DiseaseGene', ['Disease', 'Gene'])
quick_load = True
spreadsheet_names = {
'train': '../../sentence_labels_train.xlsx',
'dev': '../../sentence_labels_train_dev.xlsx',
'test': '../../sentence_labels_dev.xlsx'
}
candidate_dfs = {
key:load_candidate_dataframes(spreadsheet_names[key])
for key in spreadsheet_names
}
for key in candidate_dfs:
print("Size of {} set: {}".format(key, candidate_dfs[key].shape[0]))
label_functions = (
list(DG_LFS["DaG_DB"].values()) +
list(DG_LFS["DaG_TEXT"].values())
)
if quick_load:
labeler = LabelAnnotator(lfs=[])
label_matricies = {
key:labeler.load_matrix(session, cids_query=make_cids_query(session, candidate_dfs[key]))
for key in candidate_dfs
}
else:
labeler = LabelAnnotator(lfs=label_functions)
label_matricies = {
key:label_candidates(
labeler,
cids_query=make_cids_query(session, candidate_dfs[key]),
label_functions=label_functions,
apply_existing=(key!='train')
)
for key in candidate_dfs
}
lf_names = [
label_matricies['test'].get_key(session, index).name
for index in range(label_matricies['test'].shape[1])
]
plot_label_matrix_heatmap(label_matricies['train'].T,
yaxis_tick_labels=lf_names,
figsize=(10,8))
plot_label_matrix_heatmap(get_overlap_matrix(label_matricies['train'], normalize=True),
yaxis_tick_labels=lf_names, xaxis_tick_labels=lf_names,
figsize=(10,8), colorbar=False, plot_title="Overlap Matrix")
plot_label_matrix_heatmap(get_conflict_matrix(label_matricies['train'], normalize=True),
yaxis_tick_labels=lf_names, xaxis_tick_labels=lf_names,
figsize=(10,8), colorbar=False, plot_title="Conflict Matrix")
label_matricies['train'].lf_stats(session)
regularization_grid = pd.np.round(pd.np.linspace(0.001, 0.8, num=25), 3)
gen_ci_models = {
"{}".format(str(parameter)):train_generative_model(
label_matricies['train'],
burn_in=100,
epochs=100,
reg_param=parameter,
step_size=1/label_matricies['train'].shape[0]
)
for parameter in tqdm_notebook(regularization_grid)
}
ci_marginal_df = pd.DataFrame(pd.np.array([
gen_ci_models[model_name].marginals(label_matricies['dev'])
for model_name in sorted(gen_ci_models.keys())
]).T, columns=sorted(gen_ci_models.keys()))
ci_marginal_df['candidate_id'] = candidate_dfs['dev'].candidate_id.values
ci_marginal_df.head(2)
ci_aucs = plot_roc_curve(
ci_marginal_df.drop("candidate_id", axis=1),
candidate_dfs['dev'].curated_dsh,
model_type='scatterplot', xlim=[0,0.7], figsize=(14,8),
plot_title="Disease Associates Gene CI AUROC"
)
ci_auc_stats_df = get_auc_significant_stats(candidate_dfs['dev'], ci_aucs).sort_values('auroc', ascending=False)
ci_auc_stats_df
plot_pr_curve(
ci_marginal_df.drop("candidate_id", axis=1),
candidate_dfs['dev'].curated_dsh,
model_type='scatterplot', xlim=[0, 1], figsize=(14,8),
plot_title="Disease Associates Gene CI AUPRC"
)
from snorkel.learning.structure import DependencySelector
gen_da_models = {
"{}".format(parameter):train_generative_model(
label_matricies['train'],
burn_in=100,
epochs=100,
reg_param=parameter,
step_size=1/label_matricies['train'].shape[0],
deps=DependencySelector().select(label_matricies['train']),
lf_propensity=True
)
for parameter in tqdm_notebook(regularization_grid)
}
da_marginal_df = pd.DataFrame(pd.np.array([
gen_da_models[model_name].marginals(label_matricies['dev'])
for model_name in sorted(gen_da_models.keys())
]).T, columns=sorted(gen_da_models.keys()))
da_marginal_df['candidate_id'] = candidate_dfs['dev'].candidate_id.values
da_marginal_df.head(2)
da_aucs = plot_roc_curve(
da_marginal_df.drop("candidate_id", axis=1),
candidate_dfs['dev'].curated_dsh,
model_type='scatterplot', xlim=[0,1], figsize=(14,8),
plot_title="Disease Associates Gene DA AUROC"
)
da_auc_stats_df = get_auc_significant_stats(candidate_dfs['dev'], da_aucs).sort_values('auroc', ascending=False)
da_auc_stats_df
plot_pr_curve(
da_marginal_df.drop("candidate_id", axis=1),
candidate_dfs['dev'].curated_dsh,
model_type='scatterplot', xlim=[0, 1], figsize=(14,8),
plot_title="Disease Associates Gene DA AUPRC"
)
best_model_ci = '0.334'
best_model_da = '0.401'
test_marginals_df = pd.DataFrame(pd.np.array([
gen_ci_models[best_model_ci].marginals(label_matricies['test']),
gen_da_models[best_model_da].marginals(label_matricies['test'])
]).T, columns=['CI', 'DA'])
test_marginals_df['candidate_id'] = candidate_dfs['test'].candidate_id.values
test_marginals_df.head(2)
_ = plot_roc_curve(
test_marginals_df.drop('candidate_id',axis=1),
candidate_dfs['test'].curated_dsh,
model_type='curve', xlim=[0,0.7], figsize=(10,8),
plot_title="Disease Associates Gene Test AUROC"
)
| 0.340047 | 0.946695 |
### **Importing & Merging Data**
```
# Importing Pandas and NumPy
import pandas as pd
import numpy as np
# Importing Files in Google Colab
from google.colab import files
import io
#Visualization
import matplotlib.pyplot as plt
import seaborn as sns
%matplotlib inline
# "Choose the Files" prompt
data_to_load = files.upload()
# importing the files as done in google colab
analytics = pd.read_csv(io.BytesIO(data_to_load['hr_analytics.csv']))
```
### **Let's understand the structure of our dataframe**
```
analytics.head()
```
### **Dropping unnecessary variables**
```
analytics = analytics.drop(['Department'], axis=1)
```
### **Dummy Variable Creation**
```
salary = pd.get_dummies(analytics.salary)
salary
# Creating a dummy variable for the variable 'salary' and dropping the first one.
pm = pd.get_dummies(analytics['salary'],prefix='salary',drop_first=True)
#Adding the results to the master dataframe
analytics = pd.concat([analytics,pm],axis=1)
```
### **Dropping the repeated variables**
```
# We have created dummies for the below variables, so we can drop them
analytics = analytics.drop(['salary'], 1)
```
### **Checking the new data**
```
analytics.head()
analytics.info()
analytics.describe()
```
### **Checking for Missing Values and Inputing Them**
```
# Adding up the missing values (column-wise)
analytics.isnull().sum()
```
### **Feature Standardisation**
```
# Normalising continuous features
df = analytics[['number_project','average_montly_hours','time_spend_company']]
# defining a normalisation function
def normalize (x):
return ((x-np.min(x)) / (max(x) - min(x)))
# applying normalize ( ) to all columns
df_normalize = df.apply(normalize)
# Futa za zamani ili kusiwe na double-count
analytics = analytics.drop(['number_project','average_montly_hours','time_spend_company'], 1)
#Adding the results to the master dataframe
analytics = pd.concat([analytics,df_normalize],axis=1)
analytics.head()
```
## **Model Building**
Let's start by splitting our data into a training set and a test set.
### **Splitting Data into Training and Test Sets**
```
from sklearn.model_selection import train_test_split
# Putting feature variable to X
X = analytics[['satisfaction_level', 'last_evaluation', 'Work_accident', 'promotion_last_5years', 'salary_low',
'salary_medium', 'number_project', 'average_montly_hours', 'time_spend_company']]
# Putting response variable to y
y = analytics['left']
# Splitting the data into train and test
X_train, X_test, y_train, y_test = train_test_split(X,y, train_size=0.7,test_size=0.3,random_state=100)
```
### **Running your first Training Model**
```
from sklearn.linear_model import LogisticRegression
model = LogisticRegression()
model.fit(X_train, y_train)
```
### **Making Predictions**
```
model.predict(X_test)
```
### **Checking accuracy of the model**
```
model.score(X_test,y_test)
```
|
github_jupyter
|
# Importing Pandas and NumPy
import pandas as pd
import numpy as np
# Importing Files in Google Colab
from google.colab import files
import io
#Visualization
import matplotlib.pyplot as plt
import seaborn as sns
%matplotlib inline
# "Choose the Files" prompt
data_to_load = files.upload()
# importing the files as done in google colab
analytics = pd.read_csv(io.BytesIO(data_to_load['hr_analytics.csv']))
analytics.head()
analytics = analytics.drop(['Department'], axis=1)
salary = pd.get_dummies(analytics.salary)
salary
# Creating a dummy variable for the variable 'salary' and dropping the first one.
pm = pd.get_dummies(analytics['salary'],prefix='salary',drop_first=True)
#Adding the results to the master dataframe
analytics = pd.concat([analytics,pm],axis=1)
# We have created dummies for the below variables, so we can drop them
analytics = analytics.drop(['salary'], 1)
analytics.head()
analytics.info()
analytics.describe()
# Adding up the missing values (column-wise)
analytics.isnull().sum()
# Normalising continuous features
df = analytics[['number_project','average_montly_hours','time_spend_company']]
# defining a normalisation function
def normalize (x):
return ((x-np.min(x)) / (max(x) - min(x)))
# applying normalize ( ) to all columns
df_normalize = df.apply(normalize)
# Futa za zamani ili kusiwe na double-count
analytics = analytics.drop(['number_project','average_montly_hours','time_spend_company'], 1)
#Adding the results to the master dataframe
analytics = pd.concat([analytics,df_normalize],axis=1)
analytics.head()
from sklearn.model_selection import train_test_split
# Putting feature variable to X
X = analytics[['satisfaction_level', 'last_evaluation', 'Work_accident', 'promotion_last_5years', 'salary_low',
'salary_medium', 'number_project', 'average_montly_hours', 'time_spend_company']]
# Putting response variable to y
y = analytics['left']
# Splitting the data into train and test
X_train, X_test, y_train, y_test = train_test_split(X,y, train_size=0.7,test_size=0.3,random_state=100)
from sklearn.linear_model import LogisticRegression
model = LogisticRegression()
model.fit(X_train, y_train)
model.predict(X_test)
model.score(X_test,y_test)
| 0.427277 | 0.911061 |
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from sklearn.preprocessing import StandardScaler
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import StratifiedKFold
from sklearn.metrics import accuracy_score
from sklearn.metrics import roc_curve, auc
from matplotlib.legend_handler import HandlerLine2D
from imblearn.over_sampling import ADASYN
# TO IMPORT CVS FILE AND CONVERT THEM INTO PANDAS DATAFRAME
all_features_list_df=pd.read_csv("training_linear_all_features_list_result.csv",index_col=False)
all_features_count_df=all_features_list_df.stack().value_counts().sort_index().sort_values(ascending=False) # it returns a dataframe with the frequency for each features
# LEARNING CURVE (AUC vs incremental number of features )
# CHOOSE THE MODEL
model = RandomForestClassifier (random_state=1, n_estimators=100)
# CHOOSE THE FOLDS FOR CROSS VALIDATION
CV = 10
max_n_features_to_select = 50
all_accuracy_score_CV = []
all_roc_auc_CV = []
list_n_features = []
for first_n_features_to_select in range (max_n_features_to_select):
first_n_features_to_select+=1
print ("Computing with", first_n_features_to_select, "features:")
# To create a dataframe with N features (only from training dataset)
training_dataframe_df = pd.read_csv("training - linear after WEKA CfsSubsetEval.csv",index_col='exam')
size_mapping = {"codeletion":0,"noncodeletion":1}
training_dataframe_df["outcome"] = training_dataframe_df["outcome"].map(size_mapping)
training_feature_names = [x[2:-2] for x in [*all_features_count_df.index]]
training_selected_features = training_feature_names[:first_n_features_to_select]
training_New_dataframe = training_dataframe_df[training_selected_features]
training_New_dataframe["outcome"] = training_dataframe_df["outcome"]
training_dataframe_with_selected_features_df = training_New_dataframe
# To rename the dataframe into X_np, Y_np (numpy arrays)
X_np=(training_dataframe_with_selected_features_df.drop('outcome',axis=1)).values
Y_np=(training_dataframe_with_selected_features_df['outcome']).values
# Run classifier with cross-validation and store data (into all_roc_auc_CV)
cv = StratifiedKFold(CV)
Y_trues = []
Y_predictions = []
Y_probabilities = []
tprs = []
aucs = []
mean_fpr = np.linspace(0, 1, 100)
for train, test in cv.split(X_np, Y_np):
# StandardScaler
ss = StandardScaler()
X_train_CV_SS_np = ss.fit_transform(X_np[train])
X_test_CV_SS_np = ss.transform(X_np[test])
# Balancing X_train_CV_SS_np with ADASYN
sm = ADASYN(random_state=1)
X_train_CV_SS_BAL_np, y_train_CV_balanced = sm.fit_sample(X_train_CV_SS_np, Y_np[train])
X_for_CV_model_training = X_train_CV_SS_BAL_np
Y_for_CV_model_training = y_train_CV_balanced
# Model fitting
model.fit (X_for_CV_model_training, Y_for_CV_model_training)
pred_ = model.predict(X_test_CV_SS_np)
probas_ = model.predict_proba(X_test_CV_SS_np)
accuracy_ = accuracy_score(Y_np[test], pred_)
Y_trues.extend(Y_np[test])
Y_predictions.extend(pred_)
Y_probabilities.extend(probas_)
# Compute Accuracy
Y_trues_CV = Y_trues
predicted_CV = Y_predictions
accuracy_score_CV = accuracy_score (Y_trues_CV, predicted_CV)
print ('Accuracy (computed with Cross Validation) for', first_n_features_to_select, 'features:', round(accuracy_score_CV,3))
# Compute AUC
Y_trues_CV = Y_trues
Y_probabilities_CV = Y_probabilities
fpr_CV, tpr_CV, threshold_CV = roc_curve (Y_trues_CV, np.array(Y_probabilities_CV)[:,1])
roc_auc_CV = auc(fpr_CV, tpr_CV)
print ("AUC (computed with Cross Validation) for", first_n_features_to_select, 'features:', round(roc_auc_CV,3))
all_accuracy_score_CV.append(accuracy_score_CV)
all_roc_auc_CV.append(roc_auc_CV)
list_n_features.append(first_n_features_to_select)
# plot the learning curve
line1, = plt.plot(list_n_features, all_accuracy_score_CV, "r", label="Accuracy (Cross Validation)")
line2, = plt.plot(list_n_features, all_roc_auc_CV, "b", label="AUC (Cross Validation)")
plt.legend(handler_map={line1: HandlerLine2D(numpoints=2)})
plt.ylabel("Metric")
plt.xlabel("Number of features")
plt.show()
```
|
github_jupyter
|
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from sklearn.preprocessing import StandardScaler
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import StratifiedKFold
from sklearn.metrics import accuracy_score
from sklearn.metrics import roc_curve, auc
from matplotlib.legend_handler import HandlerLine2D
from imblearn.over_sampling import ADASYN
# TO IMPORT CVS FILE AND CONVERT THEM INTO PANDAS DATAFRAME
all_features_list_df=pd.read_csv("training_linear_all_features_list_result.csv",index_col=False)
all_features_count_df=all_features_list_df.stack().value_counts().sort_index().sort_values(ascending=False) # it returns a dataframe with the frequency for each features
# LEARNING CURVE (AUC vs incremental number of features )
# CHOOSE THE MODEL
model = RandomForestClassifier (random_state=1, n_estimators=100)
# CHOOSE THE FOLDS FOR CROSS VALIDATION
CV = 10
max_n_features_to_select = 50
all_accuracy_score_CV = []
all_roc_auc_CV = []
list_n_features = []
for first_n_features_to_select in range (max_n_features_to_select):
first_n_features_to_select+=1
print ("Computing with", first_n_features_to_select, "features:")
# To create a dataframe with N features (only from training dataset)
training_dataframe_df = pd.read_csv("training - linear after WEKA CfsSubsetEval.csv",index_col='exam')
size_mapping = {"codeletion":0,"noncodeletion":1}
training_dataframe_df["outcome"] = training_dataframe_df["outcome"].map(size_mapping)
training_feature_names = [x[2:-2] for x in [*all_features_count_df.index]]
training_selected_features = training_feature_names[:first_n_features_to_select]
training_New_dataframe = training_dataframe_df[training_selected_features]
training_New_dataframe["outcome"] = training_dataframe_df["outcome"]
training_dataframe_with_selected_features_df = training_New_dataframe
# To rename the dataframe into X_np, Y_np (numpy arrays)
X_np=(training_dataframe_with_selected_features_df.drop('outcome',axis=1)).values
Y_np=(training_dataframe_with_selected_features_df['outcome']).values
# Run classifier with cross-validation and store data (into all_roc_auc_CV)
cv = StratifiedKFold(CV)
Y_trues = []
Y_predictions = []
Y_probabilities = []
tprs = []
aucs = []
mean_fpr = np.linspace(0, 1, 100)
for train, test in cv.split(X_np, Y_np):
# StandardScaler
ss = StandardScaler()
X_train_CV_SS_np = ss.fit_transform(X_np[train])
X_test_CV_SS_np = ss.transform(X_np[test])
# Balancing X_train_CV_SS_np with ADASYN
sm = ADASYN(random_state=1)
X_train_CV_SS_BAL_np, y_train_CV_balanced = sm.fit_sample(X_train_CV_SS_np, Y_np[train])
X_for_CV_model_training = X_train_CV_SS_BAL_np
Y_for_CV_model_training = y_train_CV_balanced
# Model fitting
model.fit (X_for_CV_model_training, Y_for_CV_model_training)
pred_ = model.predict(X_test_CV_SS_np)
probas_ = model.predict_proba(X_test_CV_SS_np)
accuracy_ = accuracy_score(Y_np[test], pred_)
Y_trues.extend(Y_np[test])
Y_predictions.extend(pred_)
Y_probabilities.extend(probas_)
# Compute Accuracy
Y_trues_CV = Y_trues
predicted_CV = Y_predictions
accuracy_score_CV = accuracy_score (Y_trues_CV, predicted_CV)
print ('Accuracy (computed with Cross Validation) for', first_n_features_to_select, 'features:', round(accuracy_score_CV,3))
# Compute AUC
Y_trues_CV = Y_trues
Y_probabilities_CV = Y_probabilities
fpr_CV, tpr_CV, threshold_CV = roc_curve (Y_trues_CV, np.array(Y_probabilities_CV)[:,1])
roc_auc_CV = auc(fpr_CV, tpr_CV)
print ("AUC (computed with Cross Validation) for", first_n_features_to_select, 'features:', round(roc_auc_CV,3))
all_accuracy_score_CV.append(accuracy_score_CV)
all_roc_auc_CV.append(roc_auc_CV)
list_n_features.append(first_n_features_to_select)
# plot the learning curve
line1, = plt.plot(list_n_features, all_accuracy_score_CV, "r", label="Accuracy (Cross Validation)")
line2, = plt.plot(list_n_features, all_roc_auc_CV, "b", label="AUC (Cross Validation)")
plt.legend(handler_map={line1: HandlerLine2D(numpoints=2)})
plt.ylabel("Metric")
plt.xlabel("Number of features")
plt.show()
| 0.719285 | 0.491334 |
# Monthly traffic on English Wikipedia January 2008 - August 2020
The purpose of this notebook is to implement a analysis project that is fully reproducible by others. The goal of this notebook is to construct and analyze a dataset of monthly traffic on English Wikipedia from January 2008 to August 2020.
## Table of Content
1. [Gathering the data](#Gathering-the-data) <br>
2. [Processing the data](#Processing-the-data) <br>
3. [Analyze the data](#Analyze-the-data)
## Gathering the data
### Import useful packages, establish API endpoints, and specify parameters
```
import json
import csv
import requests
import pandas as pd
import datetime
from functools import reduce
%matplotlib inline
import matplotlib.pyplot as plt
endpoint_pagecounts = 'https://wikimedia.org/api/rest_v1/metrics/legacy/pagecounts/aggregate/en.wikipedia.org/{access-site}/monthly/2008010100/2016080100'
endpoint_pageviews = 'https://wikimedia.org/api/rest_v1/metrics/pageviews/aggregate/en.wikipedia.org/{access}/{agent}/monthly/2015070100/2020090100'
# can adjust the parameters accordingly
params_pagecounts_desktop = {"access-site" : "desktop-site"}
params_pagecounts_mobile = {"access-site" : "mobile-site"}
params_pageviews_desktop = {"access" : "desktop", "agent" : "user"}
params_pageviews_mobileapp = {"access" : "mobile-app", "agent" : "user"}
params_pageviews_mobileweb = {"access" : "mobile-web", "agent" : "user"}
# Customize these with your own information
headers = {
'User-Agent': 'https://github.com/mabelli',
'From': 'yunhongl@uw.edu'
}
```
### Retrieve corresponding data
```
def api_call(endpoint, parameters):
call = requests.get(endpoint.format(**parameters), headers=headers)
response = call.json()
return response
pagecounts_desktop = api_call(endpoint_pagecounts, params_pagecounts_desktop)
pagecounts_mobile = api_call(endpoint_pagecounts, params_pagecounts_mobile)
pageviews_desktop = api_call(endpoint_pageviews, params_pageviews_desktop)
pageviews_mobileapp = api_call(endpoint_pageviews, params_pageviews_mobileapp)
pageviews_mobileweb = api_call(endpoint_pageviews, params_pageviews_mobileweb)
```
### Store data into five json files
```
def save_json_file(data, apiname, accesstype, startmonth, endmonth):
params = {"apiname" : apiname, "accesstype" : accesstype, "startmonth" : startmonth, "endmonth": endmonth}
filename = '{apiname}_{accesstype}_{startmonth}-{endmonth}.json'
with open(filename.format(**params), 'w') as outfile:
json.dump(pagecounts_desktop, outfile)
save_json_file(pagecounts_desktop, "pagecounts", "desktop-site", "200801", "201607")
save_json_file(pagecounts_mobile, "pagecounts", "mobile-site", "200801", "201607")
save_json_file(pageviews_desktop, "pageviews", "desktop", "201507", "202008")
save_json_file(pageviews_mobileapp, "pageviews", "mobile-app", "201507", "202008")
save_json_file(pageviews_mobileweb, "pageviews", "mobile-web", "201507", "202008")
```
## Processing the data
### Convert data to pandas dataframe
```
pagecounts_desktop_df = pd.DataFrame.from_dict(pagecounts_desktop['items'])
pagecounts_mobile_df = pd.DataFrame.from_dict(pagecounts_mobile['items'])
pageviews_desktop_df = pd.DataFrame.from_dict(pageviews_desktop['items'])
pageviews_mobileapp_df = pd.DataFrame.from_dict(pageviews_mobileapp['items'])
pageviews_mobileweb_df = pd.DataFrame.from_dict(pageviews_mobileweb['items'])
```
### Drop unwanted columns and rename counts/views column
```
def drop_rename_columns(df, accesstype):
df_drop = df.drop(['access-site', 'access', 'granularity', 'project', 'agent'], axis = 1, errors = 'ignore')
if 'count' in df_drop.columns:
name = "pagecount_{accessType}_views".format(accessType = accesstype)
df_drop = df_drop.rename(columns={"count": name})
else:
name = "pageview_{accessType}_views".format(accessType = accesstype)
df_drop = df_drop.rename(columns={"views": name})
return df_drop
pagecounts_desktop_processed = drop_rename_columns(pagecounts_desktop_df, "desktop")
pagecounts_mobile_processed = drop_rename_columns(pagecounts_mobile_df, "mobile")
pageviews_desktop_processed = drop_rename_columns(pageviews_desktop_df, "desktop")
pageviews_mobileapp_processed = drop_rename_columns(pageviews_mobileapp_df, "mobileapp")
pageviews_mobileweb_processed = drop_rename_columns(pageviews_mobileweb_df, "mobileweb")
```
### Merge all dataframes into one dataframe using the timestamp column as key and fill missing data with zeros
```
def merge_two_df(df1, df2):
return df1.merge(df2, on = 'timestamp', how='outer')
merged_df = reduce(merge_two_df, [pagecounts_desktop_processed, pagecounts_mobile_processed, pageviews_desktop_processed, pageviews_mobileapp_processed, pageviews_mobileweb_processed])
merged_df.fillna(0, inplace=True)
```
### Create summary columns for pagecount and pageview, and convert timestamp column into year and month
```
merged_df['pagecount_all_views'] = merged_df.pagecount_desktop_views + merged_df.pagecount_mobile_views
merged_df['pageview_mobile_views'] = merged_df.pageview_mobileapp_views + merged_df.pageview_mobileweb_views
merged_df['pageview_all_views'] = merged_df.pageview_mobile_views + merged_df.pageview_desktop_views
merged_df['year'] = merged_df.timestamp.map(lambda timestamp: timestamp[:4])
merged_df['month'] = merged_df.timestamp.map(lambda timestamp: timestamp[4:6])
# another approach: convert to datetime format, convert to string, and fill missing digits for months
# pd.to_datetime(pagecounts_desktop_df.timestamp, format="%Y%m%d%H").map(lambda x: f'{x.month:02}')
final_df = merged_df.drop(['timestamp', 'pageview_mobileapp_views', 'pageview_mobileweb_views'], axis =1)
```
### Save the final dataframe to a csv
```
with open("en-wikipedia_traffic_200712-202008.csv", 'w') as outputfile:
final_df.to_csv(outputfile, index = False)
```
## Analyze the data
### Remove zero datapoints as we do not want to plot them in the graph
```
df_for_visualization = final_df.replace(to_replace = 0, value = pd.np.nan)
```
### Create a datetime column for plotting
```
df_for_visualization['month_year'] = df_for_visualization.year + df_for_visualization.month
df_for_visualization['month_year_converted'] = pd.to_datetime(df_for_visualization.month_year, format="%Y%m")
```
### Visualization of time versus different pageview traffic and save the figure
```
plt.figure(figsize=(18, 8))
plt.plot(df_for_visualization.month_year_converted, df_for_visualization.pageview_all_views)
plt.plot(df_for_visualization.month_year_converted, df_for_visualization.pageview_mobile_views)
plt.plot(df_for_visualization.month_year_converted, df_for_visualization.pageview_desktop_views)
plt.plot(df_for_visualization.month_year_converted, df_for_visualization.pagecount_all_views)
plt.plot(df_for_visualization.month_year_converted, df_for_visualization.pagecount_mobile_views)
plt.plot(df_for_visualization.month_year_converted, df_for_visualization.pagecount_desktop_views)
#ymin, ymax = plt.ylim()
plt.legend(["pageview all", "pageview mobile", "pageview desktop", "pagecount all", "pagecount mobile", "pagecount desktop"])
plt.xlabel("Time (Year)")
plt.ylabel("Traffic (pageviews)")
plt.title("Wikipedia Pageviews Traffic From January 2008 to August 2020")
plt.grid()
plt.savefig('visualization.jpeg')
```
|
github_jupyter
|
import json
import csv
import requests
import pandas as pd
import datetime
from functools import reduce
%matplotlib inline
import matplotlib.pyplot as plt
endpoint_pagecounts = 'https://wikimedia.org/api/rest_v1/metrics/legacy/pagecounts/aggregate/en.wikipedia.org/{access-site}/monthly/2008010100/2016080100'
endpoint_pageviews = 'https://wikimedia.org/api/rest_v1/metrics/pageviews/aggregate/en.wikipedia.org/{access}/{agent}/monthly/2015070100/2020090100'
# can adjust the parameters accordingly
params_pagecounts_desktop = {"access-site" : "desktop-site"}
params_pagecounts_mobile = {"access-site" : "mobile-site"}
params_pageviews_desktop = {"access" : "desktop", "agent" : "user"}
params_pageviews_mobileapp = {"access" : "mobile-app", "agent" : "user"}
params_pageviews_mobileweb = {"access" : "mobile-web", "agent" : "user"}
# Customize these with your own information
headers = {
'User-Agent': 'https://github.com/mabelli',
'From': 'yunhongl@uw.edu'
}
def api_call(endpoint, parameters):
call = requests.get(endpoint.format(**parameters), headers=headers)
response = call.json()
return response
pagecounts_desktop = api_call(endpoint_pagecounts, params_pagecounts_desktop)
pagecounts_mobile = api_call(endpoint_pagecounts, params_pagecounts_mobile)
pageviews_desktop = api_call(endpoint_pageviews, params_pageviews_desktop)
pageviews_mobileapp = api_call(endpoint_pageviews, params_pageviews_mobileapp)
pageviews_mobileweb = api_call(endpoint_pageviews, params_pageviews_mobileweb)
def save_json_file(data, apiname, accesstype, startmonth, endmonth):
params = {"apiname" : apiname, "accesstype" : accesstype, "startmonth" : startmonth, "endmonth": endmonth}
filename = '{apiname}_{accesstype}_{startmonth}-{endmonth}.json'
with open(filename.format(**params), 'w') as outfile:
json.dump(pagecounts_desktop, outfile)
save_json_file(pagecounts_desktop, "pagecounts", "desktop-site", "200801", "201607")
save_json_file(pagecounts_mobile, "pagecounts", "mobile-site", "200801", "201607")
save_json_file(pageviews_desktop, "pageviews", "desktop", "201507", "202008")
save_json_file(pageviews_mobileapp, "pageviews", "mobile-app", "201507", "202008")
save_json_file(pageviews_mobileweb, "pageviews", "mobile-web", "201507", "202008")
pagecounts_desktop_df = pd.DataFrame.from_dict(pagecounts_desktop['items'])
pagecounts_mobile_df = pd.DataFrame.from_dict(pagecounts_mobile['items'])
pageviews_desktop_df = pd.DataFrame.from_dict(pageviews_desktop['items'])
pageviews_mobileapp_df = pd.DataFrame.from_dict(pageviews_mobileapp['items'])
pageviews_mobileweb_df = pd.DataFrame.from_dict(pageviews_mobileweb['items'])
def drop_rename_columns(df, accesstype):
df_drop = df.drop(['access-site', 'access', 'granularity', 'project', 'agent'], axis = 1, errors = 'ignore')
if 'count' in df_drop.columns:
name = "pagecount_{accessType}_views".format(accessType = accesstype)
df_drop = df_drop.rename(columns={"count": name})
else:
name = "pageview_{accessType}_views".format(accessType = accesstype)
df_drop = df_drop.rename(columns={"views": name})
return df_drop
pagecounts_desktop_processed = drop_rename_columns(pagecounts_desktop_df, "desktop")
pagecounts_mobile_processed = drop_rename_columns(pagecounts_mobile_df, "mobile")
pageviews_desktop_processed = drop_rename_columns(pageviews_desktop_df, "desktop")
pageviews_mobileapp_processed = drop_rename_columns(pageviews_mobileapp_df, "mobileapp")
pageviews_mobileweb_processed = drop_rename_columns(pageviews_mobileweb_df, "mobileweb")
def merge_two_df(df1, df2):
return df1.merge(df2, on = 'timestamp', how='outer')
merged_df = reduce(merge_two_df, [pagecounts_desktop_processed, pagecounts_mobile_processed, pageviews_desktop_processed, pageviews_mobileapp_processed, pageviews_mobileweb_processed])
merged_df.fillna(0, inplace=True)
merged_df['pagecount_all_views'] = merged_df.pagecount_desktop_views + merged_df.pagecount_mobile_views
merged_df['pageview_mobile_views'] = merged_df.pageview_mobileapp_views + merged_df.pageview_mobileweb_views
merged_df['pageview_all_views'] = merged_df.pageview_mobile_views + merged_df.pageview_desktop_views
merged_df['year'] = merged_df.timestamp.map(lambda timestamp: timestamp[:4])
merged_df['month'] = merged_df.timestamp.map(lambda timestamp: timestamp[4:6])
# another approach: convert to datetime format, convert to string, and fill missing digits for months
# pd.to_datetime(pagecounts_desktop_df.timestamp, format="%Y%m%d%H").map(lambda x: f'{x.month:02}')
final_df = merged_df.drop(['timestamp', 'pageview_mobileapp_views', 'pageview_mobileweb_views'], axis =1)
with open("en-wikipedia_traffic_200712-202008.csv", 'w') as outputfile:
final_df.to_csv(outputfile, index = False)
df_for_visualization = final_df.replace(to_replace = 0, value = pd.np.nan)
df_for_visualization['month_year'] = df_for_visualization.year + df_for_visualization.month
df_for_visualization['month_year_converted'] = pd.to_datetime(df_for_visualization.month_year, format="%Y%m")
plt.figure(figsize=(18, 8))
plt.plot(df_for_visualization.month_year_converted, df_for_visualization.pageview_all_views)
plt.plot(df_for_visualization.month_year_converted, df_for_visualization.pageview_mobile_views)
plt.plot(df_for_visualization.month_year_converted, df_for_visualization.pageview_desktop_views)
plt.plot(df_for_visualization.month_year_converted, df_for_visualization.pagecount_all_views)
plt.plot(df_for_visualization.month_year_converted, df_for_visualization.pagecount_mobile_views)
plt.plot(df_for_visualization.month_year_converted, df_for_visualization.pagecount_desktop_views)
#ymin, ymax = plt.ylim()
plt.legend(["pageview all", "pageview mobile", "pageview desktop", "pagecount all", "pagecount mobile", "pagecount desktop"])
plt.xlabel("Time (Year)")
plt.ylabel("Traffic (pageviews)")
plt.title("Wikipedia Pageviews Traffic From January 2008 to August 2020")
plt.grid()
plt.savefig('visualization.jpeg')
| 0.47098 | 0.721007 |
# Executing DFT simulations using ASE, and post-processing the output via McStas
### Authors:
- Mads Bertelsen (ESS)
- Mousumi Upadhyay Kahaly (ELI-ALPS)
- Shervin Nourbakhsh (ILL)
- Gergely Nagy (ELI-ALPS)
This notebook demonstrates a very simple, yet powerful, integrated workflow of executing a DFT-based crystal structure relaxation, and then using the result in a simulated neutron-scattering experiment. The DFT calculation is conducted via the open-source *[Quantum Espresso](https://www.quantum-espresso.org)* software, and the neurton experiment is simulated via *[McStas](https://www.mcstas.org)*.
Two Python module is used to seamlessly interface between these softwares and the notebook. The *[Atomistic Simulation Environment](https://wiki.fysik.dtu.dk/ase/index.html)* (***ASE***), a very powerful tool for atomistic simulations in general, is used to obtain the initial structure from a public database, [*COD*](https://www.crystallography.net/cod/) and communicate with Quantum Espresso; and the [***McStasScript*** module](https://github.com/PaNOSC-ViNYL/McStasScript) is used to set up and execute the McStas simulation.
## Initial setup
```
import sys
import os
```
If your Quantum-Espresso binaries are in a local folder, add it to `$PATH` here. If they are already there, you can skip the cell below.
```
QE_bin_path = os.environ["HOME"]+"/PANOSC/bin"
os.environ['PATH']=os.environ['PATH']+":"+QE_bin_path
mcstas_outdir = "mcstas_output"
os.environ['PATH']=os.environ['PATH']+":/usr/lib64/mpich/bin:"
print(os.environ['PATH'])
```
### Set the path for the temporary files
This folder will be used for temporary files created while running the simulations.
```
tmpdir='/tmp/jupiter/'
print('Create temporary directory: '+tmpdir)
os.makedirs(tmpdir,exist_ok=True)
os.chdir(tmpdir)
os.makedirs(mcstas_outdir,exist_ok=True)
```
## Set up the input files for ASE and Quantum Espresso
ASE can take in input several file formats.
In this demo we will check that the conversion is done properly and that the output of the Quantum-Espresso (QE) simulation can be carried on with them.
In the following only CIF files will be considered as inputs for the simulation workflow
### Convert CIF to QE input file
ASE is able to convert from different formats. If you plan to run QE as a standalone package you need to use files in its input format, so you need to convert for example a CIF file into QE format. This can be done in the following way
```ase convert myfile.cif myfile.pwi```
If you run the simulation using ASE, this step is not needed since conversions are done internally and transparently
### Download the structure
First we download a selected cif file from the Crystallography Open Database. (Of course, this could also be done manually.)
```
CIF_file = '1527603.cif'
print('Downloading CIF file '+CIF_file+' from crystallography.net')
os.system("wget -c https://www.crystallography.net/cod/"+CIF_file)
```
### Download the pseudo potential for Nitrogen
Quantum Espresso needs a suitable pseudopotential. We get it from their collection at
```
pseudopotfile = 'N.pbe-n-kjpaw_psl.1.0.0.UPF'
pseudo_dir = tmpdir+"/pseudo/"
os.makedirs(pseudo_dir,exist_ok=True)
os.chdir(pseudo_dir)
os.system("wget -c https://www.quantum-espresso.org/upf_files/"+pseudopotfile)
os.system("wget -c https://raw.githubusercontent.com/PaNOSC-ViNYL/workshop2020/team2/demo/team2/N.pbe-n-radius_5.UPF")
os.chdir(tmpdir)
pseudopotfile = 'N.pbe-n-radius_5.UPF'
```
### Check the list of files in the current working directory
```
os.listdir()
```
# Setup the simulation
First, we read the CIF file and display the (initial) structure.
```
from ase import io, Atom, Atoms
atomCIF = io.read(CIF_file)
print(atomCIF)
print(atomCIF.get_positions())
from ase.visualize import view
view(atomCIF)
```
Then, we setup the Quantum Espresso calculation. The parameters correspond to the ones found in the input files of pw.x, for which the documentation is available [here](https://www.quantum-espresso.org/Doc/INPUT_PW.html).
```
from ase.calculators.espresso import Espresso
pseudopotentials={'N': pseudopotfile}
calc = Espresso(pseudopotentials=pseudopotentials,
tstress=True,
tprnfor=True,
kpts=(6, 6, 6),
ecutrho=480,
ecutwfc=60,
ibrav=0,
nat=8,
ntyp=1,
calculation='relax',
occupations='smearing',
smearing='cold',
degauss=0.001,
outdir=tmpdir,
pseudo_dir=pseudo_dir,
conv_thr=1e-7,
mixing_mode='plain',
electron_maxstep=80,
mixing_beta=0.5,
ion_dynamics='bfgs',
)
atom = atomCIF
atom.calc = calc
#atom.set_calculator(calc)
#atom.get_potential_energy()
#fermi_level = calc.get_fermi_level()
```
### Calculate the potential energy
This takes some time, about 15 minutes.
When the calculation starts, two additional files are created:
- espresso.pwi: QE input file with atomic structure and parameters
- espresso.pwo: QE output
ASE will parse the espresso.pwo file, and update the `atoms` object accordingly.
```
potential_energy = atom.get_potential_energy()
print("Total energy: {0} eV".format(potential_energy))
print("Total energy: {0} eV".format(atom.get_total_energy()))
fermi_level = calc.get_fermi_level()
print("Fermi energy: {0} eV".format(fermi_level))
```
### Compare atom positions before and after calculation
```
# first read the output of the QE calculation, index=-1 allow to read only the last set of positions (those at convergence)
atomsOUT = io.read('espresso.pwo',index=-1)
atomsOUT.get_positions() - atom.get_positions()
```
### output the result to CIF format
```
ase_outfile = 'output.cif'
hklfile=ase_outfile+'.hkl'
io.write(ase_outfile, atom)
os.listdir()
os.system('cif2hkl '+ase_outfile)
os.listdir()
```
# Neutron scattering experiment simulation
The optimized structure is now ready. From this point, the data can be used in whatever way we desire.
As an example, we use McStas to simulate a neutron-scattering experiment using the optimiized structure.
## Get McStasScript
```
os.system("git clone git@github.com:PaNOSC-ViNYL/McStas_ViNYL_concept.git")
os.chdir('McStas_ViNYL_concept')
#os.chdir(tmpdir)
os.makedirs(mcstas_outdir,exist_ok=True)
os.listdir()
```
If your McStas is installed in a different location, update the paths below accordingly.
```
import McStasCalculator
import McStasParameters
import math
from mcstasscript.interface import instr, plotter
from mcstasscript.interface import functions
my_configurator = functions.Configurator()
my_configurator.set_mcrun_path("/usr/local/bin/")
my_configurator.set_mcstas_path("/usr/local/mcstas/2.6/")
Instr = instr.McStas_instr("powder_diffractometer")
Instr.add_parameter("wavelength", value=1.2, comment="[AA]")
src = Instr.add_component("Source", "Source_Maxwell_3")
src.xwidth = 0.12
src.yheight = 0.12
src.Lmin = "wavelength*0.94" # Simulate wavelengths in small band around requested wavelength
src.Lmax = "wavelength*1.06"
src.dist = 3.0
src.focus_xw = guide_width = 0.04
src.focus_yh = guide_height = 0.08
# Set source spectrum to ILL
src.T1 = 683.7
src.I1 = 0.5874E13
src.T2 = 257.7
src.I2 = 2.5099E13
src.T3 = 16.7
src.I3 = 1.0343E12
guide = Instr.add_component("guide", "Guide_gravity", AT=[0,0,3.0], RELATIVE="Source")
guide.w1 = guide_width
guide.h1 = guide_height
guide.l = guide_length = 10 # 10 m long guide
guide.m = 3.0
guide.G = -9.82 # Gravity
Instr.add_component("guide_end", "Arm", AT=[0, 0, guide_length], RELATIVE="guide")
Instr.add_component("mono_pos", "Arm", AT=[0, 0, 0.2], RELATIVE="guide_end")
Instr.add_parameter("mono_Q", value=3.355, comment="Monochromator scattering vector length (PG) [AA^-1]")
Instr.add_declare_var("double", "mono_theta")
Instr.add_declare_var("double", "wavevector")
# Calculate wavevector and find theta from Q = 2k sin(theta)
Instr.append_initialize("wavevector = 2*PI/wavelength;")
Instr.append_initialize("mono_theta = RAD2DEG*asin(0.5*mono_Q/wavevector);")
mono = Instr.add_component("mono", "Monochromator_curved", AT=[0,0,0], RELATIVE="mono_pos")
mono.Q = "mono_Q"
mono.height = 0.1
mono.zwidth = 0.03
mono.NH = 3
mono.NV = 11
mono.RV = 1.5 # Focusing
mono.set_ROTATED([0, "mono_theta", 0], RELATIVE="mono_pos")
Instr.add_component("mono_out", "Arm", AT=[0,0,0], ROTATED=[0, "mono_theta", 0], RELATIVE="mono")
L_mon = Instr.add_component("L_mon", "L_monitor", AT=[0, 0, 1.0], RELATIVE="mono_out")
L_mon.Lmin = "wavelength*0.94"
L_mon.Lmax = "wavelength*1.06"
L_mon.filename = '"L_mon.dat"'
L_mon.xwidth = 0.1
L_mon.yheight = 0.1
L_mon.nL = 150
sample = Instr.add_component("sample", "PowderN", AT=[0, 0, 1.5], RELATIVE="mono_out")
sample.radius = 0.008
sample.yheight = 0.03
sample.reflections = '"../output.cif.hkl"'
sample.barns = 0 # output.cif.hkl cross section read as fm^2
# Wish to focus on the detector, specify height and radius for use in focusing.
detector_height = 0.2
detector_radius = 1.0
sample.d_phi = math.atan(detector_height/detector_radius)*180/3.14159
sample.tth_sign = 1.0
sample.set_SPLIT(1000)
# Set up a banana monitor to measure scattering pattern
monitor = Instr.add_component("monitor", "Monitor_nD", RELATIVE="sample")
monitor.xwidth = 2.0*detector_radius
monitor.yheight = detector_height
monitor.options = '"banana, theta limits=[10,170], bins=320"'
monitor.filename = '"banana.dat"'
%%capture # the output of McStas is verbose and unneccesary for us.
data = Instr.run_full_instrument(ncount=5E6, mpi=4,
foldername=mcstas_outdir, increment_folder_name=True)
# Instr.show_instrument() Uncomment to view instrum
plotter.make_sub_plot(data)
```
|
github_jupyter
|
import sys
import os
QE_bin_path = os.environ["HOME"]+"/PANOSC/bin"
os.environ['PATH']=os.environ['PATH']+":"+QE_bin_path
mcstas_outdir = "mcstas_output"
os.environ['PATH']=os.environ['PATH']+":/usr/lib64/mpich/bin:"
print(os.environ['PATH'])
tmpdir='/tmp/jupiter/'
print('Create temporary directory: '+tmpdir)
os.makedirs(tmpdir,exist_ok=True)
os.chdir(tmpdir)
os.makedirs(mcstas_outdir,exist_ok=True)
If you run the simulation using ASE, this step is not needed since conversions are done internally and transparently
### Download the structure
First we download a selected cif file from the Crystallography Open Database. (Of course, this could also be done manually.)
### Download the pseudo potential for Nitrogen
Quantum Espresso needs a suitable pseudopotential. We get it from their collection at
### Check the list of files in the current working directory
# Setup the simulation
First, we read the CIF file and display the (initial) structure.
Then, we setup the Quantum Espresso calculation. The parameters correspond to the ones found in the input files of pw.x, for which the documentation is available [here](https://www.quantum-espresso.org/Doc/INPUT_PW.html).
### Calculate the potential energy
This takes some time, about 15 minutes.
When the calculation starts, two additional files are created:
- espresso.pwi: QE input file with atomic structure and parameters
- espresso.pwo: QE output
ASE will parse the espresso.pwo file, and update the `atoms` object accordingly.
### Compare atom positions before and after calculation
### output the result to CIF format
# Neutron scattering experiment simulation
The optimized structure is now ready. From this point, the data can be used in whatever way we desire.
As an example, we use McStas to simulate a neutron-scattering experiment using the optimiized structure.
## Get McStasScript
If your McStas is installed in a different location, update the paths below accordingly.
| 0.321034 | 0.877529 |
```
import torch
```
### Why you need a good init
To understand why initialization is important in a neural net, we'll focus on the basic operation you have there: matrix multiplications. So let's just take a vector `x`, and a matrix `a` initiliazed randomly, then multiply them 100 times (as if we had 100 layers).
[Jump_to lesson 9 video](https://course.fast.ai/videos/?lesson=9&t=1132)
```
x = torch.randn(512)
a = torch.randn(512,512)
a.shape
for i in range(100): x = a @ x
x.mean(),x.std()
```
The problem you'll get with that is activation explosion: very soon, your activations will go to nan. We can even ask the loop to break when that first happens:
```
x = torch.randn(512)
a = torch.randn(512,512)
for i in range(100):
x = a @ x
if x.std() != x.std(): break
i
```
It only takes 27 multiplications! On the other hand, if you initialize your activations with a scale that is too low, then you'll get another problem:
```
x = torch.randn(512)
a = torch.randn(512,512) * 0.01
a
for i in range(100): x = a @ x
x.mean(),x.std()
```
Here, every activation vanished to 0. So to avoid that problem, people have come with several strategies to initialize their weight matices, such as:
- use a standard deviation that will make sure x and Ax have exactly the same scale
- use an orthogonal matrix to initialize the weight (orthogonal matrices have the special property that they preserve the L2 norm, so x and Ax would have the same sum of squares in that case)
- use [spectral normalization](https://arxiv.org/pdf/1802.05957.pdf) on the matrix A (the spectral norm of A is the least possible number M such that `torch.norm(A@x) <= M*torch.norm(x)` so dividing A by this M insures you don't overflow. You can still vanish with this)
### The magic number for scaling
Here we will focus on the first one, which is the Xavier initialization. It tells us that we should use a scale equal to `1/math.sqrt(n_in)` where `n_in` is the number of inputs of our matrix.
[Jump_to lesson 9 video](https://course.fast.ai/videos/?lesson=9&t=1273)
```
import math
x = torch.randn(512)
a = torch.randn(512,512) / math.sqrt(512)
for i in range(100): x = a @ x
x.mean(),x.std()
```
And indeed it works. Note that this magic number isn't very far from the 0.01 we had earlier.
```
1/ math.sqrt(512)
```
But where does it come from? It's not that mysterious if you remember the definition of the matrix multiplication. When we do `y = a @ x`, the coefficients of `y` are defined by
$$y_{i} = a_{i,0} x_{0} + a_{i,1} x_{1} + \cdots + a_{i,n-1} x_{n-1} = \sum_{k=0}^{n-1} a_{i,k} x_{k}$$
or in code:
```
y[i] = sum([c*d for c,d in zip(a[i], x)])
```
Now at the very beginning, our `x` vector has a mean of roughly 0. and a standard deviation of roughly 1. (since we picked it that way).
```
x = torch.randn(512)
x.mean(), x.std()
```
NB: This is why it's extremely important to normalize your inputs in Deep Learning, the intialization rules have been designed with inputs that have a mean 0. and a standard deviation of 1.
If you need a refresher from your statistics course, the mean is the sum of all the elements divided by the number of elements (a basic average). The standard deviation represents if the data stays close to the mean or on the contrary gets values that are far away. It's computed by the following formula:
$$\sigma = \sqrt{\frac{1}{n}\left[(x_{0}-m)^{2} + (x_{1}-m)^{2} + \cdots + (x_{n-1}-m)^{2}\right]}$$
where m is the mean and $\sigma$ (the greek letter sigma) is the standard deviation. Here we have a mean of 0, so it's just the square root of the mean of x squared.
If we go back to `y = a @ x` and assume that we chose weights for `a` that also have a mean of 0, we can compute the standard deviation of `y` quite easily. Since it's random, and we may fall on bad numbers, we repeat the operation 100 times.
```
mean,sqr = 0.,0.
for i in range(100):
x = torch.randn(512)
a = torch.randn(512, 512)
y = a @ x
mean += y.mean().item()
sqr += y.pow(2).mean().item()
mean/100,sqr/100
```
Now that looks very close to the dimension of our matrix 512. And that's no coincidence! When you compute y, you sum 512 product of one element of a by one element of x. So what's the mean and the standard deviation of such a product? We can show mathematically that as long as the elements in `a` and the elements in `x` are independent, the mean is 0 and the std is 1. This can also be seen experimentally:
```
mean,sqr = 0.,0.
for i in range(10000):
x = torch.randn(1)
a = torch.randn(1)
y = a*x
mean += y.item()
sqr += y.pow(2).item()
mean/10000,sqr/10000
```
Then we sum 512 of those things that have a mean of zero, and a mean of squares of 1, so we get something that has a mean of 0, and mean of square of 512, hence `math.sqrt(512)` being our magic number. If we scale the weights of the matrix `a` and divide them by this `math.sqrt(512)`, it will give us a `y` of scale 1, and repeating the product has many times as we want won't overflow or vanish.
### Adding ReLU in the mix
We can reproduce the previous experiment with a ReLU, to see that this time, the mean shifts and the standard deviation becomes 0.5. This time the magic number will be `math.sqrt(2/512)` to properly scale the weights of the matrix.
```
mean,sqr = 0.,0.
for i in range(10000):
x = torch.randn(1)
a = torch.randn(1)
y = a*x
y = 0 if y < 0 else y.item()
mean += y
sqr += y ** 2
mean/10000,sqr/10000
```
We can double check by running the experiment on the whole matrix product.
```
mean,sqr = 0.,0.
for i in range(100):
x = torch.randn(512)
a = torch.randn(512, 512)
y = a @ x
y = y.clamp(min=0)
mean += y.mean().item()
sqr += y.pow(2).mean().item()
mean/100,sqr/100
```
Or that scaling the coefficient with the magic number gives us a scale of 1.
```
mean,sqr = 0.,0.
for i in range(100):
x = torch.randn(512)
a = torch.randn(512, 512) * math.sqrt(2/512)
y = a @ x
y = y.clamp(min=0)
mean += y.mean().item()
sqr += y.pow(2).mean().item()
mean/100,sqr/100
```
The math behind is a tiny bit more complex, and you can find everything in the [Kaiming](https://arxiv.org/abs/1502.01852) and the [Xavier](http://proceedings.mlr.press/v9/glorot10a.html) paper but this gives the intuition behing those results.
|
github_jupyter
|
import torch
x = torch.randn(512)
a = torch.randn(512,512)
a.shape
for i in range(100): x = a @ x
x.mean(),x.std()
x = torch.randn(512)
a = torch.randn(512,512)
for i in range(100):
x = a @ x
if x.std() != x.std(): break
i
x = torch.randn(512)
a = torch.randn(512,512) * 0.01
a
for i in range(100): x = a @ x
x.mean(),x.std()
import math
x = torch.randn(512)
a = torch.randn(512,512) / math.sqrt(512)
for i in range(100): x = a @ x
x.mean(),x.std()
1/ math.sqrt(512)
y[i] = sum([c*d for c,d in zip(a[i], x)])
x = torch.randn(512)
x.mean(), x.std()
mean,sqr = 0.,0.
for i in range(100):
x = torch.randn(512)
a = torch.randn(512, 512)
y = a @ x
mean += y.mean().item()
sqr += y.pow(2).mean().item()
mean/100,sqr/100
mean,sqr = 0.,0.
for i in range(10000):
x = torch.randn(1)
a = torch.randn(1)
y = a*x
mean += y.item()
sqr += y.pow(2).item()
mean/10000,sqr/10000
mean,sqr = 0.,0.
for i in range(10000):
x = torch.randn(1)
a = torch.randn(1)
y = a*x
y = 0 if y < 0 else y.item()
mean += y
sqr += y ** 2
mean/10000,sqr/10000
mean,sqr = 0.,0.
for i in range(100):
x = torch.randn(512)
a = torch.randn(512, 512)
y = a @ x
y = y.clamp(min=0)
mean += y.mean().item()
sqr += y.pow(2).mean().item()
mean/100,sqr/100
mean,sqr = 0.,0.
for i in range(100):
x = torch.randn(512)
a = torch.randn(512, 512) * math.sqrt(2/512)
y = a @ x
y = y.clamp(min=0)
mean += y.mean().item()
sqr += y.pow(2).mean().item()
mean/100,sqr/100
| 0.313945 | 0.99083 |
# Modelling running power with the metrics provided by Stryd
In this notebook, we analyse the run recordings collected using Stryd power meter, and try to model the running power using the other metrics that Stryd provides.
Stryd is the state-of-the-art closed-source running power meter, that is mounted on foot, and calculates/measures power using the built-in sensors (accelerometer, gyroscope, altimeter)
```
from fitparse import FitFile
import seaborn as sns
import matplotlib.pyplot as plt
import pandas as pd
from sklearn.linear_model import LinearRegression, Ridge, Lasso, ElasticNet
from sklearn.neural_network import MLPRegressor
from sklearn.preprocessing import PolynomialFeatures, StandardScaler
from sklearn.model_selection import train_test_split, cross_validate
from sklearn.metrics import mean_absolute_error, r2_score
import numpy as np
```
Import .fit file as pandas data frame
```
def read_fit(path):
fit = FitFile(path)
def record_to_series(record):
return pd.Series({f.name: f.value for f in record.fields})
df = pd.DataFrame([record_to_series(record) for record in fit.get_messages("record")]).drop(["timestamp", "distance", "heart_rate", "enhanced_altitude", "enhanced_speed", "speed", "Form Power"], axis=1)
return df
df = pd.concat([
read_fit("../data/stryd/stryd-backgaden-1km.fit"),
read_fit("../data/stryd/stryd-up-and-down.fit"),
read_fit("../data/stryd/stryd-sport-field-circles.fit"),
read_fit("../data/stryd/stryd-sport-field-lap.fit"),
read_fit("../data/stryd/stryd-sport-field.fit"),
read_fit("../data/stryd/stryd-backgaden-0.3km.fit"),
])
```
Absolute altitude and distance are not useful for our purposes. However, changes of altitude and distance over time might be important features.
```
df.loc[:,"altitude_diff"] = df.altitude.diff()
df.loc[:,"distance_diff"] = df.Distance.diff()
df.drop(["altitude", "Distance"], axis=1, inplace=True)
df = df.dropna()
df.shape
```
We remove all the data points that have zero or close to zero power, as they bare no information for our task (under the assumption that 0 W power is measured iff there's no motion)
```
df = df[df.power > 5]
df.shape
```
# Correlation
We explore what features correlate to power -- our target variable
Observations:
* The strongest positive correlations to power can be seen for speed, followed by air power and cadence.
* Stance time shows strong negative correlation with leg spring stiffness, followed by power, cadence, vertical oscillation.
```
sns.set_context(rc={"axes.labelsize":18})
sns.pairplot(df)
sns.pairplot(df.loc[:, ["power", "cadence", "stance_time", "Speed", "Air Power", "Leg Spring Stiffness"]])
sns.set_context("talk")
display(df.describe())
sns.heatmap(df.corr(), cmap="RdBu", center=0.0)
plt.show()
```
# Modelling
## Helper functions
For the purpose of our experiments we perform 5-fold cross validation, to get less biased performance metrics.
```
def get_cv_stats(reg, X, y, name=""):
cv = pd.DataFrame(cross_validate(reg, X, y, scoring=['r2', 'neg_mean_absolute_error']))
return pd.Series({
"mae_mean": cv.test_neg_mean_absolute_error.mean(),
"mae_std": cv.test_neg_mean_absolute_error.std(),
"r2_mean": cv.test_r2.mean(),
"r2_std": cv.test_r2.std(),
}, name=name)
def get_cv_mae(reg_class, alpha, X, y):
reg = reg_class(alpha=alpha)
return get_cv_stats(reg, X, y).mae_mean
def get_cv_r2(reg_class, alpha, X, y):
reg = reg_class(alpha=alpha)
return get_cv_stats(reg, X, y).r2_mean
```
## Preprocessing
* Separate the features (`X`) and the target (`y`)
* Some ML models benefit from features being uniformly scaled, hence Standard Scaler is applied (`X_scaled` and `y_scaled`)
* From physical models, we know that power is not related to speed linearly, but rather to a square of speed. Therefore, we can transform our features into combinations of polynomial terms (with degree up to 2) -- `X_polynomial`
* In addition, `X_selected` contains hand picked features. Cadence, vertical oscillation, stance time and speed are the features that can be approximated from accelerometer data, and should be relevant for power calculations.
```
X = df.iloc[:,1:]
X_scaled = pd.DataFrame(StandardScaler().fit_transform(X), columns=X.columns)
X_selected = X_scaled.loc[:,["cadence", "vertical_oscillation", "stance_time", "Speed"]]
poly_features = PolynomialFeatures(degree=2, include_bias=False)
X_polynomial = poly_features.fit_transform(X_scaled)
feature_names = list(poly_features.get_feature_names())
for i, x in enumerate(X.columns):
feature_names = [y.replace(f"x{i}", x) for y in feature_names]
X_polynomial = pd.DataFrame(X_polynomial, columns=feature_names)
y = df.iloc[:,0]
y_scaled = StandardScaler().fit_transform(y.values.reshape(-1,1)).reshape(-1,)
```
## Linear Regression
It's the simplest regression model, that fits `y=sum(w_i * x_i) + b` minimising sum of square difference between the predicted and the target values.
Note on the performance metrics:
* Negative mean absolute error (MAE) -- higher values (or lower absolute values) are better
* R^2 score -- coefficient of determination, a proportion of the target variance that can be predicted from the input features. Values closer to 1.0 are better
Fitting the regression with the basic set of features, gives average performance of R^2 = 0.606
```
lr_naive_cv_summary = get_cv_stats(LinearRegression(), X_scaled, y, "Ordinary Least Squares")
lr_naive_cv_summary
```
Fitting with only the selected features makes the performance significantly worse, with R^2 of 0.385
```
lr_manual_cv_summary = get_cv_stats(LinearRegression(), X_selected, y, "Ordinary Least Squares (Selected)")
lr_manual_cv_summary
```
Using the polynomial features, the performance gets even worse with R^2 of 0.256
```
lr_poly_cv_summary = get_cv_stats(LinearRegression(), X_polynomial, y, "Ordinary Least Squares (Polynomial)")
lr_poly_cv_summary
```
## Ridge regression
Ridge regression can be used to attempt improving performance of the ordinary linear regression. In addition to minimising the square error, Ridge introduces a penalty for the size of the coefficients (L2 regularisation).
As opposed to the ordinary Linear Regression, Ridge requires a parameter for weight penalty (alpha). We try a range of different alpha values, and pick one that gives the best MAE on cross-validation.
The performance of Ridge with optimised alpha is slightly better than that of the original model, with R^2=0.619
```
alpha = np.arange(0.1,100.0,0.1)
r2 = [get_cv_r2(Ridge, x, X_scaled, y) for x in alpha]
sns.lineplot(alpha, r2)
plt.xlabel("alpha")
plt.ylabel("R^2")
plt.title("Original Features")
plt.show()
best_alpha = alpha[np.argmax(r2)]
best_alpha
ridge_cv_summary = get_cv_stats(Ridge(best_alpha), X_scaled, y, "Ridge")
ridge_cv_summary
```
Fitting Ridge with polynomial features gives the best performance for alpha = 11.7, however R^2 is still lower than that of the plain linear model, even though the performance is significantly better if compared to the orignial model fitted with the polynomial features
```
alpha = np.arange(0.1,20.0,0.1)
r2 = [get_cv_r2(Ridge, x, X_polynomial, y) for x in alpha]
sns.lineplot(alpha, r2)
plt.xlabel("alpha")
plt.ylabel("R^2")
plt.title("Polynomial Features")
plt.show()
best_alpha = alpha[np.argmax(r2)]
best_alpha
ridge_poly_cv_summary = get_cv_stats(Ridge(best_alpha), X_polynomial, y, "Ridge (Polynomial)")
ridge_poly_cv_summary
```
## Lasso Regression
Another improvement over the ordinary Linear Regression is Lasso. It employs L1 reguralisation, that penalises non-zero coefficients, effectively acting as feature selection.
Same as Ridge, it requires an alpha parameter.
Using the original features, we get the model, which is better than our original model, and shows R^2 of 0.638.
```
alpha = np.arange(0.1,3.0,0.01)
r2 = [get_cv_r2(Lasso, x, X_scaled, y) for x in alpha]
sns.lineplot(alpha, r2)
plt.xlabel("alpha")
plt.ylabel("R^2")
plt.title("Original Features")
plt.show()
best_alpha = alpha[np.argmax(r2)]
best_alpha
lasso_cv_summary = get_cv_stats(Lasso(best_alpha), X_scaled, y, "Lasso")
lasso_cv_summary
```
Observing the coefficients, Lasso Regression model mainly makes use of speed, leg spring stiffness and change in altitude
```
model = Lasso(best_alpha).fit(X_scaled, y)
coef = pd.Series(model.coef_, index=X_scaled.columns)
display("Weights",coef[coef.abs() > 0])
display("Intercept", model.intercept_)
```
Applying the same procedure to the polynomial features and alpha = 0.30, the performance is significantly better than that of the original model.
```
alpha = np.arange(0.1,3.0,0.1)
r2 = [get_cv_r2(Lasso, x, X_polynomial, y) for x in alpha]
sns.lineplot(alpha, r2)
plt.xlabel("alpha")
plt.ylabel("R^2")
plt.title("Polynomial Features")
plt.show()
best_alpha = alpha[np.argmax(r2)]
best_alpha
lasso_poly_cv_summary = get_cv_stats(Lasso(best_alpha), X_polynomial, y, "Lasso (Polynomial)")
lasso_poly_cv_summary
```
The highest positive weights are given to speed and change in altitude. However, it is unexpected for the cadence to have a large negative weight
```
model = Lasso(best_alpha).fit(X_polynomial, y)
coef = pd.Series(model.coef_, index=X_polynomial.columns)
display("Weights",coef[coef.abs() > 0])
display("Intercept", model.intercept_)
```
# Summary
L1 and L2 improves performance for both feature sets. And the best performing method is Lasso with the original features.
Surprisingly, feature engineering does not improve the results either. For example, even though it is expected that the power is related to the square of speed, rather than the speed itself, models does not seem to utilise it at all, bringing the performance down.
```
pd.DataFrame([
lr_naive_cv_summary, lr_manual_cv_summary, lr_poly_cv_summary,
ridge_cv_summary, ridge_poly_cv_summary,
lasso_cv_summary, lasso_poly_cv_summary,
])
```
|
github_jupyter
|
from fitparse import FitFile
import seaborn as sns
import matplotlib.pyplot as plt
import pandas as pd
from sklearn.linear_model import LinearRegression, Ridge, Lasso, ElasticNet
from sklearn.neural_network import MLPRegressor
from sklearn.preprocessing import PolynomialFeatures, StandardScaler
from sklearn.model_selection import train_test_split, cross_validate
from sklearn.metrics import mean_absolute_error, r2_score
import numpy as np
def read_fit(path):
fit = FitFile(path)
def record_to_series(record):
return pd.Series({f.name: f.value for f in record.fields})
df = pd.DataFrame([record_to_series(record) for record in fit.get_messages("record")]).drop(["timestamp", "distance", "heart_rate", "enhanced_altitude", "enhanced_speed", "speed", "Form Power"], axis=1)
return df
df = pd.concat([
read_fit("../data/stryd/stryd-backgaden-1km.fit"),
read_fit("../data/stryd/stryd-up-and-down.fit"),
read_fit("../data/stryd/stryd-sport-field-circles.fit"),
read_fit("../data/stryd/stryd-sport-field-lap.fit"),
read_fit("../data/stryd/stryd-sport-field.fit"),
read_fit("../data/stryd/stryd-backgaden-0.3km.fit"),
])
df.loc[:,"altitude_diff"] = df.altitude.diff()
df.loc[:,"distance_diff"] = df.Distance.diff()
df.drop(["altitude", "Distance"], axis=1, inplace=True)
df = df.dropna()
df.shape
df = df[df.power > 5]
df.shape
sns.set_context(rc={"axes.labelsize":18})
sns.pairplot(df)
sns.pairplot(df.loc[:, ["power", "cadence", "stance_time", "Speed", "Air Power", "Leg Spring Stiffness"]])
sns.set_context("talk")
display(df.describe())
sns.heatmap(df.corr(), cmap="RdBu", center=0.0)
plt.show()
def get_cv_stats(reg, X, y, name=""):
cv = pd.DataFrame(cross_validate(reg, X, y, scoring=['r2', 'neg_mean_absolute_error']))
return pd.Series({
"mae_mean": cv.test_neg_mean_absolute_error.mean(),
"mae_std": cv.test_neg_mean_absolute_error.std(),
"r2_mean": cv.test_r2.mean(),
"r2_std": cv.test_r2.std(),
}, name=name)
def get_cv_mae(reg_class, alpha, X, y):
reg = reg_class(alpha=alpha)
return get_cv_stats(reg, X, y).mae_mean
def get_cv_r2(reg_class, alpha, X, y):
reg = reg_class(alpha=alpha)
return get_cv_stats(reg, X, y).r2_mean
X = df.iloc[:,1:]
X_scaled = pd.DataFrame(StandardScaler().fit_transform(X), columns=X.columns)
X_selected = X_scaled.loc[:,["cadence", "vertical_oscillation", "stance_time", "Speed"]]
poly_features = PolynomialFeatures(degree=2, include_bias=False)
X_polynomial = poly_features.fit_transform(X_scaled)
feature_names = list(poly_features.get_feature_names())
for i, x in enumerate(X.columns):
feature_names = [y.replace(f"x{i}", x) for y in feature_names]
X_polynomial = pd.DataFrame(X_polynomial, columns=feature_names)
y = df.iloc[:,0]
y_scaled = StandardScaler().fit_transform(y.values.reshape(-1,1)).reshape(-1,)
lr_naive_cv_summary = get_cv_stats(LinearRegression(), X_scaled, y, "Ordinary Least Squares")
lr_naive_cv_summary
lr_manual_cv_summary = get_cv_stats(LinearRegression(), X_selected, y, "Ordinary Least Squares (Selected)")
lr_manual_cv_summary
lr_poly_cv_summary = get_cv_stats(LinearRegression(), X_polynomial, y, "Ordinary Least Squares (Polynomial)")
lr_poly_cv_summary
alpha = np.arange(0.1,100.0,0.1)
r2 = [get_cv_r2(Ridge, x, X_scaled, y) for x in alpha]
sns.lineplot(alpha, r2)
plt.xlabel("alpha")
plt.ylabel("R^2")
plt.title("Original Features")
plt.show()
best_alpha = alpha[np.argmax(r2)]
best_alpha
ridge_cv_summary = get_cv_stats(Ridge(best_alpha), X_scaled, y, "Ridge")
ridge_cv_summary
alpha = np.arange(0.1,20.0,0.1)
r2 = [get_cv_r2(Ridge, x, X_polynomial, y) for x in alpha]
sns.lineplot(alpha, r2)
plt.xlabel("alpha")
plt.ylabel("R^2")
plt.title("Polynomial Features")
plt.show()
best_alpha = alpha[np.argmax(r2)]
best_alpha
ridge_poly_cv_summary = get_cv_stats(Ridge(best_alpha), X_polynomial, y, "Ridge (Polynomial)")
ridge_poly_cv_summary
alpha = np.arange(0.1,3.0,0.01)
r2 = [get_cv_r2(Lasso, x, X_scaled, y) for x in alpha]
sns.lineplot(alpha, r2)
plt.xlabel("alpha")
plt.ylabel("R^2")
plt.title("Original Features")
plt.show()
best_alpha = alpha[np.argmax(r2)]
best_alpha
lasso_cv_summary = get_cv_stats(Lasso(best_alpha), X_scaled, y, "Lasso")
lasso_cv_summary
model = Lasso(best_alpha).fit(X_scaled, y)
coef = pd.Series(model.coef_, index=X_scaled.columns)
display("Weights",coef[coef.abs() > 0])
display("Intercept", model.intercept_)
alpha = np.arange(0.1,3.0,0.1)
r2 = [get_cv_r2(Lasso, x, X_polynomial, y) for x in alpha]
sns.lineplot(alpha, r2)
plt.xlabel("alpha")
plt.ylabel("R^2")
plt.title("Polynomial Features")
plt.show()
best_alpha = alpha[np.argmax(r2)]
best_alpha
lasso_poly_cv_summary = get_cv_stats(Lasso(best_alpha), X_polynomial, y, "Lasso (Polynomial)")
lasso_poly_cv_summary
model = Lasso(best_alpha).fit(X_polynomial, y)
coef = pd.Series(model.coef_, index=X_polynomial.columns)
display("Weights",coef[coef.abs() > 0])
display("Intercept", model.intercept_)
pd.DataFrame([
lr_naive_cv_summary, lr_manual_cv_summary, lr_poly_cv_summary,
ridge_cv_summary, ridge_poly_cv_summary,
lasso_cv_summary, lasso_poly_cv_summary,
])
| 0.671578 | 0.958693 |
# Multiple Input and Multiple Output Channels
:label:`sec_channels`
While we have described the multiple channels
that comprise each image (e.g., color images have the standard RGB channels
to indicate the amount of red, green and blue) and convolutional layers for multiple channels in :numref:`subsec_why-conv-channels`,
until now, we simplified all of our numerical examples
by working with just a single input and a single output channel.
This has allowed us to think of our inputs, convolution kernels,
and outputs each as two-dimensional tensors.
When we add channels into the mix,
our inputs and hidden representations
both become three-dimensional tensors.
For example, each RGB input image has shape $3\times h\times w$.
We refer to this axis, with a size of 3, as the *channel* dimension.
In this section, we will take a deeper look
at convolution kernels with multiple input and multiple output channels.
## Multiple Input Channels
When the input data contain multiple channels,
we need to construct a convolution kernel
with the same number of input channels as the input data,
so that it can perform cross-correlation with the input data.
Assuming that the number of channels for the input data is $c_i$,
the number of input channels of the convolution kernel also needs to be $c_i$. If our convolution kernel's window shape is $k_h\times k_w$,
then when $c_i=1$, we can think of our convolution kernel
as just a two-dimensional tensor of shape $k_h\times k_w$.
However, when $c_i>1$, we need a kernel
that contains a tensor of shape $k_h\times k_w$ for *every* input channel. Concatenating these $c_i$ tensors together
yields a convolution kernel of shape $c_i\times k_h\times k_w$.
Since the input and convolution kernel each have $c_i$ channels,
we can perform a cross-correlation operation
on the two-dimensional tensor of the input
and the two-dimensional tensor of the convolution kernel
for each channel, adding the $c_i$ results together
(summing over the channels)
to yield a two-dimensional tensor.
This is the result of a two-dimensional cross-correlation
between a multi-channel input and
a multi-input-channel convolution kernel.
In :numref:`fig_conv_multi_in`, we demonstrate an example
of a two-dimensional cross-correlation with two input channels.
The shaded portions are the first output element
as well as the input and kernel tensor elements used for the output computation:
$(1\times1+2\times2+4\times3+5\times4)+(0\times0+1\times1+3\times2+4\times3)=56$.

:label:`fig_conv_multi_in`
To make sure we really understand what is going on here,
we can (**implement cross-correlation operations with multiple input channels**) ourselves.
Notice that all we are doing is performing one cross-correlation operation
per channel and then adding up the results.
```
import torch
from d2l import torch as d2l
def corr2d_multi_in(X, K):
# First, iterate through the 0th dimension (channel dimension) of `X` and
# `K`. Then, add them together
return sum(d2l.corr2d(x, k) for x, k in zip(X, K))
```
We can construct the input tensor `X` and the kernel tensor `K`
corresponding to the values in :numref:`fig_conv_multi_in`
to (**validate the output**) of the cross-correlation operation.
```
X = torch.tensor([[[0.0, 1.0, 2.0], [3.0, 4.0, 5.0], [6.0, 7.0, 8.0]],
[[1.0, 2.0, 3.0], [4.0, 5.0, 6.0], [7.0, 8.0, 9.0]]])
K = torch.tensor([[[0.0, 1.0], [2.0, 3.0]], [[1.0, 2.0], [3.0, 4.0]]])
corr2d_multi_in(X, K)
```
## Multiple Output Channels
Regardless of the number of input channels,
so far we always ended up with one output channel.
However, as we discussed in :numref:`subsec_why-conv-channels`,
it turns out to be essential to have multiple channels at each layer.
In the most popular neural network architectures,
we actually increase the channel dimension
as we go higher up in the neural network,
typically downsampling to trade off spatial resolution
for greater *channel depth*.
Intuitively, you could think of each channel
as responding to some different set of features.
Reality is a bit more complicated than the most naive interpretations of this intuition since representations are not learned independent but are rather optimized to be jointly useful.
So it may not be that a single channel learns an edge detector but rather that some direction in channel space corresponds to detecting edges.
Denote by $c_i$ and $c_o$ the number
of input and output channels, respectively,
and let $k_h$ and $k_w$ be the height and width of the kernel.
To get an output with multiple channels,
we can create a kernel tensor
of shape $c_i\times k_h\times k_w$
for *every* output channel.
We concatenate them on the output channel dimension,
so that the shape of the convolution kernel
is $c_o\times c_i\times k_h\times k_w$.
In cross-correlation operations,
the result on each output channel is calculated
from the convolution kernel corresponding to that output channel
and takes input from all channels in the input tensor.
We implement a cross-correlation function
to [**calculate the output of multiple channels**] as shown below.
```
def corr2d_multi_in_out(X, K):
# Iterate through the 0th dimension of `K`, and each time, perform
# cross-correlation operations with input `X`. All of the results are
# stacked together
return torch.stack([corr2d_multi_in(X, k) for k in K], 0)
```
We construct a convolution kernel with 3 output channels
by concatenating the kernel tensor `K` with `K+1`
(plus one for each element in `K`) and `K+2`.
```
K = torch.stack((K, K + 1, K + 2), 0)
K.shape
```
Below, we perform cross-correlation operations
on the input tensor `X` with the kernel tensor `K`.
Now the output contains 3 channels.
The result of the first channel is consistent
with the result of the previous input tensor `X`
and the multi-input channel,
single-output channel kernel.
```
corr2d_multi_in_out(X, K)
```
## $1\times 1$ Convolutional Layer
At first, a [**$1 \times 1$ convolution**], i.e., $k_h = k_w = 1$,
does not seem to make much sense.
After all, a convolution correlates adjacent pixels.
A $1 \times 1$ convolution obviously does not.
Nonetheless, they are popular operations that are sometimes included
in the designs of complex deep networks.
Let us see in some detail what it actually does.
Because the minimum window is used,
the $1\times 1$ convolution loses the ability
of larger convolutional layers
to recognize patterns consisting of interactions
among adjacent elements in the height and width dimensions.
The only computation of the $1\times 1$ convolution occurs
on the channel dimension.
:numref:`fig_conv_1x1` shows the cross-correlation computation
using the $1\times 1$ convolution kernel
with 3 input channels and 2 output channels.
Note that the inputs and outputs have the same height and width.
Each element in the output is derived
from a linear combination of elements *at the same position*
in the input image.
You could think of the $1\times 1$ convolutional layer
as constituting a fully-connected layer applied at every single pixel location
to transform the $c_i$ corresponding input values into $c_o$ output values.
Because this is still a convolutional layer,
the weights are tied across pixel location.
Thus the $1\times 1$ convolutional layer requires $c_o\times c_i$ weights
(plus the bias).

:label:`fig_conv_1x1`
Let us check whether this works in practice:
we implement a $1 \times 1$ convolution
using a fully-connected layer.
The only thing is that we need to make some adjustments
to the data shape before and after the matrix multiplication.
```
def corr2d_multi_in_out_1x1(X, K):
c_i, h, w = X.shape
c_o = K.shape[0]
X = X.reshape((c_i, h * w))
K = K.reshape((c_o, c_i))
# Matrix multiplication in the fully-connected layer
Y = torch.matmul(K, X)
return Y.reshape((c_o, h, w))
```
When performing $1\times 1$ convolution,
the above function is equivalent to the previously implemented cross-correlation function `corr2d_multi_in_out`.
Let us check this with some sample data.
```
X = torch.normal(0, 1, (3, 3, 3))
K = torch.normal(0, 1, (2, 3, 1, 1))
Y1 = corr2d_multi_in_out_1x1(X, K)
Y2 = corr2d_multi_in_out(X, K)
assert float(torch.abs(Y1 - Y2).sum()) < 1e-6
```
## Summary
* Multiple channels can be used to extend the model parameters of the convolutional layer.
* The $1\times 1$ convolutional layer is equivalent to the fully-connected layer, when applied on a per pixel basis.
* The $1\times 1$ convolutional layer is typically used to adjust the number of channels between network layers and to control model complexity.
## Exercises
1. Assume that we have two convolution kernels of size $k_1$ and $k_2$, respectively (with no nonlinearity in between).
1. Prove that the result of the operation can be expressed by a single convolution.
1. What is the dimensionality of the equivalent single convolution?
1. Is the converse true?
1. Assume an input of shape $c_i\times h\times w$ and a convolution kernel of shape $c_o\times c_i\times k_h\times k_w$, padding of $(p_h, p_w)$, and stride of $(s_h, s_w)$.
1. What is the computational cost (multiplications and additions) for the forward propagation?
1. What is the memory footprint?
1. What is the memory footprint for the backward computation?
1. What is the computational cost for the backpropagation?
1. By what factor does the number of calculations increase if we double the number of input channels $c_i$ and the number of output channels $c_o$? What happens if we double the padding?
1. If the height and width of a convolution kernel is $k_h=k_w=1$, what is the computational complexity of the forward propagation?
1. Are the variables `Y1` and `Y2` in the last example of this section exactly the same? Why?
1. How would you implement convolutions using matrix multiplication when the convolution window is not $1\times 1$?
[Discussions](https://discuss.d2l.ai/t/70)
|
github_jupyter
|
import torch
from d2l import torch as d2l
def corr2d_multi_in(X, K):
# First, iterate through the 0th dimension (channel dimension) of `X` and
# `K`. Then, add them together
return sum(d2l.corr2d(x, k) for x, k in zip(X, K))
X = torch.tensor([[[0.0, 1.0, 2.0], [3.0, 4.0, 5.0], [6.0, 7.0, 8.0]],
[[1.0, 2.0, 3.0], [4.0, 5.0, 6.0], [7.0, 8.0, 9.0]]])
K = torch.tensor([[[0.0, 1.0], [2.0, 3.0]], [[1.0, 2.0], [3.0, 4.0]]])
corr2d_multi_in(X, K)
def corr2d_multi_in_out(X, K):
# Iterate through the 0th dimension of `K`, and each time, perform
# cross-correlation operations with input `X`. All of the results are
# stacked together
return torch.stack([corr2d_multi_in(X, k) for k in K], 0)
K = torch.stack((K, K + 1, K + 2), 0)
K.shape
corr2d_multi_in_out(X, K)
def corr2d_multi_in_out_1x1(X, K):
c_i, h, w = X.shape
c_o = K.shape[0]
X = X.reshape((c_i, h * w))
K = K.reshape((c_o, c_i))
# Matrix multiplication in the fully-connected layer
Y = torch.matmul(K, X)
return Y.reshape((c_o, h, w))
X = torch.normal(0, 1, (3, 3, 3))
K = torch.normal(0, 1, (2, 3, 1, 1))
Y1 = corr2d_multi_in_out_1x1(X, K)
Y2 = corr2d_multi_in_out(X, K)
assert float(torch.abs(Y1 - Y2).sum()) < 1e-6
| 0.80479 | 0.992229 |
# Settings
```
%env TF_KERAS = 1
import os
sep_local = os.path.sep
import sys
# sys.path.append('..' + sep_local + '..' + sep_local +'..' + sep_local + '..' + sep_local + '..'+ sep_local + '..') # For Windows import
os.chdir('..' + sep_local + '..' + sep_local +'..') # For Linux import
print(sep_local)
print(os.getcwd())
import tensorflow as tf
print(tf.__version__)
```
# Dataset loading
```
dataset_name='atari_pacman'
images_dir = '/home/azeghost/datasets/.mspacman/atari_v1/screens/mspacman' #Linux
#images_dir = 'C:\\projects\\pokemon\DS06\\'
validation_percentage = 25
valid_format = 'png'
from training.generators.file_image_generator import create_image_lists, get_generators
imgs_list = create_image_lists(
image_dir=images_dir,
validation_pct=validation_percentage,
valid_imgae_formats=valid_format,
verbose = 0
)
```
### inputs_shape changed to (EPIS_LEN, ) + image_size due to class_mode='episode' when data is loaded
```
scale=1
image_size=(160//scale, 210//scale, 3)
batch_size = 10
EPIS_LEN = 10
EPIS_SHIFT = 5
inputs_shape= (EPIS_LEN, ) + image_size
latents_dim = 3
intermediate_dim = 30
#we created new class_mode episode_flat witch returns
#da.from_array(np.reshape(batch_x, (-1,)+self.image_shape )), da.from_array(np.reshape(batch_gt, (-1,)+self.image_shape))
```
### Class_mode is episode because time dimention "TimeDistributed" of the model
```
training_generator, testing_generator = get_generators(
images_list=imgs_list,
image_dir=images_dir,
image_size=image_size,
batch_size=batch_size,
class_mode='episode',
episode_len=EPIS_LEN,
episode_shift=EPIS_SHIFT
)
import tensorflow as tf
import numpy as np
```
### #Transformative changed output shapes of Dataset Generators
from (batch_size, EPIS_LEN, ) + image_size)
to (batch_size* EPIS_LEN, ) + image_size)
```
train_ds = tf.data.Dataset.from_generator(
lambda: training_generator,
output_types=(tf.float32, tf.float32) ,
output_shapes=(tf.TensorShape((batch_size, EPIS_LEN, ) + image_size),
tf.TensorShape((batch_size, EPIS_LEN, ) + image_size)
)
)
test_ds = tf.data.Dataset.from_generator(
lambda: testing_generator,
output_types=(tf.float32, tf.float32) ,
output_shapes=(tf.TensorShape((batch_size, EPIS_LEN, ) + image_size),
tf.TensorShape((batch_size, EPIS_LEN, ) + image_size)
)
)
_instance_scale=1.0
for data in train_ds:
_instance_scale = float(data[0].numpy().max())
break
_instance_scale = 1.0
import numpy as np
from collections.abc import Iterable
if isinstance(inputs_shape, Iterable):
_outputs_shape = np.prod(inputs_shape)
inputs_shape
next(training_generator)[0].shape
```
# Model's Layers definition
```
# TdDense is for timedistributed dense model
# tdDense = lambda **kwds: tf.keras.layers.TimeDistributed(tf.keras.layers.Dense(**kwds))
# c = 3
# enc_lays = [
# tdConv(filters=c, kernel_size=3, strides=(2, 2), activation='relu'),
# tdConv(filters=2*c, kernel_size=3, strides=(2, 2), activation='relu'),
# tf.keras.layers.Flatten(),
# # No activation
# tf.keras.layers.Dense(latents_dim)
# ]
# dec_lays = [
# tf.keras.layers.Dense(units=c**3, activation=tf.nn.relu),
# tf.keras.layers.Reshape(target_shape=(1, c, c, c)),
# tdDeConv(filters=2*c, kernel_size=3, strides=(2, 2), padding="SAME", activation='relu'),
# tdDeConv(filters=c, kernel_size=3, strides=(2, 2), padding="SAME", activation='relu'),
# # No activation
# tdDeConv(filters=1, kernel_size=3, strides=(1, 1), padding="SAME"),
# ]
tdConv = lambda **kwds: tf.keras.layers.TimeDistributed(tf.keras.layers.Conv2D(**kwds))
tdDeConv = lambda **kwds: tf.keras.layers.TimeDistributed(tf.keras.layers.Conv2DTranspose(**kwds))
```
### #Transformative Dense layer units=latents_dim*EPIS_LEN meaning 30 due to uniformity between all networks target_shape=(EPIS_LEN,) + c
```
units=30
c=(image_size[0]//4, image_size[1]//6, intermediate_dim//2) # now 4x and 6x smaller since kernels are 2 and 3
enc_lays = [
tdConv(filters=units, kernel_size=3, strides=(2, 2), activation='relu'),
tdConv(filters=units//5, kernel_size=3, strides=(2, 3), activation='relu'),
tf.keras.layers.Flatten(),
# No activation
tf.keras.layers.Dense(units=latents_dim*EPIS_LEN)
]
dec_lays = [
tf.keras.layers.Dense(units=np.product((EPIS_LEN,) + c), activation=tf.nn.relu),
tf.keras.layers.Reshape(target_shape=(EPIS_LEN,) + c),
tdDeConv(filters=units//5, kernel_size=3, strides=(2, 3), padding="SAME", activation='relu'),
tdDeConv(filters=units, kernel_size=3, strides=(2, 2), padding="SAME", activation='relu'),
# No activation
tdDeConv(filters=3, kernel_size=3, strides=(1, 1), padding="SAME")
]
```
# Model definition
```
import os
model_name = 'Atari_AE_TimeDist_Conv'
#experiments_dir='..'+sep_local+'..'+sep_local+'..'+sep_local+'experiments'+sep_local + model_name
experiments_dir=os.getcwd()+ sep_local +'experiments'+sep_local + model_name
from training.autoencoding_basic.transformative.AE import autoencoder as AE
```
### #Transformative 'outputs_shape':latents_dim*EPIS_LEN,
```
variables_params = \
[
{
'name': 'inference',
'inputs_shape':inputs_shape,
'outputs_shape':latents_dim*EPIS_LEN,
'layers': enc_lays
}
,
{
'name': 'generative',
'inputs_shape':latents_dim*EPIS_LEN,
'outputs_shape':inputs_shape,
'layers':dec_lays
}
]
from os.path import abspath
from utils.data_and_files.file_utils import create_if_not_exist
_restore = os.path.join(experiments_dir, 'var_save_dir')
create_if_not_exist(_restore)
absolute = abspath(_restore)
print("Restore_dir",absolute)
absolute = abspath(experiments_dir)
print("experiments_dir",absolute)
print("Current working dir",os.getcwd())
ae = AE(
name=model_name,
latents_dim=latents_dim,
batch_size=batch_size*EPIS_LEN,
episode_len= 1,
variables_params=variables_params,
filepath=None
)
# ae.compile(metrics=None)
ae.compile()
```
# Callbacks
```
import logging
mpl_logger = logging.getLogger('matplotlib')
mpl_logger.setLevel(logging.WARNING)
from training.callbacks.sample_generation import SampleGeneration
from training.callbacks.save_model import ModelSaver
es = tf.keras.callbacks.EarlyStopping(
monitor='loss',
min_delta=1e-12,
patience=12,
verbose=1,
restore_best_weights=False
)
ms = ModelSaver(filepath=_restore)
csv_dir = os.path.join(experiments_dir, 'csv_dir')
create_if_not_exist(csv_dir)
csv_dir = os.path.join(csv_dir, model_name+'.csv')
csv_log = tf.keras.callbacks.CSVLogger(csv_dir, append=True)
absolute = abspath(csv_dir)
print("Csv_dir",absolute)
image_gen_dir = os.path.join(experiments_dir, 'image_gen_dir')
create_if_not_exist(image_gen_dir)
absolute = abspath(image_gen_dir)
print("Image_gen_dir",absolute)
```
### #Transformative latents_shape should be latents_dim* EPIS_LEN
```
sg = SampleGeneration(latents_shape=latents_dim* EPIS_LEN, filepath=image_gen_dir, gen_freq=5, save_img=True, gray_plot=True)
```
# Model Training
```
ae.fit(
x=train_ds,
input_kw=None,
steps_per_epoch=2,
epochs=2,
verbose=2,
callbacks=[ es, ms, csv_log, sg],
workers=-1,
use_multiprocessing=True,
validation_data=test_ds,
validation_steps=2
)
```
# Model Evaluation
## inception_score
```
from evaluation.generativity_metrics.inception_metrics import inception_score
is_mean, is_sigma = inception_score(ae, tolerance_threshold=1e-6, max_iteration=200)
print(f'inception_score mean: {is_mean}, sigma: {is_sigma}')
```
## Frechet_inception_distance
```
from evaluation.generativity_metrics.inception_metrics import frechet_inception_distance
fis_score = frechet_inception_distance(ae, training_generator, tolerance_threshold=1e-6, max_iteration=10, batch_size=32)
print(f'frechet inception distance: {fis_score}')
```
## perceptual_path_length_score
```
from evaluation.generativity_metrics.perceptual_path_length import perceptual_path_length_score
ppl_mean_score = perceptual_path_length_score(ae, training_generator, tolerance_threshold=1e-6, max_iteration=200, batch_size=32)
print(f'perceptual path length score: {ppl_mean_score}')
```
## precision score
```
from evaluation.generativity_metrics.precision_recall import precision_score
_precision_score = precision_score(ae, training_generator, tolerance_threshold=1e-6, max_iteration=200)
print(f'precision score: {_precision_score}')
```
## recall score
```
from evaluation.generativity_metrics.precision_recall import recall_score
_recall_score = recall_score(ae, training_generator, tolerance_threshold=1e-6, max_iteration=200)
print(f'recall score: {_recall_score}')
```
# Image Generation
## image reconstruction
### Training dataset
```
from training.generators.image_generation_testing import reconstruct_from_a_batch
from utils.data_and_files.file_utils import create_if_not_exist
save_dir = os.path.join(experiments_dir, 'reconstruct_training_images_like_a_batch_dir')
create_if_not_exist(save_dir)
reconstruct_from_a_batch(ae, training_generator, save_dir)
from utils.data_and_files.file_utils import create_if_not_exist
save_dir = os.path.join(experiments_dir, 'reconstruct_testing_images_like_a_batch_dir')
create_if_not_exist(save_dir)
reconstruct_from_a_batch(ae, testing_generator, save_dir)
```
## with Randomness
```
from training.generators.image_generation_testing import generate_images_like_a_batch
from utils.data_and_files.file_utils import create_if_not_exist
save_dir = os.path.join(experiments_dir, 'generate_training_images_like_a_batch_dir')
create_if_not_exist(save_dir)
generate_images_like_a_batch(ae, training_generator, save_dir)
from utils.data_and_files.file_utils import create_if_not_exist
save_dir = os.path.join(experiments_dir, 'generate_testing_images_like_a_batch_dir')
create_if_not_exist(save_dir)
generate_images_like_a_batch(ae, testing_generator, save_dir)
```
### Complete Randomness
```
from training.generators.image_generation_testing import generate_images_randomly
from utils.data_and_files.file_utils import create_if_not_exist
save_dir = os.path.join(experiments_dir, 'random_synthetic_dir')
create_if_not_exist(save_dir)
generate_images_randomly(ae, save_dir)
from training.generators.image_generation_testing import interpolate_a_batch
from utils.data_and_files.file_utils import create_if_not_exist
save_dir = os.path.join(experiments_dir, 'interpolate_dir')
create_if_not_exist(save_dir)
interpolate_a_batch(ae, testing_generator, save_dir)
```
### Stacked inputs outputs and predictions
```
from training.generators.image_generation_testing import predict_from_a_batch
from utils.data_and_files.file_utils import create_if_not_exist
save_dir = os.path.join(experiments_dir, 'predictions')
create_if_not_exist(save_dir)
predict_from_a_batch(ae, testing_generator, save_dir)
```
|
github_jupyter
|
%env TF_KERAS = 1
import os
sep_local = os.path.sep
import sys
# sys.path.append('..' + sep_local + '..' + sep_local +'..' + sep_local + '..' + sep_local + '..'+ sep_local + '..') # For Windows import
os.chdir('..' + sep_local + '..' + sep_local +'..') # For Linux import
print(sep_local)
print(os.getcwd())
import tensorflow as tf
print(tf.__version__)
dataset_name='atari_pacman'
images_dir = '/home/azeghost/datasets/.mspacman/atari_v1/screens/mspacman' #Linux
#images_dir = 'C:\\projects\\pokemon\DS06\\'
validation_percentage = 25
valid_format = 'png'
from training.generators.file_image_generator import create_image_lists, get_generators
imgs_list = create_image_lists(
image_dir=images_dir,
validation_pct=validation_percentage,
valid_imgae_formats=valid_format,
verbose = 0
)
scale=1
image_size=(160//scale, 210//scale, 3)
batch_size = 10
EPIS_LEN = 10
EPIS_SHIFT = 5
inputs_shape= (EPIS_LEN, ) + image_size
latents_dim = 3
intermediate_dim = 30
#we created new class_mode episode_flat witch returns
#da.from_array(np.reshape(batch_x, (-1,)+self.image_shape )), da.from_array(np.reshape(batch_gt, (-1,)+self.image_shape))
training_generator, testing_generator = get_generators(
images_list=imgs_list,
image_dir=images_dir,
image_size=image_size,
batch_size=batch_size,
class_mode='episode',
episode_len=EPIS_LEN,
episode_shift=EPIS_SHIFT
)
import tensorflow as tf
import numpy as np
train_ds = tf.data.Dataset.from_generator(
lambda: training_generator,
output_types=(tf.float32, tf.float32) ,
output_shapes=(tf.TensorShape((batch_size, EPIS_LEN, ) + image_size),
tf.TensorShape((batch_size, EPIS_LEN, ) + image_size)
)
)
test_ds = tf.data.Dataset.from_generator(
lambda: testing_generator,
output_types=(tf.float32, tf.float32) ,
output_shapes=(tf.TensorShape((batch_size, EPIS_LEN, ) + image_size),
tf.TensorShape((batch_size, EPIS_LEN, ) + image_size)
)
)
_instance_scale=1.0
for data in train_ds:
_instance_scale = float(data[0].numpy().max())
break
_instance_scale = 1.0
import numpy as np
from collections.abc import Iterable
if isinstance(inputs_shape, Iterable):
_outputs_shape = np.prod(inputs_shape)
inputs_shape
next(training_generator)[0].shape
# TdDense is for timedistributed dense model
# tdDense = lambda **kwds: tf.keras.layers.TimeDistributed(tf.keras.layers.Dense(**kwds))
# c = 3
# enc_lays = [
# tdConv(filters=c, kernel_size=3, strides=(2, 2), activation='relu'),
# tdConv(filters=2*c, kernel_size=3, strides=(2, 2), activation='relu'),
# tf.keras.layers.Flatten(),
# # No activation
# tf.keras.layers.Dense(latents_dim)
# ]
# dec_lays = [
# tf.keras.layers.Dense(units=c**3, activation=tf.nn.relu),
# tf.keras.layers.Reshape(target_shape=(1, c, c, c)),
# tdDeConv(filters=2*c, kernel_size=3, strides=(2, 2), padding="SAME", activation='relu'),
# tdDeConv(filters=c, kernel_size=3, strides=(2, 2), padding="SAME", activation='relu'),
# # No activation
# tdDeConv(filters=1, kernel_size=3, strides=(1, 1), padding="SAME"),
# ]
tdConv = lambda **kwds: tf.keras.layers.TimeDistributed(tf.keras.layers.Conv2D(**kwds))
tdDeConv = lambda **kwds: tf.keras.layers.TimeDistributed(tf.keras.layers.Conv2DTranspose(**kwds))
units=30
c=(image_size[0]//4, image_size[1]//6, intermediate_dim//2) # now 4x and 6x smaller since kernels are 2 and 3
enc_lays = [
tdConv(filters=units, kernel_size=3, strides=(2, 2), activation='relu'),
tdConv(filters=units//5, kernel_size=3, strides=(2, 3), activation='relu'),
tf.keras.layers.Flatten(),
# No activation
tf.keras.layers.Dense(units=latents_dim*EPIS_LEN)
]
dec_lays = [
tf.keras.layers.Dense(units=np.product((EPIS_LEN,) + c), activation=tf.nn.relu),
tf.keras.layers.Reshape(target_shape=(EPIS_LEN,) + c),
tdDeConv(filters=units//5, kernel_size=3, strides=(2, 3), padding="SAME", activation='relu'),
tdDeConv(filters=units, kernel_size=3, strides=(2, 2), padding="SAME", activation='relu'),
# No activation
tdDeConv(filters=3, kernel_size=3, strides=(1, 1), padding="SAME")
]
import os
model_name = 'Atari_AE_TimeDist_Conv'
#experiments_dir='..'+sep_local+'..'+sep_local+'..'+sep_local+'experiments'+sep_local + model_name
experiments_dir=os.getcwd()+ sep_local +'experiments'+sep_local + model_name
from training.autoencoding_basic.transformative.AE import autoencoder as AE
variables_params = \
[
{
'name': 'inference',
'inputs_shape':inputs_shape,
'outputs_shape':latents_dim*EPIS_LEN,
'layers': enc_lays
}
,
{
'name': 'generative',
'inputs_shape':latents_dim*EPIS_LEN,
'outputs_shape':inputs_shape,
'layers':dec_lays
}
]
from os.path import abspath
from utils.data_and_files.file_utils import create_if_not_exist
_restore = os.path.join(experiments_dir, 'var_save_dir')
create_if_not_exist(_restore)
absolute = abspath(_restore)
print("Restore_dir",absolute)
absolute = abspath(experiments_dir)
print("experiments_dir",absolute)
print("Current working dir",os.getcwd())
ae = AE(
name=model_name,
latents_dim=latents_dim,
batch_size=batch_size*EPIS_LEN,
episode_len= 1,
variables_params=variables_params,
filepath=None
)
# ae.compile(metrics=None)
ae.compile()
import logging
mpl_logger = logging.getLogger('matplotlib')
mpl_logger.setLevel(logging.WARNING)
from training.callbacks.sample_generation import SampleGeneration
from training.callbacks.save_model import ModelSaver
es = tf.keras.callbacks.EarlyStopping(
monitor='loss',
min_delta=1e-12,
patience=12,
verbose=1,
restore_best_weights=False
)
ms = ModelSaver(filepath=_restore)
csv_dir = os.path.join(experiments_dir, 'csv_dir')
create_if_not_exist(csv_dir)
csv_dir = os.path.join(csv_dir, model_name+'.csv')
csv_log = tf.keras.callbacks.CSVLogger(csv_dir, append=True)
absolute = abspath(csv_dir)
print("Csv_dir",absolute)
image_gen_dir = os.path.join(experiments_dir, 'image_gen_dir')
create_if_not_exist(image_gen_dir)
absolute = abspath(image_gen_dir)
print("Image_gen_dir",absolute)
sg = SampleGeneration(latents_shape=latents_dim* EPIS_LEN, filepath=image_gen_dir, gen_freq=5, save_img=True, gray_plot=True)
ae.fit(
x=train_ds,
input_kw=None,
steps_per_epoch=2,
epochs=2,
verbose=2,
callbacks=[ es, ms, csv_log, sg],
workers=-1,
use_multiprocessing=True,
validation_data=test_ds,
validation_steps=2
)
from evaluation.generativity_metrics.inception_metrics import inception_score
is_mean, is_sigma = inception_score(ae, tolerance_threshold=1e-6, max_iteration=200)
print(f'inception_score mean: {is_mean}, sigma: {is_sigma}')
from evaluation.generativity_metrics.inception_metrics import frechet_inception_distance
fis_score = frechet_inception_distance(ae, training_generator, tolerance_threshold=1e-6, max_iteration=10, batch_size=32)
print(f'frechet inception distance: {fis_score}')
from evaluation.generativity_metrics.perceptual_path_length import perceptual_path_length_score
ppl_mean_score = perceptual_path_length_score(ae, training_generator, tolerance_threshold=1e-6, max_iteration=200, batch_size=32)
print(f'perceptual path length score: {ppl_mean_score}')
from evaluation.generativity_metrics.precision_recall import precision_score
_precision_score = precision_score(ae, training_generator, tolerance_threshold=1e-6, max_iteration=200)
print(f'precision score: {_precision_score}')
from evaluation.generativity_metrics.precision_recall import recall_score
_recall_score = recall_score(ae, training_generator, tolerance_threshold=1e-6, max_iteration=200)
print(f'recall score: {_recall_score}')
from training.generators.image_generation_testing import reconstruct_from_a_batch
from utils.data_and_files.file_utils import create_if_not_exist
save_dir = os.path.join(experiments_dir, 'reconstruct_training_images_like_a_batch_dir')
create_if_not_exist(save_dir)
reconstruct_from_a_batch(ae, training_generator, save_dir)
from utils.data_and_files.file_utils import create_if_not_exist
save_dir = os.path.join(experiments_dir, 'reconstruct_testing_images_like_a_batch_dir')
create_if_not_exist(save_dir)
reconstruct_from_a_batch(ae, testing_generator, save_dir)
from training.generators.image_generation_testing import generate_images_like_a_batch
from utils.data_and_files.file_utils import create_if_not_exist
save_dir = os.path.join(experiments_dir, 'generate_training_images_like_a_batch_dir')
create_if_not_exist(save_dir)
generate_images_like_a_batch(ae, training_generator, save_dir)
from utils.data_and_files.file_utils import create_if_not_exist
save_dir = os.path.join(experiments_dir, 'generate_testing_images_like_a_batch_dir')
create_if_not_exist(save_dir)
generate_images_like_a_batch(ae, testing_generator, save_dir)
from training.generators.image_generation_testing import generate_images_randomly
from utils.data_and_files.file_utils import create_if_not_exist
save_dir = os.path.join(experiments_dir, 'random_synthetic_dir')
create_if_not_exist(save_dir)
generate_images_randomly(ae, save_dir)
from training.generators.image_generation_testing import interpolate_a_batch
from utils.data_and_files.file_utils import create_if_not_exist
save_dir = os.path.join(experiments_dir, 'interpolate_dir')
create_if_not_exist(save_dir)
interpolate_a_batch(ae, testing_generator, save_dir)
from training.generators.image_generation_testing import predict_from_a_batch
from utils.data_and_files.file_utils import create_if_not_exist
save_dir = os.path.join(experiments_dir, 'predictions')
create_if_not_exist(save_dir)
predict_from_a_batch(ae, testing_generator, save_dir)
| 0.414425 | 0.716959 |
# Riemann Problems and Jupyter Solutions
### by David I. Ketcheson, Randall J. LeVeque, and Mauricio del Razo Sarmina
The Github repository containing these notebooks is https://github.com/clawpack/riemann_book.
You can view html versions of these notebooks at http://www.clawpack.org/riemann_book/html/Index.html.
Parts I and II of these notebooks will also be published by SIAM as a paperback book, to appear with luck in late 2019.
## Contents
- [Preface](Preface.ipynb). Describes the aims and goals, and different ways to use the notebooks.
## Part I: The Riemann problem and its solution
1. [Introduction](Introduction.ipynb). Introduces basic ideas with some sample solutions.
1. [Advection](Advection.ipynb) The scalar advection equation is the simplest hyperbolic problem.
2. [Acoustics](Acoustics.ipynb) This linear system of two equations illustrates how eigenstructure is used.
3. [Burgers' equation](Burgers.ipynb) The classic nonlinear scalar problem with a convex flux.
4. [Traffic flow](Traffic_flow.ipynb) A nonlinear scalar problem with a nice physical interpretation.
5. [Nonconvex_scalar](Nonconvex_scalar.ipynb) More interesting Riemann solutions arise when the flux is not convex.
6. [Shallow water waves](Shallow_water.ipynb) A classic nonlinear system of two equations
7. [Shallow water with a tracer](Shallow_tracer.ipynb) Adding a passively advected tracer and a linearly degenerate field.
8. [Euler equations of compressible gas dynamics](Euler.ipynb) The classic equations for an ideal gas.
## Part II: Approximate solvers
1. [Approximate_solvers](Approximate_solvers.ipynb). Introduction to two basic types of approximations.
1. [Burgers equation](Burgers_approximate.ipynb)
2. [Shallow water](Shallow_water_approximate.ipynb)
3. [Euler approximate solvers](Euler_approximate.ipynb).
4. [Numerical comparisons](Euler_compare.ipynb)
Parts III - VI are still very much under development and updates may appear in the [Github repository](https://github.com/clawpack/riemann_book) in the future.
## Part III: Riemann problems with spatially-varying flux
1. Advection
2. [Acoustics in heterogeneous media](Acoustics_heterogeneous.ipynb)
3. [Traffic with varying road conditions](Traffic_variable_speed.ipynb)
4. [Nonlinear elasticity in a heterogeneous medium](Nonlinear_elasticity.ipynb)
5. Ideal gas shock tube with different ratio of specific heats
6. [Euler equations with Tammann equation of state](Euler_equations_TammannEOS.ipynb)
## Part IV: Source terms
1. [Traffic with an on-ramp](Traffic_with_ramps.ipynb)
## Part V: Non-classical problems
1. [Nonconvex flux for a scalar problem](Nonconvex_scalar.ipynb)
2. [Pressureless flow](Pressureless_flow.ipynb)
## Part VI: Multidimensional systems
1. [Acoustics](http://nbviewer.jupyter.org/github/maojrs/ipynotebooks/blob/master/acoustics_riemann.ipynb) *[To be improved and incorporated into this project]*
2. [Elasticity](http://nbviewer.jupyter.org/github/maojrs/ipynotebooks/blob/master/elasticity_riemann.ipynb) *[To be improved and incorporated into this project]*
3. [The Kitchen Sink: shallow water in cylindrical coordinates](Kitchen_sink_problem.ipynb)
|
github_jupyter
|
# Riemann Problems and Jupyter Solutions
### by David I. Ketcheson, Randall J. LeVeque, and Mauricio del Razo Sarmina
The Github repository containing these notebooks is https://github.com/clawpack/riemann_book.
You can view html versions of these notebooks at http://www.clawpack.org/riemann_book/html/Index.html.
Parts I and II of these notebooks will also be published by SIAM as a paperback book, to appear with luck in late 2019.
## Contents
- [Preface](Preface.ipynb). Describes the aims and goals, and different ways to use the notebooks.
## Part I: The Riemann problem and its solution
1. [Introduction](Introduction.ipynb). Introduces basic ideas with some sample solutions.
1. [Advection](Advection.ipynb) The scalar advection equation is the simplest hyperbolic problem.
2. [Acoustics](Acoustics.ipynb) This linear system of two equations illustrates how eigenstructure is used.
3. [Burgers' equation](Burgers.ipynb) The classic nonlinear scalar problem with a convex flux.
4. [Traffic flow](Traffic_flow.ipynb) A nonlinear scalar problem with a nice physical interpretation.
5. [Nonconvex_scalar](Nonconvex_scalar.ipynb) More interesting Riemann solutions arise when the flux is not convex.
6. [Shallow water waves](Shallow_water.ipynb) A classic nonlinear system of two equations
7. [Shallow water with a tracer](Shallow_tracer.ipynb) Adding a passively advected tracer and a linearly degenerate field.
8. [Euler equations of compressible gas dynamics](Euler.ipynb) The classic equations for an ideal gas.
## Part II: Approximate solvers
1. [Approximate_solvers](Approximate_solvers.ipynb). Introduction to two basic types of approximations.
1. [Burgers equation](Burgers_approximate.ipynb)
2. [Shallow water](Shallow_water_approximate.ipynb)
3. [Euler approximate solvers](Euler_approximate.ipynb).
4. [Numerical comparisons](Euler_compare.ipynb)
Parts III - VI are still very much under development and updates may appear in the [Github repository](https://github.com/clawpack/riemann_book) in the future.
## Part III: Riemann problems with spatially-varying flux
1. Advection
2. [Acoustics in heterogeneous media](Acoustics_heterogeneous.ipynb)
3. [Traffic with varying road conditions](Traffic_variable_speed.ipynb)
4. [Nonlinear elasticity in a heterogeneous medium](Nonlinear_elasticity.ipynb)
5. Ideal gas shock tube with different ratio of specific heats
6. [Euler equations with Tammann equation of state](Euler_equations_TammannEOS.ipynb)
## Part IV: Source terms
1. [Traffic with an on-ramp](Traffic_with_ramps.ipynb)
## Part V: Non-classical problems
1. [Nonconvex flux for a scalar problem](Nonconvex_scalar.ipynb)
2. [Pressureless flow](Pressureless_flow.ipynb)
## Part VI: Multidimensional systems
1. [Acoustics](http://nbviewer.jupyter.org/github/maojrs/ipynotebooks/blob/master/acoustics_riemann.ipynb) *[To be improved and incorporated into this project]*
2. [Elasticity](http://nbviewer.jupyter.org/github/maojrs/ipynotebooks/blob/master/elasticity_riemann.ipynb) *[To be improved and incorporated into this project]*
3. [The Kitchen Sink: shallow water in cylindrical coordinates](Kitchen_sink_problem.ipynb)
| 0.674587 | 0.847968 |
```
import json
with open("F:/Dataset/coco training/26/annotations/instances_default.json") as f: # Here you will put dataset to what you want to add new patient
data = json.load(f)
print(data['categories'])
IDOfAnotatedCathegory=[1,2,9,3] # For this parametrs it is Left Kidney, Right Kidney and Heart ( According to id in data['categories'])
helper=data['annotations']
counter=0
for i in range(len(helper)):
if data['annotations'][i]['category_id']==1 or data['annotations'][i]['category_id']==IDOfAnotatedCathegory[1] or data['annotations'][i]['category_id']==IDOfAnotatedCathegory[2]:
# i dont know, but for category_id==7 there are nothing, but in fact we have anotation of hearth
#print('ok', data['annotations'][i]['category_id'])
counter=counter+1
print('counter= ', counter)
print(data.keys())
print(type(data))
print(type(data["categories"]))
data_out = {}
data.keys()
data_out = dict() #new dictionary
data_out["licenses"] = data["licenses"]
data_out["info"] = data["info"]
data_out["categories"]=[]
data_out["images"] = data["images"]
data_out["annotations"] = []
data.keys()
dataFin={}
print(data['categories'])
trainingCathegories=4
for i in range(trainingCathegories):
print(i)
data_out['categories'].append(dict())
print(data_out['categories'])
def NumberOfSegmantationedElemetns(data,IDOfAnotatedCathegory):
helper=data['annotations']
counter=0
for i in range(len(helper)):
if data['annotations'][i]['category_id']==IDOfAnotatedCathegory[0] or data['annotations'][i]['category_id']==IDOfAnotatedCathegory[1] or data['annotations'][i]['category_id']==IDOfAnotatedCathegory[2] or data['annotations'][i]['category_id']==IDOfAnotatedCathegory[3]:
# i dont know, but for category_id==7 there are nothing, but in fact we have anotation of hearth
#print('ok', data['annotations'][i]['category_id'])
counter=counter+1
print('counter= ', counter)
return(counter)
NeededAnotations=NumberOfSegmantationedElemetns(data,IDOfAnotatedCathegory)
for i in range(NeededAnotations):
#print(i)
data_out['annotations'].append(dict())
#print(data_out['annotations'])
magic=data["categories"]
#print("zzzzz", data["categories"][0])
counter=0;
for i in range(len(magic)):
#print('i =', i)
#print(data["categories"][i]['name'])
if data["categories"][i]['name']=="Left Kidney" or data["categories"][i]['name']=="Liver" or data["categories"][i]['name']=="Heart" or data["categories"][i]['name']=="Right Kidney":
#print("ok")
#print(data["categories"][i])
data_out['categories'][counter]=(data["categories"][i])
#data_out['categories'][counter+1]['id']=counter
counter=counter+1
#print(data_out['categories'])
magic2=data["annotations"]
#print("zzzzz", data["annotations"][0])
counter=0;
for i in range(len(magic2)):
#print('i =', i)
#print(data["categories"][i]['name'])
if data['annotations'][i]['category_id'] in IDOfAnotatedCathegory:
#print(data['annotations'][i]['category_id'])
#print(data["categories"][i])
data_out['annotations'][counter]=(data["annotations"][i])
#data_out['categories'][counter]['id']=counter
counter=counter+1
#print(data_out['annotations'])
#code how to add new patient - start.
with open("F:\Dataset/coco training/28/annotations/instances_default.json") as f:
data2 = json.load(f) # druhy soubor dat
magic2=data2["annotations"]
#print("zzzzz", data["annotations"][0])
counter=0;
for i in range(len(magic2)):
#print('i =', i)
#print(data["categories"][i]['name'])
if data['annotations'][i]['category_id'] in IDOfAnotatedCathegory:
#print(data["categories"][i])
data_out['annotations'].append(data["annotations"][i])
#data_out['categories'][counter]['id']=counter
counter=counter+1
#print(data_out['annotations'])
data_out["images"].append(data2["images"])
#code how to add new patient - end.
with open("F:\Dataset/coco training/32/annotations/instances_default.json") as f:
data3 = json.load(f) # 3 dataset. Unfortunatelly we need to add them one by one in our queue.
magic3=data3["annotations"]
#print("zzzzz", data["annotations"][0])
counter=0;
for i in range(len(magic3)):
#print('i =', i)
#print(data["categories"][i]['name'])
if data['annotations'][i]['category_id'] in IDOfAnotatedCathegory:
#print(data["categories"][i])
data_out['annotations'].append(data["annotations"][i])
#data_out['categories'][counter]['id']=counter
counter=counter+1
#print(data_out['annotations'])
data_out["images"].append(data3["images"])
#Here you will all code for adding new information from new patient.
print(data_out.keys())
EntirePatient=json.dumps(data_out, indent=5)
with open('F:/Dataset/coco training/EntireDatasetCoco/EntireCoco.json', 'w') as json_file: #Here we make a new json file, which will be our entire dataset.
json.dump(data_out, json_file)
```
|
github_jupyter
|
import json
with open("F:/Dataset/coco training/26/annotations/instances_default.json") as f: # Here you will put dataset to what you want to add new patient
data = json.load(f)
print(data['categories'])
IDOfAnotatedCathegory=[1,2,9,3] # For this parametrs it is Left Kidney, Right Kidney and Heart ( According to id in data['categories'])
helper=data['annotations']
counter=0
for i in range(len(helper)):
if data['annotations'][i]['category_id']==1 or data['annotations'][i]['category_id']==IDOfAnotatedCathegory[1] or data['annotations'][i]['category_id']==IDOfAnotatedCathegory[2]:
# i dont know, but for category_id==7 there are nothing, but in fact we have anotation of hearth
#print('ok', data['annotations'][i]['category_id'])
counter=counter+1
print('counter= ', counter)
print(data.keys())
print(type(data))
print(type(data["categories"]))
data_out = {}
data.keys()
data_out = dict() #new dictionary
data_out["licenses"] = data["licenses"]
data_out["info"] = data["info"]
data_out["categories"]=[]
data_out["images"] = data["images"]
data_out["annotations"] = []
data.keys()
dataFin={}
print(data['categories'])
trainingCathegories=4
for i in range(trainingCathegories):
print(i)
data_out['categories'].append(dict())
print(data_out['categories'])
def NumberOfSegmantationedElemetns(data,IDOfAnotatedCathegory):
helper=data['annotations']
counter=0
for i in range(len(helper)):
if data['annotations'][i]['category_id']==IDOfAnotatedCathegory[0] or data['annotations'][i]['category_id']==IDOfAnotatedCathegory[1] or data['annotations'][i]['category_id']==IDOfAnotatedCathegory[2] or data['annotations'][i]['category_id']==IDOfAnotatedCathegory[3]:
# i dont know, but for category_id==7 there are nothing, but in fact we have anotation of hearth
#print('ok', data['annotations'][i]['category_id'])
counter=counter+1
print('counter= ', counter)
return(counter)
NeededAnotations=NumberOfSegmantationedElemetns(data,IDOfAnotatedCathegory)
for i in range(NeededAnotations):
#print(i)
data_out['annotations'].append(dict())
#print(data_out['annotations'])
magic=data["categories"]
#print("zzzzz", data["categories"][0])
counter=0;
for i in range(len(magic)):
#print('i =', i)
#print(data["categories"][i]['name'])
if data["categories"][i]['name']=="Left Kidney" or data["categories"][i]['name']=="Liver" or data["categories"][i]['name']=="Heart" or data["categories"][i]['name']=="Right Kidney":
#print("ok")
#print(data["categories"][i])
data_out['categories'][counter]=(data["categories"][i])
#data_out['categories'][counter+1]['id']=counter
counter=counter+1
#print(data_out['categories'])
magic2=data["annotations"]
#print("zzzzz", data["annotations"][0])
counter=0;
for i in range(len(magic2)):
#print('i =', i)
#print(data["categories"][i]['name'])
if data['annotations'][i]['category_id'] in IDOfAnotatedCathegory:
#print(data['annotations'][i]['category_id'])
#print(data["categories"][i])
data_out['annotations'][counter]=(data["annotations"][i])
#data_out['categories'][counter]['id']=counter
counter=counter+1
#print(data_out['annotations'])
#code how to add new patient - start.
with open("F:\Dataset/coco training/28/annotations/instances_default.json") as f:
data2 = json.load(f) # druhy soubor dat
magic2=data2["annotations"]
#print("zzzzz", data["annotations"][0])
counter=0;
for i in range(len(magic2)):
#print('i =', i)
#print(data["categories"][i]['name'])
if data['annotations'][i]['category_id'] in IDOfAnotatedCathegory:
#print(data["categories"][i])
data_out['annotations'].append(data["annotations"][i])
#data_out['categories'][counter]['id']=counter
counter=counter+1
#print(data_out['annotations'])
data_out["images"].append(data2["images"])
#code how to add new patient - end.
with open("F:\Dataset/coco training/32/annotations/instances_default.json") as f:
data3 = json.load(f) # 3 dataset. Unfortunatelly we need to add them one by one in our queue.
magic3=data3["annotations"]
#print("zzzzz", data["annotations"][0])
counter=0;
for i in range(len(magic3)):
#print('i =', i)
#print(data["categories"][i]['name'])
if data['annotations'][i]['category_id'] in IDOfAnotatedCathegory:
#print(data["categories"][i])
data_out['annotations'].append(data["annotations"][i])
#data_out['categories'][counter]['id']=counter
counter=counter+1
#print(data_out['annotations'])
data_out["images"].append(data3["images"])
#Here you will all code for adding new information from new patient.
print(data_out.keys())
EntirePatient=json.dumps(data_out, indent=5)
with open('F:/Dataset/coco training/EntireDatasetCoco/EntireCoco.json', 'w') as json_file: #Here we make a new json file, which will be our entire dataset.
json.dump(data_out, json_file)
| 0.028585 | 0.24907 |
## Tracking points through a reconstruction
A consequence of having plate reconstructions with dynamically evolving plate boundaries is that we can define how a point moves through time, beginning on one plate but subsequently being transferred to other plates. For example:
- a point within an oceanic plate that subsequently splits into two plates
- a point on an oceanic plate that reaches a subduction zone (such that the crust may get subducted, or may become accreted to the overriding plate.
A geological example could be the products of a deep plume have ended up, assuming that
1. the mantle plume produces seamounts (or thick crust) on the plate lying over it
2. these seamounts then travels with the plate, but later reaches a subduction zone where it is accreted to an active margin.
This notebook goes through the steps of defining a point at some moment in the geological past, finding out what plate it is on at this time, then tracking the motion of this point incrementally for a series of time steps, testing to see whether the point still lies within the boundaries of the plate that it started on.
##### Technical details
From a technical point of view, the process involves knowledge of:
- resolved topologies
- assigning plate ids using plate partitioning
- modifying the geometry of a feature
```
import pygplates
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
rotation_filename = 'Data/Seton_etal_ESR2012_2012.1.rot'
input_topology_filename = 'Data/Seton_etal_ESR2012_PP_2012.1.gpmlz'
topology_features = pygplates.FeatureCollection(input_topology_filename)
rotation_model = pygplates.RotationModel(rotation_filename)
# Alternative method using 'partition_geometry'
time_step = 2.
oldest_seed_time = 150.
# Empty array for storing Long/Lat of
point_longitude = []
point_latitude = []
time_list = np.arange(oldest_seed_time,time_step,-time_step)
for seed_time in time_list:
# Location of seed point for Kerguelen
#seed_geometry = pygplates.PointOnSphere(-50, 80)
# Seed point for Hawaii
seed_geometry = pygplates.PointOnSphere(19, -155)
for time in np.arange(seed_time,0.,-time_step):
#print max_time, time
# Get the plate polygons for this time
resolved_topologies = []
pygplates.resolve_topologies(topology_features, rotation_model, resolved_topologies, time)
# make plate partitioner from polygons
plate_partitioner = pygplates.PlatePartitioner(resolved_topologies, rotation_model)
# Find the plate id of the polygon that contains the point
partitioned_inside_geometries = []
plate_partitioner.partition_geometry(seed_geometry, partitioned_inside_geometries)
PlateID = partitioned_inside_geometries[0][0].get_feature().get_reconstruction_plate_id()
#print PlateID
# Get the stage rotation that will move the point from where it is at the current time
# to its location at the next time step, based on the plate id that contains the point at the
# current time
stage_rotation = rotation_model.get_rotation(time-time_step, PlateID, time, anchor_plate_id=1)
# use the stage rotation to reconstruct the tracked point from position at current time
# to position at the next time step
seed_geometry = stage_rotation * seed_geometry
print('seed time = %d, plume is within plate %i' % (seed_time, PlateID))
point_longitude.append(seed_geometry.to_lat_lon_point().get_longitude())
point_latitude.append(seed_geometry.to_lat_lon_point().get_latitude())
print('coordinates of reconstructed plume products')
print(zip(time_list,point_longitude,point_latitude))
import cartopy.crs as ccrs
import cartopy.feature as cfeature
import cartopy.io.shapereader as shpreader
%matplotlib inline
# Create figure
fig = plt.figure(figsize=(10,8))
m = fig.add_subplot(111,projection=ccrs.Orthographic(central_latitude=40., central_longitude=-160.))
# Draw coastlines, parallels, meridians.
m.coastlines(resolution='10m', color='black', linewidth=1)
m.gridlines(color='black',linestyle='--', xlocs=np.arange(-180,180,20.), ylocs=np.arange(-80,90,20))
# Plot the movement of the seedpoint
x,y = np.array(point_longitude), np.array(point_latitude)
c1 = m.scatter(x,y,c=time_list,s=50, cmap=plt.cm.gnuplot_r, transform=ccrs.PlateCarree(), edgecolor='k',zorder=1)
# Add a colorbar and show plot
cb1 = plt.colorbar(c1).set_label('Age (Ma)')
m.set_global()
plt.show()
```
|
github_jupyter
|
import pygplates
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
rotation_filename = 'Data/Seton_etal_ESR2012_2012.1.rot'
input_topology_filename = 'Data/Seton_etal_ESR2012_PP_2012.1.gpmlz'
topology_features = pygplates.FeatureCollection(input_topology_filename)
rotation_model = pygplates.RotationModel(rotation_filename)
# Alternative method using 'partition_geometry'
time_step = 2.
oldest_seed_time = 150.
# Empty array for storing Long/Lat of
point_longitude = []
point_latitude = []
time_list = np.arange(oldest_seed_time,time_step,-time_step)
for seed_time in time_list:
# Location of seed point for Kerguelen
#seed_geometry = pygplates.PointOnSphere(-50, 80)
# Seed point for Hawaii
seed_geometry = pygplates.PointOnSphere(19, -155)
for time in np.arange(seed_time,0.,-time_step):
#print max_time, time
# Get the plate polygons for this time
resolved_topologies = []
pygplates.resolve_topologies(topology_features, rotation_model, resolved_topologies, time)
# make plate partitioner from polygons
plate_partitioner = pygplates.PlatePartitioner(resolved_topologies, rotation_model)
# Find the plate id of the polygon that contains the point
partitioned_inside_geometries = []
plate_partitioner.partition_geometry(seed_geometry, partitioned_inside_geometries)
PlateID = partitioned_inside_geometries[0][0].get_feature().get_reconstruction_plate_id()
#print PlateID
# Get the stage rotation that will move the point from where it is at the current time
# to its location at the next time step, based on the plate id that contains the point at the
# current time
stage_rotation = rotation_model.get_rotation(time-time_step, PlateID, time, anchor_plate_id=1)
# use the stage rotation to reconstruct the tracked point from position at current time
# to position at the next time step
seed_geometry = stage_rotation * seed_geometry
print('seed time = %d, plume is within plate %i' % (seed_time, PlateID))
point_longitude.append(seed_geometry.to_lat_lon_point().get_longitude())
point_latitude.append(seed_geometry.to_lat_lon_point().get_latitude())
print('coordinates of reconstructed plume products')
print(zip(time_list,point_longitude,point_latitude))
import cartopy.crs as ccrs
import cartopy.feature as cfeature
import cartopy.io.shapereader as shpreader
%matplotlib inline
# Create figure
fig = plt.figure(figsize=(10,8))
m = fig.add_subplot(111,projection=ccrs.Orthographic(central_latitude=40., central_longitude=-160.))
# Draw coastlines, parallels, meridians.
m.coastlines(resolution='10m', color='black', linewidth=1)
m.gridlines(color='black',linestyle='--', xlocs=np.arange(-180,180,20.), ylocs=np.arange(-80,90,20))
# Plot the movement of the seedpoint
x,y = np.array(point_longitude), np.array(point_latitude)
c1 = m.scatter(x,y,c=time_list,s=50, cmap=plt.cm.gnuplot_r, transform=ccrs.PlateCarree(), edgecolor='k',zorder=1)
# Add a colorbar and show plot
cb1 = plt.colorbar(c1).set_label('Age (Ma)')
m.set_global()
plt.show()
| 0.571049 | 0.974264 |
[](https://pythonista.io)
## El estándar XML.
XML es el acrónimo de "Extensible Markup Language" que se puede traducir como Lenguaje Extensible de Marcadores y corresponde a un estándar general para serializar datos de diversas índoles de forma estructurada.
El estándar de XML fue publicado en 1996 por el W3C y se utiliza de forma intensiva para definir estructuras de datos.
A un documento XML se le conoce como *Elemento* y contiene estructuras de datos basada en contenidos delimitados por marcadores (markups). Dichos marcadores corresponden a etiquetas (tags) que indican el principio y el fin de la estructura que delimitan.
## El paquete *xml.etree.ElementTree*
El paquete *xml* forma parte de la Biblioteca Estándar de Python y contiene a su vez una serie de paquetes y módulos especializados en la gestión y manipulación de documentos estrcuturados.
El paquete *xml.etree.ElementTree*, se especializa en documentos XML y contiene diversas clases y funciones que se pueden utilizar para tal propósito.
```
import xml.etree.ElementTree as ET
dir(ET)
```
### La clase * Element*.
El módulo *xml.etree.elementree* contiene a la clase *Element*, la cual permite inspeccionar un documento XML mediante el el acceso a sus métodos y atributos, así como el indexado de sus elementos.
## Documento de ejemplo.
El documento localizado en [data/cursos.xml](data/cursos.xml) contiene el siguiente código:
``` xml
<?xml version="1.0"?>
<data>
<curso clave="py101" nombre="Introducción a la Programación con Python 3">
<requiere />
<categoria>Introductorio</categoria>
<version>2018</version>
<url>https://pythonista.mx/cursos/py101</url>
<tema orden="1" nombre="Introducción al lenguaje Python" />
<tema orden="2" nombre="Palabras reservadas y espacio de nombres" />
<tema orden="3" nombre="Expresiones y declaraciones" />
<tema orden="4" nombre="Números, cadenas de caracteres, tipos y operadores" />
<tema orden="5" nombre="Orientación a objetos e introspección" />
<tema orden="6" nombre="Entrada y salida estándar" />
<tema orden="7" nombre="Bloques, comentarios y condicionales" />
<tema orden="8" nombre="Ciclos, iteraciones e interrupciones de ejecución" />
<tema orden="9" nombre="Objetos tipo list y tipo tuple" />
<tema orden="10" nombre="Objetos tipo dict" />
<tema orden="11" nombre="Objetos tipo str" />
<tema orden="12" nombre="Objetos tipo set y frozenset" />
<tema orden="13" nombre="Funciones" />
<tema orden="14" nombre="Gestión de excepiones" />
<tema orden="15" nombre="Iteradores y generadores" />
<tema orden="16" nombre="Completado de elementos" />
<tema orden="17" nombre="Entrada y salida de archivos" />
<tema orden="18" nombre="Módulos y paquetes" />
<tema orden="19" nombre="Gestión de módulos y paquetes con pip" />
<tema orden="20" nombre="Creación de paquetes con setuptools" />
<tema orden="21" nombre="Entornos virtuales" />
</curso>
<curso clave="py111" nombre="Introducción a la programación orientada a objetos con Python 3">
<requiere clave="py101"/>
<categoria>Introductorio</categoria>
<version>2018</version>
<url>"https://pythonista.mx/cursos/py111"</url>
<tema orden="1" nombre="Clases e instancias" />
<tema orden="2" nombre="Atributos y métodos" />
<tema orden="3" nombre="Atributos y métodos especiales" />
<tema orden="4" nombre="Interfaces, implementaciones y encapsulamientos" />
<tema orden="5" nombre="Propiedades" />
<tema orden="6" nombre="Métodos estáticos y de clase" />
<tema orden="7" nombre="Herencias" />
<tema orden="8" nombre="Mixins" />
<tema orden="9" nombre="Clases abstractas" />
<tema orden="10" nombre="Creación de excepciones personalizadas" />
<tema orden="11" nombre="Persistencia de objetos" />
</curso>
<curso clave="py121" nombre="Introducción a manejo de datos con Python">
<requiere clave="py101"/>
<requiere clave="py111"/>
<categoria>Introductorio</categoria>
<version>2018</version>
<url>"https://pythonista.mx/cursos/py121"</url>
<tema orden="1" nombre="Documentos separados por comas (CSV)" />
<tema orden="2" nombre="Gestión de estructura basadas en JSON" />
<tema orden="3" nombre="Análisis de redes sociales con la API de Twitter" />
<tema orden="4" nombre="Análisis de transacciones HTTP con Requests"/>
<tema orden="5" nombre="Análisis de HTML con Beautifulsoup" />
<tema orden="6" nombre="Expresiones regulares" />
<tema orden="7" nombre="XML y texto estructurado" />
<tema orden="8" nombre="Adquisición de contenido web con Scrapy" />
<tema orden="9" nombre="Escritura y lectura de hojas de cáculo en Excel" />
<tema orden="10" nombre="Escritura y lectura de documentos PDF" />
<tema orden="11" nombre="Introducción a gestión de datos en MongoDB" />
<tema orden="12" nombre="Introducción a gestión de bases de datos SQL" />
<tema orden="13" nombre="Introducción al ORM SQLAlchemy" />
</curso>
</data>
```
## Adqusición de XML a partir de una cadena de caracteres.
``` python
Elementtree.fromstring()
```
## Adquisición de XML a partir de un archivo.
### Lectura de un archivo XML.
``` python
Elementtree.parse('<ruta>')
```
### Creación de un objeto *Element* a partir del archivo.
```
<objeto resultante de Elementtree.parse>.getroot()
```
**Ejemplo:**
```
%pwd
cursos = ET.parse("/opt/pythonista/py121/data/cursos.xml")
raiz = cursos.getroot()
help(raiz)
dir(raiz)
raiz.tag
```
#### Acceso a atributos.
```
for hijo in raiz:
print(hijo.tag, hijo.attrib)
```
#### Acceso al contenido de los atributos.
```
for hijo in raiz:
print(hijo.tag, hijo.attrib['clave'])
```
Los elementos dentro del objeto "Element" son indexables.
```
raiz[0][4]
raiz[0][4].attrib
print(raiz[0][1].text)
raiz[0][1]
```
### Búsqueda mediante XPath.
XPath permite realizar búsquedas dentro de un elemento XML de forma similar a como se busca en la ruta de un sistema de archivos mediante expresiones.
Puede conocer más sobre la especificaciónde XPath en https://www.w3.org/TR/xpath/.
El módulo *Elementtree* cuenta con soporte limitado para las expresiones de XPath. Puede consultar dichasd expresiones en https://docs.python.org/3/library/xml.etree.elementtree.html#xpath-support
**Ejemplo:**
```
raiz.findall('./curso')
raiz.findall('./curso/url')[1].text
for dato in raiz.findall('curso[@clave]'):
print(dato.attrib['clave'])
raiz.findall('curso[@clave="py121"]')
raiz.findall('curso[@clave="py121"]')[0].attrib
raiz.find('curso/tema[@nombre="Persistencia de objetos"]')
```
<p style="text-align: center"><a rel="license" href="http://creativecommons.org/licenses/by/4.0/"><img alt="Licencia Creative Commons" style="border-width:0" src="https://i.creativecommons.org/l/by/4.0/80x15.png" /></a><br />Esta obra está bajo una <a rel="license" href="http://creativecommons.org/licenses/by/4.0/">Licencia Creative Commons Atribución 4.0 Internacional</a>.</p>
<p style="text-align: center">© José Luis Chiquete Valdivieso. 2018.</p>
|
github_jupyter
|
import xml.etree.ElementTree as ET
dir(ET)
## Adqusición de XML a partir de una cadena de caracteres.
## Adquisición de XML a partir de un archivo.
### Lectura de un archivo XML.
### Creación de un objeto *Element* a partir del archivo.
**Ejemplo:**
#### Acceso a atributos.
#### Acceso al contenido de los atributos.
Los elementos dentro del objeto "Element" son indexables.
### Búsqueda mediante XPath.
XPath permite realizar búsquedas dentro de un elemento XML de forma similar a como se busca en la ruta de un sistema de archivos mediante expresiones.
Puede conocer más sobre la especificaciónde XPath en https://www.w3.org/TR/xpath/.
El módulo *Elementtree* cuenta con soporte limitado para las expresiones de XPath. Puede consultar dichasd expresiones en https://docs.python.org/3/library/xml.etree.elementtree.html#xpath-support
**Ejemplo:**
| 0.290578 | 0.899652 |
# COVID-19 Deaths Per Capita
> Comparing death rates adjusting for population size.
- comments: true
- author: Joao B. Duarte & Hamel Husain
- categories: [growth, compare, interactive]
- hide: false
- image: images/covid-permillion-trajectories.png
- permalink: /covid-compare-permillion/
```
#hide
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import altair as alt
%config InlineBackend.figure_format = 'retina'
chart_width = 550
chart_height= 400
```
## Deaths Per Million Of Inhabitants
Since reaching at least 1 death per million
> Tip: Click (Shift+ for multiple) on countries in the legend to filter the visualization.
```
#hide
data = pd.read_csv("https://raw.githubusercontent.com/CSSEGISandData/COVID-19/master/csse_covid_19_data/csse_covid_19_time_series/time_series_covid19_deaths_global.csv",
error_bad_lines=False)
data = data.drop(columns=["Lat", "Long"])
data = data.melt(id_vars= ["Province/State", "Country/Region"])
data = pd.DataFrame(data.groupby(['Country/Region', "variable"]).sum())
data.reset_index(inplace=True)
data = data.rename(columns={"Country/Region": "location", "variable": "date", "value": "total_cases"})
data['date'] =pd.to_datetime(data.date)
data = data.sort_values(by = "date")
data.loc[data.location == "US","location"] = "United States"
data.loc[data.location == "Korea, South","location"] = "South Korea"
data_pwt = pd.read_stata("https://www.rug.nl/ggdc/docs/pwt91.dta")
filter1 = data_pwt["year"] == 2017
data_pop = data_pwt[filter1]
data_pop = data_pop[["country","pop"]]
data_pop.loc[data_pop.country == "Republic of Korea","country"] = "South Korea"
data_pop.loc[data_pop.country == "Iran (Islamic Republic of)","country"] = "Iran"
# per habitant
data_pc = data.copy()
countries = ["China", "Italy", "Spain", "France", "United Kingdom", "Germany",
"Portugal", "United States", "Singapore", "South Korea", "Japan",
"Brazil", "Iran", 'Netherlands', 'Belgium', 'Sweden',
'Switzerland', 'Norway', 'Denmark', 'Austria', 'Slovenia', 'Greece',
'Cyprus']
data_countries = []
data_countries_pc = []
MIN_DEATHS = 10
filter_min_dead = data_pc.total_cases < MIN_DEATHS
data_pc = data_pc.drop(data_pc[filter_min_dead].index)
# compute per habitant
for i in countries:
data_pc.loc[data_pc.location == i,"total_cases"] = data_pc.loc[data_pc.location == i,"total_cases"]/float(data_pop.loc[data_pop.country == i, "pop"])
# get each country time series
filter1 = data_pc["total_cases"] > 1
for i in countries:
filter_country = data_pc["location"]== i
data_countries_pc.append(data_pc[filter_country & filter1])
#hide_input
# Stack data to get it to Altair dataframe format
data_countries_pc2 = data_countries_pc.copy()
for i in range(0,len(countries)):
data_countries_pc2[i] = data_countries_pc2[i].reset_index()
data_countries_pc2[i]['n_days'] = data_countries_pc2[i].index
data_countries_pc2[i]['log_cases'] = np.log(data_countries_pc2[i]["total_cases"])
data_plot = data_countries_pc2[0]
for i in range(1, len(countries)):
data_plot = pd.concat([data_plot, data_countries_pc2[i]], axis=0)
data_plot["trend_2days"] = np.log(2)/2*data_plot["n_days"]
data_plot["trend_4days"] = np.log(2)/4*data_plot["n_days"]
data_plot["trend_12days"] = np.log(2)/12*data_plot["n_days"]
data_plot["trend_2days_label"] = "Doubles every 2 days"
data_plot["trend_4days_label"] = "Doubles evey 4 days"
data_plot["trend_12days_label"] = "Doubles every 12 days"
# Plot it using Altair
source = data_plot
scales = alt.selection_interval(bind='scales')
selection = alt.selection_multi(fields=['location'], bind='legend')
base = alt.Chart(source, title = "COVID-19 Deaths Per Million of Inhabitants").encode(
x = alt.X('n_days:Q', title = "Days passed since reaching 1 death per million"),
y = alt.Y("log_cases:Q",title = "Log of deaths per million"),
color = alt.Color('location:N', legend=alt.Legend(title="Country", labelFontSize=15, titleFontSize=17),
scale=alt.Scale(scheme='tableau20')),
opacity = alt.condition(selection, alt.value(1), alt.value(0.1))
)
lines = base.mark_line().add_selection(
scales
).add_selection(
selection
).properties(
width=chart_width,
height=chart_height
)
trend_2d = alt.Chart(source).encode(
x = "n_days:Q",
y = alt.Y("trend_2days:Q", scale=alt.Scale(domain=(0, max(data_plot["log_cases"])))),
).mark_line(color="grey", strokeDash=[3,3])
labels = pd.DataFrame([{'label': 'Doubles every 2 days', 'x_coord': 6, 'y_coord': 4},
{'label': 'Doubles every 4 days', 'x_coord': 16, 'y_coord': 3.5},
{'label': 'Doubles every 12 days', 'x_coord': 25, 'y_coord': 1.8},
])
trend_label = (alt.Chart(labels)
.mark_text(align='left', dx=-55, dy=-15, fontSize=12, color="grey")
.encode(x='x_coord:Q',
y='y_coord:Q',
text='label:N')
)
trend_4d = alt.Chart(source).mark_line(color="grey", strokeDash=[3,3]).encode(
x = "n_days:Q",
y = alt.Y("trend_4days:Q", scale=alt.Scale(domain=(0, max(data_plot["log_cases"])))),
)
trend_12d = alt.Chart(source).mark_line(color="grey", strokeDash=[3,3]).encode(
x = "n_days:Q",
y = alt.Y("trend_12days:Q", scale=alt.Scale(domain=(0, max(data_plot["log_cases"])))),
)
plot1= (
(trend_2d + trend_4d + trend_12d + trend_label + lines)
.configure_title(fontSize=20)
.configure_axis(labelFontSize=15,titleFontSize=18)
)
plot1.save(("../images/covid-permillion-trajectories.png"))
plot1
```
Last Available Total Deaths By Country:
```
#hide_input
label = 'Deaths'
temp = pd.concat([x.copy() for x in data_countries_pc]).loc[lambda x: x.date >= '3/1/2020']
metric_name = f'{label} per Million'
temp.columns = ['Country', 'date', metric_name]
# temp.loc[:, 'month'] = temp.date.dt.strftime('%Y-%m')
temp.loc[:, f'Log of {label} per Million'] = temp[f'{label} per Million'].apply(lambda x: np.log(x))
temp.groupby('Country').last()
#hide
# Get data and clean it
data = pd.read_csv("https://raw.githubusercontent.com/CSSEGISandData/COVID-19/master/csse_covid_19_data/csse_covid_19_time_series/time_series_covid19_confirmed_global.csv", error_bad_lines=False)
data = data.drop(columns=["Lat", "Long"])
data = data.melt(id_vars= ["Province/State", "Country/Region"])
data = pd.DataFrame(data.groupby(['Country/Region', "variable"]).sum())
data.reset_index(inplace=True)
data = data.rename(columns={"Country/Region": "location", "variable": "date", "value": "total_cases"})
data['date'] =pd.to_datetime(data.date)
data = data.sort_values(by = "date")
data.loc[data.location == "US","location"] = "United States"
data.loc[data.location == "Korea, South","location"] = "South Korea"
# Population data (last year is 2017 which is what we use)
data_pwt = pd.read_stata("https://www.rug.nl/ggdc/docs/pwt91.dta")
filter1 = data_pwt["year"] == 2017
data_pop = data_pwt[filter1]
data_pop = data_pop[["country","pop"]]
data_pop.loc[data_pop.country == "Republic of Korea","country"] = "South Korea"
data_pop.loc[data_pop.country == "Iran (Islamic Republic of)","country"] = "Iran"
# per habitant
data_pc = data.copy()
# I can add more countries if needed
countries = ["China", "Italy", "Spain", "France", "United Kingdom", "Germany",
"Portugal", "United States", "Singapore","South Korea", "Japan",
"Brazil","Iran"]
data_countries = []
data_countries_pc = []
# compute per habitant
for i in countries:
data_pc.loc[data_pc.location == i,"total_cases"] = data_pc.loc[data_pc.location == i,"total_cases"]/float(data_pop.loc[data_pop.country == i, "pop"])
# get each country time series
filter1 = data_pc["total_cases"] > 1
for i in countries:
filter_country = data_pc["location"]== i
data_countries_pc.append(data_pc[filter_country & filter1])
```
## Appendix
> Warning: The following chart, "Cases Per Million of Habitants" is biased depending on how widely a country administers tests. Please read with caution.
### Cases Per Million of Habitants
```
#hide_input
# Stack data to get it to Altair dataframe format
data_countries_pc2 = data_countries_pc.copy()
for i in range(0,len(countries)):
data_countries_pc2[i] = data_countries_pc2[i].reset_index()
data_countries_pc2[i]['n_days'] = data_countries_pc2[i].index
data_countries_pc2[i]['log_cases'] = np.log(data_countries_pc2[i]["total_cases"])
data_plot = data_countries_pc2[0]
for i in range(1, len(countries)):
data_plot = pd.concat([data_plot, data_countries_pc2[i]], axis=0)
data_plot["trend_2days"] = np.log(2)/2*data_plot["n_days"]
data_plot["trend_4days"] = np.log(2)/4*data_plot["n_days"]
data_plot["trend_12days"] = np.log(2)/12*data_plot["n_days"]
data_plot["trend_2days_label"] = "Doubles every 2 days"
data_plot["trend_4days_label"] = "Doubles evey 4 days"
data_plot["trend_12days_label"] = "Doubles every 12 days"
# Plot it using Altair
source = data_plot
scales = alt.selection_interval(bind='scales')
selection = alt.selection_multi(fields=['location'], bind='legend')
base = alt.Chart(source, title = "COVID-19 Confirmed Cases Per Million of Inhabitants").encode(
x = alt.X('n_days:Q', title = "Days passed since reaching 1 case per million"),
y = alt.Y("log_cases:Q",title = "Log of confirmed cases per million"),
color = alt.Color('location:N', legend=alt.Legend(title="Country", labelFontSize=15, titleFontSize=17),
scale=alt.Scale(scheme='tableau20')),
opacity = alt.condition(selection, alt.value(1), alt.value(0.1))
).properties(
width=chart_width,
height=chart_height
)
lines = base.mark_line().add_selection(
scales
).add_selection(
selection
)
trend_2d = alt.Chart(source).encode(
x = "n_days:Q",
y = alt.Y("trend_2days:Q", scale=alt.Scale(domain=(0, max(data_plot["log_cases"])))),
).mark_line( strokeDash=[3,3], color="grey")
labels = pd.DataFrame([{'label': 'Doubles every 2 days', 'x_coord': 10, 'y_coord': 6},
{'label': 'Doubles every 4 days', 'x_coord': 28, 'y_coord': 6},
{'label': 'Doubles every 12 days', 'x_coord': 45, 'y_coord': 3},
])
trend_label = (alt.Chart(labels)
.mark_text(align='left', dx=-55, dy=-15, fontSize=12, color="grey")
.encode(x='x_coord:Q',
y='y_coord:Q',
text='label:N')
)
trend_4d = alt.Chart(source).mark_line(color="grey", strokeDash=[3,3]).encode(
x = "n_days:Q",
y = alt.Y("trend_4days:Q", scale=alt.Scale(domain=(0, max(data_plot["log_cases"])))),
)
trend_12d = alt.Chart(source).mark_line(color="grey", strokeDash=[3,3]).encode(
x = "n_days:Q",
y = alt.Y("trend_12days:Q", scale=alt.Scale(domain=(0, max(data_plot["log_cases"])))),
)
(
(trend_2d + trend_4d + trend_12d + trend_label + lines)
.configure_title(fontSize=20)
.configure_axis(labelFontSize=15,titleFontSize=18)
)
```
Last Available Cases Per Million By Country:
```
#hide_input
label = 'Cases'
temp = pd.concat([x.copy() for x in data_countries_pc]).loc[lambda x: x.date >= '3/1/2020']
metric_name = f'{label} per Million'
temp.columns = ['Country', 'date', metric_name]
# temp.loc[:, 'month'] = temp.date.dt.strftime('%Y-%m')
temp.loc[:, f'Log of {label} per Million'] = temp[f'{label} per Million'].apply(lambda x: np.log(x))
temp.groupby('Country').last()
```
This analysis was conducted by [Joao B. Duarte](http://jbduarte.com). Assitance with creating visualizations were provided by [Hamel Husain](https://twitter.com/HamelHusain). Relevant sources are listed below:
1. ["2019 Novel Coronavirus COVID-19 (2019-nCoV) Data Repository by Johns Hopkins CSSE"](https://systems.jhu.edu/research/public-health/ncov/) [GitHub repository](https://github.com/CSSEGISandData/COVID-19).
2. [Feenstra, Robert C., Robert Inklaar and Marcel P. Timmer (2015), "The Next Generation of the Penn World Table" American Economic Review, 105(10), 3150-3182](https://www.rug.nl/ggdc/productivity/pwt/related-research)
|
github_jupyter
|
#hide
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import altair as alt
%config InlineBackend.figure_format = 'retina'
chart_width = 550
chart_height= 400
#hide
data = pd.read_csv("https://raw.githubusercontent.com/CSSEGISandData/COVID-19/master/csse_covid_19_data/csse_covid_19_time_series/time_series_covid19_deaths_global.csv",
error_bad_lines=False)
data = data.drop(columns=["Lat", "Long"])
data = data.melt(id_vars= ["Province/State", "Country/Region"])
data = pd.DataFrame(data.groupby(['Country/Region', "variable"]).sum())
data.reset_index(inplace=True)
data = data.rename(columns={"Country/Region": "location", "variable": "date", "value": "total_cases"})
data['date'] =pd.to_datetime(data.date)
data = data.sort_values(by = "date")
data.loc[data.location == "US","location"] = "United States"
data.loc[data.location == "Korea, South","location"] = "South Korea"
data_pwt = pd.read_stata("https://www.rug.nl/ggdc/docs/pwt91.dta")
filter1 = data_pwt["year"] == 2017
data_pop = data_pwt[filter1]
data_pop = data_pop[["country","pop"]]
data_pop.loc[data_pop.country == "Republic of Korea","country"] = "South Korea"
data_pop.loc[data_pop.country == "Iran (Islamic Republic of)","country"] = "Iran"
# per habitant
data_pc = data.copy()
countries = ["China", "Italy", "Spain", "France", "United Kingdom", "Germany",
"Portugal", "United States", "Singapore", "South Korea", "Japan",
"Brazil", "Iran", 'Netherlands', 'Belgium', 'Sweden',
'Switzerland', 'Norway', 'Denmark', 'Austria', 'Slovenia', 'Greece',
'Cyprus']
data_countries = []
data_countries_pc = []
MIN_DEATHS = 10
filter_min_dead = data_pc.total_cases < MIN_DEATHS
data_pc = data_pc.drop(data_pc[filter_min_dead].index)
# compute per habitant
for i in countries:
data_pc.loc[data_pc.location == i,"total_cases"] = data_pc.loc[data_pc.location == i,"total_cases"]/float(data_pop.loc[data_pop.country == i, "pop"])
# get each country time series
filter1 = data_pc["total_cases"] > 1
for i in countries:
filter_country = data_pc["location"]== i
data_countries_pc.append(data_pc[filter_country & filter1])
#hide_input
# Stack data to get it to Altair dataframe format
data_countries_pc2 = data_countries_pc.copy()
for i in range(0,len(countries)):
data_countries_pc2[i] = data_countries_pc2[i].reset_index()
data_countries_pc2[i]['n_days'] = data_countries_pc2[i].index
data_countries_pc2[i]['log_cases'] = np.log(data_countries_pc2[i]["total_cases"])
data_plot = data_countries_pc2[0]
for i in range(1, len(countries)):
data_plot = pd.concat([data_plot, data_countries_pc2[i]], axis=0)
data_plot["trend_2days"] = np.log(2)/2*data_plot["n_days"]
data_plot["trend_4days"] = np.log(2)/4*data_plot["n_days"]
data_plot["trend_12days"] = np.log(2)/12*data_plot["n_days"]
data_plot["trend_2days_label"] = "Doubles every 2 days"
data_plot["trend_4days_label"] = "Doubles evey 4 days"
data_plot["trend_12days_label"] = "Doubles every 12 days"
# Plot it using Altair
source = data_plot
scales = alt.selection_interval(bind='scales')
selection = alt.selection_multi(fields=['location'], bind='legend')
base = alt.Chart(source, title = "COVID-19 Deaths Per Million of Inhabitants").encode(
x = alt.X('n_days:Q', title = "Days passed since reaching 1 death per million"),
y = alt.Y("log_cases:Q",title = "Log of deaths per million"),
color = alt.Color('location:N', legend=alt.Legend(title="Country", labelFontSize=15, titleFontSize=17),
scale=alt.Scale(scheme='tableau20')),
opacity = alt.condition(selection, alt.value(1), alt.value(0.1))
)
lines = base.mark_line().add_selection(
scales
).add_selection(
selection
).properties(
width=chart_width,
height=chart_height
)
trend_2d = alt.Chart(source).encode(
x = "n_days:Q",
y = alt.Y("trend_2days:Q", scale=alt.Scale(domain=(0, max(data_plot["log_cases"])))),
).mark_line(color="grey", strokeDash=[3,3])
labels = pd.DataFrame([{'label': 'Doubles every 2 days', 'x_coord': 6, 'y_coord': 4},
{'label': 'Doubles every 4 days', 'x_coord': 16, 'y_coord': 3.5},
{'label': 'Doubles every 12 days', 'x_coord': 25, 'y_coord': 1.8},
])
trend_label = (alt.Chart(labels)
.mark_text(align='left', dx=-55, dy=-15, fontSize=12, color="grey")
.encode(x='x_coord:Q',
y='y_coord:Q',
text='label:N')
)
trend_4d = alt.Chart(source).mark_line(color="grey", strokeDash=[3,3]).encode(
x = "n_days:Q",
y = alt.Y("trend_4days:Q", scale=alt.Scale(domain=(0, max(data_plot["log_cases"])))),
)
trend_12d = alt.Chart(source).mark_line(color="grey", strokeDash=[3,3]).encode(
x = "n_days:Q",
y = alt.Y("trend_12days:Q", scale=alt.Scale(domain=(0, max(data_plot["log_cases"])))),
)
plot1= (
(trend_2d + trend_4d + trend_12d + trend_label + lines)
.configure_title(fontSize=20)
.configure_axis(labelFontSize=15,titleFontSize=18)
)
plot1.save(("../images/covid-permillion-trajectories.png"))
plot1
#hide_input
label = 'Deaths'
temp = pd.concat([x.copy() for x in data_countries_pc]).loc[lambda x: x.date >= '3/1/2020']
metric_name = f'{label} per Million'
temp.columns = ['Country', 'date', metric_name]
# temp.loc[:, 'month'] = temp.date.dt.strftime('%Y-%m')
temp.loc[:, f'Log of {label} per Million'] = temp[f'{label} per Million'].apply(lambda x: np.log(x))
temp.groupby('Country').last()
#hide
# Get data and clean it
data = pd.read_csv("https://raw.githubusercontent.com/CSSEGISandData/COVID-19/master/csse_covid_19_data/csse_covid_19_time_series/time_series_covid19_confirmed_global.csv", error_bad_lines=False)
data = data.drop(columns=["Lat", "Long"])
data = data.melt(id_vars= ["Province/State", "Country/Region"])
data = pd.DataFrame(data.groupby(['Country/Region', "variable"]).sum())
data.reset_index(inplace=True)
data = data.rename(columns={"Country/Region": "location", "variable": "date", "value": "total_cases"})
data['date'] =pd.to_datetime(data.date)
data = data.sort_values(by = "date")
data.loc[data.location == "US","location"] = "United States"
data.loc[data.location == "Korea, South","location"] = "South Korea"
# Population data (last year is 2017 which is what we use)
data_pwt = pd.read_stata("https://www.rug.nl/ggdc/docs/pwt91.dta")
filter1 = data_pwt["year"] == 2017
data_pop = data_pwt[filter1]
data_pop = data_pop[["country","pop"]]
data_pop.loc[data_pop.country == "Republic of Korea","country"] = "South Korea"
data_pop.loc[data_pop.country == "Iran (Islamic Republic of)","country"] = "Iran"
# per habitant
data_pc = data.copy()
# I can add more countries if needed
countries = ["China", "Italy", "Spain", "France", "United Kingdom", "Germany",
"Portugal", "United States", "Singapore","South Korea", "Japan",
"Brazil","Iran"]
data_countries = []
data_countries_pc = []
# compute per habitant
for i in countries:
data_pc.loc[data_pc.location == i,"total_cases"] = data_pc.loc[data_pc.location == i,"total_cases"]/float(data_pop.loc[data_pop.country == i, "pop"])
# get each country time series
filter1 = data_pc["total_cases"] > 1
for i in countries:
filter_country = data_pc["location"]== i
data_countries_pc.append(data_pc[filter_country & filter1])
#hide_input
# Stack data to get it to Altair dataframe format
data_countries_pc2 = data_countries_pc.copy()
for i in range(0,len(countries)):
data_countries_pc2[i] = data_countries_pc2[i].reset_index()
data_countries_pc2[i]['n_days'] = data_countries_pc2[i].index
data_countries_pc2[i]['log_cases'] = np.log(data_countries_pc2[i]["total_cases"])
data_plot = data_countries_pc2[0]
for i in range(1, len(countries)):
data_plot = pd.concat([data_plot, data_countries_pc2[i]], axis=0)
data_plot["trend_2days"] = np.log(2)/2*data_plot["n_days"]
data_plot["trend_4days"] = np.log(2)/4*data_plot["n_days"]
data_plot["trend_12days"] = np.log(2)/12*data_plot["n_days"]
data_plot["trend_2days_label"] = "Doubles every 2 days"
data_plot["trend_4days_label"] = "Doubles evey 4 days"
data_plot["trend_12days_label"] = "Doubles every 12 days"
# Plot it using Altair
source = data_plot
scales = alt.selection_interval(bind='scales')
selection = alt.selection_multi(fields=['location'], bind='legend')
base = alt.Chart(source, title = "COVID-19 Confirmed Cases Per Million of Inhabitants").encode(
x = alt.X('n_days:Q', title = "Days passed since reaching 1 case per million"),
y = alt.Y("log_cases:Q",title = "Log of confirmed cases per million"),
color = alt.Color('location:N', legend=alt.Legend(title="Country", labelFontSize=15, titleFontSize=17),
scale=alt.Scale(scheme='tableau20')),
opacity = alt.condition(selection, alt.value(1), alt.value(0.1))
).properties(
width=chart_width,
height=chart_height
)
lines = base.mark_line().add_selection(
scales
).add_selection(
selection
)
trend_2d = alt.Chart(source).encode(
x = "n_days:Q",
y = alt.Y("trend_2days:Q", scale=alt.Scale(domain=(0, max(data_plot["log_cases"])))),
).mark_line( strokeDash=[3,3], color="grey")
labels = pd.DataFrame([{'label': 'Doubles every 2 days', 'x_coord': 10, 'y_coord': 6},
{'label': 'Doubles every 4 days', 'x_coord': 28, 'y_coord': 6},
{'label': 'Doubles every 12 days', 'x_coord': 45, 'y_coord': 3},
])
trend_label = (alt.Chart(labels)
.mark_text(align='left', dx=-55, dy=-15, fontSize=12, color="grey")
.encode(x='x_coord:Q',
y='y_coord:Q',
text='label:N')
)
trend_4d = alt.Chart(source).mark_line(color="grey", strokeDash=[3,3]).encode(
x = "n_days:Q",
y = alt.Y("trend_4days:Q", scale=alt.Scale(domain=(0, max(data_plot["log_cases"])))),
)
trend_12d = alt.Chart(source).mark_line(color="grey", strokeDash=[3,3]).encode(
x = "n_days:Q",
y = alt.Y("trend_12days:Q", scale=alt.Scale(domain=(0, max(data_plot["log_cases"])))),
)
(
(trend_2d + trend_4d + trend_12d + trend_label + lines)
.configure_title(fontSize=20)
.configure_axis(labelFontSize=15,titleFontSize=18)
)
#hide_input
label = 'Cases'
temp = pd.concat([x.copy() for x in data_countries_pc]).loc[lambda x: x.date >= '3/1/2020']
metric_name = f'{label} per Million'
temp.columns = ['Country', 'date', metric_name]
# temp.loc[:, 'month'] = temp.date.dt.strftime('%Y-%m')
temp.loc[:, f'Log of {label} per Million'] = temp[f'{label} per Million'].apply(lambda x: np.log(x))
temp.groupby('Country').last()
| 0.345768 | 0.886715 |
```
%matplotlib inline
import numpy as np
# charger les données
import pandas as pd
data = pd.read_csv('sources/winequality-white.csv', sep=';')
# créer la matrice de données
X = data.as_matrix(data.columns[:-1])
# créer le vecteur d'étiquettes
y = data.as_matrix([data.columns[-1]])
y = y.flatten()
# transformer en un problème de classification binaire
y_class = np.where(y<6, 0, 1)
from sklearn import model_selection
X_train, X_test, y_train, y_test = \
model_selection.train_test_split(X, y_class,
test_size=0.3)
# standardiser les données
from sklearn import preprocessing
std_scale = preprocessing.StandardScaler().fit(X_train)
X_train_std = std_scale.transform(X_train)
X_test_std = std_scale.transform(X_test)
# Créer une SVM avec un noyau gaussien de paramètre gamma=0.01
from sklearn import svm
classifier = svm.SVC(kernel='rbf', gamma=0.01)
# Entraîner la SVM sur le jeu d'entraînement
classifier.fit(X_train_std, y_train)
# prédire sur le jeu de test
y_test_pred = classifier.decision_function(X_test_std)
# construire la courbe ROC
from sklearn import metrics
fpr, tpr, thr = metrics.roc_curve(y_test, y_test_pred)
# calculer l'aire sous la courbe ROC
auc = metrics.auc(fpr, tpr)
# créer une figure
from matplotlib import pyplot as plt
fig = plt.figure(figsize=(6, 6))
# afficher la courbe ROC
plt.plot(fpr, tpr, '-', lw=2, label='gamma=0.01, AUC=%.2f' % auc)
# donner un titre aux axes et au graphique
plt.xlabel('False Positive Rate', fontsize=16)
plt.ylabel('True Positive Rate', fontsize=16)
plt.title('SVM ROC Curve', fontsize=16)
# afficher la légende
plt.legend(loc="lower right", fontsize=14)
# afficher l'image
# choisir 6 valeurs pour C, entre 1e-2 et 1e3
C_range = np.logspace(-2, 3, 6)
# choisir 4 valeurs pour gamma, entre 1e-2 et 10
gamma_range = np.logspace(-2, 1, 4)
# grille de paramètres
param_grid = {'C': C_range, 'gamma': gamma_range}
# critère de sélection du meilleur modèle
score = 'roc_auc'
# initialiser une recherche sur grille
grid = model_selection.GridSearchCV(svm.SVC(kernel='rbf'),
param_grid,
cv=5, # 5 folds de validation croisée
scoring=score)
# faire tourner la recherche sur grille
grid.fit(X_train_std, y_train)
# afficher les paramètres optimaux
print("The optimal parameters are %s with a score of %.2f" % \
(grid.best_params_, grid.best_score_))
# prédire sur le jeu de test avec le modèle optimisé
y_test_pred_cv = grid.decision_function(X_test_std)
# construire la courbe ROC du modèle optimisé
fpr_cv, tpr_cv, thr_cv = metrics.roc_curve(y_test, y_test_pred_cv)
# calculer l'aire sous la courbe ROC du modèle optimisé
auc_cv = metrics.auc(fpr_cv, tpr_cv)
# créer une figure
fig = plt.figure(figsize=(6, 6))
# afficher la courbe ROC précédente
plt.plot(fpr, tpr, '-', lw=2, label='gamma=0.01, AUC=%.2f' % auc)
# afficher la courbe ROC du modèle optimisé
plt.plot(fpr_cv, tpr_cv, '-', lw=2, label='gamma=%.1e, AUC=%.2f' % \
(grid.best_params_['gamma'], auc_cv))
# donner un titre aux axes et au graphique
plt.xlabel('False Positive Rate', fontsize=16)
plt.ylabel('True Positive Rate', fontsize=16)
plt.title('SVM ROC Curve', fontsize=16)
# afficher la légende
plt.legend(loc="lower right", fontsize=14)
from sklearn import metrics
import matplotlib.cm
kmatrix = metrics.pairwise.rbf_kernel(X_train_std, gamma=0.01)
kmatrix100 = kmatrix[:100, :100]
# dessiner la matrice
plt.pcolor(kmatrix100, cmap=matplotlib.cm.PuRd)
# rajouter la légende
plt.colorbar()
# retourner l'axe des ordonnées
plt.gca().invert_yaxis()
plt.gca().xaxis.tick_top()
# charger les données
import pandas as pd
data = pd.read_csv('sources/winequality-white.csv', sep=';')
# créer la matrice de données
X = data.as_matrix(data.columns[:-1])
# créer le vecteur d'étiquettes
y = data.as_matrix([data.columns[-1]])
y = y.flatten()
# créer un jeu d'entrainement et un jeu de test (30% des données)
from sklearn import model_selection
X_train, X_test, y_train, y_test = \
model_selection.train_test_split(X, y, test_size=0.3)
# standardiser les données
from sklearn import preprocessing
std_scale = preprocessing.StandardScaler().fit(X_train)
X_train_std = std_scale.transform(X_train)
X_test_std = std_scale.transform(X_test)
# initialiser un objet de classification par kRR
from sklearn import kernel_ridge
predicteur = kernel_ridge.KernelRidge(alpha=1.0, # valeur par défaut
kernel='rbf', # noyau Gaussien
gamma=0.01) # valeur de 1/(2 * sigma**2)
# entraîner le classifieur sur le jeu d'entrainement
predicteur.fit(X_train_std, y_train)
# prédire sur le jeu de test
y_test_pred = predicteur.predict(X_test_std)
# calculer la RMSE sur le jeu de test
from sklearn import metrics
rmse = np.sqrt(metrics.mean_squared_error(y_test, y_test_pred))
print("RMSE: %.2f" % rmse)
# créer une figure
fig = plt.figure(figsize=(6, 6))
# Compter, pour chaque paire de valeurs (y, y') où y est un vrai score et y' le score prédit,
# le nombre de ces paires.
# Ce nombre sera utilisé pour modifier la taille des marqueurs correspondants
# dans un nuage de points
sizes = {}
for (yt, yp) in zip(list(y_test), list(y_test_pred)):
if sizes in (yt, yp):
sizes[(yt, yp)] += 1
else:
sizes[(yt, yp)] = 1
keys = sizes.keys()
# afficher les prédictions
plt.scatter([k[0] for k in keys],
[k[1] for k in keys],
s=[sizes[k] for k in keys],
label='gamma = 0.01: RMSE = %0.2f' % rmse)
# étiqueter les axes et le graphique
plt.xlabel('Vrai score', fontsize=16)
plt.ylabel(u'Score prédit', fontsize=16)
plt.title('kernel Ridge Regression', fontsize=16)
# limites des axes
plt.xlim([2.9, 9.1])
plt.ylim([2.9, 9.1])
# afficher la légende
plt.legend(loc="lower right", fontsize=12)
# valeurs du paramètre C
alpha_range = np.logspace(-2, 2, 5)
# valeurs du paramètre gamma
gamma_range = np.logspace(-2, 1, 4)
# grille de paramètres
param_grid = {'alpha': alpha_range, 'gamma': gamma_range}
# score pour sélectionner le modèle optimal
score = 'neg_mean_squared_error'
# initialiser la validation croisée
grid_pred = model_selection.GridSearchCV(kernel_ridge.KernelRidge(kernel='rbf'),
param_grid,
cv=5,
scoring=score)
# exécuter la validation croisée sur le jeu d'entraînement
grid_pred.fit(X_train_std, y_train)
# prédire sur le jeu de test avec le modèle sélectionné
y_test_pred_cv = grid_pred.predict(X_test_std)
# calculer la RMSE correspondante
rmse_cv = np.sqrt(metrics.mean_squared_error(y_test, y_test_pred_cv))
```
|
github_jupyter
|
%matplotlib inline
import numpy as np
# charger les données
import pandas as pd
data = pd.read_csv('sources/winequality-white.csv', sep=';')
# créer la matrice de données
X = data.as_matrix(data.columns[:-1])
# créer le vecteur d'étiquettes
y = data.as_matrix([data.columns[-1]])
y = y.flatten()
# transformer en un problème de classification binaire
y_class = np.where(y<6, 0, 1)
from sklearn import model_selection
X_train, X_test, y_train, y_test = \
model_selection.train_test_split(X, y_class,
test_size=0.3)
# standardiser les données
from sklearn import preprocessing
std_scale = preprocessing.StandardScaler().fit(X_train)
X_train_std = std_scale.transform(X_train)
X_test_std = std_scale.transform(X_test)
# Créer une SVM avec un noyau gaussien de paramètre gamma=0.01
from sklearn import svm
classifier = svm.SVC(kernel='rbf', gamma=0.01)
# Entraîner la SVM sur le jeu d'entraînement
classifier.fit(X_train_std, y_train)
# prédire sur le jeu de test
y_test_pred = classifier.decision_function(X_test_std)
# construire la courbe ROC
from sklearn import metrics
fpr, tpr, thr = metrics.roc_curve(y_test, y_test_pred)
# calculer l'aire sous la courbe ROC
auc = metrics.auc(fpr, tpr)
# créer une figure
from matplotlib import pyplot as plt
fig = plt.figure(figsize=(6, 6))
# afficher la courbe ROC
plt.plot(fpr, tpr, '-', lw=2, label='gamma=0.01, AUC=%.2f' % auc)
# donner un titre aux axes et au graphique
plt.xlabel('False Positive Rate', fontsize=16)
plt.ylabel('True Positive Rate', fontsize=16)
plt.title('SVM ROC Curve', fontsize=16)
# afficher la légende
plt.legend(loc="lower right", fontsize=14)
# afficher l'image
# choisir 6 valeurs pour C, entre 1e-2 et 1e3
C_range = np.logspace(-2, 3, 6)
# choisir 4 valeurs pour gamma, entre 1e-2 et 10
gamma_range = np.logspace(-2, 1, 4)
# grille de paramètres
param_grid = {'C': C_range, 'gamma': gamma_range}
# critère de sélection du meilleur modèle
score = 'roc_auc'
# initialiser une recherche sur grille
grid = model_selection.GridSearchCV(svm.SVC(kernel='rbf'),
param_grid,
cv=5, # 5 folds de validation croisée
scoring=score)
# faire tourner la recherche sur grille
grid.fit(X_train_std, y_train)
# afficher les paramètres optimaux
print("The optimal parameters are %s with a score of %.2f" % \
(grid.best_params_, grid.best_score_))
# prédire sur le jeu de test avec le modèle optimisé
y_test_pred_cv = grid.decision_function(X_test_std)
# construire la courbe ROC du modèle optimisé
fpr_cv, tpr_cv, thr_cv = metrics.roc_curve(y_test, y_test_pred_cv)
# calculer l'aire sous la courbe ROC du modèle optimisé
auc_cv = metrics.auc(fpr_cv, tpr_cv)
# créer une figure
fig = plt.figure(figsize=(6, 6))
# afficher la courbe ROC précédente
plt.plot(fpr, tpr, '-', lw=2, label='gamma=0.01, AUC=%.2f' % auc)
# afficher la courbe ROC du modèle optimisé
plt.plot(fpr_cv, tpr_cv, '-', lw=2, label='gamma=%.1e, AUC=%.2f' % \
(grid.best_params_['gamma'], auc_cv))
# donner un titre aux axes et au graphique
plt.xlabel('False Positive Rate', fontsize=16)
plt.ylabel('True Positive Rate', fontsize=16)
plt.title('SVM ROC Curve', fontsize=16)
# afficher la légende
plt.legend(loc="lower right", fontsize=14)
from sklearn import metrics
import matplotlib.cm
kmatrix = metrics.pairwise.rbf_kernel(X_train_std, gamma=0.01)
kmatrix100 = kmatrix[:100, :100]
# dessiner la matrice
plt.pcolor(kmatrix100, cmap=matplotlib.cm.PuRd)
# rajouter la légende
plt.colorbar()
# retourner l'axe des ordonnées
plt.gca().invert_yaxis()
plt.gca().xaxis.tick_top()
# charger les données
import pandas as pd
data = pd.read_csv('sources/winequality-white.csv', sep=';')
# créer la matrice de données
X = data.as_matrix(data.columns[:-1])
# créer le vecteur d'étiquettes
y = data.as_matrix([data.columns[-1]])
y = y.flatten()
# créer un jeu d'entrainement et un jeu de test (30% des données)
from sklearn import model_selection
X_train, X_test, y_train, y_test = \
model_selection.train_test_split(X, y, test_size=0.3)
# standardiser les données
from sklearn import preprocessing
std_scale = preprocessing.StandardScaler().fit(X_train)
X_train_std = std_scale.transform(X_train)
X_test_std = std_scale.transform(X_test)
# initialiser un objet de classification par kRR
from sklearn import kernel_ridge
predicteur = kernel_ridge.KernelRidge(alpha=1.0, # valeur par défaut
kernel='rbf', # noyau Gaussien
gamma=0.01) # valeur de 1/(2 * sigma**2)
# entraîner le classifieur sur le jeu d'entrainement
predicteur.fit(X_train_std, y_train)
# prédire sur le jeu de test
y_test_pred = predicteur.predict(X_test_std)
# calculer la RMSE sur le jeu de test
from sklearn import metrics
rmse = np.sqrt(metrics.mean_squared_error(y_test, y_test_pred))
print("RMSE: %.2f" % rmse)
# créer une figure
fig = plt.figure(figsize=(6, 6))
# Compter, pour chaque paire de valeurs (y, y') où y est un vrai score et y' le score prédit,
# le nombre de ces paires.
# Ce nombre sera utilisé pour modifier la taille des marqueurs correspondants
# dans un nuage de points
sizes = {}
for (yt, yp) in zip(list(y_test), list(y_test_pred)):
if sizes in (yt, yp):
sizes[(yt, yp)] += 1
else:
sizes[(yt, yp)] = 1
keys = sizes.keys()
# afficher les prédictions
plt.scatter([k[0] for k in keys],
[k[1] for k in keys],
s=[sizes[k] for k in keys],
label='gamma = 0.01: RMSE = %0.2f' % rmse)
# étiqueter les axes et le graphique
plt.xlabel('Vrai score', fontsize=16)
plt.ylabel(u'Score prédit', fontsize=16)
plt.title('kernel Ridge Regression', fontsize=16)
# limites des axes
plt.xlim([2.9, 9.1])
plt.ylim([2.9, 9.1])
# afficher la légende
plt.legend(loc="lower right", fontsize=12)
# valeurs du paramètre C
alpha_range = np.logspace(-2, 2, 5)
# valeurs du paramètre gamma
gamma_range = np.logspace(-2, 1, 4)
# grille de paramètres
param_grid = {'alpha': alpha_range, 'gamma': gamma_range}
# score pour sélectionner le modèle optimal
score = 'neg_mean_squared_error'
# initialiser la validation croisée
grid_pred = model_selection.GridSearchCV(kernel_ridge.KernelRidge(kernel='rbf'),
param_grid,
cv=5,
scoring=score)
# exécuter la validation croisée sur le jeu d'entraînement
grid_pred.fit(X_train_std, y_train)
# prédire sur le jeu de test avec le modèle sélectionné
y_test_pred_cv = grid_pred.predict(X_test_std)
# calculer la RMSE correspondante
rmse_cv = np.sqrt(metrics.mean_squared_error(y_test, y_test_pred_cv))
| 0.430626 | 0.643917 |
# Tutorial: display on-sky footprint
`Telescope` instance provides a function to calculate the detector footprints on the sky. This notebook demonstrates how to use the function.
```
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
import pickle as pkl
import warpfield as w
from astropy.coordinates import SkyCoord, Longitude, Latitude, Angle
from astropy.time import Time
import astropy.units as u
```
An artificial source set is used in this notebook. Load the coordinate file and convert to the `SkyCoord` instance. The artificial sources are distributed around the Galactic center. The telescope is pointed toward the Galactic center too. The position angle of the telescope is set to 0.0 deg in the galactic coordinate.
```
coo = np.loadtxt('sample.coo')
src = SkyCoord(coo[:,0]*u.deg, coo[:,1]*u.deg, frame='galactic')
```
Create a `Telescope` instance. Here, we define a telescope with four InGaAs detectors tiled in 2×2.
```
from warpfield.InGaAs import get_jasmine
```
The telescope is pointed toward the Galactic center, and the position angle of the telescope is aligned north-up.
```
pointing = SkyCoord(0.0*u.deg,0.0*u.deg, frame='galactic')
position_angle = Angle(0.0, unit='degree')
jasmine = get_jasmine(pointing, position_angle)
```
The focal plane of the telescope is illustrated. The red rectangles are the InGaAs detectors. The gray rectangle is an available region of the focal plane.
```
jasmine.display_focal_plane()
```
The figure below shows the on-sky distribution of the artifical sources with the detector footprints.
```
ax = w.display_sources(pointing, src, title='J shape')
jasmine.overlay_footprints(ax, color='C2', label='footprint')
plt.show()
```
Here is another example with a different telescope. Point-spread functions at the four corners of the focal plane can be degraded in the latest design of JASMINE. Thus, the available focal plane is truncated to an octagonal shape. Set the option `octagonal` true to obtain the truncated optics.
```
pointing = SkyCoord(0.0*u.deg,0.0*u.deg, frame='galactic')
position_angle = Angle(0.0, unit='degree')
jasmine = get_jasmine(pointing, position_angle, octagonal=True)
```
The layout of the focal plane is illustrated. The available region is truncated.
```
jasmine.display_focal_plane()
```
The detector footprints on the sky are truncated as well.
```
ax = w.display_sources(pointing, src, title='J shape')
jasmine.overlay_footprints(ax, color='C2', label='footprint')
plt.show()
```
|
github_jupyter
|
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
import pickle as pkl
import warpfield as w
from astropy.coordinates import SkyCoord, Longitude, Latitude, Angle
from astropy.time import Time
import astropy.units as u
coo = np.loadtxt('sample.coo')
src = SkyCoord(coo[:,0]*u.deg, coo[:,1]*u.deg, frame='galactic')
from warpfield.InGaAs import get_jasmine
pointing = SkyCoord(0.0*u.deg,0.0*u.deg, frame='galactic')
position_angle = Angle(0.0, unit='degree')
jasmine = get_jasmine(pointing, position_angle)
jasmine.display_focal_plane()
ax = w.display_sources(pointing, src, title='J shape')
jasmine.overlay_footprints(ax, color='C2', label='footprint')
plt.show()
pointing = SkyCoord(0.0*u.deg,0.0*u.deg, frame='galactic')
position_angle = Angle(0.0, unit='degree')
jasmine = get_jasmine(pointing, position_angle, octagonal=True)
jasmine.display_focal_plane()
ax = w.display_sources(pointing, src, title='J shape')
jasmine.overlay_footprints(ax, color='C2', label='footprint')
plt.show()
| 0.578448 | 0.989811 |
<a href="https://colab.research.google.com/github/danio2010/ON2022/blob/main/ObliczeniaNaukowe_lab1.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
```
```
#LAB nr 1
Plan:
* Git/GitHub
* wprowadzenie do Jupyter/Colab
* Przypomnienie pythona
Zadanie 1. Założyć konto na GitHub (lub zalogować się)
Zadanie 2. Utworzyć repozytorium o nazwie "Obliczenia_naukowe"
Zadanie 3. Utworzyć w CoLab plik zad1.3.ipynb o treści "Test wysyłania", zapisać go na swoim GoogleDrive, a kopię zapisać w GitHub
(https://towardsdatascience.com/google-drive-google-colab-github-dont-just-read-do-it-5554d5824228)
Zadanie 4. Utworzyć w swoim repozytorium plik zad1.4.txt o dowolnej treści bezpośrednio w GitHub
Zadanie 5. Dodaj nowe zagadnienie (issue) do repozytorium https://github.com/danio2010/ON2022
Zadanie 6. Określ ile razy modyfikowany był plik ON_lab1.ipynb i wpisz tę liczbę to pliku zad1.6.txt
### Praca w konsoli
Wejść do docelowego katalogu
```git
git config --global user.name <nazwa>
git config --global user.mail <e-mail>
git clone <adres ssh repozytorium na git> #należy skonfigurować klucze ssh lub gpg. Przy użyciu wersji https trzeba podawać token przy push
git add <plik>
git commit -m <wiadomość>
git push
```
```
print('cześć')
```
```
```
### Możliwości pól komentarza
* Nagłówki
```markdown
# Sekcja 1
# Sekcja 2
## Podskecja sekcji 2
# Sekcj
```
* Wyróżnianie tekstu
Składnia | Efekt
--- | ---
`**bold text**` | **bold text**
`*italicized text*` or `_italicized text_` | *italicized text*
`~~strikethrough~~` | ~~strikethrough~~
* Bloki z kodem python
````
```python
print("a")
```
````
```python
print("a")
```
* Listy
1. pierwszy
1. drugi
1. trzeci
* LaTeX
$\lim_{n\to\infty}$
# Nagłówek 1
## Pod nim
# Sekcja 2
## Podesekcja
### Zadanie
### Zadanie 2
**pogrubienie**
```
2+2
2**3
3+5>6
import math
math.sin(30)
math.log(4,2)
```
### Zadanie domowe lab1
Przygotuj w CoLab dokument, który będzie zawierał tytuł oraz cztery sekcje (nazwij je dowolnie). W pierwszej sekcji umieść skrypt, który zapyta Cię o imię, a następnie przywita Cię po imieniu. W drugiej skecji wstaw (w polu komentarza) kod tego skryptu. W trzeciej umieść dowolny obrazek.
W czwartej sekcji napisz **funkcję** bezargumentową o nazwie listownik(), która
* zapyta o długośc listy, która musi znaleźć się w przedziale $[10,20]$ (przypilnuj użytkownika)
* wygeneruje i wydrukuje listę o odpowiedniej długości z liczbami losowymi przedziału $[0,10]$
* **zwróci** liczbę unikalnych wartości w wygenerowanej liście
W rozwiązaniu można korzystać z funkcji pomocniczych.
Nazwij plik zadDomowe.ipynb i umieść w swoim repozytorium.
```
```
|
github_jupyter
|
```
#LAB nr 1
Plan:
* Git/GitHub
* wprowadzenie do Jupyter/Colab
* Przypomnienie pythona
Zadanie 1. Założyć konto na GitHub (lub zalogować się)
Zadanie 2. Utworzyć repozytorium o nazwie "Obliczenia_naukowe"
Zadanie 3. Utworzyć w CoLab plik zad1.3.ipynb o treści "Test wysyłania", zapisać go na swoim GoogleDrive, a kopię zapisać w GitHub
(https://towardsdatascience.com/google-drive-google-colab-github-dont-just-read-do-it-5554d5824228)
Zadanie 4. Utworzyć w swoim repozytorium plik zad1.4.txt o dowolnej treści bezpośrednio w GitHub
Zadanie 5. Dodaj nowe zagadnienie (issue) do repozytorium https://github.com/danio2010/ON2022
Zadanie 6. Określ ile razy modyfikowany był plik ON_lab1.ipynb i wpisz tę liczbę to pliku zad1.6.txt
### Praca w konsoli
Wejść do docelowego katalogu
### Możliwości pól komentarza
* Nagłówki
* Wyróżnianie tekstu
Składnia | Efekt
--- | ---
`**bold text**` | **bold text**
`*italicized text*` or `_italicized text_` | *italicized text*
`~~strikethrough~~` | ~~strikethrough~~
* Bloki z kodem python
print("a")
```python
print("a")
2+2
2**3
3+5>6
import math
math.sin(30)
math.log(4,2)
| 0.552781 | 0.948202 |
# Imports and setup
## Imports
```
import re, os, sys, shutil
import shlex, subprocess
import glob
import pandas as pd
import panedr
import numpy as np
import MDAnalysis as mda
import nglview
import matplotlib.pyplot as plt
import parmed as pmd
import py
import scipy
from scipy import stats
from importlib import reload
from thtools import cd
from paratemp import copy_no_overwrite
from paratemp import geometries as gm
from paratemp import coordinate_analysis as ca
import paratemp.para_temp_setup as pts
import paratemp as pt
from gautools import submit_gaussian as subg
from gautools.tools import use_gen_template as ugt
```
## Common functions
```
def plot_prop_PT(edict, prop):
fig, axes = plt.subplots(4, 4, figsize=(16,16))
for i in range(16):
ax = axes.flat[i]
edict[i][prop].plot(ax=ax)
fig.tight_layout()
return fig, axes
def plot_e_props(df, labels, nrows=2, ncols=2):
fig, axes = plt.subplots(nrows, ncols, sharex=True)
for label, ax in zip(labels, axes.flat):
df[label].plot(ax=ax)
ax.set_title(label)
fig.tight_layout()
return fig, axes
def plot_rd(univ): # rd = reaction distance
univ.calculate_distances(rd=(20,39))
fig, axes = plt.subplots(1, 2, figsize=(12, 4))
univ.data.rd.plot(ax=axes[0])
univ.data.rd.hist(ax=axes[1], grid=False)
print(f'reaction distance mean: {univ.data.rd.mean():.2f} and sd: {univ.data.rd.std():.2f}')
return fig, axes
def plot_hist_dist(univ, name, indexes=None):
if indexes is not None:
kwargs = {name: indexes}
univ.calculate_distances(**kwargs)
fig, axes = plt.subplots(1, 2, figsize=(12, 4))
univ.data[name].plot(ax=axes[0])
univ.data[name].hist(ax=axes[1], grid=False)
print(f'{name} distance mean: {univ.data[name].mean():.2f} and sd: {univ.data[name].std():.2f}')
def get_solvent_count_solvate(proc):
for line in proc.stdout.split('\n'):
m = re.search(r'(?:atoms\):\s+)(\d+)(?:\s+residues)', line)
if m:
return int(m.group(1))
else:
raise ValueError('Solvent count not found.')
def set_solv_count(n_gro, s_count,
res_name='DCM', prepend='unequal-'):
"""
Remove solvent residues from the end of a gro file to match s_count
This assumes all non-solvent molecules are listed in the input gro
file before the solvent residues.
"""
bak_name = os.path.join(os.path.dirname(n_gro),
prepend+os.path.basename(n_gro))
copy_no_overwrite(n_gro, bak_name)
with open(n_gro, 'r') as in_gro:
lines = in_gro.readlines()
for line in lines[2:]:
if res_name in line:
non_s_res_count = resid
break
else:
resid = int(line[:5])
res_count = s_count + non_s_res_count
# TODO check reasonability of this number
box = lines.pop()
while True:
line = lines.pop()
if int(line[:5]) > res_count:
continue
elif int(line[:5]) == res_count:
atom_count = line[15:20]
lines.append(line)
break
elif int(line[:5]) < res_count:
raise ValueError("Desired res "
"count is larger than "
"line's resid.\n" +
"res_count: {}\n".format(res_count) +
"line: {}".format(line))
lines[1] = atom_count + '\n'
lines.append(box)
with open(n_gro, 'w') as out_gro:
for line in lines:
out_gro.write(line)
def get_solv_count_top(n_top, res_name='DCM'):
"""
Return residue count of specified residue from n_top"""
with open(n_top, 'r') as in_top:
mol_section = False
for line in in_top:
if line.strip().startswith(';'):
pass
elif not mol_section:
if re.search(r'\[\s*molecules\s*\]', line,
flags=re.IGNORECASE):
mol_section = True
else:
if res_name.lower() in line.lower():
return int(line.split()[1])
def set_solv_count_top(n_top, s_count,
res_name='DCM', prepend='unequal-'):
"""
Set count of res_name residues in n_top
This will make a backup copy of the top file with `prepend`
prepended to the name of the file."""
bak_name = os.path.join(os.path.dirname(n_top),
prepend+os.path.basename(n_top))
copy_no_overwrite(n_top, bak_name)
with open(n_top, 'r') as in_top:
lines = in_top.readlines()
with open(n_top, 'w') as out_top:
mol_section = False
for line in lines:
if line.strip().startswith(';'):
pass
elif not mol_section:
if re.search(r'\[\s*molecules\s*\]', line,
flags=re.IGNORECASE):
mol_section = True
else:
if res_name.lower() in line.lower():
line = re.sub(r'\d+', str(s_count), line)
out_top.write(line)
```
# Get charges
Calculate RESP charges using Gaussian through [submit_gaussian](https://github.com/theavey/QM-calc-scripts/blob/master/gautools/submit_gaussian.py) for use with GAFF.
```
d_charge_params = dict(opt='SCF=tight Test Pop=MK iop(6/33=2) iop(6/42=6) iop(6/50=1)',
func='HF',
basis='6-31G*',
footer='\ng16.gesp\n\ng16.gesp\n\n')
l_scripts = []
s = subg.write_sub_script('01-charges/TS2.com',
executable='g16',
make_xyz='../TS2.pdb',
make_input=True,
ugt_dict={'job_name':'GPX TS2 charges',
'charg_mult':'+1 1',
**d_charge_params})
l_scripts.append(s)
s = subg.write_sub_script('01-charges/R-NO2-CPA.com',
executable='g16',
make_xyz='../R-NO2-CPA.pdb',
make_input=True,
ugt_dict={'job_name':'GPX R-NO2-CPA charges',
'charg_mult':'-1 1',
**d_charge_params})
l_scripts.append(s)
l_scripts
subg.submit_scripts(l_scripts, batch=True, submit=True)
```
# Parameterize molecule in GAFF with ANTECHAMBER and ACPYPE
Note, ACPYPE was installed from [this repository](https://github.com/alanwilter/acpype), which seems to be from the original author, though maybe not the one who put it [onto pypi](https://github.com/llazzaro/acpype).
## For the catalyst:
Use antechamber to create mol2 file with Gaussian ESP charges (though wrong atom types and such, for now):
antechamber -i R-NO2-CPA.gesp -fi gesp -o R-NO2-CPA.mol2 -fo mol2
Use ACPYPE to use this mol2 file (and it's GESP charges) to generate GROMACS input files:
acpype.py -i R-NO2-CPA.mol2 -b CPA-gesp --net_charge=-1 -o gmx -d -c user
## For the reactant:
antechamber -i TS2.gesp -fi gesp -o TS2.mol2 -fo mol2
acpype.py -i TS2.mol2 -b GPX-ts --net_charge=1 -o gmx -c user
Then the different molecules can be combined using [ParmEd](https://github.com/ParmEd/ParmEd).
```
gpx = pmd.gromacs.GromacsTopologyFile('01-charges/GPX-ts.acpype/GPX-ts_GMX.top', xyz='01-charges/GPX-ts.acpype/GPX-ts_GMX.gro')
cpa = pmd.gromacs.GromacsTopologyFile('01-charges/CPA-gesp.acpype/CPA-gesp_GMX.top', xyz='01-charges/CPA-gesp.acpype/CPA-gesp_GMX.gro')
for res in gpx.residues:
if res.name == 'MOL':
res.name = 'GPX'
for res in cpa.residues:
if res.name == 'MOL':
res.name = 'CPA'
struc_comb = gpx + cpa
struc_comb
struc_comb.write('gpx-cpa-dry.top')
struc_comb.save('gpx-cpa-dry.gro')
```
## Move molecules
In VMD, the molecules were moved so that they were not sitting on top of each other.
## Solvate
As before, using DCM parameters and solvent box from virtualchemistry.org.
```
f_dcm = py.path.local('~/GROMACS-basics/DCM-GAFF/')
f_solvate = py.path.local('02-solvate/')
sep_gro = py.path.local('gpx-cpa-sep.gro')
boxed_gro = f_solvate.join('gpx-cpa-boxed.gro')
box = '3.5 3.5 3.5'
solvent_source = f_dcm.join('dichloromethane-T293.15.gro')
solvent_top = f_dcm.join('dichloromethane.top')
solv_gro = f_solvate.join('gpx-cpa-dcm.gro')
top = py.path.local('../params/gpxTS-cpa-dcm.top')
verbose = True
solvent_counts, key = dict(), 'GPX'
with f_solvate.as_cwd():
## Make box
cl = shlex.split(f'gmx_mpi editconf -f {sep_gro} ' +
f'-o {boxed_gro} -box {box}')
proc = subprocess.run(cl, universal_newlines=True,
stdout=subprocess.PIPE,
stderr=subprocess.STDOUT)
outputs[key+'_editconf'] = proc.stdout
proc.check_returncode()
## Solvate
cl = shlex.split(f'gmx_mpi solvate -cp {boxed_gro} ' +
f'-cs {solvent_source} -o {solv_gro}')
proc = subprocess.run(cl, universal_newlines=True,
stdout=subprocess.PIPE,
stderr=subprocess.STDOUT)
outputs[key+'_solvate'] = proc.stdout
proc.check_returncode()
solvent_counts[key] = get_solvent_count_solvate(proc)
if verbose:
print(f'Solvated system into {solv_gro}')
struc_g_c = pmd.load_file('gpx-cpa-dry.top')
struc_dcm = pmd.load_file(str(f_dcm.join('dichloromethane.top')))
struc_g_c_d = struc_g_c + solvent_counts['GPX'] * struc_dcm
struc_g_c_d.save(str(top))
```
## Minimize
```
ppl = py.path.local
f_min = ppl('03-minimize/')
f_g_basics = py.path.local('~/GROMACS-basics/')
mdp_min = f_g_basics.join('minim.mdp')
tpr_min = f_min.join('min.tpr')
deffnm_min = f_min.join('min-out')
gro_min = deffnm_min + '.gro'
with f_min.as_cwd():
## Compile tpr
if not tpr_min.exists():
cl = shlex.split(f'gmx_mpi grompp -f {mdp_min} '
f'-c {solv_gro} '
f'-p {top} '
f'-o {tpr_min}')
proc = subprocess.run(cl, universal_newlines=True,
stdout=subprocess.PIPE,
stderr=subprocess.STDOUT)
outputs[key+'_grompp_em'] = proc.stdout
proc.check_returncode()
if verbose:
print(f'Compiled em tpr to {tpr_min}')
elif verbose:
print(f'em tpr file already exists ({tpr_min})')
## Run minimization
if not gro_min.exists():
cl = shlex.split('gmx_mpi mdrun '
f'-s {tpr_min} '
f'-deffnm {deffnm_min} ')
proc = subprocess.run(cl, universal_newlines=True,
stdout=subprocess.PIPE,
stderr=subprocess.STDOUT)
outputs[key+'_mdrun_em'] = proc.stdout
# TODO Get the potential energy from this output
proc.check_returncode()
if verbose:
print(f'Ran {key} em to make {gro_min}')
elif verbose:
print(f'em output gro already exists (gro_min)')
```
## Equilibrate
```
f_equil = ppl('04-equilibrate/')
plumed = f_equil.join('plumed.dat')
mdp_equil = f_g_basics.join('npt-298.mdp')
tpr_equil = f_equil.join('equil.tpr')
deffnm_equil = f_equil.join('equil-out')
gro_equil = deffnm_equil + '.gro'
gro_input = gro_min
with f_equil.as_cwd():
## Compile equilibration
if not tpr_equil.exists():
cl = shlex.split(f'gmx_mpi grompp -f {mdp_equil} '
f'-c {gro_input} '
f'-p {top} '
f'-o {tpr_equil}')
proc = subprocess.run(cl, universal_newlines=True,
stdout=subprocess.PIPE,
stderr=subprocess.STDOUT)
outputs[key+'_grompp_equil'] = proc.stdout
proc.check_returncode()
if verbose:
print(f'Compiled equil tpr to {tpr_equil}')
elif verbose:
print(f'equil tpr file already exists ({tpr_equil})')
## Run equilibration
if not gro_equil.exists():
cl = shlex.split('gmx_mpi mdrun '
f'-s {tpr_equil} '
f'-deffnm {deffnm_equil} '
f'-plumed {plumed}')
proc = subprocess.run(cl, universal_newlines=True,
stdout=subprocess.PIPE,
stderr=subprocess.STDOUT)
outputs[key+'_mdrun_equil'] = proc.stdout
proc.check_returncode()
if verbose:
print(f'Ran {key} equil to make {gro_equil}')
elif verbose:
print(f'equil output gro already exists (gro_equil)')
```
## Setup and submit parallel tempering (PT)
```
f_pt = ppl('05-PT/')
template = f_pt.join('template-mdp.txt')
index = ppl('index.ndx')
sub_templ = f_g_basics.join('sub-template-128.sub')
d_sub_templ = dict(tpr_base = 'TOPO/npt',
deffnm = 'PT-out',
name = 'GPX-PT',
plumed = plumed,
)
scaling_exponent = 0.025
maxwarn = 0
start_temp = 298.
verbose = True
skip_existing = True
jobs = []
failed_procs = []
for key in ['GPX']:
kwargs = {'template': str(template),
'topology': str(top),
'structure': str(gro_equil),
'index': str(index),
'scaling_exponent': scaling_exponent,
'start_temp': start_temp,
'maxwarn': maxwarn}
with f_pt.as_cwd():
try:
os.mkdir('TOPO')
except FileExistsError:
if skip_existing:
print(f'Skipping {key} because it seems to '
'already be done.\nMoving on...')
continue
with cd('TOPO'):
print(f'Now in {os.getcwd()}\nAttempting to compile TPRs...')
pts.compile_tprs(**kwargs)
print('Done compiling. Moving on...')
print(f'Now in {os.getcwd()}\nWriting submission script...')
with sub_templ.open(mode='r') as templ_f, \
open('gromacs-start-job.sub', 'w') as sub_s:
[sub_s.write(l.format(**d_sub_templ)) for l in templ_f]
print('Done.\nNow submitting job...')
cl = ['qsub', 'gromacs-start-job.sub']
proc = subprocess.run(cl,
stdout=subprocess.PIPE,
stderr=subprocess.STDOUT,
universal_newlines=True)
if proc.returncode == 0:
output = proc.stdout
jobs.append(re.search('[0-9].+\)', output).group(0))
print(output, '\nDone.\nMoving to next...')
else:
print('\n\n'+5*'!!!---'+'\n')
print(f'Error with calling qsub on {key}')
print('Command line input was', cl)
print('Check input and try again manually.'
'\nMoving to next anyway...')
failed_procs.append(proc)
print('-----Done-----\nSummary of jobs submitted:')
for job in jobs:
print(job)
```
The energies from the simulations can be read in as a pandas DataFrame using [panedr](https://github.com/jbarnoud/panedr) and then analyzed or plotted to check on equilibration, convergence, etc.
```
e_05s = dict()
for i in range(16):
e_05s[i] = panedr.edr_to_df(f'05-PT/PT-out{i}.edr')
fig, axes = plot_prop_PT(e_05s, 'Pressure')
```
# Setup for several systems/molecules at once
Working based on what was done above (using some things that were defined up there as well
## Get charges
```
l_scripts = []
s = subg.write_sub_script('01-charges/TS1.com',
executable='g16',
make_xyz='../TS1protonated.mol2',
make_input=True,
ugt_dict={'job_name':'GPX TS1 charges',
'charg_mult':'+1 1',
**d_charge_params})
l_scripts.append(s)
s = subg.write_sub_script('01-charges/TS3.com',
executable='g16',
make_xyz='../TS3protonated.mol2',
make_input=True,
ugt_dict={'job_name':'GPX TS3 charges',
'charg_mult':'+1 1',
**d_charge_params})
l_scripts.append(s)
s = subg.write_sub_script('01-charges/anti-cat-yamamoto.com',
executable='g16',
make_xyz='../R-Yamamoto-Cat.pdb',
make_input=True,
ugt_dict={'job_name':
'yamamoto catalyst charges',
'charg_mult':'-1 1',
**d_charge_params})
l_scripts.append(s)
l_scripts
subg.submit_scripts(l_scripts, batch=True, submit=True)
```
Copied over the g16.gesp files and renamed them for each molecule.
## Make input files
Loaded amber/2016 module (and its dependencies).
antechamber -i TS1.gesp -fi gesp -o TS1.mol2 -fo mol2
acpype.py -i TS1.mol2 -b TS1-gesp --net_charge=1 -o gmx -d -c user
There was a warning for assigning bond types.
antechamber -i TS3.gesp -fi gesp -o TS3.mol2 -fo mol2
acpype.py -i TS3.mol2 -b TS3-gesp --net_charge=1 -o gmx -d -c user
Similar warning.
antechamber -i YCP.gesp -fi gesp -o YCP.mol2 -fo mol2
acpype.py -i YCP.mol2 -b YCP-gesp --net_charge=-1 -o gmx -d -c use
No similar warning here.
```
ts1 = pmd.gromacs.GromacsTopologyFile(
'01-charges/TS1-gesp.acpype/TS1-gesp_GMX.top',
xyz='01-charges/TS1-gesp.acpype/TS1-gesp_GMX.gro')
ts3 = pmd.gromacs.GromacsTopologyFile(
'01-charges/TS3-gesp.acpype/TS3-gesp_GMX.top',
xyz='01-charges/TS3-gesp.acpype/TS3-gesp_GMX.gro')
ycp = pmd.gromacs.GromacsTopologyFile(
'01-charges/YCP-gesp.acpype/YCP-gesp_GMX.top',
xyz='01-charges/YCP-gesp.acpype/YCP-gesp_GMX.gro')
for res in ts1.residues:
if res.name == 'MOL':
res.name = 'TS1'
for res in ts3.residues:
if res.name == 'MOL':
res.name = 'TS3'
for res in ycp.residues:
if res.name == 'MOL':
res.name = 'YCP'
ts1_en = ts1.copy(pmd.gromacs.GromacsTopologyFile)
ts3_en = ts3.copy(pmd.gromacs.GromacsTopologyFile)
ts1_en.coordinates = - ts1.coordinates
ts3_en.coordinates = - ts3.coordinates
sys_ts1 = ts1 + ycp
sys_ts1_en = ts1_en + ycp
sys_ts3 = ts3 + ycp
sys_ts3_en = ts3_en + ycp
sys_ts1.write('ts1-ycp-dry.top')
sys_ts3.write('ts3-ycp-dry.top')
sys_ts1.save('ts1-ycp-dry.gro')
sys_ts1_en.save('ts1_en-ycp-dry.gro')
sys_ts3.save('ts3-ycp-dry.gro')
sys_ts3_en.save('ts3_en-ycp-dry.gro')
```
## Move molecules
I presume I will again need to make the molecules non-overlapping, and that will be done manually in VMD.
## Box and solvate
```
f_dcm = py.path.local('~/GROMACS-basics/DCM-GAFF/')
f_solvate = py.path.local('37-solvate-anti/')
box = '3.7 3.7 3.7'
solvent_source = f_dcm.join('dichloromethane-T293.15.gro')
solvent_top = f_dcm.join('dichloromethane.top')
solv_gro = f_solvate.join('gpx-cpa-dcm.gro')
ts1_top = ppl('../params/ts1-ycp-dcm.top')
ts3_top = ppl('../params/ts3-ycp-dcm.top')
l_syss = ['TS1', 'TS1_en', 'TS3', 'TS3_en']
verbose = True
solvent_counts = dict()
for key in l_syss:
sep_gro = ppl(f'{key.lower()}-ycp-dry.gro')
if not sep_gro.exists():
raise FileNotFoundError(f'{sep_gro} does not exist')
boxed_gro = f'{key.lower()}-ycp-box.gro'
solv_gro = f'{key.lower()}-ycp-dcm.gro'
with f_solvate.ensure_dir().as_cwd():
## Make box
cl = shlex.split(f'gmx_mpi editconf -f {sep_gro} ' +
f'-o {boxed_gro} -box {box}')
proc = subprocess.run(cl, universal_newlines=True,
stdout=subprocess.PIPE,
stderr=subprocess.STDOUT)
outputs[key+'_editconf'] = proc.stdout
proc.check_returncode()
## Solvate
cl = shlex.split(f'gmx_mpi solvate -cp {boxed_gro} ' +
f'-cs {solvent_source} -o {solv_gro}')
proc = subprocess.run(cl, universal_newlines=True,
stdout=subprocess.PIPE,
stderr=subprocess.STDOUT)
outputs[key+'_solvate'] = proc.stdout
proc.check_returncode()
solvent_counts[key] = get_solvent_count_solvate(proc)
if verbose:
print(f'Solvated system into {solv_gro}')
# min_solv_count = min(solvent_counts.values())
min_solv_count = 328 # want to match with syn calculations
if min(solvent_counts.values()) < min_solv_count:
raise ValueError('At least one of the structures has <328 DCMs.\n'
'Check and/or make the box larger')
for key in l_syss:
solv_gro = f'{key.lower()}-ycp-dcm.gro'
with f_solvate.as_cwd():
set_solv_count(solv_gro, min_solv_count)
struc_ts1 = pmd.load_file('ts1-ycp-dry.top')
struc_ts3 = pmd.load_file('ts3-ycp-dry.top')
struc_dcm = pmd.load_file(str(f_dcm.join('dichloromethane.top')))
struc_ts1_d = struc_ts1 + min_solv_count * struc_dcm
struc_ts1_d.save(str(ts1_top))
struc_ts3_d = struc_ts3 + min_solv_count * struc_dcm
struc_ts3_d.save(str(ts3_top))
```
## Minimize
```
f_min = ppl('38-relax-anti/')
f_min.ensure_dir()
f_g_basics = py.path.local('~/GROMACS-basics/')
mdp_min = f_g_basics.join('minim.mdp')
d_tops = dict(TS1=ts1_top, TS1_en=ts1_top, TS3=ts3_top, TS3_en=ts3_top)
for key in l_syss:
solv_gro = ppl(f'37-solvate-anti/{key.lower()}-ycp-dcm.gro')
tpr_min = f_min.join(f'{key.lower()}-min.tpr')
deffnm_min = f_min.join(f'{key.lower()}-min-out')
gro_min = deffnm_min + '.gro'
top = d_tops[key]
with f_min.as_cwd():
## Compile tpr
if not tpr_min.exists():
cl = shlex.split(f'gmx_mpi grompp -f {mdp_min} '
f'-c {solv_gro} '
f'-p {top} '
f'-o {tpr_min}')
proc = subprocess.run(cl, universal_newlines=True,
stdout=subprocess.PIPE,
stderr=subprocess.STDOUT)
outputs[key+'_grompp_em'] = proc.stdout
proc.check_returncode()
if verbose:
print(f'Compiled em tpr to {tpr_min}')
elif verbose:
print(f'em tpr file already exists ({tpr_min})')
## Run minimization
if not gro_min.exists():
cl = shlex.split('gmx_mpi mdrun '
f'-s {tpr_min} '
f'-deffnm {deffnm_min} ')
proc = subprocess.run(cl, universal_newlines=True,
stdout=subprocess.PIPE,
stderr=subprocess.STDOUT)
outputs[key+'_mdrun_em'] = proc.stdout
# TODO Get the potential energy from this output
proc.check_returncode()
if verbose:
print(f'Ran {key} em to make {gro_min}')
elif verbose:
print(f'em output gro already exists (gro_min)')
```
Made index file (called index-ycp.ndx) with solutes and solvent groups.
## SA equilibration
```
f_pt = ppl('38-relax-anti/')
template = ppl('33-SA-NPT-rest-no-LINCS/template-mdp.txt')
index = ppl('../params/index-ycp.ndx')
scaling_exponent = 0.025
maxwarn = 0
start_temp = 298.
nsims = 16
verbose = True
skip_existing = True
jobs = []
failed_procs = []
for key in l_syss:
d_sub_templ = dict(
tpr = f'{key.lower()}-TOPO/npt',
deffnm = f'{key.lower()}-SA-out',
name = f'{key.lower()}-SA',
nsims = nsims,
tpn = 16,
cores = 128,
multi = True,
)
gro_equil = f_min.join(f'{key.lower()}-min-out.gro')
top = d_tops[key]
kwargs = {'template': str(template),
'topology': str(top),
'structure': str(gro_equil),
'index': str(index),
'scaling_exponent': scaling_exponent,
'start_temp': start_temp,
'maxwarn': maxwarn,
'number': nsims,
'grompp_exe': 'gmx_mpi grompp'}
with f_pt.as_cwd():
try:
os.mkdir(f'{key.lower()}-TOPO/')
except FileExistsError:
if (os.path.exists(f'{key.lower()}-TOPO/temperatures.dat') and
skip_existing):
print(f'Skipping {key} because it seems to '
'already be done.\nMoving on...')
continue
with cd(f'{key.lower()}-TOPO/'):
print(f'Now in {os.getcwd()}\nAttempting to compile TPRs...')
pts.compile_tprs(**kwargs)
print('Done compiling. Moving on...')
print(f'Now in {os.getcwd()}\nWriting submission script...')
lp_sub = pt.sim_setup.make_gromacs_sub_script(
f'gromacs-start-{key}-job.sub', **d_sub_templ)
print('Done.\nNow submitting job...')
cl = shlex.split(f'qsub {lp_sub}')
proc = subprocess.run(cl,
stdout=subprocess.PIPE,
stderr=subprocess.STDOUT,
universal_newlines=True)
if proc.returncode == 0:
output = proc.stdout
jobs.append(re.search('[0-9].+\)', output).group(0))
print(output, '\nDone.\nMoving to next...')
else:
print('\n\n'+5*'!!!---'+'\n')
print(f'Error with calling qsub on {key}')
print('Command line input was', cl)
print('Check input and try again manually.'
'\nMoving to next anyway...')
failed_procs.append(proc)
print('-----Done-----\nSummary of jobs submitted:')
for job in jobs:
print(job)
```
!!! Need to check distance on restraint !!!
### Check equilibration
```
e_38s = dict()
for key in l_syss:
deffnm = f'{key.lower()}-SA-out'
e_38s[key] = dict()
d = e_38s[key]
for i in range(16):
d[i] = panedr.edr_to_df(f'38-relax-anti/{deffnm}{i}.edr')
for key in l_syss:
d = e_38s[key]
fig, axes = plot_prop_PT(d, 'Volume')
```
The volumes seem to look okay.
Started high (I did remove some solvents and it hadn't relaxed much), dropped quickly, then seemed to grow appropriately as the temperatures rose.
None seems to have boiled.
```
for key in l_syss:
d = e_38s[key]
fig, ax = plt.subplots()
for key in list(d.keys()):
ax.hist(d[key]['Total Energy'], bins=100)
del d[key]
```
|
github_jupyter
|
import re, os, sys, shutil
import shlex, subprocess
import glob
import pandas as pd
import panedr
import numpy as np
import MDAnalysis as mda
import nglview
import matplotlib.pyplot as plt
import parmed as pmd
import py
import scipy
from scipy import stats
from importlib import reload
from thtools import cd
from paratemp import copy_no_overwrite
from paratemp import geometries as gm
from paratemp import coordinate_analysis as ca
import paratemp.para_temp_setup as pts
import paratemp as pt
from gautools import submit_gaussian as subg
from gautools.tools import use_gen_template as ugt
def plot_prop_PT(edict, prop):
fig, axes = plt.subplots(4, 4, figsize=(16,16))
for i in range(16):
ax = axes.flat[i]
edict[i][prop].plot(ax=ax)
fig.tight_layout()
return fig, axes
def plot_e_props(df, labels, nrows=2, ncols=2):
fig, axes = plt.subplots(nrows, ncols, sharex=True)
for label, ax in zip(labels, axes.flat):
df[label].plot(ax=ax)
ax.set_title(label)
fig.tight_layout()
return fig, axes
def plot_rd(univ): # rd = reaction distance
univ.calculate_distances(rd=(20,39))
fig, axes = plt.subplots(1, 2, figsize=(12, 4))
univ.data.rd.plot(ax=axes[0])
univ.data.rd.hist(ax=axes[1], grid=False)
print(f'reaction distance mean: {univ.data.rd.mean():.2f} and sd: {univ.data.rd.std():.2f}')
return fig, axes
def plot_hist_dist(univ, name, indexes=None):
if indexes is not None:
kwargs = {name: indexes}
univ.calculate_distances(**kwargs)
fig, axes = plt.subplots(1, 2, figsize=(12, 4))
univ.data[name].plot(ax=axes[0])
univ.data[name].hist(ax=axes[1], grid=False)
print(f'{name} distance mean: {univ.data[name].mean():.2f} and sd: {univ.data[name].std():.2f}')
def get_solvent_count_solvate(proc):
for line in proc.stdout.split('\n'):
m = re.search(r'(?:atoms\):\s+)(\d+)(?:\s+residues)', line)
if m:
return int(m.group(1))
else:
raise ValueError('Solvent count not found.')
def set_solv_count(n_gro, s_count,
res_name='DCM', prepend='unequal-'):
"""
Remove solvent residues from the end of a gro file to match s_count
This assumes all non-solvent molecules are listed in the input gro
file before the solvent residues.
"""
bak_name = os.path.join(os.path.dirname(n_gro),
prepend+os.path.basename(n_gro))
copy_no_overwrite(n_gro, bak_name)
with open(n_gro, 'r') as in_gro:
lines = in_gro.readlines()
for line in lines[2:]:
if res_name in line:
non_s_res_count = resid
break
else:
resid = int(line[:5])
res_count = s_count + non_s_res_count
# TODO check reasonability of this number
box = lines.pop()
while True:
line = lines.pop()
if int(line[:5]) > res_count:
continue
elif int(line[:5]) == res_count:
atom_count = line[15:20]
lines.append(line)
break
elif int(line[:5]) < res_count:
raise ValueError("Desired res "
"count is larger than "
"line's resid.\n" +
"res_count: {}\n".format(res_count) +
"line: {}".format(line))
lines[1] = atom_count + '\n'
lines.append(box)
with open(n_gro, 'w') as out_gro:
for line in lines:
out_gro.write(line)
def get_solv_count_top(n_top, res_name='DCM'):
"""
Return residue count of specified residue from n_top"""
with open(n_top, 'r') as in_top:
mol_section = False
for line in in_top:
if line.strip().startswith(';'):
pass
elif not mol_section:
if re.search(r'\[\s*molecules\s*\]', line,
flags=re.IGNORECASE):
mol_section = True
else:
if res_name.lower() in line.lower():
return int(line.split()[1])
def set_solv_count_top(n_top, s_count,
res_name='DCM', prepend='unequal-'):
"""
Set count of res_name residues in n_top
This will make a backup copy of the top file with `prepend`
prepended to the name of the file."""
bak_name = os.path.join(os.path.dirname(n_top),
prepend+os.path.basename(n_top))
copy_no_overwrite(n_top, bak_name)
with open(n_top, 'r') as in_top:
lines = in_top.readlines()
with open(n_top, 'w') as out_top:
mol_section = False
for line in lines:
if line.strip().startswith(';'):
pass
elif not mol_section:
if re.search(r'\[\s*molecules\s*\]', line,
flags=re.IGNORECASE):
mol_section = True
else:
if res_name.lower() in line.lower():
line = re.sub(r'\d+', str(s_count), line)
out_top.write(line)
d_charge_params = dict(opt='SCF=tight Test Pop=MK iop(6/33=2) iop(6/42=6) iop(6/50=1)',
func='HF',
basis='6-31G*',
footer='\ng16.gesp\n\ng16.gesp\n\n')
l_scripts = []
s = subg.write_sub_script('01-charges/TS2.com',
executable='g16',
make_xyz='../TS2.pdb',
make_input=True,
ugt_dict={'job_name':'GPX TS2 charges',
'charg_mult':'+1 1',
**d_charge_params})
l_scripts.append(s)
s = subg.write_sub_script('01-charges/R-NO2-CPA.com',
executable='g16',
make_xyz='../R-NO2-CPA.pdb',
make_input=True,
ugt_dict={'job_name':'GPX R-NO2-CPA charges',
'charg_mult':'-1 1',
**d_charge_params})
l_scripts.append(s)
l_scripts
subg.submit_scripts(l_scripts, batch=True, submit=True)
gpx = pmd.gromacs.GromacsTopologyFile('01-charges/GPX-ts.acpype/GPX-ts_GMX.top', xyz='01-charges/GPX-ts.acpype/GPX-ts_GMX.gro')
cpa = pmd.gromacs.GromacsTopologyFile('01-charges/CPA-gesp.acpype/CPA-gesp_GMX.top', xyz='01-charges/CPA-gesp.acpype/CPA-gesp_GMX.gro')
for res in gpx.residues:
if res.name == 'MOL':
res.name = 'GPX'
for res in cpa.residues:
if res.name == 'MOL':
res.name = 'CPA'
struc_comb = gpx + cpa
struc_comb
struc_comb.write('gpx-cpa-dry.top')
struc_comb.save('gpx-cpa-dry.gro')
f_dcm = py.path.local('~/GROMACS-basics/DCM-GAFF/')
f_solvate = py.path.local('02-solvate/')
sep_gro = py.path.local('gpx-cpa-sep.gro')
boxed_gro = f_solvate.join('gpx-cpa-boxed.gro')
box = '3.5 3.5 3.5'
solvent_source = f_dcm.join('dichloromethane-T293.15.gro')
solvent_top = f_dcm.join('dichloromethane.top')
solv_gro = f_solvate.join('gpx-cpa-dcm.gro')
top = py.path.local('../params/gpxTS-cpa-dcm.top')
verbose = True
solvent_counts, key = dict(), 'GPX'
with f_solvate.as_cwd():
## Make box
cl = shlex.split(f'gmx_mpi editconf -f {sep_gro} ' +
f'-o {boxed_gro} -box {box}')
proc = subprocess.run(cl, universal_newlines=True,
stdout=subprocess.PIPE,
stderr=subprocess.STDOUT)
outputs[key+'_editconf'] = proc.stdout
proc.check_returncode()
## Solvate
cl = shlex.split(f'gmx_mpi solvate -cp {boxed_gro} ' +
f'-cs {solvent_source} -o {solv_gro}')
proc = subprocess.run(cl, universal_newlines=True,
stdout=subprocess.PIPE,
stderr=subprocess.STDOUT)
outputs[key+'_solvate'] = proc.stdout
proc.check_returncode()
solvent_counts[key] = get_solvent_count_solvate(proc)
if verbose:
print(f'Solvated system into {solv_gro}')
struc_g_c = pmd.load_file('gpx-cpa-dry.top')
struc_dcm = pmd.load_file(str(f_dcm.join('dichloromethane.top')))
struc_g_c_d = struc_g_c + solvent_counts['GPX'] * struc_dcm
struc_g_c_d.save(str(top))
ppl = py.path.local
f_min = ppl('03-minimize/')
f_g_basics = py.path.local('~/GROMACS-basics/')
mdp_min = f_g_basics.join('minim.mdp')
tpr_min = f_min.join('min.tpr')
deffnm_min = f_min.join('min-out')
gro_min = deffnm_min + '.gro'
with f_min.as_cwd():
## Compile tpr
if not tpr_min.exists():
cl = shlex.split(f'gmx_mpi grompp -f {mdp_min} '
f'-c {solv_gro} '
f'-p {top} '
f'-o {tpr_min}')
proc = subprocess.run(cl, universal_newlines=True,
stdout=subprocess.PIPE,
stderr=subprocess.STDOUT)
outputs[key+'_grompp_em'] = proc.stdout
proc.check_returncode()
if verbose:
print(f'Compiled em tpr to {tpr_min}')
elif verbose:
print(f'em tpr file already exists ({tpr_min})')
## Run minimization
if not gro_min.exists():
cl = shlex.split('gmx_mpi mdrun '
f'-s {tpr_min} '
f'-deffnm {deffnm_min} ')
proc = subprocess.run(cl, universal_newlines=True,
stdout=subprocess.PIPE,
stderr=subprocess.STDOUT)
outputs[key+'_mdrun_em'] = proc.stdout
# TODO Get the potential energy from this output
proc.check_returncode()
if verbose:
print(f'Ran {key} em to make {gro_min}')
elif verbose:
print(f'em output gro already exists (gro_min)')
f_equil = ppl('04-equilibrate/')
plumed = f_equil.join('plumed.dat')
mdp_equil = f_g_basics.join('npt-298.mdp')
tpr_equil = f_equil.join('equil.tpr')
deffnm_equil = f_equil.join('equil-out')
gro_equil = deffnm_equil + '.gro'
gro_input = gro_min
with f_equil.as_cwd():
## Compile equilibration
if not tpr_equil.exists():
cl = shlex.split(f'gmx_mpi grompp -f {mdp_equil} '
f'-c {gro_input} '
f'-p {top} '
f'-o {tpr_equil}')
proc = subprocess.run(cl, universal_newlines=True,
stdout=subprocess.PIPE,
stderr=subprocess.STDOUT)
outputs[key+'_grompp_equil'] = proc.stdout
proc.check_returncode()
if verbose:
print(f'Compiled equil tpr to {tpr_equil}')
elif verbose:
print(f'equil tpr file already exists ({tpr_equil})')
## Run equilibration
if not gro_equil.exists():
cl = shlex.split('gmx_mpi mdrun '
f'-s {tpr_equil} '
f'-deffnm {deffnm_equil} '
f'-plumed {plumed}')
proc = subprocess.run(cl, universal_newlines=True,
stdout=subprocess.PIPE,
stderr=subprocess.STDOUT)
outputs[key+'_mdrun_equil'] = proc.stdout
proc.check_returncode()
if verbose:
print(f'Ran {key} equil to make {gro_equil}')
elif verbose:
print(f'equil output gro already exists (gro_equil)')
f_pt = ppl('05-PT/')
template = f_pt.join('template-mdp.txt')
index = ppl('index.ndx')
sub_templ = f_g_basics.join('sub-template-128.sub')
d_sub_templ = dict(tpr_base = 'TOPO/npt',
deffnm = 'PT-out',
name = 'GPX-PT',
plumed = plumed,
)
scaling_exponent = 0.025
maxwarn = 0
start_temp = 298.
verbose = True
skip_existing = True
jobs = []
failed_procs = []
for key in ['GPX']:
kwargs = {'template': str(template),
'topology': str(top),
'structure': str(gro_equil),
'index': str(index),
'scaling_exponent': scaling_exponent,
'start_temp': start_temp,
'maxwarn': maxwarn}
with f_pt.as_cwd():
try:
os.mkdir('TOPO')
except FileExistsError:
if skip_existing:
print(f'Skipping {key} because it seems to '
'already be done.\nMoving on...')
continue
with cd('TOPO'):
print(f'Now in {os.getcwd()}\nAttempting to compile TPRs...')
pts.compile_tprs(**kwargs)
print('Done compiling. Moving on...')
print(f'Now in {os.getcwd()}\nWriting submission script...')
with sub_templ.open(mode='r') as templ_f, \
open('gromacs-start-job.sub', 'w') as sub_s:
[sub_s.write(l.format(**d_sub_templ)) for l in templ_f]
print('Done.\nNow submitting job...')
cl = ['qsub', 'gromacs-start-job.sub']
proc = subprocess.run(cl,
stdout=subprocess.PIPE,
stderr=subprocess.STDOUT,
universal_newlines=True)
if proc.returncode == 0:
output = proc.stdout
jobs.append(re.search('[0-9].+\)', output).group(0))
print(output, '\nDone.\nMoving to next...')
else:
print('\n\n'+5*'!!!---'+'\n')
print(f'Error with calling qsub on {key}')
print('Command line input was', cl)
print('Check input and try again manually.'
'\nMoving to next anyway...')
failed_procs.append(proc)
print('-----Done-----\nSummary of jobs submitted:')
for job in jobs:
print(job)
e_05s = dict()
for i in range(16):
e_05s[i] = panedr.edr_to_df(f'05-PT/PT-out{i}.edr')
fig, axes = plot_prop_PT(e_05s, 'Pressure')
l_scripts = []
s = subg.write_sub_script('01-charges/TS1.com',
executable='g16',
make_xyz='../TS1protonated.mol2',
make_input=True,
ugt_dict={'job_name':'GPX TS1 charges',
'charg_mult':'+1 1',
**d_charge_params})
l_scripts.append(s)
s = subg.write_sub_script('01-charges/TS3.com',
executable='g16',
make_xyz='../TS3protonated.mol2',
make_input=True,
ugt_dict={'job_name':'GPX TS3 charges',
'charg_mult':'+1 1',
**d_charge_params})
l_scripts.append(s)
s = subg.write_sub_script('01-charges/anti-cat-yamamoto.com',
executable='g16',
make_xyz='../R-Yamamoto-Cat.pdb',
make_input=True,
ugt_dict={'job_name':
'yamamoto catalyst charges',
'charg_mult':'-1 1',
**d_charge_params})
l_scripts.append(s)
l_scripts
subg.submit_scripts(l_scripts, batch=True, submit=True)
ts1 = pmd.gromacs.GromacsTopologyFile(
'01-charges/TS1-gesp.acpype/TS1-gesp_GMX.top',
xyz='01-charges/TS1-gesp.acpype/TS1-gesp_GMX.gro')
ts3 = pmd.gromacs.GromacsTopologyFile(
'01-charges/TS3-gesp.acpype/TS3-gesp_GMX.top',
xyz='01-charges/TS3-gesp.acpype/TS3-gesp_GMX.gro')
ycp = pmd.gromacs.GromacsTopologyFile(
'01-charges/YCP-gesp.acpype/YCP-gesp_GMX.top',
xyz='01-charges/YCP-gesp.acpype/YCP-gesp_GMX.gro')
for res in ts1.residues:
if res.name == 'MOL':
res.name = 'TS1'
for res in ts3.residues:
if res.name == 'MOL':
res.name = 'TS3'
for res in ycp.residues:
if res.name == 'MOL':
res.name = 'YCP'
ts1_en = ts1.copy(pmd.gromacs.GromacsTopologyFile)
ts3_en = ts3.copy(pmd.gromacs.GromacsTopologyFile)
ts1_en.coordinates = - ts1.coordinates
ts3_en.coordinates = - ts3.coordinates
sys_ts1 = ts1 + ycp
sys_ts1_en = ts1_en + ycp
sys_ts3 = ts3 + ycp
sys_ts3_en = ts3_en + ycp
sys_ts1.write('ts1-ycp-dry.top')
sys_ts3.write('ts3-ycp-dry.top')
sys_ts1.save('ts1-ycp-dry.gro')
sys_ts1_en.save('ts1_en-ycp-dry.gro')
sys_ts3.save('ts3-ycp-dry.gro')
sys_ts3_en.save('ts3_en-ycp-dry.gro')
f_dcm = py.path.local('~/GROMACS-basics/DCM-GAFF/')
f_solvate = py.path.local('37-solvate-anti/')
box = '3.7 3.7 3.7'
solvent_source = f_dcm.join('dichloromethane-T293.15.gro')
solvent_top = f_dcm.join('dichloromethane.top')
solv_gro = f_solvate.join('gpx-cpa-dcm.gro')
ts1_top = ppl('../params/ts1-ycp-dcm.top')
ts3_top = ppl('../params/ts3-ycp-dcm.top')
l_syss = ['TS1', 'TS1_en', 'TS3', 'TS3_en']
verbose = True
solvent_counts = dict()
for key in l_syss:
sep_gro = ppl(f'{key.lower()}-ycp-dry.gro')
if not sep_gro.exists():
raise FileNotFoundError(f'{sep_gro} does not exist')
boxed_gro = f'{key.lower()}-ycp-box.gro'
solv_gro = f'{key.lower()}-ycp-dcm.gro'
with f_solvate.ensure_dir().as_cwd():
## Make box
cl = shlex.split(f'gmx_mpi editconf -f {sep_gro} ' +
f'-o {boxed_gro} -box {box}')
proc = subprocess.run(cl, universal_newlines=True,
stdout=subprocess.PIPE,
stderr=subprocess.STDOUT)
outputs[key+'_editconf'] = proc.stdout
proc.check_returncode()
## Solvate
cl = shlex.split(f'gmx_mpi solvate -cp {boxed_gro} ' +
f'-cs {solvent_source} -o {solv_gro}')
proc = subprocess.run(cl, universal_newlines=True,
stdout=subprocess.PIPE,
stderr=subprocess.STDOUT)
outputs[key+'_solvate'] = proc.stdout
proc.check_returncode()
solvent_counts[key] = get_solvent_count_solvate(proc)
if verbose:
print(f'Solvated system into {solv_gro}')
# min_solv_count = min(solvent_counts.values())
min_solv_count = 328 # want to match with syn calculations
if min(solvent_counts.values()) < min_solv_count:
raise ValueError('At least one of the structures has <328 DCMs.\n'
'Check and/or make the box larger')
for key in l_syss:
solv_gro = f'{key.lower()}-ycp-dcm.gro'
with f_solvate.as_cwd():
set_solv_count(solv_gro, min_solv_count)
struc_ts1 = pmd.load_file('ts1-ycp-dry.top')
struc_ts3 = pmd.load_file('ts3-ycp-dry.top')
struc_dcm = pmd.load_file(str(f_dcm.join('dichloromethane.top')))
struc_ts1_d = struc_ts1 + min_solv_count * struc_dcm
struc_ts1_d.save(str(ts1_top))
struc_ts3_d = struc_ts3 + min_solv_count * struc_dcm
struc_ts3_d.save(str(ts3_top))
f_min = ppl('38-relax-anti/')
f_min.ensure_dir()
f_g_basics = py.path.local('~/GROMACS-basics/')
mdp_min = f_g_basics.join('minim.mdp')
d_tops = dict(TS1=ts1_top, TS1_en=ts1_top, TS3=ts3_top, TS3_en=ts3_top)
for key in l_syss:
solv_gro = ppl(f'37-solvate-anti/{key.lower()}-ycp-dcm.gro')
tpr_min = f_min.join(f'{key.lower()}-min.tpr')
deffnm_min = f_min.join(f'{key.lower()}-min-out')
gro_min = deffnm_min + '.gro'
top = d_tops[key]
with f_min.as_cwd():
## Compile tpr
if not tpr_min.exists():
cl = shlex.split(f'gmx_mpi grompp -f {mdp_min} '
f'-c {solv_gro} '
f'-p {top} '
f'-o {tpr_min}')
proc = subprocess.run(cl, universal_newlines=True,
stdout=subprocess.PIPE,
stderr=subprocess.STDOUT)
outputs[key+'_grompp_em'] = proc.stdout
proc.check_returncode()
if verbose:
print(f'Compiled em tpr to {tpr_min}')
elif verbose:
print(f'em tpr file already exists ({tpr_min})')
## Run minimization
if not gro_min.exists():
cl = shlex.split('gmx_mpi mdrun '
f'-s {tpr_min} '
f'-deffnm {deffnm_min} ')
proc = subprocess.run(cl, universal_newlines=True,
stdout=subprocess.PIPE,
stderr=subprocess.STDOUT)
outputs[key+'_mdrun_em'] = proc.stdout
# TODO Get the potential energy from this output
proc.check_returncode()
if verbose:
print(f'Ran {key} em to make {gro_min}')
elif verbose:
print(f'em output gro already exists (gro_min)')
f_pt = ppl('38-relax-anti/')
template = ppl('33-SA-NPT-rest-no-LINCS/template-mdp.txt')
index = ppl('../params/index-ycp.ndx')
scaling_exponent = 0.025
maxwarn = 0
start_temp = 298.
nsims = 16
verbose = True
skip_existing = True
jobs = []
failed_procs = []
for key in l_syss:
d_sub_templ = dict(
tpr = f'{key.lower()}-TOPO/npt',
deffnm = f'{key.lower()}-SA-out',
name = f'{key.lower()}-SA',
nsims = nsims,
tpn = 16,
cores = 128,
multi = True,
)
gro_equil = f_min.join(f'{key.lower()}-min-out.gro')
top = d_tops[key]
kwargs = {'template': str(template),
'topology': str(top),
'structure': str(gro_equil),
'index': str(index),
'scaling_exponent': scaling_exponent,
'start_temp': start_temp,
'maxwarn': maxwarn,
'number': nsims,
'grompp_exe': 'gmx_mpi grompp'}
with f_pt.as_cwd():
try:
os.mkdir(f'{key.lower()}-TOPO/')
except FileExistsError:
if (os.path.exists(f'{key.lower()}-TOPO/temperatures.dat') and
skip_existing):
print(f'Skipping {key} because it seems to '
'already be done.\nMoving on...')
continue
with cd(f'{key.lower()}-TOPO/'):
print(f'Now in {os.getcwd()}\nAttempting to compile TPRs...')
pts.compile_tprs(**kwargs)
print('Done compiling. Moving on...')
print(f'Now in {os.getcwd()}\nWriting submission script...')
lp_sub = pt.sim_setup.make_gromacs_sub_script(
f'gromacs-start-{key}-job.sub', **d_sub_templ)
print('Done.\nNow submitting job...')
cl = shlex.split(f'qsub {lp_sub}')
proc = subprocess.run(cl,
stdout=subprocess.PIPE,
stderr=subprocess.STDOUT,
universal_newlines=True)
if proc.returncode == 0:
output = proc.stdout
jobs.append(re.search('[0-9].+\)', output).group(0))
print(output, '\nDone.\nMoving to next...')
else:
print('\n\n'+5*'!!!---'+'\n')
print(f'Error with calling qsub on {key}')
print('Command line input was', cl)
print('Check input and try again manually.'
'\nMoving to next anyway...')
failed_procs.append(proc)
print('-----Done-----\nSummary of jobs submitted:')
for job in jobs:
print(job)
e_38s = dict()
for key in l_syss:
deffnm = f'{key.lower()}-SA-out'
e_38s[key] = dict()
d = e_38s[key]
for i in range(16):
d[i] = panedr.edr_to_df(f'38-relax-anti/{deffnm}{i}.edr')
for key in l_syss:
d = e_38s[key]
fig, axes = plot_prop_PT(d, 'Volume')
for key in l_syss:
d = e_38s[key]
fig, ax = plt.subplots()
for key in list(d.keys()):
ax.hist(d[key]['Total Energy'], bins=100)
del d[key]
| 0.294722 | 0.740515 |
```
import os, sys
from itertools import chain
import datasets
import random
import torch
from transformers import GPT2LMHeadModel, GPT2Config, GPT2Tokenizer
from torch.utils.data import Dataset, DataLoader
from sklearn.metrics import accuracy_score, f1_score
from greenformer import auto_fact
import numpy as np
from tqdm import tqdm
import hashlib
def count_param(module, trainable=False):
if trainable:
return sum(p.numel() for p in module.parameters() if p.requires_grad)
else:
return sum(p.numel() for p in module.parameters())
```
# Init Model
```
model = GPT2LMHeadModel.from_pretrained('gpt2-large')
tokenizer = GPT2Tokenizer.from_pretrained('gpt2-large')
count_param(model)
```
# Apply partial factorization to GPT2 model
```
# Only factorize last one-third of transformer layers of the GPT2 model
factorizable_submodules = list(model.transformer.h[-(model.config.n_layer // 3):])
%%time
fact_model = auto_fact(model, rank=384, deepcopy=True, solver='svd', num_iter=20, submodules=factorizable_submodules)
count_param(fact_model)
```
# Speed test on CPU
### Test Inference CPU
```
%%timeit
with torch.no_grad():
y = model(torch.zeros(2, 64, dtype=torch.long))
%%timeit
with torch.no_grad():
y = fact_model(torch.zeros(2, 64, dtype=torch.long))
```
# Speed test on GPU
### Move models to GPU
```
model = model.cuda()
fact_model = fact_model.cuda()
```
### Test Inference GPU
```
x = torch.zeros(2,64, dtype=torch.long).cuda()
%%timeit
with torch.no_grad():
y = model(x)
%%timeit
with torch.no_grad():
y = fact_model(x)
```
# Prepare Dataset and DataLoader
```
class SSTDataset(Dataset):
# Static constant variable
NUM_LABELS = 2
def __init__(self, data_split, exp_args, *args, **kwargs):
self.data_split = data_split
self.exp_args = exp_args
if data_split == 'train':
self.dataset = datasets.load_dataset('sst')['train']
elif data_split == 'validation':
self.dataset = datasets.load_dataset('sst')['validation']
elif data_split == 'test':
self.dataset = datasets.load_dataset('sst')['test']
else:
raise ValueError(f'Invalid dataset split: `{data_split}`')
def __getitem__(self, index):
label = np.round(self.dataset[index]['label'])
text = self.dataset[index]['sentence']
return text, label
def __len__(self):
return self.dataset.num_rows
def generate_prompt(texts_by_labels, labels, test_samples):
prompts = []
for label_1 in labels:
pos_samples = texts_by_labels[label_1]
neg_samples = []
prefix = ""
for label_2 in labels:
if label_1 != label_2:
neg_samples = neg_samples + texts_by_labels[label_2]
all_samples = pos_samples + neg_samples
random.shuffle(all_samples)
for sample in all_samples:
text, label = sample["text"], sample["label"]
if label != label_1:
prefix = prefix + text + "=>" + label_1 + "=false\n"
else:
prefix = prefix + text + "=>" + label_1 + "=true\n"
prompts.append([prefix, label_1])
few_shot_prompts = []
for sample in test_samples:
prompt_per_label = []
for prompt in prompts:
prefix, label = prompt
new_prompt = prefix + sample["text"] + "=>" + label + "="
prompt_per_label.append(new_prompt)
few_shot_prompts.append(prompt_per_label)
return few_shot_prompts
def generate_sst_dataset(k_shot):
texts_by_labels = {}
IDX_TO_LABELS = {}
train_dataset = SSTDataset('train', None)
test_dataset = SSTDataset('test', None)
IDX_TO_LABELS = {0: "negative", 1: "positive"}
for i in range(len(train_dataset)):
text, label = train_dataset[i]
if IDX_TO_LABELS[label] not in texts_by_labels:
texts_by_labels[IDX_TO_LABELS[label]] = []
texts_by_labels[IDX_TO_LABELS[label]].append({"text":text, "label":IDX_TO_LABELS[label]})
test_samples = []
for i in range(len(test_dataset)):
text, label = test_dataset[i]
test_samples.append({"text":text, "label":IDX_TO_LABELS[label]})
for label in texts_by_labels:
random.shuffle(texts_by_labels[label])
targets = ["negative", "positive"]
for label in texts_by_labels:
texts_by_labels[label] = texts_by_labels[label][:k_shot]
few_shot_samples = generate_prompt(texts_by_labels, targets, test_samples)
return few_shot_samples, test_samples, targets
few_shot_samples, test_samples, targets = generate_sst_dataset(10)
```
# Run In-Context Learning
```
def score_next(model, tokenizer, encoded, token):
with torch.no_grad():
# print(encoded.size(), token.size())
outputs = model(encoded)
next_token_logits = outputs.logits
def _log_softmax(x):
maxval = np.max(x)
logsum = np.log(np.sum(np.exp(x - maxval)))
return x - maxval - logsum
next_token_logits = next_token_logits[:,-1].squeeze()
# print(next_token_logits.size())
scores = _log_softmax(next_token_logits.cpu().detach().numpy())
del next_token_logits
return scores[int(token)]
def argmax(array):
"""argmax with deterministic pseudorandom tie breaking."""
max_indices = np.arange(len(array))[array == np.max(array)]
idx = int(hashlib.sha256(np.asarray(array).tobytes()).hexdigest(),16) % len(max_indices)
return max_indices[idx]
def logsumexp(x):
c = x.max()
return c + np.log(np.sum(np.exp(x - c)))
def normalize(x):
x = np.array(x)
return np.exp(x - logsumexp(x))
def calculate_log_prob_gpt(model, tokenizer, prefix, targets):
label2id = {}
for target in targets:
# works for single token label e.g., true or false, yes or no
# label2id[target] = tokenizer.convert_tokens_to_ids(target)
label2id[target] = tokenizer(target, truncation=True)["input_ids"][0] # only take the first token
tokenized = tokenizer(list([prefix]), truncation=True, return_tensors="pt")
input_ids = tokenized.input_ids
attention_mask = tokenized.attention_mask
input_ids = input_ids.cuda()
attention_mask = attention_mask.cuda()
with torch.no_grad():
outputs = model(input_ids=input_ids, attention_mask=attention_mask)
logits = outputs.logits.squeeze()[-1]
prob = torch.nn.functional.softmax(logits, dim=-1)
prob = prob.cpu().detach().numpy()
normalized_scores = []
for c in targets:
score = prob[label2id[c]]
normalized_scores.append(score)
pred = targets[argmax(normalized_scores)]
return pred, np.array(normalized_scores)
golds, preds = [], []
pbar = tqdm(iter(few_shot_samples), leave=True, total=len(few_shot_samples))
for id, batch in enumerate(pbar):
prompts = few_shot_samples[id]
test_sample = test_samples[id]
all_scores = []
for prompt in prompts:
pred, normalized_scores = calculate_log_prob_gpt(model, tokenizer, prompt, ["true", "false"])
all_scores.append(normalized_scores)
highest_score_idx = 0
highest_score = 0
for k in range(len(all_scores)):
if all_scores[k][0] > highest_score:
highest_score = all_scores[k][0]
highest_score_idx = k
pred = targets[highest_score_idx]
gold = test_samples[id]["label"]
golds.append(gold)
preds.append(pred)
acc = accuracy_score(preds, golds) * 100
f1 = f1_score(golds, preds, average='macro') * 100
print(f"EVAL SCORE | ACC: {acc} F1: {f1}")
golds, preds = [], []
pbar = tqdm(iter(few_shot_samples), leave=True, total=len(few_shot_samples))
for id, batch in enumerate(pbar):
prompts = few_shot_samples[id]
test_sample = test_samples[id]
all_scores = []
for prompt in prompts:
pred, normalized_scores = calculate_log_prob_gpt(fact_model, tokenizer, prompt, ["true", "false"])
all_scores.append(normalized_scores)
highest_score_idx = 0
highest_score = 0
for k in range(len(all_scores)):
if all_scores[k][0] > highest_score:
highest_score = all_scores[k][0]
highest_score_idx = k
pred = targets[highest_score_idx]
gold = test_samples[id]["label"]
golds.append(gold)
preds.append(pred)
acc = accuracy_score(preds, golds) * 100
f1 = f1_score(golds, preds, average='macro') * 100
print(f"EVAL SCORE | ACC: {acc} F1: {f1}")
```
|
github_jupyter
|
import os, sys
from itertools import chain
import datasets
import random
import torch
from transformers import GPT2LMHeadModel, GPT2Config, GPT2Tokenizer
from torch.utils.data import Dataset, DataLoader
from sklearn.metrics import accuracy_score, f1_score
from greenformer import auto_fact
import numpy as np
from tqdm import tqdm
import hashlib
def count_param(module, trainable=False):
if trainable:
return sum(p.numel() for p in module.parameters() if p.requires_grad)
else:
return sum(p.numel() for p in module.parameters())
model = GPT2LMHeadModel.from_pretrained('gpt2-large')
tokenizer = GPT2Tokenizer.from_pretrained('gpt2-large')
count_param(model)
# Only factorize last one-third of transformer layers of the GPT2 model
factorizable_submodules = list(model.transformer.h[-(model.config.n_layer // 3):])
%%time
fact_model = auto_fact(model, rank=384, deepcopy=True, solver='svd', num_iter=20, submodules=factorizable_submodules)
count_param(fact_model)
%%timeit
with torch.no_grad():
y = model(torch.zeros(2, 64, dtype=torch.long))
%%timeit
with torch.no_grad():
y = fact_model(torch.zeros(2, 64, dtype=torch.long))
model = model.cuda()
fact_model = fact_model.cuda()
x = torch.zeros(2,64, dtype=torch.long).cuda()
%%timeit
with torch.no_grad():
y = model(x)
%%timeit
with torch.no_grad():
y = fact_model(x)
class SSTDataset(Dataset):
# Static constant variable
NUM_LABELS = 2
def __init__(self, data_split, exp_args, *args, **kwargs):
self.data_split = data_split
self.exp_args = exp_args
if data_split == 'train':
self.dataset = datasets.load_dataset('sst')['train']
elif data_split == 'validation':
self.dataset = datasets.load_dataset('sst')['validation']
elif data_split == 'test':
self.dataset = datasets.load_dataset('sst')['test']
else:
raise ValueError(f'Invalid dataset split: `{data_split}`')
def __getitem__(self, index):
label = np.round(self.dataset[index]['label'])
text = self.dataset[index]['sentence']
return text, label
def __len__(self):
return self.dataset.num_rows
def generate_prompt(texts_by_labels, labels, test_samples):
prompts = []
for label_1 in labels:
pos_samples = texts_by_labels[label_1]
neg_samples = []
prefix = ""
for label_2 in labels:
if label_1 != label_2:
neg_samples = neg_samples + texts_by_labels[label_2]
all_samples = pos_samples + neg_samples
random.shuffle(all_samples)
for sample in all_samples:
text, label = sample["text"], sample["label"]
if label != label_1:
prefix = prefix + text + "=>" + label_1 + "=false\n"
else:
prefix = prefix + text + "=>" + label_1 + "=true\n"
prompts.append([prefix, label_1])
few_shot_prompts = []
for sample in test_samples:
prompt_per_label = []
for prompt in prompts:
prefix, label = prompt
new_prompt = prefix + sample["text"] + "=>" + label + "="
prompt_per_label.append(new_prompt)
few_shot_prompts.append(prompt_per_label)
return few_shot_prompts
def generate_sst_dataset(k_shot):
texts_by_labels = {}
IDX_TO_LABELS = {}
train_dataset = SSTDataset('train', None)
test_dataset = SSTDataset('test', None)
IDX_TO_LABELS = {0: "negative", 1: "positive"}
for i in range(len(train_dataset)):
text, label = train_dataset[i]
if IDX_TO_LABELS[label] not in texts_by_labels:
texts_by_labels[IDX_TO_LABELS[label]] = []
texts_by_labels[IDX_TO_LABELS[label]].append({"text":text, "label":IDX_TO_LABELS[label]})
test_samples = []
for i in range(len(test_dataset)):
text, label = test_dataset[i]
test_samples.append({"text":text, "label":IDX_TO_LABELS[label]})
for label in texts_by_labels:
random.shuffle(texts_by_labels[label])
targets = ["negative", "positive"]
for label in texts_by_labels:
texts_by_labels[label] = texts_by_labels[label][:k_shot]
few_shot_samples = generate_prompt(texts_by_labels, targets, test_samples)
return few_shot_samples, test_samples, targets
few_shot_samples, test_samples, targets = generate_sst_dataset(10)
def score_next(model, tokenizer, encoded, token):
with torch.no_grad():
# print(encoded.size(), token.size())
outputs = model(encoded)
next_token_logits = outputs.logits
def _log_softmax(x):
maxval = np.max(x)
logsum = np.log(np.sum(np.exp(x - maxval)))
return x - maxval - logsum
next_token_logits = next_token_logits[:,-1].squeeze()
# print(next_token_logits.size())
scores = _log_softmax(next_token_logits.cpu().detach().numpy())
del next_token_logits
return scores[int(token)]
def argmax(array):
"""argmax with deterministic pseudorandom tie breaking."""
max_indices = np.arange(len(array))[array == np.max(array)]
idx = int(hashlib.sha256(np.asarray(array).tobytes()).hexdigest(),16) % len(max_indices)
return max_indices[idx]
def logsumexp(x):
c = x.max()
return c + np.log(np.sum(np.exp(x - c)))
def normalize(x):
x = np.array(x)
return np.exp(x - logsumexp(x))
def calculate_log_prob_gpt(model, tokenizer, prefix, targets):
label2id = {}
for target in targets:
# works for single token label e.g., true or false, yes or no
# label2id[target] = tokenizer.convert_tokens_to_ids(target)
label2id[target] = tokenizer(target, truncation=True)["input_ids"][0] # only take the first token
tokenized = tokenizer(list([prefix]), truncation=True, return_tensors="pt")
input_ids = tokenized.input_ids
attention_mask = tokenized.attention_mask
input_ids = input_ids.cuda()
attention_mask = attention_mask.cuda()
with torch.no_grad():
outputs = model(input_ids=input_ids, attention_mask=attention_mask)
logits = outputs.logits.squeeze()[-1]
prob = torch.nn.functional.softmax(logits, dim=-1)
prob = prob.cpu().detach().numpy()
normalized_scores = []
for c in targets:
score = prob[label2id[c]]
normalized_scores.append(score)
pred = targets[argmax(normalized_scores)]
return pred, np.array(normalized_scores)
golds, preds = [], []
pbar = tqdm(iter(few_shot_samples), leave=True, total=len(few_shot_samples))
for id, batch in enumerate(pbar):
prompts = few_shot_samples[id]
test_sample = test_samples[id]
all_scores = []
for prompt in prompts:
pred, normalized_scores = calculate_log_prob_gpt(model, tokenizer, prompt, ["true", "false"])
all_scores.append(normalized_scores)
highest_score_idx = 0
highest_score = 0
for k in range(len(all_scores)):
if all_scores[k][0] > highest_score:
highest_score = all_scores[k][0]
highest_score_idx = k
pred = targets[highest_score_idx]
gold = test_samples[id]["label"]
golds.append(gold)
preds.append(pred)
acc = accuracy_score(preds, golds) * 100
f1 = f1_score(golds, preds, average='macro') * 100
print(f"EVAL SCORE | ACC: {acc} F1: {f1}")
golds, preds = [], []
pbar = tqdm(iter(few_shot_samples), leave=True, total=len(few_shot_samples))
for id, batch in enumerate(pbar):
prompts = few_shot_samples[id]
test_sample = test_samples[id]
all_scores = []
for prompt in prompts:
pred, normalized_scores = calculate_log_prob_gpt(fact_model, tokenizer, prompt, ["true", "false"])
all_scores.append(normalized_scores)
highest_score_idx = 0
highest_score = 0
for k in range(len(all_scores)):
if all_scores[k][0] > highest_score:
highest_score = all_scores[k][0]
highest_score_idx = k
pred = targets[highest_score_idx]
gold = test_samples[id]["label"]
golds.append(gold)
preds.append(pred)
acc = accuracy_score(preds, golds) * 100
f1 = f1_score(golds, preds, average='macro') * 100
print(f"EVAL SCORE | ACC: {acc} F1: {f1}")
| 0.593609 | 0.740831 |
# Introduction to Named entity recognition
Named entity recognition (NER) also known as named entity extraction, and entity identification is the task of tagging an entity is the task of extracting which seeks to extract named entities from unstructured text into predefined categories such as names, medical codes, quantities or similar.
The most common variant is the [CoNLL-20003](https://www.clips.uantwerpen.be/conll2003/ner/) format which uses the categories, person (PER), organization (ORG) location (LOC) and miscellaneous (MISC), which for example denote cases such nationalies. For example:
*Hello my name is $Kenneth_{PER}$ I live in $Trøjborg_{LOC}$ and work at $AU_{ORG}$.*
This is for example the tagset used by the Danish spaCy model DaCy.
```
# !pip install dacy
import dacy
nlp_da = dacy.load("small")
doc = nlp_da("Mit navn er Kenneth, jeg bor i Aarhus og arbejder i på Center for Humanities Computing")
displacy.render(doc, style="ent")
```
## Other tagsets
More extensive tagsets exist such as the Ontonotes 5, which for instance include Geopolitical entities (GPE), dates, nationalities and religous groups (NORP), and more. This is for example the one used by the English Spacy model:
```
import spacy
nlp = spacy.load("en_core_web_lg")
doc = nlp("Hello my name is Kenneth I live in Denmark and work at Aarhus University, I am Danish and today is monday 25th.")
from spacy import displacy
displacy.render(doc, style="ent")
```
## Tagging standards
There exist different tag standards for NER. The most used one is the IOB-format which frames the task as token classification denoting inside, outside and beginning of a token. Where outside is denotes as *"O"*, i.e. not an entity. Alternatively, *B-\** indicates the start of an entity (i.e. *B-ORG* for the *Aarhus* in *Aarhus University*), while *I-\** indicate the continuation of a token (e.g. University).
```
for t in doc:
if t.ent_type:
print(t, f"{t.ent_iob_}-{t.ent_type_}")
else:
print(t, t.ent_iob_)
```
## Variations of NER
While NER is currently framed as above this formulating does contain some limitations. For instance the entity Aarhus University really refers to both the location Aarhus, the University within Aarhus, thus nested NER (NNER) argues that it would be more correct to tag it in a nested fashion as \[\[$Aarhus_{LOC}$\] $University$\]$_{ORG}$ (Plank, 2020). Other task also include named entity linking. Which is the task of linking an entity to e.g. a wikipedia entry, thus you have to both know that it is indeed an entity and which entity it is (if it is indeed a defined entity).
|
github_jupyter
|
# !pip install dacy
import dacy
nlp_da = dacy.load("small")
doc = nlp_da("Mit navn er Kenneth, jeg bor i Aarhus og arbejder i på Center for Humanities Computing")
displacy.render(doc, style="ent")
import spacy
nlp = spacy.load("en_core_web_lg")
doc = nlp("Hello my name is Kenneth I live in Denmark and work at Aarhus University, I am Danish and today is monday 25th.")
from spacy import displacy
displacy.render(doc, style="ent")
for t in doc:
if t.ent_type:
print(t, f"{t.ent_iob_}-{t.ent_type_}")
else:
print(t, t.ent_iob_)
| 0.137996 | 0.970521 |
_Lambda School Data Science, Unit 2_
# Regression 1 Sprint Challenge
Your Sprint Challenge has two parts.
To demonstrate mastery on each part, do all the required, numbered instructions.
To earn a score of "3" for the part, also do the stretch goals.
## Part 1. Predict avocado sales 🥑
For part 1 of your challenge, you'll use historical data on avocado sales across the nation!
Run this code cell to load the dataset. It has weekly sales volume (total number of avocados sold) from January 4, 2015 through March 25, 2018. The data comes directly from retailers' cash registers based on actual retail sales of Hass avocados in multiple US markets.
```
import pandas as pd
avocado_url = 'https://drive.google.com/uc?export=download&id=1ljE-LhCA9CCFvSIJYJ1ewX8JmdDpfuhi'
avocado = pd.read_csv(avocado_url)
assert avocado.shape == (169, 2)
avocado.head()
```
### Required
#### Use the Prophet library to:
1. Fit a model and forecast avocado sales through March 25, 2020
2. Plot the forecast
3. Plot the forecast components
4. Do time series cross-validation, with these parameters: `horizon='90 days', period='45 days', initial='730 days'`
5. Plot the cross-validation Mean Absolute Percentage Error: `metric='mape'`
_For Part 1 of this Sprint Challenge, you are **not** required to calculate or plot baselines, or do train/test split._
You can be confident you've done it correctly when your plots look similar to these:
<img src="https://i.imgur.com/PoyVGp7.png" width="400" align="left">
```
avocado = avocado.rename(columns={'Date':'ds', 'Total Volume': 'y'})
avocado.head()
from fbprophet.diagnostics import cross_validation
import warnings
warnings.simplefilter(action='ignore', category=FutureWarning)
m = Prophet(daily_seasonality=False)
m.fit(avocado)
from fbprophet.diagnostics import cross_validation
df_cv = cross_validation(m, horizon='90 days', period='45 days', initial='730 days')
future = m.make_future_dataframe(periods=730)
forecast = m.predict(future)
import matplotlib.pyplot as plt
fig1 = m.plot(forecast)
fig2 = m.plot_components(forecast)
fig3 = plot_cross_validation_metric(df_cv, metric='mape')
from fbprophet.diagnostics import performance_metrics
performance_metrics(df_cv).head()
```
### Stretch Goal
- Adjust your forecasts with Prophet's options for changepoints, holidays, and fourier order. Improve the cross-validation MAPE.
```
m = Prophet(daily_seasonality=False, yearly_seasonality=20)
#m.add_country_holidays(country_name = 'US') #-> Worse results
m.fit(avocado)
from fbprophet.diagnostics import cross_validation
df_cv = cross_validation(m, horizon='90 days', period='45 days', initial='730 days')
performance_metrics(df_cv).head()
```
## Part 2. Predict NYC apartment rent 🏠💸
For part 1 of your challenge, you'll use a real-world data with rent prices for a subset of apartments in New York City!
Run this code cell to load the dataset:
```
import pandas as pd
rent_url = 'https://drive.google.com/uc?export=download&id=1lVVPmh-WYGb_0Gs_lOv22i02-150qDdZ'
rent = pd.read_csv(rent_url)
assert rent.shape == (48295, 7)
rent.head()
```
### Required
1. Begin with baselines for regression. Calculate the mean absolute error and $R^2$ score for a mean baseline. _(You can use the whole dataset, or a test set, either way is okay here!)_
2. Do train/test split. Use data from April & May 2016 to train. Use data from June 2016 to test.
3. Choose any two features.
4. Use scikit-learn to fit a Linear Regression model on the train data.
5. Apply the model to predict rent prices for the test data.
6. Get regression metrics MAE and $R^2$ for the test data.
7. Get the model's coefficients and intercept.
### Stretch Goals
- Try at least 3 different feature combinations. (You don't have to use two features, you can choose any number of features.)
- Get regression metrics RMSE, MAE, and $R^2$, for both the train and test data.
```
from sklearn.metrics import mean_absolute_error, r2_score, mean_squared_error
rent['mean_baseline'] = rent['price'].mean()
y = rent['price']
mae = mean_absolute_error(y, rent['mean_baseline'] )
r2 = r2_score(y, rent['mean_baseline'])
print(f'MAE: {mae}\tR^2: {r2}')
from sklearn.linear_model import LinearRegression
features = ['bathrooms', 'bedrooms']
target = 'price'
model = LinearRegression()
train = rent[(rent['month'] == 4)|(rent['month'] == 5)]
test = rent[rent['month'] == 6]
model.fit(train[features], train[target])
y = test[target]
y_pred = model.predict(test[features])
mae = mean_absolute_error(y, y_pred)
r2 = r2_score(y, y_pred)
print(f'MAE: {mae}\tR^2: {r2}')
print('Coefficients: ', model.coef_)
print('Intercept: ', model.intercept_)
#Stretch
from itertools import combinations
import numpy as np
#Let's try all 3-feature combinations
results = []
features = ['bathrooms', 'bedrooms', 'latitude', 'longitude']
for feats in combinations(features, 3):
model = LinearRegression()
model.fit(train[list(feats)], train[target])
y_pred = model.predict(test[list(feats)])
mae = mean_absolute_error(y, y_pred)
mse = mean_squared_error(y, y_pred)
rmse = np.sqrt(mse)
r2 = r2_score(y, y_pred)
mean_squared_error
print(f'Features: {feats}', )
print(f'MAE: {mae}\tR^2: {r2}\tRMSE: {rmse}\n')
results.append({'features': feats, 'MAE': mae, 'R^2': r2, 'RMSE': rmse})
#And the winner is:
best = min(results, key=lambda x: x['RMSE'])
print(f'Best(RMSE) 3-Feature Combination: {best["features"]}')
```
|
github_jupyter
|
import pandas as pd
avocado_url = 'https://drive.google.com/uc?export=download&id=1ljE-LhCA9CCFvSIJYJ1ewX8JmdDpfuhi'
avocado = pd.read_csv(avocado_url)
assert avocado.shape == (169, 2)
avocado.head()
avocado = avocado.rename(columns={'Date':'ds', 'Total Volume': 'y'})
avocado.head()
from fbprophet.diagnostics import cross_validation
import warnings
warnings.simplefilter(action='ignore', category=FutureWarning)
m = Prophet(daily_seasonality=False)
m.fit(avocado)
from fbprophet.diagnostics import cross_validation
df_cv = cross_validation(m, horizon='90 days', period='45 days', initial='730 days')
future = m.make_future_dataframe(periods=730)
forecast = m.predict(future)
import matplotlib.pyplot as plt
fig1 = m.plot(forecast)
fig2 = m.plot_components(forecast)
fig3 = plot_cross_validation_metric(df_cv, metric='mape')
from fbprophet.diagnostics import performance_metrics
performance_metrics(df_cv).head()
m = Prophet(daily_seasonality=False, yearly_seasonality=20)
#m.add_country_holidays(country_name = 'US') #-> Worse results
m.fit(avocado)
from fbprophet.diagnostics import cross_validation
df_cv = cross_validation(m, horizon='90 days', period='45 days', initial='730 days')
performance_metrics(df_cv).head()
import pandas as pd
rent_url = 'https://drive.google.com/uc?export=download&id=1lVVPmh-WYGb_0Gs_lOv22i02-150qDdZ'
rent = pd.read_csv(rent_url)
assert rent.shape == (48295, 7)
rent.head()
from sklearn.metrics import mean_absolute_error, r2_score, mean_squared_error
rent['mean_baseline'] = rent['price'].mean()
y = rent['price']
mae = mean_absolute_error(y, rent['mean_baseline'] )
r2 = r2_score(y, rent['mean_baseline'])
print(f'MAE: {mae}\tR^2: {r2}')
from sklearn.linear_model import LinearRegression
features = ['bathrooms', 'bedrooms']
target = 'price'
model = LinearRegression()
train = rent[(rent['month'] == 4)|(rent['month'] == 5)]
test = rent[rent['month'] == 6]
model.fit(train[features], train[target])
y = test[target]
y_pred = model.predict(test[features])
mae = mean_absolute_error(y, y_pred)
r2 = r2_score(y, y_pred)
print(f'MAE: {mae}\tR^2: {r2}')
print('Coefficients: ', model.coef_)
print('Intercept: ', model.intercept_)
#Stretch
from itertools import combinations
import numpy as np
#Let's try all 3-feature combinations
results = []
features = ['bathrooms', 'bedrooms', 'latitude', 'longitude']
for feats in combinations(features, 3):
model = LinearRegression()
model.fit(train[list(feats)], train[target])
y_pred = model.predict(test[list(feats)])
mae = mean_absolute_error(y, y_pred)
mse = mean_squared_error(y, y_pred)
rmse = np.sqrt(mse)
r2 = r2_score(y, y_pred)
mean_squared_error
print(f'Features: {feats}', )
print(f'MAE: {mae}\tR^2: {r2}\tRMSE: {rmse}\n')
results.append({'features': feats, 'MAE': mae, 'R^2': r2, 'RMSE': rmse})
#And the winner is:
best = min(results, key=lambda x: x['RMSE'])
print(f'Best(RMSE) 3-Feature Combination: {best["features"]}')
| 0.713931 | 0.964888 |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.